Note: Descriptions are shown in the official language in which they were submitted.
CA 02831300 2013-02-28
WO 2012/031052 PCT/US2011/050108
MEDICAL INFORMATION NAVIGATION ENGINE (MINE) SYSTEM
BACKGROUND OF THE INVENTION
Cross Reference to Related Applications
This application claims priority to U.S. Patent Application No. 61/379,228, by
Ansari et al.,
entitled "MEDICAL INFORMATION NAVIGATION ENGINE (MINE) SYSTEM", filed on
September 1, 2010.
Field of the Invention
The invention relates generally to medical information engine, and
particularly to
management and consolidation of medical information.
Description of the Prior Art
Despite rapid growth of innovation in other fields in recent decades, the
world of medical
information, including patient medical records, billing, and a host of other
information, has enjoyed
little to no useful consolidation, reliability, or ease-of-access, leaving
medical professionals,
hospitals, clinics, and even insurance companies with many issues, such as
unreliability of medical
information, uncertainty of diagnosis, lack of standard, and a slew of other
related problems.
One of the challenges facing those in the medical or related areas is the
number of sources of
information, the great amount of information from each source, and
consolidation of such
information in a manner that renders it meaningful and useful to those in the
field in addition to
patients. Obviously, this has contributed to increased medical costs and is
perhaps largely attributed
to the field suffering from an organized solution to better aid the medical
professionals, to better aid
those requiring more reliable patient history and those requiring more control
and access over such
information.
CA 02831300 2013-02-28
WO 2012/031052 PCT/US2011/050108
Currently, when a patient sees various medical professionals over the years,
there is no
method for universally tracking recommendations, thoughts, prescriptions,
diagnosis. This hinders
the job of insurance companies in making certain requisite determinations,
physicians making
decisions that directly impact the health of the patient, and hospitals and
other medical institutions
who similarly rely but do not have the benefit of the requisite information,
not to mention the
patient.
Further, there are problems in the current medical system that are associated
with patient
identity in that due to the exposure of a patient to various medical
associations/professionals over
the years and the possibility of various ways of identifying the same patient,
patients' records and
identity are oftentimes compromised, creating a slew of problems both for the
patient as well as
those treating the patient.
Further, privacy of a patient's health records is not currently reliably
maintained, as there are
too many cases of health record compromises. Additionally, patient control of
access to medical
information is nearly nonexistent. Additionally, secure and remote access of
medical information is
currently lacking.
Therefore, what is needed is a method and apparatus for managing medical
information in a
manner that is beneficial, reliable, portable, flexible, and efficiently
usable to those in the medical
field, including patients.
CA 02831300 2013-02-28
WO 2012/031052 PCT/US2011/050108
SUMMARY OF THE INVENTION
To overcome the limitations in the prior art described above, and to overcome
other
limitations that will become apparent upon reading and understanding the
present specification, the
present invention discloses a method and a corresponding structure for
transacting medical
information.
Briefly, a method of transacting medical information is disclosed to include
receiving
medical information from a medical sources, identifying, mapping, and
consolidating the received
medical information by a back-end medical processor, presenting access to
specific relevant data,
based on a user's security privileges, within the identified, mapped, and
consolidated medical
information, based on user-specific functions or roles by a front-end medical
processor, and
generating user-customized processed medical information to a plurality of
users, with at least a
portion of the user-customize processed medical information being provided to
each of the plurality -
of users based on its relevancy to each user's specific function or role and
each user's associated
security privileges.
These and other objects and advantages of the invention will no doubt become
apparent to
those skilled in the art after having read the following detailed description
of the preferred
embodiments illustrated in the several figures of the drawing.
CA 02831300 2013-02-28
WO 2012/031052 PCT/US2011/050108
= IN THE DRAWINGS
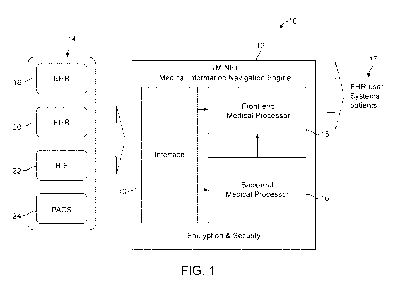
Fig. 1 shows a medical system 10, in accordance with an embodiment of the
invention.
Fig. 2 shows further details of the MINE 12 of Fig. 1, in accordance with an
embodiment
of the invention.
Fig. 3 shows an exemplary embodiment implementing the system 10 using various
devices.
Fig. 4 shows further details of the system 10, in accordance with an
embodiment of the
invention.
Fig. 5 shows a block diagram of the indexing, querying, and searching
functions of the
MINE 12, in accordance with a method of the invention.
Fig. 6 shows a block diagram of the query module, in accordance with an
apparatus
and/method of the invention.
Fig. 7 shows a block diagram of the relevant blocks/functions of the
presentation module
of the module 30 of Fig. 2.
Fig. 8 shows further details of the medical lexical search engine 310, in
accordance with
an embodiment and method of the invention.
Fig. 9 shows further details of the MATRIX concept search engine 320 of Fig.
5, in
accordance with an embodiment of the invention.
Figs. 10-19 show various screen shots of search results, in accordance with
exemplary
embodiments of the invention.
Fig. 20 shows further details of the visualizer 201, in accordance with an
embodiment of
the invention.
Fig. 21 shows a computer display or screen shot of reconciled data, in
accordance with an
exemplary output processed by the visualizer 201.
-4-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
DETAILED DESCRIPTION OF THE PREFERRED EMBODIMENT
In the following description of the embodiments, reference is made to the
accompanying
drawings that form a part hereof, and in which is shown by way of illustration
of the specific
embodiments in which the invention may be practiced. It is to be understood
that other
embodiments may be utilized because structural changes may be made without
departing from
the scope of the invention.
Referring now to Fig. 1, medical system 10, in accordance with an embodiment
of the
invention. The system 10 is shown to include medical source 14, a medical
information
navigation engine (MINE) 12, and medical information consumers (also referred
to herein as
"output" or "medical output") 17. The medical source 14 are shown to include
an electronic
health record (EHR) 18, EHR 20, health information exchange (HIE) 22, and a
picture archiving
and communication system (PACS) 24. The MINE 12 is shown to include interface
13, a back-
end medical processor 16, and a front-end medical processor 15.
"Medical information", as used herein, refers to any health-related
information, including
but not limited to patient medical records, patient entered information, care
team entered
information, healthcare device generated information, and billing information.
The source 14 generally provides various medical information to the MINE 12.
For
example, the EHRs 18 and 20 each may provide information such as medical
records and billing,
the HIE 22 may provide information such as medical records, and the PACS 24
may provide
information such as diagnostic imaging and reports.
The medical information consumers 17, which may be made of a host of entities
or
individuals, such as patients, clinics, medical institutions, health
organization, and any other
medical-related party, use information that is provided by the processor 15 of
MINE 12 and that
-5-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
can, by way of example, consist of patients, medical systems, medical
organization
administrators, medical researchers, and/or UR users. For example. user-
customized processed
medical information is provided by the processor 15 to a number of users
within the medical
information consumers 17. In this case, the processor 15 generates user-
customized processed
medical information to a plurality of users, with at least a portion of the
user-customize
processed medical information being provided to each of the users based on the
relevancy of the
portion being provided of each user's specific function or role and each
user's associated
security privileges.
The processor 16, in some embodiments, indexes identifies, maps, and
consolidates
medical information, received from the interface 13, and tags this
information, and determines to
reconcile the tagged information. In some methods and embodiments, information
that is
extracted from images is tagged to enhance recall of search queries. Indexing,
at least in part,
processes document and converts them into formats that allows for quick
searching across a large
collection of documents.
The information in the MINE 12 is encrypted and secure to ensure privacy of
sensitive
medical information.
It is understood that the sources 14 of Fig. 1 includes merely some examples
of the
sources that communicate with the MINE 12 and that other sources, known to
those in the field,
are contemplated. Similarly, the output 17 may be used by those or entities
not discussed herein
but that are contemplated and within the scope and spirit of the invention.
The interface 13 serves to receive information that is in various forms, such
as but not
limited to text, html, CCD, CCR, HL7 and any other type or formatted
information. The
interface 13 then provides to the processors 15 and 16 information, as needed.
-6-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
The processor 16 receives some of the medical information that the interface
13
processes and performs certain tasks to process it, such as indexing, semantic
meta-tagging, and
reconciliation. Indexing takes processed documents and converts them into
formats that make it
easy to quickly search across a large collection of documents. Semantic meta-
tagging embeds
information into the medical information that is relevant thereto and that can
be later used to
search for certain information for the purpose of reconciliation and search,
among many others.
One aspect of consolidation, reconciliation and deduplication, generally
refers to
removing of redundant patient medical records, such as, multiple records for
the same individual
appearing as though the records are for different individuals or multiple data
elements that are
recorded similarly but slightly differently in the different sources. In this
case, the processor 16
recognizes that the records belong to a single individual or are the same data
and just recorded
differently and automatically consolidates them. The patient or a user of the
system 10 may also
manually perform reconciliation. Whether or not reconciliation is performed is
advantageously
determined by the processor 16.
The processor 16 outputs the indexed, tagged and reconciled information to the
processor
15. The foregoing tasks are a generalization and further details of each are
provided below.
The processor 15 performs certain tasks on the information provided by the
interface 13
and the processor 16, which include query, search, presentation, and quality
checking. The
output of the processor 15 is the output of the MINE 12, or output 17.
The MINE 12, through the processor 15, in some embodiments and methods,
invites
members of a medical care team to join it thereby allowing distributed user-
organized care
teams.
-7-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
Querying, as performed by the processor 15, is the ability to receive, as
input, a free text
query, from a user, (i.e., a query without any restrictions on the structure)
¨ and converting the
free text query into commands to a medical search engine, such as Medical
Lexical Search
Engine and the MATRIX (Medical Application Terminology Relationship IndeX)
Concept
Search Engine. using a sophisticated query processing engine optimized to work
with medical
queries. The results of the search engine are sent to the presentation display
planner ¨ which
decides the most relevant presentation given the user's organization and role
(e.g. the provider,
search query program, a healthcare administrator, a study administrator, and
the patient). The
presentation discussed below, receives such information. In some embodiments
and methods,
the medical information or user information is processed to suggest relevant
queries.
Search, as performed by the processor 15, is built around the concept of Zero-
Click
Relevance ¨ or the ability to get to all the relevant information an actor in
the healthcare system
requires by typing in just a single query. The search engine, within the
processor 15, performing
the search comprises an indexing and searching, as will become apparent
shortly. Optionally,
search results may be securely embedded into third party programs. In some
embodiments,
searching involves determining presenting (also referred to herein as
"providing") access to
specific relevant data based on a search query, the patient, and the user's
specific function and/or
role and security privileges. A user may be within the output 17 and security
privileges are
either determined by the MINE 12 or by the patient'or both. Information,
uploaded to the MINE
12, by users, such as output 14, in some embodiments, is searched by the
processor 15. The
uploaded information may include information such as but not limited to status
posts, records,
and images. Such user-uploaded information is routed automatically to the
output 17, as needed.
-8-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
Some aspects of the search are now discussed relevant to an example. Assuming,
by way
of example, that general practitioner Dr. Smith's new patient, Joan Sample,
has a complaint of
chest pain. Joan has brought several ACCDs and a 600-page pdf file of her
chart. She has seen a
cardiologist who uses NextGen and a G.I. specialist whose charts are in a e-
MDs, and has visited
the emergency room. Dr. Smith uses the search of the various methods and
embodiments of the
invention to efficiently assemble the relevant information he needs. Dr. Smith
selects Joan
Sample as the pateient and enters the clinical context "chest pain" n the
search bar of a screen
presented by the MINE 12 (examples of such screens are shown in subsequent
figures herein).
He is presented with relevant lab results, such as CKMB and Amulase, relevant
diagnostic
results, such s EKG and chest CT scan, and all progress notes and consult
reports in which
concepts relevant to chest pain, like "GERD" and "Holier monitor", are
mentioned. Two distinct
types of searches are combined, in accordance with a method and embodiment of
the invention,
to retrieve information medically relevant to Joan's complaint: 1) Lexical
search, whre text in
the patient record is searched for occurrences of the search term, its
variants and synonyms; and
2) Medical concept search, where data that is medically related to the search
term is retrieved.
Medical concept search finds relevant structured data with standardized codes,
such as lab
results, and text results, such as progress notes, which include terms
medically related to the
search term. In Joan's case, a search for "chest pain" returns a CKMB lab
result and a reference
to a chest CT scan. Accordingly and advantageously, the Lexical and Medical
concept search
solves Dr. Smiths' information overload problem by returning information in
the chart most
relevant to Joan's chest pain complaint. Further, in some embodiments, the
presentation,
discussed shortly, presents a united view of Joan's history by reconciling and
de-duplicating data
-9-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
from multiple sources that may be coded and described differently. Redundant
data is
automatically reconciled even if it is described differently by differently
sources.
Presentation, as performed by the processor 15, is displaying health
information to the
requesting user in a way that reduces the number of clicks and maximizes the
amount of
meaningful information delivered based on the interpreting the intent of the
user query.
Quality checking, as performed by the processor 15, is checking of the quality
of medical
information provided by various sources, i.e. source 14, by the patients,
structured data, and
unstructured data, in a Wiki-like mannered setting whereby the users can help
maintain and
improve the quality of information displayed. The foregoing tasks, performed
by the processor
15, are further described in detail below. Additionally, the users or patients
may make comments
regarding medical information, in a Wiki-like manner.
In summary, the MINE 12 transacts medical information including the interface
13
receiving medical information from a number of medical sources (such as within
the source 14)
for processing. identifying, mapping, and consolidating by the medical
processor 16, providing
access to specific relevant data, based on a user's security privileges,
within the identified,
mapped, and consolidated medical information, based on user-specific functions
or roles,
performed by the processor 15, and generating user-customized processed
medical information
to a number of users, such as within the output 17, with at least a portion of
the user-customized
processed medical information being provided to each of the users based on its
relevancy to each
user's specific function or role and each user's associated security
privileges.
Fig. 2 shows further details of the system 10, particularly the MINE 12
thereof. That is,
the processor 16 is shown to include an indexing and metal tagging module 34,
which includes
an indexing module and a meta tagging module (both of which are not shown in
Fig. 2 in the
-10-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
=
interest of clarity), which may be a module, as shown in Fig. 2 or two
physically separate
modules. The processor 16 is further shown to include a reconciliation and de-
duplication
module 36, which also can be broken out into two modules, a reconciliation
module and a de-
duplication module, and a code and semantic mapping module 38, which also may
be a single
module or multiple modules. The modules 34, 36, and 38 communicate with one
another.
The processor 15 is shown to include a presentation and quality checking
module 30,
which may be a single module or broken out into two modules, and a query and
search module
32, which also may be separate or combined modules. The modules 30 and 32
communicate
with each other and with the modules of the processor 16. The interface 13
communicates with
modules of both processors 15 and 16 and is essentially a gateway into the
MINE 12 from the
sources 14.
The foregoing modules may be software programs, executed by a computer or
computing
engine of suitable sorts, or may be implemented in hardware.
Fig. 3 shows an exemplary embodiment implementing the system 10 using various
devices. That is, thc medical systcm 30 is analogous to the system 10 and is
shown to include
the sources 14 coupled to communicate, securely, through the secure
communication link 42, to
the interface 13. The link 42 may be any suitable communication channel
allowing information,
of various formats and types, to be transferred to the interface 13 in a
secure and encrypted
fashion. Exemplary communication channels of which the link 42 is made include
the Internet,
VPN connections over the Internet, private dedicated digital lines such as TI,
T3, El, E3,
SONET, and other fiber optic formats.
The interface 13, in some embodiments, is a software program that executes on
one or
more servers 32, which can be a server of any kind of suitable computing
engine, such as
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
personal computer (PC). The servers 32 receive secure information through the
link 42 from the
sources 14. The processor 16, in some embodiments, includes a database 36 and
one or more
servers 34, which may be any suitable computing engine, similar to the servers
32, including but
not limited to PCs or servers.
The database 36 and servers 34 perform the tasks discussed above relative to
the
processor 16 and the display 40 and servers 38 perform the tasks discussed
above relative to the
processor 15 though these processors may and often perform additional tasks
related to medical
information, some examples of which are presented and discussed below and the
rest of which
are contemplated and achieve the various advantages, results and functions
presented herein.
The processor 15, in some embodiments, includes display and visualization 40
executing
on one or more servers 38, which may be any suitable computing engine, similar
to the servers
32, including but not limited to PCs or servers. The display 40 is used to
construct presentation
and display information to users, such as the patient's records, billing
information, and other
types of medical information. The display 40, in some embodiments, also
performs processing
of sonic of thc functions of the processor 15.
As shown in Fig. 3, the servers 32 are coupled to the database 36 and the
servers 34, and
to the display 40 and the servers 38 and the database 36 and servers 34 are
coupled to the display
40 and the servers 38.
In some embodiments, the interface 13, servers 32, database 36, servers 34,
display 40,
and servers 38 are remotely located relative to the sources 14 and in some
embodiments,
remotely located relative to one another. Further, they are considered a part
of the Internet cloud
where, performing their tasks in a manner known as cloud-computing. However,
other manner
of achieving the functions and advantages of the invention, including various
other of
-12-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
implementation, not shown in Fig. 3 or other Figures herein and/or not
discussed are
contemplated.
Fig. 4 shows further details Of the system 10, in accordance with an
embodiment of the
invention. The system 10 is a block diagram of thereof with arrows shown
linking the blocks to
one another. It is understood that these arrows represent data flow between
blocks and merely
examples of some of the data flows and not all-inclusive thereof.
The system 10 is shown to include medical sources 14, MINE 100, which is
analogous to
MINE 12 of Fig. I with additional details shown, medical output 17, medical
source 14, and
medical input 125. The source 14 is analogous to the source 14 of Fig. 1
except that it is shown
to include electronic medical record 103, HIE 101, healthcare applications 107
and an adhoc data
block 108. The output 17 is analogous to the output 17 of Figs. 1 and 2 except
that it is shown to
include the patient 105, the health care (HC) provider 104, the EMR 103, the
HIE portal 102,
healthcare Apps 107, Standards Orgs 111, and Health Orgs/Payers 110. The input
125 is shown
to include the medical terms 106 and the medical knowledge 109, which are
input to the MINE
100.
The MINE 100 is shown to include a standard parser 207, a document uploader
208, an
adhoc document parser 209, a mapper 204, a relevancy matrix 205, a medical
knowledge
extractor 206, a mapped data objects block 203, a business intelligence block
210, a template
library manager block 211, an Application Programming Interface (API) 102, a
search module
200, and a visualizer module 201, which are some of the blocks/functions
performed by the
MINE 100 while others are contemplated to achieve the various functions and
advantages
discussed herein.
-13-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
The search module 200 performs functions discussed hereinabove and other
functions
that will become further clear with subsequent discussions. Similarly, the
visualizer module 201
performs the functions discussed above with further details provided in
subsequent discussions.
The parser 209, the uploader 208, and the parser 207 provide the capability of
the MINE 100 to
accept information, in a variety of different formats such as a collection of
medical records, in
standard and ad hoc forms, by the source 101. For example, the adhoc data
block 108 may
provide information to the MINE 100 in ad hoc form, which the parser 209
decodes or parses for
=use by other blocks of the MINE 100.
The extractor 206, the matrix 205, and the mapper 204 serve to collectively
collect
medical knowledge and identify, extract and map multidimensional relationships
within them.
Multidimensional relationships are between different types of medical data
such as concepts,
labs, problems, vital, medications, allergies as well as different types of
relationships between
the medical data such as "has comorbidity with- and "cures" for a drug. These
multidimensional
relationships are embodied in the relevancy matrix 205. The block 203 serves
to combine
medical rccords with their relevancy to specific medical concepts. For
example, the specific
medical concept of diabetes is related to the labs for hemoglobin ale and
glucose results, to the
medical concept of diabetic retinopathy, the diagnosis of foot sores, the
medications of
Metformin and insulin among other things. The module 201 serves to perform the
functions of
relevancy and reconciliation and presentation, as described hereinabove. The
business
intelligence block 210 can mine the collected data for trends and predictive
capabilities that
might be of interest to large health organizations and payers 110. The
collected data can be
mined/analyzed/observed/reported on, in aggregate form. Other organizations
that could make
use of the data are any organizations interested in monitoring or required to
report quality
-14-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
metrics such as integrated delivery networks (IDNs), healthplans and/or
accountable care
organizations (ACO). An example of business intelligence queries includes
selecting an arbitraty
cohort population for search of quality measures such as "show me all
patients' information
within my panel with diabetes with hemoglobin ale high in the last six
months".
The sources 14 provides medical information to the MINE 100, through a secure
link, as
noted previously. Within the medical information received by the MINE 100
generally is
medical data or medical records. Before it is received by the MINE 100,
medical data is created
in a multitude of systems including the EMR 103, healthcare applications 107,
and many other
ad hoc data sources 108 (such as but not limited to the application, MicroSoft
Word). The data,
which is typically aggregated in a HIE 101 and EMRs 103 can also be received
by MINE 100.
Structured data is increasingly becoming available in standard `xml' formats
such as continuity
of care records (CCR), continuity of care documents (CCD), clinical data
architecture (CDA),
and other HL7 related standards. Ad hoc data is available in multiple formats
such as word, pdf,
fax, and image data. The collection of medical records from multiple sources
creates a serious
data duplication problem in the current medical environment. Much of the data
from these
sources may represent the same type of information or even the same instance
of information
with slightly different variations or completeness (e.g. Patient name with and
without the middle
initial or a drug labeled as a generic or brand name with or without a start
date). The MINE 100
advantageously reconciles the same data instances for reconciliation purposes
from different
sources and identifies similar data instances for comparison purposes.
As opposed to typical health information exchange (HIE) where patient
information is
available to all the participants in the system, in the ystem 10, patient data
is shared in a
granular patient centric style to accommodate privacy and security. on a per
patient "need to
-15-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
know" basis based on the roles and needs of the users and their organizations.
Examples of the
"need to know" basis of information distribution include the following: (I)
Some organizations
such as those with healthcare providers may receive patient demographics in
addition to the other
medical information. (2) Other organizations such as sponsors may receive only
the problem that
the healthcare provider is interested in. (3) Still other organizations, such
as study clients may
just receive a count of patients that could fit a study candidate criteria and
the be able to message
those patients anonymously. The whole user interface presented by the
presentation module 30
can change based on the rights and type of user. Patient sharing is performed
on a patient or even
more granular basis. Examples of further granularity include type of data such
as labs, meds,
allergies, billing data or medical concepts such as mental health and AIDS
(PC1). Organizations
can share data with each other on an as needed basis this is similar in
concept to sending a fax
containing medical information for a given patient from one organization to
another. The
receiving party (organization or healthcare provider 104), health
organization/payers 110 in Fig.
4, can accept the sharing (similar to filing the received data into an
existing or new patient chart)
or reject the sharing request. In this manner, patient care teams are created
in an ad hoc
distributed way based on care requirements for each particular patient. In
embodiments where
security is a concern, the sharing is only performed between verified
organizations (where
verification is provided by peer organizations), is auditable by organization
administrators and
by the patients themselves. Patients also advantageously have the option to
add/remove members
from their care teams which can also include their relatives, PHRs, and
digital devices and
applications that generate, monitor, or consume healthcare data (PC2).
The collection of medical records in the standard and ad hoc forms is made
possible by
the parser 209, the uploader 208, and the parser 207 of the MINE 100. The
document parser 209
-16-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
parses the ad hoc data and whenever possible intelligently categorizes
elements of the ad hoc
data into structured elements. In some embodiments, natural language
processing, neural
network, user assisted templates, and other techniques are employed to
intelligently categorize
these elements. Intelligent categorization is the process of taking a segment
of input text and
assigning data category and structure based on the data category
probabilistically to the segment
of text. For example, the text "ale was reported abnormal" ¨ would be assigned
the category "lab
result" to the term "ale" and associate the term abnormal as the result flag
to the term "al c" with
some probability. Another example could be "stress test reported no cardiac
malfunction" would
associate the categories "problem" for the term "stress" and "cardiac
malfunction", and the
category "procedure" for the term "stress test". It would also associate
negation to the term
"cardiac malfunction". The document parser 209 then optionally presents the
user with its
analysis of the potential structured data for review and editing, through the
output 103. The
document parser 209 creates a standard XML document such as CCD or CCR formats
that can
be uploaded to the rest of MINE 100 through doc uploader 208. The document
parser 209 also
can provide users with a graphics user interface (GUI) that will allow them to
easily map/identify
structured elements in specific types of ad hoc documents to simplify the
repeat processing of
such documents. For an example, a user could bring up a document on the
document parser and
highlight the tag PatID: and indicate that the patient ID will follow this
tag.
In some embodiments, the document uploader 208 is a computer program that is
executed
in any security domain. These security domains can be cloud based services, a
hospital, a clinic,
a personal computer and other suitable forum. The document uploader 208
receives standard
XML documents such as CCD or CCR, from the parser 209 or directly from the
medical sources
14, and uses this information to verify user information, and securely
(usually via SSL) uploads
-17-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
the document to a desired destination within the MINE 100. MINE 100 maintains
two main
public environments (a product demonstration environment and a production
environment) and
multiple engineering environments. The type of document uploader employed is
optional and a
design choice. For example, the particular document uploader 208 employed in
the MINE 100
depends upon the connectivity required. Documents can be sent to the document
uploader 208
via a variety of methods including but not limited to IHE protocols, web
services, shared folders,
and drag-n-drop browser interfaces.
In some embodiment, the standard parser 207 is a computer program that is
typically
executed in a device in the Internet cloud but can, in other embodiments, be
executed within a
private network environment. The standard parser 207 parses and categorizes
standard elements
from the XML and creates data objects for specific standard elements. Most
uncategorized data
is stored as plain text into an ad hoc document object.
Additionally, the document parser 209 and the standard parser 207
advantageously
identify and remove duplicate patients as well as duplicate documents.
Identification of
duplicate patients is done by comparing patient identification, as received by
parser 209, from
the source 101, to known patients with the MINE 100 and a determination of a
threshold number
of matches of the patient's received identification. For example, a patient
whose social security
matches a known patient record within the MINE 100 may be determined to be the
same patient
or a patient whose last name partially matches another last name and has other
matching
information, may be declared a duplicate patient. Patients can be matched
based on similarities
between demographic, clinical, billing, or any other relevant patient records
within MINE 100.
The medical knowledge extractor 206 saves medical terms 106 (medical
dictionaries,
medical ontologies), medical knowledge 109, in digital formats, and medical
knowledge
-18-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
gathered from the mapped data objects 203. The medical knowledge extractor 206
creates
relationships between different medical concepts found in the medical
knowledge. These
relationships are stored in a series tables called relevancy matrix 205.
The relevancy matrix 205 are a set of granular tables of medical terms, their
synonyms,
additional search terms that are of interest and a set of multidimensional
relationships that
indicate the relevancy of each of the terms (apixions) to other terms
(apixion) along various
dimensions such as "lab for", "cures", "has reported negative interaction"
etc..., to medical
professionals. The elements in the columns and rows of these tables are called
"apixions", also
referred to herein as "atomic healthcare related terms". As is appreciated,
the set of granular
tables grows over time as additional medical terms are identified by the MINE
100.
The relevancy matrix 205 includes tables that are organized at various
information levels
including but not limited to global information, specialty specific
information, standards
organization specific information, user specific information, disease or
problem specific
information, and patient specific information. Advantageously, users
selectively and
independently adjust their versions of the apixion table by performing a
search and then adding
or removing elements that they feel are relevant or not relevant to the intent
of the search they
conducted. The relevancy matrix 205 can process, aggregate, and propagate
these user inputs to
the other information levels.
The mapper 204 receives the structured and ad hoc medical information and the
type (e.g.
medicine. allergy, labs, notes ...) of information, from the parser 207, and
uses the relevancy
matrix 205 to generate mapped data objects 203 that include the medical
information, the type of
medical information and granular terms to which they are related (i.e., the
related apixions).
Apixions can be used to help identify the same data instances from different
sources for
-19-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
reconciliation purposes and to identify similar data instances for comparison
purposes. The
mapper 204 also consists of a set of flexible rules that can be used to
identify exact matches of
data instances and "close" matches of data instances. The mapper 204 also
records the result of
these rules in the mapped data objects 203. Exact matches can be defined as
mapped data objects
203 that are identical in the defined fields of interest. Close matches can be
defined as mapped
data objects 203 that meet some matching threshold on the fields of interest.
The search module 200 is analogous to the search module of the module 32 of
Fig. 2 and
receives the mapped data objects 203, the relevancy matrix 205 and search
input from the user to
generate lists of items for each type of medical information. Federated
(cached or real-time
retrieved data from multiple sources) data is assembled at run time to create
a unified view of the
patient's clinical history via a set of mappings or rules. Data from multiple
sources are either
collected in real-time via standards or proprietary query mechanisms or are
retrieved from a
previously indexed cache of such data. The data from these multiple sources
are then combined
for a single unified view. What is shown in the unified view depends on and is
formed by the
data mapper 204 (via de-duplication rules and the data mappings rules) and
visualizer 201 which
handles the organization rules, rights and privacy rules and the user's role.
The search module
200 also ranks the list of items based on their relevancy to various factors
such as the user input,
a query, relevancy matrix, past user behavior, user role, privacy settings of
the patient data, or
knowledge gained from history of medical information. These results are passed
onto both the
visualizer 201 and the API 202.
To summarize, the data from multiple sources is indexed and meta-tagged by the
data
mapper 204. This data set is then consumed by the visualizer 201. The
visualizer presents the
-20-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
same data differently to different end users (or output 17) based on who the
user is and what role
the user plays in the healthcare spectrum.
In Fig. 4, the API 202, the document uploader 208, and the ad hoc document
parser 209
are included in the interface 13 of Fig. 2. The medical terms 106, the medical
knowledge 109,
the search 200, the visualizer 201, the mapped objects block 203, the mapper
204, the relevancy
matrix 205, the medical knowledge extractor 206, and the business intelligence
block 210 are a
part of the processor 15, and the medical terms 106, the medical knowledge
109, the medical
knowledge extractor 206, the visualizer 201, the mapped data objects 203, the
mapper 204, the
relevancy matrix 205, the standard parser 207, the ad hoc document parser 209,
and the template
library manager 211 are a part of the processor 16, and the HIE 102, the EMR
103, the HC
provider 104, the patient 105, and the health orgs/payers 110 are a part of
the outputs 17.
Document and medical data object search ranking is now described. To make
medical
data objects 203 matching search conditions (Search results 305) even more
relevant, MINE
makes use of a mathematical concept we call Concept Proximity Model (CPM). The
CPM
algorithm returns a numerical value indicating the relevancy between a pair of
documents or
medical data objects 203, or between a medical concept and a medical data
object 203. The
relevancy value can be expressed in various fashions including but not limited
to as an Euclidian
distance between two points in a multi-dimensional space, or in angular
distance, from -Ito 1,
indicating the cosine of the angle separating two vectors with the same
dimensions.
To illustrate the use of CPM in relevancy ranking, the following is an example
of how a
relevancy metric is computed and retuned by CPM that can then be used as part
of the search
strategy. For any given document CPM calculates a numeric address based on the
medical terms
(Apixions) in the Relevancy Matrix 205. The numerical address, which can be
expressed as a
-21-
CA 02831300 2013-02-28
WO 2012/031052 PCT/US2011/050108
string of numbers or the coordinates of a point in a multi-dimensional space,
can be calculated
based on various attributes of the medical data in each document (e.g.
presence of conditions,
symptoms, abnormal labs, etc.) as they relate to Apixions. For instance, let
us assume there are
only the following three Apixions provided to CPM: medical conditions A, B and
C. If a search
string inputted by the user is semantically related to conditions A and C, the
search criteria can
be expressed as a vector, S= [1, 0, 1]. A document that indicates a patient
diagnosed with
conditions B can be expressed at a vector, D1=[0, 1, 0]. A second document
that indicates a
patient diagnosed with condition A can be expressed at a vector, D2=[1, 0, 0].
The angular
distance, AD, between each document pair can be expressed as a normalized dot
product
between each vector pairs. In case of the above three vectors, the angular
distance between
vectors S and D1, would be greater than the angular distance between vectors
DI and D2. This
ranking strategy suggests that in case of the above search criteria, document
2 should receive
higher ranking and hence be listed closer to the top of the list of search
results, than document I.
This ranking is particularly useful in cases where the keyword search
indicates that condition A
is found in both documents, but is mentioned in document 1 in a way that rules
out the condition,
as opposed td document 2, that indicates the diagnosis of the condition has
been established. In
this case, a search strategy using only keywords may not accurately establish
the relevance of
each document to the search term and may list document 1 above document 2. A
simple example
would be a document for a healthy patient explicitly listing negatives for
each condition (e.g. "no
chest pain", "no fever"), which would be incorrectly presented as being more
relevant to these
conditions than a document in which they are diagnosed positively for a
condition.
Reconciliation:
-22-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
For any given patient or document MINE can calculate a numeric address based
on the
medical terms (Apixions) in the Relevancy Matrix 205. The numerical address,
which can be
expressed as a string of numbers or the coordinates of a point in a multi-
dimensional space, can
be calculated based on various attributes of the medical data (e.g. presence
of conditions,
symptoms, abnormal labs, etc.) as they relate to Apixions. For instance, let
us assume MINE has
only the following three Apixions: medical conditions A, B and C. A patient
record, RI,
indicating that the patient has been diagnosed to have condition A and C can
be expressed as a
vector. [I, 0, 1]. Another patient record, R2. that indicates a patient
diagnosed with conditions B
and C can be expressed at a vector, [0, 1, 1]. If we assume that 1) the
patient records are
comprehensive, 2) all three medical conditions A, B and C to have been
accurately diagnosed
and 3) the conditions are chronic, then a system comparing the two vectors can
conclude with a
certain level of certainty that patient records, RI and R2 do not belong to
the same patient, even
though the patients may share the same name.
Such a numerical address has a certain level of uniqueness for patients with a
fair amount
of data points particularly if the patient's record contains many data points
that deviate from the
norm (healthy) and therefore can be used as a parameter in addition to the
patient's ID in order to
establish a unique patient identification. The same strategy can potentially
be applied to
reconciling duplicate documents and data points.
Community-based template review, approval and sharing system is now described.
As
described above the template lib manager 211 and the visualizer 201 control
the display,
position, size and behavior of widgets based on pre-defined default and also
user-customizable
settings. Once an optimal display configuration has been reached, the desired
configuration
(referred to as a "Template") settings can be submitted to an online community
for them to
-23-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
review, test and if desired, rate or even certify the Template. The profiles
of the online
community and their Template submissions can either be kept private or made
public via
configurable privacy settings. All users within a community network would have
access to the
submitted Templates and can test, write reviews or rate the published
templates. A decision body
or board within a given user network has the ability to certify a Template,
making it visible to all
users within the community that the Template has received certification.
Certification description
can have many attributes including date, name of the certification body and
other pertinent
information.
The capability of "Did You Know?" is now explained. The system searches the
history of
the patient and displays information that may indicate significant risk for
the patient or may have
a significant influence on the diagnostic process or treatment plan, based on
the patient's "current
circumstances". A patient's "current circumstances" includes the patient's
current problems,
medications, allergies, available measured information (labs, diagnostics,
etc) and any active
complaints or symptoms they may have. For example, a patient who is
complaining of sore
throat and fever might have past splenectomy information displayed by the "Did
You Know?"
feature, since there is a relation between these circumstances and a serious
complication due to
past splenectomy.
The capability of graphical ongoing display and update is now explained. The
system
has the ability to display time series information in a variety of graphical
formats. Once a piece
of information is dragged onto one of these tools, then all data of that type
is automatically
displayed. As new results are added to the system, they are automatically
added to the graphical
display. In this case, time series data is any kind o information that can be
associated with a date
or date range. An example could be a laboratory measurement of Hemoglobin Al c
that is
-24-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
initially dragged to a trend plot tool. All Hemoglobin Ale laboratory results
for the patient will
be made available to the trend plot for display. Following this, any new
Hemoglobin Al c results
would be automatically added to the trend plot display.
Further details of the functions of the processor 16 of Fig. 2 are now
provided.
Indexing and Meta -raving:
Fig. 5 shows a block diagram of the indexing, querying, and searching
functions of the
MINE 12, in accordance with a method of the invention. An indexer 340 is shown
to receive
documents from the document processor 330 and to provide indexed documents to
the encrypted
index module 350, to be encrypted. The medical query processing engine 302
receives a free
text query 303 and processes the same and provides the processed query to the
medical lexical
search engine 310 and to the matrix concept search engine 320. The engines 310
and 320 also
receive the encrypted document from the module 350 and provide a medical
search engine
output and a matrix search engine output, respectively, to the presentation
display planner 304.
The planner 304 ultimately provides search results 305 as its output. The
engine 302 also
provides output to the planner 304.
The document processor 330, indexer 340, and module 350 are generally located
within
the processor 16, in one embodiment of the invention. The engines 302, 310,
and 320, and the
planner 304 are located within the processor 15, in one embodiment of the
invention.
The document processor 330 receives community patient data 200.0 and patient-
provided
patient data 200Ø1 for processing. The data 200.0 is provided from sources
other than the
patient whereas the data 200Ø1 is provided by the patient.
-25-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
The engine 302 converts free form natural language queries, free text query
303, into
calls to the engines 310 and 320. The engine 310, in a predetermined
collection of medical data
objects (such as text documents, structured or ad hoc), finds all objects
having the keyword term
match (perfect or loose) anywhere in the content of the medical data object
and filters them using
other keywords (?? What is this trying to say?). The engine 320, for a
predetermined collection
of medical terms, finds a list of medically related terms, identified through
a knowledge base
comprising medical terms, semantic relations, and strength of the relations.
The collection of
medical terms is dynamically changing according to user-input and otherwise,
as the system 10
gains medical knowledge. The planner 304 converts the output of the engines
310 and 320 into a
unified display plan for viewing by a user and this becomes the search results
305 ¨medical data
objects that can be matched to either keyword or concepts.
Two data flows are shown in Fig. 5. Indexing flow follows the flow of data
from the
200.0 and/or 200Ø1 to the processor 330, the indexer 340, and the module
350, to the engines
310 and 320. Search flow follows the flow of data from receipt of the query
303 to the engine
302 to the engines 310 and 320 to thc planner 304 to generate the search
results 305.
In the search flow, the engines 302, 310, and 320 are used to process
documents and
convert them into formats that make them easy to quickly search and retrieve
from a large
collection of documents. The indexing flow allows for reliable and automatic
replication of
documents across multiple physical machines. Indexing also supports large
throughput (large
number of documents getting indexed per minute) because it can split the work
load over a large
number of machines, such as multiple processors. The indexer 340 converts the
incoming
healthcare data into a form that makes the querying faster. The indexer 340
also stores the
-26-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
contents of the data in a completely encrypted format so that in case of data
theft, the protected
health information (PHI) is not revealed.
The module 350 stores patient data, which it receives from the indexer 340, in
the form of
an indexed document, in an encrypted format that is also easily searchable and
generates
encrypted indices. This encryption protects patient data from data theft. In
alternative
embodiments, the encrypted indices, generated by the module 350, may be stored
in multiple
processors and queried independently ¨ thus they are fault tolerant and
protect PHI.
The search engine, of the various embodiments of the invention, is built
around the
concept of Zero-Click Relevance ¨ or the ability to get all the relevant
information a provider
requires by entering a single query. That is, all relevant information,
required by a medical
provider, may be retrieved upon receipt of a single query.
As described previously relative to Fig. 5, the search engine consists of two
distinct
components ¨ namely the indexing flow and the search flow. The indexing flow
consumes
patient data from the community (in multiple formats including CDA templates,
CCR, CCD,
PDF, images, etc..) as well as data provided by the patients themselves.
Patient-provided data
can come in the form of patient uploaded material such as status updates or
images, videos and
documents or can be from health monitoring devices connected to the intemet.
The indexing
flow processes each of the documents received by the system and converts them
into structures
that make the searching of this data efficient (Indexer 200.2). These
structures are stored in a
completely encrypted fashion to protect patient information.
The search flow takes as input a free text query (i.e., a query without any
restrictions on the
structure) ¨ and converts the free text query into commands to the Medical
Lexical Search
Engine and the MATRIX (Medical Application Terminology Relationship IndeX)
Concept
-27-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
Search Engine using a sophisticated query processing engine optimized to work
with medical
queries. The results of the search engine are sent to the presentation display
planner (304) ¨
which decides the most relevant presentation given the users role and
privileges and their
organizations roles and privileges (e.g. provider, administrator, quality
manager, and the patient)
and the search query.
Query:
Fig. 6 shows a block diagram of the query module, in accordance with an
apparatus and
method of the invention. The query module 31, also referred to herein as
"medical query
processing engine", is a part of the module 32 of Fig. 2 and is shown to have
the free text query
303 received by the query tokenization module 370, which is shown to provide
tokenized query
to the semantic interpretation block 371, which, in turn, provides interpreted
semantic (semantic
what? Results?) to the query translation block 372. The block 372 provides
query translation to
the lexical search queries block 373 and the cone* search queries block 374.
The engine 31
converts free form natural language queries, or the free text query 303, into
input that can be
used by the engines 310 and 320 by first tokenizing the query 303, performed
by the block 370,
using a large dictionary of medical terms to identify all possible
interpretations of the terms in
the query. An interpretation for a query consists of all the medical terms
that were found in the
query along with their medical types. Thus, tokenizing a query, in some
embodiments; uses a
dictionary of medical terms to identify all possible interpretations of the
terms in the query. It
also consists of any relation that can be recognized between the medical terms
in the query based
on function words such as "with" and "of'.
Each interpretation is scored based on the surrounding context, for example.,
other
words, medical terms and relations present in the query, or the knowledge
provided by the
-28-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
application to the query processor such as the history of previous queries and
user responses.
After scoring all interpretations that are generated, the interpretation with
the highest score is
chosen as the most likely interpretation of the query. The most likely
interpretation of the query
is translated into a lexical search query 373 and a concept search query 374
and sent to the
lexical search engine 310 and the concept search engine 320.
The most likely interpretation is then transformed into calls to the engines
373 and 374
for lexical and concept search, respectively. Once the results are returned
from the searches the
query processing engine annotates the returned results with information about
the query so that it
can send it to the display planner to create an appropriate display plan.
Presentation:
Fig. 7 shows a block diagram of the relevant blocks/functions of the
presentation module of
the module 30 of Fig. 2. A presentation display planner 304 is shown to
receive input from a
presentation personalizer 306 and to provide search results 305 that are in
the form of medical
data objects matching search conditions. The search results 305 receives user
actions 307 and
serves as input to the relevancy metrics block 309, which is shown to provide
input to the
presentation personalizer 306.
The planner 304 receives, as input the annotated results for a query, from the
personalizer
306, and converts them into an ordered (ranked) list of results to be
presented to a user. The
ranking component includes exhaustive medical knowledge available in MATRIX to
decide the
relevance of data objects. Each result can be different in its presentation.
The planner 306 also
records user actions 307 on the output of the search results such as user
clicks and dwell times.
This information is logged internally for computing relevancy metrics and to
provide
-29-
CA 02831300 2013-02-28
WO 2012/031052 PCT/US2011/050108
personalization. For example, a result containing laboratory results can be
displayed as a trend
line over time whereas results containing problems can be displayed with
special markers
indicating the onset of the problems, related medications and procedures for
the problem. A
problem or condition that has been cured can be displayed differently from a
present problem to
indicate that the problem is no longer active. Apart from creating display
plans for each result,
determining its styling, content, and presentation (for example, graph vs.
highlighted text) the
display planner also determines the global order of results. For certain types
of queries (for
example, queries about analytes and lab tests) certain types of results might
be prioritized over
others.
The planner 304 uses logged relevancy metrics along with user data to
personalize the
presentation for the user and the query. On every query and user this data is
captured and stored.
A periodically running background task,(is this restriction necessary?) the
personalizer 306,
analyzes the relevancy metrics captured for each user and query and converts
them internally to a
personalization model which is applied when presenting the search results to
the user at query
time. This process creates a feedback loop increasing the relevancy of results
returned for each
user.
Search:
Medical Lexical search, as performed by the engine 310, extends a traditional
text search
engine by implementing a medical term analyzer, which makes the search engine
aware of the
specifics of medical terminology. For example, search for "heart attack"
should find only
occurrences of the whole term and disregard occurrences of "heart" and
"attack" as separate
-30-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
words. Another example: search for "Alpha-1 -Fetoprotein" should match "0-1-
Fetoprotein"
too.
Search can be performed using each of three following basic search algorithms:
appearing in the searched text in original order. The searched text and search
words may
be stemmed, stripped of stop-words and converted to the lower case before
matching.
= Loose match ¨ Finds all occurrences of a given set of search terms in a
collection of
documents appearing in any order in the search text within N positions of each
other.
Here, N is a matrix of numeric values, which give the maximum and. the minimum
amount of characters that may appear between any pair of search terms.
= String match ¨ The third search algorithm, used by the medical lexical
search engine 310
matches strings of characters as opposite to whole words. Since this search
can find
matches to part of a word, it is useful for the auto-complete function and the
search
suggestion box in the user interface.
Fig. 8 shows further details of the engine 310, in accordance with an
embodiment and
method of the invention. The search terms 303 and the search document
collection 203 are
received, the former by the process search term into query objects block 311,
and the latter by
the document processor 330. The processor 330 processes the searched document
received from
the collection 203 and sends the same to the medical language based spelling
error correction
313, which corrects misspellings and sends the corrected text to the
Positional Search algorithm
-31-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
312 for identification of positions of terms within the text in the document
collection, with the
final search results 305 of medical data objects matching the search. The
search terms 303 is
received by the block 311, which processes the search term into a query
object.
Fig. 9 shows further details of the engine 320 of Fig. 5, in accordance with
an
embodiment of the invention. The input query 303 is received by the medical
query processing
engine 300, which processes the received query using the relevancy matrix 205
and outputs from
the processed query a set of matching medical concepts block 322. The
relevancy matrix 205
uses the matching medical concepts block 322 and filters it to generate only
the relevant medical
concepts (block 321). The relevant medical concepts block 321obtained as the
output received
from the matrix 205 is fed back to the medical query processing engine 300 The
engine 300
further receives searched collection of documents 203, which in combination
with the relevant
medical concepts block 321, is used to generate search results 325. The search
results 325
includes medical data objects relevant to the search term, as previously
discussed.
The engine 300 is knowledge-based and includes a table of medical terms,
concepts,
relational links between concepts and numeric values (relevancy scores)
providing a quantitative
measure of the strength of the relational links. This collection of numeric
values is referred to
herein as the relevancy matrix 205. The normalized relevancy score has a range
between 0 and 1,
with 0 being completely unrelated (orthogonal) and 1 been highly related (e.g.
synonymous). It is
noted that in other embodiments, the opposite or a different range may be
employed, for
example, 1 may be used to reference unrelated and 0 may denote highly related.
The relevancy
scores are obtained through a collection of machine-learning algorithms that
operate over
collections of data obtained by a variety of sources, including, but not
limited to, data generated
by experts and medical practitioners including clinical data stored within the
system, data
-32-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
obtained from usage statistics and data obtained by converting publicly
available medical texts
into knowledge usable within the matrix.
In order to identify all the concepts relevant to the search term, the
knowledge base is
queried with the search term to find all the concepts that are related to the
search term. When all
the matching concepts are found the relevant items can be selected by
filtering the list of returned
concepts based on their relevancy score. The threshold for filtering is a
relevancy search
parameter that determines the stringency of the algorithm, from selecting only
synonyms to
finding all remotely related concepts.
The identified list of relevant medical concepts is passed as an input to the
medical query
processing engine which is then able to generate relevant lexical queries for
the lexical search
engine 320 to return the search results 325.
An additional level of complexity in identifying relations between medical
terms is
created by negative semantic constructs. For example, occurrence of the words
"diabetes ruled
out" in the context of a medical document needs to be interpreted by the
relevancy engine as a
negative relevancy value for diabetes. From the semantic algorithm
perspective, two main
problems associated with negations are detection and interpretation. Negation
detection
algorithm relies on the collection of negative semantic constructs found in
the medical texts and
falling into several categories, from simple negation, like "no fever", to
more complex patterns
like "no sufficient evidence of infection". Multiple negations, as well as
double negations present
a significant challenge to the automatic detection algorithm (for example:
"ruled out structural
abnormalities, injury, but not inflammation")(Are we trying to make the
assertion that we solve
this difficult problem? Perhaps we just state that these are examples of
negation that the system
-33-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
handles . The negation detection algorithm uses a machine learning model to
learn structural
patterns in sentences to identify if the phrases in the sentence are negated
or not.
Interpretation of negative relevance values depends on the specific use case.
When
application requires only detection of relations between concepts with no
discrimination for their
type, negative relevancy values can be used as absolute values. However,
building decision
support elements requires understanding of the semantic interactions between
medical data
objects, so interpretation of negative relevancy values becomes critical.
Figs. 10-19 show various screen shots of search results, in accordance with
exemplary
embodiments of the invention.
Fig. 10 shows a screen shot of an exemplary search result 325. Patient Joan
Sample's
information and lab results are presented showing ranges of her various test
results with these
results characterized by the relevancy matrix, in accordance with the
discussion above. For
example, calcium is noted having a relevancy value of 0 while glucose is noted
to have a
relevancy value of 1Ø
Fig. 11 shows a screen shot of an exemplary output from Apixio Search 200 as
presented
by the visualizer 201..
Fig. 12 shows a screen shot of an exemplary page of a display to the user with
default
status showing. At the start, the page is empty of most/all widgets, featuring
a welcome widget.
Visible items include Input bar and associated tabs (Patients, Favorites,
History), a Settings link,
and a Sign Out button. Upon the user typing in the input, a drop-down of
potential patient
names, conditions, or related searches appears. Patient matches listed in the
drop-down include
matching on first-name, last-name, and nicknames/psuedonames (ie. "John Doc
Feb 5") if
necessary. (birthdate, ID?) Down-key begins navigating the drop-down list. Up
and Down keys
-34-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
can step through the list, where the current item is highlighted visually.
Similarly, mouse-over on
the list highlights drop down choices. As shown in Fig. 13, a click (touch)
selects the item, thus
populating the input-field.
If the user begins or resumes typing characters or presses backspace, the
stepping process
stops and the functional process resumes to compiling and narrowing the list
of choices. If the
list exceeds 10 items (desktop) and 6 items on mobile devices, the list
accommodates pagination
elements. The bottom strip of the panel should display a "next..." feature.
When the "next..."
feature is used/accessed, the top-most segment displays (changes to) a
"prey..." feature. The
Next and Prey (displayed at the bottom and top respectively) should not be
accessible by up and
down arrow. However a Right or Left arrow key-press should activate/flash
their appearance
and paginate the list forward (right-arrow) and backward (left-arrow). On
touch devices, a press
of either Next or Prey, paginates forward and backward along the list. As with
known search
standards (Google, Yahoo, etc), each keystroke narrows the field of possible
pre-selected
choices. If no matches result, the list will disappear.
List choices that arc already in a corresponding panel (such as an admitted
patient, or
prior condition patient has been seen for) render prefixed with a star. If the
Health Care Provider
(FICP) enters a query item with no matching records, a search results widget
or pane in general
the page should list all related possibilities in a list format.
An exemplary item selection is as follows:
Tab key or Enter key press selects the highlighted item and locks the
parameter as an
object. A mouse-click on a list item selects the highlighted item. A click or
touch selects
the list item.
Once an input item is selected from a drop-down, the corresponding input
overrides (or
completes) to match the official listing, and changes visually becoming blue,
appended
by a + symbol to indicate the item is an object.
-35-
CA 02831300 2013-02-28
WO 2012/031052 PCT/US2011/050108
The patient-search object can then be operated on.
I. Search/Input Objects
Once search or input item is selected, it transforms visually into an object.
As objects,
input field items can be operated on.
= Dragging the Patient-object to the Patients tab adds the patient to the
Patients tab.
= Dragging the Complaints-object to the Favorites tab adds the item to the
favorites
list.
= Dragging objects to an operational widget may act upon that object (as a
phrase or
parameter that defines the widget.
General Search/Input box behavior is now discussed, as follows with an
exemplary
screen shot shown in Fig. 14:
= If patient has not been identified, then the patient matches or
organization/cohort/panel matches (for population search) are prioritized in
the
dropdown list.
= Once patient name or population group is locked/identified, the drop-down
list
features medical-terms and NOT patients or population groups.
= Backspace over any single item, mid-phrase or upon the plus-sign or other
delimeter,
will unlock that Object (rendering in gray/default) and redefine the items on
the page.
= Backspace deletes within the terms and phrase, character by character.
= Escape will bring back the last object
= Double clicking on a word selects the entire phrase (multiple words
comprising the
term).
= Unrecognized words and terms become a "Keyword Object" and are featured
in
orange after being operated on by a search.
Tabs and panels, are shown in an exemplary screen shot, in Fig. 15, and are as
follows.
Clicking on a tab opens the corresponding panel (using a Nuery animation) and
changes focus
to the list (auto-selecting the first one), as shown in Fig. 16. In this mode,
typing keystrokes
jumps to closest matching list-item. 'Up' and 'Down' arrow steps through the
list. 'Tab' or
'Enter' selects the highlighted favorite item, thus populating the
search/input box. 'Esc' (escape)
closes the panel and returns focus to the existing search/input. After a panel
opens, its tab
features a reversed triangle icon indicating it will close. Click/touch of the
tab or Close icon, or
press of the Esc key will close the panel and return focus to the
Search/Input. Clicking outside
-36-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
the panel will also close the panel. Selection of a panel list's item closes
the panel and returns
focus to the Search/Input. The panels features an 'Edit' button for a list
management capability,
as shown in Fig. 16.
Panel edit mode is shown in Fig. 17 and operates as follows. Clicking the
'Edit' button
transforms the panel to feature 'move' icons and 'delete' icons after each
item in the list, as
shown in Fig. 18. Delete icons are grayed-out by default.
Click on the 'Delete' icons toggle the icon between gray and red modes.
Dragging
(mouse or touch) of the list item repositions the item in the list. A 'Delete'
activator icon and an
'Edit' icon replace the 'Edit' button. Click of the 'Delete Activator' icon
deletes all items
toggled "on" in the list, and the list stays open. Click of the 'Edit' icon
ends the list management
mode (and returns to view of normal open panel).
The 'Patients Tab (Panel)' is as follows. The panel contains a list of
patients the user has
dragged to the Patients tab, ranked in the order they have been added (last
item goes at the end).
This list functions as a short-list of patients the HCP is monitoring closely.
Features scheduled-
patients in chronological order (listing all the days patients). Patients
already seen should be
subdued (visually less apparent). Upcoming patients should be visually
neutral. Admitted
patients (i.e. checked-in by clinical staff), should stand out visually
prominent (be highlighted).
Selection of an item from the Patients panel replaces all items in the
Search/Input box and locks
in the Patient parameter.
The favorites tab (panel) is as follows. Features a list of chief-complaints
and disorder
items is selected by the FICP for quick access. If the Patient Parameter has
been locked in,
features a list of chief-complaints the patient has been seen for.
-37-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
The History Tab (List) features a list of complaints the patient has
previously been seen for,
listed chronologically with the most recent on the top. The settings feature
is used by clicking
the Settings link to activate a lightbox popup with the following parameters
for the HCP to set
their preferences. Table I below is an exemplary table of the setting link,
activated.
______________________________________________________________
Setting Function Options
;
Patient name order
' Auto-signout Sets the time parameter Off, 1-
minute, 3-minutes, 5-minutes,
RFID departure
Future settings
Language Sets the language TBD
TABLE 1
An alternate view is to provide suggested searches based on multiple
parameters such as
the user's specialty, their historical searches, the patient's data and any
other parameters that can
be used to predict the user's desired search intent. This dropdown could also
show other items
such as favorite searches. This search is modality is shown in Fig. 19.
Visualizer:
The search 200 module takes the mapped data objects 203, the relevancy matrix
205 and
search input from the user and creates lists of items for each type of medical
information. The
search 200 also ranks the list of items based on their relevancy to the user
input. These results are
passed onto both the visualizer 201 and the API 202.
The visualizer 201 takes the search context input from the user in a natural
language text
box and provides widgets that react to the search context to provide specific
mapped data objects
203 that are relevant to the context. In addition the widget content, the
layout of the widgets
themselves can be context driven. The layouts can be standardized by the
template lib manager
211. Examples of context include but are not limited to patient, problems,
diseases, specialty,
-38-
=
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
laterality, and demographics. There are multiple types of widgets, those with
access to protected
health information (PHI) and those without access to protected health
information. Example of a
PHI Widget would be a widgets that brings up meds, labs, allergies, problems,
diagnostic
reports, office notes and so on. The PHI Router 406 is responsible for routing
the authorized data
to each of the widgets. These types of widgets can receive the mapped data
objects 203 in
addition to context information, they can then combine the received
information with other local
or web based information to produce unique context relevant display for the
users. Example of
non-PHI Widgets include widgets that display sponsorship, encyclopedia, or ad
information in
response to the given non-PHI context such as problems, or diseases.
The visualizer 201 also makes it easy bring up frequently requested contexts
(e.g. favorite
patients, diseases or problems). Further details of the visualizer 201 GUI are
found in the UX
design document. The visualizer 201 also takes the search result by type and
performs additional
type based processing such as data reconciliation between multiple items of
the same type from
different sources. The displayed text and coding system of data presented by
the visualizer 201
can be customized based on viewing user's preferences. The data reconciliation
can be
automated as well as user assisted. Depending upon the type of data and the
type of matching,
the reconciliation can be performed at import and persisted in the database or
can be performed
for display purposes only by the visualizer 201. Mapped data objects 203 are
reconciled by the
visualizer 201 in multiple ways based on whether they are exact matches of the
same data
instance, a potentially "close" match of a given data instance, or are
identified as a the same type
of data for comparison purposes. Rules for data matching and discrepancy
flagging can be based
on organizational or locally defined preferences, rules and/or mappings. Exact
matches can be
combined on display without flagging. Depending upon the data type close
matches are either
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
combined with a flag indicating a reconciliation occurred or are flagged (or
marked) to indicate a
potential reconciliation could occur.
The visualizer 201 can also take input from the user (healthcare provider 104)
as to the
level of relevancy of the information that they are interested in. The
visualizer's 201 layout can
also be customized by the user and by standards organizations who can use the
template library
manger 211 to manage and publish context specific layouts. The template
library manager 211
can manage layouts at the individual, group, specialty, and standards
organization level.
Fig. 20 shows further details of the visualizer 201, in accordance with an
embodiment of
the invention. The term "widget", as used herein, refers to a developer-
created entity that
populates the module spaces in the page, including all code, and its
integrating components. A
developer.is a third party or a user of the MINE 100. The arrows linking the
blocks of Fig. 20
show the major data flows, other connections between the modules and data flow
in the reverse
directions are anticipated. The functions of the blocks of Fig. 20 are as
follows:
I. The Device Manager 401 manages the major device specific layout and
functionality
differences.
2. The Layout Manager 402 manages the location, size, and repositioning of
the widgets.
3. The Widget Manager 403 manages the content of each of the widgets. Key
functionality
includes:
3.1. Display Merger 411 functionality combines multi-sourced items for a
simple view of the
data, it can take into account the among other things type of data, the
sources, and who is
viewing the data.
3.2. Flagger 412 provides visual feedback on the state of merged items
-40-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
3.2.1. No pill is displayed if multi-sourced items are considered the same by
organizational specific rules.
3.2.2. Gray pill is displayed if multi-sourced items are considered similar by
the
organizational specific rules.
3.2.3. Orange pill is displayed if the multi-sourced items are merged
manually.
3.2.4. Other colors could be used for further annotation of merged items.
3.3. Search highlighter 413 highlights items that are relevant to the search
performed
3.4. Widget collapse manager 414 collapses widgets that are do not return
relevant
information
3.5. Relevancy visualizer 415 provides mechanisms to gather specialty/user
specific
feedback on the relevancy of items by display lower relevancy items in a
compact form
such as a comma separated list and allowing the user increase its display
relevancy by
clicking on it
3.6. Details visualizer 416 provides the ability to see all the details of an
item by clicking on
it, hovering over it, or other user gestures or actions
4. Widget Navigation 404 allows users to rapidly get to the widget of
interest and may affect
the search bar/query content
5. Overlay Manager 405 displays large drill down data (eg. office notes or
images) in the main
window as an overlay. Overlays can contain one object at a time or can contain
multiple
objects for comparison purposes.
6. PI-II Router 406 routes data from the mapped data objects 203 to the
appropriate widgets
based on the type of widget and the type of information it needs to display.
The PHI router
can discriminate between may types of indexed, mapped, and consolidated data
and takes
-41-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
into account the user's role and organization and the security policies
governing each discrete
medical data item and its meta-tags. .
The API 202 exposes the search bar and search results to third party programs
as well as
allows third parties to publish their data in widgets presented by the
visualizer 201. As an
example, the search bar and search results can be embedded securely into third
party programs
using either a REST or SOAP APIs. Examples of security mechanisms include
encryption,
authentication credentials, context, and tokens. Third party developers can
take context
information provided by the search API (for example by a secure SOAP API) and
combine it
with their own data to produce customized widgets and data in the visualizer
201.
Fig. 21 shows a computer display or screen shot of the consolidated
(reconciled and de-
duplicated data), in accordance with an exemplary output processed by the
mapper 204 and
presented by the visualizer 201. The flag, for example, a pill shaped icon
with a number in it,
indicates the number of records that were merged. Clicking on the flag allows
the user to see the
merged records, change the one that is displayed, the master record, and/or
unmcrge the records.
Mapped data objects 203 that are merged or umnerged by users can be propagated
to all users
within an organization, or on the Care Team of the patient, thereby building
an improved Wiki-
styleview of the patient. This propagation of user edits can also be performed
across
organizations.
Although the invention has been described in terms of specific embodiments, it
is
anticipated that alterations and modifications thereof will no doubt become
apparent to those
skilled in the art. It is therefore intended that the following claims be
interpreted as covering all
such alterations and modification as fall within the true spirit and scope of
the invention.
-42-
CA 02831300 2013-02-28
WO 2012/031052
PCT/US2011/050108
What is claimed is:
-43-