Note: Descriptions are shown in the official language in which they were submitted.
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
BIOMARKERS FOR DIABETES, OBESITY, AND/OR
HYPERTENSION
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application claims the benefit of priority to U.S. Provisional
Application No. 60/958,125, filed July 3, 2007, the contents of which are
incorporated by reference herein in their entirety.
FIELD OF THE INVENTION
[0002] The invention is in the field of medical screening and diagnosis.
BACKGROUND OF THE INVENTION
[0003] Human blood plasma is the most complex human-derived proteome. At
the same time, plasma is a valuable informative proteome for medical
diagnosis.
The attraction of plasma for disease diagnosis lies in two characteristics:
the ease by
which it can be safely obtained and the fact that it comprehensively samples
the
human phenotype (Anderson, et al., Mol. Cell Proteomics (2002) 1.11:845-867).
[0004] Approximately half of the total protein mass in plasma is accounted by
one protein (albumin), while the top ten proteins together make up 90% of the
total
(Anderson, J. Physiol. (2005) 563.1:23-60). This enormous dynamic range
(nearly
orders of magnitude between the high abundance and very low abundance
proteins) is currently outside the range of available technologies in
proteomics.
SUMMARY OF THE INVENTION
[0005] The invention is based, at least in part, on the discovery that
proteins
present in plasma at low concentrations can be used as biomarkers for
diabetes,
obesity, and/or hypertension. This discovery was exploited to develop the
invention,
which, in one aspect, features a method of identifying a subject having, or at
risk of
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
developing, diabetes, obesity, and/or hypertension. The method includes the
steps of
determining the level of a biomarker (i) in a test biological sample obtained
from the
subject, and (ii) in a control biological sample of like tissue derivation
from a control
subject not having, or at risk of developing, diabetes, obesity, and/or
hypertension;
and comparing the level of the biomarker in the test sample and in the control
sample; a different level of the biomarker in the test sample relative to the
control
sample being indicative that the subject has, or is at risk of developing,
diabetes,
obesity, and/or hypertension.
[0006] In some embodiments, a level of the biomarker in the test sample that
is
increased relative to the control sample is indicative that that subject has,
or is at risk
of developing, diabetes, obesity, and/or hypertension. In other embodiments, a
level
of the biomarker in the test sample that is decreased relative to the control
sample is
indicative that that subject has, or is at risk of developing, diabetes,
obesity, and/or
hypertension.
[0007] In some embodiments, the biomarker is one or more of actin alpha
skeletal protein, alpha-2-antiplasmin, alpha-2-HS-glycoprotein,
angiotensinogen,
apolipoprotein All, apolipoprotein AIV, apolipoprotein B 100, apolipoprotein
CI,
apolipoprotein CII, apolipoprotein CIII, apolipoprotein D, apolipoprotein E,
apolipoprotein-L1, apolipoprotein M, beta-2-glycoprotein I, carboxypeptidase
B2,
complement Clq subcomponent B chain, complement Clr subcomponent,
complement Cls, complement C2, complement C5, complement component C7,
complement component C8 alpha chain, complement component C8 beta chain,
complement component C8 gamma chain, complement factor B, complement factor
H-related protein 1, complement factor H-related protein 2, complement factor
I,
haptoglobin-related protein, insulin-like growth factor binding protein,
kininogen-1,
lumican, N-acetylmuramoyl-L-alanine amidase, plasma protease C1 inhibitor,
plasma retinol-binding protein, plasminogen, platelet basic protein,
serotransferrin,
serum amyloid A-4 protein, serum amyloid P-component, serum
paraoxonase/arylesterase 1, tetranectin, thyroxin binding globulin, Vitamin D-
binding protein, or zinc-alpha-2-glycoprotein.
[0008] In certain embodiments, the test biological sample is a test plasma
sample and the control biological sample is a control plasma sample. In other
2
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
embodiments, the test biological sample is a test serum sample and the control
biological sample is a control serum sample.
[0009] In some embodiments, determining the level of the biomarker includes
measuring the protein level and/or the RNA level of the biomarker. In some
embodiments, the RNA is mRNA.
[0010] In other embodiments, determining the level of the biomarker in the
test
sample and in the control sample includes removing from the samples proteins
present at high levels. In one embodiment, the method includes the step of
removing high abundance proteins from the test biological sample, resulting in
a
depleted test biological sample, and removing high abundance proteins from the
control biological sample, resulting in a depleted control biological sample.
In some
embodiments, the high abundance proteins are removed using immuno-depletion.
In
some embodiments, the high abundance proteins albumin, IgG, IgA, transferrin,
antitrypsin, and/or haptoglobin are removed. In some embodiments, immuno-
depleting the samples includes using a multiple affinity removal system.
[0011] In some embodiments, the method further includes separating
components of the depleted biological samples. In particular embodiments, the
method includes separating the depleted test biological sample into at least a
first
fraction including glycosylated polypeptides and at least a second fraction
including
non-glycosylated polypeptides, and separating the depleted control biological
sample into at least a third fraction including glycosylated polypeptides and
at least a
fourth fraction including non-glycosylated polypeptides. In some embodiments,
the
depleted biological samples are separated using chromatography. In one
embodiment, a multi-lectin column is used.
[0012] In yet other embodiments, the method further includes subjecting the
fractions to digestion. In one embodiment, the glycosylated polypeptides in
the
fractions are digested. In another embodiment, the non-glycosylated
polypeptides in
the fractions are digested. In particular embodiments, the method includes
digesting
with trypsin the non-glycosylated polypeptides in the second fraction,
resulting in a
test tryptic digest, and digesting with trypsin the non-glycosylated
polypeptides in
the fourth fraction, resulting in a control tryptic digest. In another
embodiment, the
method includes digesting with trypsin the glycosylated polypeptides in the
first
3
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
fraction, resulting in a test tryptic digest, and digesting with trypsin the
glycosylated
polypeptides in the third fraction, resulting in a control tryptic digest.
[0013] In other embodiments, the method further includes separating the
components of the tryptic digests. In one embodiment, the components of the
tryptic
digests are separated using chromatography. In some embodiments, the
components
of the tryptic digests are separated using two dimensional chromatography or
liquid
chromatography-mass spectrometry (LC/MS). In one embodiment, the method
includes subjecting to LC/MS the test tryptic digest, resulting in a test
polypeptide
profile, and subjecting to LC/MS the control tryptic digest, resulting in a
control
polypeptide profile. In some embodiments, the LC/MS includes reversed phase
chromatography.
[0014] In another aspect, the disclosure features a method of determining the
level of a biomarker in a test plasma sample relative to a control plasma
sample.
The method includes the steps of obtaining a test plasma sample from a test
subject
and a control plasma sample from a control subject; immuno-depleting albumin,
IgG, IgA, transferrin, antitrypsin, and haptoglobin from the test plasma
sample,
resulting in a depleted test plasma sample, and immuno-depleting albumin, IgG,
IgA, transferrin, antitrypsin, and haptoglobin from the control plasma sample,
resulting in a depleted control plasma sample; and separating the depleted
test
plasma sample into a first fraction including glycosylated polypeptides and a
second
fraction including non-glycosylated polypeptides, and separating the depleted
control plasma sample into a third fraction including glycosylated
polypeptides and
a fourth fraction including non-glycosylated polypeptides.
[0015] In some embodiments, the method further includes digesting with trypsin
the non-glycosylated polypeptides in the second fraction, resulting in a test
tryptic
digest, and digesting with trypsin the non-glycosylated polypeptides in the
fourth
fraction, resulting in a control tryptic digest. In other embodiments, the
method
alternatively includes digesting with trypsin the glycosylated polypeptides in
the
first fraction, resulting in a test tryptic digest, and digesting with trypsin
the
glycosylated polypeptides in the third fraction, resulting in a control
tryptic digest.
4
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
In yet other embodiments, the method includes digesting with trypsin the non-
glycosylated polypeptides and the glycosylated polypeptides in the fractions.
[0016] In some embodiments, the method further includes subjecting to LC/MS
the test tryptic digest, resulting in a test polypeptide profile, and
subjecting to
LC/MS the control tryptic digest, resulting in a control polypeptide profile;
and
comparing the level of the biomarker in the test polypeptide profile to the
level of
the biomarker in the control polypeptide profile.
[0017] In some embodiments, the biomarker is one or more of actin alpha
skeletal protein, alpha-2-antiplasmin, alpha-2-HS-glycoprotein,
angiotensinogen,
apolipoprotein All, apolipoprotein AIV, apolipoprotein B 100, apolipoprotein
CI,
apolipoprotein CII, apolipoprotein CIII, apolipoprotein D, apolipoprotein E,
apolipoprotein-L1, apolipoprotein M, beta-2-glycoprotein I, carboxypeptidase
B2,
complement Clq subcomponent B chain, complement Clr subcomponent,
complement Cls, complement C2, complement C5, complement component C7,
complement component C8 alpha chain, complement component C8 beta chain,
complement component C8 gamma chain, complement factor B, complement factor
H-related protein 1, complement factor H-related protein 2, complement factor
I,
haptoglobin-related protein, insulin-like growth factor binding protein,
kininogen-1,
lumican, N-acetylmuramoyl-L-alanine amidase, plasma protease C1 inhibitor,
plasma retinol-binding protein, plasminogen, platelet basic protein,
serotransferrin,
serum amyloid A-4 protein, serum amyloid P-component, serum
paraoxonase/arylesterase 1, tetranectin, thyroxin binding globulin, Vitamin D-
binding protein, or zinc-alpha-2-glycoprotein.
[0018] In certain embodiments, immuno-depleting the samples includes a
multiple affinity removal system. In other embodiments, separating the
depleted
samples includes using a multi-lectin column. In yet other embodiments, the
LC/MS includes reversed phase chromatography.
[0019] In some embodiments, a level of the biomarker in the test plasma sample
that is different relative to the control plasma sample is indicative that the
test
subject has, or is at risk of developing, diabetes, obesity, and/or
hypertension. In
some embodiments, a level of the biomarker in the test plasma sample that is
increased relative to the control plasma sample is indicative that the test
subject has,
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
or is at risk of developing, diabetes, obesity, and/or hypertension. In other
embodiments, a level of the biomarker in the test plasma sample that is
decreased
relative to the control plasma sample is indicative that the test subject has,
or is at
risk of developing, diabetes, obesity, and/or hypertension.
[0020] In another aspect, the disclosure features a method of determining the
level of a biomarker in a test serum sample relative to a control serum
sample. In
some embodiments, the method includes the steps of obtaining a test serum
sample
from a test subject and a control serum sample from a control subject; immuno-
depleting albumin, IgG, IgA, transferrin, antitrypsin, and haptoglobin from
the test
serum sample, resulting in a depleted test serum sample, and immuno-depleting
albumin, IgG, IgA, transferrin, antitrypsin, and haptoglobin from the control
serum
sample, resulting in a depleted control serum sample; and separating the
depleted
test serum sample into a first fraction including glycosylated polypeptides
and a
second fraction including non-glycosylated polypeptides, and separating the
depleted control serum sample into a third fraction including glycosylated
polypeptides and a fourth fraction including non-glycosylated polypeptides.
[0021] In some embodiments, the method further includes digesting with trypsin
the non-glycosylated polypeptides in the second fraction, resulting in a test
tryptic
digest, and digesting with trypsin the non-glycosylated polypeptides in the
fourth
fraction, resulting in a control tryptic digest. In other embodiments, the
method
alternatively includes digesting with trypsin the glycosylated polypeptides in
the
first fraction, resulting in a test tryptic digest, and digesting with trypsin
the
glycosylated polypeptides in the third fraction, resulting in a control
tryptic digest.
In yet other embodiments, the method includes digesting with trypsin the non-
glycosylated polypeptides and the glycosylated polypeptides in the fractions.
[0022] In some embodiments, the method further includes subjecting to LC/MS
the test tryptic digest, resulting in a test polypeptide profile, and
subjecting to
LC/MS the control tryptic digest, resulting in a control polypeptide profile;
and
comparing the level of the biomarker in the test polypeptide profile to the
level of
the biomarker in the control polypeptide profile.
6
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
[0023] In some embodiments, the biomarker is one or more of actin alpha
skeletal protein, alpha-2-antiplasmin, alpha-2-HS-glycoprotein,
angiotensinogen,
apolipoprotein All, apolipoprotein AIV, apolipoprotein B 100, apolipoprotein
CI,
apolipoprotein CII, apolipoprotein CIII, apolipoprotein D, apolipoprotein E,
apolipoprotein-L1, apolipoprotein M, beta-2-glycoprotein I, carboxypeptidase
B2,
complement Clq subcomponent B chain, complement Clr subcomponent,
complement Cls, complement C2, complement C5, complement component C7,
complement component C8 alpha chain, complement component C8 beta chain,
complement component C8 gamma chain, complement factor B, complement factor
H-related protein 1, complement factor H-related protein 2, complement factor
I,
haptoglobin-related protein, insulin-like growth factor binding protein,
kininogen-1,
lumican, N-acetylmuramoyl-L-alanine amidase, plasma protease C1 inhibitor,
plasma retinol-binding protein, plasminogen, platelet basic protein,
serotransferrin,
serum amyloid A-4 protein, serum amyloid P-component, serum
paraoxonase/arylesterase 1, tetranectin, thyroxin binding globulin, Vitamin D-
binding protein, or zinc-alpha-2-glycoprotein.
[0024] In certain embodiments, immuno-depleting the samples includes a
multiple affinity removal system. In other embodiments, separating the
depleted
samples includes using a multi-lectin column. In yet other embodiments, the
LC/MS includes reversed phase chromatography.
[0025] In some embodiments, a level of the biomarker in the test serum sample
that is different relative to the control serum sample is indicative that the
test subject
has, or is at risk of developing, diabetes, obesity, and/or hypertension. In
some
embodiments, a level of the biomarker in the test serum sample that is
increased
relative to the control serum sample is indicative that the test subject has,
or is at risk
of developing, diabetes, obesity, and/or hypertension. In other embodiments, a
level
of the biomarker in the test serum sample that is decreased relative to the
control
serum sample is indicative that the test subject has, or is at risk of
developing,
diabetes, obesity, and/or hypertension.
[0026] In another aspect, the disclosure features a method of identifying a
biomarker for diabetes, obesity, and/or hypertension. The method includes the
steps
7
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
of obtaining a test plasma sample from a subject having or at risk of
developing
diabetes, obesity, and/or hypertension; obtaining a control plasma sample from
a
subject not having or not at risk of developing diabetes, obesity, and/or
hypertension; immuno-depleting albumin, IgG, IgA, transferrin, antitrypsin,
and
haptoglobin from the test sample and from the control sample, resulting in a
depleted test sample and a depleted control sample; separating the depleted
test
sample into a first fraction including test glycosylated polypeptides and a
second
fraction including test non-glycosylated polypeptides; separating the depleted
control sample into a third fraction including control glycosylated
polypeptides and
a fourth fraction including control non-glycosylated polypeptides;
independently
digesting the test non-glycosylated polypeptides in the second fraction and
the
control non-glycosylated polypeptides in the fourth fraction with trypsin,
resulting in
a test tryptic digest and a control tryptic digest; independently subjecting
the test
tryptic digest and the control tryptic to LC/MS, resulting in a test
polypeptide profile
and a control polypeptide profile; and comparing the test polypeptide profile
and the
control polypeptide profile; a polypeptide present at a level in the test
polypeptide
profile that is different than in the control polypeptide profile being
indicative of the
polypeptide as a biomarker for diabetes, obesity, and/or hypertension.
[0027] In some embodiments, a level of the biomarker in the test polypeptide
profile that is increased relative to the control polypeptide profile is
indicative of the
polypeptide as a biomarker for diabetes, obesity, and/or hypertension. In
other
embodiments, a level of the biomarker in the test polypeptide profile that is
decreased relative to the control polypeptide profile is indicative of the
polypeptide
as a biomarker for diabetes, obesity, and/or hypertension.
[0028] In some embodiments, immuno-depleting the samples includes a
multiple affinity removal system. In other embodiments, separating the
depleted
samples includes using a multi-lectin column. In yet other embodiments, the
LC/MS includes reversed phase chromatography.
[0029] In another aspect, the disclosure features a method of identifying a
biomarker for diabetes, obesity, and/or hypertension. The method includes the
steps
of obtaining a test plasma sample from a subject having or at risk of
developing
8
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
diabetes, obesity, and/or hypertension; obtaining a control plasma sample from
a
subject not having or not at risk of developing diabetes, obesity, and/or
hypertension; immuno-depleting albumin, IgG, IgA, transferrin, antitrypsin,
and
haptoglobin from the test sample and from the control sample, resulting in a
depleted test sample and a depleted control sample; separating the depleted
test
sample into a first fraction including test glycosylated polypeptides and a
second
fraction including test non-glycosylated polypeptides; separating the depleted
control sample into a third fraction including control glycosylated
polypeptides and
a fourth fraction including control non-glycosylated polypeptides;
independently
digesting the test glycosylated polypeptides in the first fraction and the
control
glycosylated polypeptides in the third fraction with trypsin, resulting in a
test tryptic
digest and a control tryptic digest; independently subjecting the test tryptic
digest
and the control tryptic to LC/MS, resulting in a test polypeptide profile and
a control
polypeptide profile; and comparing the test polypeptide profile and the
control
polypeptide profile; a polypeptide present at a level in the test polypeptide
profile
that is different than in the control polypeptide profile being indicative of
the
polypeptide as a biomarker for diabetes, obesity, and/or hypertension.
[0030] In some embodiments, a level of the biomarker in the test polypeptide
profile that is increased relative to the control polypeptide profile is
indicative of the
polypeptide as a biomarker for diabetes, obesity, and/or hypertension. In
other
embodiments, a level of the biomarker in the test polypeptide profile that is
decreased relative to the control polypeptide profile is indicative of the
polypeptide
as a biomarker for diabetes, obesity, and/or hypertension.
[0031] In some embodiments, immuno-depleting the samples includes a
multiple affinity removal system. In other embodiments, separating the
depleted
samples includes using a multi-lectin column. In yet other embodiments, the
LC/MS includes reversed phase chromatography.
[0032] In another aspect, the disclosure features a method of identifying a
biomarker for diabetes, obesity, and/or hypertension. The method includes the
steps
of obtaining a test serum sample from a subject having or at risk of
developing
diabetes, obesity, and/or hypertension; obtaining a control serum sample from
a
9
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
subject not having or not at risk of developing diabetes, obesity, and/or
hypertension; immuno-depleting albumin, IgG, IgA, transferrin, antitrypsin,
and
haptoglobin from the test sample and from the control sample, resulting in a
depleted test sample and a depleted control sample; separating the depleted
test
sample into a first fraction including test glycosylated polypeptides and a
second
fraction including test non-glycosylated polypeptides; separating the depleted
control sample into a third fraction including control glycosylated
polypeptides and
a fourth fraction including control non-glycosylated polypeptides;
independently
digesting the test non-glycosylated polypeptides in the second fraction and
the
control non-glycosylated polypeptides in the fourth fraction with trypsin,
resulting in
a test tryptic digest and a control tryptic digest; independently subjecting
the test
tryptic digest and the control tryptic to LC/MS, resulting in a test
polypeptide profile
and a control polypeptide profile; and comparing the test polypeptide profile
and the
control polypeptide profile; a polypeptide present at a level in the test
polypeptide
profile that is different than in the control polypeptide profile being
indicative of the
polypeptide as a biomarker for diabetes, obesity, and/or hypertension.
[0033] In some embodiments, a level of the biomarker in the test polypeptide
profile that is increased relative to the control polypeptide profile is
indicative of the
polypeptide as a biomarker for diabetes, obesity, and/or hypertension. In
other
embodiments, a level of the biomarker in the test polypeptide profile that is
decreased relative to the control polypeptide profile is indicative of the
polypeptide
as a biomarker for diabetes, obesity, and/or hypertension.
[0034] In some embodiments, immuno-depleting the samples includes a
multiple affinity removal system. In other embodiments, separating the
depleted
samples includes using a multi-lectin column. In yet other embodiments, the
LC/MS includes reversed phase chromatography.
[0035] In another aspect, the disclosure features a method of identifying a
biomarker for diabetes, obesity, and/or hypertension. The method includes the
steps
of obtaining a test serum sample from a subject having or at risk of
developing
diabetes, obesity, and/or hypertension; obtaining a control serum sample from
a
subject not having or not at risk of developing diabetes, obesity, and/or
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
hypertension; immuno-depleting albumin, IgG, IgA, transferrin, antitrypsin,
and
haptoglobin from the test sample and from the control sample, resulting in a
depleted test sample and a depleted control sample; separating the depleted
test
sample into a first fraction including test glycosylated polypeptides and a
second
fraction including test non-glycosylated polypeptides; separating the depleted
control sample into a third fraction including control glycosylated
polypeptides and
a fourth fraction including control non-glycosylated polypeptides;
independently
digesting the test glycosylated polypeptides in the first fraction and the
control
glycosylated polypeptides in the third fraction with trypsin, resulting in a
test tryptic
digest and a control tryptic digest; independently subjecting the test tryptic
digest
and the control tryptic to LC/MS, resulting in a test polypeptide profile and
a control
polypeptide profile; and comparing the test polypeptide profile and the
control
polypeptide profile; a polypeptide present at a level in the test polypeptide
profile
that is different than in the control polypeptide profile being indicative of
the
polypeptide as a biomarker for diabetes, obesity, and/or hypertension.
[0036] In some embodiments, a level of the biomarker in the test polypeptide
profile that is increased relative to the control polypeptide profile is
indicative of the
polypeptide as a biomarker for diabetes, obesity, and/or hypertension. In
other
embodiments, a level of the biomarker in the test polypeptide profile that is
decreased relative to the control polypeptide profile is indicative of the
polypeptide
as a biomarker for diabetes, obesity, and/or hypertension.
[0037] In some embodiments, immuno-depleting the samples includes a
multiple affinity removal system. In other embodiments, separating the
depleted
samples includes using a multi-lectin column. In yet other embodiments, the
LC/MS includes reversed phase chromatography.
[0038] In another aspect, the disclosure features a method of selecting a
subject
for gastric bypass surgery, the method including the steps of determining the
level of
a biomarker (i) in a test biological sample obtained from a candidate subject,
and (ii)
in a control biological sample of like tissue derivation from a control
subject not
having, or at risk of developing, diabetes, obesity, and/or hypertension;
comparing
the level of the biomarker in the test sample and in the control sample; and
selecting
11
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
the candidate subject as a subject for gastric bypass surgery if the level of
the
biomarker in the test sample is different relative to the control sample.
[0039] In some embodiments, the biomarker is one or more of actin alpha
skeletal protein, alpha-2-antiplasmin, alpha-2-HS-glycoprotein,
angiotensinogen,
apolipoprotein All, apolipoprotein AIV, apolipoprotein B 100, apolipoprotein
CI,
apolipoprotein CII, apolipoprotein CIII, apolipoprotein D, apolipoprotein E,
apolipoprotein-L1, apolipoprotein M, beta-2-glycoprotein I, carboxypeptidase
B2,
complement Clq subcomponent B chain, complement Clr subcomponent,
complement Cls, complement C2, complement C5, complement component C7,
complement component C8 alpha chain, complement component C8 beta chain,
complement component C8 gamma chain, complement factor B, complement factor
H-related protein 1, complement factor H-related protein 2, complement factor
I,
haptoglobin-related protein, insulin-like growth factor binding protein,
kininogen-1,
lumican, N-acetylmuramoyl-L-alanine amidase, plasma protease C1 inhibitor,
plasma retinol-binding protein, plasminogen, platelet basic protein,
serotransferrin,
serum amyloid A-4 protein, serum amyloid P-component, serum
paraoxonase/arylesterase 1, tetranectin, thyroxin binding globulin, Vitamin D-
binding protein, or zinc-alpha-2-glycoprotein.
[0040] In certain embodiments, the test biological sample is a test plasma
sample and the control biological sample is a control plasma sample. In other
embodiments, the test biological sample is a test serum sample and the control
biological sample is a control serum sample.
[0041] In some embodiments, determining the level of the biomarker includes
measuring the protein level and/or the RNA level of the biomarker. In some
embodiments, the RNA is mRNA.
[0042] In other embodiments, determining the level of the biomarker in the
test
sample and in the control sample includes removing from the samples proteins
present at high levels. In one embodiment, the method includes the step of
removing high abundance proteins from the test biological sample, resulting in
a
depleted test biological sample, and removing high abundance proteins from the
control biological sample, resulting in a depleted control biological sample.
In some
embodiments, the high abundance proteins are removed using immuno-depletion.
In
12
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
some embodiments, the high abundance proteins albumin, IgG, IgA, transferrin,
antitrypsin, and/or haptoglobin are removed. In some embodiments, immuno-
depleting the samples includes using a multiple affinity removal system.
[0043] In some embodiments, the method further includes separating
components of the depleted biological samples. In particular embodiments, the
method includes separating the depleted test biological sample into at least a
first
fraction including glycosylated polypeptides and at least a second fraction
including
non-glycosylated polypeptides, and separating the depleted control biological
sample into at least a third fraction including glycosylated polypeptides and
at least a
fourth fraction including non-glycosylated polypeptides. In some embodiments,
the
depleted biological samples are separated using chromatography. In one
embodiment, a multi-lectin column is used.
[0044] In yet other embodiments, the method further includes subjecting the
fractions to digestion. In one embodiment, the glycosylated polypeptides in
the
fractions are digested. In another embodiment, the non-glycosylated
polypeptides in
the fractions are digested. In particular embodiments, the method includes
digesting
with trypsin the non-glycosylated polypeptides in the second fraction,
resulting in a
test tryptic digest, and digesting with trypsin the non-glycosylated
polypeptides in
the fourth fraction, resulting in a control tryptic digest. In another
embodiment, the
method includes digesting with trypsin the glycosylated polypeptides in the
first
fraction, resulting in a test tryptic digest, and digesting with trypsin the
glycosylated
polypeptides in the third fraction, resulting in a control tryptic digest.
[0045] In other embodiments, the method further includes separating the
components of the tryptic digests. In one embodiment, the components of the
tryptic
digests are separated using chromatography. In some embodiments, the
components
of the tryptic digests are separated using two dimensional chromatography or
liquid
chromatography-mass spectrometry (LC/MS). In one embodiment, the method
includes subjecting to LC/MS the test tryptic digest, resulting in a test
polypeptide
profile, and subjecting to LC/MS the control tryptic digest, resulting in a
control
polypeptide profile. In some embodiments, the LC/MS includes reversed phase
chromatography.
13
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
[0046] The following figures are presented for the purpose of illustration
only,
and are not intended to be limiting.
BRIEF DESCRIPTION OF THE DRAWINGS
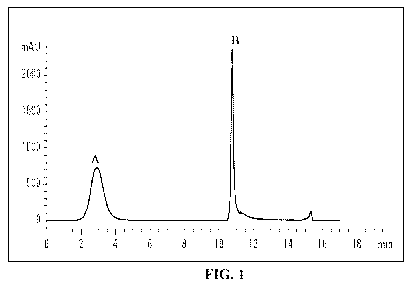
[0047] FIG. 1 is a graphic representation of a chromatogram obtained during
the
depletion of a plasma sample. Peak A corresponds to flow-through fraction (low
&
medium abundance proteins) and peak B corresponds to bound fraction (high
abundance proteins).
[0048] FIG. 2 is a graphic representation of a chromatogram obtained during
the
desalting and fractionation of a trypsin digest on a C18 column. Peak A
corresponds
to salts that were removed by the column, and peak B corresponds to peptides
that
were also separated from the undigested/partially digested proteins (peak C).
[0049] FIG. 3 is a graphic representation of the peak areas obtained at 30%
organic mobile phase elution versus the amount of protein digested with
trypsin.
[0050] FIG. 4 is a graphic representation of the relative protein
concentrations of
angiotensinogen in plasma samples from various patient groups.
[0051] FIG. 5 is a graphic representation of the relative protein
concentrations of
apolipoprotein C1 in plasma samples from various patient groups.
DETAILED DESCRIPTION OF THE INVENTION
[0052] Unless otherwise defined, all technical and scientific terms used
herein
have the same meaning as commonly understood by one of ordinary skill in the
art
to which this invention belongs. Although methods and materials similar or
equivalent to those described herein can be used in the practice or testing of
the
present invention, suitable methods and materials are described below. All
publications, patent applications, patents, and other references mentioned
herein,
including GenBank database sequences, are incorporated by reference in their
entirety. In case of conflict, the present specification, including
definitions, will
control. In addition, the materials, methods, and examples are illustrative
only and
not intended to be limiting.
14
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
[0053] Other features and advantages of the invention will be apparent from
the
following detailed description, and from the claims.
Definitions
[0054] As used herein, the term "biological sample" refers to a sample
obtained
from an organism or from components (e.g., cells) of an organism. The sample
may
be of any biological tissue or fluid, for example, a sample derived from a
patient.
Such samples include, but are not limited to, blood, blood cells (e.g., white
cells),
plasma, tissue or fine needle biopsy samples, urine, peritoneal fluid, and
pleural
fluid, or cells there from. Biological samples may also include sections of
tissues
such as frozen sections taken for histological purposes.
[0055] As used herein, the term "biomarker" of a disease or condition refers
to a
gene or a gene product that is up- or down-regulated in a biological sample of
a
subject having the disease or condition relative to a biological sample from
like
tissue derivation, which gene or gene product is sufficiently specific to the
disease or
condition that it can be used, optionally with other genes or gene products,
to
identify or detect the disease or condition. Generally, a biomarker is a gene
or a
gene product that is characteristic of the disease or condition.
[0056] The term "protein" is used interchangeably herein with the terms
"peptide" and "polypeptide".
[0057] As used herein, a "subject" is a mammal, e.g., a human, mouse, rat,
guinea pig, dog, cat, horse, cow, pig, or non-human primate, such as a monkey,
chimpanzee, baboon or rhesus.
[0058] The present disclosure relates, at least in part, to methods of
identifying
individuals having, or at risk of developing, diabetes, obesity, and/or
hypertension.
The present disclosure is based, at least in part, on the identification of
proteins that
are differentially expressed in diabetic, obese and/or hypertensive subjects
relative to
normal subjects.
Measuring Biomarkers
[0059] The methods described herein include the detection of the level of
markers, e.g., to identify subjects having, or at risk of developing,
diabetes, obesity,
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
and/or hypertension. Such markers can be detected at the RNA or protein level,
e.g.,
in samples described herein, using methods well known to those of skill in the
art.
Such assay methods include, but are not limited to, immunoassays, radio-
immunoassays, competitive-binding assays, Western Blot analysis, ELISA assays,
and immunofluorescence assays. Other assay methods include spectroscopy, such
as mass spectroscopy. These markers are detected after separation from a
biological
sample.
Separation Techniques
[0060] To evaluate the presence of biomarkers in biological samples, e.g.,
plasma, the proteins in the biological samples can be separated to facilitate
analysis
using various separation techniques. Separation techniques include, but are
not
limited to, column chromatography, filtration, ultrafiltration, salt
precipitation,
solvent precipitation, solvent extraction, distillation, immunoprecipitation,
SDS-
polyacrylamide gel electrophoresis, isoelectric point electrophoresis,
dialysis, and
recrystallization.
[0061] For chromatography, affinity chromatography, ion-exchange
chromatography, hydrophobic chromatography, gel filtration, reverse phase
chromatography, adsorption chromatography, and such may be used (see, e.g.,
Strategies for Protein Purification and Characterization: A Laboratory Course
Manual. Ed, Daniel R. Marshak et al., Cold Spring Harbor Laboratory Press
(1996)).
These chromatography procedures can also be liquid chromatography, such as
HPLC and FPLC.
[0062] In some instances, the presence of biomarkers in a biological sample
can
be measured by optionally modifying or partially degrading the proteins in a
biological sample, for example, by treating the biological sample with an
appropriate
protein modification enzyme before separation. Such a modification or partial
degradation can be utilized when, for example, the proteins in a biological
sample
are not easily separated. Such protein modification enzymes include, for
example,
trypsin, chymotrypsin, lysylendopeptidase, protein kinase, and glucosidase.
[0063] In certain instances, multidimensional separation techniques, such as
tryptic peptide fractionation using reversed phase and ion exchange LC, or
protein
16
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
pre-fractionation methods, like ion exchange, size exclusion, hydrophobic
interaction and various affinity methods, can be used (Martosella, et al., J.
Proteome
Res. (2005) 4:1522-1537). One nonlimiting example of a pre-fractionation
method
includes removing high abundance proteins to reduce the dynamic range of
protein
levels in biological fluids to better match that of the analytical platform.
[0064] A variety of depletion methods for specific removal of high abundance
proteins from bodily fluids can be used (see, e.g., Govorukhina, et al., J.
Chromatogr. A (2003) 1009:171-178). A nonlimiting example is the multiple
affinity removal system (MARS, Agilent, Palo Alto, CA), which utilizes an
affinity
column. This column can deplete albumin, IgG, IgA, transferrin, haptoglobin
and
antitrypsin in human plasma (Ogata, et al., J. Proteome Res. (2005) 4:837-845;
Bjorhall, et al., Proteomics (2005) 5:307-317). The MARS column can deplete
these proteins from 30-40 l of plasma at a time and can be regenerated up to
200
times.
[0065] Another separation technique that can be used in the method disclosed
herein involves using a combination of three lectins in the form of a multi
lectin
column (M-LAC). This affinity column can capture and enrich fractions, e.g.,
glycoprotein fractions, in plasma. In some instances, fractions can be
subjected to
LC-MS after tryptic digestion (Yang, et al., J. Chromategr. A (2004) 1053:79-
88).
Methods of Diagnosis
[0066] The methods described herein can be used to identify a subject having,
or
at risk of developing, diabetes, obesity, and/or hypertension. The methods
described
herein include obtaining a biological sample from a subject having, or at risk
of
developing, diabetes, obesity, and/or hypertension, and a sample from a
subject not
having, or not at risk of developing, these conditions. The biological sample
can be,
e.g., urine, blood, serum, plasma, saliva, semen, a vaginal secretion, or
cerebrospinal
fluid. In some instances, the biological sample is a plasma sample.
[0067] Any marker described herein, or identified using a method described
herein, can be used as a marker to identify a subject having, or at risk of
developing,
diabetes, obesity, and/or hypertension. For example, the level of one or more
of the
following markers can be measured: actin alpha skeletal protein, alpha-2-
17
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
antiplasmin, alpha-2-HS-glycoprotein, angiotensinogen, apolipoprotein All,
apolipoprotein AIV, apolipoprotein B 100, apolipoprotein CI, apolipoprotein
CII,
apolipoprotein CIII, apolipoprotein D, apolipoprotein E, apolipoprotein-L1,
apolipoprotein M, beta-2-glycoprotein I, carboxypeptidase B2, complement Clq
subcomponent B chain, complement Clr subcomponent, complement Cls,
complement C2, complement C5, complement component C7, complement
component C8 alpha chain, complement component C8 beta chain, complement
component C8 gamma chain, complement factor B, complement factor H-related
protein 1, complement factor H-related protein 2, complement factor I,
haptoglobin-
related protein, insulin-like growth factor binding protein, kininogen-1,
lumican, N-
acetylmuramoyl-L-alanine amidase, plasma protease C1 inhibitor, plasma retinol-
binding protein, plasminogen, platelet basic protein, serotransferrin, serum
amyloid
A-4 protein, serum amyloid P-component, serum paraoxonase/arylesterase 1,
tetranectin, thyroxin binding globulin, Vitamin D-binding protein, or zinc-
alpha-2-
glycoprotein. If the protein or RNA level of one or more of these markers is
different relative to the control level, the subject can be classified as
having, or at
risk of developing, diabetes, obesity, and/or hypertension. The Swiss Protein
Accession Numbers for the markers described herein are listed in Tables 2-7.
[0068] Angiotensinogen is associated with essential hypertension. Essential
hypertension is a complex disease influenced by different genetic and
environmental
factors. Initial studies done on hypertensive siblings and case-control
studies
indicated the important role of the angiotensinogen gene for the
predisposition to
essential hypertension, preeclampsia and obesity-related hypertension (Brand,
et al.,
Herz. (2000), 25:15-25). Adipose tissue is an important source of
angiotensinogen.
A local renin-angiotensinogen system (RAS) is present in human adipose tissue
and
may act as a distinct system from plasma RAS. In obese patients, increased
secretion of angiotensinogen from adipose tissue with a resulting increase in
plasma
levels of angiotensin II may be a step in the development of hypertension
(Ailhaud,
et al., Int. J. Obes. Relat. Metab. Disord. (2000), 24:S33-35).
[0069] Apolipoprotein C1 overexpression is associated with decreased
particulate uptake of Apo B-containing lipoproteins, which may lead to
increased
levels of several potentially atherogenic species, including cholesterol-
enriched
18
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
VLDL, IDL, and LDL (Jong, et al., J. Clin. Invest. (1996) 98:2259-2267). This
in
turn may lead to high blood pressure. The effects of Apo C1 overexpression on
hepatic and peripheral insulin sensitivity have been studied in a mouse model,
where
obese ob/ob mice with mild over expression of Apo C1 were generated and
resulted
in hepatic steatosis and severe hepatic insulin resistance (Muurling, et al.,
J. Lip.
Res. (2004) 45:9-16).
[0070] Once a subject is identified as having, or at risk of developing,
diabetes,
obesity, and/or hypertension, the subject can be treated with an appropriate
therapy
for the condition. Such therapies include traditional therapies known in the
art, e.g.,
insulin therapy or gastric bypass surgery.
[0071] The invention is further illustrated by the following examples. The
examples are provided for illustrative purposes only. They are not to be
construed
as limiting the scope or content of the invention in any way.
EXAMPLES
EXAMPLE 1
Identification of Proteins Differentially Expressed in Diabetic, Obese, and/or
Hypertensive Subjects Relative to Normal Subjects
Protocol
A. Samples and Reagents
[0072] Human plasma samples were obtained from Bioreclamation Inc.,
(Hicksville, New York) and Trypsin (sequencing grade) was purchased from
Promega (Madison, WI). Agarose bound Concanavalin A, Jacalin and Wheat germ
agglutinin lectins were obtained from Vector labs (Burlingame, CA). Disposable
polypropylene columns were obtained from Pierce biotechnology, Inc.,
(Rockford,
IL) and 5 kDa Amicon molecular weight cut off filters were purchased from
Millipore (Billerica, MA).
[0073] Sodium phosphate, sodium chloride, ultra pure
(hydroxymethyl)aminomethane hydrochloride, sodium azide, urea, glycine,
guanidine hydrochloride, dithiothreitol, ammonium bicarbonate, iodoacetamide,
19
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
manganese chloride tetrahydrate, calcium chloride, methyl-a-mannopyrannoside,
N-
acetyl-glucosamine, methyl-a-glucopyrannoside, Bovine fetuin, and galactose
were
purchased from Sigma-Aldrich (St. Louis, MO). Concentrated hydrochloric acid,
trifluoroacetic acid, glacial acetic acid and HPLC grade acetonitrile were
purchased
from Fisher scientific (Fair lawn, NJ), and formic acid (acid free) was
purchased
from MP Biochemicals, Inc., (Solon, OH). HPLC grade water used in all
experiments was from J.T Baker (Bedford, MA). Discovery BIO wide pore C18
cartridges were from (3 m particle diameter, 4.6 mm X 30 mm) Supelco
(Bellefonte, PA).
B. Sample Preparation for Proteomic Analysis
[0074] Human plasma samples (received frozen in dry ice from Bioreclamation,
Inc.) were selected from females; plasma was obtained from four groups of
individuals: (1) Normal (healthy); (2) Obese; (3) Obese, Diabetic and non-
Hypertensive; and (4) Obese, Diabetic and Hypertensive females. Table 1
summarizes the characteristics of the patients selected for this study and
supplied by
Bioreclamation Inc. Samples were randomized for proteomic analysis to protect
data from any variables or biases. Two samples were processed from each of
these
pools and were also analyzed in duplicate at the MS level to give four
measurements
per pool. Each group consisted of five individuals (5 ml of plasma each) and a
plasma sample pool for each of the four groups was made by combining 3 ml from
each of the five individuals within a group. The remaining 2 ml of sample from
each individual was stored separately at -70 C for the analysis of
individuals at a
later stage.
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
Table 1. Patient a Characteristics
Characteristics Normal Obese Obese+Diabetic Obese+diabetic
+Hypertensive
1. Gender Female Female Female Female
2. Age in years 38 8.51 36 10.99 44 10.26 43 14.78
ran e
3. Body Mass < 30kg/m > 30kg/m > 30kg/m >30kg/m
Index
4. Treatments for
1. Diabetes 0% 0% 100% 60%
II. Hypertension. 0% 0% 0% 60%
a Five individual plasma samples in each group were pooled for the study
C. Depletion of Six Most Abundant Proteins in Human Plasma Using the
Multiple Affinity Removal Column (MARS)
[0075] Plasma was depleted of six abundant proteins using a 4.6 mm x 100 mm
MARS column (Agilent, Palo Alto, CA). The depletion was performed at room
temperature (25 C) on an IntegralTM Analytical Workstation (Perseptive
Biosystems,
Inc. Cambridge, MA) by using manual injection mode. To remove any
particulates,
a micro filter was attached before the column. Protein elution was monitored
at a
wavelength of 280 nm during the chromatography fractionation process. The
loading capacity of the MARS column was about 30-40 l of human plasma. The
amount of plasma processed per sample was 70 l; two separate samples, each
consisting of 35 l, were injected onto the depletion column. These human
plasma
samples (35 l) were diluted to 250 l using binding buffer (supplied by the
manufacture). An internal standard was added to each sample; 2.5 l from 10
mg/ml solution of bovine fetuin was added to each sample. The final
concentration
of bovine fetuin was 10 pmol/35 l of plasma.
[0076] The column was initially equilibrated with the binding buffer (12 ml),
and the sample was loaded at a low flow rate (0.5 ml/min) for 2.5 min. The
flow
rate was then set at 1.0 ml/min for the remainder of the run. The unbound
proteins
were removed by washing the column with an additional 10 ml of binding buffer.
Then the bound six high abundance proteins were released with elution buffer
supplied by the manufacturer. The eluted proteins were collected and the
column
21
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
was immediately neutralized by adding binding buffer (10 ml). Each run cycle
took
32 min of total run time. Both depleted (flow-through fraction) and plasma
abundant proteins (bound fraction) were collected and stored at -20 C until
further
analysis.
[0077] The volume of the depleted fractions prepared from each sample was
concentrated to 200 l using Amicon 5 kDa molecular weight filters (Millipore,
MA) and buffer exchanged three times (3 ml each) with a buffer containing 25
mM
Tris, 0.1 M NaC1, 0.05% NaN3 at pH 7.4. These samples, together with the bound
fractions from the depletion step, were subjected to a Bradford protein assay
using
bovine serum albumin (BSA) as the standard, according to the manufacturer's
instructions (Pierce, Rockford, IL).
D. Affinity Capture and Enrichment of Glycoproteins From Depleted
Human
Plasma Using Multi Lectin Affinity Chromatography (M-LAC)
[0078] Disposable polypropylene columns with 5 ml capacity (Pierce
Biotechnology Inc, Rockford, IL) were used in this procedure. Each sample was
loaded onto a new column. The M-LAC column was prepared from a stock solution
of lectins. Equal amounts of a 50% slurry of agarose-bound Concanavalin A,
Jacalin and Wheat germ agglutinin lectins were mixed together to obtain a
total of 1
ml M-LAC column, ready for affinity capture of glycosylated plasma
glycoproteins.
[0079] To the depleted human plasma samples (300 l) were added 3 l of a
solution of 10 mM Ca2+ and 10 mM Mn2+ions, just before multi-lectin affinity
chromatography. The M-LAC column was equilibrated with 12 ml of binding
buffer (25 mM Tris, 0.1 M sodium chloride, 1 mM calcium chloride, 1 mM
manganese chloride, 0.05% sodium azide at pH 7.4). The plasma sample was
loaded onto the column slowly and incubated at room temperature for 15 min.
After
the incubation period, unbound non-glycosylated proteins were washed with 4 ml
of
binding buffer and collected directly in to an Amicon 5 kDa molecular weight
filter
(Millipore, MA) containing 125 l of 5 mM EDTA. Bound glycoproteins were then
eluted with buffer (0.5 M sodium chloride, 25 mM Tris, 0.2 M methyl-a-
mannopyrannoside, 0.2 M methyl- a-glucopyrannoside, 0.8 M galactose, 0.5 M N-
acetyl-glucosamine, 0.05 % sodium azide at pH 7.4) into another Amicon filter
22
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
containing EDTA. Both fractions were concentrated to 100 l. The filters were
washed with another 100 l solution of 10 mM Tris, pH 7.4, and aliquots of
combined bound and unbound fractions were subjected to a Bradford protein
assay
to determine the total amount of glycoproteins and non-glycoproteins present
in the
samples.
E. Tryptic Digestion of Non-Glycosylated Proteins
[0080] The non-glycosylated fractions obtained from the M-LAC were
concentrated to 50 l using 5 kDa Amicon molecular weight cut off filters, and
6 M
guanidine chloride (150 l) was then added to denature the proteins. This
sample
was reconcentrated to 50 l and reduced using a solution of 5 mM
dithiothreitol
(DTT) followed by incubation at 60 C for 40 min. The reduced samples were
brought to room temperature and alkylated by adding 15 mM iodoacetamide (1.5
l
of 0.5 M iodoacetamide) and incubated in the dark for 30 min. At the end of
the
incubation period, 5 l of 0.1 M DTT was added to stop the alkylation. The
samples
were diluted with 200 l of 50 mM ammonium bicarbonate solution, pH 8, and
trypsin (Promega, Madison, WI) was added at a ratio of 1:25 (w/w). The samples
were incubated at 37 C for 18 hr. Next, additional trypsin (1:25 w/w) was
added,
and the samples were held at 37 C for 2 to 4 hr to maximize protein
digestion.
Finally, the digestion was terminated by adding 1% formic acid gradually,
until the
pH was below 4.
F. Reversed Phase C18 Sample Clean-Up Before Nano LC MS/MS
Analysis
[0081] Before analyzing the digested protein samples using LC/MS, the salts
present in the samples (such as guanidium hydrochloride and ammonium
bicarbonate) were removed on a C18 cartridge (Bio C18, 3 m, 4.6 mm X 30 mm).
Desalting was performed on a Shimadzu SPD 10A high performance liquid
chromatography system (Norwell, MA).
[0082] The injection volume was 300 l and a full loop injection mode was
used. Both 280 and 214 nm were monitored during the desalting process. The
total
chromatographic procedure took 16 min.
23
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
[0083] The chromatographic column was equilibrated with a high aqueous
mobile phase composition (98% of mobile phase A). After loading the sample,
the
salts were removed with an isocratic wash. Then the organic mobile phase was
increased to 30% using 0.07% trifluoroacetic acid in acetonitrile, mobile
phase B,
where the peptides were eluted from the column. The peptides were collected
into
eppendorf tubes and the organic mobile phase was increased to 90% B to elute
the
partially digested or undigested proteins. The peptides eluted at 30% organic
mobile
phase were collected (2 ml) and concentrated to 20 l using a LABCONCO Freeze
dryer (Kansas City, MO) at -90 C and stored at -70 C until LC/MS analysis.
G. Nano LC-MS/MS Analysis
[0084] The LC/MS and LC/MS/MS experiments were performed on an Ettan
MDLC system (GE Healthcare, Piscataway, NJ) coupled to a Finnigan linear ion
trap mass spectrometer (Thermoelectron, San Jose, CA). The capillary column
used
for LC/MS/MS analysis (150 mm X 0.075 mm) was from New Objective (Woburn,
MA) and slurry packed in house with 5 m, 200 A pore size Magic C18 stationary
phase (Michrom Bioresources, Auburn, CA). The flow rate for sample separation
was 200 nl/min. Mobile phase A was 0.1 % formic acid in water and mobile phase
B was 0.1 % formic acid in acetonitrile. Routinely, 2.5 l of each sample
corresponding to approximately 4 g of total proteins was injected onto the
column
using the MDLC autosampler. The following gradient was used for all analyses:
0% B (minimum equilibration for 30 min); followed by linear gradient to 40% B
over 160 mins; then to 90% B over 20 mins; and then constant 90% B for 20
mins.
All separations were performed at ambient temperature. The ion transfer tube
of the
linear ion trap was held at 245 C; the normalized collision energy was 35%
for
MS/MS. The spray voltage was set at 2.0 W. The mass spectrometer was operated
in the data dependant mode to switch automatically between MS and MS/MS
acquisitions using MS acquisition software (Xcalibur 1.4, Thermo Electron, San
Jose, CA). Each MS full scan (mass range of m/z 400 to m/z 2000) was followed
by
seven MS/MS scans of the seven most intense peaks. A precursor ion was
excluded
from further LTQ MS/MS analysis for 2 min.
24
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
H. Data Processing and Analysis
[0085] Peptides and proteins were identified by automated searching of all MS
and MS/MS spectra against spectra of theoretical fragmentations of a human
protein
database (Swiss-Prot, uploaded in Sep 2005) using Sequest algorithm
incorporated
into the BioWorks software, Version 3.2, (Thermoelectron, San Jose, CA). Only
the
peptides resulting from tryptic cleavages were searched and two trypsin missed
cleavages were allowed. Carbamidomethylation of cysteines was included in the
search parameters. The Sequest results were filtered using correlation score
(Xcorr)
values (Xcorr 1.9, 2.2, 3.75 for singly, doubly and triply charged peptides,
respectively) and validated using Protein ProphetTM software by applying a
minimum 95% probability, and protein identifications were based on at least
two
peptides. The identified protein list was sorted again to exclude any proteins
that
were identified with fewer than two unique peptides. The proteins that had a
difference of two fold or more (by spectral counts) (Liu, et al., Anal. Chem.
(2004),
76:4193-4201), between the normal and the disease pools were identified.
However,
in some cases, a single peptide was detected for a protein within a given
group, and
are referred to as "singletons". Proteins identified by a single peptide were
accepted
only where other groups showed a significant difference in spectral counts.
The
proteins that came under the above category were selected for the peak area
quantitation. Peak area quantitation was done manually using at least two
peptides
per protein. These peak areas were normalized using a peak area of a selected
peptide obtained for the internal standard, bovine fetuin, which was carried
through
the entire analysis procedure. Normalization was performed to correct any
sample
losses and process variations (e.g., variations in injection volumes) that
might have
happened during the analysis of the sample set.
Results
[0086] The MARS column removed six abundant proteins in human plasma
(albumin, immunoglobulin G, immunoglobulin A, transferrin, antitrypsin and
haptoglobin) efficiently (Figure 1). This was assessed by the number of
peptides
identified during the LC-MS/MS analysis of the depleted sample. After
depletion of
the high abundance proteins, the protein yield in the flow through fraction
was 9.7 %
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
of the total proteins. 70 l of plasma containing 3.5-4.0 mg of total protein
were
initially used, and at the end of the depletion step, the total protein
content in the
flow-through fraction was 0.35-0.4 mg.
[0087] The M-LAC column consisted of Concanavalin A (Con A), Jacalin
(JAC) and Wheat Germ Agglutinin (WGA) lectins separately cross-linked to
agarose beads. This combination of lectins recognizes the most common sugar
residues found in human plasma: Con A, JAC and WGA recognize a-mannose,
galactose, N-acetyl glucosamine and sialic acid, respectively (Sharon et al.,
Glycobiology (2004) 14:53R-62R; Yang et al., J. Biol. Chem. (1993) 268:5905-
5910). The M-LAC column was used as a fractionation method in this approach to
simplify the sample mixture into two fractions, namely glycosylated proteins
and
non-glycosylated proteins. The column was run under gravity conditions, and a
fresh column was used for each sample. The divalent ions, Ca2+ and Mn2+, were
added to the samples before the fractionation step to facilitate binding of
plasma
glycoproteins to Con A.
[0088] From the amount of total proteins loaded on to the M-LAC column,
about 56% of the proteins were in the flow-through fraction. Some of the
proteins in
this fraction have been identified as potential glycoproteins. The tryptic
digests of
non-glycosylated fractions were then analyzed by LC/MS.
[0089] The non-glycosylated protein fraction was subjected to tryptic
digestion
and then "cleaned" before LC/MS analysis using a C18 column and a gradient
system of organic and aqueous mobile phase as described above. This procedure
separated the peptides from any undigested or partially digested proteins and
removed salt. Both 280 nm and 214 nm were monitored during the desalting
process (Figure 2).
[0090] This desalting method was further explored to better control the amount
of sample loaded onto the LC-MS column. Figure 3 shows the correlation between
the peak area measured at 214 nm (at 30% organic mobile phase) against the
total
protein content loaded onto the desalting column (Bradford assay, before
digestion).
A linear correlation between the protein amount and the peak area was found.
Therefore, peak area was used to normalize the amount of protein that was
loaded
26
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
onto the LC/MS column to control for sample losses and for variable trypsin
digestions.
[0091] While the spectral count measurement were used as an initial measure of
the amount of a given peptide in a sample, peak areas were used for further
study of
differences in protein amounts between different clinical samples. The peak
area
was measured in an extracted ion chromatogram at a predetermined precursor ion
mass and retention time window. The retention time window that was assigned
was
0.5 min. Smoothing was also applied before integrating the peak areas to
facilitate
manual integration. The quantitative information obtained from one peptide was
compared with data obtained from a second peptide from the same protein to
ensure
the accuracy of the quantitation. The peptides that were quantitated were
unique for
that particular protein, and at least two peptides were quantitated per
protein in all
the samples. In addition, a known amount of an internal standard (bovine
fetuin)
was spiked into the samples before the depletion step, and peak area of a
selected
fetuin peptide was used to normalize the sample amounts. After the peak areas
of
peptides of a given protein were normalized, a threshold for differential
protein
abundance was set at greater than two-fold change between the normal and the
disease samples.
[0092] The peptide spectra obtained from the LTQ mass spectrometer were
searched against tryptic peptide sequences of human protein database using the
Sequest algorithm incorporated in to the BioWorks software, Version 3.2,
(Thermoelectron, San Jose, CA). Stringent filtering criteria was used to
minimize
false peptide assignments, such as a minimum of 95% probability, a mass
accuracy
of 1.5 Da and Xcorr values of 1.9, 2.2 and 3.75 for +1, +2 and +3 charged
ions,
respectively. A total of 257 proteins were identified with high confidence.
From the
aggregated list of proteins identified in the four pools, 156 proteins were
found that
were identified with at least two unique peptides.
[0093] Tables 2 to 7 summarize the list of proteins identified as
differentially
expressed between the normal and disease groups.
27
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
0
O O 7 W O W 4q O O V (h 4q
N O. (h N N O (h CO N N (h N N N~ (h V N
0 O
O R 3
R d 3
R
O
C N
O O~ O V V N (h
O W
N
N O ~
Q O N
N d O O O N N CO O~ O I~ O N O(h
I
a a O m ('~) O O N~ (+) m O O('~) V V ('~)
d R E
E
d O
O.
y ._
fl- a a O N~ O O N O N O~ (h CO O O O(h
O = E
O O R
z C
y:6 N (h CO W (h W (h W V (h CO I~ CO CO
R CL
W
N O. ~
Q O N
N a O ~3- CO N CO O (h 12 CO V CO W W
E
N R
00
CV
V N
N O d ~ N ~ (h Q) N I~ N~ V Q) V ~~~
a 1
E
y y R
y y y
O
fl
fl- a V W O m ~ co M(~'~) O I~ I~ ~ I~
O E
O O R
Z (/)
L
U
N
~
U
N
N
C L~ a
LD E
cu 0 O O
d ~ fl C
N O
~ N O C 0 i
E O
O O Q C Q(6 V
N C N O N ~ Q
N U ~ U~~ N O
fn E
0 O Q U U O O ~ C Q N N `O ~,
N U N Q
O Q V Q 0 Q Q~ ~ O CO S -~
~ ~N 76 > O _= N Q O' O O O L U N O
Q m U U U `~ o - >
O m
~ N N N N N N C C C C C C 0 N
(B N N N N N N N O
Q Q Q Q Q Q E E E E E E
0) o 0 0 o o 0 0 0 0 0 0- m -
N m a a a a a a a a a a a~ E
E o o o o o E E E E E E~ N ~ b
D R U Q Q Q Q Q O O O O O O N N O
Z Q Q Q Q Q Q U U U U U U~ d Cn N
N
C
~
O _ O
a y Z
O c
nj a O I- LO (O (O M O(O 00 LO M
0 M N LO LO LO M M V ~f) 00 LO LO LO (O
N N I- (O (O (O I- O_ (O 0) 00 M
N d 00 (O V CV N N O O O(O ~f) LO ~f) I~ LO
(0 ~3 v(O O O O O O O O O M O M O N N
H (/) d d d d d d d d d d 0- 0- 0- 0- 0- d
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
N
O O V M O LO O M I~ O W O O
N O. V N O) V N CO N V LO N N N
0 d O
O R 3
0 O V ~O ~O N(h R >
N O. O O O O O O R Q
O
o 2 3
> C N
R ; O Lo
im fl
R W
o ~ N O. &
O N
y'o
; N ~O CO I~ N
t~6 d W
.
M
O N O O O N O O CO O O N O O R
f`=) N (/~
N d O CO N CO I~ (h
E U
y a a O c') O O N LO c") c") V V O
C N R ~6
N E
N a a V(h O V CO (h :6 Z VI
~ R E N E fl- d O Lo O N N O N O O O O O
Z O E
O O R
N =- Z
fl- =6 d (h I~ N Q) N O
w C 1
O E
O R p
z a'6 N V M O) LO N V LO LO CO M N
N
'm .-
R fl- W
N N O
o
C N > y
y;6 N O N (h N Q O N
R ~- W
N
N O ~
> y M O
Q O N N a M LO W V M W N CO I~ W LO O
E
~ N VI
N a O ~O O O (h O lm
C _
O l~6 C. O N N
> ~ N p. d LO O I- M O V V c") c") V O N
lm U E
.~
d y R
N o VI
~ 0
a a O O O CO O O R_
-
E y
6 a
dm (/~ O= r- a V L~O M O) N O ~ LO LO LO c') V
p O
z o
O N N(h N N
fl- ~ a N
O E U
o z w y N
m
E
m
x
n fn U L
C N C
A) E
U
U a
a) U O O y
C ~ m
y C LL m
N N Q ~ U 0 Q Q (Np V o E U 0 N C O O ~ Q U O N
O O Q O Q t6 ~ U i O E O
LL C O' N L f i~ C 0 0 ~ U O i N C 0 L T.C
(Oi~ =N O m V O R O
E fn N N O t6 d tp Q ~ N N Q N O
O c m :o U fl- a~ o a~ m U U~ o~~
U U o o co o
d N N N N a)
O N O E O~ O aL.. L O O O N N N N N
p~ E L 0 ~ E O ~ O O O N N N N Y tp X
R - a o ~~ a~ m a a a a a a a- E E O
3 z ¾¾ U U a a E ~ ~o o o E E E E~ 2
O R U Q Q Q O O O O N R N L
O ~ z Q Q Q Q U U U U a o
0
4 C O
o d z
a Z C O
a a o o z
w uNi rn v} ~ v ~ a ~ O M V_ V(O (O M 00 Lo 6)
N a) co v m ~ ~ N N M ~(p (p (O O (O 00 ~(~
m Q a O d a a d d M V N (O ~~ I-
=3 vy (O O O O O O M O N
H N¾ d d d d d d d d d d d d
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
N
0 M V O
O I[J I[J M I[J M (V
N O. O O O O O O O O O O O
d O
O 2
R ¾
d 3
O
N.a
f6 fl- W
d
P
O N
N ~~[J O N V N I~ M O V r
+~
6
I
.V
a a ~ ~ ~ co CO co m v~~
d R E
N y
d O
fl. C
y ._
~- a N M N V c0 O m ~[J O
O E
O N
Z (n
c a~ ~
y N C P
R
c
. [J O r O m O O ~ E
(fl
d R
O. y fl- 0) O N O ~[J M O N
(n
N
~ N E ~ N (C O U
_ ~ N N ~ ~ N U ~ 1 0 /)
E O E ~ p
OMO R a a cq cq >
0
~
c o
a a
N N O ~ [J I~ I~
N N ~ I~ ~[J O d I~ I~ I~ ~[J I~ I~
U N N m ~ N O N ~[J N N
m 3 co O O O o o~ O O O m O O
~ m¾ a a C3 a C3 a a a a a a
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
0
O 7 7 7 O c0 M O O I- I~ Lq OLq
C N N N V N V N V N V
O
o R
iR v
o o
c v c
G1 .6 O~ V V O N
a W
N
N C a
O 1~/1
M
N a~ O~ O N CO O I~ O O N O O
E
> R
N ~
c
a a c0 ~~ M O~ M O M O V V V
E
d R 6
N ~ y
d O
a HE
y a~ N~ o Z O o N
C N C p
y'6 Gl M~ CO O V m M O
a W
N
O VI
2 M~~ c0 c0 M~~~ W
C
E
N R
N O y
O1 a
C ~
a~ m O 6~ CO O~ M O O c0 I~
E
N d y
~ T
C C
d =_
a.6 G1 c0 M O ITH CO c0 CO M
C C
o ~
z (%
m
N L
U
C
N
~ U
E
w X
LL
> E
O
N
N O
T p U O
-O Q O C O_ (6 U
y O
U U
O C N
~- - (6
0 ~ n O O_ n N OA N O
O
N C N Q CO D U~ p O~ >
U) C C C_ C U U ~ O V N X
0 ~ o Q5 _ IE 2 W o
O
2 2
E E E Y - ~- ~6 r
-O o N O O O o ~-
N N ~ O_ O_ O_ O_ O_ O_ O_ ~ U E E N
E d) O O O O E E E n U
16 O_ O_ O_ O_ O O O N .
Z Q Q Q Q U U U J (n N
p
O d z
o
~q a VI O I~ V V CO M CO c0 V ~ 6~
~ VI VI N ~~ M V ~~ c0 ~ O
VI G1 O I~ CO CO O CO c0 c0 M
~ U CO V N N O~~ ~ I~ ~
H U3) Q d d d d d d d d d d d d d
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
N
0
O N N V V ~[J V [J O C') V C) O O N O O 1J O
N O. O O O O O O O O O O O O O O O O O O O O
0 O
o R 3
R d 3
C N
y:6 O O~ N ~ ~ ~ W CO V M M ~[J I~ N O V
R C- W
N
N O ~
Q O N
N N O W ~ CO I~ N O ~[J V O O V I~ C) O C)
O R
N
p N O O N M M ~[J
fl- w O W W V I~ N W CO M V CO w
d R E
E
d O
O. s
fl- a N~ O N I~ V N I~ CO V (") ~ V W O O V O
O E
0 O R
Z
f/)
0 N
y;6 N V O M O M N O M M O O O O V O M
R C- W
N
N O. e
Q O N
N a~J CO W CO ~ M O V O C) O O O O O O O~
C
N
a~ o y C)
~ 0
c s-
N N ~ C) N~ O N N O N V O O O O O O W O M
C
N r E
N d y
N fl-
~
O.
y ._
fl- '6 d V O O W N O N O N V O O O C) O O V O N
O E
N 0
O R
Z fn
a
~
Ll
>
~ N U Q V
N Q ~ C N U
N Q L Q
= o
-6 O L N 20 2
Q U U
U V N y N~ O~ Q N N N
a~ fl- w O O ~ U U U U U
0 0 U U N _ N C C C = Q y C Q
N N o- Q Q ~- m Q Q Q O
N~
~D J V
O Q w~ o m U o 0 0 m o o ¾ 0
~- C C C C C Q-6 U U fl- 2-6
~ m a~ a~ a~ a~ a~ o Q _ _
CO o 0 0 0 0~, a~ a~ a~ ) a) ~~~~~
2 0 N Q Q Q Q Q O T N N N N ~ 0
m a a a a a N o a a a a o 2 2
E
fl- Q Q Q Q Q N t6 0 0 0 O t6 N t6 N N ~
~ z ¾¾¾¾¾¾ m c) c) c) c) c) x z a a co co
0
0
Z 0
o d Z
a o c
0
~ a N ~[J N O O ~ ~ O~ I~ W O O 0 I~ ~[J I~ N N V
N N CO ~[J W V V V V } ~J ~J CO W M V I~ W V ~J I~
N N I~ CO O CO V V I~ C) C) C) ~[J I~ d I~ I~ I~ ~[J V I~
t6 3 0 0 0 o O O O O O O O O O
0 0 Oc1O O O M O O
~ m Q~ a a a a a c1 a a a c1 a a a a a a a
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
[0094] As an initial screen, the spectral count (number of peptide sequencing
events) was averaged. Based on these results, a further study was performed of
selected proteins by peak area quantitation, as described above.
[0095] Based on the results described herein, angiotensinogen was identified
as
a biomarker for hypertension. Angiotensinogen is the precursor of biologically
active angiotensin I and II. The renin-angiotensinogen system plays a role in
the
regulation of extracellular fluid volume and blood pressure in the body. To
determine the relative abundance of angiotensinogen in the various sample
pools,
the following two angiotensinogen peptides were quantified by peak area
measurements. The peptide R.SLDFTELDVAAEK.I (SEQ ID NO:1) (with a +2
charge, a precursor ion mass of 720.4 amu, and a retention time of 95.1 min)
and the
peptide K.ALQDQLVLVAAK.L (SEQ ID NO:2) (with a +2 charge, a precursor ion
mass of 635.5 amu, and a retention time of 87.8 min) were quantitated. As
shown in
Figure 4, angiotensinogen was overexpressed in hypertensive patients and was
also
slightly elevated in obese patients compared to the normal pool.
[0096] Apolipoprotein (Apo) C1 is a 6.6-kDa protein present in plasma and
associated with lipoproteins. ApoCl is an inhibitor of lipoprotein binding to
the
LDL receptor, LDL receptor-related protein, and the VLDL receptor. It also is
the
major plasma inhibitor of cholesteryl ester transfer protein, and may
interfere
directly with fatty acid uptake.
[0097] Based on the results described herein, apolipoprotein C1 was identified
as a biomarker for diabetes, obesity, and hypertension. To determine the
relative
abundance of apolipoprotein C1 in the various sample pools, the following two
apolipoprotein C1 peptides were quantified by peak area measurements: the
peptide
K.MREWFSETFQK.V (SEQ ID NO:3) (with a +2 charge, a precursor ion mass of
745.49 amu, and a retention time of 85.6 min) and the peptide R.EWFSETFQK.V
(SEQ ID NO:4) (with a +2 charge, a precursor ion mass of 602.2 amu, and a
retention time of 90.5 min) were quantitated.
As shown in Figure 5, apolipoprotein C1 was present at high levels in plasma
in
obese, diabetic and hypertensive patients.
33
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
[0098] In the results described herein, the increase of Apo C1 was mirrored in
the levels of related apolipoprotein CIII and apolipoprotein B, as well as
insulin-like
growth factor (IgF) binding protein.
[0099] The results described herein demonstrate that by using a combination of
six protein depletion and multi-lectin affinity chromatography, the complexity
of the
human plasma proteome can be reduced to allow identification of protein
biomarkers for disease studies by proteomic analysis. With the use of the two
column approach, the non-glycoprotein fraction was depleted in terms of both
the
major plasma protein albumin as well as glycoproteins of high abundance such
as
alpha-2-macroglobulin. The utility of this approach to a disease study by the
identification of proteins such as angiotensinogen and apolipoprotein Cl,
which
were shown to be present at higher levels in obese, diabetic and hypertensive
patients, was demonstrated.
EXAMPLE 2
Reversal of Differential Expression of Proteins in Diabetic Subjects
Following Gastric Bypass Surgery
Protocol
[0100] 24 Caucasian women are used in this study: 8 diabetics (18-55 years of
age, BMI of 35-50, 3 blood samples each - 24 total); 8 euglycemic (18-55 years
of
age, BMI of 35-50, 3 blood sample each - 24 total); and 8 "normal" controls
(lean
and euglycemic, 1 sample each - 8 total). Plasma samples are obtained from
each
patient and the samples are analyzed as described in Example 1. The patients
then
undergo routine gastric bypass surgery, e.g., silastic ring Roux-en-Y gastric
bypass
or a Fobi pouch operation. After the surgery, plasma samples are obtained from
each patient 6 days, 1 month, 3 months, and 6 months following the surgery. On
the
morning of the surgery and 6 days later, a short intravenous glucose tolerance
test is
performed on each patient, and insulin resistance is measured on the morning
of
surgery, 6 days later, and again at 3, 6, 9, and 12 months after surgery (see,
e.g.,
Wickremesekera et al., Obesity Surgery (2005) 15:474-48 1).
[0101] The samples are then analyzed as described in Example 1. Briefly, the
six most abundant proteins are depleted using MARS, glycosylated proteins are
34
CA 02692970 2009-12-30
WO 2009/006568 PCT/US2008/069145
separated from nonglycosylated proteins by M-LAC, nonglycosylated proteins are
subjected to tryptic digestion, and the tryptic digests are analyzed by nano
LC-
MS/MS.
Results
[0102] Proteins are identified that are differentially expressed between
diabetic
and control patients before gastric bypass surgery. These include
angiotensinogen
and apolipoprotein Cl. Following gastric bypass surgery, insulin resistance is
improved in the diabetic patients, which is correlated with changes in the
levels of
proteins, identified as differentially expressed in diabetic patients before
surgery
(including angiotensinogen and apolipoprotein C1), to about the same levels
measured in control patients.
OTHER EMBODIMENTS
[0103] It is to be understood that while the invention has been described in
conjunction with the detailed description thereof, the foregoing description
is
intended to illustrate and not limit the scope of the invention, which is
defined by the
scope of the appended claims. Other aspects, advantages, and modifications are
within the scope of the following claims.