Note: Descriptions are shown in the official language in which they were submitted.
DEMANDE OU BREVET VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVET COMPREND
PLUS D'UN TOME.
CECI EST LE TOME 1 DE 4
CONTENANT LES PAGES 1 A 200
NOTE : Pour les tomes additionels, veuillez contacter le Bureau canadien des
brevets
JUMBO APPLICATIONS/PATENTS
THIS SECTION OF THE APPLICATION/PATENT CONTAINS MORE THAN ONE
VOLUME
THIS IS VOLUME 1 OF 4
CONTAINING PAGES 1 TO 200
NOTE: For additional volumes, please contact the Canadian Patent Office
NOM DU FICHIER / FILE NAME
NOTE POUR LE TOME / VOLUME NOTE:
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
METHODS FOR IDENTIFYING RISK OF OSTEOARTHRITIS AND
TREATMENTSTHEREOF
Field of the Invention
[0001] The invention relates to genetic methods for identifying risk of
osteoarthritis and treatments
that specifically target such diseases.
Back r~ ound
[0002] Osteoarthritis (OA) is a chronic disease usually affecting weight-
bearing synovial joints.
There are approximately 20 million Americans affected by OA and it is the
leading cause of disability in
the United States. In addition to extensive human suffering, OA also accounts
for nearly all knee
replacements and more than half of all hip replacements in the United States.
Despite its prevalence,
OA is poorly understood and there are few treatments available besides anti-
inflammatory drugs and
joint replacement.
[0003] Osteoarthritis (OA) is a disease caused by degeneration of articular
cartilage and subsequent
joint deformation. In addition to risk factors like body weight, joint injury
and age, there is a strong
hereditary component to OA, reflected by high heritability estimates from twin
studies. So far, few of
the genes responsible for this genetic component have been identified.
Summary
[0004] It has been discovered that certain polymorphic variations in human
genomic DNA are
associated with osteoarthritis. In particular, polymorphic variants in loci
containing PADI2, APOB;'
ILIRL2, ILIRL1, WASPIP, ADAMTS2, BhES, TM7SF3, L~XLl, CASPR~ ahd APOL3 regions
and
other regions in Table B of human genomic DNA have been associated with risk
of osteoarthritis.
[0005] Thus, featured herein are methods for identifying a subject at risk of
osteoarthritis and/or a
risk of osteoarthritis in a subject, which comprise detecting the presence or
absence of one or more
polymorphic variations associated with osteoarthritis in or around the loci
described herein in a human
nucleic acid sample. In an embodiment, two or more polymorphic variations are
detected in two or
more regions of which one is the PADI2, APOB, IL1RL2, ILIRLl, WASPIP, ADAMTS2,
BYES,
TM7SF3, LO~YZl, CASPR4 or APOL3 region or other region in Table B. In certain
embodiments, 3 or
more, or 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 or 20 or
more polymorphic variants are
detected.
[0006] Also featured are nucleic acids that include one or more polymorphic
variations associated
with occurrence of osteoarthritis, as well as polypeptides encoded by these
nucleic acids. In addition,
provided are methods for identifying candidate therapeutic molecules for
treating osteoarthritis, as well
as methods for treating osteoarthritis in a subject by identifying a subject
at risk of osteoarthritis and
treating the subject with a suitable prophylactic, treatment or therapeutic
molecule.
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0007] Also provided are compositions comprising a cell from a subject having
osteoarthritis or at
risk of osteoarthritis and/or a PADI2, APOB, ILIRL2, IL1RL1, WASPIP, ADAMTS2,
BhES, TM7SF3,
LOXLl, CASPR4 or APOL3 nucleic acid or other nucleic acid referenced in Table
B, with a RNAi,
siRNA, antisense DNA or RNA, or ribozyme nucleic acid designed from a PADI2,
APOB, ILIRL2,
ILIRLl, WASPIP, ADAMTS2, BYES, TM7SF3, LOXLl, CASPR4 orAPOL3 nucleotide
sequence or
other nucleotide sequence referenced in Table B. In an embodiment, the RNAi,
siRNA, antisense DNA
or RNA, or ribozyme nucleic acid is designed from a PADI2, APOB, ILIRL2,
ILIRLl, WASPIP,
ADAMTS2, BT~ES, TM7SF3, LOXLl, CASPR4 or APOL3 nucleotide sequence or other
nucleotide
sequence referenced in Table B that includes one or more polymorphic
variations associated with
osteoarthritis, and in some instances, specifically interacts with such a
nucleotide sequence. Further,
provided are arrays of nucleic acids bound to a solid surface, in which one or
more nucleic acid
molecules ofthe array have a PADI2, APOB, ILIRL2, ILIRL1, WASPIP, ADAMTS2,
BT~ES, TM7SF3,
LOXL1, CASPR4 or APOL3 nucleotide sequence or other nucleotide sequence
referenced in Table B, or
a fragment or substantially identical nucleic acid thereof, or a complementary
nucleic acid of the
foregoing. Featured also are compositions comprising a cell from a subject
having osteoarthritis or at
risk of osteoarthritis and/or a PADI2, .APOB, IL1RL2, ILIRL1, WASPIP, ADAMTS2,
BT~ES, TM7SF3,
LOXLl, CASPR4 or APOL3 polypeptide or other polypeptide referenced in Table B,
with an antibody
that specifically binds to the polypeptide. In an embodiment, the antibody
specifically binds to an
epitope in the polypeptide that includes a non-synonymous amino acid
modification associated with
osteoarthritis (e.g., results in an amino acid substitution in the encoded
polypeptide associated with
osteoarthritis). In certain embodiments, the antibody selectively binds to an
epitope in the PADl2,
APOB, ILIRL2, IL1RL1, WASPIP, ADAMTS2, BITES, TM7SF3, LOXL1, CASPR4 orAPOL3
polypeptide, or other polypeptide referenced in Table B, having an amino acid
associated with
osteoarthritis. Thus, featured is an antibody that binds an epitope having an
amino acid encoded by
rs1367117, rs1041973 and/or rs398829, such as a isoleucine or threonine
encoded by rs1367117 (e.g., a
threonine at position 98 in anAPOB polypeptide), a glutamic acid or alanine
encoded by rs1041973
(e.g., an alanine at position 78 in a ILIRLI polypeptide), a valine or
isoleucine encoded by rs398829
(e.g., a valine at position 245 in a AD~1MTS2 polypeptide), at the
corresponding position in the
polypeptide.
Brief Description of the Drawings
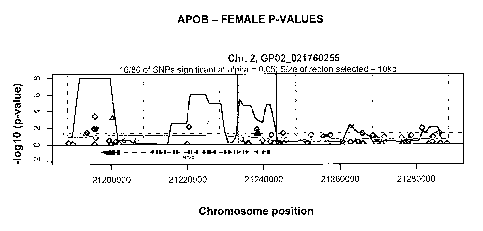
[0008] Figures lA-1J show proximal SNPs in a 100-kb window in PADI2, APOB,
IL1RL2,
ILIRLI, WASPIP, ADAMTS2, BhES, TM7SF3, LOXLl, CASPR4 and APOL3 regions of
genomic DNA,
respectively, that were compared between pools of cases and controls. The x-
axis corresponds to their
chromosomal position and the y-axis to the test P-values (shown on the -logio
scale). The continuous
bold line presents the results of a goodness-of fit test for an excess of
significance (compared to 0.05) in
a 10 kb sliding window assessed at 1 kb increments.
2
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
Detailed Description
[0009] It has been discovered that polyrnorphic variants in a locus containing
a PADI2, APOB,
ILIRL2, ILIRL1, WASPIP, ADAMTS2, BT~ES, TM7SF3, LOXLI, CASPR4 orAPOL3 region
are
associated with occurrence of osteoarthritis in subjects. Thus, detecting
genetic determinants associated
with an increased risk of osteoarthritis occurrence can lead to early
identification of a predisposition to
osteoarthritis and early prescription of preventative measures. Also,
associating a PADI2, APOB,
ILIRL2, ILIRLl, WASPIP, ADAMTS2, BITES, TM7SF3, LOXLl, CASPR4 orAPOL3
polymorphic
variant and other variants referenced in Table B with osteoarthritis has
provided new targets for
screening molecules useful in treatments of osteoarthritis.
Osteoarthritis and Sample Selection
[0010] Osteoarthritis (OA), or degenerative joint disease, is one of the
oldest and most common
types of arthritis. It is characterized by the breakdown of the joint's
cartilage. Cartilage is the part of the
joint that cushions the ends of bones, and its breakdown causes bones to rub
against each other, causing
pain and loss of movement. Type II collagen is the main component of
cartilage, comprising 15-25% of
the wet weight, approximately half the dry weight, and representing 90-95% of
the total collagen
content in the tissue. It forms fibrils that endow cartilage with tensile
strength (Mayne, R. Arthritis
Rhuem. 32:241-246 (1989)).
[0011] Most commonly affecting middle-aged and older people, OA can range from
very mild to
very severe. It affects hands and weight-bearing joints such as knees, hips,
feet and the back. Knee OA
can be as disabling as any cardiovascular disease except stroke.
[0012] Osteoarthritis affects an estimated 20.7 million Americans, mostly
after age 45, with
women more commonly affected than men. Physicians make a diagnosis of OA based
on a physical
exam and history of symptoms. X-rays are used to confirm diagnosis. Most
people over 60 reflect the
disease on X-ray, and about one-third have actual symptoms.
[0013] There are many factors that can cause OA. Obesity may lead to
0steoarthritis of the knees.
In addition, people with joint injuries due to sports, work-related activity
or accidents may be at
increased risk of developing OA.
[0014] Genetics has a role in the development of OA too. Some people may be
born with defective
cartilage or with slight defects in the way that joints fit together. As a
person ages, these defects may
cause early cartilage breakdown in the joint or the inability to repair
damaged or deteriorated cartilage
in the joint.
[0015] Inclusion or exclusion of samples for an osteoarthritis pool may be
based upon the
following criteria: ethnicity (e.g., samples derived from an individual
characterized as Caucasian);
parental ethnicity (e.g., samples derived from an individual of British
paternal and maternal descent);
relevant phenotype information for the individual (e.g., case samples derived
from individuals
diagnosed with specific knee, hand or hip osteoarthritis (OA); case samples
recruited from an OA knee
replacement clinic). Control samples may be selected based on relevant
phenotype information for the
3
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
individual (e.g,, derived from individuals free of OA at several sites (knee,
hand, hip etc)); and no
family history of OA and/or rheumatoid arthritis. Additional phenotype
information collected for both
cases and controls may include age of the individual, gender, family history
of OA, diagnosis with
osteoarthritis (joint location of OA (e.g., knee, hips, hands and spine), date
of primary diagnosis, age of
individual as of primary diagnosis), knee history (current symptoms, any major
knee injury,
menisectomy, knee replacement surgery, age of surgery), HRT history,
osteoporosis diagnosis.
[0016] Based in part upon selection criteria set forth above, individuals
having osteoarthritis can be
selected for genetic studies. Also, individuals having no history of
osteoarthritis often are selected for
genetic studies, as described hereafter.
Pol~phic Variants Associated with Osteoarthritis
[0017] A genetic analysis provided herein linked osteoarthritis with
polymorphic variant nucleic
acid sequences in the human genome. As used herein, the term "polymorphic
site" refers to a region in
a nucleic acid at which two or more alternative nucleotide sequences are
observed in a significant
number of nucleic acid samples from a population of individuals. A polymorphic
site may be a
nucleotide sequence of two or more nucleotides, an inserted nucleotide or
nucleotide sequence, a
deleted nucleotide or nucleotide sequence, or a microsatellite, for example. A
polymorphic site that is
two or more nucleotides in length may be 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15 or more, 20 or more,
30 or more, 50 or more, 75 or more, 100 or more, 500 or more, or about 1000
nucleotides in length,
where all or some of the nucleotide sequences differ within the region. A
polymorphic site is often one
nucleotide in length, which is referred to herein as a "single nucleotide
polymorphism" or a "SNP."
[0018] Where there are two, three, or four alternative nucleotide sequences at
a polymorphic site,
each nucleotide sequence is referred to as a "polymorphic variant" or "nucleic
acid variant." Where two
polymorphic variants exist, for example, the polymorphic variant represented
in a minority of samples
from a population is sometimes referred to as a "minor allele" and the
polymorphic variant that is more
prevalently represented is sometimes referred to as a "major allele." Many
organisms possess a copy of
each chromosome (e.g., humans), and those individuals who possess two major
alleles or two minor
alleles are often referred to as being "homozygous" with respect to the
polymorphism, and those
individuals who possess one major allele and one minor allele are normally
referred to as being
"heterozygous" with respect to the polymorphism. Individuals who are
homozygous with respect to one
allele are sometimes predisposed to a different phenotype as compared to
individuals who are
heterozygous or homozygous with respect to another allele.
[0019] In genetic analysis that associate polymorphic variants with
osteoarthritis, samples from
individuals having osteoarthritis and individuals not having osteoarthritis
often are allelotyped and/or
genotyped. The term "allelotype" as used herein refers to a process for
determining the allele frequency
for a polymorphic variant in pooled DNA samples from cases and controls. By
pooling DNA from each
group, an allele frequency for each SNP in each group is calculated. These
allele frequencies are then
compared to one another. The term "genotyped" as used herein refers to a
process for determining a
4
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
genotype of one or more individuals, where a "genotype" is a representation of
one or more
polymorphic variants in a population.
[0020] A genotype or polymorphic variant may be expressed in terms of a
"haplotype," which as
used herein refers to two or more polymorphic variants occurring within
genomic DNA in a group of
individuals within a population. For example, two SNPs may exist within a gene
where each SNP
position includes a cytosine variation and an adenine variation. Certain
individuals in a population may
carry one allele (heterozygous) or two alleles (homozygous) having the gene
with a cytosine at each
SNP position. As the two cytosines corresponding to each SNP in the gene
travel together on one or
both alleles in these individuals, the individuals can be characterized as
having a cytosine/cytosine
haplotype with respect to the two SNPs in the gene.
[0021] As used herein, the term "phenotype" refers to a trait which can be
compared between
individuals, such as presence or absence of a condition, a visually observable
difference in appearance
between individuals, metabolic variations, physiological variations,
variations in the function of
biological molecules, and the like. An example of a phenotype is occurrence of
osteoarthritis.
[0022] Researchers sometimes report a polymorphic variant in a database
without determining
whether the variant is represented in a significant fraction of a population.
Because a subset of these
reported polymorphic variants are not represented in a statistically
significant portion of the population,
some of them are sequencing errors and/or not biologically relevant. Thus, it
is often not known
whether a reported polymorphic variant is statistically significant or
biologically relevant until the
presence of the variant is detected in a population of individuals and the
frequency of the variant is
determined. Methods for detecting a polymorphic variant in a population are
described herein,
specifically in Example 2. A polymorphic variant is statistically significant
and often biologically
relevant if it is represented in 5% or more of a population, sometimes 10% or
more, 15% or more, or
20% or more of a population, and often 25% or more, 30% or more, 35% or more,
40% or more, 45% or
more, or 50% or more of a population.
[0023] A polymorphic variant may be detected on either or both strands of a
double-stranded
nucleic acid. Also, a polymorphic variant may be located within an intron or
exon of a gene or within a
portion of a regulatory region such as a promoter, a S' untranslated region
(UTR), a 3' UTR, and in
DNA (e.g., genomic DNA (gDNA) and complementary DNA (cDNA)), RNA (e,g., mRNA,
tRNA, and
rRNA), or a polypeptide. Polymorphic variations may or may not result in
detectable differences in
gene expression, polypeptide structure, or polypeptide function.
[0024] It was determined that polymorphic variations associated with an
increased risk of
osteoarthritis existed in SEQ ID NO: 1-13 or a nucleotide sequence referenced
in Table B. In certain
embodiments, polymorphic variants at positions rs910223, rs1367117, rs1024791,
rs1041973,
rs1465621, rs398829, rs1018810, rs1484086, rs242392, rs8818, rs1395486,
rs512294 and/or rs132659
in the human genome were associated with an increased risk of osteoarthritis,
and in specific
embodiments, the corresponding allele in the right-most column in Table B for
each position is
associated with an increased ride of osteoarthritis. In other embodiments
polymorphic variants at
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
positions rs1367117, rs1041973 and rs398829 were associated with an increased
risk of osteoarthritis,
and in specific embodiments, a threonine encoded by rs1367117, an alanine
encoded by rs1041973, and
a valine encoded by rs398829 were associated with an increased risk of
osteoarthritis.
[0025] Polymorphic variants in and around the APOB locus were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
2 selected from the
group consisting of 238, 294, 295, 347, 1425, 4891, 5087, 7041, 7121, 7219,
7443, 7485, 10939, 11367,
11571, 11839, 12551, 12646, 13469, 14913, 15205, 15246, 15695, 17473, 17610,
17828, 18130, 18281,
18623, 18890, 21561, 23100, 23872, 24581, 24582, 24983, 27540, 30846, 31415,
31453, 31899, 37000,
38681, 39287, 42951, 45648, 46222, 46687, 47020, 47593, 48513, 49723, 49986,
53018, 53296, 53547,
53899, 53916, 53933, 54305, 55327, 55895, 56143, 56640, 58486, 59576, 63048,
64008, 64018, 64859,
65995, 66905, 67183, 67942, 68101, 68521, 68664, 68988, 69178, 72143, 74183,
74312, 74407, 75518,
76153, 77398, 77615, 79092, 80000, 80125, 80595, 81061, 81151, 81918, 83072,
83137, 83235, 83263,
83279, 83280, 83533, 86856, 87186, 87189, 87727, 87978, 89129, 89556, 89702,
90233, 93060, 94779,
95367, 95844, 95942, 96884 , 96938, 97627, 97777, 97871, 98746 and 99663.
Polymorphic variants at
the following positions in SEQ ID NO: 2 in particular were associated with an
increased risk of
osteoarthritis: 7219, 7485, 11839, 31899, 37000, 48513, 49986, 56640, 74407,
77398, 93060 and
97627. In particular, the following polymorphic variants in SEQ ID NO: 2 were
associated with risk of
osteoarthritis: an adenine at position 7219, a guanine at position 7~1-85, an
adenine at position 11839, a
thymine at position 31899, an adenine at position 37000, a cytosine at
position 48513, a guanine at
position 49986, a guanine at position 56640, a cytosine at position 74407, a
guanine at position 77398,
an adenine at position 93060 and an adenine at position 97627. A threonine at
amino acid position 98 in
an APOB polypeptide was associated with increased risk of osteoarthritis
(i.e., an isoleucine to threonine
non-synonymous variation).
[0026] Polymorphic variants in and around the IL1RL2 locus were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
3 selected from the
group consisting of 225, 509, 860, 874, 939, 1483, 1798, 2189, 2215, 2282,
2340, 2963, 3369, 3481,
3564, 3653, 4860, 4941, 4975, 5321, 5346, 5541, 5633, 6007, 6317, 6378, 6382,
6426, 6479, 6641,
6703, 6705, 7963, 8525, 8526, 8598, 8624, 8883, 8980, 13578, 16135, 16141,
16642, 16931, 17004,
17009, 17010, 18713, 18853, 20783, 21335, 22180, 22268, 22285, 25378, 25906,
26015, 26475, 26798,
27042, 27649, 27827, 27873, 28122, 28202, 28232, 28240, 29546, 29748, 30054,
30646, 31149, 36912,
36936, 37184, 39064, 39343, 40868, 40917, 41113, 47343, 47806, 47911, 48009,
48621, 49245, 49247,
49299, 49302, 49514, 49626, 49791, 50010, 50294, 51482, 51556, 51855, 51956
,52155, 52448, 52458,
52511, 52607, 54049, 54224, 54567, 55052, 55857, 55941, 56120, 56349, 56727,
57232, 58806, 61181,
63808, 64526, 64865, 64928, 64966, 65080, 65690, 66228, 66982, 72511, 74170,
74264, 74333, 74502,
74741, 75321, 82558, 85366, 85469, 86485, 87687, 89463, 89660, 95718 and
95821. Polymorphic
variants at the following positions in SEQ ID NO: 3 in particular were
associated with an increased risk
of osteoarthritis: 2215, 3369, 16642, 20783, 52155, 55052, 55941, 74333,
74741, 85366, 85469, 87687,
89660 and 95718, where specific embodiments are directed to position 52155. In
particular, the
6
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
following polymorphic variants in SEQ ID NO: 3 were associated with risk of
osteoarthritis: an adenine
at position 2215, a deletion at position 3369, a deletion at position 16642, a
cytosine at position 20783, a
cytosine at position 52155, a cytosine at position 55052, a cytosine at
position 55941, a thymine at
position 74333, an adenine at position 74741, a deletion at position 85366, a
thymine at position 85469,
a thymine at position 87687, an adenine at position 89660 and a cytosine at
position 95718.
[0027] Polymorphic variants in and around the ILIRLl locus were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
4 selected from the
group consisting of 207, 6019, 6414, 7341, 10984, 12351, 13335, 16584, 16737,
23897, 24057, 25145,
25300, 26262, 26312, 26589, 27302, 27358, 27451, 27552, 30731, 32085, 32139,
33184, 42382, 42569,
44823, 45217, 45548, 45601, 45722, 45967 ,47367, 47642, 48126, 49218, 49274,
49433, 49610, 51282,
51466, 53757, 53960, 54031, 54574, 55679, 56100, 56182, 59817, 60533, 60656,
72209, 72778, 74293,
77335, 78029, 78374, 78421, 78434, 79174, 79397, 79562, 79700, 79730, 79904,
79920, 79938, 79972,
80125, 80368, 83484, 85536, 85829, 86425, 88083, 88770, 90622, 90924, 91634,
92029, 95152, 95348,
96145, 96793, 97015, 97064, 9771 l, 97855 and 98708. Polymorphic variants at
the following positions
in SEQ ID NO: 4 in particular were associated with an increased risk of
osteoarthritis: 6414, 51282,
54574, 78374, 92029 and 96793, where specific embodiments are directed to
position 54574. In
particular, the following polymorphic variants in SEQ ID NO: 4 were associated
with risk of
osteoarthritis: an adenine at position 6414, an adenine at positoin 51282, a
cytosine at position 54574, a
thymine at position 92029 and an adenine at position 96793.
[0028] Polymorphic variants in and around the WASPIP locus were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
5 selected from the
group consisting of 209, 5908, 7460, 7733, 7855, 7904, 8869, 9480, 13820,
15152, 17713, 17804,
18220, 19083, 19123, 19605, 20247, 20592, 21907, 23273, 23299, 23623, 23669,
23844, 24190, 24486,
24896, 25118, 30551, 30844, 30900, 30942, 31699, 32081, 35078, 36196, 36541,
38356, 45578, 49634,
49774, 51119, 51181, 51652, 54467, 55762, 55999, 57865, 66613, 68377, 69754,
72859, 76512, 76717,
77722, 80998, 82033, 89658, 89960, 94155 and 95679. Polymorphic variants at
the following positions
in SEQ ID NO: 5 in particular were associated with an increased risk of
osteoarthritis: 19083, 30900,
38356, 76512 and 94155, where specific embodiments are directed to positions
30900, 76512 and/or
94155. In particular, the following polymorphic variants in SEQ ID NO: 5 were
associated with risk of
osteoarthritis: a thymine at position 19083, a guanine at position 30900, an
adenine at position 38356,
an adenine at position 76512 and an adenine at position 94155.
[0029] Polymorphic variants in and around the ADAMTS2 locus were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
6 selected from the
group consisting of 210, 3608, 3609, 4318, 5593, 5629, 5639, 5640, 8943,
17968, 19887, 21034, 21085,
21596, 23379, 23432, 24007, 26121, 26273, 26755, 27411, 27710, 27842, 28379,
29603, 31232,
31504, 32583, 32794, 32840, 33044, 33150, 33218, 33513, 33959, 34486, 36289,
36570, 38247, 38477,
38518, 38529, 38667, 39781, 39856, 39927, 40506, 41869, 42452, 44788, 46059,
46846, 47712, 48796,
49441, 49602, 49723, 50050, 50171, 50477, 50818, 50833, 50881, 50882, 51386,
51534, 52317, 52368,
7
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
52970, 53023, 53356, 53882, 54553, 55475, 55530, 55691, 55848, 55879, 56316,
56911, 57320, 57391,
57437, 57478, 57500, 59111, 59333, 59715, 59804, 59851, 59929, 60052, 60240,
60359, 60381, 60456,
60724, 60875, 60968, 60978, 60998, 61557, 62091, 62645, 62943, 63131, 63145,
63406, 63427, 63554,
63661, 64093, 64153, 64409, 64544, 65257, 65626,65739 , 66392, 66720, 69177,
69336, 69636, 69823,
69928, 70547, 70633, 71805, 72181, 72200, 72474, 72567, 72973, 73468, 73889,
75730, 75970, 76114,
76342, 76449, 76465, 76791, 78042, 80758, 80778, 81356, 81576, 81689, 81759,
81950, 82562, 83591,
83700, 83821, 83842, 83923, 83929, 84021, 84175, 84417, 84747, 85746, 86129,
86335, 87315, 87648,
87764, 87770, 88221, 90474, 91148, 91150, 91160, 91733, 91772, 91785, 93140,
93148, 96080, 96157,
96313, 96759, 97026, 97320, 97732, 98713, 99707, 99959, 100009, 100020,
100065, 100086, 101270,
101276, 101371, 101376, 101439, 101820, 102392, 102602, 102604, 102896,
189104, 189134 and
189205. Polymorphic variants at the following positions in SEQ ID NO: 6 in
particular were associated
with an increased risk of osteoarthritis: 5640, 33150, 38247, 38529, 46846,
49723, 50050, 63427,
73889, 189104 and rs428901, where specific embodiments are directed to
positions 46846, 73889,
189104 and/or rs428901. In particular, the following polymorphic variants in
SEQ ID NO: 6 were
associated with risk of osteoarthritis: a cytosine at position 5640, a
cytosine at position 33150, an
adenine at position 38247, a thymine at position 38529, an adenine at position
46846, a cytosine at
position 49723, a cytosine at position 50050, a cytosine a position 63427, a
guanine at position 73889, a
thymine at position 189104, and an adenine at position rs428901.
[0030] Polymorphic variants in and around the BT~ES locus were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
7 selected from the
group consisting of 241, 801, 899, 2091, 2290, 2440, 4959, 7914, 7969, 7972,
10831, 12399, 13841,
14461, 14680, 16808, 18231, 18394, 18505, 18684, 19257, 20263, 20656, 21499,
21563, 21612, 21834,
22406, 22408, 22685, 23303, 23306, 25139, 25211, 25364, 25381, 25414, 25835,
26214, 27224, 27526,
27934, 28550, 29015, 29879, 29979, 30030, 30585, 31753, 31934, 33227, 33228,
35172, 36901, 36921,
36932, 37061, 37570, 38745, 38970, 39725, 40070, 40460, 41470, 41562, 41956,
42047, 42280, 42358,
42629, 43075, 43387, 43393, 43438, 44115, 44537, 45642, 46629,47496, 47515,
48329, 48862, 48908,
49038, 49080, 50204, 50404, 50426, 50531, 50840, 50964, 50971,51378, 52610,
53906, 53951, 54111,
54149, 55563, 55999, 58415, 58961, 60447, 61377, 61528, 61606, 62140, 62461,
63826, 64950, 65076,
66121, 66406, 67051, 68860, 69014, 70796, 72325, 73414, 75258, 76347, 76839,
77358, 77822, 77946,
80002, 80024, 80285, 80397, 82075, 82153, 83981, 84184, 85089, 85288, 85330,
85581, 85642, 86433,
86904, 88391, 89042, 90828, 92676, 92881, 94227, 94585, 94616, 94712, 94738,
95253, 95522, 95869
and 97856. Polymorphic variants at the following positions in SEQ ID NO: 7 in
particular were
associated with an increased risk of osteoarthritis: 25414, 25835, 38970,
41470, 44115, 47496, 49038,
50204, 50840, 50964, 50971, 53906, 54149, 58415, 70796, 72325, 75258, 77822,
80002, 85288, 85581,
86904, 90828, 94616, 94712, 95869 and 97856. In particular, the following
polymorphic variants in
SEQ )D NO: 7 were associated with risk of osteoarthritis: an adenine at
position 25414, a cytosine at
position 25835, an adenine at position 38970, an adenine at position 41470, an
adenine at position
44115, a guanine at position 47496, a cytosine at position 49038, an adenine
at position 50204, a
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
thymine at position 50840, a cytosine at position 50964, a cytosine at
position 50971, an adenine at
position 53906, a guanine at position 54149, a guanine at position 58415, a
thymine at position ?0796, a
guanine at position 72325, a cytosine at position 75258, an adenine at
position 77822, an adenine at
position 80002, an adenine at position 85288, an adenine at position 85581, a
guanine at position 86904,
a guanine at position 90828, an adenine thymine adenine adenine sequence at
position 94616, a cytosine
at position 94712, a guanine at position 95869 and a cytosine at position
97856.
[0031] Polymorphic variants in and around the TM7SF3 locus were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
8 selected from the
group consisting of 230, 231, 5330, 6334, 11372, 11456, 11501, 13393, 16666,
17596, 19710, 19800,
20297, 20967, 32514, 33159, 37600, 41259, 41329, 50060, 53292, 53393, 56417,
56435, 58847, 59595,
59661, 60355, 60407, 62357, 68230, 68516, 69055, 72603, 73928, 85897 and
91554. Polymorphic
variants at the following positions in SEQ ID NO: 8 in particular were
associated with an increased risk
of osteoarthritis: 56435, 59595, 53292, 33159 and 41329, with specific
embodiments directed to
positions 56435 and/or 59595. In particular, the following polymorphic
variants in SEQ ID NO: 8 were
associated with risk of osteoarthritis: a thymine thymine repeat at position
56435, a thymine at position
59595, a cytosine at position 53292, a guanine at position 33159 and a thymine
at position 41329.
[0032] Polymorphic variants in and around the LOXLI locus were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
10 selected from the
group consisting of 213, 249, 1824, 2057, 2306, 2869, 3976, 4288, 4290, 4434,
5298, 5467, 8486, 8487,
8831, 9036, 9058, 9131, 9732, 9862, 10191, 10270, 16167, 17620, 17751, 17764,
17787, 19401 ~ 21021,
21902, 22173, 22416, 22653, 24945, 2501 l, 28563, 48574, 48710, 48880, 50194,
56343, 56455, 56729,
56759, 56895, 57036, 57702, 62515, 62629, 63501, 63547, 64876, 65073, 67149,
67549, 71660, 71906
and 71911. A polymorphic variant at position 65073 in SEQ ID NO: 10, often a
guanine, in particular
was associated with. an increased risk of osteoarthritis.
[0033] Polymorphic variants in and around the ~'ASPR4 locus were tested for
association with
osteoanhritis. These include polymorphic variants at positions in SEQ ID NO:
11 selected from the
group consisting of 205, 866, 4212, 5934, 11486, 16969, 22509, 22796, 28097,
28626, 28853, 28873,
30155, 30827, 31956, 32404, 32944, 35205, 35227, 35781, 41052, 45051, 46039,
47276, 47678, 47716,
51014, 54408, 54596, 56853, 61851, 62016, 62461, 68257, 69793, 73976, 73999,
74053, 75315 75729,
76466, 77216, 77217, 79239, 80825, 81060, 81097, 81426, 84787, 84896, 85165,
86502, 86753, 86941,
88787 and 95598. Polymorphic variants at the following positions in SEQ ID NO:
11 in particular were
associated with an increased risk of osteoarthritis: 47716 and 69793. In
particular, the following
polymorphic variants in SEQ ID NO: 11 were associated with risk of
osteoarthritis: an adenine at
position 47716 and a thymine at position 69793.
[0034] Polymorphic variants in and around the APOL3 locus were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
13 selected from the
group consisting of 201, 425, 1095, 2201, 7879, 8395, 8461, 9503, 10304,
10695, 16300, 16444 17591,
17988, 19116, 19358, 20300, 20669, 20891, 21451, 21978, 22785, 24248, 24770,
24844, 25066, 25096,
9
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
25309, 25344, 25529, 25537, 25554, 27963, 28134, 28356, 29648, 29986, 30217,
30267, 30315, 30585,
30724, 30897, 30931, 31080, 31246, 31373, 31463, 31467, 32188, 32288, 32520,
32594, 32657, 32677,
32764, 32784, 32830, 32872, 33121, 33348, 33952, 34184, 34361, 35026, 35192,
35600, 36033, 36289,
38869, 39629, 40530, 41621, 42379, 42802, 42865, 43644, 45051, 45828, 45829,
46257, 47286, 47427,
47963, 48013, 48229, 48282, 48376, 48404, 49900, 52699, 52897, 53414, 53487,
54112, 55492, 59766,
60307, 60701, 60952, 61401, 62379, 62870, 62879, 63499, 64284, 64408, 64760,
65230, 66127, , 6634,
66686, 66694, 67113, 67257, 67403, 67609, 68418, 68610, 69629, 70024, 70848,
71428, 71553, 71633,
71768 ,71769, 73039, 73325, 73412, 73547, 73769, 73806, 74467, 74472, 74473,
74482, 74494, 74592,
74670, 74672, 74714, 74723, 74749, 74861, 74892, 74893, 75176, 75705, 75989,
76027, 77949, 77974,
78167, 78310, 78415, 78575, 78590, 78709, 78875, 79864, 81316, 81320, 81409,
81737, 81843, 82102,
82833, 83461, 83624, 83660, 83701, 83708, 83782, 85707, 85717, 86486, 86833,
87115, 87234, 87479,
87561, 87604, 87674, 87958, 87992, 88019, 88074, 88079, 88115, 88118, 88120,
88135, 88142 ,88143,
88149, 88340, 88344, 88512, 88521, 88650, 88827, 89230, 89236, 90754, 90984,
91110, 92026, 92954,
93375, 93794, 94937, 95068, 96188, 97092 and 98812. Polymorphic variants at
the following positions
in SEQ m NO: 13 in particular were associated with an increased risk of
osteoarthritis: 203 00, 46257,
87958, 89236, 30267, 32657, 36289, 38869, 45051, 54112, 60307, 63499, 20891,
52699, 71768, with
specific embodiments directed to position 46257. In particular, the following
polymorphic variants in
SEQ ID NO: 13 were associated with risk of osteoarthritis: an adenine at
position 20300, a thymine at
position 46257, an adenine at position 89236, a guanine at position 30267, an
adenine at position 32657,
a cytosine at position 36289, a guanine at position 38869, a thymine at
position 45051, a guanine at
position 54112, an adenine at position 60307, a thymine at position 63499, a
guanine at position 20891,
a guanine at position 52699, and a cytosine at position 71768.
[0035] Based in part upon analyses summarized in Figures lA-1J, regions with
significant
association have been identified in regions associated with osteoarthritis.
Any polymorphic variants
associated with osteoarthritis in a region of significant association can be
utilized for embodiments
described herein. For example, polymorphic variants in a region spanning
positions 21233000 to
21243000 (approximately 10,000 nucleotides in length) in aAPOB locus, a region
spanning
chromosome positions 102456500 to 102471500 (approximately 15,000 nucleotides
in length) in a
ILIRL2 locus, a region spanning chromosome positions 102570000 to 102583000
(approximately
13,000 nucleotides in length) in a ILIRLI locus, a region spanning chromosome
positions 175647734 to
175655734 (approximately 8,000 nucleotides in length) in a WASPIP locus, a
region spanning
chromosome positions 178746000 to 178751000 (approximately 5,000 nucleotides
in length) in a
ADAMTS2 locus, a region spanning chromosome positions 105595000 to 105615000
(approximately
20,000 nucleotides in length) in a BT~ES locus, in a region approximately
14,000 nucleotides in length
spanning chromosome positions 27052000 to 27066000 in a TM7SF3 locus, a region
spanning
chromosome positions 71957600 to 71962600 (approximately 5,000 nucleotides in
length) in a LOXLI
locus, a region spanning chromosome positions 76221000 to 76226000
(approximately 5,000
nucleotides in length) in a CASPR4 locus, and a region approximately 5000
nucleotides in length and
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
spanning chromosome positions 3 4828750 and 34833750 in an APOL3 locus, have
significant
association (chromosome positions are within NCBI's Genome build 34).
Additional Polymorphic Variants Associated with Osteoarthritis
[0036] Also provided is a method for identifying polymorphic variants proximal
to an incident,
founder polymorphic variant associated with osteoarthritis. Thus, featured
herein are methods for
identifying a polymorphic variation associated with osteoarthritis that is
proximal to an incident
polymorphic variation associated with osteoarthritis, which comprises
identifying a polymorphic variant
proximal to the incident polymorphic variant associated with osteoarthritis,
where the incident
polymorphic variant is in a PADI~, APOB, ILIRL2, ILIRLI, WASPIP, ADAMTS2,
BYES, Ti117SF3,
LOXL1, CASPR4 or APOL3 nucleotide sequence or other nucleotide sequence
referenced in Table B.
The nucleotide sequence often comprises a polynucleotide sequence selected
from the group consisting
of (a) a polynucleotide sequence of SEQ ID NO: 1-13 or referenced in Table B;
(b) a polynucleotide
sequence that encodes a polypeptide having an amino acid sequence encoded by a
polynucleotide
sequence of SEQ ID NO: 1-13 or referenced in Table B; and (c) a polynucleotide
sequence that encodes
a polypeptide having an amino acid sequence that is 90% or more identical to
an amino acid sequence
encoded by a nucleotide sequence of SEQ ID NO: 1-13 or referenced in Table B
or a polynucleotide
sequence 90% or more identical to the polynucleotide sequence of SEQ ID NO: 1-
13 or referenced in
Table B. The presence or absence of an association of the proximal polymorphic
variant with
osteoarthritis then is determined using a known association method, such as a
method described in the
Examples hereafter. In an embodiment, the incident polymorphic variant is a
polymorphic variant
associated with osteoarthritis described herein. In another embodiment, the
proximal polymorphic
variant identified sometimes is a publicly disclosed polymorphic variant,
which for example, sometimes
is published in a publicly available database. In other embodiments, the
polymorphic variant identified
is not publicly disclosed and is discovered using a known method, including,
but not limited to,
sequencing a region surrounding the incident polymorphic variant in a group of
nucleic samples. Thus,
multiple polymorphic variants proximal to an incident polymorphic variant are
associated with
osteoarthritis using this method.
[0037] The proximal polymorphic variant often is identified in a region
surrounding the incident
polymorphic variant. In certain embodiments, this surrounding region is about
50 kb flanking the first
polymorphic variant (e.g. about 5 O kb 5' of the first polymorphic variant and
about 50 kb 3' of the first
polymorphic variant), and the region sometimes is composed of shorter flanking
sequences, such as
flanking sequences of about 40 kb, about 30 kb, about 25 kb, about 20 kb,
about 15 kb, about 10 kb,
about 7 kb, about 5 kb, or about 2 kb 5' and 3' of the incident polymorphic
variant. In other
embodiments, the region is composed of longer flanleing sequences, such as
flanking sequences of about
55 kb, about 60 kb, about 65 kb, about 70 kb, about 75 kb, about 80 kb, about
85 kb, about 90 kb, about
95 kb, or about 100 kb 5' and 3' of the incident polymorphic variant.
11
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0038] In certain embodiments, polymorphic variants associated with
osteoarthritis are identified
iteratively. For example, a first proximal polymorphic variant is associated
with osteoarthritis using the
methods described above and then another polymorphic variant proximal to the
first proximal
polymorphic variant is identified (e.g., publicly disclosed or discovered) and
the presence or absence of
an association of one or more other polymorphic variants proximal to the first
proximal polymorphic
variant with osteoarthritis is determined.
[0039] The methods described herein are useful for identifying or discovering
additional
polymorphic variants that may be used to further characterize a gene, region
or loci associated with a
condition, a disease (e.g., osteoarthritis), or a disorder. For example,
allelotyping or genotyping data
from the additional polymorphic variants may be used to identify a functional
mutation or a region of
linkage disequilibrium. In certain embodiments, polymorphic variants
identified or discovered within a
region comprising the first polymorphic variant associated with osteoarthritis
are genotyped using the
genetic methods and sample selection techniques described herein, and it can
be determined whether
those polymorphic variants are in linkage disequilibrium with the first
polymorphic variant. The size of
the region in linkage disequilibrium with the first polymorphic variant also
can be assessed using these
genotyping methods. Thus, provided herein are methods for determining whether
a polymorphic variant
is in linkage disequilibrium with a first polymorphic variant associated with
osteoarthritis, and such
information can be used in prognosis/diagnosis methods described herein.
Isolated Nucleic Acids
[0040] Featured herein are isolated PA.17I2, APOB, ILIRL2, IL1RL1, WASPIP,
ADAMTS2, BITES,
TM7SF3, LOXL1, CASPR4 or APOL3 nucleic acid variants depicted in SEQ ID NO: 1-
13, SEQ ID NO:
14-36 or referenced in Table B, and substantially identical nucleic acids
thereof. A nucleic acid variant
may be represented on one or both strands in a double-stranded nucleic acid or
on one chromosomal
complement (heterozygous) or both chromosomal complements (homozygous).
[0041] ADAMTS2 exists in two forms, a "long" form comprising a molecule
approximately 130
kDa in length (e.g., SEQ ID NO: 21 for cDNA sequence and SEQ ID NO: 44 for
amino acid sequence),
and a "short" form comprising a molecule approximately 70 kDa in length (e.g.,
SEQ ID NO: 22 for
cDNA sequence and SEQ ID NO: 45 for amino acid sequence). Provided herein are
polynucleotide
sequences encoding both the short and long forms of ADAMTS2.
[0042] As used herein, the term "nucleic acid" includes DNA molecules (e.g., a
complementary
DNA (cDNA) and genomic DNA (gDNA)) and RNA molecules (e.g., mRNA, rRNA, siRNA
and
tRNA) and analogs of DNA or RNA, for example, by use of nucleotide analogs.
The nucleic acid
molecule can be single-stranded and it is often double-stranded. The term
"isolated or purified nucleic
acid" refers to nucleic acids that are separated from other nucleic acids
present in the natural source of
the nucleic acid. For example, with regard to genomic DNA, the term "isolated"
includes nucleic acids
which are separated from the chromosome with which the genomic DNA is
naturally associated. An
"isolated" nucleic acid is often free of sequences which naturally flank the
nucleic acid (i. e., sequences
12
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
located at the 5' and/or 3' ends of the nucleic aci d) in the genomic DNA of
the organism from which the
nucleic acid is derived. For example, in various embodiments, the isolated
nucleic acid molecule can
contain less than about 5 kb, 4 kb, 3 kb, 2 kb, 1 lcb, 0.5 kb or 0.1 kb of 5'
and/or 3' nucleotide sequences
which flank the nucleic acid molecule in genomic DNA of the cell from which
the nucleic acid is
derived. Moreover, an "isolated" nucleic acid molecule, such as a cDNA
molecule, can be substantially
free of other cellular material, or culture medimn when produced by
recombinant techniques, or
substantially free of chemical precursors or other chemicals when chemically
synthesized. As used
herein, the term "gene" refers to a nucleotide sequence that encodes a
polypeptide.
[0043] Also included herein are nucleic acid fragments. These fragments often
have a nucleotide
sequence identical to a nucleotide sequence of SEQ ID NO: 1-13 or referenced
in Table B, a nucleotide
sequence substantially identical to a nucleotide sequence of SEQ ID NO: 1-13
or referenced in Table B,
or a nucleotide sequence that is complementary to the foregoing. The nucleic
acid fragment may be
identical, substantially identical or homologous to a nucleotide sequence in
an exon or an intron in a
nucleotide sequence of SEQ ID NO: 1-13 or referenced in Table B, and may
encode a domain or part of
a domain of a polypeptide. Sometimes, the fragment will comprises one or more
of the polymorphic
variations described herein as being associated with osteoarthritis. The
nucleic acid fragment is often
50, 100, or 200 or fewer base pairs in length, and is sometimes about 300,
400, 500, 600, 700, 800, 900,
1000, 1100, 1200, 1300, 1400, 1500, 2000, 3000, 4000, 5000, 10000, 15000, or
20000 base pairs in length.
A nucleic acid fragment that is complementary to a nucleotide sequence
identical or substantially identical
to a nucleotide sequence in SEQ ID NO: 1-13 or referenced in Table B and
hybridizes to such a nucleotide
sequence under stringent conditions is often referred to as a "probe." Nucleic
acid fragments often include
one or more polymorphic sites, or sometimes have an end that is adjacent to a
polymorphic site as described
hereafter.
[0044] An example of a nucleic acid fragment is an oligonucleotide. As used
herein, the term
"oligonucleotide" refers to a nucleic acid comprising about 8 to about 50
covalently linked nucleotides,
often comprising from about 8 to about 35 nucleotides, and more often from
about 10 to about 25
nucleotides. The backbone and nucleotides with in an oligonucleotide may be
the same as those of
naturally occurring nucleic acids, or analogs or derivatives of naturally
occurring nucleic acids,
provided that oligonucleotides having such analogs or derivatives retain the
ability to hybridize
specifically to a nucleic acid comprising a targeted polymorphism.
Oligonucleotides described herein
may be used as hybridization probes or as components of prognostic or
diagnostic assays, for example,
as described herein.
[0045] Oligonucleotides are typically synthesized using standard methods and
equipment, such as
the ABITM3900 High Throughput DNA Synthesizer and the EXPEDITETM 8909 Nucleic
Acid
Synthesizer, both of which are available from Applied Biosystems (Foster City,
CA). Analogs and
derivatives are exemplified in U.S. Pat. Nos. 4,469,863; 5,536,821; 5,541,306;
5,637,683; 5,637,684;
5,700,922; 5,717,083; 5,719,262; 5,739,308; 5,7'73,601; 5,886,165; 5,929,226;
5,977,296; 6,140,482;
WO 00/56746; WO 01/14398, and related publications. Methods for synthesizing
oligonucleotides
13
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
comprising such analogs or derivatives are disclosed, for example, in -the
patent publications cited above
and in U.S. Pat. Nos. 5,614,622; 5,739,314; 5,955,599; 5,962,674; 6,1 17,992;
in WO 00/75372; and in
related publications.
[0046] Oligonucleotides may also be linked to a second moiety. The second
moiety may be an
additional nucleotide sequence such as a tail sequence (e.g., a polyadenosine
tail), an adapter sequence
(e.g., phage M13 universal tail sequence), and others. Alternatively, the
second moiety may be a non-
nucleotide moiety such as a moiety which facilitates linleage to a solid
support or a label to facilitate
detection of the oligonucleotide. Such labels include, without limitation, a
radioactive label, a
fluorescent label, a chemiluminescent label, a paramagnetic label, and the
like. The second moiety may
be attached to any position of the oligonucleotide, provided the
oligonucleotide can hybridize to the
nucleic acid comprising the polymorphism.
Uses for Nucleic Acid Sequence
[0047] Nucleic acid coding sequences may be used for diagnostic purposes for
detection and
control of polypeptide expression. Also, included herein are oligonucleotide
sequences such as
antisense RNA, small-interfering RNA (siRNA) and DNA molecules and ribozymes
that function to
inhibit translation of a polypeptide. Antisense techniques and RNA
interference techniques are known
in the art and are described herein.
[0048] Ribozymes are enzymatic RNA molecules capable of catalyzing the
specific cleavage of
RNA. The mechanism of ribozyme action involves sequence specific hybridization
of the ribozyme
molecule to complementary target RNA, followed by endonucleolytic cleavage.
For example,
hammerhead motif ribozyme molecules may be engineered that specifically and
efficiently catalyze
endonucleolytic cleavage of RNA sequences corresponding to or corriplementary
to PADI2, APOB,
ILIRL2, ILIRLl, WASPIP, ADAMTS2, BhES, TM7SF3, LOXLl, CAS'PR4 orAPOL3
nucleotide
sequences or other nucleotide sequences referenced in Table B. Specific
ribozyme cleavage sites within
any potential RNA target are initially identified by scanning the target
molecule for ribozyme cleavage
sites which include the following sequences, GUA, GUU and GUC. Once
identified, short RNA
sequences of between fifteen (15) and twenty (20) ribonucleotides
corresponding to the region of the
target gene containing the cleavage site may be evaluated for predicts d
structural features such as
secondary structure that may render the oligonucleotide sequence unsuitable.
The suitability of
candidate targets may also be evaluated by testing their accessibility to
hybridization with
complementary oligonucleotides, using ribonuclease protection assays.
[0049] Antisense RNA and DNA molecules, siRNA and ribozyrnes may be prepared
by any
method known in the art for the synthesis of RNA molecules. These include
techniques for chemically
synthesizing oligodeoxyribonucleotides well known in the art such as solid
phase phosphoramidite
chemical synthesis. Alternatively, RNA molecules may be generated by ih vit~~o
and in vivo
transcription of DNA sequences encoding the antisense RNA molecule. Such DNA
sequences may be
incorporated into a wide variety of vectors which incorporate suitable RNA
polymerase promoters such
14
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
as the T7 or SP6 polymerase promoters. Alternatively, mtisense cDNA constructs
that synthesize
antisense RNA constitutively or inducibly, depending on the promoter used, can
be introduced stably
into cell lines.
[0050] DNA encoding a polypeptide also may have a number of uses for the
diagnosis of diseases,
including osteoarthritis, resulting from aberrant expression of a target gene
described herein. For
example, the nucleic acid sequence may be used in hybridization as says of
biopsies or autopsies to
diagnose abnormalities of expression or function (e.g., Southern or Northern
blot analysis, in situ
hybridization assays).
[0051] In addition, the expression of a polypeptide during embryonic
development may also be
determined using nucleic acid encoding the polypeptide. As addressed, infra,
production of functionally
impaired polypeptide is the cause of various disease states, such as
osteoarthritis. In situ hybridizations
using polypeptide as a probe may be employed to predict problems related to
osteoarthritis. Further, as
indicated, infra, administration of human active polypeptide, recorrlbinantly
produced as described
herein, may be used to treat disease states related to functionally irrlpaired
polypeptide. Alternatively,
gene therapy approaches may be employed to remedy deficiencies of functional
polypeptide or to
replace or compete with dysfunctional polypeptide.
Expression Vectors, Host Cells, and Genetically Engineered Cells
[0052] Provided herein are nucleic acid vectors, often expressi on vectors,
which contain a PADI2,
APOB, IL1RL2, ILIRL1, WASPIP, ADAMTS2, BhES, TM7SF3, LOxLl, CASPR4 orAPOL3
nucleotide
sequence or other nucleotide sequence referenced in Table B, or a
substantially identical sequence
thereof. As used herein, the term "vector" refers to a nucleic acid molecule
capable of transporting
another nucleic acid to which it has been linked and can include a p lasmid,
cosmid, or viral vector. The
vector can be capable of autonomous replication or it can integrate into a
host DNA. Viral vectors may
include replication defective retroviruses, adenoviruses and adeno-associated
viruses for example.
[0053] A vector can include a PADI2, APOB, ILIRL2, ILIRLI, WASPIP, ADAMTS2,
BYES,
TM7SF3, LOXLl, CASPR4 or APOL3 nucleotide sequence or other nucleotide
sequence referenced in
Table B in a form suitable for expression of an encoded target polypeptide or
target nucleic acid in a
host cell. A "target polypeptide" is a polypeptide encoded by a PADI2, APOB,
ILIRL~, ILIRLI,
WASPIP, ADAMTS2, BYES, TM7SF3, LO~Ll, CASPR4 or APOL3 nucleotide sequence or
other
nucleotide sequence referenced in Table B, or a substantially identical
nucleotide sequence thereof. The
recombinant expression vector typically includes one or more regul atory
sequences operatively linked
to the nucleic acid sequence to be expressed. The term "regulatory sequence"
includes promoters,
enhancers and other expression control elements (e.g., polyadenylation
signals). Regulatory sequences
include those that direct constitutive expression of a nucleotide sequence, as
well as tissue-specific
regulatory and/or inducible sequences. The design of the expression vector can
depend on such factors
as the choice of the host cell to be transformed, the level of expression of
polypeptide desired, and the
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
like. Expression vectors can be introduced into host cells to produce target
polypeptides, including
fusion polypeptides.
[0054] Recombinant expression vectors can be designed for expression of target
p olypeptides in
prokaryotic or eukaryotic cells. For example, target polypeptides can be
expressed in E. coli, insect
cells (e.g., using baculovirus expression vectors), yeast cells, or mammalian
cells. Suitable host cells
are discussed further in Goeddel, Gene Expression Technology: Methods in
Enzymology 185,
Academic Press, San Diego, CA (1990). Alternatively, the recombinant
expression vector can be
transcribed and translated in vitro, for example using T7 promoter regulatory
sequences and T7
polymerase.
[0055] Expression of polypeptides in prokaryotes is most often carried out in
E. coli with vectors
containing constitutive or inducible promoters directing the expression of
either fusion or non-fusion
polypeptides. Fusion vectors add a number of amino acids to a polypeptide
encoded therein, usually to
the amino terminus of the recombinant polypeptide. Such fusion vectors
typically serve three purposes:
1) to increase expression of recombinant polypeptide; 2) to increase the
solubility of the recombinant
polypeptide; and 3) to aid in the purification of the recombinant polypeptide
by acting as a ligand in
affinity purification. Often, a proteolytic cleavage site is introduced at the
junction of the fusion moiety
and the recombinant polypeptide to enable separation of the recombinant
polypeptide from the fusion
moiety subsequent to purification of the fusion polypeptide. Such enzymes, and
their cognate
recognition sequences, include Factor Xa, thrombin and enterokinase. Typical
fusion expression
vectors include pGEX (Pharmacia Biotech Inc; Smith ~ Johnson, Gene 67: 31-40
(198; 8)), pMAL (New
England Biolabs, Beverly, MA) and pRITS (Pharmacia, Piscataway, NJ) which fuse
glutathione S-
transferase (GST), maltose E binding polypeptide, or polypeptide A,
respectively, to the target
recombinant polypeptide.
[0056] Purified fusion polypeptides can be used in screening assays and to
generate antibodies
specific for target polypeptides. In a therapeutic embodiment, fusion
polypeptide expressed in a
retroviral expression vector is used to infect bone marrow cells that are
subsequently transplanted into
irradiated recipients. The pathology of the subject recipient is then examined
after sufficient time has
passed (e.g., six (6) weeks).
[0057] Expressing the polypeptide in host bacteria with an impaired capacity
to pr-oteolytically
cleave the recombinant polypeptide is often used to maximize recombinant
polypeptidc expression
(Gottesman, S., Gebze Expressioh Technology: Methods in Enzyfraology, Academic
Press, Say Diego,
California I85: 119-128 (1990)). Another strategy is to alter the nucleotide
sequence of the nucleic
acid to be inserted into an expression vector so that the individual codons
for each amino acid are those
preferentially utilized in E. coli (Wada et al., Nueleic Acids Res. 20: 2111-
2118 (1992]). Such
alteration of nucleotide sequences can be carried out by standard DNA
synthesis techniques.
[0058] When used in mammalian cells, the expression vector's control functions
axe often provided
by viral regulatory elements. For example, commonly used promoters are derived
from polyoma,
Adenovirus 2, cytomegalovirus and Simian Virus 40. Recombinant mammalian
expression vectors are
16
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
often capable of directing expression of the nucleic acid in a particular cell
type (e.g,, tis sue-specific
regulatory elements are used to express the nucleic acid). Non-limiting
examples of suitable tissue-
speciFc promoters include an albumin promoter (liver-specific; Pinkert et al.,
Genes Dev. 1: 268-277
(1987)), lymphoid-specific promoters (Calame & Eaton, Adv. Imnaunol. 43: 235-
275 (1988)),
promoters of T cell receptors (Winoto & Baltimore, EMBO J. 8: 729-733 (1989))
promoters of
immunoglobulins (Banerji et al., Cell 33: 729-740 (1983); Queen & Baltimore,
Cell 33. 741-748
(1983)), neuron-specific promoters (e.g., the neurofilament promoter; Byrne &
Ruddle, P~oe. Natl.
Acad. Sci. USA 86.' 5473-5477 (1989)), pancreas-specific promoters (Edlund et
al., Sciehce 230: 912-
916 (1985)), and mammary gland-specific promoters (e.g., milk whey promoter;
U.S. Patent No.
4,873,316 and European Application Publication No. 264,166). Developmentally-
regulated promoters
are sometimes utilized, for example, the murine hox promoters (I~essel &
Gruss, Seience 249: 374-379
(1990)) and the a-fetopolypeptide promoter (Campes & Tilghman, Genes Dev. 3:
537-5 46 (1989)).
[0059] APADI2, APOB, IL1RL2, ILIRLl, WASPIP, ADAMTS2, BITES, TM7SF3, LOdTLl,
CASPR4 or APOL3 nucleic acid or other nucleic acid referenced in Table B also
may be cloned into an
expression vector in an antisense orientation. Regulatory sequences (e.g.,
viral promoters and/or
enhancers) operatively linked to a PADI2, APOB, ILIRL2, ILIRLl, WASPIP,
ADAMTS~, BT~ES,
TM7SF3, LOXLI, CASPR4 or APOL3 nucleic acid or other nucleic acid referenced
in Table B cloned in
the antisense orientation can be chosen for directing constitutive, tissue
specific or cell type specific
expression of antisense RNA in a variety of cell types. Antisense expression
vectors can be in the form
of a recombinant plasmid, phagemid or attenuated virus. For a discussion of
the regulation of gene
expression using antisense genes see, e.g., Weintraub et al., Antisense RNA as
a molecu..lar tool for
genetic analysis, Reviews - Trends in Genetics, Vol. 1(1) (1986).
[0060] Also provided herein are host cells that include a PADI2, APOB, ILIRL2,
IL1RL1,
WASPIP, ADAMTS2, BIIES, TM7SF3, LOXLl, CASPR4 or APOL3 nucleotide sequence or
other
nucleotide sequence referenced in Table B within a recombinant expression
vector or a fragment of such
a nucleotide sequence which facilitate homologous recombination into a
specific site of -the host cell
genome. The terms "host cell" and "recombinant host cell" are used
interchangeably herein. Such
terms refer not only to the particular subject cell but rather also to the
progeny or potential progeny of
such a cell. Because certain modifications may occur in succeeding generations
due to either mutation
or environmental influences, such progeny may not, in fact, be identical to
the parent ce 11, but are still
included within the scope of the term as used herein. A host cell can be any
prokaryotic or eukaryotic
cell. For example, a target polypeptide can be expressed in bacterial cells
such as E. colt, insect cells,
yeast or mammalian cells (such as Chinese hamster ovary cells (CHO) or COS
cells). ether suitable
host cells are known to those skilled in the art.
[0061] Vectors can be introduced into host cells via conventional
transformation or transfection
techniques. As used herein, the terms "transformation" and "transfection" are
intended to refer to a
variety of art-recognized techniques for introducing foreign nucleic acid
(e.g., DNA) int~ a host cell,
17
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
including calcium phosphate or calcium chloride co-precipitation,
transduction/infection, DEAE-
dextran-mediated transfection, lipofection, or electroporation.
[0062] A host cell provided herein can be used to produce (i. e., express) a
target polypeptide: or a
substantially identical polypeptide thereof. Accordingly, further provided are
methods for producing a
target polypeptide using host cells described herein. In one embodiment, the
method includes culturing
host cells into which a recombinant expression vector encoding a target
polypeptide has been introduced
in a suitable medium such that a target polypeptide is produced. In another
embodiment, the method
further includes isolating a target polypeptide from the medium or the host
cell.
[0063] Also provided are cells or purified preparations of cells which include
a PADI2, APOB,
ILIRL2, ILIRLl, WASPIP, ADAMTS2, BYES, TM7SF3, LOXLl, CASPR4 orAPOL3
transgene, or other
transgene in Table B, or which otherwise misexpress target polypeptide. Cell
preparations can consist
of human or non-human cells, e.g., rodent cells, e.g., mouse or rat cells,
rabbit cells, or pig cells. In
preferred embodiments, the cell or cells include a PADI2, APOB, ILIRL2,
ILIRLl, WASPIP,
ADAMTS2, BhES, TM7SF3, LOXLl, CASPR4 or APOL3 transgene or other transgene
referenced in
Table B (e.g., a heterologous form of a PADI2, APOB, ILIRL2, IL1RL1, WASPIP,
ADAMTS2, BTIES,
TM7SF3, LOXLl, CASPR4 or APOL3 gene or other gene referenced in Table B, such
as a human gene
expressed in non-human cells). The transgene can be misexpressed, e.g.,
overexpressed or
underexpressed. In other preferred embodiments, the cell or cells include a
gene which misexpress an
endogenous target polypeptide (e.g., expression of a gene is disrupted, also
known as a knockout,. Such
cells can serve as a model for studying disorders which are related to mutated
or mis-expressed alleles
or for use in drug screening. Also provided are human cells (e.g., a
hematopoietic stem cells)
transfected with a PADI2, APOB, ILIRL2, ILIRLI, WASPIP, ADAMTS2, BT~ES,
TM7SF3, LOXL 1,
CASPR4 or APOL3 nucleic acid or other nucleic acid referenced in Table B.
[0064] Also provided are cells or a purified preparation thereof (e.g., human
cells) in which an
endogenous PAD12, APOB, ILIRL2, IL1RLI, WASPIP, ADAMTS2, BYES, TM7SF3, LOXL1,
CAS'PR4
or APOL3 nucleic acid or other nucleic acid referenced in Table B is under the
control of a regulatory
sequence that does not normally control the expression of the endogenous gene.
The expression
characteristics of an endogenous gene within a cell (e.g., a cell line or
microorganism) can be modified
by inserting a heterologous DNA regulatory element into the genome of the cell
such that the inserted
regulatory element is operably linked to the corresponding endogenous gene.
For example, an
endogenous corresponding gene (e.g., a gene which is "transcriptionally
silent," not normally expressed,
or expressed only at very low levels) may be activated by inserting a
regulatory element which is
capable of promoting the expression of a normally expressed gene product in
that cell. Techniques such
as targeted homologous recombinations, can be used to insert the heterologous
DNA as described in,
e.g., Chappel, US 5,272,071; WO 91/06667, published on May 16, 1991.
1~
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
Trans~enic Animals
[0065] Non-human transgenic animals that express a heterologous target
polypeptide (e.g.,
expressed from a PADI2, APOB, ILIRL2, ILIRLl, WASPIP, ADAMTS2, BYES, TM7SF3,
LOXLI,
CASPR4 or APOL3 nucleic acid or other nucleic acid referenced in Table B, or
substantially identical
sequence thereof) can be generated. Such animals are useful for studying the
function and/or activity of
a target polypeptide and for identifying and/or evaluating modulators of the
activity of PADI2, APOB,
ILIRL2, ILIRLl, WASPIP, ADAMTS2, BhES, TM7SF3, LOXL1, CASPR4 orAPOL3 nucleic
acids,
other nucleic acids referenced in Table B, and encoded polypeptides. As used
herein, a "transgenic
animal" is a non-human animal such as a mammal (e.g., a non-human primate such
as chimpanzee,
baboon, or macaque; an ungulate such as an equine, bovine, or caprine; or a
rodent such as a rat, a
mouse, or an Israeli sand rat), a bird (e.g., a chicken or a turkey), an
amphibian (e.g., a frog, salamander,
or newt), or an insect (e.g., Drosophila naela~ogaster), in which one or more
of the cells of the animal
includes a transgene. A transgene is exogenous DNA or a rearrangement (e.g., a
deletion of
endogenous chromosomal DNA) that is often integrated into or occurs in the
genome of cells in a
transgenic animal. A transgene can direct expression of an encoded gene
product in one or more cell
types or tissues of the transgenic animal, and other transgenes can reduce
expression (e.g., a knockout).
Thus, a transgenic animal can be one in which an endogenous nucleic acid
homologous to a PADI2,
APOB, ILIRL2, ILIRL1, WASPIP, ADAMTS2, BhES, TM7SF3, LOXL1, CASPR~ orAPOL3
nucleic
acid or other nucleic acid referenced in Table B has been altered by
homologous recombination between
the endogenous gene and an exogenous DNA molecule introduced into a cell of
the animal (e.g., an
embryonic cell of the animal) prior to development of the animal.
[0066] Intronic sequences and polyadenylation signals can also be included in
the transgene to
increase expression efficiency of the transgene. One or more tissue-specific
regulatory sequences can
be operably linked to a PADI2, APOB, ILIRL2, ILIRLl, WASPIP, ADAMTS2, BhES,
TM7SF3, LOXLI,
CASPR4 or APOL3 nucleotide sequence or other nucleotide sequence referenced in
Table B to direct
expression of an encoded polypeptide to particular cells. A transgenic founder
animal can be identified
based upon the presence of a PADI2, APOB, ILIRL2, IL1RL1, WASPIP, ADAMTS2,
BYES, TM7SF3,
LOXLl, CASPR4 or APOL3 nucleotide sequence or other nucleotide sequence
referenced in Table B in
its genome and/or expression of encoded mRNA in tissues or cells of the
animals. A transgenic founder
animal can then be used to breed additional animals carrying the transgene.
Moreover, transgenic
animals carrying a PADI2, APOB, ILIRL2, IL1RL1, WASPIP, ADAMTS2, BhES, TM7SF3,
LOXL1,
CASPR~ or APOL3 nucleotide sequence or other nucleotide sequence referenced in
Table B can further
be bred to other transgenic animals carrying other transgenes.
[0067] Target polypeptides can be expressed in transgenic animals or plants by
introducing, for
example, a PADI2, APOB, ILIRL2, ILIRLI, WASPIP, ADAMTS2, BTpES, TM7SF3, LOXLl,
CASPR4 or
APOL3 nucleic acid or other nucleic acid referenced in Table B into the genome
of an animal that
encodes the target polypeptide. In preferred embodiments the nucleic acid is
placed under the control of
19
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
a tissue specific promoter, e.g., a milk or egg specific promoter, and
recovered from the milk or eggs
produced by the animal. Also included is a population of cells from a
transgenic animal.
Tar-eg t Polypeptides
[0068] Also featured herein are isolated target polypeptides, which are
encoded by a PADl2,
APOB, ILIRL2, IL1RL1, WASPIP, ADAMTS2, BITES, TM7SF3, LO~Ll, CASPR4 orAPOL3
nucleotide
sequence or a nucleotide sequence referenced in Table B (e.g., SEQ ID NO: 14-
36 or a sequence
referenced in Table B), or a substantially identical nucleotide sequence
thereof. Examples of PAD12,
APOB, ILIRL2, IL1RL1, WASPIP, ADAMTS2, BhES, TM7SF3, LOXLl, CASPR~ orAPOL3
polypeptides are set forth in SEQ ID NO: 37-55. The term "polypeptide" as used
herein includes
proteins and peptides. An "isolated" or "purified" polypeptide or protein is
substantially free of cellular
material or other contaminating proteins from the cell or tissue source from
which the protein is derived,
or substantially free from chemical precursors or other chemicals when
chemically synthesized. In one
embodiment, the language "substantially free" means preparation of a target
polypeptide having less
than about 30%, 20%, 10% and more preferably 5% (by dry weight), of non-target
polypeptide (also
referred to herein as a "contaminating protein"), or of chemical precursors or
non-target chemicals.
When the target polypeptide or a biologically active portion thereof is
reeombinantly produced, it is also
preferably substantially free of culture medium, specifically, where culture
medium represents less than
about 20%, sometimes less than about 10%, and often less than about 5% of the
volume of the
polypeptide preparation. Isolated or purified target polypeptide preparations
are sometimes 0.01
milligrams or more or 0.1 milligrams or more, and often 1.0 milligrams or more
and 10 milligrams or
more in dry weight. In certain embodiments, the APOL3 polypeptide or
polypeptide fragment has
APOL3 biological activity, for example, apolipoprotein activity.
[0069] Further included herein are target polypeptide fragments. The
polypeptide fragment may be
a domain or part of a domain of a target polypeptide. The polypeptide fragment
may have increased,
decreased or unexpected biological activity. The polypeptide fragment is often
50 or fewer, 100 or fewer,
or 200 or fewer amino acids in length, and is sometimes 300, 400, 500, 600,
700, or 900 or fewer amino
acids in length. In certain embodiments, the polypeptide fragment sometimes is
amino acids 90-396 of
SEQ D? NO: 53; amino acids 19-325 of SEQ ID NO: 54; or amino acids 1-196 of
SEQ ID NO: 55. Shown
in Table A below are examples of polypeptide fragments, where approximate
amino acid positions are
shown in parenthesis (e.g., a Pellino domain starts at about amino acid 3 and
ends at about amino acid 412).
Amino acid sequences can be accessed using information in Table B and in SEQ
117 NO: 37-55.
TABLE A
RS ID Locus SEQ SignalDomain (amino acid ranges)
ID
- NO. Peptide
910223 PAD12 37 none
1367117 PO~ 38 1-27 Apolipoprotein B48 mature peptide
(1-2151)
Lipoprotein amino terminal region
(46-597)
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
RS ID Locus SEQ SignalDomain (amino acid ranges)
ID
NO. Peptide
ATPase involved in DNA repair (2077-2583)
1024791 IL1RL2 39 1-19 Immunoglobulin C-2 Type (36-100;
137-197)
TIR Domain (385-535)
Neural cell adhesion molecule L1
(<53->295)
Transmembrane Domain (336-358)
1465621 WASPIP 43 none WASP-interacting protein VRP1/WIP
(14->63)
1018810 BYES 46 none Popeye protein conserved region
(123-266)
242392 PEL12 48 none Pellino (3-412)
8818 LOXL1 49 none Lysyl oxidase (370-574)
1395486 CASPR4 50 none Neurexin IV domain (3-1308)
F5/8 type C domain (57-177)
Laminin G domains (374-524; 475->750;
797-941; 1037-
1176)
51 none Neurexin IV domain (1->721)
F5/8 type C domain (29-149)
Laminin G domains (169-314; 346-496;
579->662)
512294 GPR50 52 none 7 transmembrane receptor (rhodopsin
family) (45..294)
Microtubule-associated protein
dynactin DCTN1/Glued
(462..>587)
Syndecan domain (485..>595)
[0070] Interleukin 1 receptor-like 1 isoform 1 (SEQ ID NO: 40) is a member of
the interleukin 1
receptor family with no known ligand (orphan receptor). ILIRLI exists in
soluble (SEQ 117 NO: 41-42)
and transmembrane forms, suggesting that it may have ligand or ligand
scavenging activity. In an
embodiment, ILIRLI protein agents may be administered to treat or prevent the
occurrence of OA.
IL1RL1 protein agents include ILIRLI polypeptides or fragments thereof that
have ILIRLI ligand
activity (e.g., recombinant polypeptides of SEQ ID NO: 41-42). In a related
embodiment, ILIRLI
protein agents include ILIRLl polypeptides or fragments thereof that have
IL1RL1 ligand scavenging
activity (e.g., recombinant polypeptide of SEQ ID NO: 40). Isolated ILIRLI
polypeptides featured
herein include the full-length polypeptide, the mature polypeptide (i.e., the
polypeptide without the
signal sequence MGFWILAILTILMYSTAA) or a polypeptide fragment containing a
domain or part of
a IL1RL1 domain. The polypeptide fragment may have increased, decreased or
unexpected biological
activity.
[0071] In another embodiment, provided herein are ADAMTS~ polypeptides having
an ADAMTSZ
activity (e.g., a zinc binding activity, a metalloprotease activity, a
procollagen II processing or synthesis
activity, or a collagen II synthesis activity in vitro or in vivo). In certain
embodiments, the polypeptides
are ADAMTS2 proteins including at least one propeptide domain, at least one
metalloproteinase domain,
at least one disintegrin-like domain, at least one, two, three, and often four
thrombospondin domains,
and sometimes having a ADAMTS2 activity, e.g., a ADAMTS2 activity as described
herein. ADAMTS2
21
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
polypeptides and fragments thereof often have biological activity, such as
excising the N-propeptide of
type II procollagens. Methods for monitoring and quantifying this biological
activity are known (e.g.,
Colige et al., J. Biol. Chem. 270: 16724-16730 (1995)).
[0072] Human ADAMTS2 protein (SEQ ID NO: 44-45) includes a signal sequence of
about 29
amino acids (from amino acid 1 to about amino acid 29 of SEQ ID NO: 44-45).
The ADAMTS2 protein
without the signal sequence can be approximately 1182 amino acid residues in
length (from about
amino acid 30 to amino acid 1211 of SEQ ID NO: 44) or approximately 485 amino
acid residues in
length (from about amino acid 30 to amino acid 514 of SEQ ID NO: 45). Human
ADAMTS2 protein
includes a "pro" region homologous to the reprolysin family propeptide, which
is typically post-
translationally cleaved upon conversion of the inactive (or pro-domain
containing) protein to the
catalytically active metalloprotease. The prodomain region of human ADAMTS2
protein corresponds to
about amino acids 30 to 251, 30 to 252, 30 to 253, 30 to 254, 30 to 255, 30 to
256, 30 to 257, 30 to 258
or 30 to 259 of SEQ ID NO: 44-45, where it is understood that the active form
ofADAMTS2 does not
contain the propeptide domain.
[0073] Upon cleavage, catalytically active mature protein can be approximately
960, 959, 958,
957, 956, 955, 954, 953 or 952 amino acids in length (from about amino acid
252, 253, 254, 255, 256,
257, 258, 259 or 260 to amino acid 1211 of SEQ ID NO: 44) or approximately
261, 260, 259, 258, 257,
256, 255, 254 or 253 amino acid residues in length (from about amino acid 252,
253, 254, 255, 256,
257, 258, 259 or 260 to amino acid 514 of SEQ ID NO: 45).
[0074] Human ADAMTS2 contains the following regions or other structural
features: a signal
sequence at about amino acids 1-29 of SEQ ID NO: 44-45; a reprolysin family
propeptide domain
located at about amino acid residues 30 to 251, 30 to 252, 30 to 253, 30 to
254, 30 to 255, 30 to 256, 30
to 257, 30 to 258 or 30 to 259 of SEQ ID NO: 44-45; a zinc-metalloprotease
catalytic domain at about
amino acids 251 to 479, 252 to 479, 253 to 479, 254 to 479, 255 to 479, 256 to
479, 257 to 479, 258 to
479 or 259 to 479 of SEQ ID NO: 44-45; a disintegrin domain at about amino
acids 480 to 560 of SEQ
ID NO: 44; a cysteine-rich domain at about amino acids 618 to 722 of SEQ ID
NO: 44; four
thrombospondin motifs-2 motifs at about amino acids 561 to 616, 854 to 912,
914 to 971, and 975 to
1029 of SEQ ID NO: 44; and eight N-glycosylation sites located at about amino
acids 112, 251, 949,
993, 1031, 1098, 1145, and 1150 of SEQ ID NO: 44.
[0075] In other embodiments, provided are methods of increasing the synthesis
of procollagen II
comprising providing or administering to individuals in need of increasing
levels of type II collagen the
pharmaceutical or physiologically acceptable composition comprising active
human ADAMTS2 protein
or fragment thereof, where ADAMTS2 polypeptide fragments having activity are
selected from amino
acids 252-1211, 253-1211, 254-1211, 255-1211, 256-1211, 257-1211, 258-1211,
259-1211 or 260-1211
of SEQ ID NO: 4, where it is understood that the active form ofADAMTS2 does
not contain the
propeptide domain.
[0076] Substantially identical target polypeptides may depart from the amino
acid sequences of
target polypeptides in different manners. For example, conservative amino acid
modifications may be
22
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
introduced at one or more positions in the amino acid sequences of target
polypeptides. A "conservative
amino acid substitution" is one in which the amino acid is replaced by another
amino acid having a
similar structure and/or chemical function. Families of amino acid residues
having similar structures
and functions are well known. These families include amino acids with basic
side chains (e.g., lysine,
arginine, histidine), acidic side chains (e.g., aspartic acid, glutamic acid),
uncharged polar side chains
(e.g., glycine, asparagine, glutamine, serine, threonine, tyrosine, cysteine),
nonpolar side chains (e.g.,
alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine,
tryptophan), beta-branched side
chains (e.g., threonine, valine, isoleucine) and aromatic side chains (e.g.,
tyrosine, phenylalanine,
tryptophan, histidine). Also, essential and non-essential amino acids may be
replaced. A "non-
essential" amino acid is one that can be altered without abolishing or
substantially altering the biological
function of a target polypeptide, whereas altering an "essential" amino acid
abolishes or substantially
alters the biological function of a target polypeptide. Amino acids that are
conserved among target
polypeptides are typically essential amino acids. In certain embodiments, the
polypeptide includes one
or more non-synonymous polymorphic variants associated with osteoarthritis, as
described above (e.g.,
a threonine encoded by rs1367117 in an AP~B polypeptide).
[0077] Also, target polypeptides may exist as chimeric or fusion polypeptides.
As used herein, a
target "chimeric polypeptide" or target "fusion polypeptide" includes a target
polypeptide linked to a
non-target polypeptide. A "non-target polypeptide" refers to a polypeptide
having an amino acid
sequence corresponding to a polypeptide which is not substantially identical
to the target polypeptide,
which includes, for example, a polypeptide that is different from the target
polypeptide and derived
from the same or a different organism. The target polypeptide in the fusion
polypeptide can correspond
to an entire or nearly entire target polypeptide or a fragment thereof. The
non-target polypeptide can be
fused to the N-terminus or C-terminus of the target polypeptide.
[0078] Fusion polypeptides can include a moiety having high affinity for a
ligand. For example,
the fusion polypeptide can be a GST-target fusion polypeptide in which the
target sequences are fused
to the C-terminus of the GST sequences, or a polyhistidine-target fusion
polypeptide in which the target
polypeptide is fused at the N- or C-terminus to a string of histidine
residues. Such fusion polypeptides
can facilitate purification of recombinant target polypeptide. Expression
vectors are commercially
available that already encode a fusion moiety (e.g., a GST polypeptide), and a
nucleotide sequence in
SEQ ID NO: 1-13 or referenced in Table B, or a substantially identical
nucleotide sequence thereof, can
he cloned into an expression vector such that the fusion moiety is linked in-
frame to the target
polypeptide. Further, the fusion polypeptide can be a target polypeptide
containing a heterologous
signal sequence at its N-terminus. In certain host cells (e.g., mammalian host
cells), expression,
secretion, cellular internalization, and cellular localization of a target
polypeptide can be increased
through use of a heterologous signal sequence. Fusion polypeptides can also
include all or a part of a
serum polypeptide (e.g., an IgG constant region or human serum albumin).
[0079] Target polypeptides can be incorporated into pharmaceutical
compositions and
administered to a subject in vivo. Administration of these target polypeptides
can be used to affect the
23
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
bioavailability of a substrate of the target polypeptide and may effectively
increase target polypeptide
biological activity in a cell. Target fusion polypeptides may be useful
therapeutically for the treatment
of disorders caused by, for example, (i) aberrant modification or mutation of
a gene encoding a target
polypeptide; (ii) mis-regulation of the gene encoding the target polypeptide;
and (iii) aberrant post-
translational modification of a target polypeptide. Also, target polypeptides
can be used as immunogens
to produce anti-target antibodies in a subject, to purify target polypeptide
ligands or binding partners,
and in screening assays to identify molecules which inhibit or enhance the
interaction of a target
polypeptide with a substrate.
[0080] In addition, polypeptides can be chemically synthesized using
techniques known in the art
(See, e.g., Creighton, 1983 Proteins. New York, N.Y.: W. H. Freeman and
Company; and Hunkapiller
et al., (1984) Nature July 12 -18;310(5973):105-11). For example, a relative
short fragment can be
synthesized by use of a peptide synthesizer. Furthermore, if desired, non-
classical amino acids or
chemical amino acid analogs can be introduced as a substitution or addition
into the fragment sequence.
Non-classical amino acids include, but are not limited to, to the D-isomers of
the common amino acids,
2,4-diaminobutyric acid, a-amino isobutyric acid, 4-aminobutyric acid, Abu, 2-
amino butyric acid, g-
Abu, e-Ahx, 6-amino hexanoic acid, Aib, 2-amino isobutyric acid, 3-amino
propionic acid, ornithine,
norleucine, norvaline, hydroxyproline, sarcosine, citrulline, homocitrulline,
cysteic acid, t-butylglycine,
t-butylalanine, phenylglycine, cyclohexylalanine, b-alanine, fluoroamino
acids, designer amino acids
such as b-methyl amino acids, Ca-methyl amino acids, Na-methyl amino acids,
and amino acid analogs
in general. Furthermore, the amino acid can be D (dextrorotary) or L
(levorotary).
[0081] Polypeptides and polypeptide fragments sometimes are differentially
modified during or
after translation, e.g., by glycosylation, acetylation, phosphorylation,
amidation, derivatization by
known protecting/blocking groups, proteolytic cleavage, linkage to an antibody
molecule or other
cellular ligand, etc. Any of numerous chemical modifications may be carried
out by known techniques,
including but not limited, to specific chemical cleavage by cyanogen bromide,
trypsin, chymotrypsin,
papain, V8 protease, NaBH4; acetylation, formylation, oxidation, reduction;
metabolic synthesis in the
presence of tunicamycin; and the like. Additional post-translational
modifications include, for example,
N-linked or O-linked carbohydrate chains, processing of N-terminal or C-
terminal ends), attachment of
chemical moieties to the amino acid backbone, chemical modifications of N-
linked or O-linked
carbohydrate chains, and addition or deletion of an N-terminal methionine
residue as a result of
prokaryotic host cell expression. The polypeptide fragments may also be
modified with a detectable
label, such as an enzymatic, fluorescent, isotopic or affinity label to allow
for detection and isolation of
the polypeptide.
[0082] Also provided are chemically modified derivatives of polypeptides that
can provide
additional advantages such as increased solubility, stability and circulating
time of the polypeptide, or
decreased immunogenicity (see e.g., U.S. Pat. No: 4,179,337. The chemical
moieties for derivitization
may be selected from water soluble polymers such as polyethylene glycol,
ethylene glycol/propylene
glycol copolymers, carboxymethylcellulose, dextran, polyvinyl alcohol and the
like. The polypeptides
24
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
may be modified at random positions within the molecule, or at predetermined
positions within the
molecule and may include one, two, three or more attached chemical moieties.
[0083] The polymer may be of any molecular weight, and may be branched or
unbranched. For
polyethylene glycol, the preferred molecular weight is between about 1 kDa and
about 100 kDa (the
term "about" indicating that in preparations of polyethylene glycol, some
molecules will weigh more,
some less, than the stated molecular weight) for ease in handling and
manufacturing. Other sizes may be
used, depending on the desired therapeutic profile (e.g., the duration of
sustained release desired, the
effects, if any on biological activity, the ease in handling, the degree or
lack of antigenicity and other
known effects of the polyethylene glycol to a therapeutic protein or analog).
[0084] The polymers should be attached to the polypeptide with consideration
of effects on
functional or antigenic domains of the polypeptide. There are a number of
attachment methods available
to those skilled in the art (e.g., EP 0 401 384 (coupling PEG to G-CSF) and
Malik et al. (1992) Exp
Hematol. September;20(8):1028-35 (pegylation of GM-CSF using tresyl
chloride)). For example,
polyethylene glycol may be covalently bound through amino acid residues via a
reactive group, such as
a free amino or carboxyl group. Reactive groups are those to which an
activated polyethylene glycol
molecule may be bound. The amino acid residues having a free amino group may
include lysine
residues and the N-terminal amino acid residues; those having a free carboxyl
group may include
aspartic acid residues, glutamic acid residues and the C-terminal amino acid
residue. Sulfhydryl groups
may also be used as a reactive group for attaching the polyethylene glycol
molecules. For therapeutic
purposes, the attachment sometimes is at an amino group, such as attachment at
the N-terminus or
lysine group.
[0085] Proteins can be chemically modified at the N-terminus. Using
polyethylene glycol as an
illustration of such a composition, one may select from a variety of
polyethylene glycol molecules (by
molecular weight, branching, and the like), the proportion of polyethylene
glycol molecules to protein
(polypeptide) molecules in the reaction mix, the type of pegylation reaction
to be performed, and the
method of obtaining the selected N-terminally pegylated protein. The method of
obtaining the N-
terminally pegylated preparation (i.e., separating this moiety from other
monopegylated moieties if
necessary) may be by purification of the N-terminally pegylated material from
a population of pegylated
protein molecules. Selective proteins chemically modified at the N-terminus
may be accomplished by
reductive alkylation, which exploits differential reactivity of different
types of primary amino groups
(lysine versus the N-terminal) available for derivatization in a particular
protein. Under the appropriate
reaction conditions, substantially selective derivatization of the protein at
the N-terminus with a
carbonyl group containing polymer is achieved.
Substantially Identical Nucleic Acids and Polypeptides
[0086] Nucleotide sequences and polypeptide sequences that are substantially
identical to a PADI2,
APOB, ILIRL2, ILIRL1, WASPIP, ADAMTS2, BYES, TM7SF3, LOdYLl, CASPR4 orAPOL3
nucleotide
sequence or other nucleotide sequence referenced in Table B and the target
polypeptide sequences
2,5
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
encoded by those nucleotide sequences, respectively, are included herein. The
term "substantially
identical" as used herein refers to two or more nucleic acids or polypeptides
sharing one or more
identical nucleotide sequences or polypeptide sequences, respectively.
Included are nucleotide
sequences or polypeptide sequences that are 55% or more, 60% or more, 65% or
more, 70% or more,
75% or more, 80°!0 or more, 85% or more, 90% or more, 95% or more (each
often within a 1%, 2%, 3%
or 4% variability) identical to a PADl2, APOB, ILIRL2, ILIRLl, WASPIP,
ADAMTS2, BYES, TM7SF3,
LOXLl, CASPR4 or APOL3 nucleotide sequence, or other nucleotide sequence
referenced in Table B, or
the encoded target polypeptide amino acid sequences. One test for determining
whether two nucleic
acids are substantially identical is to determine the percent of identical
nucleotide sequences or
polypeptide sequences shared between the nucleic acids or polypeptides.
[0087] Calculations of sequence identity are often performed as follows.
Sequences are aligned for
optimal comparison purposes (e.g., gaps can be introduced in one or both of a
first and a second amino
acid or nucleic acid sequence for optimal alignment and non-homologous
sequences can be disregarded
for comparison purposes). The length of a reference sequence aligned for
comparison purposes is
sometimes 30% or more, 40% or more, 50% or more, often 60% or more, and more
often 70% or more,
80% or more, 90°!0 or more, or 100% of the length of the reference
sequence. The nucleotides or amino
acids at corresponding nucleotide or polypeptide positions, respectively, are
then compared among the
two sequences. When a position in the first sequence is occupied by the same
nucleotide or amino acid
as the corresponding position in the second sequence, the nucleotides or amino
acids are deemed to be
identical at that position. The percent identity between the two sequences is
a function of the number of
identical positions shared by the sequences, taking into account the number of
gaps, and the length of
each gap, introduced for optimal alignment of the two sequences.
[0088] Comparison of sequences and determination of percent identity between
two sequences can
be accomplished using a mathematical algorithm. Percent identity between two
amino acid or
nucleotide sequences can be determined using the algorithm of Meyers & Miller,
CABIOS 4: 11-17
(1989), which has been incorporated into the ALIGN program (version 2.0),
using a PAM120 weight
residue table, a gap length penalty of 12 and a gap penalty of 4. Also,
percent identity between two
amino acid sequences can be determined using the Needleman & Wunsch, J. Mol.
Biol. 48: 444-453
(1970) algorithm which has been incorporated into the GAP program in the GCG
software package
(available at the http address www.gcg.com), using either a Blossum 62 matrix
or a PAM250 matrix,
and a gap weight of 16, 14, 12, 10, 8, 6, or 4 and a length weight of 1, 2, 3,
4, 5, or 6. Percent identity
between two nucleotide sequences can be determined using the GAP program in
the GCG software
package (available at http address www.gcg.com), using a NWSgapdna.CMP matrix
and a gap weight
of 40, 50, 60, 70, or 80 and a length weight of 1, 2, 3, 4, 5, or 6. A set ~f
parameters often used is a
Blossum 62 scoring matrix with a gap open penalty of 12, a gap extend penalty
of 4, and a frameshift
gap penalty of 5.
[0089] Another manner for determining if two nucleic acids are substantially
identical is to assess
whether a polynucleotide homologous to one nucleic acid will hybridize to the
other nucleic acid under
26
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
stringent conditions. As use herein, the term "stringent conditions" refers to
conditions for
hybridization and washing. Stringent conditions are known to those skilled in
the art and can be found
in Current Pr~otoeols in Molecular Biology, John Wiley & Sons, N.Y. , 6.3.1-
6.3.6 (1989). Aqueous
and non-aqueous methods are described in that reference and either can be
used. An example of
stringent hybridization conditions is hybridization in 6X sodium
chloride/sodium citrate (SSC) at about
45°C, followed by one or more washes in 0.2X SSC, 0.1% SDS at
50°C. Another example of stringent
hybridization conditions are hybridization in 6X sodium chloride/sodium
citrate (SSC) at about 45°C,
followed by one or more washes in 0.2X SSC, 0.1% SDS at 55°C. A further
example of stringent
hybridization conditions is hybridization in 6X sodium chloride/sodium citrate
(SSC) at about 45°C,
followed by one or more washes in 0.2X SSC, 0.1% SDS at 60°C. Often,
stringent hybridization
conditions are hybridization in 6X sodium chloride/sodium citrate (SSC) at
about 45°C, followed by
one or more washes in 0.2X SSC, 0.1% SDS at 65°C. More often,
stringency conditions are O.SM
sodium phosphate, 7% SDS at 65°C, followed by one or more washes at
0.2X SSC, 1% SDS at 65°C.
[0090] An example of a substantially identical nucleotide sequence to a
nucleotide sequence in
SEQ ID NO: 1-13 or referenced in Table B is one that has a different
nucleotide sequence but still
encodes the same polypeptide sequence encoded by the nucleotide sequence in
SEQ ID NO: 1-13 or
referenced in Table B. Another example is a nucleotide sequence that encodes a
polypeptide having a
polypeptide sequence that is more than 70% or more identical to, sometimes
more than 75% or more,
80% or more, or 85% or more identical to, and often more than 90% or more and
95% or more identical
to a polypeptide sequence encoded by a nucleotide sequence in SEQ ID NO: 1-13
or referenced in Table
B.
[0091] Nucleotide sequences in SEQ ID NO: 1-13 or referenced in Table B and
amino acid
sequences of encoded polypeptides can be used as "query sequences" to perform
a search against public
databases to identify other family members or related sequences, for example.
Such searches can be
performed using the NBLAST and XBLAST programs (version 2.0) of Altschul et
al., J. Mol. Biol.
215: 403-10 (1990). BLAST nucleotide searches can be performed with the NBLAST
program, score =
100, wordlength = 12 to obtain nucleotide sequences homologous to nucleotide
sequences in SEQ ID
NO: 1-13, SEQ ID NO: 14-36 or referenced in Table B. BLAST polypeptide
searches can be performed
with the XBLAST program, score = 50, wordlength = 3 to obtain amino acid
sequences homologous to
polypeptides encoded by the nucleotide sequences of SEQ ff~ NO: 14-36 or
referenced in Table B. To
obtain gapped alignments for comparison purposes, Gapped BLAST can be utilized
as described in
Altschul et al., Nucleic Acids Res. 25(17): 3389-3402 (1997). When utilizing
BLAST and Gapped
BLAST programs, default parameters of the respective programs (e.g., XBLAST
and NBLAST) can be
used (see the http address www.ncbi.nlm.nih.gov).
[0092] A nucleic acid that is substantially identical to a nucleotide sequence
in SEQ ID NO: 1-13
or referenced in Table B may include polymorphic sites at positions equivalent
to those described herein
when the sequences are aligned. For example, using the alignment procedures
described herein, SNPs
27
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
in a sequence substantially identical to a sequence in SEQ ID NO: 1-13 or
referenced in Table B can be
identified at nucleotide positions that match (i.e., align) with nucleotides
at SNP positions in each
nucleotide sequence in SEQ ID NO: 1-13 or referenced in Table B. Also, where a
polymorphic
variation results in an insertion or deletion, insertion or deletion of a
nucleotide sequence from a
reference sequence can change the relative positions of other polymorphic
sites in the nucleotide
sequence.
[0093] Substantially identical nucleotide and polypeptide sequences include
those that are naturally
occurring, such as allelic variants (same locus), splice variants, homologs
(different locus), and
orthologs (different organism) or can be non-naturally occurring. Non-
naturally occurring variants can
be generated by mutagenesis techniques, including those applied to
polynucleotides, cells, or organisms.
The variants can contain nucleotide substitutions, deletions, inversions and
insertions. Variation can
occur in either or both the coding and non-coding regions. The variations can
produce both
conservative and non-conservative amino acid substitutions (as compared in the
encoded product).
Orthologs, homologs, allelic variants, and splice variants can be identified
using methods known in the
art. These variants normally comprise a nucleotide sequence encoding a
polypeptide that is 50% or
more, about 55% or more, often about 70-75% or more or about 80-85% or more,
and sometimes about
90-95% or more identical to the amino acid sequences of target polypeptides or
a fragment thereof.
Such nucleic acid molecules can readily be identified as being able to
hybridize under stringent
conditions to a nucleotide sequence in SEQ ID NO: 1-13 or referenced in Table
B or a fragment of this
sequence. Nucleic acid molecules corresponding to orthologs, homologs, and
allelic variants of a
nucleotide sequence in SEQ ID NO: 1-13 or referenced in Table B can further be
identified by mapping
the sequence to the same chromosome or locus as the nucleotide sequence in SEQ
ID NO: 1-13 or
referenced in Table B.
[0094] Also, substantially identical nucleotide sequences may include codons
that are altered with
respect to the naturally occurring sequence for enhancing expression of a
target polypeptide in a
particular expression system. For example, the nucleic acid can be one in
which one or more codons are
altered, and often 10% or more or 20% or more of the codons are altered for
optimized expression in
bacteria (e.g., E. coli.), yeast (e.g., S. eervesiae), human (e.g., 293
cells), insect, or rodent (e.g., hamster)
cells.
Methods for Identifying Risk of Osteoarthritis
[0095] Methods for prognosing and diagnosing osteoarthritis are included
herein. These methods
include detecting the presence or absence of one or more polymorphic
variations in a nucleotide
sequence associated with osteoarthritis, such as variants in or around the
loci set forth herein, or a
substantially identical sequence thereof, in a sample from a subject, where
the presence of a
polymorphic variant described herein is indicative of a risk of
osteoarthritis. Determining a risk of
osteoarthritis sometimes refers to determining whether an individual is at an
increased risk of
osteoarthritis (e.g., intermediate risk or higher risk).
28
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0096] Thus, featured herein is a method for identifying a subject who is at
risk of osteoarthritis,
which comprises detecting an aberration associated with osteoarthritis in a
nucleic acid sample from the
subject. An embodiment is a method for detecting a risk of osteoarthritis in a
subject, which comprises
detecting the presence or absence of a polymorphic variation associated with
osteoarthritis at a
polymorphic site in a nucleotide sequence in a nucleic acid sample from a
subject, where the nucleotide
sequence comprises a polynucleotide sequence selected from the group
consisting of: (a) a nucleotide
sequence of SEQ ID NO: 1-13 or referenced in Table B; (b) a nucleotide
sequence which encodes a
polypeptide consisting of an amino acid sequence encoded by a nucleotide
sequence of SEQ ID NO: 1-
13 or referenced in Table B; (c) a nucleotide sequence which encodes a
polypeptide that is 90% or more
identical to an amino acid sequence encoded by a nucleotide sequence of SEQ ID
NO: 1-13 or
referenced in Table B, or a nucleotide sequence about 90% or more identical to
a nucleotide sequence of
SEQ ID NO: 1-13 or referenced in Table B; and (d) a fragment of a nucleotide
sequence of (a), (b), or
(c) comprising the polymorphic site; whereby the presence of the polymorphic
variation is indicative of
a predisposition to osteoarthritis in the subject. In certain embodiments,
polymorphic variants at the
positions described herein are detected for determining a risk of
osteoarthritis, and polymorphic variants
at positions in linkage disequilibrium with these positions are detected for
determining a risk of
osteoarthritis. As used herein, the terms "SEQ ID NO: 1-13" and other
nucleotide sequences
"referenced in Table B" refers to individual sequences in SEQ ID NO: 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12
or 13 or any individual sequence referenced in Table B, or any individual
nucleic acid sequence
provided in the sequence listing, including SEQ ID NO: 14-36 each sequence
being separately
applicable to embodiments described herein.
[0097] Risk of osteoarthritis sometimes is expressed as a probability, such as
an odds ratio,
percentage, or risk factor. Risk often is based upon the presence or absence
of one or more polymorphic
variants described herein, and also may be based in part upon phenotypic
traits of the individual being
tested. Methods for calculating risk based upon patient data are well known
(see, e.g., Agresti,
Categorical Data Analysis, 2nd Ed. 2002. Wiley). Allelotyping and genotyping
analyses may be
carried out in populations other than those exemplified herein to enhance the
predictive power of the
prognostic method. These further analyses are executed in view of the
exemplified procedures
described herein, and may be based upon the same polymorphic variations or
additional polymorphic
variations.
[0098] In certain embodiments, determining the presence of a combination of
two or more
polymorphic variants associated with osteoarthritis in one or more genetic
loci (e.g., one or more genes)
of the sample is determined to identify, quantify and/or estimate, risk of
osteoarthritis. The risk often is
the probability of having or developing osteoarthritis. The risk sometimes is
expressed as a relative risk
with respect to a population average risk of osteoarthritis, and sometimes is
expressed as a relative risk
with respect to the lowest risk group. Such relative risk assessments often
are based upon penetrance
values determined by statistical methods, and are particularly useful to
clinicians and insurance
companies for assessing risk of osteoarthritis (e.g., a clinician can target
appropriate detection,
29
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
prevention and therapeutic regimens to a patient after determining the
patient's risk of osteoarthritis, and
an insurance company can fine tune actuarial tables based upon population
genotype assessments of
osteoarthritis risk). Risk of osteoarthritis sometimes is expressed as an odds
ratio, which is the odds of a
particular person having a genotype has or will develop osteoarthritis with
respect to another genotype
group (e.g., the most disease protective genotype or population average). In
related embodiments, the
determination is utilized to identify a subject at risk of osteoarthritis. In
an embodiment, two or more
polymorphic variations are detected in two or more regions in human genomic
DNA associated with
increased risk of osteoarthritis, such as a locus containing a PADI2, APOB,
IL1RL2, ILIRLI, WASPIP,
ADAMTS2, BhES, TM7SF3, LOXLl, CASPR4 or APOL3 or other locus referenced in
Table B, for
example. In certain embodiments, 3 or more, or 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20,
25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 80, 90, 100 or more polymorphic
variants are detected in the
sample. In specific embodiments, polymorphic variants are detected in a PAD12,
APOB, ILIRL2,
ILIRL1, WASPIP, ADAMTS2, BYES, TM7SF3, LOXLl, CASPR4 or APOL3 region or other
region
referenced in Table B, for example. In another embodiment, polymorphic
variants are detected at two
or three positions in a nucleotide sequence of SEQ ID NO: 1-13 or referenced
in Table B. In certain
embodiments, polymorphic variants are detected at other genetic loci (e.g.,
the polymorphic variants can
be detected in a PADI2, APOB, ILIRL2, ILIRLl, WASPIP, ADAMTS2, BT~ES, TM7SF3,
LOXLl,
CASPR4 or APOL3 nucleotide sequence or other nucleotide sequence referenced in
Table B in addition
to other loci or only in other loci), where the other loci include but are not
limited to those described in
patent applications 60/559,011; 60/559,202; 60/559,203; 60/559,042;
60/559,275; 60/559,040 and
60/559,225, each of which is entitled "Methods for Identifying Risk of
Osteoarthritis and Treatments
Thereof," each of which was filed on 1 April 2004 and each of which is
incorporated herein by
reference in its entirety in jurisdictions allowing incorporation by
reference.
[0099] Results from prognostic tests may be combined with other test results
to diagnose
osteoarthritis. For example, prognostic results may be gathered, a patient
sample may be ordered based
on a determined predisposition to osteoarthritis, the patient sample is
analyzed, and the results of the
analysis may be utilized to diagnose osteoarthritis. Also osteoarthritis
diagnostic method can be
developed from studies used to generate prognostic methods in which
populations are stratified into
subpopulations having different progressions of osteoarthritis. In another
embodiment, prognostic
results may be gathered, a patient's risk factors for developing
osteoarthritis (e.g., age, weight,
occupational history, race, diet) analyzed, and a patient sample may be
ordered based on a determined
predisposition to osteoarthritis.
[0100] The nucleic acid sample typically is isolated from a biological sample
obtained from a
subject. For example, nucleic acid can be isolated from blood, saliva, sputum,
urine, cell scrapings, and
biopsy tissue. The nucleic acid sample can be isolated from a biological
sample using standard
techniques, such as the technique described in Example 2. As used herein, the
term "subject" refers
primarily to humans but also refers to other mamrnals such as dogs, cats, and
ungulates (e.g., cattle,
sheep, and swine). Subjects also include avians (e.g., chickens and turkeys),
reptiles, and fish (e.g.,
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
salmon), as embodiments described herein can be adapted to nucleic acid
samples isolated from any of
these organisms. The nucleic acid sample may be isolated from the subject and
then directly utilized in
a method for determining the presence of a polymorphic variant, or
alternatively, the sample may be
isolated and then stored (e.g., frozen) for a period of time before being
subjected to analysis.
[0101] The presence or absence of a polymorphic variant is determined using
one or both
chromosomal complements represented in the nucleic acid sample. Determining
the presence or
absence of a polymorphic variant in both chromosomal complements represented
in a nucleic acid
sample from a subject having a copy of each chromosome is useful for
determining the zygosity of an
individual for the polymorphic variant (i. e., whether the individual is
homozygous or heterozygous for
the polymorphic variant). Any oligonucleotide-based diagnostic may be utilized
to determine whether a
sample includes the presence or absence of a polymorphic variant in a sample.
For example, primer
extension methods, ligase sequence determination methods (e.g., U.S. Pat. Nos.
5,679,524 and
5,952,174, and WO 01/27326), mismatch sequence determination methods (e.g.,
U.S. Pat. Nos.
5,851,770; 5,958,692; 6,110,684; and 6,183,958), microarray sequence
determination methods,
restriction fragment length polymorphism (RFLP), single strand conformation
polymorphism detection
(SSCP) (e.g., U.S. Pat. Nos. 5,891,625 and 6,013,499), PCR-based assays (e.g.,
TAQMAN~ PCR
System (Applied Biosystems)), and nucleotide sequencing methods may be used.
[0102] Oligonucleotide extension methods typically involve providing a pair of
oligonucleotide
primers in a polymerise chain reaction (PCR) or in other nucleic acid
amplification methods for the
purpose of amplifying a region from the nucleic acid sample that comprises the
polymorphic variation.
One oligonucleotide primer is complementary to a region 3' of the polymorphism
and the other is
complementary to a region 5' of the polymorphism. A PCR primer pair may be
used in methods
disclosed in U.S. Pat. Nos. 4,683,195; 4,683,202, 4,965,188; 5,656,493;
5,998,143; 6,140,054; WO
01/27327; and WO 01/27329 for example. PCR primer pairs may also be used in
any commercially
available machines that perform PCR, such as any of the GENEAMP~ Systems
available from Applied
Biosystems. Also, those of ordinary skill in the art will be able to design
oligonucleotide primers based
upon a PAD12, APOB, IL1RL2, ILIRLI, WASPIP, ADAMTS2, BTdES, TM7SF3, LOXLI,
CASPR4 or
APOL3 nucleotide sequence or other nucleotide sequence referenced in Table B
using knowledge
available in the art.
[0103] Also provided is an extension oligonucleotide that hybridizes to the
amplified fragment
adjacent to the polymorphic variation. As used herein, the term "adjacent"
refers to the 3' end of the
extension oligonucleotide being often 1 nucleotide from the 5' end of the
polymorphic site, and
sometimes 2, 3, 4, 5, 6, 7, 8, 9, or 10 nucleotides from the 5' end of the
polymorphic site, in the nucleic
acid when the extension oligonucleotide is hybridized to the nucleic acid. The
extension
oligonucleotide then is extended by one or more nucleotides, and the number
and/or type of nucleotides
that are added to the extension oligonucleotide determine whether the
polymorphic variant is present.
Oligonucleotide extension methods are disclosed, for example, in U.S. Pat.
Nos. 4,656,127; 4,851,331;
5,679,524; 5,834,189; 5,876,934; 5,908,755; 5,912,118; 5,976,802; 5,981,186;
6,004,744; 6,013,431;
31
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
6,017,702; 6,046,005; 6,087,095; 6,210,891; and WO 01/20039. Oligonucleotide
extension methods
using mass spectrometry are described, for example, in U.S. Pat. Nos.
5,547,835; 5,605,798; 5,691,141;
5,849,542; 5,869,242; 5,928,906; 6,043,031; and 6,194,144, and a method often
utilized is described
herein in Example 2.
[0104] A microarray can be utilized for determining whether a polymorphic
variant is present or
absent in a nucleic acid sample. A microarray may include any oligonucleotides
described herein, and
methods for making and using oligonucleotide microarrays suitable for
diagnostic use are disclosed in
U.S. Pat_ Nos. 5,492,806; 5,525,464; 5,589,330; 5,695,940; 5,849,483;
6,018,041; 6,045,996;
6,136,541; 6,142,681; 6,156,501; 6,197,506; 6,223,127; 6,225,625; 6,229,911;
6,239,273; WO
00/52625; WO 01/25485; and WO 01/29259. The microarray typically comprises a
solid support and
the oligonucleotides may be linked to this solid support by covalent bonds or
by non-covalent
interactions. The oligonucleotides may also be linked to the solid support
directly or by a spacer
molecule. A microarray may comprise one or more oligonucleotides complementary
to a polymorphic
site set forth herein.
[0105] A lcit also may be utilized for determining whether a polymorphic
variant is present or
absent in a nucleic acid sample. A kit often comprises one or more pairs of
oligonucleotide primers
useful for amplifying a fragment of a nucleotide sequence of SEQ ID NO: 1-13
or referenced in Table B
or a substantially identical sequence thereof, where the fragment includes a
polymorphic site. The kit
sometimes comprises a polymerizing agent, for example, a thermostable nucleic
acid polymerase such
as one disclosed in U.S. Pat. Nos. 4,889,818 or 6,077,664. Also, the kit often
comprises an elongation
oligonucleotide that hybridizes to a PADI2, APOB, ILIRL2, ILIRL1, WASPIP,
ADAMTS2, BYES,
TM7SF3, LO~'Ll, CASPR4 or APOL3 nucleotide sequence or other nucleotide
sequence referenced in
Table B in a nucleic acid sample adjacent to the polymorphic site. Where the
kit includes an elongation
oligonucleotide, it also often comprises chain elongating nucleotides, such as
dATP, dTTP, dGTP,
dCTP, and dITP, including analogs of dATP, dTTP, dGTP, dCTP and dITP, provided
that such analogs
are substrates for a thermostable nucleic acid polymerase and can be
incorporated into a nucleic acid
chain elongated from the extension oligonucleotide. Along with chain
elongating nucleotides would be
one or more chain terminating nucleotides such as ddATP, ddTTP, ddGTP, ddCTP,
and the like. In an
embodiment, the kit comprises one or more oligonucleotide primer pairs, a
polymerizing agent, chain
elongating nucleotides, at least one elongation oligonucleotide, and one or
more chain terminating
nucleotides. Kits optionally include buffers, vials, microtiter plates, and
instructions for use.
[0106] An individual identified as being at risk of osteoarthritis may be
heterozygous or
homozygous with respect to the allele associated with a higher risk of
osteoarthritis. A subject
homozygous for an allele associated with an increased risk of osteoarthritis
is at a comparatively high
risk of osteoarthritis, a subject heterozygous for an allele associated with
an increased risk of
osteoarthritis is at a comparatively intermediate risk of osteoarthritis, and
a subject homozygous for an
allele associated with a decreased risk of osteoarthritis is at a
comparatively low risk of osteoarthritis. A
32
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
genotype may be assessed for a complementary strand, such that the
complementary nucleotide at a
particular position is detected.
[0107] Also featured are methods for determining risk of osteoarthritis and/or
identifying a subject
at risk of osteoarthritis by contacting a polypeptide or protein encoded by a
PADI2, APOB, ILIRL2,
ILIRLl, WASPIP, ADAMTS2, BITES, TM7SF3, LOXLl, CASPR4 or AI'OL3 nucleotide
sequence or
other nucleotide sequence referenced in Table B from a subject with an
antibody that specifically binds
to an epitope associated with increased risk of osteoarthritis in the
polypeptide (e.g., an epitope
comprising a valine at position 245 in an IRILRI polypeptide).
Applications of Prognostic and Diagnostic Results to Pharmaco~enomic Methods
[0108] Pharmacogenomics is a discipline that involves tailoring a treatment
for a subject according
to the subject's genotype as a particular treatment regimen may exert a
differential effect depending
upon the subject's genotype. For example, based upon the outcome of a
prognostic test described
herein, a clinician or physician may target pertinent information and
preventative or therapeutic
treatments to a subject who would be benefited by the information or treatment
and avoid directing such
information and treatments to a subject who would not be benefited (e.g., the
treatment has no
therapeutic effect and/or the subject experiences adverse side effects).
[0109] The following is an example of a pharmacogenomic embodiment. A
particular treatment
regimen can exert a differential effect depending upon the subject's genotype.
Where a candidate
therapeutic exhibits a significant interaction with a major allele and a
comparatively weak interaction
with a minor allele (e.g., an order of magnitude or greater difference in the
interaction), such a
therapeutic Typically would not be administered to a subject genotyped as
being homozygous for the
minor allele, and sometimes not administered to a subject genotyped as being
heterozygous for the
minor allele. In another example, where a candidate therapeutic is not
significantly toxic when
administered to subjects who are homozygous for a major allele but is
comparatively toxic when
administered to subjects heterozygous or homozygous for a minor allele, the
candidate therapeutic is not
typically administered to subjects who are genotyped as being heterozygous or
homozygous with
respect to the minor allele.
[0110] The methods described herein are applicable to pharmacogenomic methods
for preventing,
alleviating or treating osteoarthritis. For example, a nucleic acid sample
from an individual may be
subjected to a prognostic test described herein. Where one or more polymorphic
variations associated
with increased risk of osteoarthritis are identified in a subject, information
for preventing or treating
osteoarthritis and/or one or more osteoarthritis treatment regimens then may
be prescribed to that
subject.
[0111] In certain embodiments, a treatment or preventative regimen is
specifically prescribed
and/or administered to individuals who will most benefit from it based upon
their risk of developing
osteoarthritis assessed by the methods described herein. Thus, provided are
methods for identifying a
subject predisposed to osteoarthritis and then prescribing a therapeutic or
preventative regimen to
33
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
individuals identified as having a predisposition. Thus, certain embodiments
are directed to a method
for reducing osteoarthritis in a subject, which comprises: detecting the
presence or absence of a
polymorphic variant associated with osteoarthritis in a nucleotide sequence in
a nucleic acid sample
from a subject, where the nucleotide sequence comprises a polynucleotide
sequence selected from the
group consisting of (a) a nucleotide sequence of SEQ ID NO: 1-13 or referenced
in Table B; (b) a
nucleotide sequence which encodes a polypeptide consisting of an amino acid
sequence encoded by a
nucleotide sequence of SEQ ID NO: 1-13 or referenced in Table B; (c) a
nucleotide sequence which
encodes a polypeptide that is 90% or more identical to an amino acid sequence
encoded by a nucleotide
sequence of SEQ ID NO: 1-13 or referenced in Table B, or a nucleotide sequence
about 90% or more
identical to a nucleotide sequence of SEQ ID NO: 1-13 or referenced in Table
B; and (d) a fragment of a
polynucleotide sequence of (a), (b), or (c); and prescribing or administering
a treatment regimen to a
subj ect from whom the sample originated where the presence of a polymorphic
variation associated
with osteoarthritis is detected in the nucleotide sequence. In these methods,
predisposition results may
be utilized in combination with other test results to diagnose osteoarthritis.
[0112] Certain preventative treatments often are prescribed to subjects having
a predisposition to
osteoarthritis and where the subject is diagnosed with osteoarthritis or is
diagnosed as having symptoms
indicative of an early stage of osteoarthritis. The treatment sometimes is
preventative (e.g., is
prescribed or administered to reduce the probability that osteoarthritis
arises or progresses), sometimes
is therapeutic, and sometimes delays, alleviates or halts the progression of
osteoarthritis. Any known
preventative or therapeutic treatment for alleviating or preventing the
occurrence of osteoarthritis is
prescribed and/or administered. For example, the treatment often is directed
to decreasing pain and
improving joint movement. Examples of OA treatments include exercises to keep
joints flexible and
improve muscle strength. Different medications to control pain, including
corticosteroids and
nonsteroidal anti-inflammatory drugs (NSAIDs, e.g., Voltaren); cyclooxygenase-
2 (COX-2) inhibitors
(e.g_ , Celebrex, Vioxx, Mobic, and Bextra); monoclonal antibodies (e.g.,
Remicade); tumor necrosis
factor inhibitors (e.g., Enbrel); or injections of glucocorticoids, hyaluronic
acid or chondrotin sulfate
into joints that are inflamed and not responsive to NSAIDS. Orally
administered chondroitin sulfate
also may be used as a therapeutic, as it may increase hyaluronic acid levels
and viscosity of synovial
fluid, and decrease collagenase levels in synovial fluid. Also, glucosamine
can serve as an OA
therapeutic as delivering it into joints may inhibit enzymes involved in
cartilage degradation and
enhance the production of hyaluronic acid. For mild pain without inflammation,
acetaminophen may be
used. Other treatments include: heat/cold therapy for temporary pain relief;
joint protection to prevent
strain or stress on painful joints; surgery to relieve chronic pain in damaged
joints; and weight control to
prevent extra stress on weight-bearing joints.
[0113] As therapeutic approaches for treating osteoarthritis continue to
evolve and improve, the
goal of treatments for osteoarthritis related disorders is to intervene even
before clinical signs first
manifest. Thus, genetic markers associated with susceptibility to
osteoarthritis prove useful for early
diagnosis, prevention and treatment of osteoarthritis.
34
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0114] As osteoarthritis preventative and treatment information can be
specifically targeted to
subjects in need thereof (e.g., those at risk of developing osteoarthritis or
those in an early stage of
osteoarthritis), provided herein is a method for preventing or reducing the
risk of developing
osteoarthritis in a subject, which compris es: (a) detecting the presence or
absence of a polymorphic
variation associated with osteoarthritis at a polymorphic site in a nucleotide
sequence in a nucleic acid
sample from a subject; (b) identifying a subject with a predisposition to
osteoarthritis, whereby the
presence of the polymorphic variation is indicative of a predisposition to
osteoarthritis in the subject;
and (c) if such a predisposition is identifi ed, providing the subject with
information about nethods or
products to prevent or reduce osteoarthritis or to delay the onset of
osteoarthritis. Also provided is a
method of targeting information or advertising to a subpopulation of a human
population based on the
subpopulation being genetically predisposed to a disease or condition, which
comprises: (a) detecting
the presence or absence of a polymorphic variation associated with
osteoarthritis at a polyrnorphic site
in a nucleotide sequence in a nucleic acid sample from a subject; (b)
identifying the subpopulation of
subjects in which the polymorphic variation is associated with osteoarthritis;
and (c) providing
information only to the subpopulation of subjects about a particular product
which may be obtained and
consumed or applied by the subject to help prevent or delay onset of the
disease or condition.
[0115] Pharmacogenomics methods also may be used to analyze and predict a
response to
osteoarthritis treatment or a drug. For example, if pharmacogenomics analysis
indicates a likelihood
that an individual will respond positively to osteoarthritis treatment with a
particular drug, the drug may
be administered to the individual. Conversely, if the analysis indicates that
an individual is likely to
respond negatively to treatment with a particular drug, an alternative course
of treatment may be
prescribed. A negative response may be defined as either the absence of an
efficacious response or the
presence of toxic side effects. The response to a therapeutic treatment can be
predicted in a background
study in which subjects in any of the following populations are genotyped: a
population that responds
favorably to a treatment regimen, a population that does not respond
significantly to a treatment
regimen, and a population that responds adversely to a treatment regimen
(e.g., exhibits one or more
side effects). These populations are provided as examples and other
populations and subpopulations
may be analyzed. Based upon the results of these analyses, a subject is
genotyped to predict whether he
or she will respond favorably to a treatment regimen, not respond
significantly to a treatment regimen,
or respond adversely to a treatment regimen.
[0116] The tests described herein also are applicable to clinical drug trials.
One or more
polymorphic variants indicative of response to an agent for treating
osteoarthritis or to side effects to an
agent for treating osteoarthritis may be identified using the methods
described herein. Thereafter,
potential participants in clinical trials of such an agent may be screened to
identify those individuals
most likely to respond favorably to the drug and exclude those likely to
experience side effects. In that
way, the effectiveness of drug treatment rnay be measured in individuals who
respond positively to the
drug, without lowering the measurement as a result of the inclusion of
individuals who are unlikely to
respond positively in the study and without risking undesirable safety
problems.
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0117] Thus, another embodiment is a method of selecting an individual for
inclusion in a clinical
trial of a treatment or drug comprising the steps of (a) obtaining a nucleic
acid sample from an
individual; (b) determining the identity of a polymorphic variation which is
associated with a positive
response to the treatment or the drug, or at least one polymorphic variation
which is associated with a
negative response to the treatment or the drug in the nucleic acid sample, and
(c) including the
individual in the clinical trial if the nucleic acid sample contains said
polymorphic variation associated
with a positive response to the treatment or the drug or if the nucleic acid
sample lacks said
polymorphic variation associated with a negative response to the treatment or
the drug. In addition, the
methods described herein for selecting an individual for inclusion in a
clinical trial of a treatment or
drug encompass methods with any further limitation described in this
disclosure, or those following,
specified alone or in any combination. The polymorphic variation may be in a
sequence selected
individually or in any combination from the group consisting of (i) a
nucleotide sequence of SEQ ID
NO: 1-13 or referenced in Table B; (ii) a nucleotide sequence which encodes a
polypeptide consisting
of an amino acid sequence encoded by a nucleotide sequence of SEQ ID NO: 1-13
or referenced in
Table B; (iii) a nucleotide sequence which encodes a polypeptide that is
90°!° or more identical to an
amino acid sequence encoded by a nucleotide sequence of SEQ ID NO: 1-13 or
referenced in Table B,
or a nucleotide sequence about 90% or more identical to a nucleotide sequence
of SEQ ID NO: 1-13 or
referenced in Table B; and (iv) a fragment of a polynucleotide sequence of
(i), (ii), or (iii) comprising
the polymorphic site. The including step (c) optionally comprises
administering the drug or the
treatment to the individual if the nucleic acid sample contains the
polymorphic variation associated with
a positive response to the treatment or the drug and the nucleic acid sample
lacks said biallelic marker
associated with a negative response to the treatment or the drug.
[0118] Also provided herein is a method of partnering between a
diagnostic/prognostic testing
provider and a provider of a consumable product, which comprises: (a) the
diagnostic/prognostic
testing provider detects the presence or absence of a polymorphic variation
associated with osteoarthritis
at a polymorphic site in a nucleotide sequence in a nucleic acid sample from a
subject; (b) the
diagnostic/prognostic testing provider identifies the subpopulation of
subjects in which the polymorphic
variation is associated with osteoarthritis; (c) the diagnostic/prognostic
testing provider forwards
information to the subpopulation of subjects about a particular product which
may be obtained and
consumed or applied by the subject to help prevent or delay onset of the
disease or condition; and (d)
the provider of a consumable product forwards to the diagnostic test provider
a fee every time the
diagnostic/prognostic test provider forwards information to the subject as set
forth in step (c) above.
Compositions Comprising Osteoarthritis-Directed Molecules
[0119] Featured herein is a composition comprising a cell from a subject
having osteoarthritis or at
risk of osteoarthritis and one or more molecules specifically directed and
targeted to a nucleic acid
comprising a PADI2, APOB, ILIRL2, ILIRLl, WASPIP, ADAMTS2, BT~ES, TM7SF3,
LOXLl, CASPR4
orAPOL3 nucleotide sequence, other nucleotide sequence referenced in Table B,
or an encoded amino
36
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
acid sequence referenced herein. Such directed molecules include, but are not
limited to, a compound
that binds to a PADI2, APOB, IL1RL2, ILIRLl, WASPIP, ADAMTS2, BYES, TM7SF3,
LOXLI,
CASPR4 or APOL3 nucleotide sequence, or other nucleotide sequence referenced
in Table B, or
encoded amino acid sequence; a RNAi or siRNA molecule having a strand
complementary or
substantially complementary to a PADI2, APOB, ILIRL2, ILIRLl, WASPIP, ADAMTS2,
BYES,
TM7SF3, LOXLI, CASPR4 or APOL3 nucleotide sequence or other nucleotide
sequence referenced in
Table B (e.g., hybridizes to a PADI2, APOB, ILLRL2, IL1RL1, WASPIP, ADAMTS2,
BYES, TM7SF3,
LOXLl, CASPR4 or APOL3 nucleotide sequence or other nucleotide sequence
referenced in Table B
under conditions of high stringency); an antisensa nucleic acid complementary
or substantially
complementary to an RNA encoded by a PADI2, APOB, IL1RL2, ILIRLI, WASPIP,
ADAMTS2, BYES,
TM7SF3, LO~YLl, CASPR4 or APOL3 nucleotide sequence or other nucleotide
sequence referenced in
Table B (e.g., hybridizes to a PADI2, APOB, ILLRL2, ILIRLI, WASPIP, ADAMTS2,
BYES, TM7SF3,
LOXL1, CASPR4 or APOL3 nucleotide sequence or other nucleotide sequence
referenced in Table B
under conditions of high stringency); a ribozyme that hybridizes to a PADI2,
APOB, ILIRL2, IL1RL1
WASPIP, ADAMTS2, BYES, TM7SF3, LOXLI, C~4SPR4 or APOL3 nucleotide sequence or
other
nucleotide sequence referenced in Table B (e.g., hybridizes to a PADI2, APOB,
IL1RL2, ILIRLl,
WASPIP, ADAMTS2, BYES, TM7SF3, LOXLI, CASPR4 or APOL3 nucleotide sequence or
other
nucleotide sequence referenced in Table B under conditions of high
stringency); a nucleic acid aptamer
that specifically binds a polypeptide encoded by a PADI2, APOB, ILIRL2,
ILIRLl, WASPIP,
ADAMTS2, BYES, TM7SF3, LOXLl, CASPR4 or APOL3 nucleotide sequence or other
nucleotide
sequence referenced in Table B; and an antibody that specifically binds to a
polypeptide encoded by a
PADI2, APOB, ILIRL2, ILIRL1, WASPIP, ADAlIsITS2, BYES, TM7SF3, LO~Zl, CASPR4
orAPOL3
nucleotide sequence or other nucleotide sequence referenced in Table B or
binds to a nucleic acid
having such a nucleotide sequence. In an embodiment, the antibody selectively
binds to an epitope
comprising an amino acid encoded by rs1367117, rs1041973 and rs398829. In
specific embodiments,
the osteoarthritis directed molecule interacts with a nucleic acid or
polypeptide variant associated with
osteoarthritis, such as variants referenced herein. In other embodiments, the
osteoarthritis directed
molecule interacts with a polypeptide involved in a signal pathway of a
polypeptide encoded by a
PADI2, APOB, ILIRL2, ILIRLl, WASPIP, ADAMTS2, BYES, TM7SF3, LOXL1, CASPR4
orAPOL3
nucleotide sequence or other nucleotide sequence referenced in Table B, or a
nucleic acid comprising
such a nucleotide sequence.
[0120] Compositions sometimes include an adjuvant known to stimulate an immune
response, and
in certain embodiments, an adjuvant that stimulates a T-cell lymphocyte
response. Adjuvants are
known, including but not limited to an aluminum adjuvant (e.g., aluminum
hydroxide); a cytokine
adjuvant or adjuvant that stimulates a cytokine response (e.g., interleukin
(IL)-12 and/or gamma-
interferon cytokines); a Freund-Type mineral oil adjuvant emulsion (e.g.,
Freund's complete or
incomplete adjuvant); a synthetic lipoid compound; a copolymer adjuvant (e.g.,
TitreMax); a saponin~
Quil A; a liposome; an oil-in-water emulsion (e.g_, an emulsion stabilized by
Tween 80 and pluronic
37
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
polyoxyethlene/polyoxypropylene block copolymer (Syntex Adjuvant Formulation);
TitreMax;
detoxified endotoxin (MPL) and mycobacterial cell wall components (TDW, CWS)
in 2% squalene
(Ribi Adjuvant System)); a muramyl dipeptide; an immune-stimulating complex
(ISCOM, e.g., an Ag-
modified saponin/cholesterol micelle that forms stable cage-like structure);
an aqueous phase adjuvant
that does not have a depot effect (e.g., Gerbu adj uvant); a carbohydrate
polymer (e.g., AdjuPrime); L-
tyrosine; a manide-oleate compound (e.g., Montanide); an ethylene-vinyl
acetate copolymer (e.g., Elvax
40W1,2); or lipid A, for example. Such compositions are useful for generating
an immune response
against osteoarthritis directed molecule (e.g., an I~LA-binding subsequence
within a polypeptide
encoded by a PADI2, APOB, ILIRL2, ILIRLI, WASPIP, ADAMTS2, BYES, TM7SF3,
LOXLI, CASPR4
or APOL3 nucleotide sequence). In such methods, a peptide having an amino acid
subsequence of a
polypeptide encoded by a PAD12, APOB, ILIRL2, ILIRLl, WASPIP, ADAMTS~, BYES,
TM7SF3,
LOXLl, CASPR4 or APOL3 nucleotide sequence is delivered to a subject, where
the subsequence binds
to an HLA molecule and induces a CTL lymphocyte response. The peptide
sometimes is delivered to
the subject as an isolated peptide or as a minigerie in a plasmid that encodes
the peptide. Methods for
identifying HLA-binding subsequences in such polypeptides are known (see e.g.,
publication
W002/20616 and PCT application US98/01373 for methods of identifying such
sequences).
[0121] The cell may be in a group of cells cultured i~ vitro or in a tissue
maintained in vitro or
present in an animal in vivo (e.g., a rat, mouse, ape or human). In certain
embodiments, a composition
comprises a component from a cell such as a nuc leic acid molecule (e.g.,
genomic DNA), a protein
mixture or isolated protein, for example. The aforementioned compositions have
utility in diagnostic,
prognostic and pharmacogenomic methods described previously and in
therapeutics described hereafter.
Certain osteoarthritis directed molecules are described in greater detail
below.
Compounds
[0122] Compounds can be obtained using any of the numerous approaches in
combinatorial library
methods known in the art, including: biological libraries; peptoid libraries
(libraries of molecules having
the functionalities of peptides, but with a novel, non-peptide backbone which
are resistant to enzymatic
degradation but which nevertheless remain bioactive (see, e.g., Zuckermann et
al., J. Med. Chem.37:
2678-85 (1994)); spatially addressable parallel solid phase or solution phase
libraries; synthetic library
methods requiring deconvolution; "one-bead one-compound" library methods; and
synthetic library
methods using affinity chromatography selection. Biological library and
peptoid library approaches are
typically limited to peptide libraries, while the other approaches are
applicable to peptide, non-peptide
oligomer or small molecule libraries of compourids (Lam, Anticancer Drug Des.
12: 145, (1997)).
Examples of methods for synthesizing molecular libraries are described, for
example, in DeWitt et al.,
Proc. Natl. Acad. Sci. U.S.A. 90: 6909 (1993); Erb et al., Proc. Natl. Acad.
Sci. USA 91: 11422 (1994);
Zuckermann et al., J. Med. Chem. 37: 2678 (1994); Cho et al., Science 261:
1303 (1993); Carrell et al.,
Angew. Chem. Int. Ed. Engl. 33: 2059 (1994); Carell et al., Angew. Chem. Int.
Ed. Engl. 33: 2061
(1994); and in Gallop et al., J. Med. Chem. 37: 1233 (1994).
38
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0123] Libraries of compounds may be presented in solution (e.g,, Houghten,
Biotechniques 13:
412-421 (1992)), or on beads (Lam, Nature 354: 82-84 ( 1991)), chips (Fodor,
Nature 364: 555-556
(1993)), bacteria or spores (Ladner, United States Patent No. 5,223,409),
plasmids (Cull et al., Proc.
Natl. Acad. Sci. USA 89: 1865-1869 (1992)) or on phaga (Scott and Smith,
Science 249: 386-390
(1990); Devlin, Science 249: 404-406 (1990); Cwirla et al., Proc. Natl. Acad.
Sci. 87: 6378-6382
(1990); Felici, J. Mol. Biol. 222: 301-310 (1991); Ladner supra.).
[0124] A compound sometimes alters expression and sometimes alters activity of
a polypeptide
target and may be a small molecule. Small molecules include, but are not
limited to, peptides,
peptidomimetics (e.g., peptoids), amino acids, amino acid analogs,
polynucleotides, polynucleotide
analogs, nucleotides, nucleotide analogs, organic or inorganic compounds
(i.e., including heteroorganic
and organometallic compounds) having a molecular weight less than about 10,000
grams per mole,
organic or inorganic compounds having a molecular weight less than about 5,000
grams per mole,
organic or inorganic compounds having a molecular weight less than about 1,000
grams per mole,
organic or inorganic compounds having a molecular weight less than about 500
grams per mole, and
salts, esters, and other pharmaceutically acceptable forms of such compounds.
Antisense Nucleic Acid Molecules, Ribozymes, RNAi, siRNA and Modified Nucleic
Acid Molecules
[0125] An "antisense" nucleic acid refers to a nucle otide sequence
complementary to a "sense"
nucleic acid encoding a polypeptide, e.g., complementary to the coding strand
of a double-stranded
cDNA molecule or complementary to an mRNA sequence. The antisense nucleic acid
can be
complementary to an entire coding strand, or to a portion thereof or a
substantially identical sequence
thereof. In another embodiment, the antisense nucleic acid molecule is
antisense to a "noncoding
region" of the coding strand of a nucleotide sequence (e,g,, 5' and 3'
untranslated regions in SEQ ID
NO: 1-13 or a nucleotide sequence referenced in Table $).
[0126] An antisense nucleic acid can be designed such that it is complementary
to the entire coding
region of an mRNA encoded by a nucleotide sequence (e.g., SEQ ID NO: 1-13, SEQ
ID NO: 14-36 or a
nucleotide sequence referenced in Table B), and often the antisense nucleic
acid is an oligonucleotide
antisense to only a portion of a coding or noncoding region of the mRNA. For
example, the antisense
oligonucleotide can be complementary to the region surrounding the translation
start site of the mRNA,
e.g., between the -10 and +10 regions of the target gene nucleotide sequence
of interest. An antisense
oligonucleotide can be, for example, about 7, 10, 15, 20, 25, 30, 35, 40, 45,
50, 55, 60, 65, 70, 75, 80, or
more nucleotides in length. The antisense nucleic acids, which include the
ribozymes described
hereafter, can be designed to target a PADI2, APOB, IL11zL2, ILIRLl, WASPIP,
ADAMTS2, BT~ES,
TM7SF3, LOXLI, CASPR~ orAPOL3 nucleotide sequence, often a variant associated
with
osteoarthritis, or a substantially identical sequence thereof. Among the
variants, minor alleles and major
alleles can be targeted, and those associated with a higher risk of
osteoarthritis are often designed,
tested, and administered to subjects.
39
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0127] An antisense nucleic acid can be constructed using chemical synthesis
and enzymatic
ligation reactions using standard procedures. For example, an antisense
nucleic acid (e.g., an antisense
oligonucleotide) can be chemically synthesized using naturally occurring
nucleotides or variously
modified nucleotides designed to increase the biological stability of the
molecules or to increase the
physical stability of the duplex formed between the antisense and sense
nucleic acids, e.g.,
phosphorothioate derivatives and acridine substituted nucleotides can be used.
Antisense nucleic acid
also can be produced biologically using an expression vector into which a
nucleic acid has been
subcloned in an antisense orientation (i.e., RNA transcribed from the inserted
nucleic acid will be of an
antisense orientation to a target nucleic acid of interest, described further
in the following subsection).
[012] When utilized as therapeutics, antisense nucleic acids typically are
administered to a subject
(e.g., by direct injection at a tissue site) or generated in situ such that
they hybridize with or bind to
cellular mRNA and/or genomic DNA encoding a polypeptide and thereby inhibit
expression of the
polypeptide, for example, by inhibiting transcription and/or translation.
Alternatively, antisense nucleic
acid molecules can be modified to target selected cells and then are
administered systemically. For
systemic administration, antisense molecules can be modified such that they
specifically bind to
receptors or antigens expressed on a selected cell surface, for example, by
linking antisense nucleic acid
molecules to peptides or antibodies which bind to cell surface receptors or
antigens. Antisense nucleic
acid molecules can also be delivered to cells using the vectors described
herein. Sufficient intracellular
concentrations of antisense molecules are achieved by incorporating a strong
promoter, such as a pol II
or pol III promoter, in the vector construct.
[0129] Antisense nucleic acid molecules sometimes are alpha-anoneric nucleic
acid molecules.
An alpha-anomeric nucleic acid molecule forms specific double-stranded hybrids
with complementary
RNA in which, contrary to the usual beta-units, the strands run parallel to
each other (Gaultier et al.,
Nucleic Acids. Res. 15: 6625-6641 (1987)). Antisense nucleic acid molecules
can also comprise a 2'-0-
methylribonucleotide (moue et al., Nucleic Acids Res. 15: 6131-6148 ( 1987))
or a chimeric RNA-DNA
analogue (moue et al., FEBS Lett. 215: 327-330 (1987)). Antisense nucleic
acids sometimes are
composed of DNA or PNA or any other nucleic acid derivatives described
previously.
[0130] In another embodiment, an antisense nucleic acid is a ribozyme. A
ribozyme having
specificity for a PADI2, APOB, ILIRL2, ILIRLI, WASPIP, ADAMTS2, BYES, TM7SF3,
LOXL1,
CASPR4 or APOL3 nucleotide sequence or other nucleotide sequence referenced in
Table B can include
one or more sequences complementary to such a nucleotide sequence, and a
sequence having a known
catalytic region responsible for mRNA cleavage (see e.g., U.S. Pat. No_
5,093,246 or Haselhoff and
Gerlach, Nature 334: 585-591 (1988)). For example, a derivative of a
Tetrahymena L-19 IVS RNA is
sometimes utilized in which the nucleotide sequence of the active site is
complementary to the
nucleotide sequence to be cleaved in a mRNA (see e.g., Cech et al. U.S .
Patent No. 4,987,071; and Cech
et al. U.S. Patent No. 5,116,742). Also, target mRNA sequences can ba used to
select a catalytic RNA
having a specific ribonuclease activity from a pool of RNA molecules Csee
e.g., Bartel ~ Szostak,
Science 261: 1411-1418 (1993)).
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0131] Osteoarthritis directed molecules include in certain embodiments
nucleic acids that can
form triple helix structures with a PADI2, APOB, ILIRL2, ILIRLl, WASPIP,
ADAMTS2, BhES,
TM7SF3, LOXL1, CASPR4 or APOL3 nucleotide sequence or other nucleotide
sequence referenced in
Table B, or a substantially identical sequence thereof, especially one that
includes a regulatory region
that controls expression of a polypeptide. Gene expression can be inhibited by
targeting nucleotide
sequences complementary to the regulatory region of a nucleotide sequence
referenced herein or a
substantially identical sequence (e.g., promoter and/or enhancers) to form
triple helical structures that
prevent transcription of a gene in target cells (see e.g., Helene, Anticancer
Drug Des. 6(6): 569-84
(1991); Helene et al., Ann. N.Y. Acad. Sci. 660: 27-36 (1992); and Maher,
Bioassays 14(12): 807-15
(1992). Potential sequences that can be targeted for triple helix formation
can be increased by creating a
so-called "switchback" nucleic acid molecule. Switchback molecules are
synthesized in an alternating
5'-3', 3'-5' manner, such that they base pair with first one strand of a dup
lex and then the other,
eliminating the necessity for a sizeable stretch of either purines or
pyrimidines to be present on one
strand of a duplex.
[0132] Osteoarthritis directed molecules include RNAi and siRNA rmcleic acids.
Gene expression
may be inhibited by the introduction of double-stranded RNA (dsRNA), which
induces potent and
specific gene silencing, a phenomenon called RNA interference or RNAi_ See,
e.g., Fire et al., US
Patent Number 6,506,559; Tuschl et al. PCT International Publication No . WO
01/75164; Kay et al.
PCT International Publication No. WO 03/010180A1; or Bosher JM, Labouesse, Nat
Cell Biol 2000
Feb;2(2):E31-6. This process has been improved by decreasing the size of the
double-stranded RNA to
20-24 base pairs (to create small-interfering RNAs or siRNAs) that "switched
off' genes in mammalian
cells without initiating an acute phase response, i.e., a host defense
mechanism that often results in cell
death (see, e.g., Caplen et al. Proc Natl Acad Sci U S A. 2001 Aug 14;98
17):9742-7 and Elbashir et al.
Methods 2002 Feb;26(2):199-213). There is increasing evidence of post-
rtranscriptional gene silencing
by RNA interference (RNAi) for inhibiting targeted expression in mammalian
cells at the mRNA level,
in human cells. There is additional evidence of effective methods for
inhibiting the proliferation and
migration of tumor cells in human patients, and for inhibiting metastatic
cancer development (see, e.g.,
U.S. Patent Application No. US2001000993183; Caplen et al. Proc Natl Acad Sci
U S A; and
Abderrahmani et al. Mol Cell Biol 2001 Nov21(21):7256-67).
[0133] An "siRNA" or "RNAi" refers to a nucleic acid that forms a double
stranded RNA and has
the ability to reduce or inhibit expression of a gene or target gene when the
siRNA is delivered to or
expressed in the same cell as the gene or target gene. "siRNA" refers to short
double-stranded RNA
formed by the complementary strands. Complementary portions of the siRNA that
hybridize to form
the double stranded molecule often have substantial or complete identity to
the target molecule
sequence. In one embodiment, an siRNA refers to a nucleic acid that has
substantial or complete
identity to a target gene and forms a double stranded siRNA.
[0134] When designing the siRNA molecules, the targeted region often is
selected from a given
DNA sequence beginning 50 to 100 nucleotides downstream of the start c odon.
See, e.g., Elbashir et
41
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
al,. Methods 26:199-213 (2002). Initially, 5' or 3' UTRs and regions nearby
the start codon were
avoided assuming that UTR-binding proteins and/or translation initiation
complexes may interfere with
binding of the siRNP or RISC endonuclease complex. Sometimes regions of the
target 23 nucleotides
in length conforming to the sequence motif AA(N19)TT (N, an nucleotide), and
regions with
approximately 30% to 70% G/C-content (often about 50% G/C-content) often are
selected. If no
suitable sequences are found, the search often is extended using the motif
NA(N21 ). The sequence of
the sense siRNA sometimes corresponds to (N19) TT or N21 (position 3 to 23 of
the 23-nt motif),
respectively. In the latter case, the 3' end of the sense siRNA often is
converted to TT. The rationale
for this sequence conversion is to generate a symmetric duplex with respect to
the sequence composition
of the sense and antisense 3' overhangs. The antisense siRNA is synthesized as
the complement to
position 1 to 21 of the 23-nt motif. Because position 1 of the 23-nt motif is
not recognized sequence-
specifically by the antisense siRNA, the 3'-most nucleotide residue of the
antisense siRNA can be
chosen deliberately. However, the penultimate nucleotide of the antisense
siRNA (complementary to
position 2 of the 23-nt motif) often is complementary to the targeted
sequence. For simplifying
chemical synthesis, TT often is utilized. siRNAs corresponding to the target
motif NAR(N17)YNN,
where R is purine (A,G) and Y is pyrimidine (C,U), often are selected.
Respective 21 nuc leotide sense
and antisense siRNAs often begin with a purine nucleotide and can also be
expressed from pol III
expression vectors without a change in targeting site. Expression of RNAs from
pol III promoters often
is efficient when the first transcribed nucleotide is a purine.
[0135] The sequence of the siRNA can correspond to the full length target
gene, or a subsequence
thereof. Often, the siRNA is about 15 to about 50 nucleotides in length (e.g.,
each complementary
sequence of the double stranded siRNA is 15-50 nucleotides in length, and the
double stranded siRNA
is about 15-50 base pairs in length, sometimes about 20-30 nucleotides in
length or about 20-25
nucleotides in length, e.g., 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30
nucleotides in length. The siRNA
sometimes is about 21 nucleotides in length. Methods of using siRNA are well
known in -the art, and
specific siRNA molecules may be purchased from a number of companies including
Dharmacon
Research, Inc.
[0136] Antisense, ribozyme, RNAi and siRNA nucleic acids can be altered to
form modified
nucleic acid molecules. The nucleic acids can be altered at base moieties,
sugar moieties or phosphate
backbone moieties to improve stability, hybridization, or solubility of the
molecule. For example, the
deoxyribose phosphate backbone of nucleic acid molecules can be modified to
generate peptide nucleic
acids (see Hyrup et al., Bioorganic & Medicinal Chemistry 4 (1): 5-23 (1996)).
As used herein, the
terms "peptide nucleic acid" or "PNA" refers to a nucleic acid mimic such as a
DNA mirrzic, in which
the deoxyribose phosphate baclcbone is replaced by a pseudopeptide backbone
and only the four natural
nucleobases are retained. The neutral backbone of a PNA can allow for specific
hybridization to DNA
and RNA under conditions of low ionic strength. Synthesis of PNA oligomers can
be performed using
standard solid phase peptide synthesis protocols as described, for example, in
Hyrup et al. ~ (1996) supra
and Perry-O'Keefe et al., Proc. Natl. Acad. Sci. 93: 14670-675 (1996).
42
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0137] PNA nucleic acids can be used in prognostic, diagnostic, and
therapeutic applications. For
example, PNAs can be used as antisense or antigene agents for sequence-
specific modulation of gene
expression by, for example, inducing transcription or translation arrest or
inhibiting replication. PNA
nucleic acid molecules can also be used in the analysis of single base pair
mutations in a gene, (e.g., by
PNA-directed PCR clamping); as "artificial restriction enzymes" when used in
combination with other
enzymes, (e.g., Sl nucleases (Hyrup (1996) supra)); or as probes or primers
for DNA sequencing or
hybridization (Hyrup et al., (1996) supra; Perry-O'Keefe supra).
[0138] In other embodiments, oligonucleotides may include other appended
groups such as
peptides (e.g., for targeting host cell receptors in vivo), or agents
facilitating transport across cell
membranes (see e.g., Letsinger et al., Proc. Natl. Acad. Sci. USA 86: 6553-
6556 (1989); Lemaitre et al.,
Proc. Natl. Acad. Sci. USA 84: 648-652 (1987); PCT Publication No. W088/09810)
or the blood-brain
barrier (see, e.g., PCT Publication No. W089/10134). In addition,
oligonucleotides can be modified
with hybridization-triggered cleavage agents (See, e.g., Krol et al., Bio-
Techniques 6: 958-976 [ 1988))
or intercalating agents. (See, e.g., Zon, Pharm. Res. 5: 539-549 (1988) ). To
this end, the
oligonucleotide may be conjugated to another molecule, (e.g., a peptide,
hybridization triggered cross-
linking agent, transport agent, or hybridization-triggered cleavage agent).
[0139] Also included herein are molecular beacon oligonucleotide primer and
probe molecules
having one or more regions complementary to a PAD12, APOB, ILIRL2, ILIRLl,
WASPIP, AD~1MTS2,
BT~ES, TM7SF3, LOXI,1, C'ASPR4 or APOL3 nucleotide sequence or other
nucleotide sequence
referenced in Table B, or a substantially identical sequence thereof, two
complementary regions one
having a fluorophore and one a quencher such that the molecular beacon is
useful for quantifying the
presence of the nucleic acid in a sample. Molecular beacon nucleic acids are
described, for example, in
Lizardi et al., U.S. Patent No. 5,854,033; Nazarenko et al., U.S. Patent No.
5,866,336, and Lival~ et al.,
U.S. Patent 5,876,930.
Antibodies
[0140] The term "antibody" as used herein refers to an immunoglobulin molecule
or
immunologically active portion thereof, i.e., an antigen-binding portion.
Examples of immunologically
active portions of immunoglobulin molecules include Flab) and F(ab')2
fragments which can be
generated by treating the antibody with an enzyme such as pepsin. An antibody
sometimes is a
polyclonal, monoclonal, recombinant (e.g., a chimeric or humanized), fully
human, non-human (e.g.,
murine), or a single chain antibody. An antibody may have effector function
and can fix complement,
and is sometimes coupled to a toxin or imaging agent.
[0141] A full-length polypeptide or antigenic peptide fragment encoded by a
nucleotide sequence
referenced herein can be used as an immunogen or can be used to identify
antibodies made with other
immunogens, e.g., cells, membrane preparations, and the like. An antigenic
peptide often includes at
least 8 amino acid residues of the amino acid sequences encoded by a
nucleotide sequence referenced
herein, or substantially identical sequence thereof, and encompasses an
epitope. Antigenic peptides
43
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
sometimes include 10 or more amino acids, 15 or more amino acids, 20 or more
amino acids, or 30 or
more amino acids. Hydrophilic and hydrophobic fragments of polypeptides
sometimes are used as
immunogens.
[0142] Epitopes encompassed by the antigenic peptide are regions located on
the surface of the
polypeptide (e.g., hydrophilic regions) as well as regions with high
antigenicity. For example, an Emini
surface probability analysis of the human polypeptide sequence can be used to
indicate the regions that
have a particularly high probability of being localized to the surface of the
polypeptide and are thus
likely to constitute surface residues useful for targeting antibody
production. The antibody may bind an
epitope on any domain or region on polypeptides described herein.
[0143] Also, chimeric, humanized, and completely human antibodies are useful
for applications
which include repeated administration to subjects. Chimeric and humanized
monoclonal antibodies,
comprising both human and non-human portions, can be made using standard
recombinant DNA
techniques. Such chimeric and humanized monoclonal antibodies can be produced
by recombinant
DNA techniques known in the art, for example using methods described in
Robinson et al International
Application No. PCT/US86/02269; Akira, et al European Patent Application
184,187; Taniguchi, M.,
European Patent Application 171,496; Morrison et al European Patent
Application 173,494; Neuberge=r
et al PCT International Publication No. WO 86/01533; Cabilly et al U.S. Patent
No. 4,816,567; Cabilly
et al European Patent Application 125,023; Better et al., Science 240: 1041-
1043 (1988); Liu et al.,
Proc. Natl. Acad. Sci. USA 84: 3439-3443 (1987); Liu et al., J. Immunol. 139:
3521-3526 (1987); Sun
et al., Proc. Natl. Acad. Sci. USA 84: 214-218 (1987); Nishimura et al., Canc.
Res. 47: 999-1005
(1987); Wood et al., Nature 314: 446-449 (1985); and Shaw et al., J. Natl.
Cancer Inst. 80: 1553-1559
(1988); Morrison, S. L., Science 229: 1202-1207 (1985); Oi et al.,
BioTechniques 4: 214 (1986);
Winter U.S. Patent 5,225,539; Jones et al., Nature 321: 552-525 (1986);
Verhoeyan et al., Science 239:
1534; and Beidler et al., J. Immunol. 141: 4053-4060 (1988).
[0144] Completely human antibodies are particularly desirable for therapeutic
treatment of human
patients. Such antibodies can be produced using transgenic mice that are
incapable of expressing
endogenous immunoglobulin heavy and light chains genes, but which can express
human heavy and
light chain genes. See, for example, Lonberg and Huszar, Int. Rev. Immunol.
13: 65-93 (1995); and
U.S. PatentNos. 5,625,126; 5,633,425; 5,569,825; 5,661,016; and 5,545,806. In
addition, companies
such as Abgenix, Inc. (Fremont, CA) and Medarex, Inc. (Princeton, NJ), can be
engaged to provide
human antibodies directed against a selected antigen using technology similar
to that described above.
Completely human antibodies that recognize a selected epitope also can be
generated using a technique
referred to as "guided selection." In this approach a selected non-human
monoclonal antibody (e.g., a
murine antibody) is used to guide the selection of a completely human antibody
recognizing the same
epitope. This technology is described for example by Jespers et al.,
Bio/Teclmology 12: 899-903
( 1994).
[0145] An antibody can be a single chain antibody. A single chain antibody
(scFV) can be
engineered (see, e.g., Colcher et al., Ann. N Y Acad. Sci. 880: 263-80 (1999);
and Reiter, Clin. Cancer
44
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
Res. 2: 245-52 (1996)). Single chain antibodies can be dimerized or
multimerized to generate
multivalent antibodies having specificities for different epitopes of the same
target polypeptide.
[0146] Antibodies also may be selected or modified so that they exhibit
reduced or no ability to
bind an Fc receptor. For example, an antibody may be an isotype or subtype,
fragment or other mutant,
which does not support binding to an Fc receptor (e.g., it has a mutagenized
or deleted Fc receptor
binding region).
[0147] Also, an antibody (or fragment thereof) may be conjugated to a
therapeutic moiety such as a
cytotoxin, a therapeutic agent or a radioactive metal ion. A cytotoxin or
cytotoxic agent includes any
agent that is detrimental to cells. Examples include taxol, cytochalasin B,
gramicidin D, ethidium
bromide, emetine, mitomycin, etoposide, tenoposide, vincristine, vinblastine,
colchicin, doxorubicin,
daunorubicin, dihydroxy anthracin dione, mitoxantrone, mithramycin,
actinomycin D, 1
dehydrotestosterone, glucocorticoids, procaine, tetracaine, lidocaine,
propranolol, and puromycin and
analogs or homologs thereof. Therapeutic agents include, but are not limited
to, antimetabolites (e.g.,
methotrexate, 6-mercaptopurine, 6-thioguanine, cytarabine, 5-fluorouracil
decarbazine), alkylating
agents (e.g., mechlorethamine, thiotepa chlorambucil, melphalan, carmustine
(BCNU) and lomustine
(CCNU), cyclophosphamide, busulfan, dibromomannitol, streptozotocin, mitomycin
C, and cis-
dichlorodiamine platinum (II) (DDP) cisplatin), anthracyclines (e.g.,
daunorubicin (formerly
daunomycin) and doxorubicin), antibiotics (e.g., dactinomycin (formerly
actinomycin), bleomycin,
mithramycin, and anthramycin (AMC)), and anti-mitotic agents (e.g.,
vincristine and vinblastine).
[0148] Antibody conjugates can be used for modifying a given biological
response. For example,
the drug moiety may be a protein or polypeptide possessing a desired
biological activity. Such proteins
may include, for example, a toxin such as abrin, ricin A, pseudomonas
exotoxin, or diphtheria toxin; a
polypeptide such as tumor necrosis factor, gamma-interferon, alpha-interferon,
nerve growth factor,
platelet derived growth factor, tissue plasminogen activator; or, biological
response modifiers such as,
for example, lymphokines, interleukin-1 ("IL,-1"), interleukin-2 ("II,-2"),
interleukin-6 ("IL,-6"),
granulocyte macrophage colony stimulating factor ("GM-CSF"), granulocyte
colony stimulating factor
("G-CSF"), or other growth factors. Also, an antibody can be conjugated to a
second antibody to form
an antibody heteroconjugate as described by Segal in U.S. Patent No.
4,676,980, for example.
[0149] An antibody (e.g., monoclonal antibody) can be used to isolate target
polypeptides by
standard techniques, such as affinity chromatography or immunoprecipitation.
Moreover, an antibody
can be used to detect a target polypeptide (e.g., in a cellular lysate or cell
supernatant) in order to
evaluate the abundance and pattern of expression of the polypeptide.
Antibodies can be used
diagnostically to monitor polypeptide levels in tissue as part of a clinical
testing procedure, e.g., to
determine the efficacy of a given treatment regimen. Detection can be
facilitated by coupling (i.e.,
physically linking) the antibody to a detectable substance (i.e., antibody
labeling). Examples of
detectable substances include various enzymes, prosthetic groups, fluorescent
materials, luminescent
materials, bioluminescent materials, and radioactive materials. Examples of
suitable enzymes include
horseradish peroxidase, alkaline phosphatase, (3-galactosidase, or
acetylcholinesterase; examples of
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
suitable prosthetic group complexes include streptavidin/biotin and
avidin/biotin; examples of suitable
fluorescent materials include umbelliferone, fluorescein, fluorescein
isothiocyanate, rhodamine,
dichlorotriazinylamine fluorescein, dansyl chloride or phycoerythrin; an
example of a luminescer>Et
material includes luminol; examples of bioluminescent materials include
luciferase, luciferin, and.
aequorin, and examples of suitable radioactive material include l2sh 1311, sss
or 3H. Also, an antibody
can be utilized as a test molecule for determining whether it can treat
osteoarthritis, and as a therapeutic
for administration to a subject for treating osteoarthritis.
[0150] An antibody can be made by immunizing with a purified antigen, or a
fragment thereof,
e.g., a fragment described herein, a membrane associated antigen, tissues,
e.g., crude tissue preparations,
whole cells, preferably living cells, lysed cells, or cell fractions.
[0151] Included herein are antibodies which bind only a native polypeptide,
only denatured or
otherwise non-native polypeptide, or which bind both, as well as those having
linear or conformational
epitopes. Conformational epitopes sometimes can be identified by selecting
antibodies that bind to
native but not denatured polypeptide. Also featured are antibodies that
specifically bind to a
polypeptide variant associated with osteoarthritis.
Methods for Identifyin~ Candidate Therapeutics for Treating Osteoarthritis
[0152] Current therapies for the treatment of osteoarthritis have limited
efficacy, limited
tolerability and significant mechanism-based side effects, and few of the
available therapies adequately
address underlying defects. Current therapeutic approaches were largely
developed in the absenc a of
defined molecular targets or even a solid understanding of disease
pathogenesis. Therefore, provided
are methods of identifying candidate therapeutics that target biochemical
pathways related to the
development of osteoarthritis.
[0153] Thus, featured herein are methods for identifying a candidate
therapeutic for treating
osteoarthritis. The methods comprise contacting a test molecule with a target
molecule in a system. A
"target molecule" as used herein refers to a PADI2, APOB, IL1RL2, ILIRLl,
WASPIP, ADAMTS~,
BIdES, TM7SF3, LOXLI, CASPR4 or APOL3 nucleic acid or other nucleotide
sequence referenced in
Table B, a substantially identical nucleic acid thereof, or a fragment
thereof, and an encoded
polypeptide of the foregoing. The methods also comprise determining the
presence or absence o~ an
interaction between the test molecule and the target molecule, where the
presence of an interaction
between the test molecule and the nucleic acid or polypeptide identifies the
test molecule as a candidate
osteoarthritis therapeutic. The interaction between the test molecule and the
target molecule may be
quantified.
[0154] Test molecules and candidate therapeutics include, but are not limited
to, compounds,
antisense nucleic acids, siRNA molecules, ribozymes, polypeptides or proteins
encoded by a PAI~I2,
APOB, ILIRL2, ILIRLl, WASPIP, ADAMTS2, BhES, TM7SF3, LO~Ll, CASPR~ orAPOL3
nucleotide
sequence or other nucleotide sequence referenced in Table B , or a
substantially identical sequence or
fragment thereof, and immunotherapeutics (e.g., antibodies and HLA-presented
polypeptide fragments).
46
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
A test molecule or candidate therapeutic may act as a modulator of target
molecule concentration or
target molecule function in a system. A "modulator" may agonize (i.e., up-
regulates) or antagonize
(i.e., down-regulates) a target molecule concentration partially or completely
in a system by affecting
such cellular functions as DNA replication and/or DNA processing (e.g., DNA
methylation or DNA
repair), RNA transcription and/or RNA processing (e.g., removal of intronic
sequences and/or
translocation of spliced mRNA from the nucleus), polypeptide production (e.g.,
translation of the
polypeptide from mRNA), and/or polypeptide post-translational modification
(e.g., glycosylation,
phosphorylation, and proteolysis of pro-polypeptides). A modulator may also
agonize or antagonize a
biological function of a target molecule partially or completely, where the
function may include
adopting a certain structural conformation, interacting with one or more
binding partners, ligand
binding, catalysis (e.g., phosphorylation, dephosphorylation, hydrolysis,
methylation, and
isomerization), and an effect upon a cellular event (e.g., effecting
progression of osteoarthritis). Any
modulator may be utilized, such as a peptidyl arginine deiminase modulator
(e.g., PADI2 likely is a
peptidyl arginine deiminase) described in WO-09851784 and WO0244360A2 or an
apolipoprotein (e.g.,
APOB includes an apolipoprotein domain) modulatory compound (e.g., WO-
2004017969, WO-
03002533, US 6,369,075, WO-02098839, WO-02098871, WO-00177077, WO-00153260, WO-
00105767), antibody (e.g., WO-9600903A1, US 6,309,844 and US 5,330,910) or
antisense molecule
(e.g., W003011887A2 and W003097662A1).
[0155] As used herein, the term "system" refers to a cell free in vitro
environment and a cell-based
environment such as a collection of cells, a tissue, an organ, or an organism.
A system is "contacted"
with a test molecule in a variety of manners, including adding molecules in
solution and allowing them
to interact with one another by diffusion, cell injection, and any
administration routes in an animal. .As
used herein, the term "interaction" refers to an effect of a test molecule on
test molecule, where the
effect sometimes is binding between the test molecule and the target molecule,
and sometimes is an
observable change in cells, tissue, or organism.
[0156] There are many standard methods for detecting the presence or absence
of interaction
between a test molecule and a target molecule. For example, titrametric,
acidimetric, radiometric,
NMR, monolayer, polarographic, spectrophotometric, fluorescent, and ESR assays
probative of a target
molecule interaction may be utilized. Examples of G protein-coupled receptor
assays are known, fog
example, and are described in WO-0242461 and WO-04013285.
[0157] ADAMTS2 activity and/or ADAMTS2 interactions can be detected and
quantified using
assays known in the art. For example, an immunoprecipitation assay or a kinase
activity assay that
employs a lcinase-inactivated MEI~ can be utilized. Kinase inactivated MEI~s
are known in the art, such
as a MEI~ that includes the mutation I~97M. In these assays, mammalian cells
(e.g., COS or NIH-3T3)
are transiently transfected with constructs expressing ADAMTS2, and in
addition, the cells are co-
transfected with oncogenic RAS or SRC or both. Oncogenic RAS or SRC activates
ADAMTS2 kina_se
activity. ADAMTS2 is immunoprecipitated from cell extracts using a monoclonal
antibody (e.g., 9E 10)
or a polyclonal antibody (e.g., from rabbit) specific for a unique peptide
from ADAMTS2. ADAMI'S'2 is
47
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
then resuspended in assay buffer containing GST-Mekl or GST-Mek2 and/or GST-
ERI~2. In addition,
[gamma 3zP] ATP can be added to detect and/or quantify phosphorylation
activity. Samples are
incubated for 5-30 minutes at 30°C, and then the reaction is terminated
by addition of EDTA. The
samples are centrifuged and the supernatant fractions are collected.
Phosphorylation activity is detected
using one of two methods: (i) activity of GST-ERK2 kinase can be measured
using MBP (myelin basic
protein, a substrate for ERIC) as substrate, or (ii) following incubation of
immunoprecipitated ADAMTS2
in reaction buffer containing GST-ERIC and [gamma 32P] ATP, transfer of
labeled ATP to kinase-dead
ERK can be quantified by a phosphor-imager or densitometer following PAGE
separation of
polypeptide products (phosphorylated and non-phosphorylated forms). These
types of assays are
described in Weber et al., Oncogene 19: 169-176 (2000); Mason et al., EMBO J.
18: 2137-2148 (1999);
Marais et al., J. Biol. Chem. 272: 4378-4383 (1997); Marais et al., EMBO J.
14: 3136-3145 (1995).
[0158] As noted above, ADAMTS2 includes a domain having metalloprotease
activity, and
modulators of such activity are known. Examples of such modulators are set
forth in W003063~62A2;
WO-09937625; WO-09918076; WO-09838163; WO-09837877; W09947550A1; W00177092A1;
WO0040577A1; W09942436A1; W09838163A1; W09837877A1; W004014379A1;
W003106381A2; WO03014098A1; W003014092A1 and W002096426A1.
[0159] Test molecule/target molecule interactions can be detected and/or
quantified using assays
known in the art. For example, an interaction can be determined by labeling
the test molecule and/or
the target molecule, where the label is covalently or non-covalently attached
to the test molecule or
target molecule. The label is sometimes a radioactive molecule such as l2sh
1311, ssS or 3H, which can be
detected by direct counting of radioemission or by scintillation counting.
Also, enzymatic labels such as
horseradish peroxidase, alkaline phosphatase, or luciferase may be utilized
where the enzymatic label
can be detected by determining conversion of an appropriate substrate to
product. In addition, presence
or absence of an interaction can be determined without labeling. For example,
a microphysiometer
(e.g., Cytosensor) is an analytical instrument that measures the rate at which
a cell acidifies its
environment using a light-addressable potentiometric sensor (LAPS). Changes in
this acidificati on rate
can be used as an indication of an interaction between a test molecule and
target molecule (McConnell,
H. M. et al., Science 257: 1906-1912 (1992)).
[0160] In cell-based systems, cells typically include a PADI2, APOB, ILIRL2,
ILIRLl, T~YAS'PIP,
ADAMTS~, BYES, TM7SF3, LOXL1, CASPR4 or APOL3 nucleic acid or other nucleotide
sequence
referenced in Table B, an encoded polypeptide, or substantially identical
nucleic acid or polypeptide
thereof, and are often of mammalian origin, although the cell can be of any
origin. Whole cells, cell
homogenates, and cell fractions (e.g., cell membrane fractions) can be
subjected to analysis. Where
interactions between a test molecule with a target polypeptide are monitored,
soluble and/or membrane
bound forms of the polypeptide may be utilized. Where membrane-bound forms of
the polypeptide are
used, it may be desirable to utilize a solubilizing agent. Examples of such
solubilizing agents include
non-ionic detergents such as n-octylglucoside, n-dodecylglucoside, n-
dodecylmaltoside, octanoyl-N-
methylglucamide, decanoyl-N-methylglucamide, Triton~ X-100, Triton~ X-114,
Thesit~,
48
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
Isotridecypoly(ethylene glycol ether)n, 3-[(3-cholamidopropyl)dimethylamminio]-
1-propane sulfonate
(CHAPS), 3-[(3-cholamidopropyl)dimethylamminio]-2-hydroxy-1-propane sulfonate
(CHAPSO), or N-
dodecyl-N,N-dimethyl-3-ammonio-1-propane sulfonate.
[0161] An interaction between a test molecule and target molecule also can be
detected by
monitoring fluorescence energy transfer (FET) (see, e.g., Lakowicz et al.,
U.S. Patent No. 5,631,169;
Stavrianopoulos et al. U.S. Patent No. 4,868,103). A fluorophore label on a
first, "donor" molecule is
selected such that its emitted fluorescent energy will be absorbed by a
fluorescent label on a second,
"acceptor" molecule, which in turn is able to fluoresce due to the absorbed
energy. Alternately, the
"donor" polypeptide molecule may simply utilize the natural fluorescent energy
of tryptophan residues.
Labels are chosen that emit different wavelengths of light, such that the
"acceptor" molecule label may
be differentiated from that of the "donor". Since the efficiency of energy
transfer between the labels is
related to the distance separating the molecules, the spatial relationship
between the molecules can be
assessed. In a situation in which binding occurs between the molecules, the
fluorescent emission of the
"acceptor" molecule label in the assay.should be maximal. An FET binding event
can be conveniently
measured through standard fluorometric detection means well known in the art
(e.g., using a
fluorimeter).
[0162] In another embodiment, determining the presence or absence of an
interaction between a
test molecule and a target molecule can be effected by monitoring surface
plasmon resonance (see, e.g.,
Sjolander & Urbaniczk, Anal. Clzem. 63: 2338-2345 (1991) and Szabo et al.,
Curr. Opin. Struet. Biol. S:
699-705 (1995)). "Surface plasmon resonance" or "biomolecular interaction
analysis (BIA)" can be
utilized to detect biospecific interactions in real time, without labeling any
of the interactants (e.g.,
BIAcore). Changes in the mass at the binding surface (indicative of a binding
event) result in
alterations of the refractive index of light near the surface (the optical
phenomenon of surface plasmon
resonance (SPR)), resulting in a detectable signal which can be used as an
indication of real-time
reactions between biological molecules.
[0163] In another embodiment, the target molecule or test molecules are
anchored to a solid phase,
facilitating the detection of target molecule/test molecule complexes and
separation of the complexes
from free, uncomplexed molecules. The target molecule or test molecule is
immobilized to the solid
support. In an embodiment, the target molecule is anchored to a solid surface,
and the test molecule,
which is not anchored, can be labeled, either directly or indirectly, with
detectable labels discussed
herein.
[0164] It may be desirable to immobilize a target molecule, an anti-target
molecule antibody,
and/or test molecules to facilitate separation of target molecule/test
molecule complexes from
uncomplexed forms, as well as to accommodate automation of the assay. The
attachment between a test
molecule and/or target molecule and the solid support may be covalent or non-
covalent (see, e.g., U.S.
Patent No. 6,022,688 for non-covalent attachments). The solid support may be
one or more surfaces of
the system, such as one or more surfaces in each well of a microtiter plate, a
surface of a silicon wafer, a
surface of a bead (see, e.g., Lam, Nature 354: 82-84- (1991)) that is
optionally linked to another solid
49
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
support, or a channel in a microfluidic device, for example. Types of solid
supports, linker molecules
for covalent and non-covalent attachments to solid supports, and methods for
immobilizing nucleic
acids and other molecules to solid supports are well known (see, e.g., U.S.
Patant Nos. 6,261,776;
5,900,481; 6,133,436; and 6,022,688; and WIPO publication WO 01/18234).
[0165] In an embodiment, target molecule may be immobilized to surfaces via
biotin and
streptavidin. For example, biotinylated target polypeptide can be prepared
from biotin-NHS (N-
hydroxy-succinimide) using techniques known in the art (e.g., biotinylation
kit, Pierce Chemicals,
Rockford, IL), and immobilized in the wells of streptavidin-coated 96 well
plates (Pierce Chemical). In
another embodiment, a target polypeptide can be prepared as a fusion
polypeptide. For example,
glutathione-S-transferaseltarget polypeptide fusion can be adsorbed onto
glutathione sepharose beads
(Sigma Chemical, St. Louis, MO) or glutathione derivitized microtiter plates,
which are then combined
with a test molecule under conditions conducive to complex formation (e.g., at
physiological conditions
for salt and pH). Following incubation, the beads or microtiter plate wells
are yvashed to remove any
unbound components, or the matrix is immobilized in the case of beads, and
complex formation is
determined directly or indirectly as described above. Alternatively, the
complexes can be dissociated
from the matrix, and the level of target molecule binding or activity is
determined using standard
techniques.
[0166] In an embodiment, the non-immobilized component is added to the coated
surface
containing the anchored component. After the reaction is complete, unreacted
components are removed
(e.g., by washing) under conditions such that a significant percentage of
complexes formed will remain
immobilized to the solid surface. The detection of complexes anchored on the
solid surface can be
accomplished in a number of manners. Where the previously non-immobilized
component is pre-
labeled, the detection of label immobilized on the surface indicates that
complexes were formed. Where
the previously non-immobilized component is not pre-labeled, an indirect label
can be used to detect
complexes anchored on the surface, e.g., by adding a labeled antibody specific
for the immobilized
component, where the antibody, in turn, can be directly labeled or indirectly
labeled with, e.g., a labeled
anti-Ig antibody.
[0167] In another embodiment, an assay is performed utilizing antibodies that
specifically bind
target molecule or test molecule but do not interfere with binding of the
target molecule to the test
molecule. Such antibodies can be derivitized to a solid support, and unbound -
target molecule may be
immobilized by antibody conjugation. Methods for detecting such complexes ~ in
addition to those
described above for the GST-immobilized complexes, include immunodetection of
complexes using
antibodies reactive with the target molecule, as well as enzyme-linked assays
which rely on detecting an
enzymatic activity associated with the target molecule.
[0168] Cell free assays also can be conducted in a liquid phase. In such an
assay, reaction products
are separated from unreacted components, by any of a number of standard
techniques, including but not
limited to: differential centrifugation (see, e.g., Rivas, G., and Minton,
Ti~es~ds Biochern Sci Aug;18(8):
284-7 (1993)); chromatography (gel filtration chromatography, ion-exchange
chromatography);
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
electrophoresis (see, e.g., Ausubel et al., eds. Current Protocols in
Molecular Biology , J. Wil ey: New
York (1999)); and immunoprecipitation (see, e.g., Ausubel et al., eds.,
supra). Media and
chromatographic techniques are known to one skilled in the art (see, e.g.,
Heegaard, JMoI. Recognit.
Winter; 11 (1-6): 141-8 (1998); Hage & Tweed, J. Clzromatog~. B Biomed. Sci.
Appl. Oct 10; 699 (1-2):
499-525 (1997)). Further, fluorescence energy transfer may also be
conveniently utilized, as described
herein, to detect binding without further purification of the complex from
solution.
[0169] In another embodiment, modulators of target molecule expression are
identified. For
example, a cell or cell free mixture is contacted with a candidate compound
and the expression of target
mRNA or target polypeptide is evaluated relative to the level of expression of
target mRNA or target
polypeptide in the absence of the cmdidate compound. When expression of target
mRNA or target
polypeptide is greater in the presence of the candidate compound than in its
absence, the candidate
compound is identified as an agonist of target mRNA or target polypeptide
expression. Alternatively,
when expression of target mRNA or target polypeptide is less (e.g., less with
statistical significance) in
the presence of the candidate compound than in its absence, the candidate
compound is identified as an
antagonist or inhibitor of target mRNA or target polypeptide expression. The
level of target rnRNA or
target polypeptide expression can be determined by methods described herein.
[0170] In another embodiment, binding partners that interact with a target
molecule are detected.
The target molecules can interact with one or more cellular or extracellular
macromolecules, such as
polypeptides in vivo, and these interacting molecules are referred to herein
as "binding partne:rs."
Binding partners can agonize or antagonize target molecule biological
activity. Also, test mo lecules
that agonize or antagonize interactions between target molecules and binding
partners can be useful as
therapeutic molecules as they can up-regulate or down-regulated target
molecule activity in vzvo and
thereby treat osteoarthritis.
[0171] Binding partners of target molecules can be identified by methods known
in the art. For
example, binding partners may be identified by lysing cells and analyzing cell
lysates by elec-trophoretic
techniques. Alternatively, a two-hybrid assay or three-hybrid assay can be
utilized (see, e.g., U.S.
Patent No. 5,283,317; Zervos et al., Cell 72:223-232 (1993); Madura et al., J.
Biol. ChenZ. 2~SS: 12046-
12054 (1993); Bartel et al., BioteclZniques 14: 920-924 (1993); Iwabuchi et
al., Oncogene ~: 1693-1696
(1993); and Brent W094/10300). A two-hybrid system is based on the modular
nature of most
transcription factors, which consist of separable DNA-binding and activation
domains. The assay often
utilizes two different DNA constructs. In one construct, a PAD12, APOB,
ILIRL2, ILIRLI, WASPIP,
ADAMTS2, BVES, TM7SF3, LOXLI, CASPR4 or APOL3 nucleic acid or other nucleic
acid referenced
in Table B (sometimes referred to as the "bait") is fused to a gene encoding
the DNA binding domain of
a known transcription factor (e.g., GAL-4). In another construct, a DNA
sequence from a library of
DNA sequences that encodes a potential binding partner (sometimes referred to
as the "prey") is fused
to a gene that encodes an activation domain of the known transcription factor.
Sometimes, a PADI2,
APOB, ILIRL2, IL1RL1, WASPIP, ADAMTS2, BVES, TM7SF3, LOXLl, CASPR4 orAPOL3
nucleic
acid or other nucleic acid referenced in Table B can be fused to the
activation domain. If the "bait" and
51
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
the "prey" molecules interact in vivo, the DNA-binding and activation domains
of the transcription
factor are brought into close proximity. This proximity allows transcription
of a reporter gene (e.g.,
LacZ) which is operably linked to a transcriptional regulatory site responsive
to the transcription factor.
Expression of the reporter gene can be detected and cell colonies containing
the functional traps cription
factor can be isolated and used to identify the potential binding partner.
[0172] In an embodiment for identifying test molecules that antagonize or
agonize complex
formation between target molecules and binding partners, a reaction mixture
containing the target
molecule and the binding partner is prepared, under conditions and for a time
sufficient to allow
complex formation. The reaction mixture often is provided in the presence or
absence of the test
molecule. The test molecule can be included initially in the reaction mixture,
or can be added a-t a time
subsequent to the addition of the target molecule and its binding partner.
Control reaction mixtures are
incubated without the test molecule or with a placebo. Formation of any
complexes between the target
molecule and the binding partner then is detected. Decreased formation of a
complex in the reaction
mixture containing test molecule as compared to in a control reaction mixture
indicates that the
molecule antagonizes target molecule/binding partner complex formation.
Alternatively, increased
formation of a complex in the reaction mixture containing test molecule as
compared to in a control
reaction mixture indicates that the molecule agonizes target molecule/binding
partner complex
formation. In another embodiment, complex formation of target molecule/binding
partner can be
compared to complex formation of mutant target molecule/binding partner (e.g.,
amino acid
modifications in a target polypeptide). Such a comparison can be important in
those cases where it is
desirable to identify test molecules that modulate interactions of mutant but
not non-mutated target gene
products.
[0173] The assays can be conducted in a heterogeneous or homogeneous format.
In heterogeneous
assays, target molecule and/or the binding partner are immobilized to a solid
phase, and complexes are
detected on the solid phase at the end of the reaction. In homogeneous assays,
the entire reaction is
carried out in a liquid phase. In either approach, the order of addition of
reactants can be varied to
obtain different information about the molecules being tested. For example,
test compounds that
agonize target molecule/binding partner interactions can be identified by
conducting the reaction in the
presence of the test molecule in a competition format. Alternatively, test
molecules that agonize
preformed complexes, e.g., molecules with higher binding constants that
displace one of the
components from the complex, can be tested by adding the test compound to the
reaction mixture after
complexes have been formed.
[0174] In a heterogeneous assay embodiment, the target molecule or the binding
partner is
anchored onto a solid surface (e.g., a microtiter plate), while the non-
anchored species is labeled, either
directly or indirectly. The anchored molecule can be immobilized by non-
covalent or covalent
attachments. Alternatively, an immobilized antibody specific for the molecule
to be anchored can be
used to anchor the molecule to the solid surface. The partner of the
immobilized species is exposed to
the coated surface with or without the test molecule. After the reaction is
complete, unreacted
52
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
components are removed (e.g., by washing) such that a significant portion of
any complexes formed
will remain immobilized on the solid surface. Where the non-immobilized
species is pre-labeled, the
detection of label immobilized on the surface is indicative of complex. Where
the non-immobilized
species is not pre-labeled, an indirect label can be used to detect complexes
anchored to the surface;
e.g., by using a labeled antibody specific for the initially non-immobilized
species. Depending upon the
order of addition of reaction components, test compounds that inhibit complex
formation or that disrupt
preformed complexes can be detected.
[0175] In another embodiment, the reaction can be conducted in a liquid phase
in the presence or
absence of test molecule, where the reaction products are separated from
unreacted components, and the
complexes are detected (e.g., using an immobilized antibody specific for one
of the binding components
to anchor any complexes formed in solution, and a labeled antibody specific
for the other partner to
detect anchored complexes). Again, depending upon the order of addition of
reactants to the liquid
phase, test compounds that inhibit complex or that disrupt preformed complexes
can be identified.
[0176] In an alternate embodiment, a homogeneous assay can be utilized. For
example, a
preformed complex of the target gene product and the interactive cellular or
extracellular binding
partner product is prepared. One or both of the target molecule or binding
partner is labeled, and the
signal generated by the labels) is quenched upon complex formation (e.g., U.S.
Patent No. 4,109,496
that utilizes this approach for immunoassays). Addition of a test molecule
that competes with and
displaces one of the species from the preformed complex will result in the
generation of a. signal above
background. In this way, test substances that disrupt target molecule/binding
partner complexes can be
identified.
[0177] Candidate therapeutics for treating osteoarthritis are identified from
a group of test
molecules that interact with a target molecule. Test molecules are normally
ranked according to the
degree with which they modulate (e.g., agonize or antagonize) a function
associated with the target
molecule (e.g., DNA replication and/or processing, RNA transcription and/or
processing, polypeptide
production and/or processing, and/or biological function/activity), and then
top ranking modulators are
selected. Also, pharmacogenomic information described herein can determine the
rank of a modulator.
The top 10% of ranked test molecules often are selected for further testing as
candidate therapeutics,
and sometimes the top 15%, 20%, or 25% of ranked test molecules are selected
for further testing as
candidate therapeutics. Candidate therapeutics typically are formulated for
administration to a subject.
Therapeutic Formulations
[0178] Formulations and pharmaceutical compositions typically include in
combina_-tion with a
pharmaceutically acceptable carrier one or more target molecule modulators.
The modulator often is a
test molecule identified as having an interaction with a target molecule by a
screening method described
above. The modulator may be a compound, an antisense nucleic acid, a ribozyme,
an antibody, or a
binding partner. Also, formulations may comprise a target polypeptide or
fragment thereof in
combination with a pharmaceutically acceptable carrier, where the polypeptide
or fragment sometimes
53
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
has an APOL3 biological activity (e.g., apolipoprotein activity), and
sometimes includes all or part of an
apolipoprotein domain.
[0179] Formulations or pharmaceutical compositions typically include in
combination with a
pharmaceutically acceptable carrier, a compound, an antisense nucleic acid, a
ribozyme, an antibody, a
binding partner that interacts with an ADAMTS2 polypeptide, a ADAMTS2 nucleic
acid, or a fragment
thereof. The formulated molecule may be one that is identified by a screening
method described above.
Also, formulations may comprise a ADAMTS2 polypeptide or fragment thereof,
where the ADAMTS2
polypeptide contains an isoleucine at position 245 of SEQ ID NO: 44, and a
pharmaceutically
acceptable carrier. Also, formulations may comprise an active ADAMTS2
polypeptide or fragment
thereof, where ADAMTS2 polypeptide fragments having activity are selected from
amino acids 252-
1211, 253-1211, 254-1211, 255-1211, 256-1211, 257-1211, 258-1211, 259-1211 or
260-1211 of SEQ
ID NO: 44, where it is understood that the active form of ADAMTS2 does not
contain the propeptide
domain. As used herein, the term "pharmaceutically acceptable carrier"
includes solvents, dispersion
media, coatings, antibacterial and antifungal agents, isotonic and absorption
delaying agents, and the
like, compatible with pharmaceutical administration. Supplementary active
compounds can also be
incorporated into the compositions.
[0180] As used herein, the term "pharmaceutically acceptable carrier" includes
solvents,
dispersion media, coatings, antibacterial and antifungal agents, isotonic and
absorption delaying agents,
and the lilee, compatible with pharmaceutical administration. Supplementary
active compounds can also
be incorporated into the compositions. Pharmaceutical compositions can be
included in a container,
pack, or dispenser together with instructions for administration.
[0181] A pharmaceutical composition typically is formulated to be compatible
with its intended
route of administration. Examples of routes of administration include
parenteral, e.g., intravenous,
intradermal, subcutaneous, oral (e.g., inhalation), transdermal (topical),
transmucosal, and rectal
administration. Solutions or suspensions used for parenteral, intradermal, or
subcutaneous application
can include the following components: a sterile diluent such as water for
injection, saline solution, fixed
oils, polyethylene glycols, glycerin, propylene glycol or other synthetic
solvents; antibacterial agents
such as benzyl alcohol or methyl parabens; antioxidants such as ascorbic acid
or sodium bisulfate;
chelating agents such as ethylenediaminetetraacetic acid; buffers such as
acetates, citrates or phosphates
and agents for the adjustment of tonicity such as sodium chloride or dextrose.
pH can be adjusted with
acids or bases, such as hydrochloric acid or sodium hydroxide. The parenteral
preparation can be
enclosed in ampoules, disposable syringes or multiple dose vials made of glass
or plastic.
[0182] Oral compositions generally include an inert diluent or an edible
carrier. For the purpose of
oral therapeutic administration, the active compound can be incorporated with
excipients and used in the
form of tablets, troches, or capsules, e.g., gelatin capsules. Oral
compositions can also be prepared
using a fluid carrier for use as a mouthwash. Pharmaceutically compatible
binding agents, and/or
adjuvant materials can be included as part of the composition. The tablets,
pills, capsules, troches and
the like can contain any of the following ingredients, or compounds of a
similar nature: a binder such as
54
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
microcrystalline cellulose, gum tragacanth or gelatin; an excipient such as
starch or lactose, a
disintegrating agent such as alginic acid, Primogel, or corn starch; a
lubricant such as magnesium
stearate or Sterotes; a glidant such as colloidal silicon dioxide; a
sweetening agent such as sucrose or
saccharin; or a flavoring agent such as peppermint, methyl salicylate, or
orange flavoring.
l[0183] Pharmaceutical compositions suitable for injectable use include
sterile aqueous solutions
(where water soluble) or dispersions and sterile powders for the
extemporaneous preparation of sterile
injectable solutions or dispersion. For intravenous administration, suitable
carriers include
physiological saline, bacteriostatic water, Cremophor ELTM (BASF, Parsippany,
NJ) or phosphate
buffered saline (PBS). In all cases, the composition must be sterile and
should be fluid to the extent that
easy syringability exists. It should be stable under the conditions of
manufacture and storage and must
be preserved against the contaminating action of microorganisms such as
bacteria and fungi. The
carrier can be a solvent or dispersion medium containing, for example, water,
ethanol, polyol (for
example, glycerol, propylene glycol, and liquid polyethylene glycol, and the
like), and suitable mixtures
thereof. The proper fluidity can be maintained, for example, by the use of a
coating such as lecithin, by
the maintenance of the required particle size in the case of dispersion and by
the use of surfactants.
Prevention of the action of microorganisms can be achieved by various
antibacterial and antifungal
agents, for example, parabens, chlorobutanol, phenol, ascorbic acid,
thimerosal, and the like. In many
cases, it will be preferable to include isotonic agents, for example, sugars,
polyalcohols such as
mannitol, sorbitol, sodium chloride in the composition. Prolonged absorption
of the injectable
compositions can be brought about by including in the composition an agent
which delays absorption,
for example, aluminum monostearate and gelatin.
[0184] Sterile injectable solutions can be prepared by incorporating the
active compound in the
required amount in an appropriate solvent with one or a combination of
ingredients enumerated above,
as required, followed by filtered sterilization. Generally, dispersions are
prepared by incorporating the
active compound into a sterile vehicle which contains a basic dispersion
medium and the required other
ingredients from those enumerated above. In the case of sterile powders for
the preparation of sterile
injectable solutions, the preferred methods of preparation are vacuum drying
and freeze-drying which
yields a powder of the active ingredient plus any additional desired
ingredient from a previously sterile-
filtered solution thereof.
[0185] For administration by inhalation, the compounds are delivered in the
form of an aerosol
spray from pressured container or dispenser which contains a suitable
propellant, e.g., a gas such as
carbon dioxide, or a nebulizer.
[0186] Systemic administration can also be by transmucosal or transdermal
means. For
transmucosal or transdermal administration, penetrants appropriate to the
barrier to be permeated are
used in the formulation. Such penetrants are generally known in the art, and
include, for example, for
transmucosal administration, detergents, bile salts, and fusidic acid
derivatives. Transmucosal
administration can be accomplished through the use of nasal sprays or
suppositories. For transdermal
administration, the active compounds are formulated into ointments, salves,
gels, or creams as generally
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
known in the art. Molecules can also be prepared in the form of suppositories
(e.g., with conventional
suppository bases such as cocoa butter and other glycerides) or retention
enemas for rectal delivery.
[0187] In one embodiment, active molecules are prepared with carriers that
will protect the
compound against rapid elimination from the body, such as a controlled release
formulation, including
implants and microencapsulated delivery systems. Biodegradable, biocompatible
polymers can be used,
such as ethylene vinyl acetate, polyanhydrides, polyglycolic acid, collagen,
polyorthoesters, and
polylactic acid. Methods for preparation of such formulations will be apparent
to those skilled in the
art. Materials can also be obtained commercially from Alza Corporation and
Nova Pharmaceuticals,
Inc. Liposomal suspensions (including liposomes targeted to infected cells
with monoclonal antibodies
to viral antigens) can also be used as pharmaceutically acceptable carriers.
These can be prepared
according to methods known to those skilled in the art, for example, as
described in U.S. Patent
No. 4,522,811.
[0188] It is advantageous to formulate oral or parenteral compositions in
dosage unit form for ease
of administration and uniformity of dosage. Dosage unit form as used herein
refers to physically
discrete units suited as unitary dosages for the subject to be treated; each
unit containing a
predetermined quantity of active compound calculated to produce the desired
therapeutic effect in
association with the required pharmaceutical carrier.
[0189] Toxicity and therapeutic efficacy of such compounds can be determined
by standard
pharmaceutical procedures in cell cultures or experimental animals, e.g., for
determining the LDso (the
dose lethal to 50% of the population) and the EDso (the dose therapeutically
effective in 50% of the
population). The dose ratio between toxic and therapeutic effects is the
therapeutic index and it can be
expressed as the ratio LDso/EDso. Molecules which exhibit high therapeutic
indices are preferred.
While molecules that exhibit toxic side effects may be used, care should be
taken to design a delivery
system that targets such compounds to the site of affected tissue in order to
minimize potential damage
to uninfected cells and, thereby, reduce side effects.
[0190] The data obtained from the cell culture assays and animal studies can
be used in
formulating a range of dosage for use in humans. The dosage of such molecules
lies preferably within a
range of circulating concentrations that include the EDso with little or no
toxicity. The dosage may vary
within this range depending upon the dosage form employed and the route of
administration utilized.
For any molecules used in the methods described herein, the therapeutically
effective dose can be
estimated initially from cell culture assays. A dose may be formulated in
animal models to achieve a
circulating plasma concentration range that includes the ICSO (i.e., the
concentration of the test
compound which achieves a half maximal inhibition of symptoms) as determined
in cell culture. Such
information can be used to more accurately determine useful doses in humans.
Levels in plasma may be
measured, for example, by high performance liquid chromatography.
[0191] As defined herein, a therapeutically effective amount of protein or
polypeptide (i.e., an
effective dosage) ranges from about 0.001 to 30 mg/kg body weight, sometimes
about 0.01 to 25 mg/kg
body weight, often about 0.1 to 20 mg/kg body weight, and more often about 1
to 10 mg/kg, 2 to 9
56
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
mg/kg, 3 to 8 mg/kg, 4 to 7 mg/kg, or 5 to 6 mg/kg body weight. The protein or
polypeptide can be
administered one time per week for between about 1 to 10 weeks, sometimes
between 2 to 8 weeks,
often between about 3 to 7 weeks, and more often for about 4, 5, or 6 weeks.
The skilled artisan will
appreciate that certain factors may influence the dosage and timing required
to effectively treat a
subject, including but not limited to the severity of the disease or disorder,
previous treatments, the
general health and/or age of the subject, and other diseases present.
Moreover, treatment of a subject
with a therapeutically effective amount of a protein, polypeptide, or antibody
can include a single
treatment or, preferably, can include a series of treatments.
[0192] With regard to polypeptide formulations, featured herein is a method
for treating
osteoarthritis in a subject, which comprises contacting one or more cells in
the subject with a first
polypeptide, where the subject comprises a second polypeptide having one or
more polymorphic
variations associated with cancer, and where the first polypeptide comprises
fewer polymorphic
variations associated with cancer than the second polypeptide. The first and
second polypeptides are
encoded by a nucleic acid which comprises a nucleotide sequence in SEQ ID NO:
1-13 or referenced in
Table B; a nucleotide sequence which encodes a polypeptide consisting of an
amino acid sequence
encoded by a nucleotide sequence referenced in SEQ ID NO: 1-13 or referenced
in Table B; a
nucleotide sequence which encodes a polypeptide that is 90% or more identical
to an amino acid
sequence encoded by a nucleotide sequence of SEQ ID NO: 1-13 or referenced in
Table B and a
nucleotide sequence 90% or more identical to a nucleotide sequence in SEQ ID
NO: 1-13 or referenced
in Table B. The subject often is a human.
[0193] For antibodies, a dosage of 0.1 mg/kg of body weight (generally 10
mg/kg to 20 mg/kg) is
often utilized. If the antibody is to act in the brain, a dosage of 50 mg/kg
to 100 mg/kg is often
appropriate. Generally, partially human antibodies and fully human antibodies
have a longer half life
within the human body than other antibodies. Accordingly, lower dosages and
less frequent
administration is often possible. Modifications such as lipidation can be used
to stabilize antibodies and
to enhance uptake and tissue penetration (e.g., into the brain). A method for
lipidation of antibodies is
described by Cruikshank et al., J. Acquired Inamune Defieiency Syndromes and
Human Retnovinology
14:193 (1997).
[0194] Antibody conjugates can be used for modifying a given biological
response, the drug
moiety is not to be construed as limited to classical chemical therapeutic
agents. For example, the drug
moiety may be a protein or polypeptide possessing a desired biological
activity. Such proteins may
include, for example, a toxin such as abrin, ricin A, pseudomonas exotoxin, or
diphtheria toxin; a
polypeptide such as tumor necrosis factor, alpha-interferon, beta-interferon,
nerve growth factor, platelet
derived growth factor, tissue plasminogen activator; or, biological response
modifiers such as, for
example, lymphokines, interleukin-1 ("IL-1"), interleukin-2 ("IL-2"),
interleukin-6 ("IL-6"),
granulocyte macrophage colony stimulating factor ("GM-CSF"), granulocyte
colony stimulating factor
("G-CSF"), or other growth factors. Alternatively, an antibody can be
conjugated to a second antibody
to form an antibody heteroconjugate as described by Segal in U.S. Patent No.
4,676,980.
57
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0195] For compounds, exemplary doses include milligram or microgram amounts
of the
compound per kilogram of subject or sample weight, for example, about 1
microgram per kilogram to
about 500 milligrams per kilogram, about 100 micrograms per kilogram to about
5 milligrams per
kilogram, or about 1 microgram per kilogram to about 50 micrograms per
kilogram. It is understood
that appropriate doses of a small molecule depend upon the potency of the
small molecule with respect
to the expression or activity to be modulated. When one or more of these small
molecules is to be
administered to an animal (e.g., a human) in order to modulate expression or
activity of a polypeptide or
nucleic acid described herein, a physician, veterinarian, or researcher may,
for example, prescribe a
relatively low dose at first, subsequently increasing the dose until an
appropriate response is obtauied.
In addition, it is understood that the specific dose level for any particular
animal subject will depend
upon a variety of factors including the activity of the specific compound
employed, the age, body
weight, general health, gender, and diet of the subject, the time of
administration, the route of
administration, the rate of excretion, any drug combination, and the degree of
expression or activity to
be modulated.
[0196] With regard to nucleic acid formulations, gene therapy vectors can be
delivered to a subject
by, for example, intravenous injection, local administration (see, e.g., U.S.
Patent 5,328,470) or by
stereotactic injection (see e.g., Chen et al., (1994) Pr~oc. Natl. Acad. Sci.
USA 91:3054-3057).
Pharmaceutical preparations of gene therapy vectors can include a gene therapy
vector in an acceptable
diluent, or can comprise a slow release matrix in which the gene delivery
vehicle is imbedded.
Alternatively, where the complete gene delivery vector can be produced intact
from recombinant cells
(e.g., retroviral vectors) the pharmaceutical preparation can include one or
more cells which produce the
gene delivery system. Examples of gene delivery vectors are described herein.
Therapeutic Methods
[0197] A therapeutic formulation described above can be administered to a
subject in need of a
therapeutic for inducing a desired biological response. Therapeutic
formulations can be administered by
any of the paths described herein. With regard to both prophylactic and
therapeutic methods of
treatment, such treatments may be specifically tailored or modified, based on
knowledge obtained from
pharmacogenomic analyses described herein.
[0198] As used herein, the term "treatment" is defined as the application or
administration of a
therapeutic formulation to a subject, or application or administration of a
therapeutic agent to an isolated
tissue or cell line from a subject with the purpose to cure, heal, alleviate,
relieve, alter, remedy,
ameliorate, improve or affect osteoarthritis, symptoms of osteoarthritis or a
predisposition towards
osteoarthritis. A therapeutic formulation includes, but is not limited to,
small molecules, peptides,
antibodies, ribozymes and antisense oligonucleotides. Administration of a
therapeutic formulation can
occur prior to the manifestation of symptoms characteristic of osteoarthritis,
such that osteoarthritis is
prevented or delayed in its progression. The appropriate therapeutic
composition can be determined
based on screening assays described herein.
58
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0199] As discussed, successful treatment of osteoarthritis can be brought
about by techniques that
serve to agonize target molecule expression or function, or alternatively,
antagonize target molecule
expression or function. These techniques include administration of modulators
that include, but are not
limited to, small organic or inorganic molecules; antibodies (including, for
example, polyclonal,
monoclonal, humanized, anti-idiotypic, chimerie or single chain antibodies,
and Fab, F(ab')Z and Fab
expression library fragments, scFV molecules, and epitope-binding fragments
thereof); and peptides,
phosphopeptides, or polypeptides.
[0200] Further, antisense and ribozyme molecules that inhibit expression of
the target gene can
also be used to reduce the level of target gene expression, thus effectively
reducing the level of target
gene activity. Still further, triple helix molecules can be utilized in
reducing the level of target gene
activity. Antisense, ribozyme and triple helix molecules are discussed above.
It is possible that the use
of antisense, ribozyme, and/or triple helix molecules to reduce or inhibit
mutant gene expression can
also reduce or inhibit the transcription (triple helix) and/or translation
(antisense, ribozyme) of mRNA
produced by normal target gene alleles, such that the concentration of normal
target gene product
present can be lower than is necessary for a normal phenotype. In such cases,
nucleic acid molecules
that encode and express target gene polypeptides exhibiting normal target gene
activity can be
introduced into cells via gene therapy method. Alternatively, in instances in
that the target gene
encodes an extracellular polypeptide, it can be preferable to eo-administer
normal target gene
polypeptide into the cell or tissue in order to maintain the requisite level
of cellular or tissue target gene
activity.
[0201] Another method by which nucleic acid molecules may be utilized in
treating or preventing
osteoarthritis is use of aptamer molecules specific for target molecules.
Aptamers are nucleic acid
molecules having a tertiary structure which permits them to specifically bind
to ligands (see, e.g.,
Osborne, et al., Cur. Opin. Chem. Biol. l (1): 5-9 (1997); and Patel, D. J.,
Curs. Opitz. Chem. Biol.
Jun; l (1): 32-46 (1997)).
[0202] Yet another method of utilizing nucleic acid molecules for
osteoarthritis treatment is gene
therapy, which can also be referred to as allele therapy. Provided herein is a
gene therapy method for
treating osteoarthritis in a subject, which comprises contacting one or more
cells in the subject or from
the subject with a nucleic acid having a first nucleotide sequence (e.g., the
first nucleotide sequence is
identical to or substantially identical to a nucleotide sequence of SEQ ID NO:
1-13 or other nucleotide
sequence referenced in Table B). Genomic DNA in the subject comprises a second
nucleotide sequence
having one or more polymorphic variations associated with osteoarthritis
(e.g., the second nucleotide
sequence is identical to or substantially identical to a nucleotide sequence
of SEQ ID NO: 1-13 or other
nucleotide sequence referenced in Table B). The first and second nucleotide
sequences typically are
substantially identical to one another, and the first nucleotide sequence
comprises fewer polymorphic
variations associated with osteoarthritis than the second nucleotide sequence.
The first nucleotide
sequence may comprise a gene sequence that encodes a full-length polypeptide
or a fragment thereof.
The subject is often a human. Allele therapy methods often are utilized in
conjunction with a method of
59
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
first determining whether a subject has genomic DNA that includes polymorphic
variants associated
with osteoarthritis.
[0203] In a certain embodiment, the method often comprises supplementing
arthritis-associated
ADAMTS2 polypeptide with a non-arthritis-associated ADAMTS2 polypeptide or
fragment thereof,
where the non-arthritis-associated form ofADAMTS2 contains an isoleucine at
position 245 of SEQ ID
NO: 44 having enzymatic activity. The arthritis-associated ADAMTS2 polypeptide
sometimes contains
a valine at position 245 of SEQ ID NO: 44 having an altered enzymatic activity
varying from the non-
arthritis-associated polypeptide.
[0204] In an embodiment, provided is a method of increasing the synthesis of
procollagen II
comprising providing or administering to individuals in need of increasing
levels of type II collagen the
pharmaceutical or physiologically acceptable composition comprising active
human ADAMTS2 protein
or fragment thereof, where ADAMTS2 polypeptide fragments having activity are
selected from amino
acids 252-1211, 253-1211, 254-1211, 255-1211, 256-1211, 257-1211, 258-1211,
259-1211 or 260-1211
of SEQ B7 NO: 44, where it is understood that the active form ofADAMTS2 does
not contain the
propeptide domain.
[0205] In another embodiment, provided herein is a method of increasing the
synthesis of
procollagen II comprising providing or administering to individuals in need of
increasing levels of type
II collagen the pharmaceutical or physiologically acceptable composition
comprising an enzyme or
molecule capable of cleaving ADAMTS2 propeptide, e.g., a furin-type
endopeptidase or N-
ethylmaleimide described herein
[0206] In another allele therapy embodiment, provided herein is a method which
comprises
contacting one or more cells in the subject or from the subject with a
polypeptide encoded by a nucleic
acid having a first nucleotide sequence (e.g., the first nucleotide sequence
is identical to or substantially
identical to the nucleotide sequence of SEQ ID NO: 1-13 or other nucleotide
sequence referenced in
Table B). Genomic DNA in the subject comprises a second nucleotide sequence
having one or more
polymorphic variations associated with osteoarthritis (e.g., the second
nucleotide sequence is identical
to or substantially identical to a nucleotide sequence of SEQ ID NO: 1-13 or
other nucleotide sequence
referenced in Table B). The first and second nucleotide sequences typically
are substantially identical
to one another, and the first nucleotide sequence comprises fewer polymorphic
variations associated
with osteoarthritis than the second nucleotide sequence. The first nucleotide
sequence may comprise a
gene sequence that encodes a full-length polypeptide or a fragment thereof.
The subject is often a
human.
[0207] k'or antibody-based therapies, antibodies can be generated that are
both specific for target
molecules and that reduce target molecule activity. Such antibodies may be
administered in instances
where antagonizing a target molecule function is appropriate for the treatment
of osteoarthritis.
[0208] In circumstances where stimulating antibody production in an animal or
a human subject by
injection with a target molecule is harmful to the subject, it is possible to
generate an immune response
against the target molecule by use of anti-idiotypic antibodies (see, e.g.,
Herlyn, Ann. Med.; 31 (1): 66-78
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
(1999); and Bhattacharya-Chatterjee & Foon, Cancer Treat. Res.; 94.' S1-68
(1998)). Introducing an
anti-idiotypic antibody to a mammal or human subject often stimulates
production of anti-anti-idiotypic
antibodies, which typically are specific to the target molecule. Vaccines
directed to osteoarthritis also
may be generated in this fashion.
[0209] In instances where the target molecule is intracellular and whole
antibodies are used,
internalizing antibodies may be preferred. Lipofectin or liposomes can be used
to deliver the antibody
or a fragment of the Fab region that binds to the target antigen into cells.
Where fragments of the
antibody are used, the smallest inhibitory fragment that binds to the target
antigen is preferred. For
example, peptides having an amino acid sequence corresponding to the Fv region
of the antibody can be
used. Alternatively, single chain neutralizing antibodies that bind to
intracellular target antigens can
also be administered. Such single chain antibodies can be administered, for
example, by expressing
nucleotide sequences encoding single-chain antibodies within the target cell
population (see, e.g.,
Marasco et al., Px~oc. Natl. Acad. Sci. USA 90: 7889-7893 (1993)).
[0210] Modulators can be administered to a patient at therapeutically
effective doses to treat
osteoarthritis. A therapeutically effective dose refers to an amount of the
modulator sufficient to result
in amelioration of symptoms of osteoarthritis. Toxicity and therapeutic e~cacy
of modulators can be
determined by standard pharmaceutical procedures in cell cultures or
experimental animals, e.g., for
determining the LDSO (the dose lethal to 50% of the population) and the EDSO
(the dose therapeutically
effective in 50% of the population). The dose ratio between toxic and
therapeutic effects is the
therapeutic index and it can be expressed as the ratio LDSO/EDso. Modulators
that exhibit large
therapeutic indices are preferred. While modulators that exhibit toxic side
effects can be used, care
should be taken to design a delivery system that targets such molecules to the
site of affected tissue in
order to minimize potential damage to uninfected cells, thereby reducing side
effects.
[0211] Data obtained from cell culture assays and animal studies can be used
in formulating a
range of dosages for use in humans. The dosage of such compounds lies
preferably within a range of
circulating concentrations that include the EDso with little or no toxicity.
The dosage can vary within
this range depending upon the dosage form employed and the route of
administration utilized. For any
compound used in the methods described herein, the therapeutically effective
dose can be estimated
initially from cell culture assays. A dose can be formulated in animal models
to achieve a circulating
plasma concentration range that includes the ICSO (i. e., the concentration of
the test compound that
achieves a half maximal inhibition of symptoms) as determined in cell culture.
Such information can be
used to more accurately determine useful doses in humans. Levels in plasma can
be measured, for
example, by high performance liquid chromatography.
[0212] Another example of effective dose determination for an individual is
the ability to directly
assay levels of "free" and "bound" compound in the serum of the test subject.
Such assays may utilize
antibody mimics and/or "biosensors" that have been created through molecular
imprinting techniques.
Molecules that modulate target molecule activity are used as a template, or
"imprinting molecule", to
spatially organize polymerizable monomers prior to their polymerization with
catalytic reagents. The
61
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
subsequent removal of the imprinted molecule leaves a polymer matrix which
contains a repeated
"negative image" of the compound and is able to selectively rebind the
molecule under biological assay
conditions. A detailed review of this technique can be seen in Ansell et al.,
Cur~etzt Opihioh in
Biotechnology 7.' 89-94 (1996) and in Shea, Trends in Polymer Science 2: 166-
173 (1994). Such
"imprinted" affinity matrixes are amenable to ligand-binding assays, whereby
the immobilized
monoclonal antibody component is replaced by an appropriately imprinted
matrix. An example of the
use of such matrixes in this way can be seen in Vlatakis, et al., Natut~e 361:
645-647 (1993). Through
the use of isotope-labeling, the "free" concentration of compound which
modulates target molecule
expression or activity readily can be monitored and used in calculations of
ICSO. Such "imprinted"
affinity matrixes can also be designed to include fluorescent groups whose
photon-emitting properties
measurably change upon local and selective binding of target compound. These
changes readily can be
assayed in real time using appropriate flberoptic devices, in turn allowing
the dose in a test subject to be
quickly optimized based on its individual ICSO. An example of such a
"biosensor" is discussed in I~riz
et al., Analytieal Chemistry 67: 2142-2144 (1995).
[0213] The examples set forth below are intended to illustrate but not limit
the invention.
Examples
[0214] In the following studies a group of subjects was selected according to
specific parameters
relating to osteoarthritis. Nucleic acid samples obtained from individuals in
the study group were
subjected to genetic analysis, which identified associations between
osteoarthritis and certain
polymorphic variants in the following genes: PADI2, APOB, ILIRL~, ILIRL1,
WASPIP, ADAMTS2,
BT~ES, TM7SF3, PELl2, LO~L'LI, CASPR4, GPR50 or APOL3 (herein referred to as
"Targets"). The
polymorphisms were genotyped again in two replication cohorts consisting of
individuals selected for
OA. In addition, SNPs proximal to the incident polymorphism in APOB, IL1RL2,
IL1RL1, WASPIP,
ADAMTS2, BYES, TM7SF3, LOXLl, CASPR4 and APOL3 regions were identified and
allelotyped in
OA case and control pools. Methods are described for producing target
polypeptides encoded by the
nucleic acids of Table B in vitro or in vivo, which can be utilized in methods
that screen test molecules
for those that interact with target polypeptides. Test molecules identified as
interactors with target
polypeptides can be screened further as osteoarthritis therapeutics.
Example 1
Samples and Poolin S~ trateaies
Sample Selection
[0215] Blood samples were collected from individuals diagnosed with knee
osteoarthritis, which
were referred to as case samples. Also, blood samples were collected from
individuals not diagnosed
with knee osteoarthritis as gender and age-matched controls. A database was
created that listed all
62
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
phenotypic trait information gathered from individuals for each case and
control sample. Genomic
DNA was extracted from each of the blood samples for genetic analyses.
DNA Extraction from Blood Samples
[0216] Six to ten milliliters of whole blood was transferred to a 50 ml tube
containing 27 ml of red
cell lysis solution (RCL). The tube was inverted until the contents were
mixed. Each tube was
incubated for 10 minutes at room temperature and inverted once during the
incubation. The tubes were
then centrifuged for 20 minutes at 3 000 x g and the supernatant was carefully
poured off. 100-200 p,l of
residual liquid was left in the tube and was pipetted repeatedly to resuspend
the pellet in the residual
supernatant. White cell lysis solution (WCL) was added to the tube and
pipetted repeatedly until
completely mixed. While no incubation was normally required, the solution was
incubated at 37°C or
room temperature if cell clumps were visible after mixing until the solution
was homogeneous. 2 ml of
protein precipitation was added to the cell lysate. The mixtures were vortexed
vigorously at high speed
for 20 sec to mix the protein precipitation solution uniformly with the cell
lysate, and then centrifuged
for 10 minutes at 3000 x g. The supernatant containing the DNA was then poured
into a clean 15 ml
tube, which contained 7 ml of 100°fo isopropanol. The samples were
mixed by inverting the tubes gently
until white threads of DNA were visible. Samples were centrifuged for 3
minutes at 2000 x g and the
DNA was visible as a small white pellet. The supernatant was decanted and 5 ml
of 70% ethanol was
added to each tube. Each tube was inverted several times to wash the DNA
pellet, and then centrifuged
for 1 minute at 2000 x g. The ethanol was decanted and each tube was drained
on clean absorbent
paper. The DNA was dried in the tube by inversion for 10 minutes, and then
1000 wl of 1X TE was
added. The size of each sample was estimated, and less TE buffer was added
during the following DNA
hydration step if the sample was smaller. The DNA was allowed to rehydrate
overnight at room
temperature, and DNA samples were stored at 2-8°C.
[0217] DNA was quantified by placing samples on a hematology mixer for at
least 1 hour. DNA
was serially diluted (typically 1:80, 1:160, 1:320, and 1:640 dilutions) so
that it would be within the
measurable range of standards. 125 ~l of diluted DNA was transferred to a
clear U-bottom microtitre
plate, and 125 pl of 1X TE buffer was transferred into each well using a
multichannel pipette. The
DNA and 1X TE were mixed by repeated pipetting at least 15 times, and then the
plates were sealed. 50
pl of diluted DNA was added to wells AS-H12 of a black flat bottom microtitre
plate. Standards were
inverted six times to mix them, and then 50 p,l of 1X TE buffer was pipetted
into well A1, 1000 ng/ml
of standard was pipetted into well A2, 500 ng/ml of standard was pipetted into
well A3, and 250 ng/ml
of standard was pipetted into well A4. PicoGreen (Molecular Probes, Eugene,
Oregon) was thawed and
freshly diluted 1:200 according to the number of plates that were being
measured. PicoGreen was
vortexed and then SOp.I was pipetted into all wells of the black plate with
the diluted DNA. DNA and
PicoGreen were mixed by pipetting repeatedly at least 10 times with the
multichannel pipette. The plate
was placed into a Fluoroskan Ascent Machine (microplate fluorometer produced
by Labsystems) and
63
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
the samples were allowed to incubate for 3 minutes before the machine was run
using filter pairs 485
nm excitation and 538 nm emission wavelengths. Samples having measured DNA
concentrations of
greater than 450 ng/p,l were re-measured for conformation. Samples having
measured DNA
concentrations of 20 ng/p.l or less were re-measured for confirmation.
Poolin Strate ies - Discoverer Cohort
[0218] Samples were derived from the Nottingham knee OA family study (IJK)
where index cases
were identified through a knee replacement registry. Siblings were approached
and assessed with knee
x-rays and assigned status as affected or unaffected. In all 1,157 individuals
were available. In order to
create same-sex pools of appropriate sizes, 335 unrelated female individuals
with OA from the
Nottingham OA sample were selected for the case pool. The control pool was
made up of unrelated
female individuals from the St. Thomas twin study (England) with normal knee x-
rays and without
other indications of OA, regardless of anatomical location, as well as lacking
family history of OA. The
St. Thomas twin study consists of Caucasian, female participants from the St.
Thomas' Hospital,
London, adult-twin registry, which is a voluntary registry of >4,000 twin
pairs ranging from 18 to
76 years of age. The female case samples and female control samples are
described further in Table 1
below.
[0219] A select set of samples from each group were utilized to generate
pools, and one pool was
created for each group. Each individual sample in a pool was represented by an
equal amount of
genomic DNA. For example, where 25 ng of genomic DNA was utilized in each PCR
reaction and
there were 200 individuals in each pool, each individual would provide 125 pg
of genomic DNA.
Inclusion or exclusion of samples for a pool was based upon the following
criteria: the sample was
derived from an individual characterized as Caucasian; the sample was derived
from an individual of
British paternal and maternal descent; case samples were derived from
individuals diagnosed with
specific knee osteoarthritis (OA) and were recruited from an OA knee
replacement clinic. Control
samples were derived from individuals free of OA, family history of OA, and
rheumatoid arthritis.
Also, sufficient genomic DNA was extracted from each blood sample for all
allelotyping and
genotyping reactions performed during the study. Phenotype information from
each individual was
collected and included age of the individual, gender, family history of OA,
general medical information
(e.g., height, weight, thyroid disease, diabetes, psoriasis, hysterectomy),
joint history (previous and
current symptoms, joint-related operations, age at onset of symptoms, date of
primary diagnosis, age of
individual as of primary diagnosis and order of involvement), and knee-related
findings (crepitus,
restricted passive movement, bony swelling/deformity). Additional knee
information included knee
history, current symptoms, any major knee injury, menisectomy, knee
replacement surgery, age of
surgery, and treatment history (including hormone replace therapy (HRT)).
Samples that met these
criteria were added to appropriate pools based on disease status.
64
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0220] The selection process yielded the pools set forth in Table 1, which
were used in the studies
that follow:
TALE 1
Female case Female control
Pool size 335 335
(Number)
Pool Criteria
(ex: case/control)control case
Mean Age 57 95
21 69
(ex: years) , .
Example 2
Association of Pol.~phic Variants with Osteoarthritis
[0221] A whole-genome screen was performed to identify particular SNPs
associated with
occurrence of osteoarthritis. As described in Example 1, two sets of samples
were utilized, which
included samples from female individuals having knee osteoarthritis
(osteoarthritis cases), and samples
from female individuals not having knee osteoarthritis (female controls). The
initial screen of each pool
was performed in an allelotyping study, in which certain samples in each group
were pooled. By
pooling DNA from each group, an allele frequency for each SNP in each group
was calculated. These
allele frequencies were then compared to one another. Particular SNPs were
considered as being
associated with osteoarthritis when allele frequency differences calculated
between case and control
pools were statistically significant. SNP disease association results obtained
from the allelotyping study
were then validated by genotyping each associated SNP across all samples from
each pool. The results
of the genotyping then were analyzed, allele frequencies for each group were
calculated from the
individual genotyping results, and a p-value was calculated to determine
whether the case and control
groups had statistically significant differences in allele frequencies for a
particular SNP. When the
genotyping results agreed with the original allelotyping results, the SNP
disease association was
considered validated at the genetic level.
SNP Panel Used for Genetic Anal_
[0222] A whole-genome SNP screen began with an initial screen of approximately
25,000 SNPs
over each set of disease and control samples using a pooling approach. The
pools studied in the screen
are described in Example 1. The SNPs analyzed in this study were part of a set
of 25,488 SNPs
confirmed as being statistically polymorphic as each is characterized as
having a minor allele frequency
of greater than 10°fo. The SNPs in the set reside in genes or in close
proximity to genes, and many
reside in gene exons. Specifically, SNPs in the set are located in exons,
introns, and within 5,000 base-
pairs upstream of a transcription start site of a gene_ In addition, SNPs were
selected according to the
following criteria: they are located in ESTs; they are located in Locuslink or
Ensembl genes; and they
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
are located in Genomatix promoter predictions. SNPs in the set were also
selected on the basis of even
spacing across the genome, as depicted in Table 2.
[0223] A case-control study design using a whole genome association strategy
involving
approximately 28,000 single nucleotide polymorphisms (SNPs) was employed.
Approximately 25,000
SNPs were evenly spaced in gene-based regions of the human genome with a
median inter-marker
distance of about 40,000 base pairs. Additionally, approximately 3,000 SNPs
causing amino acid
substitutions in genes described in the literature as candidates for various
diseases were used. The case-
contr-of study samples were of female Caucasian origin (British paternal and
maternal descent) 670
individuals were equally distributed in two groups: female controls and female
cases. The whole
genome association approach was first conducted on 2 DNA pools representing
the 2 groups.
Significant markers were confirmed by individual genotyping.
TABLE 2
General Statistics Suacin~ Statistics
Total # of SNPs 25,488 Median 37,058 by
# of Exonic SNPs >4,335 (17%)Minimum* 1,000 by
# SNPs with refSNP20,776 (81%)Maximum* 3,000,000
ID by
Gene Coverage >10,000 Mean 122,412 by
Chromosome CoverageAll Std Deviation 373,325 by
*E.~cludes outliers
Allelotypi~n and Genotyping Results
[0224] The genetic studies summarized above and described in more detail below
identified allelic
variants associated with osteoarthritis, which are summarized in Table B.
Assay for Veri ,ping, Alleloty~ing, and Genotyping SNPs
[0225] A MassARRAYTM system (Sequenom, Inc.) was utilized to perform SNP
genotyping in a
high-throughput fashion. This genotyping platform was complemented by a
homogeneous, single-tube
assay method (hMETM or homogeneous MassEXTENDTM (Sequenom, Inc.)) in which two
genotyping
primers anneal to and amplify a genomic target surrounding a polymorphic site
of interest. A third
primer (the MassEXTENDTM primer), which is complementary to the amplified
target up to but not
including the polymorphism, was then enzymatically extended one or a few bases
through the
polyrnorphic site and then terminated.
[0226] For each polymorphism, SpectroDESIGNERTM software (Sequenom, Inc.) was
used to
generate a set of PCR primers and a MassEXTENDTM primer which where used to
genotype the
polymorphism. Other primer design software could be used or one of ordinary
skill in the art could
manually design primers based on his or her knowledge of the relevant factors
and considerations in
designing such primers. Table 3 shows PCR primers and Table 4 shows extension
primers used for
analyzing polymorphisms. The initial PCR amplification reaction was performed
in a 5 p.l total volume
66
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
containing 1X PCR buffer with 1.5 mM MgClz (Qiagen), 200 p,M each of dATP,
dGTP, dCTP, dTTP
(Gibco-BRL), 2.5 ng of genomic DNA, 0.1 units of HotStar DNA polymerase
(Qiagen), and 200 nM
each of forward and reverse PCR primers specific for the polymorphic region of
interest.
TABLE 3: PCR Primers
SNP Forward PCR primer Reverse PCR primer
Reference
rs910223ACGTTGGATGACAGAGTGTCAGGGCTCAGAACGTTGGATGTGGTTTTTCCAGTGTCTTAC
rs1367117ACGTfGGATGTTGGTTTTCTTCAGCAAGGCACGTTGGATGAGCTTCATCCTGAAGACCAG
rs1024791ACGTTGGATGGTGTAAGGGACTGCAGATACACGTTGGATGAAACAGAACCAGGAGGTTGG
rs1041973ACGTTGGATGGGGACTTCTGACAATACAGGACGTTGGATGAATCGTGTGTTTGCCTCAGG
rs1465621ACGTTGGATGTTCTCCTCCCATTCTTCCTGACGTTGGATGGCGGGACTAGAAGTAGATTC
rs398829ACGTTGGATGTAGTCATCGTCCGCAGCATGACGTTGGATGAAGACGGTGTCCTCTCCTTG
rs1018810ACGTTGGATGTGCTGCTCCCATTfCTCATGACGTTGGATGAAGGAGTAGAGACCTTGCTG
rs1484086ACGTTGGATGTGTCACTCT1'CGGAAGTCTCACGTTGGATGCATGTACAGGGCATTCACAG
rs242392ACGTTGGATGTGTTTGGGCTGCTGTGGCTCTACGTTGGATGACCACTTCTCACGGTfACTG
rs8818 ACGTTGGATGAATCTCTCCCCTTCCAAAGCACGTTGGATGTCCCTGTGGTTTTCATCCAC
rs1395486ACGTTGGATGCTCATTTATTTCATGTTCACACGTTGGATGTGCTGGAATAATGATfGTTG
rs512294ACGTTGGATGTCTTGCTACCCACCTCCGAGACGTTGGATGAGAGCTCATGAGGGAATGGG
rs132659ACGTTGGATGGGCCCATAGTGGGTCATAACACGTTGGATGGTGGGGTGAGTGCCCAAAAG
~
[0227] Samples were incubated at 95°C for 15 minutes, followed by 45
cycles of 95°C for 20
seconds, 56°C for 30 seconds, and 72°C for 1 minute, finishing
with a 3 minute final extension at 72°C.
Following amplification, shrimp alkaline phosphatase (SAP) (0.3 units in a 2
p,l volume) (Amersham
Pharmacia) was added to each reaction (total reaction volume was 7 pl) to
remove any residual dNTPs
that were not consumed in the PCR step. Samples were incubated for 20 minutes
at 37°C, followed by
minutes at 85°C to denature the SAP.
[0228] Once the SAP reaction was complete, a primer extension reaction was
initiated by adding a
polymorphism-specific MassEXTENDTM primer cocktail to each sample. Each
MassEXTENDTM
cocktail included a specific combination of dideoxynucleotides (ddNTPs) and
deoxynucleotides
(dNTPs) used to distinguish polymorphic alleles from one another. Methods for
verifying, allelotyping
and genotyping SNPs are disclosed, for example, in U.S. Pat. No. 6,258,538,
the content of which is
hereby incorporated by reference. In Table 4, ddNTPs are shown and the fourth
nucleotide not shown is
the dNTP.
TABLE 4: Extension Primers
SNP Extend Probe Termination
Reference Mix
rs910223GGGTCTGCACTG GTCCCA ACT
rs1367117AGCCATACACCTCTTfCAGG ACT
rs1024791CTGGCTGATGTCAGAAAGCA ACG
rs 1041973ATACCAGAATCAGCAACT ACT
rs1465621CCATTCTTCCTGACATTCGCC CGT
rs398829TGGCGTGCTCCTCTAGGA ACG
rs1018810CTGCTTTTATACATGCCACAC ACT
rs1484086CTCTTCGGAAGTCTCTTTCTCA ACT
rs242392CTGCTGTGGCTCTACTGGT ACG
~
67
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
SNP Extend Probe Termination
Reference Mix
rs8818 AGCCCCCAACCCACAGGCA ACT
rs1395486TTTCATGTTCACAAAAAATCTTCT ACG
rs512294 AGCTGGAGAGCAAACCACC ACT
rs 132659AGAACTCCCCAAATCGTCCT ACG
[0229] The MassEXTENDTM reaction was performed in a total volume of 9 ~,1,
with the addition of
1X ThermoSequenase buffer, 0.576 units of TherrnoSequenase (Amersham
Pharmacia), 600 nM
MassEXTENDTM primer, 2 mM of ddATP and/or ddCTP and/or ddGTP and/or ddTTP, and
2 mM of
dATP or dCTP or dGTP or dTTP. The deoxy nucleotide (dNTP) used in the assay
normally was
complementary to the nucleotide at the polymorphic site in the amplicon.
Samples were incubated at
94°C for 2 minutes, followed by 55 cycles of 5 seconds at 94°C,
5 seconds at 52°C, and 5 seconds
at 72°C.
[0230] Following incubation, samples were desalted by adding 16 pl of water
(total reaction
volume was 25 ~1), 3 mg of SpectroCLEANTM sample cleaning beads (Sequenom,
Inc.) and allowed to
incubate for 3 minutes with rotation. Samples were then robotically dispensed
using a piezoelectric
dispensing device (SpectroJETTM (Sequenom, Inc.)) onto either 96-spot or 384-
spot silicon chips
containing a matrix that crystallized each sample (SpectroCHIPTM (Sequenom,
Inc.)). Subsequently,
MALDI-TOF mass spectrometry (Biflex and Autoflex MALDI-TOF mass spectrometers
(Bruker
Daltonics) can be used) and SpectroTYPER RTT"'i software (Sequenom, Inc.) were
used to analyze and
interpret the SNP genotype for each sample.
Genetic Analysis
[0231] Minor allelic frequencies for the polymorphisms set forth in Table B
were verified as being
10% or greater using the extension assay described above in a group of samples
isolated from 92
individuals originating from the state of Utah in the United States, Venezuela
and France (Coriell cell
repositories).
[0232] Genotyping results are shown for female pools in Table 5. In Table 5,
"AF" refers to allelic
frequency; and "F case" and "F control" refer to female case and female
control groups, respectively.
TABLE 5: Genotyping Results
SNP Reference P-~alue
F c se F co trol
rs910223 A = 0.148 A = 0.099 . 0
0069
G=0.852 G=0.901 .
rs1367117 A = 0.339 A = 0.402 0
0181
G=0.661 G=0.598 .
rs1024791 G = 0.129 G = 0.088 0
0158
A=0.871 A=0.912 .
rs1041973 A = 0.189 A = 0.233 0
0539
C=0.811 C=0.767 .
rs1465621 T = 0.071 T = 0.107 0
0194
A = 0.929 A = 0.893 .
rs398829 G = 0.740 G = 0.652 0.0002
68
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
A = 0.260 A = 0.348
rs 1018810 A = 0.142 A = 0.094 0
0063
G = 0.858 G = 0.906 .
rs1484086 T = 0.821 T = 0.753 0
0027
C=0.179 C=0.247 .
rs242392 C = 0.100 C = 0.139 0
0272
T = 0.900 T = 0.861 .
rs8818 G = 0.158 G = 0.213 0
0105
C=0.842 C=0.787 .
rs1395486 C = 0.115 C = 0.158 0
0231
T = 0.885 T = 0.842 .
rs512294 A = 0.078 A = 0.124 0
0054
G=0.922 G=0.876 .
rs132659 C = 0.675 C = 0.589 0
0015
T=0.325 T=0.411 .
[0233] All of the single marker alleles set forth in Table B were considered
validated, since the
genotyping data agreed with the allelotyping data and each SNP significantly
associated with
osteoarthritis. Particularly significant associations with osteoarthritis are
indicated by a calculated p-
value of less than 0.05 for genotype results.
Example 3
Association of Pol morphic Variants with Osteoarthritis in Replication Cohorts
[0234] The single marker polymorphisms set forth in Table B were genotyped
again in two
replication cohorts consisting of individuals selected for OA.
Sample Selection and Poolin Strategies -Replication Sample 1
[0235] A second case control sample (replication sample #1) was created by
using 100 Caucasian
female cases from Chingford, UK, and 148 unrelated female cases from the St.
Thomas twin study.
Cases were defined as having Kellgren-Lawrence (KL) scores of at least 2 in at
least one knee x-ray. In
addition, 199 male knee replacement cases from Nottingham were included. (For
a cohort description,
see the Nottingham description provided in Example 1). The control pool was
made up of unrelated
female individuals from the St. Thomas twin study (England) with normal knee x-
rays and without
other indications of OA, regardless of anatomical location, as well as lacking
family history of OA. The
St. Thomas twin study consists of Caucasian, female participants from the St.
Thomas' Hospital,
London, adult-twin registry, which is a voluntary registry of >4,000 twin
pairs ranging from 18 to
76 years of age. The replication sample 1 cohort was used to replicate the
initial results. Table 6 below
summarizes the selected phenotype data collected from the case and control
individuals.
TABLE 6
Phenotype Female cases (n=248):Male cases (n=199):Female controls
(n=313):
median (range)/ median (range)/ mean (range)/ (n,%)
(n,%) (n,%)
Age 59 (39- 73) 66 (45- 73) 55 (50- 72)
Height (cm)162 (141- 178) 175 (152- 198) 162 (141- 176)
69
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
Phenotype Female cases (n=248):Male cases (n=199):Female controls
(n=313):
median (range)/ median (range)/ mean (range)/ (n,%)
(n,%) (n,%)
Weight (kg) 68 (51- 123) 86 (62- 127) 64 (40- 111)
Body mass 2g (18-44) 29 (21-41) 24 (18-46)
index
(Ecglm )
Kellgren- 0 (63, 26%), 1
(20, 8%), 2
Lawrence* (105, 43%), 3 (58,NA NA
left 23%), 4
knee (1, 0%)
Kellgren- 0 (43, 7%), 1 (18,
7%), 2
Lawrence* (127, 52%), 3 (57,NA NA
right 23%), 4
knee (1, 0%)
KL* >2 both No (145, 59%), NA NA
Yes (101,
knees 41 %)
KL* >2 eitherNo (0, 0%), Yes NA NA
(248, 100%)
knee
* 0: normal, 1: doubtful, 2: definite osteophyte (bony protuberance), 3: joint
space narrowing
(with or without osteophyte), 4: joint deformity
Sample Selection and Poolin Sg trate~ies - Replication Sample 2
[0236] A third case control sample (replication sample #2) was created by
using individuals with
symptoms of OA from Newfoundland, Canada. These individuals were recruited and
examined by
the umatologists. Affected joints were x-rayed and a f nal diagnosis of
definite or probable OA was
made according to American College of Rheumatology criteria by a single
rheumatologist to avoid any
inter-examiner diagnosis variability. Controls were recruited from volunteers
without any symptoms
from the musculoskeletal system based on a normal joint exam performed by a
rheumatologist. Only
cases with a diagnosis of definite OA were included in the study. Only
individuals of Caucasian origin
were included. The cases consisted of 228 individuals with definite knee OA,
106 individuals with
definite hip OA, and 74 individuals with hip OA.
TABLE 7
Phenotype Case Control
Age at Visit 62.7 52.5
Sex (Female/Male) 227/119 174/101
Knee OA Xray: No 35% (120) 80% (16)
Unknown 1% (4) 0% (0)
Yes 64% (221 ) 20% (4)
Hip OA Xray: No 63% (215) 80% (16)
Unknown 2% (7) 0% (0)
Yes 35% (121 ) 20% (4)
Assay for Verifying, Allelotypin~, and Genotyping SNPs
[0237] Genotyping of the replication cohorts described in Tables 6 and 7 was
performed using the
same methods used for the original genotyping, as described herein. A
MassARRAYTM system
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
(Sequenom, Inc.) was utilized to perform SNP genotyping in a high-throughput
fashion. This
genotyping platform was complemented by a homogeneous, single-tube assay
method (hMETM or
homogeneous MassEXTENDTM (Sequenom, Inc.)) in which two genotyping primers
anneal to and
amplify a genomic target surrounding a polymorphic site of interest. A third
primer (the
MassEXTENDTM primer), which is complementary to the amplified target up to but
not including the
polymorphism, was then enzymatically extended one or a few bases through the
polymorphic site and
then terminated.
[0238] For each polymorphism, SpectroDESIGNERTM software (Sequenom, Inc.) was
used to
generate a set of PCR primers and a MassEXTENDTM primer which where used to
genotype the
polymorphism. Other primer design software could be used or one of ordinary
skill in the art could
manually design primers based on his or her knowledge of the relevant factors
and considerations in
designing such primers. Table 3 shows PCR primers and Table 4 shows extension
probes used for
analyzing (e.g., genotyping) polymorphisms in the replication cohorts. The
initial PCR amplification
reaction was performed in a 5 p,l total volume containing 1 X PCR buffer with
1.5 mM MgClz (Qiagen),
200 ~,M each of dATP, dGTP, dCTP, dTTP (Gibco-BRL), 2.5 ng of genomic DNA, 0.1
units of HotStar
DNA polymerase (Qiagen), and 200 nM each of forward and reverse PCR primers
specific for the
polymorphic region of interest.
[0239] Samples were incubated at 95°C for 15 minutes, followed by 45
cycles of 95°C for 20
seconds, 56°C for 30 seconds, and 72°C for 1 minute, finishing
with a 3 minute final extension at 72°C.
Following amplification, shrimp alkaline phosphatase (SAP) (0.3 units in a 2
~,l volume) (Amersham
Pharmacia) was added to each reaction (total reaction volume was 7 ~,l) to
remove any residual dNTPs
that were not consumed in the PCR step. Samples were incubated for 20 minutes
at 37°C, followed by
minutes at 85°C to denature the SAP.
[0240] Once the SAP reaction was complete, a primer extension reaction was
initiated by adding a
polymorphism-specific MassEXTENDTM primer cocktail to each sample. Each
MassEXTENDTM
cocktail included a specific combination of dideoxynucleotides (ddNTPs) and
deoxynucleotides
(dNTPs) used to distinguish polymorphic alleles from one another. Methods for
verifying, allelotyping
and genotyping SNPs are disclosed, for example, in LT.S. Pat. No. 6,258,538,
the content of which is
hereby incorporated by reference. In Table 7, ddNTPs are shown and the fourth
nucleotide not shown is
the dNTP.
[0241] The MassEXTENDTM reaction was performed in a total volume of 9 ~,1,
with the addition of
1X ThermoSequenase buffer, 0.576 units of ThermoSequenase (Amersham
Pharmacia), 600 nM
MassEXTENDTM primer, 2 mM of ddATP and/or ddCTP and/or ddGTP and/or ddTTP, and
2 mM of
dATP or dCTP or dGTP or dTTP. The deoxy nucleotide (dNTP) used in the assay
normally was
complementary to the nucleotide at the polymorphic site in the amplicon.
Samples were incubated at
94°C for 2 minutes, followed by 55 cycles of 5 seconds at 94°C,
5 seconds at 52°C, and 5 seconds
at 72°C.
71
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0242] Following incubation, samples were desalted by adding 16 ~1 of water
(total reaction
volume was 25 wl), 3 mg of SpectroCLEANTM sample cleaning beads (Sequenom,
Inc.) and allowed to
incubate for 3 minutes with rotation. Samples were then robotically dispensed
using a piezoelectric
dispensing device (SpectroJETTM (Sequenom, Inc.)) onto either 96-spot or 384-
spot silicon chips
containing a matrix that crystallized each sample (SpectroCI-i11'TM (Sequenom,
Inc.)). Subsequently,
MALDI-TOF mass spectrometry (Biflex and Autoflex MALDI-TOF mass spectrometers
(Bruker
Daltonics) can be used) and SpectroTYPER RTTM software (Sequenom, Inc.) were
used to analyze and
interpret the SNP genotype for each sample.
Genetic Anal
[0243] Genotyping results for replication cohorts #1 and #2 are provided in
Tables 8 and 9,
respectively.
TABLE 8
Replication Meta-analysis
rslD #1 Disc. + Rep
(Mixed #1
MaleIFemale P-value
cases
and Female
controls)
AF OA Con
AF OA
Cas Delta
P-value
rs910223 0.87 0.86 0.01 0.650 0.1800
rs1367117 0.67 0.64 0.03 0.182 0.9900
rs1024791 0.87 0.87 -0.01 0.718 0.5900
rs1041973 0.77 0.79 -0.02 0.357 Not calculated
rs1465621 0.89 0.91 -0.02 0.209 0.0095
rs398829 0.30 0.28 0.02 0.307 0.0260
rs1018810 0.91 0.89 0.02 0.289 0.0062
rs1484086 0.23 0.20 0.03 0.287 0.0077
rs242392 0.87 0.87 0.00 0.927 0.2400
rs8818 0.78 0.81 -0.03 0.259 0.0150
rs1395486 0.87 0.88 -0.01 0.492 0.0390
rs512294 0.89 0.88 0.00 0.909 0.3600
rs132659 0.38 0.34 0.04 0.128 .0077
TABLE 9
Replication Meta-analysis
rslD #2 (Newfoundland) Disc. + Rep
(MaleIFemale #2
cases Not Done
and controls)
AF OA Con
AF OA
Cas Delta
P-value
rs910223 0.86 0.86 0.001 0.974
rs1367117 0.64 0.69 -0.049 0.081
rs1024791 0.87 0.87 0.006 0.767
rs 1041973 0.78 0.79 -0.016 0.510
rs1465621 0.92 0.92 0.003 0.837
rs398829 0.27 0.28 -0.013 0.627
rs1018810
rs1484086 0.23 0.21 0.026 0.280
_
rs242392 0.88 0.88 -0.005 0.813
72
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
Replication Meta-analysis
rslD #2 (Newfoundland) Disc. + Rep
(Male/Female #2
cases and Not Done
controls)
AF OA Con
AF OA Cas
Delta P-value
rs8818 0.85 0.82 0.034 0.127
rs1395486 0.86 0.85 0.015 0.486
rs512294 0.90 0.93 -0.037 0.021
rs132659 0.36 0.36 -0.001 0.973
I
[0244] To combine the evidence for association from multiple sample
collections, a meta-analysis
procedure was employed. The allele frequencies were compared between cases and
controls within the
discovery sample, as well as within the replication cohort #1 using the
DerSimian-Laird approach
(DerSimonian, R. and N. Laird. 1986. Meta-analysis in clinical trials. Control
Clin Trials 7: 177-188.)
[0245] The absence of a statistically significant association in one or more
of the replication
cohorts should not be interpreted as minimizing the value of the original
finding. There are many
reasons why a biologically derived association identified in a sample from one
population would not
replicate in a sample from another population. The most important reason is
differences in population
history. Due to bottlenecks and founder effects, there may be common disease
predisposing alleles
present in one population that are relatively rare in another, leading to a
lack of association in the
candidate region. .Also, because common diseases such as arthritis-related
disorders are the result of
susceptibilities in many genes and many environmental risk factors,
differences in population-specific
genetic and environmental backgrounds could mask the effects of a biologically
relevant allele. For
these and other reasons, statistically strong results in the original,
discovery sample that did not replicate
in one or more of the replication samples may be further evaluated in
additional replication cohorts and
experimental systems.
[0246] APOS, ILIRL2, WASPIP, BT~ES, LO.~'Ll and C'ASPR4 regions were analyzed
further, as
shown in the examples below. PADI2, described above, is a peptidyl arginine
deiminase enzyme, type
II, that converts arginine residues within proteins to citrulline residues _
This gene is one of four known
PADI genes that encode enzymes that catalyze conversion of arginine to
citrulline in proteins.
Individuals with rheumatoid arthritis (RA) frequently have autoantibodies to
citrullinated peptides,
suggesting the involvement of the peptidylarginine deiminases citrullinating
enzymes in RA (van
Venrooij et al., A~t7~y~itis ReS.;2(4):249-51. Epub 2000 May 24).
[0247] Pellino homolog 2 from Drosophila (PELI2) is a a member of the Pellino
gene family,
which are involved in Toll-like signalling pathways. Pellino-2 associates with
the pelle-like kinase/1L
1R-associated kinase protein to couple the pelle-like kinase/IL-1R-associated
kinase protein to IL-1- or
LPS-dependent signaling. PELI2 may act as a downstream effector of interleukin
receptor signaling
and may play a ro le in inflammation-mediated Osteoarthritis. Pathway members
downstream of PELF
may be targetable (e.g., interleukin receptors).
73
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0248] G protein-coupled receptor 50 (GPRSO) is a member of the G protein-
coupled receptor
family. GPR50 has significant homology to melatonin receptors and was isolated
by PCR of human
genomic DNA with degenerate primers based on conserved regions of melatonin
receptors.
Example 4
APOB Proximal SNPs
[0249] It has been discovered that rs1367117 is associated with occurrence of
osteoarthritis in
subjects. The polymorphic variant lies within the APOB gene and codes for a
I98T amino acid change.
The guanine allele of SNP rs1367117 is associated with osteoarthritis (see
Table 5) and codes for a
threonine at position 98 (see, for example, amino acid sequence in SEQ ID NO:
38).
[0250] Apolipoprotein B (ApoB) is the main apolipoprotein of chylomicrons and
low density
lipoproteins (LDL). ApoB binds to sulfated proteoglycans, especially
chondroitin and dermatan sulfate,
that are components of cartilage (Camejo et. al., AtlZerosclerosis. 1998
Aug;13 9(2):205-22). This may
contribute to inflarnmation/joint damage by lipoprotein oxidation products. In
addition, increased levels
of ApoB is seen as a risk factor for osteonecrosis (Miyanishi et. al., Ann
Rheum Dis. 1999
Aug;58(8):514-6) _ Lipoprotein deposition has been noted in inflammatory
(rheumatoid) arthritis and
may play a role in inflammation mediated osteoarthritis. ApoB function can be
modulated by addition
of an antibody or a decoy receptor for ApoB. Examples of antibodies and small
molecules that
specifically interact with ApoB are described in U.S. Patent Nos. 6,107,045;
6,309,844; 5,330,910; and
6,369,075.
[0251] One hundred twenty-two additional allelic variants proximal to rs
1367117 were identified
and subsequently allelotyped in osteoarthritis case and control sample sets as
described in Examples 1
and 2. The polymorphic variants are set forth in Table 10. The chromosome
positions provided in
column four of Table 10 are based on Genome "Build 34" of NCBI's GenBanlc.
TABLE 10
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
2
rs1318006 2 238 21188688 C/T
rs1318005 2 294 21188744 C/T
rs1318004 2 295 21188745 A/G
rs1318003 2 347 21188797 A/C
rs4327259 2 1425 21189875 A/C
rs6756501 2 4891 21193341 C/T
rs6725189 2 5087 21193537 G/T
rs4665709 2 7041 21195491 A/G
rs4665710 2 7121 21195571 A/C
rs4371387 2 7219 21195669 A/G
rs952274 2 7443 21195893 G/T
rs952275 2 7485 21195935 G/T
rs1801695 2 10939 21199389 A/G
rs1042034 2 11367 21199817 A/G
rs1801702 2 11571 21200021 C/G
74
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo- Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
2
rs10420312 11839 21200289 A/G
rs26783782 12551 21201001 A/G
rs26783792 12646 21201096 A/G
rs18004792 13469 21201919 G/C
rs18017012 14913 21203363 A/G
rs43625892 15205 21203655 G/T
rs57429042 15246 21203696 A/G
rs17998122 15695 21204145 G/A
rs21632042 17473 21205923 G/T
rs6762102 17610 21206060 A/G
rs10420062 17828 21206278 A/C
rs18016962 18130 21206580 AlG
rs693 2 18281 21206731 C/T
rs10419742 18623 21207073 C/G
rs10419682 18890 21207340 C/T
rs5684132 21561 21210011 C/T
rs28547262 23100 21211550 A/T
rs28547252 23872 21212322 A/C
rs20009982 24581 21213031 A/T
rs20009972 24582 21213032 A/T
rs4971662 24983 21213433 C/T
rs5629562 27540 21215990 A/T
rs75893002 30846 21219296 C/T
rs37919802 31415 21219865 G/T
rs37919812 31453 21219903 A/G
rs18017002 31899 21220349 T/C
rs6798992 37000 21225450 A/G
rs10419522 38681 21227131 C/G
rs67277062 39287 21227737 C/T
rs67192072 42951 21231401 A/T
rs14695132 45648 21234098 C/T
rs18004782 46222 21234672 C/T
rs5506192 46687 21235137 A/G
rs67520262 47020 21235470 A/G
rs5798262 47593 21236043 C/T
rs5973312 48513 21236963 C/T
rs13671162 49723 21238173 A/G
rs13671172 49986 21238436 A/G
rs18004802 53018 21241468 C/G
rs18004812 53296 21241746 C/T
rs9341972 53547 21241997 A/G
rs16257642 53899 21242349 C/T
rs16257142 53916 21242366 G/T
rs15603572 53933 21242383 A/C
rs6173142 54305 21242755 G/T
rs5471862 55327 21243777 A/T
rs5895662 55895 21244345 C/T
rs5882452 56143 21244593 CIT
rs5859672 56640 21245090 G/T
rs75627772 58486 21246936 A/G
rs75758402 59576 21248026 G/T
rs75676532 63048 21251498 A/G
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
2
rs65480102 64008 21252458 A/G
rs65480112 64018 21252468 C/T
rs934198 2 64859 21253309 A/C
rs634292 2 65995 21254445 G/T
rs10031772 66905 21255355 A/G
rs67261152 67183 21255633 A/G
rs481069 2 67942 21256392 C/T
rs13671152 68101 21256551 A/G
rs666126 2 68521 21256971 A/G
rs75660302 68664 21257114 ClG
rs75901352 68988 21257438 A/G
rs67185132 69178 21257628 C/G
rs515135 2 72143 21260593 A/G
rs13671142 74183 21262633 C/G
rs563290 2 74312 21262762 C/T
rs562338 2 74407 21262857 C/T
rs581411 2 75518 21263968 A/G
rs580889 2 76153 21264603 A/G
rs548145 2 77398 21265848 AlG
rs668948 2 77615 21266065 A/G
rs594677 2 79092 21267542 C/T
rs571468 2 80000 21268450 G/T
rs46654922 80125 21268575 A/C
rs622236 2 80595 21269045 G/T
rs541041 2 81061 21269511 C/T
rs540156 2 81151 21269601 A/G
rs13671132 81918 21270368 C/T
rs18970842 83072 21271522 C/T
rs18970832 83137 21271587 C/T
rs478588 2 83235 21271685 C/T
rs664894 2 83263 21271713 A/T
rs15942862 83279 21271729 A/G
rs74221682 83280 21271730 C/G
rs565202 2 83533 21271983 C/T
rs14299742 86856 21275306 G/T
rs58297692 87186 21275636 -/TATA
rs30565752 87189 21275639 -/ATAT
rs67081682 87727 21276177 A/T
rs67567432 87978 21276428 C/T
rs21955982 89129 21277579 A/G
rs75672172 89556 21278006 C/T
rs568938 2 89702 21278152 AIG
rs666416 2 90233 21278683 A/G
rs67613002 93060 21281510 A/G
rs58297702 94779 21283229 -/T
rs14299732 95367 21283817 A/G
rs14299722 95844 21284294 A/G
rs67562842 95942 21284392 A/G
rs749988 2 96884 21285334 C/T
rs749987 2 96938 21285388 A/G
rs754524 2 97627 21286077 A/C
rs754523 2 97777 21286227 C/T
76
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
in SEQ
rs# some ID NO: 2 Position Variants
rs6754302 97871 21286321 A/C
rs6000122 98746 21287196 AlG
rs6143032 99663 21288113 A/G
Assay for Veri in~Land Allelotyping SNPs
[0252] The methods used to verify and allelotype the 122 proximal SNPs of
Table 10 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 11 and Table 12, respectively.
TABLE 11
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1318006ACGTTGGATGTCTCATGGCCCATCCAAGGCACGTTGGATGAAGGAGCCCATGAAGGCAGC
rs1318005ACGTTGGATGACAGCCTTGGATGGGCCATGACGTTGGATGTCTCCCAGTCTGGTGGAAAG
rs1318004ACGTTGGATGACAGCCTTGGATGGGCCATGACGTTGGATGTCTCCCAGTCTGGTGGAAAG
rs1318003ACGTTGGATGTTTCCACCAGACTGGGAGACACGTTGGATGAGTGCCCAGCACAGAGTCTT
rs4327259ACGTTGGATGAACAAGCTTGCTCAGCCACTACGTTGGATGTGTGTTCTGTCCAGGAAGAG
rs6756501ACGTTGGATGATGCATTCATTCGCTGTTTGAGGTTGGATGGAGATCAATGAGAAAAATAGG
rs6725189ACGTTGGATGAAGAACAATAGAGAGGGCCGACGTTGGATGAGTATTGACTGCCTTGGTTC
rs4665709ACGTTGGATGGCACAACCTCATAGATGTGGACGTTGGATGCCACCTCCATCATTGTGGAT
rs4665710ACGTTGGATGAATCCACAATGATGGAGGTGACGTTGGATGGATAACTCACTCACTATCACG
rs4371387ACGTTGGATGTAAAAGTGTGTAGCACCTCCACGTTGGATGTCATGGCAGAAGTTAAAGGG
rs952274ACGTTGGATGCAGAAGGGTGACATGCATTGACGTTGGATGCTCATATCCAGATTCACCCC
rs952275ACGTTGGATGCACAATGCATGTCACCCTTCACGTTGGATGGACACTCTCTTTGCTGAAGG
rs1801695ACGTTGGATGGAAATTATTTTCTTCGTCGACGTTGGATGTGCTCAGGAAATAATTAAA
rs1042034ACGTTGGATGATCCAAGATGAGATCAACACACGTTGGATGGGCATAGGTTTTCTTTCAAC
rs1801702ACGTTGGATGTTTTGATAAATCTTTCAACACGTTGGATGCTAATAGATGTAATCTCGA
rs1042031ACGTTGGATGGTTTGATGGCTTGGTACGAGACGTTGGATGTTTCCCCGGAAACTGGAATC
rs2678378ACGTTGGATGGTTTTCAGTCCTAGGAAGGCACGTTGGATGTATCACATGCCCCAGAAAGG
rs2678379ACGTTGGATGCTTCCTAGGACTGAAAACTGACGTTGGATGTGGGCTCCAACTTGCCTTTT
rs1800479ACGTTGGATGAAGGGTATGGAGATGAAGAACGTTGGATGACCTTATACCTTTTGAAA
rs1801701ACGTTGGATGCTTGGTCATTGGAAAGCTCGACGTTGGATGGTGGCCCTGAATGCTAACAC
rs4362589ACGTTGGATGCTGCAGGGCACTTCCAAAATACGTTGGATGTATATGCGTTGGAGTGTGGC
rs5742904ACGTTGGATGATTTTGGAAGTGCCCTGCAGACGTTGGATGCTATTGCTAGTGAGGCCAAC
rs1799812ACGTTGGATGTTGTGGTGCCCTCTAATTTACGTTGGATGCATCTTCATCTGTCATTGA
rs2163204ACGTTGGATGTTTGGACTCTCCTTTGGCAGACGTTGGATGGCTGACATAGGGAATGGAAC
rs676210ACGTTGGATGCCCAACTCTCAACCTTAATGACGTTGGATGAATTGTGTGTGAGATGTGGG
rs1042006ACGTTGGATGCAGCATCTGGTCAATGGTTCACGTTGGATGACACCTTCCACATTCCTTCC
rs1801696ACGTTGGATGTGCTAAGAACCTTACTGACACGTTGGATGGCCCAATCTTGGATAGAAT
rs693 ACGTTGGATGCAGCATCTTTGGCTCACATGACGTTGGATGTCCTGCTGAATGTCCATTTG
rs1041974ACGTTGGATGTACTTTGAGAAATTGGTTGGACGTTGGATGGTTAACATCTTCAATGAATG
rs1041968ACGTTGGATGTCAGCTACTTCAAAATCCCCACGTTGGATGGGCTATTGATGTTAGAGTGC
rs568413ACGTTGGATGGAGACTGGGTTGTTTCCAAGACGTTGGATGCCACAAGAATACGTTCACAC
rs2854726ACGTTGGATGCTCTAGCTTAACAGCAAGCCACGTTGGATGGCAAATTCTCCCTCTGACTG
rs2854725ACGTTGGATGCATTCAGCTTTGTGTAACTGACGTTGGATGTTTCCAAAGACTGTATAAGG
rs2000998ACGTTGGATGTGAACCATCCTTGTATCTGGACGTTGGATGTGGCACCAATGATTTTGTCC
rs2000997ACGTTGGATGTGGCACCAATGATTTTGTCCACGTTGGATGTGAACCATCCTTGTATCTGG
rs497166ACGTTGGATGTCCCAAAGTGCTGGGATTACACGTTGGATGAAATCCAACTGGACATGCGC
rs562956ACGTTGGATGTAACAGTCTTACCACACGGCACGTTGGATGATAAGGGAAAGTCTCCCTGG
77
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs7589300ACGTTGGATGTACCACGTATGTTGAGTGAGACGTTGGA-fGCCCTTACTCTATGATTACTGC
rs3791980ACGTTGGATGTCTGGAGAGATCATCTTTGGACGTTGGATGCTACCTAGCTACCTCAAATC
rs3791981ACGTTGGATGTGTTTTGAGAATGAAGAAACACGTTGGATGGTTCTTAGGTATTTTTTGGG
rs1801700ACGTTGGATGGACCCGACTCGTGGAAGAAACGTTGGATGTCGCTAGGAGTGGGGTCCA
rs679899ACGTTGGATGCTGAAGTCCATGACAGTTGGACGTTGGATGTTGTGGCTTCCCATATTGCC
rs1041952ACGTTGGATGCATGGAGCAGTTAACTCCAGACGTTGGATGTCTGGATCATCAGTGATGGC
rs6727706ACGTTGGATGGCACCACTTATTGAAAAGGGACGTTGGATGCACATACTTACAGTCAACGG
rs6719207ACGTTGGATGGTCCCAGTTGTAACCATGTCACGTTGGATGGGAATCCAGACTTGTCTGAG
rs1469513ACGTTGGATGCTTTTCTGCACAAGGACTCCACGTTGGATGACTCCACTTCATGGGATGAG
rs1800478ACGTTGGATGTGACGGTAAAGTGAGTGGAGACGTTGGATGCCCGTGTTGAATACATGTGG
rs550619ACGTTGGATGGCAAACACAGGTGAAGCATCACGTTGGATGGGCTTATCAGGTTGGGTCTA
rs6752026ACGTTGGATGCAGAAGGGAAGCAGGTTTTCACGTTGGATGCAGAAATGATGCCCCTCTTG
rs579826ACGTTGGATGAAAGTGCTGGGACTACAGGCACGTTGGATGATATGGGTGGAGAACAGAGC
rs597331ACGTTGGATGACACTCTCTCAGAAAGTTCCACGTTGGATGGTATGGTGATCAGATCAGAG
rs1367116ACGTTGGATGCAAGAAGTTTAAAGCATGAGACGTTGGATGATCATCAAAAAGAGAGAAGC
rs1367117ACGTTGGATGTTGGTTTTCTTCAGCAAGGCACGTTGGATGAGCTTCATCCTGAAGACCAG
rs1800480ACGTTGGATGCCGAGAAGGGCACTCAGCCACGTTGGATGCGCCGGCCGCGCATTCCCA
rs1800481ACGTTGGATGATCTGAAGAAGGCACCCCTGACGTTGGATGAAGCGTCTTCAGTGCTCTGG
rs934197ACGTTGGATGTGACTGGTCACTCACCAGACACGTTGGATGATCCTGATCAGAATCTGTGG
rs1625764ACGTTGGATGCAGAGGCATCGAGCGCTGGACGTTGGATGGACAGGACACGTCATGTTCC
rs1625714ACGTTGGATGAATTCCACTACCGCTGATTCACGTTGGe~TGATCGTTTCCTTCTCTTCTAG
rs1560357ACGTTGGATGGTCCCTGAAATTCCACTACCACGTTGGATGATTTCCCACCGGAAGCTTCA
rs617314ACGTTGGATGCAGTCTTCACCAGTAGCTTGACGTTGGATGTTGCAGAAGTCAGTGTGTGC
rs547186ACGTTGGATGCAGTTCAGGGAAGACTTGCCACGTTGGATGGAGAGGACTGTCACCATCTC
rs589566ACGTTGGATGCCCAGCAGACCAATATTCTGACGTTGGATGGGTATAGCTGAATGGTGCAG
rs588245ACGTTGGATGGCTCCAAAATCTCATCTGGCACGTTGGATGAGCTTCTGGGCATCATTTGC
rs585967ACGTTGGATGTGACAGGGAATCAGAGTCACACGTTGGATGCCACCTACTGCACTGAATCT
rs7562777ACGTTGGATGTTGGAGATTGCTCTTTGGGCACGTTGGATGTGACCTCAGGTTATCCACAC
rs7575840ACGTTGGATGCATAGACTGTCCATCACAGGACGTTGGATGGGTGTCAGAAAAACTTCCAC
rs7567653ACGTTGGATGAAAGTGGTGATGGATGCCTGACGTTGGATGGGGAGCAAATAGCTCATCTG
rs6548010ACGTTGGATGGCCTGGATTCGGGTTTTTAAACGTTGGATGCTATAAGCTGCTTATCAGAG
rs6548011ACGTTGGATGCTATAAGCTGCTTATCAGAGACGTTGGATGGCCTGGATTCGGGTTTTTAA
rs934198ACGTTGGATGACATGGAAGGAGGATGAGTGACGTTGGATGAGGTAGGACCCTCATGATTG
rs634292ACGTTGGATGGAGGCTTGTTTATGGCACAGACGTTGGATGCGTGCTTTTTCTCAAGTGCC
rs1003177ACGTTGGATGTACACAGACCCAGAAGATACACGTTGGATGGATGCATGAACAAAGGAAGC
rs6726115ACGTTGGATGATACAGATAAGGCACTTGGCACGTTGGATGAGGGAACTGAACGTGAAAGG
rs481069ACGTTGGATGTTTGAACTTCCTGAATGGTGACGTTGGATGATTGTGAGGGTTTACTTTCC
rs1367115ACGTTGGATGGGTTTGGAACAACTGATTGGACGTTGGA-T'GGGTAGGGAAATACTTTCAACG
rs666126ACGTTGGATGTTCTGCAGGATTCATCTCTCACGTTGGATGTTTTGTATGCCAGGTTAAGG
rs7566030ACGTTGGATGGATACAGAAGAGAGTGGTGGACGTTGGATGAGACTTGAGCCTTCAATGGC
rs7590135ACGTTGGATGACTGGTCTTAGGGTTACACCACGTTGGATGACAAAGCACCTGCTCCAAGA
rs6718513ACGTTGGATGCTTCCCTAGGTCTGAAGAACACGTTGGATGGCTTCTTTAGTGCCAAAGAG
rs515135ACGTTGGATGGGCTTACAGCCAAGTAACAGACGTTGGATGACCATCTTGTTACTGCACAG
rs1367114ACGTTGGATGGTTGGAGAATTATTTGCAGGACGTTGGATGGTGTGTGTGTATTTGTGTTTG
rs563290ACGTTGGATGGGGAAAATGCTGCAATGAACACGTTGGATGTCTGGGTATTCATCCAGAAG
rs562338ACGTTGGATGACCCAAGATGTAGAAACAGCACGTTGGATGCCATGGTTTGCATACATCAC
rs581411ACGTTGGATGACCTGGTGTGCTTAACTGTTACGTTGGATGGACAAGTGAAAAAGTTGGGC
rs580889ACGTTGGATGTGGGCTGACTCTCTTATCTCACGTTGGATGCCTCTGAACTGCAATAAGCC
rs548145ACGTTGGATGGAAGGAGGATGGTCAGAAACACGTTGGATGAGCTGTATCTCCCCTTTGTG
rs668948ACGTTGGATGATTGGAATAGGAAGGGCATGACGTTGGATGCTCTATCGTAATGGGGAAAAG
rs594677ACGTTGGATGGACTTGGTATTGAACAGGACACCTTGGATGTAGCAGGCATTTGCACTTTG
rs571468ACGTTGGATGGTGATGAAATTAAGGCCAGGACGTTGGATGATCTCACTGTTTCTCCAGGG
78
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer pCR primer
rs4665492ACGTTGGATGAGTGCGTCACTTCTATTGACACGTTGGATGCCAACAAGCATGTAAGTCAC
rs622236ACGTTGGATGCGCTTTTCTGTACTGTTTGAGACGTTGGATGTCCCTTGTCACTACAAAGAC
rs541041ACGTTGGATGGAGAGGAAAAGGTCACATTCACGTTGGATGATGCAGTAAGAGTAAGTGGC
rs540156ACGTTGGATGCTTGTCTTTGAAATTCCATAGACGTTGGATGCTCTCCTCCATGAATAATTAC
rs1367113ACGTTGGATGATAATACTGCAGGAGGACAGACGTTGGATGAGAACAAATGTCCTTCTCTG
rs1897084ACGTTGGATGCTTCATCCTCTTAAAAGGTCACGTTGGATGCACAAAACTATGAAACTTCC
rs1897083ACGTTGGATGGTTCAACCTATCATTTTCTTCACGTTGGATGTAACTCAATATGGATTAGAC
rs478588ACGTTGGATGATCTCTTGAACCCAAGAGATACGTTGGATG'f-GTTTAAGGTTTATGTCTTG
rs664894ACGTTGGATGCTTGAACCCAAGAGATGGAGACGTTGGATGTGGATTCTCTTTCTGCTGCC
rs1594286ACGTTGGATGCTTGAACCCAAGAGATGGAGACGTTGGATGTGAATTCTCTTTCTGCTGCC
rs7422168ACGTTGGATGCTTGAACCCAAGAGATGGAGACGTTGGATGTGAATTCTCTTTCTGCTGCC
rs565202ACGTTGGATGGCAAAGGCAATTCCATGGAGACGTTGGATGCTCGCAGCCTATGTCTTGTT
rs1429974ACGTTGGATGCTTCATTCTGGTCTGATTTCAACGTTGGATGGAAAGAATTCTATCAAGAAG
rs5829769ACGTTGGATGGTTGGAGCAGATGTTAAGGGACGTTGGATGGATCATGCTTCTGCCTTAAG
rs3056575ACGTTGGATGGTTGGAGCAGATGTTAAGGGACGTTGGATGGATCATGCTTCTGCCTTAAG
rs6708168ACGTTGGATGATGGTTACAGTAGCACCCTGACGTTGGATGTTTTTTACGGCAGCCTGAGC
rs6756743ACGTTGGATGTGGAATCGCAAGTGTAAGTGACGTTGGATGT-fGCACATGTATCCCAGAAC
rs2195598ACGTTGGATGATGGGCAAAGACTTCTTGACACGTTGGATGTGCTGTCAGAAGCTCTTTAG
rs7567217ACGTTGGATGCTCAAAACTCTTCTGGCCTCACGTTGGATGAACAGATGCTGGAGAGGATG
rs568938ACGTTGGATGCTCCTCAGCTAAATATCCAGACGTTGGATGAAAGTGGCAAAGTACTTGGC
rs666416ACGTTGGATGACCCTTTGAAACTGAGGTGGACGTTGGATGTCAGAAGTCCTTAGGACTGC
rs6761300ACGTTGGATGCCTACGAAGTAATTTTTCTCCACGTTGGATGCTATATTGAATGACAAGAGG
rs5829770ACGTTGGATGCACCTAACTGAGAATACACAGACGTTGGATGGCTGTAATTTCCTTAGTGGC
rs1429973ACGTTGGATGAAATATGGCTTGAACCCAGGACGTTGGATGTGGAGTGCAGTGGCACGATCT
rs1429972ACGTTGGATGCTTTCTTTGCTAACCACTGCACGTTGGATGCAGAATCTCTCTGAAAGCTG
rs6756284ACGTTGGATGTGGGATTATAGGCATGAGCCACGTTGGATGT'TCAGCTTTCAGAGAGATTC
rs749988ACGTTGGATGTTTTCTATTTGCATCTACTGACGTTGGATGGTGACAAAACAAACCAAAGTC
rs749987ACGTTGGATGGTCTTCAAATATAGTATGGCACGTTGGATGATTTCCAGGGTTTGACTTTG
rs754524ACGTTGGATGGACTTTCTGGGATTTCTCATCACGTTGGATGCTTCCACTCTAAGCCTTAAG
rs754523ACGTTGGATGGTATTTGCAAAGTAGGTGACACGTTGGATGTCTTGAAAGTGAAAGCCTCC
rs675430ACGTTGGATGATGAGCATGACACAACAACCACGTTGGATGAGGTATCTTCAGAGACACAG
rs600012ACGTTGGATGACTCCAGCCTGGGAGACAGAACGTTGGATGGCCTTGAACTTACACTCAAG
rs614303ACGTTGGATGCAAAACTCACATTCTTTGACACGTTGGATG'1-TTAAATTCCTGCCATGCAC
TABLE 12
dbSNP Extend Term
rs# Primer Mix
rs1318006 CCCTGACCTGTCACAGGG ACG
rs1318005 ATGAGAGCCCACCTCCTGT ACT
rs1318004 TGAGAGCCCACCTCCTGTA ACG
rs1318003 ACTGGGAGACTCACAGGGA ACT
rs4327259 GCCACTGGTCCAGCACAG ACT
rs6756501 GCCACTTTCTCCTCCTGCT ACG
rs6725189 AGAGAGGGCCGACTGCTG CGT
rs4665709 GTCCCCACCCAAATCTCAC ACT
rs4665710 GGCGGATTTCTCCTTTGGTG CGT
rs4371387 GTTCCAGCCATGTAGGTTGT ACT
rs952274 GGGTGACATGCATTGTGATTT CGT
rs952275 I CCTTCTGCTCAAAAACTTTTACI ACT
79
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs1801695 ATTATTTTCTTCGTCGCAATGG ACG
rs1042034 GAGATCAACACAATCTTCA ACT
rs1801702 GATAAATCTTTCAACAGTTCC ACT
rs1042031 TTGGTACGAGTTACTCAA ACT
rs2678378 AGTCCTAGGAAGGCTTTAATTT ACG
rs2678379 AGTCAGGAAATGACAGATAGG ACT
rs1800479 GGTATGGAGATGAAGAAAATCA ACT
rs1801701 AAAGACCCAGAATGAATC ACG
rs4362589 GGGCACTTCCAAAATTGATGAT CGT
rs5742904 CCTGCAGCTTCACTGAAGAC ACG
rs1799812 GGTGCCCTCTAATTTGTACTG ACG
rs2163204 GCTGCGATACCTGCTTCGT ACT
rs676210 AAGTTCCTGACCTTCACATAC ACG
rs1042006 CTGATGATCTTTACTTTCATTTCACT
rs1801696 GAACCTTACTGACTTTGCA ACT
rs693 GGCCAAATTCCGAGAGAC ACG
rs1041974 GTTGGATTTATTGATGATGCTGTACT
rs1041968 TTTGACATGCTCAAGAAC ACT
rs568413 TGGCGTAGAGACCCATCA ACT
rs2854726 AGCCTGTAGTCAATAACGCC CGT
rs2854725 AGCTTTGTGTAACTGGGTAAC ACT
rs2000998 TATCTGGTTTTGATCACCACAT CGT
rs2000997 CAGGATTAAACAGAAGTTCCAA CGT
rs497166 AGTGCTGGGATTACAGGTGT ACT
rs562956 CGGCTTCTCCTCTTATTTCTG CGT
rs7589300 AAGGTCCCTGACCTTTGAAC ACT
rs3791980 GGAAAATTAATATTTTCCCCCC CGT
rs3791981 GAGAATGAAGAAACAATAGCTC ACG
rs1801700 GACTCGTGGAAGAAGTTGGT ACT
rs679899 AAGTTGAGATTCTTTCAGA ACT
rs1041952 CAGAACTCAAGTCTTCAATCCT ACT
rs6727706 TCCCTAGTGTATGTTTTTGTCA ACT
rs6719207 TGTAACCATGTCAACAGTAGC CGT
rs1469513 CAAGCCTCTGGCCTTTGAAG ACT
rs1800478 CATACACGGTATCCTATGGAG ACT
rs550619 GTGGCCAGGACTCCTCAAT ACT
rs6752026 GGGAAGCAGGTTTTCCTTTAC ACG
rs579826 TGAGCCACCAGGTCCAGC ACG
rs597331 CTCTCAGAAAGTTCCCAACAC ACT
rs1367116 TTAAAGGAACCTAACTAGGGAA ACT
rs1367117 AGCCATACACCTCTTTCAGG ACT
rs1800480 GGCACTCAGCCCCGCAG ACT
rs1800481 TCTCAGACCCTGAGGCGC ACG
rs934197 CTGCATCCCCCTTCTCTCT ACG
rs1625764 CATCGAGCGCTGGCTGAAG ACG
rs1625714 TCCAGCTGGGCAGAGGCA ACT
rs1560357 CCACTACCGCTGATTCCCT CGT
rs617314 GTAGCTTGTTACATCTGGGG ACT
rs547186 GGGAAGACTTGCCAAAGACC I CGT
~0
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs589566 TCTGAGTTTAGTGCTGTTCAC ACT
rs588245 AGCCTATCTCGTTTCTGCCT ACT
rs585967 CTATGAAGTCTAACTGGGCTG ACT
rs7562777 ATGGTGCCTCGTGCCTGTA ACT
rs7575840 TCACAGGGAAAGCCAGGAAT ACT
rs7567653 ACTTCATTAATAACATCGCCGT ACT
rs6548010 GGTTTTTTGGTATACACATATTCACT
rs6548011 AAGGATAGAAAAAATATAGTCCCACT
rs934198 AGGAGGATGAGTGGGGAGA ACT
rs634292 CTTGTTTATGGCACAGAAGATG ACT
rs1003177 CACCATTTATGCAGGGCTAG ACT
rs6726115 CTGGTACTTGGTTAATAGTCC ACT
rs481069 CAGGACCCCAGCCCCCA ACT
rs1367115 TGGATTAGTGAATGGGAGGG ACT
rs666126 GCAGGATTCATCTCTCCATATA ACG
rs7566030 TGCCTGCCCCAACCCTCT ACT
rs7590135 CACCAGGCTGTTTTAGCAGC ACG
rs6718513 AAGAACAAAAAGAGGATTGGGA ACT
rs515135 ACAGCCAAAATGGAACCAAAG ACT
rs1367114 TTGCAGGTCACTTTTTTAAAGTTACT
rs563290 AACACAGAAATGCAGATATCTC ACG
rs562338 CATTGTCTTGACAGATGAATGC ACT
rs581411 TGATAGAGACAGTTATCAATTTCACT
rs580889 TCTCCGGCTGGGCCGTC ACT
rs548145 AGAAACAATGACAGAATACTAAGACT
rs668948 GGCATGCTGTCTCCTCTGC ACT
rs594677 GTATTGAACAGGACTGAGTAAT ACG
rs571468 GAAGAGAAGGCTGGCGCC CGT
rs4665492 CCTATAGATAAGACTTTTATTCCAACT
rs622236 GTGAATGAATGAATGAATGAACCCGT
rs541041 CTATTCATGTTTCAGGGCCCA ACG
rs540156 TACGAGTATATGTATACATTTGCACT
rs1367113 GGCTAGATAGGGAAGTGGG ACT
rs1897084 TCTTAAAAGGTCTTTTGCAAAGAACT
rs1897083 TCTATATTTTCTTTTGGAAGTTTCACT
rs478588 CTGGGCAGCAGAAAGAGAAT ACG
rs664894 GCCAAGATCATGCCACTGC CGT
rs1594286 ATGGAGGTTGCAGTGAGCC ACT
rs7422168 GATGGAGGTTGCAGTGAGC ACT
rs565202 CAGGAACAATTGGAAGTCTACA ACG
rs1429974 CTGGTCTGATTTCAGTTGCC ACT
rs5829769 GAGGATATATTCCAGGAGATATACGT
rs3056575 CAGAGGATATATTCCAGGAGA CGT
rs6708168 CCCTGCTTCTCAGTACCAAA CGT
rs6756743 CGCAAGTGTAAGTGATCAAAG ACG
rs2195598 ACTAAAACACCAAAAGCAATGG ACG
rs7567217 ACTCTTCTGGCCTCATCTAC ACT
rs568938 CCTCACACAAAACACCAGAAC ACT
rs666416 GCCTGTCCCACTGGGCC ACG
81
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs6761300 GGAATTCTTCAATAATGACAACAACT
rs5829770 CTTGATAACATGTACCAAAAAAAACGT
rs1429973 CTTGAACCCAGGAGGCAGA ACT
rs1429972 GCTAACCACTGCAGCTCCT ACG
rs6756284 GGCATGAGCCACCGCGC ACG
rs749988 TCTATTTGCATCTACTGAATTTTTACG
rs749987 CGAATAAGGAGCTATCTGTGA ACG
rs754524 TAGAAAACAAGCTATACATTCATAACT
rs754523 TGCAAAGTAGGTGACAATTGC ACG
rs675430 GTGAAAAATGAACAGATTTGTCCACT
rs600012 CTGGGAGACAGAGCGAGATT ACG
rs614303 CTTTGACAATACATGAGCCCT I ACG
Genetic Analysis
[0253] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 13.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (A1 AF
= 1-A2 AF). For
example, the SNP rs1318006 has the following case and control allele
frequencies: case A1 (C) = 0.494;
case A2 (T) = 0.506; control Al (C) = 0.460; and control A2 (T) = 0.540, where
the nucleotide is
provided in paranthesis. Some SNPs are labeled "untyped" because of failed
assays.
TABLE 13
dbSNP PositionChromosomeAl/A2 F A2 F A2 F p-
rs# in position AlleleCase Control Value
SEQ D AF AF
NO:
rs1318006238 21188688 C/T 0.506 0.540 0.326
rs1318005294 21188744 C/T 0.044 0.034 0.643
rs1318004295 21188745 A/G
rs1318003347 21188797 A/C
rs43272591425 21189875 A/C 0.962 0.965 0.865
rs67565014891 21193341 C/T 0.195 0.141 0.061
rs67251895087 21193537 G/T 0.317 0.250 0.036
rs46657097041 21195491 A/G 0.683 0.757 0.014
rs46657107121 21195571 A/C 0.206 0.209 0.926
rs43713877219 21195669 A/G 0.579 0.688 0.0001
rs952274 7443 21195893 G/T 0.163 0.123 0.158
rs952275 7485 21195935 G/T 0.234 0.319 0.013
rs180169510939 21199389 A/G 0.047 0.071 0.319
rs104203411367 21199817 A/G 0.191 0.182 0.743
rs180170211571 21200021 C/G
rs104203111839 21200289 A/G 0.686 0.785 0.001
rs267837812551 21201001 A/G
rs267837912646 21201096 A/G 0.693 0.714 0.466
rs180047913469 21201919 G/C 0.144 0.130 0.687
rs180170114913 21203363 A/G 0.090 0.116 0.314
rs436258915205 21203655 G/T
rs574290415246 21203696 A/G
rs179981215695 21204145 G/A
rs216320417473 21205923 G/T
rs676210 17610 21206060 A/G 0.186 0.177 0.758
rs104200617828 21206278 A/C
82
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP positionChromosomeAllA2 F A2 F A2 F p-
rs# in position Allele Case Control Value
SEQ ID AF AF
NO:
rs180169618130 21206580 A/G
rs693 18281 21206731 C/T 0.494 0.537 0.208
rs104197418623 21207073 CIG
rs104196818890 21207340 C/T
rs568413 21561 21210011 C/T
rs285472623100 21211550 A/T
rs285472523872 21212322 A/C
rs200099824581 21213031 A/T
rs200099724582 21213032 A/T
rs497166 24983 21213433 C/T
rs562956 27540 21215990 A/T
rs758930030846 21219296 C/T
rs379198031415 21219865 G/T
rs379198131453 21219903 A/G 0.964 0.968 0.849
rs180170031899 21220349 TIC 0.832 0.897 0.008
rs679899 37000 21225450 A/G 0.378 0.474 0.004
rs104195238681 21227131 ClG
rs672770639287 21227737 C/T
rs671920742951 21231401 A/T
rs146951345648 21234098 ClT 0.477 0.534 0.079
rs180047846222 21234672 C/T
rs550619 46687 21235137 A/G 0.053 0.062 0.656
rs675202647020 21235470 A/G
rs579826 47593 21236043 C/T 0.069 0.063 0.817
rs597331 48513 21236963 C/T 0.435 0.512 0.014
rs136711649723 21238173 A/G
rs136711749986 21238436 A/G 0.431 0.367 0.049
rs180048053018 21241468 C/G 0.978 NA NA
rs180048153296 21241746 C/T 0.100 0.082 0.487
rs934197 53547 21241997 A/G 0.338 0.398 0.075
rs162576453899 21242349 C/T
rs162571453916 21242366 G/T
rs156035753933 21242383 A/C
rs617314 54305 21242755 G/T 0.977 0.971 0.741
rs547186 55327 21243777 A/T 0.468 0.490 0.528
rs589566 55895 21244345 C/T 0.386 0.377 0.780
rs588245 56143 21244593 C/T 0.425 0.398 0.397
rs585967 56640 21245090 G/T 0.724 0.781 0.046
rs756277758486 21246936 A/G
rs757584059576 21248026 G/T 0.436 0.408 0.422
rs756765363048 21251498 A/G 0.918 0.910 0.739
rs654801064008 21252458 A/G 0.293 0.345 0.081
rs654801164018 21252468 C/T 0.530 0.482 0.135
rs934198 64859 21253309 A/C 0.526 0.484 0.225
rs634292 65995 21254445 G/T 0.456 0.492 0.256
rs100317766905 21255355 A/G
rs672611567183 21255633 A/G 0.293 0.342 0.119
rs481069 67942 21256392 C/T 0.138 0.104 0.167
rs136711568101 21256551 A/G 0,421 0.408 0.693
rs666126 68521 21256971 A/G 0.500 0.530 0.388
rs756603068664 21257114 C/G 0.397 0.416 0.536
rs759013568988 21257438 A/G 0.268 0.324 0.082
rs671851369178 21257628 CIG
rs515135 72143 21260593 A/G 0.726 0.747 0.455
rs136711474183 21262633 C/G
rs563290 74312 21262762 C/T 0.667 0.690 0.516
rs562338 74407 21262857 C/T 0.482 0.578 0.006
rs581411 75518 21263968 A/G 0.162 0.157 0.839
rs580889 76153 21264603 A/G 0.127 0.111 0.487
rs548145 77398 21265848 A/G 0.709 0.765 0.049
rs668948 77615 21266065 A/G 0.133 0.127 0.805
rs594677 79092 21267542 C/T
rs571468 80000 21268450 G/T 0.455 0.502 0.169
~3
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in position Allele Case Control Value
SEQ iD AF AF
NO:
rs466549280125 21268575 A/C 0.274 0.327 0.088
rs622236 80595 21269045 G/T
rs541041 81061 21269511 C/T 0.779 0.791 0.694
rs540156 81151 21269601 A/G 0.237 0.277 0.237
rs136711381918 21270368 C/T 0.394 0.366 0.370
rs189708483072 21271522 C/T
rs189708383137 21271587 C/T 0.279 0.326 0.139
rs478588 83235 21271685 C/T
rs664894 83263 21271713 A/T 0.319 0.343 0.467
rs159428683279 21271729 A/G
rs742216883280 21271730 CIG
rs565202 83533 21271983 C/T 0.483 0.514 0.373
rs142997486856 21275306 G/T 0.583 0.535 0.189
rs582976987186 21275636 -/TATA
rs305657587189 21275639 -/ATAT
rs670816887727 21276177 AIT 0.610 0.563 0.163
rs675674387978 21276428 C/T 0.051 0.051 0.978
rs219559889129 21277579 A/G
rs756721789556 21278006 C/T 0.100 0.087 0.547
rs568938 89702 21278152 A/G 0.177 0.150 0.304
rs666416 90233 21278683 A/G 0.421 0.364 0.093
rs676130093060 21281510 A/G 0.271 0.348 0.012
rs582977094779 21283229 -/T 0.036 0.037 0.971
rs142997395367 21283817 A/G
rs142997295844 21284294 A/G 0.422 0.443 0.533
rs675628495942 21284392 A/G 0.155 0.114 0.133
rs749988 96884 21285334 CIT
rs749987 96938 21285388 A/G
rs754524 97627 21286077 A/C 0.248 0.306 0.044
rs754523 97777 21286227 C/T 0.567 0.512 0.113
rs675430 97871 21286321 A/C 0.352 0.345 0.812
rs600012 98746 21287196 A/G
rs614303 99663 21288113 A/G 0.722 0.730 0.805
[0254] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure lA for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in Figure
1 A can be determined by consulting Table 13. For example, the left-most X on
the left graph is at
position 21188688. By proceeding down the Table from top to bottom and across
the graphs from left
to right the allele frequency associated with each symbol shown can be
determined.
(0255] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). The black line
84
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a l0kb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-8 were truncated at that
value.
[0256] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 5
ILIRL2 Proximal SNPs
[0257] It has been discovered that rs1024791, which lies within the ILIRL2
gene, is associated
with occurrence of osteoarthritis in subjects. Interleukin-1 receptor-like 2
is a member of the interleukin
1 receptor family. ILIRL2 inhibits IL-1 activity and contains immmunoglobulin
domains. This gene
and four other interleulcin 1 receptor family genes, including interleukin 1
receptor, type I (ILIRI),
interleukin 1 receptor, type II (IL1R2), interleukin 1 receptor-like 1
(IL1RL1), and interleukin 18
receptor 1 (IL18R1), form a cytokine receptor gene cluster in a region mapped
to chromosome 2q12.
ILIRL2 may mediate inflammatory responses that can contribute to the
development of OA. ILIRL2
biological activity can be modulated by addition of an antibody, a recombinant
binding partner, a
binding agent, or a recombinant ILIRL2 protein or functional fragment thereof.
[025] One hundred forty additional allelic variants proximal to rs 1024791
were identified and
subsequently allelotyped in osteoarthritis case and control sample sets as
described in Examples 1 and 2.
The polymorphic variants are set forth in Table 14. The chromosome positions
provided in column
four of Table 14 are based on Genome "Build 34" of NCBI's GenBank.
TABLE 14
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
3
rs39173042 225 102409525 G/T
rs20417472 509 102409809 C/T
rs39173052 860 102410160 C/T
rs37712002 874 102410174 C/T
rs39173062 939 102410239 A/G
rs39173072 1483 102410783 G/T
rs39173082 1798 102411098 C/T
rs39173102 2189 102411489 A/T
rs39173112 2215 102411515 A/G
rs39173122 2282 102411582 ClG
rs39173132 2340 640 C/T
1
02
411
~rs39173142 ~ 2963 ~ _ A/C
~ _
_
_
102412263
~
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
3
rs39173162 3369 102412669 -/T
rs31718452 3481 102412781 A/G
rs31718462 3564 102412864 G/T
rs39173172 3653 102412953 -/TC
rs39173182 4860 102414160 A/G
rs39173192 4941 102414241 A/T
rs39173202 4975 102414275 A/C
rs39173212 5321 102414621 A/G
rs39173222 5346 102414646 A/G
rs39173232 5541 102414841 A/G
rs39173242 5633 102414933 C/G
rs39173252 6007 102415307 G/T
rs37321342 6317 102415617 C/G
rs37321332 6378 102415678 A/G
rs21107262 6382 102415682 C/T
rs39173262 6426 102415726 C/T
rs39173272 6479 102415779 C/G
rs39173282 6641 102415941 C/T
rs37321312 6703 102416003 C/T
rs37321302 6705 102416005 C/T
rs39173292 7963 102417263 G/T
rs39173302 8525 102417825 G/T
rs39173312 8526 102417826 A/T
rs39173442 8598 102417898 C/T
rs39173322 8624 102417924 A/T
rs39173332 8883 102418183 A/T
rs39173342 8980 102418280 G/T
rs10300212 13578 102422878 G/T
rs22411322 16135 102425435 G/T
rs22411312 16141 102425441 G/T
rs38350362 16642 102425942 -/TGG
rs19975042 16931 102426231 A/G
rs18052322 17004 102426304 A/G
rs19716962 17009 102426309 C/T
rs19716952 17010 102426310 AIG
rs37711992 18713 102428013 C/T
rs19223032 18853 102428153 C/T
rs32137342 20783 102430083 C/T
rs19975032 21335 102430635 A/G
rs15586492 22180 102431480 C/T
rs15586482 22268 102431568 A/C
rs15586472 22285 102431585 C/T
rs15586462 25378 102434678 C/T
rs18825142 25906 102435206 C/G
rs18825132 26015 102435315 A/G
rs867770 2 26475 102435775 A/G
rs23102352 26798 102436098 A/T
rs870684 2 27042 102436342 A/G
rs37711972 27649 102436949 A/G
rs37711962 27827 102437127 A/T
rs38212072 27873 102437173 A/G
rs37711952 28122 102437422 A/G
~6
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
3
rs37711942 28202 102437502 A/G
rs37711932 28232 102437532 A/C
rs37711922 28240 102437540 G/T
rs37552902 29546 102438846 G/T
rs38212062 29748 102439048 A/G
rs23026232 30054 102439354 A/T
rs37552892 30646 102439946 G/T
rs19223022 31149 102440449 A/C
rs21107252 36912 102446212 A/C
rs14653262 36936 102446236 C/G
rs28714582 37184 102446484 C/T
rs20803102 39064 102448364 C/T
rs19222892 39343 102448643 G/T
rs19222902 40868 102450168 C/G
rs19222912 40917 102450217 A/G
rs19222922 41113 102450413 A/C
rs38155172 47343 102456643 A/T
rs22411302 47806 102457106 A/G
rs19222952 47911 102457211 A/G
rs19222942 48009 102457309 C/T
rs23026222 48621 102457921 C/G
rs23102402 49245 102458545 C/G
rs10247922 49247 102458547 CIG
rs38361122 49299 102458599 -/CTCT
rs30749692 49302 102458602 -/AGAG
rs9179942 49514 102458814 CIT
rs20417532 49626 102458926 G/T
rs20417522 49791 102459091 A/G
rs10247912 50010 102459310 A/G
rs10247902 50294 102459594 A/G
rs9955152 51482 102460782 AIG/T
rs9955142 51556 102460856 A/G
rs19222932 51855 102461155 A/G
rs37552872 51956 102461256 C/T
rs37295642 52155 102461455 A/G
rs37711882 52448 102461748 A/G
rs37711872 52458 102461758 C/T
rs37711862 52511 102461811 C/T
rs37711852 52607 102461907 A/G
rs23102412 54049 102463349 A/C
rs23026212 54224 102463524 A/C
rs23026202 54567 102463867 A/G
rs37711842 55052 102464352 C/T
rs38341612 55857 102465157 -/C
rs37552862 55941 102465241 C/G
rs37552852 56120 102465420 A/G
rs19975022 56349 102465649 C/T
rs37711822 56727 102466027 A/G
rs38361112 57232 102466532 -/CT
rs37711812 58806 102468106 C/T
rs9557542 _61_181 102470481 C/T
rs23026122 ~ 63808 _ ~ A/G
( 102473108
~7
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
3
rs37552842 64526 102473826 A/T
rs3821 2 64865 102474165 A/G
205
rs38155112 64928 102474228 C/T
rs22870412 64966 102474266 A/C
rs22870402 65080 102474380 A/G
rs22870392 65690 102474990 C/T
rs37552832 66228 102475528 AlG
rs37552822 66982 102476282 A/G
rs18123262 72511 102481811 A/G
rs15586262 74170 102483470 A/T
rs15586252 74264 102483564 C/T
rs15586242 74333 102483633 C/T
rs15586232 74502 102483802 A/T
rs10351312 74741 102484041 A/C
rs21106612 75321 102484621 C/T
rs142O0932 82558 102491858 A/G
rs30749712 85366 102494666 -/TTG
rs13453022 85469 102494769 C/T
rs14200922 86485 102495785 G/T
rs13453012 87687 102496987 C/T
rs23102422 89463 102498763 G/T
rs231 2 89660 102498960 A/G
0243
rs18825102 95718 102505018 C/T
rs18825112 95821 102505121 A!G
Assay for Verifying and Allelotypin~ SNPs
[0259] The methods used to verify and allelotype the 140 proximal SNPs of
Table 14 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 15 and Table 16, respectively.
TABLE 15
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs3917304ACGTTGGATGCAGAGAAGATAAGGAATGAGACGTTGGATGAAGGAAAATTACCCTAAACC
rs2041747ACGTTGGATGGGGAAGACTATTACAGGTATGACGTTGGATGTAGGAGCAACTAACACTTGC
rs3917305ACGTTGGATGGTTGTGAAGGAGAGGTCATGACGTTGGATGCGAAAGCCTCTACTGGTTTC
rs3771200ACGTTGGATGCTGGTTTCCTACTGCTCATCACGTTGGATGAGTGCTTTGCAGGTGTTGTG
rs3917306ACGTTGGATGCACCTGCAAAGCACTTTGTCACGTTGGATGTGCATTGTGTTCTCCATGGG
rs3917307ACGTTGGATGCTGTAGTAAGATTCCATGACACGTTGGATGACCCAAGTAATGAGGAAGTG
rs3917308ACGTTGGATGCAGTGACTTCTGATGTCCTCACGTTGGATGAAGTTAGGTCTGGTACATTG
rs3917310ACGTTGGATGGAGAAGAACTAAATGGAAGGACGTTGGATGGGGAAGAACTGATATCTTCA
rs3917311ACGTTGGATGCCATAGATTCATTTGGGGAAGACGTTGGATGGAGAAGAACTAAATGGAAGG
rs3917312ACGTTGGATGCCATACAAACACTGACTCTCACGTTGGATGGAAGATATCAGTTCTTCCCC
rs3917313ACGTTGGATGCACCATGACTATACTTGGTCACGTTGGATGTCAGTGTTTGTATGGGTGTG
rs3917314ACGTTGGATGGGCCTGCATTCAGACAATATACGTTGGATGGAAACTTCATAGAATGCACC
rs3917316ACGTTGGATGAGTATTCTTGTATATGCCACACGTTGGATGGTTAGGAGATGTAGAAGATG
rs3171845ACGTTGGATGGAACTCATTAGGCTGAATATCACGTTGGATGACAGATGCTCTAAATACCTG
gg
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP . Forward Reverse
rs# PCR primer PCR primer
rs3171846ACGTTGGATGTGTCCTATTCATCACAGAGCACGTTGGATGCTGCCTCAACATTCATATTGG
rs3917317ACGTTGGATGTCTCAGCCCTGAATTCTATCACGTTGGATGGACTAGATCTTCATGCATCAG
rs3917318ACGTTGGATGAAAAGCCTTGTGTGGCTTTGACGTTGGATGGTCTGAAAAACAGGAAGCAC
rs3917319ACGTTGGATGGTGCTTCCTGTTTTTCAGACACGTTGGATGAAGCCTGATGTTTCTCTGAC
rs3917320ACGTTGGATGCGTAAAGAAAAGCAGAAGACACGTTGGATGTTGCTCTTCAGATGAACCAC
rs3917321ACGTTGGATGAGGAGAAACTGCAAAGAGAGACGTTGGATGACAGGAGGCACCTAAAGAAC
rs3917322ACGTTGGATGAGTCAGCATGAGGCATAACCACGTTGGATGAGCATGGAGAAGTTGCCAAG
rs3917323ACGTTGGATGACTTCAGAGTAGAGGGCTTGACGTTGGATGAAGTGCTGGGATTATAGGCG
rs3917324ACGTTGGATGATCACCAGAGGTCAGGAGTTACGTTGGATGCCACCATGCCTAGCTCATTT
rs3917325ACGTTGGATGTAGTTAAGTCATCCACAGCCACGTTGGATGTGTCAGTCTCACTTTGCCTG
rs3732134ACGTTGGATGTTAATGCTTTCCTCCCTGGCACGTTGGATGTAGGGAGCTGTTCCTCCAAA
rs3732133ACGTTGGATGAAGGATGGTTCATGTGTGGGACGTTGGATGTTACGTCTTTGGAGGAACAG
rs2110726ACGTTGGATGAAGGATGGTTCATGTGTGGGACGTTGGATGTACGTCTTTGGAGGAACAGC
rs3917326ACGTTGGATGTGCACAGCCCACACATGAACACGTTGGATGTTCAGCTCCTGAACAGGTGG
rs3917327ACGTTGGATGAAAGCATGGGCTTCAGCTCCACGTTGGATGATGCCGCTCTTCTGTCATCC
rs3917328ACGTTGGATGTAGGCAAAGGAGGAGGAAGGACGTTGGATGTGTGTGAATTCCCAGGTTGG
rs3732131ACGTTGGATGAGGCCTTCTCGCATTTTCTCACGTTGGATGTCCCAGAGACTGTGGAATTG
rs3732130ACGTTGGATGTCCCAGAGACTGTGGAATTGACGTTGGATGAGGCCTTCTCGCATTTTCTC
rs3917329ACGTTGGATGAAGTCAAAGGAAGTTCACGGACGTTGGATGGTGCAAAGTTATTCCCCATC
rs3917330ACGTTGGATGTAAGCCAATAGCCTCTGACCACGTTGGATGAACAAGGTGAGGAGACCTTC
rs3917331ACGTTGGATGAACAAGGTGAGGAGACCTTCACGTTGGATGTAAGCCAATAGCCTCTGACC
rs3917344ACGTTGGATGAGAGTTCTTCCTGTTGTGGGACGTTGGATGTAGAAGAAGGGAGTTAGGGC
rs3917332ACGTTGGATGATTGGCTTAACAGTGAGCCCACGTTGGATGAGAAGCAAATGAGCAGAGGG
rs3917333ACGTTGGATGTAAGAAGGAGGCACTGACTGACGTTGGATGGCTGTCCAAAATGCATGCTC
rs3917334ACGTTGGATGGACTCAGACTCTAAGCCAACACGTTGGATGAGTCAGTGCCTCCTTCTTAC
rs1030021ACGTTGGATGCATTGCTTCATGTTCTTACCACGTTGGATGAAAACTGGGCATAACCTCTC
rs2241132ACGTTGGATGAGGAGGATGGGCGAGGAGTAACGTTGGATGTCTGGACACCAGCCTGCTTC
rs2241131ACGTTGGATGAGGAGGATGGGCGAGGAGTAACGTTGGATGCTGTCAGGTGGCAGAAGCAG
rs3835036ACGTTGGATGTTCCGCGGAAGAGGAAACAGACGTTGGATGTCACCTCCAAGCTCAAAGGC
rs1997504ACGTTGGATGCCTGTAATCCCAGTACTTTGACGTTGGATGTGTTAGCCAGGATGGTCTAG
rs1805232ACGTTGGATGTTGAGTAGCTGGGACTACAGACGTTGGATGTAACACGGTGAAACCCCGTC
rs1971696ACGTTGGATGTAGACCATCCTGGCTAACACACGTTGGATGTTGAGTAGCTGGGACTACAG
rs1971695ACGTTGGATGTTGAGTAGCTGGGACTACAGACGTTGGATGTAGACCATCCTGGCTAACAC
rs3771199ACGTTGGATGTGAATAACACAGGCCTGCTGACGTTGGATGGCTTGACCTGAATAGACAGC
rs1922303ACGTTGGATGGTGGGGCCTGAATAAAACACACGTTGGATGTAAGGTCATGCAAGCCAGTG
rs3213734ACGTTGGATGAACCCACTGTTTTTTATAGGACGTTGGATGTGACTGCTAGCTAACTAATC
rs1997503ACGTTGGATGAAAACTCATGACCCAGAGGGACGTTGGATGGCACAGGCTAGTCATTTGAG
rs1558649ACGTTGGATGTGCATGGTGGTTCATGCCTGACGTTGGATGAATCTTGCTATGATGCCCAG
rs1558648ACGTTGGATGAGATTTCCTACAACCTTGTGACGTTGGATGAGGTACATTTTATACCCACC
rs1558647ACGTTGGATGGAAAAATGTGGTCAATCTCACACGTTGGATGCAACCTTGTGTTGAACTTTG
rs1558646ACGTTGGATGGGCCTTGGTTAGAGTTTAGGACGTTGGATGGCTTTAGGTTGGCATAAATGG
rs1882514ACGTTGGATGTTCCTTTCCTGTCCATCCTGACGTTGGATGCAGAGTTGAGGTACTGGAAG
rs1882513ACGTTGGATGAAAGTAGAGAGGTCAGGTGGACGTTGGATGGGGCATTACACTTTTCCACC
rs867770ACGTTGGATGGCAGGTGGTGTATTTCAGAGACGTTGGATGACACTGCAGAAGTAGCTTGC
rs2310235ACGTTGGATGGAGCTGGAATAGGGAATCAGACGTTGGATGGCCATTATCCAGAACCTCTG
rs870684ACGTTGGATGCCCAAATTACTCCTCAGCACACGTTGGATGAGAGCGCGAAGTAACTTCAG
rs3771197ACGTTGGATGTAAGCAGTTCCAGTCCACAGACGTTGGATGCCTTTGCTTACCTAAGACTG
rs3771196ACGTTGGATGCCTTTAAACTACACAGCAACACGTTGGATGAGAAGCTTTCTGAGCAAGAG
rs3821207ACGTTGGATGAAAACCATGAAGAGGAGACGACGTTGGATGGCAACTAAAGGATCTTTCTC
rs3771195ACGTTGGATGGTGGACGCTATTGTTCTTAACACGTTGGATGTAAACTCTCAATGAGCTTGG
rs3771194ACGTTGGATGATCTTAAAGTTCAGCCTTGCACGTTGGATGATAATGTTCCAGTGGATCAG
rs3771193ACGTTGGATGGTTCCAGTGGATCAGAATAGACGTTGGATGTTAAAGTTCAGCCTTGCAGC
~9
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs3771192ACGTTGGATGGGGTTCATTCTTTCTTTCAAGACGTTGGATGATAGCAAAGCGACAGAATGG
rs3755290ACGTTGGATGCCCAATTACACTTTCTGCACACGTTGGATGTGATCACTGTTCAGACCTTC
rs3821206ACGTTGGATGAGAGTGGCCTACATGAGTTGACGTTGGATGCCTCCTGCAAAAAACTGACC
rs2302623ACGTTGGATGGAATACTTAGAAACCTGTGTGACGTTGGATGATCTGTTGTCTTCCAGTTAG
rs3755289ACGTTGGATGTCCAGAACTCTGAGCTCTGCACGTTGGATGCCTCAGCCTTCATTGTCGTG
rs1922302ACGTTGGATGGAGATCTTTCACTTCTTTGGACGTTGGATGGCCACACATAAAACCATATC
rs2110725ACGTTGGATGATTCTCTCCCCAAGCTATACACGTTGGATGCAATAACCAGGTTTGTGACC
rs1465326ACGTTGGATGTGTGTTTGGAAAAACCAATGACGTTGGATGTTTACAGAGTTCCAGGAGGG
rs2871458ACGTTGGATGAGATCCCCATAGGGATCCACACGTTGGATGCACACTTCAGAGTACTAGGG
rs2080310ACGTTGGATGGAATGATCCATTCCAGGGTGACGTTGGATGGACATCATGTTACCTGTGCC
rs1922289ACGTTGGATGTGAGTTTGGTCATTGCTACGACGTTGGATGATACAGGCCATGACCTACTC
rs1922290ACGTTGGATGACACCCAGTTTCCAGCTTTGACGTTGGATGCTTCGGCTCTCTGGTGTTTT
rs1922291ACGTTGGATGGACTTCTCTGCTACCACAACACGTTGGATGCTCATGGGGAGAGGAATCAA
rs1922292ACGTTGGATGATATTACCTCACAATGCAAGACGTTGGATGATGCTTATTGATCCTTTTCC
rs3815517ACGTTGGATGACAATGGTTGTCCTGGAAGGACGTTGGATGAATAGCCCCCTAGGCAAATG
rs2241130ACGTTGGATGGAGAAATGGATCTTACTGCTCACGTTGGATGCAATCCCACCTATCACATAG
rs1922295ACGTTGGATGGTTATATCATGAGCCATCGGACGTTGGATGGTGTCATTTCCAGTGTTTGC
rs1922294ACGTTGGATGCGGGCATACAAAGCAAACACACGTTGGATGACTGTCTTCCCTAAGAGTCC
rs2302622ACGTTGGATGTACTCCAGTGGGTTACACACACGTTGGATGGCATTTAGAGTCACTGCTCC
rs2310240ACGTTGGATGAAATTCAAGTCCTCTCTCTTACGTTGGATGGTGGTTTACCAAGACAGTTG
rs1024792ACGTTGGATGGCTGTGTGGTTTACCAAGACACGTTGGATGCCACACACGTGCGTGTCAAA
rs3836112ACGTTGGATGACGCACGTGTGTGGCTAGCTAACGTTGGATGGATGTATGCAAGCATAGG
rs3074969ACGTTGGATGACGTGTGTGGCTAGCTACATACGTTGGATGGGCTTTAGCTTGATGTATGC
rs917994ACGTTGGATGCCTCCCTTAGAATTGCAGTGACGTTGGATGAAGCAGAGAATGTGCACACC
rs2041753ACGTTGGATGTCCACATGTTGCAACCCAAGACGTTGGATGTAACTGTGAGTGAGCACAGC
rs2041752ACGTTGGATGGAACCTCTTAGAGGTACCAGACGTTGGATGTCCTTCTCCATCACTTTCCC
rs1024791ACGTTGGATGGTGTAAGGGACTGCAGATACACGTTGGATGAAACAGAACCAGGAGGTTGG
rs1024790ACGTTGGATGAGAAAATTCCAGCTGATTCTACGTTGGATGGACTCCTGCCCTACACTTTAA
rs995515ACGTTGGATGTGGGATGGAAATCGCTATTGACGTTGGATGTGTCCCAACCTAGAAGTTTG
rs995514ACGTTGGATGGCTTGGACTTGGCCTCAGAAACGTTGGATGCCAATAGCGATTTCCATCCC
rs1922293ACGTTGGATGGGACAGAGCTAAGGTTATAGACGTTGGATGGATTCAAATCTGGAGGTGTC
rs3755287ACGTTGGATGAAATTGGGTGTGCTCTTCCGACGTTGGATGGACTACTACCAGCCTTCAAC
rs3729564ACGTTGGATGCCTGAGTCCCTCTGAATGTAACGTTGGATGTGCCTTCGAGAGTACTGATG
rs3771188ACGTTGGATGAATCCAATCCTGGGCACTTGACGTTGGATGAGAGTAGAGGATGAGGAAGC
rs3771187ACGTTGGATGAGAGTAGAGGATGAGGAAGCACGTTGGATGAATCCAATCCTGGGCACTTG
rs3771186ACGTTGGATGAAGTGCCCAGGATTGGATTGACGTTGGATGGAGTAAGTCCCAATGCAGCC
rs3771185ACGTTGGATGATCTTGAGGCCCAAGATTTCACGTTGGATGGGCACCAAATGTGTTCTTAG
rs2310241ACGTTGGATGACCTTCTCCAGCTGGTTCTGACGTTGGATGTGGGAGTCCAGCTGTTCAAC
rs2302621ACGTTGGATGCGTCTACCACCGGAAACTAGACGTTGGATGGGAAACAAGTCAGCTCCTGG
rs2302620ACGTTGGATGGTCTCTGTAGAATGGAAGGCACGTTGGATGTGGCTGTGTCTGTTGTGTAC
rs3771184ACGTTGGATGTCTCTCTAGGCCCTGTACTTACGTTGGATGACTTGGTTTGATCTCTCTCC
rs3834161ACGTTGGATGAGGGAAACTGGTTGTCTGAGACGTTGGATGCAAAGCAAGCACTTGATGCC
rs3755286ACGTTGGATGGCATCAAGTGCTTGCTTTGCACGTTGGATGCAAGTTAGTGAATAGCCACG
rs3755285ACGTTGGATGTGCAGATGCCAGAGCCAAAAACGTTGGATGACCTGAAGTGCTGCTAGTAC
rs1997502ACGTTGGATGCGTATTCTTCCTGGAAGCTCACGTTGGATGTCACTGACAGAGTCAGTGAG
rs3771182ACGTTGGATGGCCAACACACAGAGATATTACACGTTGGATGGTATGTGTGCATTTTGTGATG
rs3836111ACGTTGGATGTCTACCCCGACTTGTTTTCCACGTTGGATGGGCTAAAACGAAGACAAGCC
rs3771181ACGTTGGATGTTCCTTCTCCAAAAGTTCAGACGTTGGATGGCCAGAGGATTTTTTTTCCG
rs955754ACGTTGGATGGTGATGTGGCCAGAAATGAGACGTTGGATGTATCCTCCTGCTTCAGCTTG
rs2302612ACGTTGGATGTGACAAACCTCGTGTCCTCCACGTTGGATGAAGGTGTCGGCCGTTTCCTC
rs3755284ACGTTGGATGGCTGCTCAGAATTCTGGTTGACGTTGGATGACCCTTCCATGTTTGAGAGG
rs3821205ACGTTGGATGATGCCATCCTAAGACCACAGACGTTGGATGCTTAGTAAGCAGTCAGTGGG
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs3815511ACGTTGGATGTACCACCCATCGCCTGTGAAACGTTGGATGGTGGTCTTAGGATGGCATGG
rs2287041ACGTTGGATGTGAAAGTCCATCCCACACTGACGTTGGATGTGTGGTCTTAGGATGGCATG
rs2287040ACGTTGGATGATAAAGAGTGGACCAATGTCACGTTGGATGTTATGTTCCAAGGTGACCTC
rs2287039ACGTTGGATGTTCACAGGCACACCCTTCAGACGTTGGATGAGCCACAGTGTGGGGAGAGT
rs3755283ACGTTGGATGTTCTTGCTGCATTGCATCCCACGTTGGATGGGAGAGAGAAATCGAGATGC
rs3755282ACGTTGGATGGAGGACCAAGCAAGATGAAGACGTTGGATGATATTTTGGCAGGCCAGCTC
rs1812326ACGTTGGATGTTCAAGTGATTCTACTGCCGACGTTGGATGAACCCCCGTCTCTACTAAAC
rs1558626ACGTTGGATGACCTCCAAGCATGATCTCAGACGTTGGATGTGGTTTTCCCTTGGTACTCG
rs1558625ACGTTGGATGTCAGCAAAGCAGGACCGACCACGTTGGATGTGAGATCATGCTTGGAGGTC
rs1558624ACGTTGGATGGGAAAGAACGGCCTGTCTTCACGTTGGATGATCCACAGGGTTCGTGTTGT
rs1558623ACGTTGGATGAAGTCCCAAACCCAAGTGAGACGTTGGATGTTAGGAAGCGAAGGAAAAAC
rs1035131ACGTTGGATGACTCTTCCTACCTTGATGGCACGTTGGATGTAGGCTTCAGGATTGGATGG
rs2110661ACGTTGGATGTCCCTCCAAAACCCACCTTTACGTTGGATGTGGATGGTGACACCTTCATG
rs1420093ACGTTGGATGAAGAAATTTAAAGCCCAGAGACGTTGGATGTATCTCAATAGAGGCTCTAC
rs3074971ACGTTGGATGAAACAAACTGAACCGCTAGGACGTTGGATGCAGCGTTCTTCTGGGTATTT
rs1345302ACGTTGGATGGGTAATCAGAAAACAGAGTCACGTTGGATGTGCCAGTAGAAGTACAGTAG
rs1420092ACGTTGGATGGTGCTCAGAGATGGTTAAACACGTTGGATGACTGCACCCTAGTTGATTTG
rs1345301ACGTTGGATGGCTCAAGTCTGGAGAAATGAACGTTGGATGCATGGTTGGATTTTGTGTTG
rs2310242ACGTTGGATGCCACCACTCAAACCTTTGTCACGTTGGATGGACAGCAAGAGTGAAACTCC
rs2310243ACGTTGGATGTGTAGCTAAGCACTATAGCGACGTTGGATGGCTCCTTCTAGATATGCATG
rs1882510ACGTTGGATGCTCGCTAGTCACTGGAGCTGACGTTGGATGAAGTCCAGGTGGACCCTGGT
rs1882511ACGTTGGATGAAGGAACTGTCAGGGCCATGACGTTGGATGAATGGTGCAACTGCCTTGGG
TABLE 16
dbSNP Extend Term
rs# Primer Mix
rs3917304 GGTTACTAATGGTGGTTTTCTCTGACT
rs2041747 ATGCTAAGAGTTATTCACATTTTGACT
rs3917305 GGAGATCCTTGTCCCATAGAT ACT
rs3771200 TACTGCTCATCTATGGGACAA ACT
rs3917306 GCACTTTGTCATCTGCCCCA ACT
rs3917307 AAGTTTGAAATGCCATTTCCTCT ACT
rs3917308 TAGTCTTACCCTATGCATCATCA ACT
rs3917310 ATGGAAGGATATACAATGTTCAT CGT
rs3917311 ATTCATTTGGGGAAGAACTGATA ACG
rs3917312 TACAAACACTGACTCTCACTTGTAACT
rs3917313 CTTGGTCCTTTACAGTTCCCT ACT
rs3917314 GGATACTAATGTACAAAGCAATGAACT
rs3917316 ATTTTAGAAACCCTCTTAGTAAAACGT
rs3171845 TGAATATCATTGTTTTCTAA ACT
rs3171846 ATCACAGAGCAAGGCCTA CGT
rs3917317 AGTTTAAACAAAGGAGAGAGAGA ACT
rs3917318 GTGTG GCTTTGGTTCAGGAG ACT
rs3917319 GTTGAGGTCATTAATGAAAACGT CGT
rs3917320 GAAGACTGATTATCATTTTAGTC CGT
rs3917321 ACGTGCCTCTCGGGTAGC ACT
rs3917322 CCATAAGACAGGAGGCACC ACG
rs3917323 GGGAAGATCTTTTAAAAAGGCA I ACT
91
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs3917324 TCAGGAGTTCGAGACCAGC ACT
rs3917325 CCTGTAGAGTCACTGACCC ACT
rs3732134 TTCCTCCCTGGCATGACCAT ACT
rs3732133 CAAGGGACATTGCAGACGGA ACT
rs2110726 TGCAAGGGACATTGCAGA ACG
rs3917326 CCCACACATGAACCATCCTTCC ACG
rs3917327 GCTTCAGCTCCTGAACAGGTG ACT
rs3917328 GGAGGAAGGGTGCAGGCAA ACT
rs3732131 TCTCGCATTTTCTCTAGCTGATC ACT
rs3732130 GGATGTTCTGAATTTTGGTAAAATACG
rs3917329 CTTCTTCCTCCAGAATTCAAC CGT
rs3917330 TCCCCACAACAGGAAGAACT CGT
rs3917331 TGAGGAGACCTTCTGCAGAG CGT
rs3917344 GAGTGGAGGTCAGAGGCTAT ACG
rs3917332 ACAGTGAGCCCTAACTCCC CGT
rs3917333 GGGTGTCATCTCTGACCATC CGT
rs3917334 TCAGACTCTAAGCCAACCTGCCA CGT
rs1030021 CTTTTAAATTTTGCCAGTTTTGC ACT
rs2241132 CGGTGGGGACCGCGTGG ACT
rs2241131 CGGCGGCGGTGGGGACC ACT
rs3835036 GCGGAAGAGGAAACAGAGAACCA ACT
rs1997504 GCGGGCGGATCACGAGG ACG
rs1805232 CGCCCGCCACCGCGCCC ACT
rs1971696 ACATTAAAAAAATTAGCCGGGC ACT
rs1971695 TACAGGCGCCCGCCACC ACT
rs3771199 TGACTGTGGTCAGCTGGAAA ACT
rs1922303 GGGGCCTGAATA,AAACACATCTGTACT
rs3213734 TTTAAGGCAGAATTGGTAAAGAAAACT
rs1997503 AGAGGGGTGTGCTGGCAGGC ACT
rs1558649 GGTGGTTCATGCCTGTAATCC ACG
rs1558648 TGTTGAACTTTGTATTATAAGCC ACT
rs1558647 GGTACATTTTATACCCACCAAA ACG
rs1558646 CTTGGTTAGAGTTTAGGGCACAT ACT
rs1882514 GGATTCACGTGTCCATCACTT ACT
rs1882513 GTGGGCTAATTCCAGTTAAGA ACG
rs867770 AGAAGTAGCTTGCCCTGAGAGC ACG
rs2310235 GGGAATCAGTCAGAAAGTAATA CGT
rs870684 CACAGTGGTTTTGGGTCCC ACG
rs3771197 GTTCCAGTCCACAGAATTTAGT ACT
rs3771196 CTACACAGCAACTAAAGGATC CGT
rs3821207 AAGAGGAGACGAGCATCAGA ACT
rs3771195 TTAAATCTTGTTAGTGAGACATTAACG
rs3771194 TGTCGCTTTGCTATAACTTAGACTACT
rs3771193 GTTATAGCAAAGCGACAGAATG ACT
rs3771192 CATCTTAAAGTTCAGCCTTGCA ACT
rs3755290 TGCACTTATCAAGCATTGGAC ACT
rs3821206 GGAAGGAAGACTTCATGGAG ACT
rs2302623 GAAACCTGTGTGATCCCTAG CGT
rs3755289 TCAGCTGGAAGGCCCGCA ~CT
92
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs1922302 TTAATTCCTAGGTATTTAATTTCGACT
rs2110725 CATTTTACAGAGTTCCAGGAGGG ACT
rs1465326 GGCTCTGTTTCTGACAATAACCAGACT
rs2871458 GGATCCACACCACCCAGAA ACT
rs2080310 GGTGGATCAGAAGTGCAGGT ACT
rs1922289 CATTGCTACGTTGAGTATGAG ACT
rs1922290 CCCAGTTTCCAGCTTTGGATATACACT
rs1922291 TCTGCTACCACAACTTTTCCA ACG
rs1922292 ACCTCACAATGCAAGATATATTA CGT
rs3815517 GCCACTTGCCCCTTGTGG CGT
rs2241130 GATCTTACTGCTCTCAGGGAT ACT
rs1922295 GCCTTCAAAGCTTAATGCCC ACG
rs1922294 GTTCTTTGCTATACTAAACAAGC ACT
rs2302622 CACACTGTTCAGAGTGTTCAAAACACT
rs2310240 TGCAAACACACACACACACACA ACT
rs1024792 CGTGTCAAACACACACACACACA ACT
rs3836112 TGGCTAGCTACATGCAAGAG ACT
rs3074969 TGGCTAGCTACATGCAAGAG ACT
rs917994 CAGTGAP;TAGGGATCTGTGC ACT
rs2041753 CCCATGTGCTCAGGGTGAG ACT
rs2041752 CTTAGAGGTACCAGAGAGAGA ACT
rs1024791 CTGGCTGATGTCAGAAAGCA ACG
rs1024790 CACAGAGAGGTTGAGTGACA ACT
rs995515 CTATTGGTCAGCTTCAGTCTAT ACT
rs995514 ACTTGGCCTCAGAATCCTTC ACT
rs1922293 GCTTCTCCATTTGACTTCCTTA ACG
rs3755287 GGTGTGCTCTTCCGTGAATTCGC ACT
rs3729564 TTCCAATTTCATTCTCTTTTAGCTACG
rs3771188 TGTGAGAACCCCTCACTTCA ACT
rs3771187 TCTGTCTTATGATTGAAGTGAG ACT
rs3771186 CGGTGTGTGGTGCAGTGC ACT
rs3771185 AGGCCCAAGATTTCTCATTTACT ACT
rs2310241 CAGCTGGTTCTGCTGCCC ACT
rs2302621 GGGCTCTGCAGACTTTTACTC ACT
rs2302620 CTGTAGAATGGAAGGCACTTCG ACT
rs3771184 CCCTGTACTTGGTGCCTGAAG ACT
rs3834161 GTTGTCTGAGAACGTTTTATGGG ACT
rs3755286 AGTACGGTTGTTGCCCACAT ACT
rs3755285 ACCCCCTCCCCATGCCC ACT
rs1997502 TCCTGGAAGCTCAGGCCCC ACT
rs3771182 GTTCTCGTAGACAGAGCTGT ACT
rs3836111 CCTTGGTTTCCCTTTGATCACT CGT
rs3771181 TCAGAAACATAAGAAACTTATGAAACT
rs955754 GCCAGAAATGAGAATTAAAGGCAGACT
rs2302612 GTAGCAAGGTGTGTGCTGC ACT
rs3755284 TGTCTAAAAGAGAGAGAAAAGG CGT
rs3821205 CCTCTGGCTCCCTCTCTC ACT
rs3815511 GGCACAGCACCTCCTAACC ACG
rs2287041 CATCCCACACTGGGTACCA I CGT
93
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs2287040 TGGACCAATGTCAAGTCGAG ACT
rs2287039 CAGAGAGGACACGTCCCC ACT
rs3755283 CCTATTATTTCATTAGGAATTAGTACT
rs3755282 CATGTGAAAAGTGCTTGGCAAAC ACG
rs1812326 AGGTGCATGCCACCACACT ACG
rs1558626 TTCAGGCTAGTTTCACCCGA CGT
rs1558625 GCAGGACCGACCCTCCCT ACG
rs1558624 GGCCTGTCTTCAGGGCTC ACT
rs1558623 AAGTGAGGGCTCCAGCGAT CGT
rs1035131 GATGGCACATCTCTAGAAAAG CGT
rs2110661 GTCTCTCCTCAGATATGAGCC ACG
rs1420093 TTTAAAGCCCAGAGATTTTAAAAAACT
rs3074971 CTAGGAAAAAAGAAAGGCAACA CGT
rs1345302 GAAAACAGAGTCTTTACCAATC ACT
rs1420092 AGAGATGGTTAAACAGGCACA ACT
rs1345301 CACAAGTTTACACCTTTTCTTTA ACT
rs2310242 CTCTATAACCTTACAAATGTTATTCGT
rs2310243 TGCAGTTTGGGACACAAAGG ACG
rs1882510 AAAACTGAGCTGGGCCTGC ACT
rs1882511 GGGAGGCATTCAGGGATCA I ACG
Genetic Anal,
[0260] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 17.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calcularted by subtracting the A2 allele frequency from 1 (A1 AF
= 1-A2 AF). For
example, the SNP rs3917304 has the following case and control allele
frequencies: case A1 (G) = 0.431;
case A2 (T) = 0.569, control Al (G) = 0.450; and control A2 (T) = 0.550, where
the nucleotide is
provided in paranthesis. Some SNPs are labeled "untyped" because of failed
assays.
TABLE 17
dbSNP position ChromosomeAl/A2 F A2 F A2 F p-
rs# in position Allele Case Control Value
SEQ D AF AF
NO:
rs3917304225 102409525G/T 0.569 0.550 0.460
rs2041747509 102409809C/T 0.027 0.023 0.800
rs3917305860 102410160C/T
rs3771200874 102410174C/T 0.467 0.473 0.809
rs3917306939 102410239A/G
rs39173071483 102410783G/T
rs39173081798 102411098C/T
rs39173102189 102411489A/T
rs39173112215 102411515A/G 0.945 0.964 0.193
rs39173122282 102411582CIG
rs39173132340 102411640C/T
rs39173142963 102412263A/C 0.025 0.028 0.881
rs39173163369 102412669-/T 0.785 0.856 0.004
rs31718453481 102412781A/G 0.904 0.894 0.624
94
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in position Allele Case Control Value
SEQ D AF AF
NO:
rs31718463564 102412864G/T
rs39173'173653 102412953-/TC 0.320 0.325 0.824
rs39173184860 102414160A/G 0.151 0.151 0.978
rs39173'194941 102414241A/T
rs39173204975 102414275A/C 0.936 0.946 0.585
rs39173215321 102414621A/G
rs39173225346 102414646A/G 0.978 unt ed NA
rs39173235541 102414841A/G 0.977 unt ed NA
rs39173245633 102414933C/G
rs39173256007 102415307G/T 0.029 0.030 0.901
rs37321346317 102415617C/G
rs37321336378 102415678A/G
rs21107266382 102415682C/T 0.320 0.318 0.944
rs39173266426 102415726C/T
rs39173276479 102415779C/G
rs39173286641 102415941C/T 0.898 0.891 0.706
rs37321316703 102416003C/T 0.047 0.036 0.434
rs37321306705 102416005C/T
rs39173297963 102417263G/T 0.070 0.081 0.473
rs39173308525 102417825G/T
rs39173318526 102417826A/T
rs39173448598 102417898C/T
rs39173328624 102417924A/T 0.224 0.209 0.473
rs39173338883 102418183A/T
rs39173348980 102418280G/T
rs103002113578 102422878GIT 0.160 0.183 0.255
rs224113216135 102425435G/T 0.604 0.631 0.385
rs224113116141 102425441G/T 0.451 0.477 0.282
rs383503616642 102425942-/TGG 0.424 0.463 0.112
rs199750416931 102426231A/G
rs180523217004 102426304A/G
rs197169617009 102426309C/T
rs197169517010 102426310A/G
rs377119918713 102428013C/T 0.299 0.291 0.726
rs192230318853 102428153C/T
rs321373420783 102430083C/T 0.826 0.860 0.099
rs199750321335 102430635A/G 0.830 0.806 0.281
rs155864922180 102431480C/T
rs155864822268 102431568A/C 0.127 0.142 0.439
rs155864722285 102431585C/T 0.824 0.825 0.955
rs155864625378 102434678C/T 0.576 0.580 0.886
rs188251425906 102435206C/G 0.547 0.556 0.730
rs188251326015 102435315A/G 0.500 0.513 0.574
rs867770 26475 102435775A/G
rs231023526798 102436098A/T 0.608 0.573 0.252
rs870684 27042 102436342A/G 0.687 0.685 0.931
rs377119727649 102436949A/G 0.534 0.544 0.676
rs377119627827 102437127A/T 0.171 0.189 0.558
rs38212 27873 102437173A/G 0.029 0.033 0.751
07
rs377119528122 102437422A/G 0.342 0.326 0.480
rs377119428202 102437502A/G 0.474 0.465 0.725
rs37711 28232 102437532A/C
93
rs37711 28240 102437540G/T
92
rs375529029546 102438846G/T 0.348 0.329 0.428
rs38212 29748 102439048A/G 0.914 0.920 0.803
06
rs230262330054 102439354A/T 0.261 0.263 0.948
rs375528930646 102439946G/T 0.429 0.442 0.621
rs192230231149 102440449A/C 0.574 0.539 0.166
rs2110T2536912 102446212A/C
rs146532636936 102446236C/G 0.592 0.613 0.413
rs287145837184 102446484C/T 0.068 0.059 0.549
rs208031039064 102448364C/T 0.258 0.256 0.926
rs192228939343 102448643G/T 0.593 0.593 0.976
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP PositionChromosomeAl/A2 F A2 F A2 F p-
rs# in position Allele Case Control Value
SEQ D AF AF
NO:
rs192229040868 102450168 C/G 0.577 0.595 0.489
rs192229140917 102450217 A/G 0.344 0.358 0.549
rs192229241113 102450413 A/C 0.226 0.221 0.874
rs381551747343 102456643 A/T 0.291 0.291 0.984
rs224113047806 102457106 A/G 0.112 0.088 0.153
rs192229547911 102457211 A/G 0.362 0.349 0.594
rs192229448009 102457309 C/T 0.075 0.065 0.581
rs230262248621 102457921 CIG
rs231024049245 102458545 C/G
rs102479249247 102458547 C/G
rs383611249299 102458599 -/CTCT 0.374 0.360 0.568
rs307496949302 102458602 -/AGAG 0.361 0.353 0.747
rs917994 49514 102458814 C/T 0.289 0.304 0.544
rs204175349626 102458926 G/T 0.330 0.329 0.981
rs204175249791 102459091 A/G 0.492 0.528 0.176
rs102479150010 102459310 A/G
rs102479050294 102459594 A/G 0.771 0.776 0.828
rs995515 51482 102460782 A/G/T 0.312 0.310 0.91
T
rs995514 51556 102460856 A/G 0.393 0.420 0.246
rs192229351855 102461155 A/G 0.597 0.608 0.653
rs375528751956 102461256 C/T 0.869 0.885 0.458
rs372956452155 102461455 A/G 0.331 0.315 0.511
rs377118852448 102461748 A/G
rs377118752458 102461758 C/T 0.280 0.258 0.332
rs377118652511 102461811 C/T 0.764 0.813 0.048
rs377118552607 102461907 A/G 0.429 0.395 0.160
rs231024154049 102463349 A/C 0.424 0.406 0.462
rs230262154224 102433524 A/C 0.323 0.340 0.473
rs230262054567 102433867 A/G 0.103 0.092 0.512
rs377118455052 102434352 C/T 0.779 0.809 0.173
rs383416155857 102435157 -/C 0.062 0.069 0.674
rs375528655941 102435241 CIG 0.786 0.817 0.150
rs375528556120 102435420 A/G 0.184 0.174 0.619
rs199750256349 102485649 C/T 0.580 0.564 0.559
rs377118256727 102466027 A/G 0.101 0.085 0.352
rs383611157232 102466532 -/CT 0.138 0.113 0.154
rs377118158806 102468106 C/T
rs955754 61181 102470481 C/T 0.194 0.172 0.29'1
rs230261263808 102473108 A/G 0.135 0.120 0.456
rs375528464526 102473826 A/T 0.757 0.789 0.147
rs382120564865 102474165 A/G 0.831 0.832 0.992
rs381551164928 102474228 C/T 0.022 unt ed NA
rs228704164966 102474266 A/C 0.118 0.100 0.346
rs228704065080 102474380 A/G 0.518 0.536 0.462
rs228703965690 102474990 C/T 0.975 0.970 0.752
rs375528366228 102475528 A/G
rs375528266982 102476282 A/G 0.312 0.295 0.452
rs181232672511 102481811 A/G 0.343 0.297 0.054
rs155862674170 102483470 A/T 0.536 0.551 0.643
rs155862574264 102483564 C/T 0.661 0.697 0.128
rs155862474333 102483633 C/T 0.322 0.278 0.074
rs155862374502 102483802 A/T 0.303 0.273 0.200
rs103513174741 102484041 A/C 0.543 0.595 0.046
rs211066175321 102484621 C/T 0.430 0.413 0.485
rs142009382558 102491858 A/G 0.381 0.388 0.826
rs307497185366 102494666 -/TTG 0.438 0.479 0.096
rs134530285469 1024J4769 C/T 0.428 0.397 0.223
rs142009286485 102495785 G/T 0.792 0.793 0.965
rs134530187687 102496987 C/T 0.514 0.477 0.139
rs231024289463 102498763 G/T 0.108 0.114 0.804
rs231024389660 1024J8960 A/G 0.490 0.523 0.194
rs188251095718 102505018 C/T 0.617 0.667 0.075
rs188251195821 102505121 A/G 0.664 0.652 0.599
96
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0261] The IL1RL2 proximal SNPs were also allelotyped in the replication
cohorts using the
methods described herein and the primers provided in Tables 15 and 16. The
replication allelotyping
results for replication cohort #1 and replication cohort #2 are provided in
Tables 18 and 19,
respectively.
TABLE 18
dbSNP PositionChromosom Al/A2 F A2 F A2 F p-
rs# in a Allele Case Control Value
SEQ D position AF AF
NO:
rs3917304 225 102409525 G/T 0.599 0.592 0.843
rs2041747 509 102409809 C/T 0.021 0.026 0.845
rs3917305 860 102410160 C/T
rs3771200 874 102410174 C/T 0.442 0.482 0.207
rs3917306 939 102410239 A/G
rs3917307 1483 102410783 G/T
rs3917308 1798 102411098 C/T
rs3917310 2189 102411489 A/T
rs3917311 2215 102411515 AIG 0.933 0.974 0.042
rs3917312 2282 102411582 C/G
rs3917313 2340 102411640 C/T
rs3917314 2963 102412263 A/C 0.036 0.038 0.918
rs3917316 3369 102412669 -/T 0.904 0.963 0.072
rs3171845 3481 102412781 A/G 0.898 0.882 0.610
rs3171846 3564 102412864 G/T
rs3917317 3653 102412953 -/TC 0.313 0.323 0.759
rs3917318 4860 102414160 A/G 0.149 0.142 0.803
rs3917319 4941 102414241 A/T
rs3917320 4975 102414275 A/C 0.921 0.930 0.749
rs3917321 5321 102414621 A/G
rs3917322 5346 102414640 A/G
rs3917323 5541 10241484'1A/G
rs3917324 5633 102414933 C/G
rs3917325 6007 10241530 G/T 0.033 0.040 0.716
rs3732134 6317 10241561 C/G
7
rs3732133 6378 102415678 A/G
rs2110726 6382 102415682 C/T 0.334 0.339 0.880
rs3917326 6426 102415726 C/T
rs3917327 6479 102415779 C/G
rs3917328 6641 10241594'1C/T 0.885 0.867 0.523
rs3732131 6703 102416003 C/T 0.045 0.022 0.224
rs3732130 6705 102416005 C/T
rs3917329 7963 102417263 G/T 0.068 0.091 0.296
rs3917330 8525 102417825 G/T
rs3917331 8526 102417826 A/T
rs3917344 8598 102417898 C/T
rs3917332 8624 102417924 A/T 0.203 0.195 0.785
rs3917333 8883 102418183 A/T
rs3917334 8980 10241828 G/T
O
rs1030021 13578 102422878 G/T 0.148 0.174 0.325
rs2241132 16135 102425435 G/T 0.604 0.595 0.815
rs2241131 16141 10242544'1G/T 0.452 0.464 0.696
rs3835036 16642 102425942 -/TGG 0.402 0.479 0.017
rs1997504 16931 10242623'1AlG
rs1805232 17004 102426304 A/G
rs1971696 17009 102426309 C/T
rs1971695 17010 10242631 A/G
O
rs3771199 18713 102428013 C/T 0.317 0.310 0.818
rs1922303 18853 102428153 C/T
rs3213734 20783 102430083 C/T 0.824 0.892 0.012
rs1997503 21335 102430635 A/G 0.838 0.790 0.114
97
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP PositionChromosomeAl/A2 F A2 F A2 F p-
rs# in position Allele Case Control Value
SEQ D AF AF
NO:
rs155864922180 102431480C/T
rs155864822268 102431568A/C 0.125 0.164 0.121
rs155864722285 102431585C/T 0.834 0.831 0.895
rs155864625378 102434678C/T 0.547 0.561 0.672
rs188251425906 102435206C/G 0.538 0.542 0.905
rs188251326015 102435315A/G 0.471 0.497 0.414
rs867770 26475 102435775A/G
rs231023526798 102436098AIT 0.562 NA 0.608
rs870684 27042 102436342A/G 0.657 0.680 0.509
rs377119727649 102436949A/G 0.502 0.534 0.351
rs377119627827 102437127A/T 0.171 0.189 0.558
rs382120727873 102437173A/G 0.033 0.038 0.821
rs377119528122 102437422A/G 0.374 0.342 0.311
rs377119428202 102437502A/G 0.493 0.480 0.696
rs377119328232 102437532A/C
rs377119228240 102437540G/T
rs375529029546 102438846G/T 0.364 0.346 0.602
rs382120629748 102439048A/G 0.940 NA 0.914
rs230262330054 102439354A/T 0.267 0.268 0.984
rs375528930646 102439946G/T 0.417 0.451 0.281
rs 192230231149 102440449A/C 0.600 0.559 0.245
rs211072536912 102446212A/C
rs146532636936 102446236C/G 0.573 0.614 0.296
rs287145837184 102446484C/T 0.085 0.070 0.530
rs208031039064 102448364C/T 0.277 0.268 0.776
rs192228939343 102448643G/T 0.580 0.576 0.924
rs192229040868 102450168C/G 0.558 0.579 0.556
rs192229140917 102450217A/G 0.322 0.348 0.401
rs192229241113 102450413A/C 0.235 un ed NA
rs381551747343 102456643A/T 0.310 0.312 0.950
rs224113047806 102457106A/G 0.110 0.068 0.071
rs 192229547911 102457211A/G 0.378 0.364 0.695
rs192229448009 102457309C/T 0.061 0.055 0.799
rs230262248621 102457921C/G
rs231024049245 102458545C/G
rs102479249247 102458547C/G
rs383611249299 102458599-/CTCT 0.407 0.378 0.382
rs307496949302 102458602-/AGAG 0.385 0.362 0.497
rs917994 49514 102458814C/T 0.271 0.281 0.757
rs204175349626 102458926G/T 0.357 0.342 0.672
rs204175249791 102459091A/G 0.459 0.511 0.155
rs102479150010 102459310A/G
rs102479050294 102459594A/G 0.781 0.773 0.769
rs995515 51482 102460782A/G/T 0.331 0.323 0.825
rs995514 51556 102460856A/G 0.373 0.412 0.221
rs192229351855 102461155A/G 0.568 0.597 0.376
rs375528751956 102461256C/T 0.867 0.907 0.138
rs372956452155 102461455A/G 0.362 0.320 0.212
rs377118852448 102461748AIG
rs377118752458 102461758C/T 0.308 0.276 0.288
rs377118652511 102461811C/T 0.761 0.847 0.003
rs377118552607 102461907A/G 0.445 0.385 0.069
rs231024154049 102463349A/C 0.446 0.400 0.161
rs230262154224 102463524A/C 0.304 0.326 0.499
rs230262054567 102463867A/G 0.100 0.074 0.236
rs377118455052 102464352C/T 0.785 0.853 0.014
rs383416155857 102465157-lC 0.068 0.081 0.596
rs375528655941 102465241C/G 0.791 0.850 0.038
rs375528556120 102465420A/G 0.194 0.173 0.446
rs199750256349 102465649C/T 0.604 0.577 0.536
rs377118256727 102466027A/G 0.107 0.070 0.117
rs383611157232 102466532-/CT 0.137 0.090 0.048
I
rs377118158806 102468106C/T I
98
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP PositionChromosomeAl/A2 F A2 F A2 F p-
rs# in position Allele Case Control Value
SEQ )aD AF AF
NO:
rs955754 61181 102470481C/T 0.209 0.160 0.084
rs2302612 63808 102473108A/G 0.138 0.111 0.331
rs3755284 64526 102473826A/T 0.754 0.829 0.010
rs3821205 64865 102474165A/G 0.799 0.814 0.594
rs3815511 64928 102474228C/T
rs2287041 64966 102474266A/C 0.113 0.074 0.143
rs2287040 65080 102474380A/G 0.493 0.521 0.386
rs2287039 65690 102474990C/T 0.970 0.962 0.703
rs3755283 66228 102475528A/G
rs3755282 66982 102476282A/G 0.327 0.312 0.636
rs1812326 72511 102481811A/G 0.362 0.299 0.067
rs1558626 74170 102483470A/T 0.558 unt ed
rs1558625 74264 102483564C/T 0.635 0.683 0.137
rs1558624 74333 102483633C/T 0.350 0.278 0.024
rs1558623 74502 102483802A/T 0.323 0.281 0.204
rs1035131 74741 102484041A/C 0.513 0.598 0.026
rs2110661 75321 102484621C/T 0.449 0.412 0.237
rs1420093 82558 102491858A/G 0.390 unt ed
rs3074971 85366 102494666-ITTG 0.398 0.485 0.006
rs1345302 85469 102494769C/T 0.468 0.392 0.036
rs1420092 86485 102495785G/T 0.810 0.808 0.958
rs1345301 87687 102496987C/T 0.554 0.470 0.016
rs2310242 89463 102498763G/T 0.110 unt ed
rs2310243 89660 102498960A/G 0.452 0.529 0.031
rs1882510 95718 102505018C/T 0.597 0.688 0.022
rs1882511 95821 102505121A/G 0.684 0.657 0.373
TABLE 19
dbSNP PositionChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
Figure AF AF
2
rs3917304 225 102409525G/T 0.531 0.483 0.236
rs2041747 509 102409809CIT 0.034 unt ed
rs3917305 860 102410160C/T
rs3771200 874 102410174C/T 0.500 0.460 0.282
rs3917306 939 102410239A/G
rs3917307 1483 102410783G/T
rs3917308 1798 102411098C/T
rs3917310 2189 102411489A/T
rs3917311 2215 102411515A/G 0.959 0.947 0.574
rs3917312 2282 102411582C/G
rs3917313 2340 102411640C/T
rs3917314 2963 102412263A/C
rs3917316 3369 102412669-/T 0.633 0.687 0.176
rs3171845 3481 102412781A/G 0.912 0.913 0.964
rs3171846 3564 102412864G/T
rs3917317 3653 102412953-/TC 0.329 0.329 0.999
rs3917318 4860 102414160A/G 0.153 0.165 0.696
rs3917319 4941 102414241A/T
rs3917320 4975 102414275A/C 0.955 0.971 0.463
rs3917321 5321 102414621A/G
rs3917322 5346 102414646A/G
rs3917323 5541 102414841A/G
rs3917324 5633 102414933C/G
rs3917325 6007 102415307G/T 0.023 unt ed
rs3732134 6317 102415617C/G
rs3732133 6378 102415678A/G
rs2110726 6382 102415682CIT 0.301 0.285 0.632
rs3917326 6426 102415726C/T
rs3917327 6479 102415779C/G
rs3917328 6641 102415941C/T 0.915 0.929 0.621
99
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP PositionChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
Figure AF AF
2
rs37321316703 102416003C/T 0.049 0.058 0.670
rs37321306705 102416005C/T
rs39173297963 102417263G/T 0.073 0.067 0.798
rs39173308525 102417825G/T
rs39173318526 102417826A/T
rs39173448598 102417898ClT
rs39173328624 102417924A/T 0.251 0.231 0.534
rs39173338883 102418183A/T
rs39173348980 102418280G/T
rs103002113578 102422878G/T 0.176 0.197 0.489
rs224113216135 102425435G/T unt 0.688 NA
ed
rs224113116141 102425441G/T 0.451 0.498 0.204
rs383503616642 102425942-/TGG 0.453 0.439 0.715
rs199750416931 102426231A/G
rs180523217004 102426304A/G
rs197169617009 102426309C/T
rs197169517010 102426310A/G
rs377119918713 102428013C/T 0.277 0.262 0.665
rs192230318853 102428153C/T
rs321373420783 102430083C/T 0.827 0.809 0.573
rs199750321335 102430635A/G 0.821 0_ 832 0.740
rs155864922180 102431480C/T
rs155864822268 102431568A/C 0.130 0.105 0.368
rs155864722285 102431585C/T 0.810 0_ 815 0.861
rs155864625378 102434678ClT 0.613 0_ 608 0.893
rs188251425906 102435206C/G 0.558 0_ 578 0.630
rs188251326015 102435315A/G 0.537 0_ 539 0.952
rs867770 26475 102435775A/G
rs231023526798 102436098A/T 0.589 0_ 019
rs870684 27042 102436342A/G 0.726 0_ 693 0.392
rs377119727649 102436949A/G 0.574 0_ 561 0.725
rs377119627827 102437127A/T
rs382120727873 102437173A/G 0.023 0_026 0.884
rs377119528122 102437422A/G 0.303 0 _ 301 0.952
rs377119428202 102437502A/G 0.450 0 _ 442 0.832
rs377119328232 102437532A/C
rs377119228240 102437540G/T
rs375529029546 102438846G/T 0.328 0 _ 302 0.452
rs382120629748 102439048A/G 0.889 0_026
rs230262330054 102439354A/T 0.254 0 _ 255 0.962
rs375528930646 102439946G/T 0.444 0_429 0.744
rs 192230231149 102440449A/C 0.541 0 _ 507 0.364
rs211072536912 102446212A/C
rs146532ti36936 102446236C/G 0.616 0_612 0.919
rs287145837184 102446484C/T 0.046 0_041 0.775
rs208031039064 102448364C/T 0.235 0 _238 0.933
rs192228939343 102448643G/T 0.611 0_618 0.845
rs192229040868 102450168C/G 0.601 0 _619 0.631
rs192229140917 102450217A/G 0.372 0 _374 0.961
rs 192229241113 102450413A/C 0.215 0 _ 221 0.827
rs381551747343 102456643A/T 0.268 0 _257 0.766
rs224113047806 102457106A/G 0.115 0 _ 119 0.854
rs192229547911 102457211A/G 0.342 0 _325 0.632
rs192229448009 102457309C/T 0.092 0 _081 0.677
rs230262248621 102457921C/G
rs231024049245 102458545C/G
rs102479249247 102458547C/G
rs383611249299 102458599-/CTCT 0.332 0 _332 0.999
rs307496949302 102458602-/AGAG 0.330 0 _339 0.822
rs917994 49514 102458814C/T 0.312 0.339 0.456
rs204175349626 102458926G/T 0.296 0.310 0.737
rs204175249791 102459091A/G 0.534 ~ _556 0.587
rs102479150010 102459310A/G
rs102479050294 102459594A/G 0.759 0.780 0.498
100
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP PositionChromosomeAl/A2 F A2 F A2 F p-
rs# in Position AlleleCase Control Value
Figure AF AF
2
rs995515 51482 102460782 A/G/T 0.288 0.288 0.992
rs995514 51556 102460856 A/G 0.417 0.434 0.657
rs192229351855 102461155 A/G 0.634 0.625 0.806
rs375528751956 102461256 C/T 0.873 0.850 0.471
rs372956452155 102461455 A/G 0.291 0.308 0.643
rs377118852448 102461748 A/G
rs377118752458 102461758 C/T 0.246 0.231 0.677
rs377118652511 102461811 C/T 0.766 0.759 0.850
rs377118552607 102461907 AIG 0.409 0.410 0.972
rs231024154049 102463349 AIC 0.396 0.416 0.591
rs230262154224 102463524 A/C 0.347 0.363 0.667
rs230262054567 102463867 A/G 0.107 0.121 0.605
rs377118455052 102464352 CIT 0.772 0.740 0.364
rs383416155857 102465157 -/C 0.054 0.051 0.860
rs375528655941 102465241 CIG 0.781 0.766 0.641
rs375528556120 102465420 AIG 0.172 0.175 0.897
rs199750256349 102465649 C/T 0.550 0.543 0.849
rs377118256727 102466027 AIG 0.094 0.109 0.562
rs383611157232 102466532 -!CT 0.139 0.148 0.750
rs377118158806 102468106 C/T
rs955754 61181 102470481 C/T 0.173 0.190 0.571
rs230261263808 102473108 A/G 0.132 0.135 0.909
rs375528464526 102473826 A/T 0.760 0.726 0.332
rs382120564865 102474165 A/G 0.873 0.859 0.629
rs381551164928 102474228 C/T
rs228704164966 102474266 A/C 0.124 0.141 0.517
rs228704065080 102474380 A/G 0.550 0.559 0.802
rs228703965690 102474990 C/T
rs375528366228 102475528 A/G
rs375528266982 102476282 A/G 0.293 0.268 0.452
rs181232672511 102481811 A/G 0.320 0.294 0.453
rs155862674170 102483470 A/T 0.541 unt ed
rs 155862574264 102483564 C/T 0.694 0.719 0.473
rs155862474333 102483633 C/T 0.285 0.279 0.865
rs155862374502 102483802 A/T 0.277 0.261 0.615
rs103513174741 102484041 A/C 0.581 0.590 0.795
rs211066175321 102484621 C/T 0.405 0.414 0.800
rs142009382558 102491858 A/G 0.384 unt ed
rs307497185366 102494666 -/TTG 0.488 0.469 0.619
rs134530285469 102494769 C/T 0.378 0.406 0.437
rs142009286485 102495785 G/T 0.769 0.768 0.980
rs134530187687 102496987 C/T 0.464 0.487 0.531
rs231024289463 102498763 G/T 0.120 unt ed
rs231024389660 102498960 A/G 0.537 0.514 0.548
rs188251095718 102505018 C/T 0.642 0.635 0.875
rs188251195821 102505121 A/G 0.639 0.644 x.871
[0262] Allelotyping results were considered particularly significant with a
calculatedL p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure 1B for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 1 O) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the gxaphs in Figure
1B can be determined by consulting Table 17. For example, the left-most X on
the left graph is at
101
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
position 102409525. By proceeding down the Table from top to bottom and across
the graphs from left
to right the allele frequency associated with each symbol shown can be
determined.
[0263] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
BrookslCole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a l Okb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-8 were truncated at that
value.
[0264] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 6
ILIRLI Region Proximal SNPs
[0265] It has been discovered that SNP rs1041973 in Interleukin 1 receptor-
like 1 isoform 1
(ILIRLI) is associated with occurrence of osteoarthritis in subjects.
Interleukin 1 receptor-like 1
isoform 1 is a member of the interleukin 1 receptor family with no known
ligand (orphan receptor).
ILIRLl exists in both a soluble and transmembrane form, suggesting that it may
have ligand or lig~.nd
scavenging activity. Studies of the similar gene in mouse suggested that this
receptor can be induced by
proinflammatory stimuli. This gene and four other interleukin 1 receptor
family genes, including
interleulcin 1 receptor, type I (IL1R1), interleukin 1 receptor, type II
(IL1R2), interleukin 1 receptor-like
2 (IL1RL2), and interleukin 18 receptor 1 (IL18R1), form a cytokine receptor
gene cluster.
[0266] Ninety-one additional allelic variants proximal to rs 1041973 were
identified and
subsequently allelotyped in osteoarthritis case and control sample sets as
described in Examples 1 and 2.
The polymorphic variants are set forth in Table 20. The chromosome positions
provided in column
four of Table 20 are based on Genome "Build 34" of NCBI's GenBank.
TABLE 20
dhSNP Position Chromosome Allele
in
rs# ChromosomeSEQ ID Position variants
NO: 4
rs884517 2 207 102527857 c/t
rs14769842 6019 102533669 a/
102
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position Chromosome Allele
rs# Chromosomein Position Variants
SEQ ID
NO: 4
rs951774 2 6414 102534064 a/c
rs20417372 7341 102534991 a/
rs14200912 10984 102538634 a/
rs21106602 12351 102540001 c/
rs13623472 13335 102540985 a/
rs30739682 16584 102544234 -/t t /t
t a
rs40904732 16737 102544387 c/
rs15586222 23897 102551547 c/t
rs15586212 24057 102551707 c/t
rs15586202 25145 102552795 a/
rs15586192 25300 102552950 a/c
rs950881 2 26262 102553912 a/c
rs950880 2 26312 102553962 /t
rs13623462 26589 102554239 c/t
rs19681712 27302 102554952 a/
rs18132992 27358 102555008 a/t
rs18132982 27451 102555101 c/
rs19681702 27552 102555202 c/t
rs974389 2 30731 102558381 c/t
rs971764 2 32085 102559735 a/
rs14200892 32139 102559789 a/
rs14200882 33184 102560834 a/
rs14201032 42382 102570032 /t
rs14201022 42569 102570219 a/
rs19974672 44823 102572473 c/t
rs19974662 45217 102572867 c/
rs13623502 45548 102573198 c/
rs23102202 45601 102573251 a/
rs13623492 45722 102573372 c/
rs37552782 45967 102573617 a/
rs37711802 47367 102575017 a/c
rs37711792 47642 102575292 a/c
rs985523 2 48126 102575776 c/t
rs10419732 49218 102576868 a/c
rs32143632 49274 102576924 -/a
rs873022 2 49433 102577083 /t
rs37711772 49610 102577260 a/c
rs37321292 51282 102578932 a/
rs14201012 51466 102579116 a/
rs12905 2 53757 102581407 a/
rs37711752 53960 102581610 a/t
rs38212042 54031 102581681 c/
rs21602032 54574 102582224 c/t
rs19461312 55679 102583329 a/
rs10540962 56100 102583750 c/t
rs22870382 56182 102583832 c/t
rs19216222 59817 102587467 a/
rs18612462 60533 102588183 a/
rs18612452 60656 102588306 a/
rs37552762 72209 102599859 a/
rs22870372 72778 102600428 a/
rs14200992 74293 102601943 c/
103
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP ChromosomePosition Chromosome Allele
rs# in Position Variants
SEQ ID
NO: 4
rs3771174 2 77335 102604985 a/
rs1420098 2 78029 102605679 a/
rs1362348 2 78374 102606024 c/
rs1882348 2 78421 102606071 a/t
rs1558627 2 78434 102606084 c/t
rs2058622 2 79174 102606824 c/t
rs3836110 2 79397 102607047 -/
rs3771172 2 79562 102607212 a/
rs3771171 2 79700 102607350 a/
rs3771170 2 79730 102607380 a/t
rs2160202 2 79904 102607554 c/t
rs2058623 2 79920 102607570 a/
rs3771167 2 79938 102607588 c/t
rs3771166 2 79972 102607622 c/t
rs1974675 2 80125 102607775 c/t
rs1465321 2 80368 102608018 a/
rs2041740 2 83484 102611134 c/t
rs3771164 2 85536 102613186 a/t
rs2270298 2 85829 102613479 c/t
rs2270297 2 86425 102614075 a/
rs2041739 2 88083 102615733 a!
rs2080289 2 88770 102616420 c/t
rs3821203 2 90622 102618272 a/
rs3771162 2 90924 102618574 alt
rs3213733 2 91634 102619284 It
rs3213732 2 92029 102619679 c/t
rs1035130 2 95152 102622802 a/
rs3752659 2 95348 102622998 clt
rs3755274 2 96145 102623795 c/t
rs2241117 2 96793 102624443 a/
rs2241116 2 97015 102624665 /t
rs881890 2 97064 102624714 c/t
rs3771161 2 97711 102625361 /t
rs3771160 2 97855 102625505 a/c
rs3771159 2 98708 102626358 a/
rs1420104 2 not ma not ma ed c/t
ed
rs2041738 2 not ma not ma ed a/c
ed
Assay for Verifying and Allelotypin~ SNPs
[0267] The methods used to verify and allelotype the 91 proximal SNPs of Table
20 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 21 and Table 22, respectively.
TABLE 21
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs884517ACGTTGGATGCATTTTCTGGTGTGACTCCCACGTTGGATGATGTTCGGTCACTTGTGAGC
rs1476984ACGTTGGATGTGAGAGAGTTGAAGAATGGGACGTTGGATGCCAAGAAGTGATTTCCTTCC
104
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs951774ACGTTGGATGTCAGCCAGAGGTCTTTACTCACGTTGGATGTTAGAAGTCTCTTGGGTGGG
rs2041737ACGTTGGATGGAGATGGAGTTTCCCTCTTGACGTTGGATGAAACCAAGAGGTGGAGGTTG
rs1420091ACGTTGGATGCACCCCTATTATAAAACCCACACGTTGGATGACCAGAAATGGCATCTATGG
rs2110660ACGTTGGATGTCTCTCCGAGATGAGGAATCACGTTGGATGGTGATCTCCTCAGTACTCTG
rs1362347ACGTTGGATGTTCTTTGGTAATGAGGTAGGACGTTGGATGTGCTTGCCCTCTATTTATGG
rs3073968ACGTTGGATGGAATGATGAGGAAGGAAGGGACGTTGGATGTAAAGCCACATGTTCACCCG
rs4090473ACGTTGGATGTAGTGTGTTTCACTCTTCCCACGTTGGATGTCAAGCACCTCTGTTAACTC
rs1558622ACGTTGGATGATACTTCCTGGTTTTCTGGGACGTTGGATGGGCTCAAAGTCATCACCCAA
rs1558621ACGTTGGATGACAGTGGCGATGCCAACATTACGTTGGATGCCTGTAGTAGGACCCTACTG
rs1558620ACGTTGGATGTTGCAGGTGTCTGGTGATAGACGTTGGATGAGTTTGCCTTTCTTCATGGC
rs1558619ACGTTGGATGCCCTAATTAGGATTCCGCACACGTTGGATGCTCCATCACACTTTGACTGC
rs950881ACGTTGGATGCTTATCTCAGTCTGCCAGTGACGTTGGATGGGTGAGTGAATTAGTCCTGG
rs950880ACGTTGGATGTGCCAAAGACAATCAAATCCACGTTGGATGCACTCACCTCTGATTTCTAG
rs1362346ACGTTGGATGTTCCTCAGGTTCACCAAGAGACGTTGGATGTCCCGAACCTCATCTCATAC
rs1968171ACGTTGGATGAATGTTTCAGCCCAGCATGGACGTTGGATGATCTCCTGACCTCATGATCC
rs1813299ACGTTGGATGAATTCCAGCACTTTGGGAGGACGTTGGATGTTTCACCGTGTTAGCCAGGA
rs1813298ACGTTGGATGTTACTGCAAGCTCCACCTCCACGTTGGATGATTAACTGGGCGTGGTGGTG
rs1968170ACGTTGGATGAGCTTGCAGTAAGCCCAGATACGTTGGATGTGTTAGGGTAATTACAGTGC
rs974389ACGTTGGATGCTCTAGCCCAATATGTCTCCACGTTGGATGACTGGAGATGTGAACCCATC
rs971764ACGTTGGATGGAGATGATGGAGATTAAGAGGACGTTGGATGAGTTGTTTGACTTCGGACTG
rs1420089ACGTTGGATGAGACAGCACATATCAATGACACGTTGGATGTATTGTGCGGTTCGCTATAG
rs1420088ACGTTGGATGGGATGACTGTCAAAAACATCACGTTGGATGTAATTTTTCAGGAGCAAGGC
rs1420103ACGTTGGATGTCCATTGGAATATGACCTCCACGTTGGATGCCAGGCACATGAGCTATATC
rs1420102ACGTTGGATGGATTGGTCAGGAACTCAAACACGTTGGATGTGGGTTGCTTCTAGCTATTG
rs1997467ACGTTGGATGTGAATTTCAGTGAGTCAGGCACGTTGGATGTGAGGGGAAAAAAAACATCC
rs1997466ACGTTGGATGATAGGCACATACAGGATTTCACGTTGGATGCTCCCTTTTCAGATTAATCTC
rs1362350ACGTTGGATGGAGAACATTCTCTATACCAGACGTTGGATGTGCCTGAATAGTGAGAAGCC
rs2310220ACGTTGGATGGGTTGAAACCAGACTTGCTGACGTTGGATGCAGCCTAATCTCTGGTATAG
rs1362349ACGTTGGATGCAATACTCTGTGGTACTTATCACGTTGGATGTAAACAGTCTTATCCTTGGG
rs3755278ACGTTGGATGAGTGCTGAATAGGTTTGTTCACGTTGGATGGCCTAGTTTAAGAATGAATGC
rs3771180ACGTTGGATGGTCAACATCAAGAATTCTTAGACGTTGGATGCCTGAAATTTGATTTGTGGC
rs3771179ACGTTGGATGGTCTTCATAATTCATGATTGACGTTGGATGTCTTAAAATATAAGGGGAAG
rs985523ACGTTGGATGTCCTATGGAAGTTTTGGGTCACGTTGGATGCTGCGAAGTAGCATGATAAC
rs1041973ACGTTGGATGGGGACTTCTGACAATACAGGACGTTGGATGAATCGTGTGTTTGCCTCAGG
rs3214363ACGTTGGATGCAGGCAATCAACCACTGAAGACGTTGGATGCTGCAGTTGCTGATTCTGGT
rs873022ACGTTGGATGCCTAGTCCTTTCTGGAACAGACGTTGGATGATCCCTGCAACTGTAAATCC
rs3771177ACGTTGGATGAAGGTTAGAAGCCCCTTTTCACGTTGGATGGGCTGGAATTAAGAACAAAC
rs3732129ACGTTGGATGCTAATTCAAAGCCACATCTGACGTTGGATGTAAGTTAGCATTCAGATTGC
rs1420101ACGTTGGATGCAACATTTATGTACACCATAGACGTTGGATGTTAGTAATACTCATTGGATT
rs12905 ACGTTGGATGCTCCCAGCAAACAGGAACAGACGTTGGATGATCAAGACAATGGGAATGGC
rs3771175ACGTTGGATGAAAGAGCACAAAAGAACACGACGTTGGATGTTATGAACTCCCTCTGTGTC
rs3821204ACGTTGGATGCATGTTGTAAGCATGGTCCGACGTTGGATGACTTTACCACCCTCGCTAAC
rs2160203ACGTTGGATGACACAGACCCAAACCATACCACGTTGGATGTTCCCGTGTGTTCCATGTAC
rs1946131ACGTTGGATGGGGAACTCAGGGTTTAACACACGTTGGATGTACACTCATCACTCCTCAGG
rs1054096ACGTTGGATGATCAAGGTGCTATGTGAGGGACGTTGGATGAAAGCAGGAGTACACAAGGG
rs2287038ACGTTGGATGAATGTCCCTGGTTACCTATGACGTTGGATGACAAATAAGCTAGAAGGAGC
rs1921622ACGTTGGATGGCCACTTCTTAATTCTGTCCACGTTGGATGATTTCAGCTAGTGCCTATGG
rs1861246ACGTTGGATGCACAAGCTCTTCACCTCTTCACGTTGGATGTGGCTGAGGAGAAGTGTAAC
rs1861245ACGTTGGATGTGCTGCCTTCAATGTGTGACACGTTGGATGAGGAAAGGTCAGAGGACATG
rs3755276ACGTTGGATGCCAGCACTCACTAACATGTGACGTTGGATGAAACTCATATGGGCAGCCAC
rs2287037ACGTTGGATGCAGATTCAGCCAAAGCTTTCACGTTGGATGAAAAATCTGTGTGCCAGAAG
rs1420099ACGTTGGATGTTACACACTCTCCAGAGGTGACGTTGGATGAAAGCTTCTAGCTGCCTGAG
105
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs3771174ACGTTGGATGACCCAGATTCTCTGGCTTTGACGTTGGATGTACCACAAGTGCCGAAAGAG
rs1420098ACGTTGGATGGGGACGTGAAGTACAAGATACGTTGGATGGGAGACCAAAAAAAGTTACC
rs1362348ACGTTGGATGCATGTCATAGGAAGAGTAGGACGTTGGATGTCAGCAACTCAAATATGCAG
rs1882348ACGTTGGATGCCTACTCTTCCTATGACATGACGTTGGATGCCCTAAAAGGAAATCCTATC
rs1558627ACGTTGGATGCCCTAAAAGGAAATCCTATCACGTTGGATGCCTACTCTTCCTATGACATG
rs2058622ACGTTGGATGCTGTGAAACCTTGGTAGCACACGTTGGATGTTTCTGATGCCTGGGAGTTC
rs3836110ACGTTGGATGACTCACAAATGGGGTAAAGGACGTTGGATGTGCCTTCATTCAATCAGGAG
rs3771172ACGTTGGATGCAGAAGCAAATGGCATTGGCACGTTGGATGCCATTGTTGCTTCCTAAGCC
rs3771171ACGTTGGATGAGGGTAGCAGATAGGAGATGACGTTGGATGAAGCTGCTTCTCTCCTCATC
rs3771170ACGTTGGATGCAAGGCCATTGTCAAAGCTGACGTTGGATGGTGTCCCAGAGTGGATATTG
rs2160202ACGTTGGATGAGCAGTATTTACTGCAGATGACGTTGGATGCCCACATCAAACTGCAAAGG
rs2058623ACGTTGGATGATTTACTGCAGATGTGTGTGACGTTGGATGTGTTCACTGATAGATCCCAC
rs3771167ACGTTGGATGCTAACTTAAGTGTGTAACCCACGTTGGATGCTAACGGGAAATTTTCAGGTG
rs3771166ACGTTGGATGGTGAACAGACTTTACACCTGACGTTGGATGCCTCAGTGGCATTTGATTAT
rs1974675ACGTTGGATGACTAAGAAGGAAGGGGATACACGTTGGATGGTACATTTCCCTCTACCTTC
rs1465321ACGTTGGATGTCACAGCTTTGGGTCAGTTGACGTTGGATGTCAACAACACACTGCACCTG
rs2041740ACGTTGGATGCATCCATGTCCCTACAAAAGACGTTGGATGAAAGCTCTTATACACCATGG
rs3771164ACGTTGGATGCCTGTGACATGTATGGAAATGACGTTGGATGTCAAATCCATAGGTACACTC
rs2270298ACGTTGGATGTGAAGTAGTGTTCTCTCTCACGTTGGATGAATATGAGCACTGTAGCTGC
rs2270297ACGTTGGATGTTTCCTGCCAAAAAGAAAGGACGTTGGATGGACCACACCACTAGTTCAAA
rs2041739ACGTTGGATGTAGACCCTGAAGTTTCCCACACGTTGGATGCACCTAGAGGTTCCTTTTGC
rs2080289ACGTTGGATGTGGAGAATGTCAACTGAGTCACGTTGGATGATACAAACAAGAGGCCATGG
rs3821203ACGTTGGATGTCAAAGACAAAGGGCAGGAGACGTTGGATGGGATCCAGAGAAAGGTAGTC
rs3771162ACGTTGGATGTGAGTGGAGTACAGTGAGACACGTTGGATGTGGCACTGCACTTTCTGAGA
rs3213733ACGTTGGATGTGAAAGCACCTTGTATCTGGACGTTGGATGCATCTTCCTCTGCCTTTTAG
rs3213732ACGTTGGATGGTCAGGTTAAAAGTGGCAACACGTTGGATGTGACACTGGATACACATTTC
rs1035130ACGTTGGATGTTAGGATCCGATCCATTTTCACGTTGGATGCTCTGCTTTGCTGAATGAAG
rs3752659ACGTTGGATGTGCATAATGCGTCCACCTAGACGTTGGATGGGCTGATGTGTATTTTGGGC
rs3755274ACGTTGGATGTATCAAAGGTGTGTGCACCCACGTTGGATGAGGGGTAGAAAACCACAGTG
rs2241117ACGTTGGATGTGGCTGGAAGATCATGATGCACGTTGGATGCCCCAAGTTGTTAGGAAGAG
rs2241116ACGTTGGATGAATGCAGGCAACATCACAGCACGTTGGATGAGTAGGCTCTGTTCGTTACC
rs881890ACGTTGGATGATGCCATTTGCCTTCTGGAGACGTTGGATGTCTCAGGGTAACGAACAGAG
rs3771161ACGTTGGATGCCATCAGGTGAGCACTGAAAACGTTGGATGTCATTGCCTCCTGAACTTGG
rs3771160ACGTTGGATGAGAAATGGCTGTGACTGGAGACGTTGGATGTATCCAGGGAGTTGATGGTG
rs3771159ACGTTGGATGCAGGTGATGGTCCAACAAAGACGTTGGATGTGCTGTGGTCCACTCACTTG
rs1420104ACGTTGGATGTATTCTGGAGGCTGAGGTGGACGTTGGATGTGGAGTGCAGTGGTGTGATC
rs2041738ACGTTGGATGTGGTGAAACCCCATCTCTACACGTTGGATGTTTCAAGCTATTCTCCTGCC
TABLE 22
dbSNP Extend Term
rs# Primer Mix
rs884517 GGTGTGACTCCCAGACCAA ACT
rs1476984 ATGGGTAGTTAATGGTGGAAATTTACT
rs951774 CAAAGTAGTTGACTTGTCTTTCT ACT
rs2041737 CCAGGCTAGTGCAGTGGC ACT
rs1420091 CCCACATTATATTGTCATTACTTTACG
rs2110660 ATGAGGAATCAGAGCTGGGA ACT
rs1362347 GTAATGAGGTAGGAATAATATTG ACT
rs3073968 GGCAATTGTGTGTGTGTGTG CGT
rs4090473 CTTACTCCTATTCCAAAGTTCA ACT
rs1558622 ACTGCAAGGGAGAGCCCC I ACT
106
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs1558621 AGTGTGTGTGTGTGCGTGC ACT
rs1558620 GTCTGGTGATAGTTGGGTGC ACG
rs1558619 GATTCCGCACATCCTATGCCT ACT
rs950881 GATGGTTTGTGCCTCTGGTC ACT
rs950880 ATTTAAGAATGCTTTCGTCATAAGACT
rs1362346 GAATATCTATGCCCACCAGAT ACG
rs1968171 GCCCAGCATGGTGGCTCA ACG
rs1813299 GTGGATCATGAGGTCAGGAG CGT
rs1813298 GCCTCAGCCTCCCGAGTA ACT
rs1968170 AGCCTGGGTGACAGAGCC ACT
rs974389 GTCTCCTGAATTTCAGAAGCA ACT
rs971764 GTCAAGGTAAAAACATTATTGTG ACG
rs1420089 GCACATATCAATGACAAGACTA ACT
rs1420088 CATGTTATGTAACTCTGAGTTC ACT
rs1420103 GAATATGACCTCCAGAAGGCAA ACT
rs1420102 GAACTCAAACAAATACTTGGACACACG
rs1997467 TTCAGTGACTCTCACAATAAGC ACG
rs1997466 AAGAAAAAGCTGGTTCAATGAG ACT
rs1362350 ACATTCTCTATACCAGAGATTAGGACT
rs2310220 CTGAACTTCAAAGTCAAGCTTTT ACG
rs1362349 CTGTGGTACTTATCATTAACATCAACT
rs3755278 ACTCGGAATTCTTTTACATTTGGTACT
rs3771180 CATCAAGAATTCTTAGTACATGATACT
rs3771179 TATGTTAGTAAATTTCTATGTTGGACT
rs985523 CATATAGCTTTCACAATGATCATGACG
rs1041973 ATACCAGAATCAGCAACT ACT
rs3214363 GAGCAGGGTGAAAGAAGATGGG ACT
rs873022 TTCTAGGAATACTATCAGGTTGA ACT
rs3771177 TTTTCACCTACTAGAGGCCC CGT
rs3732129 GCCACATCTGTTCTTTATTCTTT ACG
rs1420101 CCATCACAAAGCCTCTCATTA ACT
rs12905 AGACAGCAAACAACATCC ACG
rs3771175 CACAAAAGAACACGTTCAGTTT CGT
rs3821204 TAAGCATGGTCCGTTCTATAC ACT
rs2160203 CCACACACATTATCATTGTTA ACT
rs1946131 TTAACACTCTTTGGCTATTTGACAACT
rs1054096 TCCATCCAGCCTGCCCAC ACG
rs2287038 TACCTATGTGTTTGAATTATCTTCACT
rs1921622 GAAAGAGGACTTAAAAATTGATGAACT
rs1861246 CTTCACCTCTTCTTTTTCAGTC ACG
rs1861245 CTGGAATGGTTTTCTACTTCC ACG
rs3755276 GTGTGTATGCATGTGTTCGC ACT
rs2287037 ACAAAAGTGTGCCTATCTTATGAAACT
rs1420099 GGTGGGAGGTTGATAATTGAAA ACT
rs3771174 CTGACCATCATCTACCCAGG ACT
rs1420098 ACGTGAAGTACAAGATTCTTCA ACT
rs1362348 GAGTAGGAAAGAAAAGGATGTG ACT
rs1882348 TCCTATGACATGAAATACATTCT CGT
rs1558627 AAGCAGAGAGAGATAAACTTATT ACG
107
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs2058622 AAACCTTGGTAGCACTTCTGT ACT
rs3836110 AACAAACACCGCCCCCCC CGT
rs3771172 GCATTGGCCATCTTTCTGATA ACG
rs3771171 GAGGTGTCCCAGAGTGGATA ACG
rs3771170 CAAAGCTGCTTCTCTCCTCA CGT
rs2160202 TATACACATATGTGTTCTAACTTAACT
rs2058623 ACTTAGGTGTGTAACCCTTTG ACG
rs3771167 CTTTGTAGTTTGATGTGGGATCTACT
rs3771166 ACTTTACACCTGAAAATTTCCC ACT
rs1974675 GAAGGGGATACAAAAGGGATA ACT
rs1465321 CAGTTGGCCTCAGTGTTAACCC ACG
rs2041740 GAACTCATGCTTTTTTATGGCTGACG
rs3771164 GACATGTATGGAAATGTGTGTG CGT
rs2270298 CTCTCTCTCTGCATGTGTGT ACT
rs2270297 AGCCAAGTAGAGGAGCACC ACT
rs2041739 CTCCTGAGTTCCTGTGAATAC ACT
rs2080289 TCTCAGGACTCCACTCAAATGTCACT
rs3821203 GGCAGGAGGCAATTTCGGT ACT
rs3771162 CAGTGAGACTCAGGAGTGC CGT
rs3213733 TGTATCTGGTTTTCTCTCACTCAACT
rs3213732 CAACATTCAAAAAATGGCACTCTTACG
rs1035130 TCCGATCCATTTTCTTCCCC ACT
rs3752659 CCTAGGGTATGGCCACTATAATTAACG
rs3755274 CACCCAACTATAAAGAAAGACCTCACG
rs2241117 ATCATGATGCTAAGTTGAAAATATACT
rs2241116 TCAAGCATTTTAAACATGTGAATTCGT
rs881890 TGCCTTCTGGAGTCCTGTAA ACT
rs3771161 GTGAGCACTGAAAAACTTTAAGAACT
rs3771160 GCCAGAAAGCTGTGATTTCCA ACT
rs3771159 CCAACAAAGATTTGAGCCCC ACT
rs1420104 CTGGGAGGTGGAGACTGCA ACT
rs2041738 AAAAATACAAAAATTAGCTGGGCI ACT
Genetic Anal.
[0268] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 23.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (A1 AF
= 1-A2 AF). For
example, the SNP rs951774 has the following case and control allele
frequencies: case A1 (A) = 0.24;
case A2 (C) = 0.76; control A1 (A) = 0.20; and control A2 (C) = 0.80, where
the nucleotide is provided
in paranthesis. Some SNPs are labeled "untyped" because of failed assays.
108
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
TABLE 23
dbSNP Position ChromosomeAllA2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 4
rs884517 207 102527857C/T
rs1476984 6019 102533669A/G 0.83 0.83 0.973
rs951774 6414 102534064A/C 0.76 0.80 0.099
rs2041737 7341 102534991A/G 0.38 0.32 0.146
rs1420091 10984 102538634A/G 0.33 0.35 0.388
rs2110660 12351 102540001C/G 0.41 0.40 0.753
rs1362347 13335 102540985A/G 0.83 0.83 0.895
/TGTG/T
rs3073968 16584 102544234GTGAG 0.48 0.48 0.878
rs4090473 16737 102544387C/G 0.42 0.43 0.633
rs1558622 23897 102551547C/T 0.40 0.39 0.879
rs1558621 24057 102551707C/T 0.32 0.31 0.795
rs1558620 25145 102552795A/G 0.37 0.37 0.998
rs1558619 25300 102552950A/C 0.46 0.47 0.556
rs950881 26262 102553912A/C 0.75 0.74 0.636
rs950880 26312 102553962G/T 0.45 0.48 0.285
rs1362346 26589 102554239C/T
rs1968171 27302 102554952A/G 0.43 0.43 0.891
rs1813299 27358 102555008A/T
rs1813298 27451 102555101C/G
rs1968170 27552 102555202C/T 0.65 0.65 0.941
rs974389 30731 102558381C/T 0.41 0.42 0.734
rs971764 32085 102559735A/G 0.45 0.44 0.738
rs1420089 32139 102559789A/G 0.16 0.19 0.099
rs1420088 33184 102560834A/G 0.41 0.40 0.869
rs1420103 42382 102570032G/T 0.68 0.68 0.952
rs 1420102 42569 102570219A/G 0.48 0.46 0.349
rs1997467 44823 102572473C/T
rs1997466 45217 102572867C/G 0.46 0.46 0.693
rs1362350 45548 102573198C/G 0.48 0.46 0.475
rs2310220 45601 102573251A/G 0.40 0.41 0.480
rs1362349 45722 102573372C/G 0.41 0.42 0.893
rs3755278 45967 102573617A/G 0.07 0.08 0.876
rs3771180 47367 102575017A/C 0.91 0.90 0.669
rs3771179 47642 102575292A/C 0.08 0.08 0.986
rs985523 48126 102575776C/T 0.17 0.13 0.064
rs1041973 49218 102576868A/C
rs3214363 49274 102576924-/A
rs873022 49433 102577083G/T 0.53 0.56 0.321
rs3771177 49610 102577260A/C 0.33 0.31 0.278
rs3732129 51282 102578932A/G 0.46 0.50 0.127
rs1420101 51466 102579116A/G 0.55 0.57 0.257
rs12905 53757 102581407A/G 0.30 0.27 0.262
rs3771175 53960 102581610A/T 0.84 0.82 0.174
rs3821204 54031 102581681C/G 0.26 0.23 0.222
rs2160203 54574 102582224C/T 0.21 0.26 0.033
rs1946131 55679 102583329A/G 0.73 0.74 0.710
rs1054096 56100 102583750C/T 0.69 0.65 0.137
rs2287038 56182 102583832C/T 0.98 0.95 0.207
rs1921622 59817 102587467A/G 0.40 0.43 0.218
rs1861246 60533 102588183A/G 0.22 0.18 0.068
rs1861245 60656 102588306A/G 0.35 0.37 0.377
rs3755276 72209 102599859A/G 0.51 0.48 0.355
rs2287037 72778 102600428A/G 0.49 0.53 0.195
rs1420099 74293 102601943C/G 0.58 0.56 0.416
rs3771174 77335 102604985A/G
rs1420098 78029 102605679A/G 0.33 0.32 0.532
rs1362348 78374 102606024C/G 0.02 0.03 0.590
109
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP ~ PositionChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 4
rs1882348 78421 102606071A/T 0.36 0.35 0.596
rs1558627 78434 102606084C/T 0.62 0.65 0.219
rs2058622 79174 102606824C/T 0.57 0.59 0.528
rs3836110 79397 102607047-/G 0.72 0.73 0.856
rs3771172 79562 102607212A/G 0.28 0.25 0.261
rs3771171 79700 102607350A/G
rs3771170 79730 102607380A/T 0.24 0.23 0.533
rs2160202 79904 102607554C/T 0.55 0.62 0.061
rs2058623 79920 102607570A/G 0.67 0.68 0.631
rs3771167 79938 102607588C/T
rs3771166 79972 102607622C/T 0.55 0.53 0.624
rs1974675 80125 102607775C/T 0.57 0.55 0.470
rs1465321 80368 102608018A/G 0.27 0.26 0.614
rs2041740 83484 102611134C/T 0.26 0.25 0.622
rs3771164 85536 102613186A/T 0.76 0.73 0.197
rs2270298 85829 102613479C/T 0.23 0.21 0.329
rs2270297 86425 102614075A/G 0.60 0.60 0.900
rs2041739 88083 102615733A/G 0.43 0.40 0.235
rs2080289 88770 102616420C/T 0.56 0.59 0.322
rs3821203 90622 102618272A/G 0.58 0.62 0.194
rs3771162 90924 102618574A/T 0.30 0.28 0.260
rs3213733 91634 102619284G/T 0.76 0.73 0.287
rs3213732 92029 102619679C/T 0.44 0.42 0.507
rs1035130 95152 102622802A/G 0.58 0.61 0.234
rs3752659 95348 102622998C/T 0.80 0.80 0.957
rs3755274 96145 102623795C/T 0.26 0.25 0.549
rs2241117 96793 102624443A/G 0.71 0.75 0.077
rs2241116 97015 102624665G/T 0.16 0.15 0.469
rs881890 97064 102624714C/T
rs3771161 97711 102625361G/T 0.70 0.68 0.348
rs3771160 97855 102625505A/C
rs3771159 98708 102626358A/G 0.38 0.40 0.294
rs1420104 not ma not ma C/T
ed ed
rs2041738 not ma not ma A/C
ed ed
(0269] The ILIRLl proximal SNPs were also allelotyped in the replication
cohorts using the
methods described herein and the primers provided in Tables 21 and 22. The
replication allelotyping
results for replication cohort #1 and replication cohort #2 are provided in
Tables 24 and 25,
respectively.
TABLE 24
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position AlleleCase Control Value
SEQ ID AF AF
NO: 4
rs884517 207 102527857 C/T
rs14769846019 102533669 A/G 0.82 0.81 0.878
rs951774 6414 102534064 A/C 0.76 0.82 0.024
rs20417377341 102534991 A/G 0.36 0.32 0.382
rs142009110984 102538634 A/G 0.32 0.35 0.340
rs211066012351 102540001 C/G 0.39 0.39 0.951
rs136234713335 102540985 A/G 0.83 0.83 0.766
/TGTG/T
rs307396816584 102544234 GTGAG 0.47 0.48 0.822
rs409047316737 102544387 C/G 0.40 0.41 0.663
rs155862223897 102551547 C/T 0.38 0.38 0.943
rs155862124057 102551707 C/T 0.33 0.32 0.631
rs155862025145 102552795 A/G 0.34 0.34 0.957
rs155861925300 102552950 A/C 0.44 0.47 0.368
110
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position _AlleleCase Control Value
SEQ ID NO: AF AF
4
rs950881 26262 102553912A/C 0.76 0.74 0.476
rs950880 26312 102553962G/T 0.42 0.47 0.199
rs136234626589 102554239C/T
rs196817127302 102554952A/G 0.43 0.44 0.641
rs181329927358 102555008A/T
rs181329827451 102555101C/G
rs196817027552 102555202C/T 0.64 0.65 0.797
rs974389 30731 102558381C/T 0.39 0.41 0.678
rs971764 32085 102559735A/G 0.47 0.46 0.710
rs142008932139 102559789AIG 0.16 0.21 0.075
rs142008833184 102560834A/G 0.41 0.40 0.869
rs142010342382 102570032G/T 0.69 0.72 0.268
rs142010242569 102570219A/G 0.50 0.47 0.329
rs199746744823 102572473C/T
rs199746645217 102572867C/G 0.49 0.47 0.675
rs136235045548 102573198C/G 0.51 0.47 0.308
rs231022045601 102573251A/G 0.40 0.44 0.282
rs136234945722 102573372C/G 0.42 0.43 0.730
rs375527845967 102573617A/G 0.08 0.08 0.902
rs377118047367 102575017A/C 0.93 0.92 0.591
rs377117947642 102575292A/C 0.08 0.08 0.936
rs985523 48126 102575776C/T 0.17 0.13 0.156
rs104197349218 102576868A/C
rs321436349274 102576924-/A
rs873022 49433 102577083G/T 0.51 0.56 0.138
rs377117749610 102577260A/C 0.36 0.32 0.125
rs373212951282 102578932A/G 0.43 0.50 0.048
rs142010151466 102579116A/G 0.50 0.55 0.132
rs12905 53757 102581407A/G 0.33 0.28 0.127
rs377117553960 102581610A/T 0.86 0.83 0.217
rs382120454031 102581681C/G 0.29 0.23 0.071
rs216020354574 102582224C/T 0.19 0.26 0.016
rs194613155679 102583329A/G 0.72 0.73 0.771
rs105409656100 102583750C/T 0.70 0.65 0.079
rs228703856182 102583832C/T 0.93 NA 0.975
rs192162259817 102587467AlG 0.37 0.41 0.260
rs186124660533 102588183A/G 0.22 0.15 0.031
rs186124560656 102588306A/G 0.34 0.39 0.149
rs375527672209 102599859A/G 0.53 0.46 0.072
rs228703772778 102600428A/G 0.45 0.51 0.069
rs142009974293 102601943C/G 0.59 0.55 0.312
rs377117477335 102604985A/G
rs142009878029 102605679A/G 0.35 0.32 0.328
rs136234878374 102606024C/G 0.02 NA 0.025
rs188234878421 102606071A/T 0.40 0.37 0.399
rs155862778434 102606084C/T 0.64 0.69 0.118
rs205862279174 102606824C/T 0.59 0.62 0.491
rs383611079397 102607047-/G 0.74 0.75 0.625
rs377117279562 102607212A/G 0.31 0.27 0.200
rs377117179700 102607350A/G
rs377117079730 102607380A/T 0.22 0.20 0.346
rs216020279904 102607554C/T 0.55 0.60 0.217
rs205862379920 102607570A/G 0.69 0.72 0.266
rs377116779938 102607588C/T
rs377116679972 102607622C/T 0.57 unt ed NA
rs197467580125 102607775C/T 0.58 0.54 0.297
rs146532180368 102608018A/G 0.25 0.23 0.471
rs204174083484 102611134C/T 0.25 0.22 0.450
rs377116485536 102613186A/T 0.77 0.72 0.073
rs227029885829 102613479C/T 0.25 0.22 0.324
rs227029786425 102614075A/G 0.63 0.64 0.589
rs204173988083 102615733A/G 0.44 0.40 0.157
rs208028988770 102616420C/T 0.53 0.58 0.114
rs382120390622 102618272A/G 0.55 0.61 0.104
111
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position AlleleCase Control Value
SEQ ID AF AF
NO: 4
rs377116290924 102618574 A/T 0.34 0.29 0.261
rs321373391634 102619284 G/T 0.77 0.73 0.260
rs321373292029 102619679 C/T 0.48 0.41 0.026
rs103513095152 102622802 A/G 0.55 0.60 0.152
rs375265995348 102622998 C/T 0.81 0.80 0.760
rs375527496145 102623795 C/T 0.25 0.22 0.319
rs224111796793 102624443 A/G 0.72 0.80 0.024
rs224111697015 102624665 G/T 0.18 NA NA
rs881890 97064 102624714 C/T
rs377116197711 102625361 G/T 0.71 0.66 0.146
rs377116097855 102625505 A/C
rs377115998708 102626358 A/G 0.38 0.42 0.175
rs1420104not ma not ma C/T
ed ed
rs2041738not ma not ma A/C
ed ed
TABLE 25
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 4
rs884517 207 102527857C/T
rs14769846019 102533669A/G 0.84 0.85 0.759
rs951774 6414 102534064A/C 0.77 0.75 0.730
rs20417377341 102534991A/G 0.40 NA
rs142009110984 102538634A/G 0.34 0.35 0.819
rs211066012351 102540001C/G 0.43 0.42 0.665
rs136234713335 102540985A/G 0.82 0.83 0.576
/TGTG/T
rs307396816584 102544234GTGAG 0.49 0.49 0.997
rs409047316737 102544387C/G 0.44 0.45 0.732
rs155862223897 102551547C/T 0.42 0.42 0.976
rs155862124057 102551707C/T 0.31 0.31 0.926
rs155862025145 102552795A/G 0.42 0.42 0.867
rs155861925300 102552950A/C 0.48 0.48 0.930
rs950881 26262 102553912A/C 0.73 0.73 0.955
rs950880 26312 102553962G/T 0.48 0.49 0.919
rs136234626589 102554239C/T
rs196817127302 102554952A/G 0.43 0.42 0.717
rs181329927358 102555008A/T
rs181329827451 102555101C/G
rs196817027552 102555202C/T 0.67 0.66 0.830
rs974389 30731 102558381C/T 0.44 0.45 0.857
rs971764 32085 102559735A/G 0.43 0.42 0.845
rs142008932139 102559789A/G 0.15 0.16 0.809
rs142008833184 102560834A/G
rs142010342382 102570032G/T 0.68 0.63 0.178
rs142010242569 102570219A/G 0.45 0.44 0.722
rs199746744823 102572473C/T
rs199746645217 102572867C/G 0.44 0.43 0.805
rs136235045548 102573198ClG 0.45 0.46 0.890
rs231022045601 102573251A/G 0.38 0.37 0.661
rs136234945722 102573372C/G 0.41 0.40 0.762
rs375527845967 102573617A/G 0.07 0.07 0.984
rs377118047367 102575017A/C 0.88 0.87 0.812
rs377117947642 102575292A/C 0.07 0.07 0.868
rs985523 48126 102575776C/T 0.16 0.13 0.270
rs104197349218 102576868A/C
rs321436349274 102576924-/A
rs873022 49433 102577083G/T 0.57 0.56 0.868
rs377117749610 102577260A/C 0.29 0.29 0.988
rs373212951282 102578932A/G 0.51 0.52 0.795
rs142010151466 102579116A/G 0.60 0.61 0.864
rs12905 53757 102581407A/G 0.26 0.26 0.994
112
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 4
rs377117553960 102581610A/T 0.82 0.80 0.444
rs382120454031 102581681C/G 0.22 0.23 0.769
rs216020354574 102582224C/T 0.23 0.25 0.732
rs194613155679 102583329A/G 0.75 0.77 0.723
rs105409656100 102583750C/T 0.66 0.65 0.807
rs228703856182 102583832C/T 0.97 0.00
rs192162259817 102587467A/G 0.44 0.46 0.472
rs186124660533 102588183AIG 0.23 0.24 0.824
rs186124560656 102588306A/G 0.36 0.34 0.590
rs375527672209 102599859A/G 0.48 0.51 0.423
rs228703772778 102600428A/G 0.55 0.54 0.941
rs142009974293 102601943C/G 0.58 0.58 0.904
rs377117477335 102604985A/G
rs142009878029 102605679A/G 0.30 0.31 0.827
rs136234878374 102606024C/G 0.04 -0.01
rs188234878421 102606071A/T 0.31 0.31 0.968
rs155862778434 102606084C/T 0.60 0.59 0.839
rs205862279174 102606824C/T 0.56 0.55 0.961
rs383611079397 102607047-/G 0.70 0.69 0.643
rs377117279562 102607212A/G 0.24 0.22 0.675
rs377117179700 102607350A/G
rs377117079730 102607380A/T 0.26 0.27 0.700
rs216020279904 102607554C/T unt 0.64 NA
ed
rs205862379920 102607570A/G 0.65 0.62 0.389
rs377116779938 102607588C/T
rs377116679972 102607622C/T 0.53 0.53 0.820
rs197467580125 102607775C/T 0.55 0.56 0.842
rs146532180368 102608018A/G 0.29 0.30 0.781
rs204174083484 102611134C/T 0.28 0.30 0.658
rs377116485536 102613186A/T 0.73 0.74 0.905
rs227029885829 102613479C/T 0.19 0.18 0.654
rs227029786425 102614075A/G 0.57 0.53 0.249
rs204173988083 102615733A/G 0.42 0.41 0.892
rs208028988770 102616420C/T 0.61 0.60 0.840
rs382120390622 102618272A/G 0.62 0.63 0.927
rs377116290924 102618574A/T 0.26 0.25 0.621
rs321373391634 102619284G/T 0.75 0.74 0.728
rs321373292029 102619679C/T 0.39 0.45 0.176
rs103513095152 102622802A/G 0.62 0.63 0.792
rs375265995348 102622998C/T 0.79 0.80 0.826
rs375527496145 102623795C/T 0.27 0.29 0.618
rs224111796793 102624443A/G 0.70 0.67 0.480
rs224111697015 102624665G/T 0.15 0.15 0.849
rs881890 97064 102624714C/T
rs377116197711 102625361G/T 0.68 0.70 0.681
rs377116097855 102625505A/C
rs377115998708 102626358A/G 0.37 0.37 0.970
rs1420104not ma not ma C/T
ed ed
rs2041738not ma not ma A/C
ed ed
[0270] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure 1 C for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in Figure
1 C can be determined by consulting Table 23. For example, the left-most X on
the left graph is at
113
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
position 102527857. By proceeding down the Table from top to bottom and across
the graphs from left
to right the allele frequency associated with each symbol shown can be
determined.
[0271] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a l Okb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10'8 were truncated at that
value.
[0272] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 7
WASPIP Region Proximal SNPs
(0273] It has been discovered that rs1465621 in the untranslated region (LJTR)
of the WASPIP gene
is associated with occurrence of osteoarthritis in subjects. This gene encodes
a protein that plays a role
in actin cytoskeleton organization. The encoded protein binds to a region of
Wiskott-Aldrich syndrome
protein that is frequently mutated in Wiskott-Aldrich syndrome, an X-linked
recessive disorder.
Impairment of the interaction between these two proteins may contribute to the
disease. Alternative
transcript variants exist for this gene. Biological activity of WASPIP or a
pathway member downstream
of WASPIP (e.g., IL-2) can be modulated by addition of an antibody, a
recombinant binding partner, a
binding agent, or a recombinant WASPIP or downstream pathway member protein or
functional
fragment thereof.
[0274] Sixty-one additional allelic variants proximal to rs1465621 were
identified and
subsequently allelotyped in osteoarthritis case and control sample sets as
described in Examples 1 and 2.
The polymorphic variants are set forth in Table 26_ The chromosome positions
provided in column four
of Table 26 are based on Genome "Build 34" of NCBI's GenBank.
114
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
TABLE 26
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
5
rs18644552 209 175603909 C/T
rs19717632 5908 175609608 C/T
rs934269 2 7460 175611160 A/G
rs934270 2 7733 175611433 A/G
rs20333092 7855 175611555 A/G
rs20333102 7904 175611604 A/C
rs934271 2 8869 175612569 G/T
rs934272 2 9480 175613180 C/T
rs18971102 13820 175617520 C/T
rs20333112 15152 175618852 A/G
rs10100272 17713 175621413 A/G
rs10100282 17804 175621504 C/T
rs28845022 18220 175621920 C/T
rs14301772 19083 175622783 C/T
rs14301782 19123 175622823 C/G
rs30437792 19605 175623305 -/GTAAA
rs15497422 20247 175623947 G/T
rs30437812 20592 175624292 -/CCCCC
rs20333132 21907 175625607 C/T
rs7739 2 23273 175626973 C/T
rs11482 2 23299 175626999 A/C
rs30879072 23623 175627323 G/T
rs23588882 23669 175627369 A/T
rs10460362 23844 175627544 A/T
rs32050602 24190 175627890 A/G
rs15327 2 24486 175628186 C/T
rs14301792 24896 175628596 A/C
rs14301802 25118 175628818 C/G
rs21632362 30551 175634251 C/G
rs32173512 30844 175634544 -/GAGA
rs23038912 30900 175634600 A/G
rs38159692 30942 175634642 AlG
rs22886222 31699 175635399 A/G
rs22886232 32081 175635781 G/T
rs10443352 35078 175638778 A/G
rs22886242 36196 175639896 A/T
rs10605112 36541 175640241 A/C
rs13672182 38356 175642056 A/G
rs13672172 45578 175649278 A/G
rs14656212 49634 175653334 A/T
rs14656222 49774 175653474 G/T
rs21158722 51119 175654819 AlG
rs14656232 51181 175654881 A/G
rs14695212 51652 175655352 C/T
rs18644512 54467 175658167 C/G
rs14301832 55762 175659462 A/G
rs14301822 55999 175659699 A/G
rs14301812 57865 175661565 A/C
115
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
5
rs19916012 66613 175670313 A/G
rs23588902 68377 175672077 C/T
rs21158752 69754 175673454 C/T
rs14301852 72859 175676559 A/G
rs22174292 76512 175680212 A/G
rs30499092 76717 175680417 -/AT
rs14301842 77722 175681422 ClT
rs22783212 80998 175684698 A/G
rs21158742 82033 175685733 C/T
rs20333152 89658 175693358 C/T
rs20333142 89960 175693660 A/G
rs19916002 94155 175697855 A/G
rs18644532 95679 175699379 A/G
Assay for Verifying and Allelotypin~ SNPs
(0275] The methods used to verify and allelotype the 61 proximal SNPs of Table
26 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 27 and Table 28, respectively.
TABLE 27
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1864455ACGTTGGATGACAGGTGTGCAGTGAATGTCACGTTG GATGTCAGCAGTTGTCCCATCTTC
rs1971 ACGTTGGATGAATGATTTACTTGAGGCCGGACGTTG GATGTCTCAAACTCCTGACCTCTG
763
rs934269ACGTTGGATGAAGTCCCTAGGACTACAGGTACGTTG GATGTGGGCAACATAGCAAGACCC
rs934270ACGTTGGATGATGATCTGCCCTGTTCTTGCACGTTG GATGAGGTGCAATCTACTCACCAG
rs2033 ACGTTGGATGCCATAGCTTCCTCACACAACACGTTG GATGTTCTCCTTGCAGACAAGGTG
309
rs2033 ACGTTGGATGATGAGTCTCTGTGAGTTGAGACGTTG GATGTTGTGTGAGGAAGCTATGGC
310
rs934271ACGTTGGATGCCTGAAATGCCAAGAAGAATGACGTTG GATGATTCTTGCTACATAGTCAGG
rs934272ACGTTGGATGAGTCTTGCTTCTCTTCACACACGTTG GATGACTAAGAGGTATTTGGGTGC
rs1897110ACGTTGGATGTCAGCATCCCAAAGTGCTAGACGTTG GATGTAAAAATCGGCTGGGTGTGG
rs2033 ACGTTGGATGCGGGACTCTGTGTTAACAAGACGTTG GATGGAGTTACAAGATGCTGGAGC
311
rs1010 ACGTTGGATGGCCGTCTCTGTTGTGAGAAGACGTTGGATGAATTCCTCTCTGACTCTTTC
027
rs1010 ACGTTGGATGGTAACTTAAGGCCTCACAGCACGTTGGATGGACTGAAAGAGTCAGAGAGG
028
rs2884502ACGTTGGATGGAAATCCCCATGTCAGAATCACGTTG GATGTGAACAGTACAAAGGAAGGG
rs1430177ACGTTGGATGGCCAGACCCTGTCTCAAATAACGTTG GATGTGAGTAGCTAGGAGTATAGG
rs1430178ACGTTGGATGTATTTGAGACAGGGTCTGGCACGTTG GATGTGAGCCCTGGAATTCAAGAC
rs3043 ACGTTGGATGAGTTCCTCAACTACTGTTTGACGTTG GATGCCCACATGATTTAATGGAGC
779
rs1549742ACGTTGGATGTGAGACACTGTGCCTAGCTGACGTTG GATGGGTCCAGGTTTTGTGATGTC
rs3043781ACGTTGGATGATAATAAATAGTTAGAAGCCACGTTGGATGAGAAGCTAATTAAGCTCAAG
rs2033 ACGTTGGATGAAGCCGTGCACTCACAAATCACGTTG GATGACCACCTACAAAGCTTCTGG
313
rs77 ACGTTGGATGTGATGACACAGATAGCAAAATGTGACGTTGGATGTTCCCTCCTTATAGTCAAGGACC
39
rs11482 ACGTTGGATGAAATGTTGGCATGAAATTAATTTTACGTTGGATGTGTGTCTGTTTACATAGTGCATG
rs3087907ACGTTGGATGGAACACTGAGTTTTAATACTGACGTTGGATGAATCAGAGCTTACATGTGTG
rs2358888ACGTTGGATGAATCAGAGCTTACATGTGTGACGTTG GATGGAGGTGAATGTTAAAATACTG
rs1046036ACGTTGGATGCAAAGTTGCCATTCATCCAGACGTTGGATGAGGGTGTAGGTGTATTAATG
rs3205060ACGTTGGATGAAGCCAACACTTTGCCAAGCACGTTGGATGTCCTCTCTCCTCTACCATTC
116
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs15327 ACGTTGGATGGGGTTGGTrTCTTGGTAGCAACGTTGGATGCCTAAACATfGTATCATGGTITCA
rs1430179ACGTTGGATGAGACTAGGAAGGCTTGGTAGACGTTGGATGGGTTCCCTTCTfCTfCCATG
rs1430180ACGTTGGATGCTfCAAAGTACCAAGGTCAGACGTTGGATGCAGGCTITCCATfTGTTTCC
rs2163236ACGTTGGATGTTGAGTAGCCTGAGTGACACACGTTGGATGTAGATGGCTCCAAAGGGTfC
rs3217351ACGTTGGATGGTAACGAAAGGCACAGAATGACGTTGGATGTAGCACTTCCAGCTTTTCTG
rs2303891ACGTTGGATGACCACAGACATCAGTGCTAGACGTTGGATGCAGTGTACTAATTCGTGACC
rs3815969ACGTTGGATGGAAGTGCTACAAAGGTCACGACGTTGGATGGCTGGATCCTAATCACTCTC
rs2288622ACGTTGGATGGGCCTGGAGCAAAAAAAGACACGTTGGATGCATCAGCTGTACACCAATGG
rs2288623ACGTTGGATGGAAATTATTTfAGGTCTTCAGACGTi'GGATGTATACATCACAGAAACATGC
rs1044335ACGTTGGATGCTACTCAGTGTCCTCATCTCACGTfGGATGTTfAAGTGGCACACGACACG
rs2288624ACGTrGGATGCATAGGCTGTAGAAGTTGGGACGTTGGATGT'TGTTGGTCCTTCTTGGGAG
rs1060511ACGTTGGATGTCCCTATGAAGAGAAATGCCACGTTGGATGCTGATGGTTCTTIT1'CCTTTC
rs1367218ACGTTGGATGTTGTGAGCCGCTTTTCAAACACGTTGGATGCATGCAAAACACTTTfTCAG
rs1367217ACGTTG GATGGAGCTGTAACTAAAAAGGGTGACGTTGGATGGTGTATATTGCCAAAGATGC
rs1465621ACGTrGGATGTTCTCCTCCCATTCTTCCTGACGTTGGATGG CGGGACTAGAAGTAGATTC
rs1465622ACGTTGGATGGGTCTTfGAGTGCTCCAAACACGTTGGATGAGAATGTCAGGTGGAAAGCA
rs2115872ACGTTGGATGTAGACCGCCCACTTfGAATGACGTrGGATGAAGACACTGCTGGACTTGTC
rs1465623ACGTTGGATGGGATCCAGCAGATTCTCCATACGTTGGATGAGTGGGCGGTCTAGAAAATG
rs1469521ACGTTGGATGTGGTCCTAGGAGACGTCTGAACGTTGGATGAGGACTGGGTGCCTGTGTTA
rs1864451ACGTfGGATGCTGTATGTGAAAACAAAAGCCACGTTGGATGTCTTTACTfGGTGTGTTGAC
rs1430183ACGT'fGGATGCATGTCATTCTGTAGTGTGGACGTTGGATGTCCTTGGGATCAAGAAAGTG
rs1430182ACGTfGGATGAATGTI'GCTAAAAGTAACCCACGTTGGATGATCTTfTTGGGGAAAAGAAG
rs1430181ACGTTGGATGAAGCTCCTAGCCAGTCTfAGACGTTGGATGTTATTTTGGCGGGGAGTAGG
rs1991601ACGTTGGATGATCCTCAACAGATCTGGTTCACGTTGGATGTCTGGTGATGGCTTGTGATC
rs2358890ACGTTGGATGTCAGAGTAGAGTTACTCCAGACGTfGGATGCATGATGCAGCTATTCTGTG
rs2115875ACGTTGGATGCAGACCCTTTITTCTAGATCACGTTGGATGACTATTfTTGAAGTAGTGTG
rs1430185ACGTfGGATGATCTGAGCCTAGACCTfAACACGTfGGATGGGGAATGAATACAACAGTGC
rs2217429ACGTTGGATGTGCACAAAATTAGCCACAGCACGTfGGATGAGTGACCGTTTCTGTGTGTT
rs3049909ACGTfGGATGCAAAAGCAGGAATGCCTTGGACGTfGGATGGGGTCACAACTGCTGTTITC
rs1430184ACGTt-GGATGAATTAGCAATGGCTCTCTCCACGTTGGATGCCTAAAAACACAGTTGCTCC
rs2278321ACGTTGGATGCAGACAGCAGGTAGATGAACACGTTGGATGTCTGGAAAAGAGAGACAGCC
rs2115874ACGTTGGATGCAGTGGACTfAAGAGAGGAGACGTTGGATGGGTTCAGGTACCTGAAAAGC
rs2033315ACGTTGGATGGTCAAGGTAGTTGAGAGTATTACGTTGGATGCAATGACAAAAAGCAATTTC
rs2033314ACGTTGGATGCATCTfTCTTAATGGTCTTGGACGTTGGATGATGCAGAGTCACATfCCATG
rs1991600ACGTrGGATGTTTCGTCATCAGTCAGAAGGACGTTGGATGCTGGTTCCT1'TTTTTGGGAG
rs1864453ACGT'fGGATGAGATAGGAATGACTGCCAAGACGTTGGATGAGGTGACTTCATCTCTTTCC
TABLE 28
dbSNP Extend Term
rs# Primer Mix
rs1864455 TCCTTTTCTCTCAGTTCCCC ACT
rs1971763 AGCACTTTGGGAGGCCAAGG ACG
rs934269 GCACGCCACCACACTCGG ACG
rs934270 CCCTGTTCTTGCTCCTGCTfCTT ACT
rs2033309 ACAACACAAAGAAGGGTfGTTA ACG
rs2033310 GGGTGGGAAATCTGCTGAG ACT
rs934271 GCATAATTTTTCAGGGAGGCAG ACT
rs934272 TGCTTCTCTTCACACTTATAAG ACG
rs1897110 GCATCCCAAAGTGCTAGGATTACAI ACT
117
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs203331 CTTCCAGGAGGTGCGATGAG ACT
'I
rs1010027 TCTGTTGTGAGAAGATGCGC ACT
rs1010028 ACAGCTGTTGGGCTCACAG ACT
rs2884502 TGCCTAGTTAATTTGCTTTCCT ACT
rs1430177 CCCTGTCTCAAATAAATTTTAAAAACT
rs1430178 GACAGGGTCTGGCTATGTTGTC ACT
rs3043779 ACTGTTTGTTGATGATTTGAATAAACT
rs1549742 GCCTAGCTGGGGCTTCAAGTTA CGT
rs3043781 TAGAAGCCAACCCCCCCC ACT
rs2033313 CCCTGTGAGGCCATAGACAA ACT
rs7739 CTGTTTACATAGTGCATG ACT
rs11482 CTTATAGTCAAGGACCGT CGT
rs3087907 CAATATAAAATAAGAGGTGAATGTACT
rs2358888 GCTTACATGTGTGTTTTTT CGT
rs1046036 CATTCATCCAGAATAGATTGTTTTCGT
rs3205060 TTTGCCAAGCTTGTTATA ACG
rs15327 GGTAGCATCTCCCAGTAA ACG
rs1430179 GAGGGGAAAAAAGTCAGGAAAA ACT
rs1430180 AAGTACCAAGGTCAGAAATTGATTACT
rs2163236 AGTCCAGGCTTCTTGCCTG ACT
rs3217351 AGGCACAGAATGAAAGAGAGA ACT
rs2303891 TAGAAGTTTACAGAAAAGCTGGAAACT
rs3815969 TTAGTACACTGACATATATACAG ACT
rs2288622 CTTACATCCACATTCCATTACC ACT
rs2288623 TTTTAGGTCTTCAGAAGAACAAAGACT
rs1044335 GAAATATTGGTCCCACTTTCC ACG
rs2288624 GACTCGCAGGTAAATAGAGCT CGT
rs1060511 CCCAAAAAAAGTGGAAAA CGT
rs1367218 CTTTTCAAACACGATGGAGCAC ACT
rs1367217 AACTAAAAAGGGTGATTTCACTATACT
rs1465621 CCATTCTTCCTGACATTCGCC CGT
rs1465622 CAAACATAAGGTTGACCCCC CGT
rs2115872 TTTGAATGGGACTCTTCC ACT
rs1465623 TCCATACATGAGAGCTGCTG ACG
rs1469521 TAGGAGACGTCTGACTCCAA ACT
rs1864451 GAAAACAAAAGCCTTTTCTGTC ACT
rs1430183 ATTCTGTAGTGTGGGCCCTA ACT
rs1430182 GTAACCCTTAAATACTATCATAC ACG
rs1430181 CTAGCCAGTCTTAGTGATGTT CGT
rs1991601 AGCTCGCCTCAGCCTACAA ACT
rs2358890 GTCCAGAACACCATAATCCC ACT
rs2115875 TTTTTTCTAGATCAGCACTGTTCAACT
rs1430185 CTAGACCTTAACTCCAATTTATA ACG
rs2217429 AGTCCTTGGTTTATGAACATTTG ACT
rs3049909 TTTATGTTATGCACATGCAGAC ACT
rs1430184 CATAAAACCAACTTATTAATCCC ACG
rs2278321 GCTCACAGGCTTTGTAACATC ACT
rs2115874 GGGGAGATCTGCCATCTCCTGG ACT
rs20333~ GGTAGTTGAGAGTATTGTGAGA I ACG
118
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs2033314 GTCTTGGTTTAATATCACTCCT ACT
rs1991600 TAAAGGGGAAAAAAAAGCTCTAA ACT
rs1864453 CTGCCAAGTTGAATACTGAGTT ACT
Genetic Anal
[0276] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 29.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A 1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (A1 AF
= 1-A2 AF). For
example, the SNP rs1971763 has the following case and control allele
frequencies: case A1 (C~ = 0.456;
case A2 (T) = 0.544; control A1 (C) = 0.444-; and control A2 (T) = 0.556,
where the nucleotide is
provided in paranthesis. Some SNPs are labeled "untyped" because of failed
assays.
TABLE 29
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 5
rs1864455 209 175603909C/T
rs1971763 5908 175609608C/T 0.544 0.556 0.630
rs934269 7460 175611160A/G
rs934270 7733 175611433A/G
rs2033309 7855 175611555A/G 0.158 0.172 0.502
rs2033310 7904 175611604A/C 0.428 0.423 0.845
rs934271 8869 175612569G/T
rs934272 9480 175613180C/T
rs1897110 13820 175617520C/T
rs2033311 15152 175618852A/G
rs1010027 17713 175621413A/G
rs1010028 17804 175621504C/T 0.448 0.449 0.965
rs2884502 18220 175621920C/T
rs1430177 19083 175622783C/T 0.051 0.309 0.0001
rs1430178 19123 175622823C/G
rs3043779 19605 175623305-/GTAAA
rs1549742 20247 175623947G/T
rs3043781 20592 175624292-ICCGCC
rs2033313 21907 175625607C/T
rs7739 23273 175626973C/T 0.057 0.042 0.371
rs11482 23299 175626999A/C 0.934 0.935 0.958
rs3087907 23623 175627323G/T 0.427 0.425 0.918
rs2358888 23669 175627369A/T 0.083 0.064 0.245
rs1046036 23844 175627544A/T
rs3205060 24190 175627890A/G 0.478 0.483 0.859
rs15327 24486 175628186C/T 0.901 0.917 0.336
rs1430179 24896 175628596A/C
rs1430180 25118 175628818C/G
rs2163236 30551 175634251C/G 0.956 0.955 0.994
rs3217351 30844 175634544-/GAGA 0.481 0.487 0.823
rs2303891 30900 175634600A/G 0.750 0.687 0.006
rs3815969 30942 175634642A/G 0.232 0.239 0.771
rs2288622 31699 175635399A/G 0.863 0.828 0.082
rs2288623 32081 175635781G/T 0.081 0.106 0.134
rs1044335 35078 175638778A/G 0.105 0.115 0.550
rs2288624 36196 175639896A/T 0.901 0.871 0.117
rs1060511 36541 175640241A/C 0.968 0.979 0.413
119
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 5
rs1367218 38356 175642056A/G 0.931 0.958 0.068
rs1367217 45578 175649278A/G 0.027 0.020 0.648
rs1465621 49634 175653334A/T
rs1465622 49774 175653474G/T 0.084 0.108 0.161
rs2115872 51119 175654819A/G 0.483 0.500 0.500
rs1465623 51181 175654881A/G
rs1469521 51652 175655352C/T 0.433 0.435 0.953
rs 186445154467 175658167C/G 0.316 0.315 0.970
rs1430183 55762 175659462A/G 0.972 0.970 0.930
rs1430182 55999 175659699A/G 0.711 0.691 0.366
rs1430181 57865 175661565A/C 0.939 0.943 0.836
rs1991601 66613 175670313A/G 0.754 0.713 0.062
rs2358890 68377 175672077C/T 0.404 0.443 0.109
rs2115875 69754 175673454C/T 0.633 0.620 0.613
rs 143018572859 175676559A/G 0.768 0.750 0.445
rs2217429 76512 175680212A/G 0.428 0.489 0.028
rs3049909 76717 175680417-/AT 0.161 0.200 0.064
rs1430184 77722 175681422C/T 0.025 unt ed NA
rs2278321 80998 175684698A/G
rs2115874 82033 175685733C/T 0.729 0.698 0.179
rs2033315 89658 175693358C/T 0.649 0.663 0.542
rs2033314 89960 175693660A/G 0.697 0.692 0.835
rs1991600 94155 175697855A/G 0.526 0.576 0.048
rs1864453 95679 175699379A/G 0.675 0.672 0.883
[0277] The WASPIP proximal SNPs were also allelotyped in the replication
cohorts using the
methods described herein and the primers provided in Tables 27 and 28. The
replication allelotyping
results for replication cohort #1 and replication cohort #2 are provided in
Tables 30 and 31,
respectively.
TABLE 30
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in PositionAllele Case Control Value
SEQ ID AF AF
NO: 5
rs1864455209 175603909C/T
rs19717635908 175609608C/T 0.472 0.509 0.276
rs934269 7460 175611160A/G
rs934270 7733 175611433A/G
rs20333097855 175611555A/G 0.179 0.186 0.784
rs20333107904 175611604A/C 0.428 0.405 0.493
rs934271 8869 175612569G/T
rs934272 9480 175613180C/T
rs189711013820 175617520C/T
rs203331115152 175618852A/G
rs101002717713 175621413A/G
rs101002817804 175621504C/T 0.447 0.465 0.579
rs288450218220 175621920C/T
rs143017719083 175622783C/T 0.051 0.098 0.138
rs143017819123 175622823C/G
rs304377919605 175623305/GTAAA
rs154974220247 175623947G/T
rs304378120592 175624292/CCCCC
rs203331321907 175625607C/T
rs7739 23273 175626973C/T 0.076 0.053 0.342
rs11482 23299 175626999A/C 0.919 0.919 0.996
rs308790723623 175627323G/T 0.422 0.390 0.348
rs235888823669 175627369A/T 0.104 0.074 0.204
rs104603623844 175627544A/T
120
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position ~111e1eCase Control Value
SEQ ID AF AF
NO: 5
rs3205060 24190 175627890A/G 0.501 0.472 0.391
rs15327 24486 175628186C/T 0.883 0.904 0.370
rs1430179 24896 175628596A/C
rs1430180 25118 175628818C/G
rs2163236 30551 175634251C/G 0.976 unt ed 0.921
rs3217351 30844 175634544-/GAGA 0.514 0.480 0.329
rs2303891 30900 175634600A/G 0.780 0.699 0.009
rs3815969 30942 175634642A/G 0.183 0.213 0.426
rs2288622 31699 175635399A/G 0.856 0.818 0.201
rs2288623 32081 175635781G/T 0.083 0.112 0.216
rs1044335 35078 175638778A/G 0.113 0.115 0.959
rs2288624 36196 175639896A/T 0.908 0.872 0.215
rs1060511 36541 175640241A/C 0.971 unt ed 0.945
rs1367218 38356 175642056A/G 0.952 0.947 0.824
rs1367217 45578 175649278A/G 0.020 unt ed NA
rs1465621 49634 175653334A/T
rs1465622 49774 175653474G/T 0.077 0.118 0.108
rs2115872 51119 175654819A/G 0.493 0.499 0.861
rs1465623 51181 175654881A/G
rs1469521 51652 175655352C/T 0.453 0.427 0.436
rs1864451 54467 175658167C/G 0.302 0.321 0.556
rs1430183 55762 175659462A/G 0.959 0.962 0.903
rs1430182 55999 175659699A/G 0.727 0.678 0.114
rs 143018157865 175661565A/C 0.942 0.940 0.943
rs1991601 66613 175670313A/G 0.773 0.722 0.081
rs2358890 68377 175672077C/T 0.389 0.443 0.111
rs2115875 69754 175673454C/T 0.639 0.601 0.267
rs1430185 72859 175676559A/G 0.790 0.774 0.586
rs22'1742976512 175680212A/G 0.412 0.504 0.029
rs3049909 76717 175680417-/AT 0.144 0.193 0.079
rs1430184 77722 175681422C/T
rs2278321 80998 175684698A/G
rs2115874 82033 175685733C/T 0.744 0.703 0.169
rs2033315 89658 175693358C/T 0.675 0.695 0.533
rs2033314 89960 175693660A/G 0.726 0.703 0.529
rs1991600 94155 175697855A/G 0.467 0.566 0.005
rs1864453 95679 175699379A/G 0.702 0.680 0.468
TALE 31
dbSNP Position Chromosome~1/A2 F A2 F A2 F p-
rs# in positionAllele Case Control Value
SEQ D AF AF
NO:
rs1864455209 175603909C/T
rs19717635908 175609608C/T 0.635 0.629 0.879
rs934269 7460 175611160A/G
rs934270 7733 175611433A/G
rs20333097855 175611555A/G 0.131 0.149 0.576
rs20333107904 175611604A/C 0.428 0.452 0.548
rs934271 8869 175612569G/T
rs934272 9480 175613180C/T
rs189711013820 175617520C/T
rs203331115152 175618852A/G
rs101002717713 175621413A/G
rs101002817804 175621504C/T 0.449 0.424 0.503
rs288450218220 175621920C/T
rs143017719083 175622783C/T unt ed 0.642 NA
rs143017819123 175622823C/G
rs304377919605 175623305-/GTAAA
rs154974220247 175623947G/T
rs304378120592 175624292-ICCCCC
rs203331321907 175625607C/T
121
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP positionChromosomeAl/A2 F A2 F A2 F p-
rs# in position Allele Case Control Value
SEQ D AF AF
NO:
rs7739 23273 175626973C/T 0.032 0.023 0.700
rs 11482 23299 175626999A/C 0.953 0.960 0.743
rs308790723623 175627323G/T 0.435 0.479 0.295
rs235888823669 175627369A/T 0.055 0.048 0.731
rs104603623844 175627544A/T
rs320506024190 175627890A/G 0.449 0.500 0.197
rs15327 24486 175628186C/T 0.923 0.937 0.552
rs143017924896 175628596A/C
rs143018025118 175628818C/G
rs216323630551 175634251C/G 0.923 unt ed
rs321735130844 175634544-/GAGA 0.439 0.496 0.125
rs230389130900 175634600A/G 0.712 0.667 0.208
rs381596930942 175634642A/G 0.294 0.281 0.705
rs228862231699 175635399A/G 0.872 0.843 0.309
rs228862332081 175635781G/T 0.078 0.096 0.444
rs104433535078 175638778A/G 0.094 0.117 0.366
rs228862436196 175639896A/T 0.894 0.869 0.356
rs106051136541 175640241A/C
rs136721838356 175642056A/G 0.903 0.976 0.001
rs136721745578 175649278A/G 0.035 0.021 0.504
rs146562149634 175653334A/T
rs146562249774 175653474G/T 0.092 0.093 0.959
rs211587251119 175654819A/G 0.471 0.502 0.397
rs146562351181 175654881A/G
rs146952151652 175655352C/T 0.408 0.447 0.282
rs186445154467 175658167C/G 0.334 0.306 0.420
rs143018355762 175659462A/G
rs143018255999 175659699A/G 0.691 0.711 0.547
rs143018157865 175661565A/C 0.936 0.946 0.651
rs199160166613 175670313A/G 0.730 0.700 0.372
rs235889068377 175672077C/T 0.423 0.444 0.566
rs211587569754 175673454C/T 0.625 0.650 0.510
rs143018572859 175676559A/G 0.740 0.714 0.462
rs221742976512 175680212A/G 0.447 0.464 0.644
rs304990976717 175680417-/AT 0.184 0.212 0.386
rs143018477722 175681422C/T
rs227832180998 175684698A/G
rs211587482033 175685733C/T 0.709 0.691 0.605
rs203331589658 175693358C/T 0.616 0.614 0.962
rs203331489960 175693660A/G 0.660 0.674 0.679
rs199160094155 175697855A/G 0.602 0.590 0.749
rs186445395679 175699379A/G 0.641 0.659 0.631
[0278] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure 1D for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in Figure
1D can be determined by consulting Table 29. For example, the left-m~st X on
the left graph is at
position 175603909. By proceeding down the Table from top to bottom and across
the graphs from left
to right the allele frequency associated with each symbol shown can be
determined.
122
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0279] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between SNP
s. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a lOkb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-$ were truncated at that
value.
[0280] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 8
ADAMTS2 Region Proximal SNPs
[0281] It has been discovered that SNP rs398829 in ADAMTS2 is associated with
occurrence of
osteoarthritis in subjects. This gene encodes a disintegrin and
metalloproteinase with thrombospondin
motifs-2 (ADAMTS2), which is a member of the ADAMTS protein family. Members of
the family share
several distinct protein modules, including a propeptide region, a
metalloproteinase domain, a
disintegrin-like domain, and a thrombospondin type 1 (TS) motif. ADAMTS~ is
involved in collagens 1,
2 and 5 N-terminal processing, (type II collagen is the major form in
cartilage). Mutations in this gene
cause Ehlers-Danlos syndrome type VIIC, a recessively inherited connective-
tissue disorder that causes
loose joints and fragile skin. Mild loss of function may exacerbate physical j
oint damage leading to a
predisposition to OA and incorrectly processed collagen can act dominantly to
inhibit self assembly of
fibrils. Alternative splicing of the gene generates 2 transcript variants. The
short transcript encodes a
protein, which has no significant procollagen N-peptidase activity.
[0282] Two hundred-nine additional allelic variants proximal to rs398829 were
identified and
subsequently allelotyped in osteoarthritis case and control sample sets as des
cribed in Examples 1 and 2.
The polymorphic variants are set forth in Table 32. The chromosome positi ons
provided in column
four of Table 32 are based on Genome "Build 34" of NCBI's GenBank.
TABLE 32
dhSNP Chromo-Position Chromosome Allele
in SEQ
rs# some ID NO: 6 Position Variants
rs22782215 210 178695460 c/t
123
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
6
rs16503585 3608 178698858 c/
rs16438185 3609 178698859 c/
rs37339165 4318 178699568 clt
rs16249335 5593 178700843 a/
rs16248575 5629 178700879 c/t
rs16248325 5639 178700889 a/
rs16248295 5640 178700890 c/t
rs21611715 8943 178704193 a/c
rs15304995 17968 178713218 a/
rs888764 5 19887 178715137 a/
rs873987 5 21034 178716284 a/
rs40786995 21085 178716335 c/t
rs870311 5 21596 178716846 a/
rs16438175 23379 178718629 a/c
rs16438165 23432 178718682 a/c
rs16503555 24007 178719257 a/c
rs888763 5 26121 178721371 a/
rs18622125 26273 178721523 alt
rs11105145 26755 178722005 a/t
rs37976005 27411 178722661 c/t
rs37976025 27710 178722960 /t
rs37976035 27842 178723092 c/t
rs37768195 28379 178723629 c/t
rs252076 5 29603 178724853 c/t
rs252075 5 31232 178726482 c/
rs252074 5 31504 178726754 a/
rs252068 5 32583 178727833 c/
rs252069 5 32794 178728044 a/
rs194040 5 32840 178728090 c/t
rs252070 5 33044 178728294 c/t
rs37976065 33150 178728400 a/c
rs171667 5 33218 178728468 a/
rs187539 5 33513 178728763 clt
rs38368345 33959 178729209 /tatcaaactaccatga
as
rs252071 5 34486 178729736 a/
rs252072 5 36289 178731539 c/t
rs252073 5 36570 178731820 c/t
rs379589 5 38247 178733497 a/t
rs20524725 38477 178733727 a/c
rs20524715 38518 178733768 c/t
rs20524705 38529 178733779 c/t
rs20524695 38667 178733917 a/
rs37976085 39781 178735031 c/t
rs37976095 39856 178735106 c/t
rs38226015 39927 178735177 c/t
rs153131 5 40506 178735756 a/
rs751546 5 41869 178737119 c/
rs22799795 42452 178737702 c/t
rs252060 5 44788 178740038 c/t
rs37976105 46059 178741309 a/c
124
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
6
rs1940395 46846 178742096 al
rs1687735 47712 178742962 a/t
rs2520615 48796 178744046 c/t
rs1875375- 49441 178744691 cl
rs2520625 49602 178744852 a/t
rs24312555 49723 178744973 a/c
rs37976125 50050 178745300 c/t
rs37976135 50171 178745421 c/t
rs6141145 50477 178745727 c/t
rs2520635 50818 178746068 c/t
rs2520(i45 50833 178746083 c/t
rs2520655 50881 178746131 a/
rs4505025 50882 178746132 a/
rs4392525 51386 178746636 clt
rs2520665 51534 178746784 c/t
rs4579575 52317 178747567 a/
rs37976145 52368 178747618 c/t
rs4235525 52970 178748220 a/
rs3988295 53023 178748273 a/
rs4166465 53356 178748606 a/
rs1874505 53882 178749132 /t
rs3378075 54553 178749803 c/t
rs3378065 55475 178750725 a/c
rs13964385 55530 178750780 a/
rs13964375 55691 178750941 c/t
rs24118115 55848 178751098 a/c
rs28988135 55879 178751129 c/
rs1892565 56316 178751566 a/
rs1730725 56911 178752161 a/c
rs3378055 57320 178752570 a/
rs1914155 57391 178752641 c/t
rs1800455 57437 178752687 clt
rs1892555 57478 178752728 c/
rs6527665 57500 178752750 c/t
rs4667505 59111 178754361 /t
rs4424065 59333 178754583 a/
rs6624075 59715 178754965 a/
rs5929715 59804 178755054 a/
rs4571875 59851 178755101 a/
rs4594905 59929 178755179 c/t
rs4596685 60052 178755302 c/t
rs4626465 60240 178755490 c/t
rs4582725 60359 178755609 /t
rs4634555 60381 178755631 a/
rs6758805 60456 178755706 c/t
rs8106175 60724 178755974 c/
rs4641565 60875 178756125 c/t
rs4580835 60968 178756218 a/
rs4673335 60978 178756228 c/
rs4653815 60998 178756248 c/t
rs4663635 61557 178756807 c/t
rs24570995 62091 178757341 c/t
125
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
6
rs4639015 62645 178757895 c/t
rs4656215 62943 178758193 a/c
rs4637245 63131 178758381 a/t
rs4652425 63145 178758395 /t
rs4674195 63406 178758656 a/
rs4561355 63427 178758677 c/
rs4645365 63554 178758804 c/t
rs4618985 63661 178758911 a/
rs3895585 64093 178759343 al
rs4667525 64153 178759403 c/t
rs4556555 64409 178759659 c/
rs4634355 64544 178759794 c/t
rs21749715 65257 178760507 c/t
rs19799795 65626 178760876 a/
rs4118045 65739 178760989 a/
rs16238855 66392 178761642 c/t
rs16438115 66720 178761970 c/t
rs4344305 69177 178764427 a/t
rs1875385 69336 178764586 /t
rs2520675 69636 178764886 a/
rs4593195 69823 178765073 a/
rs4672895 69928 178765178 c/t
rs4626445 70547 178765797 c/t
rs4587525 70633 178765883 c/t
rs7083205 71805 178767055 a/c
rs4579545 72181 178767431 c/
rs24118105 72200 178767450 c/t
rs30846875 72474 178767724 -/at
rs69638 5 72567 178767817 c/
rs4554525 72973 178768223 a/
rs4648505 73468 178768718 a/
rs4314725 73889 178769139 a/
rs24118095 75730 178770980 c/t
rs24570945 75970 178771220 a/
rs24570955 76114 178771364 a/
rs22617405 76342 178771592 c/t
rs11091805 76449 178771699 a/
rs11091795 76465 178771715 c/t
rs11091785 76791 178772041 a/c
rs4569095 78042 178773292 a/
rs4691245 80758 178776008 a/
rs4680395 80778 178776028 c/t
rs4670175 81356 178776606 a/c
rs4692905 81576 178776826 a/
rs4690905 81689 178776939 c/t
rs4695685 81759 178777009 /t
rs4683865 81950 178777200 c/
rs4693495 82562 178777812 a/c
rs4690995 83591 178778841 c/t
rs4568685 83700 178778950 a/
rs4653895 83821 178779071 c/
rs4638925 83842 178779092 c/
126
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
6
rs4685485 83923 178779173 /t
rs6546125 83929 178779179 a/c
rs4685425 84021 178779271 c/
rs4692625 84175 178779425 c/t
rs7083235 84417 178779667 a/
rs4690895 84747 178779997 c/
rs4693965 85746 178780996 c/
rs4687235 86129 178781379 c/t
rs4676045 86335 178781585 a/
rs3388745 87315 178782565 c/
rs3388755 87648 178782898 a/
rs13858035 87764 178783014 a/c
rs13858045 87770 178783020 c/
rs3388765 88221 178783471 c/t
rs1898035 90474 178785724 a/c
rs4522155 91148 178786398 /t
rs6411705 91150 178786400 It
rs5843985 91160 178786410 /t
rs3853305 91733 178786983 c/t
rs4295385 91772 178787022 a/c
rs3712295 91785 178787035 c/t
rs4608745 93140 178788390 a/t
rs6461215 93148 178788398 a/t
rs4682625 96080 178791330 a/
rs4678635 96157 178791407 c/
rs1914345 96313 178791563 a/c
rs20547825 96759 178792009 c/t
rs4684995 97026 178792276 a/c
rs1802875 97320 178792570 c/
rs3388775 97732 178792982 a/t
rs6506655 98713 178793963 c/
rs1934195 99707 178794957 a/c
rs1802885 99959 178795209 cl
rs1868345 100009 178795259 a/
rs1892665 100020 178795270 c/
rs1892675 100065 178795315 a/c
rs1709375 100086 178795336 c/
rs4632635 101270 178796520 c/
rs4632625 101276 178796526 /t
rs4604545 101371 178796621 c/t
rs4604555 101376 178796626 c/
rs4605055 101439 178796689 c/t
rs9313165 101820 178797070 c/t
rs4634315 102392 178797642 c/
rs4615425 102602 178797852 a/
rs4635575 102604 178797854 a/c
rs1914535 102896 178798146 c/t
rs22712125 189104 178884354 c/t
rs4620095 189134 178884384 c/t
rs22712115 189205 178884455 a/
rs3964745 Not ma Not ma ed a/c
ed
rs4289015 Not mappedNot mapped ~ a/t
127
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
in SEQ
rs# some ID NO: Position Variants
6
rs452300 5 Not ma Not ma ed /t
ed
rs670256 5 Not ma Not ma ed /t
ed
Assay for Verif~n~ and AllelotYpin~ SNPs
[0283] The methods used to verify and allelotype the 209 proximal SIvTPs of
Table 32 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 33 and Table 34, respectively.
TABLE 33
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs2278221ACGTTGGATGTCTCATGGGCCACCACAAACACGTTGGATGTATGCTCCTGTCACCGGCAT
rs1650358ACGTTGGATGTGGATGGCTCCATGTTCTTGACGTTGGATGAAGTGCTGGGATTACAGGTG
rs1643818ACGTTGGATGCTGGGATTACAGGTGTGAACACGTTGGATGTGGATGGCTCCATGTTCTTG
rs3733916ACGTTGGATGCCGAGCAGGCTGTAGTGTTGACGTTGGATGCTTTGTACCACCTGGAACAG
rs1624933ACGTTGGATGAGGCTGGTCTCAAACTCCTGACGTTGGATGTAACAAAAAGTTGGCCGTGC
rs1624857ACGTTGGATGTGAGGTCAGGAGTTTGAGACACGTTGGATGGCCACCAAGCCAGACTAAGT
rs1624832ACGTTGGATGTGAGGTCAGGAGTTTGAGACACGTTGGATGGCCACCAAGCCAGACTAAGT
rs1624829ACGTTGGATGTGAGGTCAGGAGTTTGAGACACGTTGGATGGCCACCAAGCCAGACTAAGT
rs2161171ACGTTGGATGCCCGTCACCACTTTATTTCCACGTTGGATGAGAGTGGATCCAGTCTGCAG
rs1530499ACGTTGGATGACTCCAAGATTTCCCATTTCACGTTGGATGTTCTGTGTTCCACCCTATGG
rs888764ACGTTGGATGTAGTTGAATGTTGTATTGGCACGTTGGATGACCGTGATAAACACAGAATG
rs873987ACGTTGGATGGCTGTTAATCATGTGTCGGGACGTTGGATGATTTGGCCACATCACCAGAC
rs4078699ACGTTGGATGGTACCGTGGATTCTTTTAGGACGTTGGATGGT-ATTGGAAAAGAGCAGAGAC
rs870311ACGTTGGATGTCAGGGCTCCAGTGTTGAAGACGTTGGATGAAAAGGAGGAGTGCCCTGTG
rs1643817ACGTTGGATGATGGGAAACTCCTGGTCCTGACGTTGGATGAAAATGCAAGCCGCCACCTG
rs1643816ACGTTGGATGTTTTCTCCCCTTTCTAGCCCACGTTGGATGTTGGCATGAGAGATGGACAG
rs1650355ACGTTGGATGTCAACAGCAACAAAACCAAAACGTTGGATGTTAAATAGGTCAGAGGGTTG
rs888763ACGTTGGATGAAGAGGAAGAGACATACCAGACGTTGGATGAACAACATGGACTCAGGCTG
rs1862212ACGTTGGATGGGCCACATTTTAAAACAAGGGACGTTGGATGTCCCCTGAGGTTCCTATAAG
rs1110514ACGTTGGATGTGCCCACGTTCCATGTTCAGACGTTGGATGA-f'CACTGTAGCCCCTTCCTG
rs3797600ACGTTGGATGCCTTCCTGTCACCTCCTTTGACGTTGGATGG GAAGTGACTGCTGAGCTG
rs3797602ACGTTGGATGAGAAACAGGGACTGGCTGTGTACGTTGGATGAGCAGGCTCCGGGAAGTATG
rs3797603ACGTTGGATGCACCCATCCATCATGATGTCACGTTGGATGTGCTACCTCAAAACAGTGGG
rs3776819ACGTTGGATGCAAGCACCATTCCATTGCACACGTTGGATGAAATGAGGATTGCAGTCCCC
rs252076ACGTTGGATGACTTCTGACTTCAGGTGATCACGTTGGATGTATAGGAACGAAAGAAAGCC
rs252075ACGTTGGATGTGGGAGCATTTGCAGGCATGACGTTGGATGAAGCCTCAGATGGTTCGGAG
rs252074ACGTTGGATGTTGCGATGGCCTCCTGGCTACGTTGGATGAAGTTGAGGGCTCCGGAGCA
rs252068ACGTTGGATGGGGTAGGAAGGGTTTAAGCACGTTGGATGG CAGCCCCTCAATTCTTTAG
rs252069ACGTTGGATGTGCCCATTTCCTGTTATTCCACGTTGGATGT-T'TGGACTTGCCGTGCAACT
rs194040ACGTTGGATGTGCCCATTTCCTGTTATTCCACGTTGGATGT-fTGGACTTGCCGTGCAACT
rs252070ACGTTGGATGCTCAAGGACATTGTCCCTGGACGTTGGATGG GAGAAGCAGCTCTCCTTTC
rs3797606ACGTTGGATGGTTTCCCCAAACAAGAGAGCACGTTGGATGG GAAATGTTCAAAGCCGCAG
rs171667ACGTTGGATGGGGAAACACATTGTAATGCGACGTTGGATGCCTTCCTCATTGTCTATTCC
rs187539ACGTTGGATGAGCCACCCCAACCTTCAGGAACGTTGGATG'1-TGCTCCTGGACATGGTTTT
rs3836834ACGTTGGATGAAGAAACGTGACTCTTGCTCACGTTGGATGTAGTAATTCTGATCCTGGCC
rs252071ACGTTGGATGGCTTCAACCTGAAACAACCCACGTTGGATGGGGATATTCCCACTCTGAG
rs252072ACGTTGGATGTTGTTTCCCCAAAGGCGACGACGTTGGATGTGTGTTTTCCAGAGCTGGAG
12~
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs252073ACGTTGGATGGGGAAAGGCCGAGAAAAGTCACGTTGGATGACAAGCTCAGCAGAGTTCCA
rs379589ACGTTGGATGAAACACGGGAGTACTGAGCAACGTTGGATGTTGTTAGCTGTCTGTCCGTC
rs2052472ACGTTGGATGAACCAGCTCAAGGATCACCCACGTTGGATGAAAGGAGACGGTCAGCTGTC
rs2052471ACGTTGGATGACAGCTGACCGTCTCCTTTGACGTTGGATGCCCGTCCTGGACAAGCTTTT
rs2052470ACGTTGGATGACAGCTGACCGTCTCCTTTGACGTTGGATGCCCGTCCTGGACAAGCTTTT
rs2052469ACGTTGGATGAGGGAAAGATATCGCACGCGACGTTGGATGAGTGAACAACTGCTCGCCTC
rs3797608ACGTTGGATGTGCTTTGCCTTGGCTTCTGCACGTTGGATGTGCACTAAGGGAGTGAGTGG
rs3797609ACGTTGGATGTGCAGAAGCCAAGGCAAAGCACGTTGGATGACAGCATTTGGAGTCCCCTG
rs3822601ACGTTGGATGAGGTCAGTGAGGCCTGAGATACGTTGGATGTGTCTGGCCTGAAGATCGAG
rs153131ACGTTGGATGTAATCACGTGTCCTGATCCCACGTTGGATGAGCTGTCCTCAGT-CATGTTC
rs751546ACGTTGGATGTCCTGCTCTGCCGTTCTACAACGTTGGATGATCAGCTCAAAGGACCGGTG
rs2279979ACGTTGGATGTATTGCTACCAGGAACACGTAACGTTGGATGAAAAAGGGGCCACTTCAGGG
rs252060ACGTTGGATGTGGCCAGAGCCCGTGTTTCACGTTGGATGCGGCCAATCCCATCTCTATG
rs3797610ACGTTGGATGAAAAGCTTCTCCCTTGGGTGACGTTGGATGCAAGTAGGGCAGAAACTCAG
rs194039ACGTTGGATGAAAGTGCTGGGATTACAGGCACGTTGGATGTGCTGGGAGAAGACATTCAC
rs168773ACGTTGGATGTGGTGCACTGGAGATATCACACGTTGGATGGATCCCTATCCTA.CCTCTTC
rs252061ACGTTGGATGTGTCACACTCCTCTTGTAAGACGTTGGATGCTGTCCTTCCATGCTTTTGC
rs187537ACGTTGGATGCGAGGATGTCATGCTAAGTGACGTTGGATGGTACCTCGCATAA.GTGGATC
rs252062ACGTTGGATGAAGCACATTCATGTGGCTGGACGTTGGATGCTGAAACTCAATG GGCACAG
rs2431255ACGTTGGATGGGTGAAGACGGTGACTTATGACGTTGGATGCTGGTGTCCTTGAAGAACTG
rs3797612ACGTTGGATGAGTGAGGACGCAGGGCATTCACGTTGGATGAGCGTGGGCGAGGGAGATAA
rs3797613ACGTTGGATGATCAGAGGCAGAGACCCCCCACGTTGGATGGGGTGTCCTGCAGAGGCGG
rs614114ACGTTGGATGGGTTGGAGGATGTCTAGAACACGTTGGATGGGCTGGATCACTAGGGTTTG
rs252063ACGTTGGATGTTGGAATTACAGTCCGATGGACGTTGGATGCTGAGAGACTGAAAAGCACA
rs252064ACGTTGGATGCTGAGAGACTGAAAAGCACAACGTTGGATGTTGGAATTACAGTCCGATGG
rs252065ACGTTGGATGAAAACTAAGGCTCAGAGGACACGTTGGATGTGGGCTTGGAATT-ACAGTCC
rs450502ACGTTGGATGATGAGAAAACCAAGGCTCAGACGTTGGATGCTGGGCTTGGAATTACAGTC
rs439252ACGTTGGATGATCTCCTGACCTCGTGATCCACGTTGGATGTCATAATAACGGGCGGGTGC
rs252066ACGTTGGATGTTTCCTCTTGACCGGTCTTGACGTTGGATGTAAAACGAATTCTGCCGATG
rs457957ACGTTGGATGTTCACGTGCATTAGAGCGAGACGTTGGATGAATTCCCTCCCCAATTCCTC
rs3797614ACGTTGGATGACTGCGAGCTTTAAGGAGGGACGTTGGATGCCAAACAGAAGCCCCTTTTC
rs423552ACGTTGGATGGCAGGACCTCGATGTTGTAGACGTTGGATGATCCTAGAGGAGCACGCCAAC
rs398829ACGTTGGATGTAGTCATCGTCCGCAGCATGACGTTGGATGAAGACGGTGTCCTCTCCTTG
rs416646ACGTTGGATGGCTGGGTCTCTCACAGTCTCACGTTGGATGAGACAGGCACCTCTGTGACTT
rs187450ACGTTGGATGAGAAGGCAGGGACGATATCCACGTTGGATGACCAAGATGAACCCCTCTGT
rs337807ACGTTGGATGTCACCCAGTGCTGACAGCAGACGTTGGATGATGCTGGGATGCCATGGGTC
rs337806ACGTTGGATGAATTAAGAGATGGGGCCACCACGTTGGATGGCCCTGTGTGTTTTGTCTCC
rs1396438ACGTTGGATGTACCTTCTGGTGCCAGAATGACGTTGGATGCCTGGAGACAAAACACACAG
rs1396437ACGTTGGATGTAAAAACTCTGCCTGCTCGGACGTTGGATGTCCAGACATTCCCCGTAGGA
rs2411811ACGTTGGATGGAGGGATGCTCTAGAACATAACGTTGGATGCTGAATTTCACC'T-GAAATGG
rs2898813ACGTTGGATGTCCTCACCCACTTTGCCTTTACGTTGGATGATCGTGATAATTTTGGGGTG
rs189256ACGTTGGATGCTCCCTATAGCAAGGCTCTAACGTTGGATGTTAACCCAGGCCATGAAGAG
rs173072ACGTTGGATGAGCTGGAGATCTCTTTGCTCACGTTGGATGCTAAAACAGGATCGCTCTGG
rs337805ACGTTGGATGGAAACAAACCAAGGAGCAGGACGTTGGATGATGTGGACAACGTTGGACTC
rs191415ACGTTGGATGAATTACATGACTCGGACAAGACGTTGGATGTGCTGGTGAAGTACAGAAGG
rs180045ACGTTGGATGGTCCCAGGTTTTCTGTTCTCACGTTGGATGTGTACTTCACCACCACTGAG
rs189255ACGTTGGATGAGGTTGCAGACTCAGTCCCAACGTTGGATGGGGTGATTTGCGGGAATGAG
rs652766ACGTTGGATGGGGTGATTTGCGGGAATGAGACGTTGGATGACCATCCCACGA.TGCTCCC
rs466750ACGTTGGATGTATCTCCTTAAATGCCTTGGACGTTGGATGTGACCAGGAGGAGTTAAAAC
rs442406ACGTTGGATGTGACAAGGTCACGTGTTCTGACGTTGGATGCCAGACAAGTCTGATACAGC
rs662407ACGTTGGATGCCACAGTCACCATTACTGAGACGTTGGATGCTTGAGCCATGAGTGGAATG
rs592971ACGTTGGATGGGAAGCATTTCTTTGACTGCACGTTGGATGATTCCATCTCATCGCTCAAG
129
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs457187ACGTTGGATGTGTGAGATGAGGAGTATCTGACGTTGGATGGCAGTCAAAGAAATGCTTCC
rs459490ACGTTGGATGACAGATACTCCTCATCTCACACGTTGGATGGGGAGTTTTGCTGTTATAGC
rs459668ACGTTGGATGGCTTCATTTACTGAGGTCTTCACGTTGGATGTGAATGTTCAACGACTACAC
rs462646ACGTTGGATGCAATTATTCGACGGAGATTAACGTTGGATGCTCCTCCAAATGAATCAAGAA
rs458272ACGTTGGATGATGCCTCCTCATTGTCATTCACGTTGGATGCCCAACAAAGTGATTCCAAC
rs463455ACGTTGGATGATGCCTCCTCATTGTCATTCACGTTGGATGCCCAACAAAGTGATTCCAAC
rs675880ACGTTGGATGCAGCTCCATTGATCTGTTTCACGTTGGATGAAGAATGACAATGAGGAGGC
rs810617ACGTTGGATGTGATCTCAGCTTACCACAGCACGTTGGATGATGCCTGTAATCCCAGCTAC
rs464156ACGTTGGATGCAGATCCAAGAATATGTGGGACGTTGGATGTTCTAGAAAGGAGCCAAATC
rs458083ACGTTGGATGTGTTGTTTCTTCCCCTCCTGACGTTGGATGTGGCTCCTTTCTAGAATCCC
rs467333ACGTTGGATGCTTGTTATTTCTTCCCCTCCACGTTGGATGTTGGCTCCTTTCTAGAATCC
rs465381ACGTTGGATGACTTGCCCATCTGTTTCCAGACGTTGGATGACAAGCCTCTAAGGATAGGG
rs466363ACGTTGGATGAAGTGACCCTGAGGTGATGGACGTTGGATGTGAAGACAGTTCACCCCGTG
rs2457099ACGTTGGATGTCTCCTTACACTGCCAGCGTACGTTGGATGCACTGTATTGCTACTTGAGC
rs463901ACGTTGGATGAGAGTGCCAAGTGCAAAAGGACGTTGGATGTGTCTTGCGTCTGTGTATCC
rs465621ACGTTGGATGGGAAGTCATGGAAGTGCTAGACGTTGGATGAAAGAGCCCTAGGCTTGGAA
rs463724ACGTTGGATGAGTGTGCCTGTCTGCCCTCAACGTTGGATGAAGGGCAGATGGCACACTTG
rs465242ACGTTGGATGAGTGTGCCTGTCTGCCCTCAACGTTGGATGAAGGGCAGATGGCACACTTG
rs467419ACGTTGGATGAGTCCCCAAAACGTAAGTCCACGTTGGATGAGTCTAATTCCCTGAGCCTC
rs456135ACGTTGGATGAGTCTAATTCCCTGAGCCTCACGTTGGATGACGTAAGTCCTAATGACCGC
rs464536ACGTTGGATGTGCTCCAGGCTTTGGTCCTCACGTTGGATGAATTAGACTAAGGCCATGATG
rs461898ACGTTGGATGGGGAATACACAGCCACAGAGACGTTGGATGAGGTCAACGGGAACAAGGTC
rs389558ACGTTGGATGGCAGTCCTGACAGTTCTCTAACGTTGGATGTTTTTCTCCCTGAAGCATGG
rs466752ACGTTGGATGGGCCTTCTCTCCTTTAGTGCACGTTGGATGAGTCCTGACAGTTCTCTAAA
rs455655ACGTTGGATGCTATTTGCACCCCATATGGCACGTTGGATGAACACACAGCATCAGGTTCC
rs463435ACGTTGGATGTTCAGCCATAGCTGGATTTGACGTTGGATGCTCTGCTGGGAAAATGTGAC
rs2174971ACGTTGGATGAACACAACTTCCCCTTCGTCACGTTGGATGTGAATCCTTGGAGGTGAGTG
rs1979979ACGTTGGATGTGGCTGTCAGCACCCCACTTACGTTGGATGCCCAAAGGAAGGGAGAATTC
rs411804ACGTTGGATGCAGATGACAGGCGGAAAATCACGTTGGATGAGGCTTCCAGATGATGTCCA
rs1623885ACGTTGGATGAATCAGCTAGGAAGAGCCTGACGTTGGATGTTCCTGACCCCTCTAGGTCAG
rs1643811ACGTTGGATGCAGGGCCCTGGTACTTTCAGACGTTGGATGCATGGTGGTGATTGCACCTG
rs434430ACGTTGGATGTCCAGGAGTTCACTGTAGAGACGTTGGATGCACATGCATACATTCATCAC
rs187538ACGTTGGATGACATGGGGCTTGGCAAAATGACGTTGGATGCACCTGCTCAGAAGTAGCAT
rs252067ACGTTGGATGAGAATTGCTGTGGTGTGAGGACGTTGGATGTTTTTCTTGGGAGCTGTCGC
rs459319ACGTTGGATGCCATCTCTCTGACCTAGACAACGTTGGATGGCTCCAAGGAAAATTGGGAG
rs467289ACGTTGGATGGGCCCTCTTGGCTTGTCTTTACGTTGGATGAGGCAGTGTGCCCTCTCATC
rs462644ACGTTGGATGATGATGTGGGTGAGCCCTTGACGTTGGATGTAACACTCAGCACGCACCAG
rs458752ACGTTGGATGCACCCACATCATGTGCGCTTACGTTGGATGCCCTTCTCTACCCAGCACTT
rs708320ACGTTGGATGAAACCAGCCTGGCTAACATGACGTTGGATGACAGGTGCCTGCTATCATAC
rs457954ACGTTGGATGAACCAGACCTTGACTGATGGACGTTGGATGCCTCATACAAGTAGCCAAGG
rs2411810ACGTTGGATGGCTTAACCAGACCTTGACTGACGTTGGATGAGTGTAAGGATATCCACGGC
rs3084687ACGTTGGATGATCCCTTGAGCCAGAGATTCACGTTGGATGATGTCCTGTGCACACACAAG
rs69638ACGTTGGATGTGCTCATTGCTGTCCTCATCACGTTGGATGAGAAGAAAGGTGTGCAGTGG
rs455452ACGTTGGATGAGTGATGATGAGCCTGCTGGACGTTGGATGTCAGGTTCCCTCTCTGTGTC
rs464850ACGTTGGATGTCTCTCTGTGCTCCAGACCAACGTTGGATGTGGGCTGAGATTTCTGTGGG
rs431472ACGTTGGATGAACCAGTGTGGGTGTGAAGCACGTTGGATGAGAGACTGCATCAGGCAGGA
rs2411809ACGTTGGATGAGCGCATAAGTGACCACCAGACGTTGGATGGCACTCACAGGGCATTGATG
rs2457094ACGTTGGATGTTACTGTCACCTTGGGTCTCACGTTGGATGGGAAGTCTGTATAGACGCAG
rs2457095ACGTTGGATGTTATCAAGGCCTGCGCAGTGACGTTGGATGACTCCTGACCTCAGGCAATC
rs2261740ACGTTGGATGATCGTGCCACTGCACTCCAGACGTTGGATGTCATCTTTTGGTAGCCCCCC
rs1109180ACGTTGGATGCCAGGCCTGTATTGCACATCACGTTGGATGAGAATGCGTGTGCATGTGGG
rs1109179ACGTTGGATGTGTAATGGTATGCAGACCCCACGTTGGATGGAGTGCCGTATTTGTCCTTC
130
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1109178ACGTTGGATGGCAAACAACAACAGCAACAGACGTTGGATGAAGTGTGGATTTGTGCAGAC
rs456909ACGTTGGATGTAGCTGCTTCATCTGTAAAGACGTTGGATGGGCACTTTACCGATCTACTC
rs469124ACGTTGGATGACTTGGACACACATAGGCTGACGTTGGATGTGAAATGCTCAGGGTGTGTG
rs468039ACGTTGGATGTGAAATGCTCAGGGTGTGTGACGTTGGATGAGGACTTGGACACACATAGG
rs467017ACGTTGGATGGTCTAGCTGCCACTAAACAGACGTTGGATGATGTGCCAAGAGGCTTTGAG
rs469290ACGTTGGATGTGCCCTTTGTGTGCTCAGAGACGTTGGATGTCCCTCTGTGCTGTGTTTGG
rs469090ACGTTGGATGACTTGTCTTCAGGTGCTTGGACGTTGGATGGATGGTTAGTCTCCTGGTTC
rs469568ACGTTGGATGAGCACCTCTGGCTTTCATTGACGTTGGATGATTCACCAGGAAATCCCAAC
rs468386ACGTTGGATGTAATCCCAGCCCTTTGGAAGACGTTGGATGTATGGAGACAGGGTTTTACC
rs469349ACGTTGGATGTTAGAGACAGAGTCTCACTCACGTTGGATGTTGATCCCAGGAGTTCAAGG
rs469099ACGTTGGATGTTGGAGCTGCTCTAGTTCTCACGTTGGATGTGAAAACCGGGACTCAGCTC
rs456868ACGTTGGATGACAGAGCAGGGAGCTGCGGTACGTTGGATGATTCACCCCCAGCTACTGTG
rs465389ACGTTGGATGAGGCTTTGTAGACAGCTCCCACGTTGGATGTGCCAGTGCTCTGAGTATGC
rs463892ACGTTGGATGAGGCTTTGTAGACAGCTCCCACGTTGGATGTGCCAGTGCTCTGAGTATGC
rs468548ACGTTGGATGACTGGAAGGGAACATGCAAGACGTTGGATGCCTGGATGCCCTTTATAGAC
rs654612ACGTTGGATGACTGGAAGGGAACATGCAAGACGTTGGATGTGGATGCCCTTTCTAGACAC
rs468542ACGTTGGATGGCCTCCATTTTCCTTCTCACACGTTGGATGTGTCTAGAAAGGGCATCCAG
rs469262ACGTTGGATGTTCTGAGCTGAACGAAGCAGACGTTGGATGGGTCAGGGATCCTTTGATGC
rs708323ACGTTGGATGCACATACTATACAGGTCACCACGTTGGATGGAGGGAGAAGATGTTGTGAA
rs469089ACGTTGGATGTTTGGAAGTACCACCTCAGCACGTTGGATGAATGGAAGGAAGGATCAGCC
rs469396ACGTTGGATGAGTGACTCCAATGAGGGAACACGTTGGATGTCTCACACCACTGATCCTTC
rs468723ACGTTGGATGTGTGGATCTTGCTGTTTGGGACGTTGGATGTATTGGCATCGCGTATCAGG
rs467604ACGTTGGATGACTCCTGCCATTAAACTCTCACGTTGGATGCTTGGCTTAACTTACAAGGG
rs338874ACGTTGGATGCCCCACCACAGCCACTGGGACGTTGGATGAAGGGCCTTGCCCCACCCAA
rs338875ACGTTGGATGTGCTGTCTTGCTCGCGTGTGACGTTGGATGACACTGGATATGTCAGGGTC
rs1385803ACGTTGGATGTCACCACCATTCCAGAAGTGACGTTGGATGACCTTCCTTATTGCTGTGGC
rs1385804ACGTTGGATGTCACCACCATTCCAGAAGTGACGTTGGATGACCTTCCTTATTGCTGTGGC
rs338876ACGTTGGATGTTAGGGCTGGGTGGAGGAAGACGTTGGATGTCCAACTCCCAGTGACAGAG
rs189803ACGTTGGATGCCTCCAGTTTCTCTCTTCTGACGTTGGATGATCCTGGATTAGCCAGATGG
rs452215ACGTTGGATGTAGCTCTATTCTTCCACCCCACGTTGGATGAGCGAGACTCCGTCTCAAAA
rs641170ACGTTGGATGATAGCTCTATTCTTCCACCCACGTTGGATGAGCGAGACTCCGTCTCAAAA
rs584398ACGTTGGATGTTCCTGTGAGCTATAGAAACACGTTGGATGCGAGACTCCGTCTCAAAAAAA
rs385330ACGTTGGATGTTGCCCCAACTATTGTCCTGACGTTGGATGGGTTTCCCAGACAGTGTTTG
rs429538ACGTTGGATGTATTATCTGCAGACACCTGGACGTTGGATGATCTCATTCCCACCCTCTTC
rs371229ACGTTGGATGTATTATCTGCAGACACCTGGACGTTGGATGATCTCATTCCCACCCTCTTC
rs460874ACGTTGGATGGTCCTGCGGCTAAAAATTCCACGTTGGATGGGGCAGGTCAACTAGAAAAC
rs646121ACGTTGGATGGGGCAGGTCAACTAGAAAACACGTTGGATGGTCCTGCGGCTAAAAATTCC
rs468262ACGTTGGATGGCCAGGTTTCGAAAGTTAGGACGTTGGATGTGGGTTGGTCATGCGGTAAC
rs467863ACGTTGGATGTTTCGAAACCTGGCTGATGGACGTTGGATGTGCCACTGTCAGAAGACAAG
rs191434ACGTTGGATGCCAGCTGAAACACTAGACAGACGTTGGATGAGCTGAAGAGGTCTTTCTCC
rs2054782ACGTTGGATGAAAAAAGCAGGCCTCAGACCACGTTGGATGTCTGACTCTCATCTGCAGAC
rs468499ACGTTGGATGCTCCAGGAGGGACACTACGTACGTTGGATGTGGCCAGCTTCTCCTCGATG
rs180287ACGTTGGATGTTGTCTGCAGAATTACCTATACGTTGGATGGAAAAAGAAAAAAAAATCAG
rs338877ACGTTGGATGCGTGGATGGAAATTTACATTACGTTGGATGTTCTTTGGATCAATGTTGCC
rs650665ACGTTGGATGCCCATCTTACTCTATGATCTCACGTTGGATGAAAGTGCTGGGATTATAGGC
rs193419ACGTTGGATGGCAAATCCAAAGACACAGGGACGTTGGATGATGTTTTCATCACCCCAGTG
rs180288ACGTTGGATGTGTGACCTGGTAGCTTAGAGACGTTGGATGTTGTAGGAGGTCAGAAGAGG
rs186834ACGTTGGATGTAAGCTACCAGGTCACACACACGTTGGATGAGTTGATAGGAGAGTCAGGC
rs189266ACGTTGGATGTAAGCTACCAGGTCACACACACGTTGGATGAGTTGATAGGAGAGTCAGGC
rs189267ACGTTGGATGCCTCATTGTGCCCTGTTGTGACGTTGGATGCTCTGCCTGACTCTCCTATC
rs170937ACGTTGGATGCCTATCAACTGTTGATGGCGACGTTGGATGTTCCTCATTGTGCCCTGTTG
rs463263ACGTTGGATGTACTGGACCCCTTTGCACAGACGTTGGATGTGCCCATGCTCATGTGTTGG
131
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs463262ACGTTGGATGTGCCCATGCTCATGTGTTGGACGTTGGATGACCCCTTTGCACAGATGCTG
rs460454ACGTTGGATGAAGAAGGACCGTGTCAGAGAACGTTGGATGACATGAGCATGGGCAGGTAC
rs460455ACGTTGGATGACATGAGCATGGGCAGGTACACGTTGGATGAAGAAGGACCGTGTCAGAGA
rs460505ACGTTGGATGACCGTGGACAGCGTCTCTGAACGTTGGATGTGCTCTGAGGGCAGAACAAG
rs931316ACGTTGGATGATGCACACACCCATGGTCAGACGTTGGATGCGGTTCACTCCAGCATTTCC
rs463431ACGTTGGATGTCACCACAGCCCATGGGGAACGTTGGATGTTTGAAACTCACAATGTGGG
rs461542ACGTTGGATGTGATGAAGGCCAAGAATGCTACGTTGGATGTGTGTCCAGAACGTCAGGTG
rs463557ACGTTGGATGTGATGAAGGCCAAGAATGCTACGTTGGATGTGTGTCCAGAACGTCAGGTG
rs191453ACGTTGGATGCATCCAACAGCTCTGTCTGCACGTTGGATGACCCATCTGTAGCGCATCAG
rs2271212ACGTTGGATGAGCTTCCCCGGAGGCAACGAACGTTGGATGTGCAGGTCTCGGCCAAAGAC
rs462009ACGTTGGATGCAGGCTCCTCCTCGTTGCCACGTTGGATGTTGGTGTCCCACGTGGTGT
rs2271211ACGTTGGATGTCGTACCCCTGCTCTGGACGACGTTGGATGACTGACGCCCAGGGCCGCTT
rs396474ACGTTGGATGTGGGAGTTGGAGATGATGAGACGTTGGATGTTCCTCAGATCCCAGTCAAG
rs428901ACGTTGGATGTCAGTGACAGAGCGAGACTCACGTTGGATGGGGCTCGATAATGTAGCCAT
rs452300ACGTTGGATGAGCACAAGCTGAAGAGGTCTACGTTGGATGAGGAGAGAAGTGCACAGATC
rs670256ACGTTGGATGTAGCTCTATTCTTCCACCCCACGTTGGATGAGCGAGACTCCGTCTCAAAA
TABLE 34
dbSNP Extend Term
rs# Primer Mix
rs2278221 CAAACGCTGAGGAGAAGCC ACT
rs1650358 AAGAGACAAAAGGCCGGGC ACT
rs1643818 TACAGGTGTGAACCACCGC ACT
rs3733916 AGGCTGTAGTGTTGACAGAC ACG
rs1624933 GTCTCAAACTCCTGACCTCA ACT
rs1624857 AGACCAGCCTGGCCAACAT ACT
rs1624832 GGCCAACATGGTGAAACCC ACG
rs1624829 TGGCCAACATGGTGAAACCCT ACT
rs2161171 TGGAATAAGAGCCCTGCAGTGG ACT
rs1530499 CCCCTGCCCCAGCCACAGGAA ACT
rs888764 ATGTTGTATTGGCTATATTTGTCAACG
rs873987 AAAACCTAAAAGAATCCACGGTAACG
rs4078699 GACACATGATTAACAGCAAACAATACT
rs870311 AAGGGCGTGACGGCCCC ACT
rs1643817 GAAAGGGGAGAAAAGATTATCCCCGT
rs1643816 AGGACCAGGAGTTTCCCATTTT ACT
rs1650355 GAATCAATGAAGAAGAGAGCTT ACT
rs888763 GGTCAGGAGGCAGAGGGA ACT
rs1862212 GGGGTGAAAGGGAGCAGGG CGT
rs1110514 CAGGCCCCAGGTGAGGAA CGT
rs3797600 CTTTGTTGGTTAACCAAACCC ACG
rs3797602 GCTGACAGCTCCGGACATG ACT
rs3797603 TGTCATTCTCCTTGTGAACCCTCACT
rs3776819 CCATTCCATTGCACCTGCATG ACT
rs252076 CAAAGTGCTGGGATTGCAGG ACG
rs252075 GAGCATTTGCAGGCATGCCCTCTACT
rs252074 CTGGGTGGCTGCTGGGC ACG
rs252068 GGAAGGGTTTAAGCAAGGAG ACT
rs252069 TGAGCACCTACTATGGGCTAG ~ ACT
132
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs194040 ATTCCATATCTTCAAAGTGATTCAACG
rs252070 CCTGGGCTTCCCCTCCC ACG
rs3797606 AGCCCTTGGCCTCTCTCC ACT
rs171667 CGCCTTTTGCTTATGCAAAGA ACG
rs187539 AACCTTCAGGAAAGTTCCCAT ACT
rs3836834 TCAAAATATCAAACTACCATGAAAACG
rs252071 ACCCTGAGACACAGGGACT ACT
rs252072 GCTGGGTCACACTCGCGGA ACG
rs252073 GCCGAGAAAAGTCAGGGATTCT ACT
rs379589 CGGGAGTACTGAGCACCCAGG CGT
rs2052472 CCCCACTGTGACTATCTCCAC ACT
rs2052471 GTCTCCTTTGGCTGCCAAG ACT
rs2052470 TGCCAAGGCCCTGTCCTC ACT
rs2052469 CGCGGGGAAGTACTCGGC ACT
rs3797608 GTCCTCCTGTTCTGAGGCCC ACT
rs3797609 GGCAGAGCGGATGGCCTG ACG
rs3822601 GTGAGGCCTGAGATGAGAACC ACG
rs153131 TCCCCATACTCCTGTGCTC ACG
rs751546 CCGTTCTACAGCGGTTAAGA ACT
rs2279979 GGCCACCAGACAGATGTAAG ACT
rs252060 CGTGTTTCGGCAGAGGTGA ACT
rs3797610 CTTCTCCCTTGGGTGATGTGTT ACT
rs194039 CCACCGTGCCGGGACATTTTTTTTACT
rs168773 ACTGGAGATATCACGGGAGC CGT
rs252061 CCAGCTGGTCACAGGGCTCCC ACG
rs187537 TCATGCTAAGTGAAATAAGCCA ACT
rs252062 GCCACCACCGTCCACAGA CGT
rs2431255 CTGTATATTTCACCGCAATTAAAAACT
rs3797612 GGCATTCATCGTCAGGGCAA ACG
rs3797613 CCGCCGCCGGTCTCCCA ACG
rs614114 GAACGTTCTCTCACTTTTGCC ACT
rs252063 CTCCCGTCCTCTGAGCCTT ACT
rs252064 GAAAACTAAGGCTCAGAGGAC ACT
rs252065 TGGAAAAGGCGAGGCCTGGAGT ACG
rs450502 GGAAAAGGCGAGGCCTGGAGTT ACT
rs439252 GCCTCCCAAAGTGCTGGGATTA ACG
rs252066 TAGCCCTCTGGAGCCCAG ACG
rs457957 GGGCCCCTCCTTAAAGCTC ACT
rs3797614 TGGCCCTCGCTCTAATGCA ACG
rs423552 CTCGATGTTGTAGTCATCGTC ACG
rs398829 TGGCGTGCTCCTCTAGGA ACG
rs416646 CTCAGCAGGTCTGATCCATC ACT
rs187450 GGGCAGACTCCCCAGGAT ACT
rs337807 GCAGGCCACTCGGTGGAC ACT
rs337806 CCACCCCAGGGGTAGCCC ACT
rs1396438 GGCAGGCAGGTGGCCTG ACT
rs1396437 CGGCAGAAGCAGCCTCAAGA ACG
rs2411811 ACATAATTTCCAAATTTCACCCC CGT
rs2898813 GGTCCTGGGTGGAGGGAT ACT
133
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs189256 AGCAAGGCTCTATTTGGGGA ACT
rs173072 CTTTGCTCACATCGTGGCCAAA ACT
rs337805 GGAGCAGGAAAATTACATGACT ACG
rs191415 GAGTCCAACGTTGTCCACAT ACT
rs180045 ACTTGTTTCTACAATTCTCATTC ACG
rs189255 GACTCAGTCCCAGGTTTTCT ACT
rs652766 AGAAAACCTGGGACTGAGTCT ACT
rs466750 TCCTTAAATGCCTTGGTTGGCAATACT
rs442406 TTCTGGCTGTTGGGTTTGAAC ACT
rs662407 AAACATCTGAAATTAAAAGCACC ACT
rs592971 AGCATTTCTTTGACTGCTCTTTCAACT
rs457187 GGAGTATCTGTTCCTTGTGG ACT
rs459490 TTGAACATAGGAATAACCCGC ACT
rs459668 GTCTTCTTTTGTGTTTTTGGAGA ACG
rs462646 ATTATTCGACGGAGATTATTTGACACT
rs458272 ATTATTTTTCTGTCTGGTGTGG ACT
rs463455 CCTCCTCATTGTCATTCTTTTTC ACT
rs675880 CTTTCATGACATTGACACAACTACACT
rs810617 CCACAGCCTCCGCCTCCC ACT
rs464156 GGGTTTCCAGGTTAAAATGGC ACT
rs458083 CTCCTGCTCTGCCTATCCTT ACT
rs467333 ATTTCTTCCCCTCCTGCTCT ACT
rs465381 GCCTCCCACAGTTCCCTTGTT ACT
rs466363 GTGATGGCTCTGCACCAGA ACG
rs2457099 AGCGTGTGCCAGCTCTCC ACT
rs463901 GACACAATTCAGAGCGACTTAC ACT
rs465621 AAGTGCTAGAAGAAAATGTAGC ACT
rs463724 CCTTGCGCCATCCCCTAG CGT
rs465242 TGTGCCCATCCCCCCCTT ACT
rs467419 AACGTAAGTCCTAATGACCGCCC ACG
rs456135 CCCTCTCCTCTTCTGGGCA ACT
rs464536 GCTTTGGTCCTCCTGAGCC ACG
rs461898 CACAGAGCGACTCTCTCTTGGTT ACT
rs389558 CTGACAGTTCTCTAAACTCCCA ACG
rs466752 TTCCTTTTTCTCCCTGAAGCA ACG
rs455655 CACCCCATATGGCTCATGGG ACT
rs463435 GGGAAGGAGGTACTTAGCAG ACG
rs2174971 GTGCCACTCTCCAGCGGCC ACG
rs1979979 TTGCCGGCCCCCACCTC ACG
rs411804 GAAAATCCCTGTCACCAGTC ACG
rs1623885 CTTGGCTGCAGCACCCCA ACT
rs1643811 GCCCTGGTACTTTCAGCTCCCT ACG
rs434430 GTGTGCATGTGTGTGCCTG CGT
rs187538 TAAACGGGCCAAAAACGCCTAT ACT
rs252067 GCGCCTACGGATGTCAGG ACT
rs459319 CATGTTGAACAGAGAGAAACGGTCACG
rs467289 TCACTGAGAAATATTTTGCTCCC ACT
rs462644 GGTGAGCCCTTGGCTGTG ACG
rs458752 TAAAGCGCTCTTACAAATCAACA ACT
134
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs708320 TGGTGAAATCCTGTCTCTACTAAACGT
rs457954 CACCGTTTCTTATAATGCAGCC ACT
rs2411810 GGGGACGTTACTTCTTTTCAC ACG
rs3084687 ATTTATATATGTGTGTGTACACATACT
rs69638 CCCATTGGCTGTCCTGGAA ACT
rs455452 CCTCAACCCCAGATGCCCTC ACG
rs464850 ACTCCTGCCTGAGTGTCTC ACT
rs431472 GTGAAGCGGAAGGAGACTC ACG
rs2411809 CTGCACACCCTCTGCACAG ACG
rs2457094 TGGCTGGCACCACTGCACTGC ACT
rs2457095 TGGCTCATGCTTCTAATCCCA ACT
rs2261740 CTGCACTCCAGCCTGGGC ACT
rs1109180 ACATCAGTGACAGTGTAATGGTA ACG
rs1109179 TATGCAGACCCCCTCCCC ACT
rs1109178 AACAACAGCAACAGAAATGAAG ACT
rs456909 CGATTCCCACGCGTGTCTG ACG
rs469124 CCTGGCTCCATTGGTGTGAA ACT
rs468039 CCTTCACACCAATGGAGCCAG ACT
rs467017 CTGCCACTAAACAGATGAGAA ACT
rs469290 ATTTCTGGGCCCAAAGTCCA ACT
rs469090 CCAATTGTTCCAGCCACTCCC ACT
rs469568 TGATATTGCTTGCTTGGGTCTTAGACT
rs468386 GGTCAAGAATTCAAGAGCAGC ACT
rs469349 GTGCAGTGGCACGATCCTA ACT
rs469099 GCAGGTGGAACCGCAGAC ACT
rs456868 GGAGCTGCGGTGACTCCC ACT
rs465389 CCCTGGCACTCGCAGACC ACT
rs463892 AGCTCCCCCCGCACCAC ACT
rs468548 AAGGGAACATGCAAGCAAAGACTCACT
rs654612 TGCAAGCAAAGACTCGAATGA ACT
rs468542 TCACTCACTTGATTCCTGCCATC ACT
rs469262 CACTGTGGGATTTCCAGCAGA ACT
rs708323 TATACAGGTCACCCATTTAAAGT ACT
rs469089 CCTCGGCCTTCCCCAGCT ACT
rs469396 AATGAGGGAACCTGCAGTTTAAGAACT
rs468723 CAGACCCCATGCCTTGCC ACT
rs467604 GAGTTTCCTCCTCTTTCACAA ACT
rs338874 CACAGCCACTGGGGAGTAG ACT
rs338875 TTGCTCGCGTGTGCCAGCAAAT ACG
rs1385803 AAGTGGAATTCTCATGGCAGAT ACT
rs1385804 CATTCCAGAAGTGGAATTCTCATGACT
rs338876 AGGAAGGTGCTCCGGCCT ACG
rs189803 TGCTTCCCCCTTCCCCCT CGT
rs452215 TCTATTCTTCCACCCCCATCTTT ACT
rs641170 CTATTCTTCCACCCCCATCT ACT
rs584398 CTCTTATATAGCTCTATTCTTCC CGT
rs385330 AGGTGTCTGCAGATAATACATT ACG
rs429538 CCTGGGGCACAGGACAATA ACT
rs371229 I GACAATAGTTGGGGCAAGAC ACT
135
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs460874 ACAAAACTATCCTTCAAAAATACACGT
rs646121 GTTTTTGTTTCTCTGAAAGTGTCTCGT
rs468262 CACCCAACTACTTGCTCCC ACG
rs467863 GCTGATGGGAGGCCAATGT ACT
rs191434 GTCCAGAGATCCTGCTCACT CGT
rs2054782 CCCCCTCCATCACCTCCC ACG
rs468499 GTGAGCCAGCAATTCTCCTA ACT
rs180287 CAATGATCAGAACTCAGAGGTTTTACT
rs338877 AGAGATAAATTTCCAGTGTGAG CGT
rs650665 AGACATCCCGGCCGGGC ACT
rs193419 CCAAAGACACAGGGAGTAGATTA ACT
rs180288 GAGAATATTCTTGTGGGCTTAAT ACT
rs186834 CCAGGTCACACACACACTC ACG
rs189266 CACACACTCCCTCTCACTGT ACT
rs189267 TTCTGTGCATCTTTGACGCCATC CGT
rs170937 GATGGCGTCAAAGATGCACA ACT
rs463263 CCCCTTTGCACAGATGCTG ACT
rs463262 GGGGAGCAGCCAGTTCCTA ACT
rs460454 AGAGGCTGGGGACAGAGAA ACT
rs460455 GGTACCCACCAGTCTCCTTCT ACT
rs460505 CAGCGTCTCTGACACGGTC ACG
rs931316 GGTCAGAGCAGACACATCCACAT ACG
rs463431 CCCATGGGGAGCACCAAG ACT
rs461542 TGGGAGCTCCCGGGATATTGCC ACG
rs463557 GCTCCCGGGATATTGCCCA ACT
rs191453 CTGGGCTGGGGCCCTGC ACT
rs2271212 CGAGGAGGAGCCTGGCAG ACG
rs462009 CTCCTCGTTGCCTCCGGG ACT
rs2271211 GACGTAGCTGCCGACACCA ACG
rs396474 CTGGTGGCCCATCTATCCTGG ACT
rs428901 GAGCGAGACTCCGTCTCAA CGT
rs452300 CTGAAGAGGTCTTTCTCCTTCC CGT
rs670256 TTCTTCCACCCCCATCTTTG ACT
Genetic Anal,
[0284] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 35.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (Al AF
= 1-A2 AF). For
example, the SNP rs2278221 has the following case and control allele
frequencies: case A1 (C) = 0.36;
case A2 (T) = 0.64; control A1 (C) = 0.37; and control A2 (T) = 0.63, where
the nucleotide is provided
in paranthesis. Some SNPs are labeled "untyped" because of failed assays.
136
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
TABLE 35
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 6
rs2278221 210 178695460C/T 0.64 0.63 0.770
rs1650358 3608 178698858C/G
rs1643818 3609 178698859C/G
rs3733916 4318 178699568C/T
rs1624933 5593 178700843A/G 0.69 0.71 0.255
rs1624857 5629 178700879C/T 0.79 0.81 0.574
rs1624832 5639 178700889A/G 0.41 0.44 0.203
rs1624829 5640 178700890C/T 0.89 0.93 0.044
rs2161171 8943 178704193A/C
rs1530499 17968 178713218A/G 0.39 0.39 0.861
rs888764 19887 178715137A/G
rs873987 21034 178716284AlG
rs4078699 21085 178716335C/T 0.56 0.54 0.374
rs870311 21596 178716846A/G 0.51 0.50 0.590
rs1643817 23379 178718629A/C 0.27 NA NA
rs1643816 23432 178718682A/C
rs1650355 24007 178719257A/C
rs888763 26121 178721371A/G 0.40 0.42 0.390
rs1862212 26273 178721523A/T 0.55 0.54 0.753
rs1110514 26755 178722005A/T 0.29 0.28 0.572
rs3797600 27411 178722661C/T 0.56 0.57 0.738
rs3797602 27710 178722960GlT 0.65 0.64 0.564
rs3797603 27842 178723092C/T
rs3776819 28379 178723629C/T 0.46 0.46 0.850
rs252076 29603 178724853C/T 0.46 0.48 0.519
rs252075 31232 178726482C/G 0.35 0.36 0.859
rs252074 31504 178726754A/G 0.35 0.34 0.816
rs252068 32583 178727833C/G 0.47 0.48 0.656
rs252069 32794 178728044A/G 0.28 0.27 0.626
rs194040 32840 178728090C/T 0.31 0.32 0.665
rs252070 33044 178728294C/T 0.58 0.57 0.573
rs3797606 33150 178728400A/C 0.88 0.88 0.684
rs171667 33218 178728468A/G 0.48 0.51 0.166
rs187539 33513 178728763C/T 0.33 0.34 0.652
/TATCA
rs3836834 33959 178729209AACTAC
CATGAA
A
rs252071 34486 178729736A/G 0.30 0.31 0.666
rs252072 36289 178731539C/T 0.49 0.50 0.677
rs252073 36570 178731820C/T
rs379589 38247 178733497A/T 0.59 0.63 0.096
rs2052472 38477 178733727A/C 0.05 0.06 0.508
rs2052471 38518 178733768C/T 0.89 0.88 0.459
rs2052470 38529 178733779C/T 0.83 0.80 0.125
rs2052469 38667 178733917A/G 0.83 0.80 0.172
rs3797608 39781 178735031C/T 0.06 0.07 0.578
rs3797609 39856 178735106C/T 0.05 0.05 0.812
rs3822601 39927 178735177C/T 0.08 0.08 0.802
rs153131 40506 178735756A/G 0.76 0.77 0.944
rs751546 41869 178737119C/G 0.93 0.92 0.585
rs2279979 42452 178737702C/T 0.93 0.92 0.436
rs252060 44788 178740038C/T 0.81 0.82 0.760
rs3797610 46059 178741309A/C 0.17 0.17 0.858
rs194039 46846 178742096A/G 0.41 0.47 0.035
rs168773 47712 178742962A/T 0.35 0.38 0.266
rs252061 48796 178744046C/T 0.21 0.19 0.508
rs187537 49441 178744691C/G
rs252062 49602 178744852A/T 0.95 0.95 0.960
rs2431255 49723 178744973A/C 0.24 0.19 0.034
rs3797612 50050 178745300C/T 0.38 0.43 0.036
137
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position .ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID NO: AF AF
6
rs379761350171 178745421C/T 0.21 0.21 0.941
rs614114 50477 178745727C/T 0.50 0.53 0.387
rs252063 50818 178746068C/T 0.57 0.55 0.313
rs252064 50833 178746083C/T 0.52 0.52 0.806
rs252065 50881 178746131A/G 0.22 0.22 0.857
rs450502 50882 178746132A/G
rs439252 51386 178746636C/T
rs252066 51534 178746784C/T 0.19 0.18 0.618
rs457957 52317 178747567A/G 0.67 0.70 0.172
rs379761452368 178747618C/T
rs423552 52970 178748220A/G 0.90 0.92 0.215
rs398829 53023 178748273A/G
rs416646 53356 178748606A/G 0.56 0.57 0.650
rs187450 53882 178749132G/T
rs337807 54553 178749803C/T 0.55 0.59 0.208
rs337806 55475 178750725A/C 0.11 0.10 0.925
rs139643855530 178750780A/G 0.56 0.54 0.494
rs139643755691 178750941C/T
rs241181155848 178751098A/C
rs289881355879 178751129C/G
rs189256 56316 178751566A/G 0.19 0.19 0.988
rs173072 56911 178752161A/C
rs337805 57320 178752570A/G 0.25 0.24 0.657
rs191415 57391 178752641C/T
rs180045 57437 178752687C/T 0.51 0.47 0.211
rs189255 57478 178752728C/G 0.15 0.12 0.273
rs652766 57500 178752750C/T 0.57 0.61 0.213
rs466750 59111 178754361G/T 0.35 0.33 0.493
rs442406 59333 178754583A/G 0.57 0.59 0.420
rs662407 59715 178754965A/G 0.31 0.27 0.102
rs592971 59804 178755054A/G
rs457187 59851 178755101A/G 0.23 0.24 0.842
rs459490 59929 178755179C/T 0.21 0.20 0.604
rs459668 60052 178755302C/T 0.20 0.19 0.648
rs462646 60240 178755490C/T 0.43 0.43 0.905
rs458272 60359 178755609G/T 0.22 0.20 0.523
rs463455 60381 178755631AIG 0.25 0.24 0.644
rs675880 60456 178755706C/T 0.63 0.65 0.591
rs810617 60724 178755974C/G
rs464156 60875 178756125C/T 0.34 0.34 0.892
rs458083 60968 178756218A/G 0.80 0.82 0.499
rs467333 60978 178756228C/G 0.11 0.12 0.369
rs465381 60998 178756248C/T
rs466363 61557 178756807C/T 0.31 0.34 0.358
rs245709962091 178757341C/T 0.44 0.44 0.956
rs463901 62645 178757895C/T 0.43 0.45 0.395
rs465621 62943 178758193A/C 0.62 0.63 0.534
rs463724 63131 178758381A/T 0.09 0.08 0.523
rs465242 63145 178758395G/T
rs467419 63406 178758656A/G 0.65 0.66 0.647
rs456135 63427 178758677C/G 0.79 0.80 0.686
rs464536 63554 178758804C/T 0.36 0.34 0.296
rs461898 63661 178758911A/G 0.30 0.32 0.411
rs389558 64093 178759343A/G 0.24 0.26 0.325
rs466752 64153 178759403C/T 0.35 0.37 0.446
rs455655 64409 178759659C/G 0.87 0.89 0.536
rs463435 64544 178759794C/T 0.68 0.66 0.428
rs217497165257 178760507C/T 0.52 0.51 0.695
rs197997965626 178760876A/G 0.07 0.06 0.692
rs411804 65739 178760989A/G 0.78 0.78 0.976
rs162388566392 178761642C/T 0.82 0.80 0.492
rs164381166720 178761970C/T 0.24 0.24 0.924
rs434430 69177 178764427A/T
rs187538 ~ -69336 L 178764586I-.-G~ I f
13~
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 6
rs252067 69636 178764886 A/G 0.21 0.23 0.606
rs459319 69823 178765073 A/G 0.19 0.20 0.640
rs467289 69928 178765178 C/T 0.26 0.26 0.988
rs462644 70547 178765797 C/T 0.59 0.58 0.914
rs458752 70633 178765883 C/T 0.18 0.20 0.513
rs708320 71805 178767055 A/C
rs457954 72181 178767431 C/G 0.71 0.73 0.327
rs2411810 72200 178767450 C/T 0.28 0.26 0.252
rs3084687 72474 178767724 -lAT 0.13 0.12 0.884
rs69638 72567 178767817 C/G 0.54 0.52 0.449
rs455452 72973 178768223 A/G 0.59 0.60 0.733
rs464850 73468 178768718 A/G 0.11 0.09 0.249
rs431472 73889 178769139 A/G 0.33 0.34 0.713
rs2411809 75730 178770980 C/T
rs2457094 75970 178771220 A/G 0.71 0.73 0.383
rs2457095 76114 178771364 A/G 0.74 0.76 0.551
rs2261740 76342 178771592 C/T 0.35 0.36 0.702
rs1109180 76449 178771699 A/G
rs1109179 76465 178771715 C/T
rs1109178 76791 178772041 A/C 0.46 0.45 0.820
rs456909 78042 178773292 A/G 0.55 0.53 0.444
rs469124 80758 178776008 A/G
rs468039 80778 178776028 C/T
rs467017 81356 178776606 A/C 0.33 0.32 0.665
rs469290 81576 178776826 A/G 0.57 0.57 0.871
rs469090 81689 178776939 C/T 0.82 0.83 0.387
rs469568 81759 178777009 G/T 0.38 0.38 0.888
rs468386 81950 178777200 C/G
rs469349 82562 178777812 A/C
rs469099 83591 178778841 C/T 0.66 0.63 0.264
rs456868 83700 178778950 A/G
rs465389 83821 178779071 C/G
rs463892 83842 178779092 C/G
rs468548 83923 178779173 G/T
rs654612 83929 178779179 A/C
rs468542 84021 178779271 C/G
rs469262 84175 178779425 C/T 0.45 0.47 0.405
rs708323 84417 178779667 A/G 0.73 0.69 0.138
rs469089 84747 178779997 C/G
rs469396 85746 178780996 C/G 0.38 0.37 0.817
rs468723 86129 178781379 C/T 0.37 0.38 0.754
rs467604 86335 178781585 A/G 0.34 0.32 0.504
rs338874 87315 178782565 C/G 0.43 0.44 0.879
rs338875 87648 178782898 A/G 0.48 0.50 0.289
rs1385803 87764 178783014 A/C
rs1385804 87770 178783020 C/G
rs338876 88221 178783471 C/T 0.39 0.39 0.889
rs189803 90474 178785724 A/C
rs452215 91148 178786398 G/T
rs641170 91150 178786400 G/T
rs584398 91160 178786410 G/T
rs385330 91733 178786983 C/T
rs429538 91772 178787022 A/C
rs371229 91785 178787035 C/T
rs460874 93140 178788390 A/T 0.74 0.71 0.351
rs646121 93148 178788398 A/T 0.93 0.94 0.687
rs468262 96080 178791330 A/G
rs467863 96157 178791407 C/G
rs191434 96313 178791563 A/C
rs2054782 96759 178792009 C/T 0.44 0.42 0.353
rs468499 97026 178792276 A/C
rs180287 97320 178792570 C/G
rs338877 97732 178792982 A/T 0.04 0.04 0.863
rs650665 98713 178793963 C/G
139
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 6
rs193419 99707 178794957~A/C
rs180288 99959 178795209C/G
rs186834 100009 178795259A/G
rs189266 100020 178795270C/G
rs189267 100065 1787 95315A/C
rs170937 100086 178795336C/G
rs463263 101270 178796520C/G
rs463262 101276 1787 96526G/T
rs460454 101371 178796621C/T
rs460455 101376 178796626C/G
rs460505 101439 178796689C/T
rs931316 101820 178797070C/T
rs463431 102392 1787 97642ClG
rs461542 102602 178797852A/G
rs463557 102604 178797854A/C
rs191453 102896 178798146C/T 0.11 0.14 0.123
rs2271212189104 178884354C/T 0.65 0.57 0.003
rs462009 189134 1788 84384C/T
rs2271211189205 1788 84455A/G
rs396474 Not ma Not ma A/C
ed ed
rs428901 Not ma Not ma A/T 0.64 0.72 0.015
ed ed
rs452300 Not ma Not ma G/T
ed ed
rs670256 Not ma Not ma G/T
ed ed
[0285] The ADAMTS2 proximal SNPs were also allelotyped in the replication
cohorts using the
methods described herein and the primers provided in Tables 33 and 34. The
replication allelotyping
results for replication cohort #1 and replication cohort #2 are provided in
Tables 36 and 37,
respectively.
TABLE 36
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position AlleleCase Control Value
SEQ ID AF AF
NO: 6
rs2278221210 178695460 C/T 0.64 0.62 0.624
rs16503583608 1786 98858C/G
rs16438183609 1786 98859C/G
rs37339164318 178699568 C/T
rs16249335593 178700843 A/G 0.65 0.69 0.322
rs16248575629 178700879 C/T 0.81 unt ed NA
rs16248325639 178700889 A/G 0.38 0.42 0.265
rs16248295640 178700890 C/T 0.87 unt ed NA
rs21611718943 1787 04193A/C
rs 153049917968 178713218 A/G 0.39 0.40 0.765
rs888764 19887 178715137 A/G
rs873987 21034 178716284 A/G
rs407869921085 178716335 C/T 0.55 0.54 0.733
rs870311 21596 178716846 A/G 0.50 0.50 0.828
rs164381723379 178718629 A/C 0.27 unt ed
rs164381623432 178718682 A/C
rs165035524007 178719257 A/C
rs888763 26121 178721371 A/G 0.40 0.40 0.816
rs186221226273 178721523 A/T 0.55 0.55 0.936
rs111051426755 178722005 A/T 0.29 0.29 0.997
rs379760027411 178722661 C/T 0.57 0.58 0.604
rs379760227710 178722960 G/T 0.64 0.63 0.879
rs379760327842 178723092 C/T
rs377681928379 178723629 C/T 0.47 0.46 0.889
rs252076 29603 178724853 C/T 0.46 0.49 0.410
rs252075 31232 178726482 C/G 0.35 0.37 0.572
140
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 6
rs252074 31504 178726754 A/G 0.35 0.35 0.914
rs252068 32583 178727833 C/G 0.48 0.48 0.853
rs252069 32794 178728044 A/G 0.29 0.28 0.765
rs194040 32840 178728090 C/T 0.31 0.33 0.450
rs252070 33044 178728294 C/T 0.57 0.58 0.609
rs3797606 33150 178728400 A/C 0.87 0.91 0.119
rs171667 33218 178728468 A/G 0.45 0.50 0.125
rs187539 33513 178728763 C/T 0.33 0.34 0.709
/TATCA
rs3836834 33959 178729209 AACTAC
CATGAA
A
rs252071 34486 178729736 A/G 0.30 0.32 0.566
rs252072 36289 178731539 C/T 0.48 0.51 0.400
rs252073 36570 178731820 C/T
rs379589 38247 178733497 A/T 0.59 0.65 0.035
rs2052472 38477 178733727 A/C 0.04 0.06 0.493
rs2052471 38518 178733768 C/T 0.87 0.88 0.697
rs2052470 38529 178733779 C/T 0.84 0.78 0.036
rs2052469 38667 178733917 A/G 0.84 0.79 0.086
rs3797608 39781 178735031 C/T 0.06 0.07 0.530
rs3797609 39856 178735106 C/T 0.04 0.05 0.841
rs3822601 39927 178735177 C/T 0.08 0.08 0.904
rs153131 40506 178735756 A/G 0.77 0.77 0.964
rs751546 41869 178737119 C/G 0.94 0.92 0.265
rs2279979 42452 178737702 C/T 0.94 0.92 0.238
rs252060 44788 178740038 C/T 0.82 0.80 0.553
rs3797610 46059 178741309 A/C 0.16 0.18 0.459
rs194039 46846 178742096 A/G 0.43 0.45 0.589
rs168773 47712 178742962 A/T 0.34 0.35 0.845
rs252061 48796 178744046 C/T 0.23 0.22 0.884
rs187537 49441 178744691 C/G
rs252062 49602 178744852 A/T 0.98 0.96 0.310
rs2431255 49723 178744973 A/C 0.24 0.19 0.108
rs3797612 50050 178745300 C/T 0.42 0.46 0.254
rs3797613 50171 178745421 C/T 0.19 0.21 0.576
rs614114 50477 178745727 C/T 0.52 0.54 0.717
rs252063 50818 178746068 C/T 0.55 0.57 0.537
rs252064 50833 178746083 C/T 0.52 0.50 0.609
rs252065 50881 178746131 A/G 0.21 0.25 0.234
rs450502 50882 178746132 A/G
rs439252 51386 178746636 C/T
rs252066 51534 178746784 C/T 0.20 0.20 0.883
rs457957 52317 178747567 A/G 0.66 0.71 0.162
rs3797614 52368 178747618 C/T
rs423552 52970 178748220 A/G 0.90 0.92 0.380
rs398829 53023 178748273 A/G
rs416646 53356 178748606 A/G 0.58 0.59 0.915
rs187450 53882 178749132 G/T
rs337807 54553 178749803 C/T 0.60 NA NA
rs337806 55475 178750725 A/C 0.10 0.10 0.997
rs1396438 55530 178750780 A/G 0.52 0.57 0.188
rs1396437 55691 178750941 C/T
rs2411811 55848 178751098 A/C
rs2898813 55879 178751129 CIG
rs189256 56316 178751566 A/G 0.21 0.20 0.852
rs173072 56911 178752161 A/C
rs337805 57320 178752570 A/G 0.24 0.24 0.950
rs191415 57391 178752641 C/T
rs180045 57437 178752687 C/T 0.47 0.46 0.918
rs189255 57478 178752728 C/G 0.14 0.13 0.764
rs652766 57500 178752750 C/T 0.59 0.61 0.570
rs466750 59111 178754361 G/T 0.38 0.37 0.606
141
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 6
rs442406 59333 178754583A/G 0.56 0.57 0.882
rs662407 59715 178754965A!G 0.32 0.27 0.134
rs592971 59804 178755054A/G
rs457187 59851 178755101A/G 0.23 0.25 0.451
rs459490 59929 178755179C/T 0.22 0.21 0.671
rs459668 60052 178755302C/T 0.20 0.19 0.712
rs462646 60240 178755490C/T 0.42 0.44 0.439
rs458272 60359 178755609G/T 0.21 0.21 0.755
rs463455 60381 178755631A/G 0.25 0.25 0.783
rs675880 60456 178755706C/T 0.62 0.63 0.741
rs810617 60724 178755974C/G
rs464156 60875 178756125C/T 0.32 0.34 0.541
rs458083 60968 178756218A/G 0.80 0.82 0.499
rs467333 60978 178756228C/G 0.10 0.13 0.243
rs465381 60998 178756248C/T
rs466363 61557 178756807C/T 0.31 0.34 0.494
rs245709962091 178757341C/T 0.45 0.45 0.997
rs463901 62645 178757895C/T 0.46 0.46 0.852
rs465621 62943 178758193A/C 0.64 0.63 0.853
rs463724 63131 178758381A/T 0.09 0.08 0.737
rs465242 63145 178758395G/T
rs467419 63406 178758656A/G 0.64 0.65 0.694
rs456135 63427 178758677C/G 0.79 0.76 0.339
rs464536 63554 178758804C/T 0.36 0.34 0.553
rs461898 63661 178758911A/G 0.31 0.33 0.727
rs389558 64093 178759343A/G 0.27 0.28 0.762
rs466752 64153 178759403C/T 0.34 0.38 0.223
rs455655 64409 178759659C/G 0.87 unt ed NA
rs463435 64544 178759794C/T 0.65 0.65 0.973
rs217497165257 178760507C/T 0.49 0.51 0.476
rs197997965626 178760876A/G 0.08 0.07 0.579
rs411804 65739 178760989A/G 0.77 0.79 0.420
rs162388566392 178761642C/T 0.81 0.78 0.451
rs164381166720 178761970C/T 0.26 0.25 0.715
rs434430 69177 178764427A/T
rs187538 69336 178764586G/T
rs252067 69636 178764886A/G 0.22 0.22 0.978
rs459319 69823 178765073A/G 0.19 0.22 0.245
rs467289 69928 178765178C/T 0.26 0.29 0.377
rs462644 70547 178765797C/T 0.58 0.56 0.637
rs458752 70633 178765883C/T 0.18 0.23 0.129
rs708320 71805 178767055A/C
rs457954 72181 178767431C/G 0.69 0.73 0.143
rs241181072200 178767450C/T 0.28 0.23 0.083
rs308468772474 178767724-/AT 0.12 0.13 0.767
rs69638 72567 178767817C/G 0.53 0.49 0.157
rs455452 72973 178768223A/G 0.58 0.61 0.313
rs464850 73468 178768718A/G 0.13 0.10 0.171
rs431472 73889 178769139A/G 0.32 0.39 0.048
rs241180975730 178770980C/T
rs245709475970 178771220A/G 0.70 0.75 0.157
rs245709576114 178771364A/G 0.74 0.75 0.707
rs226174076342 178771592C/T 0.34 unt ed NA
rs110918076449 178771699A/G
rs110917976465 178771715C/T
rs110917876791 178772041A/C 0.47 0.48 0.715
rs456909 78042 178773292A/G 0.56 0.54 0.537
rs469124 80758 178776008A/G
rs468039 80778 178776028C/T
rs467017 81356 178776606A/C 0.33 0.31 0.480
rs469290 81576 178776826A/G 0.63 0.66 0.427
rs469090 81689 178776939C/T 0.80 0.83 0.300
rs469568 81759 178777009G/T 0.39 0.43 0.234
rs468386 81950 178777200C/G
142
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 6
rs469349 82562 178777812A/C
rs469099 83591 178778841C/T 0.66 0.60 0.066
rs456868 83700 178778950A/G
rs465389 83821 178779071C/G
rs463892 83842 178779092C/G
rs468548 83923 178779173G/T
rs654612 83929 178779179A/C
rs468542 84021 178779271C/G
rs469262 84175 178779425C/T 0.46 0.50 0.232
rs708323 84417 178779667A/G 0.72 0.66 0.071
rs469089 84747 178779997C/G
rs469396 85746 178780996C/G 0.37 0.35 0.522
rs468723 86129 178781379C/T 0.39 0.41 0.495
rs467604 86335 178781585A/G 0.33 0.30 0.303
rs338874 87315 178782565C/G 0.44 0.46 0.628
rs338875 87648 178782898A/G 0.49 0.54 0.106
rs138580387764 178783014A/C
rs138580487770 178783020C/G
rs338876 88221 178783471C/T 0.38 0.36 0.609
rs1898p3 90474 178785724A/C
rs452215 91148 178786398G/T
rs641170 91150 178786400G/T
rs584398 91160 178786410G/T
rs385330 91733 178786983C/T
rs429538 91772 178787022A/C
rs371229 91785 178787035C/T
rs460874 93140 178788390A/T 0.74 0.69 0.118
rs646121 93148 178788398A/T 0.93 0.95 0.477
rs468262 96080 178791330A/G
rs467863 96157 178791407C/G
rs191434 96313 178791563A/C
rs205478296759 178792009C/T 0.45 0.42 0.514
rs468499 97026 178792276A/C
rs180287 97320 178792570C/G
rs338877 97732 178792982A/T 0.04 0.04 0.781
rs650665 98713 178793963C/G
rs193419 99707 178794957A/C
rs180288 99959 178795209C/G
rs186834 100009 178795259A/G
rs189266 100020 178795270C/G
rs189267 100065 178795315A/C
rs170937 100086 178795336C/G
rs463263 101270 178796520C/G
rs463262 101276 178796526G/T
rs460454 101371 178796621C/T
rs460455 101376 178796626C/G
rs460505 101439 178796689C/T
rs931316 101820 178797070C/T
rs463431 102392 178797642C/G
rs461542 102602 178797852A/G
rs463557 102604 178797854A/C
rs191453 102896 178798146C/T 0.15 0.19 0.139
rs2271212189104 178884354C/T 0.64 0.58 0.072
rs462009 189134 178884384C/T
rs2271211189205 178884455A/G
rs396474 Not ma Not ma A/C
ed ed
rs428901 Not ma Not ma AIT 0.66 unt ed NA
ed ed
rs452300 Not ma Not ma G/T
ed ed
rs670256 Not ma Not ma G/T
ed ed
143
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
TABLE 37
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in PositionAllele Case Control Value
SEQ ID AF AF
NO: 6
rs2278221210 178695460C/T 0.64 0.64 0.837
rs16503583608 178698858C/G
rs16438183609 178698859C/G
rs37339164318 178699568CIT
rs16249335593 178700843A/G 0.73 0.75 0.447
rs16248575629 178700879C/T 0.78 0.81 0.289
rs16248325639 178700889A/G 0.44 0.47 0.423
rs16248295640 178700890C/T 0.90 0.93 0.294
rs21611718943 178704193A/C
rs153049917968 178713218A/G 0.39 0.36 0.499
rs888764 19887 178715137A/G
rs873987 21034 178716284A/G
rs407869921085 178716335C/T 0.57 0.54 0.316
rs870311 21596 178716846A/G 0.52 0.50 0.579
rs164381723379 178718629A/C
rs164381623432 178718682A/C
rs165035524007 178719257A/C
rs888763 26121 178721371A/G 0.40 0.44 0.264
rs186221226273 178721523A/T 0.56 0.53 0.529
rs1 'I 26755 178722005A/T 0.30 0.27 0.381
10514
rs379760027411 178722661C/T 0.55 0.54 0.840
rs379760227710 178722960G/T 0.68 0.65 0.534
rs379760327842 178723092C/T
rs377681928379 178723629C/T 0.45 0.47 0.662
rs252076 29603 178724853C/T 0.46 0.46 0.986
rs252075 31232 178726482C/G 0.36 0.34 0.666
rs252074 31504 178726754A/G 0.35 0.33 0.604
rs252068 32583 178727833C/G 0.47 0.48 0.648
rs252069 32794 178728044A/G 0.27 0.26 0.640
rs194040 32840 178728090C/T 0.31 0.30 0.734
rs252070 33044 178728294C/T 0.61 0.55 0.157
rs379760633150 178728400A/C 0.91 0.83 0.005
rs171667 33218 178728468A/G 0.51 0.52 0.674
rs187539 33513 178728763C/T 0.32 0.33 0.836
/TATCA
rs383683433959 178729209AACTAC
CATGAA
A
rs252071 34486 178729736A/G 0.30 0.30 0.942
rs252072 36289 178731539C/T 0.50 0.49 0.684
rs252073 36570 178731820C/T
rs379589 38247 178733497A/T 0.60 0.61 0.981
rs205247238477 178733727A/C 0.06 0.06 0.856
rs205247138518 178733768C/T 0.91 0.86 0.079
rs205247038529 178733779C/T 0.82 0.83 0.828
rs205246938667 178733917A/G 0.82 0.82 0.983
rs379760839781 178735031C/T 0.06 0.06 0.969
rs379760939856 178735106C/T 0.05 0.05 0.879
rs382260139927 178735177C/T 0.07 0.08 0.838
rs'15313140506 178735756A/G 0.76 0.76 0.981
rs751546 41869 178737119C/G 0.91 0.92 0.526
rs227997942452 178737702C/T 0.92 0.92 0.906
rs252060 44788 178740038C/T 0.81 0.85 0.157
rs379761046059 178741309A/C 0.18 0.16 0.593
rs'19403946846 178742096A/G 0.39 0.49 0.005
rs'16877347712 178742962A/T 0.37 0.43 0.098
rs252061 48796 178744046C/T 0.19 0.15 0.164
rs'18753749441 178744691C/G
rs252062 49602 178744852A/T 0.93 0.95 0.290
rs243125549723 178744973A/C 0.23 0.19 0.201
rs379761250050 178745300C/T 0.32 0.38 0.102
144
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ I~ AF AF
NO: 6
rs379761350171 178745421C/T 0.23 NA
rs614114 50477 178745727CIT 0.48 0.51 0.423
rs252063 50818 178746068C/T 0.60 0.51 0.011
rs252064 50833 178746083C/T 0.51 0.56 0.265
rs252065 50881 178746131A/G 0.22 0.18 0.175
rs450502 50882 178746132AIG
rs439252 51386 178746636C/T
rs252066 51534 178746784C/T 0.18 0.16 0.451
rs457957 52317 178747567A/G 0.67 0.68 0.728
rs379761452368 178747618C/T
rs423552 52970 178748220A/G 0.89 0.91 0.398
rs398829 53023 178748273A/G
rs416646 53356 178748606AIG 0.54 0.55 0.643
rs187450 53882 178749132G/T
rs337807 54553 178749803C/T 0.49 0.59 0.009
rs337806 55475 178750725A/C 0.11 0.10 0.889
rs139643855530 178750780A/G 0.61 0.50 0.007
rs139643755691 178750941CIT
rs241181155848 178751098A/C
rs289881355879 178751129C/G
rs189256 56316 178751566A/G 0.17 0.17 0.923
rs173072 56911 178752161A/C
rs337805 57320 178752570AIG 0.27 0.25 0.582
rs191415 57391 178752641C/T
rs180045 57437 178752687C/T 0.56 0.48 0.115
rs189255 57478 178752728C/G 0.16 0.12 0.168
rs652766 57500 178752750CIT 0.55 0.61 0.231
rs466750 59111 178754361G/T 0.31 0.28 0.473
rs442406 59333 178754583A/G 0.58 0.63 0.209
rs662407 59715 178754965A/G 0.30 0.28 0.449
rs592971 59804 178755054A/G
rs457187 59851 178755101A/G 0.23 0.21 0.402
rs459490 59929 178755179C/T 0.20 0.19 0.708
rs459668 60052 178755302C/T 0.21 0.20 0.821
rs462646 60240 178755490C/T 0.44 0.41 0.460
rs458272 60359 178755609G/T 0.22 0.20 0.524
rs463455 60381 178755631A/G 0.23 0.22 0.629
rs675880 60456 178755706C/T 0.65 0.67 0.564
rs810617 60724 178755974C/G
rs464156 60875 178756125C/T 0.37 0.34 0.439
rs458083 60968 178756218A/G
rs467333 60978 178756228C/G 0.11 0.11 0.902
rs465381 60998 178756248C/T
rs466363 61557 178756807C/T 0.32 0.34 0.547
rs245709962091 178757341C/T 0.43 0.43 0.974
rs463901 62645 178757895C/T 0.39 0.43 0.342
rs465621 62943 178758193A/C 0.59 0.64 0.195
rs463724 63131 178758381A/T 0.09 0.07 0.539
rs465242 63145 178758395G/T
rs467419 63406 178758656A/G 0.66 0.67 0.752
rs456135 63427 178758677C/G 0.79 0.85 0.029
rs464536 63554 178758804C/T 0.36 0.32 0.332
rs461898 63661 178758911A/G 0.28 0.31 0.423
rs389558 64093 178759343A/G 0.20 0.23 0.311
rs466752 64153 178759403C/T 0.36 0.35 0.781
rs455655 64409 178759659C/G NA 0.72 NA
rs463435 64544 178759794C/T 0.72 0.68 0.230
rs217497165257 178760507C/T 0.56 0.51 0.142
rs197997965626 178760876A/G 0.05 0.05 0.993
rs411804 65739 178760989A/G 0.80 0.77 0.343
rs162388566392 178761642C/T 0.84 0.84 0.819
rs164381166720 178761970C/T 0.22 0.23 0.847
rs434430 69177 178764427A/T
rs187538 69336 178764586G/T
145
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in PositionAllele Case Control Value
SEQ ID AF AF
NO: 6
rs252067 69636 178764886_ 0.21 0.24 0.369
A/G
rs459319 69823 178765073A/G 0.18 0.15 0.353
rs467289 69928 178765178C/T 0.27 0.22 0.179
rs462644 70547 178765797C/T 0.60 0.61 0.609
rs458752 70633 178765883C/T 0.18 0.15 0.271
rs708320 71805 178767055A/C
rs457954 72181 178767431C/G 0.72 0.72 0.882
rs241181072200 178767450C/T 0.29 0.30 0.630
rs308468772474 178767724-/AT 0.13 0.11 0.509
rs6 9638 72567 178767817ClG 0.54 0.57 0.440
rs455452 72973 178768223A/G 0.60 0.58 0.499
rs464850 73468 178768718A/G 0.10 0.09 0.839
rs431472 73889 178769139A/G 0.35 0.27 0.025
rs241180975730 178770980C/T
rs245709475970 178771220A/G 0.71 0.70 0.792
rs245709576114 178771364A/G 0.75 0.76 0.602
rs226174076342 178771592C/T 0.36 0.36 0.924
rs11 0918076449 178771699A/G
rs110917976465 178771715C/T
rs110917876791 178772041A/C 0.45 0.42 0.420
rs456909 78042 178773292A/G 0.53 0.51 0.598
rs469124 80758 178776008A/G
rs468039 80778 178776028C/T
rs467017 81356 178776606A/C 0.34 0.35 0.762
rs469290 81576 178776826A/G 0.49 0.44 0.223
rs469090 81689 178776939C/T 0.83 0.84 0.883
rs469568 81759 178777009G/T 0.36 0.30 0.115
rs468386 81950 178777200C/G
rs469349 82562 178777812A/C
rs469099 83591 178778841C/T 0.65 0.67 0.560
rs456868 83700 178778950A/G
rs465389 83821 178779071C/G
rs463892 83842 178779092C/G
rs468548 83923 178779173G/T
rs654612 83929 178779179A/C
rs468542 84021 178779271C/G
rs469262 84175 178779425C/T 0.45 0.43 0.762
rs708323 84417 178779667A/G 0.74 0.74 0.899
rs469089 84747 178779997C/G
rs469396 85746 178780996C/G 0.39 0.42 0.569
rs468723 86129 178781379C/T 0.36 0.34 0.573
rs467604 86335 178781585A/G 0.35 0.36 0.763
rs338874 87315 178782565C/G 0.42 0.40 0.564
rs338875 87648 178782898A/G 0.46 0.45 0.701
rs138580387764 178783014A/C
rs138580487770 178783020C/G
rs338876 88221 178783471C/T 0.41 0.44 0.580
rs189803 90474 178785724A/C
rs452215 91148 178786398G/T
rs641170 91150 178786400G/T
rs584398 91160 178786410G/T
rs385330 91733 178786983C/T
rs429538 91772 178787022A/C
rs371229 91785 178787035C/T
rs460874 93140 178788390A/T 0.73 0.75 0.550
rs646121 93148 178788398A/T 0.93 0.92 0.697
rs468262 96080 178791330A/G
rs467863 96157 178791407C/G
rs191434 96313 178791563A/C
rs205478296759 178792009C/T 0.43 0.40 0.473
rs468499 97026 178792276A/C
rs180287 97320 178792570C/G
rs338877 97732 178792982A/T 0.04 0.04 0.928
rs6 5066598713 178793963C/G
146
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 ~ F F A2 F p-
rs# in Pos Allele A2 Control Value
SEQ ID itio Case AF
NO: 6 n AF
rs 19341999707 _ A/C
_
178794957
rs 18028899959 178795209C/G
rs186834 100009 178795259A/G
rs189266 100020 178795270C/G
rs 189267100065 178795315A/C
rs170937 100086 178795336C/G
rs463263 101270 178796520C/G
rs463262 101276 178796526G/T
rs460454 101371 178796621C/T
rs460455 101376 178796626C/G
rs460505 101439 178796689C/T
rs 931316101820 178797070C/T
rs463431 102392 178797642C/G
rs461542 102602 178797852A/G
rs463557 102604 178797854A/C
rs 191453102896 178798146C/T 0.06 0.06 0.929
rs2271212189104 178884354C/T 0.66 0.56 0.012
rs462009 189134 178884384C/T
rs2271211189205 178884455A/G
rs396474 Not ma Not ma A/C
ed ed
rs428901 Not ma Not ma A/T 0.61 0.72 0.002
ed ed
rs452300 Not ma Not ma G/T
ed ed
rs670256 Not ma Not ma G/T
ed ed
[0286] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure lE for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in Figure
lE can be determined by consulting Table 35. For example, the left-most ~ on
the left graph is at
position 17 8695460. By proceeding down the Table from top to bottom and
across the graphs from left
to right the allele frequency associated with each symbol shown can be
determined.
[0287] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a lOkb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-$ were truncated at that
value.
147
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0288] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 9
BVES Proximal SNPs
[0289] It was discovered that rs 1018810, an intronic SNP in the BYES gene, is
associated with
occurrence of osteoarthritis in subjects. BYES was identified as a blood
vessel epicardial substance.
Sequence analysis predicted 3 transmembrane helices with an extracellular C
terminus. Northern blot
analysis revealed that expression of an approximately 5.5-kb BYES transcript
is restricted to skeletal
muscle and adult and fetal heart. BYES is highly expressed in osteoarthritic
cartilage according to EST
database analysis, and may play a role in chondrocyte and/or bone cell
development. BYES biological
activity may be modulated by addition of an antibody, a recombinant binding
partner, a binding agent,
or a recombinant BVES protein or functional fragment thereof.
[0290] One hundred fifty-four additional allelic variants proximal to rs
1018810 were identified
and subsequently allelotyped in osteoarthritis case and control sample sets as
described in Examples 1
and 2. The polymorphic variants are set forth in Table 38. The chromosome
positions provided in
column four of Table 38 are based on Genome "Build 34" of NCBI's GenBank.
TABLE 38
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
7
rs24000806 241 105557091 A/G
rs69302096 801 105557651 A/G
rs221628 6 899 105557749 A/G
rs221629 6 2091 105558941 C/G
rs221630 6 2290 105559140 C/T
rs221631 6 2440 105559290 A/G
rs11492846 4959 105561809 G/T
rs221633 6 7914 105564764 C/G
rs423366 6 7969 105564819 A/G
rs436460 6 7972 105564822 C/T
rs22110106 10831 105567681 C/T
rs379908 6 12399 105569249 C/T
rs11492856 13841 105570691 C/T
rs73411946 14461 105571311 C/T
rs715153 6 14680 105571530 C/T
rs221634 6 16808 105573658 A/T
rs77573076 18231 105575081 C/T
rs221fi356 18394 105575244 C/T
rs41454186 18505 105575355 G/T
rs221636 6 18684 105575534 A/T
rs31859586 19257 105576107 C/T
rs49466546 20263 105577113 A/T
148
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
7
rs2216376 20656 105577506 A/C
rs2216386 21499 105578349 A/G
rs2216396 21563 105578413 AIC
rs6435456 21612 105578462 C/G
rs2216406 21834 105578684 C/T
rs39576966 22406 105579256 A/T
rs39955546 22408 105579258 A/T
rs74535026 22685 105579535 A/T
rs11904716 23303 105580153 C/T
rs2216416 23306 105580156 C/G
rs2216426 25139 105581989 A/G
rs11904726 25211 105582061 C/T
rs11904736 25364 105582214 A/G
rs1864046 25381 105582231 A/C
rs2216436 25414 105582264 A/T
rs2216446 25835 105582685 C/T
rs12034756 26214 105583064 A/G
rs2216456 27224 105584074 A/G
rs1702776 27526 105584376 A/G
rs2216466 27934 105584784 C/T
rs2216476 28550 105585400 C/T
rs2216486 29015 105585865 A/G
rs2216496 29879 105586729 G/T
rs2216506 29979 105586829 A/G
rs11492876 30030 105586880 A/G
rs2216516 30585 105587435 C/T
rs77625916 31753 105588603 C/G
rs77485556 31934 105588784 C/T
rs58788336 33227 105590077 -/T
rs58788346 33228 105590078 -/T
rs2216526 35172 105592022 C/T
rs2216536 36901 105593751 A/G
rs2216546 36921 105593771 A/G
rs2216556 36932 105593782 A/G
rs2216566 37061 105593911 C/T
rs2216576 37570 105594420 C/T
rs2216586 38745 105595595 G/T
rs1100656 38970 105595820 A/T
rs2216596 39725 105596575 C/T
rs2216606 40070 105596920 A/C
rs77428216 40460 105597310 C/G
rs2216626 41470 105598320 A/G
rs77484266 41562 105598412 A/G
rs69114946 41956 105598806 A/G
rs69398466 42047 105598897 A/T
rs3684716 42280 105599130 A/G
rs4301906 42358 105599208 A/G
rs4551146 42629 105599479 C/G
rs4059566 43075 105599925 C/T
rs58788356 43387 105600237 -/A
rs14738146 43393 105600243 G/T
rs4232726 43438 105600288 C/T
149
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO: 7
rs4138066 441 15 105600965 A/G
rs49466556 44537 105601387 A/G
rs69156326 45642 105602492 A/G
rs20957236 46629 105603479 A/G
rs74500786 47496 105604346 A/G
rs74530716 47515 105604365 A/C
rs90188906 48329 105605179 A/G
rs74509446 48862 105605712 C/G
rs77486576 48908 105605758 A/G
rs10131376 49038 105605888 C/T
rs58788366 49080 105605930 -/T
rs19814806 50204 105607054 A/T
rs19814796 50404 105607254 A/G
rs30351876 50426 105607276 -/TTA
rs74539936 50531 105607381 C/T
rs20011196 50 840 105607690 C/T
rs20011186 50964 105607814 C/T
rs20011176 50971 105607821 C/T
rs69404336 51 378 105608228 C/T
rs13187466 52610 105609460 A/C
rs7630996 53906 105610756 A/T
rs58788376 53951 105610801 -/C
rs9647316 54111 105610961 A/C
rs9647306 54149 105610999 G/T
rs69218696 555f>3 105612413 C/G
rs39450296 55999 105612849 C/T
rs49457156 58415 105615265 C/G
rs77752526 58 961 105615811 C/G
rs77420986 60447 105617297 C/T
rs37572896 61 377 105618227 A/G
rs69054586 61 528 105618378 A/G
rs37572906 61606 105618456 C/G
rs22752896 62 1 40 105618990 A/G
rs49457166 62461 105619311 C/T
rs69226386 63 826 105620676 C/T
rs77395726 64950 105621800 G/T
rs69011876 65 076 105621926 G/T
rs49466566 66 1 21 105622971 C/T
rs13380206 66406 105623256 C/T
rs77714726 67 051 105623901 A/C
rs69262606 68 860 105625710 C/T
rs69266276 69 014 105625864 C/T
rs49466576 70 796 105627646 C/T
rs65712186 72325 105629175 G/T
rs74499446 73414 105630264 A/C
rs9521756 75258 105632108 C/G
rs18902286 76 347 105633197 A/G
rs19332376 76 839 105633689 A/C
rs13380196 77 358 105634208 A/G
rs74531276 77 822 105634672 A/G
rs73815516 77 946 105634796 G/T
rs6571219I - ~ _ $0002 ~ 105636852
6 -
150
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
7
rs65712206 80024 1 05636874 A/G
rs21850176 80285 1 05637135 A/G
rs15917206 80397 1 05637247 C/G
rs69250466 82075 1 05638925 C/T
rs69404236 82153 1 05639003 A/G
rs11902746 83981 1 05640831 A/G
rs11902766 84184 1 05641034 A/G
rs15917196 85089 1 05641939 C/T
rs19332366 85288 1 05642138 A/G
rs69052026 85330 1 05642180 C/T
rs12091506 85581 1 05642431 A/T
rs11902776 85642 1 05642492 A/G
rs69262786 86433 1 05643283 A/G
rs11902786 86904 1 05643754 A/G
rs46264636 88391 1 05645241 A/G
rs69246206 89042 105645892 C/T
rs11902806 90828 1 05647678 G/T
rs45575526 92676 1 05649526 C/T
rs69327116 92881 1 05649731 C/T
rs16861406 94227 1 05651077 G/T
rs11902816 94585 1 05651435 A/G
rs23081626 94616 105651466 -/ATAA
rs11902826 94712 1 05651562 C/G
rs17659076 94738 1 Q5651588 A/G
rs58788386 95253 105652103 -/G
rs11902836 95522 1 05652372 A/G
rs11902846 95869 1 05652719 G/T
rs11902856 97856 1 05654706 C/T
Assay for Verifying and Allelotypin~ SNPs
[0291] The methods used to verify and allelotype the 154 proximal SNPs of
Table 38 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 39 and Table 40, respectively.
TABLE 39
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs2400080ACGTTGGATGGTGCCCAGCAAGTGATGATAACGTTGGATGACAGAGCAAGACTCCATCTC
rs6930209ACGTTGGATGGCTCTGTGGTGCATATTTACACGTTGGATGGGTTCTCTCACTTAACTGTG
rs221628ACGTTGGATGAGTGAGAGAACCAAATGTTGACGTTGGATGCCAGTTTTGGCTTCATTTGC
rs221629ACGTTGGATGTCTGTCCATTTCTCCCTCTGACGTTGGATGGCTGATTCTTGGCAAAAGGC
rs221630ACGTTGGATGTCCTTCTCATTGCTGTGTAGACGTTGGATGTCATGTGCAAGAGCCAAAAG
rs221631ACGTTGGATGCACTGGCCCTCTATAAATGCACGTTGGATGCCAGCCCCCTGCATTATTAT
rs1149284ACGTTGGATGGATGAGAAATTAACTAGACACACGTTGGATGGTCCATTTGGTTTTCATTTG
rs221633ACGTTGGATGCTTAACAATTTGTCTTGGAGACGTTGGATGAGCCACATATACCAAAAAAC
rs423366ACGTTGGATGAGCCACATATACCAAAAAACACGTTGGATGGAGATCTTTGCATGTCAATAC
rs436460ACGTTGGATGAGCCACATATACCAAAAAACACGTTGGATGGAGATCT't-f'GCATGTCAATAC
151
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs2211010ACGTTGGATGTTTTTTGAGACAGAGTCTCGACGTTGGATGTTTGCAGTGAGCTGAGATTG
rs379908ACGTTGGATGTGAGTGGGCAAAATGGTTCCACGTTGGATGCTCTCCTGCAGACACATCAA
rs1149285ACGTTGGATGCCAAATACATTTATGACTCCACGTTGGATGGAGAGAGATTCCATCTCAAA
rs7341194ACGTTGGATGCTGTAGAAACCAGCTAAACTGACGTTGGATGCTGACTAGACTCTGACTTTC
rs715153ACGTTGGATGTT'f-f'GTTGAATATTCGCTGCACGTTGGATGCTTCCATATAGAAAGGATTCC
rs221634ACGTTGGATGTGCCCATAACATCTAGAGCCACGTTGGATGTTGGTCCTGTTAGGTTTCGG
rs7757307ACGTTGGATGTGCTTAAGTTGAACAGTGCCACGTTGGATGGCAAAGTCTCCAAACATTTCC
rs221635ACGTTGGATGGGCAGCACAGACAGTAAATGACGTTGGATGTGCAGGTATTCATGCTAGGC
rs4145418ACGTTGGATGTGCATTGCCAGTCTCTTAGCACGTTGGATGGGCCTTCTAGTGAAGACTAG
rs221636
rs3185958ACGTTGGATGGACACAGATCATACAACCACACGTTGGATGAGCATCAAACTCTGTCTTAC
rs4946654ACGTTGGATGATGTAGTCAGAAGAGTGGTCACGTTGGATGGGTACTGATAAAATTTGCCC
rs221637ACGTTGGATGCAATCGTAGCTTACTGTGGGACGTTGGATGCTGTAGTCCAGCTACTCAAG
rs221638ACGTTGGATGCACACCTGGCTGAAAATCTTAACGTTGGATGTGGTTATTTCTAGGCGATGG
rs221639ACGTTGGATGCCCGCATGTGTATGTATCTCACGTTGGATGCCCATCGCCTAGAAATAACC
rs643545ACGTTGGATGAAAATCACCCGCATGTGTATACGTTGGATGCGCCTAGAAATAACCATTAGC
rs221640ACGTTGGATGTAATCCCAGCACTTTGGGAGACGTTGGATGTTTCACCATGTTAGCCAGGC
rs3957696ACGTTGGATGAACCAGTATGTTGCCCTTTCACGTTGGATGCCAGGCAGTCCAAATTAATTC
rs3995554ACGTTGGATGAACCAGTATGTTGCCCTTTCACGTTGGATGCCAGGCAGTCCAAATTAATTC
rs7453502ACGTTGGATGCTCCAAGGTTGGAGTTTGTGACGTTGGATGT'rfCTGAGCTCCTCAGCATC
rs1190471ACGTTGGATGATATGTGGCCCGATGATCTCACGTTGGATGCCTCCCAAAGTGCTAGGATT
rs221641ACGTTGGATGCCTCCCAAAGTGCTAGGATTACGTTGGATGATATGTGGCCCGATGATCTC
rs221642ACGTTGGATGTCTTCCACCATGATTGTGAGACGTTGGATGAGACATACCTGAGACTGGAC
rs1190472ACGTTGGATGTGTCCAGTCTCAGGTATGTCACGTTGGATGGCCCAGCTAAGGTTTTGTAG
rs1190473ACGTTGGATGTTGATCACACCACTGCACTCACGTTGGATGCCCCAATGAAGAAGTCTTGC
rs186404ACGTTGGATGTTGATCACACCACTGCACTCACGTTGGATGCCCCAATGAAGAAGTCTTGC
rs221643ACGTTGGATGCCCCAATGAAGAAGTCTTGCACGTTGGATGGAGACACAGTGAGACTGTCA
rs221644ACGTTGGATGGTGTCTTTCCTAGCTAGCTCACGTTGGATGTTACAGATGGGTTCAGGGAG
rs1203475ACGTTGGATGTAATCCCAGCTACTTGGGAGACGTTGGATGACAATCTCGGCTCACTGCAA
rs221645ACGTTGGATGTGTTTTTCATCTGCCCAATGACGTTGGATGGCTGCTGTTAAGGACCACAT
rs170277ACGTTGGATGACAAGGAAGTTCTGAACCTCACGTTGGATGTTTTGGATCAAGAGGTGACC
rs221646ACGTTGGATGAATTGGCTCTTCTCTCTGCCACGTTGGATGTTACAGCAGAAATGGCTGGA
rs221647ACGTTGGATGTTCCCAGCTCCTTTCCTTAGACGTTGGATGTTCCTAAGAAAATGCCCCTC
rs221648ACGTTGGATGATCATGCCACTGCACTCCAGACGTTGGATGTTAGGTCTCCAGGACGACAG
rs221649ACGTTGGATGGACAGGATGAAGAAGAAGGCACGTTGGATGTCTTGCTATTCGCCAAGGAC
rs221650ACGTTGGATGTAATATCCAGGATCCAGCTGACGTTGGATGTTGAACCCCTGAACTCAAGC
rs1149287ACGTTGGATGATGGAGGTCTCACCATGTTCACGTTGGATGTAGCACTTTGGGAGGCCAAG
rs221651ACGTTGGATGGGAGGATCACTTGAATCCAGACGTTGGATGAGACAGGTTCTTGCTCTGTT
rs7762591ACGTTGGATGATCTCTGCTCAC'rGCAGCTTACGTTGGATGAAATTAGCCAGGTGTGGTGG
rs7748555ACGTTGGATGTTGGGATTACAGGTGTGAGCACGTTGGATGCCCACTGCTTCACTTGACTA
rs5878833ACGTTGGATGACACTGTCTACACTGCCTTCACGTTGGATGACCTGACTTCAAAGGTCCTG
rs5878834ACGTTGGATGACACTGTCTACACTGCCTTCACGTTGGATGACCTGACTTCAAAGGTCCTG
rs221652ACGTTGGATGTACTTTCTACTCAGGGAAGGACGTTGGATGAGTTTACACGCGCATAAGAC
rs221653ACGTTGGATGGTTTCACTGTGTTAGCCAGGACGTTGGATGTAATCCCAGCACTCTGGGAG
rs221654ACGTTGGATGGAGATCAAGACCATCCTGGCACGTTGGATGAGTAGCTGGGACTACAGGCA
rs221655ACGTTGGATGGTCAGGAGATCAAGACCATCACGTTGGATGCCGCGCCCAGCTAATTTTTT
rs221656ACGTTGGATGAGATGGAGTTTCACTCTGTCACGTTGGATGAATCCAGGAGGTGGAGTTTG
rs221657ACGTTGGATGAGAACTCTTCCATCCTTGAC~1CGTTGGATGTTCTGCTTTAGTGCATCCAG
rs221658ACGTTGGATGCCAGCTGAGTTCAGCATTTGACGTTGGATGACACCCATATCTTCGCTACC
rs110065ACGTTGGATGTGACATGCTCATAGCCCTTGACGTTGGATGAGATCAGCTGTCATTCACTG
rs221659ACGTTGGATGCGAAACACAACCTCTACTTCACGTTGGATGCAGGTAAGGAAATTAAGGCAC
rs221660ACGTTGGATGAATATGATGGAAACCAGGGCACGTTGGATGTCTTAGCTCTCTTGAGTGTG
152
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs7742821ACGTTGGATGAGCTCTTGGGAAGTTCTCACACGTTGGATGCCCAACTCTCTCACCTATAC
rs221662ACGTTGGATGGACAATGGGTTAAATGTTGGGACGTTGGATGAAGTGCTTTGAGTTTCTGAG
rs7748426ACGTTGGATGATTCACCCTCACCACATCTGACGTTGGATGCCACCCCTCTCTGTTTTCTT
rs6911494ACGTTGGATGTCAATGGTACAGAAGGCCAGACGTTGGATGAACCCCTCGCTTGGAATTAG
rs6939846ACGTTGGATGTCCTCAAAGCTGGGCTTTCTACGTTGGATGAGACAAAAGGATCACCTGCC
rs368471ACGTTGGATGCCCCTAATACATCCAAAACCACGTTGGATGACCAGGCAAACCTGTAGAAG
rs430190ACGTTGGATGTCTCTGGAAGATAGTTGGGCACGTTGGATGACTTCTACAGG'tTtGCCTGG
rs455114ACGTTGGATGCCCAGAAAATTGATTCTTAGACGTTGGATGACAGAAGTCTTTTCCTGATC
rs405956ACGTTGGATGAAACTCCAAGTCAAGGACCCACGTTGGATGAAAGGTGTCCACTGTTTCGC
rs5878835ACGTTGGATGCTGTCTTCCAGAGTCTTGAGACGTTGGATGTACATCCACTATGTACCCAC
rs1473814ACGTTGGATGGTTAAAGAACCACAGAAGGCACGTTGGATGTACATCCACTATGTACCCAC
rs423272ACGTTGGATGCACAGAAGGCCTTAAAAACCACGTTGGATGTCACGTTGCATTCCTGTATC
rs413806ACGTTGGATGCTGACAGATTTCACATCGTGACGTTGGATGGTTCCAGAGGATGAACAAAC
rs4946655ACGTTGGATGCTAAAGAGTAGCTTTGGCTTGACGTTGGATGTTTTGTACGCTTTGCCTGAG
rs6915632ACGTTGGATGGTCGTGATCTTGACTCACTGACGTTGGATGGCCTGTAATCCCAGTTACTC
rs2095723ACGTTGGATGTGTGCTCTCTCATGCCAGTAACGTTGGATGCTGTATAAAATACCTTCAGG
rs7450078ACGTTGGATGGCCATCACCTCCAGATAATTACGTTGGATGAAGGCAGGAGGATCTCTTGA
rs7453071ACGTTGGATGAATCCCAGCACT'(-fGGGAGGACGTTGGATGTATGTTGCCCAGGCTCGTCT
rs1018810ACGTTGGATGTGCTGCTCCCATTTCTCATGACGTTGGATGAAGGAGTAGAGACCTTGCTG
rs7450944ACGTTGGATGATTCAGCCACTACACCTCAGACGTTGGATGGTTGTTCTACAGGACAAACC
rs7748657ACGTTGGATGAGAGAGAGATGGAAAGGGAGACGTTGGATGTCGAATCACGATCTGAACAG
rs1013137ACGTTGGATGATTACAAGCAGTGTCACTCCACGTTGGATGGGGTTAATGAATAGGTGGAAC
rs5878836ACGTTGGATGTTTGGTATGGAGTGACACTGACGTTGGAT~CCAATGATAATCTCCAGTGTC
rs1981480ACGTTGGATGCGACTGTCTTCCTTCTGCAGACGTTGGATGTGCTGCACTTCCCTACTCTT
rs1981479ACGTTGGATGTGAGTAGCTAGAACTACAGGACGTTGGATGATCACTGCAGCCTTAAACTC
rs3035187ACGTTGGATGTGAGTAGCTAGAACTACAGGACGTTGGATGATCACTGCAGCCTTAAACTC
rs7453993ACGTTGGATGTGACAAAGTGAGACCAACTCACGTTGGATGTGGGAGATCACCTTTCATAC
rs2001119ACGTTGGATGGCTTCTTTAGGTCTTCATTTCACGTTGGATGTGAGTTTGTGTTAAAAGCTC
rs2001118ACGTTGGATGGGTCCAGCCAAAAAACAACCACGTTGGATGAGGCTGGAATTTACAAGGCC
rs2001117ACGTTGGATGGTCCAGCCAAAAAACAACCCACGTTGGATGAGGCTGGAATTTACAAGGCC
rs6940433ACGTTGGATGTTGTGAGCTACCTCATTCACACGTTGGATGCAACATCTGGGTTATTTGTG
rs1318746ACGTTGGATGTAAGCTGGTGCTTATTTCAGACGTTGGATGGGTGGCCAATAAACATAAGC
rs763099ACGTTGGATGGAGGCAAGTTGTGAAAGACCACGTTGGATGGGCCCTTGAAGTTTTCTCAG
rs5878837ACGTTGGATGTCACCAGCCGTATTCATCAGACGTTGGATGTGAAAGACCTTCTGCCCATC
rs964731ACGTTGGATGGGAAATCATACCCCCTTTCCACGTTGGATGTGAGGGATACTTGAGCTCTG
rs964730ACGTTGGATGCACTCTGGCAAAGGGATTTAACGTTGGATGGTAGGAAAGCAGAAAGGTAC
rs6921869ACGTTGGATGTAGTAGAGACAGGGTTTCACACGTTGGATGTACTTGGGAGGCTAAGATGG
rs3945029ACGTTGGATGCTCTTCCTGTAAATCTTGCCACGTTGGATGAGAGAAAGGCTGAACACATG
rs4945715ACGTTGGATGCTCAAGGGACAGTCATTGAGACGTTGGATGGTCAGGGTGCTCATGAATTG
rs7775252ACGTTGGATGGACTAGGGATTGGATTTTGGACGTTGGA'f-GTTTCTTCATCCAGCTATGGC
rs7742098ACGTTGGATGGAAGAAAACCAGAAAACTGGCACGTTGGAT-GAAGAACTTCGTTCTTTCCCC
rs3757289ACGTTGGATGGCGATTTTATTTTGTAGTACAGACGTTGGATGAATACTTGTGCCTCAAGAAG
rs6905458ACGTTGGATGAGGAATATCAGCCT'ITT'GGGACGTTGGATGGCTCTTCTAACAGAAGTGACC
rs3757290ACGTTGGATGTAACAATGCCAGCACAACAGACGTTGGAT'GTGCTCCAGAGTTAAT1-t'GTC
rs2275289ACGTTGGATGTTGAAAAGGAACTCAGTGGCACGTTGGAT'GGTCCAGTTAGTCTTCTGAAC
rs4945716ACGTTGGATGTAGAGCCTCACTGTGTTACCACGTTGGATGAATTCTGGCACTTTGGGAGG
rs6922638ACGTTGGATGGCTTAGTGTCTGTGCTTTTGACGTTGGA'I-GCCTGCTGTTTCATTTTGAGG
rs7739572ACGTTGGATGGTTTTAAGAGACATTGGGTGACGTTGGAT-GTCTATTTGGACCATGCATTC
rs6901187ACGTTGGATGTCAGCACAGACCCTTAAATGACGTTGGAT-GGGCTTTTTTTCTCACCCACC
rs4946656ACGTTGGATGTGGCCCAGACGATATAAAGGACGTTGGATGATTAAGCTCCCCACTTAGGC
rs1338020ACGTTGGATGTCTGTGGTCAACAACAGTCCACGTTGGATGCATCTCAGGCAGGATATAGC
r rs7771472ACGTTGGATGTTACCTGAAGGTGAATCTAG~ ACGTTGGAT-GGTACAAACCTTTTGGAAAAC;
l
153
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs6926260ACGTTGGATGTACCACAGTGCTGGGATTACACGTTGGATGCGTAGAGTAGTGCATTGTGC
rs6926627ACGTTGGATGAGGTGTGCACCCATTATCCAACGTTGGATGGGATACTATACCCATTTACTC
rs4946657ACGTTGGATGCCAGGTAGAATTATTATGGGACGTTGGATGCCACCATTAAATCACTGTATC
rs6571218ACGTTGGATGCGCACGACACCTTATTAAAGACGTTGGATGTTCGACAATAGGTAACTGGC
rs7449944ACGTTGGATGAACTTTGTCGGCCCTGGCGGACGTTGGATGCCAGCGAGGAGGGACAGAG
rs952175ACGTTGGATGATGCTCTGCCAGCCTTTTTTACGTTGGATGTCAAAACAGCTGGTAGGGAC
rs1890228ACGTTGGATGTTAAGGCATTCCCATATCCTACGTTGGATGCCCAGATGTATGAATAGTAGC
rs1933237ACGTTGGATGGGGTTCAAGCAATTCCTGTCACGTTGGATGCAAAAATTAGCCCGGTGTGG
rs1338019ACGTTGGATGTATGTGTGTCACAAAGGGAGACGTTGGATGCCTGCAGAATCTACAACATG
rs7453127ACGTTGGATGCATCACCTCAGATAGTTACCACGTTGGATGGTGACTCCAGTTAGCTATAC
rs7381551ACGTTGGATGAGTTTGTACCCTTTGACCACACGTTGGATGGTTGA,ACTCACAGAAACAGAG
rs6571219ACGTTGGATGGACAGTACTGAAAGTCTTCGACGTTGGATGCTTCT'fCCTATCTGATTTGG
rs6571220ACGTTGGATGTCCTATCTGATTTGGAAGGCACGTTGGATGAACAAGACGAGAGTGTCTTG
rs2185017ACGTTGGATGATGTGGGAAGATCACTTGAGACGTTGGATGAGCCCGCTAATTGTCATAT
rs1591720ACGTTGGATGTTGGAATTACAGGTGTGAGCACGTTGGATGACAAGCCCACAGCTAACATC
rs6925046ACGTTGGATGGCTTGCTTTTTGAGACAGGGACGTTGGATGTAGAGGCTGTAGTGAGCTGT
rs6940423ACGTTGGATGGTGCTGGGATTACAGATGTGACGTTGGATGCCCTGTCTCAAAAAGCAAGC
rs1190274ACGTTGGATGCATTTAGTCTCTGAGGACAACACGTTGGATGCCTTT-CTAACCACTAAATACC
rs1190276ACGTTGGATGCTGTAATCCCAGCACTTTGGACGTTGGATGTAGTAGAGACTGGCTTTCAC
rs1591719ACGTTGGATGCTCACACATTCCCCTGAAAGACGTTGGATGCTGTCAGAAACTGCTCTGTC
rs1933236ACGTTGGATGCCAAGTCATTTGAAACCTTCACGTTGGATGTAAGCTCAGAAAATGGCATC
rs6905202ACGTTGGATGGTATTACAGTGTGAATCAGGACGTTGGATGCCCATTCAACATCAATTTTC
rs1209150ACGTTGGATGTCCTCCAGAAACTTTTGACCACGTTGGATGGGCCTTTATTACTTGCTACC
rs1190277ACGTTGGATGATCATGTGCTAAGCACCACGACGTTGGATGCCCTCCAGGTCAAAAGTTTC
rs6926278ACGTTGGATGTAGAACTCCCAGGCTCAAGAACGTTGGATGTATTAGCTGGGTGTAGTGGC
rs1190278ACGTTGGATGAGATACTGAGAAGGGTAGTCACGTTGGATGGTGCTACTGAATACTAGATC
rs4626463ACGTTGGATGAGAAATTGCCAACCAGCCTCACGTTGGATGGGTCCAGAAGCAAGACAAAG
rs6924620ACGTTGGATGAGAACAATGCCTGGCACATGACGTTGGATGTGACAGAGTGAGACTCTGTC
rs1190280ACGTTGGATGTAGAAAGTGCCATCCAATGCACGTTGGATGACAAACTAGGCAGACAGTAC
rs4557552ACGTTGGATGCGTCCTTTACATAACCCCAGACGTTGGATGCATTTTCTCGGTGACCTAGG
rs6932711ACGTTGGATGATCACCTGCTCAAGGTCATCACGTTGGATGGATGGTGCATTTGCATGCAG
rs1686140ACGTTGGATGAGAAAGAACCCTAGTTGGAGACGTTGGATGGAACATAGTCTGCATGTGATC
rs1190281ACGTTGGATGCACTTTTTTGCTACAACCTCACGTTGGATGATC'f-CTTGCATTTATTCTAC
rs2308162ACGTTGGATGCACTTnTTGCTACAACCTCCACGTTGGATGGCAT'CAAGTAACTGCACATT
rs1190282ACGTTGGATGTATGTGGACAGTAGCAACCCACGTTGGATGAGAC-TCAGGAGTTGCTTCTC
rs1765907ACGTTGGATGCTTTCTTGAGAAGCAACTCCACGTTGGATGGGGAGAATGAAATTCCACTT
rs5878838ACGTTGGATGCCCTGTCATTCAAGGCATAGACGTTGGATGTTGC'TCAGCATCGCTACATC
rs1190283ACGTTGGATGCCCACTGACCTACAATATATACGTTGGATGGACAGATTGAAGATGGCTAG
rs1190284ACGTTGGATGATCTTTCAAAACTGCCAGACACGTTGGATGGCCAGTGGATTTCAGTTGTT
rs1190285ACGTTGGATGACTTGAGTCACAGACATAGCI ACGTTGGATGGGCTCTTGATTATTTTCTGC
TABLE 40
dbSNP Extend T erm
rs# Primer Mix
rs2400080 TCCTTTACTTTACCTITITfTCCACG
rs6930209 GATTTTTATGCAAATATCAGATGAACT
rs221628 AAGAATAGACATATTTGTAGATCAACT
rs221629 TCTCCCTCTGGCCCAACTG ACT
rs221630 GACAGGTGATGGCTTGGGA ACG
I I
154
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs221631 TGTCAAAATGGAAAGATGATTAATACT
rs1149284 CTAGACACATTGTCTGCTAGT ACT
rs221633 AACAATTTGTCTTGGAGATCTTTACT
rs423366 ACCAAAAAACATTTTGCAGATAGACG
rs436460 ATATACCAAAAAACATTTTGCAGAACG
rs2211010 GAGACAGAGTCTCGCTCTGT ACG
rs379908 TGGATAACACAGTGCATACCA ACG
rs1149285 ATTTATGAAGCACAAAGAACAACACT
rs7341194 ACTGGAAAAATTTTTTTCCTTTGTACT
rs715153 GCCCTCTAGTGGGCTTAATG ACT
rs221634 GCCAGGATGACCCCAAAATA CGT
rs7757307 CCCATAATTCTTTAAACTAAATACACT
rs221635 ACAGTAAATGAAGGACATTGGC ACG
rs4145418 TAGCCCTGTAAGCTGATC CGT
rs221636
rs3185958 CCACATCTTAAAGAGGCTGTT ACT
rs4946654 GCCTATTGAAGAAATCATTTTAGACGT
rs221637 ACTTGAGCGATCCTCCCAC CGT
rs221638 CCTGGCTGAAAATCTTAAAAAAAACT
rs221639 GTGTGTGTGTGTGTGTAACCA ACT
rs643545 CACCCGCATGTGTATGTATCT ACT
rs221640 TCGTCTGAAGTCAGGAGTTC ACT
rs3957696 TCTCTCTCTCTCTCTCTCAC CGT
rs3995554 TCTCTCTCTCTCTCTCACAC CGT
rs7453502 GAGTTTGTGTTTTAAAGAACTTTTCGT
rs1190471 AAGAGTGATAAATGACCAGGC ACT
rs221641 GAGATGTGAGCCACTGCGC ACT
rs221642 CCAACCATGTGGAACTGTGA ACT
rs1190472 TTATCAACAGCATGAAAACGGA ACG
rs1190473 CTGCACTCCAGCCCGGGA ACT
rs186404 GGAGACACAGTGAGACTGTC CGT
rs221643 AATGAAGAAGTCTTGCATTTCTTCGT
rs221644 CTAGCTCCAAGCCAGGTTAT ACT
rs1203475 GCAGGAGAATCGCTTGAACC ACG
rs221645 CAGACCTCAAAGTGGTCAAGA ACT
rs170277 GACCCTTGCTAGCACTCAGA ACG
rs221646 CAGGCAAACAGGTCCAGAG ACG
rs221647 AGCTCCTTTCCTTAGGTTATC ACT
rs221648 GGCAACGGAGTGAGACCC ACG
rs221649 AAGAAGAAGGCTGGGAGAAC ACT
rs221650 GGCACAGTGGCTCACACTT ACT
rs1149287 TCCCAGGCTGGTCTTGAAC ACG
rs221651 AGCTGCAATGAGCTGTGATCG ACG
rs7762591 CTTCCGTCTCCTGAGTTCCA ACT
rs7748555 CAGGTGTGAGCCACCATGC ACG
rs5878833 GCCTTCTGGCCA~ ACG
rs5878834 GCCTTCTGGCCATTTTTTTTTT ACG
rs221652 TACTCAGGGAAGGATGTTACA ACG
rs221653 I TGTGTTAGCCAGGATGGTCT ACG
155
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs221654 AAGACCATCCTGGCTAACAC ACT
rs221655 GCTAACACGGTGAAACCCC ACT
rs221656 CAGGCTGGAGTGTAGTGGC ACT
rs221657 CACTTCCTCCCTCCGACTC ACG
rs221658 TCAGCATTTGTGGGCTGCC ACT
rs110065 CTCCTTGCTGGTTGTGGCA CGT
rs221659 ATGAATTCTATCTGTGCGACC ACG
rs221660 GGAAACCAGGGCTTTTTTTTTTACT
rs7742821 ATTTCCATTTGTGTTGAGTCCTACT
rs221662 GAAATAAAAAGGAATCACACCCACT
rs7748426 CACATCTGTACTATTATTTCTACTACT
rs6911494 AGGCCAGGCTAACTGGGG ACG
rs6939846 GTGGCCATGACAGTTGCAG CGT
rs368471 TTATATTTCAAGGGAATGCTCTTACT
rs430190 GCCTCTGGGCAAATTTCTGA ACG
rs455114 TTTTTACAGTTGGGAGGCAGA ACT
rs405956 AAGACTGGGACAGCAGCGA ACT
rs5878835 GAACCACAGAAGGCCTTAAAAACGT
rs1473814 GAACCACAGAAGGCCTTAAAAACGT
rs423272 GTGGGTACATAGTGGATGTAT ACT
rs413806 ACAGATTTCACATCGTGGTACTCACG
rs4946655 GTAGCTTTGGCTTGTGCACC ACT
rs6915632 CTTGACTCACTGCAACCTCA ACT
rs2095723 TCTGTCCTCACACAGCATTTT ACG
rs7450078 CGCTATGTTGCCCAGGCTC ACT
rs7453071 CCAAGGCAGGAGGATCTCT ACT
rs1018810 CTGCTTTTATACATGCCACAC ACT
rs7450944 GGGCTCCCTTTCCATCTCT ACT
rs7748657 GTAGTGGCTGAATGCGATGT ACT
rs1013137 CACTCCATACCAAATTAAATATACACG
rs5878836 GTGACACTGCTTGTAATTCTG CGT
rs1981480 TACAATGGCAGTGACCCAGA CGT
rs1981479 CTACAGGCCTGCACCACGA ACG
rs3035187 ATGCCTGGCATTTTTTTTTT1T1-1'CGT
rs7453993 GTGAGACCAACTCCCATCC ACG
rs2001119 CTTACAAAAGCTTCTGTGCCATACT
rs2001118 CAGCCAAAAAACAACCCTAAAAACT
rs2001117 AAAAACAACCCTAAAAAGGAAGAACT
rs6940433 TGCCAAGAGGCACATTTTCC ACT
rs1318746 AGGCTACTAAGTATATTTGATTTTACT
rs763099 AAAGACCTTCTGCCCATCCA CGT
rs5878837 CGTATTCATCAGCAACAGCC ACT
rs964731 ATACCCCCTTTCCTTCAGTAT ACT
rs964730 TGAGGGATACTTGAGCTCTGT ACT
rs6921869 GTCTCGAGCTCCTGGCCT ACT
rs3945029 ATTAGCAGCCTCCTCCACTA ACT
rs4945715 CTTCTCTTTCCTCCTTTTCATCACT
rs7775252 TTGAGAATTATTCCCGGTAATTAACT
rs7742098 I CCAGAAAACTGGCTTTGCCTTI ACT
156
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs3757289 AAAAATTCCACAGAGATGATGG ACT
rs6905458 CCTCTCAGAAGTGTGCCAG ACG
rs3757290 GACTGACTCTCTCCCCAAAA ACT
rs2275289 AGGAACTCAGTGGCATGTAC ACG
rs4945716 CTCACTGTGTTACCCAGGCT ACT
rs6922638 GTGCTTTTGTTTCTTCTCATACTACT
rs7739572 AGACATTGGGTGTTTCTCTTTT ACT
rs6901187 CTGACACATAGCTGCCAGAG ACT
rs4946656 CTGTTGAAGAGCAAAGTTAACA ACG
rs1338020 GCAAGACATTCTGAATAGTGC ACT
rs7771472 GAAGGTGAATCTAGGGAATGAA CGT
rs6926260 GTAAGCCACTGTGTCCAGC ACG
rs6926627 AAGGCAGAGCAGGGTCCC ACT
rs4946657 GTTTCATGTTGTATCTCTCTGT ACT
rs6571218 CCTTATTAAAGAGAGAGAGAGA ACT
rs7449944 GGCGGCAGCTGCTTGTTC ACT
rs952175 CTGGGCGCACTGCAACCT ACT
rs1890228 CCATATCCTGGGCTATGTGT ACG
rs1933237 CTTAGCCTCCAGAGTAGCTG ACT
rs1338019 GTCACAAAGGGAGAACTCAAA ACG
rs7453127 CTACTCTCTTAGCAAATTCAGTTACT
rs7381551 TTCCCACCCTTCAGCCCC ACT
rs6571219 CGCTGGGGCAGAAAAAGAAA ACG
rs6571220 TTCTTTTTCTGCCCCAGCGA ACT
rs2185017 CAACACAGTGAGCAGTGAGA ACG
rs1591720 GTGTGAGCCACCATGCCCA ACT
rs6925046 GACAGGGTCTTGCTCTGTC ACT
rs6940423 GGATTACAGATGTGAGCCAC ACG
rs1190274 GGACAACACTTTTAAAGGTACT ACT
rs1190276 CCAGCACTTTGGGAGGCC ACT
rs1591719 TTGAATCTCTTTTTAGAGTATGGACT
rs1933236 ATTTCCTGATTCACACTGTAATAACG
rs6905202 GAAATTITfCACGTTTTGAAGGTACG
rs1209150 TGACCTGGAGGGAGAAAAAG CGT
rs1190277 GCTAAGCACCACGGAGATAC ACT
rs6926278 CTCCCACCTCAGCCTCCC ACG
rs1190278 GGGTAGTCGGTAAAGGGGA ACG
rs4626463 AGGGACTTTCCACACTAACC ACT
rs6924620 TAAATATTCATTGCATAGAAGGAAACT
rs1190280 ATGCTGCATGTATATTTATGGC ACT
rs4557552 ACCCCAGTACTTCCTCTCC ACG
rs6932711 GTCATCACTCCCGCAGTTCA ACG
rs1686140 CCCTTCCTTTGGAAAACTGG ACT
rs1190281 TTTAAATGTGCAGTTACTTGATGACG
rs2308162 CCTCCAGTGAAAGCAATTATTT CGT
rs1190282 GCTGAGAATACTTGCTGGCT ACT
rs1765907 TGAGAAGCAACTCCTGAGTC ACG
rs5878838 TGCCAATTAGCACTGAAP,AAAGACT
rs1190283 I CTACAAAATTCGTTACTACATACACT
157
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs1190284 CAGACGTGGCAGCAGAGTAA ACT
rs1190285 GTCACAGACATAGCCATTTAGAI ACT
I
Genetic Analysis
[0292] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 41.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (A1 AF
= 1-A2 AF). For
example, the SNP rs1474555 has the following case and control allele
frequencies: case Al (C) = 0.64;
case A2 (T) = 0.36; control A1 (C) = 0.70; and control A2 (T) = 0.30, where
the nucleotide is provided
in paranthesis. Some SNPs are labeled "untyped" because of failed assays.
TABLE 41
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position AlleleCase Control Value
SEQ ID AF AF
NO: 7
rs2400080241 105557091A/G
rs6930209801 105557651A/G
rs221628899 105557749A/G 0.716 0.755 0.216
rs2216292091 105558941C/G 0.775 0.801 0.338
rs2216302290 105559140C/T 0.066 0.049 0.465
rs2216312440 105559290A/G 0.147 0.137 0.686
rs11492844959 105561809G/T
rs2216337914 105564764C/G 0.094 0.091 0.911
rs4233667969 105564819A/G 0.392 0.418 0.448
rs4364607972 105564822C/T 0.186 0.175 0.720
rs221101010831 105567681C/T
rs37990812399 105569249C/T 0.773 0.809 0.242
rs114928513841 105570691C/T
rs734119414461 105571311C/T
rs71515314680 105571530C/T
rs22163416808 105573658A/T 0.330 0.314 0.630
rs775730718231 105575081C/T
rs22163518394 105575244C/T
rs414541818505 105575355G/T 0.380 0.377 0.929
rs22163618684 105575534A/T 0.807 0.829 0.458
rs318595819257 105576107C/T
rs494665420263 105577113A/T
rs22163720656 105577506A/C 0.879 0.901 0.409
rs22163821499 105578349A/G 0.089 0.072 0.427
rs22163921563 105578413A/C 0.934 0.951 0.537
rs64354521612 105578462C/G 0.824 0.842 0.486
rs22164021834 105578684C/T
rs395769622406 105579256A/T
rs399555422408 105579258A/T
rs745350222685 105579535A/T
rs119047123303 105580153C/T
rs22164123306 105580156C/G 0.070 0.053 0.415
rs22164225139 105581989A/G 0.868 0.869 0.987
rs119047225211 105582061C/T 0.227 0.191 0.244
rs119047325364 105582214A/G 0.722 0.742 0.521
rs18640425381 105582231A/C
rs22164325414 105582264A/T 0.550 0.766 -0.0001
rs22164425835 10_5582685C/T 0.695_ 0.774 0.007
rs1203475~ 26214 ~ 105583064A/G
~
158
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in PositionAllele Case Control Value
SEQ ID AF AF
NO: 7
rs22164527224 105584074A/G 0.066 0.048 0.344
rs17027727526 105584376A/G 0.840 0.882 0.137
rs22164627934 105584784C/T 0.866 0.897 0.244
rs22164728550 105585400C/T 0.844 0.884 0.102
rs22164829015 105585865A/G 0.865 0.891 0.341
rs22164929879 105586729G/T 0.102 0.081 0.359
rs22165029979 105586829A/G 0.856 0.887 0.192
rs114928730030 105586880A/G
rs22165130585 105587435C/T 0.177 un ed NA
rs776259131753 105588603C/G
rs774855531934 105588784C/T 0.670 0.712 0.199
rs587883333227 105590077-/T 0.140 0.113 0.338
rs587883433228 105590078-/T 0.142 0.114 0.309
rs22165235172 105592022C/T 0.172 0.120 0.064
rs22165336901 105593751A/G
rs22165436921 105593771A/G
rs22165536932 105593782A/G
rs22165637061 105593911C/T
rs22165737570 105594420C/T 0.924 0.953 0.218
rs22165838745 105595595G/T 0.043 0.028 0.421
rs11006538970 105595820A/T 0.834 0.894 0.031
rs22165939725 105596575C/T 0.048 0.027 0.347
rs22166040070 105596920A/C 0.841 0.878 0.133
rs774282140460 105597310C/G
rs22166241470 105598320A/G 0.778 0.879 0.0001
rs774842641562 105598412A/G
rs691149441956 105598806A/G 0.043 0.032 0.652
rs693984642047 105598897A/T
rs36847142280 105599130A/G 0.150 0.104 0.074
rs43019042358 105599208A/G 0.053 0.033 0.386
rs45511442629 105599479C/G 0.059 0.027 0.100
rs40595643075 105599925C/T 0.132 0.089 0.063
rs587883543387 105600237-/A
rs147381443393 105600243G/T 0.126 unt ed NA
rs42327243438 105600288C/T 0.023 unt ed NA
rs41380644115 105600965A/G 0.837 0.895 0.037
rs494665544537 105601387A/G 0.062 0.033 0.128
rs691563245642 105602492A/G
rs209572346629 105603479A/G
rs745007847496 105604346A/G 0.261 0.163 0.001
rs745307147515 105604365A/C
rs101881048329 105605179A/G
rs745094448862 105605712C/G
rs774865748908 105605758A/G 0.972 unt ed NA
rs101313749038 105605888C/T 0.699 0.785 0,006
rs587883649080 105605930-/T
rs198148050204 105607054A/T 0.880 0.946 0.012
rs 198147950404 105607254A/G 0.052 0.035 0.453
rs303518750426 105607276-/TTA 0.033 unt ed NA
rs745399350531 105607381C/T 0.170 0.135 0.222
rs200111950840 105607690C/T 0.176 0.122 0.033
rs200111850964 105607814C/T 0.793 0.883 0.001
rs200111750971 105607821C/T 0.575 0.650 0.035
rs694043351378 105608228C/T
rs131874652610 105609460A/C 0.140 0.089 0.171
rs76309953906 105610756A/T 0.865 0.922 0.029
rs587883753951 105610801-/C 0.423 0.463 0.215
rs96473154111 105610961A/C 0.865 0.926 0.089
rs96473054149 105610999G/T 0.903 0.951 0.022
rs692186955563 105612413C/G
rs394502955999 105612849C/T 0.972 0.976 0.820
rs494571558415 105615265C/G 0.057 0.021 0.048
rs777525258961 105615811C/G 0.027 unt ed NA
rs774209860447 105617297C/T
159
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 ~ F A2 F A2 F p-
rs# in Position AlleleCase C Value
SEQ ID AF ont
NO: 7 ro
I AF
rs375728961377 105618227A/G _
_
_
rs690545861528 105618378A/G 0.045 0.023 0.345
rs375729061606 105618456C/G
rs227528962140 105618990A/G
rs494571662461 105619311C/T
rs692263863826 105620676C/T 0.086 0.054 0.120
rs773957264950 105621800G/T 0.920 0.931 0.613
rs690118765076 105621926G/T 0.054 0.026 0.122
rs494665666121 105622971C/T
rs133802066406 105623256C/T 0.109 0.077 0.145
rs777147267051 105623901A/C 0.035 unt ed NA
rs692626068860 105625710C/T 0.921 0.952 0.196
rs692662769014 105625864C/T
rs494665770796 105627646C/T 0.224 0.136 0.001
rs657121872325 105629175G/T 0.589 0.677 0.011
rs744994473414 105630264A/C
rs952175 75258 105632108C/G 0.650 0.730 0.007
rs189022876347 105633197A/G 0.046 0.028 0.426
rs193323776839 105633689A/C 0.925 0.953 0.175
rs133801977358 105634208A/G 0.888 0.930 0.101
rs745312777822 105634672A/G 0.415 0.534 0.002
rs738155177946 105634796G/T 0.026 unt ed NA
rs657121980002 105636852A/G 0.837 0.903 0.017
rs657122080024 105636874A/G 0.464 unt ed NA
rs218501780285 105637135A/G 0.066 0.036 0.196
rs159172080397 105637247C/G 0.027 unt ed NA
rs692504682075 105638925C/T
rs694042382153 105639003A/G 0.024 0.029 0.840
rs119027483981 105640831A/G 0.067 0.041 0.183
rs119027684184 105641034A/G
rs159171985089 105641939C/T
rs193323685288 105642138A/G 0.892 0.942 0.046
rs690520285330 105642180C/T 0.888 0.909 0.435
rs120915085581 105642431A/T 0.862 22 .023
0.9 0
rs119027785642 105642492A/G 0.158 _ _
_ _
0.118 0.098
rs692627886433 105643283p~G __ . _
rs119027886904 105643754A/G 0.211 0.147 0.030
rs462646388391 105645241A/G 0.067 0.050 0.383
rs692462089042 105645892C/T
rs119028090828 105647678G/T 0.890 0.948 0.008
rs455755292676 105649526C/T 0.033 0.025 0.736
rs693271192881 105649731C/T
rs168614094227 105651077G/T
rs119028194585 105651435A/G 0.914 0.950 0.140
rs230816294616 105651466-/ATAA0.127 0.072 0.035
rs119028294712 105651562C/G 0.879 0.937 0.009
rs176590794738 105651588A/G 0.095 0.058 0.143
rs587883895253 105652103-/G
rs119028395522 105652372A/G 0.054 0.032 0.245
rs119028495869 105652719G/T 0.858 0.921 0.005
rs119028597856 105654706C/T 0.908 0.957 0.017
rs6931398 A/G
[0293] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values axe indicated
in bold. The allelotyping p-
values were plotted in Figure 1F for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
160
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in Figure
1F can be determined by consulting Table 41. For example, the left-most X on
the left graph is at
position 105557091. By proceeding down the Table from top to bottom and across
the graphs from left
to right the allele frequency associated with each symbol shown can be
determined.
[0294] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a l Okb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-8 were truncated at that
value.
[0295] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 10
TM7SF3 Region Proximal SNPs
[0296] It has been discovered that SNP rs1484086 in TM7SF3 is associated with
occurrence of
osteoarthritis in subjects. TM7SF3 is an orphan receptor and is a member of
the superfamily of 7-
transmembrane domain proteins, one of the largest superfamilies of cell
surface proteins. Members of
this family include receptors for a variety of ligands, such as peptides,
hormones, and ions, and for
external sensory stimuli, such as odorants and light. Many 7-transmembrane
molecules are able to
recruit small G proteins, suggesting that they can transduce external signals
to the cytoplasm.
[0297] Thirty-seven additional allelic variants proximal to rs1484086 were
identified and
subsequently allelotyped in osteoarthritis case and control sample sets as
described in Examples 1 and 2.
The polymorphic variants are set forth in Table 42. The chromosome positions
provided in column
four of Table 42 are based on Genome "Build 34" of NCBI's GenBank.
161
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
TABLE 42
dbSNP ChromosomePositionChromosome Allele
rs# in Position Variants
SEQ ID
NO:
8
rs105870112 230 27004780 a/c
rs11613 12 231 27004781 c/
rs900743 12 5330 27009880 /t
rs212909112 6334 27010884 a/c
rs15556 12 11372 27015922 c/t
rs105372412 11456 27016006 /t
rs9699 12 11501 27016051 a/
rs4856 12 13393 27017943 c/t
rs378231412 16666 27021216 a/
rs208773612 17596 27022146 c/t
rs206837212 19710 27024260 al
rs206837112 19800 27024350 a/
rs118452912 20297 27024847 a/
rs210125612 20967 27025517 /t
rs155225712 32514 27037064 c/t
rs156558512 33159 27037709 a/
rs408080012 37600 27042150 a/
rs138865812 41259 27045809 a/
rs378231612 41329 27045879 c/t
rs148408612 50060 27054610 c/t
rs148408712 53292 27057842 c/t
rs229154912 53393 27057943 a/
rs156558412 56417 27060967 c/t
rs307116612 56435 27060985 -/tt
rs190765212 58847 27063397 a/t
rs375911912 59595 27064145 c/t
rs375912012 59661 27064211 /t
rs378231712 60355 27064905 c/t
rs382516612 60407 27064957 a/c
rs187219112 62357 27066907 a/
rs8628 12 68230 27072780 /t
rs187219312 68516 27073066 a/
rs104283312 69055 27073605 c/t
rs2476 12 72603 27077153 c/
rs197620612 73928 27078478 a/
rs138865912 85897 27090447 c/t
rs118452812 91554 27096104 a/
Assay for Verifyin$ and Allelotypin_~ SNPs
[0298] The methods used to verify and allelotype the 37 proximal SNPs of Table
42 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 43 and Table 44, respectively.
162
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
TABLE 43
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1058701ACGTTGGATGCTTCTCTCCAGTCCATGTTGACGTTGGATGGTAACAAGTCCGAAGGATTC
rs11613 ACGTTGGATGTCCAAGTGCCTTCTCTCCAGACGTTGGATGTCCTTGATGCATCTCGACAC
rs900743 ACGTTGGATGAACAGATCCGGAACTf~fTTTACGTTGGATGGGCTCATAGAACCCTTTnT
rs2129091ACGTTGGATGTTAAAATTGGTATGGTCTCCACGTTGGATGGTTTTGCACTAAGAAGAGAC
rs15556 ACGTTGGATGTGTAGGGACAAAGTTATATGGAAACGTTGGATGTGTCTTTACAGTCTTCACTGGCA
rs1053724ACGTTGGATGCCATATAACTTTGTCCCTACACGTTGGATGAAAATACAGTGCAGGTGACC
rs9699 ACGTTGGATGTTTCAGTGTTACTGAAGTTAATTACGTTGGATGCAACATAGAAAATATAAAACAATTT
rs4856 ACGTTGGATGTTGTAAACCAAAAGGTAATTTCTCAACGTTGGATGGATGACCAAAAGGTTGGTGG
rs3782314ACGTTGGATGAATGCTGCACAACTTAGGCCACGTTGGATGAGCAGTCAGCCTATTCTTGC
rs2087736ACGTTGGATGCAGTGAGACTCTGTCTCTACACGTTGGATGTACATCAGCCTCCCAAGTAG
rs2068372ACGTTGGATGCTAGAGAGGGTTGCTCTATGACGTTGGATGCCAGCTTCAGCTTTCTGTAG
rs2068371ACGTTGGATGATTGACAAGCCAAGCCTGTCACGTTGGATGATAGAGCAACCCTCTCTAGG
rs1184529ACGTTGGATGTTCCTTTCCTGGTGGTTTGCACGTTGGATGGGTGTCAGTCTTGTAATCAG
rs2101256ACGTTGGATGCTGGGTTTTTTTGGTGTGTTGACGTTGGATGCTTGTACTGTATCTCAATTTC
rs1552257ACGTTGGATGCAATACCTTAGCTCACCTACACGTTGGATGCATTAGGGAAAATCTGGATC
rs1565585ACGTTGGATGGCTAGAATATACCACAGGTGACGTTGGATGTCACTGAAAATGGATCAGCC
rs4080800ACGTTGGATGGGGTTCAAGCAATTCTCCTGACGTTGGATGAAAATTAGCCAGGTGTGGTG
rs1388658ACGTTGGATGGAAAAGAGAGAGGATGGGTGACGTTGGATGCAGGAGCTTAAGAACCACTG
rs3782316ACGTTGGATGGTCTTGCTGTAATGTCATTAGACGTTGGATGCTTCTCTTTTTTAACTAGATC
rs1484086ACGTTGGATGTGTCACTCTTCGGAAGTCTCACGTTGGATGCATGTACAGGGCATTCACAG
rs1484087ACGTTGGATGGCATGGAATTTTTACACCCCACGTTGGATGTTCCCTCTCAAACTCTGTAG
rs2291549ACGTTGGATGACTAAAGCGCCTCTTTGCTGACGTTGGATGCATTAAGCCGCTAGTTCCAC
rs1565584ACGTTGGATGGAAAAGGCAAGTTATGATGCACGTTGGATGAGTATCCATTCCTCTGAGTC
rs3071166ACGTTGGATGGAAAAGGCAAGTTATGATGCACGTTGGATGAGTATCCATTCCTCTGAGTC
rs1907652ACGTTGGATGCCATATTTCCAGTACCTAGCACGTTGGATGGGTTGATAAATATGTGCCAG
rs3759119ACGTTGGATGCTAAACTGTTCTTAGTGCCGACGTTGGATGGGGAATTTCAAAGAGTGTGG
rs3759120ACGTTGGATGCGGCACTAAGAACAGTTTAGAACGTTGGATGGGTTTTTATGACTGTAGCAAC
rs3782317ACGTTGGATGGGAAGCTCTTGAAGCTGTAGACGTTGGATGGCCATTGAGAAATCCTGAGC
rs3825166ACGTTGGATGACCAGACTCTCAACTTAGCCACGTTGGATGTGGCAGGTGGGTTATTCTTG
rs1872191ACGTTGGATGTTTATGCCGAAGCCCTGTCTACGTTGGATGTAAAGCAAGGGAGGAAAGGG
rs8628 ACGTTGGATGCAAGCCTTACCAACAATTACAGAAACGTTGGATGGCTTATCAAGAGTGAAAATAGAAGA
rs1872193ACGTTGGATGAATGCCACCTCTAAGAGGCAACGTTGGATGCTCAAGCCAAAGGAGAAGAC
rs1042833ACGTTGGATGTCATCCAACCCTTGGATCTCACGTTGGATGATACAGCCCCAGGAAATAGC
rs2476 ACGTTGGATGAGGAGCAGGTGACGTTAATGACGTTGGATGATTGACTAGCCACCAGGAAG
rs1976206ACGTTGGATGAGCTGGGATTGGATTACAGGACGTTGGATGATGGAGAAACCCCGTCTCTA
rs1388659ACGTTGGATGACCTCAGCCTTCCACATTTGACGTTGGATGAGCAGGAGGAAACTTTTGGG
rs1184528ACGTTGGATGTGAGAATCGCTTGAACCCAGACGTTGGATGTTTGAATCAGAGTCTCCCTC
TABLE 44
dbSNP Extend Term
rs# Primer Mix
rs1058701 CCAGTCCATGTTGAGGTGC ACT
rs11613 CCAGTCCATGTTGAGGTG ACT
rs900743 ATCCGGAACTTTTTTTTTGAAATTACT
rs2129091 ATTTAATTTGAAAAGTGCTTACCCACT
rs15556 GTTTGTTATTTTCTATAC ACG
rs1053724 CATCAAGTTTCAGTGTTA CGT
rs9699 TACAGAATAAGCAGTAAA ACG
rs4856 CAAGTATTTGGAAATAAG ~ ACT
163
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs3782314 ACAAATATATGAGAACTCCTCTTTACT
rs2087736 ACTCTGTCTCTACTAAAAATAAAAACT
rs2068372 TGGCTTCATGGCACCACTG ACT
rs2068371 AGCCAAGCCTGTCACTGGCCCT ACT
rs1184529 TCCTGGTGGTTTGCCACTTA ACT
rs2101256 GTTTTTTTGGTGTGTTGATATGTAACT
rs1552257 AGCTCACCTACGAAAATGAATAAACG
rs1565585 TGTCAGACTGCACTACAT ACT
rs4080800 ATTCTCCTGTCTCAGCCTCC ACG
rs1388658 GAGAGGATGGGTGAAATAAGG ACT
rs3782316 TGTAATGTCATTAGGAAGAAACAACG
rs1484086 CTCTTCGGAAGTCTCTTTCTCA ACT
rs1484087 TTTACACCCCCAAATCTAGAG ACT
rs2291549 CCTCTTTGCTGCCCAGTGG ACT
rs1565584 TGATGCAATAAGTATATATAGTACACT
rs3071166 TAGTACGTGGCTTTTTTTTTTTTTCGT
rs1907652 TCCAGTACCTAGCACCTAAC CGT
rs3759119 TTAGTGCCGGGATGAATAACT ACT
rs3759120 AAATTGCAACTGTGAGTATTAAAGACT
rs3782317 AGAATAACCCACCTGCCAAAT ACT
rs3825166 ACTTAGCCTACATTTGAAAAGGGACT
rs1872191 CTCAGTCCGCTCCCCACTT ACG
rs8628 TTAAACACTATGACACAT ACG
rs1872193 AGAGGCAGGACACTAGCC ACG
rs1042833 CCCTTGGATCTCTTTGAG ACG
rs2476 GTGACGTTAATGGGACAGCT ACT
rs1976206 TACAGGCGCCCACCACCA ACG
rs1388659 CCACATTTGGTAAGTTTTGACATACT
rs1184528 ~ AGAGGTTGCAGAGAGCCAAGATCACT
I
Genetic Anal,
[0299] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 45.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (A1 AF
= 1-A7, AF). For
example, the SNP rs900743 has the following case and control allele
frequencies: case A1 (G) = 0.92;
case A2 (T) = 0.08; control A1 (G) = 0.90; and control A2 (T) = 0.10, where
the nucleotide is provided
in paranthesis. Some SNPs are labeled "untyped" because of failed assays.
TABLE 45
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
in SEQ
rs# ID NO: 8 Position Allele Case C__ont_rolValue
AF AF
rs1058701 230 27004780 A/C
rs11613 231 27004781 C/G
rs900743 5330 27009880 G/T 0.08 0.10 0.381
164
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ Position AlleleCase Control Value
ID NO: 8 AF AF
rs2129091 6334 27010884 A/C 0.19 0.20 0.646
rs15556 11372 27015922 C/T 0.74 0.74 0.923
rs1053724 11456 27016006 G/T 0.14 0.15 0.592
rs9699 11501 27016051 A/G 0.67 0.68 0.595
rs4856 13393 27017943 C/T 0.10 0.09 0.474
rs3782314 16666 27021216 A/G 0.20 0.22 0.278
rs2087736 17596 27022146 C/T
rs2068372 19710 27024260 A/G 0.11 0.10 0.634
rs2068371 19800 27024350 A/G
rs1184529 20297 27024847 A/G
rs2101256 20967 27025517 G/T
rs1552257 32514 27037064 C/T 0.67 0.67 0.969
rs1565585 33159 27037709 A/G 0.69 0.65 0.064
rs4080800 37600 27042150 A/G
rs1388658 41259 27045809 A/G 0.35 0.38 0.362
rs3782316 41329 27045879 C/T 0.47 0.46 0.740
rs1484086 50060 27054610 C/T
rs1484087 53292 27057842 C/T 0.35 0.38 0.127
rs2291549 53393 27057943 A/G
rs1565584 56417 27060967 C/T 0.66 0.69 0.327
rs3071166 56435 27060985 -/TT 0.52 0.47 0.042
rs1907652 58847 27063397 A/T 0.64 0.64 0.984
rs3759119 59595 27064145 C/T 0.08 0.05 0.042
rs3759120 59661 27064211 G/T 0.15 0.14 0.861
rs3782317 60355 27064905 C/T 0.08 0.07 0.780
rs3825166 60407 27064957 A/C 0.83 0.84 0.660
rs1872191 62357 27066907 A/G 0.04 0.05 0.555
rs8628 68230 27072780 G/T 0.59 0.61 0.411
rs1872193 68516 27073066 A/G 0.92 0.91 0.669
rs1042833 69055 27073605 C/T 0.85 0.86 0.605
rs2476 72603 27077153 C/G 0.81 0.82 0.392
rs1976206 73928 27078478 A/G 0.16 0.15 0.781
rs1388659 85897 27090447 C/T 0.49 0.50 0.692
rs1184528 91554 27096104 AlG 0.20 0.17 0.228
[0300] The TM7SF3 proximal SNPs were also allelotyped in the replication
cohorts using the
methods described herein and the primers provided in Tables 43 and 44. The
replication allelotyping
results for replication cohort #1 and replication cohort #2 are provided in
Tables 46 and 47,
respectively.
TABLE 46
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ Position Allele Case Control Value
ID NO: 8 AF AF
rs1058701230 27004780 A/C
rs11613 231 27004781 C/G
rs900743 5330 27009880 G/T 0.07 0.11 0.258
rs21290916334 27010884 A/C 0.18 0.20 0.450
rs15556 11372 27015922 C/T 0.73 0.74 0.858
rs105372411456 27016006 G/T 0.15 0.15 0.969
rs9699 11501 27016051 A/G 0.65 0.68 0.290
rs4856 13393 27017943 C/T 0.11 0.10 0.735
rs378231416666 27021216 A/G 0.18 0.22 0.171
rs208773617596 27022146 C/T
rs206837219710 27024260 A/G 0.11 0.11 0.867
rs206837119800 27024350 A/G
rs118452920297 27024847 A/G
rs210125620967 27025517 G/T
rs155225732514 27037064 C/T 0.70 0.66 0.239
rs156558533159 27037709 A/G 0.67 0.65 0.599
165
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ Position Allele Case Control Value
ID NO: AF AF
8
rs4080800 37600 27042150 A/G
rs1388658 41259 27045809 A/G 0.36 NA NA
rs3782316 41329 27045879 C/T 0.45 0.49 0.230
rs1484086 50060 27054610 C/T
rs1484087 53292 27057842 C/T 0.32 0.40 0.023
rs2291549 53393 27057943 A/G
rs1565584 56417 27060967 C/T 0.61 0.65 0.297
rs3071166 56435 27060985 -/TT 0.55 0.45 0.003
rs1907652 58847 27063397 A/T 0.62 NA 0.638
rs3759119 59595 27064145 C/T 0.09 0.04 0.025
rs3759120 59661 27064211 G/T 0.16 0.15 0.651
rs3782317 60355 27064905 C/T 0.07 0.07 0.986
rs3825166 60407 27064957 A/C 0.84 0.84 0.945
rs1872191 62357 27066907 A/G 0.04 0.06 0.562
rs8628 68230 27072780 G/T 0.58 0.64 0.060
rs1872193 68516 27073066 A/G 0.92 0.91 0.957
rs1042833 69055 27073605 C/T 0.85 0.86 0.746
rs2476 72603 27077153 C/G 0.81 0.84 0.397
rs1976206 73928 27078478 A/G 0.17 0.16 0.788
rs1388659 85897 27090447 C/T 0.47 0.50 0.283
rs1184528 91554 27096104 A/G 0.21 0.16 0.170
TABLE 47
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ Position AlleleCase ~ Value
ID NO: AF Control
8 AF
rs1058701230 27004780 A/C
rs11613 231 27004781 C/G
rs900743 5330 27009880 G/T 0.10 0.09 0.763
rs21290916334 27010884 A/C 0.21 0.20 0.896
rs15556 11372 27015922 C/T 0.75 0.75 0.974
rs105372411456 27016006 G/T 0.12 0.15 0.373
rs9699 11501 27016051 A/G 0.69 0.67 0.615
rs4856 13393 27017943 C/T 0.10 0.08 0.413
rs378231416666 27021216 A/G 0.22 0.22 0.944
rs208773617596 27022146 C/T
rs206837219710 27024260 A/G 0.11 0.10 0.561
rs206837119800 27024350 A/G
rs118452920297 27024847 A/G
rs210125620967 27025517 G/T
rs 155225732514 27037064 C/T 0.63 0.68 0.247
rs156558533159 27037709 A/G 0.73 0.66 0.029
rs408080037600 27042150 A/G
rs138865841259 27045809 A/G 0.33 0.38 0.217
rs378231641329 27045879 C/T 0.51 0.42 0.036
rs148408650060 27054610 C/T
rs148408753292 27057842 C/T 0.38 0.36 0.575
rs229154953393 27057943 A/G
rs156558456417 27060967 C/T 0.73 0.74 0.635
rs307116656435 27060985 -/TT 0.48 0.49 0.727
rs190765258847 27063397 A/T 0.66 -0.02
rs375911959595 27064145 CIT 0.06 0.05 0.669
rs375912059661 27064211 G/T 0.13 0.14 0.881
rs378231760355 27064905 C/T 0.08 0.07 0.655
rs382516660407 27064957 A/C 0.82 0.84 0.546
rs187219162357 27066907 A/G 0.04 0.04 0.894
rs8628 68230 27072780 G/T 0.61 0.56 0.234
rs187219368516 27073066 A/G 0.93 0.91 0.528
rs104283369055 27073605 C/T 0.85 0.86 0.682
rs2476 72603 27077153 C/G 0.80 0.81 0.829
rs197620673928 27078478 A/G 0.14 0.14 0.834
rs138865985897 27090447 C/T 0.51 0.48 0.499
rs118452891554 27096104 A/G 0.18 0.18 0.993
166
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
[0301] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure 1G for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis provides the negative
logarithm (base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in Figure
1 G can be determined by consulting Table 45. For example, the left-most X on
the left graph is at
position 27004780. By proceeding down the Table from top to bottom and across
the graphs from left
to right the allele frequency associated with each symbol shown can be
determined.
[0302] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a lOkb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-$ were truncated at that
value.
[0303] The exons and introns of the genes in the covered region are plotted
below each graph at the
appropriate chromosomal positions. The gene boundary is indicated by the
broken horizontal line. The
exon positions are shown as thick, unbroken bars. An arrow is place at the 3'
end of each gene to show
the direction of transcription.
Exam In a 11
LOXLI Region Proximal SNPs
[0304] It has been discovered that rs8818 in the untranslated region (UTR) of
the lysyl oxidase-like
1 (LO~~LI) gene is associated with occurrence of osteoarthritis in subjects.
LOXLl is a Lysyl oxidase-
like protein that catalyzes the cross-linking of collagen via lysine residues.
Deficiency of the related
protein, lysyl oxidase, causes a form of Ehlers-Danlos syndrome. LOXLI likely
is a secreted protein
and its biological activity may be modulated by addition of an antibody, a
recombinant binding partner,
a binding agent, or a recombinant LOXLI protein or functional fragment
thereof.
[0305] Fifty-eight additional allelic variants proximal to rs912428 were
identified and subsequently
allelotyped in osteoarthritis case and control sample sets as described in
Examples 1 and 2. The
167
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
polymorphic variants are set forth in Table 48. The chromosome positions
provided in column four of
Table 48 are based on Genome "Build 34" of NCBI's GenBank.
TABLE 48
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
10
rs104866115 213 71935363 G/T
rs382594215 249 71935399 C/T
rs155043615 1824 71936974 C/T
rs155043815 2057 71937207 C/T
rs155043915 2306 71937456 A/T
rs216524115 2869 71938019 C/T
rs155043315 3976 71939126 A/C
rs305631415 4288 71939438 -/TC
rs241520415 4290 71939440 A/C
rs199231415 4434 71939584 C/G
rs144010115 5298 71940448 A/G
rs228941415 5467 71940617 A/G
rs241520515 8486 71943636 C/G
rs289980715 8487 71943637 A/T
rs893815 15 8831 71943981 C/G
rs305634215 9036 71944186 -/AG
rs407728415 9058 71944208 A/G
rs893816 15 9131 71944281 C/T
rs893817 15 9732 71944882 A/G
rs893818 15 9862 71945012 A/G
rs893819 15 10191 71945341 A/G
rs893820 15 10270 71945420 C/T
rs230471915 16167 71951317 C/T
rs100150715 17620 71952770 G/T
rs153016715 17751 71952901 C/T
rs153016815 17764 71952914 C/T
rs153016915 17787 71952937 C/T
rs230472015 19401 71954551 C/T
rs230472115 21021 71956171 A/C
rs893821 15 21902 71957052 C/T
rs750460 15 22173 71957323 C/T
rs230472215 22416 71957566 C/T
rs144010215 22653 71957803 A/G
rs8898 15 24945 71960095 ClG
rs3522 15 25011 71960161 C/T
rs241520615 28563 71963713 C/T
rs198452615 48574 71983724 C/G
rs198452515 48710 71983860 C/T
rs303165315 48880 71984030 -/TTG
rs241518715 50194 71985344 C/T
rs2507 15 56343 71991493 A/G
rs228941115 56455 71991605 C/T
rs320207715 56729 71991879 C/T
rs228941215 56759 71991909 A/G
rs228941315 56895 71992045 A/G
rs106108215 57036 71992186 ClG
168
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
10
rs227760015 57702 71992852 C/G
rs73485415 62515 71997665 C/T
rs241518815 62629 71997779 C/G
rs321469515 63501 71998651 -/C
rs381619715 63547 71998697 C/T
rs381619815 64876 72000026 C/G
rs230471515 65073 72000223 C/G
rs230127215 67149 72002299 C/T
rs230127315 67549 72002699 C/T
rs378456315 71660 72006810 A/C
rs378456115 71 72007056 C/T
906
rs37845601 15 _ 72007061 A/C
~ __ ~
71911 ~
Assay for Verifying and Allelotypin_g SNPs
[0306] The methods used to verify and allelotype the 58 proximal SNPs of Table
48 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 49 and Table 50, respectively.
TABLE 49
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1048661ACGTTGGATGTTGCTGGGAGACGGAGGTGACGTTGGATGATTCGGCTfTGGCCAGGTGC
rs3825942ACGTfGGATGTAGGTGCTGGCGAAGGCCGAAACGTTGGATGACCTCCGTCTCCCAGCAAC
rs1130133ACGTTGGATGACCAAGTCAGGGAGACCGCGACGTTGGATGAGCGGAACGGCGCGCAGCA
rs1550436ACGTTGGATGGCCAAAAAAACTCAGTAACGACGTTGGATGGTTCATTACAGATAGTITfGC
rs1550437ACGTTGGATGTTGGGCCTTCCCAAGAGGAGACGTTGGATGAGAGCCCCAGCTGTGGACA
rs1550438ACGTTGGATGAGTCAGCCCTTGTCACAGTAACGTTGGATGCATGAGGACACAGTGGAAAG
rs1550439ACGTTGGATGATTCTCTGCTCCCCATTGAGACGTTGGATGTATACTCTGAGGCACTGGAG
rs2165241ACGT1'GGATGTAGAAGACCCACTGACTTGGACGTfGGATGGGGCAGAGAAAACTGAGCTC
rs1550433ACGTTGGATGATAGCAGGAGTGGTCACATCACGTTGGATGTAGCAAATCCTTGAAGAGAG
rs3056314ACGTTGGATGTCTCTCCTGGCCTCTGATTGACGTfGGATGCCTGACGTGTGTCCTCTATC
rs2415204ACGTTGGATGGTTCTCTCCTGGCCTCTGATACGTfGGATGCCTGACGTGTGTCCTCTATC
rs1992314ACGTTGGATGTTTGTCCTAAAGGCCCTGAGACGTTGGATGAGATAAACCCCTGCAGTCTG
rs1440101ACGTTGGATGAAAAGTCAGCAAGTGAGCTCACGTTGGATGTTAATTCCCAGGTCCTAGCC
rs2289414ACGTTGGATGTTGCTTATCTfGTACACCTCACGTfGGATGCTCACCCTGTCACAACCAGT
rs2415205ACGTTGGATGTGATGCTfCAGTTACTCCAGACGTTGGATGTGTGGGCAGCGTAAGTTTTG
rs2899807ACGTTGGATGTGATGCTTCAGTTACTCCAGACGTTGGATGTGTGGGCAGCGTAAGTTTTG
rs893815ACGTTGGATG CACCCTTTTACAGCACTCACACGTTGGATGATCCCTTCTGTGAGTCAAGC
rs3056342ACGTTGGATGTAAGGATCAGTAGGCAGGTCACGTTGGATGATAGCTGGGAATTCCAGGAC
rs4077284ACGTfGGATGTAAGGATCAGTAGGCAGGTCACGTTGGATGATAGCTGGGAATfCCAGGAC
rs893816ACGTTGGATGATfGCCCACAGAATCAAGCCACG1TGGATGTTCTGGAAGGCTAGGTAAGG
rs893817ACGTTGGATGAAACAGGTGAGGTGTGGACGACGTfGGATGAGAAATCTGTTCCCTCCTGC
rs893818ACGTTGGATGTTTTAGGAGCTGTTCAGTTCACGTTGGATGTGGGAGAATTTCCTGACTGC
rs893819ACGTTGGATGCTGTCACACTGACTCTt'GGGACGTTGGATGATGGTCTT'fGTCCTCCGGTT
rs893820ACG'TTGGATGAGAGTCAGTGTGACAGGTTCACGTfGGATGTTCTATATCCTGGCTCTGCC
rs2304719ACGTTGGATGTTTCATCAGTGAGCCTTGCCACGTTGGATGCCTfGTATAGTGAGGTACAG
rs1001507I ACGT1'GGATGAGAATCCTGCAAAACTGGAGACGTTGGATGTGCAGCATGTGAACTGGCAC
169
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1530167ACGTTGGATGAAACATCCTCCTTCCCTCTGACGTTGGATGGCCTAGAACCTAGACCCTTA
rs1530168ACGTTGGATGCAAAACATCCTCCTTCCCTCACGTTGGATGGACCCTTATGGTTfCCCATG
rs1530169ACGTTGGATGTGTGCTGAGCTGAACAGAAGACGTTGGATGGGAATCTGTCTATGTCTGGG
rs2304720ACGTTGGATGATGCTGGGTTCTGGTGTCACACGTTGGATGATAGGCTGTGCTGCAGGGAC
rs2304721ACGTTGGATGCTCAAGTGATGCCTCAGATGACGTTGGATGCTGAAAGAAGCTI'CAGCCTC
rs893821ACGTTGGATGTGGATTAACTAGGGTAGGGCACGTTGGATGGAAAGTTGCATCCCTGCATC
rs750460ACGTTGGATGATGTTTCCCTAGAGCTAGAGACGTTGGATGCTCAGCTCCTCATTACTGCA
rs2304722ACGTTGGATGTrACCACCTTCTCTGGTGAGACGTTGGATGGAGGAAGAAGAGAAACAGGG
rs1440102ACGTTGGATGAGTAAGAGTTrGCCCACCACACGTTGGATGTGACCTAAAGTGCAGGTATC
rs8818 ACGTTGGATGAATCTCTCCCCTTCCAAAGCACGTTGGATGTCCCTGTGGTTTTCATCCAC
rs3522 ACGTTGGATGAACAACACTGTAGAGAAAAGTGAAACGTTGGATGACGTGGATGAAAACCACAGG
rs2415206ACGTrGGATGCACCTTGAGGTGAAACAGACACGTTGGATGTTACTTAGTAGACCCCGAGG
rs1984526ACGTTGGATGATCCTTTGTTCTTGAAACAGACGTTGGATGGGATI'ACAAACGTGAGCCAC
rs1984525ACGTTGGATGCAGCTGGGATTACAGGTATGACGTTGGATGACCAACATGGTGAAACCCTG
rs3031653ACGTTGGATGATAAACGTrAAGCTCAGTTGACGTTGGATGP~AAAAAAAAGTGAAAGTCG
rs2415187ACGTTGGATGTTCTATGAGTTACTTGACACACGTTGGATGGTGTCTTTATCTGACTAGTG
rs2507 ACGTTGGATGGCTGCTCCCAAAGATT'TCTGACGTTGGATGTAAGAAGCACAGAACGCAGG
rs2289411ACGTTGGATGCTGTGGCGAAGTTACCTGGGACGTTGGATGTGCTCCTTCCCATGCCCAAT
rs3202077ACGTTGGATGACAGTGGTTCTCTGGACAAGACGTTGGATGTCTCCTCCTGGAATCACACC
rs2289412ACGTTGGATGGGACAAAGCCTTGTCCAGAGACGTTGGATGATGAATGGAGGCTGCAGGAG
rs2289413ACGTTGGATGTTGGCTGACTTTCCAGAGCCACGTTGGATGTGCAGATGAACACCTCCTCC
rs1061082ACGTTGGATGGGCCCTGCTATGCAGAGAGACGTTGGATGAGGTCGCCCTTCACCTTCAG
rs2277600ACGTTGGATGTAGTGAGGTCCAGGAAGTAGACGTTGGATGCCTGCTACCAGTTCAATGTC
rs734854ACGTTGGATGATAACTCCAAAGGCCATGTGACGTTGGATGCAGACCACAGAGATGAAAAG
rs2415188ACGTTGGATGAAAGTTGACAAAGCCCTTTCACGTTGGATGAGGAAACTGTCTGTCCTTGG
rs3214695ACGTTGGATGACACTTGCCCAAGTTCACTCACGTTGGATGTACATCTGCAGGTGAGAGCA
rs3816197ACGTTGGATGGTGAACTTGGGCAAGTGTACACGTTGGATGAGATfGAGAGCCCTGAGAAG
rs3816198ACGTTGGATGTAGGGTCATGGGGCTTfGGACGTTGGATGGGCTGATAAGAGCCGAGGAC
rs2304715ACGTTGGATGGTGAGTGGCCGCCTGGCACACGTTGGATGTCCTCGGAGGCAGAGATTCG
rs2301272ACGTfGGATGATGATACCCAAGGAGTGTGCACGTrGGATGTCAGCAACTTCCCATCACTC
rs2301273ACGTTGGATGACCTACCGCTGACTTACGGACGTTGGATGACGGATGAATGGATCAAAG
rs3784563ACGTTGGATGAATGTGGTCTGCAGATATGCACGTTGGATGAAACTTACTATCCACCTGCG
rs3784561ACGTT'GGATGATGACCACAATTTATGCTGCACGTTGGATGTGCAAAGATGATTCTGCAGC
rs3784561ACGTTGGATGCAGTAAGGCTGGATTCTAGGACGTTGGATGGCTGCCTGGTGTTAATGGTT
rs3784560I ACGTTGGATGGCTGGATTCTAGGATCAGAGACGTTGGATGACATTCTCAGATAGCGCTGC
TABLE 50
dbSNP Extend Term
rs# Primer Mix
rs1048661 GGAGACGGAGGTGCGGGCC CGT
rs3825942 GAGACCGAGGAGGCGGAG ACG
rs1130133 GGCCGGTACACGCTGCC ACG
rs1550436 AAAAAACTCAGTAACGGAGATAA ACT
rs1550437 TTCACCCCCTGAAAAGCCAGA ACT
rs1550438 GTAGCCCTGTCTGCTAACAGCAT ACT
rs1550439 CTCCCCATTGAGGTTGCTG CGT
rs2165241 CCAGGCATGCCTCTGCCA ACT
rs1550433 GGTCACATCGAGGGAGCC ACT
rs3056314 TGGCCTCTGATTGGCCATG ACT
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs2415204 CTCCTGGCCTCTGATTGGCCA ACT
rs1992314 AAGGCCCTGAGGAGCTACA ACT
rs1440101 CTCGTCACCACATCTGTAACA ACG
rs2289414 TTTATTCACTCATfCATTTGGTC ACT
rs2415205 CTCAGGCCCTGCACAGTGA ACT
rs2899807 CTCAGGCCCTGCACAGTG CGT
rs893815 ACAGCACTCACCTGTCCAC ACT
rs3056342 CACACCCCAACCTTTTTTACCCC ACT
rs4077284 GGCAGGTCTCTGGCAGCA ACG
rs893816 CAGAGTGGCAGCTAAAGCC ACT
rs893817 GGTGTGGACGAGCAATGGGAA ACT
rs893818 AGCCCTCTCACAACCCCTACAGA ACG
rs893819 CACCCTGTCCTCCTGCTCAA ACG
rs893820 ACAGGTTCCTCCTACTGTGC ACG
rs2304719 CAGGAGGGGAGGGGAGCAAG ACG
rs1001507 GGCCCTCTGAGATCATTTCAA ACT
rs1530167 CTGTTCAGCTCAGCACACC ACT
rs1530168 CAGTTAAATCCTGCCCTTCTGTTCACT
rs1530169 TGAACAGAAGGGCAGGATTTAAC ACG
rs2304720 TGTGCCCCAACCCCCCC ACG
rs2304721 TCAGATGCTGCCTCTGCTC ACT
rs893821 GCCAGCTTTATTTGCAGAACATCTACT
rs750460 CAGAGAGGTTGGATCCTGCC ACG
rs2304722 CTCTGGTGAGCAGTTGAGG ACG
rs1440102 GCAGGCAAGGCCACCTGA ACT
rs8818 AGCCCCCAACCCACAGGCA ACT
rs3522 TATAAAATGGGGTCTGGC ACT
rs2415206 GAAACAGACCCCCACCCC ACG
rs1984526 AGCATAAAGGTGAAAGATGGGCC ACT
rs1984525 GGATTACAGGTATGCACCACCA ACG
rs3031653 AAGCTCAGTTGTGGCTCCAAACAAACT
rs2415187 TCTTTTTAAAAAACTACACCAGGTACG
rs2507 TGACTCATCTGCCAGCTC ACG
rs2289411 GGGATCCTGGCTGGCCC ACT
rs3202077 CTGGACAAGGCTTTGTCCAT ACG
rs2289412 GCCTTGTCCAGAGAACCACT ACT
rs2289413 CAAGCCTGGCACCAAGCC ACG
rs1061082 CTATGCAGAGAGCTGCGGC ACT
rs2277600 GGAAGTAGGCGCTTTGGGTG ACT
rs734854 ACTCCAAAGGCCATGTGTCTTAACACG
rs2415188 GGGGTGCTGTTAGGGCAGCC ACT
rs3214695 CGCTfGGCAGCTGTCGTG ACT
rs3816197 CTTGGGCAAGTGTACCTTACG ACG
rs3816198 CCCCAGAGCCAGCCAGC ACT
rs2304715 CCGCCTGGCACGGCGGA ACT
rs2301272 TGTGCTAGGACAAGATCCTAGCT ACT
rs2301273 GCTGACTTACGGTAAAGCGG ACT
rs3784563 TGACCACAATT1'ATGCTGCCA ACT
rs3784561 GCAGGTGGATAGTAAGTTTCCA I ACT
171
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs3784561 GCTGGATTCTAGGATCAGAGACAACT
rs3784560 CTAGGATCAGAGACAGGTAG ACT
Genetic Analysis
[0307] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 51.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (A1 AF
= 1-A2 AF). For
example, the SNP rs1048661 has the following case and control allele
frequencies: case A1 (G) = 0.725;
case A2 (T) = 0.275; control A1 (G) = 0.767; and control A2 (T) = 0.233, where
the nucleotide is
provided in paranthesis. Some SNPs are labeled "untyped" because of failed
assays.
TABLE 51
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ Position Allele Case Control Value
ID NO: 10 AF AF
rs1048661213 71935363 G/T 0.275 0.233 0.077
rs3825942249 71935399 C/T 0.107 0.148 0.056
rs15504361824 71936974 C/T 0.401 0.420 0.470
rs15504382057 71937207 C/T
rs15504392306 71937456 A/T
rs21652412869 71938019 C/T 0.427 0.430 0.883
rs15504333976 71939126 A/C
rs30563144288 71939438 -/TC
rs24152044290 71939440 A/C 0.176 0.177 0.982
rs19923144434 71939584 C/G 0.599 0.601 0.938
rs14401015298 71940448 A/G
rs22894145467 71940617 A/G
rs24152058486 71943636 C/G
rs28998078487 71943637 A/T 0.951 0.956 0.863
rs893815 8831 71943981 C/G
rs30563429036 71944186 -/AG 0.290 0.292 0.927
rs40772849058 71944208 A/G 0.358 0.358 0.985
rs893816 9131 71944281 C/T 0.517 0.515 0.928
rs893817 9732 71944882 A/G 0.162 0.158 0.819
rs893818 9862 71945012 A/G 0.311 0.313 0.920
rs893819 10191 71945341 A/G 0.637 0.642 0.866
rs893820 10270 71945420 C/T 0.901 0.910 0.605
rs230471916167 71951317 C/T 0.320 0.299 0.387
rs100150717620 71952770 G/T 0.910 0.916 0.709
rs153016717751 71952901 C/T
rs153016817764 71952914 ClT
rs153016917787 71952937 C/T 0.209 0.203 0.779
rs230472019401 71954551 C/T 0.942 0.947 0.724
rs230472121021 71956171 A/C 0.798 0.814 0.519
rs893821 21902 71957052 C/T 0.113 0.116 0.879
rs750460 22173 71957323 C/T 0.473 0.438 0.176
rs230472222416 71957566 CIT 0.744 0.747 0.926
rs144010222653 71957803 A/G
rs8818 24945 71960095 C/G
rs3522 25011 71960161 C/T 0.424 0.441 0.472
rs241520628563 71963713 C/T 0.376 0.366 0.731
rs198452648574 71983724 ClG 0.593 unt ed NA
rs198452548710 71983860 C/T
rs303165348880 71984030 -/TTG
172
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ Position Allele Case Control Value
ID NO: 10 AF AF
rs241518750194 71985344 C/T
rs2507 56343 71991493 A/G 0.655 0.653 0.924
rs228941156455 71991605 C/T
rs320207756729 71991879 C/T
rs228941256759 71991909 A/G 0.971 0.968 0.855
rs228941356895 71992045 A/G 0.972 0.972 0.997
rs106108257036 71992186 C/G
rs227760057702 71992852 C/G
rs734854 62515 71997665 C/T 0.381 0.379 0.915
rs241518862629 71997779 ClG 0.532 0.538 0.832
rs321469563501 71998651 -!C 0.308 0.300 0.751
rs381619763547 71998697 C/T 0.327 0.311 0.512
rs381619864876 72000026 ClG 0.598 0.584 0.575
rs230471565073 72000223 ClG 0.660 0.643 0.534
rs230127267149 72002299 C/T 0.974 0.972 0.853
rs230127367549 72002699 C/T 0.952 0.966 0.409
rs378456371660 72006810 A/C 0.495 0.508 0.590
rs378456171906 72007056 C/T 0.470 0.466 0.872
rs378456071911 72007061 A/C
[0308] The LOXLI proximal SNPs were also allelotyped in the replication
cohorts using the
methods described herein and the primers provided in Tables 49 and 50. The
replication allelotyping
results for replication cohort # 1 and replication cohort #2 are provided in
Tables 52 and 53,
respectively.
TABLE 52
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ PositionAllele Case Control Value
II? NO: AF AF
10
rs1048661213 , 71935363G/T 0.250 0.252 0.953
rs3825942249 71935399C/T 0.126 0.141 0.539
rs15504361824 71936974C/T 0.397 0.405 0.845
rs15504382057 71937207C/T
rs15504392306 71937456A/T
rs21652412869 71938019C/T 0.429 0.425 0.894
rs15504333976 71939126A/C
rs30563144288 71939438-/TC
rs24152044290 71939440A/C 0.162 unt ed 0.176
rs19923144434 71939584C/G 0.583 0.594 0.756
rs14401015298 71940448A/G
rs22894145467 71940617A/G
rs24152058486 71943636C/G
rs28998078487 71943637A/T 0.939 unt ed NA
rs893815 8831 71943981C/G
rs30563429036 71944186-/AG 0.317 0.311 0.846
rs40772849058 71944208A/G 0.372 0.365 0.881
rs893816 9131 71944281C/T 0.510 0.518 0.793
rs893817 9732 71944882A/G 0.178 0.170 0.784
rs893818 9862 71945012A/G 0.327 0.320 0.818
rs893819 10191 71945341A/G 0.610 unt ed NA
rs893820 10270 71945420C/T 0.874 0.903 0.218
rs230471916167 71951317C/T 0.309 0.289 0.537
rs100150717620 71952770G/T 0.908 0.924 0.525
rs153016717751 71952901C/T
rs153016817764 71952914C/T
rs153016917787 71952937C/T 0.237 0.202 0.249
rs230472019401 71954551C/T 0.935 0.944 0.661
rs230472121021 71956171A/C 0.759 0.823 0.091
rs893821 21902 71957052C/T 0.114 0.122 0.778
173
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position~.inChromosomeAl/A2 F A2 F A2 F p-
rs# SEQ Position Allele Case Control Value
ID NO: 10 AF AF
rs750460 22173 71957323 C/T 0.469 0.440 0.433
rs230472222416 71957566 C/T 0.729 0.746 0.572
rs144010222653 71957803 A/G
rs8818 24945 71960095 C/G
rs3522 25011 71960161 C/T 0.416 0.440 0.454
rs241520628563 71963713 C/T 0.362 unt ed NA
rs198452648574 71983724 C/G 0.593 unt ed
rs198452548710 71983860 C/T
rs303165348880 71984030 -/TTG
rs241518750194 71985344 C/T
rs2507 56343 71991493 A/G 0.676 0.653 0.471
rs228941156455 71991605 C/T
rs320207756729 71991879 C/T
rs228941256759 71991909 A/G 0.964 0.954 0.626
rs228941356895 71992045 A/G 0.963 0.959 0.833
rs106108257036 71992186 ClG
rs227760057702 71992852 C/G
rs734854 62515 71997665 C/T 0.403 0.383 0.531
rs241518862629 71997779 C/G 0.555 0.564 0.809
rs321469563501 71998651 -/C 0.289 0.300 0.721
rs381619763547 71998697 C/T 0.304 0.308 0.904
rs381619864876 72000026 ClG 0.601 0.598 0.922
rs230471565073 72000223 C/G 0.649 0.678 0.457
rs230127267149 72002299 C/T 0.966 0.959 0.752
rs230127367549 72002699 C/T 0.935 0.946 0.649
rs378456371660 72006810 A/C 0.502 0.516 0.685
rs378456171906 72007056 C/T 0.438 0.471 0.319
rs378456071911 72007061 A/C
TABLE 53
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ Position AlleleCase Control Value
ID NO: 10 AF AF
rs1048661213 71935363 G/T 0.307 0.203 0.007
rs3825942249 71935399 C/T 0.084 0.159 0.031
rs15504361824 71936974 C/T 0.406 0.445 0.274
rs15504382057 71937207 C/T
rs15504392306 71937456 A/T
rs21652412869 71938019 C/T 0.423 0.439 0.669
rs15504333976 71939126 A/C
rs30563144288 71939438 -/TC
rs24152044290 71939440 A/C 0.200 unt ed
rs19923144434 71939584 C/G 0.618 0.612 0.854
rs14401015298 71940448 A/G
rs22894145467 71940617 A/G
rs24152058486 71943636 C/G
rs28998078487 71943637 A/T 0.965 0.956 0.737
rs893815 8831 71943981 C/G
rs30563429036 71944186 -/AG 0.257 0.264 0.833
rs40772849058 71944208 A/G 0.341 0.345 0.905
rs893816 9131 71944281 C/T 0.526 0.509 0.655
rs893817 9732 71944882 A/G 0.142 0.139 0.895
rs893818 9862 71945012 A/G 0.290 0.302 0.712
rs893819 10191 71945341 A/G 0.671 0.642 0.431
rs893820 10270 71945420 C/T 0.934 0.922 0.681
rs230471916167 71951317 C/T 0.334 0.316 0.613
rs100150717620 71952770 G/T 0.911 0.903 0.741
rs153016717751 71952901 C/T
rs153016817764 71952914 C/T
rs153016917787 71952937 C/T 0.173 0.203 0.360
rs230472019401 71954551 C/T 0.951 0.952 0.952
rs230472121021 71956171 A/C 0.848 0.799 0.150
174
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ Position AlleleCase Control Value
ID NO: 10 AF AF
rs89382121902 71957052 C/T 0.112 0.106 0.829
rs75046022173 71957323 C/T 0.478 0.435 0.242
rs230472222416 71957566 C/T 0.764 0.748 0.626
rs144010222653 71957803 A/G
rs8818 24945 71960095 C/G
rs3522 25011 71960161 C/T 0.435 0.444 0.814
rs241520628563 71963713 C/T 0.394 0.366 0.419
rs198452648574 71983724 C/G
rs198452548710 71983860 C/T
rs303165348880 71984030 -/TTG
rs241518750194 71985344 C/T
rs2507 56343 71991493 A/G 0.630 0.653 0.509
rs228941156455 71991605 C/T
rs320207756729 71991879 C/T
rs228941256759 71991909 A/G 0.979 unt ed
rs228941356895 71992045 A/G
rs106108257036 71992186 C/G
rs227760057702 71992852 C/G
rs73485462515 71997665 C/T 0.354 0.372 0.611
rs241518862629 71997779 C/G 0.502 0.497 0.897
rs321469563501 71998651 -/C 0.331 0.300 0.367
rs381619763547 71998697 C/T 0.357 0.317 0.259
rs381619864876 72000026 C/G 0.594 0.562 0.416
rs230471565073 72000223 C/G 0.674 0.587 0.020
rs230127267149 72002299 C/T
rs230127367549 72002699 C/T 0.973 unt ed
rs378456371660 72006810 A/C 0.485 0.496 0.777
rs378456171906 72007056 C/T 0.511 0.459 0.174
rs378456071911 72007061 A/C
[0309] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure 1H for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in Figure
lE can be determined by consulting Table 51. For example, the left-most X on
the left graph is at
position 71935363. By proceeding down the Table from top to bottom and across
the graphs from left
to right the allele frequency associated with each symbol shown can be
determined.
[0310] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoothcr through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J-M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a l Okb sliding window with lkb step sizes. Within each
window, a chi-square
175
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-$ were truncated at that
value.
[0311] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 12
CASPR4 Region Proximal SNPs
[0312] It has been discovered that rs1395486 in the cell recognition protein
CASPR4 gene is
associated with occurrence of osteoarthritis in subjects. This gene product
belongs to the neurexin
family, members of which function in the nervous system as cell adhesion
molecules and receptors.
Like other neurexin proteins, CASPR4 contains epidermal growth factor repeats
and laminin G domains.
In addition, it includes an FS/8 type C domain, discoidin/neuropilin- and
fibrinogen-like domains, and
thrombospondin N-terminal-like domains. Alternative splicing of this gene
results in 2 transcript
variants encoding different isoforms. CASPR4 biological activity can be
modulated by addition of an
antibody, a recombinant binding partner, a binding agent, or a recombinant
CASPR4 protein or
functional fragment thereof.
[0313] Fifty-six additional allelic variants proximal to rs1395486 were
identified and subsequently
allelotyped in osteoarthritis case and control sample sets as described in
Examples 1 and 2. The
polymorphic variants are set forth in Table 54. The chromosome positions
provided in column four of
Table 54 are based on Genome "Build 34" of NCBI's GenBank.
TABLE 54
dbSNP Chromo- Position ChromosomeAllele
rs# some in SEQ Position Variants
ID NO:
11
rs189675316 205 76177855 C/T
rs397445116 866 76178516 C/T
rs182077016 4212 76181862 C/T
rs142875316 5934 76183584 C/T
rs722229 16 11486 76189136 C/T
rs385175416 16969 76194619 A/G
rs234043016 22509 76200159 A/G
rs234043116 22796 76200446 AIG
rs115941516 28097 76205747 C/T
rs150683616 28626 76206276 C/T
rs150683716 28853 76206503 C/T
rs150683816 28873 76206523 C/T
rs966668 16 30155 76207805 A/G
rs191124516 30827 76208477 C/T
rs150683916 31956 76209606 C/T
rs150684016 32404 76210054 ~C~
176
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo- Position ChromosomeAllele
rs# some in SEQ Position Variants
ID NO:
11
rs187627516 32944 76210594 A/G
rs191124216 35205 76212855 A/G
rs191124316 35227 76212877 C/T
rs981231 16 35781 76213431 C/T
rs150682916 41052 76218702 C/T
rs150683316 45051 76222701 A/G
rs139548616 46039 76223689 C/T
rs150683216 47276 76224926 A/G
rs150683016 47678 76225328 CIT
rs968537 16 47716 76225366 A/G
rs150681616 51014 76228664 A/G
rs150682816 54408 76232058 A/G
rs150682716 54596 76232246 C/T
rs154296916 56853 76234503 CIG
rs139548416 61851 76239501 A/G
rs187627416 62016 76239666 A/G
rs187627316 62461 76240111 C/T
rs150682216 68257 76245907 C/G
rs150682016 69793 76247443 C/T
rs150681916 73976 76251626 A/C
rs150681816 73999 76251649 A/T
rs150681716 74053 76251703 A/G
rs139548816 75315 76252965 A/G
rs222153416 75729 76253379 G/T
rs191124416 76466 76254116 A/G
rs213562416 77216 76254866 C/T
rs213562316 77217 76254867 G/T
rs150683516 79239 76256889 C/G
rs150683416 80825 76258475 A/G
rs199565316 81060 76258710 C/G
rs199565216 81097 76258747 A/C
rs139548716 81426 76259076 G/T
rs394708316 84787 76262437 C/T
rs150682516 84896 76262546 A/T
rs150682416 85165 76262815 C/G
rs156711816 86502 76264152 C/G
rs103968316 86753 76264403 CIT
rs287977716 86941 76264591 C/T
rs187627216 88787 76266437 C/T
rs303587816 95598 76273248 -/AGAGC
Assa ~~for-Verifyin~ and Allelotypin~ SNPs
[0314] The methods used to verify and allelotype the 56 proximal SNPs of Table
54 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 55 and Table 56, respectively.
177
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
TABLE 55
dbSNP Fo rward Reverse
rs# PCR primer PCR primer
rs1896753ACGTTGGATGTTTGAAGAGAGGGACTAGAGACGTTGGATGGAAAATGAACTGGAATGGGG
rs3974451ACGTTGGATGTTGCATAAGGTGTGAGGAAGACGTTGGATGAATGGTGTTGGGA.AAACTGG
rs1820770ACGTTGGATGCTTGGAACCAACCCAAATGCACGTTGGATGGGCTGCATAGTAT-fCCACAG
rs1428753ACGTTGGATGCAATAGCTATCTCCTACTTGACGTTGGATGGATGCTTTGTATTGACAACC
rs722229ACGTTGGATGGAAGGAGGCTCACTATTTCCACGTTGGATGGGCTAGGGTAGCAAACATCA
rs3851754ACGTTGGATGAGGTTTGGAGAATGCCAACTACGTTGGATGAGATTGAATCAGA-fGGACTG
rs2340430ACGTTGGATGATGGCCTTCCAAAGATGTTCACGTTGGATGCATCTACAATCCCAATATGCC
rs2340431ACGTTGGATGTTTGTGCAACCTCTGCAAGCACGTTGGATGAGATGTCAGCAGGATGCATG
rs1159415ACGTTGGATGGCTTTCCAATGATTTGGGAGACGTTGGATGCTGGGTCTTCCTAATGTGTT
rs1506836ACGTTGGATGCCTGGGCACAGATTCATTTCACGTTGGATGCTGCAGCGACCTTTCATTCA
rs1506837ACGTTGGATGCTGACATTGAGCTAGTCTTTCACGTTGGATGGTAGTTGGTGAATTTGGTGG
rs1506838ACGTTGGATGGTAGTTGGTGAATTTGGTGGACGTTGGATGGACATTGAGCTAGTCTTTCC
rs966668ACGTTGGATGCACTTCATAGTGTGAAAAGTCACGTTGGATGCCAGTAAATGCAAGATTTTCC
rs1911245ACGTTGGATGAACAACTAGGCAATTCGGTGACGTTGGATGCCATCAGAAGTAAACCGTTTC
rs1506839ACGTTGGATGCCAAATTTTGCTTTGTTAGACACGTTGGATGTGCACAATTCAAGTGAAGTC
rs1506840ACGTTGGATGGGAAGAATGACCTTGTGTGGACGTTGGATGAGCTGTGAGTGAGGATGATG
rs1876275ACGTTGGATGAACTGTTCTCTGCCCTTTGGACGTTGGATGTTCACGGACATAAGGGAAGG
rs1911242ACGTTGGATGGTTCCCTAAGTACTTTAGAAACGTTGGATGCTCTGCAAAGCAATAAGCTAC
rs1911243ACGTTGGATGCTTATAATTCAGTTCCCTAAGACGTTGGATGGCAATAAGCTACCAAAATAG
rs981231ACGTTGGATGATGCTAACCTGTCTAAATCCACGTTGGATGTAGTGCTCTGGACTAGAAAG
rs1506829ACGTTGGATGTGGAAAGTTGCAATTCCCTGACGTTGGATGCCATCTTAAAACCATGCGAG
rs1506833ACGTTGGATGGTTTTATCTGGTTCCTTACAGACGTTGGATGGCTGTATACGTACTTTAAAC
rs1395486ACGTTGGATGCTCATTTATTTCATGTTCACACGTTGGATGTGCTGGAATAATGATTGTTG
rs1506832ACGTTGGATGGGTAATGGTCATAAGAATGCCACGTTGGATGGAGCTCAATTAGCATCTCTC
rs1506830ACGTTGGATGCAACAGTAAAGGCATGAAAGACGTTGGATGCATTGGACTATCAAAAAGTG
rs968537ACGTTGGATGATTATTTGGTGGGAAGAGGGACGTTGGATGAAATGTTACGTAGGCCAAAC
rs1506816ACGTTGGATGTACATATGACCACTGTTTCCACGTTGGATGCTAAGCAGGGAAGTAGTAAG
rs1506828ACGTTGGATGGAGCTTTTTCCATTAGACCCACGTTGGATGGTTGAAAATCAGACAAGGGC
rs1506827ACGTTGGATGAATGCGCTATATCTGATGACACGTTGGATGAACCCATTTCTTAGCCAGAG
rs1542969ACGTTGGATGCAGATTACAGCCAAGTTTGCACGTTGGATGGGTTTGAATTCCCAAGACAG
rs1395484ACGTTGGATGCAAGCTCACATAACACAGGCACGTTGGATGAAGAGATGCCGCGATTTTGG
rs1876274ACGTTGGATGGGTATCTGATCATCTGCCTGACGTTGGATGGGGATTGATTGACAAGGAG
rs1876273ACGTTGGATGTGGAAGAAACATAGCTCCTGACGTTGGATGAAAATCCCTCCAGTGTTTGC
rs1506822ACGTTGGATGTTCTCCAGATCTGCAAACAGACGTTGGATGGTAATGAGAGAAGTAGAGGC
rs1506820ACGTTGGATGTTCTATATATGTGTGTGTGCACGTTGGATGTTAGGGTTCTCTAGAAAGAC
rs1506819ACGTTGGATGTGAGGGAATTGTGTCTGCAGACGTTGGATGGCCAGAGAGGCTAGAAATTG
rs1506818ACGTTGGATGAGGGCTGCTTAGCATTTCACACGTTGGATGAGATCAGAGAGCAATGGTCC
rs1506817ACGTTGGATGCCTCTTTCTCGTGCTTTCTCACGTTGGATGCTCAGATCCTTGG CCAATTC
rs1395488ACGTTGGATGGACACTTGAATGCATCACCGACGTTGGATGGGTGACTTCTGTGACATTGC
rs2221534ACGTTGGATGTAATGCAGGTCTCAAGTGCCACGTTGGATGCAAATCAGACTGAGTCGCTG
rs1911244ACGTTGGATGACCTGTATTCCTGTTCCAGGACGTTGGATGCAACATTCTACTTCCTGGGC
rs2135624ACGTTGGATGGTACGCCCTACTCTCATATCACGTTGGATGAGCTCTTAATTCCATGGCAG
rs2135623ACGTTGGATGGTACGCCCTACTCTCATATCACGTTGGATGAGCTCTTAATTCCATGGCAG
rs1506835ACGTTGGATGAATT-AGCTGGACATGGTGGCACGTTGGATGTCAAGTGAACCTCCAACCTC
rs1506834ACGTTGGATGACATTTTCCCAGCACTGTCCACGTTGGATGCTCACTCCTACTCTGAGTAC
rs1995653ACGTTGGATGCCAGCCTTCTGTTACTCTTGACGTTGGATGCTGTCCTCATGGTGTTTCCA
rs1995652ACGTTGGATGCGTGTTACAACCTGTAATGCACGTTGGATGACATAAATATGGCCCCTGTC
rs1395487ACGTTGGATGAAAAGCTTTAGGTGCCACAGACGTTGGATGGCTTGTGTTACTT-f'AGCTAC
rs3947083ACGTTGGATGAAGGTGGGCTCTTTATAGTGACGTTGGATGGAGGTGTGATGGT-fATGTTTC
rs1506825ACGTTGGATGCCTGCATATGATGTTCTGTGACGTTGGATGTAGCAGCTTTCGGTGTATAG
178
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1506824ACGTTGGATGAGCAATGGATTCAAATGCTCACGTTGGATGCACTGGTCGATGAAAAATAC
rs1567118ACGTTGGATGTCGGCCAATCTGTCCAAATGACGTTGGATGAATTGTCCCCGTTTCCACAG
rs1039683ACGTTGGATGTGATGTGTGGAGGCATGTTGACGTTGGATGACAGGCAACAACTGCCAAAG
rs2879777ACGTTGGATGCTAATCATGTGCGATGAGGGACGTTGGATGAAGAAGAGATGGGCCATAGT
rs1876272ACGTTGGATGTTCTTTGTCTGGAGTGGGAGACGTTGGATGGGTTCCAACACTAGCAGTTC
rs3035878ACGTTGGATGTTCTACAAGGAGCTGTGTAGACGTTGGATGCTGACTGGTAAATTCACGAC
TABLE 56
dbSNP Extend Term
rs# Primer Mix
rs1896753 GGAATT1-AATTTGGTGCCTCTTCAACT
rs3974451 TTCAGT-TTCAGCTTTCTGCATA ACG
rs1820770 GAACCAACCCAAATGCCCATCA ACT
rs1428753 TAACATT-f'ACTGATAGAATAAAGCACT
rs722229 TTCCCTGCAGAAAATGAGACA ACT
rs3851754 AACTCACACACACACACAGAA ACT
rs2340430 CGTTG GGACCTATAGGTATG ACT
rs2340431 CTCTGCAAGCTGGAAAGGAC ACT
rs1159415 TATGTTTAGGAACATTTTCCTAACACT
rs1506836 GTCTCACAGCTTGAAGATGC ACG
rs1506837 CATTGAGCTAGTCTTTCCTCTGT ACG
rs1506838 GTTGGTGAATTTGGTGGAGAATCTACT
rs966668 TCATAGT-GTGAAAAGTCTAAAAAAACT
rs1911245 TTCCTCTTTTTCAGACAAAATTG ACG
rs1506839 AATTTTGCTTTGTTAGACCTTAGGACG
rs1506840 GCTGGTGTCCTGTGAAATTG ACG
rs1876275 TCTTGGTTCAGGTATCACCTA ACG
rs1911242 TAGAAAAATTTGCCTTTTGAGAAAACG
rs1911243 TAATTCA.GTTCCCTAAGTACTTTAACT
rs981231 CCTGTCTAAATCCATTTGATTAAAACT
rs1506829 GATCTAAATAGCTACTGGGAAA ACT
rs1506833 TCTGGT'>-CCTTACAGAAACACTTAACG
rs1395486 TTTCATGTTCACAAAAAATCTTCTACG
rs1506832 GGTCATAAGAATGCCATTATTCT ACG
rs1506830 AATAATATGTTTGGCCTACGTAA ACG
rs968537 AGGGAGGTAAGAGTCAACAGTAA ACT
rs1506816 ATATGACCACTGTTTCCTCATTT ACT
rs1506828 CCATTAGACCCCTTAGCATAT ACG
rs1506827 TGACAA'1-AGAAACTAAGACAAATAACT
rs1542969 GCCAAGTTTGCATCTTTCATGT ACT
rs1395484 AACACAGGCACAGCTGTGAT ACT
rs1876274 CTAATTCACAAATATTCCCTTACTACT
rs1876273 TAGCTCCTGGCCCTACCAT ACT
rs1506822 CTGCAAACAGGATCACTGCT ACT
rs1506820 ATATACAGAACACACACACACA ACG
rs1506819 TCTGCAGGAGCACGGACC CGT
rs1506818 GGCCAAGGATCTGAGGGAA CGT
179
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs1506817 TGCTTTCTCTAGGGCTGCTT ACT
rs1395488 TGAATGCATCACCGGAGGAT ACT
rs2221534 CTCAAGTGCCTATCTATCATG CGT
rs1911244 TTCCAGGTTAGAATTCCAGAGAT ACG
rs2135624 CTCTCATATCAATTCTCCCTGTT ACG
rs2135623 CTCTCATATCAATTCTCCCTGT ACT
rs1506835 GACATGGTGGCAAATTCCTGTA ACT
rs1506834 ACTGTCCCA'1-TCACTGTCATAAACT
rs1995653 TTCTGTTACTC-TTGATCAGAATGCACT
rs1995652 TAATGCTTTTA~'GAACTTAGTTGTACT
rs1395487 CTTTAGGTGCCACAGAAGATA CGT
rs3947083 GGGCTCTTTA'rAGTGTATTTTCCTACG
rs1506825 ATACTGTGAGAAAGATGAAGGT CGT
rs1506824 CAAATGCTCAAATATCAATATGTGACT
rs1567118 TGTCCAAATGGCAATGTTGGT ACT
rs1039683 GGAGGCATGTT-GGAACTTACAGACACT
rs2879777 GAGGGGTGGTCACACAGC ACT
rs1876272 CTGGAGTGGGAGACAGGGT ACG
rs3035878 TGTGTAGCTAAATGTTGAGCAGAGACT
~
Genetic Anal, skis
[0315] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 57.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (A1 AF
= 1-A2 AF). For
example, the SNP rs1896753 has the following case arid control allele
frequencies: case A1 (A) = 0.79;
case A2 (T) = 0.21; control A1 (A) = 0.81; and control A2 (T) = 0.19, where
the nucleotide is provided
in paranthesis. Some SNPs are labeled "untyped" because of failed assays.
TABLE 57
dbSNP Position ChromosomeA.1JA2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 11
rs1896753 205 76177855 C/T
rs3974451 866 76178516 C/T
rs1820770 4212 76181862 C/T
rs1428753 5934 76183584 C/T 0.486 0.467 0.459
rs722229 11486 76189136 C/T
rs3851754 16969 76194619 A/G 0.287 0.300 0.569
rs2340430 22509 76200159 A/G 0.488 0.523 0.155
rs2340431 22796 76200446 A/G 0.030 0.028 0.844
rs1159415 28097 76205747 C/T 0.480 0.477 0.919
rs1506836 28626 76206276 C/T 0.401 0.404 0.891
rs1506837 28853 76206503 C/T 0.394 0.396 0.933
rs1506838 28873 76206523 C/T 0.334 0.343 0.727
rs966668 30155 76207805 A/G
rs1911245 30827 76208477 C/T 0.836 0.824 0.631
rs1506839 31956 76209606 C/T 0.434 0.436 0.936
rs1506840 32404 76210054 C/T 0.382 0.381 0.993
180
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 11
rs1876275 32944 76210594 A/G 0.463 0.461 0.918
rs1911242 35205 76212855 A/G 0.419 0.410 0.703
rs1911243 35227 76212877 C/T
rs981231 35781 76213431 C/T 0.451 0.430 0.510
rs1506829 41052 76218702 C/T 0.393 0.379 0.576
rs1506833 45051 76222701 A/G 0.509 0.530 0.378
rs1395486 46039 76223689 C/T
rs1506832 47276 76224926 A/G 0.518 0.516 0.949
rs1506830 47678 76225328 C/T 0.036 0.031 0.710
rs968537 47716 76225366 A/G 0.243 0.275 0.175
rs1506816 51014 76228664 A/G 0.392 0.369 0.348
rs1506828 54408 76232058 A/G 0.418 0.413 0.816
rs1506827 54596 76232246 C/T 0.432 0.449 0.477
rs1542969 56853 76234503 C/G
rs1395484 61851 76239501 A/G 0.417 0.441 0.349
rs1876274 62016 76239666 A/G 0.381 0.369 0.629
rs1876273 62461 76240111 C/T 0.382 0.364 0.445
rs150(i82268257 76245907 C/G 0.355 0.351 0.855
rs1506820 69793 76247443 C/T 0.326 0.256 0.054
rs1506819 73976 76251626 A/C 0.446 0.424 0.358
rs1506818 73999 76251649 A/T 0.126 0.145 0.465
rs1506817 74053 76251703 A/G 0.186 0.199 0.570
rs1395488 75315 76252965 A/G 0.489 0.499 0.689
rs2221534 75729 76253379 G/T 0.450 0.431 0.455
rs1911244 76466 76254116 A/G 0.493 0.491 0.960
rs2135624 77216 76254866 C/T
rs2135623 77217 76254867 G/T 0.034 0.032 0.899
rs1506835 79239 76256889 C/G 0.549 0.538 0.666
rs1506834 80825 76258475 A/G 0.390 0.392 0.958
rs1995653 81060 76258710 C/G 0.396 0.402 0.783
rs1995652 81097 76258747 A/C 0.436 0.435 0.979
rs1395487 81426 76259076 G/T 0.505 0.504 0.975
rs3947083 84787 76262437 C/T 0.373 0.366 0.773
rs1506825 84896 76262546 A/T 0.412 0.398 0.569
rs1506824 85165 76262815 C/G 0.444 0.414 0.242
rs1567118 86502 76264152 C/G 0.032 0.024 0.557
rs1039683 86753 76264403 C/T 0.382 0.373 0.707
rs2879777 86941 76264591 C/T 0.269 0.279 0.676
rs1876272 88787 76266437 C/T
rs3035878 95598 76273248 -/AGAGC0.978 unt ed NA
[0316] The CASPR4 proximal SNPs were also allelotyped in the replication
cohorts using the
methods described herein and the primers provided in Tables 55 and 56. The
replication allelotyping
results for replication cohort #1 and replication cohort #2 are provided in
Tables 58 and 59,
respectively.
TABLE 58
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ Position AlleleCase Control Value
ID NO: AF AF
11
rs1896753205 76177855 C/T
rs3974451866 76178516 C/T
rs18207704212 76181862 C/T
rs14287535934 76183584 C/T 0.463 0.474 0.756
rs722229 11486 76189136 C/T
rs385175416969 76194619 A/G 0.283 0.309 0.375
rs234043022509 76200159 A/G 0.494 0.519 0.477
rs234043122796 76200446 A/G 0.035 0.028 0.748
rs115941528097 ~ 76205747 C/T 0.436 0.472 __
~ ~ ~ ~ ~ 0.287
181
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ PositionAllele Case Control Value
ID NO: AF AF
11
rs150683628626 76206276C/T 0.392 0.401 0.786
rs150683728853 76206503C/T 0.388 0.399 0.727
rs150683828873 76206523C/T 0.318 0.327 0.778
rs966668 30155 76207805A/G
rs191124530827 76208477C/T 0.825 0.821 0.896
rs150683931956 76209606C/T 0.450 0.441 0.817
rs150684032404 76210054C/T 0.379 0.383 0.926
rs187627532944 76210594A/G 0.469 0.470 0.986
rs191124235205 76212855A/G 0.437 0.415 0.514
rs191124335227 76212877C/T
rs981231 35781 76213431C/T 0.449 0.414 0.415
rs150682941052 76218702C/T 0.398 0.394 0.894
rs150683345051 76222701A/G 0.515 0.544 0.393
rs139548646039 76223689C/T
rs150683247276 76224926A/G 0.526 0.511 0.720
rs150683047678 76225328C/T 0.053 0.039 0.488
rs968537 47716 76225366A/G 0.241 0.298 0.045
rs150681651014 76228664A/G 0.379 0.370 0.771
rs150682854408 76232058A/G 0.416 0.429 0.706
rs150682754596 76232246C/T 0.428 0.435 0.836
rs154296956853 76234503C/G
rs139548461851 76239501A/G 0.418 0.459 0.208
rs187627462016 76239666A/G 0.384 0.382 0.942
rs 187627362461 76240111C/T 0.393 0.360 0.271
rs150682268257 76245907C/G 0.353 0.368 0.637
rs150682069793 76247443C/T 0.288 unt ed NA
rs150681973976 76251626A/C 0.453 0.424 0.378
rs150681873999 76251649AIT 0.149 NA 0.126
rs150681774053 76251703A/G 0.195 0.212 0.573
rs139548875315 76252965A/G 0.490 0.490 1.000
rs222153475729 76253379G/T 0.446 0.433 0.711
rs191124476466 76254116A/G 0.495 0.480 0.646
rs213562477216 76254866C/T
rs213562377217 76254867G/T 0.027 0.030 0.896
rs150683579239 76256889C/G 0.563 0.556 0.848
rs150683480825 76258475A/G 0.377 0.388 0.722
rs199565381060 76258710C/G 0.381 0.395 0.675
rs199565281097 76258747A/C 0.435 0.423 0.750
rs139548781426 76259076G/T 0.505 0.500 0.874
rs394708384787 76262437C/T 0.367 0.370 0.929
rs150682584896 76262546A/T 0.406 0.397 0.798
rs150682485165 76262815C/G 0.446 0.413 0.361
rs156711886502 76264152C/G 0.029 0.023 0.776
rs103968386753 76264403C/T 0.376 0.365 0.729
rs287977786941 76264591C/T 0.265 0.278 0.669
rs187627288787 76266437C/T
0.972 untyped
rs303587895598 76273248/AGAGC NA
TABLE 59
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ Position AlleleCase Control Value
ID NO: AF AF
11
rs1896753205 76177855 C/T
rs3974451866 76178516 C/T
rs18207704212 76181862 C/T
rs14287535934 76183584 C/T 0.515 0.457 0.124
rs72222911486 76189136 C/T
rs385175416969 76194619 A/G 0.292 0.286 0.868
rs234043022509 76200159 A/G 0.480 0.531 0.169
rs234043122796 76200446 A/G 0.024 0.027 0.900
rs115941528097 76205747 CIT 0.535 0.485 0.252
182
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in SEQ PositionAlleleCase Co, ntrolValue
ID NO: 11 AF AF
rs150683628626 76206276C/T 0.412 0.410 _
0.947
rs150683728853 76206503C/T 0.402 0.391 0.768
rs150683828873 76206523C/T 0.355 0.368 0.734
rs96666830155 76207805A/G
rs191124530827 76208477C/T 0.849 0.828 0.569
rs150683931956 76209606C/T 0.414 0.428 0.746
rs150684032404 76210054C/T 0.384 0.379 0.905
rs187627532944 76210594A/G 0.456 0.447 0.805
rs191124235205 76212855A/G 0.397 0.402 0.892
rs191124335227 76212877C/T
rs98123135781 76213431C/T 0.454 0.455 0.971
rs150682941052 76218702C/T 0.386 0.356 0.424
rs150683345051 76222701A/G 0.500 0.509 0.811
rs139548646039 76223689C/T
rs150683247276 76224926A/G 0.508 0.524 0.689
rs150683047678 76225328C/T
rs96853747716 76225366A/G 0.246 0.237 0.806
rs150681651014 76228664A/G 0.408 0.367 0.284
rs150682854408 76232058A/G 0.421 0.387 0.358
rs150682754596 76232246C/T 0.436 0.471 0.346
rs154296956853 76234503C/G
rs139548461851 76239501A/G 0.416 0.413 0.938
rs187627462016 76239666A/G 0.376 0.350 0.447
rs187627362461 76240111C/T 0.367 0.370 0.924
rs150682268257 76245907C/G 0.358 0.325 0.355
rs150682069793 76247443C/T 0.373 0.256 0.007
rs150681973976 76251626A/C 0.438 0.424 0.703
rs150681873999 76251649A/T 0.139 -0.013
rs150681774053 76251703A/G 0.174 0.178 0.897
rs139548875315 76252965A/G 0.487 0.512 0.505
rs222153475729 76253379G/T 0.455 0.429 0.463
rs191124476466 76254116A/G 0.489 0.509 0.581
rs213562477216 76254866C/T
rs213562377217 76254867G/T 0.042 0.035 0.748
rs150683579239 76256889C/G 0.531 0.510 0.562
rs150683480825 76258475A/G 0.407 0.397 0.787
rs199565381060 76258710C/G 0.414 0.413 0.984
rs199565281097 76258747A/C 0.437 0.455 0.629
rs139548781426 76259076G/T 0.506 0.512 0.869
rs394708384787 76262437C/T 0.379 0.359 0.559
rs150682584896 76262546A/T 0.419 0.399 0.579
rs150682485165 76262815C/G 0.442 0.414 0.471
rs156711886502 76264152C/G 0.036 0.025 0.574
rs103968386753 76264403C/T 0.389 0.385 0.910
rs287977786941 76264591C/T 0.275 0.280 0.883
rs187627288787 76266437C/T
rs303587895598 76273248- untyped 0.980
/AGAG NA
C
[0317] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
ire bold. The allelotyping p-
values were plotted in Figure lI for the discovery cohort. The position of
each SNP on the chromosome
is presented on the x-axis. The y-axis gives the negative logarithm (base 10]
of the p-value comparing
the estimated allele in the case group to that of the control group. The minor
allele frequency of the
control group for each SNP designated by an X or other symbol on the grapras
in Figure lI can be
determined by consulting Table 57. For example, the left-most X on the left
graph is at position
1S3
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
76177855. By proceeding down the Table from top to bottom and across the
graphs from left to right
the allele frequency associated with each symbol shown can be determined.
[0318] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapte=r 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). Th a black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a lOkb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a rtest wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-8 were truncated at that
value.
[0319] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at -the 3' end
of each gene to show the direction of transcription.
Example 13
APOL3 Region Proximal SNPs
[0320] It has been discovered that SNP rs132659 inAPOL3 is associated with
occurrence of
osteoarthritis in subjects. APOL3 belongs to the high density lipoprotein
family that plays a central role
in cholesterol transport. The cholesterol content of membranes is important in
cellular proce sses such
as modulating gene transcription and signal transduction both in the adult
brain and during
neurodevelopment. It has been shown that the APOL1-APOL4 gene cluster on
chromosome 22 exists
only in primates (humans and African green monkeys) and not in dogs, pigs, or
rodents, suggesting that
this gene cluster has arisen recently in evolution (Monajemi et. al.,
Genofnies 79: 539-546, 2002). Six
transcript variants encoding three different isoforms have been identified.
[0321] Expression of this gene is upregulated by tumor necrosis factor-alpha
in endothe lial cells
lining the normal and atherosclerotic iliac artery and aorta (Horrevoets et.
al., Blood 93: 3418-3431,
1999). APOL3 is genetically linked to OA and may play a role in the
pathophysiology of OA brought
about by inflammation. APOL3 is likely inhibited by a small molecule inhibitor
or by specific
antibodies. APOL3 activity may be increased in a subject by administeringAPOL3
recombinant protein
or a functional fragment thereof.
[0322] Two hundred-nineteen additional allelic variants proximal to rs132659
were identified and
subsequently allelotyped in osteoarthritis case and control sample sets as
described in Examples 1 and 2.
184
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
The polymorphic variants are set forth in Table 60. The chromosome positions
provided in column
four of Table 60 are based on Genome "Build 34" of NCBI's GenBank.
TABLE 60
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
13
rs388881822 201 34781551 c/t
rs201060522 425 34781775 a/
rs743919 22 1095 34782445 /t
rs100813422 2201 34783551 a/c
rs132607 22 7879 34789229 a/
rs147602922 8395 34789745 c/t
rs147603022 8461 34789811 c/t
rs241338022 9503 34790853 c/t
rs205160922 10304 34791654 /t
rs241338122 10695 34792045 c/t
rs189460422 16300 34797650 a/
rs189460522 16444 34797794 /t
rs132609 22 17591 34798941 c/t
rs132610 22 17988 34799338 -/a
rs132611 22 19116 34800466 -/t
rs132612 22 19358 34800708 c/t
rs100879022 20300 34801650 a/
rs23085 22 20669 34802019 a/t
rs105161 22 20891 34802241 a/
rs132613 22 21451 34802801 c/t
rs132614 22 21978 34803328 c/t
rs132615 22 22785 34804135 c/
rs132617 22 24248 34805598 c/t
rs386572422 24770 34806120 c/t
rs201965722 24844 34806194 a/
rs386572522 25066 34806416 /t
rs201936422 25096 34806446 c/t
rs200838322 25309 34806659 a/
rs398600222 25344 34806694 a/c
rs388894222 25529 34806879 a/t
rs388894322 25537 34806887 a/
rs388894422 25554 34806904 a/c
rs132618 22 27963 34809313 a/t
rs132619 22 28134 34809484 /t
rs382734622 28356 34809706 a/
rs132620 22 29648 34810998 -/a
rs132621 22 29986 34811336 a/
rs80575 22 30217 34811567 /t
rs80578 22 30267 34811617 a/
rs80577 22 30315 34811665 a/
rs80578 22 30585 34811935 c/t
rs80579 22 30724 34812074 a/c
rs80580 22 30897 34812247 c/t
rs132622 22 30931 34812281 c/t
rs132623 22 31080 34812430 /t
rs 13262422 31246 34812596 c/t
rs132625 22 31373 34812723 a/t
185
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
13
rs13262622 31463 34812813 a/
rs13262722 31467 34812817 a/
rs180767222 32188 34813538 /t
rs13262822 32288 34813638 c/t
rs13262922 32520 34813870 a/t
rs13263022 32594 34813944 a/c
rs13263122 32657 34814007 a/c
rs13263222 32677 34814027 a/
rs13263322 32764 34814114 c/t
rs13263422 32784 34814134 al
rs13263522 32830 34814180 c/t
rs13263622 32872 34814222 c/t
rs12960322 33121 34814471 a/c
rs13263722 33348 34814698 /t
rs378851822 33952 34815302 c/
rs13263822 34184 34815534 c/
rs13263922 34361 34815711 a/t
rs13264022 35026 34816376 a/
rs13264122 35192 34816542 a/
rs13264222 35600 34816950 a/t
rs13264322 36033 34817383 c/t
rs13264422 36289 34817639 c/t
rs13264522 38869 34820219 a/
rs201732922 39629 34820979 a/t
rs73919822 40530 34821880 c/t
rs13264722 41621 34822971 c/t
rs209746522 42379 34823729 c/t
rs210591522 42802 34824152 c/t
rs13264822 42865 34824215 t/c
rs13264922 43644 34824994 a/
rs 13265022 45051 34826401 c/t
rs13265122 45828 34827178 a/c
rs13265222 45829 34827179 a/t
rs80584 22 46257 34827607 c/t
rs13265322 47286 34828636 a/c
rs91633422 47427 34828777 c/t
rs 13265422 47963 34829313 c/t
rs 13265522 48013 34829363 c/t
rs13265622 48229 34829579 c/t
rs383468422 48282 34829632 -/a
rs13265722 48376 34829726 -/
rs91633522 48404 34829754 a/
rs93265922 49900 34831250 c/t
rs13266022 52699 34834049 It
rs13266122 52897 34834247 a/
rs13266222 53414 34834764 a/
rs13266322 53487 34834837 alt
rs13266422 54112 34835462 /t
rs13266722 55492 34836842 a/
rs 13267022 59766 34841116 c/t
rs13267122 60307 34841657 a/
rs13267222 60701 34842051 alg
~
186
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
13
rs13267322 60952 34842302 al
rs13267422 61401 34842751 c/t
rs13267522 62379 34843729 c/t
rs80585 22 62870 34844220 c/t
rs80586 22 62879 34844229 a/
rs13267622 63499 34844849 a/t
rs13267722 64284 34845634 -/a
rs13267822 64408 34845758 a/
rs13268022 64760 34846110 a/
rs13268122 65230 34846580 a/
rs13268322 66127 34847477 a/
rs226959422 66634 34847984 c/t
rs13268422 66686 34848036 a/
rs13268522 66694 34848044 c/
rs13268622 67113 34848463 a/
rs13268722 67257 34848607 a/
rs13268822 67403 34848753 a/
rs13268922 67609 34848959 a/
rs13269022 68418 34849768 -/a
rs13269122 68610 34849960 c/
rs13269222 69629 34850979 c/t
rs13269322 70024 34851374 a/
rs13269422 70848 34852198 a/
rs13269522 71428 34852778 c/
rs196626622 71553 34852903 c/t
rs196626722 71633 34852983 a/
rs10680822 71768 34853118 a/c
rs13269622 71769 34853119 a/
rs223982922 73039 34854389 a/
rs228515422 73325 34854675 a/
rs223983022 73412 34854762 a/c
rs223983122 73547 34854897 c/t
rs386572222 73769 34855119 a/t
rs386572322 73806 34855156 a/
rs398599622 74467 34855817 c/t
rs398599722 74472 34855822 c/t
rs398599822 74473 34855823 a/
rs398599922 74482 34855832 c/t
rs398600022 74494 34855844 a/c
rs241338222 74592 34855942 a/
rs241338322 74670 34856020 /t
rs241338422 74672 34856022 !t
rs241338522 74714 34856064 /t
rs241338622 74723 34856073 a/t
rs189460622 74749 34856099 a/
rs91633622 74861 34856211 c/
rs91633722 74892 34856242 c/t
rs91633822 74893 34856243 c/t
rs13269722 75176 34856526 a/
rs12781 22 75705 34857055 a/
rs105398322 75989 34857339 a/
rs 1053982I 32 ~ 76027 ~ 34857377 ~ a/g
-
187
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
13
rs222716722 77949 34859299 a/
rs222716822 77974 34859324 c/t
rs13270022 78167 34859517 c/t
rs307536422 78310 34859660 -/ct
rs222716922 78415 34859765 c/t
rs209746622 78575 34859925 c/t
rs209746722 78590 34859940 c/t
rs241338722 78709 34860059 c/t
rs13270122 78875 34860225 c/t
rs13270222 79864 34861214 c/t
rs13270322 81316 34862666 c/t
rs226959522 81320 34862670 a/
rs226959622 81409 34862759 c/t
rs13270422 81737 34863087 c/t
rs200746822 81843 34863193 a/
rs13270522 82102 34863452 clt
rs200770622 82833 34864183 c/t
rs 13270622 83461 34864811 c/t
rs13270722 83624 34864974 c/t
rs13270822 83660 34865010 c/
rs13270922 83701 34865051 a/t
rs13271022 83708 34865058 al
rs13271122 83782 34865132 c/t
rs13271222 85707 34867057 a/
rs13271322 85717 34867067 a/
rs13271422 86486 34867836 c/t
rs13271622 86833 34868183 a/
rs13271722 87115 34868465 c/t
rs13271822 87234 34868584 a/
rs13271922 87479 34868829 /t
rs13272022 87561 34868911 a/
rs13272122 87604 34868954 a/
rs13272222 87674 34869024 c/t
rs13272322 87958 34869308 a/
rs13272422 87992 34869342 -/
rs13272522 88019 34869369 a/
rs13272622 88074 34869424 c/
rs13272722 88079 34869429 c/
rs13272822 88115 34869465 a/
rs13272922 88118 34869468 c/
rs13273022 88120 34869470 a/
rs13273122 88135 34869485 -/ctcat
rs13273222 88142 34869492 /t
rs13273322 88143 34869493 /t
rs14057522 88149 34869499 aca/t
rs13273422 88340 34869690 a/
rs13273522 88344 34869694 /t
rs80587 22 88512 34869862 c/
rs13273622 88521 34869871 c/t
rs13273722 88650 34870000 c/
rs13273822 88827 34870177 c/t
rs180767322 89230 34870580 a/
1~~
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Chromo-Position Chromosome Allele
rs# some in SEQ Position Variants
ID NO:
13
rs201470022 89236 34870586 a/
rs132739 22 90754 34872104 /a
rs181202322 90984 34872334 a/
rs181202422 91110 34872460 a/
rs200559022 92026 34873376 c/t
rs132740 22 92954 34874304 c/t
rs398600122 93375 34874725 -ltt c
rs241339022 93794 34875144 c/t
rs132743 22 94937 34876287 c/
rs132744 22 95068 34876418 c/t
rs241339122 96188 34877538 a/
rs132749 22 97092 34878442 clt
rs132750 22 98812 34880162 c/t
rs132741 22 not ma not ma ed a/c
ed
rs241338822 not ma not ma ed a/t
ed
rs2413389~ 22 not mappednot mapped c/~
~ ~ ~
Assay for Verifying and AllelotXping SNPs
[0323] The methods used to verify and allelotype the 219 proximal SNPs of
Table 60 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 61 and Table 62, respectively.
TABLE 61
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs3888818ACGTTGGATGTGAGGTCAGGAGTTTGAGACACGTTGGATGCCATGCCCAGCTAATTTTCG
rs2010605ACGTTGGATGTGTGCTTTTTATGTCTTAGGACGTTGGATGGACTTTTAGAAGAAAAGTAC
rs743919ACGTTGGATGTTCTTCACCAAGCCCTCTTCACGTTGGATGCCCAACACACACAAAGATGG
rs1008134ACGTTGGATGACATATCCGGGCATCTTTTCACGTTGGATGCATCCACAGGATGCAATATC
rs132607ACGTTGGATGTAGTTTGCAGGTCACAAGGGACGTTGGATGGGAAGGAAGACCCAAACAGC
rs1476029ACGTTGGATGTGGGTGACAGAGATCCTTACACGTTGGATGAAACTCAGGGAAACGGACTC
rs1476030ACGTTGGATGATCCTAGCACTGGGATTTGGACGTTGGATGCGTTTCCCTGAGTTTCACTG
rs2413380ACGTTGGATGATGATACTGAGTCCAGGAGGACGTTGGATGAAAGGCTACTTCTTGCTCAC
rs2051609ACGTTGGATGTAATCCCAGCACTTTGGGAGACGTTGGATGACAGACGGGGTTTCATCATG
rs2413381ACGTTGGATGAGGGCTGCAGTAGAAAAGCGACGTTGGATGACGATGCGTGTGCCGACAG
rs1894604ACGTTGGATGAAGTGCTGCTGCAAAAAGAGACGTTGGATGTTCTCCACTTCCATTCTGTG
rs1894605ACGTTGGATGTGATGAGAGATGCAGATCGCACGTTGGATGTATTCAGAATTCAGCCTGCG
rs132609ACGTTGGATGGCATTTGAAAGGTCCGTATCACGTTGGATGCCCAAATCTGTCTTTTAGCC
rs132610ACGTTGGATGTTCTACAAGAGCTAGGGACCACGTTGGATGGATCTATTGCTGCTTAGGCC
rs132611ACGTTGGATGGCCTTCTTTACTCTGTCCTCACGTTGGATGGGTTTTCTTTCAGGTCCTCC
rs132612ACGTTGGATGAAAAATCTTCCCGCTACCTCACGTTGGATGTTCTGTGAGCTTTCTCTCTG
rs1008790ACGTTGGATGGTGAGGTCCCTTTTATGATGACGTTGGATGATAACAGCCCCTGACAGATG
rs23085ACGTTGGATGAAACCGTGCCAGCTGAGGATACGTTGGATGGTCGGCAAGGAAGAGGAATC
rs105161ACGTTGGATGAAGGAGCGGGAAATCTTTTGACGTTGGATGGTAGGAGGCTGAAATGCTAG
rs132613ACGTTGGATGATGTAAAACCAATGGCCTCCACGTTGGATGAAGCTTCAGATTGTTCACCC
rs132614ACGTTGGATGCAAGAGCCCTGCTTTGTGAGACGTTGGATGTCCTGCACCAGCAGAGATGA
rs132615ACGTTGGATGAAATCTGGAGGCTTGGTGACACGTTGGATGTGAGCATTCACATGGGACAG
rs132617ACGTTGGATGGAGAAGAAGAGTGTGTGTGCACGTTGGATGACAGCCACCTGAATTTGTGC
~rs3865724~ ACGTTGGATGTTCCTGGATAATTCCCATTCACGTTGGATGGCTGGATCACTGAAGAAGTA
1~9
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs2019657ACGTTGGATGACGCCAGAACATTGTGTTTCACGTTGGATGGTGCCAGAAACATTCAAAGC
rs3865725ACGTTGGATGAATATAGAACTGCTGGGCGCACGTTGGATGTGACTTAGGAGAGGTTCTGG
rs2019364ACGTTGGATGTGACTTAGGAGAGGTTCTGGACGTTGGATGAATATAGAACTGCTGGGCGC
rs2008383ACGTTGGATGTGATTCTAGGAGCAGGACTGACGTTGGATGACATGGGTGACCCTATCAAG
rs3986002ACGTTGGATGCTTCTGTCCTTCTCTGTGTCACGTTGGATGCAGGCAGAGGATTTGTTTGG
rs3888942ACGTTGGATGCTGGGCTTTTGTGCTAAGAGACGTTGGATGGGGCCAATTTGCCCCATAAA
rs3888943ACGTTGGATGCTGGGCTTTTGTGCTAAGAGACGTTGGATGTTGGGCCAATTTGCCCCATA
rs3888944ACGTTGGATGATACAGCCCTTGCCACTATGACGTTGGATGTTGAAGACATGGAAGCAGGG
rs132618ACGTTGGATGAATCCGTGCCATCAGGCAAGACGTTGGATGCCTGCAATCGTTCTCTCTGC
rs132619ACGTTGGATGTCATCAGCAGAAGCTGAAGCACGTTGGATGGGTGTGATGTCACGCATAAC
rs3827346ACGTTGGATGAAGAGGTCCACAGAGGCTGACGTTGGATGAAAACAAGACCAGCAAGGGA
rs132620ACGTTGGATGTCACATTAGATCAGGAAGCCACGTTGGATGTTAGGCCAGTTTAGCAGAAA
rs132621ACGTTGGATGCTTCAAATCTGCAACTGGTGACGTTGGATGGATAGCTTAAGGACTCAGAG
rs80575ACGTTGGATGGCTGCACATGAACTCTCAAGACGTTGGATGTGACATGTGACAGTGAGACC
rs80576ACGTTGGATGTGAAGCTGTCACCTGCTAAGACGTTGGATGAACTCTCAAGCCACTTGACG
rs80577ACGTTGGATGAGTCCATAAGAGGTTCCATGACGTTGGATGAACTAATGCCTTAGCAGGTG
rs80578ACGTTGGATGACTGTTTCCCTGACAGCATGACGTTGGATGTGTAGAACAGAAGAGGGTCC
rs80579ACGTTGGATGTGGGAGTAGGGTGAGAAGAGACGTTGGATGACTCACTGGTCCTCTGCAAG
rs80580ACGTTGGATGTTCAATCAGATGGGCGTGTGACGTTGGATGGATGGCATCATGCTACTTGG
rs132622ACGTTGGATGTATGTCTTGGAGACTGGGACACGTTGGATGACCTGCTGTTCATTCTCAGG
rs132623ACGTTGGATGAGCTCTGTCCAACTCCATTCACGTTGGATGCTGAGGAACTGCACAAACAC
rs132624ACGTTGGATGTGCTGGGATTACAGGCATGAACGTTGGATGTCAAAGAAAGTCCTGCTGGG
rs132625ACGTTGGATGTTTCACGCCATTCTCCTGCCACGTTGGATGCGATGAAACCCCGTCTGTAC
rs132626ACGTTGGATGTGGAGTGCAGTGGCATGATCACGTTGGATGGCAGGAGAATGGCGTGAAAC
rs132627ACGTTGGATGAGGAGAATGGCGTGAAACCGACGTTGGATGAGACAGAGTCTTGCTTGTCC
rs1807672ACGTTGGATGGTGTGCTACAGCCTAAATGGACGTTGGATGAATACCCCATGTGACAGCTG
rs132628ACGTTGGATGTATAGACTGAGTTGTGTGCCACGTTGGATGTCCTTAAAGGCTCAATCTCC
rs132629ACGTTGGATGCTCTCTCCCTGTCTCTCTTTACGTTGGATGTGTGTCCTCACATGGCCTTC
rs132630ACGTTGGATGTTCCAAGGTGAAGGTGCCAGACGTTGGATGAAGGCCATGTGAGGACACAG
rs132631ACGTTGGATGGGTGGCTCCAACAACTGATGACGTTGGATGATCAACCCTGCTGGCACCTT
rs132632ACGTTGGATGCTTGGAATTTTTGCCTCCAGACGTTGGATGTCAGGATGCCTTAGTAAAAC
rs132633ACGTTGGATGAGAAGAGTGATTCACCAGGGACGTTGGATGGGAAAGCTCACTTTCTGGTG
rs132634ACGTTGGATGAAGTGCCATGGTGCTTTGTGACGTTGGATGGAAAGCATGGTGGAAAGCTC
rs132635ACGTTGGATGAATAGGCACATGGCAGAAGGACGTTGGATGCACCAGAAAGTGAGCTTTCC
rs132636ACGTTGGATGAAGCGTTTGACAATAGGCACACGTTGGATGAAAGTGAGCTTTCCACCATG
rs129603ACGTTGGATGGTGTCATATTGACACAGATTGACGTTGGATGAGGGTGTATATATATATACCC
rs132637ACGTTGGATGGCATCTTAGTACACAGCAGGACGTTGGATGTTCCCAAATCCCTGCAAACC
rs3788518ACGTTGGATGAATCCTTCAGAAGGGCTTGGACGTTGGATGGCCGCGTTATTAAACCACAG
rs132638ACGTTGGATGCATCCTTTCAGTGAAGGAGGACGTTGGATGTTGCCAAGGCAACTCAGTGA
rs132639ACGTTGGATGACACCTGGGCAAACAAAAGCACGTTGGATGAAGTTCCCCATAGTTGGCAG
rs132640ACGTTGGATGTAAGAAGCTCCAGGTGACACACGTTGGATGAAAAGAGTGACTCAGCGTCC
rs132641ACGTTGGATGAGGGTCAGCTGGGAGCAGAACGTTGGATGAGGGCTGAGAGAGGAGGTTG
rs132642ACGTTGGATGAAGAAGCAAGCCTACCTGAGACGTTGGATGAAACGAACCCTTCCAGTCAG
rs132643ACGTTGGATGATCACAGACACCCAGTACACACGTTGGATGACGTTCTGACAATGACCTGG
rs132644ACGTTGGATGGCATAGAGTGCAAGACACAGACGTTGGATGGGGCTCCACTCCCTTAAATA
rs132645ACGTTGGATGTGAAGGCAAACAGTACAAGAACGTTGGATGAAGTTAACCAAGTGTTTAC
rs2017329ACGTTGGATGCCTTCCCAATTAAAAGCAGCACGTTGGATGGGGCAACAAGAGTGAAATTC
rs739198ACGTTGGATGAAACTTTGGTCTCCACAACCACGTTGGATGTGAGTTTGTCTAAAGACCGG
rs132647ACGTTGGATGCCTCACTACAGAAACCATGGACGTTGGATGAACTCAACTGGTTCAACCAC
rs2097465ACGTTGGATGGAATTGACCAAACTGCAGGCACGTTGGATGAGGGTTGAAGCTGGATACTG
rs2105915ACGTTGGATGAACCCAGGAGTTCAGGACAAACGTTGGATGGGGAACTACAAGTGCATCAC
rs132648ACGTTGGATGGTGGCTCAGGGCTGTAATTCACGTTGGATGTGTCCTGAACTCCTGGGTTC
190
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs132649ACGTTGGATGGTCCTCCCCAGTCTTATTACACGTTGGATGATTCAGAGGTTAGCTGGCTC
rs132650ACGTTGGATGAAAGTGCTGGGATTACAGGCACGTTGGATGCTAAATCTCCTGCCATAGGG
rs132651ACGTTGGATGAGGTCAGGTGTTGACCTTCCACGTTGGATGAGCAGGGTAGGGCATCCTAA
rs132652ACGTTGGATGAGCAGGGTAGGGCATCCTAACACGTTGGATGAGGTCAGGTGTTGACCTTCC
rs80584 ACGTTGGATGCATGGAGTCCTGTGATCTACACGTTGGATGAAACTGAGGCCATGGGAGAT
rs132653ACGTTGGATGCAGCCGTGCATCTGCATAATACGTTGGATGCACTTTCCCTTTTGGGTTCC
rs916334ACGTTGGATGAAACAGGATGCTTCCCAGCCACGTTGGATGCTGCTCTTGGATCAGCAGGA
rs132654ACGTTGGATGTAAGGAAGTGTCCAGAAGCCACGTTGGATGAAATGATCCTCCTGCCTCAG
rs132655ACGTTGGATGACTTTTCCAGGTGAAGGTAGACGTTGGATGTTGTCGAACGCCATACCTGT
rs132656ACGTTGGATGTCACTAGAAGCAAGGAACCCACGTTGGATGCCCAGGTTGACTGAACAAAG
rs3834684ACGTTGGATGAGCCCTTGTTCACTAGAAGCACGTTGGATGGGTGGATGTGGGAGTAAAAG
rs132657ACGTTGGATGGGCTGACTGACAATTACCTGACGTTGGATGAGGGCTCTGAGCTTTTCAAG
rs916335ACGTTGGATGGATGACGAGAAAAAGGTGGGACGTTGGATGATTTGAAGGATGCAGTCTTG
rs932659ACGTTGGATGGGCCCATAGTGGGTCATAACACGTTGGATGGTGGGGTGAGTGCCCAAAAG
rs132660ACGTTGGATGTACATGTGGTTGTACCCTCCACGTTGGATGCTGGCATGGTTTTACCCATC
rs132661ACGTTGGATGCTTCGCAGAAATCATTCCGCACGTTGGATGCCAAAATGCAAGCTCAAGGC
rs132662ACGTTGGATGCATCTCTTAAATGGGCCAGGACGTTGGATGTTGAAAGCCACAGCCTCATG
rs132663ACGTTGGATGCCACTAAACGGATTGAGATCACGTTGGATGTGGCTTTCAACCAGCAACTC
rs132664ACGTTGGATGATCATGCCACTGCAATCCAGACGTTGGATGGCATATGTGACTGCTTCCTC
rs132667ACGTTGGATGCTGGAGAAATCAAATAGAGAGACGTTGGATGTGTACAGCTTTTGACAGTTG
rs132670ACGTTGGATGTAAGGTCGGGAGTTCAAGACACGTTGGATGACGCCCGGCTGATTTTGTAT
rs132671ACGTTGGATGGTGAGCCATACCATCACATCACGTTGGATGCTGTAGTAAAGGTCTGGTCG
rs132672ACGTTGGATGCTCCCCAATAAGCTCAACACACGTTGGATGCTGTTAGGGCAATGAAAGGC
rs132673ACGTTGGATGTGAGTAGTTGGTGTGAGTGGACGTTGGATGCAATGGATGAAGCTGATCCC
rs132674ACGTTGGATGAACTGTAGTCCCAGCTACTCACGTTGGATGTAGCTCTATCACTCATGCTG
rs132675ACGTTGGATGGTAGAGCAGATGTGCAATGGACGTTGGATGTCCTAACCATCTGCCTTGTG
rs80585 ACGTTGGATGCTGTTGTTCCAACACTTCACACGTTGGATGGGTCTGCTACTAGAATTCAG
rs80586 ACGTTGGATGGTAAGTGTAAGAAGGTCTGCACGTTGGATGCAAGGCATAATATTCTGACC
rs132676ACGTTGGATGCAAACATTCTGCAGAAAGCGACGTTGGATGAAGCGTGTTGCTGAGAAATG
rs132677ACGTTGGATGCTCTGTTACAAAATGAAGGGACGTTGGATGGCTATCTAGGCTAAAAATCCC
rs132678ACGTTGGATGAAGGCACTGAAAATGCCTAGACGTTGGATGGGAATCCAGATGCTTACATG
rs132680ACGTTGGATGGCCTTAGCTATCATGTTCTCACGTTGGATGGCGTGTTTAAGGCAATTCTC
rs132681ACGTTGGATGTACTGAAGCCTGAGACTAGCACGTTGGATGCTAGCAGAAACTAACCGAGC
rs132683ACGTTGGATGTTACCCTATGGTAATGGCAGACGTTGGATGACTGATTAGTACAGGAAGGG
rs2269594ACGTTGGATGTGGCATGGCTAAAAGGACAGACGTTGGATGGATTGTTCTGATGCCCAGTG
rs132684ACGTTGGATGCCTTTTAGCCATGCCATTCGACGTTGGATGTCAGTGTAAAACGTGCCACC
rs132685ACGTTGGATGTCAGTGTAAAACGTGCCACCACGTTGGATGCCTTTTAGCCATGCCATTCG
rs132686ACGTTGGATGCAGAATATCCACGTCAGGTGACGTTGGATGGACAGCTTAGGACTATGTGC
rs132687ACGTTGGATGCAACTGTAAGCAGCCCATTGACGTTGGATGCTGACGGTGCAAATGGATAC
rs132688ACGTTGGATGAGTACTACAGGACGTGCTTGACGTTGGATGGGTCGCCTCATATATGGTAG
rs132689ACGTTGGATGTACTGGGACAGTCTGCTTTCACGTTGGATGACTTTACAGTGCTGGAGCAG
rs132690ACGTTGGATGTGTTTTGCTTTGCGCTCTCCACGTTGGATGTCTGCAACCAACTCTTTGGG
rs132691ACGTTGGATGGTCAAAGCCAGGCATTTGTCACGTTGGATGCTGTCATCTTGTGGAAAGGG
rs132692ACGTTGGATGGAATCTAAGCCAGCTGTTGGACGTTGGATGGGAGCATCATGTGGATTCCT
rs132693ACGTTGGATGGCCAGAAGAAAAGAGTGTGGACGTTGGATGATTCTGCATGTGGAACGTCC
rs132694ACGTTGGATGATAGAGACTGAGAGCTGCAGACGTTGGATGCAGAACAAAGCAGGAAGCTC
rs132695ACGTTGGATGGCCTCTCTCTATGACTACACACGTTGGATGTTCACAGCAGGGAACTCTTC
rs1966266ACGTTGGATGTGATTGTACAAGGCAGACCCACGTTGGATGTGTAAGCACCTGCATTCAGC
rs1966267ACGTTGGATGTAACTCACAGACCATGAGGGACGTTGGATGGGAGGAAAGCACAGCAGAAT
rs106808ACGTTGGATGAACAAGGCAGATCCTTCCCGACGTTGGATGATGGTTCCTGAAGAGCAGTG
rs132696ACGTTGGATGATGGTTCCTGAAGAGCAGTGACGTTGGATGAACAAGGCAGATCCTTCCCG
rs2239829ACGTTGGATGTGTCTTTGTCGTTCGGATGGACGTTGGATGAAAGAGCGAAACTCCGTCTC
191
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs2285154ACGTTGGATGTGAACTCAAATGATCCGCCCACGTTGGATGAAGAACCCTTTTCGACTGGG
rs2239830ACGTTGGATGAAACCCTAATGGGAAGCCTCACGTTGGATGTGTGGTAGCAAGCAGTTGAC
rs2239831ACGTTGGATGAGAACAGTCACTGACCCAAGACGTTGGATGGCTCCACACACTTTGATTCC
rs3865722ACGTTGGATGCCACTGTACTGCTAGTATTGACGTTGGATGACCTGCTCTAGTTTTCATCC
rs3865723ACGTTGGATGCCTGCATTTGATGCAATTCCACGTTGGATGGTTTCTGTTTCTGTTGCTTGC
rs3985996ACGTTGGATGACATGGGTGACCCTATCAAGACGTTGGATGTGATTCTAGGAGCAGGACTG
rs3985997ACGTTGGATGCAAGAATTTCTCCCGGCATCACGTTGGATGTGATTCTAGGAGCAGGACTG
rs3985998ACGTTGGATGTGATTCTAGGAGCAGGACTGACGTTGGATGCAAGAATTTCTCCCGGCATC
rs3985999ACGTTGGATGTGATTCTAGGAGCAGGACTGACGTTGGATGCAAGAATTTCTCCCGGCATC
rs3986000ACGTTGGATGCAGGCAGAGGATTTGTTTGGACGTTGGATGCTTCTGTCCTTCTCTGTGTC
rs2413382ACGTTGGATGAAACAAATCCTCTGCCTGGGACGTTGGATGAAAAGCCCAGAGCCTTCATG
rs2413383ACGTTGGATGCTGGGCTTTTGTGCTAAGAGACGTTGGATGGGGCCAAGTTGACCCATAAA
rs2413384ACGTTGGATGCTGGGCTTTTGTGCTAAGAGACGTTGGATGGGGCCAAGTTGACCCATAAA
rs2413385ACGTTGGATGAATGGTCTTCGCTGATACACACGTTGGATGATGGAAGCCGGTGTTTGATG
rs2413386ACGTTGGATGTTTTATGGGTCAACTTGGCCACGTTGGATGTTCCAAAATGGTCTTCGCTG
rs1894606ACGTTGGATGTTTTATGGGTCAACTTGGCCACGTTGGATGAGACTCCTGCAAAAGCTTCC
rs916336ACGTTGGATGGGATGAGGGTATTTGCTGTCACGTTGGATGGGCCTGTATGTAGGTTGAAG
rs916337ACGTTGGATGAGATCAGAAGGGCCTGTATGACGTTGGATGATTTGCTGTCTGGCTGTCTC
rs916338ACGTTGGATGATTTGCTGTCTGGCTGTCTCACGTTGGATGAGATCAGAAGGGCCTGTATG
rs132697ACGTTGGATGCATGAGGAAGAGAAGTCAGGACGTTGGATGACACTGACTGATGACTGAGC
ACGTTGGATGGAGCCAGAAAATTAACTGAAAA
rs12781ACGTTGGATGTTACACACAGGGCACTCAGCGC
rs1053983ACGTTGGATGCCATCATCAAGAAGCCACTGACGTTGGATGTGTCTTACCAGCATCCACTC
rs1053982ACGTTGGATGGGAGAGTGGATGCTGGTAAGACGTTGGATGCGGTTGAATGTCCTTCCAAG
rs2227167ACGTTGGATGCTCCTGAGTGTATGGACATCACGTTGGATGGGCATTAAGGGACATTCTGC
rs2227168ACGTTGGATGCCAATTGGAGGCATTAAGGGACGTTGGATGCTCCTGAGTGTATGGACATC
rs132700ACGTTGGATGAATGTGGTGTCTGGCTCCACACGTTGGATGCTCAGCCCTGCTGTAAATGG
rs3075364ACGTTGGATGGAACAGCAGTTTAGGGAGTGACGTTGGATGCGAAGCCTTTCTATGGACTC
rs2227169ACGTTGGATGAGGTAAGTAAGCTGCCTTTCACGTTGGATGTTCAGAGCTTCATAGAGAGC
rs2097466ACGTTGGATGCTGGGATTACAAGCATGAGCACGTTGGATGCTGCATAAATCACAGAGCTG
rs2097467ACGTTGGATGATCTCCTGACCTTGTGATCCACGTTGGATGATTCTTTTCAAGGCCGGGCG
rs2413387ACGTTGGATGAAGTAGCTGGGACTACAGGCACGTTGGATGTAACACGGTGAAACCCCGTC
rs132701ACGTTGGATGGTGGCATATCTATGTTGTACACGTTGGATGGCGAGACTCCATCTCAAAAA
rs132702ACGTTGGATGGAAGCTCACCCAGTTAAGGAACGTTGGATGCCCCTGTAACAACAATCCTG
rs132703ACGTTGGATGCTTGACCTGATCAATGTGTGACGTTGGATGTTTGTGCAGTTCCTCAGAAG
rs2269595ACGTTGGATGAGAAGTTCAGGAAAAGGGCCACGTTGGATGCAGCAGGACTTTCTTTGGGA
rs2269596ACGTTGGATGAGGTGCTCAGTTAGCGTTACACGTTGGATGTCCCAAAGAAAGTCCTGCTG
rs132704ACGTTGGATGATATTCTTCCTGCACTGCTGACGTTGGATGATCTCCCCGGGCTAGTTTTC
rs2007468ACGTTGGATGAGGTTACCTGGGCAATTCAGACGTTGGATGGAAAATCCTGCTGACTAGCG
rs132705ACGTTGGATGTTTTGATGGAGGCACCAGTGACGTTGGATGTCTCCAAATACGGTCACTGG
rs2007706ACGTTGGATGCCCAGGAATTTACATAAGGGACGTTGGATGTTGAACATAGCAAGAGTGAG
rs132706ACGTTGGATGAAGGATCAGTGCTGAGGGTCACGTTGGATGATTCCTCCTGCTGGTCATGG
rs132707ACGTTGGATGAATCCTTAGGAAGGGCTGGGACGTTGGATGAGCTGGCCCCGTTAGTAAAC
rs132708ACGTTGGATGTCTTGTTTCAGAGGGAGAGCACGTTGGATGTCTCAGCCAATCCCAGAATC
rs132709ACGTTGGATGTGAGTCCTGTCCAAGATGAGACGTTGGATGAGCCCTTCCTAAGGATTCTG
rs132710ACGTTGGATGTGAGTCCTGTCCAAGATGAGACGTTGGATGCCTAAGGATTCTGGGATTGG
rs132711ACGTTGGATGTCATCTTGGACAGGACTCAGACGTTGGATGTTGCCATGGCAACCAAGTCA
rs132712ACGTTGGATGGTCTTCAAGGCTGAGTGAGCACGTTGGATGACTCCACGTGGCCTCTCTTG
rs132713ACGTTGGATGAAGGCTGAGTGAGCCCCAACACGTTGGATGACTCCACGTGGCCTCTCTT
rs132714ACGTTGGATGACACGGTGAAACCCCTTCTCACGTTGGATGAGTAGCTGGGACTACAGGTG
rs132716ACGTTGGATGTGGATTTGCAATGAGGAGTCACGTTGGATGTCAATGACTGTGCTCTACTC
rs132717ACGTTGGATGAATGTGGGCAGTTTTACGTGACGTTGGATGGATGGACCTTAGGGTGTTTC
192
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs132718ACGTTGGATGTCAGAGGGTATCAACATCTCACGTTGGATGTGGGCATCTTCATATACTGC
rs132719ACGTTGGATGATACCCTCAGTTGTACCCAGACGTTGGATGCTGAACAAAGGAGAAGGAGG
rs132720ACGTTGGATGATACTGGGTACAACTGAGGGACGTTGGATGCCTCTCCACCTTTTCCTAAC
rs132721ACGTTGGATGCAGCTACAAAGTTGCTAATGGACGTTGGATGTCTTATTTGTACCCTCCCTC
rs132722ACGTTGGATGACAATGGTAATGCTTGGAGCACGTTGGATGGGGAGGGTACAAATAAGATG
rs132723ACGTTGGATGACACAGATGTCTGTCTTCTGACGTTGGATGATACTCCCCTGGTGAATGCT
rs132724ACGTTGGATGCCCCATGCAACAAGGGTAAAACGTTGGATGTGTCCCTTACAGCAAGAAGC
rs132725ACGTTGGATGCCCCATGCAACAAGGGTAAAACGTTGGATGTGTCCCTTACAGCAAGAAGC
rs132726ACGTTGGATGGGGTCACACAGTGAACAAAGACGTTGGATGTTCTTGCTGTAAGGGACAGG
rs132727ACGTTGGATGGGGTCACACAGTGAACAAAGACGTTGGATGTTCTTGCTGTAAGGGACAGG
rs132728ACGTTGGATGAGTTCTACTGGCTCATGGTGACGTTGGATGTTCGCCTTCTTCCTGCTTTG
rs132729ACGTTGGATGAGTTCTACTGGCTCATGGTGACGTTGGATGTTCGCCTTCTTCCTGCTTTG
rs132730ACGTTGGATGTTCGCCTTCTTCCTGCTTTGACGTTGGATGAGTTCTACTGGCTCATGGTG
rs132731ACGTTGGATGTTTGTTCACTGTGTGACCCCACGTTGGATGTCCTGGTCCTCCCAGTTCTA
rs132732ACGTTGGATGGCCAAGGCAACCATCTCAACACGTTGGATGTGCAGCTCATCACAAGCGTC
rs132733ACGTTGGATGGCAACCATCTCAACACCATGACGTTGGATGTGCAGCTCATCACAAGCGTC
rs140575ACGTTGGATGTGCAGCTCATCACAAGCGTCACGTTGGATGCCATCTCAACACCATGAGCC
rs132734ACGTTGGATGGGATACTGACTGTTAGCCTCACGTTGGATGCGGAATTGACCAACTGGTAG
rs132735ACGTTGGATGGGATACTGACTGTTAGCCTCACGTTGGATGCGGAATTGACCAACTGGTAG
rs80587ACGTTGGATGTCTGAGCCAAGCTCACCAGAACGTTGGATGTTTTCCTGCCCAAGGAGGAG
rs132736ACGTTGGATGTTTTCCTGCCCAAGGAGGAGACGTTGGATGAAGCTCACCAGATGCAGACG
rs132737ACGTTGGATGTCTAGGCAGCAATGAGCTAGACGTTGGATGTGCTCCTCCTGAGAAATCAC
rs132738ACGTTGGATGTGAAGCCTGTAATCCCAGTGACGTTGGATGCATAGAGACAGCATCTCCTG
rs1807673ACGTTGGATGTTGCAGTGAGCAGAGATTGCACGTTGGATGGTGAAATCTGAGTCGTGGTC
rs2014700ACGTTGGATGTTGCAGTGAGCAGAGATTGCACGTTGGATGGTGAAATCTGAGTCGTGGTC
rs132739ACGTTGGATGGGAATCAAAGAAGGTGGAGGACGTTGGATGTGGTTGTGGCCAGACCATAA
rs1812023ACGTTGGATGAGCAGGAGGGAGGGAGCAATACGTTGGATGGACCTCCCTCCATCTCCTTA
rs1812024ACGTTGGATGTGTCAGAGGAAGATCCCTTGACGTTGGATGCCTCATAGAGCTATTGCGAG
rs2005590ACGTTGGATGGGAGGCAATGCCTGATTTTGACGTTGGATGTGCTTCCACCACCTGGAAAA
rs132740ACGTTGGATGTTCATTCTTCTTGTGCACAGACGTTGGATGCTTGACTGGTACCTAACAATG
rs3986001ACGTTGGATGTTTTCTGACTTGGCATCACCACGTTGGATGCACAAAGTATTCCACCTTCC
rs2413390ACGTTGGATGATAAATTCGTGGCTGAGCTCACGTTGGATGATCTTGTGGCATAAGGAGTC
rs132743ACGTTGGATGACAGGGAGAAAGTGAGGAAGACGTTGGATGCATCCTGTTTCCCCTAAAGG
rs132744ACGTTGGATGGGGTCTGTTTCAGGAGCATGACGTTGGATGTCTATGGCTGATGGCCACAG
rs2413391ACGTTGGATGGGGCAAAAGCAGAAATACTGACGTTGGATGCCCTCAAACCCTGTTTTCTG
rs132749ACGTTGGATGCCATGCACTCTCTAGTACTCACGTTGGATGTGTGGCCTTGGGGAAATGAT
rs132750ACGTTGGATGTCCTGTGCCTGTGGAAACTCACGTTGGATGGGTTCTCCAGGTGGCAAAAG
rs132741ACGTTGGATGCTACAATTTATCCGCACTAGACGTTGGATGGCCAAGTCAGAAAAATGAGAG
rs2413388ACGTTGGATGTACAGAATTCAGACCAACCCACGTTGGATGGCCCTGAGATTTGATTTTCC
rs2413389ACGTTGGATGGCTAGAATCTCATAACAGACGACGTTGGATGGCGTCCTACTATGATTTGTC
TABLE 62
dbSNP Extend Term
rs# Primer Mix
rs3888818 TGAGACCAGCCTAGCCAAC ACT
rs2010605 TGGTCCCAAGATATTCTATAGA ACT
rs743919 CTCACCTAAGGACTGCCTCT ACT
rs1008134 CCGGGCATCTTTTCTTCCATC ACT
rs132607 GCAGGCAGGACAGCATGTG ACT
rs1476029 CTTAGAGGCTATATTAAGACCA ACG
I
193
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs1476030 TTTGGCAATACTGGCCTATTC ACT
rs2413380 TCCAGGAGGGAAGACAACC ACT
rs2051609 CGCTTGAGGTCAGGACCAG CGT
rs2413381 GCGTGTTGCCAACAGCCTC ACT
rs1894604 AAATGGGAGGGAATGTTGGC ACG
rs1894605 TGCAGATCGCAACTGAGCG ACT
rs132609 GTTTCTCAGAGGATCAGGGA ACT
rs132610 GGTGTGAGATTTGGAGACTTT ACG
rs132611 CTCTGTCCTCTAGCCCCC ACT
rs132612 ATCTTCCCGCTACCTCAAGAGT ACT
rs1008790 CATAATCACAAGTCCTATGATTA ACT
rs23085 AGGATGACCATGGCAAGGAA CGT
rs105161 GGCCCTGGCAGGAAACAG ACG
rs132613 CCAATGGCCTCCACTGGC ACT
rs132614 CGGCCACAGCGCTGCCC ACG
rs132615 GCTTTCAGAACAACGGTAGAA ACT
rs132617 AAGAGTGTGTGTGCAGTAGCAAG ACG
rs3865724 TCACTTAAGCTTTGAATGTTTCTGACT
rs2019657 GCCAGAACATTGTGTTTCATTTGTACG
rs3865725 GGCAAGAGATACAGAATGCACA CGT
rs2019364 GTATCTCTTGCCCCTGCTC ACG
rs2008383 GAAGGACAGAAGGCTGATGC ACG
rs3986002 TCCTTCTCTGTGTCACTCCT CGT
rs3888942 ACATGGAAGCAGGGGTTTGA CGT
rs3888943 AGCAGGGGTTTGATGAAATCT ACT
rs3888944 CATCCTCCACATTGGGCCAA ACT
rs132618 AATCTCAGCTGGAAGTGG CGT
rs132619 TGCAACCAGCATTGACCG CGT
rs3827346 GAGGCTGCACCATCTCCAA ACG
rs132620 GGAAGCCTTTATTCAGGATTGT ACT
rs132621 TCAAATCTGCAACTGGTGTCAGAAACT
rs80575 GAACTCTCAAGCCACTTGAC ACT
rs80576 GCTAAGGCATTAGTTTGGCTGG ACG
rs80577 TGAAATTGCACATGGCATTGG ACG
rs80578 ATGCCTGGGAACTGGGGC ACT
rs80579 GGGAGGCACTGAGGGCATGAAA ACT
rs80580 CTGAGAATGAACAGCAGGTCA ACT
rs132622 ATAGTAGTTCAATCAGATGGGC ACT
rs132623 ATTCCAGCCTCTCTGTGTTCTG ACT
rs132624 ACAGGCATGAGCCGCTGC ACT
rs132625 GGCCACCGCATCCGGCTA CGT
rs132626 AGTGGCATGATCTCGGCCCAC ACG
rs132627 GAGGCGGAGGTTGCGGTG ACT
rs1807672 CATTGAGAATAAGGTGGTTCGA ACT
rs132628 CCTCCAAATTCATATACTGAGACCACG
rs132629 TCTCTCTCTCTCACACAC CGT
rs132630 CTGCTCCTGGCTTACAGAG ACT
rs132631 TCACAGTTCTGGAGGCAAAAA ACT
rs132632 I TGGAATTTTTGCCTCCAGAACTGTACT
194
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs132633 AGTGATTCACCAGGGAAGTGCCA ACG
rs132634 GGTGCTTTGTGGAGGAACC ACT
rs132635 ATTTCCCGTACATGGGGAGAAA ACT
rs132636 TGACAATAGGCACATGGCAG ACT
rs129603 GCTGCCATCCTAAACACATCTA ACT
rs132637 ACACAGCAGGATTACTGCCCAGA ACT
rs3788518 TGGGAGGCTCAAGGAAGAAACTCTACT
rs132638 GGAAAAAATAAAAGCAAAATACCCACT
rs132639 AAAGCAAACAGGCCTTCAGAA CGT
rs132640 GGTGACACAGAGAAGACGTGGC ACT
rs132641 GGGAGGTCAGAGGTCGGG ACT
rs132642 GTCACTGAGAGACTTTCC CGT
rs132643 GACACCCAGTACACACTGGCT ACT
rs132644 TTTGGAATGAGGAGTCATTTACA ACT
rs132645 CTCAACAGTAAGCAAGATTTAAA ACT
rs2017329 TGATGTTCAGATTTTCCTTTTTTTCGT
rs739198 TGGTCTCCACAACCTCTTATC ACT
rs132647 AAACCATGGAAGTCTCTAGAGTCAACT
rs2097465 CAAACTGCAGGCTTGCCCAG ACT
rs2105915 CAAGCCTGGGCAGCATAGCAC ACT
rs132648 AATTCCCGTGCTAATGCACG ACT
rs132649 CAGTCTTATTACTTTTGTACGAGGACT
rs132650 CATGAGCCACCGTGCCAG ACT
rs132651 CCTCAGGGTTTTTCACCTGCCT ACT
rs132652 AGGGCATCCTAACCCCCTA CGT
rs80584 ATCTACCTGCTCAACTTCCTGA ACG
rs132653 AATAACCAGACACGTTCTCCAG ACT
rs916334 GTCCAGCAGCACCCTTGGT ACG
rs132654 CGACAAGAGCAGGTCTGGAAC ACT
rs132655 CAGAAGAACCCACATAAGGAA ACT
rs132656 TCTTTGTCTTTTACTCCCACATCCACT
rs3834684 CACTAGAAGCAAGGAACCCCC ACT
rs132657 TACCTGACAATCACCCCCC CGT
rs916335 TCAGGTAATTGTCAGTCAGCC ACT
rs132659 AGAACTCCCCAAATCGTCCT ACG
rs132660 CCCCAGAGTGGGCTTTTCT ACT
rs132661 CCGCTCTCCCTCTGAGAGT ACT
rs132662 TGATCTGAGTTTACAGGTGAG ACT
rs132663 TGAGATCTGTCTCAGACGCA CGT
rs132664 CTGGGCTAGAGAGGGAGAC ACT
rs132667 CTTTAACTTTTGCTCACAAGAGT ACT
rs132670 AGACCAGCCTGATCAACATG ACT
rs132671 ACATCAATAGGCCTAAAAATCGTTACT
rs132672 GAAACTTGAAATTCCTTGAGAAATACT
rs132673 GTGTGAGTGGGAAGCCTCC ACG
rs132674 GGAGTTGGAGGCTGTAGTAA ACT
rs132675 AGATGTGCAATGGAATTTGGCAA ACT
rs80585 AGGCATAATATTCTGACCATTAAGACG
rs80586 I GGTCTGCTACTAGAATTCAGAAACG
195
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs132676 ATCCCTTAATATTGCATAGGAC CGT
rs132677 GGGTTGAAGTACTATGCTAGT CGT
rs132678 AGGTTAGTTCATGTAACTCCAT ACT
rs132680 TTTTATTTTAGCTTGAGCTTTTCAACG
rs132681 CTAGCTCTAAATCACATTCTGC ACG
rs132683 CAGGCCCATACCCAAAATATGCT ACT
rs2269594 TGGCTAAAAGGACAGATAGAG ACT
rs132684 GACACTAAGAGCGGTGAGAC ACT
rs132685 CGTGCCACCCAACTGGAGA ACT
rs132686 TGTGCATCTTATGGTGTACCA ACG
rs132687 TCGTTACCCCCATTCTATCC ACT
rs132688 ACAGGACGTGCTTGAAAGAG ACG
rs132689 TGGCGATGGCCTCTGCTC ACT
rs132690 GCTCTCCTTGCTTCAAAAAAAAA CGT
rs132691 TGACCTATCCTGCTTCAGGT ACT
rs132692 CGAAGTGTGTTAGCTCATGAC ACT
rs132693 GAGTGTGGACACCAGGTCA ACT
rs132694 CACCTTAGGAATGGCAGCTTC ACG
rs132695 TATGACTACACATGCTGGCAAAC ACT
rs1966266 GCAGACCCCTAACTCTAATTTG ACG
rs1966267 CACTGAGTTATGAGTACTCAAC ACT
rs106808 GATCCTTCCCGAGGACACC ACT
rs132696 CAGGCTGCCTGGAAGGAGA ACT
rs2239829 GATGGCTGGATTCATAACAGGTAAACT
rs2285154 AGTGCTGGGATTACAGGCAT ACT
rs2239830 CAGTCACCTGAATTTGTGCTTATTACT
rs2239831 GTCACTGACCCAAGCTATCCTC ACG
rs3865722 CAATTGCAAGCAACAGAAACAGAACGT
rs3865723 TCCACTGTACTGCTAGTATTG ACG
rs3985996 AGAATTTCTCCCGGCATCAG ACG
rs3985997 TCTCCCGGCATCAGCCTTC ACG
rs3985998 GGGAGGAGTGACACAGAGAAGGA ACG
rs3985999 GGGGTACTGGGAGGAGTGACAC ACT
rs3986000 GCAGGACTGGGGTACTGG ACT
rs2413382 CAGGGGAACTCAGGCCACA ACT
rs2413383 AGCCATTGAAGACATGGAAGCC ACT
rs2413384 ATTGAAGACATGGAAGCCGG ACT
rs2413385 GGCCAGCTCTTCCTCCAC CGT
rs2413386 CTTGGCCCAATGTGGAGGA CGT
rs1894606 CCTAGCGGCAAGGGCTGT ACT
rs916336 AGGGTATTTGCTGTCTGGCTGTCTACT
rs916337 AAGGGCCTGTATGTAGGTTGAA ACT
rs916338 CCCTCTATGTCTCATGGATTTTCCACG
rs132697 CCTAGGGGAGCCCATATATCA ACT
rs12781 AGACAGCTCGAGAGATCC ACT
rs1053983 GCTGACTCAGATACACCCC ACG
rs1053982 GATGCTGGTAAGACAGGG ACG
rs2227167 CGTCAAAATCAAGTGCAAA ACT
rs2227168 ~ATTAAGGGACATTCTGC ~ AC~
196
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs132700 ATCCTGTCTGTCATTGGCGTT ACT
rs3075364 GTTTAGGGAGTGGTTTTTGAAAG CGT
rs2227169 TGTCCTTTATTGGTACAGGGAAGAACT
rs2097466 TACAAGCATGAGCCACCGC ACT
rs2097467 TCCCAAAGTGCTGGGATTACA ACT
rs2413387 TACAGGCACTCACCACCAC ACT
rs132701 TGTACAAAACATATTTAACCTTGAACT
rs132702 CACCCAGTTAAGGAAAAATTCCT ACT
rs132703 GATCAATGTGTGTTCCCGGA ACT
rs2269595 GCCCCAGACAGCATCTCC ACT
rs2269596 TTGCTGGCAAGAGACCAGG ACT
rs132704 GGGCTGCCTGGAGGAGG ACG
rs2007468 TGGGCAATTCAGCCACACGCAC ACT
rs132705 TATAGACTGAATTGTGTGCCC ACG
rs2007706 GGAATTTACATAAGGGTCTATAG ACT
rs132706 GAACCCCCTCCACTGCCC ACT
rs132707 GCTCTCCCTCTGAAACAAGATG ACT
rs132708 GAGAGCTTCTTCCTTGGCC ACT
rs132709 CAGGGAAGATTAGAAGCTGAGAGCCGT
rs132710 GGGCAGGGAAGATTAGAAGC ACG
rs132711 TTTGCTGTCCAGGGCGGC ACG
rs132712 AACCCAGACGGAGGTGGC ACG
rs132713 GCCCCAACGGAACCCAGA ACG
rs132714 CCCCTTCTCTACTGAAAATACAAAACT
rs132716 TGAGGAGTCATTTACCATGAG ACG
rs132717 GCAGTTTTACGTGAAGGAGG ACT
rs132718 GTTTTATACCTAGAGCCACACT ACT
rs132719 TACCCAGTATTTCTTAACTTCC CGT
rs132720 CAGCTACAAAGTTGCTAATGG ACT
rs132721 ATGTTAGGAAAAGGTGGAGAG ACT
~
rs132722 CCTAACTGGGATGGGCCTGAA ACT
rs132723 TTCTGGGGCCCCCATGCA ACT
rs132724 ACCCAGTCCTGGGCAGCA ACT
rs132725 GGGAGTATGCAGAGGGGC ACT
rs132726 CAGTGAACAAAGCAGGAAGAAGG ACT
rs132727 GTCACACAGTGAACAAAGCAGGAAACT
rs132728 CATGGTGTTGAGATGGTTGCC ACG
rs132729 GCTCATGGTGTTGAGATGGTT ACT
rs132730 GTGACCCCCTAGGCCAAGGCA ACT
rs132731 GGCAACCATCTCAACACCAT ACT
rs132732 AACCATCTCAACACCATGAGCCA ACT
rs132733 ATCTCAACACCATGAGCCAG ACT
rs140575 GCTCTCTCCTGGTCCTCC ACG
rs132734 AGCCTCAACTAGGACACA ACT
rs132735 TCAACTAGGACACAGTGC ACT
rs80587 GCCAAGCTCACCAGATGCAGA ACT
rs132736 AGGGAGCTGCTTTGCTGAAA ACT
rs132737 CCTGCAGCCTGGGTGACA ACT
rs132738 GGCCAGGAGTTCAAGACAGCCTG ACT
~
197
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Extend Term
rs# Primer Mix
rs1807673 GGGCAACAGAGCGAGACTCC ACT
rs2014700 GAGCGAGACTCCATCTCA ACT
rs132739 GGGAGGTGACCTGGAGCC ACG
rs1812023 GCAATCAGACTCAAGCCTGG ACT
rs1812024 GGGATGGTGTGACCTCCC ACG
rs2005590 CAATGCCTGATTTTGTCACTGAAC ACT
rs132740 GGCATATGTGCATTTGTCTGAG ACG
rs3986001 TCCTTTTTCTAAACCCCTGCAA ACT
rs2413390 GGCTGAGCTCAAGGTTTTAAA ACT
rs132743 GGAGAAAGTGAGGAAGAAAATTA ACT
rs132744 TGGGGTTACAGTTGGTCATAACC ACT
rs2413391 TGATATGTTCAGCGGTGCAC ACT
rs132749 TCTTGATGTTTCTCCTATCCC ACG
rs132750 CCTGTGGAAACTCAGCAGC ACG
rs132741 GCACTAGATATTGAATTCTTTCC ACT
rs2413388 CAACCCCGTGACTGGAGATTC CGT
rs2413389 TTTCTCTCTCTAGTACTCTATTT ACT
Genetic Analysis
[0324] AlleloTyping results from the discovery cohort are shown for cases and
controls in Table 63.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (Al AF
= 1-A2 AF). For
example, the SNP rs2010605 has the following case and control allele
frequencies: case A1 (A) = 0.19;
case A2 (G) = 0.81; control A1 (A) = 0.18; and control A2 (G) = 0.82, where
the nucleotide is provided
in paranthesis. Some SNPs are labeled "untyped" because of failed assays.
TALE 63
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 13
rs3888818 201 34781551 C/T
rs2010605 425 34781775 A/G 0.81 0.82 0.782
rs743919 1095 34782445 G/T 0.10 0.11 0.502
rs1008134 2201 34783551 A/C
rs132607 7879 34789229 A/G 0.11 0.11 0.813
rs1476029 8395 34789745 C/T 0.15 0.15 0.983
rs1476030 8461 34789811 C/T 0.36 0.37 0.708
rs2413380 9503 34790853 C/T 0.29 0.29 0.900
rs2051609 10304 34791654 G/T
rs2413381 10695 34792045 C/T
rs1894604 16300 34797650 A/G 0.08 0.08 0.759
rs1894605 16444 34797794 G/T 0.08 0.09 0.468
rs132609 17591 34798941 C/T 0.68 0.67 0.777
rs 132610 17988 34799338 -/A
rs132611 19116 34800466 -/T 0.14 0.15 0.863
rs132612 19358 34800708 C/T 0.23 0.23 0.951
rs1008790 20300 34801650 A/G 0.03 0.10 0.007
rs23085 20669 34802019 A/T 0.31 0.32 0.738
rs105161 20891 34802241 A/G 0.76 0.77 0.826
rs132613 21451 34802801 C/T 0.80 0.81 0.619
rs132614 21978 34803328 C/T 0.16 0.14 0.434
198
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAllA2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 13
rs132615 22785 34804135 C/G 0.32 0.31 0.740
rs132617 24248 34805598 C/T 0.35 0.36 0.825
rs386572424770 34806120 C/T 0.65 0.65 0.940
rs201965724844 34806194 A/G 0.20 0.20 0.857
rs386572525066 34806416 G/T
rs201936425096 34806446 C/T 0.40 0.39 0.767
rs200838325309 34806659 A/G 0.18 0.17 0.665
rs398600225344 34806694 A/C
rs388894225529 34806879 A/T
rs388894325537 34806887 A/G
rs388894425554 34806904 A/C
rs132618 27963 34809313 A/T 0.43 0.43 0.934
rs132619 28134 34809484 G/T
rs382734628356 34809706 A/G 0.84 0.84 0.976
rs132620 29648 34810998 -/A 0.29 0.29 0.879
rs 13262129986 34811336 A/G 0.32 0.31 0.867
rs80575 30217 34811567 G/T 0.27 0.27 0.948
rs80576 30267 34811617 A/G 0.26 0.25 0.443
rs80577 30315 34811665 A/G 0.26 0.23 0.191
rs80578 30585 34811935 C/T 0.49 0.48 0.548
rs80579 30724 34812074 A/C 0.23 0.25 0.574
rs80580 30897 34812247 C/T 0.31 0.31 0.878
rs132622 30931 34812281 C/T 0.29 0.30 0.943
rs132623 31080 34812430 G/T 0.60 0.59 0.806
rs132624 31246 34812596 C/T 0.36 0.37 0.772
rs132625 31373 34812723 A/T
rs132626 31463 34812813 A/G 0.89 0.84 0.082
rs132627 31467 34812817 A/G 0.12 0.11 0.836
rs180767232188 34813538 G/T 0.30 0.30 0.974
rs132628 32288 34813638 C/T 0.25 0.24 0.691
rs132629 32520 34813870 A/T 0.04 0.06 0.250
rs132630 32594 34813944 A/C 0.75 0.75 0.978
rs 13263132657 34814007 A/C 0.72 0.73 0.509
rs132632 32677 34814027 A/G 0.66 0.65 0.798
rs 13263332764 34814114 C/T 0.34 0.33 0.796
rs 13263432784 34814134 A/G 0.45 0.42 0.317
rs132635 32830 34814180 C/T 0.41 0.40 0.772
rs132636 32872 34814222 C/T 0.41 0.44 0.192
rs129603 33121 34814471 A/C
rs132637 33348 34814698 G/T 0.09 0.09 0.628
rs378851833952 34815302 C/G 0.17 0.19 0.297
rs132638 34184 34815534 C/G 0.56 0.58 0.509
rs132639 34361 34815711 A/T 0.13 0.12 0.561
rs132640 35026 34816376 A/G 0.32 0.30 0.388
rs132641 35192 34816542 A/G 0.48 0.51 0.287
rs132642 35600 34816950 A/T 0.15 0.14 0.732
rs132643 36033 34817383 C/T 0.44 0.46 0.360
rs132644 36289 34817639 C/T 0.53 0.58 0.075
rs1$2645 38869 34820219 A/G 0.19 0.18 0.572
rs201732939629 34820979 A/T 0.39 0.40 0.915
rs739198 40530 34821880 C/T 0.70 0.70 0.878
rs132647 41621 34822971 C/T 0.23 0.23 0.957
rs209746542379 34823729 C/T 0.54 0.51 0.344
rs210591542802 34824152 C/T 0.57 0.56 0.468
rs132648 42865 34824215 T/C
rs132649 43644 34824994 A/G 0.21 0.22 0.579
rs132650 45051 34826401 C/T 0.34 0.31 0.248
rs132651 45828 34827178 A/C
rs132652 45829 34827179 A/T
rs80584 46257 34827607 C/T 0.81 0.75 0.043
rs132653 47286 34828636 A/C 0.17 0.15 0.312
rs916334 47427 34828777 C/T 0.34 0.36 0.345
rs132654 47963 34829313 C/T 0.54 0.56 0.439
rs132655 48013 34829363 C/T 0.41 0.41 0.838
199
CA 02561742 2006-09-28
WO 2005/100604 PCT/US2005/010912
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID NO: AF AF
13
rs132656 48229 34829579 C/T 0.37 0.36 0.813
rs3834684 48282 34829632 -/A 0.21 0.22 0.480
rs132657 48376 34829726 -/G 0.49 0.50 0.719
rs916335 48404 34829754 AIG 0.38 0.36 0.509
rs132659 49900 34831250 CIT
rs132660 52699 34834049 G/T 0.37 0.38 0.754
rs132661 52897 34834247 A/G 0.27 0.29 0.291
rs132662 53414 34834764 A/G 0.61 0.58 0.186
rs132663 53487 34834837 A/T 0.26 0.29 0.167
rs132664 54112 34835462 GIT 0.25 0.29 0.098
rs132667 55492 34836842 A/G 0.35 0.36 0.559
rs132670 59766 34841116 C/T
rs132671 60307 34841657 A/G 0.49 0.53 0.145
rs132672 60701 34842051 A/G 0.23 0.22 0.716
rs132673 60952 34842302 A/G 0.41 0.37 0.184
rs132674 61401 34842751 C/T 0.32 0.31 0.476
rs132675 62379 34843729 C/T 0.38 0.35 0.188
rs80585 62870 34844220 CIT 0.28 0.27 0.525
rs80586 62879 34844229 A/G 0.66 0.66 0.966
rs132676 63499 34844849 A/T 0.26 0.23 0.177
rs132677 64284 34845634 -/A 0.69 0.69 0.873
rs132678 64408 34845758 A/G 0.46 0.48 0.395
rs132680 64760 34846110 A/G 0.20 0.20 0.995
rs132681 65230 34846580 A/G 0.24 0.24 0.901
rs132683 66127 34847477 A/G 0.19 0.19 0.851
rs2269594 66634 34847984 C/T 0.70 0.67 0.332
rs132684 66686 34848036 A/G 0.21 0.20 0.756
rs132685 66694 34848044 C/G 0.30 0.28 0.553
rs132686 67113 34848463 A/G 0.46 0.48 0.398
rs132687 67257 34848607 A/G 0.96 0.96 0.767
rs132688 67403 34848753 A/G 0.24 0.23 0.553
rs132689 67609 34848959 A/G 0.61 0.63 0.564
rs132690 68418 34849768 -/A 0.16 0.17 0.672
rs132691 68610 34849960 C/G 0.52 0.52 0.976
rs132692 69629 34850979 C/T 0.63 0.62 0.800
rs132693 70024 34851374 A/G 0.58 0.58 0.868
rs132694 70848 34852198 A/G 0.17 0.16 0.583
rs132695 71428 34852778 C/G 0.23 0.22 0.616
rs1966266 71553 34852903 C/T 0.49 0.47 0.413
rs1966267 71633 34852983 A/G 0.40 0.41 0.773
rs106808 71768 34853118 A/C 0.68 0.67 0.617
rs132696 71769 34853119 A/G
rs2239829 73039 34854389 A/G 0.34 0.36 0.510
rs2285154 73325 34854675 A/G
rs2239830 73412 34854762 A/C 0.49 0.50 0.841
rs2239831 73547 34854897 C/T 0.52 0.50 0.564
rs3865722 73769 34855119 A/T 0.57 0.56 0.861
rs3865723 73806 34855156 A/G 0.59 0.58 0.722
rs3985996 74467 34855817 C/T 0.30 0.29 0.861
rs3985997 74472 34855822 C/T 0.89 0.90 0.527
rs3985998 74473 34855823 A/G
rs3985999 74482 34855832 C/T 0.19 0.19 0.968
rs3986000 74494 34855844 A/C
rs2413382 74592 34855942 A/G 0.61 0.59 0.618
rs2413383 74670 34856020 G/T
rs2413384 74672 34856022 G/T
rs2413385 74714 34856064 G/T 0.70 0.70 0.944
rs2413386 74723 34856073 A/T 0.70 0.71 0.816
rs1894606 74749 34856099 A/G
rs916336 74861 34856211 C/G
rs916337 74892 34856242 C/T
rs916338 74893 34856243 CIT 0.40 0.40 0.939
rs132697 75176 34856526 A/G 0.59 0.61 0.418
rs12781 75705 34857055 A/G
200
DEMANDE OU BREVET VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVET COMPREND
PLUS D'UN TOME.
CECI EST LE TOME 1 DE 4
CONTENANT LES PAGES 1 A 200
NOTE : Pour les tomes additionels, veuillez contacter le Bureau canadien des
brevets
JUMBO APPLICATIONS/PATENTS
THIS SECTION OF THE APPLICATION/PATENT CONTAINS MORE THAN ONE
VOLUME
THIS IS VOLUME 1 OF 4
CONTAINING PAGES 1 TO 200
NOTE: For additional volumes, please contact the Canadian Patent Office
NOM DU FICHIER / FILE NAME
NOTE POUR LE TOME / VOLUME NOTE: