Note: Descriptions are shown in the official language in which they were submitted.
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
UNIVERSAL MATRIX
RELATED APPLICATIONS
This application claims priority to US provisional serial no. 60/945,164 filed
June 20,
2007, and also to US provisional serial no. 60/929,314 filed June 21, 2007,
and also to US
provisional serial no. 61/047,642 filed Apri124, 2008, all of which are hereby
incorporated
by reference in their entireties.
FEDERAL FUNDING STATEMENT
Various embodiments described herein were funded by the federal government
under
the following grants: Air Force Office Scientific Research (AFOSR grant:AFOSR
FA9550-
05-1-0054) and NSF/NSEC (grant: EEC-0647560). The government has certain
rights in the
invention.
BACKGROUND
Microarrays and nanoarrays are important commercial developments. The use of
microarrayed-patterned biomolecules, such as DNA, proteins, and cells, has led
to extensive
and significant advances in fields such as geonomics and proteomics, with
applications to
many areas of medical and biological research; see for example Miller, et al.,
Microarray
Technology and Its Applications=, Springer; New York (2005). Within current
microarray
technology, a need exists to decrease spot-size to the nanometer regime, thus
increasing the
density of combinatorial libraries. This can not only increase the number of
interactions one
can simultaneously monitor, but also decrease the amount of costly reagents
necessary for
example to sequence an organism's DNA or screen interactions. With the advent
of powerful
new nanolithographic methods, such as dip-pen nanolithography (DPN) printing
or patterning
(see for example Piner et al., Science, 283, 661-663 (1999)), there is now the
ability to reduce
the feature size in such 1-dimensional or 2-dimensional arrays to their
physical limit, the size
of the structures from which they are made of and the size of the structures
they are intended
to interrogate; see for example Rosi et al., Nanostructures Chemical Reviews,
105, 1547-
1562 (2005). Such massive miniaturization not only allows one to increase the
density of
combinatorial libraries, increase the sensitivity of such structures in the
context of a
biodiagnostic event, and reduce the required sample analyte volume, but also
allows one to
carryout studies not possible with the more conventional microarray format.
In order to achieve the potential DPN may offer to the field of patterned
biomolecuie
arrays, simple and/or robust techniques can be developed for the direct-write
patterning of
1
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
biomolecules at the nanometer scale. Also, massively parallel, multiplexed
patterning of
biomolecules is desirable. Conventional methods of biomolecule patterning by
DPN are
generally limited to a single ink composition, be it oligonucleotide or
protein. Multiple-ink
DPN patterns require first patterning a single component biomolecule, then
performing a
lengthy alignment procedure before patterning a second biomolecule. One
prominent
technical challenge in creating multiplexed biomolecule patterns deposited in
a massively
parallel format resulting from the different diffusion rates inherently
associated with different
biomolecules; see for example Lee et al., J. Am. Chem. Soc. 125, 5588-5589
(2003); Lim et
al., Angew. Chem. Int. Ed. 42, 2309-2312 (2003). Previous advances in this
area have been
made, but needs yet exist, particularly for commercial applications. One
potential limitation
is the chemical modification of a tip such as an AFM tip for reproducible tip
coating.
Different biomolecules may require a specific modification, which can lead to
compatibilitv
issues. The second is in the context of parallel DPN printing. Biological
molecules can have
different transport properties, which can lead to heterogeneous surface
features from tip-to-
tip, and in some cases, cannot be deposited at all. Finally, denaturation and
loss of biological
activity potentially can be an issue. In order to bypass these potential
limitations, a method
that can equalize the transport rates while preserving the biological activity
of the molecules
is desirable.
SUMMARY
Embodiments provided herein include methods of making, methods of using,
devices,
compositions, and the like.
One embodiment provides a method that comprises: (i) providing a tip and a
substrate
surface, (ii) disposing a patterning composition at the end of the tip, (iii)
depositing at least
some of the patterning composition from the tip to the substrate surface to
form a deposit
disposed on the substrate surface, wherein the patterning composition
comprises at least one
patterning species, at least one carrier that is different from the patterning
species, and at least
one additive different from the patterning species and the carrier.
Another embodiment provides a method for using a chemical additive comprising:
co-
mixing the additive with a patterning composition comprising at least one
patterning species
different from the additive and at least one carrier different from the
additive and the
patterning species to promote patterning of the patterning species when
disposed on the end
of a tip and deposited onto a substrate surface.
2
WASH_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
Another embodiment describes a method to control the diffusion rate of
biomolecules
for direct-write dip-pen nanolithography (DPN) using agarose as a carrier ink.
The nature of
the agarose gel matrix is modified with a number of different additives,
resulting in an
additional method to control spot size, besides contact time, humidity or
temperature. By
selecting an appropriate type and concentration of additive, the diffusion of
two or more
different biomolecules may be regulated, resulting in similar spot size for
two different
biomolecule inks, with similar or substantially the same contact time.
BRIEF DESCRIPTION OF THE DRAWINGS
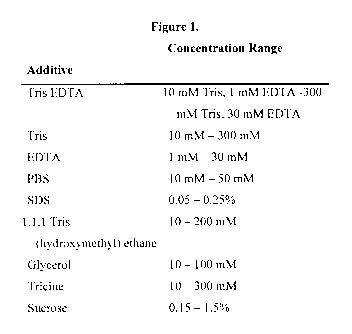
Figure 1 illustrates additives and their concentration ranges that can be used
for accelerating
effects of agarose/biomolecule-based Inks.
Figure 2 provides microscope image of agarose/biomolecule inked AFM tips. (a)
Agarose/DNA inked tip without any additives. (b) Agarose/DNA inked tip with
1.5%
sucrose additive.
Figure 3 shows ink diffusion rates of DNA/agarose/Tris-EDTA buffer. A) AFM
micrograph
of a typical diffusion experiment patterned at 60% humidity with dwell times
from 0.01 -
10s. B) Corresponding fluorescence image. C) Ink diffusion curves showing
difference in
diffusion rates between a 30 x Tris-EDTA (TE) (300 mM Tris, 30 mM EDTA) and 1
x TE
(10 mM Tris, 1 mM EDTA).
Figure 4 shows Spot Size vs. Dwell time for DNA/agarose/30 mM Glycerol and
cholera toxin
protein/agarose/30 mM glycerol inks showing similar diffusion rates for the
two inks.
Figure 5 provides fluorescence micrographs of DNA hybridization. A) Spots of
DNA
deposited by DPN using a agarose/glycerol ink. B) Negative control after
introduction of a
non-complimentary sequence. C) Positive signal showing hybridization with
complementary
sequence.
Figure 6 shows fluorescence micrographs of proteins depicting biorecognition.
A) Spots of
cholera toxin protein using an agarose/glycerol ink. B) Negative control after
introduction of
non-cholera toxin reactive Cy 5 labeled IgG antibodies. C) Positive signal
showing
biorecognition of anti-cholera toxin labeled with Alexa Fluor 488 with
immobilized cholera
toxin proteins.
3
WASH_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
Figure 7 shows AFM contact mode tips coated with agarose matrix in one
embodiment.
Figure 8 provides fluorescent microscopy images of Cy3-labeled DN nanoarrays
fabricated
by DPN multipen-array.
Figure 9 (a) and (b) show fluorescent microscopy images of Cy3-labelled DNA
nanoarrays,
each comprise 216 spots, fabricated by DPN multipen-arrays.
Figure 10 provides fluorescent microscopy images of cholera toxin protein
(Alex 594-
labeled) fabricated by DPN multipen array.
DETAILED DESCRIPTION
INTRODUCTION
All references cited herein are hereby incorporated by reference in their
entirety.
Priority US provisional serial no. 60/945,164 filed June 20, 2007, and
priority US
provisional serial no. 60/929,314 filed June 21, 2007, and priority US
provisional serial no.
61/047,642 filed April 24, 2008, are all hereby incorporated by reference in
their entireties
including claims, figures, working examples, and other descriptive
embodiments.
Copending application US serial no. to Mirkin et al. "Matrix
Assisted Ink Transport," filed on same day as present application, is hereby
incorporated by
reference including figures, claims, working examples, and other descriptive
embodiments.
Copending application US serial no. to Mirkin et al. "Universal
Matrix," filed on same day as present application, is hereby incorporated by
reference
including figures, claims, working examples, and other descriptive
embodiments.
Herein, a variety of novel approaches are demonstrated. In one embodiment, a
polysaccharide and a chemical additive are utilized as part of a patterning
composition. The
polysaccharide can be for example agarose, while the chemical additive can be
for example a
hygroscopic molecule such as a carbohydrate such as for example sucrose. For
example, the
polysaccharide can function as a bio-compatible ink carrier to pattern
directly a variety of
molecules and species, including biological molecules (e.g., oligonucleotides
and proteins)
onto a surface with dip-pen nanolithography (DPN) printing, while the chemical
additive can
4
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
function as simultaneously a hygroscopic agent preventing the carrier from
drying out and an
agent that modifies the physical properties of the carrier.
The use of polysaccharide as an ink carrier is desirable because it allows for
a
convenient biocompatible environment that can preserve the structure and
bioactivity of the
biomolecules being delivered. When using the preferred polysaccharide and
chemical
additive as a part of a patterning composition, surface modification of the
AFM tips prior to
DPN printing can be carried out if helpful but is optional. Furthermore, the
rate of deposition
of the embedded biomolecules can be controlled by controlling the diffusion
rate of the
carrier matrix via selecting a desirable combination of the carrier matrix and
the additive.
This allows for a simultaneous deposition of different biomolecules at a
controllable rate
using parallel DPN printing. One can also apply this approach for the delivery
of biological
molecules that are more difficult to pattern.
DPN printing, including instrumentation, materials, and methods, is generally
known
in the art. For practice of the various embodiments described herein,
lithography,
microlithography, and nanolithography instruments, pen arrays, active pens,
passive pens,
inks, patterning compounds, kits, ink delivery, software, and accessories for
direct-write
printing and patterning can be obtained from Nanolnk, Inc., Chicago, IL.
Softwares include
INKCAD and NSCRIPTOR softwares (Nanolnk, Chicago, IL), providing user
interfaces for
lithography design and control. E-Chamber can be used for environmental
control. Dip Pen
NanolithographyTM and DPNTm are trademarks of Nanolnk, Inc.
The following patents and co-pending applications related to direct-write
printing
with use of cantilevers, tips, and patterning compounds are hereby
incorporated by reference
in their entirety and can be used in the practice of the various embodiments
described herein,
including inks, patterning compounds, software, ink delivery devices, and the
like:
U.S. Patent No. 6,635,311 to Mirkin et al.., which describes fundamental
aspects of
DPN printing including inks, tips, substrates, and other instrumentation
parameters and
patterning methods;
U.S. Patent No. 6,827,979 to Mirkin et al.., which further describes
fundamental
aspects of DPN printing including software control, etching procedures,
nanoplotters, and
complex and combinatorial array formation.
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
U.S. patent publication number 2002/0122873 Al published September 5, 2002
("Nanolithography Methods and Products Produced Therefor and Produced
Thereby"), which
describes aperture embodiments and driving force embodiments of DPN printing.
U.S. regular patent application, serial no. 10/366,717 to Eby et al.., filed
February 14,
2003 ("Methods and Apparatus for Aligning Patterns on a Substrate"), which
describes
alignment methods for DPN printing (published October 2, 2003 as
2003/0185967).
U.S. regular patent application, serial no. 10/375,060 to Dupeyrat et al..,
filed
February 28, 2003 ("Nanolithographic Calibration Methods"), which describes
calibration
methods for DPN printing.
U.S. Patent Publication 2003/0068446, published April 10, 2003 to Mirkin et
al..
("Protein and Peptide Nanoarrays"), which describes nanoarrays of proteins and
peptides;
U.S. Regular Patent Application, Ser. No. 10/307,515 filed Dec. 2, 2002 to
Mirkin et
al.. ("Direct-Write Nanolithographic Deposition of Nucleic Acids from
Nanoscopic Tips"),
which describes nucleic acid patterning (PCT /US2002/038252 published June 12,
2003).
U.S. Regular Patent Application, Ser. No. 10/320,721 filed Dec. 17, 2002 to
Mirkin et
al.. ("Patterning of Solid State Features by Direct-Write Nanolithographic
Printing"), which
describes reactive patterning and sol gel inks (now published August 28, 2003
as
2003/0162004).
US Patent Nos. 6,642,129 and 6,867,443 to Liu et al.. ("Parallel, Individually
Addressible Probes for Nanolithography"), describing active pen arrays.
U.S. Patent Publication 2003/0007242, published January 9, 2003 to Schwartz
("Enhanced Scanning Probe Microscope and Nanolithographic Methods Using
Same").
U.S. Patent Publication 2003/0005755, published January 9, 2003 to Schwartz
("Enhanced Scanning Probe Microscope").
U.S. Patent Application 10/637,641 filed August 11, 2003, now published as
2004/0101469, describing catalyst nanostructures and carbon nanotube
applications.
U.S. Patent Application 10/444,061 filed May 23, 2003, now published as
2004/0026681 published February 12, 2004, and US patent publication
2004/0008330
6
WASH_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
published January 15, 2004, describing printing of proteins and conducting
polymers
respectively.
U.S. Patent Application 10/647,430 filed August 26, 2003, now US Patent No.
7,005,378, describing conductive materials as patterning compounds.
U.S. Patent Application 10/689,547 filed October 21, 2003, now published as
2004/0175631 on September 9, 2004, describing mask applications including
photomask
repair.
U.S. Patent Application 10/705,776 filed November 12, 2003, now published as
2005/0035983 on February 17, 2005, describing microfluidics and ink delivery.
U.S. Patent Application 10/788,414 filed March 1, 2004, now published as
2005/0009206 on January 13, 2005 describing printing of peptides and proteins.
U.S. Patent Application 10/893,543 filed July 19, 2004, now published as
2005/0272885 on December 8, 2005, describing ROMP methods and combinatorial
arrays.
U.S. Patent Application 11/056,391 filed February 14, 2005, now published as
2005/0255237 published on November 17, 2005, describing stamp tip or polymer
coated tip
applications.
U.S. Patent Application 11/065,694 filed February 25, 2005, now published as
2005/0235869 on October 27, 2005, describing tipless cantilevers and flat
panel display
applications.
US Patent publication 2006/001,4001 published January 19, 2006 describing
etching
of nanostructures made by DPN methods.
WO 2004/105046 to Liu & Mirkin published December 2, 2004 describes scanning
probes for contact printing
US Patent Publication 2007/0129321 to Mirkin describing virus arrays.
See also two dimensional nanoarrays described in, for example, US Patent
Publication
2008/0105042 to Mirkin et al., filed March 23, 2007, which is hereby
incorporated by
reference in its entirety.
7
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
All references cited above are incorporated by reference in their entirety,
and the
teachings therein can be adapted for use with the various embodiments
described herein.
DPN methods are also described in Ginger et al.., "The Evolution of Dip-Pen
Nanolithography," Angew. Chem. Int. Ed. 43, 30-45 (2004), including
description of high-
throughput parallel methods.
Direct write methods, including DPN printing and pattern transfer methods, are
described in for example Direct- Write Technologies, Sensors, Electronics, and
Integrated
Power Sources, Pique and Chrisey (Eds) (2002).
The direct-write nanolithography instruments and methods described herein are
particularly of interest for use in preparing bioarrays, nanoarrays, and
microarrays based on
peptides, proteins, nucleic acids, DNA, RNA, viruses, biomolecules, and the
like. See, for
example, US Patent No. 6,787,313 for mass fabrication of chips and libraries;
5,443,791 for
automated molecular biology laboratory with pipette tips; 5,981,733 for
apparatus for the
automated synthesis of molecular arrays in pharmaceutical applications.
Combinatorial
arrays can be prepared. See also, for example, US Patent Nos. 7,008,769;
6,573,369; and
6,998,228 to Henderson et al..
Scanning probe microscopy is reviewed for example in Bottomley, Anal. Chem.
70,
425R-475R (1998). Also, scanning probe microscopes are known in the art
including probe
exchange mechanisms as described in, for example, US Patent No. 5,705,814
(Digital
Instruments).
Patterning compositions can be formulated and adapted for transfer and
deposition
from the tip to a substrate surface. The compositions can comprise two or more
components
including one or more polysaccharides, one or more patterning species, and one
or more
chemical additives. The patterning composition can be formulated to exclude
components
and amounts of components that would interfere with the deposition process,
wherein the
patterning composition comprises the ingredients needed to carry out a
successful result.
Patterning compositions can be dried, partially or fully, on the tip before
the deposition step.
If desired, surfactants in an ink formulation can be used. See for example US
Patent
Publication No. 2006/0242740 to Collier et al.., which is hereby incorporated
by reference in
its entirety.
8
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
PATTERNING COMPOSITION - BIOMOLECULE
The patterning composition can be in the form of an ink. It can comprise one
or more
patterning species. The patterning species can be molecular or particulate or
colloid. It can
be synthetic or natural. It can be polymeric, oligomeric, or non-polymeric. It
can be a small
molecule. Biomolecular applications are particular of note. For example, the
patterning
species can be a biomolecule (wherein water is not a biomolecule). The
patterning species
can be a biopolymer. The patterning species can comprise polymerized or
repeating units of
nucleic acid or amino acid units. Patterning species can be for example
oligonucleotides,
DNA, RNA, protein, peptide, sugar, carbohydrate, and the like. The patterning
species can
be used such that it is not adapted synthetically for interaction with a
substrate surface. For
example, it can be a natural species such as for example a natural protein.
Alternatively, the
patterning species can be used such that it is adapted synthetically for
interaction with a
substrate surface. For example, an end group can be functionalized to bond to
the surface.
This can be represented by, for example, R-X or R-(X), wherein R is a
patterning species tiiat
has been functionalized with group X, and n is the number of groups X, which
can be for
example 1-10, or 1-5, or 1-3.
Non-biological compounds which can serve as patterning species include for
example
particulate materials, nanostructured materials, organic compounds, inorganic
compounds,
polymers, synthetic polymers, compounds which chemisorb to metals (e.g., gold)
such as
thiols and sulfides, and the like.
PROTEIN MOLECULES
The patterning species can comprise proteinaceous material and proteins and
peptides.
Proteinaceous materials include for example antibodies, enzymes, and the like.
In the peptide and protein embodiments, the nanoarrays can be prepared
comprising
various kinds of chemical structures comprising peptide bonds. These include
peptides,
proteins, oligopeptides, and polypeptides, be they simple or complex. The
peptide unit can
be in combination with non-peptide units. The protein or peptide can contain a
single
polypeptide chain or multiple polypeptide chains. Higher molecular weight
peptides are
preferred in general although lower molecular weight peptides including
oligopeptides can be
used. The number of peptide bonds in the peptide can be, for example, at least
three, ten or
less, at least 100, about 100 to about 300, or at least 500.
9
WASH4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
Proteins are particularly preferred. The protein can be simple or conjugated.
Examples of conjugated proteins include, but are not limited to,
nucleoproteins, lipoproteins,
phosphoproteins, metalloproteins and glycoproteins.
Proteins can be functional when they coexist in a complex with other proteins,
polypeptides or peptides. The protein can be a virus, which can be complexes
of proteins
and nucleic acids, be they of the DNA or RNA types. The protein can be a shell
to larger
structures such as spheres or rod structures.
Proteins can be globular or fibrous in conformation. The latter are generally
tough
materials that are typically insoluble in water. They can comprise a
polypeptide chain or
chains arranged in parallel as in, for example, a fiber. Examples include
collagen and elastin.
Globular proteins are polypeptides that are tightly folded into spherical or
globular shapes
and are mostly soluble in aqueous systems. Many enzymes, for example, are
globular
proteins, as are antibodies, some hormones and transport proteins, such as
serum albumin and
hemoglobin.
Proteins can be used which have both fibrous and globular properties, like
myosin and
fibrinogen, which are tough, rod-like structures but are soluble. The proteins
can possess
more than one polypeptide chain, and can be oligomeric proteins, their
individual
components being called protomers. The oligomeric proteins usually contain an
even number
of polypeptide chains, not normally covalently linked to one another.
Hemoglobin is an
example of an oligomeric protein.
Types of proteins that can be incorporated include, but are not limited to,
enzymes,
storage proteins, transport proteins, contractile proteins, protective
proteins, toxins,
hormones, and structural proteins.
Examples of enzymes include, but are not limited to ribonucleases, cytochrome
c,
lysozymes, proteases, kinases, polymerases, exonucleases, and endonucleases.
Enzymes and
their binding mechanisms are disclosed, for example, in Enzyme Structure and
Mechanism,
2"d Ed., by Alan Fersht, 1977, including in Chapter 15 the following enzyme
types:
dehydrogenases, proteases, ribonucleases, staphyloccal nucleases, lysozymes,
carbonic
anhydrases, and triosephosphate isomerase.
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
Examples of storage proteins include, but are not limited to ovalbumin,
casein,
ferritin, gliadin, and zein.
Examples of transport proteins include, but are not limited to hemoglobin,
hemocyanin, myoglobin, serum albumin, (31-lipoprotein, iron-binding globulin,
and
ceruloplasmin.
Examples of contractile proteins include, but are not limited to myosin,
actin, dynein.
Examples of protective proteins include, but are not limited to antibodies,
complement proteins, fibrinogen, and thrombin.
Examples of toxins include, but are not limited to, Clostridium botulinum
toxin,
diptheria toxin, cholera toxin proteins, Alexa Fluor 594 modified cholera
toxin proteins,
snake venoms, and ricin.
Examples of hormones include, but are not limited to, insulin,
adrenocorticotrophic
hormone and insulin-like growth hormone, and growth hormone.
Examples of structural proteins include, but are not limited to, viral-coat
proteins,
glycoproteins, membrane-structure proteins, a-keratin, sclerotin, fibroin,
collagen, elastin,
and mucoproteins.
Natural or synthetic peptides and proteins can be used. Proteins that can be
used, for
example, are prepared by recombinant methods.
Examples of preferred proteins include immunoglobulins, IgG (rabbit, human,
mouse,
and the like), Protein A/G, fibrinogen, fibronectin, lysozymes, streptavidin,
avdin, ferritin,
lectin (Con. A), and BSA. Rabbit IgG and rabbit anti-IgG, bound in sandwhich
configuration
to IgG are useful examples.
Spliceosomes and ribozomes and the like can be used.
A wide variety of proteins are known to those of skill in the art and can be
used. See,
for instance, Chapter 3, "Proteins and their Biological Functions: A Survey,"
at pages 55-66
of BtocxENtlsTxY by A. L. Lehninger, 1970, which is incorporated herein by
reference.
11
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
Additional proteins are described below in the working examples, including
labeled
proteins and fluorescently labeled proteins. Proteins can include cholera
toxin subunit B and
trypsin inhibitor.
NUCLEIC ACID PATTERNING SPECIES
In nucleic acid embodiments, the nucleic acid is not particularly limited. For
example, the nucleic acid can be synthetically made, modified to include, for
example,
functional groups tailored for chemisorption or covalent bonding to the
substrate, as well as
naturally occurring. It can be of low, medium, or high molecular weight,
oligomeric or
polymeric. It can be single-, double-, or even triple-stranded. The nucleic
acid can be based
on deoxyribonucleic acid (DNA), ribonucleic acid (RNA), or combinations
thereof. The
structure of nucleic acids is generally described in, for example, Calladine
and Drew,
Understanding DNA, The Molecule and How it Works, 2'd Ed., 1997.
General types of nucleic acid that can be patterned include, for example, DNA,
RNA,
PNA, CNA, RNA, HNA, p-RNA, oligonucleotides, oligonucleotides of DNA,
oligonucleotides of RNA, primers, A-DNA, B-DNA, Z-DNA, polynucleotides of DNA,
polynucleotides of RNA, T-junctions of nucleic acids, domains of non-nucleic
acid polymer-
nucleic acid block copolymers, and combinations thereof. Additional general
types of
nucleic acids include, for example, viral RNA or DNA, a gene associated with a
disease,
bacterial DNA, fungal DNA, nucleic acid from a biological source, nucleic acid
which is a
product of a polymerase chain reaction amplification, nucleic acid contacted
with
nanoparticles, and nucleic acid double-stranded and hybridized with the
oligonucleotides on
the nanoparticles resulting in the production of a triple-stranded complex.
In general, the nucleic acid can be any of a group of organic substances found
in cells
and viruses that play a central role in the storage and replication of
hereditary information
and in the expression of this information through protein synthesis. Purines,
pyrimidines,
carbohydrates, and phosphoric acid generally characterize the fundamental
organic
substances of a nucleic acid. Purines and pyrimidines are nucleotides, a
nucleoside in which
the primary hydroxy group of either 2-deoxy-D-ribose or of D-ribose is
esterified by
orthophosphoric acid. A nucleoside is a compound in which a purine or
pyrimidine base is
bound via a N-atom to C-1 replacing the hydroxy group of either 2-deoxy-D-
ribose or of D-
ribose, but without any phosphate groups. The common nucleosides in biological
systems
are adenosine, guanosine, cytidine, and uridine (which contain ribose) and
deoxyadenosine,
12
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
deoxyguanosine, deoxycytidine and thymidine (which contain deoxyribose). Thus,
a purine
base may be an adenine nucleotide or a guanine nucleotide. A pyrimidine base
may be
thymine nucleotide, a cytosine nucleotide, or a uracil nucleotide.
The sequence of a nucleic acid may be random or specific so as to encode a
desired
amino acid structure. For instance, a group of three nucleotides may comprise
a codon. One
codon comprises an amino acid. The coding region of a nucleic acid comprises
codons.
The nucleic acid can exist freely or can be bound to peptides or proteins to
form
nucleoproteins in discreet bundles or structured forms such as, for example,
chromosomes. A
nucleic acid also can exist in single-stranded or double-stranded forms. A
nucleic acid may
also be linear, circular, or supercoiled. Nucleic acid may be isolated
directly from a cell or
organelle. A plasmid or cloning vector are also examples of nucleic acids.
The nucleic acid can be made up of nucleotides, each containing a carbohydrate
sugar
(deoxyribose), a phosphate group, and mixtures of nitrogenous purine- and
pyrimidine- bases.
The sugar may be of a cyclic or acyclic form. DNA comprises only thymine and
cytosine
pyrimidines and no uracil. DNA may be isolated from a cell as genomic,
nuclear, or
mitochondrial DNA, or made synthetically (i.e., by chemical processes).
A gene present in a cell typically comprises genomic DNA made up of exonic and
intronic stretches of DNA. The exonic stretches comprises nucleotides that
comprise codons
that encode amino acids, whereas the intronic stretches of DNA comprise
nucleotides that
likely do not comprise codons that encode amino acids. The nucleotide sequence
of purines
and pyrimidines determine the sequences of amino acids in the polypeptide
chain of the
protein specified by that gene.
DNA may also be isolated as complementary or copy DNA (cDNA) synthesized from
an RNA template by the action of RNA-dependent DNA polymerase. For example,
the
cDNA can be about 100-800mer strands from PCR amplification. If the RNA
template has
been processed to remove introns, the cDNA will not be identical to the gene
from which the
RNA was transcribed. Thus, cDNA may comprise a stretch of nucleotides that are
largely
exonic in nature.
When in double-stranded form, the two DNA strands form a double helix. In this
helix, each nucleotide in one strand is hydrogen bonded to a specific
nucleotide on the other
13
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
strand. Thus, in DNA, adenine bonds with thymine and guanine bonds with
cytosine. The
ability of nucleotides present in each strand to bind to each other determines
that the strand,:
will be complementary, e.g., that for every adenine on one strand there will
be a thymine on
the other strand.
RNA can be generally similar to DNA, but contains the sugar ribose instead of
deoxyribose and the base uracil instead of thymine. RNA can be single-stranded
or double-
stranded and is transcribed from a cell's DNA. An RNA molecule may form a
hairpin loop
or other double-stranded structures. RNA may be template RNA, messenger RNA
(mRNA),
total RNA, or transfer RNA (tRNA). polysome. RNA-DNA hybrid molecules can be
deposited according to the present invention. Furthermore, protein-nucleic
acids, or "peptide
nucleic acids" ("PNA") also may be used.
The binding properties exhibited between complementary nucleotides can make
nucleic acids useful as probes that can bind to other nucleic acids. Nucleic
acids can be
labelled and used as probes. By any one of a number of standard labelling
techniques,
nucleic acid probes can be used to detect, by hybridization, another nucleic
acid. The
hybridization can be visualized or detected if the label is, for example, a
fluorescent,
radioactive, or enzymatic label. Thus, a nucleic acid of the present invention
also can be
labelled, or modified so as to comprise a detectable entity, like a
fluorescent marker or tag, a
gold particle, streptavidin, digoxigenin, a magnetic bead, or other markers
known to the
skilled artisan. See, for example, U.S. Patent No. 4,626,501 ("Labeled DNA")
to Landes,
which is hereby incorporated by reference in its entirety.
Nucleotides and nucleic acids also can be modified so that it is protected
against
nucleic acid degradation. For instance, a nucleic acid may be encapsulated
within a
liposome. Alternatively, a thiol group may be incorporated into a
polynucleotide, such as
into an RNA or DNA molecule, by replacing the phosphorous group of the
nucleotide. When
so incorporated into the "backbone" of a nucleic acid, a thiol can prevent
cleavage of the
DNA at that site and, thus, improve the stability of the nucleic acid
molecule.
U.S. Patent No. 5,965,721 to Cook et al.. is also incorporated by reference in
its
entirety, disclosing oligonucleotides, which can be patterned and can have
improved nuclease
resistance and improved cellular uptake.
14
WASH_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
Thus, the bioavailability of a nucleic acid treatment in vivo may be improved
by
modifying the nucleic acid as described. For instance, a modified nucleic acid
formulation
may have an increased half-life and/or be retained in plasma for longer
periods of time than
non-modified nucleic acids. A formulation of nucleic acid and polyethylene
glycol, for
instance, may also increase the half-life of the nucleic acid in vivo, as
could any known slow-
release nucleic acid formulation. Thus, modifying a nucleic acid may increase
the
effectiveness of the nucleic acid in vivo and/or its bioavailability.
The size of a nucleic acid can range considerably, from the size of a few
nucleotides,
to an oligonucleotide, or probe, to a polynucleotide, gene, chromosome
fragment to entire
chromosomes and genomes. For instance, a single- or double-stranded nucleic
acid may be at
least 10-, 20-, 30-, 40-, 50-, 60-, 70-, 80-, 90, or 100-nucleotides or base
pairs (bp) in length.
Larger still, a nucleic acid may be at least 0.2 kb, 0.3 kb, 0.4 kb, 0.5 kb,
0.6 kb, 0.7 kb, 0.8
kb, 0.9 kb, or 1.0 kb in size. Indeed, a nucleic acid for use in the present
invention can be at
least 1 kb, 2 kb, 3 kb, 4 kb, 5 kb, 6 kb, 7 kb, 8 kb, 9 kb, or 10 kb or larger
in size. One
preferred size range is 1-2 kb. The nucleic acid can be a chain of varying
length of
nucleotides and are typically called polynucleotides or oligonucleotides. An
oligonucleotide
is an oligomer generally resulting from a linear sequences of nucleotides. The
oligonucleotide can comprise, for example, about 2 to about 100, about 2 to
about 20, about
to about 90, or about 15 to about 35 nucleotides. In oligonucleotide arrays,
about 25-mer
oligonucleotides can be used. Another particular range is about 60- to about
80-mers, which
are relatively long oligonucleotides.
Microarray methods, including selection of nucleic acid, probling, labeling,
and
detection, are described in U.S. Patent Nos. 6,379,932 and 6,410,231 (Incyte
Genomics) and
can be used. These patents are incorporated by reference in their entirety.
Although these
references mention dip pen nanolithographic methods, they do not suggest how
or provide
guidance on how dip pen nanolithographic methods can be used to make improved
nanoarrays as described herein.
A compound comprising a single nucleotide can also be used as ink. Mixtures of
nucleic acids can be used, and different spots on an array can comprise
different nucleic
acids.
WASH_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
A nucleic acid for deposition may be formulated or mixed with other elements
prioi
to, or after direct write deposition onto a substrate surface. Thus, an "ink"
of the present
invention may comprise other chemicals, compounds, or compositions for
deposition onto a
substrate surface in addition to a desired nucleic acid sample. Solvent and
salt can be used to
apply the nucleic acid to the tips. Surfactants can also be used. For
instance, proteins,
polypeptides, and peptides may be deposited along with a desired nucleic acid
onto a
substrate surface.
Nucleic acid arrays, and the types of nucleic acids used therein, are
described for
example in A Primer of Genome Science, G. Gibson and S. Muse, 2002, Chapters 3-
4 (pages
123-181), which is hereby incorporated by reference. This reference, for
example, describes
both cDNA microarrays and oligonucleotide arrays, labeling, hybridization, and
statistical
analysis. cDNA arrays can be used for monitoring the relative levels of
expression of
thousands of genes simultaneously. PCR-amplified cDNA fragments (ESTs) can be
spotted
and probed against fluorescently or radioactively labeled cDNA. The intenstity
of the signal
observed can be assumed to be in proportion to the amount of transcript
present in the RNA
population being studied. Differences in intensity reflect differences in
transcript level
between treatments. Statistical and bioinformatic analyses can then be
performed, usually
with the goal of generating hypotheses that may be tested with established
molecular
biological approaches. Current cDNA microarrays, however, can have an upper
limit of
15,000 elements and are unable to represent the complete set of genes present
in higher
eukaryotic genomes. The advantages and disadvantages of oligonucleotide versus
cDNA
microarrays are described in the aforementioned A Primer of Genome Science and
can be
used in constructing nucleic acid nanoarrays as described herein.
Oligonucleotides are also described in the working examples hereinbelow
including
labeled oligonucleotides and fluorolabeled oligonucleotides.
PATTERNING COMPOSITION - SOLVENT
The patterning composition can comprise one or more solvents. The solvent can
be
for example water including pure water, distilled water, deionized water, and
the like. It can
be a buffered solvent. The pH can be varied for the application. The solvent
can be one or
more organic solvents. Mixtures of solvent compounds can be used. Examples
include
alcohols, ethers, alkanes, esters, aromatics, as known in the art.
16
WASH_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
PATTERNING COMPOSITION - CARRIER
The patterning composition can comprise one or more carriers. A carrier can
function
for example to facilitate transport of the patterning species or to
encapsulate the patterning
species. The carrier can be synthetic or natural, and is preferably
hydrophilic. For example,
the carrier can be a polysaccharide, and polysaccharides are generally known
in the art. See
for example Hohinski, Modem Concepts in Biochemistry, 4th Ed., 1983, Chapter 7
(discussing carbohydrates and various saccharides and polysaccharides). For
example,
polysaccharides can be starch, cellulose, or unbranched polysaccharides such
as agarose.
Agarose is a naturally occurring polysaccharide gel, which has been
demonstrated to be
biocompatible, biologically inert, and has low intrinsic florescence; see for
example Mateo et
al., Enzyme and Microbial Technology (2006). It can be obtained from for
example seaweed
or some species of red algae. Alternatively, it may also be synthesized. It is
frequently used
as a solidifying agent or supporting medium in microbial culture. It can also
be used for gel
electrophoresis. It can be for example cast in a tube or slab form.
Agarose can undergo a thermo-reversible gelation mechanism, where the gelation
temperature is generally about 30 C. This relatively low temperature can
allow agarose to be
mixed with for example biomolecules while still in the sol state at
physiological
temperatures. The ratio of agarose to biomolecules can generally be any ratio,
such as for
example 1:3, 1:1, or 3:1. An agarose matrix used as a carrier can function as
a reservoir of
the patterning species with the probe and ensures constant delivery of the
patterning
composition on to the substrate for greater than 480 points per tip (see
Figure 7).
PATTERNING COMPOSITION - CHEMICAL ADDITIVE
Chemical additives can be used to control physical and/or chemical properties
of the
carrier matrix in the patterning composition. In general, hygroscopic
materials are preferred
as additives, as they can help prevent the carrier from drying out. One
attractive feature of
the additives is that the additives can modify the properties of the carrier
such as the cross-
link density of the polymer chain, thereby making the carrier for example less
viscous.
Without wishing to be bound by any particular theory, an additive such as a
carbohydrate
such as sucrose can function as an agent to reduce the degree of aggregation
in the carrier gel,
resulting in for example a smaller correlation length, which can be associated
with smaller
cross-section radii of the polymer chains in carrier matrix or gel.
17
WASH_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
For example, tris buffer such as Tris(hydroxymethyl) aminomethane Ethylene
Diamine Tetraacetic Acid ("Tris-EDTA") and sucrose both have the effect of
breaking up
agarose bundles into a finer gel structure; see for example Normand et al.,
Carbohydrate
Polymers (2003). These hygroscopic materials also absorb moisture from the
air, keeping the
agarose gel hydrated. In practice, the use of additives provide a parameter to
control the
diffusion rate and transport properties of the gel from AFM tip being
deposited onto the
substrate surface, in addition to parameters such as humidity, temperature and
contact time.
The type of additive need not be confined to carbohydrate or a buffer. Any
material
with the desired properties described above can be used. For example, the
additive can be a
lipid or a sugar alcohol such as glycerol. Glycerol comprises three hydroxyl
groups which
upon esterification with one, two or three fatty acids forms monoglycerides,
diglycerides and
triglycerides respectively. If one of the fatty acids is replaced with a sugar
or a phosphate the
resulting compound is a glycolipid or a phospholipid respectively. The fatty
acids can be
unsaturated, saturated, monounsaturated or polyunsaturated.
TIPS AND INSTRUMENTATION
Instrumentation to execute patterning by transferring materials from tip to
substrate
surface are known in the art. See for example products from Nanolnk, Inc.
(Skokie, IL). See
also for example US Patent Nos. 6,827,979; 6,642,129; 6,867,443; 7,008,769;
6,573,369; and
6,998,228. For example, the tip can be a nanoscopic tip. The tip for example
can be a
scanning probe microscope tip or an atomic force microscope tip. The tip can
be a solid tip;
or the tip can be a hollow tip. The hollow tip can comprise an aperature and
can delivery
flow paths for delivering ink compositions to the end of the tip. The tip can
comprise, for
example, an inorganic surface or an organic surface. Tips can be made from
hard materials
through, for example, microfabrication. Sharpening of tips can be carried out.
After tip fabrication, the tip can be used as is, although the tip can be
cleaned first
when used as is. The tip can be also surface modified if desired after
fabrication. For
example, an organic coating can be added to an inorganic tip surface.
The tip can comprise a tip surface, including an inorganic tip surface, which
has not
been modified by organic material.
18
WASH_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
Tips can be made from materials known in the AFM art, including silicon
nitride,
silicon, and other hard materials.
The tip can be disposed on a cantilever, as known in the art, including at an
end of a
cantilever or near the end of a cantilever.
The tips can be if desired relatively long tips having for example a length of
at least 5
microns, or at least 10 microns.
The tip can be part of an array of tips, so that a plurality of tips can be
provided. For
moving in the z-direction with respect to the surface, the tips can move
together in a passive
mode or can be moved individually in an active or actuated mode. Hence, in the
depositing
step, the tip can be passively used, or can be used as an actuated tip. The
actuation
mechanism can be for example thermal or electrostatic or piezoresistive. One-
dimensional
array of tips can be used; or two-dimensional array of tips can be used. In
particular, arrays
can be used which have large numbers of tips. See for example US Patent
Application serial
no. 11/690,738 filed March 23, 2007 to Mirkin et al.., which is hereby
incorporated by
reference in its entirety including the Lenhart Small paper (Lenhart et al.,
Small 3, no. 1, 71-
75 (2007)).
Instrumentation methods are known in the art to move tips, and tips disposed
on
cantilevers, in the x, y, and z-directions with respect to the surface.
Instrumentation can be adapted to allow for heating of tips. See for example
US
Patent Publication No. 2006/0242740 to Sheehan et al..
SUBSTRATE AND SUBSTRATE SURFACE
A wide variety of substrates can be used which present surfaces for
deposition.
Substrates can be those used to prepare microarrays in the art. Substrates can
be polymeric,
glass, ceramic, composite, metal, semiconductor, oxides, silicon, and the
like. The substrate
can be monolithic, one piece, or can comprise layers disposed on each other.
The substrate
can comprise an inorganic or an organic surface coating. A monolayer coating
can be used.
The surface can be functionalized with organic functional groups or organic
material. For
example, the substrate can comprise an inorganic material surface modified
with an organic
material. The substrate can be for example a biomolecule.
19
WASH_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
The substrate surface can be adapted to covalently bond to or chemisorb to one
or
more components of the patterning composition. For example, the substrate
surface can be
an electrophilic surface. The substrate surface can be adapted to be reactive
with functional
groups in the patterning species. For example, amino groups in a protein can
react with
succinimide. Or a thiol group or compound can chemisorb to gold. For example,
aldehyde-
modified substrate can also be used as a reactive support for the
immobilization of amine-
modified or amine-containing biomolecules via imine formation. Once the
encapsulated
biomolecules are deposited onto the substrate from the ADM tip, the agarose
gel matrix can
be dried by the exposure to the air and removed by washing with MilliQ water.
If fluorescent detection is used, the substrate and patterning can be adapted
to
minimize or avoid quenching of the fluorescence.
Substrates can be pre-patterned as needed to provide boundaries for and
designate
spaces for the deposition zones.
DEPOSITION
The tip and the substrate surface can be moved with respect to each other so
that
deposition of the patterning composition occurs and material is transferred
from the tip to the
surface to form a deposit. In some cases, a meniscus may be present to
facilitate deposition.
The tip the tip is in position for deposition can be controlled as desired.
In some cases, heat can be used to facilitate deposition. Tips and cantilevers
supporting tips can be heated, or the environment around the deposition area
can be heated.
An environmental chamber can be used to control humidity, temperature,
atmospheric gases,
and other parameters. For example, the deposition can be carried out at a
relative humidity
sufficient, e.g., sufficiently high, to allow the deposition to occur. In some
cases, higher
relative humidity may activate or speed up deposition. The deposition can be
carried out at a
relative humidity of for example at least 30%, or at least 50%, or at least
70%.
If the carrier exhibits a gel-liquid crystal transition temperature, the
deposition
temperature can be above this temperature, e.g., 10 C or more above the gel-
liquid crystal
transition temperature.
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
The deposition step can be carried out by contacting the tip with the surface,
wherein
the tip is held stationary in the xy plane with respect to the surface.
Alternatively, the
deposition step can be carried out by contacting the tip with the surface,
wherein the tip is riot
held stationary in the xy plane with respect to the surface, but rather the
tip is moving.
The contact time during the spotting/depositing can vary between for example 7
and
seconds, resulting in features of about 200 to 500 nm in diameter. AFM probes
that can
be used can have a spring constant k ranging from for example about 0.3 to
about 2 N/m2.
Aldehyde-substrates can be fabricated by vapor deposition of
trimethoxysilylalkylaldehyde
onto activated glass slides.
If scanning probe instrumentation is used, such as AFM instrumentation, a
variety of
modes for use can be used including for example contact mode, noncontact mode,
or tapping
mode or intermittent contact mode.
After a short incubation period in which the gel forms, AFM tips may be
immediately
coated by directly dipping the tips into the gel-ink, by inkwells, or by
placing a drop of the
gel-ink on a solid substrate and lowering the tips into the gel by an AFM or
other controlled
mechanics. The sticky, viscous nature of the agarose gel-ink can allo for
minimal to none tip
modification for its inking procedures.
ACTIVATION AND RATE OF DEPOSITION
The additive can activate or increase the rate of deposition of the patterning
composition comprising a carrier such as agarose. For example, in some
embodiments, the
patterning composition will not substantially leave the tip without the
additive, or the amount
leaving the tip may be too small to detect, or take too long to be
commercially useful.
Detecting deposition can be carried out by for example fluorescence detection
or scanning
probe methods.
DEPOSIT
The deposit can be formed in a variety of shapes and patterns. A pattern can
be found
in a single deposit, or in a series of separate deposits. The deposit can be
for example a dot or
a line. The line can be straight or curved. The deposit can be characterized
by a line width or
a dot diameter. For example, the dot diameter or the line width can be about
10 nm to about
21
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
20 microns, or about 50 nm to about 10 microns, or about 100 nm to less than
about one
micron.
The deposit can be also characterized by a height. For example, the height can
be
about 1 nm to about 1 micron, or about 10 nm to about 750 nm, or about 100 nm
to about 500
nm.
The distance between deposits can reflect high resolution and can be for
example
about 50 microns or less, or about 10 microns or less, or about 1 micron or
less, or about 15
nm to about ten microns, or about 100 nm to about one micron. The distance
between
deposit can be measured as an edge-to-edge distance or a distance between
center points
(e.g., centers of dots).
The deposit can be treated by for example washing to remove one or more
components including carrier, patterning species, additives, or a combination
thereof. The
deposition and washing can be adapted so that the molecules in the patterning
composition
are not removed. All or substantially all of the carrier can be removed, or if
desired some
lipid can be retained if washing is adapted accordingly.
ARRAYS
Also provided herein are articles including arrays, wherein the array
comprises a
substrate and one or more deposits disposed on the substrate surface. The
deposits can be
formed by methods described herein.
APPLICATIONS
Applications include microarrays and nanoarrays, including biological arrays,
and the
known applications of such arrays. For example, the development of direct
patterning and
nanopatterning methods for protein-based nanostructures is important for
researchers
working in the areas of proteomics, and theranostics. Such methods would allow
one to
generate multi-component biological nanostructures of proteins,
oligonucleotides, and
viruses. Other applications include the development of biological microarrays
and
nanoarrays for high-throughput genomic and proteomic analysis, exploring
biomolecular
interactions on the nanoscale with larger biological entities (i.e. eukaryotic
cells, viruses,
bacteria, and spores), and for biosensing and medical diagnostics.
22
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
NON-LIMITING WORKING EXAMPLES
A series of non-limiting working examples are provided.
MATERIALS AND METHODS
The following materials and techniques were employed in several embodiments of
the
non-limiting working examples:
Materials
A variety of biomolecules, such as DNA or proteins, were patterned by dip-pen
nanolithography on activated glass surfaces (either aldehyde, or NHS activated
slides) using
the agarose matrix as a carrier ink. The ink comprised a 1:1 mixture of about
0.3% agarose
(for a total concentration of about 0.15%) and biomolecules, such as DNA or
proteins at
various concentrations in different embodiments. A variety of chemical
additives were also
included in the final ink composition (Figure 1). The purposes of the chemical
additives were
at leasttwo-fold. First, they generally comprise hygroscopic materials, which
prevent the
agarose matrix from drying out on the AFM tip (see Figure 2). Second, they
modify the
nature of the agarose gel and thus accelerate the ink diffusion rate from tip
to substrate
surface. Though only a few chemical additives have been examined, the use of
additives to
control the diffusion rate may be extended to other chemicals that modify the
nature of the
agarose matrix.
Oligonucleotides were either purchased from Integrated DNA Technologies, Inc.,
San
Diego, USA or synthesized on an Expidite DNA synthesizer using precursors
obtained from
Glen Research, USA. The probe sequence employed for DPN studies had the
sequence 5'-
NH2-(CH2)6-GTG CAC CTG ACT CCT GTG GAG-Cy3-3'. Probe concentrations were
about 15 to about 100 M. The complimentary sequence was of the form 5'-Cy5-
CTC CAC
AGG AGT CAG GTG CAC-3'. The random sequence was of the form 5'-Cy5-TCA TAG
TGT GGA CCC CTA GCA-3'. Cholera toxin protein Alexa Fluor 594 (1 mg/ml) was
purchased from Molecular Probes. Anti-cholera toxin IgG antibodies were
purchased from
Biodesign International and modified with Alexa Fluor 488 (Molecular Probes).
Donkey
anti-goat IgG antibodies (Cy5) were obtained from R & D Systems, USA. Tris-
EDTA
(Sigma) additives were in the concentration range 10 mM Tris, 1 mM EDTA (lx)
to 300 mM
Tris, 30 mM EDTA (30 x). Glycerol (Sigma) additives were used in the range of
about 20 to
23
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
about 100 mM. All buffers and biomolecule solutions were prepared using 18.2
MS2=cm
distilled water (MilliQ). The following chemicals were used as received:
Trimethoxysilylalkylaldehyde (United Chemical Technologies, Inc.) agarose low
melting
(Fisher Biotech, BP165-25), Tris (hydroxymethyl) amine (Sigma),
ethylenediamine-
tetraacetic acid (Sigma), 1,1,1 Tris (hydroxymethyl) ethane (Sigma), Tricine
buffer solution
(Sigma), sucrose (Sigma), ethanolamine (Sigma), Codelink activated glass
slides (GE
Healthcare).
Modification glass slides.
Codelink activated glass slides from GE Healthcare were used as received.
These
slides have been coated with a 3-D chemistry that comprises a hydrophilic
polymer
containing amine-reactive groups, which can facilitate attachment of molecules
such as
protein to the slides. For the aldehyde modification, clean microscope cover
glass (Fisher
Scientific) was activated in oxygen plasma for 15 minutes, and immediately
submitted to
vapor deposition of trimethoxysilylalkylaldehyde for 20 minutes. Following
monolayer
formation, the substrates were rinsed with ethanol to remover any excess
silanes and dried
with N2.
Ink Preparation
A 0.3% Agarose gel was prepared by dissolving agarose in MilliQ water by
heating
for 2 minutes in a microwave oven. The gel in sol form was mixed with a
solution of
biomolecules with or without additives in a 1:1 ratio.
Dip-Pen Nanolithography
DPN was preformed on an NScriptor (Nanoink, Skokie, IL, USA) in contact mode
using multi-pen 1-dimensional array M-tips (spring constant 0.5 N/m) in air at
about 60%
humidity. Tips were inked in M-tip inkwells by dipping 3-5 times. Dwell times
(i.e., time
during which the tip remains immersed in the ink) varied from about 0.01 to
about 20 s.
Fluorescence Microscopy
Fluorescent images were obtained with a Carl Zeiss, Axiovert 200M
epifluorescent
microscope.
24
WAS H_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
Hybridization on Substrate Surface
DNA patterns on Codelink slides were allowed to react for a minimum of 8 hours
at
50 % humidity and washed with PBS (5 minutes) under vigorous agitation and
rinsed with
water to remove residual agarose. The reaction time can vary, for example in
the range of 6
to about 9 hours. These substrates were then blocked with 50 mM ethanolamine
in Tris-
EDTA at a pH 8 for 1 hour at 50 C. For hybridization, a 5'-Cy5-labeled
oligonucleotide was
diluted to 1 M in 4 x SSC containing 0.02 % SDS and a drop of that solution
was applied to
the surface of the modified glass slide. A coverslip was mounted gently on the
top of the
solution, and the substrates were transferred to the hybridization oven at 45
C for 8 hours.
The unhybridized probes were removed by washing with vigorous agitation in a 1
x SSC with
0.01 % SDS solution for 5 minutes at hybridization temperature, 0.1 x SSC with
0.01 % SDS
for 5 minutes at room temperature, and subsequent washing in water for 5
minutes.
Biorecognition on the Substrate Surface
Protein patterns on Codelink sides were allowed to react for a minimum of 8
hours at
50 % humidity and then washed with PBS (5 minutes) under vigorous agitation
and rinsed
with water to remove residual agarose. Either anti-cholera toxin IgG
antibodies (Alexa Fluor
488) or donkey anti-goat (Cy5) was diluted to a concentration of 100 g/mL in
1 x PBS, 1%
BSA, 0.25 % Tween-20. A drop of this solution was applied to the surface of
the substrate
and a coverslip gently mounted on top. The substrate was subsequently
transferred to an
oven at 37 C for 45 minutes. The un-reacted probes were removed by washing
with
vigorous agitation in lx PBS, 1% BSA, 0.25% Tween-20 for 5 minutes at room
temperature
and thereafter rinsed with water.
EXAMPLE 1 Effect of Tris-EDTA Buffer on Diffusion Rate
In one embodiment, the diffusion rate may be accelerated by the use of a Tris-
EDTA
buffer. Without the buffer, a pure DNA/agarose ink was found to be almost
immobile, and
would not diffuse from the AFM tip to the substrate. The ratio of Tris to EDTA
can bne of
any value. In one embodiment, with a concentration of 10 mM Tris, 1 mM EDTA,
the
diffusion rate of a DNA/agarose ink was measured to be approximately 0.0035
m/s2 on
aldehyde modified glass substrates. When the concentration of buffer was
increased to 300
mM Tris, 30 mM EDTA, the diffusion rate was increased to 0.2 m/s2 (see Figure
3) on the
same aldehyde modified substrate.
WASH_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
EXAMPLE 2 Effect of Glycerol on Diffusion Rate
In one embodiment, glycerol was used as an additive in a patterning ink
composition
for DPN. With no additive, a DNA/agarose ink was not found to diffuse from AFM
tip to an
NHS activated Codelink glass slide substrate. In contrast, with no additive, a
cholera toxin
protein/agarose ink was found to migrate from tip to substrate. Thus. the
diffusion
characteristics of the two inks were different. However, witha 30 mM of
glycerol, the
diffusion rate of both the DNA/agarose ink and cholera toxin protein/agarose
ink were
modified to be approximately equal, varying from 0.4 - 0.7 m2/s (see Figure
4).
EXAMPLE 3 Bioactivity Studies
In one embodiment, the bioactivity of oligonucleotides and proteins deposited
on
Codelink activated glass slide substrates using 30 mM glycerol as an additive
in the ink was
probed by fluorescence. A single strand of mutant B-globin with a 5' amine and
3' Cy3
fluorophore was spotted onto a Codelink activated glass substrate by DPN
(Figure 5).
Fluorescent microscopy images of Cy3-labeled DNA nanoarrays, each comprising
216 spots,
can be fabricated by DPN multipen-arrays (see Figures 9(a) and (b)). After a
non-
complementary, random sequence oligonucleotide strand with a 5' Cy5
modification was
introduced, the Cy3 signal of the immobilized DNA was detected, though no Cy5
signal was
observed. The same substrate was then challenged with the complementary
sequence
modified at the 5' end with Cy5, and a positive signal was clearly detected.
Thus, the
oligonucleotides in an agarose carrier immobilized on the Codelink activated
glass slide and
remained bioactive. The negative control with a random sequence showed that
the
interaction between oligonucleotides spotted using agarose as a carrier ink
remains sequence
specific, and that there was no interaction between a random oligonucleotide
sequence and
any residual agarose that may or may not be on the surface.
In addition, the bioactivity of spotted proteins was probed using the same
fluorescence method outlined for DNA. An ink composed of Alexa Fluor 594
modified
cholera toxin proteins, agarose, and 30 mM glycerol was deposited onto
Codelink activated
glass substrates by DPN. Incubation with a Cy5 labeled IgG antibody non-
specific towards
cholera toxin resulted in a negative control. However, when the same substrate
was
incubated with Alexa Fluor 488 modified IgG antibodies specific to cholera
toxin, a positive
signal was observed for both the cholera toxin proteins, and antibodies. Thus,
proteins
26
WASH_4221716.1
CA 02691295 2009-12-15
WO 2009/035739 PCT/US2008/067234
deposited using agarose as a carrier matrix remained active and retained
specificity after Dl'N
(see Figure 6 and Figure 10).
27
WAS H_4221716.1