Note: Descriptions are shown in the official language in which they were submitted.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
1
TITLE OF THE INVENTION
METHODS AND KITS FOR DIAGNOSING AND/OR PROGNOSING OSTEOARTHRITIS
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This
application is PCT application No. PCT/CA2012/00* filed on October 15, 2012
and
published in English under PCT Article 21(2), which claims benefit of U.S.
provisional application Serial No.
61/547,275, filed on October 14, 2011. All documents above are incorporated
herein in their entirety by reference.
FIELD OF THE INVENTION
[0002] The
present invention relates to the degradation of joints, and more particularly
to the
prognosis and/or diagnosis of osteoarthritis (OA).
REFERENCE TO SEQUENCE LISTING
[0003]
Pursuant to 37 C.F.R. 1.821(c), a sequence listing is submitted herewith as an
ASCII compliant
text file named 13200.18_5T25.txt, created on October *, 2012 and having a
size of *** kilobytes. The content of
the aforementioned file is hereby incorporated by reference in its entirety.
BACKGROUND OF THE INVENTION
[0004] The
etiology of OA, the most common form of arthritis, remains unclear
notwithstanding
the multiplicity of factors that have been considered in primary OA (1, 2). At
present, it has become increasingly
evident that the majority of OA genetic susceptibility loci cannot be
attributed only to structural genes or genes
regulating bone mass (3-5). These studies have also highlighted the great
heterogeneity and differences in the
degree of OA heritability between different joint sites (e.g., hand versus
knee) and gender. This is also reflected
by the multiplicity of loci identified in OA linkage studies and their
discrepancies. Moreover, the functional
importance of these susceptibility loci has yet to be confirmed and
illustrates our incomplete knowledge of the
biology of OA.
[0005]
Diagnosis of OA is generally made based on history and clinical examination to
observe
signs and symptoms associated with OA such as joint swelling, joint
tenderness, decreased range of motion in
joints, visible joint damage (i.e., bony growths), etc. X-rays are typically
used to confirm the diagnosis of
osteoarthritis. The typical changes seen on X-ray include: joint space
narrowing, subchondral sclerosis (increased
bone formation around the joint), subchondral cyst formation, and osteophytes.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
2
[0006] There
is a need for novel methods and kits for the assessment of the risk of
development,
progression and/or severity of OA.
[0007] The
present description refers to a number of documents, the content of which is
herein
incorporated by reference in their entirety.
SUMMARY OF THE INVENTION
[0008] In
accordance with an aspect of the present invention, there is provided a method
of
determining whether a subject (e.g., asymptomatic or diagnosed) is at risk of
developing (e.g., first symptoms or
more severe symptoms) osteoarthritis, said method comprising: determining the
cellular localization of a
Prohibitin-1 (PHB1) polypeptide and/or Small Ubiquitin-like Modifier (SUMO)
polypeptide, and/or increased
expression or activity of Ubc9 polypeptide in a blood cell sample from said
subject; and determining whether said
subject is at risk of developing osteoarthritis based on the cellular
localization of a PHB1 polypeptide and/or
SUMO polypeptide. In that context, OA patients exhibiting stronger nuclear
accumulation of PHB1 and/or SUMO-
1, and/or SUM02 and/or SUM03 sumoylated proteins and/or Ubc9 expression or
activity present a greater risk of
disease aggravation (disease staging).
[0009] In
accordance with another aspect of the present invention, there is provided a
method of
determining whether a subject is at risk of developing osteoarthritis (OA),
said method comprising: determining
the cellular localization of a Prohibitin-1 (PHB1) polypeptide and/or Small
Ubiquitin-like Modifier (SUMO)
polypeptide, in a cell sample from said subject; and determining whether said
subject is at risk of developing OA
based on the cellular localization of a PHB1 polypeptide and/or SUMO
polypeptide.
[0010] In an
embodiment, the above-mentioned method further comprises determining whether
the PHB1 polypeptide and/or SUMO polypeptide nuclear concentration is higher
in the subject blood cell sample
relative to that in a control blood cell sample; wherein a higher PHB1
polypeptide and/or SUMO polypeptide
nuclear concentration in the subject cell sample is indicative that the
subject is at risk of developing osteoarthritis.
[0011] In a
specific embodiment, said method further comprises determining whether the
PHB1
polypeptide and/or SUMO polypeptide nuclear concentration is higher in the
subject blood cell sample relative to
that in a control blood cell sample; wherein a higher PHB1 polypeptide and/or
SUMO polypeptide nuclear
concentration in the subject cell sample is indicative that the subject is at
risk of developing OA.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
3
[0012] In a
specific embodiment, said method further comprises determining the cellular
localization of a promyelocytic leukemia (PML) polypeptide, in the cell sample
from said subject, wherein a higher
level of co-localization of a SUMO-1 and/or SUMO-2 and/or SUMO-3 polypeptide
and the PML polypeptide in
nuclear bodies of the cell from said subject is indicative that the subject is
at risk of developing OA.
[0013] In
another specific embodiment, said cell sample (e.g., blood cell sample) is a
peripheral
blood mononuclear cell (PBMC) sample. In another specific embodiment, said
SUMO polypeptide is a SUMO-1
polypeptide.
[0014] In another specific embodiment, said SUMO polypeptide is a SUMO-2
polypeptide.
[0015] In another specific embodiment, said SUMO polypeptide is a SUMO-3
polypeptide.
[0016] In another specific embodiment, a higher level of the SUMO polypeptide
in nuclear bodies of the cell
from said subject is indicative that the subject is at risk of developing OA.
[0017] In another specific embodiment, said method comprises: determining
whether the level of co-localization
of the SUMO-1 polypeptide and the PHB1 polypeptide in the nuclear bodies is
higher relative to that in a control
cell; wherein a higher level of co-localization of a SUMO-1 polypeptide and a
PHB1 polypeptide in nuclear bodies
of the cell from said subject is indicative that the subject is at risk of
developing OA.
[0018] In accordance with another aspect of the present invention, there is
provided a method of determining
whether a subject is at risk of developing osteoarthritis (OA), said method
comprising: determining the level of an
enzyme involved in the sumoylation of protein in a cell sample from said
subject; and determining whether said
subject is at risk of developing OA based on the level of said enzyme in said
cell sample.
[0019] In a specific embodiment, said method further comprises determining
whether the level of said enzyme is
higher in the subject sample relative to that in a control cell sample,
wherein a higher level of said enzyme in the
subject cell sample is indicative that the subject is at risk of developing
OA.
[0020] In another specific embodiment, said enzyme is ubiquitin-like protein
sumo conjugating enzyme (UBC9).
[0021] In
accordance with another aspect of the present invention, there is provided a
method of
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
4
determining whether a subject is at risk of developing osteoarthritis, said
method comprising: determining whether
the level of a SUMO polypeptide in nuclear bodies of a cell from said subject
is higher relative to that in a control
cell; wherein a higher level of a SUMO polypeptide in nuclear bodies of the
cell from said subject is indicative that
the subject is at risk of developing osteoarthritis.
[0022] In
accordance with another aspect of the present invention, there is provided a
method of
determining whether a subject is at risk of developing osteoarthritis, said
method comprising: determining whether
the level of co-localization of a SUMO-1 polypeptide and a PHB1 polypeptide in
nuclear bodies of a cell from said
subject is higher relative to that in a control cell; wherein a higher level
of co-localization of a SUMO-1 polypeptide
and a PHB1 polypeptide in nuclear bodies of the cell from said subject is
indicative that the subject is at risk of
developing osteoarthritis.
[0023] In
accordance with another aspect of the present invention, there is provided a
method of
determining whether a subject is at risk of developing osteoarthritis (OA),
said method comprising: determining
whether the level of co-localization of a SUMO-1 and/or SUMO-2 and/or SUMO-3
polypeptide and a PML
polypeptide in nuclear bodies of a cell from said subject is higher relative
to that in a control cell; wherein a higher
level of co-localization of a SUMO-1 and/or SUMO-2 and/or SUMO-3 polypeptide
and a PML polypeptide in
nuclear bodies of the cell from said subject is indicative that the subject is
at risk of developing OA.
[0024] In
accordance with another aspect of the present invention, there is provided a
method of
determining whether a subject is at risk of developing osteoarthritis, said
method comprising: determining whether
(i) the amount of PML nuclear bodies in a cell from said subject is higher
relative to that in a control cell and/or (ii)
the size of PML nuclear bodies in a cell from said subject is larger relative
to that in a control cell; wherein a
higher amount and/or larger size of PML nuclear bodies in the cell from said
subject is indicative that the subject
is at risk of developing osteoarthritis.
[0025] In
accordance with another aspect of the present invention, there is provided a
method of
determining whether a subject is at risk of developing osteoarthritis, said
method comprising: determining whether
the level of an enzyme involved in sumoylation (e.g., ubiquitin-like protein
SUMO conjugating enzyme (UBC9)) in
a cell sample from said subject; and determining whether said subject is at
risk of developing osteoarthritis based
on the level of said enzyme involved in sumoylation in said cell sample. In an
embodiment, the above-mentioned
method further comprises determining whether the level of said enzyme is
higher in the subject cell sample
relative to that in a control cell sample; wherein a higher level of said
enzyme in the subject cell sample is
indicative that the subject is at risk of developing osteoarthritis.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
[0026] In an
embodiment, the above-mentioned method further comprises determining whether
the level of said enzyme is higher in the subject sample relative to that in a
control cell sample; wherein a higher
level of said enzyme in the subject cell sample is indicative that the subject
is at risk of developing OA. In another
embodiment, said enzyme is ubiquitin-like protein SUMO conjugating enzyme
(UBC9).
[0027] In
another embodiment, of the above-mentioned methods, said cell is an articular
chondrocyte, a growth plate chondrocyte, an osteoblast, a skeletal myoblast,
synoviocyte or a blood cell.
[0028] In another embodiment, of the above-mentioned methods, said cell sample
is an articular chondrocyte
sample, a growth plate chondrocyte sample, an osteoblast sample, a skeletal
myoblast sample, a synoviocyte
sample or a blood cell sample.
[0029] In another embodiment, said cell or cell sample is a peripheral blood
mononuclear cell (PBMC) sample.
[0030] In another embodiment, said cell or cell sample is a leucocytes sample.
[0031] In
accordance with another aspect of the present invention, there is provided a
method of
determining whether a subject is at risk of developing osteoarthritis, said
method comprising: determining the
level of PHB1 in a blood sample from said subject; and determining whether
said subject is at risk of developing
osteoarthritis based on the level of PHB1 in said blood sample; wherein a
lower level of PHB1 in the subject
blood sample is indicative that the subject is at risk of developing
osteoarthritis.
[0032] In an
embodiment, the above-mentioned methods further comprise identifying a subject
suspected of having osteoarthritis (OA).
[0033] In an
embodiment, the above-mentioned methods further comprise identifying a subject
suspected of having primary osteoarthritis (OA).
[0034] In an
embodiment of the above-mentioned methods, the OA is knee joint arthritis, hip
joint
arthritis or temporo-mandibular joint arthritis. In an embodiment of the above-
mentioned methods, the OA is knee
joint arthritis. In an embodiment of the above-mentioned methods, the OA is
hip joint arthritis. In an embodiment
of the above-mentioned methods, the OA is primary OA.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
6
[0035] The method of any one of claims 1 to 20, wherein the determining of
whether the subject is at risk of
developing OA determines whether the subject is at risk of developing a more
severe primary OA symptoms at a
future time.
[0036] In
accordance with another aspect of the present invention, there is provided a
method of
selecting a compound, said method comprising (a) contacting a test compound
with a blood cell sample; and (b)
determining a PHB1 polypeptide and/or SUMO polypeptide nuclear localization in
the blood cell; wherein the test
compound is selected if the PHB1 polypeptide and/or SUMO polypeptide nuclear
localization in the cell sample is
decreased in the presence of the test compound relative to in the absence
thereof.
[0037] In
accordance with another aspect of the present invention, there is provided a
method of
selecting a compound, said method comprising (a) contacting a test compound
with a cell sample; and (b)
determining a level of a SUMO polypeptide in nuclear bodies in the cell;
wherein the test compound is selected if
the level of SUMO polypeptide in nuclear bodies is decreased in the presence
of the test compound relative to in
the absence thereof.
[0038] In
accordance with another aspect of the present invention, there is provided a
method of
selecting a compound, said method comprising (a) contacting a test compound
with a cell sample; and (b)
determining a level of co-localization of a SUMO-1 polypeptide and a PHB1
polypeptide in nuclear bodies in the
cell; wherein the test compound is selected if the level of co-localization of
SUMO-1 polypeptide and PHB1
polypeptide in nuclear bodies is decreased in the presence of the test
compound relative to in the absence
thereof.
[0039] In
accordance with another aspect of the present invention, there is provided a
method of
selecting a compound, said method comprising (a) contacting a test compound
with a cell sample; and (b)
determining (i) an amount of promyelocytic leukemia (PML) nuclear bodies in
the cell and/or (ii) the size of PML
nuclear bodies in the cell; wherein the test compound is selected if the
amount and/or size of PML nuclear bodies
is decreased in the presence of the test compound relative to in the absence
thereof.
[0040] In
accordance with another aspect of the present invention, there is provided a
method of
selecting a compound, said method comprising (a) contacting a test compound
with a cell sample; and (b)
determining a level of an enzyme involved in sumoylation (e.g., UBC9) in the
cell sample; wherein the test
compound is selected if the level of said enzyme in the cell sample is
decreased in the presence of the test
compound relative to in the absence thereof.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
7
[0041] In
accordance with another aspect of the present invention, there is provided a
method of
selecting a compound, said method comprising (a) administering a test compound
to a subject; and (b)
determining a level of PHB1 in a blood sample from said subject; wherein the
test compound is selected if the
level of PHB1 in the blood sample is increased in the presence of the test
compound relative to in the absence
thereof.
[0042] In
another specific embodiment, the selected test compound is potentially useful
in the
treatment of primary osteoarthritis.
[0043] In a
specific embodiment of the methods, the osteoarthritis is knee joint
arthritis, hip joint
arthritis or temporo-mandibular joint arthritis. In another specific
embodiment, the osteoarthritis is knee joint
arthritis. In another specific embodiment, the osteoarthritis is hip joint
arthritis. In another embodiment, the
osteoarthritis is primary osteoarthritis.
[0044] In an
embodiment, the above-mentioned cell is an articular chondrocyte, a growth
plate
chondrocyte, an osteoblast, a skeletal myoblast, a synoviocyte or a blood
cell. In a further embodiment, the blood
cell is a peripheral blood mononuclear cell (PBMC).
[0045] In a specific embodiment of the methods, the subject is a woman.
[0046] In
accordance with another aspect of the present invention, there is provided a
kit
comprising a ligand specific to a Prohibitin-1 (PHB1) polypeptide and/or Small
Ubiquitin-like Modifier (SUMO)
polypeptide, and/or UBC9 polypeptide and instructions to use the ligand to
predict whether a subject is at risk for
developing osteoarthritis.
[0047] In a specific embodiment of the kit, the kit comprises at least two of
a ligand specific to a Prohibitin-1
(PHB1) polypeptide, a ligand specific to a Small Ubiquitin-like Modifier
(SUMO) polypeptide (SUMO 1, 2 and/or
3), and a ligand specific to a UBC9 polypeptide. In a specific embodiment of
the kit, the kit comprises a ligand
specific to a Prohibitin-1 (PHB1) polypeptide, a ligand specific to a Small
Ubiquitin-like Modifier (SUMO)
polypeptide, and a ligand specific to a UBC9 polypeptide. In a specific
embodiment the ligand is a ligand specific
to a Prohibitin-1 (PHB1) polypeptide and/or Small Ubiquitin-like Modifier
(SUMO) polypeptide, and/or UBC9
polypeptide.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
8
[0048] The articles "a," an and the are used herein to refer to one or to more
than one (i.e., to at least one)
of the grammatical object of the article.
[0049] The
terms "including" and "comprising" are used herein to mean, and re used
interchangeably with, the phrases "including but not limited to and
"comprising but not limited to.
[0050] The
terms such as are used herein to mean, and is used interchangeably with, the
phrase such as but not limited to.
[0051] As used
herein the term "osteoarthritis" refers to a form of arthritis involving the
deterioration of the cartilage that cushions the ends of bones within joints.
It is also called degenerative arthritis,
degenerative joint disease or hypertrophic arthritis. This term includes early
onset of osteoarthritis. Worldwide,
osteoarthritis is the most common joint disorder. In western countries,
radiographic evidence of this disease is
present in the majority of persons by 65 years of age and in about 80 percent
of persons more than 75 years of
age (33). Approximately 11 percent of persons more than 64 years of age have
symptomatic osteoarthritis of the
knee (34).
[0052] As used
herein the terms "early onset of osteoarthritis" refer to a form of
osteoarthritis that
either is first diagnosed at 40 years of age or earlier or that leads to knee
joint replacement of the subject before
he is 55 years old.
[0053] As used
herein the terms "risk of developing osteoarthritis" refers to a
predisposition of a
subject of presenting primary OA symptoms and/or more severe primary OA
symptoms at a future time (disease
staging). Similarly, the "risk of developing osteoarthritis in a joint where
Pitx1 is normally expressed" refers to a
risk for a subject of presenting primary OA symptoms, and/or more severe
primary OA symptoms at a future time
in a joint where Pitx1 is normally expressed.
[0054] As used
herein the terms "primary OA" when used to qualify knee/hip joint OA refer to
knee/hip joint OA due to a disease or degeneration for instance as opposed to
secondary knee/hip joint OA
resulting from trauma, joint overuse, obesity, etc.
[0055] As used
herein the term "subject" is meant to refer to any mammal including human,
mice, rat, dog, cat, pig, monkey, horse, etc. In a particular embodiment, it
refers to a human. In another particular
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
9
embodiment, it refers to a horse and more specifically a racing horse.
[0056] As used
herein the terms "predisposition for developing a disease or condition" refers
to a
predisposition of a subject of presenting symptoms of the disease or condition
and/or more severe symptoms of
the disease or conditions at a future time.
[0057] As used
herein the terms "control sample" are meant to refer to a sample that does not
come from a subject known to suffer from the disease or disorder or from the
subject under scrutiny but before
the subject had the disease or disorder. In methods of diagnosing a
predisposition of a subject to develop a
disease or disorder, the sample may also come from the subject under scrutiny
at an earlier stage of the disease
or disorder. The term "control sample" may also refer to a pre-determined,
control value recognized in the art or
established based on levels measured in one or a group of control subjects.
The corresponding control
level/value may be adjusted or normalized for age, gender, race, or other
parameters. The control level can thus
be a single number/value, equally applicable to every patient individually, or
the control level can vary, according
to specific subpopulations of patients.
[0058] As used
herein the term "cell sample" is meant to refer to a sample containing any
type of
cell wherein, in a subject affected by OA, PHB1, SUMO (SUMO-1 and/or SUMO-2
and/or SUMO-3) and/or UBC9
pathologically accumulates in the cell nuclei (e.g., in nuclear bodies).
Without being so limited, it includes articular
chondrocytes, growth plate chondrocytes, osteoblasts, skeletal myoblasts,
synoviocytes, blood cells (e.g.,
PBMCs). As used herein the term "articular chondrocyte" is meant to refer to
chondrocytes found in joints.
[0059] As used
herein, the term "blood cell sample" refers to a sample containing cells
normally found in blood, and includes for example peripheral blood mononuclear
cells (PBMCs) as well as
particular cell types such as lymphocytes (T cells, B cells, NK cells),
monocytes, basophils, and dendritic cells, or
any mixture thereof. In an embodiment, the above-mentioned blood cell sample
may be submitted to one or more
cell depletion or enrichment steps, so as to enrich the sample in one or more
cell types of interest.
[0060] As used
herein the term "blood sample" is meant to refer to a sample derived from
blood,
and include for example whole blood, or to a fraction thereof, such as serum,
plasma and the like. It also refers to
any sample that may be obtained following one or more purification,
enrichment, and/or treatment steps using
blood (obtained by venous puncture, for example) as starting material. In an
embodiment, the blood sample is a
plasma sample.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
[0061] As used
herein the term "not clinically diagnosed with osteoarthritis" is meant to
refer to a
subject that was never diagnosed with OA using a clinical method such as an
imaging method like X-ray, and
magnetic resonance imaging (MRI). In particular, for diagnosing hip OA, a
current clinical method recommended
by the American College of Rheumatology includes hip pain and at least 2 of
the following 3 features: ESR<20
mm/hour; radiographic femoral or acetabular osteophytes; and radiographic
joint space narrowing (superior, axial,
and/or medial). In particular, for diagnosing knee OA, there are three methods
currently recommended by the
American College of Rheumatology Clinical and laboratory method: knee
pain and at least 5 of the following
9 features: age > 50 years, stiffness < 30 minutes, crepitus, bony tenderness,
bony enlargement, no palpable
warmth, ESR <40 mm/hour, RF <1:40; and SF OA; 2) Clinical and radiographic:
knee pain, and at least 2 of the
following 3 features, Age > 50 years; stiffness < 30 minutes; crepitus; +
osteophytes; and 3) Clinical: knee pain
and at least 3 of the following 6 features: age > 50 years, stiffness <30
minutes, crepitus, bony tenderness, bony
enlargement, no palpable warmth.
[0062] As used
herein the terminology "purified", "isolated", "purification" or "isolation"
in the
expressions "purified polypeptide", "isolated polypeptide", "isolated
protein", "purified complexes", "isolated
complexes" or "tandem affinity purification" means altered by the hand of man"
from its natural state (i.e. if it
occurs in nature, it has been changed or removed from its original
environment) or it has been synthesized in a
non-natural environment (e.g., artificially synthesized). These terms do not
require absolute purity (such as a
homogeneous preparation) but instead represents an indication that it is
relatively more pure than in the natural
environment. For example, a protein/peptide naturally present in a living
organism is not "purified" or "isolated",
but the same protein separated (about 90-95% pure at least) from the
coexisting materials of its natural state is
"purified" or "isolated" as this term is employed herein.
[0063]
Sumoylation is a post-translational modification in which a molecule called
SUMO (Small
Ubiquitin-like MOdifier) is covalently but reversibly linked to a lysine
residue in a process similar to ubiquitination.
SUMO proteins are ubiquitous in eukaryotes and highly conserved from yeast to
humans. Generally, sumoylation
seems to have an inhibitory effect on gene transcription and it was proposed
that sumoylation could act on
various transcription factors to promote their interaction with co-repressors
(Gill G. Curr.Opin.Genet.Dev. 2005;
15:536-541). In vertebrates, there are four isoforms of SUMO proteins named
SUMO-1 to SUMO-4 (Gill 2005,
supra, Figures 19 to 22). SUMOs are attached to their target proteins in a
three-step process implying an
activation enzyme El, a conjugation enzyme E2 and an E3 ligase. In humans, El
is composed of two subunits
(SAE1/SEA2), the unique conjugation E2 enzyme is called UBC9 and there are at
least five known E3 ligases:
PIAS1 (protein inhibitor of activated signal transducer), which is the
prototype of a family that encompasses three
additional members (PIAS3, PIASy and PIASx), Pc2 (human Polycomb member 2) and
Ran-BP2. There also
exist at least seven SUMO-specific proteases in humans named SENP-1 to SENP-8.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
11
[0064] In an
embodiment, the above-mentioned SUMO polypeptide is a SUMO-1, SUMO-2,
SUMO-3 and/or SUMO-4 polypeptide. In an embodiment, the above-mentioned SUMO
polypeptide is a SUMO-1,
SUMO-2, and/or SUMO-3 polypeptide. In a further embodiment, above-mentioned
SUMO polypeptide is a
SUMO-1 polypeptide.
[0065] In an
embodiment, the above-mentioned enzyme involved in sumoylation is an
activation
enzyme El, a conjugation enzyme E2 and/or an E3 ligase. In a further
embodiment, the above-mentioned
enzyme involved in sumoylation is a conjugation enzyme E2, in a further
embodiment UBC9.
Diagnostic or prognostic methods
[0066] A
method for diagnosing or screening for the presence of a disease or disorder
or a
predisposition for developing the disease or disorder in a subject ("risk of
developing") , which disease or disorder
is characterized by an aberrant amount, activity, protein composition,
intracellular localization and/or formation of
a complex, comprising the steps of: (1) comparing the amount of, activity of,
protein composition of, intracellular
localization (e.g., in nuclear bodies such as PML nuclear bodies) of, and/or
formation of said complex (e.g.,
SUMO-1 and/or -2 and/or -3 with at least another protein (e.g., PML, PHB1)) in
a sample from the subject with
that in a control sample, wherein a difference in said amount, activity,
protein composition of, intracellular
localization and/or formation of said complex as compared to that in the
control sample is indicative that the
subject has the disease or disorder or a predisposition for developing the
disease or condition. A comparison of
amount, activity, protein composition, intracellular localization and/or
formation of a complex of certain proteins
between various OA patients may also provide means of classifying/stratifying
the patients. Hence for example,
when comparing two OA subjects, detecting a higher level of PHB1 and/or SUMO-1
and/or SUMO-2 and/or
SUMO-3 and/or UBC9 in the first OA subject than in the second OA subject is an
indication that the first OA
subject has a higher risk of developing OA or a risk of developing a more
severe OA form than the second OA
subject.
[0067] In a
specific embodiment, the control sample is selected from a sample from the
subject
at an earlier stage of the disease or disorder or before the subject had the
disease. In another embodiment, the
control sample is from a different subject that does not have the disease or
disorder or predisposition to develop
the disease or condition.
[0068] The
amount and/or localization of PHB1, SUMO (e.g., SUMO-1) and/or UBC9 may be
determined using any known method in the art. In an embodiment, the amount
and/or localization of PHB1,
SUMO (e.g., SUMO-1) and/or UBC9 is determined at the protein/polypeptide
level, for example using a molecule
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
12
capable of specifically binding to a PHB1, SUMO (e.g., SUMO-1) or UBC9
polypeptide. PHB1, SUMO (e.g.,
SUMO-1) or UBC9 polypeptide expression levels may be determined using any
standard methods known in the
art. Non-limiting examples of such methods include Western blot, tissue
microarray, immunoblot, enzyme-linked
immunosorbant assay (ELISA), radioimmunoassay (RIA), immunoprecipitation,
surface plasmon resonance,
chemiluminescence, fluorescent polarization, phosphorescence,
immunohistochemical analysis,
immunofluorescence, matrix-assisted laser desorption/ionization time-of-flight
(MALDI-TOF) mass spectrometry,
microcytometry, microscopy, fluorescence-activated cell sorting (FACS), flow
cytometry, and assays based on a
property of the protein including but not limited to DNA binding, ligand
binding, or interaction with other protein
partners.
[0069] In an
embodiment, the molecule capable of specifically binding to a PHB1, SUMO
(e.g.,
SUMO-1) or UBC9 polypeptide is an antibody specifically binding to, or
specifically recognizing, a PHB1, SUMO
(e.g., SUMO-1) or UBC9 polypeptide.
[0070] As used
herein, the term "antibody" refers to an antibody that specifically binds to
(interacts with) a protein of interest (PHB1, SUMO (1, 2 and/or 3) and/or
UBC9) and displays no substantial
binding to other naturally occurring proteins other than the ones sharing the
same antigenic determinants. The
term antibody or immunoglobulin is used in the broadest sense, and covers
monoclonal antibodies (including full
length monoclonal antibodies), polyclonal antibodies, multispecific
antibodies, and antibody fragments so long as
they exhibit the desired biological activity. Antibody fragments comprise a
portion of a full length antibody,
generally an antigen binding or variable region thereof. Examples of antibody
fragments include Fab, Fab',
F(ab')2, and Fv fragments, diabodies, linear antibodies, single-chain antibody
molecules, single domain antibodies
(e.g., from camelids), shark NAR single domain antibodies, and multispecific
antibodies formed from antibody
fragments. Antibody fragments can also refer to binding moieties comprising
CDRs or antigen binding domains
including, but not limited to, VH regions (VH, VH-VH), anticalins,
PepBodiesTM, antibody-T-cell epitope fusions
(Troybodies) or Peptibodies. Additionally, any secondary antibodies, either
monoclonal or polyclonal, directed to
the first antibodies would also be included within the scope of this
invention.
[0071] In
general, techniques for preparing antibodies (including monoclonal antibodies
and
hybridomas) and for detecting antigens using antibodies are well known in the
art (Campbell, 1984, In
"Monoclonal Antibody Technology: Laboratory Techniques in Biochemistry and
Molecular Biology", Elsevier
Science Publisher, Amsterdam, The Netherlands) and in Harlow et al., 1988 (in:
Antibody A Laboratory Manual,
CSH Laboratories).
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
13
[0072] The
present invention also relates to methods for the determination of the level
of
expression of transcripts or translation product of a gene such as SUMO, PHB1
or UBC9. The present invention
therefore encompasses any known method for such determination including real
time PCR and competitive PCR,
in situ PCR, SAGE, Northern blots, in situ hybridization, Southern blot,
nuclease protection, plaque hybridization
and slot blots.
[0073] The
present invention also concerns isolated nucleic acid molecules including
probes. In
specific embodiments, the isolated nucleic acid molecules have no more than
300, or no more than 200, or no
more than 100, or no more than 90, or no more than 80, or no more than 70, or
no more than 60, or no more than
50, or no more than 40 or no more than 30 nucleotides. In specific
embodiments, the isolated nucleic acid
molecules have at least 20, or at least 30, or at least 40 nucleotides. In
other specific embodiments, the isolated
nucleic acid molecules have at least 20 and no more than 300 nucleotides. In
other specific embodiments, the
isolated nucleic acid molecules have at least 20 and no more than 200
nucleotides. In other specific
embodiments, the isolated nucleic acid molecules have at least 20 and no more
than 100 nucleotides. In other
specific embodiments, the isolated nucleic acid molecules have at least 20 and
no more than 90 nucleotides. In
other specific embodiments, the isolated nucleic acid molecules have at least
20 and no more than 80
nucleotides. In other specific embodiments, the isolated nucleic acid
molecules have at least 20 and no more
than 70 nucleotides. In other specific embodiments, the isolated nucleic acid
molecules have at least 20 and no
more than 60 nucleotides. In other specific embodiments, the isolated nucleic
acid molecules have at least 20
and no more than 50 nucleotides. In other specific embodiments, the isolated
nucleic acid molecules have at
least 20 and no more than 40 nucleotides. In other specific embodiments, the
isolated nucleic acid molecules
have at least 20 and no more than 30 nucleotides. In other specific
embodiments, the isolated nucleic acid
molecules have at least 30 and no more than 300 nucleotides. In other specific
embodiments, the isolated nucleic
acid molecules have at least 30 and no more than 200 nucleotides. In other
specific embodiments, the isolated
nucleic acid molecules have at least 30 and no more than 100 nucleotides. In
other specific embodiments, the
isolated nucleic acid molecules have at least 30 and no more than 90
nucleotides. In other specific embodiments,
the isolated nucleic acid molecules have at least 30 and no more than 80
nucleotides. In other specific
embodiments, the isolated nucleic acid molecules have at least 30 and no more
than 70 nucleotides. In other
specific embodiments, the isolated nucleic acid molecules have at least 30 and
no more than 60 nucleotides. In
other specific embodiments, the isolated nucleic acid molecules have at least
30 and no more than 50
nucleotides. In other specific embodiments, the isolated nucleic acid
molecules have at least 30 and no more
than 40 nucleotides.
[0074] Probes
of the invention can be utilized with naturally occurring sugar-phosphate
backbones as well as modified backbones including phosphorothioates,
dithionates, alkyl phosphonates and a-
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
14
nucleotides and the like. Modified sugar-phosphate backbones are generally
known (62,63). Probes of the
invention can be constructed of either ribonucleic acid (RNA) or
deoxyribonucleic acid (DNA), and preferably of
DNA.
[0075] The
types of detection methods in which probes can be used include Southern blots
(DNA
detection), dot or slot blots (DNA, RNA), and Northern blots (RNA detection).
Although less preferred, labeled
proteins could also be used to detect a particular nucleic acid sequence to
which it binds. Other detection
methods include kits containing probes on a dipstick setup and the like.
[0076] As used
herein the terms "detectably labeled" refer to a marking of a probe in
accordance
with the presence invention that will allow the detection of the mutation of
the present invention. Although the
present invention is not specifically dependent on the use of a label for the
detection of a particular nucleic acid
sequence, such a label might be beneficial, by increasing the sensitivity of
the detection. Furthermore, it enables
automation. Probes can be labeled according to numerous well known methods
(64). Non-limiting examples of
labels include 3H, 1403 32p3 and 355. Non-limiting examples of detectable
markers include ligands, fluorophores,
chemiluminescent agents, enzymes, and antibodies. Other detectable markers for
use with probes, which can
enable an increase in sensitivity of the method of the invention, include
biotin and radionucleotides. It will become
evident to the person of ordinary skill that the choice of a particular label
dictates the manner in which it is bound
to the probe.
[0077] As
commonly known, radioactive nucleotides can be incorporated into probes of the
invention by several methods. Non-limiting examples thereof include kinasing
the 53 ends of the probes using
gamma 32P ATP and polynucleotide kinase, using the Klenow fragment of Pol I of
E. coli in the presence of
radioactive dNTP (e.g. uniformly labeled DNA probe using random
oligonucleotide primers in low-melt gels),
using the 5P6/T7 system to transcribe a DNA segment in the presence of one or
more radioactive NTP, and the
like.
[0078] The
present invention also relates to methods of selecting compounds. As used
herein
the term "compound" is meant to encompass natural, synthetic or semi-synthetic
compounds, including without
being so limited chemicals, macromolecules, cell or tissue extracts (from
plants or animals), nucleic acid
molecules, peptides, antibodies and proteins.
[0079] The
present invention also relates to arrays. As used herein, an "array" is an
intentionally
created collection of molecules which can be prepared either synthetically or
biosynthetically. The molecules in
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
the array can be identical or different from each other. The array can assume
a variety of formats, e.g., libraries of
soluble molecules; libraries of compounds tethered to resin beads, silica
chips, or other solid supports.
[0080] As used
herein "array of nucleic acid molecules is an intentionally created collection
of
nucleic acids which can be prepared either synthetically or biosynthetically
in a variety of different formats (e.g.,
libraries of soluble molecules; and libraries of oligonucleotides tethered to
resin beads, silica chips, or other solid
supports). Additionally, the term "array" is meant to include those libraries
of nucleic acids which can be prepared
by spotting nucleic acids of essentially any length (e.g., from 1 to about
1000 nucleotide monomers in length)
onto a substrate. The term "nucleic acid" as used herein refers to a polymeric
form of nucleotides of any length,
either ribonucleotides, deoxyribonucleotides or peptide nucleic acids (PNAs),
that comprise purine and pyrimidine
bases, or other natural, chemically or biochemically modified, non-natural, or
derivatized nucleotide bases. The
backbone of the polynucleotide can comprise sugars and phosphate groups, as
may typically be found in RNA or
DNA, or modified or substituted sugar or phosphate groups. A polynucleotide
may comprise modified nucleotides,
such as methylated nucleotides and nucleotide analogs. The sequence of
nucleotides may be interrupted by non-
nucleotide components. Thus the terms nucleoside, nucleotide, deoxynucleoside
and deoxynucleotide generally
include analogs such as those described herein. These analogs are those
molecules having some structural
features in common with a naturally occurring nucleoside or nucleotide such
that when incorporated into a nucleic
acid or oligonucleotide sequence, they allow hybridization with a naturally
occurring nucleic acid sequence in
solution. Typically, these analogs are derived from naturally occurring
nucleosides and nucleotides by replacing
and/or modifying the base, the ribose or the phosphodiester moiety. The
changes can be tailor made to stabilize
or destabilize hybrid formation or enhance the specificity of hybridization
with a complementary nucleic acid
sequence as desired.
[0081] As used
herein "solid support", "support", and "substrate" are used interchangeably
and
refer to a material or group of materials having a rigid or semi-rigid surface
or surfaces. In many embodiments, at
least one surface of the solid support will be substantially flat, although in
some embodiments it may be desirable
to physically separate synthesis regions for different compounds with, for
example, wells, raised regions, pins,
etched trenches, or the like. According to other embodiments, the solid
support(s) will take the form of beads,
resins, gels, microspheres, or other geometric configurations.
[0082] Any
known nucleic acid arrays can be used in accordance with the present
invention. For
instance, such arrays include those based on short or longer oligonucleotide
probes as well as cDNAs or
polymerase chain reaction (FOR) products (52). Other methods include serial
analysis of gene expression
(SAGE), differential display, (53) as well as subtractive hybridization
methods (54), differential screening (DS),
RNA arbitrarily primer (RAP)-PCR, restriction endonucleolytic analysis of
differentially expressed sequences
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
16
(READS), amplified restriction fragment-length polymorphisms (AFLP).
[0083]
"Stringent hybridization conditions" and "stringent hybridization wash
conditions" in the
context of nucleic acid hybridization experiments such as Southern and
Northern hybridization are sequence
dependent, and are different under different environmental parameters. The Tm
is the temperature (under defined
ionic strength and pH) at which 50% of the target sequence hybridizes to a
perfectly matched probe. Specificity is
typically the function of post-hybridization washes, the critical factors
being the ionic strength and temperature of
the final wash solution. For DNA-DNA hybrids, the Tm can be approximated from
the equation of Meinkoth and
Wahl, 1984; Tm 81.5 C + 16.6 (log M) +0.41 (%GC) ¨ 0.61 (% form) ¨ 500/L;
where M is the molarity of
monovalent cations, %GC is the percentage of guanosine and cytosine
nucleotides in the DNA, % form is the
percentage of formamide in the hybridization solution, and L is the length of
the hybrid in base pairs. Tm is
reduced by about 1 C for each 1% of mismatching; thus, Tm, hybridization,
and/or wash conditions can be
adjusted to hybridize to sequences of the desired identity. For example, if
sequences with >90% identity are
sought, the Tm can be decreased 10 C. Generally, stringent conditions are
selected to be about 5 C lower than
the thermal melting point I for the specific sequence and its complement at a
defined ionic strength and pH.
However, severely stringent conditions can utilize a hybridization and/or wash
at 1, 2, 3, or 4 C lower than the
thermal melting point I; moderately stringent conditions can utilize a
hybridization and/or wash at 6, 7, 8, 9, or
C lower than the thermal melting point I; low stringency conditions can
utilize a hybridization and/or wash at
11, 12, 13, 14, 15, or 20 C lower than the thermal melting point I. Using the
equation, hybridization and wash
compositions, and desired T, those of ordinary skill will understand that
variations in the stringency of
hybridization and/or wash solutions are inherently described. If the desired
degree of mismatching results in a T
of less than 45 C (aqueous solution) or 32 C (formamide solution), it is
preferred to increase the SSC
concentration so that a higher temperature can be used. An extensive guide to
the hybridization of nucleic acids
is found in Tijssen, 1993. Generally, highly stringent hybridization and wash
conditions are selected to be about
5 C lower than the thermal melting point Tm for the specific sequence at a
defined ionic strength and pH.
[0084] An
example of highly stringent wash conditions is 0.15 M NaCI at 72 C for about
15
minutes. An example of stringent wash conditions is a 0.2X SSC wash at 65 C
for 15 minutes (see 64 for a
description of SSC buffer). Often, a high stringency wash is preceded by a low
stringency wash to remove
background probe signal. An example medium stringency wash for a duplex of,
e.g., more than 100 nucleotides,
is 1X SSC at 45 C for 15 minutes. An example low stringency wash for a duplex
of, e.g., more than 100
nucleotides, is 4-6X SSC at 40 C for 15 minutes. For short probes (e.g., about
10 to 50 nucleotides), stringent
conditions typically involve salt concentrations of less than about 1.5 M,
more preferably about 0.01 to 1.0 M, Na
ion concentration (or other salts) at pH 7.0 to 8.3, and the temperature is
typically at least about 30 C and at least
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
17
about 60 C for long robes (e.g., >50 nucleotides). Stringent conditions may
also be achieved with the addition of
destabilizing agents such as formamide. In general, a signal to noise ratio of
2X (or higher) than that observed for
an unrelated probe in the particular hybridization assay indicates detection
of a specific hybridization. Nucleic
acids that do not hybridize to each other under stringent conditions are still
substantially identical if the proteins
that they encode are substantially identical. This occurs, e.g., when a copy
of a nucleic acid is created using the
maximum codon degeneracy permitted by the genetic code.
[0085] Very
stringent conditions are selected to be equal to the Tm for a particular
probe. An
example of stringent conditions for hybridization of complementary nucleic
acids which have more than 100
complementary residues on a filter in a Southern or Northern blot is 50%
formamide, e.g., hybridization in 50%
formamide, 1 M NaCI, 1% SDS at 37 C, and a wash in 0. 1X SSC at 60 to 65 C.
Exemplary low stringency
conditions include hybridization with a buffer solution of 30 to 35%
formamide, 1 M NaCI, 1% SDS (sodium
dodecyl sulphate) at 37 C, and a wash in 1X to 2X SSC (20X SSC = 3.0 M
NaCl/0.3 M trisodium citrate) at 50 to
55 C. Exemplary moderate stringency conditions include hybridization in 40 to
45% formamide, 1.0 M NaCI, 1%
SDS at 37 C, and a wash in 0.5X to 1X SSC at 55 to 60 C.
[0086] Washing
with a solution containing tetramethylammonium chloride (TeMAC) could allow
the detection of a single mismatch using oligonucleotide hybridization since
such mismatch could generate a
C difference in the annealing temperature. The formulation to determine the
washing temperature is Tm ( C)
=1-682 (L-1) + 97 where L represents the length of the oligonucleotide that
will be used for the hybridization.
[0087] The
present invention also encompasses arrays to detect and/or quantify the
nuclear
localization of proteins including PHB1, SUMO and UBC9. Such arrays include
protein micro- or macroarrays, gel
technologies including high-resolution 2D-gel methodologies, possibly coupled
with mass spectrometry (55),
imaging system at the cellular level such as microscopy combined with a
fluorescent labeling system.
[0088] The
present invention also includes the use of tissue biopsy to determine the
nuclear
accumulation of PHB1, SUMO and UBC9 within articular chondrocytes, growth
plate chondrocytes, osteoblasts,
skeletal myoblasts and synoviocytes. For instance, cartilage biopsy could be
performed during arthroscopy
procedure to assess OA or its progression by immunofluorescence microscopy to
determine the nuclear
localization of PHB1, SUMO and UBC9. This method could be useful for instance
when arthroscopy procedure is
required to establish a clinical diagnostic. Alternatively, a muscle biopsy in
lower limbs could be used to test
whether or not PHB1, SUMO and UBC9 are accumulated in the nuclei of myoblasts.
This method would
advantageously be less invasive than a regular arthroscopy. The determination
of the cellular localization or
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
18
concentration of a protein as disclosed herein (e.g., PHB1, SUMO and/or UBC9)
is typically performed either by
a) preparing a nuclear extract of a subject sample and determining
concentration of PHB1, SUMO and UBC9; or
by (b) determining the localization of PHB1, SUMO and UBC9 by
immunohistochemistry. Cellular localization or
concentration of these molecules may also be detected by other imaging or
detection methods enabling the
visualization and quantification of biomolecules, such as flow cytometry.
[0089] The
present invention relates to a kit for diagnosing OA and/or predicting whether
a
subject is at risk of developing OA comprising an isolated nucleic acid, a
protein or a ligand such as an antibody
in accordance with the present invention. For example, a compartmentalized kit
in accordance with the present
invention includes any kit in which reagents are contained in separate
containers. Such containers include small
glass containers, plastic containers or strips of plastic or paper. Such
containers allow the efficient transfer of
reagents from one compartment to another compartment such that the samples and
reagents are not cross-
contaminated and the agents or solutions of each container can be added in a
quantitative fashion from one
compartment to another. Such containers will include a container which will
accept the subject sample (DNA
genomic nucleic acid, cell sample or blood samples), a container which
contains in some kits of the present
invention, the probes used in the methods of the present invention, containers
which contain enzymes, containers
which contain wash reagents, and containers which contain the reagents used to
detect the extension products.
The present invention also relates to a kit comprising the antibodies which
are specific to PHB1, SUMO and/or
UBC9. Kits of the present invention may also contain instructions to use these
probes and or antibodies to
diagnose OA or predict whether a subject is at risk of developing OA.
[0090] As used
herein, the term "ligand" broadly refers to natural, synthetic or semi-
synthetic
molecules. The term "molecule" therefore denotes for example chemicals,
macromolecules, cell or tissue
extracts (from plants or animals) and the like. Non limiting examples of
molecules include nucleic acid molecules,
peptides, antibodies, carbohydrates and pharmaceutical agents. The ligand
appropriate for the present invention
can be selected and screened by a variety of means including random screening,
rational selection and by
rational design using for example protein or ligand modeling methods such as
computer modeling. The terms
"rationally selected" or "rationally designed" are meant to define compounds
which have been chosen based on
the configuration of interacting domains of the present invention. As will be
understood by the person of ordinary
skill, macromolecules having non-naturally occurring modifications are also
within the scope of the term "ligand".
For example, peptidomimetics, well known in the pharmaceutical industry and
generally referred to as peptide
analogs can be generated by modeling as mentioned above.
[0091] Other
objects, advantages and features of the present invention will become more
apparent upon reading of the following non-restrictive description of specific
embodiments thereof, given by way
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
19
of example only with reference to the accompanying drawings.
BRIEF DESCRIPTION OF THE DRAWINGS
[0092] The patent or application file contains at least one drawing
executed in color. Copies of
this patent or patent application publication with color drawing(s) will be
provided by the Office upon request and
payment of the necessary fee.
[0093] In the appended drawings:
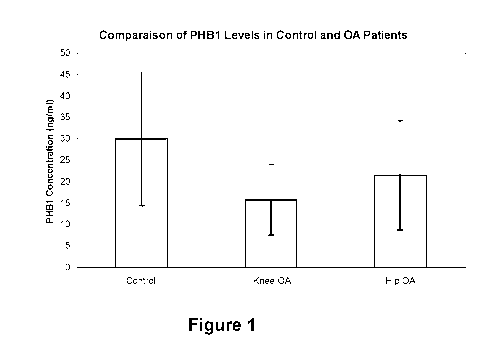
[0094] Figure 1 shows mean levels of PHB1 in plasma samples obtained
from knee (n = 43) and
hip (n = 44) osteoarthritis patients and age-matched healthy subjects (n =
31). Values were generated with in-
house kit;
[0095] Figure 2 shows data analysis of plasmatic PHB1 levels (PHB).
Descriptive values of
PHB1 plasma levels are shown by sex and health group expressed in ng/ml;
[0096] Figure 3 shows Statistical analysis system (SAS). Output
statistical analysis using data
for women only as an interaction exist between levels of PHB1 and sex. In the
logistic model used, the outcome
is OA or Healthy status and the predictor is PHB1 alone or in combination with
co-variate(s);
[0097] Figure 4 shows the distribution of PHB1 in the cytosol (C-X) and
nucleus (N-X) of human
lymphocytes from two OA patients and a control subject. Proteins were resolved
by SDS-PAGE and PHB1
protein was detected using an anti-PHB1 antibody. GAPDH was used as a
cytoplasmic marker. The demographic
and clinical data corresponding to each patient tested are indicated in the
table. The Kellgren-Lawrence (KL)
score is a radiographic score used to differentiate the severity of OA (from 1
to 4, wherein 4 corresponds to the
most severe form of OA; N/A = data not available). Early stage of OA is
defined as patients exhibiting a KL score
2, while late stage OA patients is defined as those exhibiting a KL score 3;
[0098] Figure 5 shows the accumulation of PHB1 in clusters in
lymphocyte nucleus. (A) PHB1
(green) (arrows point to examples of PHB-1 agglomerates/clusters (i.e. PHB1
positive nuclear bodies)) was
visualized by immunofluorescence using confocal microscopy and pictures were
taken at an optical section
localising to the center of the nuclear region. Upper panels represent
representative examples of agglomerates
in lymphocytes derived from a healthy control (left) and a subject having
osteoarthritis (OA, right) affected patient.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
Lower panels correspond to higher magnification of lymphocytes derived from a
healthy donor (left) or an OA
patient (right) immunostained for PHB1. Arrows indicate nuclear
aggregate/clusters of PHB1 seen in these cells.
(B) Quantification of the number of PHB1 agglomerates (paler) per lymphocyte
nucleus (darker) per patient (OA,
n=3; Healthy, n=4). Represented is the average of number of aggregate/clusters
per cell per patient with its
associated standard error. Three to thirty cells were analysed per patient and
three or four patients were analysed
per group (*P<0.01). (C) Frequency distribution for the cells analysed in (B).
The Y-axis represents the number of
cells with the given number of aggregate/clusters (X-axis). Overlay is a
polynomial regression curve for each
patient group. Images were obtained with a ZeissTM microscope (LSM 510 META)
and its associated LSM
acquisition software (Release 2.5) using a 63X objective. Images were exported
as Tiff files for quantitative
analysis. The number of PHB1 nuclear aggregates, regardless of their size, was
quantified manually per cell per
patient. These data were reported either as frequency distribution i.e. as
numbers of cells that have a given
number of nuclear aggregates/clusters per patient group (C) or as the average
means number of
aggregates/clusters per cell in patient groups (B) with its associated
standard error. Based on the frequency
distribution, the Poisson frequency distribution per patient group was
calculated using Microsoft ExcelTM
spreadsheet (Microsoft Office 2007). To facilitate the visual comparison
between the curves, these curves were
calculated for 100 cells. Comparison among patient groups for the average
number of PHB1 nuclear aggregates
was performed using the procedure GLM of the SAS software v8.02 (Cary, NC);
[0099] Figure
6 shows that the signal intensity for PHB1, which denotes protein levels, is
increased in whole leucocytes of OA patients (lymphocytes and monocytes). (A):
shows the sensitivity value of
the microscope camera (gain detector) for each sample analysed. All samples
were obtained from women. The
lower the sensitivity value, the brighter the signal. For healthy subject
(n=9) and for OA patients (n=7). Horizontal
bars represent average value in each group (Healthy subjects = 1015; OA
subjects = 947). Analysis of the gain
detector values was performed using logistic regression with the software SAS
v9.2. (B): shows a Table
summarizing the covariate parameters. (C): shows the analysis of the gain
detector values was performed using
logistic regression with the software SAS v9.2. Represented is the p value for
the gain detector variable adjusted
for the indicated co-variable;
[00100] Figure
7 shows that the proportion of leucocytes nuclei expressing low levels of PHB1
is
decreased in OA patients (n=5) vs. controls (healthy subjects) (n=3);
[00101] Figure
8 shows a comparison between the number of nuclear PHB1 agglomerates in
leucocytes of control subjects (n=5) and OA patients (n=6);
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
21
[00102] Figure 9 shows that PHB1 mostly accumulates in nucleus of OA
chondrocytes. (A) PHB1
immunostaining performed on cartilage sections from control (top panels) or OA
(bottom panels) subjects. Arrows
show cells with a positive nuclear PHB1 signal. Cartilage sections were
counterstained with Harris Modified
Hematoxylin. (B) Quantification of the percentage of cells from control
subjects (n=3) and OA subjects (n=5)
showing PHB1 nuclear signal. Asterisks represent a significant increase in
PHB1 nuclear signal (Mann-Whitney U
test: * p<0.05). (C) Cytoplasmic (C) and nuclear (N) protein extracts of
primary chondrocytes from one control
subject (n=1) and three different OA patients (n=3). Proteins were resolved by
SDS-PAGE and PHB1 protein was
detected using anti-PHB1 antibody. GAPDH, F1-ATPase and Lamin A/C were used as
cytoplasmic, mitochondrial
and nuclear marker respectively. (D) lmmunofluorescence against PHB1 and
TOM20, a mitochondrial marker, on
primary chondrocytes from one control subject (n=1) and OA subjects (n=2).
PHB1 signal appears in green and
TOM20 signal in red. DAPI was used as a DNA marker. (E) PHB1 signal
quantification of immunofluorescence
results. Data is presented as the percentage of the signal which co-localizes
with DAPI signal when compared
with the total PHB1 signal. (F) Real time RT-PCR against PHB1 gene in
chondrocytes from four healthy subjects
(n=4) and nineteen OA patients (n=19) showing that PHB1 gene expression (RNA)
is not increased in OA
subjects. Black marks represent the median value for each group; (G) Western
blot for total levels of PHB1 in
leucocyte. Alpha-tubulin is used as loading control;
[00103] Figure
10 shows the distribution of PHB1 in Cytosol (C-X) and Nucleus (N-X) of human
articular chondrocytes from OA patients (n=6) and a control subject (n=1).
Proteins were resolved by SDS-PAGE
and PHB1 protein was detected using anti-PHB1 antibody. GAPDH and Lamin NC
were used as cytoplasmic
and nuclear marker respectively. Demographic and clinical data corresponding
to each patient tested are
indicated below. The Kellgren-Lawrence (KL score) is a radiographic score used
to differentiate the severity of
OA (1 to 4), where 4 score corresponds to the most severe OA form; N/A data
not available); Early stage of OA is
defined as patients exhibiting a KL score 2, while late stage OA patients is
defined as those exhibiting a KL
score 3;
[00104] Figure
11 shows the increase in nuclear accumulation of PHB1 in knee joint articular
chondrocytes of aging STR-ORT mice (n=?), a mouse model of osteoarthritis.
Osteoarthritis symptoms in STR-
ORT mice are known to occur at about week 30. (A) Microphotographs of STR-ORT
mice knee sections stained
with Safranin 0 to visualize the proteoglycan content and the overall knee
cartilage structure (M : meniscus; SB;
subchondral bone). Mice were aged from 8 to 16 weeks. (B) Representative
immunohistochemistry pictures for
PHB1 (darker) on paraffin embedded knee sections of STR-ORT mice. Large arrows
show cells with nuclear
accumulation of PHB1. Small arrows show PHB1 positive cells with no
accumulation in the nucleus. Dash line
represents the border between cartilage and the subchondral bone (SB). (C)
Graphical representation of the
results depicted in B (STR-ORT n=7 and Control n=1). (D) qPCR analysis of
Pitx1 expression in knees sections
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
22
of STR-ORT mice (n=5) and control (n=5);
[00105] Figure
12 shows Pitx1 gene repression by PHB1 in 028/12 chondrocytes cell line. PITX1
mRNA (A) or proteins (B) level from 028/12 cells stably overexpressing Flag-
PHB1 or vector alone. In A, real time
RT-PCR was performed against Pitx1 gene. Data is presented as PITX1 mRNA
relative quantification and error
bars represents standard deviation of triplicates (paired t test: *p<0.01; **
p<0.05). In B, immunoblots of FLAG
epitope and PITX1. 3-tubuline protein was used as endogenous control. (C-D)
Luciferase assays in 028/12 cell
line transiently transfected with PITX1 (-3895/+61 bp)-promoter-luciferase
reporters or with luciferase plasmid
containing smaller fragments (in C, fragment -3034/+61, -1577/+61 bp, -729/+61
bp, -524/+61, -374/+61 bp or -
84/+61 bp and in D, fragment -729/+61). In C, cells were co-transfected with
Flag-PHB1 expressing vector or an
empty control vector. In D, cells were co-transfected with Flag-PHB1
expressing vector (Vector/PHB) or an empty
control vector (VectorNector) and with pBabe plasmid expressing ER fused to
E2F1 (E2F1/PHB) or the empty
control vector (E2F1Nector), and induced with 4-hydroxytamoxifen (OHT) for
24h. Data represents mean and
standard deviation of three independent experiments. Asterisks represent a
significant decrease in luciferase
activity (paired t test: * p<0.05; ** p<0.01) compared to control cells. (E)
ChIP assay showing the preferential co-
localization of PHB1 on the distal promoter elements of the human PITX1 gene.
Real-time PCR analysis was
performed after chromatin immunoprecipitation assay against PHB1. Different
primers were used to amplify
specific PITX1 promoter regions. Data is presented as DNA relative
quantification compared to the mean amount
of DNA present after immunoprecipitation of control cells.
[00106] Figure
13 shows the rescue of Pitx1 expression in OA articular chondrocytes through
PHB1 inhibition. Real time RT-PCR performed against PITX1 gene in chondrocytes
from four different OA
patients (n=4) transfected with control siRNA or PHB1 siRNA. Data represents
mean and standard deviation of
three independent experiments. RQ= Relative Quantity. Asterisks represent a
significant increase between
control and siPHB1 transfected cells (paired t test: *p<0,02);
[00107] Figure
14 shows that specific SUMO proteins accumulate in nuclei of OA articular
chondrocytes. (A) lmmunoblot of PHB-1 and three SUMOs (PanSumos) were
performed using nuclear extract (N)
and cytoplasmic extract (C) of articular chondrocytes from a healthy subject
(traumatic case non-arthrosic) (n=1),
a RA patient (n=1) and two OA patients (n=2). (B) lmmunofluorescence staining
against SUMO-1 and SUMO-2/3
(the same antibody detect both SUMO-2 and 3) carried out on articular
chondrocytes of OA patients and control
subjects. Representative staining are shown. OA chondrocytes show a strong
nuclear accumulation of SUMO-1
and SUMO-2/3 proteins in the nuclear bodies;
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
23
[00108] Figure
15 shows that the SUMO-1 protein strongly accumulates in the nuclei of
articular
chondrocytes of OA patients compared to healthy subjects. (A) lmmunoblot of
PHB1 and SUMO-1 were
performed using nuclear (N) and cytoplasmic (C) extracts from a healthy
patient (n=1) and three OA patients
(n=3). Chondrocytes of OA patients show nuclear accumulation of PHB1 and an
increase in total sumoylation in
the nuclear fraction. (B) lmmunofluorescences (IF) against PHB1 and SUMO-1
carried out on the articular
chondrocytes of patient 0A1 1. Upper panel show healthy chondrocytes and lower
panels show affected
chondrocytes;
[00109] Figure
16 shows that PHB1 co-localizes with SUMO-1 in nuclear bodies of OA articular
chondrocytes. Double lmmunofluorescence stainings against PHB1 and SUMO-1 (A)
and PHB1 and SUMO-2/3
(B) were performed on articular chondrocytes of OA patients and control
subjects. In OA patients the SUMOs
proteins accumulate in nuclear bodies, while control subjects show little or
no accumulation of SUMOs in nuclear
bodies. PHB1 is co-localized with SUMO-1 while no co-localization was found
with SUMO-2/3 in the nuclei of OA
chondrocytes (panel B);
[00110] Figure
17 shows that SUMO proteins accumulate in PML nuclear bodies in OA articular
chondrocytes. Double immunofluorescence stainings against PML nuclear bodies
(PML positive nuclear bodies)
and SUMO-1 (A), and PML nuclear bodies and SUMO-2/3 (B) were performed on
articular chondrocytes of
patients OA and control. In OA chondrocytes, nuclear accumulation of SUMO-2/3
proteins is localized almost
solely in PML nuclear bodies while the accumulation of SUMO-1 is in all
nucleus including PML nuclear bodies. In
OA chondrocytes, PML nuclear bodies are different in size and sometimes they
adopt a ring structure, as
indicated by an arrow (Panel A). SUMO-1 and SUMO-2/3 both are co-localized in
PML in these structures but
only in OA chondrocytes;
[00111] Figure
18 shows that PML and PHB1 in human articular chondrocytes from OA patients
do not co-localize in nuclei. Double fluorescence staining of OA and control
human articular chondrocytes with
antibodies against PML and PHB1 shows that PHB1 is accumulated mostly in
nuclei of OA chondrocytes like
PML although it does not co-localize with PML nuclear bodies;
[00112] Figure
19 shows SUMO-1 expression in leucocytes from 22 weeks old C57BI/6 (n= 1) (a
mouse not presenting OA symptoms) or STR-ort male mice (n= 1) (a mouse model
of osteoarthritis). Leucocytes
were isolated from blood samples and immunostained for SUMO-1. Nuclei were
stained with Draq5. Leucocytes
(lymphocytes and monocytes) were isolated by Ficoll gradient, centrifuged (300
g during 6 minutes) on 8-well
slides coated with poly-D-lysine. Cells were washed twice in PBS and fixed
in 4% Paraformaldehyde at Room
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
24
temperature for 7 minutes, permeabilised in 0.3% Triton X for 5 minutes at
room temperature, wells were
removed, then blocked in 5% BSA/PBS for 1-2 hr and incubated with primary
antibodies overnight at 40. Primary
antibodies are washed 3-4 times and cells are incubated with secondary
antibodies for lh at room temperature,
washed 2-3 times, incubated with Draq5 and Hoescht for 5 minutes, washed 3
times and mount with
ProlongGoldTM antifade reagent (Invitrogen). Slides were let to dry overnight,
the images were captured with
Zeiss confocal microscope;
[00113] Figure
20 shows an in silico analysis of putative sumoylation sites and SUMO-binding
sites in human PHB1 protein ;
[00114] Figure
21 shows that PHB1 cannot be sumoylated by SUMO-1 in vitro. An in vitro
sumoylation assay in the presence of SUMO-1, El and E2 enzymes, ATP and
purified GST-PHB1 protein
indicated that PHB1 cannot be sumoylated in vitro. GST and GST-RanGapl
proteins were used as negative and
positive controls respectively. (A) The purified GST and GST fusion proteins
were analyzed by SDS-PAGE
followed by a Coomassie blue staining. (B) Four times less protein GST were
used for the test compared to the
fusion proteins. The products of in vitro sumoylation assay were analyzed by
immunoblot against PHB1 and
RanGapl. The asterisk (*) represents the sumoylated GST-RanGapl;
[00115] Figure
22 shows that PHB1 can bind SUMO-1 proteins via a SBM (SUMO-binding
module). (A) Diagram represents various PHB1 constructs generated for the
study. Wild type PHB1, a mutant in
which the nuclear signal of export was deleted (PHB1_,ANES), or was replaced
by a nuclear localization signal
(PHBl_NLS), and a mutant where a putative SUMO-binding module was deleted
(PHBLASBM). All PHB1
constructs have triple Flag-tag at the N-terminal. (B) 028/12 cells were
infected with each construct in order to
produce stable lines. The nuclear proteins (X-N) and cytoplasmic proteins (X-
C) were isolated and analyzed by
immunoblot. (C) Co-IP assays with anti-C-myc antibody demonstrated that PHB1
interacts with SUMO-1 through
the SBM. SUMO-1 was tagged with c-myc;
[00116] Figure
23 shows that UBC9 expression is increased in knee joint OA cartilage and
correlates with disease severity. Left panels are Safranin-O staining and
represent the proteoglycan content
which decreases with the severity of OA. Right panels represent IHC
experiments performed with anti-Ubc9
antibody where staining intensity also correlates with disease progression;
[00117] Figure
24 shows representative immunohistological sections showing UBC9 proteins in
normal cartilage (B, D) and knee joint OA (C, E) sections. (A) represents the
mean value of UBC9 proteins
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
detected by IHC in superficial and deep zone of normal knee cartilage (n=3)
and knee joint OA cartilage sections
(n=9). In brief, three sections of each specimen were examined (40x Leica DM R
Microscope) from either the
superficial zone of the cartilage, scored, and the resulting data integrated
as a mean for each specimen. The final
results were expressed as the percentage of chondrocytes staining positive for
the antigen (cell score) with the
maximum score being 100%. Each slide was subjected to evaluation by two
observers with >95% degree of
agreement. Panels B and C correspond to superficial zones of normal and OA
cartilage respectively and panels
D and E represent the deep zones of normal and OA cartilage respectively;
[00118] Figure
25 shows that sumoylation stabilizes PHB1 and promotes its nuclear
accumulation
in U2OS cells. (A) U2OS cells were transfected with the pLPC-3xFlag-PHB1 alone
or co-transfected with different
components of the sumoylation pathway. (B) The nuclear proteins (X-N) as well
as total proteins (X-T) were
isolated from cells transfected with pLPC-3xFlag-PHB1, PHB1-NLS or PHB1-ASBM
in presence or absence of
myc-SUMO-1 and UBC9. (C) The cells transfected with the empty vector pLPC-
3xFlag or containing PHB1 and
PHB1-NLS constructs were treated with the MG132 for 4 hours, then the total
proteins were extracted;
[00119] Figure
26 shows human PHB1 mRNA nucleotide (A, obtained from
giI6031190IrefINM_002634.2) (SEQ ID NO: 1) and amino acid sequence (B,
obtained from
gi14505773IrefINP_002625.1) (SEQ ID NO: 2);
[00120] Figure
27 shows human SUMO-1 precursor mRNA nucleotide (obtained from NCBI
Reference Sequence NM_003352.4) (SEQ ID NO: 3) and amino acid sequence
(obtained from NCBI Reference
Sequence: NP_003343.1) (SEQ ID NO: 4);
[00121] Figure
28 shows human SUMO-2 precursor mRNA nucleotide (obtained from NCBI
Reference Sequence NM_006937.3) (SEQ ID NO: 5) and amino acid sequence
(obtained from NCBI Reference
Sequence: NP_008868.3) (SEQ ID NO: 6);
[00122] Figure
29 shows human SUMO-3 precursor mRNA nucleotide (obtained from NCBI
Reference Sequence NM_006936.2) (SEQ ID NO: 7) and amino acid sequence
(obtained from NCBI Reference
Sequence: NP_008867.2) (SEQ ID NO: 8);
[00123] Figure
30 shows human SUMO-4 precursor mRNA nucleotide (obtained from NCBI
Reference Sequence NM_001002255.1) (SEQ ID NO: 9) and amino acid sequence
(obtained from NCBI
Reference Sequence: NP_001002255.1) (SEQ ID NO: 10); and
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
26
[00124] Figure
31 shows human UBC9 mRNA nucleotide (obtained from NCBI Reference
Sequence NM_006936.2) (SEQ ID NO: 11) and amino acid sequence (obtained from
NCBI Reference Sequence:
NP_008867.2) (SEQ ID NO: 12).
DESCRIPTION OF ILLUSTRATIVE EMBODIMENTS
Sumoylation and PML bodies
[00125] PML
nuclear bodies (PML-NBs) are highly dynamic micro-nuclear structures composed
solely of proteins. The main component of PML-NBs is the PML protein
(promyelocytic leukemia protein), of
which there are seven isoforms in humans (Condemine et al., 2006). So far, the
PML-NBs have been associated
with several functions such as cell cycle regulation, regulation of gene
transcription, response to DNA damage,
senescence and apoptosis (Bernardi and Pandolfi, 2007).
[00126] The
present invention is illustrated in further details by the following non-
limiting
examples.
EXAMPLE 1
Materials and methods
Derivation of human articular chondrocytes
[00127]
Articular cartilage from OA knee patients was collected, cut into small pieces
and washed
twice in sterile PBS 1X pH 7.4 (phosphate buffer saline: 0.137 M NaCI, 8.1 mM
Na2HPO4, 2.7 mM KCI , 1.5 mM
KH2PO4). For each patient, some pieces were fixed in a solution of
paraformaldehyde (PFA) 4% v / v, embedded
in paraffin blocks and stored for histological analysis. The pieces of
remaining cartilage were incubated for one
hour at 37 C with shaking in D-MEM (Dulbecco's modified Eagle's medium 1X:
Wisent Inc., St-Bruno, Quebec,
Canada)) containing 10% (v / v) FBS ( FCS: Gibco BRL, Burlington, Ontario,
Canada), 1% pen-strep and 1mg/m1
pronase (Sigma-Aldrich, Oakville, ON, Canada) and then digested for 4 to 6
hours at 37 C with stirring presence
of 2mg/m1 collagenase (Sigma-Aldrich, Oakville, ON, Canada) diluted in D-MEM
supplemented with FBS and
pen-strep. The digested tissue was passed through a sieve sterile, and then
centrifuged at 215 x g for 10
minutes. The pellets of chondrocytes were resuspended in a small volume of
culture medium and counting the
number and cell viability was performed using the Vi-CeIITM XR Cell Viability
analysis (Beckman Coulter:
Mississauga, ON, Canada). The cells were then placed in primary culture at
high density (2x106 cells) in T-75
flasks and then placed in 10 cm or kneaded LabteksTM according to the desired
use. The primary chondrocytes
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
27
were then either frozen and stored in liquid nitrogen in a solution containing
10% FBS DMSO, or maintained in
culture in the first passage for immediate use.
Cell lines
[00128] All
chondrocytes of patients and cell lines, MCF-7 and U2OS were cultured in D-MEM
(Dulbecco's modified Eagle's medium 1X: Wisent Inc., St-Bruno, Quebec, Canada)
supplemented with 10% (v / v)
FBS (FCS: Gibco BRL, Burlington, Ontario, Canada) and 100 units / ml
penicillin and 10Oug/m1 streptomycin
(Gibco BRL, Burlington, Ontario, Canada). C28/I2 cells, a line of human
chondrocytes were cultured in medium
containing a mixture of D-MEM and F12 (Gibco BRL, Burlington, Ontario, Canada)
in a 1:1 ratio, supplemented
with 10% FBS (Gibco BRL, Burlington, Ontario, Canada) and penicillin and
10Oug/m1 100unites/m1 streptomycin
(Gibco BRL, Burlington, Ontario, Canada). C28/I2 cells were generously
provided by the group of Dr. Mary B.
Goldring (Cornell University, New York) and MCF-7 cells were provided by the
group of Dr. Andre Tremblay
(Research Centre of the CHU Ste-Justine). All cells were grown at 37 C in an
incubator containing 5% CO2 and
95% air and culture medium was changed every 3 to 4 days.
Plasmids and constructs
[00129] The
different PHB-1 mutants were constructed from a clone of the commercial wild-
type
prohibitin (Origen) and cloned into the retroviral expression vector PLPC-
3xFlag (Calabrese et al., 2009), to mark
the protein with a triple Flag epitope in the N-terminal. Four constructs were
made with a wild type (PHB-1) and
three mutants: PHB1-ASBM including a putative binding site of SUMO proteins
(residues 76-79) was deleted;
PHB1-ANES, lacking the signal nuclear export (residues 257-272) and PHB1-NLS,
where the NES was replaced
by a nuclear localization signal (NLS). The nucleotide sequences of various
primers used are shown in Table I
below. Plasmids pCDNA3-Myc-SUMO-1, pcDNA-Myc-SUMO-3 and pCDNA-HA-SUMO-2 were
provided by the
laboratory of Dr. Christopher K. Glass (University of California, San Diego).
UBC9 plasmid was provided by the
team of Dr. Muriel Aubry (University of Montreal, Montreal). The various
plasmids were transformed into DH5a
strain of E. coli by heat shock of 45 seconds at 42 C, followed by incubation
for 16-18 hours at 37 C with
shaking in 450 ml of medium 2YXT (12.5 g yeast extract, 12.5 g bacto-
tryptone/L of water) supplemented with 50
ml of saline (0.17 M KH2PO4, 0.72 M K2HPO4). The various plasmids were then
isolated and purified by cesium
gradients.
Table I: Nucleotide sequence of the primers used for the construction of
various protein fusions for the trials of
sumoylation in vitro and for the overexpression of proteins. S: sense, AS:
antisense.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
28
Constructs Plasmids PRIMERS
GCGAATTCTGCTGCCAAAGTGTTTGAGTCCATTGGC 3'
PHB-1 pLPC-3xFlag S: (SEQ ID NO: *)
AS: 5' GCCTCGAGTCACTGGGGCAGCTGAGGA 3' (SEQ ID NO: *)
5' GCCTCGAGTCAGCCCACTTTGCGCTTTTTCTTGGGG (SEQ
PHB-1-NLS pLPC-3xFlag AS: ID NO: *)
TTCCGAGAGCGTGAGAGCTGGTA 3' (SEQ ID NO: *)
5' GCCTCGAGTCAGTTCCGAGAGCGTGAGAGCTGGTA 3'
PHB-1-ANES pLPC-3xFlag AS: (SEQ ID NO: *)
5' CCACGTAATACTGGTAGCAAAGATTTACAGAATGTC 3'
PHB-1-ASBM pLPC-3xFlag S: (SEQ ID NO: *)
5' GCTACCAGTATTACGTGGTCGAGAACGGCAGTCA 3' (SEQ
AS: ID NO: *)
5' ACGGATCCCCGCCTCGGAAGACATTGCCAAG 3' (SEQ ID
GST-RanGap1 pGEX-5X-3 S: NO: *)
5' ATGAATTCGACCTTGTACAGCGTCTGCAGCAG 3' (SEQ ID
AS: NO:*)
Immunohistochemistry
[00130] The cartilage slices, mounted on SuperFrostTM slides (Fisher
Scientific, Hampton, NH,
USA) were deparaffinized by soaking slides in three successive baths of
toluene, rehydrated in four baths of
alcohol at 100%, 90%, 70% and 50%, and washed in a water bath and a 1X PBS pH
7.4 bath, at 5 minutes per
bath. The slides were then heated 20 minutes at 65 C in a solution of sodium
citrate, 0.01 mM, pH 6.0, then
washed 5 minutes with PBS 1X. The slides were then incubated for 30 minutes in
a solution of 1X PBS containing
0.3% Triton TM and washed 3 times 5 minutes with PBS 1X. After a 30 min.
incubation in methanol containing 2%
H202, slides were placed in a humidity chamber and the sections were incubated
for 1 hour at room temperature
in a blocking solution containing normal horse serum (ABC VectasinTM kit,
Vector Laboratories, Curlingame, CA).
The sections were then incubated overnight in blocking solution containing
primary antibody (anti-PHB1: Ab-2,
Neomarker), washed 3 times in 1 x PBS, then incubated 45 minutes in the
presence of biotinylated secondary
antibody diluted in blocking solution, and washed again three times with PBS
1X. After incubation with avidin-
biotin complex (ABC VectasinTM kit, Vector Laboratories, Curlingame, CA), the
labeling was revealed using the
system diaminobenzidine (DAB) (Dako Diagnostics Canada Inc., Mississauga, ON,
Canada) according to the
manufacturer's instructions, giving a brownish color to the expressed protein.
The sections were then stained with
Harris hematoxylin (Fisher Scientific, Hampton, NH, USA) and mounted with a
coverslip using PermountTM
(Fisher Scientific, Hampton, NH, USA).
Extraction of total protein
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
29
[00131] One
petri dish of confluent cells per condition were washed twice in cold 1X PBS,
then
harvested and lysed in lysis buffer (20 mM Tris-HCI pH 7.5, 150 mM NaCI, 1 mM
EDTA, 1 mM EGTA, 1%
Triton TM X-100, 2.5 mM sodium pyrophosphate, 1 mM 3-glycerophosphate)
containing in addition to a cocktail of
protease inhibitors 1X (Roche, Indianapolis, IN, United Kingdom) and 25 mM NEM
(Sigma-Aldrich, Oakville, ON,
Canada ). After 30 to 60 minutes of incubation at 4 C with shaking, the
protein lysates were collected after
centrifugation for 15 minutes at 11,200 x g.
Separation of nuclear and cytoplasmic proteins
[00132] Two or
three petri dishes of confluent cells (5x106 cells / petri dish) by condition
were
washed twice in 1 x PBS (10.1 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCI, 137 mM
NaCI) cold, scraped and
transferred into 1.5 ml tubes. After a centrifugation at 100 x g for 5
minutes, the cell pellets were resuspended in
300 pl of hypotonic lysis buffer (10 mM HEPES pH 7.9, 1.5 mM MgC12, 10 mM KCI,
1% NP-40, 0.5 mM DTT)
supplemented with a cocktail 1X of protease inhibitors (Roche, Indianapolis,
IN, United Kingdom) and 25 mM
NEM (Sigma-Aldrich, Oakville, ON, Canada), incubated on ice for 25 minutes by
vortexing every 3 to 4 minutes.
The lysates were centrifuged at 4 C for 5 minutes at 1200 x g to obtain a
pellet containing the cell nuclei. The
supernatants containing the cytoplasmic proteins were transferred to new 1.5
ml tubes and recentrifuged a
second time at 4 C for 10 minutes at 1200 x g to remove remaining debris and
minimize contamination by
nuclear proteins. The supernatants were transferred back into new 1.5 ml
tubes. The nuclei pellets were
resuspended in 8 ml of nuclear lysis buffer (50 mM Tris-HCI pH 7.6, 2 mM EDTA,
2 mM EGTA, 1 mM DTT,
protease inhibitor cocktail 1X (Roche, Indianapolis, IN, United Kingdom), 25
mM NEM (Sigma-Aldrich, Oakville,
ON, Canada) containing 0.1% TritonTm X-100 and placed on 2 ml of sucrose
cushion (nuclear lysis buffer
containing 30% w / v sucrose) in tubes of 15 ml. The samples were then
centrifuged at 4 C for 50 minutes at
3500 x g in a SorvallTM Legend RT centrifuge. The buffer was decanted to leave
the bottom of the tubes that the
pellets of nuclei purified which were resuspended in 50 to 100 pl 4X Laemlli
buffer (0.52 M Tris-HCI pH 6.8,
6.85% SDS, 3.3% 3-mercaptoethanol, 20% glycerol) and boiled for 5 minutes.
After quantification of proteins by a
Bradford assay (Bio-Rad, Hercules, CA, United States), 50 mg of cytoplasmic
and nuclear proteins were
separated by SDS-PAGE and analyzed by Western blot.
Immuno / Co-immunoprecipitation assays
[00133] U205
cells were transfected with pCMV4-Myc-sumo1 in the presence of PLPC-3xFlag-
PHB-1 or PLPC-3xFlag-PHB1-ASBM (15 g total DNA) by using calcium phosphate
precipitation. The culture
medium was changed 24 hours post-transfection and cells were harvested 48
hours after transfection. The cells
were washed twice in cold 1X PBS, then harvested and lysed in lysis buffer (20
mM Tris-HCI pH 7.5, 150 mM
NaCI, 1 mM EDTA, 1 mM EGTA, 1% Triton TM X-100, 2.5 mM sodium pyrophosphate, 1
mM 3-glycerophosphate)
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
containing in addition to a cocktail of protease inhibitors 1X (Roche,
Indianapolis, IN, United Kingdom) and 25 mM
NEM (Sigma-Aldrich, Oakville, ON, Canada). After 30 to 60 minutes of
incubation at 4 C with shaking, the
protein lysates were harvested by centrifugation for 20 min at 11,200 x g. The
immunoprecipitations were
performed overnight at 4 C in the presence of 1 to 2 mg of total protein and
the primary antibody. The following
antibodies were used: anti-PHB1 (N-20, Santa Cruz), anti-c-myc (MAB8865,
Millipore). The immunoprecipitates
were collected by following a 1-hour incubation at 4 C in the presence of
protein A / G SepharoseTM (Amersham
Biosciences Corp., Qc, Canada) and washed 3 times with the lysis buffer, 1
time in 1 x PBS and 1 time with
water. The precipitates were eluted in 70p1 3X Laemlli buffer, boiled for 5
minutes and 35 pl were used for
analysis by Western blot.
GST pull-down
[00134] Nucleic
acid encoding PHB-1 and RanGap1 proteins were first cloned into the vector
pGEX-5X-3. The different GST-fusion proteins were produced in E. coli strain
BL21. Each of the plasmids,
including the empty vector, was transformed by heat shock of 45 seconds at 42
C. Bacteria containing each of
the plasmids were grown at 37 C in 400 ml of 2YXT medium (16g / L tryptone,
10 g / L yeast extract, 5 g / L
NaCI) to an optical density of 0.8 at 600 nm, then induced in the presence of
0.4 mM IPTG for 4 hours at 30 C.
[00135] The
bacterial pellets from a 250 ml culture were resuspended in 3 ml of STE buffer
(10
mM Tris-HCI pH 8, 1 mM EDTA, 100 mM NaCI) supplemented with DTT (5 mM), PMSF
(1 mM) and a cocktail of
protease inhibitors (Roche, Indianapolis, IN, United Kingdom). Then, 1 mg/ml
of lysozyme (Sigma-Aldrich,
Oakville, ON, Canada) was added. After a 30-45 minutes incubation on ice, 1.5%
sarcosyl (Sigma-Aldrich,
Oakville, ON, Canada) was added and sonication was performed (5 times, 10
seconds per tube). Cell lysates
were then transferred into 2 ml tubes and centrifuged 10 minutes at 11,200 x g
at 4 C. The supernatants were
transferred to new tubes and 120 pl of 2 ml of glutathione beads / 50%
SepharoseTM (Amersham Biosciences
Corp., Qc, Canada) were added to each tube. After incubation with agitation at
4 C for 2 hours, the beads were
washed two times in NETN buffer (10 mM Tris-HCI pH 8, 1 mM EDTA, 100 mM NaCI,
0.5% NP-40), once in
NETN buffer 500 mM NaCI and again once in the NETN buffer. The beads were
finally resuspended in an equal
volume of 1X PBS supplemented with protease inhibitors (Roche, Indianapolis,
IN, United Kingdom). 5 pl of
beads were then analyzed by SDS-PAGE followed by staining with Coomassie blue.
Sumoylation assays
[00136] GST-
PHB1 fusion proteins purified by GST pull-down were used as substrate for the
testing of sumoylation by SUMO-1. GST and GST-RanGap1 proteins were used as
negative and positive
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
31
controls, respectively. Each reaction was performed in a total volume of 20 pl
in a reaction buffer containing 20
mM Hepes pH 7.5 and 5 mM MgC12 in the presence of 7.5 mg / ml of El enzyme, 50
pg/ml of E2 enzyme,
50pg/m1 of Sumo-1 and 20 mM ATP for 1 hour at 37 C. All reagents were
obtained commercially (LAE Biotech
International) and used according to the manufacturer's instructions. For each
reaction, 5 pl were then separated
on SDS-PAGE gel and analyzed by Western blot.
Western blots
[00137] All
protein extracts were separated by SDS-PAGE on acrylamide gels using the Mini-
Protean TM 11 (BioRad, Hercules, CA). The gels consisted of a stacking gel
consisting of 4% acrylamide (v / v) in
0.5 M Tris buffer pH 6.8 and a resolving gel containing between 8 and 12.5% of
acrylamide (v / v) and 1.5 M Tris
buffer pH 8.8. The migration of proteins was carried out at room temperature
at a voltage of 120 volts. The
proteins were then transferred to PVDF membranes (Millipore) for 90 minutes at
a voltage of 100 volts. Once the
transfer was complete, the membranes were pretreated by incubation for a few
seconds in methanol, followed by
a one hour incubation in blocking solution (1 x PBS, 0.02% TweenTm-20, 10%
milk fat-free). Following three 15-
minute wash in PBST (1X PBS, 0.02% TweenTm-20), the membranes were incubated
overnight at 4 C in the
presence of the primary antibody (Table II) diluted in a solution of PBST
containing 3% BSA (bovine serum
albumin: Bioshop) and 0.02% sodium azide. The next day the membranes were
incubated in the presence of
secondary antibodies coupled to peroxidase (Thermo Scientific, Rockford,
United Kingdom) diluted in PBST
containing 5% non-fat milk for one hour at room temperature. After a 1-hour
wash in PBST, signals were revealed
using the ECL reagent (enhanced chemiluminescence substrate: PerkinElmer,
Watlham, MA, United Kingdom)
and detected on an autoradiography film (Amersham Biosciences Corp.., QC,
Canada). To make a second
immunoblotting on the same membrane, the membranes were incubated in a
solution containing 25 mM glycine
pH 2.0 and 1% SDS (sodium dodecyl sulfate) for 45 minutes at room temperature
with agitation to remove
antibodies already present. The same steps were then repeated from the
blocking step.
Table II: Antibodies used for Western blots
Primary antibodies Origin Company
anti-PHB-1 (Ab-1) monoclonal, mouse -- Lab vision
anti-Sumo-1 (#4930) polyclonal, rabbit -- Cell Signaling
anti-PanSumo (#AP1299a) polyclonal, rabbit -- Abgent
anti-Lamine A/C (#2032) polyclonal, rabbit -- Cell signaling
anti-GAPDH (V-18) polyclonal, goat Santa cruz
anti-Flag (M2) monoclonal, mouse -- Sigma
anti-RanGapl monoclonal, mouse -- Zymed
anti-GST (M1) polyclonal, rabbit -- Millipore
Immunofluorescence
[00138]
Chondrocytes of OA patients and healthy subjects at the first passage were
grown in 8-
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
32
well LabTek at a rate of 10 000 cells per well. After 24 to 48 hours of
incubation, the cells were washed two times
in 1X PBS, fixed in a solution of 3.7% paraformaldehyde, and permeabilized in
1X PBS containing 0.1% Triton TM
X-100 for 10 minutes. The cells were then incubated for 30 minutes in blocking
solution (PBSA) containing 1 x
PBS supplemented with 1% bovine serum albumin (BSA: BioShop, Burlington, ON,
Canada). Subsequently, the
cells were incubated with primary antibodies (Table III below) diluted in PBSA
for 2 hours at 37 C. The
secondary antibodies (Alexa fluorTM, Invitrogen, Eugene, Oregon, United
States) diluted in PBSA were then
applied for 1 hour at 37 C. After 3 washes in 1 x PBS, the slides were
mounted using an adhesive containing
DAPI (Prolong G0IdTM: Invitrogen, Eugene, Oregon, United States) and then
observed by confocal microscopy.
Table III: Antibodies used for immunofluorescence
Primary antibodies Origin Company
anti-PHB1 (Ab-1) monoclonal, mice Lab vision
anti-Sumo-1 (A21C7) monoclonal, rabbit Cell Signaling
anti-Sumo-2/3 (18H8) monoclonal, rabbit Cell Signaling
anti-PM L (PG-M3) monoclonal, mice Santa Cruz
Anti-PHB1 (#2413-1) monoclonal, rabbit Epitomics
Anti-PHB1 (#11-14-10) monoclonal, mice NeoMarkers
Confocal microscopy
[00139] The
slides were observed using a ZeissTM LSM 510 Meta confocal microscope (Zeiss
Canada, Toronto, ON, Canada) at a magnification of 630X. The images were then
analyzed using the ZeissTM
LSM image browser software.
EXAMPLE 2
PHB1 levels in plasma samples obtained from osteoarthritis patients and age-
matched healthy subjects
[00140] Figure
1 depicts results from experiments performed on blood samples (plasma) from
knee (n=43, demographic characteristics depicted in Table IV below) and hip
(n=44, demographic characteristics
depicted in Table V below) osteoarthritis patients and age-matched healthy
subjects (n=31, demographic
characteristics depicted in Table VI), which show that mean plasma levels of
the pitx1 repressor protein PHB1 are
significantly lower in osteoarthritis patients. The source of circulating PHB1
may be PHB1 shed from the plasma
membrane (Mielenz D et al. J Immunol 2005; 174(6):3508-3517), or released from
adipocytes and possibly other
cells in lipid droplets (Brasaemle DL et al. J Biol Chem 2004; 279(45):46835-
46842). The Kellgren-Lawrence (KL)
radiographic score also presented and differentiates the severity of OA in
Table V (from 1 to 4, wherein 4
corresponds to the most severe form of OA; N/A = data not available). Early
stage of OA is defined as patients
exhibiting a KL score 2, while late stage OA patients was defined as those
exhibiting a KL score 3.
Table IV. Demographic characteristics of recruited subjects for HIP.
Patient Random
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
33
ID
Gender Age (Years) BMI
0A-1 Female 77 31.05
0A-3 Female 57 32.46
0A-5 Male 76 28.98
0A-6 Female 69 28.54
0A-9 Female 76 21.87
0A-10 Female 55 22.98
0A-20 Female 66 28.65
0A-21 Male 45 NA
0A-24 Male 50 32.55
0A-30 Male 79 30.83
0A-31 Female 71 29.09
0A-32 Female 57 25.91
0A-35 Female 55 27.4
0A-38 Female 77 29.51
0A-39 Male 60 35.98
0A-40 Male 70 29.76
0A-42 Female 66 33.17
0A-43 Male 69 21.79
0A-44 Male 66 58.13
0A-46 Male 48 29.04
0A-51 Male 59 27.04
0A-53 Female 58 21.63
0A-54 Female 37 16.13
0A-72 Male 62 28.08
0A-75 Male 52 28.68
0A-76 Male 63 33.19
0A-77 Female 55 41.23
0A-78 Female 73 31.58
0A-79 Female 76 22.03
0A-80 Male 47 34.81
0A-93 Female 75 25.53
0A-94 Female 77 34.88
0A-96 Female 66 27.47
0A-97 Male 40 31.43
0A-102 Male 48 28.08
0A-110 Female 87 26.84
CA 02843631 2014-01-30
WO 2013/053065 PCT/CA2012/050723
34
0A-111 Male 82 31.8
0A-118 Male 64 28.7
0A-120 Male 49 25.28
0A-121 Male 56 30.08
0A-122 Male 56 35.92
0A-123 Female 55 20.7
0A-127 Male 11 28.66
0A-128 Female 13 32.87
0A-285 Female 13 23.04
Table V. Demographic characteristics of recruited subjects- Knee
Patient Random
ID Characteristic
Gender Age (Years) KL Score BMI
0A-2 Female 77 2 25.51
0A-11 Female 54 3 41.86
0A-14 Female 76 3 41.11
0A-16 Male 59 4 33.63
0A-26 Female 75 4 40.64
0A-33 Male 78 1 25.77
0A-37 Female 65 4 33.67
0A-57 Female 77 2 28.51
0A-59 Male 57 1 34.08
0A-64 Male 74 3 36.5
0A-66 Female 72 1 29.68
0A-95 Male 66 1 36.06
0A-103 Female 72 2 28.12
0A-104 Female 70 3 29.39
0A-105 Female 68 3 38.39
0A-106 Female 81 3 27.94
0A-112 Male 70 2 33.7
0A-134 Female 72 2 35.49
0A-139 Female 69 3 36.26
0A-140 Female 78 4 23.82
0A-152 Male 69 NA 22.85
0A-153 Male 53 1 29.38
0A-155 Female 74 4 31.14
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
0A-158 Male 58 1 29.06
0A-159 Male 67 2 36.93
0A-165 Male 65 3 30
0A-186 Female 59 2 30.11
0A-187 Female 66 1 32.03
0A-188 Male 58 3 32.17
0A-206 Female 61 3 34.44
0A-208 Female 58 1 27.58
0A-209 Female 70 2 29.39
0A-210 Female 77 2 23.61
0A-211 Male 64 2 29.51
0A-212 Female 74 3 22.65
0A-223 Female 66 2 43
0A-225 Male 79 3 25.61
0A-227 Male 54 3 37.87
0A-228 Female 59 4 35.49
0A-239 Female 80 4 29.03
0A-266 Female 47 1 38.08
0A-267 Male 55 2 30.38
0A-281 Female 78 3 26.67
Table VI. Demographic characteristics of recruited subjects-Control
Patient Random
ID
Gender Age (Years) BMI
CTRL-4 Male 61 31.67
CTRL-7 Female 46 21.33
CTRL-8 Male 32 26.87
CTRL-23 Female 19 23.53
CTRL-27 Female 51 28.12
CTRL-60 Female 19 21.29
CTRL-61 Female 49 23.87
CTRL-68 Male 36 25.53
CTRL-69 Male 47 28.36
CTRL-71 Female 37 23.24
CTRL-73 Female 49 17.84
CTRL-74 Female 43 22.03
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
36
CTRL-83 Female 52 22.71
CTRL-84 Male 46 24.03
CTRL-86 Female 52 19.43
CTRL-87 Female 49 21.9
CTRL-90 Female 38 23.04
CTRL-91 Female 37 NA
CTRL-92 Male 25 NA
CTRL-98 Female 57 21.09
CTRL-99 Female 32 NA
CTRL-100 Male 50 NA
CTRL-108 Female 55 28.28
CTRL-109 Female 42 NA
CTRL-114 Female 65 NA
CTRL-131 Female 24 NA
CTRL-136 Female 77 24.43
CTRL-137 Female 41 24
CTRL-143 Male 18 NA
CTRL-316 Male 24 NA
CTRL-329 Male 32 NA
EXAMPLE 3
PHB1 levels in plasma samples obtained from osteoarthritis patients, men and
women, healthy subjects
and subjects having rheumatoid arthritis
[00141]
Plasmatic PHB1 levels were determined in a group of 231 patients. Plasma was
isolated
from peripheral blood by centrifugation and frozen at -800 until analysed.
ELISA analysis was performed as per
vendor protocol (Uscnk (www.uscnk.us), Prohibitin kit. Protocol manual 7th
edition revised in November 2011)
[00142] Results
are presented in Figure 2 and presents the values of PHB1 plasma levels by sex
and health group. Plasma levels of PHB1 was obtained using Uscnk ELISA kit for
PHB1. Analysis of the values
was performed using Proc Logistic in SAS v9.2. Procedure Proc means was used
to calculate the average
(mean), minimum, maximum, median, and the 95% confidence interval of the PHB1
values for healthy subjects,
OA and RA for man and women. Figure 2 shows a statistically significant
decrease in circulating PHB1 of OA
patients as compared to control subjects.
[00143]
Statistical analysis system (SAS) is is a software made by the SAS institute:
http://www.sas.com/company/about/history.html. SAS output analysis were
performed using data for women only
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
37
as an interaction exist between levels of PHB1 and sex. In the logistic model
used, the outcome is OA or Healthy
status and the predictor is PHB alone or in combination of co-variate(s) (see
Figure 3). These analyses were
performed with the Proc Logistic procedure in SAS v9.2. Logistic models were
used to analyse the significance
of PHB1 plasmatic levels on the prediction of OA vs healthy status. These
analyses were performed in a step-
wise manner. First with PHB1 levels alone and then in presence of covariates
(age and BMI) as these two
covariates that are also associated with the occurrence of OA. Reported is the
p values for PHB1 and covariates.
The fact that the results obtained with OA men did not show a statistically
significant difference vs. control
subjects may be explained by the small sample size and/or that the origin of
the arthrosis was secondary
(trauma) rather than primary (genetic predisposition).
EXAMPLE 4
Distribution of PHB1 in the cytosol and nucleus of lymphocytes from OA
patients
[00144] Figures
4 and 5 depict results from Western blot (Figure 4) and immunofluorescence
(Figure 5) experiments that show that PHB1 is found at higher levels in the
nucleus of human lymphocytes from
two OA patients, relative to lymphocytes from a control subject. The KL
radiographic score was also used to
differentiate the severity of OA. Early stage of OA was defined as patients
exhibiting a KL score 2, while late
stage OA patients was defined as those exhibiting a KL score 3 (Figure 4).
Figure 5 shows the accumulation of
PHB1 in clusters in lymphocyte nucleus. lnsupport with an increase in nuclear
PHB1 levels (Figure 4), an
increase in the number of nuclear agglomeration in OA cells indicates an
increase quantity of nuclear PHB1 in
these cells.
EXAMPLE 5
PHB1 level is higher in leucocytes nuclei from OA patients
[00145]
Leucocytes (lymphocytes and monocytes) obtained from healthy subjects or OA
patients
were isolated by FicollTM gradient, centrifuged onto eight-wells chamber
slides (coated with poly-D-Lysine) and
immunostained for PHB1. Nuclei were counterstained with Drag5 and Hoechst.
Confocal images of PHB1
staining were obtained by adjusting the focal plane (less than 1 micron thick)
at the center of the nuclear signal.
For each sample, gain detector (equivalent to exposure time) for PHB1 signal
was adjusted such that only a few
pixels of the brightest cells were saturated (this was done using the
"palette" function of the image acquisition
software). The intensity of the PHB1 signal was measured indirectly by
adjusting settings of the microscope
camera to generate images with the same saturation levels. Then the settings
used were compared between
healthy subjects and OA patients.
[00146] For
this approach 9 controls and 7 OA patients matched for age, BMI and sex (all
women)
were used. Data is represented in Figure 6. As shown in Figure 6, PHB1 signal
is increased in leucocytes from
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
38
OA patients.
[00147] The
statistical analyses were performed using a logistic model (using SAS v9.2)
for which
the subject condition (OA vs healthy) was predicted from the values of gain
detector adjusted with the covariate
listed in Table 3 of Figure 6. Since both the mean age and the percentage of
subject with family history are very
similar in both groups, these two variables could be omitted in the final
analysis to obtain the simplest model in
which only BMI is included as a covariate. No interactions were observed among
the listed variable. Using that
simple model, the significance of the gain detector is 0.0782, which is close
to significance.
[00148] In
addition, the percentage of leucocytes nuclei expressing low levels of PHB1
was
determined (Figure 7). For this set of experiment, the gain detector was kept
constant and the percentage of cell
with non-detectable signal for PHB was calculated out of a minimum of 20 cells
(3 healthy subjects and 5 OA
patients were used in this analysis).
[00149] The
results presented in Figures 6 and 7 suggest that nuclear levels of PHB1 are
lower in
healthy subjects than in OA diagnosed patients.
EXAMPLE 6
The number of nuclear PHB1 agglomerates is similar in control and
osteoarthritis patients
[00150] Figure
8 shows the analysis of the average number of nuclear PHB1 agglomerates in
leucocytes nucleus in control subjects (n=5) and OA subjects (n=6).
[00151] For
these analyses the number of nuclear agglomerates was manually counted for a
minimum of 20 cells per patient from confocal images.
[00152] Figure
8 shows that there is no significant difference in the average number of PHB1
agglomerates in leucocytes from control and OA subjects.
EXAMPLE 7
PHB1 level is higher in chondrocytes nuclei from OA patients
[00153] PHB1 expression levels were compared with its cellular localization in
normal and OA articular
chondrocytes by IHC assays (Figure 9A). This experiment revealed a strong
positive signal for PHB1 in the nuclei
in a majority of OA patients, which correlates with Pitx1 repression. Indeed,
PHB1 mostly accumulated in nuclei
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
39
from OA chondrocytes (60% of total PHB1 in the nuclei), whereas it was mainly
present in the cytoplasm of
control chondrocytes (10% in the nuclei), and this in both superficial and
deep zones of articular cartilage (Figure
9B).
[00154] To
confirm these results, subcellular localisation of PHB1 was investigated by
isolation of
nuclear and cytosolic extracts using a saccharose gradient and verified by
measuring through immunoblotting the
relative presence of PHB1 in the two cellular fractions isolated from control
and OA primary chondrocytes
cultures. PHB1 was detected in both fractions in OA chondrocytes whereas in
control chondrocytes, it was
detectable in the cytoplasmic fraction only (Figure 90 and Figure 10). GAPDH,
F1-ATPase, and Lamin NC were
used as cytoplasmic, mitochondrial, and nuclear markers respectively, and they
were used to ensure that there
was no contamination between the different fractions. Next, the nuclear
localization of PHB1 was compared in
control and OA chondrocytes by indirect immunofluorescence against PHB1.
Results showed a stronger signal
for PHB1 in nuclei from OA chondrocytes compared to nuclei from control
chondrocytes (Figures 9D-E). It is clear
from Figure 10 that the more severely affected subjects 0A224 et 0A241 ( KL4)
and subject 0A49 ( KL3) show
the most significant PHB1 nuclear accumulation. Subject 0A237 (KL3) also shows
a higher PHB1 nuclear
accumulation than 0A240 (KL2) and ctrl.
[00155] Since PHB1 features both MTS (mitochondrial targeting sequence) and
NES (nuclear export signal)
domains, it was investigated whether PHB1 nuclear accumulation in OA articular
chondrocytes was due to
mutations affecting those domains by direct sequencing and no mutations were
found (data not shown).
[00156] Finally, to determine whether nuclear accumulation of PHB1 in OA
chondrocytes was due to an
increased PHB1 expression, quantitative real-time RT-PCR analysis was
performed to quantify PHB1 expression,
as well as Western blot analysis of whole cell extracts. No variation in PHB1
expression levels between control
and OA groups was detected (Figures 9F-G). Taken together, these results
showed that PHB1 accumulates
specifically in nuclei of OA articular chondrocytes, and this, without any
change in PHB1 expression.
EXAMPLE 8
PHB1 level is higher in knee joint articular chondrocytes nuclei of aging STR-
ORT mice
[00157] The
results presented in Figure 11 show that there is an increase in nuclear
accumulation
of PHB1 in knee joint articular chondrocytes of aging STR-ORT mice, a mouse
model of osteoarthritis.
Osteoarthritis symptoms in STR-ORT mice are known to occur at about week 30.
Between weeks 8 and 16,
osteoarthritis symptoms have not yet occurred (e.g., loss of proteoglycans in
joints has not yet occurred, Figure
11A) while PHB1 level has already increased (Figures 12B and C). These
experiments show that chondrocytes
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
nuclear PHB1 level in the mouse model starts increasing before osteoarthritis
symptoms appear.
EXAMPLE 9
PHB1 represses Pitx1 expression essentially by acting at the distal region of
the promoter
[00158] Figure
120 depicts results demonstrating that over-expression of PHB1 in co-
transfection
assays with a Pitx1-promoter-luciferase construct -3895/+61 harboring a distal
E2F-like site showed a strong
repression of luciferase reporter gene (fragments -38951+61 and smaller Pitx1
promoter fragments -30341+61, -
15771+61, -7291+61, -524/+61 and -374/+61), thus showing that PHB1 represses
Pitx1 expression essentially by
acting at the distal region of the promoter. Figure 12D shows that PHB1 blocks
the effect of E2F1 with the Pitx1-
promoter-luciferase reporter construct -38951+61. These results strongly
suggest that the binding of Pitx1
repressor proteins to the E2F site of the Pitx1 promoter inhibits Pitx1
expression, which in turn leads to the
development/progression of osteoarthritis.
EXAMPLE 10
Rescue of Pitx1 expression in OA articular chondrocytes through PHB1
inhibition
[00159]
Treatment of chondrocytes from OA patients with a PHB1 siRNA, but not with a
control
siRNA, leads to a rescue of Pitx1 expression (Figure 13). Knock down of PHB1
indicated a 3 to 25 fold increase
in PITX1 expression in articular chondrocytes derived from OA patients.
EXAMPLE 11
SUMO proteins accumulate in nuclei of OA articular chondrocytes
[00160] Figure
14A compares PHB1 and Pan Sumo expression in cytoplasmic extract (C) and
nuclear extract (N) of articular chondrocytes of a non arthrosic subject (Ctrl
3), a rheumatoid arthritis subject (RA)
and 2 OA patients. Figure 14B shows that OA chondrocytes display a strong
nuclear accumulation of SUMO-1
and SUMO-2/3 proteins in the nuclear bodies.
EXAMPLE 12
SUMO-1 protein accumulates and co-localizes with PHB1 in the nuclei of
articular chondrocytes of OA
patients
[00161]
lmmunoblot analysis using nuclear and cytosolic extracts purified from human
articular
chondrocytes of OA patients and control subjects against PHB1 and SUMO-1
revealed a band around 43 kDa
(MW of PHB1 is 32 kDa and SUMO-1 is 11.6 kDa) which is consistent with the
molecular weight observed with
anti-PHB1 antibody at the same position, suggesting that PHB1 was sumoylated
in Ail (Figures 16A and 16B).
Sumoylated proteins are generally difficult to detect by Western blot because
the reagents and steps for sample
preparation alter the sumoylation and reduce their detection. Nevertheless,
the 43 kDa band is visible in the
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
41
0A11 patient which may be more severely affected than the two other patients.
lmmunofluorescence (IF)
experiments also revealed an increased accumulation of SUMO-1 and SUMO-2/3
proteins in nuclei of articular
chondrocytes of OA subject Ail (Figure 15B, 16A and 16B).
EXAMPLE 13
SUMO proteins accumulate in PML nuclear bodies in OA articular chondrocytes
[00162] To
study the intracellular localization of PHB1, SUMO-1, SUMO 2/3 and PML nuclear
bodies in OA patients, IF staining combined with confocal microscopy of human
articular chondrocytes in OA
patients and control subjects were performed with corresponding antibodies.
Double fluorescence staining with
antibodies anti-PHB1 and anti-SUMO-1, indicates that both proteins were co-
localized in nuclei of human articular
chondrocytes in OA patients whereas in control subject, PHB1 and SUMO-1 is
mostly located in the cytosol
(Figure 16A). Double fluorescence staining of OA and control human articular
chondrocytes with antibodies
against SUMO-2/3 and PHB1 did not show a nuclear co-localization despite the
fact that both proteins are
accumulated in the nuclei (Figure 16B). In contrast, double fluorescence
staining with antibodies anti-PML and
SUMO-1 and SUMO-2/3 indicate that all these proteins were co-localized in
nuclei of human articular
chondrocytes of OA patients (Figure 17). Furthermore, in OA chondrocytes, PML
nuclear bodies are different in
size and sometimes they adopt a ring structure, as indicated by an arrow
(Figure 17A, upper right panel).
[00163] Similar
IF staining experiments showed that PHB1 does not colocalize with PML nuclear
bodies (See Figure 18).
EXAMPLE 14
SUM01 immunofluorescence in mice leucocytes
[00164] An
assay was performed to determine whether STR-ort mice which develop OA with
age
(as do human) show an increase in SUM01 in their leucocytes. Leucocytes were
isolated from peripheral blood
of 22 weeks old male mice (C57BI/6 or STR-ort) by ficoll gradient. Blood was
obtained by intracardiac ponction
and collected in EDTA tubes and kept at RT for less than 1 hr. before
leucocytes isolation. Following their
isolation on Ficoll gradiant, cells were washed in RPMI containing antibiotics
and anti-mycotics but no serum.
Cells were kept in this medium for about 20-30 min. (time to collect cells
from all ficoll
gradients+centrifugation+calculating cell concentration) before being plated
on poly-D-lysine coated glass 8-well
chamber slides and centrifuged at 300g for 6 minutes, rinsed (2 quick washes)
in PBS and fixed in 4% PFA for 7
minutes at room temperature. Cells were immunostained for SUMO-1 (or UBC9).
Nuclei were counterstained with
Draq5. Images were captured using a confocal microscope. Field of view were
selected based on the Draq5
signal, focal plane was adjusted to the center of nuclei and then SUMO-1
signal was captured. Manual cell count
was done using lmageJTM cell count tool.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
42
[00165] As
shown in Figure 19, the proportion of cells expressing strong positive signal
for
SUM01 in 22 weeks old STR-ort male is 10X that of 22 weeks old C57BI/6 male.
[00166] Similar
experiments were conducted with 7 weeks old male mice. All cells have at least
some SUMO-1 signal, but the number of cells
displaying clear SUMO-1 signal (that is mainly
cytoplasmic/membrane ) is 1 out of 31 cells in C57BI/6 mice and 9 out of 33
cells in STR/ort mice (Data not
shown(..
EXAMPLE 15
Assessment of PHB1 /SUMO-1 interaction and PHB1 sumoylation by SUMO-1 in vitro
[00167] Putative sumoylation and SUMO-binding sites in human PHB1 are
depicted in Figure 20.
[00168] To
determine whether PHB1 is directly sumoylated, a classical in vitro
sumoylation assay
using RanGap1 as a positive sumoylation control was performed. No evidence
that PHB1 is sumoylated directly
was found using this assay (Figure 21), although PHB1 is able to bind to SUMO-
1 proteins in stably-transfected
028/12 cells (Figure 22). Deletion of the SUMO-binding module (SBM, residues
76-79) of PHB1 significantly
reduced its nuclear accumulation in transfected cells.
EXAMPLE 16
Increased UBC9 expression in human knee joint OA cartilage and correlation
with disease progression
[00169] The
contribution of UBC9, the E2 ligase involved in the sumoylation pathway, was
investigated. The level of UBC9 protein was increased in knee joint of OA
patients when compared to matched
non-OA controls (Figure 23 and Figure 24) and correlates with disease severity
as evidenced by the intensity of
UBC9 IHC labelling (Figure 25, see also Figure 23). Overexpression of UBC9
showed that it stabilises PHB1 and
promotes its nuclear accumulation in transfected U2OS cells (Figure 25).
However, this mechanism is probably
indirect, as the SBM of PHB1 is involved, as evidenced by the fact that its
removal in the mutant PHBLASBM
abrogated the effect of UBC9 (Figure 25B).
[00170]
Although the present invention has been described hereinabove by way of
specific
embodiments thereof, it can be modified, without departing from the spirit and
nature of the subject invention as
defined in the appended claims.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
43
REFERENCES
1. Loughlin,J. Genetic epidemiology of primary osteoarthritis. Curr. Opin.
Rheumatol. 13, 111-116 (2001).
2. Spector,T.D. & MacGregor,A.J. Risk factors for osteoarthritis: genetics.
Osteoarthritis. Cartilage. 12
Suppl A, S39-S44 (2004).
3. Peach,C.A., Carr,A.J., & Loughlin,J. Recent advances in the genetic
investigation of osteoarthritis.
Trends Mol. Med. 11, 186-191 (2005).
4. Reginato,A.M. & Olsen,B.R. The role of structural genes in the
pathogenesis of osteoarthritic disorders.
Arthritis Res. 4, 337-345 (2002).
5. Loughlin,J. Polymorphism in signal transduction is a major route through
which osteoarthritis
susceptibility is acting. Curr. Opin. Rheumatol. 17, 629-633 (2005).
6. Drouin,J., Lanctot,C., & Tremblay,J.J. La famille Ptx des facteurs de
transcription a homeodomaine.
Medecine/Sciences 14, 335-339 (1998).
7. Drouin,J., Lamolet,B., Lamonerie,T., Lanctot,C., & Tremblay,J.J. The PTX
family of homeodomain
transcription factors during pituitary developments. Mol. Cell Endocrinol.
140, 31-36 (1998).
8. Lanct6t,C., Lamolet,B., & Drouin,J. The bicoid-related homeoprotein Ptx1
defines the most anterior
domain of the embryo and differentiates posterior from anterior lateral
mesoderm. Development 124,
2807-2817 (1997).
9. Lanct6t,C., Moreau,A., Chamberland,M., Tremblay,M.L., & Drouin,J.
Hindlimb patterning and mandible
development require the Ptx1 gene. Development 126, 1805-1810 (1999).
10. Messeguer,X. et al. PROMO: detection of known transcription regulatory
elements using species-
tailored searches. Bioinformatics. 18, 333-334 (2002).
11. Farre,D. et al. Identification of patterns in biological sequences at
the ALGGEN server: PROMO and
MALGEN. Nucleic Acids Res. 31, 3651-3653 (2003).
12. Tommasi,S. & Pfeifer,G.P. Constitutive protection of E2F recognition
sequences in the human thymidine
kinase promoter during cell cycle progression. J. Biol. Chem. 272, 30483-30490
(1997).
13. Muller,H. et al. E2Fs regulate the expression of genes involved in
differentiation, development,
proliferation, and apoptosis. Genes Dev. 15, 267-285 (2001).
14. Zheng,N., Fraenkel,E., Pabo,C.O., & Pavletich,N.P. Structural basis of
DNA recognition by the
heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13, 666-674
(1999).
15. Takahashi,Y., Rayman,J.B., & Dynlacht,B.D. Analysis of promoter binding
by the E2F and pRB families
in vivo: distinct E2F proteins mediate activation and repression. Genes Dev.
14, 804-816 (2000).
16. Bremner, R. et al. Direct transcriptional repression by pRB and its
reversal by specific cyclins. Mol. Cell
Biol. 15, 3256-3265 (1995).
17. Hamel,P.A., Gill,R.M., Phillips,R.A., & Gallie,B.L. Transcriptional
repression of the E2-containing
promoters EllaE, c-myc, and RB1 by the product of the RB1 gene. Mol. Cell
Biol. 12, 3431-3438 (1992).
18. Lam,E.W. & Watson,R.J. An E2F-binding site mediates cell-cycle
regulated repression of mouse B-myb
transcription. EMBO J 12, 2705-2713 (1993).
19. McClung,J.K., Jupe,E.R., Liu,X.T., & Dell'Orco,R.T. Prohibitin: potential
role in senescence,
development, and tumor suppression. Exp. Gerontol. 30, 99-124 (1995).
20. Dell'Orco,R.T., McClung,J.K., Jupe,E.R., & Liu,X.T. Prohibitin and the
senescent phenotype. Exp.
Gerontol. 31, 245-252 (1996).
21. Rastogi,S. et al. Prohibitin facilitates cellular senescence by
recruiting specific corepressors to inhibit
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
44
E2F target genes. Mol. Cell Biol. 26, 4161-4171 (2006).
22. Campisi,J. Cellular senescence as a tumor-suppressor mechanism. Trends
Cell Biol. 11, S27-S31
(2001).
23. Martin,J.A. & Buckwalter,J.A. The role of chondrocyte senescence in the
pathogenesis of osteoarthritis
and in limiting cartilage repair. J Bone Joint Surg Am 85-A Suppl 2, 106-110
(2003).
24. Wang,S., Fusaro,G., Padmanabhan,J., & Chellappan,S.P. Prohibitin co-
localizes with Rb in the nucleus
and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene 21, 8388-
8396 (2002).
25. Altman,R. et al. Development of criteria for the classification and
reporting of osteoarthritis. Classification
of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee
of the American
Rheumatism Association. Arthritis Rheum. 29, 1039-1049 (1986).
26. Roy-Beaudry,M. et al. Endothelin 1 promotes osteoarthritic cartilage
degradation via matrix
metalloprotease 1 and matrix metalloprotease 13 induction. Arthritis Rheum.
48, 2855-2864 (2003).
27. Mankin,H.J., Dorfman,H., Lippiello,L., & Zarins,A. Biochemical and
metabolic abnormalities in articular
cartilage from osteo-arthritic human hips. II. Correlation of morphology with
biochemical and metabolic
data. J. Bone Joint Surg. Am. 53, 523-537 (1971).
28. Moreau,A., Yotov,W.V., Glorieux,F.H., & St Arnaud,R. Bone-specific
expression of the alpha chain of the
nascent polypeptide- associated complex, a coactivator potentiating c-Jun-
mediated transcription. Mol.
Cell Biol. 18, 1312-1321 (1998).
29. Deeks,J.J. & Altman,D.G. Sensitivity and specificity and their
confidence intervals cannot exceed 100%.
BMJ 318, 193-194 (1999).
30. Mishra S., Murphy L.C., The Prohibitins: emerging roles in diverse
functions. J Cell Mol Med. 10(2): 353-
63 (2006).
31. Nijtmans LG, Artal SM, Grivell LA, Coates PJ. The mitochondrial PHB
complex: roles in mitochondrial
respiratory complex assembly, ageing and degenerative disease. Cell Mol Life
Sci 2002; 59(1):143-155.
32. Mishra S, Murphy LC, Nyomba BL, Murphy U. Prohibitin: a potential
target for new therapeutics. Trends
Mol Med 2005; 11(4):192-197.
33. Lawrence RC, Hochberg MC, Kelsey JL, McDuffie FC, Medsger TA Jr, Felts
WR, et al. Estimates of the
prevalence of selected arthritic and musculoskeletal diseases in the United
States. J Rheumatol
1989; 16:427-41.
34. Felson DT, Zhang Y. An update on the epidemiology of knee and hip
osteoarthritis with a view to
prevention. Arthritis Rheum 1998;41:1343-55.
35. Xavier Messeguer, Ruth Escudero, Domenec Farre, Oscar Nunez, Javier
Martinez, M.Mar Alba.
PROMO: detection of known transcription regulatory elements using species-
tailored searches.
Bioinformatics, 18, 2, 333-334, 2002;
36. Domenec Farre, Roma Roset, Mario Huerta, Jose E. Adsuara, Llorenc
Roselle), M.Mar Alba, Xavier
Messeguer. Identification of patterns in biological sequences at the ALGGEN
server: PROMO and
MALGEN. Nucleic Acids Res, 31, 13, 3651-3653, 2003.
37. Gamble SC, Odontiadis M, Waxman J et al. Androgens target prohibitin to
regulate proliferation of
prostate cancer cells. Oncogene 2004; 23(17):2996-3004.
38. Sun L, Liu L, Yang XJ, Wu Z. Akt binds prohibitin 2 and relieves its
repression of MyoD and muscle
differentiation. J Cell Sci 2004; 117(Pt 14):3021-3029.
39. Wang S, Zhang B, Faller DV. Prohibitin requires Brg-1 and Brm for the
repression of E2F and cell
growth. EM BO J 2002; 21(12):3019-3028.
40. Montano MM, Ekena K, age-Mourroux R, Chang W, Martini P,
Katzenellenbogen BS. An estrogen
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
receptor-selective coregulator that potentiates the effectiveness of
antiestrogens and represses the
activity of estrogens. Proc Natl Acad Sci U S A 1999; 96(12):6947-6952.
41. Kurtev V, Margueron R, Kroboth K, Ogris E, Cavailles V, Seiser C.
Transcriptional regulation by the
repressor of estrogen receptor activity via recruitment of histone
deacetylases. J Biol Chem 2004;
279(23):24834-24843.
42. Rastogi S, Joshi B, Fusaro G, Chellappan S. Camptothecin induces
nuclear export of prohibitin
preferentially in transformed cells through a CRM-1-dependent mechanism. J
Biol Chem 2006;
281(5):2951-2959.
43. Wang S, Nath N, Fusaro G, Chellappan S. Rb and prohibitin target
distinct regions of E2F1 for
repression and respond to different upstream signals. Mol Cell Biol 1999;
19(11):7447-7460.
44. Wang S, Zhang B, Faller DV. BRG1/BRM and prohibitin are required for
growth suppression by estrogen
antagonists. EMBO J 2004; 23(11):2293-2303.
45. Wang S, Fusaro G, Padmanabhan J, Chellappan SP. Prohibitin co-localizes
with Rb in the nucleus and
recruits N-CoR and HDAC1 for transcriptional repression. Oncogene 2002;
21(55):8388-8396.
46. Becher S, Achatz G, Schmitz ML, Lamers MC. Prohibitin and prohibitone
are contained in high-
molecular weight complexes and interact with alpha-actinin and annexin A2.
Biochimie 2002; 84:1205-
1218.
47. Rastogi S, Joshi B, Dasgupta P, Morris M, Wright K, Chellappan S.
Prohibitin facilitates cellular
senescence by recruiting specific corepressors to inhibit E2F target genes.
Mol Cell Biol 2006;
26(11):4161-4171.
48. Rasmussen RK, Ji H, Eddes JS et al. Two-dimensional electrophoretic
analysis of mixed lineage kinase
2 N-terminal domain binding proteins. Electrophoresis 1998; 19(5):809-817.
49. Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin
induces the transcriptional activity
of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem
2003; 278(48):47853-
47861.
50. Rajalingam K, Wunder C, Brinkmann V et al. Prohibitin is required for
Ras-induced Raf-MEK-ERK
activation and epithelial cell migration. Nat Cell Biol 2005; 7(8):837-843.
51. Wang S, Nath N, Adlam M, Chellappan S. Prohibitin, a potential tumor
suppressor, interacts with RB and
regulates E2F function. Oncogene 1999; 18(23):3501-3510.
52. Lyons P., 2003. Advances in spotted microarray ressources for
expression profiling. Briefings in
Functionnal Genomics and Proteomics 2, 21-30.
53. Ding G. and Cantor C.R., 2004. Quantitative analysis of nucleic acids -
the last few years of progress. J
Biochem Biol 37, 1-10.
54. Scheel J., Von Brevern M.C., Horlein A., Fisher A., Schneider A., Bach
A. 2002. Yellow pages to the
transcriptome. Pharmacogenomics 3, 791-807.
55. Lopez M.F., Pluskal M.G., 2003. Protein micro- and macroarrays:
digitizing the proteome. J
Chromatography B 787, 19-27.
56. Wood, W.I. et al. 1985. Base composition-independent hybridization in
tetramethylammonium chloride:
A method for oligonucleotide screening of highly complex gene library. Proc.
Natl. Acad. Sci. USA 82.
1585-1588.
57. Kolonin et al. 2006., Nature Medicine Advance online publication of
June issue.
58. United States Patent 5,401,635.
59. Abecasis GR, Cookson WO., 2000. GOLD--graphical overview of linkage
disequilibrium. Bioinformatics
16:182-3.
CA 02843631 2014-01-30
WO 2013/053065
PCT/CA2012/050723
46
60. Ott, J. 2002. Predicting the range of linkage disequilibrium. Proc.
Nat! Acad. Sci. USA, 97, 2-3.
61. Camper-Kirby D, Welch S, Walker A et al. Myocardial Akt activation and
gender: increased nuclear
activity in females versus males. Circ Res 2001; 88(10):1020-1027.
62. Miller, 1988. Ann. Reports Med. Chem. 23:295.
63. Moran etal., 1987. Nucleic Acids Res., 14:5019.
64. Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning:
A Laboratory Manual, Cold Spring
Harbor Laboratory Press, Cold Spring Harbor, NY.
65. Lanctot et al., 1999. Development 126, 1805-1810.