Note: Descriptions are shown in the official language in which they were submitted.
- 1 -
TITLE: METHODS AND KITS FOR SEPARATING NUCLEIC ACIDS BY
SIZE
RELATED APPLICATION
[001] This application claims priority benefits from US Provisional Patent
Application No. 62/310,368 and entitled "Method and Kits for Separating
Nucleic Acids by
Size".
FIELD OF INVENTION
[002] The present invention relates to methods and kits suitable for the
size selective
isolation of nucleic acids using silicon carbide. The methods and kits of the
present
invention are particularly useful for isolating RNA and DNA molecules having a
desired
minimum size during the preparation of a sequencing library.
BACKGROUND
[003] For many applications in molecular biology, and particularly
applications in
global gene expression analysis, it is often necessary to separate desired
nucleic acid
sequences from other non-desired sequences. Often, this separation is based on
size
differences in the nucleic acids. One such global gene expression analysis
application¨
wherein smaller undesired nucleic acids must be separated from larger, desired
nucleic
acids¨is in massive parallel sequencing, also known as next generation
sequencing. Next
generation sequencing (NGS) involves the sequencing of a large number of reads
(as much as
over 40 billion) per instrument run.
[004] There are many different platforms that can be used for next
generation
sequencing, including Roche 454, Roche GS FLX Titanium, Illumina MiSeq,
Illumina
HiSeq, Illumina Genome Analyzer IIX, Life Technologies SOLiD4, Life
Technologies Ion
Proton, Complete Genomics, Helicos Biosciences Heliscope, and Pacific
Biosciences SMRT.
All of the different platforms for next generation sequencing follow the same
general
procedure, namely preparing the purified RNA or DNA into a sequencing library,
followed
Date recue/ date received 2022-02-18
CA 2961177 2017-03-17
- 2 -
by massive parallel sequencing of relatively short sequences and subsequent
bioinformatics
to de-multiplex samples, align, annotate and aggregate reads, among other
things.
10051 In order to generate a sequencing library, multiple enzymatic
reactions are
required to modify and/or amplify the original input nucleic acid (RNA or
DNA). In
particular, fragments of nucleic acids, usually in the form of RNA or DNA
oligonucleotides,
are added to both the 5' and 3' end of the template nucleic acid targeted for
sequencing. The
best quality libraries will ensure that all the resulting reads obtained are
dedicated to the
targeted RNA or DNA template. However, in many cases, the usable read number
is
drastically reduced due to various contaminants being incorporated into the
library, including
adapter monomers as well as adapter-adapter ligation products. In addition, as
these
contaminants are of nucleic acid-origin, their presence in the final library
preparation could
negatively affect the accuracy of library quantification and the subsequent
amount of library
loaded onto the sequencing platform.
[006] Different purification technologies have been utilized to tackle the
issue of
removing unligated adapter monomers and adapter-adapter ligation products from
NGS
libraries. Some methods are directed at removing the excessive adapters prior
to cDNA
synthesis and PCR amplification, while others are directed at purifying the
desired library
containing the inserts of interest for sequencing from gels based on size.
However, all of
these current methods have shortcomings. For example, gel purification usually
requires a
lengthy workflow and subsequent purification and will add an extensive amount
of time to
the library preparation (sometimes overnight). In addition, most of the
existing purification
systems may not be able to resolve small size differences at the low nucleic
acid molecular
weight range (such as in the case of small RNA sequence ligation steps).
SUMMARY OF INVENTION
10071 Disclosed are methods and kits suitable for the size selective
isolation of
nucleic acids using silicon carbide.
CA 2961177 2017-03-17
-3-
10081 In one aspect, disclosed is a method for isolating nucleic acids
having a size
above a desired cut-off size from a nucleic acid containing sample, comprising
the steps of:
a) combining the sample with a binding buffer, alcohol and silicon carbide to
provide
a binding mixture, wherein the nucleic acids having a size above the cut-off
size bind
to the silicon carbide, and wherein the cut-off size is determined by the
alcohol
concentration of the binding mixture;
b) separating the bound nucleic acids from the remaining sample;
c) optionally, washing the bound nucleic acids; and
d) eluting the bound nucleic acids from the silicon carbide.
10091 This size selective isolation method can be employed in a variety of
applications, and in particular, the preparation of sequencing libraries, such
as next
generation sequencing libraries.
100101 In one embodiment of the disclosed size selective isolation method,
the
alcohol is ethanol, isopropanol or methanol.
100111 In another embodiment, the alcohol is ethanol and the ethanol
concentration of
the binding buffer and/or the binding mixture is at least about 10% (v/v) and
the cut-off size
is about 43 nucleotides or larger. In a further embodiment, the ethanol
concentration of the
binding buffer and/or the binding mixture is at least about 25% (v/v) and the
cut-off size is
about 22 nucleotides or larger. In a still further embodiment, the ethanol
concentration of the
binding buffer and/or the binding mixture is at least about 50% (v/v) and the
cut-off size is
about 10 nucleotides or larger.
100121 In another embodiment, the silicon carbide can be provided in a
column, such
as a spin column. In a further embodiment, the silicon carbide can be provided
as slurry.
CA 2961177 2017-03-17
-4-
100131 In another embodiment, the nucleic acid can be a single stranded
RNA, a
double stranded RNA, a single stranded DNA, a double stranded DNA or a double
stranded
RNA/DNA hybrid.
[0014] In another embodiment, the nucleic acid containing sample may
comprise: i)
extracted nucleic acids, which have optionally been subject to mechanical or
enzymatic
treatment; ii) amplification reaction products; and/or iii) ligation reaction
products. In a
further embodiment, the amplification reaction products can be polymerasc
chain reaction
(PCR) products. In a still further embodiment, the ligation reaction products
are adaptor
ligation products, wherein the adaptor ligation products are nucleic acids
flanked by 5' and/or
3' adapters; adapter monomers; and/or adapter-adapter ligation products
[0015] In another embodiment, the nucleic acid containing sample is
obtained during
the preparation of a sequencing library. In a further embodiment, the nucleic
acid containing
sample is obtained during the preparation of a next generation sequencing
library and the
nucleic acid containing sample is an adapter ligation sample comprising
nucleic acids flanked
by 5' and/or 3' adapters; adapter monomers; and/or adapter-adapter ligation
products.
[0016] In another embodiment, the method can be used for isolating adapter-
ligated
RNA molecules from an adapter ligation sample and for removing adapter
monomers and
adapter-adapter ligation products based on the larger size of the adapter-
ligated RNA
molecules, and wherein step a) comprises contacting the adapter ligation
sample with the
binding buffer, alcohol and silicon carbide and binding adapter ligated RNA
molecules to the
silicon carbide, wherein under the used alcohol concentration, the adapter
monomers and
adapter-adapter ligation products substantially do not bind to the silicon
carbide. In a further
embodiment, the cut-off size lies above the size of adapter monomers and above
the size of
expected adapter-adapter ligation products and wherein the cut-off size is at
least 10
nucleotides, at least 15 nucleotides, at least 20 nucleotides, at least 25
nucleotides or at least
30 nucleotides above the size of expected adapter-adapter ligation products.
[0017] In another aspect, disclosed is a method for preparing a small RNA
library
suitable for next generation sequencing, wherein said method comprises:
CA 2961177 2017-03-17
- 5 -
a) isolating small RNA molecules from a sample;
b) performing a 3' adapter ligation step to provide a 3' adaptor ligation
reaction mixture comprising single stranded small RNA molecules that are
flanked 3' by adapters;
c) isolating single stranded small RNA molecules having a fragment size
above a predetermined cut-off size in order to remove 3' adapter monomers
and adapter-adapter ligation products, wherein said size selection step
comprises:
i) combining the 3 'adaptor ligation reaction mixture with a binding
buffer, alcohol and silicon carbide to provide a binding mixture,
wherein the single stranded small RNA molecules having a fragment
size above the cut-off size bind to the silicon carbide, and wherein the
cut-off size is determined by the alcohol concentration of the binding
mixture;
ii) separating the bound single stranded small RNA molecules from the
remaining ligation reaction mixture;
iii) optionally, washing the bound single stranded small RNA
molecules; and
iv) eluting the bound single stranded small RNA molecules from the
silicon carbide;
d) performing a 5' adapter ligation step to provide a 5'adaptor ligation
reaction mixture comprising single stranded small RNA molecules that are
flanked 3' and 5' by adapters;
c) optionally, performing a step of isolating single stranded small RNA
molecules having a fragment size above a predetermined cut-off size in order
CA 2961177 2017-03-17
- 6 -
to remove 5' adapter monomers and adapter-adapter ligation products,
wherein said size selection step comprises:
i) combining the 5'adaptor ligation reaction mixture with a binding
buffer, alcohol and silicon carbide to provide a binding mixture,
wherein the single stranded small RNA molecules having a fragment
size above the cut-off size bind to the silicon carbide, and wherein the
cut-off size is determined by the alcohol concentration of the binding
mixture;
ii) separating the bound single stranded small RNA molecules from the
remaining ligation reaction mixture;
iii) optionally, washing the bound single stranded small RNA
molecules; and
iv) eluting the bound single stranded small RNA molecules from the
silicon carbide;
0 reverse transcribing single stranded RNA molecules flanked with 5' and 3'
adaptors to provide single stranded cDNA molecules; and
g) amplifying the single stranded cDNA molecules by limited cycle PCR to
incorporate an index sequence.
[0018] In one embodiment of the disclosed methods for preparing a small RNA
library suitable for next generation sequencing, the alcohol is ethanol,
isopropanol or
methanol.
[0019] In another embodiment, the alcohol is ethanol and the ethanol
concentration of
the binding buffer and/or the binding mixture is at least about 10% (v/v) and
the cut-off size
is about 43 nucleotides or larger. In a further embodiment, the ethanol
concentration of the
binding buffer and/or the binding mixture is at least about 25% (v/v) and the
cut-off size is
about 22 nucleotides or larger. In a still further embodiment, the ethanol
concentration of the
CA 2961177 2017-03-17
- 7 -
binding buffer and/or the binding mixture is at least about 50% (v/v) and the
cut-off size is
about 10 nucleotides or larger.
[0020] In another embodiment, the silicon carbide can be provided in a
column, such
as a spin column. In a further embodiment, the silicon carbide can be provided
as slurry.
[0021] In another aspect, disclosed is a method for preparing a small RNA
library
suitable for next generation sequencing, wherein said method comprises:
a) isolating small RNA molecules from a sample;
b) performing a 5' adapter ligation step to provide a 5' adaptor ligation
reaction mixture comprising single stranded small RNA molecules that are
flanked 5' by adapters;
c) isolating single stranded small RNA molecules having a fragment size
above a predetermined cut-off size in order to remove 5' adapter monomers
and adapter-adapter ligation products, wherein said size selection step
comprises:
i) combining the 5'adaptor ligation reaction mixture with a binding
buffer, alcohol and silicon carbide to provide a binding mixture,
wherein the single stranded small RNA molecules having a fragment
size above the cut-off size bind to the silicon carbide, and wherein the
cut-off size is determined by the alcohol concentration of the binding
mixture;
ii) separating the bound single stranded small RNA molecules from the
remaining ligation reaction mixture;
iii) optionally, washing the bound single stranded small RNA
molecules; and
CA 2961177 2017-03-17
- 8 -
iv) eluting the bound single stranded small RNA molecules from the
silicon carbide;
d) performing a 3' adapter ligation step to provide a 3'adaptor ligation
reaction mixture comprising single stranded small RNA molecules that arc
flanked 3' and 5' by adapters;
e) optionally, performing a step of isolating single stranded small RNA
molecules having a fragment size above a predetermined cut-off size in order
to remove 3' adapter monomers and adapter-adapter ligation products,
wherein said size selection step comprises:
i) combining the 3'adaptor ligation reaction mixture with a binding
buffer, alcohol and silicon carbide to provide a binding mixture,
wherein the single stranded small RNA molecules having a fragment
size above the cut-off size bind to the silicon carbide, and wherein the
cut-off size is determined by the alcohol concentration of the binding
mixture;
ii) separating the bound single stranded small RNA molecules from the
remaining ligation reaction mixture;
iii) optionally, washing the bound single stranded small RNA
molecules; and
iv) eluting the bound single stranded small RNA molecules from the
silicon carbide;
f) reverse transcribing single stranded RNA molecules flanked with 5' and 3'
adaptors to provide single stranded cDNA molecules; and
g) amplifying the single stranded cDNA molecules by limited cycle PCR to
incorporate an index sequence.
CA 2961177 2017-03-17
-9-
100221 In one embodiment of the disclosed method for preparing a small RNA
library
suitable for next generation sequencing, the alcohol is ethanol, isopropanol
or methanol.
[0023] In another embodiment, the alcohol is ethanol and the ethanol
concentration of
the binding buffer and/or the binding mixture is at least about 10% (v/v) and
the cut-off size
is about 43 nucleotides or larger. In a further embodiment, the ethanol
concentration of the
binding buffer and/or the binding mixture is at least about 25% (v/v) and the
cut-off size is
about 22 nucleotides or larger. In a still further embodiment, the ethanol
concentration of the
binding buffer and/or the binding mixture is at least about 50% (v/v) and the
cut-off size is
about 10 nucleotides or larger.
[0024] In another embodiment, the silicon carbide can be provided in a
column, such
as a spin column. In a further embodiment, the silicon carbide can be provided
as slurry.
[0025] In another aspect, disclosed is a kit for the selective binding of
nucleic acids
having a size above a desired cut-off size, comprising: a) a binding buffer to
be combined
with ethanol to provide an ethanol concentration of about 1 to about 50%
(v/v); b) a wash
solution; c) an elution solution; d) silicon carbide, and c) instructions for
adjusting the
ethanol concentration to selectively bind nucleic acids having a size above
the desired cut-off
size; wherein the cut-off size is at least about 10 nucleotides.
[0026] In one embodiment, the silicon carbide can be provided in a column,
such as a
spin column. In a further embodiment, the silicon carbide can be provided as
slurry.
100271 In another embodiment, the ethanol concentration is at least about
10% (v/v)
and the cut-off size is about 43 nucleotides or larger. In a further
embodiment, the ethanol
concentration is at least about 25% (v/v) and the cut-off size is about 22
nucleotides or larger.
In a still further embodiment, the ethanol concentration is about 50% (v/v)
and the cut-off
size is about 10 nucleotides or larger.
CA 2961177 2017-03-17
- 10 -
BRIEF DESCRIPTION OF THE DRAWINGS
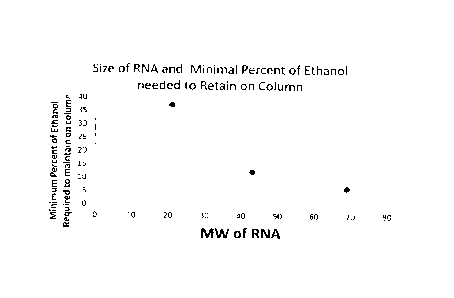
[0028] Figure 1 is a graph comparing the size of RNA molecules captured by
a SiC
column using different binding buffers having varying concentrations of
ethanol.
[0029] Figure 2 is a graph comparing the size of RNA molecules captured by
a SiC
column using different binding buffers having varying concentrations of
isopropanol.
100301 Figure 3 is a line graph comparing the amount of FAM-cel-miR-39-RA3
(43
nt) and Hex-RA3 (21 nt) captured by a SiC column using different binding
buffers having
varying concentrations of ethanol.
100311 Figure 4 is a line graph comparing the amount of RA5-cel-miR-39-RA3
(69
nt) and RA3 (21 nt) captured by a SiC column using different binding buffers
having varying
concentrations of ethanol.
[0032] Figure 5 compares the predominant PCR products obtained for HeLa RNA
input (left panel) and for plasma RNA input (right panel), with and without
adaptor cleanup
prior to PCR amplification.
100331 Figure 6, panel A compares the predominant PCR products obtained for
a
small RNA library prepared using a blocking oligonucleotide for 3' adapter
cleanup (left
panel) and a SiC based size exclusion 3' adapter cleanup method as disclosed
herein (right
panel) prior to PCR amplification.
[0034] Figure 6, panel B is a Venn diagram generated using miRNA with reads
equal
or more than 5 RPM (reads per millions) and which illustrates that both the
blocking
oligonucleotide and SiC based size exclusion cleanup methods shared the same
diversity of
200 microRNAs and with the SiC based size exclusion method yielding more
unique
miRNAs.
CA 2961177 2017-03-17
- 1 1 -
DESCRIPTION
(00351 It has been unexpectedly found that the size of nucleic acids bound
to silicon
carbide (SiC) can be controlled by the alcohol concentration used during the
binding step.
This finding allows for size selective binding of nucleic acids, and in
particular, small nucleic
acids such as microRNAs (miRNAs), from a sample containing different sized
nucleic acids.
The cut-off size (e.g. the minimum desired size) for the targeted nucleic
acids can be set by
selecting the appropriate alcohol concentration for the binding step.
Generally, the higher the
concentration of alcohol used during the binding step, the lower the cut-off
size of the nucleic
acids that will be selectively bound to the SiC. Size selective binding of
nucleic acids to SiC
can be used in various applications, including as part of a cleanup step in
the preparation of
sequencing libraries. It has been further surprisingly found that use of a SiC
based cleanup
step in the preparation of a small RNA sequencing library may enhance the
number of
different miRNA incorporated into the sequencing library as compared to prior
art cleanup
methods.
Method for isolating nucleic acids based on size
100361 Disclosed is a method for isolating nucleic acids having a size
above a desired
cut-off size from a nucleic acid containing sample. The method comprises the
steps of:
a) combining the sample with a binding buffer, alcohol and SiC to provide a
binding mixture, wherein nucleic acids having a size above the desired cut-off
size are selectively bound to the SiC and wherein the cut-off size for
selective
binding to the SiC is determined by the alcohol concentration of the binding
mixture;
b) separating the bound nucleic acids from the remaining sample;
c) optionally, washing the bound nucleic acids; and
d) eluting the bound nucleic acids from the SiC.
CA 2961177 2017-03-17
- 12 -
10037] As used herein, "size" refers to the length of the nucleic acid,
expressed either
in terms of the number of nucleotides (nt) or the number of base pairs (bp) in
the context of
double stranded nucleic acids. The "desired cut-off size" refers to the
minimum size of the
target nucleic acids to be isolated. For example, if the targeted nucleic
acids are those having
a length >100nt, then the "desired cut-off size" would be 100nt and the
concentration of
alcohol adjusted accordingly to affect selective binding of those nucleic
acids having a length
>100nt.
100381 The disclosed size selective isolation method can be used to
selectively enrich
the population of target nucleic acids that are the desired minimum size
and/or selectively
deplete the population of non-target nucleic acids that are shorter than the
desired minimum
size. This is accomplished by selecting the appropriate alcohol concentration
for the binding
step, as the alcohol concentration determines the cut-off size for the nucleic
acids that will
selectively bind to the SiC. Nucleic acids having a size above the cut-off
size will efficiently
bind to the SiC, while nucleic acids having a size below the cut-off size are
predominantly
not bound and are not recovered by the SiC. Therefore, nucleic acids shorter
than the cut-off
size are efficiently depleted and nucleic acid molecules having the desired
minimum size are
enriched.
[00391 In a preferred embodiment, the disclosed size selective isolation
method can
be used specifically to isolate adapter-ligated RNA molecules from an adapter
ligation
sample and to remove adapter monomers and adapter-adapter ligation products
based on the
larger size of the adapter-ligated RNA molecules, wherein step a) comprises
contacting the
adapter ligation sample with the binding buffer, alcohol and silicon carbide
and binding
adapter ligated RNA molecules to the silicon carbide, wherein under the used
alcohol
concentration, the adapter monomers and adapter-adapter ligation products
substantially do
not bind to the binding matrix. In this preferred embodiment, the cut-off size
lies above the
size of adapter monomers and above the size of expected adapter-adapter
ligation product.
Preferably, the cut-off size is at least 10 nt, at least 15 nt, at least 20
nt, at least 25 nt or at
least 30 nt above the size of expected adapter-adapter ligation products.
CA 2961177 2017-03-17
- 13 -
[0040] The removal of undesirable small nucleic acids achieved with the
disclosed
size selective isolation method does not need to be a complete removal. For
various
applications, including applications associated with library preparation for
next generation
sequencing, it is sufficient to deplete small nucleic acid molecules to an
extent such that their
interference with downstream applications, such as sequencing, is reduced. As
will be
described in greater detail below, it is not necessary that 100% of the small
nucleic acids are
removed during utilization of the disclosed size selective isolation method in
the preparation
of sequencing libraries.
Nucleic acid containing samples
100411 The nucleic acid containing sample may comprise different types of
nucleic
acids and nucleic acids of different sizes (i.e. lengths). The nucleic acid
containing sample
may comprise single stranded RNA, double stranded RNA, single stranded DNA,
double
stranded DNA, and/or a double stranded RNA/DNA hybrid. Preferably, the nucleic
acids are
single stranded RNA or double stranded RNA/DNA hybrids.
[0042] The nucleic acid containing sample can be of various origins,
including
biological samples and artificial samples. The nucleic acid containing sample
may comprise
extracted nucleic acids (e.g. from nucleic acids derived from cells, tissues,
bodily fluids) or
synthetic nucleic acids. The extracted or synthetic nucleic acids can be
further processed by
way of various mechanical (e.g. shearing) and enzymatic treatments (e.g.
treatment with a
proteinase, lipases, DNase, RNase as appropriate).
[0043] The nucleic acid containing sample may comprise nucleic acids
obtained after
an enzymatic reaction, including but not limited to amplification reactions,
ligation reactions,
and in particular, adapter ligation reactions. The nucleic acid containing
sample may
comprise amplification products (e.g. PCR products) and/or adaptor ligation
products (e.g.
nucleic acids flanked by 5' and/or 3' adapters; adapter monomers; adapter-
adapter ligation
products).
CA 2961177 2017-03-17
- 14 -
[00441 The nucleic acid containing sample can be obtained during the
preparation of
a sequencing library, in particular, during the preparation of a next
generation sequencing
(NGS) library. According to one preferred embodiment, the nucleic acid
containing sample
is an adapter ligation sample obtained as a result of an adapter ligation step
during library
preparation for next generation sequencing. The sample may comprise (i) single
stranded
RNA molecules that are ligated to 5' and/or 3' adapters, (ii) adapter monomers
and (iii)
adapter-adapter ligation products including adapter dimers.
Size selective binding
[0045] In one embodiment, the nucleic acid containing sample can be
combined with
a binding buffer, an alcohol and SiC slurry to provide a binding mixture. The
SiC slurry can
be prepared with a typical industrial preparation of SiC, which is composed of
about 97.8%
silicon carbide and small amounts of silicon dioxide, silicon, iron, aluminum
and carbon.
SiC is available in a variety of grit sizes or grades, with each grade having
a different average
particle size. The SiC slurry can be prepared using any grade of SiC and an
appropriate
liquid carrier, such as PBS buffers (e.g. IX PBS, pH 7) and Tris buffers (e.g.
10 mM Tris, pH
7). The SiC can have a grit size between 500-2500 (diameter ca. 1-10 urn),
preferably a grit
size between 2000-2500 and even more preferably, a grit size of 2500. The SiC
slurry can be
prepared in various ratios of SiC to liquid carrier, with a preferred ratio
being between 30%
and 70% (w/v), and even more preferred ratio being 50% (w/v).
100461 In order to isolate nucleic acids having a size above the desired
cut-off size
from the nucleic acid containing sample, the alcohol concentration of the
binding mixture
needs to be adjusted to the appropriate concentration. The alcohol
concentration used will
depend on the desired cut-off size for the target nucleic acid and the alcohol
used.
[0047] The alcohol concentration in the binding mixture can be adjusted
using any
alcohol known in the art. Examples of suitable alcohols include are but not
limited to
ethanol, isopropanol and methanol. To achieve size selective binding of the
target size
nucleic acids to the SiC, the alcohol concentration of the binding mixture can
be adjusted to a
concentration of between 1-50% (v/v), more preferably between 10-40% (v/v).
and even
CA 2961177 2017-03-17
- 15 -
more preferably between 20-30% (v/v) depending on the desired cut-off size.
The
appropriate alcohol concentration can be easily determined by titrating the
alcohol
concentration. It will be appreciated that one only needs to determine the
minimum alcohol
concentration necessary for preferential SiC binding of the target size
nucleic acids (e.g. the
amount of bound target size nucleic acids is greater than the amount of bound
non-target size
nucleic acids), as downstream applications, such as sequencing, do not require
the complete
removal of non-target size nucleic acids.
10048] As shown in Figure 1, when ethanol is used to adjust the alcohol
concentration of the binding mixture, the use of a minimum ethanol
concentration of about
6% (v/v) can provide a cut-off size of about 69 nt. Use of a minimum ethanol
concentration
of about 13 % (v/v) can provide a cut-off size of about 43 nt. Use of a
minimum ethanol
concentration of about 38 % (v/v) can provide a cut-off size of about 21 nt.
100491 In general, ethanol concentrations of at least 10% (v/v) can be used
to
selectively bind nucleic acids that are 43 nt and larger; ethanol
concentrations of at least 25%
(v/v) can be used to selectively bind nucleic acids that are 22 nt and larger;
and ethanol
concentrations of at least 50% (v/v) can be used to selectively bind nucleic
acids that are 10
nt and larger.
100501 As shown in Figure 2, isopropanol can be preferably used for
selectively
removing very small nucleic acids. Use of a minimum isopropanol concentration
of about
33% (v/v) can provide a cut-off size of about 20 nt. Use of a minimum
isopropanol
concentration of about 60% (v/v) can provide a cut-off size of about 10 nt.
Use of a
minimum isopropanol concentration of about 75% (v/v) can provide a cut-off
size of about 5
nt.
100511 In general, isopropanol concentrations of at least 10% (v/v) can be
used to
selectively bind nucleic acids that are 20 nt and larger; isopropanol
concentrations of at least
25% (v/v) can be used to selectively bind nucleic acids that are 10 nt and
larger; and
isopropanol concentrations of at least 50% (v/v) can be used to selectively
bind nucleic acids
that are 5 nt and larger.
CA 2961177 2017-03-17
- 16 -
10052] The appropriate amount of alcohol (e.g. the amount which will yield
the
desired alcohol concentration in the binding mixture) can be added to the
binding buffer and
the sample prior to combining the binding buffer and the sample with the SiC
slurry to
provide the binding mixture. Alternatively, the alcohol concentration can be
adjusted to the
appropriate concentration after the binding buffer, sample and the SiC slurry
have been
combined together to provide the binding mixture.
[0053] The size selective binding step can be performed under low salt
conditions
and slightly acidic to neutral pH conditions of about pH 4-7. The nucleic
acids contained in
the sample having a size above the desired cut-off size will come into contact
to with the SiC
and selectively bind to the SiC particles. The SiC particles with the bound
nucleic acids can
then be separated from the liquid phase (which contains the smaller unbound
nucleic acids)
through pelleting by centrifugation, or by passing the SiC particles through a
solid support
column or through gravity settling. Once the bound SiC particles have been
separated, the
size selected nucleic acids can optionally be washed with an appropriate low
salt wash
solution (for example, 1-100 mM Tris.HC1, MOPS or HEPES with 0-100 mM NaCl or
KCl)
to remove materials not selectively bound to the solid support. The bound size
selected
nucleic acids can then be eluted using an appropriate low salt elution
solution (for example,
1-10 mM Tris.HC1 or water) under slightly basic to neutral pH conditions of
about p11 7-9
and collected for downstream applications.
100541 In another embodiment of the method, the SiC can be used in a column
format. The term "column" as used herein describes a container having at least
two
openings. Thereby, a solution and/or sample can pass through said column. A
SiC slurry as
described above (see paragraph [0024]), can be packed into a column of any
size, from small
spin columns all the way to large chromatography columns operating through the
use of
gravity or pumps. The choice of column size will depend on the volume of the
nucleic acid
containing sample to be processed.
100551 For embodiments employing a SiC column, the alcohol concentration
can be
adjusted to the appropriate concentration (see e.g. paragraphs [0046] to
[0052]) using any
alcohol as described above (see e.g. paragraph [0047]). For example, the
appropriate amount
CA 2961177 2017-03-17
- 17 -
of alcohol can be added to the binding buffer and the sample. The resulting
combination of
the binding buffer, the alcohol and the sample can then be introduced into the
SiC column to
provide the binding mixture.
[0056] For example, a nucleic acid containing sample can be mixed with a
suitable
amount of a low salt binding buffer having a slightly acidic to neutral pH of
about pH 4-7,
the appropriate amount of alcohol can be added and the resulting mixture can
be loaded into
a spin column using a pipette. The spin column is centrifuged causing the
sample to travel
through the spin column. The nucleic acids contained in the sample having a
size above the
desired cut off size will come into contact with the SiC and selectively bind
to the SiC, while
any nucleic acids below the desired cut-off size will be removed in the
flowthrough. The
size selected bound nucleic acids can be optionally washed with an appropriate
low salt wash
solution (for example, 1-100 mM Tris.HC1, MOPS or HEPES with 0-100 mM NaCl or
KCl)
to remove materials not selectively bound to the solid support. The size
selected bound
nucleic acids can then be eluted from the SiC by passing an appropriate low
salt elution
solution (for example, 1-10 mM Tris.HC1 or water) tinder slightly basic to
neutral pH
conditions of about pH 7-9, and the eluted nucleic acids can be collected for
downstream
applications.
Preparation of sequencing libraries
[0057] The disclosed size selective isolation method is particularly suited
for
performing size selection steps (e.g. removal of non-target nucleic acids and
contaminants)
during the preparation of a sequencing library for next generation sequencing.
The disclosed
isolation method can be integrated into existing work-flows for preparing next
generation
sequencing libraries, in particular, small RNA libraries, wherein adapter
ligation steps are
usually performed in the early stages of library preparation.
[0058] Through the application of the disclosed size selective isolation
method during
library preparation, the library preparation process can be greatly improved
by eliminating
the lengthy procedure of having to gel purify the library prior to sequencing
in order to
remove adapter monomers and adapter-adapter ligation products. Prior art gel
purification
CA 2961177 2017-03-17
- 18 -
steps typically take 1 day to perform, whereas use of the disclosed size
selective isolation
method for cleaning up adapter monomers and adapter-adapter ligation products
during
library preparation may take less than 1 hour.
10059] To prepare a small RNA library for next generation sequencing,
adapters are
ligated to the 5' and/or 3' ends of single stranded RNA fragments. The
specific design of the
adapters will depend on the next generation sequencing platform to be used,
and for the
purposes of this discussion, any adaptors useful for preparing sequencing
libraries for next
generation sequencing can be used. The adapter sequences provide a known
sequence
composition allowing for annealing of primers for cDNA generation, as well as
for
amplification and addition of an index during subsequent steps of library
preparation.
[0060] For efficient adapter ligation during library preparation, the
adapters are
usually used in excess during the adapter ligation step. Typically, during
small RNA library
preparation for next generation sequencing, the 3' adapter is ligated first.
Therefore, excess
amounts of the adapter are added to the purified small RNA in order to ensure
that all small
RNA molecules present will bind to an adapter. Thus, after 3' adapter ligation
an RNA
containing sample is provided which comprises RNA molecules that are ligated
at the 3' end
to an adapter, as well as unligated adapter monomers and adapter-adapter
ligation products
including adapter dimers. At the next step in the library preparation, the 5'
adapter is then
ligated to the small RNA molecules, which already have a ligated 3' adapter.
Again, excess
amounts of the 5' adapter are added to ensure that all the RNA molecules will
be flanked by
both a 3' and a 5' adapter.
100611 A problem with conventional library preparation methods lies in the
fact that
the 5' adapter can ligate to the 3' adapter monomers, thereby resulting in
ligation products
that can become incorporated into that library and that will diminish the
sequencing power of
the subsequent sequencing reaction. Therefore, these unligated adapter
monomers and
adapter-adapter products should be removed before starting the sequencing run.
[0062] Common prior art clean up methods for removing of library
contaminants
(e.g. PEG/bead based methods, gel based purifications) can be time consuming
and labour
CA 2961177 2017-03-17
- 19 -
intensive. Furthermore, the majority of the prior art methods are performed as
the last step
before starting the sequencing of the small RNA library, as such, the
formation of 5' adapter-
3'adapter ligated products are not avoided. These problems are avoided by
incorporating the
disclosed size selective isolation method into the library preparation method.
[0063] In contrast to the prior art library preparation methods, the
disclosed method
for preparing a small RNA library suitable for next generation sequencing
quickly removes
library contaminants (e.g. 1 day for gel based purification vs. about 1 hour
for the disclosed
size selective isolation method). Further in contrast to prior art methods,
the disclosed
method for preparing a small RNA library suitable for next generation
sequencing comprises
a size based cleanup step that is performed after the first 3' adapter
ligation in a small RNA
sequencing library preparation process rather than as an end step in the
library preparation.
By employing a size selective isolation step early on in the preparation of
the library, it is
possible to avoid and/or reduce the formation of 5' adapter-3'adapter ligated
products. The
size based cleanup step allows for the removal of excess unligated 3'adapters
and reduces the
likelihood of the formation of 5' adapter-3'adapter ligated products without
any RNA insert.
[0064] It will be apparent to those skilled in the art that the size
selective isolation
cleanup step can also be performed after the 5' adapter ligation step as well.
Furthermore, it
will be apparent to those skilled in the art that the method can also be used
for size selective
RNA isolation when the 5' adapter is ligated to the RNA prior to the 3'
adapter, and again
the size selective RNA isolation can be performed after the 5' adaptor
ligation and/or after
the subsequent 3' adapter ligation.
[0065] In a preferred embodiment, disclosed is a method for preparing a
small RNA
library suitable for next generation sequencing, the method comprising the
steps of:
a) isolating small RNA molecules from a sample;
b) performing a 3' adapter ligation step to provide a 3' adaptor ligation
reaction mixture comprising single stranded small RNA molecules that are
flanked 3' by adapters;
CA 2961177 2017-03-17
- 20 -
c) isolating single stranded small RNA molecules having a fragment size
above a predetermined cut-off size in order to remove 3' adapter monomers
and adapter-adapter ligation products, wherein said size selection step
comprises:
i) combining the 3'adaptor ligation reaction mixture with a binding
buffer, alcohol and silicon carbide to provide a binding mixture,
wherein the single stranded small RNA molecules having a fragment
size above the cut-off size bind to the silicon carbide, and wherein the
cut-off size is determined by the alcohol concentration of the binding
mixture;
ii) separating the bound single stranded small RNA molecules from the
remaining ligation reaction mixture;
iii) optionally, washing the bound single stranded small RNA
molecules; and
iv) eluting the bound single stranded small RNA molecules from the
silicon carbide;
d) performing a 5' adapter ligation step to provide a 5'adaptor ligation
reaction mixture comprising single stranded small RNA molecules that are
flanked 3' and 5' by adapters;
e) optionally, performing a step of isolating single stranded small RNA
molecules having a fragment size above a predetermined cut-off size in order
to remove 5' adapter monomers and adapter-adapter ligation products,
wherein said size selection step comprises:
i) combining the 5'adaptor ligation reaction mixture with a binding
buffer, alcohol and silicon carbide to provide a binding mixture,
wherein the single stranded small RNA molecules having a fragment
CA 2961177 2017-03-17
- 21 -
size above the cut-off size bind to the silicon carbide, and wherein the
cut-off size is determined by the alcohol concentration of the binding
mixture;
ii) separating the bound single stranded small RNA molecules from the
remaining ligation reaction mixture;
iii) optionally, washing the bound single stranded small RNA
molecules; and
iv) eluting the bound single stranded small RNA molecules from the
silicon carbide;
f) reverse transcribing single stranded RNA molecules flanked with 5' and 3'
adaptors to provide single stranded cDNA molecules; and
g) amplifying the single stranded cDNA molecules by limited cycle PCR to
incorporate an index sequence.
100661 The size selection steps can be performed using either a SiC slurry
or a SiC
column as described in greater detail above (see e.g. paragraphs [0045] to
[0056]).
100671 In another preferred embodiment, disclosed is a method for preparing
a small
RNA library suitable for next generation sequencing, wherein said method
comprises:
a) isolating small RNA molecules from a sample;
b) performing a 5' adapter ligation step to provide a 5' adaptor ligation
reaction mixture comprising single stranded small RNA molecules that are
flanked 5' by adapters;
c) isolating single stranded small RNA molecules having a fragment size
above a predetermined cut-off size in order to remove 5' adapter monomers
CA 2961177 2017-03-17
- 22 -
and adapter-adapter ligation products, wherein said size selection step
comprises:
i) combining the 5'adaptor ligation reaction mixture with a binding
buffer, alcohol and silicon carbide to provide a binding mixture,
wherein the single stranded small RNA molecules having a fragment
size above the cut-off size bind to the silicon carbide, and wherein the
cut-off size is determined by the alcohol concentration of the binding
mixture;
ii) separating the bound single stranded small RNA molecules from the
remaining ligation reaction mixture;
iii) optionally, washing the bound single stranded small RNA
molecules; and
iv) eluting the bound single stranded small RNA molecules from the
silicon carbide;
d) performing a 3' adapter ligation step to provide a 3'adaptor ligation
reaction mixture comprising single stranded small RNA molecules that are
flanked 3' and 5' by adapters;
e) optionally, performing a step of isolating single stranded small RNA
molecules having a fragment size above a predetermined cut-off size in order
to remove 3' adapter monomers and adapter-adapter ligation products,
wherein said size selection step comprises:
i) combining the 3'adaptor ligation reaction mixture with a binding
buffer, alcohol and silicon carbide to provide a binding mixture,
wherein the single stranded small RNA molecules having a fragment
size above the cut-off size bind to the silicon carbide, and wherein the
CA 2961177 2017-03-17
- 23 -
cut-off size is determined by the alcohol concentration of the binding
mixture;
ii) separating the bound single stranded small RNA molecules from the
remaining ligation reaction mixture;
iii) optionally, washing the bound single stranded small RNA
molecules; and
iv) eluting the bound single stranded small RNA molecules from the
silicon carbide;
f) reverse transcribing single stranded RNA molecules flanked with 5' and 3'
adaptors to provide single stranded cllNA molecules; and
g) amplifying the single stranded cDNA molecules by limited cycle PCR to
incorporate an index sequence.
100681 The size selection steps can be performed using either a SiC slurry
or a SiC
column as described in greater detail above (see e.g. paragraphs [0045] to
[0056]).
Kits for the selective binding of nucleic acids
100691 In a further embodiment, provided are kits for the selective binding
of nucleic
acid molecules having a size above a desired cut-off size, comprising: a) a
binding buffer to
be combined with an alcohol, preferably ethanol, to provide an alcohol
concentration of
about 1 to about 50% (v/v); b) SiC, c) a wash solution; and d) an elution
solution, wherein
the cut-off size is at least 5 nucleotides.
100701 The kit can be provided with printed instructions that describe the
appropriate
amount of alcohol to be added to the binding buffer, sample and SiC to achieve
the
appropriate alcohol concentration in the binding mixture to affect selective
binding of nucleic
acids above the desired cut-off size. The SiC can be provided in a slurry
format or in a
column format. Details regarding the SiC slurry or column, as well as details
regarding the
CA 2961177 2017-03-17
- 24 -
addition of the alcohol to the binding mixture to achieve a desired alcohol
concentration for
selective binding were described in detail above in conjunction with the
isolation method.
100711 In a preferred embodiment, the disclosed kits can be used for
isolating
adapter-ligated RNA molecules from an adapter ligation sample and for removing
adapter
monomers and adapter-adapter ligation products based on the larger size of the
adapter-
ligated RNA molecules for use in preparing a small RNA sequencing library.
100721 Although the invention has been described with reference to
illustrative
embodiments, it is to be understood that the invention is not limited to these
precise
embodiments, and that various changes and modification are to be intended to
be
encompassed in the appended claims.
EXAMPLES
100731 These examples arc described for the purposes of illustration and
are not
intended to limit the scope of the invention.
Example 1 - Separation of adapter-microRNA ligated molecules from adapter
monomers
100741 To mimic the steps during a small RNA library preparation, synthetic
oligos
containing Adapter-microRNA ligated molecules were designed. The example used
was
based on a C. elegans mircoRNA (cel-miR-39) (miRBase Accession: NI10000010)
with the
sequence of:
5' UCACCGGGUGUAAAUCAGCUUG 3' (SEQ ID NO: 1)
The synthetic Adapter-microRNA ligated molecule was generated by adding the
Illumina
TruSeq 3' Adapter sequence (RA3)
5' TGGAATTCTCGGGTGCCAAGG 3'(SEQ ID NO: 2)
to the 3' end of ccl-miR-39, resulting in a final product of
CA 2961177 2017-03-17
- 25 -
5' UCACCGGGUGUAAAUCAGCUUGTGGAATTCTCGGGTGCCAAGG 3'
(SEQ ID NO: 3),
mimicking the product after the 3' adapter ligation step in the small RNA
sequencing library
preparation. Different fluorophores were added to the aforementioned molecules
(PAM for
cel-miR-39-RA3 ligated product and Hex for RA3 monomer) for subsequent
tracking and
quantification.
[0075] In the example depicted in Figure 3, each synthetic oligo (FAM-cel-
mi R-39-
RA3 and Ilex-RA3) was adjusted to a concentration of 1 luM and mixed at a 1:1
ratio. 20 AL
of the mixture was then mixed with 180 uL of a Binding Solution (RNA Clean-Up
and
Concentration Micro-Elute Kit, Cattf 61000, Norgen Thorold, Canada). The
resulting
mixture was adjusted to a final volume of 400 u.1_, by adding various amounts
of water and/or
ethanol to give a final ethanol concentration of 0%, 6.25%, 12.5%, 25%, 37.5%
and 50%
(v/v). The oligo-ethanol mix was then applied to a spin column containing 5 mg
SiC by
centrifugation at 6,000 RPM (¨ 3,500 x g) for 1 minute. The column was washed
twice with
600 uL of a Wash Solution (RNA Clean-Up and Concentration Micro-Elute Kit,
Cat# 61000,
Norgen Thorold, Canada) by centrifugation at 14,000 RPM (¨ 14,000 x g) for 1
minute. The
column was further dried by centrifugation at 14,000 RPM (-14,000 x g) for 2
minutes. The
RNA was then eluted with 10 uL of an Elution Solution (RNA Clean-Up and
Concentration
Micro-Elute Kit, Cat# 61000, Norgen Thorold, Canada) by first centrifugation
at 2,000 RPM
(¨ 200 x g) for 1 minute, followed by centrifugation at 14,000 RPM (¨ 14,000 x
g) for 2
minutes.
100761 The presence of each synthetic oligo was then evaluated on a QIAGEN
Rotor
Gene Q (Q1AGEN, Toronto, Canada) at the excitation and absorption wavelengths
specific
for FAM and Hex, respectively. As shown in Figure 3, both cel-miR-39-RA3 (43
bp) and
the RA3 (21 bp) were effectively captured by the column at ¨ 50 % (v/v)
ethanol. However,
as ethanol % (v/v) decreased, the amount of RA3 (21 bp) captured by the column
drastically
decreased, while a high amount of cel-miR-39-RA3 (43 bp) was still captured
(data points of
25% (v/v) and 37.5% (v/v) ethanol), giving maximum separation. As ethanol %
(v/v) further
CA 2961177 2017-03-17
- 26 -
decreased, the cel-miR-39-RA3 captured by the column started to decrease but
still
maintained a higher level than RA3 at as little as ¨ 10% (v/v) ethanol.
Example 2 - Separation of mieroRNA ligated at both S' and 3' ends frotn
adapter
monomers
100771 To mimic the final modified RNA product during the library
preparation,
synthetic oligos containing microRNA with both 5' and 3' ends ligated with
adapters were
designed. The example used was based on a C. elegons mircoRNA (cel-miR-39)
(miRBase
Accession: MI0000010) with the sequence
5' UCACCGGGUGUAAAUCAGCUUG 3' (SEQ ID NO: 1).
The synthetic 5'/3'-ligated molecule was generated by adding the Illumina
TruSeq 3'
Adapter sequence (RA3)
5' TGGAATTCTCGGGTGCCAAGG 3'(SEQ ID NO: 2)
to the 3' end of cel-miR-39 and Illumina TruSeq 5' Adapter sequence (RA5)
5' GUUCAGAGUUCUACAGUCCGACGAUC 3'(SEQ ID NO: 4)
to the 5' end of the same cel-miR-39 resulting in a final product of
5'GUUCAGAGUUCUACAGUCCGACGAUCUCACCGGGUGUAAAUCAGCUU
GTGGAATTCTCGGGTGCCAAGG 3' (SEQ ID NO: 5).
Different fluorophores were added to the aforementioned molecules (Cy5.5 for
RA5-cel-
miR-39-RA3 ligated product and Hex for RA3 monomer) for subsequent tracking
and
quantification.
100781 In the example depicted in Figure 4, each synthetic oligo (Cy5.5 for
RA5-cel-
miR-39-RA3 and Hex-RA3) was adjusted to a concentration of 1 11M and mixed at
a 1:1
ratio. 20 ji.L of the mixture was then mixed with 180 jut of a Binding
Solution (RNA Clean-
Up and Concentration Micro-Elute Kit, Cat# 61000, Norgen Thorold, Canada). The
resulting
CA 2961177 2017-03-17
- 27 -
mixture was adjusted to a final volume of 400 jiL by adding various amounts of
water and/or
ethanol to give a final ethanol concentration of 0%, 6.25%, 12.5%, 25%, 37.5%
and 50%
(v/v). The oligo-ethanol mix was then applied to a spin column containing 5 mg
of SiC by
centrifugation at 6,000 RPM (¨ 3,500 x g) for 1 minute. The column was washed
twice with
600 IA of a Wash Solution (RNA Clean-Up and Concentration Micro-Elute Kit,
Cat# 61000,
Norgen Thorold, Canada) by centrifugation at 14,000 RPM (¨ 14,000 x g) for 1
minute. The
column was further dried by centrifugation at 14,000 RPM (¨ 14,000 x g) for 2
minutes. The
RNA was then eluted with 10 uL of an Elution Solution by first centrifugation
at 2,000 RPM
(¨ 200 x g) for I minute, followed by centrifugation at 14,000 RPM (¨ 14,000 x
g) for 2
minutes.
100791 The presence of each synthetic oligo was then evaluated on a Qiagen
Rotor
Gene Q at the excitation and absorption wavelengths specific for Cy5.5 and
Hex,
respectively. As shown in Figure 4, both RA5-cel-miR-39-RA3 (69 bp) and the
RA3 (21
bp) were effectively captured by the column at ¨ 50 % (v/v) ethanol. However,
as ethanol %
(v/v) decreased, the amount of RA3 (21 bp) captured by the column drastically
decreased,
while a high amount of cel-miR-39-RA3 (43 bp) was still captured (data points
of 25% and
37.5% ethanol), giving maximum separation. As ethanol % (v/v) further
decreased, the
RA5cel-miR-39-RA3 captured by the column started to decrease but still
maintained a higher
level than RA3 at as little as ¨ 10% (v/v) ethanol.
Example 3 - Effect of 3' adapter cleanup on final small RNA-sequence library
product
100801 To test the effect of adapter cleanup on the subsequent small RNA-
sequence
library product, libraries were prepared using a standard small RNA library
procedure with or
without the use of the SiC-based size separation technology. About 1 jig of I
leLa total RNA
or ¨ 10 ng of human plasma RNA was used as an input. The RNA was first ligated
at the 3'
end in a 20 jt1.., reaction using T4 RNA Ligase 2 (deleted mutant, Cat# T4RL2T-
100, MC
Lab, San Francisco, United States) and a pre-adenylated RA3 adapter. The
ligation reaction
was then subjected to size separation as described in Example 1, using 25%
(v/v) ethanol as
final binding concentration. A control without separation/cleanup was
performed in parallel.
The purified product was then subjected to 5' adapter ligation using T4 RNase
Ligase 1
CA 2961177 2017-03-17
- 28 -
(Cat# T4RL1-100, MC Lab, San Francisco, United States) and RA5 adapter. The
final
ligated product (RA5-microRNA-RA3) was then subjected to cDNA synthesis using
a M-
MIN reverse transcriptase (Norgen TruScript Reverse Transcriptase, Cat#54440,
Norgen,
Thorold, Canada) and reverse primer complementary to the RA3 sequence. This
was
followed by a limited (15) cycle PCR amplification to enrich the cDNA and also
to attach the
indexing (barcode) sequences. The indexed libraries were then subjected to PCR
clean-up
using Norgen's PCR Purification Kit (Norgen, Thorold, Canada) and resolved on
an Agilent
High Sensitivity DNA Chip (Cat# 5067-4626, Agilent, Santa Clara, United
States).
[0081] As shown in Figure 5, without the 3' adapter cleanup removal, the
predominant PCR product for HeLa RNA input was a 120-122 bp DNA, corresponding
to
product from 5' adapter - 3' adapter ligation without any microRNA insert.
Similarly, a 120-
122 bp product was observed in the plasma RNA input product. In contrast, with
the 3'
adapter removal using SiC columns, a predominant PCR product of 142-144 bp was
obtained, reflecting roughly a 20 bp difference, which is the size of the
microRNA insert.
This 142-144 bp PCR product was observed as the predominant product in both
inputs, with
little to no detection of the 120-122 bp product.
Example 4- Comparison of the effect of 3' adapter cleanup using an established
blocking
oligo method and the SiC-based column removal method on the diversity of small
RNA-
sequence library products
[00821 To test the effect of adapter cleanup on the miRNA diversity of the
subsequent small RNA-sequence library product, libraries were prepared using a
standard
small RNA library procedure with the use of the SiC-based size separation
technology for 3'
adapter cleanup and compared to the use of a blocking oligonucleotide for 3'
adapter cleanup
(as illustrated in US Patent No. 8,883,421).
[0083] Approximately 10 ng of human plasma RNA was used as the input for
each
reaction. The RNA was first ligated at the 3' end in a 20 1.11 reaction using
T4 RNA Ligase 2
(deleted mutant, Cat# T4RL2T-100, MC Lab, San Francisco, United States) and a
pre-
adenylated RA3 adapter. For 3' adapter cleanup, one ligation reaction was then
subjected to
CA 2961177 2017-03-17
- 29 -
size separation as described in Example 1, using 25% (v/v) ethanol as final
binding
concentration. For 3' adapter cleanup of the second reaction using blocking
oligos, a reverse
primer complementary to the RA3 sequence was added to the 3' adaptor ligation.
The
control blocking method reaction and the column-based removal method reaction
of the
current invention were then subjected to 5' adapter ligation using T4 RNase
Ligase 1 (Cat#
14RL1-100, MC Lab, San Francisco, United States) and RA5 adapter. The final
ligated
products (RA5-microRNA-RA3) were then subjected to cDNA synthesis using a M-
MLV
reverse transcriptase (Norgen TruScript Reverse Transcriptase, Cat#54440,
Norgen, Thorold,
Canada) and reverse primer complementary to the RA3 sequence. This was
followed by a
limited (15) cycle PCR amplification to enrich the cDNA and also to attach the
indexing
(barcode) sequences. The indexed libraries were then subjected to clean-up
using Norgen's
PCR Purification Kit (Cattt 14400, Norgen, Thorold, Canada) and resolved on an
Agilent
High Sensitivity DNA Chip (Cat# 5067-4626, Agilent, Santa Clara, United
States).
100841 As shown in Figure 6, Panel A, both the control/oligo blocking
method and
the SiC column removal method of the current invention yielded a predominant
PCR product
of 142-151 bp, typical of the small RNA diversity of plasma RNA. The libraries
were
sequenced on an Illumina MiScq sequencer at 50 cycles single read. The
resulting reads
were mapped to microRNAs using execRpt small RNA-seq Pipeline
(genboree.org/theCommons/projectslexrna-tools-may2014/wiki/Small_RNA-
seq_Pipeline).
10085] The Venn diagram (Figure 6, Panel B) generated using miRNA with
reads
equal or more than 5 RPM (reads per millions) showed that both methods shared
the same
diversity of 200 miRNAs. Interestingly, the library generated using the SiC-
based column
removal method for the 3' adapter cleanup yielded more unique miRNAs (62) than
that of
the control method based on using blocking oligos (27). This showed that while
the SiC-
based column removal method does not alter the diversity of miRNA
incorporated, it may
further enhance the number of different miRNA incorporated into a small RNA
sequencing
library.