Note: Descriptions are shown in the official language in which they were submitted.
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE I)E CETTE DEMANDE OU CE BREVETS
COMPRI~:ND PLUS D'UN TOME.
CECI EST ~.E TOME 1 DE 2
NOTE: Pour les tomes additionels, veillez contacter 1e Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME 1 OF 2
NOTE: For additional vohxmes please contact the Canadian Patent Oi~ice.
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
Identification of the precise.amino acid sequence of the epitope recognized
by the potent neutralizing human anti-HIV-1 monoclonal antibody IgG1b12
FIELD OF THE INVENTION
The present invention relates generally to the field of medical treatments.
BACKGROUND OF THE INVENTION
Global eradication of HIV will likely not occur with some miracle drug, but
with a vaccine. ' The development of a safe, effective HIV vaccine has eluded
scientists for over a decade. As the standard approaches to vaccine design are
being exhausted, it is evident that in order to combat such a complex virus,
newer
technologies may hold the key to a successful vaccine. Until very recently,
biology
and mass spectrometry have been two facets of science without overlap. In the
last few years, it has become apparent that the tools of mass spectrometry may
be
exploited to determine the specificities of the immune system receptors- from
antibodies to toll like receptors- in order to gain information on the human
immune
response (3,4). . This information can be applied to vaccine and therapeutics
design, reagent development and so on.
In order to understand how to combat the HIV virus, we must study cases
where individuals have combated HIV relatively successfully. In 1991, Burton,
et
al (5) cloned a Glade B HIV-1-specific Fab fragments from an~ antibody library
from
the B cells of an HIV positive American male who had remained asymptomatic for
over 6 years. They were then able to show that a certain monoclonal Fab
antibody, b12, was a potently neutralizing antibody (6). Several years later,
the
crystal structure of the whole IgG1 b12 molecule was resolved. It was
determined
that the docking site of IgG1b12 blocks the CD4 binding site of the HIV-1
surface
glycoprotein termed gp120 (7). Gp120 amino acids involved in gp120-CD4
binding, as determined by x-ray crystallography, include Asp 368 (fig 3.
D362), Glu
370 (G364) and Trp 427 (W484) (8). Gp120 amino acids determined to be
involved in gp120-IgG1b12 binding are, according to the sequence in figure 3,
6371, D373, P374, 1376 and Y389.
The neutralizing capabilities of IgG1b12 have been described by many
groups for many strains of HIV-1, and under many conditions (refs. 6,9-24). In
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
2
vitro and ex vivo neutralization assays have been ~ performed on primary
isolates
and lab-adapted strains from Glades A, B, C, D, ' E, F and O (refs. 11, 16-
24).
Mouse (refs. 12, 14) and macaque (15) studies have shown that IgG1b12 can
protect these animals from HIV-7 and SHIV challenges, respectively. In
September of 2002, Lewis et al showed that neutralizing antibody could be
found
in the serum of mice receiving the IgG1b12 antibody gene delivered to the
muscle
by a recombinant adeno-associated .virus (13). It has been shown that IgG1b12
can block HIV-1 attachment to CD4+ cells (10), as well as dendritic cell
infection
and transfer to T cells (9). There is no doubt that IgG1b12 is an invaluable
tool for
vaccine research and development.
The abilities of a vaccine to elicit immune responses that block viral
infection of target cells and/or replication within these cells are ~ critical
to its
success. Antibodies are capable of combating invading virus in many ways.
When HIV-1 exits an infected cell, it acquires its envelope from that cell's
membrane. Gp120 is therefore expressed on the surface of infecfie~ cells
containing replicating virus. Gp120 may also exist on the surface as a result
of
HIV fusion to the cell membrane. Antibody can bind to the gp120 and mediate
antibody dependant cellular cytotoxicity (ADCC), or compliment-dependant
cytotoxicity (CDC) of the infected cell, or potentially block viral release.
Perhaps
the most important and exciting function that a protective antibody may have
is~ its
capacity to yield sterilizing immunity. Antibodies can bind to surface
proteins on
the virus and specifically block virus particles required for cell invasion.
Higher
affinity antibodies will remain tightly attached to the viral surface, out-
competing for
binding by the target cell receptor molecules) and are in general more
powerfully
neutralizing. Antibody-bound viruses can also trigger complement-mediated
virolysis or phagocytosis (25): Virus that has entered the body is unable to
infect
target cells because neutralizing antibody mops up free virus which is then
cleared
by normal mechanisms. Infection cannot be established, and the host remains
healthy. In contrast, cytotoxic T cells can only be specifically activated
after an
infection has been established and cells begin presenting antigen. Sterilizing
immunity does not result in this case, and it is importanfi to remember that
once
HIV has infected cells, it can begin to mutate and evolve to escape the immune
response. Antibody-focused vaccine researchers strive to create a vaccine that
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
3
will elicit a sterilizing antibody response. It is also important to induce
cross clade-
specific antibody responses so that the vaccine recipient is protected from
infection
by HIV of any Glade. ,
One of the most important HIV-related phenomena to have been discovered
in the last ten years is the existence of individuals who are resistant to HIV
infection. This model of protection may hold the secret to the specific and/or
innate immune responses required to successfully block HIV infection. Our
group
has identified a group of Kenyan female sex workers who, despite repeated
exposure to HIV, remain uninfected (26). HIV-1 gp120-specific IgA has been
isolated from the cervix of these women. The cervical IgA not only neutralizes
HIV, but it can also inhibit the transcytosis of HIV across human epithelial
cells
(28). These women are exposed to HIV through heterosexual contact, therefore
HIV initially comes into contact with cells of the genital tract: The virus
must pass
through epithelial cells via transcytosis in order to establish an infection.
It is
therefore plausible that neutralizing, transcytosis-inhibiting antibody may
play a
crucial role in HIVresistance in these women. Any vaccine that could educe
such
antibodies may provide sterilizing immunity to its recipients.
The IgG1b12 antibody was cloned from an HIV+ donor who had been HIV+
for over 6 years. Why had he remained AIDS-free for over 6 years? It is very
possible that his immune response was effective at combating the disease,
' keeping the virus 'at bay' for an extended period of time. HIV researchers
continue
to hash out the mechanism by which some individuals become long-term non-
progressors. It is possible that the existence of potently neutralizing
antibodies
aids in harnessing the infection. The IgG1b12 epitope specificity may provide
information, in the form of a marker, about those individuals who will not
progress
quickly to AIDS. Knowledge of what comprises a neutralizing epitope for
antibodies may be applicable to clinical settings as well. For instance, if
antibodies
from patient 'X' recognized a specific sequence in the HIV envelope protein,
they
have an increased chance of being a long-term non-progressor. Doctors could
use this information to tailor drug regimens specifically for each patient.
SUMMARY OF THE INVENTION
According to a first aspect of the invention, there is provided a purified
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
4
polypeptide, the amino acid sequence of which comprises at least 6 contiguous
residues of any one of SEQ ID No. 1-6.
According to a~ second aspect of the invention, there is provided a method of
immunizing an individual against HIV infection comprising administering to an
individual a purified polypeptide, the amino acid sequence of which comprises
at
least 6 contiguous residues of any one of SEQ ID No. 1-6.
According to a third aspect of the invention, there is provided the use of a
purified polypeptide as a vaccine, the amino acid sequence of which comprises
at
least 6 contiguous residues of any one of SEQ ID No. 1-6.
According to a fourth aspect of the invention, there is provided the use of a
purified polypeptide as a medicament, the amino acid sequence of which
comprises at least 6 contiguous residues of any one of SEQ ID No: 1-6.
According to a fifth aspect of the invention, there is provided a method of
preparing an immune globulin .efFective against Human Immunodeficiency virus
comprising:
vaccinating a plurality of donors with a purified polypeptide, the amino acid
sequence of which comprises at least 6 contiguous residues of any one of SEQ
ID
No. 1-6;
isolating plasma from each of said donors after a period of time sufficient to
allow production of antibodies against said polypeptide;
pooling the plasma; and
preparing an immune globulin from the pooled plasma.
According to a sixth aspect of the invention, there is provided a method of
determining a course of treatment for an individual infected with human o
immunodeficiency virus comprising: ,
screening . a sample from an individual infected with human
immunodeficiency virus for antibodies binding to a purified polypeptide, the
amino
acid sequence of which comprises at least 6 contiguous residues of any one of
SEQ ID No. 1-6,
wherein presence of antibodies against said polypeptide indicates that a
less aggressive treatment is needed. ,
According to a seventh aspect of the invention, there is provided a method
of treating an individual infected or suspected of being infected by human
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
immunodeficiency virus comprising administering to said individual a
therapeutically effective amount of a purified polypeptide, the amino acid
sequence
of which comprises at least.6 contiguous residues of any one of SEQ ID No. 1-
6.
According to an eighth aspect of the invention, there is provided a method
5 of treating an individual infected or suspected of being infected by human
immunodeficiency virus comprising administering to said individual a purified
polypeptide, the amino acid sequence of which comprises at least 6 contiguous
residues of any one of SEQ ID No. 1-6. .
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1. MALDI QqTOF mass spectrometry results of HIV-1 gp120 epitope
mapping. IIIB (a) or MN (b) gp120 was bound to IgG1b12 antibody and was
digested overnight with Glu-C endopeptidase. Protein fragments not bound by
IgG1b12 were washed away. Antibody-bound fragments protected from digestion
' were analyzed by mass spectrometry. The size of the first peak, 1807
(mass/charge, or m/z), corresponds to the N-terminal sequence
ATTTLFCASDAKAYDTE (as determined by the theoretical digest program). 1867,
the second peak, corresponds to the sequence KLWVT\/YYGVPVWK. The third
peak, 2097 corresponds to.the sequence.TEKLV'JVTVYYGVPVWKE.
' Figure 2. MALDI QqTOF mass spectrometry results of gp120 epitope
mapping. IIIB (a) or MN (b) gp120 was bound to IgG1b12 antibody and was
digested overnight with trypsin endopeptidase. Protein fragments not bound by
IgG1b12 were washed away. Antibody-bound fragments protected from digestion
were analyzed by mass spectrometry. The size of the peak 1357 corresponds to
the N-terminal sequence EATTTLFCASDAK. The peaks at 1609 corresponds to
the N-terminal sequence LWVTVYYGVPVWK.
Figure 3. Amino acid sequence of HIV-1 gp120 IIIB (Immunodiagnostics,
Inc.). Amino acids identified by mass spectrometry are underlined.
Figure 4. Amino acid sequence of HIV-1 gp120 MN (Immunodiagnostics,
Inc.). Amino acids identified by mass spectrometry are underlined. Bold G
(glycine) is the unknown putative amino acid change G-~T (threonine)
recognized
by the exquisite sensitivity of mass spectrometry (2097 peak).
Figure 5. MALDI QqTOF mass spectrometry results of HIV-1 gp120 epitope
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
6
mapping. Amino terminus and CD4 binding site gp120 peptides were bound to
IgG1b12 antibody.
Fi. uq_ re 6. Mass spectrometry results of 2 hour N-.term peptide digestion
with
trypsin. The peak at m/z=1737 corresponds to the amino acids that remained
bound to the IgG1 b12 after trypsin digest. The 1737 peak. corresponds to the
digested fragment amino acid sequence KLVWTVYYGVPVWK.
Figure 7. Consensus sequence for IgG1b12 binding.
Figure 8. Epitope mapping confirmation that the IgG1 b12 epitope on glu-C
digested gp120 is variable region-specific. Gp120 MN was incubated with either
IgG1 b12 (A) or KZ52 control (B) antibodies linked to Sepharose beads. The
antigen-antibody complex was digested with the endoprotease glu-C, washed and
tested by MALDI QqTOF mass spectrometry for bound epitopes.
Fi uq re 9. Epitope excision mapping by trypsin digestion of gp120 confirms
that the IgG1 b12 epitope recognition by gp120 is variable region-specific.
Gp120
MN was incubated with either (a) IgG1 b12 or (b) KZ52 control antibodies
linked to
Sepharose beads: Unbound antigen was washed away and bound gp120 was
digested with the endoprotease trypsin, washed and tested for antigen-antibody
interactions.
Fiqure 10. Western blot of IgG1 b12 shows that IgG1 b12 binds whole,
denatured gp120. Soluble gp120 was resolved on SDS-PAGE gel and b12
binding was detected by cheriiiluminescence. The first lane shows b12 binding
to
soluble gp120, lane 2 shows reactivity of the secondary antibody alone vs.
gp120
(negative control), lane 3 shows b12 binding to BSA (negative control).
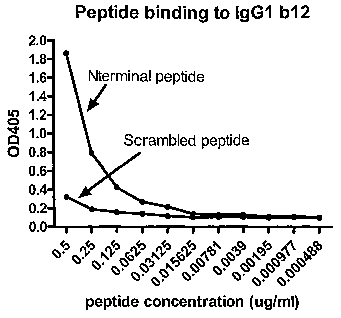
Figure 11. IgG1b12 recognizes synthetic peptide sequence from the amino
terminus of gp120, and not a scrambled version of the same peptide. ELISA
plates were coated with IgG1 b12, biotinylated N-term and scrambled peptides
were added and tested for binding. Representative data from one of three
experiments is shown. a
Figure 12. The binding of N terminal peptide to IgG1 b12, can be blocked by
soluble gp120. ELISA plates were coated with IgG.1 b12. Soluble gp120 was
added then biotinylated peptide was added and tested for binding.
Representative
data from one of 3 experiments is shown.
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
7
Figure 13. Antigen recognized by differentially immunized mice. Four
groups of 4 mice were immunized on five separate occasions with adjuvant plus
PBS alone, gp120, N terminal peptide, or scrambled peptide. Serum was
collected
and tested for antibody recognition of gp120 (A), N terminal peptide (B), and
scrambled peptide (C) by indirect ELISA. Representative data from one of ' 3
experiments is shown.
DESCRIPTION OF THE PREFERRED EMBODIMENTS
Unless defined otherwise, all technical and scientific terms used herein
have the same meaning as commonly understood by one of ordinary skill in the
art.
to which the invention belongs. Although any methods and materials similar or
equivalent to those described herein can be used in the practice or testing of
the
present, invention, the preferred methods and materials are now described. All
publications mentioned hereunder are incorporated herein by reference.
DEFINITIONS
As used herein, "effective amount" refers to the administration of an amount
of a given compound that achieves the desired effect.
As used herein, "purified" does not require absolute purity but is instead
intended as a relative definition. For example, purification of starting
material or
natural material to at least one order of magnitude, preferably two or three
orders
of magnitude is expressly contemplated as falling within the definition of
"purified".
As used herein, the term "isolated" requires that the material be removed
from its original environment.
As used herein, the term "treating" in its various grammatical forms refers to
preventing, curing, reversing, attenuating, alleviating, minimizing,
suppressing or
halting the deleterious effects of a disease state, disease progression,
disease
causitive agent other abnormal condition.
As described herein, the region recognized by IgG1b12, which is the most
potent antibody yet described which is capable of neutralizing HIV-1, has been
identified. Furthermore; the neutralizing ability of IgG1b12 is likely
involved in
protective immune responses, to HIV-1 and this can be induced in others to
generate protective HIV-1 specific responses. For example, the sequence of
this
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
8
particular epitope could be used in blocking HIV inflection. Knowledge of
when/how responses to this epitope develop may also be useful in tailoring
alternate therapeutic interventions, as discussed below.
As discussed below, the minimal epitopes as defined by Glu-C digestion
are:
LWVTVYYGVPVWKE and ATTTLFCASDAK
while the minimal epitopes as defined by Trypsin digestion are:
LVWTVYYGVPVWK and ~EATTTLFCASDAK
This leads to . a consensus sequence for IgG1 b12 binding of
LWVTVYYGVPVWKEATTTLFCASDAK (SEQ ID No. 1, shown in Figure 7) and a
sequence of GVPVWKEATTTL (SEQ ID No. 2). As can be seen in Figure 7, the
sequence of this region varies somewhat in different strains and Glades.
In one embodiment ofi the invention, there is provided an isolated and/or
purified polypepfide, the amino acid sequence of the polypeptide comprised of
or
consisting essentially of 6 or more consecutive residues of~SEQ ID No. 1 or
SEQ
ID No. 2, that is, LWVTVYYGVPVWKEATTTLFCASDAK or GVPVWKEATTTL or a .
variant thereof, for example, as shown in Figure 7. In other embodiments, the
polypeptide may consist of 7 or more consecutive residues, 8 or more
consecutive
residues, 9 or more consecutive residues or 10 or more consecutive residues of
SEQ ID No. 1 or SEQ ID No. 2, that is, LWVTVYYGVPVWKEATTTLFCASDAK or
GVPVWKEATTTL or a variant thereof.
As will be apparent to one of skill in the art, as used herein, "variant
thereof'
refers to peptides derived from or based on the amino acid sequence firom the
same region of gp120 from a different Glade or isolate of HIV that act as a
neutralizing peptide, as discussed below. Examples of such variants are shown
in
Figure 7. Other potential variants can readily be determined using means known
in
the art and any suitable database containing gp120 sequences.
Thus, in another~embodiment of the invention, there is provided an isolated
and/or purified polypeptide, the amino acid sequence of the polypeptide
comprised
of or consisting essentially of 6 or more consecutive residues of SEQ ID No. 3
or
SEQ I D No. 4, that is,
LWVTVYYGVPVW(E/K/R)(E/D)A(E/T/N/K/D/A)(T/P)(TlP/V)LFCASDAK or
GVPVW(E/K/R)(E/D)A(E/T/N/K/D/A)(T/P)(T/P/V)L or a variant thereof, for
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
9
example, as shown in Figure 7. In other embodiments, the polypeptide may
consist
of 7 or more consecutive residues, 8 or more consecutive residues, 9 or more
consecutive residues or 10 or more consecutive residues of SEQ ID No. 3 or SEQ
I D No. 4, that is,
LWVTVYYGVPVW(E/K/R)(E/D)A(E/T/N/K/D/A)(T/P)(T/P/V)LFCASDAK or
GVPVW(E/K/R)(E/D)A(E/T/N/K/D/A)(T/P)(T/PIV)L or a variant thereof.
In yet another embodiment of the invention, there is provided ,an isolated
and/or purified polypeptide, the amino acid sequence of the polypeptide
comprised
of or consisting essentially of 6 or more consecutive residues of SEQ ID No.
5. or
SEQ ID No. 6, that is,
LWVTVYYGVPVW(E/K/R)(E/D)A(E/T/N/D)(T/P)(T/P)LFCASDAK or
GVPVW(E/K/R)(E/D)A(E/T/N/D)(T/P)(T/P)L or a variant thereof, for example, as
. shown in Figure 7. In other embodiments, the polypeptide may consist of 7 or
more
consecutive residues, 8 or more consecutive residues, 9 or more consecutive
residues or 10 or more consecutive residues of SEQ ID No. 5 or SEQ ID No. 6,
that is, LWVTVYYGVPVW(E/K/R)(E/D)A(E/T/N/D)(T/P)(T/P)LFCASDAK or
GVPVW(E/K/R)(E/D)A(E/T/N/D)(T/P)(TlP)L or a variant thereof.
In yet another embodiment, there is provided an isolated and/or purified
polypeptide consisting of or consisting essentially of SEQ ID No. 1, SEQ ID
No. 2,
SEQ ID No. 3, SEQ ID No. 4, SEQ ID No. 5 or SEQ ID No. 6.
Furthermore, it is of . note that It is well known in the art that some
modifications' and changes can be made in the structure of a polypeptide
without
substantially altering the biological function of that peptide, to obtain a
biologically
equivalent polypeptide. In one aspect of the invention, the above-described
peptides may include peptides that differ by conservative amino acid
substitutions.
The peptides of the present invention also extend , to biologically equivalent
peptides that differ by conservative amino acid substitutions. As used herein,
the
term "conserved amino acid substitutions" refers to the substitution of one
amino
acid for another at a given .location in the peptide, where the substitution
can be
made without substantial loss of the relevant function, in this case, the
folding of
the epitope. In making such changes, substitutions of like amino acid residues
can
be made on~the basis of relative similarity of side-chain substituents, for
example,
their size, charge, hydrophobicity, hydrophilicity,, and the like, and such
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
substitutions may be assayed for their effect on the function of the peptide
by
routine testing. It is of. note. that one of skill in the art would anticipate
that
unconserved .or not highly conserved amino acids are more likely candidates
for
substitution without loss of function.
5 . In some embodiments, conserved amino acid substitutions may be made
where an amino acid residue is substituted for another having a similar
hydrophilicity value (e.g., within a value of plus or minus 2.0), where the
following
may be an amino acid having a hydropathic index of about -1.6 such as Tyr (-
1.3)
or Pro (-1.6)s are assigned to amino acid residues (as detailed in United
States
10 Patent No. 4,554,101, incorporated herein by reference): Arg (+3.0); Lys
(+3.0);
Asp (+3.0); Glu (+3.0); Ser (+0.3); Asn (+0.2); Gln (+0.2); Gly (0); Pro (-
0.5); Thr (-
0.4); Ala (-0.5); His (-0.5); Cys (-1.0); Met (-1.3); Val (-1.5); Leu (-1.8);
Ile (-1.8);
Tyr (-2.3); Phe (-2.5); and Trp (-3.4).
In alternative embodiments, conserved amino acid substitutions may be
made where an amino acid residue is substituted for another having a similar
hydropathic index'(e.g., within a value of plus or minus 2.0). In such
embodiments,
each amino acid residue may be assigned a hydropathic index on the basis of
its
hydrophobicity and charge'characteristics, as follows: Ile (+4.5); Val (+4.2);
Len
(+3.8); Phe (+2.8); Cys (+2.5); Met (+1.9); Ala (+1.8); Gly (-0.4); Thr (-
0.7); Set (
0.8); Trp (-0.9); Tyr (-1.3); Pro (-1.6); His (-3.2); Glu (-3.5); Gln ~(-3.5);
Asp (-3.5);
Am (-3.5); Lys (-3.9); and Arg (-4.5).
In alternative embodiments, conserved amino acid substitutions may be
made where an amino acid residue is substituted for another in the same class,
where the amino acids are divided into non-polar, acidic, 'basic and neutral
classes, as follows: non-polar: Ala, Val, Len, Ile, Phe, Trp, Pro, Met;
acidic: Asp,
Glu; basic: Lys, Arg, His; neutral: Gly, Ser, Thr, Cys, Asn, Gln, Tyr.
As will be apparent to one of skill in the art, it is also possible that the
b12
antibodies are recognizing a conformational site within the above-described
sequence, for example, a conformational epitope formed by non-adjacent, that
is,
non-contiguous residues. As such, in these embodiments, the peptide may
comprise 2 or more contiguous amino acids from a first region of SEQ ID No. 1
separated by a linker of variable sequence to 2 or more contiguous amino acids
from a second region of SEQ ID No. 2. As will be appreciated by one of skill
in the
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
11
art, the linker does not necessarily need to correspond verbatim to the
intervening
residues between the two regions but should be such that the confirmation of
the
residues within the conformational epitope is approximated. That is, in other
embodiments, there is provided a peptide comprising 2 or more amino acids from
the N-terminus region of any one of the peptides according to SEQ ID No. 1, 3
or 5
fused to 2 or more amino acids from the C-terminal region of any one of the
peptides according to SEQ ID No. 1, 3 or 5 separated by a linker of a length
that
corresponds to the number of amino acids separating the N-terminal region
amino
acids from the C-terminal region amino acids in the native sequence.
In another embodiment of the invention, there is provided a method of
immunizing an individual against HIV infection comprising administering to an
individual an effective amount the isolated or purified polypeptide described
above.
As will~be appreciated by one of skill in the art, the effective amount is an
amount
sufficient to induce an immune response within the individual. It is of note
that
when the purified polypeptide is used as a vaccine, the preparation may
include at
least one suitable excipient.
In these embodiments, administration of the polypeptide to an individual, for
example, a human, results in the individual obtaining sterilizing immunity
against
the human immunodeficiency virus, as discussed below. Specifically,
immunization
will result in the production of neutralizing antibodies against the above'-
described
polypeptide under subsequent challenge. That is, on subsequent exposure to
gp120, antibodies will be produced which will bind to gp120 at the site
required for
binding to CD4, thereby preventing viral infection of T-cells and will also
target the
viral particles for removal by antibody dependant cellular ~ cytotoxicity or
complement dependent cytotoxicity. ~ .
In other embodiments, any one of the above-described peptides is
administered to an individual infected with or suspected of being infected
with
Human Immunodeficiency virus. As discussed above, immunization will promote
the production of neutralizing antibodies, which will in turn bind to gp120,
thereby
preventing viral infection of T-cells. Thus, immunization in these embodiments
will
slow disease progression by decreasing the rate of viral infection. As will be
apparent to one of skill in the art, the immunization may be combined with
other
anti-HIV compounds, for' example, azidothymidine (AZT), lamivudine (3TC),
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
12
dideoxyinosine (ddi), dideoxycytidine. (ddc) and ritonavir, as well as other
reverse
transcriptase and protease inhibitors.
In yet other embodiments, an immune globulin effective against Human
Immunodeficiency virus may be prepared by vaccinating a plurality of donors
with
any one of the above-described isolated or purified polypeptides; isolating
plasma
from each of said donors after a period of time sufficient to allow production
of
antibodies against said polypeptide; pooling the plasma; and preparing an
immune
globulin from the pooled. plasma using means known in the art. As will be
appreciated by one of skill in the art, the immune globulin preparation may be
used
as a treatment for individuals having been~recently infected or suspected of
having
been infected with human immunodeficiency virus. That is, antibodies within
the
immune globulin preparation will bind to gp120, preventing binding to CD4 and
targeting the viral particles for removal as discussed above.
In another embodiment of the invention, there is provided a method of
treating an individual infected or suspected of being infected by human
immunodeficiency virus comprising administering to said individual a
therapeutically effective amount, of any one of the above-described purified
polypeptides. In these embodiments, the above-described polypeptide interacts
with CD4, effectively acting as a decoy substrate and preventing or greatly
reducing gp120 binding to CD4 by occupying CD4 binding sites. As will be
apparent to one of skill in the art, this treatment may be combined with other
treatments known in the . art as well as for example the immune globulin
preparation described above.
In another embodiment of the invention, there is provided a method of
a 25 determining a course of treatment for an individual infected with human
immunodeficiency virus comprising screening a sample from an individual
infected
with human immunodeficiency virus for antibodies binding to any one of the
above
described purified polypeptides, wherein presence of antibodies against said
polypeptide indicates that a less aggressive treatment is needed.
Specifically, as
discussed herein, the presence of antibodies against this region of gp120 has
been shown to result in non-progression of the disease. As a consequence,
individuals having natural immunity against this specific region of gp120 may
not
need to be treated aggressively, as discussed below.
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
13
In some embodiments, any one of the above-described polypeptides may
be combined with a suitable carrier peptide .known in the art.
In some embodiments, the purified polypeptide or immune globulin may be
combined with other compounds or compositions known in the art such that the
is
a pharmaceutical composition in the form of, for example, a pill, tablet,
liquid, film
or coating using means known in the art and as discussed below.
It is of note that the purified polypeptide or immune globulin
discussed above may be prepared to be administered in .a variety of ways, for
example, topically, orally, intravenously, intramuscularly, subcutaneously,
intraperitoneally, intranasally or by local or systemic intravascular infusion
using
means known in the art and~as discussed below.
In some embodiments, the above-described pharmaceutical composition
may be combined with a pharmaceutically or pharmacologically acceptable
carrier,
excipient or diluent, either biodegradable or non-biodegradable. Exemplary
examples ~ of carriers include, but are by no means limited to, for example,
polyethylene-vinyl acetate), copolymers of lactic acid and glycolic acid,
poly(lactic
acid), gelatin, collagen matrices, polysaccharides, poly(D,L lactide),
poly(malic
acid), poly(caprolactone), celluloses, albumin, starch, casein, dextran,
polyesters,
ethanol, mathacrylate, polyurethane, polyethylene, vinyl polymers, glycols,
mixtures thereof and the like. Standard excipients include gelatin, casein,
lecithin,
gum acacia, cholesterol, tragacanth, sfearic acid, benzalkonium chloride,
calcium
stearate, glyceryl monostearate, cetostearyl alcohol, cetomacrogol emulsifying
wax, sorbitan esters, polyoxyethylene alkyl ethers, polyoxyethylene castor oil
derivatives, polyoxyethylene sorbitan fatty acid esters, polyethylene glycols,
polyoxyethylene stearates, colloidol silicon dioxide, phosphates, sodium
dodecylsulfate, carboxymethylcellulose calcium, carboxymethylcellulose sodium,
methylcellulose, .. ..hydroxyethylcellulose, hydroxypropylcellulose,
hydroxypropylmethycellulose phthalate, noncrystalline cellulose, magnesium
aluminum silicate, triethanolamine, polyvinyl alcohol, polyvinylpyrrolidone,
sugars
and starches. See, for example, Reminaton: The Science and Practice of
.Pharmacy, 1995, Gennaro ed.
As will be apparent to orie knowledgeable in the art, specific carriers
and carrier combinations known in the art may be selected based on their
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
14
properties and release characteristics in view of the intended use.
Specifically, the
carrier may be pH-sensitive, thermo-sensitive, thermo-gelling, arranged for
sustained release or a quick burst. In some embodiments, carriers of different
classes may be used in combination for multiple effects, for example, a quick
burst
followed by sustained release.
In other embodiments; the above-described pharmaceutical
composition at concentrations or dosages described above may be encapsulated
for delivery. Specifically, the pharmaceutical composition may be encapsulated
in
biodegradable microspheres, microcapsules, microparticles, or nanospheres. The
delivery vehicles may be composed of, for example, hyaluronic acid,
polyethylene
glycol, poly(lactic acid), gelatin, poly(E-caprolactone), or a poly(lactic-
glycolic) acid
polymer. Combinations may also be used, as, for example, gelatin nanospheres
may be coated with a polymer of poly(lactic-glycolic) acid. As will be
apparent to
one knowledgeable in the art, these and other suitable delivery vehicles may
be
prepared according to protocols known in the art and utilized for delivery of
the.
Alternatively, the delivery vehicle, may be suspended in saline and used as a
nanospray for aerosol dispersion.
The above-described pharmaceutical compounds at therapeutically
effective dosages would therefore reduce the spread of an HIV infection by
accomplishing at least one of the following: decreasing viral load, preventing
or
limiting the rate of viral infection and preventing further infection by the
virus.
The kits of the invention comprise one or more containers comprising
a purified polypeptide or immune globulin as described above, a suitable
excipient
as described herein and a set of instructions, generally written instructions
although electronic storage media (e.g., magnetic diskette or optical disk)
containing instructions are also acceptable, relating to the use and dosage of
the
for the intended treatment. The instructions included with the kit generally
include
information as to dosage, dosing schedule, and route of administration for the
intended treatment. The containers of the glandular kallikrein may be unit
doses,
bulk packages (e.g., multi-dose packages) or sub-unit doses.
The invention will now be described by way of examples; however, the
invention is not limited by the examples.
Full length, purified IgG1b12 antibody was contributed by Carlos Barbas, III
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
(The Scripps Research .Institute, La Jolla-, CA, USA). The antibody was linked
to
cyanogen-bromide activated sepharose beads (Sigma-Aldrich, Oakville, Ontario).
Epitope. excision was performed to include potential conformational epitopes.
The
antibody-bead mixture was incubated with either MN or IIIB HIV-1 gp120
5 (ImmunoDiagnostics, Inc. Woburn, MA.) or synthetic HIV-1 gp120 peptides
(United
Biochemical Research, Inc, Seattle, Washington, USA) for 2 hours under
physiological conditions. After several washes, trypsin (Calbiochem-
Novabiochem
Corporation, San Diego; California, USA). or Glu-C (Roche Diagnostics Canada.
Laval, Quebec) enzyme digests were performed. Unbound, digested material was
10 washed away. Antibody-bound fragments protected from digestion were
analyzed
by mass spectrometry. Bead + antibody + peptide complex were spotted on a gold
plate along with DHB matrix (Sigma-Aldrich, Oakville, Ontario) (3,4). The
sample
was analyzed on the prototypic QqTOF mass spectrometer by matrix-assisted
laser desorption/ionization.
15 Theoretical MN and IIIB trypsin and Glu-C digests were performed on
ProMac, which provided molecular masses.for each possible fragment created by
enzyme digestion.
As shown in Figure 1, IIIB (a) or MN (b) gp120 was bound to IgG1b12
antibody and was digested overnight with Glu-C endopeptidase. Protein
fragments not bound by IgG1 b12 were washed away. Antibody-bound fragments
protected from digestion were analyzed by mass spectrometry: The size of the
first peak, 1807 (mass/charge, or m/z), corresponds to the N-terminal sequence
ATTTLFCASDAKAYDTE (as determined by the theoretical digest program). 1867,
the second peak, corresponds to the sequence KLWVTVYYGVPVWK. The third
peak, 2097 corresponds to the sequence TEKLWVTVYYGVPVWKE.
This is the first experiment describing a region other than the CD4-binding
site as the IgG1b12 epitope. The CD4-binding site epitope expected peak size
is
2067.01 m/z with the sequence QFGNNKTIIFKQSSGGDPE. This epitope is
clearly missing, implying that it may not be involved in the specific
interactions
between IgG1b12 and gp120.
As can be seen in Figure 2, IIIB (a) or MN (b) gp120 was bound to IgG1b12
antibody and was digested overnight with ' trypsin endopeptidase. Protein
fragments not bound by IgG1b12 were washed away. Antibody-bound fragments
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
16
protected from digestion were analyzed by mass spectrometry. The size of the
peak 1357 corresponds to the N-terminal sequence EATTTLFCASDAK. The peak
at 1609 corresponds to the N-terminal sequence LWVTVYYGVPVWK.
This indicates that the amino terminus is the source of the IgG1b12 epitope.
The mass spectrometer has unequivocally identified the amino terminus as the
epitope. The trypsin digest mass spectrometry results strengthens the results
seen in the first figure.
Figure 3 shows the amino acid sequence of HIV-1 gp120 IIIB
(Immunodiagnostics, Inc.). Amino acids identified by mass spectrometry are
underlined.
Figure 4 shows the amino acid sequence of HIV-1 gp120 MN
(Immunodiagnostics, Inc.). Amino acids identified by mass spectrometry are
underlined. Bold G (glycine) is the unknown putative amino acid change G-~T
(threonine) recognized by the exquisite sensitivity of mass spectrometry (2097
peak).
It appears that IgG1 b12 specifically interacts with amino acids within a 34
amino acid sequence near the amino terminus of HIV-1 gp120. It is unlikely
that
the antibody recognizes the entire linear sequence, because linear epitopes
are
normally half this length. The digestive cut between the 1867/1807 and
1609/1357
peaks suggests that this- part of the peptide is not bound to the. antibody,
and
therefore exposed to digestion. Interaction probably occurs between IgG1 b12
and
amino acids from both sequences.
Thus, as described above, mass spectrometry epitope mapping has .
identified the gp120 sequence involved in IgG1 b12-binding. E. O. Saphire et
al
' (Science, 2001 ) initially used the computer program AutoDock to predict the
gp120
amino acids involved in gp120-CD4 interactions. The software predicted that
amino acids Ser3ss, Aspasa, Ile3'~, Tyr384, and Val43o. According to our gp120
sequences IIIB and MN, these amino acids correspond to Ser3'o, Asp3'3, Ile3'6,
and
likely Val4ss for the IIIB sequence, and Ser34~, Asp3'~, 11e34', and likely
Val4os in the
MN sequence. Saphire et al then confirmed their results by alanine mutation
studies. These amino acids are near the CD4-binding site,, and therefore it is
assumed that IgG1b12 physically blocks the CD4 binding site. Although both of
these methods of epitope mapping are commonly accepted and practiced, it is
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
17
important to note that conclusions drawn from both these experiments about
epitope sequence are purely based on inference. The computer, program is
entirely theoretical and the mutation studies only provide information on
amino
acids that play a role in binding, though this role may be conformational, and
not
direct contact. Our mass spectrometry epitope mapping experiments gave much
different results than those of Saphire's group. Through mass spectrometry, we
have identified the N-terminal gp120 sequence
LV1IVTVYYGVPVWKEATTTLFCASDAK as the sequence containing the amino
acids involved in IgG1b12 binding. According to the ImmunoDiagnostics IIIB
gp120 sequence, these amino acids fall between Leu34 and Lys59, and the amino
acids Leu6 and Lys3~ in the MN sequence.
Because, as discussed above, previous experiments have suggested that
residues near the CD4 binding site may be important for IgG1 b12-gp120
interactions, further studies are required. It has been proposed that IgG1 b12
may
interact with an isoleucine residue that would otherwise be cleaved off our
trypsin
digested CD4 binding site peptide (...SSGGDPEL..) (7). Epitope mapping
experiments were performed using synthetic peptides. One peptide, the "N-term"
(amino terminus) peptide was tested for it's binding to IgG1b12. The N-term
sequence is as follows: KLWVTVYYGVPVWKEATTTLFCASDAKAYDTE.. A
second peptide covered the CD4 binding site, including the isoleucine (I)
residue.
The sequence QFGNNKTIIFKQSSGGDPEIVTHSFNCGGE was tested for
antibody binding, and these results were measured by mass spectrometry, as
shovyn in Figure 5.
With the peptides as starting material, IgG1b12 still specifically recognizes
the amino terminus and not the CD4 binding site. ~ A 2-hour trypsin digest
confirmed. the identity of our peptide (figure 6). The 1737 peak corresponds
to the
digested fragment amino acid sequence KLVVVTVYYGVPVWK.
Through mass spectrometry epitope mapping, using two different strains of
HIV-1 envelope, two different endopeptidases, and synthetic peptides, it is
clear
that the IgG1b12-gp1.20 binding interface is not as was previously proposed.
Though it is possible that the amino acids previously implicated in IgG1 b12
binding
play an important role in bond formation, it is likely that this role is
merely
conformational. Mass spectrometry identification of epitopes is based strictly
on
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
18
identifying peptide regions that bind to antibody, and gives proof of those
interactions. Most other antibody epitope mapping strategies rely on inference
based on indirect observations. The epitope mapping experiments performed
thoroughly identified the amino terminal sequence
(TEKLWVTVYYGVPVWKEATTTLFCASDAKAYDTE) as the fragment that
contains individual residues involved in IgG1b12-gp120 interactions.
Thus, as discussed above, the region recognized by IgG1 b12, which is the
most potent antibody yet described which is capable of neutralizing HIV-1 in
multiple assays, has been identified. Furthermore, the neutralizing ability of
IgG1b12 is likely involved in protective immune responses to HIV-1 and this
can be
induced in others to generate protective HIV-1 specific responses.
Furthermore,
knowledge of the sequence of this particular epitope could be used in blocking
HIV
infection. Knowledge of when/how responses to this epitope develop may be
useful in tailoring alternate therapeutic interventions.
IgG1 KZ52 (Ebola glycoprotein-specific) antibody (28) was provided by Dr.
Dennis R. Burton (The Scripps Research Institute, La Jolla, CA, USA). To
confirm
the sequence of the peptide peaks identified by epitope excision mass
spectrometry, tandem mass spectrometry (MS/MS) was performed. One ~,g of
soluble gp120 MN (ImmunoDiagnostics, Inc., Woburn, MA.) was digested with
either trypsin or glu-C and subjected to matrix-assisted laser
desorptionlionization
quadrupole time of flight mass spectrometry (MALDI QqTOF).
Antigen capture ELISA was' carried out by first coating 96 well plates
(NUNC, Mississauga, ON) with 2.5~g/ml IgG1 b12 at 4°C overnight. Plates
were
washed with phosphate buffered saline (PBS) 0.05% Tween 20 and blocked with
PBS containing 0.17% bovine serum albumin. Plates were washed and incubated
at 4°C overnight with serially diluted biotinylated peptides (Nterm and
scrambled
peptides) corresponding to the amino terminal region of gp120 N-term (biotin-
LWVTVYYGVPVWKEATTTLFCASDAK)~ and a scrambled peptide control (biotin-
VWCAPLVYWTSTGELAVDKFVTATYK) (United Biochemical Research, Inc.,
Seattle, WA.). Plates were washed and incubated at 37°C for 45
minutes with
streptavidin alkaline phosphatase (Jackson ImmunoResearch Laboratories, Inc.
West Grove, PA.) then washed again and incubated with MgCl2 diethanolamine
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
19
substrate buffer plus alkaline phosphatase yelloviv (pNPP) (Sigma, Oakville,
ON.) at
room temperature. Plates were read on a Spectramax Plus (Molecular Devices,
Sunnyvale, CA) at 405rim. Competition ELISAs were performed similar to the
antigen capture test, but serially diluted soluble gp120 IIIB
(ImmunoDiagnostics,
Inc., Woburn, MA.) was added to the plate for 1 hour at 37°C, non-
bound gp120
was washed away, and then 20~,g/ml peptide was added as above.
Western blot analysis was carried out as follows. One ~,g of gp120 or BSA
was run on a 7.5% SDS-PAGE minigel. The gel was blotted onto a nitrocellulose
membrane by Transblot semi-dry transfer cell (Bio-Rad, Mississauga, ON.),
blocked, and IgG1 b12 was added at 0.25~,g/ml and incubated for 2 hours at
37°C.
The blot was washed with PBS Tween 20 and HRP-sheep anti-human antibody
(The Binding Site, San Diego, CA.) was incubated on the blots. Blots were
washed and detected by ECL Advance western blot detection system (Amersham
Bioscience, Baie d'Urfe, PQ.).
For immunogenicity studies, 4 groups of 4 BALB/c mice (16 total) were
immunized intraperitoneally (i.p.) 5 times over 2 months. The first group of
mice
received PBS plus Freund's adjuvant in each immunization, and the second group
received 10-50~.g gp120. The third and fourth groups received 10~,g
biotinylated
peptide (group 3 received N-term, group 4, scrambled) linked to avidin.
Biotinylated peptides were linked to avidin (Zymed Laboratories, Inc., San
Francisco, CA) by incubating them at a 1:1 molecular ratio for 30 minutes at
37°C
before inoculation. .
ELISAs to detect Ab responses in mice were carried out by coating 96 well
plates with 2.5~g/ml gp120 or 5wg/ml biotinylated peptide overnight at
4°C. Blood
was obtained by venous tail puncture, serum was obtained by collecting the
supernatant of blood that had been incubated at 4°C for one hour and
centrifuged
in an Eppendorf microcentrifuge (Centrifuge 5417C, Brinkmann Instruments, Ltd.
Mississauga, ON) twice for 30 minutes at maximum speed. Serum was diluted at
1/50 down in doubling dilutions and incubated on plates for 2 hours at
37°C,
washed, and HRP-goat anti-mouse secondary antibody (Southern Biotechnology
Associates, Inc., Birmingham, AL) was added. ABTS substrate (Roche
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
Diagnostics Canada, Laval, PQ) was used for detection. Plates were read at
405nm on the Spectramax Plus.
In order to confirm that the mass spectrometry epitope mapping
experiments specifically revealed peptides involved in interactions with the
variable
5 region of IgG1 b12, control experiments were performed: A non-HIV specific
anti
Ebola isotype matched antibody KZ52 was used for such experiments. The KZ52
antibody shares the same constant region and framework region as IgG1 b12, but
differs at the variable region (Fv) responsible for binding antigen. This
first set of
experiments was performed using glu-C endoprotease to digest whole gp120
10 bound to antibodies. Figure 8 (a) shows epitope excision mapping results of
IgG1
b12 and KZ52 revealing the N-terminal specificity of IgG1 b12 (m/z value
1806.8,
1866.0 and .2096.1 ); whereas KZ52 (b) lacks these peaks. The masses of the 3
peaks correspond to the earlier identified linear sequence
TEKLWVTVYYGVPVWKEATTTLFCASDAK located near the amino terminus of
15 gp120.
Figure 9 confirms the results observed in Figure 8. Epitope excision
mapping using the endoprotease trypsin shows specific peaks for IgG1 b12 (a)
at
1357 and 1609 (roughly), whereas the KZ52 antibody (b) revealed no specific
gp120 peptide epitopes. These peaks correspond to the earlier identified
linear
20 sequence KLWVTVYYGVPVWKEATTTLFCASDAK, again demonstrating the
recognition of IgG1 b12 to the amino terminus of gp120.
MALDI QqTOF mass spectrometry epitope mapping identifies peptide
masses that can then be assigned an amino acid sequence based on a theoretical
digest of gp120. In order to confirm the predicted sequences observed upon
trypsin and glu-C digestion, digested soluble gp120 was subjected to MS/MS
sequencing. Soluble gp120, and not antibody-bound gp120 fragments, were used
for confirmation because MALDI QqTOF epitope excision mapping yields peptide
quantities high enough to be detected, but too low to sequence. Through tandem
mass spectrometry, the gp120 peak masses 1357, 1609 (trypsin digest) and 2097
(glu-C digest) sequences were confirmed. This confirmation came by matching
more than 50% of the theoretical peaks generated by MS/MS to the actual ones:
Although many amino acids in the sequences of the peaks 1807' and .1867 were
able to be verified, their numbers of matching peaks were less than 50%. These
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
21
results support the predicted sequence of LWVTVYYGVPVWKEATTTLFCASDAK
for the gp120 amino-terminal epitope of IgG1 b12.
To confirm the linear .nature of the IgG1 b12 epitope, we conducted a
Western blot analysis of the ability of IgG1 b12 to bind denatured gp120.
Analysis
of the blot shows IgG1 b12-gp120 recognition bands at approximately 116kDa
(monomeric gp120) and 230kDa (corresponding to dimeric gp120) in Figure 10,
. ' lane 1. No detectable bands were observed in the second lane, consisting
of
secondary antibody (sheep anti-human IgG) only. No detectable bands were
observed In the third lane, consisting of IgG1 b12 reaction to an irrelevant
antigen
(BSA- 66kDa).
To confirm that IgG1 b12 recognizes the amino terminal sequence of
gp120, ELISA experiments were carried out. A biotinylated peptide matching the
identified sequence was tested for binding by antigen capture ELISA. As a
control,
a biotinylated peptide consisting of the same amino acid sequence as the amino
terminal sequence but in a random order (scrambled peptide) was also tested
for
binding. Results show that the gp120 peptide bound IgG1 b12, and this binding
was dose dependant. The binding of the scrambled peptide was significantly
lower, and did show dose-dependant binding as did the N-term peptide. This
experiment strengthens the evidence that IgG1 b12 recognizes N-term sequence
on gp120.
To substantiate the evidence that IgG1 b12 targets the amino terminus of
gp120, we assessed the ability of whole gp120 to compete with peptide for
binding
to IgG1 b12 by ELISA. We incubated plate-bound IgG1 b12 with soluble gp120
before the addition of biotinylated peptide. As evident in figure 11, the
addition of
soluble gp120 blocks peptide binding, and that this binding is gp120 dose
dependant.
To determine the immunogenicity of the Nterm peptide, we performed
mouse immunizations. Four groups of mouse were given i.p. injections of
adjuvant
alone, gp120, amino-terminal peptide, or scrambled peptide. Immunizations were
administered at days 1, 15, 30, 45 and 60. Testing of the sera collected at
day 67
shows that the serum of mice immunized with the N-terminal peptide contains
antibodies capable of binding both whole gp120 and N-terminal peptide (figure
13,
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
22
A and. B respectively) compared to control mice. The N terminal peptide was
therefore immunogenic in the mouse model.
The mass spectrometry control experiments (Figures 8 and 9) validate the
mass spectrometry results that IgG1 b12 specifically .recognizes gp120 at a
'5 sequence located near the amino terminus of the whole gp120 molecule. Since
.
the control KZ52 antibody. and IgG1 b12 share the same constant region (Fc),
but
differ in their variable regions (Fv), the differences in their recognition of
gp120 by
mass spectrometry are attributable only to specific interactions occurring at
the
antibody Fv. In this set of experiments, the control antibody used, IgG1 KZ52
recognizes specifically Ebola glycoprotein, and not the HIV glycoprotein
gp120.
The negative IgG1 KZ52 mass spectrometry results indicate that the nature of
the
interaction between IgG1 b12 and gp120 are highly specific and localized at
the
antibody variable region. These results ~ solidify and confirm the original
mass
spectrometry results.
MALDI QqTOF epitope mapping yields peptide peaks that large enough to
detect, but too small to sequence. We therefore confirmed the sequence through
MS/MS of soluble gp120, a commonly utilized practice. By MS/MS, both trypsin
peaks corresponding to the sequences LWVTVYYGVPVWK (1357) and
EATTTLFCASDAK (1609) were confirmed, and the glu-C digested peak
TEKLWVTVYYGVPVWKE (2097) was also confirmed. Taken together these data
confirm that the MALDI QqTOF identified peaks are present in soluble, digested
gp120 and correspond to. the epitopes recognized by IgG1 b12.
Previously; it has been suggested that the gp120 epitope for IgG1 b12 is
conformational (7). Contrary to the accepted idea that IgG1 b12 binds a
conformation-dependant epitope, the mass spectrometry mapping uncovered an
amino acid sequence that was, in fact, linear and located at the amino
terminus of
gp120. It has yet to be shown whether IgG1 b12 binds denatured gp120. The
binding of IgG1 b12 to denatured gp120 suggests that the previously described
IgG1 b12-CD4 binding site interactions are incorrect. While the mass
spectrometry results do not prove that gp120 conformation is irrelevant to
IgG1
b12 binding, it is suggested. The Western blot experiment was performed to
back
up the evidence that IgG1 b12 binds a linear portion of gp120. That these
results
were positive corroborates with our mass spectrometry findings. While this
does
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
23
not disprove a conformational component to the interaction of b12 and gp120,
what it suggests in conjunction with the mass spectrometry is that there is
enough
strength in the interactions between b12 and the linear portion at the amino
terminus of gp120 to maintain binding over many stringent washes and even
endoprotease digestion. We argue that the Nterm sequence on gp120 is the true
epitope recognized by IgG1 b12.
To provide further data that supports the IgG1 b12 interaction with the
amino terminus of gp120 we tested that ability of IgG1 b12 to bind a
biotinylated N-
term synthetic peptide via ELISA. The antigenicity of the N terminal peptide
was
tested. As the synthetic peptide was biotinylated, we used an antigen capture
ELISA to detect antibody-peptide interactions. ELISA plates were coated with
IgG1 b12 and biotinylated peptide was added and detected for directly. The
Nterm
peptide bound IgG1 b12 4 times better than the scrambled peptide, in a dose-
dependant manner. The scrambled peptide results did not dilute out as did the
N
terminal peptide signal, indicating non-specific background interactions. We
then
performed an ELISA where peptide binding to IgG1 b12 was first blocked by the
addition of gp120. Addition of gp120 inhibited N-term peptide IgG1 b12
interactions in a dose dependednt manner suggesting that gp120 specifically
interferes with N-term peptide binding. This confirms that the area of the
variable
region on IgG1 b12 that binds gp120 overlaps the area that binds the Nterm
peptide. Antigen capture and blocking tests further strengthen the suggestion
that
IgG1. b12 binds specifically to the amino terminal sequence
LWVTVYYGVPVWKEATTTLFCASDAK.
The goal of the mouse immunizations was to test the immunogenicity of the
peptide in an animal model - that is to determine if the peptide can elicit
antibodies
iri an in vivo situation. Mice are commonly and easily used as a first step
for
testing the capability of an antigen to elicit responses in a live animal
model. Our
mouse data shows that amino terminal peptide was able to elicit antibodies
that
were able to bind both N terminal peptide and whole gp120. Background-level
responses only were seen on the scrambled peptide-coated plates. The N
terminal peptide is immunogenic. The positive mouse data also suggests that
the
Nterm peptide can be exploited as a vaccinogen.
Given that the amino terminus is the binding site of a powerfully neutralizing
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
24
anti-HIV antibody, it follows that immunization with that amino-terminal
sequence
will elicit antibody production, and that these antibodies will be HIV-
neutralizing.
IgG1 b12-like antibodies should block HIV infection of cells in neutralization
assays. A future direction will be to perform neutralization assays with the
mouse
serum generated as above.. Briefly, cells will be incubated with dilutions of
HIV-1
and mouse serum and infection will be measured by p24 and gal production. We
expect that like the monoclonal antibody IgG1 b12, the mouse serum will block
HIV-1 infection in vitro. The next step will be to adapt the N-term peptide
for
human testing and eventually human vaccine phase trials.
The serum generated above will be tested for potency of neutralization of
live HIV-1 in vitro cell culture assays. Basically, serum generated above will
be
incubated with laboratory strains and primary isolates of HIV-1 before the
virus is
used to infect a variety of susceptible cell lines. The ability to block HIV-1
infection
will be determined via HIV-1 p24 protein production and compared to non-
specific
control antibody preparations. We can also test for the ability of antibodies
specific
for the described epitope to inhibit transcytosis (the ability of HIV-1
viruses to pass
. across stratified cell layers) as previously described (27). Basically the
ability of
antibodies to block this process will be compared to non-specific controls
antibodies.
If antibody responses against the described epitope appear to be capable of
generating protective responses in vitro (tested above), the ability of fihe
peptide to
protect against HIV infection will be assessed in one of two in vivo animal
models
currently used in HIV-1~ challenge studies (14, 15). Basically animals will be
vaccinated with the appropriate adjuvant a number of times. They will then be
challenged with live HIV-1 and their susceptibility to infection will be
assessed at
the appropriate time for the animal model involved.
For the HIV-1 neutralization assays, briefly, TZM-b1 cells, a cell line
derivative expressing CD4, GXCR4, and CCRS, and firefly luciferase upon
infection
with HIV were seeded at a density of 3 x 103 cells/96-well plate (J. BioL
Chem.
2005; 280: 4095-4101 ). The next day cells were treated with mouse serum at
different concentrations, and one thousand infectious units/well (as
determined on
TZM-b1 cells) of HIV-1 strains IIIB, SF162, and QH0692 were used to challenge
the cells, and 2 days later the cells were lysed and the activity of firefly
luciferase
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
activity was determined (Steady-Glo luciferase system, Promega). Because of
the
induction of firefly luciferase upon infection, the reduction of the relative
light units
detectable correlates with the inhibition of infection by mouse serum. The
viability
of the cells was not affected by the addition of serum. The serum dilutions
are
5 listed in table 2.
Neutralization results show that the serum of mice immunized with the
Nterm peptide can block HIV-1 infection of cells in vitro. The Nterm-specific
serum
neutralized' HIV-1 strain IIIB 18.6 times better that the scrambled peptide-
specific
serum and 8 times better than mock immunization serum. These results indicate
10 that the N-terminal peptide can elicit HIV-1-specific antibodies that can
block
infection in an in vitro model.
Minimal to moderate neutralization was noted against the other two HIV
strain tested. The slightly high background results seen in the PBS-immunized
groups are not surprising, as mouse serum historically displays high
background
15 levels in this assay (D. Montefiori, personal' communication). These
results are a
good predictor of the serum antibodies' neutralizing capabilities in vivo.
Binding of synthetic peptides to b12 will assessed by ELISA and other EIA
based methods as described and will be done to confirm that this epitope binds
to
the b12 Mab under approachable physiologic conditions in vivo. This
information
20 will be used to design a panel of synthetic peptides that can be used to
assess the
minimal inhibitory peptide epitope, or peptide sequence that will inhibit
binding of
b12 to the described epitope. These minimal epitopes will be assessed for
their
ability to intertere with b12/gp120 binding using ELISA based methods.
The ability of the minimal inhibitory peptide epitope (identified above) to
25 block HIV-1 infection will be determined essentially as previously
described, but
using peptides corresponding to the identified epitope rather than serum, or
MAb's
to inhibit HIV-1 infection of the target cells.
The ability of the minimal inhibitory peptide epitope (identified above) to
block HIV-1 transcytosis will. be determined essentially as previously
described, but
using peptides corresponding to the identified epitope rather than serum, or
MAb's
to inhibit HIV-1 transcytosis across target cells.
One further application of the invention is detecting the presence of Ab
capable of recognizing described epitope as a means of correlating or
analyzing
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
26
HIV disease progression and/or resistance to infection by HIV-1. This would be
the
basis for determining if knowledge of reactivity to the described epitope
(diagnostic
usage) could be useful in altered therapeutic intervention. For example, in a
well-
described cohort of sex workers from Nairobi, Kenya we have identified groups
of
HIV-1 infected individuals who progress to AIDS at different rates. The
presence of
humoral antibody responses to the described epitope may act as a marker for
disease progression (i.e. individuals who have this. reactivity may be more
likely to
progress at a slower rate #o AIDS and death). This may be a useful diagnostic
tool
to aid in the treatment and prescription using HIV-1 anti-retroviral drugs. We
will
assess reactivity to this epitope in 3 groups of HIV infected individuals, HIV
rapid
progressors (those who develop AIDS within 3 years), normal progressors (AIDS
within 5-7 years), and long-term non-progressors (no AIDS for >10 years).
While the preferred embodiments of the invention ~ have been described
above, it will be recognized and understood that various modifications may be
made therein, and the appended claims are intended to cover all such
modifications which may fall within the spirit and scope of the invention.
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
27
References:
1. Papac, D.'I., Hoyes, J., and K. B. Tourer. 1994. Epitope mapping of the
gastrin-
releasing peptide/anti-bombesin monoclonal antibody complex by proteolysis
followed by matrix-assisted laser desorption ionization mass spectrometry.
Protein
Sci. 3(9):1485-92.
2. Parker, C. E.; Papac, D.I., Trojak, 'S. K., and K. B. Tourer. 1996. Epitope
mapping by mass spectrometry Determination of an epitope on HIV-1111B p26
recognized by a monoclonal antibody. J. Immunol. 157:198-206.
3. Parker, C. E., and K. B. Tourer. 2000. Epitope mapping by a combination of
epitope excision and MALDI-MS. Methods in Molecular Biology, Vol. 146: Protein
and Peptide Analysis. New Mass Spectrometric Applications. J.R. Chapman, Ed.
. Humana Press, Totowa, NJ. 185-199.
4. Hochleitner, E. 0., Gorny, M. K., Zolla-Pazner, S., and K. B. Tourer. 2000.
Mass
spectrometric characterization of a discontinuous epitope of the HIV protein
HIV-
gp120 recognized by the human monoclonal antibody 1331A. J. Immunol.
164:4156-61.
5. Collet, T. A., Roben, P., O'Kennedy, R., Barbas III, C. F., Burton D. R.,
and R.
A. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human
immunodeficiency virus from combinatorial libraries of asymptomatic
seropositive
individuals. Proc. Natl. Acad. Sci. USA. 88:10134-7.
6. Barbas III, C. F., Bjorling, E., Chiodi, F., Dunlop, N., Cababa, D., Jones,
T. M.,
Zebedee, S. L., Persson, M. A. A., Nara, P. L., Norrby, E., and D. R. Burton.
1992.
Reombinant human Fab fragments neutralize human type 1 immunodeficiency
virus in vitro. Proc. Natl. Acad. Sci. USA. 89:9339-43.
7. Saphire, E. O., Parren, P. W., Pantophlet, R., Zwick, M. B., Morris, G. M.,
Rudd,
P. M., Dwek, R. A., Stanfield, R. L., Burton, D. R., and I. A. Wilson. 2001
Crystal
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
28
structure of a neutralizing human IGG against HIV-1: a template for vaccine
design. Science. 10;293(5532):1155-9.
8. Kwong, P.' D., Wyatt, R., Robinson, J., Sweet, R. W., Sodroski, J., and W.
A.
Hendrickson. 1998 Structure of an HIV gp120~envelope~glycoprotein in complex
with the CD4 receptor and a neutralizing human antibody. Nature.
18;393(6686):648-59.
9. Frankel, S: S., Steinman, R. M., Michael, N. L., Ratto Kim, S., Bhardwaj,
N.,
Pope, M., Louder, M. K., Ehrenberg, P. K., Parren, P. W. H. I., Burton, D. R.,
Katinger, H., VANCott, Robb, M. L., ~Birx, D. L., and J. R. Mascola. 1998.
Neutralizing monoclonal antibodies block human immunodeficiency virus type, I
infection of dendritic cells and transmission to T cells. J. Virol.
72(12):9788-94.
10. Ugolini, S., Mondor, I., Parren, P. W. H. I., Burton, D. R., Katinger, H.,
Tilley,
S. A., Klasse, P. J., and Q. J. Sattenau. 1997. Inhibition of virus attachment
to
CD4+ target cells is a major mechanism of T cell line-adapted HIV-1
neutralization.
J. Exp. Med. 186(8):1287-98.
11. Gaudin, M. C., Allaway, G. P. Maddon, P. L., Barbas III, C. F., Burton, D.
R.,
and R. A. Koup. 1996. Effective ex vivo neutralization of human
immunodeficiency
virus type 1 in plasma by recombinant immunoglobulin molecules. J. Virol.
70(4):2586-92.
12. Gaudin, M. C., Weir, R.~ Barbas III, C. F., Burton, D. R., and R. A. Koup.
1997.
Passive immunization with a human monoclonal antibody protects hu-PBL-SCID
mice against challenge by primary isolates of HIV-1.
13. Lewis, A. D., Chen, R., Montefiori, D. C., Johnson, P. R., and K. R.
Clark.
2002. Generation of neutralizing activity against human immunodeficiency virus
type 1 in serum by antibody gene transfer. J. Virol. 76(17):8769-75.
14. Parren, P. W. H. I., Ditzel, H. J., Gulizia, R. J., Binley, J. M., Barbas
III, C. F.,
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
29
Burton, D. R., and D. E. Mosier. 1995. Protection against infection in hu-PBL-
SCID
mice by passive immunization with a neutralizing human monoclonal antibody
against the gp120 CD4-binding site. AIDS 9:F1-F6.
15. Parren, P. W. H. L, Marx, P. A., Hessell, A. J., Luckay, A., Harouse, J.,
Cheng-
Mayer, C., Moore, J. P., and D. R. Burton. 2001. Antibody protects macaques
against vaginal challenge with a pathogenic R5 simian/human immunodeficiency
virus at serum levels giving complete neutralization in vitro. J. Virol.
75(17):8340-7.
16. Crawford, J. M., Earl, P. L., Moss, B., Reimamm, K. A., Wyand, M. S.,
Manson, K. H., Bilska, M., ~hou, Z. T., Pauza, C. D., Parren, P. W. H. I.,
Burton, D.
R., Sodrowski, J. G., Letvinj N. L., and D. C. Montefiori. 1999.
Characterization of
primary isolate-like variants of simian-human immunodeficiency virus. J.
Virol.
73(12):10199-207.
17. Kessler II, J. A., McKenna, P. M., Emini, E. A., Chan, C. P., Patel, M.
D.,
Gupta, S. K., Mark III, G. E., Barbas III, C. F., Burton, D. R., and A. J.
Conley.
1997. Recombinant human monoclonal antibody IgG1b12 neutralizes diverse
human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retro.
13(7):575-82.
18. Moore, J. P., Cao, Y., Quing, L., Sattentau, Q. J., Pyati, J., Koduri, R.,
Robinson, J., Barbas III, C. F., Burton, D. R., and D. H. Ho. 1995. Primary
isolates
of human immunodeficiency virus type 1 are relatively resistant to
neutralization is
not predicted by studies with monomeric gp120. J. Virol. 69(1):101-9:
19. Parren, P. W. H. I., Fisicaro, P., Labrijn, A. F., Binley, J. M., Yang, W-
P..
Ditzel, H. J., Barbas III, C. F., and D. R. Burton. 1996. In vitro antigen
challenge of
human antibody libraries for vaccine evaluation: the human immunodeficiency
virus type 1 envelope. J. Virol. 73(13):9046-50.
20. Trkola, A., Pomales, A. B., Yuan, H., Korber, B., Maddon, P. J., Allaway,
G.
P., Katinger, H., Barbas III, C. F., Burton, D. R., Ho, D. D., and J. P.
Moore. 1995 .
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
Cross-c;ade neutralization of primary isolates of human immundeficiency virus
type 1 by human monoclonal antibodies and tetrameric .CD4-IgG. J. Virol.
69(11):6609-17.
5 21. Verrier, F., Nadas, A., Gorny, M. K., and 'S. Zolla-Pazner. 2001.
Additive
effects characterize the interaction of antibodies involved in neutralization
of the
dualtropic human immunodeficiency virus type 1 isolate 89.6. J. Virol.
75(19):9177-
86.
10 22. Xiang, S-H., Kwong. P. D., Gupta, R., Rizzuto, C.D., Casper, D. J.,
Wyatt, R.,
Wang, L., Hendrickson, W. A., Doyle, M. L., and J. Sodrowski. 2002 Mutagenic
stabilization and/or disruption of a CD4-bound state reveals distinct
conformations
of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J.
Virol.
76(19):9888-99.
23. Xu W., Smith-Franklin, B. A., Li, P. L., Wood, C., He, J., Du, Q., Bhat,
'G. J.,
Kankasa C., Katinger, H., .Cavacini, L. A., Posner, M. R., Burton D. R., Chou,
T. C.,
and R. M. Ruprect. 2001. Potent neutralization of primary ' human
immunodeficiency virus Glade C isolates with a synergistic combination of
human'
monoclonal antibodies raised against Glade B. J. Hum. V,irol. 4(2):55-61.
24. Zwick, M. B., Wang, M., Poignard, P., Stiegler, G., Katinger, H., Burton,
D. R.,
and P. H. H. I. Parren. 2001. Neutralization synergy of human immunodeficiency
. virus type 1 primary isolates by cocktails of broadly neutralizing
antibodies. J. Virol.
75(24):12198-208.
25. Burton, D. R. 2002. Antibodies, viruses and vaccines. Nat. Rev. Immunol.
2:706-13.
26. Fowke, K. R., Nagelkerke, N. J., Kimani, J., Simonsen, J. N., Anzala, A.
O.,
Bwayo, J. J., MacDonald, K. S., Ngugi, E. N., and F. A. Plummer. 1996.
Resistance to HIV-1 infection among persistently seronegative prostifiutes in
Nairobi, Kenya. Lancet 16;348(9038):1347-51.
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
31
27. Devito, C., Hinkula, J., Kaul, R., Lopalco, L., Bwayo, J. J., Plummer, F.,
Clerici,
M., and K. Broliden. 2000. .Mucosal and plasma IgA from HIV-exposed
seronegative individuals neutralize a primary HIV-1 isolate. AIDS 14(13):1917-
20
28. Toshiaki Maruyama, Luis L. Rodriguez, Peter B. Jahrling, Anthony Sanchez,
Ali S. Khan, Stuart T. Nichol, C. J. Peters, Paul W. H. I. Parren, and Dennis
R.
Burton. J. Virol. 1999. Ebola virus can be effectively neutralized by antibody
produced in natural human infection. 73: 6024-6030.
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
32
Table 1. Tandem mass spectrometry sequencing confirms 3 of the 5 sequences
derived from epitope mapping. Peaks 1357, 1609, 1807, 1867 and 2097 were
selected from digested gp120 complete spectra for MS/MS on the MALDI QqTOF.
The a, b. and y ionic fragmentation peptide masses were measured and percent
match .was calculated. Tabulated are the y ion fragment matches, which are
representative of the three forms of fragmentation.
Trypsinsequence y ion matched sequences% sequence
match match
1357 EATTTLFCASDAKE,EAT,EATT,EATTTL,EATTTLF,EATTTLFC,54 yes
EATTTLFCA,EATTTLFCAS,EATTTLFCASDA,
EATTTLFCASDAK
1609 LWVTVYYGVPVWKL,LWV,LWVT,LWVTV,LWVTVYLWVTVY,67 yes
LWVTVYY,LWVTVYYG,LWVTVYYGV,
LWVTVYYGVP,LWVTVYYGVPV, .
LWVTVYYGVPVW,LWVTVYYGVPVWK
glu-c sequence y ion matched sequences% sequence
match match
'1$07 ATTTLFCASDAKAYDTEA,ATTTL,ATTTLFCA,ATTTLFCASDAK,25 n0
ATTTLFCASDAKAYD
1867 KLWVTVYYGVPVWKEK,KL,KLWVTVYYG,KLWVTVYYGVP,3$ n0
KLWVTVYYGVPVW
2097 TEKLWVTVYYGVPVWKET,TEK,TEKLWV,TEKLWVT,TEKLVTV,66 yes
TEKLWVTVY,TEKLWVTVYY,TEKLWVTVYYG,
TEKLWVTVYYGV,TEKLWVTVYYGVP,
TEKLWVTVYYGVPV,TEKLWVTVYYGVPVW,
TEKLWVTVYYGVPVWK
CA 02562385 2006-10-06
WO 2005/097822 PCT/CA2005/000547
33
Table 2. Neutralization assay results for mice immunized with the N terminal
epitope. Serum was isolated and from 4 groups of 4 mice immunized with PBS
(P), gp120(G), N terminal peptide (N), or Scrambled peptide (S), then pooled
by
group. Neutralizing ability was tested for both serum collected from a pre-
y immunization bleed (P0, G0, NO, SO), and post immunization (P1, G1, N1, S1).
Neutralization ability was tested against 3 strains of HIV-1 (IIIB, SF162.LS
and
QH0692.4) in TZM-b1 cells.
'Values are the serum dilution at which relative luminescence units (RLUs)
uvere
reduced 50% compared to virus control wells (no test sample).
r '' ~~ i -~s1?f ,{.~?~ d . :~; ~~ ~ s p ~~'
~~,< ga b 1e .k"n n, ' 8.~ ~ ''..~~
~~ ~,_ ~~IIIB SF~162 . ,~ .,.f
_~ ~.. ~ ~ ~ sG~H069
~ , .r~
34 <20 61
P1 ~ 228 <20 42
, ~.
.
GO... .3 3
G1 108 26 49
~ '._
_.aNO _ w~ 2p <20
49
N 1 1, 826 39 88
A ~
_
. <20 X20
..45
S1 ~ 98 ~ <20 I 21
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPRI~:ND PLUS D'UN TOME.
CECI EST L,E TOME 1 DE 2
NOTE: Pour les tomes additionels, veillez contacter 1e Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME 1 OF 2
NOTE: For additional valumes please contact the Canadian Patent Office.