Note: Descriptions are shown in the official language in which they were submitted.
CA 02911709 2015-11-10
RETINOIC ACID RECEPTOR GAMMA (RARG) GENE POLYMORPHISMS
PREDICTIVE OF ANTHRACYCLINE-INDUCED CARDIOTOXICITY (ACT)
FIELD OF THE INVENTION
This invention relates to the field of genetic markers for adverse drug
reactions. More
specifically, methods and compositions useful for identifying individuals that
may be at risk
for an adverse drug reaction.
CROSS REFERENCE TO RELATED APPLICATIONS
This application claims the benefit of U.S. Provisional Patent Application
Serial No.
62/077,702 filed on 10 November 2014, entitled "RETINOIC ACID RECEPTOR GAMMA
(RARG) GENE POLYMORPHISMS PREDICTIVE OF ANTHRACYCLINE-INDUCED
CARDIOTOXICITY (ACT)".
BACKGROUND
Adverse drug reactions (ADRs) are a significant cause of illness,
hospitalization and death
for both children and adults in the Western world (LAZAROU et al. JAMA 1998;
PIRMOHAMED et al. BMJ 2004). Estimates suggest that 15% of hospitalized
children
experience an ADR. Those that do survive the ADR may be left disabled
(MITCHELL et al.,
1988 Pediatrics 82:24-9; MARTINEZ-MIR et al., 1999. Br J Clin Pharmacol 47
:681-8).
Many approved drugs used in children are untested in pediatric populations.
While it is
known that children metabolize drugs differently than adults, in many cases
pediatric
dosage forms are not available. This is of particular concern with
chemotherapy drugs,
which may frequently be supplied as a single-dose package, and in combination
with other
agents, excipients and the like. Pediatric populations also represent a more
varied
population, and this increased variability may be due to developmental
differences in the
normal expression of drug metabolism genes.
Genetic factors are involved in variability in drug response ¨ ranging from 20-
95% in some
studies. Age, sex, body weight, health, medical history and the like may be
accounted for,
but patient genotype is largely an unknown factor (EVANS et al. 2003. NEJM
348:538-
549; WEINSHILBOUM 2003. NEJM 348:529-537).
Anthracyclines are used as cytotoxic agents in chemotherapeutic protocols in
both children
and adults, for a variety of neoplasms. Over 70% of childhood cancers are
treated with
anthracyclines. However, clinical utility of anthracyclines is limited by
anthracycline-
1
CA 02911709 2015-11-10
induced cardiotoxicity (ACT). This manifests as asymptomatic cardiac
dysfunction in 57%
of children and as congestive heart failure in 16-20% of children. Some
genetic risk factors
have been identified, but much of the variability in the susceptibility to ACT
remains
unaccounted for, suggesting the existence of additional genetic factors. ACT
may be
characterized by reduced ventricular wall thickness and mass, indicative of
decreased
cardiac muscle and depressed ventricular contractility. Increased and
cumulative dose,
nature of the particular anthracycline, administration route, age, sex and
prior radiation
treatment may affect onset and severity of cardiotoxicity. Administration of
enalapril,
dexrazoxane or antioxidants such as vitamin E, coenzyme Qio, carnitine, or
glutathione,
for example may be beneficial in preventing or reducing cardiac injury during
chemotherapy. Other agents that may be administered to reduce anthracycline
cardiotoxicity are described (WOUTERS et al. 2005. Br. J Hematol 131:561-578).
Examples of anthracyclines and anthracycline analogues include daunorubicin,
doxorubicin, idarubicin and epirubicin. For example, anthracyclines may be
used in the
treatment of solid and hematologic cancers, such as breast cancer, acute
myeloid leukemia,
acute lymphoblastic leukemia, multiple myeloma, Hodgkin's disease, non-
Hodgkin's
lymphoma, sarcoma, renal cancer and liver cancer.
Dose limits have been empirically set in the clinic, above which the
cardiotoxicity is deemed
to be unacceptable. Subclinical and clinical cardiotoxicity may occur below
these doses
(JOHNSON 2006. Seminars in Oncology 33:S33-7o) and affect current and
subsequent
therapeutic regimens. Liposomal anthracycline compositions may demonstrate
reduced
cardiotoxicity (EWER et al. 2004. Seminars in Oncology 31:161-181).
Proteomic methods have been developed for early detection of drug-induced
cardiotoxicity
(PETRICOIN et al. 2004. Toxicol Pathol 32:122-30).
Some polymorphisms in NAD(P)H oxidase are associated with anthracycline-
induced
cardiotoxicity (WOJNOWSKI et al. 2005. Circulation 112:3754-3762). Similarly,
polymorphisms have been identified that associated with ACT in pediatric
populations
(W02012162812 and W02008058394).
Genotype has been shown to alter response to therapeutic interventions.
Genentech's
HERCEPTIN was not effective in its overall Phase III trial but was shown to
be effective
in a genetic subset of subjects with human epidermal growth factor receptor 2
(HER2)-
positive metastatic breast cancer. Similarly, Novartis' GLEEVECC) is only
indicated for the
2
CA 02911709 2015-11-10
subset of chronic myeloid leukemia subjects who carry a reciprocal
translocation between
chromosomes 9 and 22.
SUMMARY
This invention is based in part on the identification that the particular
nucleotide (allele) or
genotype at the site of a given RARG SNP may be associated with an increased
likelihood of
cardiotoxicity ('risk genotype') or a decreased likelihood of cardiotoxicity
('decreased risk
genotype').
This invention is also based in part on the surprising discovery that n2229774
SNP is
useful in predicting a subject's risk of cardiotoxicity following
anthracycline treatment.
Furthermore, SNPs in linkage disequilibrium (i.e. rs11170479; rs11170481;
rs7334991.74 and
rs57789211) are also useful in predicting a subject's risk of cardiotoxicity
following
anthracycline treatment. Whereby the subjects having a decreased risk genotype
are less
likely to experience cardiotoxicity and subjects having a risk genotype are
more likely to
experience cardiotoxicity from the same treatment.
In accordance with one aspect of the invention, methods are provided for
selecting human
subjects for anthracycline compound administration, the method including:
(a) performing an amplification reaction using a nucleic acid sample from a
subject to
amplify polymorphic site: n2229774 or a polymorphic site in linkage
disequilibrium
thereto selected from one or more of the following: rs1117o479; rs111704.84
rs73309171;
and rs57789211;
(b) performing a sequencing reaction using the amplified nucleic acid from
(a) to
determine whether the subject has a risk genotype selected from the following:
r52229774
A/A; or rs2229774 A/G; or a reduced risk genotype rs2229774 GIG or the
corresponding
genotype at a polymorphic site in linkage disequilibrium to n2229774 selected
from one or
more of the following: rs11170479; rs11170481; rs733091.71; and rs57789211;
and
(c) identifying the subject as having a risk genotype or a reduced risk
genotype.
In accordance with a further aspect of the invention, methods are provided for
assisting in
the identification of human subjects at risk for cardiotoxicity from
anthracycline compound
administration, the method including:
(a) performing an amplification reaction using a nucleic acid sample from
a subject to
amplify polymorphic site m2229774 or a polymorphic site in linkage
disequilibrium thereto
selected from one or more of the following: rs11170479; rs11170481;
rs73309171; and
3
CA 02911709 2015-11-10
rs57789211;
(b) performing a sequencing reaction using the amplified nucleic acid from
(a) to
determine whether the subject has a risk genotype selected from the following:
rs2229774
A/A; or r52229774 A/G; or a reduced risk genotype n2229774 GIG or the
corresponding
genotype at a polymorphic site in linkage disequilibrium to n2229774 selected
from one or
more of the following: rs11170479; rs11170481; rs73309171; and rs57789211; and
(c) identifying the subject as having a risk genotype or a reduced risk
genotype.
In accordance with a further aspect of the invention, methods are provided for
treating a
neoplastic disease in a human subject in need thereof, the method including:
(a) administering one or more anthracycline compounds to a subject having a
reduced
risk genotype n2229774 G/G;
(b) administering one or more anthracycline compounds and heart function
monitoring
or a cardioprotective agent or both to subjects with a risk genotype selected
from the
following: n2229774 A/A; and m2229774 A/G;
(c) administering one or more anthracycline compounds in conjunction with a
non-
anthracycline anti-neoplastic compound and heart function monitoring or a
cardioprotective agent or both to subjects with a risk genotype selected from
the following:
r52229774 A/A; and m2229774 A/G; or
(d) administering one or more non-anthracycline compounds to subjects with
a risk
genotype selected from one or more of the following: n2229774 A/A; and
rs2229774 A/G.
In accordance with a further aspect of the invention, methods are provided for
diagnosing a
predisposition for cardiotoxicity risk in a human subject from anthracycline
administration, the method including: a) determining an identity for one or
more of the
following single nucleotide polymorphisms (SNPs) in a biological sample from
the subject:
S2229774 or a polymorphic site in linkage disequilibrium thereto selected from
one or
more of the following: r51117o479; rs1117o481; rs73309171; and rs57789211; and
b) making
a cardiotoxicity risk determination based on the prevalence of risk alleles in
the subject
sample.
In accordance with a further aspect of the invention, uses of an anthracycline
compound
are provided having a cardiotoxicity risk for the treatment of a subject,
wherein the subject
treated has a reduced cardiotoxicity risk genotype at polymorphic site:
m2229774 or a
polymorphic site in linkage disequilibrium thereto selected from one or more
of the
following: rs1117o479; rs1117o481; rs733o9171; and rs57789211; for the
subject, where the
subject is a candidate for anthracycline administration.
4
CA 02911709 2015-11-10
In accordance with a further aspect of the invention, there is provided a use
of one or more
anthracycline compounds or one or more non-anthracycline compounds for the
treatment
of a neoplastic disease in a human subject in need thereof, wherein the
treatment depends
on the risk genotype as follows: (a) a subject having a reduced risk genotype
rs2229774
G/G would be selected for treatment with one or more anthracycline compounds;
(b) a
subject having a risk genotype selected from the following: r52229774 A/A; and
1'52229774
A/G would be selected for treatment with one or more anthracycline compounds
and heart
function monitoring or a cardioprotective agent or both; (c) a subject having
a risk
genotype selected from the following: r52229774 A/A; and rs2229774 A/G would
be
selected for treatment with one or more anthracycline compounds in conjunction
with a
non-anthracycline anti-neoplastic compound and heart function monitoring or a
cardioprotective agent or both; or (d) a subject having a risk genotype
selected from one or
more of the following: n2229774 A/A; and r52229774 A/G would be selected for
treatment
with one or more non-anthracycline compounds.
In accordance with a further aspect of the invention anthracyclines are
provided for use in a
method for treating a neoplastic disease in a subject in need there of, the
method including:
(a) selecting a subject having a reduced risk of developing cardiotoxicity,
wherein
cardiotoxicity is based on the identity of a single nucleotide polymorphism
(SNP) at
polymorphic site: r52229774 or a polymorphic site in linkage disequilibrium
thereto
selected from one or more of the following: rs1117o479; n1117 481; rs73309171;
and
rs57789211; and
(b) administering said subject one or more anthracyclines.
The method may further include selecting a treatment regimen based on the
subject's
cardiotoxicity risk status, as follows: (i) a subject with a reduced risk
genotype is
administered the anthracycline compound; (ii) a subject with a risk genotype
is
administered the anthracycline compound and is given heart function monitoring
or a
cardioprotective agent or both; (iii) a subject with a risk genotype is
administered the
anthracycline compound in conjunction with a non-anthracycline anti-neoplastic
compound and is given heart function monitoring or a cardioprotective agent or
both; (iv) a
subject with a risk genotype is administered a non-anthracycline anti-
neoplastic
compound.
The anthracycline may be selected from one or more of the following:
daunorubicin,
daunomycin, rubidomycin, doxorubicin, idarubicin, epirubicin, mitoxantrone,
carminomycin, esorubicin, quelamycin, aclarubicin, esorubicin, zorubicin,
pirarubicin,
5
CA 02911709 2015-11-10
amrubicin, iododoxorubicin, mitoxantrone and valrubicin.
The non-anthracycline anti-neoplastic compound may be selected from one or
more of:
cyclophosphamide, ifosphamide, fluorouracil, paclitaxel, vincristine,
cisplatin, streptozocin,
and docetaxel.
The anthracycline compound may be doxorubicin. The polymorphic site may be
rs2229774
and wherein the risk genotype is rs2229774 A/A or r52229774 A/G and the
reduced risk
genotype is rs2229774 G/G. The cardioprotective agent may be dexrazoxane. The
non-
anthracycline anti-neoplastic compound may be selected from one or more of:
cyclophosphamide, ifosphamide, fluorouracil, paclitaxel, vincristine,
cisplatin, streptozocin,
and docetaxel. The neoplastic disease may be selected from: breast cancer,
acute myeloid
leukemia, acute lymphoblastic leukemia, multiple myeloma, Hodgkin's disease,
non-
Hodgkin's lymphoma, sarcoma, renal cancer and liver cancer.
The method may further include administering the anthracycline in accordance
with the
subject's risk of developing cardiotoxicity.
The identity of a single nucleotide polymorphism may be determined by one or
more of the
following techniques: restriction fragment length analysis; sequencing; micro-
sequencing
assay; hybridization; invader assay; gene chip hybridization assays;
oligonucleotide ligation
assay; ligation rolling circle amplification; 5' nuclease assay; polymerase
proofreading
methods; allele specific PCR; matrix assisted laser desorption ionization time
of flight
(MALDI-TOF) mass spectroscopy; ligase chain reaction assay; enzyme-amplified
electronic
transduction; single base pair extension assay; and reading sequence data.
In accordance with a further aspect of the invention, uses of an anthracycline
in the
manufacture of a medicament for the treatment of neoplastic disease, wherein
the subjects
treated may have a reduced cardiotoxicity risk genotype at one or more of the
following
RARG polymorphic sites: rs2229774; rs1117o479; rs1117o481; rs73309171; and
rs57789211.
The anthracycline may be selected from one or more of the following:
anthracycline
antibiotics such as daunorubicin (daunomycin, rubidomycin), doxorubicin,
idarubicin,
epirubicin, mitoxantrone, carminomycin, esorubicin, quelamycin, aclarubicin,
esorubicin,
zorubicin, pirarubicin, amrubicin, iododoxorubicin, mitoxantrone and
valrubicin or other
anthracycline compounds described herein.
6
CA 02911709 2015-11-10
The method may further include obtaining a biological sample or samples from a
subject or
subjects. The method may further include administering the anthracycline in
accordance
with the subject's risk of developing cardiotoxicity.
The identity of a single nucleotide polymorphism may be determined by one or
more of the
following techniques: restriction fragment length analysis; sequencing; micro-
sequencing
assay; hybridization; invader assay; gene chip hybridization assays;
oligonucleotide ligation
assay; ligation rolling circle amplification; 5' nuclease assay; polymerase
proofreading
methods; allele specific PCR; matrix assisted laser desorption ionization time
of flight
(MALDI-TOF) mass spectroscopy; ligase chain reaction assay; enzyme-amplified
electronic
transduction; single base pair extension assay; and reading sequence data.
In accordance with a further aspect of the invention, there are provided two
or more
oligonucleotides or peptide nucleic acids of about in to about 400 nucleotides
that
hybridize specifically to a sequence contained in a human target sequence
consisting of a
subject's cardiotoxicity associated gene sequence, a complementary sequence of
the target
sequence or RNA equivalent of the target sequence and wherein the
oligonucleotides or
peptide nucleic acids are operable in determining the presence or absence of
two or more
polymorphism(s) in the cardiotoxicity associated gene sequence selected from
of the
following polymorphic site m2229774 or a polymorphic site in linkage
disequilibrium
thereto.
In accordance with a further aspect of the invention, a kit is provided for
determining a
genotype at one or more of the following polymorphic sites: rs2229774;
rs11170479;
rs11170481; rs73309171; and rs57789211; in a subject to assess the subject's
risk of
cardiotoxicity following anthracycline administration, by distinguishing
alternate
nucleotides at the polymorphic site; or a labeled oligonucleotide having
sufficient
complementary to the polymorphic site so as to be capable of hybridizing
distinctively to
said alternate. The kit may further include an oligonucleotide or a set of
oligonucleotides
operable to amplify a region including the polymorphic site. The kit may
further include a
polymerization agent. The kit may further include instructions for using the
kit to
determine genotype.
In accordance with another aspect of the invention, there is provided a
commercial package
containing, as active pharmaceutical ingredient, use of anthracycline, or a
pharmaceutically
acceptable salt thereof, together with instructions for its use for the
curative or prophylactic
treatment of a neoplastic disease in a subject, wherein the subject treated
has a reduced
7
CA 02911709 2015-11-10
risk polymorphism in one or more of the following polymorphic sites:
rs2229774;
rs11170479; rs11170481; rs73309171; and rs57789211.
BRIEF DESCRIPTION OF THE DRAWINGS
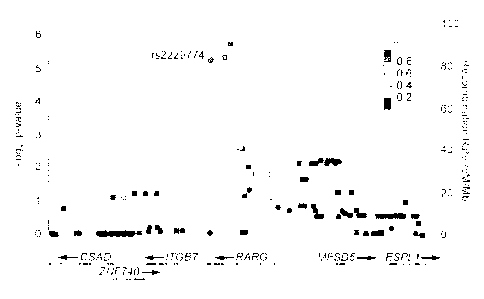
FIGURE 1: shows a pharmacogenetic association with susceptibility to
anthracycline-
induced cardiotoxicity is situated within RARG, wherein the association
results are shown
for genotyped (circles) and imputed (squares) SNPs along with recombination
rates for a
122 kb region of chromosome 12q13.13 and wherein each point represents the
nominal P-
value (left y-axis) for the stage 1 cohort. P-values are from logistic
regression analysis using
an additive model, adjusted for age, dose, tumour type (ALL, Ewing's sarcoma
and
rhabdomyosarcoma) and cardiac radiation therapy. SNPs are coloured according
to their
pairwise correlation (r2) with r52229774 (purple circle) using the tow Genomes
CEU
reference population. Overlaid are the recombination rates (right y-axis) for
estimating
putative recombination hotspots also based upon the Iwo Genomes CEU
population.
FIGURE 2: shows a functional characterization of RARGS427L. (a),
Transcriptional
activation of luciferase coupled to an optimized retinoic acid response
element (RARE) by
transiently transfected RarG (WT) or RARGS427L in HEK293T cells. Data are
presented
as the average s.e.m. in aggregate (n=48) from three separate experiments of
sixteen
replicates each. Data represent the fold induction in luminescence upon ATRA
treatment
compared to untreated samples. ** denotes P value <0.005 using a t-test
analysis. Inset,
Immunoblot of 2on HEK293T lysate generated 48 hours post-transfection with
empty
vector (negative), or the indicated construct using anti-DDK 4C5 (top panel)
and anti-
GAPDH (bottom panel) antibodies. Untagged wild type RARG has an estimated
molecular
weight of 50.4 kDa. Molecular sizes are indicated on the left. (b), Relative
Top2b expression
in untransfected or RARG-transfected H9c2 cells in the presence or absence of
ATRA. Data
are presented as the average s.e.m. in aggregate (n=6) from two separate
experiments of
three replicates each. ' denotes P value <0.0001 using a one-way ANOVA
analysis. (c),
Repression of Top2b expression in H9c2 cells transfected with RarG WT or RARG
8427L
compared to untransfected cells. Data, normalized to the relative expression
of the
transfected RARG construct, are presented as the average + s.e.m. in aggregate
(n=12)
from two separate experiments of six replicates each. *** denotes P value
<o.000i using a
t-test analysis.
8
CA 02911709 2015-11-10
DETAILED DESCRIPTION
1. Definitions
In the description that follows, a number of terms are used extensively, the
following
definitions are provided to facilitate understanding of the various
embodiments of the
invention.
An "anthracycline compound" or "anthracycline" or "anthracycline derivatives"
or
"anthracycline analogues" as used herein is typically an anthraquinone core
attached to a
carbohydrate moiety and derivative thereof (see for example, FAN et al. J.
Org. Chem.
(2007) 72:2917-2928; Goodman and Gilman's The Pharmacological Basis of
Therapeutics
8th edition editors Alfred Goodman Gilman, Theodore Rall, Alan Nies, Palmer
Taylor.
Pergamon Press. 1990 pg 1241-1244). For example, include anthracycline
antibiotics such
as daunorubicin (daunomycin, rubidomycin), doxorubicin, idarubicin,
epirubicin,
mitoxantrone, carminomycin, esorubicin, quelamycin, aclarubicin, esorubicin,
zorubicin,
pirarubicin, amrubicin, iododoxorubicin, detorubicin, marcellomycin,
rodorubicin, and
valrubicin. Alternatively, the anthracycline may be selected from daunorubicin
and
doxorubicin.
As used herein "anthracycline-induced cardiotoxicity" or "ACT" is defined
based on
CTCAEv3 (Common Terminology Criteria for Adverse Events ¨ see Cancer Therapy
Evaluation Program - Common Terminology Criteria for Adverse Events- Version 3
in
edition 2003) as early- or late-onset left ventricular dysfunction measured by
echocardiogram (shortening fraction, SF) and/or symptoms requiring
intervention. We
used a more stringent threshold of SF..26% at any time during or after
anthracycline
therapy to better differentiate between cardiotoxicity cases and controls. To
exclude
transient acute cardiotoxicity, echocardiograms obtained <21 days after a dose
of
anthracyclines were excluded. Control patients were required to have normal
echocardiograms (SF3o%) during and after therapy, with a follow-up of >5 years
after
completion of anthracycline therapy. Doxorubicin equivalents were used to
calculate
cumulative anthracycline doses (Altman A.J. editor, Children's Oncology Group.
Supportive care of children with cancer: current therapy and guidelines from
the Children's
Oncology Group. Baltimore: Johns Hopkins University Press; 2004.412 p.p.).
"Genetic material" includes any nucleic acid and can be a deoxyribonucleotide
or
ribonudeotide polymer in either single or double-stranded form.
9
CA 02911709 2015-11-10
A nucleotide represented by the symbol M may be either an A or C, a nucleotide
represented by the symbol W may be either an T/U or A, a nucleotide
represented by the
symbol Y may be either an C or T/U, a nucleotide represented by the symbol S
may be
either an G or C, while a nucleotide represented by the symbol R may be either
an G or A,
and a nucleotide represented by the symbol K may be either an G or T/U.
Similarly, a
nucleotide represented by the symbol V may be either A or G or C, while a
nucleotide
represented by the symbol D may be either A or G or T, while a nucleotide
represented by
the symbol B may be either G or C or T, and a nucleotide represented by the
symbol H may
be either A or C or T. A nucleotide represented by the symbol N may be an A or
G or T or C.
A "polymorphic site" or "polymorphism site" or "polymorphism" or "single
nucleotide
polymorphism site" (SNP site) or single nucleotide polymorphism" (SNP) as used
herein is
the locus or position with in a given sequence at which divergence occurs. A
"polymorphism" is the occurrence of two or more forms of a gene or position
within a gene
(allele), in a population, in such frequencies that the presence of the rarest
of the forms
cannot be explained by mutation alone. The implication is that polymorphic
alleles confer
some selective advantage on the host. Polymorphic sites have at least two
alleles, each
occurring at frequency of greater than 1%, and may be greater than 10% or 20%
of a
selected population. Polymorphic sites may be at known positions within a
nucleic acid
sequence or may be determined to exist. Polymorphisms may occur in both the
coding
regions and the noncoding regions (for example, promoters, introns or
untranslated
regions) of genes. Polymorphisms may occur at a single nucleotide site (SNPs)
or may
involve an insertion or deletion as described herein.
A "risk genotype" as used herein refers to an allelic variant (genotype) at
one or more of the
following polymorphic sites: rs2229774; rs1117o479; rs1117o481; rs73309171;
and
rs57789211; as described herein, as being indicative of an increased
likelihood of
cardiotoxicity following administration of an anthracycline. The risk genotype
may be
determined for either the haploid genotype or diploid genotype, provided that
at least one
copy of a risk allele is present. Risk genotype may be an indication of an
increased risk of
cardiotoxicity. Subjects having one copy (heterozygotes) or two copies
(homozygotes) of
the risk allele are considered to have the "risk genotype" even though the
degree to which
the subjects is at risk cardiotoxicity may increase, depending on whether the
subject is a
homozygote rather than a heterozygote. Such "risk genotypes" may be selected
from the
following: rs2229774 A/A; and rs2229774 A/G; or a polymorphic sites in linkage
disequilibrium thereto (risk genotype on the reverse strand; T and C on the
forward
strand).
CA 02911709 2015-11-10
A "decreased risk genotype" as used herein refers to an allelic variant
(genotype) at
polymorphic site rs2229774 or a polymorphic site in linkage disequilibrium
thereto, for the
subject as described herein, as being indicative of a decreased likelihood of
cardiotoxicity
following administration of an anthracycline. "Decreased risk genotype" or
"reduced risk
genotypes" may be and a "reduced risk genotype" may be n2229774 GIG; or a
polymorphic
site in linkage disequilibrium thereto (risk genotype on the reverse strand; T
and C on the
forward strand).
A "clade" is a group of haplotypes that are closely related phylogenetically.
For example, if
haplotypes are displayed on a phylogenetic (evolutionary) tree a clade
includes all
haplotypes contained within the same branch.
The pattern of a set of markers along a chromosome is referred to as a
"Haplotype".
Accordingly, groups of alleles on the same small chromosomal segment tend to
be
transmitted together. Haplotypes along a given segment of a chromosome are
generally
transmitted to progeny together unless there has been a recombination event.
Absence of a
recombination event, haplotypes can be treated as alleles at a single highly
polymorphic
locus for mapping.
As used herein "haplotype" is a set of alleles of closely linked loci on a
chromosome that
tend to be inherited together. Such allele sets occur in patterns, which are
called
haplotypes. Accordingly, a specific SNP or other polymorphism allele at one
SNP site is
often associated with a specific SNP or other polymorphism allele at a nearby
second SNP
site or other polymorphism site. When this occurs, the two SNPs or other
polymorphisms
are said to be in Linkage Disequilibrium (LD) because the two SNPs or other
polymorphisms are not just randomly associated (i.e. in linkage equilibrium).
In general, the detection of nucleic acids in a sample depends on the
technique of specific
nucleic acid hybridization in which the oligonucleotide is annealed under
conditions of
"high stringency" to nucleic acids in the sample, and the successfully
annealed
oligonucleotides are subsequently detected (see for example Spiegelman, S.,
Scientific
American, Vol. 210, p. 48 (1964)). Hybridization under high stringency
conditions
primarily depends on the method used for hybridization, the oligonucleotide
length, base
composition and position of mismatches (if any). High-stringency hybridization
is relied
upon for the success of numerous techniques routinely performed by molecular
biologists,
such as high-stringency PCR, DNA sequencing, single strand conformational
polymorphism analysis, and in situ hybridization. In contrast to Northern and
Southern
11
CA 02911709 2015-11-10
hybridizations, these aforementioned techniques are usually performed with
relatively
short probes (e.g., usually about 16 nucleotides or longer for PCR or
sequencing and about
40 nucleotides or longer for in situ hybridization). The high stringency
conditions used in
these techniques are well known to those skilled in the art of molecular
biology, and
examples of them can be found, for example, in Ausubel et al., Current
Protocols in
Molecular Biology, John Wiley & Sons, New York, N.Y., 1998.
"Oligonucleotides" as used herein are variable length nucleic acids, which may
be useful as
probes, primers and in the manufacture of microarrays (arrays) for the
detection and/or
amplification of specific nucleic acids. Such DNA or RNA strands may be
synthesized by
the sequential addition (5'-3' or 3'-5') of activated monomers to a growing
chain, which may
be linked to an insoluble support. Numerous methods are known in the art for
synthesizing
oligonucleotides for subsequent individual use or as a part of the insoluble
support, for
example in arrays (BERNFIELD MR. and ROTTMAN FM. J. Biol. Chem. (1967)
242(18):4134-43; SULSTON J. et al. PNAS (1968) 6o(2):409-415; GILLAM S. et al.
Nucleic
Acid Res.(1975) 2(5):613-624; SONORA GM. et al. Nucleic Acid Res.(199o)
18(11):3155-9;
LASHKARI DA. et al. Proc Nat Acad Sci (1995) 92(17)7912-5; MCGALL G. et al.
PNAS
(1996) 93(24):13555-6o; ALBERT TJ. et a/. Nucleic Acid Res.(2003) 31(7):e35;
GAO X. et
al. Biopolymers (2004) 73(5):579-96; and MOORCROFT MJ. et al. Nucleic Acid
Res.(2005) 33(8):e75). In general, oligonucleotides are synthesized through
the stepwise
addition of activated and protected monomers under a variety of conditions
depending on
the method being used. Subsequently, specific protecting groups may be removed
to allow
for further elongation and subsequently and once synthesis is complete all the
protecting
groups may be removed and the oligonucleotides removed from their solid
supports for
purification of the complete chains if so desired.
"Peptide nucleic acids" (PNA) as used herein refer to modified nucleic acids
in which the
sugar phosphate skeleton of a nucleic acid has been converted to an N-(2-
aminoethyl)-
glycine skeleton. Although the sugar-phosphate skeletons of DNA/RNA are
subjected to a
negative charge under neutral conditions resulting in electrostatic repulsion
between
complementary chains, the backbone structure of PNA does not inherently have a
charge.
Therefore, there is no electrostatic repulsion. Consequently, PNA has a higher
ability to
form double strands as compared with conventional nucleic acids, and has a
high ability to
recognize base sequences. Furthermore, PNAs are generally more robust than
nucleic
acids. PNAs may also be used in arrays and in other hybridization or other
reactions as
described above and herein for oligonucleotides.
12
CA 02911709 2015-11-10
An "addressable collection" as used herein is a combination of nucleic acid
molecules or
peptide nucleic acids capable of being detected by, for example, the use of
hybridization
techniques or by any other means of detection known to those of ordinary skill
in the art. A
DNA microarray would be considered an example of an "addressable collection".
In general the term "linkage", as used in population genetics, refers to the
co-inheritance of
two or more nonallelic genes or sequences due to the close proximity of the
loci on the same
chromosome, whereby after meiosis they remain associated more often than the
50%
expected for unlinked genes. However, during meiosis, a physical crossing
between
individual chromatids may result in recombination. "Recombination" generally
occurs
between large segments of DNA, whereby contiguous stretches of DNA and genes
are likely
to be moved together in the recombination event (crossover). Conversely,
regions of the
DNA that are far apart on a given chromosome are more likely to become
separated during
the process of crossing-over than regions of the DNA that are close together.
Polymorphic
molecular markers, like SNPs, are often useful in tracking meiotic
recombination events as
positional markers on chromosomes.
Furthermore, the preferential occurrence of a disease gene in association with
specific
alleles of linked markers, such as SNPs or other polymorphisms, is called
"Linkage
Disequilibrium" (LD). This sort of disequilibrium generally implies that most
of the disease
chromosomes carry the same mutation and the markers being tested are
relatively close to
the disease gene(s).
For example, in SNP-based association analysis and LD mapping, SNPs can be
useful in
association studies for identifying polymorphisms, associated with a
pathological
condition, such as sepsis. Unlike linkage studies, association studies may be
conducted
within the general population and are not limited to studies performed on
related
individuals in affected families. In a SNP association study the frequency of
a given allele
(i.e. SNP allele) is determined in numerous subjects having the condition of
interest and in
an appropriate control group. Significant associations between particular SNPs
or SNP
haplotypes and phenotypic characteristics may then be determined by numerous
statistical
methods known in the art.
Association analysis can either be direct or LD based. In direct association
analysis,
potentially causative SNPs may be tested as candidates for the pathogenic
sequence. In LD
based SNP association analysis, SNPs may be chosen at random over a large
genomic
region or even genome wide, to be tested for SNPs in LD with a pathogenic
sequence or
13
CA 02911709 2015-11-10
pathogenic SNP. Alternatively, candidate sequences associated with a condition
of interest
may be targeted for SNP identification and association analysis. Such
candidate sequences
usually are implicated in the pathogenesis of the condition of interest. In
identifying SNPs
associated with cardiotoxicity, candidate sequences may be selected from those
already
implicated in the pathway of the condition or disease of interest. Once
identified, SNPs
found in or associated with such sequences, may then be tested for statistical
association
with an individual's prognosis or susceptibility to the condition or to the
side effect of a
medication.
For an LD based association analysis, high density SNP maps are useful in
positioning
random SNPs relative to an unknown pathogenic locus. Furthermore, SNPs tend to
occur
with great frequency and are often spaced uniformly throughout the genome.
Accordingly,
SNPs as compared with other types of polymorphisms are more likely to be found
in close
proximity to a genetic locus of interest. SNPs are also mutationally more
stable than
variable number tandem repeats (VNTRs) and short tandem repeats (STRs).
In population genetics linkage disequilibrium refers to the "preferential
association of a
particular allele, for example, a mutant allele for a disease with a specific
allele at a nearby
locus more frequently than expected by chance" and implies that alleles at
separate loci are
inherited as a single unit (Gelehrter, T.D., Collins, F.S. (1990). Principles
of Medical
Genetics. Baltimore: Williams & Wilkens). Accordingly, the alleles at these
loci and the
haplotypes constructed from their various combinations serve as useful markers
of
phenotypic variation due to their ability to mark clinically relevant
variability at a particular
position (see Akey, J. et al. Eur J Hum Genet (2001) 9:291-300; and Zhang, K.
et al.
(2002). Am J Hum Genet. 71:1386-1394). This viewpoint is further substantiated
by
Khoury et al. ((1993). Fundamentals of Genetic Epidemiology. New York: Oxford
University Press at p. 16o) who state, "[w]henever the marker allele is
closely linked to the
true susceptibility allele and is in [linkage] disequilibrium with it, one can
consider that the
marker allele can serve as a proxy for the underlying susceptibility allele."
As used herein "linkage disequilibrium" (LD) is the occurrence in a population
of certain
combinations of linked alleles in greater proportion than expected from the
allele
frequencies at the loci. For example, the preferential occurrence of a disease
gene in
association with specific alleles of linked markers, such as SNPs, or between
specific alleles
of linked markers, are considered to be in LD. This sort of disequilibrium
generally implies
that most of the disease chromosomes carry the same mutation and that the
markers being
tested are relatively close to the disease gene(s). Accordingly, if the
genotype of a first locus
14
CA 02911709 2015-11-10
is in LD with a second locus (or third locus etc.), the determination of the
allele at only one
locus would necessarily provide the identity of the allele at the other locus.
When
evaluating loci for LD those sites within a given population having a high
degree of linkage
disequilibrium (i.e. an absolute value for r2 0.5) are potentially useful in
predicting the
identity of an allele of interest (i.e. associated with the condition of
interest). A high degree
of linkage disequilibrium may be represented by an absolute value for r2 >
o.6.
Alternatively, a high degree of linkage disequilibrium may be represented by
an absolute
value for r2 0.7 or by an absolute value for r2 ._. 0.8. Additionally, a high
degree of linkage
disequilibrium may be represented by an absolute value for r2> 0.85 or by an
absolute
value for r2 .. 0.9 or by an absolute value for r2 0.95. Accordingly, two SNPs
that have a
high degree of LD may be equally useful in determining the identity of the
allele of interest
or disease allele. Therefore, we may assume that knowing the identity of the
allele at one
SNP may be representative of the allele identity at another SNP in LD.
Accordingly, the
determination of the genotype of a single locus can provide the identity of
the genotype of
any locus in LD therewith and the higher the degree of linkage disequilibrium
the more
likely that two SNPs may be used interchangeably.
LD may be useful for genotype-phenotype association studies. For example, if a
specific
allele at one SNP site (e.g. "A") is the cause of a specific clinical outcome
(e.g. call this
clinical outcome "B") in a genetic association study then, by mathematical
inference, any
SNP (e.g. "C") which is in significant LD with the first SNP, will show some
degree of
association with the clinical outcome. That is, if A is associated (¨) with B,
i.e. A¨B and
C¨A then it follows that C¨B. Of course, the SNP that will be most closely
associated with
the specific clinical outcome, B, is the causal SNP ¨ the genetic variation
that is
mechanistically responsible for the clinical outcome. Thus, the degree of
association
between any SNP, C, and clinical outcome will depend on LD between A and C.
Until the mechanism underlying the genetic contribution to a specific clinical
outcome is
fully understood, LD helps identify potential candidate causal SNPs and also
helps identify
a range of SNPs that may be clinically useful for prognosis of clinical
outcome or of
treatment effect. If one SNP within a gene is found to be associated with a
specific clinical
outcome, then other SNPs in LD will also have some degree of association and
therefore
some degree of prognostic usefulness.
Polymorphisms in linkage disequilibrium may be identified, for example, using
the
Haploview program (BARRETT JC. et al. Bioinformatics (2005) 21(2):263-5) and
the LD
function in the Genetics Package in R (R Core Development Group, 2005 - R
Development
CA 02911709 2015-11-10
Core Team (www.R-project.org). Linkage Disequilibrium between markers may be
defined
using r2 whereby all SNPs available on Hapmap.org (phase II) (cohort H), all
SNPs
genotyped internally using the Illumina Goldengate assay (cohort I) and SNPs
may be
sequenced using the Sequenom Iplex Platform (cohort S) for genes of interest.
A minimum
r2 of 0.5 may be used as the cutoff to identify LD SNPs.
Numerous sites have been identified as polymorphic sites associated
cardiotoxicity
following anthracycline administration (see FIGURE 1).
TABLE 1. Single Nucleotide Polymorphisms Associated with Anthracycline-Induced
Cardiotoxicity
SNP SNP Odds
AD
Gene Position Alleles ( P - Ratio
SNP ID Chr RP
Symbol (BUILD reverse
V value (95%CI
35) strand)
6.0(2.9
rs2229774 4.1X10-8
RARG 53605545 12 T/C (A/G) (A) ¨ 12.5)
T/C 3.1x10- 5.9
(2.9
rs11170479 53610627
RARG 12 10 - 11.8)
G/A 7.5x10- 5.5
(2.8
rs11170481 53611791
RARG 12 10 - 11.0)
C/T 3.8x10- 6.1
(2.9
S73309171 53606565
RARG 12 8 ¨ 12.9)
C/T 3.8x10- 6.1
(2.9
rs57789211 53609992
RARG 12 8 ¨ 12.9)
Adverse Drug Reaction Predictive Variant (ADRPV)
Chromosome (Chr)
TABLE 2. below shows the flanking sequences for the SNPs described in TABLE 1
providing their rs designations and corresponding SEQ ID NO designations. Each
polymorphism is shown within the flanking sequence, and the polymorphism is
identified
in bold.
16
CA 02911709 2015-11-10
TABLE 2. Sequence for Cardiotoxicity-Associated Polymorphisms with SEQ ID NO
designations
SNP SEQ
Alleles.ID
Gene Mnor
Sybol
SNP ID (* GENOMIC SEQUENCE NO: m Allele
reverse
strand)
CTTAATCCGA GAGATGCTGG 1
AGAACCCTGA AATGTTTGAG
GATGACTCCT
rs222974
GCAGCCTGGT CCCCACCCCA
TIC ATGCCTCTAG CGAGGATGAG
RARG (A/G) A GTTCCTGGGG
GTAAGTGTGT GTGTGTGTGT 2
GTGTG
rs11170479 TIC
GCGCGCGCGC GCGCGCGCGT
RARG GTGGT
ACAATGGGAT CCTAGAACCC 3
TCACT
rs11170481 G/A A
GATTCTGGCA CAGACACACA
RARG GCCAG
ACACACACAC ACACACACAC 4
ACACA
rs73309171 C/T
ACTTGGAATT GTGCTGAATT
RARG AAAAA
AGTCACTCTC TGTCTCTTTG 5
GTATA
rs57789211 C/T
CTTTGTCAGG TAGTCTACTT
RARG CCCAC
It will be appreciated by a person of skill in the art that further linked
polymorphic sites
and combined polymorphic sites may be determined. A haplotype of the above
genes can
be created by assessing polymorphisms in normal subjects using a program that
has an
expectation maximization algorithm (for example PHASE). A constructed
haplotype of
these genes may be used to find combinations of SNPs that are in LD with the
tag SNPs
(tSNPs) identified herein. Accordingly, the haplotype of an individual could
be determined
by genotyping other SNPs or other polymorphisms that are in LD with the tSNPs
identified
herein. Single polymorphic sites or combined polymorphic sites in LD may also
be
genotyped for assessing subject risk of cardiotoxicity following anthracycline
treatment.
It will be appreciated by a person of skill in the art that the numerical
designations of the
positions of polymorphisms within a sequence are relative to the specific
sequence and the
orientation of the strand being read (i.e. forward or reverse). Also the same
positions may
be assigned different numerical designations depending on the way in which the
sequence
is numbered and the sequence chosen. Furthermore, sequence variations within
the
17
CA 02911709 2015-11-10
population, such as insertions or deletions, may change the relative position
and
subsequently the numerical designations of particular nucleotides at and
around a
polymorphic site. For example, the sequences represented by accession numbers
NM 0 3786, Y17151, BC104952, BCo5o37o, AF15400f all comprise ABCC3 nucleotide
sequences, but may have some sequence differences and numbering differences
between
them. Furthermore, one of skill in the art will appreciate that a variety of
sequencing,
amplification, extension, genotyping or hybridization primers or probes may be
designed to
specifically identify the polymorphisms described in TABLE 2, and the
sequences flanking
the various polymorphisms as provided herein are illustrative examples. One of
skill in the
art will also appreciate that a variety of sequencing, amplification,
extension, genotyping or
hybridization primers or probes adjacent to, complimentary to, or overlapping
with the
sequences provided in TABLE 2, may be developed or designed for the
identification of
the polymorphisms described herein, without going beyond the scope of various
embodiments of the invention as described herein.
One example of a partial gene sequence is a human ABCC3 gene sequence
illustrated as
GenBank accession # NM 003786. The genomic sequence of the human ABCC3 gene
(NC 00 017.9 nucleotides 45979561-46185071) further includes 5' and 3'
untranslated
sequences, introns and the like. Sequence databases with this information,
such as
GenBank, operated by the National Centre for Biotechnology Information (NCBI)
store
such information in a retrievable format, and are publicly accessible. A
person of skill in
the art will appreciate the various methods and tools that may be used to
access such
information, in a context suitable to their particular application of aspects
described herein.
Polymorphic sites in SEQ ID NO:1-5 are identified by their variant designation
(i.e. M, W,
Y, S, R, K, V, B, D, H or by "¨" for a deletion, a "+"or for example "G" etc.
for an insertion).
An "rs" prefix designates a SNP in the database is found at the National
Center for
Biotechnology Information (NCBI) SNP database. The "rs" numbers are the NCBI
rsSNP
ID form.
The Sequences given in TABLE 2 (SEQ ID NO:1-5) above may be useful to a person
of skill
in the art in the design of primers and probes or other oligonudeotides or
PNAs for the
identification of polymorphisms as described herein.
An "allele" is defined as any one or more alternative forms of a given gene.
In a diploid cell
or organism the members of an allelic pair (i.e. the two alleles of a given
gene) occupy
18
CA 02911709 2015-11-10
corresponding positions (loci) on a pair of homologous chromosomes and if
these alleles
are genetically identical the cell or organism is said to be "homozygous", but
if genetically
different the cell or organism is said to be "heterozygous" with respect to
the particular
gene.
A "gene" is an ordered sequence of nucleotides located in a particular
position on a
particular chromosome that encodes a specific functional product and may
include
untranslated and untranscribed sequences in proximity to the coding regions
(5' and 3' to
the coding sequence). Such non-coding sequences may contain regulatory
sequences
needed for transcription and translation of the sequence or introns etc. or
may as yet to
have any function attributed to them beyond the occurrence of the SNP of
interest.
A "genotype" is defined as the genetic constitution of an organism, usually in
respect to one
gene or a few genes or a region of a gene relevant to a particular context
(i.e. the genetic loci
responsible for a particular phenotype).
A "phenotype" is defined as the observable characters of an organism. In gene
association
studies, the genetic model at a given locus can change depending on the
selection pressures
(i.e., the environment), the population studied, or the outcome variable
(i.e., the
phenotype).
A similar observation would be seen in a gene association study with the
hemoblobin, beta
gene (HBB) with mortality as the primary outcome variable. A mutation in the
HBB gene,
which normally produces the beta chain subunit of hemoglobin (B allele),
results in an
abnormal beta chain called hemoglobin S (S allele; Allison A (1955) Cold
Spring Harbor
Symp. Quant. Biol. 20:239-255). Hemoglobin S results in abnormal sickle-shaped
red
blood cells which lead to anemia and other serious complications including
death. In the
absence of malaria, a gene association study with the HBB gene would suggest a
codominant model (survival(BB) > survival (BS) > survival (SS)). However, in
the presence
of marlaria, a gene association study with the HBB gene would suggest a
heterozygote
advantage model (survival(BB) < survival(BS) > survival(SS)).
A "single nucleotide polymorphism" (SNP) occurs at a polymorphic site occupied
by a
single nucleotide, which is the site of variation between allelic sequences.
The site is
usually preceded by and followed by highly conserved sequences of the allele
(e.g.,
sequences that vary in less than 1/100 or 1/1000 members of the populations).
A single
nucleotide polymorphism usually arises due to substitution of one nucleotide
for another at
19
CA 02911709 2015-11-10
the polymorphic site. A "transition" is the replacement of one purine by
another purine or
one pyrimidine by another pyrimidine. A "transversion" is the replacement of a
purine by a
pyrimidine or vice versa. Single nucleotide polymorphisms can also arise from
a deletion
(represented by "-" or "del") of a nucleotide or an insertion (represented by
"+" or "ins" or
"I") of a nucleotide relative to a reference allele. Furthermore, a person of
skill in the art
would appreciate that an insertion or deletion within a given sequence could
alter the
relative position and therefore the position number of another polymorphism
within the
sequence. Furthermore, although an insertion or deletion may by some
definitions not
qualify as a SNP as it may involve the deletion of or insertion of more than a
single
nucleotide at a given position, as used herein such polymorphisms are also
called SNPs as
they generally result from an insertion or deletion at a single site within a
given sequence.
A "subject", as used herein, refers to a patient or test subject, for example
a human patient.
The subject may have been previously diagnosed with a neoplastic disorder, or
may be
suspected of having a neoplastic disorder and thus may be a candidate for a
chemotherapeutic regimen. The subject may be selected as part of a general
population
(for example a 'control' subject), or may be selected as part of a particular
ethnic, gender,
age or genetic subgroup of a population, or may be excluded from selection as
part of a
particular ethnic, gender, age or genetic subgroup of a population. Patients
and test
subjects, whether control or not, may be generally referred to as a subject.
As used herein, the terms "cancer" or "neoplastic condition" or "neoplastic
disorder" or
"neoplastic disease" refer to a proliferative disorder caused or characterized
by the
proliferation of cells which have lost susceptibility to normal growth
control. A "cancer" or
neoplastic condition" or "neoplastic disorder" or "neoplastic disease" may
include tumors
and any other proliferative disorders. Cancers of the same tissue type usually
originate in
the same tissue, and may be divided into different subtypes based on their
biological
characteristics. Four general categories of cancers are carcinoma (epithelial
tissue
derived), sarcoma (connective tissue or mesodermal derived), leukemia (blood-
forming
tissue derived) and lymphoma (lymph tissue derived). Over 200 different types
of cancers
are known, and every organ and tissue of the body may be affected. Specific
examples of
cancers that do not limit the definition of cancer may include melanoma,
leukemia,
astrocytoma, glioblastoma, retinoblastoma, lymphoma, glioma, Hodgkins'
lymphoma and
chronic lymphocyte leukemia. Examples of organs and tissues that may be
affected by
various cancers include pancreas, breast, thyroid, ovary, uterus, testis,
prostate, thyroid,
pituitary gland, adrenal gland, kidney, stomach, esophagus or rectum, head and
neck, bone,
nervous system, skin, blood, nasopharyngeal tissue, lung, urinary tract,
cervix, vagina,
CA 02911709 2015-11-10
exocrine glands and endocrine glands. Alternatively, a cancer may be
multicentric or of
unknown primary site (CUPS).
As used herein, a "therapeutic regimen" refers to a chemotherapeutic regimen
or a
radiotherapy regimen, or a combination thereof.
As used herein, a "chemotherapeutic regimen" or "chemotherapy" refers to the
use of at
least one chemotherapy agent to destroy cancerous cells. There are a myriad of
such
chemotherapy agents available for treating cancer. Chemotherapy agents may be
administered to a subject in a single bolus dose, or may be administered in
smaller doses
over time. A single chemotherapeutic agent may be used (single-agent therapy)
or more
than one agent may be used in combination (combination therapy). Chemotherapy
may be
used alone to treat some types of cancer. Alternatively, chemotherapy may be
used in
combination with other types of treatment, for example, radiotherapy or
alternative
therapies (for example immunotherapy) as described herein. Additionally, a
chemosensitizer may be administered as a combination therapy with a
chemotherapy
agent.
As used herein, a "chemotherapeutic agent" or "chemotherapeutic agent" refers
to a
medicament that may be used to treat cancer, and generally has the ability to
kill cancerous
cells directly. Examples of chemotherapeutic agents include alkylating agents,
antimetabolites, natural products, hormones and antagonists, and miscellaneous
agents.
Examples of alternate names are indicated in brackets. Examples of alkylating
agents
include nitrogen mustards such as mechlorethamine, cyclophosphamide,
ifosfamide,
melphalan (L-sarcolysin) and chlorambucil; ethylenimines and methylmelamines
such as
hexamethylmelamine and thiotepa; alkyl sulfonates such as busulfan;
nitrosoureas such as
carmustine (BCNU), semustine (methyl-CCNU), lomustine (CCNU) and streptozocin
(streptozotocin); DNA synthesis antagonists such as estramustine phosphate;
and triazines
such as dacarbazine (DTIC, dimethyl-triazenoimidazolecarboxamide) and
temozolomide .
Examples of antimetabolites include folic acid analogs such as methotrexate
(amethopterin); pyrimidine analogs such as fluorouracin (5-fluorouracil, 5-FU,
5FU),
floxuridine (fluorodeoxyuridine, FUdR), cytarabine (cytosine arabinoside) and
gemcitabine; purine analogs such as mercaptopurine (6-mercaptopurine, 6-MP),
thioguanine (6-thioguanine, TG) and pentostatin (2'-deoxycoformycin,
deoxycoformycin),
cladribine and fludarabine; and topoisomerase inhibitors such as amsacrine.
Examples of
natural products include vinca alkaloids such as vinblastine (VLB) and
vincristine; taxanes
such as paclitaxel and docetaxel (Taxotere); epipodophyllotoxins such as
etoposide and
21
CA 02911709 2015-11-10
teniposide; camptothecins such as topotecan or irinotecan; antibiotics such as
dactinomycin (actinomycin D), bleomycin, mitomycin (mitomycin C);
anthracycline
antibiotics such as daunorubicin (daunomycin, rubidomycin), doxorubicin,
idarubicin,
epirubicin; enzymes such as L-asparaginase; and biological response modifiers
such as
interferon alpha and interleukin 2. Examples of hormones and antagonists
include
luteinising releasing hormone agonists such as buserelin;
adrenocorticosteroids such as
prednisone and related preparations; progestins such as hydroxyprogesterone
caproate,
medroxyprogesterone acetate and megestrol acetate; estrogens such as
diethylstilbestrol
and ethinyl estradiol and related preparations; estrogen antagonists such as
tamoxifen and
anastrozole; androgens such as testosterone propionate and fluoxymesterone and
related
preparations; androgen antagonists such as flutamide and bicalutamide; and
gonadotropin-releasing hormone analogs such as leuprolide. Examples of
miscellaneous
agents include thalidomide; platinum coordination complexes such as cisplatin
(cis-DDP),
carboplatin, oxaliplatin, tetraplatin, ormiplatin, iproplatin or satraplatin;
anthracenediones
such as mitoxantrone; substituted ureas such as hydroxyurea; methylhydrazine
derivatives
such as procarbazine (N-methylhydrazine, MIH); adrenocortical suppressants
such as
mitotane (o,p'-DDD) and aminoglutethimide; RXR agonists such as bexarotene; or
tyrosine
kinase inhibitors such as imatinib. Alternate names and trade-names of these
and
additional examples of chemotherapeutic agents, and their methods of use
including dosing
and administration regimens, will be known to an individual versed in the art,
and may be
found in, for example "The Pharmacological basis of therapeutics", loth
edition.
HARDMAN HG., LIMBIRD LE. editors. McGraw-Hill, New York, or in "Clinical
Oncology",
3rd edition. Churchill Livingstone/ Elsevier Press, 2004. ABELOFF, MD. editor.
2. General Methods
Once a subject is identified as a candidate for anthracycline administration,
then genetic
sequence information may be obtained from the subject to determine the risk of
cardiotoxicity for the subject. Genetic sequence information may be obtained
from a
subject by any of several methods. For example, a biological sample comprising
genetic
material with a sequence or sequences of interest, may be obtained from the
subject, for
example a blood sample, a saliva sample, a hair sample including a follicle,
skin scraping,
such as a cheek scraping and the like. Or alternatively genetic sequence
information may
already have been obtained from the subject. For example, a subject may have
already
provided a biological sample for other purposes or may have even had their
genetic
sequence determined in whole or in part and stored for future use. Genetic
sequence
information may be obtained in numerous different ways and may involve the
collection of
a biological sample that contains genetic material, particularly, genetic
material containing
22
CA 02911709 2015-11-10
the sequence or sequences of interest. Many methods are known in the art for
collecting
biological samples and extracting genetic material from those samples. Genetic
material
can be extracted from blood, tissue, hair and other biological material. There
are many
methods known to isolate DNA and RNA from biological material. Typically, DNA
may be
isolated from a biological sample when first the sample is lysed and then the
DNA is
separated from the lysate according to any one of a variety of multi-step
protocols, which
can take varying lengths of time. DNA isolation methods may involve the use of
phenol
(Sambrook, J. et al., "Molecular Cloning", Vol. 2, pp. 9.14-9.23, Cold Spring
Harbor
Laboratory Press (1989) and Ausubel, Frederick M. et al., "Current Protocols
in Molecular
Biology", Vol. 1, pp. 2.2.1-2.4.5, John Wiley & Sons, Inc. (1994)). Typically,
a biological
sample is lysed in a detergent solution and the protein component of the
lysate is digested
with proteinase for 12-18 hours. Next, the lysate is extracted with phenol to
remove most of
the cellular components, and the remaining aqueous phase is processed further
to isolate
DNA. In another method, described in Van Ness et al. (U.S. Pat. # 5,130,423),
non-
corrosive phenol derivatives are used for the isolation of nucleic acids. The
resulting
preparation is a mix of RNA and DNA.
Other methods for DNA isolation utilize non-corrosive chaotropic agents. These
methods,
which are based on the use of guanidine salts, urea and sodium iodide, involve
lysis of a
biological sample in a chaotropic aqueous solution and subsequent
precipitation of the
crude DNA fraction with a lower alcohol. The resulting nucleic acid sample may
be used
`as-is' in further analyses or may be purified further. Additional
purification of the
precipitated, crude DNA fraction may be achieved by any one of several
methods,
including, for example, column chromatography (Analects, (1994) Vol 22, No. 4,
Pharmacia
Biotech), or exposure of the crude DNA to a polyanion-containing protein as
described in
Koller (U.S. Pat. # 5,128,247).
Yet another method of DNA isolation, which is described by Botwell, D. D. L.
(Anal.
Biochem. (1987) 162:463-465) involves lysing cells in 6M guanidine
hydrochloride,
precipitating DNA from the lysate at acid pH by adding 2.5 volumes of ethanol,
and
washing the DNA with ethanol.
Numerous other methods are known in the art to isolate both RNA and DNA, such
as the
one described by CHOMCZYNSKI (U.S. Pat. # 5,945,515), whereby genetic material
can be
extracted efficiently in as little as twenty minutes. EVANS and HUGH (U.S.
Pat. #
5,989,431) describe methods for isolating DNA using a hollow membrane filter.
23
CA 02911709 2015-11-10
The level of expression of specific nucleic acids such as mRNAs or microRNAs,
copy
number of a gene, or the degree of heterozygosity for a polymorphism may also
be
determined once the nucleic acid sample has been obtained. Quantitative and
semi-
quantitative methods are known in the art, and may be found in, for example
AUSUBEL,
supra; SAMBROOK, supra or Harrison's Principles of Internal Medicine 15th ed.
BRAUNWALD et al. eds. McGraw-Hill.
Once a subject's genetic material has been obtained from the subject it may
then be further
be amplified by Reverse Transcription Polymerase Chain Reaction (RT-PCR),
Polymerase
Chain Reaction (PCR), Transcription Mediated Amplification (TMA), Ligase chain
reaction
(LCR), Nucleic Acid Sequence Based Amplification (NASBA) or other methods
known in
the art, and then further analyzed to detect or determine the presence or
absence of one or
more polymorphisms or mutations in the sequence of interest, provided that the
genetic
material obtained contains the sequence of interest. Particularly, a person
may be
interested in determining the presence or absence of a polymorphism in a
cardiotoxicity
associated gene sequence, as described herein.
Detection or determination of a nucleotide identity, or the presence of one or
more single
nucleotide polymorphism(s) (SNP typing), may be accomplished by any one of a
number
methods or assays known in the art. Many DNA typing methodologies are useful
for use in
the detection of SNPs. The majority of SNP genotyping reactions or assays can
be assigned
to one of four broad groups (sequence-specific hybridization, primer
extension,
oligonucleotide ligation and invasive cleavage). Furthermore, there are
numerous methods
for analyzing/detecting the products of each type of reaction (for example,
fluorescence,
luminescence, mass measurement, electrophoresis, etc.). Furthermore, reactions
can occur
in solution or on a solid support such as a glass slide, a chip, a bead, etc.
In general, sequence-specific hybridization involves a hybridization probe,
which is capable
of distinguishing between two DNA targets differing at one nucleotide position
by
hybridization. Usually probes are designed with the polymorphic base in a
central position
in the probe sequence, whereby under optimized assay conditions only the
perfectly
matched probe target hybrids are stable and hybrids with a one base mismatch
are
unstable. A strategy which couples detection and sequence discrimination is
the use of a
molecular beacon", whereby the hybridization probe (molecular beacon) has 3'
and 5'
reporter and quencher molecules and 3' and 5' sequences which are
complementary such
that absent an adequate binding target for the intervening sequence the probe
will form a
hairpin loop. The hairpin loop keeps the reporter and quencher in close
proximity
24
CA 02911709 2015-11-10
resulting in quenching of the fluorophor (reporter) which reduces fluorescence
emissions.
However, when the molecular beacon hybridizes to the target the fluorophor and
the
quencher are sufficiently separated to allow fluorescence to be emitted from
the fluorophor.
Similarly, primer extension reactions (i.e. mini sequencing, nucleotide-
specific extensions,
or simple PCR amplification) are useful in sequence discrimination reactions.
For example,
in mini sequencing a primer anneals to its target DNA immediately upstream of
the SNP
and is extended with a single nucleotide complementary to the polymorphic
site. Where
the nucleotide is not complementary, no extension occurs.
Oligonucleotide ligation assays require two sequence-specific probes and one
common
ligation probe per SNP. The common ligation probe hybridizes adjacent to a
sequence-
specific probe and when there is a perfect match of the appropriate sequence-
specific
probe, the ligase joins both the sequence-specific and the common probes.
Where there is
- not a perfect match the ligase is unable to join the sequence-specific and
common probes.
Probes used in hybridization can include double-stranded DNA, single-stranded
DNA and
RNA oligonucleotides, and peptide nucleic acids. Hybridization methods for the
identification of single nucleotide polymorphisms or other mutations involving
a few
nucleotides are described in the U.S. Pat. 6,270,961; 6,025,136; and
6,872,530. Suitable
hybridization probes for use in accordance with the invention include
oligonucleotides and
PNAs from about 10 to about 400 nucleotides, alternatively from about 20 to
about 200
nucleotides, or from about 30 to about loo nucleotides in length.
A unimolecular segment amplification method for amplifying nucleic acids is
described in
US patent 5854033. A rolling circle replication reporter system may be used
for
identification of polymorphisms or mutations.
An invasive cleavage method employs an "InvaderTM" (Applied Biosystems) probe
and
sequence-specific probes to hybridize with the target nucleic acid, usually
DNA, with an
overlap of one nucleotide. When the sequence specific probe is an exact match
to the site of
polymorphism, the overlapping probes form a structure that is specifically
cleaved by a
FLAP endonuclease, Release of the 5' end of the allele-specific probe may be
detected by
known methods as described. See for example, Lu, M., et al. J. Am. Chem. Soc.
2001, 124,
7924 ¨ 7931; Lyamichev, etal. 1999. Nature Biotech. 17, 292 ¨ 296; Landegren
et al. 1998.
Genome Research, 8, 769 ¨ 776; Brookes, 1999. Gene 234, 177 ¨ 186; Chen, et
al. 2004. J.
Am. Chem. Soc. 126, 3016-3017; Wang, D.G., etal. Science 1998, 280, 1077 ¨
1082. The
TaqMan' assay (Applied Biosystems) exploits the 5' exonuclease activity of the
Taq
CA 02911709 2015-11-10
polymerase to displace and cleave an oligonucleotide probe hybridized to the
target nucleic
acid, usually DNA, generating a fluorescent signal. See, for example U.S.
Patents
4,683,202, 4,683,195, and 4,965,188.
5' exonuclease activity or TaqManTm assay (Applied BiosystemsTM) is based on
the 5'
nuclease activity of Taq polymerase that displaces and cleaves the
oligonucleotide probes
hybridized to the target DNA generating a fluorescent signal. It is necessary
to have two
probes that differ at the polymorphic site wherein one probe is complementary
to the
'normal' sequence and the other to the mutation of interest. These probes have
different
fluorescent dyes attached to the 5' end and a quencher attached to the 3' end
when the
probes are intact the quencher interacts with the fluorophor by fluorescence
resonance
energy transfer (FRET) to quench the fluorescence of the probe. During the PCR
annealing
step the hybridization probes hybridize to target DNA. In the extension step
the 5'
fluorescent dye is cleaved by the 5' nuclease activity of Taq polymerase,
leading to an
increase in fluorescence of the reporter dye. Mismatched probes are displaced
without
fragmentation. The presence of a mutation in a sample is determined by
measuring the
signal intensity of the two different dyes.
The Illumina Golden GateTM Assay uses a combined oligonucleotide ligation
assay/ allele-
specific hybridization approach (SHEN R etal. Mutat Res 2005573:70-82). The
first series
of steps involve the hybridization of three oligonucleotides to a set of
specific target SNPs;
two of these are fluorescently-labelled allele-specific oligonucleotides
(AS0s) and the third
a locus-specific oligonucleotide (LSO) binding 1-20 bp downstream of the ASOs.
A second
series of steps involve the use of a stringent polymerase with high 3'
specificity that extends
only oligonucleotides specifically matching an allele at a target SNP. The
polymerase
extends until it reaches the LSO. Locus-specificity is ensured by requiring
the
hybridization of both the ASO and LSO in order that extension can proceed.
After PCR
amplification with universal primers, these allele-specific oligonucleotide
extension
products are hybridized to an array which has multiple discretely tagged
addresses (in this
case 1536 addresses) which match an address embedded in each LSO. Fluorescent
signals
produced by each hybridization product are detected by a bead array reader
from which
genotypes at each SNP locus may be ascertained.
It will be appreciated that numerous other methods for sequence discrimination
and
detection are known in the art and some of which are described in further
detail below. It
will also be appreciated that reactions such as arrayed primer extension mini
sequencing,
tag microarrays and sequence-specific extension could be performed on a
microarray. One
26
CA 02911709 2015-11-10
such array based genotyping platform is the microsphere based tag-it high
throughput
genotyping array (BORTOLIN S. et al. Clinical Chemistry (2004) 50(11): 2028-
36). This
method amplifies genomic DNA by PCR followed by sequence-specific primer
extension
with universally tagged genotyping primers. The products are then sorted on a
Tag-It array
and detected using the Luminex xMAPTm system.
Mutation detection methods may include but are not limited to the following:
Restriction Fragment Length Polymorphism (RFLP) strategy ¨ An RFLP gel-based
analysis
can be used to indicate the presence or absence of a specific mutation at
polymorphic sites
within a gene. Briefly, a short segment of DNA (typically several hundred base
pairs) is
amplified by PCR. Where possible, a specific restriction endonuclease is
chosen that cuts
the short DNA segment when one polymorphism is present but does not cut the
short DNA
segment when the polymorphism is not present, or vice versa. After incubation
of the PCR
amplified DNA with this restriction endonuclease, the reaction products are
then separated
using gel electrophoresis. Thus, when the gel is examined the appearance of
two lower
molecular weight bands (lower molecular weight molecules travel farther down
the gel
during electrophoresis) indicates that the DNA sample had a polymorphism was
present
that permitted cleavage by the specific restriction endonuclease. In contrast,
if only one
higher molecular weight band is observed (at the molecular weight of the PCR
product)
then the initial DNA sample had the polymorphism that could not be cleaved by
the chosen
restriction endonuclease. Finally, if both the higher molecular weight band
and the two
lower molecular weight bands are visible then the DNA sample contained both
polymorphisms, and therefore the DNA sample, and by extension the subject
providing the
DNA sample, was heterozygous for this polymorphism;
For example the Maxam-Gilbert technique for sequencing (MAXAM AM. and GILBERT
W.
Proc. Natl. Acad. Sci. USA (1977) 74(4):56o-564) involves the specific
chemical cleavage of
terminally labelled DNA. In this technique four samples of the same labeled
DNA are each
subjected to a different chemical reaction to effect preferential cleavage of
the DNA
molecule at one or two nucleotides of a specific base identity. The conditions
are adjusted
to obtain only partial cleavage, DNA fragments are thus generated in each
sample whose
lengths are dependent upon the position within the DNA base sequence of the
nucleotide(s)
which are subject to such cleavage. After partial cleavage is performed, each
sample
contains DNA fragments of different lengths, each of which ends with the same
one or two
of the four nucleotides. In particular, in one sample each fragment ends with
a C, in
another sample each fragment ends with a C or a T, in a third sample each ends
with a G,
and in a fourth sample each ends with an A or a G. When the products of these
four
27
CA 02911709 2015-11-10
reactions are resolved by size, by electrophoresis on a polyacrylamide gel,
the DNA
sequence can be read from the pattern of radioactive bands. This technique
permits the
sequencing of at least 100 bases from the point of labeling. Another method is
the dideoxy
method of sequencing was published by SANGER et al. (Proc. Natl. Acad. Sci.
USA (1977)
74(12):5463-5467). The Sanger method relies on enzymatic activity of a DNA
polymerase
to synthesize sequence-dependent fragments of various lengths. The lengths of
the
fragments are determined by the random incorporation of dideoxynucleotide base-
specific
terminators. These fragments can then be separated in a gel as in the Maxam-
Gilbert
procedure, visualized, and the sequence determined. Numerous improvements have
been
made to refine the above methods and to automate the sequencing procedures.
Similarly,
RNA sequencing methods are also known. For example, reverse transcriptase with
dideoxynucleotides have been used to sequence encephalomyocarditis virus RNA
(ZIMMERN D. and KAESBERG P. Proc. Natl. Acad. Sci. USA (1978) 75(9):4257-
4261).
MILLS DR. and KRAMER FR. (Proc. Natl. Acad. Sci. USA (1979) 76(5):2232-2235)
describe the use of QI3 replicase and the nucleotide analog inosine for
sequencing RNA in a
chain-termination mechanism. Direct chemical methods for sequencing RNA are
also
known (PEATTIE DA. Proc. Natl. Acad. Sci. USA (1979) 76(4):1760-1764). Other
methods
include those of Donis-Keller etal. (1977, Nucl. Acids Res. 4:2527-2538),
SIMONCSITS A.
etal. (Nature (1977) 269(5631):833-836), AXELROD VD. etal. (Nucl. Acids
Res.(1978)
5(10):3549-3563), and KRAMER FR. and MILLS DR. (Proc. Natl. Acad. Sci. USA
(1978)
75(11):5334-5338). Nucleic acid sequences can also be read by stimulating the
natural
fluoresce of a cleaved nucleotide with a laser while the single nucleotide is
contained in a
fluorescence enhancing matrix (U.S. Pat. # 5,674,743); In a mini sequencing
reaction, a
primer that anneals to target DNA adjacent to a SNP is extended by DNA
polymerase with a
single nucleotide that is complementary to the polymorphic site. This method
is based on
the high accuracy of nucleotide incorporation by DNA polymerases. There are
different
technologies for analyzing the primer extension products. For example, the use
of labeled
or unlabeled nucleotides, ddNTP combined with dNTP or only ddNTP in the mini
sequencing reaction depends on the method chosen for detecting the products.
Probes used in hybridization can include double-stranded DNA, single-stranded
DNA and
RNA oligonucleotides, and peptide nucleic acids. Hybridization methods for the
identification of single nucleotide polymorphisms or other mutations involving
a few
nucleotides are described in the U.S. Pat. 6,270,961; 6,025,136; and
6,872,530. Suitable
hybridization probes for use in accordance with the invention include
oligonucleotides and
PNAs from about io to about 400 nucleotides, alternatively from about 20 to
about 200
nucleotides, or from about 30 to about too nucleotides in length.
28
CA 02911709 2015-11-10
A template-directed dye-terminator incorporation with fluorescent polarization-
detection
(TDI-FP) method is described by FREEMAN BD. et al. (J Mol Diagnostics (2002)
4(4):209-215) for large scale screening.
Oligonucleotide ligation assay (OLA) is based on ligation of probe and
detector
oligonucleotides annealed to a polymerase chain reaction amplicon strand with
detection
by an enzyme immunoassay (VILLAHERMOSA ML. J Hum Virol (2001) 4(5):238-48;
ROMPPANEN EL. Scand J Clin Lab Invest (2001) 61(2):123-9; IANNONE MA. etal.
Cytometry (2000) 39(2):131-40).
Ligation-Rolling Circle Amplification (L-RCA) has also been successfully used
for
genotyping single nucleotide polymorphisms as described in QI X. et al.
Nucleic Acids Res
(2001) 29(22):E116.
5' nuclease assay has also been successfully used for genotyping single
nucleotide
polymorphisms (AYDIN A. etal. Biotechniques (2001) (4):920-2, 924, 926-8.).
Polymerase proofreading methods are used to determine SNPs identities, as
described in
WO 0181631.
Detection of single base pair DNA mutations by enzyme-amplified electronic
transduction
is described in PATOLSKY F et al. Nat Biotech. (2001) 19(3):253-257.
Gene chip or microarray technologies are also known for single nucleotide
polymorphism
discrimination whereby numerous polymorphisms may be tested for simultaneously
on a
single array (for example: EP 1120646; and GILLES PN. et al. Nat.
Biotechnology (1999)
17(4):365-7o).
Matrix assisted laser desorption ionization time of flight (MALDI-TOF) mass
spectroscopy
is also useful in the genotyping single nucleotide polymorphisms through the
analysis of
microsequencing products (HAFF LA. and SMIRNOV IP. Nucleic Acids Res. (1997)
25(18):3749-50; HAFF LA. and SMIRNOV IP. Genome Res. (1997) 7:378-388; SUN X.
et
al. Nucleic Acids Res. (2000) 28 e68; BRAUN A. etal. Clin. Chem. (1997)
43:1151-1158;
LITTLE DP. etal. Eur. J. Clin. Chem. Clin. Bioehem. (1997) 35:545-548; FEI Z.
etal.
Nucleic Acids Res. (2000) 26:2827-2828; and BLONDAL T. et al. Nucleic Acids
Res.
(2003) 31(24):e155).
29
CA 02911709 2015-11-10
Sequence-specific PCR methods have also been successfully used for genotyping
single
nucleotide polymorphisms (HAWKINS JR. et a/. Hum Mutat (2002) 19(5):543-553).
Alternatively, a Single-Stranded Conformational Polymorphism (SSCP) assay or a
Cleavase
Fragment Length Polymorphism (CFLP) assay may be used to detect mutations as
described herein.
US 7,074,597 describes methods for multiplex genotyping using solid phase
capturable
dideoxynucleotides and mass spectrometry. Nucleotide identity is detected at a
specific site
of a nucleic acid sample by contacting DNA-primer complex with labeled
dideoxynucleotides (ddNTPs) to generate labeled single base extended (SBE)
primer. The
identifying ddNTP may be within the SBE primer.
Multiplex analysis of PCR-amplified products may also be used to detect
specific SNPs.
Reporting DNA sequences comprising a fluorophore on a 5' end may be used to
combine a
multiplex PCR amplification reaction with microsphere based hybridization (US
7,083,951). Other multiplex detection methods include BeadArrayTM and similar
hybridization-based methods, for example, those described in US Patent Nos.
6,429,027,
6,396,995, 6,355,431.
Microarray or 'gene chips' of oligonucleotides may be used for SNP
discrimination.
Oligonucleotides may be nucleic acids or modified nucleic acids, including
PNAs, and may
be 'spotted' onto a solid matrix, such as a glass or plastic slide.
Alternatively,
oligonucleotides may be synthesized in situ on the slide. See, for example,
GAO et al. 2004.
Biopolymers 73:579-596; US 5,445,934; US 5,744,305, US 5,800,992, US
5,796,715.
Alternatively, if a subject's sequence data is already known, then obtaining
may involve
retrieval of the subjects nucleic acid sequence data (for example from a
database), followed
by determining or detecting the identity of a nucleic acid or genotype at a
polymorphic site
by reading the subject's nucleic acid sequence at the one or more polymorphic
sites.
Once the identity of a polymorphism(s) is determined or detected an indication
may be
obtained as to the subject's risk of cardiotoxicity following anthracycline
administration.
Methods for predicting a subject's risk of cardiotoxicity following
anthracycline
administration may be useful in making decisions regarding the administration
of
anthracycline(s).
CA 02911709 2015-11-10
TREATMENT
Anthracycline compounds (for example, doxorubicin) may be used to treat a
variety of
cancers in children and adults. In a given therapeutic regimen, the
anthracycline
compound may be administered alone or in combination with other
chemotherapeutic
agents in various doses and compositions, depending on the type of cancer, age
of subject,
health of subject, body mass, etc. The choice of dose, chemotherapeutic agents
or
combinations, methods of administration and the like will be known to those
skilled in the
art. Further, methods of assessing response to treatment and side effects are
also known.
For example, heart function in a subject suspected of experiencing
cardiotoxicity may be
assessed by various methods including medical history, electrocardiogram (ECG)
monitoring, endomyocardial biopsy, radionuclide angiography (MUGA scan) or
LVEF
monitoring with serial echo or exercise stress testing, or other methods that
may be
dependent on the age and condition of the subject, as are known in the art.
Early signs of
cardiotoxicity may include persistent reduction in the voltage of the QRS
wave,
prolongation of the systolic time interval, or reduction of LVEF as determined
by echo or
MUGA. A reduction of 10% to below the lower limit of normal, 20% at any level,
or an
absolute LVEF 45% indicates deterioration of cardiac function.
Response to a therapeutic regimen may be monitored. Tumor staging provides a
method to
assess the size and spread of a tumor in response to a treatment regimen. The
TNM tumor
staging system uses three components to express the anatomic extent of
disease: T is a
measure of the local extent of tumor spread (size), N indicates the presence
or absence of
metastatic spread to regional lymph nodes, and M specifies the presence or
absence of
metastatic spread to distant sites. The combination of these classifications
combine to
provide a stage grouping. Clinical TNM (cTNM) defines the tumor based on
clinical
evidence. Pathologic TNM (pTNM) defines the tumor based on examination of a
surgically
resected specimen.
Changes in tumor size may be observed by various imaging methods known to
physicians
or surgeons in the field of oncology therapy and diagnostics. Examples of
imaging methods
include positron emission tomography (PET) scanning, computed tomography (CT)
scanning, PET/CT scanning, magnetic resonance imaging (MRI), chemical shift
imaging,
radiography, bone-scan, mammography, fiberoptic colonoscopy or ultrasound.
Contrast
agents, tracers and other specialized techniques may also be employed to image
specific
types of cancers, or for particular organs or tissues, and will be known to
those skilled in
the art. Changes in rate of metastasis may also be observed by the various
imaging
methods, considering particularly the appearance, or frequency of appearance,
of tumors
31
CA 02911709 2015-11-10
distal to the primary site. Alternatively, the presence of tumor cells in
lymph nodes
adjacent and distal to the primary tumor site may also be detected and used to
monitor
metastasis.
A subject may be tested for a cardiotoxicity-associated polymorphism before
undergoing a
therapeutic regimen involving an anthracycline compound. If a subject's
genotype includes
a cardiotoxicity-associated polymorphism, this may indicate that the subject
is at a risk for
cardiotoxicity when an anthracycline compound is administered.
A subject at risk for cardiotoxicity may be administered a therapeutic regimen
involving an
anthracycline compound and the cardiac function monitored as described. If a
decrease in
cardiac function is identified, the therapeutic regimen may be altered to
decrease the dose
of the anthracycline compound, eliminate the dose of the anthracycline
compound, or
increase the dose of a second chemotherapeutic agent in the therapeutic
regimen.
Examples of chemotherapeutic agents that may be used in combination with an
anthracycline compound in a therapeutic regimen may include, for example,
cyclophosphamide, Ifosphamide, fluorouracil, Paclitaxel, vincristine,
cisplatin,
streptozocin, docetaxel, and the like.
A subject at risk for cardiotoxicity may also be administered a therapeutic
regimen
involving an anthracycline compound and the cardiac function monitored as
described.
The therapeutic regimen may be supplemented to include a cardioprotective
agent.
Examples of cardioprotective agents are known in the art, and may include
those described
by Wouters et al. 2005. Br. J Hematol 131:561-578). For example, Dexrazoxane
is a
cardioprotective agent and is approved for use in conjunction with doxorubicin
to reduce
the incidence and severity of cardiomyopathy associated with doxorubicin
administration.
Alternatively, a subject at risk for cardiotoxicity may be administered a
therapeutic regimen
that does not involve an anthracycline compound and the cardiac function
monitored as
described.
Alternatively, a subject at risk for cardiotoxicity may be administered a
cardioprotective
agent in conjunction with a therapeutic regimen.
32
CA 02911709 2015-11-10
GENE
Detailed information relating to the sequence, expression patterns, molecular
biology, etc
of these and related genes in both Homo sapiens and in other model species is
known, and
may be found at, for example Entrez Gene (NCBI ) and references therein.
Retinoic acid receptor, gamma (RARG) [Homo sapiensRalternative designations
RARC;
NR1B3) maps to chromosome 22q13. A representative human RARG sequence may be
found in GenBank under accession number NT o29419.11 BG740799. RARG encodes a
retinoic acid receptor that belongs to the nuclear hormone receptor family.
Retinoic acid
receptors (RARs) act as ligand-dependent transcriptional regulators and when
bound to
ligands, RARs activate transcription by binding as heterodimers to the
retinoic acid
response elements (RARE) found in the promoter regions of the target genes.
Unbound
RARs repress transcription of their target genes. RARs are involved in various
biological
processes, including limb bud development, skeletal growth, and matrix
homeostasis.
METHODS
Study Design
In stage 1, a total of 280 European Canadian patients (32 cases and 248
controls) were
used as the discovery cohort and genotyped on the Illumina Infinium Human Omni
ExpressTM panel (740K), to perform a GWAS for ACT (stage 1). Markers that
reached P <
i.oxio-5 (ref. (Welter, MacArthur et al. 2014)) in the discovery cohort were
tested for
replication in 96 European Dutch patients (22 cases and 74 controls) in stage
2. RARG
rs2229774 was further tested for association with ACT in non-European
populations in
stage 3. We also performed a combined analysis for all European patients
(stage 1 and
stage 2) and an overall association test for rs2229774 using all populations
(stages 1-3). In
addition, we performed genetic fine mapping analyses of the associated region
and
functionally characterized n2229774 in vitro.
Genetic ancestry was self-reported and ascertained by principal component
analysis (PCA)
using the EIGENSTRAT method, with the GWAS Illumina 740K (Canadian patients),
Illumina 4.5K (Dutch patients) and Illumina 8K (USA patients) SNP genotype
data sets.
Detailed Analysis of Clinical Data
This study was approved by the individual ethics committees/institutional
review boards of
the universities and institutions where patients were enrolled. Written
informed
consent/assent was obtained from patients/parents/legal guardians in
accordance with the
Helsinki Declaration as revised in 2008.
33
CA 02911709 2015-11-10
Patients' medical records were reviewed prior to genotyping by a clinical
pharmacologist, a
cardiologist, an oncologist and an ADR surveillance clinician, who reviewed
the
echocardiogram test results and other clinical information. The demographic,
clinical and
therapeutic information was extracted from these medical records and included
the
following data: demographics, disease characteristics, chemotherapy,
diagnostic
echocardiograms to document baseline and follow-up cardiac function and any
cardiac
compromise and its severity and any symptoms and/or signs consistent with ACT.
All
patients were children 18 years at cancer diagnosis, who received
anthracyclines as part
of their chemotherapy protocol, had normal cardiac function before
anthracycline
chemotherapy and were recruited from outpatient clinics and inpatient units.
ACT was
monitored by echocardiograms (or comparable cardiac imaging) according to the
"The
Children's Oncology Group Long-Term Follow-Up Guidelines for Survivors of
Childhood,
Adolescent, and Young Adult Cancers". The recommended frequency of
echocardiograms
(or comparable cardiac imaging) is every year, every two years or every 5
years post-
treatment depending on the age at treatment, radiation with potential impact
to the heart
and the cumulative anthracycline dose. Grading of ACT based on this detailed
clinical
characterization was performed using the Cancer Therapy Evaluation Program,
Common
Terminology Criteria for Adverse Events version 3 (CTCAEv3) as previously
described
(Visscher, Ross et al. 2012; Visscher, Ross et al. 2013). Patients with
serious ACT were
defined as those with grade 2 or higher CTCAE impairment of cardiac function
after
treatment with anthracycline.
ACT was defined as early- or late-onset left ventricular dysfunction assessed
by
echocardiogram measurements using shortening fraction [SF]) and/or symptoms
(dyspnea, orthopnea, and/or fatigue) and/or signs (edema, hepatomegaly, and/or
rales) of
cardiac compromise requiring intervention based on the CTCAEv3. Due to
variability in
echocardiographic measurements a conservative threshold of SF 24% was used to
define
asymptomatic cases who developed ACT. All cases had grade 2 or higher ACT.
Also, grade
2-4 ACT is the point at which anthracycline chemotherapy protocols recommend
clinical
intervention such as heart failure treatment, halting or reducing
anthracycline doses and
switching to alternate treatments. Early-onset chronic ACT was defined as
developing less
than 1 year after start of treatment(Rodvold, Rushing et al. 1988; Kremer, van
Dalen et al.
2001) while late-onset chronic ACT was defined as developing more than 1 year
after start
of treatment (Lipshultz, Colan et al. 1991; Lipshultz, Lipsitz et al. 1995)
since ventricular
dysfunction, heart failure, and arrhythmias can occur years or even decades
after the
discontinuation of anthracycline therapy(Lipshultz, Colan et al. 1991;
Lipshultz, Lipsitz et
al. 1995). To exclude transient acute ACT, only echocardiograms obtained 21
days after
34
CA 02911709 2015-11-10
an anthracycline dose were considered. To exclude cardiotoxicity unrelated to
anthracycline chemotherapy patients with no baseline echocardiogram were
excluded from
the study. Controls had no signs or symptoms of cardiac compromise at study
participation
(grade o). Due to the delayed onset of ACT in some patients, SF 30% with 5
years of
follow-up after the end of anthracycline treatment was used to define
controls. The
cumulative anthracycline exposure was calculated using the doxorubicin
isotoxic equivalent
(Altman 2004) and cumulative dose stratification into low-to-moderate 250
mg/m2) and
at high (> 250 mg/m2) anthracycline exposure was performed as previously
described
(Mulrooney, Yeazel et al. 2009; Blanco, Sun et al. 2012; Wang, Liu et al.
2014) where
appropriate. Radiation therapy included significant radiation exposure to the
heart or
surrounding tissue. This included mantle and mediastinal radiation, whole lung
radiation,
whole or upper abdominal radiation, left sided flank radiation and total body
irradiation.
Study Populations
Canadian Patient Populations: We genotyped 434 pediatric oncology patients
treated
with anthracyclines that were recruited from 13 pediatric oncology units from
across
Canada between February 2005 and April 2011 (Visscher, Ross et al. 2012;
Visscher, Ross
et al. 2013). A total of 13 samples failed quality control (details of quality
control procedure
described below). In 421 remaining patients (average call rate = 99.5%), we
performed
principal component analysis (PCA) with GWAS IlluminaTM 740K SNP genotype data
set to
determine the population structure and defined four distinct population
clusters comprised
of European, African, East Asian and Aboriginal Canadian patients. Patients
with genetic
ancestry that fell outside of these four population clusters were excluded
from further
analysis. We excluded an additional 27 patients with CTCAE grade 1 toxicity
(shortening
fraction: 24% < SF < 30%). A total of 97 patients were excluded and 337
patients were
available for further analyses: Europeans (280 patients; 32 cases and 248
controls),
Africans (n = ii patients; 2 cases and 9 controls), East Asians (n = 31
patients; 8 cases and
23 controls) and Aboriginal Canadians (n = 15 patients; 4 cases and ii
controls).
Dutch Patient Population: We recruited 128 pediatric oncology patients treated
with
anthracyclines from Emma Children's Hospital/Academic Medical Centre in
Amsterdam,
the Netherlands, between July 2009 and April 2011 (Visscher, Ross et al. 2012;
Visscher,
Ross et al. 2013). We performed PCA using the Illumina 4.5K SNP genotype data
set
generated for these patients (Visscher et al. 2015, Pharmacogenomies in the
press) and
excluded 14 patients not genetically matching European ancestry. Based on the
detailed
clinical assessment of the 114 patients, we excluded another 18 patients with
CTCAE grade 1
CA 02911709 2015-11-10
toxicity (shortening fraction: 24% < SF < 30%). A total of 96 patients (22
cases and 74
controls) were then available for further analysis.
USA Patient Population: We recruited 164 pediatric oncology patients treated
with
anthracyclines from Lucile Packard Children's Hospital at Stanford (Palo Alto,
USA)
between December 2008 and October 2010. We performed a detailed clinical
assessment
of these patients and excluded 10 patients who had missing or abnormal
baseline
echocardiogram readings, 8 patients with CTCAE grade 1 toxicity (shortening
fraction: 24%
< SF < 30%), and 102 patients with insufficient cardiac follow-up data (either
SF>30% but
less than 5 year follow-up (97 patients), or SF<24% but <21 days from end of
treatment (5
patients)). Principal component analysis identified four clusters of the
remaining patients
aligning with their self-reported ancestries of Hispanic (specifically
reported as "White
Hispanic"), Asian, African-American, and European (specifically reported as
"White Non-
Hispanic"). The cluster of 23 Hispanic patients (5 cases and 18 controls) was
selected for
analysis in stage 3.
Molecular Genetic Methods and GWAS Quality Control Procedure (QC)
DNA Extraction and Molecular Genotyping
Genotyping experiments were conducted at the Canadian Pharmacogenomics Network
for
Drug Safety (CPNDS) genotyping core facility, Child & Family Research
Institute, The
University of British Columbia, Vancouver, BC, Canada. All patients provided a
biologic
specimen for DNA extraction. De-identified genomic DNA was extracted using the
QIAampTM DNA purification system (QiagenTm, Toronto, Ontario, Canada) and
quantified
by Quant-iT PicoGreenTm assay (InvitrogenTM, Eugene, OR, USA), according to
the
manufacturer's protocols. Whole Genome Amplification (WGA) was performed prior
to
genotyping for all samples available at a low concentration (< 50 ng/ I)
and/or low volume
(< 20 D. The laboratory assistants were blinded to the case-control status of
the patients
genotyped in the study. To ensure the accuracy of all genotyping results,
multiple positive
and negative controls and replicate samples were included in all genotyping
assays and
plates. The concordance of genotype calls between replicate genotyped samples
was 100%.
The genome-wide association study (GWAS) was performed using the Illumina
Infinium
HumanOmniExpressTM assay (740K), according to the manufacturer's instructions
(IlluminaTm, San Diego, CA, USA). This assay provides high sample throughput
and
coverage of common variants. Genotypes were called with the Illumina Genome
StudioTm
software package and the SNPs were clustered using the IlluminaTM 7401(
cluster file. All
samples in the study population were used to determine cluster boundaries in
order to
36
CA 02911709 2015-11-10
maximize clustering accuracy. A detailed GWAS quality control procedure was
performed
for all SNPs and samples prior to analysis. The quality control analysis was
performed by
date of genotyping and by plate to check for systematic errors in the
generated data set. No
systematic errors were found. The quality control procedure was performed with
the
Illumina Genome Studio software package.
The Dutch and Hispanic patients were genotyped for the GWAS candidate variants
using
TaqManTm SNP genotyping assays (Applied BiosystemsTM, Streetsville, ON,
Canada),
according to the manufacturer's protocols. The top replicated SNP from the
Illumina
Infinium HumanOmniExpressTM assay was re-genotyped in loo randomly selected
patients
from the discovery patient population using the TaqmanTm SNP genotyping assay.
The
concordance rate of genotype calls between the two assays was 100%.
Importing GWAS raw data and pre-QC Steps
The raw data was initially imported and clustered using the IlluminaTM 740K
cluster file.
Sample statistics were subsequently updated. Next, an iterative
genotyping/cluster QC
process was performed using the highest-quality samples in our specific
dataset (all
samples with initial call rates < 99% were temporarily removed). A sequence of
QC filters
was then applied to both SNPs and samples as described below.
GWAS Genetic Markers (SNPs) Detail Quality Control Procedure
The quality control for genotype data was performed in Illumina Genome
StudioTM
software package. We used a combination of thresholds for various quality
control metrics
with visual inspection of cluster plots for markers at the boundaries of the
thresholds.
Poorly-clustering markers (call rates < 95%) were filtered out for re-
clustering. Y
chromosome and mtDNA SNPs were excluded. Poorly-clustered SNPs were re-
clustered
and their new cluster positions evaluated using various quality control
metrics. Newly
defined cluster positions were either left alone, manually edited, or dropped
altogether
using the following quality control metrics available in Illumina Genome
StudioTM.
1. SNP Call Rates: Re-clustered SNPs with low call rates (< 0.95) were
manually edited
and the ambiguous ones were excluded.
2. Cluster separation: Re-clustered SNPs with poor cluster separation < 0.3,
were
visually reviewed and SNPs with cluster separation from the threshold where
the
clusters were no longer separable by eye were excluded.
3. Mean Normalized Intensity: Low intensity re-clustered markers k 0.25) were
removed.
37
CA 02911709 2015-11-10
4. Heterozygote clusters shifted too close to a homozygote cluster: SNPs with
an AB T Mean < 0.2 or o.8 were visually inspected and appropriate cutoff for
exclusion of poor quality markers determined based on how close the
heterozygote
cluster was to the homozygotes. Poor quality re-clustered markers were then
excluded
based on the pre-determined cutoff.
5. Heterozygous Excess: X chromosome loci were ignored here. First, re-
clustered
markers with heterozygous excess 0.16 were
selected and further re-clustered
amongst themselves. Then all SNPs with heterozygous excess < -0.3 and > 0.2
were
excluded.
6. False Homozygotes: Using R dev 13.05 and AA or BB T Dev 0.05 respectively,
SNPs with multiple minor-allele homozygote clusters in the R dimension and
SNPs
with multiple clusters on the T axis called together as homozygotes were
excluded. Also,
markers where all three clusters were so close to each other that they get
called as a
single heterozygous cluster were excluded.
7. Reproducibility/Replication Errors: All SNPs with 1 or more errors were
evaluated and any remaining markers with > 3 replication errors were excluded.
8. Call Frequency: All remaining SNPs with call frequency < o.95 were
excluded.
9. Hardy-Weinberg Equilibrium: Deviation of the genotype distributions from
Hardy-Weinberg equilibrium (HWE) was tested in control patients. All SNPs with
Fisher's exact test for HWE P-value < i.oxio-4 were excluded.
10. Previously excluded samples were included at this point and the sample
statistics
generated.
We implemented the same quality-control procedures such as call rates and HWE
from the
GWAS, in the replication cohorts. Nine SNPs in the replication studies had
call rates of
>90%, while one SNP had a call rate <90% and was subsequently excluded from
further
analysis.
GWAS Sample Quality Control Procedures
Quality control for DNA samples was per-formed with SVS/HelixTree 8.l.1TM.
Samples
were excluded if they had a call rate below 95%, if the reported and
genotypically inferred
genders did not match, if the ancestry departs from the expected homogenous
genetic
ancestry (principal component analysis via EigenstratTM (Visscher, Ross et al.
2009)), and
if they were related (identity by descent estimation).
GWAS Association Testing Quality Control
X and Y chromosomes and mitochondria SNPs were excluded from the association
analyses
in keeping with recent GWAS quality control practices. A total of 657,694 SNPs
from the
38
CA 02911709 2015-11-10
GWAS were retained after QC. Cluster plots for all GWAS associated SNPs were
visually
inspected.
Functional Assays
Constructs, cells and reagents: A Myc-DDK-tagged mammalian expression clone of
human RARG was purchased from OrigeneTM (Rockville, MD). The rs2229774 SNP was
introduced into this expression vector using the Quikchange IITm kit according
to the
manufacturer's specifications (Agilent Technologies, Mississauga, ON) with
mutagenic
primers. Transfections were performed with X-tremeGENE 9T1 (RocheTM, Laval,
PQ) or
EffecteneTM (QiagenTM, Toronto, ON) reagents according to the manufacturer's
specifications. HEK293T and H9c2(2-1) (ATCC CRL-1446) cells were purchased
from
ATCC (CedarlaneTm, Burlington, ON) and routinely cultured in DMEM supplemented
with
10% FBS, looU/m1 Penicillin, loong/m1 Streptomycin, with additional o.25 g/m1
Amphotericin B for H9C2 cells. RARG transcriptional regulation was assayed
using the
Cignal RARE ReporterTM luciferase kit (QiagenTM, Toronto, ON). Luminescence
assays
were developed using the Dual-Glo Luciferase Assay SystemTM according to the
manufacturer's specifications (PromegaTM, Madison, WI) and read on a POLARstar
OmegaTM plate reader (BMG LabtechTm). For real-time RT-PCR (qPCR) experiments
RNA
was purified from H9c2 cells using the Ambion Purelink RNATM mini kit with
PurelinkTM
homogenizers and PurelinkTM on-column DNAse digestion according to the
manufacturer's
specifications (Life TechnologiesTm, Burlington, ON). cDNA was generated using
the
Invitrogen Superscript IIITM first strand synthesis kit according to the
manufacturer's
specifications (Life TechnologiesTm, Burlington, ON). Rat Top2b, rat Hprti and
human
RARG gene expression was measured using validated Applied Biosystems TaqManTm
assays Rn01537914_mi, Rno152784o_ml, and Hso1559234 mi, respectively (Life
Technologies, Burlington, ON). qPCR was performed on the PikoReal 96 Real-Time
PCRTM
system (Thermo ScientificTM) and relative gene expression was calculated by
the AACq
method using the instrument software. The anti-DDK 4C5 antibody (catalog #
TA5ooll)
was purchased from OrigeneTM (Rockville, MD) and the anti-GAPDH antibody
(catalog #
MAB374) was purchased from MilliporeTM (Etobicoke, ON).
RarG transcriptional regulation assay: To assess whether general RARG activity
was affected by the rs2229774 variant, we used a reporter construct fused to
an optimized
RARETM element (Cignal Reporter SystemTM, SABiosciencesTm). The proprietary
constituents of the Cignal reporter system impart a technical limitation for
the use of a
39
CA 02911709 2015-11-10
robustly transfectable cell line. Accordingly we used HEK293T cells that have
been
validated and recommended by the manufacturer. HEK293T cells were reverse co-
transfected in a 96-well plate with song (per well) of either empty vector
(pcDNA3.1.),
RARG WT or RARG5427L expression constructs and the Cignal RARETM reporter
Luciferase
kit constructs according to the manufacturer's specifications. After 20-24
hours of
transfection cells were washed with PBS and the medium was replaced with 750
Cignal
AssayTM medium (OptiMemTm supplemented with 1 % charcoal-stripped FBS, o.imM
NEAA, imM sodium pyruvate, looU/m1 Penicillin and 1oop.g/m1 Streptomycin)
containing
ipM ATRA or DMSO control for 6 hours. Firefly and Renilla luminescence was
measured
as indicated above. Firefly luciferase/Renilla luciferase ratios (L/R) were
calculated for
each well and converted to a Relative response ratio (RRR) = [(L/R)sample
(L/R)negative
control]/[(L/R)positive control ¨ (L/R)negative control] to allow sample
comparison between
experiments. The fold induction of ATRA-treated versus untreated samples was
calculated
using the corrected RRR values (subtraction of empty vector RRR values) for
RARG WT-
and RARGs427L-transfected samples.
Relative gene expression studies in rat cardiomyoblasts: 7.5 x 104 H9c2 cells
were seeded into each well of a 6-well dish in DMEM supplemented with 10% FBS.
The
following day, cells were transfected with wg RARG WT or RARGs427L expression
constructs for 7 hours before replacing with fresh medium. Where required
250nM ATRA
was added to cells 24 hours post-transfection. Cells were grown for 48 hours
post-
transfection then total RNA was immediately purified and used for cDNA
synthesis. qPCR
reactions were conducted in a 100 reaction volume that consisted of sal 2X
TaqmanTm
Universal Master MixTM, 0.50 TaqmanTm probe and 411 cDNA with standard cycling
conditions according to the manufacturer's specifications. Relative gene
expression was
calculated using Hprti as a housekeeping gene. To calculate the fold
repression of Top2b
expression in RarG-transfected H9c2 cells, relative Top2b expression in
untransfected cells
was divided by expression levels in transfected cells and normalized to the
relative
expression of the appropriate RARG construct.
Statistical Methods
We performed statistical analyses using SVS/HelixTree 8.1.1 (Golden HelixTM,
Bozeman,
MT, USA), R 3.1.0 for Statistical ComputingTm, SPSS Version 18.0 (IBMTm,
Armonk, NY),
Quanto, HaploviewTm (Barrett, Fry et al. 2005), LocusZoomTm(PruimTm, Welch et
al.
201.0), Epi InfoTM Version 7.1.3, Comprehensive Meta-Analysis, BEAGLE 4TM
(ref.
CA 02911709 2015-11-10
(Browning and Browning 2007)), GraphPad Prism S.oaTM and PikoReal version 21TM
software packages. All statistical tests were 2-sided. Baseline quantitative
and qualitative
variables were analyzed with Wilcoxon-Mann-Whitney U test and Fisher exact
test,
respectively. Genetic associations were tested by logistic regression with an
additive model
and adjusted for appropriate clinical covariates unless indicated otherwise,
e.g. where
logistic regression was not possible. Covariates for logistic regression were
derived from
each study cohort and represented relevant baseline (clinical and demographic)
differences
between cases and controls. Covariates for the European Canadian patients
(stage 1)
analysis included age at start of treatment, cumulative anthracycline dose,
tumor type
(acute lymphoblastic leukemia, Ewing's sarcoma and rhabdomyosarcoma) and
cardiac
radiation therapy. Cumulative anthracycline dose was included as a covariate
for the
European Dutch patient (stage 2) analysis. Covariates for the combined
European patient
(stage i and 2) and overall combined (stages 1-3) analyses included age at
start of
treatment, cumulative anthracycline dose, tumor type (acute lymphoblastic
leukemia,
Ewing's sarcoma and rhabdomyosarcoma) and cardiac radiation therapy. All
effect sizes
(odds ratios) were calculated for the minor allele for each SNP.
We performed genetic fine mapping analysis by genotype imputation of
additional SNPs
not present on the GWAS genotyping platform using BEAGLE 4TM with LD and
haplotype
information from CEU woo Genomes population as the reference population. We
imputed
1,005,286 additional variants on Chromosome 12 region containing RARG. The
quality
control metric (BEAGLETM allelic R2) was calculated for all imputed SNPs. We
then
examined evidence of additional genetic associations with ACT in this region
based on
SNPs with imputed BEAGLETM allelic R2 0.5, using logistic regression adjusted
for stage 1
covariates. LD analyses (r2 and D') of the imputed variants were conducted
using the moo
Genomes CEU reference population.
To control for Type I error, we implemented screening thresholds based upon
multiple
testing correction for all genetic association analyses. A threshold of P<1x10-
5, indicative of
putative genetic associations (Welter, MacArthur et al. 2014) was implemented
for the
stage i analysis. For the stage 2 replication analysis in European Dutch
patients, a
Bonferroni-adjusted threshold of P < 0.006 (replication testing of SNPs from 9
LD blocks)
was implemented. Further stage 3 replication in independent non-European
populations
was tested using a threshold of P < 0.05 (1 replicated SNP). Potentially
confounding clinical
risk factors were identified at P <0.05.
41
CA 02911709 2015-11-10
To prevent spurious (obscured, false positive and false negative) genetic
associations, all
patients within each study population shared the same genetic ancestry, which
was self-
reported and ascertained by principal component analysis (PCA). Then for each
study
cohort, we computed the genomic inflation factor (XGc) to verify the presence
of intra-ethnic
fine-scale population structures or admixtures or inflation of the test
statistics due to
population stratification(Devlin and Roeder 1999): Stage i European-Canadian
patient
discovery cohort (based on 657,694 SNPs) ¨ XGc = 1.021; stage 2 European Dutch
patient
replication cohort (based on 4516 SNPs) ¨ XGc = 1.014; and stage 3 Non-
European patient
replication cohort (based upon 7798 mutual SNPs available to all patients) ¨
XGc = 0.941.
We performed meta-analyses of all European patients (stages 1-2) and of all
study
populations (stages 1-3) using SVS/HelixTree 8.l.1TM and calculated the
heterogeneity by
Cochran's Q statistics to assess the diversity across the different study
populations using
Comprehensive Meta Analysis software. The Manhattan plot of ¨log10 P values
and the
quantile-quantile distribution were generated using SVS/HelixTree 8.l.1TM. The
regional
association plot for the associated genomic region was created using
LocusZoomTM.
Linkage disequilibrium plots were created using HaploviewTM and color coded as
follows:
white (D' < 1, LOD < 2); blue (D' = 1, LOD < 2); pink shading (D' < 1, LOD 2);
bright red
(D' = 1, LOD 2).
Statistical analyses of the functional data provided in FIGURE 2 shows that
all data
(FIGURE 2a¨c) were normally distributed when assessed by the D'Agostino &
Pearson
omnibus normality test. Variation coefficients were estimated as: FIGURE 2a ¨
RARG
WT (23.77%), RARGs4271- (31.28%); FIGURE 2h ¨ untransfected (9.30%), RARG WT
(9.37%), RARG WT + ATRA (7.33%); FIGURE 2C ¨ RARG WT (16.48%), RARG54271-
(22.69%). F tests showed the variances between groups significantly differed
in FIGURE
2b and 2c.
42
CA 02911709 2015-11-10
EXAMPLES
Example 1: RARG m2229774 is associated with Anthracycline-associated
cardiotoxicity (ACT)
We recruited well-phenotyped patients treated with anthracyclines for
childhood cancer
(.18 years at treatment) from 13 pediatric oncology centers across Canada.
Clinical
information was used to assess cardiac function, define cases and controls,
and determine
important baseline differences between these groups. Cases were defined as
exhibiting
shortening fractions [SF] of 5_24% or signs and symptoms of cardiac compromise
requiring
intervention based on CTCAEv3, while controls had SF 3o% and no symptoms of
cardiac
compromise for at least 5 years after treatment(Visscher, Ross et al. 2012;
Visscher, Ross et
al. 2013). A stage 1 discovery analysis was performed in Canadian patients of
European
ancestry (280 patients; 32 cases and 248 controls) as shown in TABLE 3.
Compared with
controls, cases were significantly older at the start of treatment, had higher
cumulative
anthracycline exposure (dose), were less likely to have acute lymphoblastic
leukemia (ALL),
but more likely to have rhabdomyosarcoma or Ewing's sarcoma, and receive
radiotherapy
to the heart (RT) (P<o.o5). Age, dose and RT are established risk factors for
ACT and after
verifying that the associations with ALL, Ewing's sarcoma and rhabdomyosarcoma
tumor
types were not exclusively explained by the cumulative anthracycline dose
(remained
associated with ACT after accounting for dose), these six clinical factors
were included as
covariates for logistic regression analyses. Notably, the limited sample size
may have
precluded our detection of other significant clinical differences between
cases and controls.
Patients were genotyped using an Illumina HumanOmniExpressTm (740K SNP) assay
with
657,694 SNPs passing quality control assessment, conferring a Bonferroni-
adjusted
multiple testing correction threshold of P<7.6xio-8. The GWAS discovery
analysis was
performed using logistic regression adjusted for age, dose, ALL, Ewing's
sarcoma,
rhabdomyosarcoma, and RT (as shown in TABLE 4). Analysis of the test
statistics (XGc =
1.021), suggested that they were not influenced by cryptic population
stratification (as
shown in TABLE 4). Eighteen variants tagging nine distinct linkage
disequilibrium (LD)
blocks (r2>o.9 and D'>0.9 in the l000 Genomes CEU reference dataset) with
P<i.oxio-5
were prioritized for further analysis in an independent patient population (as
shown in
TABLE 4).
43
TABLE 3: shows the baseline characteristics of the patients of European
Ancestry (Discovery and Replication Patient Populations).
Stage 1- Discovery Canadian Patient Stage 2 - Replication Dutch Patient
Combined European Patient Population
Population Population
(n = 376 patients)
Patient Characteristicsa (n = 280 patients) (n =
96 patients)
Cases Controls Cases
Controls Cases Controls
(n = 32) (n = 248) P (n = 22)
(n = 74) P (n = 54) (n = 322) P
Age at the Start of Treatment 9.0 (2.5 - 14) 4 (2 -
7.5) 0.004e 7.5 (5 - 12) 11 (6 - 14) 0.14 8.5 (4 - 14)
5 (2 - 10) 0.007
Age in yrs, median (IQ range)
Gender, female/male (13/0 female) 17/15 (53.1) 112/136 (45.2)
0.45 10/12 (45.5) 36/38 (48.6) 0.81 27/27 (50) 148/174 (46) 0.66
Cumulative Anthracycline Exposure 260 (177.5 - 175 (140-
407.5 (270- 277.5 (180- 281.5 (200- 200 (150 -
Dose') in mg/m2, median 0.011
0.010 <0.0001
365) 295) 480) 364)
450) 300)
0
(Interquartile range)
Chemotherapy (Anthracycline
01..)
Doxorubicin 25 (78.1) 178 (71.8) 0.53 13 (59.1)
40 (54.1) 0.81 38 (70.4) 218 (67.7) 0.75 ko
1-,
Daunorubicin 2 (6.3) 26 (10.5) 0.75 1 (4.5) 7
(9.5) 0.68 3 (5.6) 33 (10.2) 0.45
.4
Doxorubicin plus daunorubicin 2 (6.3) 37 (14.9) 0.28 0
(0) 4 (5.4) 0.57 2 (3.7) 41 (12.7) 0.063 0
ko
Doxorubicin plus other 0(0) 1(0.4) 1.0 2(9.1)
7(9.5) 1.0 2(3.7) 8(2.5) 0.64 1..)
Daunorubicin plus other 3(9.4) 6 (2.4) 0.071 0 (0)
0(0) 0.57 3(5.6) 6(1.9) 0.13 0
1-,
Epirubicin 0 (0) 0 (0) 1.0 5 (22.7)
13 (17.6) 0.55 5 (9.3) 13 (4) 0.16 Ln
1
Epirubicin plus other 0 (0) 0 (0) 1.0 1 (4.5) 2
(2.7) 0.55 1 (1.9) 2 (0.6) 0.37 1-,
1-,
1
Otherb 0(0) 0(0) 1.0 0(0) 1(1.4)
1.0 0(0) 1(0.3) 1.0 1-,
Primary Diagnosis (Tumor type), no.
0
Acute Lymphoblastic Leukemia 5(15.6) 105 (42.3) 0.0035 5(22.7)
13 (17.6) 0.55 10 (18.5) 118 (36.6) 0.0085
Acute Myelogenous Leukemia 3 (9.4) 8 (3.2) 0.12 0 (0) 7
(9.5) 0.35 3 (5.6) 15 (4.7) 0.73
Other Leukemia 0(0) 4 (1.6) 1.0 0 (0)
1(1.4) 1.0 0 (0) 5(1.6) 1.0
Hodgkin's Lymphoma 4 (12.5) 19(7.7) 0.31 1(4.5)
8(10.8) 0.68 5(9.3) 27(8.4) 0.79
Non-Hodgkin's Lymphoma 3(9.4) 23(9.3) 1.0 6(27.3)
16 (21.6) 0.57 9(16.7) 39 (12.1) 0.38
Osteosarcoma 0 (0) 11(4.4) 0.62 0 (0) 9 (12.2)
0.11 0 (0) 20 (6.2) 0.093
Rhabdomyosarcoma 2(6.3) 2 (0.8) 0.066 3(13.6)
3 (4.1) 0.13 5(9.3) 5(1.6) 0.0073
Ewing's sarcoma 5 (15.6) 8 (3.2) 0.0095 4
(18.2) 5 (6.8) 0.20 9 (16.7) 13 (4) 0.0015
Other sarcoma 1(3.1) 3(1.2) 0.39 0(0)
1(1.4) 1.0 1(1.9) 4(1.2) 0.54
Hepatoblastoma 2 (6.3) 11(4.4) 0.65 0 (0) 0 (0)
1.0 2 (3.7) 11(3.4) 1.0
Neuroblastoma 1(3.1) 28 (11.3) 0.22 0(0) 0(0)
1.0 1(1.9) 28(8.7) 0.099
Wilms Tumor 6(18.8) 26 (10.5) 0.23 3(13.6)
11 (14.9) 1.0 9(16.7) 37 (11.5) 0.27
44
TABLE 3: Continued
12 (37.5) 40 (16.1) 0.0068 6 (27.3)
18 (24.3) 0.78 18 (33.3) 58 (18.0) 0.016
Rariinthpranv invnlvinn thp hPartd
Use of cardioprotectants, no. (%) 2(6.3) 7(2.8) 0.27 0(0.0)
2 (2.7) 1.0 2(3.7) 9(2.8) 0.66
Duration of Follow-up in years, 7.5 (2.5- 9 (7- 12)
0.33 22 (19 -25) 17 (14 -22) 0.012 15.5 (7 - 22) 10 (7 - 15) 0.021
a Age, dose and duration of follow-up were analyzed by Wilcoxon-Mann-Whitney U
test. Gender, anthracycline type, tumor type, radiotherapy involving the
heart and use of cardioprotectant were analyzed by Fisher exact test. b
Cumulative anthracycline dose in doxorubicin isotoxic equivalent doses. C
Other
anthracycline type included idarubicin, epirubicin or mitoxantrone. d Includes
mantle and mediastinal radiation, whole lung radiation, whole or upper
abdominal radiation, left sided flank radiation and total body irradiation. e
Bold font indicates statistically significant P-value (P < 0.05) and
covariates for
logistic regression.
ci
0
0
0
=
TABLE 4: shows Genome-Wide Association Study (GWAS) of ACT in Patients of
European Ancestry: Pharmacogenomic Discovery and Replication Analyses.
Stage 1 - Discovery Canadian Patient Stage 2 - Replication Dutch Patient
Population
Population
n = 280 (32 cases; 248 controIs)a
n = 96 (22 cases; 74 controls)b
ChGenomic Min Odds Ratio MAF
MAF Odds Ratio MAP MAF
Variantg Positiong Function P
rd Region or (95%Ce (Cases)
(Cont. P (95%CI) (Case (Contro
Allel )
s) Is)
rs6895189 5 13430225 CTIVND2 INTG C 2.4 x 10-6 6.1 (2.8 - 0.359
0.105 N/Af N/A N/A N/A
I DNAH5 13.3)
rs7731918 5 13397992 CTNND2 INTG A 4.0 x 10-6
5.9, (2.7 - 0.355 0.105 0.98 1.0 (0.34 - 0.114
0.115 Cl
I pNAH5 12.9)
3.0)
o
CTNND2 5.812.7) (2.6 -
"
rs2081944 5 13405946 INTG A 4.8 x io-6 0.355
0.105 LD g LD LD LD ko
1 DNAH5
1-,
1-,
-.3
o
rs10085086 5 13424707 CTNND2 INTG C 3.6 x 10-6 5.9 (2.7 - 0.355
0.103 LD LD LD LD ko
I DNAH5 13.0)
n.)
o
rs15736 21 44273858 WDR4 NONS-C A 2.6 x 10-6
4.4 (2.2 - 8.7) 0.703 0.366 0.44 0.76 (0.38 - 0.386 0.431
ol
1.5)
1
1-,
rs6586252 21 44276387 WDR4 INTRON A
2.6 x 10-6 4.4 (2.2 - 8.8) 0.703 0.367 LD LD LD LD
1
rs8133752 21 44271989 WDR4 INTRON A
3.3 x 10-6 4.4 (2.2 - 8.6) 0.703 0.370 LD LD LD LD
0
rs4381672 18 22712791 ZNF521 INTRON A 2.9 x 10-6
4.3 (2.2 - 8.3) 0.597 0.342 0.78 0.89 (0.40 - 0.341 0.372
2.0)
S4275929 18 22706688 ZNF52/ INTRON C 2.8 x 10-6 4.3 (2.2 - 8.5)
0.594 0.343 LD LD LD LD
rs4519409 18 22722077 ZNF521 INTRON A
5.6 x 10-6 4.8 (2.3 - 0.516 0.313 LD LD LD LD
10.0)
rs358224h 4 22860785 GBA3 I INTG A 3.3 x 10-6
4.2 (2.2 - 8.1) 0.500 0.249 0.47 1.4 (0.56 - 0.289 0.217
PPARGC
3.5)
GBA3 I
rs412218 4 22843458 RGC INTG C 5.7 x 10-6 4 (2.1 - 7.5)
0.531 0.270 LD LD LD LD
PPA
IA
46
TABLE 4: Continued
rs11946006 4 22850202 GBA3 I INTG G 9.1 x 10-6 3.9
(2.1 - 7.4) 0.531 0.282 LD LD LD LD
PPARG
rs7676830i 4 23169854 GBA3 I INTG G
5.5 x 10-6 4.8 (2.3 - 9.8) 0.500 0.262 0.057 2.3 (0.96 - 0.364
0.243
PPARG
5.4)
rs2282889 7 21476188 SP4 INTRON A
4.4 x 10-6 0.2 (0.088 - 0.234 0.446 0.75 0.9 (0.44 - 0.364
0.401
0.44)
1.8)
rS22297743 12 53605545 RARG NONS-C A 5.0 x 10-6 7 (2.9 -17)
0.297 o.o8i 0.0043 4.1 (1.5 - 0.25 0.061
11.5)
o
rs9323880 14 93129810 R/N3 INTRON A 6.8 x 10-6 4.2
(2.1 - 8.2) 0.594 0.348 0.17 1.6 (0.82 - 0.50 0.372
3.1)
rs7042745 9 27248177 NCR1VA INTRON A
7.5 x 10-6 4.5 (2.2 - 8.9) 0.484 0.222 0.65
0.85 (0.41 - 0.295 0.319 0
00032
1.8) o
tv
a Covariates for the Logistic regression were age at treatment, cumulative
anthracycline exposure, radiotherapy involving the heart and tumour type l0
I-,
(acute lymphoblastic leukemia, rhabdomyosarcoma and Ewing's sarcoma).
.4
b Covariate for the Logistic regression was cumulative anthracycline exposure.
0
l0
c Variants with P < 1.0 x to-bin the discovery GWAS analysis'.
n.)
5' Chr, chromosome.
0
1-,
e Chromosomal positions in the GRCH37.p13.
ol
1
f Not Applicable; call rates for this SNP were < 90% in the Stage 2 cohort.
1-,
g LD (r2> 0.9 and D' > 0.9 in the CEU component of HapMap), therefore only 9
of 18 variants were genotyped in the Stage 2 - Replication Dutch patient 1
1-,
population.
0
The call rate for this SNP was 92% in the replication cohort.
' Tags a distinct LD block in this genomic region.
3 Bold font indicates statistically significant SNP after multiple testing
correction (discovery P < 1.ox10-5 and replication P < 0.05/9 LD blocks =
o.006).
INTG = INTERGENIC; NUNS-C = NONSYN-CODING; INTRON = INTRON; and Cont. =
Control
47
TABLE 5: shows the association of RARG rs2229774 with Anthracycline-induced
Cardiotoxicity in Childhood Cancer Patients.
Biomarker Pharmacogenomic Analyses
Adjusted Logistic Regression Genotypic Test
(Additive Model)
MAF MAF
SNP Gene Function Study Population P
Odds Ratio (95%CI) P
Cases Controls
NON-SYN Stage 1 ¨ Discovery GWASa
rs2229774 RARG 0.297 0.081 5.0 x
10 7.0 (2.9 ¨ 17.0) 4.1 x 10-'
CODING Canadian European Patient
Stage 2 ¨ Replicationb
0
P
0.25 0.061 0.0043
4.1 (1.5 ¨ 11.5) 0.0042
o
Dutch European Patients
N)
ko
1¨`
All European Patientsa 0.278 0.076 7.8 x
10-8 5.4 (2.9 ¨ 10.3) 1.2 x 10-9
¨.1
0
ko
Stage 3 ¨ Replication
0.158 0 N/A`
N/A 1.2x10-4 N)
o
Non-European Patients
Ui
I
1¨`
All Populations'
1
European and Non-European 0.247 0.064 5.9 x
10 4.7 (2.7 ¨ 8.3) 4.3x10-11
0
Patients
a Covatiates for the Logistic regression were age at treatment, cumulative
anthracycline exposure, radiotherapy involving the heart and incidence of
acute
lymphoblastic leukemia, rhabdomyosarcoma and Ewing's sarcoma.
b Covari ate for the Logistic regression was cumulative anthracycline
exposure.
' Not applicable, rs2229774 absent in controls.
48
TABLE 6: shows Top GWAS Associations by Cumulative Anthracycline Exposure.
Pharmacogenomic Associations of Anthracycline Low-to-moderate
Anthracycline Exposure High Anthracycline Exposure
Cardiotoxicity (5. 250 mg/ml
(>250 mg/m2)
Stage 1: Discovery" Stage 2: Replication"
Stage 1: Discovery Stage 2: Replication"
Variantb Genomic Regions Function n = 184 patients n =
38 patients n = 96 patients n = 58 patients
(16 cases; 168 controls) (5 cases; 33 controls)
(16 cases; 80 controls) (17 cases; 41 controls)
rs7731918 CTNND2 I DNAH5 INTERGENIC 0.00016 0.70
0.0010 0.85
o
P
rs15736 WDR4 NONSYN-CODING 0.0011 0.62
0.00029 0.59
o
N)
rs4381672 ZNF521 INTRON 6.9 x 10-5 0.29
0.021 0.56 ts)
1-`
1-`
-.1
rs358224 GBA3 I PPARGC1A INTERGENIC 0.00020 0.37
0.0065 0.69 0
ts)
rs7676830' GBA3 I PPARGC1A INTERGENIC 9.1 x 105 0.0240
0.0076 0.30 "
0
1-`
(xi
rs2282889 SP4 INTRON 0.0037 0.84
0.00041 (0.05)h 1
1-`
1-`
l
rs2229774 RARG NONSYN-CODING 0.00041 0.0036
0.0021 0.084
1-`
0
rs9323880 R1N3 INTRON 5.8 x 107 0.76
0.31 0.15
rs7042745 NCRNA00032 INTRON 0.0011 0.84
0.0011 0.67
Stratification by cumulative anthracycline exposure as previously
performed'''.
Variants with P< 1.0 x 10 in the discovery GWAS analysis'.
P-values are for logistic regression (additive model) with adjustment for
covariates.
Covariates for the Logistic regression were age at treatment, cumulative
anthracycline exposure, radiotherapy involving the heart and acute
lymphoblastic leukemia tumor type.
Covariate for the Logistic regression was cumulative anthracycline exposure.
Covariates for the Logistic regression were age at treatment, cumulative
anthracycline exposure, radiotherapy involving the heart and tumor type (acute
lymphoblastic leukemia,
ha bdomyosarcoma and Ewing's sarcoma).
Tags a distinct LD block in this genomic region.
Regression failed due to absence of variant in cases in this stratified dose,
genotypic testing P-value shown.
49
CA 02911709 2015-11-10
In stage 2, we genotyped one GWAS candidate variant per LD block in an
independent
Dutch population of childhood cancer patients of European ancestry (96
patients; 22 cases
and 74 controls). In this cohort cases and controls were similarly matched
with the
exception that cases had higher cumulative anthracycline exposure (TABLE 3).
Genetic
associations were tested using logistic regression with adjustment for
cumulative
anthracycline dose. Of the 9 candidate variants tested, only rs2229774, a
nonsynonymous
coding variant (p.Ser427Leu) in RARG (Retinoic Acid Receptor Gamma), was
replicated
(P=o.0o42, OR=4.1 (1.5-11.5); FIGURE 1 and TABLE 5).
Given that the Stage 1 and 2 cohorts differed in some patient characteristics,
e.g.
anthracycline dose, the replication analysis merits circumspect
interpretation. To address
this we performed additional genetic association analyses by logistic
regression with the
prioritized GWAS variants in subsets of cases and controls stratified by low-
to-moderate
( .25o mg/m2) and high (>250 mg/m2) anthracycline exposure (TABLE 6). RARG
rs2229774 was associated with ACT in stages 1, 2 and the combined analysis at
both low-to-
moderate (P=4.1x10-4, P=o.0036, and P=9.8x10-6 , respectively) and high
anthracycline
doses (P=o.0021, P=o.o84, and P=8.7xio-4, respectively). By contrast, none of
the other
top GWAS candidate SNPs were significantly associated with ACT in the
replication cohort,
even at low-to-moderate anthracycline exposure. Logistic regression analyses
in the Stage 1
cohort indicated that RARG rs2229774 was similarly associated with early-onset
chronic
ACT (n=16 cases; P=2.8x10-4; OR=7.3 (2.3-22.9)) and late-onset chronic ACT
(n=r6 cases;
P=5.oxio-3; OR=5.8 (1.7-19.4)).
In stage 3, we examined the association of RARG rs2229774 with ACT in a third
cohort of
non-European patients representative of different ancestries (80 patients; 19
cases and 61
controls; TABLE 7). Due to the absence of RARG rs2229774 in controls, logistic
regression
with adjustment for population stratification and other clinical covariates
could not be
performed in this cohort (TABLE 8). Instead the association of RARG 1.52229774
with
ACT in this cohort was analyzed by genotypic test. RARG 1.52229774 was highly
associated
with ACT (P=1.2xio-4; TABLE 1) in the stage 3 cohort, and in each of the four
non-
European populations separately (African, Aboriginal Canadians, Hispanic, and
East Asian;
TABLE 8). Notably, genotypic association testing reached genome-wide
significance in the
discovery and combined cohorts (P<5.oxio-8; TABLE 1).
TABLE 7: shows assessment of Baseline Characteristics in Non-European Patient
Populations.
Patient
Aboriginal Canadians -
Hispanic USA -Stanford African - CPNDS
East Asian - CPNDS
Characteristicsa
CPNDS
(n = 23 patients) (n = 11 patients)
(n = 31 patients)
(n = 15 patients)
Cases Controls Cases Controls Cases
Controls Cases Controls
(n= 5) (n = 18) P (n = 2) (n = 9) P (n = 8)
(n = 23) P (n = 4) (n = 11) P
Age at the Start of
Treatment 14.0 (12.5- .,
6.o (2.5 - 3.5 (1.5 -
5.5 (3 - 12) 0.01.9e 4.5 (4 - 5) 4.o (1.5 - 7.0) 0.73 3.5 (0.5 - 8)
0.32 , 4 (2.5 - 7) 0.49
Age in yrs, median
17.5) 9.5) 5.5)
(Interquartile
Gender,
4/0
female/male (% 1/4 (20.0) 7/11 (38.9) 0.62 0/2 (o)
6/3 (66.7) 1.0 5/3 (62.5) 11/12 (47.8)
0.69 (loo.o) 6/5 (54.5) 0.23
Cumulative
0
Anthracycline
200 (141- 162.5 (150 - 319.5 (240 -24o (114 -
_ 300.5 (270.0 - 290 (162.5 , 25o (137.5 150 (135 -
Exposure Doseb in o.8o o.5o
0.40 0.66 o
245) 300) 399) 382.5) 362.5)
- 360) - 330) 245) N.)
mg/m2, median
ko
1-,
(Interquartile
1-,
=4
0
Anthracycline typec,
ko
Doxorubicin 2 (40) 13 (72.2) 0.30 2 (100) 6 (66.7)
1.0 5 (62.5) 14 (60.9) 1.0 2 (50) 9 (81.8)
0.52 N.)
0
Daunorubicin 1(20) 2(11.1) 0.54 o(o) 2(22.2) 1.0 1(12.5)
1(4.3) 0.46 2(50) 1(9.1) 0.15 1-,
ol
Doxorubicin plus 2 (40) 3 (16.7) 0.29 o (o) 1 (11.1)
1.0 o (o) 5 (21.7) 0.29 o (o) 1 (9.1) 1.0 I
1-,
Daunorubicin o (o) o (o) 1.0 o (o) o (o) 1.0
2(25) 2(8.7) 0.27 o(o) o(o) 1.0 1-,
1
1-,
Doxorubicin plus
0
daunorubicin plus o (o) o (o) 1.0 o (o) o (o)
1.0 o (o) 1 (4.3) 1.0 o (o) o (o) 1.0
others
Primary Diagnosis,
Acute 2 (40) 13 (72.2) 0.30 1 (5o) 3 (33.3)
1.0 o (o) 8 (34.8) 0.07 2 (50) 5 (45.5) 1.0
Acute 1 (20) 2 (11.1) 0.54 0 (o) o (o) 1.0
2 (25) 2 (8.7) 0.27 1 (25) o (o) 0.27
Other Leukemia 2 (40) 3 (16.7) 0.29 o (o) o (o) Lo
1 (12.5) o (o) 0.26 0 (0) 0 (0) 1.0
Hodgkin's o (o) o (o) 1.0 o (o) 1 (11.1) 1.0
o (o) o (o) 1.0 o (o) 1 (9.1) 1.0
Non-Hodgkin's o (o) o (o) 1.0 o (o) 1 (11.1) 1.0
2 (25) 3 (13) 0.58 o (o) 1 (9.1) 1.0
Osteosarcoma o (o) o (o) 1.0 o (o) 1 (11.1) 1.0
o (o) 3 (13) 0.55 o (o) 1 (9.1) 1.0
Rhabdomyosarco o (o) o (o) 1.0 1 (5o) o (o) o.18
o (o) 1 (4.3) 1.0 o (o) o (o) 1.0
Ewing's sarcoma o (o) o (o) 1.0 o (o) o (o) to o (o)
o (o) Lo o (o) 1 (9.1) 1.0
Hepatoblastoma o (o) o (o) 1.0 o (o) o (o) 1.0
2 (25) 2 (8.7) 0.27 0 (0) 1 (9.1) 1.0
51
TABLE 7: Continued
Neuroblastoma o(o) o (o) 1.0 o (o) o (o) 1.0 1
(12.5) 3 (13) 1.0 o (o) 1 (9.1) 1.0
Wilms Tumor o (o) o (o) 1.0 o (o) 3 (33.3) t.o
o (0) 1 (4.3) 1.0 1 (25) o (0) 0.27
Radiotherapy
involving heartd,
no. (%) Data not available for all patients o (0) 2 (22.2)
1.0 0 (0) 0 (0) 1.0 1 (25.0) 0 (0) 0.27
Use of
cardioprotectants,
no. (%) Data not available for all patients o (o) o (o)
1.0 o (0) 2 (8.7) 1.0 0 (0) 0 (0) 1.0
Duration of Follow-
up in years median
(range) 4 (3 -4) 6 (5 - 7) 10.5 (9 -
12) 8 (6.5 ¨ 10.5) 0.33 8.5 (4 - 14) 7 (6.5 - 8) 0.84 7 (2 - 19) 7 (6.5 -
lo) 0.85
a Age, dose and follow-up were analyzed by Wilcoxon-Mann-Whitney U test.
Gender, anthracycline type, tumor type, radiotherapy involving the
heart, and use of cardioprotectant were analyzed by Fisher exact test. b
Cumulative anthracycline dose in doxorubicin isotoxic equivalent doses. c o
Other anthracycline type included idarubicin, epirubicin or mitoxantrone.
d Radiotherapy involving the heart include: mantle and mediastinal radiation,
whole lung radiation, whole or upper abdominal radiation, left sided 0
1..)
flank radiation and total body irradiation
ko
1-,
e Bold indicates statistically significant P-value (P < o.o5).
.4
0
ko
iv
0
1-,
(xi
1
1-,
1-,
1
1-,
0
52
TABLE 8: shows association of RARG rs2229774 with ACT in Non-European
Populations.
Africans Hispanics East Asians
Aboriginal Canadians Combined
n = 11 patients n = 23 patients n = 31 patients n
= 15 patients n = 80 patients
(2 cases; 9 controls) (5 cases;18 controls) (8
cases; 23 controls) (4 cases; 11 controls) (19 cases; 61 controls)
MAFa'b
Expected 11.0% 5.0% 0%
Unreported N/A
Range 6.0% - 16.0% 3.0% - 8.0% 0%
Unreported N/A
Genetic Association
Observed MAF
25.0% vs. 0 /0 20.0% vs. 0% 6.3% vs. 0%
25.0% vs. 0% 15.8% vs. 0%
(Cases vs. Controls)
0
Pc 0.026 0.052 0.085
0.012 1.2x10-4
0
MAF are from http://www.1000genomes.org.
(xi
b Abbreviations: MAP, minor allele frequency; N/A, not applicable.
P-values are for genotypic association tests.
53
TABLE 9: shows the fine mapping of genetic association signals in the RARG
gene region.
Biomarker Pharmacogenomic Analyses
Adjusted Logistic regression (additive model)b
LD with MAF MAF
Odds Ratio Conditional Analysis
SNP rs-ID` Position Type Source
rs2229774 on rs2229774
Cases Controls
(95%CI)
rs11170481 53611791 Intronic Imputed 1.00(0.84) 0.313
0.081 1.7x10-6 7.0 (3.0 ¨ 16.6) 0.10
rs73309171 53606565 Intronic Imputed 1.00 (0.55) 0.281
0.071 4.1x10-6 7.5 (3.0¨ 18.4) 0.42
rs57789211 53609992 Intronic Imputed 1.00 (0.84) 0.281
0.071 4.1x10-6 7.5 (3.0¨ 18.4) 0.42
rs2229774 53605545 Nonsyn Genotyped 1.00(1.00) 0.297 0.081
5.0x10-6 7.0 (2.9 ¨ 17.0)
a1,005,286 additional variants on Chr12 imputed into stage 1 cohort using the
CEU component of the 1000 Genomes populations as a reference.
bCovariates for logistic regression analysis (additive model) were age at
treatment, cumulative anthracycline exposure, radiotherapy involving the heart
and
incidence of acute lymphoblastic leukemia, rhabdomyosarcoma, Ewing's sarcoma
and rs2229774 where indicated. n.)
'Association analyses for imputed SNPs were restricted to those with BEAGLE
allelic R2 > 0.5.
n.)
dChromosomal positions in the GRCH37.p13.
0
(xi
'Calculated using the CEU component of 1000 Genomes reference population.
0
54
TABLE 10: shows stage 1 Discovery Analysis - Results for Previous ACT-
associated Regions.
MAF
MAF
IQ,
Marker Chra Positionb Gene Function P-
Odds Ratio Minor Referen
(248
valueb (95%C1)
Allele µ''` ce
Cases)
Controls)
rs17583889 2 138746039 HNMT INTRON 0.008 2.4 (1.3 -
4.5) A 0.344 0.175 5,6
ABCC2/ NONSYN-
rs8187710 10 101611294 0.021 4.3 (1.4 -
13.8) A 0.078 0.046 7,8
MRP2 CODING
rs2868177 7 75589903 POR INTRON 0.016 2.1 (1.1 -
4.0) G 0.438 0.312 9
rs13240755 7 75606109 POR INTRON 0.033 2.0 (1.0 -
3.7) G 0.453 0.349 9
rs4732513 7 75607608 POR INTRON 0.041 1.9 (1.0 -
3.6) G 0.466 0.348 9
NONSYN-
rs2232228 16 69143577 HAS3 0.18 0.67 (0.36 -
1.2) G 0.375 0.427 2 0
CODING
ABCC1/
0
rs3743527 16 16235681 UTR 0.24 0.65 (0.30 -
1.4) A 0.172 0.204 10 "
MRP1
ko
1-,
1-,
rs13058338 22 37632770 RA C2 INTRON 0.28
0.68 (0.34 - 1.4) T 0.188 0.245 7,8 --.1
0
l 0
rs10836235 11 34460704 CAT INTRON 0.46 0.70 (0.26 -
1.9) A 0.109 0.118 11
0
1-,
SYN-
rs1695 11 67352689 GSTP1 0.46 1.3 (0.68 -
2.4) G 0.344 0.349 12-14 1
CODING
1-,
1-,
1
SYN-
3,13,15,1
rs1056892 21 37518706 CBR3 0.64 0.85 (0.42 -
1.7) A 0.344 0.351 1-,
CODING
6
ABCC1/ NONSYN-
rs246221 16 16138322 0.68 1.1 (0.60 -
2.2) G 0.281 0.274 10
MRP1 CODING
NONSYN-
rs1799945 6 26091179 HFE 0.68 0.84 (0.37 -
1.9) G 0.125 0.151 8,17
CODING
SYN-
rs4673 16 88713236 CYBA 0.81 1.1 (0.59 -
2.0) A 0.371 0.356 7,18
CODING
'Abbreviations: Chr, chromosome; MAF, minor allele frequency.
b Chromosomal positions in the GRCH37.p13.
b P-values and odds ratios (95%C1) are for logistic regression analysis
(additive model) with adjustment for age at treatment,
cumulative anthracycline exposure, radiotherapy involving the heart and
incidence of acute lymphoblastic leukemia,
rhabdomyosarcoma and Ewing's sarcoma.
CA 02911709 2015-11-10
The stage 3 cohort, comprised of four distinct populations, was assessed for
population
stratification using the genomic inflation factor and principal component
analysis. PCs i¨lo
did not significantly differ between cases and controls (P>0.14) and the Xcc
of 0.941 suggested
no confounding effect of population stratification on the genetic association
of rs2229774 with
ACT in this cohort. In line with the reported high incidence of ACT in studies
conducted in
India (Agarwala, Kumar et al. 2000) and Pakistan(Shaikh, Saleem et al. 2013),
and in African-
American patients in the USA (Krischer, Epstein et al. 1997; Hasan, Dinh et
al. 2004), RARG
p52229774 is more frequent in South Asian (22%) and African populations (11%)
compared to
European (6%) and Hispanic (5%) populations. By contrast, rs2229774 is very
rare in East
Asian (0%) populations (woo Genomes). It will be interesting to correlate the
frequency of
n2229774 with the incidence of ACT in children of different ancestries as data
on multiethnic
studies become available for comparison.
A meta-analysis of all study populations (456 patients; 73 cases and 383
controls) using
logistic regression adjusted for age, dose, ALL, RT, Ewing's sarcoma and
rhabdomyosarcoma
showed that RARG rs2229774 was significantly associated with ACT (P-5.9x10-8,
OR=4.7
(2.7-8.3)), which surpassed the Bonferroni-adjusted multiple testing
correction threshold,
with no significant heterogeneity across the GWAS and two independent
replication analyses
(Pheterogeneity=0.402). Overall, rs2229774-carriers had significantly
increased odds of developing
ACT compared to non-carriers (OR=5.2 (3.0-9.0); P=5.9x10-10).
Example 2: Functional validation of the RARGs427L variant
To fine-map the rs2229774 association with ACT, we imputed variants on
chromosome 12
using the stage 1 cohort and the woo Genomes CEU reference population, and
tested their
association with ACT by logistic regression (FIGURE 1). The linkage
disequilibrium (D')
based upon the moo Genomes CEU population for this region was similarly
demonstrated that
the associated haplotype is localized to RARG (data not shown). Of the
1,005,286 imputed
SNPs, three were associated with ACT at P<I.0x10-5 (rsiii70481, rs73309171 and
rs57789211;
TABLE 9). These were all located in introns of RARG, highly correlated with
each other
(D' r2-0=54), and in high linkage disequilibrium (LD) with rs2229774
(D'=1.0o,
r20.55). Since the ACT associations of the imputed variants were similar to
rs2229774, and
logistic regression with conditioning on n2229774 abolished these associations
(TABLE 9),
the non-synonymous variant r52229774 was prioritized for functional
characterization to gain
biological insight into its association with ACT.
56
CA 02911709 2015-11-10
Retinoic acid receptors bind to DNA regulatory sequences termed retinoic acid
response
elements (RARE) and transcriptionally co-regulate downstream gene expression
in response to
their agonist all-trans retinoic acid (ATRA)(Chambon 1996). Notably, RARG can
both activate
and repress transcription in response to ATRA(Tang, Chen et al. 2011). To
explore the
functional properties of rs2229774 (i.e. RARGs4271-), we first examined its
transactivation of
RARG regulatory elements and then identified a putative role in the
dysregulation of a critical
gene involved in the development of ACT (see Online Methods). In HEK293T cells
expressing
RARGs4271-, the ATRA-inducible transcriptional activation of a RARE-coupled
luciferase
reporter was significantly reduced compared to wild type RARG-expressing cells
(FIGURE
2a). Immunoblot analysis verified that both wild type and variant RARG
proteins were
detected at similar levels in HEK293T cell lysate (FIGURE 2a inset). The
significant 17%
decrease in RARG activity conferred by the rs2229774 variant may be an
underestimate since
endogenous retinoic acid receptors were present in this assay. These results
suggested that
dysregulation of an RARG-regulated gene might underlie the association of
rs2229774 with
ACT.
Anthracyclines are mechanistically and genetically linked with Topoisomerase
II beta (Top2b).
Anthracyclines exert their anticancer activity by binding and inhibiting
Topoisomerase II
(Minotti, Menna et al. 2004). In addition, Top2b is necessary for the
development of ACT in a
murine model (Zhang, Liu et al. 2012), while in a rat cardiomyoblast (H9c2)
cell line, Top2b
levels are decreased by the ACT cardioprotectant, dexrazoxane (Lyu, Kerrigan
et al. 2007).
RARG expression has been reported as "particularly high" in the heart (Nuclear
Receptor
Signaling Atlas) and is highly induced in murine cardiac cells following
cardiac injury (Bilbija,
Haugen et al. 2012). Since RARG has been shown to bind to the Top2b promoter
(Delacroix,
Moutier et al. 2010) it was one potential candidate of RARGs4271-
dysregulation. We found that
Top2b expression in H9c2 cells was significantly decreased when human RARG was
added,
and this effect was further exacerbated with the addition of ATRA (FIGURE 2b).
By contrast,
the RARGs427L variant did not repress Top2b expression as effectively as wild
type RARG
(FIGURE 2c). Taken together these results demonstrate that RARG represses
Top2b
expression and that rat cardiomyoblasts carrying rs2229774 express higher
basal levels of
Top2b, consistent with an increased susceptibility to ACT.
We have identified a novel genetic biomarker for ACT, RARG rs2229774, using a
three-stage
genetic association study combined with biological functional analyses. This
discovery, despite
originating from a relatively small number of patients, is likely due to its
large effect size. In
57
CA 02911709 2015-11-10
general, sample size limitation remains a major challenge in pediatric cancer
pharmacogenomic studies (McLeod 2013; Wheeler, Maitland et al. 2013), where
the need for a
homogenous study population and well defined clinical phenotypes results in
reduced
numbers of study patients, particular the number of affected individuals.
However, clinically
relevant genetic markers of severe ADRs such as ACT, abacavir-induced
hypersensitivity
reaction, and carbamazepine-induced Stevens-Johnson syndrome, are expected to
have large
effect sizes and therefore, the number of patients, particularly the number of
affected
individuals, required to uncover these genetic associations from large-scale
genome screens,
has been shown to be in the range of 10-100 patients(Nelson, Bacanu et al.
2009) - similar to
the number in this study.
Our study was 8o% powered to detect genome-wide associations of P.5.5 x io-8
with per-allele
012.4.5 and MAFo.io. There are likely additional genetic contributions to ACT
including
previously reported associations (Wojnowski, Kulle et al. 2005; Blanco,
Leisenring et al. 2008;
Rajic, Aplenc et al. 2009; Rossi, Rasi et al. 2009; Windsor, Strauss et al.
2011; Blanco, Sun et
al. 2012; Cascales, Sanchez-Vega etal. 2012; Lubieniecka, Liu etal. 2012;
Semsei, Erdelyi etal.
2012; Visscher, Ross et al. 2012; Volkan-Salanci, Aksoy et al. 2012; Armenian,
Ding et al.
2013; Cascales, Pastor-Quirante et al. 2013; Lipshultz, Lipsitz et al. 2013;
Lubieniecka,
Graham et al. 2013; Sachidanandam, Gayle et al. 2013; Visscher, Ross et al.
2013; Vivenza,
Feola et al. 2013; Wang, Liu et al. 2014) that were not uncovered in the
current GWAS,
potentially owing to the strict ACT case definition and resulting small
patient numbers used in
this study. For example, ABCC2/MRP2, HNMT and POR, although not reaching
genome-wide
significance in this study, exhibited strong trends of association (P<o.o5;
TABLE io) in the
same direction of effect as previously reported(Wojnowski, Kulle et al. 2005;
Visscher, Ross et
al. 2012; Armenian, Ding et al. 2013; Lubieniecka, Graham et al. 2013;
Sachidanandam, Gayle
et al. 2013). This underscores the complementary nature of GWAS and candidate
gene
association studies in identifying variants associated with pharmacogenomic
traits.
In agreement with in silico predictions, our in vitro studies demonstrate that
rs2229774 causes
a relatively tolerated amino acid substitution that results in a moderate, but
significant,
reduction in RARE transcriptional activation. We established a genetic
interaction between
RARG and Top2b, where expression of the latter is significantly repressed by
RARG in the
presence of ATRA. Notably, the treatment of acute promyelocytic leukemia with
high
cumulative doses of anthracycline and ATRA has resulted in a significantly
lower incidence of
ACT (Ortega, Madero et al. 2005; Pellicori, Calicchia et al. 2012). We further
showed that
58
CA 02911709 2015-11-10
rs2229774 causes derepression of Top2b in cardiomyoblasts, directly linking
this variant with
ACT. These data are consistent with a model where rs2229774 carriers have
higher basal levels
of TOP2B in cardiomyocytes, conferring increased susceptibility to
cardiotoxicity when treated
with anthracyclines. Nevertheless, despite rs2229774 encoding a non-synonymous
variant, it is
plausible that the rs2229774 haplotype confers increased susceptibility to ACT
through a
regulatory mechanism. To our knowledge a comprehensive analysis of RARG-
regulated gene
expression in cardiomyocytes has not been reported and it is possible that
additional genes in
cardiomyocytes may contribute to the development of ACT when dysregulated in
RARG
1.52229774 carriers. For example, ATRA-regulated gene expression in
cardiomyocytes is
important for cardiac development and the progression of cardiomyocyte
hypertrophy (Palm-
Leis, Singh et at. 2004; Arima, Shiotsugu et al. 2005; Simandi, Balint et at.
2010), though the
specific contribution of RARG to these processes is unknown.
The identification of RARG rs2229774, genetically and functionally linked to
ACT, provides a
clinical tool that may be used to predict genetic risk and improve ACT risk
stratification. ACT is
a clinically significant ADR, and carriers of 1.52229774 have 5-fold increased
odds of ACT. This
novel finding merits further exploration of the role of RARG in ACT and of the
clinical utility of
RARG 1.52229774 predictive testing to inform ACT risk assessment.
Agarwala, S., R. Kumar, et al. (2000). "High incidence of adriamycin
cardiotoxicity in children
even at low cumulative doses: role of radionuclide cardiac angiography." J
Pediatr Surg
35(12): 1786-1789.
Altman, A. J. (2004). Supportive care of children with cancer: current therapy
and guidelines
from the Children's Oncology Group. Baltimore, John Hopkins University Press.
Arima, K., J. Shiotsugu, et al. (2005). "Global analysis of RAR-responsive
genes in the
Xenopus neurula using cDNA microarrays." Dev Dyn 232(2): 414-431.
Armenian, S. H., Y. Ding, et al. (2013). "Genetic susceptibility to
anthracycline-related
congestive heart failure in survivors of haematopoietic cell transplantation."
Br J
Haematol 163(2): 205-213.
Barrett, J. C., B. Fry, et at. (2005). "Haploview: analysis and visualization
of LD and haplotype
maps." Bioinformatics 21(2): 263-265.
Bilbija, D., F. Haugen, et al. (2012). "Retinoic acid signalling is activated
in the postischemic
heart and may influence remodelling." PLoS One 7(9): e44740.
Blanco, J. G., W. M. Leisenring, et al. (2008). "Genetic polymorphisms in the
carbonyl
reductase 3 gene CBR3 and the NAD(P)H:quinone oxidoreductase 1 gene NQ01 in
59
CA 02911709 2015-11-10
patients who developed anthracycline-related congestive heart failure after
childhood
cancer." Cancer 112(12): 2789-2795.
Blanco, J. G., C. L. Sun, et al. (2012). "Anthracycline-related cardiomyopathy
after childhood
cancer: role of polymorphisms in carbonyl reductase genes--a report from the
Children's Oncology Group." J Clin Oncol 30(13): 1415-1421.
Browning, S. R. and B. L. Browning (2007). "Rapid and accurate haplotype
phasing and
missing-data inference for whole-genome association studies by use of
localized
haplotype clustering." Am J Hum Genet 81(5): 1084-1097.
Cascales, A., F. Pastor-Quirante, etal. (2013). "Association of anthracycline-
related cardiac
histological lesions with NADPH oxidase functional polymorphisms." Oncologist
18(4):
446-453.
Cascales, A., B. Sanchez-Vega, et al. (2012). "Clinical and genetic
determinants of
anthracycline-induced cardiac iron accumulation." Int J Cardiol 154(3): 282-
286.
Chambon, P. (1996). "A decade of molecular biology of retinoic acid
receptors." FASEB J
10(9): 940-954.
Delacroix, L., E. Moutier, et al. (2010). "Cell-specific interaction of
retinoic acid receptors with
target genes in mouse embryonic fibroblasts and embryonic stem cells." Mol
Cell Biol
30(1): 231-244.
Devlin, B. and K. Roeder (1999). "Genomic control for association studies."
Biometrics 55(4):
997-1004.
Hasan, S., K. Dinh, et al. (2004). "Doxorubicin cardiotoxicity in African
Americans." J Natl
Med Assoc 96(2): 196-199.
Kremer, L. C., E. C. van Dalen, et al. (2001). "Anthracycline-induced clinical
heart failure in a
cohort of 607 children: long-term follow-up study." J Clin Oncol 19(1): 191-
196.
Krischer, J. P., S. Epstein, etal. (1997). "Clinical cardiotoxicity following
anthracycline
treatment for childhood cancer: the Pediatric Oncology Group experience." J
Clin
Oncol 15(4): 1544-1552.
Lipshultz, S. E., S. D. Colan, et al. (1991). "Late cardiac effects of
doxorubicin therapy for acute
lymphoblastic leukemia in childhood." N Engl J Med 324(12): 808-815.
Lipshultz, S. E., S. R. Lipsitz, et al. (2013). "Impact of hemochromatosis
gene mutations on
cardiac status in doxorubicin-treated survivors of childhood high-risk
leukemia."
Cancer 119(19): 3555-3562.
Lipshultz, S. E., S. R. Lipsitz, et al. (1995). "Female sex and drug dose as
risk factors for late
cardiotoxic effects of doxorubicin therapy for childhood cancer." N Engl J Med
332(26): 1738-1743.
Lubieniecka, J. M., J. Graham, etal. (2013). "A discovery study of
daunorubicin induced
cardiotoxicity in a sample of acute myeloid leukemia patients prioritizes P450
oxidoreductase polymorphisms as a potential risk factor." Front Genet 4: 231.
Lubieniecka, J. M., J. Liu, et al. (2012). "Single-nucleotide polymorphisms in
aldo-keto and
carbonyl reductase genes are not associated with acute cardiotoxicity after
daunorubicin chemotherapy." Cancer Epidemiol Biomarkers Prey 21(11): 2118-
2120.
Lyu, Y. L., J. E. Kerrigan, et al. (2007). "Topoisomerase IIbeta mediated DNA
double-strand
breaks: implications in doxorubicin cardiotoxicity and prevention by
dexrazoxane."
Cancer Res 67(18): 8839-8846.
McLeod, H. L. (2013). "Cancer pharmacogenomics: early promise, but concerted
effort
needed." Science 339(6127): 1563-1566.
Minotti, G., P. Menna, et al. (2004). "Anthracyclines: molecular advances and
pharmacologic
developments in antitumor activity and cardiotoxicity." Pharmacol Rev 56(2):
185-229.
Mulrooney, D. A., M. W. Yeazel, et al. (2009). "Cardiac outcomes in a cohort
of adult survivors
of childhood and adolescent cancer: retrospective analysis of the Childhood
Cancer
Survivor Study cohort." BMJ 339: b4606.
CA 02911709 2015-11-10
Nelson, M. R., S. A. Bacanu, et al. (2009). "Genome-wide approaches to
identify
pharmacogenetic contributions to adverse drug reactions." Pharmacogenomics J
9(1):
23-33.
Ortega, J. J., L. Madero, et al. (2005). "Treatment with all-trans retinoic
acid and
anthracycline monochemotherapy for children with acute promyelocytic leukemia:
a
multicenter study by the PETHEMA Group." J Clin Oncol 23(30): 7632-7640.
Palm-Leis, A., U. S. Singh, et al. (2004). "Mitogen-activated protein kinases
and mitogen-
activated protein kinase phosphatases mediate the inhibitory effects of all-
trans
retinoic acid on the hypertrophic growth of cardiomyocytes." J Biol Chem
279(52):
54905-54917.
Pellicori, P., A. Calicchia, et al. (2012). "Subclinical anthracycline
cardiotoxicity in patients
with acute promyelocytic leukemia in long-term remission after the AIDA
protocol."
Congest Heart Fail 18(4): 217-221.
Pruim, R. J., R. P. Welch, et al. (2010). "LocusZoom: regional visualization
of genome-wide
association scan results." Bioinformatics 26(18): 2336-2337.
Rajic, V., R. Aplenc, et al. (2009). "Influence of the polymorphism in
candidate genes on late
cardiac damage in patients treated due to acute leukemia in childhood." Leuk
Lymphoma 50(10): 1693-1698.
Rodvold, K. A., D. A. Rushing, et al. (1988). "Doxorubicin clearance in the
obese." J Clin Oncol
6(8): 1321-1327.
Rossi, D., S. Rasi, etal. (2009). "Analysis of the host pharmacogenetic
background for
prediction of outcome and toxicity in diffuse large B-cell lymphoma treated
with R-
CHOP21." Leukemia 23(6): 1118-1126.
Sachidanandam, K., A. A. Gayle, et al. (2013). "Unexpected doxorubicin-
mediated
cardiotoxicity in sisters: possible role of polymorphisms in histamine n-
methyl
transferase." J Oncol Pharm Pract 19(3): 269-272.
Semsei, A. F., D. J. Erdelyi, et al. (2012). "ABCC1 polymorphisms in
anthracycline-induced
cardiotoxicity in childhood acute lymphoblastic leukaemia." Cell Biol Int
36(1): 79-86.
Shaikh, A. S., A. F. Saleem, et al. (2013). "Anthracycline-induced
cardiotoxicity: prospective
cohort study from Pakistan." BMJ Open 3(11): e003663.
Simandi, Z., B. L. Balint, etal. (2010). "Activation of retinoic acid receptor
signaling
coordinates lineage commitment of spontaneously differentiating mouse
embryonic
stem cells in embryoid bodies." FEBS Lett 584(14): 3123-3130.
Tang, Q., Y. Chen, etal. (2011). "A comprehensive view of nuclear receptor
cancer cistromes."
Cancer research 71(22): 6940-6947.
Visscher, H., C. J. Ross, et al. (2009). "Application of principal component
analysis to
pharmacogenomic studies in Canada." Pharmacogenomics J 9(6): 362-372.
Visscher, H., C. J. Ross, et al. (2012). "Pharmacogenomic prediction of
anthracycline-induced
cardiotoxicity in children." J Clin Oncol 30(13): 1422-1428.
Visscher, H., C. J. Ross, et al. (2013). "Validation of variants in SLC28A3
and UGT1A6 as
genetic markers predictive of anthracycline-induced cardiotoxicity in
children." Pediatr
Blood Cancer 60(8): 1375-1381.
Vivenza, D., M. Feola, etal. (2013). "Role of the renin-angiotensin-
aldosterone system and the
glutathione S-transferase Mu, Pi and Theta gene polymorphisms in
cardiotoxicity after
anthracycline chemotherapy for breast carcinoma." Int J Biol Markers 28(4):
e336-
347.
Volkan-Salanci, B., H. Aksoy, et al. (2012). "The relationship between changes
in functional
cardiac parameters following anthracycline therapy and carbonyl reductase 3
and
glutathione S transferase Pi polymorphisms." J Chemother 24(5): 285-291.
61
CA 02911709 2015-11-10
Wang, X., W. Liu, et al. (2014). "Hyaluronan synthase 3 variant and
anthracycline-related
cardiomyopathy: a report from the children's oncology group." J Clin Oncol
32(7): 647-
653.
Welter, D., J. MacArthur, et al. (2014). "The NHGRI GWAS Catalog, a curated
resource of
SNP-trait associations." Nucleic Acids Res 42(Database issue): Mom-1006.
Wheeler, H. E., M. L. Maitland, et al. (2013). "Cancer pharmacogenomics:
strategies and
challenges." Nat Rev Genet 14(1): 23-34.
Windsor, R. E., S. J. Strauss, et al. (2011). "Germline genetic polymorphisms
may influence
chemotherapy response and disease outcome in osteosarcoma: A Pilot Study."
Cancer
118(7): 1856.
Wojnowski, L., B. Kulle, et al. (2005). "NAD(P)H oxidase and multidrug
resistance protein
genetic polymorphisms are associated with doxorubicin-induced cardiotoxicity."
Circulation 112(24): 3754-3762.
Zhang, S., X. Liu, et al. (2012). "Identification of the molecular basis of
doxorubicin-induced
cardiotoxicity." Nat Med 18(11): 1639-1642.
62