Note: Descriptions are shown in the official language in which they were submitted.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
METHODS AND COMPOSITIONS FOR REMOVING CELLS FROM COLLECTION
MATERIAL
100011 This application claims the benefit of the filing date of Provisional
Application
No. 60/838,687, filed Aug. I 8, 2006, entitled "Methods and Kits for Eluting
Sperm" this
entire disclosure is hereby incorporated by referencc into the present
disclosure.
BACKGROUND
[0002] DNA analysis has proven to be one of the most valuable and reliable
tools to help
acquit or convict suspects of a crime, identify individuals that are related,
or even help
diagnose disease.
100031 DNA is present in all kinds of cells including blood, hair, skin,
saliva, bone, sperm,
etc. At the crime scene, the suspect's DNA can be obtained from cells found
from the
environment or even on the victim. The cells can be collected on suitable
collection material,
(e.g., swab, gauze, paper, etc.) and then sent to the forensic.laboratory for
analysis. In the
forensic lab, typically the scientist elutes cells fi-om the collection
material, extracts DNA from
the eluted cells and then analyzes the DNA to generate a profile to see if it
matches the
suspect's DNA profile.
100041 One of the most time consuming tasks for the laboratory personnel is
removing cells
from collection material so that DNA can be extracted. This task becomes even
more labor
intensive when the cells have adhered to the collection material for months or
when few cells
were collected. For example, even with the most careful criminal investigation
at the crime
scene sometimes only very few cell are available to be analyzed. These cells
may be eluted by
soaking the evidence containing the cells in an overnight solution, such as
phosphate buffered
saline (PBS) or citrate buffer, accompanied by periodic agitation to recover
cells. Many times
no meaningful profile can be generated because too few cells or no cells can
be eluted from
the collection material.
100051 Other times, particularly in rape and murder cases, there are mixtures
of different cell
types (e.g., cells from the victim, perpetrator, and/or surrounding
environment), which may
cause further time consuming steps in separating cells and confound test
results.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
2
100061 Differential extraction (DE) is conimonly used to separate cells (e.g.,
sperm cells,
vaginal epithelial cells, plant cells, bacterial cells, etc.) from the
collection material. Currcnt
DE extraction protocols utilize proteinase K and an anionic detergent to
selectively lyse
vaginal epithelial cells, and later, centrifugation is employed causing the
spenn cells to be
pelleted, while vaginal epithelial cells i-emain in the supernatant for
removal. Pelleted sperm
cells are then resuspended in dithiothreitol (DTT), which lyses the sperm
cells by breaking
down the protein disulfide bridges that make up sperm nuclear membranes to
release DNA.
Conventional DE extraction protocols suffer from the disadvantages of
substantial cell lysis,
overnight incubation time and low cell recovery (e.g., 16-30% recovery).
100071 Other modifications to DE procedures have been developed that utilize
protein
precipitation to extract DNA. For example, the addition of 6 M NaC l to a
proteinase K-
digested cell extract followed by vigorous shaking and centrifugation results
in protein
precipitation so that the supernatant containing the DNA portion of cell
extract can then be
isolated and later amplified using standard PCR protocols. These modifications
eliminate
some wash steps,' however, they still have low cell recovery.
100081 Conventional alternatives to the DE method focus on the separation of
cells before the
DNA is extracted. These conventional alte atives utilize laser capture
microdissection or
micro-device based sorting of cells to separate cells and later extract DNA.
Although these
methods may effectively sort cells, often they may utilize costly equipment
and/or involve
many steps that are cumbersome. 100091 Other alternatives to DE use sperm
binding proteins, glycopeptides, lecitins,
derivatives ofN-acetylglucosamine, triazine dyes, inhibitors of
glycosyltransferase, or
monoclonal antibodies specific for epitopes that can bind and/or precipitate
the sperm from the
collection material. I-lowever, these methods can be cumbersome and do not
increase
recovery of the cells from the collection material.
100101 Attempts have been made to increase cell recovery from the collection
material by
digesting it with cellulases (e.g., enzymes obtained from Aspergilhis nige,-,
7)-ichoderina
reesei or Ti=ichodernia viride) to release intact cells from the collection
material.
Unfortunately, the cellulases are used at high concentrations to digest the
collection material,
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
3
which often causes substantial lysis of cells and little improvement in cell
recovery (e.g., 15-
21 % recovery).
100111 New methods and compositions for removing cells from collection
material are
needed. Methods and compositions that reduce time needed to i-emove cells from
the
collection material as well improve recovery of cells from the collection
material are needed.
SUMMARY
100121 Methods and compositions are provided for removing cells from
collection material.
In various embodiments, the methods and compositions provided reduce the time
needed to
remove cells from the collection material as well as improve cell recovery
from the collection
material.
100131 In various embodiments, the methods and compositions allow cells to be
removed
from samplcs thought to contain few or no cells -- because cells could not be
eluted using
conventional prior art methods. In various embodiments, the methods and
compositions
provided allow elution of intact cells without substantial lysis of the cells
or substantial
digestion of the collection material.
100141 In various embodiments, the methods and compositions allow DNA profiles
to be
generated for an individual where none were previously generated -- because
sufficient
numbers of cells could not be eluted using conventional prior art methods.
100151 In various embodiments, a method for removing sperm cells adhered to a
material is
provided, comprising: contacting the material with at least a first and second
reagent, and an
effective amount of an enzyme so as to remove the sperm cells from the
material without
substantially lysing the sperm cells and without substantially digesting the
material.
100161 In various embodiments, a method for removing sperm cells and
epithelial cells
associated with a collection material is provided, comprising: (a) contacting
the collection
material with at least a first reagent to substantially remove the epithelial
cells associated with
the collection material; (b) contacting the collection material with at least
a second reagent and
an enzyme to aid in substantially removing the sperm cells from the collection
material,
without substantially lysing the sperm cells and without substantially
digesting the collection
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
4
material; and (c) sonicating the collection material to substantially reniove
the sperm cells
associated with the collection material.
100171 A kit is provided for removing spei-m cells and epithelial cells
adhered to a material,
the kit comprising: a) a first reagent having a pH of about 7.0-8.0; b) a
second i-eagent having
a pH of about 8.0-9.0; and c) about 0.0010 pg to about 0.5pg of proteinase K.
100181 Additional features and advantages of various embodiments will be set
forth in part in
the description that follows, and in part will be apparent from the
description, or may be
learned by practice of various embodiments. The objectives and other
advantages of various
embodiments will be realized and attained by means of the elements and
combinations
particularly pointed out in the description and appended claims.
BRIEF DESCRIPTION OF THE DRAWINGS
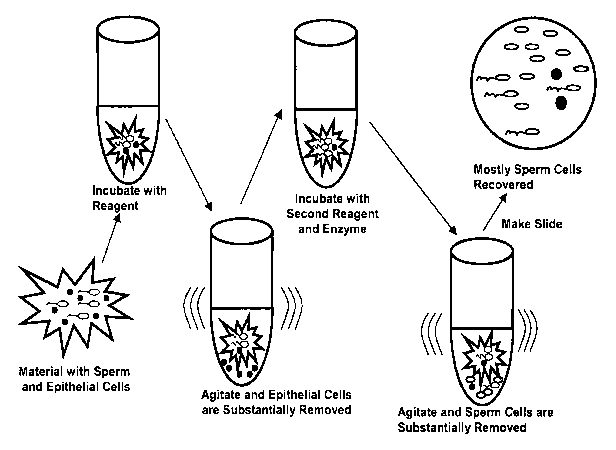
10019) Figure 1 illustrates an embodiment of the method of renioving cells
from a collection
material. The collection material is incubated with a reagent (e.g., PBS, beta-
amylase, etc.)
and agitated (e.g., vortexing, centrifuging, etc.) to remove epithelial cells.
Subsequently, a
second reagent (e.g., Tris-HCL, a citrate buffer, etc.) and an enzyme (e.g.,
proteinase K) are
incubated with the collection material and agitation causes substantial
removal of mostly the
head of the sperm cells. A slide is prepared from the re-suspended pellet and
examined
showing that mostly sperm cells are removed without substantially lysing the
sperm cells and
digesting the collection material.
100201 Figure 2 illustrates an embodiment of the method of removing cells from
a collection
material. This embodiment employs a sonication step to remove sperm from the
collection
material. Greater than 50% of sperm cells are removed from the collection
material without
substantially lysing the sperm cells and digesting the collection material.
100211 Figure 3 illustrates a GenoTyper electropherogram showing an
acceptable DNA
profile generated from the sperm eluted from a swab utilizing an embodiment of
the method
described herein.
100221 Figure 4 illustrates a GenoTyper electropherogram showing an
acceptable DNA
profile generated from the sperm eluted from a swab utilizing an embodiment of
the method
described herein. This figure illustrates a predominantly female with partial
male profile.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
100231 It is to be understood that the figures are not drawn to scale. Furthei-
, the relation
between objects in a figure may not be to scale, and may in fact have a
reverse relationship as
to size. The figures arc intended to bring understanding and clarity to the
structure of each
object shown, and thus, some features may be exaggerated in order to illusti-
ate a specific
feature of a structure.
DETAILED DESCRIPTION
100241 For the purposes of this specification and appended claims, unless
otherwise indicated,
all numbers expressing quantities of ingredients, percentages or propoi-tions
of materials,
reaction conditions, and other numerical values used in the specification and
claims, are to be
understood as being modified in all instances by the term "about_"
Accordingly, unless
indicated to the contrary, the numerical parameters set forth in the following
specification and
attached claims are approximations that may vary depending upon the desired
properties
sought to be obtained by the present invention. At the very least, and not as
an attempt to limit
the application of the doctrine of equivalents to the scope of the claims,
each numerical
parameter should at least be construed in light of the number of reported
significant digits and
by applying ordinary rounding techniques.
100251 Notwithstanding that the numerical ranges and parameters setting forth,
the broad
scope of the invention are approximations, the numerical values set forth in
the specific
examples are reported as precisely as possible. Any numerical value, however,
inherently
contains certain errors necessarily resulting from the standard deviation
found in their
respective testing measurements. Moreover, all ranges disclosed herein are to
be understood
to encompass any and all subranges subsumed therein. For example, a range of"1
to 10"
includes any and all subranges between (and including) the minimum value of I
and the
maximum value of 10, that is, any and all subranges having a minimum value of
equal to or
greater than 1 and a maximum value of equal to or less than 10, e.g., 5.5 to
10.
100261 It is noted that, as used in this specification and the appended
claims, the singular
forms "a," "an," and "the," include plural referents unless expressly and
unequivocally limited
to one referent. Thus, for example, reference to "a reagent" includes one or
more reagents.
100271 Reference will now be made in detail to certain embodiments of the
invention,
examples of which arc illustrated in the accompanying drawings. While the
invention will be
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
6
described in conjunction with the illustrated embodiments, it will be
understood that they are
not intcnded to limit the invention to those embodiments. On the contrary, the
invention is
intended to cover all alternatives, modifications, and equivalents, which may
be included
within the invention as defined by the appended claims.
100281 The headings below are not meant to limit the disclosure in any way;
embodinients
under any one heading may be used in conjunction with embodiments under any
other
heading.
10029] In various embodiments, a method for removing sperm cells adhered to a
collection
material is provided, comprising: contacting the collection material with at
least a first and
second reagent, and an effective amount of an enzyme so as to remove the sperm
cells from
the collection material without substantially lysing the sperm cells and
without substantially
digesting the collection material.
Material
[0030] The collection material can be any material that can have sperm
associated with the
material. As used herein, "associated with" or "attached" or "adhered"
encompasses
interactions including, but not limited to, adhesion, non-covalent bonding,
covalent bonding,
ionic bonding, hydrogen bonding, Van der Waals interactions, chemisorption,
physisorption,
hydrophobic interaction, hydrophilic interaction or a combination thereof,
which the sperm
cells have with the material.
10031] The material can be evidence or a piece of evidence from the crime
scene (e.g_,
clothing, carpet, linen, sheet, paper, victim's remains, etc.) containing the
sperm cells. The
material can also be a collection material to gather the cells. Material
includes, but is not
limited to, cotton, cellulose, nitrocellulose, carboxymethylcellulose,
hydrophilic polymers,
polytetrafluoroethylene, porous ceramics, Dacron , rayon, nylon, resin, or a
combination
thereof.
[0032] Cells from the environment or crime scene may be found on the surface
or embedded
in the material. These cells include, but are not limited to, bacterial cells,
viral cells, yeast
cells, fungal cells, plant cells, animal cells (e.g., human, ovine, equine,
bovine, porcine, foul,
canine or feline cells). These cells may be contained in blood, vaginal fluid,
semen, saliva,
urine, teeth, bone, hair, body tissue, or a combination thereof.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
7
100331 The cells can be from any source, for example, they can be biological,
medical or
forensic sainples. In various embodiments, the cells are obtained from vaginal
swabs, or
semen samples from rape victims, or semen samples from soiled clothing.
100341 The material may have sperm and other cell types embedded in the
material or on a
surface of the material. In one embodiment, the material includes mixtures of
different cells.
In other embodiments, the material includes sperm cells and epithelial cells.
In other
embodiments, the material includes only sperm cells.
100351 Cells embedded in or on a surface of the material can be eluted or
removed from the
material without substantially lysing the sperm cells and without
substantially digesting the
material. "Without substantially lysing" includes embodiments where some of
the sperm cells
may undergo mild disruption, however, more than 50% of the sperm cells remain
intact. For
example, often times when the material has sperm cells associated with it, on
removal of
sperm cells from the material, the cells will lose their tails, which mostly
reinain on or in the
material. Notwithstanding the loss of the tail, the head of the sperm cell
(which contains
DNA) will mostly be removed by the methods herein and the sperm cell will
remain intact.
When the tail of the sperm cell remains on or in the material, this is not
considered to be lysis
of the sperm cell. "Lysis" as used herein generally refers to the physico-
chemical disruption
of the structural components (e.g., cell membrane, structural proteins, etc.)
of the sperm cell.
Lysis will cause the sperm cell membrane to disrupt releasing the cell
contents (e.g., DNA
from the nucleus). The methods and compositions employed herein allow removal
of cells
without substantial lysis of sperm cells other than loss of the tail of the
sperm cell. After
removal of the sperm cell from the material, DNA will be extracted from the
sperm cell in one
or more steps.
[0036] In various embodiments, more than 75% of the sperm cells are removed
from the
material without lysis of the sperm cell other than loss of the tail. In
various embodiments,
more than 90% of the sperm cells are removed from the material without lysis
of the cell other
than loss of the tail.
100371 "Without substantially digesting" includes embodiments where some of
the material
may undergo enzymic breakdown, however, more than 50% of the material remains
undigested. For example, cellulases (e.g., enzymes obtained from Aspergillits
niger,
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
8
Ti-ichodei-ma reesei or Trichodei-ma vir-ide) are often employed to degrade a
cotton swab, but
they also may lyse spenn cells as well. The methods and composition of the
present invention
are designed to reduce or prevent digestion of the material so that sperm
cells can be eluted
from the material substantially intact. For example, lower concentrations of
the enzyme are
employed that do not substantially digest or cause enzymatic breakdown of the
material or
lyse the cell membrane. In various embodiments, more than 75% of the material
remains
undigested by the enzyme. In various embodiments, morc than 95% of the
material remains
undigested by the enzyme.
100381 Mechanical, chemical, thermal methods, or a combination thereof may be
utilized in
the methods described to remove cells from the material. However, these
methods are
perfonmed so as not to cause substantial lysis of sperm cells. For example,
ultrasonication is a
mechanical method that may be perfon.ned on sperm cells. Such ultrasonication,
if performed
for long periods of time, may lead to substantial sperm cell lysis. Similarly,
chemical
methods, which are non-mechanical methods, can be utilized to remove sperm
cells. For
example, chemical methods utilizing an acid, a base, a detergent, a solvent, a
chaotropic agent,
ctc. may also cause substantial lysis or disruption of sperm cells if used at
a high enough
concentration or for a long enough period of time. The methods and composition
of the
present invention are designed to reduce or prevent sperm cell lysis so that
the sperm cell can
be eluted from the material substantially intact.
Reagents
100391 The cells may be substantially removed from the material by contacting
it with a first
reagent and a second reagent. "Reagent" refers to an agent, in various
embodiments, an
eluent, which is used to affect or modify the material and/or cells so as to
aid in their removal
from the material. The elution characteristics of an eluent can depend, for
example, on pH,
ionic strength, hydrophobicity, degree of chaotropism, detergent strength and
temperature.
100401 Contacting the material to remove cells from the materials can be
accomplished by,
e.g., bathing, soaking, dipping, rinsing, spraying, and/or washing the
material with the eluent.
Typically, the eluent can be at a temperature of between 0 C to l 00 C.
[0041] Any suitable eluent can be used to wash the material one or more times.
For example,
each of the one or more washes optionally includes an identical or a different
elution condition
relative to a preceding wash. Elution conditions typically differ according
to, e.g., pH,
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
9
buffering capacity, ionic strength, a water structure characteristic,
detergent type, detergent
strength, hydrophobicity, concentration of the eluent or the like.
100421 In various embodiments, an aqueous solution is used as the eluent.
Exemplary
aqueous solutions include, but are not limited to, TE, HEPES (2-[4-(2-
hydroxyethyl)-1-
piperazinyl]ethanesulfonic acid), MES (2-morpholinoethanesulfonic acid),
sodium acetate
buffer, sodium citrate buffer, sodium phosphate buffer, a Tris buffer (e.g.,
Tris-HCL),
phosphate buffered saline (PBS), sodium phosphate, potassium phosphate, sodium
chloride,
potassium chloride, glycerol, calcium chloride or a combination thereof. In
various
embodiments, the buffer concentration can be from about l mM to 100 mM.
100431 In various embodiments, the pI-I of the eluent can be adjusted to the
range of about 7.0
to 9.0 with the addition of a suitable acid or base. For example, acetic acid,
citric acid, or
sodium bicarbonate can be added to adjust pH. In various embodiments, the
first reagent has a
pH of about 7.0-8.0 and the second reagent has a pH of about 8.0-9Ø
100441 In various embodiments, the reagent may contain one or more detergents
capable of
solubilizing the cells from the material. Suitable detergents include for
example
polyoxyethylene sorbitan fatty acid ester such as Tween 20, Tween 40, Tween
60, Tween 80,
and Tween 85, an alkylaryl polyether alcohol such as Triton X 100, non-ionic
and ionic
detergents such as sodium dodecylsulfate (SDS), carbohydrate based detergents
such as
octylglycoside, and combinations thereof. The detergent is advantageously
added to reach a
concentration of between about 0.05% (v/v) and 10% (v/v), such as about 0.1%
(v/v).
100451 In various embodiments, an enzyme can be added to elute cells. The
enzyme can be
beta-amylase, a cellulase or a proteinase (e.g., serine proteinase) or
combinations thereof. In
various embodiments, the enzyme is in solution with a storage buffer, and the
storage buffer
(containing the enzyme) contacts the material. In various embodiments, the
collection
material can be incubated with the enzyme at room temperature for about 30
minutes to 4
hours.
100461 Examples of suitable proteinases include proteinase, proteinase K,
pepsin, trypsin,
chymotrypsin, and the like. The minimum amount of proteinase is generally
about 0.00025 pg
and the maximum amount of proteinase is generally about 0.5 pg. Any of the
above minima
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
and maxima can be combined to provide a range for the proteinase that aids in
removal of the
sperm cells.
100471 The term "effective amount" as used herein refers to that amount of the
enzyme that is
effective to aid in removing sperm cells without substantially digesting the
material and/or
causing substantial lysis of cells.
100481 In various embodiments, about 0.00025 pg to about 0.5 pg of proteinase
K is used.
Proteinase K is an endolytic protease that cleaves peptide bonds at the
carboxylic sides of
aliphatic, aromatic or hydrophobic amino acids, which may make up the
structural
components of various cell types or the material. Proteinase K may contain a
suitable storage
buffer (e.g., 50mM Tris-HCI (pH 8_0), 5mM CaC12 and 50% (v/v) glycerol)_
Proteinase K is
active with or without the presence of SDS, EDTA and urea. Proteinase-K is
available from
various manufacturers (e.g., New England BioLabs) at a wide variety of
concentrations (e.g.,
20mg/mi concentration).
100491 Generally, the first reagent, second reagent, material, and/or enzyme
can be added
separately or together in any order in the same or different vessels. In
various embodiments,
the material is added to the first and second reagent and/or enzyme to elute
cells. Incubation
time of the material with the one or more reagents and/or enzymes can be for
at least one or
more minutes. Additions of reagents, enzymes and/or material usually take
place at
temperatures between room temperature and 90 C, with or without agitation.
Examples of
types of agitation include, but are not limited to, shaking, stirring,
centrifuging, vibrating,
vortexing, sonicating or any other type of mechanical blending.
100501 In various embodiments, after the first reagent contacts the material
containing
epithelial and sperm cells, the material is vortexed and/or centrifuged and a
pellet containing
epithelia] cells is formed. The material is removed from the container and
transferred to
another container where a second reagent and an enzyme contact the material.
The material
(containing predominantly now sperm cells) is vortexed and/or centrifuged and
the pellet is
resuspended in a buffer to recover the sperm cells.
100511 In various embodiments, the vortexing and centrifuge conditions (e.g.,
time, speed,
temperature, etc.) can be determined by those skilled in the art. Typically,
time periods, which
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
11
do not substantially lyse the sperm cells (e.g., vortex about 5 seconds to
about 2 minutes,
centrifuge from about 12 seconds to about 5 minutes at 13,200 rpms).
100521 Figure I illustrates an enibodiment of the method of removing cells
from a collection
material. The collection material having sperm cells and epithelial cells is
added to a first
reagent (e.g., PBS, beta-amylase, etc.) and the collection material is later
agitated (e.g.,
vortexing, centrifuging, etc.) to remove epithelial cells. Subsequently, the
collection material
is transferred to a new tube and incubated with a second reagent (e.g., Tris-
HCL, a citrate
buffer, etc.) and an enzyme (e.g., proteinase K). Later the tube is agitated
causing substantial
removal of mostly the head of the sperm cells from the material (seen as a
pellet in the tube).
The pellet is resuspended in a buffer. A slide is prepared from the re-
suspended pellet and
examined, which shows mostly intact sperm cells. Thus, sperm cells are removed
from the
collection material without substantially lysing the sperm cells and digesting
the collection
material. After removal of the sperm cells from the material, the DNA can be
extracted from
the sperm cells and a DNA profile generated using methods known in the art.
100531 Sonication
In various embodiments, after the material is contacted with the second
reagent, the material is
subjected to sonication, where sound wave energy creates shear forces to help
remove sperm
cells from the material. In various embodiments, the method employs direct
sonication where
the sonotrode is placed directly in contact with the liquid (e.g., reagent)
and high-energy
ultrasound in either a continuous fashion or in pulses (e.g., 1-30 minutes) is
generated to
induce the cells to be removed from the material. In various embodiments, the
container
holding the material is placed in a bath and the bath is subjected to bath
sonication (e.g., about
20 to about 50 kI-Iz), where ultrasound waves are transmitted through the bath
to aid in the
removal of cells from the material.
100541 Typically, sonication periods, which do not substantially lyse the
sperm cells, can be
from about 2 minutes to about I hour. In various embodiments, the sonicator
utilized is Mini
ULTRAsonikT"' from Ney Dental, Inc. However, other sonicators from other
manufacturers
can be used.
100551 In various embodimcnts, the material (containing both epithelial and
sperm cells) is
contacted with a first reagent comprising PBS at a pH of about 7.4 in a
centrifuge tube. The
tube is vortexed and centrifuged. The material containing predominantly sperm
cells is
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
12
removed from the tube while the supernatant and pellet containing the
epithelial cells remains.
The material is then contacted with a second reagent comprising Tris-HCI
buffer having a pH
of about 8.5, and proteinase K. The material is vortexed, centrifuged and
placed in a
sonication bath. The resulting product is vortexed and centrifuged and the
pellet containing
predominantly sperm cells is re-suspended in buffer and examined by light
microscopy.
Mostly intact sperm cells will be seen on the slide utilizing various spermac
stains known in
the art.
100561 Since more intact sperm cells are removed from the material, there is a
higher purity of
the sample recovered and less chance of contamination by other cell types (non-
sperm cells).
Thus, when DNA is extracted from the sample, a more robust and reliable DNA
profile can be
generated because the source of the DNA is from a sample of high purity
spermatozoa. In
various embodiments, on recovery, the sperm cells are essentially pure, which
means that the
sperm cells are free other cell types, for example, the sperm cells are at
least 90% free, at least
95% free or at least 98% free of such other cell types.
100571 In various embodiments, greater than 50%, greater than 75%, greater
than 95%, or
substantially all of the sperm cells are removed from the collection material.
In various
embodiments, the sperm cells removed are counted by any method known in the
art, such as
for example, spectrophotometrically, using cell counters, using hematology
analyzers,
using flow cytometry, or microscopically or the like. In various embodiments
of a
microscopic count, a portion of the sperm are placed on a microscope slide,
stained, and
counted under 200X - 400X magnification using a light microscope. Success
rates of the
recovery of the sperm can be determined by any method known in the art, such
as for
example, using cell counters, using hematology analyzers, using flow
cytometry, or
microscopically or the like. In one embodiment, a predetermined amount of
spenmatozoa is
placed onto a cotton-tipped applicator swab and an aliquot of the eluted sperm
pellet is
counted. For example if 2000 sperm are placed on a cotton swab and the sperm
elution
procedure produces a 50 l pellet and a 5 l aliquot of the pellet yields a
count of 180, the
percent recovery is calculated as follows: 180 (count) X 10 (fraction of
pellet counted)/ 2000
(total sperm applied to swab = 90%. It will be understood by one of ordinary
skill in the art
that other techniques can be used to detennine the cell recovery.
100581 Figure 2 illustrates an embodiment of the method of removing cells from
a collection
material. The collection material having sperm cells and epithelial cells is
added to a first
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
13
reagent (e.g., PBS, beta-amylase, etc.) and the collection material is later
agitated (e.g.,
vortexing, centrifuging, etc.) to remove epithelial cells. Subsequently, the
collection material
is transferred to a new tube and incubated with a second reagent (e.g., Tris-
HCL, a citrate
buffer, etc.) and an enzyme (e.g., proteinase K). Later the tube is sonicated
and agitated
causing substantial removal of mostly the head-ol'the sperm cells from the
material (seen as a
pellet in the tube). The pellet is resuspended in a buffer. A slide is
prepared from the re-
suspended pellet and examined, which shows mostly intact sperm cells. Thus,
sperm cells are
removed from the collection matcrial without substantially lysing the sperm
cells and
digesting the collection material. After removal of the sperm cells from the
material, the DNA
can be extracted from the sperm cells and a DNA profile generated using
methods known in
the art.
(0059] The present method allows elution of cells without the need for
expensive
electrophoretic devices, pressure gradients, and/or expensive reagents for
elution of cells from
the material. For example, expensive reagents such as sperm binding proteins,
glycopeptides,
lecitins, derivatives of N-acetylglucosamine, triazine dyes, or inhibitors of
glycosyltransferase,
monoclonal antibodies specific for epitopes, or the like are not required to
elute cells from the
material.
(0060] Once the cells are recovered, the DNA can be extracted from the cells
by lysing the
cell membrane to release the DNA by methods known in the art. For example,
cell lysis can
be carried out according to a method or combination of methods selected from,
but not limited
to, mechanical disruption, chemical treatment or enzymatic digestion, such as
grinding,
hypotonic lysis, proteinase digestion, phenol-chloroform extraction, Chelex
extraction, EZ-
1 extraction (Qiagen), , detergent lysis, electroporation, ultrasound,
sonication, or change in
ionic concentration. In one embodiment, after elution, cell lysis can be
carried out with a
reagent or combination of reagents, including but not limited to, Tris-HCI,
NaCI, Na.)EDTA,
EGTA, SDS, proteinase, proteinase K, TNE, N-lauroyl-sarcosine, sarkosyl,
Triton, sodium
pyrophosphate, glycerophosphate, leupeptin, DTT, EGTA, MgC12, KCI, NaF, Sodium
valdalate, sodium molybdate, B-glycerophosphate, RIPA buffer (1 % NP-40,
Triton X-100, 1%
sodium deoxycholate, 0.1 % SDS, 0.15 molar NaCI, 0.01 molar sodium phosphate,
pH 7.2, 1%
Trasylol), TWEEN, or the like, or combinations thereof.
(00611 Once the DNA is extracted it can be isolated by methods known in the
art, such as for
example, separating the DNA on gels. One common method involves capturing the
DNA on a
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
14
support and eluting the nucleic acids or utilizing anion exchangers, or
chromatographic
techniques known in the art.
DNA Profile
100621 After isolation, a DNA profile can be generated utilizing methods known
in the art.
These methods include Restriction Fragment Length Polymoiphism (RFLP)
analysis, Short
Tandem Repeat (STR) analysis using specific primers and labeled probes
specific for the STR,
and DNA amplification and typing of HLA-DQA I loci and polymarker loci, or
using single
nucleotide polymorphisms (SNPs).
100631 Restriction fragment-length polymorphism (RFLP) analysis generates DNA
fragments
of different length by restriction endonucleolytic digestion. The RFLP
approach in general
entails: (i) extraction and isolation of DNA, (ii) digestion of the DNA into
fragments using a
restriction endonuclease, (iii) electrophoretic separation of the fragments,
based on size, for
example, by agarose gel electrophoresis, (iv) denaturing the double- stranded
DNA fragments,
for example in a high pH environment; (v) transferring the single-stranded
molecules out of
the gel onto a membrane support, for example, by capillary action; (vi)
hybridizing the
immobilized DNA fragments with specifically labeled DNA probes; and (vii)
detection of the
hybrid products by any mcthod known in the art, for example by autoradiography
or chemi-
luminescence, etc.
10064] One method to generate a DNA profile involves utilizing genetic markers
known as
variable number tandem repeats (VNTRs) or minisatellites. The VNTRs are
tandemly
repeated sequences (usually 9-80 bases in length per repeat unit) that exhibit
variation in the
number of repeats for alleles within and among individuals. Following
digestion with a
restriction enzyme, the length of each fragment is determined by the number of
repeats
contained within each fragment. Many VNTR loci used for human identity testing
exhibit
more than 100 types in a population. In fact, such a high degree
ofpolymorphism is exhibited
that the typing of five to eight markers is sufficient to differentiate most,
if not all, unrelated
individuals. In other words, a multiple locus VNTR profile is extremely rare.
More
importantly, profiling VNTR loci currently provides one of the best avenues to
exclude a
suspect who has been falsely associated with an evidentiary sample. The loci
alleles should
generally fall in a size range that is greater than 500 bp and less than
20,000 bp. The loci
routinely used for a profile are D1S7, D2S44, D4S110, D10S28, and D17S79.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
100651 A subclass of variable number tandem repeats (VNTRs) is the short
tandem repeat
(STR), or microsatellite, loci. The STR loci contain tandemly repeated
sequences, each of
which is 2 to 7 bp in length. Loci containing repeat sequences comprising 4 bp
(or
tetranucleotides) are used routinely for human identification and, in some
cases, 5 bp repeat
STRs are used. These repeat sequence loci are abundant in the human genome and
are highly
polymorphic. The number of alleles at a tetranucleotide repeat STR locus
typically ranges
from 5 to 20. STR loci are amenable to amplification by PCR.
100661 In various embodiments, STR typing can be accomplished using one or
more of the
thirteen STR loci: CSFIPO, FGA, I'HOI, TPOX, vWA, D3S 1358, D5S818, D7S820,
D8S 1179, D 13S317, D 16S539, D 18 S51, and D2l S l 1, that make up the core
loci for use in the
national DNA databank, (Combined DNA Index System - CODIS). Loci containing
repeat
sequences containing 4 bp (e.g., D2S1338 and D19S433) can also be employed in
DNA
profiling.
100671 After the DNA fragments from the sperm cells are isolated and separated
by
electrophoresis, specific fluorescent dyes may be detected and an
electropherogram generated
indicating the DNA profile. Figure 3 illustrates a GenoTyper electropherogram
showing an
acceptable DNA profile generated fi-om the sperm eluted utilizing an
embodiment of the
method described herein. The fluorescent dye label color and relative PCR
product size range
for various loci are shown. Figure 4 illustrates a GenoTyper electropherogram
showing an
acceptable DNA profile generated from the sperm eluted from a swab utilizing
an embodiment
of the method described herein. This figure illustrates a predominantly female
with partial
male profile.
(0068] Kits
The present invention also provides kits for removing sperm cells and
epithelia] cells adhered
to a material. The kit generally includes a first reagent (e.g., PBS) having a
pH of about 7.0-
8.0; b) a second reagent (e.g., Tris-HCL) having a pH of about 8.0-9.0; and c)
about 0.0010 g
to about 0.5 g of proteinase K.
100691 In addition, kits also generally include instructions (e.g., in the
form of a label or a
separate insert) for removing sperm cells and epithelial cells adhered to a
material. The
instructions may also include other operational parameters. For example, the
kit may have
standard instructions informing users how to wash the material. In another
embodiment, the
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
16
kit may have instructions for collecting the spenn and/or epithelial cells.
Optionally, the kit
may further include a standard or control information for comparison with
information derived
from test samples.
100701 Having now generally described the invention, the same may be more
readily
understood through the following reference to the following examples, which
are provided by
way of illustration and are not intended to limit the present invention unless
specified.
100711 EXAMPLES
The ability to perform forensic DNA analysis in criminal investigations has
become an
important part of solving numerous types of crime. DNA testing has become
especially
important in criminal cases involving sexual assault and rape. In these types
of cases it is
necessary to collect spermatozoa from the evidence (e.g., underwear, clothing,
bed linens, etc.)
or from a post-coital collection device, such as a cotton-tipped applicator
swab. Currently, the
most common method of cellular recovery from these types of samples involves
soaking the
evidence (or a portion thereof) in a neutral, isotonic solution, such as
phosphate buffered
saline (PBS) or citrate buffer, accompanied by periodic agitation such as
vortexing.
Oftentimes, in order to achieve optimal recovery, the samples require an
overnight incubation
in the soaking solution. Recent studies have indicatcd that less than 10% of
the spermatozoa
present on a cotton swab are actually recovered using this type of procedure.
Using the
current method, some sexual assault specimens are inconrectly characterized as
negative for
sperm and therefore insufficient for DNA testing. Some of these samples in
fact do contain an
adequate number of sperm, and if properly screened and eluted from the
collection device,
could yield a suitable DNA profile. Due to the importance of accurately
assessing the
presence of sperm, it is imperative that an improved method be developed that
is aimed at
recovering significantly more spermatozoa from the swab. Improving this
recovery step
should lead to a larger number of successful profiles generated and therefore
a larger number
of forensic cases solved.
100721 In an effort to address the concerns of reporting samples as negative
due to poor
sperm recovery from the sexual assault swab or material, a series of
experiments was
undertaken to increase the overall recovery of spermatozoa from these types of
samples.
Based on the literature and our forensic experience with these types of
samples, a list of
factors has been created that might increase the forensic scientist's ability
to elute sperm from
cotton swabs. The factors examined in this study were as follows 1) the type
and pH of the
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
17
buffer system used, 2) incubation time and temperature, 3) the use of various
enzymes to aid
in sperm i-emoval 4) and a more stringent mechanical method of agitation to
help remove
sperm adhered to the swab. Initial experiments indicated that Tris-HCI was a
better alternative
to PBS, the use of Proteinase K at low concentrations could increase the
number of sperm
recovered from mock sexual assault samples, and sonication alone could
increase the recovery
4-fold. With this information in hand, optimization of each of the factors
listed above was
undertaken to develop a new improved method of sperm elution.
(0073] Demonstration of need:
The sperm elution method currently used by a number of laboratories (protocol
listed in
Appendix 1) consistently yields between-3-8% recovery of sperm. By way of
example, the
following table demonstrates the results of a comparison between this method
of elution
versus the new method being presented in this validation. In this example, 20
I of a semen
sample diluted 1:1000 was applied to four cotton swabs. Two of the swabs were
subjected to
the current method of elution and two were subjected to the new method
(protocol in appendix
I]). Five 1 of a 40p1 sperm elution pellet were used to determine the sperm
count for each
swab. The total number of sperm placed on each swab was - 650.
Results:
~''` C_U NT ME;~TH_O_p
NEW~METHOQs_,,~
SAMPLE 1 NEG -
SAMPLE 2 - 61
SAMPLE 3 2 -
SAMPLE 4 - 46
Conclusion: Using the current inethod Sample #1 would have been missed as a
positive
specimen. The new method is vastly superior to the currently approved method.
The
following examples will demonstrate how this type of improvement in recovery
of sperm can
be achieved.
(0074] Validation Examples:
All of the examples to validate the new method utilized mock sexual assault
samples except
for the non-probative casework samples. The mock samples were manufactured by
Orchid
Cellmark using buccal swabs taken from female employees. To each of these
swabs varying
amounts of a commercially available spenn sample (pre-washed cells - not
containing seminal
fluid) were added and allowed to dry. For the reproducibility studies shown in
Example I 1 an
accurate assessrnent of sperm counts was made (see this section for detail).
For Examples -1-
10, an attempt was made to target a specific number of sperm cells, however
the actual
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
18
number of sperm applied to each swab is likely somewhat lower than anticipated
due to
problcros inhcrent in dealing with diluted spcrm.
100751 Example 1
Purpose: To optimize the pH of 0.05M Tris-HCI for use in the spei-m elution
method.
Method:
= 750 l of 0.05M Tris-HCI was added to each of the swab samples and vortexed
for 20
sec. The samples were centrifuged long enough to reach 13,200 rpm
(approximately
12 sec.) and were incubated at room temperature for 1 hr. The samples were
vortexed
again for 20 sec. They were quick spun again for 12 sec. The swabs were placed
in a
spin basket and centrifuged at 13,200 rpm for 5 min. The basket was placed in
a new
tube and the supernatant removed. The pellet left in the original tube
included ---40N1
of supematant. The pellet was resuspended using a 20 1 pipet. A 5 1 aliquot
was
placed on a microscope slide and heat fixed for 5 min. at 65 C. The slides
were
stained with Christmas tree stain - red dye for 15 min, rinsed with water;
green dye for
15 sec, and rinsed with ethanol.
= Sample size: n=1
Results:
y
:Sam fe IP e~~r
Buff4s_ and=OH , Sam le; Numb..e..r S erount
'. _ Recove, ~
31 NV Tris 6.06 0 0%
32 NV Tris 6.98 0 0%
33 NV Tris 7.50 2 1%
34 NV Tris 8.05 3 2%
35 NV Tris 8.55 7 4%
36 NV Tris 8.97 3 2%
Conclusion: The optimal pH for 0.05M Tris-HCI is 8.5 although the rate of
recovery is very
low. Optimization of the other factors being studied in conjunction with Tris-
HCI should
increase the recovery rate.
100761 Example 2
Purpose: To optimize the amount of ProK needed for sperm elution.
Method:
= The method for Example 2 includes the addition of varying amounts of ProK
and a 5
min sonication step after the one-hour incubation step.
= 750 1 of 0.05M Tris-HCI pH 8.5 were added to the samples along with 15 1 of
ProK
at concentrations of 0.001, 0.0005, 0.00025, 0.0001 mg/m1.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
19
= The samples were centi-ifuged long enough to reach 13,200 rpm (approximately
12
sec.) and were incubated at room temperature for I hr. The samples were then
sonicated for 5 min. in a Mini ULTRAsonikT"' (Ney Dental, Inc.). Following
sonication, the samples were vortexed again for 20 sec. They were quick spun
again
for 12 sec. The swabs were placed in a spin basket and centrifuged at 13,200
rpm for
min. The baskct was placcd in a ncw tube and the supcrnatant rcmoved. The
pellet
left in the original tube included --40 l of supernatant. The pellet was
resuspended
using a 20p1 pipet. A 5p1 aliquot was placed on a microscope slide and heat
fixed for
5 min. at 65 C. The slides were stained with Christmas tree stain - red dye
for 15 min,
rinsed with water; green dye for 15 sec, and rinsed with ethanol.
= Sample size: n=1
Results:
y
~
~Sam .le. Number= -~ Sam~~ID Pr,o~K m/ml USperm-Count fteco~e .
60 JH 0.001 75 43%
61 JH 0.0005 58 33%
62 TB 0.00025 41 23%
63 TB 0.0001 6 3%
Conclusion: The optimal ProK amount added to the incubation solution is 15Et1
of
0.001 mg/ml. Therefore, the final concentration of ProK in this solution is 20
pg/ l.
100771 Example 3
Purpose: To optimize the concentration of Tris-HCI at pH 8.5 followed by 5 min
of
sonication.
Method:
= The procedure for Example 3 is the same as for Example 2 (see above) except
that no
ProK was added in this example.
= Sample size: n=1
Results:
y
~~~>a=Ã _ ~<- ~. -~. . . _ ::: ~ ~ ~=t ~~.~ ; =
Sam~Ie Number _~Sa .!e ID ATiris:t Conc . '; . : wPetlet sie ny ~S
50 KV 0.01 M 70 I 8 8%
51 KV 0.05M 50 I 4 3%
52 KV 0.1M 40 I 4 2%
53 KV 0.5M 50 I 3 2%
100781 Conclusion: The optimal concentration of Tris-HCI at the optimal pH of
8.5 is
0.0] M. Although the recovery rate is low, it is the highest recovery seen for
samples using
only the Tris-HCI buffer without the addition of ProK.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
100791 Example 4
Purpose: To determine the proper incubation temperature for the new elution
method. The
optimal ProK concentration was detei-mined to be 0.001 mg/ml at a Tris-HCI
concentration of
0.5M. This experiment is also designed to verity that the optimal ProK
concentration of 0.001
mg/mi is still effective at a Tris-HCI concentration of 0.01 M.
Method:
= 750pl of 0.05M and 0.01 M Tris pH 8.5 and 15 l of 0.001 mg/ml ProK were
added to
the samples and voi-texed for 20 sec. The samples were centrifuged for 12 sec
and
were incubated in a 37 watei- bath or at room temperature for I hr. The
remainder of
the procedure was the sanie as Examples 2 and 3 above_
= Sample size: n=1
Results:
y y
~ ~sr~~~~r ~~_
Tm= 's?C u tKtReco_v_er,
S.am IelNumber~ . S ' am .le. ID
79 CJ RT 0.05M 0.001 18 10%'*
80 CJ RT 0.01 M 0.001 25 14%`*
81 CD 37 C 0.05M 0.001 18 10%**
82 CD 37 C 0.01 M 0.001 15 9%*'
Conclusion: The difference in Tns-HCl concentration does not impact the
function of the
ProK enzyme. The procedure seems to work equally well at 37 C or at room
temperature. It
would be best to continue performing this assay at room temperature to avoid
using the water
bath.
** Recovery in this example is lower than expected most likely due to fewer
numbers of
spen:n initially added to the mock swabs than was anticipated. The targeted
number of sperm
added to each swab was - 1400, but was probably much lower due to inherent
difficulties
dealing with diluted sperm samples. More accurate sperm counts were determined
for the
reproducibility experiments shown in Example 11.
100801 Example 5
Purpose: To detetmine the optimal incubation time for the assay at room
temperature.
Method:
= 750p1 of 0.01 M Tris-HCI and 15 1 of 0.001 mg/ml ProK were added to the
samples
and vortexed for 20 sec. The samples were centrifuged and incubated at room
temperature for the designated amount of time. The remainder of the procedure
was
carried out as in Example 2.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
21
= Sample size: n=l.
Results:
y
~ 1:5 v.1 iy,t,~..~~r,~-te+ .~ #n' e~*Z s.u5'_ =~z ~ a ~ . . ;.., . : .~ . .
,. .: - . :.:. . . _x = -:'. - ' ,t .+.. S ''=s:~~,
=:;Pro: K = T.em . ~ T~me~~ ~~ ~ C"punt~~ fRecove_
103 UA 0.01 M Tris 8.5 0.001 RT lhr 16 32%
104 UA 0.01M Tris 8.5 0.001 RT 2hr 17 34%
105 UA 0.01 M Tris 8.5 0.001 RT 4hr 45 90%
106 UA 0.01M Tris 8.5 0.001 RT overnight 50 100%
Conclusion: Using 0.01 M Tris-HCI atpH 8.5 and 0.001 mg/ml of ProK, incubation
for 4 hrs
at room temperature is working very well. An almost full recovcry of sperm can
be observed
after long incubation periods of either four hrs or overnight. Optimizing the
sonication time
may help decrease the incubation time so that samples can be processed more
efficiently.
10081) Example 6
Purpose: To determine the optimal sonication time. A 1 hr 45 min incubation
time will be
used to help determine how much of an improvement sonication can provide. If a
four hr or
overnight incubation were used, the impact of the sonication could not be
properly gauged
since the recovery at these incubation times is already extremely high.
Method:
= 750 l of 0.O1M Tris-HCI pH 8.5 and l 5 1 of 0.001mg/ml ProK were added to
the
samples and vortexed for 20 sec. The samples were centrifuged for 12 sec to
reach
13,200 rpm and were incubated at room temperature for 1 hr and 45 min. The
remainder of the procedure was carried out as above (see Example 2).
= Sample size: n=2
Results:
~
~Y Sonlcatlon
~=
ample , _ Sam _le ID ~Buffer r- pH~~ ~Pro=K ; Tem . <~ l~TAme i . ~ time:
~Gouna r-Rec.o_v_e ,
129 DB 0.01 M Tris 8.5 0.001 RT 1:45 1 min 41 82%
130 DB 0.01 M Tris 8.5 0.001 RT 1:45 1 min 32 64%
131 DB 0.01 M Tris 8.5 0.001 RT 1:45 2.5min 38 76%
132 DB 0.01M Tris 8.5 0.001 RT 1:45 2.5min 40 80%
133 DB 0.01 M Tris 8.5 0.001 RT 1:45 5min 19 38%
134 DB 0-01 M Tris 8.5 0.001 RT 1:45 5min 11 22%
135 DB 0-01 M Tris 8.5 . 0.001 RT 1:45 10min 23 46%
136 DB 0.01 M Tris 8.5 0.001 RT 1:45 10min 13 26%
100821 Conclusion: The highest average percent recovery is observed after 2.5
min of
sonication. Literature and previous experience have shown that the water
inside the sonicator
can increase in temperature after extended periods of use. This increase in
temperature can
diminish the intensity of the sonic waves making the sonication less
efficient. Therefore, it
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
22
was necessary to change the water after every sonication. This is a very
imponant fact that
needs to be stressed to the personnel that will be performing this task.
100831 Example 7
Purpose: To determine the optimal incubation time at room temperature using a
sonication
time of 2.5 min.
Method:
= 750p1 of 0.01 M Tris-HCI pH 8.5 and l 5pl of 0.001 mg/ml ProK were added to
the
samples and vortexed for 20 sec. The samples were centrifuged for 12 sec to
reach
13,200 rpm and were incubated at room temperature for the designated amounts
of
time. The remainder of the procedure was carried out as above (see Example 2).
Results:
y
~ : ,~ ontc~on ~ t~
Szam Ie Buff er,~ pH ,.N . Pr_o,K Tem ~tme
137 KZ 0.01 M Tris 8.5 0.001 RT lHr 2.5min 32 64%
138 KZ 0.01M Tris 8.5 0.001 RT 1Hr 2.5min 41 82%
139 KZ 0.01 M Tris 8.5 0.001 RT 2Hr 2.5min 59 118%
140 KZ 0.01 M Tris 8.5 0.001 RT 2Hr 2.5min 48 96%
141 KZ 0.01 M Tris 8.5 0.001 RT 3Hr 2.5min 16 32%
142 KZ 0.01 M Tris 8.5 0.001 RT 3Hr 2.5min 42 84%
Conclusion: The optimal incubation time is two hrs. followed by a 2.5 min.
sonication.
100841 ADDITIONAL STUDIES
Observation:
By optimizing every aspect of the assay the number of sperm recovered from the
swabs has
increased significantly. Due to the fact that many sexual assault samples
contain a large
number of epithelial cells, often the ability to visualize eluted sperm cells
on the microscope
slide is difficult due to the over abundance of epithelial cells compared to
spermatozoa. We
have postulated that pre-treatment of the sexual assault swab prior to the new
improved sperm
elution method may assist in removing many of the epithelial cells. Doing this
would help
create a less dense cell pellet following elution thereby making the
visualization of sperm
much easier. In addition, this pre-treatment might possibly lead to cleaner
STR profiles.
Various enzymes and buffers will be tested to determine their effectiveness at
removing
epithelial cells without eluting the sperm cells. An example of one such
experiment is shown
below.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
23
[0085] Example 8
Purpose: To determine if a pre-treatment method could be used to significantly
decrease the
number of epithelia] cells observed on the slide without decreasing the
recoveiy of-the sperm
cells.
Method:
= Performed pre-treatment with various solutions (referred to as MO-Lite)
followed by
the new sperm elution method (referred to as MO-Classic) on 4 mock sexual
assault
samples:
= Samples 210 and 211 were pre-treated with Beta-amylase Barley as one MO-Lite
solution.
= Samples 212 and 213 were pre-treated with lX PBS (pH 7.4) as a second MQ-
Lite
solution.
= Add 7S0ul of either MO-Lite buffer to the swab tubes and incubate at RT for
1 hour.
= Following the 1 hr. incubation, the samples were placed at 95 C to
inactivate the ProK.
In this example only samples 211 and 213 were placed at 95 C for 5m1n,
vortexed for
20 sec, and quick spun.
= The swab was transferred into a spin basket and centrifuged for 5min at
13,200 rpm.
The swab was removed from the MO-Lite buffer and incubated in MO-Classic using
the normal procedure for 2 hours at RT. The sample was then vortexed for 20
scc,
quick spun, placed in a spin basket and centrifuged for 5 min. at 13,200 rpm.
The
swab was placed into a new tube and stored. A slide was made from the sperm
pellet
as previously described.
Results:
~
{ "~~s
>'~T'rgeted Number,~,~ Mo-Lite Buffer Spe~m Sperm '
Sam ~le . ~Sam Ie;ID~ f~of~S erm Used~ ~~ ZA ~Cou.nt~~ ~ Reco_ve . ~
210 PN 400 count Am lase Barley 25 50%
211 PN 400 count Am lase Barley 4 12%
212 PN 400 count PBS 32 53%
213 PN 400 count PBS 12 24%
[0086] Conclusion: Pre-treating the sexual assault swabs with PBS does not
negatively
impact the ability to recover sperm from the swab. Analysts that perform sperm
search
routinely commented that the slides were much easier to read following pre-
treatment with
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
24
MO-Lite. The incubation at 95"C fot- 5 min was determined to be detrimental
and was
eliminated from the protocol.
100871 Example 9
Purpose: To detern-iine if pre-treatment with PBS (MO-Lite) has any negative
effect on
pcrforming P30 assays.
Method:
= 5 l of semen was added to cotton swabs; samples were allowed to dry for 2
weeks.
= Pretreated with MO-Lite (1XPBS) and MO-Classic on these samples.
= Performed P30 from MO-Lite supernatant and MO-Classic supernatant using the
standard Orchid Cellmark P30 protocol.
= Sample size: n = 4
Results:
y
' ~ t San m Ie*111D~ ., Sam le, ~
.' Coun 1P30
... ...._...... n .- . .._ ...... u... N
218 PN 5 I semen Many POS
219 PN 5 I semen Many POS
220 PN 5 I semen Many POS
221 PN 5 1 semen Many POS
Conclusion: P30 assays performed on the supernatant from either the MO-Lite or
MO-
Classic treatments were unaffected.
100881 Example 10
Purpose: To determine if DNA isolated from sperm eluted from cotton swabs
using the new
sperm elution method will yield acceptable DNA profiles.
Method:
= - 400 sperm placed on cotton swab
= Sperm were eluted from the swab using the protocol as described above with
MO-Lite
Buffer followed by MO-Classic Buffer.
= DNA was extracted from the eluted sperrn pellet using known DNA extraction
methods.
= The DNA was amplified using the Identifiler kit (ABI) under standard, full-
strength
conditions.
= Figure 3 illustrates a GenoTyper electropherogram showing an acceptable DNA
profile generated from the sperm eluted utilizing an embodiment of the method
described herein.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
Conclusion: DNA extracted from cells eluted using the new sperm extraction
protocol
described herein yielded acceptable DNA profiles.
100891 Observation:
During the course of these validation studies, a great deal of day-to-day
fluctuation in the
number of spetm present in the same stock solution of washed and diluted sperm
was
observed. In order to obtain fairly consistent and reliable sperm counts, it
was necessary to
prepare a small stock solution of diluted sperm. Analysts would sample this
stock solution to
prepare new mock sexual assault samples and perfonm a sperm count at the same
time.
Predetermined sperm counts were considered unreliable even if the counts were
determined on
the same day as the swabs were being produced. All sperm counts were performed
at the
exact same time of swab production.
100901 Example ll
Purpose: To detenmine if the new method of elution yields a reproducible
number of sperm.
Method:
= 10 mock sexual assault samples were subjected to the new spe-m elution
procedure.
= The swabs were prepared by making a stock solution of sperm and adding 1 O l
of this
solution to each swab. In between making each swab, a 5 l aliquot was applied
to a
microscope slide for staining and counting.
~ ~ s ~ ~ , ~* ~ ~aTargeted Sperm ` 2 Slide~."~Courtt Totai N~'u"mber' of~ ~"
i`Sample~ Co nt on~`~Swti (uted from Sperm`
Sam le a~ed on 5 I a ~ 's~w b ,~ ~ ~Recove.r,ed~~h ~ fo:RecovBy a El 1 PN
660 46 414 63%
2 PN 546 32 323 59%
3 PN 572 44 449 79%
4 PN 498 45 405 81%
5 PN 538 41 414 77%
6 PN 494 36 400 81%
7 PN 514 32 352 68%
8 PN 432 36 403 93%
9 PN 440 29 319 73%
10 PN 314 26 289 92%
Conclusion: The recovery of sperm ranged from 59% - 93% for an average
recovery of
76.5%. This rcprescnts a 15-20-fold improvement in sperm recovery compared to
the current
method.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
26
100911 Example 12 - Non-Probative Evidence Samples
Purpose: Determine if sperm from actual case work samples can be eluted using
this
procedure. (Note: Many of the samples examined in this study have already been
through
sperm elution and some have actually been through differential extraction)
Method:
In the first set of cases, 6 sexual assault samples from a total of 5 cases
that were previously
processed using the old method were examined using Ihe new sperm elution
method. Since
these samples had already been through a previous sperm elution, no pre-
treatment with MO-
Lite was performed.
= Place the swab in a microcentrifuge tube containing 750 1 of 0.01 M Tris-HCI
and
15p1 of 0.001 mg/ml of Proteinase K.
= Vortex for 25 seconds and quick spin. Incubate the samples for 2 hours at
RT. After
the first hour, briefly vortex the samples and then allow the samples to
continue
incubation for the remainder of the 2 hr duration.
= Sonicate the samples for 2.5 min, vortex for 25 sec, and quick spin to get
the contents
to the bottom of the tube.
= Transfer the swab into spin basket, centrifuge for 5 min. at 13,200 rpm.
= Remove the swab and the supematant; leave the pellet in 40 1 buffer
solution.
= Using a 5 l aliquot from the sperm pellet make and stain the slide and
perform sperm
search.
= Perform differential extraction according to current protocol and extract
the sperm
pellet using EZ-1 extraction protocol (Qiagen).
= Quantitate the DNA from the sperm pellet using Quantifiler-Y (ABI).
= Amplify DNA using the Identifiler kit (ABI) according to standard protocol.
Results:
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
27
QUANT Y !
Si41MPLE'ID F%RESULTS " PROFILE~~, RESULTS t RESULT~S PROFILE :~
.=:".
.:.
NEW~ :~ ~PROVEp~
:_<.=. ~
:. ~ METtHOD~d~ ~METH.O,D~'.~ M,ETHOD~~~~ z ~:.. 1~CLMETHOD x ._. . ,..
NO
AL016-01.02.2 NEG PROFILE RARE 0.0008n / I NO PROFILE
NO PARTIAL
AL021-01.01.2 NEG PROFILE ~RARE 0.0018n / I PROFILE
NO
AL022-01.01.2 NEG PROFILE NEG 0 nglpl DID NOT AMP.
NO
AL024-01.01.2 NEG PROFILE NEG 0 n/ I DID NOT AMP.
NO AL035-01.01.2 NEG PROFILE RARE . 0 n/ 1 NO PROFILE
NO
AL035-02.01.2 NEG PROFILE NEG 0 n/ 1 DID NOT AMP.
In thc second set of cases, 19 sexual assault samples from a total of 10 cases
that had been
previously extracted using the old elution method were examined using the new
sperm elution
method. The procedure was performed exactly as with the first set of non-
probative casework
samples above. Some of these cases had previously yielded positive sperm
results and some
had also yielded full or partial DNA profiles.
Results:
SAMP~E ID RESULTS PROFILE RESULTS = RESULTS PROFILE' Sa+
4.0
IMNROVED`= n +>~ ~ Q ~ ~ ~ ~ ~ METHOD''
4 f a~ ` ~ LD 4D K~ OLD M TIiOD METHOP ¾
~
.:., _ ...,..,. . . _ . saesxi
NO
MA035-01.01.1 RARE PROFILE RARE 0 ng/pt DID NOT AMP
NO NOT
MA036-01.01.1 RARE PROFILE NEG EXTRACTED DID NOT AMP
MA036.02.01.1 RARE FULL RARE 0.005 n/of FULL
MA031-01.01.1 RARE FULL = OCCASIONAL 0.039 n/ 1 FULL
MA031-02.01.1 RARE FULL MANY 0.19 n/=I,".' FULL
NO NOT
MA031-03.01.1 RARE PROFILE NEG EXTRACTED DID NOT AMP
NO NOT
MA031-04.01.1 RARE PROFILE NEG EXTRACTED DID NOT AMP
MA017-01.01.1 RARE PARTIAL RARE 0 n/ I DID NOT AMP
NOT
MA023-01.01.1 RARE PARTIAL NEG EXTRACTED DID NOT AMP
MA024-01.01.1 FEW FULL OCCASIONAL 0.007.n / I FULL
NO
MA025-01 _01.1 RARE PROFILE RARE 0 n/ I DID NOT AMP
NO
MA043-01.01.1 RARE PROFILE RARE 0 n/ I DID NOT AMP
MA043-02.-1.1 RARE FULL FEW 0.006 n/ I FULL
NO
MA013-01.01.1 RARE PROFILE RARE 0 n/ I DID NOT AMP
NO
MA056-01.01.1 RARE PROFILE RARE 0 n/ 1 DID NOT AMP
NO
MA056-02.01.1 RARE PROFILE RARE 0 ng/pl DID NOT AMP
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
28
100921 Conclusion: The new sperm elution method appears to work well on actual
case
work samples as evidenced by the results above. In every situation where a
full profile had
been seen previously, the new sperm elution method also yielded a full
profile, in spite of the
fact that these swabs had previously been subjected to the current sperm
elution method, as
well as, differential DNA extraction. In addition, there were a few cases
where the original
swabs were considered to be negative for sperm and the new sperm elution
technique showed
a few sperm were actually present.
100931 Summary:
Following an observation that the current sperm clution mcthod was very
inefficient (3-8%) at
eluting sperm from cotton swabs, our Research and Development team set out to
investigate a
number of factors that might improve our ability to elute sperm from sexual
assault swabs.
The following factors were examined: Salt concentration, pH, various
proteases, several
caotropic agents, incubation times and temperatures, and sonication. Based on
our studies and
the findings of this validation, we have selected the following general
procedure for sperm
elution from cotton swabs. For a detailed procedure, see Appendix 2.
= Pre-treatment of swab in PBS (pH7.4) for 30 min.
= 750 N1 of 0.01 M Tris-HCI buffer pH 8.5
= 0.020 ng/ l Proteinase K
= Two hour incubation at room temperature
= 2_5 min. sonication
100941 Together, these factors have increased the sperm recovery from cotton
swabs from <
10% using the current method to > 70%.
(0095] Example 13
Titrate the amount of ProK (0.1 mg/ml, 0.05mg/mi, 0.025mg/mi, and 0.01 mg/ml)
= Add 750pl of PBS buffer pH 7.4 (really IX buffer pH 6.8) and 15pl of Pro K
added.
The samples were vortexed for a 20 sec. count, centrifuged for 12 sec. and
were
incubated at 37 C or 56 C for 1 hour. The samples were vortexed again for a 20
sec.
count. They were quick spun again for 12 sec. The swabs were placed in a spin
basket, using a pipet tip. They were centrifuged at 13,200 for 5 min. The
basket was
placed in a new tube and the supernatant removed. The pellet left in the
original tube
included -40p1 of supernatant. The pellet was resuspended using a 20 1 pipet.
A 5 l
aliquot was placed on a microscope slide and heat fixed for 5 minutes at 65 C.
The
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
29
slides were stained with the red dye for 15 min., rinsed with water, stained
with the
green dye for 15 sec., and rinsed with ethanol.
;M.r:37~+'r ple~~" ~~>.,e:rwn~ ~ ,.r Sa~. rc R 4a --.rs~.~ya-~~w;~.v 7a^~+ i~n
.. . ~ _
,~- ,= *.. , ,.
Number~ m le,_ID,~ P,_,o,K~and Tem ,õ S ecm C
`~Sam ount Recove:f .>
13 VW 0.1, 37 3 2%
14 VW 0.1, 56 6 3%
15 MN 0.05, 37 10 6%
16 MN 0.05, 56 11 6%
17 VW 0.025, 37" 9 5%
18 VW 0.025, 56 3 2%
19 VW 0.01, 37 12 7%
20 VW 0.01, 56 6 3%
100961 Example 14
6/27/06
Redo pH range in PBS 7.5-8_5 with and without sonication
~ Sam~ple Sample,Buffer,
~
t, ~Numbel r. : ~ID -`` a and~pH~ ~ ~Sonication ~ ~ermRecoye_.
21 MK PBS 7.5 No 0 0%
22 MK PBS 8.0 No 4 2%
23 MK PBS 8.5 No 3 2%
24 MK PBS 7.5 Yes 4 2%
25 MK PBS 8.0 Yes 23 13%
26 MK PBS 8.5 Yes 15 9%
100891 Example 15
PBS pH 7.5, incubate with 0.01 mg/ml Prok for I hour at 37 C with and without
sonication
_ p ~ m Ie~iD~ _~1 , $perm "
and~P,~roK S_o..nic_ation C.o,unt RecoYe,
27 JC PBS, 7.5, 0.01m /mI Prok No 26 15%
28 JC PBS, 7.5, 0.01 m/ml Prok No 14 8%
29 JC PBS, 7.5, 0.01 m/mI Prok Yes 56 32%
30 JC PBS, 7.5, 0.01m /ml Prok Yes 72 41%
100901 Example 16
6/28/06
PBS pH 8.0 with I hour incubation at 37 C with 0.01 mg/ml and 0.005mg/ml ProK
minute sonication with and without vortex
= 750p1 of PBS were added to the samples along with 15 1 of ProK. The samples
were
vortexed for a 20 sce. count and centrifuged for 12 sec. and were incubated at
37 C for
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
I hour. The samples were vortexed again for a 20 sec. count. They were quick
spun
for 12 sec and sonicated for 5 minutes. The tubes were vortexed again for 20
sec. and
quick spun for 12 sec. The swabs were placed in a spin basket, using a pipet
tip. They
were centrifuged at 13,200 for 5 min. The basket was placed in a new tube and
the
supernatant removed. The pellet left in the original tube included --40 I of
supematant. The pellet was resuspended using a 20 1 pipet. A 5p1 aliquot was
placed
on a microscope slide and heat fixed for 5 minutes at 65 C. The slides were
stained
with the red dye for ] 5 min., rinsed with water, stained with the green dye
for 15 sec.,
and rinsed with ethanol.
erm~Co,unt~ ~Rec,o_xer
Sa p :'Nu b`e~ ~~Sam~ le D`~ "Pro; K~ 9Vortex~
37 TB 0.01 No 49 28%
38 TB 0.01 Yes 72 41%
39 TB 0.005 No 48 27%
TB 0.005 Yes 51 29%
100921 Example 17
0.05M Tris-HCI pH 8_5 with 1 hour incubation at 37 C with 0.01 and 0.005mg/m1
ProK 5
minute sonication with and without vortex
~ 750 l of Tris were added to the samples along with 15p1 of ProK. The samples
were
vortexed for a 20 sec. count and centrifuged for 12 sec. and were incubated at
37 C for
l hour. The samples were vortexed for a 20 sec. count. They were quick spun
for 12
sec and sonicated for 5 minutes. The tubes wcre vortexed again for 20 sec. and
quick
spun for 12 sec. The swabs were placed in a spin basket, using a pipet tip.
They were
centrifuged at 13,200 for 5 min. The basket was placed in a new tube and the
supernatant removed. The pellet left in the original tube included -40p1 of
supernatant. The pellet was resuspended using a 20pl pipet. A 5pl aliquot was
placed
on a microscope slide and heat fixed for 5 minutes at 65 C. The slides were
stained
with the red dye for 15 min., rinsed with water, stained with the green dye
for 15 sec.,
and rinsed with ethanol.
x, 3 eim Count ,. Re.c.o,v_e ,~
~``~~ ~~~ ~ le ID~ V,orte:
;;$am te;Number Opu'
41 JH 0.01 No 32 18%
42 JH 0.01 Yes 67 38%
43 JH 0.005 No 96 55%
44 JH 0.005 Yes 102 58%
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
31
100931 Example 18
6/29/06
PBS pH 8 room temperature incubation at I hr, 2hr, 4hr, 8hr, and ovemight with
sonication
and vortexing
= 750 1 of PBS were added to the samples and vortexed for a 20 sec. count. The
samples were centrifuged for 12 sec. and were incubated at room temperature
according to their specific length of time. The samples were vortexed for a 20
sec.
count. They were quick spun for 12 sec and sonicated for 5 minutes. The tubes
were
vortexed again for 20 sec. and quick spun for 12 sec. The swabs were placed in
a spin
basket, using a pipet tip_ They were centrifuged at 13,200 for 5 min. The
basket was
placed in a new tube and the supernatant removed. The pellet left in the
original tube
included -40 1 of supernatant. The pellet was resuspended using a 20 1 pipet.
A 5 l
aliquot was placed on a microscope slide and heat fixed for 5 minutes at 65 C_
The
slides were stained with the red dye for 15 min., rinsed with water, staincd
with the
green dye for 15 sec., and rinsed with ethanol.
~s~' Sample ! g ~mpte ~~ "IncubaUon ~
rM
, WPeile,txsize~.,.,,~ ntr Er' õeco_v_e-,
45 JN lhr 40 I 22 13%
46 JN 2hr 40 I 9 5%
47 JN 4hr 70 I 8 8%
48 JN 8hr 70 I 3 3%
49 JN overnight 50 I 17 12%
100951 Example 19
0.05M Tris pH 8.5 with 0.0025 and 0.001mg/ml ProK incubate at 37 C for 1 hour
with
sonication and vortexing.
= 750p1 of Tris were added to the samples along with 15p1 of ProK. The samples
were
vortexed for a 20 sec. count and centrifuged for 12 sec. and were incubated at
37 C for
1 hour. The samples were vortexed for a 20 sec. count. They were quick spun
for 12
sec and sonicated for 5 minutes. The tubes were vortexed again for 20 sec. and
quick
spun for 12 sec. The swabs were placed in a spin basket, using a pipet tip.
They were
centrifuged at 13,200 for 5 min. The basket was placed in a new tube and the
supernatant removed. The pellet left in the original tube included ---40 1 of
supernatant. The pellet was resuspended using a 20 1 pipet. A 5 1 aliquot was
placed
on a microscope slide and heat fixed for 5 minutes at 65 C. The slides were
stained
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
32
with the red dye for 15 min., rinsed with water, stained with the green dye
for 15 sec.,
and rinsed with ethanol.
Pro.,K. S erm ;Count :;, Reccw.er
,Sam le>Number Sam Ie IDn }.
54 JC 0.0025 97 55%
55 JC 0.001 111 63%
100961 Example 20
PBS pH 8.0 with 0.01 mg/ml ProK incubate at 37 C (]hr, 2hr, 4hr, overnight)
with sonication
and vortexing
= 750p1 of PBS wei-e added to the samples along with 15 1 of ProK and vortexed
for a
20 sec. count. The samples were centrifuged for 12 sec. and were incubated at
37
according to their specific length of tinie. The samples were vortexed for a
20 sec.
count. They were quick spun for 12 sec and sonicated for 5 minutes. The tubes
were
vortexed again for 20 sec_ and quick spun for 12 sec. The swabs were placed in
a spin
basket, using a pipet tip. They were centrifuged at 13,200 for 5 min. The
basket was
placed in a new tube and the supernatant removed. The pellet left in the
original tube
included -40 l of supernatant. The pellet was resuspended using a 20p1 pipet.
A 5pl
aliquot was placed on a microscope slide and heat fixed for 5 minutes at 65 C.
The
slides were stained with the red dye for 15 min., rinsed with water, stained
with the
green dye for 15 sec., and rinsed with ethanol.
ia~-~~'^_#`"x:'~u" ts1#= s . - s -
;,~ 144T ntron,;Time _` ~S m'Co nt ~_t Recove~,=jg
.~MSam Ie Number t Sample ID
56 KV lhr 64 37%
57 CS 2hr 22 13%
58 PN 4hr 0%
59 JN overnight 26 15%
100981 Example 21
7/3/06
Exp. 12
0.05M Tris pH 8.5 with ProK (0.005, 0.0025, 0.001mg/mI) 1 hour incubation at
37 C with
vortex and sonication
= 750p1 of Tris were added to the samples along with 15 l of ProK. The samples
were
vortexed for a 20 sec. count and centrifuged for 12 sec. and were incubated at
37 C for
1 hour. The samples were vortexed for a 20 sec. count. They were quick spun
for 12
sec and sonicated for 5 minutes using a foam insert. The tubes were vortexed
again
for 20 sec. and quick spun for 12 sec. The swabs were placed in a spin basket,
using a
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
33
pipet tip. They were centrifuged at 13,200 for 5 min. The basket was placed in
a new
tube and the supernatant removed. For samples 67-69 the supernatant was added
to
the swab in the spin basket and spun again for 5 minutes. The pellet left in
the original
tube included -50 1 of supernatant. The pellet was resuspended using a 20pl
pipet. A
5N1 aliquot was placed on a microscope slide and heat fixed for 5 minutes at
65 C.
The slides were stained with the red dye for 15 min., rinsed with water,
stained with
the green dye for 15 sec., and rinsed with ethanol.
. . . .: ':.n w>. .. . ; r *~ ~c.ti% ' i .
; Sam le Nuinber :Sam Ie.ID, bExt.r,a Rnse =~S` erm Gount'~ ecover,
64 AM 0.005 No 25 14%
65 AM 0.0025 No 0%
66 AM 0.001 No 0%
67 AM 0.005 Yes 12 7%
68 AM 0.0025 Yes 0%
69 AM 0.001 Yes 0%
100991 Example 22
Tris pH 8.5 (0.05M, 0.01 M, 0.005M 0.0025M, 0.001 M) 1 hour incubation at room
temperature with no ProK with vortexing and sonication
= 750p1 of Tris were added to the samples and vortexed for a 20 sec. count.
The
samples were centrifuged for 12 sec. and were incubated at room temperature
for I
hour. The samples were vortexed for a 20 sec. count. They were quick spun for
12
sec and sonicated for 5 minutes with a foam insert. The tubes were vortexed
again for
20 sec. and quick spun for 12 sec. The swabs were placed in a spin basket,
using a
pipet tip. They were centrifuged at 13,200 for 5 min. The basket was placed in
a new
tube and the supernatant removed. The pellet left in the original tube
included -40 1
of supematant. The pellet was resuspended using a 20 1 pipet. A 5 1 aliquot
was
placed on a microscope slide and heat fixed for 5 minutes at 65 C. The slides
were
stained with the red dye for 15min., rinsed with water, stained with the green
dye for
15 sec., and rinsed with ethanol.
i'6~mF~^ . . ..~-q"'~,. r i', Serm..C,-~.out..ni= .:.." '~^-'s~~t. <. [e
~~Sam _le. Number . ~S.am.Ie,ID , Conc. ~, Recove
70 SB 0.05m 4 2%
71 SB 0.01M 7 4%
72 SB 0.005M 4 2%
73 SB 0.0025M 9 5%
74 SB 0.001 M 6 3%
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
34
1001001 Example 23
7/5/06
0.05M Tris pH 8.5 below 0.001 nig/ml ProK 1 hour incubation @ 37 C with
sonication and
vortex, using new concentrations of ProK
= 750p I of Tris were added to the samples and vortexed for a 20 sec. count.
The
samples were centrifuged for 12 sec. and were incubated at room temperature
for I
hour. The samples were vortexed for a 20 sec. count. They were quick spun for
12
sec and sonicated for 5 minutes using a foam insert. The tubes were vortexed
again
for 20 sec. and quick spun for 12 sec. The swabs were placed in a spin basket,
using a
pipet tip. They were centrifuged at 13,200 for 5 min. The basket was placed in
a new
tube and the supernatant removed. The pellet left in the original tube
included -40 1
of supernatant. The pellet was resuspended using a 20 1 pipet. A 5pl aliquot
was
placed on a microscope slide and heat fixed for 5 minutes at 65 C. The slides
were
stained with the red dye for 15 min., rinsed with water, stained with the
green dye for
15 sec., and rinsed with ethanol.
= Sample 75 was re-sonicated without using the foam insert and the slide
remade.
.. ~~ ~ ~ - - ~
z
~ ~ -S..am _Ie. Number , ,nSam Iea1D~ ro K ~, ~S . er,m~Count R Recov,e ~xig
75 CJ 0.001 12 7%
76 CJ 0.0005 9 5%
77 CJ 0.00025 9 5%
78 CJ 0.0001 19 11%
75-Remake CJ 0.001 20 11%
100102] Example 24
7/6/05
Used samples with sperm counts of 340-360_ Samples 91-94 contain newly
prepared Tris
= 750 1 of buffer and 15p1 ProK wcre added to the samples and vortexed for a
20 sec.
count. The samples were centrifuged in the hood and were incubated at the
temperature and time specified. The samples with ProK were incubated at 95 C
for 10
min. The samples were vortexed for a 20 sec. count. They were quick spun in
the
hood and sonicated for 5 minutes using a foam insert. The swabs were placed in
a
spin basket, using a pipet tip. They were centrifuged at 13,200rpm for 5 min.
The
basket was placed in a new tube and the supematant removed. The pellet left in
the
original tube included -40 1 of supernatant. The pellet was resuspended using
a 20 1
pipet. A 5 l aliquot was placed on a microscope slide and heat fixed for 5
minutes at
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
65 C. The slides were stained with the red dye for 15min., rinsed with water,
stained
with the green dye for 15 sec., and rinsed with ethanol.
~~
Sam ,le Sa D,e Buff,er ~ r H:` P o~K Tem~ Trm'e ',Coui~
83 PN PBS 8 0.001 56 C 30min 5 10%
84 PN PBS 8 0.05 56 C 30min 21 42%
85 PN 0.05M TRIS 8.5 0.001 56 C 30min 5 10%
86 PN 0.05M TRIS 8.5 0.05 56 C 30min 2 4%
Room
87 PBS 8 Temp lHr 2 4%
88 PBS 8 fridge 23Hr Inc. 0%
Room
89 0.05M TRIS 8.5 Temp lHr 1 2%
90 0.05M TRIS 8.5 fridge 23Hr 2 4%
'91 PBS 8 0.001 41 lHr 7 14%
92 PBS 8 0.05 41 lHr 29 58%
93 0.05M TRIS 8.5 0.001 41 lHr 31 62%
94 0.05M TRIS 8.5 0.05 41 lHr 28 56%
Room
95 PBS 8 0.05 Tem lHr 19 38%
Room
96 Jena PBS 8 0.001 Temp lHr 1 2%
Room
97 Jena 0.01 M TRIS 8.5 0.05 Temp lHr 22 44%
Room
98 Jena 0.01 M TRIS 8.5 0.05 Temp lHr 14 28%
Room
98-1 Jena 0.01M TRIS 8.5 0.05 Tem 2Hr 37 74%
Room
98-2 Jena 0.01M TRIS 8.5 0.05 Temp 2Hr 17 34%
1001031 Example 25
0.05M Tris pN 8.5 with 0.001 mg/ml ProK at 41 C for 5 min, 15 min, 30 min, 1
hr.
~ 750 l of buffer and 15 1 ProK were added to the samples and vortexed for a
20 sec.
count. The samples were centrifuged in the hood and were incubated at 41 C
for the
designated time. The samples were incubated at 95 C for 10 min. The samples
were
vortexed for a 20 sec. count. They were quick spun in the hood and sonicated
for 5
minutes using a foam insert. The swabs were placed in a spin basket, using a
pipet tip.
They were centrifuged at 13,200 rpm for 5 min. The basket was placed in a new
tube
and the supematant removed. The pellet left in the original tube included -40
l of
supernatant. The pellet was resuspended using a 20 1 pipet. A 5 l aliquot was
placed
on a microscope slide and heat fixed for 5 minutes at 65 C. The slides were
stained
with the red dye for 15 min., rinsed with water, stained with the green dye
for 15 sec.,
and rinsed with ethanol.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
36
;.Sam le :Number "'Sam le"tD";,~ Time'>' 'nvS e~m Count~~'_~r Reco er,~`~~
99 JLC 5min 2 4%
100 JLC 15min 2 4%
101 JLC 30min 3 6%
102 JLC lhr 11 22%
1001041 Example 26
7/7/06
= 750 I of buffer and 15 1 ProK were added to the samples and vortexed for a
20 sec.
count. The samples were centrifuged in the hood and were incubated at
designated
time and temperature. The samples were incubated at 95 C for 10 min. The
samples
wei-e vortexed for a 20 sec. count. They were quick spun in the hood and
sonicated for
minutes using a foam insert. The swabs were placed in a spin basket, using a
pipet
tip. They were centnfuged at 13,200 rpm for 5 min. The basket was placed in a
new
tube and the supernatant removed. The pellet left in the original tube
included --40 1
of supernatant. The pellet was resuspended using a 20 1 pipet. A 5 1 aliquot
was
placed on a microscope slide and heat fixed for 5 minutes at 65 C. The slides
were
stained with the red dye for 15 min., rinsed with water, stained with the
green dye for
sec., and rinsed with ethanol.
_. _ ._ .'~ '. " ~=
: Sam le "I Sain le ID~~ :~ . Buf(er x, sI; H ~}~ Pro,K '~ eTemp.. ,_ - Time
~Gount ~ g iReco.v,e ~
9
0.01M Room
103 UA Tris 8.5 0.001 Tem lhr 16 32%
0.01 M Room
104 UA Tris 8.5 0.001 Tem 2hr 17 34%
0.01 M Room
105 UA Tris 8.5 0.001 Temp 4hr 45 90%
0.01M Room overnig
106 UA Tris 8.5 0.001 Temp ht 50 100%
0.01 M Room
107 RF Tris 8.5 0.05 Tem lhr 24 48%
0.01 M Room
108 RF Tris 8.5 0.05 Temp 2hr 16 32%
0.01 M Room
109 RF Tris 8.5 0.05 Tem 4hr 17 34%
0.01 M Room overnig
110 RF Tris 8.5 0.05 Temp ht 20 40%
Room
111 JLH PBS 8 0.05 Temp lhr 3 6%
Room
112 JLH PBS 8 0_05 Temp 2hr 22 44%
Room
113 JLH PBS 8 0.05 Temp 4hr 8 16%
Room overnig
114 JLH PBS 8 0.05 Temp ht 24 48%
Room
115 AJ PBS 8 0.01 Temp lhr 1 2%
Room
116 AJ PBS 8 0.01 Tem 2hr 5 10%
117 AJ PBS 9 0.01 Room 4hr 3 6%
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
37
= . _ _ _ .
r= ~~~
;,. <Sam le ' Sam le ID' ; Buffer .. H Prok-;' ~Tem Time ;~1Count. = Recove
Temp c
Room overnig
118 AJ PBS 10 0.01 Temp ht 7 14%
0.05M
119 SW Tris 8.5 0.001 41 C 1_5hr 8 16%
0.05M
120 SW Tris 8.5 0.001 41 C 2hr 11 22%
0.05M
121 sw Tris 8.5 0.001 41 C 4hr 6 12%
0.05M overnig
122 SW Tris 8.5 0.001 41 C ht 9 18%
Samples after 2hr incubation were sonicated after changing the water.
1001051 Example 27
7/10/06
0.01 M Tris pH 8.5 with 30 1 0.0001 mg/ml ProK at room temperature
= 750 1 of buffer, 30 1 of ProK. Vortex 20sec_ Quick spin for l2seconds to
reach
13,200 rpm. Incubate at room temperature for designated time. Incubate at 95 C
for
minutes. Vortex 20sec. Quick spin for 12seconds to reach 13,200 rpm. Sonicate
for 5minutes using foam insert and switching water after each sonication.
Transfer to
spin basket and spin at 13,200rpm for 5 minutes. Make a slide.
~K .3 ' ~19NÃ+''T-'~~
Sam L.e Sam Ie;ID~ ~Buffer~N2',H~<MP~o .. .`: Temp Thme Count~ l~e'o o.very
123 KB 0.01 M Tris 8.5 0.001 Room Temp lhr 13 26%
124 KB 0.01 M Tris 8.5 0.001 Room Temp 2hr 9 18%
125 KB 0.01 M Tris 8.5 0.001 Room Temp 3hr 22 44%
126 KB 0.01 M Tris 8.5 0.001 . Room Temp 4hr 23 46%
1001061 Example 28
0.01 M Tris pH 8.5 with 15p1 of ProK at 0.0001 mg/ml at room temperature
= 750 1 of buffer, 15 1 of ProK. Vortex 20sec. Quick spin for l2seconds to
reach
13,200 rpm. Incubate at room temperature for designated time. Incubate at 95 C
for
10 minutes. Vortex 20sec. Quick spin for 12seconds to reach 13,200 rpm.
Sonicate
for 5minutes using foam insert and switching water after each sonication.
Transfer to
spin basket and spin at 13,200rpm for 5 minutes. Make a slide.
.- , . . .
Sam le ~S m~I~ID_: ; Buff.er~i d.! P~r~K~ Time Count~ Reco~e ;
127 KB 0.01M Tris 8.5 0.0001 Room Temp lhr 2 4%
128 KB 0.01 M Tris 8.5 0.0001 Room Temp 2hr 1 2%
1001091 Example 29
0.01 M Tris pH8.5 with 30p1 of 0.001 mg/ml ProK at room temperature
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
38
= 750p1 of buffer, 30p1 ofProK_ Voi-tex 20 sec. Quick spin for 12 seconds to
reach
13,200 rpm. Incubate at room temperatul-e for designated time. Incubate at 95
C for
minutes. Vortex 20 sec. Quick spin f'or 12 seconds to reach 13,200 rpm.
Sonicate
for 2.5min. using foam insert and switching water aftcr each sonication.
Transfer to
spin basket and spin at 13,200rpm for 5 minutes. Make a slide.
a~ ~ Sample. .'. :, r
Sam te ? ~,_ < ID.?' :.: But(er pH Pro, K>' Temp::: Time_ CCouni ~QC.OVe r
0.01 M Room
143 CS Tris 8.5 0.001 Temp lHr 2.5min 32 64%
0.01 M Room
144 CS Tris 8.5 0.001 Temp lHr 2.5min 23 46%
1001101 Example 30
7/11/06
0.01 M Tri s pH 8.5 with 15 1 ProK 0.001 mg/ml at room temperature
= 750p1 of buffer, 15p1 of ProK. Vortex 20sec. Incubate at room temperature
for 30min.
Quick spin for 12seconds to reach 13,200 rpm. Incubate at room temperature for
1:30hr. Incubate at 95 C for 10 minutes. Vortex 20sec. Quick spin for
12seconds to
reach 13,200 rpm. Sonicate for designated time using foam insert. Transfer to
spin
basket and spin at 13,200rpm for 5 minutes. Make a slide.
e~Sample $o'nicaUon"~ ~' ``õ 7
Sam le õID. uHer, H& Pr:o K ,F Tem :' ' ~T~me~ time, ~ z a Count, eeove
0.01 M Room
145 EBW Tris 8.5 0.001 Temp 2Hr 0 30 60%
0.01 M Room
146 EBW Tris 8.5 0.001 Temp 2Hr 0 23 46%
0.01 M Room
147 EBW Tris 8.5 0.001 Temp 2Hr 30sec 44 88%
0.01 M Room
148 EBW Tris 8.5 0.001 Temp 2Hr 30sec 33 66%
0.01 M Room
149 EBW Tris 8.5 0.001 Temp 2Hr 1 min 20 40%
0.01 M Room
150 EBW Tris 8.5 0.001 Temp 2Hr lmin 38 76%
0.01 M Room
151 RS Tris 8.5 0.001 Temp 2Hr 1.5min dense
0.01 M Room
152 RS Tris 8.5 0.001 Temp 2Hr 1.5min dense
0.01 M Room
153 RS Tris 8.5 0.001 Temp 2Hr 2min dense
0.01 M Room
154 RS Tris 8.5 0.001 Temp 2Hr 2min dense
0.01 M Room
155 RS Tris 8.5 0.001 Temp 2Hr 2.5min dense
0.01 M Room
156 RS Tris 8.5 0.001 Temp 2Hr 2.5min dense
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
39
1001111 Example 31
MO Classic on previously tested swabs
= 750 1 of buffer, l 5 1 of ProK. Voi-tex 20 sec. Quick spin for 12 seconds to
reach
13,200 rpm. Incubate at room temperature for 2 hr. Incubate at 95 C for 10
minutes.
Voi-tex 20 sec. Quick spin for 12 seconds to reach 13,200 rpm. Sonicate for
2.5min
using foam insert. Transfer to spin basket and spin at 13,200rpm for 5
minutes. Make
a slide.
,4 . ~= . ;- _
~ ~.~eyi us Condibons,- : Count~,; rReeove ~
.$am le ~Prev~ous~ $ ~ z_
157 115 PBS with 0.01 ProK 1 hr. 4 8%
158 139 MO Classic 10 20%
159 140 MO Classic 6 12%
Conclusion: Previous treatment with PBS and ProK does not yield good recovery
using the
MO Classic method. Previous treatment with MO Classic removed almost all the
sperm and
when re-treated with the same method, only a small aniount of sperm could be
removed.
1001121 Example 32
MO Classic on previously tested swabs
= 750 1 of buffer, 15 1 of ProK. Vortex 20 sec. Quick spin for 12 seconds to
reach
13,200 rpm. Incubate at room temperature for 2 hr. Incubate at 95 C for 10
minutes.
Vortex 20 sec. Quick spin for 12 seconds to reach 13,200 rpm. Sonicate for
2.5min
using foam insert. Transfer to spin basket and spin at 13,200rpm for 5
minutes. Make
a slide.
Sam~ _)e` WRevious~# g4 Pce F''~'~~'~Co tl t~ons~ .,. Coun.t. Reco e~
__
160 31 0.05M Tris pH 6 1 hr. 96 55%
161 32 0.05M Tris pH 7 1 hr. 85 49%
1001131 Example 33
MO Classic on previously tested swabs
= 750 1 of buffer, 15 1 of 0.001 mg/ml ProK. Vortex 20 sec. Quick spin for 12
seconds
to reach 13,200 rpm. Incubate at room temperature for I hr. Incubate at 95 C
for 10
minutes. Vortex 20 sec. Quick spin for 12 seconds to reach 13,200 rpm.
Sonicate for
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
I min using foam insert. Transfer to spin basket and spin at 13,200rpm for 5
minutes.
Make a slide.
.. .._.. ~.a ., :vC 'v C a ount;~;yf t. ~ r::. rl :
am Prev~ousa# Pr, ev~ousConditions: ' z Reco~e
162 127 Tris with 0.001 ProK 1 hr. 9 18%
163 128 Tris with 0.OO1ProK 2hr. 3 6%
1001141 Example 34
7/12/06
Perform second step on sample treated with 0-amylase Barley
= Make stock 5mg/50m1. 750 1 of solution. Vortex 20 sec. Quick spin for 12
sec.
Incubate at 56 C for 30 min. Incubate 95 C for 10 min.
= 750p1 of buffer, 15p1 of 0_001mg/ml ProK. Vortex 20 sec. Quick spin for 12
seconds
to reach 13,200 rpm. Incubate at room temperature for 2 hr. Incubate at 95 C
for 10
minutes. Vortex 20 sec. Quick spin foi- 12 seconds to reach 13,200 rpm.
Sonicate for
2.5 min using foam insert. Transfer to spin basket and spin at 13,200rpm for 5
minutes. Make a slide.
~Sam _le ~Jff
Coupt. : ~R~'re~~~
Barie 0 0%
1001151 Example 35
Old and New sperm stock from 6/21/06 using existing second step (MO Classic)
= 750 1 of buffer, 15 1 of 0.001mg/mi ProK. Vortex 20 sec. Quick spin for 12
seconds
to reach 13,200 rpm. Incubate at room temperature for 2 hr. Incubate at 95 C
for 10
minutes. Vortex 20 sec. Quick spin for 12 seconds to reach 13,200 rpm.
Sonicate for
2.5 min using foam insert. Transfer to spin basket and spin at 13,200rpm for 5
minutes. Make a slide.
a ~~-"_ F _ ~r: ~~ ~~-~
Pr:evious ~ P r"evious Condrtrons r ~Count~~~R ecov.e;.
165 1X PBS pH 6.83 overnight 75 43%
166 1X PBS pH 6.83 overnight 91 52%
1001161 Example 36
New swabs prepared 7/7/06
Sonication Example. 0.01 M Tris pH 8.5 with I or 2 hour incubation at room
temperature
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
41
= 750p1 of buffer, 15p1 of 0.001mg/ml ProK. Vortex 20 sec. Quick spin for 12
seconds
to reach 13,200 rpm_ Incubate at room temperature for I or 2 hr. Incubate at
95 C for
minutes. Vortex 20 sec. Quick spin for 12 seconds to reach 13,200 rpm.
Sonicate
for designated time using foam insert. Transfer to spin basket and spin at
13,200rpm
for 5 minutes. Make a slide.
'
- ~ 7Somcatron
r .' ~ ~ ~i B11ffeC H Pr, ; Terti .,, ;sr Tline
~Sam le.; . ID.i .... . P_. ~ o K =;Gount. ~sRecov_e : ::`_.
0.01 M Room
167 CS Tris 8.5 0.001 Temp lHr 0 20 40%
0.01 M Room
168 CS Tris 8.5 0.001 Temp lHr 1 min 6 12%
0.01 M Room
169 CS Tris 8.5 0.001 Temp lHr 2.5min 8 16%
0.01 M Room
170 CS Tris 8.5 0.001 Temp 2Hr 0 5 10%
0.01 M Room
171 CS Tris 8.5 0.001 Temp 2Hr 1min 4 8%
0.01 M Room
172 CS Tris 8.5 0.001 Temp 2Hr 2.5min 3 9%
0.01 M Room
173 JK Tris 8.5 0_001 Temp lHr 0 2 4%
0.01 M Room
174 JK Tris 8.5 0.001 Temp lHr 1 min 2 4%
0.01 M Room
175 JK Tris 8.5 0_001 Temp lHr 2.5min 2 4%
0.01 M Room
176 JK Tris 8.5 0_001 Temp 2Hr 0 4 10%
0_01 M Room
177 JK Tris 8_5 0.001 Temp 2Hr lmin 2 5%
0.01 M Room
178 JK Tris 8.5 0.001 Temp 2Hr 2.5min 7 18%
1001171 Example 37
Perform MO Classic as second step after performing first step with 0.01 M Tris
pH 8.5 with
15p1 of 0.0001 mg/ml ProK without 95 C incubation on samples prepared 6/30/06
= 750p1 of buffer, 15 1 of 0.0001 mg/m1 ProK. Vortex 20 sec. Quick spin for 12
seconds to reach 13,200 rpm. Incubate at room temperature for 1 hr. Vortex 20
sec.
Quick spin for 12 seconds to reach 13,200 rpm. Sonicate for 2.5min. with foam
insert.
Transfer to spin basket and spin at 13,200rpm for 5 minutes.
= 750 1 of buffer, l5 l of 0.001mg/ml ProK. Vortex 20 sec. Quick spin for
12seconds
to reach 13,200 rpm. Incubate at room temperature for 2 hr. Incubate at 95 C
for 10
minutes. Vortex 20sec. Quick spin for 12 seconds to reach 13,200 rpm. Sonicate
for
2.5min using foam insert. Transfer to spin basket and spin at 13,200rpm for 5
minutes. Make a slide.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
42
ount,, Rec ve 6W
179 UA 4 15%
180 UA 3 11%
1001181 Example 38
Perform MO Classic as second step after performing first step with B-amylase
Barley without
95 C incubation with samples from 6/30/06
= Make a 1:10 dilution of the stock 5mg/50ml. 750 1 of solution. Vortex 20
sec. Quick
spin for 12 sec. Incubate at 56 C for 30 min. Vortex 20 sec. Quick spin 12
sec.
Transfer to spin basket and spin at 13,200 rpm for 5 minutes. Make a slide.
= Place swab in tube with 750 1 of buffer, 15 l of 0.001 mg/ml ProK. Vortex 20
sec.
Quick spin for 12 seconds to reach 13,200 rpm. Incubate at room temperature
for 2 hr.
Incubate at 95 C for 10 minutes. Vortex 20 sec. Quick spin for 12 seconds to
reach
13,200 rpm. Sonicate for 2.5 min using foam insert. Transfer to spin basket
and spin
at 13,200rpm for 5 minutes. Make a slide.
+. +~yS ,~=~ .~a+~zs~a=.:ar_. ;r,~'y'i_ .
Sam le~ < Sam Ie;ID ,?~Ste = a,CountA ~Rec".o_ve,
Barley PN First 0 0%
Barley PN Second 4 8%
1001191 Example 39
Perform MO Classic as second step on sample 71
= 750p1 of buffer, 15 1 of 0.001mg/mi ProK. Vortex 20 sec. Quick spin for 12
seconds
to reach 13,200 rpm. Incubate at room temperature for 2 hr. Incubate at 95 C
for 10
minutes. Vortex 20 sec. Quick spin for 12 seconds to reach 13,200 rpm.
Sonicate for
2.5min using foam insert. Transfer to spin basket and spin at 13,200rpm for 5
minutes. Make a slide.
mg : -~ ~~~ ~~e,t~:z~
Sa le r,ev,Io,us,S_am . le4# Pr~evlo_us Treatment ~Count RecoAv;
181 71 0.01 M Tris H 8.5 for 1 hr 11 6%
1001201 Example 40
7/14/06
Sonication Example with samples prepared 6/30/06 using new ProK and new 0.01 M
Tris
= 750p1 of buffer, 15 1 of 0.001mg/ml ProK. Vortex 20 sec. Quick spin for 12
seconds
to reach 13,200 rpm. Incubate at room temperature for 1 or 2 hr. Incubate at
95 C for
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
43
minutes. Vortex 20 sec. Quick spin for 12 seconds to reach 13,200 rpm.
Sonicate
for dcsignatcd time using foam insert. Transfer to spin basket and spin at
13,200rpm
for 5 minutes. Make a slide.
~* SamPle ~`; }ti . r ~ TM~r ~,S.onicaLOn x ~
': Sample?,: ~v 3 ID ufferi..: ~~1 N ,~ P_rotlC_~ Temp~ ; ATime- e- `a~
~Counf~; 6.Reiovery: '
0.01 M Room
182 RF Tris 8.5 0.001 Temp lHr 0 14 28%
0.01 M Room
183 RF Tris 8.5 0.001 Temp lHr 1 min 12 24%
0.01 M Room
184 CS Tris 8.5 0.001 Temp lHr 2.5min 28 56%
0_01 M Room
185 CS Tris 8.5 0.001 Temp 2Hr 0 31 62%
0.01 M Room
186 LC Tris 8.5 0.001 Temp 2Hr 1min 13 26%
0.01 M Room
187 LC Tris 8.5 0.001 Temp 2Hr 2.5min 13 26%
0.01 M Room
188 PR Tris 8.5 0.001 Temp lHr 0 28 56%
0.01 M Room
189 PR Tris 8.5 0.001 Temp lHr 1 min 78 156%
0.01 M Room
190 PR Tris 8.5 0.001 Temp lHr 2.5min 64 128%
0.01 M Room
191 PR Tris 8.5 0.001 Temp 2Hr 0 28 56%
0.01 M Room
192 PR Tris 8.5 0.001 Temp 2Hr 1min 54 108%
0.01 M Room
193 PR Tris 8.5 0.001 Temp 2Hr 2.5min 53 106%
100122J Example 41
Perform MO Classic as second step on samples previously tested with PBS at
different pHs
0 750 1 of buffer, 15 1 of 0.001 mg/ml ProK. Vortex 20 sec. Quick spin for 12
seconds
to reach 13,200 rpm. Incubate at room temperature for 2hr. Incubate at 95 C
for 10
minutes. Vortex 20 sec. Quick spin for 12 seconds to reach 13,200 rpm.
Sonicate for
2.5 min using foam insert. Transfer to spin basket and spin at 13,200rpm for 5
minutes. Make a slide.
~~~~~ "
;,~Sam le , Pr.evtous~#x Previous~.Condi.tions; Gount Reco~e.,.
194 1 PBS pH 6 88 50%
195 2 PBS pH 7 137 78%
196 3 PBS pH 7.5 71 41%
197 4 PBS pH 8 121 69%
198 5 PBS pH 8.5 75 43%
199 6 PBS pH 9 51 29%
1001231 Example 42
Perform MO Classic on samples prepared 7/7/06 with new ProK and new 0.O1M Tris
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
44
Sam le. Sam le ID < ~~~C_ount; ~~Recover ,:,t
200 PN 1 2%
201 PN 2 4%
1001241 Example 43
= Make a 1:1000 dilution of the Barley stock 5mg/50m1. 750 1 of solution.
Vortex 20
sec. Quick spin for 12 sec. Incubate at 56 C or room temperature for 1 hour.
Vortex
20sec. Quick spin 12 sec. Transfer to spin basket and spin at 13,200 rpm for 5
minutes.
= Place swab in new tube with 750 1 ofbuffer, 15 1 of 0.001 mg/ml ProK. Vortex
20
sec. Quick spin for 12 seconds to reach 13,200 rpm. Incubate at room
temperature for
2hr. Incubate at 95 C for 10 minutes. Vortex 20 sec. Quick spin for 12 seconds
to
reach 13,200 rpm. Sonicate for 2.5 min using foam insert. Transfer to spin
basket and
spin at 13,200rpm for 5 minutes_ Make a slide.
~Sam le : Sam le}I..D iiSte Incu .b...ation~Tem _ r Gount Recover-.
Barley SW First 56 2 4%
Barley SW Second room temperature 38 76%
Barley SW First room temperature 2 4%
Barley SW Second room temperature 17 34%
New Sperm 3
Prepared spenm stock to make new swab samples.
Took l 0 1 of -30,000 sperm/ I stock and diluted it in 990 1 of 1 X PBS pH 7.4
from Cambex.
A 5 l aliquot was placed on a slide and counted. The count yielded
>2,000spenm/5 1.
New Sperm 3 Dilution
= The sperm was diluted by taking 250 1 of the above solution and mixing it
with 750 1
of 1 X PBS from Cambrex. 5 l of the dilution was blotted onto buccal swabs. A
5 l
aliquot was placed on a slide to count after a set of 14 swabs was made. The
solution
was also vortexed after the 14 samples.
1001251 Example 44
7/17/06
Performed MO Classic on different types of samples to determine if the sperm
counts are
accurate.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
~~'~~33'wF-
Sam _! Sam IEl~. &4 ~' Actual~Amount on wGo..
Rec_o_.y.e,
Sam le : Ie~ID~= unt
202 PN 2 I Forensic Semen Undetermined 0 0%
203 PN 2 1 Forensic Semen Undetermined 0 0%
204 PN 1200 count 60 4 53%
205 PN 1200 count 60 1 13%
206 AJ 400 count 400 17 34%
207 PN 150 count 1500 1 5%
208 JK 1600 count 1600 81 41%
209 KV 1600 count 1600 16 8%
1001261 Example 45
7/18/06
New Sperm Dilution 2
Prepare new sperm sample for reproducibility study.
= Vortex New Spenn 3 stock for 20 sec. Sonicate for 1 minute. Dilute 400P1 of
the
stock with 600 1 of IX PBS from Cambrex. Vortex 20 sec_ Sonicate 2 min. Make a
slide 3 spots 5 l each. Continue to aliquot 5 1 of sample into a new 1.5m]
tube. After
30 samples -2min., make another slide 3 spots 5 1 each. Continue to aliquot
the
sample in 5pl increments onto the tube. When at appr. 0.5m1, make a new slide
I spot,
vortex the sample 20 sec. and make another spot. Continue aliquoting in 5 )
increments into the tube until almost all the sample is consumed. Make another
slide 3
spots.
= Make 10 swabs by placing 16 1 of the above dilution 2. Make 4 swabs by
placing
20p 1 of the above solution.
1001281 Example 46
7/28/06
= Experiment about vortexing before or after sonication
= 750u1 of 0.01 M Tris and 15u1 of 0.001 mg/ml ProK add to tubes. 4 samples:
214 with
vortex before 5 min sonication, 215 vortex after 2.5 min sonicaton; 216 no
sonicaton;
217 vortex after 5 min sonication. The sperm count add to each swab is read on
the
slide when preparing the samples.
...
0Sam le ~Sam le ID~ ~ Sam le, -,; Count Re_cover,
214 PN 160 count 6 49%
215 PN 120 count 9 90%
216 PN 80 count 4 67%
217 PN 160 count 8 70%
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
46
1001301 Example 47
Purpose: To determine if the addition of 1% SDS to the sperm elution solution
will
improve the recovery rate of sperm.
Method:
= Target of - 400 sperm placed on cotton swab
= Sperm were eluted from the swab using the same protocol as described in
Experiment 12 of the previous validation (p. 11) with MO-Lite Buffer followed
by
MO-Classic. In this experiment, 1% SDS was added to the MO-Classic. The MO-
Classic solution consisted of 7501A1 Tris-HCI, pH 8.5 with 1% SDS, and 15 l of
0.001 mg/ml Pro K.
~Sam~,le.: õ H~ o;K~'~SDS :~ ~Time~ ,Co,u ts~ ~Recove ~
353 PN 0.01M Tris 8.5 0.001 1% lhr 52 41%
354 PN 0.01 M Tris 8.5 0.001 1% 2hr 70 54%
Conclusion: The number of sperm recovered using MO-Classic with 1% SDS does
not
appear to be accurate due to the difficulty of heat fixing and staining the
slide with 1%
SDS present in the eluted sperm pellet. It is quite likely that some sperm are
getting
washed off of the slide during the washing of the red and green dyes.
1001311 Example 48
Purpose: To determine the impact of washing the sperm pellet to remove most of
the 1%
SDS present in the pellet. This wash step should theoretically help in the
making and
reading of the slides.
Method:
= Pipette - 2500 sperm (as diluted semen) on the cotton swab
= Sperm were eluted from the swab using the protocol as described above with
MO-
Lite and MO-Classic w/ l% SDS.
= The swab and spin basket are removed from the SR (sample remains) tube and
placed in the SE (sperm elution fraction) tube containing the sperm pellet.
Add
600 1 of Tris-HCI pH 8.5 buffer onto the swab to rinse (IX). Spin the SE tube
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
47
containing the spin basket for 5 min at 13,500 rpm. Remove the spin basket and
place it back in the SR tube; discard the supernatant leaving approximately 50
l of
solution to resuspend the pellet. An aliquot can be removed for slide
preparation.
y
0_1 ~ 73,L` .~r "' . 0.
F}$ample ,_,: , SampleilD ;~SDS, iTrHCI~Rmse, C.~.~ount ~ V_Recovry
367 PN 0.01M Tris 8.5 0.001 1% 600N1 121 80%
370 PN 0.01 M Tris 8.5 0_001 1% 600 I 139 87%
373 PN 0.01 M Tris 8.5 0.001 1% 600 I 131 86%
Conclusion: Rinsing the eluted sperm pellet one time with Tris-HCI
successfully removed
most of the SDS based on our ability to properly stain and visualize the
eluted sperm and
on the improved rate of sperm recovery.
1001321 Example 49
Purpose: To determine the reproducibility of sperm elution using the MO-
Classic buffer
containing I % SDS.
Method:
= 5u1 of I:100 dilution of semen were added to each swab sample.
= The number of sperm present on each swab was approximately 2500.
= 9 samples were subjected to the elution procedure using Tris-HCl with 1% SDS
followed by a I X rinse step of Tris-HCI w/o SDS.
Conclusion: The recovery of sperm ranged from 74% - 99% for an average
recovery of
87.6%. This represents an 1 I.1 % improvement over the elution buffer without
I % SDS as
shown in Experiment 1 I (shown in Table AA below).
.
1001331 Example 50
Purpose: To determine if pre-soaking the swab in MO-Lite or eluting with MO-
Classic
1% SDS buffer has any negative effect on the performance of P30 assays.
Method:
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
48
= Perform P30 on the MO-Lite supematant and MO-Classic l% SDS supematant of
samples 353 and 354 from experiment 13 using the standard Orchid Cellmark P30
protocol.
= Also perform P30 on the supernatants of samples 353 and 354 after elution in
the
MO-Classic l%'SDS buffer followed by a 1 X Tris-HCl wash of the sperm pellet.
In the table below, EE = epithelial-enhanced fraction, SE = sperm elution, and
SE-I X=
sperm elution rinsed once with 600 p1 Tris-HCl (pH 8.5) (shown in Table BB
below).
~ -rex+ Y
Table Targeted Sperm t Sl~de,Count ?~.LD lufin Factar-
~ s~Yl sKV 1l .~Sl-~-.ft!õ.S. := t+~ ':^'i .` ~y~t3 k...
qAy` "~~ ~ Count,on`Swab (Eluted from (Based on size Tota Numbe" ot W
. .3 .-.
Sample~ }~ample ID ~~~:.(b~ased on S NI)K sw~abj of cell pell~et)~
~3peer~m.Recovered eco~very~
1 ~ PN 2500 98 19 1862 74%
2 PN 2500 139 17 2363 95%
3 PN 2500 131 17.6 2306 92%
4 PN 2500 127 15.6 1981 79%
PN 2500 117 17 1989 79%
6 PN 2500 169 14.6 2467 99%
7 PN 2500 127 17 2159 86%
8 PN 2500 135 17 2295 92%
9 PN 2500 143 16 2288 91%
, 4
Sam le T ,j < ~Sam~leklD~ ~S erm~Count~~ APx30
353-EE PN 400 POS
353-SE PN 400 NEG
353-SE-IX PN 400 NEG
354-EE PN 400 POS
354-SE PN 400 NEG
354-SE-1X PN 400 NEG
Conclusion: The addition of 1% SDS in the elution buffer results in a complete
loss of
P30 activity in the supematant following sperm elution. Rinsing once with Tris-
HCl has
no reparative effect on P30 results. The MO-Lite supematant (epithelial-
enhanced
fraction) will be used to test for P30 in future tests.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
49
1001341 Example 51
Purpose: To determine if testing for P30 on the EE (epithelial-enhanced)
fraction
following MO-Lite elution produces equal or similar results when compared to
the
standard way of performing P30 following PBS elution with vortexing.
~
Tarnrget~spefm~ ":_ ~ ~ ~ ~ ~~ ,,~: =
Sample Sampie,lq Volume; ,.Dilution f,~ >added',; ~<Total count~~ Method,._
P30=; '
378, CYN 90 1: I;to..1000 A50 MO=Lite POS.."..;
379 CYN 10 I 1 to 10000 30 24 MO-Lite NEG
380 CYN 10NI 1 to 100000 4 0 MO-Lite NEG
. . ..,,,.=.:....
1'to:1000 <.: 450 . _: e. ; Noslide. . A_#',.. Vqrtex ~POSw,,,
382 CYN 10NI 1 to 10000 30 No slide Vortex NEG
383 CYN 10N1 1 to 100000 4 No slide Vortex NEG
S .~ ~ e;y .'' x~y .õr~t &<,. . 'r'-"~ ~=: . .- ' .Y~~" ~'2~`~ ~
erm
4 Target sp
~ Sample Sam: , Ie, ID ,zVo. Iume~~,Y ution P ~ 'added." Method ~ P30
396 ` PN 10 I : 1 to 1000. :: 1 465 :VORTEX
397 PN 10 I 1 to 2500 251 VORTEX NEG
398 PN 10 I 1 to 5000 137 VORTEX NEG
399 PN 10 I I to 7500 43 VORTEX NEG
400 PN 10 I 1 to 10000 21 VORTEX NEG
... . =., ,. .. _ ,
1 to 1000; ,.:,
: 465. MO Life -POS
402 PN 10 I 1 to 2500 251 MO-Lite NEG
403 PN 10 I 1 to 5000 137 MO-Lite NEG
404 PN 10 1 1 to 7500 43 MO-Lite NEG
405 PN 10 I 1 to 10000 21 MO-Lite NEG
Conclusion: The EE (epithelial-enhanced) fraction yields similar results for
P30 analysis
when compared to the standard PBS elution with vortexing.
1001351 Example 52
Purpose: To determine the purity of the MO-Lite fraction when a large number
of sperm
is present on the cotton swab.
Method:
= 250K sperm were applied to each of four cotton swabs
0 Soak each swab in 750 pl MO-Lite buffer for 30 minutes
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
= Prepare pellet from eluted cells, remove 5 l aliquot, and determine number
of
sperm present
= Add cell lysis buffer containing DTT to the cell pellet and incubate
overnight at
56 C
= Perform STR analysis
Results:
T ge' t~Sp n : xT talCo~u~t_~~~
`~e `s-r~ -* 1~ ~s~ ~'- :' s.''`~.4g'r .. ~ ~ < Number~ XSam ,ie~ID, Aded=.~ r
Reco~eredt Reco~e Y_~~~ Profle
Male
374 PN 250,000 2350 < 1% No
375 PN 250,000 2260 < 1% No
376 PN 250,000 2490 < 1% Partial
377 PN 250,000 2430 < 1 /a Partial
Example of electropherogram - Sample #377 - Predominantly female with partial
male
profile present are shown in Figure 4.
Conclusion: Even in the presence of significant amounts of sperm present on
the cotton
swab, the amount of sperm present in the MO-Lite elution is small (- I%) and
does not
significantly impact the outcome of the STR profiles obtained from this
fraction.
However, rather than referring to the MO-Lite fraction as the epithelial
fraction, Orchid
Cellmark will now refer to this fraction as the epithelial-cnhanced (EE)
fraction.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
51
APPENDIX I
Standard Sperm Elution Method - Vortexing Method
1. Aliquot the swab into 1.5u1 tube.
2. Add 750u1 of PBS into the tube.
3. Vortex the sample for approximately 15-30 seconds.
4. Quick spin the tube.
5. Incubate the tube in the fridge overnight.
6. Vortex the sample for approximately 15-30seconds.
7_ Quick spin to pull the liquid to the bottom of the tube.
8. Transfer the swab sample into the spin basket.
9. Centrifuge for 5minutes at 13,200rpm.
10. Remove about 725u1 of the supernatant without disturbing the pellet.
11. Mix the pellet in the 25u1 volume using the pipette and drawing the sample
up and
down several times and while swirling.
12. Pipette 5ul of the resuspended pellet onto a microscope slide.
13. Heat fix the slide by placing it in a 65 C oven for --20min.
14. Stain and examine the slide.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
52
APPENDIX 2
New Sperm Elution Method
I Tum on the 65 C oven befoi-e you begin the procedure to ensure it will be
ready when needed.
2. Place 750 1 of 1 X PBS buffer pH 7.4 (MO-Lite buffer) in a 1.5m1 centrifuge
tube; the tube is labeled as Epithelial Fraction (EF) pellet.
3. Aliquot the swab into the tube breaking the swab as close to the cotton
bulb as
possible.
4. Vortex the tubes, two at a time, for 20 seconds on a vortex with dial set
at 8.
5. Quick spin the tubes in a microcentrifuge for long enough to reach 13,200
rpm
(approximatcly 12 seconds).
6. Incubate the samples at room temperature for 30 minutes.
7. Transfer the swab into a spin basket.
8. Centrifuge the tube with the spin basket for 5 minutes at 13,200 rpm.
9. Remove the spin basket from the tube and place it in a new tube. Make sure
to
have a bleach soaked towel to clean your gloves between samples.
10. Remove the supernatant from the EF pellet making sure to leave 40-50 1.
I L In another 1.5m) microcentrifuge tube, place 750 1 of 0.0IM Tris-HCI
buffer
pH 8.5, add 15 l of 0.001mg/ml Pro K (MO-Classic).
12. Vortex the tube briefly to mix the two reagents; label this tube as Sperm
Fraction (SF) pellet.
13. Transfer the swab from the spin basket to tube labeled SF. Vortex the tube
for
20 seconds.
14. Quick spin the tube in a centrifuge for 12 seconds as before.
15. Incubate the sample at room temperature for 2 hours; voi-tex for 25 sec
after I
hr incubation; shake down the contents of'the tube and incubate for another
hr.
16. Sonicate the tubes, 2-3 at a time, by placing them in the foam insert and
into
the sonicator for 2:30 minutes. After a 5 minute period, change out the water
to
prevent it from heating up_
17. Vortex the tubes, two at a time, for 20 seconds and transfer into spin
baskets.
18. Centrifuge the tubes with the spin baskets for 5 minutes at 13,200 rpm.
19. Remove the spin basket from the tube and place it in a new tube. Make sure
to
have a bleach soaked towel to clean your gloves between samples.
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
53
20. Remove the supernatant from the pellet making sure to leave at least 40-50
1
depending on the size of the pellet. Place the supernatant in a new tube.
21. Resuspend the pellet by vortexing and mixing the sample up and down using
a
pipette.
22. Using the same pipette tip, place 5 i of the sample on a blue microscope
slide
and heat fix for 5 minutes at 65 C. Place the slide in the oven immediately
after
putting the samples on the slide.
23. Stain the slide by adding the red dye for 15-20 minutes. Rinse with water.
Stain with the green dye for 15 seconds and rinse with ethanol_ Make sure to
hold
the slide with the longest edge parallel to the floor while rinsing. Let the
slide air
dry on a rack.
24. Slides are ready to be viewed under the microscope.
Lot Numbers
0.01 M Tris-HC1 pH 8.5
0.001 mg/mI ProK
Red dye
Green dye
CA 02661604 2009-02-23
WO 2008/021502 PCT/US2007/018274
54
1001341 It will be apparent to those skilled in the art that various
modifications and variations
can be made to various embodiments described herein without departing from the
spirit or
scope of the teachings herein. Thus, it is intended that various embodiments
cover other
modifications and variations of various embodiments within the scope of the
present
teachings.