Note: Descriptions are shown in the official language in which they were submitted.
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
BLOOD TEST PROTOTYPES AND METHODS FOR THE DETECTION OF
CIRCULATING TUMOR AND ENDOTHELIAL CELLS
RELATED APPLICATIONS
[0001] This application claims benefit of U.S. Provisional Patent
Application Serial No. 60/516,571, filed on October 31, 2003, from which
priority is
sought and the disclosure of which is herein incorporated by reference.
BACKGROUND OF THE INVENTION
1. Field of the Invention
[0002] The present invention generally relates to an improved cell
adhesion matrix ("CAM") and an improved cell isolation device for separating
target cells such as tumor, fetal and angiogenic cells from blood or other
tissue
fluid samples such as ascites, scrape and smear specimens. More particularly,
the present invention relates to a CAM system that may be used to selectively
isolate cell, for example, target cancer cells with metastatic potential
and/or
endothelial progenitor cells that display invadopodia.
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
2. Description of the Related Art
Circulating Tumor Cells (CTC) And Cancer Detection
[0003] Malignant tumors of epithelial tissues are the most common form
of cancer and are responsible for the majority of cancer-related deaths.
Because
of progress in the surgical treatment of these tumors, mortality is linked
increasingly to early metastasis and recurrence, which is often occult at the
time of
primary diagnosis (Racila et al., 1998; Pantel et al., 1999). For example, the
remote anatomical location of the pancreas and other gastro-intestinal (GI)
organs
makes it unlikely that pancreatic and other GI cancers will be detected before
they
have invaded neighboring structures and grown to tumors larger than 1-cm
(Compton, 2003; Flatmark et al., 2002; Koch et al., 2001; Liefers et al.,
1998;
Matsunami et al., 2003; Nomoto et al., 1998; Pantel et al., 1999; Walsh and
Terdiman, 2003; Weihrauch, 2002). Even with respect to breast cancers, 12-37%
of small tumors of breast cancer (<1 cm) detected by mammography already have
metastasized at diagnosis (Chadha M et al., 1994; Wilhelm MC et al., 1991 ).
[0004] Evidence has accumulated in the literature showing that epithelial
tumor cells found in the circulation represent the earliest sign of metastasis
formation and that circulating tumor cells ("CTC") can be considered an
independent diagnostic for cancer progression of carcinomas (Beitsch and
Clifford,
2000; Brandt et al., 2001; Feezor et al., 2002; Fehm et al., 2002; Ghossein et
al.,
1999; Glaves, 1983; Karczewski et al., 1994; Koch et al., 2001; Liefers et
al.,
1998; Luzzi et al., 1998; Matsunami et al., 2003; Molnar et al., 2001; Wang et
al.,
2
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
2000; Weitz et al., 1999; Wharton et al., 1999; Racila et al., 1998; Pantel et
al.,
1999). Given the same, reliable procedures to isolate cancer cells from the
bloodstream would have significant impact in both clinical diagnostic and
therapeutic applications of cancer (Racila et al., 1998; Pantel et al., 1999).
A new
tumor staging, called Stage Mi, has been proposed to indicate the presence of
tumor cells in the circulation of patients with cancers. The staging warrants
the
development of a blood test that could detect circulating tumor cells (CTC).
The
cancer research field awaits novel tumor cell enrichment methods that can
increase detection sensitivity, advantageously by at least one order of
magnitude
(Pantel et al., 1999), over existing methods.
Circulating Endothelial Progenitor Cells Anaioaenesis And Cardio-Vascular Risk
[0005] Endothelial-cell injury is an important stimulus for the
development of atherosclerotic plaque (Ross, 1993). Circulating endothelial
progenitor cells ("CEC") that can be isolated from the mononuclear cell
fraction of
the peripheral blood, bone marrow, and cord blood, have been identified
(Asahara
et al., 1997; Hill et al., 2003) as indicative of endothelial-cell injury.
Laboratory
evidence suggests that these cells express a number of endothelial-specific
cell-
surface markers and exhibit numerous endothelial properties. It has been noted
that when these cells are injected into animal models with ischemia, they are
rapidly incorporated into sites of neovascularization.
[0006] In a pilot study, Hill et al., 2003 found that a low CEC level was
associated with cardiovascular risk factors and with brachial reactivity. It
has been
3
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
suggested that endothelial injury in the absence of sufficient CEC might
affect the
progression of cardiovascular disease. This early-phase study pointed to the
potential of CEC in diagnosis and treatment of cardiovascular diseases. CEC
might contribute to endothelial repair by providing a circulating pool of
cells to
promote angiogenesis (Szmitko et al., 2003). Thus, CEC may be a negative
predictor of the risk of cardiovascular diseases. An efficient enrichment
method
for CEC would be very useful therefore in pre-diagnosis of and management of
cardiovascular disease.
Cell Heteroaeneity And Current Cell Separation Technologies
[0007] Tumor and endothelial progenitor cells circulating in the blood (a
heterogeneous source of cells) are rare. These cells can be hard to purify for
analysis. In cancer patients, the number of CTC or exfoliated abnormal cells
(neoplastic cells) in blood is generally very small compared to the number of
non-
neoplastic cells. Therefore, the detection of exfoliated abnormal cells by
routine
cytopathology is often limited. Further, exfoliated cells are frequently
highly
heterogeneous being composed of many different cell types (interestingly, many
of
the genes initially reported to be differentially expressed in exfoliated
cells have
actually turned out to be expressed by non-tumor cells instead). Compounding
this heterogeneicity problem , the frequency of neoplastic cells present in
each
clinical specimen is variable, which biases and complicates the quantification
of
differential gene expression in randomized mixed population. Apoptotic and
necrotic cells are common in larger tumors, peripheral blood and ascites.
These
4
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
cells do not contain high quality RNA and thus present technical problems for
molecular analyses (Karczewski et al., 1994).
[0008] A number of cell enrichment methods for circulating tumor and
endothelial progenitor cells have been described:
[0009] a) Microdissection can be used to isolate rare tumor
cells one by one (Suarez-Quian et al., 1999). This method typically has
several limitations: (1 ) the subsequent sample processing is complicated,
(2) cell viability cannot readily be established, and (3) selection of the
cells
to be dissected is based mainly on morphological criteria, which has a high
frequency of giving rise to false-positive results.
[00010] b) Physical characteristics of tumor cells, such as
shape, size, density or electrical charge, can also be used (Vona et al.,
2000). Several density gradient centrifugation methods have been
developed to enrich tumor cells in nucleated blood cells (devoid of mature
red blood cells). Density gradient centrifugation methods can achieve 500
to 1,000-fold cell enrichment. The enriched tumor cells can then be
subjected to molecular analysis using highly sensitive assays such as
immunocytochemistry and reverse transcriptase polymerase chain reaction
(RT-PCR) which may be used to amplify putative tumor markers or epithelial
markers such as prostate specific antigen (PSA) mRNA or cytokeratin 19
mRNA (Peck et al., 1998). However, these methods may not effectively
s
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
enrich viable tumor cells from normal cells. That is, 500 - 1,000 fold cell
enrichment is often found to be relatively modest enrichment which
generates substantial background noise adversely affecting further
molecular analysis. In addition, enrichment methods based on physical
separation techniques are often cumbersome, lengthy, and involve steps
(e.g. more than 2-3 rounds of centrifugation) that can result in cellular
damage.
[00011] c) Antibody-based techniques are a more recent
development. Immunoaffinity methods include affixing an antibody to a
physical carrier or fluorescent label. Sorting steps can then be used to
positively or negatively enrich for the desired cell type after the antibody
binds to its target present on the surface of the cells of interest. Such
methods include affinity chromatography, particle magnetic separation,
centrifugation, or filtration, and flow cytometry (including fluorescence
activated cell sorting; FACS).
(1 ) Flow cytometry or a fluorescence activated cell
sorter ("FACS") detects and separates individual cells
one-by-one from background cells. In model
experiments, this method can detect breast carcinoma
cells (Gross et al., 1995) and endothelial progenitor
cells (Hill et al., 2003) in the mononuclear cell fraction
that had been enriched from the peripheral blood by
6
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
density gradient centrifugation. Furthermore, FACS
can detect naturally occurring breast and prostate
tumor cells in blood after an enrichment step using
antibody-coated magnetic microbeads (Racila et al.,
1998; Beitsch and Clifford, 2000). However, cells that
exist in clusters or clumps are discarded during the
FACS process, and in some instances, for example,
ovarian cancer, most of the cells are present as
aggregates, making FACS CTC or CEC detection
highly ineffective.
(2) Approaches based on antibody-coated microbeads
can use magnetic fields (Racila et at, 1998), column
chromatography, centrifugation, filtration or FACS to
achieve separation. Despite its great power for
enrichment, there are also inherent limitations
associated with all of the antibody-based cell
separation methods. The most serious one is that
cancer cells usually express putative tumor-specific
antigens to variable degrees (Sabile et al., 1999);
hence it is easy to lose a large and potentially non-
random subset of tumor cells during the collection.
Antibodies also tend to bind with significant non-
7
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
specific affinity to damaged cells, leading to their co-
purification with the cells of interest. Overall, such
antibody-based cell separation methods have a higher
than desired false-negative rate. Current antibody-
initiated magnetic separation methods have detected
CTC at much lower levels, i.e., 1 - 100 CTC per mL of
blood from patients with breast and prostate cancer
(Racila et al., 1998), or less than 50 CEC per mL of
blood of individuals at risk of cardiovascular diseases
(Hill et al., 2003; Beitsch and Clifford, 2000). There are
approximately 5 x 109 red cells and 5 x 106 white
nucleate cells present in one milliliter (mL) or gram of
blood. Therefore, it is still a challenging task to detect
the presence of thousands of cancer or endothelial
cells in one mL of blood (Gulati and Acaba, 1993).
[00012] Over the past 20 years, specialized complexes found on the
surface of invasive tumor cells that facilitate their movement from the
primary
tumor to sites of metastasis have been characterized (Aoyama and Chen, 1990;
Chen and Chen, 1987; Chen et al., 1994a; Chen et al., 1984; Chen et al.,
1994b;
Chen, 1996; Chen, 1989; Chen and Wang, 1999; Ghersi et al., 2002; Goldstein
and Chen, 2000; Goldstein et al., 1997; Kelly et al., 1994; Monsky et at,
1994;
Monsky et al., 1993; Mueller et al., 1999; Mueller and Chen, 1991; Mueller et
al.,
s
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
1992; Nakahara et al., 1996; Nakaliara et al., 1998; Nakahara et al., 1997;
Pavlaki
et al., 2002; Pineiro-Sanchez et al., 1997; Saga et at, 1988; Zucker et at,
2000;
Zukowska-Grojec et al., 1998). These complexes, which we have denoted as
"invadopodia", bind to and degrade multiple types of endothelial cell matrix
(ECM)
components. Invadopodia are not found on differentiated normal blood cells or
on
primary tumor cells, and they do not function effectively on dead or dying
cells.
Invadopodia are present in circulating endothelial progenitor cells but not in
more
than 99.999% of blood cells, and in fetal cells found in maternal blood of
pregnant
females. The present inventors have recognized an enrichment step based on
invadopodia function would powerfully serve to separate viable metastatic
tumor
cells and endothelial cells from the majority of cell types found in ascites,
blood,
and many other body fluids and would address the limitations of the other
technologies described above.
SUMMARY OF THE INVENTION
[00013] In one embodiment, there is provided CAM for isolating specific
viable target cells in a blood sample or other tissue fluid sample for use in
the
screening, diagnostic evaluation, prognosis and management of disease.
[00014] A CAM of the present invention utilizes a cell-adhesion material
about a core material to effectively promote the adhesion of target cells
including,
CTC and CEC. Useful cell-adhesion materials include blood-borne adhesion
compounds and include, without limitation, fibronectin, fibrin, heparin,
laminin,
tenascin or vitronectin, and synthetic compounds, such as synthetic
fibronectin
9
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
and laminin peptides, extra cellular matrix compounds, or fragments thereof,
combinations thereof, and the like. Useful cell-adhesion materials in a CAM
should have the ability to effectively coat the core material of the matrix
alone, or
in combination with other materials. The core preferably comprises a
chemically
non-reactive material such as, but not limited to, gelatin particles, bone
fragments,
collagen, glass beads, inert polymeric materials (such as magnetic colloid,
polystyrene, polyamide materials like nylon, polyester materials, cellulose
ethers
and esters like cellulose acetate), urethane DEAE-dextran, as well as other
natural
and synthetic materials, such as foam particles, cotton, wool, dacron, rayon,
acrylates and the like. The CAM may be applied to form a coat, such as from
about 1.0 - 1.5 mm in thickness.
[00015] For example, a CAM might comprise gelatin particle or glass
bead core materials coated with a type I collagen solution that is then
polymerized
to form a film. The film containing such porous collagen-coated beads can then
be
exposed to a sample, such as serum or whole blood containing one or more
blood-borne adhesion components that promote the adhesion of a target cell,
such
as CTC and CEC. Mood-borne adhesion materials that promote adhesion of cells
such as CTC and CEC may comprise, for example, basement membrane
components such as fibronectin, fibrin, laminin, heparin, and vitronectin,
fragments
thereof, combinations thereof, or biological mimics of these components, and
modified versions thereof as seen in extravasation or endothelial injury, and
may
be prepared by purification from natural sources or synthesized by artificial
io
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
means. A CAM may further comprise specific ligands which also recognize and
bind target cells with a high degree of sensitivity and specificity.
(00016] The CAM film may include microbeads, such as type I collagen
coated gelatin-microbeads or glass-microbeads, covered with blood borne-cell
adhesion molecules, such as those present in blood or body fluids, and a
binding
material. For example, microbeads may comprise (but are not limited to)
dehydrated gelatin particle or glass beads, with diameter in the range of 200
microns to 2,000 microns. In one embodiment, the microbeads are configured, or
of such shape and size, to create anastomosic channels allowing blood flow in
the
film.
[00017] In embodiments wherein the target cells are CTC and CEC, the
CAM film of the invention preferably has an affinity and specificity for the
target
cells, CTC and CEC, with minimal affinity for other cells, such as a small
fraction of
hematopoietic cells. The CAM film may be designed to mimic the site at the
vessel wall of arteriovenous anastomosis or loci of metastases or
cardiovascular
plaques, where extracellular matrix (ECM) components, including collagens,
proteoglycans, fibronectin, laminin, fibrin, heparin, tenascin and vitronectin
etc.,
have been modified during the process of extravasation or endothelial injury.
In
essence, the CAM composition and assay surface architecture may be designed,
using the information presented herein, to improve mimicry of the cell
microenvironment so as to enable a more maximal number of viable target cells,
such as CTC and CEC, to be recovered from whole blood. The target cells,
a
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
including CTC and CEC, isolated by the methods of this invention are typically
viable, may exhibit growth ex vivo, and may exhibit the adhesive activity
against
extracellular matrix components, ECM. Isolated CTC and CEC from blood may be
used to establish an expression profile of CTC and CEC.
(00018] A CAM of the present disclosure may be used, for example, in
the detection, diagnosis and management of cancer. The CAM may be used to
recognize and bind with high affinity and specificity to viable cancer cells,
and
therefore, the matrix may be used to isolate cancer cells from fluid samples
such
as blood samples and/or ascites fluid taken from a patient suffering with
cancer. The CAM may be used for capturing metastatic cancer cells in the
patient's sample for the diagnosis and monitoring of the disease in such
patients
inflicted with cancer. CAMs may be used to detect and isolate viable
circulating
metastatic tumor cells from all types of cancers, including, ovarian, lung
cancer
such as non-small cell and small cell lung cancer, prostatic, pancreatic,
breast
cancer, melanoma, liver, stomach, cervical, renal, adrenal, thyroid, and .
adenocarcinomas such as colorectal cancer.
[00019] Alternatively, the matrix can be used to capture endothelial cells
in blood samples for the detection, diagnosis and management of cardiovascular
disease in a patient. CAM has the ability to bind with high affinity and
selectivity to
viable endothelial cells present in the blood sample when a blood sample taken
from a patient having cardiovascular disease is contacted with the matrix.
Endothelial cells at various stages of development, including progenitor
12
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
endothelial cells, may be used in diagnosis of cardiovascular disease, such as
angiogenesis in patients inflicted with this disease.
(00020] The present invention also provides a cell isolation device
utilizing the CAM of the present invention to isolate target cells from fluid
samples
such as blood. Such device may provide, for example, an "endothelial cell
trap"
that allows for the efficient enrichment and identification of target cells,
wherein the
target cells are, for example, viable endothelial progenitor cells in the
peripheral
blood of a subject with risk of cancer andlor cardiovascular diseases. A CAM-
initiated cell isolation device may be designed to provide a one million-fold
enrichment of viable circulating tumor cells and circulating endothelial cells
from
blood.
(00021] In another embodiment, the CAM can be used to capture and
isolate target cells such as fetal cells present in the maternal circulation
of
pregnant females. The isolated cells adhering to the CAM can then be used for
analysis in prenatal diagnosis of diseases such as Down's Syndrome, Marfan's
Syndrome, Taysach's disease and others using standard procedures. Isolating
fetal cells using the present matrix allows for a safer method for prenatal
diagnosis
of disease, since the fetal cells can be isolated directly from a blood sample
and
no invasive procedures of the pregnant mother are necessary. In this and other
embodiments of the invention, the CAM enriches or increases the number of
cells
that would normally be available for analysis in a blood sample using standard
techniques of cell isolation.
13
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
[00022] Using the present disclosure, CAM cell enrichment may be
designed to have one or more of the following features: (a) a one-million-fold
enrichment of viable target cells, including CTC and CEC, from whole blood
with a
high degree of sensitivity and specificity for the target cells necessary for
the
diagnosis of disease; (~b) concurrent functional and morphological
discrimination,
for example, cell size and density, of the target cells, including CTC and
CEC,
from other normal blood and tissue cells; (c) whole blood may be used as the
starting sample or cell fractions prepared by a common density gradient
centrifugation procedure. CAM cell enrichment may be a single or multistep
process.
[00023] Further disclosed is a CAM-initiated cell isolation device that
permits efficient captures of viable target cells, including CTC and CEC, from
the
mononuclear cell population. Target cells may be fractioned from blood or
tissue
fluid samples derived from subjects inflicted with a disease such as
cardiovascular
disease or cancer, as discussed in co-pending application PCT Patent
Application
PCT/US01126735 -- claiming priority to U.S. Provisional Patent Application No.
60/231,517 (the disclosure of which is incorporated herein by reference in its
entirety). Such a device may comprise, for example, a CAM coating that is
preferably immobilized to the surface of a vessel, such as, but not limited
to, the
inner bottom surface of a tube, a surface of a slide, or the inner bottom
surface of
a Petrie dish. The matrix-coated surfaces of the CAM-initiated cell isolation
vessels are preferably designed to maximize contact for the sample when sample
14
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
is placed into the vessel. The CAM-initiated cell isolation device may make
use of
a variety of already available laboratory diagnostic vessels, for example, a
cell
culture chamber slide, a culture microtiter plate, a culture flask, etc.
[00024] The CAM-initiated cell isolation device may be rotated to more
optimally imitate blood flow to increase contact between the cells and CAM,
thus
promoting more efficient enrichment (of, for example, viable CTC and CEC).
[00025] A CAM-initiated blood device may be constructed based on the
present disclosure that is more efficient in removing viable target cells
including,
CTC from the peripheral blood of a subject sufFering with, for example, CTC
related disease, than that described in co-pending application PCT Patent
Application PCT/US01/26735 (claiming priority to U.S. Provisional Patent
Application No. 60/231,517).
[00026] The methods and CAM films described above for enrichment of
tumor cells may also readily be used as a negative filtration step for
harvested
autologous blood or bone marrow to remove cancer cells. A CAM-initiated blood
filtration device of the present disclosure may be employed to remove
contaminating cancer cells, for example, in respect of the auto transfusion of
blood
salvaged during cancer surgery, therapeutic bone marrow transplantation,
peripheral blood stem cell transplantation and aphaeresis, in which autologous
transfusions are done, Further, the described CAM-initiated blood filtration
unit
is
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
may be used to prevent full blown cancer from occurring by removing cells
capable of metastasis from the circulation.
[00027] CAM-initiated blood filtration may similarly be utilized in the
preparation of cancer-free autologous bone marrow cells intended for
replacement
after aggressive, bone-marrow chemotherapy - radiation in cancer patients.
Detection of cancerous cells may be improved by molecular amplification
techniques, and CAM-enriched cells may be used in multiplex molecular analysis
such as tests for DNA, proteins and immunological tests (as, for example,
specific
for CTC and CEC from a subject).
[00028] CAM-enriched cells and their DNAs, RNAs, proteins or antigens
may be applied to multiplex detection assays for cancer diagnostic purposes.
Cell
markers used in the multiplex CTC detection assay include, but not limited to,
the
CTC invasive phenotype [collagen ingestion and acetyl LDL uptake by the cell],
the epithelial antigens [cytokeratins, epithelial specific antigens (EpCAM,
HEA,
Muc-1, EMA, GA733-1, GA733-2, E-cadherin, EGFR, TAG12, lipocalin 2
(oncogene 24p3)], endothelial antigens [CD31/PECAM1, van Willebrand factor
(vWF), Flt-1 (a receptor for VEGF), VE-cadherin] and other tumor associated
antigens [including, but not limited to, carcinoembryonic antigen (CEA),
epidermal
growth factor receptor (EGFR), human kallikrein-2 (HK2), mucin (MUC), prostate-
specific antigen (PSA), prostate-specific membrane antigen (PMA), 13 subunit
of
human chorionic gonadotropin (13-hCG) etc.]. Markers may be applied
individually or jointly to achieve the effective identification and
enumeration of
16
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
viable tumor cells in a given volume of the blood or body fluids from a
subject. The
methods for data readouts include, but are not limited to, flow cytometry,
fluorescent microscopy, enzyme-linked immunoabsorb assay (ELISA), and
quantitative real-time RT-PCR etc.
[00029] CAM-enriched CTC cells provide sources for genetic testing for
cancer. The alterations in gene structure and function that may be genetically
tested in CTC cells include, but are not limited to, oncogenes (e.g., ERBB2,
RAS,
MYC, BCL2, etc.), tumor suppression genes (e.g., p53, APC, BR GA], BRCA2,
CDKN2A, CCND1, CDC2SA, CDC25B, KIPj, RB] etc), genes associated with
tumor progression [e.g., carcino-embryonic antigen (CEA), epidermal growth
factor
receptor (EGFR), human kallikrein-2 (HK2), mucin (MUG), prostate-specific
antigen (PSA), prostate-specific membrane antigen (PMA), 13 subunit of human
chorionic gonadotropin (13-hCG), etc.], and genes associated with metastatic
cascades [e.g., nm23 family (HJ-6) of necleoside diphosphate kinases (cell
migration), PTENlMMAC] (cell migration and focal adhesions), CADJlE-cadherin
(cell-cell adhesion), MKK4/SEKi (cellular response to stress), KISS-i
(regulation of
MIIMLP9 expression), BRlVISi (cell motility) etc]. For example, aneuploidy and
CKi9, ERB2, CEA, MUG], EGF receptor, J3-hCG alterations are useful in
diagnosis of breast cancer; pS3, Ki-ras mutations CDKN2A, LOH 3p, FHIP for
lung
cancer; p53, APC, GEA, CKi9, CK20, ER882, Ki-ras mutations for colorectal,
gastric, and pancreatic cancers; PSA, PSM, HK2 for prostate cancer; p53
mutations and microsatellite alterations for head and neck cancer. The genetic
m
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
markers may be applied individually or jointly to achieve the effective
detection of
genetic changes in a subject. The methods for data readouts include, but
limited
to, flow cytometry, fluorescent microscopy, fluorescent or color based
polymerise
chain reaction readers etc.
[00030] CAM-enriched CEC cells and their DNAs, RNAs, proteins or
antigens currently known in a specific tumor may also be applied to multiplex
CEC
detection assays for detecting subjects with risk of cardiovascular diseases.
The
cell markers used in the multiplex CEC detection assay include, but are not
limited
to, the CEC functional phenotype [acetyl LDL uptake by the cell] and
endothelial
antigens [CD3 1lPECAM- 1, van Willebrand factor (vWF), Flk-1 (a receptor for
VEGF), VE-cadherin]. The markers may be applied individually or jointly to
achieve the effective identification and enumeration of viable endothelial
cells in a
given volume of blood or body fluids from a subject. Methods for data readouts
include, but are limited to, flow cytometry, fluorescent microscopy, enzyme-
linked
immunoabsorb assay (ELISA), and quantitative real-time RT-PCR, etc. CAM-
enriched CEC cells may further provide a source for genetic testing of a
subject.
That is, alterations in gene structure and function of a subject may be
genetically
tested using the CTC cells enriched by CAM. The genetic markers may be applied
individually or jointly to achieve the effective detection of genetic changes
in a
subject.
[00031] In one embodiment, viable cells captured on the CAM can be
released readily from the device surface by the use of digestive enzymes,
is
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
including, but not limited to, collagenases, trypsin/EDTA solution (purchased
from
GIBCO), and hyaluronases by selecting appropriate core materials and cell
adhesion coatings. For example, cell adhesion molecules and collagen or
gelatin
of the CAM film may be sensitive to digestion. Enzymes that will cleave
binding
between the cells and the matrix, will release viable cells from the CAM film
into
suspension. For example, CAM-captured cells may be effectively released into
suspension using collagenase when type I collagen is the skeleton supporting
the
cell adhesion molecules.
[00032] The detection methods of the present invention may be used for
cancer diagnostic purposes, e.g. early detection, monitoring therapeutic and
surgical responses, and prognostication of cancer progression. CAM-enriched
CTC may be used, for example, to detect cancer earlier than using current
surgical
methods of isolating tumor cells, to monitor therapeutic and surgical
responses, to
improve the accuracy of cancer staging, and to determine the metastatic
potential
of the patient's tumor. These applications may be further enhanced using
additional multiplex molecular assays known to those of skill in the art, such
as
determining the genetic alterations of a subject, verifying the tissue origin
of
circulating tumor cells, measuring the molecular markers of the types of
cancer,
and determining the degree of reduction in tumor cytotoxic leukocyte count or
complement association.
[00033] Prognosis and therapeutic effectiveness may also be adjudged
by the detection assays of the present invention. For example, the count of
viable
19
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
CTC during and post therapeutic interventions) may be used to ascertain
therapeutic effectiveness. CAM-enriched CTC and associated anti-tumor host
immunity may be detected and quantified in conjunction with microscopic
imaging
and flow cytometry. Selection of chemotherapeutic regimen may be optimized by
determining those regimens that most effectively, without undue side effects,
reduce the number of viable CTC in the blood sample. Optimization of selection
of
chemotherapeutic regimen may also be performed by subjecting the CAM-
enriched CTC to a battery of chemotherapeutic regimes ex vivo. Effective doses
or
drug combinations could then be administered to that same patient. The number
of viable CTC can be determined before and after the administration of the
compound or agent. Compounds or agents that significantly reduce the number of
viable CTC after administration may be selected as promising anti-cancer
agents.
Agents exhibiting efficacy are those, which are capable of decreasing number
of
CTC, increasing cytotoxic leucocytes and complement system (host immunity),
and suppressing tumor cell proliferation.
[00034] The detection methods of the present invention may also be used
to detect whether a new compound or agent has anti-cardiovascular disease, or
other activity.
[00035] It should be noted that most CTC are dead or apoptotic in the
circulation due to the presence of host immunity to tumors, as described in co-
pending PCT Patent Application PCT/US01/26735. The viability of CTC and
tumor associated cytotoxic leukocytes, and measurements with respect to the
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
autologous complement system derived from individual donors put together an
effective means of determining host immunity against tumors. A subject may be
considered as having anti-tumor immunity, when the number of viable CTC
enriched by CAM is high in the absence of autologous plasma but low in the
presence of autologous plasma. On the other hand, a subject who loses anti-
tumor immunity would have high levels of viable CTC in the presence and
absence of autologous plasma that resist immune killing.
[00036] Viable CTC enriched from blood of cancer patients by a CAM
method may also be used in fusions with dendritic cells for anti-cancer
vaccine
development. For example, the CTC from individual patients with different
cancers
may be subjected to ex vivo culture and expansion, and the cells may be used
in
whole, or purified for specific membrane structures or for specific antigens,
to
interact with dendritic cells for the development of an effective tumor
vaccine.
[00037] Cytotoxic lymphocytes enriched by the CAM methods from blood
of cancer patients may be valuable in their own right: careful comparison of
their
gene expression profile in comparison to non-tumor associated lymphocytes may
yield valuable information concerning the type of ongoing immune reaction and
inflammation that are being mounted against the metastatic tumor cells.
Moreover, another valuable therapy approach may be to expand these cells in
vitro, for example, using IL-2, and then reintroduce them into the patients to
augment their anti-tumor immune response. This approach may have dramatic
utility in the management of melanoma and other tumors.
21
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
(00038] Embodiments of the present invention would be useful both for
diagnostic and therapeutic purposes in providing the ability to separate, for
example, the small fraction of CTC that are metastatic from the large number
of
other circulating cells in a patient's body.
[00039] Embodiments of the present invention: (1 ) can isolate specifically
viable target cells such as tumor and endothelial cells but leave alone
unrelated or
damaged cells; (2) can achieve an enrichment of over one hundred target cells
such as tumor or endothelial cells, from over five billion cells in whole
blood; (3)
can identify target cells such as "cancer cells" or "endothelial progenitor
cells" from
normal blood cells readily in the same assay format; (4) can enrich cells from
background normal blood cells that are useful in diagnosis and treatment of
patients suffering with a disease such as metastatic cancers and
cardiovascular
diseases.
BRIEF DESCRIPTION OF THE DRAWINGS
[00040] FIG. 1A depicts a front sectional view of a CAM 16-well chamber
slide whose bottom surface is coated with a CAM film, such as a fluorescently
labeled collagen film, capable of enriching circulating tumor cells and
endothelial
progenitor cells that may be used in the diagnosis of cancer and
cardiovascular
diseases;
[00041] FIG. 1B depicts a front sectional view of a CAM 96-well chamber
slide whose bottom surface is coated with a CAM film, such as a fluorescently-
22
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
labeled CAM film, comprising collagen that is capable of enriching circulating
tumor cells and endothelial progenitor cells and that may be used in the
diagnosis
of cancer and cardiovascular diseases;
[00042] FIGS. 2A, 2B and 2C depict a front sectional view of upright 7m1,
15m1 and 30m1 vacuum blood collection tubes that may be used in the diagnosis
of
diseases that are coated along their internal surface with a CAM film;
[00043] FIG. 2D depicts a front sectional view of an upright tissue culture
bottle coated along its internal surface with a CAM film that may be used in
the
diagnosis and treatment of cancer and cardiovascular diseases;
[00044] FIG. 2E depicts an enlarged front sectional view of a CAM film in
a vessel such as in FIGS. 2A-2D;
[00045] FIG. 3A depicts a front sectional view of an upright blood
collection tube with a dipstick insert coated with a CAM film;
[00046] FIG. 3B depicts a front sectional view of the dipstick of FIG. 3A;
[00047] FIG. 4A depicts a three-dimensional view of a blood filtration
cassette containing a pre-filter mesh inlet in the housing for the
introduction of the
sample to be filtered; a main filter compartment filled with cell separation
beads
coated with a thin CAM film; a post-filter mesh outlet in the housing for the
removal
of filtered blood, which may be used in conjunction with a blood filtration
system
for diagnostics, therapeutics or treatment according to the invention; and
23
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
(00048] FIG. 4B is an expanded cross-sectional view of the main filter
compartment of FIG. 4A filled with cell separation beads coated with a CAM
film
depicting the anastomosic channels formed by the cell separation beads within
the
inner confinement area.
(00049] FIG. 5 is a immunocytochemistry micrograph of leukocytes (A)
and tumor cells (B)/(C)l(D) derived from ascites of adenocarcinoma of the
ovary
enriched by a cell adhesion matrix using antibodies directed against CD45, a
pan-
leukocyte antigen, and pan-cytokeratins (B)/(C) or CD-31 (D) without (A)/(B)
and
without (C)/(D) antibody EpCA of positive-selection.
(00050] FIG. 6A-C is a real-time RT-PCR relative expression analysis of
the expression of 10 genes selected from DNA microarray clusters with respect
to
tumor cells from ascites (FIGS. 6A and 6B) and tumor cells from a solid
primary
tumor (FIGS. 6A and 6C).
DETAILED DESCRIPTION OF EMBODIMENTS OF THE INVENTION
(00051] The invention is directed to the isolation and detection of target
cells in fluid samples taken from a patient for screening, diagnosis and
management of diseases such as cancer and cardiovascular disease, and in
prenatal diagnosis.
(00052] The isolation of target cells from fluid samples taken from a
patient is facilitated by the present methods. Isolation of such cells may be
useful
in managing a disease state associated with such cells. For example, tumor and
24
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
endothelial cell identification in blood samples taken from a patient are
indicative
of metastatic cancer and cardiovascular disease, respectively. Similarly,
fetal cells
present in a pregnant female's blood, therefore, can be isolated and used in
prenatal diagnosis of disease associated with the fetus.
[00053] Embodiments of the invention involve target cell separation
including tumor, endothelial, and fetal cells separation strategy using a
functional
enrichment procedure that captures the target cells based on an adhesive
phenotypic behavior of invadopodia. This cell adhesion properties, which
manifests as the propensity to bind with tight affinity and specificity to ECM
matrices that mimic the blood vessel microenvironment, appears to be mediated
by not one specific protein, but rather by a complex of proteins including
specific
cell adhesion receptor integrins that cluster on the cell surface in
projections of
cells denoted as "invadopodia."
CAM Cell Enrichment
(00054] The tumor and endothelial cell separation strategy of CAM cell
enrichment involves using a functional enrichment procedure that captures the
target cells based on an adhesive phenotypic behavior to materials, as
characterized in detail over the past decades (Aoyama and Chen, 1990; Chen and
Chen, 1987; Chen et al., 1994a; Chen et al., 1984; Chen et al., 1994b; Chen,
1996; Chen, 1989; Chen and Wang, 1999; Ghersi et al., 2002; Goldstein and
Chen, 2000; Goldstein et al., 1997; Kelly et al., 1994; Monsky et al., 1994;
Monsky
et al., 1993; Mueller et al., 1999; Mueller and Chen, 1991; Mueller et al.,
1992;
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Nakahara et al., 1996; Nakahara et al., 1998; Nakahara et al., 1997; Pavlaki
et al.,
2002; Pineiro-Sanchez et al., 1997; Saga et al., 1988; Zucker et al., 2000;
Zukowska-Grojec et al., 1998). It has been found that cells having invadopodia
("invadopodic cells") bind with tight affinity to matrices that mimic the
blood vessel
microenvironment, especially in the perturbed state. Based on invadopodia
behavior, a functional cell enrichment step that is highly selective for
viable
metastatic tumor cells and angiogenic endothelial cells and which captures few
of
leukocytes/monocytes and red cells, and leaves in solution other cell types
may be
designed. The CAM cell enrichment assay may additionally include a negative
identificationlselection procedure using antibodies directed against the
leukocyte
common antigen CD45.
[00055] The present method employs a CAM comprising biochemically a
non-reactive core, such as collagen polymer, physically-associated with cell
adhesion molecules, in particular natural and synthetic blood-borne adhesion
molecules. A CAM preferentially is designed to permit viable tumor cells to
adhere
to the matrix while avoiding adherence to normal background cells in the
blood;
that is, allowing viable tumor cells to attach with great avidity but avoiding
attachment to normal cells (preferably including, for example, more than 99.9%
of
white cells and 99.9999% of red cells) and dead or dying tumor cells. The CAM
coating may also comprise a ligand (e.g., antibodies, fluorescent and/or
colorimetric markers, etc.) capable of reacting with one or more CAM-invading
cells. The ligand may cause a visible or non-visible (but detectable) change
in the
26
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
CAM indicative of the presence of one or more cells to be detected. Such
ligands
may alternatively in tandem be placed in a separate detection layer associated
with the CAM. A thin CAM coating is preferably immobilized to the inner bottom
surface of the cell separation unit.
[00056] Thus, CAM can be used to successfully recover viable tumor
cells from, for example, the mononucleate cell fraction of blood samples from
patients with stage I and IV non-small-cell lung cancer (NSCLC).
[00057] The CAM approach can also be used to mark tumor cells for the
purpose of identification. For example, when the CAM is prepared using
fluorescently labeled collagen, the invasive tumor cells become labeled, since
they
exhibit a propensity to digest and ingest collagen. In contrast, normal cells
leave
the CAM undisturbed.
[0005] In respect of invadopodic cells desired to be enriched, the CAM
composition and assay surface architecture may be designed to improve mimicry
of the intravascular microenvironment so that the maximal numbers of viable
desired cells are recovered from a sample, such as whole blood. More efficient
enrichment of the invadopodic cells may also be accomplished by use of a unit
rotation procedure to optimally imitate blood flow and increase contact
between
the tumor cells and CAM. In a preferred embodiment, the sample typically
should
be processed in a manner to provide for retention of the viability of the
invadopodic
cell in the sample.
27
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Cam-Initiated Cell Isolation Device
[00059] A CAM-initiated cell isolation device may comprise numerous
designs such as a cell culture chamber slide, a culture microtiter plate, or a
culture
flask, etc.
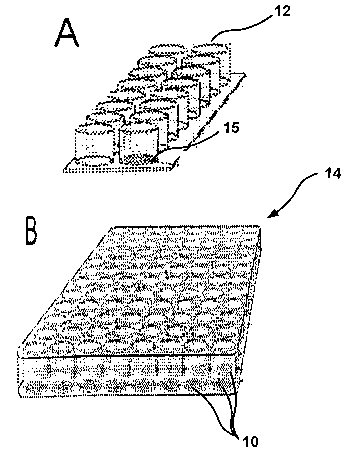
[00060] For example, the CAM-initiated cell isolation device may, as
shown in FIG. 1, comprise a plurality of wells (12) in a unit array (14)
having a
CAM (10) at the bottom of one or more wells (12). FIG. 1A illustrates a 13-
well
microarray while FIG. 1 B illustrates a 96-well microarray. The CAM-initiated
cell
isolation device may comprise a blood collection tube of various shapes (16,
18,
22) which may or may not be fitted with a cap (20) or a container (24) such as
shown in FIG. 2, where the inner walls are coated with a CAM film (10), the
bottom
surface (26) uncoated, and fitted or not fitted with a cap (20). Preferably
such
vessels are sterilized before use. The CAM-initiated blood device may be used,
for example, to~isolate CTC and/or CEC in the CAM (30) from samples (28)
placed
in the vessel. The CAM (10) may be comprised, for example, of glass beads (34)
incorporated within a layer (30) comprising a cell adhesion material.
[00061] The CAM-initiated cell isolation device may utilize a dipstick (36)
comprising a measuring card (38) such in perspective view and sectional view
in
FIG. 3B, the surface of the measuring card (38) being coated with CAM film.
The
dipstick or measuring card is inserted in a cell separation vessel (16). The
CAM
film may be spread over the surface of a dipstick (38) and/or the inner wall
of the
tube (16) and/or cap (20).
28
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
(00062] In one embodiment, the CAM-initiated cell isolation device further
includes pre- and/or post-separation features such as filters (e~.g., Amicon
filters,
hollow filters), membranes, or gradients (such as ficoll, sucrose, etc.) that
help
separate out cell populations before the population contacts the CAM film.
[00063] Turning to FIG. 4, there is shown a three-dimensional view of a
blood filtration cassette (43) containing a pre-filter (41 ) such as a mesh
(or CAM-
coated mesh) in the housing for the introduction of the sample to be filtered,
a
main-filter compartment (40) filled with a CAM (10) and a post-filter (42)
outlet in
the housing. FIG.4B is an expanded cross-sectional view of the main-filter
compartment (40) filled with CAM (10).
[00064] In one embodiment, the CAM film of the CAM-initiated cell
isolation device comprises collagen-coated microbeads, advantageously with a
diameter in the range of 200 microns to 2,000, microns configured to create
anastomosic channels allowing blood flow in the film. Whole blood in this
blood
filtration unit may be incubated at about 37°C and rotated to imitate
blood flow that
increases contact between cells and CAM and supports efficient enrichment of
viable cells from blood. Blood containing target cells such as tumor and
endothelial progenitor cells may be stored in a CAM-initiated enrichment
device for
extended periods of time ranging from 4 to 48 hours to add efficiency of
enrichment.
29
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
[00065] Three parameters may need to be addressed in designing a
CAM-initiated cell isolation device and system: (i) the CAM composition and
assay surface architecture to improve mimicry of the tumor intravascular
microenvironment so that maximal numbers of viable tumor cells are recovered
from whole blood; (ii) the unit rotation procedure to optimally imitate blood
flow,
increase contact between the tumor cells and CAM, and promote more efficient
enrichment of viable tumor cells; and (iii) the blood process mode to improve
retention of tumor cell viability in the blood samples.
[00066] The positive CTC selection method described above to enrich
tumor cells may also be used as a negative filtration step for harvested
autologous
blood or bone marrow to remove cancer cells. The CAM-initiated blood
filtration
method of the invention thus may be employed in respect of the autotransfusion
of
blood salvaged during cancer surgery, therapeutic bone marrow transplantation,
and peripheral blood stem cell transplantation and aphaeresis. The described
CAM-initiated blood filtration unit may also be used to prevent full blown
cancer.
from occurring by removing cells capable of metastasis from the circulation.
[00067] Specificity and sensitivity control experiments may be performed
to optimize an assay's tumor cell enrichment efficiency. Significant variables
include: (a) the viability of the exogenously added tumor cell lines after
capture by
CAM, (b) the conditions that most effectively enrich and isolate viable tumor
cells,
and (c) the cell processing mode that leads to complete elution of the cells
from
the CAM film.
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Example 1
CTC and CEC from Blood
[00068] Whole blood may be placed in a CAM blood
collection unit, such as a blood collection tube (FIGS. 2 and 3). The
tube may be incubated at about 37°C and rotated to imitate blood
flow so as to increase contact between cells and CAM. Blood may
be collected in the presence of anticoagulants, i.e., Anticoagulant
Citrate Dextrose solution USP (ACD, Baxter Healthcare Corporation,
Deerfield, IL) plus 50 units of lithium heparin per mE, to prevent
clotting in the CAM blood test unit. The sealed CAM-blood tube may
be placed on a roller and rotated at 5-30 cycles per minute at about
37°C, and then incubated for 1-3 hours for cell attachment to occur.
Example 2
Specificity and Sensitivity Control
[00069] Human tumor cell lines of different tumor origins
may be chosen for use in performing specificity and sensitivity
control experiments. For examples, the human colon tumor cell line
SW-480, human gastric tumor cell line RF-48, several breast tumor
cell lines, human malignant melanoma line LO?C, and several ovarian
tumor cell lines may be used. Tumor cell lines may be purchased
from American Type Culture Collection (Manassas, VA). All cell lines
should be confirmed to be negative for Mycoplasma infection. The
tumor cell lines should be examined for: (a) high affinity binding to
CAM within one hour after plating; (b) high proliferation rate; and (c)
the tumor cell lines should be readily and stably (100%) fluorescently
labeled with red or green fluorescent dyes prior to use or
transformed with an expression plasmid for green fluorescent protein
(GFP) in order to be able to visualize the tumor cells directly at the
31
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
end of the enrichment procedures. Control normal blood will be
seeded with known numbers of the green fluorescence labeled or
GFP-expressing fluorescent human tumor cells and subjected to the
CAM cell enrichment methods, to assess their comparable
efficiencies.
[00070] Whole blood from a healthy donor or cord blood
derived from umbilical cords may be obtained through the National
Disease Research Interchange (Philadelphia). Immediately after
reception, blood should be supplemented with Anticoagulant Citrate
Dextrose solution USP (ACD, Baxter Healthcare Corporation,
Deerfield, IL) plus lithium heparin to prevent clotting that often occurs
during further experimental manipulations. Normal blood does not
contain cells with cancer characteristics. Thus, the tumor cells spiked
into these blood samples should be the only ones recovered in this
test for specificity and sensitivity.
[00071 ] Cord blood or blood samples from healthy
individuals may be seeded with known numbers of fluorescently-
labeled, i.e., fluorescent dye pre-labeled or GFP-tagged tumor cells.
The mixed blood samples of 3 mL aliquots may be transferred to
CAM assay units for tumor cell enrichment. Suspended blood cells
may be removed. When, for example, as type I collagen is the
skeleton supporting the CAM film, the CAM-captured cells may be
released into suspension using collagenase. To determine the
number of control viable tumor cells from cord blood, for example,
approximately 3,000 GFP-tumor cells may be spiked into 3 mL of
cord blood (approximately 15,000,000,000 blood cells) or cell culture
complete medium (containing 15% human serum) and subjected to
CAM enrichment. Cells recovered from medium would indicate the
number of actual viable tumor cells. The ratio, (cell number
32
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
recovered from cord blood) / (cell number recovered from medium),
signifies the efficiency of the assay. The percent recovery of viable
tumor cells from cord blood as compared to medium may be used to
determine optimal conditions for CAM enrichment assay. These
conditions include period of time for incubation of CAM-blood tubes
(e.g., 1 - 3 hours), rotation speed (e.g., 5 - 30 cycles per minute), and
length of time of storing blood to retain cell viability (e.g., 4 - 48
hours). The presence of extremely large numbers of background
blood cells would prevent direct contact of cancer cells with the CAM
surface and diminish detection sensitivity of the CAM method. The
CAM film of the blood collection tube advantageously is designed to
maximize surtace contact areas of CAM to tumor cells. Length of
cell incubation time is also important, as CAM depends on differential
adhesion of tumor cells than hematopoietic cells.
Examule 3
Determination of Cell Viability in a CAM-Blood Filtration Unit
v. Slood Collection Tube
[00072] Another problem is the cell viability of the blood
samples, which may vary during transportation to the research
laboratory. Increasing the time of storage may be expected to
damage cells in the blood. To determine if tumor cells in the CAM
blood unit can stay viable during shipping, 3,000 GFP-tumor cells
were spiked into 3 mL of cord blood and control medium containing
15% human serum (Sigma). Each aliquot was stored at 4°C for
series of time (4, 6, 8, 12, 16, 24, 36 and 48 hours). Each aliquot
was then captured by CAM and the percent recovery of GFP-tumor
cells by CAM determined. For each time point, four duplicate
33
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
experiments were performed, and percent recoveries determined.
The results showed that CAM-captured tumor cells survived better
than suspended cells in blood.
[00073] CAM-enriched cells may be counted by any
means known to those of ordinary skill in the art, including
microscopic and flow cytometric methods (see below for detailed
methods). For cell enrichment experiments, preliminary data
obtained by microscopic counting suggest the recovery rate
increases with spike dosage, roughly following a logistic curve.
Using a CAM-initiated cell isolation device of the present disclosure,
one can obtain approximately 40% recovery of the GFP-LOX human
malignant melanoma cells spiked into cord blood when there is
greater than 1,000 GFP-LOX cells per mL of blood in the initial
sample, with a variability of approximately 10%.
Strategy For Enumeration And Validation Of Viable Tumor Cells In Blood Of A
Subject By Flow C ometry
[00074] In a clinical laboratory, labeled tumor cells can be measured by
multi-parameter flow cytometric cell analyzer using FITC labeled collagen
(green)
to detect invasive tumor cells, PE labeled anti-CD45 leukocyte common antigen
antibody (red) to detect and exclude leukocytes, and 7-AAD to exclude dead
cells.
This automatic cellular analysis can be validated by a parallel and
independent
microscopic evaluation using microscopy, for example, with cell lineage
markers
including antibodies directed against epithelial, endothelial and
hematopoietic
antigens.
[00075] Enumeration of invasive tumor cells in blood by flow cytometry
may be accomplished by multi-parameter flow cytometric cell analyzer using,
for
34
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
example: (a) FITC labeled collagen that would be ingested by tumor cells
(green)
to detect invasive tumor cells, and (b) PE-labeled anti-CD45 leukocyte common
antigen antibody (red) to detect and exclude leukocytes contaminated in the
cell
population. For example, tumor cells captured by CAM and co-isolated normal
blood cells may be post-stained with phycoerythrin (PE)-conjugated CD45
antibody and dead-cell nucleic acid dye 7-AAD. Labeled cell sample may be
aspirated and analyzed, for example, on a FACSCalibur flow cytometer (Becton
Dickinson). Criteria for data analysis may include, among other factors: (a)
size
defined by forward light scatter, (b) granularity defined by orthogonal light
scatter,
(c) negative events of dead 7-AAD cells, (d) negative events of PE-labeled
CD45
mAb normal cells, and (e) positive events of the FITC-tumor cells.
[00076 As would be understood by one of ordinary skill in the art, there
are several cytometric methods of discriminating apoptotic and dead cells from
alive cells in heterogeneous clinical specimens (e.g., using FITC-libeled
annexin V
and propidium iodide). For example, to incorporate the cell viability test
into the
multiparameter flow cytometry of CAM purified cells, one may use 7-amino-
actinomycin D (7-AAD, Molecular Probes) to label dead cells in a fixed CAM
cell
population. 7-AAD can be excited by the 488 nm argon laser line and emits in
the
far red range of the spectrum. 7-AAD spectral emission can be separated from
the
emissions of FITC and PE (OLIVER et al., 1999). The fluorescence parameters
allow characterization of dead cells (7-AAD), viable and invasive tumor cells
(FITC-collagen) and leukocytes (PE-CD45) in a subset of CAM purified blood
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
cells. Freshly labeled cells may be delivered to the flow lab for immediate
counting or stored in suspension, for example, at 4°C for ~ 1 - 3 days.
The
FACSCalibur flow cytometer may be configured to count 2 - 4 cell samples per
hour.
[00077] In a typical blood sample obtained from an individual with cancer
or cardiovascular diseases, the circulating tumor and endothelial cells are
vastly
outnumbered (in the range of over a million-fold) by the normal hematopoietic
cells.
[00078] While the embodiments described are not limited to any particular
hypothesis, the present inventors postulate that:
(a) During the earliest stage of cancer progression, metastatic
cells start emerging from primary tumors; these cells exhibit
an invasive behavior,
(b) Tumor cell populations from blood that are indicative of the
presence of a cancer will enable early diagnosis and further
molecular analysis, and
(c) There are diagnostic sets of genes present in both circulating
and primary tumor cells that can be used to: resolve the
tissue-site origin of circulating tumor cells, determine a
specific cancer subtype, and predict the metastatic potential of
a patient with a high degree of confidence.
36
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Microscopic Characterization of the Cells Enriched by CAM Culture Method
[00079] A high yield, CAM culture may be performed in parallel as an
independent CAM method to validate the tumor cells enriched by CAM and
counted by flow cytometry. The CAM culture method can be readily augmented
with microscopy and immunocytochemistry using cell lineage or putative tumor
markers. Microscopy can be used to identify the CTC enriched from blood by CAM
as possessing the following features denoted Co+ / Epi+ l Endo+ I Leu- ; the
CEC
as Co- I Epi- / Endo+ I Leu-; tumor-associated lymphocytes as Co- / Epi- /
Endo- l
Leu+. Specifically, the CTC are:
1 ) Positive fluorescence from ingested and concentrated TRITC-
labeled collagen fragments (Co+; the proclivity to degrade and
ingest ECM is one of the hallmarks of invasive and metastatic
cells).
2) Positive immunocytochemical detection for the epithelial-
specific markers, including cytokeratins and epithelial
membrane antigens (BerEP4, EpCAM, GA733 and Muc-I)
(Epi+).
3) Positive immunocytochemical detection for the endothelial
specific markers, including CD31, van Willebrand factor (vWF)
and VEGF receptor (Endo+).
37
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
4) Negative immunocytochemical detection for markers of the
leukocytelmonocyte lineages, including CD45, CD14 and
CD68; negative for leukocyte-like cytology (L_eu-).
[00080] The antibody labeling design of the CAM cell chamber method, in
combination with differential interference contrast (DIC) bright field and use
of a
triple fluorescent filter, employable for example on a Nikon Eclipse E300
inverted
fluorescent microscope, provide a powerful multiplex means of characterizing
tumor cells in each microscopic field. In the same fluorescence microscopic
field,
TRITC-collagen labeling of invasive cells is seen as red fluorescence, FITC-
cell
type marker as green florescence and Hoechst 33258 nuclear dye as blue-
fluorescence, whereas APAAP stained cell type marker is shown as red color in
DIC bright light. Images may be stored in a computer hard drive and the number
of color-or fluorescence-labeled cells in a sample may be counted with the aid
of
software such as Metamorph image analysis software (Universal Imaging
Corporation).
[00081] Slides with the CAM-enriched and labeled cells may be scanned
under fluorescent light microscopy for positive tumor cells.
Multiplex molecular analysis of CAM-enriched cells: Microarray and Real-time
RT-
PCR
[00082] The expression levels of mRNAs expected to be present
specifically in circulating tumor cells versus those expected to be present in
leukocytes may be used as a measure of the degree to which enrichment is
successful. The percentage of tumor cells in a given cell population may be
38
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
validated using expression of epithelial (GA733-1 ) and leukocyte (CD45)
markers,
using tumor cell lines and leukocyte cell samples as positive controls.
[00083] Real-time RT-PCR may be performed using, for example, the
Roche Light Cycler on cell samples purified from blood samples. Real-time PCR
quantification of the epithelial marker GA733-1 and the leukocyte marker CD45
relative to i~-actin may be performed. The epithelial marker GA733-1 is
expected
to be expressed at high levels in the pure tumor cell subsets and tumor cell
lines
but not in leukocytes. In turn, the leukocyte marker CD45 should be detected
in
the leukocyte samples and impure tumor cell populations but not in tumor cell
lines
nor in pure tumor cell samples. Observation of a substantial GA733-1 signal in
the
tumor cell sample recovered can be interpreted as demonstrating that the CAM
enrichment procedure returns a cell pool in which tumor-characteristic markers
can easily and reproducibly be measured. It is also important to determine the
level of CD45 signal in each CAM tumor cell set to indicate degrees of
contamination of leukocytes. If substantial contamination is observed, then
one
may conclude that, for example, a CD45 negative-selection step may be
necessary to test and incorporate into the final protocol.
[00084] The molecular basis of most solid cancers is not understood. In
each clinical specimen, carcinoma cells are variable in number and
pathological
types; carcinoma cells are also surrounded by numerous types and number of
normal cells. Furthermore, tumor cells alter their gene expression profiles
during
progression and metastasis. The CAM cell enrichment methods offer viable tumor
39
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
cell populations that are available for the molecular analysis of the tumor
cells ex
vivo using DNA microarray and real-time RT-PCR analyses. These viable tumor
cell populations can enable a broad investigation into finding genes commonly
expressed in the tumor cells derived from primary tumors and blood, and genes
that are specifically expressed in the tumor cells of specific epithelial
cancers. As
seen in Table 1 and 2, the present cell separation method has allowed for the
characterization of tumor cells isolated from blood samples using microarrays
and
RT-PCR technologies. The data show the characteristic gene expression for
specific tumor cell types.
Table 1. Histo-pathological information of cell samples and their original
clinical specimens
CategorySampleSite Histology . GradeStageMicro-ReaITime
arra PCR
Tumor A01 Ovary Serous adenocarcinoma3 IIICv v
Cells
from A02 Ovary Serous adenocarcinoma3 IIICv
Ascites
AO3 Ovary Serous adenocarcinoma3 IIICv v
Pimary
A04 eritoneal Serous adenocarcinoma3 IIICv v
A05 Ovar Mixed clear cell,
y papillary and 3 IIICv v
endometrioid adenocarcinoma
A06 Ovary Serous adenocarcinoma3 IIIC v
A07 Ovary Serous adenocarcinoma3 IIIC v
A08 Ovary Serous adenocarcinoma3 IIIC v
A09 Ovary Serous adenocarcinoma3 IIIC v
AO10 Ovary Serous adenocarcinomaN/A IIIC v
A011 Ovary Serous adenocarcinoma3 IIIC v
A012 Ovary Serous adenocarcinoma3 IIIC v
A013 Ovary Serous adenocarcinoma3 IIIC v
AO14 Ovary Serous adenocarcinoma3 IIIC v
A015 Primary Serous adenocarcinomaN/A IIIC v
eritoneal
A016 Ovary Clear cell adenocarcinoma3 IIIC v
I A017 Ovary ~ Clear cell adenocarcinoma3 IIIC
~ ~ ~
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
A018 Ovary Clear cell adenocarcinoma3 IIIC v
AO19 Ovary Clear cell adenocarcinoma3 IIIC v
A020 Ovary Clear cell adenocarcinoma3 IIIC v
AU1 EndometriumSerous adenocarcinoma3 IVB v v
AU2 EndometriumSerous adenocarcinoma3 IVB v
AU3 EndometriumSerous adenocarcinoma3 IVB v v
AU4 EndometriumSerous adenocarcinoma3 IVB v v
AU5 EndometriumSerous adenocarcinoma3 IVB v v
AU6 EndometriumSerous adenocarcinoma3 IVB v
ll Li CL1 OVCAR3 v v
C
ne CL2 SKOV3 v v
e
T01 Ovary Serous adenocarcinoma3 IIIC v v
T02 Ovary Serous adenocarcinoma3 IV v v
T03 Ovary Serous adenocarcinoma3 IIIG v v
T04 Ovary Serous adenocarcinoma3 IIIC v v
T05 Ovary Serous adenocarcinoma3 IIIC v
T06 Ovary Serous adenocarcinoma3 IIIC v
T07 Ovary Serous adenocarcinoma3 IIIC v
Tumor T08 Ovary Serous adenocarcinoma3 IIIC v
Cell
from T~g Ovary Serous adenocarcinoma3 IIIC v
Primary
Tumors T010 Ovary Serous adenocarcinoma3 IIIG v
TG1 Ovary Granulosa Aduet-IIC v
TG2 Ovary Granulosa Aduet-IIC v v
TG3 Ovary Granulosa Aduet-IIC v v
TG4 Ovary Granulosa Adplet-IIC v v
FB1 Head & Neck v
FB2 Head & Neck v
FB3 Ovary Fibroma BenignBenignv v
F84 Ovary Serous adenocarcinoma3 IV v v
FibroblastsFB5 Ovary Serous adenocarcinoma3 IIIC v
FB6 Ovary Mixed clear cell, 3 IIIC v v
papillary and
endometrioid adenocarcinoma
FB7 Ovary Serous adenocarcinoma3 IIIC v
FB8 Ovary Serous adenocarcinoma3 IIIC v
FB9 Ovary Clear cell adenocarcinoma3 IIC v
LeukocytesLE1 Ovary Serous adenocarcinoma3 IV _
v
LE2 Ovary Serous adenocarcinoma3 IV v v
LE3 Ovary Serous adenocarcinoma3 lIIC v
LE4 Ovary Serous adenocarcinoma3 IIIC v
LE5 Ovary Serous adenocarcinoma3 IV v
LE6 Ovary Serous adenocarcinoma3 IIIC v v
LE7 Ovary Serous adenocarcinoma3 IV v v
41
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
LE8 Ovary Serous adenocarcinoma3 IIIC v
~
LE9 Ovary Serous adenocarcinoma3 IIIC v
LE10 Ovary Serous adenocarcinoma3 IIIC v
LE11 Ovary Serous adenocarcinoma3 IIIC v
LE12 Pimary Serous adenocarcinoma3 IIIC v v
eritoneal
Pimary
LE13 eritoneal Serous adenocarcinoma3 IIIC v v
LE14 Ovary Mixed clear cell, 3 IIIC v v
papillary and
endometrioid adenocarcinoma
LE15 EndometriumSerous adenocarcinoma3 IVB v v
LE16 Ovary Serous adenocarcinoma3 IC v
LE17 Ovary Serous adenocarcinoma3 IC . v
LE18 Ovary Serous adenocarcinoma3 IIIC v
LE19 Ovary Serous adenocarcinoma3 IIIC v
LE20 Ovary Serous adenocarcinoma3 IIIC v
LE21 Ovary Serous adenocarcinoma3 IIIC v
LE22 Ovary Serous adenocarcinoma3 IIIC v
LE23 Ovary Serous adenocarcinoma3 IIIC v
LE24 Primary Serous adenocarcinomaN/A IIIC v
peritoneal
LE25 Primary Serous adenocarcinoma3 IV v
eritoneal
LE26 Primary Serous adenocarcinoma3 IV v
peritoneal
Among the 77 total cell samples, 41 cell samples were examined by DNA
microarray; 63 cell samples by real-time RT-PCR; 27 cell samples by both DNA
microarray and real-time RT-PCR.
Table 2A. 126 genes up-regulated in different types of tumor cells enriched
from ovarian and uterine tumor specimens
Probe Gene Description UniGene
Bank
Common
977 at 235402 E-cadherinH. sapiens gene encoding E-cadherin
s
38324 AD000684LISCH7 liver-specific bHLH-Zip transcription
at factor
- LISCH7
575 at M93036 GA733-2 GA733-2
s
266 at L33930 CD24 CD24 (small cell lung carcinoma
s - cluster 4 Hs.375108
- antigen)
291 at J04152 M1 S1 GA733-1
s
35276at AB000712hCPE-R claudin 4 Hs.5372
42
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
34674at X58079 S100A1 S100 calcium binding proteinHs.433503
A1
35207at X76180 SCNN1A sodium channel, nonvoltage-gatedHs.446415
- 1
alpha
33904 AB000714hRVP1 claudin 3 Hs.25640
at
32821at AI762213LCN2 lipocalin 2 (oncogene 24p3)Hs.204238
38783at J05581 MCNAA mucin 1, transmembrane Hs.89603
700 at mucin 1, transmembrane
s
38784-g MCNAA mucin 1, transmembrane Hs.89603
at
J05581
38482at AJ011497CLDN7 claudin 7 Hs.278562
2011 034584 BIK BCL2-interacting killer Hs.155419
s (apoptosis-
at inducing)
-
-
37909at L34155 LamA3 laminin, alpha 3 Hs.83450
38086at AB007935KIAA0466 immunoglobulin superfamily,Hs.81234
member 3
37483 AB018287KIAA0744 histone deacetylase 9 Hs.116753
at
33572at 078722 Zpf165 zinc finger protein 165 Hs.55481
33282at 042408 LAD ladinin 1 Hs.18141
39951at L20826 PLS1 plastin 1 (I isoform) Hs.203637
36929at 017760 LAMB3 Homo sapiens laminin S B3
chain (LAM)
38051at X76220 MAL H.sapiens MAL gene axon
- 1 (and joined
CDS).
34775 AF065388TSPAN-1 tetraspan 1 Hs.38972
at
36869at X69699 PAXB paired box gene 8 Hs.308061
33323r X57348 H.sapiens mRNA (clone 9112).
at
668 at L22524 MMP7 Human matrilysin gene
s_
41610 AB011105KIAA0533 laminin, alpha 5 Hs.11669
at
34348at 078095 SPINT2 serine protease inhibitor, Hs.31439
Kunitz type, 2
1898 L24203 TRIM29 tripartite motif-containingHs.82237
at 29
40425at M57730 B61 ephrin-A1 Hs.399713
34213at AB020676KIAA0869 KIBRA protein Hs.434243
927 at J05582 MUC1 Human pancreatic mucin mRNA,
s complete cds.
-
-
41286at X77753 TROP-2 tumor-associated calcium Hs.23582
- signal
transducer 2
1585 M34309 ERBB3 v-erb-b2 erythroblastic Hs.306251
at leukemia viral
onco ene homolog 3 (avian)
889 M73780 ITGB8 integrin, beta 8 Hs.355722
at
863-g_at 004313 SERPINB5 serine (or cysteine) proteinaseHs.55279
inhibitor,
Glade B (ovalbumin), member
5
40218at 060808 CDS CDP-diacylglycerol synthase
(phosphatidate cytidylyltransferase)Hs.380684
1
35280at 215008 LAMC2 laminin, gamma 2 Hs.54451
41377 J05428 UGT2B7 UDP glycosyltransferase Hs.10319
f 2 family,
at polypeptide B7
35148at AC005954TJP3 Tight junction protein 3
37286at AB002341KIAA0343 neuronal cell adhesion moleculeHs.7912
43
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
38489at M60047 HBp17 heparin-binding growth factorHs.1690
p binding
rotein
40434at 097519 PODXL podocalyxin-like Hs.16426
31792at M20560 ANX3 annexin A3 Hs.442733
37920at 070370 Bft paired-like homeodomain transcriptionHs.84136
- factor 1
34771at AF035959PAP2-g phosphatidic acid phosphataseHs.24879
type 2C
36591at X06956 TUBA1; Human HALPHA44 gene for alpha-tubulin
330 at Tubulin, alpha1, isoform
s 44
41660at AL031588CELSR1 Cadherin
36890at AF001691PPL periplakin Hs.192233
31610at 021049 DD96 membrane-associated protein Hs.431099
17
33128s W68521 CST6 cystatin E/M Hs.139389
at
32139at Y09538 ZNF185 zinc finger protein 185 (LIMHs.16622
domain)
41352at X62822 SIAT1 sialyltransferase 1 (beta-galactosideHs.2554
- alpha-2,6-sialyltransferase)
33272_at AA829286SAA1 serum amyloid A1 Hs.332053
408 X54489 MGSA Human gene for melanoma growth
at stimulatory activity (MGSA).
-
35281at 031201 LAMC2 Human laminin gamma2 chain
gene
/1 A11A!\
41376 i at J05428 UGT2B7 UDP glycosyltransferase 2 family, Hs.10319
-- polypeptide B7
40705 at AF103905 EPAC Rap1 guanine-nucleotide-exchange factor Hs.8578
- directly activated by cAMP
35444 at AC004030 Homo sapiens DNA from chromosome
- 19. cosmid F21856
1886 053476 Wnt7a wingless-type MMTV integrationHs.72290
at site
- family, member 7A
40679at 027699 SLC6A12 solute carrier family 6 Hs.82535
- (neurotransmitter
transporter, betaine/GABA),
member 12
37533_r_atD86980 fCIAA0227fCIAA0227 protein Hs.79170
35023_at 000803 FRK fyn-related kinase Hs.89426
36292at 007225 P2RY2 purinergic receptor P2Y, Hs.339
- G-protein
coupled, 2
40217s 065887 CDS1 CDP-diacylglycerol synthaseHs.380684
- at (phosphatidate cytidylyltransferase)
- 1
1887_g_at 053476 Wnt7a wingless-type MMTV integrationHs.72290
f site
amily, member 7A
36105at M18728 CEACAM6 Carcinoma embryonic antigen-related
- cell
adhesion molecule 6
39912at AB006179HS6ST1 heparan sulfate 6-O-sulfotransferaseHs.380792
1
35577at AF027866SERPINB7 serine (or cysteine) proteinaseHs.138202
- inhibitor,
Glade B (ovalbumin), member
7
40314at AJ002309SYNGR3 synaptogyrin 3 Hs.435277
142 075308 hTAF11130TAF4 RNA polymerase II, Hs.24644
at TATA box
- binding protein (TBP)-associated
factor
41066at AF071219SCGB2A1 secretoglobin, family 2A, Hs.97644
member 1
44
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
39575 AF052143MOT8 transmembrane protein Hs.25924
at S HREW1
36010 010492 MOX1 mesenchyme homeo box 1 Hs.438
at
157 065011 PRAME preferentially expressed Hs
at antigen in 30743
melanoma .
38515 X51801 BMP7 bone morphogenetic protein
at 7 (osteogenic
Hs.170195
p rotein 1)
32558 AB021868PIAS3 protein inhibitor of activatedHs.435761
at STAT3
34703 AA151971
f at
32163 f at AA216639
31885 M64572 PTPN3 protein tyrosine phosphatase,Hs.405666
at non-
receptor type 3
1812
s at
41587_g FGF18 fibroblast growth factors.87191
at 18 H
AF075292
39579 089916 CLDN10 claudin 10 Hs.26126
at
39016 L42611 KRT6E keratin Hs.446417
r at 6 E
41790 AL031230ALDH5A1
at
39882 066035 DDP translocase of inner Hs.125565
at mitochondria)
- membrane 8 homolo A (yeast)
40717 AB001928CTSL2 cathepsin L2 H s.87417
at
40710 D86322 CLGN calmegin Hs.86368
at
881 M35198 ITGB6 integrin, beta 6 Hs.57664
at
1317 X70040 RON macrophage stimulating Hs.2942
at 1 receptor
41544 AF059617SNK serum-inducible kinase Hs.398157
at
38882 AF096870EBBP tripartite motif-containings.241305
r at 16 H
1177 Dna-binding protein Ap-2
at
1603-g L33881 PRKCI protein kinase C, iota s.355476
at H
1602 L33881 PRKCI protein kinase C, iota Hs.355476
at
32262 AL049669CGI-01 CGI-01 protein Hs.19469
at
40069 AF051850SVIL supervillin Hs.163111
at
36909 X62048 Wee1 Hu WEE1 homolog (S. pombe) Hs.249441
at
2017 M64349 CCND1 cyclin D1 (PRAD1: parathyroidHs.371468
s at adenomatosis 1 )
- -
39962 059305 CDC42BPA CDC42 binding protein Hs.18586
at kinase alpha
- (DMPK-like)
38881 AF096870EBBP tripartite motif-containingHs.241305
i at 16
41359 298265 PKP3 plakophilin 3 Hs.26557
at
39556 M96803 SPTBN1 spectrin, beta, non-erythrocyticHs.205401
at 1
37902 L13278 CRYZ crystallin, zeta (quinoneHs.83114
at reductase)
35709 AF038172FLJ11149 hypothetical protein Hs.37558
at FLJ11149
36849 090920 PARG1 PTPL1-associated RhoGAP Hs.430919
at 1
35803 S82240 RhoE ras homolog gene family,Hs.6838
at member E
182 001062 ITPR3 inositol 1,4,5-triphosphate
at receptor, type 3 Hs
77515
37199 AI760932CGI-60 dynein 2 light intermediateHs.309257
at chain
37832 AL080062DKFZp5641122DKFZP5641122 protein Hs.13024
at
168 050196 ADK adenosine kinase Hs.355533
at
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
37728 X78669 ERC-55 reticulocalbin 2, EF-hand Hs.79088
r at calcium binding
- domain
41060 M74093 CCNE1 cyclin E1 Hs.244723
at
38007 L11353 NF2 neurofibromin 2 (bilateral Hs.902
at acoustic
- neuroma)
protein tyrosine phosphatase,
41781 U22815 PPFIA1 receptor Hs.128312
at type, f polypeptide (PTPRF),
interacting
protein (liprin), alpha
1
38340 AB014555KIAA0655 huntingtin interacting protein-1-relatedHs.96731
at
40004 X91868 six1 sine oculis homeobox homologHs.54416
at 1
- (Drosophila)
37143 KIAA0361 phosphoribosylformylglycinamidineHs.88139
s at synthase (FGAR amidotransferase)
AB002359
- -
34189 D31891 KIAA0067 SET domain, bifurcated 1 Hs.345058
at
40762_g SLC16A5 solute carrier family 16 Hs.90911
at a (monocarboxylic
AA705628 t cid
ransporters), member 5
41294 AJ238246SCL Homo sapiens mRNA for sarcolectin.
at
35766 M26326 KRT18 keratin 18 Hs.406013
at
40445 AF017307ERT E74-like factor 3 (ets domain
at transcription Hs.67928
- factor, epithelial-specific
)
1681 X03635 ESR1 estrogen receptor 1
at
Table 2B. 48 genes up-regulated in different types of leukocytes enriched
from ovarian and uterine tumor specimens ,
Probe Gene BankCommon Description UniGene
931 L08177 EB12 IYmPhocyte-specific G protein-coupledHs.784
at receptor
-
40520_g_at Y00638PTPRC yp~ C Protein tyrosine phosphatase,
receptor Hs.444324
40742 M16591 HCK Protein-tyrosine kinase;
at Human hemopoietic
- cell protein-t rosine kinase
(HCK) ene
38533 at J03925ITGAM integrin, alpha M Hs.172631
s
35659 U00672 IL10RA interleukin 10 receptor, Hs.327
at alpha
33641_g AIF1 allograft inflammatory factor
at 1
Y14768
35261 W07033 GMFG glia maturation factor, gammaHs.5210
at
40019 M60830 EVI2B open reading frame; Human
at EVI2B3P gene
38796 X03084 C1QB complement component 1, q Hs.8986
at subcomponent,
- beta polypeptide
37975 X04011 CYBB cytochrome b-245, beta polypeptideHs.88974
at
37011 U49392 AIF-1 allograft inflammatory factorHs.76364
at 1
39994 D10925 HM145 chemokine (C-C motif) receptorHs.301921
at 1
34660 AI142565RNASE6 ribonuclease, RNase A family,Hs.23262
at k6
35012 M81750 MNDA myeloid cell nuclear differentiationHs.153837
at antigen
46
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
39221 AF004231M1R cl-10leukocyte immunoglobulin-likeHs.306230
at receptor,
- B, member 3
subfamily
_ _
Human IgG Fc receptor I gene,
exon 6 and
37220 M63835 CD64 complete cds.
at
-
34210 N90866 CDW52 CDW52 antigen (CAMPATH-1 Hs.276770
at antigen)
38363 W60864 TYROBP TYRO protein tyrosine kinaseHs.9963
at binding protein
35926 AF004230MIR cl-7leukocyte immunoglobulin-likeHs
s at receptor, 149924
- - subfamily B, member 1 .
36889 M33195 FCER1 Fc fragment of IgE, high Hs.433300
at G affinity I, receptor for;
- gamma polypeptide
37759 051240 LAPTm5 LYsosomal-associated multispanningHs.436200
at
- membrane_protein-5
31870 X14046 CD37 CD37 antigen Hs.153053
at
40519 Y00638 PTPRC CD45, protein tyrosine phosphatase,Hs.444324
at receptor
- _ type C
40518 Y00062 PTPRC De Cprotein tyrosine phosphatase,Hs.444324
at receptor
y
p
40331 AF035819MARCO macrophage receptor with Hs.67726
at collagenous
- structure
37918 M15395 LYAM1 integrin, beta 2 Hs.375957
at
32068 062027 HNFAG09 complement component 3a receptorHs.155935
at 1
39982 D13265 MSR1 macrophage scavenger receptorHs.436887
r_at 1
-
Human macrophage mannose
receptor
at M93221 MRC1 (MRC1) gene
36908
-
31499 FCGR3B Fc fragment of IgG, low affinityHs.372679
s at Ills, receptor
X16863
- - for (CD16)
37688 M31932 FCGR2A Fc fragment of IgG, low affinityHs.352642
f at Ila, receptor
-- for (CD32)
37148 AF025533LIR-3 leukocyte immunoglobulin-likeHs.306230
at receptor,
- subfamily B, member 3
34223 M59818 G-CSFR-1colony stimulating factor Hs.381027
at 3 receptor
- (granulocyte)
39319 020158 LCP2 lymphocyte cytosolic proteinHs.2488
at 2
39857 AF044309STX11 syntaxin 11 Hs.118958
at
36879 M63193 ECGF1 endothelial cell growth factorHs.435067
at 1 (platelet-
- derived)
1665_s_at Interleukin 18
33731 AJ130718y+LAT1 solute carrier family 7 (cationicHs.194693
at amino acid
t ransporter, y+ system), member
7
39593 AI432401FGL2 fibrinogen-like 2 Hs.351808
at
Human P-selectin glycoprotein
ligand
37541 025956 SELPLG (SELPLG) elene
at
-
37099 _ ALOXSAP arachidonate 5-lipoxygenase-activatingHs.100194
at AI806222
- protein
_ M37766 MEM-102 CD48 antigen (B-cell membraneHs.901
38006_at protein)
41723 M32578 HLA-DRB1major histocompatibility Hs.308026
s at complex, class II, DR
- - beta 3
37039 J00194 HLA-DRA major histocompatibility Hs.409805
at complex, class II, DR
- alpha
47
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
35016 M13560 CD74 la-associated gamma chain; Human
at la-
- associated invariant amma-chain gene
_
38833 X00457 HLA-DPA1 major histocompatibility complex,
at class II, DP Hs
914
.
alpha 1
_
33374 L09708 C2 complement component 2 (C2) gene
at allele b,
- exons 10 throu h 18 and complete
cds.
_
36878 M60028 HLA-DQB1 major histocompatibility complex,
f at class II, DQ Hs
409934
-- .
beta 1
Table 2C. 45 genes up-regulated in different types of fibroblasts enriched
from ovarian and uterine tumor specimens
Probe Gene Description UniGene
Bank
Common
672 at J03764 PAI1 plasminogen activator inhibitor-1
1968_g_atX76079 PDGFRA platelet-derived growth Hs.74615
a factor receptor,
lpha polypeptide
659_g L12350 THBS2 thrombospondin 2 Hs.108623
at
658 at L12350 THBS2 thrombospondin 2 Hs.108623
37671 S78569 laminin laminin, alpha 4 Hs.437536
at alpha 4
chain
39945 U09278 Seprase/ Seprase, FAP alpha Hs.436852
at FAPalpha
38420 Y14690 COL5A2 collagen, type V, alpha Hs.283393
at 2
1466 S81661 IfGF fibroblast growth factor Hs.374988
s at 7
32307 V00503 COL1A2 collagen, type I, alpha Hs.232115
s at 2
32306_g_at COL1A2 collagen, type I, alpha Hs.232115
J03464 2
32305 J03464 COL1A2 collagen, type I, alpha Hs.232115
at 2
38637 L16895 LOX Human lysyl oxidase
at
36976 D21255 osf-4 cadherin 11, type 2, Hs.443435
at
2087 D21254 osf-4 cadherin 11, type 2 Hs.443435
s at
36073 U35139 NDN necdin homolog Hs.50130
at
1147 V-Erba Related Ear-3 Protein
at
32551 U03877 S1-5 EGF-containing fibulin-likeHs.76224
at extracellular
matrix protein 1
1731 M21574 PDGFRA platelet-derived growth Hs.74615
at factor receptor,
_ - alpha polypeptide
36233 AF091242PAPSS2 3~-phosphoadenosine 5'-phosphosulfateHs.274230
at synthase 2
-
32488 X14420 COL3A1 collagen, type III, alpha Hs.443625
at 1
reversion-inducing-cysteine-rich
35234 D50406 ST15 protein Hs.388918
at k with
azal motifs
34303 AL049949FLJ90798 hypothetical protein FLJ90798Hs.28264
at
33440 U19969 TCF8 Human two-handed zinc finger
at protein
- ZEB mRNA
159 at U43142 VEGFC vascular endothelial growthHs.79141
factor C
48
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
SWIISNF related, matrix
456 066619 BAF60c associated, Hs.444445
at actin dependent regulator
of chromatin,
subfamily d, member 3
33883 AB001466EFS embryonal Fyn-associated Hs.24587
at substrate
39395 AA704137THY1 Thy-1 cell surface antigenHs.134643
at
39260 059185 MCT solute carrier family Hs.351306
at 16 (monocarboxylic
acid transporters), member
4
33240 AB029018KIAA1095 likely ortholog of mouse
at semaF Hs.177635
cytoplasmic domain associated
protein 3
35347 AF093119UPH1 EGF-containing fibulin-likeHs.381870
at extracellular
- matrix protein 2
39069 AF053944AEBP1 AE binding protein 1 Hs.439463
at
581 M61916 LAMB1 laminin, beta 1 Hs.122645
at ~
37578 D25248 AFAP actin filament associatedHs.115912
at protein
33328 W28612 HEG 49b3 Human retina cDNA Hs.433452
at randomly
primed sublibrary
1934 X94216 VEGF-C vascular endothelial growthHs.79141
s at factor C
35366 M30269 NID nidogen (enactin) Hs.356624
at
31897 053445 Doc1 downregulated in ovarian Hs.15432
at cancer 1
35832 AB029000KIAA1077 sulfatase 1 Hs.409602
at
35985 AB023137KIAA0920 A kinase (PRKA) anchor Hs.42322
at protein 2
36065 AF052389LDB1 LIM domain binding 2 Hs.4980
at
39973 047926 LEPREL2 leprecan-like 2 protein Hs.46458
at
SWIISNF related, matrix
32565 066619 BAF60c associated, Hs.444445
at actin dependent regulator
of chromatin,
subfamily d, member 3
1319 X74764 TKT discoidin domain receptorHs.71891
at family,
- member 2
1834 D38449 GPR putative G protein coupledHs.37196
at receptor
35740 AL050138DKFZp586M121elastin microfibril interfacesHs.63348
at 1
Methods And Compositions For The Determination Of Host Immunity Against
Tumor
(00085] Most CTC are dead or apoptotic in the circulation due to the
presence of host immunity to tumors, as described in co-pending PCT Patent
Application PCTlUS01/26735. The CAM-initiated blood device, the viability of
CTC,
and the plasma derived from individual donors put together an effective means
of
determining host immunity against tumor. CAM-enriched CTC often form clusters
with cytotoxic leukocytes. The cell-adhesion matrix could readily isolate such
49
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
clusters of immune and cancer cell complex from patients who might exhibit
encouraging prognosis. Furthermore, soluble components of complement system
involving in tumor cytolysis could be determined by the viability of CTC in
the
presence of autologous plasma, derived from the blood of the same subject.
Thus,
the presence of tumor cytotoxic leukocytes and soluble complement system would
~be an important indicator for host immunity.
(00086] To determine the number of viable CTC in the presence of anti-
tumor cytotoxic leukocytes and complement system, whole blood or the
mononuclear cells in the presence of 10-20% autologous plasma may be
screened by way of a CAM-initiated cell isolation device. When the number of
CTC
enriched by CAM is high in the absence of autologous plasma but low in the
presence of autologous plasma, the subject could be high in anti-tumor
immunity.
On the other hand, high levels of viable CTC that resist immune killing
detected in
the presence and absence of autologous plasma would be the strongest indicator
for patients who possess a high degree of malignancy.
Example 4
CAM Positive Isolation Of Tumor Cells From Whole Blood
[00087] An exemplar protocol that might be practiced for the
isolation of tumor cells from whole blood is set forth below:
1. Preparation of cord blood: Add 3 mL of anticoagulated cord
blood (plus 300 pg of ACD and lithium heparin) spiked with a
so
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
known number of GFP-tumor cells into each tube of the CAM
blood test unit. Place the sealed CAM-blood tube on a roller and
rotate at 5-30 cycles per minute at 37°C. Incubate for 1-3 hours
for tumor cell attachment to occur.
2. Preparation of control medium: Add 3 mL of control medium
(plus 300 pl of ACD and lithium heparin) spiked with a known
number of GFP-tumor cells into each tube of the CAM blood test
unit. Place the sealed CAM-tumor tube on a roller and rotating at
5-30 cycles per minute at 37°C. Incubate for 1-3 hours for tumor
cell attachment to occur.
3. Remove blood or medium supernatants carefully by pipetting.
Wash the tubes five times in 3 mL without disturbing the CAM
film on the inner wall Washing solution (PBS/0.1 %, BSA/10%,
ACD and lithium heparin).
4. Add 1 mL of collagenase solution into each tube of CAM blood
filtration unit that has been thoroughly washed and clear of red
cells. Place the sealed CAM-blood tube on the roller and rotate
at 5 cycles per minute at 37°C. Incubate for 10 minutes, in order
to dissolve CAM and release tumor cells into suspension.
Collagenase solution (PBS, 0.3 mM CaCl2, 0.2 pg/mL type I
collagenase [Worthington Biochemical], 25 Ng/mL DNase
[Roche]).
5. Transfer the suspension to a new Bppendorf tube. Keep on ice
for immediate immunofluorescent labeling using TRITC- anti-
CD45 for microscopy or PE-anti-CD45 for flow cytometry.
Labeled tumor cells will be counted by both microscopy and flow
cyto m etry.
si
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Example 5
Fluorescent Material Containine~ CAM Film
[00088] As invadopodic cells digest and internalize ECM
matrix, if the CAM matrix is fluorescent, then the tumor cells should
become fluorescent during the enrichment process. To accomplish
this, fluorescent TRITC or FITC-type I collagen polymers are
incorporated into the CAM substrate before it is coated on the
capture vessels. A negative identification procedure may be used to
distinguish the cancer cells from leukocytes using phycoerythrmn
(PE)- or FITC or TRITC- conjugated antibodies directed against the
leukocyte common antigen CD45.
[00089] Currently, RT-PCR and immunocytochemistry
(targeted against epithelial molecules, such as CK18 and CK20
cytokeratins, GA733 epithelial membrane antigens, Muc- 1, and pan-
epithelial antigen BerEP4) are used for confirmation of the epithelial
origin of circulating tumor cells (Ghossein et al., 1999; Molnar et al.,
2001; Racila et al., 1998; Schoenfeld et at, 1997; Soeth et al., 1997;
Vlems et at, 2002; Wharton et al., 1999). Although both methods
have high detection sensitivity and have successfully been used to
resolve circulating tumor cells in blood after differential centrifugation
enrichment (approximately 500) of the mononuclear cell fraction from
whole blood, the detection rate remains low because circulating
tumor cells represent less than 100 cells per one billion of normal
cells in blood. In addition, it is not known if this approach captures
the most critical cells, since genes responsible for metastatic
progression to the circulation remain unknown. The use of anti-
epithelial antibodies-based affinity purification would result in
significant loss of tumor cells in blood.
52
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
[00090] In contrast, a one million-fold cell enrichment of
CAM, which may be performed in one step, may achieve greater
than 40% recovery of the 3,000 viable tumor cells from 15 ?C 109
blood cells.
[00091] To further improve enrichment of the targeted
cells, a multi-step cell enrichment procedure may be employed to
recover greater than 85% of tumor cells from blood. This method
involves first a density gradient centrifugation of whole blood cells to
concentrate mononuclear cells, followed by culturing these cells on
the fluorescent CAM film for an appropriate period of time, e.g., 12 -
18 hours, in order to: (a) label the tumor cells, (b) culture the tumor
cells and less than 0.1 % of leukocytes on CAM films, and (c) stain
the CAM-captured cell population with antibodies or nucleic acid
dyes. Both individual tumor cells and clumps may be readily
observed by microscopy (whereas cell clumps often generate
difficulty in flow cytometry).
[00092) A CAM blood filtration assay may be used to
isolate viable tumor cells, endothelial progenitor cells and immune
lymphocytes in the blood of patients with cancers. CAM- captured
cells will then be seeded in parallel onto a 16-well chamber slide
(Lab-Tek, Rochester, NY) coated with FITC (or TRITC)-collagen-
based CAM and cultured for 12-18 hours. Invasive tumor cells will
ingest fluorescent CAM and become labeled with FITC (or TRITC),
whereas co-purified endothelial cells and leukocytes will remain
unlabeled. In addition to the positive identification of circulating tumor
cells, isolated cells will be tested for a negative identification by
labeling TRITC (or. FITC)-CD45 or CD31 for fluorescent microscopy
or with PE-CD45 or CD3 1 for flow cytometry.
53
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Example 6
Enumeration of Isolated Cells by Flow Cytometry
[00093] Approximately 10 to 20 mL of blood per patient
may be collected in Vacutainer tubes (Becton Dickinson, green top,
lithium heparin anticoagulant, each tube holds 7-ml). Aliquots of
freshly collected blood samples may be transferred to CAM blood
test tubes or undergoing density gradient centrifugation to obtain the
mononuclear cells, and subjected to further cell enrichment and
identification on CAM. Enumeration of viable tumor cells in blood by
flow cytometry may be accomplished based on following criteria: (a)
tumors cells visualized via their ingestion of FITC labeled collagen;
(b) PE-labeling of normal blood cells may be used as a
complementary signal to identify contaminating leukocytes; (c)
negative events of dead 7-AAD cells.
[00094] FITC-collagen- or GFP-tagged tumor cells may
be captured by CAM and coisolated normal blood cells may be post-
immuno-stained with phycoerytbrin (PE)-conjugated CD45 antibody.
As little as a 500 pl sample may be aspirated and analyzed on a
FACSCalibur flow cytometer (Becton Dickinson). Data may be
acquired in listmode by using a threshold on the fluorescence of the
nucleic acid dye 7-AAD. Criteria for multi-parameter data analysis
include: (a) size defined by forward light scatter, (b) granularity
defined by orthogonal light scatter, (c) negative events of dead 7-
AAD cells, (d) positive events of the FITC-collagen- or GFP-tumor
cells, and (e) negative events of PE-labeled CD45 mAb normal cells.
[00095] To enable the enumeration of tumor cells
present in blood at frequencies below published rates of 100,000
54
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
tumor cells in 10,000,000,000 blood cells per mL of blood (Glaves et
al., 1988; Karczewski et al., 1994) by flow cytometry, the following
may be advantageously noted:
[00096] (i) The sample volume may be reduced from
3 - 20 mL to 500 pl and total cell count from 15,000,000,000 to
1,000,000 without a significant loss of tumor cells passing through
the flow cytometer in a reasonable time period (sample flow rate = 60
pllmm).
[00097] (ii) The enriched tumor cells have to be
distinguishable from normal cells co-isolated with them. The tumor
cells may be FITC-collagen- or GFP-labeled, whereas more than
99% of the co-isolated cells should be leukocytes and may be
labeled with phycoerytbrin (PE)-conjugated anti-CD45 antibody.
[00098] (iii) Tumor cells often may exist as clumps of
50 pm to 500 pm in diameter. Cell samples derived from the CAM
blood filtration and antibody-based magnetic bead methods may be
filtered through 50 pm mesh to remove large clumps before loading
into the flow cytometer. Alternatively, when clumps are cultured on
the CAM, circulating tumor cells break out from clumps and start to
invade CAM films within 12 - 18 hours. When fluorescent CAM films
are used, tumor cells enriched by the CAM method may be labeled
with fluorescent collagen and they may be suspended by
collagenases as individual cells.
ss
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Example 7
CAM Enrichment Of Tumor Cells From A Subject For Use In Flow C ometry
[00099] (1 ). Add 3 mL of anticoagulated blood (0.3 mL of
lithium heparin plus Anticoagulant Citrate Dextrose solution USP -
ACD, Baxter Healthcare Corporation, Deerfield, Jib) into each tube of
the CAM blood filtration unit coated with FITC-labeled collagen.
Place the sealed CAM-blood tube on a roller and rotate at 5 - 30
cycles per minute at 37°C. Incubate for 1 - 3 hours for tumor cell
attachment to occur.
[000100] (2). Remove non-adherent cells and
supernatants carefully by pipetting. Wash the tube five times in 3mL
solution carefully to avoid disturbing the CAM film on the inner wall.
Washing solution (PBS/O 1 % BSA 1 % ACD and lithium heparin).
[000101] (3). Add 1 mL of the complete cell culture
medium containing 15% human serum in HEPE buffer, pH 7.4 into
each CAM blood filtration unit. Place the sealed CAM-blood tube on
a roller and rotate at 5 cycles per minute at 37°C. Incubate for 9 - 15
hours to allow labeling of tumor cells with ingested FITC-type I
collagen.
[000102] (4). Remove medium supernatants carefully by
pipetting. Wash the tubes 3 times in 3 mL PBS without disturbing the
CAM film on the inner wall.
[000103] (5). Add 1 mL of collagenase solution into each
tube of CAM blood filtration unit that has been thoroughly washed.
Place the sealed CAM-blood tube on the roller and rotate at 5 cycles
per minute at 37°C. Incubate for 10 minutes, in order to dissolve
56
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
CAM and release tumor cells into suspension. Collagenase solution
(PBS, 0.3 mM CaCl2, 0.2 ~.g/mL type I collagenase [Worthington
Biochemical], 25 ~,g/mL DNase [Roche]).
[000104] (6). Transfer the suspension, 500 pl each, to one
of two Eppendorf tubes.
[000105] (7). Staining / preparation for multi-parameter
flow cytometry: Add 100 pl of fixative solution (PBS, 6%
paraformaldehyde, pH 7.2) into the 500 pl cell suspension in an
Eppendorf tube (final fixative concentration at 1 % paraformaldehyde)
and fix at 20-25 C for 10 minutes.
[000106] (8). Spin down cell pellet a t 1,000 rpm for 1
minute. Remove fixative and wash the tube 3 times in 500 pl PBS
solution. Keep on ice and add 10 Ng/mL of PE-anti-CD45 (for
marking leukocytes) and 1 pg/mL of 7-AAD (for staining dead cells),
followed by incubation for 10 mm at 4°C in the dark.
[000107] The protocol above is specified for CTC detection. For the
detection of CEC and tumor-associated lymphocytes, PE-anti-CD3 1 and PE-anti-
CD45 could be used to mark CEC and tumor-associated lymphocytes,
respectively.
Example 8
Tumor Cells Enriched By The CAM 96-Well Cell Chamber Method
For Use In Flow Cytometrr
[000108] (1 ). Preparation of the MNC fraction by density
centrifugation: Use remaining 3-15 mL of anticoagulated blood in a
Vacutainer blood collection tube (Becton Dickinson, green top,
lithium heparin as anticoagulant, each tube holds 7-mL). The cell
57
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
pellet is spun down at 1,000 rpm and the cells are resuspended in 5
mL PBS containing 0.5 mM EDTA. The mononucleate cell (MNC)
fraction is obtained by Ficoll-Paque density centrifugation
(Pharmacia) according to manufacturer's instruction, washed in
complete culture medium containing 15% bovine serum, and
suspended in 3-15 mL of the complete medium.
[000109] (2). Culture of the MNC fraction on a CAM 96-
well chamber slide: Seed 100 pl/well of the cell suspension (also
applicable to the cells captured by other methods such as CAM and
Dynal AAMB) onto desired wells, such 8 wells of a 96-well microtiter
plate that were coated with FITC-collagen-based CAM that have
been filled with 100 pl of complete culture medium containing 15%
bovine serum and cultured in a C02 incubator at 37°C for 12-18
hours. This step labels tumor cells by assaying their ability to digest
and internalize fluorescent collagen fragments.
[000110] (3). Non-adherent cells and supernatants are
removed carefully by pipetting, and the wells are washed 2 times in
200 pl of PBS without disturbing the CAM film on the inner wall.
Non-adherent cells consist of dead tumor cells and non-tumor blood
cells in the MNC fraction. Suspended cells can be pooled and
subjected to cell isolation for CD 19 leukocytes or stem cells.
[000111] (4). Add 100 pl of collagenase solution (PBS, 0.3
mM CaCl2, 0.2 pg/mL type I collagenase [Worthington Biochemical],
25 pg/mL DNase [Roche]) into each well of the 8-well row of the 96-
well CAM blood unit that has been thoroughly washed. The
adherent cells are Incubate for 10 minutes, in order to dissolve CAM
and release bound tumor cells into suspension.
sa
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
[000112] (5). Transfer the suspension from the 8-well, 800
pl total, to Eppendorf tubes.
[000113] (6). Add 200 ~,1 of fixative solution (PBS, 10%
paraformaldehyde, pH 7.2) into the 800 pl cell suspension in an
Eppendorf tube (final fixative concentration at 2% paraformaldehyde)
and fix at 20 - 25°C for 10 minutes.
[000114] (7). Spin down cell pellet at 1,000 rpm for 1
minute, remove the fixative and wash the tube 3 times in 500 pl PBS
solution. Keep cell pellet on ice and add 10 pg/mL of PE-anti-CD45,
CD 14 and CD68 (for marking leukocytes, monocytes, macrophages)
and 1 pg/mL of 7-AAD (for staining dead cells), followed by
incubation for 10 minutes at 4°C in the dark.
[000115] The protocol above is specified for CTC detection. For the
detection of CEC and tumor-associated lymphocytes, PE-anti-CD31 and PE-anti-
CD45 could be used to mark CEC and tumor-associated lymphocytes,
respectively.
Example 9
Microscopic Characterization Of Tumor Cells Enriched BY
The CAM 16-Well Cell Chamber Method
[000116] (1 ) Preparation of the cellular and plasma
fractions by low speed. 750 rpm for 5 mm, centrifugation: Spin
down cell pellet in 3 - 7 mL of anticoagulated blood in a Vacutainer
blood collection tube (Becton Dickinson, green top, lithium heparin
as anticoagulant, each tube holds 7-ml) at 750 rpm for 5 mm or
1,000 rpm for 3 mm. Transfer the plasma from the supernatant of the
centrifuged blood, 120 pl total, to an Eppendorf tube that are filled
59
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
with 680 pl of anticoagulated complete culture medium containing
15% bovine serum [called the plasma medium: 15% plasma from a
specific donor, in 10% anticoagulant (ACD and lithium heparmn) and
75% complete culture medium]. The rest of plasma is stored in 0.5
pL aliquots.
[000117] (2) Preparation of the M7NC fraction by density
centrifugation: Cells will be resuspended in 5 mL PBS containing 0.5
mM EDTA. Mononucleate cell (MINC) fraction are obtained by Ficoll-
Paque density centrifugation (Pharmacia) according to
manufacturer's instruction, washed in complete culture medium
containing 15% bovine serum, and suspended in same volume of the
complete medium as blood prior to fractionation.
[000118] (3) Preparation of a CAM 16-well chamber slide
pre-incubated with complete culture media with and without 15%
plasma from each specific donor: Into each well of the upper 8-wells
of a 16-well chamber slide (in 96-well microtiter plate format; Lab-
Tek, Rochester, NY) coated with TRITC-collagen-based CAM, seed
100 ~I of the complete culture medium and 10% anticoagulant. Into
each well of the lower 8-wells of a 16-well chamber slide (in 96-well
microtiter plate format; Lab-Tek, Rochester, NY) coated with TRITC
collagen-based CAM, seed 100 pl of the complete culture medium
and 10% anticoagulant, and 15% individual plasma [the plasma
medium 15% plasma from a specific donor, in 10% anticoagulant
(CDA + heparin), prepared in procedure 1].
(000119] (4) Culture of the MNC fraction on a CAM 16-
well chamber slide: Seed 100 g,1 of the cell suspension (also
applicable to the cells captured by other methods such as CAM and
Dynal AAMB) onto each well of a 16-well chamber slide (in 96-well
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
microtiter plate format; Lab-Tek, Rochester, NY) coated with TRITC-
collagen-based CAM that have been filled with 100 pl of complete
culture medium containing 15% bovine serum and cultured in a C~2
incubator at 37°C for 12 - 18 hours. This step labels tumor cells by
assaying their ability to digest and internalize fluorescent collagen
fragments.
[000120] (5) Non-adherent cells and supernatants are
removed carefully by pipetting. Non-adherent cells consist of dead
tumor cells and non-tumor blood cells in the MNC fraction.
[000121] (6) Antibody and nucleic acid staining: Add 200
pl of fixative solution (PBS, 3.7% paraformaldehyde, pH 7.2) into
each well of CAM labeling chamber unit and incubate at 20 - 25°C
for 10 minutes. The fixative is removed and cells in the wells are
washed 3 times in 200 pl of PBS solution and kept on ice for
immediate immuno-labeling using blue-fluorescent Hoechst 33342
nuclear dye and green-fluorescent FITC- anti-von Willebrand factor
(marking an endothelial phenotype) for fluorescent microscopy, and
red-color APAAP- anti-ESA (cytokeratins, EMA etc epithelial
markers, hematopoietic cell markers CD45/CD14/CD68/CD19/CDB,
or other endothelial cell markers CD31, fit-1, etc.) for DIC bright field
microscopy.
[000122] The protocol above is specified for CTC detection. For the
detection of CEC and tumor-associated lymphocytes, anti-CD31 and anti-CD45
could be used to mark CEC and tumor-associated lymphocytes, respectively, and
then used to generate cRNA probes.
61
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Example 10
Tumor Cells Enriched By The CAM 96-Well Cell Chamber Method For Use In
Real-Time RT-PCR And DNA Microarray Molecular Anal ses
[000123] (1 ) Preparation of the MNC fraction by density
centrifugation [Parallel to Example 7 Protocol above]: Use remaining
3-15 mL of anticoagulated blood in a Vacutainer blood collection
tube (Becton Dickinson, green top, lithium heparin as anticoagulant,
each tube holds 7-mL). Spin down cell pellet at 1,000 rpm. Cells are
resuspended in 5 mL ~ PBS containing 0.5 mM EDTA and the
mononucleate cell (MNC) fraction is obtained by Ficoll-Paque density
centrifugation (Pharmacia) according to manufacturer's instruction,
washed in complete culture medium containing 15% bovine serum,
and suspended in 3-15 mL of the complete medium.
[000124] (2) Culture of the MNC fraction on a CAM 96-
well chamber slide: Seed 100 pl/well of the cell suspension (also
applicable to the cells captured by other methods such as CAM and
Dynal AAMB) onto the remaining 88 wells of a 96-well microtiter
plate that were coated with type I-collagen-based CAM that have
been filled with 100 NI of complete culture medium containing 15%
bovine serum and cultured in a C02 incubator at 37°C for 12 - 18
hours.
[000125] (3) Non-adherent cells and supernatants are
removed carefully by pipetting. Wash the wells 3 times in 200 pl of
PBS without disturbing the CAM film on the inner wall. Non-adherent
cells consist of dead tumor cells and non-tumor blood cells in the
MNC fraction. Suspended cells can be pooled and subjected to cell
isolation for CD 19 leukocytes or stem cells.
62
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
[000126] (4) Isolation of RNA for CAM-captured cells:
Add 10 pLlwell of Trizol reagent into each well of the 88-well row of
the 96-well CAM blood unit that has been thoroughly washed. Total
RNA is extracted using Trizol reagent (Invitrogen, Carlsbad, CA),
followed by clean up on a RNeasy spin column (Qiagen, Inc.,
Valencia, CA).
Example 11
Immunocytochemistry Using Cell Type Antibody Markers
To Validate Purity of Cell Fractions
[000127] Immunocytochemistry using cell type antibody
markers was used to validate the purity of cell fractions. The upper
two panels of FIG. 5 show immuno-cytochemical identification of
leukocytes (Leu) and tumor cells (Epi) enriched by CAM from ascites
of serous adenocarcinoma of the ovary, using antibodies directed
against CD45, a pan-leukocyte antigen (left panel, Leu, red), and
antibodies against pan-cytokeratins, epithelial antigens (right panel,
Epi, red). The lower two panels show immunocytochemical
identification of pure tumor cells enriched by CAM and followed by
antibody EpCAM positive-selection. Tumor cells labeled with
antibodies against pan-cytokeratins now predominate (left panel, Epi,
red). Note that some EpCAM antibody-Dynal beads are visible on
tumor cells. A few (2%) of the pure tumor cells were labeled with
antibodies directed against CD31 (right panel, Endo, red), an
endothelial surface antigen. Nuclei were stained blue as a universal
cell marker using Hoechst 33342 nuclear staining after
permeablizing the plasma membrane with non-ionic detergents.
(Picture size, 331 pm x 239 pm.)
63
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Example 12
Real-Time RT-PCR Anal sis
[000128] Real-time RT-PRC analysis may be used to
further elucidate the genetic basis for one or more cancers. RT-PCR
analysis may also be used to validate microarray data.
(000129] Quantitative real-time RT-PCR was used to
measure the expression of 10 genes selected from DNA microarray
clusters that were specific for the seven cell populations
representative of 63 cell samples purified (FIG. 5A). (A) Quantitative
real-time RT-PCR analysis of five genes up-regulated among the
different tumor cell types (MMP7, mucin 1, GA733-1, lipocalin 2 and
cytokeratin 18); four gene up-regulated among leukocytes (CD45,
autotaxin, C?CCR4 and SDF-1 ); one gene up-regulated among
fibroblasts (type I collagen) on all 63 cell samples. (B) Quantitative
real-time RT-PCR analysis of the ten genes differentially regulated
among the seven cell groups. Bar graphic plot is used to
demonstrate the typical gene expression patterns of different cell
groups as well as fluctuations of expression levels within and
between cell groups. For each gene, relative expression is
compared with the mean fold expression (normalized to f~-actin) of
numbers of cell samples in each group. Error bars, SE of the
means.
[000130] Of the four different types of tumor cells isolated
by a CAM-initiated cell separation device, the five up-regulated
genes were found to be highly expressed in most adenocarcinoma
cell samples enriched from ovarian and uterine tumor specimens
(FIG. 6A - 6C). Expression differences beteen different types of cell
64
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
groups for tumor cell-, leukocyte- and fibroblast-associated genes
were also seen to be similar between DNA microarray data and real-
time RT-PCR data. These results suggest that most array probe
sets are likely to accurately measure the levels of the intended
transcript within a complex mixture of transcripts.
[000131] It will be appreciated that various of the above-disclosed
and other features and functions or alternatives thereof may be desirably
combined into many other different systems or applications. Also, it will be
appreciated that various presently unforeseen or unanticipated alternatives,
modifications, variations or improvements therein may be subsequently
made by those skilled in the art which are also intended to be encompassed
by the following claims.
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
REFERENCES
Aoyaina,A. and Chen,W.-T. (1990). A 170-kDa membrane-bound protease is
associated with the expression of invasiveness by human malignant melanoma
cells. Proc. Nati. Acad. Sci. U. S. A. 87, 8296-8300.
Asahara,T., Murohara,T., Sullivan,A., Silver,M., Van der Zee,R, Li,T.,
Witzenbichler,B., Schatteman,G., and Isner,J.M. (1997). Isolation of putative
progenitor endothelial cells for angiogenesis. Science 275, 964-967.
Beitsch,P.D. and Clifford,E. (2000). Detection of carcinoma cells in the blood
of
breast cancer patients. American Journal of Surgery 180, 446-448.
Brandt,B.H., Scbmidt,H., de Angelis,G., and Zanker,K.S. (2001 ). Predictive
laboratory diagnostics in oncology utilizing blood-borne cancer cells--current
best
practice and unmet needs. Cancer Letters 162 Supp, S11-S16.
Brugger,W., Bubring,H.J., Grnnebach,F., VogeI,W., KauI,S., Muller,R,
Brummendorf,T.H., ZiegIer,B.L., Rappold,l., Brossart,P., Scheding,S., and
Kutnz,L. (1999). Expression of MUC-I epitopes on normal bone marrow:
implications for the detection of micrometastatic tumor cells. J. Chin. Oncol.
17,
1535-1544.
66
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Chadha M, Chabon AB, Friedmann P, and Vikram B (1994). Predictors of axillary
lymph node metastases in patients with TI breast cancer. A multivariate
analysis.
Cancer 73, 350-353.
Chen,J.M. and Chen,W.T. (1987). Fibronectin-degrading proteases from the
membranes of transformed cells. Cell. 48, 193-203.
Chen,W.T. (1989). Proteolytic activity of specialized surface protrusions
formed at
rosette contact sites of transformed cells. J. Exp. Zool. 251, 167-185.
Chen,W.T. (1996). Proteases associated with invadopodia, and their role in
degradation of extracellular matrix. Enzyme Protein 49, 59-71.
Chen,W.T., Lee,C.C., (ioldstein,L., Bemier,S., Liu,C.H., Lin,C.Y., Yeh,Y.,
Monsky,W.L., KeIIy,T., Dai,M., and Mueller,S.C. (1994b). Membrane proteases as
potential diagnostic and therapeutic targets for breast malignancy. Breast
Cancer
Res. Treat. 31, 217-226.
Chen,W.T., Olden,K~, Bernard,B.A., and Chu,F.F. (1984). Expression of
transformation-associated protease(s) that degrade fibronectin at cell contact
sites. J. Cell Biol 98, 1546-1555.
67
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Chen,W.T. and Wang,J.Y. (1999). Specialized surface protrusions of invasive
cells, invadopodia and lamelhipodia, have differential MTI-MMP, MMP-2, and
TIMP-2 localization. [Review] (52 refs]. Annals of the New York Academy of
Sciences 878, 36 1-371:
Chen,W.T., Yeh,Y., and Nakahara,H. (1994a). An in vitro cell invasion assay:
determination of cell surface proteolytic activity that degrades extracellular
matrix.
J. Tiss. Cult. Meth. 76, 177-181.
Compton,C.C. (2003). Colorectal Carcinoma: Diagnostic, Prognostic, and
Molecular Features. Mod Pathol 76, 376.
Feezor,R.J., Copeland,E.M., III, and Hochwald,S.N. (2002). Significance of
Micrometastases in Colorectal Cancer. Ann Surg Oncol 9, 944-953.
Fehm,T., Sagalowsky,A., Clifford,E., Beitsch,P., Saboorian,H., Euhus,D.,
Meng,S.,
Morrison,L., Tucker,T., Lane,N., Ghadimi,B.M., Hesehneyer-Haddad,K., Ried,T.,
Rao,C., and Ubr,J. (2002). Cytogenetic Evidence That Circulating Epithelial
Cells
in Patients with Carcinoma Are Malignant. Clinical Cancer Research 8, 2073.
Flatmnark,K., Bjornland,K., Johannessen,H.O., Hegstad,E., Rosales,R,
HarkIau,L.,
Solhaug,J.H., Faye,R S., Soreide,0., and Fodstad,0. (2002). linmunomagnetic
68
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Detection of Micrometastatic Cells in Bone Marrow of Colorectal Cancer
Patients.
Clinical Cancer Research 8,444-449.
Ghersi,G., Dong,H., Goldstein,L.A., Yeh,Y., Hakkinen,L., Larjava,H.S., and
Chen,W.T. (2002). Regulation of fibroblast migration on collagenous matrix by
a
cell surface peptidase complex. J. Biol. Chem. 277, 29231-29241.
Ahossein,RA., Bhattacharya,S., and Rosai,J. (1999). Molecular detection of
micrometastases and circulating tumor cells in solid tumors. Chin Cancer Res
5,
1950-1960.
Glaves,D. (1983). Correlation between circulating cancer cells and incidence
of
metastases. British Journal of Cancer 48, 665-673.
Glaves,D., Huben,RP., and Weiss,L. (1988). Haematogenous dissemination of
cells from human renal adenocarcinomas. British Journal of Cancer 57, 32-35.
Goldstein,L.A. and Chen,W.T. (2000). Identification of an alternatively
spliced
seprase mRNA that encodes a novel intracellular isoform. J. Biol. Chem. 275,
2554-2559.
69
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Goldstein,L.A. . . . and Chen,W.T. (1997). Molecular cloning of seprase: A
seine
integral membrane protease from human melanoma. Biochimica et Biophysics
Acts 7367, 11-19.
Gross,H., Verwer,B., Houck,D., Hoffinan,RA., and Recktenwald,D. (1995). Model
Study Detecting Breast Cancer Cells in Peripheral Blood Mononuclear Cells at
Frequencies as Low as 10-7. PNAS 92, 537-547.
Gulati,S.C. (1993). Questioning the role of purging in BMT. [Review] [11
refs].
Stem Cells 7 7, 249-251.
Gulati,S.C. and Acaba,L. (1993). Rationale for purging in autologous stem cell
transplantation. [Review] [17 refs]. Journal of Hematotherapy 2, 467-471.
HiII,J.M., Zalos,G., Halcox,J.P.J., Schenke,W.H., Waclawiw,M.A., Quyyumi,A.A.,
and FinkeI,T. (2003). Circulating Endothehial Progenitor Cells, Vascular
Function,
and Cardiovascular Risk. N Engl J Med 348, 593-600.
Karczewski,D.M., Lema,M.J., and Giaves,D. (1994). The efficiency of an
autotransfusion system for tumor cell removal from blood salvaged during
cancer
surgery. Anesthesia & Analgesia 78, 1131-1135.
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
KeIIy,T., Mueller,S.C., Yeh,Y., and Chen,W.T. (1994). Invadopodia promote
proteolysis of a wide variety of extracellular matrix proteins. J.~ Cell
Physiol. 158,
299-308.
Koch,M., Weitz,J., KienIe,P., Benner,X, Willeke,F., Lebnert,T., Herfarth,C.,
and
Knebel Doeberitz,M. (2001 ). Comparative Analysis of Tumor Cell Dissemination
in
Mesenteric, Central, and Peripheral Venous Blood in Patients With Colorectal
Cancer. Arch Suing 136, 85.
Liefers,G.J., Cleton-Jansen,A.M., van de VeIde,C.J., Hermans,J., van
Krieken,J.H., Comeisse,C.J., and Tollenaar,RA. (1998). Micrometastases and
survival in stage II colorectal cancer. N. Engl. J Med. 339, 223-228
Luzzi,K.J., MacDonaId,I.C., Schinidt,E.E., Kerkvliet,N., Morris, VL,
Chambers,A.F.,
and Groom,A.C. (1998). Multistep nature of metastatic inefficiency: dormancy
of
solitary cells after successful extravasation and limited survival of early
micrometastases. Am J Pathol 153, 865-873.
Matsunami,K, Nakamura,T., Oguma,H., Kitamura,Y., and Takasaki,K. (2003).
Detection of Bone Marrow Micrometastasis in Gastric Cancer Patients by
Immunomagnetic Separation. Ann Suing Oncol 10, 171.
71
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Molnar,B., Ladanyi,A., Tanko,L., Sreter,L., and Tulassay,~. (2001 ).
Circulating
Tumor Cell Clusters in the Peripheral Blood of Colorectal Cancer Patients.
Clinical
Cancer Research 7, 4030.
Monsky,W.L., KeIIy,T., Lin,C.Y., Yeh,Y., Stetler-Stevenson,W.G., Mueller,S.C.,
and Chen,W.-T. (1993). Binding and localization of M(r) 72,000 matrix
metalloproteinase at cell surface invadopodia. Cancer Res. 53, 3 159-3164.
Monsky,W.L., Lin,C.-Y., Aoyama,N, KeIIy,T., Mueller,S.C., Akiyama,S.K, and
Chen,W.-T. (1994). A potential marker protease of invasiveness, seprase, is
localized on invadopodia of human malignant melanoma cells. Cancer Res. 54,
5702-5710.
Mueller,S.C. and Chen,W.T. (1991 ). Cellular invasion into matrix beads:
localization of beta 1 integrins and fibronectin to the invadopodia. J. Cell
Sci 99, 2
13-225.
Mueller,S.C., Ghersi,G., Akiyama,S.K., Sang,Q.X., Howard,L., Pineiro-
Sanchez,M., Nalcahara,H., Yeh,Y., and Chen,W.-T. (1999). A novel protease-
docking function of integrin at invadopodia. J. Biol. Chem. 274, 24947-24952.
7a
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Mueller,S.C., Yeh,Y., and Chen,W.-T. (1992). Tyrosine phosphorylation of
membrane proteins mediates cellular invasion by transformed cells. J. Cell
Biol
119, 1309-1325.
Nakahara,H., Howard,L., Thompson,E.W., Sato,H., Seiki,M., Yeh,Y., and
Chen,W.T. (1997). Transniembrane/ cytoplasmic domain-mediated membrane
type 1-matrix metalloprotease docking to invadopodia is required for cell
invasion.
Proc. Natl. Acad. Sci. U. S. A. 94, 7959-7964.
Nakahara,H., Mueller,S.C., Nomizu,M., Yamada,Y., Yeh,Y., and Chen,W.T.
(1998). Activation ofbetal integrin signaling stimulates tyrosine
phosphorylation
ofp190RhoGAP and membrane-protrusive activities at invadopodia. Journal of
Biological Chemistry 273, 9-12.
Nakahara,H., Nomizu,M., Akiyama,S.K, Yamada,Y., Yeh,Y., and Chen,W.-T.
(1996). A mechanism for regulation of melanoma invasion. Ligation of
alpha6betal
integrin by laminin G peptides. J. Biol. Chem. 271, 27221-27224.
Nomoto,S., Nakao,A., Ando,N., Takeda,S., Kasai,Y., Inoue,S., Kaneko,T., and
Takagi,H. (1998). Clinical application of K-ras oncogene mutations in
pancreatic
carcinoma: detection of micrometastases. Seminars in Surgical Oncology 15, 40-
46.
73
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Olivier, et al. (1999). A rapid single-laser flow cytometric method for
discrimination
of early apoptotic cells in a heterogenous cell population. Br J Haematol 104,
530-
537.
PanteI,K., Cote,RJ., and Fodstad,O. (1999). Detection and clinical importance
of
micrometastatic disease. J. Natl. Cancer Inst. 91, 1113-1124.
Pavlaki,M., Cao,J., Hymowitz,M., Chen,W.T., Bahou,W., and Zucker,S. (2002). A
Conserved Sequence within the Propeptide Domain of Membrane Type 1 Matrix
Metalloproteinase Is Critical for Function as an Intramolecular Chaperone.
Journal
of Biological Chemistry 277, 2740-2749.
Peck,K., Sher,Y.P., Shih,J.Y., Roffier,S.R, Wu,C.W., and Yang,P.C. (1998).
Detection and quantitation of circulating cancer cells in the peripheral blood
of lung
cancer patients. Cancer Res. 58, 2761-2765.
Pineiro-Sanchez,M.L., Goldstein,L.A., Dodt,J., Howard,L., Yeh,Y., Tran,H.,
Argraves,W.S., and Chen,W.T. (1997). Identification of the 170-kDa melanoma
membrane-bound gelatinase (seprase) as a seine integral membrane protease. J.
Biol. Chem. 272, 7595-7601; Correction (1998) J. Biol. Chem. 273, 13366.
Racila,E., Euhus,D., Weiss,A.J., Rao,C., McConneII,J., Terstappen,L.W., and
Uhr,J.W. (1998). Detection and characterization of carcinoma cells in the
blood.
74
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Proceedings of the National Academy of Sciences of the United States of
America
95, 45894594.
RiII,D.R., Santana,V.M., Roberts,W.M., Nilson,T., Bowman,L.C., Krance,R.A.,
HesIop,H.E., Moen,RC., JihIe,J.N., and Brenner,M.K. (1994). Direct
demonstration
that autologous bone marrow transplantation for solid tumors can return a
multiplicity of tumorigenic cells. Blood 84, 380-383.
Ross,R. (1993). The pathogenesis of atherosclerosis: a perspective for the
1990s
Nature 352, 80 1-809.
Sabile,A., Louha,M., Bonte,E., Poussin,K., Vona,G., Mejean,A., Chretien,Y.,
Bougas,L., Lacour,B., Capron,F., Roseto,A., Brechot,C., and Paterlini-
Binechot,P.
(1999). Efficiency of Ber-EP4 antibody for isolating circulating epithelial
tumor cells
before RT-PCR detection. American Journal of Clinical Pathology 112, 171-178.
Saga,S., Chen,W.T., and Yamada,K.M. (1988). Enhanced fibronectin receptor
expression in Rous sarcoma virus- induced tumors. Cancer. Res. 48, 5510-5513.
Schoenfeld,A., Kruger,ICH., Gomm,J., Sinnett,H.D., Gazet,J.C., Sacks,N.,
Bender,
HG, Luqmani,Y., and Coombes,RC. (1997). The detection of micrometastases in
the peripheral blood and bone marrow of patients with breast cancer using
7s
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
immunobistochemistry and reverse transcriptase polymerase chain reaction for
keratin 19. European Journal of Cancer 33, 854-861.
Soeth,E., Vogel,l., Roder,C., JuIiI,H., Marxsen,J., Kruger,U., Henne-Bruns,D.,
Kremer,B., and Kalthoff,H. (1997). Comparative analysis of bone marrow and
venous blood isolates from gastrointestinal cancer patients for the detection
of
disseminated tumor cells using reverse transcription PCR. Cancer Research 57,
3106.
Suarez-Quian,C.A., Goldstein,S.R., Pohida,T., Smith,P.D., Peterson,J.l.,
Welluer,
E, Ghany,M., and Bonner,RF. (1999). Laser capture microdissection of single
cells
from complex tissues. BioTechniques 26, 328-335.
Szmitko,P.E., Fedak,P.W.M., WeiseI,RD., Stewart,D.J., Kutryk,M.J.B., and
Verma,S: (2003). Endothelial Progenitor Cells: New Hope for a Broken Heart.
Circulation 707, 3093-3100.
Vlems,F.A., Diepstra,J.H.S., Cornelissen,I.M.H.A., Ruers,T.J.M.,
Ligtenberg,M.J.L., Punt,C.J.A., Van Krieken,J.H.J.M., Wobbes,T., and Van
Muijen,G.N.P. (2002). Limitations of cytokeratin 20 RT-PCR to detect
disseminated tumour cells in blood and bone marrow of patients with colorectal
cancer: expression in controls and dowuregulation in tumour tissue. Mol Pathol
55,
156.
76
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Vona,G., Sabile,X, Louha,M., Sitruk,V., Romana,S., Schutze,K., Capron,F.,
Franco,D., Pazzagli,M., Vekemans,M., Lacour,B., Brechot,C., and Paterlini-
Brechot,P. (2000). Isolation by Size of Epithelial Tumor Cells : A New Method
for
the Immunomorphological and Molecular Characterization of Circulating Tumor
Cells. Am J Pathol 756, 57-63.
WaIsh,J.M.E. and Terdiman,J.P. (2003). Colorectal Cancer Screening: Clinical
Applications. JAMA: The Journal of the American Medical Association 289, 1297.
Wang,~.P., Eisenberger,MA, Carducci,MA, Partin,A.W., Scher, Hi, and Ts'o,P.O.
(2000). Identification and characterization of circulating prostate carcinoma
cells.
Cancer 88, 2787-2795.
Weihrauch,M.R (2002). Immunomagnetic Enrichment and Detection of
Micrometastases in Colorectal Cancer: Correlation With Established Clinical
Parameters. [Report]. J. Chin. Oncol. 20, 4338-4343.
Weitz,J., KienIe,P., Magener,A., Koch,M., Schrodel,A., Willeke,F.,
Autschbach,F.,
Lacroix,J., Lehnert,T., Herfarth,C., and Doeberitz,M.v.K. (1999). Detection of
Disseminated Colorectal Cancer Cells in Lymph Nodes, Blood and Bone Marrow.
Clinical Cancer Research 5, 1830.
77
CA 02544373 2006-05-O1
WO 2005/043121 PCT/US2004/036177
Wharton,R.Q., Jonas,S.K., Glover,C., Khan,Z.A.J., Klokouzas,A., Quinn,H.,
Henry,M., and AIIenMersh,T.G. (1999). Increased Detection of Circulating Tumor
Cells in the Blood of Colorectal Carcinoma Patients Using Two Reverse
Transcription-PCR Assays and Multiple Blood Samples. Clinical Cancer Research
5, 4158.
Wilhelm MC, Edge SB, Cole DD, deParedes E, and Frierson HF Jr (1991).
Nonpalpable invasive breast cancer. Am Surg. 213, 600-603.
~ucker,S., Cao,J., and Chen,W.T. (2000). Critical appraisal of the use of
matrix
metalloproteinase inhibitors in cancer treatment. Oncogene 79, 6642-6650.
~ukowska-Grojec,Z., Karwatowska,P., Rose,W., Rone,J., Movafagh,S., Ji,H.,
Yeh,Y., Chen,W.-T., Kleinman,H.K., Grouzrnann,E., and Grant,D.S. (1998).
Neuropeptide Y: a novel angiogenic factor from the sympathetic nerves and
endothelium. Circ. Res. 83, 187-195.
78