Note: Descriptions are shown in the official language in which they were submitted.
CA 02595040 2007-08-03
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPREND PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
NOTE: Pour les tomes additionels, veillez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME OF _2
NOTE: For additional volumes please contact the Canadian Patent Office_
CA 02595040 2007-08-03
N '7/30074 PCT/US97/0229f
ISOLATION AND USE OF SH3 BINDING PEPTIDES
1. Field of the Invention
The present invention relates to SH3 binding peptides
having a broad range of binding specificities. That is,
certain members of the SH3 binding peptides disclosed bind
with approximately the same facility with SH3 domains derived
from different SH3 domain-containing proteins. Other
members, in contrast, bind with a much greater degree of
affinity for specific SH3 domains. The SH3 binding peptides
are obtained from random peptide libraries that are also
phage-displayed. Methods are described of obtaining the
phage clones that bind to the SH3 domain targets and of
determining their relevant nucleotide sequences and
consequent primar.y amino acid sequence of the binding
peptides. The resulting SH3 binding proteins are useful in a
number of ways, including, but not.limited to, providing a
method of modulating signal transduction pathways at the
cellular level, of modulating oncogenir, protein activity or
of providing lead compounds for development of drugs with the
ability to modulate broad classes, as well as specific
classes, of proteins involved in signal transduction.
2. Background of the Invention
2.1. 6rc and the 6H3 Domain
Among a number of proteins involved in eukaryotic cell
signaling, there is a common sequence motif called the SH3
domain. It is 50-70 amino acids in length, moderately
conserved in primary structure, and can be present from one
to several times in a large number of proteins involved in
signal transduction and in cytoskeletal proteins.
The protein pp60c-src represents a family of at :Least
nine non-receptor protein tyrosine kinases (NR-PTKs).
Members of this family share an overall structural
organization comprising a series of catalytic and non=-
catalytic domains. In Src, a 14-amino-acid myristylation
- 1 -
CA 02595040 2007-08-03
WO 97/3W74 I'CT/LJS97iu,._,98
signal resides at the extreme amino-terminus, and is followed
by a unique region that is not highly conserved among family
members. Following this region are two highly conserved 60-
and 100-amino-acid regions, the Src homology (SH) domains 3
and 2, respectively. SH2 and SH3 domains have been shown to
play an important role in mediating protein-protein
interactions in a variety of signaling pathways. Koch, C.A.,
et al., in Science (1991) 252:668-74. The carboxy-terminal
half of Src contains the PTK catalytic domain, as well as a
negative regulatory tyrosine (Y527) near the carboxy
terminus. Phosphorylation of this residue (e.g., by Csk)
results in the inhibition of PTK activity. Cooper, J.A., et
al., in Science (1986) 231:1431-1434. Mutation of Y527->F
generates forms of Src with increased PTK and oncogenic
activity. Cartwright, C.A., et al., in Cell (1987) 49:83-91;
Kmiecik, T.E., et al., in Cell (1987) 49:65-73; and Piwicna-
Worms, H., et al., in Cell (1987) 75-82.
The fact that some mutations which result in increased
Src PTK and transforming activity map to the Src SH2 (Seidel-
Dugan, C., et al., in Mol. Cell. Biol. (1992) 12:1835-45; and
Hirai, H. and Varmus, H.E. in Mol. Cell. Biol. (1990)
10:1307-1318) and SH3 domains (Seidel-Dugan, C., et al.,
supra; Hirai, H. and Varmus, H.E., supra; Superti-Furga, G.,
et al., in Embo. J. (1993) 12:2625-34; and Potts, W.M., et
al., in Oncogene Res. (1988) 3:343-355) suggests a negative
regulatory role for these domains. That phosphotyrosine
residues within specific sequence contexts represent high
affinity ligands for SH2 domains suggests a model in which
the SH2 domain participates in Y527-mediated inhibition of
PTK activity by binding phosphorylated Y527, thereby locking
the kinase domain in an inactive configuration. Matsuda, M.,
Mayer, B.J., et al., in Science (1990) 248:1537-1539. This
model is supported by the observation that phosphopeptides
corresponding to the carboxy-terminal tail of Src bind
active, but not inactive, variants of Src. Roussel, R.R., et
al., in Proc. Natl. Acad. Sci. U S A (1991) 88:10696-700; and
Liu, X., et al., in 0ncoaene (1993) 8:1119-1126.
- 2 -
CA 02595040 2007-08-03
)97130074 PCTfUS97/022
The mechanism of SH3-mediated inhibition of Src PTK
activity remains unclear. There is evidence that pY527-
mediated inhibition of Src PTK activity involves the SH3
domain as well as the SH2 domain. Okada, M., Howell, et al.,
in J. Biol. Chem. (1993) 268:18070-5; Murphy, S.M., et al.,
in Mol. Cell. Biol. (1993) 13:5290-300; and Superti-Furga,
G., et al., supra. Although these effects are thought to be
a consequence of SH3-mediated protein-protein interactions,
precisely how the Src SH3 domain exerts its negative
regulatory effect is unclear. Identification of high
affinity ligands for the Src SH3 domain could help resolve
these issues.
2.2. Protein Tyrosine Kinases and The Smmune Response
1.5 Src-related tyrosine kinases are expressed in a variety
of cell types including those of the immune system
(lymphocytes, T cells, B cells, and natural killer cells) and
the central nervous system (neural cells, neurons,
oligodendrocytes, parts of the cerebellum, and the like).
Umemori, H. et al.; in Brain Res. Mol. Brain Res. (19.92) Dec.
16(3-4):303-310. Their presence in these cells and tissues
and their interaction with specific cell surface receptors
and immunomodulatory proteins (such as T cell antigen
receptor, CD14, CD2, CD4, CD40 or CD45) suggest that these
kinases serve an important role in the signalling pathways of
not only the central nervous system but of the immune system,
as well. See, e.g., Ren, C.L. et al., in J. Exp. Med. (1994)
179(2):673-680 (signal transduction via CD40 involves
activation of Lyn kinase); Donovan, J.A. and Koretzky, G.A.,
in J. Am. Soc. Nephrol. (1993) 4(4):976-985 (CD45, the immune
response, and regulation of Lck and Fyn kinases); and Carmo,
A.M. et al., in Eur. J. Immunol. (1993) 23(9):2196-2201
(physical association of the cytoplasmic domain of CD2 with
p561ck and p59fyn).
For instance, mice with disruptions in their Src-like
genes, Hck and Fgr, possess macrophages with impaired
phagocytic activity or exhibit a novel immunodeficiency
- 3 -
CA 02595040 2007-08-03
/O 97/30074 PCT/US97/0:'.,4y8
characterized by an increased susceptibility to infection
with Listeria monocytogenes. Lowell, C.A. et al., in Genes
Dev. (1994) 8(4):387-398. Also, it has been shown that
bacterial lipopolysaccharide (LPS) activates CD14-associated =
p561yn, p68hck, and p59c-fgr, while inducing the production
of lymphokines, such as TNF-alpha, IL-1, IL-6, and IL-8.
Inhibition of the protein tyrosine kinases blocks production
of TNF-alpha and IL-1.
2.3. SH3 Binding Peptides
As mentioned above, it has long been suspected that SH3
domains are sites of protein-protein interaction, but it has
been unclear what SH3 domains actually bind. Efforts to
identify ligands for SH3 domains have led to the
characterization of a number of SH3-binding proteins,
including 3BP1 and 2 (Ren, R., Mayer, et al., in Science
(1993) 259:1157-61); SOS (Olivier, J.P., et al., in Cell
(1993) 73:179-91; and Rozakis-Adcock, M., et al., in Nature
(1993) 363:83-5), p85 PI-3~ Kinase (Xingquan, L., et al., in
Mol. Cell. Biol. (1993) 13:5225-5232), dynamin (Gout, 1., et
al., in Cell (1993) 75:25-36), AFAP-110 (Flynn, D.C., et al.,
in Mol. Cell. Biol.(1993 ) 13 : 7892=-7900) , and CD42 (Barfod,
E.T., et al., in J. Biol. Chem. (1993) 268:26059-26062).
These proteins tend to possess short, proline-rich stretches
of amino acids, some of which have been directly implicated
in SH3 binding. A variety of consensus sequences have been
proposed, although the similarity among proline-rich regions
of different SH3-binding proteins tends to be fairly low.
Also, attempts to build consensus sequences are likely
complicated by the incorporation of data from proteins that
bind different SH3 domains.
Thus, Cicchetti, P., et al., in Science (1992) 257:803-
806, published their work relating to the isolation and
sequencing of two naturally-occurring proteins that could be
bound in vitro by the SH3 domain of the abl oncogene prc>duct.
These workers found that SH3 domains bind short, proline-rich
regions of such proteins. Subsequently, this same group
- 4 -
CA 02595040 2007-08-03
J 97/30074 PCT/US97/02292s
disclosed further results (Ren, R. et al., supra) in which
the SH3 binding sites of the SH3 binding proteins were
localized to "a nine- or ten-amino acid stretch rich in
proline residues." A consensus sequence incorporating the
features of the SH3 binding sites of four SH3 binding
proteins was proposed: XPXXPPPYXP (SEQ ID NO:1), wherein X
indicates a position in the amino acid sequence which is not
conserved among the four SH3 binding proteins, P represents
proline, and V indicates a hydrophobic amino acid residue,
such as P or L.
The screening of complex random peptide libraries has
been used to identify peptide epitopes for monoclonal (Scott,
J.K. and Smith, G.P. in Science (1990) 249:386-390) and
polyclonal (Kay, B.K., et al., in Gene (1993) 128:59-65)
antibodies, as well as peptide ligands for a variety of
proteins, including streptavidin (Devlin, J.J., et al., in
Science (1990) 249:404-406; and Lam, K., et al., in Nature
(1991) 354:82-84), the endoplasmic reticulum chaperone BiP
(Blond-Elguindi, S., et al., in Cell (1993) 75:71i-728), and
CaM (Dedman, J.R., et al., in J. Biol. Chem. (1993)
268:23025-23030).
Recently, Chen, J.K. et al., in J. Am. Chem. Soc. (1993)
115:12591-12592, described ligands for the SH3 domain of
phosphatidylinositol 3-kinase (PI-3' Kinase) which were
isolated from a biased combinatorial library. A "biased"
library is to be distinguished from a "random" library in
that the amino acid residue at certain positions of the
synthetic peptide are fixed, i.e., not allowed to vary in a
random fashion. indeed, as stated by these research workers,
screening of a "random" combinatorial library failed to yield
suitable ligands for a PI-3' Kinase SH3 domain probe. The
binding affinities of these unsuitable ligands was described
as weak, >100 M, based on dissociation constants.measured by
the Biosensor System (BIAcore).
More recently, Yu, et al. (Yu, H., et al., in Cell
(1994) 76:933-945) used a "biased" synthetic peptide library
of the form XXXPPXPXX (SEQ ID NO:2), wherein X represents any
- 5 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/02198
amino acid other than cysteine, to identify a series of
peptides which bind the Src and PI-3' Kinase SH3 domains.
The bias was accomplished by fixing the proline residues at
the specific amino acid positions indicated for the "random"
peptide. As stated previously, without this bias, the
technique disclosed fails to identify any SH3 domain-binding
peptides.
A consensus sequence, based on 13 binding peptides was
suggested: RXLPPRPXX (SEQ ID NO:3), where X tends to be a
l0 basic residue (like R, K or H). The binding affinities of
several SH3 binding peptides were disclosed as ranging from
8.7 to 30 M. A "composite" peptide, RKLPPRPRR (SEQ ID
NO:4), was reported to have a binding affinity of 7.6 M.
This value compares favorably to the binding affinity of the
peptide, VPPPVPPRRR (SEQ ID NO:5), to the N-terminal SH3
domain of Grb2. See, Kraulis, P.J. J. Appl. Crvstallocrr.
(1991) 24:946. Recognizing the limitations of their
technique, Chen and co-workers, supra, stated that their
results "illustrate the utility of biased combinatorial
libraries for ligand discovery in systems where there is some
general knowledge of the ligand-binding characteristics; of
the receptor" (emphasis added).
Yu and co-workers, supra, further described an SH3
binding site consensus sequence, XpOPpXP (SEQ ID NO:6),
wherein X represents non-conserved residues, 0 represents
hydrophobic residues, P is proline, and p represents residues
that tend to be proline. A consensus motif of RXLPPRPXX (SEQ
ID NO:7), where X represents any amino acid other than
cysteine, was proposed for ligands of PI-3' Kinase SH3
domain. A consensus motif of RXLPPLPRO (SEQ ID NO:8), where
0 represents hydrophobic residues, was proposed for ligands
of Src SH3 domain. Still, the dissociation constants
reported for the 9-mer peptides ranged only from-about 8-70
M and selectivity between one type of SH3 domain and another
was relatively poor, the KDs differing by only about a factor
of four.
- 6 -
CA 02595040 2007-08-03
J 97/30074 PCT/US971022S.,
Hence, there remains a need to develop techniques for
the identification of Src SH3 binding peptides which do not
rely on such "biased" combinatorial peptide libraries that
are limited to a partially predetermined set of amino acid
sequences. Indeed, the isolation of SH3 binding peptides
from a"random" peptide library has not been achieved
successfully before now. Furthermore, particular peptides
having much greater binding affinities, whether general or
more selective binding for specific SH3 domains, remain to be
.lo identified. Binding peptides specific for particular SH3
domains are useful, for example, in modulating the activity
of a particular SH3 domain-containing protein, while leaving
others bearing an SH3 domain unaffected. Still; the more
promiscuous general binding peptides are useful for the
modulation of a broad spectrum of SH3 domain-containing
proteins.
The present invention relates to such SH3 binding
peptides, methods for their identification, and compositions
comprising same. In particular, peptides comprising
particular sequences of amino acid residues are disclosed
which were isolated from random peptide libraries. In the
present invention, clones were isolated from a phage-
displayed random peptide library which exhibited strong
binding affinities for SH3 domain-containing protein targets.
Some of these protein targets, include Abl, Src, Grb2, PLC-6,
PLC--y, Ras GAP, Nck, and p85 PI-3' Kinase. From the
nucleotide sequence of the binding phage, the amino acid
sequence of the peptide inserts has been deduced. Synthetic
peptides having the desired amino acid sequences are shown to
bind the SH3 domain of the target proteins. In particu:Lar,
synthetic peptides combining a core consensus sequence and
additional amino acid residues flanking the core sequence are
especially effective at binding to particular target protein
SH3 domains. The SH3 binding peptides disclosed herein can
be utilized in a number of ways, including the potential
modulation of oncogenic protein activity in vivo. These
peptides also serve as useful leads in the production of
- 7 -
CA 02595040 2007-08-03
VO 97/30074 PCT/US97/0. s
peptidomimetic drugs that modulate a large class of proteins
involved in signal transduction pathways and oncogenesis.
3. Summary of the Invention
Accordingly, three phage-displayed random peptide
libraries were screened-for isolates that bind to bactei-ial
fusion proteins consisting of the Src homology region 3 (SH3)
and glutathione S-transferase (GST). DNA sequencing of the
isolates showed that they contained sequences that resemble
the consensus motif, RPLPPLP (SEQ ID NO:9), within their 8,
22, or 36 amino acid long random regions. When peptides were
synthesized corresponding to the pIII inserts of the SH3-
binding phage, they bound to the GST fusions of the SH3
domains of Src and the Src-related proteins, such as Yes, but
not of Grb2, Crk, Abl, or PLCT1. The synthesized peptides
bind quite well to the Src SH3 domain and act as potent
competitors of natural Src SH3 interactions in cell lysates.
For instance, these peptides can compete with radiolabelled
proteins from cell lysates in binding to immobilized Src:=-GST,
with an apparent ICso of 1-10 gM. When a peptide, beariing the
consensus sequence RPLPPLP (SEQ ID NO:9) was injected into
Xenopus laevis oocytes, it accelerated the rate of
progesterone-induced maturation. These results demonstrate
the utility of phage-displayed random peptide libraries in
identifying SH3-binding peptide sequences and that such
identified peptides exhibit both in vivo and in vitro
biological activity.
Thus, it is an object of the present invention to
provide peptides having at least nine and up to forty-five
amino acid residues, including an amino acid sequence of the
formula, R-2-L-P-5-6-P-8-9 (SEQ ID NO:10), positioned
anywhere along the peptide, in which each number represents
an amino acid residue, such that 2 represents any amino acid
residue except cysteine, 5 and 6 each represents a
hydrophobic amino acid residue, 8 represents any amino acid
residue except cysteine, and 9 represents a hydrophilic amino
acid residue except cysteine, each letter being the standard
- 8 -
CA 02595040 2007-08-03
97/34074 PCT/US97/0225
one-letter symbol for the corresponding amino acid, said
peptide exhibiting a binding affinity for the SH3 domain of
Src, provided that said peptide is not R-P-L-P-P-L-P-T-S (SEQ
ID No:11). In a particular embodiment of the present
invention, the peptides also exhibit a binding affinity for
the SH3 domain of Src-related proteins, including Yes, Fyn,
Lyn, Lck, Hck and Fgr.
The present invention also contemplates SH3 domain-
binding peptides that further comprise a C-terminal-flanking
amino acid sequence of the formula 10, 10-11, 10-11-12, 10-
11-12-13 (SEQ ID N0:12) or 10-11-12-13-14 (SEQ ID NO:13), in
which each number represerits any amino acid residue except
cysteine, such that 10 is bound to 9 by a peptide bond.
Furthermore, peptides are also provided which further
comprise an N-terminal-flanking amino acid sequence of the
formula 1', 2'-1', 3'-2'-1' or 4'-3'-2'-1' (SEQ ID NO:14) in
which each number represents any amino acid residue except
cysteine, such that 1' is bound to R by a peptide bond.
Thus, in a particular embodiment, a peptide is disclosed
having at least thirteen and up tc forty-five amino acid
residues, including an amino acid sequence of the formula,
3'-2'-1'-R-2-L-P-5-6-P-8-9-10 (SEQ ID NO:15), positioned
anywhere along the peptide, in which each number represents
an amino acid residue, such that 3', 2', 1', 2, 8, and 10
each represents any amino acid residue except cysteine, 5 and
6 each represents a hydrophobic amino acid residue, and 9
represents a hydrophilic amino acid residue except cysteine,
each letter being the standard one-letter symbol for the
corresponding amino acid, said peptide exhibiting a binding
affinity for the SH3 domain of Src.
The present invention also seeks to provide new
consensus sequences or motifs that reflect variations in SH3
domain binding selectivities or specificities. The present
invention also contemplates conjugates of the SH3 binding
peptides and a second molecule or chemical moiety. This
second molecule may be any desired substance whose delivery
to the region of the SH3 domain of a particular protein (or
- 9 -
CA 02595040 2007-08-03
75361-88
cell containing the protein) is sought. Possible target
cells include, but are not limited to, neural cells, immurle
cells (e.g., T cells, B cells, natural killer cells, and the
like), osteoclasts, platelets, epidermal cells, and the
like, which cells express Src, Src-related proteins, and
potentially, other SH3 domain-containing proteins. In this
manner, the modulation of the biological activity of
proteins bearing an SH3 domain can be accomplished.
In one aspect, the invention provides a peptide
that binds to the SH3 domain of Cortactin, said peptide
comprising an amino acid sequence selected from the group
consisting of: PVKPPLPAKPWWLPPL (SEQ ID NO: 167);
YPQFRPPVPPKPSLMQ (SEQ ID NO: 168); VTRPPLPPKPGHMADF
(SEQ ID NO: 169); VSLGLKPPVPPKPMQL (SEQ ID NO: 170);
YKPEVPARPIWLSEL (SEQ ID NO: 171); and GAGAARPLVPKKPLFL
(SEQ ID NO: 172).
In another aspect, the invention provides a method
of identifying an inhibitor of the binding between a first
molecule comprising an SH3 domain and a second molecule that
binds to the SH3 domain, wherein the second molecule is a
peptide as described above, the method comprising:
incubating one or more compounds from which it is desired to
select such an inhibitor with the first molecule and the
second molecule under conditions conducive'to binding, arid
detecting the one or more compounds that inhibit binding of
the first molecule to the second molecule.
In another aspect, the invention provides a method
of identifying a compound that affects the binding of a
molecule comprising an SH3 domain and a ligand of the SH3
domain, wherein the ligand is a peptide as described
- 10 -
CA 02595040 2007-08-03
75361-88
above, the method comprising: (a) contacting the SH3 domain
and the ligand under conditions conducive to binding in the
presence of a candidate compound and measuring the amount of
biriding between the SH3 domain and the ligand; and (b)
comparing the amount of binding measured in step (a) with
the amount of binding known or determined to occur between
the molecule and the ligand in the absence of the candidate
compound, where a difference between the amount of binding
measured in step (a) and the amount of binding known or
determined to occur between the molecule and the ligand in
the absence of the candidate compound indicates that the
candidate compound is a compound that affects the binding of
the molecule comprising an SH3 domain and the ligand.
Other methods and compositions consistent with the
objectives of the present invention are likewise disclosed.
In particular, a method is disclosed of modulating the
activity of Src or Src-related proteins comprising
administering a composition comprising an effective amount
of a peptide of the present invention and a carrier,
preferably a pharmaceutically acceptable carrier. In a
specific embodiment, the contemplated method results in the
inhibition of the activity of Src or Src-related proteins.
Alternatively, the method is effective to activate Src or
Src-related proteins.
In yet another embodiment, a method is disclosed
of identifying a peptide having a region that binds to ari
SH3 domain comprising: (a) providing an immobilized target
protein comprising an SH3 domain; (b) incubating the
immobilized target protein with an aliquot taken from a
random peptide library; (c) washing unbound library peptides
from the immobilized target protein; (d) recovering the
peptide bound to the immobilized target protein; and
- l0a -
CA 02595040 2007-08-03
75361-88
(e) determining the primary sequence of the SH3 domain-
binding peptide.
Moreover, a method is disclosed of imaging cells,
tissues, and organs in which Src or Src-related proteins are
expressed, which comprises administering an effective amount
of a composition comprising an SH3 domain-binding peptide
conjugated to detectable label or an imaging agent.
Other objectives of the present invention will
become apparent to one of ordinary skill in the art after
- 10b -
CA 02595040 2007-08-03
97130074 PCTIUS97/022yo
consideration of the above disclosure and the following
detailed description of the preferred embodiments.
The invention also provides assays for identifying a
compound that affects the binding between a first molecule
s comprising an SH3 domain and a second molecule that binds to
the SH3 domain comprising incubating one or more candidate
compounds from which it is desired to select such a compound
with the first molecule and the second molecule under
conditions conducive to binding and detecting the one or more
lo compounds that affect binding of the first molecule to the
second molecule.
Also provided are kits for performing such assays
comprising a first molecule comprising an SH3 domain and a
second molecule that binds to the SH3 domain.
4. Brief Description of the Ficxures
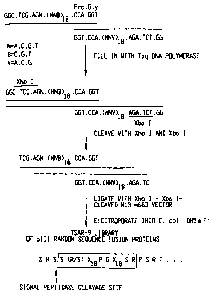
FIG. I illustrates a scheme for the generation of a
random 36 amino acid peptide library (TSAR-9; e.g., SEQ ID
NO:16). Oligonucleotides were synthesized (SEQ ID NOS:17-
18), converted into double-stranded DNA, cleaved with
restriction enzymes (SEQ ID NOS:19-20), and cloned into the
M13 vector, m663. The random peptide region encoded by the
oligonucleotides is shown in the box (SEQ ID NO:16) and is
situated at the N-terminus of mature protein III (SEQ ID
NO:21). SEQ ID NO:22 includes the three amino acids
preceding the signal peptidase cleavage site.
FIG. 2 illustrates a scheme for the generation of a
random 22 amino acid peptide library (TSAR-12; e.g., SEQ ID
NO:23). Oligonucleotides were synthesized (SEQ ID NOS:24-
25), converted into double-stranded DNA, cleaved with
restriction enzymes (SEQ ID NOS:26-27), and cloned into the
M13 vector, m663. The random peptide region encoded by the
oligonucleotides is shown in the box (SEQ ID NO:23) and is
situated at the N-terminus of mature protein III (SEQ ID
NO:28). SEQ ID NO:29 includes the three amino acids
preceding the signal peptidase cleavage site.
- 11 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/U,..198
FIG. 3 illustrates a scheme for the generation of a
random 8 amino acid peptide library (RBC; SEQ ID NO:30).
oligonucleotides were synthesized (SEQ ID NOS:31-32),
converted into double-stranded DNA, cleaved with restriction
enzymes (SEQ ID NOS:33-34), and cloned into the M13 vector,
m663. The random peptide region (SEQ ID N0:30) is flanked by
cysteine residues and is situated at the N-terminus of mature
protein III (SEQ ID NO:35).
FIG. 4 illustrates the possible origin of one class of
io double-insert R8C recombinants (e.g., encoding SEQ ID N0:36).
Double-stranded oligonucleotides (e.g., SEQ ID NO:37) may
have ligated in a head-to-head fashion at the Xba I SitE'
prior to cloning in the Xho I- Xba I cleaved M13 vector.
FIG. 5 shows a list of random peptide recombinants (SEQ
ID NOS:38-61 and 106) isolated by the method of the present
invention and the displayed peptide sequence. The aminc> acid
sequences are aligned to highlight the core sequences. The
flanking sequences are shown to the N-terminal and C-terminal
ends of the core sequence. SEQ ID NOS:38-61 are shown in
order from top to bottom except that SSCDHTLGLGWCGSRSTRQLPIPP
TTTRPSR is SEQ ID N0:106 and RPLPPLP is SEQ ID N0:9.
T12.Src3.1 is a Class II ligand (See Section 6.14.5).
FIG. 6 graphically illustrates the relative bindincf
affinities of selected phage clones for various SH3 domains.
The results indicate that certain amino acid sequences
provide generic SH3 domain binding, while others can provide
greater selectivity for the SH3 domain of Src. Still other
clones exhibit Src SH3 domain preferential binding.
FIG. 7 shows the binding of synthetic peptides (SEQ ID
NOS:9 and 62-70) representing Src SH3-selected phage inserts
to Src SH3-GST fusion target (shaded columns) over background
GST binding (unshaded columns) relative to the core pept:ide
RPLPPLP (SEQ ID NO:9) and proline-rich peptide segments
derived from naturally occurring proteins. Bound
biotinylated peptide was detected with streptavidin-alkaline
phosphatase ELISA. Each point was performed in triplicate;
average absorbance at 405 nm is presented. Error bars
- 12 -
CA 02595040 2007-08-03
75361-88
represent SD. SEQ ID NOS:62-70 are shown in order from top
to bottom except that RPLPPLP is SEQ ID NO:9.
FIG. 8 illustrates the relative specificity of selected
peptides (SEQ ID NOS:9 and 62-70) for SH3 domains derived
from different proteins. In particular, the binding
affinities of the peptides for the SH3 domains of the
following protein fusion targets were tested: Src SH3-GST,
Yes SH3-GST, Grb2-GST, Crk SH3-GST, Abl SH3-GST, PLCTl
SH2SH3-GST. Bound biotinylatecl peptide was detected with
lo streptavidin-alkaline phosphatase. Each point was performed
in triplicate; values are average signal (absorbance at 405
nm) above GST background, with error bars representing
standard deviation. Hatched bars indicate saturation of the
ELISA signal. SEQ ID NOS:62-70 are shown in order from top
to bottom except that RPLPPLP is SEQ T_D NO:9.
FIG. 9 presents the results of competition experiments
in which selected peptides were found to in.h.ibit the binding
of proteins from ce11 lysates to immobilized Src SH3-GST or
Abl SH3-GST protein fusion targets.
FIG. 10 presents a graph illustrating the increased rate
of progesterone-induced maturation of oocytes injected with
an SH3 domain-binding peptide, VLKRPLPIPPVTR (SEQ ID NO:64),
of the present invention. Briefly, Stage VI oocyte.s were
prepared and injected as previously described (see, Kay,
B.K., in Methods in Cell Biol. (1991) 36:663-669). Oocytes
were injected with 40 nL of 100 M test peptide or water.
After injection, the oocytes were placed in 2 g/mL
progesterone (Sigma, St. Louis, MO) and scored hourly for
germinal vesicle breakdown (GVBD). LAPPKPPLPEGEV is SEQ ID
NO:70.
FIG. 11 shows the results of fluorescence experiments in
which certain peptides, Panel A = VLKRPLPIPPVTR (SEQ ID
NO:64), Panel B = GILAPPVPPRNTR (SEQ ID ND:63), Panel C
RSTPRPLPPLPTTR (SEQ ID N0:67), of the invention were shown to
localize within cellular compartments thought to contain Src
or Src-related proteins.
- 13 -
CA 02595040 2007-08-03
-O 97/30074 PCT/US97/0:._ J
FIG. 12 illustrates a scheme for the generation of a
biased peptide library. Oligonucleotides were synthesized
(SEQ ID NOS:162-163), converted into double-stranded DNA (SEQ
ID NO:454), cleaved with restriction enzymes XhoI and XbaI
(SEQ ID NOs:455-456), and cloned into the mBAX vector (SEQ ID
NOs:457-458), described further below in the Examples
section. The biased peptide region (SEQ ID NO:459) is
situated at the N-terminus of mature piII protein.
CTAGACGTGTCAGT is a portion of SEQ ID NO:162. ACTGACACGT is
a portion of SEQ ID NO:454. TCGAGGCACAG is a portion of SEQ
ID NO:454.
FIG. 13 illustrates the peptide sequence encoded in the
mBAX vector situated at the N-terminus of mature piII
protein. TCCTCGAGTATCGACATGCCTTAGACTGCTAGCACTATGTACAACATGCTT
CATCGCAACGAGCCA is SEQ ID NO:460. SSIDMP*TASTMYNM LHRNEP is
SEQ ID NO:461. GGTGGGAGGAAGTTGAGCCCGCCCGCCAACGA
CATGCCGCCCGCCCTCCTGAAGAGGTCTAGA is SEQ ID N0:464"..
GGRKLSPPANDMPPALLKRSR is SEQ ID NO:463.
FIG. 14 illustrates the relative binding of SH3-selected
phage clones to various SH3 domains. Two clones (A and B)
representing each consensus motif were assayed for binding to
1 g of each immobilized GST-SH3 fusion protein. Bound phage
were detected by anti-phage ELISA. Sequences of peptides
displayed by each clone are aligned with their respective
consensus motifs. Invariant proline residues are underlined.
Solid bars, specific binding; open bars, cross-reactive
binding. Values are average OD90S SD (N =3).
5. Detailed Description of the Invention
5.1. General Considerations
The present invention relates to peptides that exhibit a
binding affinity for an SH3 domain, which domain has been
found to be present in a number of physiologically
significant proteins. In particular, peptides are disclosed
which exhibit general binding characteristics to the SH3
domains found in a group of proteins, including but not
limited to Abl, Src, Grb2, PLC-6, PLC-y, Ras GAP, Nck, and
- 14 -
CA 02595040 2007-08-03
)97/30074 PCT1US97/0225
p85 PI-3' Kinase. Preferred peptides exhibit selective, if
not specific, binding affinity for the SH3 domain of Src. As
described herein, the peptides of the present invention
include a core sequence, preferably a consensus seqeunce, and
additional amino acid residues that flank the core sequence.
These peptides, including the methods for their
identification, are described in greater detail, below.
Thus, in a specific embodiment of the invention,
peptides are provided which have at least nine and up to
lo about forty-five amino acid residues, including an amino acid
sequence resembling the formula,
R-2-L-P-5-6-P-8-9 (SEQ ID NO:10),
positioned anywhere along the peptide. In the above-
mentioned formula, each number represents an amino acid
residue, such that 2 represents any amino acid residue except
cysteine, 5 and 6 each represents a hydrophobic amino acid
residue, 8 represents any amino acid residue except cysteine,
and 9 represents a hydrophilic amino acid residue except
cysteine. Each letter used in the formulas herein represent
the standard one-letter symbol for the corresponding amino
acid. When the peptide is a 9-mer, the peptide
R-P-L-P-P-L-P-T-S (SEQ ID NOill) is excluded. The peptides
of particular interest are those that exhibit a binding
affinity for the SH3 domain of Src and Src-related proteins,
including Yes, Fyn, Lyn, Lck, Hck and Fgr. Preferably, the
peptides of the invention exhibit a binding affinity for the
SH3 domain of Src, which is at least three-fold, more
preferably at least four-fold, most preferably at least about
five-fold greater than that exhibited by the peptide RPLPPLP
(SEQ ID NO:9). In still other embodiments, the peptides
exhibit a binding affinity for the SH3 domain of Src which is
at least ten-fold greater than that exhibited by the peptide
RPLPPLP (SEQ ID NO:9).
In specific embodiments, peptides are disclosed in which
the various amino acid residues at the indicated positions
may independently have the following preferred identities: 2
is a P, R, A, L, Q, E or S, more preferably P or R; 5
- 15 -
CA 02595040 2007-08-03
WO 97130074 PCT/US97/l._ A
represents a P, M, I or L, more preferably P or M; 6 is a P,
L, I or V, more preferably P or L; 8 is a T, R, P, I, N, E,
v, s, A, G or L, more preferably T or R; and 9 is a T, R, S,
H or D, more preferably T or R. Despite the preference for
hydrophobic amino acid residues at 5 and 6, in some cases it
may be desirable to have hydrophilic amino acid residues at
these positions. Specifically, amino acid residue 5 may be a
T, R or S, and amino acid residue 6 may be a T or R.
Likewise, while a hydrophilic amino acid residue is preferred
at position 9, in some instances a hydrophobic residue, such
as a P or A, may be desirable.
The present invention also contemplates SH3 domain-
binding peptides with a minimum length of 10, 11, 12, 13, 14,
or more amino acids. Such peptides contain additional
15 amino acid residues flanking the core sequence of
R-2-L-P-5-6-P (SEQ ID NO:71) either at the C-terminal end,
the N-terminal end or both. Thus, for example, such peptides
include those that further comprise a C-terminal-flanking
amino acid sequence of the formula 10, 10-11, 10-11-12, 10-
11-12-13 (SEQ ID NO:12) or 10-11-12-13-14 (SEQ ID NO:13), in
which each number represents ariy amino acid residue except
cysteine, such that the amino acid residue 10 is bound to the
amino acid residue 9 by a peptide bond. In that case,
specific embodiments include an amino acid residue 10 which
is T, R, L, S, D, P, A or N, preferably T or R, an amino acid
residue 11 which is R, P, A, Q, S or T, preferably R or P, an
amino acid residue 12 which is P, S, R or T, preferably P or
S, an amino acid residue 13 which is P, S, R, F, H or T,
preferably P or S, and an amino acid residue 14 which is S,
R, G or T, preferably, S or R.
Furthermore, peptides are also provided which further
comprise an N-terminal-flanking amino acid sequence of the
formula 1', 2'-1', 3'-2'-1' or 4'-3'-2'-1' (SEQ ID NO:14) in
which each number represents any amino acid residue except
cysteine, such that 1' is bound to R by a peptide bond. In
such a case, specific embodiments are provided in which the
amino acid residue 1' is T, P, S, N, F, W, K, H, Q or G,
- 16 -
CA 02595040 2007-08-03
-097/30074 PCT/US97/0227o
preferably T or P, wherein the amino acid.residue 2' is S, T,
G, P, R, Q, L, A or H, preferably S or T, wherein the amino
acid residue 3' is R, S, P, G, A, V, Y or L, preferably S or
T, and wherein the amino acid residue 4' is R, S, V, T, G, L
or F, preferably R or S.
In a particular embodiment, a peptide is disclosed
having at least thirteen and up to forty-five amino acid
residues, including an amino acid sequence of the formula,
3'-2'-1'-R-2-L-P-5-6-P-8-9-10 (SEQ ID NO:15), positioned
anywhere along the peptide, in which each number represents
an amino acid residue, such that 3', 2', 1', 2, 8,.and 10
each represents any amino acid residue except cysteine, 5 and
6 each represents a hydrophobic amino acid residue, and 9
represents a hydrophilic amino acid residue except cysteine,
each letter being the standard one-letter symbol for the
corresponding amino acid, said peptide exhibiting a binding
affinity for the SH3 domain of Src. Preferred 13-mers
include, but are not limited to, those having an amino acid
residue 5 which is a P or M, an amino acid residue 1' which
is T, P, S or N, an amino acid residue 2' which is S or T, ari
amino acid residue 3' which is R or S, and an amino acid
residue 10 which is T or R. In all the SH3 domain-binding
peptides described herein, the prohibition against the use of
the hydrophilic amino acid residue cysteine (C) does not
extend beyond the 7-mer "core" sequence and the additional
amino acid residues Tlanking the core up to a total (core +
flanking) of about 20 amino acids. That is, the occasional
use of a cysteine is not absolutely prohibited. What should
be kept in mind is that the potential for the formation of
intramolecular disulfide bonds, to form a cyclic structure,
be minimized as much as possible. Applicants have found that
cyclized structures appear to be disfavored, at least with
potential binding peptides of less than about 15.amino acid
residues in length. The concern for the formation of
cyclized structures comprising the core sequence diminishes
with increasing s.ize of the peptide. Presumably, a large
- 17 -
CA 02595040 2007-08-03
.JO 97130074 PCT/US97/02~. 3
enough structure, though cyclic, may allow the critical core
sequence to adopt a more or less linear conformation.
In particular, specific peptides are disclosed which
exhibit binding affinities to SH3 domains. These include the
peptides, RSTPRPLPMLPTTR (SEQ ID NO. 62), RSTPRPLPPLPTTR (SEQ
ID NO. 67), GILAPPVPPRNTR (SEQ ID NO. 63), VLKRPLPIPPVTR (SEQ
ID NO. 64), GPHRRLPPTPATR (SEQ ID NO. 65), and ANPSPATRPLPTR
(SEQ ID NO. 66).
Phage clones are also disclosed, along with the amino
lo acid sequences that are responsible for SH3 domain binding.
These phage clones are identified in Figure 5.
In other embodiments of the present invention, SH3
domain-=binding peptides are contemplated which have a total
of 11, 13, 14, 18, 20, 22, 23, 25, 30, 36, 38 or 45 amino
acid residues.
The peptides of the present invention, having been
disclosed herein, may be prepared by any number of
practicable methods, including but not limited to solution-
phase synthesis, solid-phase synthesis, protein expression by
a 16-ransformed host, cleavage from a naturally-derived,
synthetic or semi-synthetic polypeptide, or a combination of
these techniques.
The SH3 binding peptides exhibit a wide range of
biological activity which includes the enhancement (or
inhibition, depending on the particular peptide or the nature
of the peptide's target molecule, in this case a protein
bearing an SH3 domain) of the natural function or biological
activity of the peptide's target molecule. For example, the
interaction of the binding peptide of the present invention
could result in the modulation of the oncogenic activity of
the target molecule bearing the SH3 domain. If the target
molecule has, in turn, a natural binding partner or ligand,
the peptides of the present invention may also exhibit
antagonistic or agonistic activity in relation to the
biological activity of the natural binding partner.
Thus, it is an object of the present invention to
provide a method of activating Src or Src-related protein
- le -
CA 02595040 2007-08-03
,97/30074 PCT/US97/0224
tyrosine kinases by administering an effective amount of the
SH3 domain-binding peptides generally described herein. The
intensity of the immune response can thus be stimulated, for
, example, by the increased production of certain lymphokines,
such as TNF-alpha and interleukin-l. As is generally known
to those of ordinary skill in the art, a more intense immune
response may be in order in certain conditions, such as in
combating a particularly tenacious infection, viral or
otherwise, or a malignancy.
Furthermore, in a specific embodiment of the present
invention, a conjugate compound is contemplated which
comprises the peptide of the present invention and a second
chemical moiety. The second chemical moiety can be selected
from a wide variety of chemical compounds including the
peptide itself. Typically, however, the second chemical
moiety is selected to be other than the peptide of the
present invention, including but not limited to an amino
acid, a peptide other than an SH3 binding peptide of the
present invention, a polypeptide or protein (i.e., the
conjugate is a fusion protein), a nucleic acid, a nucleoside,
a glycosidic residue (i.e., any sugar or carbohydrate), a
label or image-enhancing agent (including metals, isotopes,
radioisotopes, chromophores, fluorophores (such as FITC,
TRITC, and the like), and enzyme substrates), a drug
(including synthetic, semisynthetic, and naturally-occurring
compounds), small molecules (e.g., biotin, hormones, factors)
and the like.
The peptide of the present invention can be conjugated
to the second chemical moiety either directly (e.g., through
appropriate functional groups, such as an amine or carboxylic
acid group to form, for example, an amine, imine, amide,
ester, acyl or other carbon-carbon bond) or indirectly
through the intermediacy of a linker group (e.g., an
aliphatic or aromatic polyhydroxy, polyamine, polycarboxylic
acid, polyolefin or appropriate combinations thereof).
Moreover, the term "conjugate," as used herein, is also meant
to encompass non-covalent interactions, including but not
- 19 -
CA 02595040 2007-08-03
75361-88
limited to ionic,, affinity or other complexation
interactions. Preferably, such other non-covalent
interactions provide definable, most preferably, isolatable
chemical conjugate species.
As described further herein, the peptides of the present
invention have been shown to localize within certain cellular
compartments which contain Src or Src-related proteins.
Consequently, the above-described conjugate can be utilized
as a delivery system for introduction of a drug to cells,
1o tissues or organs that include SH3 domain-containing
proteins.
It should also be pointed out that the present invention
seeks to provide a recombinant construct comprising a nucleic
acid or its complement that includes codons or nucleotide
15 sequences encoding a peptide having a region that binds to an
SH3 domain, preferably the Src SH3 domain. The recombinant
nucleic acid may be a DNA or RNA polynucleotide.
in a specific embodiinent, the present invention
contemplates a recombinant construct which is a transforming
20 vector. Such vectors include those well knowP.- to those of
ordinary skill in the art, which effect the transfer or
expression of the nucleotide sequence after introduction to a
host, such as recombinant plasmid, phage or yeast artificial
chromosome. These vectors may be closed circular loops or
25 they may be linearized. The vectors contemplated include
those that exist extrachromosomally after host transformation
or transfection, as well'as those that integrate within or
even displace portions of the host chromosome. The vectors
may be introduced to the cell with the help of transfection
3o aids or techniques well-known in the art. For example, these
aids or techniques may take the form of electroporatior., use
of calcium chloride, calcium phosphate, DEAE dextran,
TN
liposomes or polar lipid reagents known as LIPOFECTIN or
TM
LIPOFECTAMINE = In addition, the present invention
35 contemplates the direct introduction of the desired nucleic
acid to the host cell, for instance, by injection.
- 20 -
CA 02595040 2007-08-03
75361-88
Transformed host cells are also obtained by the methods
of the present invention which are capable of reproducing the
po.lynucleotide sequences of interest and/or expressing the
corresponding peptide products. A variety of hosts are
contemplated, including prokaryotic and eukaryotic hosts. In
particular, bacterial, viral, yeast, animal, and plant cells
are potentially transformable hosts. Thus, a transfornied host
cell may be obtained and carl produce, preferably secrete, a
peptide having a region that binds to an SH3 domain by carrying
out a method comprising (a) providing an expression vector,
pref erably a secretory expression vector, cornprising a
nucleotide sequence encoding at least one copy of a peptide
having a region that binds to an SH3 domain; and (b)
introducing the vector to a competent host cell.
15The peptides, thus produced, may then be introduced to
cells, tissues, -organs, or administered to the subject for
the purpose of modulating the biochemical activity of the SH3
domain-containing proteins present therein. Accordingly, in
specific embodiments of the present invention, c:ompositior.s
2o are provided which comprise an SH3 domain-binding peptide,
including a core sequence and flanking sequences, and a
suitable carrier.
The compositions contemplated by the present invention
may also include other components, from those that facilitate
25 the introduction or administration of the compositions to
those that have their own innate activity, such as a
prophylactic, a diagnostic or a therapeutic action. Such
.innate activity may be distinct from that of the peptides of
the present invention or be complementary thereto. In any
3o event, the compositions of the present invention include
those that are suitable .for administration into mammals,
including humans. Preferably, the compositions (including
necessarily the carrier) of the present invention are
sterile, though others may need only be cosmetically,
35 agriculturally or pharmaceutically acceptable. Still other
compositions may be adapted for veterinary use.
- 21 -
CA 02595040 2007-08-03
'NO 97/30074 PCT/US97/0iz98
The compositions, including the drug delivery systems
described herein, are contemplated to be administered in a
variety of ways, such as parenterally, orally, enterally,
topically or by inhalation. The compositions may also be
adminstered intranasally, opthalmically or intravaginally.
Furthermore, the compositions of the invention can take
several forms, such as solids, gels, liquids, aerosols or
patches.
In another embodiment of the present invention a method
is provided of identifying a peptide having a region that
binds to an SH3 domain comprising: (a) providing an
immobilized target protein comprising an SH3 domain; (b)
incubating the immobilized target protein with an aliquot
taken from a phage-displayed random peptide library, which
library includes peptides having a random sequence of >8
amino acid residues; (c) washing unbound phage from the
immobilized target protein; (d) recovering the phage bound to
the immobilized target protein; and (e) determining the
relevant nucleotide sequence of said binding phage nucleic
acid and deducing the primary sequence corresponding to the
SH3 domain-binding peptide. Preferably, the method further
comprises amplifying the titer of the recovered phage and
repeating the steps of incubation, washing and recovery to
provide SH3 domain-binding peptide-enriched phage.
Any other mode by which the peptide library, random or
otherwise, can be "displayed" can be utilized in the present
invention, however. Moreover, the present applicants believe
that longer random peptide sequences (e.g., >6 amino acid
residues, preferably >10, and most preferably, >12) provide
not only much greater diversity but also a richer degree of
secondary structure conducive to binding activity. If t:he
random region of the peptide is less than or equal to an 8=
mer, it should preferably not be cyclized.
- 22 -
CA 02595040 2007-08-03
.:O 97/30074 PCT/US97/022.
5.2. Preparation of Random Peptide Libraries
The preparation and characterization of the preferred
phage-displayed random peptide libraries have been described
elsewhere. See, for example, Kay, B.K. et al. in Gene (1992)
128:59-65, for a description of the preparation of the phage-
displayed random peptide library known as TSAR-9, more below.
In particular, by cloning degenerate oligonucleotides of
fixed length into bacteriophage vectors, recombinant
libraries of random peptides can be generated which are
expressed at the amino-terminus of the pIII protein on the
surface of M13 viral particles. (There are 3-5 copies of the
piII-fusion on the surface of each particle.) Phage display
offers several conveniences: first, the expressed peptides
are on the surface of the viral particles and accessible for
interactions; second, the recombinant viral particles are
stable (i.e., can be.frozen, exposed to pH extremes); third,
the viruses can be amplified; and fourth, each viral particle
contains the DNA encoding the recombinant genomQ.
Consequently, these libraries can be screened by isolating
viral particles that bind to targets. The isolates can be
grown up overnight, and the displayed peptide sequence
responsible for binding can be deduced by DNA sequencing.
These libraries have approximately >108 different
recombinants, and nucleotide sequencing of the inserts
suggests that the expressed peptides are indeed random in
amino acid sequence. These libraries are referred to herein
as TSAR libraries, where TSAR stands for Totally gynthetic
Affinity Reagents. The preparation of the TSAR libraries are
described further below.
5.3. SH3 Binding Clones And Their Characteristics
Accordingly, peptides have been isolated from an
unconstrained random peptide library which exhibit a binding
affinity for SH3 domains. Furthermore, the binding
affinities exhibited by the disclosed peptides differ in
their selectivities with certain peptides showing comparable
binding affinities for SH3 domains derived from different
- 23 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/u._98
proteins, while others manifest greater affinities for
specific SH3 domains.
The amino acid sequence of various peptides isolated by
the present method are listed in Figure 5. As can be seen
from this list, certain groups of SH3 domain binding peptides
are isolated from three separate random peptide libraries,
each based on a different type of random peptide insert, all
displayed at the amino-terminus of the piII protein on the
surface of M13 viral particles. Ten clones were isolated
1o from the R8C library, seven from the TSAR-12 library, and
seven from the TSAR-9 library. The sequences are presented
to highlight the particular amino acid residues believed to
bind directly to the SH3 domain, as well as to point out the
remaining amino acid resiudes of the random insert and the
viral flanking sequences and complementary site amino acid
residues common to each group of clones. The frequency with
which each particular clone is found in each library is also
indicated in Figure 5. Thus, clones T12.SRC3.1 and
T12.SRC3.2 are by far the most abundant clones found among
the three libraries.
Interestingly,=all the binding peptides are found to
have the proline-rich amina acid residue motif, which is
apparently responsible for binding, the motif being loc:ated
predominantly at the C-terminal end of the insert, although
each clone also contains an insert at the N-terminal end.
The significance of this observation is not presently
understood, although this finding may indicate the possible
importance of the C-terminal viral flanking sequences in SH3
domain binding.
Indeed, a synthetic peptide bearing only the core
consensus sequence RPLPPLP (SEQ ID NO:9) was less effective
in binding to target SH3 domains than synthetic peptides that
also included additional amino acid residues flanking the
core sequences. Thus, 13-mers and 14-mers having the
sequences RSTPRPLPMLPTTR (SEQ ID NO:62), RSTPRPLPPLPTTR (SEQ
ID NO:67), GILAPPVPPRNTR (SEQ ID NO:63), GPHRRLPPTPATR (SEQ
ID NO:65), and VLKRPLPIPPVTR (SEQ ID NO:64) have been
- 24 -
CA 02595040 2007-08-03
D 97/30074 PCT1US97/02'__
prepared and shown to bind to SH3 domains, such as those of
Src and Yes, much more avidly than the 7-mer, RPLPPLP (SEQ ID
NO:9). The 13-mer ANPSPATRPLPTR (SEQ ID NO:66) has been
shown to have binding affinities comparable to the core
consensus sequence. In each case, the 13-mers comprise a 7-
mer "core" sequence plus additional amino acid residues
flanking same, some of which additional amino acid residues
are contributed by the viral flanking sequences.
Thus, in one embodiment of the present invention, a 7-
mer core includes a consensus motif of the formula RXLPO~P
(SEQ ID NO:71), wherein R is arginine, L is leucine, P is
proline, X represents any amino acid except cysteine and 0
represents a hydrophobic amino acid residue. By "hydrophobic
amino acid residue," the applicants mean to include F, Y, W,
V, A, I, L, P or M, each letter representing the standard
one-letter designation for the corresponding amino acid
residue.
Furthermore, a preferred 9-mer peptide comprising two
additional amino acids on the C-terminal end of the core
sequence is envisioned having a consensus motif of the
formula RXLPOOPX~ (SEQ ID NO:10). In this preferred 9-mer
consensus motif, the symbol ~ represents a hydrophilic amino
acid residue, except cysteine. By "hydrophilic amino acid
residue," the applicants mean to include K, R, H, D, E, N, Q,
T, S or C, and the other symbols are as defined above. For
the purposes of the present invention, a glycine residue (G)
may be considered either a hydrophobic or a hydrophilic amino
acid residue. The one-letter symbols B and Z, which stand
for N or D and Q or E, respectively, are considered
hydrophilic amino acid residues.
Particular 13-mer peptides of the present invention
include those listed, below. It is noted, however, that not
all the following 13-mer peptides correlate strictly to or
comply with the preferred 9-mer consensus motif, described
above. Those peptides that do not comply (indicated in
italics, with the non-complying amino acid residues
underscored) can, thus, be described as "resembling" those
- 25 -
CA 02595040 2007-08-03
75361-88
that do comply (indicated in normal type) with the preferred
9-mer consensus motif: PGFRELPPLPPSR (SEQ ID NO:72),
AQSRPLPIPPETR (SEQ ID NO:73), VLKRPLPIPPVTR (SEQ ID NO:64),
PPNSPLPPLPTHL (SEQ ID NO:74), TGRGPLPPLPNDS (SEQ ID NO:75),
YSTRPVPPII'RPS (SEQ ID NO:76), SHKSRLPPLPTRP (SEQ ID NO:77),
YRFRALPSPPSAS (SEQ ID NO:78), GPHRRLPPTPATR (SEQ ID NO:65),
LAQRQLPPTPGRD (SEQ ID NO:79), ALQRRLPSTPPPA (SEQ ID NO:80),
PATRPLPTRPSRT (SEQ ID NO:81), YSTRPLPSRPSRT (SEQ ID NO:82),
XPGRILLLPSEPR (SEQ ID NO:83), SGGILAPPVPPRN (SEQ ID NO:84),
RSTRPLPILPRTT (SEQ ID NO:85), STPRPLPMLPTTR (SEQ ID NO:86),
STNRPLPMIPTTR (SEQ ID NO:87), RSTRPLPSLPITT (SEQ ID NO:88),
STSRPLPSLPTTR (SEQ ID NO:89), RSTRSLPPLPPTT (SEQ ID NO:90),
RSTRQLPIPPTTT (SEQ ID NO:91), STPRPLPLIPTTP (SEQ ID NO:92),
RSTRPLPPTPLTT (SEQ ID NO:93), and RSTRPQPPPPITT (SEQ ID
NO:94). Accordingly, other peptides not specifically
disclosed, which either, comply with or "resemble" the
preferred 9-mer consensus motif, can be readily envisioned by
those of ordinary skill in the art and are considered to be
equivalent to those that are specifically disclosed above.
In particular, non-compliance at positions 1(S, G, and I, in
place of R, are tolerated), 3 (V, A, and Q, in place of L,
are tolerated), 4 (L, in place of P, is tolerated), 5
(hydrophilic amino acid residues, S, R, and T, are tolerated
in place of a hydrophobic amino acid residue), 6 (hydrophilic
amino acid residues, R and T, are tolerated in place of a
hydrophobic amino acid residue), 7(T, and S, in place of P,
are tolerated), and 9 (P and A are tolerated in place of a
hydrophilic amino acid residue) have been observed.
5.3.1. Binding specificities
It has been discovered that certain binding
peptides disclosed have greater relative binding affinity
for one SH3 domain over another. Referring now to Figure 8,
the relative binding affinities of the various peptides
described above toward different SH3 domain targets are
graphically presented. As one can see, the relative binding
affinities of the respective peptides can differ by orders of
- 26 -
CA 02595040 2007-08-03
75361-88
magnitude. Thus, as shown in Figure 8, the peptide
GPHRRLPPTPATR (SEQ ID N0:65), having the relevant sequence of
the phage clone identified as T12.SRC3.3, is specific to Src
family SH3 domains, including, but not limited to, Src, Y'es,
Lck, Hck, Fgr, Fyn, and Lyn. This SH3 binding peptide has
little affinity for SH3 domains derived from PLCy or Grb2.
On the other hand, the peptide GILAPPVPPRNTR (SEQ ID NO:63),
corresponding -to the relevant sequence of the phage clone
TI2.SRC3.1, which is one of the most abundant binding clones
lo found by the present method, binds generically to a broad
range of SH3 domains, including Src, PLC-y, and Grb2..
On an intermediate level, a peptide, VLKRPLPIPPVTR
(SEQ ID NO: 64), corresponding to the relevant sequence of
the phage clone T12.SRC3.6, which is Src preferential, has
also been uncovered; that is, this peptide
exhibits strong binding af f ini.ties for members of the Src
family, some binding affinities for Grb2 proteins, but little
binding affinities for PLCy domains. The peptide
ANPSPATRPLPTR (SEQ ID'NO:66), corresponding to the relevant
2o sequence of the phage clone T12.SRC3.2, also exhibits Src
family specificity similar to GPHRRLPPTPATR (SEQ ID N0:65).
The peptides RSTPRPLPMLPTTR (RBC.YES3.5; SEQ ID NO:62) and
RSTPRPLPPLPTTR (representative consensus motif; SEQ ID NO:67)
are highly specific for SH3 domain of Src, Yes, and. other
Src-related proteins.
5.4. Further Discussion of Binding Experiments
At the outset it is apparent that the binding affinity
of certain peptides to the SH3 domain of Src and Src-related
proteins is governed by more than just the presence of the
preferred core consensus sequences, RPLPPLP (SEQ ID NO:9) or
RPLPMLP (SEQ ID NO:95; i.e., RPLP(P/M)LP, SEQ ID NO:96).
Thus, while the synthetic peptides RSTPRPLPMLPTTR
(R8C.YES3.5; SEQ ID NO:62) and RSTPRPLPPLPTTR (consensus;
(SEQ ID NO:67) exhibit a strong specific binding affinity for
Src SH3, the other synthetic peptides tested also exhibited
an avid binding affinity to SH3 domains relative to the 7-
- 27 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/0~.498
mer, RPLPPLP (SEQ ID NO:9). These other peptides,
GILAPPVPPRNTR (SEQ ID NO:63), VLKRPLPIPPVTR (SEQ ID NO:64),
GPHRRLPPTPATR (SEQ ID NO:65), and ANPSPATRPLPTR (SEQ ID
NO:66), sport core sequences and flanking sequences that do
not closely adhere to the preferred core consensus sequences.
Thus, these results suggest that binding affinity tc SH3
domains is governed to a large extent by the nature of the
amino acid residues flanking the core 7-mer sequence.
The binding characteristics of Src SH3-selected peptides
was determined using synthetic biotinylated peptides
corresponding to the sequences displayed by Src SH3-selected
phage. These biotinylated peptides were assayed for direct
binding to immobilized Src SH3-GST. Each of the five
library-derived peptides tested were found to bind to Src
SH3-GST and Yes SH3-GST over background (Figure 8).
Furthermore, a strong correlation was observed between the
similarity of a given peptide to the preferred core consensus
sequence RPLP(P/M)LP (SEQ ID NO:96) and the pept'Lde's
af f ini.ty for Src SH3 -GST . The core sequence of the clone
T12.SRC3.1 (GILAPPVPPRNTR; SEQ ID NO:63) appears to provide
more generic SH3 domain-binding characteristics.
Experiments comparing the relative binding of various
phage clones to SH3 domains taken from a variety of proteins
demonstrated the preference of these clones for Src and Src-
related SH3 domains over SH3 domains taken from other
proteins.
It was further found that while the 7-mer having the
consensus sequence RPLPPLP (SEQ ID NO:9) bound to Src SH3-GST
only weakly, peptides comprising the consensus sequence
flanked by residues encoded by one of the Src SH3-selected
clones (RBC.YES3.5), RSTP (SEQ ID NO:97) at the N-terminal
end and TTR at the C-terminal end, bound significantly better
than any of the peptides tested (Figure 7). Thus, as stated
previously, sequences that flank the RPLP(P/M)LP (SEQ ID
NO:96) core appear to be important contributors to SH3
binding. It is further surmised that a peptide having or
resembling the sequence RSTPAPPVPPRTTR (SEQ ID NO:98) should
- 28 -
CA 02595040 2007-08-03
vrt) 97/30074 PCT/US97/022925
exhibit strong but generic binding to a variety of SH3
domains.
Similarly, it is observed that most of the Src SH3-
binding motifs are located near the carboxy-terminus of the
random peptides, adjacent to sequences which are fixed in
every clone (Figure 5). The exceptional clones tend to
possess sequences that resemble motifs that include fixed
flanking sequences. This clustering contrasts with previous
results, in which binding motifs are distributed throughout
the random peptide. Kay, B.K., et al., in Gene (1993)
128:59-65.
The binding of the library-derived Src SH3-binding
peptides was compared to that of peptides corresponding to
proline-rich regions of natural proteins. Peptides
i5 corresponding to SH3-binding regions in human PI-3' Kinase
(KISPPTPKPRPPRPLPV; SEQ ID NO:69) and human SOS1.20
(GTVEPVPPPVPPRRRPESA; SEQ ID NO:68), as well a.s a proline-
rich region of the cytoskeletal protein vinculin
(LAPPKPPLPEGEV; SEQ ID NO:70), bound Src SH3 iau.-h less well
than the library-derived peptides (Figure 7).
As mentioned above, the relative specificity of binding
was explored. Thus, the relative binding of Src SH3-selected
peptides to equal amounts of GST fusions to SH3 domains from
different proteins was determined (Figure 8). While all of
the library-derived peptides bound the Src and Yes SH3
domains almost equally well, none of the peptides (with the
exception of peptide T12.SRC3.1, the most divergent peptide
tested) bound the SH3 domains of Grb2, Crk, Abl or PLC71
appreciably. Thus, the library-derived peptides, in contrast
with a peptide derived from SOS1, exhibit SH3 binding that is
relatively specific for Src-family members.
Next, it was determined whether the binding to the Src
SH3 domain was qualitatively like the interactions of the SH3
domain and natural proteins found in cell lysates. Thus;
radiolabeled proteins were prepared from NIH 3T3 cell lysates
and chromatographed over Src SH3-GST immobilized on
glutathione linked Sepharose. SDS-PAGE shows that a number
- 29 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/02298
of proteins can be affinity purified in this manner. The
synthesized peptides bind quite well to the Src SH3 domain,
as they can compete the binding of radiolabeled proteins from
cell lysates to immobilized Src-GST, with an ICso of 1-10 mM
5(Figure 9). In conclusion, the peptides can efficiently
block the interaction of cellular proteins with Src Sri3 in
vitro.
Moreover, Xenopus laevis oocytes injected with mRNA
encoding constitutively active Src undergo progesterone-
1o induced maturation at an accelerated rate relative to oocytes
injected with water or c-Src mRNA. Unger, T.F. and Steele,
R.E. in Mol. Ce11.Bio1. (1992) 12:5485-5498. To explore the
ability of the library-derived Src SH3-binding peptides to
exert a biochemical effect in vivo, the influence of the
15 peptides on the maturation of Xenopus laevis oocytes was
examined. Hence, stage VI oocytes were injected with
peptide, exposed to progesterone, and scored for germinal
vesicle breakdowr.. Figure 10 shows that the rate of
maturation was accelerated by approximately one hour when
20 oocytes were injected with the SH3-binding peptide consisting
of RPLPPLP (SEQ ID NO:9) flanked by residues from clone
T12.SRC3.6 (VLKRPLPIPPVTR; SEQ ID NO:64), but not wii:h water
or a peptide corresponding to a proline-rich segment of
vinculin (LAPPKPPLPEGEV; SEQ ID NO:70) as controls. The
25 magnitude of this effect is roughly equivalent to that seen
with injection of mRNA encoding constituitively active Src.
See, e.g., Figure 3B in Unger, T.F. and Steele, R.E., supra.
This result suggests that the library-derived Src SH3-binding
peptide is effectively relieving an inhibitory effect of the
30 Src SH3 domain upon Src PTK activity. This model is
consistent with a number of studies which have demonstrated
an inhibitory effect of the Src SH3 domain upon Src kinase
and transforming activity. See, e.g., Okada, M., et a:l.,,
supra; Murphy, S.M., et al., supra; and Superti-Furga, G., et
35 al., supra.
- 30 -
CA 02595040 2007-08-03
. O 97/30074 PCT/US971022yo
5.5. Diagnostic And Therapeutic Agents Based On SH3
Binding Peptides and Additional Methods of
Their Use
As already indicated above, the present invention also
seeks to provide diagnostic, prophylactic, and therapeutic
agents based on the SH3 binding peptides described herein.
In one embodiment, diagnostic agents are provided,
preferably in the form of kits, comprising an SH3 domain-
binding peptide and a detectable label conjugated to said
peptide directly, indirectly or by complexation, said peptide
comprising: (i) a core sequence motif of the formula RXLPOOP
(SEQ ID NO:71), wherein X represents any amino acid except
cysteine and 0 represents a hydrophobic amino acid residue,
including F, Y, W, V, A, I, L, P, M or G, each letter
representing the standard one-letter designation for the
corresponding amino acid residue; and (ii) two or more
additional amino acid residues flanking said core sequence at
its C-terminal end, N-terminai end or both.
The diagnostic agents of the present invention can be
used to detect the presence of SH3 domains of a generic or
specific type in cells, tissues or organs either in vitro or
in vi.vo. For in vivo applications, the diagnostic agent is
preferably mixed with a pharmaceutically acceptable carrier
for administration, either enterally, parenterally or by some
other route dictated by the needs of the particular
application.
In a particular embodiment, for example, an assay based
on a fusion product is contemplated which comprises a Src SH3
domain-binding peptide of the invention and a substrate for
deregulated or "activated" Src. For instance, a muscle
biopsy, taken from a subject suspected of being infected by
the Rous sarcoma virus, can be treated with an effective
amount of the fusion product. By subsequent analysis of the
degree of conversion of the substrate, one can potentially
detect infection by the Rous sarcoma virus in the subject,
particularly mammals, especially chickens. The presence of
the retrovirus, which causes the expression of deregulated or
- 31 -
CA 02595040 2007-08-03
NO 97130074 PCT/US97/02298
"activated" Src, may thus be indicated by unusually high
levels of Src as revealed by large amounts of the converted
substrate. See, for example, Paxton, W.G. et al., in
Biochem. Biophys. Res. Commun. (1994) 200(1):260-267
(detection of phosphorylated tyrosine and serine residues of
angiotensin II AT1 receptor, a substrate of Src family
tyrosine kinases); another suitable substrate may be the
protein p68 (Fumagalli, S. et al., in Nature (1994)
368(6474):871-874; Taylor, S.J. and Shalloway, D., in Ibid.
at 867-871.
Alternatively, the enzyme can be isolated by selective
binding to a form of the SH3 domain-binding peptides of the
present invention (e.g., biotin-peptide conjugate). After
isolation of the protein-peptide conjugate complex (e.g., on
a column comprising streptavidin), the activity of the enzyme
can then be assayed by conventional methods to determine its
level of protein kinase activity which can be taken as an
indication of the presence of the deregulated or "act.;.vated"
form of the enzyme. An assay for Src kinase has been
described by Klinz and Maness, in Neuroprotocols (a c;cmpanion
to Neuroscience)(1992) 1(3):224-231.
Moreover, the diagnostic agents of the invention can
also serve as imaging agents of cells, tissues or organs,
especially those that contain proteins with an SH3 domain.
For example, neural cells (e.g., neurons, other areas of the
brain), osteoclasts, osteoblasts, platelets, immune cells,
and other dividing cells are known to express or contain
proteins with SH3 domains. Thus, an image can be taken of
portions of the body to serve as a baseline for subsequent
images to detect physiologic or biochemical changes in the
subject's body. For instance, changes in the condition of
cellular levels of Src or a transformation of the cellular
Src to an "activated" form may be detected using the
diagnostic or imaging agents of the present invention.
Accordingly, it has been demonstrated that an SH3-
binding peptide tagged with a fluorescence emitter can
provide an image of the cytoskeleton. The images are
- 32 -
CA 02595040 2007-08-03
)97130074 PCTr[JS97/0225
presented in Figure 11. As can be seen from Figure 11,
panels A, B, and C show the fluorescence image that is
obtained on treating NIH 3T3 fibroblasts with SH3 domain-
binding peptides modified to include a fluorescent tag. in
sharp contrast, panel D shows only a dark image that is
produced when the cells are treated with a proline-rich
seyment of vinculin as a control.
In another embodiment, an SH3 domain-binding peptide-
horseradish immunoperoxidase complex or related
lo immunohistochemical agent could be used to detect and
quantitate specific receptor molecules in tissues, serum or
body fluids. In particular, the present invention provides
useful diagnostic reagents for use in immunoassays, Southern
or Northern hybridization, and in situ assays. Accordingly,
the diagnostic agents described herein may be suitable for
use in vitro or in vivo.
in addition, the diagnostic or imaging agent of the
present invention is not limited by the nature of the
detectable label. Hence, the diagnostic agent may contc.in
one or more such labels including, but not limited to,
radioisotope, fluorescent tags, paramagnetic subscatices,
heavy metals, or other image-enhancing agents. Those of
ordinary skill in the art would be familiar with the range of
label and methods to incorporate or conjugate them into the
SH3 domain-binding peptide to form diagnostic agents.
In yet a further embodiment, pharmaceutical compositions
are provided comprising an SH3 domain-binding peptide and a
pharmaceutically acceptable carrier. In a specific
embodiment of the invention, the pharmaceutical composition
is useful for the modulation of the activity of SH3 domain-
containing proteins. By "modulation" is meant either
inhibition or enhancement of the activity of the protein,
target. Accordingly, a pharmaceutical composition is
disclosed comprising an SH3 domain-binding peptide and a
pharmaceutically acceptable carrier, said peptide comprising:
(i) a 9-mer sequence motif of the formula RXLPO~PX~ (SEQ ID
No:lO), wherein X represents any amino acid except cysteine,
- 33 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/02t98
(p represents a hydrophobic amino acid residue, and wherein
is a hydrophilic amino acid residue except cysteine, each
letter representing the standard one-letter designation for
the corresponding amino acid residue; and, optionally, (ii)
additional amino acid residues flanking the 9-mer sequence at
its C-terminal end, N-terminal end or both, up to a total of
45 amino acid residues, including said 9-mer sequence.
Preferably, the peptide comprises at least one, more
preferably at least two, and most preferably at least three
additional amino acids flanking the 9-mer sequence.
As stated above, the therapeutic or diagnostic agents of
the invention may also contain appropriate pharmaceutically
acceptable carriers, diluents and adjuvants.. Such
pharmaceutical carriers can be sterile liquids, such as water
and oils including those of petroleum, animal, vegetable or
synthetic origin, such as peanut oil, soybean oil, mineral
oil, sesame oil and the like. Water is a preferred carrier
when the pharmaceutical composition is administered
intravenously. Saline solutions and aqueous dextrose and
glycerol solutions can also be employed as liquid carriers,
particularly for injectable solutions. Suitable
pharmaceutical excipients include starch, glucose, lactose,
sucrose, gelatin, malt, rice, flour, chalk, silica gel,
magnesium carbonate, magnesium stearate, sodium stearate,
glycerol monostearate, talc, sodium chloride, dried skim
milk, glycerol, propylene, glycol, water, ethanol and the
like. These compositions can take the form of solutions,
suspensions, tablets, pills, capsules, powders, sustained-
release formulations and the like. Suitable pharmaceutical
carriers are described in "Remington's Pharmaceutical
Sciences" by E.W. Ntartin.
Such compositions will contain an effective therapeutic
amount of the active compound together with a suitable amount
of carrier so as to provide the form for proper
administration to the subject. While intravenous injection
is a very effective form of administration, other modes can
be employed, including but not limited to intramuscular,
- 34 -
CA 02595040 2007-08-03
J 97/30074 PCT/US97/0229o
intraperitoneal, and subcutaneous injection, and oral, nasal,
enteral, and parenteral administration.
The therapeutic agents and diagnostic agents of the
instant invention are used for the treatment and/or diagnosis
of animals, and more preferably, mammals including humans, as
well as dogs, cats, horses, cows, pigs, guinea pigs, mice and
rats. Accordingly, other methods contemplated in the present
invention, include, but are not limited to, a method of
modulating, i.e., inhibiting or enhancing, bone resorption in
a mammal (see, e.g., Hall, T.J., in Biochem. Biophys. Res.
Commun. (1994) 199(3):1237-44), a method of disrupting
protein tyrosine kinase-mediated signal transduction pathways
or a method of regulating the processing, trafficking or
translation of RNA in a cell by introducing or administering
an effective amount of an SH3 domain-binding peptide of the
present invention (see, e.q., Taylor, S.J. and Shalloway, D.,
supra).
The diagnostic or therapeutic agents of the present
invention can be modified by attachment to soluble
macromolecules such as proteins, polysaccharides, or
synthetic polymers. For example, the peptide could be
coupled to styrene-maleic acid copolymers (see, e.g.,
Matsumura and Maeda, Cancer Res. (1986) 46:6387),
methacrylamide copolymers (Kopececk and Duncan, J. Controlled
Release (1987) 6:315), or polyethylene glycol (PEG) (e.g.,
Hershfield and Buckley, N. Engl. J. Med. (1987) 316:589; Ho
et al., Drug Metab. Dispos. (1986) 14:349; Chua et al., Ann.
Intern. Med. (1988) 109:114). The agents, if desired,
are further targeted by attachment to an antibody, especially
a monoclonal antibody. Such antibodies include but are not
limited to chimeric, single chain, Fab fragments, and Fab
expression libraries. In one embodiment the agent is coupled
to the macromolecule via a degradable linkage so that it will
be released in vivo in its active form.
In another embodiment, the therapeutic or diagnostic
agent may be delivered in a vesicle, in particular a
liposome. See, Langer, Science (1990) 249:1527-1533; Treat
- 35 -
CA 02595040 2007-08-03
WO 97130074 PCT/US97l02298
et al., in Liposomes in the Therapy of Infectious Disease
and Cancer, Lopez-Berestein and Fidler (eds.), Liss, New York
(1989) pp. 353-365; Lopez-Berestein,,ibid., pp. 317-327.
In yet another embodiment, the therapeutic or in vivo
diagnostic agent can be delivered in a controlled release
system. In one embodiment, a pump may be used (see Langer,
supra; Sefton, CRC Crit. Ref. Biomed. Eng. (1987) 14:201;
Buchwald et al., SurgerV (1980) 88:507; Saudek et al., N.
Engi. J. Med. (1989) 321:574). In another embodiment,
polymeric materials may be used (see Medical Applications of
Controlled Release, Langer and Wise (eds.), CRC Pres., Boca
Raton, Florida, 1974; Controlled Drug Bioavailability, Drug
Product Design and Performance, Smolen and Ball (eds.) Wiley,
New York 1984; Raner and Peppas, J. Macromol. Sci. Rev.
Macromol. Chem. (1983) 23:61; see, also, Levy et al., Science
(1985) 228:190; During et al., Ann. Neurol. (1989) 25:351;
Howard et al., J. Neurosurg. (1989) 71:105). In a preferred
embodiment, a controlled release system may be placed next to
the therapeutic target, thus requiring only a fraction of the
systemic dose (see, e.g., Goodson, in Medical Applications of
Controlled Release, supra, (1984) 2:115-138). It will be
recognized by one of ordinary skill in the art that a
particular advantage of the invention is that a peptide will
not be subject to the problems of denaturation and
aggregation associated with proteins held in the warm, most
environment of a body in a controlled release system.
Other controlled release systems are discussed in the
review by Langer, in Science (1990) 249:1527-1533.
5.6. Identification of Compounds that Affect
Binding of S83 Domain-containing Proteins and
their Ligands
A common problem in the development of new drugs is that
of identifying a single, or a small number, of compounds that
possess a desirable characteristic from among a background of
a large number of compounds that lack that desired
characteristic. This problem arises both in the testing of
- 36 -
CA 02595040 2007-08-03
WO 97/3(}074 PCT/US97/02298
compounds that are natural products from plant, animal, or
microbial sources and in the testing of man-made compounds.
Typically, hundreds, or even thousands, of compounds are
randomly screened by the use of in vitro assays such as those
that monitor the compound's effect on some enzymatic activity
or its ability to bind to a reference substance such as a
receptor or other protein.
The compounds which pass this original screening test
are known as "lead" compounds. These lead compounds are tYien
put through further testing, including, eventually, in vivo
testing in animals and humans, from which the promise shown
by the lead compounds in the original in vitro tests is
either confirmed or refuted. See Remington's Pharmaceutical
Sciences, 1990, A.R. Gennaro, ed., Chapter 8, pages 60-62,
Mack Publishing Co., Easton, PA; Ecker and Crooke, 1995,
Bio/Technology 13:351-360.
There is, of course, a contiriuaJ. need for new compounds
to be tested. in the,in vitro assays that make up the first
testing step described above. There is also a continual need
for new assays by which the pharmacological activities cf
these compounds may be tested. It is an object of the
present invention to provide such new assays to determine
whether a candidate compound is capable of affecting the
binding between a protein or polypeptide containing an SH3
domain and a ligand of the SH3 domain. A compound capable of
affecting this binding would be useful as a means of
modulating the pharmacological activity of proteins or
polypeptides containing the SH3 domain. The present
invention provides suitable ligands for SH3 domains for use
in such assays. Such assays can be performed where the SH3
domains include, but are not limited to, SH3 domains from
Cortactin, Nck, Abl, PLCT, Src, p53bp2, Crk, Yes, and Grb2.
The present invention provides methods of identifying a
compound that affects the binding of a molecule comprising an
SH3 domain and a ligand of the SH3 domain. The effect on
binding can be an increase or decrease in total amount of
- 37 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/02298
binding or in affinity of bidning. Preferably, the effect is
an inhibition (reduction in or loss of binding).
Accordingly, the invention provides a method of
identifying an inhibitor of the binding between a first
molecule comprising an SH3 domain=and a second molecule that
binds to the SH3 domain comprising incubating one or more
compounds from which it is desired to select such an
inhibitor with the first molecule and the second molecule
under conditions conducive to binding and detecting the one
or more compounds that inhibit binding of the first molecule
to the second molecule.
In a particular embodiment of the above-described
metnod, the second molecule is obtained by:
(i) screening a peptide library with the SH3 domain to
obtain peptides that bind the SH3 domain;
(ii) determining a conseiisus sequence for the peptides
obtained in step (i);
(iii) producing a peptide comprising the consensus
sequence;
wherein the second molecule comprises the peptide
comprising the consensus sequence.
In another embodiment, the second molecule is obtained
by:
(i) screening a peptide library with the SH3 domain to
obtain peptides that bind the SH3 domain;
(ii) determining a consensus sequence for the peptides
obtained in step (i);
(iii) searching a database to identify amino acid
sequences that resemble the consensus sequence of step (ii);
(iv) producing a peptide comprising an amino acid
sequence identified in step (iii);
wherein the second molecule comprises the peptide
comprising an amino acid sequence identified in step (iii).
Second molecules that bind SH3 domains can be obtained
by, e.g., the use of diversity libraries, such as random or
combinatorial peptide or nonpeptide libraries which cari be
screened for molecules that specifically bind to SH3 domains.
- 38 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/02298
Many libraries are known in the art that can be used, e.g.,
chemically synthesized libraries, recombinant (e.g., phage
display libraries), and in vitro translation-based libraries.
Examples of chemically synthesized libraries are
described in Fodor et al., 1991, Science 251:767-773;
Houghten et al., 1991, Nature 354:84-86; Lam et al., 1991,
Nature 354:82-84; Medynski, 1994, Bio/Technology 12:709-710;
Gallop et al., 1994, J. Medicinal Chemistry 37(9):1233-1251;
Ohlmeyer et al., 1993, Proc. Natl. Acad. Sci. USA
90:10922-10926; Erb et al., 1994, Proc. Nati. Acad. Sci. USA
91:11422-11426; Houghten et al., 1992, Biotechniques 13:412;
Jayawickreme et al., 1994, Proc. Natl. Acad. Sci. USA
91:1614-1618; Salmon et al., 1993, Proc. Natl. Acad. Sci. USA
90:11708-11712; PCT Publication No. WO 93/20242; and Brenner
and Lerner, 1992, Proc. Natl. Acad. Sci. USA 89:5381-5383.
Examples of phage display libraries are described in
Scott and Smith, 1990, Science 249:386-390; Devlin et al.,
1990, Science, 249:404-406; Christian, R.B., et al., 1992, J.
Mol. Biol. 227:711-718); Lenstra, 1992, J. ImmunoJ.. Meth.
152:14.9-157; Kay et al., 1993, Gene 128:59-65; and PCT
Publication No. WO 94/18318 dated August 18, 1954.
In vitro translation=-based libraries include but are not
limited to those described in PCT Publication No. WO 91/05058
dated April 18, 1991; and Mattheakis et al., 1994, Proc.
Natl. Acad. Sci. USA 91:9022-9026.
By way of examples of nonpeptide libraries, a
benzodiazepine library (see e.g., Bunin et al., 1994, Proc.
Natl. Acad. Sci. USA 91:4708-4712) can be adapted for use.
Peptoid libraries (Simon et al., 1992, Proc. Natl. Acad. Sci.
USA 89:9367-9371) can also be used. Another example of a
library that can be used, in which the amide functionalities
in peptides have been permethylated to generate a chemically
transformed combinatorial library, is described by Ostresh et
al. (1994, Proc. Natl. Acad. Sci. USA 91:11138-11142).
Screening the libraries can be accomplished by any of a
variety of commonly known methods. See, e.g., the following
references, which disclose screening of peptide libraries:
- 39 -
CA 02595040 2007-08-03
.NO 97/30074 Pr T1US97/02298
Parmley and Smith, 1989, Adv. Exp. Med. Biol. 251:215-218;
Scott and Smith, 1990, Science 249:386-390; Fowlkes et al.,
1992; BioTechniques 13:422-427; Oldenburg et al., 1992, Proc.
Natl. Acad. Sci. USA 89:5393-5397; Yu et al., 1994, Cell
76:933-945; Staudt et al., 1988, Science 241:577-580; Bock et
al., 1992, Nature 355:564-566; Tuerk et al., 1992, Proc.
Natl. Acad. Sci. USA 89:6988-6992; Ellington et al., 1992,
Nature 355:850-852; U.S. Patent No. 5,096,815, U.S. Patent
No. 5,223,409, and U.S. Patent No. 5,198,346, all to Ladner
et al.; Rebar and Pabo, 1993, Science 263:671-673; and PCT
Publication No. WO 94/18318.
In a specific embodiment, screening can be carried out
by contacting the library members with an SH3 domain
immobilized on a solid phase and harvesting those library
members that bind to the SH3 domain. Examples of such
screening methods, termed "panning" techniques are described
by way of example in Parmley and Smith, 1988,. Gene
73:305-318; Fowlkes et al., 1992, BioTechniques 13:422-427;
PCT Publication No. WO 94/18318; and in references cited
hereinabove.
In another embodiment, the two-hybrid system for
selecting interacting proteiiis in yeast (Fields and Song,
1989, Nature 340:245-246; Chien et al., 1991, Proc. Natl.
Acad. Sci. USA 88:9578-9582) can be used to identify
molecules that specifically bind to SH3 domains.
A typical assay of the present invention consists of at
least the following components: (1) a molecule (e.g., protein
or polypeptide) comprising an SH3 domain; (2) a ligand of the
SH3 domain; (3) a candidate compound, suspected of having the
capacity to affect the binding between the protein containing
the SH3 domain and the ligand. The assay components may
further comprise (4) a means of detecting the binding of the
protein comprising the SH3 domain and the ligand. Such means
can be e.g., a detectable label affixed to the protein, the
ligand, or the candidate compound.
In another specific embodiment, the invention provides a
method of identifying a compound that affects the binding of
- 40 -
CA 02595040 2007-08-03
a molecule comprising an SH3 domain and a ligand of the SH3
domain comprising:
(a) contacting the SH3 domain and the ligand under
conditions conducive to binding in the presence of a
candidate compound and measuring the amount of binding
between the SH3 domain and the ligand;
(b) comparing the amount of binding in step (a) with the
amount of binding known or determined to occur between the
molecule and the ligand in the absence of the candidate
lo compound, where a difference in the amount of binding between
step (a) and the amount of binding known or determined to
occur between the molecule and the ligand in the absence of
the candidate compound ind.icates that the candidate compound
is a compound that affects the binding of the molecule
25 comprising an SH3 domain and the ligand.
One or more components of the above-described assay of
the invention, e.g., a first molecule comprising an SH3
domain and a second molecule th.at binds to the SH3 domain,
may be provided as a kit comprising one or more containers.
20 in one embodiment, the assay comprises allaw:ing the
protein or polypeptide containing an SH3 domain to contact
the ligand of the SH3 domain in the presence and in the
absence of the candidate compound under conditions such that
binding of the ligand to the protein containing an SH3 domain
25 will occur unless that binding is disrupted or prevented by
the candidate compound. By detecting the amount of binding
of the ligand to the protein containing an SH3 domain in the
presence of the candidate compound and comparing that amount
of binding to the amount of binding of the ligand to the
30 protein or polypeptide containing an SH3 domain in the
absence of the candidate compound, it is possible to
determine whether the candidate compound affects the binding
and thus is a useful lead compound for the modulation of the
activity of proteins containing the SH3 domain. The effect
35 of the candidate compound may be to either increase or
decrease the binding.
- 41 -
CA 02595040 2007-08-03
.VO 97/30074 PCT/US97/02298
One version of an assay suitable for use in the present
invention comprises binding the protein containing an SH3
domain to a solid support such as the wells of a microtiter
plate. The wells contain a suitable buffer and other
substances to ensure that conditions in the wells permit the
binding of the protein or polypeptide containing an SH3
domain to its ligand. The ligand and a candidate compound
are then added to the wells. The ligand is preferably
labeled, e.g., it might be biotinylated or labeled with a
radioactive moiety, or it might be linked to an enzyme, e.g.,
alkaline phosphatase. After a suitable period of incubation,
the wells are washed to remove any unbound ligand and
compound. Tf the candidate compound does not interfere with
the binding of the protein or polypeptide containing an SH3
domain to the labeled ligand, the labeled ligand will bind to
the protein or polypeptide containing an SH3 domain in the
well. This binding can then be detected. If the candidate
compound interferes with the binding of the protein or
polypeptide containing an SH3 domain and the lGbeled ?.:igdnd,
label will not be present in the wells, or will be preseiit to
a lesser degree than is the case when compared to control
wells that contain the protein or polypeptide containing an
SH3 domain and the labeled ligand but to which no candidate
compound is added. Of course,=it is possible that the
presence of the candidate compound will increase the binding
between the protein or polypeptide containing an SH3 domain
and the labeled ligand. Alternatively, the ligand can be
affixed to solid substrate during the assay.
The present invention provides ligands capable of
binding SH3 domains that are suitable for incorporation into
assays such as those described above. Ligands provided by
the present invention include those SH3 domain-binding amino
acid sequences disclosed in Tables 1-13 below and proteins or
polypeptides containing those amino acid sequences. Also
provided are nucleic acids encoding the SH3 domain-binding
amino acid sequences disclosed in Tables 1-13 below.
- 42 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/02.498
6. EXAMPLES
6.1. Preparation of the TSAR-9 Library
6.1.1. Synthesis and Assembly of
Oligonucleotides
Figure 1 shows the formula of the oligonucleotides and
the assembly scheme used in construction of the TSAR-9
library. The oligonucleotides were synthesized with an
applied Biosystems 380a synthesizer (Foster City, CA), and
the full-length oligonucleotides were purified by HPLC.
Five micrograms of each of the pair of oligonucleotides
were mixed together in buffer (10 mM Tris-HC1, pH 8.3, 15 mM
KCl, 0.001% gelatin, 1.5 mM magnesium chloride), with 0.1 %
Triton X-100, 2 mM dNTP's, and 20 units of Tag DNA
polymerase. The assembly reaction mixtures were incubated at
71 C for 30 seconds and then 30 C for 30 seconds; this
cycle was repeated 60 times. It should be noted that the
assembly reaction is not PCR, since a denaturation step was
not used. Fill-in reactions were carried out in a thermal
cycling, device (Ericomp, LaJolla, CA) with the following
protocol: 30 seconds at 72 C, 30 secorids at 30 C, repeated
for 60 cycles. The lower temperature allows for annealing of
the six base complementary region between the two sets of the
oligonucleotide pairs. The reaction products were
phenol/chloroform extracted and ethanol precipitated.
Greater than 90% of the nucleotides were found to have been
converted to double stranded synthetic oligonucleotides.
After resuspension in 300 L of buffer containing 10 mM
Tris-HCI, pH 7.5, 1 mM EDTA (TE buffer), the ends of the
oligonucleotide fragments were cleaved with Xba I and Xho I
(New England BioLabs, Beverly, MA) according to the
supplier's recommendations. The fragments were purified by
4% agarose gel electrophoresis. The band of correct size was
removed and electroeluted, concentrated by ethanol
precipitation and resuspended in 100 L TE buffer.
Approximately 5% of the assembled oligonucleotides can be
expected to have internal Xho I or Xba I sites; however, only
the full-length molecules were used in the ligation step of
- 43 -
CA 02595040 2007-08-03
:/O 97130074 PCT/US97/02298
the assembly scheme. The concentration of the synthetic
oligonucleotide fragments was estimated by comparing the
intensity on an ethidium bromide stained gel run along with
appropriate quantitated markers. All DNA manipulations not
described in detail were performed according to Maniatis,
supra.
To demonstrate that the assembled enzyme digested
oligonucleotides could be ligated, the synthesized DNA
fragments were examined for their ability to self-ligate.
The digested fragments were incubated overnight at 18 C in
ligation buffer with T4 DNA ligase. When the ligation
products were examined by agarose gel electrophoresis, a
concatamer of bands was visible upon ethidium bromide
staining. As many as five different unit length concatamer
bands (i.e., dimer, trimer, tetramer, pentamer, hexamer) were
evident, suggesting that the synthesized DNA fragments were
efficient substrates for ligation.
6.1.2. Construction of Vectors
The construction of the M13 derived phage vectors useful
for expressing a TSAR library has been recently described
(Fowikes, D. et al. BioTech_ (1992} 13:422-427). To express
the TSAR-9 library, an M13 derived vector, m663, was
constructed as described in Fowikes. The m663 vector
contains the pIII gene having a c-myc-epitope, i.e., as a
stuffer fragment, introduced at the mature N-terminal end,
flanked by Xho I and Xba I restriction sites (see also,
Figure I of Fowikes).
6.1.3. Expression of the T8AR-9 Library
The synthesized oligonucleotides were then ligated to
Xho I and Xba I double-digested m663 RF DNA containing, the
piII gene (Fowlkes) by incubation with ligase overnight at 12
C. More particularly, 50 ng of vector DNA and 5 ng of the
digested synthesized DNA and was mixed together in 50 L
ligation buffer (50 mM Tris, pH 8.0, 10 mM MgClz, 20 mM DTT,
0.1 mM ATP) with T4 DNA ligase. After overnigh.t ligation at
- 44 -
CA 02595040 2007-08-03
75361-88
12 C, the DNA was concentrated by ethanol precipitation and
washed with 70% ethanol. The ligated DNA was then introduced
TM
into E. coli (DH5aF'; GIBCO BRL, Gaithersburg, MD) by
electroporation.
A small aliquot of the electroporated cells was plated
and the number of plaques counted to determine that 108
recombinants were generated. The library of E. coli cells
containing recombinant vectors was plated at a high density
(-400,000 per 150 mM petri plate) for a single amplification
l0 of the recombinant phage. After 8 hr, the recombinant
bacteriophage were recovered by washing each plate for 18 hr
with SMG buffer (100 mM NaCl, 10 mM Tris-HC1, pH 7.5, 10 mM
MgC121 0.05o gelatin) and after the addition of glycerol to
50% were frozen at -80 C. The TSAR-9 library thus formed
had a working titer of -2 x 1011 pfu/ml. -
6.2. Preparation of the TSAR-12 Library
Figure 2 shows the formula for the synthetic
oligonucleotides and the assembly scheme used in the
2oconstruction of the TSAR-12 library. As shown in Fi-gure 2,
the TSAR-12 library was prepared substantially the same as
the TSAR-9 library described in Section 6.1 above with the
following exceptions: (1) each of the variant non-predicted
oligonucleotide sequences, i.e., NNB, was 30 nucleotides in
length, rather than 54 nucleotides; (2) the restriction sites
included at the 5' termini of the variant, non-predicted
sequences were Sal I and Spe 1, rather than Xho I and Xba I;
and (3) the invariant sequence at the 3' termini to aid
annealing of the two strands was GCGGTG and CGCCAC rather
than CCAGGT and GGTCCA (5' to 3').
After synthesis including numerous rounds of annealing
and chain extension in the presence of dNTP's and Tag DNA
polymerase, and purification as described above in Section
6.1.1, the synthetic double stranded, oligonucleotide
fragments were digested with Sal I and Spe I restriction
enzymes and ligated with T4 DNA ligase to the nucleotide
sequence encoding the M13 pIII gene contained in the m663
- 45 -
CA 02595040 2007-08-03
75361-88
vector to yield a library of TSAR-expression vectors as
described in Sections 6.1.2 and 6.1.3. The ligated DNA was
then introduced into E. coli (DH5aF'; GIBCO BRLM,
Gaithersburg, MD by electroporation. The library of E. coli
cells were plated at high density (-400,000 per 150 mm petri
plate) for amplification of the recombinant phage. After
about 8 hr, the recombinant bacteriophage were recovered by
washing, for 18 hr with SMG buffer and after the addition of
glycerol to 50% were frozen at -80 C.
The TSAR-12 library thus formed had a working titer of
-2 x 1011 pfu/mL.
6.3. Characterization of the TSAR-9 and -12
Libraries
The inserted synthetic oligonucleotides for each of the
TSAR. libraries, described inSections 6.1 and 6.2 above, had
a potential coding complexity of 20'6 (--3.0"') and 2020,
respectively, and since -=1014 molecules were used in each
transformation experiment, each member of these TSAR
libraries should be unique. After plate amplification the
library solution oz= stock has 109 copies of each member/mL.
It was observed that very few (<10%) of the inserted
oligonucleotide sequences characterized so far in both of the
libraries have exhibited deletions or insertions. This is
likely a reflection of the accuracy assembling the
oligonucleotides under the conditions used and the fact that
certain types of mutations (i.e., frame-shifts) would not be
tolerated as piII an essential protein for phage propagation.
In order to determine whether any coding bias existed in
the variant non-predicted peptides expressed by these
libraries, perhaps due to biases imposed in vitro during
synthesis of the oligonucleotides or in vivo during
expression by the reproducing phage, inserts were sequenced
as set forth below.
- 46 -
CA 02595040 2007-08-03
75361-88
6.3.1. Characterization of T6A.R-9 Library
Inserted synthetic oligonucleotide fragments of 23
randomly chosen isolates were examined from the TSAR-9
library. Individual plaques were used to inoculate I ml of
2XYT broth containing E. co1.i (DH5aF') cells and the cultures
were allowed to grow overnight at 37 C with aeration. DNA
was isolated from the culture supernatants according to
Maniati.s, supra. Twenty-three individual isolates were
sequenced according to the method of Sanger (Proc. Natl.
;.0 Acad. Sci. USA (1979) 74:5463-5467) using as a primer the
oligonucleotide 5'-AGCGTAACGATCTCCCG (SEQ ID NO. 99), which
is 89 nucleotides downstream of the pIZI gene cloning site of
the m663 vector used to express the TSARS.
Nucleotide sequences and their encoded am.ino acid
sequences were analyzed with the MacVectorMcomputer program
(IBI, New Haven, CT). The Microsoft.EnCEL program was used'
to evaluate amino acid frequencies. Such analyses showed
that the nucleotide codons coding for and hence most amino
acids, occurred at the expected frequency in the TSAR-9
library o.f expressed proteins. The notable exceptions were'
glutamine and tryptophan, which were over- and under-
represented, respectively.
It is of interest to note the paucity of TAG stop codons
in the inserts, i.e., only 2 of -200 isolates characterized
contained a TAG stop codon. About half [1-(47J48)36] of the
phage inserts were expected -to have at least one TAG codon in
view of the assembly scheme used. However, most of the TAG-
bearing phage appear to have been lost from the library, even
though the bacterial host was supE. This may be a
.3o consequence of suppression being less than 100% effective.
The amino acids encoded by the inserted double stranded
synthesized oligonucleotide sequences, excluding the fixed
PG-encoding centers, were concatenated into a single sequence
and the usage frequency determined for each amino acid using
TM
the Microsoft EXCEL program. These frequencies were compared
to that expected from the assembly scheme of the
oligonucleotides, and the divergence from expected values
- 47 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/0,LA98
represented by the size of the bars above and below the
baseline. Chi square analysis was used to determine the
significance of the deviations. The majority of amino acids
were found to occur at the expected frequency, with the
notable exceptions that glutamine and tryptophan were
somewhat over- and under-represented, respectively. Thus,
except for the invariant Pro-Gly, any position could have any
amino acid; hence, the sequences are unpredicted or random.
6.3.2. Characterization of TSAR-12 Library
Approximately 10 randomly chosen inserted
oligonucleotides from the TSAR-12 library were examined by
DNA sequencing as described above in Section 6.3.1. The
isolates were chosen at random from the TSAR-12 library and
prepared for sequencing, as were the TSAR-9 isolates.
Analysis showed that except zor the invariant Gly any
position could have any amino acid; hence, the sequences are
unpredicted or random.
6.4. Preparation of R8C Library
Referring now to Figure 3, two oligonucleotides were
synthesized on an Applied Biosystems Model 380a machine with
the sequence 5'-
TGACGTCTCGAGTTGTNNKNNKNNKNNKNNKNNKNNKNNKTGTGGATCTAGAAGGATC-3'
ISEQ ID NO:31) and 5'-GATCCTTCTAGATCC-3' (SEQ ID NO:32),
where N is an equimolar ratio of deoxynucleotides A, C, G,
and T, and K is an equimolar ratio of G and T. Fifty pmol of
each oligonucleotide was incubated at 42 C for 5 min, then
37 C for 15 min, in 50 L of Sequenase1'''' buffer (U.S.
Biochemicals, Cleveland, OH) with 0.1 g/ L acetylated BSA,
and 10 mM DTT. After annealing, 10 units of SequenaseT" (U.S.
Biochemicals) and 0.2 mM of each dNTP were added and
incubated at 37 C for 15 min. The sample was then heated at
65 C for 2 hr, digested with 100 units of both Xho I and Xba
I(New England BioLabs, Beverly, MA), phenol extracted,
ethanol precipitated, and resolved on a 15% non-denaturing
polyacrylamide gel. The assembled, digested fragment was gel
- 48 -
CA 02595040 2007-08-03
75361-88
purif ied prior to ligation. The vector, m663 (Fowlkes, D. et
al. Biotech. (1992) 13:422-427), was prepared by digestion
with Xho 'I and Xba I, calf alkaline phosphatase (Boehringer
Mannheim, Indianapolis, IN) treatment, phenol eytracted, and
purified by agarose gel electrophoresis. To ligate, 20 g
vector was combined with 0.2 g insert in .3 mL with T4 DNA
ligase (Boehringer Mannheim), according to the manufacturer.
After removal of the protein and buffer by phenol extraction
and ethanol precipitation, the ligated DNA was electroporated
1o into XL1-Blue E. coli (Stratagene, San Diego, CA) and plated
for eight hours at 37 C. To recover the recombinant phage,
the top agar was collected with a spatula, mixed with an
equal volume of 100 mM NaC1, 10 mM MgClz, and 50 mM Tr.is-HCI
(pH7.5), and disrupted by two passes through ar. 18-gauge
syringe needle. The bacterial cells were removed by
centrifugation, and phage particles were coll.ected.by
polyethylene glycol precipitation and stored at -70 C in -25%
glycerol. The library had 10E total recombinants and a
working titer of 6 x 101=ptu/mL.
Members of the library were checked for inserte .by the
polymerase chain reaction (Saiki, et al. Science (1988) -
239:487-491). Individual plaques on a petri plate were
touched with a sterile toothpick and the tip was stirred into
2xYT with- F'E. coli bacteria and incubated overnight-at 37 C
with aeration. Five microliters of the phage supernatant
were then transferred to new tubes containing buffer (67 mM
Tris-HC1, pH 8.8/10 mM (3= mercaptoethanol/16.-6 mM ammonium
sulfate/6.7 mM EDTA/50 gg bovine serum albumin per.mL), 0.1
mM deoxynucleotide triphosphates, and 1.25 units of Tag DNA
polymerase (Boehringer Mannheim, Indianapolis, IN) with lDo
pmoles of oligonucleotide primers. The primers flanked the
cloning site in gene III of m663 (5'-TTCACCTCGAAAGCAAGCTG-3'
(SEQ ID N0:100) and 5'-CCTCATAGTTAGCGTA.ACG-3' (SEQ ID
N0:101)). The assembly reactions were incubated at 94 C for
1 min, 56 C for 2 min, and 72 C for 3 min; this cycle was
repeated 24 times. The reaction products were then resolved
Tll
by electrophoresis on a NuSieve 2.0% agarose gel (FMC,
- 49 -
CA 02595040 2007-08-03
75361-88
Rockland, ME). Gels revealed that for 20 plaques tested, all
were recombinant and had single inserts of the expected size.
Based on the sample size of the library, it was
anticipated that 100% of the recombinants had single inserts.
However, all of the SH3-binding phage isolated from the R8C
library had double-inserts. Such phage are presumed rare
(i.e., <5%) within the library, yet because the SH3-binding
peptide appears to need to be linear they were selected for
by our screening methods. Most likely they were formed
lo during the generation of the library; one scenario is that
the inserts ligated together to form head-to-head dimers and
that they were subsequently cloned into m663 DNA by ligation
with the vector's Xho T sticky end and by illegitimate
ligation with the vector's Xba I site (.see, Figure 4).
6.5. Preparation Of Target-Coated Microtiter Wells
6.5.1. Preparation Of GST-SH3 Fusion
Proteins
The preparation of Src-GST fusion protein was first
described by Smith and Johnson, in,Gene (1968) 67:31.
Briefly, pGEX-derived (Pharmac.ia, Piscataway, NJ) constructs
expressing GST fusion proteins containing the SH3 domains of
Src, Grb2, Crk, Abl, or PLCy were obtained from 'Dr. Channing
Der (University of North Carolina at Chapel Hill); a
construct expressing the SH3 domain of Yes was obtained from
Dr. Marius Sudol (Rockefeller University). The use of the
pGEX bacterial expression vector for the production of GST-
SH3 fusion proteins is well-known to those in the art. See,
e.g., Cicchetti, P. et al., in Science (1992) 257:803-806.
Briefly, the.coding region for a particular SH3 domain can be
fused in-frame at the Bam HI site of pGEX-2T. Thus, fusion
proteins were prepared as per the manufacturer's
instructions, and quantified by Coomassie Blue staining of
SDS-polyacrylamide gels. Microtiter wells were coated with
5-20 g GST-SH3 fusion protein in 100 mM NaHCO3, pH 8.5,
blocked with 100 mM NaHCO3 (pH 8..5) 1% BSA, and washed. All
- 50 -
CA 02595040 2007-08-03
75361-88
washes consisted of five applications of 1XPBS, 0.1% TweenTM
20, 0.1% BSA (Buffer A). Where appropriate, the amount of
protein bound to each well was quantified with an anti-GST
antibody-based ELISA (Pharmacia, Piscataway, NJ), and with a
GST-binding phage, isolated during'the course of this work.
6.5.2. Coating of Microtiter Wells
Bacterially expressed Src SH3 glutathione-S-transferase
(Src-GST) fusion protein was purified from bacterial lysates
using glutathione agarose 4B (Pharmacia), according to the
manufacturer's instructions. Bound Src-GST fusion protein
was eluted from the glutathione agarose with 10 mM
glutathione in PBS. Microtiter wells were then coated with
Src-GST fusion protein (1-10 g/well, in 50 mM NaHCO3, pH 8.5)
overnight at 4 C. To block non-specific binding of phage,
100 L 1% BSA in 100"mM NaHCG3, pH 8.5, was added to each well
and allowed to incubate at room temperature for 1 hour. The
wells were then washed five times with 200 L PBS, 0.1% TweenTM
20, 0.1% BSA (Buffer A).
6.6. Biopanning And Subsequent Characterization Of
Phage-Displayed Random Peptide Libraries With
Src-GST Fusion Protein As Target Molecule
6.6.1. Isolation of Src SH3-Binding Phage
Library screens were performed as previously described.
Kay, B.K., et al., in Gene. (1993) 128:59-65. Briefly, 1 X
1011 pf u TSAR 9, TSAR 12, or RBC phage in Buffer A were
incubated in a Src SH3-GST-coated well for 2 hours. The wells
were washed, and bound phage were eluted with 100 4L.50 mM
glycine-HC1 (pH 2.2), transferred to a new well, and
neutralized with 100 mL 200 mM NaHPO4 (pH 7.0). Recovered
phage were used to infect 1 x 109 DH5aF' E. coli cells in 20
mL 2xYT; the infected cells were grown overnight, resulting
in a 1000- to 10,000-fold amplification of phage titer.
Amplified phage were panned twice more, as above, excepting
the amplification step. Binding phage recovered after the
third round of panning were plated at a low density on a lawn
- 51 -
CA 02595040 2007-08-03
WO 97/30074 PCTIUS97/02298
of DH5aF' E. coli cells to yield isolated plaques for clonal
analysis. Isolated plaques were used to produce small
cultures from which phage stocks and DNA were recovered for
phage binding experiments and dideoxy sequencing (Sanger, F.,
et al., in Proc. Natl. Acad. Sci. USA (1977) 74:5463-5467),
respectively. Clones were confirmed as binding the SH3
domain by applying equal titers of phage to wells containing
Src SH3-GST or GST alone, and titering the number of eluted
particles from each well, or detecting bound phage with an
anti-phage antibody-based ELISA (Pharmacia).
indeed, the ability of isolated phage clones to bind to
several SH3 domains derived from a variety of different
proteins can be investigated by the maizner described above.
GST-SH3 fusion proteins containing SH3 domains from a variety
of different proteins are bound to microliter wells. An
aliquot of the aforementioned phage stocks (50 gL) is
introduced into wells containing the different GST-SH3 fusion
proteins. After room temperature incubation for 1-2 hours,
the liquid contents of the microtiter plates are removed, and
the wells are washed 5 times with 200 L Buffer A. Bound
phage are eluted with 100 gL 50 mM glycine (pH 2.2),
transferred to a new well, and neutralized with 100 L 200 mM
NaHPOq (pH 7.0). The phage are diluted 10 -3- to 10-6-f old, and
aliquots are plated onto lawns of DH5aF' E. coli cells to
establish the number of plaque forming units in the output
sample. From these experiments, the relative specificity of
different Src SH3 binding clones for SH3 domains derived from
other proteins is determined.
6.6.2. Phage ELISA and Nucleotide
Sequencing
To evaluate the binding of isolates to various targets
proteins, enzyme-linked-immuno-assays (ELISA) were also
performed. Bacterial cultures were infected with phage
isolates and cultured overnight in 2XYT at 37 C. The cells
were spun down and 25 mL of supernatant was added to
microtiter plate wells coated with 50 L of protein (1 mg/mL
- 52 -
CA 02595040 2007-08-03
753 6 1 -8a
.in 10 0 mM NaHCD3,-pH -8 . 4; overnight a~=t 4 C or for a few hours
at room temperature) and blocked (=1 mg/mL BSA in 100 m.M
NaHCO3, pH 8.4; for about one hour). The phage are incubated
in the well with 25 L of PBS-0.1% Tween7.20 at RT for 2 hr.
The wells are then washed multiple times over 30 minutes. To
each well is added 50 uL of polyclonal anti-phage antibody
conjugated to horseradish peroxidase. The antibody is
TM
diluted 1:3000 in BSA-PBS--Tween 20; it was obtained from
Pharmacia (Piscataway, NJ; catalog number 27-9402-01). After
30 minutes, the wells are washed again with BSA-PBS-Tweeri 20
for -20 minutes. Finally, 100 L of ABTS reagent (Pharmacia,
with H202) are added to each well for the development of
color. Plates are read with a plate reader (Molecular
Devices, Menlo Park, CA) at 405 nm wavelength.
The nucleotide sequence of the relevant segments of the
Src SH3 binding clones (or phage clones that bind to SH3
domains of other proteins) were sequenced using standard
methods. Sanger, F., et al., in.Proc. Natl. Acad. Sc.i. USA
(1977) 74:5463-5467. The oligo primer 51-AGCGTAACGATCTAAA--3'
(SEQ ID ND:102) was used, which is 89 nucleotides downstream
of the gene III cloning site of M13 m666. The nucleotide
sequences were analyzed with the MacVector computer prograir.
(IBI, New Haven, CT, USA). From this nucleotide sequence
information the primary .sequence of each Src SH3 binding
peptide was deduced. The corresponding synthetic peptides
were then prepared by techniques well known in the art with
or without flanking sequences. Indeed, these synthetic
peptides have been shown to bind to SH3 domain targets, with
those possessing the phage flanking amino acid residues
3o exhibiting greater binding affinity.
6.7.In Vitro Peptide Binding Assays
Peptides were obtained from Research Genetics
(Birmingham, AL), Chiron Mimotopes (Victoria, Australia),- or
synthesized by conventional techniques by Dr. J. Mark Carter
of Cytogen Corporation (Princeton, NJ). Peptide purity was
assessed by HPLC and/or mass spectrometry. Biotinylated
- 53 -
CA 02595040 2007-08-03
peptides were synthesized with either a KSGSG (SEQ ID NO:loi)
or a GSGS (SEQ ID NO:104) peptide linker (a spacer) between
the biotin and the N-terminus of the peptide. Binding
experiments were performed as above, excepting the use of 10
M peptide instead of phage. Bound biotinylated peptide was
detected with streptavidin conjugated to alkaline phosphatase
(Sigma Chemical Co., St. Louis, MO). After one hour
incubation period at room temperature, the wells were washed,
and a solution of 3 mM p-nitrophenyl-phosphate (US
Biochemicals, Cleveland, OH) in 50 mM NaCO3 (pH 9.8), and 50
mM MgCl2 was added and color allowed to develop. Signals were
read with an ELISA plate reader (Molecul.ar Devices, Menlo
Park, CA) at 405 nm wavelength. Binding experiments were
performed in triplicate. The results are presented in
Figures 7 and S.
6.8. Peptide Competi.tion of G8T-SH3 Affinity
Precipitations of Cell Lysates
Labeled proteins are prepared by incubating a cul-ture of
HeLa cells overnight with. >-100 Ci/mL 'SS-methioriine. The
cells are then washed and lysed with mild detergent. This
mixture of radioactive proteins is incubated with Src-GST
fusion protein that has been immobilized on glutathionF-
linked Sepharose beads (Pharmacia, Piscataway, NJ). After
several hours of tumbling, the beads are pelleted gently by
low-speed centrifugation, and the supernatant is discarded.
The beads are then resuspended into a slurry in PBS-0.1o
Tweer20, pelleted, and washed several additional times.
Finally, a 2% SDS solution is added to the sample, which is
then boiled at 100 C for 3 minutes. Afterward, the sample
is centrifuged, and the supernatant loaded on a 100
polyacrylamide SDS gel for electrophoresis. After the
proteins have been resolved, the gel is fixed, dried down,
and exposed to X-ray film for autoradiography or phosphor
plates for scanning by a Molecular Dynamics Phosphorimager.
The ability of Src SH3 to bind certain 3SS-labeled
proteins is examined for competability with exogenous
- 54 -
CA 02595040 2007-08-03
75361-88
peptides. Synthetic peptides corresponding to phage-
displayed inserts and motifs are added at the time that the
lysate is incubated with the Src-GST fusion protein
immobilized on glutathione-linked sepharose beads. The SH3
binding peptides block binding of all or some of the labeled
proteins while negative control peptides (unrelated peptide
sequences) do not. The amount of competition is quantified
and correlated with the amount of added SH3-domain binding
peptides.
Alternatively., NIH 3T3 cells were grown in Dulbecco's
Modified Eagle Medium (DME) + 10% fetal calf serum (FCS) + B0
Ci/mL Tran35Slabel (ICN), washed with PBS, lysed in RIPA
buffer, and pelleted. Supernatant from 1.5 x 106 cells was
precleared with 100 g glutathione-agarose-immobilized GST.
The supernatant was then incubated with 10 g glutathione-
agarose-immobilized GST-SH3 fusion protein with or without
added test peptide in a final volume of 250 L. Pel.leted
beads were washed with 1 mL each of RIPA, RIPA + 1%
deoxycholate + 0.1% SDS, and PBS; resuspended in 50 uL
SDS-PAGE sample buffer, boiled, and subjected to SDS-PAGE
(7.5%). Labeled proteins were detected by phosprcorimaging
(Molecular Dynamics). The results are presented in Figure 9.
6.9. Peptide Competition of GST-SIi3 Affinity
Precipitations of PI-3' Rinase From Cell
Lysates
It is possible to follow the precipitation of PI-3'
Kinase by Src from cell lysates in the presence or absence of
SH3-binding peptides.. HeLa cells are lysed with detergent
and the protein mixtures are incubated for several hours with
the Src-GST fusion protein immobilized on glutathione-linked
Sepharose beads. After several hours of tumbling, the beads
are pelleted gently by low-speed centrifugation and the
supernatant is discarded. The beads are then resuspended
into a slurry in PBS-0.1% TweeriM20, pelleted, and washed
several additional times. Finally, an SDS solution is added
to the sample, which is then boiled at 100 C for 3 minutes.
- 55 -
CA 02595040 2007-08-03
.NO 97/30074 PC'r /US97/024A
Subsequently, the sample is centrifuged, and the supernatant
is loaded on a 10% polyacrylamide SDS gel for
electrophoresis. After the proteins have been resolved, the
gel is blotted to nitrocellulose or nylon (i.e., western
blot). The filter is then probed with a PI-3' Kinase
antibody (monoclonal and polyclonal antibodies are available
from Upstate Biotechnology Incorporated, Lake Placid, NY) and
an enzyme-linked secondary antibody. The amount of PI-3'
Kinase is then quantitated.
The ability of Src SH3 to bind PI-3' Kinase is examined
for competability with exogenous peptides. Synthetic
peptides corresponding to phage-displayed inserts and motifs
are added at the time that the lysate is incubated with the
Src-GST fusion protein that has been immobilized on
glutathione-linked sepharose beads. Ten-fold and one
hundred-fold molar excess of peptides are used relative to
SH3 proteins. The SH3 binding peptides block binding of the
PI-3' Kinase as detected on western blot.s while negative
control peptides (unrelated peptide sequences) do not. The
amounz of competition is quantified and correlated with the
amount of added SH3-domain binding peptides.
6.10. in Vivo Association Of SH3-Binding
Peptides With 8H3-Domains Of Proteins
To demonstrate association of the SH3-binding peptides
with SH3-domains of proteins inside cells, the SH3-binding
peptides are tagged and localized in cells. For example,
Bar-Sagi et al., in Cell (1993) 74:83-91, have shown that
SH3-binding proteins localize to the cytoskeleton when
expressed in cells. Thus, the SH3 domain-binding peptides of
the present invention can serve as cellular targetting
signals (e.g., to the cytoskeleton). Accordingly, the
peptides are tagged with biotin and, subsequently, injected
into cells. Alternatively, one can transfect into cells a
recombinant plasmid that expresses a fusion protein
comprising of the SH3-binding peptide and the green
fluorescent protein (GFP, Chalfie et al., in Science (1994)
- 56 -
CA 02595040 2007-08-03
75361-88
263:802-805). The location of the biotinylated peptide or
the GFP fusion protein is then assayed with FITC-labeled
streptavidin in paraformaldehyde-fixed cells or by direct
fluorescence in living cells, respectively. Localization of
the SH3-binding peptides to the cytoskeleton demonstrates
that the SH3-binding peptides can bind SH3-domain proteins in
vivo. In addition, focal adhesions, which are rich in Src,
are also sites of potential subeellular localization of SH3-
binding peptides.
Thus, NIH 3T3 f ibroblasts were cultured in vitro on
glass coverslips coated with fibronectin. After two days of
growth at 37 C, the cells were fixed for one hour at room
temperature in the presence of 2% paraformaldehyde (pH 7.5).
The coverslips were washed with PBS-O.lo Tween 20 several
times to remove the fixative. Next, the coverslips were
dipped into acetone (chilled at -2C "C) fcr approximately 20
seconds and allowed to air-dry. The coverslips were washed
TM'
again with PBS-0.1% Tween .20, containing BSA (I mg/mL), and
incubated for 2 hours at room temperature with different
biotinylated peptides in PBS-0.1%.Tween720. The coverslips
were washed and then incubated with 1 mg/mL streptavidin-Cy3
(Jackson Immunoresearch Co., West Grove, PA) for 1 hour at
room temperature. Finally, the coverslips were washed in
PBS-0.1o Tween 20, mounted in a glycerol solution on a glass
slide, and viewed with a Nikon optiphot epifluorescence
microscope and a 60x oil immersion lens.
The results are presented in Figure 11, in which panel A
displays cells stained with the conjugate biotin-spacer-
VLKRPLPIPPVTR (SEQ ID N0:64); panel B exhibits cells stained
with the conjugate, biotin-spacer-GILAPPVPPRNTR (SEQ ID
No:63); panel C shows cells stained with the long consensus
peptide, biotin-spacer-RSTPRPLPPLPTTR (SEQ ID N0:67); and
panel D shows cells stained with the.proline-rich vinculin
peptide conjugate, biotin-spacer-LAPPKPPLPEGEV (SEQ ID
N0:70). The "spacer" sequence is KSGSG (SEQ ID N0:103). As
shown in Figure 11, the panels in which SH3 domain-binding
peptides were used present a bright display of fluorescence
- 57 -
CA 02595040 2007-08-03
,O 97/30074 PCT/US97/02_
activity that is in sharp contrast to the relatively "dark"
features of panel D (non-SH3 domain binding vinculin
segment). These results demonstrate further the ability of
the SH3 domain-binding peptides of the present invention to
localize to protein targets (e.g., Src and Src-related
proteins) within cells and provide an image thereof.
6.11. In Vivo Modulation Of Src In Oocytes With
SH3-Binding Peptides
Wheii Xenopus laevis oocytes are injected with mRNA
encoding deregulated Src, there are dramatic cytological and
biochemical changes in the oocyte (Unger, T.F. and Steele,
R.E., in Mol. Cell. Biol. (1992) 12:5485-5498). The
applicants have obtained plasmids for generating wild type
and deregulated Src mRNA, which are available from Dr. Robert
Steele (University of California at Irvine). Synthetic SH3-
binding peptides are injected into oocytes that have been
previously injected with Src mRNA. The state of the
cytoskeleton is inspected visua'Lly by observing the
arrangement of cortical pigment granules under a dissecting
microscope. The state of phosphorylation of several proteins
is examined by western blotting with an anti-phosphotryosine
monoclonal antibody (4G10; Upstate Biotechnology
Incorporated), as described in Unger and Steele, above.
6.12. Progesterone-induced X. laevis Oocyte
Maturation
Segments of adult ovary were removed surgically and
incubated in 0.1% collagenase type D (Boehringer Mannheim,
Indianapolis, IN) in Ca2+-free OR2 (82.5 mM NaCl, 2.5 mM KC1,
1.0 mM MgC121 1.0 mM Na2HPO9, 5.0 mM HEPES, and 3.8 mM NaOH,
pH 7.6). Oocytes were then washed 3-5 times with OR2
containing 1.0 mM CaC12 and allowed to recover in OR2
overnight at 18 C. Stage VI oocytes were injected with 40 nL
of 100 mM peptide or water. After injection, the oocytes
were placed in OR2 with 2 mg/mL progesterone (Sigma, St
- 58 -
CA 02595040 2007-08-03
40 97/30074 PCT/US97/02298
Louis, MO) and incubated at 20 C. Oocytes were scored at
hourly time points for germinal vesicle breakdown (GVBD).
Figure 10 presents the results of this experiment. As
shown by the graph, oocytes injected with the SH3 domain-
binding peptide VLKRPLPIPPVTR (SEQ ID NO:64) exhibit a faster
rate of progesterone-induced germinal vesicle breakdown
relative to oocytes that had been injected with water or with
the proline-rich vinculin peptide, LAPPKPPLPEGEV (SEQ ID
NO:70). These results parallel those of Unger and Steele,
supra, wherein oocytes injected with deregulated or active
Src RNA matured at a faster rate than oocytes injected with
water or wild-type Src mRNA (See Figrure 3B of the Unger and
Steele article).
The present results obtained with Src SH3 domain-binding
peptides suggest that these peptides modulate the biochemical
activity of "cellular" Src; in particular, it is proposed
that at least some of the Src SH3 domain-binding peptides of
the present invention upregulate the biochemical activity of
"cellular" Src, which may be downregulated,or inhibited in
its normal state. Hence, the administration of the SH3
domain-binding peptides of the present invention can
constitute a novel method of modulating the activity of Src
or Src-related proteins. Specifically, certain of these
peptides are able to activate Src-family proteins.
6.13. In Vivo Antagonism Of Src In Src
Transformed Cells With B83-Binding
Peptides
The coding regions for SH3-binding peptides are cloned
into vectors that direct their expression in animal cells. A
bipartite gene -i.s constructed, encoding a protein with c-myc
epitope and SH3-binding peptide, which is transcribed from a
strong constitutive promoter (e.g., SV40, CMV, HSV TK,
calmodulin). The vector is introduced into either normal or
Src-transformed cells via transfection (e.g.,
electroporation, calcium phosphate, liposomes, DEAE dextran).
Transfected cells express the bipartite gene transiently in
- 59 -
CA 02595040 2007-08-03
097130074 PCTIUS97/02
culture. To create stable transformed cell lines, the vector
carries a selectable marker (e.g., neomycin resistance) or
transfection is performed in the presence of excess plasmid
carrying a selectable marker (e.g., neomycin resistance) and
cells selected for the marker. Transfected cells are stained
by immunofluorescence to detect expression of the bipartite
protein. The hybridoma 9E10 secretes a monoclonal antibody
that is highly specific for the c-myc epitope (EQKLISEEDLN
[SEQ ID NO:105]; see, Evan, G.A. et al., in Mol. Cell. Biol.
(1985) 5:3610-3616). This antibody is used in
immunofluorescence experiments to demonstrate that the
bipartite protein is expressed inside the cells, and in some
cases, localized to subcellular structures enriched in SH3
domain bearing proteins.
There are several controls used in these experiments.
First, cells are transfected with vectors that do not have
the SH3-binding peptide coding region. Second, normal (non-
trarisformed) cells are transfected. Third, cells transformed
by oncogenes other than Src are used in the transfection
experiments. Fourth, cells are stained with other monoclonal
antibodies that do not recognize the c-myc epitope.
Transfected cells are examined for any changes in cell
shape, behavior, and metabolism as a consequence of
expressing the SH3 binding peptides. Cell shape is examined
by phase contrast microscope at several times after
transfection; in particular, the flatness of the cells, their
adhesion to the substrate, and the degree of cell ruffling
are monitored. Cell division rates, cell migration, and
contact inhibition are also observed over time. Finally, the
amount of phosphorylated tyrosine in transfected cells is
quantitated by phosphoaminoacid analysis and with an anti-
phosphotryosine monoclonal antibody (4G10; Upstate
Biotechnology Incorporated) in western blotting experiments.
- 60 -
CA 02595040 2007-08-03
3 97/30074 PCT/US97102298
6.14. Distinct Ligand Preferences of various
SIi3 Domains
6.14.1. Preparation of PXXP (SEQ ID No: 161)
Biased Peptide Libraries
Using procedures similar to those described in Sections
6.1 and 6.4 and also described in Sparks, A. B., et al., in
Methods in Enzymolocrv, (1995) 255:498-509, oligonucleotide
inserts were constructed according to the schematic provided
in FIG. 12. The two synthetic oligonucleotides (5'-
ctgtgcctcgagk(nnk)6cca(nnk)2cca(nnk)6tctagacgtgtcagt-3' (SEQ
ID NO:162) and 5'-actgacacgtctaga-3'(SEQ ID NO:163), where
k=g+t and n=g+a+t+c) were annealed and filled in with
Sequenase (Amersham, Arlington Heights, Il). The inserts
were then digested with Xho I and Xba I and were ligated into
gene III of the mBAX vector.
The mBAX vector was created by generating cloning sites
in aene III of the M13mp18 vector (Messing, J., 1991,
"Cloning in M13 phage or how to use biciogy at its best,"
Gene 100, 3-12) in the manner of Fowlkes et al., 1992,
Biotechniques 13, 422-427. The mBAX vector displays a
peptide sequence at the N-terminus of the mature pIII protein
that encodes the epitope for the mouse monoclonal antibody
7E11 (see FIG. 13); it includes the stop codon TAG in the
coding region, which is suppressed in E. coli carrying
suppressor tRNA gene mutations known as supE or supF. There
are no other stop codons in the mBAX genome. The mBAX vector
also carries a segment of the alpha fragment of
0-galactosidase; bacterial cells expressing the omega
fragment of Q-galactosidase that are infected by a
bacteriophage that expresses the alpha fragment convert the
clear XGal substrate into an insoluble blue precipitate.
Thus, plaques of such bacteriophage on such cells appear
blue.
Recombinant mBAX molecules can be distinguished easily
from non-recombinant molecules due to the TAG codon in the
XhoI - XbaI segment in gene III of mBAX. When recombinants
are generated by replacing the Xho I - Xba I fragment with
- 61 -
CA 02595040 2007-08-03
oligonucleotides encoding random peptides, the recombinants
can be grown in bacteria with (e.g., DH5aF') or without
(e.g., JS5) suppressor tRNA mutant genes. On the other hand,
the non-recombinant mBAX molecules fail to produce plaques on
bacterial lawns where the bacteria (e.g., JS5) lack such
suppressor genes. This is because in JSS, the TAG codon
serves as a stop codon to yield a truncated pIII molecule
during translation; since pIII is an essential protein
component of viable M13 viral particles, no plaques will
form.
The ligated DNA was electroporated into JS5 E. coli and
recombinant phage were propagated on two hundred 100 mm 2xYT
+ 0.8% agar plates as described in Sambrook, J., Frisch, E,
F., & Maniatis, T. (1989) Molecular Cloning: A Laboratory
Manual (Cold Spring Harbor Laboratory, Plainview, NY)
(Sambrook et al.). To minimize the recovery of sibling
clones during affinity purification of binding phage, six
distinct library fractions were prepared by dividing the
plates into six roughly equal groups. Each fraction was
2o treated separately in all subsequent manipulations. Phage
,particles were harvested from each fraction by diffusion into
100 ml PBS (137 mM NaCl, 2.7 mM KC1, 4.3 mM Na:2HPOq, 1.4 mM
KHZPO,), concentrated by polyethylene glycol precipitation as
in Sambrook et al. (1989, supra), and resuspended in 10 ml
PBS + 10% glycerol. Each fraction contained approximately
5x10' un:.que recombinants, for a total library complexity of
approximately 3x108. The resulting phage-displayed library
contained peptides of the form XSPXXPX6 (SEQ ID NO:164), where
X represents any amino acid.
6.14.2. Affinity purification of SFI3-binding
phage
Library screens were performed as described in Sparks,
A. B., et al., in Methods in Enzymolocty, (1995) 255:498-509.
Briefly, wells of an ELISA microtiter plate were coated with
10 g GST-SH3 fusion protein in 100 mM NaHCO3 (pH 8.5) for 3
hours and blocked with SuperblockM(Pierce, Rockford, IL) for
- 62 -
CA 02595040 2007-08-03
75361-88
1 hour. Approximately 5 x 1011 infectious particles from each
library fraction were diluted in 200 l PBS + 0.1% TweerT20
and incubated in a GST-SH3-coated well for 3 hours. The
wells were washed five times with PBS + 0.1% TweenM'20, and
bound phage were eluted with 50 mM glycine-HC1 (pH 2.2).
Recovered phage were propagated in 10 ml 2xYT media and 100
l of a saturated DH5aF' E. coli culture and affinity
purified twice more as above. Affinity purified phage were
plated onto 2xYT + 0.8% agar plates to yield isolated plaques
l0 from which clonal phage stocks and DNA were produced. Phage
binding was confirmed by incubating equal amounts of a clonal
phage stock in wells coated with 1 g GST-SH3 or GST. The
wells were washed :'ive times with PBS + 0.1% TweenM20, and
bound phage were detected by anti-phage ELISA according to
the manufacturer's instructions (Pharmacia, Piscataway, NJ).
Clones with strong SH3-binding activity were selected for
further analysis. The sequences of peptides displayed by
these clones were determined by DNA sequencing of phage
inserts.
6.14.3. Preparation of GST-SH3 fusion
proteins
Constructs encoding GST fusions to the Grb2 N-terminal
(Grb2 N, aa 1-58), Nck N-terminal (Nck N, aa 1-68), Nck
middle (Nck M, aa 101-166), Nck C-terminal (Nck C, aa 191-
257), p53bp2 (aa 454-530), or Src (aa 87-143) SH3 domains
were generated by PCR cloning of the appropriate cDNAs into
pGEX-2T (Pharmacia, Piscataway, NJ; a general reference for
the pGEX vectors is Smith, D. B., & Johnson, K. S. (1988)
Gene 67, 31-40). The integrity of the constructs was
confirmed by DNA sequencing. pGEX-derived constructs
expressing GST fusions to the SH3 domains of Yes, Cortactin,
Crk, Abl, and PLC-y were kindly provided by M. Sudol
(Rockefeller University), J. T. Parsons (University of
Virginia at Charlottsville), M. Matsuda (Tokyo, Japan), A. M.
Pendergast (Duke University), and S. Earp (University of'
North Carolina at Chapel Hill), respectively. Alternatively,
- 63 -
CA 02595040 2007-08-03
WO 97/30074 PCTIUS97/02298
the GST-SH3 fusion proteins for Yes, Cortactin, Crk, Abl, and
PLC-y could have been prepared as above for Grb2 N, Nck N, Nck
M, Nck C, p53bp2, and Src, using published sequence
information for these proteins. See, e.g., Suen et al.,
(1993) Mol. Cell. Biol. 13, 5500-5512 (Grb2); Lehmann et al.,
(1990) Nucleic Acids Res. 18, 1048 (Nck); Iwabuchi et al.,
(1994) Proc. Natl. Acad. Sci. USA 91, 6098-6102 (p53bp2);
Takeya et al., (1983) Cell 32, 881-890 (Src); Sudol et al.,
(1988) Nucleic Acids Res. 16, 9876 (Yes); Wu et al., (1991)
Mol. Cell. Biol. 11, 5113-5124 (Cortactin); Matsuda et al.,
(1992) Mol. Cell. Biol. 12, 3482-3489 (Crk); Shtivelman et
al., (1986) Cell 47, 277-284 (Abl); Burgess et al., (1990)
Mol. Cell. Bio1. 10, 4770-4777 (PLCy). GST-SH3 fusion
proteins were prepared as described in Smith, D. B., &
Johnson, K. S. (1988) Gene 67, 31-40. The integrity and
purity of the fusion proteins were confirmed by SDS-PAGE.
Protein concentrations were determined usinq a the BioRad
protein assay (BioRad, Hercules, CA).
6.14.4. 8H3 Domain Binding Peptides and
Consensus Sequences
The use of second generation or biased peptide
libraries, which fix all or part of the PXXP (SEQ ID NO:161)
consensus motif for SH3 domain binding peptides and randomize
flanking residues, has defined additional sequence-residues
exhibiting selective SH3 domain binding.
Tables 1-5, below, list some of the relevant amino acid
sequences obtained when the biased peptide library described
in Section 6.14.1 was screened with GST-SH3 fusion proteins.
The underscored amino acid residues in Tables 1-5 indicate
the fixed positions. Also, indicated for each set of new
binders is a "consensus" sequence, which seeks to include the
additional features gleaned from the new binding peptides.
The symbol "O" in the consensus sequences of Tables 1-5
represents a hydrophobic residue. The symbol x in the
consensus sequences of Tables 1-5 represents any amino acid.
- 64 -
CA 02595040 2007-08-03
) 97l30074 PGT/US97/0229-
For the Nck SH3 domain binding clones, a GST-SH3 fusion
protein containing the middle SH3 domain of Nck was used.
lQ
20
30
- 65 -
CA 02595040 2007-08-03
WO 97/30(}74 nCT/LJS97/02298
TABLE 1 CORTACTIN SH3-BINDING PEPTIDES
SEQ. ID NO.
PXXP.CORT.M1/2/3.PP SSLLGPPVPPKPQTLFSFSR 107
PXXP.CORT.M4.PP SRLGEFSKPPIPQKPTWMSR 108
PXXP.CORT.N2.PP SRTERPPLPQRPDWLSYSSR 109
PXXP.CORT.N3.PP.INC SREPDWLCPNCPLLLRSDSR 110
PXXP.CORT.01/2/3.PP SSSSHNSRPPLPEKPSWLSR 111
PXXP.CORT.04.PP SRLTPQSKPPLPPKPSAVSR 112
CONSENSUS KPP~PxKPxW 113
R
20
30
- 66 -
CA 02595040 2007-08-03
wO 97/30074 PCT1US97/02298
TABLE 2 NCK SH3-BINDING PEPTIDES
SEQ. ID NO.
= PXXP.NCK.Q1/4.PP SSLGVGWKPLPPMRTASLSR 114
PXXP.NCK.Q2/3.PP.INC SSVGFADRPRPPLRVESLSR 115
= PXXP.NCK.RI.PP.INC SSAGILRPPEKPXRSFSLSR 116
PXXP.NCK.R2.PP SSPYTGDVPIPPLRGASLSR 117
PXXP.NCK.R3.PP SSLMGSWPPVPPLRSDSLSR 118
PXXP.NCK.R4.PP SSIGEDTPPSPPTRRASLSR 119
PXXP.NCK.S1/4.PP SRSLSEVSPKPPIRSVSLSR 120
PXXP.NCK.S2.PP.INC SSVSEGYSPPLPPRSTSLSR 121
PXXP.NCK.S3.PP SSSFTLAAPTPPTRSLSLSR 122
PXXP.NCK.Ti.PP SSPPYELPPRPPNRTVSLSR 123
PXXP.NCK.T2.PP SRVVDGLAPPPPVRLSSLSR 124
FXXP.NCK.T3.PP.INC SSLGYSGAPVPPHRxSSLSR 125
PXXP.NCK.T4.PP SSISDYSRPPPPVRTLSLSR 126
CONSENSUS oxxxxxPxPPORSxSL 127
T
25
35
- 67 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/02298
TABLE 3 ABL SH3 BINDING PEPTIDES
IFF - SEQ. ID NO.
PXXP.ABL.G1/2.PP SRGPRWSPPPVPLPTSLDSR 128
PXXP.ABL.G3/4.PP SSPPDYAAPAIPSSLWVDSR 129
PXXP.ABL.H1/3/4.PP SSPPHWAPPAPPAMSPPISR 130
PXXP.ABL.H2.PP.INC SSDRCWECPPWPAGGQRGSR 131
PXXP.ABL.I1/2/3.PP SSPPKFSPPPPPYWQLHASR 132
PXXP.ABL.I4.PP SSPPSFAPPAAPPRHSFGSR 133
PXXP.ABL.JI.PP SSAPKKPAPPVPMMAHVMSR 134
PXXP.ABL.J2.PP.INC SSPTYPPPPPPDTAKGASR 135
PXXP.ABL.J3.PP.INC SSPPXXXPPPIPNSPQVLSR 136
PXXP.ABL.J4.PP SSPPTWTPPKPPGWGVVFSR 137
PXXP.ABL.LI.PP SSAPTWSPPALPNVAKYKSR 138
PXXP.ABL.L2/3.PP SSIKGPRFPVPPVPLNGVSR 139
PXXP.ABL.L4.PP SSPPAWSPPHRPVAFGSTSR 140
CONSENSUS PPxWxPPPOP 141
TABLE 4 PLCT SH3-BINDING PEPTIDES
SEQ. ID NO.
PXXP.PLCy.PI.PP SSNiKVHNFPLPPLPSYETSR 142
PXXP.PLCT.P2.PP SRVPPLVAPRPPSTLNSLSR 143
PXXP.PLCy.PE.PP.INC SSLYWQHGPDPPVGAPQLSR 144
PXXP.PLCy.P4.PP SSHPLNSWPGGPFRHNLSSR 145
- 68 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/02298
TABLE 5 SRC SH3-BINDING PEPTIDES
SEQ. ID NO.
PXXP.SRC.AI.PP SSRALRVRPLPPVPGTSLSR 146
PXXP.SRC.A2.PP SSFRALPLPPTPDNPFAGSR 147
PXXP.SRC.A3.PP SRDAPGSLPFRPLPPVPTSR 148
PXXP.SRC.A4.PP SSISQRALPPLPLMSDPASR 149
PXXP.SRC.Bi.PP SSPAYRPLPRLPDLSVIYSR 150
pXXP.SRC.B2/3/PP SSFINRRLPALPPDNSLLSR 151
PXXP.SRC.B4.PP SRLTGRPLPALPPPFSDFSR 152
PXXP.SRC.CI.PP SRMKDRVLPPIPTVESAVSR 153
PXXP.SRC.C2.PP.INC SSLYSAIAPDPPPRNSSSSR 1'54
PXXP.SRC.C3.PP SSLASRPLPLLPNSAPGQSR 155
PXXP.SRC.Dl.PP SSLTSRPLPDIPVRPSKSSR 156
PXXP.SRC.D2.PP.INC SSLKWRALPPLPETDTPYSk 157
PXXF.SRC.D3.PP SSNTNRLPPPTPllGLDVRSR 178
PXXP.SRC.D4.PP SSLQSRPLPLPPQSSYPISR 159
CONSENSUS RPLPPLP 9
In addition to the consensus sequence shown in Table 5,
the amino acid sequences of the inserts from the Src SH3 '
domain-binding phage isolated from the PXXP (SEQ ID NO:161)
biased peptide library described in Section 6.14.1 also give
rise to the consensus sequence LXXRPLPXViP (SEQ ID NO:165), as
shown in Table 6, below. In the consensus sequence
LXXRPLPXOP (SEQ ID NO:165), 0 represents aliphatic amino acid
residues (A, V, L, I, P); X represents any amino acid.
- 69 -
CA 02595040 2007-08-03
WO 97/30074 PCT/-i1S97/02298
TABLE 6
Src SH3 Binding Peptides
LASRPLPLLPNSAPGQ (a portion of SEQ ID NO:155)
LTGRPLPALPPPFSDF (a portion of SEQ ID NO:152)
PAYRPLPRLPDLSVIY (a portion of SEQ ID NO:150)
RALRVRPLPPVPGTSL (a portion of SEQ ID NO:146)
DAPGSLPFRPLPPVPT (a portion of SEQ ID NO:148)
LKWRALPPLPETDTPY (a portion of SEQ ID NO:157)
ISQRALPPLPLMSDPA (a portion of SEQ ID NO:149)
LTSRPLPDIPVRPSKS (a portion of SEQ ID N0:156)
NTNRPLPPTPDGLDVR (a portion of SEQ ID NO:158)
MKDRVLPPIPTVESAV (a portion of SEQ ID NO:153)
LQSRPLPLPPQSSYPI (a portion of SEQ ID NO:159)
FINRRLPALPPDNSLL (a portion of SEQ ID NO:151)
FRALPT.,PPTPDNPFAG (a portion of SEQ ID NO:147)
LYSAIAPDPPPRNSSS= (a portion of SEQ ID NO:154)
LSXRPLP$OP = CONSENSUS (SEQ. ID NO:165)
In Table 6, ik represents aliphatic amino acid residues
(A, V, L, I, P); X represents any aminc acid; = putative
class II peptide (see Section 6.14.5). Invariant proline
residues are underlined.
Another consensus sequence that can be derived from the
amino acid sequences of the inserts from the Src SH3 domain-
binding phage is:
LX1XZRPLPX30PX4X5 (SEQ ID NO: 454)
where 0 represents aliphatic amino acid residues (A, V,
L, I, P) and Xõ X2 , X3 , X4, and X5 represent any amino acid;
except that if
X3 = P, ~= L, X4 = P, and XS = P, then:
where X1 = F, then X2 is not H or R; or
where X. = S, then XZ is not R, H, A, N, T, G, V, M, or
W; or
where X1 = C, then X2 is not S or G; or
- 70 -
CA 02595040 2007-08-03
..O 97130074 PCT/US97/02298
where X, = R, then X2 is not T or F; or
where X1 = A, then X2 is not R, Q, N, S, or L; or
where X1 = Q, then X2 is not M; or
. where X1 = L, then X2 is not R; or
where X1 = I, then X2 is not A; or
where X1 = P, then X2 is not P, W, or R; or
where X1 = G, then X2 is not S or R; or
where X, = T, then X2 is not T.
In addition to the consensus sequence shown in Table 1,
the amino acid sequences of the inserts from the cortactin
SH3 domain-binding phage isolated from the PXXP (SEQ ID
NO:'.61) biased peptide library described in Section 6.14.1
also give rise to the consensus sequence +PPOPXKPXWL (SEQ ID
NO:166), as shown in Table 7, below.
30
- 71 -
CA 02595040 2007-08-03
JVO 97/30074 PCT/US97/0*4..4
TABLE 7
cortactin SH3 Binding Peptides
LTPQSKPPLPPKPSAV (a portion of SEQ ID NO:112)
SSHNSRPPLPEKPSWL (a portion of SEQ ID NO:111)
PVKPPLPAKPWWLPPL (SEQ ID NO:167)
TERPPLPQRPDWLSYS (a portion of SEQ ID NO:109)
LGEFSKPPIPQKPTWM (a portion of SEQ ID NO:108)
YPQFRPPVPPKPSLMQ (SEQ ID NO:168)
VTRPPLPPKPGHMADF (SEQ ID NO:169)
VSLGLKPPVPPKPMQL (SEQ ID NO:170)
LLGPPVPPKPQTLFSF (a portion of SEQ ID NO:107)
YKPEVPARPIWLSEL (SEQ ID NO:171)
GAGAARPLVPKKPLFL (SEQ ID NO:172)
+PPOP%KP%WL = CONSENSUS (SEQ ID NO:166)
In Table 7, + represents-basic amino acid residues (R,
K); ~ represents aliphatic amino acid residues (A, V, L, 1,
P); X represents any amino acid. Invariant proline residues
are underlined.
In addition to the consensus sequence shown in Table 3,
the amino acid sequences of the inserts from the Abl SH3
doma.in-binding phage isolated from the PXXP (SEQ ID NO:161)
biased peptide library described in Section 6.14.1 also give
rise to the consensus sequence PPXBXPPP~P (SEQ ID NO:173), as
shown in Table 8, below.
35
- 72 -
CA 02595040 2007-08-03
WO 97130074 PCT/US97/02298
TABLE 8
Abl SH3 Binding Peptides
PPWWAPPPIPNSPQVL (SEQ..ID NO:174)
PPKFSPPPPPYWQLHA (a portion of SEQ ID NO:132)
PPHWAPPAPPAMSPPI (a portion of SEQ ID NO:130)
PPTWTPPKPPGWGVVF (a portion of SEQ ID NO:137)
PPSFAPPAAPPRHSFG (a portion of SEQ ID NO:133)
PTYPPPPPPDTAKGA# (a portion of SEQ ID NO:135)
GPRWSPPPVPLPTSLD (a portion of SEQ ID NO:128)
APTWSPPALPNVAKYK (a portion of SEQ ID NO:138)
PPDYAAPAIPSSLWVD (a portion of SEQ ID NO:129)
IKGPRFPVPPVPLNGV (a portion of SEQ ID NO:139)
PPAWSPPHRPVAFGST (a portion of SEQ ID NO:140)
APKKPAPPVPbiMAHVM (a portion of SEQ ID NO:134)
PP89XPPPOP = CONSENSUS (SEQ ID NO:173)
In Table 8, B represents aromatic amino acid residues; 2o represents
alip:hatic amino acid residues (A, V, L, I, P); X
represents any amino acid. Invariant proline residues are
underlined.
# This clone contained a three nucleotide deletion in the
random peptide coding sequence.
The amino acid sequences of the inserts from the PLCy
SH3 domain-binding phage isolated from the PXXP (SEQ ID
NO:161) biased peptide library described in Section 6.14.1
give rise to the consensus sequence PPVPPRPXXTL (SEQ ID
NO:175), as shown in Table 9, below.
- 73 -
CA 02595040 2007-08-03
WO 97l30074 PCT/US97l02298
TABLE 9
PLC7 SH3 Binding Peptides
MPPPVPPRPPGTLQVA (SEQ ID NO:176)
LSYSPPPVPPRPDSTL (SEQ ID NO:177)
VLAPPVPPRPGNTFFT (SEQ ID NO:178)
YRPPVAPRPPSSLSVD (SEQ ID NO:179)
LQCPDCPRVPPRPIPI (SEQ ID NO:180)
VPPLVAPRPPSTLNSL (a portion of SEQ ID NO:143)
LTPPPFPKRPRWTLPE (SEQ ID NO:181)
YWPHRPPLAPPQTTLG (SEQ ID NO:182)
PPVPPRPXXTL = CONSENSUS (SEQ ID NO:175)
In Table 9, the symbol X represents any amino acid.
invariant proline residues are underlined.
The PXXP (SEQ ID NO:161) biased peptide librarv
described in Section 6.14.1 was also used to obtain phagE:
clones that specifically bound the SH2 domain from the p53bp2
protein. The amino acid sequences of the peptides expressed
by the p53bp2 SH3 domain-binding phage are shown in Table 10
below.
35
- 74 -
CA 02595040 2007-08-03
.dO 97130074 PCT/US97/022>o
TABLE 10
p53bp2 SH3 Binding Peptides
YDASSAPQRPPLPVRKSRP (SEQ ID NO:183)
EYVNASPERPPIPGRKSRP (SEQ ID NO:184)
WNGIAIPGRPEIPPRASRP (SEQ ID NO:185)
SMIFIYPERPSPPPRFSRP (SEQ ID NO:186)
GVEEWNPERPQIPLRLSRP (SEQ ID NO:187)
WVVDSRPDIPLRRSLP (SEQ ID NO:188)
VVPLGRPEIPLRKSLP (SEQ ID NO:189)
GGTVGRPPIPERKSVD (SEQ ID NO:190)
YSHAGRPEVPPRQSKP (SEQ ID NO:191)
FSAAARPDIPSRASTP (SEQ ID NO:192)
LYIPKRPEVPPRRHEA (SEQ ID NO:193)
NNISARPPLPSRQNPP (SEQ ID NO:194)
MAGTPRPAVPQRMNPP (SEQ ID NO:195)
RPXOPiGR+SXP = CONSENSUS (SEQ ID NO : 19 6)
In Table 10, + represents basic amino acid residues (R,
K); 0 represents aliphatic amino acid residues (A, V, L, I,
P); X represents any amino acid. Znvariant proline or
flanking residues are underlined.
The PXXP (SEQ ID NO:161) biased peptide library
described in Section 6.14.1 was also used to obtain phage
clones that specifically bound the SH3 domain from the N
terminal portion of the Crk protein. The amino acid
sequences of the peptides expressed by the Crk N terminal SH3
domain-binding phage are shown in Table 11 below.
- 75 -
CA 02595040 2007-08-03
,JVO 97/30074 PCT/US97/02~98
TABLE 11
Crk N BH3 Binding Peptides
GQPAGDPDPPPLPAKF .(SEQ ID NO:197)
FEQTGVPLLPPKSFKY (SEQ ID NO:198)
IFGDPPPPIPMKGRSL (SEQ ID NO:199)
SNQGSIPVLPIKRVQY (SEQ ID NO:200)
NYVNALPPGPPLPAKN (SEQ ID NO:201)
SSDPERPVLPPKLWSV (SEQ ID NO:202)
HFGPSKPPLPIKTRIT (SEQ ID NO:203)
DWKVPEPPVPKLPLKQ (SEQ ID NO:204)
ATSEGLPILPSKVGSY (SEQ ID NO:205)
NANVSAPRAPAFPVKT (SEQ ID NO:206)
EMVLGPPVPPKRGTVV (SEQ ID NO:207)
AGSRHPPTLPPKESGG (SEQ ID NO:208)
SVAADPPRLPAKSRPQ (SEQ ID NO:209)
*P*LPV,K = CONSENSUS (SEQ ID NO = 210 )
In Table 11, ~ represents aliphatic amino acid residues
(A, V, L, I, P). Invariant proline residues are underlined.
The present invention provides a purified peptide that
binds to the SH3 domain of Crk, the purified peptide
comprising the amino acid sequence OPV/LPOK (SEQ ID NO:210),
where 0 represents aliphatic amino acid residues (A, V, L, I,
P),-with the proviso that the peptide does not comprise the
amino acid sequence WNERQPAPALPPKPPKPT (SEQ ID NO:456).
The PXXP (SEQ ID NO:161) biased peptide. library
described in Section 6.14.1 was also used to obtain phage
clones that specifically bound the SH3 domain from the Yes
protein. The amino acid sequences of the peptides expressed
by the Yes SH3 domain-binding phage are shown in.Table 12
below.
~-76 -
CA 02595040 2007-08-03
J 97/30074 PCT/US97/0225,.
TASLE 12
Yes SH3 Binding Peptides
ITMRPLPALPGHGQIH (SEQ ID NO:211)
LPRRPLPDLPMA.AGKG (SEQ ID NO:212)
LGSRPLPPTPRQWPEV (SEQ ID NO:213)
STIRPLPAIPRDTLLT (SEQ ID NO:214)
RSGRPLPPIPEVGHNV (SEQ ID NO:215)
IGSRPLPWTPDDLGSA (SEQ ID NO:216)
LAQRELPGLPAGAGVS (SEQ ID NO:217)
IPGRALPELPPQRALP (SEQ ID NO:218)
FVGRELPPTPRTVIPW (SEQ ID NO:219)
DPRSALPALPLTPLQT (SEQ ID NO:220)
SPHDVLPALPDSHSKS (SEQ ID NO:221)
tJrBXRPLPBLP = CONSENSUS (SEQ ID NO:222)
In Table 12, 0 represents aliphatic amino acid residues
(A, V, L, I, P); X represents any amino acid. Invariant
proline residues are underlined.
:inother consensus sequence that can be derived from the
amino acid sequences of the inserts from the Yes SH3 domain-
binding pha.ge is:
OX1XZRPLPX3LPX4X5 (SEQ ID NO : 4 55 )
where 0 represents aliphatic amino acid residues (A, V,
L, I, P) and Xl, X2, X3, X4, and XS represent any amino acid;
except that if
X3 = P, X4 = P, and X. = P, then :
when = L,
where X. = F, then X. is not H or R; or
where X1 = S, then XZ is not R, H, A, N, T, G, V, M, or
W; or
where X; = C, then XZ is not S or G; or
where X1 = R, then X2 is not T or F; or
where X1 = A, then X2 is not R, Q, N, S, or L; or
where X1 = Q, then X2 is not M; or
where X1 = L, then X2 is not R; or
- 77 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/0,4z98
where X1 = I, then X2 is not A; or
where X, = P, then X2 is not P, W, or R; or
where X1 = G, then X2 is not S or R; or
where X, = T, then X2 is not T; and
when = P,
where X, = A, then X2 is not R; or
where X1 = S, then X2 is not R or Y; or
where X1 = M, then X2 is not S; or
where X1 = V, then X2 is not G; or
where X, = R, then X2 is not S; or
where X1 = I, then X2 is not R; and
when = A,
where Xy = A, then X2 is not K; and
when V,
where Xl = A, then X2 is not C or Q; or
where X1 = P, then X2 is not P; and
when ~ = I,
where X1 = G, then X2 is not H.; or
where X; = T, then X, is not S; o.r.
where X1 = R, then X7 is not S.
The present invention alao provides a purified peptide
that binds to the SH3 domain of Yes, the purified peptide
comprising the amino acid sequence V'X1X2RPLPX3LPX4XS (SEQ ID
NO:455), where 0 represents aliphatic amino acid residues (A,
V, L, I, P) and Xl , XZ , X3 , X4, and X5 represent any amino
acid, with the proviso that the peptide does not comprise the
amino acid sequence AGDRPLPPLPYNPKS (SEQ ID NO:457).
The PXXP (SEQ ID N0:161) biased peptide library,
described in Section 6.14.1 was also used to obtain phage
clones that specifically bound the SH3 domain from the N
terminal portion of the Grb2 protein. The amino acid
sequences of the peptides expressed by the Grb2 N terminal
SH3 domain-binding phage are shown in Table 13 below. These
sequences can be arranged into three groups of sequences that
have different, but related, consensus sequences. An overall
- 78 -
CA 02595040 2007-08-03
w U 97/30074 PCTIUS97/02298
consensus sequence, +6DXPLPXLP (SEQ ID NO:223), can be
derived for the three groups.
10
20
30
- 79 -
CA 02595040 2007-08-03
WO 97130074 PCTYUS97l02298
TABLE 13
Grb2 N SH3 Binding Peptides
KWDSLLPALPPAFTVE (SEQ ID NO:224)
RWDQVLPELPTSKGQI (SEQ ID NO:225)
RFDFPLPTHPNLQKAH (SEQ ID NO:226)
RLDSPLPALPPTVMQN (SEQ ID NO:227)
RWGAPLPPLPEYSWST (SEQ ID NO:228)
YWDMPLPRLPGEEPSL (SEQ ID NO:229)
RFDYNLPDVPLSLGTA (SEQ ID NO:230)
TKKPNAPLPPLPAYMG (SEQ ID NO:231)
KWDLDLPPEPMSLGNY (SEQ ID NO:232)
+ODXPLPXLP = CONSENSUS (SEQ ID NO:223)
YYQRPLPPLPLSHFES (SEQ ID NO:234)
YYRKPLPNLPRGQTDD (SEQ ID NO:235)
YFDKPLPESPGALMSL (SEQ ID NO:236)
YFSRALPGLPERQEAH (SEQ ID NO:2371)
YOX+PLPXLP = CONSENSUS (SEQ ID NO:238)
SLWDPLPPIPQSKTSV (SEQ ID NO:239)
SYYDPLPKLPDPGDLG (SEQ ID NO:240)
KLYYPLPPVPFKDTKH (SEQ ID NO:241)
DPYDAL=PETPSMKASQ (SEQ ID NO:242)
ODPLPXLP = CONSENSUS (SEQ ID NO:243)
+BDXPLPXLP = OVERALL CONSENSUS (SEQ ID NO:223)
In Table 13, + represents basic amino acid residues (R,
K); B represents aromatic amino acid residues; X represents
any amino acid. Invariant proline residues are underlined.
- 80 -
CA 02595040 2007-08-03
97/30074 PCT/US97/0229b
6.14.5. 8H3 Ligand Binding Orientation
Peptide ligands bound to SH3 domains have been shown to
assume a left-handed polyproline type II (PPII) helix
conformation (Yu, H., Chen, J. K., Feng, S., Dalgarno, D. C.,
Brauer, A. W., & Schreiber, S. L. (1994) Cell 76, 933-45).
SH3 ligands are pseudo-symmetrical and may therefore bind in
one of two opposite orientations (Feng, S., Chen, J. K., Yu,
H., Simon, J. A., & Schreiber, S. L. (1994) Science 266,
1241-7) (Feng et al.). Feng et al., supra, have demonstrated
that peptides that bind in one or the other orientation share
different consensus motifs. Specifically, ligands that bind
in the Class I or Class II orientation conform to the
consensus +pYPpYP (SEQ ID NO:244) or YPpYPp+ (SEQ ID NO:245)
respectively, where uppercase positions represent conserved
residues that contact the SH3 domain and confer specificity,
and lowercase positions represent scaffolding residues that
tend to be proline.
According to this model, we predict that the peptides
selected by the Src, Yes, Abl, and Grr2 N SH3 domains bind in
the Class I orientation, whereas the peptides selected by the
Cortactin, p53bp2, PLCT, and.Crk N SH3 domains bind in the
Class II orientation (see Table 14). Interestingly, most of
the SH3 ligand consensus motifs identified in this work
contain additional conserved residues flanking the SH3-
binding core defined by Feng et al., supra. Furthermore,
these conserved residues are situated N- and C-terminal of
.the SH3-binding core in Class I and C1ass.II motifs,
respectively, and are therefore predicted to interact with
equivalent regions of their target SH3 domains (see Table
14).
- 81 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US9710.Lt98
TABLE 14
Alignment of SH3 ligand consensus motifs
SEQ ID NO:
Class I +DoPpoP 244
Src LXXRPLPXOP 165
Yes ~XXRPLPXLP 222
Abl PPXBXPPPOP 173
Grb2 N +ODXPLPXLP 223
YBXRPLPXLP 246
BDPLPXLP 243
Class II ViPpV/PD+ 245
Cortactin +PP~PXRPXWL 166
p53bp2 RPX~POR+SXP 196
PLC-y XPPVPPRPXXTL 247
Crk N V/POLPOR 210
In Table 14, each SH3 ligand consensus motif was
assigned to class I or II based on its agreement with the
class I or II consensus motif. Highly (>90%) conserved
pesitions in each SH3 ligand consensus motif are listed in
boldface and were interpreted as GH3 contact residues.
+ represents basic amino acid residues (K, R); ~ represents
aliphatic amino acid residues (A, V, L, I, P); 6 represents
aromatic amino acid residues; X represents any amino acid;
lower case p represents residues that tend to be proline.
The Src SH3 domain is capable of binding both Class I
and Class II peptides'Feng et al., supra. Although Class I
peptides predominate in the population of Src SH3 ligands
selected from the PXXP (SEQ ID NO:161) library, one clone
conforms well to the Class II consensus (see Table 6).
Previously, Sparks, A. B., Quilliam, L. A., Thorn, J. M.,
Der, C. J., & Kay, B. K. (1994) J. Biol. Chem. 269, 23853-6
and Yu, H., Chen, J. K., Feng, S., Dalgarno, D. C., Brauer,
A. W., & Schreiber, S. L. (1994) Cell 76, 933-45 had isolated
Class II Src SH3 ligands sharing the consensus PP~PPR (SEQ ID
NO:248). Similarly, whereas the Grb2 N SH3 domain has been
- 82 -
CA 02595040 2007-08-03
.197/30074 PCT/US97/02298
shown to bind peptides from SOS with the Class II consensus
sequence PPOPPR (SEQ ID NO:248) (Rozakis-Adcock, M., Fernley,
R., Wade, J., Pawson, T., & Bowtell, D. (1993) Nature 363,
= 83-5), we have isolated Grb2 N SH3 ligands that conform to
the Class I consensus (see Table 14). Thus, both the Src and
the Grb2 N SH3 domains apparently have the capacity to bind
both Class I and Class II peptide ligands.
6.14.6. SH3 Ligand Binding Characteristics
To explore further the capacity of SH3 domains.to
discriminate between different SH3 ligands, we investigated
the binding of phage expressing various peptide ligands to a
pane'i of SH3 domains. Equal titer.; of clonal phage stocks
were incubated in microtiter wells coated with different GST-
SH3 fusion proteins. The wells were washed several times,
and bound phage were detected with an anti-phage antibody
(see Fig. 14). Positive ELISA signals were equivalent to
those obtained with previously characterized Src SH3-binding
clones (Sparks, A. B., Quilliam, L. A., Thorn, J. M., Der, C.
J., & Kay, B. K. (1994) J. Biol. Chem. 269, 23853-6) and are
indicative of SH3:peptide affinities in the 5 to 75 M range
(Yu, H., Chen, J. K., Feng, S., Dalgarno, D. C., Brauer, A.
W., & Schreiber, S. L. (1994) Cell 76, 933-945; Rickles, R.
J., Botfield, M. C., Weng, Z., Taylor, J. A., Green, O. M.,
Brugge, J. S., & Zoller, M. J. (1994) EMBO J. 13, 5598-604).
Whereas the Src, Yes, Crk, and Grb2 N SH3 domains cross-
reacted with a few phage clones selected with other SH3
domains, the Abl, Cortactin, p53bp2, and PLCy SH3 domains
displayed considerable specificity. Significantly, only 33
of 220 potential instances of cross-reactivity were observed,
suggesting that SH3 selectivity is the rule rather than the
exception.
Each instance of cross-reactivity may be explained by
similarities between the sequences of the peptides and the
ligand preferences of the cross-reactive SH3 domains. For
example, Crk SH3 cross-reacted with three phage clones
selected with other SH3 domains; each of these clones
- 83 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/0,Lt98
coincidentally expressed peptides conforming to the Crk SH3
preferred ligand consensus motif. Similarly, the cross-
reactivity observed between the Src, Yes, and Grb2 SH3
domains and clones selected by other SH3 domains within this
group may be a consequence of the fact that these SH3 domains
prefer the same proline-rich core. Finally, the Src and Yes
SH3 domains cross-reacted with the PLCT SH3 ligand
MPPPVPPRPPGTL (a portion of SEQ ID NO:176), which contains
the Class II Src SH3-binding sequence PPVPPR (SEQ ID NO:249).
Taken together, these data demonstrate the capacity of SH3
domains to discern subtle differences in the primary
structure of potential ligands.
6.15. Use of Consensus Sequences to Determine
the Amino Acid Sequences Responsible for
i5 Binding in Proteins that are Known to
Bind SH3 Domains
There are many proteins that are known tc, bind Sh3
domains but for which the specific sequences of those
proteins that are responsible for binding to SH3 domains are
2 0 not known. The consensus sequences shown above in Tables 1-
13 can be used to search databases (e.g., GenBank) containing
the amino acid sequences of those proteins in order to
determine which sequences are responsible for the binding of
those proteins to SH3 domains. This was done for a number of
25 known SH3 domain binding proteins and sequences resembling
the consensus sequences of Tables 1-13 were identified. The
results are shown in Table 15. For comparison, also shown in
Table 15 are the amino acid sequences that had previously
been demonstrated to be responsible for SH3 domain binding
30 for a number of proteins.
- 84 -
CA 02595040 2007-08-03
WO 97130074 PCT/US97/02298
TABLE 15
= SEQ ID NO: Reference
Brc BH3 Class I LBBRPLPXOP 165
. Hs AFAP-110 (62-73) PPQMPLPEIPQQ 250 1
(76-87) PPDNGPPPLPTS 251 1
Hs CDC42 GAP (250-261) TAPKPMPPRPPL 252 2
Hs hnRNP K (302-313)*SRARNLPLPPPP 253 3
Mm p62 (328-339) TVTRGVPPPPTV 254 3
Hs PI3K p85 (90-101)* RPPRPLPVAPGS 255 9
Hs Shc p52 (296-307) VRKQMLPPPPCP 256 3
Src 8H3 Class II PP}GPPR 248
Hs Dynamin (810-820) GGAPPVPSRPG 257 6
(827-837), GPPPQVPSRPN 258 6
(838-848) RAPPGVPSRSG 259 6
Hs hnRNP K (308-318)* PLPPPPPPRGG 260 3
virn p62 (294-304) APPPPPVPRGR 261 3
Hs Paxillin (42-52) AVPPP17PPPPS 262 10
Hs P13K p85 (302-312)* QPAPALPPKPP 263 9
HS Shb (50-60) GGPPPGPGRRG 264 11
(103-113) TKSPPQPPRPD 265 11
Yes BH3 ~BBRPIP%LP 222
Hs Yap65- (240-251)_PVKQPPPLAPQS 266 4
Ab1 8H3 PPXBBPPPOP 173
Mm 3BP-1 (265-276)*RAPTMPPPLPPV 267 12
Mm 3BP-2 (200-211)*YPPAYPPPPVPV 268 12
Dm Ena (350-361) PGPGYGPPPVPP 269 5
PLCT SIi3 PPVPPRPXXTL 175
Hs Dynamin (812-823) APPVPSRPGASP 270 6
(829-840) PPQVPSRPNRNR 271 6
- 85 -
CA 02595040 2007-08-03
WO 97130074 PCT/US97/02298
SEQ ID NO: Reference
Hs c-Cbl (493-504) LPPVPPRLDLLP 272 7
Crk N 8H3 POLPOK = 210
Hs Abl (524-533)*QAPELPTKTR 273 13
(568-577)*VSPLLPRKER 274 13
(758-767) EKPALPRKRA 275 13
Hs C3G (282-291)*PPPALPPKKR 276 14
(452-461)*TPPALPEKKR 277 14
(539-548)*KPPPLPEKKN 278 14
(607-616)*PPPALPPKQR 279 14
Grb2 N SH3 Class I +ODXPLPXLP 233
YBX+PLPXLP 238
BDPLPXLF 243
Hs c-Cbl (560-571) PQRFPLPCTPGD 280 P
(589-600) W,T.,PRPIPKVPVS 2~31 8
Grb2 N SH3 Class II PPP4,PPR 282
Hs Abl (523-533)* LQAPELPTKTR 283 13
(567-577)* AVSPLLPRKER 284 13
(609-619)* KTAPTPPKRSS 285 13
Hs c-Cbl (491-501) ASLPPVPPRLD 286 8
Hs Dynamin(810-820) GGAPPVPSRPG 287 6
(827-837) GPPPQVPSRPN 288 6
(838-848) RAPPGVPSRSG 289 6
Hs SOSl (1148-1158)* PVPPPVPPRRR 290 15
(1177-1187) DSPPAIPPRQP 291 15
(1209-1219)* ESPPLLPPREP 292 15
(1287-1297)* IAGPPVPPRQS 293 15
Rn Synapsin'I(592-602) NLPEPAPPRPS 294 16
(670-680) PPGPAGPIRQA 295 16
-- 86 -
CA 02595040 2007-08-03
~. v 97130074 PCT/US97/02298
In Table 15, + represents basic amino acid residues (R,
K); ~ represents aliphatic amino acid residues (A, V, L, I,
P); B represents aromatic amino acid residues; X represents
any amino acid. * represents amino acid sequences previously
demonstrated to bind their respective SH3 domains. Residues
within the sequences that agree with the most highly
conserved residues of the consensus motifs are shown in bold.
Each entry shows an abbreviation of the name of the SH3
domain binding protein and the species from which it was
derived. The amino acid positions in the mature proteins of
the sequences shown are indicated in parentheses. For more
details, see the reference listed for each protein.
Reference 1 is Flynn, D. C., Leu, T. H., Reynolds, A.
B., & Parsons, J. T. (1993) Mol Ce11 Biol 13, 7892-7900.
Reference 2 is Barfod, E. T., Zheng, Y., Kuang, W, J.,
Hart, M. J., Evans, T., Cerione, R. A., & Ashkenazi, A.
(1993) J Biol Chem 268, 26059-62.
Reference 3 is Weng, Z., Thomas, S. M., Rickles, R. J.,
Taylor, J. A.,. Brauer, A. W., Seidel.-Dugan, C., Michael, W.
M., Dreyfuss, G:, & Brugge, J. S. (1994) Mol Cell Biol 14r
4509-21.
Reference 4 is Sudol, M. (1994) Oricogene 9, 2145-52.
Reference 5 is Gertler, F. B., Comer, A. R., Juang, J.
L., Ahern, S. M., Clark, M. J., Liebi, E. C., & Hoffmann, F.
M. (1995) Genes Dev 9, 521-33.
Reference 6 is Gout, I., Dhand, R., Hiles, I. D., Fry,
M. J., Panayotou, G., Das, P., Truong, 0., Totty, N. F.,
Hsuan, J., Booker, G, W. & et al. (1993) Cell 75, 25-36.
Reference 7 is Rivero-Lezcano,=O. M., Sameshima, J. H.,
Marcilla, A., & Robbins, K. C. (1994) J Biol Chem 269, 17363-
6.
Reference 8 is Odai, H., Sasaki, K., Iwamatsu, A.,
Hanazono, Y., Tanaka, T., Mitani, K., Yazaki, Y.-& Hirai, H.
(1995) J Biol Chem 270, 10800-5.
Reference 9 is Kapeller, R., Prasad, K. V., Janssen, 0.,
Hou, W., Schaffhausen, B. S., Rudd, C. E., & Cantley, L. C.
(1994) J Biol Chem 269, 1927-33.
- 87 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/02298
Reference 10-is Weng, Z., Taylor, J. A., Turner, C. E.,
Brugge, J. S., & Seidel-Dugan, C. (1993) J Bio1 Chem 268,
14956-63.
Reference 11 is Karlsson, T., Songyang, Z., Landgren,
E., Lavergne, C., Di-Fiore, P. P., Anafi, M., Pawson, T.,
Cantley, L. C., Claesson-Welsh, L., & Welsh, M. (1995)
Oncogene 10, 1475-83.
Reference 12 is Ren, R., Mayer, B. J., Cicchetti, P., &
Baltimore, D. (1993) Science 259, 1157-61.
Reference 13 is Ren, R., Ye, Z. S., & Baltimore, D.
(1994) Genes Dev 8, 783-95.
Reference 14 is Knudsen, B. S., Feller, S. M., &
Hanafusa, H. (1994) J Biol Chem 269, 32781-7.
Reference 15 is Rozakis-Adcock, M., Fernley, R., Wade,
J., Pawson, T., & Bowtell, D. (1993) Nature 363, 83-5.
Reference 16 is McPherson, P, S., Czernik, A, J.,
Chilcote, T, J., Onofri, F., Benfenati, F., Greengard, P.,
Schlessi.nger, J., & De-Camilli, P. (1994) Proc Nat1 Acad Sci
USA 91, 6486-90.
The sequences shown in Table 15 are useful in that they
can be used as ligands in the assays for the identification
of compounds that affect binding of SH3 domain-containing
proteins and their ligands that is described above in Section
5.6.
6.16. Use of Consensus Sequences to Identify
Amino Acid Sequences Resembling SH3
Domain-binding Sequences in Proteins that
are Not Known to Bind SH3 Domains
The consensus sequences shown above in Tables 1-13 can
be used to search databases (e.g., GenBank) containing the
amino acid sequences of proteins that are not known to bind
to SH3 domains. In this way, a large number of proteins not
previously suspected of containing amino acid sequences that
bind SH3 domains have been shown to contain such sequences.
The portions of the amino acid sequences of these proteins
that resemble one or more of the consensus motifs of Tables
1-13 are shown below in Table 16. The SH3 domain-binding
- 88 -
CA 02595040 2007-08-03
wO 97/30074 PCT/US97/02298
sequences of the proteins shown in Table 16 can be used as
ligands in the assays for the identification of compounds
that affect binding of SH3 domain-containing proteins and
= their ligands that are described above in Section 5.6.
15
25
35
- 89 -
CA 02595040 2007-08-03
WO 97130074 PCT/US97I02298
TABLE 16
I,OCUS ACCESSION M'S DESCRIPTION SEQUENCE
ABL DROME P00522 TYROSINE-PROTEIN KINASE DRO 132 146 LLQSRPLPHIPAGST C!96) 1 u
DASH/AB
1380 1395 QIQQKPAVPHKPPLND (297) i 2
ABPI YEAST P15891 ACTIN BINDING PROTEIN SAC 514 528 SSAAPPPPPRRATPE (298) 1
ACES HUMAN P22303 IACETYLCHOLINESTERASE HOM 73 E7 MGPRRFLPPEPKQPW (299) 2
PRECURSOR
ACM4 HUMAN P0E173 I MUSCARINIC HOM 276 290 PPPALPPPPRPVADK p00) 3 2
ACETYLCHOLINE RECEPi
ACRO HUMAN P10323 1ACROSIN PRECURSOR (EC HOM 329 343 QPPPRPLPPRPPAAQ (301) 1 2
1
3.4.21.10
1O RAT 642 656 PNLRRGLPQVPYFSL (302) 2
AGIE RAT Q00900 AGIE-BPI (A DNA-BINDING PROTEIN
ANUR HUMA.N PI0275 ANDROGEN RECEPTOR HOM 368 385 ALAGPPPPPPPPHPHARI (303)
AOFB HUMAN P2733E AMINE OXIDASE (FLAVIN- HOM 480 494 TFLERHLPSVPGLLR (304) 2
CONTAINING)
AP2 HUMAN P05549 TRANSCRIPlION FACTOR HOM 52 68 DFQPPYFPPPPYQPIYPQ (305) 2
_ AP2
ATF3 HUMAN P18847 CYCLIC-AMP-DEPENDENT HOM 57 'P1 CFCHRPLPVPPGSLV (306) I
' ITRANSCRIPT
BIAR HUMAN POB588 IBETA=1 ADRENERGIC HOM 2E2 296 APAPPPGPPRPAAAA f3071 0 0
1 5 - RECEPTOR.
83AR HUMAN PIKdS IBE7A=3 ADRF.NERGIC HUM 361 375 CRCGRRLPPEPCAM (308) 2
RECEPrOR.
I APOPiOSIS REGULATOR rõAL 33 47 GF-DRPPVPPAPAPAA (3091
LL:L2 CHlCX Q00709
BCL-2.
I8NI1 PROTEIN (SYNTHETIC
BMI YFA51 P41832 SAC 1242 12i6 PPPPPhPVPAKLFG4l310) ~ 4 0
LETHAL
CADM MGUtE P33146 IMUSCLE-CADHERIN (M- MUS 645 659 PQPHP,=/LPiSPSDIA (3111 3
CADHERINI
20 CM R PI:; P25117 CALCifONIN RECEPTOR SUS 14 26 IFLNRP:_PVLPDSAD (3.21 1
PRECURSOR
CB: HUMAN P22681 I PROTO.ONCOGENE C=CB)-. HOM 490 504 QASSLPPVPPRLDLLP (313) I
536 555 PPTLRDLPPPPPPDRPYSVG 2 2 2
(3W
559 S73 RPQRRPLPCTPGDCP pi3) 2
I
CCB3 RABIT QQ2343 BRAIN CALCIUM CHANNEL ORY 19 33 SDQGRNLPGTPVPAS (316) 3
' BII-1 PR
2 5 2100 2114 RHSRRQLPPVPPKPRPI.L (317) 1 1 1 0
CCAA RABR Q02344 BRAIN CALCIUM CHANNE:. ORY 19 33 SDQGRNLPGTPVPAS (!IB) 3
BIt 2 PRO
CG2A BOVIN P30274 02/MITOTIC=SPECIFIC BOS 56 70 NDEYVPVPPWKANNK (319) 5
CYCLIN A
CICI RAT P35324 CHLORIDE CHANNEL RAT 724 741 QTPiPPPPPPPPLPPQFP(320)
PROTEIN=SKELE ,
CBCS_HUMAN P22460 POTASSIUM CHANNEL HOM 60 74 DSGVRPLPPLPDPGV (32!) 0
PROTEIN KVI.5
71 8S DPGVRPLPPLPEELP(322) 0
CINC RAT P153E9 SODIUM CHANNEL PROTEINRAT 1723 1739 LNiGPPYCDPNLPNSNG Cl23) --
CARDIAO
CPl2 RAB(1 P00187 CYTOCHROME P450 IA2 (EC ORY 238 252 FPILRYLPNRPLQRF (324) 3
1-14.14
CP7S SOLME P37120 1 CYTOCHROME P450 LXXVA SpL 30 4( SWRRRKLPPGPEGWP (325) 2
(EC 1.14
CPC7 RAT F0S179 1CYTOCHROME P450 IIC7 (EC RAT 23 37 SSRRRKLPPGPTPLP (!26) 2
~ 1.14
3.5 CPCB HUMAN P10631 CYTOCHROME P450 IICB (EC HOM 23 37 SCRRRKLPPGFTPLP (327)
2
1.14.
CPCK_MACFA P33262 CYTDCHROME P450 I(C20 MAC 23 37 SSGRRKLPPGPTPLP (328) 2
(EC 1.14
- 90 -
CA 02595040 2007-08-03
v..! 97/30074 PCT/US97/02298
LOCUS ACCESSION /"S DESCRIPI70N SEQUENCE a
CPCM RAT P19225 CYTOCHROME P430 IIC22 RAT 23 37 HHVRRKLPPOPTPLP (329) 2
1 1 (EC 1. 14
CPT"1 MOUSE P27786 CYTOCHROME P450 XVIIAI MUS 25 39 AKFPRSLPFLPLVGS (330) 2
1 1 (P{SO-C
CR2_MOUSE P19070 COMPLEMENT RECEPPOR MUS 22 38 NARKPYYSLPIVPGTVL 031) 3
TYPE 2 PREG
CI'KI YbASI. QO1957 CTD KINASE ALPHA SUBUNR' (EC ? SAC 30 4 QSLARPPPPKRIRTD
(332) 3 1 3
CXA3_BOVIN P41987 GAPIUNCI'ION ALPHA-3 805 287 301 ASPARALPGPPHPRR (333) 2 3 3
PROTEIN
CYA3_RAT P21932 ADENYLATE CYCLASE. RAT 829 843 TDSRLPLVPSKYSMT (334) 4
OLFACTIVE TY
CYGR HUMAN Q02846 RETINAL GUANYLYL HOM 15 31 OLCGPAWWAPSLPRLPR (335) 3
CYCLASE PRECUR
CY(J_HUMAN P35663 CYLICIN (FRAGMENT). HOM 571 387 LCWCKMPPPPPKPRYAP (336; 2 3
2
CYR(; MOUSE P34902 CYTOKINE RECEPTOR MUS 283 298 WLERMPPIPP'KNLED (737) 5 2
COMMONGAMMA
DCD_HUMAN P20711 AROMATIC-L-AMINO-ACID HOM 31 47 PDVEPGYLRPLIPAAAP (338) 3
DECARBOXY
DMD dUMAN P115S DYSTROPH.N HOM 700 714 QEELPPPPPOKKRQI (339) I
DPOD BOV-N P28339 DNA POLYMERASE DELTA BOS 104 118 VAPARPLPGAPPPSQ (340) I
CATALYTIC
DRA_HUMAN P40879 DRA PROTEIN (DOWN- HOM 319 335 GDMNPGFOPPITPDVET (741) 3
REGULATED IN
DY15 DROME P13496 tSO KD DYNEIN-ASSUCIATED DRO 1250 1261 ARSARRLPSWPPl'LD
(342) 3
' POLYPE
DYNt HUMAN Q05193 DYNAMM-1. HOM 809 823 LGGAPPVPSR?GASP (343) 1 1
E7iC DROME P13055 ECDYSONE-IHDLCIBLc DRO 39B 413 VMRPPPPPPPFKVKr(A (34t) 3 3 1
PROTEIN E7S-
5117 601 MRH=GF.GLPS"P(:H'SS (yt5) 3
EGR2 HUMAN P11161 EARLY GR')WTii RESK)N.SE 140M :13 t27 HL'i37PF. PPPFYS r.a.
/3~61
- PROTeiN
ELKI Mo(!iE P419t9 PROTe'IN E! K-1 (FRAGMENT) MUS 1" 178 PQPQPP)PPRPASVL (30=
1
ENL HUMAN Q03111 ENI. PROTEIN. HOM 272 286 PPPPPPPPPi:ASSKR 0+8: 1 2
452 4E7 LPSREPPPPQKPPPN (Y!9) 2
EPI5 HUMAN PCA66 EPIDERMAL GROWTH HOM 763 T18 KSEDE'PPALPPKIGTP 050) 3 0
- FACTOR RECEtTOR
ERB3 HIIMAN P21860 ERBB-3 RECEPTOR PROTEIN- HOM 1204 1218 RRHSPPHPPRPSSLE
(751) 4 2 1
- TYROSIN
EZRI HUMAN P1911 P23714 ~~ (P81) (CY7'OVILLIN) ~M 465 6-ry VkfTAPPPPPPPVYEP
(352)
- (VILLI
FAK HUMAN Q05397 FOCAL ADHFSION KINASE NOM 183 197 KEGERALPSH9Ct.AN (353)
- (EC 2.7.1
FASL_MOUSE P41047 FAS ANTIGEN LIGAND MUS 41 55 DQRRPPPPPPPVSPL (]54) 3
TYROSINE-PROTEINKINASE
FGR FSVGR POOS44 ~~O FEL 9 23 VCRPRPLPPLPPIAM pSS)
FOR1 MOUSE Q05859 FORMIN 4(UMB MUS 65S 669 PPLIPPPPPL.PPGLG (!56)
- DEFORMITY PRO('EIII
U1 700 CPVSPPPPPPPPPPPPVPPS
C357)
699 712158) O58)
721 740 NSGGPPPPPPPPPPPPGLAP
(359)
FOSB_MOUSE PL3346 POSB PROTEIN MUS 253 269 GWLLPPPPPPPLPFQSS (360)
FOSB CHICK P11939 PSS-C-FOS PROTO GAL 239 254 LMTEAPPAVPPKEPSG (361) 3 0
ONCOGENE PROTEIN
FSH DROME PI3709 Pi3710 FEMALE STERILE DRO 4 20 SEPPPRYEPPVEPVNGI (362) 2
" HOMEOTIC PROTEIN
033 RATE P03432 GENE 33 POLYPEPTIDE RAT 146 160 DRSSRPLPPLPISED (363)
0
- 91 -
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/u,4298
LOCUS ACCESSION N'S DESCRIPfION
I SEQUENCE N~8 a
281 295 IPPRVPIPPRPAKPD (364) 3 3 1 3
GLLI_HUMAN P10071 GLI3 PROTEIN HOM 789 804 MFPRLNPILPPKAPAV (365) 4 3 1
986 1000 AAPPRLLPPLPTCYG (366) I
GTPA BOVIN P09851 GTPASE-ACTIVATIGN 805 IZ7 141 GGGFPPLPPPPPQLP (367)
PROTEIN (GAP)
HMEIMOUSE P090d5 HOMEOBOX PROTEIN MUS 72 yl LPHPPPPPPPPPPPPPQHL 1
ENGRAILED4 (M (368)
HMOC DROME P22810 IHOMEOTIC PROTEIN DRO 453 467 SAPQRPMPPNRPSPP (369) 4 1 2
ORTHODENTICLE
HS21_HUMAN P04792 1 HEAT SHOCK 27 KD PROTEI HOM 48 64 GSSWPGYVRPLPPAAIE 070) 4
2
HXA4 CHICK P17277 HOMHOBOX PROTEIN HOX- GAL 42 39 HPHAPPPPPPPPPPHLHA (371)
A4 (CHOX-1
I27
I I i41 GASPPPPPPAKGHPG p72) 3 5
HXAA HUMAN P31260 I HOMEOBOX PROTEIN HOX- HOM :23 !37 PQQQPPPPPQPPQPA (373)
A10 (HOX-)
HXB2 HUMAN P14652 P17485 P109 HOMEOBOX PROTEIN HOX- HOM 75 91
GPALPPPPPPPLPAAPP (374)
' 13 82 (HOX=2H
HXB3 HUMAN P1165! P17484 HOMEOBOX PROTEIN HOX- HOM 280 296 HSMTPSYECPSPPAFGK
(373) 4
1133 (HOX=2G
HXB4 HUMAN P17483 HOMEOBOX PROTEIN HOX- HOM 69 91 RDPC7PPPPPPPPPPPPPPPGLSP
- B4 (HOX-2 (376)
HXC4 HIIMAN P09017 HOMEOBOX PROTEIN HOX- HOM SO 61 QELYPPPPPRPSYPE 1377) 1
~ C4 (HOX-3
BiPI BOVIN P2A391 INSULIN-LIKE GROWTH BOX 83 97 GLSCRALPGEPRPLH (978) 3
' FACTOR BIND
IDF. HUMAN P74735 INSULIN=DF,GRADING HOM 9?! 1009 TfiFKRGLP:.'PLVKP {379) 3
' ENZYME(EC3
IEF4 HUMAIv P31948 TRANSFORMATION. MOM 195 211 EIATPTPPPPPKKI:TKP (310) 3 ?
- SENSTTIVE PROTEI
IHBB RAT P1749) INHIBIN BETA B CHAIN RAT 35 49 SPAAPF'PFoPPGAPQ (381;
- PRECURiOR
IRS) HUMAN PiS568 INSULIN RECEPTOR HOM 1197 1211 PEPQPPP-PPPHOPL (792)
SUBSTRATE=( (I
ISP3 SCHPO P40899 SEXUAL DIFFERENTIATION ~H 39 55 QHQQPI'YWYPPPPPRHH (3~3) 2 3
~ PROCESS
)UND CHICK P2792) TRANSCRIPTION FACTOR GAL 203 218 PRLPPPPPPPL KDFPQ r38/) 4 ~
JUN-D
KICH H(JMAN P3579D CHOLINE KINASE (EC MOM
53 67 ALALPPPPPLPLYLP 085)
KUS YEAST P40494 MOBABLE SAC 744 759 KDKSRPPRPPPKPLBL (386) 2
SERINElTHREONI NE-PROTE
KIRI_HUMAN QOtTlI SER(NE/fHREONINE- MOM 430 464 VDQQRPNIPNRWFSD (387) 3 1
PROTEIN KINASE
KIR4 HUMAN P36897 SERINER'HREONINE- HO~i1 447 46) EQKLRPNIPNRWQSC (SIE) I
~ PROTEIN KINASE
KRAF CAEEL Q07292 RAF HOMOLOO CAE 456 473 LDAQRPRPPQKPHNED 089) 2
SERINE?HREONINE-P
MAPA_Rr1T P34926 MICROTUBULE-ASSOCIATED RAT 1412 1826 VPKDRPLPPAPLSPA 090)
PR07'ElN
2421 2477 GELSPSFLNPPLPPSTD (391) 2
3 ~ MAPB_MOUSE P14873 MICROTUBULE-ASSOCIATED PROTEIN MUS 520 535
DLTGQVPTPPVKQVKL (392) 5
MIS HUMAN P03971 MUELLERIAN INHIBR'ING HOM 266 280 LDTVPFPPPRPSAEL (393) 2
~ FACTOR
387 401 AAELRSLPGLPPATA (394) 2
MPKI_XENLA QOSl16 DUAL SPECIFICITY XEN 296 300 ELAPRPRPPGRPISS (395) 3 0 3
MrI'OGEN-ACTIVA
MPK2-HUMAN P36507 DUAL SPECIFICITY HOM 293 307 SISPRPRPPORPVSG (396) 3 0 3
MITOGEN-ACTIVA
MYBB CHICK QD3237 MYB-RELATED PROTEIN B GAL 512 326 YGPIRPLPQTPHl.EE (397) 2
MYSA'CAEEL PI2844 MYOSINE HEAVY CHAIN A CAE 561 ST! LGKHPNFQKPKPPKGKQ (398) 4
(MHC A)
_ 92 _
CA 02595040 2007-08-03
WO 97/30074 PCT/US97/02298
LOCUS ACCESSION I'S DF.SCRIPl7ON
SEQUENCE y y
I
MYSB CAEEL P02566 MYOSINE HEAVY CHAIN B CAE 559 575 LGKHPNFEKPKPPKGKQ (399) 4
(MHC B)
MYSC CAEEL P12944 MYOSINE HEAVY CHAIN C CAE 562 S78 LGKHPNFEKPKPPKGKQ (400) 4
(MHC C)
MYSD_CAEEL P02367 MYOSINE HEAVY CHAIN D CAE 556 572 LGKHPNFEKPKPPKGKQ (401) 4
(MHC D) 5 NCFI HUMAN PI4399 I NEUTROPHIL CYTOSOL NOM 359 'T! SKPQPAVPPRPSADL
(402) 2
FACTORIfN
NEU_RA7 P06494 NEU ONCOGENE RAT 560 374 VSDKRCi.PCHPECQP (403) 3
PRECURSOR (EC 2.7.
NG3 DROME P10140 NEW-GLUE PROTEIN 3 DRO 33 47 LRLPPPLPPRPRQPL 4401) 0
PRECURSOR(
NME4MOUSE Q03391 GLUTAMATE (NMDA) MUS 90l 915 PPAKPPPPPQPLPSP (405)
- RECEPTOR SUBU
OIF HJMAN PS0774 OSTEOINDUCTIVE FACTOR HOM 177 1922 NQLLKLPVLPPKLTLF 1406) 3
_
PRECURSOR
P118 HUMAN P42336 PHOSPHATIDYLINOSfCOL 3- HOM 709 323 SNLPLPI.PPKKTR11/407) 4
' KINASE (
P2B1 HUMAN P16298 SERINE?HREONINE HOM 7 25 ARAAPPPPPPPPPPPGADR 3
- PROTEIN PHOSPH (406)
PS7 CH1CK P10360 CELLULAR TUMOR GAL 4S 62 EPSDPPPPPPPPPi.P1.AA (409)
AN77GEN P33.
MA HUMAN P27986 PHOSPHATIDYLINOSriOL 3- HUM 99 103PRPPRPLPVAPGSSK (410) 1
KINASE
PlSB BOVIN P23726 PHOSPHATIDYLINOS1i0L l BOS 90 103 PRGPRPLI7'ARPRDGP (411) 2
3 0
KINASE
290 30 EQEVAPPALPPKPPKT 1412) 2 0
PFTA RJ. f. Q0163I PROTEIN PAT 18 il OPEQP'"i 1PPPPPPAtXP (413)
FARNESYLTRANSFERASE AL
PRGR_!It.MAN P06401 PROGESTERONE RECRPTOR HUM -19433 i:;PpPP1.PPKA'YPSA (41a)
1
1PR) (FOR
1'1t : DR(?MR' P29617 PROTEIN PROSPERO. ORO 1076 10fK: YHPQPPPPPPPMMPV (4131
PRPB 11JM!.f: PL2114 PROUNE-IUCH PEP-i'IDE 0-3. HOM 17 ].: QPI=GOCiI-
VPPPPPPPYG (416) 2
P7Nt HUMaN PI11031 PRM'EIN-TYROSINE HCN !02 316 PPe'kIPPPPRPP1iRI 1417) 3 3 2
2
' PHOSPHATASE 1
Prr.: HI.MAN P26(115 PROTEIN-TYROSINE HOM W n74 C1.T'ERNLP'YPLDIV 1419) 3
' PHOSPHATASE P
P7N4 HLMAN P29074 PROTEIN-TYROSINE HOM 4S7 47i itiDGkPP_%I.PPKQSKK 1419) 3
PHOSPHATASE ME
PTFI DROME P3S9St IPROTEIN=TYROSINE DRO 143C Ia16 FiM2'D?O% PNPPQiLV (420) 4
PHOSPHATASE 10
PTPK MOUSE P3.S&? 1 PROTEIN-TYROSINE MUS 60 76 SAQEP:IYLPPEMPQGSY (421) 2
~ PHOSPHATASE KA
RA01 IIUNAN P3:241 RADIXIN. HOM 466 43! VMSAPPPPPPPPVIPP (422)
RB IiUMAN P06400 IRE'i INOBLASTOMA- HOM 19 33 EPPAPPPPPPPEEDP (423)
ASSOCIATED PROTE
ROG_HUMAN P36159 HETEROGENEOUS NUCLEAR HOU 97 106 GRRGPPPPPPSRGPP (424) 4 1 2
R OPIUCLE
ROK HUMAN QlTf1M HETEROGENEOUS NUCLEAR ~M 267 281 GRGGRPMPPSRRDYD 1425) 3 1
- RB)ONUCLE
HOM 301 321 GSRARNLPLPPPPPPRGGDL 3 1 1
(426)
ROL HUMAN P14366 HETEROGENEOUS NUCLEAR HOM 326 M6 SRYGPQYGHPPPPPPPPEYGP 3
~ RIBONUCLE (r_*7-
RRGI HUMAN P13631 RETINOIC ACID RECEPfOR HOM 76 90 SSPSPPPPPRVYKPC (428) 2 2
-
~ GAMMA-I
RRG2 HUMAN P22932 REI7NOIC ACID RECBPTOR NOM 65 79 SSPSPPPPPRVYKPC (429) 2 -
GAMMA=2
RRXB HUMAN P2i702 RETINOIC ACID RECEPtOR HOM ]M311 GSGAPPPPPMPPPPL (430)
- RXR-BETA
RRXC HUMAN P26703 IRETINOIC ACID RECEPTOR GSGAPPPPPMPPPPL (431)
- RXR=BETA
3 5 RYNR HUMAN P21b17 RYANODINE RECEPTOR. HOM PKKQAPPSPPPKKEEA (432) 4
SKELETAL MU
SHC HUMAN P29353 SHC TRANSFORMING HOM RKQMPPPPPCPGREL (4331
PROTEINS 46.E
93
CA 02595040 2007-08-03
WO 97/30074 PCT/fJS97/02298
LOCUS ACCESSION N'S DESCRIPTION SEQUENCE N~~9 qn~'
SLPI DROME P32030 FORK HEAD DOMAIN DRO 242 258 GAPAPSYGYPAVPFAAA (434) 3
TRANSCRIPTION
SON OF SEVENLES5
SOS DROME P36675 DRO 1339 1353 RAVPPPLPPRRKERT (435) 0 1
PROTEIN
1377 1391 ELSPPPIPPRLNHST (436) 0
ST20 YEAST 003497 SERINEITHREONINE- SAC 533 $47 EQPLPP(PPTKSKTS (437)
PROTEIN KINASE
SL'F DROME 1,25991 SUPPRESSOR OF FORkED DRO 229 243 KGLNRNLPAVPPTLT (438) 2
PROTEIN.
SXLFDROME P19339 SEX-LETHAL PROTEIN, DRO 308 322 PANVPPPPPQPPAHM (439)
- FEMALE-SPEC
TACT HUMAN P402G0 T-CELL SURFACE PROTEIN HOM 538 553 PPPFKPPPPPIKYTCI (440) 1
4
- TACTILE
TGFls HUMAN P22064 TRANSFORMING GROWTH HOM 440 434 KSTHPPPLPAKEEPV 1441) 3
FACTOR BETA
TIE7-MOUSE Q02858 TYROSINE-PROTEIN KINASE MUS 725 739 SHELRTLPHSPASAD (442) 3
RECEPTOR
T16 MOUSE= Pt5920 IMMUNE StIPPRESSOR. MUS 81 96 EGEASPPAPPLKHVLE (443)
FACTOR J6B7.
TLL DROME P18102 TAILLESS PROTEIN. DRO 214 228 ALATRALPPfPPLMA (444) 2
TOPI HUMAN P11387 DNA TOPOISOMERASE I(EC HOM 221 237 EHKGPVFAPPYEPLPEN (445) 3
5.99.1.
TOPA HUMAN P113U DNA TOPOISOMERASE 11. HOM 133 849 QRVEPEWYIPIIPMVLI (446) 3
- ALPHA ISO
TOPB, HUMAN Q02880 DNA TOPOISOMERASE ll, HOM 855 871 QRVEPEWYIPIIPMVLI (447) 3
BETA ISOZ
IRAI HUMAN P3t708 SEX-DETERMINING CAE 1069 1090 PEDDPIYALPPPPPPPAPPRRR 1 3
TRANSFORMER PRO (44i8)
TRrI HUH-.N PI38U5 TROPONIN T. SLOW NOM A2 57 SRPVVPPI.IPPKIPEG (449) 3
- SKELETAL MUSCLE _
XAI XENl.1 P23507 XA=1 PROTEIN PRECUP.SOR. Xa*e 22 39 GEDSPVFRPPSPPMGPS (430)
3
121 136 FRTGRPLLPIKPEHGR (43!i
Z0 _HUMAN Q071S7 TIGHT JUNCTION PROTEIN HOM 1410 1.424 IQATPPPPPLPSQYA (452)
ZO= I.
Z fX C NICK Q04584 ZYXIN. tiAL 120 234 AFPSPPPPPPPMFDE (453) 1
35
- 94 -
CA 02595040 2007-08-03
75361-88
in Tabie 16, locus and accession number refer to the
entries' names and accession numbers in GenBank or the Swiss-
Prot database. The two numbers immediately to the left of
the displayed sequences refer to the amino acid positions of
the displayed sequences in the mature proteins. The leftmost
of these two numbers refers to the starting amino acid number
of the displayed sequence in the mature protein. The numbers
in parentheses immediately to the right. of the displayed
sequences refer to the sequences' SEQ ID NOs: The eight
.1o colunins to the extreme right of Table 16 show. the
discrepancies between the displayed sequences and the
consensus motifs of Tables 6-15. The leftmost Src column
refers to Class I motif s; the rightmost Src column refers to
Class II motifs.
zt should be apparent to one of ordinary skill that many
other embodiments of the px=esent invention can be
contemplated beyond the prFferred embodiments described above
but which other embodiments nevertheless fall ta-ithtn the
scope and spirit of the p,: e.sent invention. zIence, the
20 present invention should not be construed to be limitpd to
the preferred embodiments described herein, which serve only
to illustrate the present invention, but only by the claims
that follow.
Also, numerous references are cited throughout the
25 specification.
35
- 95 -
CA 02595040 2007-08-03
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPREND PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
NOTE: Pour les tomes additionels, veillez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME OF _2
NOTE: For additional volumes please contact the Canadian Patent Office.