Note: Descriptions are shown in the official language in which they were submitted.
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
1
METABOLICALLY ENGINEERED MICROORGANISM USEFUL
FOR THE PRODUCTION OF 1,2-PROPANEDIOL
The present invention concerns a metabolically engineered micro-organism and
its
use for the preparation of 1,2-propanediol.
1,2-propanediol or propylene glycol, a C3 dialcohol, is a widely-used
chemical. It is
a component of unsaturated polyester resins, liquid detergents, coolants, anti-
freeze and de-
icing fluids for aircraft. Propylene glycol has been increasingly used since
1993-1994 as a
replacement for ethylene derivatives, which are recognised as being more toxic
than
propylene derivatives.
1,2-propanediol is currently produced by chemical means using a propylene
oxide
hydration process that consumes large amounts of water. Propylene oxide can be
produced
by either of two processes, one using epichlorhydrin, and the other
hydroperoxide. Both
routes use highly toxic substances. In addition, the hydroperoxide route
generates by-
products such as tert-butanol and 1-phenyl ethanol. For the production of
propylene to be
profitable, a use must be found for these by-products. The chemical route
generally
produces racemic 1,2-propanediol, whereas each of the two stereoisomers (R)1,2-
propanediol and (S)1,2-propanediol are of interest for certain applications.
The disadvantages of the chemical processes for the production of 1,2-
propanediol
make biological synthesis an attractive alternative. Two routes have been
characterized for
the natural production of 1,2-propanediol from sugars by microorganisms.
In the first route 6-deoxy sugars (e.g. L-rhamnose or L-fucose) are cleaved
into
dihydroxyacetone phosphate and (S)-lactaldehyde, which can be further reduced
to (S)- 1,2-
propanediol (Badia et al, 1985). This route is functional in E. coli, but can
not yield an
economically feasible process due to the elevated cost of the deoxyhexoses.
The second route is the metabolism of common sugars (e.g. glucose or xylose)
through the glycolysis pathway followed by the methylglyoxal pathway.
Dihydroxyacetone
phosphate is converted to methylglyoxal that can be reduced either to
lactaldehyde or to
acetol. These two compounds can then undergo a second reduction reaction
yielding 1,2-
propanediol. This route is used by natural producers of (R)-1,2-propanediol,
such as
Clostridium sphenoides and Thermoanaerobacter thermosaccharolyticum.
Clostridium
sphenoides has been used to produce 1,2-propanediol at a titer of 1,58 g/l
under phosphate
limited conditions (Tran Din and Gottschalk, 1985). Thermoanaerobacter
thermosaccharolyticum has also been investigated for the production of 1,2-
propanediol
(Cameron and Cooney, 1986, Sanchez-Rivera et al, 1987). The best performances
obtained
were a titer of 9 g/l and a yield from glucose of 0,2 g/g. However, the
improvement of the
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
2
performances obtained with these organisms is likely to be limited due to the
shortage of
available genetic tools.
PRIOR ART
Cameron et al (1998) have investigated the use of E. coli as a platform for
metabolic engineering for the conversion of sugars to 1,2-propanediol. Their
theoretical
analysis showed that the upper limit of the realistic product yield
(considering mass
balances and production of energy for growth) is significantly different
depending on the
culture conditions. Under anaerobic conditions, acetate will be produced as a
by-product in
order to recycle the reduced co-factors and the best yield shall be limited to
1 mole of 1,2-
propanediol per mole of glucose (0,42 g/g). Under aerobic conditions,
recycling of co-
factors shall be ensured by the respiratory chain using oxygen as terminal
electron acceptor
and it could become possible to produce 1,2-propanediol without the production
of by-
products. Under these conditions, yield could reach at best 1.42 moUmol (0,6
g/g).
Considering the maximum titer of 1,2-propanediol, Cameron et al discussed its
dependence
on product and by-product toxicity. 1,2-propanediol is significantly less
toxic than 1,3-
propanediol and E. coli exhibits a residual growth rate of 0.5 h-1 with 100
g/l 1,2-
propanediol. The inhibition of growth is more likely to be due to the by-
product acetate
that is known to be highly growth inhibiting. Development of an anaerobic
process for the
production of 1,2-propanediol with high titers and yields will have to address
the acetate
issue. Conversion of acetate into acetone, which is less inhibitory and easily
removed in
situ has been proposed (WO 2005/073364).
Several investigations for genetic modifications of E. coli in order to obtain
a 1,2-
propanediol producer using simple carbon sources have been done by the group
of
Cameron (Cameron et al, 1998, Altaras and Cameron, 1999, Altaras and Cameron,
2000)
and the group of Bennett (Huang et al, 1999, Berrios-Rivera et al, 2003).
These studies rely
on the one hand on the expression of one or several enzymatic activities in
the pathway
from dihydroxyacetone phosphate to 1,2-propanediol and on the other hand on
the removal
of NADH and carbon consuming pathways in the host strain. The best results
obtained by
the group of Cameron are production of 1.4 g/1 1,2-propanediol in anaerobic
flask culture
with a yield of 0.2 g/ g of glucose consumed. When extrapolated in anaerobic
fed-batch
fermenter, the production was 4.5 g/1 1,2-propanediol with a yield of 0.19 g/g
from
glucose, far from their theoretical expectations. These performances have been
obtained
with the overexpression of the methylglyoxal synthase gene of E. coli (mgs),
the glycerol
dehydrogenase gene of E. coli (gldA) and the 1,2-propanediol oxidoreductase
gene of
E. coli (fucO) in a strain lacking the gene coding for lactate dehydrogenase
(ldhA). Results
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
3
obtained with the same approach but with lower titers and yields are also
described in the
patents US 6,087,140, US 6,303,352 and WO 98/37204.
The group of Bennett also used an E. coli host strain lacking ldhA for the
overexpression of the mgs gene from Clostridium acetobutylicum and the gldA
gene from
E. coli. Flask cultures under anaerobic conditions gave a titer of 1.3 g/l and
a yield
of 0.12 g/g whereas microaerobic cultures gave a titer of 1.4 g/l with a yield
of 0.13 g/g.
At this stage, all these results are not better than those obtained with the
species T.
thermosaccharolyticum.
The catabolism of glucose trough the glycolysis pathway in E. coli results in
two
triose phosphate molecules, dihydroxyacetone phosphate (DHAP) and
glyceraldehyde 3
phosphate, after the cleavage of fructose 1,6 bisphosphate. These two triose
phosphate
molecules can be interconverted by the triose phosphate isomerase activity. It
is generally
recognized that DHAP is converted to GA3P and the two GA3P originating from
glucose
are further catabolized.
The glyceraldehyde 3-phosphate dehydrogenase, also called GAPDH, is one of the
key enzymes involved in the glycolytic conversion of glucose to pyruvic acid.
GAPDH
catalyzes the following reaction:
Glyceraldehyde 3-phosphate + phosphate + NAD+ -> 1,3-biphosphoglycerate + NADH
+ H+
The gene encoding this enzyme was cloned in 1983 in E. coli (Branlant et al.,
Gene,
1983) and named "gap". Later another gene encoding a product having the same
enzymatic
activity was identified and named gapB (Alefounder et al., Microbiol., 1987).
Characterization of E. coli strains with deleted gapA and gapB genes have
shown that
gapA is essential for glycolysis although gapB is dispensable (Seta et al., J.
Bacter., 1997).
A microorganism with a down regulated gapA gene was reported in patent
application
WO 2004/033646 for the production of 1,3-propanediol from glucose by
fermentation.
The inventors of the present application have shown that 2 factors in
combination
are required to obtain an increase of the 1,2-propanediol yield:
- an improved activity of the biosynthesis pathway of 1,2-propanediol, and
- an attenuation of the GAPDH activity.
The inventors demonstrate also that increasing intracellular
phosphoenolpyruvate
concentration or using an alternative sugar transport system can further boost
the 1,2-
propanediol production by fermentation of a micro-organism.
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
4
DESCRIPTION OF THE INVENTION
The invention is related to a microorganism useful for the production of 1,2-
propanediol from a carbon source, wherein said microorganism is characterized
by :
a) an improved activity of the biosynthesis pathway from dihydroxyacetone
phosphate to 1,2-propanediol, and
b) an attenuated activity of the glyceraldehyde 3-phosphate dehydrogenase
The improved activity of the biosynthesis pathway from DHAP to 1,2-propanediol
is obtained by increasing the activity of at least one enzyme involved in said
biosynthetic
pathway. This can be obtained by increasing the expression of the gene coding
for said
enzyme and in particular the expression of at least one gene selected among
mgsA, yqhD,
yafB, ycdW, yqhE, yeaE, yghZ, yajO, tas, ydjG, ydbC, gldA and fucO.
Preferentially, the
expression of the three genes mgsA, yqhD and gldA is increased.
In a further aspect of the invention, the Entner-Doudoroff pathway is
eliminated by
deleting either the edd or eda gene or both. Furthermore, the synthesis of
unwanted by-
products is attenuated by deleting the genes coding for enzymes involved in
synthesis of
lactate from methylglyoxal (such as gloA, aldA, aldB), lactate from pyruvate
(ldhA),
formate (pflA, pflB), ethanol (adhE) and acetate (ackA, pta, poxB).
The glyceraldehyde 3 phosphate activity is attenuated in order to redirect a
part of
the available glyceraldehyde 3 phosphate toward the synthesis of 1,2-
propanediol via the
action of the enzyme triose phosphate isomerase. The yield of 1,2-propanediol
over
glucose can then be greater than 1 mole/mole. However, due to the reduced
production of
phosphoenolpyruvate (PEP), the PEP-dependent sugar import system will be
negatively
impacted. Therefore, in one aspect of the invention, the efficiency of the
sugar import is
increased, either by using a sugar import independent of PEP like the one
encoded by galP,
or by providing more PEP to the sugar-phosphotransferase system. This is
obtained by
eliminating the pathways consuming PEP like pyruvates kinases (encoded by the
pykA and
pykF genes) and/or by promoting the synthesis of PEP e. g. by overexpressing
the ppsA
gene coding for PEP synthase.
Additionally, it is valuable for the enzyme converting pyruvate into acetyl-
coA to
be resistant to high concentrations of NADH found under anaerobic conditions.
This can
be obtained by a specific mutation in the lpd gene. Finally, in order to spare
NADH for the
reduction of acetol into 1,2-propanediol, the arcA and the ndh genes can be
deleted.
The microorganism used for the preparation of 1,2-propanediol is selected
among
bacteria, yeasts and fungi, but is preferentially from the species Escherichia
coli or
Clostridium acetobutylicum.
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
It is also an object of the present invention to provide a process for the
production
of 1,2-propanediol by cultivating the modified microorganism in an appropriate
growth
medium and by recovering and purifying the 1,2-propanediol produced.
5 BRIEF DESCRIPTION OF THE DRAWINGS
The accompanying drawing that is incorporated in and constitutes a part of
this
specification exemplifies the invention and together with the description,
serves to explain
the principles of this invention.
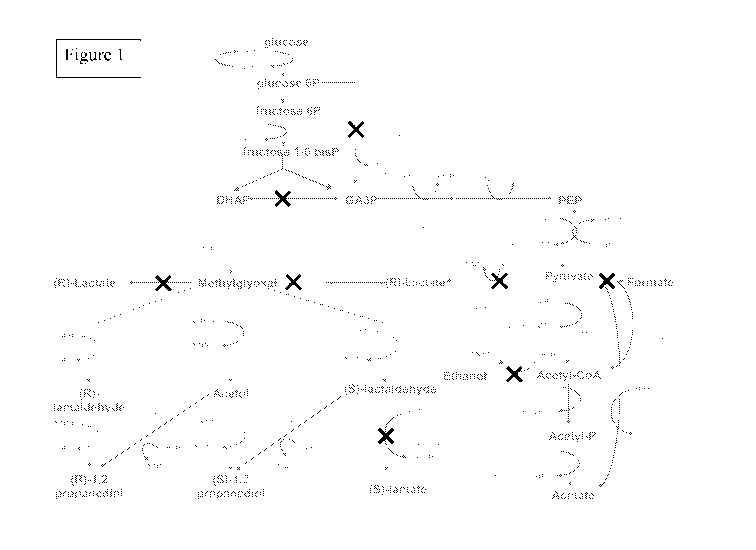
Figure 1 depicts the genetic engineering of central metabolism in the
development
of a 1,2-propanediol production system from carbohydrates.
DETAILED DESCRIPTION OF THE INVENTION
As used herein the following terms may be used for interpretation of the
claims and
specification.
According to the invention the terms `culture', `growth' and `fermentation'
are used
interchangeably to denote the growth of bacteria in an appropriate growth
medium
containing a simple carbon source.
The term `carbon source' according to the present invention denotes any source
of
carbon that can be used by those skilled in the art to support the normal
growth of a micro-
organism, and which can be hexoses, pentoses, monosaccharides, disaccharaides,
oligosaccharides, starch or its derivatives, hemicelluloses, glycerol and
combinations
thereof.
The term "useful for the production of 1,2-propanediol" denotes that the
microorganism produces said product of interest, preferably by fermentation.
Fermentation
is a classical process that can be performed under aerobic, microaerobic or
anaerobic
conditions.
The phrase "attenuation of the activity of an enzyme" refers to a decrease of
the
activity of the enzyme of interest in the modified strain compared to the
activity in the
initial strain before any modification. The man skilled in the art knows
numerous means to
obtain this result. Possible examples include:
- Introduction of a mutation into the gene, decreasing the expression level of
this
gene, or the level of activity of the encoded protein.
- Replacement of the natural promoter of the gene by a low strength promoter,
resulting in a lower expression.
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
6
- Use of elements destabilizing the corresponding messenger RNA or the
protein.
- Deletion of the gene if no expression at all is needed.
The term "expression" refers to the transcription and translation of a gene
sequence
leading to the generation of the corresponding protein product of the gene.
Advantageously, the activity of the glyceraldehyde 3-phosphate dehydrogenase
is
less than 30% of the activity observed in an unmodified strain under the same
conditions,
more preferably less than 10%.
The term "improved activity of the biosynthesis pathway from dihydroxyacetone
phosphate to 1,2-propanediol" means that at least one of the enzymatic
activities involved
in the pathway is improved (see below).
Advantageously, the microorganism of the invention is genetically modified to
increase the activity of at least one enzyme involved in the biosynthetic
pathway from
dihydroxyacetone phosphate to 1,2-propanediol.
Preferentially, the increase of the activity of an enzyme is obtained by
increasing
the expression of the gene coding for said enzyme.
To obtain an overexpression of a gene of interest, the man skilled in the art
knows
different methods such as:
- Replacement of the endogenous promoter with a stronger promoter
- Introduction into the microorganism of an expression vector carrying said
gene of
interest.
- Introducing additional copies of the gene of interest into the chromosome
Several techniques are currently used for introducing DNA into a bacterial
strain. A
preferred technique is electroporation, which is well known to those skilled
in the art.
Advantageously, at least one gene of interest is overexpressed, selected
among:
mgsA, yajB, yeaE, yghZ, yqhE, yqhD, ydhF, ycdW, yajO, ydjG, ydbC, tas, gldA
and fuco.
The mgsA gene codes for methylglyoxal synthase catalysing the conversion of
DHAP into methylglyoxal. The genes yajB, yeaE, yghZ, yqhE, yqhD, ydhF, ycdW,
yajO,
ydjG, ydbC, tas encode enzymatic activities able to convert methylglyoxal into
acetol. The
gldA gene encodes glycerol dehydrogenase, which catalyses the conversion of
acetol into
1,2-propanediol. The fucO gene encodes 1,2-propanediol oxidoreductase
catalysing the
conversion of lactaldehyde into 1,2-propanediol.
A preferred microorganism harbours modifications leading to the overexpression
of
three genes of particular interest : mgsA, yqhD and gldA.
Preferentially, in the microorganism according to the invention, at least one
gene
involved in the Entner-Doudoroff pathway is attenuated. The Entner-Doudoroff
pathway
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
7
provides an alternative way to degrade glucose to glyceraldehyde-3-phosphate
and
pyruvate besides glycolysis. The attenuation of the Entner-Doudoroff pathway
assures that
most or at best all glucose is degraded via glycolysis and be used for the
production of 1,2-
propanediol.
In particular at least one of the two genes of this pathway edd or eda is
attenuated.
The term `attenuation of the expression of a gene' according to the invention
denotes the partial or complete suppression of the expression of a gene, which
is then said
to be `attenuated'. This suppression of expression can be either an inhibition
of the
expression of the gene, the suppression of an activating mechanism of the
gene, a deletion
of all or part of the promoter region necessary for the gene expression, or a
deletion in the
coding region of the gene. Preferentially, the attenuation of a gene is
essentially the
complete deletion of that gene, which gene can be replaced by a selection
marker gene that
facilitates the identification, isolation and purification of the strains
according to the
invention. A gene is preferentially inactivated by the technique of homologous
recombination as described in Datsenko, K.A. & Wanner, B.L. (2000) "One-step
inactivation of chromosomal genes in Escherichia coli K-12 using PCR
products". Proc.
Natl. Acad. Sci. USA 97: 6640-6645.
Preferentially, in the microorganism according to the invention, at least one
enzyme
involved in the conversion of methylglyoxal into lactate is attenuated. The
purpose of this
attenuation is that the available methylglyoxal is used by the cell machinery
essentially for
the synthesis of 1,2-propanediol (see figure 1).
Genes involved in the conversion of methylglyoxal into lactate are in
particular:
- Genes encoding for enzymes having glyoxalase activity, such as the gloA
gene coding for glyoxalase I, catalysing the synthesis of lactoyl
glutathione from methylglyoxal;
- the aldA and aldB genes coding for a lactaldehyde dehydrogenase
(catalysing the synthesis of (S) lactate from (S) lactaldehyde).
The expression of one or more of these genes is advantageously attenuated in
the
initial strain. Preferentially the gene gloA is completely deleted.
In the microorganism of the invention, it is preferable that at least one
enzyme
involved in the synthesis of by-products such as lactate, ethanol and formate
is attenuated.
In particular, it is advantageous to attenuate the gene ldhA coding for
lactate
dehydrogenase catalysing the synthesis of lactate from pyruvate, and the gene
adhE coding
for alcohol-aldehyde dehydrogenase catalysing the synthesis of ethanol from
acetyl-CoA.
Similarly, it is possible to force the micro-organism to use the pyruvate
dehydrogenase complex to produce acetyl-CoA, C02 and NADH from pyruvate,
instead
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
8
of acetyl-CoA and formate. This can be achieved by attenuating the genes pflA
and pflB
coding for pyruvate formate lyase.
In another specific embodiment of the invention, the synthesis of the by-
product
acetate is prevented by attenuating at least one enzyme involved in its
synthesis. It is
preferable to avoid such acetate synthesis to optimize the production of 1,2-
propanediol.
To prevent the production of acetate, advantageously the expression of at
least one
gene selected among ackA, pta and poxB is attenuated. These genes all encode
enzymes
involved in the different acetate biosynthesis pathways (see figure 1).
Preferentially, in the microorganism according to the invention, the
efficiency of
sugar import is increased. A strong attenuation of the expression of the gapA
gene resulting
in a decrease of the carbon flux in the GAPDH reaction by more than 50%, this
will result
in the synthesis of less than 1 mole of phosphoenolpyruvate (PEP) per mole of
glucose
imported. PEP is required by the sugar-phosphotransferase system (PTS)
normally used for
the import of simple sugars into the cell, since import is coupled to a
phospho-transfer
from PEP to glucose yieding glucose-6-phosphate. Thus reducing the amount of
PEP will
negatively impact on sugar import.
In a specific embodiment of the invention, the sugar might be imported into
the
microorganism by a sugar import system independent of phosphoenolpyruvate. The
galactose-proton symporter encoded by the gene galP that does not involve
phosphorylation can be utilized. In this case the imported glucose has to be
phosphorylated
by glucose kinase encoded by the glk gene. To promote this pathway, the
expression of at
least one gene selected among galP and glk is increased. As a result the PTS
becomes
dispensable and may be eliminated by attenuating at least one gene selected
among ptsH,
ptsl or crr.
In another specific embodiment of the invention, the efficiency of the sugar-
phosphotransferase system (PTS) is increased by increasing the availability of
the
metabolite phosphoenopyruvate. Due to the attenuation of the gapA activity and
of the
lower carbon flux toward pyruvate, the amount of PEP in the modified strain of
the
invention could be limited, leading to a lower amount of glucose transported
into the cell.
Various means exist that may be used to increase the availability of PEP in a
strain
of microorganism. In particular, a mean is to attenuate the reaction PEP --->
pyruvate.
Preferentially, at least one gene selected among pykA and pykF, coding for the
pyruvate
kinase enzyme, is attenuated in said strain to obtain this result. Another way
to increase the
availability of PEP is to favour the reaction pyruvate -> PEP, catalyzed by
the
phosphoenolpyruvate synthase by increasing the activity of the enzyme. This
enzyme is
encoded by the ppsA gene. Therefore,preferentially in the microorganism, the
expression
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
9
of the ppsA gene is preferentially increased. Both modifications can be
present in the
microorganism simultaneously.
Especially under anaerobic or microaerobic conditions, it is advantageous that
the
pyruvate dehydrogenase complex (PDC), converting pyruvate into acetyl-coA has
low
sensitivity to inhibition by NADH. Lower sensitivity is defined with reference
to the
sensitivity of the unmodified enzyme. Such characteristic can be obtained by
introducing a
specific mutation in the lpd gene (coding for the sub-unit lipoamide
dehydrogenase of the
PDC) resulting in the replacement of alanine 55 in the protein sequence of the
enzyme with
the residue valine.
Under anaerobic or microaerobic conditions, availability of NADH for the
reduction of the precursors into 1,2-propanediol is advantageously increased.
This is
obtained by alleviating the repression on the tricarboxylic acid cycle
mediated by the
global regulator ArcA (encoded by the arcA gene). NADH concentration in the
cell can
also be increased by inactivating the NADH dehydrogenase II encoded by the
gene ndh.
Therefore, preferably, at least one gene selected among arcA and ndh is
attenuated.
Preferentially the microorganism according to the invention is selected among
bacteria, yeasts or fungi. More preferentially, the microorganism is selected
from the group
consisting of Enterobacteriaceae, Bacillaceae, Clostridiaceae,
Streptomycetaceae and
Corynebacteriaceae. Even more preferentially, the microorganism is either
Escherichia
coli or Clostridium acetobutylicum.
Another object of the invention is a method for preparing 1,2-propanediol,
wherein
a microorganism such as described previously is grown in an appropriate growth
medium
containing a simple carbon source, and the produced 1,2-propanediol is
recovered. The
production of 1,2-propanediol is performed under aerobic, microaerobic or
anaerobic
conditions.
The culture conditions for the fermentation process can be readily defined by
those
skilled in the art. In particular, bacteria are fermented at temperatures
between 20 C and
55 C, preferably between 25 C and 40 C, and preferably at about 35 C for C.
acetobutylicum and at about 37 C for E. coli.
This process can be carried out either in a batch process, in a fed-batch
process or
in a continuous process.
`Under aerobic conditions' means that oxygen is provided to the culture by
dissolving the gas into the liquid phase. This could be obtained by (1)
sparging oxygen
containing gas (e.g. air) into the liquid phase or (2) shaking the vessel
containing the
culture medium in order to transfer the oxygen contained in the head space
into the liquid
phase. Advantages of the fermentation under aerobic conditions instead of
anaerobic
conditions is that the presence of oxygen as an electron acceptor improves the
capacity of
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
the strain to produce more energy in form of ATP for cellular processes.
Therefore the
strain has its general metabolism improved.
Micro-aerobic conditions are defined as culture conditions wherein low
percentages
of oxygen (e.g. using a mixture of gas containing between 0.1 and 10% of
oxygen,
5 completed to 100% with nitrogen), is dissolved into the liquid phase.
Anaerobic conditions are defined as culture conditions wherein no oxygen is
provided to the culture medium. Strictly anaerobic conditions are obtained by
sparging an
inert gas like nitrogen into the culture medium to remove traces of other gas.
Nitrate can be
used as an electron acceptor to improve ATP production by the strain and
improve its
10 metabolism.
The term `appropriate growth medium' according to the invention denotes a
medium of known molecular composition adapted to the growth of the micro-
organism.
For example a mineral culture medium of known set composition adapted to the
bacteria
used, containing at least one carbon source. In particular, the mineral growth
medium for
E. coli can thus be of identical or similar composition to M9 medium
(Anderson, 1946,
Proc. Natl. Acad. Sci. USA 32:120-128), M63 medium (Miller, 1992; A Short
Course in
Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and
Related
Bacteria, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York)
or a
medium such as that defined by Schaefer et al. (1999, Anal. Biochem. 270: 88-
96), and in
particular the minimum culture medium named MPG described below:
K2HPO4 1.4 g/1
Nitrilo Triacetic Acid 0.2 g/l
trace element solution* 10 ml/1
(NH4)2SO4 1 g/1
NaC1 0.2 g/l
NaHCO3 0.2 g/l
M SO4 0.2 g/l
glucose 20 to 100 g/l
NaNO3 0.424 g/l
thiamine 10 mg/1
FeSO4, 7H20 50 mg/l
yeast extract 4 g/l
The pH of the medium is adjusted to 7.4 with sodium hydroxide.
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
11
*trace element solution : Citric acid 4.37 g/L, MnSO4 3 g/L, CaC1z 1 g/L,
CoC1z,
2H20 0.1 g/L, ZnSO4, 7H20 0.10 g/L, CuSO4, 5H20 10 mg/L, H3B03 10 mg/L,
NazMoO4
8.31 mg/L.
In a specific embodiment of the invention, the method is performed with a
strain of
E. coli grown in a medium containing a simple carbon source that can be
arabinose,
fructose, galactose, glucose, lactose, maltose sucrose or xylose. An
especially preferred
simple carbon source is glucose.
In another specific embodiment of the invention, the method is performed with
a
strain of C. acetobutylicum grown in a medium containing a simple or a complex
carbon
source.
The growth medium for can thus be of identical or similar composition to
Clostridial Growth Medium (CGM, Wiesenbom et al., Appl. Environm. Microbiol.,
54 :
2717-2722) or a mineral growth medium as given by Monot et al. (Appl.
Environm.
Microbiol., 44: 1318-1324) or Vasconcelos et al. (J. Bacteriol., 176 : 1443-
1450).
The carbon source used for the culture of C. acetobutylicum is either a simple
or a
complex carbon. The simple carbon source can be arabinose, fructose,
galactose, glucose,
lactose, maltose sucrose or xylose. An especially preferred simple carbon
source is
glucose. The complex carbon source can be starch or hemicellulose. An
especially
preferred complex carbon source is starch.
Advantageously the recovered 1,2-propanediol is furthermore purified. The man
skilled in the art knows various means for recovering and purifying the 1,2-
propanediol.
The invention is described above, below and in the Examples with respect to
E. coli. Thus the genes that can be attenuated, deleted or over-expressed for
the initial and
evolved strains according to the invention are defined mainly using the
denomination of
the genes from E. coli. However, this designation has a more general meaning
according to
the invention, and covers the corresponding genes in other micro-organisms.
Using the
GenBank references of the genes from E. coli, those skilled in the art can
determine
equivalent genes in other organisms than E. coli.
The means of identification of the homologous sequences and their percentage
homologies are well-known to those skilled in the art, and include in
particular the BLAST
programmes that can be used on the website http://www.ncbi.nlm.nih.~4ov/BLAST/
with
the default parameters indicated on that website. The sequences obtained can
be exploited
(aligned) using for example the programmes CLUSTALW
(http://www.ebi.ac.uk/clustalw/), with the default parameters indicated on
these websites.
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
12
The PFAM database (protein families database of alignments and hidden Markov
models http://www.san~4er.ac.uk/Software/Pfaml) is a large collection of
alignments of
protein sequences. Each PFAM makes it possible to visualise multiple
alignments, view
protein domains, evaluate distributions among organisms, gain access to other
databases
and visualise known protein structures.
COGs (clusters of orthologous groups of proteins
http://www.ncbi.nlm.nih.gov/COG/) are obtained by comparing protein sequences
derived
from 66 fully sequenced unicellular genomes representing 44 major phylogenetic
lines.
Each COG is defined from at least three lines, making it possible to identify
ancient
conserved domains.
REFERENCES in the order of citation in the text
l. Badia J, Ros J, Aguilar J (1985), J. Bacteriol. 161: 435-437.
2. Tran Din K and Gottschalk G (1985), Arch. Microbiol. 142: 87-92
3. Cameron DC and Cooney CL (1986), Bio/Technology, 4: 651-654
4. Sanchez-Rivera F, Cameron DC, Cooney CL (1987), Biotechnol. Lett. 9: 449-
454
5. Altaras NE and Cameron DC (1999), Appl. Environ. Microbiol. 65: 1180-1185
6. Cameron DC, Altaras NE, Hoffman ML, Shaw AJ (1998), Biotechnol. Prog. 14:
116-125
7. Altaras NE and Cameron DC (2000), Biotechnol. Prog. 16 : 940-946
8. Huang K, Rudolph FB, Bennett GN (1999), Appl. Environ. Microbiol. 65: 3244-
3247
9. Berrios-Rivera SJ, San KY, Bennett GN (2003), J. Ind. Microbiol.
Biotechnol. 30:
34-40
10. Branlant G, Flesch G, Branlant C (1983), Gene, 25: 1-7
11. Alefounder PR and Perham RN (1989), Mol. Microbiol., 3: 723-732
12. Seta FD, Boschi-Muller F, Vignais ML, Branlant G (1997), J. Bacteriol.
179:
5218-5221
13. Datsenko KA and Wanner BL (2000), Proc. Natl. Acad. Sci. USA 97: 6640-6645
14. Anderson EH (1946), Proc. Natl. Acad. Sci. USA 32:120-128
15. Miller (1992), A Short Course in Bacterial Genetics: A Laboratory Manual
and
Handbook for Escherichia coli and Related Bacteria, Cold Spring Harbor
Laboratory Press, Cold Spring Harbor, New York
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
13
16. Schaefer U, Boos W, Takors R, Weuster-Botz D (1999), Anal. Biochem. 270:
88-96
17. Wiesenbom DP, Rudolph RB, Papoutsakis ET (1987), Appl. Environ.
Microbiol.,
54 : 2717-2722
18. Monot F, Martin JR, Petitdemange H, Gay R (1982), Appl. Environ.
Microbiol. 44:
1318-1324
19. Vasconcelos I, Girbal L, Soucaille P (1994), J. Bacteriol. 176: 1443-1450
20. Lemer CG and Inouye M (1990), Nucleic Acids Res. 18: 4631
EXAMPLES
Example 1: Construction of modified strains of E. coli MG1655 Ptrcl6-gapA::cm
(pME101VB01 yqhD-mgsA-gldA), E. coli MG1655 Ptrc16-gapA::cm (pME101VB01-
yafB-mgsA-gldA) and E. coli MG1655 Ptrc16-gapA::cm (pME101VB01 yqhE-mgsA-
gldA)
To increase the production of 1,2-propanediol different combinations of genes
were
expressed from the plasmid pMElOlVB01 using the trc promoter.
a) Construction of modified strains of E. coli MG1655 (pME101VB01 yqhD-mgsA-
gldA) , MG1655 (pME101VB01 yafB-mgsA-gldA) and MG1655 (pME101VB01 yqhE-
mgsA-gldA)
Construction of plasmid pME101VB01
The plasmid pMElOlVB01 was derived from plasmid pMEIOI and harbored a multiple
cloning site containing recognition site sequences specific for the rare
restriction
endonucleases Nhel, SnaBI, PacI, BglII, AvrII, SacII and Agel following by the
adc
transcription terminator of Clostridium acetobutylicum ATCC824.
For the expression from a low copy vector the plasmid pME 1 O l was
constructed as
follows. The plasmid pCL1920 (Lemer & Inouye, 1990, NAR 18, 15 p 4631 -
GenBank
AX085428) was PCR amplified using the oligonucleotides PMElOlF and PMElOlR and
the BstZl71-Xmnl fragment from the vector pTrc99A (Amersham Pharmacia Biotech,
Piscataway, N.J) harboring the lacI gene and the trc promoter was inserted
into the
amplified vector.
PME101F (SEQ ID NO 1):
ccgacagtaagacgggtaagcctg
PME101R (SEQ ID NO 2):
agcttagtaaagccctcgctag
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
14
A synthetic double-stranded nucleic acid linker comprising the multicloning
site and adc
transcriptional terminator was used to generate pMElOlVB01. Two 100 bases
oligonucleotides that complement flanked by Ncol or HindIII digested
restriction sites
were annealed. The 100-base pair product was subcloned into Ncol / HindIII
digested
plasmid pME101 to generate pMElOlVB01.
pME101VB01 1, consisting of 100 bases (SEQ ID NO 3):
catgggcta cg tac~4tattaattaaa~4atctcctagg~4act~ cacc"tTAAAAATAAGAGTTACCTTAAAT
GGTAACTCTTATTTTTTTAggcgcgcca
pMElOlVB01 2, consisting of 100 bases (SEQ ID NO 4):
agcttggcgcgccTAAAAAAATAAGAGTTACCATTTAAGGTAACTCTTATTTTTAacc"
t~4a~4ctcccta"a~4atctttaattaatac~4tagctagcc
with:
- a region (underlined lower-case letters) corresponding to the multicloning
site
- a region (upper-case letters) corresponding to the adc transcription
terminator
(sequence 179847 to 179814) of Clostridium acetobutylicum ATCC 824 pSOLl
(NC_001988).
Construction of plasmids for expression of different combinations of genes of
the
biosynthetic pathway of 1,2- propanediol (pME101VB01 yqhD-mgsA-gldA
pME101VB01 yafB-mgsA-gldA and pME101VB01 yqhE-mgsA-gldA)
The different genes were PCR amplified from genomic DNA of E. coli MG1655
using the
oligonucleotides given in Table 1.
Table 1: oligonucleotides used for amplification of genes of 1,2-propanediol
pathway
Gene name Names of SEQ ID Homology with gene Restriction sites
oligos
yqhD yqhDR2 N 5 3153369-3153400 BspHI added
yqhDF2 N 6 3154544- 3154475 BspHI removed
1VheI added
mgsA mgsAF N 7 1026268-1026248 SnaBI added
mgsAR N 8 1025780-1025800 BglII added
gldA g1dAF N 9 4136631-4136612 AvrII added
g1dAR N 10 4135512-4135530 SacI added
yafB yafB F2 N 11 229167-229190 NcoI added
yafB R N 12 229970-229950 Nhel added
yqhE yqhE F N 13 3154641-3154661 Ncol added
yqhE R N 14 3155464-3155444 Nhel added
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
The PCR amplified fragments were cut with the restriction enzymes mentioned in
Table 1
and cloned into the restriction sites of the plasmid pME101 VBOI. The
following plasmids
were built: pME101VB01 yqhD-mgsA-gldA, pME101VB01 yafB-mgsA-gldA and
pME101VB01 yqhE-mgsA-gldA.
5
The plasmids were then introduced into the strain E. coli MG1655.
b) Construction of a modified strain of E. coli MG1655 Ptrc16-gapA::cm
The replacement of the natural gapA promoter with the synthetic short Ptrcl6
promoter
10 (SEQ ID NO 15 : gagct tg tgacgattaatcatccggctcgaataatgtgtgg) into the
strain E. coli
MG1655 was made by replacing 225 pb of upstream gapA sequence with FRT-CmR-FRT
and an engineered promoter. The technique used was described by Datsenko, K.A.
&
Wanner, B.L. (2000).
The two oligonucleotides used to replace the natural gapA promoter according
to the
15 Protocol 1 are given in Table 2.
Protocol 1: Introduction of a PCR product for recombination and selection of
the
recombinants
The oligonucleotides chosen and given in Table 2 for replacement of a gene or
an
intergenic region were used to amplify either the chloramphenicol resistance
cassette from
the plasmid pKD3 or the kanamycin resistance cassette from the plasmid pKD4
(Datsenko,
K.A. & Wanner, B.L. (2000). The PCR product obtained was then introduced by
electroporation into the recipient strain bearing the plasmid pKD46 in which
the system
Red (õõexo) expressed greatly favours homologous recombination. The antibiotic-
resistant transformants were then selected and the insertion of the resistance
cassette was
checked by PCR analysis with the appropriate oligonucleotides given in Table
3.
The resulting strain was named E. coli MG1655 Ptrcl6-gapA::cm.
The 3 plasmids were introduced separately into the strain E. coli MG1655
Ptrcl6-
gapA::cm.
Table 2 : oligonucleotides used for replacement of a chromosomal region by
recombination
with a PCR product in the strain E. coli MG1655
Region name Names of oligos SEQ ID Homology with chromosomal
region
gapA promoter Ptrc-gapAF N 16 1860478-1860536
(Ptrcl6-gapA) Ptrcl6-gapAR N 17 1860762-1860800
edd and eda genes DeddF N 18 1932582-1932501
DedaR N 19 1930144-1930223
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
16
gloA gene GLOAD f N 20 1725861- 1725940
GLOA D R N 21 1726268-1726189
aldA gene A1dA D f N 22 1486256- 1486336
aldAD r N 23 1487695- 1487615
aldB gene A1dB D f N 24 3752603-3752682
aldBD r N 25 3754141-3754062
ldhA gene D1dhAF N 26 1440865- 1440786
D1dhAR N 27 1439878- 1439958
pflAB gene DpflB r N 28 952315-952236
DpflAf N 29 949470-949549
adhE gene DadhE r N 30 1297344-1297264
DadhEf N 31 1297694-1297773
ackA-pta genes DackAF N 32 2411494-2411573
DptaR N 33 2414906-2414830
poxB gene DpoxBF N 34 908557-908635
DpoxBR N 35 910262-910180
pykA gene DpykAF N 36 1935756-1935836
DpykAR N 37 1755129-1755051
pykF gene DpykFF N 38 1753689-1753766
DpykFR N 39 1755129-1755051
Table 3 : oligonucleotides used for checking the insertion of a resistance
cassette or the
loss of a resistance cassette
Region name Names of oligos SEQ ID Homology with chromosomal
re ion
gapA promoter yeaAF N 40 1860259-1860287
(Ptrcl6-gapA) gapAR N 41 1861068-1861040
edd and eda genes eddF N 42 1932996-1932968
edaR N 43 1929754-1929777
gloA gene NemAQd N 44 1725331 to 1725361
Rnt Cr N 45 1726795 to 1726765
aldA gene Ydc F C f N 46 1485722 to 1485752
gapCCr N 47 1488225 to 1488195
aldB gene aldB C f N 48 3752056 to 3752095
YiaYCr N 49 3754674 to 3754644
ldhA gene ldhAF N 50 1439724 to 1439743
ldhAR N 51 1441029 to 1441007
pflAB gene pflAB 1 N 52 948462 to 948491
pflAB 2 N 53 953689 to 983660
adhE ychGf N 54 1294357 to1294378
adhECr N 55 1297772 to 1297749
ackA pta genes B2295 N 56 2410900 to 2410919
YfcCR N 57 2415164 to 2415145
poxB gene poxBF N 58 908475 to 908495
poxBR N 59 910375 to 910352
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
17
pykA gene pykAF N 60 1935338 to 1935360
pykAR N 61 1937425 to 1937401
pykF gene pykFF N 62 1753371 to 1753392
pykFR N 63 1755518 to 1755495
Example 2: Construction of modified strains of E. coli MG1655 Ptrcl6-gapA,
Aedd-
eda, OgloA, ApykA, ApykF (pME101VB01 yqhD-mgsA-gldA), (pJB137-PgapA ppsA),
E. coli MG1655 Ptrcl6-gapA, Aedd-eda, OgloA, ApykA, OpykF (pME101VB01 yafB-
mgsA-gldA), (pJB137-PgapA-ppsA) and E. coli MG1655 Ptrcl6-gapA , Aedd-eda,
OgloA, ApykA, ApykF (pME101VB01 yqhE-mgsA-gldA), (pJB137-PgapA-ppsA) able to
produce 1,2-propanediol with high yield.
The genes edd-eda were inactivated in strain E. coli MG1655 by inserting a
kanamycin
antibiotic resistance cassette and deleting most of the genes concerned using
the technique
described in Protocol 1 with the oligonucleotides given in Table 2. The strain
obtained was
named MG1655 Dedd-eda: : km.
This deletion was transferred in strain E. coli MG1655 Ptrcl6-gapA::cm
according to
Protocol 2.
Protocol 2 : Transduction with phage Pl for deletion of a gene
The deletion of the chosen gene by replacement of the gene by a resistance
cassette
(kanamycin or chloramphenicol) in the recipient E. coli strain was performed
by the
technique of transduction with phage Pl. The protocol was in two steps, (i)
the preparation
of the phage lysate on the strain MG1655 with a single gene deleted and (ii)
the
transduction of the recipient strain by this phage lysate.
Preparation of the phage lysate
- Seeding with 100 l of an overnight culture of the strain MG1655 with a
single gene
deleted of 10 ml of LB + Cm 30 g/ml + glucose 0.2% + CaC1z 5 m1VI.
- Incubation for 30 min at 37 C with shaking.
- Addition of 100 l of phage lysate Pl prepared on the wild type strain
MG1655
(approx. 1 x 109 phage/ml).
- Shaking at 37 C for 3 hours until all cells were lysed.
- Addition of 200 l of chloroform, and vortexing.
- Centrifugation for 10 min at 4500 g to eliminate cell debris.
- Transfer of supematant in a sterile tube and addition of 200 1 of
chloroform.
- Storage of the lysate at 4 C
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
18
Transduction
- Centrifugation for 10 min at 1500 g of 5 ml of an overnight culture of the
E. coli recipient strain in LB medium.
- Suspension of the cell pellet in 2.5 ml of MgSO4 10 mM, CaC1z 5 mM.
- Control tubes: 100 l cells
100 l phages Pl of the strain MG1655 with a single gene deletion.
- Tube test: 100 l of cells + 100 l phages Pl of strain MG1655 with a single
gene
deletion.
- Incubation for 30 min at 30 C without shaking.
- Addition of 100 l sodium citrate 1 M in each tube, and vortexing.
- Addition of 1 ml of LB.
- Incubation for 1 hour at 37 C with shaking.
- Plating on dishes LB + Cm 30 g/ml after centrifugation of tubes for 3 min
at
7000 rpm.
- Incubation at 37 C overnight.
The antibiotic-resistant transformants were then selected and the insertion of
the
deletion was checked by a PCR analysis with the appropriate oligonucleotides.
The resulting strain was named E. coli MG1655 Ptrcl6-gapA::cm, Dedd-eda::km.
The antibiotic resistance cassettes were then eliminated according to Protocol
3.
Protocol 3 : Elimination of resistance cassettes
The chloramphenicol and/or kanamycin resistance cassettes were eliminated
according to the following technique. The plasmid pCP20 carrying the FLP
recombinase
acting at the FRT sites of the chloramphenicol and/or kanamycin resistance
cassettes were
introduced into the recombinant strains by electroporation. After serial
culture at 42 C, the
loss of the antibiotics resistance cassettes was checked by PCR analysis with
the
oligonucleotides given in Table 3.
The strain MG1655 OgloA: : cm was built according to Protocol 1 with the
oligonucleotides given in Table 2 and this deletion was transferred in the
strain previously
built according to Protocol 2. The resulting strain was named E. coli MG1655
Ptrcl6-
gapA, Dedd-eda,OgloA::cm.
The gene pykA was inactivated into the previous strain by inserting a
kanamycin
antibiotic resistance cassette according to Protocol 1 with the
oligonucleotides given in
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
19
Table 2. The resulting strain was named E. coli MG1655 Ptrc16-gapA, Dedd-
eda, OgloA: : cm, OpykA: : km.
The antibiotic resistance cassettes were then eliminated according to Protocol
3.
The gene pykF was inactivated by inserting a chloramphenicol antibiotic
resistance
cassette according to Protocol 1 with the oligonucleotides given in Table 2.
The resulting
strain was named E. coli MG1655 Ptrcl6-gapA, Dedd-eda,OgloA, OpykA, ApykF::cm.
The antibiotic resistance cassette was then eliminated according to Protocol
3.
At each step, the presence of all the deletions previously built was checked
using
the oligonucleotides given in Table 3.
To increase the production of phosphoenolpyruvate the ppsA gene was expressed
from the
plasmid pJB 137 using the gapA promoter. For the construction of plasmid
pJB137-PgapA-
ppsA, the gene ppsA was PCR amplified from genomic DNA of E. coli MG1655 using
the
following oligonucleotides:
1. gapA-ppsAF, consisting of 65 bases (SEQ ID NO 64)
ccttttattcactaacaaatagctggtggaatatATGTCCAACAATGGCTCGTCACCGCTGGTGC
with:
- a region (upper-case letters) homologous to the sequence (1785106-1785136)
of
the gene ppsA (1785136 to 1782758), a reference sequence on the website
http://genolist.pasteur.fr/Colibril), and
- a region (lower letters) homologous to the gapA promoter (1860794- 1860761).
2. ppsAR, consisting of 43 bases (SEQ ID NO 65)
aatcgcA~cttGAATCCGGTTATTTCTTCAGTTCAGCCAGGC
with:
- a region (upper letters) homologous to the sequence (1782758-1782780) the
region of the gene ppsA (1785136 to 1782758)
- a restriction site HindIIl (underlined letters)
At the same time the gapA promoter region of the E. coli gene gapA was
amplified using
the following oligonucleotides:
1. gapA-ppsAR, consisting of 65 bases (SEQ ID NO 66)
GCACCAGCGGTGACGAGCCATTGTTGGACATatattccaccagctatttgttagtgaataaaagg
with:
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
- a region (upper-case letters) homologous to the sequence (1785106 -1785136)
of
the gene ppsA (1785136 to 1782758), and
- a region (lower letters) homologous to the gapA promoter (1860794 -
1860761).
5 2. gapAF, consisting of 33 bases (SEQ ID NO 67)
ACGTCCCGGGcaagcccaaaggaagagtgaggc
with:
- a region (lower letters) homologous to the gapA promoter (1860639 -
1860661).
10 - a restriction site Smal (underlined letters)
Both fragments were subsequently fused using the oligonucleotides ppsAR and
gapAF
(Horton et al. 1989 Gene 77:61-68). The PCR amplified fragment were cut with
the
restriction enzymes HindIIl and Smal and cloned into the HindIII/Smal sites of
the vector
15 pJBJ37 (EMBL Accession number: U75326) giving vector pJB137-PgapA ppsA.
The different pMElOlVB01 plasmids and pJBl37-PgapA ppsA were introduced into
the
strain E. coli MG1655 Ptrcl6-gapA, Dedd-eda,OgloA, ApykA, ApykF. The strains
obtained
were named respectively E. coli MG1655 Ptrcl6-gapA, Dedd-eda,OgloA, ApykA,
ApykF,
20 pME101VB01 yqhD-mgsA-gldA, pJBl37-PgapA ppsA (strain 1), E. coli MG1655
Ptrc16-
gapA, Dedd-eda,,AgloA, ApykA, ApykF, pME101VB01 yafB-mgsA-gldA, pJB137-PgapA-
ppsA (strain 2) and E. coli MG1655 Ptrcl6-gapA, Dedd-eda,OgloA, OpykA, ApykF,
pME101VB01 yqhE-mgsA-gldA, pJBl37-PgapA ppsA (strain 3).
Example 3: Construction of a modified strains of E. coli MG1655 Ptrcl6-gapA,
Aedd-
eda, OgloA, DaldA, DaldB, OZdhA, ApflAB, AadhE, DackA pta, ApoxB, ApykA, ApykF
(pME101VB01 yqhD-mgsA-gldA), (pJB137-PgapA ppsA), E. coli MG1655 Ptrc16-
gapA , Aedd-eda, OgloA, DaldA, DaldB, OZdhA, ApflAB, AadhE, DackA pta, ApoxB,
OpykA, ApykF (pME101VB01 yafB-mgsA-gldA), (pJB137-PgapA-ppsA) and E. coli
MG1655 Ptrcl6-gapA , Aedd-eda, OgloA, DaldA, DaldB, OldhA, ApflAB, AadhE,
DackA pta, ApoxB, ApykA, OpykF (pME101VB01 yqhE-mgsA-gldA), (pJB137-PgapA-
ppsA) able to produce 1,2-propanediol with a yield higher than 1 mole / mole
glucose.
The strains MG1655 AaldA::km , MG1655 AaldB::cm, MG1655 ApflAB::km
MG1655 AadhE::cm, MG1655 AackA-pta::cm are built according to Protocol 1 with
the
oligonucleotides given in Table 2 and these deletions are transferred in the
strain
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
21
previously built according to Protocol 2. When necessary, the antibiotic
resistance
cassettes are eliminated according to Protocol 3.
The gene ldhA and the gene poxB are inactivated in the strain previously built
by
inserting a chloramphenicol antibiotic resistance cassette according to
Protocol 1 with
the oligonucleotides given in Table 2. When necessary, the antibiotic
resistance
cassettes are eliminated according to Protocol 3.
At each step, the presence of all the deletions previously built is checked
using the
oligonucleotides given in Table 3.
The resulting strain is named E. coli MG1655 Ptrcl6-gapA, Dedd-eda,OgloA,
DaldA, DaldB, OldhA, ApflAB, AadhE, DackA pta, ApoxB, OpykA, ApykF.
The differents pMElOlVB01 plasmids and pJBl37-PgapA ppsA are introduced
into the strain E. coli MG1655 Ptrcl6-gapA, Dedd-eda,OgloA, DaldA,DaldB,
OldhA,
ApflAB, DadhE, DackA pta, ApoxB, ApykA, ApykF. The strains obtained are named
respectively E. coli MG1655 Ptrcl6-gapA, Dedd-eda,OgloA, DaIdA,DaldB, OldhA,
ApflAB,
AadhE, DackA pta, ApoxB, OpykA, ApykF, pME 101 VB01 yqhD-mgsA-gldA, pJB 137-
PgapA ppsA, E. coli MG1655 Ptrcl6-gapA, Dedd-eda,OgloA, DaldA,DaldB, OldhA,
ApflAB, DadhE, DackA pta, ApoxB, OpykA, ApykF, pME101VB01 yafB-mgsA-gldA,
pJBl37-PgapA ppsA and E. coli MG1655 Ptrcl6-gapA, Dedd-eda,OgloA, DaldA,DaldB,
OldhA, ApflAB, AadhE, DackA pta, ApoxB, ApykA, ApykF, pMElOlVB01 yqhE-mgsA-
gldA, pJBl37-PgapA ppsA.
Example 4: Comparison of the different strains for 1,2-propanediol
production under aerobic conditions.
The strains obtained as described in example 2 (strains 1, 2 and 3) and the
control
strains (control 1: MG1655 pMElOlVB01-yqhD-mgsA-g1dA, control 2 : MG1655
pME101VB01-yafB-mgsA-g1dA, control 3 : MG1655 pME101VB01-yqhE-mgsA-g1dA
and control 4 : MG1655 Ptrcl6-gapA, Dedd-eda,AgloA, ApykA, ApykF) were
cultivated
in an Erlenmeyer flask assay under aerobic conditions in minimal medium with
glucose as
carbon source. The culture was carried out at 34 C or 37 C and the pH was
maintained by
buffering the culture medium with MOPS. At the end of the culture, 1,2-
propanediol,
acetol and residual glucose in the fermentation broth were analysed by HPLC
and the
yields of 1,2-propanediol over glucose and 1,2-propanediol + acetol over
glucose were
calculated. The best strain is then selected for a fermenter fed-batch
culture.
CA 02679987 2009-09-03
WO 2008/116848 PCT/EP2008/053438
22
Strain 1,2- Acetol 1,2- 1,2-
propanediol titer titer (g/1) propanediol yield propanediol +
(g/1) (g/g glucose) acetol yield (g/g
glucose)
Control 1 0.02 0 0.004 0.004
Control2 0 0 0 0
Control 3 0.01 0 0.002 0.002
Control4 0.05 0.34 0 0.04
Strain 1 2.25 1.40 0.14 0.23
Strain 2 1.64 1.31 0.10 0.18
Strain 3 0.77 0.47 0.06 0.10
Example 5: Production of 1,2-propanediol in fed-batch culture with the best
strain.
The best strain selected in the previous experiment is cultivated in a 21
fermenter
using a fed-batch protocol.
The temperature of the culture is maintained constant at 37 C and the pH is
permanently adjusted to values between 6.5 and 8 using an NH4OH solution. The
agitation
rate is maintained between 200 and 300 rpm during the batch phase and is
increased to up
to 1000 rpm at the end of the fed-batch phase. The concentration of dissolved
oxygen is
maintained at values between 30 and 40% saturation by using a gas controller.
When the
optical density reaches a value between three and five, the fed-batch is
started with an
initial flow rate between 0.3 and 0.5 ml/h and a progressive increase up to
flow rate values
between 2.5 and 3.5 mllh. At this point the flow rate is maintained constant
for 24 to 48
hours. The medium of the fed is based on minimal media containing glucose at
concentrations between 300 and 500 g/l.