Note: Descriptions are shown in the official language in which they were submitted.
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
A PROKARYOTIC-TYPE ISOCITRATE DEHYDROGENASE AND ITS
APPLICATION FOR IMPROVING NITROGEN UTILIZATION IN TRANSGENIC
PLANTS
Field of the Invention
moon The invention relates generally to plants with improved nitrogen
utilization and
stress tolerance, more specifically, to heterologous expression of an
isocitrate dehyrogenase
(ICDH) enzyme in plants, including the overexpression and characterization of
a prokaryotic-
based isocitrate dehyrogenase that improves stress tolerance and nitrogen
uptake, metabolism or
both. The invention also includes stacking of the icdh gene with one or more
other transgenes to
improve nitrogen utilization and/or stress tolerance.
Background of the Invention
1-00021 Plants require nitrogen during their vegetative and reproductive
growth phases.
Nitrogen is made available to the plant through soil mineralization, the
application of nitrogen
fertilizer, or both. It has been estimated, however, that between 50 and 70
percent of nitrogen
applied to crops is lost from the plant-soil system [Peoples. M.B. et al.,
"Minimizing Gaseous
Losses of Nitrogen," In Nitrogen Fertilizer in the Environment (Bacon, P.E.,
ed.) Marcel Dekker,
pp. 565-606 (1995)]. Nitrogen is one of the most expensive plant nutrients to
supply, nitrogen
fertilizer is not always available at a reasonable cost, and excessive
application of nitrogen
fertilizer can result in environmental challenges. Corn is an example of an
agronomically
important plant that often requires nitrogen fertilizers to perform at its
genetic potential.
1110031 Native ICDH can exist in the mitochondria, chloroplast and cytosol,
with each
having a different physiological impact although the catalytic action may be
similar. In general.
ICDH1 is found in the cytosol and ICDH2 is found in the chloroplast.
1-00041 For co-factor reducing power, ICDH can use either nicotinamde
adenine dinucleotide
(NAD+) or nicotinamde adenine dinucleotide phosphate (NADP+), depending on
which
metabolic pathway it is active. Some publications indicate that the main
function of ICDH may
be to generate reducing power (NADH, NADPH) for other metabolic reactions, for
example, in
the I3-oxidation of unsaturated fatty acids. Other theories include the
suggestion that the reaction
product, 2-oxyglutarate (OG), could be used to support amino acid synthesis
via the GOGAT
1
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
cycle (Hodges, M. Enzyme redundancy and the importance of 2-oxoglutarate in
plant ammonium
assimilation. J. Exp. Botany (2002), 53, 905). In addition, the over-
expression of the ICDH
enzyme in a stack with another gene or genes may allow the effective
utilization of the additional
carbon skeletons. A previous study of transgenic tobacco plants that
overexpressed a
mitochondrial icdh gene was focused on redox pathways and did not mention nor
evaluate any
possible impact on nitrogen utilization (Gray, G., Villarimo, A., Whitehead,
C., McIntosh, L.
Transgenic Tobacco (Nicotiana tabacum L.) Plants with Increased Expression
Levels of
Mitochondrial NADV--dependent Isocitrate Dehydrogenase: Evidence Implicating
this Enzyme
in the Redox Activation of the Alternative Oxidase, Plant and Cell Physiology
2004; 45, 1413-
1425).
[00051 Regulation of NAD- and NADP-dependent isocitrate dehydrogenases (NAD-
ICDH,
EC 1.1.1.41 and NADP_ICDH. EC 1.1.1.42) is complex due to expression,
substrates,
compartments and post-translational regulation. While it is unclear which ICDH
version
generates OG for amino acids, any such OG would have to be in, or enter, the
chloroplast where
nitrogen is assimilated into amino acids. The literature suggests that plant
cytostolic versions of
ICDH are homodimers with subunits of approximately 47 kD. Mitochondrial ICDH
is suspected
to have more subunits. Bacterial versions of ICDH may be monomeric and have
been
considered to overcome the typical regulation of expression and function that
occurs with plant
ICDH in plants, that is, phosphorylation may inactivate the homodimer.
[00061 Cytostolic NADP-specific ICDH catalyzes the conversion of citrate to
oxoglutarate.
One strategy is to design a construct containing a gene encoding a monomeric
prokaryotic-type
isocitrate dehydrogenase gene (icdh), and to direct overexpression of ICDH in
the cytoplasm of
plants. The expressed ICDH enzyme will enhance the plant's ability to utilize
available nitrogen
via an enhanced flow of carbon into the nitrogen assimilatory mechanism. Here,
we describe the
overexpression and characterization of asynthetic icdh gene based on selection
from among
bacterial icdh sequences and optimized for expression in corn, and the
stacking of a icdh genes
with other transgenes.
Summary of the Invention
[00071 The present invention relates to transgenic plants that have
increased nitrogen use
efficiency, stress tolerance, or both, that have been transformed using a
novel vector construct
2
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
including an icdh nucleic acid sequence that modulates nitrogen use in plants.
A variety of icdh
nucleic acid sequences were identified for use with the present invention from
the several
bacterial and plant genomic sequencing projects that have been archived in
public databases
from which sequences that encode ICDH enzymes with robust activity could be
selected. These
candidate icdh sequences were then screened to deselect those that had a
relatively high content
of poly A regions, which can be inhibitory to expression in plants. The
sequence chosen to
exemplify these icdh sequences was then codon-optimized for expression in
maize (SEQ ID No
1). The invention also includes stacking an icdh gene with one or more
heterologous genes so as
to induce the over-expression of the ICDH enzyme in combination with nitrogen
assimilatory
enzymes. The invention also relates to isolated vectors for transforming
plants and to antibodies
for detecting expression of the nucleotide sequence(s) of interest in the
transformed plants. The
invention also relates to methods of expressing in plants the nucleic acid
molecules
corresponding to the nucleic acid sequences that modulate nitrogen use in
plants.
[00081 Specifically, vectors for transforming plants and bacterial cells
have been
constructed using the nucleotide sequences SEQ ID NO: 1 and 3, as well as
combinations,
variants, fragments, and complements thereof. These vectors include a 5' DNA
promoter
sequence and a 3' terminator sequence, wherein the nucleic acid sequence, the
DNA promoter
sequence, and the terminator sequence are operatively coupled to permit
transcription of the
nucleotide sequence. In some embodiments, the promoter sequence may be a
constitutive plant
promoter or a tissue specific promoter.
[00091 The invention also includes polyclonal antibodies, comprising
polyclonal antibodies
to a polypeptide encoded by nucleotide sequences SEQ ID NO: 1 and 3 and
combinations
thereof.
[000101 The invention also includes plants transformed with a nucleotide
sequences SEQ ID
NO: 1 and 3, as well as combinations, variants and fragments thereof. The
plant is selected from
the group consisting of corn (maize), sorghum, wheat, sunflower, tomato,
crucifers, peppers,
potato, cotton, rice, soybean, sugarbeet, sugarcane, tobacco, barley, and
oilseed rape, Brassica
sp., alfalfa, rye, millet, safflower, peanuts, sweet potato, cassava, coffee,
coconut, pineapple, citrus
trees, cocoa, tea, banana, avocado, fig, guava, mango, olive, papaya, cashew,
macadamia, almond,
oats, vegetables, grasses (such as turf grasses, forage grasses, or pasture
grasses), ornamentals, trees
(such as fruit trees, nut trees, pulp trees, oil palms) and conifers. The
invention also includes a
3
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
component part of such plants, plant seed produced from such plants, and a
plant seed
transformed with a vector construct of the present invention.
[0010] The invention also includes a host cell transformed with a
nucleotide sequence
selected from SEQ ID NO: 1 and 3, and combinations thereof. The host cell may
be a bacterial
cell or a plant cell.
[0011] The invention also includes a method of expressing a nucleic acid
molecule that
modulates nitrogen in a plant, said method comprising the steps of providing a
transgenic plant
or plant seed transformed with a vector construct according to the present
invention, and growing
the transgenic plant or a plant grown from the transgenic plant seed under
conditions effective to
express the nucleic acid molecule in said transgenic plant or said plant grown
from the transgenic
plant seed. Growing of the transgenic plant is effective in increasing
nitrogen uptake of said
transgenic plant or said plant grown from the transgenic plant seed, and/or in
increasing
efficiency of nitrogen utilization of said transgenic plant or said plant
grown from the transgenic
plant seed, and/or alleviating a limitation such that yield is increased in
said transgenic plant or
said plant grown from the transgenic plant seed. The invention also includes
the foregoing
methods wherein a transgenic plant is provided or a transgenic seed is
provided. The invention
also includes the foregoing method wherein the plant is selected from the
group consisting of
corn (maize), sorghum, wheat, sunflower, tomato, crucifers, peppers, potato,
cotton, rice,
soybean, sugarbeet, sugarcane, tobacco, barley, and oilseed rape, Brassica
sp., alfalfa, rye, millet,
safflower, peanuts, sweet potato, cassava, coffee, coconut, pineapple, citrus
trees, cocoa, tea,
banana, avocado, fig, guava, mango, olive, papaya, cashew, macadamia, almond,
oats,
vegetables, grasses (such as turf grasses, forage grasses, or pasture
grasses), ornamentals, trees
(such as fruit trees, nut trees, pulp trees, oil palms) and conifers.
[0012] The invention also includes a method of improving the stress
tolerance of a plant by
expressing a nucleic acid molecule modulated by nitrogen in a plant, said
method comprising the
steps of providing a transgenic plant or plant seed transformed with a vector
construct according
to the present invention and growing the transgenic plant or a plant grown
from the transgenic
plant seed under conditions effective to express the nucleic acid molecule in
said transgenic plant
or said plant grown from the transgenic plant seed.
[0013] The invention also includes a method of altering the morphology of a
plant by
expressing a nucleic acid molecule modulated by nitrogen in a plant, said
method comprising the
4
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
steps of providing a transgenic plant or plant seed transformed with a vector
construct according
to the present invention and growing the transgenic plant or a plant grown
from the transgenic
plant seed under conditions effective to express the nucleic acid molecule in
said transgenic plant
or said plant grown from the transgenic plant seed.
[0014] The invention also includes a vector construct, comprising a
nucleotide sequence
encoding the ICDH amino acid sequence including SEQ ID NO: 2 and 4, and
combinations
thereof, a 5' DNA promoter sequence, and a 3' terminator sequence, wherein the
nucleotide
sequence, the DNA promoter sequence, and the terminator sequence are
operatively coupled to
permit transcription of the nucleotide sequence.
[0015] The invention also includes a vector construct comprising a
nucleotide sequence that
modulates nitrogen in a plant, wherein said nucleotide sequence is selected
from SEQ ID NO: 1
and 3, and combinations thereof; a nucleotide sequence having at least 85%
sequence identity to
the corresponding nucleotide sequence of SEQ ID NO: 1 and 3, and combinations
thereof,
wherein said nucleotide sequence modulates nitrogen in a plant; a nucleotide
sequence selected
from those encoding the ICDH amino acid sequences SEQ ID NO: 2 and 4, and
combinations
thereof; and, a nucleotide sequence encoding an amino acid sequence having at
least 85%
sequence identity to the amino acid sequence of SEQ ID NO: 2 and 4, and
combinations thereof;
wherein said nucleotide sequence modulates nitrogen in a plant, wherein said
construct further
comprises a 5' DNA promoter sequence and a 3' terminator sequence, wherein the
nucleotide
sequence, the DNA promoter sequence, and the terminator sequence are
operatively coupled to
permit transcription of the nucleotide sequence.
Brief Description of the Figures
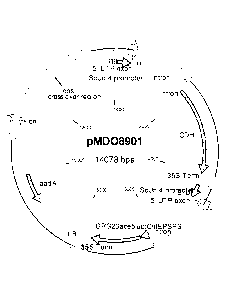
[0016] Fig. 1 is a vector map for the plasmid pMD08901, wherein the Main
elements in the
plasmid (clockwise from top) are: Right Border, ScUbi4 promoter, 5' UTR exon,
intron, icdh
gene. 35S terminator, ScUbi4 promoter, 5' UTR exon, intron, chloroplast
transit peptide from
EPSPS, nagk gene, 35S terminator, ScUbi4 promoter, 5' UTR exon, intron,
chloroplast transit
peptide from EPSPS, elyphosate tolerance SM (GRG23ac35), 35S terminator, Left
Border.
[0017] Fig. 2 is a vector map for the plasmid pMD08902, wherein the main
elements in the
plasmid (clockwise from top) are: Right Border, ScUbi4 promoter, 5' UTR exon,
intron, icdh
gene. 35S terminator, ScUbi4 promoter, 5' UTR exon, intron, chloroplast
transit peptide from
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
EPSPS, nagk gene, 35S terminator, ScUbi4 promoter, 5' UTR exon, intron,
glyphosate tolerance
SM, 35S terminator, Left Border.
Detailed Description of Preferred Embodiments
[0018] The development of plant varieties that use nitrogen more
efficiently will reduce the
need for excessive inputs of nitrogen, save production costs for farmers,
benefit farmers in
developing countries who do not have access to fertilizer inputs, and reduce
environmental
contamination associated with the application of excessive nitrogen
fertilizers. One approach
that has been used in the development of plant varieties with improved
nitrogen utilization relies
on conventional plant breeding techniques. However, such approaches have had
variable success
due to lack of specification in the genetic recombination.
[0019] There is a need to develop plant cultivars that absorb and use
nitrogen more
efficiently. Plant scientists have adopted the shorthand term nitrogen use
efficiency (NUE), and
a variety of methods of measuring and evaluating NUE have been developed
[Craswell, E.T. and
Godwin, D.C. (1984) The efficiency of nitrogen fertilizers applied to cereals
grown in different
climates. In Advances in Plant Nutrition (Vol. 1) (Tinker, P.B. and Lauchli,
A., eds), pp. 1-55,
Praeger Publishers; Steenbjer2, F. and Jakobsen, S.T. (1963) Plant nutrition
and yield curves.
Soil Sci. 95,69-90; Siddiqi, M.Y. and Glass, D.M. (1981) Utilization index: a
modified
approach to the estimation and comparison of nutrient utilization efficiency
in plants. J. Plant
Nutr. 4,289-302; Moll, R.H. et al. (1982) Analysis and interpretation of
factors which contribute
to efficiency of nitrogen utilization. Agron. J. 74,562-564]. There are
differences in the specific
definitions, and context of use. For example, some definitions are based on
total biomass while
others are based on the weight of grain yielded. Another set of definitions
uses the efficiency of
extracting nitrogen from the soil. The efficiency with which applied nitrogen
is used to improve
grain yield may be measured by agronomic efficiency (AE), the product of
physiological
efficiency and utilization efficiency, or NUEg which is the product of uptake
efficiency and
utilization efficiency. Other definitions take physiological factors into
account.
[0020] As used in this specification, the term nitrogen use efficiency, or
NUE, is defined to
include a measurable change in any of the main nitrogen metabolic pool sizes
in the assimilation
pathways (for example, may include a measurable change in one or more of the
following:
nitrate, nitrite, ammonia, glutamic acid, aspartic acid, dutamine, asparagine,
lysine, leucine,
6
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
threonine, methionine, glycine, tryptophan, tyrosine, total protein content of
a plant part, total
nitrogen content of a plant part, and/or chlorophyll content), or where the
plant is shown to
provide the same or elevated biomass or harvestable yield at lower nitrogen
fertilization levels,
or where the plant is shown to provide elevated biomass or harvestable yields
at the same
nitrogen fertilization levels when compared to a plant that has not been
transformed with a
nitrogen-modulating nucleic acid construct of the invention. A "measurable
change" can include
an increase or a decrease in the amount of any component ("metabolic pool") of
the nitrogen
assimilation pathway. A change can include either a decrease or an increase in
one or more
metabolic pools in the pathway, or a decrease in one or more pools with a
concomitant increase
in one or more other pool(s), such as when one intermediate in the nitrogen
assimilation pathway
is being utilized for the purpose of generating another intermediate or
product of the pathway.
For example, in the conversion of glutamate to glutamine, the level of
glutamate may decrease
while the level of glutamine may increase. Thus, while not being bound by any
particular theory
or mechanism, any change in one or more of these pools indicates that nitrogen
is being utilized
more efficiently by the plant.
[0021] An increase in nitrogen utilization efficiency can be associated
with about a 5%, about
a 10%, 15%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 100%, 125%, 150%, about a
200%
or greater measurable change in any of the main nitrogen metabolic pool sizes
in the assimilation
pathway. In one embodiment, the transgenic plants of the invention have an
increased nitrogen
uptake from the environment when compared to a plant that does not contain a
nitrogen-
modulating sequence of the invention. By -nitrogen modulating sequence" it is
intended to
mean a nucleotide or amino acid sequence that modulates NUE, by way of non-
limiting
example: either by generating an enzyme that impacts NUE, or by generating a
protein that
interacts with the components involved in NUE, or by generating a protein that
impacts the
internal homeostatic signal cascade regulating NUE, or by a combination of
these mechanisms
that results in a measurable change in N uptake, N assimilation, N metabolism,
N transport, N
utilization, N storage, or a combinations of these. The present invention
further provides a
method of improving stress tolerance in a plant by expressing one or more
nitrogen-modulating
nucleotide sequences within the plant. In one embodiment, the nitrogen-
modulating nucleotide
sequence is SEQ ID NO: 1, or variants and fragments thereof. In another
embodiment, the
nitrogen-modulating nucleotide sequence is a nucleotide sequence that encodes
SEQ ID NO: 2,
7
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
or variants and fragments thereof. In another embodiment, the nitrogen-
modulating nucleotide
sequence is a nucleotide sequence that encodes SEQ ID NO 1 plus SEQ ID NO: 2,
or variants
and fragments thereof, respectively.
[0022] As used herein, the term "stress" or "stress condition" refers to
the exposure of a plant,
plant cell, or the like, to a physical, environmental, biological or chemical
agent or condition that
has an adverse effect on metabolism, growth, development, propagation and/or
survival of the
plant (collectively "growth"). A stress can be imposed on a plant due, for
example, to an
environmental factor such as water (e.g., flooding, drought, dehydration),
anaerobic conditions
(e.g., a low level of oxygen), abnormal osmotic conditions, salinity or
temperature (e.g., hot/heat,
cold, freezing, frost), a deficiency of nutrients such as nitrogen, phosphate,
potassium, sulfur,
micronutrient, or exposure to pollutants, or by a hormone, second messenger or
other molecule.
Anaerobic stress, for example, is due to a reduction in oxygen levels (hypoxia
or anoxia)
sufficient to produce a stress response. A flooding stress can be due to
prolonged or transient
immersion of a plant, plant part, tissue or isolated cell in a liquid medium
such as occurs during
monsoon, wet season, flash flooding or excessive irrigation of plants, or the
like. A cold stress or
heat stress can occur due to a decrease or increase, respectively, in the
temperature from the
optimum range of growth temperatures for a particular plant species. Such
optimum growth
temperature ranges are readily determined or known to those skilled in the
art. Dehydration
stress can be induced by the loss of water, reduced turgor, or reduced water
content of a cell,
tissue, organ or whole plant. Drought stress can be induced by or associated
with the deprivation
of water or reduced supply of water to a cell, tissue, organ or organism.
Saline stress (salt stress)
can be associated with or induced by a perturbation in the osmotic potential
of the intracellular or
extracellular environment of a cell. Osmotic stress also can be associated
with or induced by a
change, for example, in the concentration of molecules in the intracellular or
extracellular
environment of a plant cell, particularly where the molecules cannot be
partitioned across the
plant cell membrane.
[0023] An improvement in stress tolerance can be assessed by any
quantitative or qualitative
measure of plant performance under a given stress condition and is relative to
the performance of
a plant grown under the same stress conditions that has not been transformed
with a nitrogen-
modulating sequence of the invention. Thus, the plants may exhibit improved
nitrogen contents,
altered amino acid or protein compositions, altered carbohydrate composition,
altered oil
8
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
composition, vigorous growth characteristics, increased vegetative yields or
better seed yields
and qualities. These plants may be identified by examining any of following
parameters: 1) the
rate of growth, measured in terms of rate of increase in fresh or dry weight;
2) vegetative yield of
the mature plant, in terms of fresh or dry weight; 3) the seed or fruit yield;
4) the seed or fruit
weight; 5) the total nitrogen content of the plant; 6) the total nitrogen
content of the fruit or seed;
7) the free amino acid content of the plant; 8) the free amino acid content of
the fruit or seed; 9)
the total protein content of the plant; 10) the total protein content of the
fruit or seed; 11)
measurable change in carbohydrates or oils. The procedures and methods for
examining these
parameters are well known to those skilled in the art. These methods may
involve enzymatic
assays and immunoassays to measure enzyme/protein levels; assays to measure
the amino acid
composition. free amino acid pool or total nitrogen content of various plant
tissues; measurement
of growth rates in terms of fresh weight gains over time; or measurement of
plant yield in terms
of total dry weight and/or total seed weight.
Transformation of Bacterial or Plant Cells
[0024] Provided herein are novel nucleotide sequences that modulate
nitrogen utilization
efficiency in plants. Also provided are amino acid sequences of the proteins
of the invention,
that may be nitrogen-modulating or modulated by nitrogen concentration.
[0025] The nitrogen-modulating nucleotide sequences of the invention may be
modified to
obtain or enhance expression in plant cells. The nitrogen-modulating sequences
of the invention
may be provided in expression cassettes for expression in the plant of
interest. -Plant expression
cassette" includes DNA constructs that are capable of resulting in the
expression of a protein
from an open reading frame in a plant cell. The cassette will include in the
5'-3' direction of
transcription, a transcriptional initiation region (i.e., promoter) operably-
linked to a DNA
sequence of the invention, and a transcriptional and translational termination
region (i.e.,
termination region) functional in plants. The cassette may additionally
contain at least one
additional gene to be co-transformed into the organism, such as a selectable
marker gene or a
stacked gene of different function. Alternatively, the additional gene(s) can
be provided on
multiple expression cassettes. Such an expression cassette is provided with a
plurality of
restriction sites for insertion of the nitrogen-modulating sequence to be
under the transcriptional
regulation of the regulatory regions.
9
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
[0026] By "promoter" is intended a nucleic acid sequence that functions to
direct
transcription of a downstream coding sequence. The promoter, together with
other transcriptional
and translational regulatory nucleic acid sequences (also termed as "control
sequences"), are
necessary for the expression of a DNA sequence of interest. Preferably, the
promoter is one that
is known to stimulate transcription in the organism into which the nucleotide
sequence of the
invention is being introduced.
[0027] The promoter may be native or analogous, or foreign or heterologous,
to the plant host
and/or to the DNA sequence of the invention. Additionally, the promoter may be
the natural
sequence or alternatively a synthetic sequence. Where the promoter is "native"
or "homologous"
to the plant host, it is intended that the promoter is found in the native
plant into which the
promoter is introduced. Where the promoter is "foreign" or "heterologous" to
the DNA
sequence of the invention, it is intended that the promoter is not the native
or naturally occurring
promoter for the operably linked DNA sequence of the invention. lieterologous"
generally
refers to the nucleic acid sequences that are not endogenous to the cell or
part of the native
genome in which they are present, and have been added to the cell by
infection, transfection,
microinjection, electroporation, microprojection, or the like. By "operably
linked" is intended a
functional linkage between a promoter and a second sequence, wherein the
promoter sequence
initiates and mediates transcription of the DNA sequence corresponding to the
second sequence.
Generally. "operably linked" means that the nucleic acid sequences being
linked are contiguous,
including exons and introns and, where necessary to join two protein coding
regions, contiguous
and in the same reading frame.
[0028] In one embodiment, the promoter is a constitutive promoter. Suitable
constitutive
promoters for use in plants include: the promoters from plant viruses, such as
the peanut
chlorotic streak caulimovirus (PC1SV) promoter (U.S. Pat. No. 5,850,019); the
35S promoter
from cauliflower mosaic virus (CaMV) (Odell etal. (1985) Nature 313:810-812);
promoters of
Chlorella virus methyltransferase genes (U.S. Pat. No. 5,563,328) and the full-
length transcript
promoter from figwort mosaic virus (FMV) (U.S. Pat. No. 5,378,619); the
promoters from such
genes as rice actin (McElroy etal. (1990) Plant Cell 2:163-171); ubiquitin
(Christensen et al.
(1989) Plant Mol. Biol. 12:619-632 and Christensen etal. (1992) Plant Mol.
Biol. 18:675-689),
including the TrpPro5 promoter (U.S. Patent Application No. 10/377,318; filed
March 16, 2005);
pEMU (Last etal. (1991) Theor. Appl. Genet. 81:581-588): MAS (Velten et al.
(1984) EMBO J.
3:2723-2730); maize H3 histone (Lepetit et al. (1992) Mol. Gen. Genet. 231:276-
285 and
Atanassova etal. (1992) Plant J. 2(3):291-300); Brassica napus ALS3 (PCT
application WO
97/41228); and promoters of various Agrobacterium genes (see U.S. Pat. Nos.
4,771,002;
5,102,796; 5,182,200; and 5,428,147).
[0029] In another embodiment, the promoter is a tissue-specific promoter. A
list of
commonly-used tissue-specific promoters can be found in Reviewed in Moore et
al. (2006) Plant
J. 45(4):651-683.
[0030] Often, such constructs will also contain 5' and 3' untranslated
regions. Such constructs
may contain a "signal sequence" or "leader sequence" to facilitate co-
translational or post-
translational transport of the peptide of interest to certain intracellular
structures such as the
chloroplast (or other plastid), endoplasmic reticulum, or Golgi apparatus, or
to be secreted. For
example, the gene can be engineered to contain a signal peptide to facilitate
transfer of the
peptide to the endoplasmic reticulum. By "signal sequence" is intended a
sequence that is known
or suspected to result in co-translational or post-translational peptide
transport across the cell
membrane. In eukaryotes, this typically involves secretion into the Golgi
apparatus, with some
resulting glycosylation. By "leader sequence" is intended any sequence that
when translated,
results in an amino acid sequence sufficient to trigger co-translational
transport of the peptide
chain to a sub-cellular organelle. Thus, this includes leader sequences
targeting transport and/or
glycosylation by passage into the endoplasmic reticulum, passage to vacuoles,
plastids including
chloroplasts, mitochondria, and the like. It may also be preferable to
engineer the plant
expression cassette to contain an intron, such that mRNA processing of the
intron is required for
expression.
[0031] By "3' untranslated region" is intended a nucleotide sequence
located downstream of a
coding sequence. Polyadenylation signal sequences and other sequences encoding
regulatory
signals capable of affecting the addition of polyadenylic acid tracts to the
3' end of the mRNA
precursor are 3' untranslated regions. By "5' untranslated region" is intended
a nucleotide
sequence located upstream of a coding sequence.
[0032] Other upstream or downstream untranslated elements include
enhancers. Enhancers
are nucleotide sequences that act to increase the expression of a promoter
region. Enhancers are
well known in the art and include, but are not limited to, the SV40 enhancer
region and the 35S
enhancer element.
11
CA 2906278 2017-11-23
[0033] The termination region may be native with the transcriptional
initiation region, may be
native with the nitrogen-modulating sequence of the present invention, or may
be derived from
another source. Convenient termination regions are available from the Ti-
plasmid of A.
tumefaciens, such as the octopine synthase and nopaline synthase termination
regions, or the
potato proteinase inhibitor II sequence (PinII) as described in Liu et al.
(2004) Acta Biochim
Biophys Sin 36(8):553-558. See also Guerineau et al. (1991) Mol. Gen. Genet.
262:141-144;
Proudfoot (1991) Cell 64:671-674; Sanfacon et al. (1991) Genes Dev. 5:141-149;
Mogen etal.
(1990) Plant Cell 2:1261-1272; Munroe etal. (1990) Gene 91:151-158; Ballas
etal. (1989)
Nucleic Acids Res. 17:7891-7903; and Joshi et al. (1987) Nucleic Acid Res.
15:9627-9639.
[0034] Where appropriate, the gene(s) may be optimized for increased
expression in the
transformed host cell. That is, the genes can be synthesized using host cell-
preferred codons for
improved expression, or may be synthesized using codons at a host-preferred
codon usage
frequency. Generally, the GC content of the gene will be increased. See, for
example, Campbell
and Gown i (1990) Plant Physiol. 92:1-11 for a discussion of host-preferred
codon usage.
Methods are known in the art for synthesizing host-preferred genes. See, for
example, U.S.
Patent Nos. 6,320,100; 6,075,185; 5,380,831; and 5,436,391, U.S. Published
Application Nos.
20040005600 and 20010003849, and Murray etal. (1989) Nucleic Acids Res. 17:477-
498.
[0035] In one embodiment, the nucleic acids of interest are targeted to the
chloroplast for
expression. In this manner, where the nucleic acid of interest is not directly
inserted into the
chloroplast, the expression cassette will additionally contain a nucleic acid
encoding a transit
peptide to direct the gene product of interest to the chloroplasts. Such
transit peptides are known
in the art. See, for example, Von Heijne et al. (1991) Plant Mol. Biol. Rep.
9:104-126; Clark et
al. (1989) J. Biol. Chem. 264:17544-17550; Della-Cioppa etal. (1987) Plant
Physiol. 84:965-
968; Romer etal. (1993) Biochern. Biophys. Res. Commun. 196:1414-1421; and
Shah etal.
(1986) Science 233:478-481.
[0036] The nucleic acids of interest to be targeted to the chloroplast may
be optimized for
expression in the chloroplast to account for differences in codon usage
between the plant nucleus
and this organelle. In this manner, the nucleic acids of interest may be
synthesized using
chloroplast-preferred codons. See, for example, U.S. Patent No. 5,380,831.
12
CA 2906278 2017-11-23
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
[0037] Typically this "plant expression cassette" will be inserted into a
"plant transformation
vector." By "transformation vector" is intended a DNA molecule that is
necessary for efficient
transformation of a cell. Such a molecule may consist of one or more
expression cassettes, and
may be organized into more than one "vector" DNA molecule. For example, binary
vectors are
plant transformation vectors that utilize two non-contiguous DNA vectors to
encode all requisite
cis- and trans-acting functions for transformation of plant cells (Hellens and
Mullineaux (2000)
Trends in Plant Science 5:446-451). "Vector" refers to a nucleic acid
construct designed for
transfer between different host cells. "Expression vector" refers to a vector
that has the ability to
incorporate, integrate and express heterologous DNA sequences or fragments in
a foreign cell.
[0038] This plant transformation vector may be comprised of one or more DNA
vectors
needed for achieving plant transformation. For example, it is a common
practice in the art to
utilize plant transformation vectors that are comprised of more than one
contiguous DNA
segment. These vectors are often referred to in the art as "binary vectors."
Binary vectors as
well as vectors with helper plasmids are most often used for Agrobacterium-
mediated
transformation, where the size and complexity of DNA segments needed to
achieve efficient
transformation is quite large, and it is advantageous to separate functions
onto separate DNA
molecules. Binary vectors typically contain a plasmid vector that contains the
cis-acting
sequences required for T-DNA transfer (such as left border and right border),
a selectable marker
that is engineered to be capable of expression in a plant cell, and a
"nucleotide sequence of
interest" (a nucleotide sequence engineered to be capable of expression in a
plant cell for which
generation of transgenic plants is desired). Also present on this plasmid
vector are sequences
required for bacterial replication. The cis-acting sequences are arranged in a
fashion to allow
efficient transfer into plant cells and expression therein. For example, the
selectable marker gene
and the gene of interest are located between the left and right borders. Often
a second plasmid
vector contains the trans-acting factors that mediate T-DNA transfer from
Agrobacterium to
plant cells. This plasmid often contains the virulence functions (Vir genes)
that allow infection of
plant cells by Agrobacterium, and transfer of DNA by cleavage at border
sequences and vir-
mediated DNA transfer, as is understood in the art (Hellens and Mullineaux
(2000) Trends in
Plant Science, 5:446-451). Several types of Agrobacterium strains (e.g.
LBA4404, GV3101,
EHA101, EHA105, etc.) can be used for plant transformation. The second plasmid
vector is not
13
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
necessary for transforming the plants by other methods such as
microprojection, microinjection,
electroporation, polyethylene glycol, etc.
Altered or Improved Variants Useful in the Constructs of the Invention
[0039] It is recognized that nucleotide and amino acid sequences useful in
the present
invention may be altered by various methods, and that these alterations may
result in sequences
encoding proteins with amino acid sequences different than that encoded by the
nitrogen-
modulating sequences disclosed herein.
[0040] Nucleotide sequences useful in the present invention include the
sequences set forth in
SEQ ID NO: 1 and 3. and combinations, variants, fragments, and complements
thereof. As used
herein, the term -nucleotide sequence" or -nucleic acid molecule" is intended
to include DNA
molecules (e.g., cDNA or genomic DNA) and RNA molecules (e.g., mRNA) and
analogs of the
DNA or RNA generated using nucleotide analogs. The nucleic acid molecules can
be single-
stranded or double-stranded, but preferably are double-stranded DNA. By
"complement" is
intended a nucleotide sequence that is sufficiently complementary to a given
nucleotide sequence
such that it can hybridize to the given nucleotide sequence to thereby form a
stable duplex. The
corresponding amino acid sequences for the nitrogen-modulating proteins
encoded by these
nucleotide sequences are set forth in SEQ ID NO: 2 and 4, as well as
combinations, variants and
fragments thereof. The invention also encompasses the use of nucleic acid
molecules comprising
nucleotide sequences encoding partial-length nitrogen-modulating proteins, and
complements
thereof.
[0041] Nucleic acid molecules that are fragments of these nitrogen-
modulating nucleotide
sequences are also useful in the present invention. By "fragment" is intended
a portion of a
nucleotide sequence encoding a nitrogen-modulating protein. A fragment of a
nucleotide
sequence may encode a biologically active portion of a nitrogen-modulateing
protein, or it may
be a fragment that can be used as a hybridization probe or PCR primer using
methods disclosed
below. Nucleic acid molecules that are fragments of a nitrogen-modulating
nucleotide sequence
comprise at least about 15, 20, 50, 75, 100, 200, 300, 350, or at least about
400 contiguous
nucleotides, or up to the number of nucleotides present in a full-length
nitrogen-modulating
nucleotide sequence disclosed herein depending upon the intended use. By
"contiguous"
nucleotides is intended nucleotide residues that are immediately adjacent to
one another.
14
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
[0042] Polypeptides that are fragments of these nitrogen-modulating
polypeptides are also
useful in the present invention. By "fragment" is intended a portion of an
amino acid sequence
encoding a nitrogen-modulating protein as set forth SEQ ID NO: 2 and/or 4, and
that retains
nitrogen utilization efficiency. A biologically active portion of a nitrogen-
modulating protein
can be a polypeptide that is, for example, 10, 25, 50, 100, 125, 150, 175,
200, 250, 300, 350, 400
or more amino acids in length. Such biologically active portions can be
prepared by recombinant
techniques and evaluated for nitrogen utilization efficiency. As used here, a
fragment comprises
at least 8 contiguous amino acids of SEQ ID NO: 2 and/or 4. The invention
encompasses other
fragments, however, such as any fragment in the protein greater than about 10,
20, 30, 50, 100,
150, 200, 250, 300, 350, or 400 amino acids.
[0043] The invention also encompasses the use of variant nucleic acid
molecules, or variant
amino acid sequences, in the methods and compositions of the inventions.
"Variants" of the
nitrogen-modulating nucleotide sequences include those sequences that encode a
nitrogen-
modulating protein disclosed herein but that differ conservatively because of
the degeneracy of
the genetic code, as well as those that are sufficiently identical as
discussed above. Naturally
occurring allelic variants can be identified with the use of well-known
molecular biology
techniques, such as polymerase chain reaction (PCR) and hybridization
techniques as outlined
below. Variant nucleotide sequences also include synthetically derived
nucleotide sequences
that have been generated, for example, by using site-directed mutagenesis but
which still encode
the nitrogen-modulating proteins disclosed in the present invention as
discussed below. Variant
proteins useful in the present invention are biologically active, that is they
retain the desired
biological activity of the native protein, that is, nitrogen utilization
efficiency and/or improved
stress tolerance.
[0044] By "variants" is intended proteins or polypeptides having an amino
acid sequence that
is at least about 60%, 65%, about 70%, 75%, 80%, 85%, or 90%, 91%, 92%, 93%,
94%, 95%,
96%, 97%, 98% or 99% identical to the amino acid sequences of SEQ ID NO: 2
and/or 4.
Variants also include polypeptides encoded by a nucleic acid molecule that
hybridizes to a
nucleic acid molecule of SEQ ID NO: 1 and/or 3, or a complement thereof, under
stringent
conditions. Variants include polypeptides that differ in amino acid sequence
due to mutagenesis.
Variant proteins encompassed by the present invention are biologically active,
that is they
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
continue to possess the desired biological activity of the native protein,
that is, retain nitrogen
utilization efficiency and/or improved stress tolerance.
[0045] Preferred nitrogen-modulating proteins useful in the present
invention are encoded by
a nucleotide sequence sufficiently identical to a nucleotide sequence of SEQ
ID NO: 1 and/or 3.
The term "sufficiently identical" is intended an amino acid or nucleotide
sequence that has at
least about 60% or 65% sequence identity, about 70% or 75% sequence identity,
about 80% or
85% sequence identity, or about 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or
99%
sequence identity compared to a reference sequence using one of the alignment
programs
described herein using standard parameters. One of skill in the art will
recognize that these
values can be appropriately adjusted to determine corresponding identity of
proteins encoded by
two nucleotide sequences by taking into account codon degeneracy, amino acid
similarity,
reading frame positioning, and the like.
[0046] To determine the percent identity of two amino acid sequences or of
two nucleic acids,
the sequences are aligned for optimal comparison purposes. The percent
identity between the
two sequences is a function of the number of identical positions shared by the
sequences (i.e.,
percent identity = number of identical positions/total number of positions
(e.g., overlapping
positions) x 100). In one embodiment, the two sequences are the same length.
The percent
identity between two sequences can be determined using techniques similar to
those described
below, with or without allowing gaps. In calculating percent identity,
typically exact matches
are counted.
[0047] The determination of percent identity between two sequences can be
accomplished
using a mathematical algorithm. A nonlimiting example of a mathematical
algorithm utilized for
the comparison of two sequences is the algorithm of Karlin and Altschul (1990)
Proc. Natl.
Acad. Sci. USA 87:2264, modified as in Karlin and Altschul (1993) Proc. Natl.
Acad. Sci. USA
90:5873-5877. Such an algorithm is incorporated into the BLASTN and BLASTX
programs of
Altschul et al. (1990) J. Mol. Biol. 215:403. BLAST nucleotide searches can be
performed with
the BLASTN program, score = 100, wordlength = 12, to obtain nucleotide
sequences
homologous to nitrogen-modulating nucleic acid molecules of the invention.
BLAST protein
searches can be performed with the BLASTX program, score = 50, wordlength = 3,
to obtain
amino acid sequences homologous to nitrogen-modulating protein molecules of
the invention.
To obtain gapped alignments for comparison purposes, Gapped BLAST can be
utilized as
16
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
described in Altschul et al. (1997) Nucleic Acids Res. 25:3389. Alternatively,
PSI-Blast can be
used to perform an iterated search that detects distant relationships between
molecules. See
Altschul et al. (1997) supra. When utilizing BLAST, Gapped BLAST, and PSI-
Blast programs,
the default parameters of the respective programs (e.g., BLASTX and BLASTN)
can be used.
See www.ncbi.nlm.nih.gov. Another non-limiting example of a mathematical
algorithm utilized
for the comparison of sequences is the ClustalW algorithm (Higgins et al.
(1994) Nucleic Acids
Res. 22:4673-4680). ClustalW compares sequences and aligns the entirety of the
amino acid or
DNA sequence, and thus can provide data about the sequence conservation of the
entire amino
acid sequence. The ClustalW algorithm is used in several commercially
available DNA/amino
acid analysis software packages, such as the ALIGNX module of the Vector NTI
Program Suite
(Invitrogen Corporation, Carlsbad, CA). After alignment of amino acid
sequences with
ClustalW, the percent amino acid identity can be assessed. A non-limiting
example of a software
program useful for analysis of ClustalW alignments is GENEDOCTM. GENEDOCTM
(Karl
Nicholas) allows assessment of amino acid (or DNA) similarity and identity
between multiple
proteins. Another non-limiting example of a mathematical algorithm utilized
for the comparison
of sequences is the algorithm of Myers and Miller (1988) CABIOS 4:11-17. Such
an algorithm is
incorporated into the ALIGN program (version 2.0), which is part of the GCG
sequence
alignment software package (available from Accelrys, Inc., 9865 Scranton Rd.,
San Diego,
California, USA). When utilizing the ALIGN program for comparing amino acid
sequences, a
PAM120 weight residue table, a gap length penalty of 12, and a gap penalty of
4 can be used.
[0048] A preferred program is GAP version 10, which used the algorithm of
Needleman and
Wunsch (1970) I Mol. Biol. 48:443-453. GAP Version 10 may be used with the
following
parameters: % identity and % similarity for a nucleotide sequence using GAP
Weight of 50 and
Length Weight of 3, and the nwsgapdna.cmp scoring matrix; % identity and %
similarity for an
amino acid sequence using GAP Weight of 8 and Length Weight of 2, and the
BLOSUM62
Scoring Matrix. Equivalent programs may also be used. By "equivalent program"
is intended
any sequence comparison program that, for any two sequences in question,
generates an
alignment having identical nucleotide or amino acid residue matches and an
identical percent
sequence identity when compared to the corresponding alignment generated by
GAP Version 10.
[0049] The skilled artisan will further appreciate that changes can be
introduced by mutation
into the nucleotide sequences of the invention thereby leading to changes in
the amino acid
17
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
sequence of the encoded nitrogen-modulating protein, without altering the
biological activity of
the protein. Thus, variant isolated nucleic acid molecules can be created by
introducing one or
more nucleotide substitutions, additions, or deletions into the corresponding
nucleotide sequence
disclosed herein, such that one or more amino acid substitutions, additions or
deletions are
introduced into the encoded protein. Mutations can be introduced by standard
techniques, such
as site-directed mutagenesis and PCR-mediated mutagenesis. Such variant
nucleotide sequences
are also encompassed by the present invention.
[0050] For example, conservative amino acid substitutions may be made at one
or more
predicted, preferably nonessential amino acid residues. A "nonessential" amino
acid residue is a
residue that can be altered from the wild-type sequence of a nitrogen-
modulating protein without
altering the biological activity, whereas an "essential" amino acid residue is
required for
biological activity. A "conservative amino acid substitution" is one in which
the amino acid
residue is replaced with an amino acid residue having a similar side chain.
Families of amino
acid residues having similar side chains have been defined in the art. These
families include
amino acids with basic side chains (e.g., lysine, arginine, histidine), acidic
side chains (e.g.,
aspartic acid, glutamic acid), uncharged polar side chains (e.g., glycine,
asparagine, glutamine,
serine, threonine, tyrosine, cysteine), nonpolar side chains (e.g., alanine,
valine, leucine,
isoleucine, proline, phenylalanine, methionine, tryptophan), beta-branched
side chains (e.g.,
threonine, valine, isoleucine) and aromatic side chains (e.g., tyrosine,
phenylalanine, tryptophan,
histidine). Amino acid substitutions may be made in nonconserved regions that
retain function.
In general, such substitutions would not be made for conserved amino acid
residues, or for amino
acid residues residing within a conserved motif, where such residues are
essential for protein
activity. However, one of skill in the art would understand that functional
variants may have
minor conserved or nonconserved alterations in the conserved residues.
Examples of residues
that are conserved and that may be essential for protein activity include, for
example, residues
that are identical between all proteins contained in an alignment of similar
or related sequences
known to be involved in nitrogen assimilation. Examples of residues that are
conserved but that
may allow conservative amino acid substitutions and still retain activity
include, for example,
residues that have only conservative substitutions between all proteins
contained in an alignment
of similar or related sequences known to be involved in nitrogen assimilation.
18
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
[0051] Alternatively, variant nucleotide sequences can be made by
introducing mutations
randomly along all or part of the coding sequence, such as by saturation
mutagenesis, and the
resultant mutants can be screened for ability to confer nitrogen utilization
efficiency to identify
mutants that retain activity. Following mutagenesis, the encoded protein can
be expressed
recombinantly, and the activity of the protein can be determined using
standard assay techniques.
[0052] Using methods such as PCR, hybridization, and the like,
corresponding nitrogen-
modulating sequences can be identified, such sequences having substantial
identity to the
sequences of the invention. See, for example, Sambrook J., and Russell, D.W.
(2001) Molecular
Cloning: A Laboratory Manual. (Cold Spring Harbor Laboratory Press, Cold
Spring Harbor,
NY) and Innis, etal. (1990) PCR Protocols: A Guide to Methods and Applications
(Academic
Press, NY). In a hybridization method, all or part of the nitrogen-modulating
nucleotide
sequence can be used to screen cDNA or genomic libraries. Methods for
construction of such
cDNA and genomic libraries are generally known in the art and are disclosed in
Sambrook and
Russell, 2001, supra.
[0053] Variants and fragments of the nucleotide or amino acid sequences of
the present
invention generally will encode protein fragments that retain the biological
activity of the full-
length nitrogen-modulating protein; i.e., retain nitrogen utilization
efficiency. By "retains
nitrogen utilization efficiency" is intended that the variant or fragment will
have at least about
30%, at least about 50%. at least about 70%, or at least about 80% of the
nitrogen utilization
efficiency and/or stress tolerance of the full-length nitrogen-modulating
protein disclosed herein
as SEQ ID NO: 2 and/or 4, or the full-length nitrogen-modulating nucleotide
sequence disclosed
herein as SEQ ID NO: 1 and/or 3. Methods for monitoring nitrogen utilization
efficiency include
detecting a change in any of the main nitrogen metabolic pool sizes in the
assimilation pathways
(for example, a measurable change in nitrate, nitrite, ammonia, glutamic acid,
aspartic acid,
glutamine, asparagine, lysine, leucine, threonine, methionine, glycine,
tryptophan, tyrosine, total
protein content of a plant part, total nitrogen content of a plant part,
and/or chlorophyll content)
or detecting the ability of a plant to provide the same or elevated yield at
lower nitrogen
fertilization levels, or the ability of a plant to provide elevated yields at
the same nitrogen
fertilization levels when compared to a plant that does not contain or express
a nitrogen-
modulating sequence of the invention. The designation of "same" or "lower"
nitrogen
fertilization levels refers to the level of nitrogen generally applied to a
plant not expressing a
19
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
nitrogen-modulating sequence of the invention. Sufficient nitrogen levels are
known in the art
for the majority, if not all, plant varieties of interest. Additional guidance
may be found in, for
example, Hewitt (1966) Sand and Water Culture Methods Used in the Study of
Plant Nutrition,
2nd ed., Farnham Royal (B uck s ) , Commonwealth Agricultural Bureaux; and.
Hewitt (1975)
Plant Mineral Nutrition, London, English University Press.
[0054] The polypeptide sequences useful in the present invention may be
altered in various
ways including amino acid substitutions, deletions, truncations, and
insertions. Methods for such
manipulations are generally known in the art. For example, amino acid sequence
variants of the
nitrogen-modulating proteins disclosed herein can be prepared by mutations in
the nucleotide
sequences. This may also be accomplished by one of several forms of
mutagenesis and/or in
directed evolution. In some aspects, the changes encoded in the amino acid
sequence will not
substantially affect function of the protein. Such variants will possess the
desired nitrogen
utilization efficiency. However, it is understood that the ability of the
nitrogen-modulating
sequences of the invention to alter or improve nitrogen utilization may be
further improved by
one use of such techniques upon the compositions of this invention. For
example, one may
express the nucleotide sequences disclosed herein in host cells that exhibit
high rates of base
misincorporation during DNA replication, such as XL-1 Red (Stratagene, La
Jolla, CA). After
propagation in such strains, one can isolate the DNA (for example by preparing
plasmid DNA, or
by amplifying by PCR and cloning the resulting PCR fragment into a vector),
transform it into
plants as described elsewhere herein, and measure nitrogen utilization
efficiency.
[0055] Alternatively, alterations may be made to the protein sequence of
many proteins at the
amino or carboxy terminus without substantially affecting activity. This can
include insertions,
deletions, or alterations introduced by modern molecular methods, such as PCR,
including PCR
amplifications that alter or extend the protein coding sequence by virtue of
inclusion of amino
acid encoding sequences in the oligonucleotides utilized in the PCR
amplification. Alternatively,
the protein sequences added can include entire protein-coding sequences, such
as those used
commonly in the art to generate protein fusions. Such fusion proteins are
often used to (1)
increase expression of a protein of interest, (2) introduce a binding domain,
enzymatic activity,
or epitope to facilitate either protein purification, protein detection, or
other experimental uses
known in the art, or, (3) target secretion or translation of a protein to a
subcellular organelle, such
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
as the periplasmic space of gram-negative bacteria, or the endoplasmic
reticulum of eukaryotic
cells, the latter of which often results in glycosylation of the protein.
[0056] Variant nucleotide and amino acid sequences of the present invention
also encompass
sequences derived from mutagenic and recombinogenic procedures such as DNA
shuffling.
With such a procedure, one or more different nitrogen-modulating protein
coding regions can be
used to create a new nitrogen-modulating protein possessing the desired
properties. In this
manner, libraries of recombinant polynucleotides are generated from a
population of related
sequence polynucleotides comprising sequence regions that have substantial
sequence identity
and can be homologously recombined in vitro or in vivo. For example, using
this approach,
sequence motifs encoding a domain of interest may be shuffled between the
nitrogen-modulating
sequence useful in the present invention and other known nitrogen-modulating
sequences to
obtain a new sequence coding for a protein with an improved property of
interest, such as
improved nitrogen utilization. Strategies for such DNA shuffling are known in
the art. See, for
example, Stemmer (1994) Proc. Nail. Acad. Sci. USA 91:10747-10751; Stemmer
(1994) Nature
370:389-391; Crameri et al. (1997) Nature Biotech. 15:436-438; Moore et al.
(1997) J. Mol.
Biol. 272:336-347; Zhang etal. (1997) Proc. Natl. Acad. Sci. USA 94:4504-4509;
Crameri et al.
(1998) Nature 391:288-291; and U.S. Patent Nos. 5,605,793 and 5,837,458.
Plant Transformation
[0057] Methods of the invention involve introducing one or more nitrogen-
modulating
nucleotide sequences into a plant. In some embodiments, only one of the
nitrogen-modulating
sequences disclosed herein is introduced into the plant. In other embodiments,
at least 2, at least
3, at least 4, or more of the sequences are introduced. Where multiple
sequences are introduced,
each of the nucleotide sequences is non-identical. Two nucleotide sequences
are considered non-
identical if they differ in at least one nucleotide position. Thus, non-
identical nucleotide
sequences include two or more different nucleotide sequences that each encodes
the same amino
acid sequence (e.g., one or more has been optimized for expression in the
plant), as well as two
or more different nucleotide sequences that encode at least two different
amino acid sequences.
[0058] By "introducing" it is intended to present to the plant one or more
constructs
comprising the one or more nitrogen-modulating sequences in such a manner that
the
construct(s) gain(s) access to the interior of a cell of the plant. The
methods of the invention do
21
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
not require that a particular method for introducing a nucleotide construct to
a plant is used, only
that the nucleotide construct(s) gain(s) access to the interior of at least
one cell of the plant.
Methods for introducing nucleotide constructs into plants are known in the art
including, but not
limited to, stable transformation methods, transient transformation methods,
and virus-mediated
methods.
[0059] In general, plant transformation methods involve transferring
heterologous DNA into
target plant cells (e.g. immature or mature embryos, suspension cultures,
undifferentiated callus,
protoplasts, etc.), followed by applying a maximum threshold level of
appropriate selection
(depending on the selectable marker gene) to recover the transformed plant
cells from a group of
untransformed cell mass. Explants are typically transferred to a fresh supply
of the same
medium and cultured routinely. Subsequently, the transformed cells are
differentiated into
shoots after placing on regeneration medium supplemented with a maximum
threshold level of
selecting agent (i.e., antibiotics, such as spectinomycin and kanamycin). The
shoots are then
transferred to a selective rooting medium for recovering rooted shoot or
plantlet. The transgenic
plantlet then grow into mature plant and produce fertile seeds (e.g. Hiei et
al. (1994) The Plant
Journal 6:271-282; Ishida et al. (1996) Nature Biotechnology 14:745-750).
Explants are
typically transferred to a fresh supply of the same medium and cultured
routinely. A general
description of the techniques and methods for generating transgenic plants are
found in Ayres
and Park (1994) Critical Reviews in Plant Science 13:219-239 and Bommineni and
Jauhar
(1997) Maydica 42:107-120. Since the transformed material contains many cells,
both
transformed and non-transformed cells are present in any piece of subjected
target callus or
tissue or group of cells. The ability to kill non-transformed cells and allow
transformed cells to
proliferate results in transformed plant cultures. Often, the ability to
remove non-transformed
cells is a limitation to rapid recovery of transformed plant cells and
successful generation of
transgenic plants. Molecular and biochemical methods can then be used to
confirm the presence
of the integrated heterologous gene of interest in the genome of transgenic
plant.
[0060] Generation of transgenic plants may be performed by one of several
methods,
including but not limited to introduction of heterologous DNA by Agrobacteri
urn into plant cells
(Agrobacteriurn-mediated transformation), bombardment of plant cells with
heterologous foreign
DNA adhered to particles, and various other non-particle direct-mediated
methods (e.g. Hiei et
al. (1994) The Plant Journal 6:271-282; Ishida et al. (1996) Nature
Biotechnology 14:745-750;
22
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
Ayres and Park (1994) Critical Reviews in Plant Science 13:219-239; Bommineni
and Jauhar
(1997) Maydica 42:107-120) to transfer DNA.
[0061] Methods for transformation of chloroplasts are known in the art.
See, for example,
Svab et al. (1990) Proc. Natl. Acad. Sci. USA 87:8526-8530; Svab and Maliga
(1993) Proc. Natl.
Acad. Sci. USA 90:913-917; Svab and Maliga (1993) EMBO .1. 12:601-606. The
method relies
on particle gun delivery of DNA containing a selectable marker and targeting
of the DNA to the
plastid genome through homologous recombination. Additionally, plastid
transformation can be
accomplished by transactivation of a silent plastid-borne transgene by tissue-
preferred expression
of a nuclear-encoded and plastid-directed RNA polymerase. Such a system has
been reported in
McBride et al. (1994) Proc. Natl. Acad. Sci. USA 91:7301-7305.
[0062] Transformation of bacterial cells is accomplished by one of several
techniques known
in the art, including but not limited to electroporation or chemical
transformation (see, for
example, Ausubel, ed. (1994) Current Protocols in Molecular Biology, John
Wiley and Sons,
Inc., Indianapolis, IN). Markers conferring resistance to toxic substances are
useful in
identifying transformed cells (having taken up and expressed the test DNA)
from non-
transformed cells (those not containing or not expressing the test DNA).
[0063] In one aspect of the invention, the nucleotide sequences of the
invention are useful as
markers to assess transformation of bacterial or plant cells. In this manner,
transformation is
assessed by monitoring nitrogen utilization efficiency as described above.
[0064] Transformation of plant cells can be accomplished in similar
fashion. By "plant" is
intended whole plants, or component parts including plant organs (e.g.,
leaves, stems, roots, etc.),
seeds, plant cells, propagules, embryos and progeny of the same. Plant cells
can be
differentiated or undifferentiated (e.g. callus, suspension culture cells,
protoplasts, leaf cells, root
cells, phloem cells, pollen). "Transgenic plants" or "transformed plants" or
"stably transformed"
plants or cells or tissues refer to plants that have incorporated or
integrated exogenous nucleic
acid sequences or DNA fragments into the plant cell. By "stable
transformation" is intended that
the nucleotide construct introduced into a plant integrates into the genome of
the plant and is
capable of being inherited by progeny thereof.
[0065] The cells that have been transformed may be grown into plants in
accordance with
conventional ways. See, for example, McCormick etal. (1986) Plant Cell Reports
5:81-84.
These plants may then be grown, and either pollinated with the same
transformed strain or
23
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
different strains, and the resulting hybrid having constitutive expression of
the desired
phenotypic characteristic identified. Two or more generations may be grown to
ensure that
expression of the desired phenotypic characteristic is stably maintained and
inherited and then
seeds harvested to ensure expression of the desired phenotypic characteristic
has been achieved.
In this manner, the present invention provides transformed seed (also referred
to as "trans genic
seed") having a nucleotide construct of the invention, for example, an
expression cassette of the
invention, stably incorporated into their genome.
Methods to increase plant yield by modulating nitrogen utilization
[0066] Methods for increasing plant yield are provided. The methods
comprise introducing
into a plant or plant cell a nitrogen-modulating nucleotide sequence disclosed
herein such that an
increase in nitrogen utilization efficiency corresponds to an increase in
plant yield. As defined
herein, the "yield" of the plant refers to the quality and/or quantity of
biomass, and/or
harvestable yield, produced by the plant. By "biomass" is intended any
measured plant product
(e.g., any component part of a plant, such as seed, stalk, root, grain, leaf,
etc.). An increase in
biomass production is any improvement in the yield of the measured plant
product. An increase
in harvestable yield is a higher weight of a plant component that is easily
collected using known
harvest methods, or an increase in the compositional amount of a compound of
interest in the
harvested part: a nonlimiting example, being the amount of an amino acid, such
as lysine, that is
harvested per unit land area. Increasing plant yield or harvestable yield has
several commercial
applications. For example, increasing plant leaf biomass may increase the
yield of leafy
vegetables for human or animal consumption. Additionally, increasing leaf
biomass can be used
to increase production of plant-derived pharmaceutical or industrial products.
An increase in
yield can comprise any statistically significant increase including, but not
limited to, at least a
1% increase, at least a 3% increase, at least a 5% increase, at least a 10%
increase, at least a 20%
increase, at least a 30%, at least a 50%, at least a 70%, at least a 100% or a
greater increase in
plant yield compared to the yield of a plant into which a nucleotide sequence
that modulates use
of nitrogen of the invention has not been introduced.
Plants
24
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
[0067] The present invention may be used for transformation of any plant
species, including, but
not limited to, monocots and dicots. Examples of plants of interest include,
but are not limited to,
corn (maize), sorghum, wheat, sunflower, tomato, crucifers, peppers, potato,
cotton, rice,
soybean, sugarbeet, sugarcane, tobacco, barley, and oilseed rape, Brassica
sp., alfalfa, rye, millet,
safflower, peanuts, sweet potato, cassava, coffee, coconut, pineapple, citrus
trees, cocoa, tea,
banana, avocado, fig, guava, mango, olive, papaya, cashew, macadamia, almond,
oats, vegetables,
grasses (such as turf grasses, forage grasses, or pasture grasses),
ornamentals, trees (such as fruit
trees, nut trees, pulp trees, oil palms) and conifers.
[0068] Vegetables include, but are not limited to, onions, tomatoes,
lettuce, green beans, lima
beans, peas, and members of the genus Curcumis such as cucumber, cantaloupe,
and muskmelon.
Ornamentals include, but are not limited to, azalea, hydrangea, hibiscus,
roses, tulips, daffodils,
petunias, carnation, poinsettia, and chrysanthemum. Preferably, plants of the
present invention are
crop plants (for example, maize, sorghum, wheat, sunflower, tomato, crucifers,
peppers, potato,
cotton, rice, soybean, sugarbeet, sugarcane, tobacco, barley, oilseed rape,
etc.).
[0069] This invention is particularly suitable for any member of the
monocot plant family
including, but not limited to, maize, rice, barley, oats, wheat, sorghum, rye,
sugarcane, pineapple,
yams, onion, banana, coconut, and dates.
Evaluation of Plant Transformation
[0070] Following introduction of heterologous foreign DNA into plant cells,
the
transformation or integration of heterologous gene in the plant genome is
confirmed by various
methods such as analysis of nucleic acids, proteins and metabolites associated
with the integrated
gene.
[0071] PCR analysis is a rapid method to screen transformed cells, tissue
or shoots for the
presence of incorporated nucleotide sequences at the earlier stage before
transplanting into the
soil (Sambrook and Russell (2001) Molecular Cloning: A Laboratory Manual, Cold
Spring
Harbor Laboratory Press, Cold Spring Harbor, NY). PCR is carried out using
oli2onucleotide
primers specific to the gene of interest or Agrobacterium vector background,
etc.
[0072] Plant transformation may be confirmed by Southern blot analysis of
genomic DNA
(Sambrook and Russell, 2001, supra). In general, total DNA is extracted from
the transformant,
digested with appropriate restriction enzymes, fractionated in an agarose gel
and transferred to a
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
nitrocellulose or nylon membrane. The membrane or "blot" is then probed with,
for example,
radiolabeled 32P target DNA fragments to confirm the integration of the
introduced gene in the
plant genome according to standard techniques (Sambrook and Russell, 2001,
supra).
[0073] In Northern analysis, RNA is isolated from specific tissues of
transformant,
fractionated in a formaldehyde agarose eel, blotted onto a nylon filter
according to standard
procedures that are routinely used in the art (Sambrook and Russell, 2001,
supra). Expression of
RNA encoded by the nucleotide sequence of the invention is then tested by
hybridizing the filter
to a radioactive probe derived from a polynucleotide of the invention, by
methods known in the
art (Sambrook and Russell, 2001, supra)
[0074] Western blot and biochemical assays and the like may be carried out
on the transgenic
plants to determine the presence of protein encoded by the nitrogen-modulating
gene by standard
procedures (Sambrook and Russell, 2001, supra) using antibodies that bind to
one or more
epitopes present on the nitrogen-modulating protein. For example, the
polyclonal antibodies
generated by the methods of the present invention can be used to detect the
presence of a
nitrogen-modulating protein.
Antibodies
[0075] Antibodies to the polypeptides useful in the present invention, or
to variants or
fragments thereof, are also encompassed. Methods for producing antibodies are
well known in
the art (see, for example. Harlow and Lane (1988) Antibodies: A Laboratory
Manual, Cold
Spring Harbor Laboratory, Cold Spring Harbor, N.Y.; U.S. Patent No.
4,196,265).
EXPERIMENTAL
I. icdh Gene
Materials and Methods
[0076] Using genomic tools, multiple searches were made among the plant and
bacterial icdh
genes in Genbank. The target focus was on cytosolic, monomeric, NADP+-
dependent versions,
and with a particular "AT" pattern in the polyA signals that would not be
inhibitory to expression
in plants. This work eventually led to a sequence from an Azotobacter species
that had a fit with
the criteria. The sequence of the selected bacterial isocitrate dehydrogenase
gene (icdh) was
26
CA 02906278 2015-09-14
WO 2014/142946
PCT/US2013/031913
codon optimized for expression in maize. A synthetic gene encoding an ICDH
enzyme was
generated (SEQ ID NO: 1):
ATGAGCACCCCCAAGATCATCTACACCTTGACAGATGAGGCGCCGGCGCTGGCCACCTAC
AGCTTGCTGCCCATCATCAAGGCTTTCACTGGAAGCTCAGGCATTGCTGTGGAAACAAGG
GACATCTCCCTTGCTGGAAGGCTGATCGCCACCTTCCCAGAATATTTGACAGACACCCAG
AAGATCTCTGATGATCTTGCTGAGCTGGGGAAGCTGGCCACCACGCCAGATGCCAACATC
ATCAAGCTGCCAAACATCTCTGCTTCAGTTCCTCAGCTGAAGGCCGCCATCAAGGAACTC
CAGCAGCAAGGCTACAAGCTGCCAGATTATCCAGAAGAACCAAAAACAGACACAGAGAAG
GATGTCAAGGCAAGATATGACAAGATCAAGGGCAGCGCCGTCAACCCCGTGCTGAGAGAA
GGAAATTCAGACCGCCGCGCGCCGCTCTCCGTCAAGAACTATGCAAGGAAGCATCCTCAC
AAGATGGGCGCCTGGAGCGCCGACAGCAAGAGCCATGTTGCTCACATGGACAATGGAGAT
TTCTATGGATCAGAGAAGGCGGCGCTGATTGGTGCTCCTGGAAGCGTCAAGATTGAGCTG
ATCGCCAAGGATGGAAGCAGCACCGTGCTGAAGGCCAAGACATCAGTTCAAGCTGGAGAG
ATCATCGACAGCTCCGTGATGAGCAAGAATGCTCTGAGGAACTTCATTGCTGCCGAGATT
GAAGATGCCAAGAAGCAAGGAGTGCTGCTCTCCGTCCACCTCAAGGCCACCATGATGAAG
GTTTCAGATCCCATCATGTTTGGCCAGATTGTITCAGAGTTCTACAAGGATGCTCTCACC
AAGCATGCTGAGGTGCTGAAGCAGATTGGATTTGATGTCAACAATGGCATTGGAGATCTC
TATGCAAGGATCAAGACCCTACCAGAAGCAAAGCAGAAGGAGATTGAAGCTGACATCCAA
GCTGTTTATGCTCAAAGGCCGCAGCTGGCAATGGTGAACAGCGACAAGGGCATCACCAAC
CTCCATGTTCCTTCTGATGICATCGTCGACGCCTCCATGCCGGCCATGATCAGAGATTCA
GGGAAGATGTGGGGGCCAGATGGCAAGCTGCATGACACCAAGGCCGTCATCCCAGATCGC
TGCTATGCTGGCGTCTACCAGGTGGTGATTGAAGATTGCAAGCAGCATGGCGCCITCGAC
CCAACAACAATGGGCTCAGTTCCAAATGTTGGGCTGATGGCGCAGAAGGCAGAAGAATAT
GGAAGCCATGACAAGACCTTTCAGATCCCTGCTGATGGCGTCGTCCGCGTCACTGATGAA
AGCGGCAAGCTGCTGCTGGAGCAATCAGTGGAAGCTGGAGACATCTGGAGGATGTGCCAA
GCAAAGGATGCTCCCATCCAAGATTGGGTGAAGCTCGCCGTCAACAGGGCGCGCGCCACC
AACACGCCGGCGGTGTTCTGGCTGGACCCAGCAAGGGCTCATGATGCTCAGGTGATCGCC
AAGGTGGAGAGATATCTAAAGGATTATGACACCTCCGGCCTGGACATCAGGATCTTGTCG
CCGGTGGAAGCAACAAGGTICTCCTTGGCAAGGATCAGAGAAGGAAAGGACACCATCTCA
GTGACAGGAAATGTGCTGAGGGACTACCTCACCGACCTCTTCCCCATCATGGAGCTGGGC
ACCTCCGCCAAGATGCTCTCCATTGTTCCTCTGATGAGCGGCGGCGGCCTCTTTGAAACT
GGAGCTGGAGGATCAGCGCCCAAGCATGITCAGCAGTTCCTGGAAGAAGGCTACCTCAGA
TGGGACAGCCTTGGAGAGTTCCTGGCGCTCGCCGCCTCCTTGGAGCATCTTGGAAATGCC
TACAAGAACCCAAAGGCGCTGGTGCTGGCCTCCACCCTAGATCAAGCTACTGGCAAGATC
CIGGACAACAACAAGAGCCCAGCAAGGAAGGTIGGTGAGATCGACAACAGAGGAAGCCAC
TTCTACCTGGCGCTCTACTGGGCTCAAGCTCTTGCTGCTCAAACAGAGGACAAGGAGCTA
CAAGCTCAGTTCACCGGCATTGCCAAGGCGCTGACAGACAATGAAACAAAAATTGTTGGA
GAGCTGGCTGCTGCTCAAGGAAAGCCGGIGGACATTGCTGGCTACTACCATCCAAACACC
GACCTCACCAGCAAGGCCATCAGGCCATCTGCCACCTTCAATGCTGCTCTGGCGCCGCTG
27
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
GCATAGTAAGG
[0077] The icdh DNA sequence shown above encodes the following ICDH protein
sequence
(SEQ ID NO:2) (741 amino acids):
mstpkiivtl tdeapalaty sllpiikaft gssgiavetr dislagrlia tfpeyltdtq
kisddlaelg klattpdani iklpnisasv pqlkaaikel qqqgyklpdy peepktdtek
dvkarydkik gsavnpvlre gnsdrrapls vknyarkhph kmgawsadsk shvahmdngd
fygsekaali gapgsvkiel iakdgsstvl kaktsvgage iidssvmskn alrnfiaaei
edakkqgvll svhlkatmmk vsdpimfgqi vsefykdalt khaevlkqig fdvnngigdl
yariktlpea kqkeieadiq avyaqrpqla mvnsdkgitn lhvpsdvivd asmpamirds
gkmwgpdgkl hdtkavipdr cyagvyqvvi edckqhgafd pttmgsvpnv glmaqkaeey
gshdktfqip adgvvrvtde sgkllleqsv eagdiwrmcq akdapiqdwv klavnrarat
ntpavfwldp arandaqvia kverylkdyd tsgldirils pveatrfsla riregkdtis
vtgnvirdyl tdlfpimelg tsakmlsivp lmsggglfet gaggsapkhv qqfleegylr
wdslgeflal aaslehlgna yknpkalvla stldqatgki ldnnkspark vgeidnrgsh
fylalywaqa laaqtedkel qaqftgiaka ltdnetkivg elaaaqgkpv diagyyhpnt
dltskairps atfnaalapl a
[0078] Vector Construction for Overexpression of ICDH
The open reading frame described in the previous section was introduced into a
vector for
plant expression. The vector also contains a gene encoding a glyphosate
tolerant EPSPS enzyme
(GRG23ace5) that was used as the selectable marker during maize
transformation. Expression of
each these genes was controlled by the ScUbi4 promoter to produce robust
expression in maize.
A vector map of this vector, denominated pMD08901, is shown in Fig. 1.
Plant Transformation
[0079] The pMD08901vector was used to carry out Agrobacteri urn-mediated
transformation
of maize. Following vector construction and transformation of Agrobacterium,
the vectors were
confirmed by Southern blot by methods known in the art. Positive Agrobacteri
urn strains that
passed these tests were then grown on a solid medium to produce cell counts
for large-scale
transformation experiments.
28
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
[0080] The vector pMD08901was introduced into an Agrobacterium tumefaciens
strain by
electroporation. The formation of the recombinant vector, pMD08901, was
confirmed by
Southern blot hybridization of this Agrobacierium strain. The selection agent
for these
experiments was glyphosate
[0081] The Agrobacterium strain harboring the cointegrate can be used to
transform plants,
for example, by the PureIntro method (Japan Tobacco, Inc.).
Western Blot Analysis
[0082] Expression of icdh in these plants was examined by generating
antibodies that bind
specifically to the ICDH protein. Briefly, the icdh gene was sub cloned into
the vector pRSFlb
(Novagen) to allow overexpression of the ICDH protein in E. coli following
IPTG induction.
The vector also introduces a 6xHis tag at the N-terminus of the protein.
Following protein
overexpression, the ICDH protein was purified by cobalt column chromatography
and the
identity of the purified protein was confirmed by N-terminal sequencing. The
purified protein
was then used to immunize rabbits, with serum collection beginning 42 days
after immunization.
[0083] Next, the ICDH antiserum was used to assess protein expression in
the transgenic
maize plants by Western blot analysis. Leaf samples were taken from individual
plants
following 4 weeks of growth in the greenhouse, and protein extracts were
prepared by grinding
the plant material in water. Protein concentration in each extract was
determined by Bradford
assay, and 25 ug of each extract was separated on polyacrylamide gels with a 4-
12% gradient.
The separated proteins were transferred to nitrocellulose and then probed with
the rabbit
antiserum at a 1:5000 dilution. Following wash steps, the nitrocellulose was
contacted with goat
anti-rabbit conjugated with horseradish peroxidase (1:10,000 dilution), and
antibody complexes
were visualized using ECL detection reagents (GE Healthcare). At the TO stage
most icdh events
were found to be expressing ICDH. Two events had particularly good expression
levels and were
promoted to the Ti stage.
Maize Nitrogen Analysis
[0084] A series of assays that quantify nitrogen intermediates in plants
have been developed.
These nitrogen assay methods are described in a previous patent filing (WO
2008/051608
"Plants with improved nitrogen utilization and stress tolerance"). These
assays were utilized here
29
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
to analyze a total of 10 transgenic plants containing the icdh gene. Each of
the plants was
sampled (leaf) following 4 weeks of growth in soil in a greenhouse. These leaf
samples were
processed to determine their nitrate, asparagine, glutamine, aspartic acid,
glutamic acid,
ammonium, total amino acid, chlorophyll, and total protein levels. Included
alongside in the
analysis were plants that were transformed with a construct containing only
the GRG23ace5
selectable marker (no icdh nor nagk). These plants were likewise sampled at 4
weeks and are
referred to as "non-GOI" plants. The results of the nitrogen assays carried
out on both types of
plants are shown below in Table 1.
Table 1 - Nitrogen levels, ICDH vs. non-GOT maize events, 4 weeks after
transfer to soil
,
Aspartic Acid Glutamic Acid Asparagine
Total Chlorophyll Ammonium Total Amino Total Protein
0
Plant # GOI (pg/g) (pg/g) (pg/g) Glutamine
(pg/g) (a+b) (mg/g) (pg/g) Acids (mg/g) (mg/g) b.)
o
15091 ICDH 177.3 1240.2 1692.0 5175.0 0.086
755.795 285.830 16.990
.6.
15092 ICDH 451.7 1161.9 1898.3 2055.2 0.007
533.265 234.620 25.471
4-
15093 ICDH 82.9 962.6 450.0 1344.0 0.061
502.607 157.230 24.621
15094 ICDH 201.3 1417.4 148.9 2144.1 0.074
670.780 166.334 19.411 .6.
01
15095 ICDH 174.9 1176.2 1403.1 3009.6 0.066
714.197 196.974 22.753
15096 ICDH 51.2 1435.5 2006.0 2630.9 0.052
626.751 302.167 20.094
15097 ICDH 213.6 1213.7 1364.2 2357.3 0.054
634.482 207.681 24.317
15098 ICDH 256.4 1762.7 3595.3 5148.4 0.357
1822.651 448.316 42.052
15099 ICDH 82.4 848.0 1946.7 2503.9 0.064
590.783 201.124 19.159
15100 ICDH 208.4 1562.0 2226.4 4765.9 0.061
1140.701 303.099 19.038
15101 ICDH 167.6 914.3 1184.2 2873.7 0.049
542.457 267.801 18.433
15122 ICDH 269.2 1081.6 1424.1 6786.6 0.061
574.964 265.796 35.271
15123 ICDH 322.1 1713.5 1625.9 7988.8 0.062
24.671 375.517 23.669 R
15124 ICDH 296.5 1262.3 1138.7 6023.9 0.077
367.674 237.945 29.045 N,
,..
0
15125 ICDH 387.2 2006.6 1652.1 7789.2 0.079
926.448 289.497 30.555 0,
r.
0
rl,
0
IC 1 Control non-GOI 537.8 596.3 2335.0 3516.7 0.154
984.302 362.225 28.635 ,..
,
I-'
IC 2 Control non-GOI 101.6 473.7 1490.3 2485.0 0.157
505.712 264.265 34.564 A
103 Control non-GOI 153.6 914.9 1799.3 2191.5 0.066
534.599 297.756 34.019
IC 4 Control non-GOI 59.7 526.2 313.0 659.3 0.082
347.337 183.701 24.077
:.=
Avg 213.2 627.8 1484.4 2213.1 0.114
984.3 277.0 30.3
Std Dev 219.8 197.9 855.4 1181.5 0.048
273.6 74.3 5.0
ocl
el
cA
N.)
o
,--
c...)
-Z,
(...)
,-
o
,-,
(...,
31
..
... ,
4
These data demonstrate that the synthetic gene we designed encodes a
functional ICDH enzyme.
Maize Plants Containing icdh Gene Showed Differences Over Controls
[01001 Events 15122 and 15125 were selected and progressed onto the T1 stage.
The TI plants were
sampled and evaluated as described previously. The results are set out in
Tables 2-4.
Table 2 ¨ TO Event Promoted to T1 Testing Due to Elevated Glutamine
15122 GO1 Aspartic Glutamic Asparagine Glutamine Total
Ammonium Total Total
Plant Acid Acid Gig/g)
(ttg/g) Chlorophyll (nig) Amino Protein
# ( ug/g) (ug/g)
(a+b) Acids (mg/g)
(mg/g) (mg/g)
2 ICDH 331 2537 575 2551 0.197 566 229
18.6
3 ICDH 456 2299 647 2105 0.144 477 254
18.1
ICDH 563 2880 220 2106 0.450 294 170 12.9
7 ICDH 1151 2350 671 2628 0.243 351 217
22.5
8 ICDH 664 2707 466 3826 0.115 413 202
14.0
9 ICDH 446 3155 89 1156 0.148 406 163
27.9
Avg 602 2655 445 2396 0.216 418 206
19.0
Std 292 328 240 875 0.123 95 35
5.6
Dev
Table 3 ¨ TO Event Promoted to T1 Testing Due to Elevated Aspartic Acid,
Glutamic Acid and
Glutamine
15125 GOI Aspartic Glutamic Asparagine Glutamine Total Ammonium Total Total
Plant Acid Acid
(tig/g) (ttg/g) Chlorophyll ( g/g) Amino Protein
# (tig/g) (lig/g) (a+b) (mg/g)
Acids (mg/g)
(mg/g)
1 ICDH 478 2929 140 3961 0.166 609 204
21.6
2 ICDH 827 1912 855 4177 0.207 391 256
19.3
3 ICDH 838 2406 416 4364 0.157 613 251
14.2
4 ICDH 865 24410 606 6268 0.284 376 192
28.2
5 ICDH 391 2564 167 2447 0.184 535 221
17.3
ICDH 328 3010 376 ' 2128 0.209 417 345 14.0
Avg 621 6205 427 3891 0.201 490 245
19.1
Std 248 8927 271 1494 0.046 109 55
5.3
Dev
'
32
CA 2906278 2019-11-05
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
Table 4 ¨ Controls
Control GOI Aspartic Glutamic Asparagine Glutamine Total
Ammonium Total Total
Plant # Acid Acid ( g/g) ( g/g) Chlorophyll (
g/g) Amino Protein
(p g/g) ( g/g) (a+b) Acids
(mg/g)
(mg/g) (mg/g)
1 253 2351 1759 2412 0.298 567 235 22.0
2 739 2154 3488 3989 0.089 452 226 17.4
3 297 1767 373 2532 0.107 408 197 21.2
4 905 2233 941 3237 0.145 314 190 17.2
394 3037 3346 5262 0.155 504 252 19.5
6 789 2535 4166 9084 0.133 459 295 28.9
Avg 563 2346 2346 4419 0.154 451 232 21.0
Std 281 424 1538 2516 0.074 86 38 4.3
Dey
[0101] The data for Ti plants indicated lower asparagine levels for the
transgenic events (both
the average and for most plants). Individual plants had results that differed
from the control; for
example, event 15122 plant #5, had higher glutamic acid and higher chlorophyll
but lower amino
acids and lower protein levels compared to the controls.
[0102] In Event 15125, plant #4 had much a higher level of glutamic acid. The
recorded number
was so high as to appear that it might be an outlier or a sampling error.
However, the same plant
had higher chlorophyll and higher protein levels compared to the control (the
chlorophyll and
protein measurements are taken from a separate sample than the glutamine),
therefore all the
elevated levels could not have been due to the same experimental error, even
if one assumed
there was an error for glutamine.
[0103] Overall the data indicates some potential effects on the measured pool
sizes (e.g. lower
asparagine) and others for individual plants (e.g. chlorophyll). Lower
asparagine could be due to
more N incorporation into other pools resulting in less N for internal
transport (via the transport
amino acid, asparagine).
II. icdh Gene + nagk Gene
Vector construction for overexpression of bacterial icdh + nagk
[0104] The ICDH vector pMD08901 was further modified to add a gene encoding an
arginine-
insensitive N-acetylglutamate kinase (NAGK) protein. This new vector
(pMD08902; Fig. 2) is
designed to introduce overexpression of both ICDH and NAGK proteins to further
improve
nitrogen utilization in plants.
33
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
[0105] The DNA sequence encoding NAGK, SEQ ID NO: 3 set out below, includes a
chloroplast transit peptide (CTP) obtained from the 5' end of the enolpyruvyl
shikimate
phosphate synthase (EPSPS) gene from the algae Chlamydornonas reinhardlii. The
sequence of
the CTP is distinguished from the nagk gene in that the nagk gene is shown in
boldface type:
SEQ ID NO: 3
ATGCAGCTGCTCAACCAGCGGCAGGCGCTGCGGCTGGGAAGAAGCTCCGCCAGCAAGAAC
CAGCAGGTGGCGCCGCTGGCATCAAGGCCGGCAAGCAGCCTCTCCGTCTCCGCCICCTCC
GTGGCGCCGGCGCCGGCCTGCTCGGCGCCGGCCGGCGCCGGCCGCCGCGCCGTGGTGGTG
CGCGCCTCCGCCACCAAGGAGAAGGTGGAGGAGCTCACCATCCAGATGCTGCATGAGGT
GAT GGT GATCAAGT GC GGC GGCAGCAT GC TGGAGCAGC TGC C GGAGAGC T TCTACAACA
AGC TGGC GAC GC TGCAAGCAGAAGGAAGAAGCAT CGT CAT T GTT CAT GGAGGAGGGCC G
GC CAT CAACCAGAT GC TGGAGCAGC TGAAGAT T GAGC CAAC C TT C T CAAAT GGGC TGAG
GGT GACAGAT GAGC CAACAAT GCAAGC T GTGGAGAT GGTGC T CT CAGGGC C CAT CAACA
AGC TGGT GGT GAGGAAGC T GC TGCACGC C GGC GGCAAGGCAT GGGGC CT CAGC GGCGT G
GAT GGAAGCC T GCT GCAAGC T GT TGAGAAGACTCAAGGCCTCGGCCTGGTGGGCAGCAT
CAC CGT GGTGGATCAAGC GC C GC T C CAGC TGC T GCT GAGCAATGGC TACAT CC C GGTGG
T GT CT C C CAT C GCC GT CT CAGAAGATGGAAGAACAAGATACAAC T GCAAC GCC GACAC C
GT C GC C GGCGC CAT TGCT T CAGC T C TC GGCGC CAAGCAGC T GCT GAT GC T CAC T
GATGT
T C C TGGCATC T GGGCAGAAAATGAGCT GGGAGAGAAGCAGC T GC T GC CGAC GGT GACAA
CAGAAGATAT T CAGC T GAT GATGAAGAAC CAGAT CAT CAC C GGC GGCAT GATC C C CAAG
GT GCAAGCGGC GCT GGAT GC T CTAGCT CAAGGAGTT CAAGAAGT GGT GAT C TGCAAAGG
AGAAGC T GAGAC GC T GGAC GGCGT GGT GAAGGGCAT GGCC GT CGGCACC T C CAT C TCC G
CCGAGATGAGCAGAGGACAAGAT TCTCAAGCCT T CAT CAGCAACAAGGT GT GAGG
[0106] This amino acid sequence of NAGK (after removal of the CTP), SEQ ID NO:
4, is shown
here (277 amino acids):
SEQ ID NO: 4:
MLHEVMVIKC GGSMLEQLPE SFYNKLATLQ AEGRSIVIVH GGGPAINQML EQLKIEPTFS
NGLRVTDEPT MQAVEMVLSG PINKLVVRKL LHAGGKAWGL SGVDGSLLQA VEKTQGLGLV
GSITVVDQAP LQLLLSNGYI PVVSPIAVSE DGRTRYNCNA DTVAGAIASA LGAKQLLMLT
DVPGIWAENE LGEKQLLPTV TTEDIQLMMK NQIITGGMIP KVQAALDALA QGVQEVVICK
GEAETLDGVV KGMAVGTSIS AEMSRGQDSQ AFISNKV
Maize transformation with vector pMD08902
34
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
[0107] The plant vectors pMD08902 was transformed into Agrobacterium and
subsequently
entered into plant transformation experiments, as previously described, to
introduce the stacked
genes into the maize genome. The selection agent for these experiments was
glyphosate.
Western Blot Analysis
[0108] As before, the generated polyclonal antibodies were used in Western
blots to determine if
the transgenic protein was being expressed. At the TO stage most icdh events
were found to be
expressing ICDH. Two events were promoted to the Ti stage and each of these
had good
expression levels.
Nitrogen Assays. TO Events
[0109] A series of assays that quantify nitrogen intermediates in plants have
been developed.
These nitrogen assay methods are described in the previous section. Briefly,
each of the plants
was sampled (leaf) following 4 weeks of growth in soil in a greenhouse. These
leaf samples
were processed to determine their nitrate, asparagine, glutamine, aspartic
acid, glutamic acid,
ammonium, total amino acid, chlorophyll and total protein levels. Included
alongside in the
analysis were plants that were transformed with a construct containing only
the glyphosate
selectable marker (no icdh, nagk, genes) and are referred to as control "non-
GOI" plants.
[0110] The TO data is shown in Table 5.
Table 5 - Nitrogen levels, ICDH + NAGK vs. non-GOI maize events, 4 weeks after
transfer to soil
,
Aspartic Acid Glutamic Acid Asparagine
Total Chlorophyll Ammonium Total Amino Total Protein 0
Plant # GOI (pg/g) (pg/g) (pg/g) Glutamine
(pg/g) (a+b)(mg/g) (pg/g) Acids (mg/g) (mg/g) b.)
0
15102 ICDH + NAGK 146.0 785.7 1230.0 1464.9 0.092
754.605 216.736 24.521
.6.
15103 ICDH + NAGK 187.1 372.6 973.5 3279.8 0.127
836.415 268.941 43.110
4-
15104 ICDH + NAGK 318.6 905.9 2202.9 7399.3 0.370
1087.857 605.138 53.679
15105 ICDH + NAGK 264.6 1082.6 2941.9 11934.4 0.134
865.201 440.129 25.618 .6.
01
15106 ICDH + NAGK 194.3 438.0 1766.1 10241.0 0.144
724.948 390.051 43.614
15107 ICDH + NAGK 122.4 481.7 1683.7 7154.5 0.126
582.000 282.809 30.601
15108 ICDH + NAGK 222.0 27.8 1839.7 4568.3 0.152 700.611
295.594 35.903
15109 ICDH + NAGK 466.7 603.5 1699.4 8891.9 0.281
851.859 347.156 28.974
15110 ICDH + NAGK 124.8 475.6 996.7 6484.5 0.114
654.786 236.433 37.768
IC 1 Control non-GO! 537.8 596.3 2335.0 3516.7
0.154 984.3 362.2 28.6
R
IC 2 Control non-GO! 101.6 473.7 1490.3 2485.0
0.157 505.7 264.3 34.6 0
N,
IC 3 Control non-G01 153.6 914.9 1799.3 2191.5
0.066 534.6 297.8 34.0 ,..
0
IC 4 Control non-G01 59.7 526.2 313.0 659.3
0.082 347.3 183.7 24.1 0,
r.
..,
Avg 213.2 627.8 1484.4 2213.1 0.114
593.0 277.0 30.3 .
Std Dev 219.8 197.9 855.4 1181.5 0.048
273.6 74.3 5.0 0
L7,
0
,..
,
1-'
A
*0
el
,-i
.....,
c,
,...,
,....)
µ..z
.
,....e
36
[0111] Compared to the control (and standard deviation), one TO event (15109)
had higher levels
of aspartic acid. Several events had higher glutamine levels, higher total
amino acids and higher
protein compared to the control. From these results the events 15105 (elevated
Glu, Asp, Gln,
total AAs) and 15106 (elevated Gln, total AAs, protein) were selected for T1
stage evaluations.
T1 Results
[0112] For each of the selected events, six T1 plants were produced, grown and
sampled as
described previously. Assays were performed and the results are shown in Table
6.
37
CA 2906278 2019-11-05
Table 6 ¨ Nitrogen levels, ICDH + NAGK vs. non-GOI maize events, 4 weeks after
transfer to soil
Aspartic Acid Glutamic Acid Asparagine Total Chlorophyll
Ammonium Total Amino Total Protein 0
Plant # GOI ( g/g) (pg/g) (pg/g) Glutamine (pg/g)
(a+b)(mg/g) (pg/g) Acids (mg/g) (mg/g) n.1
o
15105, #5 ICDH + NAGK= 947 2789 1269 13837
0.544 930 555 18.7 4.
15105, #13 ICDH + NAGK 827 1809 298 3163 0.312
619 339 22.0
15105,#14 - ICDH + NAGK 430 1825 = 371 2818
= 0.384 611 511 17.6 tz'
4.
15105,#15 ICDH + NAGK 785 3149 335 6217 0.214
460 423 21.7 c,
15105,#17 ICDH + NAGK 274 1006 388 2632 0.609
425 368 .. 19.9
15105,#18 .1CDH + NAGK 453 . 2030 320 2523
0.233 485 . 247 17.1 .
::::,::.,......:..:.,.,.,. . . .. . .. . . ....,:i...i,,.,.,..,..,-
,:i..zi,ww,.,..,.:.:.:.:. . . .. ..,,w.,.,..:.:.,:.:.::. .
.,,.,i...::E.,:.,.,.,......:..:.,.:.:.:.,.,,,,4wim.,.,:.:.:.:.:0383,::,.,.::43:
::i:,.,.,....:.:.:.:.:.:.,.:.,..,.i.i.wawa.,.,.,.....:.:.:.:.:.:.:..i...i.:,w;.
.w.:.:.:.:.
AVg:M:','Ea:i:]:i:]:ia:i:iiin:i?..:i?..:i:]:i:]:ini:i:in:Egini:i:in:i?..:ii:.:0
1.9*.Ai?..:i:]:i:]:ip:i:i:i:i:ii:ZI.Dt.',.iiiili?..:i:]:=.i:i:iC:4.97bi:i:in:i?
..:i:]:i:]:i:i.1 ..C:549an:i:i:in&:]:i:]:i:i.1
.2ii:i:i:in:i?..:i:]:W50.V.Mi:i:i:ini:]:i:g4QVii]iii]ii:]iini:i:in:i?..:i:]:ilt
5ii&:]:i:]:iq
Std Dev 442 : 766 : 380 4455 :
0.163 185 114 2.1
15106, #5 ICDH + NAGK 722 3797 3858 10471
0.674 963 425 17.9
15106, #9 ICDH + NAGK 184 2469 549 2556 0.279
600 260 13.9
15106,#10 ICDH + NAGK 373 1408 3319 10395
0.234 773 458 21.0
R
15106, #11 ICDH + NAGK 1463 4102 4382 9148 0.383
637 632 22.1 .
15106,#13 ICDH + NAGK 544 3437 3294 5693 0.223
884 416 23.3 .
15106,#17 ICDH + NAGK 591 ... 2331 ... ..
2790 .... .. 5572 .. .. 0.255 ..... .. . 815 ... . 501
...... 30..4.......... ,
0,
0.4.i.delli!1!1!1!1811!1!1!1!1!1!1,11,1!1!1!1111,440011!1111000.1131,40.0tilt.1
1Øt00.g.M.211!0Ø111......::!.'::1#01111111111111!4:4111111 ilii!i!NN21
.011!1!1!1!1!1!1!1!1!1!
Std Dev 442 1027 1333 3198 0.173 140
121 5.5 .
,
Aspartic Acid Glutamic Acid Asparagine Total Chlorophyll
Ammonium Total Amino Total Protein
Plant # GOI (pg/g) (pg/g) (pg/g) Glutamine (pg/g) (a
+b) (mg/g) (pg/g) Acids (mg/g) (mg/g)
:Control 1 253 ' 2351 1759 2412 0.298 567
' 235 = 22.0 :
Control 2 739 2154 3488 3989 0.089 452
226 , 17.4
Control 3 297 1767 373 2532 0.107 408
197 = 21.2
Control 4 905 2233 941 3237 0.145 314
190 17.2
Control 5 394 3037 3346 5262 0.155 ,
504 252 19.5
od
Control 6 789 2535 4166 9084 0.133 459
295 28.9 el
2 56
Avg
i*i:i:i::.:.:...........¨.......,:momo::::::x.,....:::...:::,.....:::::::::,...
.:::::*ioi:i:i:i:i::i:ioi*i:.:.,:¨:-......:*ioi:i:i
i:i:i:i:ioi*i:::.....:::.:...:::
:.:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::fi!!3.M=:
::::::::?i.$40..nm:::
0!N!2.4<4tM::::::V:A:4.1%::::::::::AIP?.;:g:::::::=:A5;=:::
:::::::H:P.:.r4M:::::::::::2%k::::::::
Std Dev 281 424 1538 2516 0.074 86
38 4.3 cn
n.)
1--,
c...)
-1'
(....)
1-,
1-
,....,
38
[0113] Compared to the control average (and Standard Deviation), the averages
showed that
15105 had lower asparagine, higher chlorophyll, and higher total amino acids;
15106 had higher
glutamate, higher chlorophyll, and higher total amino acids.
[0114] Since the stack ICDH + NAGK appeared to have several positive effects,
especially in
the later nitrogen-containing metabolites (e.g. chlorophyll, total amino
acids), we used previous
data on NAGK effects (United States Patent Application Serial No. 12/916,854,
filed November
1, 2010 and compared that to ICDH alone and ICDH + NAGK. The results are shown
in Table 7
(data are A of control).
Table 7¨ Data on NAGK Alone, ICDH Alone and ICDH + NAGK
Aspartic Glutamic Asparagine Glutamine Total Ammonium
Total Total
Acid Acid ( g/g) ( g/g) Chlorophyll (pig/g) Amino Protein
g/g) (ptg/g) (a+b) (mg/g) Acids (mg/g)
NAGK 162% 155% 191% 140% 90% 120% 124% 105%
Alone
ICDH 105% 111% 18% 65% 131% 102% 98% 87%
Alone
ICDH 112% 107% 75% 141% 234% 152% 184% 97%
NAGK
[0115] We excluded the one plant (15125 #4) from the ICDH values since it
seemed to be a high
outlier. The results show a pattern where: (1) NAGK alone tends to impact the
intermediate N-
metabolites (Asp, Asn, Glu, Gin); (2) ICDH alone impacts chlorophyll; and (3)
ICDH + NAGK
shows impacts on chlorophyll and total amino acids. This stack effect
indicates that N was
assimilated and moved towards end-products to a greater extent than with
either gene alone.
[0116]
[0117] The foregoing description and drawings comprise illustrative
embodiments of the present
inventions. The foregoing embodiments and the methods described herein may
vary based on
the ability, experience, and preference of those skilled in the art. Merely
listing the steps of the
method in a certain order does not constitute any limitation on the order of
the steps of the
method. The foregoing description and drawings merely explain and illustrate
the invention, and
the invention is not limited thereto. Those skilled in the art who have the
disclosure before them
39
CA 2906278 2017-11-23
CA 02906278 2015-09-14
WO 2014/142946 PCT/US2013/031913
will be able to make modifications and variations therein without departing
from the scope of the
invention.
CA 02906278 2015-09-14
WO 2014/142946
PCT/US2013/031913
SEQUENCE LISTING
SEQ ID NO: 1
ATGAGCACCCCCAAGATCATCTACACCT TGACAGATGAGGCGCCGGCGCTGGCCACCTAC
ACCTTCCTGCCCATCATCAAGGCT TTCACTGGAAGCTCAGCCATTGCTGTGGAAACAAGG
GACATCTCCCT TGCTGGAAGGCTGATCGCCACCTTCCCAGAATAT T TGACAGACACCCAG
AAGATCTCTGATGATCTTGCTGAGCTGGGGAAGCTGGCCACCACGCCAGATGCCAACATC
ATCAAGCTGCCAAACATCTCTGCT TCAGT TCCTCAGCTGAAGGCCGCCATCAAGGAACTC
CAGCAGCAAGGCTACAAGCTGCCAGAT TA TCCAGAAGAACCAAAAACAGACACAGAGAAG
GAT GT CAAGGCAAGATATGACAAGATCAAGGGCAGCGCCGT CAACCCCGT GCT GAGAGAA
GGAAAT TCAGACCGCCGCGCGCCGCTCTCCGTCAAGAACTATGCAAGGAAGCATCCTCAC
AAGAT GGGCGCC TGGAGCGCCGACAGCAAGAGCCAT GT TGCTCACATGGACAATGGAGAT
T TCTATGGATCAGAGAAGGCGGCGCTGAT TGGTGCTCCTGGAAGCGTCAAGATTGAGCTG
AT CGCCAAGGAT GGAAGCAGCACCGT GC T GAAGGCCAAGACATCAGT TCAAGCT GGAGAG
ATCATCGACAGCTCCGTGATGAGCAAGAA TGCTCTGAGGAACTTCATTGCTGCCGAGAT T
GAAGATGCCAAGAAGCAAGGAGTGCTGCTCTCCGTCCACCTCAAGGCCACCATGATGAAG
GT T TCAGATCCCATCATGT T TGGCCAGAT TGT T TCAGAGT TCTACAAGGATGCTCTCACC
AAGCATGCTGAGGTGCTGAAGCAGATTGGATT TGATGTCAACAATGGCAT TGGAGATCTC
TAT GCAAGGAT CAAGACCC TACCAGAAGCAAAGCAGAAGGAGAT T GAAGC T GACATCCAA
GCTGTT TATGCTCAAAGGCCGCAGCTGGCAATGGTGAACAGCGACAAGGGCATCACCAAC
CTCCATGTTCCTTCTGATGTCATCGTCGACGCCTCCATGCCGGCCATGATCAGAGATTCA
GGGAAGAT GT GGGGGCCAGAT GGCAAGC T GOAT GACACCAAGGCCGTCAT CCCAGATCGC
TGCTATGCTGGCGTCTACCAGGTGGTGAT TGAAGAT TGCAAGCAGCATGGCGCCT TCGAC
CCAACAACAATGGGCTCAGT TCCAAAT GT TGGGCTGATGGCGCAGAAGGCAGAAGAATAT
GGAAGCCATGACAAGACCT T TCAGATCCCTGCTGATGGCGTCGTCCGCGTCACTGATGAA
AGCGGCAAGC T GOT GC T GGAGCAATCAGT GGAAGCT GGAGACATC T GGAGGAT GI GCCAA
GCAAAGGATGCTCCCATCCAAGAT TGGGTGAAGCTCGCCGTCAACAGGGCGCGCGCCACC
AACACGCCGGCGGTGT TCTGGCTGGACCCAGCAAGGGCTCATGATGCTCAGGTGATCGCC
AAGGTGGAGAGATATCTAAAGGAT TATGACACCTCCGGCCTGGACATCAGGATCT TGTCG
CCGGTGGAAGCAACAAGGT T CTCC T TGGCAAGGATCAGAGAAGGAAAGGACACCATCT CA
GTGACAGGAAATGTGCTGAGGGACTACCTCACCGACCTCT TCCCCATCATGGAGCTGGGC
ACC TCCGCCAAGAT GC TOT CCAT T GT T CO TOT GAT GAGCGGCGGCGGCCTCT T T GAAACT
GGAGCTGGAGGATCAGCGCCCAAGCATGT TCAGCAGT TCCTGGAAGAAGGCTACCTCAGA
TGGGACAGCCT TGGAGAGT TCCTGGCGCTCGCCGCCTCCTTGGAGCATCT TGGAAATGCC
TACAAGAACCCAAAGGCGC T GGT GC T GGCCTCCACCC TAGAT CAAGCTAC T GGCAAGAT C
CTGGACAACAACAAGAGCCCAGCAAGGAAGGT T GOT GAGAT CGACAACAGAGGAAGCCAC
T TO TACO T GGCGCTC TACT GGGCT CAAGC TOT T GOT GC TCAAACAGAGGACAAGGAGC TA
CAAGCTCAGT TCACCGGCAT TGCCAAGGCGCT GACAGACAAT GAAACAAAAAT T GT TGGA
GAGCT GGCT GC T GC T CAAGGAAAGCCGGT GGACAT T GC T GGC TAO TACCAT CCAAACACC
GACCT CA CCAGCAAGGCCAT CAGGCCAT CT GCCACC T T CAAT GOT GCTCT GGCGCCGCTG
GCATAGTAAGG
41
CA 02906278 2015-09-14
WO 2014/142946
PCT/US2013/031913
SEQ ID NO: 2
MSTPKIIYTL TDEAPALATY SLLPIIKAFT GSSGIAVETR DISLAGRLIA TFPEYLTDTQ
KISDDLAELG KLATTPDANI IKLPNISASV PQLKAAIKEL QQQGYKLPDY PEEPKTDTEK
DVKARYDKIK GSAVNPVLRE GNSDRRAPLS VKNYARKHPH KMGAWSADSK SHVAHMDNGD
FYGSEKAALI GAPGSVKIEL IAKDGSSTVL KAKTSVQAGE IIDSSVMSKN ALRNFIAAEI
EDAKKQGVLL SVHLKATMMK VSDPIMFGQI VSEFYKDALT KHAEVLKQIG FDVNNGIGDL
YARIKTLPEA KQKETEADIQ AVYAQRPQLA MVNSDKGITN LHVPSDVIVD ASMPAMIRDS
GKMWGPDGKL HDTKAVIPDR CYAGVYQVVI EDCKQHGAFD PTTMGSVPNV GLMAQKAEEY
GSHDKTFQIP ADGVVRVTDE SGKLLLEQSV EAGDIWRMCQ AKDAPIQDWV KLAVNRARAT
NTPAVFWLDP ARAHDAQVIA KVERYLKDYD TSGLDIRILS PVEATRFSLA RIREGKDTIS
VTGNVLRDYL TDLFPIMELG TSAKMLSIVP LMSGGGLFET GAGGSAPKHV QQFLEEGYLR
WDSLGEFLAL AASLEHLGNA YKNPKALVLA STLDQATGKI LDNNKSPARK VGEIDNRGSH
FYLALYWAQA LAAQTEDKEL QAQFTGIAKA LTDNETKIVG ELAAAQGKPV DIAGYYHPNT
DLTSKAIRPS ATFNAALAPL A
SEQ ID NO: 3
AT GCAGC TGC T CAACCAGCGGCAGGCGC T GCGGCTGGGAAGAAGC T CCGCCAGCAAGAAC
CAGCAGGT GGC GCC GC T GGCAT CAAGGCC GGCAAGCAGCCT C T CCGTCT CC GCC T CCT CC
GT GGCGCCGGCGCCGGCCT GCTCGGCGCCGGCCGGCGCCGGCCGCCGCGCCGT GGTGGT G
CGCGCCTCCGCCACCAAGGAGAAGGTGGAGGAGCTCACCATCCAGATGCTGCATGAGGTG
AT GGT GATCAAGTGCGGCGGCAGCATGC T GGAGCAGC T GCCGGAGAGCT TCTACAACAAG
CTGGCGACGCTGCAAGCAGAAGGAAGAAGCATCGTCAT TGT T CAT GGAGGAGGGCCGGCC
AT CAACCAGAT GCT GGAGCAGCT GAAGAT TGAGCCAACCTTCTCAAATGGGCTGAGGGTG
ACAGAT GAGCCAACAAT GCAAGC T CT GGAGAT GGT GC T CT CAGGGCCCAT CAACAAGC T G
GT GGT GAGGAAGCT GC TGCACGCCGGCGGCAAGGCAT GGGGCCTCAGCGGCGT GGATGGA
AGCCT GC TGCAAGC T GT TGAGAAGACT CAAGGCCTCGGCCT GGTGGGCAGCAT CACCGT G
GT GGAT CAAGCGCCGC TCCAGCT GC TGC T GAGCAAT GGCTACATCCCGGT GGT GTCTCCC
AT C GCCGTCT CAGAAGAT GGAAGAACAAGATACAAC T GCAAC GCC GACACC GT C GCCGGC
GCCAT T GC T T CAGC TC TCGGCGCCAAGCAGC T GC TGAT GC T CAC T GATGT T CC T GGCAT
C
TCGGCACAAAATGACCTGGGAGAGAAGCAGCTGCTGCCGACCGTGACAACAGAACATAT T
CAGCT GATGAT GAAGAACCAGAT CATCACCGGCGGCAT GAT CCCCAAGGT GCAAGCGGCG
CTGGATGCTCTAGCTCAAGGAGT T CAAGAAGT GGT GAT CT GCAAAGGAGAAGCT GAGACG
CT GGAC GGCGT GGT GAAGGGCAT GGCCGT CGGCACC T CCAT C T CC GCCGAGAT GAGCAGA
GGACAAGATTCTCAAGCCT T CAT CAGCAA CAAGGTGTGAGG
SEQ ID NO: 4:
MLHEVMVIKC GGSMLEQLPE SFYNKLATLQ AEGRSIVIVH GGGPAINQML EQLKIEPTFS
NGLRVTDEPT MQAVEMVLSG PINKLVVRKL LHAGGKAWGL SGVDGSLLQA VEKTQGLGLV
GSITVVDQAP LQLLLSNGYI PVVSPIAVSE DGRTRYNCNA DTVAGAIASA LGAKQLLMLT
DVPGIWAENE LGEKQLLPTV TTEDIQLMMK NQIITGGMIP KVQAALDALA QGVQEVVICK
GEAETLDGVV KGMAVGTSIS AEMSRGQDSQ AFISNKV
42