Note: Descriptions are shown in the official language in which they were submitted.
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
IMETI1Q'0 AND QUANTITATING MULTIPLE SUBCELLULAR
COMPONENTS
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This application claims benefit of U.S. Provisional Patent Application
Serial Number 60/612,067, filed September 22, 2004, the disclosures of which
is incorporated
by reference herein in its entirety to the extent not contrary to the present
disclosure.
BACKGROUND OF THE INVENTION
[0002] The invention relates to a method for detecting and quantitating
multiple
subcellular components of cells using immunostaining and fluorescence-labeled
in situ
hybridization techniques. In particular, the combination of immunostaining
with in situ
hybridization allows for the detection of subcellular components in cells,
such as fetal
hemoglobin in maternal blood samples. The method is useful in prenatal and/or
pre-implantation
diagnosis of genetic diseases.
[0003] . A number of techniques exist for the staining and analysis of cells
and
their components. The ability to simultaneously apply a number of such
techniques is highly
advantageous for the detailed investigation of specimens in diagnosis of
genetic disease has been
of special interest. However, combination of prior art techniques have not
given any advantages
over the single techniques applied alone. Of particular interest, for example,
the ability to
simultaneously apply immunostaining and fluorescent in situ hybridization
(FISH) analysis to a
biological specimen offers the potential to obtain quantitative data on, for
example, specific
protein and nucleic acid components of the same cell at the same time.
However, traditional or
standard immunostaining and FISH protocols are mutually exclusive. The harsh
conditions
required for successful FISH analysis are not generally compatible with the
retention of
significant recognizable antigen, or with the persistence of stable antibody
based signal for
proper detection of the cellular component. Therefore, there is a need to
develop better
techniques in the diagnosis of genetic disease using genetic targeting with
visualization and
quantitation techniques.
SUMMARY OF THE INVENTION
[0004] A single continuous method for the preparation of a biological sample
for
immunostaining and in situ hybridization analysis is provided.
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
Ryla~,tqehod for identifying multiple cellular components in a cell is
provided which method comprises:
reacting a cell sample with at least one antibody, wherein each antibody binds
to a
specific cellular component and generates a unique fluorescent signal;
treating said cell sample by in situ hybridization using one or more nucleic
acid
probes; wherein each nucleic acid probe is constructed to hybridize with a
target nucleic acid
sequence in said cell and generates a unique fluorescent signal;
generating one or more images of said reacted and treated cell sample; and
detecting and analyzing in said image(s) fluorescent signals corresponding to
both
said antibody and said nucleic acid probe.
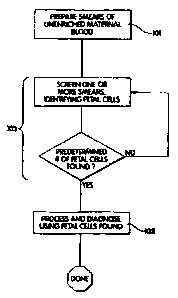
BRIEF DESCRIPTION OF THE DRAWINGS
[0010] [0006] In the accompanying drawings, in which like reference
designations indicate like elements:
Figure 1 is a flow chart summarizing the method of one embodiment of the
invention;
Figure 2 is a block diagram of an analysis system used in one embodiment of
one aspect
of the invention;
Figure 3 is a flow chart of stage I leading to detecting the first signal;
Figure 4A and 4B taken together are a flow chart of stage 111eading to
detecting the first
signal;
Figure 5 is a flow chart of detection of the second signal;
Figure 6 is a schematic representation of a variation of an apparatus
illustrating one
embodiment of the invention, using a continuous smear technique;
Figure 7 is a block diagram of an analysis and reagent dispensing system used
in one
embodiment of one aspect of the invention;.
Figure 8 is showing an outline of one embodiment of the invention wherein a
multiple
objective microscopy system;
Figure 9 is an image "composition" method;
Figure 10 is a flowchart of the calibration steps of one embodiment of the
invention;
-2-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
Aq..of the preprocessing steps of one embodiment of the invention;
and
Figures 12A and 12B are a flowchart of the main processing steps of one
embodiment of
the invention.
FIG. 13 is a photomicrograph of a combined immunostaining and FISH analysis of
cells
prepared with the method of the invention as described in Example 1 to
identify fetal
hemoglobin by immunostaining and the X and Y chromosomes using FISH in the
cells.
DETAILED DESCRIPTION OF THE INVENTION
[0007] In embodiments illustrated herein, there is provided a method for
detecting
and quantitating subcellular components of cells in a cell sample. The method
can be applied to
a variety of biological samples containing cells, for example, a blood sample,
and in particular
for the diagnosis of genetic disease in maternal blood.
[0008] In one embodiment, the method comprises producing a fluorescent signal
generated from one or more antibodies from immunostaining which signals are
unique to each
antibody used and persist following subsequent treatment of the cell sample
for fluorescent in
situ hybridization (FISH) analysis. In one embodiment, the methods comprises
selecting a
desired or unique fluorophore for the FISH probe utilized, which allows
discrete visualization
and quantitation of each and all fluorescent signals produced, both
immunohistochemical and
FISH signals fluorescent from the cell sample.
[0009] In one embodiment, a method of operating a computer system to detect
whether a genetic condition defined by at least one target nucleic acid is
present in a sample. The
method involves the use of probes and digitized images of the probes
hybridized to a sample,
together with counting objects and analysis of a statistical expectation to
detect whether the
genetic condition is present. The counting may involve, for example, counting
the number of
times a genetic abnormality is detected and comparing that count to a
statistical expectation of
the abnormality in a particular tissue type, cell type or sample. The counting
may involve
counting the number of times a genetic abnormality occurs and comparing that
count to the
number of times a cell type occurs in the same sample or to the number of
times a normal nucleic
acid occurs in the same sample. The counting may involve counting the number
of times more
than one different genetic abnormality occurs in a single cell. The computer
system also may be
used to identify cell type, count cells, examine cell morphology, etc. and
compare or correlate
-3-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
thisllinfdxirfat~cilt~;;w~til;~ri~ t+of the genetic abnormality. Various
diagnostic analysis can be
carried out.
[0011] In one embodiment, it is provided a method of operating a computer
system to detect whether a genetic condition defined by at least one target
nucleic acid is
present in a fixed sample, the method comprising: receiving a digitized image,
preferably a
color image, of the fixed sample, which has been subjected to fluorescence in
situ hybridization
under conditions to specifically hybridize a fluorophor-labeled probe to a
target nucleic acid and
fluorescent immunostaining to detect first objects of interest; processing the
image in a
computer to separate first objects, for example, a cell component; determining
first objects of
interest displaying probe associated with the target nucleic acid within
specific predetermined
characteristics; counting the first objects of interest having probe signals;
and analyzing the
count of the first objects, for example cells, with respect to a statistical
expectation to detect
whether the genetic condition is present. This method is applicable to many
genetic conditions,
including wherein the genetic condition is human trisomy 21. In addition to
the foregoing, it
will be understood that the statistical expectation can be based on a tissue
type, for example.
The computer can be used to identify the tissue type of a cell being examined,
but the tissue
type also can be known.
[0012] In some embodiments, the step of receiving further includes a step of
producing an image file of red, green and blue pixels representative of red,
green and blue
intensities at respective pixel locations within the color image received. In
some embodiments,
the step of processing further includes steps of manually selecting a
plurality of pixels within
the background; determining color intensity value ranges corresponding to the
portion of the
background; and identifying as the background those areas of the image having
color intensity
values within the ranges determined. In some embodiments, before the step of
measuring, there
may be processing in the computer to filter the color image to make color
intensity values of
dark pixels in the color image lighter and to make color intensity values of
light pixels in the
color image darker. The step of filtering may further comprise passing the
color image through
a hole filling filter; passing the filled color image through an erosion
filter; performing a
separate operation on the eroded filled color image, to define outlines around
areas; selecting
pixels within the outlines by performing a logical NOT operation; and
performing a logical
AND operation between the selected pixels and the filled color image.
[0013] In some embodiments, the genetic condition is further defined by a
ratio of
the target nucleic acid to a second nucleic acid. Then, the method further
includes identifying
second objects having specific predetermined characteristics associated with
the second nucleic
-4-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
'le.ciU;= 'aAe9 ~UoUMIng s&.cxihd11df:bjbibts identified; wherein analyzing
the count of first objects
includes finding a ratio of the count of first objects to the count of second
objects. In some
embodiments, the target nucleic acid defines a dominant trait and the second
nucleic acid
defines a corresponding recessive trait. The method in those embodiments may
include
indicating the genetic condition as possessing the dominant trait, possessing
the recessive trait,
or possessing the dominant trait and carrying the recessive trait depending on
the ratio found.
When the target nucleic acid is a rearrangement of the second nucleic acid,
the method may
further include selecting the probe to hybridize with a break region between
rearranged and
non-rearranged nucleic acids. Finally, the method may include indicating the
genetic condition
as a severity level related to the ratio found.
[0014] According to one embodiment of the invention, there is provided a
computer software product comprising: a computer readable storage medium
having fixed
therein a sequence of computer instructions directing a computer system to
count occurrences of
a target substance in a cell-containing sample which has been labeled with a
target-specific
fluorophor, the instructions directing steps of : receiving a digitized color
image of the
fluorophor-labeled sample; obtaining a color image of the fluorophor-labeled
sample; separating
objects of interest from background in the color image; measuring parameters
of the objects of
interest so as to enumerate object having specific characteristics; and
analyzing the enumeration
of objects with respect to a statistically expected enumeration to determine
the genetic
abnormality. The instructions can be made to implement all of the variations
on the methods
described above.
[0015] According to another embodiment of the invention, there is provided an
apparatus for analyzing an image of a cell-containing sample which has been
labeled with a
target-specific fluorophor, comprising: a computer system on which image
processing software
executes; and a storage medium in which is fixed a sequence of image
processing instructions
including receiving a digitized color image of the fluorophor-labeled sample,
obtaining a color
image of the fluorophor-labeled sample, separating objects of interest from
background in the
color image, measuring parameters of the objects of interest so as to
enumerate object having
specific characteristics, and analyzing the enumeration of objects with
respect to a statistically
expected enumeration to determine the genetic abnormality. Again, the
instructions can be
varied to implement all the variations described above.
100161 In yet another embodiment, there is provided a computer-implemented
method of processing body fluid or tissue sample image data, the method
comprising creating a
subset of a first image data set representing an image of a body fluid or
tissue sample taken at a
-5-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
,, ... ., .. ,,. õ ,,..,
~4~,~d#representing a candidate blob which may contain a rare cell
creating a subset of a second image data set representing an image of the
candidate blob taken at
a second magnification, the subset of the second data set representing the
rare cell, storing the
subset of the second data set in a computer memory, measuring size and color
parameters of the
objects of interest so as to identify objects having specific predetermined
characteristics
associated with the target nucleic acid, counting the objects identified in
the step of measuring,
and analyzing the count of objects with respect to a statistically expected
count to detect
whether the genetic abnormality is present.
In one embodiment, there is provided a method including the step of measuring,
processing in the computer to filter the color image to make color intensity
values of dark pixels
in the color image lighter and to make color intensity values of light pixels
in the color image
darker. Filtering may include the steps of passing the color image through a
hole filling filter;
passing the filled color image through an erosion filter; performing a
separate operation on the
eroded filled color image, to define outlines around areas; selecting pixels
within the outlines by
performing a logical NOT operation, and performing a logical AND operation
between the
selected pixels and the filled color image.
[0017] In one embodiment, a subset of a first image data set can be created by
observing an optical field of a monolayer of cells from a body fluid or tissue
sample using a
computerized microscopic vision system to detect a signal indicative of the
presence of a rare
cell. In one embodiment, the method can further produce an image file of red,
green and blue
pixels representative of red, green and blue intensities at respective pixel
locations within the
color image received. According to some aspects of the invention, the
processing further
includes manually selecting a plurality of pixels within the background;
determining color
intensity value ranges corresponding to the portion of the background; and
identifying as the
background those areas of the image having color intensity values within the
ranges determined.
In one embodiment, the signal can be measured to determine whether it is a
significant signal
level. The first and/or the second image data subsets can be transformed into
a representation
that is more suitable for control and processing by a computer as described
herein. the image
data is transformed from, for example, a Red Green Blue, (RGB) signal into an
Hue
Luminescence Saturation (HLS) signal. Filters and/or masks are utilized to
distinguish those
cells that meet preselected criteria and eliminate those that do not, and thus
identify, for
example, rare cells.
[0018] In another embodiment of the invention, there is provided a method of
operating a laboratory service, the method comprising steps of receiving a
body fluid or tissue
-6-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
tissue sample smear, immunostaining object of interest in the
smear with a fluorescent immunostain; treating the smear with a fluorescent
probe designed to
hybridize with nucleic acid sequences of diagnostic significance; operating a
computerized
microscope so that a software program automatically identifies objects of
interest having
hybridized nucleic acid sequences of diagnostic significance based on
fluorescent signals
generated by the immunostain and nucleic acid probes.
[0019] In yet another embodiment of the invention, there is provided computer
software product including a computer-readable storage medium having fixed
therein a
sequence of instructions which when executed by a computer direct performance
of steps of
detecting objects of interest having nucleic acid sequences of diagnostic
significance. The steps
encompass: creating a subset of a first image data set representing an image
of a body fluid or
tissue sample taken at a first magnification, the subset representing a
candidate blob which may
contain an object of interest, such as a cell or rare cell (less than 1 in
10,000 cells), creating a
subset of a second image data set representing an image of the candidate blob
taken at a second
magnification, the subset of the second data set representing the object of
interest, storing the
subset of the second data set in a computer memory, measuring fluorescence
associated with a
fluorescent nucleic acid probe directed to a nucleic acid sequence of
diagnostic interest that is
associated with objects of interest so as to identify objects having
predetermined characteristics
associated with the target nucleic acid; counting the objects identified in
the step of measuring;
and analyzing the count of objects with respect to a statistically expected
count to detect
whether the genetic abnormality is present.
[0020] According to one embodiment of the invention, there is provided a
method
of preparing a sample of cells for a diagnostic procedure. The sample of cells
is obtained and
fixed as a monolayer on a substrate, the sample of cells including a rare cell
which is present in
the sample at no greater than one in every 10,000 cells (i.e. no greater than
0.01 %). The
monolayer is immunostained with a fluorescent immunostain directed to the rare
cell and then
treated with a fluorescent probe directed to a nucleic acid sequence
associated with a disease
sate or abnormality. An optical field covering at least a portion of the
sample of cells is
observed using a computerized microscopic vision system for fluorescent
signals indicative of
the presence of a rare cell and the nucleic acid sequence of interest. Each
signal is detected, and
coordinates where the signals are detected are identified, for the diagnostic
procedure. The
count of rare cells displaying the nucleic acid sequence associated with a
disease state or
abnormality may be used to make a diagnosis. A tentative diagnosis may be
automatically
made by the computerized microscopic system. In one embodiment the rare cell
is present at no
-7-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
~gr~ater~.~liaa~L1D.0~=1'%a:(t~~.l~tt~li&1115. In other embodiments the rare
cell is present at no greater than
0.0001%, 0.00001% or even 0.000001%.
[0021] In another embodiment of the invention, the rare cell type to be
detected
and diagnosed is a cancer cell found in a sample of cells or tissue from an
animal or patient. The
sample can be blood or other body fluid containing cells or a tissue biopsy.
As an illustration of
this embodiment, cancer cell markers described in Section 5,infia, e.g, GM4
protein, telomerase
protein or nucleic acids, and p53 proteins or nucleic acids, may be used in
the generation of the
first or second signal, in a manner to be determined by the specific
application of the invention.
[0022] In one embodiment of the invention, when the rare cell type is present
in
the sample, the method of the invention detects the rare cell type at a
frequency of no less than
80%. In other embodiments, the detection frequencies are no less than 85%,
90%, 95% and
99%.
[0023] According to one embodiment of the invention, there is provided a
method
of preparing a sample of blood for a diagnostic procedure, which includes:
preparing a smear of
a sample of unenriched maternal blood containing a naturally present
concentration of fetal
cells; treating said smear with a fluorescent immunostain directed to said
fetal cells; treating
said smear with fluorescent nucleic acid probes directed to nucleic acid
sequences of interest;
observing an optical field covering a portion of the smear using a
computerized microscopic
vision system for a fluorescent signal indicative of the presence of a fetal
cell; and identifying,
fetal cells having nucleic acid sequences of interest by way of fluorescent
signal from said
nucleic acid probes.
[0024] In one embodiment, the signal is further processed to represent
morphological measurements of the rare cell. In another embodiment, the cells
are treated with
a label to enhance the optical distinction of rare cells from other cells. In
this embodiment, the
signal can be, for example, from a label which selectively binds to the rare
cells. In another
embodiment, the diagnostic procedure involves moving to the coordinates
identified and
magnifying the optical field until the image is of an isolated rare cell.
[0025] In some embodiments, the optical field is stepped over a sequence of
portions of the cells covering substantially all of the cells. This may be
achieved, for example,
by moving the cells on the substrate under control of the computerized
microscopic vision
system relative to a lens of the computerized microscopic vision system. In
another
embodiment, the coordinates at which the first signal was obtained are
identified, and then the
rare cell at those coordinates specifically is contacted after the coordinates
have been identified.
-8-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
[002'6].,:::ii,,:::IrY.;l 6il'i6 embodiments, the diagnostic signal can be
used to identify the rare
cell. In other embodiments, a locating signal can be used to identify the rare
cell, and the
diagnostic signal is obtained after the cell is located.
[0027] In one embodiment, the rare cell is present in the sample at no greater
than
one in every 10,000 cells (i.e., no greater than 0.01% of the cells). In other
embodiments, the
rare cell is present at no greater than 0.001%, 0.00001% or even 0.000001%. In
one
particularly important embodiment, the rare cell is a fetal cell in a sample
of cells from maternal
blood. Preferably the sample contains only a naturally present concentration
of fetal cells which
can be no greater than 0.001 %, 0.0001 %, 0.00001 %, 0.000001 % or even
0.0000001 %.
[0028] In any of the foregoing embodiments, the cells can be prepared on, for
example a microscope slide or the substrate may have a coordinate system that
can be calibrated
to the substrate so that coordinates of the rare cell identified in one step
can be returned to later
in another step. Likewise, the substrate in embodiments has a length that is
10 times its width,
the substrate being substantially elongated in one direction. The length can
even be 20 times the
width. The substrate can be a flexible film, and in one important embodiment,
is an elongated
flexible film that can carry a relatively large volume of cells, such as would
be provided from a
relatively large volume of smeared maternal blood. In any of the foregoing
embodiments, the
fluorescent signal from the immunostain and the fluorescent signal from the
nucleic acid probe
can be selected whereby they do not mask one another when both are present.
[0029] According to embodiments, such methods may employ unenriched or
enriched samples, e.g., maternal blood containing naturally present fetal
cells.
[0030] The invention will be better understood upon reading the following
detailed
description of the invention and of various exemplary embodiments of the
invention, in
connection with the accompanying drawings. While the detailed description
explains the
invention with respect to fetal cells, a rare cell type, and blood as the body
fluid or tissue
sample, it will be clear to those skilled in the art that the invention can be
applied to and, in fact,
encompasses diagnosis based on any cell type and any body fluid or tissue
sample, particularly
where the sample is deposited as a monolayer of cells on a substrate.
[0031] Body fluids and tissue samples that fall within the scope of the
invention
include but are not limited to blood, tissue biopsies, spinal fluid, meningeal
fluid, urine, alveolar
fluid, etc.. For those tissue samples in which the cells do not naturally
exist in a monolayer, the
cells can be dissociated by standard techniques known to those skilled in the
art. These
techniques include but are not limited to trypsin, collagenase or dispase
treatment of the tissue.
-9-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
[0032].:::a~ .:::RThIldri,d=,6mbodiment, the invention is used to detect and
diagnose fetal cells.
The fluorescent immunostain may be used in an exemplary embodiment to indicate
cell identity.
For example, the immunostain may be a fluorescent dye bound to an antibody
against the
hemoglobin c-chain, i.e., embryonal hemoglobin, for example. Additionally, a
metric of each
cell's similarity to the characteristic morphology of nucleated erythrocytes,
discerned using cell
recognition algorithms may be employed to define cell identity.
[0033] Diagnosing can be based on the nucleic acid probe signal (or on a
combination of a immunostain signal and nucleic probe signal).
[0034] In an exemplary embodiment, FISH comprises hybridizing the denatured
test DNA of the rare cell type, e. g. a fetal cell, with a denatured
dioxygenin (DIG)-labeled
genomic probe. The samples containing the test DNA are washed and allowed to
bind to an
anti-DIG antibody coupled to a fluorophore. Optionally, a second layer of
fluorophore (e.g.
FITC) is added by incubation with fluorophore-conjugated anti-Fab antibodies.
In one
embodiment, FISH comprises hybridizing the denatured DNA of the rare cell with
a
fluorescently labeled probe comprising DNA sequence(s) homologous to a
specific target DNA
region (s) directly labeled with a particular fluorophore.
[0035] Automated sample analysis may be performed by an apparatus and method
of distinguishing in an optical field objects of interest from other objects
and background. An
example of an automated system is disclosed in our U. S. Patent No. 5,352,613,
issued October
4,1994. Furthermore, once an object has been identified, the color, i.e., the
combination of the
red, green, blue components for the pixels that comprise the object, or other
parameters of
interest relative to that object can be measured and stored.
[0036] Automated sample analysis and diagnosis of a genetic condition may
proceed as follows: (i) receiving a digitized color image of the fixed sample,
which has been
subjected to fluorescence in situ hybridization under conditions to
specifically hybridize a
fluorophor-labeled probe to the target nucleic acid; (ii) processing the color
image in a computer
to separate objects of interest from background in the color image; (iii)
measuring parameters of
the objects of interest identifying objects having specific characteristics;
(iv) counting the
objects identified; and (v) analyzing the count of objects with respect to a
statistically expected
count to determine the genetic condition. The method is useful for diagnosing
genetic
conditions associated with an aberration in chromosomal number and/or
arrangement. Thus, for
example, the invention can be used to detect chromosomal rearrangements by
using a
combination of labeled probes which detect the rearranged chromosome segment
and the
chromosome into which the segment is translocated. More generally, as well as
trisomy, genetic
-10-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
arhpliflCa~ioiis ~rid ~e~h"ali~geY~ents including translocations, deletions
and insertions can be
detected using a method embodying this aspect of the invention in connection
with properly
selected fluorescent probes.
[0037] As used herein, "genetic abnormalities" refers to an aberration in the
number and/or arrangement of one or more chromosomes with respect to the
corresponding
number and/or arrangement of chromosomes obtained from a healthy subject, i.
e., an individual
having a normal chromosome complement. Genetic abnormalities include, for
example,
chromosomal additions, deletions, amplifications, translocations and
rearrangements that are
characterized by nucleotide sequences of, typically, as few as about 15 base
pairs and as large as
an entire chromosome. Genetic abnormalities also include point mutations.
[0038] The method is useful for determining one or more genetic abnormalities
in
a fixed sample, i. e., a sample attached to a solid support which preferably
has been treated in a
manner to preserve the structural integrity of the cellular and subcellular
components contained
therein. Methods for fixing a cell containing sample to a solid support, e.
g., a glass slide, are
well known to those of ordinary skill in the art.
[0039] The sample may contain at least one target nucleic acid, the
distribution of
which is indicative of the genetic abnormality. By "distribution", it is meant
the presence,
absence, relative amount and/or relative location of the target nucleic acid
in one or more
nucleic acids (e. g., chromosomes) known to include the target nucleic acid.
In one
embodiment, the target nucleic acid is indicative of a trisomy 21 and, thus,
the method is useful
for diagnosing Down's syndrome. In an embodiment, the sample intended for
Down's syndrome
analysis is derived from maternal peripheral blood. More particularly, cells
are isolated from
peripheral blood according to standard procedures, the cells are attached to a
solid support
according to standard procedures (see, e.g., the Examples) to permit detection
of the target
nucleic acid.
[0040] Fluorescence in situ hybridization refers to a nucleic acid
hybridization
technique which employs a fluorophor-labeled probe to specifically hybridize
to and thereby,
facilitate visualization of, a target nucleic acid. Such methods are well
known to those of
ordinary skill in the art and are disclosed, for example, in U. S. Patent No.
5,225,326; U. S.
patent application serial no. 07/668,751; PCT WO 94/02646, the entire contents
of which are
incorporated herein by reference. In general, in situ hybridization is useful
for determining the
distribution of a nucleic acid in a nucleic acid-containing sample such as is
contained in, for
example, tissues at the single cell level. Such techniques have been used for
karyotyping
applications, as well as for detecting the presence, absence and/or
arrangement of specific genes
-11-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
ms.%or karyotyping, the cells in the sample typically are allowed to
proliferate until metaphase (or interphase) to obtain a " metaphase- spread"
prior to attaching the
cells to a solid support for performance of the in situ hybridization
reaction.
[0041] Briefly, fluorescence in situ hybridization involves fixing the sample
to a
solid support and preserving the structural integrity of the components
contained therein by
contacting the sample with a medium containing at least a precipitating agent
and/or a
crosslinking agent. Exemplary agents useful for "fixing" the sample are
described in the
Examples. Alternative fixatives are well known to those of ordinary skill in
the art and are
described, for example, in the above-noted patents and/or patent publications.
[00421 In situ hybridization may be performed by denaturing the target nucleic
acid so that it is capable of hybridizing to a complementary probe contained
in a hybridization
solution. The fixed sample may be concurrently or sequentially contacted with
the denaturant
and the hybridization solution. Thus, in one embodiment, the fixed sample is
contacted with a
hybridization solution which contains the denaturant and at least one
oligonucleotide probe. The
probe has a nucleotide sequence at least substantially complementary to the
nucleotide sequence
of the target nucleic acid. The hybridization solution may optionally contains
one or more of a
hybrid stabilizing agent, a buffering agent and a selective membrane pore-
forming agent.
Optimization of the hybridization conditions for achieving hybridization of a
particular probe to
a particular target nucleic acid is well within the level of the person of
ordinary skill in the art.
[0043] In reference to a probe, the phrase "substantially complementary"
refers to
an amount of complementarity that is sufficient to achieve the purposes of the
invention, i. e.,
that is sufficient to permit specific hybridization of the probe to the
nucleic acid target while not
allowing association of the probe to non-target nucleic acid sequences under
the hybridization
conditions employed for practicing the invention. Such conditions are known to
those of
ordinary skill in the art of in situ hybridization.
[0044] The genetic abnormalities for which the invention is useful include
those
for which there is an aberration in the number and/or arrangement of one or
more chromosomes
with respect chromosomes obtained from an individual having a normal
chromosome
complement. Exemplary chromosomes that may be detected by the present
invention include
the human X chromosome, the Y chromosome and chromosomes 13,18 and 21. For
example,
the target nucleic acid can be an entire chromosome, e.g., chromosome 21,
wherein the presence
of three copies of the chromosome ("the distribution" of the target nucleic
acid) is indicative of
the genetic abnormality, Down's syndrome). Exemplary probes that are useful
for specifically
hybridizing to the target nucleic acid (e. g. chromosome) are probes which can
be located to a
-12-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
4$4 ncqstic of a genetic abnormality. See e. g., Harrison's Principles of
Internal Medicine, 12th edition, ed. Wilson et al., McGraw Hill, N. Y., N. Y.
(1991).
[0045] One embodiment of the invention is directed to the prenatal diagnosis
of
Down's syndrome by detecting trisomy 21 (discussed below) in fetal cells
present in, for
example, maternal peripheral blood, placental tissue, chorionic villi,
amniotic fluid and
embryonic tissue. However, the method of the invention is not limited to
analysis of fetal cells.
Thus, for example, cells containing the target nucleic acid may be eukaryotic
cells (e. g., human
cells, including cells derived from blood, skin, lung, and including normal as
well as tumor
sources); prokaryotic cells (e. g., bacteria) and plant cells. According to
one embodiment, the
invention is used to distinguish various strains of viruses. According to this
embodiment, the
target nucleic acid may be in a non-enveloped virus or an enveloped virus
(having a non-
enveloped membrane such as a lipid protein membrane). See, e.g., Asgari supra.
Exemplary
viruses that can be detected by the present invention include a human
immunodeficiency virus,
hepatitis virus and herpes virus.
[0046] The oligonucleotide probe may be labeled with a fluorophor (fluorescent
"tag" or "label") according to standard practice. The fluorophor can be
directly attached to the
probe (i. e., a covalent bond) or indirectly attached thereto (e.g., biotin
can be attached to the
probe and the fluorophor can be covalently attached to avidin; the biotin-
labeled probe and the
fluorophor-labeled avidin can form a complex which can function as the
fluorophor-labeled
probe in the method of the invention).
[0047] Fluorophors that can be used in accordance with the method and
apparatus
of the invention are well known to those of ordinary skill in the art. These
include 4,6-
diamidino-2phenylindole (DIPA), fluorescein isothiocyanate (FITC) and
rhodamine. See, e. g.,
the Example. See also U.S. Patent No. 4,373,932, issued February 15,1983 to
Gribnau et al., the
contents of which are incorporated herein by reference, for a list of
exemplary fluorophors that
can be used in accordance with the methods of the invention. The existence of
fluorophors
having different excitation and emission spectrums from one another permits
the simultaneous
visualization of more than one target nucleic acid in a single fixed sample.
As discussed below,
exemplary pairs of fluorophors can be used to simultaneously visualize two
different nucleic
acid targets in the same fixed sample.
[0048] The distribution of the target nucleic acid is indicative of the
genetic
abnormality. See e. g., Asgari supra. The genetic abnormalities that may be
detected include
mutations, deletions, additions, amplifications, translocations and
rearrangements. For example,
a deletion can be identified by detecting the absence of the fluorescent
signal in the optical field.
-13-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
1r,gp,~etXc sequence, a population of probes are prepared that are
complementary to a target nucleic acid which is present in a normal cell but
absent in an
abnormal one. If the probe (s) hybridize to the nucleic acid in the fixed
sample, the sequence
will be detected and the cell will be designated normal with respect to that
sequence. However,
if the probes fail to hybridize to the fixed sample, the signal will not be
detected and the cell
will be designated as abnormal with respect to that sequence. Appropriate
controls are included
in the in situ hybridization reaction in accordance with standard practice
known to those of
ordinary skill in the art.
[0049] A genetic abnormality associated with an addition of a target nucleic
acid
can be identified, for example, by detecting binding of a fluorophor-labeled
probe to a
polynucleotide repeat segment of a chromosome (the target nucleic acid). To
detect an addition
of a genetic sequence (e.g., trisomy 21), a population of probes are prepared
that are
complementary to the target nucleic acid. Hybridization of the labeled probe
to a fixed cell
containing three copies of chromosome 21 will be indicated as discussed in the
Examples.
[0050] Amplifications, mutations, translocations and rearrangements may be
identified by selecting a probe which can specifically bind to a break point
in the nucleic acid
target between a normal sequence and one for which amplification, mutation,
translocation or
rearrangement is suspected and performing the above-described procedures. In
this manner, a
fluorescent signal can be attributed to the target nucleic acid which, in
turn, can be used to
indicate the presence or absence of the genetic abnormality in the sample
being tested. The
probe may have a sequence that is complementary to the nucleic acid sequence
across the break
point in a normal individual's DNA, but not in an abnormal individual's DNA.
Probes for
detecting genetic abnormalities are well known to those of ordinary skill in
the art.
[0051] An innovative feature of an embodiment of a computer controlled system
that may be utilized is an array of two or more objective lenses having the
same optical
characteristics. The lenses are arranged in a row and each of them has its own
z-axis movement
mechanism, so that they can be individually focused. This system may be
equipped with a
suitable mechanism so that the multiple objective holder can be exchanged to
suit the same
variety of magnification needs that a common single-lens microscope can cover.
[0052] Each objective may be connected to its own CCD camera. Each camera
may be connected to an image acquisition device. For each optical field
acquired, the computer
may record its physical location on the microscopical sample. This may be
achieved through the
use of a computer controlled x-y mechanical stage. The image provided by the
camera is
digitized and stored in the host computer memory.
-14-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
puter may keep track of the features of the objectives-array in use
as well as the position of the motorized stage. The stored characteristics of
each image can be
used in fitting the image in its correct position in a virtual patchwork, i.e.
"composed" image, in
the computer memory.
[0054] The host computer system may be driven by software system that controls
all mechanical components of the system through suitable device drivers. The
software may
comprise image composition algorithms that compose the digitized image in the
computer
memory and supply the composed image for processing to further algorithms.
Through image
decomposition, synthesis and image processing specific features particular to
the specific
sample may be detected.
[0055] In one embodiment both the immunostain signals and probe signals are
detected simultaneously. The signals may be processed separately (with signals
from different
fluorophores for the immunostain and probe also being processed separately).
In an
embodiment, the simultaneous presence of both immunostain and probe signals at
a single set of
coordinates or even a single signal which results from the interaction of two
components (e. g. a
quenching of a signal by a partner' signal') may be used for diagnostic
purposes.
[0056] Generally the materials and techniques used to generate the immunostain
signal should not interfere adversely with the materials and techniques used
to generate the
second probe (to an extent which compromises unacceptably the diagnosis), and
visa versa.
Nor should immunostain or probe damage or alter the cell characteristics
sought to be measured
to an extent that compromises unacceptably the diagnosis. Finally, any other
desirable or
required treatment of the cells should generally not interfere with the
materials or techniques
used to generate the first and second signals to an extent that compromises
unacceptably the
diagnosis. Within those limits, any suitable generators of the first and
second signals may be
used.
[0057] In one embodiment of the invention, when a rare cell type is to be
detected,
the method of the invention detects the rare cell type at a frequency of no
less than 80%. In
other embodiments, the detection frequencies are no less than 85%, 90%, 95%
and 99%.
[0058] While the of single fluorophores for the tagging of an individual
allele may
create an upper limit as to the number of mutations that can be tested
simultaneously, the use of
combinatorial chemistry may be employed to the number of allele specific
mutations that can be
tagged and detected simultaneously. Chromosomal abnormalities that fall within
the scope of
the invention include but are not limited to Trisomy 21,18,13 and sex
chromosome aberrations
such as XXX, XXY, XYY. With the use of combinatorial chemistry, the methods of
the
-15-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
n6rit','-oAkhnbeYisdd:,fd idfdgnose a multitude of rearrangements, including
translocations
iv
observed in genetic disorders and cancer. Mendelian disorders that fall within
the scope of the
invention include but are not limited to cystic fibrosis, hemochromatosis,
hyperlipidemias,
Marfan syndrome and other heritable disorders of connective tissue,
hemoglobinopathies, Tay-
Sachs syndrome or any other genetic disorder for which the mutation is known.
The use of
combinatorial chemistry dyes allows for the simultaneous tagging and detection
of multiple
alleles thus making it possible to establish the inheritance of predisposition
of common
disorders, e. g. asthma and/or the presence of several molecular markers
specific for cancers, e.
g., prostate, breast, colon, lung, leukemias, lymphomas, etc.
[0059] One use of the invention is in the field of cancer. Cancer cells of
particular
types often can be recognized morphologically against the background of
noncancer cells. The
morphology of cancer cells therefore can be used as the first signal. Heat
shock proteins also are
markers expressed in most malignant cancers. Labeled antibodies, such as
fluorescently-tagged
antibodies, specific for heat shock proteins can be used to generate the first
signal. Likewise,
there are antigens that are specific for particular cancers or for particular
tissues, such as
Prostate Specific Antigen, and antibodies specific for cancer or tissue
antigens, such as Prostate
Specific Antigen can be used to generate a first signal for such cancer cells.
[0060] Thus, rare cancer cells in a background of other cells can be
identified and
characterized according to the invention. The characterization may include a
confirmation of a
diagnosis of the presence of the cancer cell, a determination of the type of
cancer, a
determination of cancer risk by determining the presence of a marker of a
genetic change which
relates to cancer risk, etc.
[0061] Markers of genetic changes enable assessment of cancer risk. They
provide
information on exposure to carcinogenic agents. They can detect early changes
caused by
exposure to carcinogens and identify individuals with a particularly high risk
of cancer
development. Such markers include LOH on chromosome 9 in bladder cancer, and
chromosomelp deletions and chromosome 7,17 and 8 gains/losses detected in
colorectal
tumorigenesis.
[0062] Development of lung cancer requires multiple genetic changes.
Activation
of oncogenes includes K-ras and myc. Inactivation of tumor suppressor genes
includes Rb, p53
and CDKN2. Identification of specific genes undergoing alteration is useful
for the early
detection of cells destined to become malignant and permits identification of
potential targets
for drugs and gene-based therapy.
-16-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
=====,~n.=~~t~e~r=rn~ning trisomy, the invention contemplates determining the
~ == ~ ,,;;;ll . ==.;; ~ .=;;;~
presence of trisomy in a single cell, and/or determining the frequency of
single cells with
trisomy in a population of cells (which could be done without knowing which
cells are trisomic;
i. e. total number of cells counted and total number of chromosomes counted).
The existence of
trisomy or the risk of a condition associated with trisomy then could be
evaluated.
[0064] Important is the recognition that signals can be counted and be
compared to
other information (e. g. other signal counts, statistical information about
predicted signal
frequency for different tissue types, etc.) so as to yield relevant diagnostic
information.
[0065] The invention also has been described in connection with identifying a
pair
of signals, one which identifies a target rare cell such as a fetal cell and
another which is useful
in evaluating the state of the cell such as a fetal cell having a genetic
defect. It should be
understood that according to certain embodiments, only a single signal need be
detected. For
example, where a fetal cell carries a Y chromosome and the diagnosis is for an
abnormality on
the Y chromosome, then the signal which identifies the genetic abnormality can
be the same as
that which identifies the fetal cell. As another example, a single signal can
be employed in
circumstances where the observed trait is a recessive trait. A pair of signals
also can be used to
detect the presence of two alleles or the existence of a condition which is
diagnosed by the
presence of two or more mutations in different genes. In these circumstances
the pair of signals
(or even several signals) can identify both the phenotype and the cell having
that phenotype.
Such embodiments will be apparent to those of ordinary skill in the art.
6. EXEMPLARY EMBODIMENTS
EXAMPLE 1
[0066] The following procedure for analyzing blood samples for the presence of
cells containing fetal hemoglobin using an immunostaining technique and to
determine the
presence of the X and Y chromosomes in the same cells by a fluorescent-labeled
in situ
hybridization technique.
[0067] Cells are deposited on a solid support suitable for microscopic
analysis and
fixed with methanol. Following air drying, cells are rinsed in phosphate
buffered saline and
further fixed in 2% formaldehyde in phosphate buffered saline. Cells are then
washed
sequentially in phosphate buffered saline, followed by Tris-buffered saline,
pH 7.6 containing
Tween 20. Following removal of excess liquid, blocking agent is added and the
slides
incubated in a humidified chamber. After the blocking solution is removed, a
dilution of
primary antibody in blocking agent is added and the cells incubated for 30 to
120 minutes in a
-17-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
hur:: y~~~~f tibody solution is then removed and the cells rinsed several
times in
Tris-buffered saline pH 7.6 containing Tween 20. Excess liquid is removed, and
a dilution of
anti-mouse secondary antibody in blocking agent is added, and the cells are
incubated in a
humidified chamber for 30 to 120 minutes. The antibody solution is then
removed and the cells
again rinsed several times in Tris-buffered saline, pH 7.6 containing Tween
20. After removal
of excess fluid, a fresh, filtered solution of HNPP/Fast Red dye in Alkaline
phosphatase buffer
is added and the cell sample is incubated for 10 minutes. The staining
solution is removed and
the cells rinsed in Tris-buffered saline, pH 7.6, containing Tween 20,
followed by a solution
of DAPI in Tris-buffered saline pH 7.6 containing Tweeng 20. The cells are
rinsed twice in
Tris-buffered saline, pH 7.6 containing Tween(g 20 and then in standard saline
citrate, excess
liquid removed and the cells are air dried. The cells are then incubated in
pre-warmed
0.005%pepsin at 37 C for 5 minutes. The cells are then washed in 50 mM MgC12
in phosphate
buffered saline for 5 minutes, then twice in phosphate buffered saline, excess
liquid removed
and the cells dried. A solution of fluorescently labeled FISH probe, such as
DNA and or RNA,
in hybridization is then added, a coverslip applied on top of the slide
containing the cells, and
then cells incubated at 74 C for 2.5 minutes, then at 37 C for 4 to 16 hours
in a humidified
chamber. The coverslip is removed and the cells washed in 0.4X standard saline
citrate at room
temperature for 2 minutes. Excess liquid is removed and the cells air dried
and mounted for
microscope observation and analysis.
EXAMPLE 2 -
APPARATUS
[00681 The block diagram of Figure 1 shows the basic elements of an embodiment
system suitable for embodying this aspect of the invention. The basic elements
of such system
include an X-Y stage 201, a mercury light source 203, a fluorescence
microscope 205 equipped
with a motorized objective lens turret (nosepiece) 207, a color CCD camera
209, a personal
computer (PC) system 211, and one or two monitors 213,215.
[0069] The individual elements of the system can be custom built or purchased
offthe-shelf as standard components. Each element will now be described in
somewhat greater
detail.
[0070] The X-Y stage 201 can be any motorized positional stage suitable for
use
with the selected microscope 205. Preferably, the X-Y stage 201 can be a
motorized stage that
can be connected to a personal computer and electronically controlled using
specifically
compiled software commands. When using such an electronically controlled X-Y
stage 201, a
-18-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
rc_plugged into an expansion bus of the PC 211 connects the stage 201
=,+ ,,,, =õi ,,,., + , ,,,., ,,,=c~ ,.=u p~
to the PC 211. The stage 201 should also be capable of being driven manually.
Electronically
controlled stages such as described here are produced by microscope
manufacturers, for
example including Olympus (Tokyo, Japan), as well as other manufacturers, such
as LUDL
(NY, USA).
[0071] The microscope 205 may be, for example, any fluorescence microscope
equipped with a reflected light fluorescence illuminator 203 and a motorized
objective lens
turret 207 with a 20x and an oil immersion 60x or 63x objective lens,
providing a maximum
magnification of 600x. The motorized nosepiece 207 is preferably connected to
the PC 211 and
electronically switched between successive magnifications using specifically
compiled software
commands. When using such an electronically controlled motorized nosepiece
207, a nosepiece
controller circuit card plugged into an expansion bus of the PC 211 connects
the stage 201 to
the PC 211. The microscope 205 and stage 201 are set up to include a mercury
light source 203,
capable of providing consistent and substantially even illumination of the
complete optical field.
[0072] The microscope 205 produces an image viewed by the camera 209. The
camera 209 can be any color 3-chip CCD camera or other camera connected to
provide an
electronic output and providing high sensitivity and resolution. The output of
the camera 209 is
fed to a frame grabber and image processor circuit board installed in the PC
211. A camera
found to be suitable is the SONY 930 (SONY, Japan).
[0073] Various frame grabber systems can be used in connection with the
present
invention. The frame grabber can be, for example a combination of the MATROX
IM-CLD
(color image capture module) and the MATROX IM-640 (image processing module)
set of
boards, available from MATROX (Montreal, CANADA). The MATROX IM-640 module
features on-board hardware supported image processing capabilities. These
capabilities
compliment the capabilities of the MATROX IMAGINGLIBRARY (MIL) software
package.
Thus, it provides extremely fast execution of the MIL based software
algorithms. The
MATROX boards support display to a dedicated SVGA monitor. The dedicated
monitor is
provided in addition to the monitor usually used with the PC system 211. Any
monitor SVGA
monitor suitable for use with the MATROX image processing boards can be used.
One
dedicated monitor usable in connection with the invention is a ViewSonic 4E
(Walnut Creek,
CA) SVGA monitor. In order to have sufficient processing and storage
capabilities available,
the PC 211 can be any INTEL PENTIUM-based PC having at least 32 MB RAM and at
least
2GB of hard disk drive storage space. The PC 211 preferably further includes a
monitor. Other
-19-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
thap,1111spgic,=,,~~~t~~~~; yi,scribed herein, the PC 211 is conventional, and
can include
keyboard, printer or other desired peripheral devices not shown.
[0074] The PC 211 may execute a smear analysis software program compiled in
MICROSOFT C++ using the MATROX IMAGING LIBRARY (MIL). MIL is a software
library of functions, including those which control the operation of the frame
grabber 211 and
which process images captured by the frame grabber 211 for subsequent storage
in PC 211 as
disk files. MIL comprises a number of specialized image processing routines
particularly
suitable for performing such image processing tasks as filtering, object
selection and various
measurement functions. The smear analysis software program may run as a
WINDOWS 95
application. The program prompts and measurement results are shown on the
computer monitor
213, while the images acquired through the imaging hardware 211 are displayed
on the
dedicated imaging monitor 215.
[0075] In order to process microscopic images using the smear analysis
program,
the system is first calibrated. Calibration compensates for day to day
variation in performance
as well as variations from one microscope, camera, etc., to another. During
this phase a
calibration image is viewed and the following calibration parameters are set:
= the color response of the system;
= the dimensions or bounds of the area on a on a slide containing a smear to
be scanned for
fetal cells ;
= the actual dimensions of the optical field when using magnifications 20x and
60x (or
63x); and
= the minimum and maximum fetal nuclear area when using magnifications 20x and
60x (or 63x).
DETECTION OF AN OBJECT IDENTIFICATION SIGNAL
[0076] The detection algorithm may operate in two stages. The first may be a
prescan stage I, illustrated in embodiment the flow chart of Figure 2, where
possible fetal cell
positions are identified using a low magnification and high speed. The 20x
objective may be,
for example, selected and the search of fetal cells can start:
= The program moves the automated stage (Figure 2,201) to a preset starting
point, for
example one of the corners of a slide containing a smear (Step 301).
= The x-y position of the stage at the preset starting point is recorded (Step
303) optical
field.
-20-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
= al quired (Step 305) using the CCD camera 209 and transferred to the
PC 211 as an RGB (Red/Green/Blue) image.
= The RGB image is transformed (Step 307) to the ILLS
(Hue/Luminance/Saturation)
representation.
= The Hue component is binary quantized (Step 309) as a black and white image
so that
pixels with Hue values ranging between 190 and 255 are set to 0 (black)
representing
interesting areas (blobs), while every other pixel value is set to 255 (white,
background).
The blobs represent possible fetal cell nuclear areas.
= The area of each blob in the binary quantized image is measured. If, at 20x
magnification, it is outside a range of about 20 to 200 pixels in size, the
blob's pixels are
set to value 255 (background); they are excluded from further processing
(Steps
311,313,315 and 317).
= Then the coordinates of each blob's center of gravity (CG) are calculated
(Step 3 19),
using a custom MATROX function. The center of gravity of a blob is that point
at which
a cut-out from a thin, uniform density sheet of material of the blob shape
would balance.
These coordinates are stored in a database along with the z-y position of the
current
optical field, so the blob can be located again at the next processing stage
using higher
magnification.
= Additional optical fields are processed similarly, recording the x-y
position of each
succeeding optical field, until the complete slide are is covered (Steps 321
and 323).
[0077] Stage 11, illustrated in embodiment flow chart of Figs. 3A and 3B,
includes
the final fetal cell recognition process:
= 63x magnification is selected (Step 401).
= The program moves the automated stage (Figure 2,201) so that the coordinates
of the first
position of a CG found earlier, which is possible fetal cell nuclear area, is
at the center of
the optical field (Step 403).
= The optical field is acquired using the CCD camera (Figure 2,209) and
transferred to the
computer as an RGB image (Step 405).
= The RGB image is transformed to the HLS model (Step 407).
= The program then generates a Luminance histogram (Step 409) by counting the
number
of pixels whose Luminance value equals each possible value of Luminance. The
counts
are stored as an array of length 256 containing the count of pixels having a
grey-level
value corresponding to each index into the array.
-21-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
= !,149 the Luminance distribution curve (Step 411), as represented
by the values stored in the array, and locates the last peak. It has been
found that this
peak includes pixel values that represent plasma area in the image. The
function that
analyzes the Luminance distribution curve: calculates a 9-point moving average
to
smooth the curve; calculates the tangents of lines defined by points
grey-level values distant; calculates the slopes of these lines in degrees;
finds the
successive points where the curve has zero slope and sets these points (grey-
levels) as -1
if they represent a minimum (valley in the curve) or I if they represent a
maximum (peak
in the curve); then finds the locations of peaks or valleys in the curve by
finding the
position of a 1 ora-1 in the array of grey-level values.
= The program then sets as a cut-off value the grey-level value of pixels
lying in the valley
of the Luminance distribution which occurs before the last peak of the
distribution (Step
413).
= Using this cut-off value, the program then produces (Step 415) a second
binary quantized
image. This is a black-and-white image in which pixels corresponding to pixels
in the
Luminance image having grey-level values lower than the cut off point are set
to 255
(white) and pixels corresponding to pixels in the Luminance image having grey-
level
values higher than the cut off point are set to 0 (black). The white blobs of
this image are
treated as cells while the black areas are treated as non-cellular area.
= A closing filter is applied (Step 417) to the second binary quantized image;
in this way
holes, i. e., black dots within white regions, are closed.
= The program now measures the area of the cells. If the area of any of the
cells is less than
200 pixels then these cells are excluded, i. e. the pixels consisting these
cells are set to
pixel value 255 (black) (Step 419).
= A hole fill function, found in the MIL, is applied to the remaining blobs
(Step 412).
= The resulting binary quantized image, after processing, is a mask whose
white regions
denote only cells.
= Red blood cells are now distinguished from white blood cells based on the
Saturation
component of the HLS image. The mask is used to limit processing to only the
cell areas.
= The program now counts the number of pixels whose Saturation value is each
possible
value of Saturation. The counts are stored as an array of length 256
containing the count
of pixels having a grey-level value corresponding to each index into the array
(Step 423).
-22-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
i= õ ~ppr~y,i,a~alyzes (Step 425) the Saturation distribution curve, as
represented by
the values stored in the array, and locates the first peak. This peak includes
pixel values
that represent areas contained in white blood cells.
= The grey-level value that coincides with the first minimum (valley) after
the peak is set as
a cut-off point (Step 427).
= Using this cut-off value the program produces (Step 429) a third binary
quantized image.
Pixels corresponding to pixels in the Saturation image having grey-level
values higher
than the cut-off point are set to 255 (white). They constitute red blood cell
areas. Pixels
corresponding to pixels in the Saturation image having grey-level values lower
than the
cutoff point are set to 0 (black). The white blobs of this third binary
quantized image are
seeds for areas that belong to red blood cells.
= A closing filter is applied (Step 431) to the third binary quantized image;
in this way
holes, i. e., black dots within white regions, are closed.
= A hole fill function, found in the MIL, is applied (Step 433) to the
remaining blobs.
= The resulting binary quantized image, after processing, is a new mask that
contains only
white blood cells.
= An erase border blob function of MIL is now applied (Step 435) to the
remaining blobs,
removing those which include pixels coincident with a border of the image
area. Such
blobs cannot be included in further processing as it is not known how much of
the cell is
missing when it is coincident with a border to the image area.
= An erosion filter is applied 6 times to this mask; thus any connected blobs
(white blood
cell seeds) are disconnected (Step 437).
= A "thick" filter is applied 14 times (Step 439). The "thick" filter is
equivalent to a dilation
filter. That is, it increases the size of a blob by successively adding a row
of pixels at the
periphery of the blob. If a growing blob meets an adjacent blob growing next
to it, the
thick filter does not connect the two growing blobs. Thus adjacent blobs can
be
separated.
= The first binary quantized mask (containing all the cells) and the third
binary quantized
mask (containing the separated seeds of white blood cells) are combined with a
RECONSTRUCTFROMSEED MIL operator. A fourth mask thus constructed contains
blobs (cells) copied from the first mask that are allowed by the third mask
and therefore
represent white blood cells (Step 441).
= The blobs in the fourth mask are measured for their area and compactness:
Area (A) is
the number of pixels in a blob; Compactness is derived from the perimeter (p)
and area
-23-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
,~ a.bl;ob~ ts,-a,ual to: p2/4 (A). The more convoluted the shape, the bigger
the
value. A circle has the minimum compactness value (1.0). Perimeter is the
total length of
edges in a blob, with an allowance made for the staircase effect which is
produced when
diagonal edges are digitized (insidecorners are counted as 1.414, rather than
2.0). Blobs
are retained in the fourth mask only if their area is between 1000 and 8000
pixels and
they have a compactness less than 3, thus allowing for cells with relatively
rough outline.
Blobs that touch the border of the image are excluded from further processing
(Step 443).
= The fourth mask is applied to the Hue component in the following manner
(Steps 445,
447,449 and451) :
= Pixels from the Hue component are copied to a new image retaining their Hue
value,
provided that their coordinates coincide with white (255) pixels in the "mask"
; all other
pixels in the new image are set to 0 (black) (Step 445).
= The pixel values in each of the contiguous non-0 pixel areas, i. e., those
blobs
corresponding to images of red cells, are checked for values between 190 and
255.
The number of such pixels in each blob is counted (Step 447).
= If there are more than 200 such pixels, the blob represents a nucleated red
blood cell.
The coordinates of the center of gravity of each such cell are stored. The
mask is binary
quantized so that all pixels having non-0 values are set to 255 (white); and
the mask is
stored as a separate Tagged Image File Format (TIFF) file (Step 449).
= The program moves to the next stored coordinates for a possible fetal cell
which do not
coincide with any of the coordinates stored during the previous step. The
entire process is
repeated until a preset number of nucleated red blood cells have been
identified. The
results, including the nucleated red blood cell coordinates and the names of
the respective
mask files, along with various characteristic codes for the blood slide are
stored in a
result text file. The nucleated red blood cells whose coordinates are stored
are the fetal
cells sought (Step451).
[0078] After the object of interest, such as the fetal cells, are identified,
the second
signal is generated, for example by in situ PCR or PCR in situ hybridization
or FISH, as
described above.
DETECTION OF THE DIAGNOSTIC SIGNAL
[0079] A smear including in situ PCR or PCR in situ hybridization treated
cells is
positioned on the stage (Figure 2,201). If necessary calibration steps are
taken, as before.
Calibration permits the software to compensate for day to day variation in
performance as well
-24-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
~~~cope, camera, etc. to another. Detection of the diagnostic signal in
an embodiment method may proceed as shown in the flow chart of Figure 4, as
follows:
= Magnification objective 60x (63x) is chosen (Step 501).
= The x-y stage is moved to the first fetal cell position according to data
from the result file
compiled from detection of the first signal, as described above (Step 503).
= The optical field is acquired using the CCD camera (Figure 2,209) and
transferred to the
computer (Figure 2,211) as an RGB image (Step 505).
= The RGB image is transformed to the HLS model (Step 507).
= The TIFF file containing the black and white mask is loaded as a separate
image (Step
509).
= The pixels of the Hue component not corresponding to white areas in the mask
arc set to
0 (black) (Step 511).
= The remaining areas, which represent fetal cells, are searched for pixel
values
corresponding to a signal produced following PCR. For example, the signal may
be a
color which arises due to the presence of alkaline phosphatase, i. e., red.
The non black
areas of the Hue component are searched for pixel values ranging from 0 to 30
(Step
513).
= The stage is moved to the next non-processed fetal cell and the above
process is repeated
(Step 515).
[0080] The PC 211 executes a software program called SIMPLE which controls
operation of the frame grabber and image processor circuit 217. SIMPLE also
processes images
captured by frame grabber and image processor circuit 217 and subsequently
stores images and
processed data in PC 211 as disk files. SIMPLE provides an icon-based
environment with
specialized routines particularly suitable for performing such image
processing tasks as
filtering, object selection and measurement. Most of the SIMPLE tasks are
directed by a human
operator using a pointing device connected to PC 211, such as a mouse or
trackball (not shown).
[0081] In order to process images using SIMPLE, a number of image calibration
steps must first be taken. In an embodiment, a new slide properly stained
using the
fluorescence in situ hybridization (FISH) technique is placed under the
fluorescence
microscope. The objects of interest which are to be recognized, i. e., the
nuclear or
chromosomal areas, have specific chromatic features. Multiple targets can be
delineated
simultaneously in a particular specimen by combining fluorescence detection
procedures. That
is, if different targets are labeled with different fluorophors that fluoresce
at different
wavelengths, then the software program can be made to separately identify
objects emitting the
-25-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
Qyõ}ded full color.:information is available in the image. Targets with
.=== =., ,= ,.=., ==.== ~ .,, ,. ~
differing affinities for different fluorophors may be differentiated by the
color combinations
emitted. Each target may emit at wavelengths corresponding to two or more
fluorophors, but the
intensity of each may differ, for example. Thus, all three color components of
the microscopic
images are used during processing.
[0082] For each new specimen inserted under the microscope, a preprocessing
procedure is first executed. The flowchart of Fig. 11 shows the preprocessing
steps of this
embodiment of the present invention. Preprocessing may be used to permit the
software to
compensate for specimen-to-specimen variations.
[0083] In one embodiment, the slide containing the FISH-treated cells is
positioned into the X-Y stage201. The X-Y stage 201 is moved to an initial
observation position
found to contain a rare cell. A processing loop is executed repeatedly until
either a
predetermined number of the rare cells of a particular type have been
measured. In the
application for which the present embodiment is intended, identifying multiple
targets of
chromosomal DNA, the loop is executed until 20-100 nuclei have been processed.
Data
representing the measurement of the chromosomal areas within those nuclei may
be collected in
an ASCII file.
[0084] The filtering steps 12000 may operate on a pixel-by-pixel basis, as
follows.
In step 12001, a hole filling filter is applied to the image. This filter,
available through the
SIMPLE language, determines when dark holes have appeared within the lighter
fluorescent
chromosomes by searching for dark areas within light objects. Those areas are
lightened up.
The output of the hole filling filter is held in a temporary image file 12101,
as well as being
used as the input to the erosion filter, step 12003. Erosion filtering, also
available through the
SIMPLE language, replaces the center pixel of a small kernel with the darkest
pixel in the
kernel.. The kernel used is 3 x 3 may be used. A separate operation, step
12005 is next
performed, to grow the objects until they meet, but do not merge. This step
also creates outlines,
defining the edges of all the objects. A logical NOT operation, step 12007,
causes the pixels
within the outlines to become selected rather than the outlines. Finally, in
step 12009, the result
of step 12007 is logically ANDed with the stored temporary image file12101.
This causes only
those pixels which are defined in both the temporary image file12101 and the
output of step
12007 to be retained.
[0085] If a combination of fluorescence detection procedures is used, more
than
two chromosomal areas may be detected per nucleus. Therefore, it is possible
to recognize two
chromosomal areas relative to chromosomes 21, another two relative to
chromosome 18, one
-26-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
~rel~tj~~ti~ FI'?Cii. nd one relative to chromosome Y, enabling the discovery
of
possible numerical aberrations detected by the enumeration of hybridization
signals. The
enumeration of the hybridization signals may be executed after completing the
measurement of
20100 nuclei through an application program external to SIMPLE, compiled using
CLIPPER
(COMPUTER ASSOCIATES, CA). This program reads the measurement results ASCII
file
and classifies the chromosomal areas detected according to their RGB color
combination.
When two or more different fluorophors are used in combination, different
combinations of
RGB color values may be used to distinguish different targets, some targets of
which may be
labeled by more than one fluorophor. For example, targets may be stained with
red and green
fluorophors, but one target may receive fluorophors to emit 30% red and 70%
green, another
target may receive fluorophors to emit 70% red and 30% green, while a third
target may receive
fluorophors to emit only red. The three targets may be distinguished on the
basis of their
relative emissions. If the number of signals indicative of a chromosomal area
corresponding to
a specific chromosome, e. g., chromosome 21, is greater than two to an
operator-selected
statistically significant level, then a report is issued identifying an
increased likelihood for
trisomy 21 in the specific sample.
[0086] Although the present invention has been described in connection with
the
clinical detection of chromosomal abnomialities in a cell-containing sample,
the image
processing methods disclosed herein has other clinical applications. For
example, the image
processing steps described can be used to automate a urinalysis process. When
the techniques of
the present application are combined with those of Application Serial No.
08/132,804, filed
October 7,1993, a wide variety of cell types can be visualized and analyzed,
based on their
morphology. Cell morphology can be observed for the purpose of diagnosing
conditions for
which cell morphology has been correlated to a physiological condition. Such
conditions are
known to those of skill in the art. See, e. g., Harrison, supra. Various cell
characteristics and
abnormalities may be detected based on these techniques. Finally, it should be
noted that the
particular source of the sample is not a limitation of the present invention,
as the sample may be
derived from a blood sample, a serum sample, a urine sample or a cell sample
from the uterine
cervix. The cell visualization and image analysis techniques described herein
may be used for
any condition detectable by analysis of individual cells, either by morphology
or other
characteristics of the isolated cells.
[0087] Antibodies specific for human fetal hemoglobin (Research
Diagnostics Inc., NJ) and for embryonic epsilon hemoglobin chain (Immuno-Rx,
GA) are
commercially available and can be used as fluorescently labeled antibodies or
a fluorescent
-27-
CA 02580961 2007-03-20
WO 2006/036735 PCT/US2005/033964
s~gpp:;c" ipe geqc~.~t-9'P y.1pse of a fluorescently labeled secondary
antibody. Fluorescent light
can be produced by other types of stains or labels for rare cells, as known in
the art. Fluorescent
staining of the type required for this processing step is known in the art,
and will not be
discussed in further detail.
[0088] Computer and image processing"technologies are constantly changing.
Newer technologies which meet the needs of the above-described methods and
apparatus, while
not specifically described here, are clearly contemplated as within the
invention. For example,
certain conventional pixel and image file formats are mentioned above, but
others may also be
used. Image files may be compressed using JPEG or GIF techniques now known in
the art or
other techniques yet to be developed. Processing may be performed in an RGB
color description
space instead of the HLS space currently used. Other color spaces may also be
used, as desired
by the skilled artisan, particularly when detection of a sought-after
characteristic is enhanced
thereby.
[0089] The present invention has now been described in connection with a
number
of particular embodiments thereof. Additional variations should now be evident
to those skilled
in the art, and are contemplated as falling within the scope of the invention,
which is limited
only by the claims appended hereto and equivalents thereof.
[0090] In embodiments of the invention, there is illustrated an example for
analysis of subcellular components of cells for the detection of for example,
chromosomal
abnormalities in prenatal and pre-implantation genetic diagnosis, or the sex
chromosomes of
embryonal or fetal cells.
[0091] FIG. 13 is a photomicrograph of a combined immunostaining and FISH
analysis of cells for the presence of fetal hemoglobin and the identification
of X and Y
chromosomes in the cells. Fetal hemoglobin is present in the sample as shown
by the orange
fluorescent signal detected from the cells and throughout the cytoplasm of the
cell in the lower
right quadrant of the figure. X and Y chromosomes are shown as green aqua red
fluorescent
dots, respectively, in the nucleus of the cells.
-28-