Note: Descriptions are shown in the official language in which they were submitted.
CA 02768046 2012-02-07
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPREND PLUS D'UN TOME.
CECI EST LE TOME DE
NOTE: Pour les tomes additionels, veillez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME OF
NOTE: For additional volumes please contact the Canadian Patent Office.
CA 02768046 2012-02-07
78864-257D
1
NUCLEIC ACID MOLECULES ENCODING PLANT CELL CYCLE PROTEINS
AND USES THEREFOR
Related Applications
This application claims priority to U.S. provisional patent application serial
number
60/204,045, filed May 12, 2000. The contents of this provisional patent
application are
incorporated herein by reference in their entirety.
to This application is a division of application 2,408,038 filed May 14, 2001.
Background of the Invention
Cell division plays a crucial role during all phases of plant development. The
continuation of organogenesis and growth responses to a changing environment
require
precise spatial, temporal, and developmental regulation of cell division.
t5 The basic mechanisms controlling the progression through the cell cycle
appear to
be conserved in all higher eukaryotes, although the temporal and spatial
control of cell
division can differ largely, between organisms. Plants have unique
developmental features
which are not found in either animals or fungi. First, due to the presence of
a rigid cell
wall, plant cells cannot move and consequently organogenesis is dependent on
cell division
20 and cell expansion at the site of formation of new organs. Secondly, cell
divisions are
confined to specialized regions, called meristems. These meristems
continuously produce
new cells which, as they move away from the meristem, become differentiated.
The
meristem identity itself can change from a vegetative to a reproductive phase,
resulting- in
the formation of flowers. Thirdly, plant development is largely post-
embryonic. During
25' embryogenesis, the main developmental event is the establishment of the
root-shoot axis.
Most plant growth occurs after germination, by iterative development at the
meristems.
Lastly, as a consequence of the sessile life of plants, development and cell
division are, to a
large extent, influenced by environmental factors such as light, gravity,
wounding,
nutrients, and stress conditions. All these features are reflected in a plant-
specific
3o regulation of the factors controlling cell division.
The unparalleled potential of plants for continuous organogenesis and plastic
growth also relies on the competent or active state of the cell division
apparaturs. The
discovery of a common mechanism underlying the regulation of the cell cycle in
yeasts
and animals has led to efforts to extend these findings to the plant kingdom
and is leading
35 to research aimed at converting the gathered knowledge into useful traits
introduced in
transgenic plants.
When eukaryotic cells and, thus, also plant cells divide they go through a
highly
ordered sequence of events collectively termed as the "cell cycle." Briefly,
DNA
replication or synthesis (S) and mitotic segregation of the chromosomes (M)
occur with
40 intervening gap phases (G 1 and G2) and the phases follow the sequence G 1-
S-G2-M. Cell
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-2-
division is completed after cytokinesis, the last step of the M-phase. Cells
that have exited
the cell cycle and have become quiescent are said to be in the GO phase. Cells
at the GO
stage can be stimulated to reenter the cell cycle at the GI phase. The
transition between
the different phases of the cell cycle are basically driven by the sequential
activation/inactivation of a kinase (called "cyclin-dependent kinase", "CDC"
or "CDK")
by different agonists.
Proteins called cyclins are required for kinase activation. Cyclins are also
important for targeting the kinase activity to a given subset of substrate(s).
Other factors
regulating CDK activity include CDK inhibitors (CKIs or ICKs, KIPs, CIPs,
INKs), CDK.
activating kinase (CAK) and CDK phosphatase (CDC25) (Mironov et al. (1999)
Plant Cell
11, 509-522 and Won K. et al. (1996) EMBO J. 15, 4182-4193).
Summary of the Invention
The present invention is based, at least in part, on the discovery of novel
plant
nucleic acid molecules and polypeptides encoded by such nucleic acid
molecules, referred
to herein as "cell cycle proteins" or "CCP." The CCP nucleic acid and
polypeptide
molecules of the present invention are useful as modulating agents in
regulating cell cycle
progression in, for example, plants. Accordingly, in one aspect, this
invention provides
isolated nucleic acid molecules encoding CCP polypeptides, as well as nucleic
acid
fragments suitable as primers or hybridization probes for the detection of CCP-
encoding
nucleic acids.
In one embodiment, a CCP nucleic acid molecule of the invention is at least
60%,
65%, 70%, 75%, 80%, 85%, 90%, 95%, 98% or more identical to the nucleotide
sequence
(e.g., to the entire length of the nucleotide sequence) of SEQ ID NO:1-66 or
228-239, or a
complement thereof.
In a preferred embodiment, the isolated nucleic acid molecule includes the
nucleotide sequence shown in SEQ ID NO:1-66 or 228-239, or a complement
thereof In
another preferred embodiment, an isolated nucleic acid molecule of the
invention encodes
the amino acid sequence of a plant CCP polypeptide.
Another embodiment of the invention features nucleic acid molecules,
preferably
CCP nucleic acid molecules, which specifically detect CCP nucleic acid
molecules relative
to nucleic acid molecules encoding non-CCP polypeptides. For example, in one
embodiment, such a nucleic acid molecule is at least 15, 20, 25, 30, 40, 50,
100, 150, 200,
250, 300, 350, 400, 450, 500, 550, 600, 650, 700, 750, or 800 nucleotides in
length and
hybridizes under stringent conditions to a nucleic acid molecule comprising
the nucleotide
sequence shown in SEQ ID NO:I-66'\or 228-239, or a complement thereof
In other preferred embodiments, the nucleic acid molecule encodes a naturally
occurring allelic variant of a plant CCP polypeptide, wherein the nucleic acid
molecule
CA 02768046 2012-02-07
= WO 01/85946 PCT/IBOI/01307
-3-
hybridizes to the nucleic acid molecule of SEQ ID NO:1-66 or 228-239 under
stringent
conditions.
Another embodiment of the invention provides an isolated nucleic acid molecule
which is antisense to a CCP nucleic acid molecule, e.g., the coding strand of
a CCP nucleic
acid molecule.
Another aspect of the invention provides a vector comprising a CCP nucleic
acid
molecule. In certain embodiments, the vector is a recombinant expression
vector. In
another embodiment, the invention provides a host cell containing a vector of
the
invention. The invention also provides a method for producing a CCP
polypeptide, by
culturing in a suitable medium a host cell of the invention, e.g., a plant
host cell such as a
host monocot plant cell (e.g., rice, wheat or corn) or a dicot host cell
(e.g., Arabidopsis
thaliana, oilseed rape, or soybeans) containing a recombinant expression
vector, such that
the polypeptide is produced.
Another aspect of this invention features isolated or recombinant CCP
polypeptides. In one embodiment, an isolated CCP polypeptides has one or more
of the
following domains: a "cyclin destruction box", a "cyclin box motif I", a
"cyclin box motif
2", a "CDC2 motif', a "CDK phosphorylation site", a "nuclear localization
signal", a "Cy-
like box", an "Rb binding domain", a "DEF domain", a "DNA binding domain", a
"DCB1
domain", a "DCB2 domain" and/or a "SAP domain".
In a preferred embodiment, a CCP polypeptide includes at least one or more of
the
following domains: a "cyclin destruction box", a "cyclin box motif 1 ", a
"cyclin box motif
2", a "CDC2 motif', a "CDK phosphorylation site", a "nuclear localization
signal", a "Cy-
like box", an "Rb binding domain", a "DEF domain", a "DNA binding domain", a
"DCB I
domain", a "DCB2 domain" and/or a "SAP domain", and has an amino acid sequence
at
least about 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 98%, 99% or more
identical to the amino acid sequence of SEQ ID NO:67-132, 205, 211, 215-216,
or 220-
227.
In another preferred embodiment, a CCP polypeptide includes at least one or
more
of the following domains: a "cyclin destruction box", a "cyclin box motif I",
a "cyclin box
motif 2", a "CDC2 motif', a "CDK phosphorylation site", a "nuclear
localization signal", a
"Cy-like box", an "Rb binding domain", a "DEF domain", a "DNA binding domain",
a
"DCB I domain", a "DCB2 domain" and/or a SAP domain and has a CCP activity (as
described herein).
In yet another preferred embodiment, a CCP polypeptide includes one or more of
the following domains: a "cyclin destruction box", a "cyclin box motif 1 ", a
"cyclin box
motif 2", a "CDC2 motif', a "CDK phosphorylation site", a "nuclear
localization signal", a
"Cy-like box", an "Rb binding domain", a "DEF domain", a "DNA binding domain",
a
"DCBl domain", a "DCB2 domain" and/or a SAP domain and is encoded by a nucleic
acid
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-4-
molecule having a nucleotide sequence which hybridizes under stringent
hybridization
conditions to a nucleic acid molecule comprising the nucleotide sequence of
SEQ ID
NO:1-66 or 228-239.
In another embodiment, the invention features fragments of the polypeptide
having
the amino acid sequence of SEQ ID NO:67-132, 205, 211, 215-216, or 220-227,
wherein
the fragment comprises at least 15 amino acids (e.g., contiguous amino acids)
of the amino
acid sequence of SEQ ID NO:67-132, 205, 211, 215-216, or 220-227. In another
embodiment, a CCP polypeptide has the amino acid sequence of SEQ ID NO:67-132,
205,
211, 215-216, or 220-227.
In another embodiment, the invention features a CCP protein which is encoded
by a
nucleic acid molecule consisting of a nucleotide sequence at least about 50%,
55%, 60%,
65%, 70%, 75%, 80%, 85%, 90%, 95%, 98%, 99% or more identical to a nucleotide
sequence of SEQ ID NO:1-66 or 228-239, or a complement thereof. This invention
further
features a CCP polypeptide, which is encoded by a nucleic acid molecule
consisting of a
nucleotide sequence which hybridizes under stringent hybridization conditions
to a nucleic
acid molecule comprising the nucleotide sequence of SEQ ID NO: 1-66 or 228-
239, or a
complement thereof'
In another embodiment the invention provides transgenic plants (e.g., monocot
or
dicot plants) containing an isolated nucleic acid molecule of the present
invention. For
example, the invention provides transgenic plants containing a recombinant
expression
cassette including a plant promoter operably linked to an isolated nucleic
acid molecule of
the present invention. The present invention also provides transgenic seed
from the
transgenic plants. In another embodiment the invention provides methods of
modulating,
in a transgenic plant, the expression of the nucleic acids of the invention.
The proteins of the present invention or portions thereof, e.g., biologically
active
portions thereof, can be operatively linked to a non-CCP polypeptide (e.g.,
heterologous
amino acid sequences) to form fusion proteins. The invention further features
antibodies,
such as monoclonal or polyclonal antibodies, that specifically bind
polypeptide of the
invention, preferably CCP polypeptide. In addition, the CCP polypeptide or
biologically
active portions thereof can be incorporated into pharmaceutical compositions,
which
optionally include pharmaceutically acceptable carriers.
In another aspect, the present invention provides a method for detecting the
presence of a CCP nucleic acid molecule, polypeptide in a biological sample by
contacting
the biological sample with an agent capable of detecting a CCP nucleic acid
molecule,
polypeptide such that the presence of a CCP nucleic acid molecule, polypeptide
is detected
in the biological sample.
In another aspect, the present invention provides a method for detecting the
presence of CCP activity in a biological sample by contacting the biological
sample with
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-5-
an agent capable of detecting an indicator of CCP activity such that the
presence of CCP
activity is detected in the biological sample.
In another aspect, the invention provides a method for modulating CCP activity
comprising contacting a cell capable of expressing CCP with an agent that
modulates CCP
activity such that CCP activity in the cell is modulated. In one embodiment,
the agent
inhibits CCP activity. In another embodiment, the agent stimulates CCP
activity. In one
embodiment, the agent is an antibody that specifically binds to a CCP
polypeptide. In
another embodiment, the agent modulates expression of CCP by modulating
transcription
of a CCP gene or translation of a CCP mRNA. In yet another embodiment, the
agent is a
nucleic acid molecule having a nucleotide sequence that is antisense to the
coding strand of
a CCP mRNA or a CCP gene.
In one embodiment, the methods of the present invention are used to increase
crop
yield, improve the growth characteristics of a plant (such as growth rate or
size of specific
tissues or organs in the plant), modify the architecture or morphology of a
plant, improve
tolerance to environmental stress conditions (such as drought, salt,
temperature, nutrient or
deprivation), or improve tolerance to plant pathogens (e.g., pathogens that
abuse the cell
cycle) by modulating CCP activity in a cell. In one embodiment, the CCP
activity is
modulated by modulating the expression of a CCP nucleic acid molecule. In yet
another
embodiment, the CCP activity is modulated by modulating the activity of a CCP
polypeptide. Modulators of CCP activity include, for example, a CCP nucleic
acid or
polypeptide.
The present invention also provides diagnostic assays for identifying the
presence
or absence of a genetic alteration characterized by at least one of (i)
aberrant modification
or mutation of a gene encoding a CCP polypeptide; (ii) mis-regulation of the
gene; and (iii)
aberrant post-translational modification of a CCP polypeptide, wherein a wild-
type form of
the gene encodes a protein with a CCP activity.
In another aspect the invention provides methods for identifying a compound
that
binds to or modulates the activity of a CCP polypeptide, by providing an
indicator
composition comprising a CCP polypeptide having CCP activity, contacting the
indicator
composition with a test compound, and determining the effect of the test
compound on
CCP activity in the indicator composition to identify a compound that
modulates the
activity of a CCP polypeptide. The identified compounds may be used as
herbicides or
plant growth regulators.
Other features and advantages of the invention will be apparent from the
following
detailed description and claims.
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
-6-
Brief Description of the Drawings
Figure 1 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP1. The complete nucleotide sequence (Figure IA)
corresponds
to nucleic acids 1 to 1715 of SEQ ID NO:39. The complete amino acid sequence
(Figure
1B) corresponds to amino acids 1 to 460 of SEQ ID NO: 105. Underlined in
Figure IA and
Figure lB are the partially characterized nucleotide (SEQ ID NO: 1) and
predicted partial
amino acid (SEQ ID NO:67) sequence, respectively. Further indicated in Figure
IA are
the stop and start codons (both in black shaded boxes) which are part of the
primers (grey
shaded boxes) used to amplify the coding region of CCP1 by PCR. The SEQ ID NOs
of
the primers used can be found in Table III. Indicated in Figure 1 B are the
cyclin
destruction box (black shaded box) and the cyclin box motifs 1 and 2 (both in
gray shaded
boxes).
Figure 2 depicts the cDNA sequence of the Arabidopsis thaliana CCP2. The
complete nucleotide sequence corresponds to nucleic acids 1 to 2195 of SEQ ID
NO:40.
Underlined is the partially characterized nucleotide (SEQ ID NO:2) sequence.
Nucleotide
sequence differences between SEQ ID NO:40 and SEQ ID NO:2 are depicted.
Indicated
are the stop and start codons (both in black shaded boxes) which are part of
the primers
(grey shaded boxes) used to amplify the coding region of CCP2 by PCR. SEQ ID
NOs of
the primers used can be found in Table III.
Figure 3 depicts the predicted amino acid sequence of the Arabidopsis thaliana
CCP2. The complete amino acid sequence corresponds to amino acids 1 to 664 of
SEQ ID
NO:106. Underlined is the predicted partial amino acid (SEQ ID NO:68)
sequence.
Figure 4 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP3. The complete nucleotide sequence (Figure 3A)
corresponds
to nucleic acids 1 to 1413 of SEQ ID NO:41. The complete amino acid sequence
(Figure
3B) corresponds to amino acids 1 to 450 of SEQ ID NO:69. Underlined in Figure
3A and
Figure 3B are the partially characterized nucleotide (SEQ ID NO:3) and
predicted partial
amino acid (SEQ ID NO:69) sequences, respectively. Indicated in Figure 3A are
the stop
and start codons (both in black shaded boxes) which are part of the primers
(grey shaded
boxes) used to amplify the coding region of CCP3 by PCR. SEQ ID NOs of the
primers
used can be found in Table III. Nucleotide sequence differences between SEQ ID
NO:41
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-7-
and SEQ ID NO:3 are depicted Indicated in Figure 3B are the cyclin destruction
box (black
shaded box) and the cyclin box motifs 1 and 2 (both in gray shaded boxes).
Figure 5 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP4. The complete nucleotide sequence (Figure 5A)
corresponds
to nucleic acids 1 to 672 of SEQ ID NO:4. The complete amino acid sequence
(Figure 5B)
corresponds to amino acids 1 to 223 of SEQ ID NO:70. Indicated in Figure.5A
are stop
and start codon (both in black shaded boxes) which are part of the primers
(grey shaded
boxes) used to amplify the coding region of CCP4 by PCR. SEQ ID NOs of the
primers
used can be found in Table III. Indicated in Figure 5B is the CDK
phosphorylation site
(black shaded box).
Figure 6 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP5. The complete nucleotide sequence (Figure 6A)
corresponds
to nucleic acids 1 to 1287 of SEQ ID NO:5. The complete amino acid sequence
(Figure
6B) corresponds to amino acids 1 to 429 of SEQ ID NO:71. Indicated in Figure
6A are the
stop and start codons (both in black shaded boxes) which are part of the
primers (grey
shaded boxes) used to amplify the coding region of CCP5 by PCR. SEQ ID NOs of
the
primers used can be found in Table III. Indicated in Figure 6B are the cyclin
destruction
box (black shaded box) and the cyclin box motifs I and 2 (both in gray shaded
boxes).
Figure 7depicts the cDNA sequence of the Arabidopsis thaliana CCP6. The
complete nucleotide sequence corresponds to nucleic acids 1 to 2766 of SEQ ID
NO:42.
Underlined is the partially characterized nucleotide (SEQ ID NO:6) sequence.
Indicated
are the stop and start codons (both in black shaded boxes) which are part of
the primers
(grey shaded boxes) used to amplify the coding region of CCP6 by PCR. SEQ ID
NOs of
the primers used can be found in Table III. Nucleotide sequence differences
between SEQ
ID NO:42 and SEQ ID NO:6 are depicted.
Figure 8 depicts the predicted amino acid sequence of the Arabidopsis thaliana
CCP6. The complete amino acid sequence corresponds to amino acids 1 to 901 of
SEQ
ID NO:108. Underlined is the predicted partial amino acid (SEQ ID NO:72)
sequence.
Figure 9 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP7/CCP8. The complete nucleotide sequence (Figure 9A)
corresponds to nucleic,acids 1 to 1260 of SEQ ID NO:43. The complete amino
acid
sequence (Figure 9B) corresponds to amino acids 1 to 358 of SEQ ID NO:109.
Underlined
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
in Figure 9A and Figure 9B are the partially characterized nucleotide (SEQ ID
NO:7) and
predicted partial amino acid (SEQ ID NO:73) sequence, respectively. Italic
sequences in
Figure 9A and Figure 9B correspond to the partially characterized nucleotide
(SEQ ID
NO:8) and amino acid (SEQ ID NO:74) sequence, respectively, of another clone
found
independently to interact with an AtE2F protein in a yeast two-hybrid screen.
Indicated in
Figure 9A are the stop and start codons (both in black shaded boxes) which are
part of the
primers (grey shaded boxes) used to amplify the coding region of CCP7/8 by
PCR. SEQ
ID NOs of the primers used can be found in Table III. Nucleotide sequence
differences
between SEQ ID NO:43 and SEQ ID NO:7-8 are depicted.
Figure 10 depicts the eDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP9. The complete nucleotide sequence (Figure 1OA)
corresponds
to nucleic acids 1 to 1308 of SEQ ID NO:9. The complete amino acid sequence
(Figure
I OB) corresponds to amino acids 1 to 436 of SEQ ID NO:75. Indicated in Figure
1 OA are
the stop and start codons (both in black shaded boxes) which are part of the
primers (grey
shaded boxes) used to amplify the coding region of CCP9 by PCR. SEQ ID NOs of
the
primers used can be found in Table III. Indicated in Figure I OB are the
cyclin destruction
box (black shaded box) and the cyclin box motifs 1 and 2 (both in gray shaded
boxes).
Figure 11 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP10. The complete nucleotide sequence (Figure I IA)
corresponds to nucleic acids I to 1006 of SEQ ID NO: 10. The complete amino
acid
sequence (Figure 11B) corresponds to amino acids 1 to 254 of SEQ ID NO:76.
Indicated in
Figure 1IA are the stop and start codons (both in black shaded boxes) which
are part of the
primers (grey shaded boxes) used to amplify the coding region of CCP10 by PCR.
SEQ ID
NOs of the primers used can be found in Table III.
Figure 12 depicts the eDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP 11. The complete nucleotide sequence (Figure 12A)
corresponds to nucleic acids 1 to 653 of SEQ ID NO:44. Indicated in Figure 12A
are the
stop and start codons (both in black shaded boxes) which are part of the
primers (grey
shaded boxes) used to amplify the coding region of CCPI 1 by PCR. SEQ ID NOs
of the
primers used can be found in Table III. However, during prediction of the open
reading
frame a frame shift was introduced which effected the CCP11 open reading
frame. The
stop codon indicated in italics in a black shaded box is the putative correct
stop codon.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-9-
The amino acid sequence in Figure 12B corresponds to amino acids 1 to 86 of
SEQ ID
NO:77, the protein encoded by the initially identified open reading frame of
SEQ ID
NO:11. The putative correct complete amino acid sequence in Figure 12C
corresponds to
amino acids 1 to 98 of SEQ ID NO: 110.
Figure 13 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP12/13. The complete nucleotide sequence (Figure 13A)
corresponds to nucleic acids 1 to 1266 of SEQ ID NO:45. The complete amino
acid
sequence (Figure 13B) corresponds to amino acids 1 to 385 of SEQ ID NO:111.
Double
underlined in Figure 13A and Figure 13B are the partially characterized 3'
nucleotide
(SEQ ID NO:12) and C-terminal predicted partial amino acid (SEQ ID NO:78)
sequence,
respectively. Single underlined in Figure 13A and Figure 13B are the partially
characterized 5' nucleotide (SEQ ID NO: 13) and N-terminal predicted partial
amino acid
(SEQ ID NO:79) sequences, respectively. Indicated in Figure 13A are the stop
and start
codons (both in black shaded boxes) and the primers (grey shaded boxes) used
to amplify
the coding region of CCP12/13 by PCR. SEQ ID NOs of the primers used can be
found in
Table III. Nucleotide sequence differences between SEQ ID NO:45 and SEQ ID
NO:12 are
depicted.
Figure 14 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP14. The complete nucleotide sequence (Figure 14A)
corresponds to nucleic acids 1 to 1520 of SEQ ID NO:46. The complete amino
acid
sequence (Figure 14B) corresponds to amino acids I to 465 of SEQ ID NO: 112.
Underlined in Figure 14A and Figure 14B are the partially characterized
nucleotide (SEQ
ID NO:14) and predicted partial amino acid (SEQ ID NO:80) sequence,
respectively.
Indicated in Figure 14A are the stop and start codons (both in black shaded
boxes) which
are part of the primers (grey shaded boxes) used to amplify the coding region
of CCP14 by
PCR. SEQ ID NOs of the primers used can be found in Table III.
Figure 15 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP 15. The complete nucleotide sequence (Figure 15A)
corresponds to nucleic acids I to 1142 of SEQ ID NO:47. The complete amino
acid
sequence (Figure 1B) corresponds to amino acids I to 313 of SEQ ID NO: 113.
Underlined
in Figure 15A and Figure 15B are the partially characterized nucleotide (SEQ
ID NO:15)
and predicted partial amino acid (SEQ ID NO: 81) sequence, respectively.
Indicated in
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-10-
Figure 15A are the stop and start codons (both in black shaded boxes) which
are part of the
primers (grey shaded boxes) used to amplify the coding region of CCP15 by PCR.
SEQ
ID NOs of the primers used can be found in Table III. Nucleotide sequence
differences
between SEQ ID NO:47 and SEQ ID NO:15 are depicted. Indicated in Figure 15B
are the
PSTTLRE motif (boxed) characteristic for the subclass of plant PSTTLRE CDC2
kinases.
Further indicated in Figure 15B are three CDC2 motifs (black shaded box, grey
shaded box
and double underlined). Other residues conserved in CDC2s are underscored by
`*'
(residues in common with ProDom domain PD198850), `+' (residues in common with
ProDom domain PD015684), `-` (residues in common with ProDom domain PD063669),
and ` 1' (residues in common with ProDom domain PD 195780).
Figure 16 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP 16. The complete nucleotide sequence (Figure 16A)
corresponds to nucleic acids 1 to 1189 of SEQ ID NO:48. The complete amino
acid
sequence (Figure 16B) corresponds to amino acids 1 to 292 of SEQ ID NO: 114.
Indicated
in Figure 16A are the stop and the three possible start codons (all in black
shaded boxes)
and the primers (grey shaded boxes) used to amplify the coding region of CCP16
by PCR.
SEQ ID NOs of the primers used can be found in Table III. Nucleotide sequence
differences between SEQ ID NO:48 and SEQ ID NO: 16 are depicted. Indicated in
Figure
16B are the DNA binding domain (black shaded box), DEF domain (grey shaded
box),
DCB 1 domain (single underlined) and DCB2 domain (double underlined), all
domains
characteristic for a DP protein.
Figure 17 depicts the CDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP17. The complete nucleotide sequence (Figure 17A)
corresponds to nucleic acids 1 to 794 of SEQ ID NO: 17. The complete amino
acid
sequence (Figure 17B) corresponds to amino acids 1 to 173 of SEQ ID NO:83.
Indicated in
Figure 17A are the stop and start codons (both in black shaded boxes) which
are part of the
primers (grey shaded boxes) used to amplify the coding region of CCP17 by PCR.
SEQ
ID NOs of the primers used can be found in Table III.
Figure 18 depicts the CDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP18. The complete nucleotide sequence (Figure 18A)
corresponds to nucleic acids I to 805 of SEQ ID NO:49. The complete amino acid
sequence (Figure 18B) corresponds to amino acids 1 to 165 of SEQ ID NO:115.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-11-
Underlined in Figure 15A and Figure 15B are the partially characterized
nucleotide (SEQ
ID NO:18) and predicted partial amino acid (SEQ ID NO:84) sequence,
respectively.
Indicated in Figure 18A are the stop and start codons (both in black shaded
boxes) which
are part of the primers (grey shaded boxes) used to amplify the coding region
of CCP18 by
PCR. SEQ ID NOs of the primers used can be found in Table III.
Figure 19 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP 19. The complete nucleotide sequence (Figure 19A)
corresponds to nucleic acids 1 to 1152 of SEQ ID NO:19. The complete amino
acid
sequence (Figure 1B) corresponds to amino acids 1 to 383 of SEQ ID NO:85.
Indicated in
Figure 19A are the stop and start codons (both in black shaded boxes) which
are part of the
primers (grey shaded boxes) used to amplify the coding region of CCP19 by PCR.
SEQ
ID NOs of the primers used can be found in Table III.
Figure 20 depicts the cDNA sequence of the Arabidopsis thaliana CCP20/21. The
complete nucleotide sequence corresponds to nucleic acids 1 to 1539 of SEQ ID
NO:50.
Underlined are the partially characterized 5' nucleotide (SEQ ID NO:20)
sequence and the
partially characterized 3' nucleotide (SEQ ID NO:2 1). Indicated in Figure 20
are the stop
and start codons (both in black shaded boxes) which are part of the primers
(grey shaded
boxes) used to amplify the coding region of CCP20/21 by PCR. SEQ ID NOs of the
primers used can be found in Table III. Nucleotide sequence differences
between SEQ ID
NOs:20-21 and SEQ ID NO:50 are depicted.
Figure 21 depicts the predicted amino acid sequence. of the Arabidopsis
thaliana
CCP20/21. The complete amino acid sequence corresponds to amino acids 1 to 432
of
SEQ ID NO: 116. Underlined are the partially characterized N-terminal
predicted partial
amino acid (SEQ ID NO:50) sequence and the partially characterized C-terminal
amino
predicted partial acid (SEQ ID NO: 87) sequence. Indicated are further
differences in
amino acid sequence between SEQ ID NO:87 and SEQ ID NO:116.
Figure 22 depicts the cDNA sequence of the Arabidopsis thaliana CCP22. The
complete nucleotide sequence corresponds to nucleic acids I to 1977 of SEQ ID
NO:5 1.
Underlined is the partially characterized nucleotide (SEQ ID NO:22). Indicated
are the
stop and start codons (both in black shaded boxes) which are part of the
primers (grey
shaded boxes) used to amplify the coding region of CCP22 by PCR. SEQ ID NOs of
the
primers used can be found in Table Ill.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-12-
Figure 23 depicts the predicted amino acid sequence of the Arabidopsis
thaliana
CCP22. The complete amino acid sequence corresponds to amino acids 1 to 559 of
SEQ
ID NO: 117. Underlined is the predicted partial amino acid (SEQ ID NO:88)
sequence.
Figure 24 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP23. The complete nucleotide sequence (Figure 24A)
corresponds to nucleic acids 1 to 525 of SEQ ID NO:52. Indicated in Figure 24A
are the
stop and start codons (both in black shaded boxes) which are part of the
primers (grey
shaded boxes) used to amplify the coding region of CCP23 by PCR. SEQ ID NOs of
the
primers used can be found in Table III. Nucleotide sequence differences
between SEQ ID
NOs:23 and SEQ ID NO:52 are depicted. The amino acid sequence in Figure 24B
corresponds to amino acids 1 to 98 of SEQ ID NO:89. The complete amino acid
sequence
in Figure 24C corresponds to amino acids 1 to 86 of SEQ ID NO:118.
Figure 25 depicts the cDNA sequence of the Arabidopsis thaliana CCP24. The
complete nucleotide sequence corresponds to nucleic acids 1 to 2610 of SEQ ID
NO:53.
Underlined is the partially characterized nucleotide (SEQ ID NO:24). Indicated
are the
stop and start codons (both in black shaded boxes) which are part of the
primers (grey
shaded boxes) used to amplify the coding region of CCP24 by PCR. SEQ ID NOs of
the
primers used can be found in Table III.
Figure 26 depicts the predicted amino acid sequence of the Arabidopsis
thaliana
CCP24. The complete amino acid sequence corresponds to amino acids 1 to 784 of
SEQ
ID NO: 119. Underlined is the predicted partial amino acid (SEQ ID NO:90)
sequence.
Figure 27 depicts the cDNA sequence of the Arabidopsis thaliana CCP25. The
complete nucleotide sequence corresponds to nucleic acids 1 to 2235 of SEQ ID
NO:54.
Underlined is the partially characterized nucleotide (SEQ ID NO:25) sequence.
Indicated
are stop and start codon (both in black shaded boxes) which are part of the
primers (grey
shaded boxes) used to amplify the coding region of CCP25 by PCR. SEQ ID NOs of
the
primers used can be found in Table III.
Figure 28 depicts the predicted amino acid sequence of the Arabidopsis
thaliana
CCP25. The complete amino acid sequence corresponds to amino acids I to 724 of
SEQ
ID NO:120. Underlined is the predicted partial amino acid (SEQ ID NO:91)
sequence.
Figure 29 depicts the eDNA sequence of the Arabidopsis thaliana CCP26. The
complete nucleotide sequence corresponds to nucleic acids 1 to 4002 of SEQ ID
NO:55.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-13-
Underlined is the partially characterized nucleotide (SEQ ID NO:26) sequence.
Indicated
are stop and start codon (both in black shaded boxes) which are part of the
primers (grey
shaded boxes) used to amplify the coding region of CCP26 by PCR. SEQ ID NOs of
the
primers used can be found in Table III. Nucleotide sequence differences
between SEQ ID
NOs:26 and SEQ ID NO:55 are depicted.
Figure 30 depicts the predicted amino acid sequence.of the Arabidopsis
thaliana
CCP26. The complete amino acid sequence corresponds to amino acids I to 1313
of SEQ
ID NO:121. Underlined is the predicted partial amino acid (SEQ ID NO:92)
sequence.
Amino acid sequence differences between SEQ ID NOs:92 and SEQ ID NO:121 are
depicted.
Figure 31 depicts the eDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP27. The complete nucleotide sequence (Figure 3 1A)
corresponds to nucleic acids 1 to 1251 of SEQ ID NO:56. The complete amino
acid
sequence (Figure 31 B) corresponds to amino acids 1 to 310 of SEQ ID NO:122.
Underlined in Figure 31A and Figure 31B are the partially characterized
nucleotide (SEQ
ID NO:27) and predicted partial amino acid (SEQ ID NO:93) sequence,
respectively.
Indicated in Figure 31 A are the stop and start codons (both in black shaded
boxes) which
are part of the primers (grey shaded boxes) used to amplify the coding region
of CCP27 by
PCR. SEQ ID NOs of the primers used can be found in Table III. Nucleotide
sequence
differences between SEQ ID NO:27 and SEQ ID NO:56 are depicted in Figure 31A.
Figure 32 depicts the eDNA sequence of the Arabidopsis thaliana CCP28. The
complete nucleotide sequence corresponds to nucleic acids 1 to 2955 of SEQ ID
NO:56.
Underlined is the partially characterized nucleotide (SEQ ID NO:28) sequence.
Indicated
are the stop and start codons (both in black shaded boxes) which are part of
the primers
(grey shaded boxes) used to amplify the coding region of CCP28 by PCR. SEQ ID
NOs of
the primers used can be found in Table III. Nucleotide sequence differences
between SEQ
ID NO:28 and SEQ ID NO:57 are depicted.
Figure 33 depicts the predicted amino acid sequence of the Arabidopsis
thaliana
CCP28. The complete amino acid sequence corresponds to amino acids 1 to 964 of
SEQ
ID NO:123. Underlined is the predicted partial amino acid (SEQ ID NO:94)
sequence.
Figure 34 depicts the eDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP29. The complete nucleotide sequence (Figure 34A)
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
-14-
corresponds to nucleic acids 1 to 546 of SEQ ID NO:29. The complete amino acid
sequence (Figure 34B) corresponds to amino acids 1 to 181 of SEQ ID NO:95.
Indicated in
Figure 34A are the stop and start codons (both in black shaded boxes) which
are part of the
primers (grey shaded boxes) used to amplify the coding region of CCP29 by PCR.
SEQ ID
NOs of the primers used can be found in Table III.
Figure 35 depicts the cDNA sequences and predicted amino acid sequences of the
Arabidopsis thaliana CCP30. The complete nucleotide sequence (Figure 35A)
corresponds to nucleic acids 1 to 492 of SEQ ID NO:30. Indicated in Figure 35A
are the
stop and start codons (both in black shaded boxes) , the complete sense primer
and part of
the antisense primer (grey shaded boxes) used to amplify the coding region of
CCP30 by
PCR. SEQ ID NOs of the primers used can be found in Table III. However, after
sequencing of the PCR product a sequence error in SEQ ID NO:30 was detected
(boxed
nucleotide `a' in Figure 35A not present) which caused a frame shift
effectuating the
CCP30 open reading frame. The putative correct cDNA sequence is given in
Figure 35B
(nucleic acids 1 to 865 of SEQ ID NO:58) wherein the three putative start
codons are
marked by a black shaded box. The originally identified start codon is
indicated in bold
letters. The stop codon is unaltered. The amino acid sequence in Figure 35C
corresponds
to amino acids 1 to 163 of SEQ ID NO:96, the protein encoded by the initially
identified
open reading frame of SEQ ID NO:30. The putative correct complete amino acid
sequence
in Figure 35D corresponds to amino acids 1 to 222 of SEQ ID NO:124 which
comprises
the longest possible open reading frame. The Met residues corresponding to the
three
possible start codons in SEQ ID NO:58 (Figure 35B) are bold faced.
Figure 36 depicts the cDNA sequence of the Arabidopsis thaliana CCP3 1. The
complete nucleotide sequence corresponds to nucleic acids 1 to 723 of SEQ ID
NO:31.
Indicated in Figure IA are the stop and start codons (both in black shaded
boxes).
Figure 37 depicts the predicted amino acid sequence of the Arabidopsis
thaliana
CCP3 1. The complete amino acid sequence corresponds to amino acids 1 to 148
of SEQ
ID NO:125.
Figure 38 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP32. The complete nucleotide sequence (Figure 38A)
corresponds to nucleic acids I to 426 of SEQ ID NO:60. The complete amino acid
sequence (Figure 38B) corresponds to amino acids 1 to 70 of SEQ ID NO:126.
Underlined
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-15-
in Figure 38A is the partially characterized nucleotide (SEQ ID NO:32)
sequence.
Indicated in Figure 38A are the stop and start codons (both in black shaded
boxes) which
are part of the primers (grey shaded boxes) used to amplify the coding region
of CCP32 by
PCR. SEQ ID NOs of the primers used can be found in Table III. Figure 38C
gives the
originally erroneously predicted amino acid sequence of CCP32 (amino acids I
to 38 of
SEQ ID NO:98).
Figure 39 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP33. The complete nucleotide sequence (Figure 39A)
corresponds to nucleic acids 1 to 1442 of SEQ ID NO:61. The complete amino
acid
sequence (Figure 39B) corresponds to amino acids 1 to 385 of SEQ ID NO:127.
Indicated
in Figure 39A are the stop and start codons (both in black shaded boxes) which
are part of
the primers (grey shaded boxes) used to amplify the coding region of CCP33 by
PCR. SEQ
ID NOs of the primers used can be found in Table III. Indicated in Figure 39B
are the
DNA binding domain (black shaded box), DEF domain (grey shaded box), DCBI
domain
(single underlined) and DCB2 domain (double underlined), all domains
characteristic for a
DP protein.
Figure 40 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP34. The complete nucleotide sequence (Figure 40A)
corresponds to nucleic acids 1 to 1506 of SEQ ID NO:62. The complete amino
acid
sequence (Figure 40B) corresponds to amino acids 1 to 437 of SEQ ID NO:128.
Underlined in Figure 40A and Figure 40B are the partially characterized
nucleotide (SEQ
ID NO:34) and predicted partial amino acid (SEQ ID NO:62) sequence,
respectively.
Indicated in Figure 40A are the stop and start codons (both in black shaded
boxes) which
are part of the primers (grey shaded boxes) used to amplify the coding region
of CCP34 by
PCR. SEQ ID NOs of the primers used can be found in Table III.
Figure 41 depicts the cDNA sequence of the Arabidopsis thaliana CCP35. The
complete nucleotide sequence corresponds to nucleic acids I to 2631 of SEQ ID
NO:63.
Underlined is the partially characterized nucleotide (SEQ ID NO:35) sequence.
Indicated
are the stop and start codons (both in black shaded boxes) and of the primers
(grey shaded
boxes) used to amplify the coding region of CCP35 by PCR. SEQ ID NOs of the
primers
used can be found in Table III. Nucleotide sequence differences between SEQ ID
NO:33
and SEQ ID NO:63 are depicted.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-16-
Figure 42 depicts the predicted amino acid sequence of the Arabidopsis
thaliana
CCP35. The complete amino acid sequence corresponds to amino acids 1 to 749 of
SEQ
ID NO:129. Underlined is the predicted partial amino acid (SEQ ID NO:101)
sequence.
Figure 43 depicts the cDNA sequence of the Arabidopsis thaliana CCP36. The
complete nucleotide sequence corresponds to nucleic acids 1 to 2743 of SEQ ID
NO:64.
Underlined is the partially characterized nucleotide (SEQ ID NO:36) sequence.
Indicated
are the stop and start codons (both in black shaded boxes). Nucleotide
sequence differences
between SEQ ID NO:36 and SEQ ID NO:64 are depicted.
Figure 44 depicts the predicted amino acid sequence of the Arabidopsis
thaliana
CCP36. The complete amino acid sequence corresponds to amino acids 1 to 742 of
SEQ
ID NO:130. Underlined is the predicted partial amino acid (SEQ ID NO:102)
sequence.
Figure 45 depicts the cDNA sequence of the Arabidopsis thaliana CCP37. The
complete nucleotide sequence corresponds to nucleic acids 1 to 2959 of SEQ ID
NO:65.
Underlined is the partially characterized nucleotide (SEQ ID NO:37) sequence.
Indicated
are the stop and start codons (both in black shaded boxes) and primers (grey
shaded boxes)
used to amplify the coding region of CCP45 by PCR. SEQ ID NOs of the primers
used can
be found in Table III.
Figure 46 depicts the predicted amino acid sequence of the Arabidopsis
thaliana
CCP37. The complete amino acid sequence corresponds to amino acids I to 911 of
SEQ
ID NO: 131. Underlined is the predicted partial amino acid (SEQ ID NO: 103)
sequence.
Indicated in a black shaded box is a SAP-like domain.
Figure 47 depicts the cDNA sequence and predicted amino acid sequence of the
Arabidopsis thaliana CCP38. The complete nucleotide sequence (Figure 47A)
corresponds to nucleic acids 1 to 1295 of SEQ ID NO:66. The complete amino
acid
sequence (Figure 47B) corresponds to amino acids 1 to 357 of SEQ ID NO:132.
Underlined in Figure 47A and Figure 47B are the partially characterized
nucleotide (SEQ
ID NO:38) and predicted partial amino acid (SEQ ID NO:104) sequence,
respectively.
Indicated in Figure 47A are the stop and start codons (both in black shaded
boxes) which
are part of the primers (grey shaded boxes) used to amplify the coding region
of CCP38 by
PCR. SEQ ID NOs of the primers used can be found in Table III.
Figure 48 depicts phosphorylation of the Arabidopsis thaliana CCP4 by CDKs.
The protein CDC2bDN-IC26M (SEQ ID NO:70) contains a consensus CDK
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-17-
phosphorylation site (TPWK, residues 54-57 of SEQ ID NO:263). The
corresponding gene
(SEQ ID NO:4) was expressed in E. coli and the protein was purified from the
crude
extracts. The purified protein was subsequently shown to be phosphorylated by
CDKs in
an in vitro CDK phosphorylation assay. -: no IC26M added; +: IC26M added.
Figure 49 schematically represents the domain organization of AtE2Fa and
AtE2Fb. The DNA-binding domain (DB), the dimerization domain (DIM), the marked
box
(MB), and the Rb-binding domain (RB) are indicated by marked boxes, the N-
terminal
domains are indicated by open boxes. Numbering on the right refers to the
amino acid
sequence contained in the different AtE2F constructs, which were used in the
in vitro
binding assays.
Figure 50 depicts AtDPa in vitro interactions with AtE2Fa and AtE2Fb. The c-
myc-tagged AtDPa (c-myc-AtDPa) was in vitro translated and used as control.
The lower
migrating proteins observed in the case of c-rnyc-AtDPa are most probably due
to initiation
of translation at internal methionine codons (panel A, unnumbered left lane).
The c-myc-
AtDPa was in vitro co-translated with HA-AtE2Fb (panels A and B, lane 1), HA-
AtE2Fa
(panels B, lane 2), the C-terminal deleted form of HA-AtE2Fb (panels A and B,
lane 3),
HA-AtE2Fa 1-420 (panels A and B, lane 4) and the N-terminal truncated form of
HA-
AtE2Fa 162-485 (panels A and B, lane 5) as indicated. Numbers in the case of
the mutant
AtE2Fs refer to the amino acid sequence contained in these constructs (see
Figure 49). An
aliquot of each sample was analyzed directly by SDS-PAGE and autoradiographed
(panel
A; total IVT, total in vitro translation). Another aliquot of the same samples
was subjected
to immunoprecipitation with anti-c-myc monoclonal antibodies (panel B), lanes
are
indicated by numbering. The position of c-myc-AtDPa proteins are marked by
arrows in
both panels. Molecular mass markers are indicated at the left.
Figure 51 shows AtDPb in vitro interactions with AtE2Fa and AtE2Fb. The c-myc-
tagged AtDPb (c-myc-AtDPb, panels A and B, lane 2) and the HA-tagged AtE2Fb
(HA-
AtE2Fb, panels A and B, lane 1) were in vitro translated and used as controls.
The lower
migrating proteins observed in the case of c-myc-AtDPb are most probably due
to initiation
of translation at internal methionine codons (panel A, lane 2). The c-myc-
AtDPb was in
vitro co-translated with HA-AtE2Fb (panels A and B, lane 3), HA-AtE2Fa (panels
A and
B lane 4), HA-AtE2Fa 1-420 (panels A and B, lane 5) and the N-terminal
truncated form
of HA-AtE2Fa 162-485 (panels A and B, lane 6) as indicated. Numbers in the
case of the
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-18-
mutant AtE2Fs refer to the amino acid sequence contained in these constructs
(see Figure
49). An aliquot of each sample was analyzed directly by SDS-PAGE and
autoradiographed
(panel A; total IVT, total in vitro translation). Another aliquot of the same
samples was
subjected to immunoprecipitation with anti-c-myc monoclonal antibodies (panel
B), lanes
are indicated by numbering. The c-myc-AtDPb (panels A and B, lanes 2-6;
indicated with
`y') co-migrated almost exactly with the mutant HA-AtE2Fa 1-420 (panels A and
B, lane
5; indicated with `x') and HA-AtE2Fa 162-485 (panels A and B, lane 6;
indicated with `z')
in the gel system. These polypeptides as well as the position of c-myc-AtDPa
and c-myc-
AtDPb proteins are marked by arrows marked with `y', `x' and `z', respectively
(cfr.
supra). Molecular mass markers are indicated at the left.
Figure 52 schematically represents AtDPa and mutants. The DNA-binding domain
(DB) and the dimerization domain (DIM) are indicated by marked boxes, N- and C-
terminal regions are indicated by open boxes. Numbering on the right side
refers to the
amino acid sequence contained in the different AtDP constructs, which were
used in the in
vitro binding assays.
Figure 53 schematically represents AtDPb and mutants. The DNA-binding domain
(DB) and the dimerization domain (DIM) are indicated by marked boxes, N- and C-
terminal regions are indicated by open boxes. Numbering on the right side
refers to the
amino acid sequence contained in the different AtDP constructs, which were
used in the in
vitro binding assays.
Figure 54 shows the mapping of regions in AtDPa required for in vitro binding
to
AtE2Fb. HA-AtE2Fb was co-translated with series of c-myc-AtDPa mutants. An
aliquot of
each sample was analyzed directly by SDS-PAGE and autoradiographed (panel A).
Another aliquot of the same samples was subjected to immunoprecipitation with
anti-HA
(panel B) or anti-c-myc (panel C) monoclonal antibodies. The c-myc-AtDPa
mutants are
marked by dots. Positions of the HA-AtE2Fb proteins are indicated by arrows.
Molecular
mass markers are indicated at the left.
Figure 55 shows the mapping of regions in AtDPb required for in vitro binding
to
AtE2Fb. HA-AtE2Fb was co-translated with series c-myc-AtDPb mutants. An
aliquot of
each sample was analyzed directly by SDS-PAGE and autoradiographed (panel A).
Another aliquot of the same samples was subjected to immunoprecipitation with
anti-HA
(panel B) or anti-c-myc (panel C) monoclonal antibodies. The c-myc-AtDPb
mutants are
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-19-
marked by dots. Positions of the HA-AtE2Fb proteins are indicated by arrows.
Molecular
mass markers are indicated at the left.
Figure 56 shows the mapping of regions in AtDPb required for in vitro binding
to
AtE2Fb. HA-AtE2Fb was co-translated with c-myc-AtDPb 182-263. Because of the
small
size of this protein, it was hardly detectable when it was directly analyzed
by SDS-PAGE
(data not shown). An aliquot of this sample was subjected to
immunoprecipitation with
anti-c-myc monoclonal antibodies. The c-myc-AtDP mutant is marked by dots.
Position of
the HA-AtE2Fb protein is indicated by an arrow. Molecular mass markers are
indicated at
the left.
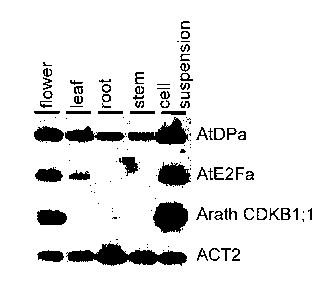
Figure 57 shows organ- and cell cycle-specific expression of AtE2Fa and AtDPa.
Tissue-specific expression of AtDPa and AtE2Fa genes. cDNA prepared from the
indicated tissues was subjected to semi-quantitative RT-PCR analysis. The
Arath;CDKB 1; 1 gene was used as a marker for highly proliferating tissues.
The actin 2
gene (ACT2) was used as loading control.
Figure 58 shows organ- and cell cycle-specific expression of AtE2Fa and AtDPa.
Co-regulated cell cycle phase-dependent transcription of AtE2Fa and AtDPa. The
cDNA
was prepared from partially synchronized Arabidopsis cells harvested at the
indicated time
point after removal of the cell cycle blocker was subjected to semi-
quantitative RT-PCR
analysis. Histone H4 and Arath;CDKB 1;1 were used as markers for S and G2/M
phase,
respectively, and ROC5 and Arath;CDKA;I as loading controls.
Figure 59 is a photographic representation of Northern blotting analysis of
DPa
expression in independent Arabidopsis thaliana DPa overexpressing lines (lines
16-27 as
indicated) and one untransformed control line (indicated by Q.
Figure 60 describes the molecules defined in SEQ ID NOs: 199-204 and 240-290.
Detailed Description of the Invention
The present invention is based, at least in part, on the discovery of novel
molecules,
referred to herein as "cell cycle proteins" or "CCP " nucleic acid and
polypeptide
molecules. The CCP molecules of the present invention were identified based on
their
ability, as determined using yeast two-hybrid assays (described in detail in
Example 1), to
interact with proteins involved in the cell cycle, such as plant cyclin
dependent kinases
(e.g., a dominant negative form of CDC2b, CDC2bAt.N161), cyclin dependent
kinase
subunits referred herein as "CKS" (such as CKS 1 At), cyclin dependent kinase
inhibitors
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-20-
referred to herein as "CKI" (such as CKI4), PHO80-like proteins referred to
herein as
"PLP", E2F, and different domains of kinesin-like proteins referred to herein
as "KLPNT.
Because of their ability to interact with (e.g., bind to) the cyclin dependent
kinases, the CCP molecules of the present invention may modulate, e.g.,
upregulate or
downregulate, the activity of plant CDKs, such as CDC2a or CDC2b; CKSs, CKIs,
PLPs
and KLPNTs. Furthermore, because of their ability to interact with (e.g., bind
to) the
aforementioned proteins which are proteins involved in cell cycle regulation,
the CCP
molecules of the present invention may also play a role in or function in cell
cycle
regulation, e.g., plant or animal cell cycle regulation.
As used herein, the term "cell cycle protein" includes a polypeptide which is
involved in controlling or regulating the cell cycle, or part thereof, in a
cell, tissue, organ
or whole organism. Cell cycle proteins may also be capable of binding to,
regulating, or
being regulated by cyclin dependent kinases, such as plant cyclin dependent
kinases, e.g.,
CDC2a or CDC2b, or their subunits. The term cell cycle protein also includes
peptides,
polypeptides, fragments, variant, homologs, alleles or precursors (e.g., pre-
proteins or pro-
proteins) thereof.
As used herein, the term "cell cycle" includes the cyclic biochemical and
structural
events associated with growth, division and proliferation of cells, and in
particular with the
regulation of the replication of DNA and mitosis. The cell cycle is divided
into periods
called: Go, Gap, (G,), DNA synthesis (S), Gape (G2), and mitosis (M). Normally
these four
phases occur sequentially, however, the cell cycle also includes modified
cycles wherein
one or more phases are absent resulting in modified cell cycle such as
endomitosis,
acytokinesis, polyploidy, polyteny, and endoreduplication.
As used herein, the term "plant" includes reference to whole plants, plant
organ
(e.g., leaves, stems, roots), plant tissue, seeds, and plant cells and progeny
thereof. Plant
cell, as used herein includes, without limitation, seeds, e.g., seed
suspension cultures,
embryos, meristematic regions, callus tissue, leaves, roots, shoots,
gametophytes,
sporophytes, pollen, and microspores. The class of plants which can be used in
the
methods of the invention is generally as broad as the class of higher plants
amenable of
transformation techniques, including both monocotyledonous and dicotyledonous
plants.
Particularly preferred plants are Arabidopsis thaliana, rice, wheat, maize,
tomato, alfalfa,
oilseed rape, soybean, cotton, sunflower or canola. The term plant also
includes
monocotyledonous (monocot) plants and dicotyledonous (dicot) plants including
a fodder
or forage legume, ornamental plant, food crop, tree, or shrub selected from
the list
comprising Acacia spp., Acer spp., Actinidia spp.,Aesculus spp., Agathis
australis, Albizia
amara, Alsophila tricolor, Andropogon spp., Arachis spp, Areca catechu,
Astelia fragrans,
Astragalus titer, Baikiaea pluri/uga, Betula spp., Brassica spp., Bruguiera
gymnorrhiza,
Burkea africana, Butea fi=ondosa, Cadabafarinosa, Calliandra spp, Camellia
sinensis,
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-21-
Canna indica, Capsicum spp., Cassia spp., Centroema pubescens, Chaenomeles
spp., Cinnamomum cassia, Coffea arabica, Colophospermum mopane, Coronillia
varia,
Cotoneaster serotina, Crataegus spp., Cucumis spp., Cupressus spp., Cyathea
dealbata,
Cydonia oblonga, Cryptomeriajaponica, Cymbopogon spp., Cynthea dealbata,
Cydonia
oblonga, Dalbergia monetaria, Davallia divaricata, Desmodium spp., Dicksonia
squarosa,
Diheteropogon amplectens, Dioclea spp, Dolichos spp., Dorycnium rectum,
Echinochloa
pyramidalis, Ehrartia spp., Eleusine coracana, Eragrestis spp., Erythrina
spp., Eucalyptus
spp., Euclea schimperi, Eulalia villosa, Fagopyrum spp., Feijoa sellowiana,
Fragaria spp.,
Flemingia spp, Freycinetia banksii, Geranium thunbergii, Ginkgo biloba,
Glycine
javanica, Gliricidia spp, Gossypium hirsutum, Grevillea spp., Guibourtia
coleosperma,
Hedysarum spp., Hemarthia altissima, Heteropogon contortus, Hordeum vulgare,
Hyparrhenia rufa, Hypericum erectum, Hyperthelia dissoluta, Indigo incarnata,
Iris spp.,
Leptarrhena pyrolifolia, Lespediza spp., Lettuca spp., Leucaena leucocephala,
Loudetia
simplex, Lotonus bainesii, Lotus spp., Macrotyloma axillare, Malus spp.,
Manihot
esculenta, Medicago sativa, Metasequoia glyptostroboides, Musa sapientum,
Nicotianum
spp., Onobrychis spp., Ornithopus spp., Oryza spp., Peltophorum afr-icanum,
Pennisetum
spp., Persea gratissima, Petunia spp., Phaseolus spp., Phoenix canariensis,
Phormium
cookianum, Photinia spp., Picea glauca, Pinus spp., Pisum sativum, Podocarpus
totara,
Pogonarthriafleckii, Pogonarthria squarrosa, Populus spp., Prosopis cineraria,
Pseudotsuga menziesii, Pterolobium stellatum, Pyrus communis, Quercus spp.,
Rhaphiolepsis umbellata, Rhopalostylis sapida, Rhus natalensis, Ribes
grossularia, Ribes
spp., Robinia pseudoacacia, Rosa spp., Rubus spp., Salix spp., Schyzachyrium
sanguineum, Sciadopitys verticillata, Sequoia sempervirens, Sequoiadendron
giganteum,
Sorghum bicolor, Spinacia spp., Sporobolusfimbriatus, Stiburus alopecuroides,
Stylosanthos humilis, Tadehagi spp, Taxodium distichum, Themeda triandra,
Trifolium
spp., Triticum spp., Tsuga heterophylla, Vaccinium spp., Vicia spp. Vitis
vinifera, Watsonia
pyramidata, Zantedeschia aethiopica, Zea mays, amaranth, artichoke, asparagus,
broccoli,
brussel sprout, cabbage, canola, carrot, cauliflower, celery, collard greens,
flax, kale, lentil,
oilseed rape, okra, onion, potato, rice, soybean, straw, sugarbeet, sugar
cane, sunflower,
tomato, squash, and tea, amongst others, or the seeds of any plant
specifically named
above or a tissue, cell or organ culture of any of the above species.
The cell cycle proteins of the present invention are involved in cell cycle
regulation
which is largely, but not completely, similar in plants and animals.
Accordingly, the
nucleic acid molecules and polypeptide of the invention, or derivatives
thereof, may be
used to modulate the cell cycle in a plant or an animal such as by modulating
the activity
or level or expression of CCP, altering the rate of the cell cycle or phases
of the cell cycle,
and entry into and out of the various cell cycle phases. In plants, the
molecules of the
present invention may be used in agriculture to, for example, improve the
growth
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
-22-
characteristics of plant such as growth rate or size of specific tissues or
organs, the
architecture or morphology of the plant, increase crop yield, improve
tolerance to
environmental stress conditions (such as drought, salt, temperature, or
nutrient
deprivation), improve tolerance to plant pathogens that abuse the cell cycle
or as targets to
facilitate the identification of inhibitors or activators of CCPs that may be
useful as
phytopharmaceuticals such as herbicides or plant growth regulators.
As used herein, the term "cell cycle associated disorders" includes a
disorder,
disease or condition which is caused or characterized by a misregulation
(e.g.,
downregulation or upregulation), abuse, arrest, or modification of the cell
cycle. In plants
cell cycle associated disorders include endomitosis, acytokinesis, polyploidy,
polyteny,
and endoreduplication which may be caused by external factors such as
pathogens
(nematodes, viruses, fungi, or insects), chemicals, environmental stress
(e.g., drought,
temperature, nutrients, or UV) resulting in for instance neoplastic tissue
(e.g., galls, root
knots) or inhibition of cell division/proliferation (e.g., stunted growth).
Cell cycle
associated disorders in animals include proliferative disorders or
differentiative disorders,
such as cancer, e.g., melanoma, prostate cancer, cervical cancer, breast
cancer, colon
cancer, or sarcoma.
The present invention is based, at least in part, on the discovery of novel
molecules,
referred to herein as CCP protein and nucleic acid molecules, which comprise a
family of
molecules having certain conserved structural and functional features. The
term "family"
when referring to the protein and nucleic acid molecules of the invention is
intended to
mean two or more proteins or nucleic acid molecules having a common structural
domain
or motif and having sufficient amino acid or nucleotide sequence homology as
defined
herein. Such family members can be naturally or non-naturally occurring and
can be from
either the same or different species. For example, a family can contain a
first protein of
plant, e.g. Arabidopsis, origin, as well as other, distinct proteins of plant,
e.g., Arabidopsis,
origin or alternatively, can contain homologues of other plants, e.g., rice,
or of non-plant
origin. Members of a family may also have common functional characteristics.
In one embodiment of the invention, a CCP protein of the present invention is
identified based on the presence of at least one or more of the following
domains:
A. Cyclin destruction box
As used herein, the term "Cyclin destruction box" includes a domain of 9-10
amino
acid residues in length which typically contains the following consensus
pattern:
R - X2 - L - X2 - [IN] -X,_2 - N (SEQ ID NO:267),
wherein X can be any amino acid, X,, is a stretch of n Xs, X".m is a stretch
of n to in Xs,
and wherein [IN] means that an Ile or Val residue can occur at that position.
SEQ ID
NO:267 depicts the minimal consensus sequence of the cyclin destruction box
and
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-23-
underlies the ubiquitin-mediated proteolytic destruction of the cyclins
bearing this motif
(Yamano et al. (1998), EMBO J. 17: 5670-5678; Renaudin et al. (1998) in Plant
Cell
Division (Francis, Dudits and Inze, eds.), Portland Press Research Monograph,
Portland
Press Ltd. London (1998), pp 67-98).
B. Cyclin box motif 1
As used herein, the term "Cyclin box motif 1" includes a domain of 8 amino
acid
residues in length and which typically contains the following consensus
pattern:
MRXIL[I/V]DW (SEQ ID NO:268),
wherein X can be any amino acid and wherein [I/V] means that an Ile or Val
residue can
occur at that position. This motif forms part of the helix H1 of the first
cyclin fold and is
the best conserved motif in the cyclinA/B family (Renaudin et al. (1998) in
Plant Cell
Division (Francis, Dudits and Inze, eds.), Portland Press Research Monograph,
Portland
Press Ltd. London (1998), pp 67-98).
C. Cyclin box motif 2
As used herein, the term "Cyclin box motif 2" includes a domain of 8 amino
acid
residues in length and which typically contains the following consensus
pattern:
KYEE - X3 - P (SEQ ID NO:269),
wherein X can be any amino acid and wherein X,, is a stretch of n Xs. This
motif forms
part of the helix H3 of the first cyclin fold wherein the 2 acidic residues
are part of the
CDK binding site (Renaudin et al. (1998) in Plant Cell Division (Francis,
Dudits and Inze,
eds.), Portland Press Research Monograph, Portland Press Ltd. London (1998),
pp 67-98).
D. CDC2 motifs
As used herein, the term "CDC2 motifs" includes domains of about 9-12 amino
acid residues in length and which typically contain one of the following
consensus
patterns:
GXG -X2_ GXVY (SEQ ID NO:270)
HRDXK-X2- NXL (SEQ ID NO:271)
D-X1.2-[W/Y]SXG -X4- E (SEQ ID NO:272)
wherein wherein X can be any amino acid, X,, is a stretch of n Xs, X,,_m is a
stretch of n to
in Xs, and wherein [W/Y] means that an Trp or Tyr residue can occur at that
position.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-24-
E. CDK phosphorylation site
As used herein the term "CDK phosphorylation site" includes a domain of about
5-
7 amino acids in length and which contains one or more of the following
consensus
domains:
TPX,_,[R/K] (SEQ ID NO:273)
SPX[R/K] (SEQ ID NO:274)
SPX(Hu) (SEQ ID NO:275)
SP(Hu)X (SEQ ID NO:276)
with Hu being a hydrophobic uncharged amino acid (M, I, L, V) and X any amino
acid.
The foregoing are typically found in cyclin-dependent kinase substrates such
as histone
kinase, transcription factors such as E2F or transcription regulators like Rb.
CDK
phosphorylation sites are described in, for example, Tamrakar et al. 2000,
Frontiers Biosci
5, d121-137.
CCP proteins of the present invention comprising a CDK phosphorylation site
can
be mutated in said CDK phosphorylation site such that said CCP proteins are no
longer
able to be phosphorylated on the CDK phosphorylation site. Mutations of a CDK
phosphorylation site include all mutations of the ser or thr residue in any of
SEQ ID
NOs:273-276 into a non-phosphorylatable amino acid residue, e.g., an ala or
glu residue.
Mutation of one or more CDK phosphorylation site(s) in a CCP protein of the
invention is
expected to modulate modifications of the CCP protein by CDKs and, thus, to
modulate
the biological or biochemical function of the CCP protein.
F. E Nuclear localisation signal (NLS)
As used herein the term "nuclear localization signal" or "NLS" includes a
domain
conferring to a protein comprising the NLS domain the ability to be imported
into the
nucleus and to, for example, accumulate within the nucleus. NLS domains
include one or
more of the following concensus patterns:
PKKKRKV (SEQ ID NO:277)
KRX,QKKKK (SEQ ID NO:278)
KRPRP (SEQ ID NO:279)
PAAKRVKLD (SEQ ID NO:280)
NLS domains have been found in the SV40 T antigen, in nucleoplasmin (bipartite
NLS), in a Adeno EIA, and in c-Myc. NLS domains are described in, for example,
Laskey
et al. (1998) Biochem. Soc. Trans. 26, 561-567.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-25-
G. Cy-like boxes
As used herein, the term "Cy-like box" includes a domain of 3-6 amino acid
residues in length with has the consensus motif R-X-X-F (SEQ ID NO:281) with X
being
any amino acid and one of two Xs preferably being a hydrophobic residue.
H. Rb binding domain
As used herein, the term "Rb binding domain" includes a domain which when
present in a protein confers to the protein the ability to bind the Rb
protein. Rb binding
domains include one or more of the following concensus paterns:
LXCXE (SEQ ID NO:282)
LXSXE (SEQ ID NO:283)
DYX7EX3DLFD (SEQ ID NO:284)
DYX6DX4DMWE (SEQ ID NO:285)
=15
Rb binding domains have been found in D-cyclins, in protein phosphatase 1, in
human E2F-1, and in plant E2F. Rb binding domains are described in, for
example, Rubin
et al. (1998) Frontiers Biosci 3, d1209-1219; Phelps et al. (1992) J. Virol.
66, 2418-2427,
and Cress et al. (1993) Mol. Cell Biol. 13, 6314-6325.
1. DEF Domain
As used herein the term "DEF domain" includes a protein domain which is
required
for the formation of heterodimers between DP proteins and E2F proteins. DEF
domains
comprise the following concensus pattern:
[D/N/-] [Q/E]KNIR[R/G]RV [Y/D]DALNV [L/F]MA[M/I/L/-] [N/D]
[V/I] I [S/A] [K/R] [D/E]KKEI [K/Q/R/-] W [R/K/I] GLP
(SEQ ID NO:286)
J. DNA Binding Domain
As used herein the term "DNA binding domain" includes a domain which is
involved in the binding of DP proteins and/or DP-E2F heterodimers to DNA. DNA
binding domains include the following concensus pattern:
[G/N][K/R]GLR[H/Q]FS[M/V][K/M][UV]X(e-17)C[E/Q]K[V/L][Q/E/-][S/-]XK[G/K]-
[R/I/-]TT[S/-]Y[N/K]EVADE[L/I] [V/I] [A/S] [E/D]F
(SEQ ID NO:287)
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-26-
DNA binding domains are described in, for example, Hao et al. (1995) J. Cell
Sci.108,
2945-2954; Bandara et al. (1993) EMBO J. 12, 4317-4324; and Girling et al.
(1994) Mol.
Biol. Cell5, 1081-1092.
K. DCB 1 Domain:
As used herein the term "DCB 1 domain" includes a protein domain which is
conserved among DP proteins and has the following concensus patterns:
[R/S] [I/V]X[Q/K]KX3[L/S]XE
(SEQ ID NO:288)
[R/S ] [I/V ]X[Q/K] KX3 [L/S]XE [L/M]X2.3 [Q/H]X4_SNL [V/I/M] [Q/E] RN
(SEQ ID NO:289)
DCB 1 domains are described in, for example, Hao et al. (1995) J. Cell Sci.
108, 2945-
2954; Bandara et al. (1993) EMBO J. 12, 4317-4324; and Girling et al. (1994)
Mol. Biol.
Cell5, 1081-1092.
L. DCB2 Domain:
As used herein the term "DCB2 domain" includes a protein domain which is
conserved among DP proteins and has the following concensus patern:
[L/I]PFI[L/I] [V/L]XTX3.,[T/V]VX12_,4FX34F [E/S] [Hu]HDDX2[V/I]L[R/K]XM
(SEQ ID NO:290)
DCB2 domains are described in, for example, Hao et al. (1995) J Cell Sci. 108,
2945-
2954; Bandara et al. (1993) EMBO J. 12, 4317-4324; and Girling et al. (1994)
Mol. Biol.
Cell 5, 1081-1092.
M. SAP Domain:
As used herein the term SAP motif includes a protein domain of about 35 amino
acid residues which is found in a variety of nuclear proteins involved in
transcription,
DNA repair, DNA processing or apoptotic chromatin degradation. It was named
after SAF-
A/B, Acinus and PIAS, three proteins known to contain it. The SAP motif
reveals a
bipartite distribution of strongly conserved hydrophobic, polar and bulky
amino acids
separated by a region that contains a glycine. The SAP domain has been
proposed to be a
DNA-binding motif (Aravind and Koonin (2000) Trends Biochem. Sci. 25:112-114).
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-27-
Isolated CCP proteins of the present invention have an amino acid sequence
sufficiently identical to the amino acid sequence of SEQ ID NO:67-132, 205,
211, 215-
216, or 220-227 or are encoded by a nucleotide sequence sufficiently identical
to SEQ ID
NO:1-66 or 228-239. As used herein, the term "sufficiently identical" refers
to a first
amino acid or nucleotide sequence which contains a sufficient or minimum
number of
identical or equivalent (e.g., an amino acid residue which has a similar side
chain) amino
acid residues or nucleotides to a second amino acid or nucleotide sequence
such that the
first and second amino acid or nucleotide sequences share common structural
domains or
motifs and/or a common functional activity. For example, amino acid or
nucleotide
sequences which share common structural domains have at least 30%, 40%, or 50%
homology, preferably 60% homology, more preferably 70%-80%, and even more
preferably 90-95% homology across the amino acid sequences of the domains and
contain
at least one and preferably two structural domains or motifs, are defined
herein as
sufficiently identical. Furthermore, amino acid or nucleotide sequences which
share at
least 30%, 40%, or 50%, preferably 60%, more preferably 70-80%, or 90-95%
homology
and share a common functional activity are defined herein as sufficiently
identical.
As used interchangeably herein, an "CCP activity", "biological activity of CCP
" or
"functional activity of CCP", refers to an activity exerted by a CCP protein,
polypeptide or
nucleic acid molecule on a CCP responsive cell or tissue, or on a CCP protein
substrate, as
determined in vivo, or in vitro, according to standard techniques. In one
embodiment, a
CCP activity is a direct activity, such as an association with a CCP-target
molecule. As
used herein, a "target molecule" or "binding partner" is a molecule with which
a CCP
protein binds or interacts in nature, such that CCP-mediated function is
achieved. A CCP
target molecule can be a non-CCP molecule or a CCP protein or polypeptide of
the present
invention, e.g., a plant cyclin dependent kinase, such as CDC2b. In an
exemplary
embodiment, a CCP target molecule is a CCP ligand. Alternatively, a CCP
activity is an
indirect activity, such as a cellular signaling activity mediated by
interaction of the CCP
protein with a CCP ligand. The biological activities of CCP are described
herein. For
example, the CCP proteins of the present invention can have one or more of the
following
activities: (1) they may interact with a non-CCP protein molecule, e.g., a CCP
ligand; (2)
they may modulate a CCP-dependent signal transduction pathway; (3) they may
modulate
the activity of a plant cyclin dependent kinase, such as CDC2a, CDC2b, or
CDC2c, and (4)
they may modulate the cell cycle.
Accordingly, another embodiment of the invention features isolated CCP
proteins
and polypeptides having a CCP activity. Preferred proteins are CCP proteins
having at
least one or more of the following domains: a "cyclin destruction box", a
"cyclin box motif
1 ", a "cyclin box motif 2", a "CDC2 motif', a "CDK phosphorylation site", a
"nuclear
localization signal", a "Cy-like box", an "Rb binding domain", a "DEF domain",
a "DNA
CA 02768046 2012-02-07
WO 01/85916 PCT/IBO1/01307
-28-
binding domain", a "DCB 1 domain", a "DCB2 domain" and/or a SAP domain, and,
preferably, a CCP activity.
Additional preferred proteins have at least one or more of the following
domains: a
"cyclin destruction box", a "cyclin box motif I", a "cyclin box motif 2", a
"CDC2 motif',
a "CDK phosphorylation site", a "nuclear localization signal", a "Cy-like
box", an "Rb
binding domain", a "DEF domain", a "DNA binding domain", a "DCB 1 domain", a
"DCB2 domain" and/or a SAP domain and are, preferably, encoded by a nucleic
acid
molecule having a nucleotide sequence which hybridizes under stringent
hybridization
conditions to a nucleic acid molecule comprising the nucleotide sequence of
SEQ ID
NO:1-66 or 228-239.
The sequences of the present invention are summarized below, in Table I.
TABLE I:
SEQ SEQ SEQ SEQ
Bait Homolog/ motif ID ID ID ID
CCP Clone function NO: NO: NO: NO:
Molecule Name partia full- partial full-
I length Protei length
DNA DNA n Protei
n
CCPI CDC2bD CDC2bAt. Novel CYCB2;3 cyclin box 1 39 67 105
N-IC 19 N161 motifs 1
and 2;
cyclin
destruction
box
CCP2 CDC2bD CDC2bAt. ARR2 2 40 68 106
N-IC20 N161
CCP3 CDC2bD CDC2bAt. novel A-type cyclin box 3 41 69 107
N-IC21 N161 cyclin motifs 1
and 2;
cyclin
destruction
box
CCP4 CDC2bD CDC2bAt. CDK 4 4 70 70
N-IC26M N161 phospho-
rylation
site
CCP5 CDC2bD CDC2bAt. ArathCYCB2 cyclin box 5 5 71 71
N-IC39 N161 ;I motifs 1
and 2;
cyclin
CA 02768046 2012-02-07
= WO 01/85946 PCT/IBOI/01307
-29-
destruction
box
CCP6 CDC2bD CDC2bAt. 6 42 72 108
N-IC57 N161
CCP7 CDC2bD CDC2bAt. AJH2-COP9 7 43 73 109
N-IC62 N161
CCP8 E2F3ca55 E2F3 N- 8 43 74 109
terminal
CCP9 CDC2bD CDC2bAt. Arath cyclin box 9 9 75 75
N-IC9 N161 CYCA2;2 motifs 1
and 2;
cyclin
destruction
box
CCP10 CKSBCO CKS I At 10 10 76 76
01
CCP11 CKSBCO CKS1At gibberellin- 11 44 77 110
11 regulated
protein
GASA1
precursor
CCP12 CKSBC9 CKS1At 12 45 78 111
8-7
(Cterm)
CCP13 CKSBC9 CKS 1 At 13 45 79 111
8-7
(Nterm)
CCP14 CKSBC1 CKS1At 14 46 80 112
03-19
(Cterm)
CCP15 CKSBC1 CKS1At PSTTLRE-type CDC2 15 47 81 113
99-20 CDK motifs
CCP16 E2F5BB E2F5 DPa DNA-binding 16 48 82 114
C1 dimerisati domain; DEF
domain; DCBI
on domain and DCB2
domain
CCP17 FL67BC4 CKI4 17 17 83 83
-2
CCP18 FL67BC1 CK14 RNA 18 49 84 115
2-17 polymerase B
transcription
factor 3
CCP19 JUT1 PLPI 19 19 85 85
CCP20 JUT2 PLPI 20 50 86 116
CCP21 JUT3 PLP1 21 50 87 116
CCP22 JUT6 PLPI Submergence 22 51 88 117
induced
CA 02768046 2012-02-07
WO 01/85946 PCT/!B01/01307
-30-
protein2 of
Oryza sativa
CCP23 kbpl KLPNTI HSF1 23 52 89 118
36-508aa
(motor
domain)
KLPNT2
(TH65)
73-186 as
(neck
domain)
CCP24 kbp3 KLPNTI 24 53 90 119
(427-
867aa)
stalk
domain
CCP25 kbp6 KLPNT2 25 54 91 120
(TH65)
73-186 as
neck
domain
CCP26 kbp9 KLPNT2 AtKLPNTI 26 55 92 121
(TH65)
73-186 as
neck
domain
CCP27 kbpll KLPNT2 27 56 93 122
(TH65)
73-186 as
neck
domain
CCP28 kbpl2 KLPNT2 28 57 94 123
(TH65)
73-186 as
neck
domain
CCP29 kbp13 KLPNT2 29 29 95 95
(TH65)
73-186 as
neck
domain
CCP30 kbpl5 KLPNT2 Centromere/ 30 58 96 124
(TH65) microtubule
73-186 as binding
neck protein CBF5
domain from yeast
CC-P31 kbp20 KLPNT2 VL'91 C 31 59 97 125
CA 02768046 2012-02-07
WO 01/85946 PCT/1B01/01307
-31-
(TH65) calmodulin
73-608 as from yeast
stalk
domain
CCP32 E2F5BB E2175 32 60 98 126
C16 dimerizati
on
CCP33 DPb / DNA-binding 33 61 99 127
domain; DEF
domain; DCBI
and DCB2
domain
CCP34 E2F3cal E2173 N- 34 62 100 128
terminal
CCP35 E2F3ca2 E2F3 N- 35 63 101 129
terminal
CCP36 E2F3ca9 E21`3 N- 36 64 102 130
terminal
CCP37 E2F3cal2 E2F3 N- SAP 37 65 103 131
terminal domain
CCP38 E2F3ca5O E2F3 N- 38 66 104 132
terminal
Detailed studies of interactions between AtDPs (a and b forms, SEQ ID NO: 114
and SEQ ID NO:127 , respectively) and AtE2Fs (a and b forms; GenBank accession
numbers AJ294534 and AJ294533, respectively) revealed that the regions of
AtDPa and
AtDPb involved in the binding of AtE2Fb are different.
Binding of AtDPa to AtE2Fb requires at least the AtDPa dimerization domain and
the whole (or possibly part of) the C-terminal domain of AtDPa. The N-terminal
domain
and the DNA-binding domain of AtDPa do not seem to contribute to the
interaction of
AtDPa with AtE2Fb (Examples 11, 12, Table 5, Figure 54).
Binding of AtDPb to AtE2Fb, however, only requires an intact AtDPb
dimerization
domain. Neither the region including the N-terminal and DNA-binding domains of
AtDPb, nor the C-terminal region of AtDPb seem to contribute to the
interaction of AtDPb
with AtE2Fb (Examples 11, 12, Table 5, Figure 55). These observations indicate
that
modulating the formation of specific E2F/DP-complexes may be useful in
modulating cell
cycle traversal and the regulation thereof.
AtDPa and AtDPb, respectively, do not form homodimers but both interact with
either AtE2Fa or AtE2Fb (Example 12, Table 5). In reciprocal experiments it
was shown
that the N-terminal domain of AtE2Fa is not required for binding AtDPa or
AtDPb.
Likewise, the Rb-binding domains of AtE2Fa and AtE2Fb, respectively, do not
seem to'
contribute to the binding to either AtDPa or AtDPb. The region of AtE2Fa
encompassing
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-32-
the dimerization domain and the marked box is sufficient for binding to AtDPa
and AtDPb
(Examples 11, 12, Fig. 50, Fig. 51, Table 5). The dimerization domain of
AtE2Fs appears
to be sufficient for binding to AtDPs.
Accordingly, it is shown herein for the first time (for plant DPs and plant
E2Fs)
that the minimal DP and E2F proteins or corresponding coding DNA sequences
that can be
used in modifying E2F/DP-related processes, e.g., regulation of gene
expression by
E2F/DP, include:
(A) Plant DP dimerization domain with or without (part of) the C-terminal DP
domain. These domains include the proteins AtDPa143-292 and AtDPal43-213
I o (numbering indicates the amino acids included in said fragment relative to
the full-length
AtDPa protein) set forth in SEQ ID NO:221 and SEQ ID NO:222, respectively. The
coding sequences corresponding to the foregoing amino acid sequences are set
forth in
SEQ ID NO:232 and SEQ ID NO:233, respectively. Also included are the
corresponding
regions of the AtDPb protein characterized by AtDPb182-385 and AtDPb182-263
(parts of
the full-length AtDPb protein). The foregoing regions of AtDPb are set forth
in SEQ ID
NO:216 and SEQ ID NO:215, respectively, and the coding sequences corresponding
thereto are set forth in SEQ ID NO:231 and SEQ ID NO:230, respectively. The
AtDPb1-
263 domain (SEQ ID NO:223) and the corresponding AtDPa1-214 domain (SEQ ID
NO:220) encoded by the nucleic acid sequences SEQ ID NO:234 and SEQ ID NO:239,
respectively, can also be used. Further included are nucleic acid sequences
hybridizing to
SEQ ID NOs:229-234 or SEQ ID NO:239 or encoding a protein at least 50%, 55%,
60%,
65%, 70%, 75%, 80%, 85%,90%,95%,98% or more identical to SEQ ID NOs:211, 215-
216 and 220-223.
(B) Plant E2F dimerization domain with or without (part of) the marked box.
These domains include the proteins AtE2Fa232-282, AtE2Fa232-352 and AtE2Fa226-
356
set forth in SEQ ID NO:224, SEQ ID NO:225 and SEQ ID NO:205, respectively. The
corresponding coding DNA sequences are set forth in SEQ ID NO:235, SEQ ID
NO:236
and SEQ ID NO:228, respectively. Also included are the corresponding regions
of the
AtE2Fb protein characterized by AtE2Fbl94-243 and AtE2Fb194-311 set forth in
SEQ ID
NO:226 and SEQ ID NO:227, respectively. The corresponding coding DNA sequences
are
set forth in SEQ ID NO:237 and SEQ ID NO:238, respectively. Further included
are
nucleic acid sequences hybridizing to SEQ ID NO:228 or SEQ ID NOs:235-238 or
encoding a protein at least 70%, 75%, 80%, 85%, 90%, 95%, 98% identical to SEQ
ID
NO:205 or SEQ ID NOs:224-227.
(C) Full-length plant DP and plant E2F proteins or corresponding DNA sequences
may also be used to modify said E2F/DP-related processes. Furthermore, plant
DP and
plant E2F proteins or corresponding DNA sequences, or parts thereof, can be
used either
separately or in combination to modify said E2F/DP-related processes. This is
underscored
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-33-
by the demonstration that AtDPs and AtE2Fs are co-expressed in actively
dividing cells
and in at least some plant tissues (Example 13 and Figures 57 and 58).
Various aspects of the invention are described in further detail in the
following
subsections:
1. Isolated Nucleic Acid Molecules
One aspect of the invention pertains to isolated nucleic acid molecules that
encode
CCP proteins or biologically active portions thereof, as well as nucleic acid
fragments
sufficient for use as hybridization probes to identify CCP-encoding nucleic
acids (e.g.,
CCP mRNA) and fragments for use as PCR primers for the amplification or
mutation of
CCP nucleic acid molecules. As used herein, the term "nucleic acid molecule"
is intended
to include DNA molecules (e.g., cDNA or genomic DNA) and RNA molecules (e.g.,
mRNA) and analogs of the DNA or RNA generated using nucleotide analogs. The
nucleic
acid molecule can be single-stranded or double-stranded, but preferably is
double-stranded
DNA.
An "isolated" nucleic acid molecule is one which is separated from other
nucleic
acid molecules which are present in the natural source of the nucleic acid.
For example,
with regards to genomic DNA, the term "isolated" includes nucleic acid
molecules which
are separated from the chromosome with which the genomic DNA is naturally
associated.
Preferably, an "isolated" nucleic acid is free of sequences which naturally
flank the nucleic
acid (i.e., sequences located at the 5' and 3' ends of the nucleic acid) in
the genomic DNA
of the organism from which the nucleic acid is derived. For example, in
various
embodiments, the isolated CCP nucleic acid molecule can contain less than
about 5 kb,
4kb, 3kb, 2kb, 1 kb, 0.5 kb or 0.1 kb of nucleotide sequences which naturally
flank the
nucleic acid molecule in genomic DNA of the cell from which the nucleic acid
is derived.
Moreover, an "isolated" nucleic acid molecule, such as a cDNA molecule, can be
substantially free of other cellular material, or culture medium when produced
by
recombinant techniques, or substantially free of chemical precursors or other
chemicals
when chemically synthesized.
A nucleic acid molecule of the present invention, e.g., a nucleic acid
molecule
having the nucleotide sequence of SEQ ID NO:1-66 or 228-239, or a portion
thereof, can
be isolated using standard molecular biology techniques and the sequence
information
provided herein. For example, using all or portion of the nucleic acid
sequence of SEQ ID
NO:1-66 or 228-239, as a hybridization probe, CCP nucleic acid molecules can
be isolated
using standard hybridization and cloning techniques (e.g., as described in
Sambrook, J.,
Fritsh, E. F., and Maniatis, T. Molecular Cloning: A Laboratory Manual. 2nd,
ed., Cold
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO]/01307
-34-
Spring Harbor Laboratory, Cold Spring Harbor Laboratory Press, Cold Spring
Harbor,
NY, 1989).
Moreover, a nucleic acid molecule encompassing all or a portion of SEQ ID NO:1-
66 or 228-239 can be isolated by the polymerase chain reaction (PCR) using
synthetic
oligonucleotide primers designed based upon the sequence of SEQ ID NO: 1-66 or
228-
239, respectively.
A nucleic acid of the invention can be amplified using cDNA, mRNA or
alternatively, genomic DNA, as a template and appropriate oligonucleotide
primers
according to standard PCR amplification techniques. The nucleic acid so
amplified can be
cloned into an appropriate vector and characterized by DNA sequence analysis.
Furthermore, oligonucleotides corresponding to CCP nucleotide sequences can be
prepared
by standard synthetic techniques, e.g., using an automated DNA synthesizer.
In a preferred embodiment, an isolated nucleic acid molecule of the invention
comprises the nucleotide sequence shown in SEQ ID NO:1-66 or 228-239.
In another preferred embodiment, an isolated nucleic acid molecule of the
invention
comprises a nucleic acid molecule which is a complement of the nucleotide
sequence
shown in SEQ ID NO:1-66 or 228-239, or a portion of any of these nucleotide
sequences.
A nucleic acid molecule which is complementary to the nucleotide sequence
shown in
SEQ ID NO:1-66 or 228-239, is one which is sufficiently complementary to the
nucleotide
sequence shown in SEQ ID NO:1-66 or 228-239, respectively, such that it can
hybridize to
the nucleotide sequence shown in SEQ ID NO:1-66 or 228-239, respectively,
thereby
forming a stable duplex.
In still another preferred embodiment, an isolated nucleic acid molecule of
the
present invention comprises a nucleotide sequence which is at least about 50%,
55%, 60%,
65%, 70%, 75%, 80%, 85%, 90%, 95%, 98% or more homologous to the nucleotide
sequence (e.g., to the entire length of the nucleotide sequence) shown in SEQ
ID NO: 1-66
or 228-239, or a portion of any of these nucleotide sequences.
Moreover, the nucleic acid molecule of the invention can comprise only a
portion
of the nucleic acid sequence of SEQ ID NO:1-66 or 228-239, for example a
fragment
which can be used as a probe or primer or a fragment encoding a biologically
active
portion of a CCP protein. The nucleotide sequence determined from the cloning
of the
CCP gene allows for the generation of probes and primers designed for use in
identifying
and/or cloning other CCP family members, as well as CCP homologues from other
species. The probe/primer typically comprises substantially purified
oligonucleotide. The
oligonucleotide typically comprises a region of nucleotide sequence that
hybridizes under
stringent conditions to at least about 12 or 15, preferably about 20 or 25,
more preferably
about 30, 35, 40, 45, 50, 55, 60, 65, or 75 consecutive nucleotides of a sense
sequence of
SEQ ID NO: 1-66 or 228-239, or of a naturally occurring allelic variant or
mutant of SEQ
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-35-
ID NO:1-66 or 228-239. In an exemplary embodiment, a nucleic acid molecule of
the
present invention comprises a nucleotide sequence which is at least 100, 150,
200, 250,
300, 350, 400, 450, 500, 550, 600, 650, 700, 750, or 800 nucleotides in length
and
hybridizes under stringent hybridization conditions to a nucleic acid molecule
of SEQ ID
NO:1-66 or 228-239.
Probes based on the CCP nucleotide sequences can be used to detect transcripts
or
genomic sequences encoding the same or homologous proteins. In preferred
embodiments,
the probe further comprises a label group attached thereto, e.g., the label
group can be a
radioisotope, a fluorescent compound, an enzyme, or an enzyme co-factor. Such
probes
can be used as a part of a diagnostic test kit for identifying cells or
tissues which
misexpress a CCP protein, such as by measuring a level of a CCP-encoding
nucleic acid in
a sample of cells from a subject e.g., detecting CCP mRNA levels or
determining whether
a genomic CCP gene has been mutated or deleted.
A nucleic acid fragment encoding a "biologically active portion of a CCP
protein"
can be prepared by isolating a portion of the nucleotide sequence of SEQ ID
NO:1-66 or
228-239, which encodes a polypeptide having a CCP biological activity (the
biological
activities of the CCP proteins are described herein), expressing the encoded
portion of the
CCP protein (e.g., by recombinant expression in vitro) and assessing the
activity of the
encoded portion of the CCP protein.
The invention further encompasses nucleic acid molecules that differ from the
nucleotide sequence shown in SEQ ID NO:1-66 or 228-239, due to the degeneracy
of the
genetic code and, thus, encode the same CCP proteins as those encoded by the
nucleotide
sequence shown in SEQ ID NO:1-66 or 228-239. In another embodiment, an
isolated
nucleic acid molecule of the invention has a nucleotide sequence encoding a
CCP protein.
In addition to the CCP nucleotide sequences shown in SEQ ID NO:1-66 or 228-
239, it will be appreciated by those skilled in the art that DNA sequence
polymorphisms
that lead to changes in the amino acid sequences of the CCP proteins may exist
within a
population (e.g., an Arabidopsis or rice plant population). Such genetic
polymorphism in
the CCP genes may exist among individuals within a population due to natural
allelic
variation. As used herein, the terms "gene" and "recombinant gene" refer to
nucleic acid
molecules which include an open reading frame encoding an CCP protein,
preferably a
plant CCP protein, and can further include non-coding regulatory sequences,
and introns.
Such natural allelic variations include both functional and non-functional CCP
proteins
and can typically result in 1-5% variance in the nucleotide sequence of a CCP
gene. Any
and all such nucleotide variations and resulting amino acid polymorphisms in
CCP genes
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-36-
that are the result of natural allelic variation and that do not alter the
functional activity of a
CCP protein are intended to be within the scope of the invention. Differences
in preferred
codon usage are illustrated below for Agrobacterium tuinefaciens (a
bacterium),
Arabidopsis thaliana, Medicago sativa (two dicotyledonous plants) and Oryza
sativa (a
monocotyledonous plant). These examples were extracted from
http://www.kazusa.orjp/codon. For example, the codon GGC (for glycine) is the
most
frequently used codon in A. tumefaciens (36.2 %o), is the second most
frequently used
codon in O. sativa but is used at much lower frequencies in A. thaliana and M
sativa (9 %o
and 8.4 %o , respectively). Of the four possible codons encoding glycine the
GGC codon is
most preferably used in A. tumefaciens and O. sativa. However, in A. thaliana
the GGA
(and GGU) codon is most preferably used, whereas in M. sativa the GGU (and
GGA)
codon is most preferably used.
Moreover, nucleic acid molecules encoding other CCP family members and, thus,
which have a nucleotide sequence which differs from the CCP sequences of SEQ
ID NO: I -
66 or 228-239 are intended to be within the scope of the invention. For
example, another
CCP cDNA can be identified based on the nucleotide sequence of the plant CCP
molecules
described herein. Moreover, nucleic acid molecules encoding CCP proteins from
different
species, and thus which have a nucleotide sequence which differs from the CCP
sequences
of SEQ ID NO:1-66 or 228-239 are intended to be within the scope of the
invention. For
example, a human CCP eDNA can be identified based on the nucleotide sequence
of a
plant CCP.
Nucleic acid molecules corresponding to natural allelic variants and
homologues of
the CCP cDNAs of the invention can be isolated based on their homology to the
CCP
nucleic acids disclosed herein using the cDNAs disclosed herein, or a portion
thereof, as a
hybridization probe according to standard hybridization techniques under
stringent
hybridization conditions.
Accordingly, in another embodiment, an isolated nucleic acid molecule of the
invention is at least 15, 20, 25, 30 or more nucleotides in length and
hybridizes under
stringent conditions to the nucleic acid molecule comprising the nucleotide
sequence of
SEQ ID NO:1-66 or 228-239. In other embodiment, the nucleic acid is at least
30, 50, 100,
150, 200, 250, 300, 350, 400, 450, 500, 550, or 600 nucleotides in length. As
used herein,
the term "hybridizes under stringent conditions" is intended to describe
conditions for
hybridization and washing under which nucleotide sequences at least 30%, 40%,
50%, or
60% homologous to each other typically remain hybridized to each other.
Preferably, the
conditions are such that sequences at least about 70%, more preferably at
least about 80%,
even more preferably at least about 85% or 90% homologous to each other
typically
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-37-
remain hybridized to each other. Such stringent conditions are known to those
skilled in
the art and can be found in Current Protocols in Molecular Biology, John Wiley
& Sons,
N.Y. (1989), 6.3.1-6.3.6. A preferred, non-limiting example of stringent
hybridization
conditions are hybridization in 6X sodium chloride/sodium citrate (SSC) at
about 45 C,
followed by one or more washes in 0.2 X SSC, 0.1% SDS at about 45 C, followed
by one
or more washes in 0.2 X SSC, 0.1% SDS at 50 C, preferably at 55 C, more
preferably at
60T, and even more preferably at 65 C. Ranges intermediate to the above-
recited values,
e.g., at 60-65 C or at 55-60 C are also intended to be encompassed by the
present
invention. Preferably, an isolated nucleic acid molecule of the invention that
hybridizes
under stringent conditions to the sequence of SEQ ID NO:1-66 or 228-239
corresponds to
a naturally-occurring nucleic acid molecule. As used herein, a "naturally-
occurring"
nucleic acid molecule refers to an RNA or DNA molecule having a nucleotide
sequence
that occurs in nature (e.g., encodes a natural protein).
In addition to naturally-occurring allelic variants of the CCP sequences that
may
exist in nature, the skilled artisan will further appreciate that changes can
be introduced by
mutation into the nucleotide sequences of SEQ ID NO:1-66 or 228-239, thereby
leading to
changes in the amino acid sequence of the encoded CCP proteins, without
altering the
functional ability of the CCP proteins. For example, nucleotide substitutions
leading to
amino acid substitutions at "non-essential" amino acid residues can be made in
the
sequence of a CCP protein. A "non-essential" amino acid residue is a residue
that can be
altered from the wild-type sequence of CCP without altering the biological
activity,
whereas an "essential" amino acid residue is required for biological activity.
For example,
amino acid residues that are conserved among the CCP proteins of the present
invention,
are predicted to be particularly unamenable to alteration. Furthermore,
additional amino
acid residues that are conserved between the CCP proteins of the present
invention and
other CCP family members are not likely to be amenable to alteration.
Accordingly, another aspect of the invention pertains to nucleic acid
molecules
encoding CCP proteins that contain changes in amino acid residues that are not
essential
for activity.
An isolated nucleic acid molecule encoding a CCP protein homologous to the CCP
proteins of the present invention can be created by introducing one or more
nucleotide
substitutions, additions or deletions into the nucleotide sequence of SEQ ID
NO: 1-66 or
228-239, such that one or more amino acid substitutions, additions or
deletions are
introduced into the encoded protein. Mutations can be introduced into SEQ ID
NO: 1-66 or
228-239 by standard techniques, such as site-directed mutagenesis and PCR-
mediated
mutagenesis. Preferably, conservative amino acid substitutions are made at one
or more
predicted non-essential amino acid residues. A "conservative amino acid
substitution" is
one in which the amino acid residue is replaced with an amino acid residue
having a
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-38-
similar side chain. Families of amino acid residues having similar side chains
have been
defined in the art. These families include amino acids with basic side chains
(e.g., lysine,
arginine, histidine), acidic side chains (e.g., aspartic acid, glutamic acid),
uncharged polar
side chains (e.g., glycine, asparagine, glutamine, serine, threonine,
tyrosine, cysteine),
nonpolar side chains (e.g., alanine, valine, leucine, isoleucine, proline,
phenylalanine,
methionine, tryptophan), beta-branched side chains (e.g., threonine, valine,
isoleucine) and
aromatic side chains (e.g., tyrosine, phenylalanine, tryptophan, histidine).
Thus, a
predicted nonessential amino acid residue in a CCP protein is preferably
replaced with
another amino acid residue from the same side chain family. Alternatively, in
another
embodiment, mutations can be introduced randomly along all or part of a CCP
coding
sequence, such as by saturation mutagenesis, and the resultant mutants can be
screened for
CCP biological activity to identify mutants that retain activity. Following
mutagenesis of
SEQ ID NO: 1-66 or 228-239, the encoded protein can be expressed recombinantly
and the
activity of the protein can be determined. Another alternative embodiment
comprises
targeted in vivo gene correction or modification which can be achieved by
chimeric
RNA/DNA oligonucleotides (e.g., Yoon et al. (1996), Proc. Natl. Acad. Sci. USA
93,
2071-2076; Arntzen et al. (1999) W099/07865).
In a preferred embodiment, a mutant CCP protein can be assayed for the ability
to:
(1) regulate transmission of signals from cellular receptors, e.g. hormone
receptors; (2)
control cell cycle checkpoints, e.g. entry of cells into mitosis; (3) modulate
the cell cycle;
(4) modulate cell death, e.g., apoptosis; (5) modulate cytoskeleton function,
e.g. actin
bundling; (6) phosphorylate a substrate; (7) create dominant negative or
dominant positive
effects in transgenic plants; (8) interact with other cell cycle control
proteins in, e.g. a yeast
two hybrid assay; (9) modulate CDK activity (e.g., cyclin-CDK activity); (10)
regulate
cyclin-CDK complex assembly; (11) regulate the commitment of cells to divide,
e.g., by
integrating mitogenic and antimitogenic signals; (12) regulate cell cycle
progression; (13)
regulate DNA replication and/or DNA repair; (14) modulate gene transcription,
e.g.,
regulate E2F/DP-dependent transcription of genes; (15) regulate cyclin
degradation; (16)
modulate cell cycle withdrawal and/or cell differentiation; (17) control organ
(e.g., plant
organ) and/or organism (e.g., plant organism) size; (18) control organ (e.g.,
plant organ)
and/or organism (e.g., plant organism) growth or growth rate; and (19)
regulate
endoreduplication.
In addition to the nucleic acid molecules encoding CCP proteins described
above,
another aspect of the invention pertains to isolated nucleic acid molecules
which are
antisense thereto. An "antisense" nucleic acid comprises a nucleotide sequence
which is
complementary to a "sense" nucleic acid encoding a protein, e.g.,
complementary to the
coding strand of a double-stranded cDNA molecule or complementary to an mRNA
sequence. Accordingly, an antisense nucleic acid can hydrogen bond to a sense
nucleic
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-39-
acid. The antisense nucleic acid can be complementary to an entire CCP coding
strand, or
only to a portion thereof. In one embodiment, an antisense nucleic acid
molecule is
antisense to a "coding region" of the coding strand of a nucleotide sequence
encoding
CCP. The term "coding region" refers to the region of the nucleotide sequence
comprising
codons which are translated into amino acid residues. In another embodiment,
the
antisense nucleic acid molecule is antisense to a "noncoding region" of the
coding strand of
a nucleotide sequence encoding CCP. The .term "noncoding region" refers to 5'
and 3'
sequences which flank the coding region that are not translated into amino
acids (i.e., also
referred to as 5' and 3' untranslated regions).
Given the coding strand sequences encoding CCP disclosed herein, antisense
nucleic acids of the invention can be designed according to the rules of
Watson and Crick
base pairing. The antisense nucleic acid molecule can be complementary to the
entire
coding region of CCP mRNA, but more preferably is an oligonucleotide which is
antisense
to only a portion of the coding or noncoding region of CCP mRNA. For example,
the
antisense oligonucleotide can be complementary to the region surrounding the
translation
start site of CCP mRNA. An antisense oligonucleotide can be, for example,
about 5, 10,
15, 20, 25, 30, 35, 40, 45 or 50 nucleotides in length. An antisense nucleic
acid of the
invention can be constructed using chemical synthesis and enzymatic ligation
reactions
using procedures known in the art. For example, an antisense nucleic acid
(e.g., an
antisense oligonucleotide) can be chemically synthesized using naturally
occurring
nucleotides or variously modified nucleotides designed to increase the
biological stability
of the molecules or to increase the physical stability of the duplex formed
between the
antisense and sense nucleic acids, e.g., phosphorothioate derivatives and
acridine
substituted nucleotides can be used. Examples of modified nucleotides which
can be used
to generate the antisense nucleic acid include 5-fluorouracil, 5-bromouracil,
5-chorouracil,
5-iodouracil, hypoxanthine, xantine, 4-acetylcytosine, 5-
(carboxyhydroxylmethyl) uracil,
5-carboxymethylaniinomethyl-2-thiouridine, 5-carboxymethylaminomethyluracil,
dihydrouracil, beta-D-galactosylqueosine, inosine, N6-isopentenyladenine, 1-
methylguanine, 1-methylinosine, 2,2-dimethylguanine, 2-methyladenine, 2-
methylguanine,
3-methylcytosine, 5-methylcytosine, N6-adenine, 7-methylguanine, 5-
methylaminomethyluracil, 5-methoxyaminomethyl-2-thiouracil, beta-D-
mannosylqueosine, 5'-methoxycarboxymethyluracil, 5-methoxyuracil, 2-methylthio-
N6-
isopentenyladenine, uracil-5-oxyacetic acid (v), wybutoxosine, pseudouracil,
queosine, 2-
thiocytosine, 5-methyl-2-thiouracil, 2-thiouracil, 4-thiouracil, 5-
methyluracil, uracil-5-
oxyacetic acid methylester, uracil-5-oxyacetic acid (v), 5-methyl-2-
thiouracil, 3-(3-amino-
3-N-2-carboxypropyl) uracil, (acp3)w, and 2,6-diaminopurine. Alternatively,
the antisense
nucleic acid can be produced biologically using an expression vector into
which a nucleic
acid has been subcloned in an antisense orientation (i.e., RNA transcribed
from the inserted
CA 02768046 2012-02-07
WO 01/8-5946 PCT/IBOI/01307
-40-
nucleic acid will be of an antisense orientation to a target nucleic acid of
interest, described
further in the following subsection). Preferably, production of antisense
nucleic acids in
plants occurs by means of a stably integrated transgene comprising a promoter
operative in
plants, an antisense oligonucleotide, and a terminator.
Other known nucleotide modifications include methylation, cyclization and
'caps'
and substitution of one or more of the naturally occurring nucleotides with an
analog such
as inosine. Modifications of nucleotides include modifications generated by
the addition to
nucleotides of acridine, amine, biotin, cascade blue, cholesterol, Cy3 , Cy5 ,
Cy5.5
Dabcyl, digoxigenin, dinitrophenyl, Edans, 6-FAM, fluorescein, 3'-glyceryl,
HEX, IRD-
700, IRD-800, JOE, phosphate psoralen, rhodamine, ROX, thiol (SH), spacers,
TAMRA,
TET, AMCA-S , SE, BODIPY , Marina Blue , Pacific Blue , Oregon Green ,
Rhodamine
Green , Rhodamine Red , Rhodol Green and Texas Red . Polynucleotide backbone
modifications include methylphosphonate, 2'-OMe-methylphosphonate RNA,
phosphorothiorate, RNA, 2'-OMeRNA. Base modifications include 2-amino-dA, 2-
aminopurine, 3'-(ddA), 3'dA(cordycepin), 7-deaza-dA, 8-Br-dA, 8-oxo-dA, N6-Me-
dA,
abasic site (dSpacer), biotin dT, 2'-OMe-5Me-C, 2'-OMe-propynyl-C, 3'-(5-Me-
dC), 3'-
(ddC), 5-Br-dC, 5-I-dC, 5-Me-dC, 5-F-dC, carboxy-dT, convertible dA,
convertible dC,
convertible dG, convertible dT, convertible dU, 7-deaza-dG, 8-Br-dG, 8-oxo-dG,
06-Me-
dG, S6-DNP-dG, 4-methyl-indole, 5-nitroindole, 2'-OMe-inosine, 2'-dl, 06-
phenyl-dI, 4-
methyl-indole, 2'-deoxynebularine, 5-nitroindole, 2-aminopurine, dP(purine
analogue),
dK(pyrimidine analogue), 3-nitropyrrole, 2-thio-dT, 4-thio-dT, biotin-dT,
carboxy-dT, 04-
Me-dT, 04-triazol dT, 2'-OMe-propynyl-U, 5-Br-dU, 2'-dU, 5-F-dU, 5-1-dU, 04-
triazol dU.
The antisense nucleic acid molecules of the invention are typically introduced
into
a plant or administered to a subject or generated in situ such that they
hybridize with or
bind to cellular mRNA and/or genomic DNA encoding a CCP protein to thereby
inhibit
expression of the protein, e.g., by inhibiting transcription and/or
translation. The
hybridization can be by conventional nucleotide complementarity to form a
stable duplex,
or, for example, in the case of an antisense nucleic acid molecule which binds
to DNA
duplexes, through specific interactions in the major groove of the double
helix. An
example of a route of introduction or administration of antisense nucleic acid
molecules of
the invention include transformation in a plant or direct injection at a
tissue site in a
subject. Alternatively, antisense nucleic acid molecules can be modified to
target selected
cells and then administered systemically. For example, for systemic
administration,
antisense molecules can be modified such that they specifically bind to
receptors or
antigens expressed on a selected cell surface,=e.g., by linking the antisense
nucleic acid
molecules to peptides or antibodies which bind to cell surface receptors or
antigens. The
antisense nucleic acid molecules can also be delivered to cells using the
vectors described
herein. To achieve sufficient intracellular concentrations of the antisense
molecules,
CA 02768046 2012-02-07
WO 01/85946 PCT/1B01/01307
-41-
vector constructs in which the antisense nucleic acid molecule is placed under
the control
of a constitutive promoter or a strong pol II or pol III promoter are
preferred.
In yet another embodiment, the antisense nucleic acid molecule of the
invention is
an a-anomeric nucleic acid molecule. An a-anomeric nucleic acid molecule forms
specific
double-stranded hybrids with complementary RNA in which, contrary to the usual
R-units,
the strands run parallel to each other (Gaultier et al. (1987) Nucleic Acids.
Res. 15:6625-
6641). The antisense nucleic acid molecule can also comprise a 2'-o-
methylribonucleotide
(Inoue et al. (1987) Nucleic Acids Res. 15:6131-6148) or a chimeric RNA-DNA
analogue
(Inoue et al. (1987) FEBS Lett. 215:327-330).
In another embodiment, the antisense nucleic acid molecule further comprises a
sense nucleic acid molecule complementary to the antisense nucleic acid
molecule. Gene
silencing methods based on such nucleic acid molecules are well known to the
skilled
artisan (e.g., Grierson et al. (1998) WO 98/53083; Waterhouse et al. (1999) WO
99/53050).
In still another embodiment, an antisense nucleic acid of the invention is a
ribozyme. Ribozymes are catalytic RNA molecules with ribonuclease activity
which are
capable of cleaving a single-stranded nucleic acid, such as an mRNA, to which
they have a
complementary region. Thus, ribozymes (e.g., hammerhead ribozymes (described
in
Haselhoff and Gerlach (1988) Nature 334:585-591)) can be used to catalytically
cleave
CCP mRNA transcripts to thereby inhibit translation of CCP mRNA. A ribozyme
having
specificity for a CCP-encoding nucleic acid can be designed based upon the
nucleotide
sequence of a CCP cDNA disclosed herein (i.e., SEQ ID NO:1-66 or 228-239). For
example, a derivative of a Tetrahymena L- 19 IVS RNA can be constructed in
which the
nucleotide sequence of the active site is complementary to the nucleotide
sequence to be
cleaved in a CCP-encoding mRNA. See, e.g., Cech et al. U.S. Patent No.
4,987,071; and
Cech et al. U.S. Patent No. 5,116,742. Alternatively, CCP mRNA can be used to
select a
catalytic RNA having a specific ribonuclease activity from a pool of RNA
molecules. See,
e.g., Bartel, D. and Szostak, J.W. (1993) Science 261:1411-1418.
The use of ribozymes for gene silencing in plants is known in the art (e.g.,
Atkins et
al. (1994) WO 94/00012; Lenne et al. (1995) WO 95/03404; Lutziger et al.
(2000) WO
00/00619; Prinsen et al. (1997) WO 97/13865 and Scott et al. (1997) WO/
97/38116).
Alternatively, CCP gene expression can be inhibited by targeting nucleotide
sequences complementary to the regulatory region of the CCP (e.g., the CCP
promoter
and/or enhancers) to form triple helical structures that prevent transcription
of the CCP
gene in target cells. See generally, Helene, C. (1991) Anticancer Drug Des.
6(6):569-84;
Helene, C. et al. (1992) Ann. N. Y. Acad. Sci. 660:27-36; and Maher, L.J.
(1992) Bioassays
14(12):807-15.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-42-
In yet another embodiment, the CCP nucleic acid molecules of the present
invention can be modified at the base moiety, sugar moiety or phosphate
backbone to
improve, e.g., the stability, hybridization, or solubility of the molecule.
For example, the
deoxyribose phosphate backbone of the nucleic acid molecules can be modified
to generate
peptide nucleic acids (see Hyrup B. et al. (1996) Bioorganic & Medicinal
Chemistry 4 (1):
5-23). As used herein, the terms "peptide nucleic acids" or "PNAs" refer to
nucleic acid
mimics, e.g., DNA mimics, in which the deoxyribose phosphate backbone is
replaced by a
pseudopeptide backbone and only the four natural nucleobases are retained. The
neutral
backbone of PNAs has been shown to allow for specific hybridization to DNA and
RNA
under conditions of low ionic strength. The synthesis of PNA oligomers can be
performed
using standard, solid phase peptide synthesis protocols as described in Hyrup
B. et al.
(1996) supra; Perry-O'Keefe et al. Proc. Natl. Acad. Sci. 93: 14670-675.
PNAs of CCP nucleic acid molecules can be used for increasing crop yield in
plants or in therapeutic and diagnostic applications. For example, PNAs can be
used as
antisense or antigene agents for sequence-specific modulation of gene
expression by, for
example, inducing transcription or translation arrest or inhibiting
replication. PNAs of
CCP nucleic acid molecules can also be used in the analysis of single base
pair mutations
in a gene, (e.g., by PNA-directed PCR clamping); as'artificial restriction
enzymes' when
used in combination with other enzymes, (e.g., S1 nucleases (Hyrup B. (1996)
supra)); or
as probes or primers for DNA sequencing or hybridization (Hyrup B. et al.
(1996) supra;
Perry-O'Keefe supra).
In another embodiment, PNAs of CCP can be modified, (e.g., to enhance their
stability or cellular uptake), by attaching lipophilic or other helper groups
to PNA, by the
formation of PNA-DNA chimeras, or by the use of liposomes or other techniques
of drug
delivery known in the art. For example, PNA-DNA chimeras of CCP nucleic acid
molecules can be generated which may combine the advantageous properties of
PNA and
DNA. Such chimeras allow DNA recognition enzymes, (e.g., RNAse H and DNA
polymerases), to interact with the DNA portion while the PNA portion would
provide high
binding affinity and specificity. PNA-DNA chimeras can be linked using linkers
of
appropriate lengths selected in terms of base stacking, number of bonds
between the
nucleobases, and orientation (Hyrup B. (1996) supra). The synthesis of PNA-DNA
chimeras can be performed as described in Hyrup B. (1996) supra and Finn P.J.
et al.
(1996) Nucleic Acids Res. 24 (17): 3357-63. For example, a DNA chain can be
synthesized on a solid support using standard phosphoramidite coupling
chemistry and
modified nucleoside analogs, e.g., 5'-(4-methoxytrityl)amino-5'-deoxy-
thymidine
phosphoramidite, can be used as a between the PNA and the 5' end of DNA (Mag,
M. et al.
(1989) Nucleic Acid Res. 17: 5973-88). PNA monomers are then coupled in a
stepwise
manner to produce a chimeric molecule with a 5' PNA segment and a 3' DNA
segment
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
-43-
(Finn P.J. et al. (1996) supra). Alternatively, chimeric molecules can be
synthesized with
a 5' DNA segment and a 3' PNA segment (Peterser, K.H. et al. (1975) Bioorganic
Med.
Chem. Lett. 5: 1119-11124).
In other embodiments, the oligonucleotide may include other appended groups
such
as peptides (e.g., for targeting host cell receptors in vivo), or agents
facilitating transport
across the cell membrane (see, e.g., Letsinger et al. (1989) Proc. Natl. Acad.
Sci. US.
86:6553-6556; Lemaitre et al. (1987) Proc. Natl. Acad. Sci. USA 84:648-652;
PCT
Publication No. W088/098 10) or the blood-brain barrier (see, e.g., PCT
Publication No.
W089/10134). In addition, oligonucleotides can be modified with hybridization-
triggered
cleavage agents (See, e.g., Krol et al. (1988) Bio-Techniques 6:958-976) or
intercalating
agents. (See, e.g., Zon (1988) Pharm. Res. 5:539-549). To this end, the
oligonucleotide
may be conjugated to another molecule, (e.g., a peptide, hybridization
triggered cross-
linking agent, transport agent, or hybridization-triggered cleavage agent).
II. Isolated CCP Proteins and Anti-CCP Antibodies
One aspect of the invention pertains to isolated CCP proteins (e.g., the amino
acid
sequences set forth in SEQ ID NO:67-132, 205, 211, 215-216, or 220-227) and
biologically active portions thereof, as well as polypeptide fragments
suitable for use as
immunogens to raise anti-CCP antibodies. In one embodiment, native CCP
proteins can
be isolated from cells or tissue sources by an appropriate purification scheme
using
standard protein purification techniques. In another embodiment, CCP proteins
are
produced by recombinant DNA techniques. Alternative to recombinant expression,
a CCP
protein or polypeptide can be synthesized chemically using standard peptide
synthesis
techniques.
An "isolated" or "purified" protein or biologically active portion thereof is
substantially free of cellular material or other contaminating proteins from
the cell or tissue
source from which the CCP protein is derived, or substantially free from
chemical
precursors or other chemicals when chemically synthesized. The language
"substantially
free of cellular material" includes preparations of CCP protein in which the
protein is
separated from cellular components of the cells from which it is isolated or
recombinantly
produced. In one embodiment, the language "substantially free of cellular
material"
includes preparations of CCP protein having less than about 30% (by dry
weight) of non-
CCP protein (also referred to herein as a "contaminating protein"), more
preferably less
than about 20% of non-CCP protein, still more preferably less than about 10%
of non-CCP
protein, and most preferably less than about 5% non-CCP protein. When the CCP
protein
or biologically active portion thereof is recombinantly produced, it is also
preferably
substantially free of culture medium, i.e., culture medium represents less
than about 20%,
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-44-
more preferably less than about 10%, and most preferably less than about 5% of
thl
volume of the protein preparation.
The language "substantially free of chemical precursors or other chemicals"
includes preparations of CCP protein in which the protein is separated from
chemical
precursors or other chemicals which are involved in the synthesis of the
protein. In one
embodiment, the language "substantially free of chemical precursors or other
chemicals"
includes preparations of CCP protein having less than about 30% (by dry
weight) of
chemical precursors or non-CCP chemicals, more preferably less than about 20%
chemical
precursors or non-CCP chemicals, still more preferably less than about 10%
chemical
precursors or non-CCP chemicals, and most preferably less than about 5%
chemical
precursors or non-CCP chemicals.
Biologically active portions of a CCP protein include peptides comprising
amino
acid sequences sufficiently homologous to or derived from the amino acid
sequence of the
CCP protein, which include less amino acids than the full length CCP proteins,
and exhibit
at least one activity of a CCP protein. Typically, biologically active
portions comprise a
domain or motif with at least one activity of the CCP protein. A biologically
active
portion of a CCP protein can be a polypeptide which is, for example, at least
10, 25, 50,
100 or more amino acids in length.
To determine the percent identity of two amino acid sequences or of two
nucleic
acid sequences, the sequences are aligned for optimal comparison purposes
(e.g., gaps can
be introduced in one or both of a first and a second amino acid or nucleic
acid sequence for
optimal alignment and non-homologous sequences can be disregarded for
comparison
purposes). In a preferred embodiment, the length of a reference sequence
aligned for
comparison purposes is at least 30%, preferably at least 40%, more preferably
at least 50%,
even more preferably at least 60%, and even more preferably at least 70%, 80%,
or 90% of
the length of the reference sequence. The amino acid residues or nucleotides
at
corresponding amino acid positions or nucleotide positions are then compared.
When a
position in the first sequence is occupied by the same amino acid residue or
nucleotide as
the corresponding position in the second sequence, then the molecules are
identical at that
position (as used herein amino acid or nucleic acid "identity" is equivalent
to amino acid or
nucleic acid "homology"). The percent identity between the two sequences is a
function
of the number of identical positions shared by the sequences, taking into
account the
number of gaps, and the length of each gap, which need to be introduced for
optimal
alignment of the two sequences.
The comparison of sequences and determination of percent identity between two
sequences can be accomplished using a mathematical algorithm. In a preferred
embodiment, the percent identity between two amino acid sequences is
determined using
the Needleman and Wunsch (J. Mol. Biol. (48):444-453 (1970)) algorithm which
has been
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
-45-
incorporated into the GAP program in the GCG software package (available at
http://www.gcg.com), using either a Blosum 62 matrix or a PAM250 matrix, and a
gap
weight of 16, 14, 12, 10, 8, 6, or 4 and a length weight of 1, 2, 3, 4, 5, or
6. In yet another
preferred embodiment, the percent identity between two nucleotide sequences is
determined using the GAP program in the GCG software package (available at
http://www.gcg.com), using a NWSgapdna.CMP matrix and a gap weight of 40, 50,
60,
70, or 80 and a length weight of 1, 2, 3, 4, 5, or 6. A preferred, non-
limiting example of
parameters to be used in conjunction with the GAP program include a Blosum 62
scoring
matrix with a gap penalty of 12, a gap extend penalty of 4, and a frameshift
gap penalty of
5.
In another embodiment, the percent identity between two amino acid or
nucleotide
sequences is determined using the algorithm of E. Meyers and W. Miller
(Comput. Appl.
Biosci., 4:11-17 (1988)) which has been incorporated into the ALIGN program
(version
2.0 or version 2.OU), using a PAM120 weight residue table, a gap length
penalty of 12 and
a gap penalty of 4.
The nucleic acid and polypeptide sequences of the present invention can
further be
used as a "query sequence" to perform a search against public databases to,
for example,
identify other family members or related sequences. Such searches can be
performed using
the NBLAST and XBLAST programs (version 2.0) of Altschul, et al. (1990) J.
Mol. Biol.
215:403-10. BLAST nucleotide searches can be performed with the NBLAST
program,
score = 100, wordlength =12 to obtain nucleotide sequences homologous to
Kinase and
Phosphatase nucleic acid molecules of the invention. BLAST protein searches
can be
performed with the XBLAST program, score = 100, wordlength = 3, and a Blosum62
matrix to obtain amino acid sequences homologous to Kinase and Phosphatase
polypeptide
molecules of the invention. To obtain gapped alignments for comparison
purposes,
Gapped BLAST can be utilized as described in Altschul et al., (1997) Nucleic
Acids Res.
25(17):3389-3402. When utilizing BLAST and Gapped BLAST programs, the default
parameters of the respective programs (e.g., XBLAST and NBLAST) can be used.
See
http://www.ncbi.nlm.nih.gov.
The invention also provides CCP chimeric or fusion proteins. As used herein, a
CCP "chimeric protein" or "fusion protein" comprises a CCP polypeptide
operatively
linked to a non-CCP polypeptide. An "CCP polypeptide" refers to a polypeptide
having an
amino acid sequence corresponding to CCP, whereas a "non-CCP polypeptide"
refers to a
polypeptide having an amino acid sequence corresponding to a protein which is
not
substantially homologous to the CCP protein, e.g., a protein which is
different from the
CCP protein and which is derived from the same or a different organism. Within
a CCP
fusion protein the CCP polypeptide can correspond to all or a portion of a CCP
protein. In
a preferred embodiment, a CCP fusion protein comprises at least one
biologically active
CA 02768046 2012-02-07
WO 01/85946 PCT/1B01/01307
-46-
portion of a CCP protein. In another preferred embodiment, a CCP fusion
protein
comprises at least two biologically active portions of a CCP protein. Within
the fusion
protein, the term "operatively linked" is intended to indicate that the CCP
polypeptide and
the non-CCP polypeptide are fused in-frame to each other. The non-CCP
polypeptide can
be fused to the N-terminus or. C-terminus of the CCP polypeptide or can be
inserted within
the CCP polypeptide. The non-CCP polypeptide can, for example, be (histidine)6-
tag,
glutathione S-transferase, protein A, maltose-binding protein, dihydrofolate
reductase,
Tag=100 epitope (EETARFQPGYRS; SEQ ID NO: 199), c-myc epitope (EQKLISEEDL;
SEQ ID NO:200), FLAG -epitope (DYKDDDK; SEQ ID NO:201), lacZ, CMP
(calmodulin-binding peptide), HA epitope (YPYDVPDYA; SEQ ID NO:202), protein C
epitope (EDQVDPRLIDGK; SEQ ID NO:203) or VSV epitope (YTDIEMNRLGK; SEQ
ID NO:204).
For example, in one embodiment, the fusion protein is a GST-CCP fusion protein
in which the CCP sequences are fused to the C-terminus of the GST sequences.
Such
fusion proteins can facilitate the purification of recombinant CCP.
In another embodiment, the fusion protein is a CCP protein containing a
heterologous signal sequence at its N-terminus. In certain host cells (e.g.,
plant or
mammalian host cells), expression and/or secretion of CCP can be increased
through use
of a heterologous signal sequence.
The CCP fusion proteins of the invention can be incorporated into
pharmaceutical
compositions and administered to a plant or a subject in vivo. The CCP fusion
proteins can
be used to affect the bioavailability of a CCP substrate. Use of CCP fusion
proteins may
be useful agriculturally for the increase of crop yields or therapeutically
for the treatment
of cellular growth related disorders, e.g., cancer. Moreover, the CCP-fusion
proteins of the
invention can be used as immunogens to produce anti-CCP antibodies in a
subject, to
purify CCP ligands and in screening assays to identify molecules which inhibit
the
interaction of CCP with a CCP substrate, e.g., a kinase such as CDC2b.
Preferably, a CCP chimeric or fusion protein of the invention is produced by
standard recombinant DNA techniques. For example, DNA fragments coding for the
different polypeptide sequences are ligated together in-frame in accordance
with
conventional techniques, for example by employing blunt-ended or stagger-ended
termini
for ligation, restriction enzyme digestion to provide for appropriate termini,
filling-in of
cohesive ends as appropriate, alkaline phosphatase treatment to avoid
undesirable joining,
and enzymatic ligation. In another embodiment, the fusion gene can be
synthesized by
conventional techniques including automated DNA synthesizers. Alternatively,
PCR
amplification of gene fragments can be carried out using anchor primers which
give rise to
complementary overhangs between two consecutive gene fragments which can
subsequently be annealed and reamplified to generate a chimeric gene sequence
(see, for
CA 02768046 2012-02-07
WO 01/8.5946 PCT/IBO1/01307
-47-
example, Current Protocols in Molecular Biology, eds. Ausubel et al. John
Wiley & Sons:
1992). Moreover, many expression vectors are commercially available that
already encode
a fusion moiety (e.g., a GST polypeptide). A CCP-encoding nucleic acid can be
cloned
into such an expression vector such that the fusion moiety is linked in-frame
to the CCP
protein.
The present invention also pertains to variants of the CCP proteins which
function
as either CCP agonists (mimetics) or as CCP antagonists. Variants of the CCP
proteins
can be generated by mutagenesis, e.g., discrete point mutation or truncation
of a CCP
protein. An agonist of the CCP proteins can retain substantially the same, or
a subset, of
the biological activities of the naturally occurring form of a CCP protein. An
antagonist of
a CCP protein can inhibit one or more of the activities of the,naturally
occurring form of
the CCP protein by, for example, competitively modulating a cellular activity
of a CCP
protein. Thus, specific biological effects can be elicited by treatment with a
variant of
limited function. In one embodiment, treatment of a subject with a variant
having a subset
of the biological activities of the naturally occurring form of the protein
has fewer side
effects in a subject relative to treatment with the naturally occurring form
of the CCP
protein.
In one embodiment, variants of a CCP protein which function as either CCP
agonists (mimetics) or as CCP antagonists can be identified by screening
combinatorial
libraries of mutants, e.g., truncation mutants, of a CCP protein for CCP
protein agonist or
antagonist activity. In one embodiment, a variegated library of CCP variants
is generated
by combinatorial mutagenesis at the nucleic acid level and is encoded by a
variegated gene
library. A variegated library of CCP variants can be produced by, for example,
enzymatically ligating a mixture of synthetic oligonucleotides into gene
sequences such
that a degenerate set of potential CCP sequences is expressible as individual
polypeptides,
or alternatively, as a set of larger fusion proteins (e.g., for phage display)
containing the set
of CCP sequences therein. There are a variety of methods which can be used to
produce
libraries of potential CCP variants from a degenerate oligonucleotide
sequence. Chemical
synthesis of a degenerate gene sequence can be performed in an automatic DNA
synthesizer, and the synthetic gene then ligated into an appropriate
expression vector. Use
of a degenerate set of genes allows for the provision, in one mixture, of all
of the
sequences encoding the desired set of potential CCP sequences. Methods for
synthesizing
degenerate oligonucleotides are known in the art (see, e.g., Narang, S.A.
(1983)
Tetrahedron 39:3; Itakura et al. (1984) Annu. Rev. Biochem. 53:323; Itakura et
al. (1984)
Science 198:1056; Ike et al. (1983) Nucleic Acid Res. 11:477.
In addition, libraries of fragments of a CCP protein coding sequence can be
used to
generate a variegated population of CCP fragments for screening and subsequent
selection
of variants of a CCP protein. In one embodiment, a library of coding sequence
fragments
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-48-
can be generated by treating a double stranded PCR fragment of a CCP coding
sequence
with a nuclease under conditions wherein nicking occurs only about once per
molecule,
denaturing the double stranded DNA, renaturing the DNA to form double stranded
DNA
which can include sense/antisense pairs from different nicked products,
removing single
stranded portions from reformed duplexes by treatment with Si nuclease, and
ligating the
resulting fragment library into an expression vector. By this method, an
expression library
can be derived which encodes N-terminal, C-terminal and internal fragments of
various
sizes of the CCP protein.
Several techniques are known in the art for screening gene products of
combinatorial libraries made by point mutations or truncation, and for
screening cDNA
libraries for gene products having a selected property. Such techniques are
adaptable for
rapid screening of the gene libraries generated by the combinatorial
mutagenesis of CCP
proteins. The most widely used techniques, which are amenable to high through-
put
analysis, for screening large gene libraries typically include cloning the
gene library into
replicable expression vectors, transforming appropriate cells with the
resulting library of
vectors, and expressing the combinatorial genes under conditions in which
detection of a
desired activity facilitates isolation of the vector encoding the gene whose
product was
detected. Recrusive ensemble mutagenesis (REM), a new technique which enhances
the
frequency of functional mutants in the libraries, can be used in combination
with the
screening assays to identify CCP variants (Arkin and Yourvan (1992) Proc.
Natl. Acad.
Sci. USA 89:7811-7815; Delgrave et al. (1993) Protein Engineering 6(3):327-
331).
In one embodiment, cell based assays can be exploited to analyze a variegated
CCP
library. For example, a library of expression vectors can be transfected into
a cell line
which ordinarily synthesizes and secretes CCP. The transfected cells are then
cultured
such that CCP and a particular mutant CCP are secreted and the effect of
expression of the
mutant on CCP activity in cell supernatants can be detected, e.g., by any of a
number of
enzymatic assays. Plasmid DNA can then be recovered from the cells which score
for
inhibition, or alternatively, potentiation of CCP activity, and the individual
clones further
characterized.
An isolated CCP protein, or a portion or fragment thereof, can be used as an
immunogen to generate antibodies that bind CCP using standard techniques for
polyclonal
and monoclonal antibody preparation. A full-length CCP protein can be used or,
alternatively, the invention provides antigenic peptide fragments of CCP for
use as
immunogens. The antigenic peptide of CCP comprises at least 8 amino acid
residues and
encompasses an epitope of CCP such that an antibody raised against the peptide
forms a
specific immune complex with CCP. Preferably, the antigenic peptide comprises
at least
10 amino acid residues, more preferably at least 15 amino acid residues, even
more
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-49-
preferably at least 20 amino acid residues, and most preferably at least 30
amino acid
residues.
Preferred epitopes encompassed by the antigenic peptide are regions of CCP
that
are located on the surface of the protein, e.g., hydrophilic regions.
A CCP immunogen typically is used to prepare antibodies by immunizing a
suitable subject, (e.g., rabbit, goat, mouse or other mammal) with the
immunogen. An
appropriate immunogenic preparation can contain, for example, recombinantly
expressed
CCP protein or a chemically synthesized CCP polypeptide. The preparation can
further
include an adjuvant, such as Freund's complete or incomplete adjuvant, or
similar
immunostimulatory agent. Immunization of a suitable subject with an
immunogenic CCP
preparation induces a polyclonal anti-CCP antibody response.
Accordingly, another aspect of the invention pertains to anti-CCP antibodies.
The
term "antibody" as used herein refers to immunoglobulin molecules and
immunologically
active portions of immunoglobulin molecules, i.e., molecules that contain an
antigen
binding site which specifically binds (immunoreacts with) an antigen, such as
CCP.
Examples of immunologically active portions of immunoglobulin molecules
include F(ab)
and F(ab')2 fragments which can be generated by treating the antibody with an
enzyme
such as pepsin. The invention provides polyclonal and monoclonal antibodies
that bind
CCP. The term "monoclonal antibody" or "monoclonal antibody composition", as
used
herein, refers to a population of antibody molecules that contain only one
species of an
antigen binding site capable of immunoreacting with a particular epitope of
CCP. A
monoclonal antibody composition thus typically displays a single binding
affinity for a
particular CCP protein with which it immunoreacts.
Polyclonal anti-CCP antibodies can be prepared as described above by
immunizing
a suitable subject with a CCP immunogen. The anti-CCP antibody titer in the
immunized
subject can be monitored over time by standard techniques, such as with an
enzyme linked
immunosorbent assay (ELISA) using immobilized CCP. If desired, the antibody
molecules directed against CCP can be isolated from the mammal (e.g., from the
blood)
and further purified by well known techniques, such as protein A
chromatography to
obtain the IgG fraction. At an appropriate time after immunization, e.g., when
the anti-
CCP antibody titers are highest, antibody-producing cells can be obtained from
the subject
and used to prepare monoclonal antibodies by standard techniques, such as the
hybridoma
technique originally described by Kohler and Milstein (1975) Nature 256:495-
497) (see
also, Brown et al. (1981) J. Immunol. 127:539-46; Brown et al. (1980) J Biol.
Chem
.255:4980-83; Yeh et al. (1976) Proc. Natl. Acad. Sci. USA 76:2927-31; and Yeh
et al.
(1982) Int. J Cancer 29:269-75), the more recent human B cell hybridoma
technique
(Kozbor et al. (1983) Immunol Today 4:72), the EBV-hybridoma technique (Cole
et al.
(1985), Monoclonal Antibodies and Cancer Therapy, Alan R. Liss, Inc., pp. 77-
96) or
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-50-
trioma techniques. The technology for producing monoclonal antibody hybridomas
is well
known (see generally R. H. Kenneth, in Monoclonal Antibodies: A New Dimension
In
Biological Analyses, Plenum Publishing Corp., New York, New York (1980); E. A.
Lerner
(1981) Yale J. Biol. Med., 54:387-402; M. L. Gefter et al. (1977) Somatic Cell
Genet.
3:231-36). Briefly, an immortal cell line (typically a myeloma) is fused to
lymphocytes
(typically splenocytes) from a mammal immunized with a CCP immunogen as
described
above, and the culture supernatants of the resulting hybridoma cells are
screened to
identify a hybridoma producing a monoclonal antibody that binds CCP.
Any of the many well known protocols used for fusing lymphocytes and
immortalized cell lines can be applied for the purpose of generating an anti-
CCP
monoclonal antibody (see, e.g., G. Galfre et al. (1977) Nature 266:55052;
Gefter et al.
Somatic Cell Genet., cited supra; Lerner, Yale J. Biol. Med., cited supra;
Kenneth,
Monoclonal Antibodies, cited supra). Moreover, the ordinarily skilled worker
will
appreciate that there are many variations of such methods which also would be
useful.
Typically, the immortal cell line (e.g., a myeloma cell line) is derived from
the same
mammalian species as the lymphocytes. For example, murine hybridomas can be
made by
fusing lymphocytes from a mouse immunized with an immunogenic preparation of
the
present invention with an immortalized mouse cell line. Preferred immortal
cell lines are
mouse myeloma cell lines that are sensitive to culture medium containing
hypoxanthine,
aminopterin and thymidine ("HAT medium"). Any of a number of myeloma cell
lines can
be used as a fusion partner according to standard techniques, e.g., the P3-
NS1/1-Ag4-1,
P3-x63-Ag8.653 or Sp2/O-Agl4 myeloma lines. These myeloma lines are available
from
ATCC. Typically, HAT-sensitive mouse myeloma cells are fused to mouse
splenocytes
using polyethylene glycol ("PEG"). Hybridoma cells resulting from the fusion
are then
selected using HAT medium, which kills unfused and unproductively fused
myeloma cells
(unfused splenocytes die after several days because they are not transformed).
Hybridoma
cells producing a monoclonal antibody of the invention are detected by
screening the
hybridoma culture supernatants for antibodies that bind CCP, e.g., using a
standard ELISA
assay.
Alternative to preparing monoclonal antibody-secreting hybridomas, a
monoclonal
anti-CCP antibody can be identified and isolated by screening a recombinant
combinatorial
immunoglobulin library (e.g., an antibody phage display library) with CCP to
thereby
isolate immunoglobulin library members that bind CCP. Kits for generating and
screening
phage display libraries are commercially available (e.g., the Pharmacia
Recombinant
Phage Antibody System, Catalog No. 27-9400-01; and the Stratagene
SurJZAPTMPhage
Display Kit, Catalog No. 240612). Additionally, examples of methods and
reagents
particularly amenable for use in generating and screening antibody display
library can be
found in, for example, Ladner et al. U.S. Patent No. 5,223,409; Kang et al.
PCT
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-51-
International Publication No. WO 92/18619; Dower et al. PCT International
Publication
No. WO 91/17271; Winter et al. PCT International Publication WO 92/20791;
Markland et
al. PCT International Publication No. WO 92/15679; Breitling et al. PCT
International
Publication WO 93/01288; McCafferty et al. PCT International Publication No.
WO
92/01047; Garrard et al. PCT International Publication No. WO 92/09690; Ladner
et al.
PCT International Publication No. WO 90/02809; Fuchs et al. (1991)
Bio/Technology
9:1370-1372; Hay et al. (1992) Hum. Antibod. Hybridomas 3:81-85; Huse et al.
(1989)
Science 246:1275-1281; Griffiths et al. (1993) EMBO J 12:725-734; Hawkins et
al. (1992)
J. Mol. Biol. 226:889-896; Clarkson et al. (1991) Nature 352:624-628; Gram et
al. (1992)
Proc. Natl. Acad. Sci. USA 89:3576-3580; Garrad et al. (1991) Bio/Technology
9:1373-
1377; Hoogenboom et al. (1991) Nuc. Acid Res. 19:4133-4137; Barbas et al.
(1991) Proc.
Natl. Acad. Sci. USA 88:7978-7982; and McCafferty et al. Nature (1990) 348:552-
554.
Additionally, recombinant anti-CCP antibodies, such as chimeric and humanized
monoclonal antibodies, comprising both human and non-human portions, which can
be
made using standard recombinant DNA techniques, are within the scope of the
invention.
Such chimeric and humanized monoclonal antibodies can be produced by
recombinant
DNA techniques known in the art, for example using methods described in
Robinson et al.
International Application No. PCT/US86/02269; Akira, et al. European Patent
Application
184,187; Taniguchi, M., European Patent Application 171,496; Morrison et al.
European
Patent Application 173,494; Neuberger et al. PCT International Publication No.
WO
86/01533; Cabilly et al. U.S. Patent No. 4,816,567; Cabilly et al. European
Patent
Application 125,023; Better et al. (1988) Science 240:1041-1043; Liu et al.
(1987) Proc.
Natl. Acad. Sci. USA 84:3439-3443; Liu et al. (1987) J. Immunol. 139:3521-
3526; Sun et
al. (1987) Proc. Natl. Acad. Sci. USA 84:214-218; Nishimura et al. (1987)
Canc. Res.
47:999-1005; Wood et al. (1985) Nature 314:446-449; and Shaw et al. (1988) J.
Natl.
Cancer Inst. 80:1553-1559); Morrison, S. L. (1985) Science 229:1202-1207; Oi
et al.
(1986) BioTechniques 4:214; Winter U.S. Patent 5,225,539; Jones et al. (1986)
Nature
321:552-525; Verhoeyan et al. (1988) Science 239:1534; and Beidler et al.
(1988) J.
Immunol. 141:4053-4060.
An anti-CCP antibody (e.g., monoclonal antibody) can be used to isolate CCP by
standard techniques, such as affinity chromatography or immunoprecipitation.
An anti-
CCP antibody can facilitate the purification of natural CCP from cells and of
recombinantly produced CCP expressed in host cells. Moreover, an anti-CCP
antibody
can be used to detect CCP protein (e.g., in a cellular lysate or cell
supernatant) in order to
evaluate the abundance and pattern of expression of the CCP protein. These
antibodies can
also be used, for example, for the immunoprecipitation and immunolocalization
of proteins
according to the invention as well as for the monitoring of the synthesis of
such proteins,
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-52-
for example, in recombinant organisms, and for the identification of compounds
interacting
with the protein according to the invention.
Anti-CCP antibodies can be used diagnostically to monitor protein levels in
tissue
as part of a clinical testing procedure, e.g., to, for example, determine the
efficacy of a
given treatment regimen. Detection can be facilitated by coupling (i.e.,
physically linking)
the antibody to a detectable substance. Examples of detectable substances
include various
enzymes, prosthetic groups, fluorescent materials, luminescent materials,
bioluminescent
materials, and radioactive materials. Examples of suitable enzymes include
horseradish
peroxidase, alkaline phosphatase, -galactosidase, or acetylcholinesterase;
examples of
suitable prosthetic group complexes include streptavidin/biotin and
avidin/biotin; examples
of suitable fluorescent materials include umbelliferone, fluorescein,
fluorescein
isothiocyanate, rhodamine, dichlorotriazinylamine fluorescein, dansyl chloride
or
phycoerythrin; an example of a luminescent material includes luminol; examples
of
bioluminescent materials include luciferase, luciferin, and aequorin, and
examples of
suitable radioactive material include 1251, 1311, 35S or 3H.
III. Computer Readable Means
The CCP nucleotide sequences of the invention (e.g., SEQ ID NO:1-66 or 228-
239)
or amino acid sequences of the invention (e.g., SEQ ID NO:67-132, 205, 211,
215-216, or
220-227) are also provided in a variety of mediums to facilitate use thereof.
As used
herein, "provided" refers to a manufacture, other than an isolated nucleic
acid or amino
acid molecule, which contains a nucleotide or amino acid sequences of the
present
invention. Such a manufacture provides the nucleotide or amino acid sequences,
or a
subset thereof (e.g., a subset of open reading frames (ORI's)) in a form which
allows a
skilled artisan to examine the manufacture using means not directly applicable
to
examining the nucleotide or amino acid sequences, or a subset thereof, as they
exist in
nature or in purified form.
In one application of this embodiment, a nucleotide or amino acid sequence of
the
present invention can be recorded on computer readable media. As used herein
"computer
readable media" includes any medium that can be read and accessed directly by
a
computer. Such media include, but are not limited to: magnetic storage media,
such as
floppy discs, hard disc storage medium, and magnetic tape; optical storage
media such a
CD-ROM; electrical storage media such as RAM and ROM; and hybrids of these
categories such as magnetic/optical storage media. The skilled artisan will
readily
appreciate how any of the presently known computer readable mediums can be
used to
create a manufacture comprising computer readable medium having recorded
thereon a
nucleotide or amino acid sequence of the present invention.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-53-
As used herein "recorded" refers to a process of storing information on
computer
readable medium. The skilled artisan can readily adopt any of the presently
known
methods for recording information on a computer readable medium to generate
manufactures comprising the nucleotide or amino acid sequence information of
the present
invention.
A variety of data storage structures are available to a skilled artisan for
creating a
computer readable medium having recorded thereon a nucleotide or amino acid
sequence
of the present invention. The choice of the data storage structure will
generally be based
on the means chosen to access the stored information. In addition, a variety
of data
processor programs and formats can be used to store the nucleotide sequence
information
of the present invention on computer readable medium. The sequence information
can be
represented in a word processing text file, formatted in commercially-
available software
such as WordPerfect and Microsoft Word, or represented in the form of an ASCII
file,
stored in a database application, such as DB2, Sybase Oracle, or the like. The
skilled
artisan can readily adapt any number of dataprocessor structuring formats
(e.g., text file or
database) in order to obtain computer readable medium having recorded thereon
the
nucleotide sequence information of the present invention.
By providing the nucleotide or amino acid sequences of the invention in
computer
readable form, the skilled artisan can routinely access the sequence
information for a
variety of purposes. For example, one skilled in the art can use the
nucleotide or amino
acid sequences of the invention in computer readable form to compare a target
sequence or
target structural motif with the sequence information stored within the data
storage means.
Search means are used to identity fragments or regions of the sequences of the
invention
which match a particular target sequence or target motif.
As used herein, a "target sequence" can be any DNA or amino acid sequence of
six
or more nucleotide or two or more amino acids. A skilled artisan can readily
recognize
that the longer a target sequence is, the less likely a target sequence will
be present as a
random occurrence in the database. The most preferred sequence length of a
target
sequence is from about 10 to 100 amino acids or form about 30 to 300
nucleotide residues.
However, it is well recognized that commercially important fragments, such as
sequence
fragments involved in gene expression and protein processing, may be shorter
length.
As used herein, "a target structural motif," or "target motif," refers to any
rationally
selected sequence or combination of sequences in which the sequence(s) are
chosen based
on a three-dimensional configuration which is formed upon the folding of the
target motif.
There are a variety of target motifs known in the art. Protein target motifs
include, but are
not limited to, enzyme active sites and signal sequences. Nucleic acid target
motifs
include, but are not limited to, promoter sequences, hairpin structures and
inducible
expression elements (protein binding sequences).
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-54-
Computer software is publicly available which allows a skilled artisan to
access
sequence information provided in a computer readable medium for analysis and
comparison to other sequences. A variety of known algorithms are disclosed
publicly and
a variety of commercially available software of conducting search means are
and can be
used in the computer-based systems of the present invention. Examples of such
software
include, but are not limited to, MacPatter (EMBL), BLASTN and BASTX (NCBIA).
For example, software which implements the BLAST (Altschul et al. (1990) J.
Mol. Biol. 215:403-410) and BLAZE (Brutlag et al. (1993) Comp. Chein. 17:203-
207)
search algorithms on a Sybase system can be used to identify open reading
frames (ORFs)
l0 of the sequences of the invention which contain homology to ORFs or
proteins from other
libraries. Such ORFs are protein encoding fragments and are useful in
producing
commercially important proteins such as enzyme used in various reactions and
in the
production of commercially useful metabolites.
IV. Recombinant Expression Vectors and Host Cells
Another aspect of the invention pertains to vectors, preferably expression
vectors,
containing a nucleic acid encoding a CCP protein (or a portion thereof). As
used herein,
the term "vector" refers to a nucleic acid molecule capable of transporting
another nucleic
acid to which it has been linked. One type of vector is a "plasmid", which
refers to a
circular double stranded DNA loop into which additional DNA segments can be
ligated.
Another type of vector is a viral vector, wherein additional DNA segments can
be ligated
into the viral genome. Certain vectors are capable of autonomous replication
in a host cell
into which they are introduced (e.g., bacterial vectors having a bacterial
origin of
replication and episomal mammalian vectors). Other vectors (e.g., non-episomal
mammalian vectors) are integrated into the genome of a host cell upon
introduction into
the host cell, and thereby are replicated along with the host genome.
Moreover, certain
vectors are capable of directing the expression of genes to which they are
operatively
linked. Such vectors are referred to herein as "expression vectors". In
general, expression
vectors of utility in recombinant DNA techniques are often in the form of
plasmids. In the
present specification, "plasmid" and "vector" can be used interchangeably as
the plasmid is
the most commonly used form of vector. However, the invention is intended to
include
such other forms of expression vectors, such as viral vectors (e.g.,
replication defective
retroviruses, adenoviruses and adeno-associated viruses), which serve
equivalent functions.
The recombinant expression vectors of the invention comprise a nucleic acid of
the
invention in a form suitable for expression of the nucleic acid in a host
cell, e.g., a plant
cell, which means that the recombinant expression vectors include one or more
regulatory
sequences, selected on the basis of the host cells to be used for expression,
which is
operatively linked to the nucleic acid sequence to be expressed. Within a
recombinant
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-55-
expression vector, "operably linked" is intended to mean that the nucleotide
sequence of
interest is linked to the regulatory sequence(s) in a manner which allows for
expression of
the nucleotide sequence (e.g., in an in vitro transcription/translation system
or in a host cell
when the vector is introduced into the host cell). The term "regulatory
sequence" is
intended to includes promoters, enhancers and other expression control
elements (e.g.,
polyadenylation signals). Such regulatory sequences are described, for
example, in
Goeddel; Gene Expression Technology: Methods in Enzymology 185, Academic
Press,
San Diego, CA (1990). Regulatory sequences include those which direct
constitutive
expression of a nucleotide sequence in many types of host cell and-those which
direct
expression of the nucleotide sequence only in certain host cells (e.g., tissue-
specific
regulatory sequences). It will be appreciated by those skilled in the art that
the design of
the expression vector can depend on such factors as the choice of the host
cell to be
transformed, the level of expression of protein desired, and the like. The
expression
vectors of the invention can be introduced into host cells to thereby produce
proteins or
peptides, including fusion proteins or peptides, encoded by nucleic acids as
described
herein (e.g., CCP proteins, mutant forms of CCP proteins, fusion proteins, and
the like).
The vectors of the invention comprise a selectable and/or scorable marker.
Selectable marker genes useful for the selection of transformed plant cells,
callus, plant
tissue and plants are well known to those skilled in the art and comprise, for
example,
antimetabolite resistance as the basis of selection for dhfr, which confers
resistance to
methotrexate (Reiss, Plant Physiol. (Life Sci. Adv.) 13 (1994), 143-149); npt,
which
confers resistance to the aminoglycosides neomycin, kanamycin and paromycin
(Herrera-
Estrella, EMBO J. 2 (1983), 987-995) and hygro, which confers resistance to
hygromycin
(Marsh, Gene 32 (1984), 481-485). Additional selectable genes have been
described,
namely trpB, which allow cells to utilize indole in place of tryptophan; hisD,
which allows
cells to utilize histinol in place of histidine (Hartman, Proc. Natl. Acad.
Sci. USA 85
(1988), 8047); mannose-6-phosphate isomerase which allows cells to utilize
maimose (WO
94/20627) and ODC (ornithine decarboxylase) which confers resistance to the
ornithine
decarboxylase inhibitor, 2-(difluoromethyl)-DL-ornithine, DFMO (McConlogue,
1987, In:
Current Communications in Molecular Biology, Cold Spring Harbor Laboratory
ed.) or
deaminase from Aspergillus terreus which confers resistance to Blasticidin S
(Tamura,
Biosci. Biotechnol. Biochem. 59 (1995), 2336-2338).
Useful scorable markers are also known to those skilled in the art and are
commercially available. Advantageously, the marker is a gene encoding
luciferase
(Giacomin, Pl. Sci. 116 (1996), 59-72; Scikantha, J. Bact. 178 (1996), 121),
green
fluorescent protein (Gerdes, FEBS Lett. 389 (1996), 44-47) or 13-glucuronidase
(Jefferson,
EMBOJ. 6 (1987), 3901-3907). This embodiment is particularly useful for simple
and
rapid screening of cells, tissues and organisms containing a vector of the
invention.
CA 02768046 2012-02-07
WO 01/85946 PCT/JBO1/01307
-56-
A "plant promoter" is a promoter capable of initiating transcription in plant
cells.
Exemplary plant promoters include, but are not limited to, those that are
obtained from
plants, plant viruses, and bacteria. Preferred promoters may contain
additional copies of
one or more specific regulatory elements, to further enhance expression and/or
to alter the
spatial expression and/or temporal expression of a nucleic acid molecule to
which it is
operably connected. For example, copper-responsive, glucocorticoid-responsive
or
dexamethasone-responsive regulatory elements may be placed adjacent to a
heterologous
promoter sequence driving expression of a nucleic acid molecule to confer
copper
inducible, glucocorticoid-inducible, or dexamethasone-inducible expression
respectively,
on said nucleic acid molecule. Examples of promoters under developmental
control
include promoters that preferentially initiate transcription in certain
tissues, such as leaves,
roots, seeds, endosperm, embryos, fibers, xylem vessels, tracheids, or
sclerenchyma. Such
promoters are referred to as "tissue preferred." Promoters which initiate
transcription only
in certain tissue are referred to as "tissue specific." A "cell type" specific
promoter
primarily drives expression in certain cell types in one or more organs, for
example,
vascular cells in roots or leaves. An "inducible" promoter is a promoter which
is under
environmental control. Examples of environmental conditions that may effect
transcription by inducible promoters include anaerobic conditions or the
presence of light.
Tissue specific, tissue preferred, cell type specific, and inducible promoters
constitute the
class of "non-constitutive" promoters. A "constitutive" promoter is a promoter
which is
active under most environmental conditions.
Another aspect of the invention pertains to host cells into which a
recombinant
expression vector of the invention has been introduced. The terms "host cell"
and
"recombinant host cell" are used interchangeably herein. It is understood that
such terms
refer not only to the particular subject cell but to the progeny or potential
progeny of such a
cell. Because certain modifications may occur in succeeding generations due to
either
mutation or environmental influences, such progeny may not, in fact, be
identical to the
parent cell, but are still included within the scope of the term as used
herein.
A host cell can be any prokaryotic or eukaryotic cell. For example, a CCP
protein
can be expressed in plant cells, bacterial cells such as E. coli, insect
cells, yeast or
mammalian cells (such as Chinese hamster ovary cells (CHO) or COS cells).
Other
suitable host cells are known to those skilled in the art.
Vector DNA can be introduced into prokaryotic or eukaryotic cells via
conventional transformation or transfection techniques. As used herein, the
terms
"transformation" and "transfection" are intended to refer to a variety of art-
recognized
techniques for introducing foreign nucleic acid (e.g., DNA) into a host cell,
including
calcium phosphate or calcium chloride co-precipitation, DEAF-dextran-mediated
transfection, lipofection, or electroporation. Suitable methods for
transforming or
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
-57-
transfecting host cells can be found in Sambrook, et al. (Molecular Cloning: A
Laboratory
Manual. 2nd, ed., Cold Spring Harbor Laboratory, Cold Spring Harbor Laboratory
Press,
Cold Spring Harbor, NY, 1989), and other laboratory manuals.
Means for introducing a recombinant expression vector of this invention into
plant
tissue or cells include, but are not limited to; transformation using CaCl2
and variations
thereof, in particular the method described by Hanahan (J. Mol.Biol. 166, 557-
560, 1983),
direct DNA uptake into protoplasts (Krens et al, Nature 296: 72-74, 1982;
Paszkowski et
al, EMBO J. 3:2717-2722, 1984), PEG-mediated uptake to protoplasts (Armstrong
et al,
Plant Cell Reports 9: 335-339, 1990) microparticle bombardment,
electroporation (Fromm
et al., Proc. Natl. Acad. Sci. (USA) 82:5824-5828, 1985), microinjection of
DNA
(Crossway et al., Mol. Gen. Genet. 202:179-185, 1986), microparticle
bombardment of
tissue explants or cells (Christou et al, Plant Physiol 87: 671-674, 1988;
Sanford,
Particulate Science and Technology 5: 27-37, 1987), vacuum-infiltration of
tissue with
nucleic acid, or in the case of plants, T-DNA-mediated transfer from
Agrobacterium to the
plant tissue as described essentially by An et al.( EMBO J 4:277-284, 1985),
Herrera-
Estrella et al. (Nature 303: 209-213, 1983a; EMBO J. 2: 987-995, 1983b; In:
Plant Genetic
Engineering, Cambridge University Press, N.Y., pp 63-93, 1985), or inplanta
method
using Agrobacterium tumefaciens such as that described by Bechtold et al.,
(C.R. Acad.
Sci. (Paris, Sciences de la vie/Life Sciences)316: 1194-1199, 1993), Clough et
al (Plant J.
16: 735-743, 1998), Trieu et al. (Plant J. 22:531-541, 2000) or Kloti
(WO0l/12828,
2001). Methods for transformation of monocotyledonous plants are well known in
the art
and include Agrobacterium-mediated transformation (Cheng et al. (1997) WO
97/48814;
Hansen (1998) WO 98/54961; Hiei et al. (1994) WO 94/00977; Hiei et al. (1998)
WO
98/17813; Rikiishi et al. (1999) WO 99/04618; Saito et al. (1995) WO
95/06722),
microprojectile bombardment (Adams et al. (1999) US 5,969,213; Bowen et al.
(1998) US
5,736,369; Chang et al. (1994) WO 94/13822; Lundquist et al. (1999) US
5,874,265/US
5,990,390; Vasil and Vasil (1995) US 5,405,765; Walker et al. (1999) US
5,955,362),
DNA uptake (Eval et al. (1993) WO 93/181,168), microinjection of Agrobacterium
cells
(von Holt 1994 DE 4309203), sonication (Finer et al. (1997) US 5,693,512) and
flower-dip
or in planta- transformation (Kloti, WO01/12828, 2001).
The vector DNA may further comprise a selectable marker gene to facilitate the
identification and/or selection of cells which are transfected or transformed
with a genetic
construct. Suitable selectable marker genes contemplated herein include the
ampicillin
resistance (Amp`), tetracycline resistance gene Tc'), bacterial kanamycin
resistance gene
(Kan'), phosphinothricin resistance gene, neomycin phosphotransferase gene
(nptIl),
hygromycin resistance gene, (3-glucuronidase (GUS) gene, chloramphenicol
acetyltransferase (CAT) gene, green fluorescent protein (gfp) gene (Haseloff
et al, 1997),
and luciferase gene.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-58-
For microparticle bombardment of cells, a microparticle is propelled into a
cell to
produce a transformed cell. Any suitable ballistic cell transformation
methodology and
apparatus can be used in performing the present invention. Exemplary apparatus
and
procedures are disclosed by Stomp et al. (U.S. Patent No. 5,122,466) and
Sanford and
Wolf (U.S. Patent No. 4,945,050). When using ballistic transformation
procedures, the
gene construct may incorporate a plasmid capable of replicating in the cell to
be
transformed. Examples of microparticles suitable for use in such systems
include I to 5
gm gold spheres. The DNA construct may be deposited on the microparticle by
any
suitable technique, such as by precipitation.
A whole plant may be regenerated from the transformed or transfected cell, in
accordance with procedures well known in the art. Plant tissue capable of
subsequent
clonal propagation, whether by organogenesis or embryogenesis, may be
transformed with
a gene construct of the present invention and a whole plant regenerated
therefrom. The
particular tissue chosen will vary depending on the clonal propagation systems
available
for, and best suited to, the particular species being transformed. Exemplary
tissue targets
include leaf disks, pollen, embryos, cotyledons, hypocotyls, megagametophytes,
callus
tissue, existing meristematic tissue (e.g., apical meristem, axillary buds,
and root
meristems), and induced meristem tissue (e.g., cotyledon meristem and
hypocotyl
meristem).
The term "organogenesis", as used herein, includes a process by which shoots
and
roots are developed sequentially from meristematic centres.
The term "embryogenesis", as used herein, includes a process by which shoots
and
roots develop together in a concerted fashion (not sequentially), whether from
somatic
cells or gametes.
Preferably, the plant is produced according to the methods of the invention by
transfecting or transforming the plant with a genetic sequence, or by
introducing to the
plant a protein, by any art-recognized means, such as microprojectile
bombardment,
microinjection, Agrobacterium-mediated transformation (including in planta
transformation), protoplast fusion, or electroporation, amongst others. Most
preferably the
plant is produced by Agrobacterium-mediated transformation.
Agrobacterium-mediated transformation or agrolistic transformation of plants,
yeast,
moulds or filamentous fungi is based on the transfer of part of the
transformation vector
sequences, called the T-DNA, to the nucleus and on integration of said T-DNA
in the
genome of said eukaryote.
The term "Agrobacterium" as used herein, includes a member of the
Agrobacteriaceae, more preferably Agrobacterium or Rhizobacterium and most
preferably
Agrobacterium tumefaciens.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-59-
The term "T-DNA", or "transferred DNA", as used herien, includes the
transformation vector flanked by T-DNA borders which is, after activation of
the
Agrobacterium vii- genes, nicked at the T-DNA borders and is transferred as a
single
stranded DNA to the nucleus of an eukaryotic cell.
As used herein, the terms "T-DNA borders", "T-DNA border region", or "border
region" include either right T-DNA borders (RB) or left T-DNA borders (LB),
which
comprise a core sequence flanked by a border inner region as part of the T-DNA
flanking
the border and/or a border outer region as part of the vector backbone
flanking the border.
The core sequences comprise 22 bp in case of octopine-type vectors and 25 bp
in case of
nopaline-type vectors. The core sequences in the right border region and left
border region
form imperfect repeats.
As used herein, the term "T-DNA transformation vector" or "T-DNA vector"
includes any vector encompassing a T-DNA sequence flanked by a right and left
T-DNA
border consisting of at least the right and left border core sequences,
respectively, and used
for transformation of any eukaryotic cell.
As used herein, the term "T-DNA vector backbone sequence" or "T-DNA vector
backbone sequences" includes all DNA of a T-DNA containing vector that lies
outside of
the T-DNA borders and, more specifically, outside the nicking sites of the
border core
imperfect repeats.
The present invention includes optimized T-DNA vectors such that vector
backbone integration in the genome of a eukaryotic cell is minimized or
absent. The term
"optimized T-DNA vector" as used herein includes a T-DNA vector designed
either to
decrease or abolish transfer of vector backbone sequences to the genome of a
eukaryotic
cell. Such T-DNA vectors are known to the one of skill in the art and include
those
described by Hanson et al. (1999) and by Stuiver et al. (1999 - W09901563).
The current invention clearly considers the inclusion of a DNA sequence
encoding
a CCP, homologue, analogue, derivative or immunologically active fragment
thereof as
defined supra, in any T-DNA vector comprising binary transformation vectors,
super-
binary transformation vectors, co-integrate transformation vectors, Ri-derived
transformation vectors as well as in T-DNA carrying vectors used in agrolistic
transformation.
As used herein, the term "binary transformation vector" includes a T-DNA
transformation vector comprising: a T-DNA region comprising at least one gene
of interest
and/or at least one selectable marker active in the eukaryotic cell to be
transformed; and
a vector backbone region comprising at.least origins of replication active in
E. coli and
Agrobacterium and markers for selection in E. coli and Agrobacterium.
Alternatively,
replication of the binary transformation vector in Agrobacterium is dependent
on the
presence of a separate helper plasmid. The binary vector pGreen and the helper
plasmid
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-60-
pSoup form an example of such a system (Hellens et al. (2000), Plant Mol.
Biol. 42, 819-
832; http://www.pgreen.ac.uk).
The T-DNA borders of a binary transformation vector can be derived from
octopine-type or nopaline-type Ti plasmids or from both. The T-DNA of a binary
vector is
only transferred to a eukaryotic cell in conjunction with a helper plasmid. As
used herein,
the term "helper plasmid" includes a plasmid that is stably maintained in
Agrobacterium
and is at least carrying the set of vir genes necessary for enabling transfer
of the T-DNA.
The set of vir genes can be derived from either octopine-type or nopaline-type
Ti plasmids
or from both.
As used herein, the term "super-binary transformation vector" includes a
binary
transformation vector additionally carrying in the vector backbone region a
vir region of
the Ti plasmid pTiBo542 of the super-virulent A. tumefaciens strain A281
(EP0604662,
EP0687730). Super-binary transformation vectors are used in conjunction with a
helper
plasmid.
As used herein, the term "co-integrate transformation vector" includes a T-DNA
vector at least comprising: a T-DNA region comprising at least one gene of
interest and/or
at least one selectable marker active in plants; and a vector backbone region
comprising at
least origins of replication active in Escherichia coli and Agrobacterium, and
markers for
selection in E. coli and Agrobacterium, and a set of vir genes necessary for
enabling
transfer of the T-DNA. The T-DNA borders and the set of vir genes of the T-DNA
vector
can be derived from either octopine-type or nopaline-type Ti plasmids or from
both.
The term "Ri-derived plant transformation vector" includes a binary
transformation
vector in which the T-DNA borders are derived from a Ti plasmid and the binary
transformation vector being used in conjunction with a `helper' Ri-plasmid
carrying the
necessary set of vir genes.
The terms "agrolistics", "agrolistic transformation" or "agrolistic transfer"
include a
transformation method combining features of Agrobacterium-mediated
transformation and
of biolistic DNA delivery. As such, a T-DNA containing target plasmid is co-
delivered
with DNA/RNA enabling in planta production of VirDI and VirD2 with or without
VirE2
(Hansen and Chilton 1996; Hansen et al. 1997; Hansen and Chilton 1997 -
W09712046).
A host cell of the invention, such as a prokaryotic or eukaryotic host cell in
culture,
can be used to produce (i.e., express) a CCP protein. Accordingly, the
invention further
provides methods for producing a CCP protein using the host cells of the
invention. In one
embodiment, the method comprises culturing the host cell of invention (into
which a
recombinant expression vector encoding a CCP protein has been introduced) in a
suitable
medium such that a CCP protein is produced. In another embodiment, the method
further
comprises isolating a CCP protein from the medium or the host cell.
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
-61-
The host cells of the invention can also be used to produce transgenic plant
or non-
human transgenic animals in which exogenous CCP sequences have been introduced
into
their genome or homologous recombinant plants or animals in which endogenous
CCP
sequences have been altered. Such plants and animals are useful for studying
the function
and/or activity of a CCP and for identifying and/or evaluating modulators of
CCP activity.
Trangenic Plants
As used herein, "transgenic plant" includes a plant which comprises within its
genome a heterologous polynucleotide. Generally, the heterologous
polynucleotide is
stably integrated within the genome such that the polynucleotide is passed on
to successive
generations. The heteroglogous polynucleotide may be integrated into the
genome alone
or as part of a recombinant expression cassette. "Transgenic" is used herein
to include any
cell, cell line, callus, tissue, plant part or plant, the genotype of which
has been altered by
the presence of heterologous nucleic acid including those transgenics
initially so altered as
well as those created by sexual crosses as asexual propagation from the
initial transgenic.
The term "transgenic" as used herein does not encompass the alteration of the
genome
(chromosomal or extra-chromosomal) by conventional plant breeding methods or
by
naturally occurring event such as random cross-fertilization, non-recombinant
viral
infection, non-recombinant bacterial transformation, non-recombinant
transposition, or
spontaneous mutation.
A transgenic plant of the invention can be created by introducing a CCP-
encoding
nucleic acid into the plant by placing it under the control of regulatory
elements which
ensure the expression in plant cells. These regulatory elements may be
heterologous or
homologous with respect to the nucleic acid molecule to be expressed as well
with respect
to the plant species to be transformed. In general, such regulatory elements
comprise a
promoter active in plant cells. These promoters can be used to modulate (e.g.
increase or
decrease) CCP content and/or composition in a desired tissue. To obtain
expression in all
tissues of a transgenic plant, preferably constitutive promoters are used,
such as the 35 S
promoter of CaMV (Odell, Nature 313 (1985), 810-812) or promoters from such
genes as
rice actin (McElroy et al. (1990) Plant Cell 2:163-171) maize H3 histone
(Lepetit et al.
(1992) Mol. Gen. Genet 231:276-285) or promoters of the polyubiquitin genes of
maize
(Christensen, Plant Mol. Biol. 18 (1982), 675-689). In order to achieve
expression in
specific tissues of a transgenic plant it is possible to use tissue specific
promoters (see, e.g.,
Stockhaus, EMBO J. 8 (1989), 2245-2251 or Table II, below).
CA 02768046 2012-02-07
WO 01/85946 PCT/11301/01307
-62-
Table II:
GENE SOURCE EXPRESSION REFERENCE
PATTERN
a-amylase (Amy32b) aleurone Lanahan, M.B., e t a!., Plant Cell 4:203-
211, 1992; Skriver, K., et al. Proc. Natl.
Acad. Sci. (USA) 88: 7266-7270, 1991
cathepsin (3-like gene aleurone Cejudo, F.J., et al. Plant Molecular Biology
20:849-856, 1992.
Agrobacteriwn rhizogenes ro1B cambium Nilsson et al., Physiol. Plant. 100:456-
462,
1997
PRP genes cell wall http://salus.medium.edu/mmg/tiemey/html
barley Itrl promoter endosperm
synthetic promoter endosperm Vicente-Carbajosa et al., Plant J. 13: 629-
640, 1998.
AtPRP4 flowers http://salus.medium.edu/mmg/tiemey/html
chalene synthase (chsA) flowers Van der Meer, et a!., Plant Mol. Biol. 15,
95-109, 1990.
LAT52 anther Twell et al Mol. Gen Genet. 217:240-245
(1989)
apetala-3 flowers
chitinase fruit (berries, grapes, etc) Thomas et al. CSIRO Plant Industry,
Urrbrae, South Australia, Australia;
http://winetitles.com.au/gwrdc/csh95-l.html
rbcs-3A green tissue (eg leaf) Lam, E. et al., The Plant Cell 2: 857-866,
1990.; Tucker et al., Plant Physiol. 113:
1303-1308, 1992.
leaf-specific genes leaf Baszczynski, et al., Nucl. Acid Res. 16:
4732, 1988.
AtPRP4 leaf http://salus.medium.edu/mmg/tierney/html
Pinus cab-6 leaf Yamamoto et al., Plant Cell Physiol.
35:773-778, 1994.
SAM22 senescent leaf Crowell, et al., Plant Mol. Biol. 18: 459-
466, 1992.
R. japonicum nif gene nodule United States Patent No. 4, 803, 165
B. japonicum nifH gene nodule United States Patent No. 5, 008, 194
GmENOD40 nodule Yang, et al., The Plant J. 3: 573-585.
PEP carboxylase (PEPC) nodule Pathirana, et al., Plant Mol. Biol. 20: 437-
450, 1992.
leghaemoglobin (Lb) nodule Gordon, et al., J. Exp. Bot. 44: 1453-1465,
1993.
Tungro bacilliform vines gene phloem Bhattacharyya-Pakrasi, et al, The Plant
J. 4:
71-79, 1992.
sucrose-binding protein gene plasma membrane Grimes, et al., The Plant Cell
4:1561-
1574,1992.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-63-
pollen-specific genes pollen; microspore Albani, et al., Plant Mol. Biol. 15:
605,
1990; Albani, et al., Plant Mol. Biol. 16:
501, 1991)
Zm13 pollen Guerrero at al Mol. Gen. Genet. 224:161-
168(1993)
apg gene microspore Twell et al Sex. Plant Reprod. 6:217-224
(1993)
maize pollen-specific gene pollen Hamilton, of al., Plant Mol. Biol. 18: 211-
218, 1992.
sunflower pollen-expressed gene pollen Baltz, et al., The Plant J. 2: 713-721,
1992.
B. napus pollen-specific gene pollen;anther; tapetum Arnoldo, et a!., J. Cell.
Biochem.,
Abstract No. Y101, 204, 1992.
root-expressible genes roots Tingey, et a!., EMBO J. 6: 1, 1987.
tobacco auxin-inducible gene root tip Van der Zaal, of al., Plant Mol. Bioi.
16,
983, 1991.
3-tubulin root Oppenheimer, et al., Gene 63: 87, 1988.
tobacco root-specific genes root Conkling, et al., Plant Physiol. 93: 1203,
1990.
B. napus G1-3b gene root United States Patent No. 5, 401, 836
SbPRPI roots Suzuki et a!., Plant Mol. Biol. 21: 109-
119,1993.
AtPRPI; AtPRP3 roots; root hairs http://salus.medium.edu/mmg/tierney/html
RD2 gene root cortex http://www2.cnsu.edu/ncsu/research
TobRB7 gene root vasculature http://www2.cnsu.edu/ncsu/research
AtPRP4 leaves; flowers; lateral http://salus.medium.edu/mmg/tierney/html
root primordia
seed-specific genes seed Simon, et al., Plant MoL Biol. 5: 191,
1985; Scofield, et al., J. Biol. Chem.
262: 12202, 1987.; Baszczynski, et a!.,
Plant Mol. Biol. 14:633, 1990.
Brazil Nut albumin seed Pearson, et al., Plant Mol. Biol. 18: 235-
245, 1992.
legumin seed Ellis, et a!., Plant Mol. Biol. 10: 203-214,
1988.
glutelin (rice) seed Takaiwa, of al., Mol. Gen. Genet. 208:
15-22, 1986; Takaiwa, et al., FEBS
Letts. 221: 43-47, 1987.
zein seed Matzke et a! Plant Mol Biol, 14(3):323-
32 1990
CA 02768046 2012-02-07
WO 01/85946 PCT/1801/01307
-64-
napA seed Stalberg, et al, Planta 199: 515-519,
1996.
sunflower oleosin seed (embryo and dry Cummins, at at, Plant Mol. Biol. 19:
seed) 873-876, 1992
LEAFY shoot meristem Weigel et al., Cell 69:843-859, 1992.
Arabidopsis thaliana knatl shoot meristem Accession number AJ 131822
Malus domestica knl shoot meristem Accession number Z71981
CLAVATAI shoot meristem Accession number AF049870
stigma-specific genes stigma Nasrallah, at al., Proc. Natl. Acad. Sci.
USA 85: 5551, 1988; Trick, et at, Plant
Mol. Biol. 15: 203, 1990.
class I patatin gene tuber Liu et al., Plant Mol. Biol. 153:386-395,
1991.
blz2 endosperm EP99106056.7
PCNA rice meristem Kosugi et a!, Nucleic Acids Research
19:1571-1576, 1991; Kosugi S. and
Ohashi Y, Plant Cell 9:1607-1619,
1997.
The promoters listed in the foregoing table are provided for the purposes of
exemplification only and the present invention is not to be limited by the
list provided
therein. Those skilled in the art will readily be in a position to provide
additional promoters
that are useful in performing the present invention. The promoters listed may
also be
modified to provide specificity of expression as required.
Known are also promoters which are specifically active in tubers of potatoes
or in
seeds of different plants species, such as maize, Vicia, wheat, barley and the
like. Inducible
promoters may be used in order to be able to exactly control expression under
certain
environmental or developmental conditions such as pathogens, anaerobia, or
light. Examples
of inducible promoters include the promoters of genes encoding heat shock
proteins or
microspore-specific regulatory elements (W096/16182). Furthermore, the
chemically
inducible Tet-system may be employed (Gatz, Mol. Gen. Genet. 227 (1991); 229-
237).
Further suitable promoters are known to the person skilled in the art and are
described, e.g.,
in Ward (Plant Mol. Biol. 22 (1993), 361-366). The regulatory elements may
further
comprise transcriptional and/or translational enhancers functional in plants
cells.
Furthermore, the regulatory elements may include transcription termination
signals, such as a
poly-A signal, which lead to the addition of a poly A tail to the transcript
which may improve
its stability.
CA 02768046 2012-02-07
WO 01/85946 PCTIIB01/01307
-65-
In the case that a nucleic acid molecule according to the invention is
expressed in the
sense orientation, the coding sequence can be modified such that the protein
is located in any
desired compartment of the plant cell, e.g., the nucleus, endoplasmatic
reticulum, the
vacuole, the mitochondria, the plastids, the apoplast, or the cytoplasm.
Methods for the introduction of foreign DNA into plants are also well known in
the
art. These include, for example, the transformation of plant cells or tissues
with T-DNA
using Agrobacterium tumefaciens or Agrobacterium rhizogenes, the fusion of
protoplasts,
direct gene transfer (see, e.g., EP-A 164 575), injection, electroporation,
biolistic methods
like particle bombardment, pollen-mediated transformation, plant RNA virus-
mediated
transformation, liposome-mediated transformation, transformation using wounded
or
enzyme-degraded immature embryos, or wounded or enzyme-degraded embryogenic
callus
and other methods known in the art. The vectors used in the method of the
invention may
contain further functional elements, for example "left border"- and "right
border"-
sequences of the T-DNA of Agrobacterium which allow for stably integration
into the
plant genome. Furthermore, methods and vectors are known to the person skilled
in the art
which permit the generation of marker free transgenic plants, i.e., the
selectable or scorable
marker gene is lost at a certain stage of plant development or plant breeding.
This can be
achieved by, for example, cotransformation (Lyznik, Plant Mol. Biol. 13
(1989), 151-161;
Peng, Plant Mol. Biol. 27 (1995), 91-104) and/or by using systems which
utilize enzymes
capable of promoting homologous recombination in plants (see, e.g.,
W097/08331;
Bayley, Plant Mol. Biol. 18 (1992), 353-361); Lloyd, Mol. Gen. Genet. 242
(1994), 653-
657; Maeser, Mol. Gen. Genet. 230 (1991), 170-176; Onouchi, Nucl. Acids Res.
19
(1991), 6373-6378). Methods for the preparation of appropriate vectors are
described by,
e.g., Sambrook (Molecular Cloning; A Laboratory Manual, 2nd Edition (1989),
Cold
Spring Harbor Laboratory Press, Cold Spring Harbor, NY).
Suitable strains of Agrobacterium tumefaciens and vectors, as well as
transformation of Agrobacteria, and appropriate growth and selection media are
described
in, for example, GV3 101 (pMK90RK), Koncz, Mol. Gen. Genet. 204 (1986), 383-
396;
C58C1 (pGV 3850kan), Deblaere, Nucl. Acid Res. 13 (1985), 4777; Bevan,
Nucleic. Acid
Res. 12(1984), 8711; Koncz, Proc. Natl. Acad. Sci. USA 86 (1989), 8467-8471;
Koncz,
Plant Mol. Biol. 20 (1992), 963-976; Koncz, Specialized vectors for gene
tagging and
expression studies. In: Plant Molecular Biology Manual Vol 2, Gelvin and
Schilperoort
(Eds.), Dordrecht, The Netherlands: Kluwer Academic Publ. (1994), 1-22; EP-A-
120 516;
Hoekema: The Binary Plant Vector System, Offsetdrukkerij Kanters B.V.,
Alblasserdam
(1985), Chapter V, Fraley, Crit. Rev. Plant. Sci., 4, 1-46; An, EMBO J. 4
(1985), 277-287).
Although the use of Agrobacterium tumefaciens is preferred in the method of
the invention,
other Agrobacterium strains, such as Agrobacterium rhizogenes, may be used,
for example, if
a phenotype conferred by said strain is desired.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-66-
Methods for the transformation using biolistic methods are known to the person
skilled in the art; see, e.g., Wan, Plant Physiol. 104 (1994), 37-48; Vasil,
Bio/Technology 11
(1993), 1553-1558 and Christou (1996) Trends in Plant Science 1, 423-431.
Microinjection
can be performed as described in Potrykus and Spangenberg (eds.), Gene
Transfer To Plants.
Springer Verlag, Berlin, NY (1995).
The transformation of most dicotyledonous plants may be performed using the
methods described above or using transformation via biolistic methods as,
e.g., described
above as well as protoplast transformation, electroporation of partially
permeabilized cells, or
introduction of DNA using glass fibers.
In general, the plants which are modified according to the invention may be
derived
from any desired plant species. They can be monocotyledonous plants or
dicotyledonous
plants, preferably they belong to plant species of interest in agriculture,
wood culture or
horticulture interest, such as crop plants (e.g., maize, rice, barley, wheat,
rye, oats), potatoes,
oil producing plants (e.g., oilseed rape, sunflower, pea nut, soy bean),
cotton, sugar beet,
sugar cane, leguminous plants (e.g., beans, peas), or wood producing plants,
preferably trees.
The present invention also relates to a transgenic plant cell which contains
(preferably
stably integrated into its genome) a nucleic acid molecule of the present
invention linked to
regulatory elements which allow expression of the nucleic acid molecule in
plant cells. The
presence and expression of the nucleic acid molecule in the transgenic plant
cells leads to the
synthesis of a CCP protein and may lead to physiological and phenotypic
changes in plants
containing such cells.
Transformed plant cells which are derived by any of the above transformation
techniques can be cultured to regenerate a whole plant which possesses the
transformed
genotype. Such regeneration techniques often rely on manipulation of certain
phytohormones in a tissue culture growth medium, typically relying on a
biocide and/or
herbicide marker which has been introduced with a polynucleotide of the
present
invention.
Plant cells transformed with a plant expression vector can be regenerated,
e.g., from
single cells, callus tissue or leaf discs according to standard plant tissue
culture techniques.
It is well known in the art that various cells, tissues, and organs from
almost any plant can
be successfully cultured to regenerate an entire plant. Plant regeneration
from cultured
protoplasts is described in Evans et al., Protoplasts Isolation and Culture,
Handbook of
Plant Cell Culture, Macmillilan Publishing Company, New York, pp. 124-176
(1983); and
Binding, Regeneration of Plants, Plant Protoplasts, CRC Press, Boca Raton, pp.
21-73
(1985).
Transformed plant cells, calli or explant can be cultured on regeneration
medium in
the dark for several weeks, generally about I to 3 weeks to allow the somatic
embryos to
mature. Preferred regeneration media include media containing MS salts, such
as PHI-E
CA 02768046 2012-02-07
WO 01/85946 PCT/1B01/01307
-67-
and PHI-F media. The plant cells, calli or explant are then typically cultured
on rooting
medium in a light/dark cycle until shoots and roots develop. Methods for plant
regeneration are known in the art and preferred methods are provided by Kamo
et al., (Bot.
Gaz. 146(3):324-334, 1985), West et al., (The Plant Cell 5:1361-1369. 1993),
and Duncan
et al. (Planta 165:322-332, 1985).
Small plantlets can then be transferred to tubes containing rooting medium and
allowed to grow and develop more roots for approximately another week. The
plants can
then be transplanted to soil mixture in pots in the greenhouse.
The regeneration of plants containing the foreign gene introduced by
Agrobacterium from leaft explants can be achieved as described by Horsch et
al., Science,
227:1229-1231 (1985). In this procedure, transformants are grown in the
presence of a
selection agent and in a medium that induces the regeneration of shoots in the
plant species
being transformed as described by Fraley et al., Proc. Natl. Acad. Sci, U.S.A.
80:4803
(1983). This procedure typically produces shoots within two to four weeks and
these
transformant shoots are then transferred to an appropriate root-inducing
medium
containing the selective agent and an antibiotic to prevent bacterial growth.
Transgenic
plants of the present invention may be fertile or sterile.
Regeneration can also be obtained from plant callus, explants, organs, or
parts
thereof. Such regeneration techniques are described generally in Klee et al.,
Ann. Rev. of
Plant Phys., 38:467-486(1987). The regeneration of plants from either single
plant
protoplasts or various explants is well known in the art. See, from example,
Methods for
Plant Molecular Biology, A. Weissbach and H. Weissback, eds., Academic Press,
Inc., San
Diego, Calif. (1988). This regeneration and growth process includes the steps
of selection
of transformant cells and shoots, rooting ht transformant shoots and growth of
the plantlets
in soil. For maize cell culture and regeneration see generally, The Maize
Handbook,
Freeling and Walbot, Eds., Springer, New York (1994); Corn and Corn
Improvement, 3`d
edition, Sprague and Dudley Eds., American Society of Agronomy, Madison,
Wisconsin
(1988).
One of skill will recognize that after the recombinant expression cassette is
stably
incorporated in transgenic plants and confirmed to be operable, it can be
introduced into
other plants by sexual crossing. Any of a number of standard breeding
techniques can be
used, depending upon the species to be crossed.
In vegetatively propagated crops, mature transgenic plants can be propagated
by the
taking of cuttings or by tissue culture techniques to produce multiple
identical plants.
Selection of desirable transgenics is made and new varieties are obtained and
propagated
vegetatively for commercial use. In seed propagated crops, mature transgenic
plants can
be self crossed to produce a homozygous inbred plant. The inbred plant
produces seed
containing the newly introduced heterologous nucleic acid. These seeds can be
grown to
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-68-
produce plants that would produce the selected phenotype, (e.g., altered cell
cycle content
or composition).
Parts obtained from the regenerated plant, such as flowers, seeds, leaves,
branches,
fruit and the like are included in the invention, provided that these parts
comprise cells
comprising the isolated nucleic acid of the present invention. Progeny and
variants, and
mutants of the regenerated plants are also included within the scope of the
invention,
provided that these parts comprise the introduced nucleic acid sequences.
Transgenic plants expressing the selectable marker can be screened for
transmission of the nucleic acid of the present invention by, for example,
standard
immunoblot and DNA detection techniques. Transgenic lines are also typically
evaluated
on levels of expression of the heterologous nucleic acid. Expression at the
RNA level can
be determined initially to identify and quantitate expression-positive plants.
Standard
techniques for RNA analysis can be employed and include PCR amplification
assays using
oligonucleotide primers designed to amplify only the heterologous RNA
templates and
solution hybridization assays using heterologous nucleic acid-specific probes.
The RNA-
positive plants can then analyzed for protein expression by Western immunoblot
analysis
using the specifically reactive antibodies of the present invention. In
addition, in situ
hybridization and immunocytochemistry according to standard protocols can be
done using
heterologous nucleic acid specific polynucleotide probes and antibodies,
respectively, to
localize sites of expression within transgenic tissue. Generally, a number of
transgenic
lines are usually screened for the incorporated nucleic acid to identify and
select plants
with the most appropriate expression profiles.
A preferred embodiment of the invention is a transgenic plant that is
homozygous
for the added heterologous nucleic acid; i. e., a transgenic plant that
contains two added
nucleic acid sequences, one gene at the same locus on each chromosome of a
chromosome
pair. A homozygous transgenic plant can be obtained by sexually mating
(selfing) a
heterozygous transgenic plant that contains a single added heterologous
nucleic acid,
germinating some of the seed produced and analyzing the resulting plants
produced for
altered cell division relative to a control plant (i.e., native, non-
transgenic). Back-crossing
to a parental plant and out-crossing with a non-transgenic plant are also
contemplated.
The present invention also relates to transgenic plants and plant tissue
comprising
transgenic plant cells according to the invention. Due to the (over)expression
of a CCP
molecule, e.g., at developmental stages and/or in plant tissue in which they
do not naturally
occur, these transgenic plants may show various physiological, developmental
and/or
morphological modifications in comparison to wild-type plants.
Therefore, part of this invention is the use of the CCP molecules to modulate
the
cell cycle and/or plant cell division and/or growth in plant cells, plant
tissues, plant organs
and/or whole plants. To the scope of the invention also belongs a method for
influencing
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-69-
the activity of CDKs such as CDC2a, or CDC2b, CKSs, CKIs, PLPs and KLPNTs in a
plant cell by transforming the plant cell with a nucleic acid molecule
according to the
invention and/or manipulation of the expression of the molecule.
Furthermore, the invention also relates to a transgenic plant cell which
contains
(preferably stably integrated into its genome) a nucleic acid molecule of the
invention or
part thereof, wherein the transcription and/or expression of the nucleic acid
molecule or
part thereof leads to reduction of the synthesis of a CCP. In a preferred
embodiment, the
reduction is achieved by an anti-sense, sense, ribozyme, co-suppression and/or
dominant
mutant effect. The reduction of the synthesis of a protein according to the
invention in the
transgenic plant cells can result in an alteration in, e.g., cell division. In
transgenic plants
comprising such cells this can lead to various physiological, developmental
and/or
morphological changes.
In yet another aspect, the invention relates to harvestable parts and to
propagation
material of the transgenic plants of the invention which either contain
transgenic plant cells
expressing a nucleic acid molecule according to the invention or which contain
cells which
show a reduced level of the described protein. Harvestable parts can be in
principle any
useful parts of a plant, for example, flowers, pollen, seedlings, tubers,
leaves, stems, fruit,
seeds, roots etc. Propagation material includes, for example, seeds, fruits,
cuttings,
seedlings, tubers, rootstocks, and the like.
Transgenic Animals
As used herein, a "transgenic animal" is a non-human animal, preferably a
mammal, more preferably a rodent such as a rat or mouse, in which one or more
of the
cells of the animal includes a transgene. Other examples of transgenic animals
include
non-human primates, sheep, dogs, cows, goats, chickens, amphibians, and the
like. A
transgene is exogenous DNA which is integrated into the genome of a cell from
which a
transgenic animal develops and which remains in the genome of the mature
animal,
thereby directing the expression of an encoded gene product in one or more
cell types or
tissues of the transgenic animal. As used herein, a "homologous recombinant
animal" is a
non-human animal, preferably a mammal, more preferably a mouse, in which an
endogenous CCP gene has been altered by homologous recombination between the
endogenous gene and an exogenous DNA molecule introduced into a cell of the
animal,
e.g., an embryonic cell of the animal, prior to development of the animal.
A transgenic animal of the invention can be created by introducing a CCP-
encoding
nucleic acid into the male pronuclei of a fertilized oocyte, e.g., by
microinjection, retroviral
infection, and allowing the oocyte to develop in a pseudopregnant female
foster animal.
The CCP cDNA sequence of SEQ ID NO: 1-66 or 228-239 can be introduced as a
transgene into the genome of a non-human animal. Alternatively, a nonhuman
homologue
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
-70-
of a human CCP gene, such as a mouse or rat CCP gene, can be used as a
transgene.
Alternatively, a CCP gene homologue, such as another CCP family member, can be
isolated based on hybridization to the CCP cDNA sequences of SEQ ID NO: 1-66
or 228-
239 (described further in subsection I above) and used as a transgene.
Intronic sequences
and polyadenylation signals can also be included in the transgene to increase
the efficiency
of expression of the transgene. A tissue-specific regulatory sequence(s) can
be operably
linked to a CCP transgene to direct expression of a CCP protein to particular
cells.
Methods for generating transgenic animals via embryo manipulation and
microinjection,
particularly animals such as mice, have become conventional in the art and are
described,
for example, in U.S. Patent Nos. 4,736,866 and 4,870,009, both by Leder et
al., U.S.
Patent No. 4,873,191 by Wagner et al. and in Hogan, B., Manipulating the Mouse
Embryo,
(Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 1986). Similar
methods
are used for production of other transgenic animals. A transgenic founder
animal can be
identified based upon the presence of a CCP transgene in its genome and/or
expression of
CCP mRNA in tissues or cells of the animals. A transgenic founder animal can
then be
used to breed additional animals carrying the transgene. Moreover, transgenic
animals
carrying a transgene encoding a CCP protein can further be bred to other
transgenic
animals carrying other transgenes.
V. Agricultural, Phytopharmaceutical and Pharmaceutical Compositions
The CCP nucleic acid molecules, CCP proteins, and anti-CCP antibodies (also
referred to herein as "active compounds") of the invention can be incorporated
into
compositions useful in agriculture and in plant cell and tissue culture. Plant
protection
compositions can be prepared by conventional means commonly used for the
application
of, for example, herbicides and pesticides. For example, certain additives
known to those
skilled in the art stabilizers or substances which facilitate the uptake by
the plant cell, plant
tissue or plant may be used.
The CCP nucleic acid molecules, CCP proteins, and anti-CCP antibodies (also
referred to herein as "active compounds") of the invention can also be
incorporated into
pharmaceutical compositions suitable for administration into animals. Such
compositions
typically comprise the nucleic acid molecule, protein, or antibody and a
pharmaceutically
acceptable carrier. As used herein the language "pharmaceutically acceptable
carrier" is
intended to include any and all solvents, dispersion media, coatings,
antibacterial and
antifungal agents, isotonic and absorption delaying agents, and the like,
compatible with
pharmaceutical administration. The use of such media and agents for
pharmaceutically
active substances is well known in the art. Except insofar as any conventional
media or
agent is incompatible with the active compound, use thereof in the
compositions is
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
-71-
contemplated. Supplementary active compounds can also be incorporated into the
compositions.
The nucleic acid molecules of the invention can be inserted into vectors and
used as
gene therapy vectors. Gene therapy vectors can be delivered to a plant or
subject by, for
example, injection, local administration (see U.S. Patent 5,328,470) or by
stereotactic
injection (see e.g., Chen et al. (1994) Proc. Natl. Acad. Sci. USA 91:3054-
3057). The
agricultural or pharmaceutical preparation of the gene therapy vector can
include the gene
therapy vector in an acceptable diluent, or can comprise a slow release matrix
in which the
gene delivery vehicle is imbedded. Alternatively, where the complete gene
delivery vector
can be produced intact from recombinant cells, e.g., retroviral vectors, the
agricultural or
pharmaceutical preparation can include one or more cells which produce the
gene delivery
system.
The agricultural and pharmaceutical compositions can be included in a
container,
pack, or dispenser together with instructions for administration.
VI. Uses and Methods of the Invention
The nucleic acid molecules, proteins, protein homologues, and antibodies
described
herein can be used in one or more of the following methods: a) agricultural
uses (e.g., to
increase plant yield and to develop phytopharmaceuticals); b) screening
assays; c)
predictive medicine (e.g., diagnostic assays, prognostic assays, monitoring
clinical trials);
d) methods of treatment (e.g., phytotherapeutic, therapeutic and
prophylactic); e)
transcriptomics; f) proteomics; g) metabolomics; h) ligandomics; and i)
pharmacogenetics
or pharmacogenomics. The isolated nucleic acid molecules of the invention can
be used,
for example, to express CCP protein (e.g., via a recombinant expression vector
in a host
cell or in gene therapy applications), to detect CCP mRNA (e.g., in a
biological sample) or
a genetic alteration in a CCP gene, and to modulate CCP activity, as described
further
below. The CCP proteins can be used to treat disorders characterized by
insufficient or
excessive production of a CCP substrate or production of CCP inhibitors. In
addition, the
CCP proteins can be used to screen for naturally occurring CCP substrates, to
screen for
drugs or compounds which modulate CCP activity, as well as to treat disorders
characterized by insufficient or excessive production of CCP protein or
production of CCP
protein forms which have decreased or aberrant activity compared to CCP wild
type
protein. Moreover, the anti-CCP antibodies of the invention can be used to
detect and
isolate CCP proteins, regulate the bioavailability of CCP proteins, and
modulate CCP
activity.
CA 02768046 2012-02-07
WO 01185946 PCT/IBO1/01307
-72-
A. Agricultural Uses:
In another embodiment of the invention, a method is provided for modifying
cell
fate and/or plant development and/or plant morphology and/or biochemistry
and/or
physiology comprising the modification of expression in particular cells,
tissues or organs
of a plant, of a genetic sequence encoding a CCP, e.g., a CCP operably
connected with a
plant-operable promoter sequence.
Modulation of the expression in a plant of a CCP or a homologue, analogue or
derivative thereof as defined in the present invention can produce a range of
desirable
phenotypes in plants, such as, for example, the modification of one or more
morphological,
biochemical, or physiological characteristics including: (i) modification of
the length of the
G1 and/or the S and/or the G2 and/or the M phase of the cell cycle of a plant;
(ii)
modification of the G1/S and/or S/G2 and/or G2/M and/or M/G I phase transition
of a plant
cell; (iii) modification of the initiation, promotion, stimulation or
enhancement of cell
division; (iv) modification of the initiation, promotion, stimulation or
enhancement of
DNA replication;(v) modification of the initiation, promotion, stimulation or
enhancement
of seed set and/or seed size and/or seed development; (vi) modification of the
initiation,
promotion, stimulation or enhancement of tuber formation; (vii) modification
of the
initiation, promotion, stimulation or enhancement of fruit formation; (viii)
modification of
the initiation, promotion, stimulation or enhancement of leaf formation; (ix)
modification
of the initiation, promotion, stimulation or enhancement of shoot initiation
and/or
development; (x) modification of the initiation, promotion, stimulation or
enhancement of
root initiation and/or development; (xi) modification of the initiation,
promotion,
stimulation or enhancement of lateral root initiation and/or development;
(xii) modification
of the initiation, promotion, stimulation or enhancement of nodule formation
and/or nodule
function; (xiii) modification of the initiation, promotion, stimulation or
enhancement of the
bushiness of the plant; (xiv) modification of the initiation, promotion,
stimulation or
enhancement of dwarfism in the plant; (xv) modification of the initiation,
promotion,
stimulation or enhancement of senescence; (xvi) modification of stem thickness
and/or
strength characteristics and/or wind-resistance of the stem and/or stem
length; (xvii)
modification of tolerance and/or resistance to biotic stresses such as
pathogen infection;
and (xviii) modification of tolerance and/or resistance to abiotic stresses
such as drought
stress or salt stress.
Methods to effect expression of a CCP or a homologue, analogue or derivative
thereof as defined in the present invention in a plant cell, tissue or organ,
include either the
introduction of the protein directly to a cell, tissue or organ such as by
microinjection of
ballistic means or, alternatively, introduction of an isolated nucleic acid
molecule encoding
the protein into the cell, tissue or organ in an expressible format. Methods
to effect
expression of a CCP or a homologue, analogue or derivative thereof as defined
in the
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-73-
current invention in whole plants include regeneration of whole plants from
the
transformed cells in which an isolated nucleic acid molecule encoding the
protein was
introduced in an expressible format.
The present invention clearly extends to any plant produced by the inventive
method described herein, and any and all plant parts and propagules thereof.
The present
invention extends further to encompass the progeny derived from a primary
transformed or
transfected cell, tissue, organ or whole plant that has been produced by the
inventive
method, the only requirement being that the progeny exhibits the same
genotypic and/or
phenotypic characteristic(s) as those characteristic(s) that (have) been
produced in the
parent by the performance of the inventive method.
By "cell fate and/or plant development and/or plant morphology and/or
biochemistry and/or physiology" is meant that one or more developmental and/or
morphological and/or biochemical and/or physiological characteristics of a
plant is altered
by the performance of one or more steps pertaining to the invention described
herein.
"Cell fate" includes the cell-type or cellular characteristics of a particular
cell that are
produced during plant development or a cellular process therefor, in
particular during the
cell cycle or as a consequence of a cell cycle process.
The term "plant development' 'or the term "plant developmental characteristic"
or
similar terms shall, when used herein, be taken to mean any cellular process
of a plant that
is involved in determining the developmental fate of a plant cell, in
particular the specific
tissue or organ type into which a progenitor cell will develop. Cellular
processes relevant
to plant development will be known to those skilled in the art. Such processes
include, for
example, morphogenesis, photomorphogenesis, shoot development, root
development,
vegetative development, reproductive development, stem elongation, flowering,
and
regulatory mechanisms involved in determining cell fate, in particular a
process or
regulatory process involving the cell cycle.
The term "plant morphology" or the term "plant morphological characteristic"
or
similar term will, when used herein, be understood by those skilled in the art
to include the
external appearance of a plant, including any one or more structural features
or
combination of structural features thereof. Such structural features include
the shape, size,
number, position, color, texture, arrangement, and patternation of any cell,
tissue or organ
or groups of cells, tissues or organs of a plant, including the root, stem,
leaf, shoot, petiole,
trichome, flower, petal, stigma, style, stamen, pollen, ovule, seed, embryo,
endosperm,
seed coat, aleurone, fibre, fruit, cambium, wood, heartwood, parenchyma,
aerenchyma,
sieve element, phloem or vascular tissue.
The term "plant biochemistry" or the term "plant biochemical characteristic"
or
similar term will, when used herein, be understood by those skilled in the art
to include the
metabolic and catalytic processes of a plant, including primary and secondary
metabolism
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
-74-
and the products thereof, including any small molecules, macromolecules or
chemical
compounds, such as but not limited to starches, sugars, proteins, peptides,
enzymes,
hormones, growth factors, nucleic acid molecules, celluloses, hemicelluloses,
calloses,
lectins, fibres, pigments such as anthocyanins, vitamins, minerals,
micronutrients, or
macronutrients, that are produced by plants.
The term "plant physiology" or the term "plant physiological characteristic"
or
similar term will, when used herein, be understood to include the functional
processes of a
plant, including developmental processes such as growth, expansion and
differentiation,
sexual development, sexual reproduction, seed set, seed development, grain
filling, asexual
reproduction, cell division, dormancy, germination, light adaptation,
photosynthesis, leaf
expansion, fibre production, secondary growth or wood production, amongst
others;
responses of a plant to externally-applied factors such as metals, chemicals,
hormones,
growth factors, environment and environmental stress factors (e.g., anoxia,
hypoxia, high
temperature, low temperature, dehydration, light, daylength, flooding, salt,
heavy metals,
amongst others), including adaptive responses of plants to said externally-
applied factors.
The CCP molecules of the present invention are useful in agriculture. The
nucleic
acid molecules, proteins, protein homologues, and antibodies described herein
can be used
to modulate the protein levels or activity of a protein involved in the cell
cycle, e.g.,
proteins involved in the G1/S and/or the G2/M transition in the cell cycle due
to
environmental conditions, including abiotic stress such as cold, nutrient
deprivation, heat,
drought, salt stress, or biotic stress such as a pathogen attack.
Thus, the CCP molecules of the present invention may be used to modulate,
e.g.,
enhance, crop yields; modulate, e.g., attenuate, stress, e.g. heat or nutrient
deprivation;
modulate tolerance to pests and diseases; modulate plant architecture;
modulate plant
quality traits; or modulate plant reproduction and seed development.
The CCP molecules of the present invention may also be used to modulate
endoreduplication in storage cells, storage tissues, and/or storage organs of
plants or parts
thereof. The term "endoreduplication" includes recurrent DNA replication
without
consequent mitosis and cytokinesis. Preferred target storage organs and parts
thereof for
the modulation of endoreduplication are, for example, seeds (such as from
cereals, oilseed
crops), roots (such as in sugar beet), tubers (such as in potatoes) and fruits
(such as in
vegetables and fruit species). Increased endoreduplication in storage organs,
and parts
thereof, correlates with enhanced storage capacity and, thus, with improved
yield. In
another embodiment of the invention, the endoreduplication of a whole plant is
modulated.
B. Screening Assays:
The invention provides a method (also referred to herein as a "screening
assay") for
identifying modulators, i.e., candidate or test compounds or agents (e.g.,
peptides,
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-75-
peptidomimetics, small molecules or other drugs) which bind to CCP proteins,
have a
stimulatory or inhibitory effect on, for example, CCP expression or CCP
activity, or have a
stimulatory or inhibitory effect on, for example, the expression or activity
of a CCP
substrate.
In one embodiment, the invention provides assays for screening candidate or
test
compounds which are substrates of a CCP protein or polypeptide or biologically
active
portion thereof. In another embodiment, the invention provides assays for
screening
candidate or test compounds which bind to or modulate the activity of a CCP
protein or
polypeptide or biologically active portion thereof, e.g., modulate the ability
of CCP to
interact with its cognate ligand. The test compounds of the present invention
can be
obtained using any of the numerous approaches in combinatorial library methods
known in
the art, including: biological libraries; spatially addressable parallel solid
phase or solution
phase libraries; synthetic library methods requiring deconvolution; the 'one-
bead one-
compound' library method; and synthetic library methods using affinity
chromatography
selection. The biological library approach is limited to peptide libraries,
while the other
four approaches are applicable to peptide, non-peptide oligomer or small
molecule libraries
of compounds (Lam, K.S. (1997) Anticancer Drug Des. 12:145).
Examples of methods for the synthesis of molecular libraries can be found in
the
art, for example in: DeWitt et al. (1993) Proc. Natl. Acad. Sci. U.S.A.
90:6909; Erb et al.
(1994) Proc. Natl. Acad. Sci. USA 91:11422; Zuckermann et al. (1994). J. Med.
Chem.
37:2678; Cho et al. (1993) Science 261:1303; Carrell et al. (1994) Angew.
Chem. Int. Ed.
Engl. 33:2059; Carell et al. (1994) Angew. Chem. Int. Ed. Engl. 33:2061; and
in Gallop et
al. (1994) J. Med. Chem. 37:1233.
Libraries of compounds may be presented in solution (e.g., Houghten (1992)
Biotechniques 13:412-421), or on beads (Lam (1991) Nature 3 54:82-84), chips
(Fodor
(1993) Nature 364:555-556), bacteria (Ladner USP 5,223,409), spores (Ladner
USP '409),
plasmids (Cull et al. (1992) Proc Natl Acad Sci USA 89:1865-1869) or on phage
(Scott and
Smith (1990) Science 249:386-390); (Devlin (1990) Science 249:404-406);
(Cwirla et al.
(1990) Proc. Natl. Acad. Sci. 87:6378-6382); (Felici (1991) J. Mol. Biol.
222:301-310);
(Ladner supra.).
In another embodiment, an assay is a cell-based assay comprising contacting a
cell
expressing a CCP target molecule (e.g., a plant cyclin dependent kinase) with
a test
compound and determining the ability of the test compound to modulate (e.g.
stimulate or
inhibit) the activity of the CCP target molecule. Determining the ability of
the test
compound to modulate the activity of a CCP target molecule can be
accomplished, for
example, by determining the ability of the CCP protein to bind to or interact
with the CCP
target molecule, or by determining the ability of the target molecule, e.g.,
the plant cyclin
dependent kinase, to phosphorylate a protein.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-76-
The ability of the target molecule, e.g., the plant cyclin dependent kinase,
to
phosphorylate a protein can be determined by, for example, an in vitro kinase
assay.
Briefly, a protein can be incubated with the target molecule, e.g., the plant
cyclin
dependent kinase, and radioactive ATP, e.g., [y-32P] ATP, in a buffer
containing MgC12
and MnC12, e.g., 10 mM MgC12 and 5 mM MnC12. Following the incubation, the
immunoprecipitated protein can be separated by SDS-polyacrylamide gel
electrophoresis
under reducing conditions, transferred to a membrane, e.g., a PVDF membrane,
and
autoradiographed. The appearance of detectable bands on the autoradiograph
indicates that
the protein has been phosphorylated. Phosphoaminoacid analysis of the
phosphorylated
1 o substrate can also be performed in order to determine which residues on
the protein are
phosphorylated. Briefly, the radiophosphorylated protein band can be excised
from the
SDS gel and subjected to partial acid hydrolysis. The products can then be
separated by
one-dimensional electrophoresis and analyzed on, for example, a phosphoimager
and
compared to ninhydrin-stained phosphoaminoacid standards.
1s Determining the ability of the CCP protein to bind to or interact with a
CCP target
molecule can be accomplished by determining direct binding. Determining the
ability of
the CCP protein to bind to or interact with a CCP target molecule can be
accomplished, for
example, by coupling the CCP protein with a radioisotope or enzymatic label
such that
binding of the CCP protein to a CCP target molecule can be determined by
detecting the
20 labeled CCP protein in a complex. For example, CCP molecules, e.g., CCP
proteins, can
be labeled with 1251, 355, 14C, or 3H, either directly or indirectly, and the
radioisotope
detected by direct counting of radioemmission or by scintillation counting.
Alternatively,
CCP molecules can be enzymatically labeled with, for example, horseradish
peroxidase,
alkaline phosphatase, or luciferase, and the enzymatic label detected by
determination of
25 conversion of an appropriate substrate to product.
It is also within the scope of this invention to determine the ability of a
compound
to modulate the interaction between CCP and its target molecule, without the
labeling of
any of the interactants. For example, a microphysiometer can be used to detect
the
interaction of CCP with its target molecule without the labeling of either CCP
or the target
30 molecule. McConnell, H. M. et al. (1992) Science 257:1906-1912. As used
herein, a
"microphysiometer" (e.g., Cytosensor) is an analytical instrument that
measures the rate at
which a cell acidifies its environment using a light-addressable
potentiometric sensor
(LAPS). Changes in this acidification rate can be used as an indicator of the
interaction
between compound and receptor.
35 In a preferred embodiment, determining the ability of the CCP protein to
bind to or
interact with a CCP target molecule can be accomplished by determining the
activity of the
target molecule. For example, the activity of the target molecule can be
determined by
detecting induction of a cellular second messenger of the target (e.g.,
intracellular Ca2+,
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-77-
diacylglycerol, IP3, etc.), detecting catalytic/enzymatic activity of the
target an appropriate
substrate, detecting the induction of a reporter gene (comprising a target-
responsive
regulatory element operatively linked to a nucleic acid encoding a detectable
marker, e.g.,
chloramphenicol acetyl transferase), or detecting a target-regulated cellular
response.
In yet another embodiment, an assay of the present invention is a cell-free
assay in
which a CCP protein or biologically active portion thereof is contacted with a
test
compound and the ability of the test compound to bind to the CCP protein or
biologically
active portion thereof is determined. Binding of the test compound to the CCP
protein can
be determined either directly or indirectly as described above. In a preferred
embodiment,
the assay includes contacting the CCP protein or biologically active portion
thereof with a
known compound which binds CCP to form an assay mixture, contacting the assay
mixture
with a test compound, and determining the ability of the test compound to
interact with a
CCP protein, wherein determining the ability of the test compound to interact
with a CCP
protein comprises determining the ability of the test compound to
preferentially bind to
CCP or biologically active portion thereof as compared to the known compound.
In another embodiment, the assay is a cell-free assay in which a CCP protein
or
biologically active portion thereof is contacted with a test compound and the
ability of the
test compound to modulate (e.g., stimulate or inhibit) the activity of the CCP
protein or
biologically active portion thereof is determined. Determining the ability of
the test
compound to modulate the activity of a CCP protein can be accomplished, for
example, by
determining the ability of the CCP protein to bind to a CCP target molecule by
one of the
methods described above for determining direct binding. Determining the
ability of the
CCP protein to bind to a CCP target molecule can also be accomplished using a
technology such as real-time Biomolecular Interaction Analysis (BIA).
Sjolander, S. and
Urbaniczky, C. (1991) Anal. Chem. 63:2338-2345 and Szabo et al. (1995) Curr.
Opin.
Struct. Biol. 5:699-705. As used herein, "BIA" is a technology for studying
biospecific
interactions in real time, without labeling any of the interactants (e.g.,
BlAcore). Changes
in the optical phenomenon of surface plasmon resonance (SPR) can be used as an
indication of real-time reactions between biological molecules.
In an alternative embodiment, determining the ability of the test compound to
modulate the activity of a CCP protein can be accomplished by determining the
ability of
the CCP protein to further modulate the activity of a CCP target molecule
(e.g., a CCP
mediated signal transduction pathway component). For example, the activity of
the
effector molecule on an appropriate target can be determined, or the binding
of the effector
to an appropriate target can be determined as previously described.
In yet another embodiment, the cell-free assay involves contacting a CCP
protein or
biologically active portion thereof with a known compound which binds the CCP
protein
to form an assay mixture, contacting the assay mixture with a test compound,
and
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-78-
determining the ability of the test compound to interact with the CCP protein,
wherein
determining the ability of the test-compound to interact with the CCP protein
comprises
determining the ability of the CCP protein to preferentially bind to or
modulate the activity
of a CCP target molecule.
The cell-free assays of the present invention are amenable to use of both
soluble
and/or membrane-bound forms of proteins (e.g., CCP proteins or biologically
active
portions thereof). In the case of cell-free assays in which a membrane-bound
form a
protein is used it may be desirable to utilize a solubilizing agent such that
the membrane-
bound form of the protein is maintained in solution. Examples of such
solubilizing agents
include non-ionic detergents such as n-octylglucoside, n-dodecylglucoside, n-
dodecylmaltoside, octanoyl-N-methylglucamide, decanoyl-N-methylglucamide,
Triton
X-100, Triton X-114, Thesit , Isotridecypoly(ethylene glycol ether)n, 3-[(3-
cholamidopropyl)dimethylamminio]- 1-propane sulfonate (CHAPS), 3-[(3-
cholamidopropyl)dimethylamminio)-2-hydroxy-l-propane sulfonate (CHAPSO), or N-
dodecyl=N,N-dimethyl-3-ammonio-l-propane sulfonate.
In more than one embodiment of the above assay methods of the present
invention,
it may be desirable to immobilize either CCP or its target molecule to
facilitate separation
of complexed from uncomplexed forms of one or both of the proteins, as well as
to
accommodate automation of the assay. Binding of a test compound to a CCP
protein, or
interaction of a CCP protein with a target molecule in the presence and
absence of a
candidate compound, can be accomplished in any vessel suitable for containing
the
reactants. Examples of such vessels include microtitre plates, test tubes, and
micro-
centrifuge tubes. In one embodiment, a fusion protein can be provided which
adds a
domain that allows one or both of the proteins to be bound to a matrix. For
example,
glutathione-S-transferase/ CCP fusion proteins or glutathione-S-
transferase/target fusion
proteins can be adsorbed onto glutathione sepharose beads (Sigma Chemical, St.
Louis,
MO) or glutathione derivatized microtitre plates, which are then combined with
the test
compound or the test compound and either the non-adsorbed target protein or
CCP protein,
and the mixture incubated under conditions conducive to complex formation
(e.g., at
physiological conditions for salt and pH). Following incubation, the beads or
microtitre
plate wells are washed to remove any unbound components, the matrix
immobilized in the
case of beads, complex determined either directly or indirectly, for example,
as described
above. Alternatively, the complexes can be dissociated from the matrix, and
the level of
CCP binding or activity determined using standard techniques.
Other techniques for immobilizing proteins on matrices can also be used in the
screening assays of the invention. For example, either a CCP protein or a CCP
target
molecule can be immobilized utilizing conjugation of biotin and streptavidin.
Biotinylated
CCP protein or target molecules can be prepared from biotin-NHS (N-hydroxy-
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-79-
succinimide) using techniques well known in the art (e.g., biotinylation kit,
Pierce
Chemicals, Rockford, IL), and immobilized in the wells of streptavidin-coated
96 well
plates (Pierce Chemical). Alternatively, antibodies reactive with CCP protein
or target
molecules but which do not interfere with binding of the CCP protein to its
target molecule
can be derivatized to the wells of the plate, and unbound target or CCP
protein trapped in
the wells by antibody conjugation. Methods for detecting such complexes, in
addition to
those described above for the GST-immobilized complexes, include
immunodetection of
complexes using antibodies reactive with the CCP protein or target molecule,
as well as
enzyme-linked assays which rely on detecting an enzymatic activity associated
with the
CCP protein or target molecule.
In another embodiment, modulators of CCP expression are identified in a method
wherein a cell is contacted with a candidate compound and the expression of
CCP mRNA
or protein in the cell is determined. The level of expression of CCP mRNA or
protein in
the presence of the candidate compound is compared to the level of expression
of CCP
mRNA or protein in the absence of the candidate compound. The candidate
compound can
then be identified as a modulator of CCP expression based on this comparison.
For
example, when expression of CCP mRNA or protein is greater (statistically
significantly
greater) in the presence of the candidate compound than in its absence, the
candidate
compound is identified as a stimulator of CCP mRNA or protein expression.
Alternatively, when expression of CCP mRNA or protein is less (statistically
significantly
less) in the presence of the candidate compound than in its absence, the
candidate
compound is identified as an inhibitor of CCP mRNA or protein expression. The
level of
CCP mRNA or protein expression in the cells can be determined by methods
described
herein for detecting CCP mRNA or protein.
In yet another aspect of the invention, the CCP proteins can be used as "bait
proteins" in a two-hybrid assay or three-hybrid assay (see, e.g., U.S. Patent
No. 5,283,317;
Zervos et al. (1993) Cell 72:223-232; Madura et al. (1993) J. Biol. Chem.
268:12046-
12054; Bartel et al. (1993) Biotechniques 14:920-924; Iwabuchi et al. (1993)
Oncogene
8:1693-1696; and Brent W094/10300), to identify other proteins, which bind to
or interact
with CCP ("CCP-binding proteins" or "CCP-bp") and are involved in CCP
activity. Such
CCP-binding proteins are also likely to be involved in the propagation of
signals by the
CCP proteins or CCP targets as, for example, downstream elements of a CCP-
mediated
signaling pathway. Alternatively, such CCP-binding proteins are likely to be
CCP
inhibitors. Alternatively, a mammalian two-hybrid system can be used which
includes e.g.
a chimeric green fluorescent protein encoding reporter gene (Shioda et al.
2000, Proc. Natl.
Acad. Sci. USA 97, 5520-5224). Yet another alternative consists of a bacterial
two-hybrid
system using e.g. HIS as reporter gene (Joung et al. 2000, Proc. Natl. Acad.
Sci. USA 97,
7382-7387).
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-80-
The two-hybrid system is based on the modular nature of most transcription
factors,
which consist of separable DNA-binding and activation domains. Briefly, the
assay
utilizes two different DNA constructs. In one construct, the gene that codes
for a CCP
protein is fused to a gene encoding the DNA binding domain of a known
transcription
factor (e.g., GAL-4). In the other construct, a DNA sequence, from a library
of DNA
sequences, that encodes an unidentified protein ("prey" or "sample") is fused
to a gene that
codes for the activation domain of the known transcription factor. If the
"bait" and the
"prey" proteins are able to interact, in vivo, forming a CCP-dependent
complex, the DNA-
binding and activation domains of the transcription factor are brought into
close proximity.
This proximity allows transcription of a reporter gene (e.g., LacZ) which is
operably linked
to a transcriptional regulatory site responsive to the transcription factor.
Expression of the
reporter gene can be detected and cell colonies containing the functional
transcription
factor can be isolated and used to obtain the cloned gene which encodes the
protein which
interacts with the CCP protein.
This invention further pertains to novel agents identified by the above-
described
screening assays. Accordingly, it is within the scope of this invention to
further use an
agent identified as described herein in an appropriate plant or animal model.
For example,
an agent identified as described herein (e.g., a CCP modulating agent, an
antisense CCP
nucleic acid molecule, a CCP-specific antibody, or a CCP-binding partner) can
be used in
a plant or animal model to determine the efficacy, toxicity, or side effects
of treatment with
such an agent. Alternatively, an agent identified as described herein can be
used in a plant
or animal model to determine the mechanism of action of such an agent.
Furthermore, this
invention pertains to uses of novel agents identified by the above-described
screening
assays for the agricultutal and therapeutic uses described herein.
C. Detection Assays
Portions or fragments of the cDNA sequences identified herein (and the
corresponding complete gene sequences) can be used in numerous ways as
polynucleotide
reagents. For example, these sequences can be used to: map their respective
genes on a
chromosome; and, thus, locate gene regions associated with genetic disease;
identify an
individual from a minute biological sample (tissue typing); and aid in
forensic
identification of a biological sample. Once the sequence (or a portion of the
sequence) of a
gene has been isolated, this sequence can be used to map the location of the
gene on a
chromosome. This process is called chromosome mapping. Accordingly, portions
or
fragments of the CCP nucleotide sequences, described herein, can be used to
map the
location of the CCP genes on a chromosome. The mapping of the CCP sequences to
chromosomes is an important first step in correlating these sequences with
genes
associated with disease.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-81-
Briefly, CCP genes can be mapped to chromosomes by preparing PCR primers
(preferably 15-25 bp in length) from the CCP nucleotide sequences. Computer
analysis of
the CCP sequences can be used to predict primers that do not span more than
one exon in
the genomic DNA, thus complicating the amplification process. These primers
can then be
used for PCR screening of cell hybrids containing individual plant or human
chromosomes. Only those hybrids containing the plant or human gene
corresponding to
the CCP sequences will yield an amplified fragment.
Other mapping strategies which can similarly be used to map a CCP sequence to
its
chromosome include in situ hybridization (described in Fan, Y. et al. (1990)
Proc. Natl.
Acad. Sci. USA, 87:6223-27), pre-screening with labeled flow-sorted
chromosomes, and
pre-selection by hybridization to chromosome specific cDNA libraries.
Fluorescence in situ hybridization (FISH) of a DNA sequence to a metaphase
chromosomal spread can further be used to provide a precise chromosomal
location in one
step. Chromosome spreads can be made using cells whose division has been
blocked in
metaphase by a chemical such as colcemid that disrupts the mitotic spindle.
The
chromosomes can be treated briefly with trypsin, and then stained with Giemsa.
A pattern
of light and dark bands develops on each chromosome, so that the chromosomes
can be
identified individually. The FISH technique can be used with a DNA sequence as
short as
500 or 600 bases. However, clones larger than 1,000 bases have a higher
likelihood of
binding to a unique chromosomal location with sufficient signal intensity for
simple
detection. Preferably 1,000 bases, and more preferably 2,000 bases will
suffice to get good
results at a reasonable amount of time. For a review of this technique, see
Verma et al.,
Human Chromosomes: A Manual of Basic Techniques (Pergamon Press, New York
1988).
Reagents for chromosome mapping can be used individually to mark a single
chromosome or a single site on that chromosome, or panels of reagents can be
used for
marking multiple sites and/or multiple chromosomes. Reagents corresponding to
noncoding regions of the genes actually are preferred for mapping purposes.
Coding
sequences are more likely to be conserved within gene families, thus
increasing the chance
of cross hybridizations during chromosomal mapping.
Once a sequence has been mapped to a precise chromosomal location, the
physical
position of the sequence on the chromosome can be correlated with genetic map
data.
(Such data are found, for example, in V. McKusick, Mendelian Inheritance in
Man,
available on-line through Johns Hopkins University Welch Medical Library). The
relationship between a gene and a disease, mapped to the same chromosomal
region, can
then be identified through linkage analysis (co-inheritance of physically
adjacent genes),
described in, for example, Egeland, J. et al. (1987) Nature, 325:783-787.
Moreover, differences in the DNA sequences between plants affected and
unaffected with a disease associated with the CCP gene, can be determined. If
a mutation
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
-82-
is observed in some or all of the affected plants but not in any unaffected
plants, then the
mutation is likely to be the causative agent of the particular disease.
Comparison of
affected and unaffected plants generally involves first looking for structural
alterations in
the chromosomes, such as deletions or translocations that are visible from
chromosome
spreads or detectable using PCR based on that DNA sequence. Ultimately,
complete
sequencing of genes from several plants can be performed to confirm the
presence of a
mutation and to distinguish mutations from polymorphisms.
D. Predictive Medicine:
The present invention also pertains to the field of predictive medicine in
which
diagnostic assays, prognostic assays, and monitoring clinical trials are used
for prognostic
(predictive) purposes to thereby treat an individual prophylactically.
Accordingly, one
aspect of the present invention relates to diagnostic assays for determining
CCP protein
and/or nucleic acid expression as well as CCP activity, in the context of a
biological
sample (e.g., blood, serum, cells, tissue) to thereby determine whether an
individual is
afflicted with a disease or disorder, or is at risk of developing a disorder,
associated with
aberrant CCP expression or activity. The invention also provides for
prognostic (or
predictive) assays for determining whether an individual is at risk of
developing a disorder
associated with CCP protein, nucleic acid expression or activity. For example,
mutations
in a CCP gene can be assayed in a biological sample. Such assays can be used
for
prognostic or predictive purpose to thereby phophylactically treat an
individual prior to the
onset of a disorder characterized by or associated with CCP protein, nucleic
acid
expression or activity.
Another aspect of the invention pertains to monitoring the influence of agents
(e.g.,
drugs, compounds) on the expression or activity of CCP in clinical trials.
E. Methods of Treatment:
The present invention provides for both prophylactic and therapeutic methods
of
treating a subject at risk of (or susceptible to) a disorder or having a
disorder associated
with aberrant CCP expression or activity. With regards to both prophylactic
and
therapeutic methods of treatment, such treatments may be specifically tailored
or modified,
based on knowledge obtained from the field of pharmacogenomics.
"Pharmacogenomics",
as used herein, refers to the application of genomics technologies such as
gene sequencing,
statistical genetics, and gene expression analysis to drugs in clinical
development and on
the market. More specifically, the term refers the study of how a patient's
genes determine
his or her response to a drug (e.g., a patient's "drug response phenotype", or
"drug response
genotype".) Thus, another aspect of the invention provides methods for
tailoring an
individual's prophylactic or therapeutic treatment with either the CCP
molecules of the
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-83-
present invention or CCP modulators according to that individual's drug
response
genotype. Pharmacogenomics allows a clinician or physician to target
prophylactic or
therapeutic treatments to patients who will most benefit from the treatment
and to avoid
treatment of patients who will experience toxic drug-related side effects.
This invention is further illustrated by the following examples which should
not be
construed as limiting. The contents of all references, patents and published
patent
applications cited throughout this application, as well as the Figures and the
Sequence
Listing are incorporated herein by reference.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-84-
EXAMPLES
EXAMPLE 1: IDENTIFICATION OF PLANT CCP POLYPEPTIDES USING
THE TWO HYBRID SYSTEM WITH CDC2B AS A BAIT
A two-hybrid screening was performed using as bait a fusion between the GAL4
DNA-binding domain and one of the following: CDC2bAt.N 161 (GenBank accession
number D10851; residue Asp161 converted into Asnl61); CKSlAt (GenBank
accession
number AJ000016); E2Fa (=E2F5) (GenBank accession number AJ294534)
dimerization
domain (226-356aa; SEQ ID NO:205); CKI4 (SEQ ID NO:264); PLP1 (GenBank
accession number TO1601); KLPNTI (GenBank accession number ABO11479; protein
ID
number BAB 11568) motor domain (36-508 aa); KLPNTI (GenBank accession number
ABO 11479; protein ID number BAB 11568) stalk domain (427-867 aa); KLPNT2=TH65
(GenBank accession number AJ001729) neck domain (3-186 aa); KLPNT2=TH65
(GenBank accession number AJ001729) stalk domain (73-608 aa); E2Fb (=E2F3)
(GenBank accession number AJ294533) N-terminal domain (1-385 aa; SEQ ID
NO:206),
respectively
CDC2bAt.N161 is a dominant negative form of the CDC2bAt protein. The D161
residue in CDC2bAt is crucial for ATP binding and, thus, the mutation of this
residue
results in an inactive kinase. The interactions between this mutated CDK and
its substrates
and regulatory proteins are also more stabilised as a result of this mutation.
In yeast the PHO genes are part of a complex regulatory network linking
phosphate
availability with the expression of phosphatases. When phosphate levels are
high the
PH080/PHO85 cyclin/CDK complex phosphorylates a transcription factor. This
transcription factor of phosphatase genes thereby becomes inactive. The S.
cerevisiae
PH085 protein can interact with the G1 specific cyclins PCL1 and PCL2 (close
homologues to the PHO80). In a yeast strain deficient for the GI cyclins CLN1
and CLN2,
PHO80 is required for GI progression. This result suggests that PH085 is
involved in a
regulatory pathway that links the nutrient status of the cell with cell
division activity. The
five PLP of A. thaliana show similarity to the yeast cyclin-like PHO80 gene.
Kinesins use the cytoskeleton to move around vesicles, organelles, chromosomes
and the like in the cell. They can also be involved in spindle formation.
Kinesins consist
of three functional unrelated domains: the motor domain (involved in
microtubule binding;
contains the ATPase domain), the stalk region (involved in homo- or
heterodimirisation of
the kinesins), and the tail (involved in the interaction with the `substrates'
of the kinesin).
Two hybrid screens were performed using different parts of two-kinesin-related
proteins
(KLPNTI and KLPNT2 (being more than 80% identical to KLPNTI). Other
information
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-85-
obtained by the two hybrid approach is the dimerization of the kinesins: the
KLPNTI and
KLPNT2 interact (stalks and stalks-tail) with and between themselves.
Vectors and strains used were provided with the Matchmaker Two-Hybrid System
(Clontech, Palo Alto, CA). The bait was constructed by inserting the CDC2bAt.N
161
(GenBank accession number D10851; residue Aspl61 converted into Asnl6l);
CKS1At
(GenBank accession number AJ000016); E2Fa (=E2F5) (GenBank accession number
AJ294534) dimerization domain (226-356aa; SEQ ID NO:205); CKI4 (SEQ ID
NO:264);
PLP1 (GenBank accession number TO 1601); KLPNT1 (GenBank accession number
ABO11479; protein ID number BABI 1568) motor domain (36-508 aa); KLPNTI
(GenBank accession number ABOI 1479; protein ID number BAB11568) stalk domain
(427-867 aa); KLPNT2=TH65 (GenBank accession number AJ001729) neck domain (3-
186 aa); KLPNT2=TH65 (GenBank accession number AJ001729) stalk domain (73-608
aa); E2Fb (=E2F3) (GenBank accession number AJ294533) N-terminal domain (1-385
aa;
SEQ ID NO:206), respectively, into the pGBT9 vector. Bait vectors where
constructed by
introducing the PCR fragment created from the corresponding cDNA using primers
to
incorporate EcoRI and BamHI restriction enzyme sites. The PCR fragment was cut
with
EcoRI and BamHI and cloned into the EcoRI and BamHl sites of pGBT9, resulting
in the
desired plasmid. The GAL4 activation domain cDNA fusion library was
constructed as
described in De Veylder et al 1999, 208(4) p453-62 from mRNA of Arabidopsis
thaliana
cell suspensions harvested at various growing stages: early exponential,
exponential, early
stationary, and stationary phase.
For the screening a 1-liter culture of the Saccharomyces cerevisiae strain
HF7c
(MATa ura3-52 his3-200 ade2-101 lys2-801 trpl-901 leu2-3,112 gal4-542 ga180-
538
LYS2:: GAL1 UAS GALITATA-HISS URA3:: GAL417mers(3x)-CyCJTATA-LacZ) was
sequentially transformed with the bait plasmid and 20 g DNA of the library
using the
lithium acetate method (Geitz et al. (1992) supra). To estimate the number of
independent
cotransformants, 1/1000 of the transformation mix was plated on Leu- and Trp-
medium.
The rest of the transformation mix was plated on medium to select for
histidine
prototrophy (Trp-, Leu-, His-). After 5 days of growth at 30 C, the colonies
larger than 2
mm were streaked on histidine-lacking medium. At total for each screening at
least 10'
independent cotransformants were screened for there ability to grow on
histidine free
medium. Of the His+ colonies the activation domain plasmids were isolated as
described
(Hoffman and Winston, 1987, Gene 57, 267-272). The hybriZAP' inserts were PCR
amplified and the PCR fragments were digested with AluI and fractionized on a
2%
agarose gel. Plasmid DNA of which the inserts gave rise to different
restriction patterns
were electroporated into Escherichia coli XL1-Blue, and the DNA sequence of
the inserts
was determined. Extracted DNA was also used to retransform HF7c to test the
specificity
of the interaction.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-86-
Using the foregoing technique, 61 cDNAs were identified, their sequences were
determined and found to contain open reading frames termed CCPI through CCP61
(Figures 1-61).
EXAMPLE 2: EXTENSION OF CCP ENCODING POLYNUCLEOTIDES
TO FULL LENGTH OR TO RECOVER REGULATORY
ELEMENTS
The CCP encoding nucleic acid sequences (SEQ ID NO:1-66 or 228-239) are used
to design oligonucleotide primers for extending a partial nucleotide sequence
to full length
or for obtaining 5' sequences from genomic or cDNA libraries. One primer is
synthesized
to initiate extension in the antisense direction (XLR) and the other is
synthesized to extend
sequence in the sense direction (XLF). Primers allow the extension of the
known CCP
encoding sequence "outward" generating amplicons containing new, unknown
nucleotide
sequence for the region of interest. The initial primers are designed from the
cDNA using
OLIGO 4.06 Primer Analysis Software (National Biosciences), or another
appropriate
program, to be preferably 22-30 nucleotides in length, to have a GC content of
preferably
50% or more, and to anneal to the target sequence at temperatures preferably
about 68 -
72 C. Any stretch of nucleotides which would result in hairpin structures and
primer-
primer dimerizations is avoided. The original, selected cDNA libraries,
prepared from
mRNA isolated from actively dividing cells or a plant genomic library are used
to extend
the sequence; the latter is most useful to obtain 5' upstream regions. If more
extension is
necessary or desired, additional sets of primers are designed to further
extend the known
region.
Sense XLF primers can also be designed based on publicly available genomic
sequences. GENEMARK.hmm (hidden morkov model) version 2.2a software (default
parameters) can e.g. be used to predict open reading frames. The 5' end of the
predicted
open reading frame is then subsequently used to design the sense XLF primer.
Said XLF
primer and the appropriate XLR primer are then used in an RT-PCR (reverse
transcription-
polymerase chain reaction) reaction to amplify the predicted cDNA. The
resulting PCR
product is cloned in a suitable vector and subjected to DNA sequence analysis
to verify the
prediction.
Primers used to amplify coding regions of the CCPs of the invention are
designed
such that the PCR product can be cloned in the pDONR201 vector (Gateway'
cloning
system, Invitrogen). Thus, a sense primer has the attB l site (SEQ ID NO:246)
at its 5' end.
For current purposes, the attB 1 site is followed by a consensus Kozak
sequence (SEQ ID
NO:247; Kozak (1989) J Cell Biol 108:229-241; Liitck et al. (1987) EMBO J 6:43-
48).
The 3' end of the sense primer comprises the gene-specific parts as indicated
in Figures 1-
46. An antisense primer has at the 5' end the attB2 site (SEQ ID NO:248)
followed by the
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-87-
inverse complement of the gene/coding region of interest as indicated in
Figures 1-46.
Primers used for CCP amplification by PCR are given with their SEQ ID NOs in
Table 3.
The sequence of cloned CCP PCR products was or is determined using the sense
primer
prml024 (SEQ ID NO:265) and the antisense primer prml025 (SEQ ID NO:266).
TABLE-III:
PCR primers sense antisense
CCP sense + antisense primer primer
Molecule SEQ ID SEQ ID
NO: NO:
CCPI prm0733 +prm0734 133 134
CCP2 prm0663 + prm0664 135 136
CCP3 prm0705 + prm0706 137 138
CCP4 prm0659 + prm0660 139 140
CCPS prm0749 + pm-10750 141 142
CCP6 prm0707 + prm 0708 143 144
CCP7/8 prm0657 + prm0658 145 146
CCP9 prm0582 + prm0583 147 148
CCP10 prm0671 +prm0672 149 150
CCPII prm0729 + prm0730 151 152
CCP 12+ prm 1676 + prm 1677 153 154
CCP13
CCP14 prm0701 +prm0702 155 156
CCP15 prm0445 +prm0446 157 158
CCP16 prm0321 +prm0322 159 160
CCP17 pr-n0632 + prm0633 161 162
CCP18 prm0488 + prm0489 163 164
CCP19 prm0661 + prm0662 165 166
CCP20+ prm07O9 + prm0710 167 168
CCP21
CCP22 prm0711 + prm0712 169 170
CCP23 prm0819 + prmO820 171 172
CCP24 prm0739 + prm0740 173 174
CCP25 prm0741 + prm0742 175 176
CCP26 prm0703 + prm07O4 177 178
CCP27 prm0817 + prm0818 179 180
CCP28 prm07l3 + prm0714 181 182
CCP29 / / /
CCP30 prm0480 + prm0481 183 184
CCP31 prm0737 + prm0738 185 186
CCP32 prm1493 +prm1494 187 188
CCP33 prm0319 + prm0320 189 190
CCP34 prml377 + prm1378 191 192
CA 02768046 2012-02-07
WO 01/85946 PCT/IB01/01307
-88-
CCP35 prml38l + prm1382 193 194
CCP36 / / /
CCP37 prm1379 + prml380 195 196
CCP38 prm1383 + prml384 197 198
By following the instructions for the XL-PCR kit (Perkin Elmer) and thoroughly
mixing the enzyme and reaction mix, high fidelity amplification is obtained.
Beginning
with 40 pmol of each primer and the recommended concentrations of all other
components
of the kit, PCR is performed suing the Peltier Thermal Cycle (PTC200; MJ
Research,
Watertown MA) and the following parameters:
Step 1 94 C for 1 min (initial denaturation)
Step 2 65 C for 1 min
Step 3 68 C for 6 min
Step 4 94 for 15 sec
Step 5 65 C for 1 min
Step 6 68'C for 7 min
Step 7 Repeat steps 4-6 for 15 additional cycles
Step 8 94 C for 15 sec
Step 9 65 C for 1 min
Step 10 68 C for 7:15 min
Step 11 Repeat step 8-10 for 12 cycles
Step 12 72 C for 8 min
Step 13 4 C (and holding)
A 5-10 pl aliquot of the reaction mixture is analyzed by electrophoresis on a
low
concentration (about 0.6-0.8%) agarose mini-gel to determine which reactions
were
successful in extending the sequence. Bands thought to contain the largest
products were
selected and cut out of the gel. Further purification involves using a
commercial gel
extraction method such as QIAQuickT' (QIAGEN Inc). After recovery of the DNA,
Klenow enzyme was used to trim single-stranded, nucleotide overhangs creating
blunt
ends which facilitate religation and cloning. After ethanol precipitation, the
products are
redissolved in 13 gl of ligation buffer, 1 l T4-DNA ligase (15 units) and 1
gI T4
polynucleotide kinase are added, and the mixture is incubated at room
temperature for 2-3
hours or overnight at 16 C. Competent E. coli cells (in 40 l of appropriate
media) are
transformed with 3 gl of ligation mixture and cultured in 80 gil of SOC medium
(Sambrook, supra). After incubation for one hour at 37 C, the whole
transformation
mixture is plated on Luria Bertani (LB)-agar (Sambrook, supra) containing
2xCarb. The
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-89-
following day, several colonies are randomly picked from each plate and
cultured in 150 l
of liquid LB/2xCarb medium placed in an individual well of an appropriate,
commerically-
available, sterile 96-well microtiter plate. The following day, 5 l of each
overnight culture
is transferred into a non-sterile 96-well plate and after dilution 1:10 with
water, 5 l of
each sample is transferred into a PCR array. For PCR amplification, 18 gl of
concentrated
PCR reaction mix (3.3x) containing 4 units of 4Tth DNA polymerase, a vector
primer and
both of the gene specific primers used for the extension reaction are added to
each well.
Amplification is performed using the following conditions:
Step 1 94 C for 60 sec
Step 2 94 C for 20 sec
Step 3 55 C for 30 sec
Step 4 72 C for 90 sec
Step 5 Repeat steps 2-4 for an additional 29 cycles
Step 6 72 C for 180 sec
Step 7 4 C (and holding)
Aliquots of the PCR reactions are run on agarose gels together with molecular
weight
markers. The sizes of the PCR products are compared to the original partial
cDNAs, and
appropriate clones are selected, ligated into plasmid and sequenced.
EXAMPLE 3: EXPRESSION OF RECOMBINANT CCP PROTEINS IN
TRANSGENIC PLANTS
In this example, the CCP molecules of the present invention were expressed in
a
35S expression vector in transgenic plants. The CCP molecules of this
invention were
cloned using standard cloning procedures between a suitable promoter, e.g. the
CaMV35S
promoter or any promoter from e.g. Table II, and a suitable terminator, e.g.,
the NOS 3'
untranslated region. The resulting recombinant gene is subsequently cloned in
a suitable
binary vector and the resulting plant transformation vector is then
transferred to
Agrobacterium tumefaciens. Arabidopsis thaliana is transformed with this
Agrobacterium
applying the in planta flower-dip transformation method (Clough and Bent,
Plant J.
16:735-743, 1998). Transgenic plant lines are selected on a growth medium
containing the
suitable selection agent (e.g., kanamycin or Basta) or on the basis of scoring
the expression
of a screenable marker (e.g., luciferase, green fluorescent protein).
For tissue-specific expression, the CCP gene can also be expressed under
control of
the minimal 35S promoter containing UAS elements. These UAS elements are sites
for
transcriptional activation by the GAL4-VP 16 fusion protein. The GAL4-VP 16
fusion
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-90-
protein in turn is expressed under control of a tissue-specific promoter. The
UAS-CCP
construct and the GAL4-VP16 construct are combined by co-transformation of
both
constructs, subsequent transformation of single constructs or by sexual cross
of lines that
contain the single constructs. The advantage of this two-component system is
that a wide
array of tissue-specific expression patterns can be generated for a specific
transgene, by
simply crossing selected parent lines expressing the UAS-CCP construct with
various
tissue-specific GAL4-VP 16 lines. A tissue-specific promoter/CCP combination
that gives
a desired phenotype can subsequently be recloned in a single expression
vector, to avoid
stacking of transgene constructs in commercial lines.
Primary transformants are characterized by Northern and Western blotting using
1-
4 week old plantlets. Expression levels were compared with those of non-
transformed
(control) plants.
EXAMPLE 4: DOWNREGULATION OF TARGET CCP GENES IN
TRANSGENIC PLANTS
Plant genes can be specifically downregulated by antisense and co-suppression
technologies. These technologies are based on the synthesis of antisense
transcripts,
complementary to the mRNA of a given CCP gene. There are several methods
described in
literature, that increase the efficiency of this downregulation, for example
to express the
sense strand with introduced inverted repeats, rather than the antisense
strand. The
constructs for downregulation of target genes are made similarly as those for
expression of
recombinant proteins, i.e., they are fused to promoter sequences and
transcription
termination sequences (see example 3). Promoters used for this purpose are
constitutive
promoters as well as tissue-specific promoters.
EXAMPLE 5: AGROBACTERIUM-MEDIATED RICE TRANSFORMATION
Mature dry seeds of the rice japonica cultivars Nipponbare or Taipei 309 are
dehusked, sterilised and germinated on a medium containing 2,4-D (2,4-
dichlorophenoxyacetic acid). After incubation in the dark for four weeks,
embryogenic,
scutellum-derived calli are excised and propagated on the same medium.
Selected
embryogenic calluses are then co-cultivated with Agrobacterium. Widely used
Agrobacterium strains such as LBA4404 or C58 harbouring binary T-DNA vectors
can be
used. The hpt gene in combination with hygromycin is suitable as a selectable
marker
system but other systems can be used. Co-cultivated callus is grown on 2,4-D-
containing
medium for 4 to 5 weeks in the dark in the presence of a suitable
concentration of the
selective agent. During this period, rapidly growing resistant callus islands
develop. After
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-91-
transfer of this material to a medium with a reduced concentration of 2,4-D
and incubation
in the light, the embryogenic potential is released and shoots develop in the
next four to
five weeks. Shoots are excised from the callus and incubated for one week on
an auxin-
containing medium from which they can be transferred to the soil. Hardened
shoots are
grown under high humidity and short days in a phytotron. Seeds can be
harvested three to
five months after transplanting. The method yields single locus transformants
at a rate of
over 50 % (Aldemita and Hodges (1996) Planta 199:612-617; Chan et at (1993)
Plant
Mol. Biol. 22: 491-506 ; Hiei et at (1994) Plant J. 6 :271-282).
1 o EXAMPLE 6: EXPRESSION OF RECOMBINANT CCP PROTEINS IN
BACTERIAL CELLS
In this example, the CCP molecules of the present invention are expressed as a
recombinant glutathione-S-transferase (GST) fusion polypeptide in E. coli and
the fusion
polypeptide is isolated and characterized. Specifically, CCP molecules are
fused to GST
and this fusion polypeptide is expressed in E. coli, e.g., strain PEB 199.
Expression of the
GST-CCP fusion protein in PEB 199 is induced with IPTG. The recombinant fusion
polypeptide is purified from crude bacterial lysates of the induced PEB 199
strain by
affinity chromatography on glutathione beads. Using polyacrylamide gel
electrophoretic
analysis of the polypeptide purified from the bacterial lysates, the molecular
weight of the
resultant fusion polypeptide is determined.
EXAMPLE 7: EXPRESSION OF RECOMBINANT CCP PROTEINS
IN COS CELLS
To express the CCP gene of the present invention in COS cells, the peDNA/Amp
vector by Invitrogen Corporation (San Diego, CA) is used. This vector contains
an SV40
origin of replication, an ampicillin resistance gene, an E. coli replication
origin, a CMV
promoter followed by a polylinker region, and an SV40 intron and
polyadenylation site. A
DNA fragment encoding the entire CCP protein and an HA tag (Wilson et al.
(1984) Cell
37:767) or a FLAG tag fused in-frame to its 3' end of the fragment is cloned
into the
polylinker region of the vector, thereby placing the expression of the
recombinant protein
under the control of the CMV promoter.
To construct the plasmid, the CCP DNA sequence is amplified by PCR using two
primers. The 5' primer contains the restriction site of interest followed by
approximately
twenty nucleotides of the CCP coding sequence starting from the initiation
codon; the 3'
end sequence contains complementary sequences to the other restriction site of
interest, a
translation stop codon, the HA tag or FLAG tag and the last 20 nucleotides of
the CCP
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-92-
coding sequence. The PCR amplified fragment and the pCDNA/Amp vector are
digested
with the appropriate restriction enzymes and the vector is dephosphorylated
using the
CIAP enzyme (New England Biolabs, Beverly, MA). Preferably the two restriction
sites
chosen are different so that the Kinase and/or Phosphatase gene is inserted in
the correct
orientation. The ligation mixture is transformed into E. coli cells (strains
HB101, DH5a,
SURE, available from Stratagene Cloning Systems, La Jolla, CA, can be used),
the
transformed culture is plated on ampicillin media plates, and resistant
colonies are
selected. Plasmid DNA is isolated from transformants and examined by
restriction
analysis for the presence of the correct fragment.
COS cells are subsequently transfected with the CCP-pCDNA/Amp plasmid DNA
using the calcium phosphate or calcium chloride co-precipitation methods, DEAE-
dextran-
mediated transfection, lipofection, or electroporation. Other suitable methods
for
transfecting host cells can be found in Sambrook, J., Fritsh, E. F., and
Maniatis, T.
Molecular Cloning: A Laboratory Manual. 2nd, ed., Cold Spring Harbor
Laboratory,
Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989. The
expression of
the CCP polypeptide is detected by radiolabelling (35S-methionine or 35S-
cysteine
available from NEN, Boston, MA, can be used) and immunoprecipitation (Harlow,
E. and
Lane, D. Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory Press,
Cold
Spring Harbor, NY, 1988) using an HA specific monoclonal antibody. Briefly,
the cells
are labelled for 8 hours with 35S-methionine (or 35S-cysteine). The culture
media are then
collected and the cells are lysed using detergents (RIPA buffer, 150 mM NaCl,
1% NP-40,
0.1% SDS, 0.5% DOC, 50 mM Tris, pH 7.5). Both the cell lysate and the culture
media
are precipitated with an HA specific monoclonal antibody. Precipitated
polypeptides are
then analyzed by SDS-PAGE.
Alternatively, DNA containing the Kinase and/or Phosphatase coding sequence is
cloned directly into the polylinker of the pCDNA/Amp vector using the
appropriate
restriction sites. The resulting plasmid is transfected into COS cells in the
manner
described above, and the expression of the CCP polypeptide is detected by
radiolabelling
and immunoprecipitation using a CCP specific monoclonal antibody.
EXAMPLE 8: IN VITRO PHOSPHORYLATION OF CDC2bDN-IC26M BY
PLANT CDKs.
The CDC2bDN-IC26M coding region (SEQ ID NO:4) was amplified by PCR with
Pfu polymerise (Stratagene, La Jolla, CA). The PCR product was subcloned into
pET19b
(Novagen, Madison, WI), to obtain CDC2bDN-IC26MpET19b. The CDC2bDN-IC26M
gene is located downstream of a T71ac promoter, in frame with a sequence
encoding a 10-
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-93-
histidine tag followed by an enterokinase recognition site. Escherichia coli
BL21(DE3)
cells (Novagen) containing the CDC2bDN-IC26MpET19b plasmid were grown at 37 C
in
M9 medium (Sambrook and Russel, Molecular Cloning, A Laboratory Manual, 3`d
Edition,
CSHL Press, CSH New York, 2001), supplemented with 100 g/ml of ampicillin, to
obtain a cell density corresponding to an A600 of 0.6. Subsequently,
expression of the
CDC2bDN-IC26M gene was induced by addition of 0.4 mM isopropyl R-D-
thiogalactoside, and culture was continued for 4 h at 30 C.
Cells were collected in lysis buffer containing 50 mM sodium phosphate buffer,
pH
8.0, 300 mM NaCl, 0.1% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride
(PMSF)
and were lysed on ice by sonication. The extract was clarified by
centrifugation for 20
minutes at 20,000 x g. The crude extract was loaded at 4 C on a nickel-
nitrilotriacetic
acid-agarose affinity resin (Qiagen), and protein fractionation was performed
according to
the manufacturer's instructions. The fractions containing the CDC2bDN-1C26M
fusion
protein were pooled.
CDC2bDN-IC26M kinase assays were performed with CDK complexes purified
from total plant (Arabidopsis seedlings) protein extracts by p13s `'-Sepharose
affinity
binding according to Azzi et al. (Eur. J. Biochem. 203: 353-360). Briefly,
p13s `1 was
purified from an overproducing E. coli strain by chromatography in Sephacryl
S2000, and
conjugated to CNBr-activated Sepharose 4B (Pharmacia) according to the
manufacturer's
instructions. Total plant protein extracts (300 g) were incubated with 50 l
50% (v/v)
p13s `'-Sepharose beads for 2h at 4 C. The washed beads were combined with 30
1 kinase
buffer containing -1 mg/ml CDC2bDN-IC26M, 150 mM ATP and 1 Ci of [-32P]ATP
(Amersham). After 20 minutes of incubation at 30 C, samples were analysed by
SDS-
PAGE and autoradiographed.
As shown in Figure 48, the purified CDC2bDN-1C26M protein is phosphorylated
by CDKs in vitro.
EXAMPLE 9: PCR AMPLIFICATION OF AtDPb
Based on available sequence data of putative plant DP-related partial clones
from
the databank (soybean DP (A1939068), tomato DP(AW217514), and cotton DP
(AI731675)), three oligonucleotides, corresponding to the most conserved part
of the
DNA-binding and E2F heterodimerization domains (MKVCEKV, SEQ ID NO:240;
LNVLMAMD, SEQ ID NO:241 and FNSTPFEL, SEQ ID NO:242), were synthesized and
designated A (ATAGAATTCATGAAAGTTTGTGAAAAGGTG, SEQ ID NO:243), B
(ATAGAATTCCTGAATGTTCTCATGGCAATGGAT, SEQ ID NO:244) and C
(ATAGGATCCCAGCTCAAAAGGAGTGCTATTGAA, SEQ ID NO:245), respectively.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-94-
PCR was performed on an Arabidopsis/yeast two-hybrid suspension culture cDNA
library. The PCR products were purified, digested with EcoRl and BamHI, and
ligated into
pCR-XL-TOPO vector (Invitrogen). The cloned inserts were sequenced by double-
stranded dideoxy sequencing.
EXAMPLE 10; CONSTRUCTION OF AtDP and AtE2F MUTANTS, IN VITRO
TRANSCRIPTION-TRANSLATION SYSTEM AND
IMMUNOPRECIPITATION
Influenza hemagglutinin (HA)-tagged versions of the wild-type and mutant
AtE2Fa
and AtE2Fb were constructed by cloning into the pSK plasmid (Stratagene)
containing the
HA-tag (SEQ ID NO:202). The AtE2F mutants, namely AtE2Fa 1-420 (SEQ ID
NO:217),
AtE2Fa 162-485 (SEQ ID NO:218), and AtE2Fb 1-385 (SEQ ID NO:206), were
obtained
by PCR and cloned into the EcoRI and BarHI sites of HA-pSK. The c-myc (SEQ ID
NO:200)-tagged versions of wild-type and AtDP mutants (AtDPa 1-292, SEQ ID NO:
114;
AtDPa 121-292, SEQ ID NO:21 1; AtDPa 1-142, SEQ ID NO:208; AtDPa 172-292, SEQ
ID NO:213; AtDPa 121-213, SEQ ID NO:212; and AtDPb 1-385, SEQ ID NO:127;
AtDPb 182-385, SEQ ID NO:216; AtDPb 1-263, SEQ ID NO:223; AtDPb 1-193, SEQ
ID NO:214; and AtDPb 182-263, SEQ ID NO:215) were generated by PCR and cloned
into the EcoRl and Pstl sites of the pBluescript plasmid (Stratagene)
containing a double c-
myc tag. All cloning steps were carried out according to standard procedures,
and the
reading frames were verified by direct sequencing.
In vitro transcription and translation experiments were performed using the
TNT
T7-coupled wheat germ extract kit (Promega) primed with appropriate plasmids
for 90 min
at 30 C. For immunoprecipitation, 10 l of the total in vitro translated
extract (50 l) was
diluted at 1:5 in Nonidet P40 buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1%
Nonidet P40,
1 mM phenylmethylsulfonyl fluoride, 10 tg/ml leupeptin/aprotinin/pepstatin)
and
incubated for 2 h at 4 C with anti-c-myc (9E10; BabCo) or anti-HA (16B12;
BabCo)
antibodies. Protein-A-Sepharose (40 l 25% (v/v)) was added and incubated for
1 h at 4 C,
then the beads were washed four times with Nonidet P40 buffer. Immune
complexes were
eluted with 10 l 2 U sodium dodecyl sulfate (SDS) sample buffer and analyzed
by 10% or
15% SDS-PAGE and by autoradiography.
An overview of the AtDP and AtE2F fragments and their SEQ ID NOs is given in
Table 4.
CA 02768046 2012-02-07
WO 01/85946 PCT/1B01/01307
-95-
TABLE IV
CCP or partial CCP SEQ ID NO SEQ ID NO
amino acid DNA
sequence sequence
AtE2Fa 226-356 205 228
AtE2Fb 1-385 206
AtE2Fb 1-127 207
AtDPa 1-142 208
AtDPa 42-142 209
AtDPa 42-292 210
AtDPa 121-292 211 229
AtDPa 121-213 212
AtDPa 172-292 213
AtDPb 1-193 214
AtDPb 182-263 215 230
AtDPb 182-385 216 231
AtE2Fa 1-420 217
AtE2Fa 162-485 218
AtE2Fa 1-38 219
AtDPa 1-214 220 239
AtDPa 143-292 221 232
AtDPa 143-213 222 233
AtDPb 1-263 223 234
AtE2Fa 232-282 224 235
AtE2Fa 232-352 225 236
AtE2Fb 194-243 226 237
AtE2Fb 194-311 227 238
EXAMPLE 11: IN VITRO INTERACTION BETWEEN AtDPs, AtE2Fs AND
MUTANTS THEREOF ILLUSTRATED BY
IMMUNOPRECIPITATION EXPERIMENTS
The AtDPa and AtDPb can efficiently interact in vitro with AtE2Fa and AtE2Fb.
As a first step in comparing the biochemical properties of AtDPa and AtDPb,
the ability of
1o these molecules to heterodimerize with AtE2Fa and AtE2Fb was tested. For
this purpose,
the coupled in vitro transcription-translation system was used in which the c-
myc-tagged
AtDPa or AtDPb was co-expressed with the HA-tagged AtE2Fa or AtE2Fb. One part
of
each sample was resolved by SDS-PAGE (Figures 50 and 51, panels A), while
another part
was subjected to immunoprecipitation with monoclonal anti-c-myc antibodies
(Figures 50
and 51, panels B). In the absence of DP proteins, no AtE2F2a or AtE2F2b was
precipitated by the anti-c-myc antibodies (Figure 51, panel B, lane 1).
However, both HA-
CA 02768046 2012-02-07
WO 01/85946 PCT/1B01/01307
-96-
AtE2F proteins co-precipitated reproducibly with c-inyc-tagged AtDPa (Figure
50, panel
B, lanes 1 and 2) and AtDPb (Figure 51, panel B, lanes 3 and 4). Identical
results were
obtained in a reciprocal experiment with anti-HA monoclonal antibodies. These
data
revealed that both Arabidopsis DP-related proteins interacted in vitro with
the different
Arabidopsis E2F-related proteins.
The conserved dimerization domain of the AtE2Fs seemed to be important for the
interaction with the AtDPs, because mutational analysis showed that deletion
neither of the
N-terminal extension nor the C-terminal part of AtE2Fa and AtE2Fb impaired the
interaction with the DPs (Figures 50 and 51, panels B). Similar results were
obtained by
two-hybrid analysis (see Table 5 of Example 12). To test whether the
structural
requirements for heterodimerization of the AtDPs were similar to those of
their animal
homologs, several deletion mutants of AtDPa and AtDPb were constructed (for a
schematic illustration, see Figures 52 and 53), tagged with the c-myc epitope
(Figures 54
and 55, panels A). The interactions between the mutant AtDPs and AtE2Fb were
analyzed
in immunoprecipitation experiments with the specific anti-HA or anti-c-myc
antibodies
(Figures A6 and A7, panels B and C, respectively). As shown in Figures 54 and
55,
mutant AtDP proteins with deleted DNA-binding domain could bind sufficiently
to the co-
translated HA-AtE2Fb proteins (Figure 54, panel C, lane 2; and Figure 55,
panel C, lane
2). No detectable interaction was found between the AtE2Fb protein and mutant
DP
proteins containing the complete DNA-binding domain, but lacking the putative
dimerization domain (Figure 54, panel C, lane 3; Figure 55, panel C, lane 4).
Thus, the N-
terminal part of both AtDP proteins, including the conserved DNA-binding
domain, was
not sufficient for the in vitro interaction to occur. In contrast, a mutant
form of AtDPb
(amino acids 1-263; SEQ ID NO:223) could bind to AtE2Fb (Figure 55, panel C,
lane 3),
indicating that the region of AtDPb between amino acids 182 and 263 was
required for
interaction with AtE2Fb.
To confirm this hypothesis, a deletion mutant of AtDPb (182-263, SEQ ID
NO:215) was constructed and, as expected, it could bind to AtE2Fb (Figure 56).
The
requirement for the homologous dimerization domain of AtDPa for the
interaction with
3o AtE2Fb was supported by a binding assay in which the mutant AtDPa 172-292
(SEQ ID
NO:213), with the N-terminal part of the dimerization domain deleted, failed
to bind to
AtE2Fb (Figure 54, panels B and C, lanes 4). However, when the E2F-binding
activity of
the predicted dimerization domain of the AtDPa (amino acid positions 121-213,
SEQ ID
NO:212) was tested, no interaction could be detected between this region and
the AtE2Fb
protein (Figure 54, panel B, lane 5). These data indicate that other carboxyl-
terminal
regions of AtDPa are required for the stable interaction with AtE2Fb.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-97-
EXAMPLE 12: YEAST TWO-HYBRID EXPERIMENTS FOR SHOWING
INTERACTION BETWEEN DP AND E2F MUTANTS
For library screening, vectors and strains (HF7c) were provided with the
Matchmaker two-hybrid system (Clontech). The dimerization and DNA-binding
domains
of the AtE2Fa (amino acids 226-356; SEQ ID NO:205) were amplified by
polymerase
chain reaction (PCR) and subcloned in-frame with the GAL4 DNA-binding domain
of
pGBT9 (Clontech) to create the bait plasmid pGBTE2Fa226-356. Screens were
performed
as described previously (De Veylder et al. 1999; Planta 208, 453-462). A
second library
screening was performed with the AtE2Fb construct (pGBTE2Fb-Rb) lacking the Rb-
binding domain (amino acids 1-385; SEQ ID NO:206). Plasmids from interacting
clones
were isolated and sequenced.
For the yeast two-hybrid interaction experiments, a number of yeast two-hybrid
prey (in pAD-GAL424) plasmids were created by PCR amplification of fragments
from
the AtDPa (DPa 1-292, SEQ ID NO: 114; DPa 1-142, SEQ ID NO:208; DPa 42-142,
SEQ
ID NO:209; DPa 42-292, SEQ ID NO:210; DPa 121-292, SEQ ID NO:21 1; DPa 121-
213,
SEQ ID NO:212; and DPa 172-292, SEQ ID NO:213) and AtDPb (DPb 1-385, SEQ ID
NO:127; DPb 1-193, SEQ ID NO:214; DPb 182-263, SEQ ID NO:215; and DPb 182-385,
SEQ ID NO:216 ) genes and confirmed by sequencing. Different combinations
between
bait (pGBTE2Fa226-356, pGBTE2Fb-Rb, or pGBTE2Fb 1-127, SEQ ID NO:207) and
prey constructs were transformed into yeast cells and assayed for their
ability to grow on
His- minimal media after 3 days of incubation at 30 C. Bait plasmids co-
transformed with
empty pAD-GAL424 and prey plasmids co-transformed with empty pGBT9 were
assessed
along as controls for the specificity of the interaction.
An overview of the AtDP and AtE2F fragments and their SEQ ID NOs is given in
Table 4.
The results obtained were confirmed by two-hybrid interaction analysis.
pGBTE2Fa226-356 and pGBTE2Fb-Rb were co-transformed in an appropriate yeast
reporter stain with a plasmid producing the full-length AtDPa or AtDPb protein
fused to
the GAL4 transactivation domain. The specific reconstitution of GAL4-dependent
gene
expression measured as the ability to grow in the absence of histidine
confirms the
interaction between the two DP and E2F proteins (Table 5).
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
-98-
p.,
z
a 1 1 1 1 1
A a~
S "' + + Ce
N N
W N E
MN + + z z z
00 En
+ + z z z
00 O
z.
M
0.a H H H }+
z z z 40,
0
A a)
N 3
fI U
z z z
Ar
O
M V
N z z z N
~~+ + z z z y
b A~
N
N + z z z~.,b A W O
z z z
v Av 3 ~
~~ H H H M r4
X z z O
C A ., O a,
N n N c"~d -~-
Q + + z
O. Q - O
fi
W a Er ca M F E" n r E" N F E, y
r~ xi =, paw Pa~,N q ca c,Aa aoo C9 ~. .,.
R C7 N N U' N .G C7 N "." C7 0. N C' p, Mat; N
Q Q p1 GW N CLW R G.W .i 0.A dA .. 0.f c}
F a
CA 02768046 2012-02-07
WO 01/85946 PCT/IBOI/01307
99
EXAMPLE 13: RNA ISOLATION AND REVERSE TRANSCRIPTION-(RT)-
PCR ANALYSIS OF AtDP And AtE2F EXPRESSION
A. thaliana (L.) Heynh. cell suspension cultures were maintained as described
previously (Glab et al. 1994, FEBS Lett. 17, 207-211). The cells were
partially
synchronized by the consecutive addition of aphidicolin (5 g/ml) and
propyzamide (1.54
tg/ml). The aphidicolin block was left for 24 hours. The cells were washed for
1 hour in
B5 medium before the addition of propyzamide. Samples were taken at the end of
the 24
hour aphidicolin block, at the end of a 1 hour washing step, and at 1, 2, 3,
and 4 hours after
the addition of propyzamide to the culture medium. Total RNA was isolated from
the
Arabidopsis cell suspension culture according to Magyar et al. (1997), Plant
Cell 9, 223-
235, and with the Triazol reagent (Gibco/BRL) from different organs. Semi-
quantitative
RT-PCR amplification was carried out on reverse-transcribed mRNA, ensuring
that the
amount of amplified product stayed in linear proportion to the initial
template present in
the reaction. 10 l from the PCR was transferred onto Hybond-N/ membrane,
hybridized
to fluorescein-labeled gene-specific probes (Gene-Images random prime labeling
module;
Amersham Pharmacia Bio-tech), detected with the CDP-Star detection module
(Amersham), and visualized by short exposure to Kodak X-OMAT autoradiography
film.
The following primer pairs (forward and reverse) were used for the
amplification:
5' -ATAGAATTCATGTCCGGTGTCGTACGA-3' (SEQ ID NO:249, EcoRl site
underlined) and 5'-ATAGGATCCCACCTCCAATGTTTCTGCAGC-3' (SEQ ID
NO:250, BainHI site underlined) for AtE2Fa (GenBank accession number
AJ294533);
5' -ATAGAATTCGAGAAGAAAGGGCAAT CAAGA-3' (SEQ ID NO:251, RcoRI site
underlined) and 5'-ATACTGCAGAGAAATCTCGATTTCGACTAC-3' (SEQ ID
NO:252, Pstl site underlined) for AtDPa (GenBank accession number AJ29453 1);
5' -GCCACTCTCATAGGGTTCTC CATCG-3' (SEQ ID NO:253) and 5'-
GGCATGCCTCCAAGATCCTTGAAGT-3' (SEQ ID NO:254) for Arath;CDKA;1
(Genbank accesion number X57839); 5'-GGGTCTTGGTCGTTTTACTGTT-3' (SEQ ID
NO:255) and 5'-CCAAGACGATGACAACAGATACAGC-3' (SEQ ID NO:256) for
Arath;CDKB 1; 1 (Genbank accession number X57840);
5'-ATAAACTAAATCTTCGCTGAA- 3' (SEQ ID NO:257) and 5'-
CAAACGCGGATCTGAAAAACT-3' (SEQ ID NO:258) for histone H4 (Genbank
accession number M17132); 5' -TCTCTCTTCCAAATCTCC-3' (SEQ ID NO:259) and
5'-AAGTCTCT CACTTTCTCACT-3' (SEQ ID NO:260) for ROC5 (AtCYP1, GenBank
accesion number U072676) (Chou and Gasser 1997, Plant Mol. Biol. 35, 873-892);
5'- CTAAGCTCTCAAGATCAAAGGCTTA-3' (SEQ ID NO:261) and 5'-TTAACATTG
CAAAGAGTTTCAAGGT-3' (SEQ ID NO:262) for actin 2 gene (GenBank accession
number U41998) (An et al. 1996, Plant J. 10, 107-121).
CA 02768046 2012-02-07
WO 01 /85946 PCT/IB01 /01307
-100-
EXAMPLE 14: THE AtDPa And The AtE2Fa GENES ARE CO-EXPRESSED
IN A CELL CYCLE PHASE-DEPENDENT MANNER
The identification of the AtDPa in a yeast two-hybrid screen as a gene
encoding an
AtE2Fa-associating protein indicated that it might act cooperatively in the
plant cells as a
functional heterodimer: To strengthen this hypothesis, we investigated whether
both genes
were co-regulated at the transcriptional level. Tissue-specific expression
analysis revealed
that both genes were clearly up-regulated in flowers and were very strongly
transcribed in
actively dividing cell suspension cultures (Figure 57). Expression in these
tissues could be
a sign for the correlation between the actual proliferation activity of a
given tissue and the
transcript accumulation, as can be seen from the Arath;CDKB 1;1 gene. AtDPa
transcripts
were also detectable in leaf and, to a lesser extent, in root and stem
tissues, whereas
AtE2Fa transcripts were virtually undetectable in roots and stem with only
slight levels of
expression in leaf tissues. Cell cycle phase-dependent gene transcription was
studied using
an Arabidopsis cell suspension that was partially synchronized by the
sequential treatment
with aphidicolin and propyzamide. The Arabidopsis histone H4 and the
Arath;CDKB1;1
gene were included to monitor the cell cycle progression (Figure 58) (Chaubet
et al. 1996,
Plant J. 10, 425-435; Segers et al. 1996, Plant J. 10, 601-612). Bearing in
mind the partial
synchronization of the culture, it can be observed that histone H4 transcript
levels peaked
immediately after the removal of the inhibitor and decrease gradually
thereafter (Figure
58). The opposite expression pattern could be observed for the Arath;CDKB1;1
gene,
illustrating that cells entered the G2-M phases with partial synchrony. Within
this
experimental setting, the AtDPa and the AtE2Fa genes show a very similar
expression
pattern. Both exhibit higher transcript accumulation before the peak of
histone H4 gene
expression and quickly decay in the following samples (Figure 58). The
similarity in the
expression patterns of Arabidopsis AtDPa and AtE2Fa supports the possibility
that they act
cooperatively as a heterodimer during the S phase.
EXAMPLE 15: TRANSFORMATION OF ARABIDOPSIS THALLANA WITH
CaMV35S::DPa
Arabidopis plants were transformed (using the in planta flower dip method;
Clough
and Bent, Plant J. 16:735-743, 1998) with a construct containing the DPa gene
under the
control of the CaMV 35S promoter. The lines were molecularly analysed by
northern
blotting. As can be seen in Figure 59, all lines showed increased DPa levels
in comparison
with the untransformed control. Generally, two classes of lines were observed:
weakly
expressing (e.g., 16) and strongly expressing (e.g., 23) lines (see Figure
59). The plants are
subsequently analyzed for phenotypic alterations as described herein.
CA 02768046 2012-02-07
WO 01/85946 PCT/IBO1/01307
-101-
Equivalents
Those skilled in the art will recognize, or be able to ascertain using no more
than
routine experimentation, many equivalents to the specific embodiments of the
invention
described herein. Such equivalents are intended to be encompassed by the
following
claims.
CA 02768046 2012-02-07
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPREND PLUS D'UN TOME.
CECI EST LE TOME DE _2
NOTE: Pour les tomes additionels, veillez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME OF
NOTE: For additional volumes please contact the Canadian Patent Office.