Note: Descriptions are shown in the official language in which they were submitted.
CA 02855955 2014-03-11
W02013/039471
PCT/US2011/051202
TOLL-LIKE RECEPTOR 3 ANTAGONISTS FOR THE TREATMENT OF
METABOLIC AND CARDIOVASCULAR DISEASES
Field of the Invention
The present invention relates to Toll-Like Receptor 3
(TLR3) antibody antagonists, polynucleotides encoding TLR3
antibody antagonists or fragments thereof, and methods of
making and using the foregoing.
Background of the Invention
Toll-like receptors (TLRs) regulate activation of the
innate immune response and influence the development of
adaptive immunity by initiating signal transduction cascades
in response to bacterial, viral, parasitic, and in some
cases, host-derived ligands (Lancaster et al., J. Physiol.
563:945-955, 2005). The plasma membrane localized TLRs,
TLR1, TLR2, TLR4 and TLR6 recognize ligands including protein
or lipid components of bacteria and fungi. The predominantly
intracellular TLRs, TLR3, TLR7 and TLR9 respond to dsRNA,
ssRNA and unmethylated CpG DNA, respectively. Dysregulation
of TLR signaling is believed to cause a multitude of
problems, and therapeutic strategies are in development
towards this axis (Hoffman et al., Nat. Rev. Drug Discov.
4:879-880, 2005; Rezaei, Int. Immunopharmacol. 6:863-869,
2006; Wickelgren, Science 312:184-187, 2006). For example,
antagonists of TLR4 and TLRs 7 and 9 are in clinical
development for severe sepsis and lupus, respectively
(Kanzler et al., Nat. Med. 13:552-559, 2007).
TLR3 signaling is activated by dsRNA, mRNA or RNA
released from necrotic cells during inflammation or virus
infection. TLR3 activation induces secretion of interferons
and pro-inflammatory cytokines and triggers immune cell
activation and recruitement that are protective during
certain microbial infections. For example, a dominant-
negative TLR3 allele has been associated with increased
susceptibility to Herpes Simplex encephalitis upon primary
1
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
infection with HSV-1 in childhood (Zheng et al., Science
317:1522-1527, 2007). In mice, TLR3 deficiency is associated
with decreased survival upon coxsackie virus challenge
(Richer et al., PLoS One 4:e4127, 2009). However,
uncontrolled or dysregulated TLR3 signaling has been shown to
contribute to morbidity and mortality in certain viral
infection models including West Nile, phlebovirus, vaccinia,
and influenza A (Wang et al., Nat. Med. 10:1366-1373, 2004;
Gowen et al., J. Immunol. 177:6301-6307, 2006; Hutchens et
al., J. Immunol. 180:483-491, 2008; Le Goffic et al., PloS
Pathog. 2:E53, 2006).
The crystal structures of the human and murine TLR3
extracellular domains have been determined ((Bell et al.,
Proc. Natl. Acad. Sci. (USA), 102:10976-80, 2005; Choe, et
al., Science 309:581-585, 2005; Liu et al., Science, 320:379-
381, 2008). TLR3 adopts the overall shape of a solenoid
horseshoe decorated by glycans and has 23 tandem units of
leucine-rich repeat (LRR) motifs. The dsRNA binding sites
have been mapped to two distinct regions (Liu et al.,
Science, 320:379-81, 2008). The singaling assembly has been
proposed to consist of 1 dsRNA and two TLR3 extracellular
domains (Leonard et al., Proc. Natl. Acad. Sci. (USA) 105:
258-263, 2008).
TLR3 has been shown to drive pathogenic mechanisms in a
spectrum of inflammatory, immune-mediated and autoimmune
diseases including, for example, septic shock (Cavassani et
al., J. Exp. Med. 205:2609-2621, 2008), acute lung injury
(Murray et al., Am. J. Respir. Crit. Care Med. 178:1227-1237,
2008), rheumatoid arthritis (Kim et al., Immunol. Lett.
124:9-17, 2009; Brentano et al., Arth. Rheum. 52:2656-2665,
2005), asthma (Sugiura et al., Am. J. Resp. Cell Mob. Biol.
40:654-662, 2009; Morishima et al., Int. Arch. Allergy
Immunol. 145:163-174, 2008; Stowell et al., Respir. Res.
10:43, 2009), inflammatory bowel disease such as Crohn's
disease and ulcerative colitis (Zhou et al., J. Immunol.
178:4548-4556, 2007; Zhou et al., Proc. Natl. Acad. Sci.
2
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
(USA) 104:7512-7515, 2007), autoimmune liver disease (Lang et
al., J. Clin. Invest. 116:2456-2463, 2006) and type I
diabetes (Dogusan et al. Diabetes 57:1236-1245, 2008; Lien
and Zipris, Curr. Mol. Med. 9:52-68, 2009). Furthermore,
organ-specific increases in TLR3 expression have been shown
to correlate with a number of pathological conditions driven
by dysregulated local inflammatory responses such as in liver
tissue in primary biliary cirrhosis (Takii et al., Lab
Invest. 85:908-920, 2005), rheumatoid arthritis joints
(Ospelt et al., Arthritis Rheum. 58:3684-3692, 2008), and
nasal mucosa of allergic rhinitis patients (Fransson et al.,
Respir. Res. 6:100, 2005).
In necrotic conditions, the release of intracellular
content including endogenous mRNA triggers secretion of
cytokines, chemokines and other factors that induce local
inflammation, facilitate clearance of dead cell remnants and
repair the damage. Necrosis often perpetuates inflammatory
processes, contributing to chronic or exaggerated
inflammation (Bergsbaken et al., Nature Reviews 7:99-109,
2009). Activation of TLR3 at the site of necrosis may
contribute to these aberrant inflammatory processes and
generate a further pro-inflammatory positive feedback loop
via the released TLR3 ligands. Thus, TLR3 antagonism may be
beneficial in a variety of disorders involving chronic or
exaggerated inflammation and/or necrosis.
Down-modulation of TLR3 activation may also represent a
novel treatment strategy for oncologic indications including
renal cell carcinomas and head and neck squamous cell
carcinomas (Morikawa et al., Clin. Cancer Res. 13:5703-5709,
2007; Pries et al., Int. J. Mol. Med. 21:209-215, 2008).
Furthermore, the TLR3L423E allele encoding a protein with
reduced activity has been associated with protection against
advanced "dry" age-related macular degeneration (Yang et al.,
N. Engl. J. Med. 359:1456-1463, 2008), indicating that TLR3
antagonists may be beneficial in this disease.
3
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Pathologies associated with inflammatory conditions and
others, such as those associated with infections, have
significant health and economic impacts. Yet, despite
advances in many areas of medicine, comparatively few
treatment options and therapies are available for many of
these conditions.
Thus, a need exists to suppress TLR3 activity to treat
TLR3-associated conditions.
Brief Description of the Drawings
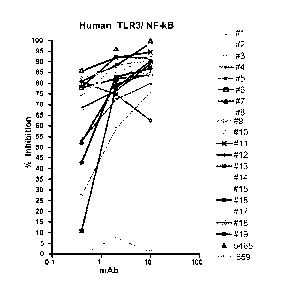
Fig. 1 shows the effect of anti-human TLR3 (huTLR3) mAbs
in an NF-KB reporter gene assay.
Figs. 2A and 2B show the effect (% inhibition) or anti-
huTLR3 mAbs in a BEAS-2B assay.
Figs. 3A and 3B show the effect of anti-huTLR3 mAbs in a
NHBE assay.
Fig. 4 shows the effect of anti-huTLR3 mAbs in a PBMC
assay.
Figs. 5A and 5B show the effect of anti-huTLR3 mAbs in a
HASM assay.
Figs. 6A, 6B and 6C show the binding of anti-huTLR3 mAbs
to TLR3 mutants.
Fig. 7A shows epitopes for mAb 15EVQ (black) and C1068 mAb
(grey) (top image) and epitope for mAb 12QVQ/QSV (black, bottom
image) superimposed on the structure of human TLR3 ECD. Fig. 7B
shows localized H/D exchange perturbation map of TLR3 ECD protein
complexed with mAb 15EVQ.
Figs. 8A and 8B show the effect of rat/mouse anti-mouse
TLR3 mAb mAb 5429 (surrogate) in A) NF-KB and B) ISRE
reporter gene assays.
Fig. 9 shows the effect of the surrogate mAbs (mAb 5429,
mAb c1811) in the MEF CXCL10/IP-10 assay.
Fig. 10 shows specificity of binding of the surrogate
mAb to TLR3. Top panel: isotype control; bottom panel: mAb
c1811.
Fig. 11 shows effect of the surrogate mAbs on penH level
in an AHR model.
4
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Fig. 12 shows effect of the surrogate mAbs on total
neutrophil numbers in BAL fluid in an AHR model.
Fig. 13 shows effect of the surrogate mAbs on CXCL10/IP-
levels in BAL fluid in an AHR model.
5 Fig. 14 shows effect of the surrogate mAb on
histopathology scores in a DSS model.
Fig. 15 shows effect of the surrogate mAb on A)
histopathology scores and B) neutrophil influx in a T-cell
transfer model.
10 Fig. 16 shows effect of the surrogate mAb on clinical
scores in a CIA model.
Fig. 17 shows effect of the surrogate mAb on the
clinical AUC scores in a CIA model.
Fig. 18 shows effect of the surrogate mAb on the
survival of C57BL/6 mice following intranasal administration
of influenza A/PR/8/34. mAb dosing began at day -1.
Fig. 19 shows effect of the surrogate mAb on clinical
scores following influenza A/PR/8/34 administration. mAb
dosing began at day -1.
Fig. 20 shows effect of the surrogate mAb on body weight
over 14 days after administration of influenza A/PR/8/34.
mAb dosing began at day -1.
Fig. 21 shows effect of the surrogate mAbs on blood
glucose levels in (A) WT DIG and (B) TLR3K0 DIG animals after
glucose challenge.
Fig. 22 shows effect of the surrogate mAb on insulin
levels in WT DIG animals.
Fig. 23 shows effect of mAb 15EVQ on (A) NTHi and (B)
rhinovirus induced CXCL10/IP-10 and CCL5/RANTES levels in
NHBE cells.
Fig. 24 shows effect of mAb 15EVQ on (A) sICAM-1 levels
and (B) viability in HUVEC cells.
Fig. 25 shows survival of animals following
administration of the surrogate mAb 3 days post infection
with influenza A.
5
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Fig. 26 shows clinical scores following administration
of the surrogate mAb 3 days post infection with influenza A.
Fig. 27 shows body weight change of animals following
administration of the surrogate mAb 3 days post infection
with influenza A.
Fig. 28 shows the molecular structure of the quaternary
complex of huTLR3 ECD with Fab 12QVQ/QSV, Fab 15EVQ and Fab
c1068 in A. in ribbon and surface representations. The TLR3
ECD is in light gray with the N-terminus labeled N; all Fab
molecules are shown in dark gray in ribbons representation.
B. The epitopes are colored light gray and labeled on the
TLR3 ECD as for the Fabs in A. In Figures 28, 29 and 30, the
Fab 12QVQ/QSV, Fab c1068 and Fab 15EVQ are abbreviated to
Fab12, Fab1068 and Fab15, respectively in the labels for
clarity.
Fig 29. Shows a mechanism of neutralization by Fab
15EVQ. A. dsRNA:TLR3 signaling unit (SU) is shown with the
Fab 15EVQ epitope highlighted (light gray) in one of the two
TLR3 ECD (light and dark gray, and labeled TLR3). The dsRNA
ligand is shown as a double helix in light gray. B. An
illustration of Fab 15EVQ binding that sterically inhibited
dsRNA binding and thus, inhibits the formation of the SU.
Binding of Fab 15EVQ, which is higher affinity, will prevent
the SU from forming or will disassemble the pre-formed SU.
Fig. 30 shows a mechanism of Fab 12QVQ/QSV and Fab c1068
and clustering of TLR3 signaling units (SU). A. Fab
12QVQ/QSV and Fab c1068 can bind (or co-bind) a single SU.
B. Model for closest clustering of two SUs on a dsRNA of
about 76 base pairs. The three epitopes are highlighted in
different molecules for clarity. C. Binding of Fab
12QVQ/QSV and Fab c1068 prevents SU clustering due to steric
clashes between the antibodies and neighboring SUs. The two
left-pointing arrows qualitatively represent different
degrees of separation of SUs due to the antibodies (bottom
arrow for Fab 12QVQ/QSV and top arrow for Fab c1068).
6
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Fig. 31 shows the correspondence between sequential,
Kabat, and Chothia numbering for exemplary antibodies. The
CDRs and HVs are highlighted in gray.
Fig. 32 shows alignment of VL of mAb 15EVQ with human
Vk1 frameworks. Chothia hypervariable loops are underlined,
paratope residues double underlined and the framework
differences highlighted in gray. The VK1 genes are *01
alleles unless otherwise indicated. Residue numbering is
sequential.
Fig. 33 shows alignment of VH of mAb 15EVQ with human
Vh5 frameworks. Sequence features indicated as in Fig. 32.
Fig. 34 shows alignment of VL of mAb 12QVQ/QSV with
human Vk3 frameworks. Sequence features indicated as in Fig.
32.
Fig. 35 shows alignment of VL and VH of mAb 15EVQ or mAb
12QVQ/QSV with human JK, A or Jh frameworks. Sequence
features indicated as in Fig. 32.
Summary of the Invention
One aspect of the invention is an isolated antibody or
fragment thereof, wherein the antibody binds toll-like
receptor 3 (TLR3) amino acid residues K416, K418, L440, N441,
E442, Y465, N466, K467, Y468, R488, R489, A491, K493, N515,
N516, N517, H539, N541, S571, L595, and K619 of SEQ ID NO: 2.
Another aspect of the invention is an isolated antibody
or fragment thereof, wherein the antibody binds toll-like
receptor 3 (TLR3) amino acid residues S115, D116, K117, A120,
K139, N140, N141, V144, K145, T166, Q167, V168, S188, E189,
D192, A195, and A219 of SEQ ID NO: 2.
Another aspect of the invention is an isolated antibody
having a heavy chain variable region and a light chain
variable region or fragment thereof, wherein the antibody
binds TLR3 having an amino acid sequence shown in SEQ ID NO:
2 with the heavy chain variable region Chothia residues W33,
F50, D52, D54, Y56, N58, P61, E95, Y97, Y100, and D100b and
7
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
the light chain variable region Chothia residues Q27, Y32,
N92, T93, L94, and S95.
Another aspect of the invention is an isolated antibody
having a heavy chain variable region and a light chain
variable region or fragment thereof, wherein the antibody
binds TLR3 having an amino acid sequence shown in SEQ ID NO:
2 with the heavy chain variable region Chothia residues N31a,
Q52, R52b, S53, K54, Y56, Y97, P98, F99, and Y100, and the
light chain variable region Chothia residues G29, S30, Y31,
Y32, E50, D51, Y91, D92, and D93.
Another aspect of the invention is an isolated antibody
reactive with TLR3, wherein the antibody has at least one of
the following properties:
a. binds to human TLR3 with a Kd fo <10 nM;
b. reduces human TLR3 biological activity in an in
vitro poly(I:C) NF-kB reporter gene assay >50% at
1 g/ml;
c. inhibits >60% of IL-6 or CXCL10/IP-10 production
from BEAS-2B cells stimulated with <100 ng/ml
poly(I:C) at 10 g/ml;
d. inhibits >50% of IL-6 or CXCL10/IP-10 production
from BEAS-2B cells stimulated with <100 ng/ml
poly(I:C) at 0.4 g/ml;
e. inhibits >50% of IL-6 production from NHBE cells
stimulated with 62.5 ng/ml poly(I:C) at 5 g/ml;
f. inhibits >50% of IL-6 production from NHBE cells
stimulated with 62.5 ng/ml poly(I:C)at 1 g/ml;
g. inhibits >20% of poly(I:C)-induced IFN-y, IL-6 or
IL-12 production by PBMC cells at 1 g/ml;
h. inhibits cynomologus TLR3 biological activity in
an in vitro NF-kB reporter gene assay with IC50 <
10 g/ml; or
i. inhibits cynomologus TLR3 biological activity in
an in vitro ISRE reporter gene assay with IC50 < 5
8
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Another aspect of the invention is an isolated antibody
reactive with TLR3 that competes for TLR3 binding with a
monoclonal antibody, wherein the monoclonal antibody
comprises the amino acid sequences of certain heavy chain
complementarity determining regions (CDRs) 1, 2 and 3, the
amino acid sequences of certain light chain CDRs 1, 2 and 3,
the amino acid sequences of certain heavy chain variable
regions (VH) or the amino acid sequence of certain light
chain variable regions (VL).
Another aspect of the invention is an isolated antibody
reactive with TLR3 comprising both a heavy chain variable
region and a light chain variable region and wherein the
antibody comprises the amino acid sequences of certain heavy
chain complementarity determining regions (CDRs) 1, 2 and 3
and the amino acid sequences of certain light chain CDRs 1, 2
and 3.
Another aspect of the invention is an isolated antibody
reactive with TLR3 comprising both a heavy chain variable
region and a light chain variable region and wherein the
antibody comprises the amino acid sequences of certain heavy
chain variable regions (VH) and the amino acid sequences of
certain light chain variable regions (VL).
Another aspect of the invention is an isolated antibody
reactive with TLR3 comprising both a heavy chain variable
region and a light chain variable region and wherein the
antibody comprises the amino acid sequence of certain heavy
chains and the amino acid sequence of certain light chains.
Another aspect of the invention is an isolated antibody
heavy chain comprising the amino acid sequence shown in SEQ
ID NO: 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32,
34, 36, 38, 40, 42, 124, 125, 126, 127, 128, 129, 159, 198,
200, 202, 164, 212, 213, 214, 215 or 216.
Another aspect of the invention is an isolated antibody
light chain comprising the amino acid sequence shown in SEQ
ID NO: 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31,
9
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
33, 35, 37, 39, 41, 122, 123, 197, 199, 201, 163, 209, 210,
211, or 225.
Another aspect of the invention is an isolated antibody
heavy chain comprising the amino acid sequence shown in SEQ
ID NO: 102, 130, 131, 132, 133, 134, 135, 160, 204, 206, 208,
220, 166 or 168.
Another aspect of the invention is an isolated antibody
light chain comprising the amino acid sequence shown in SEQ
ID NO: 155, 156, 157, 158, 203, 205, 207, 165, 167, or 227.
Another aspect of the invention is an isolated
polynucleotide encoding an antibody heavy chain comprising
the amino acid sequence shown in SEQ ID NO: 6, 8, 10, 12, 14,
16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 124,
125, 126, 127, 128, 129, 159, 198, 200, 202, 164, 212, 213,
214, 215 or 216.
Another aspect of the invention is an isolated
polynucleotide encoding an antibody light chain comprising
the amino acid sequence shown in SEQ ID NO: 5, 7, 9, 11, 13,
15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 122,
123, 197, 199, 201, 163, 209, 210, 211, or 225.
Another aspect of the invention is an isolated
polynucleotide encoding an antibody heavy chain comprising
the amino acid sequence shown in SEQ ID NO: 102, 130, 131,
132, 133, 134, 135, 160, 204, 206, 208, 220, 166 or 168.
Another aspect of the invention is an isolated
polynucleotide encoding an antibody light chain comprising
the amino acid sequence shown in SEQ ID NO: 155, 156, 157,
158, 203, 205, 207, 165, 167, or 227.
Another aspect of the invention is a pharmaceutical
composition comprising the isolated antibody of the invention
and a pharmaceutically acceptable carrier.
Another aspect of the invention is a vector comprising
at least one polynucleotide of the invention.
Another aspect of the invention is a host cell
comprising the vector of the invention.
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Another aspect of the invention is a method of making an
antibody reactive with TLR3 comprising culturing the host
cell of the invention and recovering the antibody produced by
the host cell.
Another aspect of the invention is a method of treating
or preventing an inflammatory condition comprising
administering a therapeutically effective amount of the
isolated antibody of the invention to a patient in need
thereof for a time sufficient to treat or prevent the
inflammatory condition.
Another aspect of the invention is a method of treating
or preventing a systemic inflammatory condition comprising
administering a therapeutically effective amount of the
isolated antibody of the invention to a patient in need
thereof for a time sufficient to treat or prevent the
systemic inflammatory condition.
Another aspect of the invention is a method of treating
type II diabetes comprising administering a therapeutically
effective amount of the isolated antibody of the invention to
a patient in need thereof for a time sufficient to treat type
II diabetes.
Another aspect of the invention is a method of treating
hyperglycemia comprising administering a therapeutically
effective amount of the isolated antibody of the invention to
a patient in need thereof for a time sufficient to treat the
hyperglycemia.
Another aspect of the invention is a method of treating
hyperinsulinemia comprising administering a therapeutically
effective amount of the isolated antibody of the invention to
a patient in need thereof for a time sufficient to treat the
insulin resistance.
Another aspect of the invention is a method of treating
or preventing viral infections comprising administering a
therapeutically effective amount of the isolated antibody of
the invention to a patient in need thereof for a time
sufficient to treat or prevent viral infections.
11
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Detailed Description of the Invention
All publications, including but not limited to patents
and patent applications, cited in this specification are
herein incorporated by reference as though fully set forth.
The term "antagonist" as used herein means a molecule
that partially or completely inhibits, by any mechanism, an
effect of another molecule such as a receptor or
intracellular mediator.
As used herein, a "TRL3 antibody antagonist" or an
antibody "reactive with TLR3" describes an antibody that is
capable of, directly or indirectly, substantially
counteracting, reducing or inhibiting TLR3 biological
activity or TLR3 receptor activation. For example, an
antibody reactive with TLR3 can bind directly to TLR3 and
neutralize TLR3 activity, i.e, block TLR3 signaling to reduce
cytokine and chemokine release or NF-KB activation.
The term "antibodies" as used herein is meant in a broad
sense and includes immunoglobulin or antibody molecules
including polyclonal antibodies, monoclonal antibodies
including murine, human, human-adapted, humanized and
chimeric monoclonal antibodies and antibody fragments.
In general, antibodies are proteins or peptide chains
that exhibit binding specificity to a specific antigen.
Intact antibodies are heterotetrameric glycoproteins,
composed of two identical light chains and two identical
heavy chains. Typically, each light chain is linked to a
heavy chain by one covalent disulfide bond, while the number
of disulfide linkages varies between the heavy chains of
different immunoglobulin isotypes. Each heavy and light
chain also has regularly spaced intrachain disulfide bridges.
Each heavy chain has at one end a variable domain (variable
region) (VH) followed by a number of constant domains
(constant regions). Each light chain has a variable domain
at one end (VL) and a constant domain at its other end; the
constant domain of the light chain is aligned with the first
12
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
constant domain of the heavy chain and the light chain
variable domain is aligned with the variable domain of the
heavy chain. Antibody light chains of any vertebrate species
can be assigned to one of two clearly distinct types, namely
kappa (K) and lambda (A), based on the amino acid sequences
of their constant domains.
Immunoglobulins can be assigned to five major classes,
namely IgA, IgD, IgE, IgG and IgM, depending on the heavy
chain constant domain amino acid sequence. IgA and IgG are
further sub-classified as the isotypes IgAl, IgA2, IgGI, IgG2,
IgG3 and IgG4.
The term "antibody fragments" means a portion of an
intact antibody, generally the antigen binding or variable
region of the intact antibody. Examples of antibody
fragments include Fab, Fab', F(ab')2 and Fv fragments,
diabodies, single chain antibody molecules and multispecific
antibodies formed from at least two intact antibodies.
An immunoglobulin light chain variable region or heavy
chain variable region consists of a "framework" region
interrupted by three "antigen-binding sites". The antigen-
binding sites are defined using various terms as follows: (i)
the term Complementarity Determining Regions (CDRs) is based
on sequence variability (Wu and Kabat, J. Exp. Med. 132:211-
250, 1970). Generally, the antigen-binding site has six
CDRs; three in the VH (HCDR1, HCDR2, HCDR3), and three in the
VL (LCDR1, LCDR2, LCDR3) (Kabat et al., Sequences of Proteins
of Immunological Interest, 5th Ed. Public Health Service,
National Institutes of Health, Bethesda, Md., 1991). (ii)
The term "hypervariable region", "HVR", or "HV" refers to the
regions of an antibody variable domain which are
hypervariable in structure as defined by Chothia and Lesk
(Chothia and Lesk, Mol. Biol. 196:901-917, 1987). Generally,
the antigen-binding site has six hypervariable regions, three
in VH (H1, H2, H3) and three in VL (L1, L2, L3). Chothia and
Lesk refer to structurally conserved HVs as "canonical
structures". (iii) The "IMGT-CDRs" as proposed by Lefranc
13
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
(Lefranc et al., Dev. Comparat. Immunol. 27:55-77, 2003) are
based on the comparison of V domains from immunoglobulins and
T-cell receptors. The International ImMunoGeneTics (IMGT)
database (http://www imgt org) provides a standardized
numbering and definition of these regions. The
correspondence between CDRs, HVs and IMGT delineations is
described in Lefranc et al., Dev. Comparat. Immunol. 27:55-
77, 2003. (iv) The antigen-binding site can also be
delineated based on Specificity Determining Residue Usage
(SDRU)(Almagro, Mol. Recognit. 17:132-143, 2004), where
Specificity Determining Residues (SDR), refers to amino acid
residues of an immunoglobulin that are directly involved in
antigen contact. SDRU is a precise measure of a number and
distribution of SDR for different types of antigens as
defined by analyses of crystal structures of antigen-antibody
complexes. (v) The antigen-binding site can also be defined
as the antibody paratope residues identified from crystal
structure of the antigen-antibody complex.
The term "composite sequences" as used herein means an
antigen-binding site defined to include all amino acid
residues delineated individually by Kabat, Chothia or IMGT,
or any other suitable antigen-binding site delineation.
"Chothia residues" as used herein are the antibody VL
and VH residues numbered according to Al-Lazikani (Al-
Lazikani et al., J. Mol. Biol. 273:927-48, 1997).
Correspondence between the two most used numbering systems,
Kabat (Kabat et al., Sequences of Immunological Interest, 5th
Ed. Public Health Service, NIH, Bethesda, MD, 1991) and
Chothia (Chothia and Lesk, Mol. Biol. 196:901-17, 1987) in
relation to sequential polypeptide numbering is shown in
Figure 31 for exemplary antibodies of the invention.
"Framework" or "framework sequences" are the remaining
sequences of a variable region other than those defined to be
antigen-binding site. The framework is typically divided
into four regions, FR1, FR2, FR3, and FR3, which form a
scaffold for the three antigen-binding sites in each variable
14
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
reigon. Because the antigen-binding site can be defined by
various terms as described above, the exact amino acid
sequence of a framework depends on how the antigen-binding
site was defined.
"A light chain variable region kappa 1 (VK1) framework"
or "VK1" as used herein refers to a framework having an amino
acid sequence encoded by any of the human VK1 functional
genes or alleles thereof. Exemplary functional human Vk1
genes are IGKV1-5*01, IGKV1-6*01, IGKV1-8*01, IGKV1-9*01,
IGKV1-12*01, IGKV1-13*02, IGKV1-16*01, IGKV1-17*01, IGKV1-
27*01, IGKV1-33*01, IGKV1-37*01, IGKV1-39*01, IGKV1D-8*01,
IGKV1D-12*01, IGKV1D-13*01, IGKV1D-16*01, IGKV1D-17*01,
IGKV1D-33*01, IGKV1D-37*01, IGKV1D-39*01, IGKV1D-42*01, or
IGKV1D-43*01. Nomenclature of the immunoglobulin genes is
well known.
"A light chain variable region lambda 3 (V23) framework"
or "V3" as used herein refers to a framework having an amino
acid sequence encoded by any of the human .\/.3 functional genes
or alleles thereof. Exemplary functional human .\/.3 genes are
IGLV3-1*01, IGLV3-9*01, IGLV3-10*01, IGLV3-12*01, IGLV3-
16*01, IGLV3-19*01, IGLV3-21*01, IGLV3-22*01, IGLV3-25*01,
IGLV3-27*01, and IGLV3-32*01.
"A heavy chain variable region Vh5 framework" or "Vh5"
as used herein refers to a framework having an amino acid
sequence encoded by any of the human Vh5 functional genes or
alleles thereof. Exemplary functional human Vh5 genes are
IGHV5-51*01 and IGHV5-1*01.
"A heavy chain variable region Vh6 framework" or "Vh6"
as used herein refers to a framework having an amino acid
sequence encoded by any of the human Vh6 functional genes or
alleles thereof. An exemplary functional human Vh6 gene is
IGHV6-1*01.
"A light chain kappa J-region (JK) framework" or "JK" as
used herein refers to a framework having an amino acid
sequence encoded by any of the human JK functional genes or
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
alleles thereof. Exemplary functional human Vi genes are
IGKJ1, IGKJ2, IGKJ3, IGKJ4, and IGKJ5.
"A light chain lambda J-region (A) framework" or "A" as
used herein refers to a framework having an amino acid
sequence encoded by any of the human A functional genes or
alleles thereof. Exemplary functional human A genes are
IGLJ1, IGLJ2, IGLJ3, IGLJ4, IGLJ5, IGLJ6, and IGLJ7.
"A heavy chain J-region (Jh) framework" or "Jh" as used
herein refers to a framework having an amino acid sequence
encoded by any of the human Jh functional genes or alleles
thereof. Exemplary functional human Jh genes are IGHJ1,
IGHJ2, IGHJ3, IGHJ4, IGHJ5, and IGHJ6.
"Germline genes" or "antibody germline genes" as used
herein are immunoglobulin sequences encoded by non-lymphoid
cells that have not undergone the maturation process that
leads to genetic rearrangement and mutation for expression of
a particular immunoglobulin.
"Scaffold" as used herein refers to amino acid sequences
of light or heavy chain variable regions encoded by human
germline genes. Thus, the scaffold encompasses both the
framework and the antigen-binding site.
The term "antigen" as used herein means any molecule
that has the ability to generate antibodies either directly
or indirectly. Included within the definition of "antigen"
is a protein-encoding nucleic acid.
The term "homolog" means protein sequences having
between 40% and 100% sequence identity to a reference
sequence. Homologs of human TLR3 include polypeptides from
other species that have between 40% and 100% sequence
identity to a known human TLR3 sequence. Percent identity
between two peptide chains can be determined by pairwise
alignment using the default settings of the AlignX module of
Vector NTI v.9Ø0 (Invitrogen, Carlsbad, CA). By "TLR3" is
meant human TLR3 (huTLR3) and its homologs. The nucleotide
and amino acid sequences of the full length huTLR3 are shown
in SEQ ID NOs: 1 and 2, respectively. The nucleotide and
16
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
amino acid sequences of the huTLR3 extracellular domain (ECD)
are shown in SEQ ID NOs: 3 and 4, respectively.
The term "substantially identical" as used herein means
that the two antibody or antibody fragment amino acid
sequences being compared are identical or have "insubstantial
differences". Insubstantial differences are substitutions of
1, 2, 3, 4, 5 or 6 amino acids in an antibody or antibody
fragment amino acid sequence. Amino acid sequences
substantially identical to the sequences disclosed herein are
also part of this application. In some embodiments, the
sequence identity can be about 90%, 91%, 92%, 93%, 94%, 95%,
96%, 97%, 98%, 99% or higher. Percent identity can be
determined as described above. Exemplary peptide chains
being compared are heavy or light chain variable regions.
The term "in combination with" as used herein means that
the described agents can be administered to an animal
together in a mixture, concurrently as single agents or
sequentially as single agents in any order.
The term "inflammatory condition" as used herein means a
localized response to cellular injury that is mediated in
part by the activity of cytokines, chemokines, or
inflammatory cells (e.g., neutrophils, monocytes,
lymphocytes, macrophages) which is characterized in most
instances by pain, redness, swelling, and loss of tissue
function. The term "inflammatory pulmonary condition" as
used herein means an inflammatory condition affecting or
associated with the lungs.
The term "monoclonal antibody" (mAb) as used herein
means an antibody (or antibody fragment) obtained from a
population of substantially homogeneous antibodies.
Monoclonal antibodies are highly specific, typically being
directed against a single antigenic determinant. The
modifier "monoclonal" indicates the substantially homogeneous
character of the antibody and does not require production of
the antibody by any particular method. For example, murine
mAbs can be made by the hybridoma method of Kohler et al.,
17
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Nature 256:495-497, 1975. Chimeric mAbs containing a light
chain and heavy chain variable region derived from a donor
antibody (typically murine) in association with light and
heavy chain constant regions derived from an acceptor
antibody (typically another mammalian species such as human)
can be prepared by the method disclosed in U.S. Pat. No.
4,816,567. Human-adapted mAbs having CDRs derived from a
non-human donor immunoglobulin (typically murine) and the
remaining immunoglobulin-derived parts of the molecule being
derived from one or more human immunoglobulins can be
prepared by techniques known to those skilled in the art such
as that disclosed in U.S. Pat. No. 5,225,539. Human
framework sequences useful for human-adaptation can be
selected from relevant databases by those skilled in the art.
Optionally, human-adapted mAbs can be further modified by
incorporating altered framework support residues to preserve
binding affinity by techniques such as those disclosed in
Queen et al., Proc. Natl. Acad. Sci. (USA), 86:10029-10032,
1989 and Hodgson et al., Bio/Technology, 9:421, 1991.
Fully human mAbs lacking any non-human sequences can be
prepared from human immunoglobulin transgenic mice by
techniques referenced in, e.g., Lonberg et al., Nature
368:856-859, 1994; Fishwild et al., Nature Biotechnology
14:845-851, 1996; and Mendez et al., Nature Genetics 15:146-
156, 1997. Human mAbs can also be prepared and optimized
from phage display libraries by techniques referenced in,
e.g., Knappik et al., J. Mol. Biol. 296:57-86, 2000; and
Krebs et al., J. Immunol. Meth. 254:67-84 2001. Fragments of
antibodies e.g., Fab, F(ab')2, Fd, and dAb fragments may be
produced by cleavage of the antibodies or by recombinant
engineering. For example, Fab and F(ab')2 fragments may be
generated by treating the antibodies with an enzyme such as
pepsin.
The term "epitope" as used herein means a portion of an
antigen to which an antibody specifically binds. Epitopes
usually consist of chemically active (such as polar, non-
18
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
polar or hydrophobic) surface groupings of moieties such as
amino acids or polysaccharide side chains and can have
specific three-dimensional structural characteristics, as
well as specific charge characteristics. An epitope can be
linear in nature or can be a discontinous epitope, e.g., a
conformational epitope, which is formed by a spatial
relationship between non-contiguous amino acids of an antigen
rather than a linear series of amino acids. A conformational
epitope includes epitopes resulting from folding of an
antigen, where amino acids from differing portions of the
linear sequence of the antigen come in close proximity in 3-
dimensional space.
The term "paratope" as used herein refers to a portion
of an antibody to which an antigen specifically binds. A
paratope can be linear in nature or can be discontinuous,
formed by a spatial relationship between non-contiguous amino
acids of an antibody rather than a linear series of amino
acids. A "light chain paratope" and a "heavy chain paratope"
or "light chain paratope amino acid residues" and "heavy
chain paratope amino acid residues" refer to antibody light
chain and heavy chain residues in contact with an antigen,
respectively.
The term "specific binding" as used herein refers to
antibody binding to a predetermined antigen with greater
affinity than for other antigens or proteins. Typically, the
antibody binds with a dissociation constant (KD) of 10-7 M or
less, and binds to the predetermined antigen with a KD that is
at least twofold less than its KD for binding to a non-
specific antigen (e.g., BSA, casein, or any other specified
polypeptide) other than the predetermined antigen. The
phrases "an antibody recognizing an antigen" and "an antibody
specific for an antigen" are used interchangeably herein with
the term "an antibody which binds specifically to an antigen"
or "an antigen specific antibody" e.g. a TLR3 specific
antibody. The dissociation constant can be measured using
standard procedures as described below.
19
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
The term "TLR3 biological activity" or "TLR3 activation"
as used herein refers to any activity occurring as a result
of ligand binding to TLR3. TLR3 ligands include dsRNA,
poly(I:C), and endogenous mRNA, e.g., engodenous mRNA
released from necrotic cells. An exemplary TLR3 activation
results in activation of NF-KB in response to the TLR3
ligand. NF-KB activation can be assayed using a reporter-
gene assay upon induction of the receptor with poly(I:C)
(Alexopoulou et al., Nature 413:732-738, 2001; Hacker et al.,
EMBO J. 18:6973-6982, 1999). Another exemplary TLR3
activation results in activation of interferon response
factors (IRE-3, IRE-7) in response to the TLR3 ligand. TLR3-
mediated IRE activation can be assayed using a reporter gene
driven by an interferon-stimulated response element (ISRE).
Another exemplary TLR3 activation results in secretion of
pro-inflammatory cytokines and chemokines, for example TNF-a,
IL-6, IL-8, IL-12, CXCL5/IP-10 and RANTES. The release of
cytokines and chemokines from cells, tissues or in
circulation can be measured using well-known immunoassays,
such as an ELISA immunoassay.
Conventional one and three-letter amino acid codes are
used herein as follows:
Amino acid Three-letter code One-letter code
Alanine ala A
Arginine arg
Asparagine asn
Aspartate asp
Cysteine cys
Glutamate glu
Glutamine gln
Glycine gly
Histidine his
Isoleucine ile
Leucine leu
Lysine lys
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Methionine met
Phenylalanine phe
Proline pro
Serine ser
Threonine thr
Tryptophan trp
Tyrosine tyr
Valine val V
Compositions of matter
The present invention provides antibody antagonists
capable of inhibiting TLR3 biological activity and uses of
such antibodies. Such TLR3 antagonists may have the
properties of binding TLR3 and inhibiting TLR3 activation.
Exemplary mechanisms by which TLR3 activation may be
inhibited by such antibodies include in vitro, in vivo or in
situ inhibition of ligand binding to TLR3, inhibition of
receptor dimerization, inhibition of TLR3 localization to the
endosomal compartment, inhibition of kinase activity of
downstream signaling pathways, or inhibition of TLR3 mRNA
transcription. Other antibody antagonists capable of
inhibiting TLR3 activation by other mechanisms are also
within the scope of the various aspects and embodiments of
the invention. These antagonists are useful as research
reagents, diagnostic reagents and therapeutic agents.
Antibody diversity, in a natural system, is created by
the use of multiple germline genes encoding variable regions
and a variety of somatic events. The somatic events include
recombination of variable gene segments with diversity (D)
and joining (J) gene segments to make a complete VH region,
and the recombination of variable and joining gene segments
to make a complete VL region. The recombination process
itself can be imprecise, resulting in the loss or addition of
amino acids at the V(D)J junctions. These mechanisms of
diversity occur in the developing B cell prior to antigen
exposure. After antigenic stimulation, the expressed
21
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
antibody genes in B cells undergo somatic mutation. Based on
the estimated number of germline gene segments, the random
recombination of these segments, and random VH-VL pairing, up
to 1.6x10different antibodies could be produced (Fundamental
Immunology, 3rd ed. (1993), ed. Paul, Raven Press, New York,
N.Y.). When other processes that contribute to antibody
diversity (such as somatic mutation) are taken into account,
it is thought that upwards of 1010 different antibodies could
be generated (Immunoglobulin Genes, 2nd ed. (1995), eds.
Jonio et al., Academic Press, San Diego, Calif.). Because of
the many processes involved in generating antibody diversity,
it is highly unlikely that independently derived monoclonal
antibodies with the same antigen specificity will have
identical amino acid sequences.
The invention provides novel antigen-binding sites and
immunoglobulin chains derived from human immunoglobulin gene
libraries. The structure for carrying an antigen-binding
site is generally an antibody heavy or light chain or portion
thereof, where the antigen-binding site is located to a
naturally occurring antigen-binding site as determined as
described above.
The invention provides an isolated antibody or fragment
thereof reactive with TLR3 comprising both a heavy chain and
a light chain variable region and wherein the antibody
comprises the heavy chain complementarity determining region
(CDR) amino acid sequences 1, 2 and 3 (HCDR1, HCDR2 and
HCDR3) and the light chain complementarity determining region
(CDR) amino acid sequences 1, 2 and 3 (LCDR1, LCDR2 and
LCDR3) as shown in Table la.
Table la.
22
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
SEQ ID NO:
mAb no:
HCDR1 HCDR2 HCDR3 LCDR1 LCDR2 LCDR3
16 52 88 54 49 50 51
17 58 64 60 55 56 57
18 70 77 72 67 68 69
19 82 83 84 79 80 89
1 46 47 48 43 44 45
2 52 53 54 49 50 51
3 58 59 60 55 56 57
4 61 62 60 55 56 57
5 61 64 60 55 56 63
6 61 64 60 55 56 65
7 61 64 60 55 56 66
8 70 71 72 67 68 69
9 70 73 72 67 68 69
10 70 75 72 67 68 74
11 70 77 72 67 68 76
12 70 77 72 67 68 78
13 82 83 84 79 80 81
14 82 86 84 79 80 85
15* 82 86 84 79 80 87
15** 111 112 84 109 110 113
15-1 111 114 84 109 110 113
15-2 115 112 84 109 110 113
15-3 116 112 84 109 110 113
15-4 111 117 84 109 110 113
15-5 116 118 84 109 110 113
15-6 116 112 119 109 110 113
15-7 111 112 84 120 110 113
15-8 111 112 84 121 110 113
15-9 116 118 119 109 110 113
15-10 116 112 119 79 80 226
F17 61 192 60 55 56 191
F18 70 194 72 67 68 193
F19 82 196 84 79 80 195
15* CDRs defined by IMGT
15** CDRs defined as consensus
In certain embodiments the invention provides an
isolated antibody or fragment reactive with TLR3 comprising
both a heavy chain variable region and a light chain variable
region and wherein the antibody comprises a HCDR2 amino acid
sequence as shown in SEQ ID NO: 192, wherein the HCDR2 of SEQ
ID NO: 192 is defined as shown in Formula (I):
Xaa6-I-Xaa7-Xaa8-R-S-Xaa9-W-Y-N-D-Y-A-V-S-V-K-S,
(I)
wherein
23
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Xaa6 may be Arg or Lys;
Xaa7 may be Tyr, His or Ser;
Xaa8 may be Met, Arg or Tyr; and
Xaa8 may be Lys or Arg.
In other embodiments, the invention provides an isolated
antibody or fragment reactive with TLR3 comprising both a
heavy chain variable region and a light chain variable region
and wherein the antibody comprises a HCDR2 amino acid
sequence as shown in SEQ ID NO: 194, wherein the HCDR2 of SEQ
ID NO: 194 is defined as shown in Formula (III):
I-I-Q -Xaa15-R-S-K-W-Y-N-Xaa16-Y-A-Xaa17-S-V-K-S,
(III)
wherein
Xaam may be Lys, Thr or Ile;
Xaam may be Asn or Asp; and
Xaa87 may be Val or Leu.
In other embodiments, the invention provides an isolated
antibody or fragment reactive with TLR3 comprising both a
heavy chain variable region and a light chain variable region
and wherein the antibody comprises a HCDR2 amino acid
sequence as shown in SEQ ID NO: 196, wherein the HCDR2 of SEQ
ID NO: 196 is defined as shown in Formula (V):
Xaa24-I-D-P-S-D-S-Y-T-N-Y-Xaa25-P-S-F-Q-G,
(V)
wherein
Xaa2.4 may be Phe or Arg; and
Xaa25 may be Ala or Ser.
In other embodiments, the invention provides an isolated
antibody or fragment reactive with TLR3 comprising both a
heavy chain variable region and a light chain variable region
and wherein the antibody comprises a LCDR3 amino acid
sequence as shown in SEQ ID NO: 191, wherein the LCDR3 of SEQ
ID NO: 191 is defined as shown in Formula (II):
24
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Xaal-S-Y-D¨Xaa2-Xaa3-Xaa4-Xaa5-T-V,
(II)
wherein
Xaal may be Ala, Gln, Gly or Ser;
Xaa2 may be Gly, Glu or Ser;
Xaa3 may be Asp or Asn;
Xaa4 may be Glu or Ser; and
Xaa5may be Phe, Ala or Leu.
In other embodiments, the invention provides an isolated
antibody or fragment reactive with TLR3 comprising both a
heavy chain variable region and a light chain variable region
and wherein the antibody comprises a LCDR3 amino acid
sequence as shown in SEQ ID NO: 193, wherein the LCDR3 of SEQ
ID NO: 193 is defined as shown in Formula (IV):
Xaa10-S-Y-D¨Xaau-P-Xaa12-Xaa13-Xaa,34-V,
(IV)
wherein
Xaa10 may be Gln or Ser;
Xaau may be Thr, Glu or Asp;
Xaa12 may be Val or Asn;
Xaa13 may be Tyr or Phe; and
Xaa134 may be Ser, Asn or Gln.
In other embodiments, the invention provides an isolated
antibody or fragment reactive with TLR3 comprising both a
heavy chain variable region and a light chain variable region
and wherein the antibody comprises a LCDR3 amino acid
sequence as shown in SEQ ID NO: 195, wherein the LCDR3 of SEQ
ID NO: 195 is defined as shown in Formula (VI):
Q-Q-Xaa18¨Xaa19-Xaa20-Xaa21-Xaa22-Xaa23-T,
(VI)
wherein
XaalE, may be Tyr, Gly or Ala;
Xaa19 may be Gly, Glu or Asn;
Xaa20 may be Ser or Thr;
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Xaa21 may be Val, Ile or Leu;
Xaa22 may be Ser or Leu; and
Xaa23 may be Ile, Ser, Pro or Tyr.
The invention also provides an isolated antibody or
fragment reactive with TLR3 having the heavy chain
complementarity determining region (CDR) amino acid sequences
1,2 and 3 (HCDR1, HCDR2 and HCDR3) and light chain
complementarity determining region (CDR) amino acid sequences
1, 2 and 3 (LCDR1, LCDR2 and LCDR3) as shown in Table la.
Antibodies whose antigen-binding site amino acid
sequences differ insubstantially from those shown in Table la
(SEQ ID NOs: 49-121 and 191-196) are encompassed within the
scope of the invention. Typically, this involves one or more
amino acid substitutions with an amino acid having similar
charge, hydrophobic, or stereochemical characteristics.
Additional substitutions in the framework regions, in
contrast to antigen- binding sites may also be made as long
as they do not adversely affect the properties of the
antibody. Substitutions may be made to improve antibody
properties, for example stability or affinity. One, two,
three, four, five or six substitutions can be made to the
antigen binding site. 1%, 2%, 3%, 4%, 5%, 10%, 15%, 20%,
25%, or 30% of the framework residues can be substituted, as
long as the resulting antibody retains desired properties.
Conservative modifications will produce molecules having
functional and chemical characteristics similar to those of
the molecule from which such modifications are made.
Substantial modifications in the functional and/or chemical
characteristics of the molecules may be accomplished by
selecting substitutions in the amino acid sequence that
differ significantly in their effect on maintaining (1) the
structure of the molecular backbone in the area of the
substitution, for example, as a sheet or helical
conformation, (2) the charge or hydrophobicity of the
molecule at the target site, or (3) the size of the molecule.
For example, a "conservative amino acid substitution" may
26
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
involve a substitution of a native amino acid residue with a
nonnative residue such that there is little or no effect on
the polarity or charge of the amino acid residue at that
position. Furthermore, any native residue in the polypeptide
may also be substituted with alanine, as has been previously
described for alanine scanning mutagenesis (MacLennan et al.,
Acta Physiol. Scand. Suppl. 643:55-67, 1998; Sasaki et al.,
Adv. Biophys. 35:1-24, 1998). Desired amino acid
substitutions (whether conservative or non-conservative) can
be determined by those skilled in the art at the time such
substitutions are desired. For example, amino acid
substitutions can be used to identify important residues of
the molecule sequence, or to increase or decrease the
affinity of the molecules described herein. Exemplary amino
acid substitutions are shown in Table lb.
In certain embodiments, conservative amino acid
substitutions also encompass non-naturally occurring amino
acid residues which are typically incorporated by chemical
peptide synthesis rather than by synthesis in biological
systems. Amino acid substitutions can be done for example by
PCR mutagenesis (US Pat. No. 4,683,195). Libraries of
variants can be generated using well known methods, for
example using random (NNK) or non-random codons, for example
DVK codons, which encode 11 amino acids (ACDEGKNRSYW), and
screening the libararies for variants with desired
properties, as shown in Example 1. Table lc shows
substitutions made to three parent TLR3 antibody antagonists
within the LCDR3 and HCDR2 regions to improve antibody
properties.
Depending on delineation of the antigen-binding sites,
the antigen-binding site residues of the antibodies of the
invention and subsequently the framework residues may vary
slightly for each heavy and light chain.
Table lb.
27
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
More
Original
Exemplary substitutions Conservative
residue
substitutions
Ala (A) Val, Leu, Ile Val
Arg (R) Lys, Gin, Asn Lys
Asn (N) Gin Gin
Asp (D) Glu Glu
Cys (C) Ser, Ala Ser
Gin (Q) Asn Asn
Gly (G) Pro, Ala Ala
His (H) Asn, Gin, Lys, Arg Arg
Ile (I) Leu, Val, Met, Ala, Phe, Norleucine Leu
Leu (L) Norleucine, Ile, Val, Met, Ala, Phe Ile
Lys (K) Arg, 1, 4 Diamino-butyric Acid, Gin, Asn Arg
Met (M) Leu, Phe, Ile Leu
Phe (F) Leu, Val, Ile, Ala, Tyr Leu
Pro (P) Ala Gly
Ser (S) Thr, Ala, Cys Thr
Thr (T) Ser Ser
Trp (W) Tyr, Phe Tyr
Tyr (Y) Trp, Phe, Thr, Ser Phe
Val (V) Ile, Met, Leu, Phe, Ala, Norleucine Leu
Table 2a and 2b shows the antigen-binding site residues
of exemplary antibodies of the invention delineated according
to Kabat, Chothia and IMGT, and their composite sequences.
In other embodiments, the invention provides an isolated
antibody or fragment reactive with TLR3 comprising both a
heavy chain variable region and a light chain variable region
and wherein the antibody comprises the amino acid sequences
of the heavy chain variable (VH) and the light chain variable
(VL) regions and also provides for each isolated heavy chain
variable and light chain variable region as shown in Table
3a. F17, F18 and F19 represent antibody variants comprising
consensus amino acid sequences for families 17, 18 and 19,
respectively (see Example 1).
Table lc.
28
CA 02855955 2014-03-11
WO 2013/039471 PCT/US2011/051202
Family 17 SEQ ID
LCDR3
mAb NO:
17 A SY D G D E F T V
3
4
Q E S A
6
7
consensus A,Q,G,S S Y D G,E,S D,N E,S F,A,L T V 191
Family 17 SEQ ID
HCDR2
mAb NO:
17 R I Y M R S K W YN D Y A V S V K S
3
4
5
6
7
consensus R,K I Y,H,S M,R,Y R S K,R W YN D Y A
V S V K S 192
Family 18A LCDR3 SEQ ID ----------
mAb NO:
18 QSYD SQF S F G V
8
9
Family 18B
mAb
QSYD T P V Y S V
11
12 5 0 N F Q
consensus Q,S S Y D T,E,D P V,N Y,F S,N,Q V 193
*consensus based on mAbs 10, 11, 12
Family 18A, 18B HCDR2 SEQ ID
mAb NO:
18 I IQK R SK W YNN Y A V S V K S
8
9
11
12
consensus I IQK,T,IR SK W YNN,D Y
AV,LSVKS 194
5
29
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Family 19 SEQ ID
LCDR2
mAb NO:
19 Q Q Y G S V S
13 G ESIL S
14 A E T
15 G N T L
15-1 G N T L
15-2 G N T L
15-3 G N T L
15-4 G N T L
15-5 G N T L
15-6 G N T L
15-7 G N T L
15-8 G N T L
15-9 G N T L
15-10 G N T L
consensus Q Q Y,G,A G,E,N S,T V,I,L S,L I,S,P,Y T 195
Family 19 SEQ ID
HCDR2
mAb NO:
19 F I D P SDS Y TNYAPSFQG
13
14
15.1
15.2
15.3
15.4
15.5
15.6
157
15-8
15-9
15-10
consensus F,R I D P SDS Y TNYA,SPSFQG 196
Although the embodiments illustrated in the Examples
5 comprise pairs of variable regions, one from a heavy and one
from a light chain, a skilled artisan will recognize that
alternative embodiments may comprise single heavy or light
chain variable regions. The single variable region can be
used to screen for a second variable region capable of
10 forming a two-domain specific antigen-binding fragment
capable of, for example, binding to TLR3. The screening may
be accomplished by phage display screening methods using for
example hierarchical dual combinatorial approach disclosed in
PCT Publ. No. W092/01047. In this approach, an individual
15 colony containing either a H or L chain clone is used to
infect a complete library of clones encoding the other chain
(L or H), and the resulting two-chain specific antigen-
binding domain is selected in accordance with phage display
techniques as described.
CA 02855955 2014-03-11
WO 2013/039471 PCT/US2011/051202
Table 2a.
HCDR1 HCDR2 HCDR3
mAb CDR definition
SEQID Sequence SEQ ID Sequence SEQ ID
Sequence
14 IMGT 82 GYSFTNYW 86 IDPSDSYTNY 84 ARELYQGYMDTFDS
14 Kabat NYWVG FIDPSDSYTNYAPSFQ ELYQGYMDTFDS
14 Chothia GYSFT PSDSYT LYQGYMDTFD
14 Consensus 111 GYSFTNYWVG 112 FIDPSDSYTNYAPSFQ 84
ARELYQGYMDTFDS
15 IMGT 82 GYSFTNYW 86 IDPSDSYTNY 84 ARELYQGYMDTFDS
15 Kabat NYWVG FIDPSDSYTNYAPSFQ ELYQGYMDTFDS
15 Chothia GYSFT PSDSYT LYQGYMDTFD
15 Consensus 111 GYSFTNYWVG 112 FIDPSDSYTNYAPSFQ 84
ARELYQGYMDTFDS
15-1 IMGT 82 GYSFTNYW 86 IDPSDSYTNY 84 ARELYQGYMDTFDS
15-1 Kabat NYWVG RIDPSDSYTNYAPSFQ ELYQGYMDTFDS
15-1 Chothia GYSFT PSDSYT LYQGYMDTFD
15-1 Consensus 111 GYSFTNYWVG 114 RIDPSDSYTNYAPSFQ 84
ARELYQGYMDTFDS
15-2 IMGT 82 GYSFTNYW 86 IDPSDSYTNY 84 ARELYQGYMDTFDS
15-2 Kabat NYWIG FIDPSDSYTNYAPSFQ ELYQGYMDTFDS
15-2 Chothia GYSFT PSDSYT LYQGYMDTFD
15-2 Consensus 115 GYSFTNYWIG 112 FIDPSDSYTNYAPSFQ
84 ARELYQGYMDTFDS
15-3 IMGT 82 GYSFTNYW 86 IDPSDSYTNY 84 ARELYQGYMDTFDS
15-3 Kabat NYWIS 86 FIDPSDSYTNYAPSFQ 84 ELYQGYMDTFDS
15-3 Chothia GYSFT PSDSYT LYQGYMDTFD
15-3 Consensus 116 GYSFTNYWIS 112 FIDPSDSYTNYAPSFQ
84 ARELYQGYMDTFDS
15-4 IMGT 82 GYSFTNYW 86 IDPSDSYTNY 84 ARELYQGYMDTFDS
15-4 Kabat NYWVG FIDPSDSYTNYSPSFQ ELYQGYMDTFDS
15-4 Chothia GYSFT PSDSYT LYQGYMDTFD
15-4 Consensus 111 GYSFTNYWVG 117 FIDPSDSYTNYSPSFQ 84
ARELYQGYMDTFDS
15-5 IMGT 82 GYSFTNYW 86 IDPSDSYTNY 84 ARELYQGYMDTFDS
15-5 Kabat NYWIS RIDPSDSYTNYSPSFQ ELYQGYMDTFDS
15-5 Chothia GYSFT PSDSYT LYQGYMDTFD
15-5 Consensus 116 GYSFTNYWIS 118 RIDPSDSYTNYSPSFQ
84 ARELYQGYMDTFDS
15-6 IMGT 82 GYSFTNYW 86 IDPSDSYTNY ARQLYQGYMDTFDS
15-6 Kabat NYWIS FIDPSDSYTNYAPSFQ QLYQGYMDTFDS
15-6 Chothia GYSFT PSDSYT LYQGYMDTFD
15-6 Consensus 116 GYSFTNYW IS 112 FIDPSDSYTNYAPSFQ
119 ARQLYQGYMDTFDS
15-7 IMGT 82 GYSFTNYW 86 IDPSDSYTNY 84 ARELYQGYMDTFDS
15-7 Kabat NYWVG FIDPSDSYTNYAPSFQ ELYQGYMDTFDS
15-7 Chothia GYSFT PSDSYT LYQGYMDTFD
15-7 Consensus 111 GYSFTNYWVG 112 FIDPSDSYTNYAPSFQ 84
ARELYQGYMDTFDS
15-8 IMGT 82 GYSFTNYW 86 IDPSDSYTNY 84 ARELYQGYMDTFDS
15-8 Kabat NYWVG FIDPSDSYTNYAPSFQ ELYQGYMDTFDS
15-8 Chothia GYSFT PSDSYT LYQGYMDTFD
15-8 Consensus 111 GYSFTNYWVG 112 FIDPSDSYTNYAPSFQ 84
ARELYQGYMDTFDS
15-9 IMGT 82 GYSFTNYW 86 IDPSDSYTNY 119 ARQLYQGYMDTFDS
15-9 Kabat NYWIS RIDPSDSYTNYSPSFQG QLYQGYMDTFDS
15-9 Chothia GYSFT PSDSYT LYQGYMDTFD
15-9 Consensus 116 GYSFTNYWIS 118
RIDPSDSYTNYSPSFQG 119 ARQLYQGYMDTFDS
In other embodiments, the invention provides an isolated
antibody or fragment reactive with TLR3 comprising both a
heavy chain variable region and a light chain variable region
having amino acid sequences at least 95% identical to the
variable region amino acid sequences shown in Table 3a.
In another aspect, the invention provides an isolated
antibody having certain heavy chain and light chain amino
acid sequences as shown in Table 3b.
31
CA 02855955 2014-03-11
WO 2013/039471 PCT/US2011/051202
Another aspect of the invention is isolated
polynucleotides encoding any of the antibodies of the
invention or their complement. Certain exemplary
polynucleotides are disclosed herein, however, other
polynucleotides which, given the degeneracy of the genetic
code or codon preferences in a given expression system,
encode the antibody antagonists of the invention are also
within the scope of the invention.
Table 2b.
LCDR1 LCDR2 LCDR3
mAb CDR definition SEQ ID SEQ ID SEQ ID
Sequence Sequence Sequence
NO: NO: NO:
14 !MGT 79 QSIGLY 80 AAS 85 QQAETVSPT
14 Kabat RASQSIGLYLA AASSLQS QQAETVSPT
14 Chothia SQSIGLY AAS AETVSP
14 Consensus 109 RASQSIGLYLA 110 AASSLQS 85 QQAETVSPT
!MGT 79 QSIGLY 80 AAS 87 QQGNTLSYT
15 Kabat RASQSIGLYLA AASSLQS QQGNTLSYT
15 Chothia SQSIGLY AAS GNTLSY
15 Consensus 109 RASQSIGLYLA 110 AASSLQS 113 QQGNTLSYT
15-1 !MGT 79 QSIGLY 80 AAS 87 QQGNTLSYT
15-1 Kabat RASQSIGLYLA AASSLQS QQGNTLSYT
15-1 Chothia SQSIGLY AAS GNTLSY
15-1 Consensus 109 RASQSIGLYLA 110 AASSLQS 113 QQGNTLSYT
15-2 !MGT 79 QSIGLY 80 AAS 87 QQGNTLSYT
15-2 Kabat RASQSIGLYLA AASSLQS QQGNTLSYT
15-2 Chothia SQSIGLY AAS GNTLSY
15-2 Consensus 109 RASQSIGLYLA 110 AASSLQS 113 QQGNTLSYT
15-3 !MGT 79 QSIGLY 80 AAS 87 QQGNTLSYT
15-3 Kabat RASQSIGLYLA AASSLQS QQGNTLSYT
15-3 Chothia SQSIGLY AAS GNTLSY
15-3 Consensus 109 RASQSIGLYLA 110 AASSLQS 113 QQGNTLSYT
15-4 !MGT 79 QSIGLY 80 AAS 87 QQGNTLSYT
15-4 Kabat RASQSIGLYLA AASSLQS QQGNTLSYT
15-4 Chothia SQSIGLY AAS GNTLSY
15-4 Consensus 109 RASQSIGLYLA 110 AASSLQS 113 QQGNTLSYT
15-5 !MGT 79 QSIGLY 80 AAS 87 QQGNTLSYT
15-5 Kabat RASQSIGLYLA AASSLQS QQGNTLSYT
15-5 Chothia SQSIGLY AAS GNTLSY
15-5 Consensus 109 RASQSIGLYLA 110 AASSLQS 113 QQGNTLSYT
15-6 !MGT 79 QSIGLY 80 AAS 87 QQGNTLSYT
15-6 Kabat RASQSIGLYLA AASSLQS QQGNTLSYT
15-6 Chothia SQSIGLY AAS GNTLSY
15-6 Consensus 109 RASQSIGLYLA 110 AASSLQS 113 QQGNTLSYT
15-7 !MGT QSISSY 80 AAS 87 QQGNTLSYT
15-7 Kabat RASQSISSYLA AASSLQS QQGNTLSYT
15-7 Chothia SQSISSY AAS GNTLSY
15-7 Consensus 120 RASQSISSYLA 110 AASSLQS 113 QQGNTLSYT
15-8 !MGT 79 QSIGLY 80 AAS 87 QQGNTLSYT
15-8 Kabat RASQSIGLYLN AASSLQS QQGNTLSYT
15-8 Chothia SQSIGLY AAS GNTLSY
15-8 Consensus 121 RASQSIGLYLN 110 AASSLQS 113 QQGNTLSYT
15-9 !MGT 79 QSIGLY 80 AAS 87 QQGNTLSYT
15-9 Kabat RASQSIGLYLA AASSLQS QQGNTLSYT
15-9 Chothia SQSIGLY AAS GNTLSY
15-9 Consensus 109 RASQSIGLYLA 110 AASSLQS 113 QQGNTLSYT
32
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Table 3a.
SEQ ID NO: SEQ ID NO:
mAb no: mAb no:
HV LV HV LV
16 6 5 15-1 124 41
17 8 7 15-2 125 41
18 10 9 15-3 126 41
19 12 11 154 127 41
1 14 13 15-5 128 41
2 16 15 15-6 129 41
3 18 17 15-7 42 122
4 20 19 15-8 42 123
22 21 15-9 159 41
6 24 23 15-10 129 225
7 26 25 F17 198 197
8 28 27 F18 200 199
9 30 29 F19 202 201
32 31 c1811 164 163
11 34 33 9QVQ/QSV 212 209
12 36 35 10QVQ/QSV 213 210
13 38 37 12QVQ/QSV 214 211
14 40 39 14EVQ 215 39
42 41 15EVQ 216 41
5 Exemplary antibody antagonists may be antibodies of the
IgG, IgD, IgG, IgA or IgM isotypes. Additionally, such
antibody antagonists can be post-translationally modified by
processes such as glycosylation, isomerization,
deglycosylation or non-naturally occurring covalent
10 modification such as the addition of polyethylene glycol
(PEG) moieties (pegylation) and lipidation. Such
modifications may occur in vivo or in vitro. For example,
the antibodies of the invention can be conjugated to
polyethylene glycol (PEGylated) to improve their
15 pharmacokinetic profiles. Conjugation can be carried out by
techniques known to those skilled in the art. Conjugation of
therapeutic antibodies with PEG has been shown to enhance
pharmacodynamics while not interfering with function.
(Deckert et al., Int. J. Cancer 87:382-390, 2000; Knight et
al., Platelets 15:409-418, 2004; Leong et al., Cytokine
33
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
16:106-119, 2001; Yang et al., Protein Eng. 16:761-770,
2003).
Table 3b.
Heavy chain Light chain
mAbno:
SEQ ID NO: SEQ ID NO:
14 102 155
102 156
15-1 130 156
15-2 131 156
15-3 132 156
15-4 133 156
15-5 134 156
15-6 135 156
15-7 102 157
15-8 102 158
15-9 160 156
15-10 135 227
F17 204 203
F18 206 205
F19 208 207
14EVQ 220 155
15EVQ 220 156
5429 166 165
c1811 168 167
Pharmacokinetic properties of the antibodies of the
invention could also be enhanced through Fc modifications by
10 techniques known to those skilled in the art. For example,
IgG4 isotype heavy chains contain a Cys-Pro-Ser-Cys (CPSC)
motif in the hinge region capable of forming either inter- or
intra-heavy chain disulfide bonds, i.e., the two Cys residues
in the CPSC motif may disulfide bond with the corresponding
15 Cys residues in the other heavy chain (inter) or the two Cys
residues within a given CPSC motif may disulfide bond with
each other (intra). It is believed that in vivo isomerase
enzymes are capable of converting inter-heavy chain bonds of
34
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
IgG4 molecules to intra-heavy chain bonds and vice versa
(Aalberse and Schuurman, Immunology 105:9-19, 2002).
Accordingly, since the heavy:light chain (H:L) pairs in those
IgG4 molecules with intra-heavy chain bonds in the hinge
region are not covalently associated with each other, they
may dissociate into H:L monomers that then reassociate with
H:L monomers derived from other IgG4 molecules forming
bispecific, heterodimeric IgG4 molecules. In a bispecific
IgG antibody the two Fabs of the antibody molecule differ in
the epitopes that they bind. Substituting the Ser residue in
the hinge region CPSC motif of IgG4 with Pro results in
"IgG1-like behavior," /.e., the molecules form stable
disulfide bonds between heavy chains and therefore, are not
susceptible to H:L exchange with other IgG4 molecules. In
one embodiment, the antibodies of the invention will comprise
an IgG4 Fc domain with a S to P mutation in the CPSC motif.
The location of the CPSC motif is typically found at residue
228 of a mature heavy chain but can change depending on CDR
lengths.
Further, sites can be removed that affect binding to Fc
receptors other than an FcRn salvage receptor in the
antibodies of the invention. For example, the Fc receptor
binding regions involved in ADCC activity can be removed in
the antibodies of the invention. For example, mutation of
Leu234/Leu235 in the hinge region of IgG1 to L234A/L235A or
Phe235/Leu236 in the hinge region of IgG4 to P235A/L236A
minimizes FcR binding and reduces the ability of the
immunoglobulin to mediate complement dependent cytotoxicity
and ADCC. In one embodiment, the antibodies of the invention
will comprise an IgG4 Fc domain with P235A/L236A mutations.
The location of these residues identified above is typical in
a mature heavy chain but can change depending on CDR lengths.
Exemplary antibodies having P235A/L236A mutations are
antibodies having heavy chain amino acid sequences shown in
SEQ ID NOs: 218, 219 or 220.
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Fully human, human-adapted, humanized and affinity-
matured antibody molecules or antibody fragments are within
the scope of the invention as are fusion proteins and
chimeric proteins. Antibody affinity towards an antigen may
be improved by rational design or random affinity maturation
using well-known methods such as random or directed
mutagenesis, or employing phage display libraries. For
example, substitutions can be made to the Vernier Zone
residues that mostly reside in the framework region or to the
"Affinity Determining Residues", ADRs, to modulate affinity
of an antibody (US Pat. No. 6,639,055; PCT Publ. No.
W010/045340).
Fully human, human-adapted, humanized, affinity-matured
antibody molecules or antibody fragments modified to improve
stability, selectivity, cross-reactivity, affinity,
immunogenicity or other desirable biological or biophysical
property are within the scope of the invention. Stability of
an antibody is influenced by a number of factors, including
(1) core packing of individual domains that affects their
intrinsic stability, (2) protein/protein interface
interactions that have impact upon the HC and LC pairing, (3)
burial of polar and charged residues, (4) H-bonding network
for polar and charged residues; and (5) surface charge and
polar residue distribution among other intra- and inter-
molecular forces (Worn et al., J. Mol. Biol., 305:989-1010,
2001). Potential structure destabilizing residues may be
identified based upon the crystal structure of the antibody
or by molecular modeling in certain cases, and the effect of
the residues on antibody stability can be tested by
generating and evaluating variants harboring mutations in the
identified residues. One of the ways to increase antibody
stability is to raise the thermal transition midpoint (Tm) as
measured by differential scanning calorimetry (DSC). In
general, the protein Tm is correlated with its stability and
inversely correlated with its susceptibility to unfolding and
denaturation in solution and the degradation processes that
36
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
depend on the tendency of the protein to unfold (Remmele et
al., Biopharm., 13:36-46, 2000). A number of studies have
found correlation between the ranking of the physical
stability of formulations measured as thermal stability by
DSC and physical stability measured by other methods (Gupta
et al., AAPS PharmSci. 5E8, 2003; Zhang et al., J. Pharm.
Sci. 93:3076-3089, 2004; Maa et al., Int. J. Pharm., 140:155-
168, 1996; Bedu-Addo et al., Pharm. Res., 21:1353-1361, 2004;
Remmele et al., Pharm. Res., 15:200-208, 1997). Formulation
studies suggest that a Fab Tm has implication for long-term
physical stability of a corresponding mAb. Differences in
amino acids in either framework or within the antigen-binding
sites could have significant effects on the thermal stability
of the Fab domain (Yasui, et al., FEBS Lett. 353:143-146,
1994).
The antibody antagonists of the invention may bind TLR3
with a Kd less than or equal to about 10-7, 10-8, 10-9, 10-10,
10-11 or 10-12 M. The affinity of a given molecule for TLR3,
such as an antibody can be determined experimentally using
any suitable method. Such methods may utilize Biacore or
KinExA instrumentation, ELISA or competitive binding assays
known to those skilled in the art.
Antibody antagonists binding a given TLR3 homolog with a
desired affinity can be selected from libraries of variants
or fragments by techniques including antibody affinity
maturation. Antibody antagonists can be identified based on
their inhibition of TLR3 biological activity using any
suitable method. Such methods may utilize reporter-gene
assays or assays measuring cytokine production using well
known methods and as described in the application.
Another embodiment of the invention is a vector
comprising at least one polynucleotide of the invention.
Such vectors may be plasmid vectors, viral vectors, vectors
for baculovirus expression, transposon based vectors or any
other vector suitable for introduction of the polynucleotides
37
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
of the invention into a given organism or genetic background
by any means.
Another embodiment of the invention is a host cell
comprising any of the polynucleotides of the invention such
as a polynucleotide encoding a polypeptide comprising an
immunoglobulin heavy chain variable region having the amino
acid sequence shown in SEQ ID NO: 6, 8, 10, 12, 14, 16, 18,
20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 124, 125,
126, 127, 128, 129, 159, 198, 200, 202, 164, 212, 213, 214,
215 or 216 or an immunoglobulin light chain variable region
having the amino acid sequence shown in SEQ ID NO: 5, 7, 9,
11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39,
41, 122, 123, 197, 199, 201, 163, 209, 210, 211, or 225.
Another embodiment of the invention is a host cell
comprising a polynucleotide encoding a polypeptide comprising
an immunoglobulin heavy chain having the amino acid sequence
shown in SEQ ID NO: 102, 130, 131, 132, 133, 134, 135, 160,
204, 206, 208, 220, 166 or 168, or an immunoglobulin light
chain having the amino acid sequence shown in SEQ ID NO: 155,
156, 157, 158, 203, 205, 207, 165, 167, or 227. Such host
cells may be eukaryotic cells, bacterial cells, plant cells
or archeal cells. Exemplary eukaryotic cells may be of
mammalian, insect, avian or other animal origins. Mammalian
eukaryotic cells include immortalized cell lines such as
hybridomas or myeloma cell lines such as 5P2/0 (American Type
Culture Collection (ATCC), Manassas, VA, CRL-1581), NSO
(European Collection of Cell Cultures (ECACC), Salisbury,
Wiltshire, UK, ECACC No. 85110503), FO (ATCC CRL-1646) and
Ag653 (ATCC CRL-1580) murine cell lines. An exemplary human
myeloma cell line is U266 (ATTC CRL-TIB-196). Other useful
cell lines include those derived from Chinese Hamster Ovary
(CHO) cells such as CHO-K1SV (Lonza Biologics, Walkersville,
MD), CHO-K1 (ATCC CRL-61) or DG44.
Another embodiment of the invention is a method of
making an antibody reactive with TLR3 comprising culturing a
host cell of the invention and recovering the antibody
38
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
produced by the host cell. Methods of making antibodies and
purifying them are well known in the art.
Another embodiment of the invention is a hybridoma cell
line that produces an antibody of the invention.
Another embodiment of the invention is an isolated
antibody or fragment thereof, wherein the antibody binds
toll-like receptor 3 (TLR3) amino acid residues K416, K418,
L440, N441, E442, Y465, N466, K467, Y468, R488, R489, A491,
K493, N515, N516, N517, H539, N541, S571, L595, and K619 of
SEQ ID NO: 2.
Another embodiment is an isolated antibody or fragment
thereof, wherein the antibody binds toll-like receptor 3
(TLR3) amino acid residues S115, D116, K117, A120, K139,
N140, N141, V144, K145, T166, Q167, V168, S188, E189, D192,
A195, and A219 of SEQ ID NO: 2.
Several well known methodologies can be employed to
determine the binding epitope of the antibodies of the
invention. For example, when the structures of both
individual components are known, in silico protein-protein
docking can be carried out to identify compatible sites of
interaction. Hydrogen-deuterium (H/D) exchange can be
carried out with the antigen and antibody complex to map
regions on the antigen that may be bound by the antibody.
Segment and point mutagenesis of the antigen can be used to
locate amino acids important for antibody binding. For large
proteins such as TLR3, point mutagenesis mapping is
simplified when the binding site is first localized to a
region on the protein, such as by docking, segment
mutagenesis or H/D exchange. When the structures of both
individual components are known, in silico protein-protein
docking can be carried out to identify compatible sites of
interaction. Co-crystal
structure of antibody-antigen
complex can be used to identify residues contributing to the
epitope and paratope.
Another embodiment of the invention is an isolated
antibody or fragment thereof, wherein the antibody binds TLR3
39
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
having an amino acid sequence shown in SEQ ID NO: 2 with the
heavy chain variable region Chothia residues W33, F50, D52,
D54, Y56, N58, P61, E95, Y97, Y100, and D100b, and with the
light chain variable region Chothia residues Q27, Y32, N92,
T93, L94, and S95. The heavy chain paratope and the light
chain paratope Chothia residues correspond to heavy chain
residues W33, F50, D52, D55, Y57, N59, P62, E99, Y101, Y104,
and D106 of SEQ ID NO: 216 and light chain residues Q27, Y32,
N92, T93, L94, and S95 of SEQ ID NO: 41.
Another embodiment of the invention is an isolated
antibody or fragment thereof, wherein the antibody binds TLR3
having an amino acid sequence shown in SEQ ID NO: 2 with the
heavy chain variable region Chothia residues N31a, Q52, R52b,
S53, K54, Y56, Y97, P98, F99, and Y100, and with the light
chain variable region Chothia residues G29, S30, Y31, Y32,
E50, D51, Y91, D92, and D93. The heavy chain paratope and
the light chain paratope Chothia residues correspond to heavy
chain residues N32, Q54, R56, S57, K58, Y60, Y104, P105,
F106, and Y107 of SEQ ID NO: 214 and light chain residues
G28, S29, Y30, Y31, E49, D50, Y90, D91, and D92 of SEQ ID NO:
211.
Isolated antibodies having certain paratope residues
that bind TLR3 can be made by for example grafting the
paratope residues into a suitable scaffold, assembling the
engineered scaffolds into full antibodies, expressing the
resulting antibodies, and testing the antibodies for binding
to TLR3 or for an effect on TLR3 biological activity.
Exemplary scaffolds are amino acid sequences of human
antibody variable regions encoded by human germline genes.
The scaffolds can be selected based on for example overall
sequence homology, % identity between the paratope residues,
or canonical structure class identity between the scaffold
and an exemplary antibody, such as mAb 15EVQ or mAb
12QVQ/QSV. Human antibody germline genes are disclosed in,
for example, Tomlinson et al., J. Mol. Biol 227:776-798, and
at the International ImMunoGeneTics (IMGT) database
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
(http :// www imgt org). Consensus human framework regions
can also be used, e.g., as described in U.S. Pat. No.
6,300,064. Selection of suitable scaffold can be done for
example according to methods described in PCT Publ. No.
W010/045340.
Exemplary human germline genes that can be used as
scaffolds onto which the paratope residues are grafted are
the genes encoded by the VK1, V23, Vh5, Vh6 , JK, A, and the
Jh frameworks. The germline J-regions are used in their
entirety or in part to select FR4 sequences. For example,
the mAb 15EVQ light chain paratope residues can be grafted to
a VK1 framework encoded by IGKV1-39*01 that is joined
directly to the J region sequence encoded by IGKJ1.
Sequences from other VK1 genes can also be used, and the FR4
sequences of other JK genes can be substituted in place of
IGKJ1. The mAb 15EVQ heavy chain paratope residues can be
grafted to a Vh5 framework encoded by IGHV5-51*01, followed
by about 11-13 residues, for example 12 residues,
constituting HCDR3 and the FR4 sequence encoded by IGHJ1.
The 11-13 residues span between the end of the FR3 region
("CAR") and the start of the FR4 region (WGQ for most JH
regions) and include 4 defined paratope residues from mAb
15EVQ Vh. Sequences from other Vh5 genes can also be used,
and the FR4 sequences of other Jh genes can be substituted in
place of IGJH1. In another example, the mAb 12QVQ/QSV light
chain paratope residues can be grafted to a .\/.3 framework
encoded by IGLV3-1*01 that is joined directly to the J region
sequence encoded by IGJL2. Sequences of other .\/.3 and A
genes can also be used. The length of LCDR3 is maintained at
about 9-11 residues, for example 10 residues. These about 9-
11 residues span between the end of the FR3 region ("YYC" for
most V lambda scaffolds) and the start of the FR4 region
("FGG" for most JL regions) and include 3 defined paratope
residues from mAb 12QVQ/QSV. The mAb 12QVQ/QSV heavy chain
paratope residues can be grafted to a Vh6 framework encoded
41
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
by IGHV6-1*01, followed by about 9-11 residues, for example
residues, constituting HCDR3, and the FR4 sequence encoded
by IGJH1. The about 9-11 residues span between the end of
the FR3 region ("CAR") and the start of the FR4 region (WGQ
5 for most JH regions) and include 4 defined paratope residues
from mAb 12QVQ/QSV Vh. The FR4 sequences of other Jh genes
can be substituted in place of IGHJ1. The binding to TLR3
and biological activity of the resulting antibody can be
evaluated using standard methods. Alignments of the mAb
10 15EVQ and the mAb 12QVQ/QSV light chain variable regions and
heavy chain variable regions with the exemplary VK1, Vh5, V20,
Vh6, JK, A or Jh genes are shown in Figures 32 - 35. The
paratope-grafted engineered antibodies can further be
modified by substitutions of the Vernier Zone residues or the
Affinity Determining Residues to improve antibody properties,
for example affinity, as described above. As long as the
paratope-grafted antibody retains binding to TLR3, the
framework amino acid sequence in the paratope-grafted
antibody may be 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%,
or 99% identical to the the mAb 15EVQ or 12QVQ/QSV framework
sequences.
Sequences from the antigen-binding sites can be grafted
in addition to the paratope residues using standard methods.
For example, a complete HCDR3 or LCDR3 may be grafted.
Another aspect of the invention is an isolated antibody
or fragment thereof reactive with TLR3 that competes for TLR3
binding with a monoclonal antibody, wherein the monoclonal
antibody comprises the amino acid sequences of certain heavy
chain complementarity determining regions (CDRs) 1, 2 and 3,
the amino acid sequences of certain light chain CDRs 1, 2 and
3, the amino acid sequences of certain heavy chain variable
regions (VH) or the amino acid sequence of certain light
chain variable regions (VL). Examplary monoclonal antibodies
of the invention are an isolated antibody comprising a heavy
chain variable region having an amino acid sequence shown in
SEQ ID NO: 216 and a light chain variable region amino acid
42
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
sequence shown in SEQ ID NO: 41, and an antibody comprising a
heavy chain variable region having an amino acid sequence
shown in SEQ ID NO: 214 and a light chain variable region
amino acid sequence shown in SEQ ID NO: 211.
Competition between binding to TLR3 can be assayed in
vitro using well known methods. For example, binding of MSD
Sulfo-TagTm NHS-ester -labeled antibody to TLR3 in the
presence of an unlableled antibody can be assessed by ELISA.
Exemplary antibodies of the invention are mAb 12, mAb 15 and
mAb c1811 (see Table 3a). Previously described anti-TLR3
antibodies c1068 and its derivatives (described in PCT Publ.
No. W006/060513A2), TLR3.7 (eBiosciences, cat no 14-9039) and
Imgenex IMG-315A (Imgenex IMG-315A; generated against human
TLR3 amino acids amino acids 55-70, VLNLTHNQLRRLPAAN) do not
compete with binding to TLR3 with mAbs 12, 15 or c1811 as
shown in Example 5.
Another aspect of the invention is an isolated antibody
reactive with TLR3, wherein the antibody has at least one of
the following properties:
a. binds to human TLR3 with a Kd of <10 nM;
b. reduces human TLR3 biological activity in an in vitro
poly(I:C) NF-kB reporter gene assay >50% at 1 g/ml;
c. inhibits >60% of IL-6 or CXCL5/IP-10 production from
BEAS-2B cells stimulated with <100 ng/ml poly(I:C) at 10
g/ml;
d. inhibits >50% of IL-6 or CXCL5/IP-10 production from
BEAS-2B cells stimulated with <100 ng/ml poly(I:C) at
0.4 g/ml;
e. inhibits >50% of IL-6 production from NHBE cells
stimulated with 62.5 ng/ml poly(I:C) at 5 g/ml;
f. inhibits >50% of IL-6 production from NHBE cells
stimulated with 62.5 ng/ml poly(I:C)at 1 g/ml;
g. inhibits >20% of poly(I:C)-induced IFN-y, IL-6 or IL-12
production by PBMC cells at 1 g/ml.
43
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
h. inhibits cynomologus TLR3 biological activity in an in
vitro NF-kB reporter gene assay with IC50 <10 g/ml; or
i. inhibits cynomologus TLR3 biological activity in an in
vitro ISRE reporter gene assay with IC50 <5 g/ml.
Methods of Treatment
TLR3 antagonists of the invention, for example TLR3
antibody antagonists, can be used to modulate the immune
system. While not wishing to be bound by any particular
theory, the antagonists of the invention may modulate the
immune system by preventing or reducing ligand binding to
TLR3, dimerization of TLR3, TLR3 internalization or TLR3
trafficking. The methods of the invention may be used to
treat an animal patient belonging to any classification.
Examples of such animals include mammals such as humans,
rodents, dogs, cats and farm animals. For example, the
antibodies of the invention are useful in antagonizing TLR3
activity, in the treatment of inflammation, inflammatory and
metabolic diseases and are also useful in the preparation of
a medicament for such treatment wherein the medicament is
prepared for administration in dosages defined herein.
Generally, inflammatory conditions, infection-associated
conditions or immune-mediated inflammatory disorders that may
be prevented or treated by administration of the TLR3
antibody antagonists of the invention include those mediated
by cytokines or chemokines and those conditions which result
wholly or partially from activation of TLR3 or signaling
through the TLR3 pathway. Examples of such inflammatory
conditions include sepsis-associated conditions, inflammatory
bowel diseases, autoimmune disorders, inflammatory disorders
and infection-associated conditions. It is also thought that
cancers, cardiovascular and metabolic conditions, neurologic
and fibrotic conditions can be prevented or treated by
administration of the TLR3 antibody antagonists of the
invention. Inflammation may affect a tissue or be systemic.
Exemplary affected tissues are the respiratory tract, lung,
44
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
the gastrointestinal tract, small intestine, large intestine,
colon, rectum, the cardiovascular system, cardiac tissue,
blood vessels, joint, bone and synovial tissue, cartilage,
epithelium, endothelium, hepatic or adipose tissue.
Exemplary systemic inflammatory conditions are cytokine storm
or hypercytokinemia, systemic inflammatory response syndrome
(SIRS), graft versus host disease (GVHD), acute respiratory
distress syndrome (ARDS), severe acute respiratory distress
syndrome (SARS), catastrophic anti-phospholipid syndrome,
severe viral infections, influenza, pneumonia, shock, or
sepsis.
Inflammation is a protective response by an organism to
fend off an invading agent. Inflammation is a cascading
event that involves many cellular and humoral mediators. On
one hand, suppression of inflammatory responses can leave a
host immunocompromised; hovewer, if left unchecked,
inflammation can lead to serious complications including
chronic inflammatory diseases (e.g. asthma, psoriasis,
arthritis, rheumatoid arthritis, multiple sclerosis,
inflammatory bowel disease and the like), septic shock and
multiple organ failure. Importantly, these diverse disease
states share common inflammatory mediators, such as
cytokines, chemokines, inflammatory cells and other mediators
secreted by these cells.
TLR3 activation by its ligands poly(I:C), dsRNA or
endogenous mRNA leads to activation of signaling pathways
resulting in synthesis and secretion of pro-inflammatory
cytokines, activation and recruitment of inflammatory cells,
such as macrophages, granulocytes, neutrophils and
eosinophils, cell death, and tissue destruction. TLR3
induces secretion of IL-6, IL-8, IL-12, TNF-a, MIP-1,
CXCL5/IP-10 and RANTES, and other pro-inflammatory cytokines
and chemokines implicated in immune cell recruitment and
activation, thus contributing to tissue destruction in
autoimmune and other inflammatory diseases. TLR3 ligand
endogenous mRNA is released from necrotic cells during
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
inflammation, and may result in a positive feedback loop to
activate TLR3 and perpetuate inflammation and further tissue
damage. TLR3 antagonists, such as TLR3 antibody antagonists,
may normalize cytokine secretion, reduce recruitment of
inflammatory cells, and reduce tissue damage and cell death.
Therefore, TLR3 antagonists have therapeutic potential to
treat inflammation and a spectrum of inflammatory conditions.
One example of an inflammatory condition is sepsis-
associated condition that may include systemic inflammatory
response syndrome (SIRS), septic shock or multiple organ
dysfunction syndrome (MODS). dsRNA released by viral,
bacterial, fungal, or parasitic infection and by necrotic
cells can contribute to the onset of sepsis. While not
wishing to be bound by an particular theory, it is believed
that treatment with TLR3 antagonists can provide a
therapeutic benefit by extending survival times in patients
suffering from sepsis-associated inflammatory conditions or
prevent a local inflammatory event (e.g., in the lung) from
spreading to become a systemic condition, by potentiating
innate antimicrobial activity, by demonstrating synergistic
activity when combined with antimicrobial agents, by
minimizing the local inflammatory state contributing to the
pathology, or any combination of the foregoing. Such
intervention may be sufficient to permit additional treatment
(e.g., treatment of underlying infection or reduction of
cytokine levels) necessary to ensure patient survival.
Sepsis can be modeled in animals, such as mice, by the
adminstration of D-galactosamine and poly(I:C). In such
models, D-galactosamine is a hepatotoxin which functions as a
sepsis sensitizer and poly(I:C) is a sepsis-inducing molecule
that mimics dsRNA and activates TLR3. TLR3 antagonist
treatment may increase animal survival rates in a murine
model of sepsis, and thus TLR3 antagonists may be useful in
the treatment of sepsis.
Gastrointestinal inflammation is inflammation of a
mucosal layer of the gastrointestinal tract, and encompasses
46
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
acute and chronic inflammatory conditions. Acute
inflammation is generally characterized by a short time of
onset and infiltration or influx of neutrophils. Chronic
inflammation is generally characterized by a relatively
longer period of onset and infiltration or influx of
mononuclear cells. Mucosal layer may be mucosa of the bowel
(including the small intestine and large intestine), rectum,
stomach (gastric) lining, or oral cavity. Exemplary chronic
gastrointestinal inflammatory conditions are inflammatory
bowel disease (IBD), colitis induced by environmental insults
(e.g., gastrointestinal inflammation (e.g., colitis) caused
by or associated with (e.g., as a side effect) a therapeutic
regimen, such as administration of chemotherapy, radiation
therapy, and the like), infections colitis, ischemic colitis,
collagenous or lymphocytic colitis, necrotizing
enterocolitis, colitis in conditions such as chronic
granulomatous disease or celiac disease, food allergies,
gastritis, infectious gastritis or enterocolitis (e.g.,
Helicobacter pylori-infected chronic active gastritis) and
other forms of gastrointestinal inflammation caused by an
infectious agent.
Inflammatory bowel disease (IBD) includes a group of
chronic inflammatory disorders of generally unknown etiology,
e.g., ulcerative colitis (UC) and Crohn's disease (CD).
Clinical and experimental evidence suggest that the
pathogenesis of IBD is multifactorial involving
susceptibility genes and environmental factors. In
inflammatory bowel diesase, the tissue damage results from an
inappropriate or exaggerated immune response to antigens of
the gut microflora. Several animal models for inflammatory
bowel diseases exist. Some of the most widely used models
are the 2,4,6-trinitrobenesulfonic acid/ethanol (TNBS)-
induced colitis model or the oxazalone model, which induce
chronic inflammation and ulceration in the colon (Neurath et
al., Intern. Rev. Immunol 19:51-62, 2000). Another model
uses dextran sulfate sodium (DSS), which induces an acute
47
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
colitis manifested by bloody diarrhea, weight loss,
shortening of the colon and mucosal ulceration with
neutrophil infiltration. DSS-induced colitis is
characterized histologically by infiltration of inflammatory
cells into the lamina propria, with lymphoid hyperplasia,
focal crypt damage, and epithelial ulceration (Hendrickson et
al., Clinical Microbiology Reviews 15:79-94, 2002). Another
model involves the adoptive transfer of naive CD45RBhIgh CD4 T
cells to RAG or SCID mice. In this model, donor naive T
cells attack the recipient gut causing chronic bowel
inflammation and symptoms similar to human inflammatory bowel
diseases (Read and Powrie, Curr. Protoc. Immunol. Chapter 15
unit 15.13, 2001). The administration of antagonists of the
present invention in any of these models can be used to
evaluate the potential efficacy of those antagonists to
ameliorate symptoms and alter the course of diseases
associated with inflammation in the gut, such as inflammatory
bowel disease. Several treatment options for IBD are
available, for example anti-TNF-a antibody therapies have
been used for a decade to treat Crohn's disease (Van Assche
et al., Eur. J. Pharmacol. Epub Oct 2009). However, a
significant percentage of patients are refractory to the
current treatments (Hanauer et al., Lancet 359:1541-1549,
2002; Hanauer et al., Gastroenterology 130:323-333, 2006),
and thus new therapies targeting refractory patient
populations are needed.
Another example of an inflammatory condition is an
inflammatory pulmonary condition. Exemplary inflammatory
pulmonary conditions include infection-induced pulmonary
conditions including those associated with viral, bacterial,
fungal, parasite or prion infections; allergen-induced
pulmonary conditions; pollutant-induced pulmonary conditions
such as asbestosis, silicosis, or berylliosis; gastric
aspiration-induced pulmonary conditions, immune
dysregulation, inflammatory conditions with genetic
predisposition such as as cystic fibrosis, and physical
48
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
trauma-induced pulmonary conditions, such as ventilator
injury. These inflammatory conditions also include asthma,
emphysema, bronchitis, chronic obstructive pulmonary disease
(COPD), sarcoidosis, histiocytosis, lymphangiomyomatosis,
acute lung injury, acute respiratory distress syndrome,
chronic lung disease, bronchopulmonary dysplasia, community-
acquired pneumonia, nosocomial pneumonia, ventilator -
associated pneumonia, sepsis, viral pneumonia, influenza
infection, parainfluenza infection, rotavirus infection,
human metapneumovirus infection, respiratory syncitial virus
infection and aspergillus or other fungal infections.
Exemplary infection-associated inflammatory diseases may
include viral or bacterial pneumonia, including severe
pneumonia, cystic fibrosis, bronchitis, airway exacerbations
and acute respiratory distress syndrome (ARDS). Such
infection-associated conditions may involve multiple
infections such as a primary viral infection and a secondary
bacterial infection.
Asthma is an inflammatory disease of the lung that is
characterized by airway hyperresponsiveness ("AHR"),
bronchoconstriction, wheezing, eosinophilic or neutrophilic
inflammation, mucus hypersecretion, subepithelial fibrosis,
and elevated IgE levels. Patients with asthma experience
"exacerbations", a worsening of symptoms, most commonly due
to microbial infections of the respiratory tract (e.g.
rhinovirus, influenza virus, Haemophilus influenza, etc.).
Asthmatic attacks can be triggered by environmental factors
(e.g. ascarids, insects, animals (e.g., cats, dogs, rabbits,
mice, rats, hamsters, guinea pigs and birds), fungi, air
pollutants (e.g., tobacco smoke), irritant gases, fumes,
vapors, aerosols, chemicals, pollen, exercise, or cold air.
Apart from asthma, several chronic inflammatory diseases
affecting the lung are characterized by neutrophil
infiltration to the airways, for example chronic obstructive
pulmonary disease (COPD), bacterial pneumonia and cystic
fibrosis (Linden et al., Eur. Respir. J. 15:973-977, 2000;
49
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Rahman et al., Clin. Immunol. 115:268-276, 2005), and
diseases such as COPD, allergic rhinitis, and cystic fibrosis
are characterized by airway hyperresponsiveness (Fahy and
O'Byrne, Am. J. Respir. Crit. Care Med. 163:822-823, 2001).
Commonly used animal models for asthma and airway
inflammation include the ovalbumin challenge model and
methacholine sensitization models (Hessel et al., Eur. J.
Pharmacol. 293:401-412, 1995). Inhibition of cytokine and
chemokine production from cultured human bronchial epithelial
cells, bronchial fibroblasts or airway smooth muscle cells
can also be used as in vitro models. The administration of
antagonists of the present invention to any of these models
can be used to evaluate the use of those antagonists to
ameliorate symptoms and alter the course of asthma, airway
inflammation, COPD and the like.
Other inflammatory conditions and neuropathies, which
may be prevented or treated by the methods of the invention
are those caused by autoimmune diseases. These conditions
and neuropathies include multiple sclerosis, systemic lupus
erythematous, and neurodegenerative and central nervous
system (CNS) disorders including Alzheimer's disease,
Parkinson's disease, Huntington's disease, bipolar disorder
and Amyotrophic Lateral Sclerosis (ALS), liver diseases
including primary biliary cirrhosis, primary sclerosing
cholangitis, non-alcoholic fatty liver
disease/steatohepatitis, fibrosis, hepatitis C virus (HCV)
and hepatitis B virus (HBV), diabetes and insulin resistance,
cardiovascular disorders including atherosclerosis, cerebral
hemorrhage, stroke and myocardial infarction, arthritis,
rheumatoid arthritis, psoriatic arthritis and juvenile
rheumatoid arthritis (JRA), osteoporosis, osteoarthritis,
pancreatitis, fibrosis, encephalitis, psoriasis, Giant cell
arteritis, ankylosing spondolytis, autoimmune hepatitis,
human immunodeficiency virus (HIV), inflammatory skin
conditions, transplant, cancer, allergies, endocrine
diseases, wound repair, other autoimmune disorders, airway
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
hyperresponsiveness and cell, virus, or prion-mediated
infections or disorders.
Arthritis, including osteoarthritis, rheumatoid
arthritis, arthritic joints as a result of injury, and the
like, are common inflammatory conditions which would benefit
from the therapeutic use of anti-inflammatory proteins, such
as the antagonists of the present invention. For example,
rheumatoid arthritis (RA) is a systemic disease that affects
the entire body and is one of the most common forms of
arthritis. Since rheumatoid arthritis results in tissue
damage, TLR3 ligands could be present at the site of the
inflammation. Activation of TLR3 signaling may perpetuate
inflammation and further tissue damage in the inflamed joint.
Several animal models for rheumatoid arthritis are known in
the art. For example, in the collagen-induced arthritis
(CIA) model, mice develop chronic inflammatory arthritis that
closely resembles human rheumatoid arthritis. Administration
of the TLR3 antagonists of the present invention to the CIA
model mice can be used to evaluate the use of these
antagonists to ameliorate symptoms and alter the course of
diseases.
Diabetes mellitus, diabetes, refers to a disease process
derived from multiple causative factors and characterized by
hyperglycemia (LeRoith et al., (eds.), Diabetes Mellitus,
Lippincott-Raven Publishers, Philadelphia, Pa. U.S.A. 1996),
and all references cited therein. Uncontrolled hyperglycemia
is associated with increased and premature mortality due to
an increased risk for microvascular and macrovascular
diseases, including nephropathy, neuropathy, retinopathy,
hypertension, cerebrovascular disease and coronary heart
disease. Therefore, control of glucose homeostasis is a
critically important approach for the treatment of diabetes.
Underlying defects lead to a classification of diabetes
into two major groups: type I diabetes (insulin dependent
diabetes mellitus, IDDM), which arises when patients lack
insulin-producing beta-cells in their pancreatic glands, and
51
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
type 2 diabetes (non-insulin dependent diabetes mellitus,
NIDDM), which occurs in patients with an impaired beta-cell
insulin secretion and/or resistance to insulin action.
Type 2 diabetes is characterized by insulin resistance
accompanied by relative, rather than absolute, insulin
deficiency. In insulin resistant individuals, the body
secretes abnormally high amounts of insulin to compensate for
this defect. When inadequate amounts of insulin are present
to compensate for insulin resistance and adequately control
glucose, a state of impaired glucose tolerance develops. In
a significant number of individuals, insulin secretion
declines further and the plasma glucose level rises,
resulting in the clinical state of diabetes. Adipocity-
associated inflammation has been stronly implicated in the
development of insulin resistance, type 2 diabetes,
dyslipidemia and cardiovascular disease. Obese adipose
recruits and retains macrophages and can produce excessive
pro-inflammatory cytokines including TNF-a and IL-6, free
fatty acids and adipokines, which can interfere with insulin
signaling and induce insulin resistance. TLR3 activation on
macrophages may contribute to the pro-inflammatory status of
the adipose. Several animal modes of insulin resistance are
known. For example, in a diet-induced obesity model (DIO)
animals develop hyperglycemia and insulin resistance
accompanied by weight gain. Administration of TLR3
antagonists of the present invention to the DIO model can be
used to evaluate the use of the antagonists to ameliorate
complications associated with type 2 diabetes and alter the
course of the disease.
Fatty liver disease encompasses a spectrum of liver
conditions and is typically classified as either alcoholic or
nonalcoholic. In either case, fatty liver disease ranges
from simple hepatic steatosis (lipid accumulation and
deposition) to alcoholic and non-alcoholic steatohepatitis
(ASH or NASH), which often progresses to hepatic fibrosis,
cirrhosis, and probably hepatocellular carcinoma. Alcoholic
52
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
(AFLD) and nonalcoholic fatty liver disease (NAFLD) are
histologically indistinguishable; however, by definition
NAFLD develops in patients who consume little or no alcohol.
Instead, NAFLD is frequently found in individuals with
obesity, metabolic syndrome, and type 2 diabetes and is
closely linked to insulin resistance (Utzschneider et al., J
Clin Endocrinol Metab 91:4753-4761, 2006). With the dramatic
recent increase in the prevalence of obesity and insulin
resistance, NAFLD has surpassed AFLD and viral hepatitis-
induced liver disease as the most common chronic liver
disease. It has been estimated that approximately 75% of
those with obesity have NAFLD and as many as 20% may have
NASH (Clark, J Clin Gastroenterol 40(Suppl 1):S5-S10, 2006;
Lazo et al., Semin Liver Dis 28:339-350, 2008).
Atherosclerotic coronary heart disease (CHD) represents
the major cause for death and cardiovascular morbidity in the
western world. Risk factors for atherosclerotic coronary
heart disease include hypertension, diabetes mellitus, family
history, male gender, cigarette smoke, high serum
cholesterol, high low density lipoprotein (LDL) cholesterol
levels and low high density lipoprotein (HDL) cholesterol
levels. In general, a total cholesterol level in excess of
about 225-250 mg/dl is associated with significant elevation
of risk of CHD. A variety of clinical studies have
demonstrated that elevated levels of total cholesterol or LDL
cholesterol promote human atherosclerosis. Epidemiologic
investigations have established that cardiovascular morbidity
and mortality vary directly with the level of total
cholesterol and LDL cholesterol.
Exemplary cancers may include at least one malignant
disease in a cell, tissue, organ, animal or patient,
including, but not limited to leukemia, acute leukemia, acute
lymphoblastic leukemia (ALL), B-cell or T-cell ALL, acute
myeloid leukemia (AML), chronic myelocytic leukemia (CML),
chronic lymphocytic leukemia (CLL), hairy cell leukemia,
myelodyplastic syndrome (MDS), a lymphoma, Hodgkin's disease,
53
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
a malignant lymphoma, non-Hodgkin's lymphoma, Burkitt's
lymphoma, multiple myeloma, Kaposi's sarcoma, colorectal
carcinoma, pancreatic carcinoma, renal cell carcinoma, breast
cancer, nasopharyngeal carcinoma, malignant histiocytosis,
paraneoplastic syndrome/hypercalcemia of malignancy, solid
tumors, adenocarcinomas, squamous cell carcinomas, sarcomas,
malignant melanoma, particularly metastatic melanoma,
hemangioma, metastatic disease, cancer related bone
resorption and cancer related bone pain.
Exemplary cardiovascular diseases may include
cardiovascular disease in a cell, tissue, organ, animal, or
patient, including, but not limited to, cardiac stun
syndrome, myocardial infarction, congestive heart failure,
stroke, ischemic stroke, hemorrhage, arteriosclerosis,
atherosclerosis, restenosis, diabetic atherosclerotic
disease, hypertension, arterial hypertension, renovascular
hypertension, syncope, shock, syphilis of the cardiovascular
system, heart failure, cor pulmonale, primary pulmonary
hypertension, cardiac arrhythmias, atrial ectopic beats,
atrial flutter, atrial fibrillation (sustained or
paroxysmal), post perfusion syndrome, cardiopulmonary bypass
inflammation response, chaotic or multifocal atrial
tachycardia, regular narrow QRS tachycardia, specific
arrhythmias, ventricular fibrillation, His bundle
arrhythmias, atrioventricular block, bundle branch block,
myocardial ischemic disorders, coronary artery disease,
angina pectoris, myocardial infarction, cardiomyopathy,
dilated congestive cardiomyopathy, restrictive
cardiomyopathy, valvular heart diseases, endocarditis,
pericardial disease, cardiac tumors, aordic and peripheral
aneurysms, aortic dissection, inflammation of the aorta,
occulsion of the abdominal aorta and its branches, peripheral
vascular disorders, occulsive arterial disorders, peripheral
atherosclerotic disease, thromboangitis obliterans,
functional peripheral arterial disorders, Raynaud's
phenomenon and disease, acrocyanosis, erythromelalgia, venous
54
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
diseases, venous thrombosis, varicose veins, arteriovenous
fistula, lymphederma, lipedema, unstable angina, reperfusion
injury, post pump syndrome and ischemia-reperfusion injury.
Exemplary neurological diseases may include neurologic
disease in a cell, tissue, organ, animal or patient,
including, but not limited to neurodegenerative diseases,
multiple sclerosis, migraine headache, AIDS dementia complex,
demyelinating diseases, such as multiple sclerosis and acute
transverse myelitis; extrapyramidal and cerebellar disorders
such as lesions of the corticospinal system; disorders of the
basal ganglia or cerebellar disorders; hyperkinetic movement
disorders such as Huntington's Chorea and senile chorea;
drug-induced movement disorders, such as those induced by
drugs which block CNS dopamine receptors; hypokinetic
movement disorders, such as Parkinson's disease; Progressive
supranucleo Palsy; structural lesions of the cerebellum;
spinocerebellar degenerations, such as spinal ataxia,
Friedreich's ataxia, cerebellar cortical degenerations,
multiple systems degenerations (Mencel, Dejerine-Thomas, Shi-
Drager, and Machado-Joseph); systemic disorders (Refsum's
disease, abetalipoprotemia, ataxia, telangiectasia, and
mitochondrial multisystem disorder); demyelinating core
disorders, such as multiple sclerosis, acute transverse
myelitis; and disorders of the motor unit such as neurogenic
muscular atrophies (anterior horn cell degeneration, such as
amyotrophic lateral sclerosis, infantile spinal muscular
atrophy and juvenile spinal muscular atrophy); Alzheimer's
disease; Down's Syndrome in middle age; Diffuse Lewy body
disease; Senile Dementia of Lewy body type; Wernicke-
Korsakoff syndrome; chronic alcoholism; Creutzfeldt-Jakob
disease; Subacute sclerosing panencephalitis, Hallerrorden-
Spatz disease and Dementia pugilistica.
Exemplary fibrotic conditions may include liver fibrosis
(including but not limited to alcohol-induced cirrhosis,
viral-induced cirrhosis, autoimmune-induced hepatitis); lung
fibrosis (including but not limited to scleroderma,
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
idiopathic pulmonary fibrosis); kidney fibrosis (including
but not limited to scleroderma, diabetic nephritis,
glomerular nehpritis, lupus nephritis); dermal fibrosis
(including but not limited to scleroderma, hypertrophic and
keloid scarring, burns); myelofibrosis; neurofibromatosis;
fibroma; intestinal fibrosis; and fibrotic adhesions
resulting from surgical procedures. In such a method, the
fibrosis can be organ specific fibrosis or systemic fibrosis.
The organ specific fibrosis can be associated with at least
one of lung fibrosis, liver fibrosis, kidney fibrosis, heart
fibrosis, vascular fibrosis, skin fibrosis, eye fibrosis,
bone marrow fibrosis or other fibrosis. The lung fibrosis
can be associated with at least one of idiopathic pulmonary
fibrosis, drug induced pulmonary fibrosis, asthma,
sarcoidosis or chronic obstructive pulmonary disease. The
liver fibrosis can be associated with at least one of
cirrhosis, schistomasomiasis or cholangitis. The cirrhosis
can be selected from alcoholic cirrhosis, post-hepatitis C
cirrhosis, primary biliary cirrhosis. The cholangitis is
sclerosing cholangitis. The kidney fibrosis can be
associated with diabetic nephropathy or lupus
glomeruloschelerosis. The heart fibrosis can be associated
with myocardial infarction. The vascular fibrosis can be
associated with postangioplasty arterial restenosis or
atherosclerosis. The skin fibrosis can be associated with
burn scarring, hypertrophic scarring, keloid, or nephrogenic
fibrosing dermatopathy. The eye fibrosis can be associated
with retro-orbital fibrosis, postcataract surgery or
proliferative vitreoretinopathy. The bone marrow fibrosis
can be associated with idiopathic myelofibrosis or drug
induced myelofibrosis. The other fibrosis can be selected
from Peyronie's disease, Dupuytren's contracture or
dermatomyositis. The systemic fibrosis can be systemic
sclerosis or graft versus host disease.
56
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Administration/Pharmaceutical Compositions
The "therapeutically effective amount" of the agent
effective in the treatment or prevention of conditions where
suppression of TLR3 activity is desirable can be determined
by standard research techniques. For example, the dosage of
the agent that will be effective in the treatment or
prevention of inflammatory condition such as asthma, Crohn's
Disease, ulcerative colitis or rheumatoid arthritis can be
determined by administering the agent to relevant animal
models, such as the models described herein.
In addition, in vitro assays can optionally be employed
to help identify optimal dosage ranges. Selection of a
particular effective dose can be determined (e.g., via
clinical trials) by those skilled in the art based upon the
consideration of several factors. Such factors include the
disease to be treated or prevented, the symptoms involved,
the patient's body mass, the patient's immune status and
other factors known by the skilled artisan. The precise dose
to be employed in the formulation will also depend on the
route of administration, and the severity of disease, and
should be decided according to the judgment of the
practitioner and each patient's circumstances. Effective
doses can be extrapolated from dose-response curves derived
from in vitro or animal model test systems.
In the methods of the invention, the TLR3 antagonist may
be administered singly or in combination with at least one
other molecule. Such additional molecules may be other TLR3
antagonist molecules or molecules with a therapeutic benefit
not mediated by TLR3 receptor signaling. Antibiotics,
antivirals, palliatives and other compounds that reduce
cytokine levels or activity are examples of such additional
molecules.
The mode of administration for therapeutic use of the
agent of the invention may be any suitable route that
delivers the agent to the host. Pharmaceutical compositions
of these agents are particularly useful for parenteral
57
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
administration, e.g., intradermal, intramuscular,
intraperitoneal, intravenous, subcutaneous or intranasal.
The agent of the invention may be prepared as
pharmaceutical compositions containing an effective amount of
the agent as an active ingredient in a pharmaceutically
acceptable carrier. The term "carrier" refers to a diluent,
adjuvant, excipient, or vehicle with which the active
compound is administered. Such pharmaceutical vehicles can
be liquids, such as water and oils, including those of
petroleum, animal, vegetable or synthetic origin, such as
peanut oil, soybean oil, mineral oil, sesame oil and the
like. For example, 0.4% saline and 0.3% glycine can be used.
These solutions are sterile and generally free of particulate
matter. They may be sterilized by conventional, well-known
sterilization techniques (e.g., filtration). The
compositions may contain pharmaceutically acceptable
auxiliary substances as required to approximate physiological
conditions such as pH adjusting and buffering agents,
stabilizing, thickening, lubricating and coloring agents,
etc. The concentration of the agent of the invention in such
pharmaceutical formulation can vary widely, i.e., from less
than about 0.5%, usually at or at least about 1% to as much
as 15 or 20% by weight and will be selected primarily based
on required dose, fluid volumes, viscosities, etc., according
to the particular mode of administration selected.
Thus, a pharmaceutical composition of the invention for
intramuscular injection could be prepared to contain 1 ml
sterile buffered water, and between about 1 ng to about 100
mg, e.g. about 50 ng to about 30 mg or more preferably, about
5 mg to about 25 mg, of a TLR3 antibody antagonist of the
invention. Similarly, a pharmaceutical composition of the
invention for intravenous infusion could be made up to
contain about 250 ml of sterile Ringer's solution, and about
1 mg to about 30 mg and preferably 5 mg to about 25 mg of an
antagonist of the invention. Actual methods for preparing
parenterally administrable compositions are well known and
58
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
are described in more detail in, for example, "Remington's
Pharmaceutical Science", 15th ed., Mack Publishing Company,
Easton, PA.
The antibody antagonists of the invention can be
lyophilized for storage and reconstituted in a suitable
carrier prior to use. This technique has been shown to be
effective with conventional immunoglobulins and protein
preparations and art-known lyophilization and reconstitution
techniques can be employed.
The present invention will now be described with
reference to the following specific, non-limiting examples.
Example 1
Identification and Derivation of Anti-huTLR3 Antagonist mAbs
The MorphoSys Human Combinatorial Antibody Library
(HuCALED) Gold phage display library (Morphosys AG,
Martinsried, Germany) was used as a source of human antibody
fragments and was panned against a purified TLR3 antigen
generated from the expression of amino acids 1-703 of human
TLR3 (huTLR3) (SEQ ID NO: 4) with a C-terminal poly-histidine
tag and purified by immobilized metal affinity
chromatography. Amino acids 1-703 correspond to the
predicted extracellular domain (ECD) of huTLR3. Fab
fragments (Fabs) that bound specifically to huTRL3 ECD were
selected by presenting the TLR3 protein in a variety of ways
so that a diverse set of antibody fragments could be
identified, sequenced and confirmed as unique. From
different panning strategies, 62 candidates (different V-
region sequences) were identified as unique hTLR3 ECD
binders.
The 62 candidates identified as huTLR3 ECD binders were
screened for neutralizing activity in a range of cell-based
assays relevant to identifying anti-inflammatory activity.
Using preliminary activity data (see Example 2 below), four
candidates (Fabs 16-19) defining families 16-19 were selected
from the 62 as parents for CDR maturation of heavy chain CDR2
59
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
(HCDR2) and light chain CDR3 (LCDR3). One of the parental
candidates (candidate 19) exhibited an N-linked glycosylation
site in HCDR2; a Ser to Ala (S to A) mutation was made in
this candidate to delete the site. Following CDR maturation
of the four parental candidates, a total of 15 progeny
candidates (candidates 1-15) were identified for further
characterization as described in Example 2 below. A listing
of the light and heavy chain variable regions present in each
of the 19 candidates is shown in Table 3 above. The
candidates are herein referred to as mAbs 1-19 or Fabs 1-19,
depending whether they were Fabs or cloned as full length
antibody chains (Example 3). Due to expression vector
design, the mature amino termini of the variable regions for
all candidates were QVE for heavy chain and DI for the light
chain. The preferred sequences at these termini are those in
the respective germline genes with high identity to the
candidate sequences. For families 17 and 18 the germline
sequences are QVQ for VH and SY for VL. For family 19, the
sequences are EVQ for VH and DI for VL. The SY sequence is
unique to the lambda subgroup 3 and there are reports of
heterogeneity with either S or Y as the amino terminal
residue. Thus, the QSV consensus terminus from the prominent
lambda subgroup 1 was considered a more suitable replacement
for DIE for VL of families 17 and 18. These changes were
introduced into candidates 9, 10 and 12 from family 18 and
candidates 14 and 15 from family 19. In this process, both
the VH and VL regions of these antibodies were codon
optimized. The amino acid sequences of the light chain
variable region N-terminal germline variants of candidates 9,
10 and 11 are shown in SEQ ID NO:s 209-211, and the amino
acid sequences of the heavy chain variable region N-terminal
germline variants for candidates 9, 10, 12, 14, and 15 are
shown in SEQ ID NO:s 212-216, respectively. The N-terminal
variants of the candidates are herein referred to as
candidate/mAb/Fab 9QVQ/QSV, 10QVQ/QSV, 12QVQ/QSV, 14EVQ or
15EVQ. The N-terminal germline variants were expressed as
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
mAbs and showed no effect on binding to TLR3 or in their
ability to inhibit TLR3 biological activity when compared to
their parent counterparts (data not shown).
Example 2
Determination of TLR3 Antagonist Activity in vitro
The 15 CDR-matured candidates described above were
selected as potential human therapeutics and a range of
binding and neutralizing activities were determined. The
activity assays and results for the four parental Fabs, Fabs
16-19 and 15 CDR-matured Fabs, Fabs 1-15 or their non-
germline V-region variants are described below.
Inhibition of NE-KB and ISRE Signaling Cascasde
293T cells were grown in DMEM and GlutaMax media
(Invitrogen, Carlsbad, CA) supplemented with heat-inactivated
FBS and transfected with 30 ng pNF-KB or ISRE firefly
luciferase reporter plasmids, 13.5 ng pcDNA3.1 vector, 5 ng
phRL-TK, and 1.5 ng pCDNA encoding FL TLR3 (SEQ ID NO: 2).
The phRL-TK plasmid contains the Renilla luciferase gene
driven by the HSV-1 thymidine kinase promoter (Promega,
Madion, WI). TLR3 antibodies were incubated 30-60 min.
before addition of poly(I:C) (GE Healthcare, Piscataway, NJ).
The plates were incubated 6h or 24h at 37 C before the
addition of the Dual-Glo luciferase reagent, and the plates
were read on a FLUOstar plate reader. Normalized values
(luciferase ratios) were obtained by dividing the firefly
RLUs by the Renilla RLUs. Upon stimulation with the TLR3
agonist poly (I:C) (1 g/ml), the NF-KB or ISRE signaling
cascade stimulated firefly luciferase production was
specifically inhibited by incubation of the cells with anti-
TLR3 antibodies (0.4, 2.0 and 10 pg/ml) prior to stimulation.
The results for the NF-KB assays are shown in Fig. 1 and are
expressed as inhibition of the Firefly/Renilla ratio with
5465 as the positive control (neutralizing anti-human TLR3
Mab) and an anti-human tissue factor mAb (859) as the human
61
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
IgG4 isotype control. >50% inhibition was achieved with mAb
concentrations 0.4-10 g/ml. c1068 and TLR3.7 inhibited
about 38% and 8% of TLR3 biological activity at 10 g/ml.
Similar results were obtained with the ISRE reporter gene
assay (data not shown).
Cytokine Release in BEAS-23 cells
BEAS-2B cells (SV-40 transformed normal human bronchial
epithelial cell line) were seeded in a collagen type I coated
dishes and incubated with or without anti-human TLR3
antibodies prior to addition of poly (I:C). Twenty-four
hours after treatments, supernatants were collected and
assayed for cytokine and chemokine levels using a custom
multi-plex bead assay for detection of IL-6, IL-8, CCL-2/MCP-
1, CCL5/RANTES, and CXCL1UIP-10. Results are shown in Fig.
2 as % inhibition of the individual cytokine/chemokine
following mAb treatment at 0.4, 2.0 and 10 pg/ml. 5465 is a
positive control; 859 is an isotype control.
Cytokine Release in NHBE cells
Cytokine release was also assayed in normal human
bronchial epithelial (NHBE) cells (Lonza, Walkersville, MD).
NHBE cells were expanded and transferred to collagen-coated
dishes and incubated for 48 hours after which the media was
removed and replenished with 0.2 ml of fresh media. The
cells were then incubated with or without anti-human TLR3
mAbs 60 minutes prior to the addition of poly (I:C).
Supernatants were collected after 24 hours and stored at -20 C
or assayed immediately for IL-6 levels. Results are graphed
in Fig. 3 as % inhibition of IL-6 secretion following mAb
treatment using doses between 0.001 and 50 pg/ml. 5465 is a
positive control, 859 is an isotype control. Most mAbs
inhibited at least 50% of IL-6 production at <1 g/ml, and
achieved 75% inhibition at <5 g/ml.
62
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Cytokine Release in PBMC cells
Cytokine release was also assayed in human peripheral
blood mononuclear cells (PBMC). Whole blood was collected
from human donors into heparin collection tubes to which a
Ficoll-Paque Plus solution was slowly layered underneath.
The tubes were centrifuged and the PBMCs, that formed a white
layer just above the Ficoll, were recovered and plated. The
PBMCs were then incubated with or without anti-human TLR3
mAbs prior to the addition of 25 g/ml poly(I:C). After 24
hrs, supernatants were collected and cytokine levels were
determined using Luminex technology. Results are graphed in
Fig. 4 as cumulative percentage inhibition of IFN-y, IL-12
and IL-6 using a single dose of mAb (0.4 pg/ml) with 5465 is
a positive control; hIgG4 is an isotype control.
Cytokine Release in HASM cells
Briefly, human airway smooth muscle (HASM) cells were
incubated with or without anti-human TLR3 mAbs prior to the
addition of a synergistic combination of 500 ng/ml poly(I:C)
and 10 ng/ml TNF-a. After 24 hrs, supernatants were
collected and cytokine levels were determined using Luminex
technology. Results are graphed in Fig. 5 as levels of the
chemokine CCL5/RANTES using three doses of mAb (0.4, 2 and 10
pg/ml). 5465 is a positive control; hIgG4 is an isotype
control.
The results from the in vitro assays in human cells
confirm the ability of the antibodies of the invention to
reduce cytokine and chemokines release as a result of binding
to huTLR3.
Example 3
Full-length Antibody Constructs
The four parental Fabs (candidate nos. 16-19) and 15
progeny Fabs (candidate nos. 1-15) heavy chains were cloned
onto a human IgG4 background with a S229P Fc mutation.
Candidates 9QVQ/QSV, 10QVQ/QSV, 12QVQ/QSV, 14EVQ or 15EVQ
63
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
were cloned onto a human IgG4 background with F235A/L236A and
5229P Fc mutations.
The mature full-length heavy chain amino acid sequences
are shown in SEQ ID NOs: 90-102 and 218-220 as follows:
Candidate SEQ ID NO:
16 90
17 91
18 92
19 93
1 94
2 95
3 96
4 97
5, 6, 7 98
8 99
9 100
10, 11, 12 101
13, 14, 15 102
9EVQ 218
10EVQ, 12EVQ 219
14EVQ, 15EVQ 220
For expression, these heavy chain sequences can include
an N-terminal leader sequence such as MAWVWTLLFLMAAAQSIQA
(SEQ ID NO: 103). Exemplary nucleotide sequences encoding
the heavy chain of candidates 14EVQ and 15EVQ with a leader
sequence and the mature form (without a leader sequence) are
shown in SEQ ID NOs: 104 and 105, respectively. Likewise,
for expression, the light chain sequences of the antibodies
of the invention can include an N-terminal leader sequence
such as MGVPTQVLGLLLLWLTDARC (SEQ ID NO: 106). Exemplary
nucleotide sequences encoding the light chain of codon
optimized candidate 15 with a leader sequence and the mature
64
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
form (without a leader sequence) are shown in SEQ ID NOs: 107
and 108, respectively.
Example 4
Characterization of Anti-TLR3 mAb binding
EC50 values for the binding of the mAbs to human TLR3
extracellular domain (ECD) were determined by ELISA. Human
TLR3 ECD protein was diluted to 2 g/ml in PBS and 100 1
aliquots were dispensed to each well of a 96-well plate
(Corning Inc., Acton, MA). After overnight incubation at 4 C,
the plate was washed 3 times in wash buffer consisting of
0.05% Tween-20 (Sigma-Aldrich) in PBS. The wells were
blocked with 200 1 blocking solution consisting of 2% I-
Block (Applied Biosystems, Foster City, CA) and 0.05% Tween-
20 in PBS. After blocking for 2 hours at room temperature
the plate was washed 3 times followed by addition of serial
Table 4.
Candidate no. EC50 (ng/ml)
1 17.18
2 53.12
3 23.42
4 12.77
5 19.94
6 19
7 16.13
8 18.58
9 22.61
10 15.84
11 26.33
12 25.59
13 23.51
14 33.59
15 32.64
16 43.66
17 13.8
18 9.68
19 66.54
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
dilutions of the anti-TLR3 mAb candidates 1 to 19 in blocking
buffer. The anti-TLR3 mAbs were incubated for 2 hours at
room temperature and washed 3 times. This was followed by
addition of a peroxidase-conjugated sheep anti-human IgG (GE
Healthcare, Piscataway, NJ) diluted 1:4000 in blocking
buffer, incubated for 1 hour at room temperature followed by
3 washes in wash buffer. Binding was detected by 10-15
minute incubation in TMB-S (Fitzgerald Industries
International, Inc., Concord, MA). The reaction was stopped
with 25 1 2N H2SO4 and absorbance read at 450 nm with
subtraction at 650 nm using a SPECTRA Max spectrophotometer
(Molecular Devices Corp., Sunnyvale, CA). EC50 values were
determined by non-linear regression using GraphPad Prism
software (GraphPad Software, Inc., San Diego, CA).
EC50 values were determined for binding to huTLR3 (Table
4) by incubating with 100 1 of 4-fold serial dilutions of
mAbs from 2.5 g/ml to 0.6 pg/ml. An anti-human tissue
factor mAb 859 and hu IgG4K were included as negative
controls.
Binding affinity for huTLR3 ECD was also determined by
Biacore analysis. The data (not shown) indicated that the
mAbs 1-19 had a Kd for huTLR3 ECD of less than 108 M.
Example 5
Competitive Epitope Binding
Epitope binding experiments were performed to determine
the anti-TLR3 antibody competition groups or "epitope bins".
For competitive ELISA, 5 pl (20 pg/ml) of purified human
TLR3 ECD protein generated as described in Example 1 was
coated on MSD HighBind plate (Meso Scale Discovery,
Gaithersburg, MD) per well for 2 hr at room temperature. 150
1 of 5% MSD Blocker A buffer (Meso Scale Discovery) was
added to each well and incubated for 2 hr at room
temperature. Plates were washed three times with 0.1 M HEPES
buffer, pH 7.4, followed by the addition of the mixture of
66
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
labeled anti-TLR3 mAb with different competitors. Labeled
antibodies (10 nM) were incubated with increasing
concentrations (1 nM to 2 M) of unlabeled anti-TLR3
antibodies, and then added to the designated wells in a
volume of 25 pl mixture. After 2-hour incubation with gentle
shaking at room temperature, plates were washed 3 times with
0.1 M HEPES buffer (pH 7.4). MSD Read Buffer T was diluted
with distilled water (4-fold) and dispensed at a volume of
150 p1/well and analyzed with a SECTOR Imager 6000.
Antibodies were labeled with MSD Sulfo-Tae NHS-ester
according to manufacturer's instructions (Meso Scale
Discovery).
The following anti-TLR3 antibodies were evaluated: mAbs
1-19 obtained from a MorphoSys Human Combinatorial Antibody
Library (shown in Table 3a); c1068 (described in
W006/060513A2), c1811 (rat anti-mouse TLR3 mAb produced by a
hybridoma generated from rats immunized with mouse TLR3
protein), TLR3.7 (eBiosciences, San Diego, CA, cat no 14-
9039) and IMG-315A (generated against human TLR3 amino acids
amino acids 55-70 (VLNLTHNQLRRLPAAN) from Imgenex, San Diego,
CA). For mAbs 9, 10, 12, 14 and 15, variants 9QVQ/QSV,
10QVQ/QSV, 12QVQ/QSV, 14EVQ or 15EVQ were used in this study.
Based on competiton assays, anti-TLR3 antibodies were
assigned to five distinct bins. Bin A: mAbs 1, 2, 13, 14EVQ,
15EVQ, 16, 19; Bin B: mAbs 3, 4, 5, 6, 7, 8, 9QVQ/QSV,
10QVQ/QSV, 11, 12QVQ/QSV, 17, 18; Bin C: antibody Imgenex
IMG-315A; Bin D: antibodies TLR3.7, c1068; and Bin E:
antibody c1811.
Example 6
Epitope Mapping
Representative antibodies from distinct epitope bins as
described in Example 5 were selected for further epitope
mapping. Epitope mapping was performed using various
approaches, including TLR3 segment swapping experiments,
67
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
mutagenesis, H/D exchange and in silico protein-protein
docking (The Epitope Mapping Protocols, Methods in Molecular
Biology, Volume 6, Glen E. Morris ed., 1996).
TLR3 segment swapping. TLR3 human-mouse chimeric
proteins were used to locate gross antibody binding domains
on TLR3. The human TLR3 protein extracellular domain was
divided into three segments (aa 1-209, aa 210-436, aa 437-708
according to amino acid numbering based on human TLR3 amino
acid sequence, GenBank Acc. No. NP 003256). MT5420 chimeric
protein was generated by replacing human TLR3 amino acids
210-436 and 437-708 by corresponding mouse amino acids (mouse
TLR3, GenBank Acc. No. NP 569054, amino acids 211-437 and
438-709). The MT6251 chimera was generated by replacing
human amino acids at positions 437-708 by mouse TLR3 amino
acids (mouse TLR3, GenBank Acc. No. NP 569054, amino acids
438-709). All constructs were generated in the pCEP4 vector
(Life Technologies, Carslbad, CA) using standard cloning
procedures. The proteins were transiently expressed in
HEK293 cells as V5-His6 C-terminal fusion proteins, and
purified as described in Example 1.
mAb c1068. mAb c1068 bound human TLR3 ECD with high
affinity but did not bind well to murine TLR3. c1068 lost
its ability to bind to both MT5420 and MT6251, demonstrating
that the binding site was located within the amino acids 437-
708 of the WT human TLR3 protein.
mAb 12QVQ/QSV. mAb 12QVQ/QSV bound both chimeras,
indicating that the binding site for mAb 12QVQ/QSV was
located within the amino acids 1-209 of the human TLR3
protein having a sequence shown in SEQ ID NO:2.
In silico protein-protein docking. The crystal
structure of mAb 15EVQ (see below) and the published human
TLR3 structure (Bell et al., J. Endotoxin Res. 12:375-378,
2006) were energy minimized in CHARMm (Brooks et al., J.
Computat. Chem. 4:187-217, 1983) for use as the starting
models for docking. Protein docking was carried out with
ZDOCKpro 1.0 (Accelrys, San Diego, CA), which is equivalent
68
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
to ZDOCK 2.1 (Chen and Weng, Proteins 51: 397-408, 2003) with
an angular grid of 6 degrees. Known N-linked glycosylation
site Asn residues in human TLR3 (Asn 52, 70, 196, 252, 265,
275, 291, 398, 413, 507 and 636) (Sun et al., J. Biol. Chem.
281:11144-11151, 2006) were blocked from participating in the
antibody-antigen complex interface by an energy term in the
ZDOCK algorithm. 2000 initial poses were output and
clustered and the docking poses were refined and rescored in
RDOCK (Li et al., Proteins 53:693-707, 2003). The 200 poses
with the highest initial ZDOCK scores and 200 top RDOCK poses
were visually inspected.
Crystallization of Fab 15EVQ was carried out by the
vapor-diffusion method at 20 C (Benvenuti and Mangani, Nature
Protocols 2:1633-51, 2007). The initial screening was set up
using a Hydra robot in 96-well plates. The experiments were
composed of droplets of 0.5 1 of protein solution mixed with
0.5 1 of reservoir solution. The droplets were equilibrated
against 90 pl of reservoir solution. The Fab solution in 20
mM Tris buffer, pH 7.4, containing 50 mM NaC1 was
concentrated to 14.3 mg/ml using Amicon Ultra-5 kDa cells.
The screening was performed with the Wizard I & II (Emerald
BioSystems, Bainbridge Island, WA) and in-house
crystallization screens. Fab 12QVQ/QSV was crystallized in a
similar manner.
X-ray diffraction data were collected and processed
using the Rigaku MicroMaxTv'-007HF microfocus X-ray generator
equipped with an OsmicTm VariMaxIm confocal optics, Saturn 944
CCD detector, and an X-streamTm 2000 cryocooling system
(Rigaku, Woodlands, TX). Diffraction intensities were
detected over a 270 crystal rotation with the exposure time
of 120 s per half-degree image. The X-ray data were
processed with the program D*TREK (Rigaku). The structure
was determined by the molecular replacement method using the
program Phaser or CNX (Accelrys, San Diego, CA). Atomic
positions and temperature factors were refined with REFMAC
using all data in the resolution range 15-2.2 A for Fab 15EVQ
69
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
and 50-1.9 A for Fab 12QVQ/QSV. Water molecules were added
at the (F0-F) electron density peaks using the cut-off level
of 3o. All crystallographic calculations were performed with
the CCP4 suite of programs (Collaborative Computational
Project, Number 4. 1994. The CCP4 suite: programs for
protein crystallography. Acta Crystallogr. D50:760-763).
Model adjustments were carried out using the program COOT
(Emsley et al., Acta Crystallogr. D60:2126-2132, 2004).
The resolved crystal structure of mAb 15EVQ showed that
the antibody combining site was characterized by a number of
negatively charged residues in the heavy chain (D52, D55,
E99, D106 and D109). Thus, recognition between mAb 15EVQ and
TLR3 most likely involved positively charged residues. The
protein-protein docking simulations performed suggested that
two large patches on TLR3 involving multiple positive charge
residues showed good complementarity to the antibody. The
residues on TLR3 in the interface of the TLR3 - anti-TLR3
antibody simulated complexes were R64, K182, K416, K467,
Y468, R488, R489 and K493.
Mutagenesis studies. Single and combination point
mutations were introduced into surface residues of TLR3 ECD
in the regions identified above to contain the epitopes of
mAb 12 and mAb 15EVQ and the mutant proteins were tested for
antibody binding.
The nucleotide sequence encoding human TLR3 amino acids
1-703 (the ECD), (SEQ ID NO: 4; GenBank accession number
NP 003256), was cloned using standard protocols. All mutants
were generated by site directed mutagenesis using the
Strategene Quickchange II XL kit (Stratagene, San Diego, CA)
according to the manufacturer's protocol, using the
oligonucleotides shown in Table 5a. Mutations were verified
by DNA sequencing. The proteins were expressed under a CMV
promoter as C-terminal His-tag fusions in HEK293 cells, and
purified as described in Example 1.
Binding assays. The binding activity of mAb 12QVQ/QSV
and mAb 15EVQ to human TLR3 and generated variants was
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
evaluated by ELISA. To expedite the process, mutants in the
predicted mAb 15EVQ binding site were co-expressed in HEK
cells by co-transfection of TLR3 ECD mutant containing a C-
terminal His tag with mAb 12QVQ/QSV, followed by purification
by metal affinity chromatography. The recovered sample was a
complex of the TLR3 mutant with mAb 12. This approach was
feasible because the mAb 12QVQ/QSV and mAb 15EVQ binding
sites are distant from one another; and thus, point mutations
at one site are unlikely to affect the epitope at the other
site. These complexes were used in the ELISA binding assays.
5 pl per well of 20 pg/ml wild type TLR3 ECD or mutant
proteins in PBS were coated on an MSD HighBind plate (Meso
Scale Discovery, Gaithersburg, MD). The plates were
incubated at room temperature for 60 min and blocked.
20
30
71
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Table 5a. Sequences of the sense oligonucleiotides are
shown. The anti-sense oligonucleotides with complementary
sequences were used in the mutagenesis reaction.
SeqID
Variant Oligo
NO:
R64E 5 CCTTACCCATAATCAACTCGAGAGATTACCAGCCGCCAAC 3 136
K1 82E 5 CAAGAGCTTCTATTATCAAACAATGAGATTCAAGCGCTAAAAAGTGAAG 3 137
K416E 5 CCTTACACATACTCAACCTAACCGAGAATAAAATCTCAAAAATAG 3 138
K467E/Y468A 5 GAAATCTATCTTTCCTACAACGAGGCCCTGCAGCTGACTAGGAACTC 3 139
R488/R489/K493E 5
GCCTTCAACGACTGATGCTCGAGGAGGTGGCCCTTGAGAATGTGGATAGCTCTCCTTC 3 140
T472S/R473T/N474S 5 GTACCTGCAGCTGTCTACGAGCTCCTTTGCCTTGGTCCC 3 141
N 196A 5 GAAGAACTGGATATCTTTGCCGCTTCATCTTTAAAAAAATTAGAGTTG 3 169
Q1 67A 5 GTCATCTACAAAATTAGGAACTGCGGTTCAGCTGGAAAATCTCC 3 170
K1 63E 5 CTCATAATGGCTTGTCATCTACAGAATTAGGAACTCAGGTTCAGC 3 171
K 147E 5 GAAAATTAAAAATAATCCCTTTGTCAAGCAGGAGAATTTAATCACATTAGATCTGTC 3
172
K 145E 5 GAAAATTAAAAATAATCCCTTTGTCGAGCAGAAGAATTTAATCACATTAG 3 173
V144A 5 CAGAAAATTAAAAATAATCCCTTTGCAAAGCAGAAGAATTTAATCACATTAG 3
174
N140A 5 CCAACTCAATCCAGAAAATTAAAGCTAATCCCTTTGTCAAGCAGAAG 3 175
D116R 5 CAATGAGCTATCTCAACTTTCTCGTAAAACCTTTGCCTTCTGCAC 3 176
0536K 5 GTCTTGAGAAACTAGAAATTCTCAAGTTGCAGCATAACAACTTAGCAC 3 177
D536A 5 CTTGAGAAACTAGAAATTCTCGCATTGCAGCATAACAACTTAGCAC 3 178
K619E 5 CTAAAGTCATTGAACCTTCAGGAGAATCTCATAACATCCGTTG 3 179
K619A 5 CTCTAAAGTCATTGAACCTTCAGGCGAATCTCATAACATCCGTTGAG 3 180
E570R 5 CCACATCCTTAACTTGAGGTCCAACGGCTTTGACGAG 3 181
N541A 5 GAAATTCTCGATTTGCAGCATAACGCCTTAGCACGGCTCTGGAAAC 3 182
Q538A 5 GAGAAACTAGAAATTCTCGATTTGGCGCATAACAACTTAGCACGGC 3 183
H539E 5 CTAGAAATTCTCGATTTGCAGGAAAACAACTTAGCACGGCTCTG 3 184
H539A 5 CTAGAAATTCTCGATTTGCAGGCTAACAACTTAGCACGGCTCTG 3 185
N517A 5 CATTCTGGATCTAAGCAACAACGCCATAGCCAACATAAATGATGAC 3 186
Y465A 5 GAAAATATTTTCGAAATCTATCTTTCCGCCAACAAGTACCTGCAGCTGAC 3 187
R488E 5 GCCTTCAACGACTGATGCTCGAAAGGGTGGCCCTTAAAAATG 3 188
R489E 5 CTTCAACGACTGATGCTCCGAGAGGTGGCCCTTAAAAATGTGG 3 189
K467E 5 CGAAATCTATCTTTCCTACAACGAGTACCTGCAGCTGACTAG 3 190
overnight in MSD Blocker A buffer (Meso Scale Discovery,
Gaithersburg, MD) at 4 C. The following day the plates were
washed and the MSDSulfo-tag labeled mAb 15EVQ added at
concentrations from 500 pM to 1 pM for 1.5 hours. After
washes the labeled antibody was detected using MSD Read
Buffer T and the plates were read using a SECTOR Imager 6000.
To evaluate the binding activity of mAb 12QVQ/QSV to human
TLR3 and variants, co-expression was carried out with mAb
15EVQ and binding ELISAs were performed as described for mAb
15EVQ, except that the detecting antibody was labeled mAb
12QVQ/QSV.
72
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
mAb 12QVQ/QSV: The binding site for mAb 12QVQ/QSV was
located within the amino acids 1-209 of the human TLR3
protein as determined in the segment swap studies. The
following TLR3 mutants were evaluated: D116R, N196A, N140A,
V144A, K145E, K147E, K163E, and Q167A. The wild type TLR3
and V144A mutant showed comparable binding to mAb 12QVQ/QSV
(Figure 6A). The antibody did not bind to TLR3 D116R mutant
and had significantly reduced binding affinity to the K145E
mutant. Thus, residues D116 and K145 which are closely
apposed on the surface of TLR3 were identified as key epitope
sites for mAb 12QVQ/QSV (Figure 7A).
The two critical residues of the mAb 12QVQ/QSV binding
epitope were located near the face of the dsRNA binding site
at the N-terminal segment of the TLR3 ectodomain (Pirher, et
al., Nature Struct. & Mol. Biol., 15:761-763, 2008). The
complete epitope will contain other residues in the
neighboring regions, which were not revealed by mutational
analyses performed. Not wishing to be bound to any
particular theory, it is believed that binding of mAb
12QVQ/QSV on its TLR3 epitope may directly or indirectly
interfere with dsRNA binding on TLR3 ectodomain, thereby
disrupting receptor dimerization and activation of downstream
signaling pathways.
mAb 15EVQ: The following TLR3 mutants were evaluated:
R64E, K182E, K416E, Y465A, K467E, R488E, R489E, N517A, D536A,
D536K, Q538A, H539A, H539E, N541A, E570R, K619A, K619E, a
double mutant K467E/Y468A, a triple mutant T472S/R473T/N474S,
and a triple mutant R488E/R489E/K493E. The wild type TLR3,
the R64E, K182E, K416E mutants and the triple mutant
T472S/R473T/N474S showed comparable binding to mAb 15EVQ
(Figure 6B and Table 5b). The antibody did not bind to TLR3
mutants K467E, R489E, K467E/Y468A and R488E/R489E/K493E
(Figure 6B and 6C). The remaining variants showed
intermediate binding with the R488E having the greatest
effect. All of these mutants bound to mAb 12QVQ/QSV. These
results showed that resides K467 and R489 were critical
73
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
determinants of the mAb 15EVQ epitope. Residue R488 also
contributed to the epitope. These residues were closely
apposed on the same surface of TLR3 (Figure 7A). The results
also showed that residues Y465, Y468, N517, D536, Q538, H539,
N541, E570, and K619, all on the same surface as K467, R488
and R489, contributed to the epitope. This conclusion was
further supported by the H/D exchange studies with mAb 15EVQ.
Figure 7A shows binding epitope sites for mAbs 12QVQ/QSV and
15EVQ (black) and C1068 mAb (grey) superimposed on the
structure of human TLR3. The epitope for mAb 15EVQ covers
residues Y465, K467, Y468, R488, R489, N517, D536, Q538,
H539, N541, E570, and K619.
H/D Exchange studies. For H/D exchange, the procedures
used to analyze the antibody perturbation were similar to
that described previously (Hamuro et al., J. Biomol.
Techniques 14:171-182, 2003; Horn et al., Biochemistry
45:8488-8498, 2006) with some modifications. Recombinant
TLR3 ECD (expressed from Sf9 cells with C-terminal His-tag
and purified) was incubated in a deuterated water solution
for predetermined times resulting in deuterium incorporation
at exchangeable hydrogen atoms. The deuterated TLR3 ECD was
captured on a column containing immobilized mAb 15EVQ and
then washed with aqueous buffer. The back-exchanged TLR3 ECD
protein was eluted from the column and localization of
deuterium containing fragments was determined by protease
digestion and mass spec analysis. As a reference control,
TLR3 ECD sample was processed similarly except it was exposed
to deuterated water only after capture on the antibody column
and then washed and eluted in the same manner as the
experimental sample. Regions bound to the antibody were
inferred to be those sites relatively protected from exchange
and thus contain a higher fraction of deuterium than the
reference TLR3 ECD sample. About 80% of the protein could be
mapped to specific peptides. Maps of H/D exchange
perturbation of TLR3 ECD by mAb 15EVQ are shown in Figure 7B.
Only the segment of TLR3 around the portion affected by mAb
74
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
15EVQ is shown for clarity. The remainder of the protein
extending to the amino and carboxyl termini of TLR3 ECD was
not affected appreciably.
The H/D exchange studies identified peptide segments
465YNKYLQL411, 514SNNNIANINDDML526 and 529LEKL532 of SEQ ID NO: 2 as
regions where exchange on TLR3 was particularly altered by
binding to mAb 15EVQ. By its nature, H/D exchange is a
linear mapping method and usually cannot define which
residues within the peptide segment are most affected by
antibody binding. However, the extensive overlap between the
H/D exchange and mutational results gives added confidence
that the surface shown in Figure 7A is the binding site for
mAb 15EVQ. This binding site was in same linear amino acid
sequence region as previously described for mAb c1068 (PCT
Publ. no. W006/060513A2) but it was found to be located on a
completely non-overlapping surface (Figure 7A) in agreement
with the lack of cross-competition between these antibodies.
The mAb 15EVQ binding epitope was spatially proximal to
the dsRNA binding site at the C-terminal segment on TLR3
(Bell et al., Proc. Natl. Acad. Sci. (USA) 103: 8792-8797,
2006; Ranjith-Kumar et al., J Biol Chem, 282: 7668-7678,
2007; Liu et al., Science, 320: 379-381, 2008). Not wishing
to be bound to any particular theory, it is believed that
binding of mAb 15EVQ on its TLR3 epitope causes steric
clashes with a ligand dsRNA molecule and/or the dimer
partner, preventing ligand binding and ligand-induced
receptor dimerization.
35
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Table 5b.
Variant mAb 15 Variant mAb 12
wt TLR3 ECD wt TLR3 ECD
R64E +++ D116R
K182E +++ N140A ++
K416E +++ V144A +++
Y465A ++ K145E
K467E K147E
R488E K163E ++
R489E Q167A ++
N517A ++ N196A ++
D536K ++
D536A
Q538A
H539E ++
H539A ++
N541A ++
E57OR ++
K619E ++
K619A ++
K467E/Y468A
R488/R489/K493E
T472S/R473T/N4743 +++
Example 7
Generation of variants with enhanced thermal stability
Structure-based engineering was conducted to generate
antibody variants with increased thermal stability, with
simultaneous efforts to maintain the biological activity and
minimize immunogenicity.
mAb 15EVQ was selected for engineering. To minimize
immunogenicity, only germline mutations predicted to be
beneficial based upon structural considerations were pursued.
The VL and VH sequences of mAb 15EVQ (SEQ ID NO: 41 and SEQ
ID NO: 216, respectively) were aligned with the human
germline genes using BLAST searches. The closest germline
sequences identified were GenBank Acc. No. AAC09093 and
X59318 for VH and VL, respectively. The following
76
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
differences were identified between the germline VH, VL and
those of the mAb 15EVQ VH and VL sequences: (VH) V34I, G355,
F5OR, A61S, and Q67H; (VL) G305, L31S, and A34N. The
identified sequence differences were mapped onto the crystal
structure of the mAb 15EVQ, and residues predicted to alter
packing and interface interactions were selected for
engineering. Based upon the crystal structure of the
antibody (see Example 6), potential structure destabilizing
residues were identified. (1) A small enclosed cavity was
identified in the core of VH near V34. This cavity was large
enough to accommodate a slightly larger sidechain such as
Ile. (2) E99 of VH CDR3 was buried at the VH/VL interface
without a H-bonding network. The negatively charged
carboxylate group of E99 was in a generally hydrophobic
environment with mostly van der Waals (vdw) contacts to
neighboring residues. Burying a charge group is usually
energetically unfavorable and thus has destabilizing effect.
(3) F50 of VH is a VH/VL interface residue. Its aromatic
sidechain is bulky and thus may have negative impact upon the
pairing. H-bonding and vdw packing networks for the Fv were
calculated and visually inspected in Pymol
(www:// pymol org). Buried cavities in the VH and VL domains
were computed by Caver (Petrek et al., BMC Bioinformatics,
7:316, 2006). All molecular graphics figures were prepared
in Pymol. Mutations were made to the expression vectors
encoding Fab fragments or IgG4 full human antibodies
generated as described in Example 3 using standard cloning
techniques using Quick Change II XL Site Directed Mutagenesis
Kit (Stratagene, San Diego, CA), Change-IT Multiple Mutation
Site Directed Mutagenesis Kit (USB Corporation, Cleveland,
OH) or Quick Change II Site Directed Mutagenesis Kit
(Stratagene, San Diego, CA). The reactions were performed
according to each manufacturer's recommendations. The
obtained clones were sequenced for verification, and the
resulting engineered variants were named mAbs 15-1 - 15-10
according to their modified heavy or light chain. Each
77
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
variant chain (H or L) was expressed with the wild type mAb
15EVQ L or H chain to produce antibodies, except that the
heavy chain for mAb 15-10 was from mAb 15-6. A listing of
the SEQ ID NOs: for the CDRs, variable regions of light and
heavy chains and full length heavy and light chains for mAb
15EVQ and its engineered variants is shown in Table 6. Table
7 shows primers for generation of each variant.
Table 6.
SEQ ID NO:
Candidate Heavy Light
HCDR1 HCDR2 HCDR3 LCDR1 LCDR2 LCDR3 LV HV
no: IgG4 chain
111 112 84 109 110 113 41 216 220 156
15-1 111 114 84 109 110 113 41 124 130
156
15-2 115 112 84 109 110 113 41 125 131
156
15-3 116 112 84 109 110 113 41 126 132
156
15-4 111 117 84 109 110 113 41 127 133
156
15-5 116 118 84 109 110 113 41 128 134
156
15-6 116 112 119 109 110 113 41 129 135
156
15-7 111 112 84 120 110 113 122 42 102
157
Binding of mAbs 15-1 - 15-9 to TLR3 was evaluated by
15 ELISA immunoassay. Human TLR3 ECD (100 pl of 2 pg/ml TLR3-
ECD) was bound to a black Maxisorb plate (eBioscience)
overnight at 4 C. The plates were washed and blocked, and
diluted antibodies were aliquoted at 50 pl per well in
duplicate onto the wells. The plate was incubated at RT for
2 hours shaking gently. Binding was detected using
luminescence POD substrate (Roche Applied Science, Mannheim,
Germany, Cat. No. 11 582 950 001) and goat anti-human Fc:HRP
(Jackson ImmunoResearch, West Grove, PA, Cat. No. 109-035-
098) and the plate was read in a SpectraMax plate reader
(Molecular Devices, Sunnyvale, CA).
DSC experiments were performed on a MicroCal's Auto VP-
capillary DSC system (MicroCal, LLC, Northampton, MA) in
which temperature differences between the reference and
78
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
sample cells were continuously measured, and calibrated to
power units. Samples were heated from 10 C to 95 C at a
heating rate of 60 C/hour. The pre-scan time was 15 minutes
and the filtering period was 10 seconds. The concentration
used in the DSC experiments was about 0.5 mg/ml. Analysis of
the resulting thermograms was performed using MicroCal Origin
7 software (MicroCal, LLC).
Table 7.
Candidate Seq ID
Mutants Primers
no: NO:
GCCTGGAGTGGATGGGCCGGATCGACCCCAGCG 142
15-1 HC: F50R
CGCTGGGGTCGATCCGGCCCATCCACTCCAGGC 143
AGAGGTAACTCCCGTTGCGG 144
15-2 HC: V34I
GCATCTGGCGCACCCAGCCGATCCAGTAGTTGGTGAAG 145
AGAGGTAACTCCCGTTGCGG 146
15-3 HC: V34I/G355
GCATCTGGCGCACCCAGCTGATCCAGTAGTTGGTGAAG 147
AGAGGTAACTCCCGTTGCGG 144
15-4 HC: A615/Q67H
CGCTGATGGTCACGTGGCCCTGGAAGCTAGGGCTGTAGTTGGTGTAG 148
HC: CTTCACCAACTACTGGATCAGCTGGGTGCGCCAGATGC 149
15-5 F50R/V341/G355/
A61S/Q67H CGCTGATGGTCACGTGGCCCTGGAAGCTAGGGCTGTAGTTGGTGTAG 148
HC: CGCCATGTACTACTGCGCCCGCCAGCTGTACCAGGGCTAC 150
15-6
V34I/G355/E99Q
GTAGCCCTGGTACAGCTGGCGGGCGCAGTAGTACATGGCG 151
GCCAGCCAGAGCATCAGCAGCTACCTGGCCTGGTACCAGC 152
15-7 LC: G305/L315
GCTGGTACCAGGCCAGGTAGCTGCTGATGCTCTGGCTGGC 153
AGAGGTAACTCCCGTTGCGG 144
15-8 LC: A34N
CGGGCTTCTGCTGGTACCAGTTCAGGTAGCTGCTGATGCTCTG 154
HC: CGCCATGTACTACTGCGCCCGCCAGCTGTACCAGGGCTAC 150
15-9 F50R/V341/G355
/A61S/Q67H/E99Q GTAGCCCTGGTACAGCTGGCGGGCGCAGTAGTACATGGCG 151
CAGGGCAACACCCTGCCCTACACCTTCGGCCAG 228
15-10 LC: 595P
CIGGCCGAAGGIGTAGGGCAGGGIGTTGCCCTG 229
The thermal stability (Tm) of the generated variants was
measured by DSC (Table 8). Binding of the antibody variants
to TLR3 was comparable to that of the parental antibody.
79
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Table 8. Summary of melting temperatures (TM) of the variants
and rationale for making them.
Candidate
Mutations Rationale TIVI ( C) ATIVI ( C)
no:
15EVQ WT 64.7 0
15-1 HV F5OR VH/VL interface 69.3 4.6
15-2 HV V34I VH core packing 66.9 2.2
H-bonding, VH core
15-3 HV V34I/G35S 71.2 6.5
packing
VH/VL packing, VH
15-4 HV A61S/Q67H 65.4 0.7
surface charge
F50R/V341/G35S/ VH/VL interface, H-
15-5 HV 76.2 11.5
A61S/Q67H bonding, VH core
H-bonding, VH core
15-6 HV V34I/G34S/E99Q 75 10.3
packing, removal of
L-CDR1 surface polar
15-7 LV G30S/L31S 63.1 -1.6
residues
15-8 LV A34N VUVH interface 64 -0.7
F50R/V341/G35S/ VH/VL interface, H-
15-9 HV 76 11.3
A61S/Q67H/E99Q bonding, VH core
Canonical structure
15-10 LV S95P 76.6 11.9
stabilization
Example 8
Generation of a surrogate anti-TLR3 antibody
A chimeric antagonistic rat/mouse anti-mouse TLR3
antibody, herein named mAb 5429 was generated to evaluate
effects of inhibiting TLR3 signaling in various in vivo
models, as the humanized antibodies generated in Example 1
did not have sufficient specificity or antagonist activity
for mouse TLR3. The surrogate chimeric mAb 5429 as well as
its parent rat anti-mouse TLR3 antibody c1811 inhibited mouse
TLR3 signaling in vitro, and in vivo, and ameliorated
pathogenic mechanisms in several disease models in the mouse.
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Data discussed below suggests a role for TLR3 in the
induction and perpetuation of detrimental inflammation, and
contribute to the rationale for the therapeutic use of TLR3
antagonists and TLR3 antibody antagonists, for example acute
and chronic inflammatory conditions including
hypercytokinemia, asthma and airway inflammation,
inflammatory bowel diseases and rheumatoid arthritis, viral
infections, and type II diabetes.
Generation of the surrogate mAb 5429
CD rats were immunized with recombinant murine TLR3
ectodomain (amino acids 1-703 of seq ID NO: 162, GenBank Acc.
No. NP 569054) generated using routine methods. Lymphocytes
from two rats demonstrating antibody titers specific to
murine TLR3 were fused to FO myeloma cells. A panel of
monoclonal antibodies reactive to murine TLR3 were identified
and tested for in vitro antagonist activity in the murine
luciferase reporter and murine embryonic fibroblast assays.
The hybridoma line C1811A was selected for further work.
Functional variable region genes were sequenced from mAb
c1811 secreted by the hybridoma. Cloned heavy chain and
light chain variable region genes were then respectively
inserted into plasmid expression vectors that provided coding
sequences for generating a chimeric Rat/Balb C muIgG1/K mAb
designated as mAb 5429 using routine methods. The antibodies
were expressed as described in Example 3. T he amino acid
sequences of the mAb 5429 heavy and light chain variable
regions are shown in SEQ ID NO:164 and SEQ ID NO: 163,
respectively, and the heavy and light chain full length
sequences are shown in SEQ ID NO:166 and SEQ ID NO: 165,
respectively. The heavy and light chain full length
sequences of mAb c1811 are shown in SEQ ID NO: 168 and SEQ ID
NO: 167, respectively.
81
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Characterization of mAb 5429
mAb 5429 was characterized in a panel of in vitro assays
for its neutralizing ability on TLR3 signaling. The activity
assays and results are described below.
Murine Luciferase Reporter Gene Assay
The murine TLR3 cDNA (SEQ ID NO: 161, GenBank Acc. No:
NM 126166) was amplified by PCR from murine spleen cDNA (BD
Biosciences, Bedford, MA), and cloned into the pCEP4 vector
(Life Technologies, Carslbad, CA) using standard methods.
200 1 HEK293T cells were plated in 96 well white clear-
bottom plates at a concentration of 4 x 104 cells/well in
complete DMEM, and used the following day for transfections
using Lipofectamine 2000 (Invitrogen Corp., Carslbad, CA)
using 30 ng pNF-KB firefly luciferase (Stratagene, San Diego,
CA) or 30 ng pISRE firefly luciferase (BD Biosciences,
Bedford, MA), 5 ng phRL-TK control Renilla luciferase
(Promega Corp., Madison, WI) reporter plasmids, 1.5 ng pCEP4
encoding the full-length murine TLR3, and 13.5 ng empty
pcDNA3.1 vector (Life Technologies, Carslbad, CA) to bring
the total DNA amount to 50 ng/well. 24 hours post-
transfection, the cells were incubated for 30 minutes to 1
hour at 37 C with the anti-murine TLR3 antibodies in fresh
serum-free DMEM before the addition of 0.1 or 1 pg/pl
poly(I:C). The plates were harvested after 24 hours using
the Dual-Glo Luciferase Assay System (Promega, Madison, WI).
The relative light units were measured using a FLUOstar
OPTIMA multi-detection reader with OPTIMA software (BMG
Labtech GmbH, Germany). Normalized values (luciferase
ratios) were obtained by dividing the firefly relative light
units (RLUs) by the Renilla RLUs. mAb 5429 as well as its
parent mAb c1811 and mAb 15 (Table 3a) reduced poly(I:C) -
induced NF-kB and ISRE activation in a dose-dependent fashion
(Figure 8A and 8B), demonstrating their abilities to
antagonize the activity of TLR3. IC5Os measured in the ISRE
assay were 0.5, 22, and 0.7 g/ml for mAb 5249, mAB 15 and
mAb c1811, respectively.
82
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Murine Embryonic Fibroblast (MEF) Assay
C57BL/6 MEF cells were obtained from Artis Optimus
(Opti-MEFTm C57BL/6 - 0001). The cells were plated in 96-well
flat bottom plates (BD Falcon) at 20,000 cells/well in 200 pl
MEF media (DMEM with glutamax, 10% heat inactivated-FBS, lx
NEAA, and 10 pg/ml gentamycin). All incubations were done at
37 (0/5%CO2. 24 hours after plating, mAb 5429 or mAb c1811
were added into wells. The plates were incubated with the
mAbs for 1hr, after which Poly(I:C) was added at 1 pg/ml in
each well. The supernatants were collected after a 24-hour
incubation. Cytokine levels were determined using a bead kit
(Invitrogen Corp., Carslbad, CA) to detect CXCL10/IP-10
following manufacturer's protocol. The results were graphed
using GraphPad Prism Software. Both antibodies reduced
poly(I:C)-induced CXCL10/IP-10 levels in a dose-dependent
manner, demonstrating the abilities of these antibodies to
antagonize endogenous TLR3 and inhibit TLR3 signaling (Figure
9).
Flow Cytometry- Surface Staining
C57BL/6 and TLR3 knockout (TLR3K0) (C57BL/6 background;
female, 8-12 weeks of age, Ace Animals, Inc.), 10 per group,
were dosed intraperitoneally with 1 ml of 3% Thioglycollate
medium (Sigma) and 96 hrs later, the mice were euthanized and
the peritoneum from each mouse was lavaged with 10 ml sterile
PBS. Thioglycollate-elicited peritoneal macrophages were
resuspended in PBS and cell viability was assessed using
Trypan Blue staining. Cells were pelleted by centrifugation
and resuspended in 250 pl FACS Buffer (PBS -Ca2'-Mg2', 1% heat-
inactivated FBS, 0.09% Sodium Azide) and were kept on wet
ice. The CD16/32 reagent (eBioscience) was used at 10 pg/106
cells for 10 minutes to block Fc Receptors on the
macrophages. The cells were distributed at 106 cells in 100
p1/well for surface staining. Alexa-Fluor 647 (Molecular
Probes)-conjugated mAb c1811 and mAb 1679 (rat anti-mouse
83
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
TLR3 antibody that had no TLR3 specificity, and thus used as
an isotype control) were added at 0.25 pg/106 cells and
incubated on ice in the dark for 30 minutes. The cells were
washed and resuspended in 250 pl of FACS Buffer. The
viability stain, 7-AAD (BD Biosciences, Bedford, MA), was
added at 5 p1/well no more than 30 minutes before acquisition
of samples on FACS Calibur to detect a dead cell population.
Samples were collected by the FACS Calibur using Cell Quest
Pro Software. FCS Express was used to analyze the collected
data by forming histograms.
The binding of mAb c1811 to murine thioglycollate-
elicited peritoneal macrophages from C57BL/6 and TLR3K0 mice
were evaluated by flow cytometry to determine binding
specificity. mAb 5429 was not used in this assay since the
mouse Fc region of this chimeric antibody was expected to
contribute to non-specific binding. mAb c1811 exhibited no
binding to TLR3K0 macrophages, and increased binding to the
cell surfaces of C57BL/6 peritoneal macrophages, suggesting a
specificity of the mAb for TLR3 (Figure 10). mAb 5429,
having the same binding regions as mAb c1811, is assumed to
have the same binding specificity as mAb c1811.
Example 9
TLR3 antibody antagonists protect from TLR3-mediated systemic
inflammation
Model
The Poly(I:C)-induced systemic cytokine/chemokine model
was used as a model of TLR3-mediated systemic inflammation.
In this model, poly(I:C) (PIC) delivered intraperitoneally
induced a systemic cytokine and chemokine response that was
partially TLR3-mediated.
Female C57BL/6 mice (8-10 weeks old) or female TLR3K0
mice (C57BL/6 background; 8-10 weeks old, Ace Animals, Inc.)
were given mAb 5429 at 10, 20 or 50 mg/kg in 0.5 ml PBS, mAb
c1811 at 2, 10 or 20 mg/kg in 0.5 ml PBS or 0.5 ml PBS alone
(vehicle control) subcutaneously. 24 hours after antibody
84
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
dosing, mice were given 50 g poly(I:C) (Amersham Cat. No.
26-4732 Lot no. IH0156) in 0.1 ml PBS intraperitoneally.
Retro-orbital blood was collected 1 and 4 hours after the
poly(I:C) challenge. Serum was prepared from whole blood and
analyzed for cytokine and chemokine concentrations by
Luminex.
Results
Poly(I:C) delivered intraperitoneally induced a systemic
cytokine and chemokine response that was partially TLR3-
mediated, as evidenced by the significantly reduced
production of a panel of chemokines and cytokines in the
TLR3K0 animals (Table 9A). The TLR3-dependent poly(I:C)-
induced mediators were IL-6, KC, CCL2/MCP-1 and TNF-a at 1 hr
post-poly(I:C) challenge, and IL-1a, CCL5/RANTES and TNF-a at
4 hr post-poly(I:C) challenge. Both mAb c1811 and mAb 5429
significantly reduced levels of these TLR3-dependent
mediators, demonstrating the ability of the antibodies to
reduce TLR3 signaling in vivo (Table 9B). Values in Table 9
are shown as mean cytokine or chemokine concentrations in
pg/ml of six animals/group SEM. These data suggest that
TLR3 antagonism can be beneficial in reducing excess TLR3-
mediated cytokine and chemokine levels in conditions such as
cytokine storm or lethal shock.
30
40
CA 02855955 2014-03-11
WO 2013/039471 PCT/US2011/051202
Table 9A.
C57BL/6 TLR3K0
PIC
mAb 5429 (mg/kg)
mAb c1811 (mg/kg)
1 h PIC challenge
TNFa 6.005 0.32 319.4 34.1* 9.13 4.41 43.80 10.13**
KC 129.3 9.83 2357 491.5" 152.0 21.34 432.3 90.66""
IL-6 40.91 5.66 5317 856.7" 120.1 99.99 1214 294.9""
MCP-1 84.67 18.45 694.6 127.8" 67.85 34.16
249.9 55.60""
4 h PIC challenge
IL-la 28.21 17.78 796.7 45.0* 13.94 13.84 408.5 29.91""
RANTES 20.87 1.738 4511 783.4" 36.01 4.484
706.3 84.36""
TNFa 0.10 0 561.7 81.84" 3.215 3.115 305.8 53.63""
p<0.001: One Way ANOVA to C57BL/6 PBS
**p<0.001 One Way ANOVA to C57BL/6 PIC
Table 9B.
C57BL/6
PIC
mAb 5429 (mg/kg) 50 20 10
mAb c1811 (mg/kg) 20 10 2
1 h PIC challenge
TNF-a 29.33 3.78""" 31.05
1.59""" 59.55 12.71""" 32.54 3.89""" 42.22 7.04""" 42.61 10.58*""
KC 466.3 92.35""" 440.3 10.01""" 744.6 103.1"" 637.3 151.0"""
944.2 130.9"" 919.3 231.2""
IL-6 480.2 62.88""" 375.9 46.14*"" 705.2 149.8""" 739.2
113.3""" 1047 222""" 1229 378.4"""
MCP-1 168.5 15.04"" 321.6 206.7 219.2 70.58" 184.0
14.92"" 278.3 53.57 414.9 97.17
4 h PIG challenge
IL-la 343.0 33.01""" 452.6 94.86"" 481.1 121.0" 354.8
4543""" 351.7 68.85""" 352.4 39.60"""
RANTES 1381 169.7""" 2439
308.7"" 1601 398.9""" 1303 168.0""" 1365 474.1""" 2209 402.5""
TNF-a 100.1 8.5""" 205.1 41.85""" 226.1
64.72""" 138.9 26.0""" 121.6 38.85""" 223.8 47.74"""
***p<0.001, **p<0.01, *p<0.05: One Way ANOVA statistics were compared to the
C57BL16 + PIC group
Example 10
TLR3 antibody antagonists reduce airway hyperresponsiveness
Model
Airway hyperresponsiveness was induced by Poly(I:C).
86
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Female C57BL/6 mice (12 weeks old) or female TLR3K0 mice
(C57BL/6 background; 12 weeks old, Ace Animals, Inc.) were
anesthetized with isoflurane and several doses (10-100 pg) of
poly(I:C) in 50 pl sterile PBS were administered
intranasally. Mice received three administrations of
poly(I:C) (or PBS) with a 24 hour rest period between each
administration. 24 hours following the last poly(I:C) (or
PBS) administration, lung function and airway
hyperresponsiveness to methacholine were measured using whole
body plethysmography (BUXCO system). The mice were placed
into the whole body plethysmograph chamber and allowed to
acclimate for at least 5 minutes. Following baseline
readings, mice were exposed to increasing doses of nebulized
methacholine (Sigma, St. Louis, MO). The nebulized
methacholine was administered for 2 minutes, followed by a 5-
minute data collection period, followed by a 10-minute rest
period before subsequent increasing-dose methacholine
challenges. The increased airflow resistance was measured as
Enhanced Pause (Penh) and is represented as the average Penh
value over the 5-minute recording period (BUXCO system).
Following lung function measurements, mice were euthanized
and the lungs were cannulated. Bronchoalveolar lavages (BAL)
were performed by injecting 1 ml of PBS into the lungs and
retrieving the effluent. The lung tissues were removed and
frozen. BAL fluids were centrifuged (1200 rpm, 10 min.) and
the cell-free supernatants were collected and stored at -80 C
until analysis. Cell pellets were resuspended in 200 pl PBS
for total and differential cell counts. The multiplex assay
was performed following the manufacturer's protocol and the
Multiplex Immunoassay Kit (Millipore, Billercia, MA).
Results
Previous observations demonstrated that the intranasal
administration of poly(I:C) induced a TLR3 -mediated
impairment in lung function in mice with increased enhanced
pause (PenH) measurement in whole body plethysmography
87
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
(Buxco) at baseline and an increased responsiveness to
aerosolized methacholine (an indicator of airway
hyperesponsiveness) (PCT Publ. No. W006/060513A2). This
impairment in the lung function was associated with
neutrophil recruitment into the lung, and increased levels of
pro-inflammatory cytokines/chemokines in the lung. In this
study, the effect of mAb 1811 and mAb 5429 was evaluated in
poly(I:C)-induced impairment in lung function by
administering each antibody at 50 mg/kg subcutaneously prior
to poly(I:C) challenge.
TLR3-mediated impairment of lung function was
significantly reduced by treatment of animals with TLR3
antibody antagonists prior to the poly(I:C) challenge. TLR3-
mediated increases in baseline PenH and airway sensitivity to
methacholine were prevented in the anti-TLR3 antibody-treated
animals (Figure 11). Further, TLR3-mediated recruitment of
neutrophils into the mouse lung and generation of chemokines
in the airways were reduced in the anti-TLR3 antibody -
treated animals. The neutrophil numbers (Figure 12) and the
CXCL10/IP-10 levels (Figure 13) were measured from the
collected bronchoalveolar lavage fluid (BALF). The studies
were repeated at least three times with similar results.
Data shown in Figures 11, 12 and 13 are from one
representative study. Each symbol represents a data point
from one mouse, and the horizontal bars show group means.
The study demonstrated that systemically-administed TLR3
antibody antagonists reached the lung, reduced TLR3-mediated
impairment of lung function, neutrophil infiltration into the
airway, chemokine generation and respiratory tract
inflammation in the used model. Thus, TLR3 antagonists may
be beneficial in the treatment or prevention of respiratory
diseases characterized by airway hyperresponsiveness, such as
asthma, allergic rhinitis, chronic obstructive pulmonary
disease (COPD), and cystic fibrosis.
88
CA 02855955 2014-03-11
W02013/039471
PCT/US2011/051202
Example 11
TLR3 antibody antagonists protect from inflammatory bowel
disease
Model
The DSS colitis Model was used as a model of
inflammatory bowel disease.
Female C57BL/6 mice (<8 weeks old) or female TLR3K0
mice (C57BL/6 background; <8 weeks old weighing between
16.5g and 18g, Ace Animals, Inc.) were fed gamma-irradiated
food starting on day -1. DSS (Dextran sulfate) (MP
Biomedicals, Aurora, OH, Catalog no: 160110; 35-50kDa; 18-
20% Sulfur, Lot no. 8247J) was diluted in autoclaved
acidified drinking water to a final concentration of 5%.
The DSS-water was administered for 5 days, after which it
was replaced with plain water. Mice were allowed to drink
water ad libitum throughout the study. All water bottles
were weighed every day to record water consumption. On days
0, 2, and 4 mice were dosed intraperitoneally with 5 mg/kg
(0.1 mg in 0.1 ml PBS) mAb 5429, mouse anti-TNF-a antibody,
or PBS as a control. Mice were monitored daily throughout
the study and were weighed on days 0 through 4 and day 7.
Mice were euthanized on days 2 and 7 of the study.
Abdominal cavities were opened and the ascending colons cut
where they join the cecum. Colons were collected and fixed
in 10% neutral buffered formalin. Colons were paraffin-
embedded, sectioned and H&E stained (Qualtek Molecular Labs,
Santa Barbara, CA). Colonic histopathological assessments
were done in a blinded fashion by a veterinary pathologist
as described below (PathoMetrix, San Jose, CA).
Histopathologic evaluation
Two segments of large intestine, colon and rectum were
evaluated and scored for the following changes: (i) single
cell necrosis; (ii) epithelial ulceration; (iii) epithelial
sloughing; (iv) cryptal abscess; (v) cell proliferation;
(vi) cryptal cell proliferation; (vii) granulation tissue
89
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
formation in the lamina propria; (viii) granulation tissue
in the submucosa; (ix) submucosal inflammatory cell
infiltrate, neutrophil predominant; and (x) submucosal
edema.
A single, overall score of severity was given based on
the following standards:
0 - non-existent
1 - mild, focal or occasionally found
2 - mild, multifocal
3 - moderate, frequently found but in limited areas
4 - severe, frequently found in many areas or
extensions
of the tissue submitted
5 - very severe, extends to large portions of the
tissue
submitted
Results
Previous observations demonstrated that TLR3K0 animals
showed significantly reduced histopathology compared with
wild type mice in a model of inflammatory bowel disease
induced by DSS ingestion (PCT Publ. No. W006/60513A2), thus
suggesting that TLR3 signaling plays a role in the
pathogenesis in this model. It has been reported that
commensal bacterial RNA or mammalian RNA released from
necrotic cells can act as endogenous ligands to stimulate
TLR3 signaling (Kariko et al., Immunity 23165-231175 2005;
Kariko et al., J. Biol. Chem. 279:12542-12550 2004), and
therefore TLR3 stimulation by endogenous ligands in the gut
may enhance and perpetuate inflammation in the DSS colitis
model.
Disease severity was ameliorated in DSS-exposed animals
upon treatment with anti-TLR3 antibodies, as assessed by
compound histopathology scores (Figure 14). Figure 14 shows
means, standard deviations and 95% confidence intervals for
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
disease severity scores as horizontal bars. Significant
reduction in the scores were observed in the wild type DSS-
exposed animals treated with anti-TLR3 antibodies (p < 0.05)
when compared to untreated wild type animals. DSS-exposed
TLR3K0 animals were protected from DSS-induced changes.
DSS-exposed animals receiving anti-mouse TNF-a mAb
demonstrated no improvement in histopathology in the DSS
model. Therefore, the DSS model may be useful in evaluating
therapeutics that may target the human patient population
that is non-resposive to anti-TNF-a therapies, and
neutralizing anti-TLR3 antibodies may have the potential to
provide benefit to patients with inflammatory bowel disease
who do not respond to anti-TNF-a therapies.
Model
The T cell Transfer Model was used as a model of
inflammatory bowel disease. In this model, gut inflammation
was induced in SCID mice by the transfer of a population of
regulatory T cell-devoid naive T cells from immune-competent
mice, which attack antigen-presenting cells in the gut
mucosa.
high
Naive T-cells (CD4+CD45RB T cells) were injected
intraperitoneally into SCID recipients to induce chronic
colitis. Mice were given either PBS (500 1/mouse
intraperitoneally; vehicle control), mAb 5429 (0.1 mg/mouse
intraperitoneally), or anti-TNF-a antibody (0.05 mg/mouse
intraperitoneally; positive control) beginning 48 hours
following transfer of T-cells and then twice weekly
throughout the 8 week study. At 8 weeks following T-cell
transfer (or when mice lost >15% of their original body
weight) animals were euthanized and colons removed. Colons
were fixed, paraffin-embedded and H&E stained.
Histopathology (cell infiltration, crypt abscesses,
epithelial erosion, goblet cell loss, and bowel wall
thickening) was assessed quantitatively in a blinded fashion.
91
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Results
Disease severity was ameliorated in animals that
received T-cell transfer upon treatment with anti-TLR3
antibodies, as assessed by significant reduction in the
histopathology sum of scores when compared to the control
animals (p<0.05) (Figure 15A). For the sum of scores, crypt
abscesses, ulceration, neutrophil influx, goblet cell loss,
abnormal crypts, lamina propria inflammation and transmural
involvement was assessed. Significant reduction was
observed with crypt abscesses, ulceration and neutrophil
influx (for all p< 0.05) (Figure 15B). Anti-TNF-a antibody
was used as a positive control at doses known to provide
optimal benefit.
Studies using two well known models of inflammatory
bowel diseases, the DSS and the T-cell transfer model,
demonstrated that systemically delivered TLR3 antibody
antagonists reached the gut mucosa and reduced
gastrointestinal tract inflammation induced through two
different pathogenic mechanisms. Thus, TLR3 antagonists may
be beneficial for the treatment of inflammatory bowel
diseases, including anti-TNF-a-refractory cases, and other
immune-mediated pathologies in the gastrointestinal tract.
Example 12
TLR3 antibody antagonists protect from collagen-induced
arthritis
Model
The collagen-induced arthritis (CIA) model was used as a
model of rheumatoid arthritis.
Male B1ORIII mice (6-8 weeks old, Jackson Labs) were
divided into groups of 15 per group (arthritis groups) or 4
per group (control mice). Arthritis groups were anesthetized
with Isoflurane and given injections of Type II collagen
(Elastin Products) and Freund's complete adjuvant
supplemented with M. tuberculosis (Difco) on days 0 and 15.
On day 12, mice with developing type II collagen arthritis
92
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
were randomized by body weight into treatment groups and were
dosed subcutaneously (SC) on days 12, 17, and 22 (d12, d17,
2d2) with mAb 5429 (25 mg/kg), the negative control antibody
CVAM (a recombinant mAb of no known specificity in the mouse)
(5 mg/kg) or anti-TNF-a antibody (5 mg/kg, positive control).
In addition, control groups of mice were treated with vehicle
(PBS) or dexamethasone (0.5 mg/kg, Dex, reference compound)
subcutaneously (SC) daily (QD) on days 12-25. Animals were
observed daily from days 12 through 26. Fore and Hind paws
were evaluated by a clinical scoring system (shown below).
Animals were euthanized on study day 26 and histopathology
was assessed in a blinded fashion (scoring system described
below). Efficacy evaluation was based on animal body
weights, and clinical arthritis scores. All animals survived
to study termination.
Clinical scoring criteria for fore and hind paws
0 - normal
1 - hind or fore paw joint affected or minimal diffuse
erythema and swelling
2 - hind or fore paw joints affected or mild diffuse
erythema and swelling
3 - hind or fore paw joints affected or moderate diffuse
erythema and swelling
4 - marked diffuse erythema and swelling, or =4 digit
joints affected)
5 - severe diffuse erythema and severe swelling entire
paw, unable to flex digits)
Histopathologic scoring methods for mouse joints with Type II
collagen arthritis
When scoring paws or ankles from mice with lesions of
type II collagen arthritis, severity of changes as well as
number of individual joints affected were considered. When
only 1-3 joints of the paws or ankles out of a possibility of
numerous metacarpal/metatarsal/digit or tarsal/tibiotarsal
93
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
joints were affected, an arbitrary assignment of a maximum
score of 1, 2 or 3 for parameters below was given depending
on severity of changes. If more than 2 joints were involved,
the criteria below were applied to the most severely
affected/majority of joints.
Clinical data for paw scores were analyzed using AUC for
days 1-15, and % inhibition from controls were calculated.
Inflammation
0 - normal
1 - minimal infiltration of inflammatory cells in
synovium and periarticular tissue of affected joints
2 - mild infiltration, if paws, restricted to affected
joints
3 - moderate infiltration with moderate edema, if paws,
restricted to affected joints
4 - marked infiltration affecting most areas with marked
edema
5 - severe diffuse infiltration with severe edema
Pannus
0 - normal
1 - minimal infiltration of pannus in cartilage and
subchondral bone
2 - mild infiltration with marginal zone destruction of
hard tissue in affected joints
3 - moderate infiltration with moderate hard tissue
destruction in affected joints
4 - marked infiltration with marked destruction of joint
architecture, most joints
5 - severe infiltration associated with total or near
total destruction of joint architecture, affects all
joints
Cartilage Damage
0 - normal
94
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
1 - minimal to mild loss of toluidine blue staining with
no obvious chondrocyte loss or collagen disruption in
affected joints
2 - mild loss of toluidine blue staining with focal mild
(superficial) chondrocyte loss and/or collagen
disruption in affected joints
3 - moderate loss of toluidine blue staining with
multifocal moderate (depth to middle zone) chondrocyte
loss and/or collagen disruption in affected joints
4 - marked loss of toluidine blue staining with
multifocal marked (depth to deep zone) chondrocyte loss
and/or collagen disruption in most joints
5 - severe diffuse loss of toluidine blue staining with
multifocal severe (depth to tide mark) chondrocyte loss
and/or collagen disruption in all joints
Bone Resorption
0 - normal
1 - minimal with small areas of resorption, not readily
apparent on low magnification, rare osteoclasts in
affected joints
2 - mild with more numerous areas of, not readily
apparent on low magnification, osteoclasts more numerous
in affected joints
3 - moderate with obvious resorption of medullary
trabecular and cortical bone without full thickness
defects in cortex, loss of some medullary trabeculae,
lesion apparent on low magnification, osteoclasts more
numerous in affected joints
4 - marked with full thickness defects in cortical bone,
often with distortion of profile of remaining cortical
surface, marked loss of medullary bone, numerous
osteoclasts, affects most joints
5 - severe with full thickness defects in cortical bone
and destruction of joint architecture of all joints
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Results
Dexamethasone (Dex) and anti-mouse TNF-a antibody was
used as a positive control, PBS was used as vehicle control,
and CVAM was used as a negative control antibody. All
treatments were initiated on day 12 of the study, during the
development of joint disease. Disease incidence for vehicle-
treated disease control animals was 100% by study day 22.
Negative control groups treated with vehicle or CVAM antibody
had the highest clinical scores. Significantly reduced
clinical scores were observed for the groups treated with Dex
(p<0.05 for d18-d26), 5 mg/kg anti-TNF-a antibody (p<0.05 for
d18-26), or 25 mg/kg mAb 5429 (p<0.05 for d18-d23 and d25-
d26) (Figure 16). Clinical arthritis scores expressed as
area under the curve (AUC) were significantly reduced by
treatment with 25 mg/kg mAb 5429 (43% reduction), 5 mg/kg
anti-TNF-a antibody (52%), or Dex (69%) as compared to
vehicle controls. Figure 17 shows means and standard
deviations for AUC for each group.
Histopathological effects of the treatments were also
assessed. Paw bone resorption was significantly decreased by
treatment with 25 mg/kg mAb 5429 (47% decrease) as compared
to vehicle controls. Positive control mice treated with 5
mg/kg anti-TNF-aantibody had significantly decreased paw
inflammation (33%), cartilage damage (38%), and summed paw
scores (37%). Treatment with Dex significantly reduced all
paw histopathology parameters (73% reduction of summed
scores).
These data demonstrate that TLR3 antibody antagonists
improve clinical and histopathological disease symptoms in
the CIA model, and suggest the use of TLR3 antagonists for
treatment of rheumatoid arthritis.
96
CA 02855955 2014-03-11
W02013/039471
PCT/US2011/051202
Example 14
TLR3 antibody antagonists protect from acute lethal
viral infections
Model
An influenza A virus challenge model was used as a model
of acute lethal viral infection.
On Day -1, 4, 8, and 12, female C57BL/6 mice (12 weeks
old) or female TLR3K0 mice (C57BL/6 background; 12 weeks old,
ACE Animals, inc., 15 mice per group) were dosed
subcutaneously 20 mg/kg mAb 5429, or PBS alone. On day 0,
the mice were anesthetized by isoflurane and were
intranasally administered Influenza A/PR/8/34 virus (ATCC,
Rockland, MD, Lot no. 218171), in 25 1 PBS (equivalent to
105'55CEID50). Animals were observed two times a day for
changes in body weight and survival over the period of 14
days. A clinical scoring system was used to evaluate the
level of disease progression and subtle improvements in
response to Influenza A virus treatment.
Clinical scores
0 - normal, alert and reactive, no visible signs of
illness
1 - ruffled coat, with or without slightly reduced
ambulation
2 - ruffled coat, hunched posture when walking,
reluctant ambulation, labored breathing
3 - ruffled coat, labored breathing, ataxia, tremor
4 - ruffled coat, inability to ambulate with gentle
prodding, unconsciousness, feels cold to the touch
5 - found dead
Results
Survival, daily clinical scores, and changes in body
weight were evaluated in the study. Both influenza A
infected WT mice administered mAb 5429 (20 mg/kg) and
influenza A infected TLR3K0 not receiveing mAb 5429
97
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
demonstrated a statistically significant increase in survival
(p<0.001 and p<0.01, respectively) when compared to C57BL/6
mice inoculated with the Influenza virus, indicating that
antagonism or deficiency of TLR3 can prevent influenza -
induced mortality (Figure 18). Clinical scores were
significantly reduced in the group receiving 20 mg/kg mAb
5429, as well as in the TLR3K0 group (Figure 19). The body
weight of the mice was observed over a period of 14 days
after influenza virus administration. Body weight decreased
steadily in C57BL/6 mice dosed with Influenza A virus.
However, both the C57BL/6 mice dosed with 20 mg/kg mAb 5429
and the TLR3K0 mice demonstrated significantly greater body
weight relative to the WT C57BL/6 mice inoculated with
Influenza virus (Figure 20). These results demonstrated that
TLR3 antibody antagonists reduced clinical symptoms and
mortality in an acute lethal influenza viral infection model,
and suggested that TLR3 antagonists may provide protection
for humans in acute infectious states.
Example 15
TLR3 antibody antagonists improve hyperglycemia and
reduce plasma insulin
Model
The Diet-induced obesity (DIG) model was used as a
model of hyperglycemia and insulin resistance, and obesity.
C57BL/6 WT animals (about 3 weeks old, Jackson Labs) and
TLR3K0 animals (C57BL/6 background; about 3 weeks old, Ace
Animals, Inc.) were maintained on a high fat diet for 12 to
16 weeks. Both TLR3K0 and WT C57BL/6 mice were fed either
with normal chow or high-fat diet (Purina TestDiet cat. no.
58126) consisting of 60.9% kcal fat and 20.8% kcal
carbohydrates. Mice were maintained on a 12:12-h light-dark
cycle, with water and food ad libitum. The weight of each
mouse within each group was measured weekly. mAb 5429 was
given intraperitoneally twice a week for the first week
followed by once a week dosing for total of 7 weeks. Fasting
98
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
retro-orbital blood serum samples were used for insulin
measurements at the time points indicated. Glucose tolerance
tests were performed by i.p administration of glucose at 1.0
mg/g body weight after overnight fast at week 7. In
addition, fasting insulin and glucose levels were measured.
HOMA-IR was determined from the equation based on the
levels of fasting glucose and insulin levels (12) using
following equation: HOMA-IR = ((fasting glucose (mmo1/1) x
fasting insulin (mU/1))/ 22.5 (Wallace et al., Diabetes Care
27:1487-1495õ 2004). Fasting blood glucose (BG) was
determined using glucose oxidase assay. Fasting insulin
levels were determined using the insulin rat/mouse ELISA kit
(Crystal Chem, cat. No. 90060).
Results
After 12-16 weeks on high fat diet, the WT DIG aimals
were hyperglycemic and hyperinsulinemic. Glucose tolerance
was improved in the WT DIG animals but not in the TLR3K0 DIG
animals upon treatment with mAb 5429. Significantly reduced
blood glucose levels were observed in mAb 5429 treated
animals post glucose challenge at 60, 90, 120, and 180 min
when compared to control (PBS only) (Figure 21A). About 21%
reduction in AUC was observed in the mAb 5429 treated WT DIG
animals when compared to the WT DIG mice not receiveing the
mAb. Fasting insulin levels were also reduced in the WT DIG
animals treated with mAb 5429 (Figure 22). TLR3K0 DIG
animals showed no improvement in fasting insulin upon mAb
5429 treatment. Homeostatic model assessment (HOMA) analysis
indicated improved insulin sensitivity in the WT DIG animals
treated with mAb 5429, but not in the TLR3K0 DIG animals.
The HOMA-IR values were 14.0+9.8, 8.7+4.9, 9.0+3.0 for WT
DIG, 5 mg/kg of WT DIG mAb 5429, and 20 mg/kg of WT DIG mAb
5429 animals, respectively. No effect was observed in TLR3K0
DIG animals.
The study demonstrated that TLR3 antibody antagonists
improved insulin resistance and reduced fasting glucose in
99
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
the DIG model without weight loss, suggesting that TLR3
antagonists may be beneficial for the treatment of
hyperglycemia, insulin resistance, and type II diabetes.
Example 16
TLR3 antibody antagonists protect from bacteria and
virus-induced inflammatory responses
Reagents
Nontypeable Haemophilus influenza (NTHi) strains 35,
isolated from a COPD patient with bacterial exacerbations,
was obtained from Dr. T. F. Murphy (Buffalo VA Medical
Center, Buffalo, NY). Human rhinovirus 16 was obtained from
the American Type Culture Collection (ATCC) with TCID(50)=2.8
x 107/ml.
NTHi stimulation assays
NHBE cells (Lonza, Wakersville, MD) were seeded in
Microtest 96-well tissue culture plates (BD Biosciences,
Bedford, MA) at 1 x 105/well. NTHi grown on agar plates for
16-20 hr were resuspended in growth medium at -2 x 108 cfu/ml,
treated with 100 pg/ml gentamycin for 30 min. and added at -2
x 107/well to 96-well plates containing NHBEs. After 3 hours,
supernatants were removed and replaced with fresh growth
medium with or without antibodies (0.08 to 50 g/ml final
concentration). After additional 24 hr incubation, presence
of cytokines and chemokines in cell supernatants was assayed
in triplicate with a Cytokine 25-plex AB bead kit, Human
(including IL-113, IL-1RA, IL-2, IL-2R, IL-4, IL-5, IL-6, IL-
7, IL-8, IL-10, IL12p40p70, IL-13, IL-15, IL-17, TNF-a, IFN-
a, IFN-y, GM-CSF, MIP-1a, MIP-113, IP-10, MIG, Eotaxin, RANTES
and MCP-1) (Life Technologies, Carslbad, CA) in the Luminex
100IS multiplex fluorescence analyzer and reader system
(Luminex Corporation, Austin, TX).
100
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Rhinovirus stimulation assays
NHBE cells were seeded in Microtest 96-well tissue
culture plates (BD Biosciences, Bedford, MA) at 1 x 105
cells/well. The next day, antibodies (0.08 to 50 g/ml final
concentration) were added to NHBE or BEAS-2B cells and
incubated for 1 hr, followed by addition of 10 l/well of
rhinovirus. After additional 24 hr incubation, presence of
cytokines and chemokines in cell supernatants was assayed by
luminex assays as described above.
Results
mAb 15EVO inhibited NTHi induced IP-10/CXCL10 and
RANTES/CCL5 production in a dose-dependent manner, while the
control antibody, human IgG4 (Sigma, St. Louis, MO), showed
no inhibitory effect on NTHi stimulation (Figure 23A). mAb
15EVO also inhibited rhinovirus induced CXCL10/IP-10 and
CCL5/RANTES production (Figure 23B).
Example 17
TLR3 antibody antagonists suppress inflammatory responses in
astroctyes
Methods
Normal human astrocytes from 2 donors (Lonza, Walkersville,
MD) were plated in a 24 well plate at 75,000 cells/well and
allowed to attach overnight. The next day, the astrocytes
were treated with 200 ng/ml poly(I:C) and/or 10 g/ml mAb 18
for 24 hours. Cytokines were measured by Luminex.
Results
Poly(I:C)- induced production of IL-6, IL-8, IL-12, IFN-a,
IFN-y, CXCL9/MIG, CCL3/MIP-1a, CCL4, CCL5/RANTES and
CXCL10/IP-10 were inhibited by mAb 18, as shown in Table 10.
101
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Table 10.
Donor 1 IL-6 IL-8 IL-12 IFN-y
untreated 876.0 36.8 539.7 + 32. 6 16.6 0.5
28.8 1.5 12.3 0.3
mAb 18 1011.9 57.4 1401.9 49.7 17.1 +
0.5 31.6 + 0.7 10.4 0.2
Poly(I:C) or ol 30.3 1.5 47.1 + 3.1 35.9 +
1.0
Poly(I:C) + mAb 18 2225.0 + 108.1 6104.4 259.9 16.8+ 0.9 30.5 +
1.6 11.7 0.6
Donor 2
untreated 729.1 + 7.1 248.2 4.7 14 0.5 19.5 1.8
10.5 0.5
mAb 18 779.0 + 9.8 1132.6 30.6 14.3 0.6
20.8 1.9 10.5 0.1
Poly(I:C) ol ol 25.5 0.4 36.3 + 1.9 30.8 0.9
Poly(I:C) + mAb 18 3393.3 + 107.5 8660.4 354.3 16.2 0.3 24.7 +
1.2 12.6 0.3
Donor 1 CXCL9/MIG CCL3/MIP-la CCL4 CCL5/RANTES CXCL10/IP-
10
untreated 12.6 + 0.7 21 + 0.9 14.8 + 0.7 bl** bl
mAb 18 14.8 1.7 19.5 1.5 14.8 1.1 bl bl
Poly(I:C) 78.3 4.8 1569.3 36.9 159.7 12.7 788.2 94.9
461.4 10.3
Poly(I:C) + mAb 18 18.5 1.6 31.2 1.9 13.2 0.9 bl 6.9 0.5
Donor 2
untreated 9.9 + 1.6 12.3 1.7 11.3 0.3 bl bl
mAb 18 8.9 0.7 13.2 1.5 11.1 + 0.7 bl bl
Poly(I:C) 62 3.8 1552.9 41.1 140.7 4.8 546.8
21.7 533.2 15
Poly(I:C) + mAb 18 18.3 2.7 66.6 3.8 12.1 + 0.8 bl
29.1 + 6.2
*01: over detection level
**bl: below detection level
Example 18
TLR3 antibody antagonists suppress inflammatory responses in
endothelial cells
Methods
HUVEC cells (Lonza, Walkersville, MD) were cultured in
serum-containing growth medium recommended by Lonza. Cells
were resuspended in serum-free media (Lonza, Walkersville,
MD), plated in 96-well plates at 3x105 cells/ml, and incubated
at 37 C, 5%CO2 for 24 hrs. Poly(I:C) (GE Healthcare,
Piscataway, NJ) was added at increasing concentrations (1.5
to 100 g/ml) and incubated for another 24 hours at 37 C. For
cytokine inhibition assays, mAb 15EVQ was added to the cells
at various concentrations (0 - 50 g/ml) and incubated for 30
102
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
min, after which 20 g/ml poly(I:C) was added for 24 hours.
Cell supernatants were collected and cytokine levels were
measured using the human cytokine 30-plex kit and Luminex MAP
technology (Invitrogen Corp., Carslbad, CA). To measure
sICAM-1 expession, the HUVEC cells were treated with 20 g/ml
poly(I:C) and various concentrations of mAb 15EVQ (0.8 - 50
g/ml). The cell supernatants were analyzed for sICAM-1
expression by ELISA (R&D systems). Cell viability was
measured using the CellTiterGlo kit (Promega, Madison, WI).
Results
HUVEC cells produced the following cytokines in response
to poly(I:C): IL-1RA, IL-2, IL-2R, IL-6, IL-7, CXCL8/IL-8,
IL-12 (p40p70), IL-15, IL-17, TNF-a, IFN-a, IFN-y, GM-CSF,
CCL3/MIP-la, CCL4/MIP-113, CXCL10/IP-10, CCL5/RANTES,
CCL2/MCP-1, VEGF, G-CSE, FGF-basic, and HGF (Table 11). mAb
15EVQ dose-dependently reduced levels of all cytokines
induced by poly(I:C) (Table 12). The ability of mAb 15EVQ to
reduce poly(I:C)-induced production of TNF-a, CCL2/MCP-1,
CCL5/RANTES, and CXCL10/IP-10 suggested that inhibiton of
TLR3-mediated activities may protect against leukocyte and T
cell infiltration that can lead to atherosclerosis. Also,
inhibition of VEGF by mAb 15EVQ suggested a potential benefit
of TLR3 blockade in pathologies mediated by VEGF including
angiogenesis in a variety of cancers and ocular diseases such
as age-related macular degeneration.
TNF-a and IFN-y function in leukocyte recruitment and
increase the expression of adhesion molecules on the
activated endothelium (Doukas et al., Am. J. Pathol. 145:137-
47, 1994; Pober et al., Am. J. Pathol. 133:426-33, 1988).
CCL2/MCP-1, CCL5/RANTES, and CXCL10/IP-10 have been
implicated in monocyte and T cell recruitment and contribute
to the development of atherosclerosis (Lundgerg et al., Clin.
Immunol. 2009). The generation of VEGF by endothelial cells
has been linked to abnormal tissue growth or tumors in a
103
CA 02855955 2014-03-11
WO 2013/039471 PCT/US2011/051202
variety of cancers during angiogenesis (Livengood et al.,
Cell. Immunol. 249:55-62, 2007).
Table 11.
Poly(I:C) pg/m1 IL-6 CXC L8/1-8 CCL2/MCP-1
848.8 + 50.9 12876.0 + 2314.0 11813.4 + 1420.9
5 751.3 + 2.1 11363.7 + 108.2 11365.7 + 113.1
2.5 607.1 + 91.6 10961.5 + 2200.7 11607.3 +
2155.7
1.25 419.2 + 178.4 9631.5 + 3675.8 11690.9 + 3189.9
0.63 263.8 + 81.4 6231.9 + 1568.0 9075.6 + 1152.2
0.31 183.5 + 168.3 5257.9 + 1855.0 8106.8 + 1193.1
0.16 111.9 + 72.5 4057.6 + 1127.4 6619.8 + 1728.2
no poly(I:C) 0.00 1286.6 + 300.8 1360.1 + 245.4
Poly(I:C) g/m I IL-2R IL-15 IL-17
100 784.4 + 45.4 61.3 + 12.5 43.8 + 5.3
50 718.6 + 56.8 61.3 + 12.5 47.6 + 0
25 735.7 + 23.4 56.7 + 18.9 58.3 + 4.9
12.5 650.5 + 29.8 38.8 + 6.5 39.8 + 10.9
6.25 643.4 + 39.9 34.2 + 0 32.1 + 0
3.13 681.8 + 24.3 38.8 + 6.5 43.8 + 5.3
1.56 578.6 + 10.5 29.4 + 6.7 36.1 + 5.6
no poly(I:C) 0.0 0.0 0.0
Poly(I:C) ag/m I IFNa CXCL10/IP-10 CCL4/MIP-1p
100 50.7 + 0 3803.1 + 185.5 234.5 + 19.7
50 44.9 + 1.7 2235.9 + 184.6 291.6 + 41.8
25 46.1 + 0 2403.0 + 271.9 278.7 + 4.7
12.5 41.2 + 3.5 2185.4 + 64.9 243.8 + 63.4
6.25 36.1 + 0 2100.0 + 288.1 201.9 + 46.2
3.13 40.0 + 1.8 3553.2 + 197.1 191.5 + 20.8
1.56 42.5 + 1.7 2064.3 + 242.1 165.3 + 16.3
no poly(I:C) 0.0 0.0 0.0
Poly(I:C) g/m1 RANTES TNFa VEGF
100 6266.9 + 1708.7 12.8 + 3.2 581.1 + 181.4
50 2919.7 + 119.4 11.5 + 3.2 637.9+ 47.7
25 2805.1 + 176.7 9.8 + 2.8 700.3 + 62.5
12.5 2598.6 + 68.6 7.3 + 0.9 513.2 + 73.5
6.25 2449.2 + 830.6 6.9 + 1.4 440.4 + 29.5
3.13 3117.1 + 795.7 7.3 + 0.9 393.9 + 40.2
1.56 2481.0 + 719.3 6.0 + 1.8 358.4 + 74.8
no poly(I:C) 4.9 + 4.5 1.9 + 0.4 32.1 + 8.8
concentrations shown as pg/ml
Soluble Intercellular Adhesion Molecule 1 (sICAM-1) is
generated by proteolytic cleavage and is a marker for
10 endothelial cell activation. ICAM-1 plays a key role in
leukocyte migration and activation and is upregulated on
endothelial cells and epithelial cells during inflammation
where it mediates adhesion to leukocytes via integrin
molecules LFA-1 and Mac-1. Poly(I:C)
activated the endothelial cells to upregulate sICAM-1
expression and the uregulation was reduced by treatment with
mAb 15EVQ (Figure 24A).
104
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Table 12.
mAb 15 (og/m1) 50.00 10.00 2.00 0.40 0.08 0.016 0.003
0
PIC
IL-6 177.8+ 5.6* 214.6+3.6* 359.2+ 57.6* 727.2+ 50.5*
10000+0 10000+0 10000+ 0 10000+0
CXCL8/IL-8 1040.7 + 185.9 1765.9 + 97.1 6460.3 + 3684.4 57349.5 +
6293.4 72422.8 + 88279.2 47047.5 + 52393.1 76066.5 + 11354.1 96478.0 +
122298.4
CCL2/MCP-1 1187.7 + 165.4 * 1955.4 + 72.7* 9054.4 + 4110.9* 20000 +
0.0 20000 + 0.0 20000 + 0.0 20000 + 0.0 20000 + 0.0
IL-2R 25.0 + 35.3* 0.0 + 0.0* 312.3' 137.6* 521.5 + 5.5
664.7 + 9.8 661.2 + 14.8 698.4 + 57.6 654.2 + 14.8
IL-15 0.0 + 0.0* 0.0 + 0.0* 0.0 + 0.0* 4.1 Ø0* 38.8 +
6.5 43.4 + 0.0 38.8 + 6.5 43.4 + 0.0
IL-17 1.3 + 1.8* 11.8 + 16.8 11.8 + 16.8 27.9' 6.0 47.4 +
10.4 54.3' 20.2 40.0' 0.0 51.2 '5.1
lFNc 0.9 + 1.3* 0.9 + 1.3* 19.0' 7.7* 36.1 + 0.0 44.9 +
1.7 41.2' 3.5 47.3' 1.7 40.0' 1.8
CXCL10/IP-10 0.0 + 0.0* 58.1 + 2.6* 633.0' 471.6* 1441.4' 97.1
3023.8 + 166.1 2107.5' 372.6 2346.4' 226.1 2157.4' 282.7
CCL4/MIP-1p 0.0,0.0* 0.0 + 0.0* 2.9 + 4.1 * 62.1 .7.2*
176.6 + 21.3* 227.5 + 19.9 248.3' 19.4 281.7 + 37.5
RANTES 30,0.0* 15.4 + 4.5* 201.1 + 169.1* 952.4 + 41.1 *
2454.4 + 98.5* 2698.1 .88.6* 2624.4 + 129.8* 3459.7' 181.8
TNFo 1.9 + 0.4* 1.6 + 0.0* 2.2 + 0.0* 3.4 + 0.0 6.3'
0.5 8.5' 0.0 7.6' 1.4 6.9' 2.3
VEGF 87.2 + 8.7* 28.6 + 8.7* 88.3 + 52.1 * 156.1 .6.4*
479.6 + 14.1 544.6 + 8.3 533.5 + 70.2 607.3 + 29.9
* Indicates significant p-values (less than 0.05) comparing mAb15
concentration vs. poly(I:C) alone
Values are mean (pg/m1) + SEM
This suggested that TLR3 antibody antagonists can
inhibit leukocyte trafficking and thus tissue damage caused
by the influx of inflammatory cells.
For viability assays, HUVECs were cultured, plated and
stimulated with poly(I:C) as described above. mAb 15EVQ
dose-dependently restored poly(I:C)-induced reduction in
HUVEC cell viability (Figure 24B).
Down-modulation of endothelial cell activation can
suppress excessive immune cell infiltration and reduce tissue
damage caused by cytokines that are increased during
inflammatory conditions. Inflammation and overexpression of
cytokines and adhesion molecules on endothelial cells are key
contributors to developing atherosclerosis and hypertension.
These data provide rationale for exploring the potential
benefit of TLR3 antagonists for use in diseases of the blood
vessels including vasculitides, and those featuring
endothelial dysfunction. Another disease that is affected by
inflammation and overexpressed cytokines is Kaposi's sarcoma
(KS) that is common in immunosuppressed and HIV infected
individuals and is caused by Kaposi's sarcoma herpes virus
(KSHV). VEGF and cytokine production contribute to the
survival of KS cells (Livengood et al., Cell Immunol. 249:55-
62, 2007). TLR3 antagonists could be beneficial at reducing
105
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
angiogenic risks associated with KS and other tumors and at
preventing cell viability loss and protecting endothelial
barrier integrity to prevent vascular leakage, a potentially
serious condition associated with organ failure and life-
threatening inflammatory conditions such as sepsis. TLR3
antagonism may also be beneficial in viral infections
involving endothelial cell pathology such as the viral
hemorrhagic fevers caused by members of the families
flaviviridae (e.g. Dengue, yellow fever), filoviridae (Ebola,
Marburg), bunyaviridae (e.g. Hantavirus, Nairovirus,
Phlebovirus), and arenaviridae (e.g. Lujo, Lassa, Argentine,
Bolivian, Venezuelan hemorrhagic fevers (Sihibamiya et al.,
Blood 113:714-722, 2009).
Example 20
Cross-reactivity of TLR3 antibody antagonists with
cynomolgus and murine TLR3
Activity against cynomolgus or murine TLR3 were assessed
using the ISRE reporter gene assay as described in Example 2.
The cynomolgus (SEQ ID NO: 217) and murine TLR3 cDNAs (SEQ ID
NO: 161) were amplified from whole blood and cloned into the
pCEP4 vector (Clontech), and expressed as described above.
mAb 15EVQ had IC5Os of 4.18 g/ml and 1.74 g/ml in the cyno
NF-KB and ISRE assays, respectively, compared to IC5Os of
0.44 and 0.65 g/ml in the human TLR3 NF-kB and ISRE assays,
respectively. Isotype control antibodies had no effect in
these assays.
Example 21
Therapeutic dosing of TLR3 antibody antagonists protect from
acute lethal viral infections
Example 14 descirbes prophylactic treatment (dosed on
days -1, 4, 8, and 12) with TLR3 antibody antagonists against
influenza A infection. This example demonstrates that
therapeutic dosing of TLR3 antibody antagonists (day 3 after
106
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
influenza A infection after the onset of clinical symptoms)
are efficacious in enhancing survival.
Model
An influenza A virus challenge model was used as a model
of acute lethal viral infection as described in Example 14,
except that dosing of animals with mAb 5249 was done 3 days
post infection with influenza A, and the animals dosed were 8
weeks old. Anti-mouse IgG1 isotype control mAb was from
BioLegend. The animals were dosed days 3, 7 and 11 post-
infections with influenza A.
Survival, daily clinical scores, and changes in body
weight were evaluated in the study. Both the C57B1/6 mice
administered mAb 5249 and the TLR3K0 mice demonstrated a
statistically significant increase in survival (p < 0.028 and
p < 0.001, respectively) relative to the C57BL/6 mice
inoculated with the anti-mouse IgG1 isotype control mAb and
Influenza virus (Figure 25). The clinical scores were
reduced (Figure 26) and the body weights incrased (Figure 27)
in the C57BL/6 mice dosed with mAb 5249 and in the TLR3K0
animals when compared with C57BL/6 mice dosed with anti-mouse
IgG1 isotype control mAb and Influenza A. These results
demonstrated that TLR3 antibody antagonists reduced clinical
symptoms and mortality in an acute lethal influenza viral
infection model, and suggested that TLR3 antagonists may
provide protection for humans in acute infectious states.
Example 22
Epitopes and paratopes of TLR3 antibody antagonists by X-ray
crystallography
The human TLR3 extracellular domain was crystallized in
complex with Fabs of mAb 15EVQ, mAb 12QVQ/QSV and mAb c1068.
Methods
Expression and purification of proteins
The expression and purification of the TLR3 ECD (amino
acids 1-703 of SEQ ID NO: 2) the three Fabs were as described
above.
107
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Preparation of the TLR3 ECD-three Fab quaternary complex
4 mg of human TLR3 ECD was mixed with 2.4 mg of each Fab
and incubated at 4 C for 3.5 h, corresponding to a molar
ratio of 1 TLR3 ECD:1.1 Fab. The complex was purified by
anion exchange chromatography on a MonoQ 5/50 GL column (GE
Healthcare, Piscataway, NJ), equilibrated with 20 mM Tris pH
8.5, 10% glycerol (buffer A) and eluted with 20 mM Tris pH
8.5, 10% glycerol, 1 M NaC1 (buffer B). Approximately 2.48
mg of complex in 1.74 mL was diluted to 10 mL with buffer A,
loaded onto the column at 1 mL/min and eluted with a linear
gradient of 0-40% B over 40 column volumes. Five consecutive
purification runs were performed. Fractions from peak 1 were
pooled, concentrated with an Amicon-15 mL Ultra-30000 MWCO
and a Microcon 30000 MWCO to 14.49 mg/mL in 20 mM Tris pH
8.5, 27 mM NaC1, 10 % glycerol (Extinction coefficient: A280 (1
mg/mL) = 1.31).
Crystallization
Automated crystallization screening was performed using
the Oryx4 automatic protein crystallization robot (Douglas
Instruments) dispensing equal volumes of protein and
reservoir solution in a sitting drop format using Corning
plate 3550 (Corning Inc., Acton, MA). Initial screening was
with Hampton Crystal Screen HT (HR2-130, Hampton Research,
Aliso Viejo, CA). Small crystals from several conditions
were used to generate seeds, which were then used in
Microseed-Matrix Screening (MMS). Several rounds of
refinement were performed that were based on conditions from
the initial screening that gave small crystals. Reservoir
conditions used for MMS were based on those that gave small
crystals after refinement: 18-28% polyethylene glycol (PEG)
3350, 1M LiC1, pH4.5 and 2.0-2.9 M (NH4)2504, 5% PEG400, pH
4.5, and explored pH and different additives. MMS
crystallization screening was performed using the Oryx4
automatic protein crystallization robot (Douglas Instruments)
by dispensing components in the following volume ratio: 1
108
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
protein solution: 0.25 seed stock: 0.75 reservoir solution.
Crystals diffracting to -10-A resolution grew from 0.1 M Na
acetate pH 4.5, 2.9 M (NH4)2304, 5% methyl-pentane-diol (MPD)
and 0.1 M Na acetate pH 4.5, 26% PEG3350, 1 M LiC1.
In an effort to improve the resolution of the crystals,
MMS with the above conditions was combined with additive
screening using selected components of the Hampton Additive
Screen HR2-428 (Hampton Research, Aliso Viejo, CA) in the
following volume ratio: 1 protein solution: 0.125 seed stock:
0.2 additive solution: 0.675 reservoir solution. X-ray
quality crystals of the TLR3 ECD complexed with the Fabs,
which diffract to - 5-A resolution, were obtained after
applying a combination of MMS and Additive screening from a
solution containing 0.1 M Na acetate pH 4.5, 28% PEG 3350, 1
M LiC1, and 30 mM Gly-Gly-Gly.
X-ray data collection of TLR3 ECD quaternary complex
For X-ray data collection, a crystal (size -1.0 x 0.5 x
0.1 mm3) was soaked for a few seconds in a synthetic mother
liquor (0.1 M Na acetate, pH 4.5, 28% PEG 3350, 1 M LiC1, 16%
glycerol), and flash frozen in the stream of nitrogen at 100
K. X-ray diffraction data were collected and processed using
a Rigaku MicroMaxTm-007HF microfocus X-ray generator equipped
with an OsmicTm VariMaxTm confocal optics, Saturn 944 CCD
detector, and an X-streaMm 2000 cryocooling system (Rigaku,
Woodlands, TX). Diffraction intensities were detected over a
250 crystal rotation with the exposure time of 1 min per
half-degree image to the maximum resolution of 5 A. The X-
ray data were processed with the program D*TREK (Pflugrath,
Acta Crystallographica Section D, 55:1718-1725, 1999). The
crystal belongs to the monoclinic space group C2 with unit
cell parameters: a = 214.90 A, b = 142.08 A, c = 125.04 A,
and 13 = 103.17 . The asymmetric unit contains 1 molecule of
the complex. The X-ray data statistics are given in Table
13.
109
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Table 13.
Data Collection
Space group C2
Unit cell axes (A) 214.90, 142.08, 125.04
Unit cell angles ( ) 90, 103.17, 90
Resolution (A) 30-5.0 (5.18-5.00)
No. unique reflections 15,877
(1589)
Completeness (%) 99.8 r(99.6)
Redundancy 5.2
(4.9)
Rmemea 0.121 (0.312)
<I/d> 7.1
Structure refinement
Resolution (A) 29.4-5.0
RcrydRfree(%)b 26.8/30.0
No. of reflections
Working set 15,792
Test set (5% data) 788
Rmsd from ideal values
Bond length (A) 0,007
Bond angels (c) 0.744
Number of protein atoms 15,442
Ram achandran plot
Favored regions (%) 93.1
Allowed (%) 98.8
Disallowed (9/0) 1,2
Structure Determination
The crystal structure of the TLR3 ECD - Fab 15EVQ - Fab
12QVQ/QSV - Fab c1068 was determined by molecular replacement
using Phaser (Read, Acta Crystallogr. D. Biol. Crystallogr.
57:1373-1382, 2001). The search models were TLR3 ECD
(Protein DataBank (PDB) structure ID 1ziw with all glycans
removed, Choe et al., Science 309:581-585, 2005) and the high
resolution crystal structures of the three Fabs determined
(See Table 13 for a summary of the crystal data and
refinement statistics for these Fab structures). The elbow
110
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
angle of Fab 12QVQ/QSV was found to deviate significantly
from that in the free form. A series of Fab 12QVQ/QSV models
were generated by adjusting the elbow angle at -5 intervals,
one of which was found to agree well with the electron
density. The structure refinement was carried with PHENIX
(Adams et al., J. Synchrotron Radiat. 11:53-55, 2004). The
structure was refined as rigid body domains (each V or C
domain) for the Fabs and 13 rigid segments (Definitions used
in the refinement: 30-60, 61-108,109-156,157-206,207-257,258-
307,308-363,364-415,416-464,465-514,515-570,571-618,619-687)
for the TLR3 ECD with one B factor for each Fab rigid body
and a single B for the entire TLR3 ECD.
Translation/ Libration/ Screw (TLS) refinement was
introduced for each of the Fab rigid bodies and TLR3 ECD was
divided into 2 TLS segments at residue 330 of SEQ ID NO: 2.
Glycan density was visible for 10 of the 15 N-glycosylation
sites. Carbohydrate models from the crystal structure of the
human TLR3 extracellular domain (Choe et al., Science
309:581-585, 2005, PDB structure ID: 1ziw) were then added.
The density for a short missing segment in TLR3 ECD (residues
337-342 of SEQ ID NO: 2) was visible after rigid body
refinement, and it was filled in with the corresponding
segment from the TLR3 extracellular structure 2a0z (Bell et
al., Proc. Natl. Acad. Sci. (USA) 102:10976-10980, 2005, PDB
strucutre ID: 2a0z) . The C-terminus of TLR3 ECD contained
additional density that matches that of 2a0z. This segment
(647-703 of SEQ ID NO: 2) was then replaced with (residues
647-687) of 2a0w. Thus, the TLR3 ECD model was a hybrid
between the TLR3 sturctures 1ziw and 2a0z and refined as 13
rigid body segments (amino acid range: 30-60,61-108,109-
156,157-206,207-257,258-307,308-363,364-415,416-464,465-
514,515-570,571-618,619-687).
The LCDR3 of Fab 12QVQ/QSV apparently adopted different
conformation from its free form. Multi-start simulated
annealing was carried out with standard parameters in PHENIX.
The models of this LCDR3 were visually inspected in the
111
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
electron density map and the "best-matching" conformation was
grafted onto the original model. The refinement process was
monitored by Rfree against 5% of the reflections set aside
prior to initiating the calculations. In the final round,
one B factor for each residue was included. Model inspection
and manual rebuilding of the elbow regions of the Fabs and
side chains at the protein-protein interfaces were done using
COOT (Emsley et al., Acta Crystallogr. D. Biol. Crystallogr.
60:2126-32, 2004). The final Rcryst and Rfree were 26.8% and
30.0%, respectively, for all 15,792 independent reflections
to 5.0 A. The refinement statistics are given in Tables 13
and 14.
Results
The molecular structure of the TLR3 ECD-three Fab quaternary
complex
The overall molecular structure of the complex is shown in
Figure 28. In the asymmetric unit there is one TLR3 ECD and
one molecule of each Fab. The structural model for TLR3 ECD
includes all residues from 30 to 687 of huTLR3 (SEQ ID NO:
2). For the three Fabs, all residues from their respective
unbound forms were included except solvent ions and water
molecules. The TLR3 ECD molecule is very similar to the
previously reported structures in overall topology (rmsd of
0.79 A for 1ziw, 613 Ca's, and 1.37 A for 2a0z, 595 Ca's).
The Fab structures are all identical to their respective
unbound forms except for LCDR3 of Fab 12QVQ/QSV as described
in Methods as well as the elbow regions and some side chains
at TLR3 ECD/Fab interfaces.
35
112
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
Table 1 4 .
Data collection
Fab 12QVQ/QSV Fab 15EVQ Fab c1068
Space group P21 P21 P21
Cell dimensions
a, b, c (A) 75.83, 80.35, 83.06 54.68, 74.74,64.99
82.48, 136.94,83.25
a,I3,Y( ) 90, 115.24, 90 90, 103.69, 90 90, 114.95, 90
Resolution (A) 70-2.5 (2.59-2.50) 49-2.2 (2.28-2.20) 50-1.9
(2.0-1.9)
Unique reflections 27,785 (1653) 24,439 (1859) 117,490
(5916)
Completeness (%) 88.5 9(53) 94.2 v(72.8) 89.3 (45.2)
Redundancy 4 :0 .8) 5.2 '(4.3) 3.2 '.(2)
Rmerge 0.164 (0.297) 0.088 (0.445) 0.065
(0.264)
<1/o> (unaveraged) 2.9 '(1.2) 3.8 '(1.4) 5.7 '(1.6)
Structure Refinement
Resolution (A) 15-2.5 (2.56-2.50) 15-2.2 (2.26-2.20)
75.38-1.90 (1.94-1.90)
RcrysieRfree(%)b 19.7/25.4 (30.8/40.8) 19.3/26.9 (24.6/31.1)
20.4/27.7 (39.8/51.1)
No. of reflections
Working set 26,723 23,308 111,413
Test set 882 1,008 5,917
Number of atoms
Proteins 7,046 3,705 13,421
Solvent (water, etc.) 486 333 1,779
RMSD bond lengths (A) 0.012 0.013 0.023
RMSD bond angles ( ) 1.6 1.5 2
Ramachandran plot
Favored regions (%) 92.3 96.8 97.2
Allowed (/0) 98.9 99.3 99.7
Disallowed (%) 1.1 0.7 0.3
Values for highest resolution shell are in (ys.
D, where is the intensity of the measured reflection and <I> is the rnwn
intensity
of di measurements of this reflection.
6 Reryst 11F1 IF.3,51(111Z Fob, where Fobs and Fcaic are observed and
calculated structure factors and Rfree is
calculated for a se of randomiy chosen 5% of reflections prior to refinement.
cThe Rarnachandran plot was calculated with MolProbity (Davis, LW., et al.,
Nucleic Acids Res, 32: W615-9, 2004).
The epitopes and the paratopes
The residues involved in binding between the TLR3 ECD
and the three Fabs are shown in Figure 28B. Fab 12QVQ/QSV
bound near the N-terminus of the TLR3 ECD. The
conformational epitope was composed of residues from the TLR3
LRRs 3-7 (amino acids 100-221 of SEQ ID NO: 2. The binding
of Fab 12QVQ/QSV buried approximately 928 A2 and 896 A2 on the
antigen and antibody, respectively. For Fab 12QVQ/QSV, the
crystal structure identified following TLR3 (SEQ ID NO: 2)
epitope residues: S115, D116, K117, A120, K139, N140, N141,
113
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
V144, K145, T166, Q167, V168, S188, E189, D192, A195, and
A219. For Fab 12QVQ/QSV, the crystal structure identified
following paratope residues: light chain (SEQ ID NO: 211):
G28, S29, Y30, Y31, E49, D50, Y90, D91, and D92. Heavy chain
(SEQ ID NO: 214): N32, Q54, R56, S57, K58, Y60, Y104, P105,
F106, and Y107.
Fab 15EVQ and Fab c1068 bound non-overlapping epitopes
spanning LRRs 15-23 (amino acids 406-635 of SEQ ID NO: 2)
near the C-terminus (Figure 28). Fab 15EVQ buried 1080 A2 and
1064 A2 on the antigen and antibody, respectively, whereas Fab
c1068 buried 963 A2 and 914 A2 on the antigen and antibody,
respectively. The epitope for Fab 15EVQ covers residues
K416, K418, L440, N441, E442, Y465, N466, K467, Y468, R488,
R489, A491, K493, N515, N516, N517, H539, N541, S571, L595
and K619 of TLR3 shown in SEQ ID NO: 2. For Fab 15EVQ, the
crystal structure identified following paratope residues:
light chain (SEQ ID NO: 41): Q27, Y32, N92, T93, L94, and
S95. Heavy chain (SEQ ID NO: 216): W33, F50, D52, D55, Y57,
N59, P62, E99, Y101, Y104, and D106.
For Fab c1068, the crystal structure identified
following epitope residues on TLR3 (SEQ ID NO: 2): E446,
T448, Q450, R453, R473, N474, A477, L478, P480, S498, P499,
Q503, P504, R507, D523, D524, E527, E530, and K559. For Fab
c1068, the crystal structure identified following paratope
residues light chain: H30, N31, Y32, N50, E66, S67, G68
(glyc). Heavy chain: T30, T31, Y32, W33, H35, E50, N52, N54,
N55, R57, N59, V99, M102, 1103, and T104.
Mechanisms of neutralization and implication for TLR3
function
mAb 15EVQ: The mAb 15EVQ epitope contains TLR3 residues
N517, H539 and N541, which overlap with the C-terminal dsRNA
binding site (Bell et al., Proc. Natl. Acad. Sci. USA,
103:8792-7, 2006). Thus, by not wishing to be bound by any
particulat theory, it is believed that the mAb 15EVQ competes
for TLR3 binding against its ligand and prevents ligand-
114
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
induced receptor dimerization, which is required for the
formation of the signaling unit (Liu et al., Science 320:379-
81, 2008). Figure 29 illustrates this direct competition
mechanism for mAb 15EVQ. Depending upon the antibody
concentration, this mechanism would lead to total inhibition
of poly(I:C) or dsRNA induced TLR3 activation.
mAb 12QVQ/QSV and mAb c1068: As shown in Figure 30,
these two antibodies do not have direct clashes with the
dsRNA ligand. Thus, it is unlikely that they would
neutralize TLR3 function in a similar mechanism to that of
mAb 15EVQ. The Fab fragments are also oriented away from the
ligand (Figure 30). Structurally, both mAb 12QVQ/QSV and Fab
c1068 can bind to a signaling unit (SU) without disrupting
its function. Sterically, it is unlikely that the two Fab
fragments of a mAb molecule would be able to bind
simultaneously the two TLR3 molecules in one SU, and thus
prevent dsRNA mediated TLR3 dimerization. By not wishing to
be bound by any particular theory, it is believed that
binding of mAb 12QVQ/QSV or mAb c1068 to TLR3 prevents
clustering of the signaling unit due to steric clashes
between the antibodies and neighboring signaling units.
Binding of TLR3 to dsRNA is not limited to the signaling unit
defined by the dsRNA:TLR3 complex (Liu, et al., Science, 320:
379-81, 2008). It is possible that clustering of multiple
SUs can lead to enhancement of signaling or that efficient
TLR3 signaling requires this clustering. The positioning of
mAb 12QVQ/QSV and mAb c1068 can block the clustering and
result in neutralization of TLR3 activity. The maximal
neutralization effects of antibodies would therefore be
dependent upon the degree of separation of SUs due to
antibody binding. As illustrated in Figure 30, mAb 12QVQ/QSV
would cause larger separation than mAb c1068, and this could
translate to greater potency of mAb 12QVQ/QSV. This is
consistent with observations that mAb c1068 and mAb 15EVQ can
lead to -50% and 100% TLR3 neutralization at saturation
concentrations, respectively, and mAb 12QVQ/QSV exhibits
115
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
intermediate activity. Thus, combined structural and TLR3
neturalization studies suggest a TLR3 signaling model in
which the dsRNA:TLR3 signaling units cluster to achieve
efficient signaling. mAb 12QVQ/QSV and mAb c1068 and also
define a class of antibodies that can partially modulate TLR3
signaling without interfering with ligand binding or receptor
dimerization.
Example 23
TLR3 deficiency improves lipid profiles and liver steatosis
Model
TLR3/ mice were obtained from Dr. Richard A. Flavell
(Yale University) and described previously (Shulman, J. Clin.
Invest. 106:171-6, 2000). Both TLR3 and and wild type (WT)
control mice (C57BL/6) were fed either normal chow or high
fat diet (HFD) (Purina TestDiet #58126) consisting of 60.9%
kcal fat and 20.8% kcal carbohydrates. Mice were maintained
on a 12:12-h light-dark cycle, with water and food ad
libitum. The weight of each mouse was measured weekly; the
data were presented as means SD. Liver samples were taken
for RNA isolation and histological analysis.
Plasma levels of total cholesterol (TC), HDL, LDL, and
triglycerides (TG) were measured using a clinical chemistry
instrument (Alfa Wassermann Diagnostic Technologies, West
Caldwell, NJ). Free fatty acid (FFA) levels were determined
using a NEFA kit (Wako Chemicals, Richmond, VA). Sample
preparation and assays were performed according to the
manufacturer's recommendation.
Total liver RNA was reverse-transcribed with random
hexamers by using TaqMan reverse-transcription reagents kit
(Life Technologies, Carlsbad, CA) according to manufacturer's
protocol. TaqMan probes and primers were purchased from Life
Technologies, Carlsbad, CA. Total of 35 genes involved in
lipid and glucose metabolism as well as inflammation were
116
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
examed for gene expression analysis. The relative
quantification of gene expression was performed using the 2-
DeltaDeltaCt calculations (Arocho et al., Diagn. Mol. Pathol.
15:56-61, 2006).
For histology, livers were isolated and wet weight was
determined. Liver samples were fixed in 10% neutral buffered
formalin, processed for routine paraffin sections, and HE
stained. The level of liver adiposity was determined by
histological evaluation. The statistical analysis was
performed using Mann-Whitney Test.
Results
Animals fed a HFD were hyperinsulinemic and
hyperglycemic after 26 weeks on HFD (see example 15 for 14
week data points). Lipid profiles were examined at 33 weeks.
TLR3-/- mice on chow diet had significantly decreased plasma
TG but similar TC, HDL, LDL, (Table 15) and FFA levels
compared to WT controls. On HFD at 33 weeks, WT control
animals showed elevated plasma TC, HDL, and LDL levels, while
TLR3-/- mice were partially protected (Table 15). Liver
weights (normalized to respective body weight) were reduced
-16%, p<0.05 in TLR3-/- mice fed HFD compared to those of WT
animals on HFD. Histological analysis indicated that TLR3-/-
animals on HDF also had significantly reduced lipid
accumulation in the liver parenchyma when compared to the WT
animals.
Hepatic expression of key genes involved in lipid and
cholesterol metabolism, LXRa and PPAR6, were up-regulated
(30%, p<0.05 and 186%, p<0.001, respectively) in TLR3-/- mice
fed HFD when compared to WT animals, consistent with lower
lipid levels in TLR3 deficient mice. LXRa target genes ABCA1
and SREBP1 were also upregulated (71%, p<0.01 and 131%,
p<0.001, respectively) in these animals. Cross-talk between
TLR signaling and LXR has been reported (Castrillo et al.,
Mol. Cell. 4:805-15, 2003). Upregulation of ABCA1, a
117
CA 02855955 2014-03-11
WO 2013/039471
PCT/US2011/051202
cholesterol transporter, suggests improved cholesterol
transport in HFD fed TLR3 mice.
Table 15.
Diet/Strain TO HDL LDL TG FFA
Chow Diet
WT 79.71 5.56 47.57 4.66 6.57 1.44
104.9 3.36 1.35 0.04
TLR3 75.56 3.24 75.56 3.24 47.67 2.14 5.56
0.34 91.22 4.67 1.21 0.18
p value 0.253 0.492 0.227 <0.05 0.168
High Fat Diet
WT 218.6 19.37 87.00 5.82 25.13
3.31 120.3 7.80 1.44 0.12
TLR3 120.1 120.1 10.32 59.86 4.19 9.00 0.92
123.9 2.50 1.20 0.05
p value <0.001 <0.01 <0.001 0.343 <0.05
The studies reported in this Example demonstrated that
obese TLR3 deficient animals have improved lipid and
cholesterol levels when compared to the wild type animals,
and indicate that antagonizing TLR3 signaling would be a
beneficial therapeutic approach for the treatment of
cardiovascular and metabolic diseases.
118