Note: Descriptions are shown in the official language in which they were submitted.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-1-
GENES WHOSE EXPRESSION IS INCREASED IN RESPONSE TO
STIMULATION BY CORTICOTROPIN-RELEASING HORMONE
The present invention relates generally to therapy and diagnosis of
depression. In particular this invention relates to the polypeptides as well
as to the
polynucleotides encoding these polypeptides, wherein said polypeptides are
shown
to play a central role in mediating the cellular response to corticotropin
releasing
hormone. These polypeptides and polynucleotides are useful in the diagnosis,
treatment andlor prevention of depression.
BACKGROUND OF THE INVENTION
Recent socioeconomic analyses found that depression is a leading cause of
disability
and a major risk factor for development of other diseases.~Moreover, on a
world-wide
scale depression is underdiagnosed and undertreated. Current antidepressant
drugs have
proven to be effective, but are burdened with slow onset of action and side
effects.
Above this, it is still unclear by which phaa-macological mode of action they
exert their
clinical effects. Hypothesis-driven research based upon the corticosteroid
receptor
hypothesis of depression has led to a novel concept focusing on brain
neuropeptide
receptors, specifically the corticotropin-releasing hormone (CRH) receptor as
drug
target.
Corticotropin releasing hormone (CRH), a 41-amino acid polypeptide plays a
central
.role in the regulation of the hypothalamic-pituitary-adrenal axis, mediating
the
endocrine responses to various stressors. Hypothalamic neurons release CRH
into the
hypophyseal portal system in response to stress, stimulating the secretion and
biosynthesis of pituitary adrenocorticotropin (ACTH) leading to increased
adrenal
glucocorticoid production (1). Several clinical and preclinical studies point
towards a
causal role for alterations in the CRH system in the development of depression
(2). The
first studies with CRH in humans showed that the ACTH response to CRH is
blunted in
depressed patients, reflecting a CRH receptor desensitization secondary to
continuously
increased hypothalamic CRH secretion (3;4). In support of blunted ACTH
response as
consequence of increased CRH release is the finding of elevated CRH levels in
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-2-
cerebrospinal fluid of patients with depression. Other findings strengthening
this notion
of CRH hypersecretion in the depressed state are an increased number of CRH
secreting
neurons and a decreased number of CRH receptors in suicide victims who
suffered
from depression (5;6).
For CRH two high affinity receptors have been described, CRH-R1 and CRH-R2,
both
of which exist in several splice variant forms. Activation of these receptors
by CRH
results in Gs-mediated stimulation of adenyl cyclase leading to increased
levels of
intracellular cAMP. This in itself will activate cAMP dependent protein kinase
A
(PKA) and ultimately result in increased cytosolic levels of cAMP and Ca2+.
The
increased levels of CAMP and Caz+ lead to the activation of several other
additional
kinases such as Ca2+/calmodulin-dependent kinase II (CAMKII) and p42/p44
mitogen
activated kinases (MAPK). As a result the Cap'+IcAMP response element binding
protein (CREB) is phosporylated and this in turn will regulate the
transcription of genes
containing cAMP response elements (CRE) in their promoter region. Examples of
such
genes shown to be involved in the modulation of CRH signaling include c fos,
macrophage migration-inhibitory factor gene Mzf, orphan nuclear receptors
Nurr77 and
Nacrrl .
Notwithstanding the fact that the downstream pathways for CRH activated
receptors
were extensively studied in AtT-20 cells, a cellular model of corticotrophs
and led to
the identification of a number of genes involved in the signaling cascade, a
major area
is unexplored. It was thus an object of the present invention to explore the
transcriptional response to CRH stimulation at a genome wide level in order to
identify
further genes involved in the corticotropin-releasing hormone receptor
activated gene
network. The polypeptides thus identified and the polynucleotides encoding
said
polypeptides provide new chances for drug development as drug targets through
screening techniques, or are useful in the diagnosis, prevention and/or
treatment of
depression.
SUNINIARY OF THE INVENTION
The present invention relates to the identification of a number of genes
involved in
the transcriptional response to CRH stimulation at a genome wide level. In
particular to the identification of a number of hitherto unknown genes
encoding a
protein that modulates CRH signaling, having a sequence selected from the
group
consisting of SEQ ID No. 45, SEQ ID No. 47, SEQ lD No. 49 and functional
analogs thereof.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-3-
In a further aspect the present invention relates to the recombinant use of
the
aforementioned nucleotide sequences, including vectors comprising these
sequences, host cells containing a vector to encode for one of the
aforementioned
sequences as well as transgenic non-human animals comprising a polynucleotide
or
vector according to the invention.
It is also an object of the present invention to provide a number of genes
that were
hitherto not associated with CRH signaling and accordingly useful in methods
to
identify compounds, which modulate CHR signaling response in a cell or in
diagnostic methods to identify CRH induced depression in an individual. In one
embodiment the method to identify a compound capable to alter the CRH
signaling
response in a cell comprises, contacting said cell With CRH in the presence
and
absence of said compound and determine the expression level of a
polynucleotide
comprising a nucleic acid sequence selected from the group consisting of SEQ
ID
No. l, SEQ ID No. 3, SEQ )D No. 5, SEQ ID No. 7, SEQ ID No.9, SEQ ll~ NO. 11,
SEQ ID NO 13, SEQ ID No.l5, SEQ ID No 17, SEQ ID No 19, SEQ ID No. 21,
SEQ ID No. 23, SEQ ID No. 25, SEQ ID No. 27, SEQ ID No. 29, SEQ JD NO. 31,
SEQ m No. 33, SEQ lD No. 35, SEQ ID No. 37, SEQ ID No. 39, SEQ m No. 41,
SEQ ll~ No. 43, SEQ ID No. 45, SEQ JD No. 47 or SEQ ID No. 49. In this
screening method the expression levels are typically assessed using an
oligonucleotide probe that binds to the aforementioned polynucleotides,
preferably
using array technology methods. Accordingly in a particular embodiment the
present invention provides a method to identify compounds that modulate the
CRH
signaling response in a cell said method comprising, contacting said cell with
CRH
in the presence and absence of said compound; and determine the expression
level
of the polynucleotides having the nucleic acid sequences SEQ 117 No. l, SEQ
ff~
No. 3, SEQ )D No. 5, SEQ ID No. 7, SEQ ID No.9, SEQ >D NO. 11, SEQ lD NO
13, SEQ ID No.lS, SEQ ID No 17, SEQ >D No 19, SEQ ID No. 21, SEQ ID No. 23,
SEQ m No. 25, SEQ ID No. 27, SEQ ID No. 29, SEQ ID NO. 31, SEQ ID No. 33,
SEQ ID No. 35, SEQ ID No. 37, SEQ ID No. 39, SEQ ID No. 41, SEQ ID No. 43,
SEQ ff~ No. 45, SEQ ID No. 47 and SEQ ID No. 49, wherein a change in
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-4-
expression profile of these sequences is indicative for a compound capable to
alter
the CRH signaling response in said cell.
BRIEF DESCRIPTION OF THE DRAWING
Table 1: List of proteins which modulate CRH signaling.
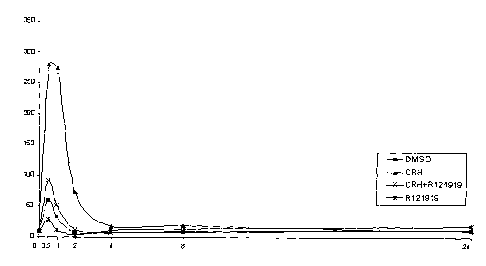
Figure 1: c-fos mRNA levels as assessed by quantitative RT-PCR normalized
against 13-
actin mRNA levels (taken as 100°Io) in AtT-20 cells treated with DMSO,
CRH,
CRH+8121919 or 8121919 for different time points (hours).
Figure 2: Correspondence analysis applied on normalized microarray data for
all time
points and treatments. Squares depict different samples whereas circles depict
genes.
Distances between squares are a measure for similarity between samples. A
positive
association of a gene with a given sample (i.e. an upregulation of that gene
in that
particular sample) results in the positioning of the gene and sample on a
common line
through the centroid (depicted by a cross). Correspondence analysis clearly
identifies
time as the major discriminator between the samples. In addition the effect of
treatment
with CRH can be identified as most prominent in the early time points.
Figure 3: A heat map depicting genes that are changed upon CRH treatment.
Values
were calculated by dividing the intensity of each sample by the intensity of
the DMSO
sample at the corresponding time point. These calculated ratio are converted
into a
color ramp using on a loge scale. In this way the different timing of
induction of
expression becomes apparent. Genes showing a 2-fold change after 30 minutes of
treatment with CRH were called "early responders", "intermediate responders"
show a
change after 1 to 2 hours of treatment and "late responders" show a response
after 2
hours or more.
Figure 4: Overview of a selection of genes induced by CRH grouped by pathway
or
function as discussed in the text. Values were calculated by dividing the
intensity of
each sample by the intensity of the DMSO sample at the corresponding time
point.
These calculated ratio are converted into a color ramp using on a loge scale
and
depicted in a heat map.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-5-
Figure 5: Induction of Rgs2 by CRH in AtT-20 cells. Induction is calculated in
comparison to levels observed in AtT-20 cells before any treatment. On top
array data
obtained for Rgs2 are shown. Below, levels of Rgs2 mRNA are shown as measured
by
quantitative RT-PCR on the same samples as used for array experiments and as
measured on a repeated experiment.
DETAILED DESCRIPTION
As used herein, the term "compound" or "agent" means a biological or chemical
compound such as a simple or complex organic molecule, a peptide, a protein or
an
oligonucleotide. A "test compound" as used herein, refers to a "compound" or
"agent" used in a method according to the invention to assess whether said
compounds modulates CRH signalling activity.
"CRH signaling" as used herein refers to the cellular changes in gene
transcription
after activation of the corticotropin releasing hormone receptor by CRH in
said
cell. It induces a CRH specific gene expression profile. Changes at the
transcriptional level can be assessed either at the protein level or at the
gene,
RNA level.
"CRH response activity" as used herein refers in general to the change of a
detectable cellular parameter as a result of the exposure of said cell to CRH.
Detectable cellular parameters include amongst others, changes in membrane
potential, changes in enzyme activity of an enzyme that modulates CRH
signalling a said cell, changes in expression levels of a protein according to
the
invention or changes in the amount of second messengers such as cGMP,
cAMP, Ca2+ ar IP3.
The term "analog" or "functional analog" refers to a modified form of protein
according to the invention in which at least one amino acid substitution has
been made such that said analog retains substantially the same biological
activity as the unmodified protein in vivo and/or ifz vitr°o.
The term "functional analog" is intended to include the "fragments,"
"variants,"
"degenerate variants," "analogs" and "homologues" or to "chemical derivatives"
of the polypeptides according to the invention. Useful chemical derivatives of
polypeptide are well known in the art and include, for example covalent
modification of reactive organic site contained within the polypeptide with a
secondary chemical moiety. Well known cross-linl~ing reagents are useful to
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-6-
react to amino, carboxyl, or aldehyde residues to introduce, for example an
affinity tag such as biotin, a fluorescent dye, or to conjugate the
polypeptide to
a solid phase surface (for example to create an affinity resin)
Variants) of polynucleotides or polypeptides, as the term is used herein, are
polynucleotides or polypeptides that differ from a reference polynucleotide or
polypeptide, respectively. A vacant of the polynucleotide may be a naturally
occurring variant such as a naturally occurring allelic variant, or it may be
a
variant that is not known to occur naturally. (1) A polynucleotide that
differs in
nucleotide sequence from another, reference polynucleotide. Generally,
differences are limited so that the nucleotide sequences of the reference and
the
variant are closely similar overall and, in many regions, identical. As noted
below, changes in the nucleotide sequence of the variant may be silent. That
is,
they may not alter the amino acids encoded by the polynucleotide. Where
alterations are limited to silent changes of this type a variant will encode a
polypeptide with the same amino acid sequence as the reference. Also as noted
below, changes in the nucleotide sequence of the variant may alter the amino
acid sequence of a polypeptide encoded by the reference polynucleotide. Such
nucleotide changes may result in amino acid substitutions, additions,
deletions,
fusions and truncations in the polypeptide encoded by the reference sequence,
as discussed above. (2) A polypeptide that differs in amino acid sequence from
another, reference polypeptide. Generally, differences are limited so that the
sequences of the reference and the variant are closely similar overall and, in
many regions, identical. A variant and reference polypeptide may differ in
amino acid sequence by one or more substitutions, additions, deletions,
fusions
and truncations, which may be present in any combination.
The terms "complementary" or "complementarity" as used herein refer to the
capacity of purine and pyrimidine nucleotides to associate through hydrogen
bonding to form double-stranded nucleic acid molecules. The following base
pairs are related by complementarily: guanine and cytosine; adenine and
thymine; and adenine and uracil. As used herein "complementary" means that
the aforementioned relationship applies to substantially all base pairs
comprising two single-stranded nucleic acid molecules over the entire length
of
said molecules. "Partially complementary" refers to the aforementioned
relationship in which one of the two single-stranded nucleic acid molecules is
shorter in length than the other such that a portion of one of the molecules
remains single-stranded.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
_7_
The term "conservative substitution" or "conservative amino acid substitution"
refers to a replacement of one or more amino acid residues) in a parent
protein
without affecting the biological activity of the parent molecule based on the
art
recognized substitutability of certain amino acids (See e.g. M. Dayhoff, In
Atlas of Protein Sequence and Structure, Vol. 5, Supp. 3, pgs 345-352, 1978).
"Fragment thereof ' refers to a fragment, piece, or sub-region of a nucleic
acid or
protein molecule whose sequence is disclosed herein, such that said fragment
comprises 5 or more amino acids, or 10 or more nucleotides that are contiguous
in the parent protein or nucleic acid molecule.
"Functional fragment" as used herein, refers to an isolated sub-region, or
fragment
of a protein disclosed herein, or sequence of amino acids that, for example,
comprises a functionally distinct region such as an active site for a
receptor.
Functional fragments may be produced by cloning technology, or as the natural
products of alternative splicing mechanims.
The term "homolog" or "homologous" describes the relationship between
different
nucleic acid molecules or amino acid sequences in which said sequences or
molecules are related by partial identity or similarity at one or more blocks
or
regions within said molecules or sequences. "Isolated nucleic acid compound"
refers to any RNA or DNA sequence, however construed or synthesized, which
is locationally distinct from its natural location.
A "nucleic acid probe" or "probe" as used herein is a labeled nucleic acid
compound which hybridizes with another nucleic acid compound. "Nucleic
acid probe" means a single stranded nucleic acid sequence that will hybridize
with a single stranded target nucleic acid sequence. A nucleic acid probe may
be an oligonucleotide or a nucleotide polymer. A "probe" will usually contain
a
detectable moiety which may be attached to the ends) of the probe or be
internal to the sequence of the probe.
The term "primer" is a nucleic acid fragment which functions as an initiating
substrate for enzymatic or synthetic elongation of, for example, a nucleic
acid
molecule.
The term "hybridization" as used herein refers to a process in which a single-
stranded nucleic acid molecule joints with a complementary strand through
nucleotide base pairing.
The term "stringency" refers to hybridization conditions. High stringency
conditions disfavor non-homologous base pairing. Low stringency conditions
have
the opposite effect. Stringency may be altered, for example, by temperature
and salt
concentration. "Stringent conditions" refers to an overnight incubation at
42°C in a
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
_$_
solution comprising 50% formamide, 5x SSC (750 mM NaCl, 75 mM sodium
citrate), 50 mM sodium phosphate (pH 7.6), 5x Denhardt's solution, 10% dextran
sulfate, and 20 ~,g/ml denatured, sheared salmon sperm DNA, followed by
washing
the filters in 0.1 x SSC at about 65°C. Further suitable hybridization
conditions are
described in the examples.
"Lower stringency conditions" include an overnight incubation at 37°C
in a solution
comprising 6X SSPE (20X SSPE = 3M NaCI; 0.2M NaH2P04; 0.02M EDTA,
pH 7.4), 0.5% SDS, 30% formamide, 100 ~g/ml salmon sperm blocking DNA;
followed by washes at 50°C with 1 X SSPE, 0.1% SDS. In addition, to
achieve
even lower stringency, washes performed following stringent hybridization can
be done at higher salt concentrations (e.g. 5X SSC). Note that variations in
the
above conditions may be accomplished through the inclusion andlor
substitution of alternate blocking reagents used to suppress background in
hybridization experiments. Typical blocking reagents include Denhardt's
reagent, BLOTTO, heparin, denatured salmon sperm DNA, and commercially
available proprietary formulations. The inclusion of specific blocl~ing
reagents
may require modification of the hybridization conditions described above, due
to problems with compatibility.
The term "fusion protein" as used herein refers to protein constructs that are
the result of combining multiple protein domains or linker regions for the
purpose
of gaining the combined functions of the domains or linker regions. This is
may be
accomplished by molecular cloning of the nucleotide sequences encoding such
domains to produce a new polynucleotide sequence that encodes the desired
fusion
protein. Alternatively, creation of a fusion protein may be accomplished by
chemically joining two proteins.
The term "linker region" or "linker domain" or similar such descriptive
terms as used herein refers to polynucleotide or polypeptide sequence that are
used
in the construction of a cloning vector or fusion protein. Functions of a
linker
region can include introduction of cloning sites into the nucleotide sequence,
introduction of a flexible component or space-creating region between two
protein
domains, or creation of an affinity tag for specific molecule interaction. A
linker
region may be introduced into a fusion protein resulting from choices made
during
polypeptide or nucleotide sequence construction.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-9-
Screening methods
The present invention relates to screening methods to identify compounds that
modulate corticotropin-releasing hormone (CRH) induced depression and stress.
It is
based on the identification of a number of genes as downstream modulators of
the CRH
activated CRH receptors. In particular this invention provides a method for
identifying
a compound capable to alter the CRH signalling response in a cell, said method
comprising;
a) contacting said cell with CRH in the presence and absence of said compound;
b) determine the change at transcriptional level of at least one protein that
modulates corticotropin releasing hormone (CRH) signaling in said cell; and
c) compare the transcriptional level of said protein in the presence and
absence of
said compound;
whereby the protein that modulates corticotropin releasing hormone (CRH)
signaling is being selected from the group consisting of SEQ ID N0.2, SEQ ID
4, SEQ ID N0.6, SEQ ID N0.8, SEQ ID NO.10, SEQ ID N0.12, SEQ ID
NO.14, SEQ ID N0.16, SEQ ID N0.18, SEQ ID N0.20, SEQ ID N0.22, SEQ
ll~ N0.24, SEQ ID N0.26, SEQ 1D N0.28, SEQ ID N0.30, SEQ m N0.32,
SEQ ID N0.34, SEQ ID N0.36, SEQ ID N0.38; SEQ D7 N0.40, SEQ ID
N0.42, SEQ ID N0.44, SEQ ID N0.46 and SEQ lD N0.48.
To determine the change of transcription at the protein level one could
determine the amount of said protein using art known techniques. For example
using separation techniques such as isoelectric focusing or SDS-page in
combination with protein staining techniques such as coomassie or silver
staining.
Alternatively, for proteins that are enzymes, the amount in a given solution
or tissue
extract can be measured or assayed in terms of the catalytic effect the enzyme
produces, that is the conversion of its substrate into reaction product. For
example,
for kinases one may assess the kinase activity using a substrate comprising
the
kinase specific phosphorylation site and by measuring the phosphorylation of
the
substrate by incorporation of radioactive phosphate into the substrate. This
assay
may be perfomed both in the presence and absence of the compound to be tested.
For proteins that are not enzymes, other quantification methods are required.
For
example transport proteins can be assayed by their binding to the molecule
they
transport and hormones and toxins by the biological effect they produce.
To assess changes in transcription at the gene level, RNA or cDNA may be used
directly for detection or may be amplified enzymatically by using PCR or other
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-10-
amplification techniques prior to analysis. Preferably said analysis method
comprises
the use of a labelled oligonucleotide probe targeted to a suitable region of
the gene.
Accordingly, in a preferred embodiment the level of gene transcription is
assessed using a probe which binds to a polynucleotide encoding an amino acid
sequence selected from the group consisting of SEQ ID N0.2, 5EQ 1D 4, SEQ >D
N0.6, SEQ ID N0.8, SEQ >D NO.10, SEQ ID N0.12, SEQ ll~ N0.14, SEQ ID N0.16,
SEQ ID N0.18, SEQ ID N0.20, SEQ ID N0.22, SEQ ID N0.24, SEQ ID N0.26, SEQ
ID NO.28, SEQ ID N0.30, SEQ ID N0.32, SEQ ID N0.34, SEQ ID N0.36, SEQ ID
N0.38, SEQ ID N0.40, SEQ ID N0.42, SEQ ID N0.44, SEQ ID N0.46 and SEQ ID
N0.48.
In another embodiment, an array of oligonucleotides probes comprising a
nucleotide sequence encoding a protein that modulates CRH signalling or
fragments
thereof can be constructed to conduct efficient screening of gene expression.
Array
technology methods are well known and have general applicability and can be
used to
address a variety of questions in molecular genetics including gene
expression, genetic
linkage, and genetic variability (see for example: M.Chee et al., Science, Vol
274, pp
610-613 (1996)). It is thus an object of the present invention to provide a
method for
identifying a compound capable to alter the CRH signalling response in a cell,
said
method comprising; contacting said cell with CRH in the presence and absence
of the
compound to be tested; and determine the level of gene transcription using an
array of
oligonucleotide probes that bind to the polynucleotides encoding the group of
polypeptides having the amino acid sequences SEQ ID N0.2, SEQ ID 4, SEQ >D
N0.6,
SEQ ID N0.8, SEQ ID N0.10, SEQ ID N0.12, SEQ ID N0.14, SEQ ID N0.16, SEQ
ID N0.18, SEQ ID N0.20, SEQ ID N0.22, SEQ ID N0.24, SEQ ID N0.26, SEQ ID
N0.28, SEQ JD N0.30, SEQ ID N0.32, SEQ ID N0.34, SEQ ID N0.36, SEQ ID
N0.38, SEQ m N0.40, SEQ ID N0.42, SEQ ID N0.44, SEQ DJ N0.46 and SEQ DJ
N0.48.
In an alternative embodiment, the method for identifying a compound capable to
alter the CRH signalling response in a cell, comprises;
a) contacting said cell with CRH in the presence and absence of said compound;
and
b) determine the expression level of a polynucleotide comprising a nucleic
acid
sequence selected from the group consisting of SEQ ID NO: 1, SEQ ID NO 3, SEQ
ID
N0.5, SEQ )D N0.7, SEQ )D N0.9, SEQ ID NO.11, SEQ TD N0.13, SEQ ZD N0.15,
SEQ ll~ N0.17, SEQ ID N0.19, SEQ 1D N0.21, SEQ ID N0.23, SEQ ID N0.25, SEQ
ID N0.27, SEQ ID NO. 29, SEQ m N0.31, SEQ ll~ N0.33, SEQ ID N0.35, SEQ ID
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-11-
NO. 37, SEQ ll~ N0.39, SEQ ID N0.41, SEQ ID N0.43, SEQ ID N0.45, SEQ ID
N0.47 or SEQ ID N0:49.
To assess changes in expression levels, RNA or cDNA may be used directly for
detection or may be amplified enzymatically by using PCR or other
amplification
techniques prior to analysis. Preferably said analysis method comprises the
use of a
labelled oligonucleotide probe targeted to a suitable region of the
polynucleotiode.
Accordingly, in a preferred embodiment the level of gene transcription is
assessed using
a probe which binds to a polynucleotide comprising a nucleic acid sequence
selected
from the group consisting of of SEQ ID NO: 1, SEQ DJ NO 3, SEQ 117 N0.5, SEQ
lD
N0.7, SEQ ID N0.9, SEQ ID N0.11, SEQ ID N0.13, SEQ ID N0.15, SEQ D7 N0.17,
SEQ ID N0.19, SEQ ID N0.21, SEQ ID N0.23, SEQ ID N0.25, SEQ ID N0.27, SEQ
ID NO. 29, SEQ ID N0.31, SEQ ID NO.33, SEQ ID N0.35, SEQ ID NO. 37, SEQ lD
N0.39, SEQ ID N0.41, SEQ ID N0.43, SEQ ID N0.45, SEQ ll~ NO.47 or SEQ 11?
N0:49.
In another embodiment, an array of oligonucleotides probes comprising a
nucleotide sequence encoding a protein that modulates CRH signalling or
fragments
thereof can be constructed to conduct efficient screening of gene expression.
In this
embodiment the invention provides a method for identifying a compound capable
to
alter the CRH signaling response in a cell, said method comprising, contacting
said cell
with CRH in the presence and absence of said compound; and determine the
expression
level of the poynucleotides having the nucleic acid sequences SEQ ID NO: 1,
SEQ ID
NO 3, SEQ ll~ N0.5, SEQ ID N0.7, SEQ ID N0.9, SEQ ID NO.11, SEQ ID N0.13,
SEQ ID N0.15, SEQ ID N0.17, SEQ ID N0.19, SEQ ID NO.21, SEQ ID N0.23, SEQ
ID NO.25, SEQ ID N0.27, SEQ ID NO. 29, SEQ ID N0.31, SEQ ID N0.33, SEQ ID
N0.35, 5EQ ID NO. 37, SEQ ID N0.39, SEQ ID N0.41, SEQ lD N0.43, SEQ ll~
N0.45, SEQ 1D N0.47 and 5EQ ID N0:49. In particular, using an array of
oligonucleotide probes that bind to the polynucleotides having the nucleic
acid
sequences SEQ ID NO: 1, SEQ ID NO 3, SEQ ID N0.5, SEQ ID N0.7, SEQ ID N0.9,
SEQ ID NO.11, SEQ B7 N0.13, SEQ ID N0.15, SEQ 117 N0.17, SEQ ID N0.19, SEQ
ID N0.21, SEQ ID N0.23, SEQ ID N0.25, SEQ ID N0.27, SEQ ID NO. 29, SEQ m
N0.31, SEQ lD N0.33, SEQ ID N0.35, SEQ ID NO. 37, SEQ ID NO.39, SEQ ID
NO.41, SEQ lD N0.43, SEQ ID N0.45, SEQ ID NO.47 and SEQ ID N0:49.
In another embodiment, an array of oligonucleotides probes comprising a
nucleotide sequence encoding a protein that modulates CRH signalling or
fragments
thereof can be constructed to conduct efficient screening of gene expression.
Array
technology methods are well known and have general applicability and can be
used to
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-12-
address a variety of questions in molecular genetics including gene
expression, genetic
linkage, and genetic variability (see for example: M.Chee et al., Science, Vol
274, pp
610-613 (1996)).
In case the genomic DNA is not used directly, mRNA may be isolated, and a
first strand cDNA synthesis carried out. A second round of DNA synthesis can
be
carried out for the production of the second strand. Subsequently by the
specific
PCR amplification an isolated cDNA can be obtained. If desired the double
stranded cDNA can be cloned into any suitable vector, for example, a plasmid,
thereby forming a cDNA libary. In analogy to the above, it is possible to
screen
cDNA libraries constructed in a bacteriophage or plasmid shuttle vector with a
labeled oligonucleotide probe targeted to any suitable region of the gene
encoding a
protein that modulates CRH signalling. See e.g. PCR Protocols: A Guide to
Method and Application, Ed. M. Innis et al., Academic Press (1990). .
Methods for constructing cDNA libraries in a suitable vector such as a plasmid
or phage for propagation in prokaryotic or eukaryotic cells are well known to
those
skilled in the art. [See e.g. Maniatis et al. SupYa). Suitable cloning vectors
are well
known and are widely available.
In a further embodiment changes in gene transcription are determined at
mRNA level. Decreased or increased expression can be measured at the RNA level
using any of the methods well known in the art for the quantitation of
polynucleotides, such as, for example; nucleic acid amplification, for
instance via
PCR, RT-PCR; RNase protection; Northern blotting and other hybridization
methods. Assay techniques that can be used to determine levels of a protein,
such as
a polypeptide of the present invention, in a sample derived from a host are
well-
known to those of skill in the art. Such assay methods include
radioimmunoassays,
competitive-binding assays, Western Blot analysis and ELISA assays. Assay
techniques that can be used to determine the presence of protein derivatives
or
variants comprise amongst others mass spectrometry.
It is thus an object of the present invention to provide a method for
identifying a
compound capable to alter the CRH signalling response in a cell, said method
comprising;
a) contacting said cell with CRH in the presence and absence of said compound;
b) determine the amount of at least one protein that modulates corticotropin
releasing hormone (CRH) signaling in said cell; and
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-13-
c) compare the amount of said protein in the presence and absence of said
compound;
whereby the protein that modulates corticotropin releasing hormone (CRH)
signaling is being selected from the group consisting of SEQ ID N0.2, SEQ ID
4, SEQ lD N0.6, SEQ ID N0.8, SEQ ID NO.10, SEQ ID N0.12, SEQ ID
N0.14, SEQ ID N0.16, SEQ ID N0.18, SEQ ID N0.20, SEQ ID N0.22, SEQ
ID N0.24, SEQ m N0.26, SEQ ID N0.28, SEQ ID NO.30, SEQ m N0.32,
SEQ ID N0.34, SEQ ll7 N0.36, SEQ ID N0.38, SEQ ID N0.40, SEQ m
N0.42, SEQ ID N0.44, SEQ ll~ N0.46 and SEQ ID NO.48.
Preferably, the method to assay the amount of protein that modulates CRH
signaling is
using an antibody which binds to a polypeptide comprising an amino acid
sequence
selected from the group consisting of SEQ ID N0.2, SEQ ID 4, SEQ ~ NO.6, SEQ
ID
N0.8, SEQ m N0.10, SEQ 117 N0.12, SEQ ID N0.14, SEQ D7 NO.16, SEQ ID
N0.18, SEQ ID NO.20, SEQ 1D N0.22, SEQ ID N0.24, SEQ 1D N0.26, SEQ ID
N0.28, SEQ m N0.30, SEQ m N0.32, SEQ ID N0.34, SEQ ID N0.36, SEQ ID
N0.38, SEQ ID N0.40, SEQ ID N0.42, SEQ ID N0.44, SEQ ID N0.46 and SEQ ID
N0.48.
Thus in another embodiment, this invention provides a monospecific antibody
immunologically reactive with a protein that modulates CRH signalling said
protein
being selected from the group consisting of SEQ ID N0.2, SEQ ID 4, SEQ ID
N0.6,
SEQ ID N0.8, SEQ ID N0.10, SEQ ID N0.12, SEQ ID N0.14, SEQ ID N0.16,
SEQ ID N0.18, SEQ lD N0.20, SEQ ID NO.22, SEQ m N0.24, SEQ ID N0.26,
SEQ ID N0.28, SEQ II7 N0.30, SEQ 117 N0.32, SEQ ID N0.34, SEQ ID N0.36,
SEQ ID N0.38, SEQ ID N0.40, SEQ 117 N0.42, SEQ lD N0.44, SEQ ID N0.46
and SEQ m N0.48. Antibodies generated against polypeptides of the present
invention may be obtained by administering the polypeptides or epitope-bearing
fragments, analogs or cells expressing these to an animal, preferably a non-
human
animal, using routine protocols. For preparation of monoclonal antibodies, any
technique which provides antibodies produced by continuous cell line cultures
can
be used. Examples include the hybridoma technique (Kohler, G. and Milstein,
C.,
NcztuYe (1975)256:495-497), the trioma technique, the human B-cell hybridoma
technique (Kozbor et ad., Imjnufz.ology Today (1983)4:72) and the EBV-
hybridoma
technique (Cole et al., MONOCLONAL ANTIBODIES AND CANCER
THERAPY, pp.77-96, Alan R. Liss, Inc., 1985).
Techniques for the production of single chain antibodies, such as those
described in U.S. Patent No.4,946,778, can also be adapted to produce single
chain
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-14-
antibodies to polypeptides of this invention. Also, transgenic mice, or other
organisms, including other mammals, may be used to express humanized
antibodies.
The above-described antibodies may be employed to isolate or to identify
clones expressing the polypeptide or to purify the polypeptides by affinity
chromatography.
Antibodies against polypeptides of the present invention may also be employed
to treat the CRH metabolism related disorders such as CRH induced stress or
depression amongst others.
To determine the amount of protein that modulates CRH signalling, the
antibodies according to the invention are used in conventional immunological
techniques. Suitable immunological techniques are well known to those skilled
in the
art and include for example, ELISA, Western Blot analysis, competitive or
sandwich
immunoassays and the like, as is otherwise well known they all depend on the
formation of an antigen-antibody immune complex wherein for the purpose of the
assay, the antibody can be detectable labeled with, e.g. radio, enzyme or
fluorescent
labels or it can be immobilized on insoluble carriers.
For example in an ELISA screening format the antibody is added to a solid
phase (for example the bottom of a microplate) which is coated with either the
protein
or a peptide fragment thereof coupled to a carrier (such as BSA), and then,
adding an
anti-immunoglobin antibody (for example when the immunization is performed in
mice, an anti-mouse immunoglobulin antibody is used, e.g. sheep-anti-mouse
immunoglobulin (Ig)) conjugated with a detectable label such as an enzyme,
preferably
horseradish peroxidase, or a radioactive isotope such as lasl.
It is thus an object of the invention to provide immunoassays for the
determination or detection of proteins that modulate CRH signalling in a
sample, the
method comprising contacting the sample with an antibody to the proteins
according to
the invention and determining whether an immune complex is formed between the
antibody and said protein. These methods can either be performed on tissue
samples
or body fluid samples and generally comprise obtaining a sample from the body
of a
subject; contacting said sample with an imaging effective amount of a
detestably
labeled antibody according to the invention; and detecting the label to
establish the
presence of proteins that modulate CRH signalling in the sample.
The measuring methods using the antibodies of the present invention are not
particularly limited. Any measuring method may be used as long as the amount
of
antibodies, antigens or the antigens~antibody complexes corresponding to the
amount of
the antigens to be measured is detected by chemical or physical means, and
calculated
from standard curves prepared by the use of standard solutions containing the
antigens
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-15-
in known amounts. For example, nephelometry, competitive methods, immunometric
methods and sandwich methods are suitably used. With respect to sensitivity
and
specificity, it is particularly preferred to use sandwich methods described
below.
In measuring methods using labelling substances, radioisotopes, enzymes,
fluorescent substances, luminous substances, etc. are used as labelling
agents.
Examples of the radioisotopes include lzsh isih 3H and ~4C. Enzymes are
usually
made detectable by conjugation of an appropriate substrate that, in turn
catalyzes a
detectable reaction. Examples thereof include, for example, beta-
galactosidase, beta
glucosidase, alkaline phosphatase, peroxidase and malate deydrogenase,
preferably
horseradish peroxidase. The luminous substances include, for example, luminol,
luminol derivatives, luciferin, aequorin and luciferase. Further, the avidin-
biotin
systems can also be used for labelling the antibodies and immunogens of the
present
invention.
Accordingly, in a further aspect, the present invention provides for a method
of
identifying and obtaining compounds that alter the CRH signalling response
activity
in a cell, comprising:
a) contacting a cell which expresses at least one protein comprising an amino
acid
sequence selected from the group consisting of SEQ II? N0.2, SEQ ID 4, SEQ ID
N0.6, SEQ ff~ N0.8, SEQ ID N0.10, SEQ ID N0.12, SEQ ZD N0.14, SEQ >D,
NO.16, SEQ ID N0.18, SEQ ID N0.20, SEQ ll~ N0.22, SEQ ID N0.24, SEQ
D7 NO.26, SEQ ID N0.28, SEQ ID N0.30, SEQ ID N0.32, SEQ ID N0.34,
SEQ JD NO.36, SEQ ll~ N0.38, SEQ ID N0.40, SEQ ID N0.42, SEQ ID
N0.44, SEQ ID N0.46 and SEQ ID N0.48, with said test compound; and
b) compare the CRH response activity of said cell in the presence and absence
of
said compound.
Changes in membrane potential can be measured using conventional
electrophysiological techniques and when they become available, using novel
high
throughput methods currently under development. Since the change in membrane
potential are normally the result of ion fluxes, as an alternative approach,
changes in
membrane potential can be measured indirectly through the change in
intracellular ion
concentrations using ion-sensitive fluorescent dyes, including fluo-3, fluo-4,
flux-5N,
fura red, Sodium Green, SBFI and other similar probes from suppliers including
Molecular Probes. Other fluorescent dyes, from suppliers including Molecular
Probes,
such as DIBAC 4 ~3~ or Di-4-Anepps can detect membrane potential changes. For
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-16-
example calcium and sodium ion fluxes can thus be characterised in real time,
using
fluorornetric and fluorescence imaging techniques, including fluorescence
microscopy
with or without laser confocal methods combined with image analysis
algorithms.
In a preferred embodiment this assay is based around an instrument called a
FLuorescence Imaging Plate Reader ((FLIPR), Molecular Devices Corporation). In
its
most common configuration, it excites and measures fluorescence emitted by
fluorescein-based dyes. It uses an argon-ion laser to produce high power
excitation at
488 nm of a fluorophore, a system of optics to rapidly scan the over the
bottom of a 96-
/384-well plate and a sensitive, cooled CCD camera to capture the emitted
fluorescence.
It also contains a 96-1384-well pipetting head allowing the instrument to
deliver
solutions of test agents into the wells of a 96-J384-well plate. The FLIPR
assay is
designed to measure fluorescence signals from populations of cells before,
during and
after addition of compounds, in real time, from all 96-/384-wells
simultaneously.
It is thus an object of the present invention to provide a FL,IPR assay used
to
screen for and characterise compounds functionally active in modulating CRH
response
in cells, said cells expressing a protein selected from the group consisting
of SEQ ff~
26, SEQ ID N0.28, SEQ ID N0.30, SEQ )D N0.32 and SEQ lD N0.34.
In an alternative embodiment, the activity of the cell may be assessed using
electrophysiological methods. Therefore, proteins modulating CRH signalling in
a
cell can be characterised using whole cell and single channel
electrophysiology.
It is thus a further object of this invention to provide a screening method to
identify compounds which modulate CRH signalling response activity in a cell,
said
method comprising;
(a) contacting a host cell expressing a protein selected from the group
consisting of
SEQ ID N0.2, SEQ ID 4, SEQ II? N0.6, SEQ II? N0.8, SEQ ID N0.10, SEQ B?
N0.12, SEQ ID N0.14, SEQ ID N0.16, SEQ 1D N0.18, SEQ ID N0.20, SEQ ID
N0.22, SEQ ID NO.24, SEQ ID N0.26, SEQ ID N0.28, SEQ ID N0.30, SEQ ID
N0.32, SEQ ll7 N0.34, SEQ >D N0.36, SEQ ID N0.38, SEQ ID N0.40, SEQ ll~
N0.42, SEQ ID N0.44, SEQ ID N0.46 and SEQ ID N0.48, with a compound to be
tested;
(b) measuring the effect of the test compound on the membrane potential of
said cell
using electrophysiological techniques; and
(c) compare the CRH response activity of said cell in the presence and absence
of
said compound. Alternatively, the host cells in the aforementioned screening
method express a protein selected from the group consisting of SEQ ID No. 26,
SEQ
ID No.28, SEQ ID No. 30, SEQ TD No. 32 and SEQ lD N0.34.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-17-
In a preferred embodiment the host cells are Xenopus oocytes and the
electrophysiological measurement consists of measuring the membrane current
using
the voltage clamp technique at distinct membrane potentials.
Changes in enzyme activity of an enzyme that modulates CRH signalling a said
cell,
can generally be measured or assayed in terms of the catalytic effect the
enzyme
produces, that is the conversion of its substrate into reaction product. For
example, for
kinases one may assess the kinase activity using a substrate comprising the
kinase
specific phosphorylation site and by measuring the phosphorylation of the
substrate.
Similarly for phosphatases one may assess the phosphatase activity using a
phosphorylated substrate and by measuring the dephosphorylation of the
substrate.
These assays may be performed both in the presence and absence of the compound
to
be tested.
It is thus an object of the present invention to provide a method for
identifying a
compound capable to alter the CRH signalling response activity in a cell, said
method
comprising;
a) contacting a mixture comprising a kinase selected from the group consisting
of SEQ ID N0.10, SEQ ID N0.12, SEQ ID NO.14, SEQ ID Nol6 and SEQ R3 NO.18,
with a source of phosphate and a suitable kinase substrate;
b) incubating said mixture in the presence or absence of a said compound and ;
measuring the level of phosphorylation of said substrate in the presence of
said
compound compared to the level of phosphorylation of said substrate in the
absence of
said test compound.
In the assay of the invention, the kinase may be provided as a protein or it
may be
provided in the assay mixture as an mRNA encoding said kinase. When the assay
comprises cell-free components, the kinase is provided as the protein. When
the assay
is conducted in the milieu of a cell, the kinase may be provided as either the
protein or
as an mRNA encoding said kinase, wherein, in order that the kinase be
available in the
assay, the mRNA is translated and kinase protein is thereby produced. It will
be
apparent from the Examples provided herein that it is a simple matter to
obtain mRNA
specifying the kinase and inject the mRNA into a cell for production of the
kinase
protein. The lcinase may also be provided by expression of a plasmid, which
encodes
the kinase protein. Standard molecular biology techniques may be used to
construct
operable plasmids encoding the kinase protein and to express the plasmid in
cells
(Sambrook, et al., 1989, In: Molecular Cloning: A Laboratory Manual, Cold
Spring
Harbor Laboratory, New Yorlc).
As discussed herein, the method of identifying a kinase modulator may be
performed
either in vitro wherein the assay mixture is cell-free, in vitro wherein live
cells are
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-18-
included in the assay, or in vivo in an animal. Thus, in one aspect of the
invention, the
mixture is contained within a eukaryotic cell and the method of the invention
may be
performed wherein some of the components of the assay mixture may be provided
exogenously to a cell my microinjection of the components therein, and some of
the
components may be endogenous in the cell.
The term "endogenous in the cell" as used herein, means that the component is
naturally
produced in the subject cell.
The term "exogenous to the cell" as used herein, means that the component is
not found
naturally in the subject cell, or is found therein at a low level, and is
added thereto.
When the method of the invention is performed using a eukaryotic cell, one or
more of
the kinase protein, the kinase substrate and the test compound may be injected
into the
eukaryotic cell prior to the incubation. The cell so injected is then
incubated under
conditions that facilitate protein kinase activity and the level of protein
kinase activity
is subsequently measured following the incubation period using the assays
described
herein.
The eukaryotic cell that is useful in the methods of the invention may be any
one of a
Xenopus laevis oocyte, a Xenopus laevis embryo cell, a mammalian cell (such as
a T
OTI/2 cell), a Drosophila melanogaster S2 cell, a Dictyostefium discoideum
cell and a
yeast cell. Still more preferably, the eukaryotic cell is the murine pituitary
corticotroph-
derived adenoma cell line cell AtT-20.
The source of phosphate for use in the methods of the invention may be any
common
source of phosphate, including, but not limited to, a nucleotide triphosphates
such as,
but not limited to, ATP or GTP. In a preferred embodiment, the phosphate
source has
bound thereon a detectable label which label is transferred with the phosphate
group to
the kinase substrate during the reaction. In this manner, phosphorylated
kinase substrate
may be distinguished from non- phosphorylated kinase substrate in that the
phosphorylated substrate will contain the detectable label whereas the non-
phosphorylated substrate will not contain the label. In another embodiment,
the
phosphate source does not have bound thereon a detectable label; instead,
phosphorylated kinase substrate may be distinguished from non- phosphorylated
kinase
substrate, for instance by recognition of one form of the substrate, but not
the other, by
an antibody.
The detectable label, which is useful in the methods of the invention may
include any
known or heretofore unknown detectable label which is transferred to the
kinase
substrate upon transfer of a phosphate group thereto as a result of protein
lcinase
activity.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-19-
Labels which are useful include, but axe not limited to, radioactive labels,
such as ~y3aP,
3iS , and non-radioactive labels, such as biotin and the like.
In another embodiment, the present invention to provide a method for
identifying a
compound capable to alter the CRH signalling response activity in a cell, said
method
comprising;
a) contacting a mixture comprising a phosphatase selected from the group
consisting of SEQ m N0.36 and SEQ m N0.38, and a suitable phosphorylated
substrate;
b) incubating said mixture in the presence or absence of a said compound and ;
measuring the level of phosphorylation of said substrate in the presence of
said
compound compared to the level of phosphorylation of said substrate in the
absence of
said test compound.
As for the kinase assay, the phosphatase in the assay of the invention, may be
provided
as a protein or it may be provided in the assay mixture as an mRNA encoding
said
phosphatase. The phosphorylated substrate is typically labeled with a
detectable
phosphate residue. Labels which axe useful include, but are not limited to,
radioactive
labels, such as ~2P, 31S , and non-radioactive labels, such as biotin and the
like. Fox
use in a phosphatase activity assay, the substrate preferably consists of a
peptide
substrate, phosphorylated at a tyrosine or serine residue, typically labeled
with ~2P. In
general, phosphorylation may be accomplished in a variety of ways. Typically,
a protein
tyrosine kinase is used. For example, a soluble EGF-receptor kinase in
combination
with . sup.32 P-labeled ATP may be used to phosphorylate a tyrosine residue on
a
peptide of the present invention. Such a phosphorylation reaction is typically
allowed to
proceed for about 2 hours at 30° C. or overnight at room temperature.
Phosphorylated peptide, hereinafter referred to as "phosphopeptide", is then
purified
from a phosphorylation reaction mixture. For example, peptide may be separated
from a
reaction mixture by addition of trichloroacetic acid and centrifugation,
whereby the
peptide remains in the supernate. The peptide is generally further purified by
column
chromatography, e.g. , on C18. Purified phosphorylated peptide may be
lyophilized and
stoxed at - 20° C. prior to use.
Following incubation, phosphopeptide which is not dephosphorylated ("non-
dephosphorylated phosphopeptide") is separated from radioactivity released by
dephosphorylation of phosphopeptide (i.e., from free radioactive phosphorus
released
by dephosphorylation). As used herein, the term "radioactive phosphorous"
includes all
forms in which a radioactive phosphorus atom may be present on a tyrosine
residue and
removed by dephosphorylation, e.g., as a phosphate group. Typically,
separation of
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-20-
non-dephosphorylated phosphopeptide from free radioactive phosphorus released
by
dephosphorylation of phosphopeptide is effected by centrifugation, following
termination of the dephosphorylation reaction by the addition of substances
including
nonradioactive phosphates and charcoal. Radioactivity in the supernate is
determined
by means well known to those of ordinary skill in the art. Based upon the
amount of
radioactivity added to the assay mixture initially via the phosphopeptide and
the amount
of radioactivity detected at the end of the assay as radioactivity released by
dephosphorylation, the phosphatase enzymatic activity of the sample assayed
may be
calculated.
It is also an embodiment of the present invention to provide a method for
identifying a
compound capable to alter the CRH signalling response activity in a cell, said
method
comprising;
a) contacting a cell which expresses at least one protein comprising an amino
acid sequence selected from the group consisting of SEQ ID N0.26, SEQ ID
N0.28,
SEQ ID N0.30, SEQ ID N0.32 and SEQ ID NO.34 with said test compound; and
b) compare the levels of a second messenger, such as cAMP, cGMP, Ca2~ or IP3
in said cell, in the presence and absence of said compound.
Levels of second messengers can be determined using art known techniques
either in
whole cells or cellular extracts comprising one of the aforementioned
proteins.
A further method to identify a compound capable to alter CRH signalling in a
cell is
based on the use of a gene, such as a reporter gene, operably linked to a gene
promoter
or regulatory sequence element thereof characterized in that said gene
promoter or
regulatory sequence element comprises a transcription factor binding site,
wherein said
transcription factor is capable of modulating CRH signalling in a cell. In a
preferred
embodiment the transcription factor capable of modulating CRH signalling in a
cell is
being selected from SEQ ID N0.2, SEQ ID N0.4, SEQ 117 N0.6 and SEQ ID N0.8.
Accordingly, the present invention provides a recombinant DNA molecule
comprising
the gene promoter region as defined above. In the said recombinant DNA
molecule, the
promoter region can be operably linked to a nucleic acid molecule encoding a
detectable product, such as a reporter gene. The term "operably linked", as
used herein,
means functionally fusing a promoter with a gene in the proper frame to
express the
gene under control of the promoter. As used herein, the term "reporter gene"
means a
gene encoding a gene product that can be identified using simple, inexpensive
methods
or reagents and that can be operably linked to the promoter region or an
active fragment
thereof. Reporter genes such as, for example, a firefly luciferase, ~-
galactosidase,
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-21-
alkaline phosphatase, the bacterial chloramphenicol acetyl transferase or
green
fluorescent protein reporter gene, can be used to determine transcriptional
activity in
screening assays according to the invention (see, for example, Goeddel (ed.),
Methods
Enzymol., Vol. 185, San Diego:Academic Press, Inc. (1990); see also Sambrook,
supra). In a preferred embodiment, the reporter gene is the firefly luciferase
gene.
The invention also provides a vector comprising the recombinant DNA molecule
as
defined above, as well as a host cell stably transformed with such a vector,
or generally
with the recombinant DNA molecule according to the invention. The term
"vector"
refers to any carrier of exogenous DNA that is useful for transferring the DNA
into a
host cell for replication and/or appropriate expression of the exogenous DNA
by the
host cell. Accordingly, in a specific embodiment said vector is an expression
vector
such as pGL3luc, pBLCATS (LMBP 2451), pGMCSFIacZ (LMBP 2979), pEGFP or
pSEAPbasic (DMB 3115),wherein LMBP and DMB numbers refer to the accession
numbers of these expression vectors at the Belgian Co-ordinated Collections of
Micro-
organisms.
In another aspect, the invention provides a method for identification of a
compound
modulating CRH signalling activity, said method comprising the steps: (i)
contacting a
candidate agent with a gene promoter region as defined above; and (ii)
determining
whether said candidate agent modulates expression of the detectable product,
such
modulation being indicative for an agent capable of modulating CRH signalling
activity. The detectable product refers either to the gene encoded protein or
to the
product of a reporter gene such as luciferase,(3-galactosidase or green
fluorescent
protein. Methods to quantify the detectable products are generally known in
the art and
include amongst others the use of a colorimetric substrate if the expression
product is
an enzyme, the use of specific antibodies in an RIA or ELISA assay or the
measurement
of the level of mRNA transcribed from genes operably linked to the promoter,
wherein
said mRNA can be measured either directly or indirectly using standard
procedures.
Preferably, the gene promoter comprising a transcription factor binding site
for a
transcription factor selected from the group consisting of SEQ DJ N0.2, SEQ ID
N0.4,
SEQ 117 N0.6 and SEQ ID N0.8.
Identification of the zzucleic acid sequezZCes encodi~tg proteins capable of
zzzodulatizZg CRH sigzzaliz2g.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-22-
In another aspect the present invention relates to isolated and purified
nucleic acid
molecules which encodes proteins capable of modulating CRH signalling, wherein
said
nucleic acid molecule is either RNA, DNA, cDNA or genomic DNA.
In particular, the present invention encompasses an isolated and purified
nucleic
acid molecule comprising a member selected from a group consisting of:
(a) a nucleic acid molecule encoding a protein that modulates CRH signalling
having at least a 70% identity to a polypeptide comprising an amino acid
sequence selected from the group consisting of SEQ m No.46 and SEQ ID
No.48 ;
(b) a nucleic acid molecule which is complementary to the polynucleotide of
(a);
(c) a nucleic acid molecule comprising at least 15 sequential bases of the
polynucleotide of (a) or (b);
(d) a nucleic acid molecule that hybridizes under stringent conditions to the
polynucleotide molecule of (a) or (b); and
(e) a nucleic acid molecule encoding a protein that modulates CRH signalling
comprising a nucleotide sequence of which is degenerated as a result of the
genetic code to a nucleotide sequence of a polynucleotide of any of (a) to
(d).
Those skilled in the art will recognize that owing to the degeneracy of the
genetic
code, numerous "silent" substitutions of nucleotide base pairs could be
introduced
into the sequences identified as SEQ ID N0:45, SEQ ID NO 47 or SEQ ID N0:49
without altering the identity of the encoded amino acids) or protein products.
All
such substitutions are intended to be within the scope of the invention.
In a further aspect, the present invention relates to Human purine permease
polynucleotides. Such polynucleotides include isolated polynucleotides
comprising
a nucleotide sequence encoding a polypeptide which has at least 70% identity,
preferably at least 80% identity, more preferably at least 90% identity, yet
more
preferably at least 95% identity, to the amino acid sequence selected from the
group
consisting of SEQ ID N0:46 and SEQ m N0:48, over the entire length said amino
acid sequence. In this regard, polypeptides which have at least 97% identity
are
highly preferred, while those with at least 98-99% are more highly preferred,
and
those with at least 99% identity are most highly preferred. Such
polynucleotides
include a polynucleotide consisting essentially of a polynucleotide sequence
selected from SEQ ff~ No.45, SEQ ll~ No.47 or SEQ m No.49.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-23-
Accordingly, in a further aspect, the present invention provides for an
isolated polynucleotide, comprising:
a) a nucleotide sequence encoding a polypeptide which has at least 70%
identity,
preferably at least 80% identity, more preferably at least 90% identity, more
preferably at least 90% identity, yet more preferably at least 90% identity,
yet more
preferably at least 95% identity, even more preferably at least 97-99%
identity to an
amino acid sequence selected from the group consisting of SEQ ID No. 46 and
SEQ
ID No.48;
b) a nucleotide sequence which has at least 70% identity, preferably at least
80%
identity, more preferably at least 90% identity, more preferably at least 90%
identity,
Yet more preferably at least 90% identity, yet more preferably at least 95%
identity,
even more preferably at least 97-99% identity, to a polynucleotide selected
from the
group consisting of SEQ >D No.45, SEQ ID No.47 and SEQ ID No.49, over the
entire length of said polynucleotide;
c) a nucleotide sequence which has at least 70% identity, preferably at least
80%
identity, more preferably at least 90% identity, more preferably at least 90%
identity,
Yet more preferably at least 90% identity, yet more preferably at least 95%
identity,
even more preferably at least 97-99% identity a polynucleotide selected from
the
group consisting of SEQ ID No.45, SEQ ID No.47 and SEQ ID No.49, over the
entire coding region of said polynucleotide; and
d) a nucleotide sequence consisting of a polynucleotide selected from the
group
consisting of SEQ ID No.45, SEQ ll~ No.47 and SEQ ID No.49.
The polynucleotides as outlined above are in particular provided for use in a
screening method or diagnostic method according to the invention. In
particular to
identify capable of modulating CRH signaling in a cell or to diagnose altered
CRH
metabolism in an individual.
Identity or similarity, as known in the art, are relationships between two or
more polypeptide sequences or two or more polynucleotide sequences, as
determined
by comparing the sequences. In the art, identity also means the degree of
sequence
relatedness between polypeptide or polynucleotide sequences, as the case may
be, as
determined by the match between strings of such sequences. Both identity and
similarity can be readily calculated (Computational Molecular Biology, Leslc,
A. M.,
ed., Oxford University Press, New York, 1988; Biocomputing: Informatics and
Genome Projects, Smith, D. W., ed., Academic Press, New York, 1993; Computer
Analysis of Sequence Data, Part I, Griffin, A. M., and Griffin, H. G., eds.,
Humana
Press, New Jersey, 1994; Sequence Analysis in Molecular Biology, von Heinje,
G.,
Academic Press, 1987; and Sequence Analysis Primer, Gribskov, M. and Devereux,
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-24-
J., eds., M Stockton Press, New York, 1991). While there exist a number of
methods to measure identity and similarity between two polynucleotide or two
polypeptide sequences, both terms are well known to skilled artisans (Sequence
Analysis in Molecular Biology, von Heinje, G., Academic Press, 1987; Sequence
Analysis Primer, Gribskov, M. and Devereux, J., eds., M 5tockton Press, New
York,
1991; and Carillo, H., and Lipman, D., (1988) SIAM J. Applied Math., 48, 1073.
Methods commonly employed to determine identity or similarity between
sequences
include, but are not limited to those disclosed in Carillo, H., and Lipman,
D., (1988)
SIAM J. Applied Math., 48, 1073. Preferred methods to determine identity are
designed to give the largest match between the sequences tested. Methods to
determine identity and similaa.-ity are codified in computer programs.
Preferred
computer program methods to determine identity and similarity between two
sequences include, but are not limited to, GCG program package (Devereux, J.,
et
al., (1984) Nucleic Acids Research 12(1), 387), BLASTP, BLASTN, and FASTA
(Atschul, S. F. et al., (1990) J. Molec. Biol. 215, 403).
The nucleic acid sequence encoding a protein capable of modulating CRH
activity, or fragment thereof, can be isolated from a tissue in which said
gene is
expressed, such as but not limited to, brain, hart, kidney, pancreas, liver
and skin.
Said sequence can also be isolated from mammals other than human and mouse.
Other cells and cell lines may also be suitable for use to isolate mammalian
purine
permease cDNA. Selection of suitable cells may be done by screening for CRH
modulating activity in cell extracts or in whole cell assays, as described
herein.
Cells that possess CRH modulating activity in any one of these assays may be
suitable for the isolation of purine permease DNA or mRNA.
Any of a variety of procedures known in the art may be used to moleculary
clone DNA encoding a protein according to the invention. In one method, mRNA
is isolated, and first strand cDNA synthesis is carried out. A second round of
DNA
synthesis can be carried out for the production of the second strand.
Subsequently
by the specific PCR amplification of DNA fragments through the design of
degenerate oligonucleotide primers from the amino acid sequence of the
purified
protein that modulates CRH signalling, an isolated cDNA can be obtained. If
desired the double-stranded cDNA can be cloned into any suitable vector, for
example, a plasmid, thereby forming a cDNA libary. Another method is to screen
cDNA libraries constructed in a bacteriophage or plasmid shuttle vector with a
labeled oligonucleotide probe targeted to any suitable region of SEQ m NO: 1,
SEQ ID NO 3, SEQ ID N0.5, SEQ II? N0.7, SEQ ID N0.9, SEQ ID N0.11, SEQ
ID N0.13, SEQ ID N0.15, SEQ ID N0.17, SEQ ID N0.19, SEQ ID N0.21, SEQ
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-25-
ID N0.23, SEQ ID N0.25, SEQ ID N0.27, SEQ ID NO. 29, SEQ ID N0.31, SEQ
ID N0.33, SEQ DJ N0.35, SEQ ID N0.37, SEQ ID N0.39, SEQ lD N0.41, SEQ
ll~ N0.43, SEQ ID N0.45, SEQ ID N0.47 or SEQ ID N0:49. See e.g. PCR
Protocols: A Guide to Method and Application, Ed. M. Innis et al., Academic
Press
(1990)..
Methods for constructing cDNA libraries in a suitable vector such as a
plasmid or phage for propagation in prokaryotic or eukaryotic cells are well
known
to those skilled in the art. [See e.g. Maniatis et al. Supra]. Suitable
cloning vectors
are well known and are widely available.
It is readily apparent to those skilled in the art that other types of
libraries, as
well as libraries constructed from other cells or cell types, may be useful
fox
isolating the nucleic acid sequences according to the invention. Other types
of
libraries include, but are not limited to, cDNA libraries derived from other
cells,
from organisms other than human and mouse, and genomic DNA libraries that
include YAC (yeast artificial chromosome) and cosmid libraries. Construction
of
genomic DNA libraries can be performed by standard techniques well known in
the
art. Well known genomic DNA library construction techniques can be found in T.
Maniatis et al. Molecular Cloning: A Laboratory Manual, 2d Ed. Chap. 14
(1989).
The skilled artisan will appreciate that, in many cases, an isolated cDNA
sequence will be incomplete, in that the region coding for the polypeptide is
short at
the 5' end of the cDNA. This is a consequence of reverse transcriptase, an
enzyme
with inherently low 'processivity' (a measure of the ability of the enzyme to
remain
attached to the template during the polymerisation reaction), failing to
complete a
DNA copy of the mRNA template during the ls' strand cDNA synthesis.
There are several methods available and well known to those skilled in the
art to obtain full-length cDNAs, or extend short cDNAs, for example those
based on
the method of Rapid Amplification of cDNA ends (RACE) (Frohman et al., 1988,
PNAS USA ~5, 8998-9002), or recent modifications of this technique,
exemplified
by the MarathonTM technology (Clontech Laboratories Inc.).
In order to clone the polynucleotide encoding aprotein according to the
invention by the above methods, the amino acid sequence of the polypeptide
encoded by said nucleic acid sequence may be necessary. To accomplish this,
the
proteins according to the invention, may be purified and partial amino acid
sequence determined by automated sequenators. It is not necessary to determine
the
entire amino acid sequence, but the linear sequence of two regions of 6 to 8
amino
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-26-
acids from the protein is determined for the production of primers for PCR
amplification of a partial DNA fragment.
Once suitable amino acid sequences have been identified, the DNA
sequences capable of encoding them are synthesized. Because the genetic code
is
degenerate, more than one codon may be used to encode a particular amino acid,
and therefore, the amino acid sequence can be encoded by any of a set of
similar
DNA oligonucleotides. Only one member of the set will be identical to the
polynucleotide sequences according to the invention and will be capable of
hybridizing to DNA encoding the desired protein, even in the presence of DNA
oligonucleotides with mismatches. DNA isolated by these methods can be used to
screen DNA libraries from a variety of cell types, from invertebrate and
vertebrate
sources, and to isolate homologous genes.
polypeptides
In a further embodiment this invention relates to a polypeptide in a
substantially pure form which modulate CRH signalling wherein said polypeptide
is
encoded by an isolated and purified nucleic acid molecule according to the
invention. In a preferred embodiment the polypeptide has the amino acid
sequence
selected from the group consisting of SEQ 1D NO 46, SEQ ID NO 48 and
functional analogs thereof
The protein according to the invention includes all possible amino acid
variants encoded by the nucleic acid according to the invention including a
polypeptide encoded by said molecule and having conservative amino acid
changes.
Those skilled in the art will recognize that theprotein which modulate CRH
signalling could be obtained by a plurality of recombinant DNA techniques
including, for example, hybridization, polymerase chain reaction (PCR)
amplification, or de novo DNA synthesis (See e.g., T. Maniatis et al.
Molecular
Cloning: A Laboratory Manual, 2d Ed. Chap. 14 (1989)).
Purified biologically active protein that modulates CRH signallings may have
several different physical forms. The polypeptides according to the invention
may
exist as full-length nascent or unprocessed polypeptides, or as partially
processed
polypeptides or combinations of processed polypeptides. The full-length
nascent
polypeptide may be post-translationally modified, amongst other, by specific
proteolytic cleavage events that result in the formation of fragments of the
full-length
nascent polypeptide. A fragment, or physical association of fragments may have
the
full biological activity associated with proteins according to the invention;
however,
the degree of CRH modulating activity may vary between individual fragments.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-27-
Also preferred in this aspect of the invention are fragments characterized by
structural or functional attributes of the polypeptide. Preferred embodiments
of the
invention in this regard include fragments that comprise alpha-helix and alpha-
helix
forming regions, beta-sheet and beta-sheet-forming regions, turn and turn-
forming
regions, coil and coil-forming regions, hydrophilic regions, hydrophobic
regions,
alpha amphipathic regions, beta amphipathic regions, flexible regions, surface-
forming regions, substrate binding region, high antigenic index regions of the
polypeptide of the invention, and combinations of such fragments. Preferred
regions
are those that mediate activities of the polypeptides of the invention. Most
highly
preferred in this regard are fragments that have a chemical, biological or
other
activity of the response regulator polypeptide of the invention, including
those with a
similar activity or an improved activity, or with a decreased undesirable
activity.
Recom.bifzafzt expressio~z of polynucleotides encoding proteifz which modulate
CRH
activity
In another embodiment polynucleotides according to the invention may be
recombinantly expressed by molecular cloning into an expression vector
containing
a suitable promoter and other appropriate transcription regulatory elements,
and
transferred into prokaryotic or eukaryotic host cells to produce a protein
that
modulates CRH signalling. Techniques for such manipulations are fully
described
in Maniatis, T, et al., su ra, and are well known in the art.
Therefore, in a further aspect this invention provides an expression vector
for
expression of a protein that modulates CRH signalling in a recombinant host,
wherein
said vector contains a nucleic acid sequence encoding a protein that modulates
CRH
signalling and functional analogs thereof. In a more preferred aspect of this
invention
this expression vector contains a nucleic acid molecule encoding a protein
that
modulates CRH signalling, having a nucleotide sequence selected from a group
consisting of: SEQ ID N0:45, SEQ ID NO 47, SEQ ID N0:49 and functional analogs
thereof or contains genomic DNA encoding a protein that modulates CRH
signalling.
Expression vectors are defined herein as DNA sequences that are required
for the transcription of cloned copies of genes and the translation of their
mRNAs in
an appropriate host. Such vectors can be used to express eulcaryotic genes in
a
variety of hosts such as bacteria including E. coli, cyanobacteria, plant
cells, insect
cells, amphibian cells, fungal cells including yeast cells, and animal cells.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-28-
Specifically designed vectors allow the shuttling of DNA between hosts
such as bacteria-yeast or bacteria-animal cells or bacteria-fungal cells or
bacteria-
invertebrate cells. An appropriately constructed expression vector may
contain: an
origin of replication for autonomous replication in host cells, selectable
markers, a
limited number of useful restriction enzyme sites, a potential for high copy
number,
and active promoters. A promoter is defined as a DNA sequence that directs RNA
polymerase to bind to DNA and initiate RNA synthesis. A strong promoter is one
that causes mRNAs to be initiated at high frequency. Expression vectors may
include, but are not limited to, cloning vectors, modified cloning vectors,
specifically designed plasmids or viruses.
The isolated and purified nucleic acid molecules, according to the invention,
encoding a protein which modulates CRH signalling may be cloned into an
expression vector for expression in a recombinant host cell. Recombinant host
cells
may be prokaryotic or eukaryotic, including but not limited to bacteria such
as E.
coli, fungal cells such as yeast, amphibian cells such as Xenopus oocytes,
mammalian cells including but not limited to cell lines of human, bovine,
porcine,
monkey and rodent origin, and insect cells including but not limited to
Drosophila-
and silkworm-derived cell lines. Cell lines derived from mammalian species
which
may be suitable and which are commercially available, include but are not
limited
to, CV-1 (ATCC CCL 70), COS-1 (ATCC CRL 1650), COS-7 (ATCC CRL 1651),
CHO-Kl (ATCC CCL 61), 3T3 (ATCC CCL 92), NIHi3T3 (ATCC CRL 1658),
HeLa (ATCC CCL 2), C127I (ATCC CRL 1616), BS-C-1 (ATCC CCL 26), MRC-
5 (ATCC CCL 171), L-cells, neuroblastoma, glial cells and HEK-293 (ATCC
CRL1573).
Therefore, in a further embodiment this invention relates to a recombinant
host cell containing a recombinantly cloned nucleic acid molecule encoding a
protein that modulates CRH signalling or functional analog thereof. In a
further
aspect the recombinant host cell according to the invention contains a nucleic
acid
molecule which is either genomic DNA or has a nucleotide sequence selected
from
a group consisting of: (SEQ ll~ N0:45); (SEQ ff~ NO 47); (SEQ TD N0:49); and
functional analogs thereof.
The expression vector may be introduced into host cells via any one of a
number of techniques including but not limited to transformation,
transfection,
protoplast fusion, lipofection, and electroporation. The expression vector-
containing cells are clonally propagated and analyzed to determine whether
they
produce a protein that modulates CRH signalling. Identification of permeases
expressing host cell clones may be done by several means, including but not
limited
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-29-
to immunological reactivity with antibodies directed against the polypeptides
according to the invention, and the presence of host cell-associated mammalian
purine permease activity.
Thus, the present invention also relates to a process for expression of
protein
that modulates CRH signalling in a recombinant host cell, comprising culturing
the
host cells according to the invention under conditions which allow expression
of the
protein that modulates CRH signalling from the expression vector as outlined
herein. The proteins of this invention may be synthesized either by direct
expression or as a fusion protein comprising the protein of interest as a
translational
fusion with another protein or peptide that may be removable by self,
enzymatic or
chemical cleavage. Therefore, in a particular embodiment this invention
provides
the proteins according to the invention wherein said polypetides are part of a
fusion
protein.
It is often observed in the production of certain peptides in recombinant
systems that expression as a fusion protein prolongs the life span, increases
the
yield of the desired peptide, or provides a convenient means of purifying the
protein. This is particularly relevant when expressing mammalian proteins in
prokaryotic hosts. A variety of peptidases (e.g. enterokinase and thrombin),
which
cleave a polypeptide at specific sites or digest, the peptides from the amino
or
carboxy termini (e.g. diaminopeptidase) of the peptide chain are known.
Furthermore, particular chemicals (e.g. cyanogen bromide) will cleave a
polypeptide chain at specific sites. The skilled artisan will appreciate the
modifications necessary to the amino acid sequence (and synthetic or semi-
synthetic
coding sequence if recombinant means are employed) to incorporate site-
specific
internal cleavage sites. See e.g., P.Carter, "Site Specific Proteolysis of
Fusion
Proteins", Chapter 13, in Protein Purification: From Molecular Mechanisms to
Lame Scale Processes, American Chemical Society, Washington, D.C. (1990).
Furthermore, one could use, e.g., a mammalian cell that already comprises in
its genome a nucleic acid molecule encoding a protein that modulates CRH
signalling as described above, but does not express the same or not in an
appropriate
manner due to, e.g., a weak promoter, and introduce into the mammalian cell a
regulatory sequence such as a strong promoter in close proximity to the
endogenous
nucleic acid molecule encoding said purine permease polypeptide so as to
induce
expression of the same.
As such a recombinant host cell containing a polynucleotide encoding a
protein which modulates CRH signalling under the control of a heterologous
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-30-
transcription and/or regulatory sequence or protein, would be another
embodiment of
this invention.
In this context the term "regulatory sequence" denotes a nucleic acid
molecule that can be used to increase the expression of the purine permease
polypeptide, due to its integration into the genome of a cell in close
proximity to the
CRH modulating protein-encoding gene. Such regulatory sequences comprise
promoters, enhancers, inactivated silencer intron sequences, 3'IJTR and/or
5'UTR
coding regions, pxotein and/or RNA stabilizing elements, nucleic acid
molecules
encoding a regulatory protein, e.g., a transcription factor, capable of
inducing or
triggering the expression of the CRH modulating protein-encoding gene or other
gene expression control elements which are known to activate gene expression
and/or increase the amount of the gene product. The introduction of said
regulatory
sequence leads to increase andlor induction of expression of polypeptides,
which
modulate CRH signalling, resulting in the end in an increased amount of said
polypeptides in the cell. Thus, the present invention is aiming at providing
de novo
andlor increased expression of polypeptides that modulate CRH signalling.
Introduction of the construct into the host cell can be effected by calcium
phosphate transfection, DEAE-dextran mediated transfection, cationic lipid
mediated transfection, electroporation, transduction, infection, or other
methods.
Such methods are described in many standard laboratory manuals, such as Davis,
Basic Methods In Molecular Biology (1986). It is specifically contemplated
that
polypeptides, which modulate CRH signalling rnay in fact be expressed by a
host
cell lacking a recombinant vector.
In addition, expression of polynucleotides according to the invention may
also be performed using in vitro produced synthetic mRNA. Synthetic mRNA or
mRNA isolated from cells capable of modulating CRH signaling can be
efficiently
translated in various cell-free systems, including but not limited to wheat
germ
extracts and reticulocyte extracts, as well as efficiently translated in cell
based
systems, including but not limited to microinjection into frog oocytes, with
microinjection into frog oocytes being generally preferred.
Transgefaic non-hu~iah animals
The present invention also relates to a method for the production of a
transgenic non-human animal, preferably transgenic mouse, comprising
introduction
of a polynucleotide or vector of the invention into a germ cell, an embryonic
cell,
stem cell or an egg or a cell derived therefrom. The non-human animal can be
used
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-31-
in accordance with a screening method of the invention described herein and
may be
a non-transgenic healthy animal, or may have a phosphate uptake or
reabsorption
disorder, preferably a disorder caused by at least one mutation in the protein
that
modulates CRH signalling. Such transgenic animals are well suited for, e.g.,
pharmacological studies of drugs in connection with mutant forms of the above
described polypeptides. Production of transgenic embryos and screening of
those can
be performed, e.g., as described by A. L. Joyner Ed., Gene Targeting, A
Practical
Approach (1993), Oxford University Press. The DNA of the embryonal membranes
of embryos can be analyzed using, e.g., Southern blots with an appropriate
probe;
see supra.
Preferably, the transgenic non-human animal of the invention further
comprises at least one inactivated wild type allele of the corresponding
mammalian
CRH modulating protein-encoding gene; see supra. This embodiment allows for
example, the study of the interaction of various mutant forms of polypeptides
according to the invention on the onset of the clinical symtoms of disease
related to
disorders in CRH metabolism. All the applications that have been herein before
discussed with regard to a transgenic animal also apply to animals carrying
two,
three or more transgenes; e.g. encoding neutral endopeptidase (NEP). It might
be
also desirable to inactivate protein that modulates CRH signalling expression
or
function at a certain stage of development and/or life of the transgenic
animal. This
can be achieved by using, for example, tissue specific, developmental and/or
cell
regulated and/or inducible promoters which drive the expression of, e.g., an
antisense or ribozyme directed against the RNA transcript encoding the protein
capable of modulating CRH signalling; see also supra. A suitable inducible
system is
for example tetracycline-regulated gene expression as described, e.g., by
Gossen and
Bujard (Proc. Natl. Acad. Sci. 89 USA (1992), 5547-5551) and Gossen et al.
(Trends
Biotech. 12 (1994), 58-62). Similar, the expression of the mutant protein that
modulates CRH signalling may be controlled by such regulatory elements.
Furthermore, the invention also relates to a transgenic mammalian cell which
contains
(preferably stably integrated into its genome) a nucleic acid molecule
according to
the invention or part thereof, wherein the transcription and/or expression of
the
nucleic acid molecule or part thereof leads to reduction of the synthesis of a
protein that modulates CRH signalling.
In a preferred embodiment, the reduction is achieved by an anti-sense, sense,
ribozyme,
co-suppression and/or dominant mutant effect. "Antisense" and "antisense
nucleotides"
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-32-
means DNA or RNA constructs which block the expression of the naturally
occurring
gene product.
The provision of the polynucleotide according to the invention opens up the
possibility to produce transgenic non-human animals with a reduced level of
the
protein as described above and, thus, with a defect in phosphate metabolism.
Techniques how to achieve this are well known to the person skilled in the
art.
These include, for example, the expression of antisense-RNA, ribozymes, of
molecules which combine antisense and ribozyme functions and/or of molecules
which provide for a co-suppression effect; see also supra. When using the
antisense approach for reduction of the amount of protein that modulates CRH
signallings in cells, the nucleic acid molecule encoding the antisense-RNA is
preferably of homologous origin with respect to the animal species used for
transformation. However, it is also possible to use nucleic acid molecules
which
display a high degree of homology to endogenously occurring nucleic acid
molecules encoding a protein that modulates CRH signalling. In this case the
homology is preferably higher than 80%, particularly higher than 90% and still
more preferably higher than 95%. The reduction of the synthesis of a protein
according to the invention in the transgenic mammalian cells can result in an
alteration in, e.g., adenine reabsorption. In transgenic animals comprising
such
cells this can lead to various physiological, developmental andlor
morphological
changes.
Thus, the present invention also relates to transgenic non-human animals
comprising
the above-described transgenic cells. These may show, for example, a
deficiency in
CRH metabolism compared to wild type animals due to the stable or transient
presence of a foreign DNA resulting in at least one of the following features:
(a) disruption of (an) endogenous genes) encoding a protein capable of
modulating CRH signalling;
(b) expression of at least on antisense RNA and/or ribozyme against a
transcript
comprising a polynucleotide of the invention;
(c) expression of a sense and/or non-translatable mRNA of the polynucleotide
of
the invention;
(d) expression of an antibody of the invention;
(e) incorporation of a functional or non-functional copy of the regulatory
sequence of the invention; or
(f) incorporation of a recombinant DNA molecule or vector of the invention.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-33-
With the polypeptides, their encoding polynucleotides and vectors of the
invention, it
is now possible to study ifz vivo and ifz vitro the efficiency of drugs in
relation to
particular mutations in protein that modulates CRH signallings of a patient
and the
affected phenotype. Furthermore, mutant forms of polypeptides of the invention
can
be used to determine the pharmacological profile of drugs and for the
identification
and preparation of further drugs which may be effective for the treatment of
disorders related to the CRH metabolism, in particular for the amelioration of
CRH
induced stress or depression.
It will thus be appreciated that the present invention also relates to a
method for
preventing, treating or ameliorating a medical condition related to a disorder
of CRH
metabolism including CRH receptor related disorders which comprises
administering to a mammalian subject a therapeutically effective amount of the
polypeptides, the polynucleotides or the vectors encoding a protein capable of
modulating CRH signalling of the present invention.
Diagfzostic Assays
This invention further relates to the use of polynucleotides of the present
invention
as diagnostic reagents. Detection of a mutated form of the gene characterised
by the
polynucleotide of SEQ ZD NO: l, SEQ ID NO 3, SEQ 117 NO.S, SEQ ID N0.7, SEQ
ID NO.9, SEQ ID NO.11, SEQ ID N0.13, SEQ ID N0.15, SEQ ID N0.17, SEQ ID
N0.19, SEQ ID N0.21, SEQ ID N0.23, SEQ ID N0.25, SEQ ID N0.27, SEQ ID
NO. 29, SEQ ID N0.31, SEQ ID N0.33, SEQ ID N0.35, SEQ ID N0.37, SEQ 1D
N0.39, SEQ ID N0.41, SEQ ID N0.43, SEQ D7 N0.45, SEQ ID N0.47 or SEQ ID
N0:49 which is associated with a dysfunction will provide a diagnostic tool
that can
add to, or define, a diagnosis of a disease, or susceptibility to a disease,
which results
from under-expression, over-expression or altered spatial or temporal
expression of
the gene. Individuals carrying mutations in the gene may be detected at the
DNA
level by a variety of techniques.
It will thus be appreciated that this invention provides a method of
diagnosing a pathological condition or a susceptibility to a pathological
condition in
a subject related to a disorder of CRH activity comprising:
(a) determining the presence or absence of a mutation in the polynucleotide
according to the invention; and
(b) diagnosing a pathological condition or susceptibility to a pathological
condition based on the presence or absence of said mutation.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-34-
Nucleic acids for diagnosis may be obtained from a subject's cells, such as
from
blood, urine, saliva, tissue biopsy or autopsy material. The genomic DNA may
be
used directly for detection or may be amplified enzymatically by using PCR or
other
amplification techniques prior to analysis. RNA or cDNA may also be used in
similar fashion. Deletions and insertions can be detected by a change in size
of the
amplified product in comparison to the normal genotype. Point mutations can be
identified by hybridizing amplified DNA to labeled mammalian purine permease
nucleotide sequences. Perfectly matched sequences can be distinguished from
mismatched duplexes by RNase digestion or by differences in melting
temperatures.
DNA sequence differences may also be detected by alterations in
electrophoretic
mobility of DNA fragments in capilary electrophoresis columns or gels, with or
without denaturing agents, or by direct DNA sequencing (e.g., Myers et al.,
Scieizce
(1985)230:1242). Sequence changes at specific locations may also be revealed
by
specific restriction endonucleases, nuclease protection assays, such as RNase
and S 1
protection or a chemical cleavage method (see Cotton et al., Proc Natl Acad
Sci USA
(1985) 85: 4397-4401). In another embodiment, an array of oligonucleotides
probes
comprising a nucleotide sequence encoding a protein capable of modulating CRH
activity or fragments thereof can be constructed to conduct efficient
screening of
e.g., genetic mutations. Array technology methods are well known and have
general
applicability and can be used to address a variety of questions in molecular
genetics
including gene expression, genetic linkage, and genetic variability (see for
example:
M.Chee et al., Science, Vol 274, pp 610-613 (1996)).
The diagnostic assays offer a process for diagnosing or determining a
susceptibility to the diseases through detection of mutations in the CRH
modulating
protein-encoding gene by the methods described. In addition, such diseases may
be
diagnosed by methods comprising determining from a sample derived from a
subject
an abnormally 'decreased or increased level of polypeptide or mRNA, as well as
by
determining from said samples the presence of protein derivatives compared to
the
normal structure. Decreased or increased expression can be measured at the RNA
level using any of the methods well known in the art for the quantitation of
polynucleotides, such as, for example; nucleic acid amplification, for
instance via
PCR, RT-PCR; RNase protection; Northern blotting and other hybridization
methods. Assay techniques that can be used to determine levels of a protein,
such as
a polypeptide of the present invention, in a sample derived from a host are
well-
known to those of skill in the art. Such assay methods include
radioimmunoassays,
competitive-binding assays, Western Blot analysis and ELISA assays. Assay
techniques that can be used to determine the presence of protein derivatives
or
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-35-
variants comprise amongst others mass spectrometry.
Thus in another aspect, the present invention provides a method of
diagnosing a pathological condition or a susceptibility to a pathological
condition in
a subject related to a disorder of prolonged CRH exposure comprising:
(a) determining the presence or amount of expression of the polypeptide or a
derivative thereof according to the invention in a biological sample; and
(b) diagnosing a pathological condition or susceptibility to a pathological
condition based on the presence or amount of expression of the polypeptide or
of a
derivative thereof.
In particular the present invention provides a method of diagnosing a CRH
induced gene expression profile in an individual, said method comprising;
a) obtaining a biological sample of said individual; and
b) determine the amount of at least one protein that modulates corticotropin
releasing hormone (CRH) signaling in said biological sample;
whereby the protein that modulates corticotropin releasing hormone (CRH)
signaling is being selected from the group consisting of SEQ ID N0.2, SEQ ID
4, SEQ ID N0.6, SEQ )D N0:8, SEQ ID N0.10, SEQ )D N0.12, SEQ ID
N0.14, SEQ m N0.16, SEQ m N0.18, SEQ m N0.20, SEQ m N0.22, SEQ
m N0.24, SEQ m N0.26, SEQ m N0.28, SEQ m N0.30, SEQ m N0.32,
SEQ JD N0.34, SEQ ID N0.36, SEQ )D N0.38, SEQ >D N0.40, SEQ >D
N0.42, SEQ m N0.44, SEQ lD N0.46 and SEQ ID N0.48. In an alternative
embodiment the method of diagnosing a CRH induced expression profile is not
limited to at least one protein according to the invention, but requires the
simultaneous assessment of the expression levels of the group of proteins
identified as being involved in CRH signaling, i.e. the proteins having the
amino
acid sequences SEQ ID N0.2, SEQ ID N0.4, SEQ ll~ N0.6, SEQ lD NO.B,
SEQ ID N0.10, SQ >D N0.12, SEQ ID N0.14, SEQ ID N0.16, SEQ )D N0.18,
SEQ )D N0.20, SEQ ID NO.22, SEQ ll~ N0.24, SEQ ID N0.26, SEQ m
N0.28, SEQ >D N0.30, SEQ ID N0.32, SEQ ID N0.34, SEQ ID N0.36, 5EQ
D7 N0.38, SEQ ID N0.40, SEQ ~ N0.42, SEQ m N0.44 and SEQ m N0.48.
Preferably the amount of said proteins is determined either at the protein
level,
preferably using antibodies that bind thereto, or at the gene transcription
level,
preferably using probes that bind to a polynucleotide encoding an amino acid
sequence
selected from the group consisting of SEQ m N0.2, SEQ ID 4, SEQ ID N0.6, SEQ m
N0.8, SEQ ll~ NO.10, SEQ ID N0.12, SEQ ID N0.14, SEQ ID NO.16, SEQ >D
N0.18, SEQ m N0.20, SEQ )~ N0.22, SEQ m N0.24, SEQ ID N0.26, SEQ m
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-36-
N0.28, SEQ ID N0.30, SEQ ID N0.32, SEQ ID N0.34, SEQ ID N0.36, SEQ ID
N0.38, SEQ ID N0.40, SEQ ID N0.42, SEQ ID N0.44, SEQ ID N0.46 and SEQ ll~
N0.48. In the simultaneous assessment of the group of proteins involved in CRH
signaling the level of gene expression is analysed using microarray
technology.
Alternatively the CRH induced gene expression profile is determined by
assessing
the level of gene transcription of a gene comprising a nucleic acid sequence
selected
from the group consisting of SEQ » NO: 1, SEQ ID NO 3, SEQ ID N0.5, SEQ ID
N0.7, SEQ 1D N0.9, SEQ ID NO.11, SEQ ID N0.13, SEQ ID N0.15, SEQ ID
N0.17, SEQ ID NO.19, SEQ ID N0.21, SEQ 1D N0.23, SEQ ID N0.25, SEQ ID
N0.27, SEQ ID NO. 29, SEQ ID N0.31, SEQ ID NO.33, SEQ ID N0.35, SEQ ll~
NO.39, SEQ ID N0.41, SEQ ID N0.43, SEQ ID N0.45, SEQ ID N0.47 0~ SEQ ID
N0:49. Methods to determine the level of gene transcription have been
described
hereinbefore and comprise in a preferred embodiment the use of a probe which
binds, preferably selectively binds to a polynucleotide selected from the
group
consisting of SEQ ID NO: 1, SEQ ID NO 3, SEQ 117 N0.5, SEQ ID N0.7, SEQ ID
N0.9, SEQ m N0.11, SEQ ll~ N0.13, SEQ lD N0.15, SEQ >D NO.17, SEQ ID
N0.19, SEQ m N0.21, SEQ ~ N0.23, SEQ 1D N0.25, SEQ ~ N0.27, SEQ gD
NO. 29, SEQ ID N0.31, SEQ ID N0.33, SEQ ID N0.35, SEQ ID N0.37, SEQ >D
N0.39, SEQ >D N0.41, SEQ ID N0.43, SEQ ID N0.45, SEQ ID N0.47 or SEQ >D
N0:49 or the complement thereof. In another embodiment, an array of
oligonucleotides probes comprising a nucleotide sequence according to the
invention
or fragments thereof can be constructed to conduct efficient screening of the
level of
gene transcription in the sample of an individual.
In a further aspect, the present invention relates to a diagnostic lit which
comprises:
(a) a polynucleotide of the present invention, preferably the nucleotide
sequence of
SEQ II7 NO: 45, SEQ ID NO: 47, SEQ ID NO: 49 or a fragment thereof;
(b) a nucleotide sequence complementary to that of (a);
(c) a polypeptide of the present invention, preferably the polypeptide of SEQ
ID
N0:46, SEQ ID NO 48 or a fragment thereof; or
(d) an antibody to a polypeptide of the present invention, preferably to the
polypeptide of SEQ ID N0:46 or SEQ ID NO 48 and optionally suitable means for
detection.
It will be appreciated that in any such kit, (a), (b), (c) or (d) may comprise
a
substantial component. Such a kit will be of use in diagnosing a disease or
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-37-
suspectability to a disease, particularly CRH metabolism related disorders
such as
CRH induced stress or depression.
The nucleotide sequences of the present invention are also valuable for
chromosome localisation. These sequences are specifically targeted to, and can
hybridize with, a particular location on an individual human chromosome. The
mapping of relevant sequences to chromosomes according to the present
invention is
an important first step in correlating those sequences with gene-associated
disease.
Once a sequence has been mapped to a precise chromosomal location, the
physical
position of the sequence on the chromosome can be correlated with genetic map
data. Such data are found in, for example, V. McKusick, Mendelian Inheritance
in
Man (available on-line through Johns Hopkins University Welch Medical
Library).
The relationship between genes and diseases that have been mapped to the same
chromosomal region are then identified through linkage analysis (coinheritance
of
physically adjacent genes). The gene of the present invention maps to human
chromosome 15.
The differences in the cDNA or genomic sequence between affected and
. unaffected individuals can also be determined. If a mutation is observed in
some or
all of the affected individuals but not in any normal individuals, then the
mutation is
likely to be the causative agent of the disease.
The nucleotide sequences of the present invention are also valuable for tissue
localisation. Such techniques allow the determination of expression patterns
of the
polypeptides according to the invention in tissues by detection of the mRNAs
that
encode them. These techniques include iu si.tu hybridziation techniques and
nucleotide amplification techniques, for example PCR. Such techniques are well
known in the art. Results from these studies provide an indication of the
normal
functions of the polypeptides in the organism.
The polypeptides of the invention or their fragments or analogs thereof, or
cells expressing them, can also be used as immunogens to produce antibodies
immunospecific for polypeptides of the present invention. The term
"immunospecific" means that the antibodies have substantially greater affinity
for
the polypeptides of the invention than their affinity for other related
polypeptides in
the prior art.
Thus in another embodiment, this invention provides a monospecific antibody
imrnunologically reactive with a mammalian purine permease. In a preferred
embodiment said antibody is immunologically reactive with a polypeptide having
an
amino acid sequence selected from a group consisting of: (SEQ m NO 46); (SEQ
lD
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-38-
N0:48); and functional analogs thereof or said antibody blocks activity of a
protein
that modulates CRH signalling.
Antibodies generated against polypeptides of the present invention may be
obtained by administering the polypeptides or epitope-bearing fragments,
analogs or
cells expressing these to an animal, preferably a non-human animal, using
routine
protocols. For preparation of monoclonal antibodies, any technique which
provides
antibodies produced by continuous cell line cultures can be used. Examples
include
the hybridoma technique (Kohler, G. and Milstein, C., Nature (1975)256:495-
497),
the trioma technique, the human B-cell hybridoma technique (Kozbor et al.,
lyramunology Today (1983)4:72) and the EBV-hybridoma technique (Cole et al.,
MONOCLONAL ANTIBODIES AND CANCER THERAPY, pp.77-96, Alan R.
Liss, Inc., 1985).
Techniques for the production of single chain antibodies, such as those
described in U.S. Patent No.4,946,778, can also be adapted to produce single
chain
antibodies to polypeptides of this invention. Also, transgenic mice, or other
organisms, including other mammals, xnay be used to express humanized
antibodies.
The above-described antibodies may be employed to isolate or to identify
clones expressing the polypeptide or to purify the polypeptides by affinity
chromatography.
Antibodies against polypeptides of the present invention may also be employed
to treat the CRH metabolism related disorders.
In a further aspect, the present invention relates to genetically engineered
soluble fusion proteins comprising a polypeptide of the present invention, or
a
fragment thereof, and various portions of the constant regions of heavy or
light
chains of immunoglobulins of various subclasses (IgG, IgM, IgD, IgE).
Preferred as
an immunoglobulin is the constant part of the heavy chain of human IgG,
particularly IgG I, where fusion takes place at the hinge region. In a
particular
embodiment, the Fc part can be removed simply by incorporation of a cleavage
sequence which can be cleaved with for instance blood clotting factor Xa.
Furthermore, this invention relates to processes for the preparation of these
fusion
proteins by genetic engineering, and to the use thereof for drug screening,
diagnosis
and therapy. A further aspect of the invention also relates to polynucleotides
encoding such fusion proteins. Examples of fusion protein technology can be
found
in International Patent Application Nos. W094i29458 and W094122914.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-39-
T72.erapeutic Utility
A further aspect of the invention relates to an immunological/vaccine
formulation (composition) which, when introduced into a mammalian host,
induces
an immunological response in that mammal to a polypeptide of the present
invention
wherein the composition comprises a polypeptide or polynucleotide of the
present
invention. The vaccine formulation may further comprise a suitable corner.
Since a
polypeptide may be broken down in the stomach, it is preferably administered
parenterally (for instance, subcutaneous, intramuscular, intravenous, or
intradermal
injection). Formulations suitable for parenteral administration include
aqueous and
non-aqueous sterile injection solutions which may contain anti-oxidants,
buffers,
bacteriostats and solutes which render the formulation isotonic with the blood
of the
recipient; and aqueous and non-aqueous sterile suspensions which may include
suspending agents or thickening agents. The formulations may be presented in
unit-
dose or mufti-dose containers, for example, sealed ampoules and vials and may
be
stored in a freeze-dried condition requiring only the addition of the sterile
liquid
carrier immediately prior to use. The vaccine formulation may also include
adjuvant
systems for enhancing the immunogenicity of the formulation, such as oil-in
water
systems and other systems known in the art. The dosage will depend on the
specific
activity of the vaccine and can be readily determined by routine
experimentation.
In still another approach, expression of the gene encoding proteins which
modulate CRH signalling can be inhibited using expression blocking techniques.
Known such techniques involve the use of antisense sequences, either
internally
generated or externally administered (see, for example, O'Connor, J.Neurochem
(1991) 56:560 ;Oligodeoxynucleotides as Antisense Inhibitors of Gene
Expression,
CRC Press, Baca Raton, FL (1988)). Alternatively, oligonucleatides which form
triple helices ("triplexes") with the gene can be supplied (see, for example,
Lee et al.,
Nucleic Acids Res (1979)6:3073; Cooney et al.., Science (1988)241:456; Dervan
et
al., Science (1991)251:1360). These oligomers can be administered per se or
the
relevant oligomers can be expressed isZ vivo. Synthetic antisense or triplex
oligonucleotides may comprise modified bases or modified bacltbones. Examples
of
the latter include methylphosphonate, phosphorothioate or peptide nucleic acid
backbones. Such backbones are incorporated in the antisense or triplex
oligonucleotide in order to provide protection from degradation by nucleases
and are
well lcnown in the art. Antisense and triplex molecules synthesised with these
and/or
other modified backbones also form part of the present invention.
In another process fox inhibiting expression of a target gene in a cell, RNA
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-40-
with partial or fully double-stranded character is introduced into the cell or
into the
extracellular environment. Inhibition is specific in that a nucleotide
sequence from a
portion of the target gene is chosen to produce inhibitory RNA. The RNA may
comprise one or more strands of polymerized ribonucleotide; it may include
modifications to either the phosphate-sugar backbone or the nucleoside. The
double-
stranded structure may be formed by a single self-complementary RNA strand or
two
complementary strands. Inhibition is sequence-specific in that the nucleotide
sequences corresponding to the duplex region of the RNA are targeted for
genetic
inhibition. RNA containing a nucleotide sequence identical to a portion of the
target
sequence is preferred. Examples of RNA inhibition technology can be found in
International Patent Application WO 99/32619.
In addition, expression of the proteins which modulate CRH signalling may be
prevented by using ribozymes specific to the mRNA sequence encoding said
protein.
Ribozymes are catalytically active RNAs that can be natural or synthetic (see
for
example Usman, N, et al., Curr. Opin. Struct. Biol (1996)6(4), 527-33.)
Synthetic
ribozymes can be designed to specifically cleave the aforementiond mRNAs at
selected positions thereby preventing translation of said mRNAs into
functional
polypeptide. Ribozymes may be synthesised with . a natural ribose phosphate
backbone and natural bases, as normally found in RNA molecules. Alternatively
the
ribozymes may be synthesised with non-natural backbones to provide protection
from ribonuclease degradation, for example, 2'-O-methyl RNA, and may contain
modified bases.
For treating abnormal conditions related to an under-expression of proteins
which modulate CRH signalling, several approaches are also available. One
approach comprises administering to a subject a therapeutically effective
amount of
a compound which activates a polypeptide of the present invention, i.e., an
agonist as
described above, in combination with a pharmaceutically acceptable earner, to
thereby alleviate the abnormal condition. Alternatively, gene therapy may be
employed to effect the endogenous production of mammalian purine permease by
the relevant cells in the subject. For example, a polynucleotide of the
invention may
be engineered for expression in a replication-defective retroviral vector, as
discussed
above. The retroviral expression construct may then be isolated and introduced
into a
paclcaging cell transduced with a retroviral plasmid vector containing RNA
encoding
a polypeptide of the present invention such that the pacleaging cell now
produces
infectious viral particles containing the gene of interest. These producer
cells may be
administered to a subject for engineering cells in vivo and expression of the
polypeptide in vivo. For an overview of gene therapy, see Chapter 20, Gefae
Therapy
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-41-
and otlaer Molecular Geyzetic-based Tl2erapeutic Approaches, (and references
cited
therein) in Human Molecular Genetics, T Strachan and A P Read, BIOS Scientific
Publishers Ltd (1996). Another approach is to administer a therapeutic amount
of a
polypeptide of the present invention in combination with a suitable
pharmaceutical
can-ier.
In a further aspect, the present invention provides for pharmaceutical
compositions comprising a therapeutically effective amount of a polypeptide,
such as
the soluble form of a polypeptide of the present invention, agonist/antagonist
peptide
or small molecule compound, in combination with a pharmaceutically acceptable
carrier or excipient. Such carriers include, but are not limited to, saline,
buffered
saline, dextrose, water, glycerol, ethanol, and combinations thereof. The
invention
further relates to pharmaceutical packs and kits comprising one or more
containers
filled with one or more of the ingredients of the aforementioned compositions
of the
invention. Polypeptides and other compounds of the present invention may be
employed alone or in conjunction with other compounds, such as therapeutic
compounds.
The composition will be adapted to the route of administration, for instance
by
a systemic or an oral route. Preferred forms of ~ systemic administration
include
injection, typically by intravenous injection. Other injection routes, such as
subcutaneous, intramuscular, or intraperitoneal, can be used. Alternative
means for
systemic administration include transmucosal and transdermal administration
using
penetrants such as bile salts or fusidic acids or other detergents. In
addition, if a
polypeptide or other compounds of the present invention can be formulated in
an
enteric or an encapsulated formulation, oral administration may also be
possible.
Administration of these compounds may also be topical and/or localized, in the
form
of patches, salves, pastes, gels, and the like.
The dosage range required depends on the choice of peptide or other
compounds of the present invention, the route of administration, the nature of
the
formulation, the nature of the subject's condition, and the judgment of the
attending
practitioner. Suitable dosages, however, are in the range of 0.1-100p,glkg of
subject.
Wide variations in the needed dosage, however, are to be expected in view of
the
variety of compounds available and the differing efficiencies of various
routes of
administration. For example, oral administration would be expected to require
higher
dosages than administration by intravenous injection. Variations in these
dosage
levels can be adjusted using standard empirical routines for optimization, as
is well
understood in the art.
Polypeptides used in treatment can also be generated endogenously in the
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-42-
subject, in treatment modalities often referred to as "gene therapy" as
described
above. Thus, for example, cells from a subject may be engineered with a
polynucleotide, such as a DNA or RNA, to encode a polypeptide ex vivo, and for
example, by the use of a retroviral plasmid vector. The cells are then
introduced into
the subject.
This invention will be better understood by reference to the Experimental
Details
that follow, but those skilled in the art will readily appreciate that these
are only
illustrative of the invention as described more fully in the claims that
follow thereafter.
Additionally, throughout this application, various publications are cited. The
disclosure of
these publications is hereby incorporated by reference into this application
to describe
more fully the state of the art to which this invention pertains.
EXPERIMENTAL PROCEDURES
Cell culture and sample prepar°atioyz - AtT-20 cells were purchased
from ATCC and
were maintained at 37°C in 5% COZ in humidified air in Dulbecco's
modified Eagle's
medium (Invitrogen Life Technologies) containing 10 % fetal bovine serum, 5%
horse
serum and 4.5 g/L D-glucose. For experiments cells were seeded in 25 cm2
flasks. The
medium was replaced 48 h later and cells were treated with either 0.1% DMSO,
1~,M
CRH (Sigma) in DMSO, 1~,M R121919 in DMSO or 1~,M CRH +1~,M R121919 in
DMSO for 0, 0.5, 1, 2, 4, 8 and 24 h in fresh medium. The incubation was
stopped by
aspirating the incubation medium and adding 3 ml Trizol (Invitrogen Life
Technologies) for lysis of the cells. Total RNA was extracted using Trizol
according to
the instructions of the manufacturer. 100~,g total RNA was further purificied
using
Rneasy lit (Qiagen) with DNAseI treatment on column.
Microar°ray Izybridi.zatiofz - cRNA was prepared as follows. Reverse
transcription was
performed on 10~g of total RNA for lh at 42°C using a T7-oligo(dT)24-
primer and
SuperscriptIl RT (Invitrogen Life Technologies). Second strand cDNA synthesis
was
done for 2 h at 16°C using Escherichia coli DNA Polymerase I, DNA
ligase and
RNAseH (Invitrogen Life Technologies). After phenol-chloroform extraction
using
phase-lock gel (Eppendorf~ in vitro transcription was performed for 6 h at
37°C using
the Bioarray high-yield RNA transcript labeling kit with Biotin labeled
ribonucleotides
(Enzo Diagnostics). cRNA samples were purified on Qiagen Rneasy columns
followed
by fragmentation for 35 min at 95°C. cRNA yields were between 50 and
100~,g.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-43-
Samples were processed on GeneChips (Affymetrix, Santa Clara, CA). In order to
check the quality of each sample, 5~,g of labeled cRNA was run on Test2-
arrays. Actual
experiments were performed on Murine Genome U74Av2 arrays, containing probe
sets
interrogating approximately 12,000 full-length mouse genes and EST clusters
from the
UniGene database (Build 74). Hybridization was performed using l5,ug cRNA for
16 h
at 45°C under continuous rotation. Arrays were stained in Affymetrix
Fluidics stations
using StreptavidinlPhycoerythrin (SAPE) followed by staining with anti-
streptavidin
antibody and a second SAPE staining. Subsequently arrays were scanned with a
HP-
Laserscanner and data were analyzed with the Microarray Suite Software
(Affymetrix).
No scaling or normalization was performed at this stage. Quality of the
experiment was
assessed based on the percentages present calls across all samples which was
on
average 47.06~2.45%. The cytoplasmic l3-actin and GAPDH 5'/3' ratios were
1.10~0.08 and 0.93~0.05 respectively.
Data afaalysis and selection of genes
Raw intensities on each array were aligned using the following global mean
algorithm:
IrnW ~ Irnw
log lava"~~ = log Ir~,v + log nv snmples _ lOg sample
# gefZes ~e# samples # geyaes
Basically this alignment sets the average intensity of one array to the
average measured
across all arrays, compensating for array to array variations in
hybridization, washing
and staining, ultimately allowing a reasonable comparison between arrays.
After
alignment polished data were analyzed using weighted spectral mapping.
Weighted
spectral mapping is an unsupervised multivariate analysis method which
includes
double-centering of the data combined with a specialized visualization
representing the
two highest principle components. Even though double-centering removes the
"size"
component of the array data, this information is reintroduced in the
visualization via the
area of the symbols representing the size of the respective samples and genes.
This
method allows the reduction of a large microarray dataset and provides means
to
visually inspect and thereby identify clusters of genes and/or subjects in the
data (7).
A more detailed analysis was carried out using the OmniViz program. All
records
which were absent in all experiments were removed and all signals less than 20
were
set to 20. Gene expression fold differences for CRH, 8121919 and CRH+8121919
treatments were calculated at each timepoint. Fox those calculations signals
at
corresponding timepoints in DMSO treated samples were used to calculate
ratios.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-44-
Quantitative RT PCR - Microarray data were confirmed using real time PCR
analysis.
First strand cDNA synthesis was performed on 0.5,ug total RNA using random
hexamer
primers and SuperscriptII RT (Invitrogen Life Technologies). Quantitative PCR
was
performed on a ABIPrism 7700 cycler (Applied Biosystems) using a Taqman PCR
kit.
Serial dilutions of cDNA were used to generate standard curves of threshold
cycles
versus the logarithms of concentration for l3-actin, c fos, Crh-Rl, Crh-R2,
Rg,r2 and the
genes of interest (see table 2 for sequences). A linear regression line
calculated from the
standard curves allowed the determination of transcript levels in RNA samples
from the
different time points.
RESULTS
Transcriptional response to CRH was studied in the CRH-R1-expressing murine
AtT-
pituitary corticotroph-derived adenoma cell line. Whereas CRH-R1 was readily
15 detectable both by real time quantitative RT-PCR (RTq) and western blot,
CRH-R2
expression could not be discerned in AtT-20 cells. In order to identify CRH-R1
specific
responses cells were exposed to lp,M CRH, lp,M CRH in the presence of l,uM of
a
CRH-R1 specific antagonist 8121919 and to 8121919 alone. Transcriptional
responses
were followed over time until 24 h after the first administration. In order to
assess
20 treatment efficacy, c fos mRNA levels were determined by RTq on RNA from
the
different treatments and time points before array experiments were carried
out. In
agreement with previous reports, exposure to CRH elicited a transient surge in
c-fos
transcription, with levels already going down after 0.5 to 1 h (see figure
1)(8;9). This
response was almost completely suppressed in the presence of 8121919.
Interestingly,
0.1°7o DMSO induced c-fos expression, however levels were between 5 to
10 times
lower compared to CRH induced expression.
All time points were analyzed on microarrays containing approximately 12000
murine
genes and ESTs. Overall analysis of the expression profiles using spectral
mapping (a
so called unsupervised method) indicated progressing time to account for most
of the
observed changes in gene expression (see figure 2). Synchronization of cell
cycle in
these cultures induced by addition of new medium and serum could possibly
account
for this phenomenon although accumulation of metabolites and progressing cell
culture
are additional contributing factors. The spectral map analysis also showed
that CRH
treated samples differed from other samples mainly in the early time points
(0.5h until
2h), with overall differences in expression becoming very small after 8h.
Because of
this obvious influence of time, expression measurements were analyzed relative
to
those observed in the corresponding time point in DMSO treated control
samples.
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-45-
Regulated genes were defined as those showing a greater than 2-fold change in
transcript levels at any one time point. Using the OmniViz Treescape view, 111
genes
that met this criterion, showing a difference in expression after treatment
with CRH
compared to treatment with the antagonist, were selected. 26 out of the 111
genes were
"early responders", showing a 2-fold change already after 30 minutes treatment
with
CRH. 32 genes were "intermediate responders", responding after 1 to 2 hours of
treatment and 53 genes were "late responders", showing a response after 2
hours or
more after treatment (see figure 3). These responses were suppressed by the
CRH-R1
antagonist 8121919. Among the early responders were known players in the
pathways
downstream of the CRH-R1 such as the transcription factors Nurrl, Nurr77, Jun-
B,
validating the assay.
Interesting novel players identified include transcription factors (e.g.
hairylenhancer-of-
split related 1 (Heyl ), nuclear factor regulated by interleukin 3 (NFIL3),
cAMP
responsive element modulator (CRElVl) and prostate specific ets transcription
factor
(Pse)), receptor and channel regulators (e.g. Ras-related GTP-binding protein
(GElI~
and receptor (calcitonin) activity modifying protein 3 (RAMP3)), secreted
peptides (e.g.
adrenomedullin, calcitonin, cholecystokinin) and proteins involved in
intracellular
signaling (e.g. regulator of G-protein signaling 2 (Rgs2), CAMP specific
phosphodiesterase 4B (Pde4b), inositol 1,4,5-triphosphate receptor 1 (IP3R1)
and the
regulatory subunit phosphatidylinositol 3-kinase, p85). Other interesting
regulated
genes comprise Period homolog Perl , fibroblast growth factor receptor 2
(Fgfr2),
serum/glucocorticoid regulated kinase and serum-inducible kinase (figure 4).
Interestingly, all responders identified according to above mentioned criteria
were up
regulated after exposure to CRH. This induction was transient and nearly all
of the
induced genes return to baseline after 4 to 8 hr. Many of the induced
transcripts encode
proteins that would exert a negative feedback on the CRH-R1 signaling (e.g.
Pde4,
Rgs2, CREM, etc...), possibly contributing to the transient nature of the
induction. In
additiob to this negative feedback, other mechanisms such as desentisation of
CRFI
through phosphorylation and internalization contribute to the transient nature
of the
transcriptional induction. In this respect it is of interest to note that
challenge with CRF
quicl~ly downregulates CRFI mRNA in rat pituitary cells. We could however not
detect
any alterations in CRFI mRNA in pituitary derived AtT-20 cells exposed to 1
~.M CRF.
Confirmation of microarray data was carried out using quantitative real time
PCR
analysis on the same samples used for hybridization experiments and on a
repeated
experiment. Levels of regulation and the time course identified by microarray
corresponded to those observed by quantitative PCR as shown for Rgs2 in figure
5. For
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-46-
those genes that have been tested, levels iof induction compared to the
untreated
samples are indicated in figure 4.
DISCUSSION
We have identified transcriptional pathways downstream of the CRH-Rl. in AtT-
20
cells using a CRH-Rl specific antagonist. Our findings are in agreement with
activation
of several second messenger such as cAMP and Ca2+ upon stimulation with CRH.
Some of the transcriptional responses can be explained by the phosphorylation
of
CREB and the subsequent transcription of genes downstream of cAMP responsive
elements. These elements have been found for example in the promoters of Perl,
Nurrl, CREM-ICER, c-Fos. Furthermore the kinetic profile of the induction of
these
genes correspond with the observed maximal transcription rate by CREB after
0.5 hr of
cAMP formation. The induction of CREM-ICER constitutes a negative feedback
mechanism in attenuating transcriptional response to cAMP. Of interest is the
reported
induction of CREM-ICER in response to acute stress in the intermediate lobe of
the
pituitary gland. Mice deficient fox CREM-ICER show a chronic increase of beta-
endorphin levels suggesting that CREM-ICER induction may be involved in the
modulation of gene expression in response to stress (10). Our results suggest
that
CREM-ICER is directly involved in the modulation of CRH signaling and as a
result,
ablation of CREM-ICER could lead to an altered response to stress signals.
Another
novel putative negative feedback regulator of CRH signaling is Rgs2. We
identified
two single CRE motifs in the promoter of the human RGS2 gene, providing a
possible
explanation for the early response behavior of this gene upon stimulation with
CRH. In
support of our findings is a recent report showing that both phosphoinositide
signaling
and cAMP induce a rapid and transient increase in Rgs2 mRNA in human
astrocytoma
and neuroblastoma cells. The Rgs2 protein is a selective inhibitor of G9a
function.
Recently it has been shown that Rgs2 reduces odorant-elicited cAMP production,
not
by acting on Ga but by directly inhibiting the activity of adenylyl cyclase
type 11I.
Although Rgs2 was originally identified as an immediate early response gene in
activated T lymphocytes, studies in Rgs2 deficient mice indicate that it also
plays a role
in the modulation of stress related behavior as these mice show increased
anxiety and
aggression (11). Also the induction of cAMP specific phosphodiesterase 4B
(Pde4b)
can be categorized under negative feedback, directly attenuating the cAMP
signal.
Another important second messenger generated upon stimulation with CRH is
Ca2+. It
has been shown that CRH triggers a steady-state depolarization stimulated
extracellular
Ca2+ entry via voltage-gated Ca2~'' channels and raises intracellular Ca2+
concentration
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-47-
through release from inositol 1,4,5-triphosphate (InsP3) sensitive Ca2+
pools(12). Both
the TnsP3 receptor and the p85 regulatory subunit of phosphatidylinositol 3-
kinase are
upregulated, possibly accounting for a compensating mechanism for prolonged
Ca2~"
signaling. Also the upregulation of the small G-protein kir/Gem points towards
an
attenuation of prolonged Ca2+ signaling. Recent studies have shown that Gem
regulates
Ca2+ channel expression at the cell membrane through the (3 auxiliary
subunits.
Increased levels of Gem have been shown to inhibit Ca2+-triggered exocytosis
and it has
been proposed that Gem could have a protective effect against Ca2+ overload.
In
addition to its role as second messenger, intracellular Ca2+ has also been
shown to play
a critical role in regulating gene expression. Of interest is the regulation
of
NFIL3/E4BP4 by calcineurin/NFAT and CaM kinase signaling, accounting for an
increase in NFIL3 mRNa levels upon CRH treatment. In B lymphocytes expression
of
NFIL3 is induced by interleukin 3 through both the Raf-mitogen-activated
protein
kinase and phosphatidylinositol 3-kinase pathways. In this cell type NFIL3
inhibits
apoptosis in synergy with Bcl-xL dependent pathways. Our data suggest a role
for
NFIL3 in prevention of apoptosis in AtT-20 cells.
CRH is the most efficacious ACTH secretagogue. Unfortunately the microarrays
that
were used did not interrogate for POMC levels. However several other
prepropeptides
mRNAs were found upregulated after CRH administration such as cholecystokinin
(CCK) and two calcitonin peptide family members, adrenomedullin (ADM) and
calcitonin (CT). Also of interest in this respect is the upregulation of
RAMP3. RAMPs
control the transport and glycosylation of the calcitonin receptor-like
receptor (CRLR).
In the case of RAMP3, it has been shown that together with CRLR it generates
an
ADM receptor. Upregulation of this gene might play a role in regulating the
responsiveness of AtT-20 cells to ADM after CRH exposure or to other
extracellular
stimuli as it is not known whether RAMP3 might regulate other G-coupled
receptors.
Although CCK is secreted by AtT-20 cells, induction of its expression by CRH
has not
been previously reported (13). Interaction between CCK and CRH has however
been
intensively studied and demonstrated in panic attacks, depression, anxiety and
gastric
emptying (14-19). Most of these experiments point towards a role for CRH in
mediating the central effects of CCK. Our data indicate that CRH in addition
might
function as a CCK secretagogue. A very similar situation to that of CCK seems
to be
case for adrenomedullin as well. It has been demonstrated that ADM is
expressed in
pituitary gland and affects basal and CRH-stimulated ACTH release in animals,
thus
suggesting its potential role in regulating the hypothalamus-pituitary~adrenal
axis(20-
23). Current expression data show that CRH induces ADM and CT. In addition
recent
findings indicate that circulating adrenomedullin is increased in Cushing's
disease, and
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-48-
the pituitary gland may represent the site of the elevated production of ADM
(24),
suggesting CRH might be inducing ADM.
In conclusion, we have unraveled part of the corticotropin-releasing hormone
receptor-1
activated gene network and have identified several novel targets of this
signaling
cascade. Our findings trigger the need for further experiments to elucidate
the function
of these transcriptional responses to CRH stimulation both on a cellular and
whole
organism level.
REFERENCES
1. De Souza EB 1995 Corticotropin-releasing factor receptors: physiology,
pharmacology,
biochemistry and role in central nervous system and immune disorders.
Psychoneuroendocrinology
20:789-819
2. Holsboer F 2001 Sh~ess, hypercortisolism and corticosteroid receptors in
depression: implications
for therapy. J Affect Disord 62:77-91
3. Holsboer F, Gerken A, Stalla GIs, Muller OA 1987 Blunted aldosterone and
ACTH release after
human CRH administration in depressed patients. Am J Psychiatry 144:229-231
4. Holsboer F, Gerken A, von Bardeleben U, Grimm W, Beyer H, Muller OA, Stalla
GIs 1986
Human corticotropin-releasing hormone in depression--correlation with
thyrotropin secretion
following thyrotropin-releasing hormone. Biol Psychiatry 21:601-611
5. Nemeroff CB, Owens MJ, Bissette G, Andorn AC, Stanley M 1988 Reduced
corticotropin
releasing factor binding sites in the frontal cortex of suicide victims. Arch
Gen Psychiatry 45:577-
579
6. Raadsheer FC, Hoogendijk WJ, Stam FC, Tilders FJ, Swaab DF 1994 Increased
numbers of
corticotropin-releasing hormone expressing neurons in the hypothalamic
paraventricular nucleus of
depressed patients. Neuroendocrinology 60:436-444
7. Wouters L, Gohlmann HW, Bijnens L, Lass SU, Molenberghs G, Lewi PJ 2002
Graphical
exploration of gene expression data:a comparative study of three multivariate
methods. Biometrics
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-49-
8. Boutillier AL, Monnier D, Lorang D, Lundblad JR, Roberts JL, Loeffler JP
1995 Corticotropin-
releasing hormone stimulates proopiomelanocortin transcription by cFos-
dependent and -
independent pathways: characterization of an AP 1 site in exon 1. Mol
Endocrinol 9:745-755
9. Boutillier AL, Sassone-Corsi P, Loeffler JP 1991 The protooncogene c-fos is
induced by
S corticotropin-releasing factor and stimulates proopiomelanocortin gene
transcription in pituitary
cells. Mol Endocrinol 5:1301-1310
10. Mazzucchelli'C, Sassone-Corsi P 1999 The inducible cyclic adenosine
monophosphate early
repressor (ICER) in the pituitary intermediate lobe: role in the stress
response. Mol Cell
Endocrinol 155:101-113
11. Oliveira-Dos-Santos AJ, Matsumoto G, Snow BE, Bai D, Houston FP, Whishaw
IQ, Mariathasan
S, Sasaki T, Wakeham A, Ohashi PS, Roder JC, Barnes CA, Siderovski DP,
Penninger JM 2000
Regulation of T cell activation, anxiety, and male aggression by RGS2. Proc
Natl Acad Sci U S A
97:12272-12277
12.. Tse A, Lee AK 2000 Voltage-gated Ca2+ channels and intracellular Ca2+
release regulate
exocytosis in identified rat corticotrophs. J Physiol 528 Pt 1:79-90.:79-90
13. Beinfeld MC 1992 CCK mRNA expression, pro-CCK processing, and regulated
secretion of
immunoreactive CCK peptides by rat insulinoma (RIN 5F) and mouse pituitary
tumor (AtT-20)
cells in culture. Neuropeptides 22:213-217
14. Coskun T, Bozkurt A, Alican I, Ozkutlu U, Kurtel H, Yegen BC 1997 Pathways
mediating CRF-
induced inhibition of gastric emptying in rats. Regul Pept 69:113-120
15. Kellner M, Wiedemann K, Yassouridis A, Levengood R, Guo LS, Holsboer F,
Yehuda R 2000
Behavioral and endocrine response to cholecystokinin tetrapeptide in patients
with posttraumatic
stress disorder. Biol Psychiatry 47:107-111
16. Geracioti TD, Jr., Ekhator NN, Nicholson WE, Arndt S, Loosen PT, Orth DN
1999 Intra- and
inter-individual correlations between cholecystokinin and corticotropin-
releasing hormone
concentrations in human cerebrospinal fluid. Depress Anxiety 10:77-80
17. Calogero AE, Nicolosi AM, Moncada ML, Coniglione F, Vicari E, Polosa P,
D'Agata R 1993
Effects of cholecystokinin octapeptide on the hypothalamic-pituitary-adrenal
axis function and on
vasopressin, prolactin and growth hormone release in humans.
Neuroendocrinology 58:71-76
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
-50-
18. Biro E, Sarnyai Z, Penke B, Szabo G, Telegdy G 1993 Role of endogenous
corticotropin-releasing
factor in mediation of neuroendocrine and behavioral responses to
cholecystokinin octapeptide
sulfate ester in rats. Neuroendocrinology 57:340-345
19. Kamilaris TC, Johnson EO, Calogero AE, Kalogeras KT, Bernardini R,
Chrousos GP, Gold PW
1992 Cholecystokinin-octapeptide stimulates hypothalamic-pituitary-adrenal
function in rats: role
of corticotropin-releasing hormone. Endocrinology 130:1764-1774
20. Shan J, Krukoff TL 2001 Intracerebroventricular adrenomedullin stimulates
the hypothalamic-
pituitary-adrenal axis, the sympathetic nervous system and production of
hypothalamic nitric
oxide. J Neuroendocrinol 13:975-984
21. Martinez V, Cuttitta F, Tache Y 1997 Central action of adrenomedullin to
inhibit gastric emptying
in rats. Endocrinology 138:3749-3755
22. Parkes DG, May CN 1995 ACTH-suppressive and vasodilator actions of
adrenomedullin in
conscious sheep. J Neuroendocrinol 7:923-929
23. Samson WK, Murphy T, Schell DA 1995 A novel vasoactive peptide,
adrenomedullin, inhibits
pituitary adrenocorticoiropin release. Endocrinology 136:2349-2352
24. Letizia C, Di Iorio R, De Toma G, Marinoni E, Cerci S, Celi M, Subioli S,
D'Erasmo E 2000
Circulating adrenomedullin is increased in patients with corticotropin-
dependent Cushing's
syndrome due to pituitary adenoma. Metabolism 49:760-763
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
1/17
SEQUENCE LISTING
SEQ ID NO.l
atgcagctgagaaaaatgcagaccatcaaaaaggagcccgcacccctagatcctaccagcagctcagacaagatgctgc
tgctgaactctgccttagct
gaggtggccgaggacctagcctcaggtgaagatttgctcctgaacgaagggagcatggggaaaaacaaatcctcggcgt
gtcggagaaaacgggaat
tcattccggacgagaagaaagacgccatgtattgggagaaacggcggaaaaacaacgaagctgccaaaagatctcggga
gaagcgccgcctcaatg
acctggttttggagaacaagctgattgccctgggagaagaaaatgccactttaaaagctgagctgctctccctgaaatt
aaagtttggtttaattagctccac
ggcgtatgcccaagaaatccagaaactcagtaattccacagctgtctactttcaggactaccagacatccaaggctgcc
gtgagctcttttgtggacgagc
1 0
atgagcctgcgatggtagccggaagttgcatctcagtcatcaagcactctccccagagctcgctctccgatgtgtcaga
ggtgtcctcggtggagcacac
tcaggaaagccccgcacagggaggctgccggagccctgagaacaagttccctgtgatcaagcaggagcccgtggagttg
gagagctttgccaggga
ggccagggaggagcggggcacgtattccacctccatctaccagagctacatgggaagctctttctccacttactcccac
tccccacccctcttgcaggtc
catgggtccactagcaactccccaagaacctcagaggccgatgagggtgtagtgggcaagtcttctgatggggaagacg
aacaacaggtccctaagg
gccccatccattctccagtggagctgcaacgggttcacgccacggtggtgaaggttccggaagtgaacccttctgcctt
accgcacaagcttcggattaa
agccaaggccatgcaggtcaaagtggaggctttggacagcgagtttgaaggcatgcagaaactctcttcacccgccgat
gcgatcgccaaaagacattt
tgacctggagaaacatggaacctcgggtatggcccattcctccctccctcctttctcagtgcaggtgacgaacattcaa
gattggtccctcaaatcggaac
actggcatcacaaagaactgagcagcaaaactcagagtagcttcaaaacaggtgtggtggaagtcaaagacggtggcta
taaggtttccgaagctgag
aatttgtatttgaagcagggaatagcaaacttatctgcagaggtggtctcgctcaagagattcatagccacacaaccga
tctcggcttcggactccaggtaa
SEQ ID No.2
MQLRKMQTIKKEPAPLDPTSSSDKMLLLNSALAEVAEDLASGEDLLLNEGSMGKNKSSACRRK
REFIPDEKKDAMYWEKRRKNNEAAKRSREKRRLNDLVLENKLIALGEENATLKAELLSLKLKFG
LISSTAYAQEIQKLSNSTAVYFQDYQTSKAAVSSFVDEHEPAMVAGSCISVIKHSPQSSLSDVSEV
SSVEHTQESPAQGGCRSPENKFPVIKQEPVELESFAREAREERGTYSTSIYQSYMGSSFSTYSHSPP
LLQVHGSTSNSPRTSEADEGV VGKSSDGEDEQQVPKGPIHSPVELQRVHAT V VKVPEVNPSALP
HKLRIKAKAMQVKVEALDSEFEGMQKLSSPADAIAKRPIFDLEKHGTSGMAHSSLPPFSVQVTNI
QDWSLKSEHWHHKELSSKTQSSFKTGVVEVKDGGYKVSEAENLYLKQGIANLSAEVVSLKRFI
ATQPISASDSR
SEQ ID No. 3
tgtccgctctgcctcccacacctagcaccccagcccgctgctgccccggtgagaacccccagcttgggccttgtcatgg
tgccagcaggtggccctgag
cttctgacaggggcctgcctatagacctgcaggcctgaggcctcagactcacactcaaggggcaagaggccctggtggc
ccacctaagagccacctct
3 5
gtccccagccctgctgccccactgatgtctgactgagacccagcagtgaccctgagctgcctgcccactgcctcctcct
ggtccctgaggttggctctgc
cgaggacggacgactcttctgaagcaggcggctaacggaagcagccccaagcctccaccgcagcatgggcagtgccagc
ccaggcctgagcaacg
tgtcccccggttgcctgctactgttcccagatgtggcaccacgaacagggacggagaaggcagcatcaggagcaatggg
ccctgagaagcaggaatg
gagtcctagtccacccgccacccctgagcagggcctgtctgctttctacctctcttactttaacatgtatcccgacgat
agcagctgggtcgccaaagtccc
cgaggcccgtgccggggaggaccacccggaggagcccgagcagtgtcccgtcattgacagccaggcctctgggagcacg
ttggatgagcactcgct
agagcaggtgcaatcgatggttgtgggcgaggtcctgaaagatattgagacggcctgcaagcttctgaacatcacagca
gaccctggggactggagcc
ctggtaacgtgcagaagtggcttttatggacagaacaccagtaccggctgcctccagcaggcaaggccttccaggagct
gggcggtaaggagctgtgc
gccatgtccgaggaacagttccgtcagcgtgcacccttgggtggggatgtactgcatgcccacctggacatctggaagt
cagcggcctggatgaagga
gaggacctcgcctgggacccttcactactgcgcctccaccagcgaggagggctggacggatggtgaggtggactcgtcg
tgctccgggcagcccatt
cacctgtggcagttcctgaaagaactgctgctcaagccccacagctatggccgcttcatccgctggctcaacaaggaga
aaggcatcttcaaaattgagg
actcagcacaggtggcccgactgtggggtgtgcgcaagaaccggccagccatgaactatgataaactaagccgctccat
ccgccagtattacaagaag
ggcatcattcgtaaacccgacatctctcagcgccttgtctaccaatttgtgcatccagtctgagagccacagagaccag
aggcctacaacctgccccagg
cagccactctctggttggcctggtcctctctgctcactctgaattcaggggctgctggtatcccagaacccaaggtccc
agatagacagccactgatctag
ggatacacatgagctctctgggtcatacacaggccccaggaagatcgagggagctagttcagcacacagggactggacc
aagtcagctcaccggaca
gtgatgtcactggtctctgctcctgccacaatcctgtaccatatctggcatggtgctaagagatgtctgtaccctgcgt
tgggaagccaggggtgccctggg
gatggataataaagacgtaagataactg
SEQ ID No.4
MGSASPGLSNVSPGCLLLFPDVAPRTGTEKAASGAMGPEKQEWSPSPPATPEQGLSAFYLSYFN
MYPDDSSWVAKVPEARAGEDHPEEPEQCPVIDSQASGSTLDEHSLEQVQSMVVGEVLKDIETAC
KLLNITADPGDWSPGNVQKWLLWTEHQYRLPPAGKAFQELGGKELCAMSEEQFRQRAPLGGD
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
2/17
VLHAHLDIWKSAAWMKERTSPGTLHYCASTSEEGWTDGEVDSSCSGQPIHLWQFLKELLLKPH
SYGRFIRWLNKEKGIFKIEDSAQVARLWGVRKNRPAMNYDKLSRSIRQYYKKGIIRKPDISQRLV
YQFVHPV
SEQ ID N0.5
cgggtcgacccacgcgtccgcccacgcgtccggcggagcttctgggttgcgggccgaaacggcaagcggatggagggcg
ctcgaacggccaggtg
tcgtgattaaattagtcagccctcagagacaggcgtcctacctcctttatccagacctcaaaagccccgttgtgcaccc
gtggtggcttcttcaccttccctg
tttcgtcctccactgtatggcccagacatgagtggtcccctagaaggggccgatgggggaggagaccccaggcccggag
aacctttttgtcctggagga
gtcccatcccctggggccccgcagcaccggccttgtccaggccccagcctggctgatgacactgatgcaaacagcaatg
gctcaagtggcaatgagtc
caacggacccgagtccaggggcgcatctcagcggagttctcatagttcctcttctggcaatggcaaggactcagctctg
ctggagaccactgagagcag
caagagtacaaactcacagagcccatccccacccagcagctccattgcctacagcctcctgagtgcgagctcagagcag
gacaacccatctaccagtg
gctgcagcagtgaacagtcagctcgagccaggacccagaaagaactcatgactgcacttcgggagctcaaacttcgact
gccaccagagcgtcgggg
caagggccgctctgggaccttggccacactgcagtacgctctggcctgtgtcaagcaggttcaggctaaccaggaatat
taccagcagtggagtctgga
ggagggtgagccttgtgccatggacatgtctacttacaccctggaggaattggagcatatcacatccgaatacacactt
cgaaaccaggacaccttctctg
tggctgtgtccttcctgacaggccggattgtctatatttcggagcaggcaggtgtcctgctgcgttgcaaacgggatgt
gtttcggggtgcccgcttctcag
agctcctggctccccaggatgtgggtgtcttctatggctctactacaccatctcgactgcccacctggggcactggcac
ctctgcaggttcaggtctcaag
gacttcacccaggaaaagtctgtcttctgccgaatcagaggaggtcctgaccgggatccagggcctcggtaccagccat
tccgcctaaccccatatgtga
ccaagattcgggtctcagatggagcccctgcacagccgtgctgcctactcattgccgagcgcatccactctggttatga
agctccccggatccctcctga
caagaggatcttcaccacccgacacacaccaagctgcctcttccaggatgtagatgaaagggctgccccactgctgggt
taccttccccaggatctcctg
ggggctccagtacttctctttctacatcctgaggaccgacccctcatgctggccattcataagaagatactgcagctgg
caggccagccctttgaccattcc
cctattcgcttctgtgctcggaacggggaatatgtcaccatggacaccagctgggccggttttgtgcacccctggagcc
gcaaggtggctttcgtgttggg
tcgccataaagtgcgcacggcacccctgaatgaggacgtcttcactcccccagcccccagcccagctccgtccctggac
tctgatatccaggagctctc
agagcagatccatcgattgctgctgcagcctgtgcacagctccagccccacggggctctgtggagttggccctctgatg
tcccctggtcctctacacagc
cctggctcctccagtgatagcaatgggggggacgctgaggggcctgggcctcctgctccagtgactttccageagatct
gtaaggatgtgcatctggtaa
agcaccagggacaacagctcttcattgaatctcgggccaagcccccaccccggccccgcctccttgctacaggtacatt
caaagccaaagtccttccct
gccagtccccaaaccccgaactggaggtggccccagttcctgaccaagcctcgttagccttggcccctgaggagccaga
gaggaaagaaacctctgg
ctgttcctaccagcagatcaactgcctggacagcatcctcaggtatttggagagctgcaacattcccagtacaaccaag
cgtaaatgtgcctcctcctcctc ~-
ctacactgcctcttcagcctctgatgatgacaagcagagggcaggtccagttcctgtgggggccaagaaagatccgtcg
tcagcaatgctgtctgggga
3 0
gggggcaactcctcggaaggagccagtggtgggaggcaccctgagcccgctcgccctggccaataaggcagagagcgtg
gtgtccgtcaccagtca
gtgtagcttcagctccaccatcgtccatgtgggagacaagaagcccccggagtcggacatcatcatgatggaagacctg
cctggcctggcccctggcc
cagcccccagtccggcccccagccccacagtagcccctgacccaaccccagatgcttatcgcccagtgggtctgaccaa
ggccgtgctgtccctgcac
acacagaaggaagagcaagccttcctcaaccgcttcagagatcttggcaggcttcgtggacttgacacctcttctgtgg
ccccctcagcccctggctgcc
accatggccccattccccctggtcgccgacaccactgccgatctaaagcaaagcgttcccgccaccaccaccaccagac
cccccggcccgaaactcc
3 5
ctgctatgtctcccatccttcacctgtgccctcttctggaccctggccacccccaccagccacgacccccttcccagca
atggtccagccctacccactcc
cagtattctcccctcgaggaggaccccagccccttccccctgcccctacatctgtgtcccctgctaccttcccttctcc
cttagtgaccccaatggtggcctt
ggtgctccctaactatctattccctaccccacctagttatccatatggggtgtcccaggcccctgttgaggggccaccc
acgcctgcttcccactcgccctc
tccatccctgcccccaccacctctcagccccccccaccgcccagactccccactgttcaactcgagatgcagctcccca
ctccagctcaatctgctgcag
cttgaggagtccccccgcacggaggggggcgctgctgcaggaggcccaggaagcagtgctgggcccctgcctcccagtg
aggagactgctgagcc
40
agaggccagattggtggaggttactgagtcgtccaatcaggatgcactttcaggctccagcgacctgctggagctactg
ctccaagaagactctcgctcg
ggcacaggctccgcagcctcaggctccctgggctctggcctgggctctgggtctggttcaggatcccacgaagggggaa
gcacctcagccagcatca
cccgcagcagtcagagcagccatacaagcaagtactttggcagcatcgactcttccgaggctgaagctggggctgctcg
ggccaggactgagcctgg
ggaccaggtcattaagtgtgtgctccaggaccccatctggctgctcatggccaatgccgaccagcgtgtcatgatgaca
taccaggtgccgtccaggga
tgcagcctctgtgctgaagcaagaccgggagaggctccgggccatgcagaaacagcagccacggttctcagaggaccag
aggcgggaactgggtg
45
ctgtgcactcctgggtccggaagggccagctgcctcgggcccttgatgtgatggcgtgtgtggactgtggcagcagcgt
tcaagatcctggccactctga
tgacccgctcttctcagaactggatggattggggctggagcccatggaagagggtggaggcgagggtggtgggtgtggt
gttggcggtggtgggggtg
atggtggtgaggaggcccagacccaaattggggctaagggttcaagctctcaggactctgccatggaggaagaagagca
aggtgggggctcatccag
eccagctttacctgcagaagaaaacagcaccagctagatccattttggggccgcttacagcagtctaatgagaggcttc
ctttcgaccatgttggggttctt
ataactcaagatacagctggaccaaccaataggaaactgccccagcttctcccaacatagggggctggacccccattac
cagcccaggcacaggagct
50
gcctctagcttcttagcagagtggaagttctcagccccatttggaggattgtccacgcccgtcccactgaggagacggg
cgggtcttcggttaaggttgct
gacaagctgctgaagtggtctgtccaaatcccagctgagcctgagtcccagtcgcagggttggggctgcacttatttat
ttgggagagacagctcactctc
ccacctcaccccaagatgggaggaggggaacctgggatctgtgtaggatccaggtccgtgaacccctagctgctccagg
gtgggggaggttggtgga
ccatggagtccctggtgctgcccctcaggtgggacccaggtgttctcagctctaccctctaccaatgacatttgtgttt
ttgatattgtgtctgttatttttttttta
atacaaaatgacaaaatgaaaaaccaaaaa
SEQ ID No.6
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
3/17
MSGPLEGADGGGDPRPGEPFCPGGVPSPGAPQHRPCPGPSLADDTDANSNGSSGNESNGPESRG
ASQRSSHSSSSGNGKDSALLETTESSKSTNSQSPSPPSSSIAYSLLSASSEQDNPSTSGCSSEQSARA
RTQKELMTALRELKLRLPPERRGKGRSGTLATLQYALACVKQVQANQEYYQQWSLEEGEPCA
MDMSTYTLEELEHITSEYTLRNQDTFSVAVSFLTGRIVYISEQAGVLLRCKRDVFRGARFSELLAP
QDVGVFYGSTTPSRLPTWGTGTSAGSGLKDFTQEKSVFCRIRGGPDRDPGPRYQPFRLTPYVTKI
RVSDGAPAQPCCLLIAERIHSGYEAPRIPPDKRIFTTRHTPSCLFQDVDERAAPLLGYLPQDLLGA
PVLLFLHPEDRPLMLAIHKKILQLAGQPFDHSPIRFCARNGEYVTMDTSWAGFVHPWSRKVAFV
LGRHKVRTAPLNEDVFTPPAPSPAPSLDSDIQELSEQIHIZLLLQPVHSSSPTGLCGVGPLMSPGPL
HSPGSSSDSNGGDAEGPGPPAPVTFQQICKDVHLVKHQGQQLFIESRAKPPPRPRLLATGTFKAK
VLPCQSPNPELEVAPVPDQASLALAPEEPERKETSGCSYQQINCLDSILRYLESCNIPSTTKRKCAS
SSSYTASSASDDDKQRAGPVPVGAKKDPSSAMLSGEGATPRKEPVVGGTLSPLALANKAESVVS
VTSQCSFSSTIVHVGDKKPPESDIIMMEDLPGLAPGPAPSPAPSPTVAPDPTPDAYRPVGLTKAVL
SLHTQKEEQAFLNRFRDLGRLRGLDTSSVAPSAPGCHHGPIPPGRRHHCRSKAKRSRHHHHQTP
RPETPCYVSHPSPVPSSGPWPPPPATTPFPAMVQPYPLPVFSPRGGPQPLPPAPTSVSPATFPSPLV
TPMVALVLPNYLFPTPPSYPYGVSQAPVEGPPTPASHSPSPSLPPPPLSPPHRPDSPLFNSRCSSPLQ
LNLLQLEESPRTEGGAAAGGPGSSAGPLPPSEETAEPEARLVEVTESSNQDALSGSSDLLELLLQE
DSRSGTGSAASGSLGSGLGSGSGSGSHEGGSTSASITRSSQSSHTSKYFGSIDSSEAEAGAARART
EPGDQVIKCVLQDPIWLLMANADQRVMMTYQVPSRDAASVLKQDRERLRAMQKQQPRFSEDQ
RRELGAVHSWVRKGQLPRALDVMACVDCGSSVQDPGHSDDPLFSELDGLGLEPMEEGGGEGG
GCGVGGGGGDGGEEAQTQIGAKGSSSQDSAMEEEEQGGGSSSPALPAEENSTS
SEQ ID No.7
gaattcggcacgagcagcgagacgccgcgcacggtgcttccccagtggagccaatcggctaacccgcgctccggcagag
tccttggcgctcgcccg
~5
ccggcgggacagaccacccgcctctggccgctctctggaccctggccgccccgagcgaagactggagcaaaatgatgct
tcaacatccaggccaggt
ctctgcctcagaagtcagtgcgaccgccattgtcccctgcctctcacctcctgggtcactggtatttgaggattttgct
aacctgacaccctttgtcaaggaa .
gagctgagattcgccatccagaataaacacctctgccatcggatgtcctctgcgctggagtcagttaccgtcaacaaca
gacccctggagatgtcagtca
ccaagtctgaggcggcccctgaagaagatgagaggaaaaggaggcggcgagaaagaaataaaattgctgctgccaagtg
tcgaaacaagaaaaag
gagaagacagagtgcctgcagaaagagtcagagaaactggagagtgtgaatgctgagctgaaggcccagattgaggagc
tgaagaatgagaaacag
catttgatatacatgctcaacctgcaccggcccacctgtatcgtccgggctcagaatggacggacaccggaagacgaga
ggaacctctttatccaacag
ataaaagaaggaacattgcagagctaagcagaggtggcacggaggcaattggggagttcttactgaatcctccttttcc
accccacaccctgaagccatt
ggaaaactggcttcctgtgcacttctagaatcccagcagccaagagccgttggggcaggagggcctgtggtgacctact
gcattgacccactctgcccc
cgagtgaaccgtggagcaggcaggagcatcctttgtctcaccaattccaggatttaggccttatcatcccggccagtct
cagatgacctagctggcccca
ggctggggtcctatgcaaagcaggatcccactaatgggattcaggcagaagtgtctaccttgataggtggggtgggacc
acatcctccactgtggctgac
aacgcccttccaagggaatatggaatgagaacattcattattgaggttgtccaatggccagggtatgctttctagaaaa
tatgctgttctgtcccagaatgac
tgtgcatagggtatccgtttcagagcctggtgttgtgctatttagatgtttgtcttgcacaacattggcatgatttttc
cgggagtttcatcagatctgatttctgag
agtctggggatctgccatggtggaaagtgcccctcaaaagcatttgtgtggccacatgaactggctggcaccaggggag
tgaaactggctgatgaccag
ctgagccactttgtgccaacagaggatggacgacacctttccctgtacccactgcagaggaagaaccctgggcacagca
gctttgtccttggctacaaac
tgttacaacgtcacacaatgaaggcacaaagtccaactttcaaagggtgtaggactccatactcagtgacagggcagga
agagccaaagataaccaca
gceacagcctgtggagaccagggttggaagccaggtgcagggccaggcatctgcattgtgggatgttaatggcactttt
gtcttgtagctattttgagatgt
ggtccagagcatttcagctgggagatctccctctggccaccaggactctggctactgttaaaatcctgatgtttctgtg
gaatcctcagtgtttaatcccactc
aatagtatcattacagttttctgtaagagaaaatattacttatttatcccagtattcctagcctgtcaacataataaat
atcggaacaaaacctggta
SEQ ID No.8
MMLQHPGQVSASEVSATAIVPCLSPPGSLVFEDFANLTPFVKEELRFAIQNKHLCHRMSSALESV
TVNNRPLEMSVTKSEAAPEEDERKRRlE2RERNKIAAAKCRNKKKEKTECLQKESEKLESVNAELK
AQIEELKNEKQHLIYMLNLHRPTCIVRAQNGRTPEDERNLFIQQIKEGTLQS
SEQ ID No.9
cccagagataagctgacgctgggcaaacccctgggggaaggttgcttcgggcaagtagtcatggctgaagcagtgggaa
tcgataaagacaaaccca
aggaggcggtcaccgtggcagtgaagatgttgaaagatgatgccacagagaaggacctgtctgatctggtatcagagat
ggagatgatgaagatgattg
ggaaacataagaacattatcaacctcctgggggcctgcacgcaggatggacctctctacgtcatagttgaatatgcatc
gaaaggcaacctccgggaata
5 5
cctccgagcccggaggccacctggcatggagtactcctatgacattaaccgtgtccccgaggagcagatgaccttcaag
gacttggtgtcctgcaccta
ccagctggctagaggcatggagtacttggcttcccaaaaatgtatccatcgagatttggctgccagaaacgtgttggta
acagaaaacaatgtgatgaag
atagcagactttggcctggccagggatatcaacaacatagactactataaaaagaccacaaatgggcgacttccagtca
agtggatggctcctgaagcc
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
4/17
ctttttgatagagtttacactcatcagagcgatgtctggtccttcggggtgttaatgtgggagatctttactttagggg
gctcaccctacccagggattcccgt
ggaggaactttttaagctgctcaaagagggacacaggatggacaagcccaccaactgcaccaatgaactgtacatgatg
atgagggattgctggcatgc
tgtaccctcacagagacccacattcaagcagttggtcgaagacttggatcgaattctgactctcacaaccaatgaggaa
tacttggatctcacccagcctct
cgaacagtattctcctagttaccccgacacaagtagctcttgttcttcaggggacgattctgtgttttctccagacccc
atgccttatgaaccctgtctgcctca
gtatccacacataaacggcagtgttaaaacatga
SEQ ID No.lO
PRDKLTLGKPLGEGCFGQV VMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEM
MKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFK
DLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGR
LPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPTN
CTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLTQPLEQYSPSYPDTSSSCS
SGDDSVFSPDPMPYEPCLPQYPHINGSVKT
SEQ ID No.l1
acccacgcgtccggccggtttcactgctcccctcagtctcttttgggctctttccgggcatcgggacgatgaccgtcaa
agccgaggctgctcgaagcac
ccttacctactccagaatgaggggaatggtagcgattctcatcgcttttatgaaacagagaaggatgggcctgaacgat
tttattcagaagattgccagcaa
cacctatgcatgcaaacacgctgaagttcagtccattttgaaaatgtcccatcctcaggagccggagcttatgaacgct
aacccctctcctccgccaagtc
cctctcaacaaatcaacctgggtccgtcctccaaccctcacgccaaaccctccgactttcacttcttgaaagtgatcgg
aaagggcagttttggaaaggttc
ttctggctaggcacaaggcagaagaagtattctatgcagtcaaagttttacagaagaaagccatcctgaagaagaaaga
ggagaagcatattatgtcaga
gcggaatgttctgttgaagaatgtgaagcaccctttcctggtgggccttcacttctcattccagaccgctgacaaactc
tactttgtcctggactacattaatg
gtggagagctgttctaccatctccagagggagcgctgcttcctggaaccacgggctcgattctacgcagctgaaatagc
cagtgccttgggctatctgca
ctccctaaacatcgtttatagagacttaaaacctgagaatattctcctagactcccaggggcacatcgtcctcactgac
tttgggctctgcaaagagaatatt
gagcataacgggacaacatctaccttctgtggcacgcctgagtatctggctcctgaggtcctccataagcagccgtatg
accggacggtggactggtggt'"
gtcttggggctgtcctgtatgagatgctctacggcctgcccccgttttatagccggaacacggctgagatgtacgacaa
tattctgaacaagcctctccagt ''
tgaaaccaaatattacaaactcggcaaggcacctcctggaaggcctcctgcagaaggaccggaccaagaggctgggtgc
caaggatgactttatggag
attaagagtcatattttcttctctttaattaactgggatgatctcatcaataagaagattacacccccatttaacccaa
atgtgagtgggcccagtgaccttcgg
3 0
cactttgatcccgagtttaccgaggagccggtccccagctccatcggcaggtcccctgacagcatccttgtcacggcca
gtgtgaaggaagcagcagaa
gccttcctcggcttctcctatgcacctcctgtggattccttcctctgagtgctcccgggatggttctgaaggacttcct
cagcgtttcctaaagtgttttccttac .
cctttggtggaggttgccagctgacagaacattttaaaagaatttgcacacctggaagcttggcagtctcgcctgcccg
gcgtggcgcgacgcagcgcg
cgctgcttgatgggagctttccgaagagcacaccctcctctcaatgagcttgtgaggtcttcttttcttctcttccttc
caacgtggtgctagctccaggcgag
cgagcgtgagagtgccgcctgagacagacaccttggtctcagttagaaggaagatgcaggtctaagaggaatccccgca
gtctgtctgagctgtgatca
agaatattctgcaatgtgccttttctgagatcgtgttagctccaaagctttttcctatcgcagagtgttcagtttgtgt
ttgtttgtttttgttttgttttgtttttcccttg
gcggatttcccgtgtgtgcagtggcgtgagtgtgctatgcctgatcacagacggttttgttgtgagcatcaatgtgaca
cttgcaggacactacaatgtggg
acattgtttgtttcttccacatttggaagataaatttatgtgtagactgttttgtaagatatagttaataactaaaacc
tattgaaacggtcttgcaatgacgagcat
tcagatgcttaaggaaagcattgctgctacaaatatttctatttttagaaagggtttttatggaccaatgccccagttg
tcagtcaaagccgttggtgttttcattg
tttaaaatgtcacctataaaacgggcattatttatgttttttttccctttgttcatattcttttgcattcctgattatt
gtatgtatcgtgtaaaggaagtctgtacattgg
gttataacactagatatttaaacttacaggcttatttgtaaaccatcattttaatgtactgtaattaacatgggttata
atatgtacaattcctcctccttaccacaca
actttttttgtgtgcgataaaccaattttggtttgcaataaaatcttgaaacct
SEQ ID No.l2
MTVKAEAARSTLTYSRMRGMVAILIAFMKQRRMGLNDFIQKIASNTYACKHAEVQSIL,KMSHP
QEPELMNANPSPPPSPSQQINLGPSSNPHAKPSDFHFLKVIGKGSFGKVLLARHKAEEVFYAVKV
LQKKAILKKKEEKHIMSERNVLLKNVKHPFLV GLHFSFQTADKLYFVLDYINGGELFYHLQRER
CFLEPRARFYAAEIASALGYLHSLNIVYRDLKPENILLDSQGHIVLTDFGLCKENIEHNGTTSTFCG
TPEYLAPEVLHKQPYDRTVDWWCLGAVLYEMLYGLPPFYSRNTAEMYDNILNKPLQLKPNITN
SARHLLEGLLQKDRTKRLGAKDDFMEIKSHIFFSLINWDDLINKKITPPFNPNVSGPSDLRHFDPE
FTEEPVPSSIGRSPDSILVTASVKEAAEAFLGFSYAPPVDSFL
SEQ ID No.l3
gcagggcgggtgagagcgccgtgaaagccgcggaacgccgtgcacctccgcgactctactacggcaagctagtccggac
gggtcgtcgtccccgc
gcgccaccagcccttggtgaaacgacagggagcgtccggcttccccagcaccgccctgcgagactcaaaacagccacac
cgcaaagcgagcctcg
ggcggaaggaggcggagcttcaggcggccccgcctccgcggaaggatacacatctccgtggtccaaaaccccggggcga
ggcggccggggcgtg
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
5/17
tgagctgctcggccagctgccgtctacgcgctttcgcgcggccaccgggcaactgcgccgcgcggctgccccgctgagc
gctcggcctcggggccgt
gggatccgccgcgctgtctgcggtcaggaagaccgccctcccgcgtccttgccggacgggtcagaggcggcaccgcacg
cgaggccacccgcgat
gctgctgtccaagttcggctccctggcgcacctctgcgggcctggcggcgtggaccacctcccagtgaagatcctacag
ccagccaaggctgacaag
gagagcttcgagaaggtgtaccaggtgggcgccgtgctgggcagcggcggcttcggcacggtctacgcgggcagccgca
tcgccgacggactccc
ggtggctgtgaagcacgtggtgaaggagcgggtgaccgagtggggcagtctcggcggagtggccgtgcccctggaggtg
gtgctgctgcgcaaggt
gggcgcggcgggcggcgcgcgcggcgtcatccgcttgctggactggttcgagcggcccgacggcttcttgttggtgctg
gagcgacccgagccggc
acaggacctcttcgacttcatcactgaacgaggcgccctggacgagccgctggcgcgtcgcttcttcgcgcaggtgctt
gccgctgtgcggcactgcca
caattgtggggtcgtgcaccgcgacatcaaggacgagaacctgctggtggacctgcgctcgggagagctgaagctcatc
gacttcggctcgggcgcg
gtgctcaaggacacggtctacactgactttgatggcacccgtgtgtacagccccccagagtggatccgatatcaccgat
atcacgggcggtctgccactg
tgtggtctctgggtgtactgctctacgacatggtgtgtggggacattccctttgagcaggatgaggagatcttgcgcgg
caggctctttttccggaggaggg
tctccccagagtgccagcagcttattgagtggtgtctctccctgaggccctcagagaggccctccctggaccaaattgc
tgcccacccctggatgctggg
gacagaggggagcgttccagagaactgtgaccttcggctttgtgccctggatactgacgacggagccagtaccacttcc
agcagtgagagcttgtgagg
aggagaaggggcctgggctcggcctagccagcgctctcccagaattgaacactttctgcctgggatgtctgctgcaaaa
gcagtgacctctgacccctg
gtgacctttgctctcggcaccgggcctgtttcctttgctttgagtgcctttttgaacgctgctccacagggcctgggtt
ttcttgagctcttctgtccaaagatgg
ctgagggctaagcaaggtcctgccctgggtggatacttgaaccagagatcccgaccctgctgctccatctcaggaggca
gccttcctgaccaagtgtgtt
tgacatggagcgccctgtggtgcccacctccaaccctccagtctcctggtgttcatctgggcatgtctgcacaagcaat
gcaacgctgggccactgctgc
ccgtctgcctccccggcacggcacggctccgcacgcaacctaagcgtgccaccacggtctcttatttatggtgtgatca
ccctggagggcgcccccgcc
ctgctggggctatttattgtttaatttatttgctgaggttcctccaagcaaccaccttctccaggcccctggggtgttg
aaagtcaaatgtggctgttgagtcca
cagacccccatcctaattcctgcacctggaggagttccccaacccccgtgtttgcgggaggaagcatttgtacagtggc
taatttaaggggagtgggaga
ccctgtcaccctgagcactctgcgctggggaggggtttaaattattgaccttgtacagtctgcttgctggctctgaaag
ctggggttgggggacagagtctc
aagcccttaatttattttagcagctgtgtttctgtgaccctggtgtgactaagcatcaggggtggggttgtataagttc
aaaagtgtgaaatgtctgaagatcat
attttttatacaggtatttcaattaaatgttttggtatataatggaaaaaaaaaaaaaaaaaaaaaaaaaaaaa
SEQ ID No.l4
MLLSKFGSLAHLCGPGGVDHLPVKILQPAKADKESFEKVYQVGAVLGSGGFGTVYAGSRIADGL
PVAVKFIVVKERVTEWGSLGGVAVPLEVVLLRKVGAAGGARGVIRLLDWFERPDGFLLVI ERPE ',
PAQDLFDFITERGALDEPLARRFFAQVLAAVRHCHNCGVVHRDIKDENLLVDLRSGELKLIDFGS
GAVLKDTVYTDFDGTRVYSPPEWIRYHRYHGRSATVWSLGVLLYDMVCGDIPFEQDEEIL,RGR
LFFRRRVSPECQQLIEWCLSLRPSERPSLDQIAAHPWMLGTEGSVPENCDLRLCALDTDDGASTT
SSSESL
SEQ ID No.lS
3 5
cctgggccccgccgcggacgcgcggagccgcctgggccgcgccggaggagggcggggagaggaccatgtgaatgtgctc
cggagctgagcgcc
aagccaagcagtgtttgaaaggaacaggatgctgatctaatcgtggcaaaaagtcagtccgaccgctggtttcgaagac
atgtggtgtatataaagtttgt
gatagttggtggaaatttgggagcttggataatgggctgtgtgcaatgtaaggataaagaagcagcgaaactgacagag
gagagggacggcagcctga
accagagctctgggtaccgctatggcacagaccccacccctcagcactaccccagcttcggcgtgacctccatcccgaa
ctacaacaacttccacgca
gctgggggccagggactcaccgtctttgggggtgtgaactcctcctctcacactgggaccctacgcacgagaggaggga
caggagtgacactgtttgt
ggcgctttatgactatgaagcacggacggaagatgacctgagttttcacaaaggagaaaaatttcaaatattgaacagc
tcggaaggagattggtgggaa
gcccgctccttgacaaccggggaaactggttacattcccagcaattacgtggctccagttgactccatccaggcagaag
agtggtactttggaaaacttgg
ccgcaaagatgctgagagacagctcctgtcctttggaaacccaagaggtacctttcttatccgcgagagccaaaccacc
aaaggtgcctactcactttcc
atccgtgattgggatgatatgaaaggggaccacgtcaaacattataaaatccgcaagcttgacaatggtggatactata
tcacaacgcgggcccagtttga
aacacttcagcaactggtacagcattactcagagaaagctgatggtttgtgttttaacttaactgtggtttcatcaagt
tgtaccccacaaacttctggattggc
taaagatgcttgggaagttgcacgtgactcgttgtttctggagaagaagctggggcaggggtgtttcgctgaagtgtgg
cttggtacctggaatggaaata
caaaagtagccataaagacccttaagccaggcaccatgtctccggagtccttcctggaggaggcgcagatcatgaagaa
gctgaagcatgacaagctg
gtgcagctctacgcggtcgtgtctgaggagcccatttacatcgtcacggagtacatgagcaaaggaagtttgcttgact
tcttaaaagatggtgaaggaag
agctctgaagttgccaaaccttgtggacatggcggcacaggttgctgcaggaatggcttacatcgagcgcatgaattat
atccacagagatctgcgatca
gcaaacattctagtggggaatggactaatttgcaagattgctgactttggattggctcggttgattgaagacaatgaat
acacagcaagacaaggtgcgaa
gtttcccattaagtggacagcccccgaagcggccctgtatggaaggttcacaatcaagtctgacgtatggtcttttgga
atcttactcacagagctggtcac
caaaggaagagtgccatacccaggcatgaacaaccgggaggtgctggagcaggtggagagaggctataggatgccctgc
ccacaggactgcccgat
ctccctgcacgagctcatgatccactgctggaaaaaggatccggaagagcgcccgaccttcgagtacttgcagggcttc
ctggaggactactttacggc
cacagagccccagtatcagcccggtgaaaacctgtgagagcctgcgcttcagacgcctcttcccgaggcctccctaccc
ctccccattagcttccaattct
gtagccagctgccccagagcaggagaaccgtccaggatcagattgcatgtgactcttgaagctgaacttccacggccct
cattaatgacacttgtccccc
agtccgaacctcctctgtgaaccatctgagacagaagcgtgttatttctcagacttggaaatgcattgtatcgatgtta
tgtcaaaggccaaacctctgttcag
tgtaaatagctgctcctgtgccaacaatcccagtgctttccttttttaaaaaagaaaaagcaaatcctatgtgatttta
actctgatttcacctgattcaactaaa
aaaaaaaaagtattattttccaaaagtggcctctttgtctaaaacaataaaattttttttcatgttttaacaaaaaaaa
aaaaaaaaaaaaaaaa
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
6/17
SEQ ID No.l6
MGCVQCKDKEAAKLTEERDGSLNQSSGYRYGTDPTPQHYPSFGVTSIPNYNNFHAAGGQGLTV
FGGVNSSSHTGTLRTRGGTGVTLFVALYDYEARTEDDLSFHKGEKFQILNSSEGDWWEARSLTT
GETGYIPSNYVAPVDSIQAEEWYFGKLGRKDAERQLLSFGNPRGTFLIRESQTTKGAYSLSIRDW
DDMKGDHVKHYKIRKLDNGGYYITTRAQFETLQQLVQHYSEKADGLCFNLTVVSSSCTPQTSG
LAKDAWEVARDSLFLEKKLGQGCFAEVWLGTWNGNTKVAIKTLKI'GTMSPESFLEEAQIMKKL
KHDKLVQLYAV VSEEPIYIVTEYMSKGSLLDFLKDGEGRALKLPNLVDMAAQVAAGMAYIERM
NYIHRDLRSANILVGNGLICKIADFGLARL1EDNEYTARQGAKFPIKWTAPEAALYGRFTIKSDV
WSFGIL.LTELVTKGRVPYPGMNNREVLEQVERGYRMPCPQDCPISLHELMIHCWKKDPEERPTF
EYLQGFLEDYFTATEPQYQPGENL
SEQ ID No.l7
ggacgtcagactagagagtagggagagagactggtgctcgagggacagggctagcccggacgcgtgtccgcgcctcgga
ggtggcaagtaggcag
tgtcgggtggcgaggcaacgatggagctcctgcggactatcacctaccagccggccgccggcaccaagatgtgcgagca
ggctctgggcaaagcttg
cggcggggactcaaagaagaagcgaccacagcagccttctgaagatgggcagccccaagcccaggtgaccccggcggcc
ccgcaccaccatcac
caccattcccactcgggacccgagatctcgcggattatagtcgaccccacgacggggaagcgctactgccggggcaaag
tgctgggcaagggtggat
ttgcaaagtgttacgaaatgacagatctgacaaacaacaaagtctacgctgcaaaaattattcctcacagcagagtagc
taaacctcatcagagggaaaa
gatcgacaaagaaatcgagcttcacagactactgcaccataagcatgtcgtgcagttttaccactactttgaagacaaa
gaaaacatttacattctcttggaa.
tactgcagtagaaggtccatggctcacatcttgaaagcaagaaaggtgttgacagagccagaagtccgatactacctca
ggcagattgtgtcaggactca
agtatcttcacgaacaagaaatcttgcacagggatctcaagctagggaactttattattaatgaagccatggagctgaa
ggtgggagactttggtttggcag
ccagactggaaccactggaacacagaaggagaacaatatgtggaaccccaaattatctctcccccgaagtcctcaacaa
acaaggacacggctgtgaa
tcagacatctgggccttaggctgtgtaatgtatacgatgctgctaggaagacctccattcgaaaccacaaatctgaaag
aaacgtacaggtgcataaggg
aagcaaggtataccatgccgtcctcattgctggcccctgctaagcacttgatagctagcatgctgtccaaaaacccaga
ggaccgccccagtttggatga
catcattcggcatgacttcttcctgcagggtttcactccggacagactctcttccagctgttgccacacagttccagat
ttccacttgtcaagcccagccaag
'
aatttctttaagaaagccgcagccgctctttttggtggcaagaaggacaaagcaagatataacgacacacacaataagg
tgictaaggaagatgaagaca
tttacaagcttcggcatgatttgaagaaagtgtcgataacccagcagcctagcaaacacagagcagacgaggagcccca
gccgcctcccactactgttg
ccagatctggaacgtccgcagtggaaaacaaacagcagattggggatgcaatccggatgatagtcagggggactctcgg
cagctgcagcagcagca
gcgaatgccttgaagacagcaccatgggaagtgttgcagacacagtggcaagagtccttcgaggatgtctagaaaacat
gccggaagctgactgtatcc
ccaaagagcagctgagcacgtcctttcagtgggtcaccaagtgggtcgactactccaacaaatatggctttgggtacca
gctctcggaccacactgttgg
cgtccttttcaacaacggggctcacatgagcctccttccggacaaaaagacagttcactattatgcggaacttggccaa
tgctctgttttcccagcaacaga
tgcccctgaacaatttattagtcaagtgacggtgctgaaatacttttctcattacatggaggagaacctcatggatggt
ggtgatctcccgagtgttactgaca
ttcgaagacctcggctctacctcctgcagtggttaaagtctgataaagccttaatgatgctcttcaatgacggcacatt
tcaggtgaatttctaccacgatcat
acaaaaatcatcatctgtaaccagagtgaagaataccttctcacctacatcaatgaggacaggatctctacaactttca
gactgacgactctgctgatgtctg
gctgttcgttagaattgaaaaatcgaatggaatatgccctgaacatgctcttacagagatgtaactgaaaacattatta
ttattattattataattatttcgagcgg
acctcatgggactcttttccactgtgagatcaacagggaagccagcggaaagatacagagcatgttagagaagtcggac
aggtggtggtacgaatacaa
ttcctctgtggcctgctggactgctggaaccagaccagcctaaggtgtagagttgactttggacaatcctgagtgtgga
gccgagtgcagttttccctgaga
tacctgtcgtgaaaaggtttatgggacagtttttcagaaagatgcattgactctgaagttctctctgttgagagcgtct
tcagttggaagacttggaactgtga
atacacttcctgaaggggagggagaagggaggttgctcccttgctgtttaaaggctacaatcagagcagcttttggctg
cttaactgtgaactatggccata
catttttttttttttggttatttttgaatacacttgtggttggaaaagtgcattccttgttaataaactttttatttat
tacagccccaagagcagtatttattatcaagat
gttctctttttttatgttgaccatttcaaactcttggcaataaagagtatgacatagaaaaaaaaaaaaaaaaaaaaaa
aaa
SEQ ID No.l8
MELLRTITYQPAAGTKMCEQALGKACGGDSKKKRPQQPSEDGQPQAQVTPAAPHHI~HI~3SHS
GPEISRIIVDPTTGKRYCRGKVLGKGGFAKCYEMTDLTNNKVYAAKIIPHSRVAKPHQREKIDKE
IELHRLLHHKHV VQFYHYFEDKENIYILLEYCSRRSMAHILKARKVLTEPEVRYYLRQIVSGLKY
LHEQEILHRDLKLGNFIINEAMELKVGDFGLAARLEPLEHRRRTICGTPNYLSPEVLNKQGHGCE
SDIWALGC VMYTMLLGRPPFETTNLKETYRCIREARYTMPSSLLAPAKHL.IASMLSKNPEDRPSL
DDIIRHDFFLQGFTPDRLSSSCCHTVPDFHLSSPAKNFFKKAAAALFGGKKDKARYNDTHNKVSK
EDEDIYKLRHDLKKVSITQQPSKHRA.DEEPQPPPTTVARSGTSAVENKQQIGDAIRMIVRGTLGS
CSSSSECLEDSTMGSVADTVARVLRGCLENMPEADCIPKEQLSTSFQWVTKWVDYSNKYGFGY
QLSDHTVGVLFNNGAHMSLLPDKKTVHYYAELGQCSVFPATDAPEQFISQVTVLKYFSHYMEE
NLMDGGDLPSVTDIRRPRLYLLQWLKSDKALMMLFNDGTFQVNFYHDHTKIIICNQSEEYLLTYI
NEDRISTTFRLTTLLMSGCSLELKNRMEYALNMLLQRCN
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
7/17
SEQ ID No.l9
aacttagctggactgcagccttctccgctggaactcgccaagccagctgatttccccatccaaagccatgaagagcggc
gtatgtctgtgcgtggtgatg
gcagtcctagctgctggcgccctggcgcagccggtagtccctgcagaagctacggaccccgtggagcagcgggcgcaag
aggcgccccgaaggca
gctgcgggctgtgctccggacggacggcgagccccgagcgcgcctgggcgcactgctagcgcgatacatccagcaggtc
cgcaaagctccttctgg
ccgcatgtccgttcttaagaacctgcagagcctggaccccagccatagaataagtgaccgggactacatgggctggatg
gattttggccggcgcagtgc
cgaggactacgaatacccatcgtagtgggccagcgtcttggccctgcttggaggaggtggaatgaggaaacaaccacac
atacgacccctcgcctcta
atgtctgacgttttgagtatctatttattaagtccccaatgtgaaatctgtccagagtgtgcaatgcagccacatctca
gcctagctgtgtggtcggaaggcag
tgtttccttcagtgactcccagacctaatgttgctatgctattaaagagatttccttctgcccccc
SEQ ID No.20
MKSGVCLCVVMAVLAAGALAQPVVPAEATDPVEQRAQEAPRRQLRAVLRTDGEPRARLGALL
ARYIQQVRKAPSGRMSVLKNLQSLDPSHRISDRDYMGWMDFGRRSAEDYEYPS
SEQ ID No.21
cttggtgacactagacagagcaactccagcgttaccgctcccgctcctggtttctcggcttctcatcgcagtcaatctt
ggactttggggttttgctactgtca
gaaggacttctttctgcttcaagtgcttgacaacgcacccctttatcagggtatcagagcatcgccacagaatgaagct
ggtttccatcaccctgatgttattg
ggttcactcgctttcctaggcgcggacactgcagggccagatactccttcgcagttccgaaagaagtggaataagtggg
cgctaagtcgtgggaagagg
gaactacaagcatccagcagctaccctacgggactcgctgatgagacgacagttcctacccagactcttgatccattcc
tggacgagcagaacacaact
ggccccctacaagccagcaatcagagcgaagcccacattcgtgtcaaacgctaccgccagagcatgaaccagggttccc
gcagcaatggatgccgct
tcgggacctgcacatttcagaaattggcccaccagatctaccagctaacagacaaagacaaggacggcatggctcccag
aaacaagatcagccctcaa
ggctatggccgccggcgccggcgttccctgctggaggtcctccggtcccggactgtggagtcctcccaggagcagacac
acacagccccaggcccct
gggcgcacatctccagactctttaggatataggtgcgggtgacagcattgaacagtcgggcgagtatcccgttggcgcc
tgcggaatcagagaacttcg
caccggggcggactgagacaatcctgcagagatctgcctggctgcccctaggggaggcagaggaacccaagaccaagcc
aggctcatgccagaaa'
ccgagacttacaggctgatactctccgggcaggggtctgagccactgccttgcccgctcataaactggtttctcacggg
gcataagcctcattactacttg
aactttccaaaacctagcgaggaacgtgcaatgcttgttgtccagccaaaggtaactatagtatttaagtttgttgctg
tcaaggtttttttttttgtaacttcaaat
atatagagatatttttgtacgttatatattgtattaagggcattttaaagtgattatattgtcaccttcccctatttta
agacgtgaatgtctcagcaaggtgtaaggt
tgtttggttccgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtaaggtggagagcgcctgattatcgcctg
tggatgaagaaaaaacattgtgtttcc
tataatctatttacataaaatatgtgatctgggaaaaagcaaaccaataaactgtctcaatgctg
SEQ ID No.22
MKLVSITLMLLGSLAFLGADTAGPDTPSQFRKKWNKWALSRGKRELQASSSYPTGLADETTVPT
QTLDPFLDEQNTTGPLQASNQSEAHIRVKRYRQSMNQGSRSNGCRFGTCTFQKLAHQIYQLTDK
DKDGMAPRNKISPQGYGRRRRRSLLEVLRSRTVESSQEQTHTAPGPWAHISRLFRI
SEQ ID No.23
tcgagcggccgcccgggcaggtccaggatcaagagtcaccgcttcgcaagcactgcctggctccatcaggatccccgca
ggctcagctccaaggcac
cgctcaccaggaaggcatcatgggcttcctgaagttctcccctttcctggttgtcagcatcttgctcctgtaccaggca
tgcagcctccaggcagtgcctttg
aggtcaatcttggaaagcagcccaggcatggccactctcagtgaagaagaagttcgcctgctggctgcactggtgcagg
actatatgcagatgaaagcc
agggagctggagcaggaggaagagcaggaggctgagggctctagcttggacagccccagatctaagcggtgtgggaatc
tgagtacctgcatgctg
ggcacgtacacacaagacctcaacaagtttcacaccttcccccaaacttcaattggggttgaagcacctggcaagaaaa
gggatgtggccaaggacttg
gagacaaaccaccaatcccattttggcaactaagctccttctctcctttctagtttccttcttgctttcttcctataac
ttgatgcatgtagttcctctctggttgctc
tccaggctattactggttgctttcctgaggcaaagaatggtatctgaaatccccagtgggtgaggagaaagtcccacag
gctaaaagagaatcacccagg
aagatggcagagagcaagggcacactcaggaagatggcagagagcaagggcagtcatctggcttcctagtagagcttct
agtcttgcttctggaagtgtt
ggttgtttgggaaataaaactattttttaaaaaaaaaaaaaaaaaaa
SEQ ID No.24
MGFLKFSPFLVVSILLLYQACSLQAVPLRSIL,ESSPGMATLSEEEVRLLAALVQDYMQMKARELE
QEEEQEAEGSSLDSPRSKRCGNLSTCMLGTYTQDLNKFHTFPQTSIGVEAPGKKRDVAKDLETN
HQSHFGN
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
8/17
SEQ H7 No. 25
aaaggcagcctgataaagctccttgtgacaggctgtcttgccagtctcccagtatgctcctcttgctctgaagtgctcc
aggattgaaaccacagcttccca
aattagcctgggaagagtgtgcggacccagcagccttttaacccgcgtcagtgcctttgctatgttcaagactgctgtt
ttggatggtgaatgctagctagc
actccatcgagacatgacagcaaaaaattctccaaaagaatttactgcttcggaatctgaggtttgcataaagactttc
aaggagcagatgcgcttggaact
tgagcttccaaagctaccaggaaacagacctacatctcccaaaatttctccacgcagttcaccaaggaattcaccatgc
tttttcagaaagttgctggtgaat
aaaagcatccgacagcggcgtcgcttcacggtggctcatacatgctttgatgtggaaaatggcccttctccaggtcgga
gcccactggaccctcaagcc
ggctcttcgtcgggactggtacttcatgccgcctttcctgggcacagccagcgcagggagtcgttcctctacgatcttg
acagcgactatgacttgtcacca
aaagcgatgtccaggaactcatcacttcccagtgagcaacacggcgatgacctgattgtcactccttttgcccaggttc
ttgccagcttgcgaagtgtaaga
1 0
aacaacttcaccctgctgacgaaccttcatggagcgccgaacaagaggtcaccagcggctagtcaggctccagtctcca
gagtcagcctgcaagagga
atcatatcagaaactagcaatggagacgctggaggaactagactggtgcctagaccagctagagaccatccagacctac
cgctctgtcagcgagatgg
cttcaaaeaagttcaaaaggatgctgaaccgggagctgacacacctctcagagatgagcagatcagggaaccaggtgtc
tgagtacatttcaaacacgtt
cttagacaagcagaacgatgtggaaatcccatctcccacgcagaaggacagggagaagaagaagaagcagcagctcatg
acccagataagtggagt
gaagaaactgatgcacagctcaagcctgaacaacacaagcatctcacgcttcgggatcaacacggaaaatgaggatcat
ctagccaaggagctggaa
gacctgaacaaatggggccttaacatcttcaatgtggctgggtactcacataatcggccccttacgtgcatcatgtatg
caatattccaggaaagagacctt
ctgaagacgtttaaaatctcatctgacacctttgtaacctacatgatgactttagaagaccattaccattctgatgtgg
catatcacaacagcctgcatgctgct
gacgtggcccagtcaactcacgttctcctttctacgccggcactggatgctgtcttcacagacctggaaatcctggctg
ccatttttgcagctgccatccatg
atgtcgatcatcctggagtctccaatcagtttctcatcaatacaaattctgaacttgctttgatgtataatgatgaatc
tgttctggaaaaccatcaccttgctgtg
ggattcaaattgctacaagaggaacactgcgacatctttcagaatcttaccaagaagcaacgccagacactcaggaaaa
tggtgattgacatggtgttgg
caactgatatgtccaaacacatgagcctcctggcagaccttaaaacaatggtagaaaccaagaaggtgacaagctccgg
tgttctcctcctggacaacta
tactgaccggatacaggttcttcgcaacatggtacactgtgcagacctgagcaaccccaccaagtccttggaattgtat
cggcaatggaccgatcgtatca
tggaggagtttttccagcagggagacaaagaacgggagaggggaatggagattagcccaatgtgtgataagcacacagc
ttctgtggaaaaatcccag
gttggtttcattgactacattgtccatccactgtgggagacctgggcagacctggttcaaccggatgctcaagatattc
tggatacactagaagataacagg
aactggtaccagagtatgataccccagagcccttccccgccactggatgagaggagcagggactgccaaggcctgatgg
agaagtttcagtttgaactg
acccttgaggaagaggattctgagggaccggaaaaggagggagaaggccacagctatttcagcagcacaaagacgcttt
gtgtgattgatccagagaa
cagggattctctggaagagactgacatagacattgcaacagaagacaagtctccgatcgacacataatctctctccctc
tgtgtggagatgaacatfccac
ccttgactgagcatgcccgctgagtggtagggtcacctaccatggccaaggcctgcacaggacaaaggccacctggcct
ttccagttacttgagtttgga
gccagaatgccaggccgtgaagcaaatagcagttccatgctgtcttgccttgcctgcaagcttggcggagacccgcagc
tgtatgtggtagtagaggcc
agttcccatcaaagctaaaatggcttgaaaacagaggacacaaagctgagagattgctctgcactaggtgttgggaagc
tgtcctgacagatgactgaac
tcactaacaacttcatctataaatctcaccacccaacccattgtctgccaacctgtgtgectttttttgtaaaatgttt
tcgcgtctttgaaatgcctgttgaatatc
tagagtttagtaccaacttctacaaacttttttgagtctttcttgaaaaacaaaaaaaaaaaaaaaaaaaaaaaaaaaa
aaaaaaaaa
SEQ ID No.26
MTAKNSPKEFTASESEVCIKTFKEQMRLELELPKLPGNRPTSPKISPRSSPRNSPCFFRKLLVNKSI
RQRRRFTVAHTCFDVENGPSPGRSPLDPQAGSSSGLVLHAAFPGHSQRRESFLYDLDSDYDLSPK
AMSRNSSLPSEQHGDDLIVTPFAQVLASLRSVRNNFTLLTNLHGAPNKRSPAASQAPVSRVSLQE
ESYQKLAMETLEELDWCLDQLETIQTYRSVSEMASNKFKRMLNRELTHLSEMSRSGNQVSEYIS
NTFLDKQNDVEIPSPTQKDREKKKKQQLMTQISGVKKLMHSSSLNNTSISRFGINTENEDHLAKE
LEDLNKWGLNIFNVAGYSHNRPLTCIMYAIFQERDLLKTFKISSDTFVTYMMTLEDHYHSDVAY
HNSLHAADVAQSTHVLLSTPALDAVFTDLEILAAIFAAAIHDVDHPGVSNQFLINTNSELALMYN
DES VLENHHLAVGFKLLQEEHCDIFQNLTKKQRQTLRKMVIDMVLATDMSKHMSLLADLKTM
VETKKVTSSGVLLLDNYTDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDKERE
RGMEISPMCDKHTASVEKSQVGFIDYIVHPLWETWADLVQPDAQDILDTLEDNRNWYQSMIPQ
SPSPPLDERSRDCQGLMEKFQFELTLEEEDSEGPEKEGEGHSYFSSTKTLCVIDPENRDSLEETDID
IATEDKSPIDT
SEQ ID No.27
5 ~
ggccgcgcgggagtctgagaatgcaaagtgccatgttcctggctgtccagcacgactgcgtacccatggacaagagtgc
aggcaacggccccaaggt
cgaggagaagcgggagaaaatgaagcggacactcttaaaccattggaagacccgtttgagctacttcttgcagaattcc
tctgctcctgggaagcccaa
aactggcaagaaaagcaaacagcaaacttttatcaagccttctcctgaggaagcgcacgtctgggcagaagcatttgat
gaactgctggccagtaaatat
gggctggctgcattcagggcgtttttaaagtccgagttctgtgaagaaaacattgaattctggttggcttgtgaagact
tcaaaaaaaccaaatcaccccaa
aaactgtcctcaaaagcaaggaaaatctataccgacttcatagagaaggaagctcccaaagagataaacatagacttcc
aaacgaaatctctgattgccc
5 5
aaaatatccaagaggctacaagtggctgcttcaccacagctcagaagagggtgtacagtttgatggagaacaattctta
tcctcggttcttggagtccgaat
tctaccaggacttatgtaaaaagccacagatcaccacggagccccatgctacatgagaccaggagtccccccacacaca
aaggacattccattctgtctc
ccaagagcaaaggctgtgacctgccagaaaaaaaaaaaaaactgaccttgaattcagcctgagtgttaggaaaacatcg
ctcagaactattgattcaatg
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
9/17
ttgggtagtgaatcaggaagtcagcaacctaggagaggctctgtgtgagaacggcttccctcactgtgtgaagaacaga
gggagggaacaggcctctg
aatgtgttcttcctccttgtcgggaaagcagagtttgagatgaaagatccgatgcaatgttgttggagcatttaaaatc
aagaggtctgggattatgtggcctt
agctagttggctgtacaccttccctaaactagtccatgttacacatagtggtgttagttctagttttaatattttagta
ctaagtaacattacaatgtttactgtgtg
caagggtgttgacgttcttaggactacagatcattagtactagtgtgtcacgtatcactgaaactgagaagtatgtttg
agttgttaaatggtgtgtgtgatgga
ccgaatgctgtgccgtgctgtagaa
SEQ ID No.28
MQSAMFLAVQHDCVPMDKSAGNGPKVEEKREKMKRTLLNHWKTRLSYFLQNSSAPGKPKTGK
KSKQQTFIKPSPEEAHVWAEAFDELLASKYGLAAFRAFLKSEFCEENIEFWLACEDFKKTKSPQK
LSSKARKIYTDFIEKEAPKEINIDFQTKSLIAQNIQEATSGCFTTAQKRVYSLMENNSYPRFLESEF
YQDLCKKPQITTEPHAT
SEQ ID No.29
aaaaagtatatgaggacaaatgtaaggcaaatgaccatggaaacagttgaatcacagcaggatcgaagtgtaacacgtt
ctgtggcagagcatagctct
gctcatatgcagactggtcaaatttctgttcctactctagctcaggtagcaacaattgcagagacagatgattctgcag
actcagaagtaattgattcgcata
aacgtagagaaattctttcacgaagaccctcatatagaaaaatactgaatgaactttcctctgatgtgcctggtattcc
caagattgaagaagaaaaatcaga
ggaagaagggacaccacctaacattgctaccatggcagtaccaactagcatatatcagactagcacggggcaatacaat
gaggagactgaccttgccc
caagtcacatggctgctgccacaggtgacatgccaacttaccagatccgagctcctactactgctttgccacaaggtgt
ggtgatggctgcctcaccagg
aagcctgcacagtccccagcaactagcagaagaagcaactcgcaagcgggagctgaggctgatgaaaaacagggaagct
gcccgggagtgtcgca
ggaagaagaaagaatatgtcaaatgtcttgaaaatcgtgtggctgtgcttgaaaatcaaaacaagaccctcattgagga
actcaaggccctcaaagacctt
tattgccataaagcagagtaactgtgtttgatttggaccttgttgactgtgaactctaatcggggcaggcgatgcagca
tcctcataatggccatgtggactt
gtagatgggtctcttaacccttgcttaagaatacagtctgctgtagagtgtgaattgggaatactgttccatgggttgg
aatgcagctcccctcacattaccaa
gcttgctctattgccaatagcatgcaacatatgttttgtttgcccttctgcttctacttttttcagggaagctgctaaa
gaatgtcgacgtcgaaagaaagagtat
gtgaagtgtcttgagagtcgagtcgcagtgctggaagttcagaacaagaagcttatagaggagcttgaaactttgaaag
acatttgctctcccaaaacaga
ttagtagaaatatttaactatgaactgattacagcatgtacagttgcttttgaatgcaatacaaatatatagccggcaa
gaattatggctttttcctttgtatcattc,
atctaactttctaaaactaacattcctaagatgctttgttgtatttaatttgctcttacctctaaggtcaattttttag
aagagacaaactcaaaaaatgtatgtaac
aaattettaaaatgaagtatttgtaagacttgttccagtcaacatatttacagttcccagtctctctgtcatgaatagt
gtcctatgcaataaaaattttgcaggttt
taagaatcattttaggaaagggtgaatcaaaggcagtgcatctctccagtagtaagataaaatcaacccatagagatac
ctcaggaaagaatgaaaggaa
gtgtatcctgatgacatgacgtgagaatagcctacaaatgaatttatgcatttatagatttttataatcgtcactttgt
aaagaaagtattgtattgctgtccttgg
gtgccacagttgaagacagttttaaatagaaccatgttggttgctctttgtactatttggtatttatttaagtatctga
gcatttactacagcttcctactatgtatgt
agtatgtgaatttctacaaaagtttgtgctctttgctgttatttaatgaaagagacaacatattttcattatctggaat
gagttccacaagtatgaatttattgctaca
ctggatcagcagccttgcaaatactgggccatttcattagaggacaacagcagggctctaggagcagagttcagtgtgg
agcacttgcctggcatgctac
3 5
atgttcagttgaaagggaagacctcaagctctgcaaatggaatggggtccaggggaagaggttagaggttagcctttgt
gctgtactaggcttcttgctgat
cgtctggagagtttctgctgatgaccctccattgtgaattcttgcaacctcaggaatgttaacgtttaaaaaacttccc
aagatgtcatttttgattttacaacttg
gatcaattttgttttgctctttggaatatagctgtgtacatttgtcacgtaggtttaggctggccttaaactcacagtt
ctcttgcctcagccttctgagtgcttgga
ttacggatgtgggccagaatatccagtttgatcaagtattcttttataaaatattactttcttttt
SEQ ID No.30
MTMETVESQQDRSVTRS VAEHSSAHMQTGQIS VPTLAQVATIAETDDSADSEVIDSHKRREILSR
RPSYRKILNELSSDVPGIPKIEEEKSEEEGTPPNIATMAVPTSIYQTSTGQYNEETDLAPSHMAAAT
GDMPTYQIRAPTTALPQGV VMAASPGSLHSPQQLAEEATRKRELRLMKNREAARECRRKKKEY
VKCLENRVAVLENQNKTLIEELKALKDLYCHKAE
SEQ ID No.31
gctgaagcgtttcctcaagcctgccggggtgggaggagaggaggaggtggtggtggtggaggaggtggaggcagagggt
ggagagagagaaagc
gcacgccgagaggaggtgtgggtgttccgctcccatcctaacggaacgagctccctcttcgcggacatgggattgccca
gcggctgctaacccctctcc
tggtcctgatcccccaaaccggcgtggctccccggtcaccaaggagctgattacaagggaccaggatttgcatccttgg
ctgggcgtccattggctaca
gagtgcctgacctgggtcaggctttccaacacggacatgtctgacaaaatgtcgagtttcctacatattggagacattt
gttctctgtatgcggagggatcta
cgaatggatttatcagcaccttaggcttggttgatgaccgttgtgttgtacagccagaagccggggaccttaacaatcc
acccaagaaattcagagactgc
ctctttaagctatgtcctatgaatcgatactccgcacagaaacagttctggaaagctgctaagcccggggccaacagca
ctacagatgcagtgctgctcaa
caaattgcatcatgctgcagacttggaaaagaagcagaatgagacagaaaacaggaaattgttggggaccgtcatccaa
tatggcaacgtgatccagct
cctgcatttgaaaagcaataaatacctgactgtgaataagaggctcccagccttgctagagaagaatgccatgagggtg
acgttggacgaggctggaaat
gaagggtcctggttttacattcaaccattttacaagcttcgctccatcggagacagtgtggtcataggcgacaaggtag
ttttgaatcctgtcaatgctggcc
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
10/17
agcctctacatgccagcagtcatcagctggtggataacccaggctgcaatgaggtcaactccgtcaactgtaatacaag
ctggaagatagtgcttttcatg
aaatggagtgataacaaagacgacattctcaaaggaggtgatgtggtgaggctcttccatgccgagcaagagaagtttc
tcacctgtgatgagcaccgg
aagaagcagcatgtgttcctgaggaccaccggcaggcagtcagccacgtcggccaccagttctaaagccctgtgggaag
tggaggtagtccagcacg
acccatgtcggggtggagctgggtactggaatagcctcttccggttcaagcacctggctacagggcattacttggctgc
agaggtagaccctgactttga
ggaagaatgcctggagtttcagccctcagtggaccctgatcaggatgcatctcggagtaggttgagaaacgcgcaagaa
aaaatggtatactctctggtc
tccgtgcctgaaggcaacgacatctcctccatctttgagctagaccccacgactctgcgtggaggtgacagccttgtcc
caaggaactcctatgtccgtct
cagacacctgtgcaccaacacctgggtacacagcacaaacatccccatcgacaaggaagaggagaagcctgtgatgctg
aaaattggtacctctcccc
tgaaggaggacaaggaagcatttgccatagttcctgtttcccctgctgaggttcgggacctggactttgccaatgatgc
cagcaaggtgctgggctccatc
gctgggaagttggaaaagggcaccatcacccagaatgagagaaggtctgtcacgaagcttttggaagacttggtttact
ttgtcacgggtggaactaact
ctggccaagacgtgcttgaagttgtcttctctaagcccaatcgagagcggcagaagctgatgagggaacagaatattct
caagcagatcttcaagctgttg
caggcccccttcacggactgcggggatggcccgatgcttcggctggaggagctgggggatcagcgccatgctcctttca
gacatatttgccgactctgct
acagggtcctgcgacactcacagcaagactacaggaagaaccaggagtacatagccaagcagtttggcttcatgcagaa
gcagattggetatgacgtg
ctggccgaagacaccatcactgccctgctccacaacaaccggaaactcctggagaagcacatcaccgcggcagagattg
acacgtttgtcagcctggt
gcgaaagaacagggagcccaggttcttggattacctctctgacctctgcgtatccatgaacaagtcaatccctgtgaca
caggagctcatctgtaaagctg
tgctcaatcecaccaatgctgacatcctgattgagaccaagctggttctttctcgttttgagtttgaaggcgtttccac
tggagagaatgctctggaagccgg
ggaggatgaggaagaggtgtggctgttctggagggacagcaacaaagagatccgtagtaagagtgtccgggaattggcg
caagatgctaaagaggg
acagaaggaagacagggacatcctcagctactacagatatcagctgaacctctttgcaaggatgtgtctggaccgccag
tacctggccatcaatgaaatc
tccgggcagctggatgttgatctcattctccgctgcatgtctgacgagaacctcccctacgacctcagggcatcctttt
gccgcctcatgcttcacatgcatg
tggaccgagatccccaagagcaggtgacacctgtgaaatatgcccgactgtggtcagaaattccctctgagatcgccat
tgatgactatgacagcagtgg
aacatccaaagatgaaattaaggagaggtttgcacagacgatggagtttgtggaggagtacctaagagatgtggtttgt
caaagattccccttctctgataa
ggagaaaaataagctcacgtttgaggttgtgaacttagccaggaatctcatatactttggtttctacaacttttctgac
cttctccgattaaccaagatcctcttg
gcaatcttagactgtgtccatgtgaccactatcttccccattagcaagatgacaaaaggagaagagaataaaggcagta
acgtgatgaggtctatccatgg
cgttggggagctgatgacccaggtggtgctgcggggaggaggcttcttgcccatgactcccatggctgcggcccctgaa
ggaaatgtgaagcaggca
gagccagagaaagaggacatcatggtcatggacaccaagttgaagatcattgaaatactccagtttattttgaatgtga
gattggattataggatctcctgcc
tcctgtgtatatttaagcgagagtttgatgaaagcaattcccagtcatcagaaacatcctccggaaacagcagccagga
agggccaagtaatgtgccagg
tgctcttgactttgaacacattgaagaacaagcggaaggcatctttggaggaagtgaggagaacacacctttggacctg
gatgaccatggtggcagaac
cttcctcagggtcctgctccacttgacaatgcatgactacccacccctggtgtctggggccctgcagctcctctttcgg
cacttcagccagaggcaggagg
tccttcaggccttcaaacaggttcaactgctggttactagccaagatgtggacaactacaaacagatcaagcaagactt
ggaccaactaaggtccattgtg
gagaagtctgagctctgggtgtacaaaggccaaggtcccgatgagcctatggacggagcctccggtgaaaatgagcata
agaaaaccgaggagggg
3 0
acgagcaagccactgaagcacgagagcaccagcagctacaactaccgagtggtgaaagagattttgattcgacttagca
agctctgcgtgcaggagag
cgcgtcggtgaggaagagccggaagcagcagcaacgactgctgaggaacatgggcgcacacgctgtggtgctggagctg
ctgcagatcccctacga
gaaggccgaagacacaaagatgcaagagatcatgcggctggctcatgaatttttgcagaatttctgtgcaggcaaccag
cagaatcaagctttgctgcat
aaacacataaacctgtttctcaagccagggatcctggaggcagtgacgatgcagcacatcttcatgaacaacttccagc
tgtgcagtgagatcaacgaga
gagtggtccagcactttgttcactgcatagagacccacggtcgaaacgtccagtatatcaagtttctccagacgattgt
caaggcagaagggaaattcatta
3 5
aaaagtgccaagacatggtcatggctgagcttgtcaactctggagaggacgtcctcgtgttctacaatgacagagcctc
tttccagactctgatccagatg
atgcggtccgagcgtgaccggatggatgagaacagccctctcatgtaccacatccatctggtggagctcttggccgtgt
gcacagagggcaagaatgtg
tacacggagatcaagtgcaactccttgctcccgctcgatgacatcgttcgtgtggtcactcatgaagactgcatccccg
aggttaagatcgcttacattaac
ttcctgaatcactgctatgtggatacggaggtggagatgaaggagatttacacaagcaaccacatgtggaagttgtttg
agaatttcctcgtggacatctgc
agggcctgtaacaacacaagcgacaggaagcacgcagactccattctggagaagtacgtcactgaaatcgtgatgagca
tcgtcaccaccttcttcagc
40
tctcccttctcagaccagagcaccactctgcagacccgccagcctgtctttgtgcaactcctgcaaggcgtgttccgag
tttaccactgcaactggctgatg
ccgagccaaaaagcctcggtggagagctgcatccgggtgctctctgacgtagccaagagccgggccatagccattcctg
ttgacctggacagccaagt
caacaacctcttcctgaagtcccacaacattgtgcagaaaacagccctgaactggcggttatcagcccgaaacgccgct
cgcagagactctgtactggc
agcatccagagactaccgaaatatcattgagaggttacaggacatcgtgtctgccctagaggaccggctcaggcccctg
gtgcaggctgagctgtctgt
gctcgtggatgttctacacagaccagaactgctcttccccgagaacacggatgccaggaggaaatgtgagagtggaggt
ttcatctgcaagctaataaaa
45
cataccaagcaactgctggaggagaatgaagagaaactatgcattaaagtcttacagaccctcagggaaatgatgacca
aagacagaggctatggaga
gaagcaaatttccattgatgaatcggaaaatgccgagctgccacaggcaccggaagctgagaactccacagagcaggag
cttgaaccaagtccaccc
ctgaggcaactggaagaccataaaaggggtgaggcactccgacaaattttggtcaaccgttactatggaaacatcagac
cttcaggaagaagagagag
ccttaccagctttggcaatggcccactatcaccaggaggacccagcaagcctggtggaggagggggaggtcctggatct
agttccacaagcaggggtg
agatgagcctggctgaggttcagtgtcacctcgacaaggagggggcctccaacctggtcatcgatctcataatgaatgc
atccagtgaccgagtattcca
50
tgaaagcattctgctggccatcgcacttctggaaggaggcaacaccaccatccagcactcgtttttctgccggctgaca
gaagataagaaatcagagaag
ttcttcaaggttttttacgatcgaatgaaggtggcccagcaggaaatcaaggcgacagtgacagtgaacaccagcgact
tgggaaacaaaaagaaagat
gatgaagtggacagggatgccccgtctcggaagaaagccaaagagcccacaacacagataacagaagaggtccgggatc
agctcctggaagcatct
gctgccaccaggaaagcctttaccaccttccggagggaggccgaccctgatgaccattaccagtctggggagggcaccc
aggctacaaccgacaaag
ccaaggatgacctagagatgagcgctgtcatcaccatcatgcagcctatcctgcgcttcctgcagctgctgtgtgaaaa
ccacaaccgagatctgcagaa
55
tttccttcgttgccaaaataataagaccaactacaatttggtgtgtgagacactgcagtttctggactgtatttgtggg
agcacaaccggaggccttggtcttc
ttggactgtacataaatgaaaagaatgtagcacttatcaaccaaaccctggagagtctgacggagtactgtcaagggcc
ttgccatgagaaccagaactg
catcgccacccacgagtccaatggcatcgatatcatcacagccctcatcctcaatgatatcaaccctctgggaaagaag
cggatggacctggtgttagaa
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
11/17
ctgaagaacaatgcttcgaagctgctactggccatcatggaaagcagacacgatagtgaaaatgcagagaggatcctgt
acaacatgaggcccaagga
gctggtggaagtgatcaagaaggcctacatgcaaggtgaagtggaatttgaggatggggagaacggtgaggatggagct
gcctcacccaggaacgtg
ggccacaacatctacatcctcgctcaccagttggctcggcataacaaagaacttcaaaccatgctgaaacctggaggcc
aggtggatggggatgaagct
ctggagttctacgcgaagcacacagcacaaattgagattgtcagactggaccggacaatggaacagatcgtcttccctg
tgcccagcatctgtgaattcct
gactaaggaatcgaaacttcgaatatattacaccacagagcgggatgagcaaggtagcaagatcaatgacttcttcctg
cgctccgaggacctctttaac
gagatgaactggcagaagaaacttcgagcccagcctgtcttgtactggtgtgcccgaaacatgtctttctggagcagca
tctccttcaacctggccgtcct
gatgaacctgctggtggcgtttttctatccatttaaaggagtgaggggaggaacactagagccacactggtcaggcctc
ctgtggacagccatgctcatct
ctctggccattgtcattgctctgcccaagccccacggcatccgggccttaattgcttctacaatcctacgactgatatt
ttcagttgggttgcagcccacactg
tttctgctgggagctttcaatgtctgcaataaaatcatcttcctgatgagctttgtgggcaactgtgggaccttcacca
gaggctaccgggccatggttctgg
atgtggagttcctctatcatttgctgtatctactcatctgtgccatgggcctcttcgtacatgagttcttctatagctt
gctgctttttgatttagtgtacagagagg
agactttgcttaatgtcattaaaagtgtcacccgcaatggacggtccatcatcttgacagcggtcctggctctgatcct
ggtttacctgttctcaattgtgggct
atctgttcttcaaggatgactttatcttggaagtagataggttgcccaatgaaacagctgttccagaaactggcgagag
tttggccaacgatttcctgtactct
gatgtgtgcagggtagagacgggggagaactgcacctctcctgcacccaaagaagagctgctccctgccgaagaaacgg
aacaggataaggaacac
acgtgtgagaccctgctcatgtgcatcgtcactgttctgagtcacgggctgcggagtgggggaggggtaggagacgtgc
tcaggaagccatccaaaga
ggagcctctgtttgctgcaagggtgatctacgacctcctcttcttcttcatggtcatcatcatcgtcctgaacctgatt
ttcggggtcatcatcgacacctttgct
gacctgaggagtgagaagcaaaagaaggaggagatcttaaaaaccacgtgcttcatctgcggcttggaaagggacaagt
ttgacaataagactgtcac
ctttgaagagcacatcaaggaagaacacaacatgtggcactatctgtgcttcatcgtgctggtgaaagtgaaggactcc
acagagtacaccgggcctga
gagttacgtggcagagatgatcagggaaagaaaccttgattggttcctcagaatgagagccatgtccctggtcagcagc
gattctgaaggggaacagaa
cgagctgaggaacctgcaggagaagctggagtctaccatgaagctggtcaccaatctttctggccagctgtcagaacta
aaggaccagatgacagaac
agaggaagcagaaacaaagaatcggccttctaggacatcctcctcacatgaatgtcaacccacagcagccggcctaggc
aaatgaggcagagggact
ctgctcagccctctgtatatcactgtcagggtgggtacggctcattggttctgatttgcccactaagggtacatgtgcg
cttagtacatttgtaaatactcagttt
tgtattgtatgtatatgattgctattctcagaggtttggactttcgtattgtaattagctctgttggcatggtgacttg
tcactcctgccaaaaatattaaaaatgcct
tttttggaaggactacagaaagtacctgatttgcacttgaaccagattatagatttaaaagtatatgacatgtattttg
tatttaaaactagaatagccagtattta
tgttttttataaaactgtgcaatacaaattatgcaatcaccataactttgtaactcctgagtgtcctaagggagtacac
atctttgaagctgatttgttgatactcg
tgtaataaatggttaaatatcaaatgctgctgctgctgccaaaattatattaatagcgagtttctggcccctgggcaat
tttgtaccttgtaattatcctatggtga
tgctgtttctcgttgctaatggcattagtgcccctgtatcctagtgataactccaggtctgtgaaccattcaaacagca
ttcattttgagaaaagcaactttagtt
tcaaggataattttaagcttcaaaattaatcatttaaagtgtttctttaagagagccatgttagaggctcacactttag
cttgaaaggagttgatgaattaatttttt
aaagggaactttttacatgacgtttggaataacagcatattgctgaccagtcagtgtcatctcccgggtgaattttgat
gtcacgttatagtcaaatgagttagc
tgatggtttctagattttcttcctctgaaccatgatgcagtaggtaagaagttattatgcgtatatacatatatacatt
catatacgacaaagtaggagctgtccc
cttaggatgcatagctgcccctagggtacgtagctgaacactgacaatggcgttcttctgaaagagccacgtttgggtt
ttatttctttgtcacatgatttctttt
ctggatgggtgcaaagtatcacaggaagtgttttctctctgtcgccttgttttgtacctgggtctcgctttactagacc
gtctctgcacaaaagtttaaaaactg
aaccgtatgcagagttccgaagcaagtcaagtttgtaaatgcatacctaaaaatatttaataaacgatgcagaatcct
SEQ ID No.32
MSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCV VQPEAGDLNNPPKKFRDCLFKLCPMNR
YSAQKQFWKAAKPGANSTTDAVLLNKLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKS
NKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQPFYKLRSIGDS V V IGDKV VLNPVNAGQ
PLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDIL,KGGDVVRLFHAEQEKFLTCD
EHRKKQHVFLRTTGRQSATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAA
EVDPDFEEECLEFQPS VDPDQDASRSRLRNAQEKMVYSLV S VPEGNDISSIFELDPTTLRGGDSLV
PRNSYVRLRHLCTNTW VHSTNIPIDKEEEKPVMLKIGTSPLKEDKEAFAIVPVSPAEVRDLDFAN
DASKVLGSIAGKLEKGTITQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMR
EQNILKQIFKLLQAPFTDCGDGPMLRLEELGDQRHAPFRHICRLCYRVLRHSQQDYRKNQEYIAK
QFGFMQKQIGYDVLAEDTITALLHNNRKLLEKHITAAEIDTFVSLVRKNREPRFLDYLSDLCVSM
NKSIPVTQELICKAVLNPTNADILIETKLVLSRFEFEGVSTGENALEAGEDEEEVWLFWRDSNKEI
RSKSVRELAQDAKEGQKEDRDILSYYRYQLNLFARMCLDRQYLAINEISGQLDVDLILRCMSDE
NLPYDLRASFCRLMLHMHVDRDPQEQVTPVKYARLWSEIPSEIAIDDYDSSGTSKDEIKERFAQT
MEFVEEYLRDVVCQRFPFSDKEKNKLTFEVVNLARNLIYFGFYNFSDLLRLTK1LLAILDCVHVT
TIFPISKMTKGEENKGSNVMRSIHGVGELMTQVVLRGGGFLPMTPMAAAPEGNVKQAEPEKEDI
MVMDTKLKIIEILQFILNVRLDYRISCLLCIFKREFDESNSQSSETSSGNSSQEGPSNVPGALDFEHI
EEQAEGIFGGSEENTPLDLDDHGGRTFLRVLLHLTMHDYPPLVSGALQLLFRHFSQRQEVLQAFK
QVQLLVTSQDVDNYKQIKQDLDQLRSIVEKSELW VYKGQGPDEPMDGASGENEHKKTEEGTSK
PLKHESTSSYNYRVVKEIL,IRLSKLCVQESASVRKSRKQQQRLLRNMGAHAVVLELLQIPYEKAE
DTKMQEIMRLAHEFLQNFCAGNQQNQALLHKHINLFLKPGILEAVTMQHIFMNNFQLCSEINER
VVQHFVHCIETHGRNVQYIKFLQTIVKAEGKFIKKCQDMVMAELVNSGEDVLVFYNDRASFQTL
IQMMRSERDRMDENSPLMYHIHLVELLAVCTEGKNVYTEIKCNSLLPLDDIVRV VTHEDCIPEV
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
12/17
KIAYINFLNHCYVDTEVEMKEIYTSNFIMWKLFENFLVDICRACNNTSDRKHADSILEKYVTEIV
MSIVTTFFSSPFSDQSTTLQTRQPVFVQLLQGVFRVYHCNWLMPSQKASVESCIRVLSDVAKSRA
IAIPVDLDSQVNNLFLKSHNIVQKTALNWRLSARNAARRDSVLAASRDYRNIIERLQDIVSALED
RLRPLVQAELSVLVDVLHRPELLFPENTDARRKCESGGFICKLIKHTKQLLEENEEKL,CIKVLQTL
S REMMTKDRGYGEKQISIDESENAELPQAPEAENSTEQELEPSPPLRQLEDHKRGEALRQILVNRY
YGNIRPSGRRESLTSFGNGPLSPGGPSKPGGGGGGPGSSSTSRGEMSLAEVQCHLDKEGASNLVI
DLIMNASSDRVFHESIL,LAIALLEGGNTTIQHSFFCRLTEDKKSEKFFKVFYDRMKVAQQEIKATV
TVNTSDLGNKKKDDEVDRDAPSRKKAKEPTTQITEEVRDQLLEASAATRKAFTTFRREADPDDH
YQSGEGTQATTDKAKDDLEMSAVITIMQPILRFLQLLCENHNRDLQNFLRCQNNKTNYNLVCET
LQFLDCICGSTTGGLGLLGLYINEKNVALINQTLESLTEYCQGPCHENQNCIATHESNGIDIITALI
LNDINPLGKKRMDLVLELKNNASKLLLAIMESRHDSENAERILYNMRPKELVEVIKKAYMQGEV
EFEDGENGEDGAASPRNVGHNIYIL,AHQLARHNKELQTMLKPGGQVDGDEALEFYAKHTAQIEI
VRLDRTMEQIVFPVPSICEFLTKESKLRIYYTTERDEQGSKINDFFL.RSEDLFNEMNWQKKLRAQ
PVLYWCARNMSFWSSISFNLAVLMNLLVAFFYPFKGVRGGTLEPHWSGLLWTAMLISLAIVIAL
PKPHGIRALIAST1I,RLIFSVGLQPTLFLLGAFNVCNKIIFLMSFVGNCGTFTRGYRAMVLDVEFLY
HLLYLLICAMGLFVHEFFYSLLLFDLVYREETLLNVIKSVTRNGRSIIL.TAVLALILVYLFSIVGYLF
FKDDFILEVDRLPNETAVPETGESLANDFLYSDVCRVETGENCTSPAPKEELLPAEETEQDKEHT
CETLLMCIVTVLSHGLRSGGGVGDVLRKPSKEEPLFAARVIYDLLFFFMVIIIVLNLIFGVIIDTFA
DLRSEKQKKEEILKTTCFICGLERDKFDNKTVTFEEHIKEEHNMWHYLCFIVLVKVKDSTEYTGP
ESYVAEMIRERNLDWFLRMRAMSLVSSDSEGEQNELRNLQEKLESTMKLVTNLSGQLSELKDQ
MTEQRKQKQRIGLLGHPPHMNVNPQQPA
SEQ ID No.33
ggcacgagccgagttggaggaagcagcggcagcggcagcggcagcggtagcggtgaggacggctgtgcagccaaggaac
cgggacagcgaag
cgacggcaggtcgcagctggatcgcaggagcctgggagctgggagcttcagaggccgctgaagcccaggctgggcagag
gaaggaagcgagccg
acccggaggtgaagctgagagtggagcgtggcagtaaaatcagacgacagatggacagtgtgacaggaacgtcagagag
gattgggcctcgctgcg
agagtcagcctggagtcaaggtgttgacaagttgctgagaaggacacgtgggaggacggtggcgcgcggagggagagcc
ctgtcttcagtcaccccg
ttgatggaggacagatggacagcagccggacggccagtcacctctcttaaacctttggatagtggtcctttgtgctctg
ctggacacctgttggggatttta
gcccattctctgaactcactttctcttaaaacgtaaactcggacggcagtgtgcgagccagctcctctgtggcagggca
ctagagctgcagacatgagtgc
agagggctaccagtacagagcactgtacgactacaagaaggagcgagaggaagacattgacctacacctgggggacata
ctgactgtgaataaaggc
tccttagtggcacttggattcagtgatggccaggaagcccggcctgaagatattggctggttaaatggctacaatgaaa
ccactggggagaggggagac
tttccaggaacttacgttgaatacattggaaggaaaagaatttcaccccctactcccaagcctcggccccctcgaccgc
ttcctgttgctccgggttcttcaa
aaactgaagctgacacggagcagcaagcgttgccccttcctgacctggccgagcagtttgcccctcctgatgttgcccc
gcctctccttataaagctcctg
gaagccattgagaagaaaggactggaatgttcgactctatacagaacacaaagctccagcaaccctgcagaattacgac
agcttcttgattgtgatgccg
cgtcagtggacttggagatgatcgacgtacacgtcttagcagatgctttcaaacgctatctcgccgacttaccaaatcc
tgtcattcctgtagctgtttacaat
gagatgatgtctttagcccaagaactacagagccctgaagactgcatccagctgttgaagaagctcattagattgccta
atatacctcatcagtgttggctta
cgcttcagtatttgctcaagcattttttcaagctctctcaagcctccagcaaaaaccttttgaatgcaagagtcctctc
tgagattttcagccccgtgcttttcag
atttccagccgccagctctgataatactgaacacctcataaaagcgatagagattttaatctcaacggaatggaatgag
agacagccagcaccagcactg
ccccccaaaccacccaagcccactactgtagccaacaacagcatgaacaacaatatgtccttgcaggatgctgaatggt
actggggagacatctcaagg
gaagaagtgaatgaaaaactccgagacactgctgatgggacctttttggtacgagacgcatctactaaaatgcacggcg
attacactcttacacctagga
aaggaggaaataacaaattaatcaaaatctttcaccgtgatggaaaatatggcttctctgatccattaaccttcaactc
tgtggttgagttaataaaccactac
cggaatgagtctttagctcagtacaaccccaagctggatgtgaagttgctctacccagtgtccaaataccagcaggatc
aagttgtcaaagaagataatatt
gaagctgtagggaaaaaattacatgaatataatactcaatttcaagaaaaaagtcgggaatatgatagattatatgagg
agtacacccgtacttcccagga
4'J
aatccaaatgaaaagaacggctatcgaagcatttaatgaaaccataaaaatatttgaagaacaatgccaaacccaggag
cggtacagcaaagaatacat
agagaagtttaaacgcgaaggcaacgagaaagaaattcaaaggattatgcataaccatgataagctgaagtcgcgtatc
agtgagatcattgacagtag
gaggaggttggaagaagacttgaagaagcaggcagctgagtaccgagagatcgacaaacgcatgaacagtattaagccg
gacctcatccagttgaga
aagacaagagaccaatacttgatgtggctgacgcagaaaggtgtgcggcagaagaagctgaacgagtggctggggaatg
aaaataccgaagatcaat
actccctggtagaagatgatgaggatttgccccaccatgacgagaagacgtggaatgtcgggagcagcaaccgaaacaa
agcggagaacctattgcg
agggaagcgagacggcactttccttgtccgggagagcagtaagcagggctgctatgcctgctccgtagtggtagacggc
gaagtcaagcattgcgtcat
taacaagactgccaccggctatggctttgccgagccctacaacctgtacagctccctgaaggagctggtgctacattat
caacacacctccctcgtgcag
cacaatgactccctcaatgtcacactagcatacccagtatatgcacaacagaggcgatgaagcgctgccctcggatcca
gttcctcaccttcaagccacc
caaggcctctgagaagcaaagggctcctctccagcccgacctgtgaactgagctgcagaaatgaagccggctgtctgca
catgggactagagctttctt
ggacaaaaagaagtcggggaagacacgcagcctcggactgttggatgaccagacgtttctaaccttatcctctttcttt
ctttctttctttctttctttctttctttc
tttctttctttctttctttctttctttctaatttaaagccacaacacacaaccaacacacagagagaaagaaatgcaaa
aatctctccgtgcagggacaaagag
gcctttaaccatggtgcttgttaacgctttctgaagctttaccagctacaagttgggactttggagaccagaaggtaga
cagggccgaagagcctgcgcct
ggggccgcttggtccagcctggtgtagcctgggtgtcgctgggtgtggtgaacccagacacatcacactgtggattatt
tcctttttaaaagagcgaatgat
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
13/17
atgtatcagagagccgcgtctgctcacgcaggacactttgagagaacattgatgcagtctgttcggaggaaaaatgaaa
caccagaaaacgtttttgttta
aacttatcaagtcagcaaccaacaacccaccaacagaaaaaaaaaaaaaa
SEQ ID No. 34
MSAEGYQYRALYDYKKEREEDIDLHLGDILTVNKGSLVALGFSDGQEARPEDIGWLNGYNETT
GERGDFPGTYVEYIGRKRISPPTPKPRPPRPLPVAPGSSKTEADTEQQALPLPDLAEQFAPPDVAP
PLLIKLLEAIEKKGLECSTLYRTQSSSNPAELRQLLDCDAASVDLEMIDVHVLADAFKRYLADLP
NPVIPVAVYNEMMSLAQELQSPEDCIQLLKKLIRLPNIPHQCWLTLQYLLKHFFKLSQASSKNLL
NARVLSEIFSPVLFRFPAASSDNTEHLIKAIEILISTEWNERQPAPALPPKPPKPTTVANNSMNNNM
SLQDAEWYWGDISREEVNEKLRDTADGTFLVRDASKMHGDYTLTPRKGGNNKLIKIFHRDGKY
GFSDPLTFNSVVELINHYRNESLAQYNPKLDVKLLYPVSKYQQDQVVKEDNIEAVGKKLHEYNT
QFQEKSREYDRLYEEYTRTSQEIQMKRTAIEAFNETIKIFEEQCQTQERYSKEYIEKFKREGNEKE
IQRIMHNHDKLKSRISEIIDSRRRLEEDLKKQAAEYREIDKRMNSIKPDLIQLRKTRDQYLMWLTQ
KGVRQKKLNEWLGNENTEDQYSLVEDDEDLPHHDEKTWNVGSSNRNKAENLLRGKRDGTFLV
RESSKQGCYACS V V VDGEVKHC VINKTATGYGFAEPYNLYSSLKELVLHYQHTSLV QHNDSLN
VTLAYPVYAQQRR
SEQ ID No.35
ggaaggatgaggcgcccgcggcggcccgggggctccgggggctccgggggctccgggggcctccggctgctggtctgcc
tgctgttgctgagcgg
ccgccccgggggctgcagcgccatcagtgcccacggctgtctgtttgaccgcagactttgttcgcatctggaagtctgt
attcaggatggcttgtttggaca
gtgccaggcaggagtggggcaggcacggcccctcttacaagtcacttccccagttctccagcgcctacaaggtgtgctc
cggcaactcatgtcccaag
gcttgtcctggcatgatgaccttacccagcatgtgatctcccaggagatggaacgcatccccaggcttcgccccccaga
gccccatccaagggacaggt
ctggtttggtgcccaggaaaccaggccctgcaggggaattgctaactcagggcaatcctactggctcctctcctgctgc
ccagggctttccaaggcctgc
agggggacggagctggggcggctccccactgtcctctctgcaggctgagttgttaccccctctcttggagcatctgcta
atgcccccacagcctccacac
cctgctctgacctatgaacctgcactgctacagccttacctcttccaccagtttggctcccgagatggctcccggggct
cagagagctcctctggggtagtt
ggtgttggtcacctgtccaaggctgaaggtcctgcactcttcagcagaagtgcctccaaggccattttggggactcact
ctggacactcttttggggacctt
acaggtccctcacctgctcaacttttccaagattcagggctgctctacatggcccaagagttgccagtgcctggcagag
cccgggcaccaaggttgcca
gagaatgggggcaacagggcagaggactcttcagagggccatgaggaggaagtactagggggtcgtggggagaagtccc
ctccccaagcagcaca
accagaattgagtctgcagagattgactgctgtactggcaggctatggagtagagctgcgtcagttgaccccggagcag
ttttctaccctcttgaccctgat
gcagttgctgcccaagggcacaggaagaaatcttgaaggggctgtaaatgttggaggagccgatgtcaagaaaacaata
caacagatgcagagagga
gacccagcagaagctctgccccccacaccctcgcttcctgggtacctcactgccagccctgcctccagcgaagttcagc
aggtgctgagccctggtttc
cctgaacctccccacacacccagccctctgggctcctcctcagtccttctggagaagaaaagtcccttgggccagagcc
agcccacagtggtgggacg
gccatcagctcgaccatcggccgaggagtatggctatatcgtcactgaccagaaacccctgagcctggtggctggagtg
aggctgctggagattctggc
tgagcacgtgcatatgtcctccggtagctttatcaacatcagtgtggtgggaccagctgtcaccttccgaatccggcac
aatgagcagaacctgtctttggc
agatgtgacccagcaagctgggctggtgaagtctgaactggaagcgcagacagggctccagattttgcagacaggggtg
ggacagagggaggaagc
agctgaagtccttccccgacaagcccatggcatatctcccatgcgctcagtgctgcttactctagtggccctggcaggc
gtcgctgggctgctagtggctt
tggcagtggccttgtgtatgcgccatcattcgagacagcgggataaggagcgcctggcagcgctggggccggagggggc
ccatggtgacactactttt
gagtaccaggacctgtgtcgccagcacatggccacaaagtccctgtttaaccgggcggagggtcagccagagccttcta
gggtgagcagtgtgtcctc
ccagttcagcgacgcggcccaggccagccccagttcccacagcagctctccatcttggtgcgaggagcccgcccaggcc
aacatggacatctccaca
ggacacatgattctggcatacatggaggatcaccttcggaaccgggaccggttggccaaggagtggcaggctctgtgcg
cctaccaagcagagccaa
acacctgtgccgccgcacaggatgagagcaacatcaagaagaaccgccatcctgacttcctaccctatgaccatgcccg
aatcaagctgaaagtggag
agcagcccttctcggagtgattacatcaacgccagccccatcatcgagcatgaccctcggatgccggcctacatagcca
cacagggaccactgtccca
caccatcgcggacttctggcagatggtgtgggagagtggctgcactgtcatcgttatgctgaccccgttggtggaggac
ggtgtcaaacagtgtgaccgc
tactggccggatgaaggatcttccctctaccacgtctatgaggtgaacctggtgtcggagcacatctggtgcgaggact
tcctggtgcggagcttctacctt
aagaacctgcagacccaggagacgcgcacgctcactcagttccacttcctcagctggccggcagagggcactccggcct
ccacccggccgctgctgg
acttccgcaggaaagtgaacaagtgctacagaggccgctcctgccccatcatagtgcactgcagtgacggtgcagggag
gacaggcacctacatcctt
attgacatggtcctgaatcgcatggccaaaggagtgaaggagattgatattgctgccaccctggagcatgtccgtgacc
agcggcctggacttgtccgtt
ctaaggaccagtttgagtttgcgctgacagccgtggcagaggaggtgaatgctatcctcaaggccctgccccagtgagc
ccccctgggcgcctcagtg
ggcatcctggcctcggctccttctgcctgtgtgagcatctgtgcacccactcttcagcccctacccatctgccaccttg
gtctgacttggccatgggagcctt
tcccaacccagtgtggaagggagtcgggagggaaggaaggggtaggctcgccctgctttatccatgctagaaccatggt
atcccatgggaagcagaca
gcaggcaaggagaggcgtggacaccggccacaggtgtgcccgagccccatcctacctgagtctctgtctccctctctgg
atatgtgcgtccccactccc
accagcctaccacctatagacaaagcagaacgaggaaaccccagctcccccaaccctgctaccactggcctgccacctt
gaccctgctcaaccttctcc
ctctagcacaagggaacatttctagaaaagtaaaatctacttttgtatcagtgtgaataaagttagtgtgttgtctgtg
c
SEQ ID No. 36
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
14/17
MRRPRRPGGSGGSGGSGGLRLLVCLLLLSGRPGGCSAISAHGCLFDRRLCSHLEVCIQDGLFGQC
QAGVGQARPLLQVTSPVLQRLQGVLRQLMSQGLSWHDDLTQHVISQEMERIPRLRPPEPHPRDR
SGLVPRKPGPAGELLTQGNPTGSSPAAQGFPRPAGGRSWGGSPLSSLQAELLPPLLEHLLMPPQP
PHPALTYEPALLQPYLFHQFGSRDGSRGSESSSGVVGVGHLSKAEGPALFSRSASKAILGTHSGHS
FGDLTGPSPAQLFQDSGLLYMAQELPVPGRARAPRLPENGGNRAEDSSEGHEEEVLGGRGEKSP
PQAAQPELSLQRLTAVLAGYGVELRQLTPEQFSTLLTLMQLLPKGTGRNLEGAVNVGGADVKK
TIQQMQRGDPAEALPPTPSLPGYLTASPASSEVQQVLSPGFPEPPHTPSPLGSSSVLLEKKSPLGQS
QPTVVGRPSARPSAEEYGYIVTDQKPLSLVAGVRLLEILAEHVHMSSGSFINISVVGPAVTFRIRH
NEQNLSLADVTQQAGLVKSELEAQTGLQILQTGVGQREEAAEVLPRQAHGISPMRSVLLTLVAL
AGVAGLLVALAVALCMRHHSRQRDKERLAALGPEGAHGDTTFEYQDLCRQHMATKSLFNRAE
GQPEPSRVSSVSSQFSDAAQA5PSSHSSSPSWCEEPAQANMDISTGHMILAYMEDHLRNRDRLAK
EWQALCAYQAEPNTCAAAQDESNIKKNRHPDFLPYDHARIKLKVESSPSRSDYINASPIIEHDPR
MPAYIATQGPLSHTIADFWQMV WESGCT VIVMLTPLVEDGVKQCDRYWPDEGSSLYHVYEVN
LVSEHIWCEDFLVRSFYLNLQTQETRTLTQFHFLSWPAEGTPASTRPLLDFRRKVNKCYRGRSCP
IIVHCSDGAGRTGTYILIDMVLNRMAKGVKEIDIAATLEHVRDQRPGLVRSKDQFEFALTAVAEE
VNAILKALPQ
SEQ ID No.37
cggcgggaggaaagcgcggtgaagccagattaggagcagcgagcacttggggacttagggccacaggacaccgcacaag
atcgaccgactttttct
ggagaaccgcagaacgggcacgctggggtcgctggggctggccatggtgatggaggtgggcatcctggacgccgggggg
ctgcgcgcgctgctgc
gagagggcgccgcgcagtgcctgttgttggattgtcgctccttcttcgctttcaacgccggccacatcgcgggctcagt
gaacgtgcgcttcagcaccat
cgtgcggcgccgcgccaagggcgccatgggcctggagcatatcgtgcccaacgctgaactgcgtggccgcctgctggcc
ggagcctaccacgccgt
.ggtgctgctggacgagcgcagcgcctccctggacggcgccaagcgcgacggcaccctggccctggccgcgggcgcgct
ctgccgagaggcgcgc
tccactcaagtcttctttctccaaggaggatatgaagcgttttcggcttcctgccctgagctgtgcagcaaacagtcca
cccccacggggctcagcctccc
cctgagtactagtgtgcctgacagtgcagaatccggatgcagctcctgtagtacccctctctacgatcaggggggccca
gtggagatcctgtccttcctgf:
acctgggcagtgcctatcacgcttctcggaaggatatgcttgacgccttgggcatcaccgccttgatcaacgtctcagc
caattgtcctaaccactttgagg
gtcactaccagtacaagagcatccctgtggaggacaaccacaaggcagacatcagctcctggttcaacgaggctattga
cttcatagactccatcaagg
atgctggagggagagtgtttgttcattgccaggccggcatctcccggtcagccaccatctgccttgcttacctcatgag
gactaaccgggtaaagctggac
gaggcctttgagtttgtgaagcagaggcggagtatcatctccccgaacttcagcttcatgggccagctgctgcagtttg
agtcccaagtgctagcccctca
ctgctctgctgaagctgggagccctgccatggctgtccttgaccggggcacctctactaccacagtcttcaacttccct
gtttccatccccgtccaccccac
gaacagtgccctgaactaccttaaaagccccatcaccacctctccaagctgctgaagggcaaggggaggtgtggagttt
cacttgccaacgggtcgcca
ctcctcctgtgggaggagcaatgcaataactctgggagaggctcatgggagctggtccttatttatttaacacccccct
caccccccaactcctcctgagtt
ccactgagttcctaagcagtcacaacaatgacttgaccgcaagacatttgctgaactcggcacattcgggaccaatata
ttgtgggtacatcaagtccctct
gacaaaacagggcagaagagaaaggactctgtttgaggcagtttcttcgcttgcctgttttttttttctagaaacttca
tgcttgacacacccaccagtattaa
ccattcccgatgacatgcgcgtatgagagtttttacctttatttatttttgtgtaggtcggtggtttctgccttcacaa
atgtcattgtctactcatagaagaaccaa
atacctcaattttgtgtttgcgtactgtactatcttgtaaatagacccagagcaggtttgctttcggcactgacagaca
aagccagtgtaggtttgtagctttca
gttatcgacagttgtatgtttgtttatttatgatctgaagtaatatatttcttcttctgtgaagacattttgttactgg
gatgactttttttatacaacagaataaattatg
acgtttctattga
SEQ ID No.38
MVMEVGILDAGGLRALLREGAAQCLLLDCRSFFAFNAGHIAGSVNVRFSTIVRRRAKGAMGLEH
IVPNAELRGRLLAGAYHAVVLLDERSASLDGAKRDGTLALAAGALCREARSTQVFFLQGGYEAF
SASCPELCSKQSTPTGLSLPLSTSVPDSAESGCSSCSTPLYDQGGPVEIL,SFLYLGSAYHASRKDM
LDALGITALINVSANCPNHFEGHYQYKSIPVEDNHKADISSWFNEAIDFIDSIKDAGGRVFVHCQA
GISRSATICLAYLMRTNRVKLDEAFEFVKQRRSIISPNFSFMGQLLQFESQVLAPHCSAEAGSPAM
AVLDRGTSTTTVFNFPVSIPVHPTNSALNYLKSPITTSPSC
SEQ ID No.39
atacccatcaaatgaagcaccaaagacaactgtgttgagcgcccctgggcctggctcattcttaatcatattgccatct
ttactgctctgattcctaggatccc
cgcaatgactctgaataatgtcaccatgcgccaaggcactgtgggcatgcagccacagcagcgctggagtatgcctgct
gatgccaggcatctgatggt
5 5
ccagaaggatccccacccctgcaacctccgaaaccgccactctactgctccggaagagcactgccggcggacgtggtcg
tccgactccacagactcg
gtcatctcttctgagtctggaaacacctactaccgagtagtgcttataggggagcaaggagtgggcaagtccaccctgg
ccaacatctttgcaggtgtgca
tgacagcatggacagcgactgtgaggtcttgggagaagatacatatgagcgtaccctggtcgttgacggagagagtgca
accattatcctcctggacatg
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
15/17
tgggaaaataagggggagaacgaatggctccatgaccactgcatgcaggtcggggatgcctatctgatcgtctactcta
tcacagaccgtgcaagcttc
gagaaggcatctgagctgaggattcagctccgcagggcccggcagacagaagacattcctataattttggttggcaaca
aaagcgacttagtgcggtgt
cgagaagtgtctgtgtcagaagggagagcttgtgctgtggtgttcgactgcaaattcatcgagacctctgcggctgtgc
agcacaacgtgaaggaactgtt
tgagggcattgagcgacaggtgcgcctgccgagggacagcaaggagaagaatgagaggaggctggcctaccagaagagg
cgggagagcattccc
aggaaagccaggcgcttctggggcaaaattgtggccaaaaacaacaagaacatggcttcaagctccaagtcaaaatcct
gccatgacctgtctgtgctct
aggcacccagtgtcacccagatgtcccttggtggacatcgttgaaggctattgggaccagtgatctatattagattgga
tacataagcattgttagacgcaa
cttcccctatggcagatggaaaccaacaggttagccttgtgggcaaccaagtgcacgggcaatgaatgagctctgtcaa
gagtcagtatttattcatagga
aaagcttgagctgctacgtggatgtctcagactcatttaagacacgcttcgggttcacatgagtttctctctcctttgg
acagcagaatatttttttcctcagtgtt
gttccatgtgatttcgaggtccttgggtcatatagaaatgtagggaaatggcggtagtttattggaaggagaagggctc
actgcatacttatatccctgaaac
gacatctttagagctggcctcatcacagttgtgactatttctgctccagtgaagagaattgttagatttgctggaaact
gaggcttactaacagtttttgtttaaa
gaccacagagattgtagacttaggagctatatggtactacttataggttcaaaaaattgtttacttatgtgtcgtagaa
gtgtttattttgaggaaactattttttttt
ttgccaaattctacttagtcaaatcatcttctatgtcttgctgttttttaaatcattaagctatcataaaatatttttt
aaaaaaatctcaactatattgataacctgcag
tgcaaaattttaaatatagtcctgtttttcccccaaaacataaacatgccccatcctttgggttgcttctgtatgccac
agctgaattatatttattattttgcaataa
ccattttatatttgataaagatatttatgagcatatttcttactgagaaaatgtctgttttattacctttttatatttt
tcaaagtattcaagtttttacctattgtcttataat
aaataaataaaatctttgaaaagg
SEQ ID No.40
MTLNNVTMRQGTVGMQPQQRWSMPADARHLMVQKDPHPCNLRNRHSTAPEEHCRRTWSSDS
TDSVISSESGNTYYRVVLIGEQGVGKSTLANIFAGVHDSMDSDCEVLGEDTYERTLVVDGESATI
ILLDMWENKGENEWLHDHCMQV GDAYLIVYSITDRASFEKASELRIQLRRARQTEDIPIILVGNK
SDLVRCREVSVSEGRACAVVFDCKFIETSAAVQHNVKELFEGIERQVRLPRDSKEKNERRLAYQ
KRRESIPRKARRFWGKIVAKNNKNMASSSKSKSCHDLSVL
SEQ ID No. 41
.tatcccgctgttgctgcaagccggctgcatcttagttggccatgaagaccccagcacagcggctgcaccttcttccac
tgttgttgctgctttgtggtgagtg
.
tgcccaggtatgcggctgcaacgagacagggatgctggagaggctgcctcgctgtgggaaagccttcgctgacatgatg
cagaaggtggctgtctgga
agtggtgcaacctgtcggagttcatcgtgtattatgaaagcttcactaactgcaccgagatggagaccaacatcatggg
ctgctactggcccaacccgctg
3 0
gcccagagcttcatcactggaatccacaggcagttcttttccaactgcacggtggacaggacccactgggaagaccccc
cggatgaagtactcatccca
ctgatcgcggttcctgtcgtgctgactgtggctatggctggcctggtggtgtggcgcagcaagcacactgatcggctgc
tgtgaggatctgctggatgga
gggccatgcctggcaggctgggagaatgttgctcagagctctgagagctggcagactcggcttctgtctggtttgcttt
ggccacaccctacctggccatg
ccaaagtcctcccgaccaggctggtgtggcccttgctgtctagcctgccgcctgctggggttcagattgtccatacttt
gctctttcttgggctagtggaaga
aagtgacaaatcccaagtttgtggaccaggcatggaaatcaactgttgctgagccccgctccccaggctcggttcccta
gtttctagccgtttcttggcaga
3 5
gtcttgctcagcctgaaccccgccccaggtcctgacccatttctagtcctgaccctgacccctgctacacttggccaga
gagggcaggcaaggtcatctg
gaagatgtggacgcccccccgcctctattcaagagactgagcacatcatttatcagacatgaaggatagcctggggtca
ttaggagccacgtgtgaccta
ctgacccacctgcctgtcctctctgtgatctgtcacgattctgtgtccagtgtgggctggagctgtggcttgtttagcc
cttcaaagacacctaccctgcaggt
agagcgtgaacctccttcttgaggggtattcctgggagtggggcgcactgagtgtgctcaagggttctgtctgctgatg
tcagttctttttgattaaagtgtct
ccttacaaaaaaaaaaaaaaaaaaaaa
SEQ ID No.42
MKTPAQRLHL,LPLLLLLCGECAQVCGCNETGMLERLPRCGKAFADMMQKVAVWKWCNLSEFI
VYYESFTNCTEMETNIMGCYWPNPLAQSFITGIHRQFFSNCTVDRTHWEDPPDEVLIPLIAVPVV
LTVAMAGLVVWRSKHTDRLL
SEQ ID No. 43
atgctcaacaaagccaagaattcaaagagtgcccagggtctggctggtcttcgaaaccttgggaacacgtgcttcatga
actcaattcttcagtgcctgag
caacacccgagagctgagagattactgcctccagaggctgtacatgcgggacctcggccacaccagcagcgctcacacg
gccctcatggaagagttt
gcaaaactaatccagaccatatggacgtcgtcccccaatgatgtggtgagcccatctgagttcaagacccagatccaga
gatatgcgccacgcttcatgg
gctataatcagcaggatgctcaggaattccttcgtttccttctggatggtctccacaatgaggtgaaccgggtggcagc
aaggcctaaggccagccctga
gacccttgatcatctccctgatgaagaaaaggggcgacagatgtggaggaagtatctggaaagggaagacagtcggatt
ggggatctcttcgttgggca
gctgaagagctccctcacatgcaccgattgtggctactgctctacagtcttcgatcccttctgggatctctcgttgccc
atcgcaaagagaggttaccctga
ggtgacgttaatggattgtatgaggctcttcaccaaagaggacatattggatggtgatgagaagccaacttgctgccgc
tgccgagccagaaaacgatgc
ataaaaaagttctctgtccagaggttcccaaagatcttggtgctccacctgaagcgattctcagaatccaggatacgaa
ccagcaagctcacaacatttgtg
aatttcccactaagagacctggacttgagagaatttgcttcagaaaacaccaaccatgctgtttacaacctgtatgctg
tgtccaatcactccggaaccacc
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
16/17
atgggaggccactatacagcctactgccgaagtccggttacaggcgaatggcacactttcaatgattccagtgtcacac
ccatgtcctccagccaagtgc
gcaccagcgacgcctatttgctcttctatgaactggccagtccaccctcccgtatgtagcattgaggagctgcggccct
tccctcttccctgtggtggcccc
acgtcctaag
SEQ ID No. 44
MLNKAKNSKSAQGLAGLRNLGNTCFMNSILQCLSNTRELRDYCLQRLYMRDLGHTSSAHTALM
EEFAKLIQTIWTSSPNDV VSPSEFKTQIQRYAPRFMGYNQQDAQEFLRFLLDGLHNEVNRVAARP
KASPETLDHLPDEEKGRQMWRKYLEREDSRIGDLFVGQLKSSLTCTDCGYCSTVFDPFWDLSLPI
AKRGYPEVTLMDCMRLFTKEDILDGDEKPTCCRCRARKRCIKKFSVQRFPKILVLHLKRFSESRI
RTSKLTTFVNFPLRDLDLREFASENTNHAVYNLYAVSNHSGTTMGGHYTAYCRSPVTGEWHTF
NDSSVTPMSSSQVRTSDAYLLFYELASPPSRM
SEQ ID No.45
gcggagcgtgagctgtgcgagcgagcgagcgcgagcatagcctgcgagcgagcagagagaaagagcgagggcaagagag
cggcgaggcgcct
gcgcgatgctcgggcccctaagcccgcggcgctgagccagccgggacggacatgcgcgggagggcgccgcggggtcccg
ctcccttgggggaat
gaaagctactggttgacttaaaaacacctgggctttacaaatttgaaggcatcccagagtggggcacaatgtcaacagc
aggagttgctgctcaggatatt
cgagtcccattaaaaactggatttctccataatggtcaggccttggggaatatgaagtcctgctggggcagtcacagtg
agtttgaaaataactttttaaatat
tgatccaataaccatggcctacaatctgaactcccctgctcaggagcacctaacaactgttggatgtgctgctcggtct
gctccagggagcggccacttctt
tgcagagtgtggtccatctccaaggtcaagcttgccccctcttgttatctcaccaagtgaaagctcgggacagcgtgaa
gaggatcaagttatgtgtggtttt
aagaaactctcagtgaatggggtctgcacttccacacctccacttacacccattaaaagctgcccttcccctttcccct
gtgcggctctgtgtgatcggggtt
ctcggccgctcccgccactgcccatctctgaagacctatgtgtggatgaggccgacagtgaggtagagcttctaaccac
cagctcagacacagacttgct
tttagaagactctgcgccttcagatttcaaatacgatgctcctggcaggcgcagcttccgtgggtgcggccagatcaac
tatgcatattttgacagcccaac
tgtttctgtggcagatcttagctgtgcatctgaccagaacagagttgttccagacccaaaccctcccccacctcaaagc
catcgcagattaaggaggtctc
actcaggaccagctgggtcatttaacaagccagccattcggatatctagctgcacacacagagcttctcctagctctga
tgaagacaagcctgaggtccct
..
cccagggttcctatacctcctaggccagcaaagccagactatagacggtggtcagcagaagtgacctccaacacctaca
gtgatgaagataggcctccc
aaagtecccccgagagaacctttgtctcggagtaactcccgtaccccaagtcctaaaagccttccgtcttacctcaatg
gggtcatgcccccaacacaga
gcttcgctcctgaccccaagtatgtcagcagcaaagccctgcagagacagagcagcgaaggatctgccaacaaggttcc
ttgcatcctgcccattattga
aaatgggaagaaggttagctcaacgcattattacttactacctgagaggccaccgtacctggacaaatatgaaaagtat
tttaaggaagcagaagaaaca
aacccaagcacccaaattcagccattacctgctgcctgtggtatggcctctgccacagaaaagctggcctccagaatga
aaatagatatgggtagccac
gggaagcgcaaacacttatcctacgtggtttctccataaatatgggggtcatgattcaacagaagttacatgggatgaa
tggctcccagttttccagtttgag
gttcgtagaacaatgtcaagtggcaaaatgaagttggtggactccgccttaatgagaaaggcttagagcagttatgagg
tgctgttatgctgggagtccct
gatctatcagcataggagaaaaaagtatgatttaaagatgtgctagggggagggaaaaatgggcaacttttacatttga
ctacattatatacctatgtataaa
3 5
agtgcggtgtaaccatagaccatagctgcaggataaccaattagtcactcttagagtaatctgtattcagaacaattca
aacaagctggaggaacagctcc
tgatagtgtgagaattgagcaaatgggagaaagcaatattgttagatcagattataaatttgttaagtttaaagattcc
tggcatacaggcctgctctataaatt
tgttttccccttccctgccagcagtcttctccatacacgacagggcgtgttctccaccaggcctgtaacatcttgttga
gatcatttctatggcccaatacttgt
cgctctggggttttgtcttgttgaggagaggacagcagtttctggaccatgttatcacctgtgtgtgtctcatatcttg
gaaattgacagatttggtgaataactt
ttccatactattcctgcttttcccatccactgaaacagcctgttgtagcaagaggctttcaagagtgcagtggagttgc
gctggccatcagtgtttggggtctg
agtttgatagactagtgcagcgatcagccatatgattgagagctactttggggatatatggtacgttgtttttgttttt
tagacttaataaaggacaacacgagc
tggtcttgtgttgctggttcctattcagtatttcctggggattgtttgctttttaagtgaaacacttctgaccaatagc
acagaacgtcttaatgccagaggtcact
tcagcatcttcctgctttgaaaactcacgctggctgcttcactgccctgagattcagtgagacacgcagtttgtgttca
gtttttacatcctctgattgtttatcttg
tgcagataaacacaaagagaaggtgcttgctagcagggacactgctgccatgtcccaacaagctgttcagtttaaactg
ctgaatgacattatttgagctat
ttaaagcttactttagtatgaactaaatgaaggttaaaacatgctttagaaaaatgcactgatctccgcactgtgtgta
cagtattggacaaaggatttattcatt
ttgttgcattattttgaatattgtcttttcattttaataaagttatattacttatttatgaaaaaaaaaaaaaaaaa
SEQ ID No. 46
MSTAGVAAQDIRVPLKTGFLHNGQALGNMKSCWGSHSEFENNFLNIDPITMAYNLNSPAQEHLT
TVGCAARSAPGSGHFFAECGPSPRSSLPPLVISPSESSGQREEDQVMCGFKKLSVNGVCTSTPPLT
PIKSCPSPFl'CAALCDRGSRPLPPLPISEDLCVDEADSEVELLTTSSDTDLLLEDSAPSDFKYDAPG
RRSFRGCGQINYAYFDSPTVSVADLSCASDQNRVVPDPNPPPPQSHRRLRRSHSGPAGSFNKPAI
RISSCTHRASPSSDEDKPEVPPRVPIPPRPAKPDYRRWSAEVTSNTYSDEDRPPKVPPREPLSRSNS
RTPSPKSLPSYLNGVMPPTQSFAPDPKYVSSKALQRQSSEGSANKVPCILPIIENGKKVSSTHYYL
LPERPPYLDKYEKYFKEAEETNPSTQIQPLPAACGMASATEKLASRMKIDMGSHGKRKHLSYVV
SP
CA 02499502 2005-03-17
WO 2004/039835 PCT/EP2003/011793
17!17
SEQ ID No.47
atggctgaacaacttcttcctcaggctttgtatttgagcaatatgcggaaagctgtgaagatacgagagagaaccccag
aagacattttcaaacctaccaat
gggatcatctatcactttaaaaccatgcaccgatacacgctggagatgttcagaacatgccagttttgcccacagttcc
gagagatcatccacaaagcactt
attgacagaagtgtccaggcttccctggaaagccagaagaagctcaactggtgtcgtgaagtcaggaagctcgtggctc
tgaaaaccaatggtgatgga
aactgcctcatgcatgcagcttgtcagtacatgtggggtgttcaggatactgacctggtcctgaggaaggccctctgca
gcacccttaaggagacagaca
ctcggaactttaaattccgctggcagctggaatctctgaaatctcaggaatttgtggaaacaggactttgctacgacac
tcggaactggaatgacgaatgg
gacaacttggtcaaaatggcatcagcagacacacctgcagcccgaagtggacttcagtacaattccctggaagaaatcc
acatatttgtcctcagcaacat
cctcagaagacccatcattgtcatttcagacaaaatgctaagaagtttggaatctggttccaattttgctcctttgaaa
gtgggtgggatttatctgcctcttcac
tggcctgcccaggagtgttacagatatcccatcgtcctaggctatgacagccagcactttgtacccctggtgaccctga
aggacagtggacctgaacttcg
cgctgttccacttgttaacagagaccggggtaggtttgaagacttaaaagttcacttcttgacagatcctgagaatgag
atgaaggaaaagcttctaaagga
gtacttgatagtgatggagatccctgtgcaaggctgggaccacggcacgactcacctgatcaacgctgcaaaattggat
gaagctaacttacccaaagaa
ataaatttggtagacgattactttgagcttgttcagcacgaatacaagaaatggcaggagaacagcgatcaggccagga
gagcggcacatgcgcagaa
ccccttggagccttccacaccccagctatcactcatggatataaaatgtgagacacccaactgtcctttcttcatgtcc
gtgaacactcagcctttatgccac
~ 5
gaatgctcagagaggcgccaaaagaatcagagcaagctcccaaagctgaactcgaagctaggccctgaaggactcccag
gcgtgggacttggctcct
caaactggagccccgaggaaaccgctggaggacctcattcagccccacccacagcacccagcctttttctcttcagtga
gaccactgcaatgaagtgca
ggagtcctgggtgcccttttactttgaatgtgcagcataatggattctgtgagcgttgccacgcccggcagattaatgc
cagccacaccgcagaccctgga
aagtgccaagcctgccttcaggatgtcactcggacctttaatggcatctgcagtacctgtttcaaaaggactacagcag
agcccagctccagcctcacttc
cagtatccctgcctcctgtcaccaacgctccaagtctgacccctcacaactcatccaaagtctcactccacactcttgc
caccggactggaaatgtctctcc
20
ttctggctgcctctcccaggctgcacggactccaggagacagagcagggacaagcaagtgcaggaaagctggctgcatg
tattttgggactccagaaa
acaagggcttttgcactctatgtttcatcgaatacagagaaaataagcagtctgttactgcctctgcgaaagctggttc
cccggcccccaggttccagaaca
atgtcccgtgcctgggcagggagtgcggcacactcggaagcaccatgtttgaagggtactgtcagaagtgtttcatcga
agctcagaaccagagattcc
atgaagcaagaagaacggaagaacagctgagatcaagccagcatagagacatgcctcgaactacacaggtagcctcaag
gctgaaatgtgcccggg
cctcctgcaagaacattctggcctgtcgcagtgaggaactctgtatggagtgccagcacctaagccaacgagtaggttc
tgtggcccaccggggtgagc
25
ccacgcctgaagagccccctaaacagcgctgccgggcccctgcttgtgatcactttggcaatgccaagtgtaatggtta
ctgcaatgagtgctaccagttc
aagcagatgtatggctaa
SEQ ID No.A.8
30 MAEQLLPQALYLSNMRKAVKIRERTPEDIFKPTNGIIYHFKTMHRYTLEMFRTCQFCPQFREIIHK
ALIDRSVQASLESQKKLNW CREVRKLV ALKTNGDGNCLMHAACQYMW GV QDTDLVLRKALC
STLKETDTRNFKFRWQLESLKSQEFVETGLCYDTRNWNDEWDNLVKMASADTPAARSGLQYN
SLEEIHIFVLSNIL,RRPIIVISDKMLRSLESGSNFAPLKVGGIYLPLHWPAQECYRYPIVLGYDSQHF
VPLVTLKDSGPELRAVPLVNRDRGRFEDLKVHFLTDPENEMKEKLLKEYLIVMEIPVQGWDHGT
35 THLINAAKLDEANLPKEINLVDDYFELVQHEYKKWQENSDQARRAAHAQNPLEPSTPQLSLMDI
KCETPNCPFFMSVNTQPLCHECSERRQKNQSKLPKLNSKLGPEGLPGVGLGSSNWSPEETAGGP
HSAPPTAPSLFLFSBTTAMKCRSPGCPFTLNVQHNGFCERCHARQINASHTADPGKCQACLQDVT
RTFNGICSTCFKRTTAEPSSSLTSSTPASCHQRSKSDPSQLIQSLTPHSCHRTGNVSPSGCLSQAART
PGDRAGTSKCRKAGCMYFGTPENKGFCTLCFIEYRENKQSVTASAKAGSPAPRFQNNVPCLGRE
40 CGTLGSTMFEGYCQKCFIEAQNQRFHEARRTEEQLRSSQHRDMPRTTQVASRLKCARASCKNIL
ACRSEELCMECQHLSQRVGSVAHRGEPTPEEPPKQRCRAPACDHFGNAKCNGYCNECYQFKQM
YG
SEQ ID No. 49
ttttttttcatacttgataaaattttatttaaaaaaaaagagaataataatttatacccttgacaaaataaaagatctt
ataatataaatgtttcttagaaaatatatga
aaagataatattacaaatattaataaatcaatattcacatgacagcaaaagtggcaatgattctacaagaaggtgagga
ggaagatgctttccggtccgca
gcaatgtctctggagaggcctcctgtcccttctttctccttcaatgaggtgtgctcetattttaagaaaacctgataca
agcagatctaatcagtttaggaagct
ggtatttatttgcaccgcaaaataatttttttacaaaaaaaattctatcaaggatcctttaaatatcaagtttcccaat
gcacttagaatacagttaaccaaatttac
aagtcttcgacttctctctggtgtagctctaccgcanggcgtgaggtattgctgaagtgagtgcgtgcgtccgtg