Note: Descriptions are shown in the official language in which they were submitted.
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 1 -
CONSTRUCTS, VECTORS AND CYANOBACTERIA FOR THE SYNTHESIS
OF FATTY ALCOHOLS, AND METHODS FOR PRODUCING FATTY ALCOHOLS
IN CYANOBACTERIA
Technical field of the invention
The present invention relates to a construct for the
synthesis of fatty alcohols in cyanobacteria, a vector
comprising the construct, a cyanobacterium comprising the
construct or transformed by the vector, and a method for
producing fatty alcohols in cyanobacteria.
Background of the invention
Currently, the sustainable development of economy and
society is increasingly restricted by energy and
environment related problems. Renewable biofuels are
considered as an effective way to solve said problems.
Technical routes for the production of bio-ethanol are
relatively well developed. However, ethanol as a fuel has
some drawbacks, namely: (1) low energy density; (2) high
volatility; (3) problems caused by its high solubility in
water, such as the increased toxicity for microorganisms
during fermentation, the high cost for the removal of water
phase during distillation separation process and the
corrosion of pipelines during transportation.
It would be desirable for a biofuel to have properties
such as high energy density, low moisture absorption, low
volatility, and/or compatibility with existing engines and
transport facilities.
Recently, biofuel components prepared from high
quality fatty acids, such as long chain fatty alcohols and
long chain biologic hydrocarbons are drawing more and more
attention.
S.K. Lee et. al. in their article titled "Metabolic
engineering of microorganisms for biofuels production:
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
2 -
from bugs to synthetic biology to fuels", published in
Current Opinion in Biotechnology, volume 19, issue 6,
December 2008 pages 556 to 563 provide a review concerning
the status and prospective of such biofuels.
Steen, E . J. , et . al . in their article titled "Microbial
production of fatty-acid-derived fuels and chemicals from
plant biomass", published in Nature , volume 463, 28 January
2010, pages 559 to 562 describe the engineering of
Escherichia coli to produce structurally tailored fatty
esters (biodiesel), fatty alcohols, and waxes directly
from simple sugars.
W02007/136762 describes the production of fatty acid
derivatives by genetically engineered microorganisms such
as E. coli and Saccharomyces cerevisiae. It is indicated
that the fatty acid derivatives can be useful as biofuels
and speciality chemicals.
At present, the microorganism systems used for studying
biofuels are primarily heterotrophic microorganisms
represented by E. coli and Saccharomyces cerevisiae.
S.A. Angermayr et. al. in their article titled "Energy
biotechnology with cyanobacteria", published in Current
Opinion in Biotechnology, volume 20, issue 3, June 2009,
pages 257 to 263, describes the possibility to fortify
photosynthetic organisms with the ability to produce
biofuels. The article describes an approach to redirect
cyanobacterial intermediary metabolism by channeling
intermediates into fermentative metabolic pathways.
J. Dexter et. al. in their article titled "Metabolic
engineering of cyanobacteria for ethanol production",
published in Energy & Environmental Science, volume 2,
issue 8, 2009, pages 857 to 864 describe the conversion
from solar energy to bioethanol (yield of 5.2 mmol/OD730/L/d)
by co-expressing the genes of pyruvate decarboxylase and
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
3 -
ethanol dehydrogenase derived from Zymomonas mobilis in
Synechocystis sp. PCC6803.
Pengcheng Fu, in his article titled "Genome-scale
modeling of Synechocystis sp. PCC 6803 and prediction of
pathway insertion", published in the Journal of Chemical
Technology & Biotechnology volume 84, issue 4, April 2009,
pages 473 to 483, describes a reconstruction of a
genome-scale Synechocystis sp. PCC 6803 metabolic network,
including 633 genes, 704 metabolites and 831 metabolic
reactions. Heterotrophic, photoautotrophic and
mixotrophic growth conditions were simulated and the
Synechocystis model was used for in silico predictions for
the ethanol fermentation pathway.
P. Lindberg et al. in their article titled "Engineering
a platform for photosynthetic isoprene production in
cyanobacteria, using Synechocystis as the model organism",
published in Metab Eng. volume 12, issue 1, October 2009,
pages 70-79 describe the genetic engineering of the
cyanobacterium synechocystis, conferring the ability to
generate volatile isoprene hydrocarbons from CO(2) and
H(2)O. Heterologous expression of the Pueraria montana
(kudzu) isoprene synthase (IspS) gene in Synechocystis
enabled photosynthetic isoprene generation in these
cyanobacteria.
S. Atsumi et al., in their article titled "Direct
photosynthetic recycling of carbon dioxide to
isobutyraldehyde", published in Nature Biotechnology, vol
27, pages 1177 to 1180 describes the use of genetically
engineered Synechococcus elongatus PCC7942 to produce
isobutyraldehyde and isobutanol directly from C02-
Productivity was increased by overexpression of ribulose
1,5-bisphosphate carboxylase/oxygenase (Rubisco).
X. Liu et al., in their article titled "Production and
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
4 -
secretion of fatty acids in genetically engineered
cyanobacteria." published in Proceedings of the National
Academy of Sciences of the USA, 29 March 2010, describe
the production and secretion of free fatty acids in
genetically modified Synechocystis sp. PCC6803.
It would be an advancement in the art to provide a method
for producing fatty alcohols and/or long chain biologic
hydrocarbons in cyanobacteria.
It may further be advantageous to construct a route for
synthesizing fatty alcohols in cyanobacteria so as to
achieve the in vivo synthesis of fatty alcohols in
microorganisms.
In addition, it would be an advancement in the art to
provide a method that allows for an improved exogenous gene
expression efficiency in cyanobacteria. Such a method
allowing highly efficient expression of exogenous genes
within cyanobacteria could be very helpful in any method
for producing fatty alcohols and/or long chain biologic
hydrocarbons in cyanobacteria.
Summary of the invention
The inventors of the present invention, for the first
time, successfully produced fatty alcohols in
cyanobacteria.
The present invention accordingly provides a construct
used for synthesizing fatty alcohols in cyanobacteria,
comprising a promoter having activity in cyanobacteria and
a fatty acyl-CoA reductase gene under the control of the
promoter.
In addition, the present invention provides a vector
comprising such a construct; a cyanobacterium comprising
the construct or transformed by the vector; and a method
for producing fatty alcohols and/or biologic hydrocarbons
in a cyanobacterium, comprising culturing a cyanobacterium
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
-
comprising the construct or transformed by the vector under
conditions suitable for the synthesis of fatty alcohols;
and extracting the desired fatty alcohols from the obtained
culture.
5 Further the present invention provides a method of
expression of exogenous genes via the use of Synechocystis
sp. 6803, the main processes of which are:
a) separately constructing a Synechocystis sp. 6803
rbcL promoter (Prbcl, 1.3 kb) and a rbc terminator sequence
(Trbc, 02 kb), a 2kb spectinomycin resistant marker gene
Q and a reporter gene lacz on a Synechocystis sp. 6803 genome
integrative plasmid platform, yielding a platform plasmid
pFQ20 which already contains the reporter gene for use in
the expression of exogenous genes and which allows genetic
integration using Synechocystis sp. 6803;
b) Assay of the effectiveness of exogenous gene
expression by the platform via submitting the transformed
pFQ20 Synechocystis sp. 6803 algal strain of which the
transformant was obtained after spectinomycin resistance
screening, to PCR genotype assay using a specific primer,
which allows detection of R- galactosidase activity of the
transformant expression platform and the exogenous gene
engineered strain of Synechocystis sp. 6803, in order to
carry out assay of the expression performance of the
Synechocystis sp. 6803 exogenous gene expression platform.
Brief description of the drawings
The invention has been illustrated by the non-limiting
following figures:
Fig. 1 represents the basic structure of plasmid pFQ9R,
in which the Omega fragment of spectinomycin resistance
gene, the Prbc promoter and the Trbc terminator are between
the upstream and downstream fragments of s1r0168 gene of
Synechocystis sp. PCC6803; and XbaI and Smal restriction
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
6 -
sites are between the promoter and the terminator.
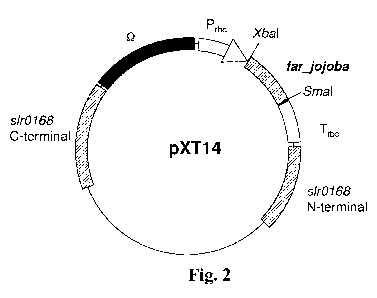
Fig. 2 represents the basic structure of plasmid pXT14,
which is obtained by cloning far gene (far jojoba) (SEQ
ID NO: 1) from Simmondsia chinensis into the plasmid pFQ9R.
Fig. 3 represents the basic structure of plasmid pXT37a,
in which the Omega fragment of spectinomycin resistance
gene, the PpetE promoter and the 1acZ gene are between the
upstream and downstream fragments of s1r0168 gene of
Synechocystis sp. PCC6803; and NdeI and EcoRI restriction
sites are at the two ends of the 1acZ gene.
Fig. 4 represents the basic structure of plasmid pXT37b,
which is similar to plasmid pXT37a, except that the
insertion direction of the fragment consisting of Omega
fragment, PpetE promoter and 1acZ gene is contrary to that
in plasmid pXT37a.
Fig. 5 represents the basic structure of plasmid pXT34,
which is obtained by cloning at3g11980 gene (SEQ ID NO:
2) from Arabidopsis thaliana into the plasmid pXT37a,
wherein the at3g11980 gene is located downstream of the
PpetE promoter .
Fig. 6 represents the basic structure of plasmid pXT51,
which is obtained by cloning far gene (far jojoba) (SEQ
ID NO: 1) from Simmondsia chinensis into the plasmid pXT37b,
wherein the far gene is located downstream of the PpetE
promoter.
Fig. 7 represents the basic structure of plasmid pLY2,
which is obtained by inserting the Omega fragment of
spectinomycin resistance gene between the upstream and
downstream fragments of s1r0168 gene of Synechocystis sp.
PCC6803, and cloning the entire construct into the vector
pUC9.
Fig. 8 illustrates the production of fatty alcohols in
the cells of the genetically engineered strain Syn-LY2
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
7 -
after 8 days of culturing (the determination results of
GC-MS), wherein C15-OH represents 1-pentadecanol (used as
internal standard), C16-OH represents 1-hexadecanol, and
C18-OH represents 1-octadecanol.
Fig. 9 illustrates the production of fatty alcohols in
the cells of the genetically engineered strain Syn-XT14
after 8 days of culturing (the determination results of
GC-MS), wherein C15-OH represents 1-pentadecanol (used as
internal standard), C16-OH represents 1-hexadecanol, and
C18-OH represents 1-octadecanol.
Fig. 10 illustrates the production of fatty alcohols
in the cells of the genetically engineered strain Syn-XT34
after 8 days of culturing (the determination results of
GC-MS), wherein C15-OH represents 1-pentadecanol (used as
internal standard), C16-OH represents 1-hexadecanol, and
C18-OH represents 1-octadecanol.
Fig. 11 illustrates the production of fatty alcohols
in the cells of the genetically engineered strain Syn-XT51
after 8 days of culturing (the determination results of
GC-MS), wherein C15-OH represents 1-pentadecanol (used as
internal standard), C16-OH represents 1-hexadecanol, and
C18-OH represents 1-octadecanol.
Fig. 12 is a photo of genetically engineered strains
cultivated in a column photo-reactor.
Description of the sequences
SEQ ID NO: 1: the sequence of fatty acyl-CoA reductase
gene from (Simmondsia chinensis) (artificially
synthesized gene).
SEQ ID NO: 2: the artificially synthesized sequence
according to at3g11980 gene of Arabidopsis thaliana.
SEQ ID NO: 3: the sequence of the Rubisco promoter
fragment Prbc at the upstream of ribulose-1,5-diphosphate
carboxylase large-subunit gene rbcLfrom Synechocystis sp.
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
8 -
PCC6803 (US National Center for Biotechnology Information
(NCBI) ID: NC000911).
SEQ ID NO: 4: the sequence of the terminator fragment
Trbe at the downstream of ribulose-1,5-diphosphate
carboxylase operator from Synechocystis sp. PCC6803 (NCBI
ID: NC000911).
SEQ ID NO: 5: the sequence of the promoter fragment PpetE
at the upstream of the plastocyanin gene petE from
Synechocystis sp. PCC6803 (NCBI ID: NC_000911).
SEQ ID NO: 6: the N-terminal sequence (also comprising
a part of the upstream sequence of the gene) of s1r0168
gene from Synechocystis sp. PCC6803 (NCBI ID: NC_000911).
SEQ ID NO: 7: the C-terminal sequence (also comprising
a part of the downstream sequence of the gene) of s1r0168
gene from Synechocystis sp. PCC6803 (NCBI ID: NC_000911).
SEQ ID NO: 8: the sequence of the Omega fragment cloned
into the plasmid pRL57 (NCBI ID: L05082).
SEQ ID NO: 9: the sequence of the 1acZ gene cloned into
the plasmid pHB1567 (NCBI ID: AP009048).
SEQ ID NO: 10: the sequence of the primer alr1524-1.
SEQ ID NO: 11: the sequence of the primer alr1524-2.
SEQ ID NO: 12: the sequence of the primer P1.
SEQ ID NO: 13: the sequence of the primer P2.
SEQ ID NO: 14: the sequence of the primer P3.
SEQ ID NO: 15: the sequence of the primer P4.
SEQ ID NO: 16: the sequence of the primer XP-1.
SEQ ID NO: 17: the sequence of the primer XP-2.
SEQ ID NO: 18: the sequence of the primer XP-3.
SEQ ID NO: 19: the sequence of the primer XP-4.
SEQ ID NO: 20: the sequence of the primer lacZ-ml.
SEQ ID NO: 21: the sequence of the primer lacZ-m2.
SEQ ID NO: 22: the sequence of the primer lacZ-m3.
SEQ ID NO: 23: the sequence of the primer M13-Rev.
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
9 -
SEQ ID NO: 24: the sequence of the primer far-1.
SEQ ID NO: 25: the sequence of the primer far-2.
SEQ ID NO: 26: the sequence of the Synechocystis sp.
6803 rbcL promoter (Prbcl, 1.3 kb).
Definition of Terms
The following terms will be understood as defined
herein unless otherwise stated. Such definitions include
without recitation those meanings associated with these
terms known to those skilled in the art.
By a "Cyanobacterium" is understood a member from the
group of photoautotrophic prokaryotic microorganisms,
which can utilize solar energy and fix carbon dioxide.
Cyanobacteria are sometimes also referred to as blue-green
algae.
By a "construct" is herein understood a segment
comprising one or more nucleic acids, for example a DNA
fragment. The construct is suitably an artificially
constructed segment of one or more nucleic acids. The
construct can be used to subclone one or more of the nucleic
acids, for example a DNA fragment, into a vector.
By a "Fatty acyl-CoA reductase" is understood an enzyme
capable of catalyzing the conversion reaction of fatty
acyl-CoA to fatty alcohols.
"Ribulose-1,5-bisphosphate carboxylase/oxygenase"
(Rubisco) is an enzyme that catalyzes the first reaction
of a so-called Calvin cycle in photosynthesis. It may
consist of two subunits and the genes encoding the two
subunits can be located in one and the same operator in
the Synechocystis sp. PCC6803 genome. In the embodiments
of the present invention, a Rubisco promoter (indicated
as Prbc in the embodiments of the present invention) may
be cloned to drive the expression of fatty acyl-CoA
carboxylase gene in cyanobacteria, and the specific
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 10 -
sequence for such a Rubisco promoter Prbc is shown in SEQ
ID NO: 3.
"Plastocyanin" (PC) is an electron carrier for
transferring electron from cytochrome b6/f complex to
photosystem I in photosynthesis, and the gene encoding it
is abbreviated as "petE". In the embodiments of the present
invention, a petE promoter (indicated as PpetE in the
embodiments of the present invention) may be cloned to drive
the expression of fatty acyl-CoA carboxylase gene in
cyanobacteria, and the specific sequence for such a
promoter PpetE is shown in SEQ ID NO: 5.
A "s1r0168 gene" is a gene in the Synechocystis sp.
PCC6803 genome, which codes for a protein with unknown
function. Previous studies proved that the deletion of this
gene does not affect the physiologic activity of cells,
so that the site of this gene has been considered as a
neutral site in Synechocystis sp. PCC6803 genome. In the
embodiments of the present invention a promoter and a fatty
acyl-CoA reductase gene may be integrated at this site by
homologous recombination so as to express exogenous fatty
acyl-CoA reductase in Synechocystis sp. PCC6803.
In the embodiments of the present invention, the term
"vector" refers to a self-replicating DNA molecule capable
of transferring a DNA fragment (for example the gene of
interest) into a recipient cell.
The term "hybridization" is intended to mean the
process during which, under suitable conditions, two
nucleic sequences bond to one another with stable and
specific hydrogen bonds so as to form a double strand. These
hydrogen bonds can form between the complementary bases
adenine (A) and thymine (T) or uracil (U), which may then
be referred to as an A-T bond; or between the complementary
bases guanine (G) and cytosine (C), which may then be
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 11 -
referred to as a G-C bond. The hybridization of two nucleic
sequences may be total (reference is then made to
complementary sequences), i.e. the double strand obtained
during this hybridization comprises only A-T bonds and C-G
bonds. Or the hybridization may be partial (reference is
then made to sufficiently complementary sequences), i.e.
the double strand obtained comprises A-T bonds and C-G bonds
allowing the double strand to form, but also bases not
bonded to a complementary base. The hybridization between
two complementary sequences or sufficiently complementary
sequences depends on the operating conditions that are used,
and in particular the stringency. The stringency may be
understood to denote the degree of homology; the higher
the stringency, the higher percent homology between the
sequences. The stringency may be defined in particular by
the base composition of the two nucleic sequences, and also
by the degree of mismatching between these two nucleic
sequences. The stringency can also depend on the reaction
parameters, such as the concentration and the type of ionic
species present in the hybridization solution, the nature
and the concentration of denaturing agents and/or the
hybridization temperature. The appropriate conditions can
be determined by those skilled in the art.
Conditions for hybridizing nucleic acid sequences to
each other can be described as ranging from low to high
stringency. Reference herein to hybridization conditions
of low stringency includes from at least about 0% v/v to
at least about 15% v/v formamide and from at least about
1 M to at least about 2 M salt for hybridization, and from
at least about 1 M to at least about 2 M salt for washing
conditions. Preferably, the temperature for hybridization
conditions of low stringency is from about 25 C, more
preferably about 30 C to about 42 C.
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 12 -
Reference herein to hybridization conditions of medium
stringency includes from at least about 16% v/v to at least
about 30% v/v formamide and from at least about 0.5 M to
at least about 0.9 M salt for hybridization, and from at
least about 0.5 M to at least about 0.9 M salt for washing
conditions.
Reference herein to hybridization conditions of high
stringency includes from at least about 31% v/v to at least
about 50% v/v formamide and from at least about 0.01 M to
at least about 0.15 M salt for hybridization, and from at
least about 0.01 M to at least about 0 .15 M salt for washing
conditions. In general, washing is carried out at Tm = 69.3
+ 0.41 (G+C) o, where Tm is in degrees Centigrade and (G+C) o
refers to the mole percentage of guanine plus cytosine;
in line with the article of J. Marmur et al. titled
"Determination of the base composition of deoxyribonucleic
acid from its thermal denaturation temperature", published
in Journal of Molecular Biology volume 5, issue 1, July
1962, pages 109-118. However, the Tm of a duplex DNA may
decrease by 1 C with every increase of 1% in the number
of mismatch base pairs in line with the article of W.M.
Bonner et al. titled " A Film Detection Method for
Tritium-Labelled Proteins and Nucleic Acids in
Polyacrylamide Gels", published in the European Journal of
Biochemistry, volume 46, issue 1, 1974, pages 83-88.
Formamide is optional in these hybridization conditions.
Accordingly, a particularly preferred non-limiting
example of a hybridization condition of low stringency is
6 x SSC (Standard Sodium Citrate) buffer, 1.0% w/v SDS
(Sodium Dodecyl Sulfate) at 25-42 C; a particularly
preferred non-limiting example of a hybridization
condition of medium stringency is 2 x SSC (Standard Sodium
Citrate) buffer, 1.0% w/v SDS (Sodium Dodecyl Sulfate) at
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 13 -
a temperature in the range 20 C to 65 C; and a particularly
preferred non-limiting example of a hybridization
conditions of high stringency is 0 . 1 x SSC (Standard Sodium
Citrate) buffer, 0.1o w/v SDS (Sodium Dodecyl Sulfate) at
a temperature of at least 65 C. An extensive guide to the
hybridization of nucleic acids can be found in Tijssen (1993)
"Laboratory Techniques in Biochemistry and Molecular
Biology - Hybridization with Nucleic Acid Probes", Part
I, Chapter 2 (Elsevier, New York); Ausubel et al., eds.
(1995) "Current Protocols in Molecular Biology", Chapter
2 (Greene Publishing and Wiley-Interscience, New York);
and/or Sambrook et al. (1989) "Molecular Cloning: A
Laboratory Manual" (2d ed., Cold Spring Harbor Laboratory
Press, Plainview, New York).
The term "identity" or "percent identity" refers to the
sequence identity between two amino acid sequences or
between two nucleic acid sequences. To determine the
percent identity of two amino acid sequences or of two
nucleic acids, the sequences are aligned for optimal
comparison purposes. The percent identity between the two
sequences is a function of the number of identical positions
shared by the sequences (i.e., percent identity = number
of identical positions/total number of positions (e.g.,
overlapping positions) x 100) . For example, a "percent
identity" is calculated by comparing two optimally aligned
sequences over the window of comparison, determining the
number of positions at which the identical nucleic acid
base or the identical amino acid residue occurs in both
sequences to yield the number of matched positions,
dividing the number of matched positions by the total number
of positions in the window of comparison (i . e . , the window
size), and multiplying the result by 100 to yield the
percentage of sequence identity.
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 14 -
Optimal alignment of sequences for comparison can be
conducted, for example, by the local homology algorithm
of Smith and Waterman (Adv. Appl Math. 2:482, 1970), by
the homology alignment algorithm of Needleman and Wunsch
(J. Mol. Biol. 48:443, 1970), by the search for similarity
method of Pearson and Lipman (Proc. Natl. Acad. Sci. USA
85:2444, 1988), by computerized implementations of these
algorithms (e.g., GAP, BESTFIT, FASTA, BLAST P, BLAST N
and TFASTA in the Wisconsin Genetics Software Package,
Genetics Computer Group, 575 Science Dr., Madison, Wis.),
and/or by manual alignment and visual inspection (see, e.g.,
Ausubel et al, Current Protocols in Molecular Biology (1995
supplement)).
Percent identities involved in the embodiments of the
present invention include at least about 60% or at least
about 65% or at least about 70% or at least about 75% or
at least about 80% or at least about 85% or at least about
90% or above, such as about 95% or about 96% or about 97%
or about 98% or about 99%, such as at least about 60%, 61%,
620, 630, 640, 650, 66%, 67%, 68%, 690, 70%, 71%, 720, 73%,
740, 750, 760, 770, 780, 790, 800, 810, 820, 830, 840, 850,
860, 870, 880, 890, 900, 910, 920, 930, 940, 950, 960, 970,
98%, 99% or 100%.
Detailed Description of the Invention
The Cyanobacteria (also known as blue-green algae) in
this invention preferably comprise a group of prokaryotic
microorganisms capable of performing plant type oxygenic
photosynthesis.
The use of cyanobacteria may have the following
advantages: (1) cyanobacteria are capable of absorbing
solar energy and fixing carbon dioxide as carbon source
for autotrophic growth, thereby having low cost for
culturing; (2) cyanobacteria are ancient microorganisms
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 15 -
and have lived on the earth for billions of years, so that
they have remarkable adaptability to the environments, and
they grow quickly; (3) cyanobacteria are convenient for
genetic manipulations, because their genetic background
is clear and genomic sequencing of many species of
cyanobacteria has been completed which facilitates the
genetic engineering of cyanobacteria. Synechocystis sp.
PCC6803 is a preferred unicellular cyanobacteria, because
for Synechocystis sp. PCC6803 the whole genome sequencing
had been completed in 1996.
The embodiments of the present invention employ a
promoter having activity in cyanobacteria. This promoter
suitably drives the expression of fatty acyl-CoA reductase
in cyanobacteria. In this manner the characteristics of
cyanobacteria as photosynthetic organism can be utilized
to absorb solar energy, fix carbon dioxide and synthesize
fatty alcohols as biofuels. One of the advantages of the
present invention is that fatty alcohols are synthesized
by using solar energy to fix carbon dioxide in the
photosynthetic microorganism cyanobacteria, wherein the
energy for synthesizing fatty alcohols is solar energy and
the carbon source is carbon dioxide. Thus, the production
of biofuels utilizing this technology would not be
restricted by the lack of raw materials, and the use of
such biofuels would not increase carbon emission, i.e.,
such biofuels are real zero emission biofuels.
In one aspect, the embodiments of the present invention
relate to a construct used for synthesizing fatty alcohols
in cyanobacteria, which may comprise a promoter having
activity in cyanobacteria as well as a fatty acyl-CoA
reductase gene under the control of the promoter.
Further, the construct may comprise a marker gene for
screening transformants of cyanobacteria, which is located
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 16 -
upstream of the promoter having activity in cyanobacteria.
Preferably such a marker gene comprises the Omega gene
as set forth in SEQ ID NO:8. However, also variants of this
Omega gene, wherein the variant has at least 80% sequence
identity, preferably at least 85% sequence identity, more
preferably at least 90% sequence identity, even more
preferably at least 95% sequence identity and most
preferably at least 99% sequence identity, with the Omega
gene can be used. Suitably such variants also have marker
activity in cyanobacteria. Further, the construct may
comprise, at the two termini thereof, the N-terminal and
C-terminal sequences of s1r0168 gene of Synechocystis sp.
PCC6803, for homologous recombination.
In a most preferred embodiment, the promoter having
activity in cyanobacteria is selected from the group
consisting of the Prbc promoter and the PpetE promoter.
However, also variants of these promoters, wherein the
variant has at least 80% sequence identity, preferably at
least 85% sequence identity, more preferably at least 90%
sequence identity, even more preferably at least 95%
sequence identity and most preferably at least 99% sequence
identity, with the Prbc promoter or the PpetE promoter can
be used. Suitably such variants also have promoting
activity in cyanobacteria.
The choice of promoter gene can be of great importance
in terms of genetic expression. For example the rbc
(ribulose-1,5-bisphosphate carboxylase/oxygenase) gene
promoter gene (Prbe) is a particularly strong promoter
within the cyanobacteria, whilst its product RuBisCO is
a main soluble protein within the cyanobacteria which plays
an important role in photosynthesis. The rbc promoter gene
(Prbe) advantageously has the effect of being a high
efficiency expression gene within cyanobacteria. An rbc
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 17 -
operon may includes the promoter gene and rbcL, rbcX and
rbcS genes. Especially for Synechocystis PCC6803, the
complete operon may further start at a 250bp position
upstream on the rbcL gene, and downstream of the rbcS stop
codon there may be a 40bp reverse complementary sequence
which acts as a termination sequence (as described in more
detail in "Construction of a Synechocystis PCC6803 mutant
suitable for the study of variant hexadecameric ribulose
biphosphate carboxylase/oxygenase enzymes", by Doron
Amichay, Ruth Levitz and Michael Gurevitz, Plant Molecular
Biology 23: 465-476, 1993, incorporated herein by
reference).
In a further preferred embodiment, the fatty acyl-CoA
reductase gene may be selected from the group consisting
of: fatty acyl-CoA reductase (far) gene from Simmondsia
chinensis, for example as set forth in SEQ ID NO: 1; and
at3g11980 gene from Arabidopsis thaliana, for example as
set forth in SEQ ID NO: 2. In addition, the fatty acyl-CoA
reductase gene may be farl gene from mouse (see for example
National Center for Biotechnology Information (NCBI) ID:
BC007178); codon-optimized farl gene from mouse; fa-r2 gene
from mouse (see for example NCBI ID: BC055759) ; or at3g56700
gene from Arabidopsis thaliana. Other suitable fatty
acyl-CoA reductase genes include: Francci3_2276 from
Frankia sp.Cc13 (see for example NCBI ID: NC_007777);
KRH_18580 from Kocuria rhizophila DC2201 (see for example
NCBI ID: NC010617); A20C104336 from Actinobacterium
PHSC20C1 (see for example NCBI ID: NZ_AAOB01000003);
HCH_05075 from Hahella chejuensis KCTC 2396 (see for
example NCBI ID: NC_007645); Maqu_2220 from Marinobacter
aquaeolei VT8 (see for example NCBI ID: NC_008740); and
RED6509889 from Oceanobacter sp. RED65 (see for example
NCBI ID: NZ_AAQH01000001). In addition, the embodiments
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 18 -
of the present invention may employ the genes having at
least 80% identity, preferably at least 85% identity, more
preferably at least 90% identity, even more preferably at
least 95% identity and most preferably at least 99% identity
to the above-mentioned genes and coding for a protein having
fatty acyl-CoA reductase activity; or the genes capable
of hybridizing with the above-mentioned genes under
stringent hybridization conditions, preferably
hybridization conditions of high stringency, and coding
for a protein having fatty acyl-CoA reductase activity.
In a further preferred embodiment, the marker gene is
the Omega fragment of spectinomycin resistance gene, for
example as set forth in SEQ ID NO: 8.
Preferably the cyanobacterium is chosen from the group
consisting of Synechococcus PCC 6301, Anabaena sp. strain
PCC 7120, Synechococcus PCC 7002 , Synechococcus elongatus
sp. strain PCC 7942 and Synechocystis sp. PCC6803. In a
most preferred embodiment, the cyanobacterium is
Synechocystis sp. PCC6803.
In another aspect, the embodiments of the present
invention may relate to a vector comprising the construct
as defined above.
Depending on the state in which the vector exists within
the cyanobacteria, this may be for example a shuttle plasmid
vector or a genomic integrative plasmid vector. Both types
of vector can play a major role in assisting natural
blastasis (mainly in single celled blue-green alga) or can
introduce one or more of the genes described herein into
the cyanobacteria via conjugal transfer. Shuttle plasmid
vectors can enter the cyanobacteria as a result of conjugal
transfer, then duplicating themselves within the cytoplasm.
Genomic integrative vectors can cause isogenesis of one
or more of the genes described herein, or even a whole operon
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 19 -
comprising one or more of such genes, within the
cyanobacteria genome via isogenic integration, with
advangtageous greater stability of expression, overcoming
the stability problems encountered with autonomous
plasmids.
Preferably, the vector is selected from the group
consisting of: plasmid pXT14, which was deposited in China
General Microbiological Culture Collection Center under
Accession Number of CGMCC 3948 on June 28, 2010, in a form
in E. coli (Eco-XT14) ; plasmid pXT34, which was deposited
in China General Microbiological Culture Collection Center
under Accession Number of CGMCC 3950 on June 28, 2010, in
a form in E. coli (Eco-XT34) ; and plasmid pXT51, which was
deposited in China General Microbiological Culture
Collection Center under Accession Number of CGMCC 3949 on
June 28, 2010, in a form in E. coli (Eco-XT51).
In another aspect, the embodiments of the present
invention may relate to a cyanobacterium comprising the
construct as defined above, or a cyanobacterium
transformed by the vector as defined above. Preferably,
the cyanobacterium is selected from the group consisting
of: cyanobacterium Syn-XT14, which was deposited in China
General Microbiological Culture Collection Center under
Accession Number of CGMCC 3894 on June 10, 2010;
cyanobacterium Syn-XT34, which was deposited in China
General Microbiological Culture Collection Center under
Accession Number of CGMCC 3895 on June 10, 2010; and
cyanobacterium Syn-XT51, which was deposited in China
General Microbiological Culture Collection Center under
Accession Number of CGMCC 3896 on June 10, 2010.
In a further aspect, the embodiments of the present
invention may relate to a method for producing fatty
alcohols in cyanobacteria, comprising: culturing a
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 20 -
cyanobacterium comprising the construct as defined above,
or a cyanobacterium transformed by the vector as described
above under conditions suitable for the synthesis of fatty
alcohols; and extracting the desired fatty alcohols from
the obtained culture.
Fatty alcohols, especially long-chain fatty alcohols,
such as 1-hexadecanol and 1-octadecanol, were successfully
produced in cyanobacteria via the embodiments of the
present invention.
If desired, these fatty alcohols may be converted to
hydrocarbons by any manner known by the person skilled in
the art to be suitable therefore. Such hydrocarbons can
include alkanes (such as hexadecane or octadecane) and/or
alkenes (such as 1-hexadecene or 1-octadecene). In a
preferred embodiment the method for producing fatty
alchols in cyanobacteria as described above may therefore
further comprise converting the fatty alcohols to
hydrocarbons, providing a method for producing
hydrocarbons.
The fatty alcohols produced via the embodiments of the
present invention and/or the hydrocarbons obtained by
converting these fatty alcohols can advantageously be used
as biofuel components and/or speciality chemicals. Such
a biofuel may advantageously have properties such as high
energy density, low moisture absorption, low volatility,
and/or compatibility with existing engines and transport
facilities. In addition such a biofuel may be considered
a real zero emission biofuel.
In another aspect, the embodiments of the present
invention may relate to a method of expression of exogenous
genes via the use of Synechocystis sp. 6803. Such a method
can be helpful in any method for producing fatty alcohols
and/or long chain biologic hydrocarbons in cyanobacteria
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 21 -
as described above. The Synechocystis sp. 6803 genome
integrative plasmid platform used in such a method may be
understood to be a kind of genomic integrative vector as
described above. In a preferred embodiment such a method
uses a Synechocystis sp. rbcL promoter (Prbcl, 1.3 kb),
a rbc terminator sequence (Trbc, 0.2 kb), a 2kb
spectinomycin resistant marker gene Q and/or a reporter
gene lacz that is obtained via cloning.
Examples
The invention is further illustrated by the following
non-limiting examples.
Example 1: Construction of vectors for the transformation
of cyanobacteria
1. Construction of the plasmid pFQ9R (figure 1)
PCR was performed by using alr1524-1
(5'-ACCTCCAGCCATTAGCG AAAC-3') and alr1524-2
(5'-CTCTCACAATTGCCCTACCT-3') as the primer pair and using
the genome of Anabaena PCC7120 as the template, and the
PCR product was cloned into the vector pMD18-T (Takara,
Catalog No .: D101A) to obtain the plasmid pQLl . Dral (Takara,
Catalog No.: D1037A) was used to digest the plasmid pRL57
(Cai Y. and Wolk C. (1990) "Use of a conditionally lethal
gene in Anabaena sp. strain PCC 7120 to select for double
recombinants and to entrap insertion sequences." J.
Bacteriol 172: starting page 3138), and the Omega fragment
of about 1. 9 kb was recovered. The plasmid pQLl was digested
with PstI (Takara, Catalog No.: D1073A), and blunt-ended
with T4 DNA polymerase (Fermentas, Catalog No.: EP0061).
The two fragments were ligated to obtain the plasmid pQL4.
PCR was performed by using P1 (5'-GCGTCGACTCACCATTTGGAC
AAAACATCAGG-3') and P2 (5'-GCTCTAGACATCTAGGTCAGTCCT
CCATAAACATTG-3') as the primer pair and using the genome
of Synechocystis sp. PCC6803 as the template, and the PCR
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 22 -
product was cloned into the vector pMD18-T to obtain the
plasmid pFQ1; PCR was performed by using P3
(5'-CCCCCGGGGTTACAGTTTTGGCAATTACT-3') and P4
(5'-CGAGCTCTTCCCCACTTAGATAAAAAATCCG-3') as the primer
pair and using the genome of Synechocystis sp. PCC6803 as
the template, and the PCR product was cloned into the vector
pMD18-T to obtain the plasmid pFQ2. SalI (Takara, Catalog
No.: D1080A) and XbaI (Takara, Catalog No.: D1093A) were
used to cut the Prbc fragment from the plasmid pFQ1; XmaI
(New England BioLabs, Catalog No. : R0180S) and Sacl (Takara,
Catalog No. : D1078A) were used to cut the Trbc fragment from
the plasmid pFQ2; the Prbc and Trbc fragments were inserted
at corresponding site of the plasmid pQL4 to obtain the
plasmid pFQ6.
The plasmid pKW1188 (Williams J. G. K.(1988)
"Construction of specific mutations in photosystem II
photosynthetic reaction center by genetic engineering
methods in Synechocystis 6803" Methods in Enzymology
167:pages 766-778) was digested with EcoRI and
self-ligated, then blunt-ended with XmaI, and then it was
self-ligated to obtain the plasmid pKW1188SL. Hindlll
(Takara, Catalog No. : D1060A) and EcoRI (Takara, Catalog
No. : D1040A) were used to digest the plasmid pFQ6, and the
Omega+Prbc+Trbc fragment was recovered; EcoRI was used to
digest the plasmid pKW1188SL; and the two fragments were
ligated to obtain the plasmid pFQ9R.
2. Construction of the plasmids pXT37a and pXT37b (figure
3 respectively figure 4)
The plasmid pHB1567 (Gao Hong, et al, (2007)
"Construction of Copper-Induced Gene Expression Platform
in Synechocystis sp. PCC6803", Acta Hydrobiologica Sinica,
Vol. 31, No. 2, pages 240-244) was digested with XbaI, the
5.4 kb fragment was recovered and self-ligated to obtain
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 23 -
the plasmid pXT24. The plasmid pXT24 was digested with NdeI
(Takara, Catalog No.: D1161A), blunt-ended with T4 DNA
polymerase and self-ligated; then, it was digested with
EcoRI, blunt-ended with T4 DNA polymerase and self-ligated
to obtain the plasmid pXT24a. PCR was performed by using
the plasmid pHB1536 (GAO Hong, et al, 2007) as the template
and using XP-1 (5'-AGTGGTTCGCATCCTCGG-3') and XP-2
(5'-ATGAATCCTTAAT CGGTACCAAATAAAAAAGGGGACCTCTAGG-3') as
well as XP-3
(5'-CCCTTTTTTATTTGGTACCGATTAAGGATTCATAGCGGTTGCC-3') and
XP-4 (5'-CCAGTGAATCCGTAATCATGGT-3') as the primer pair,
respectively, the PCR product was recovered, and
afterwards it was denatured, annealed and extended; then,
PCR was performed by using it as the template and using
XP-1 and XP-4 as the primer pair, and the PCR product was
cloned into the vector pMD18-T to obtain the plasmid pQL17.
The plasmid pQL17 was digested with BglII (Takara, Catalog
No.: D1021S) and SphI (Takara, Catalog No.: D1180A), and
the recovered fragment was ligated to pHB1536 digested with
the same enzymes to obtain the plasmid pQL18. The plasmid
pQL18 was digested with XbaI, the Omega+PpetE+lacZ fragment
was recovered and inserted at the same site of the plasmid
pXT24a to obtain the plasmid pXT36a. PCR was performed by
using the plasmid pHB1567 as the template and using lacZ-ml
(5'-ATGGTCAGGTCATGGATGAGCA-3') and lacZ-m2
(5'-AATCCCCATGTGGAAACCGT-3') as well as lacZ-m3
(5'-ACGGTTT CCACATGGGGATT-3') and M13-Rev
(5'-AGCGGATAACAATTTCACAC AGGA-3') as the primer pair,
respectively, the PCR product was recovered, and
afterwards it was denatured, annealed and extended; then,
PCR was performed by using it as the template and using
lacZ-ml and M13-Rev as the primer pair, and the PCR product
was cloned into the vector pMD18-T to obtain the plasmid
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 24 -
pXT30. The plasmid pXT30 was digested with EcoRI and EcoRV,
and the recovered fragment was ligated to pXT36a digested
with the same enzymes to obtain the plasmid pXT37b. The
plasmid pXT37b was digested with Xbal, and the two fragments
were recovered, self-ligated and screened to obtain the
plasmid pXT37a having an insertion direction contrary to
that in pXT37b.
3. Construction of the plasmid pLY2 (Figure 7)
The plasmid pRL57 was digested with Dral, and the Omega
fragment was recovered; the plasmid pKW1188SL was digested
with EcoRI, blunt-ended, and the fragment was recovered;
the two fragments were ligated to obtain the plasmid pLY2.
This plasmid was used as a control plasmid.
4. Construction of the plasmid pXT14 (figure 2)
PCR was performed by using he plasmid pXL66 (a gift from
Professor Chaitan Khosla of Standford University) as the
template and using far-1
(5'-GGGTCTAGAATGGAAGAGATGGGCAGCATC-3') and far-2 (5 '-AAA
CCCGGGATCAATTCAGGACATGTTCCACGA-3') as the primer pair,
the PCR product was recovered, digested with XbaI and Smal,
and cloned into the same site of the plasmid pFQ9R to obtain
the plasmid pXT14.
5. Construction of the plasmid pXT51 (figure 6)
The plasmid pXL66 was digested with NdeI and XhoI, the
far gene fragment of Simmondsia chinensis was recovered
and inserted into the same site of the plasmid pXT37b to
obtain the plasmid pXT51.
6. Construction of the plasmid pXT34 (figure 5)
According to the sequence of SEQ ID No: 2, at3g11980
gene of Arabidopsis thaliana was synthesized and cloned
into the plasmid pUC57 (the synthesis was conducted by
Sangon Biotech (Shanghai) Co., Ltd) to obtain the plasmid
pXT31. The plasmid pHB1567 was digested with EcoRI and XhoI,
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 25 -
and the 5.4 kb fragment was recovered; the plasmid pHB1536
was digested with XhoI and NdeI, and the 2.4 kb fragment
was recovered; the plasmid pXT31 was digested with
NdeI+EcoRI, and the at3g11980 fragment was recovered;
these three fragments were ligated to obtain the plasmid
pXT34.
A summary of the information on the plasmids and strains
used for expressing fatty acyl-CoA reductase in
Synechocystis sp. PCC6803 is provided in Table 1.
Example 2: Transformation of cyanobacteria and screening
of transformants
1. 10 mL of algae cells in logarithmic growth phase (OD730
of about 0.5-1.0) was taken, and centrifuged to collect
the cells; the cells were washed twice with fresh BG11
medium, and then resuspended in 1 mL BG11 medium (1.5 g
L-1 NaNO3r 40 mg L-1 K2HP04 = 3H20, 36 mg L-1 CaC12 = 2H20, 6 mg
L-1 citric acid, 6 mg L-1 ferric ammonium citrate, 1 mg L-1
EDTA disodium salt, 20 mg L-1 NaCO3r 2.9 mg L-1 H3BO3, 1.8
mg L-1 MnCl2 - 4H20, 0.22 mg L-1 ZnSO4 . 7H20, 0.39 mg L-1
NaMo04=2H20, 0.079 mg L-1 CuSO4=5H20 and 0.01 mg L-1
COC12=6H20).
2. 0.2 mL of cell suspension was placed in a new EP tube,
2-3 pg of the expression plasmid as listed in Table 1 was
added, and the resulting mixture was mixed well and
incubated at 30 C under an illumination condition of 30
pE m-2 s-1 for 5 hours.
3. The mixture of algae cells and DNA was applied onto a
nitrocellulose membrane on BGll plate (without antibiotics)
and cultivated at 30 C under an illumination condition of
30 pE m-2 s-1 for 24 hours. Then, the nitrocellulose membrane
was transferred to a BG11 plate containing 10 pg mL-1
spectinomycin, and further incubated at 30 C under a
condition of 30 pE m-2 s-1.
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 26 -
4. After about 5-7 days, the transformants were picked out
from the plate, and used to streak the fresh BG11 plate
(supplemented with 20 pg mL-1 spectinomycin) . After the
cells were enriched, they are inoculated into a liquid BG11
medium (containing 20 pg mL-1 spectinomycin) for
cultivation.
5. After the cells were transferred twice in a liquid medium,
the yield of fatty alcohols was measured.
Example 3: Production of fatty alcohols by the genetically
engineered cyanobacteria
1. Experimental steps:
(1) Culturing method I: shake-flask culturing. A normal
500-mL conical flask with 300 mL of liquid BG11 medium
(containing 20 pg mL-1 spectinomycin) was used for
inoculation with an initial concentration (OD730 of 0.05),
and the culturing was performed at 30 C, under an
illumination condition of 30 pE m-2 s-1 and under aeration
with air, for 7-8 days.
Culturing method II: column photo-reactor culturing.
Normal glass tubes with a height of 575 mm, a diameter of
50 mm and a liquid volume of 500 mL (loading capacity of
about 1 L) were used. The initial inoculation concentration
was OD730 of 0.5, and the culturing was performed at 30 C,
under an illumination condition of 100 pE m-2 s-1
illumination under aeration with air containing 5% CO2.
(2) 200 mL of medium was taken, algae cells were
collected by centrifugation, and resuspended in 10 mL TE
(pH8.0) buffer, and then the cells were disrupted via
ultrasonication.
(3) 40 pg pentadecanol (as internal standard) was added
to the sonicated cells, and an equivalent volume of
chloroform:methanol (v/v 2:1) was added, the resulting
mixture was mixed well and kept at room temperature for
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 27 -
0.5 hour.
(4) After centrifuging at 3,000 g for 5 minutes, the
organic phase was recovered, and dried at 55 C under blowing
with nitrogen gas.
(5) 1 mL n-hexane was added to dissolve the precipitate.
After filtration with 0.45 pm filter membrane, GC-MS
analysis was performed.
2. Experimental results:
Hexadecanol and octadecanol were detected in samples
of three strains of genetically engineered cyanobacteria:
Syn-XT14, Syn-XT34 and Syn-XT51. The total yields of
intracellular fatty alcohols under normal shake-flask
culturing conditions as shown in Table 2 were calculated
by referring to the internal standard (pentadecanol) . The
results under column photo-reactor culturing conditions
also confirmed the ability of the three strains of
genetically engineered cyanobacteria for synthesizing
fatty alcohols.
The results indicate that the genetically engineered
cyanobacteria Syn-XT14, Syn-XT34 and Syn-XT51 were capable
of producing fatty alcohols, and this process for producing
fatty alcohols can be enlarged in small scale.
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
28
-u
1-1 u) 03
U H H H
O co x x x
m >1 >1 >1
co co co
cn v)
m
U)
c
O -H -H d v d
00 +- E 03 03 1-1 Q0 rl O) H H H
O U cn r co l Q,
a4 Q4
U)
U) m U)
4) u w w
u 0
a a
p a4 a a
O a
cd
O 03
44 N x
O
Q~ Q4
Q4
cd
rH cd
U
co co co
O N O) N co N
U) _ U U
>1 >1 >1
cn rn rn
+J >1
co u
rzi U) ~4 U) H
r{ H ctj
~H U)
0 >1 U) U)
4 11 u U M
H +)
cd O O ^i
4-4
O .~ .~
U H U +-)
o o 03
O O ~
a
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
29
O (N 0)
l9 O (N
U)
0) un 0)
+1 +1 +1
4-4 - Oo Q0 M
I l~ L9 l~
4) bi q
Z N r N
cd U) M
O N r O
r1 O +1 +1 +1
cd M (N Lc)
4J b~ q m
N O
0) Ln
00 rl 00
-H
N O rl
0 +1 +1 +1
l-- 00
l9 l~
U) 00 q (N
ri
U Lc) h N
N
u-) 00
O O O O
O q0 +1 +1 +1
00 b~ q 00
>1 7~
r -I ~- Z
co O
U
U u-) O
U O O o 0 0
+1 +1 +1 +1
(N (D0 0) Q0
+~ cd rn -1 O rn
N -H'U 4 r+4 M N N M
4i
4-a cd
O 2
O 03
U) 0 n al
r -I O
O O n H
O
C) u
cd + +
+ w w N
>1 0
!14 = = !14 !1W
a~ ==
CO
co 00 00 + 03 + 03 + 4j
4 4 >1 ~o O co O co O co c)
4- 4) H co b) H bD b) N
0 o 0 0 (D o U o~
0
m ^{ E H O ^{ O ^{ O
0 U) O U) - U) - U) --
U) O
-u
N U q
N O (N Ln m 1-1 > H H H Z
i a x x x
cd N cd I I I I
H ~ +) >1 >1 >1 >1
U) U) U) U) U)
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
0 +1 +1
4) i' M +I
O N
o r~ q lq N 6l N M
M l- oo O
M N N r r
N - r r
O >1
4)
O
O
Ln LO
0 - =
H q 1-1 CD
+1 +1 +1
r O N 00 03
0 Ln r
U
N
N O
03 1-1
o q
( O q O o
00 +1 +1 +1
Hl a Z 0n 0n -1
tS CJ 00 O M
4J 0n (N
N l- l9
N M N d
O ra
q O r O
0 q +1 +1 +1
O Z
M Ln 00 >1 0 r-- CD
r-I U rn 00 00
r-I q -1 -1 M M
co 0
U O o 0 0
r -I +1 +1 +1
iS LO O (N
00 03
-H M N M r
w
N N M M
U
+~ '~ ctS
4-4
03
0 0
0 r al
r 0 r 0
n
o I
O -u
U >1 m m
rl -W `H + +
co 0 + w N
Q +' r
~
>1 N n4 nW a~ n4 CO
4) =
03 03 + 03 + 03 + +)
co cd O cd O cd U
4-a co H H N
4-4 O bi O N O N O N 4)
H E H O H O H O 'o
U) O - - ^ (
v
0
-u
= U) U) II
cn U -H r q
N O U)
> H H H Z
N n a x x x
H
U) co co
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 31 -
Example 4: Illustration of a method of expression of
exogenous genes via the use of Synechocystis sp. 6803
1) The rbcL promoter (Prbcl, 1.3 kb), the termination
sequence rbc terminator (Trbc, 0.2 kb), the 2kb
spectinomycin resistant marker gene Q and the reporter
gene lacz of Synechocystis sp. 6803 were cloned separately,
being built into a Synechocystis sp. 6803 genome
integrative plasmid platform (which relies on the EcoRI
restriction enzyme pKW1188 and which connects
automatically after removal of the C.K2 fragment, yielding
pKW1188SL), thus yielding the pFQ20 plasmid platform
Prbcl
IacZ
0 Spr
pFQ20
AprSpr rbc -
s1r0168-N 12.4kb terminator
terminer
sirO168
C-terminer
already containing the reporter gene designed for the
purposes of exogenous gene expression by Synechocystis sp.
6803 and capable of genomic integration.
2) pFQ20 was used to transform the Synechocystis sp. 6803
algal strain, the transformant being obtained by
spectinomycin resistant screening, after PCR genotype
assay using a specific primer was conducted to confirm that
there had been no error, assay of R- galactosidase
activation of the high performance expression platform and
exogenous gene genetically engineered Synechocystis sp.
6803 strain was carried out, thus confirming the expression
performance of the Synechocystis sp. 6803 exogenous gene
expression platform constructed according to this
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 32 -
invention.
Example 4a : Cloning of the Jazz gene, Prbcl and Trbc onto
a pMD18T vector
1) PCR cloning of the Prbcl and Trbc gene fragments was
carried out on Synechocystis sp. 6803; the pFQ1 plasmid
Prbcl
Sall Xbal
pFQ1
Apr
3.Ok
was obtained after Prbcl linked to the pMD18T vector; the
pFQ2 plasmid
Sma Trbc Sad
pFQ2
Apr
2.9k
was obtained after Trbc linked to the pMD18T vector. The
E.Coli (BL21 DE3) cloned LacZ gene (3.1 kb) was inserted
into the pUC19 plasmid, yielding the pQL12 plasmid.
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 33 -
IacZ
pQL12
Apr
5.8k
2) It was confirmed that there were no errors in the
nucleotide sequencing after all the plasmid fragments were
subjected to sequence analysis.
Example 4b: Cloning of the spectinomycin resistant Q gene,
Prbcl and Trbc were separately in series with the pMD18T
vector.
1) pQL4 is a vector originating in pMD18T which contains
the spectinomycin resistant Q gene.
Omega,S
PQL4
D
Using the XbaI and SalI resistant enzyme plasmid pQL4,
linear pQL4EH gene fragments were recovered; the XbaI and
SalI resistant enzyme plasmid pFQ1 was also used to recover
DNA Prbcl fragments; the pQL4EH segments and the segmented
Prbcl were connected using T4 ligase, causing conversion
of the E.Coli; spectinomycin and ampicillin were used to
perform double-resistance transformant screening, then
plasmid restriction enzyme assay was carried out to ensure
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 34 -
that there were no errors, finally yielding the pFQ5
plasmid;
2) An alternative method: Taking the Smal-Sacl
restriction enzyme pFQ2, gel extraction of the 200bp Trbc
gene fragment[s] was carried out; at the same time using
the Smal-Sacl restriction enzyme pFQ5 and a PCR product
purification test kit, purification was carried out
yielding a double enzyme resistant pFQ5EH gene fragment;
using T4 ligase, Trbc was inserted downstream in pFQ5,
converting the E.Coli, then spectinomycin and ampicillin
were used to perform double-resistance transformant
screening, after which plasmid restriction enzyme assay
was carried out to ensure that there were no errors,
yielding the pFQ6 plasmid;
Xbal Smal
Sall Prbcl
Trbc
Sad
0 Spr
pFQ6
AprSpr
5.2k
pFQ6 contained the Q , Prbcl and Trbc gene fragments;
Example 4c: Implanting the spectinomycin resistant Q gene,
Prbcl, Trbc and LacZ genes serially into a cyanobacterial
vector.
1) Structure of the expression vector and method of
processing: pKW1188 is a plasmid platform used in
Synechocystis sp. 6803 genomic integration. Using EcoRI
resistant enzyme pKW1188, linking occurred after removal
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 35 -
of the CK2 fragment, yielding pKW1188SL.
sIrO168C
EcoRl
pKW1188SL
sIrO168N Apr
5.5K
After removal of the SmaI locus from pKW1188SL the pFQ15
plasmid was obtained:
sIrO168C
EcoRl
pFQ15
sIrO168N Apr
5.5K
2) Processing of target gene: Using Hindlll-Sacs resistant
enzyme pFQ6 gel recovery, the Q, Prbcl and Trbc fragments
were obtained, reinforcement with T4 ligase was carried
out at 37 C for 30 minutes; the fragments obtained after
reinforcement were then subjected to purification using
a PCR product purification kit; after the pFQ15 vector
expression was cut open using EcoRI further reinforcement
and purification was carried out using the same methods;
after the Q, Prbcl and Trbc fragments and pFQ15 fragments
had undergone reinforcement they were linked using T4
ligase, converting E.Coli, the transformant was then
subjected to PCR assay using specific primers to confirm
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 36 -
the direction of insertion of the serially linked fragments,
allowing screening of cloned correctly inserted fragments
confirming that there were no errors regarding the
extracted plasmid resistant enzymes, the product then
being named pFQ9Forward.
Prbc Xbal
Smal
i
0 Spr rbc -
terminator
pFQ9
sIrO168 AprSpr sIrO168
N-terminer 8.OKb
C-terminer
3) Using XbaI and Smal restricted pQL12 and gel recovered
lacZ gene fragments, these were inserted into pFQ9Forward,
converting E.Coli, the lacZ fragments within the
transformant then being subjected to PCR assay using
specific primers, after it had been confirmed that there
were no errors in the extracted plasmid restricted enzyme
this was named pFQ19.
Prbc,0.3kb
IacZ
0 Spr
pFQ19
AprSpr rbc -
sIrO168-N 11.4kb terminator
terminer
sIrO168
C-terminer
Taking PCR amplified 1.3kb Prbcl, SalI and XbaI with
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 37 -
identical loci were used to replace the 0 .3kb Prbc yielding
pFQ20.
Prbcl,1.3kb
IacZ
0 Spr
pFQ20
AprSpr rbc -
sIr0168-N 12.4kb terminator
terminer
sIrO168
C-terminer
Example 4d: Conversion of the expression vector to
Synechocystis sp. 6803, genetic assay and detection of
galactosidase activity of the genetically expressed
product.
1) A natural method was used to cause pFQ20 conversion of
the wild variant of the Synechocystis sp. 6803 cell. The
transformant developed around a week later; after the
transformant was amplified in BG11 liquid containing
spectinomycin for one week the algal solution was harvested
and the genomic DNA extracted. PCR assay relying on specific
primers was used to detect whether the transformant
contained the target LacZ gene. After assay confirmed that
there were no errors, the mutated algal strain was named
Synechocystis PCC6803(FQ20).
2) Synechocystis PCC6803(FQ20) was cultured in liquid
until after the growth period, then the algal solution was
harvested and tested for(3- galactosidase activity. The
wild strain of Synechocystis PCC6803 was used as the
negative comparison 1, Synechocystis PCC6803(LY2)
was used as negative comparison 2, Synechocystis
PC6803(QL15) was used as negative comparison 2.
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 38 -
0 Spr
pLY2
AprSpr
sir0168C 7.5K
sir0168N
The Synechocystis PCC6803(HB1567) strain that exhibits
galactosidase expression activity was used as the positive
comparison.
PpetE
IacZ
Q Spr
pHB1567
sir0168-N AprSpr
terminer 10.8kb s1r0168
C-terminer
Of these, the Synechocystis PCC6803(LY2) transgenic algal
strain possessed the resistant Q gene.
3) The results demonstrate that, the galactosidase
activity of the constructed genetically engineered
Synechocystis 6803FQ20 exceeded that of the known positive
comparison under identical conditions, whilst no activity
was detected in the negative comparisons.
It will be appreciated by persons skilled in the art that
numerous variations and/or modifications may be made to
the invention as shown in the specific embodiments without
departing from the spirit or scope of the invention as
CA 02786244 2012-07-03
WO 2011/086189 PCT/EP2011/050555
- 39 -
broadly described. The present embodiments are, therefore,
to be considered in all respects as illustrative and not
restrictive.
Deposition information of the samples of
biological materials:
Strains Accession No. Deposition date
Escherichia coli DH5c - FQ20 CGMCC 3458 November 20, 2009
containing plasmid pFQ20
Synechocystis PCC6803- FQ20 CGMCC 3462 November 20, 2009
containing plasmid pFQ20
Cyanobacteria Syn-XT14 CGMCC 3894 June 10, 2010
Cyanobacteria Syn-XT34 CGMCC 3895 June 10, 1010
Cyanobacteria Syn-XT51 CGMCC 3896 June 10, 2010
E. coli Eco-XT14, CGMCC 3948 June 28, 2010
containing the plasmid pXT14
E. coli Eco-XT34, CGMCC 3950 June 28, 2010
containing the plasmid pXT34
E. coli Eco-XT51, CGMCC 3949 June 28, 2010
containing the plasmid pXT51
All of the above strains were deposited at the China General
Microbiological Culture Collection Center (CGMCC) fL st-tute of
1"[.ic..-.ob=i.oi.< gv, ]...-..:.. = se ..c: decry . c :.en = s, N . 1-
ih;est. ?e :.cbe7n
Road, Beijing ..,,.., Chin
a.