Note: Descriptions are shown in the official language in which they were submitted.
CA 02839854 2014-01-17
2D-3D RIGID REGISTRATION METHOD TO COMPENSATE FOR
ORGAN MOTION DURING AN INTERVENTIONAL PROCEDURE
TECHNICAL FIELD
[0001] The present relates to ultrasound imaging techniques, and more
particularly to an
image-based registration algorithm for 2D to 3D rigid/affine ultrasound image
registration
technique.
BACKGROUND
[0002] Prostate cancer is the second most frequently diagnosed cancer among
men in North
America [1], with prostate biopsy as the clinical standard for diagnosis.
During biopsy, the
physician systematically obtains approximately a dozen tissue samples from
different regions of
the prostate to assess disease status via histopathology analysis of the
extracted tissue. Prostate
biopsy is usually performed under two-dimensional (2D) trans-rectal ultrasound
(TRUS)
guidance by inserting a needle through the patient's rectal wall. However,
with the small size of
the biopsy cores taken, the presence of small, multi-focal cancers might
result in negative
biopsies. In fact, the false negative rate of the 2D TRUS-guided biopsy
procedure is reported to
be as high as 30% [3]. Poor visibility of cancer in TRUS images and the
limited anatomical
context available in the 2D TRUS plane make it challenging for the physician
to accurately guide
needles to suspicious locations within the prostate. With the aim of improving
the cancer
detection rate, systems have been developed [4, 5] that can plan and record
biopsy locations in a
3D TRUS image acquired at the beginning of the biopsy procedure. Target biopsy
locations can
be identified in the 3D TRUS image with the assistance of a magnetic resonance
(MR) image
acquired prior to the biopsy session, in which focal lesions are more visible.
The 3D TRUS
image can then act as a baseline image, to guide the physician to the target
biopsy locations by
augmenting the 2D TRUS planes acquired during biopsy with 3D contextual
information.
[0003] Although early reports of 3D TRUS guided systems are promising, some
limitations
have been identified that require attention [6]. Patient motion and ultrasound
probe pressure can
cause the prostate to move and deform during the biopsy procedure. This may
lead to a
1
CA 02839854 2014-01-17
misalignment between the targets identified in the initially-acquired 3D image
and their
corresponding locations within the patient's prostate as depicted by the real-
time 2D TRUS
images acquired throughout the biopsy procedure. Compensating for the prostate
motion and
deformation by registering the pre-acquired 3D image to the live 2D images
acquired throughout
the procedure is an important step toward improving the targeting accuracy of
a biopsy system.
[0004]
Previous approaches to compensation for prostate motion during biopsy have
involved
mechanical stabilization of the ultrasound probe, 3D tracking of the probe,
and the use of
biplanar or 3D transducers to continuously acquire richer image information
supporting
software-based motion compensation algorithms [4, 5, 7, 8]. The mechanically
assisted 3D
TRUS-guided biopsy system developed in our laboratory and described in detail
in [4], uses a
passive mechanical arm to track the position and orientation of the ultrasound
probe during the
biopsy procedure. The design yields a remote centre of motion positioned at
the centre of the
ultrasound probe tip that provides enhanced stability to the TRUS probe
minimizing prostate
motion. Several methods have been proposed in similar 3D TRUS-guided biopsy
systems to
register real-time TRUS images during the procedure to an initially acquired
3D image [5].
The 3D TRUS-guided biopsy system presented in Xu et al. [5] uses a magnetic
tracking method
to locate the ultrasound plane and it then performs an intermittent rigid
registration to compensate
for out-of-plane prostate motion; the registration is invoked when
misalignment is detected
visually by an operator. The magnetic tracker transform provides an
initialization for the 2D
TRUS plane within the world coordinate system in their system. In that work,
however,
registration accuracy was measured with a phantom study. Baumann et al, [7]
presented a method
relying on the simultaneous real-time acquisition of dual, orthogonal 2D TRUS
images acquired from
a 3D ultrasound probe. The same authors presented an algorithm [8] to
compensate for motion
using 3D TRUS volumes acquired continuously throughout the biopsy session.
This system
does not use any method to track ultrasound probe motion; therefore, it relies
only on the
image information for tracking and uses a coarse-to-fine image-based approach
to limit the search
space during optimization. In addition, this approach requires a special 3D
ultrasound probe with
enhanced functionality that could simultaneously acquire orthogonal 2D TRUS
planes, and image
acquisition occurs at a lower frame rate, compared to more conventional 2D
TRUS, Moreover,
2
CA 02839854 2014-01-17
compared to 2D TRUS images, orthogonal 2D planes deliver considerably more
spatial
information; registration of a single 2D TRUS plane to a 3D TRUS image is a
more challenging
problem.
[0005] To the best of our knowledge, no previous work has described and
evaluated on human
clinical images a method for the registration of 2D TRUS to 3D TRUS images for
prostate motion
compensation during biopsy. Such a technique, if properly validated, will make
it possible to
perform prostate motion compensation on 3D biopsy guidance systems that use
readily
available 2D ultrasound probes for live image acquisition throughout the
procedure, permitting
more widespread use of targeted biopsy systems and thus greater potential
impact on the patient
population. 2D-3D registration methods have been applied to several other
interventional
applications in image-guided procedures (see Markelj et al. [9]). Birkfellner
et al. [ 10]
compared the performance of several image similarity measures and optimization
techniques
for 2D-3D registration of fluoroscopic images and found that cross-correlation
is an optimal
metric for intra-modality matching. Wein et al. [11] presented a method to
compensate for
respiratory motion during abdominal biopsies and ablations under ultrasound
guidance,
optimizing local normalized cross-correlation using the Powell-Brent direction
search technique.
Although these previous successes speak to the potential feasibility of
addressing the issue of
prostate motion compensation in software using a 2D-3D intensity-based image
registration
technique, prostate appearance on TRUS and motion characteristics during
biopsy may differ
from those of other organs due to different tissue stiffness properties and
flexibility of
surrounding anatomical structures.
[0006] A number of methods for compensating for respiratory motion during
image-guided interventional procedures are known in the art. Among these are
included
breath-hold methods, gating methods (published US patent application no.
2012/0230556), and
real-time tracking methods (US patent no. 8,155,729).
[0007] Another approach taught in the art is for estimating motion of an
organ and then
transformation of the image as taught by published US patent application no.
2008/0246776.
A further approach is to incorporate a mode of respiratory motion into the
registration to
3
CA 02839854 2014-01-17
compensate for the respiratory motion and registration of pre-operative
volumetric image
dataset with the intraoperative image as disclosed in published US patent
application no.
2010/0310140.
[0008] Methods of computing a transformation linking two images is known in
the art as
taught by US patent no. 6,950,542. Methods of registration of images are also
known. US
patent nos. 7,912,259 and 7,616,836 teach the use of multiple feature masks
motion
compensation between first and second images in a temporal sequence and
deriving a
displacement between two images is known. In particular, 2D-3D registration
methods that
compensate for organ motion have been applied to several other interventional
applications
in image-guided procedures. Among these is registration of 2D images with a 3D
reference
image as disclosed in US patent no. 8,317,705. Also, Wein et al. developed a
method of
acquiring a pre-procedure ultrasound sweep over a whole liver during a breath-
hold. This
data serves as reference 3D information. The real-time ultrasound image is
tracked and a
position sensor attached to the patient's skin is employed to detect movement
due to breathing
motion of a target within the liver. Respiratory motion can be compensated
using a slice-to-
volume registration approach. The method optimizes local normalized cross
correlation
(NCC) using the Powell-Brent direction search technique.
[0009] A number of research groups and companies are working on developing
solutions
for compensating for respiratory motion during image-guided interventional
procedures,
including breath-hold methods, gating methods, and real-time tracking methods.
Breath-hold
and gating techniques have the disadvantage of increasing treatment time and
can be
uncomfortable for patients.
[00010] One known approach that is being used for radiotherapeutic treatment
of lung
cancer involves using respiratory gating to compensate for motion. The method
involves
tracking tumor motion/location in x-ray images by using a robot-mounted linear
accelerator
(Accuray Cyberknife).
4
CA 02839854 2014-01-17
[00011] Another current approach that has been developed for motion
compensation is to
track ultrasound transducers and/or magnetically tracking needle tips (Traxtal
Inc., CAS
Innovations AG, etc.). This system involves alignment of pre-operative CT or
MRI images
during the breathing phase of pre-operative CT or MRI images.
[00012] Research groups have also proposed creating pre-operative models of
the liver
motion from 4D MRI acquisitions for a patient and registering the model to
tracked 2D
ultrasound images, using PCA based methods. However, this approach is
expensive, time-.
consuming and cannot reproduce breathing irregularities which vary from
patient to patient.
[00013] 2D-3D registration methods that compensate for organ motion have been
applied
to several other interventional applications in image-guided procedures [see
Markelj et al.
paper]. Please refer to De Silva et al. paper. The respiratory motion can be
compensated for
"using a slice-to-volume registration approach" and "optimizes local
normalized cross
correlation (NCC) using the Powell-Brent direction search technique".
[00014] 3D TRUS-guided systems have been developed to improve targeting
accuracy
during prostate biopsy. However, prostate motion during the procedure is a
potential source
of error that can cause target misalignments.
BRIEF SUMMARY
[00015] We have developed a new and non-obvious 2D-3D intensity-based and
geometric
based image registration technique to compensate for prostate motion with
sufficient accuracy and
speed to be translated to clinical use for 3D biopsy guidance. Our method is
applicable to both
prostate, liver and other organs/tissues and includes features that increase
the speed of the
algorithm. In addition to medical applications, motion compensation has
applicability to
fields requiring the ability to detect and track moving objects/targets ,
including machine
vision (i.e. dynamic image analysis, object and environment modeling),
military, sensor (i.e.
object tracking), and mining.
[00016] Accordingly, there is provided a method for generating a motion-
corrected 20
image of a target, the method comprising:
CA 02839854 2014-01-17
acquiring a 3D static image of the target before an imaging procedure;
during the procedure, acquiring and displaying a plurality of 2D real time
images of
the target;
acquiring one slice of the 3D static image and registering it with at least
one 2D real
time image;
correcting the location of the 3D static image to be in synchrony with a
reference
parameter; and
displaying the reference parameter corrected 2D image of the target.
[00017] In one example, the method includes: displaying 2D real time images as
an
ultrasound video stream collected at a video frame rate of up to 30 frames per
second.
[00018] In one example, the method further comprising: matching and minimizing
target
goals or metric values for the 2D real time images.
[00019] In another example, the method described above, in which the 2D-3D
registration
is rigid/affine. Local optimization searches the minimized value which mature
of a 2D slice
inside a 3D volume image. Global optimization searches the minimized value
which mature
of a 2D slice inside a 3D volume image. Estimated values are estimated from a
few prior
output parameters of the successful 2D-3D image registrations and the priori
from last period
of respiration. The estimation can be a polynomial or Fourier series.
[00020] In another example, the method, described above, in which one slice of
the 3D
static image is matched to the correct plane as the 2D real time image.
[00021] In another example, the method, described above, in which the
reference
parameter is body movement. The 2D real time image is matched according to the
body
movement.
[00022] In another example, the method, described above, the registering of
the 2D and
3D images are done visually.
[00023] In another example, the method, described above, the registering of
the 2D and
3D images are done by identifying corresponding points in the 2D and 3D images
and
finding the best translation/rotation/shearing transform to achieve
approximate registration.
6
CA 02839854 2014-01-17
[00024] In another example, the method, described above, for each 2D real time
image:
determining the corresponding plane in the 3D static image; and
finding the corresponding 2D real time images in the 3D static image volume to
determine
which slice therein matches the 2D real time image.
[00025] In another example, the method, described above, further comprising:
minimizing errors or metric values in registering of the 2D and 3D images by
applying a local optimization method.
[00026] In another example, the method, described above, further comprising:
minimizing the errors or metric values in registering of the 2D and 3D images
by applying
Powell's optimization algorithm.
[00027] In another example, the method, described aboveõ further comprising:
[00028]
minimizing the errors or metric values in registering of the 2D and 3D images
by applying particle swarm optimization to calculate degree of matching
between the 2D and
3D images. Powell's optimization algorithm minimizes registration error
measurement by
calculating the target registration error (TRE). Powell's optimization
algorithm minimizes
registration error measurement by calculating the metric value using manually
identified
fiducials in the target.
[00029] In another example, the method, described above, the multiple initial
parameters
for 2D-3D image registration include the output parameters of the prior 2D-3D
registration;
the estimated output parameters using a group of the prior 2D-3D registration;
or the output
parameter of 2D-3D registration from last period of respiration. The particle
swarm
optimization increases the registration speed when matching large high-
resolution 2D and 3D
images comparing with other global optimization method.. Powell's optimization
algorithm
or the particle swarm optimization is continuously applied throughout the
procedure by
acquiring and registering the 2D real time images every 30-100 millisecond.
[00030] In another example, the method, described above, if the local
optimization method
fails, a global optimization method is applied, the global optimization method
being particle
swarm optimization method. The registration is carried out as a background
process to
continuously compensate for motion during the procedure.
7
CA 02839854 2014-01-17
[00031] In another example, the method, described above, a graphics processing
unit
(GPU)-accelerates the registration.
[00032] In another example, the method, described above, the target is the
liver.
[00033] In another example, the method, described above, the target is the
prostate gland.
[00034] In another example, the method, described above, the 2D and 3D images
are
TRUS images.
[00035] In another example, the method, described above, the imaging procedure
is an
interventional procedure. The interventional procedure is a biopsy procedure.
[00036] In another example, the method, described aboveõ the imaging procedure
is
remote sensing (cartography updating),
[00037] In another example, the method, described above, the imaging procedure
is
astrophotography,
[00038] In another example, the method, described above, the imaging procedure
is
computer vision in which images must be aligned for quantitative analysis or
qualitative
comparison.
[00039] In another example, the method, described above, in which the 2D-3D
registration
is non-rigid.
[00040] According to another aspect, there is provided a method for generating
a motion-
corrected 2D image of a target, the method comprising:
acquiring a 3D static image of the target before an interventional procedure;
during the procedure, acquiring and displaying a plurality of 2D real time
images of
the target;
acquiring one slice of the 3D static image and registering it with at least
one 2D real
time image;
correcting the location of the 3D static image to be in synchrony with body
motion;
and
displaying the motion corrected 2D image of the target.
8
CA 02839854 2014-01-17
[00041] According to another aspect, there is provided a system for generating
a motion-
corrected 2D image, the system comprising:
an ultrasound probe for acquiring data from a target during an interventional
procedure;
an imaging device connected to the ultrasound probe for displaying data
acquired by
the ultrasound probe;
a computer readable storage medium connected to the ultrasound probe, the
computer
readable storage medium having a non-transient memory in which is stored a set
of
instructions which when executed by a computer cause the computer to:
acquire a 3D static image of the target before the procedure;
during the procedure, acquire and display a plurality of 2D real time images
of the
target;
acquire one slice of the 3D static image and register it with at least one 2D
real time
image;
correct the location of the 3D static image to be in synchrony with body
motion; and
display the motion corrected 2D image of the target.
[00042] According to yet another aspect, there is provided a system for
generating a
motion-corrected 2D image, the system comprising:
a probe for acquiring data from a target during an imaging procedure;
an imaging device connected to the probe for displaying data acquired by the
probe;
a computer readable storage medium connected to the probe, the computer
readable
storage medium having a non-transient memory in which is stored a set of
instructions which
when executed by a computer cause the computer to:
acquire a 3D static image of the target before the procedure;
during the procedure, acquire and display a plurality of 2D real time images
of the
target;
acquire one slice of the 3D static image and register it with at least one 2D
real time
image;
9
CA 02839854 2014-01-17
correct the location of the 3D static image to be in synchrony with a
reference
parameter; and
display the reference parameter corrected 2D image of the target.
BRIEF DESCRIPTION OF THE DRAWINGS
[00043] In order that the discovery may be readily understood, embodiments of
the
invention are illustrated by way of example in the accompanying drawings.
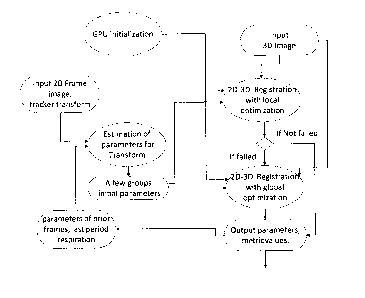
[00044] FIG. 1 is a flow diagram showing 2D-3D registration workflow;. FIG.
1(a) The
outside connections of 2D-3D registration workflow. FIG. (b) The inside of 2D-
3D
registration workflow.
[00045] FIG. 2(a), 2(b), 2(c) are histograms of TRE before and after
registration for
prostate biopsy protocol data. FIG. 2(a) shows before registration; FIG. 2(b)
shows after
registration; and FIG 2(c) showing after continuous registration after every
second;
[00046] FIG. 3 is a histogram showing TRE before registration, after
registration and after
continuous registration every second for each biopsy in biopsy prostate
protocol;
[00047] FIG. 4 are images before and after registration. The Left column
illustrates real-
time 2D TRUS images; the Middle column illustrates corresponding images before
registration assuming to prostate motion (from the transformation given by the
mechanical
tracking system); and the right column illustrates corresponding images after
registration;
[00048] FIG. 5(a), 5(b), 5(c) are graphs showing TRE as a function of time
elapsed from
the start of the biopsy. FIG.(a) TRE before registration; FIG.(b) TRE after
registration; and
FIG.(c) TRE after registering the images acquired every second;
[00049] FIG. 6(a), 6(b), 6(C) are histograms for TRE before and after
registration for
probe pressure protocol data. FIG. 6(a) shows TRE distribution before
registration; FIG. 6(b)
shows TRE distribution after registration; and FIG. 6(c) shows TRE
distribution with the best
rigid alignment for the identified fiducials;
CA 02839854 2014-01-17
[00050] FIG. 7 is a graph showing TRE as a function of metric value during the
optimization. Initial points (circles), converged (squares) and converging
points (crosses);
[00061] FIG. 8 is a graph showing TRE distributions before registration,
during
convergence and after registration;
[00052] FIG. 9 are graphs showing mean and standard deviations of normalized
cross-
correlation values for 16 image pairs of eight patients in the six-degrees-of-
freedom
transformation space, one degree-of-freedom varying at a time. The zero
location in the x-
axis corresponds to real-time 2D-TRUS frame;
[00053] FIG. 10 are graphs showing normalized cross-correlation values for a
single
image pair of a biopsy for 3 patients (each biopsy represented by a separate
line pattern) in
the six-degrees-of-freedom transformation space, one degree-of-freedom varying
at a time.
The zero location in the x-axis corresponds to real-time 2D-TRUS frame; and
[00054] FIG. 11 is a graph showing TRE as a function of distance to the probe
tip.
[00056] Further details of the discovery and its advantages will be apparent
from the
detailed description included below.
DETAILED DESCRIPTION
[00056] Ultrasound is a widely used imaging modality that is traditionally 2D.
2D
ultrasound images remove 3D volume and three-dimensional information that
allows for
determining shapes, distances, and orientations. Ultrasound is used in
medical, military,
sensor, and mining applications
[00057] In interventional oncology, ultrasound is the preferred intra-
operative image
modality for procedures, including biopsies and thermal/focal ablation
therapies in liver &
kidney, laparoscopic liver surgery, prostate biopsy and therapyõ percutaneous
liver ablationõ
and all other abdominal organs and ophthalmic intervention known to those
skilled in the art.
Some brain interventions also use ultrasound, although MR and CT are more
common.
11
CA 02839854 2014-01-17
[00058] Ultrasound allows for "live information" about anatomical changes to
be obtained
with the requirement for further radiation dose to patient or physician. For
image-guided
interventions, it can be difficult for a surgeon to navigate surgical
instruments if the target
organ is moving either due to patient motion (i.e. breathing and cardiac
motion the patient
respiratory motion) or ultrasound probe pressure (causing movement and
deformity of the
organ), in any procedure that requires a needle or needles, particularly in
ultrasound-guided
interventional procedures, it is important to be able to correct for motion of
an organ thereby
allowing the interventionist to be able to track and position/align needles
relative to the
planned trajectory, nearby vulnerable structures and to position them at their
target position
with a high degree of precision and accuracy. To gain acceptance in the
clinical practice the
speed of registration must be both accurate and fast.
[00059] 2D/3D registration is a special case of medical image registration
which is of
particular interest to surgeons. 2D/3D image registration has many potential
applications in
clinical diagnosis, including diagnosis of cardiac, retinal, pelvic, renal,
abdomen, liver, and
tissue disorders. Applications of 2D/3D registration also has applications in
radiotherapy
planning and treatment verification, spinal surgery, hip replacement,
neurointerventions, and
aortic stenting.
(00060] Target organ motion during a procedure can cause target misalignments
between
the initially acquired 3D image and their corresponding locations within the
patient's prostate
or liver as depicted by the real-time 2D ultrasound images acquired. Although
our method
was developed and tested for prostate gland and liver applications, it is
applicable to all
organs where motion compensation is required.
[00061] Accurate and fast registration to compensate for motion during
minimally
invasive interventions, such as a biopsy, is an important step to improve the
accuracy in
delivering needles to target locations within any organ.
[00062] The method steps described herein are embodied in a computer readable
storage
medium which includes a non-transient memory with a computer program stored
thereon.
The computer program represents a set of instructions to be executed by a
computer. The
12
CA 02839854 2014-01-17
computer readable storage medium is connected to the ultrasound probe and when
required
cause the computer to carry out those method steps described herein.: In
addition to a biopsy
procedure the methods described herein can also be applied to other non-
limiting
interventional procedures such as image-guided interventional procedures
including ablations
and laparoscopies and the like..
[00063] As used herein the term "imaging procedure" is intended to mean a
computerized
technique or procedure such as ultrasonography, computed tomography, magnetic
resonance
imaging, positron emission tomography, single-photon emission computed
tomography that
generates a visual representation of an object.
[00064] As used herein, the term "reference image" is intended to mean an
image which is
typically a first image, or any image that is designated as the reference to
which other images
are referenced. .A reference image can be any of the following: 3D MRI image,
3D CT
image, 3D PET image, 3D SPECT image, and 3D ultrasound image.
[00065] As used herein, the term "reference parameter" is intended to mean
body or
organ/tissue movement or motion, or any other object motion. Specifically,
reference
parameter means a value generated by the registration process that described
the "goodness"
of the registration. A typical parameter is the Normalized Cross Correlation
or Mutual
Information.
[00066] As used herein, there term "normalized cross correlation" is intended
to mean a
group of metrics including normalized cross correlation metric, Kullback-
Leibler distance
metric, Normalized Mutual Information Metric, Mean Squares Histogram Metric,
Cardinality
Match Metric, Kappa Statistics Metric, and Gradient Difference Metric.
A. Geometric-based 2D-3D rigid/affine registration
[00067] We have developed an image-based registration algorithm for 2D to 3D
rigid/affine ultrasound image registration technique to compensate for organ
motion as a
surgical instrument, such as a biopsy needle, is inserted into a target organ,
for example.
13
CA 02839854 2014-01-17
liver, and prostate gland. Specifically, this algorithm was developed for
compensating for
liver motion, but can be applied to other organs. The algorithm allows for
tracking of surgical
instruments in real-time. Speed is important for there to be any clinical use.
[00068] To perform 2D/3D registration, the 3D and 2D data are brought into
dimensional
correspondence (geometrically aligned). Registration algorithms compute
transformations to
set correspondence between the 2D image and one slide of the 3D image. The
slice is
chosen arbitrarily. Our registration algorithm computes transformations of 3D
data into 2D
with a particular application to prostate and liver, however it can be applied
to other
interventional applications in image-guided procedures including other organs
i.e. lung,
venter, breast and other fields, for example recovering the 2D tracker
information and
reconstruction 3D image from 2D frames without tracker information. Here the
tracker
information are transform parameters like rotation angles and translatory
distance of the 2D
image tracker system. 2D/3D registration also has applications in fields in
addition to
medical such as machine vision (i.e. dynamic image analysis, object and
environment modeling),
military, sensor (i.e. object tracking), and mining.
[00069] As the biopsy needle is inserted into a target organ, and if the
patient is breathing,
then the target organ will be moving. There can also be non-rigid deformation
due to
ultrasound probe pressure on the organ. The algorithm allows for the
identification of the
target on another imaging modality. The aim of the algorithm is to correct the
location of the
3D image so that it is in synchrony with those 2D ultrasound images acquired
during the
procedure. That is the 3D image must be in synchrony with body motion.
[00070] The method for generating a motion-corrected 2D image uses a
combination of
geometric-based 2D-3D rigid registration and intensity-based 2D-3D rigid
registration..
[00071] During the biopsy procedure, a 2D real-time ultrasound video stream,
which
includes intra-procedural 2D real-time live images of the target organ or
tissue, is acquired
and displayed on a computer screen (an imaging device). Typically, the
acquisition and
display of the video stream is done at a video frame rate of up to 30 frames
per second. The
target organ or tissue is typically one that is suspected of being diseased or
is known to be
14
CA 02839854 2014-01-17
diseased. A pre-procedural/interventional target image (a 3D static image)
with the target
identified, such as a tumor, is acquired at the beginning of the procedure
before the
interventionist inserts the needle into the target organ. The data sets to be
registered are
defined in coordinate systems.
[00072] An acquired 2D image is compared with one slice in the 3D image to
determine if
they match. This is done as a precaution in case the parameter of transform
changed. A
target goal is set up, and if the goals are well matched, then the function
value of the. target
goal will be minimized. Examples of the transform's parameters include
rotation, translation,
shearing and scaling. The goal here is to find the best slice inside the 3D
image. The best
slice is defined by the transform parameters, which looks like the 2D image.
[00073] The initialization phase for the algorithm involves correcting the
location of the
3D image so that it is in synchrony with body motion, caused by breathing,
heart beat and the
like, of that viewed with 2D ultrasound images acquired during the procedure.
For each 2D
image taken, the corresponding plane in the 3D volume must be found. The 3D
image can be
moved to the correct plane as the 2D image. Usually the 2D image is moved
according to
patient movement, such as breathing. At this point, the user needs to
determine which slice
in the 3D image matches the live image, i.e. the user must find the
corresponding 2D image
in the pre-acquired 3D volume, which can be problematic. We have successfully
addressed
this problem by using a geometric-based 2D-3D registration algorithm. It was
done by taking
a 3D image and extracting the 2D image from it. That is, a 2D slice (2D
extracted image) is
taken out of the 3D volume image. The 3D image and 2D extracted image are
approximately
lined up to recover a 3D contextual form of an image (also known as correct
geometrical
context). This means the 2D and 3D images appear to be aligned. The alignment
can be done
either visually or by using an algorithm such as by identifying corresponding
points in the
2D and 3D images and finding the best translation/rotation/shearing transform
to achieve
approximate registration. The resulting image is a 3D image that can be looked
at in the
same plane as the 2D real-time image.
CA 02839854 2014-01-17
[00074] The 3D image (2D extracted image) is compared to a 2D real-time image.
If the
two images do not match exactly, the plane where the 3D image and the 2D image
do not
match exactly is extracted. During registration, an image similarity metric is
optimized over a
3D transformation space. Accurate definition of similarity measure is a key
component in
image registration. To do this, minimizations for registration are performed.
Motion is
extracted in 12 degrees of freedom or less and then the plane is moved and
motion is
extracted in different angles using different image similarity metrics,
including normalized
cross-correlation, versor rigid transform or affine transform. Powell's
optimization or particle
swarm optimization is applied to calculate degree of matching. Powell's method
is used to
minimize registration error measurement. It is used for local minimizations
(i.e. will only
find a local solution). There is no guarantee that a correct match will be
found applying only
Powell's method.
[00075] In order to increase the success of the registration a few multiple
initial
parameters are applied, which for example are (a) the output parameters of
prior 2D-3D
registration results; (b) estimated parameters using a few groups output
parameters of prior
2D-3D registration; (c) the output parameters obtained in the same time of
last respiration;
and (d) the output parameter of the first successful 2D-3D registration.
[00076] The re-initialization phase. As described aboveõ Powell's method
optimization is
a local minimization, which can fail. The particle swarm optimization can be
carried out to
find the global solution in case Powell's method fails. Using the particle
swarm optimization
increases the global optimization speed. It can be calculated parallel for all
particles. The
initial parameters for the optimization of particle swarm method are the same
as those with
the Powell method. If the time of the calculation for particle swarm method
takes too long,
the estimated initial parameters will be used for this 2D frame.
[00077] Estimation of the initial parameters for of the 2D-3D registration for
each 2D
frame. Before the calculation of 2D-3D image registration, the initial
transform parameters
are estimated from the changes of known parameters y(tk) has been calculated
for a few (N)
prior frames. The estimation is done through polynomial series or Fourier
series.
16
CA 02839854 2014-01-17
[00078] f ( a i , t) = ; a.
[00079] fad = arg min E;;_l_N min lyn ¨ f(aõ 01 ,
[00080] f (bz, t) = Ef":1 b 2 exp(vi---- 32 i t)
T f
[00081] [b} = arg min ETil,,v wõ I yõ ¨ 10,, tji ,
[00082] f (ai, t) or f (hi, t) is one of estimated parameter. Wk is the weight
which can be
different with k. T is the period of the respiration. yk is one of known
registration
parameter in recorded in different time tk of respiration. N is the number of
2D frames in a
period of the respiration. I is the number of coefficient bt. t = 0 is
corresponding to current
time in which the 2D-3D registration is perfumed. We assume the parameters are
changed
independently. The estimated initial parameter is
[00083] f (bi, 01,.0 , f (Ili, Olt,-1,...
[00084] or
[00085] f(af, 0 I t.o , f (a, 0 tr. i= = =
[00086] At this point finding the best match quickly is important for the
user. Once the
user has an estimation of normalized cross-correlation, the 3D image (target)
is transformed
to the current location as that obtained from the 2D real-time image. The 3D
image is
transformed to achieve the best possible correspondence with the 2D image(s).
The
transformation is 2D/3D image-to-image registration.
[00087] The algorithm processes up to 30 frames per second. A graphics
processing unit
(GPU)-based implementation was used to improve the registration speed to
register from 2D
to 3D. The speed of registration must be fast to gain acceptance in the
clinical practice.
Affine transformation
[00088] The affine transformation has 12 parameters which as following form,
see
Eq.(8.11) of ref[20] below:
i x' MOD M01 MO2 -x - Cx -Tx C x
= M10 M11 M12 Y ¨ Cy 4- 'Ty + Cy
}
..Z' M200 M21 M22 _z - Cz
17
CA 02839854 2014-01-17
[00089] Coordinates after affine transformation:
-x 'I
e
Y
L7e
[00090] Coordinates before transformation:
x
[y]
[00091] The original point can be any point in the space. The center of the
rotation is at
_
Cx
C
Y
Cz 1
[00092] There are two kind of parameters for this transformation which are in
the
following:
[00093] 1) Affine matrix parameter:
-Moo Moi Mo2
Nilo M11 M12
-M20, M21 M22 and
[00094] 2) Translation parameters:
iTx
Ty
T,
Particle swarm optimization
[00095] For particle swarm optimization [21][22] we assume we have /
parameters:
x = fro, ..., xi, ...,xj_i}. The optimization problem is to find optimal x
with the function
f (x), i.e.
18
CA 02839854 2014-01-17
X = arc min f (x)
[00096] Assume there are N particles. The positions are of i-th particle are
x, = {x,0, ...,xzi,
[00097] Assume k is iteration index. The position of particles are updated
according to
k 1 k v k+1
2 E
where
k4-1 pry k ciricy,k _ k xlk)
Xr)c2r2 (g ,
[00098] y,k is k th iteration, th particles best(personal best) position in
the history.
y, k = arc -milt (x,)} i= canst
[00099] g,k k th iteration, i th particle's group best (group best) position
in the history.
9,k = arc min {f (x,1)) I E G 1 = 0, k
[000100] G is Group of neighbor particles. For example all particle smaller
than a fix
distance.
[000101] r1,r2 is two random variables between [0,1]. el, e2 are two constant
around 2, 13
is constant.
GPU calculate Particle swarm optimization
[000102] The functions value of
ff (x0) , f (x), ...,f (x N)}
is calculated in parallel.
[000103] The GPU calculates the 3D to 2D image normalized cross-relation
function.
19
CA 02839854 2014-01-17
B. Intensity-based 211-3D rigid registration
[000104] We also discovered an image-based registration technique to
compensate for
prostate motion by registering the live 2D TRUS images acquired during the
biopsy
procedure to a pre-acquired 3D TRUS image. The registration must be performed
both
accurately and quickly in order to be useful during the clinical procedure.
This technique, an
intensity-based 2D-3D rigid registration algorithm, optimizes the normalized
cross-
correlation metric using Powell's method, The 2D TRUS images acquired during
the
procedure prior to biopsy gun firing were registered to the baseline 3D TRUS
image acquired
at the beginning of the procedure. The accuracy was measured by calculating
the target
registration error (TRE) using manually identified fiducials within the
prostate; these
fiducials were used for validation only and were not provided as inputs to the
registration
algorithm. We also evaluated the accuracy when the registrations were
performed
continuously throughout the biopsy by acquiring and registering live 2D TRUS
images every
second. This measured the improvement in accuracy resulting from performing
the
registration as a background process, continuously compensating for motion
during the
procedure. To further validate the method using a more challenging data set,
registrations
were performed using 3D TRUS images acquired by intentionally exerting
different levels of
ultrasound probe pressures in order to measure the performance of our
algorithm when the
prostate tissue was intentionally deformed. In this data set, biopsy scenarios
were simulated
by extracting 2D frames from the 3D TRUS images and registering them to the
baseline 3D
image. A GPU-based implementation was used to improve the registration speed.
We also
studied the correlation between NCC and TREs.
[000105] The root mean square TRE of registrations performed prior to biopsy
gun firing
was found to be 1.87 0.81 mm. This was an improvement over 4.75 2.62 mm
before
registration. When the registrations were performed every second during the
biopsy, the
RMS TRE was reduced to 1.63 0.51 mm. For 3D data sets acquired under different
probe
pressures, the RMS TRE was found to be 3.18 1.6 mm. This was an improvement
from
6.89 4.1 mm before registration. With the GPU based implementation, the
registrations
CA 02839854 2014-01-17
were performed with a mean time of 1.1 s. The TRE showed a weak correlation
with the
similarity metric. However, we measured a generally convex shape of the metric
around the
ground truth, which may explain the rapid convergence of our algorithm to
accurate results.
[000106] We therefore determined that registration to compensate for prostate
motion
during 3D TRUS-guided biopsy can be performed with a measured accuracy of less
than 2
mm and a speed of 1.1 s, which is an important step towards improving the
targeting
accuracy of a 3D TRUS-guided biopsy system.
Data acquisition
[000107] We acquired images from human clinical biopsy procedures using a
mechanically assisted 3D TRUS-guided biopsy system [4] in a study approved by
the Human
Research Ethics Board of Western University. The system, using a commercially
available
end-firing 5-9 MHz TRUS transducer probe (Philips Medical Systems, Seattle,
WA),
acquired a 3D TRUS image at the beginning of the biopsy procedure, and then
acquired and
displayed 2D TRUS images at a video frame rate (7-30 frames per second) during
the biopsy
session. The mechanical encoders attached to the ultrasound probe tracked its
3D position
and orientation throughout the procedure, Using this system, we recorded
images acquired
during clinical biopsy procedures under two different protocols, in order to
obtain datasets to
test the robustness of the registration algorithm under different motion
characteristics of the
prostate. For both protocols, all 3D TRUS images were recorded prior to taking
any biopsy
tissue samples. For the first protocol (referred to hereinafter as the biopsy
protocol), we
acquired images from eight subjects. Following the standard operating
procedure for 3D
TRUS-guided biopsy in our trial, a 3D TRUS image was acquired at the start of
the biopsy
procedure, and then live 2D TRUS images were recorded at one frame per second
from the
sequence of images that follows at video frame rate. For the second protocol
(hereinafter
referred to as the probe pressure protocol), images were acquired from ten
subjects. 3D
TRUS images were acquired after applying three different probe pressures on
the prostate
gland centrally: 1) applying a medium probe pressure, similar to what a
physician usually
applies during a biopsy, 2) applying a low probe pressure that caused minimal
prostate
21
CA 02839854 2014-01-17
displacement, and 3) applying a high probe pressure that caused substantial
prostate
deformation and anterior displacement, This yielded a data set with prostate
motions and
deformations under a wide range of ultrasound probe pressures.
2D-3D registration ¨ biopsy protocol
[000108] For each of the eight subjects, we selected 1-3 2D TRUS images per
patient 1-2
seconds prior to biopsy needle insertion. This choice of 2D TRUS images was
motivated by
the fact that accurate alignment of the predefined targets with the intra-
procedure anatomy is
chiefly required immediately prior to biopsy, when a tissue sample is to be
taken from an
intended biopsy target. We analyzed 16 such images from eight subjects.
[000109] The transformation, TTr. --+ 12 given by encoders on the joints of
the linkage of
the mechanical biopsy system, maps each live 2D TRUS image, hive: R, to the
world
coordinate system of the previously acquired 3D TRUS image 'base: R where
1,ff c
and n c E. Within the 3D world coordinate system, any differences in prostate
position and
orientation between the real-time 2D TRUS images and the initially-acquired 3D
TRUS
image are due to prostate motion within the patient, gross movements of the
patient during
the procedure, and the biopsy system's tracking errors. The accuracy of the
initialization for
the prostate motion registration algorithm is based in part on tracking errors
of the biopsy
system. In the system developed by Box et al, [4], the accuracy in delivering
a needle to a
biopsy core in a phantom were found to be 1.51 0.92 mm. Registration of live
2D TRUS
images to the pre-acquired 3D image compensates for both the tracking errors
and errors due
to prostate motion.
[000110] FIG. 1 illustrates the overall workflow in our method. To reduce the
effects of
speckle, anisotropic diffusion filtering [12] (conductance parameter = 2, time
step = 0.625) of
images was used as a pre-processing step. Although there can be non-rigid
deformation of
the prostate due to ultrasound probe pressure [13], a rigid/affine alignment
can be found with
lower computational cost, so we investigated the accuracy of rigid/affine
registration in this
work to determine whether rigid registration is sufficient for the clinical
purpose of biopsy
22
CA 02839854 2014-01-17
targeting. For each 2D TRUS image, finding the corresponding plane in the pre-
acquired 3D
TRUS volume is a 2D-to-3D intra-modality rigid/affine registration problem.
Due to limited
ultrasound contrast within the prostate, reliable extraction of the boundary
and other
anatomic features is challenging. Therefore, we tested an intensity-based
registration
algorithm.
[000111] Using the mechanical tracker transform TTr, we can position and
orient the 2D
TRUS image hiõ within the 3D world coordinate system yielding a 3D image //rye
as follows:
ritifit(TrAPID luve(P4.).
where p1 C yf.
[000112] The registration of the baseline 3D image /base to hive is performed
in this 3D world
coordinate system. The objective of the registration is to find the
transformation, 7; : 1 ¨+
consisting of a six/twelve-parameter-vector given by u, that aligns
anatomically homologous
points in /base and //,õ. We used normalized cross-correlation (NCC) [15] as
the image
similarity metric that was optimized during the registration. For two images
Ii and 12, we
optimized the objective function defined as:
P argmax. MCC(F1,12; u) where
Zeirt4 tlXiltr, ra)
p
/VCCaiii2;t1) =
2
KEinia5010/}' XItomalai (T1AP#42)2)11
and Q1 and Q2 represent the subspaces of (12 c' ) containing the image domains
of Ii and
12,1.e.
23
CA 02839854 2014-01-17
C4; {P e fl1ITu-1(P E (12))
[000113] We optimized the image similarity measure given by NCC ('base,1hve.)
to obtain Tu
for each of the 16 images we acquired. We used a local optimization method
i.e. Powell's
method [16] to optimize the six/twelve dimensional search space that includes
three
translations, three rotations, and shearing. Powell's method improved the
speed of execution
and reduced the memory size of the computation, when compared with a gradient-
descent-
based method during our initial experiments.
Incremental 2D-3D registration for continuous intra-biopsy motion compensation
[000114] The registration to compensate for prostate motion can be performed
frequently
(e.g., once per second) throughout the biopsy procedure, with the frequency of
registration
limited by the time required to register a single pair of images. At a given
time point denoted
by to (time elapsed in seconds from the start of the biopsy), we initialized
the source image
for the Ilth registration with the transformation matrix obtained from
registrations at previous
time points using
Tu = rate Tut . (2)
[000115] During the nth registration, we found the parameter vector utn that
gave the
optimum NCC measure for the transformation matrix Tut,. We performed the
registration for
the complete biopsy procedure for the eight subjects described in the previous
section using
the sequence of live 2D TRUS images recorded every second from the start of
the biopsy
procedure.
2D-3D registration ¨ probe pressure protocol
24
CA 02839854 2014-01-17
[000116] 3D TRUS images acquired at different probe pressures can provide
additional
anatomical context to enhance the validation of our registration algorithm. We
denote images
acquired at low, medium and high probe pressures, respectively, as how
Imeghigh.: R
We acquired 30 such images from 10 subjects.
[000117] We set the image acquired at medium pressure, /med., as the source
image. As our
target image, we selected 2D slices (/(7,high)) from the 3D images kw and
/h,gh. For the 20
registrations performed (using the 30 3D TRUS images) mechanical tracker
transformations
(Tn.) were randomly selected from 16 frames (across 8 subjects in the biopsy
protocol)
occurring an average of 1-2 seconds prior to the firing of the biopsy gun in
real biopsy
procedures, according to
rgocht,002) htow.hlatarrr(P0) where Pt cW and 1/2 C a.
[000118] Hence, the target images are representative of live 2D TRUS images
depicting a
situation with minimal prostate motion (slice from how) and substantial
prostate motion (slice
from /h,gh). Since the physician intentionally applies different levels of
pressure during the
acquisition, the set of images contains a wide range of prostate displacements
and
deformations that are intended to represent the extremes of probe pressure
during the biopsy
procedure to challenge the registration algorithm. For each subject, we
perform registration
between images 'med.-110w and 'med. - i
high by respectively optimizing the image similarity
measures, NCC(//0.,Lned.) and NCC(ihigh,Lned) as defined above in Equation 1.
Validation
Biopsy protocol registration
[000119] The registration was validated using manually-identified
corresponding intrinsic
fiducial pairs (micro-calcifications) [13]. For the images acquired under the
biopsy protocol,
fiducials appearing in haõ, denoted by
.ase and the corresponding fiducials from lives
denoted byftive, were identified (fi,õ c vi and fiase C c2). We identified 52
fiducial pairs for
CA 02839854 2014-01-17
16 biopsies in eight patients. These fiducial pairs were used for validation
only and were not
provided as input to the registration algorithm. The target registration error
was calculated as
the root mean square (RMS) error
Ettgrit4t/4411-1fr
4bt,
ft
TRE,, (3)
N*
rim
TRElblopsy L (4)
Nb
where Nb is the number of biopsies and NI, is the number of fiducials
identified for a
particular pair of images. The TRE was estimated by first calculating RMS
values TREb
using the fiducials identified in each pair of images for each biopsy and then
calculating the
RMS value TREb,opsy for the number of biopsies performed. This approach
averaged the
contributions to the TRE from the variable number of fiducials manually
identified in each
pair of images during a biopsy. The TRE before registration was calculated
without applying
the registration transform Tu in Equation 3 to compare against TRE post
registration to assess
the improvement.
Probe pressure protocol registration
[000120] In the data set acquired under the probe pressure protocol, full 3D
anatomical
information for the whole prostate was available for both the source and
target images. We
manually identified 188 fiducials throughout the 3D volumes obtained from 10
subjects,
without limiting the fiducials to lie within the particular extracted plane
used in the
registration, The TRE was computed as
26
CA 02839854 2014-01-17
rREp
ENT30-wortd(ittuttl'" rttAkrwittottik.))2
= i CO
,
Mk
top us 2
TRE.... bia P
?MOW. "'"' 09
Np
where l f
- onedõlow,high) c. SI are the fiducials identified in "Med) km 'high.
(000121] We also computed the optimal rigid alignment using the identified
fiducials to
define the rigid transformation that yielded the minimum TRE for the given
fiducials per
patient. To do this, we found the fiducial registration error (FRE) [17] for
each set of fiducial
pairs in each patient, after transforming the fiducials with the parameters
corresponding to
the best rigid alignment. With the presence of non-rigid deformations in the
probe pressure
protocol data set, FRE gives a lower bound on the TREpressure that was
calculated using a rigid
registration.
GPU implementation
[000122] The step consuming the most computation time during execution of the
registration was the calculation of the image similarity metric during
optimization, Therefore,
we implemented the NCC calculation on an nVidia GTX 690 (Nvidia Corporation,
Santa
Clara, CA) graphics processing unit (GPU) using compute unified device
architecture
(CUDA), The normalized cross-correlation calculation is inherently
parallelizable. Instead of
using a sequential approach to transform each voxel independently, we
transformed all
voxels in the moving image in parallel during each iteration of optimization.
These
transformations were followed by 3D linear interpolation of image intensities
to resample the
moving image that was also performed within the GPU. The subsequent
calculation of the
summations in Equation 1 was also done in parallel with the GPU reduction
algorithm to
27
CA 02839854 2014-01-17
further accelerate the execution. In one iteration of the particle swarm
optimization, the
calculation for different particles are also made parallel inside the GPU.
Correlation between image similarity metric and misalignment
[000123] During registration, we optimized an image similarity metric over a
3D
transformation space. The relationship between the image similarity metric and
the amount
of misalignment not only conveys the suitability of the metric to be used in
registration, but
also it shows whether the image-similarity metric could be used as an
indicator of the
misalignment. This may be a useful feature to trigger the registration
algorithm in a system
that does not continuously compensate for motion as background process during
biopsy. To
analyze this relationship using the biopsy protocol data, we plotted the
calculated normalized
cross-correlation measures for each instance before registration, during
registration (for each
iteration during the optimization) and after registration (after the optimizer
converged) and
their corresponding TREinopsy values.
[000124] With manually identified fiducials, we should be able to find a plane
within the
3D TRUS image that yields near zero TRE. We analyzed the behaviour of
normalized cross-
correlation near this "optimum" plane by extracting 2D images lying nearby (in
terms of the
six/twelve parameters, defining 3D translation and rotation) planes in the 3D
TRUS image,
and computed the image similarity metric for the 2D TRUS image and these
nearby 2D
images from the 3D TRUS image.
[000125] Although this approach does not fully explore the six-
dimensional/twelve-
dimensional objective function, to simplify the visualization of the results,
we analyzed the
metrics by varying one degree-of-freedom at a time,
TRE as a function of distance to the probe tip
[000126] We analyzed the TRE as a function of distance of each fiducial to the
ultrasound
probe tip, to test if the registration error is larger within the regions of
the prostate close to
28
CA 02839854 2014-01-17
the ultrasound probe. Since we used a rigid/affine transformation during
registration, non-
rigid deformation of the prostate would be reflected as part of the TRE.
Ultrasound probe
pressure might cause inconsistent deformation in different regions of the
prostate, which
could lead to regionally-varying accuracy of motion compensation by a
rigid/affine
transformation.
RESULTS
A. Validation: biopsy protocol data
[000127] The TREbiopsy was calculated according to Equation 4 and its RMS
std. was
found to be 1.87 0.81 mm, after manually localising 52 fiducial pairs over 8
subjects, This
was an improvement over 4.75 2.62 mm before registration. Since these TRE
distributions
were found to be not normally distributed using one-sample Kolmogorov-Smimov
test with a
significance level p <0.0001, we tested the null hypothesis that their medians
were equal
with a non-parametric test using Prism 5.04 (Graphpad Software Inc., San
Diego, USA). The
Wilcoxon signed rank matched pairs test rejected the null hypothesis (p
<0.0001) suggesting
that there is a statistically significant difference in TREs before and after
registration. When
2D-3D registration was performed incrementally every second during the biopsy,
the
RMS std TRE was reduced to 1.63 0.51 mm The mean number of iterations
required for
convergence decreased from 5.6 to 2.75, FIG. 2 shows changes in TRE
distributions before
biopsy taken. FIG. 3 shows TRE values for each biopsy.
[000128] FIG. 4 contains two representative example images, depicting the
visual
alignment qualitatively. The post-registration TRE of these two example images
were found
to be 1.5 mm (top row) and 1.2 mm (bottom row) , which had improvements from
3.7 mm
(top row) and 5.3 mm (bottom row) before registration. Grid lines overlaid at
corresponding
locations in image space facilitate visual evaluation of the alignment of the
anatomy pre- and
post-registration.
[000129] In order to see the effect of patient motion over time during the
biopsy session, we
analyzed the TREs obtained from eight patients as a function of time elapsed
since the start
29
CA 02839854 2014-01-17
of the biopsy. According to the results shown in FIG. 5, it can be seen that
the TRE values
before and after registration have an increasing trend with the elapsed time
during the biopsy.
Weak relationships were found with correlation coefficient (r2) values of 0.23
before
registration and 0.41 after registration. When the registration was performed
every second,
the r2 value was found to be 0.37,
B. Validation: probe pressure protocol data
[000130] The RMS TRE for the data acquired under the probe pressure protocol
was 3.18
1.6 mm, This was an improvement from a 6.89 4.1 mm TRE before registration.
Note that
we used the fiducials in the whole prostate (not just the slice containing the
fiducials) in TRE
calculation as given in Equation 6. The mean value for the FRE, corresponding
to the best
rigid transform that aligns the identified fiducials, was found to be 1.85
1.2 mm. The
distribution of TRE values before registration, after registration, and after
transforming with
the best rigid alignment is shown in FIG. 6. The error in registration
includes the errors due
to non-rigid deformation occurring within prostate regions outside of the 2D
target image (as
opposed to the errors arising only due to deformation within the 2D target
image as in the
biopsy protocol) and the variability in manually locating the fiducials in 3D.
C. Speed of execution
[000131] After the GPU-accelerated implementation (nVidia GTX 690 GPIJ card
and Intel
Xeon 2.5 GHz processor) the registration was performed with mean std times
of 1.1 0,1
seconds for the biopsy protocol experiments described herein,
D.Correlation between image similarity measure and misalignment
[000132] FIG. 7 shows the relationship between the image-similarity measure
and values of
THE for each transformation obtained during the optimization iterations. The
circle points
show the values before registration, and the square points show the values
after registration
converged. The cross points depict the values during convergence. The
correlation
coefficient (r2), calculated using all points (before, during, and after
convergence) in FIG. 7,
CA 02839854 2014-01-17
was found to be 0.23. FIG. 8 shows a box plot of the TRE distributions of the
points before
registration, during convergence and after registration. While the TRE
decreases in general
during convergence, a weak correlation can be seen between image similarity
measures and
TRE from these results,
[000133] FIG. 9 shows plots of the normalized cross-correlation metric versus
out-of-plane,
in-plane rotations and translations. The solid curves represent the mean
values of the metrics
for different out-of-plane rotations and translations for 16 2D TRUS images
across eight
subjects, and the dashed curves show the values one standard deviation above
and below the
mean. The convexity of the mean curves gives an indication of the general
capture range of
the objective functions for many registrations. FIG. 10 shows the three plots
of normalized-
cross-correlation metrics similarly obtained for a single biopsy in three
patients. The
generally convex shape of 375 the functions observed in FIG. 9 and FIG. 10
encourages the
use of normalized cross-correlation during registration in compensating for
prostate motion.
[000134] FIG. 11 shows TRE as a function of the distance to the probe tip for
each
individual. The TRE tends to increase closer to the probe tip (r2 value= 0.1);
however, the
correlation between distance to the probe tip and the TRE before registration
is weak.
DISCUSSION AND CONCLUSIONS
A. Accuracy of registration
[000135] Our image registration method was validated using the fiducials
identified in
clinical images acquired during the biopsy procedures. There was a significant
improvement
of TRE after registration in both biopsy and probe pressure protocols. The
required accuracy
of the biopsy system to guide needles to target locations stems from the size
of the smallest
clinically-relevant tumours (0.5 cm3, corresponding to a spherical target with
5 mm radius)
[18]. A biopsy system with a measured RMS error of 2.5 mm in taking a sample
from the
intended target will have a probability of at least 95.4% of taking a sample
within this 5 mm
radius since 5 nun is 2 standard deviations away from the mean of the
distribution of targets
given by an system with RMS error of 2.5 mm [13]. An image-based registration
during the
31
CA 02839854 2014-01-17
procedure, while compensating for prostate motion, also corrects for tracking
errors in the
biopsy system, if any. Therefore, if the registration was performed
immediately before the
physician fires the biopsy gun to capture a tissue sample from the prostate,
the targets
identified in the pre-acquired 3D image would be aligned with the live 2D TRUS
image, with
accuracy limited by the TRE of the registration algorithm, However, the motion
and
deformation induced due to the rapid firing of the biopsy gun, which happens
during a sub-
second interval remains an error in the biopsy system that is challenging to
correct. When
targeting a predefined location, the TRE of the motion compensation algorithm
and the error
during the rapid biopsy-gun firing process, which was quantified in [19], may
accumulate
and become an important consideration.
[000136] Alignment of the targets identified in the 3D TRUS image to the live
2D TRUS
image is primarily required immediately before the physician fires the biopsy
gun,
Consequently, this registration could be integrated into the clinical workflow
by executing it
just prior to the physician aiming at target locations. However, according to
the results, both
the accuracy and speed of the registration were improved when the registration
was
performed on the 2D TRUS images acquired every second, When the baseline 3D
TRUS
image is updated more frequently, it might improve initialization of 2D TRUS
images that
follow in subsequent registrations, providing for faster convergence to a
suitably accurate
optimum. Therefore, in a clinical procedure, this algorithm can be performed
in the
background continuously compensating for motion.
.B. Change of TRE with time during biopsy
[000137] The weak positive relationship between TRE and time elapsed shown in
FIG.
5(a), suggest that the misalignment between pre-acquired and live images
increases with time
(slope of the best-fit line = 9.6 m/s). After performing the registration
just before a biopsy
sample is taken, there is still a positive relationship (slope = 4.1 m/s)
between TRE and
time. This indicates that image pairs, with higher initial misalignments
towards the end of the
biopsy procedure, were more challenging for the algorithm, In FIG. 5(c), the
slope of the
best-fit line was lower (slope 2,4 m/s) when the registrations were performed
every second,
32
CA 02839854 2014-01-17
The improved initializations when performing registrations every second may
have induced
convergence to a better solution.
C. Probe pressure protocol
[000138] In probe pressure protocol, the TRE was 1,2 mm higher than that of
the biopsy
protocol. This increase could be attributed to the use of fiducials from the
whole prostate
during validation. The best rigid transform for the selected plane may not
necessarily be the
best rigid fit for the whole prostate due to non-rigid deformations occurring
at different (out
of plane) regions of the prostate, Moreover, the high probe pressures
intentionally exerted by
the physician when acquiring these images might have caused more than the
usual
deformation that occurs during biopsy. The extreme range of probe pressures
and prostate
displacement and deformation could make accurate registration more challenging
as the
algorithm is more susceptible to local minima the further the initialization
is from target
alignment. However, the fiducial identification process was relatively more
straightforward
due to the availability of 3D contextual information in both the fixed and
moving images,
D. Correlation between similarity metric and TRE
[000139] FIG. 7 shows a weak correlation between similarity metric values
before, during
and after convergence and the TRE. The generally convex shapes observed in
FIG. 9 and
FIG.10 in metric values as a function of different amounts of introduced
translations and
rotations, suggest that the metric value could be used as a weak indicator to
the quality of the
registration.
[000140] In FIG. 11, a weak negative correlation can be seen between the TRE
and distance
to the probe tip. This suggests that near the probe tip there could be higher
non-rigid
deformation of the prostate that may not be accurately compensated with a
rigid registration
algorithm,
[000141] Accurate and quick registration to compensate for motion during
biopsy is an
important step to improve the accuracy in delivering needle to target
locations within the
33
CA 02839854 2014-01-17
prostate. We presented a 2D-to-3D rigid intensity-based registration algorithm
with a
measured error of less than 2 mm, validated on clinical human images using
intrinsic fiducial
markers, to align a 3D TRUS image (with associated prostate biopsy targets)
acquired at the
start of the procedure to 2D TRUS images taken immediately prior to each
biopsy during the
procedure. The accuracy and speed of the registration further improves when
the baseline 3D
image is updated by registering the 2D TRUS images recorded every second
during biopsy.
Using our high-speed GPU implementation (0.1 seconds total time per
registration), this
algorithm can be executed in the background during the biopsy procedure in
order to align
pre-identified 3D biopsy targets with real-time 2D TRUS images. We also
presented
evidence that image similarity metrics can be used as a weak indicator of the
amount of
prostate misalignment (with respect to the initially acquired 3D TRUS image),
and could be
used to trigger the execution of a registration algorithm when necessary.
Broader applications
[000142] In addition to medical applications, motion compensation has
applicability to
fields requiring the ability to detect and track moving objects/targets ,
including machine
vision (i.e. dynamic image analysis, object and environment modeling),
military, sensor (i.e.
object tracking), and mining. Our discovery is intended to be applicable to
these applications.
[000143] Normally, non-rigid registration is too slow for the calculations
described herein.
However, we have also discovered that we can make non-rigid registration very
fast. See
Medical Image Computing and Computer-Assisted Intervention ¨ MICCAI
2013;Computer
Science Volume 8149, 2013, pp 195-202 "Efficient Convex Optimization Approach
to 3D
Non-rigid MR-TRUS Registration"; Yue Sun, et al. It is therefore to be
understood that 2D-
3D registration can also be non-rigid.
REFERENCES
34
CA 02839854 2014-01-17
[000144] 1. Canadian Cancer Society's steering committee: Canadian cancer
statistics
2012 (2012);
[000145] 2. Howlader, N., Noone, A,M., Krapcho, M., Neyman, N., Aminou, R.,
Altekruse, S.F., Kosary, C.L., Ruhl, Jõ Tatalovich, Z., Cho, H., Mariotto, A.,
Eisner, M.P.,
Lewis, DR., Chen, H.S., Feuer, B.J., Cronin, K,A.: SEER Cancer Statistics
Review, 1975-
2009 (Vintage 2009 Populations), National Cancer Institute. Bethesda, MD,
http://seer,cancer.gov/esr/1975 2009 hops09/, based on November 2011 SEER data
submission, posted to the SEER web site, April 2012;
[000146] 3. Leite, K,R,M., Camara-Lopes, L,Hõ Dall'Oglio, M.F., Cury, J.,
Antunes,
A,A., Safludo, A., Srougi, M.: Upgrading the Gleason score in extended
prostate biopsy:
Implications for treatment choice. lm, J. Radiat. Oncol., Biol., Phys. 73(2)
(2009) 353-356;
[000147] 4. Bax, Jõ Cool, D., Gardi, L., Knight, K., Smith, D., Montreuil, J.,
Sherebrin, S.,
Romagnali, C., Fenster, A.: Mechanically assisted 3D ultrasound guided
prostate biopsy
system. Medical Physics 35(12) (2008) 5397-410.
[000148] 5. Xu, S., Kruecker, J., Turkbey, B., Glossop, N., Singh, A.K.,
Choyke, P.,
Pinto, P., Wood, B.J.: Real-time MRI-TRUS fusion for guidance of targeted
prostate
biopsies. Comput. Aided Surg. 13(5) (2008) 255-264;
[000149] 6. Cool D., Sherebrin 5õ Izawa J., Chin J., Fenster, A.: Design and
evaluation of
a 3D transrectal ultrasound prostate biopsy system. Med, Phys. 35(10) (2008)
4695-4707;
[000150] 7. Baumann, M., Mozer, P., Daanen, V., Troccaz, J.: Towards 3D
ultrasound
image based soft tissue tracking: A transrectal ultrasound prostate image
alignment system.
In: Proceedings of the 10th International Conference on Medical Image
Computing and
Computer-Assisted Intervention LNCS 4792 (Part II) (2007) 26-33;
[000151] 8. Baumann, M., Mozer, P., Daanen, V., Troccaz, J.: Prostate biopsy
assistance
system with gland deformation estimation for enhanced precision. In:
Proceedings of the
CA 02839854 2014-01-17
10th International Conference on Medical Image Computing and Computer-Assisted
Intervention LNCS 5761(12) (2009) 67-74.
[000152] 9. Markelj, P., Tomazevi, D., Likar, B., Pernu , F.: A review of
3D/2D
registration methods for image-guided interventions. Med Image Anal, 16(3)
(2010) 642-
661;
[000153] 10. Birkfellner, Wõ Figl, M., Kettenbach, J., Hummel, J., Homolka,
P.,
Schernthaner, R., Nall, T., Bergmann, H.: Rigid 2D/3D slice-to-volume
registration and its
application on fluoroscopic CT images. Medical Physics 34(1) (2007) 246-55;
[000154] 11. Wein, W., Cheng, J.Zõ Khamene, A.: Ultrasound based Respiratory
Motion
Compensation in the Abdomen, MICCAI 2008 Workshop on Image Guidance and
Computer
Assistance 500 for Soft-Tissue Interventions (2008);
[000155] 12. Perona, P., Malik, J.: Scale-space and edge detection using
anisotropie
diffusion. IEEE Transactions on Pattern Analysis Machine Intelligence 12(7)
(1990) 629-
639;
[000156] 13. Karnik, V.V., Fenster, Aõ Bax, J., Cool, D.W., Gardi, L.,
Gyacskov, I.,
Romagnoli, C., Ward, A.D,: Assessment of image registration accuracy in three-
dimensional
transrectal ultrasound guided prostate biopsy, Medical Physics 32(2) (2010)
802-813;
[000157] 14. Karnik, V.V., Fenster, Aõ Bax, Jõ Cool, Romagnoli, C., Ward,
A.D.:
Evaluation of intersession 3D-TRUS to 3D-TRUS image registration for repeat
prostate
biopsies. Medical Physics 38(4) (2011) 1832-43;
[000158] 15, Hajnal, J., Hawkes, D. J., Hill, D.: Medical Image Registration.
CRC Press
(2001);
[000159] 16. Press, W. H., Flannery, B,P., Teukolsky, S. A., Vetterling W. T.:
Numerical
Recipes in C. Cambridge University Press, second edition (1992);
36
CA 02839854 2014-01-17
[000160] 17. Fitzpatrick, J.M., West, J.B., Maurer, Jr., C. R.: Predicting
error in rigid-body
point-based registration. IEEE Trans. Med. Imaging 17(5) (1998) 694-702;
[000161] 18. Epstein J.I., Sanderson H., Carter H.B., Scharfstein D.4.:
Utility of saturation
biopsy to 515 predict insignificant cancer at radical prostectomy. Urology
66(2) (2005) 356-
360; and
[000162] 19. De Silva, T., Fenster, A., Bax, J., Romagnoli, C., Izawa, J.,
Samarabandu, J.,
Ward A.D., Quantification of prostate deformation due to needle insertion
during TRUS-
guided biopsy: comparison of hand-held and mechanically stabilized systems.
Medical
Physics 38(3) (2011) 1718-31.
[000163] 20 The ITK Software Guide, Second Edition, Updated for ITK version
2.4
[000164] 21 Riccardo Poli = James Kennedy = Tim Blackwell, Particle swarm
optimization,
Swarm Intel! (2007) 1: 33-57 DOI 10.1007/s11721-007-0002-0
[000165] 22. Yukai Hung and WeichungWang, Accelerating parallel particle swarm
optimization via GPU, Optimization Methods & Software, Vol. 27, No. 1,
February 2012,
33-51.
[000166] Although the above description relates to a specific preferred
embodiment as
presently contemplated by the inventors, it will be understood that the
invention in its broad
aspect includes mechanical and functional equivalents of the elements
described herein.
37