Note: Descriptions are shown in the official language in which they were submitted.
CA 02578463 2007-03-02
R ~
TITLE
S-ADENOSYL-L-METHIONINE SYNTHETASE PROMOTER
AND ITS USE IN EXPRESSION OF TRANSGENIC GENES IN PLANTS
FIELD OF THE INVENTION
This invention relates to a plant promoter, in particular, to an S-adenosyl-L-
methionine synthetase (SAMS) promoter and subfragments thereof and their use
in regulating
the expression of at least one heterologous nucleic acid fragment in plants.
BACKGROUND OF THE INVENTION
Recent advances in plant genetic engineering have opened new doors to engineer
plants having improved characteristics or traits, such as, resistance to plant
diseases, insect
resistance, herbicidal resistance, enhanced stability or shelf-life of the
ultimate consumer
product obtained from the plants and improvement of the nutritional quality of
the edible
portions of the plant. Thus, a desired gene (or genes) from a source different
than the plant,
but engineered to impart different or improved characteristics or qualities,
can be
incorporated into the plant's genome. This new gene (or genes) can then be
expressed in the
plant cell to exhibit the new trait or characteristic.
In order to obtain expression of the newly inserted gene in the plant cell,
the proper
regulatory signals must be present and be in the proper location with respect
to the gene.
These regulatory signals include a promoter region, a 5' non-translated leader
sequence and a
3' transcription termination/polyadenylation sequence.
A promoter is a DNA sequence that directs cellular machinery of a plant to
produce
RNA from the contiguous coding sequence downstream (3') of the promoter. The
promoter
region influences the rate, developmental stage, and cell type in which the
RNA transcript of
the gene is made. The RNA transcript is processed to produce messenger RNA
(mRNA)
which serves as a template for translation of the RNA sequence into the amino
acid sequence
of the encoded polypeptide. The 5' non-translated leader sequence is a region
of the mRNA
upstream of the protein coding region that may play a role in initiation and
translation of the
mRNA. The 3' transcription termination/polyadenylation signal is a non-
translated region
downstream of the protein coding region that functions in the plant cells to
cause termination
of the RNA transcript and the addition of polyadenylate nucleotides to the 3'
end of the RNA.
It has been shown that certain promoters are able to direct RNA synthesis at a
higher
rate than others. These are called "strong promoters". Certain other promoters
have been
shown to direct RNA production at higher levels only in particular types of
cells or tissues
and are often referred to as "tissue specific promoters". In this group, many
seed storage
protein genes' promoters have been well characterized and widely used, such as
the phaseolin
gene promoter of Phaseolus vulgaris, the helianthinin gene of sunflower, the
(3-conglycinin
gene of soybean (Chen et al., (1989) Dev. Genet. 10, 112-122), the napin gene
promoter of
Brassica napus (Ellerstrom et al, (1996) Plant Mol. Biol. 32, 1019-1027), the
oleosin gene
1
CA 02578463 2007-03-02
promoters of Brassica and Arabidopsis (Keddie et al, (1994) Plant Mol. Biol.
24, 327-340;
Li, (1997) Texas A&M Ph.D. dissertation, pp. 107-128; Plant et al, (1994)
Plant Mol. Biol.
25, 193-205). Another class of tissue specific promoters is described in, U.S.
Patent
No. 5,589,583, issued to Klee et al. on December 31, 1996; these plant
promoters are capable
of conferring high levels of transcription of chimeric genes in meristematic
tissues and/or
rapidly dividing cells. In contrast to tissue-specific promoters, "inducible
promoters" direct
RNA production in response to certain environmental factors, such as heat
shock, light,
hormones, ion concentrations etc. (Espartero et al, (1994) Plant Mol. Biol.
25, 217-227;
Gomez-Gomez and Carrasco, (1998) Plant Physiol. 117, 397-405; Holtorf et al,
(1995) Plant
Mol. Biol. 29, 637-646; MacDowell et al, (1996) Plant Physiol.111, 699-711;
Mathur et al,
(1992) Biochem. Biophys. Acta 1137, 338-348; Mett et al, (1996) Transgenlc
Res. 5,
105-113; Schoffl et al, (1989) Mol. Gen. Genet. 217, 246-253; Ulmasov et al,
(1995) Plant
Physiol. 108, 919-927).
Promoters that are capable of directing RNA production in many or all tissues
of a
plant are called "constitutive promoters". The ideal constitutive promoter
should be able to
drive gene expression in all cells of the organism throughout its development.
Expression of
many so-called constitutive genes, such as actin (McDowell et al., (1996)
Plant Physiol. 111,
699-711; Wang et al., (1992) Mol. Cell Bfol. 12, 3399-3406), and ubiquitin
(Callis et al,
(1990) J. Biol. Chem. 265, 12486-12493; Rollfinke et al, (1998) Gene 211, 267-
276) varies
depending on the tissue types and developmental stages of the plant. The most
widely used
constitutive promoter, the cauliflower mosaic virus 35S promoter, also shows
variations in
activity in different plants and in different tissues of the same plant
(Atanassova et al., (1998)
Plant Mol. Biol. 37,275-285; Battraw and Hall, (1990) Plant Mol. Biol. 15, 527-
538; Holtorf
et al., (1995) Plant Mol. Biol. 29, 637-646; Jefferson et al., (1987) EMBO J.
6, 3901-3907;
Wilmink et al., (1995) Plant Mol. Biol. 28, 949-955). The cauliflower mosaic
virus 35S
promoter is also described in U.S. Patent No. 5,106,739. The tissue-specific
expression and
synergistic interactions of sub-domains of the promoter of cauliflower mosaic
virus are
discussed in U.S. Patent No. 5,097,025, which issued to Benfey et al. on March
17, 1992. A
Brassica promoter (hsp80) that provides for constitutive expression of
heterologous genes in
a wide range of tissues and organs is discussed in U.S. Patent No. 5,612,472
which issued to
Wilson et al. on March 18, 1997.
Since the patterns of expression of a chimeric gene (or genes) introduced into
a plant
are controlled using promoters, there is an ongoing interest in the isolation
and identification
of novel promoters which are capable of controlling expression of a chimeric
gene or (genes).
SUMMARY OF THE INVENTION
This invention concerns an isolated nucleic acid fragment comprising a
promoter
wherein said promoter consists essentially of the nucleotide sequence set
forth in SEQ ID
NOs:6, 14, 15, or 16 or said promoter consists essentially of a fragment or
subfragment that is
2
CA 02578463 2007-03-02
substantially similar and functionally equivalent to the nucleotide sequence
set forth in SEQ
ID NOs:6, 14, 15, or 16.
In a second embodiment, this invention concerns a chimeric gene comprising at
least
one heterologous nucleic acid fragment operably linked to the promoter of the
invention. *
In a third embodiment, this invention concerns plants containing this chimeric
gene
and seeds obtained from such plants.
In a fourth embodiment, this invention concerns a method of increasing or
decreasing
the expression of at least one heterologous nucleic acid fragment in a plant
cell which
comprises:
(a) transforming a plant cell with the chimeric gene described above;
(b) growing fertile mature plants from the transformed plant cell of step (a);
(c) selecting plants containing the transformed plant cell wherein the
expression
of the heterologous nucleic acid fragment is increased or decreased.
In a fifth embodiment, this invention concerns an.isolated nucleic acid
fragment
comprising a constitutive plant SAMS promoter.
BRIEF DESCRIPTION OF THE DRAWINGS AND SEOUENCES
The invention can be more fully understood from the following detailed
description,
the drawings and the Sequence Descriptions that form a part of this
application. The
Sequence Descriptions contain the three letter codes for amino acids as
defined in 37 C.F.R.
1.821-1.825, which are incorporated herein by reference.
SEQ ID NO:1 is the nucleotide sequence comprising the entire cDNA insert in
clone
s2.12b06 which encodes a soybean S-adenosyl-L-methionine synthetase protein.
SEQ ID NO:2 is the nucleotide sequence comprising a soybean S-adenosyl-L-
methionine synthetase genomic DNA fragment.
SEQ ID NO:3 is the nucleotide sequence of a portion of the cDNA insert in
clone
srrlc.pk002.b21 encoding a portion of a soybean S-adenosyl-L-methionine
synthetase
protein.
SEQ ID NO:4 is a 32 base oligonucleotide primer, designated sam-5, used to
amplify
the soybean S-adenosyl-L-methionine synthetase promoter region via PCR.
SEQ ID NO:5 is a 24 base oligonucleotide primer, designated sam-6, used to
amplify
the soybean S-adenosyl-L-methionine synthetase promoter region via PCR.
SEQ ID NO:6 is the nucleotide sequence comprising a soybean S-adenosyl-L-
methionine synthetase promoter fragment produced via PCR using primers sam-5
(SEQ ID
NO:4) and sam-6 (SEQ ID NO:5).
SEQ ID NO:7 is a 22 base oligonucleotide primer, designated sam-9, used to
amplify
the soybean S-adenosyl-L-methionine synthetase promoter region via PCR.
3
CA 02578463 2007-03-02
SEQ ID NO:8 is a 19 base oligonucleotide primer, designated atps-9, used to
amplify
a chimeric gene comprising a SAMS promoter fragment and a portion of the ATP
sulfurylase
(ATPS) gene via PCR.
SEQ ID NO:9 is a 21 base oligonucleotide primer, designated cgs-8, used to
amplify a
chimeric gene comprising a SAMS promoter and a portion of the cystathionine-y-
synthase I
(CGS1) gene via PCR.
SEQ ID NO: 10 is a 20 base oligonucleotide antisense primer, designated atps-
4, used
to amplify the ATP sulfurylase transcript via RT-PCR.
SEQ ID NO:11 is a 21 base oligonucleotide antisense primer, designated cgs-10,
used
to amplify the cystathionine-y-synthase 1 transcript via RT-PCR.
SEQ ID NO: 12 is a 20 base oligonucleotide primer, designated atps-3, used to
amplify
an ATP sulfurylase cDNA via PCR.
SEQ ID NO:13 is a 23 base oligonucleotide primer, designated cgs-9, used to
amplify
a cystathionine-y-synthase 1 cDNA via PCR.
SEQ ID NO: 14 is a 2165 nucleotide sequence comprising a soybean S-adenosyl-L-
methionine synthetase genomic DNA fragment which starts at the 5' end of SEQ
ID NO:2,
and ends at the ATO translation start codon of the S-adenosyl-L-methionine
synthetase.
SEQ ID NO: 15 is a 1574 nucleotide sequence comprising a DNA fragment which
starts at the 5' end of SEQ ID NO:2, and ends at the ATG translation start
codon of the
S-adenosyl-L-methionine synthetase, and wherein a 591 nucleotide intron
sequence has been
removed.
SEQ ID NO: 16 is a 719 nucleotide sequence comprising a DNA fragment which
starts at nucleotide 4 of SEQ ID NO:6, and ends at the ATG translation start
codon of the
S-adenosyl-L-methionine synthetase, and wherein a 591 nucleotide intron
sequence has been
removed.
SEQ ID NO: 17 is a 6975 nucleotide sequence comprising plasmid pMH40A.
SEQ ID NO: 18 is a 3985 nucleotide sequence comprising a SAMS
promoter::GUS::3'
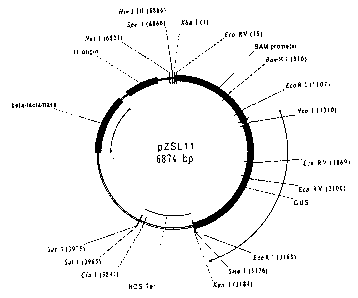
Nos DNA fragment present in plasmid pZSL 11.
SEQ ID NO:19 is a 3684 nucleotide sequence comprising a SAMS
promoter::ATPS::3' Nos DNA fragment.
SEQ ID NO:20 is a 3963 nucleotide sequence comprising a SAMS
promoter::CGS1::3' Nos DNA fragment.
Figures 1 A and I B depict Southern hybridization analyses of SAMS genes.
Soybean
genomic DNA was digested with BamHI, EcoRI, HindIII, Kpnl, and Saci , and then
the blot
was hybridized with a full length SAMS cDNA (SEQ ID NO:1) probe in Figure lA
or with a
SAMS promoter fragment (SEQ ID NO:6) probe inFigure I B.
Figure 2 depicts a SAMS genomic DNA sequence (SEQ ID NO:2) and the alignment
of the overlapping region with SAMS cDNA sequence (SEQ ID NO: 1). The 2336 bp
SAMS
4
CA 02578463 2007-03-02
genomic DNA sequence has a 191 bp region aligned with the 5' end sequence of
the SAMS
cDNA with six mismatches. The region used to make the SAMS promoter by adding
the
Ncol site at its 3' end is underlined. The translation start codon is in bold.
Figure 3 depicts the structure of the SAMS::GUS expression cassette. The SAMS
promoter was cloned into pMH40A to replace its 35S promoter. The structure of
the resulted
SAMS::GUS construct was generated by Vector NTITM software (InforMax, Inc.,
North
Bethesda, MD).
Figure 4 depicts a histochemical GUS expression analysis of transgenic
Arabidopsis
plants harboring the SAMS::GUS expression cassette. Arabidopsis tissues were
incubated at
37 C with X-Gluc overnight and dehydrated with ethanol. (A) Flower buds; (B)
leaf;
(C) Inflorescence stem and a cauline leaf; (D, E, F) developing siliques; (G)
Developing
seeds and embryos. All of the seeds were derived from GUS-positive siliques.
Genetic
segregation of the GUS gene was demonstrated by the blue funiculus of the
white seed in the
right upper corner.
Figure 5 depicts a fluorometric GUS expression assay of transgenic Arabidopsis
plants harboring the SAMS::GUS expression cassette. Triple samples of flowers,
leaves,
stems, siliques coats, young seeds, medium seeds, old seeds, and dry seeds
collected from
SAMS::GUS transgenic Arabidopsis plants were assayed for GUS activity. The
graph was
generated by Microsoft Excel and the standard deviation is indicated by the
upper part of
each column.
Figure 6 depicts a histochemical GUS transient expression analysis of SAMS
promoter in corn. The pZSL11 (SAMS::GUS) or the pMH40A (35S::GUS) plasmid DNA
was delivered into corn callus (A, C) or leaf discs (B, D), and the GUS
activity was detected
by incubation with X-Gluc overnight at 37 C. (A, B) Transformed with pZSLI I
DNA; (C,
D) Transformed with pMH40A DNA.
Figures 7(A) and 7(B) depict the presence and expression of transgenic soybean
ATPS and CGS 1 genes controlled by the SAMS promoter in transgenic Arabidopsis
plants.
Figure 7(A) is a PCR analysis. Genomic DNA of ten transgenic Arabidopsis
plants (1 to 10),
wild type Arabidopsis (a), wild type soybean (s), and plasmid DNA of SAMS::CGS
1 or
SAMS::ATPS in binary vectors (p) were used as templates in PCR with gene-
specific
primers. PCR of ten SAMS::CGS I transgenic plants with primer sam-9 which is
specific to
SAMS promoter, and primer cgs-8 which is specific to soybean CGS 1 (upper).
PCR of ten
SAMS::ATPS transgenic plants with primer sam-9 which is specific to SAMS
promoter, and
primer atps-1 which is specific to soybean ATPS gene (lower). Figure 7(B) is
an RT-PCR
analysis. Total leaf RNA of ten transgenic Arabidopsis plants (1 to 10), wild
type
Arabidopsis (a), and wild type soybean (s) were used as templates in RT-PCR
with gene-
specific primers. First strand cDNA was synthesized from a gene-specific
antisense primer
with reverse transcriptase, and then the first strand cDNA was amplified by
PCR with both
5
CA 02578463 2007-03-02
sense and antisense primers. RT-PCR of ten SAMS::CGS I transgenic plants with
primers,
cgs-9 (sense) and cgs-I0 (antisense), specific to soybean CGS 1 gene (upper).
RT-PCR of ten
SAMS::ATPS transgenic plants with primers, atps-3 (sense) and atps-4
(antisense), specific
to soybean ATPS gene (lower).
Figure 8 depicts induction of SAMS promoter activity by methionine. Seeds of
ten
transgenic Arabidopsis lines transformed with SAMS::GUS construct were
germinated on
filter papers soaked with H20, 1 x Murashige and Skoog salt, 0.01 mM, and 1 mM
methionine. Ten days old seedlings were harvested and assayed for GUS
activity. The solid
bar and hollow bar indicate, respectively, the average and the standard
variation of three
samples for each treatment.
Figure 9 depicts a northern hybridization. Soybean total RNAs from leaves,
roots,
stems, young seeds, medium seeds, old seeds, and pod coats (L, R, S, Y, M, 0,
and P) were
used to make the RNA blot which was hybridized with a full length SAMS cDNA
(SEQ ID
NO: 1) probe.
DETAILED DESCRIPTION OF THE INVENTION
In the context of this disclosure, a number of terms shall be utilized.
As used herein, an "isolated nucleic acid fragment" is a polymer of
ribonucleotides
(RNA) or deoxyribonucleotides (DNA) that is single- or double-stranded,
optionally
containing synthetic, non-natural or altered nucleotide bases. An isolated
nucleic acid
fragment in the form of DNA may be comprised of one or more segments of cDNA,
genomic
DNA or synthetic DNA.
The terms "subfragment that is functionally equivalent" and "fimctionally
equivalent
subfragment" are used interchangeably herein. These terms refer to a portion
or
subsequence of an isolated nucleic acid fragment in which the ability to alter
gene
expression or produce a certain phenotype is retained whether or not the
fragment or
subfragment encodes an active enzyme. For example, the fragment or subfragment
can be
used in the design of chimeric genes to produce the desired phenotype in a
transformed
plant. Chimeric genes can be designed for use in co-suppression or antisense
by linking a
nucleic acid fragment or subfragment thereof, whether or not it encodes an
active enzyme, in
the appropropriate orientation relative to a plant promoter sequence.
The terms "substantially similar" and "corresponding substantially" as used
herein
refer to nucleic acid fragments wherein changes in one or more nucleotide
bases does not
affect the ability of the nucleic acid fragment to mediate gene expression or
produce a
certain phenotype. These terms also refer to modifications of the nucleic acid
fragments of
the instant invention such as deletion or insertion of one or more nucleotides
that do not
substantially alter the functional properties of the resulting nucleic acid
fragment relative to
the initial, unmodified fragment. It is therefore understood, as those skilled
in the art will
appreciate, that the invention encompasses more than the specific exemplary
sequences.
6
CA 02578463 2007-03-02
Moreover, the skilled artisan recognizes that substantially similar nucleic
acid
sequences encompassed by this invention are also defined by their ability to
hybridize, under
moderately stringent conditions (for example, 0.5 X SSC, 0.1% SDS, 60 C) with
the
sequences exemplified herein, or to any portion of the nucleotide sequences
reported herein
and which are functionally equivalent to the promoter of the invention.
Preferred
substantially similar nucleic acid sequences encompassed by this invention are
those
sequences that are 80% identical to the nucleic acid fragments reported herein
or which are
80% identical to any portion of the nucleotide sequences reported herein. More
preferred
are nucleic acid fragments which are 90% identical to the nucleic acid
sequences reported
herein, or which are 90% identical to any portion of the nucleotide sequences
reported
herein. Most preferred are nucleic acid fragments which are 95% identical to
the nucleic
acid sequences reported herein, or which are 95% identical to any portion of
the nucleotide
sequences reported herein. Sequence alignments and percent similarity
calculations may be
determined using the Megalign program of the LASARGENE bioinformatics
computing
suite (DNASTAR Inc., Madison, WI). Multiple alignment of the sequences are
performed
using the Clustal method of alignment (Higgins and Sharp (1989) CABIOS. 5:151-
153) with
the default parameters (GAP PENALTY=10, GAP LENGTH PENALTY=10). Default
parameters for pairwise alignments and calculation of percent identiy of
protein sequences
using the Clustal method are KTUPLE=1, GAP PENALTY=3, WINDOW=5 and
DIAGONALS SAVED=5. For nucleic acids these parameters are GAP PENALTY=10,
GAP LENGTH PENALTY=10, KTUPLE=2, GAP PENALTY=5, WINDOW=4 and
DIAGONALS SAVED=4. A "substantial portion" of an amino acid or nucleotide
sequence
comprises enough of the amino acid sequence of a polypeptide or the nucleotide
sequence of
a gene to afford putative identification of that polypeptide or gene, either
by manual
evaluation of the sequence by one skilled in the art, or by computer-automated
sequence
comparison and identification using algorithms such as BLAST (Altschul, S. F.,
et al.,
(1993) J. Mol. Biol. 215:403-410) and Gapped Blast (Altschul, S. F. et al.,
(1997) Nucleic
Acids Res. 25:3389-3402); see also www.ncbi.nim.nih.aov/BLAST/).
"Gene" refers to a nucleic acid fragment that expresses a specific protein,
including
regulatory sequences preceding (5' non-coding sequences) and following (3' non-
coding
sequences) the coding sequence. "Native gene" refers to a gene as found in
nature with its
own regulatory sequences. "Chimeric gene" refers to any gene that is not a
native gene,
comprising regulatory and coding sequences that are not found together in
nature.
Accordingly, a chimeric gene may comprise regulatory sequences and coding
sequences that
are derived from different sources, or regulatory sequences and coding
sequences derived
from the same source, but arranged in a manner different than that found in
nature.
"Endogenous gene" refers to a native gene in its natural location in the
genome of an
organism. A "foreign" gene refers to a gene not normally found in the host
organism, but that
7
CA 02578463 2007-03-02
is introduced into the host organism by gene transfer. Foreign genes can
comprise native
genes inserted into a non-native organism, or chimeric genes. A' transgene" is
a gene that
has been introduced into the genome by a transformation procedure.
A "heterologous nucleic acid fragment" refers to a nucleic acid fragment
comprising a
nucleic acid sequence that is different from the nucleic acid sequence
comprising the plant
promoter of the invention.
"Coding sequence" refers to a DNA sequence that codes for a specific amino
acid
sequence. "Regulatory sequences" refer to nucleotide sequences located
upstream (5' non- .
coding sequences), within, or downstream (3' non-coding sequences) of a coding
sequence,
and which influence the transcription, RNA processing or stability, or
translation of the
associated coding sequence. Regulatory sequences may include, but are not
limited to,
promoters, translation leader sequences, introns, and polyadenylation
recognition sequences.
"Promoter" refers to a DNA sequence capable of controlling the expression of a
coding sequence or functional RNA. The promoter sequence consists of proximal
and more
distal upstream elements, the latter elements often referred to as enhancers.
Accordingly, an
"enhancer" is a DNA sequence which can stimulate promoter activity and may be
an innate
element of the promoter or a heterologous element inserted to enhance the
level or tissue-
specificity of a promoter. Promoters may be derived in their entirety from a
native gene, or
be composed of different elements derived from different promoters found in
nature, or even
comprise synthetic DNA segments. It is understood by those skilled in the art
that different
promoters may direct the expression of a gene in different tissues or cell
types, or at different
stages of development, or in response to different environmentaI conditions.
Promoters
which cause a gene to be expressed in most cell types at most times are
commonly referred to
as "constitutive promoters". New promoters of various types useful in plant
cells are
constantly being discovered; numerous examples may be found in the compilation
by
Okamuro and Goldberg (1989, Biochemistry ofPlants 15:1-82). It is
furtherrecognized that
since in most cases the exact boundaries of regulatory sequences have not been
completely
defined, DNA fragments of some variation may have identical promoter activity.
An "intron"
is an intervening sequence in a gene that is transcribed into RNA but is then
excised in the
process of generating the mature mRNA. The term is also used for the excised
RNA
sequences. An "exon" is a portion of the sequence of a gene that is
transcribed and is found
in the mature messenger RNA derived from the gene, but is not necessarily a
part of the
sequence that encodes the final gene product.
The "translation leader sequence" refers to a DNA sequence located between the
promoter sequence of a gene and the coding sequence. The translation leader
sequence is
present in the fully processed mRNA upstream of the translation start
sequence. The
translation leader sequence may affect processing of the primary transcript to
mRNA, mRNA
8
CA 02578463 2007-03-02
stability or translation efficiency. Examples of translation leader sequences
have been
described (Turner, R. and Foster, G. D. (1995) Molecular Biotechnology 3:225).
The '3' non-coding sequences" refer to DNA sequences located downstream of a
coding sequence and include polyadenylation recognition sequences and other
sequences
encoding regulatory signals capable of affecting mRNA processing or gene
expression. The
polyadenylation signal is usually characterized by affecting the addition of
polyadenylic acid
tracts to the 3' end of the mRNA precursor. The use of different 3' non-coding
sequences is
exemplified by Ingelbrecht et al., (1989) Plant Cell 1:671-680.
"RNA transcript" refers to a product resulting from RNA polymerase-catalyzed
transcription of a DNA sequence. When an RNA transcript is a perfect
complementary copy
of a DNA sequence, it is referred to as a primary transcript or it may be a
RNA sequence
derived from posttranscriptional processing of a primary transcript and is
referred to as a
mature RNA. "Messenger RNA" ("mRNA") refers to RNA that is without introns and
that
can be translated into protein by the cell. "cDNA" refers to a DNA that is
complementary to
and synthesized from an mRNA template using the enzyme reverse transcriptase.
The
cDNA can be single-stranded or converted into the double-stranded by using the
klenow
fragment of DNA polymerase I. "Sense" RNA refers to RNA transcript that
includes
mRNA and so can be translated into protein within a cell or in vitro.
"Antisense RNA"
refers to a RNA transcript that is complementary to all or part of a target
primary transcript
or mRNA and that blocks expression or transcripts accumulation of a target
gene (U.S.
Patent No. 5,107,065). The complementarity of an antisense RNA may be with any
part of
the specific gene transcript, i.e. at the 5' non-coding sequence, 3' non-
coding sequence,
introns, or the coding sequence. "Functional RNA" refers to antisense RNA,
ribozyme
RNA, or other RNA that may not be translated but yet has an effect on cellular
processes.
"Sense" RNA refers to RNA transcript that includes the mRNA and so can be
translated into protein by the cell. "Antisense RNA" refers to a RNA
transcript that is
complementary to all or part of a target primary transcript or mRNA and that
blocks the
expression of a target gene (U.S. Patent No. 5,107,065. The complementarity of
an antisense
RNA may be with any part of the specific gene transcript, i.e., at the 5' non-
coding sequence,
3' non-coding sequence, introns, or the coding sequence. "Functional RNA"
refers to
antisense RNA, ribozyme RNA, or other RNA that may not be translated but yet
has an effect
on cellular processes.
The term "operably linked" refers to the association of nucleic acid sequences
on a
single nucleic acid fragment so that the function of one is affected by the
other. For example,
a promoter is operably linked with a coding sequence when it is capable of
affecting the
expression of that coding sequence (i.e., that the coding sequence is under
the transcriptional
control of the promoter). Coding sequences can be operably linked to
regulatory sequences
in sense or antisense orientation.
9
CA 02578463 2007-03-02
The term "expression", as used herein, refers to the production of a
functional end-
product. Expression or overexpression of a gene involves transcription of the
gene and
translation of the mRNA into a precursor or mature protein. "Antisense
inhibition' refers to
the production of antisense RNA transcripts capable of suppressing the
expression of the
target protein. "Overexpression" refers to the production of a gene product in
transgenic
organisms that exceeds levels of production in normal or non-transformed
organisms.
"Co-suppression" refers to the production of sense RNA transcripts capable of
suppressing
the expression or transcript accumulation of identical or substantially
similar foreign or
endogenous genes (U.S. Patent No. 5,231,020). The mechanism of co-suppression
may be
at the DNA level (such as DNA methylation), at the transcriptional level, or
at post-
transcriptional level.
"Altered expression" refers to the production of gene product(s) in transgenic
organisms in amounts or proportions that differ significantly from the amount
of the gene
product(s) produced by the corresponding wild-type organisms.
"Transformation" refers to the transfer of a nucleic acid fragment into the
genome of a
host organism, resulting in genetically stable inheritance. Host organisms
containing the
transformed nucleic acid fragments are referred to as "transgenic" organisms.
The preferred
method of corn cell transformation is use of particle-accelerated or "gene
gun" transformation
technology (Klein et al. (1987) Nature (London) 327:70-73; U.S. Patent No.
4,945,050).
Standard recombinant DNA and molecular cloning techniques used herein are well
known in the art and are described more fully in Sambrook, J., Fritsch, E. F.
and Maniatis, T.,
Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press,
Cold
Spring Harbor, 1989.(hereinafter "Sambrook et al., 1989") or Ausubel, F. M.,
Brent, R.,
Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. and Struhl, K.
(eds.), Current
Protocols in Molecular Biology, John Wiley and Sons, New York, 1990
(hereinafter
"Ausubel et al., 1990").
"PCR" or "Polymerase Chain Reaction" is a technique for the synthesis of large
quantities of specific DNA segments, consists of a series of repetitive cycles
(Perkin Elmer
Cetus Instruments, Norwalk, CT). Typically, the double stranded DNA is heat
denatured,
the two primers complementary to the 3' boundaries of the target segment are
annealed at
low temperature and then extended at an intermediate temperature. One set of
these three
consecutive steps comprises a cycle.
An "expression construct" is a plasmid vector or a subfragment thereof
comprising
the instant chimeric gene. The choice of plasmid vector is dependent upon the
method that
will be used to transform host plants. The skilled artisan is well aware of
the genetic
elements that must be present on the plasmid vector in order to successfully
transform, select
and propagate host cells containing the chimeric gene. The skilled artisan
will also
recognize that different independent transformation events will result in
different levels and
CA 02578463 2007-03-02
patterns of expression (Jones et al., (1985) EMBOJ. 4:24I I-2418; De Almeida
et a1.,.(1989)
Mol. Gen. Genetics 218:78-86), and thus that multiple events must be screened
in order to
obtain lines displaying the desired expression level and pattern. Such
screening may be
accomplished by Southern analysis of DNA, Northern analysis of mRNA
expression,
Western analysis of protein expression, or phenotypic analysis.
Although the SAMS enzyme is present in most plant cell types, no SAMS promoter
capable of driving gene expression in most or all plant cell types has been
described.
Previous studies indicated that plants contain multiple SAMS genes which are
differentially
expressed in response to various stresses (Schroder et al. (1997) Plant Mol.
Biol.
33:211-222). A SAMS promoter that is preferentially active in a particular
tissue type, i.e.
vascular (Peleman et al., (1989) Plant Cell 1, 81-93; Mijnsbrugge et al.,
(1996) Plant Cell
Physiol. 37, 1108-1115), was also known. However, it was not possible to
predict, before the
studies reported herein, whether any SAMS gene was controlled by a
constitutive promoter.
It is demonstrated herein that constitutive SAMS promoters do, in fact, exist
in plants, and
that such promoters can be readily isolated and used by one skilled in the
art.
This invention concerns an isolated nucleic acid fragment comprising a
constitutive
plant SAMS promoter. This invention also concerns an isolated nucleic acid
fragment
comprising a promoter wherein said promoter consists essentially of the
nucleotide sequence
set forth in SEQ ID NOs:6, 14, 15 or 16 or said promoter consists essentially
of a fragment or
subfragment that is substantially similar and functionally equivalent to the
nucleotide
sequence set forth in SEQ ID NOs:6, 14, 15 or 16. A nucleic acid fragment that
is
functionally equivalent to the instant SAMS promoter is any nucleic acid
fragment that is
capable of controlling the expression of a coding sequence or functional RNA
in a similar
manner to the SAMS promoter. The expression patterns of the SAMS promoter are
defined
in the following paragraphs.
Northern-blot hybridization experiments indicated that SAMS gene transcripts
are
present in a variety of soybean tissues and that the abundance of SAMS gene
transcripts does
not differ greatly from tissue to tissue (Figure 9 and Example 3). Strong
expression of the
SAMS gene was also inferred by the high frequency of occurrences of cDNA
sequences with
homology to SAMS (ESTs) in a soybean cDNA sequence database created by
sequencing
random cDNAs from libraries prepared from many different soybean tissues. ESTs
encoding
SAMS can be easily identified by conducting BLAST (Basic Local Alignment
Search Tool;
Altschul, S. F., et al., (1993) J. Mol. Biol. 215:403-410; see also
www.ncbi.nlm.nih.govBLASTn searches for similarity to sequences contained in
the
BLAST "nr" database, e.g., SAMS from Oryza sativa (EMBL Accession No. Z26867)
or
SEQ ID NO:1 provided herein. SAMS homologs were among the most abundant
classes of
cDNAs found in the soybean libraries. This indicated that SAMS was a highly
expressed
11
CA 02578463 2007-03-02
gene in most soybean cell, types. The data obtained from sequencing many SAMS
ESTs also
indicated that there were several SAMS isoforms encoded by the soybean genome.
A soybean cDNA clone designated s2.12b06 was found to encode a protein which
is
very similar to the protein encoded by the eDNA to Oryza sativa SAMS (pLog
value for this
match was 61.59). The soybean cDNA clone designated s2.12b06 was completely
sequenced
(SEQ ID NO:1) and found to contain an opening reading frame which encodes a
full length
SAMS polypeptide. Southem hybridization analysis of soybean genomic DNA with
this full
length SAMS cDNA as a probe suggested that there are approximately four
related SAMS
genes in the soybean genome (Figure I A), which is consistent with the EST
sequencing data.
The soybean SAMS cDNA clone was used to isolate a soybean genomic DNA
fragment containing more than 2000 nucleotides upstream (5') of the SAMS
protein coding
sequence by hybridization of a soybean genomic DNA library to the SAMS cDNA
fragment
probe. Southern hybridization analysis of soybean genomic DNA using a 1314
base pair
DNA fragment from upstream of the SAMS protein coding sequence as a. probe
indicated that
this fragment is unique in the soybean genome (Figure IB).
The promoter activity of the soybean genomic DNA fragment upstream of the SAMS
protein coding sequence was assessed by linking the fragment to a reporter
gene, the E. coli
P-glucuronidase gene (GUS) (Jefferson (1987) Plant Mol. Biol. Rep. 5:387-405),
transforming the SAMS promoter::GUS expression cassette into Arabidopsis, and
analyzing
GUS expression in various cell types of the transgenic plants. GUS expression
was detected
in all parts of the transgenic plants that were analyzed. These results
indicated that the
nucleic acid fragment contained a constitutive promoter. Since SAMS catalyzes
the reaction
to synthesize S-adenosyl-L-methionine from methionine and ATP, free methionine
levels
might regulate SAMS promoter activity. To see if the SAMS promoter is
regulated by
external methionine, the SAMS::GUS transgenic Arabidopsis seeds were
germinated in the
presence or absence of methionine. Ten day old seedlings were analyzed for GUS
activity
according to the protocol described in Example 5. Ten independent transgenic
lines were
tested and all of them responded similarly. GUS activity was more than two-
fold higher in
seedlings germinated in the presence of methionine (Figure 8). The increased
SAMS
promoter activity in the presence of methionine may be particularly usefu.l
for efforts to
increase methionine biosynthesis via overexpression of enzymes in the
methionine
biosynthetic pathway or the sulfate assimilation pathway. It is clear from the
disclosure set
forth herein that one of ordinary skill in the art could readily isolate a
constitutive plant
SAMS promoter from any plant by performing the following procedure:
1) obtaining a SAMS cDNA from a desired plant by any of a variety of methods
well known to those skilled in the art including, but not limited to, (a)
random sequencing of
ESTs from a cDNA library and characterizing the ESTs via a BLAST search as
described
12
CA 02578463 2007-03-02
above; or (b) hybridizing a cDNA library to a known plant SAMS cDNA; or (c)
PCR
amplification using oligonucleotide primers desigined from known SAMS cDNAs;
2) obtaining a genomic DNA fragment that includes approximately 500 to 3000
nucleotides from the region 5' to a SAMS protein coding sequence, which
contains a SAMS
promoter, by hybridization of a genomic DNA library to a SAMS cDNA fragment
probe;
3) operably linking the nucleic acid fragment containing the region upstream
(5') of the SAMS protein coding sequence to a suitable reporter gene ; there
are a variety of
reporter genes that are well known to those skilled in the art, including the
bacterial GUS
gene, the firefly luciferase gene, and the green fluorescent protein gene; any
gene for which
an easy an reliable assay is available can serve as the reporter gene
4) transforming a chimeric SAMS promoter::reporter gene expression cassette
into an appropriate plant for expression of the promoter. There are a variety
of appropriate
plants which can be used as a host for transformation that are well known to
those skilled in
the art, including the dicots, Arabidopsis, tobacco, soybean, oilseed rape,
peanut, sunflower,
safflower, cotton, tomato, potato, cocoa and the monocots, corn, wheat, rice,
barley and
palm. The terms "oilseed rape" and "oilseed Brassica" are used interchangeably
herein.
5) testing for expression of a SAMS promoter in various cell typesof
transgenic
plants, e.g., leaves, roots, flowers, seeds, transfonned with the chimeric
SAMS
promoter::reporter gene expression cassette by assaying for expression of the
reporter gene
product. A constitutive SAMS promoter will produce high level expression of
the reporter in
all, or nearly all, of the plant tissues tested.
In another aspect, this invention concems a chimeric gene comprising at least
one
heterologous nucleic acid fragment operably linked to the promoter of the
present invention.
Chimeric genes can be constructed by operably linking the nucleic acid
fragment of the
invention, i.e., the SAMS promoter or a fragment or a subfragment that is
substantially
similar and functionally equivalent to any portion of the nucleotide sequence
set forth in SEQ
ID NOS:6, 14, 15 or 16, to a heterologous nucleic acid fragment. Any
heterologous nucleic
acid fragment can be used to practice the invention. The selection will depend
upon the
desired application or phenotype to be achieved. The various nucleic acid
sequences can be
manipulated so as to provide for the nucleic acid sequences in the proper
orientation.
Plasmid vectors comprising the instant chimeric genes can then be constructed.
The
choice of plasmid vector is dependent upon the method that will be used to
transform host
cells. The skilled artisan is well aware of the genetic elements that must be
present on the
plasmid vector in order to successfully transform, select and propagate host
cells containing
the chimeric gene.
The plasmid vectors or chimeric genes can be used to transform plant cells.
Transfonnation techniques are well known to those skilled in art as discussed
above. A
preferred method of plant cell transformation is the use of particle-
accelerated or "gene gun"
13
CA 02578463 2007-03-02
transformation technology (Klein et al. (1978) Nature (London) 327:70-73; U.S.
Patent
No. 4,945,050). The chimeric gene will normally be joined to a marker for
selection in plant
cells. The marker may be resistance to a biocide, particularly an antibiotic,
such as
kanamycin, G418, bleomycin, hygromycin, chloramphenicol, or the like. The
particular
marker employed will be one which will allow for selection of transformed
cells as compared
to cells lacking the heterologous nucleic acid sequence which has been
introduced. Examples
of plant cells which can be transformed using plant transformation techniques
include, but are
not limited to, monocot and dicot plant cells such as soybean, oilseed
Brassica species, corn,
peanut, rice, wheat, sunflower, safflower, cotton,cocoa,tobacco,tomato,
potato, barley, palm,
Arabidopsis and the like.
In addition to the bacterial GUS gene, two soybean genes, ATP sulfurylase
(ATPS)
and cystathionine-o-synthase I (CGS 1), were also successfully expressed by
this promoter in
transgenic Arabidopsis, as depicted in Figure 7. This further validates the
application of the
SAMS promoter of the invention in plant genetic engineering practice.
The skilled artisan will also recognize that different independent
transformation
events will result in different levels and patterns of expression of the
chimeric genes (Jones et
al., (1985) EMBOJ. 4:2411-2418; De Almeida et al., (1989) Mol, Gen. Genetics
218:78-86).
Thus, multiple events must be screened in order to obtain lines displaying the
desired
expression level and pattern. Such screening may be accomplished by northern
analysis of
mRNA expression, western analysis of protein expression, or phenotypic
analysis. Also of
interest are seeds obtained from transformed plants displaying the desired
expression prafile.
The level of activity of the SAMS promoter is comparable to that of many known
strong promoters, such as the CaMV 35S promoter (Atanassova et al., (1998)
Plant Mol.
Biol. 37:275-285; Battraw and Hall, (1990) Plant Mol. Biol. 15:527-538;
Holtorf et al.,
(1995) Plant Mol. Biol. 29:637-646; Jefferson et al., (1987) EMBO ,I. 6:3901-
3907; Wilmink
et al., (1995) Plant Mol. Biol. 28:949-955), the Arabidopsis oleosin promoters
(Plant et al.,
(1994) Plant Mol. Biol. 25:193-205; Li, (1997) Texas A&M University Ph.D.
dissertation,
pp. 107-128), the Arabidopsis ubiquitin extension protein promoters (Callis et
al., 1990), a
tomato ubiquitin gene promoter (Rolifinke et al., 1998), a soybean heat shock
protein
promoter (Schoffl et al., 1989), and a maize H3 ~histone gene promoter
(Atanassova et al.,
1998).
Expression of the chimeric genes in most plant cell makes the SAMS promoter of
the
instant invention especially useful when constitutive expression of a target
heterologous
nucleic acid fragment is required. Examples of suitable target heterologous
nucleic acid
fragments include, but are not limited to, a herbicide-resistance or pathogen-
resistance
nucleic acid fragment. Another useful feature of the constitutive plant SAMS
promoter is its
expression profile in developing seeds. The SAMS promoter of the invention is
most active
in developing seeds at early stages and gradually turns down at later stages.
Such activity is
14
CA 02578463 2007-03-02
indicated by the GUS activity detected in seeds of transgenic Arabidopsis
plants containing a
SAMS::GUS expression cassette as shown in Figures 4 and 5. The expression
profile of the
claimed SAMS promoter is different from that of many seed-specific promoters,
e.g., seed
storage protein promoters, which often provide highest activity in later
stages of development
(Chen et al., (1989) Dev. Genet. 10:112-122; Ellerstrom et al., (1996) Plant
Mol. Bfol.
32:1019-1027; Keddie et al., (1994) Plant Mol. Biol. 24:327-340; Plant et al.,
(1994) Plant
Mol. Biol. 25:193-205; Li, (1997) Texas A&M University Ph.D. dissertation, pp.
107-128).
Thus, the SAMS promoter will be a very attractive candidate when
overexpression of a gene
in embryos is desired at an early developing stage. For example, it may be
desirable to
overexpress a gene regulating early embryo development or a gene involved in
the
metabolism prior to seed maturation.
One general application of the SAMS promoter of the invention is to construct
chimeric genes that can be used in the selection of transgenic cell lines in
plant
transformation. Currently, many of the selectable marker genes for plant
transformation are
under the control of the cauliflower mosaic virus 35S promoter. Since the SAMS
promoter
of the invention is active in seedlings and callus, the appropriate selection
phase for
transgenic plants or cell lines, this promoter may be used as an alternative
to the 35S
promoter to drive the expression of selectable marker genes.
Another general application of the SAMS promoter of the invention is to
construct
chimeric genes that can be used to reduce expression of at least one
heterologous nucleic.acid
fragment in a plant cell. To accomplish this a chimeric gene designed for
cosuppression of a
heterologous nucleic acid fragment can be constructed by linking the fragment
to the SAMS
promoter of the present invention. (See U.S. Patent No. 5,231,020 for
methodology to block
plant gene expression via cosuppression.) Alternatively, a chimeric gene
designed to express
antisense RNA for a heterologous nucleic acid fragment can be constructed by
linking the
fragment in reverse orientation to the SAMS promoter of the present invention.
(See U.S.
Patent No. 5,107,065 for methodology to block plant gene expression via
antisense RNA.)
Either the cosuppression or antisense chimeric gene can be introduced into
plants via
transformation. Transformants wherein expression of the heterologous nucleic
acid fragment
is decreased or eliminated are then selected.
This invention also concerns a method of increasing or decreasing the
expression of at
least one heterologous nucleic acid fragment in a plant cell which comprises:
(a) transforming a plant cell with the chimeric genes described herein;
(b) growing fertile mature plants from the transformed plant cell of step (a);
(c) selecting plants containing a transformed plant cell wherein the
expression of
the heterologous nucleic acid fragment is increased or decreased.
Transformation and selection can be accomplished using methods well-known to
those skilled in the art including, but not limited to, the methods described
herein.
CA 02578463 2007-03-02
EXAMPLES
The present invention is further defined in the following Examples. From the
above
discussion and these Examples, one skilled in the art can ascertain the
essential characteristics
of this invention, and without departing from the spirit and scope thereof,
can make various
changes and modifications of the invention to adapt it to various usages and
conditions.
Unless otherwise stated, all parts and percentages are by weight and degrees
are
Celsius. Techniques in molecular biology were typically perfornied as
described in Ausubel,
F. M., et al., (1990, Current Protocols in Molecular Biology, John Wiley and
Sons, New
York) or Sambrook, J. et al., (1989, Molecular cloning - A Laboratory Manual,
2 d ed. Cold
Spring Harbor Laboratory Press, Cold Spring Harbor, New York).
EXAMPLE 1
Composition of cDNA Libraries: Isolation and Sequencing of cDNA Clones
cDNA libraries representing mRNAs from soybean tissues were prepared in Uni-
ZAP XRTM vectors according to the manufacturer's protocol (Stratagene, La
Jolla, CA).
Conversion of the Uni-ZAP XRTM libraries into plasmid libraries was
accomplished
according to the protocol provided by Stratagene. Upon conversion, cDNA
inserts were
contained in the plasmid vector pBluescriptTM (Stratagene). DNA was prepared
for
sequencing from randomly selected bacterial colonies containing recombinant
pBluescriptTM
plasmids either by amplifying the eDNA inserts via polymerase chain reaction
using primers
specific for vector sequences flanking the cloning site or by preparing
plasmid DNA from
cultured bacterial cells. Amplified insert DNAs or plasmid DNAs were sequenced
in dye-
primer sequencing reactions using a Perkin Elmer Model 377 fluorescent
sequencer to
generate partial cDNA sequences termed expressed sequence tags or "ESTs" (see
Adams,
M. D. et al., (1991) Science 252:1651).
EXAMPLE 2
Identification of SAMS cDNA Clones
ESTs encoding SAMS were identified by conducting BLAST (Basic Local Alignment
Search Tool; Altschul, S. F., et al., (1993) J. Mol. Biol. 215:403-410; see
also
www.ncbi.nlm.nih.govBLAST/) searches for similarity to sequences contained in
the
BLAST "nr" database (comprising all non-redundant GenBank CDS translations,
sequences
derived from the 3-dimensional structure Brookhaven Protein Data Bank, the
last major
release of the SWISS-PROT protein sequence database, EMBL, and DDBJ
databases). The
cDNA sequences obtained in Example 1 were analyzed for similarity to all
publicly available
DNA sequences contained in the "nr" database using the BLASTN algorithm
provided by the
National Center for Biotechnology Information (NCBI). The DNA sequences were
translated
in all reading frames and compared for siniilarity to all publicly available
protein sequences
contained in the "nr" database using the BLASTX algorithm (Gish, W. and
States, D. J.
(1993) Nature Genetics 3:266-272 and Altschul, S. F., et al. (1997) Nucleic
Acids Res.
16
CA 02578463 2007-03-02
25:3389-3402) provided by the NCBI. For convenience, the P-value (probability)
of
observing a match of a cDNA sequence to a sequence contained in the searched
databases
merely by chance as calculated by BLAST are reported herein as "pLog" values,
which
represent the negative of the logarithm of the reported P-value. Accordingly,
the greater the
pLog value, the greater the likelihood that the cDNA sequence and the BLAST
"hit"
represent homologous proteins.
The BLASTX search using the nucleotide sequence from clone s2.12b06 revealed
that
this nucleotide sequence encoded a protein that was similar to the protein
encoded by the
cDNA to Oryza sativa (EMBL Accession No. Z26867) S-adenosylmethionine
synthetase; the
pLog value for this match was 61.59. This cDNA clone was completely sequenced
(SEQ ID
NO: 1) and found to contain an opening reading frame ranging from nucleotides
74 to 1252
which is predicted to encode a full length SAMS polypeptide.
A high level of expression of the SAMS genes was inferred by the high
frequency of
occurrences of soybean cDNA sequences with homology to Oryza sativa SAMS
obtained
from many different cDNA libraries prepared from many different soybean cell
types.
SAMS homologs were the third most abundant class of ESTs found in the soybean
libraries.
Although the ranking might not represent a precise estimate of the relative
abundance of the
SAMS transcripts in vivo in all soybean libraries, due to the selective use of
different cDNA
libraries, it did indicate that SAMS was a highly expressed gene. The EST
sequence data
also revealed that there were several SAMS isoforms in the soybean genome.
EXAMPLE 3
S-adenosylmethionine Synthetase is Encoded by a Gene Family
Southern hybridization analysis of soybean genomic DNA with a full length SAMS
cDNA (SEQ ID NO: 1) as a probe suggested that there are at least four related
SAMS genes in
the soybean genome (Figure lA). The DNA probe for Southern hybridization was
prepared
as follows: plasmid DNA was prepared from an overnight bacteria culture in LB
broth
(GIBCO BRL, Gaithersburg, MD) using QIAprepTM miniprep kit (Qiagen, Valencia,
CA);
cDNA inserts encoding SAMS were excised by restriction enzyme digestion and
recovered
from agarose gel following electrophoretic separation using QlAquickTM gel
extraction kit
(Qiagen). The 1518 bp SAMS cDNA fragment (SEQ ID NO:1) was labeled with
digoxigenin-dUTP as a probe by random primed DNA labeling (Boehringer
Mannheim).
Twenty micrograms of soybean geneomic DNA was digested with different
restriction
enzymes and the resulted fragments were resolved on a 0.7% agarose gel. The
DNA gel was
depurinated in 0.25 M HCI, denatured in 0.5 M NaOIH/1.5 M NaC1, neutralized in
I m
Tris-Cl, pH 8.0/1.5 M NaCI, and transferred in 20x SSC (GIBCO BRL) to nylon
membrane
(Boehringer Mannheim). The Southern blot was hybridized with the SAMS cDNA-
specific
probe at 45 C overnight in Easy Hyb (Boehringer Mannheim). The blot was washed
10 minutes in 2xSSC/0.1% SDS, and 3x 10 minutes in 0.lx SSC/0.1% SDS at 65 C.
The
17
CA 02578463 2007-03-02
hybridized probe was detected with chemiluminescent reagent CDP-Star
(Boehringer
Mannheim) according to the manufacturer's protocol. Multiple bands were
detected in
BamHI, EcoRl, and HindIIl digestions (Figure IA). The large band in Kpnl and
SacI
digestions may represent more than one DNA fragment because the band is too
big for good
resolution. The hybridization patterns presented in Figure lA and the analysis
of partial
SAMS cDNA sequences from DuPont's EST database suggest that there are at least
four
copies of the SAMS gene in the soybean genome and that their sequences are
conserved.
The 1314 bp SAMS promoter fragment (SEQ ID NO:6) was labeled with
digoxigenin-dUTP also by random primed DNA labeling (Boehringer Mannheim). The
labeled SAMS promoter probe was used to hybridize the same Southern blot as
above
described. The SAMS promoter-specific probe hybridized to a single band in
each of the five
different digestions, BamHI, EcoRl, HindIIl, Kpnl, and SacI (Figure 1B). The
results
indicate that the SAMS promoter has only a single copy in soybean genome.
A northern hybridization experiment indicated that SAMS gene transcripts were
present in a variety of soybean tissues and that the abundance of SAMS gene
transcripts did
not differ greatly from tissue to tissue. Total RNAs were extracted from
soybean leaves,
stems, young seeds, medium seeds, old seeds, and pod coats using TrizolTM
Reagent
according to the manufacturer's protocol (GIBCO BRL). Ten micrograms of total
RNA were
loaded in each well of a 1.2% agarose gel containing 7% formaldehyde in Ix
MOPS buffer,
20 mM 3-[N-morpholino]propane-sulfonic acid, 5 mM sodium acetate, 1 mM EDTA,
pH 6Ø
RNA was transferred to nylon filters (Micron Separations Inc., Westborough,
MA) in I OX
SSC and crosslinked to the filters with UV light. Filters were hybridized with
probes
prepared from cDNA insert fragments in 50% deionized formamide, 5x SSPE, lx
Denhardt's
solution, 0.1 % SDS, and 100 g denatured salmon sperm DNA (Sigma, St. Louis,
MO) at
42 for 24 hours. Filters were washed in 2x SSPE and 0.1% SDS at room
temperature for
10 minutes, lx SSPE and 0.1% SDS at 65 for 10 minutes, and then in 0.Ix SSPE
and 0.1%
SDS at 65 for 10 minutes. Filters were exposed to Kodak X-ray film at -80.
The abundance
of SAMS transcripts in leaves, roots, stems, young seeds, medium seeds, old
seeds, and pod
coats can be seen in Figure 9. The weak signals observed in the hybridizations
to RNA
samples from root and young seed were attributed to underloading, because
hybridizations
with ribosomal RNAs that serve as internal controls were also relatively weak
in those
samples (data not shown). Because of the high sequence similarities among the
four SAMS
gene isoforms, this RNA gel blot was not able to indicate how the isofonrns
were distributed
in any particular tissue. However, the experiment demonstrated that all
examined soybean
tissues contained SAMS messenger RNA.
18
CA 02578463 2007-03-02
EXAMPLE 4
Cloningof the Soybean S-adenosylmethionine Synthetase Gene Promoter
The soybean full length SAMS cDNA (SEQ ID NO:1), obtained in Example 2, was
used to generate a probe to isolate a SAMS promoter. The full length SAMS cDNA
sequence consisted of 1518 bp, and it had a 74 bp 5'-untranslated region and a
PstI site at
position 296. Because the cDNA clone was harbored in a pBluescriptTM SK vector
having a
PstI site upstream of the EcoRI cloning site, digestion of the clone with Pstl
generated a
315 bp fragment of DNA. The resulting restriction fragment contained 19 bp of
vector and
cloning linker adapter sequence in addition to the 296 bp of SAMS cDNA
sequence. This
Pstl fragment was labeled with a-32P-dCTP, as described in Example 3, and used
as a probe
to screen a soybean genomic DNA library that had been constructed in a EMBL3
SP6/T7
vector (ClonTech, Palo Alto, CA). The library was plated with LE392 (ClonTech)
cells at
50,000 plaque forming units (pfu) per 150 mm NZCYM agar plate (GIBCO BRL).
Plaques
were transferred to Hybond nylon membranes, and the plaque replicas were then
denatured
and neutralized according to the manufacturer (Amersham Life Science,
Arlington Heights,
IL). The phage DNA was fixed on the membranes by W-crosslinking (Stratagene).
After
prehybridization at 65 for I hour in 0.5 M NaHPO4, pH 7.2, 1 mM EDTA, 1%=
crystalline
BSA (Sigma), and 7% SDS, the SAMS 315 bp Pstl fragment probe was denatured in
boiling
water bath for 5 minutes and added to the same hybridization solution, and was
hybridized at
65 for 24 hours. The membranes were washed in 40 mM NaHPO4, pH 7.2, 1 mM
EDTA,
0.5% crystalline BSA, and 5% SDS for 10 minutes at room temperature, and then
3x
10 minutes at 65 in 40 mM NaHPO4, pH 7.2, 1 mM EDTA, and 1% SDS. The
membranes
were exposed to Kodak X-ray film (Sigma) at -80 . Positive SAMS genomic DNA
phage
clones were suspended in SM buffer, 50 mM Tris-Cl, pH 7.5, 100 mM NaCI, 0.2%
MgSO4-7H2O, and 0.1% gelatin, and purified by a secondary screening following
the same
procedure, Twenty three strongly hybridizing plaques were identified by the
first screening
from a total of 3x105 pfu, and fifteen were later purified. DNAs were prepared
from two of
the purified phage clones (Ausubel et al., (1990) pp. 1.13.4-1.13.8), they
were digested with
BamHI, Clal, PstI, and Ncol and prepared for a Southern blot. The blot was
hybridized with
the SAMS 315 bp PstI fragment probe prepared and used as above. A single
positive
fragment of clone 1 was identified from the Clal digestion. Since the Cial
restriction site in
the cDNA clone is 843 bp from the 5' end of the full length cDNA, the 2.5 kb
Clal fragment
was expected to include about 1.7 kb of DNA upstream of the coding sequence,
which was
considered sufficient to contain the SAMS promoter.
The 2.5 kb CIaI genomic DNA fragment was cloned into pBluescriptTM KS and the
DNA insert was sequenced. The 3' end sequence of the genomic DNA fragment was
expected to match the 5' end sequence of SAMS cDNA from the 5' end to the Clal
site at
position 843. However, comparison of the genomic DNA sequence and the cDNA
sequence
19
CA 02578463 2007-03-02
revealed that the two sequences have 191 bp of overlapping sequence starting
at position 54
and ending at position 245 of the cDNA sequence (SEQ ID NO:1). The sequence of
the
2.5 kb genomic DNA clone downstream of the 191 bp overlapping region was
determined to
be derived from the cloning vector, lambda EMBL3 SP6/T7, which contributed 257
bp of
sequence to the 3' end of the 2.5 kb SAMS CIaI fragment including the Cial
cloning site.
Therefore, the soybean derived DNA in the 2.5 kb C1aI fragment is described by
the 2336 bp
DNA sequence shown in SEQ ID NO:2.
The DNA sequence of the genomic DNA in the 191 bp region (from nucleotide 2145
to the end of the sequence) was very similar to, but did not match perfectly,
the cDNA
sequence; there were six base pair mismatches in this region. This was not
surprising,
because it was known from the experiments described in Example 3 that there is
a small
family of SAMS genes in soybean. It was concluded that this genomic clone is
not derived
from the same gene from which the cDNA used as the probe was transcribed. It
was also
noted that the 53 bp at the 5' end of the cDNA did not show any similarity to
the genomic
sequence upstream of the 191 bp overlapping region (Figure 2).
A BLASTN search of the DuPont soybean EST database using the nucleotide
sequence from the soybean SAMS genomic DNA upstream of the 191 bp region
revealed
many cDNA clones that matched a 60 bp rcgion of the genomic DNA from
nucleotide 1496
to 1555. The sequence of one such cDNA, designated srrlc.pk002.b21, is shown
in SEQ ID
NO:3.
The cDNA sequence in SEQ ID NO:3 perfectly matches the genomic sequence in
SEQ ID NO:2 from nucleotide 2 to 60 of the cDNA. There follows a region of 591
nucleotides in the genomic DNA that is absent from the cDNA. Then the region
from
nucleotide 60 to 250 of the cDNA perfectly matches the 191 bp region at the 3'
end of the
genomic DNA. This indicates the presence of a 591 nucleotide intron in the
genomic DNA in
the 5' transcribed, but untranslated, region of the SAMS gene. The presence of
consensus 5'
and 3' splice junctions in the genomic DNA at the exon-intron junctions
supports this
conclusion. Thus, the 53 bp at the 5' end of the cDNA used as the probe (SEQ
ID NO:1) did
not match the genomic sequence because the genomic sequence at that positioin
in the
alignment was from the intron. However, the 53 bp at the 5' end of the cDNA of
SEQ ID
NO:1 is very similar to the 60 nucleotides at the 5' end of the cDNA of SEQ ID
NO:3,
suggesting that the gene from which SEQ ID NO: I was transcribed also contains
an intron at
the analogous position.
A 1305 bp SAMS genomic DNA fragment starting at nucleotide 856 and ending at
nucleotide 2160 of SEQ ID NO:2: was amplified by PCR from the 2.5 kb CIaI
clone. The
promoter fragment was amplified from this fragment using primers sam-5 (SEQ ID
NO:4)
and sam-6 (SEQ ID NO:5) and Pfu DNA polymerase (Stratagene).
CA 02578463 2007-03-02
CATGCCATGGCTTTATACTTCAAAAACTGCAC (SEQ ID NO:4)
GCTCTAGATCAAACTCACATCCAA (SEQ ID NO:5)
An XbaI site and an NcoI site were introduced to the 5' end and 3' end,
respectively, of the
PCR fragment by using these specifically designed primers. The Ncol site
includes the ATG
start codon of the SAMS coding region. The resulting 1314 bp fragment is shown
in SEQ ID
NO:6 and includes the SAMS promoter and the translation leader region, which
is interrupted
by the 591 nucleotide intron.
Using PCR amplification procedures and appropriate primers additional SAMS
promoter fragments can be produced from the 2336 nucleotide fragment of SEQ ID
NO:2.
These include, but are not limited to, the three fragments provided in SEQ ID
NOs: 14, 15 and
16. SEQ ID NO: 14 is a 2165 nucleotide sequence of a SAMS promoter DNA
fragment
which starts at the 5' end of the 2336 nucleotide sequence of SEQ ID NO:2 and
ends at the
ATG translation start codon of the SAMS protein. SEQ ID NO: 15 is a 1574
nucleotide
sequence of a SAMS promoter DNA fragment which starts at the 5' end of the
2336
nucleotide sequence of SEQ ID NO:2 and ends at the ATG translation start codon
of the
SAMS protein, and from which the 591 nucleotide long intron sequence has been
removed.
SEQ ID NO: 16 is a 719 nucleotide sequence of a SAMS promoter DNA fragment
which
starts at nucleotide 4 of SEQ ID NO:6 and ends at the ATG translation start
codon of the
SAMS protein, and from which the 591 nucleotide long intron sequence has been
removed.
EXAMPLE 5
Expression of the GUS Gene bv the SAMS Promoter in Arabidonsis
The activity of the soybean SAMS promoter was tested by its ability to express
the
GUS reporter gene in transgenic Arabidopsis plants carrying the SAMS
promoter::GUS::3'
Nos expression casstette. GUS refers to the E. coil 0-glucuronidase gene (GUS)
(Jefferson,
(1987) Plant Mol. Biol. Rep. 5:387-405) and 3' Nos refers to the transcription
termination
region from the nopaline synthase (Nos) gene (Depicker et al. (1982) J. Mol.
Appl. Genet.
1:561-570). The SAMS promoter fragment (SEQ ID NO:6) was digested with XbaI
and
NcoI and inserted into plasmid pMH40d (SEQ ID NO:17), which contained a 35S
promoter::GUS::3' Nos plant expression cassette. The Xbai/NcoI SAMS promoter
DNA
fragment replaced the 35S promoter of pMH40A, to form the pZSLI l plasmid
(Figure 3).
The SAMS promoter::GUS::3' Nos DNA fragment (SEQ ID NO:18) was excised from
pZSL11 by HindlII and SacI digestion and transferred into the corresponding
sites of pB1101
(ClonTech) binary vector. The cloned SAMS promoter was sequenced to verify
that no
sequence error was generated by the PCR amplification.
The SAMS::GUS expression cassette was introduced into wild typeArabidopsis
thaliana by Agrobacteria mediated transformatian. A. thaliana ecotype columbia
were
grown in 228 chamber with continuous light and transformed by
vacuum,infiltration method
21
CA 02578463 2007-03-02
using GV3 101 Agrobacteria (Bent, A. et al., (1994) Science 265:1856-1860).
Transformed
Arabidopsis seeds were selected by germination on Murashige and Skoog minimal
salt
(GIBCO BRL) plus 0.2 % phytagel (Sigma), 1% sucrose, and 100 mg/mi kanamycin.
The
kanamycin resistant seedlings were transferred into soil and grown in 228
chamber under
continuous light.
For histochemical GUS staining, plant tissues were incubated in 0.5% 5-bromo-4-
chloro-3-indoxyl-(3-D-glucuronic acid (X gluc, Biosynth AG, Switzerland) in 50
mM sodium
phosphate, pH 7.0, 10 mM EDTA, 0.5 mM potassium ferricyanide, and 0.5 mM
potassium
ferrocyanide at 378 overnight, and then chlorophyll was removed with 75%
ethanol. Pictures
were taken using a Nikon dissecting microscope. Strong GUS expression was
detected in all
the parts of the transgenic Arabidopsis plants, including flowers (Figure 4A),
leaves
(Figure 4B), stems (bolt) (Figure 4C), silique coats and developing seeds
(Figure 4D-F),
developing embryos (Figure 4G), and seedlings (not shown). The GUS staining on
leaves
and silique coats was uniform with all the veins and mesophyll tissues
similarly stained,
while staining on flowers and stems was not uniform. Although some seeds were
not stained
for GUS activity due to genetic segregation, the funiculi that connected these
seeds to the
silique coat stained positively for GUS activity (Figure 4G). These results
indicated that the
soybean SAMS promoter was a constitutive promoter and was able to function in
heterologous plant.
The GUS activities of the transgenic Arabidopsis plants were further analyzed
by a
fluorometric assay. For fluorescence analysis, plant tissues were ground in
microfuge tubes
with extraction buffer, 50 mM phosphate buffer, pH 7.0, 10 mM EDTA, 0.1 %
Triton X-100,
0.1 % N-lauroyl sarcosine, and 10 mM P-mercaptoethanol, to homogeneity. The
samples
were centrifuged at 14,000 rpm for 10 minutes, and aliquots of the supematant
were used to
determine protein concentrations by the Bradford method (Bio-Rad, Hercules,
CA) using
96 well microtiter plates read with a kinetic microplate reader (Molecular
Devices,
Sunnyvale, CA). The R-glucuronidase activities were analyzed by standard
protocol
(Jefferson et al, (1987) EMBO J. 6:3901-3907) using 96 well microtiter plates
read with
Cytofluor multiwell plate reader (PerSeptive Biosystems, Framingham, MA). Data
were
entered into a Microsoft Excel spread sheet and analyzed. Triple samples of
flower, leaf,
stem, silique coat, young seed (white), medium seed (light green), old seed
(dark green), and
dry seed from six plants were analyzed. The soybean SAMS promoter was active
in all the
tissues analyzed (Figure 5). Promoter activity varied among the six lines, as
is typically seen
among plant transformants. The basic expression patterns were similar among
all the lines,
and the average SAMS promoter activity was comparable to that of the 35S
promoter
(Battraw and Hall, (1990) Plant Mol. Biol. 1 S:527-538; Jefferson et al.,
(1987) EMBO J.
6:3901-3907; Atanassova et al., (1998) Plant Mol. Biol. 37:275-285; Holtorf et
al., (1995)
Plant Mol. Biol. 29:637-646; Wilmink et al., (1995) Plant Mol. Biol. 28:949-
955). The
22
CA 02578463 2007-03-02
SAMS promoter was very active in developing seeds, especially in early and
medium stages
of development, and the GUS specific activities are in the range of 5-40 pmole
4-Mu
(4-methylumbelliferone) per microgram protein per minute, which are comparable
to many
strong promoters (Atanassova et al., (1998) Plant Mol. Biol. 37:275-285; Comai
et al., (1990)
Plant Mol. Biol. 15:373-381; Holtorf et al., (1995) Plant Mol. Biol. 29:637-
646; Wilmink et
al., (1995) Plant Mol. Biol. 28:949-955).
EXAMPLE 6
Expression of GUS Gene by SAMS Promoter in Corn
In order to test whether the dicot SAMS promoter also worked in monocot
plants,
pZSL I 1 was introduced into corn leaf discs and cailus by gene bombardment
for transient
gene expression assay using the biolistic particle delivery system PDS-1000/He
(Bio Rad,
Hercules, CA). The pMH400 plasmid DNA (as set forth in SEQ ID NO:17), which
contained the 35S promoter and GUS reporter gene, was also introduced into
corn callus and
leaf discs by gene bombardment to serve as a positive control vector. After
incubation
overnight at 37 , bombarded tissues were stained for GUS activity. GUS
expression was
demonstrated by the blue spots on both the callus (Figure 6A) and leaf discs
(Figure 6B)
bombarded with pZSLI 1. As expected, the positive contro135S::GUS cassette was
also
expressed in both callus and leaf discs (Figure 6C, D).
EXAMPLE 7
Exnression of Methionine Biosynthesis Genes by SAMS Promoter
The SAMS promoter was fused to two soybean cDNAs, one encoding ATP
sulfurylase (ATPS) and a second encoding cystathionine-y-synthase (CGS1). The
soybean
ATPS and CGS1 cDNAs were isolated from soybean embryo cDNA libraries using the
same
procedures as described in Example I and Example 2 for isolation of soybean
SAMS cDNAs.
The coding regions and the 3' untranslated region (UTR) of soybean ATPS and
CGSI genes
were inserted into pZSLI l replacing the GUS gene. The resulting SAMS
promoter::ATPS
and SAMS promoter::CGS 1 expression cassettes, SEQ ID NO:19 and SEQ ID NO:20,
respectively, were inserted into binary vectors for Arabidopsis transformation
and
transformation was performed as described in Example 5. Transgenic Arabidopsis
plants
with soybean ATPS and CGS1 genes controlled by the SAMS promoter were analyzed
by
PCR for the presence of the transgenes and by RT-PCR for expression of the
transgenes.
Genomic DNA used for PCR analysis was prepared from Arabidopsis siliques and
leaves
using 7 M urea, 1.5 M NaC1, 50 mM Tris, pH 8.0, 20 mM EDTA, and 1% N-lauroyl-
sarcosine, followed by phenol extraction and ethanol precipitation. Primer sam-
9 (SEQ ID
NO:7) which is specific to SAMS promoter, and primers specific to the target
genes, atps-1
(SEQ ID NO:8) for the ATPS gene and cgs-8 (SEQ ID NO:9) for the CGS1 gene were
used
in PCR with Taq DNA polymerase (GIBCO BRL) to detect the existence of
SAMS::ATPS
and SAMS::CGS 1 in transgenic Arabidopsis plants.
23
CA 02578463 2007-03-02
TTCGAGTATAGGTCACAATAGG (SEQ ID NO:7)
CTTCGCTGAGGACATGGAC (SEQ ID NO:8)
GAGTTGTCGCTGTTGTTCGAC (SEQ ID NO:9)
RNA samples used for RT-PCR were prepared with TrizolTM Reagent (GIBCO BRL).
Antisense primers atps-4 (SEQ ID N0:10)and cgs-10 (SEQ ID N0:11) were used in
reverse
transcription reactions with SuperscriptIIr"' RT (GIBCO BRL) following the
vendor's
instruction.
AACACAGCATCCGCATTGCG (SEQ ID NO:10)
AGGAGTGCAGAATCAGATCAG (SEQ ID NO:11)
The first strand cDNAs were used in PCR with primer pairs atps-3 (SEQ ID
NO:12) and
atps-4 (SEQ ID NO: 10) for SAMS::ATPS transgenic plants, and cgs-9 (SEQ ID NO:
13) and
cgs-10 for SAMS::CGSI transgenic plants. PCR and RT-PCR products were resolved
by
agarose gel electrophoresis.
GCTGATCGAACCAGATGGAG (SEQ ID NO: 12)
CTGTACAGTTAAACAGTAGTTCT (SEQ ID NO:13)
All ten SAMS::CGS1 transgenic Arabidopsis harbored the SAM::CGS1 expression
cassette as revealed by PCR with SAMS::CGSI-specific primers (Figure 7A). It
was also
revealed by the same analysis that all the ten SAMS::ATPS transgenic
Arabidopsis plants
contained the SAMS::ATPS expression cassette (Figure 7A). RT-PCR analysis
detected
CGS 1 transcripts and ATPS transcripts, respectively, in most of the
transgenic plants
(Figure 713). This shows that the SAMS promoter is capable of driving
expression of a
variety of different genes in most or all cell types in transformed plants.
. EXAMPLE 8
Induction of SAMS Promoter Actiyitv by Methionine
Since SAMS catalyzes the reaction to synthesize S-adenosyl-L-methionine from
methionine and ATP, free methionine levels might regulate SAMS promoter
activity. To see
if SAMS promoter is regulated by external methionine, the SAMS::GUS transgenic
Arabidopsis seeds were germinated in the presence of either H2O, lx Murashige
and Skoog
salt (GIBCO BRL), 0.01 mM methionine (Sigma), or 1 mM methionine. Ten days old
seedlings from ten independent transgenic lines were analyzed for GUS activity
according to
the protocol described in Example 5. GUS activity for each treatment, in the
order given
above, for each transgenic line is shown in Figure 8. All lines responded
similarly to the
different treatments. Compared to the control of H2O treamtment, SAMS activity
was
24
CA 02578463 2007-03-02
induced more than two-fold by 0.01 mM free methionine and inhibited about 40%
on average
by lx MS salt. The induction effect of SAMS promoter by 1 mM methionine was
less than
that by 0.01 mM methionine, probably due to a toxic effect of the high
methionine
concentration; this toxic effect was indicated by the smaller sizes and
shorter roots of the
seedlings grown in the presence of 1 mM methionine. The toxic effect of high
levels of
methionine was even more apparent at 10 mM free methionine, since only a few
Arabidopsis
seeds were able to germinate and none survived in the presence of 10 mM free
methionine.
CA 02578463 2007-03-02
SEQUENCE LISTING
<110> E. I. du Pont de Nemours and Company
<120> S-ADENOSYL-L-METHIONINE SYNTHETASE PROMOTER AND
ITS USE IN EXPRESSION OF TRANSGENIC GENES IN PLANTS
<130> BB1205
<140>
<141>
<150> 60/113,045
<151> 1998-12-21
<160> 16
<170> Microsoft Office 97
<210> 1
<211> 1518
<212> DNA
<213> Glycine max
<400> 1
agccaagccc cactcaacca ccacaccact ctctctgctc ttcttctacc tttcaagttt 60
ttaaagtatt aagatggcag agacattcct atttacctca gagtcagtga acgagggaca 120
ccctgacaag ctctgcgacc aaatctccga tgctgtcctc gacgcttgcc ttgaacagga 180
cccagacagc aaggttgcct gcgaaacatg caccaagacc aacttggtca tggtcttcgg 240
agagatcacc accaaggcca acgttgacta cgagaagatc gtgcgtgaca cctgcaggaa 300
catcggcttc gtctcaaacg atgtgggact tgatgctgac aactgcaagg tccttgtaaa 360
cattgagcag cagagccctg atattgccca gggtgtgcac ggccacctta ccaaaagacc 420
cgaggaaatc ggtgctggag accagggtca catgtttggc tatgccacgg acgaaacccc 480
agaattgatg ccattgagtc atgttcttgc aactaaactc ggtgctcgtc tcaccgaggt 540
tcgcaagaac ggaacctgcc catggttgag gcctgatggg aaaacccaag tgactgttga 600
gtattacaat gacaacggtg ccatggttcc agttcgtgtc cacactgtgc ttatctccac 660
ccaacatgat gagactgtga ccaacgacga aattgcagct gacctcaagg agcatgtgat 720
caagccggtg atcccggaga agtaccttga tgagaagacc attttccact tgaacccctc 780
tggccgtttt gtcattggag gtcctcacgg tgatgctggt ctcaccggcc gcaagatcat 840
catcgatact tacggaggat ggggtgctca tggtggtggt gctttctccg ggaaggatcc 900
caccaaggtt gataggagtg gtgcttacat tgtgagacag gctgctaaga gcattgtggc 960
aagtggacta gccagaaggt gcattgtgca agtgtcttat gccattggtg tgcccgagcc 1020
tttgtctgtc tttgttgaca cctatggcac cgggaagatc catgataagg agattctcaa 1080
cattgtgaag gagaactttg atttcaggcc cggtatgatc tccatcaacc ttgatctcaa 1140
gaggggtggg aataacaggt tcttgaagac tgctgcatat ggacacttcg gcagagagga 1200
ccctgacttc acatgggaag tggtcaagcc cctcaagtgg gagaaggcct aaggccattc 1260
attccactgc aatgtgctgg gagtttttta gcgttgccct tataatgtct attatccata 1320
actttccacg tcccttgctc tgtgtttttc tctcgtcgtc ctcctcctat tttgtttctc 1380
ctgcctttca tttgtaattt tttacatgat caactaaaaa atgtactctc tgttttccga 1440
ccattgtgtc tcttaatatc agtatcaaaa agaatgttcc aagttaaaaa aaaaaaaaaa 1500
aaaaaaaaaa aaaaaaaa 1518
<210> 2
<211> 2336
<212> DNA
<213> Glycine max
1
CA 02578463 2007-03-02
<400> 2
atcgatagag acatgttatt cacaaaccat aaaatgatgg ctaaaattgg tgtgattgga 60
acgatatctg tttattatga tttcagggcg caaaaatgcg agtacttaat aaaattttac 120
atttaaatta gaattttttt tatcaataaa tattaattta ttagttttat tagaaatatt 180
aattagaaaa ttttgaatcc ccgatttctc ctccttttct tcgctattca tcattttcta 240
accaaaccaa tcttatatgt tcttcaaatt agaacttgaa attattaatt ataattaaac 300
tgaaaacaat ttggtatcaa ttcatataca tgcttagtaa taaaatgcga taattaattg 360
ataaatctgc aaaagatttt acaaatatct ttcagaaaaa attaataaca aattttgtcg 420
ttttcatggt gttggtctga ggaggatttg gcactataga actctcctac ggaccattct 480
ttgcacttca actaaacgat ggtcagaatt ggtggggatt ttatattcaa gcatatccct 540
ttcaaaactt cctacttact tcgtgcgttc ggtaatcggt aacattagac tttcaaaatc 600
atttttaacc cctaaacagt aaatttgaag gacaaaaata atatttttca aatttgatag 660
actatttttt ttttgtaatt tgacgaacca aaaccagatt tatcctgaat tttaggaacc 720
acagatgtaa ctaaaccaat atttatttat tttctaaaac aaaatttcat ggcagcatgc 780
ctcagcccat gaaaaaaacc ttataaaaat atctacacat tgaccattga aaagttcgtt 840
ctcccatggg taaccagatc aaactcacat ccaaacataa catggatatc tccttaccaa 900
tcatactaat tattttgggt taaatattaa tcattatttt taagatatta attaagaaat 960
taaaagattt tttaaaaaaa tgtataaaat tatattattc atgatttttc atacatttga 1020
ttttgataat aaatatattt tttttaattt cttaaaaaat gttgcaagac acttattaga 1080
catagtcttg ttctgtttac aaaagcattc atcatttaat acattaaaaa atatttaata 1140
ctaacagtag aatcttcttg tgagtggtgt gggagtaggc aacctggcat tgaaacgaga 1200
gaaagagagt cagaaccaga agacaaataa aaagtatgca acaaacaaat caaaatcaaa 1260
gggcaaaggc tggggttggc tcaattggtt gctacattca attttcaact cagtcaacgg 1320
ttgagattca ctctgacttc cccaatctaa gccgcggatg caaacggttg aatctaaccc 1380
acaatccaat ctcgttactt aggggctttt ccgtcattaa ctcacccctg ccacccggtt 1440
tccctataaa ttggaactca atgctcccct ctaaactcgt atcgcttcag agttgagacc 1500
aagacacact cgttcatata tctctctgct cttctcttct cttctacctc tcaaggtact 1560
tttcttctcc ctctaccaaa tcctagattc cgtggttcaa tttcggatct tgcacttctg 1620
gtttgctttg ccttgctttt tcctcaactg ggtccatcta ggatccatgt gaaactctac 1680
tctttcttta atatctgcgg aatacgcgtt ggactttcag atctagtcga aatcatttca 1740
taattgcctt tctttctttt agcttatgag aaataaaatc attttttttt atttcaaaat 1800
aaaccttggg ccttgtgctg actgagatgg ggtttggtga ttacagaatt ttagcgaatt 1860
ttgtaattgt acttgtttgt ctgtagtttt gttttgtttt cttgtttctc atacattcct 1920
taggcttcaa ttttattcga gtataggtca caataggaat tcaaactttg agcaggggaa 1980
ttaatccctt ccttcaaatc cagtttgttt gtatatatgt ttaaaaaatg aaacttttgc 2040
tttaaattct attataactt tttttatggc aaaaattttt gcatgtgtct ttgctctcct 2100
gttgtaaatt tactgtttag gtactaactc taggcttgtt gtgcagtttt tgaagtataa 2160
agatggcaga gacattccta ttcacctcgg agtcagtgaa cgagggacac cctgataagc 2220
tctgcgacca aatctccgat gctgtcctcg acgcttgcct cgaacaggac ccagacagca 2280
aggttgcctg cgaaacatgc accaagacca acttggtcat ggtcttcgga gagatc 2336
<210> 3
<211> 522
<212> DNA
<213> Glycine max
<220>
<221> unsure
<222> (405)
<220>
<221> unsure
<222> (509)
<220>
<221> unsure
<222> (515)
2
CA 02578463 2007-03-02
<400> 3
gaccaagaca cactcgttca tatatctctc tgctcttctc ttctcttcta cctctcaagt .60
ttttgaagta taaagatggc agagacattc ctattcacct cggagtcagt gaacgaggga 120
caccctgata agctctgcga ccaaatctcc gatgctgtcc tcgacgcttg cctcgaacag 180
gacccagaca gcaaggttgc ctgcgaaaca tgcaccaaga ccaacttggt catggtcttc 240
ggagagatca ccaccaaggc caacgttgac tacgagaaga tcgtgcgtga cacctgcagg 300
agcatcggct tcatctcaaa cgatgtggga cttgatgctg acaactgcaa ggtccttgta 360
aacattgagc agcagagccc tgatattgcc cagggcgtgc acggncacct taccaaaaga 420
cctgaagaaa ttggcgctgg tgaccaaggt cacatgtttg gctatgccac tgatgaaacc 480
ccaaaattca tgccattgag tcatgttcnt gcaancaagc tc 522
<210> 4
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: PCR Primer
<400> 4
catgccatgg ctttatactt caaaaactgc ac 32
<210> 5
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: PCR Primer
<400> 5
gctctagatc aaactcacat ccaa 24
<210> 6
<211> 1314
<212> DNA
<213> Glycine max
<400> 6
tctagatcaa actcacatcc aaacataaca tggatatctc cttaccaatc atactaatta 60
ttttgggtta aatattaatc attattttta agatattaat taagaaatta aaagattttt 120
taaaaaaatg tataaaatta tattattcat gatttttcat acatttgatt ttgataataa 180
atatattttt tttaatttct taaaaaatgt tgcaagacac ttattagaca tagtcttgtt 240
ctgtttacaa aagcattcat catttaatac attaaaaaat atttaatact aacagtagaa 300
tcttcttgtg agtggtgtgg gagtaggcaa cctggcattg aaacgagaga aagagagtca 360
gaaccagaag acaaataaaa agtatgcaac aaacaaatca aaatcaaagg gcaaaggctg 420
gggttggctc aattggttgc tacattcaat tttcaactca gtcaacggtt gagattcact 480
ctgacttccc caatctaagc cgcggatgca aacggttgaa tctaacccac aatccaatct 540
cgttacttag gggcttttcc gtcattaact cacccctgcc acccggtttc cctataaatt 600
ggaactcaat gctcccctct aaactcgtat cgcttcagag ttgagaccaa gacacactcg 660
ttcatatatc tctctgctct tctcttctct tctacctctc aaggtacttt tcttctccct 720
ctaccaaatc ctagattccg tggttcaatt tcggatcttg cacttctggt ttgctttgcc 780
ttgctttttc ctcaactggg tccatctagg atccatgtga aactctactc tttctttaat 840
atctgcggaa tacgcgttgg actttcagat ctagtcgaaa tcatttcata attgcctttc 900
tttcttttag cttatgagaa ataaaatcat ttttttttat ttcaaaataa accttgggcc 960
ttgtgctgac tgagatgggg tttggtgatt acagaatttt agcgaatttt gtaattgtac 1020
ttgtttgtct gtagttttgt tttgttttct tgtttctcat acattcctta ggcttcaatt 1080
ttattcgagt ataggtcaca ataggaattc aaactttgag caggggaatt aatcccttcc 1140
ttcaaatcca gtttgtttgt atatatgttt aaaaaatgaa acttttgctt taaattctat 1200
3
CA 02578463 2007-03-02
tataactttt tttatggcaa aaatttttgc atgtgtcttt gctctcctgt tgtaaattta 1260
ctgtttaggt actaactcta ggcttgttgt gcagtttttg aagtataacc atgg 1314
<210> 7
<211> 22
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: PCR Primer
<400> 7
ttcgagtata ggtcacaata gg 22
<210> 8
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: PCR Primer
<400> 8
cttcgctgag gacatggac 19
<210> 9
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: PCR Primer
<400> 9
gagttgtcgc tgttgttcga c 21
<210> 10
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: PCR Primer
<400> 10
aacacagcat ccgcattgcg 20
<210> 11
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: PCR Primer
<400> 11
aggagtgcag aatcagatca g 21
<210> 12
<211> 20
4
CA 02578463 2007-03-02
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: PCR Primer
<400> 12
gctgatcgaa ccagatggag 20
<210> 13
<211> 23
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: PCR Primer
<400> 13
ctgtacagtt aaacagtagt tct 23
<210> 14
<211> 2165
<212> DNA
<213> Glycine max
<400> 14
atcgatagag acatgttatt cacaaaccat aaaatgatgg ctaaaattgg tgtgattgga 60
acgatatctg tttattatga tttcagggcg caaaaatgcg agtacttaat aaaattttac 120
atttaaatta gaattttttt tatcaataaa tattaattta ttagttttat tagaaatatt 180
aattagaaaa ttttgaatcc ccgatttctc ctccttttct tcgctattca tcattttcta 240
accaaaccaa tcttatatgt tcttcaaatt agaacttgaa attattaatt ataattaaac 300
tgaaaacaat ttggtatcaa ttcatataca tgcttagtaa taaaatgcga taattaattg 360
ataaatctgc aaaagatttt acaaatatct ttcagaaaaa attaataaca aattttgtcg 420
ttttcatggt gttggtctga ggaggatttg gcactataga actctcctac ggaccattct 480
ttgcacttca actaaacgat ggtcagaatt ggtggggatt ttatattcaa gcatatccct 540
ttcaaaactt cctacttact tcgtgcgttc ggtaatcggt aacattagac tttcaaaatc 600
atttttaacc cctaaacagt aaatttgaag gacaaaaata atatttttca aatttgatag 660
actatttttt ttttgtaatt tgacgaacca aaaccagatt tatcctgaat tttaggaacc 720
acagatgtaa ctaaaccaat atttatttat tttctaaaac aaaatttcat ggcagcatgc 780
ctcagcccat gaaaaaaacc ttataaaaat atctacacat tgaccattga aaagttcgtt 840
ctcccatggg taaccagatc aaactcacat ccaaacataa catggatatc tccttaccaa 900
tcatactaat tattttgggt taaatattaa tcattatttt taagatatta attaagaaat 960
taaaagattt tttaaaaaaa tgtataaaat tatattattc atgatttttc atacatttga 1020
ttttgataat aaatatattt tttttaattt cttaaaaaat gttgcaagac acttattaga 1080
catagtcttg ttctgtttac aaaagcattc atcatttaat acattaaaaa atatttaata 1140
ctaacagtag aatcttcttg tgagtggtgt gggagtaggc aacctggcat tgaaacgaga 1200
gaaagagagt cagaaccaga agacaaataa aaagtatgca acaaacaaat caaaatcaaa 1260
gggcaaaggc tggggttggc tcaattggtt gctacattca attttcaact cagtcaacgg 1320
ttgagattca ctctgacttc cccaatctaa gccgcggatg caaacggttg aatctaaccc 1380
acaatccaat ctcgttactt aggggctttt ccgtcattaa ctcacccctg ccacccggtt 1440
tccctataaa ttggaactca atgctcccct ctaaactcgt atcgcttcag agttgagacc 1500
aagacacact cgttcatata tctctctgct cttctcttct cttctacctc tcaaggtact 1560
tttcttctcc ctctaccaaa tcctagattc cgtggttcaa tttcggatct tgcacttctg 1620
gtttgctttg ccttgctttt tcctcaactg ggtccatcta ggatccatgt gaaactctac 1680
tctttcttta atatctgcgg aatacgcgtt ggactttcag atctagtcga aatcatttca 1740
taattgcctt tctttctttt agcttatgag aaataaaatc attttttttt atttcaaaat 1800
aaaccttggg ccttgtgctg actgagatgg ggtttggtga ttacagaatt ttagcgaatt 1860
ttgtaattgt acttgtttgt ctgtagtttt gttttgtttt cttgtttctc atacattcct 1920
taggcttcaa ttttattcga gtataggtca caataggaat tcaaactttg agcaggggaa 1980
ttaatccctt ccttcaaatc cagtttgttt gtatatatgt ttaaaaaatg aaacttttgc 2040
CA 02578463 2007-03-02
tttaaattct attataactt tttttatggc aaaaattttt gcatgtgtct ttgctctcct 2100
gttgtaaatt tactgtttag gtactaactc=taggcttgtt gtgcagtttt tgaagtataa 2160
agatg 2165
<210> 15
<211> 1574
<212> DNA
<213> Glycine max
<400> 15
atcgatagag acatgttatt cacaaaccat aaaatgatgg ctaaaattgg tgtgattgga 60
acgatatctg tttattatga tttcagggcg caaaaatgcg agtacttaat aaaattttac 120
atttaaatta gaattttttt tatcaataaa tattaattta ttagttttat tagaaatatt 180
aattagaaaa ttttgaatcc ccgatttctc ctccttttct tcgctattca tcattttcta 240
accaaaccaa tcttatatgt tcttcaaatt agaacttgaa attattaatt ataattaaac 300
tgaaaacaat ttggtatcaa ttcatataca tgcttagtaa taaaatgcga taattaattg 360
ataaatctgc aaaagatttt acaaatatct ttcagaaaaa attaataaca aattttgtcg 420
ttttcatggt gttggtctga ggaggatttg gcactataga actctcctac ggaccattct 480
ttgcacttca actaaacgat ggtcagaatt ggtggggatt ttatattcaa gcatatccct 540
ttcaaaactt cctacttact tcgtgcgttc ggtaatcggt aacattagac tttcaaaatc 600
atttttaacc cctaaacagt aaatttgaag gacaaaaata atatttttca aatttgatag 660
actatttttt ttttgtaatt tgacgaacca aaaccagatt tatcctgaat tttaggaacc 720
acagatgtaa ctaaaccaat atttatttat tttctaaaac aaaatttcat ggcagcatgc 780
ctcagcccat gaaaaaaacc ttataaaaat atctacacat tgaccattga aaagttcgtt 840
ctcccatggg taaccagatc aaactcacat ccaaacataa catggatatc tcattaccaa 900
tcatactaat tattttgggt taaatattaa tcattatttt taagatatta attaagaaat 960
taaaagattt tttaaaaaaa tgtataaaat tatattattc atgatttttc atacatttga 1020
ttttgataat aaatatattt tttttaattt cttaaaaaat gttgcaagac acttattaga 1080
catagtcttg ttctgtttac aaaagcattc atcatttaat acattaaaaa atatttaata 1140
ctaacagtag aatcttcttg tgagtggtgt gggagtaggc aacctggcat tgaaacgaga 1200
gaaagagagt cagaaccaga agacaaataa aaagtatgca acaaacaaat caaaatcaaa 1260
gqgcaaaqgc tggggttggc tcaattggtt gctacattca attttcaact cagtcaacgg 1320
ttgagattca ctctgacttc cccaatctaa gccgcggatg caaacggttg aatctaaccc 1380
acaatccaat ctcgttactt aggggctttt ccgtcattaa ctcacccctg ccacccqgtt 1440
tccctataaa ttggaactca atgctcccct ctaaactcgt atcgcttcag agttgagacc 1500
aagacacact cgttcatata tctctctgct cttctcttct cttctacctc tcaaqttttt 1560
gaagtataaa gatg 1574
<210> 16
<211> 719
<212> DNA
<213> Glycine max
<400> 16
agatcaaact cacatccaaa cataacatgg atatctcctt accaatcata ctaattattt 60
tgggttaaat attaatcatt atttttaaga tattaattaa gaaattaaaa gattttttaa 120
aaaaatgtat aaaattatat tattcatgat ttttcataca tttgattttg ataataaata 180
tatttttttt aatttcttaa aaaatgttgc aagacactta ttagacatag tcttgttctg 240
tttacaaaag cattcatcat ttaatacatt aaaaaatatt taatactaac agtagaatct 300
tcttgtgagt ggtgtgggag taggcaacct ggcattgaaa cgagagaaag agagtcagaa 360
ccagaagaca aataaaaagt atgcaacaaa caaatcaaaa tcaaagggca aaggctgggg 420
ttggctcaat tggttgctac attcaatttt caactcagtc aacggttgag attcactctg 480
acttccccaa tctaagccgc ggatgcaaac ggttgaatct aacccacaat ccaatctcgt 540
tacttagggg cttttccgtc attaactcac ccctgccacc cggtttccct ataaattgga 600
actcaatgct cccctctaaa ctcgtatcgc ttcagagttg agaccaagac acactcgttc 660
atatatctct ctgctcttct cttctcttct acctctcaag tttttgaagt ataaagatg 719
<210> 17
<211> 6975
6
L
08T~ 4440b6Obb0 qbPeboaEPe obooEbobO4 OE04404eb6 bePe6EeoEe 4bbobb4460
OZT~ 6o8qleqEo6 beeo6ojooe bobqq44ebo o6oqqqeeb6 qeabbeoEeb 455o46oq6o
090C 060be3qbob 3496444045 obooeoae46 4ele6blob6 IP064bqbeo jeq6e6eP6q
000~ bebb4b4eoe booeoe4b4e eoqoeob;ob bboobe44bo e4ebb4bobb oe4eebaoeo
066Z 4Eo4Eq4e6o obPo}eab4o eee6ebbeob bqoobbqo4q oeEbePeeab b4oe4bbeeb
088Z Ebeo66oeee 65444e6086 06eeeOD464 e4b6qPbboP qaeq4baoPe 63006464ob
0Z8Z U)4Pb44404 o4ebobeo4e aoe4ebooEo P040boPbob 4ojqbjPejb qpPoqbobqo
09LZ oPa4Pboz4b oboeboooPb oqoEeE4bob oeeobeebbo bb4oeoobo4 44e4eeb6bo
00LZ Eo645ePobo o4boooe4eb 6ooeeboeeo o64qej6eb5 q64ebqbbqb obePoaoeoo
069Z eeeeEDE646 050bea ebqo 6ebeeeqqEb 06beo8qqoE obObeeobeo qoeeP66660
08SZ eeo4beo6bE 6eebabeoP4 b}oeebeeeb aobeeneeob b6obPP6oq3 jbbqqeobbP
OZSZ 44qoqo4ooP Eq3qobboqb 4obqob4o2e eb4Pb44Ebq bb4ba4Eobb 4eDeeb4e6e
096Z o656q*e5o4 ob4Pbe6ee6 qo6oe4qooo e4qPobojoo eqftoe4=4 oEEoobbbbq
006Z 4e664oebbq ee4qEoboPo oeboPobqbb qeb4obqboE PqE6ojqE66 ePeob646oe
06~Z 440eb6064P beeb4Po4bo qbb4}qob6} aP4440eqoq 4630EEeoEn OeP44P5400
08ZZ 44bEoeEoab bbeab4beob b4beo4bbao 4eobbo4bDb 0440boOoP4 34P4eb4b4b
OZZZ ebeoPbeoob PEPPoobEoe oqbobqb4oe Ebqeqoqa4e 44bbeebqbb booeeobbqo
09TZ qDoeoboo4e e545bq6Eeo bqqqoeb6bo beqoeobbee oebb4oPEa5 q4bb465eaP
00TZ eo4eb6o64E bqbob}oeeb 44babeoqb4 Pbjb64eEoo b546bq6beo 66joe6qjbq
ObOZ oqboboeooe e4b4oPbPea boba46qeab oeb4bbqboo e3.4e4eboeb b4bbb4aoeo
086T eebooboEoo eoeJ04064e e4bobPOb04 E004eabboo b4e4aee444 0q4qPb4e3a
0Z6T ;qoeqaoqbP o6EPePe6ee obboeeeebo ebaoP44e64 654eebbboo booo4eqoeb
098T eob54oeeb4 oeeboEeoPe b4bqbqq4bo oeo4e4boeq 645eeeebbb aab44Pqj6q
0081 e4booboPo4 b4ebaobeeb q4qeooboe4 e}abbobbbe o4eo6Pbb4e b4beebbea4
06LI eEjPEo4b55 46qbePeobb oeqqeoqoEo 4bbO54E604 4}bob4ob4b oqe4bobeoo
0B9T 66eobb544b 6PEebD32qE 44404beebO 535e04P4bb 404boPPObb bOb4e4lEe4
OZ9T boq4e4ebeo 64eboobolq beoqeboEeq 44qbeabbeo ob464ob4qe EobbboobPP
09ST ebeeoe44bo bobeeebb64 b644bobeo4 e644eebb4b 4oEeeebabo 4E66404beo
OOST 3420665464 oobbzEbo4o PeseEEo4ee Ebqboooeeo oooeEebEqb qoojboeq6b
066I 4EO0eeP44e 4404404044 4qoqoeebee eEoeEqPoob Eo44oeq4e4 olo444eo4o
08~T 6PbOaobOeo Pbbebe6614 4e044qE044 6EebbePqe4 P40400q400 32bEPObO44
OZ~i 034e4oP303 4eeDe060eb }ebbbPE460 ebWeo34o4 EDeb4bqeb4 4ebb46eeOb
09ZI ePPO4404bD EOOee3044b DebEEbEEEe Pbb4b34EDb PbbEbDeDDD POOOOOebb4
00ZT ebeeEoool6 b46PoEboob q040064PbPeoq:teo4e4o 66Eeebbeee Tebo54-4e04
06TT P00bleEPOe 400jo554bb eebbePEebe 4beOebbeEP b04eo440Eo q5404elobe
080T naob44eoaq qP6bo4ooqo oeeEbbbo44 4eeqeb6eeE oEeoq444oE beb44e4o6b
0Z0T beEEooEbeP bea4a4beoe qPbeePo4b4 ePEeEoo4oe 404bb4040e OPbOeobebb
096 46b4POPEOq b04404eeee beebeEoPbI Eeo4oeboeq 44qa4bebeo P420446POP =
006 ebobbloebe eb4booba4o eebPoeEqa4 Ebbe6o4eee oqqe6ePqoq bebb4eobqe
068 oobbee4o4e P64oE4oaq4 beqb6PeEee 4oqoqbe6bq 4ebebEqee4 beeobbeeoo
O8L eee4Eo44ob 44abbeeo44 Pboebb4e4b E=44e4oeq beEbeo}ebE e043444e4e
OZL oebeeebPbP aeoe2beeo4 eob4qEe4oe 5beoq4P6ee Peo46eobqE beee44bbEe
099 beea=44oo EqePeoqEbe 66Eooqo4ee oEP4bebaoo e4a3e5vebe Ea}e3}oq66
009 Pobeoboe4D a6bebebe44 b64Ebeaeoo oebqobqee6 eeeben444E e04404006E
0t5 44b6eebe64 ee4Pb4oeao eo4qbqe4oe aE6aEb4bbe eb4bbeP6o4 4beebbEebb
086 e4o4E6oe4q qo4eqDqoo4 }qeboebbqe eoe44ebE6b oooobqqqoa 404ebebPee
OZ6 eoooobeoo4 ebq6eobPoa obbeeeeooe Ebbeqoboeo 436b40E300 bEEeee06ee
09~ eooeooobeP oEeqa*obbP eqo64qqobe 6eEooobeeo 40eea4obee bebbeePaoo
00~ oqPaq4aee6 4q6beaebPo PePobeo4be eaeeoeoeqb obaEbqobeo beobebeeob
ObZ bebeoe44e4 oeoob4q4eE PbEEoeoeob 446ebbooo4 4bo6boeeeo eEobbobbEe
08T EoP4644566 54ae6jeboP 4e4E468005 qEoeoo4bbo eEqE46q54q boeloEqoee
0ZI 04e4Ea4000 eoeeoqqelb 650qEPbPob Pbe0404001 ob4q6eoeob eoEE4406e6
09 qoqeeeeeoe 0004eEoaqq Peao4ebbeb e434o64440 bee4be4oeo 4eob4e4eeb
LT <006>
pTwsPTd:aouanbas TeTOT3TI-W 3o uoTqdiaasea <~ZZ>
<0ZZ>
aouanbag TETOTJT43V <~TZ>
VNQ <ZTZ>
Z0-~0-LOOZ ~9V8LSZ0 VO
8
08L9 6044eaa044 4eeoe444ba ee44eqeeee aeel4e4eee eaeE44q4re babaeE4q4e
OZL9 EeeeapEqq4 ebqabeb}ee eeEeq;b644 e4aabbaq44 eboabq444e 66bpeqe444
0999 E6qqqqa44e qa4bba4a4e 4030eea4ae apeaeebb4a eeeaa44bq4 a4aebb4be4
0099 Ee}qqaq#a Eaaqbebbq4 bae6444aoa ba4q44qbba e6Eqeb4aaa ba4eaobb64
0659 be4bapo44b b4eb4b66E4 4eb44aEeeE eeaoaaEba4 aaeabbae44 4abq6aq;4e
0869 baa;qbb5e3 43oo34e05b 6804eee4al abeeaqbaao o444abbaab a44bapaoba
OZ69 4334400440 0044044406 04440340ba oababelaoo babeoab4qa paEqabaaeb
09~9 qbabeobabo E44bb4bb46 qbbbabbaba beE44Eabob bobe4b4ooo 606oeb4oae
00~9 a0b4beEEeb 0000444eoe obobaoaqbb 65e4eeeaee eqeeEEe6e4 44e}64ep6q
06Z9 q4p4eae4Eb ba5ebjpa4a 4544eqqbbb e34e444eob epb4qp}jE4 EEa qqqqqoa
Og19 44040plea4 aE4eeb4q54 aeebbaeaeb obbbee4eeb b6ppeeeEa5 oabjeEeeob
0ZT9 bee66eaeee eeabeb4bbb qaqqqbabpo aEoqqqaEqq 44aqeobeaq qoqe5jaeea
0909 aaeabi6a4a eaaope4b4p ba}jbEoa3E bpb44b4abo oeq4a4pb5e pa4a4apepe
0009 bab6b6alla qj6aeeeebb 44Eaqea4a6 qbeepEqqla epbeobp4ea paa6aboae4
0665 ee4ebbbae4 Eea4babbaa 0b44043643 bebaaebobb 064eqbqbeq ep6e64a3qp
088S a4beeaaeea 43e4bpbqb6 40e64b4044 44064e6eeq bo3qeaab4e 04643e4434
0Z8S a44peqeab4 aeabeabbae 44b1)qEa4ae a4ej4b;bea 6oa6644bee }beebEa4b4
09LS 3504ebaa4a 34bb044004 abE446babe eeeeeob4b4 4164ea00004 eb4eaea4be
OOLS babbEpa4eb aeEaaa44bb aa4abEaq4e aqqabb4E4b bq44ba46a; abaea4b4bb
0Y9S q6a4eobbea eqab44eaab 44b44boeEo 5ab444be4E E44beaa6aq 4belbee4bp
0855 be4obpe6bb oobqj64qEe q4E4a4bpaa 4Eaa4aabaa 4e444aeea6 4an4bb4bee
OZSS beabobeboa 565eEbboab eaobeaoeee jEeabEo4eq 4qEbeoo4ab baaea4abaE
0965 aaaebebaba ap4eb4eaob 4ob4beaaoa bb4a4Eoae4 4o666ebbba e4ebapqaeE
006S qe6pq5q5o4 baaaa3oe54 aa54qbeqeo a4ea4a6o44 4e4a454a4p 5a6ea4ole4
06~S aaeabbebqb po4Eel4a64 eeaoE4l62a eb3a4bb4qa e2eqbeb4eq e3e462eEqa
OgZS qeEaqeeeqq q4bee84eee eeq4EepI44 4aa3ebeaoo ea44o4Eb6e pEEea4e4jE
OZZS 6pblea4bbi 4qjp6bbeeq }baEa4aeEp ebaeebbqbe a3abaebqa4 b5bbaeqa34
09TS 43a4p6444a alebeebeea qaqebbeeEp epebeaba6a pqqebeabea beeab44454
00TS 44444#6}b ba6p4bb4ab aaEaaepeae epobbao4eb 44aqabp4bb q46p6peeee
060S bb04400e44 6eaabpeb4o b4a4aba64a qe466434e4 beaebbeebe aapae4abba
0866 E4aee4aobb q6646ee644 a4q6ebpap4 ob4b6abbe4 b4p4b6pbab p6eo6eq4eb
0Z66 beape4bb4a eaabeabeab bqaeaaba4e 44apbapapb ee4bbaaoee 0046e64404
0986 bo4e43ee4b baa4e44aab abqabaaeba oabea44boa aoaaeebaea b4b4b4obbb
0086 4abEeaa4ab a4qba4bbe4 b4bbaj4beo qa4e4bbe4b 4obaeo4abp 4ea4a44406
06L6 abb4babeeb bba44aaa4a 444aabaa4b 4oae4ebbaa e44abaab4o aaebaaqqb4
0896 ao4a4obab4 baqaaaaabE Ebb4aaaaaq q4babbEaae 4ebeeE4E4a ebbeapbaaa
0Z96 eeEbabb4bb ebaa4bpEa4 abaeba4eeE eEneaqeabE baeb4aaaaa abao4abbeq
09gp Eoa4444qba bbqab446o6 aob6eaepEq baaeebbEao bbeeeeabeo abbeeeeobe
OOS6 bqb4Eopebe eEbbeaboEE 4ebb6bEo4e ebeaeoa4e4 qbbae4ee4b bobbepea40
0666 ea4abeaqpj bbabebabba b4ob6a4q6o 46bo3obob4 aba4aeblae 0406040044
08~6 abaa44a4ab abb544e46a b4446bab6e bpbbbbabob oEeaobb04E Eb4ee44Ea5
OZ~6 4abeaabqba 4bqaaeee6b 604beaa444 obaaab4aea 4050b44bo6 44ee44e3Ea
09Z6 4aee4abebq 6E54eelaab 46bbb4aabp ee}b}bEeEq eobeeb6oob ebae4eoeEa
OOZ6 paea044Eeo e0406004e4 4644eeEb4b 4540044464 abpleaqb6q eaqep4ba44
pyTV epeaabbq4a eboab6ebeq 4e4baqbeb4 be4ewoa4a beb0eba4b6 a4eap4bbpE
0806 epaeeqaebq ab5pbp604p pabebqebae bqEeqbqbqa eqbb0440ee eaqebaqEbe
OZOb 4ae44b4p43 qea4b4bbab aba604244e Ee4ebbe4aE epabababp4 e4eepeaEEE
096E ebp4pbabae qpeq43Eae4 pqlepabaaa jbebp4qEb4 eq4q4qb6bq ebpbq244qe
006E 346ap6490b 4pp4bqpape 4iee4ep464 eabeE4q6oE j4eEbq4b4a 444ee4e4ea
Otg~ 4p44pb4eba b4aajb6ao6 446qao4pE6 q4Ebee44oq 44beeE4eea 66143eoeee
08L~ a44balp6pa 6pebo46064 6e6beebeee 4beb64bbqe 6eboe66eeq eea4444boe
OZLE aaae4ab4bb q4q4ba44eb 440pbobeao 44beo44oee 4bb44e54bo 6e004E04ee
099~ pabb4apeao 4pe44p6pea b6eeebbb6o b4eaq4e44e 4esa4ea44E 3b4EaboEa4
009~ aoaaeqaepb o4eaepoo64 4e4abeab44 eeobbaba4b 4ebbebb44b 04ab4E44ee
06S~ bee64a4q44 4eqaba4eoe ebbqeaeeae b4aaab4eab 4ea4eab4ba Ee4ea4aaea
086~ 4baaeabe4a ob4ae4eebb beb444aeee b4p4aeb4ap 4Eb4e4EEb4 Eb4eeab4eE
OZ6~ bbeo644a43 qa44ee3434 bab44Ebb4b 4ba6eabaab 44bbeebbaa bapubaeoap
09~~ bezEeeabeb eeaaybnpEa b444boE440 eaeeaaaEbe b4bE4eE403 e4bbb6boao
00~E a44eebbbb4 bba4aabeae 4abba4ba4e aaEababbW 04040e8ope ,~ppb~EE~E
06ZE eeabbebbbe abeabaaeee eE64bbaq4a Ee64eob54a Ebb4050eEp peo640b4o4
Z0-~0-LOOZ ~9V8LSZ0 VO
CA 02578463 2007-03-02
= ~ .
ccattcaggc tgcgcaactg ttgggaaggg cgatcggtgc gggcctcttc gctattacgc 6840
cagctggcga aagggggatg tgctgcaagg cgattaagtt gggtaacgcc agggttttcc 6900
cagtcacgac gttgtaaaac gacggccagt gccaagctga cttggtcagc ggccgcagat 6960
ttaggtgaca ctata 6975
<210> 18
<211> 3985
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:chimeric gene
<400> 18
aagctttgct ctagatcaaa ctcacatcca aacataacat ggatatcttc cttaccaatc 60
atactaatta ttttgggtta aatattaatc attattttta agatattaat taagaaatta 120
aaagattttt taaaaaaatg tataaaatta tattattcat gatttttcat acatttgatt 180
ttgataataa atatattttt tttaatttct taaaaaatgt tgcaagacac ttattagaca 240
tagtcttgtt ctgtttacaa aagcattcat catttaatac attaaaaaat atttaatact 300
aacagtagaa tcttcttgtg agtggtgtgg gagtaggcaa cctggcattg aaacgagaga 360
aagagagtca gaaccagaag acaaataaaa agtatgcaac aaacaaatca aaatcaaagg 420
gcaaaggctg gggttggctc aattggttgc tacattcaat tttcaactca gtcaacggtt 480
gagattcact ctgacttccc caatctaagc cgcggatgca aacggttgaa tctaacccac 540
aatccaatct cgttacttag gggcttttcc gtcattaact cacccctgcc acccggtttc 600
cctataaatt ggaactcaat gctcccctct aaactcgtat cgcttcagag ttgagaccaa 660
gacacactcg ttcatatatc tctctgctct tctcttctct tctacctctc aaggtacttt 720
tcttctccct ctaccaaatc ctagattccg tggttcaatt tcggatcttg cacttctggt 780
ttgctttgcc ttgctttttc ctcaactggg tccatctagg atccatgtga aactctactc 840
tttctttaat atctgcggaa tacgcgttgg actttcagat ctagtcgaaa tcatttcata 900
attgcctttc tttcttttag cttatgagaa ataaaatcac ttttttttta tttcaaaata 960
aaccttgggc cttgtgctga ctgagatggg gtttggtgat tacagaattt tagcgaattt 1020
tgtaattgta cttgtttgtc tgtagttttg ttttgttttc ttgtttctca tacattcctt 1080
aggcttcaat tttattcgag tataggtcac aataggaatt caaactttga gcaggggaat 1140
taatcccttc cttcaaatcc agtttgtttg tatatatgtt taaaaaatga aacttttgct 1200
ttaaattcta ttataacttt ttttatggct gaaatttttg catgtgtctt tgctctctgt 1260
tgtaaattta ctgtttaggt actaactcta ggcttgttgt gcagtttttg aagtataacc 1320
atggtacgtc ctgtagaaac cccaacccgt gaaatcaaaa aactcgacgg cctgtgggca 1380
ttcagtctgg atcgcgaaaa ctgtggaatt gatcagcgtt ggtgggaaag cgcgttacaa 1440
gaaagccggg caattgctgt gccaggcagt tttaacgatc agttcgccga tgcagatatt 1500
cgtaattatg cgggcaacgt ctggtatcag cgcgaagtct ttataccgaa aggttgggca 1560
ggccagcgta tcgtgctgcg tttcgatgcg gtcactcatt acggcaaagt gtgggtcaat 1620
aatcaggaag tgatggagca tcagggcggc tatacgccat ttgaagccga tgtcacgccg 1680
tatgttattg ccgggaaaag tgtacgtatc accgtttgtg tgaacaacga actgaactgg 1740
cagactatcc cgccgggaat ggtgattacc gacgaaaacg gcaagaaaaa gcagtcttac 1800
ttccatgatt tctttaacta tgccggaatc catcgcagcg taatgctcta caccacgccg 1860
aacacctggg tggacgatat caccgtggtg acgcatgtcg cgcaagactg taaccacgcg 1920
tctgttgact ggcaggtggt ggccaatggt gatgtcagcg ttgaactgcg tgatgcggat 1980
caacaggtgg ttgcaactgg acaaggcact agcgggactt tgcaagtggt gaatccgcac 2040
ctctggcaac cgggtgaagg ttatctctat gaactgtgcg tcacagccaa aagccagaca 2100
gagtgtgata tctacccgct tcgcgtcggc atccggtcag tggcagtgaa gggccaacag 2160
ttcctgatta accacaaacc gttctacttt actggctttg gtcgtcatga agatgcggac 2220
ttacgtggca aaggattcga taacgtgctg atggtgcacg accacgcatt aatggactgg 2280
attggggcca actcctaccg tacctcgcat tacccttacg ctgaagagat gctcgactgg 2340
gcagatgaac atggcatcgt ggtgattgat gaaactgctg ctgtcggctt taacctctct 2400
ttaggcattg gtttcgaagc gggcaacaag ccgaaagaac tgtacagcga agaggcagtc 2460
aacggggaaa ctcagcaagc gcacttacag gcgattaaag agctgatagc gcgtgacaaa 2520
aaccacccaa gcgtggtgat gtggagtatt gccaacgaac cggatacccg tccgcaagtg 2580
cacgggaata tttcgccact ggcggaagca acgcgtaaac tcgacccgac gcgtccgatc 2640
acctgcgtca atgtaatgtt ctgcgacgct cacaccgata ccatcagcga tctctttgat 2700
gtgctgtgcc tgaaccgtta ttacggatgg tatgtccaaa gcggcgattt ggaaacggca 2760
9
CA 02578463 2007-03-02
gagaaggtac tggaaaaaga acttctggcc tggcaggaga aactgcatca gccgattatc 2820
atcaccgaat acggcgtgga tacgttagcc gggctgcact caatgtacac cgacatgtgg 2880
agtgaagagt atcagtgtgc atggctggat atgtatcacc gcgtctttga tcgcgtcagc 2940
gccgtcgtcg gtgaacaggt atggaatttc gccgattttg cgacctcgca aggcatattg 3000
cgcgttggcg gtaacaagaa agggatcttc actcgcgacc gcaaaccgaa gtcggcggct 3060
tttctgctgc aaaaacgctg gactggcatg aacttcggtg aaaaaccgca gcagggaggc 3120
aaacaat.gaa tcaacaactc tcctggcgca ccatcgtcgg ctacagcctc ggtggggaat 3180
tccccggggg tacctaatag tgagatccaa cacttacgtt tgcaacgtcc aagagcaaat 3240
agaccacgna cgccggaagg ttgccgcagc gtgtggattg cgtctcaatt ctctcttgca 3300
ggaatgcaat gatgaatatg atactgacta tgaaactttg agggaatact gcctagcacc 3360
gtcacctcat aacgtgcatc atgcatgccc tgacaacatg gaacatcgct atttttctga 3420
agaattatgc tcgttggagg atgtcgcggc aattgcagct attgccaaca tcgaactacc 3480
cctcacgcat gcattcatca atattattca tgcggggaaa ggcaagatta atccaactgg 3540
caaatcatcc agcgtgattg gtaacttcag ttccagcgac ttgattcgtt ttggtgctac 3600
ccacgttttc aataaggacg agatggtgga gtaaagaagg agtgcgtcga agcagatcgt 3660
tcaaacattt ggcaataaag tttcttaaga ttgaatcctg ttgccggtct tgcgatgatt 3720
atcatataat ttctgttgaa ttacgttaag catgtaataa ttaacatgta atgcatgacg 3780
ttatttatga gatgggtttt tatgattaga gtcccgcaat tatacattta atacgcgata 3840
gaaaacaaaa tatagcgcgc aaactaggat aaattatcgc gcgcggtgtc atctatgtta 3900
ctagatcgat caaacttcgg tactgtgtaa tgacgatgag caatcgagag gctgactaac 3960
aaaaggtaca tcggtcgacg agctc 3985
<210> 19
<211> 3684
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:chimeric gene
<400> 19
aagctttgct ctagatcaaa ctcacatcca aacataacat ggatatcttc cttaccaatc 60
atactaatta ttttgggtta aatattaatc attattttta agatattaat taagaaatta 120
aaagattttt taaaaaaatg tataaaatta tattattcat gatttttcat acatttgatt 180
ttgataataa atatattttt tttaatttct taaaaaatgt tgcaagacac ttattagaca 240
tagtcttgtt ctgtttacaa aagcattcat catttaatac attaaaaaat atttaatact 300
aacagtagaa tcttcttgtg agtggtgtgg gagtaggcaa cctggcattg aaacgagaga 360
aagagagtca gaaccagaag acaaataaaa agtatgcaac aaacaaatca aaatcaaagg 420
gcaaaggctg gggttggctc aattggttgc tacattcaat tttcaactca gtcaacggtt 480
gagattcact ctgacttccc caatctaagc cgcggatgca aacggttgaa tctaacccac 540
aatccaatct cgttacttag gggcttttcc gtcattaact cacccctgcc acccggtttc 600
cctataaatt ggaactcaat gctcccctct aaactcgtat cgcttcagag ttgagaccaa 660
gacacactcg ttcatatatc tctctgctct tctcttctct tctacctctc aaggtacttt 720
tcttctccct ctaccaaatc ctagattccg tggttcaatt tcggatcttg cacttctggt 7B0
ttgctttgcc ttgctttttc ctcaactggg tccatctagg atccatgtga aactctactc 840
tttctttaat atctgcggaa tacgcgttgg actttcagat ctagtcgaaa tcatttcata 900
attgcctttc tttcttttag cttatgagaa ataaaatcac ttttttttta tttcaaaata 960
aaccttgggc cttgtgctga ctgagatggg gtttggtgat tacagaattt tagcgaattt 1020
tgtaattgta cttgtttgtc tgtagttttg ttttgttttc ttgtttctca tacattcctt 1080
aggcttcaat tttattcgag tataggtcac aataggaatt caaactttga gcaggggaat 1140
taatcccttc cttcaaatcc agtttgtttg tatatatgtt taaaaaatga aacttttgct 1200
ttaaattcta ttataacttt ttttatggct gaaatttttg catgtgtctt tgctctctgt 1260
tgtaaattta ctgtttaggt actaactcta ggcttgttgt gcagtttttg aagtataacc 1320
atggccactt tcttcgccca aacctccttc ccctcccact ctctctccaa aaccttcgat 1380
acccatttcg cccctgcccc gaaagtcaac gtctttgtga acttcagggc gaggaggcac 1440
gttggggtgc gagtttcgaa cgcgctgatc gaaccagatg gagggaagct cgtggagctt 1500
gtggtgacgg attttgagag ggatttgaag aagggtgagg ctctttcgtt gccgaggatc 1560
aagctctcaa ggattgacct tgagtgggtc catgtcctca gcgaaggatg ggccacaccc 1620
ctgaaaggct tcatgagaga agccgagttc ctccaaacgc ttcatttcaa ctcgctccga 1680
ctcgatgatg ggtcggtcgt gaacatgtca gtgcccatcg tgctggctat tgatgatgcg 1740
CA 02578463 2007-03-02
cagaagcatc ggatcgggga taacaaaaag gttgctcttt ttgattccaa gggagacccc 1800
gttgcaattc tcaataatat tgagatttat aagcatccta aagaagaaag aatagcccga 1860
acttggggaa ccattgcccc tggcctacct tatgttgaac aaactataac caatgctgga 1920
aattggttga ttgggggtga cctagaggtc attgaaccaa ttcagtacaa tgatggactt 1980
gatcattttc gtctatctcc ggcacaactc cgtgcagagt tcacaaggcg caatgcggat 2040
gctgtgtttg ccttccagct ccggaatcct gttcacaatg gccatgcttt gctaatgact 2100
gacacccgaa agcgccttct tgagatgggc tataagaatc ctgtcctctt gcttcatcca 2160
cttggaggct acaccaaagc tgatgatgtc ccacttgatt ggcgaatgaa gcaacatgag 2220
aaggtacttg aggatggtgt tcttgatcca gagacaactg tggtatccat attcccatct 2280
cccatgcact atgctggacc cacggaggtg cagtggcatg caaaggctag gatcaatgca 2340
ggggctaact tctatatcgt tggtcgtgac cccgcaggca tgagccatcc agttgagaaa 2400
agagatctgt atgatgctga ccatggaaag aaagtattga gcatggcacc gggactagag 2460
cgtctaaaca ttcttccttt cagggttgct gcatatgaca agactcaggg taaaatggca 2520
ttctttgacc cttcaaggcc tcaggacttc ctgttcatat caggcacaaa gatgcgcaca 2580
ctggcaagga acaaagaaag tcctcctgat ggatttatgt gccctggtgg atggaaggtg 2640
ctggttgatt actatgatag cttagtactc tcaagcaacg gcaaagtgca ggaagctgtt 2700
ccagcttaat cttgtatcat atcataatgt atatatctca tgattgggag aaaccttaag 2760
cttatgtatt ctcctgctaa gacatacttc acgaggatcc tctggcccaa tctaataata 2820
ataataaatt aaaactttgg ggaggcaaaa aaaaaaaaaa aaaaaaaaaa aactcgaggg 2880
ggggcccggt acctaatagt gagatccaac acttacgttt gcaacgtcca agagcaaata 2940
gaccacgnac gccggaaggt tgccgcagcg tgtggattgc gtctcaattc tctcttgcag 3000
gaatgcaatg atgaatatga tactgactat gaaactttga gggaatactg cctagcaccg 3060
tcacctcata acgtgcatca tgcatgccct gacaacatgg aacatcgcta tttttctgaa 3120
gaattatgct cgttggagga tgtcgcggca attgcagcta ttgccaacat cgaactaccc 3180
ctcacgcatg cattcatcaa tattattcat gcggggaaag gcaagattaa tccaactggc 3240
aaatcatcca gcgtgattgg taacttcagt tccagcgact tgattcgttt tggtgctacc 3300
cacgttttca ataaggacga gatggtggag taaagaagga gtgcgtcgaa gcagatcgtt 3360
caaacatttg gcaataaagt ttcttaagat tgaatcctgt tgccggtctt gcgatgatta 3420
tcatataatt tctgttgaat tacgttaagc atgtaataat taacatgtaa tgcatgacgt 3480
tatttatgag atgggttttt atgattagag tcccgcaatt atacatttaa tacgcgatag 3540
aaaacaaaat atagcgcgca aactaggata aattatcgcg cgcggtgtca tctatgttac 3600
tagatcgatc aaacttcggt actgtgtaat gacgatgagc aatcgagagg ctgactaaca 3660
aaaggtacat cggtcgacga gctc 3684
<210> 20
<211> 3963
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:chimeric gene
<400> 20
aagctttgct ctagatcaaa ctcacatcca aacataacat ggatatcttc cttaccaatc 60
atactaatta ttttgggtta aatattaatc attattttta agatattaat taagaaatta 120
aaagattttt taaaaaaatg tataaaatta tattattcat gatttttcat acatttgatt 180
ttgataataa atatattttt tttaatttct taaaaaatgt tgcaagacac ttattagaca 240
tagtcttgtt ctgtttacaa aagcattcat catttaatac attaaaaaat atttaatact 300
aacagtagaa tcttcttgtg agtggtgtgg gagtaggcaa cctggcattg aaacgagaga 360
aagagagtca gaaccagaag acaaataaaa agtatgcaac aaacaaatca aaatcaaagg 420
gcaaaggctg gggttggctc aattggttgc tacattcaat tttcaactca gtcaacggtt 480
gagattcact ctgacttccc caatctaagc cgcggatgca aacggttgaa tctaacccac 540
aatccaatct cgttacttag gggcttttcc gtcattaact cacccctgcc acccggtttc 600
cctataaatt ggaactcaat gctcccctct aaactcgtat cgcttcagag ttgagaccaa 660
gacacactcg ttcatatatc tctctgctct tctcttctct tctacctctc aaggtacttt 720
tcttctccct ctaccaaatc ctagattccg tggttcaatt tcggatcttg cacttctggt 780
ttgctttgcc ttgctttttc ctcaactggg tccatctagg atccatgtga aactctactc 840
tttctttaat atctgcggaa tacgcgttgg actttcagat ctagtcgaaa tcatttcata 900
attgcctttc tttcttttag cttatgagaa ataaaatcac ttttttttta tttcaaaata 960
aaccttgggc cttgtgctga ctgagatggg gtttggtgat tacagaattt tagcgaattt 1020
11
CA 02578463 2007-03-02
.y ..
tgtaattgta cttgtttgtc tgtagttttg ttttgttttc ttgtttctca tacattcctt 1080
aggcttcaat tttattcgag tataggtcac aataggaatt caaactttga gcaggggaat 1140
taatcccttc cttcaaatcc agtttgtttg tatatatgtt taaaaaatga aacttttgct 1200
ttaaattcta ttataacttt ttttatggct gaaatttttg catgtgtctt tgctctctgt 1260
tgtaaattta ctgtttaggt actaactcta ggcttgttgt gcagtttttg aagtataacc 1320
atggccgttt cgagctcgca catgcgtttc acctttgagt gccgctccga tcccgatttc 1380
tcgccccccc cgccgtcctt cgacaacctc cgccgccgaa acttccgctc ctccgcagga 1440
tccggcgcgg cgtttcacgg catctcctcc ctcatcctcc gcttccctcc caacttccag 1500
cgccagctaa gcaccaaggc gcgccgcaac tgcagcaaca tcggcgtcgc gcaaatcgtc 1560
gccgottcgt ggtcgaacaa cagcgacaac tctccggccg ccggggctcc ggcgccgccc 1620
gcggccaccg ccacggacgc cgctacggtg cctctccccg tcgtcgtcgc cgccaacgag 1680
gacgtcgttg tctccgccgc ggcagacgag aacggggctg tacagttaaa cagtagttct 1740
tattcttcat ttttgaaatc cgatgcaagc aaaacgattc atgccgctga aagactgggt 1800
aggggtattg agactgatgg aattaccacc cctgtggtta acacttctgc ctactttttt 1860
aagaaaaccg ctgatctcat tgatttcaag gagaatcgtc aagtgagtta tgaatacggg 1920
cgctatggaa acccaacgac ggtggttctg gaggagaaga taagtgcatt ggagggggcc 1980
gaatcaactg tgataatggc gtctgggatg tgtgctagcg tagtcctgtt tatggcactg 2040
gttccagctg gtggacatct tgtgaccact acggattgtt ataggaagac tagaatattc 2100
attgagactt ttcttccaaa gatggggatc acgaccactg taattgatcc agcagatgtt 2160
ggagccttgg aatctgcatt ggagcagcac aatgtgtctc tattcttcac tgagtctcct 2220
accaatccat tcctgagatg tgttgatatt aagctggttt cagagctttg ccacaagaag 2280
gggactttgc tctgtattga tggtacattt gcaactccat tgaaccagaa ggcccttgcc 2340
cttggcgctg atctgattct gcactcctta acaaaataca tgggtggaca tcatgatgtc 2400
cttggtggtt gcataagtgg ttcaattaag gtggtttcgc aaattcggac tttgcaccat 2460
gttttgggtg gtacacttaa cccgaatgct gcatacctat tcatcagagg catgaaaacg 2520
ctgcatctcc gtgtacagca gcagaattca acaggaatga ggatggccaa acttttagag 2580
gcacatccca aggtgaagcg ggtctactat ccaggcttgc cgagtcaccc tgaacatgag 2640
cttgccaaga ggcagatgac tggtttcggt ggtgttgtca gttttgagat tgatgqagat 2700
ctacatacca caataaaatt tattgattca ttgaaaatcc catatattgc ggcctcgttt 2760
ggtggctgtg agagcattgt ggatcaacct gctattttgt cttactggga tcttcctcag 2820
tcagaaaggg ccaagtacaa gatttatgac aacctggttc gcttcagctt tggagttgaa 2880
gattttgagg atttgaaggc tgatgtcctg caagctctgg aagctatata gacagttttc 2940
ctgattcacc caagtttttt tcttttataa ttgtgctatt tgtttgttat cacatctggc 3000
gattcaattg aattttgatc gtctaatgtt ctgttggaat tgtgttaaga tgaatggtct 3060
ctaatttgga tgttatgaaa cttgtgatga attgttgaaa ttgaaacctc tatttgatga 3120
aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa actcgagggg gggcccggta cctaatagtg 3180
agatccaaca cttacgtttg caacgtccaa gagcaaatag accacgnacg ccggaaggtt 3240
gccgcagcgt gtggattgcg tctcaattct ctcttgcagg aatgcaatga tgaatatgat 3300
actgactatg aaactttgag ggaatactgc ctagcaccgt cacctcataa cgtgcatcat 3360
gcatgccctg acaacatgga acatcgctat ttttctgaag aattatgctc gttggaggat 3420
gtcgcggcaa ttgcagctat tgccaacatc gaactacccc tcacgcatgc attcatcaat 3480
attattcatg cggggaaagg caagattaat ccaactggca aatcatccag cgtgattggt 3540
aacttcagtt ccagcgactt gattcgtttt ggtgctaccc acgttttcaa taaggacgag 3600
atggtggagt aaagaaqgag tgcgtcgaag cagatcgttc aaacatttgg caataaagtt 3660
tcttaagatt gaatcctgtt gccggtcttg cgatgattat catataattt ctgttgaatt 3720
acgttaagca tgtaataatt aacatgtaat gcatgacgtt atttatgaga tgggttttta 3780
tgattagagt cccgcaatta tacatttaat acgcgataga aaacaaaata tagcgcgcaa 3840
actaggataa attatcgcgc gcggtgtcat ctatgttact agatcgatca aacttcggta 3900
ctgtgtaatg acgatgagca atcgagaggc tgactaacaa aaggtacatc ggtcgacgag 3960
ctc 3963
12