Note: Descriptions are shown in the official language in which they were submitted.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
TREATMENT PREDICTION INVOLVING HMGCR PROTEIN
Technical field
The present disclosure relates to the field of breast cancer treatment
and treatment prediction. It also relates to means which may be employed in
such a treatment or treatment prediction.
Background
Cancer
Cancer is one of the most common causes of disease and death in the
western world. In general, incidence rates increase with age for most forms of
cancer. As human populations continue to live longer, due to an increase of
the general health status, cancer may affect an increasing number of
individuals. The cause of most common cancer types is still largely unknown,
although there is an increasing body of knowledge providing a link between
environmental factors (dietary, tobacco smoke, UV radiation etc) as well as
genetic factors (germ line mutations in "cancer genes" such as p53, APC,
BRCA1, XP etc) and the risk for development of cancer.
No definition of cancer is entirely satisfactory from a cell biological
point of view, despite the fact that cancer is essentially a cellular disease
and
defined as a transformed cell population with net cell growth and anti-social
behavior. Malignant transformation represents the transition to a malignant
phenotype based on irreversible genetic alterations. Although this has not
been formally proven, malignant transformation is believed to take place in
one cell, from which a subsequently developed tumor originates (the "clonality
of cancer" dogma). Carcinogenesis is the process by which cancer is
generated and is generally accepted to include multiple events that ultimately
lead to growth of a malignant tumor. This multi-step process includes several
rate-limiting steps, such as addition of mutations and possibly also
epigenetic
events, leading to formation of cancer following stages of precancerous
proliferation. The stepwise changes involve accumulation of errors
(mutations) in vital regulatory pathways that determine cell division, asocial
behavior and cell death. Each of these changes may provide a selective
Darwinian growth advantage compared to surrounding cells, resulting in a net
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
2
growth of the tumor cell population. A malignant tumor does not only
necessarily consist of the transformed tumor cells themselves but also
surrounding normal cells which act as a supportive stroma. This recruited
cancer stroma consists of connective tissue, blood vessels and various other
normal cells, e.g., inflammatory cells, which act in concert to supply the
transformed tumor cells with signals necessary for continued tumor growth.
The most common forms of cancer arise in somatic cells and are
predominantly of epithelial origin, e.g., prostate, breast, colon, urothelial
and
skin, followed by cancers originating from the hematopoetic lineage, e.g.,
leukemia and lymphoma, neuroectoderm, e.g., malignant gliomas, and soft
tissue tumors, e.g., sarcomas.
Cancer diagnostics and prognostics
Microscopic evaluation of a tissue section taken from a tumor remains
the golden standard for determining a diagnosis of cancer. For example, for
microscopic diagnosis, biopsy material from suspected tumors is collected
and examined under the microscope. To obtain a firm diagnosis, the tumor
tissue is fixated in formalin, histo-processed and paraffin embedded. From the
resulting paraffin block, tissue sections can be produced and stained using
both histochemical, i.e., hematoxylin-eosin staining, and
immunohistochemical methods. The surgical specimen is then evaluated with
pathology techniques, including gross and microscopic analysis. This analysis
often forms the basis for assigning a specific diagnosis, i.e., classifying
the
tumor type and grading the degree of malignancy, of a tumor.
Malignant tumors can be categorized into several stages according to
classification schemes specific for each cancer type. The most common
classification system for solid tumors is the tumor-node-metastasis (TNM)
staging system. The T stage describes the local extent of the primary tumor,
i.e., how far the tumor has invaded and imposed growth into surrounding
tissues, whereas the N stage and M stage describe how the tumor has
developed metastases, with the N stage describing spread of tumor to lymph
nodes and the M stage describing growth of tumor in other distant organs.
Early stages include: TO-1, NO, MO, representing localized tumors with
negative lymph nodes. More advanced stages include: T2-4, NO, MO,
localized tumors with more widespread growth and T1-4, N1-3, MO, tumors
that have metastasized to lymph nodes and T1-4, N1-3, M1, tumors with a
metastasis detected in a distant organ. Staging of tumors is often based on
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
3
several forms of examination, including surgical, radiological and
histopathological analyses. In addition to staging, there is also a
classification
system to grade the level of malignancy for most tumor types. The grading
systems rely on morphological assessment of a tumor tissue sample and are
based on the microscopic features found in a given tumor. These grading
systems may be based on the degree of differentiation, proliferation and
atypical appearance of the tumor cells. Examples of generally employed
grading systems include Gleason grading for prostatic carcinomas and the
Nottingham Histological Grade (NHG) grading for breast carcinomas.
Accurate staging and grading is crucial for a correct diagnosis and may
provide an instrument to predict a prognosis. The diagnostic and prognostic
information for a specific tumor subsequently determines an adequate
therapeutic strategy for a given cancer patient. A commonly used method, in
addition to histochemical staining of tissue sections, to obtain more
information regarding a tumor is immunohistochemical staining. IHC allows
for the detection of protein expression patterns in tissues and cells using
specific antibodies. The use of IHC in clinical diagnostics allows for the
detection of immunoreactivity in different cell populations, in addition to
the
information regarding tissue architecture and cellular morphology that is
assessed from the histochemically stained tumor tissue section. IHC can be
involved in supporting the accurate diagnosis, including staging and grading,
of a primary tumor as well as in the diagnostics of metastases of unknown
origin. The most commonly used antibodies in clinical practice today include
antibodies against cell type "specific" proteins, e.g., PSA (prostate), MelanA
(melanocytes) and Thyroglobulin (thyroid gland), and antibodies recognizing
intermediate filaments (epithelial, mesenchymal, glial), cluster of
differentiation (CD) antigens (hematopoetic, sub-classification of lympoid
cells) and markers of malignant potential, e.g., Ki67 (proliferation), p53
(commonly mutated tumor suppressor gene) and HER-2 (growth factor
receptor).
Aside from IHC, the use of in situ hybridization for detecting gene
amplification and gene sequencing for mutation analysis are evolving
technologies within cancer diagnostics. In addition, global analysis of
transcripts, proteins or metabolites all add relevant information. However,
most of these analyses still represent basic research and have yet to be
evaluated and standardized for the use in clinical medicine.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
4
Breast cancer
Breast cancer is the second most common form of cancer worldwide
and by far the most frequent cancer of women. Data from the GLOBOCAM
2002 database presented by Parkin et al. reveal 1.15 million new cases in
2002 and 0.41 million deaths during the same period (Parkin DM et al. (2005)
CA Cancer J Clin 55, 74-108). If detected at an early stage, the prognosis is
relatively good for a patient living in a developed country, with a general
five-
year survival rate of 73%, compared to 57% in a developing country. The
incidence is slowly increasing and about one in every nine women in the
10. developed world is believed to get breast cancer in her lifetime. Although
lifestyle changes related to female steroid hormones, including exposure to
exogenous hormones, affect the risk of developing breast cancer, these
factors only make up for a small fraction of the etiology, and the benefit of
preventive manipulation is believed to be low. The decreased mortality is
mainly due to earlier detection by mammography screening and the use of
modern adjuvant systemic treatment.
Treatment of breast cancer
Since its introduction in the late seventies, breast-conserving therapy,
combining breast conserving surgery and postoperative radiotherapy, has
become the primary treatment of choice in women where radical removal of
the tumor can be combined with a good cosmetic result. Mastectomy is still
preferable in some patients, i.e., women with small breasts, large tumors (> 4
cm) or multifocal/multicentric disease.
Axillary dissection is primarily performed for diagnostic purposes and
removal of at least 10 lymph nodes gives a good staging guidance with 97-
98% sensitivity (Axelsson CK et al. (1992) Eur J Cancer 28A:1415-8; Recht A
and Houlihan MJ (1995) Cancer 6(9):1491-1512). However, the next step
towards minimal surgery in the treatment of primary cancer has been the
introduction of the sentinel node biopsy technique with mapping of axillary
lymph nodes instead of axillary lymph node clearance, which is associated
with a high complication rate. This technique was introduced as a
consequence of the knowledge that most of the lymphatic drainage to the
axilla from the breast initially passes through one (or a few) lymph node(s) -
the sentinel node(s) - supporting that analysis of this lymph node may be a
sufficient indicator of axillary node status (Veronesi U et al. (2003) New
Engl
J Med 349(6): 546-53.)
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
The concept of breast cancer as a systemic disease, i.e., the presence
of disseminating micro-metastases at the time of diagnosis that may explain
treatment failure after locoregional therapy, paved the way for adjuvant
randomized trials in the 1970s, including endocrine therapy and
5 chemotherapy. Adjuvant polychemotherapy is standard treatment for
hormone-receptor negative patients with high risk of recurrence, irrespective
of nodal status. A beneficial effect on both overall- and relapse-free
survival
has been demonstrated, especially in premenopausal patients (EBCTCG
(1998) Lancet 352(9132): 930-42). For patients with hormone-responsive
disease, e.g., estrogen receptor (ER) and/or progesterone receptor (PR)
positive disease, adjuvant polychemotherapy has been delivered in
combination with endocrine therapy as sequential chemo-endocrine therapy.
Also, adjuvant chemotherapy generally induces amenorrhea, causing a
secondary endocrine effect in addition to the cytotoxic (Pagani 0 et al.
(1998)
Eur J Cancer 34(5):632-40).
Endocrine therapy has been. recommended for patients with hormone
receptor positive tumors irrespective of age, stage and menopausal status.
In hormone-responsive premenopausal patients, ovarian ablation by
surgery or irradiation, or ovarian suppression by LHRH agonists is efficient
adjuvant treatment modalities (Emens LA and Davidson NA (2003) Clin Ca
Res (1 Pt 2): 468S-94S). In postmenopausal patients, ovarian ablation has no
place, since the primary source of estrogen is not from ovarian synthesis but
from the conversion of androstenedione to estrone and estradiol in peripheral
tissues including the breast.
Tamoxifen is a selective estrogen receptor modulator (SERM) with an
agonistic effect on the ER, making it a suitable treatment for advanced breast
cancer in both pre- and postmenopausal women. Studies have shown that
five years of tamoxifen as adjuvant treatment after primary surgery reduces
the breast cancer mortality in patients with ER positive (ER+) tumors,
irrespective of lymph node status (EBCTCG (1998) Lancet 351(9114):1451-
67). While tamoxifen has a protective effect against cardiovascular disease,
the risk of developing endometrial cancer is increased, due to an agonistic
effect on the ER in the endometrium (EBCTCG (2005) Lancet
365(9472):1687-717)
Aromatase inhibitors (Als) function by inhibiting aromatase, the
enzyme converting androgens into estrogens. For example, Als can be given
as adjuvant treatment to postmenopausal women, either alone or following
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
6
tamoxifen treatment and they have been shown to significantly reduce the
mortality, possibly even more if given alone (Howell A et al. (1995) Lancet
345(8941):29-30; Ellis MJ and Rigden CE (2006) Curr Med Res Opin
22(12):2479-87; Coates AS et al. (2007) J Clin Oncol 25(5):486-92).
However, this therapy is relatively new and the long-term side effects are not
yet fully known (Buzdar A et al. (2006) Lancet Oncol 7(8):633-43), but the
most important are cardiovascular complications and osteoporosis.
Newly developed pure anti-estrogens such as fulvestrant, which
completely blocks the ER, are currently only used in advanced breast cancer
and not in the adjuvant setting (Rutqvist LE (2004) Best Pract Res Clin
Endocrinol Metab 18(1): 81-95).
Adjuvant endocrine therapy is believed to have no place in hormone
receptor negative breast cancer, although some studies indicate that some
ER negative i.e., ERa negative (ERa-), tumors respond to tamoxifen
treatment (EBCTCG (1998) Lancet 351:1451-1467)
The HER2/neu gene is overexpressed in about 20% of all, and in up to
70% of lowly differentiated, breast cancers (Berger MS et al. (1988) Cancer
Res 48(5):1238-43; Borg A et al. (1990) Cancer Res 50(14): 4332-7). Patients
with HER2 overexpressing tumors may benefit from treatment with the
monoclonal antibody trastuzumab. Experimental data support a relationship
between HER2 overexpression and resistance to endocrine treatment (Shou
J et al. (2004) J Natl Cancer Inst 96(12):926-35) while clinical data are not
consistent (Borg A et al. (1994) Cancer Left 81(2):137-44, De Placido S et al.
(2003) Clin Ca Res 9(3):1039-46, Ryden L et al. (2005) J Clin Oncol
23(21):4695-704).
Breast cancer diagnostics
Morphologic criteria are still generally considered important in the
establishment of a breast cancer diagnosis, both in situ and invasive cancer.
Among invasive breast carcinomas, invasive ductal carcinoma is the most
common tumor type (-80 %) and lobular carcinoma is the second largest
entity (-10-15%). Tubular and medullary carcinomas are other distinct types
with lower prevalence (WHO, Histological typing of breast tumors, in
International histological classification of tumors no:2, 1981, WHO, Geneva).
Breast cancer prognostics and treatment predictive factors
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
7
A correct histological classification of the tumor type may be of
prognostic relevance, since certain subtypes, such as medullary carcinomas,
in general have a more favorable prognosis. Nevertheless, assessment of the
histological grade using the Nottingham Histological Grade (NHG) system is
still a prognostic tool (Elston CW and Ellis 10 (1991) 19(5):403-10; Sundquist
M et al. (1999) Breast Cancer Res Treat 53(1):1-8).
The majority of breast cancers are hormone receptor responsive, i.e.,
express the ER and/or PR. The action of estrogen is mediated by the two
receptors ERa and ERI3. ERa and PR are routinely assessed in order to
select patients for endocrine therapy, and according to one standard, tumors
with >10 % nuclear positivity are considered positive. ERa is today
considered a predictor of tamoxifen response. However, there are studies that
indicate that PR positivity may even be a more powerful predictor of
tamoxifen response than ERa. A study of premenopausal patients
randomized to tamoxifen or no adjuvant endocrine treatment revealed that a
high expression (>75% nuclear fraction) of PR was significantly associated
with an increased recurrence-free and overall survival in tamoxifen treated
patients, irrespectively of ERa status. At a lower PR level, <75%, no positive
effect from tamoxifen treatment was observed (Stendahl M et al. (2006) Clin
Cancer Res 12:4614-18). Yu et al. recently studied the predictive value of PR
for adjuvant endocrine therapy, and found that older patients (>_ 60 years)
with
ERa+/PR+ tumors had a significantly longer disease free survival when
treated with tamoxifen than patients that received no adjuvant treatment. In
younger patients (<60 years), no significant effect was observed (Yu KD et al.
(2007) The Breast 16:307-315). Interestingly, a report from the ATAC trial
(postmenopausal women treated with arimidex, tamoxifen or in combination),
showed that the recurrence rate was halved for anastrozole-treated patients
with ERa+/PR- tumors over the follow-up period of 6 years, compared to
patients treated with tamoxifen (Dowsett M et al. (2005) J Clin Oncol
23(30):7512-7).
The role of ERR in breast cancer is not yet fully clarified, although
recent studies implicate that ERR-expression may be associated with a better
tamoxifen response (Borgquist S et al, (2008) J Clin Pathol 61(2):197-203),
particularly in ERa- tumors (Gruvberger-Saal SK et al. (2007) Clin Cancer
Res 13:1987-1994). However, determination of ERR is today generally not
considered clinically relevant.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
8
A major problem to day is that 30-40% of the ERa positive (ERa+)
patients do not respond to tamoxifen treatment (Riggins RB et al. (2007)
Cancer Letters 1:1-24, Gruvberger-Saal SK et al. (2007) Clin Cancer Res
13:1987-1994), which results in unnecessary treatment. In addition, a fraction
of the ERa- patients do respond to tamoxifen treatment, and the reason for
that is currently not known. Gruvberger-Saal suggests that ERP expression
may be a positive predictor of tamoxifen response in ERa- patients
(Gruvberger-Saal SK et al. (2007) Clin Cancer Res 13:1987-1994).
HER2 status is also assessed routinely, primarily by IHC and in cases
with moderate expression (2+), gene amplification status is determined by
fluorescence in situ hybridization (FISH) analysis. Patients with a HER2
positive tumor may benefit from treatment with trastuzumab.
Breast cancer is a truly heterogeneous disease and despite the
increasing understanding of its nature, the arsenal of available prognostic
and
treatment predictive markers is still not sufficient and some patients may
therefore receive unnecessary treatment while others may get insufficient or
even ineffective treatment. Additional molecular markers are needed in order
to better define different subgroups of breast cancer and increase the options
for tailored therapies.
Endpoint analysis
Endpoint analysis is used to evaluate trials with adjuvant treatments for
cancer as this gives information on how the patients respond to a certain
therapy. Endpoint analysis may also be useful for studies of a potential
biomarker.
Overall survival (OS) has been considered the standard primary
endpoint. OS takes in to account time to death, irrespective of cause, e.g.,
if
the death is due to cancer or not. Loss to follow-up is censored and regional
recurrence, distant metastases, second primary breast cancers, and second
other primary cancers are ignored.
To date, an increasing number of effective treatments available in
many types of cancer have resulted in the need for surrogate endpoints to
allow for a better evaluation of the effect of adjuvant treatments. Thus, the
much longer follow-up required to demonstrate that adjuvant treatments
improve OS is often complemented with other clinical endpoints that give an
earlier indication on how successful the treatment is. For these observations,
recurrence-free survival (RFS) and breast cancer-specific survival (BCSS)
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
9
may be analyzed. RFS includes time to any event related to the same cancer,
i.e., all cancer recurrences and deaths from the same cancer are events.
Distant, local and regional metastases as well as breast cancer specific death
are considered. On the other hand, second primary same cancers and other
primary cancers are ignored, as well as contralateral breast cancer. Deaths
from other cancers, non-cancer-related deaths, treatment-related deaths, and
loss to follow-up are censored observations. Breast cancer-specific survival
(BCSS) includes time to death caused by breast cancer due to the original
tumor. Both endpoints are relevant, since similarities or differences may
reflect different tumor biological behaviors. Biomarkers associated with a
locally aggressive behavior may for instance have greater impact on RFS
than BCSS, while biomarkers associated with the development of distant
metastases may be reflected in both RFS and BCSS.
Description
It is an object of some aspects of the present disclosure to provide
means, methods and/or uses useful in treatment prediction, in particular
breast cancer treatment prediction, regarding a mammalian subject having a
breast cancer.
An object of some other aspects of the present disclosure is to provide
means, methods and/or uses useful in the treatment of such a subject.
Thus, as a first configuration of a first aspect of the present invention,
there is provided a method for determining whether a mammalian subject
having a breast cancer is likely to benefit from an endocrine treatment,
comprising the steps of:
a) providing a sample earlier obtained from said subject;
b) evaluating the amount of HMGCR protein present in at least part of
said sample, and determining a sample value corresponding to said
amount;
c) comparing the sample value obtained in step b) with a reference
value; and,
if said sample value is higher than said reference value,
d) concluding that the subject is likely to benefit from an endocrine
treatment.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
Regarding step b) of the methods of the present disclosure, an
increase in the amount of HMGCR protein typically results in an increase in
the sample value, and not the other way around. However, in some
embodiments, the evaluated amount may correspond to any of a
5 predetermined number of discrete sample values. In such embodiments, a
first amount and a second, increased, amount may correspond to the same
sample value. In any case, an increase in the amount of HMGCR protein will
not result in a decrease in the sample value in the context of the present
disclosure.
10 However inconvenient, but in an equivalent fashion, the evaluated
amounts may be inversely related to sample values if the qualification
between step c) and d) is "if the sample value is lower than the reference
value" (see the method above). This applies mutatis mutandis to the other
method aspects, configurations and embodiments of the present disclosure.
In the context of the present disclosure, "likely to benefit from an
endocrine treatment" refers to a having a higher probability of survival or
recovery if undergoing an endocrine treatment than if not undergoing an
endocrine treatment. In this context, "recovery" refers to return from a
breast
cancer state to a breast cancer free state. The "survival" may be a
recurrence free survival or a breast cancer specific survival. Further, the
"recovery" may be a recurrence free recovery. Also, the "higher probability"
may be a probability benefit at five years, ten years or 15 years of at least
5
%, such as at least 10 %.
This first aspect of the present disclosure is based on the previously
unrecognized fact that the expression of HMGCR protein in a sample
obtained from a subject having a breast cancer may serve as an indicator of
response to endocrine treatment in the subject. More particularly, the
inventors have identified that, in patients suffering from breast cancer, a
correlation between values of HMGCR protein on the one hand and the
survival after endocrine treatment on the other. Typically, high HMGCR
protein values are shown herein to correlate with responsiveness to
endocrine treatment. The present disclosure, based on HMGCR protein
expression as a breast cancer treatment indicator, provides for a number of
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
11
benefits. Firstly, it provides an additional, or alternative, tool for
predicting
whether a patient is likely to respond to endocrine treatment. Secondly it
identifies a subgroup of hormone receptor negative patients that, contrary to
earlier beliefs, benefit from to endocrine treatment. Consequently, the
present
disclosure may provide for accurate treatment of a previously undertreated
group. Thirdly, the present disclosure identifies a subgroup of breast cancer
patient which generally do not benefit from endocrine treatment.
Traditionally, subjects with hormone receptor negative breast cancers,
especially ER- breast cancers, have not received systemic endocrine
treatment. However, as shown in the present disclosure, the survival rates in
the subgroup of subjects with hormone receptor negative, e.g. ER- or PR-,
ER-, PR-, or ER- and PR-, breast cancers having high HMGCR values were
improved with endocrine treatment. Consequently, the inventors have
identified new subgroups of subjects that may be treated with an endocrine
treatment. Accordingly, various aspects of the present disclosure may be
particularly relevant for the subjects having hormone receptor negative
cancers.
As a second configuration of the first aspect, there is provided a
method for determining whether a mammalian subject having a breast cancer
should undergo an endocrine treatment, comprising the steps of:
a) providing a sample earlier obtained from said subject;
b) evaluating the amount of HMGCR protein present in at least part of
said sample, and determining a sample value corresponding to said
amount;
c) comparing the sample value obtained in step b) with a reference
value; and,
if said sample value is higher than said reference value,
d) concluding that the subject should undergo an endocrine treatment.
As a third configuration of the first aspect of the present invention,
there is provided a non-treatment strategy method for a mammalian subject
having a breast cancer, comprising:
a) providing a sample earlier obtained from said subject;
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
12
b) evaluating the amount of HMGCR protein present in at least part of
said sample, and determining a sample value corresponding to said
amount;
c) comparing the sample value obtained in step b) with a reference
value; and,
if said sample value is lower than or equal to said reference value,
d) refraining from treating said subject with an endocrine treatment.
For example, the refraining of step d) may be refraining during at least
one week from the completion of steps a) - c), such as at least one month
from the completion of steps a) - c), such as at least three months from the
completion of steps a) - c), such as at least six months from the completion
of
steps a) - c), such as at least one year from the completion of steps a) - c),
such as at least two years from the completion of steps a) - c).
Alternatively, the refraining of step d) may be refraining until the next
time the method is performed or until recurrence of a breast cancer tumor.
In embodiments of configurations one to three of the first aspect, the
breast cancer may be a hormone receptor negative breast cancer, such as an
ER- or PR- breast cancer, an ER- breast cancer, a PR- breast cancer, or an
ER- and PR- breast cancer. For example, the breast cancer may be
previously diagnosed as hormone receptor negative.
In general, hormone receptor status, e.g. ER status or PR status, may
be assessed according to any method known to the skilled artisan and the
person skilled in the art knows how to obtain the ER and/or PR status of a
patient. For example, such information may be obtained from the result of a
test using the commercially available ER/PR pharmDX kit (DakoCytomation).
As another example, the method disclosed by Allred et al. (Allred et al.
(1998)
Mod Pathol 11(2), 155) may be used to obtain a total score (Allred score),
and, for both ER and PR, an Allred score of higher than two is considered
positive and an Allred score of two or lower is considered negative.
Alternatively, when classifying a sample as being positive or negative for ER
or PR, a cutoff of 10% positive cells may be used, which is a recognized limit
within the art.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
13
The person skilled in the art knows how to obtain the ER and/or PR
status of a patient. For example, such information may be obtained from the
result of a test using the commercially available ER/PR pharmDX kit
(DakoCytomation). As another example, the method disclosed by Allred et al.
(Allred et al. (1998) Mod Pathol 11(2), 155) may be used to obtain a total
score (Allred score), and, for both ER and PR, an Allred score of higher than
two is considered positive and an Allred score of two or lower is considered
negative. Alternatively, when classifying a sample as being positive or
negative for ER or PR, a cutoff of 10% positive cells may be used, which is a
recognized limit within the art.
If not otherwise stated, "ER" refers to "ERa" in the context of the
present disclosure.
As a fourth configuration of the first aspect of the present invention,
there is provided a method for determining whether a mammalian subject
having a breast cancer is likely to benefit from an endocrine treatment,
comprising the steps of:
a) providing a sample earlier obtained from said subject;
b) evaluating the amount of HMGCR protein present in at least part of
said sample, and determining a sample value corresponding to said
amount;
c) comparing the sample value obtained in step c) with a reference value
and thereby determining the HMGCR status of said subject;
d) obtaining the ER status for said subject; and
if said ER status or said HMGCR status is positive,
el) concluding that the subject is likely to benefit from an endocrine
treatment, or
if said ER status and said HMGCR status are negative,
e2) concluding that the subject is not likely to benefit from the endocrine
treatment.
However closely related and covered by the same concept, el) and
e2) provide two alternative conclusion options. Accordingly, the method of the
fourth configuration of the first aspect may answer the question whether the
subject having a breast cancer is likely to benefit from an endocrine
treatment
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
14
or the question whether the subject having a breast cancer is not likely to
benefit from an endocrine treatment. Thus, the method of the fourth
configuration of the first aspect may, but does not have to, comprise both
step
el), together with its adherent qualification ("if phrase"), and step e2),
together with its adherent qualification ("if phrase").
For example, in step c), the HMGCR status may be considered positive
if the sample value is higher than the reference value and negative if the
reference value is lower than, or equal to, the reference value.
A single sample from the subject may provide both the ER status and
the HMGCR status, for example by means of immunohistochemistry. Thus, in
embodiments of the method aspects of the present disclosure, the ER status
may be obtained from the sample of step a). Also, the ER status may be
obtained from a tissue material from which the sample of step a) was also
obtained. Consequently, the required number of biopsies or amount of
biological material from the subject may be kept low.
As a fifth configuration of the first aspect of the present disclosure,
there is provided a method for determining whether a sample value
corresponding to an amount of HMGCR protein present in at least part of a
sample earlier obtained from a mammalian subject having a breast cancer is
in favor of a decision to apply an endocrine treatment to the subject,
comprising the steps of:
a) providing the sample earlier obtained from the subject;
b) evaluating the amount of HMGCR protein present in at least part of
the sample, and determining the sample value corresponding to the
amount;
c) comparing the sample value obtained in step b) with a reference
value; and,
if the sample value is higher than the reference value,
dl) concluding that the sample value is in favor of a decision to treat
the subject with an endocrine treatment, and/or
if the sample value is lower than or equal to the reference value,
d2) concluding that the sample value is in favor of a decision to refrain
from treating the subject with an endocrine treatment.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
However closely related and covered by the same concept, dl) and
d2) of the fifth configuration of the first aspect provide two alternative
conclusion options. Accordingly, the method may answer the question
whether the sample value is in favor of a decision to refrain from treating
the
5 subject with an endocrine treatment or the question whether the sample value
is in favor of a decision to treat the subject with an endocrine treatment.
Thus,
the method of the fifth configuration of the first aspect may, but does not
have
to, comprise both step dl), together with its adherent qualification ("if
phrase"), and step d2), together with its adherent qualification ("if
phrase").
10 When deciding on a suitable treatment strategy for a patient having
breast cancer, the physician responsible for the treatment may take several
parameters into account, such as the result of an immunohistochemical
evaluation, patient age, hormone receptor status, general condition, medical
history, such as breast cancer history and hereditary characteristics, e.g.
15 whether there is a history of breast cancer in the subject's family. To be
guided in such decision, the physician may perform a HMGCR protein test, or
order an HMGCR protein test performed, according to the first aspect. In such
case, a method according to the fifth configuration of the first aspect may be
particularly relevant.
Also, the physician may instruct someone else, such as lab staff, to
perform part of a method according to the present disclosure, such as steps
a) to c), and perform the remaining, such as step d) or e), himself.
As a sixth configuration of the first aspect, there is provided a method
for determining an endocrine treatment prediction regarding a mammalian
subject having a breast cancer:
a) providing a sample from the subject;
b) evaluating the amount of HMGCR protein present in at least part of
the sample, and determining a sample value corresponding to the
evaluated amount;
c) correlating the sample value of step b) to the endocrine treatment
prediction for the subject.
In an embodiment of the above method, the sample may be an earlier
obtained sample.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
16
The correlating of step c) refers to any way of associating survival data
to the obtained sample value so as to establish the treatment prediction.
The identified correlation between high HMGCR protein expression
and responsiveness to endocrine treatment may also form a basis for the
application of a specific regimen for treatment of the subject. Thus, a first
configuration of a second aspect of the present disclosure, there is provided
a
method of treatment of a mammalian subject in need thereof, wherein said
subject has a breast cancer, comprising the steps of:
a) providing a sample from said subject;
b) evaluating the amount of HMGCR protein present in at least part of
said sample, and determining a sample value corresponding to said
amount;
c) comparing the sample value obtained in step b) with a reference
value; and,
if said sample value is higher than said reference value,
d) treating said subject with an endocrine treatment regimen.
In embodiments of the first configuration of the second aspect, the
breast cancer may be a hormone receptor negative breast cancer, such as an
ER- or PR- breast cancer, an ER- breast cancer, a PR- breast cancer, or an
ER- and PR- breast cancer. For example, the breast cancer may be
previously diagnosed as hormone receptor negative.
As a second configuration of the second aspect, there is provided a
method of treatment of a mammalian subject in need thereof, wherein said
subject has a breast cancer, comprising the steps of:
a) providing a sample from said subject;
b) evaluating the amount of HMGCR protein present in at least part of
said sample, and determining a sample value corresponding to said
amount;
c) comparing the sample value obtained in step c) with a reference value
and thereby determining the HMGCR status of said subject;
d) obtaining the ER status for said subject; and
if said ER status or said HMGCR status is positive,
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
17
el) treating said subject with an endocrine treatment regimen, or
if said ER status and said HMGCR status are negative,
e2) treating said subject with a non-endocrine treatment regimen.
However closely related and covered by the same concept, el) and
e2) of the second configuration of the second aspect provide two alternative
conclusion options. Accordingly, the method may involve an endocrine
treatment regimen or a non-endocrine treatment regimen. Thus, the method
of the second configuration of the second aspect may, but does not have to,
comprise both step e1), together with its adherent qualification ("if
phrase"),
and step e2), together with its adherent qualification ("if phrase").
Generally regarding the method of treatment aspects of the present
disclosure, the physician responsible for the treatment of the subject may
perform steps a) to c) himself or instruct someone else, such as lab staff, to
perform them.
In the context of the present disclosure, an "endocrine treatment" refers
to a systemic treatment with an anti-estrogenic effect. Further, "anti-
estrogenic effect" refers to suppression of estrogen production or inhibition
of
estrogen effects in the body. In the art, an endocrine treatment is sometimes
referred to as an anti-hormonal treatment.
The "endocrine treatment" of the present disclosure may be selected
from the group consisting of a selective estrogen receptor modulator (SERM)
treatment, an aromatase inhibitor treatment, and a steroidal estrogen receptor
antagonist treatment. The SERM treatment may for example be a treatment
selected from a toremifene, raloxifene, droloxifene, arzoxifene and tamoxifen
treatment. The aromatase inhibitor treatment may for example be selected
from anastrozole, letrozole and exemestane treatment. Further, the steroidal
estrogen receptor antagonist treatment may be a treatment with fulvestrant.
A SERM may have either an agonistic or antagonistic effect depending
on the target tissue. For example, a SERM may act as an antagonist in breast
and as an agonist in uterus.
In the Examples section of the present disclosure, various results of
treatment with the SERM tamoxifen are presented. The results show that the
level of HMGCR protein expression is relevant for predicting whether the
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
18
subjects are responsive to tamoxifen treatment. Thus, in preferred
embodiments of the present disclosure, the endocrine treatment is tamoxifen
treatment.
For example, the "non-endocrine treatment" of the present disclosure
may be selected from chemotherapies, radiation therapies and combinations
thereof. The chemotherapies comprise mono- or polychemotherapy, such as
treatment with CMF (cyclophosphamide, methotrexate, 5-fluorouracil), FEC
(fluorouracil, epirubicin, cyclofosfamid) and/or taxanes. Also, if the breast
cancer is HER2 positive (HER2+), the non-endocrine treatment may be an
anti-HER2 treatment, such as a treatment with an anti-HER2 antibody, e.g.,
trastuzumab.
In the context of the present disclosure, "a mammalian subject having
a breast cancer" refers to a mammalian subject having a primary or
secondary breast tumor or a mammalian subject which has had such tumor
removed from the breast, wherein the removal of the tumor refers to killing or
removing the tumor by any type of surgery or therapy. In the latter case, the
tumor may for example have been removed less than one year ago. For
example, a subject who has had a breast tumor removed by surgery and is
about to get adjuvant therapy is considered "having a breast cancer" in the
context of the present disclosure. "Breast tumor" includes ductal carcinoma in
situ (DCIS). In the method and use aspects of the present disclosure, "a
mammalian subject having a breast cancer" also includes the case wherein
the mammalian subject is suspected of having a breast cancer at the time of
the performance of the use or method and the breast cancer diagnosis is
established later.
In the context of the method aspects of the present disclosure, "earlier
obtained" refers to obtained before the method is performed. Consequently, if
a sample earlier obtained from a subject is provided in a method, the method
does not involve obtaining the sample from the subject, i.e., the sample was
previously obtained from the subject in a step separate from the method.
Accordingly, all methods and uses of the present disclosure, may be
performed entirely in vitro unless otherwise stated.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
19
Step b) of the methods of the above aspects involve evaluating the
amount of HMGCR protein present in at least part of the sample, and
determining a sample value corresponding to the amount. The "at least part of
the sample" refers to a relevant part, or relevant parts, of the sample for
establishing a treatment prediction or drawing conclusions regarding suitable
treatments. The person skilled in the art understands which part or parts that
are relevant under the circumstances present when performing the method.
For example, if the sample comprises tumor and non-tumor cells, the skilled
person may only consider the tumor cells, and only the cytoplasms of the
tumor cells, of the sample.
Further, in step b) an amount is evaluated and a sample value
corresponding to the amount is determined. Consequently, an exact
measurement of the amount of HMGCR protein is not required for obtaining
the sample value. For example, the amount of HMGCR protein may be
evaluated by visual inspection of a stained tissue sample and the sample
value may then be categorized, e.g., as high or low based on the evaluated
amount. The person skilled in the art understands how to perform such
evaluation and determination.
Still further, in the context of the present disclosure, the "reference
value" refers to a predetermined value which is relevant for making decisions,
or drawing conclusions, regarding the treatment or treatment prediction.
The data of the present disclosure is based on groups of human
females. Thus, in embodiments of the present disclosure, the "mammalian
subject" may be a human. Also, in embodiments of the present disclosure, the
"mammalian subject" may be a female, such as a premenopausal or
postmenopausal female. Endocrine treatment is given to both premenopausal
and postmenopausal subjects. Premenopausal females are generally
considered to be more responsive to tamoxifen treatment. Thus, in some
embodiments, in particular those wherein tamoxifen is selected as the
endocrine treatment, premenopausal human female subjects may be a
particularly relevant group.
The diagnosed or treated tumors of the present disclosure may be in
any stage. However, the subjects studied as described in Examples, section 4
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
had stage 11 invasive breast cancers. Stage 11 refers to pT2 NO MO or pT1-2
N1 MO according to the TNM staging system. Thus, in embodiments of the
present disclosure, the "breast cancer" may be a stage II breast cancer.
Also, in embodiments of the present disclosure, the "breast cancer"
5 may be a node negative or a node positive breast cancer (see e.g. fig 4).
"Node negative cancer" and "node positive cancer" refers to a cancer that has
and has not spread to the lymph nodes, respectively.
A seen in figures 4-7 and 9, the treatment predictive role of HMGCR
protein expression is particularly accentuated in node positive breast cancer.
10 Thus, in embodiments of the present disclosure, the breast cancer may be
node positive.
In embodiments of the methods of the above aspects, the sample may
be a body fluid sample, such as a sample of blood, plasma, serum, cerebral
fluid, lymph, urine or exudate. Preferably, the body fluid sample is sample of
15 blood, plasma, serum or lymph. Alternatively, the sample may be a cytology
sample or a stool sample.
The level of HMGCR protein expression may preferably be measured
intracellularly. Thus, the body fluid, cytology or stool sample may for
example
comprise cells, such as tumor cells.
20 In further embodiments of the methods of the above aspects, the
sample may be a tissue sample, such as a tumor tissue sample. As an
example, the tissue sample may be derived from a primary breast tumor,
such as an in situ or invasive carcinoma, or a secondary tumor (metastasis).
Tissue samples facilitate HMGCR protein expression analysis by means of
immunohistochemistry.
The inventors have found that the relevant HMGCR protein. expression
is primarily cytoplasmic. Thus, in embodiments of the methods of the present
disclosure, the evaluation of step b) may be limited to the cytoplasms of
cells,
such as tumor cells, of the sample. When the evaluation is limited to the
cytoplasms, only the characteristics, such as the HMGCR protein expression,
of the cytoplasms are considered in the evaluation. Such evaluation may for
example by aided by immunohistochemical staining.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
21
The inventors have found that subjects who suffer from breast cancer
and show essentially no HMGCR protein expression generally have poor
survival after endocrine treatment (see Examples, section 4 and the figures).
Consequently, the "cut-off value" determining whether the subject is "HMGCR
high" or "HMGCR low" may be zero.
Thus, in embodiments of the methods of the present disclosure, the
sample value of step b) may be either 1, corresponding to detectable HMGCR
protein in the sample, or 0, corresponding to no detectable HMGCR protein in
the sample. Consequently, in such embodiments, the evaluation of the
sample is digital: HMGCR protein is considered to be either present or not. In
the context of the present disclosure, "no detectable HMGCR protein" refers
to an amount of HMGCR protein that is so small that it is not, during normal
operational circumstances, detectable by a person or an apparatus
performing the method according to any one of the above aspects. The
"normal operational circumstances" refer to the laboratory methods and
techniques a person skilled in the art would find appropriate for performing
the invention.
Accordingly, in embodiments of the methods of the present disclosure,
the reference value of step c) may be 0. And it follows that, in further
embodiments of the methods of the present disclosure, the reference value of
step c) may correspond to a reference sample having no detectable HMGCR
protein (see below).
A sample value of HMGCR protein being higher than the reference
value, or a subject from which such sample value is obtained, is sometimes
referred to herein as "HMGCR protein high". Further, a sample value of
HMGCR protein being lower than, or equal to, the reference value, or a
subject from which such sample value is obtained, is sometimes referred to
herein as "HMGCR protein low".
In the context of the present disclosure, the terms "sample value" and
"reference value" are to be interpreted broadly. The quantification of HMGCR
protein to obtain these values may be done via automatic means, via a
scoring system based on visual or microscopic inspection of samples, or via
combinations thereof. However, it is also possible for a skilled person, such
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
22
as a person skilled in the art of histopathology, to determine the sample and
reference values merely by inspection, e.g., of tissue slides that have been
stained for HMGCR protein expression. The determination of the sample
value being higher than the reference value may thus correspond to the
determination, upon visual or microscopic inspection, that a sample tissue
slide is more densely stained and/or exhibit a larger fraction of stained
cells
than is the case for a reference tissue slide. The sample value may also be
compared to a reference value given by a literal reference, such as a
reference value described in wording or by a reference picture. Consequently,
the sample and/or reference values may in some cases be mental values that
the skilled person determines upon inspection and comparison.
For example, the skilled person may categorize a sample as being
HMGCR protein high or low, wherein the sample is categorized as high if it
contains more HMGCR protein than a previously inspected reference sample
and low if it contains less or equally much. Such evaluation may be assisted
by staining the sample, and, if necessary, a reference sample, with a staining
solution comprising e.g., antibodies selective for HMGCR protein.
A reference value, found to be relevant for making treatment decisions
regarding breast cancer subjects, for use as comparison with the sample
value from the subject, may be provided in various ways. With the knowledge
of the teachings of the present disclosure, the skilled artisan can, without
undue burden, provide relevant reference values for performing the methods
of the present disclosure.
The person performing the methods of the above aspects may, for
example, adapt the reference value to desired information. For example, the
reference value may be adapted to yield the most significant treatment
predictive information, e.g., the largest separation between the HMGCR
protein high survival curve and the HMGCRprotein low survival curve (see
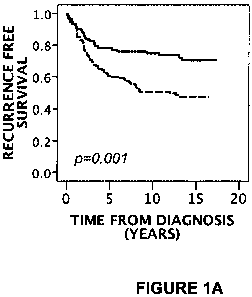
e.g., fig 1). An example of a reference value that may yield a large
separation
is an absent cytoplasmic intensity.
In embodiments of the methods of the above aspects, the reference
value may correspond to the amount of HMGCR protein expression in a
healthy tissue, such as healthy breast tissue, or stroma tissue of the subject
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
23
of the method. As another example, the reference value may be provided by
the amount of HMGCR protein expression measured in a standard sample of
normal tissue from another, comparable subject. As another example, the
reference value may be provided by the amount of HMGCR protein
expression measured in a reference sample comprising tumor cells, such as
a reference sample of tumor tissue, e.g., breast cancer tissue. The amount of
protein expression of the reference sample may preferably be previously
established. Consequently, the reference value may be provided by the
amount of HMGCR protein measured in a reference sample comprising cells
expressing a predetermined amount of HMGCR protein.
Further, the reference value may for example be provided by the
amount of HMGCR protein expression measured in a reference sample
comprising cell lines, such as cancer cell lines, expressing a predetermined,
or controlled, amount of HMGCR protein. The person skilled in the art
understands how to provide such cell lines, for example guided by the
disclosure of Rhodes et al. (2006) The biomedical scientist, p 515-520.
Consequently, in embodiments of the methods of the present
disclosure, the reference value may be a predetermined value corresponding
to the amount of HMGCR protein expression in a reference sample.
However, as discussed further below, the amount of HMGCR protein in
the reference sample does not have to directly correspond to the reference
value. The reference sample may also provide an amount of HMGCR protein
that helps a person performing the method to assess various reference
values. For example, the reference sample(s) may help in creating a mental
image of the reference value by providing a "positive" reference value and/or
a "negative" reference value.
One alternative for the quantification of HMGCR protein expression in
a sample, such as the sample earlier obtained from the subject or the
reference sample, is the determination of the fraction of cells in the sample
that exhibit HMGCR protein expression over a certain level. The fraction may
for example be: a "cellular fraction", wherein the HMGCR protein expression
of the whole cells is taken into account; a "cytoplasmic fraction", wherein
the
HMGCR protein expression of only the cytoplasms of the cells is taken into
CA 02722411 2010-10-22
WO 2009/139681 - PCT/SE2009/000066
24
account; or a "nuclear fraction", wherein the HMGCR protein expression of
only the nuclei of the cells is taken into account. The cytoplasmic fraction
may
for example be classified as < 2 %, 2 - 25 %, > 25 - 75 % or > 75 %
immunoreactive cells of the relevant cell population. The "cytoplasmic
fraction" corresponds to the percentage of relevant cells in a sample that
exhibits a positive staining in the cytoplasm, wherein a medium or distinct
and
strong immunoreactivity in the cytoplasm is considered positive and no or
faint immunoreactivity in the cytoplasm is considered negative. The person
skilled in the art of pathology understands which cells that are relevant
under
the conditions present when performing the method and may determine a
cytoplasmic fraction based on his general knowledge and the teachings of the
present disclosure. The relevant cells may for example be tumor cells.
Further, the skilled artisan understands how to perform corresponding
measurements employing the "cellular fraction" or the "nuclear fraction".
Another alternative for the quantification of HMGCR protein expression
in a sample, such as the sample earlier obtained from the subject or the
reference sample, is the determination of the overall staining intensity of
the
sample. The intensity may for example be: a "cellular intensity", wherein the
HMGCR protein expression of the whole cells is taken into account; a
"cytoplasmic intensity", wherein the HMGCR protein expression of only the
cytoplasms of the cells is taken into account, or a "nuclear intensity",
wherein
the HMGCR protein expression of only the nuclei of the cells is taken into
account. Cytoplasmic intensity is subjectively evaluated in accordance with
standards used in clinical histopathological diagnostics. Outcome of a
cytoplasmic intensity determination may be classified as: absent = no overall
immunoreactivity in the cytoplasms of relevant cells of the sample, weak =
faint overall immunoreactivity in the cytoplasms of relevant cells of the
sample, moderate = medium overall immunoreactivity in the cytoplasms of
relevant cells of the sample, or strong = distinct and strong overall
immunoreactivity in the cytoplasms of relevant cells of the sample. The
person skilled in the art understands which cells that are relevant under the
conditions present when performing the method and may determine a
cytoplasmic intensity based on his general knowledge and the teachings of
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
the present disclosure. The relevant cells may for example be tumor cells.
The determination of cytoplasmic intensity may for example be performed as
described below in the Examples, Section 4, definition of "cytoplasmic
intensity". Further, the skilled artisan understands how to perform
5 corresponding measurements employing the "cellular intensity" or the
"nuclear intensity".
The inventors have found that the cytoplasmic expression of HMGCR
protein is particularly relevant for making the treatment predictions. Thus,
in
embodiments of the methods of the above aspects, the reference value may
10 be a cytoplasmic fraction, a cytoplasmic intensity or a combination
thereof.
Accordingly, the sample value may be a cytoplasmic fraction, a cytoplasmic
intensity or a combination thereof.
Preferably, the sample value and the reference value are both the
same type of value.
15 In embodiments of the methods of the above aspects, the criterion for
the conclusion in step d) is a sample value for the cytoplasmic fraction of
HMGCR protein positive cells, i.e., a "cytoplasmic fraction", which is higher
than 0 %, such as higher than 1 %, such as higher than 2 %, such as higher
than 5 %,such as higher than 10 %, such as higher than 15 %, such as higher
20 than 20 %, such as higher than 25 %, such as higher than 30 %, such as
higher than 35 %, such as higher than 40 %, such as higher than 50 %, such
as higher than 60 %, such as higher than 70 %, such as higher than 75 %,
such as higher than 80 %, such as higher than 90 %.
In alternative or complementing embodiments of the methods of the
25 above aspects, the reference value of step c) is a cytoplasmic fraction of
95
% or lower, such as 90 % or lower, such as 85 % or lower, such as 80 % or
lower, such as 75 % or lower, such as 70 % or lower, such as 65 % or lower,
such as 60 % or lower, such as 55 % or lower, such as 50 % or lower, such
as 45 % or lower, such as 40 % or lower, such as 35 % or lower, such as 30
% or lower, such as 25 % or lower, such as 20 % or lower, such as 15 % or
lower, such as 10 % or lower, such as 5 % or lower, such as 2 % or lower,
such as 1 % or lower, such as 0 %.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
26
The inventors have realized that low cut-off values, such as a value of
0, are particularly relevant for the treatment prediction. However, they have
further noted that the examined tumor samples generally show either a
cytoplasmic fraction of higher than 50 % or a cytoplasmic fraction of 1 % or
lower, wherein the former group is associated with response to tamoxifen.
Thus, if the reference value is a cytoplasmic fraction, it is preferably 50 %
or
lower, such as 25 % or lower, such as 20 % or lower, such as 15 % or lower.
Most preferred is 10 % or lower, such as 5 % or lower, such as 2 % or lower,
such as 1 % or lower, such as 0 %.
Further, in embodiments of the methods of the above aspects, the
criterion for the conclusion in step d) may be a sample value for staining
intensity of a sample, i.e., a cytoplasmic intensity, which is higher than
absent
cytoplasmic intensity, such as higher than weak cytoplasmic intensity, such
as higher than moderate cytoplasmic intensity. In alternative or
complementing embodiments of the methods of the above aspects, the
reference value of step c) may be a moderate cytoplasmic intensity of
HMGCR protein expression or lower, such as a weak cytoplasmic intensity of
HMGCR protein expression or lower, such as an absent cytoplasmic intensity.
In the examples section, the cut-off value "absent" cytoplasmic
intensity is employed. Thus, if the reference value is a cytoplasmic
intensity, it
is preferably low, such as weak or lower. Most preferred is absent.
Further, in embodiments of the methods of the above aspects, the
reference value may be constituted of two values, wherein the criterion for
the
conclusion in step d) is a sample value being higher than any one of these
two values. An example of such a reference value is a cytoplasmic fraction of
25 % or lower and a weak cytoplasmic intensity or lower.
Alternatively, in embodiments of the methods of the above aspects, the
reference value may be a combination of a fraction value and an intensity
value, such as a cytoplasmic fraction value and a cytoplasmic intensity value.
Also, in embodiments of the methods of the above aspects, the
reference value may be a function of a cytoplasmic fraction value and a
cytoplasmic intensity value. For example, such a function may be a staining
score. The "staining score" is calculated as described in Examples, Section 3
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
27
and table 1 below. For example, the reference value may be a staining score
of 2 or lower, such as 1 or lower, such as 0.
The person skilled in the art realizes that other reference values being
an intensity value or a fraction value also fall within the scope of the
present
invention. Likewise, the person skilled in the art realizes that other
combinations of fractions and intensities also fall within the scope of the
present invention. Consequently, the reference value may involve two, and
possibly even more, criteria.
In general, the selection of a cytoplasmic intensity value and/or a
cytoplasmic fraction value as the reference value may depend on the staining
procedure, e.g., on the employed anti-HMGCR antibody and on the staining
reagents.
Guided by the present disclosure, a person skilled in the art, e.g., a
pathologist, understands how to perform the evaluation yielding a fraction,
such as a cellular, cytoplasmic or nuclear fraction, or an intensity, such as
a
cellular, cytoplasmic or nuclear intensity. For example, the skilled artisan
may
use a reference sample comprising a predetermined amount of HMGCR
protein for establishing the appearance of a certain fraction or intensity.
However, a reference sample may not only be used for the provision of
the actual reference value, but also for the provision of an example of a
sample with an amount of HMGCR protein that is higher than the amount
corresponding to the reference value. As an example, in histochemical
staining, such as in immunohistochemical staining, the skilled artisan may use
a reference sample for establishing the appearance of a stained sample
having a high amount of HMGCR protein, e.g., a positive reference.
Subsequently, the skilled artisan may assess the appearances of samples
having lower amounts of HMGCR protein, such as the appearance of a
sample with an amount of HMGCR protein corresponding to the reference
value. In other words, the skilled artisan may use a reference sample to
create a mental image of a reference value corresponding to an amount of
HMGCR protein which is lower than that of the reference sample.
Alternatively, or as a complement, in such assessments, the skilled artisan
may use another reference sample having a low amount of HMGCR protein,
CA 02722411 2010-10-22
WO 2009/139681 f q PCT/SE2009/000066
28
or lacking detectable HMGCR protein, for establishing the appearance of
such sample, e.g., as a "negative reference".
For example, if a reference value of 10 % cytoplasmic fraction is used,
two reference samples may be employed: a first reference sample having no
detectable HMGCR protein, and thus corresponding to a cytoplasmic fraction
of 0, which is lower than the reference value; and a second reference sample
having an amount of HMGCR protein corresponding to a cytoplasmic fraction
of 75 % or higher, which is higher than the reference value.
Consequently, in the evaluation, the skilled artisan may use a
reference sample for establishing the appearance of a sample with a high
amount of HMGCR protein. Such reference sample may be a sample
comprising tissue expressing a high amount of HMGCR protein, such as a
sample comprising breast tumor tissue having a pre-established high
expression of HMGCR protein.
Accordingly, the reference sample may provide an example of a strong
cytoplasmic intensity (CI). With the knowledge of the appearance of a sample
with strong Cl, the skilled artisan may then divide samples into the Cl
categories absent, weak, moderate and strong. This division may be further
assisted by a reference sample lacking detectable HMGCR protein (negative
reference), i.e., a reference sample providing an absent cytoplasmic
intensity.
Also, the reference sample may provide an example of a sample with a
cytoplasmic fraction (CF) of 75 % or higher. With the knowledge of the
appearance of a sample with more than 75 % positive cells, the skilled artisan
may then evaluate the cytoplasmic fraction of other samples having e.g., a
lower percentage of positive cells. This division may be further assisted by a
reference sample essentially lacking HMGCR protein (negative reference),
i.e., a reference sample providing a low CF (e.g., < 5%, such as < 2%), or a
CF of 0.
As mentioned above, cell lines expressing a controlled amount of
HMGCR protein may be used as the reference, in particular as a positive
reference.
On or more pictures may also be provided as the "reference sample".
For example, such a picture may be of an example of a tumor tissue slide
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
29
stained with a certain antibody during certain conditions that show a certain
cellular intensity and/or fraction. The above discussion about the "reference
sample" applies mutatis mutandis to pictures.
Further, the skilled person should recognize that the usefulness of the
methods according to the above aspects is not limited to the quantification of
any particular variant of the HMGCR protein present in the subject in
question, as long as the protein is encoded by the relevant gene and presents
the relevant pattern of expression. As a non-limiting example, the HMGCR
protein comprises, or consists of, a sequence selected from:
i) SEQ ID NO:1; and
ii) a sequence which is at least 85 % identical to SEQ ID NO:1.
In some embodiments, sequence ii) above is at least 90 % identical, at
least 91 % identical, at least 92 % identical, at least 93 % identical, at
least 94
% identical, at least 95 % identical, at least 96 % identical, at least 97 %
identical, at least 98 % identical or at least 99 % identical to SEQ ID NO:1.
As another non-limiting example, the HMGCR protein comprises, or
consists of, a sequence selected from:
i) SEQ ID NO:2; and
ii) a sequence which is at least 85 % identical to SEQ ID NO:2.
In some embodiments, sequence ii) above is at least 90 % identical, at
least 91 % identical, at least 92 % identical, at least 93 % identical, at
least 94
% identical, at least 95 % identical, at least 96 % identical, at least 97 %
identical, at least 98 % identical or at least 99 % identical to SEQ ID NO:2.
The term "% identical", as used in the context of the present disclosure,
is calculated as follows. The query sequence is aligned to the target
sequence using the CLUSTAL W algorithm (Thompson, J.D., Higgins, D.G.
and Gibson, T.J., Nucleic Acids Research, 22: 4673-4680 (1994)). The amino
acid residues at each position are compared, and the percentage of positions
in the query sequence that have identical correspondences in the target
sequence is reported as % identical. Also, the target sequence determines
the number of positions that are compared. Consequently, in the context of
the present disclosure, a query sequence that is shorter than the target
sequence can never be 100 % identical to the target sequence. For example,
a query sequence of 85 amino acids may at the most be 85 % identical to a
target sequence of 100 amino acids.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
In embodiments of the methods of the aspects above, the HMGCR
protein may be detected and/or quantified through the application to the
sample of a detectable and/or quantifiable affinity ligand, which is capable
of
selective interaction with the HMGCR protein. The application of the affinity
5 ligand is performed under conditions that enable binding of the affinity
ligand
to any HMGCR protein in the sample.
To concretize, in embodiments of the methods of the aspects above,
step b) may comprise:
b1) applying to said sample a quantifiable affinity ligand capable of
10 selective interaction with the HMGCR protein to be evaluated, said
application
being performed under conditions that enable binding of said affinity ligand
to
HMGCR protein present in said sample;
b2) removing non-bound affinity ligand; and
b3) quantifying the affinity ligand remaining in association with said
15 sample to evaluate said amount.
"Affinity ligand remaining in association with the sample" refers to
affinity ligand which was not removed in step b2), e.g., the affinity ligand
bound to the sample. Here, the binding may for example be the interaction
between antibody and antigen.
20 However, in some embodiments, the removal of non-bound affinity
ligand according to b2), e.g. the washing, may not be necessary. Thus, in
some embodiments of the methods of the aspects above, step b) may
comprise:
bI) applying to said sample a quantifiable affinity ligand capable of
25 selective interaction with the HMGCR protein to be evaluated, said
application
being performed under conditions that enable binding of said affinity ligand
to
HMGCR protein present in said sample;
bll) quantifying the affinity bound to said sample to evaluate said
amount.
30 It is regarded as within the capabilities of those of ordinary skill in the
art to select or manufacture the proper affinity ligand and to select the
proper
format and conditions for detection and/or quantification, once the connection
between HMGCR protein and treatment prediction for breast cancer is known
through the teaching of the present disclosure. Nevertheless, examples of
affinity ligands that may prove useful, as well as examples of formats and
conditions for detection and/or quantification, are given below for the sake
of
illustration.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
31
Thus, in embodiments of the present disclosure, the affinity ligand may
be selected from the group consisting of antibodies, fragments thereof and
derivatives thereof, i.e., affinity ligands based on an immunoglobulin
scaffold.
The antibodies and the fragments or derivatives thereof may be isolated
and/or mono-specific. Antibodies comprise monoclonal and polyclonal
antibodies of any origin, including murine, rabbit, human and other
antibodies,
as well as chimeric antibodies comprising sequences from different species,
such as partly humanized antibodies, e.g., partly humanized mouse
antibodies. Polyclonal antibodies are produced by immunization of animals
with the antigen of choice. Monoclonal antibodies of defined specificity can
be
produced using the hybridoma technology developed by Kohler and Milstein
(Kohler G and Milstein C (1976) Eur. J. Immunol. 6:511-519). The antibody
fragments and derivatives of the present disclosure are capable of selective
interaction with the same antigen (e.g. HMGCR protein) as the antibody they
are fragments or derivatives of. Antibody fragments and derivatives comprise
Fab fragments, consisting of the first constant domain of the heavy chain
(CH1), the constant domain of the light chain (CL), the variable domain of the
heavy chain (VH) and the variable domain of the light chain (VL) of an intact
immunoglobulin protein; Fv fragments, consisting of the two variable antibody
domains VH and VL (Skerra A and Pluckthun A (1988) Science 240:1038-
1041); single chain Fv fragments (scFv), consisting of the two VH and VL
domains linked together by a flexible peptide linker (Bird RE and Walker BW
(1991) Trends Biotechnol. 9:132-137); Bence Jones dimers (Stevens FJ et al.
(1991) Biochemistry 30:6803-6805); camelid heavy-chain dimers (Hamers-
Casterman C et al. (1993) Nature 363:446-448) and single variable domains
(Cai X and Garen A (1996) Proc. Natl. Acad. Sci. U.S.A. 93:6280-6285;
Masat L et a/. (1994) Proc. Natl. Acad. Sci. U.S.A. 91:893-896), and single
domain scaffolds like e.g., the New Antigen Receptor (NAR) from the nurse
shark (Dooley H et al. (2003) Mol. Immunol. 40:25-33) and minibodies based
on a variable heavy domain (Skerra A and Pluckthun A (1988) Science
240:1038-1041).
SEQ ID NO:1 was designed for immunizations, e.g., designed to lack
transmembrane regions to ensure efficient expression in E. coli, and to lack
any signal peptide, since those are cleaved off in the mature protein.
Consequently, an antibody or fragment or derivative thereof according to the
present disclosure may for example be one that is obtainable by a process
comprising a step of immunizing an animal, such as a rabbit, with a protein
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
32
whose amino acid sequence comprises, preferably consists of, the sequence
SEQ ID NO:1. For example, the immunization process may comprise primary
immunization with the protein in Freund's complete adjuvant. Also, the
immunization process may further comprise boosting at least two times, in
intervals of 2-6 weeks, with the protein in Freund's incomplete adjuvant.
Processes for the production of antibodies or fragments or derivatives thereof
against a given target are known in the art, and may be applied in connection
with this aspect of the present disclosure.
In the context of the present disclosure, a "mono-specific antibody" is
one of a population of polyclonal antibodies which has been affinity purified
on its own antigen, thereby separating such mono-specific antibodies from
other antiserum proteins and non-specific antibodies. This affinity
purification
results in antibodies that bind selectively to its antigen. In the case of the
present disclosure, the polyclonal antisera are purified by a two-step
immunoaffinity based protocol to obtain mono-specific antibodies selective for
the target protein. Antibodies directed against generic affinity tags of
antigen
fragments are removed in a primary depletion step, using the immobilized tag
protein as the capturing agent. Following the first depletion step, the serum
is
loaded on a second affinity column with the antigen as capturing agent, in
order to enrich for antibodies specific for the antigen (see also Nilsson P et
al.
(2005) Proteomics 5:4327-4337).
Polyclonal and monoclonal antibodies, as well as their fragments and
derivatives, represent the traditional choice of affinity ligands in
applications
requiring selective biomolecular recognition, such as in the detection and/or
quantification of HMGCR protein according to the method aspects above.
However, those of skill in the art know that, due to the increasing demand of
high throughput generation of selective binding ligands and low cost
production systems, new biomolecular diversity technologies have been
developed during the last decade. This has enabled a generation of novel
types of affinity ligands of both immunoglobulin as well as non-
immunoglobulin origin that have proven equally useful as binding ligands in
biomolecular recognition applications and can be used instead of, or together
with, immunoglobulins.
The biomolecular diversity needed for selection of affinity ligands may
be generated by combinatorial engineering of one of a plurality of possible
scaffold molecules, and specific and/or selective affinity ligands are then
selected using a suitable selection platform. The scaffold molecule may be of
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
33
immunoglobulin protein origin (Bradbury AR and Marks JD (2004) J. Immunol.
Meths. 290:29-49), of non-immunoglobulin protein origin (Nygren PA and
Skerra A (2004) J. Immunol. Meths. 290:3-28), or of an oligonucleotide origin
(Gold L et al. (1995) Annu. Rev. Biochem. 64:763-797).
A large number of non-immunoglobulin protein scaffolds have been
used as supporting structures in development of novel binding proteins. Non-
limiting examples of such structures, useful for generating affinity ligands
against HMGCR protein for use according to the present disclosure, are
staphylococcal protein A and domains thereof and derivatives of these
domains, such as protein Z (Nord K et al. (1997) Nat. Biotechnol. 15:772-
777); lipocalins (Beste G et al. (1999) Proc. Natl. Acad. Sci. U.S.A. 96:1898-
1903); ankyrin repeat domains (Binz HK et al. (2003) J. Mol. Biol. 332:489-
503); cellulose binding domains (CBD) (Smith GP et al. (1998) J. Mol. Biol.
277:317-332; Lehti6 J et al. (2000) Proteins 41:316-322); y crystallines
(Fiedler U and Rudolph R, WO01/04144); green fluorescent protein (GFP)
(Peelle B et al. (2001) Chem. Biol. 8:521-534); human cytotoxic T
lymphocyte-associated antigen 4 (CTLA-4) (Hufton SE et aL (2000) FEBS
Left. 475:225-231; Irving RA et al. (2001) J. Immunol. Meth. 248:31-45);
protease inhibitors, such as Knottin proteins (Wentzel A et al. (2001) J.
Bacteriol. 183:7273-7284; Baggio R et al. (2002) J. Mol. Recognit. 15:126-
134) and Kunitz domains (Roberts BL et al. (1992) Gene 121:9-15; Dennis
MS and Lazarus RA (1994) J. Biol. Chem. 269:22137-22144); PDZ domains
(Schneider S et al. (1999) Nat. Biotechnol. 17:170-175); peptide aptamers,
such as thioredoxin (Lu Z et al. (1995) Biotechnology 13:366-372; Klevenz B
et al. (2002) Cell. Mol. Life Sci. 59:1993-1998); staphylococcal nuclease
(Norman TC et al. (1999) Science 285:591-595); tendamistats (McConell SJ
and Hoess RH (1995) J. Mol. Biol. 250:460-479; Li R et al. (2003) Protein
Eng. 16:65-72); trinectins based on the fibronectin type III domain (Koide A
et
al. (1998) J. Mol. Biol. 284:1141-1151; Xu L eta!. (2002) Chem. Biol. 9:933-
942); and zinc fingers (Bianchi E et al. (1995) J. Mol. Biol. 247:154-160;
Klug
A (1999) J. Mol. Biol. 293:215-218; Segal DJ et al. (2003) Biochemistry
42:2137-2148).
The above-mentioned examples of non-immunoglobulin protein
scaffolds include scaffold proteins presenting a single randomized loop used
for the generation of novel binding specificities, protein scaffolds with a
rigid
secondary structure where side chains protruding from the protein surface are
randomized for the generation of novel binding specificities, and scaffolds
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
34
exhibiting a non-contiguous hyper-variable loop region used for the
generation of novel binding specificities.
In addition to non-immunoglobulin proteins, oligonucleotides may also
be used as affinity ligands. Single stranded nucleic acids, called aptamers or
decoys, fold into well-defined three-dimensional structures and bind to their
target with high affinity and specificity. (Ellington AD and Szostak JW (1990)
Nature 346:818-822; Brody EN and Gold L (2000) J. Biotechnol. 74:5-13;
Mayer G and Jenne A (2004) BioDrugs 18:351-359). The oligonucleotide
ligands can be either RNA or DNA and can bind to a wide range of target
molecule classes.
For selection of the desired affinity ligand from a pool of variants of any
of the scaffold structures mentioned above, a number of selection platforms
are available for the isolation of a specific novel ligand against a target
protein
of choice. Selection platforms include, but are not limited to, phage display
(Smith GP (1985) Science 228:1315-1317), ribosome display (Hanes J and
Pli ckthun A (1997) Proc. Natl. Acad. Sci. U.S.A. 94:4937-4942), yeast two-
hybrid system (Fields S and Song 0 (1989) Nature 340:245-246), yeast
display (Gai SA and Wittrup KD (2007) Curr Opin Struct Biol 17:467-473),
mRNA display (Roberts RW and Szostak JW (1997) Proc. NatI. Acad. Sci.
U.S.A. 94:12297-12302), bacterial display (Daugherty PS (2007) Curr Opin
Struct Biol 17:474-480, Kronqvist N et al. (2008) Protein Eng Des Sel 1-9,
Harvey BR et al. (2004) PNAS 101(25):913-9198), microbead display (Nord 0
et al. (2003) J Biotechnol 106:1-13, WO01/05808), SELEX (System Evolution
of Ligands by Exponential Enrichment) (Tuerk C and Gold L (1990) Science
249:505-510) and protein fragment complementation assays (PCA) (Remy I
and Michnick SW (1999) Proc. Natl. Acad. Sci. U.S.A. 96:5394-5399).
Thus, in embodiments of the present disclosure, the affinity ligand may
be a non-immunoglobulin affinity ligand derived from any of the protein
scaffolds listed above, or an oligonucleotide molecule.
The HMGCR protein SEQ ID NO:1 was designed to consist of a unique
sequence with low homology with other human proteins and to minimize
cross reactivity of generated affinity reagents. Consequently, in embodiments
of the present disclosure, the affinity ligand may be capable of selective
interaction with a polypeptide consisting of the sequence SEQ ID NO:1.
In Examples below, an antibody binding to an HMGCR epitope having
the sequence SEQ ID NO:4 is employed. SEQ ID NO:4 refers to the amino
acid sequence CKDNPGENARQLAR (i.e. amino acids 827-840 of EnsEMBL
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
entry no. ENSP00000287936). This antibody resulted in strong staining.
Consequently, in further embodiments of the present disclosure, the
quantifiable affinity ligand may be capable of selective interaction with an
HMGCR protein comprising, or consisting of, the sequence SEQ ID NO:4.
5 In some embodiments of present disclosure, an affinity ligand capable
of selective interaction with the HMGCR protein is detectable and/or
quantifiable. The detection and/or quantification of such an affinity ligand
may
be accomplished in any way known to the skilled person for detection and/or
quantification of binding reagents in assays based on biological interactions.
10 Thus, any affinity ligand, as described above, may be used quantitatively
or
qualitatively to detect the presence of the HMGCR protein. These "primary"
affinity ligands may be labeled themselves with various markers or may in
turn be detected by secondary, labeled affinity ligands to allow detection,
visualization and/or quantification. This can be accomplished using any one
15 or more of a multitude of labels, which can be conjugated to the affinity
ligand
capable of interaction with HMGCR protein or to any secondary affinity ligand,
using any one or more of a multitude of techniques known to the skilled
person, and not as such involving any undue experimentation.
Non-limiting examples of labels that can be conjugated to primary
20 and/or secondary affinity ligands include fluorescent dyes or metals (e.g.,
fluorescein, rhodamine, phycoerythrin, fluorescamine), chromophoric dyes
(e.g., rhodopsin), chemiluminescent compounds (e.g., luminal, imidazole) and
bioluminescent proteins (e.g., luciferin, luciferase), haptens (e.g., biotin).
A
variety of other useful fluorescers and chromophores are described in Stryer
25 L (1968) Science 162:526-533 and Brand L and Gohlke JR (1972) Annu. Rev.
Biochem. 41:843-868. Affinity ligands can also be labeled with enzymes (e.g.,
horseradish peroxidase, alkaline phosphatase, beta-lactamase),
radioisotopes (e.g., 3H, 14C 32P 35S or 1251) and particles (e.g., gold). In
the
context of the present disclosure, "particles" refer to particles, such as
metal
30 particles, suitable for labeling of molecules. Further, the affinity
ligands may
also be labeled with fluorescent semiconductor nanocrystals (quantum dots).
Quantum dots have superior quantum yield and are more photostable
compared to organic fluorophores and are therefore more easily detected
(Chan et al. (2002) Cuff Opi Biotech. 13: 40-46). The different types of
labels
35 can be conjugated to an affinity ligand using various chemistries, e.g.,
the
amine reaction or the thiol reaction. However, other reactive groups than
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
36
amines and thiols can be used, e.g., aldehydes, carboxylic acids and
glutamine.
The method aspects above may be put to use in any of several known
formats and set-ups, of which a non-limiting selection is discussed below.
In a set-up based on histology, the detection, localization and/or
quantification of a labeled affinity ligand bound to its HMGCR protein target
may involve visualizing techniques, such as light microscopy or
immunofluoresence microscopy. Other methods may involve the detection via
flow cytometry or luminometry.
A biological sample, such as a tumor tissue sample (biopsy), for
example from breast tissue, which has been removed from the subject may
be used for detection and/or quantification of HMGCR protein. The biological
sample, such as the biopsy, may be an earlier obtained sample. If using an
earlier obtained sample in a method, no steps of the method are practiced on
the human or animal body. The affinity ligand may be applied to the biological
sample for detection and/or quantification of the HMGCR protein. This
procedure enables not only detection of HMGCR protein, but may in addition
show the distribution and relative level of expression thereof.
The method of visualization of labels on the affinity ligand may include,
but is not restricted to, fluorometric, luminometric and/or enzymatic
techniques. Fluorescence is detected and/or quantified by exposing
fluorescent labels to light of a specific wavelength and thereafter detecting
and/or quantifying the emitted light in a specific wavelength region. The
presence of a luminescently tagged affinity ligand may be detected and/or
quantified by luminescence developed during a chemical reaction. Detection
of an enzymatic reaction is due to a color shift in the sample arising from
chemical reaction. Those of skill in the art are aware that a variety of
different
protocols can be modified in order for proper detection and/or quantification.
In embodiments of the methods of the above aspects, a biological
sample may be immobilized onto a solid phase support or carrier, such as
nitrocellulose or any other solid support matrix capable of immobilizing
HMGCR protein present in the biological sample applied to it. Some well-
known solid state support materials useful in the present invention include
glass, carbohydrate (e.g., Sepharose), nylon, plastic, wool, polystyrene,
polyethene, polypropylene, dextran, amylase, films, resins, cellulose,
polyacrylamide, agarose, alumina, gabbros and magnetite. After
immobilization of the biological sample, primary affinity ligand specific to
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
37
HMGCR protein may be applied, e.g., as described in Examples, Sections 3,
4 or 5, of the present disclosure. If the primary affinity ligand is not
labeled in
itself, the supporting matrix may be washed with one or more appropriate
buffers known in the art, followed by exposure to a secondary labeled affinity
ligand and washed once again with buffers to remove unbound affinity
ligands. Thereafter, selective affinity ligands may be detected and/or
quantified with conventional methods. The binding properties for an affinity
ligand may vary from one solid state support to the other, but those skilled
in
the art should be able to determine operative and optimal assay conditions for
each determination by routine experimentation.
Consequently, in embodiments of the methods of the above aspects,
the quantifiable affinity ligand of b1) may be detected using a secondary
affinity ligand capable of recognizing the quantifiable affinity ligand. The
quantification of b3) may thus be carried out by means of a secondary affinity
ligand with affinity for the quantifiable affinity ligand. As an example, the
secondary affinity ligand may be an antibody or a fragment or a derivative
thereof.
As an example, one available method for detection and/or
quantification of the HMGCR protein is by linking the affinity ligand to an
enzyme that can then later be detected and/or quantified in an enzyme
immunoassay (such as an EIA or ELISA). Such techniques are well
established, and their realization does not present any undue difficulties to
the
skilled person. In such methods, the biological sample is brought into contact
with a solid material or with a solid material conjugated to an affinity
ligand
against the HMGCR protein, which is then detected and/or quantified with an
enzymatically labeled secondary affinity ligand. Following this, an
appropriate
substrate is brought to react in appropriate buffers with the enzymatic label
to
produce a chemical moiety, which for example is detected and/or quantified
using a spectrophotometer, fluorometer, luminometer or by visual means.
As stated above, primary and any secondary affinity ligands can be
labeled with radioisotopes to enable detection and/or quantification. Non-
limiting examples of appropriate radiolabels in the present disclosure are 3H,
14C 32P, 35S or 1251. The specific activity of the labeled affinity ligand is
dependent upon the half-life of the radiolabel, isotopic purity, and how the
label has been incorporated into the affinity ligand. Affinity ligands are
preferably labeled using well-known techniques (Wensel TG and Meares CF
(1983) in: Radioimmunoimaging and Radioimmunotherapy (Burchiel SW and
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
38
Rhodes BA eds.) Elsevier, New York, pp 185-196). A thus radiolabeled
affinity ligand can be used to visualize HMGCR protein by detection of
radioactivity in vivo or in vitro. Radionuclear scanning with e.g., gamma
camera, magnetic resonance spectroscopy or emission tomography function
for detection in vivo and in vitro, while gamma/beta counters, scintillation
counters and radiographies are also used in vitro.
As a third aspect of the present disclosure, there is provided a kit for
carrying out a method according to the above aspects, which comprises:
a) a quantifiable affinity ligand capable of selective interaction with an
HMGCR protein; and
b) reagents necessary for quantifying the amount of the affinity ligand.
Various components of the kit according to the third aspect may be
selected and specified as described above in connection with the method
aspects of the present disclosure.
Thus, the kit according to the present disclosure comprises an affinity
ligand against an HMGCR protein, as well as other means that help to
quantify the specific and/or selective affinity ligand after it has bound
specifically and/or selectively to the HMGCR protein. For example, the kit
may contain a secondary affinity ligand for detecting and/or quantifying a
complex formed by the HMGCR protein and the affinity ligand capable of
selective interaction with the HMGCR protein. The kit may also contain
various auxiliary substances other than affinity ligands, to enable the kit to
be
used easily and efficiently. Examples of auxiliary substances include solvents
for dissolving or reconstituting lyophilized protein components of the kit,
wash
buffers, substrates for measuring enzyme activity in cases where an enzyme
is used as a label, target retrieval solution to enhance the accessibility to
antigens in cases where paraffin or formalin-fixed tissue samples are used,
and substances such as reaction arresters, e.g., endogenous enzyme block
solution to decrease the background staining and/or counterstaining solution
to increase staining contrast, that are commonly used in immunoassay
reagent kits.
In embodiments of the kit aspect, the affinity ligand may be selected as
described above in connection with the method aspects.
Further, in accordance with what is described above in connection with
the method aspects, the detectable affinity ligand may in embodiments of the
kit aspect comprise a label selected from the group consisting of fluorescent
dyes and metals, chromophoric dyes, chemiluminescent compounds and
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
39
bioluminescent proteins, enzymes, radioisotopes, particles and quantum dots.
Alternatively, the reagents necessary for quantifying the amount of the
affinity
ligand comprise a secondary affinity ligand capable of recognizing the
quantifiable affinity ligand. As an example, the secondary affinity ligand
capable of recognizing the quantifiable affinity ligand comprises a label
selected from the group consisting of fluorescent dyes or metals,
chromophoric dyes, chemiluminescent compounds and bioluminescent
proteins, enzymes, radioisotopes, particles and quantum dots.
The kit according to the kit aspect may also advantageously comprise
a reference sample for provision of, or yielding, the reference value to be
used for comparison with the sample value. For example, the reference may
sample comprise a predetermined amount of HMGCR protein. Such a
reference sample may for example be constituted by a tissue sample having
the predetermined amount of HMGCR protein. The tissue reference sample
may ,then be used by the person of skill in the art in the determination of
the
HMGCR expression status in the sample being studied, by manual, such as
ocular, or automated comparison of expression levels in the reference tissue
sample and the subject sample. As another example, the reference sample
may comprise cell lines, such as cancer cell lines, expressing a
predetermined, or controlled, amount of HMGCR protein. The person skilled
in the art understands how to provide such cell lines, for example guided by
the disclosure of Rhodes et al. (2006) The biomedical scientist, p 515-520. As
an example, the cell lines may be formalin fixed. Also, such formalin fixed
cell
lines may be paraffin embedded.
The wording "reference sample for provision of the reference value" is
to be interpreted broadly in the context of the present disclosure. The
reference sample may comprise an amount of HMGCR protein actually
corresponding to the reference value, but it may also comprise an amount of
HMGCR protein corresponding to a value being higher than the reference
value. In the latter case, the "high" value may be used by a person performing
the method as an upper reference (positive reference) for assessing, e.g., the
appearance of, a reference value which is lower than the "high" value. The
person skilled in the art of immunohistochemistry understands how to do such
an assessment. Further, as an alternative or a complementing example, the
skilled person may use another reference sample comprising a low amount of
HMGCR protein for provision of a "low" value in such an assessment, e.g., as
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
a negative reference. This is further discussed above in connection with the
method aspects.
Consequently, in embodiments of the kit aspect, the reference sample
may comprise an amount of HMGCR protein corresponding to the reference
5 value. As an example, the reference sample may comprise an amount of
HMGCR protein corresponding to a cytoplasmic fraction of 95 % or lower,
such as 90 % or lower, such as 85 % or lower, such as 80 % or lower, such
as 75 % or lower, such as 70 % or lower, such as 65 % or lower, such as 60
% or lower, such as 55 % or lower, such as 50 % or lower, such as 45 % or
10 lower, such as 40 % or lower, such as 35 % or lower, such as 30 % or lower,
such as 25 % or lower, such as 20 % or lower, such as 15 % or lower, such
as 10 % or lower, such as 5 % or lower, such as 2 % or lower, such as 1 % or
lower, such as 0 %.
As mentioned above, the inventors have realized that low cut-off
15 values, such as a value of 0, are particularly relevant for the treatment
prediction. However, they have further noted that the examined tumor
samples generally show either a cytoplasmic fraction of higher than 50 % or a
cytoplasmic fraction of 1 % or lower, wherein the former group is associated
with response to tamoxifen.
20 Thus, in preferred embodiments, the reference sample may comprise
an amount of HMGCR protein corresponding to a cytoplasmic fraction of 50
% or lower, such as 25 % or lower, such as 20 % or lower, such as 15 % or
lower, such as 10 % or lower, such as 5 % or lower, such as 2 % or lower,
such as 1 % or lower, such as 0 %.
25 Alternatively, or as a complement, the reference sample may comprise
an amount of HMGCR protein corresponding to a moderate cytoplasmic
intensity of HMGCR protein expression or lower, such as a weak cytoplasmic
intensity of HMGCR protein expression or lower. As shown in the attached
figures, a low cytoplasmic intensity, such as an absent cytoplasmic intensity,
30 is a relevant cut-off. Thus, in an embodiment, the reference sample may
comprise an amount of HMGCR protein corresponding to weak cytoplasmic
intensity or lower, such as an absent cytoplasmic intensity.
Further, the reference sample may comprise an amount of HMGCR
protein corresponding to a staining score of 0, 1 or 2, preferably 0.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
41
The provision of cytoplasmic fraction values or cytoplasmic intensity
values is discussed above in connection with the method aspects.
Further, in alternative or complementing embodiments of the kit aspect,
the kit may comprise a reference sample comprising an amount of HMGCR
protein corresponding to a value being higher than the reference value. In
these embodiments, the reference sample may for example comprise an
amount of HMGCR protein corresponding to a cytoplasmic fraction of 75 % or
higher and/or a strong cytoplasmic intensity of nuclear expression.
In other alternative or complementing embodiments of the kit aspect,
the kit may comprise a reference sample comprising an amount of HMGCR
protein corresponding to a value being lower than or equal to the reference
value, e.g., an absent cytoplasmic intensity and/or a cytoplasmic fraction of
<_
1 % HMGCR protein positive cells, such as 0 % HMGCR protein positive
cells.
The kit may thus comprise: a reference sample comprising an amount
of HMGCR protein corresponding to a predetermined reference value; a
reference sample comprising an amount of HMGCR protein corresponding to
a value being higher than a predetermined reference value; and/or a
reference sample comprising an amount of HMGCR protein corresponding to
a value being lower than or equal to a predetermined reference value.
Consequently, embodiments of the kit may comprise: a first reference
sample comprising an amount of HMGCR protein being higher than a
predetermined reference value; and a second reference sample comprising
an amount of HMGCR protein being lower than or equal to the predetermined
reference value.
In embodiments of the kit aspect, the reference sample may be a
tissue sample, such as a tissue sample adapted to ocular or microscopic
evaluation. As an example, the tissue reference sample may be fixated in
paraffin or buffered formalin and/or histo-processed to pm-thin sections that
are mounted on microscopic glass-slides. The tissue reference sample may
be further adapted to staining with affinity ligands, such as antibodies,
against
an HMGCR protein.
Consequently, in embodiments of the kit aspect, the reference sample
may be adapted to directly, or indirectly, provide any relevant reference
value,
such as any one of the reference values discussed above.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
42
Accordingly, further embodiments of the reference sample of the kit
aspect are discussed above in connection with the reference values and
reference samples of the method aspects.
As further discussed above, the combination of a level of HMGCR
protein expression and hormone receptor status may provide information
relevant for drawing conclusions regarding a treatment prediction breast
cancer subject.
For the reasons described above, the "breast cancer" of the third
aspect and the further aspects described below may be a hormone receptor
negative breast cancer, such as an ER- or PR- breast cancer, an ER- breast
cancer, a PR- breast cancer, or an ER- and PR- breast cancer. For example,
the breast cancer may be previously diagnosed as hormone receptor
negative.
Thus, the kit may also include means for establishing the hormone
receptor status of a subject.
Accordingly, in embodiments of the kit aspect, the kit may further
comprise:
a') a quantifiable affinity ligand capable of selective interaction with the
estrogen receptor and b') reagents necessary for quantifying the amount of
such affinity ligand; and/or
a") a quantifiable affinity ligand capable of selective interaction with the
progesterone receptor and b") reagents necessary for quantifying the amount
of such affinity ligand.
The quantifiable affinity ligands of a') and a") are provided for the
determination of the ER status and PR status, respectively.
The reagents of b), b') and b"), may be the same or different.
Quantifiable affinity ligands appropriate for steps a') and a"),
respectively, are well known to the skilled person and commercially available.
Consequently, the kit may include means for provision of the hormonal
status information necessary for drawing conclusions according to some
embodiments of the method aspects of the present disclosure.
Following the findings presented above, the inventors have realized
several uses for the HMGCR protein.
Thus, as a first configuration of a fourth aspect of the present
disclosure, there is provided a use of a HMGCR protein as an endocrine
treatment indicating marker for a mammalian subject having a breast cancer.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
43
In the context of the present disclosure, "endocrine treatment indicating
marker" refers to a something material which presence indicates a suitability
of an endocrine treatment. The marker may thus be a biomarker, such as a
human protein.
As a second configuration of the fourth aspect, there is provided a use
of a HMGCR protein, or an antigenically active fragment thereof, for the
production, selection or purification of an endocrine treatment indicating
agent
for a mammalian subject having a breast cancer.
In the context of the present disclosure, "endocrine treatment indicating
agent" refers to an agent having at least one property being valuable in an
establishment of a treatment prediction for an endocrine treatment, e.g., of a
mammalian subject having a breast cancer. For example, the indicating agent
may be capable of selective interaction with the indicating marker.
The endocrine treatment indicating agent may be an affinity ligand
capable of selective interaction with the HMGCR protein, or an antigenically
active fragment thereof. Examples of such affinity ligands are discussed
above in connection with the method aspects.
Guided by the teachings of the present disclosure, the person skilled in
the art understands how to use HMGCR protein in the production, selection or
purification of the endocrine treatment indicating agent. For example, the use
may comprise affinity purification on a solid support onto which the HMGCR
protein has been immobilized. The solid support may for example be
arranged in a column. Further, the use may comprise selection of affinity
ligands having specificity for the HMGCR protein using a solid support onto
which the polypeptide has been immobilized. Such solid support may be well
plates (such as 96 well plates), magnetic beads, agarose beads or sepharose
beads. Further, the use may comprise analysis of affinity ligands on a soluble
matrix, for example using a dextran matrix, or use in a surface plasmon
resonance instrument, such as a BiacoreTM instrument, wherein the analysis
may for example comprise monitoring the affinity for the immobilized HMGCR
protein of a number of potential affinity ligands.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
44
Also, for the production of the endocrine treatment indicating agent or
the treatment predictive agent, the HMGCR protein may be used in an
immunization of an animal.
Such use may be involved in a method comprising the steps:
i) immunizing an animal using the HMGCR protein as the antigen;
ii) obtaining serum comprising the endocrine treatment indicating
agent from the immunized animal; and, optionally,
iii) isolating the endocrine treatment indicating agent from the
serum.
Alternatively the steps following the first step may be:
ii') obtaining cells from the immunized animal, which cells comprise
DNA encoding the endocrine treatment indicating agent,
iii') fusing the cells with myeloma cells to obtain at least one clone,
and
iv') obtaining the endocrine treatment indicating agent expressed by
the clone.
In embodiments of the fourth aspect, the amino acid sequence of the
HMGCR protein may comprise a sequence selected from:
i) SEQ ID NO:1; and
ii) a sequence which is at least 85 % identical to SEQ ID NO:1.
In some embodiments, sequence ii) is at least 90 % identical, at least
91 % identical, at least 92 % identical, at least 93 % identical, at least 94
%
identical, at least 95 % identical, at least 96 % identical, at least 97 %
identical, at least 98 % identical or at least 99 % identical to SEQ ID NO:1.
Further, in embodiments of the fourth aspect the amino acid sequence
of the HMGCR protein may comprise a sequence selected from:
i) SEQ ID NO:2; and
ii) a sequence which is at least 85 % identical to SEQ ID NO:2.
In some embodiments, sequence ii) is at least 90 % identical, at least
91 % identical, at least 92 % identical, at least 93 % identical, at least 94
%
identical, at least 95 % identical, at least 96 % identical, at least 97 %
identical, at least 98 % identical or at least 99 % identical to SEQ ID NO:2.
As a fifth aspect of the present disclosure, there is provided an affinity
ligand capable of selective interaction with an HMGCR protein.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
Examples of such affinity ligands are discussed above in connection
with the method aspects.
The affinity ligand may be used for in vivo diagnosis, such as in vivo
imaging.
5 Thus, as a first configuration of the fifth aspect, there is provided an
affinity ligand capable of selective interaction with an HMGCR protein, for in
vivo use as an endocrine treatment indicating agent in a mammalian subject
having a breast cancer.
Accordingly, in an embodiment, the affinity ligand may be for use in an
10 in vivo method for establishing a prediction of an outcome of an endocrine
treatment, such as a tamoxifen treatment, of a mammalian subject having a
breast cancer, such as an ER negative breast cancer. The establishment of a
prediction of the outcome of an endocrine treatment of a subject may for
example be a determination of whether the subject is likely to benefit from
the
15 endocrine treatment. In such embodiments the affinity ligand may for
example
be labeled for enabling imaging, i.e. labeled with a detectable label.
Appropriate labels for labeling affinity ligands such as antibodies are well
known to the skilled person. The in vivo method for establishing a treatment
prediction for a mammalian subject having a breast cancer may for example
20 reveal HMGCR protein expression in a tumor in vivo, which in turn may form
the basis of a treatment decision. Various in vivo methods, labels and
detection techniques that may be used in the context of this embodiment are
further discussed above.
In a similar configuration of the fifth aspect, there is provided an affinity
25 ligand, capable of selective interaction with an HMGCR protein, for in vivo
evaluation an amount of HMGCR protein in a subject having a breast cancer.
For example, the level of HMGCR expression in the breast cancer tumor may
be evaluated.
As a sixth aspect of the present disclosure, there is provided a use of
30 an affinity ligand according to the fifth aspect as an endocrine treatment
indicating agent for a mammalian subject having a breast cancer.
Consequently, the affinity ligand may be used for indicating whether a
mammalian subject having a breast cancer would benefit from an endocrine
treatment. Such use may for example be performed in vitro, e.g., involving the
35 determination of the amount of HMGCR in at least part of a sample earlier
obtained from the subject.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
46
In the present disclosure, endocrine treatment, in particular SERM
treatment, such as tamoxifen treatment, is shown to be particularly beneficial
for those subjects who are HMGCR positive (see Examples, section 4 and the
figures). The breast cancer subjects who are HMGCR positive are a
previously unrecognized subgroup in the context of endocrine treatment.
Thus, as a seventh aspect of the present disclosure, there is provided
an endocrine treatment product for use in treatment of a mammalian subject
having a breast cancer, wherein said subject is HMCGR protein positive.
As an eighth aspect of the present disclosure, there is provided a use
of an endocrine treatment product in the manufacture of a medicament for
treatment of a mammalian subject having a breast cancer, wherein said
subject is HMCGR protein positive.
The subject is "HMGCR protein positive" if any HMGCR protein
parameter derived from said subject indicates that the subject is likely to
benefit from an endocrine treatment. For example, the subject may be
considered HMGCR protein positive if a relevant biological sample from the
subject has been found to contain an amount of HMGCR protein
corresponding to a sample value being higher than a relevant reference
value. Relevant sample and reference values are discussed above in
connection with the method aspects. Thus, the breast cancer subject may for
example be considered HMGCR protein positive if a relevant sample, such as
a tissue sample from a primary or secondary tumor, shows detectable
HMGCR protein expression in relevant parts of the sample, such in the tumor
cells. Further, the breast cancer subject may for example be considered
HMGCR protein positive if such sample contains an amount of HMGCR
corresponding to a cytoplasmic intensity which is higher than absent or a
cytoplasmic fraction which is higher than 1 %. From the present disclosure,
the person skilled in the art, such as a pathologist, understands how to
determine whether the subject is HMGCR protein positive of not.
In embodiments of the present disclosure, the "endocrine treatment
product" may be selected from the group consisting of a selective estrogen
receptor modulator (SERM), an aromatase inhibitor, and a steroidal estrogen
receptor antagonist. The SERM may for example be selected from
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
47
toremifene, raloxifene, droloxifene, arzoxifene and tamoxifen. The aromatase
inhibitor may for example be selected from anastrozole, letrozole and
exemestane. The steroidal estrogen receptor antagonist may for example be
fulvestrant. In preferred embodiments, the endocrine treatment product is a
SERM, such as tamoxifen.
Even though HMGCR expression is a powerful (independent) indicator
of response to endocrine treatment in all groups of breast cancer subjects, it
may be considered particularly relevant for the subjects having ER- breast
cancers, since they have traditionally been considered non-responsive to
such treatment.
Thus, in embodiments of the seventh and eighth aspect, the breast
cancer may be ER-.
Tamoxifen has been the mainstay of adjuvant endocrine therapy for
the last 25 years and a wealth of evidence exists supporting its role in the
treatment of breast cancer, irrespective of menopausal status. The results of
a meta-analysis have demonstrated significant reductions in both disease
recurrence (41 %), and breast cancer specific mortality (34%) when
comparing 5 years tamoxifen to no adjuvant treatment (EBCTG: (2005)
Lancet 365:1687-717).
HMGCR protein acts as a rate-limiting enzyme in the mevalonate
pathway. Although cholesterol represents the main product of this pathway, it
also produces a number of non-sterol isoprenoid side products, which have
been shown to be important regulators of angiogenesis, proliferation, and
migration (Liao JK (2002) J Clin Invest 110:285-8, Wejde J, et al (1992)
Anticancer Res 12:317-24).
Despite an ever-growing body of literature describing the anti-
neoplastic properties of statins, epidemiologic data regarding their
preventive
effect against cancer in general and breast cancer in particular remain
inconclusive. Partly in view of the teachings of the present disclosure, the
inventors believe that the mevalonate pathway plays a key role in certain
breast cancers, in particular in ER negative and/or lymph node positive
tumors.
CA 02722411 2010-10-22
WO 2009/139681 - - PCT/SE2009/000066
48
In vitro studies have demonstrated that statin induced mevalonate
depletion results in an adaptive induction of HMGCR protein expression in
chinese hamster ovary cells (Goldstein JL and Brown MS (1990) Nature
343:425-30) and MCF-7 breast cancer cells (Duncan RE, et al (2005) Cancer
Lett 224:221-8). Treatment of MCF-7 cells with mevastatin resulted in a 10- to
15-fold induction of HMGCR activity in association with a 2.5- to 3.5-fold
induction of HMGCR mRNA expression.
Based on the the fact and that statins have been shown to induce
HMGCR protein expression and the finding that increased levels of HMGCR
expression are associated with an improved response to tamoxifen in both
ER positive and ER negative tumors, the inventors conclude that a
combination of an endocrine treatment product and one or more statins is a
new therapeutic option.
Thus, as ninth aspect of the present disclosure, there are provided
products including an endocrine treatment product and a statin as a combined
preparation for simultaneous, separate or sequential use in therapy, such as
breast cancer therapy.
The combination of the two products may be provided in a "kit-of parts"
or an article of manufacture, which may comprise instruction for the
simultaneous, separate or sequential use in therapy, such as breast cancer
therapy.
Thus, as a tenth aspect of the present disclosure, the is a provided a
kit-of-parts including an endocrine treatment product and a statin.
For exmaple, such a kit-of-parts may be for use in therapy, such as
breast cancer therapy.
In embodiments of the ninth or tenth aspect, the breast cancer therapy
may be therapy of a ER- and/or lymph node positive breast cancer.
Further, as an eleventh aspect of the present disclosure, there is
provided a method of treatment of a mammalian subject in need thereof,
wherein said subject has a breast cancer, comprising simultaneous, separate
or sequential administration of a statin and an endocrine treatment product.
In embodiments of the eleventh aspect, the breast cancer may be a
ER- and/or lymph node positive breast cancer.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
Y V V V V Y
49
The statin of the present disclosure may be selected from
lipophilic/hydrophobic statins and hydrophobic statins. The
lipophilic/hydrophobic statins comprise fluvastatin, lovastatin, simvastatin,
atorvastatin and cerivastatin. The hydrophobic statins comprise pravastatin
and rosuvastatin.
Brief description of the figures
With regard to figures 1-7, tumor tissue was scored for high or low
HMGCR level, wherein a high HMGCR level is Cl = weak, moderate and
strong and a low HMGCR level is a Cl = absent. In figure 1-2, 4-5 and 7 a
solid line represents HMGCR high subjects, and a dotted line represents
HMGCR low subjects.
Figure 1 shows the results of a survival analysis based on
immunohistochemical staining of subjects diagnosed with invasive breast
carcinoma. Figure 1a shows recurrence free survival in patients treated with
adjuvant tamoxifen. Figure 1 b shows recurrence free survival in patients who
received no adjuvant endocrine treatment.
Figure 2 shows the results of a survival analysis based on
immunohistochemical staining of ER positive subjects diagnosed with
invasive breast carcinoma. A fraction score of ER > 10 % was considered
positive. Figure 2a shows recurrence free survival in patients treated with
adjuvant tamoxifen. Figure 2b shows recurrence free survival in patients who
received no adjuvant endocrine treatment.
Figure 3 shows the impact on survival if splitting subjects into groups
with different combinations of HMGCR protein expression status and ER
status. Briefly, all subjects.were split into four groups based on HMGCR
status and ER status, i.e. subjects that are HMGCR positive and ER positive,
subjects that are HMGCR positive and ER negative, subjects that are
HMGCR negative and ER positive or subjects that are HMGCR negative and
ER negative. ER positive = fraction score of > 10 % and ER negative =
fraction score of < 10 %. Figure 3a shows recurrence free survival in patients
treated with adjuvant tamoxifen. Figure 3b shows recurrence free survival in
patients who received no adjuvant endocrine treatment.
Figure 4 shows the results of a survival analysis based on
immunohistochemical-staining of subjects diagnosed with invasive breast
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
carcinoma. Figure 4a shows recurrence free survival in of node positive
subjects treated with adjuvant tamoxifen. Figure 4b shows recurrence free
survival in node negative subjects treated with adjuvant tamoxifen.
Figure 5 shows the results of a survival analysis based on
5 immunohistochemical staining of ER positive subjects diagnosed with
invasive breast carcinoma. A fraction score of ER > 10 % was considered
positive. Figure 5a shows recurrence free survival in of node positive
subjects
treated with adjuvant tamoxifen. Figure 5b shows recurrence free survival in
of node negative subjects treated with adjuvant tamoxifen.
10 Figure 6 shows the impact on survival if splitting subjects into groups
with different combinations of HMGCR protein expression status and ER
status. Briefly, all subjects were split into four groups based on HMGCR
status and ER status, i.e. subjects that are HMGCR positive and ER positive,
subjects that are HMGCR positive and ER negative, subjects that are
15 HMGCR negative and ER positive or subjects that are HMGCR negative and
ER negative. ER positive = fraction score of > 10 % and ER negative =
fraction score of > 10 %. Figure 6a shows recurrence free survival in node
positive subjects treated with adjuvant tamoxifen. Figure 6b shows recurrence
free survival in node negative subjects treated with adjuvant tamoxifen.
20 Figure 7 shows the results of a survival analysis based on
immunohistochemical staining of ER negative subjects diagnosed with breast
invasive carcinoma. A fraction score of ER expression > 10 % was
considered positive. Figure 7a shows recurrence free survival in subjects
treated with adjuvant tamoxifen. Figure 7b shows recurrence free survival in
25 subjects treated with adjuvant tamoxifen.
With regard to figures 8-12, tissue cores were scored for high or low
HMGCR level, wherein a high HMGCR level is Cl > absent and a low
HMGCR level is a Cl = absent. Further, a solid line represents a group of
subjects given adjuvant tamoxifen, and a dotted line represents patients who
30 received no adjuvant endocrine treatment.
Figure 8 shows the results of a survival analysis based on
immunohistochemical staining of subjects diagnosed with invasive breast
carcinoma. Figure 8a shows recurrence free survival in HMGCR high
patients. Figure 8b shows recurrence free survival in HMGCR low patients.
35 Figure 9 shows the results of a survival analysis based on
immunohistochemical staining of HMGCR positive and ER negative subjects
diagnosed with invasive breast carcinoma. A fraction score of ER expression
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
FNA r -Jr- LVVJ / U U U U V V
51
> 10 % was considered positive. Figure 9a shows recurrence free survival.
Figure 9b shows recurrence free survival in node positive patients.
Figure 10 shows the results of a survival analysis based on
immunohistochemical staining of subjects diagnosed with invasive breast
carcinoma. The subjects were classified as ER positive or ER negative,
wherein a fraction score of > 10 % was considered positive and a fraction
score of < 10 % was considered negative. Figure 1 Oa shows recurrence free
survival in HMGCR positive or ER positive subjects. Figure 10b shows
recurrence free survival in HMGCR negative and ER negative subjects.
Figure 11 shows the results of a survival analysis based on
immunohistochemical staining of subjects diagnosed with invasive breast
carcinoma. The subjects were classified as PR positive or PR negative,
wherein a fraction score of > 10 % was considered positive and a fraction
score of < 10 % was considered negative. Figure 1 Oa shows recurrence free
survival in HMGCR positive or PR positive subjects. Figure 10b shows
recurrence free survival in HMGCR negative and PR negative subjects.
Figure 12 shows the recurrence free survival of HMGCR positive and
PR negative subjects diagnosed with invasive breast carcinoma. A fraction
score of PR expression > 10 % was considered positive.
Examples
Generation of mono-specific antibodies against HMGCR and use thereof to
detect HMGCR in normal and cancerous samples
1. Generation of antigen
a) Materials and methods
A suitable fragment of the target protein encoded by the EnsEMBL
Gene ID ENSG00000113161 was selected using bioinformatic tools with the
human genome sequence as template (Lindskog M et al. (2005)
Biotechniques 38:723-727, EnsEMBL, www.ensembl.org). The fragment was
used as template for the production of a 140 amino acid long fragment
corresponding to amino acids 742-881 (SEQ ID NO:1) of the HMGCR protein
(SEQ ID NO:2; EnsEMBL entry no. ENSP00000287936).
A fragment of the HMGCR gene transcript containing nucleotides
2274-2693 of EnsEMBL entry number ENST00000287936 (SEQ ID NO:3),
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
52
was isolated by a SuperscriptT"" One-Step RT-PCR amplification kit with
Platinum@ Taq (Invitrogen) and a human total RNA pool panel as template
(Human Total RNA Panel IV, BD Biosciences Clontech). Flanking restriction
sites Notl and Ascl were introduced into the fragment through the PCR
amplification primers, to allow in-frame cloning into the expression vector
(forward primer: ATGGCTGGGAGCATAGGAG, reverse primer:
TCCTTGGAGGTCTTGTAAATTG). Then, the downstream primer was
biotinylated to allow solid-phase cloning as previously described, and the
resulting biotinylated PCR product was immobilized onto Dynabeads M280
Streptavidin (Dynal Biotech) (Larsson M et al. (2000) J. Biotechnol. 80:143-
157). The fragment was released from the solid support by Notl-Ascl
digestion (New England Biolabs), ligated into the pAff8c vector (Larsson M et
al, supra) in frame with a dual affinity tag consisting of a hexahistidyl tag
for
immobilized metal ion chromatography (IMAC) purification and an
immunopotentiating albumin binding protein (ABP) from streptococcal protein
G (Sjolander A et al. (1997) J. Immunol. Methods 201:115-123; Stahl S et al.
(1999) Encyclopedia of Bioprocess Technology: Fermentation, Biocatalysis
and Bioseparation (Fleckinger MC and Drew SW, eds) John Wiley and Sons
Inc., New York, pp 49-63), and transformed into E. coil BL21(DE3) cells
(Novagen). The sequences of the clones were verified by dye-terminator
cycle sequencing of plasmid DNA amplified using TempliPhi DNA sequencing
amplification kit (GE Healthcare, Uppsala, Sweden) according to the
manufacturer's recommendations.
BL21(DE3) cells harboring the expression vector were inoculated in
100 ml 30 g/I tryptic soy broth (Merck KGaA) supplemented with 5 g/I yeast
extract (Merck KGaA) and 50 mg/I kanamycin (Sigma-Aldrich) by addition of 1
ml of an overnight culture in the same culture medium. The cell culture was
incubated in a 1 liter shake flask at 37 C and 150 rpm until the optical
density
at 600 nm reached 0.5-1.5. Protein expression was then induced by addition
of isopropyl-(3-D-thiogalactopyranoside (Apollo Scientific) to a final
concentration of 1 mM, and the incubation was continued overnight at 25 C
and 150 rpm. The cells were harvested by centrifugation at 2400 g, and the
pellet was re-suspended in 5 ml lysis buffer (7 M guanidine hydrochloride, 47
mM Na2HPO4, 2.65 mM NaH2PO4, 10 mM Tris-HCI, 100 mM NaCl, 20 mM 13-
mercaptoethanol; pH = 8.0) and incubated for 2 hours at 37 C and 150 rpm.
After centrifugation at 35300 g, the supernatant containing the denatured and
solubilized protein was collected.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
53
The Hiss-tagged fusion protein was purified by immobilized metal ion
affinity chromatography (IMAC) on columns with 1 ml Talon metal (Co2+)
affinity resin (BD Biosciences Clontech) using an automated protein
purification procedure (Steen J et a/. (2006) Protein Expr. Purif. 46:173-178)
on an ASPEC XL4TM (Gilson). The resin was equilibrated with 20 ml
denaturing washing buffer (6 M guanidine hydrochloride, 46.6 mM Na2HPO4,
3.4 mM NaH2PO4, 300 mM NaCl, pH 8.0-8.2). Clarified cell lysates were then
added to the column. Thereafter, the resin was washed with a minimum of
31.5 ml washing buffer prior to elution in 2.5 ml elution buffer (6 M urea, 50
mM NaH2PO4, 100 mM NaCl, 30 mM acetic acid, 70 mM Na-acetate, pH 5.0).
The eluted material was fractioned in three pools of 500, 700 and 1300 pl.
The 700 pl fraction, containing the antigen, and the pooled 500 and 1300 pl
fractions were stored for further use.
The antigen fraction was diluted to a final concentration of 1 M urea
with phosphate buffered saline (PBS; 1.9 mM NaH2PO4, 8.1 mM Na2HPO4,
154 mM NaCI) followed by a concentration step to increase the protein
concentration using Vivapore 10/20 ml concentrator with molecular weight cut
off at 7500 Da (Vivascience AG). The protein concentration was determined
using a bicinchoninic acid (BCA) micro assay protocol (Pierce) with a bovine
serum albumin standard according to the manufacturer's recommendations.
The protein quality was analyzed on a Bioanalyzer instrument using the
Protein 50 or 200 assay (Agilent Technologies).
b) Results
A gene fragment corresponding to nucleotides 2274-2693 of the long
transcript (SEQ ID NO:3) of the HMGCR gene and encoding a peptide (SEQ
ID NO:1) consisting of amino acids 742 to 881 of the target protein HMGCR
(SEQ ID NO:2) was successfully isolated by RT-PCR from a human RNA
pool using primers specific for the protein fragment. The 140 amino acid
fragment (SEQ ID NO:1) of the target protein (SEQ ID NO:2) was designed to
lack transmembrane regions to ensure efficient expression in E. coli, and to
lack any signal peptide, since those are cleaved off in the mature protein. In
addition, the protein fragment was designed to consist of a unique sequence
with low homology with other human proteins, to minimize cross reactivity of
generated affinity reagents, and to be of a suitable size to allow the
formation
of conformational epitopes and still allow efficient cloning and expression in
bacterial systems.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
54
A clone encoding the correct amino acid sequence was identified, and,
upon expression in E. coli, a single protein of the correct size was produced
and subsequently purified using immobilized metal ion chromatography. After
dilution of the eluted sample to a final concentration of 1 M urea and
concentration of the sample to 1 ml, the concentration of the protein fragment
was determined to be 11.7 mg/ml and was 84.2 % pure according to purity
analysis.
2. Generation of antibodies
a) Materials and methods
The purified HMGCR fragment as obtained above was used as antigen
to immunize a rabbit in accordance with the national guidelines (Swedish
permit no. A 84-02). The rabbit was immunized intramuscularly with 200 pg of
antigen in Freund's complete adjuvant as the primary immunization, and
boosted three times in four week intervals with 100 pg antigen in Freund's
incomplete adjuvant.
Antiserum from the immunized animal was purified by a three-step
immunoaffinity based protocol (Agaton C et al. (2004) J. Chromatogr. A
1043:33-40; Nilsson P et al. (2005) Proteomics 5:4327-4337). In the first
step,
7 ml of total antiserum was buffered with 1 Ox PBS to a final concentration of
1x PBS (1.9 mM NaH2PO4, 8.1 mM Na2HPO4, 154 mM NaCI), filtered using a
0.45 pm pore-size filter (Acrodisc , Life Science) and applied to an affinity
column containing 5 ml N-hydroxysuccinimide-activated SepharoseTM 4 Fast
Flow (GE Healthcare) coupled to the dual affinity tag protein His6-ABP (a
hexahistidyl tag and an albumin binding protein tag) expressed from the
pAff8c vector and purified in the same way as described above for the antigen
protein fragment. In the second step, the flow-through, depleted of antibodies
against the dual affinity tag His6-ABP, was loaded at a flow rate of 0.5
ml/min
on a 1 ml Hi-Trap NHS-activated HP column (GE Healthcare) coupled with
the HMGCR protein fragment used as antigen for immunization (SEQ ID
NO:1). The His6-ABP protein and the protein fragment antigen were coupled
to the NHS activated matrix as recommended by the manufacturer. Unbound
material was washed away with 1x PBST (1x PBS, 0.1 % Tween20, pH 7.25),
and captured antibodies were eluted using a low pH glycine buffer (0.2 M
glycine, 1 mM EGTA, pH 2.5). The eluted antibody fraction was collected
automatically, and loaded onto two 5 ml HiTrapTM desalting columns (GE
Healthcare) connected in series for efficient buffer exchange in the third
step.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
The second and third purification steps were run on the AKTAxpressTM
platform (GE Healthcare). The antigen selective (mono-specific) antibodies
(msAbs) were eluted with PBS buffer, supplemented with glycerol and NaN3
to final concentrations of 40 % and 0.02 %, respectively, for long term
storage
5 at -20 C (Nilsson P et al. (2005) Proteomics 5:4327-4337).
The specificity and selectivity of the affinity purified antibody fraction
were analyzed by binding analysis against the antigen itself and against 383
other human protein fragments in a protein array set-up (Nilsson P et al.
(2005) Proteomics 5:4327-4337). The protein fragments were diluted to 40
10 pg/ml in 0.1 M urea and 1 x PBS (pH 7.4) and 50 pl of each were transferred
to the wells of a 96-well spotting plate. The protein fragments were spotted
in
duplicate and immobilized onto epoxy slides (SuperEpoxy, TeleChem) using
a pin-and-ring arrayer (Affymetrix 427). The slide was washed in 1 x PBS (5
min) and the surface was then blocked (SuperBlock , Pierce) for 30 minutes.
15 An adhesive 16-well silicone mask (Schleicher & Schuell) was applied to the
glass before the mono-specific antibodies were added (diluted 1:2000 in 1x
PBST to appr. 50 ng/ml) and incubated on a shaker for 60 min. Affinity tag-
specific IgY antibodies were co-incubated with the mono-specific antibodies in
order to quantify the amount of protein in each spot. The slide was washed
20 with 1x PBST and 1x PBS twice for 10 min each. Secondary antibodies (goat
anti-rabbit antibody conjugated with Alexa 647 and goat anti-chicken antibody
conjugated with Alexa 555, Molecular Probes) were diluted 1:60000 to 30
ng/ml in 1 x PBST and incubated for 60 min. After the same washing
procedure, as for the first incubation, the slide was spun dry and scanned
25 (G2565BA array scanner, Agilent), thereafter images were quantified using
image analysis software (GenePix 5.1, Axon Instruments).
b) Results
The quality of polyclonal antibody preparations has proven to be
30 dependent on the degree of stringency in the antibody purifications, and it
has
previously been shown that depletion of antibodies directed against epitopes
not originated from the target protein is necessary to avoid cross-reactivity
to
other proteins and background binding (Agaton C et al. (2004) J. Chromatogr.
A 1043:33-40). Thus, a protein microarray analysis was performed to ensure
35 that mono-specific polyclonal antibodies of high specificity had been
generated by depletion of antibodies directed against the His6-tag as well as
of antibodies against the ABP-tag.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
56
To quantify the amount of protein in each spot of the protein array, a
two-color dye labeling system was used, with a combination of primary and
secondary antibodies. Tag-specific IgY antibodies generated in hen were
detected with a secondary goat anti-hen antibody labeled with Alexa 555
fluorescent dye. The specific binding of the rabbit msAb to its antigen on the
array was detected with a fluorescently Alexa 647 labeled goat anti-rabbit
antibody. The protein array analysis showed that the affinity purified mono-
specific antibody against HMGCR is highly selective to the correct protein
fragment and has a very low background to all other protein fragments
analyzed on the array.
3. Tissue profiling by immunohistochemistry
a) Material and methods
In total, 576 paraffin cores containing human tissues were analyzed
using the mono-specific antibody sample obtained in Examples, section 2. All
tissues used as donor blocks for tissue microarray (TMA) production were
selected from the archives at the Department of Pathology, University
Hospital, Uppsala, in agreement with approval from the local ethical
committee. All tissue sections used for TMA analysis were examined to
determine diagnosis and to select representative areas in donor blocks.
Normal tissue was defined as microscopically normal (non-neoplastic) and
was most often selected from specimens collected from the vicinity of
surgically removed tumors. Cancer tissue was reviewed for diagnosis and
classification. All tissues were formalin fixated, paraffin embedded, and
sectioned for diagnostic purposes.
The TMA production was performed essentially as previously
described (Kononen J et al. (1998) Nature Med. 4:844-847; Kallioniemi OP et
al. (2001) Hum. Mol. Genet. 10:657-662). Briefly, a hole was made in the
recipient TMA block and a cylindrical core tissue sample from the donor block
was acquired and deposited in the recipient TMA block. This was repeated in
an automated tissue arrayer from Beecher Instrument (ATA-27, Beecher
Instruments, Sun Prairie, CA, USA) until a complete TMA design was
produced. TMA recipient blocks were baked at 42 C for 2 h prior to
sectioning.
The design of TMA:s was focused on obtaining samples from a large
range of representative normal tissues, and on including representative
cancer tissues. This has previously been described in detail in Kampf C et al.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
57
(2004) Clin. Proteomics 1:285-300. In brief, samples from 48 normal tissues
and from 20 of the most common cancer types affecting humans were
selected. In total, eight different designs of TMA blocks, each containing 72
cores of tissue with 1 mm diameter, were produced. Two of the TMA:s
represented normal tissues, corresponding to 48 different normal tissues in
triplicates from different individuals. The remaining 6 TMA:s represented
cancer tissue from 20 different types of cancer. For 17 of the 20 cancer
types,
12 individually different tumors were sampled, and for the remaining 3 cancer
types, 4 individually different tumors were sampled, all in duplicates from
the
same tumor. The TMA blocks were sectioned with 4 pm thickness using a
waterfall microtome (Leica), and placed onto SuperFrost (Roche Applied
Science) glass slides for IHC analysis.
Automated IHC was performed as previously described (Kampf C et al.
(2004) Clin. Proteomics 1:285-300). In brief, the glass slides were incubated
for 45 min in 60 C, de-paraffinized in xylene (2 x 15 min) and hydrated in
graded alcohols. For antigen retrieval, slides were immersed in TRS (Target
Retrieval Solution, pH 6.0, DakoCytomation) and boiled for 4 min at 125 C in
a Decloaking chamber (Biocare Medical). Slides were placed in the
Autostainer (DakoCytomation) and endogenous peroxidase was initially
blocked with H202 (DakoCytomation). The slides were incubated for 30 min at
room temperature with the primary antibody obtained as in Examples, Section
2, followed by incubation for 30 min at room temperature with goat anti-rabbit
peroxidase conjugated Envision . Between all steps, slides were rinsed in
wash buffer (DakoCytomation). Finally, diaminobenzidine (DakoCytomation)
was used as chromogen and Harris hematoxylin (Sigma-Aldrich) was used for
counterstaining. The slides were mounted with Pertex (Histolab).
All immunohistochemically stained sections from the eight different
TMA:s were scanned using a ScanScope T2 automated slide-scanning
systems (Aperio Technologies). In order to represent the total content of the
eight TMA:s, 576 digital images were generated. Scanning was performed at
20 times magnification. Digital images were separated and extracted as
individual tagged image file format (TIFF) files for storage of original data.
In
order to be able to handle the images in a web-based annotation system, the
individual images were compressed from TIFF format into JPEG format. All
images of immunohistochemically stained tissue were manually evaluated
under the microscope and annotated by a certified pathologist or by specially
educated personnel, subsequently verified by a pathologist.
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
58
Annotation of each different normal and cancer tissue was performed
using a simplified scheme for classification of IHC outcome. Each tissue was
examined for representativity and immunoreactivity. The different tissue
specific cell types included in each normal tissue type were annotated. For
each cancer, tumor cells and stroma were annotated. Basic annotation
parameters included an evaluation of i) subcellular localization (nuclear
and/or cytoplasmic/membranous), ii) staining intensity (SI) and iii) fraction
of
stained cells (FSC). Staining intensity was subjectively evaluated in
accordance to standards used in clinical histo-pathological diagnostics and
outcome was classified as: absent = no immunoreactivity, weak = faint
immunoreactivity, moderate = medium immunoreactivity or strong = distinct
and strong immunoreactivity. The fraction of stained cells was estimated and
classified as < 2 %, 2 - 25 %, > 25 - 75 % or > 75 % immunoreactive cells of
the relevant cell population. Based on both the intensity and fraction of
immunoreactive cells, a "staining score" was given for each tissue sample: 0
= negative, 1 = weak, 2 = moderate and 3 = strong. N.R. means that no
representative tissues were present. In detail, the staining score was given
according to the following criteria: 0 was given if SI = absent or weak and
FSC <_ 25%; 1 was given if SI = weak and FSC > 25% or if SI = moderate and
FSC <_ 25%; 2 was given if SI = moderate and FSC > 25% or if SI = strong
and FSC <_ 25% and SI = moderate; and finally 3 was given if SI = strong and
FSC > 25%. See also table 1. The skilled artisan should recognize that this
procedure is similar to a calculation of an Allred score, see e.g., Allred et
al.
(1998) Mod Pathol 11(2), 155.
Table 1: Staining score
Staining score Staining intensity Fraction of stained cells
0 absent < 2%
0 absent 2 - 25%
0 absent > 25 - 75%
0 absent > 75%
0 weak < 2%
0 weak 2 - 25%
1 weak >25-75%
1 weak > 75%
1 moderate < 2%
1 moderate 2 - 25%
2 moderate > 25 - 75%
2 moderate > 75%
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
59
2 strong < 2%
2 strong 2 - 25%
3 strong >25-75%
3 strong > 75%
b) Results
The results from tissue profiling with the mono-specific antibody
generated towards a recombinant protein fragment of the human target
protein HMGCR obtained as in Examples, Section 2 showed a particular
immunoreactivity in several normal tissues. Table 2 shows the HMGCR
protein expression pattern in normal human tissues. Using IHC and TMA
technology, 144 spots (1 mm in diameter) representing 48 different types of
normal tissue were screened for expression of HMGCR. Immunoreactivity
was observed mainly in cytoplasm of most tissues. Some cases showed
additional nuclear or membranous positivity. Strongest staining was found in
glandular epithelia. In a few cases no representative tissue (N.R.) were
observed.
Table 2: Expression pattern of HMGCR in normal tissues
Tissue type Cell type Stainin
score
Adrenal gland cortical cells 3
Appendix glandular cells 3
I m hoid tissue 1
Bone marrow bone marrow poietic cells 3
Breast glandular cells 2
Bronchus respiratory epithelial cells 2
Cerebellum cells in granular layer 2
cells in molecular layer 3
purkinje cells 3
Cerebral cortex glial cells 0
neuronal cells 3
Cervix, uterine glandular cells 3
squamous epithelial cells 1
Colon glandular cells 3
Corpus, uterine 1 cells in endometrial stroma 1
glandular cells 3
Corpus, uterine 2 cells in endometrial stroma 1
glandular cells 3
Duodenum glandular cells 3
E idid mis glandular cells 2
Esophagus squamous epithelial cells 2
SUBSTITUTE SHEET (RULE 26)
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
Fallopian tube glandular cells 2
Gall bladder glandular cells 3
Heart muscle m oc tes 2
Hi ocam us glial cells 0
neuronal cells 2
Kidney cells in lomeruli 0
cells in tubules 2
Lateral ventricle glial cells 0
neuronal cells 1
Liver bile duct cells 1
he atoc tes 1
Lung alveolar cells 2
macrophages 1
Lymph node lymphoid cells outside reaction centra 2
reaction center cells 2
Naso ha nx respiratory epithelial cells 2
Oral mucosa squamous epithelial cells 2
Ovary follicle cells N.R.
ovarian stromal cells 1
Pancreas exocrine glandular cells 3
islet cells 1
Parathyroid gland glandular cells 3
Placenta decidual cells 1
tro hoblastic cells 2
Prostate glandular cells 2
Rectum glandular cells 3
Salivary gland glandular cells 2
Seminal vesicle glandular cells 2
Skeletal muscle m oc tes 2
Skin adnexal cells N.R.
e idermal cells 2
Small intestine glandular cells 3
Smooth muscle smooth muscle cells 0
Soft tissue I mesenchymal cells 2
Soft tissue 2 mesench mal cells 2
Spleen cells in red pulp 2
cells in white pulp 2
Stomach 1 glandular cells 3
Stomach 2 glandular cells 3
Testis cells in seminiferus ducts 1
le di cells 3
Thyroid gland glandular cells 2
Tonsil I m hoid cells outside reaction centra 2
reaction center cells 1
squamous epithelial cells 2
Urinary bladder urothelial cells 2
Vagina squamous epithelial cells 1
SUBSTITUTE SHEET (RULE 26)
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
61
Vulva/anal skin squamous epithelial cells 2
HMGCR protein expression was further evaluated in tissue samples
from various cancer types. Table 3 shows the level of HMGCR expression in
12 different breast carcinoma tissues samples. All of these samples showed
representative tissue and ten showed positivity, i.e., a staining score of
higher
than zero. HMGCR expression was observed in cytoplasm.
Table 3: Expression pattern of HMGCR
Tissue sample 1 2 3 4 5 6 7 8 9 10 11 12
Staining score 3 2 2 1 1 1 1 1 1 1 0 0
4. Randomized premenopausal cohort TMA
a) Material and methods
Between 1984 and 1991, 564 pre-menopausal women with primary
breast cancer in the South and Southeast regions of Sweden were enrolled in
a multi-centre clinical trial and randomly assigned to either two years of
adjuvant tamoxifen (n=276) or a control group (n=288). The control group did
not receive adjuvant endocrine treatment, i.e. tamoxifen. The inclusion
criteria
were pre-menopausal patients, or patients younger than 50 years, with stage
II (pT2 NO MO, pT1-2 Ni MO) invasive breast cancer treated by modified
mastectomy or breast conserving surgery with axillary lymph node dissection.
Post-operative radiotherapy (50 Gy) was administered after breastconserving
surgery and all lymph node-positive patients received locoregional
radiotherapy. Less than 2% of the patients received adjuvant systemic
chemotherapy. The median follow-up time for patients without breast cancer
events was 13.9 years. Median age at diagnosis was 45 (25-57) years. The
study design is described in detail elsewhere (Ryden L et al, Eur J Cancer,
2005, 41(2): 256-64). Ethical permission was obtained from the Local Ethics
Committees at Lund and Linkoping Universities.
Tissue blocks from 500 of the 564 patients could be retrieved for TMA-
construction. All cases had been histopathologically re-evaluated on
hematoxylin and eosin stained slides. TMA:s were constructed by sampling 2
x 0.6 mm cores per case from areas representative of invasive cancer, using
an manual arraying device (MTA-1, Beecher Inc, WI, USA). Four pm sections
were dried, deparaffinized an pretreated using the PT-link system (DAKO,
Copenhagen, Denmark), then stained in a Techmate 500 (DAKO,
Copenhagen, Denmark) with a polyclonal anti-HMGCR antibody (Catalog #
SUBSTITUTE SHEET (RULE 26)
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
62
07-457, Upstate) diluted1:250, which antibody selectively interacts with a
peptide consisting of the sequence SEQ ID NO:4. IHC staining of ER and PR
had been performed previously. In line with current clinical praxis, a cut-off
at
% positive nuclei was used to define hormone receptor positivity.
5 For statistical analyses, the cytoplasmic intensity (CI) level was
evaluated, in line with what is described in Examples, Section 3 above. The
level of staining intensity of the cytoplasm was subjectively evaluated in
accordance to standards used in clinical histo-pathological diagnostics and
outcome was classified as: absent = no immunoreactivity, weak = faint
10 immunoreactivity, moderate = medium immunoreactivity or strong = distinct
and strong immunoreactivity. Based on the survival trends for all different
strata, a dichotomized variable were constructed for further statistical
analyses. Two categories were defined: "HMGCR high" corresponding to Cl =
weak, moderate and strong and "HMGCR low" corresponding to Cl = absent
This classification of samples was used for RFS analysis according to the
Kaplan-Meier estimator, and the log-rank test was used to compare survival
in different strata. ER and PR negativity was defined as < 10% positively
staining nuclei, according to current clinical guidelines in Sweden. All
statistical tests were two-sided, and p-values of < 0.05 were considered
significant. All calculations were made with the statistical package SPSS 17.0
(SPSS Inc. Illinois, USA).
b) Results
In this results section, "HMGCR" refers to HMGCR protein. For the
present study, immunohistochemical analysis of HMGCR expression could be
performed on 423 tumors. The remaining cores either did not contain invasive
cancer or had been lost during histoprocessing. Of the 423 analyzed tumors,
324 were ER positive (i.e. ERa+), defined as >10% ER nuclear fraction,
which is in line with the clinically established cut-off used for hormone
receptor assessment.
Analysis of recurrence free survival (RFS) based on high or low
HMGCR protein levels in patients treated with tamoxifen and a control group
of patients that received no adjuvant endocrine therapy are presented in fig
1 a and 1 b, respectively. This analysis reveals a significantly better RFS
for
the HMGCR high category than for the HMGCR low category as seen in
figure 1 a (p = 0.001).
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
63
Analysis of RFS based on high or low HMGCR levels in ER positive
patients of the tamoxifen treated patients and the control group are presented
in fig 2a and 2b, respectively. This analysis reveals a significantly better
RFS
for the HMGCR high category of the tamoxifen treated subjects as seen in
figure 2a.
In light of the apparent influence of HMGCR on tamoxifen response in
this cohort, the impact on RFS in strata with different combinations of
HMGCR protein expression status and ER status were analyzed. Briefly, all
subjects were split into four groups based on HMGCR and ER status, i.e.
subjects that are HMGCR positive and ER positive, subjects that are HMGCR
positive and ER negative, subjects that are HMGCR negative and ER positive
and subjects that are HMGCR negative and ER negative. The analysis
revealed that these strata were associated with differences in RFS in the
tamoxifen treated cohort (p = 0.001) (fig 3a). However very small differences
between the strata in the untreated cohort were observed (fig 3b).
Surprisingly as illustrated in figure 3a, HMGCR positive and ER negative
patients have a better outcome than HMGCR negative and ER positive
patients in the treated cohort.
Thus, a favorable effect of HMGCR expression on tamoxifen response
has been demonstrated. Further, a group of ER negative patients who may
respond to tamoxifen is identified. Consequently, HMGCR protein is an
endocrine treatment marker which may be an alternative or complement to
ER.
Next, the relationship between HMGCR expression and RFS in lymph
node positive patients treated with tamoxifen was investigated. This revealed
that HMGCR expression was associated with an improved RFS (p = 0.001) in
lymph node positive patients who received tamoxifen (fig 4a). Similar results
were obtained when analyzing the ER positive subjects of the same subgroup
(fig 5a). A trend of better survival for the HMGCR positive subjects of the
node negative subgroups was also observed (fig 4b and 5b), but these
subgroups contained too few patients to yield statistically significant
results.
The impact on RFS based on node status in strata with different combinations
of HMGCR expression and ER status in tamoxifen treated subjects was
analyzed. In node positive, tamoxifen treated subjects, HMGCR/ER status
had a highly significant impact on RFS (p = 0.000) (fig 6a), compared to
lymph node negative tamoxifen treated patients (fig 6b). Regarding the node
positive subjects, HMGCR positive and ER negative patients had a better
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
64
outcome than HMGCR negative and ER positive patients in the treated cohort
(fig 6a).
The impact on survival for tamoxifen treated subjects that are HMGCR
positive and ER negative subjects is seen in more detail in figure 7. The
positive effect on RFS based on the HMGCR level in ER negative subjects is
seen in figure 7a, and that effect appears to be further enhanced in lymph
node positive patients is seen in figure 7b.
If the parameters were changed, and the RFS analysis was based on
tamoxifen treatment of HMGCR high or low subjects, irrespective of ER
status, the results revealed a significantly improved survival upon tamoxifen
treatment for patients with HMGCR high tumors (fig 8a) in contrast to patients
with HMGCR low tumors, where tamoxifen treatment had no impact on
survival (fig 8b).
An improved survival upon tamoxifen treatment was also observed
when analyzing HMGCR positive and ER negative subjects as seen in fig 9a.
That trend appears to be further enhanced in lymph node positive patients is
seen in figure 9b. The results of fig 8 and 9 further support that HMGCR
positive subjects benefit from tamoxifen treatment and that such benefit also
exists in HMGCR positive subjects in the subgroup of ER negative patients,
which have previously been considered non-responsive to tamoxifen.
When the breast cancer subjects were divided into HMGCR positive or
ER positive and HMGCR negative and ER negative, respectively, a
significantly positive effect of tamoxifen treatment was observed in the
former
group, while no positive effect was observed in the latter (10a and 10b,
respectively).
In the literature, PR has been proposed as an alternative or
complement to ER in endocrine treatment prediction. Accordingly, its relation
to HMGCR status has been investigated. PR positive and HMGCR positive
subjects showed significant response to tamoxifen treatment, while no
response was observed in the group of PR negative and HMGCR negative
subjects (11 a and 11 b, respectively). In the group of HMGCR positive and PR
negative subjects a trend of tamoxifen impact on survival may be observed
(fig 12). Probably due to too few patients in this group the trend is not
statistically significant.
Determination of whether a breast cancer patient is likely to benefit from an
endocrine treatment and treatment of said patient
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
5) A non-limiting example
A breast cancer patient can present symptoms or signs such as a
palpable lump/tumor, secretion from the mammilla or skin deformities. A
5 proportion of breast cancers, generally without symptoms, are also detected
by screening mammography. If those tests are not conclusive for diagnosis, a
breast biopsy may be performed i.e. removal of a selected physical piece of
tissue from the suspected tumor.
Following the diagnosis of breast cancer, the tumor is normally
10 removed by surgery (e.g. by mastectomy or partial mastectomy).
To perform the treatment predictive method, a tumor tissue sample is
obtained. The tumor tissue sample may be obtained from a specimen from an
earlier surgical removal of the tumor or from a biopsy performed earlier
during
the diagnosis of the cancer.
15 For the provision of a reference sample showing a negative HMGCR
staining (negative reference), a material is taken from a tissue having no
HMGCR expression, e.g. archival material comprising tissue lacking
detectable HMGCR protein expression. The negative reference may show an
absent cytoplasmic intensity.
20 For the provision of a reference sample showing a high HMGCR
staining (positive reference), a material is taken from a tissue having high
HMGCR expression, e.g. breast cancer tissue having a pre-established high
HMGCR protein expression. The positive reference may show a strong
cytoplasmic intensity.
25 The materials (the tumor tissue sample and the reference samples) are
fixated in buffered formalin and histo-processed in order to obtain thin
sections (4 pm). Alternatively, the reference samples may be preprepared,
e.g. already fixated in buffered formalin or already mounted on slides (see
below).
30 Immunohistochemistry is performed as described in Examples, section
3. One or more sample sections from each sample are mounted on glass
slides that are incubated for 45 min in 60 C, de-paraffinized in xylene (2 x
15
min) and hydrated in graded alcohols. For antigen retrieval, the slides are
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
66
immersed in TRS (Target Retrieval Solution, pH 6.0, DakoCytomation) and
boiled for 4 min at 125 C in a Decloaking chamber (Biocare Medical). Then,
the slides are placed in the Autostainer (DakoCytomation) and endogenous
peroxidase is initially blocked with H202 (DakoCytomation). The reason for
mounting multiple sample sections may be to increase the accuracy of the
results.
A primary HMGCR specific antibody (e.g. the anti-HMGCR antibody of
Examples, section 2 or 4) is added to the slides, which are then incubated for
30 min in room temperature, followed incubation for 30 min in room
temperature with the labeled secondary antibody (e.g. goat-anti-rabbit
peroxidase conjugated Envision ). To detect the secondary antibody,
diaminobenzidine (DakoCytomation) is used as chromogen, contrasted with a
Harris hematoxylin (Sigma-Aldrich) counterstaining. Between all steps, slides
are rinsed in wash buffer (DakoCytomation). The slides are then mounted
with Pertex (Histolab).
As a tool to validate the staining procedure, two control cell-lines may
be used; e.g. one slide with cells expressing HMGCR (positive cell line) and
one slide with cells without HMGCR expression (negative cell line). The
skilled artisan understands how to provide such cell lines, for example guided
by the disclosure of Rhodes et al. (2006) The biomedical scientist, p 515-520.
The control-line slides may be simultaneously stained in the same procedure
as the breast cancer slides, i.e. incubated with the same primary and
secondary antibodies.
To obtain digital images, the breast cancer tumor slides, the reference
slides, and optionally, the slides with control cell-lines, may be scanned in
a
light microscope using a ScanScope T2 automated slide scanning system
(Aperio Technologies) at x20 magnification. However, this scanning step is
not necessary, but may make the procedure easier if, for example, the
preparation and staining of the slides and the evaluation of the cytoplasmic
intensity (see below) are performed at different locations or by different
persons.
If control cell-lines are used, these are inspected to validate the
staining procedure. If the cell-lines display staining results outside
acceptable
CA 02722411 2010-10-22
WO 2009/139681 PCT/SE2009/000066
67
criteria, e.g. staining artifacts recognized by the skilled artisan, the
staining of
the slides is considered as invalid and the whole staining procedure is
repeated with new slides. If the positive and negative cell-lines display
strong
staining intensity and no staining intensity, respectively, the staining is
considered as valid.
The stained sample slide(s) from the tumor sample is/are evaluated
manually by visual inspection, and the cytoplasmic intensity (CI) of the
breast
cancer slide(s) is/are graded as described in Examples, Section 3.
In the grading, the cytoplasmic intensity (CI) is subjectively classified as:
absent = no immunoreactivity, weak = faint immunoreactivity, moderate =
medium immunoreactivity or strong = distinct and strong immunoreactivity.
The person performing the evaluation and grading is aided by visual
inspection of the stained positive and negative reference slides.
The reference value may be an absent Cl, and in such case it is
concluded that the patient in question is likely to benefit from an endocrine
treatment if the sample value derived from the subject is higher than an
absent Cl, i.e. if the sample value is a weak, moderate or strong Cl.
The above conclusion may lead a physician to apply an endocrine
treatment. However, in some cases, such decision may depend on several
parameters. Anyhow, the fact that the patient is HMGCR positive is in favor of
a decision to treat the patient with an endocrine treatment, especially a
tamoxifen treatment.
All cited material, including but not limited to publications, DNA or
protein data entries, and patents, referred to in this application are herein
incorporated by reference.
The invention being thus described, it will be obvious that the same
may be varied in many ways. Such variations are not to be regarded as a
departure from the spirit and scope of the present invention, and all such
modifications as would be obvious to one skilled in the art are intended to be
included within the scope of the following claims.