Note: Descriptions are shown in the official language in which they were submitted.
CA 02742134 2011-05-30 -
DETECTION OF HUMAN PAPILLOMA VIRUS (HPV) UTILIZING
INVASIVE CLEAVAGE STRUCTURE ASSAYS
FIELD OF THE INVENTION
The present invention provides methods and composition related to nucleic acid
detection assays for use in basic research, clinical research, and for the
development of
clinical detection assays. In particular, the present invention provides
methods for
characterizing human papillomavirus (HPV) sequences.
BACKGROUND
Cervical cancer accounts for nearly 10% of all female cancers and is a leading
cause
of cancer among women in developing countries (Franco, E.L. et al., Can Med
Assoc J.
2001;164:1017-25). The regions with the highest incidence of the disease are
generally
those with the greatest mortality and include Central America, Africa, and the
Carribean
(Ferlay, J. et al., 1998. IARC CancerBase no. 3. Lyon:IARCPress.). Incidence
in Europe
and North America has declined precipitously over the past 50 years, possibly
due to the
advent of routine= screening by Papanicolaou (Pap) smear testing (reviewed in
Franco et al.,
ibid). Cervical cancer is one of the most preventable cancers, with survival
being directly
related to the stage of the disease at diagnosis. The 5-year survival rate is
88% for women
having initial diagnosis of localized disease as opposed to 13% for women
diagnosed with
distant disease (Report of the Gynecologic Cancers Progress Review Group,
November
2001, National Cancer Institute). More than 50% of women diagnosed with
cervical cancer
in the U.S. have not had a Pap smear in the past three years (Wright, T.C. et
al., JAMA.
2000; 283:81-6).
Pap screening remains the predominant mode of detecting cancerous and
precAncerous cervical lesions; more than 50 million women undergo Pap
screening each
year in the U.S. (Wright, T.C. etal., JAMA 2002; 287:2120-29). Despite its
widespread
use, Pap smear testing is only partially effective; some estimates place the
sensitivity of
conventional Pap smear testing at 50-60% (Lorincz, A.T. and Richart, R.M.,
(Arch Pathol
Lab Med. 2003;127:959-68; Nanda, K. etal., 2000. Ann Intern Med 132:810.;
Fahey MT,
1
CA 02742134 2011-05-30
WO 2005/030041
PCT/US2004/031680
etal. Am J Epidemiol. 1995;141:680-9; Myers ER, McCrory DC, Subramanian S, et
al.
Obstet Gynecol. 2000;96:645-52.) or 70-80% (Clavel, C. et al., 2001. Br J
Cancer
84:1616). Recent innovations in cytological screening and sampling, such as
liquid-based
tests, have improved the sensitivity of these methods to 75-95% (Lorincz, A.T.
et al. ibid;
Nanda, K. et al., ibid.; Hutchinson ML, Zahniser DJ, Sherman ME, et al.
Cancer.
1999;87:48-55.). Nonetheless, even these improved methods fail to detect a
significant
portion of abnormal, and often precancerous, cells. Once identified, patients
with atypical
squamous cells of undetermined significance (ASCUS) are subjected to various
levels of
monitoring and treatment, depending on the particular attendant risk factors
and clinical
presentation (reviewed in Wright, T.C. et al. JAMA 2002, ibid).
Human Papillomavirus (HPV) has been identified as the primary, and possibly
only,
cause of cervical cancer (Muiloz N,Bosch FX,de Sanjose S, etal., Int J Cancer
1992;52:743-9; Bosch FX, Lorincz A, Munoz N, Meijer Shah Ky., Clin Pathol
2002;55:244-65), implicated in as many as 99.7% of all cases (Wallboomers,
J.M. etal., =
1999. J Pathol 189:12-19). The HPV genome is an 8 kb, circular, double
stranded DNA
comprising 8 genes, all encoded on the same strand. As many as 200 different
HPV types
have been identified in humans (Hurd, E.M. Clin Microbiol Rev. 2003;16:1-17);
of these
approximately 40 types have been found capable of infecting the genital tract
(Munoz, N. N
Engl J Med 2003;348:518-27.). Still further classification has resulted in the
identification
of high- and low-risk viral types for development of cervical cancer.
Estimates place the
number of high-risk types between 13-19 strains, with two strains, HPV 16 and
18 together
accounting for as much as 55-85% of infections, depending on subject age and
geographical
location (Munoz, N., ibid). The predominant low-risk strains are HPV 6 and 11;
these may
lead to genital warts (reviewed in Burd, E.M.,
The elucidation of certain high risk HPV strains as the causative agents of
cervical
cancer, coupled with advances in molecular biological methods, has expanded
the spectrum
of methods available for both preventing and detecting HPV infection. Vaccines
for the
most common high-risk HPV strains are currently in clinical trials (Koutsky,
LA. et al.,
2002. NEJM 347:1645-51). Moreover, some authorities are calling for HPV DNA
screening for use in conjunction with, or in some cases, in lieu of,
conventional cytological
methods (Wright, T.C. and Schiffman, M. N. Engl. J. Med, 2003; 348: 489-90).
Various
alternative DNA-based detection methods have been introduced, including the
HYBRID
CAPTURE II (HCII) test (Digene, Gaithersburg, MD), which was been approved by
the
2
CA 02742134 2011-05-30
FDA in March, 1999. The HYBRID CAPTURE method relies on hybridization of
target
DNA to complementary RNA probes. The resultant RNA-DNA hybrids are recognized
by
surface-bound antibodies as well as antibodies conjugated to alkaline
phosphatase, allowing
generation of a chemiluminescent signal in the presence of appropriate
substrates (Lorincz,
A. T. J Obstet Gynaecol Res. 1996;22:629-36; also U.S. Pat. No. 4,908,306 and
related
patents and applications). Further alternative methods include the use of
sequence specific
=
probes for use in PCR or sandwich hybridization assays, such as those
described in U.S.
6,583,278. Other methods rely on various PCR primers for selective
amplification of
specific strains, as in U.S. Pat. No. 5,447,839 and related applications.
Still other methods
rely on in situ hybridization of sequence-specific probes to isolated cervical
cells, described
=
in WO 00/24760A1 (e.g. INFORM HPV, Ventana Medical Systems, Inc., Tuscon, AZ;
Qureshi MN etal., Diagn. Cytopathol. 2003;29:149-155).
Therefore, there exists a need for a rapid, sensitive, and highly quantitative
direct
detection assay for detecting HPV infection by high risk strains in cervical
samples. Given
the current reliance on molecular methods, it is likely that there will be an
ongoing and
increasing need for rapid, quantitative methods of detecting HPV infection.
SUMMARY OF THE INVENTION
The present invention provides compositions and methods for the detection and
characterization of sequences associated with human papillomavirus (HPV). More
particularly, the present invention provides compositions, methods and kits
for using
invasive cleavage structure assays (e.g. the INVADER assay, Third Wave
Technologies,
Madison, Wisconsin) to screen nucleic acid samples, e.g., from patients, for
the presence of
any one or more of a collection of sequences associated with HPV. The present
invention
also provides compositions, methods and kits for screening sets of different
HPV sequences
in a single reaction container. The present invention may be used to detect
integrated and/or
non-integrated viral sequences.
In other embodiments, synthetic DNA suitable for use with the methods and
compositions of the present invention is made using a purified polymerase on
multiply-
primed genomic DNA, as provided, e.g., in U.S. Patent Nos. 6,291,187, and
6,323,009, and
in PCT applications WO 01/88190 and WO 02/00934.
In these embodiments, amplification of DNA
such as genomic DNA is accomplished using a DNA polymerase, such as the highly
3
CA 02742134 2011-05-30
processive (1) 29 polymerase (as described, e.g., in US Patent Nos. 5,198,543
and 5,001,050)
in combination
with exonuclease-resistant random primers, such as hexamers.
The method is not limited by the nature of the target nucleic acid. In some
embodiments, the target nucleic acid is single stranded or double stranded DNA
or RNA. In
some embodiments, double stranded nucleic acid is rendered single stranded
(e.g., by heat)
prior to formation of the cleavage structure. In some embodiments, the source
of target
nucleic acid comprises a sample containing genomic DNA. Sample include, but
are not
limited to, tissue sections, blood, saliva, cerebral spinal fluid, pleural
fluid, milk, lymph,
sputum and semen.
In some embodiments, the present invention provides methods of detecting an
HPV
sequence or method for diagnosing cancer, comprising; a) providing; i) a
sample from a
subject; and ii) a composition comprising an oligonucleotide detection assay
(e.g. as
described herein); and b) contacting said sample with said composition such
that the
presence or absence of at least one HPV sequence is determined. In some
embodiments, the
sample is a tissue section, blood sample, mouth swab sample, saliva sample, or
other
biological fluid sample from the subject.
In some embodiments, the present invention provides a method for detecting at
least
one HPV sequence in a sample, comprising using a first and a second
oligonucleotide,
wherein the oligonucleotides are configured to form an invasive cleavage
structure with a
target sequence comprising the at least one HPV sequence. In some embodiments,
the first
oligonucleotide comprises a 5' portion and a 3' portion, wherein the 3'
portion is configured
to hybridize to the target sequence, and the 5' portion is configured to not
hybridize to the
target sequence. In some embodiments, the second oligonucleotide comprises a
5' portion
and a 3' portion, wherein the 5' portion is configured to hybridize to the
target sequence, and
wherein the 3' portion is configured to not hybridize to the target sequence.
In preferred
embodiments, the first and second oligonucleotides are selected from the group
consisting
of SEQ ID NOS. 1-5, 7-62, 64-67, 69-70, 73-116 and 122-193.
In some embodiments, the present invention provides a method for detecting the
presence or absence of HPV nucleic acid in a sample comprising providing a
sample
comprising nucleic acids and an invasive cleavage assay configured to detect
at least one
HPV nucleic acid and exposing the sample to the detection assay under
conditions such that
the at least one HPV nucleic acid can be detected, and detecting the presence
or absence of
4
CA 02742134 2011-05-30
HPV nucleic acid in a sample. In some embodiments, the detecting comprises
identifying
one or more strains of HPV present in the sample. In preferred embodiments,
the HPV
strain is selected from the group consisting of, but not limited to, HPV 16,
16Ty2, 18, 31,
33, 35, 39, 45, 51, 52, 56, 58, 58iso, 59, 66, 67, 68, 68var, 69, 70, or 82.
In some
embodiments, the nucleic acid is amplified prior to said exposure step.
In some embodiments, the present invention provides a method for detecting the
presence or absence of HPV nucleic acid in a sample comprising treating the
sample using a
first oligonucleotide and a second oligonucleotide, wherein the
oligonucleotides are
configured to form an invasive cleavage reaction and detecting the presence or
absence of
HPV nucleic acid. In particular embodiments, the oligonucleotides comprise one
or more
oligonucleotides selected from the group consisting of, but not limited to,
SEQ ID NOS. 1-
5, 7-62, 64-67, 69-70, 73-116 and 122-193. In some preferred embodiments, the
oligonucleotides individually contain one or more mismatches with target HPV
nucleic
acid. In some embodiments, the oligonucleotides are configured to hybridize to
non-HPV
nucleic acid sequences or two hybridize to two or more strains of HPV. In some
embodiments, the oligonucleotides are configured such that a stable
hybridization duplex
between one or more of the oligonucleotides and the HPV target nucleic acid is
not formed.
In some embodiments, the target nucleic acid comprises genomic DNA or mRNA.
In other embodiments, the target nucleic acid comprises synthetic DNA or RNA.
In some
preferred embodiments, synthetic DNA or RNA within a sample is created using a
purified
polymerase. In some preferred embodiments, creation of synthetic DNA using a
purified
polymerase comprises the use of PCR. In some preferred embodiments, creation
of
synthetic DNA comprises use of the methods and compositions for amplification
using
RNA-DNA composite primers (e.g., as disclosed in U.S. Patent No. 6,251,639).
In other preferred embodiments, creation of
synthetic DNA using a purified DNA polymerase suitable for use with the
methods of the
present invention comprises use of rolling circle amplification, (e.g.,as in
U.S. Pat. Nos.
6,210,884, 6,183,960 and 6,235,502 ). In
other preferred embodiments, creation of synthetic DNA comprises amplification
using
nucleic acids comprising loop-forming sequences, e.g., as described in U.S.
Patent No.
6,410,278.
In some embodiments, the present invention provides methods and kits
configured
to detect more than one HPV strain in a single reaction vessel (e.g., kits and
methods to
5
CA 02742134 2011-05-30
detect all high risk strains in four or fewer reactions). Thus, the present
invention provides
kits and methods comprising pooled detection assay components. In some
preferred
embodiments, a single oligonucleotide in the pooled detection assay components
is
configured to take part in an invasive cleavage structure in the presence of
two or more
HPV target strains. The pooled detection assay components also find use in
methods and
kits using detection technologies other than invasive cleavage technology. For
example, the
pooled detection assays for detection of HPV sequences (e.g., wherein one or
more
oligonucleotides find use in detecting multiple HPV sequences in a single
reaction)
provided in the present invention may find use in detection assays that
include, but are not
limited to, enzyme mismatch cleavage methods (e.g., Variagenics, U.S. Pat.
Nos. 6,110,684,
5,958,692, 5,851,770 ); polymerase chain
reaction; branched hybridization methods (e.g., Chiron, U.S. Pat. Nos.
5,849,481,
5,710,264, 5,124,246, and 5,624,802 );
rolling circle replication (e.g., U.S. Pat. Nos. 6,210,884, 6,183,960 and
6,235,502);
NASBA (e.g., U.S. Pat. No. 5,409,818);
molecular beacon technology (e.g., U.S. Pat. No.
6,150,097 ); E-
sensor technology (Motorola,
U.S. Pat. Nos. 6,248,229, 6,221,583, 6,013,170, and 6,063,573)
cycling probe technology (e.g., U.S. Pat. Nos. 5,403,711,
5,011,769, and 5,660,988 ); Dade
Behring signal amplification methods (e.g., U.S. Pat. Nos. 6,121,001,
6,110,677, 5,914,230,
5,882,867, and 5,792,614. );
ligase chain
reaction (Barnay Proc. Natl. Acad. Sci USA 88, 189-93 (1991)); and sandwich
hybridization methods (e.g., U.S. Pat. No. 5,288,609)
In some embodiments, the present invention provides kits or compositions
comprising a non-amplified oligonucleotide detection assay configured for
detecting at least
one HPV sequence. In other embodiments, the non-amplified oligonucleotide
detection
assay comprises first and second oligonucleotides configured to form an
invasive cleavage
structure (e.g. an INVADER assay) in combination with a target sequence
comprising said
at least one HPV sequence. In particular embodiments, the first
oligonucleotide comprises a
5' portion and a 3' portion, wherein the 3' portion is configured to hybridize
to the target
sequence, and wherein the 5' portion is configured to not hybridize to the
target sequence.
6
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
In other embodiments, the second oligonucleotide comprises a 5' portion and a
3' portion,
wherein the 5' portion is configured to hybridize to the target sequence, and
wherein the 3'
portion is configured to not hybridize to the target sequence.
In some embodiments, the present invention provides a kit comprising
oligonucleotide detection assays configured for detecting a HPV sequence,
wherein the kit
comprises one or more oligonucleotides selected from the group consisting of
SEQ ID
NOS. 1-193. In particular embodiments, the multiple HPV strains are detected
simultaneously by combining one or more of the oligonucleotides into one or
more
reactions. In preferred embodiments, none of the oligonucleotides are
completely
complementary to HPV target nucleic acid sequences. In some embodiments, the
oligonucleotides comprise sequences not completely complementary to any target
sequence
that is detected. In preferred embodiments, all high-risk HPV strains are
detected in four or
fewer reactions. In other preferred embodiments, all high-risk HPV strains can
be detected
in three or fewer reactions.
In some embodiments, the present invention provides a kit comprising
oligonucleotide detection assays configured for detecting all high-risk HPV
strains. In
preferred embodiments, the oligonucleotides are not fully complementary to
nucleic acid
sequences of the HPV strains. In further preferred embodiments, the
oligonucleotides
hybridize to multiples regions of a single HPV nucleic acid (e.g., to provide
redundancy in
detection). In even further preferred embodiments, the oligonucleotides are
selected from
the group consisting of SEQ ID NOS. 77-116 and 122-193.
In some embodiments, the detected HPV sequences are any of those found below
in
Table 1 or variants thereof. It is understood that sequences will diverge over
time and that
other HPV varieties, now know, or later discovered are readily adaptable to
the methods and
composition of the present invention, per the description herein.
TABLE 1
'strain' accession strain accession strain accession
la INIC_001356 34 X74476 74 NC_004501
la U06714 35 M74117 - 75 Y15173
2a X55964 35 NC 001529 76 Y15174
3 NC_001588 35h X74477 77 Y15175 -
- 3 X74462 36 NC_001686 80
Y15176
4 NC_001457 36 U31785 82 AB027021
7
CA 02742134 2011-05-30
W02005/030041
PCT/US2004/031680
4 X70827 37 NC_001687 82 AF293961
M17463 37 U31786 82 M3_002172
5 NCL901531 38 NC_901688- 83 -AF151983
5b D90252 38 U31787 - 83 NC_000856
5b NC_001444 39 M62849 84 AF293960
6a L41216 -39 AF548856 84 NC_002676
. 6a NC_001668 39 AF548857 85 AF131950
=
6b NC_001355 39 INK_0131535 86 AF349909
. 6 AF092932 40 NC 001589 86 NC 003115
6 NC_000904 40 X74478 87 PJ400628
7 M12588 41 NC 001354 87 NC_002627
. 7 NCL.001595 41 X56147 89 NC_004103
7 X74463 42 NC_001534 90 AY057438
8 M12737 -42 M73236 90 NC_004104
8 NC_001532 43 U12504 91 AF419318
9 NC_001596 43 Y12214 91 AF436128
9 X74464 44 NC 001689 91 NC 004085
. 10 NC 001576 44 U31788 92 AF531420
- X74465 45 NC 001590 92 NC_004500
11 J04351 45 X74479 RDURX7 U85660
11 M14119 47 M32305
11 NC_001525 47 NC 001530
12 NC_001577 48 NC 001690
. 12 X74466 48 U31789
13 NC_001349 49 NC_001591
13 X62843 49 X74480
14d NC 001578 50 NC 001691
14d X74467 50 U31790
NC_001579 51 M62877 -
. 15 X74468 51 NC_001533
16 AF125673 52 NC_001592
16 AF472508 52 X74481
16 AF472509 53 NC 001593
16' K02718 -53 X74482
16 NC_001526 54 AF436129
: 16 U89348 54 NC 001676
17 NC_001580 54 U37488
17 X74469 55 NC_001692
8
CA 02742134 2011-05-30
W02005/030041
PCT/US2004/031680
. 18 NC_001357 55 U31791
18 X05015 56 NC_001594-
18 X05349 56 X74483
19 NC_601581 57 NC_001353-
19 X74470 57 X55965 -
20 NC_001676 57b U37537
20 U31778 58 C190400
21 NC 001680 58 NCL001443
21 U31779 - 5= 9 NC_001635
22 i,X_001681 59 X77858
22 U31780 - 6= 0 NC_001693
23 NC 001682 60 U31792
23 U31781 6= 1 NC 001694
24 NC_601683 61 U31793
24 - U31782 62 U12499
25 NC_001582 63 NC_001458
25 X74471 63 X70828
26 igC_001583 64 U12495
26 X74472 65 NC_001459
27 NC_601584 65 X70829
27 X74473 66 NC 001695
28 NC 001684 66 U31794
28 U31783 67 C121208
29 -NC_001685 68 M73258
29 U31784 68 Y14591
30 NCL601585 69 AB027020
30 X74474 69 NC_002171
31 J04353 70 NC_001711
31 *NC_001527 70 U21941
32 NC_601586 71 AB040456
32 X74475 71 NC_002644
33 M12732 72 X94164
33 NC_001528 73 X94165
34 NC 001587 74 AF436130
9
CA 02742134 2011-05-30
WO 2005/030041
PCT/1552004/031680
In certain embodiments, the oligonucleotide detection assays are selected from
sequencing assays, polymerase chain reaction assays, hybridization assays,
hybridization
assays employing a probe complementary to a mutation, microarray assays, bead
array
assays, primer extension assays, enzyme mismatch cleavage assays, branched
hybridization
assays, rolling circle replication assays, NASBA assays, molecular beacon
assays, cycling
probe assays, ligase chain reaction assays, invasive cleavage structure
assays, ARMS
assays, and sandwich hybridization assays.
The present invention also provides methods of detecting target nucleic acids
through the use of probe sequences that are not completely complementary to
the target
nucleic acid. For example, the present invention provides kits and methods for
detecting a
target sequence by using mismatch probe sequences, comprising the steps of: a)
providing a
sample suspected of containing a target nucleic acid; b) exposing the sample
to one or more
oligonucleotides that contain a region that is complementary to said target
nucleic acid, said
region having a first portion completely complementary to said target nucleic
acid, a second
portion contiguous to said first portion that is not complementary to said
target nucleic acid
(e.g., a mismatch), and a third portion contiguous to said second portion that
is completely
complementary to the target nucleic acid; and c) detecting the target nucleic
acid under
conditions such that no sequences that are completely complementary to the one
or more
oligonucleotides or said region of the one or more oligonucleotides are
detected (i.e., only
sequences that are not completely complementary to the oligonucleotides or the
region of
the oligonucleotides are detected). Thus, even if the sample contains perfect
complements
to the oligonucleotides or to the region, such perfect complements are not
detected. This
can be carried out, for example, through use of two or more oligonucleotides
that, through
their coordinated action, provide specificity for the non-matched target
sequence, but do not
detect the perfect complement. The INVADER assay, methods employing ligation,
the
polymerase chain reaction, etc. are examples of methods that permit such
detection. This
can also be carried out with a single probe sequence in a hybridization method
if the probe
is of sufficient length to ensure that it is not completely complementary to
any sequence in
the sample that might be detected.
In some embodiments, the region of the one or more oligonucleotides contains
two
or more mismatches to the target nucleic acid (e.g., 3, 4, 5, 6, . . .). In
some embodiments,
the region contains no more than twenty (e.g., no more than 15, 12, 10, 9, 8,
7, 6, . . .)
contiguous nucleotides that are completely complementary to the target nucleic
acid. In
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
some embodiments, one or more of the oligonucleotides are generated by
extending a
primer in an enzymatic extension reaction using the target nucleic acid as a
template (e.g.,
in a polymerase chain reaction). In some embodiments, the target nucleic acid
is a viral
target nucleic acid (e.g., HPV). In some embodiments, the target nucleic acid
is a conserved
region of a viral genome (i.e., a region that is highly conserved between
different strains or
family members of the virus). For example, the LCR, E6, and E7 regions of the
HPV
genome contain conserved sequences.
DEFINITIONS
To facilitate an understanding of the present invention, a number of terms and
phrases are defined below:
As used herein, the terms "subject" and "patient" refer to any organisms
including
plants, microorganisms and animals (e.g., mammals such as dogs, cats,
livestock, and
humans).
As used herein, the term "INVADER assay reagents" refers to one or more
reagents
for detecting target sequences, said reagents comprising oligonucleotides
capable of
forming an invasive cleavage structure in the presence of the target sequence.
In some
embodiments, the INVADER assay reagents further comprise an agent for
detecting the
presence of an invasive cleavage structure (e.g., a cleavage agent). In some
embodiments,
the oligonucleotides comprise first and second oligonucleotides, said first
oligonucleotide
comprising a 5' portion complementary to a first region of the target nucleic
acid and said
second oligonucleotide comprising a 3' portion and a 5' portion, said 5'
portion
complementary to a second region of the target nucleic acid downstream of and
contiguous
to the first portion. In some embodiments, the 3' portion of the second
oligonucleotide
comprises a 3' terminal nucleotide not complementary to the target nucleic
acid. In
preferred embodiments, the 3' portion of the second oligonucleotide consists
of a single
nucleotide not complementary to the target nucleic acid.
In some embodiments, INVADER assay reagents are configured to detect a target
nucleic acid sequence comprising first and second non-contiguous single-
stranded regions
separated by an intervening region comprising a double-stranded region. In
preferred
embodiments, the INVADER assay reagents comprise a bridging oligonucleotide
capable of
binding to said first and second non-contiguous single-stranded regions of a
target nucleic
11
CA 02742134 2011-05-30
WO 2005/030041
PCT/US2004/031680
acid sequence. In particularly preferred embodiments, either or both of said
first or said
second oligonucleotides of said INVADER assay reagents are bridging
oligonucleotides.
In some embodiments, the INVADER assay reagents further comprise a solid
support. For example, in some embodiments, the one or more oligonucleotides of
the assay
reagents (e.g., first and/or second oligonucleotide, whether bridging or non-
bridging) is
attached to said solid support. In some embodiments, the INVADER assay
reagents further
comprise a buffer solution. In some preferred embodiments, the buffer solution
comprises a
source of divalent cations (e.g., Mn2+ and/or Mg2+ ions). Individual
ingredients (e.g.,
oligonucleotides, enzymes, buffers, target nucleic acids) that collectively
make up
INVADER assay reagents are termed "INVADER assay reagent components."
In some embodiments, the INVADER assay reagents further comprise a third
oligonucleotide complementary to a third portion of the target nucleic acid
upstream of the
first portion of the first target nucleic acid. In yet other embodiments, the
INVADER assay
reagents further comprise a target nucleic acid. In some embodiments, the
INVADER assay
reagents further comprise a second target nucleic acid. In yet other
embodiments, the
INVADER assay reagents further comprise a third oligonucleotide comprising a
5' portion
complementary to a first region of the second target nucleic acid. In some
specific
embodiments, the 3' portion of the third oligonucleotide is covalently linked
to the second
target nucleic acid. In other specific embodiments, the second target nucleic
acid further
comprises a 5' portion, wherein the 5' portion of the second target nucleic
acid is the third
oligonucleotide. In still other embodiments, the INVADER assay reagents
further comprise
an ARRESTOR molecule (e.g., ARRESTOR oligonucleotide).
In some preferred embodiments, the INVADER assay reagents further comprise
reagents for detecting a nucleic acid cleavage product. In some embodiments,
one or more
oligonucleotides in the INVADER assay reagents comprise a label. In some
preferred
embodiments, said first oligonucleotide comprises a label. In other preferred
embodiments,
said third oligonucleotide comprises a label. In particularly preferred
embodiments, the
reagents comprise a first and/or a third oligonucleotide labeled with moieties
that produce a
fluorescence resonance energy transfer (FRET) effect.
In some embodiments one or more the INVADER assay reagents may be provided
in a predispensed format (i.e., premeasured for use in a step of the procedure
without re-
measurement or re-dispensing). In some embodiments, selected INVADER assay
reagent
components are mixed and predispensed together. In preferred embodiments,
predispensed
12
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
assay reagent components are predispensed and are provided in a reaction
vessel (including
but not limited to a reaction tube or a well, as in, e.g., a microtiter
plate). In particularly
preferred embodiments, predispensed INVADER assay reagent components are dried
down
(e.g., desiccated or lyophilized) in a reaction vessel.
In some embodiments, the INVADER assay reagents are provided as a kit. As used
herein, the term "kit" refers to any delivery system for delivering materials.
In the context
of reaction assays, such delivery systems include systems that allow for the
storage,
transport, or delivery of reaction reagents (e.g., oligonucleotides, enzymes,
etc. in the
appropriate containers) and/or supporting materials (e.g., buffers, written
instructions for
performing the assay etc.) from one location to another. For example, kits
include one or
more enclosures (e.g., boxes) containing the relevant reaction reagents and/or
supporting
materials. As used herein, the term "fragmented kit" refers to delivery
systems comprising
two or more separate containers that each contains a subportion of the total
kit components.
The containers may be delivered to the intended recipient together or
separately. For
example, a first container may contain an enzyme for use in an assay, while a
second
container contains oligonucleotides. The term "fragmented kit" is intended to
encompass
kits containing Analyte specific reagents (ASR's) regulated under section
520(e) of the
Federal Food, Drug, and Cosmetic Act, but are not limited thereto. Indeed, any
delivery
system comprising two or more separate containers that each contains a
subportion of the
total kit components are included in the term "fragmented kit." In contrast, a
"combined
kit" refers to a delivery system containing all of the components of a
reaction assay in a
single container (e.g., in a single box housing each of the desired
components). The term
"kit" includes both fragmented and combined kits.
In some embodiments, the present invention provides INVADER assay reagent kits
comprising one or more of the components necessary for practicing the present
invention.
For example, the present invention provides kits for storing or delivering the
enzymes
and/or the reaction components necessary to practice an INVADER assay. The kit
may
include any and all components necessary or desired for assays including, but
not limited to,
the reagents themselves, buffers, control reagents (e.g., tissue samples,
positive and negative
control target oligonucleotides, etc.), solid supports, labels, written and/or
pictorial
instructions and product information, software (e.g., for collecting and
analyzing data),
inhibitors, labeling and/or detection reagents, package environmental controls
(e.g., ice,
desiccants, etc.), and the like. In some embodiments, the kits provide a sub-
set of the
13
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
required components, wherein it is expected that the user will supply the
remaining
components. In some embodiments, the kits comprise two or more separate
containers
wherein each container houses a subset of the components to be delivered. For
example, a
first container (e.g., box) may contain an enzyme (e.g., structure specific
cleavage enzyme
in a suitable storage buffer and container), while a second box may contain
oligonucleotides
(e.g., INVADER oligonucleotides, probe oligonucleotides, control target
oligonucleotides,
etc.).
The term "label" as used herein refers to any atom or molecule that can be
used to
provide a detectable (preferably quantifiable) effect, and that can be
attached to a nucleic
acid or protein. Labels include but are not limited to dyes; radiolabels such
as 32P; binding
moieties such as biotin; haptens such as digoxgenin; luminogenic,
phosphorescent or
fluorogenic moieties; mass tags; and fluorescent dyes alone or in combination
with moieties
that can suppress or shift emission spectra by fluorescence resonance energy
transfer
(FRET). Labels may provide signals detectable by fluorescence, radioactivity,
colorimetry,
gravimetry, X-ray diffraction or absorption, magnetism, enzymatic activity,
characteristics
of mass or behavior affected by mass (e.g., MALDI time-of-flight mass
spectrometry), and
the like. A label may be a charged moiety (positive or negative charge) or
alternatively,
may be charge neutral. Labels can include or consist of nucleic acid or
protein sequence, so
long as the sequence comprising the label is detectable.
As used herein, the term "distinct" in reference to signals refers to signals
that can be
differentiated one from another, e.g., by spectral properties such as
fluorescence emission
wavelength, color, absorbance, mass, size, fluorescence polarization
properties, charge, etc.,
or by capability of interaction with another moiety, such as with a chemical
reagent, an
enzyme, an antibody, etc.
As used herein, the terms "complementary" or "complementarity" are used in
reference to polynucleotides (i.e., a sequence of nucleotides such as an
oligonueleotide or a
target nucleic acid) related by the base-pairing rules. For example, for the
sequence" 5'-A-
G-T-3'," is complementary to the sequence " 3'-T-C-A-5'." Complementarity may
be
"partial," in which only some of the nucleic acids' bases are matched
according to the base
pairing rules. Or, there may be "complete" or "total" complementarity between
the nucleic
acids. The degree of complementarity between nucleic acid strands has
significant effects
on the efficiency and strength of hybridization between nucleic acid strands.
This is of
particular importance in amplification reactions, as well as detection methods
that depend
14
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
upon binding between nucleic acids. Either term may also be used in reference
to
individual nucleotides, especially within the context of polynucleotides. For
example, a
particular nucleotide within an oligonucleotide may be noted for its
complementarity, or
lack thereof, to a nucleotide within another nucleic acid strand, in contrast
or comparison to
the complementarity between the rest of the oligonucleotide and the nucleic
acid strand.
The term "homology" and "homologous" refers to a degree of identity. There may
be partial homology or complete homology. A partially homologous sequence is
one that is
less than 100% identical to another sequence.
As used herein, the term "hybridization" is used in reference to the pairing
of
complementary nucleic acids. Hybridization and the strength of hybridization
(i.e., the
strength of the association between the nucleic acids) is influenced by such
factors as the
degree of complementary between the nucleic acids, stringency of the
conditions involved,
and the T,, of the formed hybrid. "Hybridization" methods involve the
annealing of one
nucleic acid to another, complementary nucleic acid, i.e., a nucleic acid
having a
complementary nucleotide sequence. The ability of two polymers of nucleic acid
containing complementary sequences to find each other and anneal through base
pairing
interaction is a well-recognized phenomenon. The initial observations of the
"hybridization" process by Marmur and Lane, Proc. Natl. Acad. Sci. USA 46:453
(1960)
and Doty et al., Proc. Natl. Acad. Sci. USA 46:461 (1960) have been followed
by the
refinement of this process into an essential tool of modem biology.
The complement of a nucleic acid sequence as used herein refers to an
oligonucleotide which, when aligned with the nucleic acid sequence such that
the 5' end of
one sequence is paired with the 3' end of the other, is in "antiparallel
association." Certain
bases not commonly found in natural nucleic acids may be included in the
nucleic acids of
the present invention and include, for example, inosine and 7-deazaguanine.
Complementarity need not be perfect; stable duplexes may contain mismatched
base pairs
or unmatched bases. Those skilled in the art of nucleic acid technology can
determine
duplex stability empirically considering a number of variables including, for
example, the
length of the oligonucleotide, base composition and sequence of the
oligonucleotide, ionic
strength and incidence of mismatched base pairs.
As used herein, the term " Trn "is used in reference to the "melting
temperature."
The melting temperature is the temperature at which a population of double-
stranded
nucleic acid molecules becomes half dissociated into single strands. Several
equations for
CA 02742134 2011-05-30
WO 2005/030041 PCMJS2004/031680
calculating the Tnõ of nucleic acids are well known in the art. As indicated
by standard
references, a simple estimate of the Tn, value may be calculated by the
equation: Tr,õ = 81.5
+ 0.41(% G + C), when a nucleic acid is in aqueous solution at 1 M NaCl (see
e.g.,
Anderson and Young, Quantitative Filter Hybridization, in Nucleic Acid
Hybridization
= 5 (1985). Other references (e.g., Allawi, H.T. & SantaLucia, J., Jr.
Thermodynamics and
NMR of internal G.T mismatches in DNA. Biochemistry 36, 10581-94 (1997)
include more
sophisticated computations which take structural and environmental, as well as
sequence
characteristics into account for the calculation of Try,.
The term "gene" refers to a DNA sequence that comprises control and coding
sequences necessary for the production of an RNA having a non-coding function
(e.g., a
ribosomal or transfer RNA), a polypeptide or a precursor. The RNA or
polypeptide can be
encoded by a full length coding sequence or by any portion of the coding
sequence so long
as the desired activity or function is retained.
The term "wild-type" refers to a gene or a gene product that has the
characteristics of
that gene or gene product when isolated from a naturally occurring source. A
wild-type
gene is that which is most frequently observed in a population and is thus
arbitrarily
designated the "normal" or "wild-type" form of the gene. In contrast, the term
"modified"
,"mutant" or "polymorphic" refers to a gene or gene product which displays
modifications in
sequence and or functional properties (i.e., altered characteristics) when
compared to the
wild-type gene or gene product It is noted that naturally-occurring mutants
can be isolated;
these are identified by the fact that they have altered characteristics when
compared to the
wild-type gene or gene product.
The term "recombinant DNA vector" as used herein refers to DNA sequences
containing a desired heterologous sequence. For example, although the term is
not limited
to the use of expressed sequences or sequences that encode an expression
product, in some
embodiments, the heterologous sequence is a coding sequence and appropriate
DNA
sequences necessary for either the replication of the coding sequence in a
host organism, or
the expression of the operably linked coding sequence in a particular host
organism. DNA
sequences necessary for expression in prokaryotes include a promoter,
optionally an
operator sequence, a_ribosome binding site and possibly other sequences.
Eukaryotic cells
are known to utilize promoters, polyadenlyation signals and enhancers.
The term "oligonucleotide" as used herein is defined as a molecule comprising
two
or more deoxyribonucleotides or ribonucleotides, preferably at least 5
nucleotides, more
16
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
preferably at least about 10-15 nucleotides and more preferably at least about
15 to 30
nucleotides. The exact size will depend on many factors, which in turn depend
on the
ultimate function or use of the oligonucleotide. The oligonucleotide may be
generated in
any manner, including chemical synthesis, DNA replication, reverse
transcription, PCR, or
a combination thereof. In some embodiments, oligonucleotides that form
invasive cleavage
structures are generated in a reaction (e.g., by extension of a primer in an
enzymatic
extension reaction).
Because mononucleotides are reacted to make oligonucleotides in a manner such
that the 5' phosphate of one mononucleotide pentose ring is attached to the 3'
oxygen of its
neighbor in one direction via a phosphodiester linkage, an end of an
oligonucleotide is
referred to as the "5' end" if its 5' phosphate is not linked to the 3' oxygen
of a
mononucleotide pentose ring and as the "3' end" if its 3' oxygen is not linked
to a 5'
phosphate of a subsequent mononucleotide pentose ring. As used herein, a
nucleic acid
sequence, even if internal to a larger oligonucleotide, also may be said to
have 5' and 3'
ends. A first region along a nucleic acid strand is said to be upstream of
another region if
the 3' end of the first region is before the 5' end of the second region when
moving along a
strand of nucleic acid in a 5' to 3' direction.
When two different, non-overlapping oligonucleotides anneal to different
regions of
the same linear complementary nucleic acid sequence, and the 3' end of one
oligonucleotide
points towards the 5' end of the other, the former may be called the
"upstream"
oligonucleotide and the latter the "downstream" oligonucleotide. Similarly,
when two
overlapping oligonucleotides are hybridized to the same linear complementary
nucleic acid
sequence, with the first oligonucleotide positioned such that its 5' end is
upstream of the 5'
end of the second oligonucleotide, and the 3' end of the first oligonucleotide
is upstream of
the 3' end of the second oligonucleotide, the first oligonucleotide may be
called the
"upstream" oligonticleotide and the second oligonucleotide may be called the
"downstream"
oligonucleotide.
The term "primer" refers to an oligonucleotide that is capable of acting as a
point of
initiation of synthesis when placed under conditions in which primer extension
is initiated.
An oligonucleotide "primer" may occur naturally, as in a purified restriction
digest or may
be produced synthetically.
A primer is selected to be "substantially" complementary to a strand of
specific
sequence of the template. A primer must be sufficiently complementary to
hybridize with a
17
CA 02742134 2011-05-30
template strand for primer elongation to occur. A primer sequence need not
reflect the
exact sequence of the template. For example, a non-complementary nucleotide
fragment
may be attached to the 5' end of the primer, with the remainder of the primer
sequence being
substantially complementary to the strand. Non-complementary bases or longer
sequences
can be interspersed into the primer, provided that the primer sequence has
sufficient
complementarity with the sequence of the template to hybridize and thereby
form a template
primer complex for synthesis of the extension product of the primer.
The term "cleavage structure" as used herein, refers to a structure that is
formed by
the interaction of at least one probe oligonucleotide and a target nucleic
acid, forming a
structure comprising a duplex, the resulting structure being cleavable by a
cleavage means,
including but not limited to an enzyme. The cleavage structure is a substrate
for specific
cleavage by the cleavage means in contrast to a nucleic acid molecule that is
a substrate for
non-specific cleavage by agents such as phosphodiesterases which cleave
nucleic acid
molecules without regard to secondary structure (i.e., no formation of a
duplexed structure
is required).
The term "cleavage means" or "cleavage agent" as used herein refers to any
means
that is capable of cleaving a cleavage structure, including but not limited to
enzymes.
"Structure-specific nucleases" or "structure-specific enzymes" are enzymes
that recognize
specific secondary structures in a nucleic molecule and cleave these
structures. The
cleavage means of the invention cleave a nucleic acid molecule in response to
the formation
of cleavage structures; it is not necessary that the cleavage means cleave the
cleavage .
structure at any particular location within the cleavage structure.
The cleavage means may include nuclease activity provided from a variety of
sources including the Cleavase enzymes, the FEN-1 endonucleases (including
RAD2 and
XPG proteins), Tag DNA polymerase and E. coli DNA polymerase I. The cleavage
means
may include enzymes having 5' nuclease activity (e.g., Taq DNA polymerase
(DNAP), E.
coil DNA polymerase I). The cleavage means may also include modified DNA
polymerases having 5' nuclease activity but lacking synthetic activity.
Examples of
cleavage means suitable for use in the method and kits of the present
invention are provided
in U.S. Patent Nos. 5,614,402; 5,795,763; 5,843,669; 6,090; PCT Appin. Nos WO
98/23774; WO 02/070755A2; and W00190337A2
18
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
The term "thermostable" when used in reference to an enzyme, such as a 5'
nuclease,
indicates that the enzyme is functional or active (i.e., can perform
catalysis) at an elevated
temperature, i.e., at about 55 C or higher.
The term "cleavage products" as used herein, refers to products generated by
the
reaction of a cleavage means with a cleavage structure (i.e., the treatment of
a cleavage
structure with a cleavage means).
The term "target nucleic acid" refers to a nucleic acid molecule containing a
sequence that has at least partial complementarity with at least a probe
oligonucleotide and
may also have at least partial complementarity with an INVADER
oligonucleotide. The
target nucleic acid may comprise single- or double-stranded DNA or RNA.
The term "non-target cleavage product" refers to a product of a cleavage
reaction
that is not derived from the target nucleic acid. As discussed above, in the
methods of the
present invention, cleavage of the cleavage structure generally occurs within
the probe
oligonucleotide. The fragments of the probe oligonucleotide generated by this
target
nucleic acid-dependent cleavage are "non-target cleavage products."
The term "probe oligonucleotide" refers to an oligonucleotide that interacts
with a
target nucleic acid to form a cleavage structure in the presence or absence of
an INVADER
oligonucleotide. When annealed to the target nucleic acid, the probe
oligonucleotide and
target form a cleavage structure and cleavage occurs within the probe
oligonucleotide.
The term "INVADER oligonucleotide" refers to an oligonucleotide that
hybridizes
to a target nucleic acid at a location near the region of hybridization
between a probe and
the target nucleic acid, wherein the INVADER oligonucleotide comprises a
portion (e.g., a
chemical moiety, or nucleotide¨whether complementary to that target or not)
that overlaps
with the region of hybridization between the probe and target. In some
embodiments, the
INVADER oligonucleotide contains sequences at its 3' end that are
substantially the same
as sequences located at the 5' end of a probe oligonucleotide.
The term "cassette" as used herein refers to an oligonucleotide or combination
of
oligonucleotides configured to generate a detectable signal in response to
cleavage of a
probe oligonucleotide in an INVADER assay. In preferred embodiments, the
cassette
hybridizes to a non-target cleavage product from cleavage of the probe
oligonucleotide to
form a second invasive cleavage structure, such that the cassette can then be
cleaved.
In some embodiments, the cassette is a single oligonucleotide comprising a
hairpin
portion (i.e., a region wherein one portion of the cassette oligonucleotide
hybridizes to a
19
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
second portion of the same oligonucleotide under reaction conditions, to form
a duplex). In
other embodiments, a cassette comprises at least two oligonucleotides
comprising
complementary portions that can form a duplex under reaction conditions. In
preferred
embodiments, the cassette comprises a label. In particularly preferred
embodiments,
cassette comprises labeled moieties that produce a fluorescence resonance
energy transfer
(FRET) effect.
The term "substantially single-stranded" when used in reference to a nucleic
acid
substrate means that the substrate molecule exists primarily as a single
strand of nucleic
acid in contrast to a double-stranded substrate which exists as two strands of
nucleic acid
which are held together by inter-strand base pairing interactions.
As used herein, the phrase "non-amplified oligonucleotide detection assay"
refers to
a detection assay configured to detect the presence or absence of a particular
polymorphism
(e.g., SNP, repeat sequence, etc.) in a target sequence (e.g. genomic DNA)
that has not been
amplified (e.g. by PCR), without creating copies of the target sequence. A
"non-amplified
oligonucleotide detection assay" may, for example, amplify a signal used to
indicate the
presence or absence of a particular polymorphism in a target sequence, so long
as the target
sequence is not copied.
The term "sequence variation" as used herein refers to differences in nucleic
acid
sequence between two nucleic acids. For example, a wild-type structural gene
and a mutant
form of this wild-type structural gene may vary in sequence by the presence of
single base
substitutions and/or deletions or insertions of one or more nucleotides. These
two forms of
the structural gene are said to vary in sequence from one another. A second
mutant form of
the structural gene may exist. This second mutant form is said to vary in
sequence from
both the wild-type gene and the first mutant form of the gene.
The term "liberating" as used herein refers to the release of a nucleic acid
fragment
from a larger nucleic acid fragment, such as an oligonucleotide, by the action
of, for
example, a 5' nuclease such that the released fragment is no longer covalently
attached to
the remainder of the oligonucleotide.
The term "Km" as used herein refers to the Michaelis-Menten constant for an
enzyme
and is defined as the concentration of the specific substrate at which a given
enzyme yields
one-half its maximum velocity in an enzyme catalyzed reaction.
The term "nucleotide analog" as used herein refers to modified or non-
naturally
occurring nucleotides including but not limited to analogs that have altered
stacking
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
interactions such as 7-deaza purines (i.e., 7-deaza-dATP and 7-deaza-dGTP);
base analogs
with alternative hydrogen bonding configurations (e.g., such as Iso-C and Iso-
G and other
non-standard base pairs described in U.S. Patent No. 6,001,983 to S. Benner);
non-hydrogen
bonding analogs (e.g., non-polar, aromatic nucleoside analogs such as 2,4-
difluorotoluene,
described by B.A. Schweitzer and E.T. Kool, J. Org. Chem., 1994, 59, 7238-
7242, B.A.
Schweitzer and E.T. Kool, J. Am. Chem. Soc., 1995, 117, 1863-1872);
"universal" bases
such as 5-nitroindole and 3-nitropyrrole; and universal purines and
pyrimidines (such as
"K" and "P" nucleotides, respectively; P. Kong, et al., Nucleic Acids Res.,
1989, 17, 10373-
10383, P. Kong et al., Nucleic Acids Res., 1992, 20, 5149-5152). Nucleotide
analogs
include comprise modified forms of deoxyribonucleotides as well as
ribonucleotides.
The term "polymorphic locus" is a locus present in a population that shows
variation
between members of the population (e.g.., the most common allele has a
frequency of less
than 0.95). In contrast, a "monomorphic locus" is a genetic locus at little or
no variations
seen between members of the population (generally taken to be a locus at which
the most
common allele exceeds a frequency of 0.95 in the gene pool of the population).
The term "microorganism" as used herein means an organism too small to be
observed with the unaided eye and includes, but is not limited to bacteria,
virus, protozoans,
fungi, and ciliates.
The term "microbial gene sequences" refers to gene sequences derived from a
microorganism.
The term "bacteria" refers to any bacterial species including eubacterial and
archaebacterial species.
As used herein, the terms "high-risk HPV strains "or "high-risk HPV types"
refer to
those strains of HPV that have been found in cancers (e.g., carcinomas). These
HPV strains
include HPV types 16, 18, 30, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 67, 68,
69 and 70.
The term "virus" refers to obligate, ultramicroscopic, intracellular parasites
incapable of autonomous replication (i.e., replication requires the use of the
host cell's
machinery).
The term "multi-drug resistant" or multiple-drug resistant" refers to a
microorganism
that is resistant to more than one of the antibiotics or antimicrobial agents
used in the
treatment of said microorganism.
The term "sample" in the present specification and claims is used in its
broadest
sense. On the one hand it is meant to include a specimen or culture (e.g.,
microbiological
21
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
cultures). On the other hand, it is meant to include both biological and
environmental
samples. A sample may include a specimen of synthetic origin.
Biological samples may be animal, including human, fluid, solid (e.g., stool)
or
tissue, as well as liquid and solid food and feed products and ingredients
such as dairy
items, vegetables, meat and meat by-products, and waste. Biological samples
may be
obtained from all of the various families of domestic animals, as well as
feral or wild
animals, including, but not limited to, such animals as ungulates, bear, fish,
lagamorphs,
rodents, etc.
Environmental samples include environmental material such as surface matter,
soil,
water and industrial samples, as well as samples obtained from food and dairy
processing
instruments, apparatus, equipment, utensils, disposable and non-disposable
items. These
examples are not to be construed as limiting the sample types applicable to
the present
invention.
The term "source of target nucleic acid" refers to any sample that contains
nucleic
acids (RNA or DNA). Particularly preferred sources of target nucleic acids are
biological
samples including, but not limited to blood, saliva, cerebral spinal fluid,
pleural fluid, milk,
lymph, sputum and semen.
An oligonucleotide is said to be present in "excess" relative to another
oligonucleotide (or target nucleic acid sequence) if that oligonucleotide is
present at a
higher molar concentration that the other oligonucleotide (or target nucleic
acid sequence).
When an oligonucleotide such as a probe oligonucleotide is present in a
cleavage reaction in
excess relative to the concentration of the complementary target nucleic acid
sequence, the
reaction may be used to indicate the amount of the target nucleic acid
present. Typically,
when present in excess, the probe oligonucleotide will be present at least a
100-fold molar
excess; typically at least 1 pmole of each probe oligonucleotide would be used
when the
target nucleic acid sequence was present at about 10 finoles or less.
A sample "suspected of containing" a first and a second target nucleic acid
may
contain either, both or neither target nucleic acid molecule.
The term "reactant" is used herein in its broadest sense. The reactant can
comprise,
for example, an enzymatic reactant, a chemical reactant or light (e.g.,
ultraviolet light,
particularly short wavelength ultraviolet light is known to break
oligonucleotide chains).
Any agent capable of reacting with an oligonucleotide to either shorten (i.e.,
cleave) or
elongate the oligonucleotide is encompassed within the term "reactant."
22
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
As used herein, the term "purified" or "to purify" refers to the removal of
contaminants from a sample. For example, recombinant CLEAVASE nucleases are
expressed in bacterial host cells and the nucleases are purified by the
removal of host cell
proteins; the percent of these recombinant nucleases is thereby increased in
the sample.
As used herein the term "portion" when in reference to a protein (as in "a
portion of
a given protein") refers to fragments of that protein. The fragments may range
in size from
four amino acid residues to the entire amino acid sequence minus one amino
acid (e.g., 4, 5,
6, . . n-1).
The term "nucleic acid sequence" as used herein refers to an oligonucleotide,
nucleotide or polynucleotide, and fragments or portions thereof, and to DNA or
RNA of
genomic or synthetic origin that may be single or double stranded, and
represent the sense
or antisense strand. Similarly, "amino acid sequence" as used herein refers to
peptide or
protein sequence.
As used herein, the terms "purified" or "substantially purified" refer to
molecules,
either nucleic or amino acid sequences, that are removed from their natural
environment,
isolated or separated, and are at least 60% free, preferably 75% free, and
most preferably
90% free from other components with which they are naturally associated. An
"isolated
polynucleotide" or "isolated oligonucleotide" is therefore a substantially
purified
polynucleotide.
The term "continuous strand of nucleic acid" as used herein is means a strand
of
nucleic acid that has a continuous, covalently linked, backbone structure,
without nicks or
other disruptions. The disposition of the base portion of each nucleotide,
whether
base-paired, single-stranded or mismatched, is not an element in the
definition of a
continuous strand. The backbone of the continuous strand is not limited to the
ribose-phosphate or deoxyribose-phosphate compositions that are found in
naturally
occurring, unmodified nucleic acids. A nucleic acid of the present invention
may comprise
modifications in the structure of the backbone, including but not limited to
phosphorothioate
residues, phosphonate residues, 2' substituted ribose residues (e.g., 2'-0-
methyl ribose) and
alternative sugar (e.g., arabinose) containing residues.
The term "continuous duplex" as used herein refers to a region of double
stranded
nucleic acid in which there is no disruption in the progression of basepairs
within the duplex
(i.e., the base pairs along the duplex are not distorted to accommodate a gap,
bulge or
mismatch with the confines of the region of continuous duplex). As used herein
the term
23
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
refers only to the arrangement of the basepairs within the duplex, without
implication of
continuity in the backbone portion of the nucleic acid strand. Duplex nucleic
acids with
uninterrupted basepairing, but with nicks in one or both strands are within
the definition of a
continuous duplex.
The term "duplex" refers to the state of nucleic acids in which the base
portions of
the nucleotides on one strand are bound through hydrogen bonding the their
complementary
bases arrayed on a second strand. The condition of being in a duplex form
reflects on the
state of the bases of a nucleic acid. By virtue of base pairing, the strands
of nucleic acid
also generally assume the tertiary structure of a double helix, having a major
and a minor
groove. The assumption of the helical form is implicit in the act of becoming
duplexed.
The term "template" refers to a strand of nucleic acid on which a
complementary
copy is built from nucleoside triphosphates through the activity of a template-
dependent
nucleic acid polymerase. Within a duplex the template strand is, by
convention, depicted
and described as the "bottom" strand. Similarly, the non-template strand is
often depicted
and described as the "top" strand.
DESCRIPTION OF THE DRAWINGS
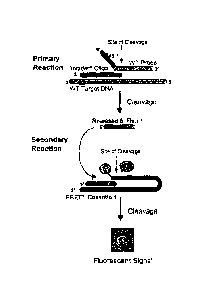
Figure 1 shows a schematic diagram of INVADER oligonucleotides, probe
oligonucleotides and FRET cassettes for detecting a two different alleles
(e.g., differing by a
single nucleotide) in a single reaction.
Figure 2 shows the results of a temperature optimization experiment carried
out in
some embodiments of the present invention.
Figure 3 shows sequences of detection assay components in some embodiments of
the present invention. Underlined portions of the sequence refer to the 5' arm
portion of
probe oligonucleotides.
Figure 4 shows results of HPV strain 16 detection experiments conducted in
some
embodiments of the present invention.
Figure 5 shows results of HPV strain 18 detection experiments conducted in
some
embodiments of the present invention.
Figure 6 shows HPV strains detected with Invader assay pools A9, A7 and A5/A6.
Figure 7 shows sequences of detection assay components in some embodiments of
the present invention. Underlined portions of the sequence refer to the 5' arm
portion of
probe oligonucleotides.
24
CA 02742134 2013-09-03
=
Figure 8 shows the quantitation of cervical sample genomic DNA using the
Oligreen
Quantitation Kit or the Alpha-Actin Invader assays.
Figure 9 shows detection of1TPV and Alpha-Actin in cervical samples conducted
in
some embodiments of the present invention.
Figure 10 shows sequences of detection assay components in some embodiments of
the present invention. Underlined portions of the sequence refer to the 5' arm
portion of
probe oligonueltotides.
DESCRIPTION OF THE INVENTION
The present invention provides means for forming a nucleic acid cleavage
structure
that is dependent upon the presence of a target nucleic acid and cleaving the
nucleic acid
cleavage structure so as to release distinctive cleavage products. 5' nuclease
activity, for
example, is used to cleave the targei-dependent cleavage structure and the
resulting
cleavage products are indicative of the presence of specific, target nucleic
acid sequences in
the sample. When two strands of nucleic acid, or oligonucleotides, both
hybridize to a
target nucleic acid strand such that they form an overlapping invasive
cleavage structure, as
described below, invasive cleavage can occur. Through the interaction of a
cleavage agent
(e.gõ a 5' nuclease) and the upstream oligonucleotide, the cleavage agent can
be made to
cleave the downstream oligonucleotide at an internal site in such a way that a
distinctive
fragment is produced. Such embodiments have been termed the INVADER assay
(Third
Wave Technologies) and are described in U.S. Patent Appl. Nos, 3,846,717,
5,985,557,
57994,069, 6,001,567, and 6,090,543, WO 97/27214 W0 98/42873, Lyamichev et
al., Nat.
13iotech., 17:292 (1999), Hall et at., PNAS, USA, 97!8272 (2000)
The INVADER assay detects hybridization of probes to a target by enzymatic
cleavage of specific structures by structure specific enzymes (See, INVADER
assays, Third
Wave Technologies; See e.g., U.S. Patent Nos. 5,846,717; 6,090,543; 6,001,567;
5,985,557;
6,090,543; 5,994,069, Lyarnichev et al., Nat. Biotech., 17:292 (1999), Hall et
al., PNAS,
USA, 97:8272 (2000), W097/27214 and W098/42873
). =
The INVADER assay detects specific DNA and RNA sequences by using stnicture-
,
specific enzymes (e.g. FEN ridonucleases) to cleave a complex formed by the
hybridization of overlapping oligonucleotide probes (See, e.g. Figure 1).
Elevated
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
temperature and an excess of one of the probes enable multiple probes to be
cleaved for
each target sequence present without temperature cycling. In some embodiments,
these
cleaved probes then direct cleavage of a second labeled probe. The secondary
probe
oligonucleotide can be 5'-end labeled with fluorescein that is quenched by an
internal dye.
Upon cleavage, the de-quenched fluorescein labeled product may be detected
using a
standard fluorescence plate reader.
The INVADER assay detects specific mutations and SNPs in unamplified, as well
as
amplified, RNA and DNA including genomic DNA. In the embodiments shown
schematically in Figure 1, the INVADER assay uses two cascading steps (a
primary and a
secondary reaction) both to generate and then to amplify the target-specific
signal. For
convenience, the alleles in the following discussion are described as wild-
type (WT) and
mutant (MT), even though this terminology does not apply to all genetic
variations. In the
primary reaction (Figure 1, panel A), the WT primary probe and the INVADER
oligonucleotide hybridize in tandem to the target nucleic acid to form an
overlapping
structure. An unpaired "flap" is included on the 5' end of the WT primary
probe. A
structure-specific enzyme (e.g. the CLEA VASE enzyme, Third Wave Technologies)
recognizes the overlap and cleaves off the unpaired flap, releasing it as a
target-specific
product. In the secondary reaction, this cleaved product serves as an INVADER
oligonucleotide on the WT fluorescence resonance energy transfer (WT-FRET)
probe to
again create the structure recognized by the structure specific enzyme (panel
A). When the
two dyes on a single FRET probe are separated by cleavage (indicated by the
arrow in
Figure 1), a detectable fluorescent signal above background fluorescence is
produced.
Consequently, cleavage of this second structure results in an increase in
fluorescence,
indicating the presence of the WT allele (or mutant allele if the assay is
configured for the
mutant allele to generate the detectable signal). In some embodiments, FRET
probes having
different labels (e.g. resolvable by difference in emission or excitation
wavelengths, or
resolvable by time-resolved fluorescence detection) are provided for each
allele or locus to
be detected, such that the different alleles or loci can be detected in a
single reaction. In
such embodiments, the primary probe sets and the different FRET probes may be
combined
in a single assay, allowing comparison of the signals from each allele or
locus in the same
sample.
If the primary probe oligonucleotide and the target nucleotide sequence do not
match perfectly at the cleavage site (e.g., as with the MT primary probe and
the WT target,
26
CA 02742134 2011-05-30
Figure 1, panel B), the overlapped structure does not form and cleavage is
suppressed. The
structure specific enzyme (e.g., CLEAVASE VIII enzyme, Third Wave
Technologies) used
cleaves the overlapped structure more efficiently (e.g. at least 340-fold)
than the non-
overlapping structure, allowing excellent discrimination of the alleles.
The probes turn over without temperature cycling to produce many signals per
target
(i.e., linear signal amplification). Similarly, each target-specific product
can enable the
cleavage of many FRET probes.
The primary INVADER assay reaction is directed against the target DNA (or RNA)
being detected. The target DNA is the limiting component in the first invasive
cleavage,
since the INVADER and primary probe are supplied in molar excess. In the
second
invasive cleavage, it is the released flap that is limiting. When these two
cleavage reactions
are performed sequentially, the fluorescence signal from the composite
reaction
accumulates linearly with respect to the target DNA amount.
In certain embodiments, the INVADER assay, or other nucleotide detection
assays,
are performed with accessible site designed oligonucleotides and/or bridging
oligonucleotides. Such methods, procedures and compositions are described in
U.S. Pat.
6,194,149, W09850403, and W00198537.
In certain embodiments, the target nucleic acid sequence is amplified prior to
detection (e.g. such that synthetic nucleic acid is generated). In some
embodiments, the
target nucleic acid comprises genomic DNA. In other embodiments, the target
nucleic acid
comprises synthetic DNA or RNA. In some preferred embodiments, synthetic DNA
within
a sample is created using a purified polymerase. In some preferred
embodiments, creation
of synthetic DNA using a purified polymerase comprises the use of PCR. In
other preferred
embodiments, creation of synthetic DNA using a purified DNA polymerase,
suitable for use
with the methods of the present invention, comprises use of rolling circle
amplification,
(e.g., as in U.S. Pat. Nos. 6,210,884, 6,183,960 and 6,235,502).
In other preferred embodiments, creation of synthetic DNA
comprises copying genomic DNA by priming from a plurality of sites on a
genomic DNA
sample. in some embodiments, priming from a plurality of sites on a genomic
DNA sample
comprises using short (e.g., fewer than about 8 nucleotides) oligonucleotide
primers. In
other embodiments, priming from a plurality of sites on a genomic DNA
comprises
extension of 3' ends in nicked, double-stranded genomic DNA (i.e., where a 3'
hydroxyl
27
CA 02742134 2011-05-30
group has been made available for extension by breakage or cleavage of one
strand of a
double stranded region of DNA). Some examples of making synthetic DNA using a
purified polymerase on nicked genomic DNAs, suitable for use with the methods
and
compositions of the present invention, are provided in U.S. Patent Nos.
6,117,634, issued
September 12, 2000, and 6,197,557, issued March 6, 2001, and in PCT
application WO
98/39485.
In some embodiments, the present invention provides methods for detecting a
target
sequence, comprising: providing a) a sample containing DNA amplified by
extension of 3'
ends in nicked double-stranded genomic DNA, said genomic DNA suspected of
containing
said target sequence; b) oligonucleotides capable of forming an invasive
cleavage structure
in the presence of said target sequence; and c) exposing the sample to the
oligonucleotides
and the agent. In some embodiments, the agent comprises a cleavage agent. In
some
particularly preferred embodiments, the method of the invention further
comprises the step
of detecting said cleavage product.
In some preferred embodiments, the exposing of the sample to the
oligonucleotides
and the agent comprises exposing the sample to the oligonucleotides and the
agent under
conditions wherein an invasive cleavage structure is formed between said
target sequence
and said oligonucleotides if said target sequence is present in said sample,
wherein said
invasive cleavage structure is cleaved by said cleavage agent to form a
cleavage product.
In some particularly preferred embodiments, the target sequence comprises a
first
region and a second region, said second region downstream of and contiguous to
said first
region, and said oligonucleotides comprise first and second oligonucleotides,
said wherein
at least a portion of said first oligonucleotide is completely complementary
to said first
portion of said target sequence and wherein said second oligonucleotide
comprises a 3'
portion and a 5' portion, wherein said 5' portion is completely complementary
to said second
portion of said target nucleic acid.
In other embodiments, synthetic DNA suitable for use with the methods and
compositions of the present invention is made using a purified polymerase on
multiply-
primed genomic DNA, as provided, e.g., in U.S. Patent Nos. 6,291,187, and
6,323,009, and
in PCT applications WO 01/88190 and WO 02/00934.
En these embodiments, amplification of DNA
such as genomic DNA is accomplished using a DNA polymerase, such as the highly
processive (1) 29 polymerase (as described, e.g., in US Patent Nos. 5,198,543
and 5,001,050,
28
CA 02742134 2013-09-03
==
) in combination
.=
with exonuclease-resistant random primers, such as hexamers.
In some embodiments, the present invention provides methods for detecting a
target
sequence, comprising: providing a) a sample containing DNA amplified by
extension of
multiple primers on genomic DNA, said genomic DNA suspected of containing said
target
sequence; b) oligonucleotides capable of forming an invasive cleavage
structure in the
presence of said taiget sequence; and 0) exposing the sample to the
oligouucleotides and the
agent In some embodiments, the agent comprises a cleavage agent. In some
preferred
embodiments, said primers are random primers. In particularly preferred
embodiments, said
primers are exonuctease resistant. In some particularly preferred embodiments,
the method
of the invention further comprises the step of detecting said cleavage
product.
In some preferred embodiments, the exposing of the sample to the
oligonucleotides .
and the agent comprises exposing the sample to the oligornicleotides and the
iigent under
conditions wherein an invasive cleavage structure is formed between said
target Sequence
and said oligonucleotides if said target sequence is present in said sample,
wherein said
invasive cleavage stmature is cleaved by said cleavage agent to form a
cleavage product.
In some preferred embodiments, the exposing of the sample to the
oligonucleotides
and the agent comprises exposing the sample to the oligonucleotides and the
agent under
= conditions wherein an invasive cleavage structure is formed between said
target sequence
and said oligonucleotides if said target sequence is present in said sample,
wherein said
invasive cleavage structure is cleaved by said cleavage agent to form a
cleavage product.
. In some particularly preferred embodiments, the target sequence
comprises a first
region and a second rcg,ion, said second region downstream of and contiguous
to said first
region, and said oligonucleotides comprise first and second oligonucleotides,
said wherein
at least a portion of said first oligonucleotide is completely complementary
to said first
portion of said target sequence and wherein said second oligonucleotide
comprises a 3'
portion and a 5' portion, wherein said 5' portion is completely
complementaryto said second
portion of said target nucleic acid.
In certain embodiments, the present invention provides kits for assaying a
pooled
sample (e.gõ a pooled blood sample) using INVADER detection reagents (e.g.
primary
probe, INVADER probe, and FRET cassette). In preferred embodiments, the kit
further
comprises instructions on how to perform the INVADER assay and specifically
how to
29
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
apply the INVADER detection assay to pooled samples from many individuals, or
to
"pooled" samples from many cells (e.g. from a biopsy sample) from a single
subject.
The present invention further provides assays in which the target nucleic acid
is
reused or recycled during multiple rounds of hybridization with
oligonucleotide probes and
cleavage of the probes without the need to use temperature cycling (i.e., for
periodic
denaturation of target nucleic acid strands) or nucleic acid synthesis (i.e.,
for the
polymerization-based displacement of target or probe nucleic acid strands).
When a
cleavage reaction is run under conditions in which the probes are continuously
replaced on
the target strand (e.g. through probe-probe displacement or through an
equilibrium between
probe/target association and disassociation, or through a combination
comprising these
mechanisms, (The kinetics of oligonucleotide replacement. Luis P. Reynaldo,
Alexander V.
Vologodskii, Bruce P. Neri and Victor I. Lyamichev. J. Mol. Biol. 97: 511-520
(2000)),
multiple probes can hybridize to the same target, allowing multiple cleavages,
and the
generation of multiple cleavage products.
In some embodiments, the detection assays of the present invention are
designed to
detect one or more HPV sequences (See, e.g., Example 5). In some embodiments,
multiple
HPV sequences are detected in a single reaction (See, e.g., Example 5, Figure
9, reactions
10-658, 10-662, 10-677 and 10-682). In some preferred embodiments, a single
oligonucleotide used in the detection assays is configured to hybridize to two
or more HPV
sequences such that multiple HF'V sequences can be detected with a single set
of detection
assay reagents (See, e.g., Example 5, Figure 9). In some embodiments, the
oligonucleotides
used in the detection assay are perfectly complementary to the intended HPV
target
sequence. In other embodiments, the oligonucleotides contain one or more
mismatches to
the HPV target sequence of interest. Mismatches find multiple uses, including,
but not
limited to, the ability to reduce hybridization efficiency (which may be
desired in some
detection assay formats), the ability to add degeneracy (e.g., to detect two
or more strains or
variants), and the ability to compensate for sequence variation that may be in
a sample. In
some embodiments, where variation at a particular nucleotide position is
identified in some
members of a tested population, multiple oligonucleotides are provides that
differ in
sequence at the position so that each variant within the population is
detected. Exemplary
detection assay components for use in invasive cleavage assays are provided in
the Example
section below for certain preferred strains of HPV.
= 30
CA 02742134 2011-05-30
WO 2005/030041 PCIYUS200-1/031680
EXAMPLES
The following examples are provided in order to demonstrate and further
illustrate
certain preferred embodiments and aspects of the present invention and are not
to be
construed as limiting the scope thereof.
=
In the experimental disclosure which follows, the following abbreviations
apply: N
(normal); M (molar); mM (millimolar); j.tM (micromolar); mol (moles); mmol
(millimoles);
mol (micromoles); nmol (nanomoles); pmol (picomoles); g (grams); mg
(milligrams); jig
(micrograms); ng (nanograms); 1 or L (liters); ml (milliliters); I
(microliters); cm
(centimeters); mm (millimeters); m (micrometers); run (nanometers); DS
(dextran sulfate);
C (degrees Centigrade); and Sigma (Sigma Chemical Co., St. Louis, MO).
Example 1
Design of oligonucleotides to detect multiple HPV strains
The objective of these experiments was to arrive at oligonucleotide designs
suitable
for use in INVADER assays comprising multiple target HPV strains. As a first
step, HPV
DNA sequences were obtained from Genbank and aligned using SEQ WEB GAP and
PRETTY programs (Accelrys, San Diego, CA). Only those regions of HPV that are
reported to remain intact following chromosomal integration were analyzed to
permit the
assays to detect both integrated and non-integrated HPV sequences. Regions of
suitable
sequence conservation were chosen for select groups of strains. In this
example, areas
within the LCR, E6, and E7 genes were found to have considerable homology
between
HPV 18 and 59.
Candidate probe oligonucleotides were designed by searching for stretches of
sequence comprising a limited number of mismatches between the two targets in
either pair.
Designs were generated to several sequences on either the sense or antisense
strands.
Suitable INVADER oligonucleotides were designed to accompany the respective
probe
oligonucleotide candidates. Initial INVADER oligonucleotide designs were
selected to
associate with only a single target, e.g. HPV 18, HPV 45, or HPV 59;
subsequent designs
hybridize to more than one HPV strain. Candidate probe sets were then
evaluated for two
types of performance criteria: (1) signal generation at the chosen reaction
temperature and
(2) limit of detection, i.e. signal over background ratios at low levels of
target DNA. Probe
=
31
CA 02742134 2011-05-30
WO 2005/030041 PCTTUS2004/031680
sets meeting desired performance cut-offs, in this case, optimal signal
generation at 63 C
and LOD of 1000 copies of HPV DNA, were then selected for further evaluation.
Temperature optimization
INVADER assays were performed in 96 well MJ Skirted microtiter plates. Plates
were incubated using either an MJ Research PTC100 Thermocycler or a
ThermoHybaid
PCR Express (Molecular Biology Instrumentation, Needham Heights, MA) and read
with
an Applied Biosystems CYTOFLUOR 4000 series multiwell plate reader.
INVADER assays to determine temperature optima of probe sets were set up by
preparing primary and secondary reaction master mixes. In these experiments,
two different
INVADER oligonucleotides were tested in combination with a single probe
oligonucleotide
in each reaction. For example, in experiments designed to test probe sets for
HPV 18 and
59, INVADER oligonucleotides for both HPV 18 and HPV 59 were included in each
reaction along with a single probe oligonucleotide designed to associate with
both strains of
HPV. These reaction mixtures were tested separately on plasmid DNA comprising
a
portion of the HPV 18 (18c1) sequence (ATCC Catalog Number: 45152D) and a
synthetic
target comprising a portion of the HPV 59 sequence (SEQ ID NO: 42). Similarly,
in
experiments designed to detect HPV 45 and 59, INVADER oligonucleotides for
both strains
were included in each reaction along with the corresponding probe
oligonucleotide and
appropriate controls.
Master mixes containing primary reaction components were assembled for each
set
of temperature optimization reactions as follows. Reactions were carried out
in parallel in
microtiter plates.
Primary Mix
(PM)
Reagent Stock Final # Rxns VollRxn Total Vol
Conc. Conc.
Invader Oligo 10 0.05 120 0.10 12
1(AM) =
Invader Oligo 10 0.05 0.10 ' 12
2 (u1V1)
32
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
Primary 10 0.5 1.00 120
Probe ( M)
MOPS (mM) 400 10 0.50 60
CLEAVASE 50 2.5 1.00 120
X enzyme
(ng/11)
MgC12 (mM) 250 12.5 1.00 120
Distilled 0 0 0.30 36
Water
4.00 l/rxn
Aliquots of 15 IA of each target at a concentration of 20 fM were placed in
the
appropriate wells of a microtiter plate and were overlaid with 20 I of
mineral oil; 20111 of
ng/Ed tRNA were used for the no target control reactions. All reactions were
run in
5 duplicate. The targets were heat denatured at 95 C for 5 minutes, cooled
to 20 C, and then
aliquots of 4 tl of the primary mix were added to each well. The microtiter
plates were
incubated for 2 hours in a ThermoHybaid thermocycler with a gradient heat
block over a
span of 10 degrees (i.e. reactions were run at 58, 58.3, 58.9, 59.8, 60.9,
62.2, 63.5, 64.9,
66.4, 67.3, 67.8, 68.1 C) and then returned to 20 C.
10 A secondary master mix (SM) was assembled as follows.
Secondary Mix
(SM)
Reagent Stock Final Vol/Rxn Total Vol
Conc. Conc.
FRET Cassette 10 0.25 0.50 62.50
Arm 1 FAM SEQ
ID NO:63 (p.M)
MOPS (mM) 400 0.91 0.05 5.68
Water ---- 2.45 306.82
3 limn
33
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
Aliquots of 3 I of secondary mix were then added to each well, and the plate
was
incubated at 63 C for 10 minutes and then cooled to 4 C prior to scanning in a
CYTOFLUOR 4000 fluorescence plate reader (Applied Biosystems, Foster City,
CA). The
settings used were: 485/20 nm excitation/bandwidth and 530/25 nm
emission/bandwidth for
FAM dye detection. Unlike typical biplex INVADER reactions, because these
assays
include only a single probe molecule, only the single corresponding FRET
cassette is
required. The instrument gain was set for each dye so that the No Target Blank
produced
between 50 ¨ 150 Relative Fluorescence Units (RFUs).
Because the optimal gain setting can vary between instruments, gain is
adjusted as
needed to give the best signal/background ratio (sample raw signal divided by
the No Target
Control signal) or No Target Control sample readings of ¨100 RFUs.
Fluorescence
microplate readers that use a xenon lamp source generally produce higher RFUs.
For
directly reading the microplates, the probe height of, and how the plate is
positioned in, the
fluorescence microplate reader may need to be adjusted according to the
manufacturer's
recommendations.
The raw data that is generated by the device/instrument is used to measure the
assay
performance (real-time or endpoint mode). The equations below provide how FOZ
(Fold
Over Zero), and other values are calculated. NTC in the equations below
represents the
signal from the No Target Control.
FOZ or Signal/No Target
FOZDyei = (RawSignalDri/NTCoyei)
Candidate probe sets were selected based on the temperature profiles generated
in
these experiments. In particular, desirable probe sets exhibit temperature
profiles on the
two targets (e.g. HPV 18 and 59) tested together that exhibit similar trends
with respect to
increase in temperature, typically a bell shaped curve with its peak at the
chosen reaction
temperature, in this case 63 C. An additional desirable feature is that the
peak not be
precipitously lower plus or minus 1 or 2 degrees from 63 C. Fewer than 30% of
the
candidate probe sets yielded suitable temperature profiles.
In order to unify reaction conditions at a single reaction temperature, probe
designs
that gave rise to similar trends in response to temperature were chosen for
further design
optimization. Redesigned probes in which probe length was altered were tested.
Figure 2
34
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
shows the results of a temperature optimization experiment carried out with
probe T3elb
(SEQ ID NO:39) and INVADER oligonucleotides designed to detect }ITV 18 (T3e4i)
(SEQ
ID NO:40) and HPV 59 (T3e6i) (SEQ ID NO:41), respectively, on both the HPV 18
plasmid and HPV 59 (T3rT59) synthetic target (SEQ ID NO:42).
Similar temperature optimization and redesign procedures were carried out for
all of
the oligonucleotides presented in Figures 3 and 7.
Limit of detection (LOD) analysis
In addition to optimizing for temperature profiles that follow the same
general trends
in response to temperature and do not present steep slopes in the immediate
vicinity of the
target reaction temperature, it is also desirable to optimize probe sets for
analytical
sensitivity or limit of detection (LOD). Measuring LOD is accomplished by
conducting
INVADER assays at a single reaction temperature while varying target
concentration.
Reactions to determine LOD of temperature optimized probe sets were set up as
follows. A dilution series of target DNAs (HPV 18 plasmid and synthetic target
SEQ ID
NO: 42) was made in 10 ng/ 1tRNA in dH20; in the example presented here,
target
amounts per assay ranged from 125 copies/rxn to 8000 copies/rxn, doubling in
each
successive reaction. Aliquots of 15 I of diluted target or 10 ng/ I tRNA in
dH20 for the no
target controls were placed in appropriate wells of a microtiter plate and
overlaid with 20
of mineral oil. All reactions were run in quadruplicate. A master mix (MM) was
made
containing buffer, CLEAVASE X enzyme, MgC12, both INVADER oligonucleotides
(SEQ
ID NOs: 40-41), primary probe T3elb (SEQ ID NO: 39) and FRET cassette
oligonucleotides (SEQ ID NO: 63) as below.
Master Mix (MM): no. 125
rxns
Reagent Stock Final Vol/ Total Vol
Conc. Conc. Rxn
FRET Cassette ( M) 10 0.25 0.5 62.5
MOPS (mM) 400 10 0.5 62.5
CLEAVASE X enzyme 50 2.5 1 125
(ng/ 1)
MgC12 (mM) 250 12.5 1 125
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
Invader oligo 1 10 0.05 0.1 12.5
(PM)
Invader oligo 2 10 0.05 0.1 12.5
(uM)
Primary Probe 10 0.5 1 125
(1M)
water N/A N/A 0.8 100
total volume 5 625
Microtiter plates were covered and incubated at 95 C for 5 minutes to denature
the
targets and then cooled to 20 C. Aliquots of 5 1 of master mix were added and
the
reactions heated to 63 C for 4 hours. Upon completion, plates were removed to
the
CYTOFLUOR plate reader and analyzed as described above. Representative results
are
presented Figure 2. These results demonstrate that the designs tested in this
experiment are
suitable for the detection of as few as 250 copies of the corresponding HPV 18
and 59
sequences.
Example 2
Design of oligonueleotides to detect HPV 16
Candidate oligonucleotide sets having a primary probe and an INVADER
oligonucleotide were designed to detect regions in both HPV 16 and HPV 31
using the
procedures described in the preceding examples. Designs were directed to the
E7 gene of
HPV. As in Example 1, different INVADER oligonucleotide sequences were tested
in
combination with a single probe sequence to find a probe set with optimal
performance
characteristics at the desired reaction temperature (63 C) and in terms of
limit of detection
(FOZ). Both temperature optimization experiments and LOD (FOZ) experiments
were
conducted as described above using 15 I of a 20 fM stock solution of HPV 16
plasmid
(ATCC Catalog Number: 45113D). A total of 24 different INVADER
oligonucleotides
were tested with SEQ ID NO: 1 (A1g3p); of these, one was chosen for use in
assays to
detect HPV 16: SEQ ID NO: 2 (Algl Oci), based on its temperature optimization
profile and
FOZ.
36
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
Co-detection of HPV 16 and a human genomic internal control sequence
In some applications, it is desirable to co-detect an internal control
sequence, for
example in order to determine whether or not there are sample inhibition
effects or operator
errors. Oligonucleotide sets were designed to detect three different human
genomic
sequences and were tested in three different biplexed INVADER assays in
combination
with SEQ ID NOs: 1 and 2 to detect the HPV 16 plasmid. The human genomic
regions
were alpha actin (Genbank accession number NM_001100), the 3' untranslated
region
(UTR) of CFTR (Genbank accession number NM_000492), and hIGF (Genbank
accession
number AY260957). The oligonucleotides used for these designs were developed
previously and optimized as described in the previous examples and were as
follows: alpha
actin probe SEQ ID NO: 64, lNVADER oligo, SEQ ID NO: 65, FRET cassette 68; 3'
UTR
CFTR probe SEQ ID NO: 66, INVADER oligo SEQ ID NO: 67, FRET cassette SEQ ID
NO: 68; hIGF probe SEQ ID NO: 69, INVADER oligo SEQ ID NO: 70, FRET cassette
SEQ ID NO: 71.
A standard curve was generated using different amounts of HPV 16 plasmid
against
a constant amount of human genomic DNA. Reactions containing 0, 250, 500,
1000, 2500,
5000, 10,000, or 20,000 copies of the HPV 16 plasmid and either 100 ng (for
hIGF and
CFTR) or 250 ng (for alpha actin) human genomic DNA. DNA was isolated using
the
Gentra PUREGENEe Autopure LS system (Gentra, Inc., Minneapolis, MN) or manual
preparation methods. All other reaction components and detection were as
described in the
previous examples except that a second FRET oligo was used in each case (for
hIGF, SEQ
ID NO: 71, red dye; for 3' UTR of CFTR and alpha actin, SEQ ID NO: 68, red
dye). The
results are presented in Figure 4 and indicate that all of the human genomic
sequences tested
were suitable for biplex detection in combination with varying levels of HPV
16 plasmid
DNA. Furthermore, these experiments demonstrate that there is no apparent
cross reactivity
between the probe sets designed to detect HPV 16 and those designed to detect
the human
genomic sequences, as evidenced by both the unchanged signal generated using
the IC
probes in the presence of variable amounts of HPV 16 DNA as well as by the
lack of
detectable signal generated using the HPV 16 probes in the absence of HPV 16
plasmid
DNA.
37
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
Example 3
Effects of genomic DNA on detection of HPV 18
Experiments were carried out to assess the effect of exogenous human genomic
DNA on detection of HPV 18. INVADER reactions were set up as follows. Serial
dilutions
of a synthetic HPV 18 target B1T18 (SEQ lD NO: 72) were made to result in
numbers of
target molecules as indicated in the X-axis of Figure 5 when 15 p.1 were
pipetted into the
appropriate wells of a microtiter plate. A second set of serial dilutions was
made
incorporating human genomic DNA, purified as described in Example 2, into each
dilution
such that each reaction contained 1 p.g of human genomic DNA. No target
controls
contained 15111 of 1Ong/ 1 tRNA in distilled water. All reactions were run in
duplicate. The
target aliquots were overlaid with 20 1 mineral oil and denatured at 95 C for
5 minutes
then chilled to 20 C. A master mix containing INVADER reaction components was
made
as follows.
Reagent Stock Final Conc. # of reactions Volume Total
Conc. /reaction volume
INVADER oligo mixture 1 0.05 10 1 10
(SEQ ID NO: 73, 74, and
75) (IN)
Primary probe (SEQ ID 10 0.5 1 10
NO: 76) B 1 b3
(11M)
FRET Cassette (SEQ ID 10 0.25 0.5 5
NO: 63) ( M)
MOPS (mM) 400 10 0.5 5
CLEAVASE X enzyme 50 2.5 1 10
(ng/0.1)
MgC12 (mM) 250 12.5 1 10
Total volume 5 50
Aliquots of 5 I of the master mix were added to each well. Reactions were
incubated at
62 C for 4 hours and then read in the CYTOFLUOR as described above.
38
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
The results are presented in Figure 5 and demonstrate that the presence of 1
Itg of
human genomic DNA does not exert a significant inhibitory effect on the
INVADER assay
designed to detect HPV 18 sequences.
Example 4
Simultaneous detection of multiple HPV strains in a single pooled reaction
In some situations, it may prove desirable to combine detection of many HPV
strains
in a single reaction vessel. For example, it may be desired to detect all high-
risk HPV
strains or all low-risk strains in a single reaction mixture. In some cases,
the output of a
pooled reaction is a qualitative answer such as a positive result, indicating
the presence of
one or more HPV strains, or a negative result, indicating the absence of HPV.
Preferred oligonucleotide designs for pooling multiple INVADER reactions in a
single well may possess the following characteristics:
The oligonucleotides do not interact with one another to promote excessive
signal
generation in the absence of a specific target. Background in the INVADER
assay
may result from fragments of certain oligonucleotides that are an intrinsic
component of some oligo synthesis mixtures. However, it is also possible for
groups of different oligonucleotides to assume structures that are recognized
and
cleaved during the INVADER assay in the absence of target.
The oligonucleotides do not interact with one another to interfere
significantly with
signal generation in the presence of a specific target.
Performance of a given oligonucleotide set is comparable when tested in the
pooled
mixture and individually.
Pooled assays are created by combining probe and INVADER oligonucleotides in
subcombinations and then assessing performance on each target by comparing
signal
generation and FOZ of the oligonucleotides in the pool to detection of that
target in
reactions containing only the probe and INVADER oligonucleotides designed to
detect it.
In the event that a given oligonucleotide set is adversely affected by
combination with other
oligo sets in a single reaction vessel, e.g. generates excessive background or
fails to
39
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
generate the expected levels of target-specific signal, in some cases it is
possible to swap in
an alternative oligonucleotide set useful for determining the presence of the
same HPV
strain. In other cases, it is possible to merely choose a different 5' arm for
a particular
probe oligonucleotide to reduce non-specific background generation or signal
inhibition. In
some cases, it is possible to detect the alternative strand of a particular
target sequence,
thereby altering the composition of the oligonucleotides and making them
suitable for
detection of the target in a pooled assay. In each case, the measure of a
successfully
performing assay is yield of statistically significant signal over background
(FOZ) in the
presence of the desired targets, e.g. 1.15 with t-test from neighbor <.05.
Ultimately, candidate oligonucleotide designs are pooled in various
combinations
and tested against a sample containing purified HPV DNA or partial HPV
sequences.
Optimally, samples of all HPV strains being tested are evaluated individually
with the
pooled oligonucleotide sets to confirm that target-specific signal is
generated for each
desired strain. Similarly, HPV negative samples or samples containing strains
of HPV not
desired to be tested (e.g. low risk strains, HPV 1, or other non-cervical
strains) are also
tested against the pools to confirm that they do not generate statistically
significant FOZ.
Example 5
Detection of multiple HPV strains in cervical samples
The methods and compositions of the present invention were used, as described
in
Example 4, to detect multiple strains of HPV in cervical samples.
INVADER oligonucleotide designs
The INVADER assay was designed to detect high-risk HPV strains including 16,
18,
31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 67, 68, and 70. Probe sets were
combined into three
pools based on HPV genetic phylogeny. Probe sets (e.g., probe and INVADER
oligonucleotides) were designed to hybridize to at least 2 different target
regions for each
HPV strain to accommodate for sequence polym. orphisms and increase analytical
sensitivity
(See, e.g., Figure 6). Multiple HPV strains are detected by each of the three
pools using the
INVADER.assay (See, e.g., Figure 6). Probe and INVADER oligonucleotide
sequences are
listed in Figure 7. The probe and INVADER oligonucleotide sequences in Figure
10 may
also be used. An internal control assay (alpha-actin) is included in each pool
to measure the
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
relative amount of genomic DNA levels in the samples and to provide a semi-
quantitative
method for HPV titer. The HPV specific probes in the A7 and A9 pools contained
arm 1
(CGCGCCGAGG; SEQ lD NOS: 85;88, 91, 94,97, 101, 105, 108, 110, and 114) and
utilized the corresponding FAM FRET cassette (Fam-TCT-Z28-
.
AGCCGGTTTTCCGGCTGAGACCTCGGCGCG-hex, SEQ ID NO: 119). The HPV
specific probes in the A5/A6 pool contained arm AH9 (GGCAGTCTGGGAGT, SEQ ID
NO: 77, 79, 81, and 83) and utilized the FAM FRET cassette (Fam-TCT-Z28-
AGCCGGTTTTCCGGCTGAGAACTCCCAGACTGCC-hex, SEQ ID NO: 120). The
alpha-actin assay contained arm 3(ACGGACGCGGAG; SEQ ID NO: 117) and utilized
the
RED FRET cassette (Red-TCT-Z28-TCGGCCITTTGGCCGAGAGACTCCGCGTCCGT-
hex, SEQ ID NO. 121).
INVADER assay reagents and methods
Preparation of genomic DNA from cervical samples: DNA was isolated from
cervical samples obtained from a clinical laboratory using PUREGENE (Gentra
Systems)
DNA Purification Kit. The extraction procedure was modified to increase DNA
yield and
purity from this type of specimen using the following procedure:
1. Remove lml of cervical specimen and transfer to 1.5 ml tube.
2. Centrifuge cells at 16000 g for 5 min.
3. Remove supernatant and resuspend pellet in Cell Lysis Solution
4. Heat lysates at 99 C for 10 minutes. Let cool to room temperature.
5. Add proteinase K and incubate at 55 for 1 hour.
6. Add 100111 of protein precipitation solution.
7. Vortex samples vigorously for 20 seconds. Place on ice for 10 minutes.
8. Centrifuge at 16000 g for 5 min.
9. Pour off the supernatant into a clean 1.5m1 tube containing 1.5 1 of
glycogen
(20mg/m1)
10. Add 300111 of 100% isopropanol.
11. Mix the sample gently by inverting 50 times.
12. Centrifuge at 16000 g for 5 mm.
13. Pour off the supernatant and drain tube on clean absorbent paper.
14. Add 500111 of 70% ethanol and invert the tube to wash the DNA pellet.
15. Centrifuge at 16000g for 2 mm. Carefully pour off supernatant.
41
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
16. Invert and drain the tube on a clean absorbant paper and allow to air dry
for 10-
15min.
17. Add 100111 of distilled water to the pellet.
18. Let sit at room temperature overnight.
10 Al aliquots of each genomic DNA sample or no target control (10 ng/1.11
tRNA)
were added to a 96 well microtiter plate. Samples were overlaid with 20 tti
mineral oil,
denatured at 99 C for 10 minutes and then cooled to 63 C. A 10111 aliquot of
the
INVADER reaction mix was then added to each well and mixed by pipetting. An
example
of what is contained in the INVADER assay reaction mix is shown below.
Component Amount Final concentrations
per (in 20 1 reaction)
reaction
MgCl2 (70mM) 4 I 14 triM MgC12
HPV Pooled Primary probes and INVADER oligos/FAM 4 ttl
FRET/MOPS 0.5 M of each probe
Stock conc. 0.05 M of each Invader
oligo
2.5 M of each probe 0.25 M FRET cassette
0.25 M of each Invader oligo 10m11 MOPS
I .25 M of FRET cassette
40mM MOPS
Alpha Actin Primary probe/INVADER oligo/RED FRET/MOPS I d 0.25 M probe
Stock conc. 0.051.IM Invader oligo
5 M probe 0.25 M FRET cassette
1 piM Invader oligo 10mM MOPS
5 M of FRET
40mM MOPS
CLEAVASE X enzyme (40ng/ 1) in CLEAVASE dilution buffer 1 I 2 ng/ I
INVADER assay reactions
Reactions were incubated at 63 C for 4 hours and then cooled to 4 C prior to
scanning in a CYTOFLUOR 4000 fluorescence plate reader (Applied Biosystems,
Foster
City, CA). The settings used were: 485/20 rim excitation/bandwidth and 530/25
rim
emission/bandwidth for FAM dye detection, and 560/20 urn excitation/bandwidth
and
620/40 nm emission/bandwidth for RED dye detection. The instrument gain was
set for
each dye so that the No Target Blank produced between 100 ¨ 250 Relative
Fluorescence
42
CA 02742134 2011-05-30
WO 2005/030041 PCT/US2004/031680
Units (RFUs). Microplates were also read in the Genios FL Plate reader (Tecan,
Research
Triangle Park, NC). The settings used were: 485/535 nm excitation/emission for
FAM dye
detection, and 560/612 nm excitation/emission for RED dye detection. The
instrument gain
was set for each dye so that the No Target Blank produced between 1000 ¨2000
Relative
Fluorescence Units (RFUs). Because the optimal gain setting can vary between
instruments, the gain was adjusted to provide the best signal/background ratio
(e.g., sample
raw signal divided by the No Target Control signal) or No Target Control
sample readings.
For directly reading the microplates, the probe height of the microplate
reader and the
positioning of the plate was adjusted according to the manufacturer's
recommendations.
The fluorescent signal from the Fam dye and the Red dye for the samples and No
Target Control (NTC) was used to calculate fold over zero (FOZ) values as
shown below.
FOZFan, dye ---- (RawSignalFarn/NTCram)
FOZRed = (RawSignalRed/NTCaed)
The Fam FOZ corresponds to the signal from the HPV assays, and the RED FOZ
corresponds to the alpha-actin signal (See, e.g., Figure 9).
Results of INVADER assays for detection ppv in Cervical Specimens
Quantification of DNA concentration in cervical samples may be achieved using
various methods. For example, DNA concentration can be measured using the
OliGreen
ssDNA Quantitation kit (Molecular probes) or the alpha-actin INVADER assay
(See, e.g.,
Figure 8). To determine the amount of DNA present in each sample using the
INVADER
Assay, a control genomic DNA sample was serially diluted to generate a
standard curve.
The alpha-actin INVADER assay standard curve was used to determine the amount
of DNA
present in each sample using a linear regression analysis. Both methods are
useful for
determining concentrations of DNA in cervical samples. Since the signal from
the alpha-
actin INVADER assay can be detected in the same well as the HPV INVADER
assays, a
separate quantitation step by OliGreen or measuring absorbance at 260 mu is
not required.
The INVADER assay was used to detect the presence or absence of HPV (e.g.,
high-
risk HPV strains) in cervical samples (See, e.g., Figure 9). Each sample was
tested in three
separate wells of a microtiter plate containing either the A5/A6, A7 or A9
INVADER
reaction mix. All wells contained the alpha-actin oligonucleotides and FRET
cassette.
Samples were considered to be HPV positive if the FAM FOZ values were greater
than 3,
HPV negative if the FAM FOZ values were less than 2, and equivocal if the FAM
FOZ
43
CA 02742134 2013-09-03
values were between 2 and 3. Of the 45 cervical samples tested, there were 21
positive
samples, 23 negative samples, and 1 equivocal sample. Four of the samples were
determined, using the methods of the present invention, to be co-infected with
multiple
HPV strains (See, e.g., Figure 9, samples 10-658, 10-662, 10-677 and 10-682).
Various modifications and
variations of the described assays of the invention will he apparent to those
skilled in the art
without departing from the scope and spirit of the invention. Although the
invention has
been described in connection with specificprefen-ed embodiments, it should be
understood
that the invention as claimed should not be unduly limited to such specific
embodiments.
Indeed, various modifications of the described modes for carrying out the
invention that are
obvious to those skilled in relevant fields are intended to be within the
scope of the
following claims_
=
=
44
CA 02742134 2011-11-03
SEQUENCE LISTING IN ELECTRONIC FORM
This description contains a sequence listing in electronic form in ASCII
text format.
A copy of the sequence listing in electronic form is available from the
Canadian Intellectual Property Office.
The sequences in the sequence listing in electronic form are reproduced in
the following Table.
SEQUENCE TABLE
<110> Third Wave Technologies, Inc.
<120> DETECTION OF HUMAN PAPILLOMA
VIRUS(HPV) UTILIZING INVASIVE CLEAVAGE
STRUCTURE ASSAYS
<130> 84012-50D
<140> CA 2,539,703
<141> 2004-09-27
<150> US 60/505,786
<151> 2003-09-25
<150> US 10/951,241
<151> 2004-09-27
<160> 193
<170> PatentIn version 3.2
<210> 1
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 1
cgcgccgagg gtccggttct gcttgacc 28
<210> 2
<211> 43
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 2
cttacactgg caacaaaagg ttacgatatt gtaatgggat ctc 43
44a
CA 02742134 2011-05-30
<210> 3
<211> 48
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 3
gtagactcac actgccaaca aaaggttacg atattgtaat tggatgtc 48
<210> 4
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 4
ggcagtctgg gagtcaacac aaacagggac cacaa 35
<210> J
<211> 54
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 5
gctccaacgg gtttcctgcg cacaatatta aacacacatt tacacgccat gtat 54
<210> 6
<211> 37
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<220>
<221> misc feature
<222> (3)..(3)
<223> The residue at this position is linked to a Z28 quencher.
<400> 6
tctagccggt tttccggctg agaactccca gactgcc 37
44b
CA 02742134 2011-05-30
<210> 7
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 7
ggcagtctgg gagtgttgta tgactatgga gcaccg 36
<210> 8
<211> 62
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 8
gctccaacgg gtttcctgct agccataatg tgatgtgtgt gtttataatt aacactgtat 60
tt 62
<210> 9
<211> 37
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 9
ggcagtctgg gagtgaagtg gacagacttt gtaaggt 37
<210> 10
<211> 55
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 10
agatggcgac accaatccgg gcacaatatt aaacacacat ttacacgcca tgtat 55
<210> 11
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
44c
CA 02742134 2011-05-30
<223> Synthetic
<400> 11
cgcgccgagg tcaagggttt ctggcacc 28
<210> 12
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 12
gtcgtttttc cttaaggtgt ctaagttttt ctgctgggta 40
<210> 13
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 13
gtcgtttgtc attaaggtgt ctaagttttt ctgctggata 40
<210> 14
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 14
cgcgccgagg gtcctttgtg tgaccgtggt 30
<210> 15
<211> 39
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 15
ggattgcgag cattacagca gctgtttctg gamaccctc 39
<210> 16
<211> 39
44d
CA 02742134 2011-05-30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 16
gaccttcgag cactccagca gctgtttttg agcaccttc 39
<210> 17
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 17
cgcgccoagg agggcaatag ggtcgcca 28
<210> 18
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 18
acaaatataa actgttgtgc tgcaaaaaat gggtt 35
<210> 19
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 19
caagtgtgct gcaagccaca aatatgggtt 30
<210> 20
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 20
cgcgccgagg gtccatctgg ccagtcca 28
44e
CA 02742134 2011-05-30
<210> 21
<211> 54
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 21
cccaaatata atcacaatgc tgatgtagta attgcttatg gcttgttctg cttc 54
<210> 22
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 22
gtagtaatca gctgtggccg gttgtgcttc 30
<210> 23
<211> 34
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 23
cgcgccgagg gaccttgtat gtcacgtgca atta 34
<210> 24
<211> 51
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 24
tgcaagaaat tgtgttagat ttatatccat gcaatgaaat agagccggtc a 51
<210> 25
<211> 48
<212> DNA
<213> Artificial Sequence
<220>
44f
CA 02742134 2011-05-30
<223> Synthetic
<400> 25
aggaaattgt attagagtta tgtccttaca atgaaataca gccggttc 48
<210> 26
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 26
cgcgccgagg gtgcacctgg agaggatg 28
<210> 27
<211> 38
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 27
agggtggaga tatgtatgct gccaaagtat tgttgcaa 38
<210> 28
<211> 38
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 28
agggtggaga tatagatgtt gccaaactat tgttgcaa 38
<210> 29
<211> 65
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 29
cctatgccta aaagctgttt tattacaagg gtggcgccac caaagttgtg caagtattgt 60
tagaa 65
44g
CA 02742134 2011-05-30
<210> 30
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 30
cgcgccgagg atgagcaatt acgtgacagc tc 32
<210> 31
<211> 59
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 31
cagccaagcg caggcgttgt tttagattta tatcctgtac caactgacct atactgctt 59
<210> 32
<211> 53
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 32
cagccaagcg caggcgtaga tttacatcct gtaccaactg acctattctg ctt 53
<210> 33
<211> 57
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 33
cagccaagcg caggcgtatt ttagatttac atcctgtacc aattgaccta ttctgct 57
<210> 34
<211> 48
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
44h
CA 02742134 2011-05-30
<400> 34
cagccaagcg caggcgttgc aacctgtaac aactgaccta cactgctt 48
<210> 35
<211> 53
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 35
cagccaagcg caggcgtaga tttgcaacca gtgacaactg atctctactg ttt 53
<210> 36
<211> 27
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 36
cgcgccgagg gagcggaacc acagcgt 27
<210> 37
<211> 46
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 37
gggccataaa taataattat cctcatgcac aactaccggc ccgacc 46
<210> 38
<211> 56
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 38
gaggaagaaa acgatgaact agatggagtt aatcatcatt tgctactagc tagacc 56
<210> 39
<211> 27
<212> DNA
<213> Artificial Sequence
44i
CA 02742134 2011-05-30
<220>
<223> Synthetic
<400> 39
cgcgccgagg gatcctcaaa gcgagcc 27
<210> 40
<217> 43
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 40
attctgtgca caaatcaggt agcttgtagg gtcgtcgtgt tgc 43
<210> 41
<211> 37
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 41
tgcacaaatc aggcagtttg taaggtcgtt gtgtagc 37
<210> 42
<211> 66
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 42
gcatggcacg ctttgaggat cctacacaac gaccatacaa actgcctgat ttgagcacaa 60
cattga 66
<210> 43
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 43
cgcgccgagg gcccactctg cgcttc 26
44j
CA 02742134 2011-05-30
<210> 44
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 44
cgcgccgagg gtaacttgcc cactctgcg 29
<210> 45
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 45
cgcgccgagg gtaacgtgcc ctctctgc 28
<210> 46
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 46
cgcgccgagg gtaacgtgcc ccctctgc 28
<210> 47
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 47
caactgctgc ttatgggtgc gttaacagta acgtc 35
<210> 48
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
44k
CA 02742134 2011-05-30
<223> Synthetic
<400> 48
gtactacagc tgcttatggg tqcgttaaca c 31
<210> 49
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 49
ggcagtctgg gagtgtacag cagaagttaa tgggc 35
<210> 50
<211> 43
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 50
gctccaacgg gtttcctgcg cagtggagac tcccttcgcg ttc 43
<210> 51
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 51
ggcagtctgg gagtgtacag cagatgttta tgggc 35
<210> 52
<211> 44
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 52
agatggcgac accaatccgg gcagaggaga cacccttcgc gttc 44
<210> 53
<211> 35
441
CA 02742134 2011-05-30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 53
ggcagtctgg gagtgtccat accgatcgcg cgatt 35
<210> 54
<211> 50
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 54
aaacggtttc aaccgaaatc ggtggatata aaaggcagtc acagtttctc 50
<210> 55
<211> 37
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 55
ggcagtctgg gagtgcaaca tccatttctc cacccta 37
<210> 56
<211> 43
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 56
cttactcatc atcctgtcca ggtgcactac aacaatactt tgc 43
<210> 57
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 57
ggcagtctgg gagtaggtag ggcacacata cca 33
44m
CA 02742134 2011-05-30
<210> 58
<211> 58
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 58
cgtcagccaa gcgcaggcgt gactaatacc acatccatta atttgtgcaa ccgaaatc 58
<210> 59
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 59
ggcagtctgg gagtagaagg caactagaac ggaca 35
<210> 60
<211> 45
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 60
tggacaccac cttgcatgac tttacaatag actgtgtcta ttgcc 45
<210> 61
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 61
ggcagtctgg gagtgcagct tattctgagt ggact 35
<210> 62
<211> 41
<212> DNA
<213> Artificial Sequence
<220>
44n
CA 02742134 2011-05-30
<223> Synthetic
<400> 62
accgaaacgg gtttatgacc gaaaacggta catataaaag c 41
<210> 63
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<220>
<221> misc feature
<222> (3)..(3)
<223> The residue at this position is linked to a Z28 quencher.
<400> 63
tctagccggt tttccggctg agacctcggc gcg 33
<210> 64
<211> 27
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 64
tccgcgcgtc caggaaccct gtgacat 27
<210> 65
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 65
ccatccaggg aagagtggcc tgttt 25
<210> 66
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
440
CA 02742134 2011-05-30
<400> 66
tccgcgcgtc ctgaagaagc accaatcatg 30
<210> 67
<211> 44
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 67
tgtacttcat gctgtctaca ctaagagaga atgagagaca caca 44
<210> 68
<211> 34
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<220>
<221> misc feature
<222> (3)..(3)
<223> The residue at this position is linked to a Z28 quencher.
<400> 68
tcttcggcct tttggccgag agaggacgcg cgga 34
<210> 69
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 69
cgcgccgagg cagcactcat ccacga 26
<210> 70
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 70
ccagcctcct tagatcacag ctccggaagt 30
44p
CA 02742134 2011-05-30
<210> 71
<211> 37
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<220>
<221> misc feature
<222> (3)..(3)
<223> The residue at this position is linked to a Z28 quencher.
<400> 71
tcttcggcct tttggccgag agagtctgcc acgtcat 37
<210> 72
<211> 73
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 72
ttcggttgca cagcaaaatg gaggattgta ggataaaatg gatgctgtaa ggtgtgcagt 60
tttataactt gat 73
<210> 73
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 73
tctgcacacc ttacagcatc cattttctcc tacaatccta 40
<210> 74
<211> 51
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 74
acaagttata aacttgcata ctacacagca tccattttcc ttataatcct a 51
44q
CA 02742134 2011-05-30
<210> 75
<211> 45
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 75
agcatttgca cattatatgg cgtccatttt ctcctttaaa tccta 45
<210> 76
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 76
cgcgccgagg ccattttgca gtgcaaccg 29
<210> 77
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 77
ggcagtctgg gagtgtacag cagatgttta tgggc 35
<210> 78
<211> 44
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 78
agatggcgac accaatccgg gcagaggaga cacccttcgc gttc 44
<210> 79
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
44r
CA 02742134 2011-05-30
<223> Synthetic
<400> 79
ggcagtctgg gagtgcttag cctgtggaag gg 32
<210> 80
<211> 50
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 80
cgttccttta gatctacatt ccaaaattta tatttggcca aaggatctgc 50
<210> 81
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 81
ggcagtctgg gagtgcagct tattctgagt ggact 35
<210> 82
<211> 41
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 92
accgaaacgg gtttatgacc gaaaacggta catataaaag c 41
<210> 83
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 83
ggcagtctgg gagtgttggt ggctgttacc g 31
<210> 84
<211> 61
44s
CA 02742134 2011-05-30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 84
gctccaacgg gtttcctgcc atcccacaat ttatatttag ctaatgggtc ctgtttttct 60
61
<210> 85
<211> 27
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 85
cgcgccgagg gatcctcaaa gcgagcc 27
<210> 86
<211> 43
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 86
attctgtgca caaatcaggt agcttgtagg gtcgtcgtgt tgc 43
<210> 87
<211> 37
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 87
tgcacaaatc aggcagtttg taaggtcgtt gtgtagc 37
<210> 88
<211> 27
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
44t
CA 02742134 2011-05-30
<400> 88
cgcgccgagg gagcggaacc acagcgt 27
<210> 89
<211> 46
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 89
gggccataaa taataattat cctcatgcac aactaccggc ccgacc 46
<210> 90
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 90
gaggaagaaa acgatgaact agatggagtt aatcatcatt tgctactagc tagacc 56
<210> 91
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 91
cgcgccgagg gattggacaa aacgatatgt atcca 35
<210> 92
<211> 34
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 92
ggtgtagcat ccttttgaca ggtaatagca acat 34
<210> 93
<211> 34
<212> DNA
<213> Artificial Sequence
44u
CA 02742134 2011-05-30
<220>
<223> Synthetic
<400> 93
ggtgtagtat ccttttgaca ggtaacagca actt 34
<210> 94
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 94
cgcgccgagg aggaagcttt acaggacagt g 31
<210> 95
<211> 69
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 95
ctgaaaccgt tgagtccagc agaaaaatta aggcacctaa ctaccaaacg aagatttcat 60
aaaatagcc 69
<210> 96
<211> 69
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 96
ctgaaaccgt tgtgtccago agaaaaatta agacacgtta ataccaaacg aagatttcat 60
caaatagcc 69
<210> 97
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
44v
CA 02742134 2011-05-30
<400> 97
cgcgccgagg ggtccggcaa tttgtatggc 30
<210> 98
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 98
gtcttgcaag gtagtgtcca gcgctgtgca cac 33
<210> 99
<211> 38
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 99
gtaatgtcat gcaatgtggt gtccaacgtc gtgcacac 38
<210> 100
<211> 34
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 100
tgtcttgcaa ggtagtgtcc agcgtcgtgc acac 34
<210> 101
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 101
cgcgccgagg gtccatctgg ccagtcca 28
<210> 102
<211> 54
<212> DNA
<213> Artificial Sequence
44w
CA 02742134 2011-05-30
<220>
<223> Synthetic
<400> 102
cccaaatata atcacaatgc tgatgtagta attgcttatg gcttgttctg cttc 54
<210> 103
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 103
tgtagtaatt agctgtggca ggttgtgctt c 31
<210> 104
<211> 58
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 104
gctccaacgg gtttcctgca gtaacaattt ggtaattggt tgtatctggt tttgcttc 58
<210> 105
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 105
cgcgccgagg gcacgttgca gccaatatg 29
<210> 106
<211> 46
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 106
cagccaagcg caggcgccca acaaatagca ttattgtgtc cctgac 46
44x
CA 02742134 2011-05-30
<210> 107
<211> 51
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 107
cagccaagcg caggcggtta ctccaacaaa tagcattatt atggccttgt c 51
<210> 108
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 108
cgcgccgagg gccacggtgt acctgcct 28
<210> 109
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 109
atgtccgtga ggcggcctag tgagc 25
<210> 110
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 110
cgcgccgagg ggttatgctt gtccagctg 29
<210> 111
<211> 59
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
44y
CA 02742134 2011-05-30
<400> 111
cagccaagcg caggcgcatt tccaacagga cgttacaata ttataattgg aggtgtctc 59
<210> 112
<211> 57
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 112
cagccaagcg caggcgctga caacaacagg taacgatatt gtaattggat gtgtccc 57
<210> 113
<211> 54
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 113
cagccaagcg caggcggcaa cacaaggtta caatattgta atgggctctg tccc 54
<210> 114
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 114
cgcgccgagg acataatcat ccgtgcttac aac 33
<210> 115
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 115
tgcctgcatg ataatagatg tttgtgcgtg cat 33
<210> 116
<211> 42
<212> DNA
<213> Artificial Sequence
44z
CA 02742134 2011-05-30
<220>
<223> Synthetic
<400> 116
gcctagaact gcctgcgtga tagtatatgt ttgttcgtgt tt 42
<210> 117
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 117
acggacgcgg agaggaaccc tgtgacat 28
<210> 118
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 118
ccatccaggg aagagtggcc tgttt 25
<210> 119
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<220>
<221> misc_feature
<222> (3)..(3)
<223> The residue at this position is linked to a Z28 quencher.
<400> 119
tctagccggt tttccggctg agacctcggc gcg 33
<210> 120
<211> 37
<212> DNA
<213> Artificial Sequence
<220>
44aa
CA 02742134 2011-05-30
<223> Synthetic
<220>
<221> misc_feature
<222> (3)..(3)
<223> The residue at this position is linked to a Z28 quencher.
<400> 120
tctagccggt tttccggctg agaactccca gactgcc 37
<210> 121
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<220>
<221> misc_feature
<222> (3)..(3)
<223> The residue at this position is linked to a Z28 quencher.
<400> 121
tcttcggcct tttggccgag agactccgcg tccgt 35
<210> 122
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 122
ggcagtctgg gagtgctgag gtttccccaa ca 32
<210> 123
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 123
ggcagtctgg gagtgctgca gtttccccaa ca 32
<210> 124
<211> 59
44bb
CA 02742134 2011-05-30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 124 ,
gctccaacgg gtttcctgca ctaccagacg tacaaattta actattagca ctgccactc 59
<210> 125
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 125
ggcagtctgg gagtggtagg agcagaccgc tt 32
<210> 126
<211> 48
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 126
cagccaagcg caggcggcct cttacgtttt gctggtgtag aggtggac 48
<210> 127
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 127
cgcgccgagg attccccttc ccccagtggc 30
<210> 128
<211> 62
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 128
tccggtgcat tatacacaag tgtgcacacg gatatacttg agcgtcctgg tactcatgta 60
44cc
CA 02742134 2011-05-30
tc 62
<210> 129
<211> 60
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 129
tccggtgcat tatacacaag tgtgcactaa tatgcttgaa acccctggca gttgtgtgtc 60
<210> 130
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 130
cgcgccgagg attccccttc ccccagtggc t 31
<210> 131
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 131
cgcgccgagg attccccttc ccccagtggc tc 32
<210> 132
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 132
cgcgccgagg gctgggttca acggtttctg g 31
<210> 133
<211> 54
<212> DNA
<213> Artificial Sequence
44dd
CA 02742134 2011-05-30
<220>
<223> Synthetic
<400> 133
gtccagctat gttgtggaat cgtcgttttt ccttaaggtg tctaggtttt tctc 54
<210> 134
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 134
cgcgccgagg tttgtgtgtc cttggtgtgc a 31
<210> 135
<211> 49
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 135
tggatgctgt caagggtgtg ccagcagctg tttctgaaga ccctgtcca 49
<210> 136
<211> 54
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 136
ggtggaggcg acagattgtg agaactacag cagatgttat ggactcacta ggaa 54
<210> 137
<211> 54
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 137
ggtggaggcg acagattgtg agactacagc atctgttttt gagcaccttg tcca 54
44ee
CA 02742134 2011-05-30
<210> 138
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 138
cgcgccgagg gatcctcaaa gcgagccat 29
<210> 139
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 139
cgcgccgagg ggatcctcaa agcgagcc 28
<210> 140
<211> 42
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 140
attctgtgca caaatcaggt agcttgtagg gtcgtcgtgt tc 42
<210> 141
<211> 37
<212> DNA
<213> Artificial sequence
<220>
<223> Synthetic
<400> 141
tgcacaaatc aggcagtttg taaggtcgtt gtgtagc 37
<210> 142
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
44ff
CA 02742134 2011-05-30
<400> 142
cgcgccgagg gagcggaacc acagcgtca 29
<210> 143
<211> 27
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 143
cgcgccgagg cgagcggaac cacagcg 27
<210> 144
<211> 45
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 144
gggccataaa taataattat cctcatgcac aactaccggc ccgaa 45
<210> 145
<211> 55
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 145
gaggaagaaa acgatgaact agatggagtt aatcatcatt tgctactagc tagaa 55
<210> 146
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 146
cgcgccgagg gattggacaa aacgatatgt atccac 36
<210> 147
<211> 31
<212> DNA
<213> Artificial Sequence
44gg
CA 02742134 2011-05-30
<220>
<223> Synthetic
<400> 147
cgcgccgagg ggtccggcaa tttgtatggc c 31
<210> 148
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 148
cgcgccgagg gccatacaaa ttgccggacc 30
<210> 149
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 149
gaatggcgcg atttcacaac cctgaagaac gc 32
<210> 150
<211> 69
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 150
agatggcgac accaatccgg agaaaaatta agacacctaa atagaaaacg aagatttcat 60
aaaatagcc 69
<210> 151
<211> 62
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 151
agatggcgac accaatccgg ctaaggcacc taacaaccaa acgaagatta cataaaatag 60
44hh
CA 02742134 2011-05-30
cc 62
<210> 152
<211> 64
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 152
agatggcgac accaatccgg aactaaggca cctaaattcc aaacgaagat ttcataaaat 60
agcc 64
<210> 153
<211> 64
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 153
agatggcgac accaatccgg aattaaggca tgttaataca aaaagaagat ttcaccaaat 60
agcc 64
<210> 154
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 154
cgcgccgagg gttgcctttg gtccatgcat 30
<210> 155
<211> 57
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 155
cagccaagcg caggcgttca ttttgtggct ctaaatgcaa tacaatgtat tgcaatc 57
44li
CA 02742134 2011-05-30
<210> 156
<211> 60
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 156
cagccaagcg caggcgctca taattttgtg gttccaaatc taaatcaatg tcacaaagtc 60
<210> 157
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 157
cgcgccgagg gcaggtacac agcctataat aca 33
<210> 158
<211> 46
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 158
gtcagccaag cgcaggcgta gcccttcgcc cagtgctctc ccatac 46
<210> 159
<211> 50
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 159
gtcagccaag cgcaggcgta cagtgccctg tgtccagtgt tctccaatgc 50
<210> 160
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
44J
CA 02742134 2011-05-30
<400> 160
cgcgccgagg acgtagagaa acccaggtgt 30
<210> 161
<211> 55
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 161
ccaagcgcag gcgtaaggcg gtcgatgtat gtcttgttgg agatcatcaa gaact 55
<210> 162
<211> 50
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 162
ccaagcgcag gcgtaaggca ggtcggtgtg tgtcctgttg gaaaccaact 50
<210> 163
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 163
cgcaccgagg acatattcat ctgtgcttac aac 33
<210> 164
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 164
tgcctgcatg ataatagatg tttgtgcgtg cat 33
<210> 165
<211> 42
<212> DNA
<213> Artificial Sequence
44kk
CA 02742134 2011-05-30
<220>
<223> Synthetic
<400> 165
gcctagaact gcctgcgtga tagtatatgt ttgttcgtgt tt 42
<210> 166
<211> 42
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 166
gtctggaact gccagcgtaa tagtaaatgc ttgtgcgtga ct 42
<210> 167
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 167
cgcgccgagg atgagcaatt acgtgacagc tc 32
<210> 168
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 168
tactagatat gaaacccgaa acaactgacc tacactgctc 40
<210> 169
<211> 42
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 169
tgttttagat ttatatcctg aaccaagtga cctattctgc tc 42
4411
CA 02742134 2011-05-30
<210> 170
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 170
cgcgccgagg gtttacgact gcgacg 26
<210> 171
<211> 38
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 171
gccgccacac ggacatctgg aaaaaaatat ggaaaact 38
<210> 172
<211> 37
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 172
ccgccacacg gacatctgta aaaaaatatg gaaacct 37
<210> 173
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 173
cgcgccgagg gcttgtccat ctggccagtc 30
<210> 174
<211> 54
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
44mm
CA 02742134 2011-05-30
<400> 174
gctccaacgg gtttcctgct gtcacaatgt agtaattgct tgtagcttgt tctt 54
<210> 175
<211> 41
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 175
acaacaggtt acaatgtagt aattagctgt ggcaggttgt t 41
<210> 176
<211> 59
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 176
gctccaacgg gtttcctgcc acacagtaac aatttggtaa ttggttgtat ctggttttt 59
<210> 177
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 177
cgcgccgagg gcacgttgca gccaatatgg 30
<210> 178
<211> 46
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 178
cagccaagcg caggcgccca acaaatagca ttattgtgtc cctgac 46
<210> 179
<211> 51
<212> DNA
<213> Artificial Sequence
44nn
CA 02742134 2011-05-30
<220>
<223> Synthetic
<400> 179
cagccaagcg caggcggtta ctccaacaaa tagcattatt atggccttgt c 51
<210> 180
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 180
cgcgccgagg gccacggtgt acctgcctc 29
<210> 181
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 181
atgtccgtga ggcggcctag tgagc 25
<210> 182
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 182
cgcgccgagg atgagcaatt gagtgacagc t 31
<210> 183
<211> 61
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 183
ggtcgtgctc caacgggttt cctttagatt tggaactcga ggcaactgac ctatactgtt 60
61
4400
CA 02742134 2011-05-30
<210> 184
<211> 61
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 184
ggtcgtgctc caacgggttt cctttagatt tgcaacctca ggcaactgac ctatactgct 60
61
<210> 185
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 185
cgcgccgagg ccagccctat taaataaatg tcaaac 36
<210> 186
<211> 60
<212> DNA
<213> Artificial Sequence
<22C>
<223> Synthetic
<400> 186
cagccaagcg caggcggcct ttaatgtata aatcgtttgg tacattttca ccaacagtat 60
<210> 187
<211> 58
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 187
cagccaagcg caggcgcctt aatatatagg tctgtaggta ctgtttcacc tacagttt 58
<210> 188
<211> 28
<212> DNA
<213> Artificial Sequence
44pp
CA 02742134 2011-05-30
<220>
<223> Synthetic
<400> 188
cgcgccgagg gtccggttat gcttgtcc 28
<210> 189
<211> 53
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 189
cagccaagcg caggcgcttg caacacaagg ttacaatatt gtaatgggct ctc 53
<210> 190
<211> 54
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 190
cagccaagcg caggcgactg acaacaaaag gaaacgatat tgtaattgga tgtc 54
<210> 191
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 191
cgcgccgagg gtccggttgt gcttgtcc 28
<210> 192
<211> 53
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 192
cagccaagcg caggcgcttg caacacaagg ttacaatatt gtaatgggct ctc 53
44qq
CA 02742134 2011-05-30
<210> 193
<211> 54
<212> DNA
<213> Artificial Sequence
<220>
<223> Synthetic
<400> 193
cagccaagcg caggcgactg acaacaaaag gaaacgatat tgtaattgga tgtc 54
44rr