Note: Descriptions are shown in the official language in which they were submitted.
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
MO-1, A Gene Associated with Morbid Obesity
1. FIELD OF THE INVENTION
[0001] The present invention relates to MO-1, a newly identified gene and gene
product associated with morbid obesity. In certain aspects, the present
invention provides
isolated MO-1 nucleic acids, MO-1 polypeptides, oligonucleotides that
hybridize to MO-1
nucleic acids, and vectors, including expression vectors, comprising MO-1
nucleic acids.
The present invention further provides isolated host cells, antibodies,
transgenic non-human
animals, compositions, and kits relating to MO-1. In other aspects, the
present invention
further provides methods of methods of detecting the presence of MO-1 nucleic
acid,
methods of screening for agents which affect MO-1 activity, and methods of
screening for
MO-1 variants.
2. BACKGROUND OF THE INVENTION
[0002] Obesity is a major risk factor for type II diabetes mellitus, heart
disease,
hypertension, the metabolic syndrome, and cancer and is increasingly prevalent
in Western
society and in developing countries. See Kopelman PG. Obesity as a medical
problem.
Nature. 2000 Apr 6;404(6778):635-43. Today, more than 1.1 billion individuals
are
overweight and more than 300 million are obese. See Hossain P, Kawar B, El
Nahas M.
Obesity and diabetes in the developing world--a growing challenge. N Engl J
Med. 2007 Jan
18;356(3):213-5. Obesity is assessed by the calculation of the body mass index
(BMI)[weight/(height)2 in kg/m2]. Individuals with a BMI >= 30 kg/m2 are
considered obese,
whereas those with a BMI >40 are morbidly obese. Despite intense scrutiny of
this
worldwide public health problem, the molecular and regulatory mechanisms which
underlie
the differences between lean and obese individuals remain largely unknown.
Obtaining a
better understanding of how energy balance is controlled should provide the
framework for
future clinical intervention and rational drug design.
[0003] In humans, the importance of genetic factors in obesity has been
clearly
defined through numerous twin, familial aggregation, and adoption studies. See
Stunkard AJ,
Sorensen TI, Hanis C, Teasdale TW, Chakraborty R, Schull WJ, Schulsinger F. An
adoption
study of human obesity. N Engl J Med. 1986 Jan 23;314(4):193-8, Stunkard AJ,
Foch TT,
Hrubec Z. A twin study of human obesity. JAMA. 1986 Jul ;256(1):51-4, Price
RA,
Stunkard AJ, Ness R, Wadden T, Heshka S, Kanders B, Cormillot A. Childhood
onset (age
less than 10) obesity has high familial risk. Int J Obes. 1990 Feb;l4(2):185-
95, and Allison
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
DB, Kaprio J, Korkeila M, Koskenvuo M, Neale MC, Hayakawa K. The heritability
of body
mass index among an international sample of monozygotic twins reared apart.
Int J Obes
Relat Metab Disord. 1996 Jun;20(6):501-6. Indeed, through these studies
heritability has
been estimated as high as 40 - 90%. See Friedman JM. Modern science versus the
stigma of
obesity. Nat Med. 2004 Jun;10(6):563-9. In the absence of rational gene
candidates,
genome-wide genetic association studies have emerged as a potentially powerful
tool. And
as may be predicted, numerous genome-wide linkage studies have identified
novel candidate
gene loci for future studies. See Rankinen T, Zuberi A, Chagnon YC, Weisnagel
SJ,
Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map:
the 2005
update. Obesity (Silver Spring). 2006 Apr;14(4):529-644. Unfortunately, these
linkage
studies have generally identified broad chromosomal regions containing scores
of candidate
genes and ESTs. Two major related problems now exist. First, the large number
of genes
within these regions need to be individually characterized. Second,
biologically plausible
gene candidates within these regions are not always intuitively obvious:
obesity-related genes
may regulate a broad spectrum of physiologic pathways, including those
governing satiety,
basal metabolic rate, and activity. In addition, novel genes or those
unrelated to the present,
limited understanding of disease pathophysiology may go undetected.
[0004] Most striking with regard to the genetic basis of obesity and providing
insights
into its molecular basis has been the identification of gene mutations causing
a number of
Mendelian obesity disorders. See Farooqi S, O'Rahilly S. Genetics of obesity
in humans.
Endocr Rev. 2006 Dec;27(7):710-18. These include leptin and leptin receptor
deficiencies,
melanocortin 4 receptor and POMC deficiencies and the pleiotropic syndromes
Prader-Willi
and Bardet-Biedl. See Bell CG, Walley AJ, Froguel P. The genetics of human
obesity. Nat
Rev Genet. 2005 Mar;6(3):221-34. Unfortunately, while each have provided
insight into the
molecular basis by which the hypothalamus controls satiety and energy
homeostasis, none
has provided insight into more common forms of obesity nor has yet provided a
useful drug
target for obesity and its comorbid features including diabetes. The present
invention is
intend to address these unmet needs.
3. SUMMARY OF THE INVENTION
[0005] The present invention is based in part on the discovery of a novel
gene, termed
MO-1, which exhibits partial structural similarity to phosphoenolpyruvate
carboxykinase,
mutations of which are associated with morbid obesity. A MO-1 cDNA has been
cloned and
sequenced, and a MO-1 amino acid sequence has been determined.
-2-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
[0006] Accordingly, in a first aspect, the present invention provides an
isolated
polypeptide comprising an amino acid sequence having at least 70% identity to
a MO-1
amino acid sequence (SEQ ID NO:1). In one embodiment, the isolated polypeptide
comprises
the amino acid sequence of SEQ ID NO: 1.
[0007] In another aspect, the invention provides an isolated nucleic acid
encoding a
polypeptide comprising an amino acid sequence having at least 70% identity to
a MO-1
amino acid sequence (SEQ ID NO: 1). In certain embodiments, the isolated
nucleic acid
encodes a polypeptide comprising the amino acid sequence of SEQ ID NO: 1. In
certain
embodiments, the isolated nucleic acid comprises a nucleic acid sequence
having at least 70%
identity to at least about 500 contiguous nucleotides selected from SEQ ID
NO:2 or the
complement thereof. In certain embodiments, the isolated nucleic acid
comprises at least
about 500 nucleotides selected from the nucleic acid sequence of SEQ ID NO:2,
or the
complement thereof. In a particular embodiment, the isolated nucleic acid
comprises the
nucleic acid sequence of SEQ ID NO:2, or the complement thereof.
[0008] In another aspect, the invention provides an isolated oligonucleotide
comprising at least about 10 consecutive nucleotides of SEQ ID NO:2 or its
complementary
strand.
[00091 In another aspect, the invention provides a vector comprising a nucleic
acid of
the invention. In certain embodiments, the vector comprises at least about 500
nucleotides
selected from the nucleic acid sequence of SEQ ID NO:2. In certain
embodiments, the
vector comprises a nucleic acid that encodes the polypeptide of SEQ ID NO: 1.
In certain
embodiments, the vector comprises the nucleic acid sequence of SEQ ID NO:2. In
a
particular embodiment, the MO-1 nucleic acid sequence in the vector is
operably linked to a
transcriptional regulatory sequence. In certain embodiments, the vector is
selected from the
group comprising a plasmid, a cosmid, a virus, and a bacteriophage. In certain
embodiments,
the vector expresses a polypeptide comprising SEQ ID NO:1 in a cell
transformed with said
vector. In certain embodiments, the polypeptide encoded by the vector can be
expressed in
adipocytes.
[0010] In another aspect, the invention provides an isolated host cell
comprising a
MO-1 nucleic acid according to the present invention. In another aspect, the
invention
provides an isolated host cell comprising a vector that expresses MO-1. In
certain
embodiments, the isolated host cell is an adipocyte.
-3-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
[0011] In another aspect, the invention provides an isolated antibody that
specifically
binds to a polypeptide comprising an amino acid sequence of SEQ ID NO: 1. In
certain
embodiments, the antibody is a polyclonal, monoclonal, single chain
monoclonal,
recombinant, chimeric, humanized, mammalian, or human antibody.
[0012] In another aspect, the invention provides a transgenic non-human
animal,
which expresses a transgenic nucleic acid encoding MO-1 polypeptide. In
certain
embodiments, the transgenic non-human animal MO-1 polypeptide comprises the
amino acid
sequence of SEQ ID NO: 1. In a particular embodiment, the transgenic non-human
animal
over- or under-expresses MO-1 polypeptide relative to wild-type expression of
MO-1
polypeptide in a human adipocyte. In one embodiment, the transgenic non-human
animal
comprises a transgenic nucleic acid having at least 70% identity to SEQ ID
NO:2, or the
complement thereof. In certain embodiments, the transgenic non-human animal is
a
mammal, including, but not limited to, a mouse, rat, rabbit, hamster, pig,
goat, or sheep.
[00131 In another aspect, the invention provides a transgenic non-human animal
whose germ cells comprise a homozygous null mutation in the endogenous nucleic
acid
sequence encoding MO-1. For example, such an animal can be one wherein the
mutation is
created by insertion of, e.g., a neomycin cassette, in reverse orientation to
MO-1 transcription
and wherein said mutation has been introdi,uced into said animal by homologous
recombination in an embryonic stem cell such that said animal does not express
a functional
MO-1 polypeptide. In certain embodiments, the transgenic non-human animal is
fertile and
transmits said null mutation to its offspring. In particular embodiments, the
transgenic non-
human animal is a mammal, including, but not limited to, a mouse, rat, rabbit,
hamster, or
sheep. In certain embodiments, the animal exhibits a phenotype associated with
mutations of
MO-1, e.g., obesity.
[0014] In another aspect, the invention provides a method of screening for
agents
which affect MO-1 activity, comprising: a) administering said agent to a cell
that expresses a
MO-1 polypeptide; and b) assessing a biological activity of the MO-1 in the
cell. In some
embodiments, the agent agonizes, e.g., increases, the MO-1 activity. In some
embodiments,
the agent antagonizes, e.g., decreases, the MO-1 activity.
[0015] In another aspect, the invention provides a method of screening for
agents
which affect MO-1 activity, comprising: a) administering said agent to a
transgenic non-
human animal according to the present invention; and b) assessing the animal
for an
-4-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
alteration in a metabolic function affected by said agent. In certain
embodiments, said
metabolic function relates to glucose or lipid metabolism.
[0016] In another aspect, the invention provides a method of detecting the
presence of
the MO-1 nucleic acid in a sample, comprising: (a) contacting the sample with
a nucleic acid
that hybridizes to the MO-i nucleic acid; and (b) determining whether the
nucleic acid binds
to a nucleic acid in the sample.
[0017] In another aspect, the invention provides a composition comprising a MO-
1
polypeptide of the invention and a pharmaceutically acceptable carrier. In
another aspect, the
invention provides a composition comprising a polypeptide having an amino acid
sequence
that comprises SEQ ID NO: 1 and a pharmaceutically acceptable carrier.
[0018] In another aspect, the invention provides a composition comprising a MO-
1-
encoding nucleic acid of the invention and a pharmaceutically acceptable
carrier. In another
aspect, the invention provides a composition comprising a polynucleotide
encoding a
polypeptide having an amino acid sequence that comprises SEQ ID NO:1 and a
pharmaceutically acceptable carrier. In certain embodiments, the
polynucleotide comprises a
nucleotide sequence of SEQ ID NO:2.
[0019] In another aspect, the invention provides a kit comprising i) an
isolated
oligonucleotide comprising at least 10 consecutive nucleotides of SEQ ID NO:2,
or its
complementary strand; and ii) a container. In certain embodiments, the kit
contains the
oligonucleotide which comprises at least 15 consecutive nucleotides of SEQ ID
NO:2 or its
complementary strand.
4. BRIEF DESCRIPTION OF THE DRAWINGS
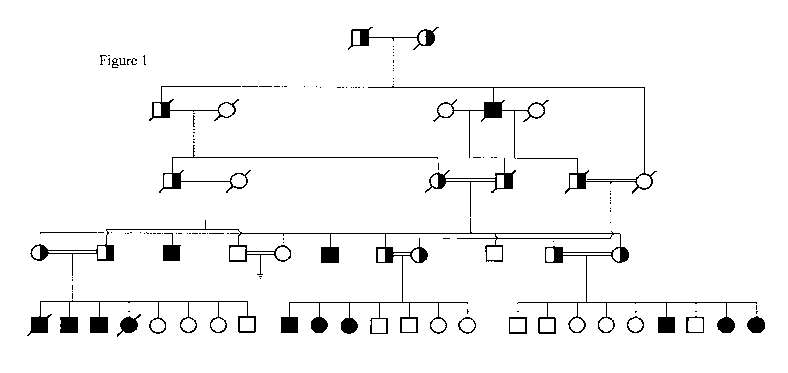
[0020] FIG. 1 presents a diagram showing the lineage of a large consanguineous
family with a high incidence of mutant MO-1.
[0021] FIG. 2 presents a diagram showing a representative vector suitable for
use in
generating a MO-1 knockout mouse.
[0022] FIG. 3 presents an exemplary PCR screen used to identify transgenic
mice
comprising a MO-1 nucleic acid.
[0023] FIG. 4 presents a diagram showing weights of mice overexpressing MO-1.
[0024] FIG. 5 presents a diagram showing glucose tolerance of transgenic mice
expressing human MO-1.
100251 FIG. 6 presents another diagram showing glucose tolerance of transgenic
mice
expressing human MO-1.
-5-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
[0026] FIG. 7A presents a diagram showing single nucleotide polymorphisms
associated with altered body mass phenotypes.
[0027] FIG. 7B presents a table showing associations between single nucleotide
polymorphisms and altered body mass phenotypes.
[0028] FIG. 8 presents a diagram showing the results of overexpression of MO-1
in
Hep3B cells.
[0029] FIG. 9 presents a diagram showing the results of inhibiting expression
of MO-
1 in Hep3B cells.
[0030] FIG. 10 presents diagram showing the results of inhibiting expression
of MO-
1 in NIH 3T3 L1 cells.
[0031] FIG. 11 presents the results of analysis of downstream effects on gene
expression of inhibiting expression of MO-1 in NIH 3T3 Ll cells.
[0032] FIG. 12 presents a diagram showing quantitatively the downstream
effects on
gene expression of inhibiting expression of MO-1 in NIH 3T3 L1 cells.
[0033] FIG. 13 presents a diagram showing expression levels of MO-1 in human
tissues.
[0034] FIG. 14 presents a diagram showing expression levels of MO-1 in
differentiating mouse 3T3 L1 cells..
[00351 FIG. 15 presents a diagram showing identification of Insulin-Regulated
Membrane Aminopeptidase as a potential binding partner for MO-1.
[0036] FIG. 16 presents a diagram showing the effects on proliferation of
inhibiting
MO-1 expression in Hep3B cells.
5. DETAILED DESCRIPTION OF THE INVENTION
[0037] This disclosure provides, for the first time, an isolated cDNA molecule
which,
when transfected into cells can produce MO-1 protein. MO-1 protein is believed
to be linked
to, inter alia, energy metabolism, e.g., glucose or lipid metabolism. This
disclosure provides
the molecule, the nucleotide sequence of this cDNA and the amino acid sequence
of MO-i
protein encoded by this cDNA.
[0038] Having herein provided the nucleotide sequence of the MO-1 cDNA,
correspondingly provided are the complementary DNA strands of the cDNA
molecule, and
DNA molecules which hybridize under stringent conditions to MO-1 cDNA
molecule, or its
complementary strand. Such hybridizing molecules include DNA molecules
differing only
by minor sequence changes, including nucleotide substitutions, deletions and
additions. Also
-6-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
comprehended by this invention are isolated oligonucleotides comprising at
least a portion of
the cDNA molecule or its complementary strand. These oligonucleotides can be
employed as
effective DNA hybridization probes or primers for use in the polymerase chain
reaction.
Such probes and primers may be particularly useful in the screening and
diagnosis of persons
genetically predisposed to obesity and other forms of metabolic dysfunction,
as the result of
MO-1 gene mutations.
[0039] Recombinant DNA vectors comprising the disclosed DNA molecules, and
transgenic host cells containing such recombinant vectors, are also provided.
Disclosed
embodiments also include transgenic nonhuman animals which over-or under-
express MO-1
protein, or over-or under-express fragments or variants of MO-1 protein.
[0040] For clarity of disclosure, and not by way of limitation, the detailed
description
of the invention hereinafter is divided into the subsections that follow. All
publications
mentioned herein are incorporated by reference to disclose and describe the
methods and/or
materials in connection with which the publications are cited.
5.1 Definitions
[0041] Unless defined otherwise, all technical and scientific terms used
herein have
the same meaning as is commonly understood by one of ordinary skill in the art
to which this
invention belongs. All patents, applications, published applications and other
publications
referred to herein are incorporated by reference in their entirety. If a
definition set forth in
this section is contrary to or otherwise inconsistent with a definition set
forth in the patents,
applications, published applications and other publications that are herein
incorporated by
reference, the definition set forth in this section prevails over the
definition that is
incorporated herein by reference.
[0042] As used herein, the singular forms "a," "an," and "the" mean "at least
one" or
"one or more" unless the context clearly dictates otherwise.
[0043] As used herein, "MO-1" refers to proteins or peptides which have an
amino
acid sequence that is identical to SEQ ID NO: 1, as well as proteins sharing
sequence
similarity, e.g., 70%, 75%, 80%, 85%, 90%, 95%, or greater percent identity,
with the amino
acid sequence of SEQ ID NO: 1. Further, these proteins have a biological
activity in common
with the polypeptide having the amino acid sequence of SEQ ID NO: 1,
including, but not
limited to, antigenic cross-reactivity, autoinhibition, phosphorylation
activity, and the like. It
is also contemplated that a MO-1 protein can have one or more conservative or
non-
conservative amino acid substitutions, or additions or deletions from the
amino acid sequence
-7-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
of SEQ ID NO:1 so long as the protein having such sequence alteration shares a
biological
activity as described above with the polypeptide of SEQ ID NO: 1. MO-1 also
includes
proteins or peptides expressed from different mutations, different spliced
forms and various
sequence polymorphisms of the MO-1 gene.
[0044] As used herein, "functional fragments and variants of MO-1" refer to
those
fragments and variants that maintain one or more functions of MO-1. It is
recognized that the
gene or cDNA encoding MO-1 can be considerably mutated without materially
altering one
or more MO-1 functions. First, the genetic code is well-known to be
degenerate, and thus
different codons may encode the same amino acids. Second, even where an amino
acid
substitution is introduced, the mutation can be conservative and have no
material impact on
the essential functions of MO-1. Third, part of the MO-1 polypeptide can be
deleted without
impairing or eliminating all of its functions. Fourth, insertions or additions
can be made in
MO-1, for example, adding epitope tags, without impairing or eliminating its
functions.
Other modifications can be made without materially impairing one or more
functions of
MO-1, for example, in vivo or in vitro chemical and biochemical modifications
which
incorporate unusual amino acids. Such modifications include, for example,
acetylation,
carboxylation, phosphorylation, glycosylation, ubiquination, labeling with
radionuclides, and
various enzymatic modifications, as will be readily appreciated by those
skilled in the art. A
variety of methods for labeling proteins and substituents or labels useful for
such purposes
are well known in the art, and include radioactive isotopes such as ligands
which bind to
labeled antiligands (e.g., antibodies), fluorophores, chemiluminescent agents,
enzymes, and
antiligands. Functional fragments and variants can be of varying length. For
example, some
fragments have at least 10, 25, 50, 75, 100, or 200 or more amino acid
residues.
[0045] As used herein, "protein" is synonymous with "polypeptide" or "peptide"
unless the context clearly dictates otherwise.
[0046] As used herein, a"MO-1 gene" refers to a gene that encodes MO-1 as
defined
herein. A mutation of MO-1 gene includes nucleotide sequence changes,
additions or
deletions, including deletion of large portions or the entire MO-1 gene, or
duplications of all
or substantially all of the gene. Alternatively, genetic expression of MO-1
can be deregulated
such that MO-1 is over or under expressed. The term "MO-1 gene" is understood
to include
the various sequence polymorphisms and allelic variations that exist within
the population.
This term relates primarily to an isolated coding sequence, but can also
include some or all of
-8-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
the flanking regulatory elements and/or intron sequences. The RNA transcribed
from a
mutant MO-1 gene is mutant MO-1 messenger RNA.
[0047] As used herein, "MO-1 cDNA" refers to a cDNA molecule which, when
transfected or otherwise introduced into cells, expresses the MO-1 protein.
The MO-1 cDNA
can be derived, for instance, by reverse transcription from the mRNA encoded
by the MO-1
gene and lacks internal non-coding segments and transcription regulatory
sequences present
in the MO-1 gene. The prototypical human MO-1 cDNA is shown as SEQ ID NO:2.
[0048] As used herein, "vector" refers to discrete elements that are used to
introduce
heterologous DNA into cells for either expression or replication thereof.
Selection and use of
such vehicles are well known within the skill of the artisan. An expression
vector includes
vectors capable of expressing DNA that are operatively linked with regulatory
sequences,
such as promoter regions, that are capable of effecting expression of such DNA
fragments.
Thus, an expression vector refers to a recombinant DNA or RNA construct, such
as a
plasmid, a phage, recombinant virus or other vector that, upon introduction
into an
appropriate host cell, results in expression of the cloned DNA. Appropriate
expression
vectors are well known to those of skill in the art and include those that are
replicable in
eukaryotic cells and/or prokaryotic cells and those that remain episomal or
those which
integrate into the host cell genome.
[0049] As used herein, "transgenic animals" refers to non-human animals,
preferably
mammals, more preferably rodents such as rats or mice, in which one or more of
the cells
includes a transgene. Other transgenic animals include primates, sheep,
rabbits, hamsters,
dogs, cows, goats, chickens, amphibians, etc. A "transgene" is exogenous DNA
that is
integrated into the genome of a cell from which a transgenic animal develops,
and which
remains in the genome of the mature animal. A "transgene" is intended to
encompass
exogenous DNA that comprises the coding sequence of a polypeptide, e.g., a MO-
1
polypeptide, as well as exogenous DNA that comprises regulatory sequences,
e.g., promoted
or enhancer sequences, that affect expression levels of an endogenous
polypeptide, e.g., a
MO-1 polypeptide.
[0050] As used herein, a "homologous recombinant animal" refers to a non-human
animal, preferably a mammal, more preferably a rodent such as a rat or mouse,
in which the
endogenous MO-1 gene has been altered by an exogenous DNA molecule that
recombines
homologously with endogenous MO-1 in a (e.g., embryonic) cell prior to
development of the
animal. Other homologous recombinant animals include rabbits, hamsters and
sheep. Host
-9-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
cells with exogenous MO-1 can be used to produce non-human transgenic animals,
such as
fertilized oocytes or embryonic stem cells into which MO-1-encoding sequences
have been
introduced. Such host cells can then be used to create non-human transgenic
animals or
homologous recombinant animals.
[0051] As used herein, the term "biological sample" includes tissues, cells
and
biological fluids isolated from a subject, as well as tissues, cells and
fluids present within a
subject.
[0052] As used herein, the term "pharmaceutically acceptable" means approved
by a
regulatory agency of the Federal or a state government, or listed in the U.S.
Pharmacopeia or
other generally recognized pharmacopeia for use in animals, and more
particularly in
humans.
[0053] As used herein, the term "carrier" refers to a diluent, adjuvant,
excipient, or
vehicle with which a therapeutic of the invention is administered. Such
pharmaceutical
carriers can be sterile liquids, such as water and oils, including those of
petroleum, animal,
vegetable or synthetic origin, such as peanut oil, soybean oil, mineral oil,
sesame oil and the
like.
[0054] As used herein, an "effective amount" of an active agent for treating a
particular disease is an amount that is sufficient to ameliorate, or in some
manner reduce the
symptoms associated with the disease. The amount may cure the disease but,
typically, is
administered in order to ameliorate the symptoms of the disease.
[0055] As used herein, "active agent" means any substance intended for the
diagnosis, cure, mitigation, treatment, or prevention of disease in humans and
other animals,
or to otherwise enhance physical and mental well being.
[0056] The terms "treatment," "treating," and the like are used herein to
generally
mean obtaining a desired pharmacological and/or physiological effect in a
subject actively
suffering from a condition. The effect may completely or partially treat a
disease or symptom
thereof and thus may be therapeutic in terms of a partial or complete cure for
a disease and/or
adverse effect attributable to the disease. "Treatment" as used herein covers
any treatment of
a disease in a mammal, particularly a human, and includes inhibiting the
disease, i.e.,
arresting its development; or relieving the disease, i.e., causing regression
of the disease. In
one example, treatment refers to treating patients with, or at risk for,
development of obesity
and related conditions. More specifically, "treatment" is intended to mean
providing a
-10-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
therapeutically detectable and beneficial effect on a patient suffering from a
metabolic
disorder.
[0057] The terms "prevent," "preventing," and the like are used herein to
generally
refer to preventing a disease from occurring in a subject which may be
predisposed to the
disease but has not yet been diagnosed as suffering from the disease. Thus,
"prevent" can
refer to prophylactic or preventative measures, wherein the object is to
prevent or slow down
(lessen) obesity or onset of obesity.
[0058] The term "about," as used herein, unless otherwise indicated, refers to
a value
that is no more than 10% above or below the value being modified by the term.
For example,
the term "about 5 g/kg" means a range of from 4.5 g/kg to 5.5 g/kg. As
another
example, "about 1 hour" means a range of from 48 minutes to 72 minutes.
5.2 Polypeptides of the Invention
[0059] The present invention provides newly identified and isolated
polypeptides
referred to in the present application as MO-1. In some embodiments, the
polypeptides are
native sequence MO-1 polypeptides. In some embodiments, the polypeptides
comprise
substantially the same amino acid sequences as found in the native MO-1
sequences. In
certain embodiments, the invention provides amino acid sequences of functional
fragments
and variants of MO-1 that comprise an antigenic determinant (i.e., a portion
of a polypeptide
that can be recognized by an antibody) or which are otherwise functionally
active, as well as
nucleic acids encoding the foregoing. MO-1 functional activity encompasses one
or more
known functional activities associated with a full-length (wild-type) MO-1
polypeptide, e.g.,
antigenicity (the ability to be bound by an antibody to a protein consisting
of the amino acid
sequence of SEQ ID NO: 1); immunogenicity (the ability to induce the
production of an
antibody that binds SEQ ID NO: 1), and so forth.
[0060] In some embodiments, the polypeptides comprise the amino acid sequences
having functionally inconsequential amino acid substitutions, and thus have
amino acid
sequences which differ from that of the native MO-1 sequence. Substitutions
can be
introduced by mutation into MO-1-encoding nucleic acid sequences that result
in alterations
in the amino acid sequences of the encoded MO-1 but do not alter MO-1
function. For
example, nucleotide substitutions leading to amino acid substitutions at "non-
essential"
amino acid residues can be made in MO-1 encoding sequences. A "non-essential"
amino
acid residue is a residue that can be altered from the wild-type sequence of
MO-1 without
altering biological activity, whereas an "essential" amino acid residue is
required for such
-11-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
biological activity. For example, amino acid residues that are conserved among
MO-1
polypeptides are predicted to be particularly unsuitable for alteration. Amino
acids for which
conservative substitutions can be made are well known in the art.
[0061] Useful conservative substitutions are shown in Table 1, "Preferred
Substitutions." Conservative substitutions whereby an amino acid of one class
is replaced
with another amino acid of the same type fall within the scope of the subject
invention so
long as the substitution does not materially alter the biological activity of
the compound. If
such substitutions result in a change in biological activity, then more
substantial changes,
indicated in Table 2 as exemplary are introduced and the products screened for
MO-1
polypeptide biological activity.
Table 1
Preferred Substitutions
Ala (A) Val, Leu, Ile Val
Arg (R) Lys, Gln, Asn Lys
Asn (N) Gln, His, Lys, Arg Gln
Asp (D) Glu Glu
Cys (C) Ser Ser
Gln (Q) Asn Asn
Glu (E) Asp Asp
Gly (G) Pro, Ala Ala
His (H) Asn, Gln, Lys, Arg Arg
Ile (I) Leu, Val, Met, Ala, Phe, Norleucine Leu
Leu (L) Norleucine, Ile, Val, Met, Ala, Phe Ile
Lys (K) Arg, Gln, Asn Arg
Met (M) Leu, Phe, Ile Leu
Phe (F) Leu, Val, Ile, Ala, Tyr Leu
Pro (P) Ala Ala
Ser (S) Thr Thr
Thr (T) Ser Ser
Trp (W) Tyr, Phe Tyr
Tyr (Y) Trp, Phe, Thr, Ser Phe
-12-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
Val (V) Ile, Leu, Met, Phe, Ala, Norleucine Leu
[0062] Non-conservative substitutions that effect: (1) the structure of the
polypeptide
backbone, such as a(3-sheet or a-helical conformation; (2) the charge; (3)
hydrophobicity; or
(4) the bulk of the side chain of the target site, can modify MO-1 polypeptide
function or
immunological identity. Residues are divided into groups based on common side-
chain
properties as denoted in Table 2. Non-conservative substitutions entail
exchanging a member
of one of these classes for another class. Substitutions may be introduced
into conservative
substitution sites or more preferably into non-conserved sites.
Table 2
Amino acid classes
Amino Acids
Class
11
hydrophobic Norleucine, Met, Ala, Val, Leu, Ile
neutral hydrophilic Cys, Ser, Thr
acidic Asp, Glu
basic Asn, Gln, His, Lys, Arg
disrupt chain conformation Gly, Pro
aromatic Trp, Tyr, Phe
[0063] The variant polypeptides can be made using methods known in the art
such as
oligonucleotide-mediated (site-directed) mutagenesis, alanine scanning, and
PCR
mutagenesis. Site-directed mutagenesis (see Carter, Biochem. J. 237:1-7
(1986); Zoller and
Smith, Methods Enzymol. 154:329-50 (1987)), cassette mutagenesis, restriction
selection
mutagenesis (Wells et al., Gene 34:315-323 (1985)) or other known techniques
can be
performed on cloned MO-1-encoding DNA to produce MO-1 variant DNA (Ausubel et
al.,
Current Protocols In Molecular Biology, John Wiley and Sons, New York (current
edition);
Sambrook et al., Molecular Cloning, A Laboratory Manual, 3d. ed., Cold Spring
Harbor
Laboratory Press, Cold Spring Harbor, New York (2001).
[0064] In certain embodiments, MO-1 used in the present invention includes MO-
1
mutants or derivatives having an amino acid.substitution with a non-classical
amino acid or
chemical amino acid analog. Non-classical amino acids include, but are not
limited to, the D-
isomers of the common amino acids, a -amino isobutyric acid, 4-aminobutyric
acid, Abu, 2-
amino butyric acid, y-Abu, s-Ahx, 6-amino hexanoic acid, Aib, 2-amino
isobutyric acid, 3-
-13-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
amino propionic acid, ornithine, norleucine, norvaline, hydroxyproline,
sarcosine, citrulline,
cysteic acid, t-butylglycine, t-butylalanine, phenylglycine,
cyclohexylalanine, 0-alanine,
fluoro-amino acids, designer amino acids such as (3-methyl amino acids, Ca-
methyl amino
acids, Na-methyl amino acids, and amino acid analogs in general.
[0065] In one embodiment, the present invention includes an isolated
polypeptide
comprising an amino acid sequence having at least 70% identity to SEQ ID NO:
1. In some
embodiments, the polypeptide comprises an amino acid sequence having at least
75%, 80%,
85%, 90%, or 95% identity to SEQ ID NO: 1. In a particular embodiment, the
isolated
polypeptide comprises the amino acid sequence of SEQ ID NO: 1.
[0066] Percent identity in this context means the percentage of amino acid
residues in
the candidate sequence that are identical (i.e., the amino acid residues at a
given position in
the alignment are the same residue) or similar (i.e., the amino acid
substitution at a given
position in the alignment is a conservative substitution, as discussed above),
to the
corresponding amino acid residue in the peptide after aligning the sequences
and introducing
gaps, if necessary, to achieve the maximum percent sequence homology. In
certain
embodiments, a MO-1 homologue is characterized by its percent sequence
identity or percent
sequence similarity with the naturally occurring MO-1 sequence. Sequence
homology,
including percentages of sequence identity and similarity, are determined
using sequence
alignment techniques well-known in the art, preferably computer algorithms
designed for this
purpose, using the default parameters of said computer algorithms or the
software packages
containing them.
[0067] Non-limiting examples of computer algorithms and software packages
incorporating such algorithms include the following. The BLAST family of
programs
exemplify a preferred, non-limiting example of a mathematical algorithm
utilized for the
comparison of two sequences (e.g., Karlin & Altschul, 1990, Proc. Natl. Acad.
Sci. USA
87:2264-2268 (modified as in Karlin & Altschul, 1993, Proc. Natl. Acad. Sci.
USA 90:5873-
5877), Altschul et al., 1990, J. Mol. Biol. 215:403-410, (describing NBLAST
and XBLAST),
Altschul et al., 1997, Nucleic Acids Res. 25:3389-3402 (describing Gapped
BLAST, and PSI-
Blast). Another preferred example is the algorithm of Myers and Miller (1988
CABIOS 4:11 -
17) which is incorporated into the ALIGN program (version 2.0) and is
available as part of
the GCG sequence alignment software package. Also preferred is the FASTA
program
(Pearson W.R. and Lipman D.J., Proc. Nat. Acad. Sci. USA, 85:2444-2448, 1988),
available
as part of the Wisconsin Sequence Analysis Package. Additional examples
include
-14-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
BESTFIT, which uses the "local homology" algorithm of Smith and Waterman
(Advances in
Applied Mathematics, 2:482-489, 1981) to find best single region of similarity
between two
sequences, and which is preferable where the two sequences being compared are
dissimilar in
length; and GAP, which aligns two sequences by finding a "maximum similarity"
according
to the algorithm of Neddleman and Wunsch (J. Mol. Biol. 48:443-354, 1970), and
is
preferable where the two sequences are approximately the same length and an
alignment is
expected over the entire length.
[0068] Examples of homologues may be the ortholog proteins of other species
including animals, plants, yeast, bacteria, and the like. Homologues may also
be selected by,
e.g., mutagenesis in a native protein. For example, homologues may be
identified by site-
specific mutagenesis in combination with assays for detecting protein-protein
interactions.
Additional methods, e.g., protein affinity chromatography, affinity blotting,
in vitro binding
assays, and the like, will be apparent to skilled artisans apprised of the
present invention.
[0069] For the purpose of comparing two different nucleic acid or polypeptide
sequences,
one sequence (test sequence) may be described to be a specific "percent
identical to" another
sequence (reference sequence) in the present disclosure. In this respect, when
the length of the test
sequence is less than 90% of the length of the reference sequence, the
percentage identity is
determined by the algorithm of Myers and Miller, Bull. Math. Biol., 51:5-37
(1989) and Myers and
Miller, Comput. Appl. Biosci., 4(1):11-17 (1988). Specifically, the identity
is determined by the
ALIGN program. The default parameters can be used.
[0070] Where the length of the test sequence is at least 90% of the length of
the
reference sequence, the percentage identity is determined by the algorithm of
Karlin and
Altschul, Proc. Natl. Acad. Sci. USA, 90:5873-77 (1993), which is incorporated
into various
BLAST programs. Specifically, the percentage identity is determined by the
"BLAST 2
Sequences" tool. See Tatusova and Madden, FEMS Microbiol. Lett., 174(2):247-
250 (1999).
For pairwise DNA-DNA comparison, the BLASTN 2.1.2 program is used with default
parameters (Match: 1; Mismatch: -2; Open gap: 5 penalties; extension gap: 2
penalties; gap
x_dropoff: 50; expect: 10; and word size: 11, with filter). For pairwise
protein-protein
sequence comparison, the BLASTP 2.1.2 program is employed using default
parameters
(Matrix: BLOSUM62; gap open: 11; gap extension: 1; x_dropoff: 15; expect:
10.0; and
wordsize: 3, with filter).
-15-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
5.3 Nucleic Acids of the Invention
[0071] In another aspect, the present invention provides newly identified and
isolated
nucleotide sequences encoding MO-1. In particular, nucleic acids encoding
native sequence
human MO-1 polypeptides have been identified and isolated.
[0072] The MO-1-encoding or related sequences provided by the instant
invention
include those nucleotide sequences encoding substantially the same amino acid
sequences as
found in native MO-1, as well as those encoded amino acid sequences having
functionally
inconsequential amino acid substitutions, and thus having amino acid sequences
which differ
from that of the native sequence. Examples include the substitution of one
basic residue for
another (i.e. Arg for Lys), the substitution of one hydrophobic residue for
another (i.e. Leu
for Ile), or the substitution of one aromatic residue for another (i.e. Phe
for Tyr, etc.).
[0073] The invention further relates to fragments of MO-1. Nucleic acids
encoding
such fragments are thus also within the scope of the invention. The MO-1 gene
and MO-1-
encoding nucleic acid sequences of the invention include human and related
genes
(homologues) in other species. In some embodiments, the MO-1 gene and MO-1-
encoding
nucleic acid sequences are from vertebrates, or more particularly, mammals. In
a preferred
embodiment of the invention, the MO-i gene and MO-1-encoding nucleic acid
sequences are
of human origin.
[0074] In one aspect, the invention provides an isolated nucleic acid encoding
a
polypeptide comprising an amino acid sequence having at least 70% identity to
SEQ ID
NO:1. In some embodiments, the nucleic acid encodes a polypeptide comprising
an amino
acid sequence having at least 75%, 80%, 85%, 90%, or 95% identity to SEQ ID
NO:1. In a
particular embodiment, the isolated nucleic acid encodes a polypeptide
comprising the amino
acid sequence of SEQ ID NO:1.
[0075] In another embodiment, the invention provides an isolated nucleic acid
comprising a nucleic acid sequence having at least 70% identity to at least
about 500
contiguous nucleotides selected from SEQ ID NO:2 or the complement thereof. In
some
embodiments, the nucleic acid comprises a nucleic acid sequence having at
least 70% identity
to at least about 500, 600, 700, 800, 900, 1000, 1100, 1200, or 1400
contiguous nucleotides
selected from SEQ ID NO:2. In some embodiments, the nucleic acid comprises a
nucleic
acid sequence having at least 75% identity to at least about 500 contiguous
nucleotides
selected from SEQ ID NO:2. In some embodiments, the nucleic acid comprises a
nucleic
acid sequence having at least 75% identity to at least about 500, 600, 700,
800, 900, 1000,
-16-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
1100, 1200, or 1400 contiguous nucleotides selected from SEQ ID NO:2. In some
embodiments, the nucleic acid comprises a nucleic acid sequence having at
least 80% identity
to at least about 500 contiguous nucleotides selected from SEQ ID NO:2. In
some
embodiments, the nucleic acid comprises a nucleic acid sequence having at
least 80% identity
to at least about 500, 600, 700, 800, 900, 1000, 1100, 1200, or 1400
contiguous nucleotides
selected from SEQ ID NO:2. In some embodiments, the nucleic acid comprises a
nucleic
acid sequence having at least 85% identity to at least about 500 contiguous
nucleotides
selected from SEQ ID NO:2. In some embodiments, the nucleic acid comprises a
nucleic
acid sequence having at least 85% identity to at least about 500, 600, 700,
800, 900, 1000,
1100, 1200, or 1400 contiguous nucleotides selected from SEQ ID NO:2. In some
embodiments, the nucleic acid comprises a nucleic acid sequence having at
least 90% identity
to at least about 500 contiguous nucleotides selected from SEQ ID NO:2. In
some
embodiments, the nucleic acid comprises a nucleic acid sequence having at
least 90% identity
to at least about 500, 600, 700, 800, 900, 1000, 1100, 1200, or 1400
contiguous nucleotides
selected from SEQ ID NO:2. In some embodiments, the nucleic acid comprises a
nucleic
acid sequence having at least 95% identity to at least about 500 contiguous
nucleotides
selected from SEQ ID NO:2. In some embodiments, the nucleic acid comprises a
nucleic
acid sequence having at least 95% identity to at least about 500, 600, 700,
800, 900, 1000,
1100, 1200, or 1400 contiguous nucleotides selected from SEQ ID NO:2. In
certain
embodiments, the isolated nucleic acid comprises at least about 500
nucleotides selected from
the nucleic acid sequence of SEQ ID NO:2, or the complement thereof. In
certain
embodiments, the isolated nucleic acid comprises at least about 500, 600, 700,
800, 900,
1000, 1100, 1200, or 1400 nucleotides selected from the nucleic acid sequence
of SEQ ID
NO:2, or the complement thereof. In a particular embodiment, the isolated
nucleic acid
comprises the nucleic acid sequence of SEQ ID NO:2, or the complement thereof.
[0076] In another aspect, the invention provides an isolated nucleic acid
comprising a
nucleic acid sequence having at least 70% identity to at least about 500
contiguous
nucleotides selected from SEQ ID NO:3 or the complement thereof. SEQ ID NO:3
presents
the genomic sequence of a human MO-1 gene, including introns and exons. In
some
embodiments, the nucleic acid comprises a nucleic acid sequence having at
least 70% identity
to at least about 500, 600, 700, 800, 900, 1000, 1100, 1200, 1500, 2000, or
2500 contiguous
nucleotides selected from SEQ ID NO:3. In some embodiments, the nucleic acid
comprises a
nucleic acid sequence having at least 75% identity to at least about 500
contiguous
-17-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
nucleotides selected from SEQ ID NO:3. In some embodiments, the nucleic acid
comprises a
nucleic acid sequence having at least 75% identity to at least about 500, 600,
700, 800, 900,
1000, 1100, 1200, 1500, 2000, or 2500 contiguous nucleotides selected from SEQ
ID NO:3.
In some embodiments, the nucleic acid comprises a nucleic acid sequence having
at least
80% identity to at least about 500 contiguous nucleotides selected from SEQ ID
NO:3. In
some embodiments, the nucleic acid comprises a nucleic acid sequence having at
least 80%
identity to at least about 500, 600, 700, 800, 900, 1000, 1100, 1200, 1500,
2000, or 2500
contiguous nucleotides selected from SEQ ID NO:3. In some embodiments, the
nucleic acid
comprises a nucleic acid sequence having at least 85% identity to at least
about 500
contiguous nucleotides selected from SEQ ID NO:3. In some embodiments, the
nucleic acid
comprises a nucleic acid sequence having at least 85% identity to at least
about 500, 600,
700, 800, 900, 1000, 1100, 1200, 1500, 2000, or 2500 contiguous nucleotides
selected from
SEQ ID NO:3. In some embodiments, the nucleic acid comprises a nucleic acid
sequence
having at least 90% identity to at least about 500 contiguous nucleotides
selected from SEQ
ID NO:3. In some embodiments, the nucleic acid comprises a nucleic acid
sequence having
at least 90% identity to at least about 500, 600, 700, 800, 900, 1000, 1100,
1200, 1500, 2000,
or 2500 contiguous nucleotides selected from SEQ ID NO:3. In some embodiments,
the
nucleic acid comprises a nucleic acid sequence having at least 95% identity to
at least about
500 contiguous nucleotides selected from SEQ ID NO:3. In some embodiments, the
nucleic
acid comprises a nucleic acid sequence having at least 95% identity to at
least about 500, 600,
700, 800, 900, 1000, 1100, 1200, 1500, 2000, or 2500 contiguous nucleotides
selected from
SEQ ID NO:3. In certain embodiments, the isolated nucleic acid comprises at
least about 500
nucleotides selected from the nucleic acid sequence of SEQ ID NO:3, or the
complement
thereof. In certain embodiments, the isolated nucleic acid comprises at least
about 500, 600,
700, 800, 900, 1000, 1100, 1200, 1500, 2000, or 2500 nucleotides selected from
the nucleic
acid sequence of SEQ ID NO:3, or the complement thereof. In a particular
embodiment, the
isolated nucleic acid comprises the nucleic acid sequence of SEQ ID NO:3, or
the
complement thereof. In certain embodiments, the nucleic acid is not a member
of a nucleic
acid library. In certain embodiments, the nucleic acid is not a member of a
genomic library.
In certain embodiments, the nucleic acid is not a member of an expression
library.
[0077] The present invention also includes nucleic acids that hybridize to or
are
complementary to the foregoing sequences. In specific aspects, nucleic acids
are provided
which comprise a sequence complementary to at least 20, 30, 40, 50, 100, 200
nucleotides or
-18-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
the entire coding region of MO-1, or the reverse complement (antisense) of any
of these
sequences. In a specific embodiment, a nucleic acid which hybridizes to a MO-1
nucleic acid
sequence (e.g., having part or the whole of sequence SEQ ID NO:2, or the
complements
thereof), under conditions of low stringency is provided.
[0078] By way of example and not limitation, procedures using such conditions
of
low stringency are as follows (see also Shilo and Weinberg, 1981, Proc. Natl.
Acad. Sci.
U.S.A. 78:6789-6792). Filters containing DNA can be pretreated for 6h at 40
C. in a
solution containing 35% formamide, 5xSSC, 50 mM Tris-HCI (pH 7.5), 5 mM EDTA,
0.1%
PVP, 0.1% Ficoll, 1% BSA, and 500 g/ml denatured salmon sperm DNA.
Hybridizations
can be carried out in the same solution with the following modifications:
0.02% PVP, 0.02%
Ficoll, 0.2% BSA, 100 g/mi salmon sperm DNA, 10% (wdvol) dextran sulfate, and
5-
20x106 cpm 32P-labeled probe can be used. Filters can be incubated in
hybridization mixture
for 18-20h at 40 C., and then washed for 1.5h at 55 C. in a solution
containing 2xSSC, 25
mM Tris-HCl (pH 7.4), 5 mM EDTA, and 0.1% SDS. The wash solution can then be
replaced with fresh solution and incubated an additional 1.5h at 60 C.
Filters may be blotted
dry and exposed for autoradiography. If necessary, filters may be washed for a
third time at
65-68 C. and re-exposed to film. Other conditions of low stringency which may
be used are
well known in the art (e.g., as employed for cross-species hybridizations).
[0079] In another specific embodiment, a nucleic acid that hybridizes to a
nucleic
acid encoding MO-l, or its reverse complement, under conditions of high
stringency is
provided. By way of example and not limitation, procedures using such
conditions of high
stringency are as follows. Prehybridization of filters containing DNA may be
carried out for
8h to overnight at 65 C. in buffer composed of 6xSSC, 50 mM Tris-HCl (pH
7.5), 1 mM
EDTA, 0.02% PVP, 0.02% Ficoll, 0.02% BSA, and 500 g/ml denatured salmon sperm
DNA.
Filters may be hybridized for 48h at 65 C. in prehybridization mixture
containing 100 g/ml
denatured salmon sperm DNA and 5-20x106 cpm of 32P-labeled probe. Washing of
filters
may be done at 37 C. for 1 h in a solution containing 2xSSC, 0.01 % PVP, 0.01
% Ficoll, and
0.01% BSA. This can be followed by a wash in 0.1xSSC at 50 C. for 45 minutes
before
autoradiography. Other conditions of high stringency that may be used are well
known in the
art.
5.3.1 Cloning of the MO-1 gene or cDNA
[0080] The present invention further provides methods and compositions
relating to
the cloning of a gene or cDNA encoding MO-1. In one embodiment of the
invention,
-19-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
expression cloning (a technique commonly known in the art), may be used to
isolate a gene
or cDNA encoding MO-1. An expression library may be constructed by any method
known
in the art. In one embodiment, mRNA (e.g., human) is isolated, and cDNA is
made and
ligated into an expression vector such that the cDNA is capable of being
expressed by the
host cell into which it is introduced. Various screening assays can then be
used to select for
the expressed MO-1 product. In one embodiment, anti-MO-1 antibodies can be
used for
selection.
[0081] In another embodiment of the invention, polymerase chain reaction (PCR)
may be used to amplify desired nucleic acid sequences of the present invention
from a
genomic or cDNA library. Isolated oligonucleotide primers representing known
MO-1-
encoding sequences can be used as primers in PCR. In certain embodiments, the
isolated
oligonucleotide primer comprises at least 10 consecutive nucleotides of SEQ ID
NO:2 or its
complimentary strand. In certain embodiments, the oligonucleotide comprises
the nucleic
acid sequence of SEQ ID NO:4. In certain embodiments, the oligonucleotide
comprises the
nucleic acid sequence of SEQ ID NO:5. In certain embodiments, the
oligonucleotide
comprises the nucleic acid sequence of SEQ ID NO:6. In certain embodiments,
the
oligonucleotide comprises the nucleic acid sequence of SEQ ID NO:7. In certain
embodiments, the oligonucleotide comprises the nucleic acid sequence of SEQ ID
NO:8. In
certain embodiments, the oligonucleotide comprises the nucleic acid sequence
of SEQ ID
NO:9. In certain embodiments, the oligonucleotide comprises the nucleic acid
sequence of
SEQ ID NO:10. The synthetic oligonucleotides may be utilized as primers to
amplify by
PCR sequences from RNA or DNA, preferably a cDNA library, of potential
interest.
Alternatively, one can synthesize degenerate primers for use in the PCR
reactions.
[0082] In the PCR reactions, the nucleic acid being amplified can include RNA
or
DNA, for example, mRNA, cDNA or genomic DNA from any eukaryotic species. PCR
can
be carried out, e.g., by use of a Perkin-Elmer Cetus thermal cycler and Taq
polymerase. It is
also possible to vary the stringency of hybridization conditions used in
priming the PCR
reactions, to allow for greater or lesser degrees of nucleotide sequence
similarity between a
known MO-1 nucleotide sequence and a nucleic acid homologue being isolated.
For cross-
species hybridization, low stringency conditions are preferred. For same-
species
hybridization, moderately stringent conditions are preferred. After successful
amplification
of a segment of a MO-1 homologue, that segment may be cloned, sequenced, and
utilized as
a probe to isolate a complete cDNA or genomic clone. This, in turn, will
permit the
-20-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
determination of the gene's complete nucleotide sequence, the analysis of its
expression, and
the production of its protein product for functional analysis. In this
fashion, additional
nucleotide sequences encoding MO-1 or MO-1 homologues may be identified.
[0083] The above recited methods are not meant to limit the following general
description of methods by which clones of genes encoding MO-1 or homologues
thereof may
be obtained.
[0084] Any eukaryotic cell potentially can serve as the nucleic acid source
for the
molecular cloning of the MO-1 gene, MO-1 cDNA or a homologue thereof. The
nucleic acid
sequences encoding MO-1 can be isolated from vertebrate, mammalian, human,
porcine,
bovine, feline, avian, equine, canine, as well as additional primate sources.
The DNA may be
obtained by standard procedures known in the art from cloned DNA (e.g., a DNA
"library"),
by chemical synthesis, by cDNA cloning, or by the cloning of genomic DNA, or
fragments
thereof, purified from the desired cell, or by PCR amplification and cloning.
See, for
example, Sambrook et al., Molecular Cloning, A Laboratory Manual, 3d. ed.,
Cold Spring
Harbor Laboratory Press, Cold Spring Harbor, New York (2001); Glover, D.M.
(ed.), DNA
Cloning: A Practical Approach, 2d. ed., MRL Press, Ltd., Oxford, U.K. (1995).
Clones
derived from genomic DNA may contain regulatory and intron DNA regions in
addition to
coding regions; clones derived from cDNA will contain only exon sequences.
Whatever the
source, the gene may be cloned into a suitable vector for propagation of the
gene.
[0085] In the cloning of the gene from genomic DNA, DNA fragments are
generated,
some of which will encode the desired gene. The DNA may be cleaved at specific
sites using
various restriction enzymes. Alternatively, one may use DNase in the presence
of manganese
to fragment the DNA, or the DNA can be physically sheared, as for example, by
sonication.
The linear DNA fragments can then be separated according to size by standard
techniques,
including but not limited to, agarose and polyacrylamide gel electrophoresis
and column
chromatography.
[0086] Once the DNA fragments are generated, identification of the specific
DNA
fragment containing the desired gene may be accomplished in a number of ways.
For
example, if a MO-1 gene (of any species) or its specific RNA is available and
can be purified
and labeled, the generated DNA fragments may be screened by nucleic acid
hybridization to
the labeled probe (Benton and Davis, Science 196:180 (1977); Grunstein and
Hogness, Proc.
Natl. Acad. Sci. U.S.A. 72:3961 (1975). Those DNA fragments with substantial
homology to
the probe will hybridize. It is also possible to identify the appropriate
fragment by restriction
-21-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
enzyme digestion(s) and comparison of fragment sizes with those expected
according to a
known restriction map if such is available. Further selection can be carried
out on the basis
of the properties of the gene.
[0087] Alternatively, the presence of the gene may be detected by assays based
on the
physical, chemical, or immunological properties of its expressed product. For
example,
cDNA clones, or DNA clones that hybrid-select the proper mRNAs, can be
selected that
produce a protein having e.g., similar or identical electrophoretic migration,
isoelectric
focusing behavior, proteolytic digestion maps, substrate binding activity, or
antigenic
properties as known for a specific MO-1. If an antibody to a particular MO-1
is available,
that MO-1 may be identified by binding of labeled antibody to the clone(s)
putatively
producing the MO-1 in an ELISA (enzyme-linked immunosorbent assay)-type
procedure.
[0088] A MO-1 or homologue thereof can also be identified by mRNA selection by
nucleic acid hybridization followed by in vitro translation. In this
procedure, fragments are
used to isolate complementary mRNAs by hybridization. Such DNA fragments may
represent available, purified DNA of another species containing a gene
encoding MO-1.
Immunoprecipitation analysis or functional assays of the in vitro translation
products of the
isolated mRNAs identifies the mRNA and, therefore, the complementary DNA
fragments
that contain the desired sequences. In addition, specific mRNAs may be
selected by
adsorption of polysomes isolated from cells to immobilized antibodies
specifically directed
against a specific MO-1. A radiolabelled MO-1-encoding cDNA can be synthesized
using
the selected mRNA (from the adsorbed polysomes) as a template. The
radiolabelled mRNA
or cDNA may then be used as a probe to identify the MO-1-encoding DNA
fragments from
among other genomic DNA fragments.
[0089] Alternatives to isolating the MO-1 genomic DNA include, but are not
limited
to, chemically synthesizing the gene sequence itself from a known sequence or
making
cDNA to the mRNA which encodes MO-1. For example RNA for the cloning of MO-1
cDNA can be isolated from cells that express a MO-1 gene. Other methods are
possible and
within the scope of the invention.
[0090] The identified and isolated MO-1 or MO-1 analog-encoding gene can then
be
inserted into an appropriate cloning vector. A large number of vector-host
systems known in
the art may be used. Possible cloning vectors include, but are not limited to,
plasmids or
modified viruses, but the vector system must be compatible with the host cell
used. Such
vectors include, but are not limited to bacteriophages such as lambda
derivatives, or plasmids
-22-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
such as pBR322, pUC plasmid derivatives, or the pBluescript vector.
(Stratagene). The
insertion into a cloning vector can, for example, be accomplished by ligating
the DNA
fragment into a cloning vector which has complementary cohesive termini.
However, if the
complementary restriction sites used to fragment the DNA are not present in
the cloning
vector, the ends of the DNA molecules may be enzymatically modified.
Alternatively, any
site desired may be produced by ligating nucleotide sequences (linkers) onto
the DNA
termini. These ligated linkers may comprise specific chemically synthesized
oligonucleotides encoding restriction endonuclease recognition sequences. In
an alternative
method, the cleaved vector and MO-1-encoding gene or nucleic acid sequence may
be
modified by homopolymeric tailing. Recombinant molecules can be introduced
into host
cells via transformation, transfection, infection, electroporation, etc., so
that many copies of
the gene sequence are generated.
[00911 In an alternative method, the desired gene may be identified and
isolated after
insertion into a suitable cloning vector in a "shotgun" approach. Enrichment
for the desired
gene, for example, by size fractionization, can be done before insertion into
the cloning
vector.
[0092] To generate multiple copies of the isolated MO-1-encoding gene, cDNA,
or
synthesized DNA sequence, host cells, for example competent strains of E.
Coli, may be
transformed with recombinant DNA molecules incorporating said sequences
according to any
technique known in the art. Thus, the gene may be obtained in large quantities
by growing
transformants, isolating the recombinant DNA molecules from the transformants
and, when
necessary, retrieving the inserted gene from the isolated recombinant DNA.
5.3.2 Expression Vectors
[0093] In still another aspect, the invention provides expression vectors for
expressing isolated MO-1-encoding sequences, e.g., cDNA sequences. Generally,
expression
vectors are recombinant polynucleotide molecules comprising expression control
sequences
operatively linked to a nucleotide sequence encoding a polypeptide. Expression
vectors can
readily be adapted for function in prokaryotes or eukaryotes by inclusion of
appropriate
promoters, replication sequences, selectable markers, etc. to result in stable
transcription and
translation of mRNA. Techniques for construction of expression vectors and
expression of
genes in cells comprising the expression vectors are well known in the art.
See, e.g.,
Sambrook et al., 2001, Molecular Cloning - A Laboratory Manual, 3d edition,
Cold Spring
Harbor Laboratory, Cold Spring Harbor, NY, and Ausubel et al., eds., Current
Edition,
- 23 -
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
Current Protocols in Molecular Biology, Greene Publishing Associates and Wiley
Interscience, NY.
[0094] Useful promoters for use in expression vectors include, but are not
limited to,
a metallothionein promoter, a constitutive adenovirus major late promoter, a
dexamethasone-
inducible MMTV promoter, a SV40 promoter, a MRP pol III promoter, a
constitutive MPSV
promoter, an RSV promoter, a tetracycline-inducible CMV promoter (such as the
human
immediate-early CMV promoter), and a constitutive CMV promoter. In one
embodiment, the
promoter is an adipocyte-specific promoter.
[0095] The expression vectors should contain expression and replication
signals
compatible with the cell in which the MO-1-encoding sequences are to be
expressed.
Expression vectors useful for expressing MO-1-encoding sequences include viral
vectors
such as retroviruses, adenoviruses and adenoassociated viruses, plasmid
vectors, cosmids,
and the like. Viral and plasmid vectors are preferred for transfecting the
expression vectors
into mammalian cells. For example, the expression vector pcDNAI (Invitrogen,
San Diego,
CA), in which the expression control sequence comprises the CMV promoter,
provides good
rates of transfection and expression into such cells.
[0096] The expression vectors can be introduced into the cell for expression
of the
MO-1-encoding sequence by any method known to one of skill in the art without
limitation.
Such methods include, but are not limited to, e.g., direct uptake of the
recombinant DNA
molecule by a cell from solution; facilitated uptake through lipofection
using, e.g., liposomes
or immunoliposomes; particle-mediated transfection; etc. See, e.g., U.S.
Patent No.
5,272,065; Goeddel et al., Methods in Enzymology, vol. 185, Academic Press,
Inc., CA
(1990); Krieger, Gene Transfer and Expression - A Laboratory Manual, Stockton
Press,
New York (1990); Ausubel et al., Current Protocols In Molecular Biology, John
Wiley and
Sons, New York (current edition); Sambrook et al., Molecular Cloning, A
Laboratory
Manual, 3d. ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New
York
(2001).
[0097] The expression vectors can also contain a purification moiety that
simplifies
isolation of the expressed protein. For example, a polyhistidine moiety of,
e.g., six histidine
residues, can be incorporated at the amino terminal end of the protein. The
polyhistidine
moiety allows convenient isolation of the protein in a single step by nickel-
chelate
chromatography. In certain embodiments, the purification moiety can be cleaved
from the
remainder of the delivery construct following purification. In other
embodiments, the moiety
-24-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
does not interfere with the function of the functional domains of the
expressed protein of the
invention and thus need not be cleaved.
5.3.3 Cells
[0098] In yet another aspect, the invention provides a cell comprising a
vector, e.g.,an
expression vector for expression of MO-1 polypeptides of the invention, or
portions thereof.
The cell is preferably selected for its ability to express high concentrations
of the MO-1
polypeptide to facilitate subsequent purification of the polypeptide. In
certain embodiments,
the cell is a prokaryotic cell, for example, E. coli. In a preferred
embodiment, the MO-1
polypeptide is properly folded and comprises the appropriate disulfide
linkages when
expressed in E. coli.
[0099] In other embodiments, the cell is a eukaryotic cell. Useful eukaryotic
cells
include, for example, plant, yeast and mammalian cells. Any mammalian cell
known by one
of skill in the art to be useful for expressing a recombinant polypeptide,
without limitation,
can be used to express the polypeptide of interest. For example, Chinese
hamster ovary
(CHO) cells can be used to express the MO-1 polypeptides of the invention. In
some
embodiments, the MO-1 polypeptide is expressed in adipocytes, e.g., human
adipocytes. In
some embodiments, the MO-1 polypeptide is labeled with a moiety to, for
example, facilitate
purification or identification, e.g., a FLAG tag, a GST tag, or a V-5 tag.
[00100] In yet other embodinients, the cell has been engineered to overexpress
MO-1.
In some embodiments, the cell expresses MO-1 at least about 10%, 20%, 30%,
40%, 50%,
60%, 70%, 80%, 90%, 100%, 150%, 200%, 300%, 400%, or 500% more than a
corresponding cell that has not been engineered to overexpress MO-1. In yet
other
embodiments, expression of MO-1 in the cell has been silenced. In certain
embodiments,
expression of MO-1 is reduced by at least about 10%, 20%, 30%, 40%, 50%, 60%,
70%,
80%, 90%, 95%, 99%, or 99.9% relative to a corresponding cell where expression
of MO-i
has not been silenced.
5.4 Antibodies
[00101] According to the invention, MO-1, or its fragments thereof, may be
used as an
immunogen to generate antibodies which immunospecifically bind MO-1
polypeptides. Such
antibodies include, but are not limited to, polyclonal, monoclonal, single
chain monoclonal,
recombinant, chimeric, humanized, mammalian, or human antibodies.
[00102] In some embodiments, antibodies to a non-human MO-1 are produced. In
certain embodiments, antibodies to mouse or rat MO-1 are produced. In other
embodiments,
- 25 -
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
antibodies to human MO-1 are produced. In another embodiment, antibodies are
produced
that specifically bind to a protein the amino acid sequence of which consists
of SEQ ID
NO:1. In another embodiment, antibodies to a fragment of non-human MO-1 are
produced.
In another embodiment, antibodies to a fragment of human MO-1 are produced. In
a specific
embodiment, fragments of MO-1, human or non-human, identified as containing
hydrophilic
regions are used as immunogens for antibody production. In a specific
embodiment, a
hydrophilicity analysis can be used to identify hydrophilic regions of MO-1,
which are
potential epitopes, and thus can be used as immunogens.
[00103] For the production of antibody, various host animals can be immunized
by
injection with native MO-1, or a synthetic version, or a fragment thereof. In
certain
embodiments, the host animal is a mammal. In some embodiments, the mammal is a
rabbit,
mouse, rat, goat, cow or horse.
[00104] For the production of polyclonal antibodies to MO-1, various
procedures
known in the art may be used. In a particular embodiment, rabbit polyclonal
antibodies to an
epitope of MO-1 encoded by a sequence of SEQ ID NO:2 or a subsequence thereof,
can be
obtained. Various adjuvants may be used to increase the immunological
response, depending
on the host species. Adjuvants that may be used according to the present
invention include,
but are not limited to, Freund's (complete and incomplete), mineral gels such
as aluminum
hydroxide, surface active substances such as lysolecithin, pluronic polyols,
polyanions,
peptides, oil emulsions, keyhole limpet hemocyanins, dinitrophenol, CpG-
containing nucleic
acids, and potentially useful human adjuvants such as BCG (bacille Calmette-
Guerin) and
Corynebacterium parvum.
[00105] For preparation of monoclonal antibodies directed toward a MO-1
polypeptide, any technique that provides for the production of antibody
molecules by
continuous cell lines in culture may be used. For example, monoclonal
antibodies may be
prepared by the hybridoma technique originally developed by Kohler and
Milstein, Nature
256:495-497 (1975), as well as the trioma technique, the human B-cell
hybridoma technique
(Kozbor et al., Immunol. Today 4:72 (1983)), or the EBV-hybridoma technique
(Cole et al.,
in Monoclonal Antibodies and Cancer Therapy, Alan R. Liss, Inc., pp. 77-96
(1985)).
[00106] Techniques for the production of single chain antibodies, as described
in U.S.
Patent 4,946,778, can also be adapted to produce single chain antibodies
specific to MO-1.
An additional embodiment of the invention utilizes the techniques described
for the
construction of Fab expression libraries (Huse et al., Science 246:1275-1281
(1988)) to allow
-26-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
rapid and easy identification of monoclonal Fab fragments with the desired
specificity for
MO-1. Antibody fragments that contain the idiotype of the molecule can be
generated by
known techniques. For example, such fragments include but are not limited to:
the F(ab'),
fragment which can be produced by pepsin digestion of the antibody molecule;
the Fab'
fragments which can be generated by reducing the disulfide bridges of the
F(ab'), fragment,
the Fab fragments which can be generated by treating the antibody molecule
with papain and
a reducing agent, and Fv fragments.
[00107] Techniques developed for the production of "chimeric" antibodies
(Morrison
et al., Proc. Natl. Acad. Sci. U.S.A. 81:6851-6855 (1984); Neuberger et al.,
Nature 312:604-
608 (1984); Takeda et al., Nature 314:452-454 (1985)) can also be used. For
example,
nucleic acid sequences encoding a mouse antibody molecule specific to MO-1 are
spliced to
nucleic acid sequences encoding a human antibody molecule.
[00108] In addition, techniques have been developed for the production of
humanized
antibodies, and such humanized antibodies to MO-1 are within the scope of the
present
invention. See, e.g., Queen, U.S. Patent No. 5,585,089 and Winter, U.S. Patent
No.
5,225,539. An immunoglobulin light or heavy chain variable region consists of
a
"framework" region interrupted by three hypervariable regions, referred to as
complementarity determining regions (CDRs). The extent of the framework region
and
CDRs have been precisely defined. See, Sequences of Proteins of Immunological
Interest,
Kabat, E. et al., U.S. Department of Health and Human Services (1983).
Briefly, humanized
antibodies are antibody molecules from non-human species having one or more
CDRs from
the non-human species and a framework region from a human immunoglobulin
molecule.
[00109] Human antibodies may be used and can be obtained by using human
hybridomas (Cote et al., Proc. Natl. Acad. Sci. U.S.A., 80:2026-2030 (1983))
or by
transforming human B cells with EBV virus in vitro (Cole et al., in Monoclonal
Antibodies
and Cancer Therapy, Alan R. Liss, pp. 77-96 (1985)).
[00110] In the production of antibodies, screening for the desired antibody
can be
accomplished by techniques known in the art, e.g. ELISA (enzyme-linked
immunosorbent
assay), RIA (radioimmunoassay) or RIBA (recombinant immunoblot assay). For
example,
to select antibodies which recognize a specific domain of MO-1, one may assay
generated
hybridomas for a product which binds to a MO-1 fragment containing such
domain. For
selection of an antibody that specifically binds a first MO-1 homologue but
which does not
-27-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
specifically bind a second, different MO-1 homologue, one can select on the
basis of positive
binding to the first MO-1 homologue and a lack of binding to the second MO-1
homologue.
[00111] Antibodies specific to a domain of MO-1 or a homologue thereof are
also
provided. The foregoing antibodies can be used in methods known in the art
relating to the
localization and activity of the MO-1 of the invention, e.g., for imaging
these proteins,
measuring levels thereof in appropriate physiological samples, in diagnostic
methods, etc.
5.5 Transgenic MO-1 Animals
[00112] Transgenic animals are useful, e.g., for identifying and/or evaluating
modulators of MO-1 activity, and, as such, for identifying modulators to be
tested for that
ability. Transgenes direct the expression of an encoded gene product in one or
more cell
types or tissues of the transgenic animal. In some embodiments, transgenes
prevent the
expression of a naturally encoded gene product in one or more cell types or
tissues (a
"knockout" transgenic animal). In some embodiments, transgenes serve as a
marker or
indicator of an integration, chromosomal location, or region of recombination
(e.g., cre/loxP
mice).
1001131 A transgenic animal can be created by introducing a nucleic acid of
the
invention into the male pronuclei of a fertilized oocyte (e.g., by
microinjection, retroviral
infection) and allowing the oocyte to develop in a pseudopregnant female
foster animal
(PFFA). The MO-1 sequences can be introduced as a transgene into the genome of
a non-
human animal. In some embodiments, the MO-1 sequence is the human MO-1
sequence
(SEQ ID NO:2). In other embodiments, a homologue of MO-1 can be used as a
transgene.
Intronic sequences and polyadenylation signals can also be included in the
transgene to
increase transgene expression. Tissue-specific regulatory sequences can be
operably-linked
to the MO-1 transgene to direct expression of MO-1 to particular cells.
Methods for
generating transgenic animals via embryo manipulation and microinjection,
particularly
animals such as mice, have become conventional in the art, e.g., Evans et al.,
U.S. Pat. No.
4,870,009 (1994); Leder and Stewart, U.S. Pat. No. 4,736,866, 1988; Wagner and
Hoppe,
U.S. Pat. No. 4,873,191 (1989). Other non-mice transgenic animals may be made
by similar
methods. A transgenic founder animal, which can be used to breed additional
transgenic
animals, can be identified based upon the presence of the transgene in its
genome and/or
expression of the transgene mRNA in tissues or cells of the animal. Transgenic
MO-1
animals can be bred to other transgenic animals carrying other transgenes.
-28-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
[00114] To create a homologous recombinant animal, a vector containing at
least a
portion of MO-1 into which a deletion, addition or substitution may be
introduced to thereby
alter, e.g., functionally disrupt MO-1 expression. In some embodiments, the
vector may
contain a neomycin cassette inserted in reverse orientation relative to MO-1
transcription to
functionally disrupt MO-1. MO-1 can be a human gene (e.g., SEQ ID NO:2 or 3),
or other
MO-1 homologue. In one approach, a knockout vector functionally disrupts the
endogenous
MO-1 gene upon homologous recombination, and thus a non-functional MO-1
protein, if any,
is expressed.
[00115] Alternatively, the vector can be designed such that, upon homologous
recombination, the endogenous MO-1 is mutated or otherwise altered but still
encodes
functional protein (e.g., the upstream regulatory region can be altered to
thereby alter the
expression of endogenous MO-1). In this type of homologous recombination
vector, the
altered portion of the MO-1 sequence is flanked at its 5'- and 3'-termini by
additional nucleic
acid sequence of MO-1 to allow for homologous recombination to occur between
the
exogenous MO-1 sequence carried by the vector and an endogenous MO-1 sequence
in an
embryonic stem cell. The additional flanking MO-1 sequence is sufficient to
engender
homologous recombination with endogenous MO-1. Typically, several kilobases of
flanking
DNA (both at the 5'- and 3'-termini) are included in the vector (see Thomas
and Capecchi,
Cell 51:503-512 (1987)).
[00116] The vector can then be introduced into an embryonic stem cell line
(e.g., by
electroporation), and cells in which the introduced MO-1 sequence has
homologously-
recombined with the endogenous MO-1 sequence are selected (Li et al., Cell
69:915-926
(1992)).
[00117] Selected cells are then injected into a blastocyst of an animal (e.g.,
a mouse) to
form aggregation chimeras (see Bradley, Teratocarcinomas and Embryonic Stem
Cells: A
Practical Approach, Oxford University Press, Inc., Oxford (1987)). A chimeric
embryo can
then be implanted into a suitable PFFA, wherein the embryo is brought to term.
Progeny
harboring the homologously-recombined DNA in their germ cells can be used to
breed
animals in which all cells of the animal contain the homologously-recombined
DNA by
germline transmission of the transgene. Methods for constructing homologous
recombination
vectors and homologous recombinant animals are described (Berns et al., WO
93/04169,
1993; Kucherlapati et al., WO 91/01140, 1991; Le Mouellic and Brullet, WO
90/11354,
1990).
-29-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
[00118] , Alternatively, transgenic animals that contain selected systems that
allow for
regulated expression of the transgene can be produced. An example of such a
system is the
cre/loxP recombinase system of bacteriophage P1 (Lakso et al., Proc. Natl.
Acad. Sci. USA
89:6232-6236 (1992)). Another recombinase system is the FLP recombinase system
of
Saccharomyces cerevisiae (O'Gorman et al., Science 251:1351-1355 (1991)). If a
cre/loxP
recombinase system is used to regulate expression of the transgene, animals
containing
transgenes encoding both the Cre recombinase and a selected protein are
required. Such
animals can be produced as "double" transgenic animals, by mating an animal
containing a
transgene encoding a selected protein to another containing a transgene
encoding a
recombinase.
[00119] Clones of transgenic animals can also be produced (Wilmut et al.,
Nature
385:810-813 (1997)). In brief, a cell from a transgenic animal can be isolated
and induced to
exit the growth cycle and enter Go phase. The quiescent cell can then be fused
to an
enucleated oocyte from an animal of the same species from which the quiescent
cell is
isolated. The reconstructed oocyte is then cultured to develop to a morula or
blastocyte and
then transferred to a PFFA. The offspring borne of this female foster animal
will be a clone
of the "parent" transgenic animal.
[00120] In certain embodiments, the transgeneic animal exhibits a phenotype
associated with altered MO-1 activity, e.g., obesity.
5.6 Methods of Screening for Modulators of MO-1 Activity
[00121] The present invention also provides methods of identifying a compound
that
modulates the activity of MO-1 in a cell or tissue of interest. A compound may
modulate
MO-1 activity by affecting, for example: (1) the number of copies of the MO-1
gene in the
cell (amplifiers and deamplifiers); (2) increasing or decreasing transcription
of the MO-1
gene (transcription up-regulators and down-regulators); (3) by increasing or
decreasing the
translation of the MO-1 mRNA into protein (translation up regulators and down
regulators);
(4) by increasing or decreasing the activity of the MO-1 protein (agonists and
antagonists), or
5) by facilitating the proper folding of the MO-1 protein (pharmacological
chaperonins). To
identify compounds that affect MO-1 at the DNA, RNA, and protein levels, cells
or
organisms are contacted with a candidate compound and the corresponding change
in MO-1
DNA, RNA or protein may be assessed. For DNA amplifiers or deamplifiers, the
amount of
MO-1 DNA may be measured. For those compounds that are transcription up-
regulators and
down-regulators, the amount of MO-1 mRNA may be measured. Alternatively, the
MO-1
-30-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
promoter sequence may be operably linked to a reporter gene, and potential
transcriptional
modulators of MO-1 may be assayed by measuring reporter gene activity in the
presence and
absence. of the compound. For translational up- and down- regulators, the
amount of MO-1
polypeptide may be measured. Alternatively, changes in MO-1 biological
activity, as
measured by the techniques described below, may be an indirect indicator of
the ability of a
compound to modulate MO-1 translation.
[00122] In one embodiment, the cell or tissue useful for the methods described
herein
expresses a MO-1 polypeptide from an endogenous copy of the MO-1 gene. In
another
embodiment, the cell or tissue expresses a MO-1 polypeptide following
transient or stable
transformation with a nucleic acid encoding a MO-1 polypeptide of the present
invention.
Any mammalian cell known by one of skill in the art to be useful for
expressing a
recombinant polypeptide, without limitation, can be used to express a MO-1
polypeptide
useful for the methods described herein.
[00123] In one embodiment, the method of identifying a compound that modulates
the
activity of MO-1 comprises determining a first level of MO-1 activity in a
cell or tissue that
expresses a MO-1 polypeptide, contacting said cell or tissue with a test
compound, then
determining a second level of MO-1 activity in said cell or tissue. A
difference in the first
level and second level of MO-1 activity is indicative of the ability of the
test compound to
modulate MO-1 activity. In one embodiment, a compound may have agonistic
activity if the
second level of MO-1 activity is greater than the first level of MO-1
activity. In certain
embodiments, agonistic activity comprises at least about a 2, 4, 6, 8, 10, or
greater fold
increase in the second level of MO-1 activity compared to the first level of
MO-1 activity. In
another embodiment, a compound may have antagonistic activity if the second
level of MO-1
activity is less than the first level of MO-1 activity. In certain
embodiments, antagonistic
activity comprises at least about a 2, 4, 6, 8, 10, or greater fold decrease
in the second level of
MO-1 activity compared to the first level of MO-1 activity.
[00124] In another embodiment, the invention provides a method of identifying
a
compound that modulates the activity of MO-1 in a cell or tissue expressing a
MO-1
polypeptide, comprising contacting said cell or tissue with a test compound
and determining a
level of MO-1 in said cell or tissue. The difference in this level and a
standard or baseline
level of MO-1 activity in a comparable cell or tissue, e.g., a control cell or
tissue not
contacted with the test compound, is indicative of the ability of said test
compound to
modulate MO-1 activity. In one embodiment, a compound may have agonistic
activity if the
-31-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
level of MO-1 activity in the cell or tissue contacted with said compound is
greater than the
level of MO-1 activity in the control cell or tissue. In certain embodiments,
agonistic activity
comprises at least about a 2-, 4-, 6-, 8-, 10-, or greater fold increase in
the level of MO-1
activity of a cell or tissue contacted with the test compound compared to the
level of MO-1
activity in the control cell or tissue. In another embodiment, a compound may
have
antagonistic activity if the level of MO-1 activity in the cell or tissue
contacted with said
compound is less than the level of MO-1 activity in the control cell or
tissue. In certain
embodiments, antagonistic activity comprises at least about a 2-, 4-, 6-, 8-,
10-, or greater
fold decrease in the level of MO-i activity of a cell or tissue contacted with
the test
compound compared to the level of MO-1 activity in the control cell or tissue.
[00125] The present invention also provides methods of identifying a compound
that
modulates the activity of MO-1 in a transgenic non-human animal which
expresses a MO-1
polypeptide, comprising administering the compound to said animal and
assessing the animal
for an alteration in metabolic function affected by the compound. Metabolic
function may be
assessed through the measurement of glucose concentrations, lipid
concentrations, mass of
the animal, and the like.
[00126] The present invention also provides methods of identifying compounds
that
specifically bind to MO-1 nucleic acids or polypeptides and thus have
potential use as
agonists or antagonists of MO-1. In certain embodiments, such compounds may
affect
glucose concentrations, lipid concentrations, mass of the animal, etc. In a
preferred
embodiment, assays are performed to screen for compounds having potential
utility as
therapies for metabolic disorders or lead compounds for drug development. The
invention
thus provides assays to detect compounds that specifically bind to MO-1
nucleic acids or
polypeptides. For example, recombinant cells expressing MO-1 nucleic acids can
be used to
recombinantly produce MO-1 polypeptides for use in these assays, e.g., to
screen for
compounds that bind to MO-1 polypeptides. Said compounds (e.g., putative
binding partners
of MO-1) are contacted with a MO-1 polypeptide or a fragment thereof under
conditions
conducive to binding, and compounds that specifically bind to MO-1 are
identified. Similar
methods can be used to screen for compounds that bind to MO-1 nucleic acids.
Methods that
can be used to carry out the foregoing are commonly known in the art.
[00127] In various embodiments, the MO-1-modulating compound is a protein, for
example, an antibody; a nucleic acid; or a small molecule. As used herein, the
term "small
molecule" includes, but is not limited to, organic or inorganic compounds
(i.e., including
-32-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
heteroorganic and organometallic compounds) having a molecular weight less
than 10,000
grams per mole, organic or inorganic compounds having a molecular weight less
than 5,000
grams per mole, organic or inorganic compounds having a molecular weight less
than 1,000
grams per mole, organic or inorganic compounds having a molecular weight less
than 500
grams per mole, organic or inorganic compounds having a molecular weight less
than 100
grams per mole, and salts, esters, and other pharmaceutically acceptable forms
of such
compounds. Salts, esters, and other pharmaceutically acceptable forms of such
compounds
are also encompassed.
[00128] By way of example, diversity libraries, such as random or
combinatorial
peptide or nonpeptide libraries can be screened for molecules that
specifically bind to MO- 1.
Many libraries are known in the art that can be used, e.g., chemically
synthesized libraries,
recombinant (e.g., phage display libraries), and in vitro translation-based
libraries.
[00129] Examples of chemically synthesized libraries are described in Fodor et
al.,
Science 251:767-773 (1991); Houghten et al., Nature 354:84-86 (1991); Lam et
al., Nature
354:82-84 (1991); Medynski, Bio/Technology 12:709-710 (1994); Gallop et al.,
J. Medicinal
Chemistry 37(9):1233-1251 (1994); Ohlmeyer et al., Proc. Natl. Acad. Sci.
U.S.A. 90:10922-
10926 (1993); Erb et al., Proc. Natl. Acad. Sci. U.S.A. 91:11422-11426 (1994);
Houghten et
al., Biotechniques 13:412 (1992); Jayawickreme et al., Proc. Natl. Acad. Sci.
U.S.A. 91:1614-
1618 (1994); Salmon et al., Proc. Natl. Acad. Sci. U.S.A. 90:11708-11712
(1993); PCT
Publication No. WO 93/20242; and Brenner and Lemer, Proc. Natl. Acad. Sci.
U.S.A.
89:5381-5383 (1992).
[00130] Examples of phage display libraries are described in Scott and Smith,
Science
249:386-390 (1990); Devlin et al., Science, 249:404-406 (1990); Christian,
R.B., et al., J.
Mol. Biol. 227:711-718 (1992)); Lenstra, J. Immunol. Meth. 152:149-157 (1992);
Kay et al.,
Gene 128:59-65 (1993); and PCT Publication No. WO 94/18318, published August
18, 1994.
In vitro translation-based libraries include but are not limited to those
described in PCT
Publication No. WO 91/05058, published April 18, 1991; and Mattheakis et al.,
Proc. Natl.
Acad. Sci. U.S.A. 91:9022-9026 (1994).
[00131] By way of examples of non-peptide libraries, a benzodiazepine library
(see
e.g., Bunin et al., Proc. Natl. Acad. Sci. US.A. 91:4708-4712 (1994)) can be
adapted for use.
Peptoid libraries (Simon et al., Proc. Natl. Acad. Sci. U.SA. 89:9367-9371
(1992)) can also
be used. Another example of a library that can be used, in which the amide
functionalities in
-33-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
peptides have been permethylated to generate a chemically transformed
combinatorial
library, is described by Ostresh et al., Proc. Natl. Acad. Sci. U.S.A.
91:11138-11142 (1994).
[00132] Screening the libraries can be accomplished by any of a variety of
commonly
known methods. See, e.g., the following references, which disclose screening
of peptide
libraries: Parmley and Smith, Adv. Exp. Med. Biol. 251:215-218 (1989); Scott
and Smith,
Science 249:386-390 (1990); Fowlkes et al., Bio/Techniques 13:422-427 (1992);
Oldenburg
et al., Proc. Natl. Acad. Sci. U.S.A. 89:5393-5397 (1992); Yu et al., Cell
76:933-945 (1994);
Staudt et al., Science 241:577-580 (1988); Bock et al., Nature 355:564-566
(1992); Tuerk et
al., Proc. Natl. Acad. Sci. U.S.A. 89:6988-6992 (1992); Ellington et al.,
Nature 355:850-852
(1992); U.S. Patent No. 5,096,815, U.S. Patent No. 5,223,409, and U.S. Patent
No.
5,198,346.; Rebar and Pabo, Science 263:671-673 (1993); and PCT Publication
No. WO
94/18318, published August 8, 1994.
[00133] In a specific embodiment, screening can be carried out by contacting
the
library members with MO-1 polypeptide (or nucleic acid) immobilized on a solid
phase and
harvesting those library members that bind to the protein (or nucleic acid).
Examples of such
screening methods, termed "panning" techniques are described by way of example
in
Parmley and Smith, Gene 73:305-318 (1988); Fowlkes et al., Bio/Techniques
13:422-427
(1992); PCT Publication No. WO 94/18318; and in references cited herein above.
[00134] In another embodiment, the two-hybrid system for selecting interacting
proteins in yeast (Fields and Song, Nature 340:245-246 (1989); Chien et al.,
Proc. Natl.
Acad. Sci. US.A. 88:9578-9582 (1991)) can be used to identify molecules that
specifically
bind to MO-1 protein or an analog thereof.
[00135] In another embodiment, screening can be carried out by creating a
peptide
library in a prokaryotic or eukaryotic cell, such that the library proteins
are expressed on the
cells' surface, followed by contacting the cell surface with MO-1 and
determining whether
binding has taken place. Alternatively, the cells are transformed with a
nucleic acid encoding
MO-1, such that MO-1 is expressed on the cells' surface. The cells are then
contacted with a
potential agonist or antagonist, and binding, or lack thereof, is determined.
In a specific
embodiment of the foregoing, the potential agonist or antagonist is expressed
in the same or a
different cell such that the potential agonist or antagonist is expressed on
the cells' surface.
[00136] In another embodiment, screening can be carried out by assessing
modulation
(e.g., an increase or decrease) of binding of MO-1 to another polypeptide. In
certain
embodiments, the polypeptide is SCP2, CYP2B6, MTO1-like or IRAP.
-34-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
[00137] As would clearly be understood by a person of ordinary skill in the
art, any
and/or all of the embodiments disclosed herein for identifying an agent, drug,
or compound
that can modulate the activity of MO-1, including such procedures that
incorporate rational
drug design, as disclosed herein, can be combined to form additional drug
screens and assays,
all of which are contemplated by the present invention.
5.7 Diagnostic Methods
[00138] The present invention also pertains to the field of predictive
medicine in which
diagnostic and prognostic assays are used for prognostic (predictive) purposes
to treat an
individual prophylactically. Accordingly, one aspect of the invention relates
to diagnostic
assays for determining MO-1 nucleic acid expression as well as MO-1 activity
in the context
of a biological sample (e.g., blood, serum, cells, tissue) to determine
whether an individual is
afflicted with a disease or disorder, or is at risk of developing a disorder.
Such a disease or
disorder may be associated with aberrant MO-1 expression or activity, and can
include, but is
not limited to, obesity or other related metabolic disorders. The invention
also provides for
prognostic assays for determining whether an individual is at risk of
developing a disorder
associated with MO-1 nucleic acid expression or activity. For example,
mutations in MO-1
can be assayed in a biological sample. Such assays can be used for prognostic
or predictive
purpose to prophylactically treat an individual prior to the onset of a
disorder characterized
by or associated with aberrant MO-1 nucleic acid expression or biological
activity.
5.7.1 Diagnostic Assays
[00139] An exemplary method for detecting the presence or absence of MO-1 in a
biological sample involves obtaining a biological sample from a subject and
contacting the
biological sample with a compound or an agent capable of detecting MO-1
nucleic acid (e.g.,
mRNA, genomic DNA) such that the presence of MO-1 is confirmed in the sample.
An
agent for detecting MO-1 mRNA or genomic DNA is a labeled nucleic acid probe
that can
hybridize to MO-1 mRNA or genomic DNA. The nucleic acid probe can be, for
example, a
full-length MO-1 nucleic acid, such as the nucleic acid of SEQ ID NOS:2 or 3,
or a portion
thereof. In some embodiments, the nucleic acid probe is an oligonucleotide of
at least 15, 30,
50, 100, 250 or 500 nucleotides in length and is sufficient to specifically
hybridize under
stringent conditions to MO-1 mRNA or genomic DNA. In certain embodiments, a
mutation
resulting in a premature stop codon at amino acid position 82 of the MO-1
protein is detected.
[00140] An agent for detecting MO-1 polypeptide can be an antibody capable of
binding to MO-1, preferably an antibody with a detectable label. Antibodies
can be
-35-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
polyclonal or monoclonal. An intact antibody or an antibody fragment, e.g., a
Fab fragment,
can be used. A labeled probe or antibody may be coupled (i.e., physically
linked) to a
detectable substance, or an indirect detection method may be employed wherein
the probe or
antibody is detected via reactivity with a directly labeled secondary reagent.
Examples of
indirect labeling include detection of a primary antibody using a
fluorescently labeled
secondary antibody, or end-labeling of a DNA probe with biotin such that it
can be detected
with fluorescently-labeled streptavidin.
1001411 The detection method of the invention can be used to detect MO-1 mRNA,
protein, or genomic DNA in a biological sample in vitro as well as in vivo.
For example, in
vitro techniques for detection of MO-1 mRNA include Northern hybridizations
and in situ
hybridizations. In vitro techniques for detection of MO-1 polypeptide include
enzyme linked
immunosorbent assays (ELISAs), Western blots, immunoprecipitations, and
immunofluorescence. In vitro techniques for detection of MO-1 genomic DNA
include
Southern hybridizations and fluorescence in situ hybridization (FISH).
Furthermore, in vivo
techniques for detecting MO-1 include introducing into a subject a labeled
anti-1VIO-1
antibody. For example, the antibody can be labeled with a radioactive marker
whose
presence and location in a subject can be detected by standard imaging
techniques.
[00142] In one embodiment, the biological sample from the subject contains
protein
molecules, and/or mRNA molecules, and/or genomic DNA molecules. In certain
embodiments, the biological sample is blood.
[00143] In another embodiment, the methods further involve obtaining a
biological
sample from a subject to provide a control, contacting the sample with a
compound or agent
to detect MO-1 mRNA or genomic DNA, and comparing the presence of MO-1 mRNA or
genomic DNA in the control sample with the presence of MO-1 mRNA or genomic
DNA in
the test sample.
[00144] In another embodiment, the methods comprise assessing a biological
activity
of MO-1. For example, lipid concentration, glucose concentration, cellular
proliferation, and
expression levels of genes whose expression is modulated by MO-1 can be
assessed.
Accordingly, in certain embodiments, decreased lipid concentrations reflects
decreased MO-1
activity. In certain embodiments, increased lipid concentrations reflects
decreased MO-1
activity. In certain embodiments, increased lipid concentrations reflects
increased MO-1
activity. In certain embodiments, decreased lipid concentrations reflects
increased MO-1
activity. In certain embodiments, decreased glucose concentrations reflects
decreased MO-1
-36-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
activity. In certain embodiments, increased glucose concentrations reflects
decreased MO-1
activity. In certain embodiments, increased glucose concentrations reflects
increased MO-1
activity. In certain embodiments, decreased glucose concentrations reflects
increased MO-1
activity.
[00145] In certain embodiments, decreased cellular proliferation reflects
decreased
MO-1 activity. In certain embodiments, increased cellular proliferation
reflects decreased
MO-1 activity. In certain embodiments, increased cellular proliferation
reflects increased
MO-1 activity. In certain embodiments, decreased cellular proliferation
reflects increased
MO-1 activity. In certain embodiments, the affected cells are adipocytes.
5.7.2 Prognostic Assays
[00146] The diagnostic methods described herein can be further utilized to
identify
subjects having, or who are at risk of developing, a disease or disorder
associated with
aberrant MO-1 expression or activity. Such a disease or disorder may include,
but is not
limited to, metabolic disorders such as, e.g., obesity. The invention provides
a method for
identifying a disease or disorder associated with aberrant MO-1 expression or
activity in
which a test sample is obtained from a subject and MO-1 nucleic acid (e.g.,
mRNA, genomic
DNA) is detected. A test sample is a biological sample obtained from a
subject. For
example, a test sample can be a biological fluid (e.g., serum), cell sample,
or tissue.
[00147] Prognostic assays can be used to determine whether a subject can be
administered a modality (e.g., an agonist, antagonist, peptidomimetic,
protein, peptide,
nucleic acid, small molecule, food, etc.) to treat a disease or disorder
associated with aberrant
MO-1 expression or activity. Such methods can be used to determine whether a
subject can
be effectively treated with an agent for a disorder. The invention provides
methods for
determining whether a subject can be effectively treated with an agent for a
disorder
associated with aberrant MO-1 expression or activity in which a test sample is
obtained and
MO-1 nucleic acid is detected (e.g., where the presence of MO-1 nucleic acid
is diagnostic
for a subject that can be administered the agent to treat a disorder
associated with aberrant
MO-i expression or activity).
[00148] The methods of the invention can also be used to detect genetic
lesions in a
MO-1 gene to determine if a subject with the genetic lesion is at risk for a
disorder, including
but not limited to obesity. Methods include detecting, in a sample from the
subject, the
presence or absence of a genetic lesion characterized by an alteration
affecting the integrity of
a gene encoding a MO-1 polypeptide, or the mis-expression of a MO-1 gene. Such
genetic
-37-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
lesions can be detected by ascertaining: (1) a deletion of one or more
nucleotides from the
MO-1 gene; (2) an addition of one or more nucleotides to the MO-1 gene; (3) a
substitution
of one or more nucleotides in the MO-1 gene; (4) a chromosomal rearrangement
of a MO-1
gene; (5) an alteration in the level of MO-1 mRNA transcripts; (6) aberrant
modification of a
MO-1 gene, such as a change in genomic DNA methylation; (7) the presence of a
non-wild-
type splicing pattern of a MO-1 mRNA transcript, (8) a non-wild-type level of
a MO-1
polypeptide; (9) allelic loss of MO-1; (10) inappropriate post-translational
modification of a
MO-1 polypeptide; and/or (11) a single-nucleotide polymorphism associated with
a particular
MO-1-related phenotype. In one embodiment, the genetic lesion is a mutation
causing
premature truncation of a MO-1 protein. There are a large number of known
assay
techniques that can be used to detect lesions in MO-1. Any biological sample
containing
nucleated cells may be used. In some embodiments, the biological sample can be
a pre-natal
sample, obtained, for example, by amniocentesis or chlorionic vili sampling
(CVS).
[00149] Detection of genetic lesions of MO-1 may employ any technique known in
the
art. In certain embodiments, lesion detection may employ a nucleic acid
probe/primer in a
polymerase chain reaction (PCR) reaction such as anchor PCR or rapid
amplification of
cDNA ends (RACE) PCR. This method may include collecting a sample from a
patient,
isolating nucleic acids from the sample, contacting the nucleic acids with one
or more nucleic
acid primers that specifically hybridize to MO-1 nucleic acid under conditions
such that
hybridization and amplification of the MO-1 sequence (if present) occurs, and
detecting the
presence or absence of an amplification product, or detecting the size of the
amplification
product and comparing the length to a control sample. It is anticipated that
PCR may be
desirable to use as a preliminary amplification step in conjunction with any
of the techniques
used for detecting mutations described herein.
[00150] Mutations in a MO-1 gene from a sample can also be identified by
alterations
in restriction enzyme cleavage patterns. For example, sample and control DNA
is isolated,
amplified (optionally), digested with one or more restriction endonucleases,
and fragment
length sizes are determined by gel electrophoresis and compared. Differences
in fragment
length sizes between sample and control DNA indicate mutations in the sample
DNA.
Moreover, the use of sequence specific ribozymes can be used to score for the
presence of
specific mutations by development or loss of a ribozyme cleavage site.
[00151] Furthermore, hybridizing a sample and control nucleic acids, e.g., DNA
or
RNA, to high-density arrays containing hundreds or thousands of
oligonucleotides probes can
-38-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
identify genetic mutations in MO-1 (see Cronin et al., Hum. Mutat. 7:244-255
(1996); Kozal
et al., Nat. Med. 2:753-759 (1996)). For example, genetic mutations in MO-1
can be
identified in two-dimensional arrays containing light-generated DNA probes as
described in
Cronin, et al., supra. Briefly, a first hybridization array of probes can be
used to scan
through long stretches of DNA in a sample and control to identify base changes
between the
sequences by making linear arrays of sequential overlapping probes. This step
allows the
identification of point mutations. This is followed by a second hybridization
array that
allows the characterization of specific mutations by using smaller,
specialized probe arrays
complementary to all variants or mutations detected. Each mutation array is
composed of
parallel probe sets, one complementary to the wild-type gene and the other
complementary to
the mutant gene.
[00152] In yet another embodiment, any of a variety of sequencing reactions
known in
the art can be used to directly sequence the MO-1 gene and detect mutations by
comparing
the sequence of the sample MO-1 sequence with the corresponding wild-type
(control)
sequence. Examples of sequencing reactions include those based on classic
techniques (see
Maxam and Gilbert, Proc. Natl. Acad. Sci USA 74:560-564 (1977); Sanger et al.,
Natl. Acad.
Sci USA 74:5463-5367 (1977)). Any of a variety of automated sequencing
procedures can be
used for performing diagnostic assays of the present invention (see Naeve et
al.,
Biotechniques 19:448-453 (1995)) including sequencing by mass spectrometry
(Cohen et al.,
Adv. Chromatogr. 36:127-162 (1996); Griffin and Griffin, Appl. Biochem.
Biotechnol.
38:147-159 (1993)).
[00153] Examples of other techniques for detecting point mutations include,
but are
not limited to, selective oligonucleotide hybridization, selective
amplification, or selective
primer extension. For example, oligonucleotide primers may be prepared in
which the
known mutation is placed centrally and then hybridized to target DNA under
conditions that
permit hybridization only if a perfect match is found (see Saiki et al.,
Nature 324:163-166
(1986); Saiki et al., Proc. Natl. Acad. Sci. USA 86:6230-6234 (1989)). Such
allele-specific
oligonucleotides are hybridized to PCR-amplified target DNA or a number of
different
mutations when the oligonucleotides are attached to the hybridizing membrane
and
hybridized with labeled target DNA.
[00154] In yet other embodiments, a single-nucleotide polymorphism (SMP)
associated with altered MO-1 expression and/or activity can be detected. In
certain
embodiments, the SNP is -171C>G, -170G>A, -79insA, IVS2 +66delCT, g.168 G>A
syn, or
-39-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
g.471A>G, syn. In certain embodiments, the SNP is -171C>G. In certain
embodiments, the
SNP is -170G>A. In certain embodiments, the SNP is -79insA. In certain
embodiments, the
SNP is IVS2 +66delCT. In certain embodiments, the SNP is g. 168 G>A syn. In
certain
embodiments, the SNP is g.471A>G, syn.
5.8 Compositions
[00155] The invention provides methods of treatment (and prophylaxis) by
administration to a subject of an effective amount of a therapeutic of the
invention, e.g., a
MO-1 polypeptide, a derivative thereof, a MO-1 nucleic acid that expresses a
MO-1
polypeptide, etc. . In a preferred aspect, the therapeutic is substantially
purified. The subject
is preferably an animal, including but not limited to animals such as cows,
pigs, horses,
chickens, cats, dogs, etc., and is preferably a mammal, and most preferably
human. In a
specific embodiment, a non-human mammal is the subject. Formulations and
methods of
administration that can be employed can be selected from among those described
herein
below.
[00156] Various delivery systems are known and can be used to administer a
therapeutic of the invention, e.g., encapsulation in liposomes,
microparticles, microcapsules,
recombinant cells capable of expressing the therapeutic, receptor-mediated
endocytosis (see,
e.g., Wu and Wu, J. Biol. Chem. 262:4429-4432 (1987)), construction of a
therapeutic
nucleic acid as part of a retroviral or other vector, etc. Methods of
introduction include but
are not limited to intradermal, intramuscular, intraperitoneal, intravenous,
subcutaneous,
intranasal, epidural, and oral routes. The compounds may be administered by
any convenient
route, for example by infusion or bolus injection, by absorption through
epithelial or
mucocutaneous linings (e.g., oral mucosa, rectal and intestinal mucosa, etc.)
and may be
administered together with other biologically active agents. Administration
can be systemic
or local. In addition, it may be desirable to introduce the pharmaceutical
compositions of the
invention into the central nervous system by any suitable route, including
intraventricular and
intrathecal injection; intraventricular injection may be facilitated by an
intraventricular
catheter, for example, attached to a reservoir, such as an Ommaya reservoir.
Pulmonary
administration can also be employed, e.g., by use of an inhaler or nebulizer,
and formulation
with an aerosolizing agent.
[00157] In a specific embodiment, it may be desirable to administer the
pharmaceutical
compositions of the invention locally to the area in need of treatment; this
may be achieved
by, for example, and not by way of limitation, local infusion during surgery,
topical
-40-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
application, e.g., in conjunction with a wound dressing after surgery, by
injection, by means
of a catheter, by means of a suppository, or by means of an implant, said
implant being of a
porous, non-porous, or gelatinous material, including membranes, such as
sialastic
membranes, or fibers.
[00158] In another embodiment, the therapeutic can be delivered in a vesicle,
in
particular a liposome (see Langer, Science 249:1527-1533 (1990); Treat et al.,
in Liposomes
in the Therapy of Infectious Disease and Cancer, Lopez-Berestein and Fidler
(eds.), Liss,
New York, pp. 317-372, 353-365 (1989)).
[00159] In yet another embodiment, the therapeutic can be delivered in a
controlled
release system. In one embodiment, a pump may be used (see Langer, supra;
Sefton, CRC
Crit. Ref. Biomed. Eng. 14:201 (1987); Buchwald et al., Surgery 88:507 (1980);
Saudek et
al., N. Engl. J. Med. 321:574 (1989)). In another embodiment, polymeric
materials can be
used (see Medical Applications of Controlled Release, Langer and Wise (eds.),
CRC Pres.,
Boca Raton, Florida (1974); Controlled Drug Bioavailability.- Drug Product
Design and
Performance, Smolen and Ball (eds.), Wiley, New York (1984); Ranger and Pewas,
J.
Macromol. Sci. Rev. Macromol. Chem. 23:61 (1983); see also Levy et al.,
Science 228:190
(1985); During et al., Ann. Neurol. 25:351 (1989); Howard et al., J.
Neurosurg. 71:105
(1989)). In yet another embodiment, a controlled release system can be placed
in proximity
of the therapeutic target, thus requiring only a fraction of the systemic dose
(see, e.g.,
Goodson, in Medical Applications of Controlled Release, supra, vol. 2, pp. 115-
138 (1984)).
Other controlled release systems are discussed in the review by Langer
(Science 249:1527-
1533 (1990)).
[00160] In a specific embodiment where the therapeutic is a nucleic acid
encoding a
protein therapeutic (e.g., SEQ ID NO: 1), the nucleic acid can be administered
in vivo to
promote expression of its encoded protein, by constructing it as part of an
appropriate nucleic
acid expression vector and administering it so that it becomes intracellular,
e.g., by use of a
retroviral vector (see U.S. Patent No. 4,980,286), or by direct injection, or
by use of
microparticle bombardment (e.g., a gene gun; Biolistic, DuPont), or coating
with lipids or
cell-surface receptors or transfecting agents, or by administering it in
linkage to a homeobox-
like peptide which is known to enter the nucleus (see e.g., Joliot et al.,
Proc. Natl. Acad. Sci.
U. S.A. 88:1864-1868 (1991)), etc. Alternatively, a nucleic acid therapeutic
can be introduced
intracellularly and incorporated within host cell DNA for expression, by
homologous
recombination. In another embodiment, the nucleic acid therapeutic can act by
altering, e.g.,
-41-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
increasing or decreasing, the expression of endogenous MO-1 from the genome of
the subject
to be treated.
[00161] The present invention also provides pharmaceutical compositions. Such
compositions comprise a therapeutically effective amount of a therapeutic, and
a
pharmaceutically acceptable carrier. Water is a preferred carrier when the
pharmaceutical
composition is administered intravenously. Saline solutions and aqueous
dextrose and
glycerol solutions can also be employed as liquid carriers, particularly for
injectable
solutions. Suitable pharmaceutical excipients include starch, glucose,
lactose, sucrose,
gelatin, malt, rice, flour, chalk, silica gel, sodium stearate, glycerol
monostearate, talc,
sodium chloride, dried skim milk, glycerol, propylene, glycol, water, ethanol
and the like.
The composition, if desired, can also contain minor amounts of wetting or
emulsifying
agents, or pH buffering agents. These compositions can take the form of
solutions,
suspensions, emulsion, tablets, pills, capsules, powders, sustained-release
formulations and
the like. The composition can be formulated as a suppository, with traditional
binders and
carriers such as triglycerides. Oral formulation can include standard carriers
such as
pharmaceutical grades of mannitol, lactose, starch, magnesium stearate, sodium
saccharine,
cellulose, magnesium carbonate, etc. Examples of suitable pharmaceutical
carriers are
described in Remington's Pharmaceutical Sciences by E.W. Martin. Such
compositions will
contain a therapeutically effective amount of the therapeutic, preferably in
purified form,
together with a suitable amount of carrier so as to provide the form for
proper administration
to the patient. The formulation should suit the mode of administration.
[00162] In a preferred embodiment, the composition is formulated in accordance
with
routine procedures as a pharmaceutical composition adapted for intravenous
administration to
human beings. Typically, compositions for intravenous administration are
solutions in sterile
isotonic aqueous buffer. Where necessary, the composition may also include a
solubilizing
agent and a local anesthetic such as lignocaine to ease pain at the site of
the injection.
Generally, the ingredients are supplied either separately or mixed together in
unit dosage
form, for example, as a dry lyophilized powder or water free concentrate in a
hermetically
sealed container such as an ampoule or sachette indicating the quantity of
active agent..
Where the composition is to be administered by infusion, it can be dispensed
with an infusion
bottle containing sterile pharmaceutical grade water or saline. Where the
composition is
administered by injection, an ampoule of sterile water for injection or saline
can be provided
so that the ingredients may be mixed prior to administration.
-42-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
[00163] The therapeutics of the invention can be formulated as neutral or salt
forms.
Pharmaceutically acceptable salts include those formed with free amino groups
such as those
derived from hydrochloric, phosphoric, acetic, oxalic, tartaric acids, etc.,
and those formed
with free carboxyl groups such as those derived from sodium, potassium,
ammonium,
calcium, ferric hydroxides, isopropylamine, triethylamine, 2-ethylamino
ethanol, histidine,
procaine, etc.
[00164] The amount of the therapeutic of the invention which will be effective
in the
treatment of a particular disorder or condition will depend on the nature of
the disorder or
condition, and can be determined by standard clinical techniques. In addition,
in vitro assays
may optionally be employed to help identify optimal dosage ranges. The precise
dose to be
employed in the formulation will also depend on the route of administration,
and the
seriousness of the disease or disorder, and should be decided according to the
judgment of the
practitioner and each patient's circumstances. However, suitable dosage ranges
for
intravenous administration are generally about 20-500 micrograms of active
compound per
kilogram body weight. Suitable dosage ranges for intranasal administration are
generally
about 0.01 pg/kg body weight to 1 mg/kg body weight. Effective doses may be
extrapolated
from dose-response curves derived from in vitro or animal model test systems.
[00165] Suppositories generally contain active ingredient in the range of 0.5%
to 10%
by weight; oral formulations preferably contain 10 % to 95% active ingredient.
5.9 Kits
[00166] The pharmaceutical compositions can be included in a kit, container,
pack, or
dispenser together with instructions for administration. When the invention is
supplied as a
kit, the different components of the composition may be packaged in separate
containers and
admixed immediately before use. Such packaging of the components separately
may permit
long-term storage without losing the active components' functions.
[00167] Kits may also include reagents in separate containers that facilitate
the
execution of a specific test, such as diagnostic tests or tissue typing. For
example, MO-1
DNA templates and suitable primers may be supplied for internal controls.
5.9.1 Containers or Vessels
[00168] The reagents included in the kits can be supplied in containers of any
sort such
that the life of the different components are preserved, and are not adsorbed
or altered by the
materials of the container. For example, sealed glass ampules may contain
lyophilized
luciferase or buffer that have been packaged under a neutral, non-reacting
gas, such as
- 43 -
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
nitrogen. Ampoules may consist of any suitable material, such as glass,
organic polymers,
such as polycarbonate, polystyrene, etc., ceramic, metal or any other material
typically
employed to hold reagents. Other examples of suitable containers include
simple bottles that
may be fabricated from similar substances as ampules, and envelopes, that may
consist of
foil-lined interiors, such as aluminum or an alloy. Other containers include
test tubes, vials,
flasks, bottles, syringes, or the like. Containers may have a sterile access
port, such as a
bottle having a stopper that can be pierced by a hypodermic injection needle.
Other
containers may have two compartments that are separated by a readily removable
membrane
that upon removal permits the components to mix. Removable membranes may be
glass,
plastic, rubber, etc.
5.9.2 Instructional Materials
[00169] Kits may also be supplied with instructional materials. Instructions
may be
printed on paper or other substrate, and/or may be supplied as an electronic-
readable medium,
such as a floppy disc, CD-ROM, DVD-ROM, Zip disc, videotape, audio tape, etc.
Detailed
instructions may not be physically associated with the kit; instead, a user
may be directed to
an internet web site specified by the manufacturer or distributor of the kit,
or supplied as
electronic mail.
5.10 Methods of Treatment
[001701 The invention provides for both prophylactic and therapeutic methods
of
treating a subject at risk for (or susceptible to) a disorder or having a
disorder associated with
aberrant MO-1 expression or activity. Exemplary disorders are characterized by
abnormal
metabolic function, including, but not limited to, diabetes, type II diabetes,
obesity, morbid
obesity, hyperglycemia, insulin resistance, hyperinsulinemia,
hypercholesterolemia,
hypertension, hyperlipoproteinemia, hyperlipidemia, hypertriglylceridemia and
dyslipidemia,
and the like.
5.10.1 Diseases and Disorders
[00171] Diseases and disorders that are characterized by increased MO-1 levels
or
biological activity may be treated with therapeutics that antagonize (i.e.,
reduce or inhibit)
activity. Antagonists may be administered in a therapeutic or prophylactic
manner.
Therapeutics that may be used include: (1) MO-1 peptides, or analogs,
derivatives, fragments
or homologues thereof; (2) Abs to a MO-1 peptide; (3) MO-1 nucleic acids; (4)
administration of antisense nucleic acid, including, for example, siRNAs, and
nucleic acids
that are "dysfunctional" (i.e., due to a heterologous insertion within the
coding sequences)
-44-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
that are used to eliminate endogenous function of MO-1 by homologous
recombination
(Capecchi, Science 244:1288-1292 (1989)); or (5) modulators (i.e., inhibitors,
agonists and
antagonists, including additional peptide mimetic of the invention or Abs
specific to MO-1)
that alter the interaction between MO-1 and its binding partner(s), e.g., IRAP
and SCP2.
[00172] Diseases and disorders that are characterized by decreased MO-1 levels
or
biological activity may be treated with therapeutics that increase (i.e., are
agonists to)
activity. Therapeutics that upregulate activity may be administered
therapeutically or
prophylactically. Therapeutics that may be used include peptides, or analogs,
derivatives,
fragments or homologues thereof; or an agonist that increases bioavailability.
Alternately, a
nucleic acid therapeutic that increases expression of MO-1 provided either
exogenously or
endogenously in the subject's genome can be used.
[00173] Increased or decreased levels can be readily detected by quantifying
peptide
and/or RNA, by obtaining a patient tissue sample (e.g., from bleed or biopsy
tissue) and
assaying in vitro for RNA or peptide levels, structure and/or activity of the
expressed
peptides (or MO-1 mRNAs). Methods include, but are not limited to,
immunoassays (e.g., by
Western blot analysis, immunoprecipitation followed by sodium dodecyl sulfate
(SDS)
polyacrylamide gel electrophoresis, immunocytochemistry, etc.) and/or
hybridization assays
to detect expression of mRNAs (e.g., Northern assays, dot blots, in situ
hybridization, and the
like).
5.10.2 Prophylactic Methods
[00174] The invention provides a method for preventing, in a subject, a
disease or
condition associated with an aberrant MO-1 expression or activity, by
administering an agent
that modulates MO-1 expression or at least one MO-1 activity. Subjects at risk
for a disease
that is caused or contributed to by aberrant MO-1 expression or activity can
be identified by,
for example, any or a combination of diagnostic or prognostic assays.
Administration of a
prophylactic agent can occur prior to the manifestation of symptoms
characteristic of the
MO-1 aberrancy, such that a disease or disorder is prevented or,
alternatively, delayed in its
progression. In a specific embodiment of the invention, ventricular muscle
cell hypertrophy
is prevented or delayed by administration of said prophylactic agent.
Depending on the type
of MO-1 aberrancy, for example, a MO-1 agonist or MO-1 antagonist can be used
to treat the
subject. The appropriate agent can be determined based on screening assays.
5.10.3 Therapeutic Methods
- 45 -
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
[00175] Another aspect of the invention pertains to methods of modulating MO-1
expression or activity for therapeutic purposes. The modulatory method of the
invention
involves contacting a cell with an agent that modulates one or more of the
activities of MO-1
activity associated with the cell. An agent that modulates MO-1 activity can
be a nucleic acid
or a protein, a naturally occurring cognate ligand of MO-1, a peptide, a MO-1
peptidomimetic, an aptamer, or other small molecule. The agent may stimulate
MO-1
activity. Examples of such stimulatory agents include active MO-1 and a MO-1
nucleic acid
molecule that has been introduced into the cell. Stimulation of MO-1 activity
is desirable in
situations in which MO-1 is abnormally down-regulated and/or in which
increased MO-1
activity is likely to have a beneficial effect.
[00176] In other embodiments, the MO-1-modulating agent inhibits MO-1
activity.
Examples of inhibitory agents include anti-MO-1 Abs, or an inhibitory nucleic
acid molecule.
For example, the nucleic acid molecule may comprise an antisense
oligonucleotide, an
aptamer, or an inhibitory/interfering RNA (e.g., a small
inhibitory/interfering RNA. Methods
for screening for, identifying and making these nucleic acid modulators are
known in the art.
[00177] In some embodiments, RNA interference (RNAi) (see, e.g. Chuang et al.,
Proc. Natl. Acad. Sci. U.S.A. 97:4985 (2000)) can be employed to inhibit the
expression of a
gene encoding MO-1. Interfering RNA (RNAi) fragments, particularly double-
stranded (ds)
RNAi, can be used to generate loss-of-MO-1 function. Methods relating to the
use of RNAi
to silence genes in organisms, including mammals, C. elegans, Drosophila,
plants, and
humans are known (see, e.g., Fire et al., Nature 391:806-811 (1998); Fire,
Trends Genet.
15:358-363 (1999); Sharp, Genes Dev. 15:485-490 (2001); Hammond, et al.,
Nature Rev.
Genet. 2:1110-1119 (2001); Tuschl, Chem. Biochem. 2:239-245 (2001); Hamilton
et al.,
Science 286:950-952(1999); Hammond et al., Nature 404:293-296 (2000); Zamore
et al.,
Cell 101:25-33 (2000); Bernstein et al., Nature 409: 363-366 (2001); Elbashir
et al., Genes
Dev. 15:188 200 (2001); Elbashir et al. Nature 411:494-498 (2001);
International PCT
application No. WO 01/29058; and International PCT application No. WO
99/32619), the
contents of which are incorporated by reference. Double-stranded RNA (dsRNA)-
expressing
constructs are introduced into a host using a replicable vector that remains
episomal or
integrates into the genome. By selecting appropriate sequences, expression of
dsRNA can
interfere with accumulation of endogenous mRNA encoding MO-1.
[00178] Modulatory methods can be performed in vitro (e.g., by culturing the
cell with
the agent) or, altematively, in vivo (e.g., by administering the agent to a
subject). As such,
-46-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
the invention provides methods of treating an individual afflicted with a
disease or disorder
characterized by aberrant expression or activity of a MO-1 or nucleic acid
molecule. In one
embodiment, the method involves administering an agent (e.g., an agent
identified by a
screening assay), or combination of agents that modulates (e.g., up-regulates
or down-
regulates) MO-1 expression or activity. In another embodiment, the method
involves
administering a MO-1 or nucleic acid molecule as therapy to compensate for
reduced or
aberrant MO-1 expression or activity.
5.10.4 Determination of the Biological Effect of the Therapeutic
[00179] Suitable in vitro or in vivo assays can be performed to determine the
effect of a
specific therapeutic and whether its administration is indicated for treatment
of the affected
tissue.
[00180] In various specific embodiments, in vitro assays may be performed with
representative cells of the type(s) involved in the patient's disorder, to
determine if a given
therapeutic exerts the desired effect upon the cell type(s). Modalities for
use in therapy may
be tested in suitable animal model systems including, but not limited to rats,
mice, chicken,
cows, monkeys, rabbits, and the like, prior to testing in human subjects.
[00181] Similarly, for in vivo testing, any of the animal model systems known
in the
art may be used prior to administration to human subjects.
5.10.5 Prophylactic and Therapeutic Uses of the Compositions of the
Invention
[00182] MO-1 nucleic acids and proteins are useful in potential prophylactic
and
therapeutic applications implicated in a variety of disorders including, but
not limited to ,
diabetes, type II diabetes, obesity, hyperglycemia, insulin resistance,
hyperinsulinemia,
hypercholesterolemia, hypertension, hyperlipoproteinemia, hyperlipidemia,
hypertriglylceridemia and dyslipidemia and the like.
[00183] As an example, a cDNA encoding MO-1 may be useful in gene therapy, and
the protein may be useful when administered to a subject in need thereof. In
some
embodiments, the MO-1 polypeptide is administered in a form that permits entry
of the MO-1
polypeptide into a cell, e.g., an adipocyte. Formulations for accomplishing
this are described
above. By way of non-limiting example, the compositions of the invention may
have
efficacy for treatment of patients suffering from obesity. In other
embodiments, the
compositions of the invention may be useful in methods of increasing the
weight of a subject,
-47-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
e.g., a subject in need of increased body mass. In some embodiments, the
compositions may
be used to treat, for example, cachexia.
[00184] MO-1 nucleic acids, or fragments thereof, may also be useful in
diagnostic
applications, wherein the presence or amount of the nucleic acid or the
protein is to be
assessed. A further use could be as an anti-bacterial molecule (i.e., some
peptides have been
found to possess anti-bacterial properties). These materials are further
useful in the
generation of Abs that immunospecifically bind to the novel substances of the
invention for
use in therapeutic or diagnostic methods.
6. EXAMPLES
[00185] The invention is illustrated by the following examples which are not
intended
to be limiting in any way.
6.1 EXAMPLE 1: Identification and Mapping of MO-1
[00186] This example describes mapping and sequencing of a gene associated
with
morbid obesity, type II diabetes heart disease, and hypertension.
[00187] A large, consanguineous multigenerational family with morbid obesity,
type II
diabetes, heart disease and hypertension was identified; Figure 1 shows the
lineage of this
family. Detailed clinical data were obtained for all kindred members,
including all 10 living
affected individuals, shown in Table 3, below. Family members were known by
history to
have normal gestational birth weights but by age 2-3, affected children had
increased BMIs.
Affected adults had average BMIs -45. Three of 12 affected individuals had
mild mental
retardation but all had normal sexual development. In addition, three
individuals had died of
coronary artery disease/myocardial infarction, three of twelve affected family
members were
type II diabetics, and eleven of twelve had hypertension. Four of six
individuals had
abnormal lipid profiles including elevated triglyerides and cholesterol.
[00188] The obesity phenotype was inherited as an autosomal recessive trait. -
Therefore, a positional-cloning strategy was used to identify the causative
gene by identifying
homozygous-by-descent regions in affected individuals. Microsatellite markers
from the
Human Screening Panel, version 9.0 (Research Genetics), were used for the
genome wide
scan. Additional markers (Research Genetics; Integrated DNA Technologies) were
obtained
to refine the critical region. The model assumed the trait to be a highly
penetrant, autosomal
recessive disorder with a disease allele frequency of 1:10,000.
[00189] A LOD score of 9.7 (0= 0) was observed within a 5.5 Mb region on the
telomeric end of chromosome 3q29 between markers D3S2418-D3S3550. Additional
-48-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
markers in this region were analyzed and the multipoint lod scores for the
identified region of
homozygosity were determined. Heterozygosity and haplotype analysis narrowed
this initial
region to the 1.6 Mb critical region between markers centromeric marker
D3S2306 and
telomeric marker D3S3550.
[00190] Inspection of the genes mapped to the region revealed a number of
candidate
genes based on their known or probable functions and expression pattern. A
number of
candidate genes including BDH1, (R)-3-hydroxybutyrate dehydrogenase (EC
1.1.1.30),
which is required for the interconversion of the two major ketone bodies
produced during
fatty acid catabolism, DLG1, involved in cell proliferation and signaling, and
PAK2, a
member of the p21 activated kinase family that link Rho GTPases to
cytoskeleton
reorganization and nuclear signaling. No homozygous mutations in any of the
candidate
genes in the region which segregated appropriately within the family were
identified.
[00191] However, an EST sequence, FLJ25426 or C3orf34, was identified that
encodes a 167 amino acid protein with a calculated MW of 19.6 kD, i.e., MO-1
(SEQ ID
NO: 1), and which shared some sequence homology to phosphoenolpyruvate
carboxylase and
carboxykinase. PCR primers were then designed to all three 3 exons and
flanking intronic
and untranslated sequences, including the two coding exons, and directly
sequenced all
affected family members. All affected individuals were homoallelic for the
same nonsense
mutation, a C-T transition in codon 82 of exon 2, which predicts premature
truncation of the
protein (R82X) and loss of greater than half of the protein sequence. This
mutated form of
MO-1 polypeptide is also contemplated to be a part of the present invention.
This nucleotide
substitution segregated appropriately within the family; unaffected parents
were heteroallelic
for the mutation while unaffected siblings and relatives were either
heteroallelic or had the
wildtype sequence. In addition, the mutation was not present in greater than
500
chromosomes from 250 unaffected, unrelated control individuals.
6.2 EXAMPLE 2: Generation of a MO-1-Snecific Monoclonal Antibody
[00192] This example describes the generation and isolation of a monoclonal
antibody
preparation that specifically binds the MO-1 polypeptide.
[00193] A monoclonal antibody was generated as follows. Mice were immunized
with
Keyhole Limplet Hemocyanin (KLH) conjugated with a peptide selected from the
MO-1
polypeptide sequence (CTRAAEQLKNNPRH; SEQ ID NO.:11). Two booster
immunizations were subsequently administered. Post-immunization serum was then
used in
-49-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
an ELISA assay against free peptide to assess the monoclonal antibody response
using
conventional techniques.
[00194] Pre-immune serum was used as negative control (data not shown). Data
from
the ELISA assay is shown as Table 4, below.
TABLE 4
Dilution Animal Number
1 2 3 4 5
1:1000 2.431 1.847 2.233 1.065 1.015
1:3000 1.553 1.296 1.326 0.525 0.785
1:9000 0.915 0.713 0.679 0.254 0.584
1:2,7000 0.245 0.311 0.477 0.202 0.285
1:8,1000 0.169 0.266 0.193 0.173 0.157
1:24,3000 0.119 0.182 0.161 0.123 0.152
Blank 0.106 0.111 0.123 0.117 0.111
Titer >27000 >81000 >27000 >9000 >27000
[00195] In Table 4, the titer with the highest dilution wherein Signal/Noise
(Sample/Blank) >=2.1 is shown in bold.
6.3 EXAMPLE 3: Generation of a MO-1-Specific Polyclonal Antibody
Preparation
[00196] This example describes the generation and isolation of a MO-1
polypeptide-
specific polyclonal antibody preparation.
[00197] A MO-1 peptide comprising amino acids 133 - 153 of the MO-1 protein
sequence (DPNFVYDIEVEFPQDDQLQSC; SEQ ID NO:21) was synthesized and
conjugated with either KLH or ovalbumin as adjuvants and injected into
rabbits. After
confirming high titers by ELISA, the serum was then tested for its ability to
detect V5-
tagged-MO-1 by Western blotting techniques. Confirmatory blots were done using
the V5
antibody. RbtA1783 from both the crude serum (1:1500 dil) and following
affinity
purification (1:200) reveals the presence of a highly prevalent band
corresponding to the
predicted size of MO-1/V5. The same extract probed with an antibody
recognizing the V5
tag also identified a similar band corresponding to the predicted size of MO-
1/V5.
6.4 EXAMPLE 4: Generation of a MO-1-Knockout Mouse
[00198] This example describes the generation and isolation of a MO-1 knockout
mouse.
[00199] First, a knockout vector was constructed and identified with a PCR-
based
screen. Ten micrograms of the targeting vector was linearized by Notl and then
transfected
-50-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
by electroporation of iTL1 IC 1 C57BL/6 embryonic stem cells. After selection
with G418
antibiotic, surviving clones were expanded for PCR analysis to identify
recombinant ES
clones.
[00200] Screening primers A1 and A2 were designed downstream of the short
homology arm (SA) outside the 3' region used to generate the targeting
construct, shown
diagramatically in Figure 2. PCR reactions using A 1 or A2 with the LAN 1
primer (located
within the Neo cassette) amplify 2.4 and 2.5 kb fragments, respectively. The
control PCR
reaction was performed using the internal targeting vector primers AT1 and
AT2, which are
located at the 3' and 5' ends, respectively, of the SA. This amplifies a
product 1.3 kb in size.
[00201] Individual clones from positive pooled samples were then screened
using the
A2 and LAN1 primers. Positive recombinant clones were identified by a 2.5 kb
PCR
fragment. Next, positive SA PCR clones were sequenced for integration using
the OUTI
primer. Clones 133, 134, 151, 152, 154, 171, 172, 173, 174, 211, 214, 241, 243
and 244 were
selected and tested for cassette integration.
[00202] Confirmation of cassette integration within the long homology arm was
performed by PCR using the 3 and UNI primers. Sequencing was performed on
purified LA
PCR DNA to confirm presence of the cassette junctions using the 3 and N7
primers. Clones
133, 151, 154, 172 and 174 were selected for expansion and reconfirmation
using the same
methods described above. Clones 133, 151, 154, 172 and 174 were successfully
reconfirmed
and were determined to be suitable for injection
[00203] The engineered embryonic stem cells are then inserted into a mouse
blastocyst
which are then implanted into the uterus of female mice, to complete the
pregnancy. The
blastocysts then contain two types of stem cells: the ones from the
contributing mouse and
the newly engineered ones. A color selection based on coat color is used to
discriminate
between the two donors (for example, black and white fur). The newborn mice
are therefore
chimeras: parts of their bodies result from the original stem cells, other
parts result from the
engineered stem cells and as such, their furs will show patches of the
different colors.
[00204] Newborn mice with the newly engineered knockout sequence incorporated
into the germ cells (egg or sperm cells) are then crossed with others of the
relevant genetic
background for offspring that are "pure". These mice now contain one
functional copy and
one "deleted" copy of the gene and are further inbred to produce mice that
carry no functional
copy of the original gene (i.e. are homozygous for the knockout) and can be
detected by PCR
or Southern blot.
-51-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
6.5 EXAMPLE 5: Generation of a MO-1-Transgenic Mouse
[00205] This example describes the generation and isolation of a transgenic
mouse
expressing the human MO-1 polypeptide.
[00206] Generation of mice. To generate the targeting construct, the human MO-
1
genomic sequence was amplified using total genomic DNA isolated from the human
Hep3B
cell line. The primers MO-1-FL-F1, which sits on the 5'UTR region on exon 2,
and MO-1-
FL-R2, which sits on the 3'UTR and includes the native STOP codon, were used
to amplify
the sequence to be inserted into the pCAGG multiple cloning site (MCS) which
utilizes the
chicken beta-actin promoter to drive ubiquitous expression of the gene of
interest. The MO-1
sequence was introduced into the vector using the vector's EcoRI site. Upon
purification, the
DNA was microinjected into FVB mouse ES cells to allow for genomic integration
of the
exogenous DNA. The cells were then transplanted into a pseudopregnant female
mouse for
normal gestation. Pups were then screened for the presence of the transgene by
Southern
blotting. MO-1 probe was generated by digesting the transgenic construct with
EcoRl and
purifying the 650 bp product. A PCR-based screen was then developed using the
MO-1-FL-
F 1/ MO-1-FL-R2 in order to facilitate ease of screening subsequent
generations (Figure 3).
Pure lines were generated by crossing transgenic founders with wild type FVB
females, thus
generating F 1 transgenic pups.
[00207] Characterization of mice. A. Weight. Two independent MO-1 transgenic
lines were compared to wild type littermates to assess differences in weight.
As shown in
Figure 4 below, overexpression of MO-1 is associated with a more lean body
mass.
[00208] B. Serum glucose and glucose tolerance testing. Glucose tolerance
testing
was performed on transgenic and wild type littermates at two distinct
developmental
timepoints and under different dietary conditions. First, on animals at 6
weeks of age.
Second, on animals at 27 weeks of age after having been placed on a high-fat
(60% fat) for
20-weeks. In both cases, and counterintuitively but consistent with the in
vitro data, the
transgenic mice had elevated serum glucose levels following an overnight fast
(Figures 5 and
6). The transgenic mice maintained consistently elevated glucose levels when
compared to
their wild type littermates. Of note, in the HF diet experiments, MO-1
transgenic mice,
despite starting at higher initial levels (trend / p = 0.07), displayed a
significantly improved
response to lowering serum glucose at 2 hours (-80 mg/dL; p <0.03). Briefly,
for both
experiments, after a 16-hour overnight fast, mice were injected
intraperitoneally with a bolus
of glucose (D-50) at a dose of 1 g glucose/ kg total body weight. Plasma
glucose was then
-52-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
measured at 15, 30, 60, and 120 minutes post injection using a glucomoter
(Freestyle Flash
Glucose Meter, Abbott Diagnostics).
6.6 EXAMPLE 6: Identification and Characterization of Single Nucleotide
Polymorphisms Associated with MO-1 Alleles
[00209] This example describes the identification and characterization of
Single
Nucleotide Polymorphisms (SNPs) associated with MO-1 alleles.
[00210] Over 500 individuals of primarily European ancestry and separated
according
to body mass index (BMI) were directly sequenced at the MO-1 gene locus on
chromosome
3q29 to identify single nucleotide polymorphisms (SNPs), useful in
diagnostic/prognostic
assays. In the figure, the MO-1 gene locus is shown with the presumed MO-1
start site in
capital letters below the diagram. Using the ATG as the start site reference
(i.e. position +1),
indiviudal SNPs and their relative positions are shown as Figure 7A. IVS =
intervening
sequence/ intronic sequence.
[00211] As an example of diagnostic/prognostic utility, SNP IVS2 +66 del CT
was
used in an association study comparing lean (BMI <25) and obese individuals
(>40) in a
sample set of approximately 300 individuals in total. As shown in Figure 7B,
the IVS2 +66
del CT SNP was present nearly 3 times more frequently in the homozygous state
in lean
individuals (Chi-square = 4.68, p < 0.05); suggestive of a protective effect
on weight gain in
this BMI range.
6.7 EXAMPLE 7: Assays Testing Effects of MO-1 Overexpression and
Silencing
[00212] This example describes assays assessing the effects of MO-1
overexpression
and silencing.
[00213] Increased expression results in decreased intracellular glucose
levels. In the
first experiment, Hep3B cells, a liver-derived human cell line with
gluconeogenic capacity
were transfected with either control (empty) or MO-1 overexpressing vectors.
The cells were
maintained in serum-free, low glucose media (low energy condition) and
supplemented with
an excess of pyruvate and lactate (known substrates of gluconeogenesis). As
shown in
Figure 8, overexpression of MO-1 results in decreased (-70%) intracellular
glucose levels.
[00214] Decreased expression results in increased intracellular glucose
levels. In this
experiment, siRNA directed against MO-1 was used to silence the expression of
endogenous
MO-1 in Hep3B cells. As shown in Figure 9, silencing of MO-1 (-50% at 48
hours) resulted
in increased intracellular glucose levels (-25%). The specificity of the siRNA
towards MO-1
-53-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
is shown by the fact that PEPCK RNA levels are unchanged following MO-1
targeted
silencing.
[00215] Effects of MO-1 silencing on preadipocyte, NIH 3t3L, differentiation.
In this
experiment, NIH 3T3 L1 cells were plated at low density. Day 0 cells were
stained and
collected for RNA. Remaining cells were transfected with siNTC (control) or
siMOl at Day
0 and induced 7 hrs later using 0.5 mM IBMX/ 1 uM Dexamethasone/ 1 uM Insulin.
Cells
were kept in induction media for 3 days. Day 3 cells were stained and
collected for RNA.
Remaining cells were re-transfected with siNTC or siMOl and media was replaced
after 7 hrs
with maintenance media containing 1 uM Insulin. Cells were left in this media
for 3 days.
[00216] At Day 6, remaining cells were again re-transfected with siNTC or
siMOl and
media was changed with fresh maintenance media (contains 1 uM Insulin) 7hrs
later. =Day 8
cells were stained and collected for RNA Successive transfections were done to
maintain
knock-down of MO-1 throughout the course of the experiment.
[00217] MO-1 expression decrease resulted in delayed adipocyte differentiation
/
adipogenesis and reduction of intracellular lipid accumulation (Figure 10). In
addition,
decreased levels of MO-1 resulted in altered expression of key adipogenic
markers of
differentiation throughout different stages of adipocyte development (Figures
11 and 12).
Shown are quantitative real-time PCR data of a number of these key regulatory
genes. These
results suggest that alterations in expression levels of MO1 can affect a
number of critical
genes and pathways involved in adipocyte differentiation and adipogenesis
[00218] In another experiment, Hep3B cells were infected with a retrovirus
expressing
an siRNA targeting MO-1 and cells stably expressing the siRNA were selected.
This resulted
in long-term reduction in MO-1 expression. Among other results, ablation of MO-
1
expression by approximately -90% resulted in increased cell proliferation
(Figure 16).
6.8 EXAMPLE 8: Assessing Expression Patterns and Localization of MO-1
Expression
[00219] This example describes the results of experiments designed to assess
expression patterns of MO-1 in tissues and within cells.
[00220] First, RNA was isolated from various human tissues and assayed by RT-
PCR
for expression of MO-1 mRNA in the tissues. Results from this assay are shown
as Figure
13. As indicated in Figure 13, MO-1 is expressed in multiple tissues,
including adipocytes,
liver, muscle and hypothalamus..
-54-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
[00221] Next, NIH 3T3 Li cells were induced to differentiate as described in
Example
7, above, and RNA was extracted once daily for 10 days. MO-1 RNA levels were
determined by qRT-PCR. Results are shown in Figure 14. Interestingly,
increased levels of
MO-1 coincide with onset of lipogenesis.
[00222] Finally, tagged MO-1 protein was expressed in Hep3B cell lines and the
pattern of expression was assessed by immunofluorescence. MO-1 protein was
expressed in
the cytoplasm and localized to the mitochondria.
6.9 EXAMPLE 9: Assessing MO-1 Protein-Protein Interactions
[00223] This example describes the results of experiments designed to assess
interactions MO-1 with other proteins.
[00224] Two complementary methods were used to identify potential MOI protein
binding partners. First, a mass spectroscopy approach identifying the
differences between
MO1-V5 tagged and empty-V5 overexpressed proteins was pursued. The schema is
shown in
Figure 15. Using this approach in Hep3B cells, 1 predominant protein was
identified: IRAP.
[00225] This interaction is believed to be of particular relevance since IRAP:
= is a member of the Zn-dependent family of membrane aminopeptidases
=contains a single TM domain
=localizes to GLUT4-containing intracellular vesicles under basal conditions
in
response to insulin:
=redistributes to the cell surface along with GLUT4 to facilitate glucose
disposal;
=cleaves extracellular peptide hormone substrates (eg. vasopressin)
=exhibits impaired translocation to cell surface in muscle, liver and adipose
tissue in
type II diabetes; and
=results in 50-80% decreased GLUT4 in muscle, liver and adipose tissue of IRAP
-/-
mice due to degradation as a result of impaired sorting and trafficking.
[00226] Next, the yeast two hybrid system was used to screen of a human liver
library.
Several binding genes were identified:
[00227] SCP2: the SCP2 gene encodes two proteins: sterol carrier protein X
(SCPx)
and sterol carrier protein 2 (SCP2), as a result of transcription initiation
from two
independently regulated promoters. The transcript initiated from the proximal
promoter
encodes the longer SCPx protein, and the transcript initiated from the distal
promoter encodes
the shorter SCP2 protein. The two proteins share a common C-terminus. SCPx is
a
peroxisome-associated thiolase that is involved in the oxidation of branched
chain fatty acids.
-55-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
SCP2 protein is an intracellular lipid transfer protein. This gene is highly
expressed in organs
involved in lipid metabolism. Of note, SCP2 plays an important role in
intracellular
movement of cholesterol and possibly other lipids. SCP2 is believed to
facilitate the transport
of cholesterol to mitochondria, where the first committed step in
steroidogenesis takes place.
[00228] CYP2B6 (GID: 82583665), encodes a member of the cytochrome P450
superfamily of enzymes. As a family, cytochrome P450 proteins are
monooxygenases which
catalyze many reactions involved in drug metabolism and synthesis of
cholesterol, steroids
and other lipids.
[00229] MTO1-like (GID: 17149038), a mitochondrial protein ubiquitously
expressed
in various tissues, but with markedly elevated expression in tissues of high
metabolic rates.
[00230] In addition, three hypothetical proteins were identified, and thus
they
themselves may play a previously unrecognized role in metabolism: GID:
14578073, GID:
158819059, and GID: 21212494.
[00231] All publications, patents and patent applications cited in this
specification are
herein incorporated by reference as if each individual publication or patent
application were
specifically and individually indicated to be incorporated by reference.
Although the
foregoing invention has been described in some detail by way of illustration
and example for
purposes of clarity of understanding, it will be readily apparent to those of
ordinary skill in
the art in light of the teachings of this invention that certain changes and
modifications may
be made thereto without departing from the spirit or scope of the appended
claims.
-56-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
Z v~ v> cn v~ w v) v~ cn cn cn cn >
Z
F- N O O O N N O (D N N 4) 4) 6) N H
_ }}Z}}}}}}}} } ~__
f0
~
= fnzZZZZZZZZZZ Z z
E
> N j
o 2ZZZZ2ZZ~Z2Z Z ML2 E
Lc)
~ Il M O 00 ~f N N(fl M 00 0) V
~ tn c MO~ ~ ~"I7cM
m ~ ~ lqt
0)
Lf')u=)00)NLO LO LO Lc)cY)LA O O)
I~ I~ f~ (D GO (O (fl (D (D o0 (fl (D (fl
I~ ~~.==-~~~~~~~ ~
C L LO
U 'O
~ I~ ~ f~ ~ N V' ~~M O O O
M~V 'IT N N M ~ M ~ M N M
r r r r r r r r r r T r r
M ~
N N (/J N cn
=
N y
~ a) C M M M `(n M L M ~ M
00 N N N cM ~ ~.-- N M N cM N
cm
+-+ C_
N~
O 4) tn fn (A fn (n tn tn (n fA fA fA V) tl)
mU-}}}}}}}}}}} } }
O O
~_ NC~ p~OMCMtn OO~ O)O) ~ OMO
m~ MqT M M~- ~ M M ~ M M M fM
O (0
NL ~MCO f- NV 1- O)tf)O -p N-p
O ~ ~ ~ ~ ~ ~ ~ ~ (0 ~ ~ ~ Lo
m ~ ~=- ~-- ~ ~ ~-- i-- e- c- ~-- ~ N 0
N
N O E
O 00 OOQ)OOMOOd)OQ)O O O
U~ < M CM ~qT Mqq C7 Mv C'M ~ If H
N
C
a)
C~ ~ ~ ~ ~ ~ ~ 2 u-
- N CM ~ tA CO I- GO O) CDI e~- 04
h-= F- F- F- F- F- H F--= H F- H F-= ~
Z Z Z Z Z Z Z Z Z Z Z Z
c _W W W W_ W_ W W_ _W W_ W_ W_ _W
F- 1- H H F- F- F- H H H H H
a aaaaaaaaaaa a Q
57
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
Primers for amplifying and sequencing genomic MO1
MO 1.1 R CTTGTTAGGAAGCCCCACAG
MO 1.2F CGCAAGGATGACACACAAAT
MO1.2R GGAAACTGAAGCTCACTGAGAGTA
MO 1.3F AATATTTTCAGGGTTCAGAGTTTTT
MO 1.3R TTGAAAAGTAACATGGTCAATCC
MO1-EX3R GGATTGACCATGTTACTTTTCAA
MO1-FL-FI TTC ATC AGA TTT CCT CTG ACT TAC CCG G
MO1-FL-R1(-TGA) GAG TGT TTG GTA TGA GAA CTC ATC AGC TG
MO 1-FL-
R2(TGA+93) GTA ACA TGG TCA ATC CCC TAT AAC CCA AC
Primers for genotyping MO1 Transgenic Mice
MO1-FL-F1 TTC ATC AGA TTT CCT CTG ACT TAC CCG G
MO 1-FL-
R2(TGA+93) GTA ACA TGG TCA ATC CCC TAT AAC CCA AC
MO1 Primers to amplify Full-length cDNA
MO1-FL-F1 TTC ATC AGA TTT CCT CTG ACT TAC CCG G
MO1-FL-R1(-TGA) GAG TGT TTG GTA TGA GAA CTC ATC AGC TG
MO 1-FL-
R2(TGA+93) GTA ACA TGG TCA ATC CCC TAT AAC CCA AC
MOI Primers to amplify R>X cDNA
(used Rev right after mut to simulate the truncated protein since
TGA cannot be used because of 3' tag (V5)
MO1-R>X trunc-
Revl CTG CCC CGA CAA GTA ACC TCG TAA
MO1-R>X trunc-
Rev2 CTG AAT AGC TTC TCT AGC TGC CTC AGG
MO1 human Realtime Primers
MO1-RT-F1 (+22) GGG ATT AGG TTT CAG CCT CCA GC
MO1-RT-R1(+196) GAA TAG CTT CTC TAG CTG CCT CAG GG
MO1-RT-F2(+70) GAA ATC AAG GGG AAA ATT CGC CAG CG
MO1-RT-R2 +246) GTT TCT GCC AGA CTC TGC CCC
MO1-RT-F3 GCA CTG CCA AGA AAT GTG GGA TTA GG
MO1-RT-R3 CTT TAA TTG TTC AGC AGC TCT GGT GC
-58-
CA 02684707 2009-10-20
WO 2008/130668 PCT/US2008/005083
MO1 mouse Realtime Primers
MOl-Mse-RT-F1 GAG TTA GGT TCC AGC CTC CAG C
MO1-Mse-RT-R1 CAG GTA ACT CTT GTG CCG TGG G
MOl-Mse-RT-F2 GAA TGA AAC CGA AGG AAA GAG CCG C
MOl-Mse-RT-R2 CGC AAA AAA ACA AAC AGC TTC TCC AGC
For pBABE-MO 1-puro construct
MO 1 pBABE-EcoRI-
F TTG CCC TTG AAT TCA GAT TTC CTC TGA CTT ACC
MO1-pBABE+V5-
SaII-R GGC TGC TCG ACG GGT TTA AAC TCA ATG G
For MO1 SNaPshot screen
MO 1-SNaP-delCT-
27F CTC TAA ATA CAG GTT CAC ATC ATG ACT
MO1-SNaP-delCT-
25R GTG TCA CAC ATC CAG TCC CTG ACA G
For synthetic siMOl generation for cloning into pSUPER-RETRO-puro
gatccccAAGAATAATCCGCGACACATTttcaagagaAAT GTG TCG CGG
siMO 1(10)-Fwd ATT ATT CTTtttttggaaa
agcttttccaaaaaAAGAATAATCCGCGACACATTtctcttgaaAAT GTG TCG
siMOl(10)-Rev CGG ATT ATT CTTggg
-59-