Note: Descriptions are shown in the official language in which they were submitted.
CA 02592705 2007-06-21
214217 31PS
METHOD FOR EVALUATING CORRELATIONS BETWEEN STRUCTURED
AND NORMALIZED INFORMATION ON GENETIC VARIATIONS BETWEEN
HUMANS AND THEIR PERSONAL CLINICAL PATIENT DATA FROM
ELECTRONIC MEDICAL PATIENT RECORDS
RELATED APPLICATIONS
This application claims the benefit of U.S. Provisional Application No.
60/813,397
(the `397 application"), filed June 14, 2006, entitled "Method For Evaluating
Correlations Between Structured And Normalized Information On Genetic
Variations
Between Humans And Their Personal Clinical Patient Data From Electronic
Medical
Patient Records." The `397 application is incorporated by reference herein in
its
entirety.
BACKGROUND OF THE INVENTION
The present invention generally relates to search and analysis of electronic
medical
record data. More particularly, the present invention relates to evaluating
correlations
between genetic and clinical information included in electronic medical
records.
Hospitals typically utilize computer systems to manage the various departments
within a hospital and data about each patient is collected by a variety of
computer
systems. For example, a patient may be admitted to the hospital for a
Transthoracic
Echo ("TTE"). Information about the patient (for example, demographics and
insurance) could be obtained by the hospital information system ("HIS") and
stored
on a patient record. This information could then be passed to the cardiology
department system (commonly known as the cardio vascular information system,
or
"CVIS"), for example. Typically the CVIS is a product of one company, while
the
HIS is the product of another company. As a result, the database between the
two
may be different. Further, information systems may capture/retain and send
different
levels of granularity in the data. Once the patient information has been
received by
the CVIS, the patient may be scheduled for a TTE in the echo lab. Next, the
TTE is
performed by the sonographer. Images and measurements are taken and sent to
the
CVIS server. The reading physician (for example, an echocardiographer) sits
down at
1
CA 02592705 2007-06-21
214217 31PS
a review station and pulls the patient's TTE study. The echocardiographer then
begins to review the images and measurements and creates a complete medical
report
on the study. When the echocardiographer completes the medical report, the
report is
sent to the CVIS server where it is stored and associated with the patient
through
patient identification data. This completed medical report is an example of
the kind
of report that could be sent to a data repository for public data mining.
Medication
instructions, such as documentation and/or prescriptions, as well as
laboratory results
and vital signs, may also be generated electronically and saved in a data
repository.
Today, medical device manufacturers and drug companies face an ever-growing
challenge in collecting clinical data on the real-life utilization of their
products. As
patient medical reports are becoming computerized, the ability to obtain real-
life
utilization data becomes easier. Further, the data is easier to combine and
analyze (for
example, mine) for greater amounts of useful information.
As medical technology becomes more sophisticated, clinical analysis may also
become more sophisticated. Increasing amounts of data are generated and
archived
electronically. With the advent of clinical information systems, a patient's
history
may be available at a touch of a button. While accessibility of information is
advantageous, time is a scarce commodity in a clinical setting. To realize a
full
benefit of medical technological growth, it would be highly desirable for
clinical
information to be organized and standardized.
Data warehousing methods have been used to aggregate, clean, stage, report and
analyze patient information derived from medical claims billing and electronic
medical records ("EMR"). Patient data may be extracted from multiple EMR
databases located at patient care provider ("PCP") sites in geographically
dispersed
locations, then transported and stored in a centrally located data warehouse.
The
central data warehouse may be a source of information for population-based
profile
reports of physician productivity, preventative care, disease-management
statistics
and research on clinical outcomes.
Current efforts to evaluate correlations between genotypic and phenotypic data
in the
human population are performed in relatively small and controlled clinical
studies
2
CA 02592705 2007-06-21
214217 31PS
using paper-based medical records. Such efforts consume considerable amounts
of
time and resources. In addition, paper-based efforts are unlikely to identify
subtle
associations between genetic variability and phenotypic susceptibility. For
example,
these efforts are unlikely to uncover subtle associations or correlations
between
genetic variability (for example, a propensity for a particular single
nucleotide
polymorphism ("SNP") or combination of SNPs) and actual phenotypic expressions
of traits associated with the genetic variability.
Current efforts to obtain such correlations and associations are also limited
by the
different syntax used in different clinical trials. In order to fully evaluate
and
understand such correlations and associations, it is often beneficial to
examine larger
amounts of data, for example from multiple clinical trials. However, genetic
and
clinical information may be recorded using different terms, or syntax, in
different
clinical trials. For example, a clinical condition or event such as a heart
attack may be
expressed or recorded as "heart attack" in one trial, as "myocardial
infarction" in
another trial, as "MI" in another trial, an "acute MI" in another trial, and
an "AMI" in
yet another trial. However, if the clinical data from two or more of these
trials were
combined (along with corresponding genetic information) in order to evaluate
correlations between one or more SNPs and the potential for a heart attack,
the
different syntax would inhibit, if not prevent, an accurate evaluation of any
such
correlations. In other words, the lack of a controlled medical vocabulary
makes it
unlikely to demonstrate conclusive evidence of such associations or
correlations due
to the variability of clinical language chosen to describe patient expression
of clinical
conditions or disease.
Therefore, there is a need for improved methods to evaluate correlations
between
genetic variations among patients and personal clinical patient data derived
from
electronic medical records in a variety of different trials.
BRIEF DESCRIPTION OF THE INVENTION
Various embodiments of the presently described invention provide a method for
evaluating correlations between genetic variations and clinical information.
The
method includes normalizing one or more of genotypic data and clinical data
3
CA 02592705 2007-06-21
214217 31PS
associated with each of a plurality of patients in a population of patients,
receiving
one or more clinical conditions from a user, selecting a subset of patients
from the
population based on the clinical conditions, and determining one or more
correlations
between at least one of the clinical conditions and one or more of the
genotypic and
clinical data for the patient subset.
Various embodiments of the presently described invention also provide a
computer-
readable storage medium comprising a set of instructions for a computer. The
instructions include a data normalization routine, a patient selection routine
and a
correlation routine. The data normalization routine is configured to normalize
one or
more of genotypic data and clinical data associated with each of a plurality
of patients
in a population of patients. The patient selection routine is configured to
select a
subset of patients from the population based on one or more clinical
conditions input
by a user. The correlation routine is configured to determine one or more
correlations
between at least one of the clinical conditions and one or more of the
genotypic and
clinical data for the subset of patients.
Various embodiments of the presently described invention also provide a method
for
determining correlations between genetic data and medical data. The method
includes
receiving genotypic data and clinical data associated with each of a plurality
of
patients from a plurality of sources, where two or more of the sources uses
different
terms to report the genotypic and/or clinical data, normalizing the genotypic
and/or
clinical data, selecting one or more patients from the plurality of patients
based on one
or more parameters, and determining a correlation between one or more of the
parameters and at least one of the genotypic and clinical data associated with
two or
more of the selected patients.
BRIEF DESCRIPTION OF SEVERAL VIEWS OF THE DRAWINGS
FIG. 1 illustrates a schematic diagram of a system for storing EMRs in
accordance
with an embodiment of the presently described technology.
FIG. 2 illustrates a schematic diagram of a data warehouse architecture in
accordance
with an embodiment of the presently described technology.
4
CA 02592705 2007-06-21
214217 31PS
FIG. 3 illustrates a schematic diagram of genetic and/or clinical data
aggregation
system in accordance with an embodiment of the presently described technology.
FIG. 4 illustrates a flowchart for a method for evaluating one or more
correlations
between genetic and clinical data in accordance with an embodiment of the
presently
described technology.
The foregoing summary, as well as the following detailed description of
certain
embodiments of the presently described technology, will be better understood
when
read in conjunction with the appended drawings. For the purpose of
illustrating the
invention, certain embodiments are shown in the drawings. It should be
understood,
however, that the present invention is not limited to the arrangements and
instrumentality shown in the attached drawings.
DETAILED DESCRIPTION OF THE INVENTION
The presently described technology provides, among other things, an improved
method for combining genetic data with more traditional clinical data of a
codified
nature and employing these data sets to come up with, and test, various
hypotheses
and correlations between diseases, traits, medical conditions/problems and
environmental factors, for example. The technology permits integration of a
data
source such as codified genetic data with a new data source such as codified
clinical
data obtained from a plurality of different sources. In doing so, different
nomenclature used by the various sources of the clinical data can be codified
so as to
permit easier comparisons between the clinical and genetic data.
FIG. 1 illustrates a schematic diagram of a system 100 for storing EMRs in
accordance with an embodiment of the presently described technology. PCP
systems
108 located at various PCP sites are connected to a network 106. The PCP
systems
108 send patient medical data (included in EMRs) to a data warehouse located
on a
data warehouse system 104. The PCP systems 108 typically include application
software to perform data extraction along with one or more storage device for
storing
the EMRs associated with patients treated at the PCP site. In addition, the
PCP
systems 108 can include PCP user systems 110 to access the EMR data, to
initiate the
CA 02592705 2007-06-21
214217 31PS
data extraction and to enter a password string to be used for encrypting a
patient
identifier.
The PCP user systems 110 can be directly attached to the PCP system 108 or the
user
systems 110 can access the PCP system 108 via the network 106. Each PCP user
system 110 can be implemented using a general-purpose computer executing a
computer program for carrying out the processes described herein. The PCP user
systems 110 can be personal computers or host attached terminals. If the PCP
user
systems 110 are personal computers, the processing described herein can be
shared by
a PCP user system 110 and a PCP system 108 by providing an applet to the PCP
user
system 110.
The storage device located at the PCP system 108 can be implemented using a
variety
of devices for storing electronic information such as a file transfer protocol
("FTP")
server. It is understood that the storage device can be implemented using
memory
contained in the PCP system 108 or it can be a separate physical device. The
storage
device contains a variety of information including an EMR database.
In addition, the system of FIG. 1 includes one or more data warehouse user
systems
102 through which an end-user can make a request to an application program on
the
data warehouse system 104 to access particular records stored in the data
warehouse.
In an example embodiment of the present invention, end-users can include PCP
staff
members, pharmaceutical company research team members and personnel from
companies that make medical products.
The data warehouse user systems 102 can be directly connected to the data
warehouse
system 104 or they can be coupled to the data warehouse system 104 via the
network
106. Each data warehouse user system 102 can be implemented using a general-
purpose computer executing a computer program for carrying out the processes
described herein. The data warehouse user systems 102 can be personal
computers or
host attached terminals. If the data warehouse user systems 102 are personal
computers, the processing described herein may be shared by a data warehouse
user
system 102 and the data warehouse system 104 by providing an applet to the
data
warehouse user system 102.
6
CA 02592705 2007-06-21
214217 31PS The network 106 can be any one or more types of known networks
including a local
area network ("LAN"), a wide area network ("WAN"), an intranet, or a global
network (for example, Internet). A data warehouse user system 102 can be
coupled to
the data warehouse system 104 through multiple networks (for example, intranet
and
Internet) so that not all data warehouse user systems 102 are required to be
coupled to
the data warehouse system 104 through the same network. Similarly, a PCP
system
108 can be coupled to the data mining host system 104 through multiple
networks (for
example, intranet and Internet) so that not all PCP systems 108 are required
to be
coupled to the data warehouse system 104 through the same network.
One or more of the data warehouse user systems 102, the PCP systems 108 and
the
data warehouse system 104 can be connected to the network 106 in a wireless
fashion
and the network 106 may be a wireless network. In an example embodiment, the
network 106 is the Internet and each data warehouse user system 102 executes a
user
interface application to directly connect to the data warehouse system 104. In
another
embodiment, a data warehouse user system 102 can execute a web browser to
contact
the data warehouse system 104 through the network 106. Alternatively, a data
warehouse user system 102 can be implemented using a device programmed
primarily
for accessing the network 106 such as WebTV.
The data warehouse system 104 can be implemented using a server operating in
response to a computer program stored in a storage medium accessible by the
server.
The data warehouse system 104 can operate as a network server (often referred
to as a
web server) to communicate with the data warehouse user systems 102 and the
PCP
systems 108. The data warehouse system 104 handles sending and receiving
information to and from data warehouse user systems 102 and PCP systems 108
and
can perform associated tasks. The data warehouse system 104 can also include a
firewall to prevent unauthorized access to the data warehouse system 104 and
enforce
any limitations on authorized access. For instance, an administrator can have
access
to the entire system and have authority to modify portions of the system and a
PCP
staff member can only have access to view a subset of the data warehouse
records for
particular patients. In an example embodiment, the administrator has the
ability to
7
CA 02592705 2007-06-21
214217 31PS
add new users, delete users and edit user privileges. The firewall can be
implemented
using conventional hardware and/or software as is known in the art.
The data warehouse system 104 also operates as an application server. The data
warehouse system 104 executes one or more application programs to provide
access
to the data repository located on the data warehouse system, as well as
application
programs to import patient data into a staging area and then into the data
warehouse.
In addition, the data warehouse system 104 can also execute one or more
applications
to create patient cohort reports and to send the patient cohort reports to the
PCP
systems 108. Processing can be shared by the data warehouse user system 102
and
the data warehouse system 104 by providing an application (for example, a java
applet) to the data warehouse user system 102. Alternatively, the data
warehouse user
system 102 can include a stand-alone software application for performing a
portion of
the processing described herein. Similarly, processing can be shared by the
PCP
system 102 and the data warehouse system 104 by providing an application to
the
PCP system 102 and alternatively, the PCP system 102 can include a stand-alone
software application for performing a portion of the processing described
herein. It is
understood that separate servers may be used to implement the network server
functions and the application server functions. Alternatively, the network
server,
firewall and the application server can be implemented by a single server
executing
computer programs to perform the requisite functions.
The storage device located at the data warehouse system 104 can be implemented
using a variety of devices for storing electronic information such as an FTP
server. It
is understood that the storage device can be implemented using memory
contained in
the data warehouse system 104 or it may be a separate physical device. The
storage
device contains a variety of information including a data warehouse containing
patient
medical data from one or more PCPs. The data warehouse system 104 can also
operate as a database server and coordinate access to application data
including data
stored on the storage device. The data warehouse can be physically stored as a
single
database with access restricted based on user characteristics or it can be
physically
stored in a variety of databases including portions of the database on the
data
warehouse user systems 102 or the data warehouse system 104. In an example
8
CA 02592705 2007-06-21
214217 31PS
embodiment, the data repository is implemented using a relational database
system
and the database system provides different views of the data to different end-
users
based on end-user characteristics.
FIG. 2 illustrates a schematic diagram of a data warehouse architecture 200 in
accordance with an embodiment of the presently described technology. Patient
data is
extracted from EMR databases located in the PCP systems 108. An EMR database
record includes medical data such as: patient name and address, medications,
allergies, observations, diagnoses, and health insurance information, for
example.
The PCP systems 108 include application software for extracting patient data
from the
EMR database. The data is then transported (for example, via Hypertext
Transfer
Protocol ("HTTP") or Secure HTTP ("HTTPS")) over the network 106 to the data
warehouse system 104.
The data warehouse system 104 includes application software to perform a data
import function 206. The data import function 206 aggregates patient data from
multiple sites and then stores the data into a staging area 208. Data received
from
multiple PCP systems 108 is normalized, checked for validity and completeness,
and
either corrected or flagged as defective. Data from multiple PCP systems 108
can
then be combined together into a relational database. Aggregation and staging
data in
the described fashion allows the data to be queried meaningfully and
efficiently,
either as a single entity or specific to each individual PCP site 108. The de-
identified
patient data is then staged into a data warehouse 210 where it is available
for
querying.
Patient cohort reports 212 are generated by application software located on
the data
warehouse system 104 and returned to the PCP systems 108 for use by the
primary
care providers in treating individual patients. Patient cohort reports 212 can
be
automatically generated by executing a canned query on a periodic basis. PCP
staff
members, pharmaceutical company research team members and personnel from
companies that make medical products may each run patient cohort reports 212,
for
example. In addition,. patient cohort reports 212 can be created by an end-
user
accessing a data warehouse user system 102 to create custom reports or to
initiate the
running of canned reports. Further, patient cohort reports 212 can be
automatically
9
CA 02592705 2007-06-21
214217 31PS
generated in response to the application software, located on the data
warehouse
system 104, determining that particular combinations of data for a patient are
stored in
the data warehouse. An example patient cohort report 212 includes all patients
with a
particular disease that were treated with a particular medication. Another
example
paiient cohort report 212 includes patients of a particular age and sex who
have
particular test results. For example, a patient cohort report 212 can list all
women
with heart disease who are taking a hormone replacement therapy drug. The
patient
cohort report 212 can list all the patients with records in the data warehouse
210 that
fit this criteria. In an example embodiment, each PCP site receives the entire
report;
in another embodiment, each PCP site can receive the report only for patients
that are
being treated at the PCP site.
FIG. 3 illustrates a schematic diagram of genetic and/or clinical data
aggregation
system 300 in accordance with an embodiment of the presently described
technology.
System 300 includes a central data warehouse 310, a plurality of data stores
320 and a
computing device 330. While seven data stores 320 are illustrated in FIG. 3,
any
number of data stores 320 can be included in system 300. For example, as few
as one
data store 320 can be included, or many more than seven data stores 320 can be
included in system 300.
In an embodiment of the presently described technology, warehouse 310 is
similar to
the data warehouse system 104 of FIG. 1. In addition, in an embodiment of the
presently described technology, one or more data stores 320 are similar to PCP
systems 108 of FIG. 1.
Warehouse 310 and each of data stores 320 comprise a storage medium 340 for
electronic data. For example, warehouse 310 and data stores 320 can each
comprise
one or more computer hard drives, server computers, or other electronic
storage
medium. In an embodiment of the presently described technology, warehouse 310
can be implemented using a server operating in response to a computer program
stored in a storage medium accessible by the server. Warehouse 310 can operate
as a
network server (often referred to as a web server) to communicate with one or
more
data stores 320.
CA 02592705 2007-06-21
214217 31PS
Computing device 330 includes any electronic device capable of carrying out
one or
more sets of instructions. For example, computing device 330 can include a
desktop
or laptop personal computer ("PC") or a mobile computing device capable of
running
one or more software applications. Computing device 330 is capable of
communicating with warehouse 310 through a wired or wireless connection. For
example, computing device 330 can be connected to warehouse 310 through one or
more networks such as a LAN, a WAN, an intranet, or a global network (for
example,
Internet). Computing device 330 can be coupled to warehouse 310 through
multiple
networks (for example, intranet and Internet).
Computing device 330 includes an input device and an output device (not
shown).
For example, computing device 330 can include a mouse, stylus, microphone
and/or
keyboard as an input device. Computing device 330 can include a computer
monitor,
liquid crystal display ("LCD") screen, printer and/or speaker as an output
device.
Computing device 330 also includes, or is in communication with, a computer-
readable memory 350. Computer-readable memory 350 can be similar or the same
as
storage medium 340. For example, computing device 330 can include a computer
hard drive, a compact disc ("CD") drive, a USB thumb drive, or any other type
of memory capable of storing one or more computer software applications. The
memory
can be included in computing device 330 or physically remote from computing
device
330. For example, the memory can be accessible by computing device 330 through
a
wired or wireless network connection.
The memory 350 accessible to computing device 330 includes a set of
instructions for
a computer (described in more detail below). The set of instructions includes
one or
more routines capable of being run or performed by computing device 330. The
set of
instructions can be embodied in one or more software applications or in
computer
code.
Data stores 320 are configured to store clinical and/or genetic data from a
plurality of
patients in a plurality of medical trials or experiments. For example, a
portion or
entirety of each data store 320 can be dedicated to the storage of clinical
and/or
11
CA 02592705 2007-06-21
214217 31PS
genetic data from a particular medical trial at a given hospital or PCP or
group of
hospitals or PCPs.
In an embodiment of the presently described technology, warehouse 310 handles
sending and receiving information to and from one or more data stores 320. In
an
embodiment, warehouse 310 can also include a firewall to prevent unauthorized
access to the data stored at warehouse 310 and enforce any limitations on
authorized
access. For instance, an administrator may have access to the entire system
and have
authority to modify portions of the system and a PCP staff member may only
have
access to view a subset of the data stored at warehouse 310 for particular
patients.
Warehouse 310 can also operate as an application server. Warehouse 310 can
execute one or more application programs to provide access to the data stored
at warehouse
310, as well as application programs to import patient data into a staging
area and
then into warehouse 310. In addition, warehouse 310 can also execute one or
more
applications to create patient cohort reports and to send the patient cohort
reports to
one or more data stores 320. Processing may be shared by warehouse 310 and one
or
more data stores 320 by providing an application (for example, java applet) to
warehouse 310. In another embodiment, warehouse 310 can include a stand-alone
software application for performing a portion of the processing described
herein. It is
understood that separate servers may be used to implement the network server
functions and the application server functions. Alternatively, the network
server,
firewall and the application server can be implemented by a single server
executing
computer programs to perform the requisite functions.
Warehouse 310 and each of data stores 320 communicate electronically over one
or
more wired or wireless links. For example, warehouse 310 and one or more data
stores 320 can communicate data over a secured or unsecured network
connection.
The network connection can be one or more networks such as a LAN, a WAN, an
intranet, or a global network (for example, Internet). One or more data stores
320 can
be coupled to the warehouse 310 through multiple networks (for example,
intranet
and Internet) so that not all data stores 320 are required to be coupled to
the
warehouse 310 through the same network.
12
CA 02592705 2007-06-21
214217 31PS
In an embodiment of the presently described technology, one or more data
stores 320
are remote from warehouse 310. In other words, one or more data stores 320 are
in a
physically and/or geographically separate location from warehouse 310.
The clinical data stored at data stores 320 includes phenotypic expressions of
a
genetic trait. In an embodiment, the phenotypic expressions are codified
according to
a coding scheme used by the PCP that stores clinical data at one or more
particular
data store(s) 320. For example, the clinical data can be stored in an EMR for
one or
more patients. The EMRs can include any codes or terms used to describe one or
more diseases, conditions, medical events and/or medical factors related to
one or
more patients. The EMRs can store data such as chronic conditions or diseases
(for
example, diabetes, heart disease, AIDS, cancer, cataracts), allergies (for
example,
allergies to pharmaceuticals or environmental factors such as smoke, dust, or
animals), past adverse reactions to medical therapeutics and/or environmental
factors,
and/or other general medical problems for each of a plurality of patients
seeking
medical treatment at a particular PCP and/or participating in a particular
medical
trial/experiment.
The genetic data stored at data stores 320 (also referred to as genotypic
data) includes
any structured information representative of genetic information. For example,
the
genetic data can include data representative of one or more SNPs for one or
more
patients. In another example, the genetic data can include data representative
of a
combination of SNPs for one or more patients. In an embodiment, the genetic
data
for one or more patients is stored in an EMR similar to, or the same EMR as,
the
clinical data for the same patients.
As described above, one problem with existing EMR systems is that different
medical
trials, hospitals, clinics and PCPs may employ different syntax or terms to
record
medical data, including clinical and genetic data. For example, a plurality of
data
stores 320 may each store genetic and/or clinical data using different
terminology or
syntax than other data stores 320. Therefore, in operation, the presently
described
technology normalizes clinical and/or genetic data so that the data (and
correlations
among the various data) can be more easily and accurately analyzed.
13
CA 02592705 2007-06-21
214217 31PS
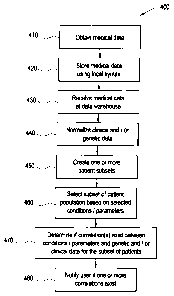
FIG. 4 illustrates a flowchart for a method 400 for evaluating one or more
correlations
between genetic and clinical data in accordance with an embodiment of the
presently
described technology. While an embodiment of the presently described
technology is
described and illustrated by FIG. 4, not all embodiments of the technology are
limited
to the exact steps described and illustrated in FIG. 4. For example, one or
more steps
may be added, removed, combined or rearranged in method 400 without departing
from the scope of the presently described invention. First, at step 410,
medical data is obtained at a hospital, clinic or other PCP. The
medical data can include clinical data and/or genetic data. For example, the
medical
data can include clinical data such as medical test results, a condition,
disease or other
medical problem, an allergy, an environmental factor (such as the fact that a
patient
lives in a household with one or more smokers, lives near power lines, etc.),
and/or a
codified phenotypic expression of a trait (which can include any of the
previously
listed clinical data).
Next, at step 420, the medical data is stored in one or more EMRs at a data
store 320
or stores 320 used by the PCP that obtained the medical data. In an embodiment
of
the presently described technology, both clinical and genetic data for
patients are
stored together in EMRs at data stores 320. In another embodiment, the
clinical data
is stored separately from genetic data in data stores 320. For example,
clinical data
for a particular patient can be stored in one EMR at a particular data store
320 and
genetic data for the same patient can be stored in a different EMR at the same
or
different data store 320.
At step 420, the medical data is stored at a plurality of data stores 320
using different
syntax or terminology. As described above, this syntax or terminology is
likely to
differ from the syntax/terminology used by a different PCP to record medical
data.
For example, different PCPs may refer to the same clinical data relating to
diabetes as
"diabetic," "diabetes," "type I diabetes," "type 1 diabetes," or "juvenile
diabetes." In
addition, different PCPs may use common terminology such as ICD-9
(International
Classification of Diseases, Ninth Revision) codes, ICD-10 codes or CPT
(Current
Procedure Terminology) codes to record medical data. In another embodiment, a
terminology common to a user or group of users of the presently described
technology
14
CA 02592705 2007-06-21
214217 31PS
can be used. For example, a particular doctor, group of physicians and/or
hospital
may have his, her or its own preferred vocabulary to be used. While common
terminologies are used as examples here, various embodiments of the presently
described technology include using proprietary codes, coding schema, syntax or
terminology.
Next at step 430, medical data is received at warehouse 310. In an embodiment
of the
presently described technology, the medical data is "pushed" by one or more
data
stores 320 to warehouse 310. For example, the medical data can be communicated
from a data store 320 to warehouse 310 without receiving a query or request at
data
store 320 from warehouse 310. The medical data can be pushed to warehouse 310
on
a periodic basis, whenever the data is obtained, or in response to a user
request, for
example.
In another embodiment, the medical data is "pulled" from one or more data
stores 320
to warehouse 310. For example, the medical data can be communicated from a
data
store 320 to warehouse 310 in response to warehouse 310 communicates a query
or
request for data to data store 320. Warehouse 310 can communicate the request
to
data store 320 on a periodic basis or in response to a user request, for
example.
Next at step 430, a part or entirety of medical data communicated to warehouse
310 is
normalized after it is received at warehouse 310. For example, all or a part
of the
clinical data and/or genetic data stored at a given data store 320 can be
normalized.
By "normalizing" it is meant that the various terms and syntax used by various
PCPs
in recording the medical data are changed or mapped to a common, controlled
medical vocabulary used for all medical data.
In another embodiment, normalizing the data can include changing or mapping
the
terms in the medical data to a vocabulary used by a subset of all users of the
presently
described technology. For example, instead of using the same common vocabulary
for all hospitals or clinics, one or more hospitals, clinics or other subset
of users can
use their own common vocabulary. In such an embodiment, the vocabulary common
only to the subset can differ from the common, controlled medical vocabulary
used by
one or more other subsets of users.
CA 02592705 2007-06-21
214217 31PS
The medical data can be normalized by mapping terms and syntax used to
describe
clinical and/or genetic data contained in an EMR to a common, controlled
vocabulary.
That is, each of several terms that can be considered synonyms and/or describe
the
same or similar phenotypic expression of a trait, medical condition, disease,
or
problem are mapped to a single code or term in a controlled vocabulary. For
example, the term "juvenile diabetes" can appear in one EMR communicated to
warehouse 310 and the term "type 1 diabetic" can appear in another EMR
communicated to warehouse 310. These terms can then be mapped, or associated
with, a term common to all synonyms for "juvenile diabetes" and "type 1
diabetic" in
the respective EMRs. Such a common term can be "type I diabetes," for example.
The mapping of terms can also be performed for any term or codes used to
describe
genetic data in an EMR.
The common terms can be provided in a list or table stored at warehouse 310.
This
list or table can also include all synonyms for the common term. Then, when
clinical
and/or genetic data is communicated in an EMR to warehouse 310, the term(s)
used to
describe the clinical and/or genetic data can be obtained from the EMR and
compared
to the synonyms included in the list or table of common terms. If a match is
found for
the term(s) used to describe the clinical and/or genetic data in the list or
table, the
common term for all synonyms associated with the clinical and/or genetic data
is then
mapped to the term(s) used to describe the clinical and/or genetic data. For
example,
a term used to describe a phenotypic expression of a trait communicated as
clinical
data in an EMR can be mapped to a common term representative of a group of
synonyms for the phenotypic expression of the trait.
In another embodiment of the presently described technology, medical data can
be
normalized by classifying terms and syntax used to describe clinical and/or
genetic
data contained in an EMR with an arbitrary term, such as a numeric or
alphanumeric
code or classification. For example, terms in the medical data can be
normalized by
codifying them with an ICD code. That is, each of several terms that can be
considered synonyms and/or describe the same or similar medical problem are
codified by assigning the terms to a single code or arbitrary term. For
example, the
term "juvenile diabetes" can appear in one EMR communicated to warehouse 310
and
16
CA 02592705 2007-06-21
214217 31PS
the term "type 1 diabetic" can appear in another EMR communicated to warehouse
310. These terms can then be codified with a numeric code that is common to a
group
of synonyms for "juvenile diabetes."
The codes or arbitrary terms can be provided in a list or table stored at
warehouse
310. This list or table can also include a group of synonyms for the code or
arbitrary
terms. Then, when a phenotypic expression of a trait is communicated in an EMR
to
warehouse 310, for example, the term used to describe the phenotypic
expression of
the trait can be obtained from the EMR and compared to the synonyms included
in the
list or table of codes/arbitrary terms. If a match is found, the EMR is then
codified
with the code common term to a group of synonyms associated with the
expression of
the trait.
Next, at step 450, one or more subsets of patients is created. The subsets can
be
created to divide up the entire population of codified clinical or medical
data into one
or more groups (that is, subsets) of patients with one or more phenotypic
expressions
of a trait, medical conditions, diseases, medical problems or environmental
conditions
in common.
These subsets can be created by a user first selecting or inputting at least
one clinical
condition. The user can input or select the condition(s) into device 330. The
clinical
conditions input by the user include one or more parameters related to the
clinical
and/or genetic data in one or more of the EMRs stored at warehouse 310. The
clinical
conditions input by the user can include any medical or genetic data, problem,
condition or disease. For example, the clinical conditions can include
diseases,
chronic ailments, disabilities, adverse reactions to medical therapeutics,
allergies,
environmental factors, and other medical problems. Environmental factors can
include any information relevant to the environment in which a patient lives
or works.
For example, the fact that a patient is a smoker, lives in a home with
smokers, works
in a smoke-filled environment, is a descendant of someone who died from
bronchogenic carcinoma, lives near power lines, and has relatives with one or
more
other clinical conditions are each examples of environmental factors. In
addition, a
patient's diet and/or pattern of exercise are other examples of environmental
factors.
17
CA 02592705 2007-06-21
214217 31PS
In another example, at step 450 a subset of patients can be created that
includes all
patients that take a particular prescription drug, such as Lipitor. Another
subset of
patients can be created that includes all patients that were checked for a
particular
medical problem using a particular laboratory or clinical test. For example, a
subset
can include all patients that have been checked for muscle breakdown using a
test that
measures muscle enzymes.
More than one clinical condition can be used to create or generate a subset.
In
continuing with the above example, a subset can be created that includes all
patients
that take a particular prescription drug and have a particular medical problem
or
laboratory test result. For example, a subset can include all patients that
take Lipitor
(at or above a certain dose, for example) and that have muscle breakdown
(measured
using a laboratory test for muscle enzymes, for example).
The clinical conditions can also include genetic data. For example, the
clinical
conditions can include one or more SNPs or one or more combinations of SNPs.
The user can input the clinical conditions using computing device 330. For
example,
the user can use an input device to type or select one or more clinical
conditions
displayed on an output device into a computer-generated list. The clinical
conditions
are used to generate a population, or group, of patients with one or more
similar or
identical clinical conditions, as described above. That is, the list of
clinical conditions
is used by computing device 330 to search through all or a subset of the EMRs
(or to
all or a subset of the data contained in one or more EMRs) to find the same or
similar
clinical conditions in the EMR(s). If a match for one or more of the clinical
conditions input by the user in one or more EMRs, those EMRs and the patients
associated with the EMRs are included in a subset of patients to be examined.
As described above, the clinical and/or genetic data included in EMRs stored
at
warehouse 310 is normalized at step 440 so that different terms used to
describe the
same or similar clinical and/or genetic data in various EMRs from various data
stores
320 are mapped to a common term or are encoded with the same or similar code.
In
this way, medical data input by different persons, hospitals, or groups using
different
18
CA 02592705 2007-06-21
214217 31PS
terms, syntax or vocabularies can easily be scanned or searched to provide a
subset of
patients with the same or similar medical or clinical conditions.
In an embodiment of the presently described technology, computing device 330
selects only those EMRs with data that matches each clinical condition
included in the
list. Therefore, if a list includes five clinical conditions and an EMR
includes data
that matches four or less of the clinical conditions, then the EMR is not
selected. On
the other hand, if a list includes five clinical conditions and an EMR
includes data that
matches all five of the clinical conditions, then the EMR is selected.
In another embodiment of the presently described technology, computing device
330
selects only those EMRs with data that matches a number of clinical conditions
included in the list that exceeds a threshold. For example, if a threshold is
set at three matches and a list includes five clinical conditions, an EMR must
include data that
matches at least three of the clinical conditions in the list. If the EMR only
includes
data that matches two or less conditions in the list, then the EMR is not
selected.
In another embodiment of the presently described technology, computing device
330
selects EMRs with data that matches a number of clinical conditions included
in the
list that meets or exceeds one of a plurality of thresholds. For example,
three
thresholds can be set at five matches (between EMR data and the list of
clinical
conditions), three matches and one match. If an EMR includes data that matches
enough clinical conditions to meet or exceed one of the thresholds, the EMR is
selected and placed into a category associated with the threshold number of
matches.
In continuing with the above example, an EMR with data that matches two
clinical
conditions is placed into the category of EMRs with data that matches at least
one, but
less than three clinical conditions; an EMR with data that matches three
clinical
conditions is placed into the category of EMRs with data that matches at least
three,
but less than five clinical conditions; and an EMR with data that matches
eight
clinical conditions is placed into the category of EMRs with data that matches
at least
five clinical conditions. By sorting the EMRs according to the number of
matches
between the EMR data and the list of clinical conditions, a user of the
presently
described technology can obtain several patient populations to select from
based on
the number of EMR data and list matches. Again continuing with the above
example,
19
CA 02592705 2007-06-21
214217 31PS
if given a set of 100 EMRs and the above thresholds, where 25 EMRs include
data
matching at least one, but less than three clinical conditions in the list, 5
EMRs
include data matching at least three, but less than five clinical conditions
in the list, 2
EMRs include data matching at least five clinical conditions, and 68 EMRs that
do
not include any data that matches any clinical condition, a user can select
the group of
25 EMRs for his/her analysis.
In another embodiment of the presently described technology, computing device
330
selects EMRs with data that matches a number of clinical conditions included
in the
list that meets or exceeds one or more of a plurality of thresholds. For
example, three
thresholds can be set at five matches (between EMR data and the list of
clinical
conditions) (referred to as "Category 5"), three matches (referred to as
"Category 3")
and one match (referred to as "Category 1"). If an EMR includes data that
matches
enough clinical conditions to meet or exceed one or more of the thresholds,
the EMR
is selected and placed into each category associated with the threshold number
of
matches that the EMR data meets or exceeds. In continuing with the above
example,
an EMR with data that matches two clinical conditions is placed into Category
1; an
EMR with data that matches three clinical conditions is placed into both
Category 1
and Category 3; and an EMR with data that matches eight clinical conditions is
placed
into Category 1, Category 3 and Category 5. By sorting the EMRs according to
the
number of matches between the EMR data and the list of clinical conditions, a
user of
the presently described technology can obtain several patient populations to
select
from based on the number of EMR data and list matches.
In an embodiment of the presently described technology, a user can input a
plurality
of lists of clinical conditions and obtain a plurality of subsets of EMRs
and/or patients
that match one or more of the lists (as described above). The user can then
use
computing device 330 to select which list(s) he or she wants to use in his or
her
analysis of the data.
In an embodiment of the presently described technology, after a user has input
a list of
clinical conditions and obtained a subset of EMRs and/or patients that match
one or
more of the lists, the user can employ the input device of computing device
330 to
change one or more clinical conditions in the list and view the corresponding
CA 02592705 2007-06-21
214217 31PS
change(s) to the subset of EMRs and/or patients that match the changed list.
This
change in the subset of EMRs and/or patients can occur in substantially real
time. By
"substantially real time," it is meant that the change in the list and/or
corresponding
change in the subset of EMRs/patients occurs and is presented to the user on
an output
device in a time period no longer than required for computing device 330,
warehouse
310 and/or data stores 320 to select and present the data. That is, no
intentional delay
is added to the selection of data that matches the changed list. By allowing a
user to
dynamically change the list and subset of EMRs/patients in this way, a user
can
quickly change one or more parameters/clinical conditions included in the list
to view
the impact on the number of EMRs/patients that match the list after the
change(s).
Once the one or more subsets of patients has been created at step 450, a user
can
select one or more of the subsets at step 460. For example, several subsets
can be
created at step 450 and one subset can be preferred (and selected) over other
subsets.
One such selected subset can be a subset with the largest number of patients
in it, for
example. In another example, a subset can be selected because it includes a
number
of patients above a threshold number of patients. The selection of a subset
can be
performed manually or automatically. For example, a user can manually select a
subset using an input device connected to computing device 330. In another
example,
a subset can be selected automatically if the number of patients in the subset
is at or
above a threshold, or has the largest number of patients in it when compared
to the
other subsets.
Next, at step 470, a determination is made as to whether any correlations
exist among
the genetic data associated with the patients in the selected subset(s). That
is, once a
subset of patients is selected, a determination is made as to whether a
statistically
significant number of the patients are associated with or have EMRs that
contain the
same or similar data. For example, a determination can be made at step 470 as
to
whether a statistically significant number of patients include the same SNP,
the same
plurality of SNPs or the same medical problem.
In an embodiment of the presently described invention, the correlation(s) are
determined or calculated between genetic data included in the subset of EMR(s)
and
one or more of the clinical conditions in the list generated at step 450. That
is, a
21
CA 02592705 2007-06-21
214217 31PS
determination is made as to whether a sufficient number of patients are
associated
with EMRs that include the same or similar genetic data. For example, if a
number of
patients exceeding a threshold have EMRs with the same SNP(s) or group(s) of
SNPs,
then a correlation is determined to exist. Such a determination is useful for
finding
correlations between medical problems, diseases, environmental factors,
allergies, for
example, and certain genetic data, such as SNPs or groups of SNPs.
In another embodiment of the presently described technology, the clinical
condition(s)
selected by a user to create a list of EMRs at step 450 is genetic data. For
example,
the user selects one or more SNPs or groups of SNPs as clinical conditions.
Then, at
step 470, a determination is made as to whether a sufficient number of
patients are
associated with EMRs that include the same or similar clinical data. For
example, if a
number of patients exceeding a threshold have EMRs with the same medical
problem,
allergy, environmental factor, or disease, then a correlation is determined to
exist.
Such a determination is useful for finding "mirror-image" correlations to
those
described above. Specifically, such a determination is useful for finding
correlations
between genetic data, such as SNPs or groups of SNPs, and certain medical
problems,
such as diseases and allergies, for example.
In an embodiment of the presently described technology, a correlation between
clinical conditions and clinical and/or genetic data is only found at step 470
if a
number of patients or EMRs exceeds a threshold. For example, if a threshold is
set at
70 and over 70 patients have or EMRs include the same or similar genetic
and/or
clinical data (as described above), then a correlation exists.
In another embodiment of the presently described technology, a correlation
between
clinical conditions and clinical and/or genetic data is only found at step 470
if a
percentage of patients or EMRs selected at step 460 exceeds a threshold. For
example, if a threshold is set at 70 percent and over 70 percent of the
patients or
EMRs selected at step 460 have or EMRs include the same or similar genetic
and/or
clinical data (as described above), then a correlation exists.
Next, at step 480, if one or more correlations is determined to exist, the
user is
provided with a notification by computing device 330 once a correlation is
found to
22
CA 02592705 2007-06-21
214217 31PS
exist. The notification can be a visual display or audible sound on an output
device
on computing device 330, for example.
In another embodiment of the presently described technology, one or more steps
in
method 400 is eliminated or performed in an order different from that
described above
and illustrated in FIG. 4. For example, step 460 can be omitted. In such an
example,
method 400 proceeds from the creation of one or more patient subsets (at step
450) to
the determination of whether any correlations exist between the genetic data
of the
patients in the subset and their associated medical problems/conditions (at
step 470),
for example.
The presently described invention provides, among other things, an automated
method
to narrow a large population of patients or EMRs to a subset determined
according to
a list of clinical conditions input by a user, where the subset of
patients/EMRs can
then be analyzed to determine if any genetic data and/or clinical data is
common to
the subset of patients/EMRs. Such a method provides a faster, more efficient
ability
to perform analysis on a large amount of genetic and clinical data. In
addition, as data
obtained from a plurality of clinical trials, PCPs, hospitals and clinics (for
example) is
normalized before analysis, correlations among patients/EMRs and clinical
and/or
genetic data can be determined even if many or all of the sources of the data
employ
different syntax to record the data.
In another embodiment of the presently described technology, step 440 occurs
before
step 430. That is, the normalization of the data stored at the various data
stores 320
occurs before the data is communicated to warehouse 310. The normalization can
be
performed by a computing device similar or identical to computing device 330
that is
connected to a data store 320. In this manner, the data included in an EMR
stored at a
data store 320 is normalized before it is received at warehouse 310 so that no
additional normalization is required.
As described above, in an embodiment of the presently described technology, a
computer-readable memory is accessible to computing device 330 and includes a
set
of instructions for a computer. The set of instructions includes one or more
routines
23
CA 02592705 2007-06-21
214217 31PS
capable of being run or performed by computing device 330. The set of
instructions
can be embodied in one or more software applications or in computer code.
The set of instructions can include a data normalization routine configured to
normalize one or more of the genotypic data and clinical data associated with
each
patient in a population of patients. As described above with respect to step
440 of
method 400, clinical data and/or genetic (or genotypic) data can be stored on
EMRs at
various data stores 320. Once a plurality of these EMRs (that can each include
different terms or syntax to describe the clinical and/or genetic data) are
received at
warehouse 310, the normalization routine can cause computing device 330 to
normalize the data. That is, the normalization routine can receive the data
and
normalize it. As described above, the normalization of the data can occur, for
example, by mapping terms used to describe the same or similar medical
conditions or
genetic information to a single common term or by codifying synonyms of the
same
or similar medical conditions or genetic information to an alphanumeric code.
In another embodiment of the presently described technology, the data
normalization
routine can be included in a second set of instructions stored on a computer-
readable
medium accessible by one or more computer devices in communication with one or
more data stores 320. As described above, the normalization of data can occur
before
the data is communicated from data store(s) 320 to warehouse 310. In such an
embodiment, the normalization routine can operate on, or cause a computing
device in
communication with a data store 320 to normalize the data before the data in
the EMR
is communicated to warehouse 310, for example.
The set of instructions can also include a patient selection routine
configured to select
a subset of patients from said population based on one or more clinical
conditions
input by a user. As described above with respect to step 450 of method 400, a
subset
of EMRs can be selected from a group of EMRs stored at warehouse 310 based on
a
plurality of clinical conditions input by a user, for example. The patient
selection
routine can operate on, or cause computing device 330 to select the subset of
EMRs
from the group of EMRs at warehouse 310.
24
CA 02592705 2007-06-21
214217 31PS
The set of instructions can also include a correlation routine configured to
determine
one or more correlations between at least one of the clinical conditions and
one or
more of the genetic and clinical data. As described above with respect to step
470 of
method 400, one or more correlations or relationships between one or more
clinical
conditions input by a user (such as a medical problem or SNP/group of SNPs,
for
example) and genetic and/or clinical data included in the EMRs selected by the
patient selection routine at step 460 can be calculated. The correlation
routine can
operate on, or cause computing device 330 to determine or calculate the
correlation(s), if any, existing between the clinical conditions and the data,
as
described above.
In an embodiment of the presently described technology, the set of
instructions can
include a notification routine configured to notify a user when one or more of
correlations calculated or determined by the correlation routine exceed one or
more
thresholds. As described above with respect to step 480 of method 400, once a
correlation is found to exist by the correlation routine, a notification is
communicated
to a user. For example, the notification routine can operate on, or cause
computing
device 330 to provide a visual display on a display device or provide an audio
notification on a speaker.
In an embodiment of the presently described technology, the set of
instructions can
include an input routine configured to alter one or more thresholds that an
amount of
match between one or more clinical conditions selected by a user and genetic
and/or clinical data in the subset of EMRs is compared against. As described
above, a user
can employ an input device of computing device 330 to change one or more
clinical
conditions in the list of clinical conditions and view any corresponding
change(s) to
the subset of EMRs and/or patients that match the changed list. For example,
the
input routine can receive input from a user in the form of the selection or de-
selection
(that is, removing one or more clinical conditions from a list of clinical
conditions
previously selected by the user) of one or more clinical conditions. The input
routine
can then operates on, or causes computing device 330 to alter the list of
clinical
conditions and, consequently, causes the patient selection routine to change
the EMRs
included in the subset of EMRs selected by the patient selection routine, for
example.
CA 02592705 2007-06-21
214217 31PS
The technical effect of the set of instructions described above is, among
other things,
to provide an automated method to narrow a large population of patients or
EMRs to a
subset determined according to a list of clinical conditions input by a user,
where the
subset of patients/EMRs can then be analyzed to determine if any genetic data
and/or
clinical data is common to the subset of patients/EMRs. The set of
instructions can
then provides a faster, more efficient ability to perform analysis on a large
amount of
genetic and clinical data. In addition, as data obtained from a plurality of
clinical
trials, PCPs, hospitals and clinics (for example) is normalized before
analysis,
correlations among patients/EMRs and clinical and/or genetic data can be
determined
even if many or all of the sources of the data employ different syntax to
record the
data, for example.
While the invention has been described with reference to example embodiments,
it
will be understood by those skilled in the art that various changes may be
made and
equivalents may be substituted for elements thereof without departing from the
scope
of the invention. In addition, many modifications may be made to adapt a
particular
situation or material to the teachings of the invention without departing from
the
essential scope thereof. Therefore, it is intended that the invention not be
limited to
the particular embodiment disclosed as the best mode contemplated for carrying
out
this invention, but that the invention will include all embodiments falling
within the
scope of the appended claims. Moreover, the use of the terms first, second,
etc. do
not denote any order or importance, but rather the terms first, second, etc.
are used to
distinguish one element from another.
In addition, while particular elements, embodiments and applications of the
present
invention have been shown and described, it is understood that the invention
is not
limited thereto since modifications may be made by those skilled in the art,
particularly in light of the foregoing teaching. It is therefore contemplated
by the
appended claims to cover such modifications and incorporate those features
that come
within the spirit and scope of the invention.
26