Note: Descriptions are shown in the official language in which they were submitted.
CA 02964161 2017-04-07
WO 2016/057903
PCT/US2015/054914
METHODS FOR DISCOVERING THERAPEUTICS THAT ALTER THE
STABILITY OF TARGET PROTEINS
RELATED APPLICATIONS
This application claims the benefit under 35 U.S.C. 119(e) of U.S.
provisional
application USSN 62/062,257, filed October 10, 2014, the entire contents of
which are
incorporated herein by reference.
FEDERALLY SPONSORED RESEARCH
This invention was made with government support under 2R01CA068490-19,
and 2R01CA076120-13 awarded by National Institute of Health. The government
has
certain rights in the invention.
BACKGROUND OF THE INVENTION
Reporter assays have been used routinely in the pharmaceutical and
biotechnology industries to identify lead compounds that affect protein
function. In the
last decade, the chemist's ability to synthesize large numbers of chemical
compounds in
a short amount of time through techniques such as combinatorial chemistry has
greatly
increased, and often, thousands to millions of compounds need to be screened
to identify
those having a desired effect on a protein of interest.
Typically, reporter assays measure the activities of one reporter protein in a
sample, but may combine multiple reporters. One strategy for co-expression of
multiple
reporters involves the design of bicistronic constructs, in which two genes
separated by
an internal ribosome entry site (IRES) sequence are expressed as a single
transcriptional
cassette (or bicistronic transcript) under the control of a common upstream
promoter
(Yen et al., Science. 2008 Nov 7;322(5903):918-23). The intervening IRES
sequence
functions as a ribosome-binding site for efficient cap-independent internal
initiation of
translation. Such a design enables transcription of both genes with IRES-
directed cap-
independent translation. This system allows for co-expression of both a
control reporter,
not expected to change upon experimental treatment, along with a test reporter
that is
normalized to the control in each test sample. However, many perturbations in
the cell
can differentially affect cap-dependent translation compared to cap-
independent
translation. Moreover, some IRESes have been shown to display variable
expression of
1
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
the downstream gene (Wong et al. Gene Ther. 2002 Mar;9(5):337-44). This leads
to
high false positives and unreliable reporter assays. Thus, there is a need for
an efficient
high-throughput approach for analysis of protein stability where nonspecific
alterations
in reporter activity are used to control for the inherent variability in cell
based protein
stability assays. This allows for reducing the error in the data required to
effectively and
efficiently run an HTS assay.
SUMMARY OF THE INVENTION
The present disclosure relates, in some aspects, to the development of a
plasmid
that can be used to efficiently monitor the stabilities of thousands of
proteins after
specific perturbations.
According to some aspects, the present disclosure provides a method to
identify a
test compound that stabilizes or destabilizes a protein of interest, the
method comprising:
(i) contacting a transformed host cell comprising a DNA plasmid with
a test
compound, wherein the plasmid comprises in operable linkage
(a) a promoter,
(b) a first internal ribosomal entry site (IRES);
(c) a nucleotide sequence encoding a first reporter protein;
(d) a second IRES; and
(e) a nucleotide sequence encoding a second reporter protein,
wherein an open reading frame (ORF) is fused to the nucleotide sequence
encoding a first reporter protein or to the nucleotide sequence encoding a
second
reporter protein and wherein said open reading frame codes for a protein of
interest;
(ii) determining ratios of fused reporter protein signal to unfused
reporter protein
signal in presence and absence of the test compound; and
(iii) identifying said test compound as a stabilizer when the ratio of
fused reporter
protein signal to unfused reporter protein signal in the presence of the test
compound is increased as compared to the ratio of fused reporter protein
signal to unfused reporter protein signal in the absence of the test compound,
and identifying said test compound as a destabilizer when the ratio of fused
reporter protein signal to unfused reporter protein signal in the presence of
the
2
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
test compound is decreased as compared to the ratio of fused reporter protein
signal to unfused reporter protein signal in the absence of the test compound.
In some embodiments, the first and second reporter proteins have
distinguishable
detectable reporter signals. In some embodiments, the first and second
reporter proteins
are enzyme proteins having distinguishable signals generated from their
products. In
some embodiments, the first and second reporter proteins are bioluminescent
proteins
having distinguishable bioluminescence signals. In some embodiments, the first
and
second reporter proteins are fluorescent proteins having distinguishable
fluorescence
signals. In some embodiments, the first and second reporter proteins are
selected from
the group consisting of renilla luciferase (Rluc) and firefly luciferase
(FLuc). In some
embodiments, the first and second reporter proteins are selected from the
group
consisting of green fluorescence protein and red fluorescence protein. In some
embodiments, the promoter is a eukaryotic promoter or a synthetic promoter. In
some
embodiments, the promoter comprises cytomegalovirus (CMV) promoter. In some
embodiments, the open reading frame is derived from an ORFeome of an organism.
In
some embodiments, the open reading frame encodes an oncoprotein. In some
embodiments, the oncoprotein is selected from the group consisting of MYC,
Ilcaros
family zinc finger protein 1 (IKZF1), Ikaros family zinc finger protein 3
(IKZF3),
Interferon regulatory factor 4 (IRF4), mutant p53, N-Ras, c-Fos, and c-Jun. In
some
embodiments, contacting a transformed host cell comprising the plasmid with a
test
compound comprises growing the transformed host cell in the presence of the
test
compound for an appropriate time.
Each of the embodiments and aspects of the invention can be practiced
independently or combined. Also, the phraseology and terminology used herein
is for
the purpose of description and should not be regarded as limiting. The use of
"including", "comprising", or "having", "containing", "involving", and
variations thereof
herein, is meant to encompass the items listed thereafter and equivalents
thereof as well
as additional items.
These and other aspects of the inventions, as well as various advantages and
utilities will be apparent with reference to the Detailed Description. Each
aspect of the
invention can encompass various embodiments as will be understood.
3
CA 02964161 2017-04-07
WO 2016/057903
PCT/US2015/054914
All documents identified in this application are incorporated in their
entirety
herein by reference.
BRIEF DESCRIPTION OF THE DRAWINGS
The accompanying drawings are not intended to be drawn to scale. In the
drawings, each identical or nearly identical component that is illustrated in
various
figures is represented by a like numeral. For purposes of clarity, not every
component
may be labeled in every drawing. In the drawings:
FIG. 1 confirms that pIRIGF constructs express in 293FT and HELA cells (FIG.
1A-C) and pUG-FIRP constructs express in U-2 OS cells (FIG. 1D). Several
different
versions of mammalian and lentiviral plasmid constructs were tested for their
ability to
generate cells (e.g. 293FT, HELA, or U-2 OS cells) expressing tagged target
proteins
(e.g., firefly or NanoLuc tag) and co-expressing a reporter luciferase (e.g.,
Renilla or
Firefly).
In FIG. 2, 293FT and HELA cells were transfected with IKZF1-firefly, IKZF3-
firefly and MYC-firefly fusion proteins and selected using puromycin and
geneticin
respectively. These pools were very unstable and lost signals in 10 to 30 days
and
generally had very small responses to IMiD's (FIG. 2A-C). Therefore,
individual clones
were isolated using a limited cloning strategy in 96-well plates. Surviving
cells were
isolated as colonies, further expanded and tested for luciferase signals and
response to
IMiDs. Clone 2B4 was identified as a strong responder to lenalidomide. HELA
cells
expressed very low levels of luciferase making isolation of HELA clones very
difficult.
Detection by western blots of firefly, IKZF1 and myc confirmed expression of
the fusion
protein and relative expression correlated with firefly luciferase signals
(FIG. 2D).
In FIG. 3 cell line clones (IKZF1-2B4, IKZF1-2B11, myc-1C3 and myc-5F2)
expressing the indicated firefly fusion protein were evaluated in the dual-glo
assay for
reproducibility. Potency of IMiD's and relative reduction in firefly
luciferase signals
confirmed the expected responses and generated data with Z' values sufficient
for
screening (FIG. 3A-D).
FIG. 4 shows pilot screen results for IKZF1 2B4 cells - Active compounds
(Prestwick collection; FIG. 4A) and NCI collection; FIG. 4B).
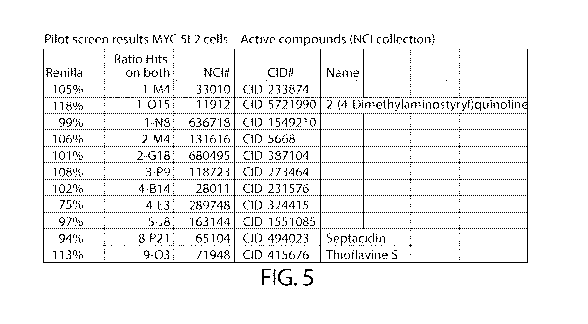
FIG. 5 shows pilot screen results for MYC 5F2 cells - Active compounds NCI
collection).
4
CA 02964161 2017-04-07
WO 2016/057903
PCT/US2015/054914
FIG. 6 confirms the hits tested on IKZF1 2B4 and MYC 5F2 cell lines (FIG. 6A-
C). Summary retest data from commercial compounds and from DTP compounds is
shown in FIG. 6D-E.
FIG. 7 shows confirmation data using Western blots. IKZF1 2B4 cell line
examples (FIG. 7A-B), MYC 5F2 example (FIG. 7C)
FIG. 8 shows further evaluations of HSP90 inhibitors. FIG. 8A demonstrates
testing of HSP 90 inhibitors CCT018159 and geldanamycin on cells transiently
transfected with the MYC-firefly fusion protein. FIG. 8B shows testing of HSP
90
inhibitors CCT018159 and geldanamycin on 293FT cells stably expressing MYC-
firefly
fusion protein. FIG. 8C shows testing of several HSP-90 inhibitors at various
doses on 5
different cell lines stably expressing the MYC-firefly fusions protein. FIG.
8D
compares the HSP90 inhibitor BIIB021 and pomalidomide on 293FT cells
transiently
expressing IKZF1-firefly fusion protein.
FIG. 9 shows an overview of the ICCB screening results. Specifically, it shows
cherry pick retests for IKZF1 ICCB screen.
FIG. 10 shows compares activity in IKZF1 vs. MYC cell lines. 133 cherry picks
in IKZF1 and MYC cell lines were tested.
FIG. 11 shows a better dose response at 16 hours for HSP90 inhibitors: BIIB021
(FIG. 11A) and PF-04929113 (FIG. 11B).
FIG. 12 shows cyclohexamide time course on 7 MYC cell lines including
cyclohexamide untagged luciferase (FIG. 12A) and cyclohexamide tagged
luciferase
(FIG. 12B).
FIG. 13 shows MG132 time course on 7 MYC cell lines including MG-132
tagged luciferase (FIG. 13A) and MG-132 untagged luciferase (FIG. 13B).
FIG. 14 shows MLN4924 time course on 7 MYC cell lines including MLN4924
tagged luciferase (FIG. 14A) and MLN4924 untagged luciferase (FIG. 14B).
FIG. 15 shows A549-MYC-firefly & H1299-MYC-firefly Western Blot
confirmation after MYC knockdown using 48 hour treatment with siRNA directed
to
MYC mRNA. The reduction in fusion protein, as observed by western blotting
with
MYC and firefly directed antibodies (FIG. 15A), is computed to the decrease in
luciferase signals (FIG. 15B). MYC antibody also detects the decrease in
endogenous
MYC.
5
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
FIG. 16 shows screening results from a commercial library of siRNA's directed
to the family of DUB enzymes including A549 (FIG. 16A), H1299 (FIG. 16B), and
HEK293T (FIG. 16C) cells expressing MYC-firefly and U2OS (FIG. 16D) cells
expressing MYC-nanoluc.
DETAILED DESCRIPTION OF THE INVENTION
The present application is based, in some aspects, on the development of a
plasmid that can be used to efficiently monitor the stabilities of thousands
of proteins
after specific perturbations. The plasmid allows for the co-expression of two
reporter
proteins, each of which is placed under the control of an IRES. In this way
both
reporters are transcribed together (i.e. are encoded by the same mRNA) and
both are
translated using an IRES. This minimizes the problem of spurious changes in
the ratio of
the two reporters caused by perturbations (e.g. compounds) that differentially
effect
IRES-dependent versus IRES-independent translation, and thus minimizes false
positives.
According to some aspects, the present disclosure provides a method to
identify a
test compound that stabilizes or destabilizes a protein of interest. The
method comprises
(i) contacting a transformed host cell comprising a DNA plasmid
with a test
compound, wherein the plasmid comprises in operable linkage
(a) a promoter,
(b) a first internal ribosomal entry site (IRES);
(c) a nucleotide sequence encoding a first reporter protein;
(d) a second IRES; and
(e) a nucleotide sequence encoding a second reporter protein,
wherein an open reading frame (ORF) is fused to the nucleotide sequence
encoding a first reporter protein or to the nucleotide sequence encoding a
second reporter
protein and wherein said open reading frame codes for a protein of interest;
(ii) determining ratios of fused reporter protein signal to unfused
reporter
protein signal in presence and absence of the test compound; and
(iii) identifying said test compound as a stabilizer when the ratio of
fused
reporter protein signal to unfused reporter protein signal in the presence of
the test
compound is increased as compared to the ratio of fused reporter protein
signal to
unfused reporter protein signal in the absence of the test compound, and
identifying said
6
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
test compound as a destabilizer when the ratio of fused reporter protein
signal to unfused
reporter protein signal in the presence of the test compound is decreased as
compared to
the ratio of fused reporter protein signal to unfused reporter protein signal
in the absence
of the test compound.
As used herein, "operable linkage" refers to a functional linkage between two
nucleic acid sequences, such as a transcription control element (e.g., a
promoter) and the
linked transcribed sequence. Thus, a promoter is in operable linkage with a
gene if it can
mediate transcription of the gene.
As used herein a "promoter" usually contains specific DNA sequences
(responsive elements) that provide binding sites for RNA polymerase and
transcriptional
factors for transcription to take place. In some embodiments, the promoter is
a
eukaryotic promoter or a synthetic promoter. Examples of promoters include,
but are not
limited to, the TATA box, the SV40 late promoter from simian virus 40,
cytomegalovirus (CMV) promoter, ubiquitin C promoter (UbC promoter) and the T7
promoter. These and other promoter sequences are well known in the art. In one
example of the invention, the promoter is a CMV promoter. In one example of
the
invention, the promoter is a UbC promoter.
As used herein, an "internal ribosomal entry site" or "IRES" is a cis acting
nucleic acid element that mediates the internal entry of ribosomes on an RNA
molecule
and thereby regulates translation in eukaryotic systems. In the methods and
compositions of the present invention, a first and a second IRES elements are
contained
in the plasmid. The first and second IRES elements permit the independent
translation
of a nucleotide sequence encoding a reporter protein and an open reading frame
fused to
a nucleotide sequence encoding another reporter protein from a single
messenger RNA.
In some embodiments, the first and second IRESs are the same (i.e., they have
identical
sequences). In some embodiments, the first and second IRESs are not the same
(i.e.,
they do not have identical sequences).
Many IRES elements have been identified in both viral and eukaryotic genomes.
In addition, synthetic IRES elements have also been developed. For example,
IRES
elements have been found in a variety of viruses including members of the
genus
Enterovirus (e.g. human poliovirus 1 (Ishii et al. (1998) J Virol. 72:2398-
405 and
Shiroki et al. (1997) J. Virol. 77:1-8), human Coxsackievirus B); Rhinovirus
(e.g.,
human rhinovirus); Hepatovirus (Hepatitis A virus); Cardiovirus
(Encephalomyocarditis
7
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
virus ECMV (nucleotides 2137-2752 of GenBank Accession No. AB041927 and Kim et
al. (1992) Mol Cell Biology 72:3636-43) and Etheirler's encephalomyelitis
virus);
Aphtovirus (Foot- and mouth disease virus (nucleotides 600-1058 of GenBank
Accession No. AF308157; Belsham et al. (1990) EMBO 77:1105-10; Poyry et al.
(2001)
RNA 7:647-60; and Stoneley et al. (2000) Nucleic Acid Research 25:687-94),
equine
rhinitis A virus, Ewuine rhinitis B); Pestivirus (e.g., Bovine viral diarrhea
virus (Poole et
al. (1995) Virology 206:150-154) and Classical swine fever virus (Rijnbrand et
al.
(1997) J. Virol 77:451-7); Hepacivirus (e.g., Hepatitis C virus (Tsukiyama-
Kohara et al.
(1992) J. Virol. 66:1476-1483, Lemon et al. (1997) Semin. Virol. 5:274-288,
and
nucleotide 1201-1812 of GenBank Accession No. AJ242654.) and GB virus B). Each
of
these references is herein incorporated by reference.
IRES elements have also been found in viruses from the family Retroviridae,
including members of the Lentivirus family (e.g., Simian immunodeficiency
virus
(Ohlmann et al. (2000) Journal of Biological Chemistry 275:11899-906) and
human
immunodeficiency virus 1 (Bucket s/. (2001) J Virol. 75:181-91); the BLV-HTLV
retroviruses (e.g., Human T-lymphotrophic virus type 1 (Attal et al. (1996)
EEES Letters
392:220-4); and the Mammalian type C retoviral family (e.g., Moloney murine
leukemia
virus (Vagner et al. (1995) J. Biol. Chem 270:20316-83), Friend murine
leukemia virus,
Harvey murine sarcoma virus, Avian retriculoendotheliosis virus (Lopez-Lastra
et al.
(1997) Hum. Gene Ther 5:1855-65), Murine leukemia virus (env RNA) (Deffaud et
al.
(2000) J. Virol. 74:846-50), Rous sarcoma virus (Deffaud et al. (2000) J.
Virol.
74:11581-8). Each of these references is herein incorporated by reference.
Eukaryotic mRNAs also contain IRES elements including, for example, BiP
(Macejak et al. (1991) Nature 355:91); Antennapedia of Drosophilia (exons d
and e) (Oh
et al. (1992) Genes and Development 6:1643-1653; c-myc; and, the X-linked
inhibitor of
apoptosis (XIAP) gene (U.S. Patent No. 6,171,821).
Various synthetic IRES elements have been generated. See, for example, De
Gregorio et al. (1999) EMBO J. 75:4865-74; Owens et al. (2001) PNAS 4:1471-6;
and
Venkatesan et al. (2001) Molecular and Cellular Biology 21:2826-37. For
additional
IRES elements known in the art, see, for example,
rangueiLinserm.fr/IRESdatabase.
In a specific embodiment, the IRES sequence is derived from
encephalomyocarditis virus (ECMV).
8
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
As used herein, a reporter protein is any protein that can be specifically
detected
when expressed (i.e, has a detectable signal when expressed), for example, via
its
fluorescence or enzyme activity. The plasmid comprises a nucleotide sequence
encoding
a first reporter protein and a nucleotide sequence encoding a second reporter
protein. An
-- open reading frame is fused either to the nucleotide sequence encoding a
first reporter
protein or to the nucleotide sequence encoding a second reporter protein. In
some
embodiments, the open reading frame is fused to the nucleotide sequence
encoding a first
reporter protein. In some embodiments, the open reading frame is fused to the
nucleotide sequence encoding a second reporter protein. This allows one to
study the
-- expression of the linked open reading frame in response to different
stimuli. As used
herein, "fused" is intended to mean that the amino acids encoded by the ORF
and the
reporter protein are joined by peptide bonds to create a contiguous protein
sequence.
Thus, the reporter protein fused to the open reading frame serves as a marker
of the
stability of the fused open reading frame. The other reporter protein that is
unfused to
-- the open reading frame (and thus does not create a contiguous protein
sequence with the
amino acids encoded by the ORF) serves as an internal control to normalize for
cell
number and expression variability.
Typically, the first and second reporter proteins have distinguishable
detectable
reporter signals. For example, the first and second reporter proteins are
enzyme proteins
-- having distinguishable signals generated from their products. In some
embodiments, the
first and second reporter proteins are bioluminescent proteins that emit light
at different
wavelengths and/or utilize different substrates. Alternatively, the first and
second
reporter proteins are fluorescent proteins that fluoresce at different
wavelengths.
Many reporter proteins known in the art may be used, including but not limited
to
-- bioluminescent proteins, fluorescent reporter proteins, and enzyme proteins
such as beta-
galactosidase, horse radish peroxidase and alkaline phosphatase that produce
specific
detectable products. The fluorescent reporter proteins include, for example,
green
fluorescent protein (GFP), cyan fluorescent protein (CFP), red fluorescent
protein (RFP)
and yellow fluorescent protein (YFP) as well as modified forms thereof e.g.
enhanced
-- GFP (EGFP), enhanced CFP (ECFP), enhanced RFP (ERFP), mCHERRY, and enhanced
YEP (EYEP).
Examples of bioluminescent proteins, such as luciferases, including but not
limited to renilla luciferase (Rluc), firefly luciferase (FLuc) and NanoLuc,
are known in
9
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
the art (see, for example, Fan, F. and Wood, K., Assay and drug development
technologies V5 #1(2007); Gupta, R. et al Nature Methods V8 #10 (2011); Nano-
Glo
Luciferase Assay System (Promega) and en.wikipedia.org/wiki/Bioluminescence.
Other non-limiting examples of reporter proteins are shown below:
Species-specific iuciferase specificity, cofactor requirements and physical
characteristics.
Size
Organism Luciferase Substrate Requires Secreted
(Oa)
firefly
Photinus pyras Northrit 61 D-luciferin Mg ATP No
lucifeaseAmerican,
Luuda
Japanese firefly (Gen 64 ji-botaru ) cruciara D-luciferin Mg, ATP
No
luciferase
Luciola hafica Italian firefly Luciferase 64 D-luciferin
Mg. ATP No
Japanese firefly (Heike)
Luciola lateralis 64 D-luciferin Mg ATP No
luciferase
Luciola rningrefica East European firefly luciferase 64 D-luciferin
Mg Al? No
Photuris pennsylvanica Pennsylvania firefly luciferase 64
D-luciferin Mg ATP No
Pyrophorus
Click beetle luciferase 64 D-luciferin Mg; ATP No
plagiophthalamus
Phrixothrix hittus Railroad worm luciferase 64 Dluciferin Mg
ATP No
Pendia luciferase 36 Coelenterazine NIA No
Rluc8 (mutant of Rendia
Renriia rennronnis 36 Coelenterazine N/A No
luciferase)
Green Rendia luciferase 33 Coelenterazine N/A No
Gsussialuciferase 20 Coelentenazine NIA Yes
Gaussia princeps
Gaussia-Dura luciferase 20 Coelenterazine NIA Yes
Cypridina nuctituca Cypridina luciferase 62
VanguliniCypridina NIA Yes
luciferin
VarguirniCypridirta
Cypadina hilgencloa Cypndina Nargula) luciferase 62 NIA Yes
luciferin
kletridia ionga kletridia luciferase 23.8 Coelenterazine
NiA Yes
Oplophorus gracilorostris OLuc 19 Coelenterazine NIA Yes
In some embodiments, the first and second reporter proteins are selected from
the
group consisting of renilla luciferase (Rluc), firefly luciferase (FLuc) and
NanoLuc. In
some embodiments, the first and second reporter proteins are selected from the
group
consisting of green fluorescence protein and red fluorescence protein.
An open reading frame is fused either to the nucleotide sequence encoding a
first
reporter protein or to the nucleotide sequence encoding a second reporter
protein. The
open reading frame is fused to the 5' or to the 3' end of the nucleotide
sequence. As
used herein, an open reading frame or ORF refers to a sequence of nucleotides
that codes
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
for a contiguous sequence of amino acids. The translated open reading frame
may be all
or a portion of a gene encoding a protein or polypeptide of interest.
The ORF of the plasmid codes for a protein of interest. As used herein, a
"protein of interest" can be any conceivable polypeptide or protein that may
be of
interest, such as to study or otherwise characterize. In some embodiments, the
ORF may
be derived from an ORFeome of an organism. A complete ORFeome contains nucleic
acids that encode all proteins of a given organism. A representative fraction
of a full
ORFeome is at least 60% of all proteins expressed by the organism. In some
embodiments, the organism is a mammal. In some embodiments, the mammal is
human.
In some embodiments, the protein of interest is a human polypeptide or
protein.
In some embodiments, the protein of interest is an oncoprotein, such as, but
not limited
to, RAS, MYC, SRC, FOS, JUN, MYB, ABL, BCL2, HOX11, HOX11L2, TALl/SCL,
LM01, LM02, EGFR, MYCN, MDM2, CDK4, GLI1, IGF2, EGFR, FLT3-ITD, TP53,
PAX3, PAX7, BCR/ABL, HER2 NEU, FLT3R, FLT3-ITD, TANI, B-RAF, E2A-
PBX1, and NPM-ALK, as well as fusion of members of the PAX and FKHR gene
families, WNT, MYC, ERK EGFR, FGFR3, CDH5, KIT, RET, Interferon regulatory
factor 4 (IRF4) and TRK. Other exemplary oncogenes are well known in the art
and
several such examples are described in, for example, The Genetic Basis of
Human
Cancer (Vogelstein, B. and Kinzler, K. W. eds. McGraw-Hill, New York, N.Y.,
1998).
In some embodiments, the protein of interest is a transcription factor. Some
examples of such transcription factors include (but are not limited to) the
STAT family
(STATs 1, 2, 3, 4, 5a, 5b, and 6) , FOS/JUN, NF KB, HIV-TAT, and the E2F
family. In
some embodiments, the protein of interest is an IKAROS family zinc finger
protein. In
some embodiments, the protein of interest is IKZF 1, IKZF2, IKZF3, IKZF4, or
IKZF5.
In some embodiments, the protein of interest is IKZF1 or IKZF3.
The nucleotide sequence encoding a reporter protein and the fused ORF are "in
frame", i.e., consecutive triplet codons of a single polynucleotide comprising
the
nucleotide sequence encoding the reporter protein and the fused open reading
frame
encode a single continuous amino acid sequence.
The methods described herein allows one to screen libraries of compounds and
identify a test compound that stabilizes or destabilizes a protein of
interest. A compound
library is a collection of stored compounds typically used in high-throughput
screening.
The library compounds may include, for example, synthesized organic molecules,
11
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
naturally occurring organic molecules, peptides, polypeptides, nucleic acid
molecules
and components thereof. Examples of compound library include, but are not
limited to,
Screen-Well Compound Libraries (Enzo Life Sciences), EXPRESS-Pick Collection
and CORE Library (Chem Bridge), National Cancer Institute Library, Prestwick
Chemical Library and Tocriscreen Compound Library Collections.
The plasmids described herein may be introduced into the host cell using any
available technique known in the art. For example, the plasmid may be
introduced into
the host cell by lipofection, calcium phosphate transfection, DEAE-dextran
mediated
transfection, electroporation, transduction, sonoporation, infection and
optical
transfection. Suitable host cells include, but are not limited to, bacterial
cells (e.g., E.
coli, Bacillus subtilis, and Salmonella typhimurium), yeast cells (e.g.,
Saccharomyces
cerevisiae and Schizosaccharomyces pombe), plant cells (e.g., Nicotiana
tabacum and
Gossypium hirsutum), and mammalian cells (e.g., CHO cells, and 3T3
fibroblasts, HEK
293 cells, U-2 OS cells).
In some embodiments, contacting a host cell transformed with the plasmid
described herein with a test compound comprises growing the transformed host
cell in
the presence of the test compound for an appropriate time. under suitable
culture
conditions. Suitable culture conditions, including the duration of the
culture, will vary
depending on the cell being cultured. However, one skilled in the art can
easily
determine the culture conditions by following standard protocols, such as
those described
in the series Methods in Microbiology, Academic Press Inc. Typically, the cell
culture
medium may contain any of the following nutrients in appropriate amounts and
combinations: salt(s), buffer(s), amino acids, glucose or other sugar(s),
antibiotics, serum
or serum replacement, and other components such as, but not limited to,
peptide growth
factors, cofactors, and trace elements. In some embodiments, the transfected
host cells
are grown in the presence of the compound for 15 mins, 30 mins, 1 hour, 2
hours, 4
hours, 6 hours, 8 hours, 10 hours, 12 hours, 14 hours, 16 hours, 18 hours, 20
hours, 24
hours, 30 hours, 48 hours, or 72 hours.
In some embodiments, a single transformed host cell is first isolated, cloned
and
expanded based on optimized responses to a control test compound and confirmed
to
provide sufficiently low error required for HTS campaigns. Selection of
appropriate
clones is aided by determining the response of the fused reporter protein of
interest
relative to the unfused reporter with the necessary response stability and
reproducibility
12
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
required for high throughput screening. Identification of useful clones is
aided by
additionally normalizing the fused reporter signals to the control unfused
reporter which
can significantly reduce the inherent error relative to measuring the response
solely from
the fused reporter. This reduction in error is critical for the identification
of a useful
-- clonal cell line that responds to a test compound with a large enough
response relative to
the response error obtained from the respective signals observed from the
treated and
untreated samples in order to provide a Z factor sufficient for high
throughput screening.
(en.wikipedia.org/wiki/Z-factor).
As used herein, "fused reporter protein signal" refers to the detectable
signal of
-- the reporter protein encoded by the nucleotide sequence that is fused to
the ORF. As
used herein, "unfused reporter protein signal" refers to the detectable signal
of the
reporter protein encoded by the nucleotide sequence that is not fused to the
ORF. The
fused and unfused reporter protein signals in the presence and absence of the
test
compound are determined using methods known in the art. Detectors such as, but
not
-- limited to, luminometers, spectrophotometers, and fluorimeters, or any
other device that
can detect changes in reporter protein activity can be used. Assay systems
known in the
art that allow for quantitation of a stable reporter signal from two reporter
genes in a
single sample can be used. Examples include, but are not limited to, Dual-Glo
Luciferase Assay System (Promega) that measures the activities of firefly and
Renilla
-- luciferases sequentially from a single sample.
After detecting the signals generated by the reporter proteins, the ratio of
the
fused reporter protein signal to unfused reporter protein signal in the
presence of the test
compound is compared to the ratio of the fused reporter protein signal to
unfused
reporter protein signal in the absence of the test compound. When the ratio of
fused
-- reporter protein signal to unfused reporter protein signal in the presence
of the test
compound is increased as compared to the ratio of fused reporter protein
signal to
unfused reporter protein signal in the absence of the test compound, the test
compound is
identified as a stabilizer of the protein of the interest. In contrast, when
the ratio of
fused reporter protein signal to unfused reporter protein signal in the
presence of the test
-- compound is decreased as compared to the ratio of fused reporter protein
signal to
unfused reporter protein signal in the absence of the test compound, the test
compound is
identified as a destabilizer of the protein of interest.
13
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
In some embodiments, the open reading frame is fused to the nucleotide
sequence
encoding a first reporter protein. In such embodiments, ratios of first
reporter protein
signal to second reporter protein signal are determined in presence and
absence of the
compound. The test compound is identified as a stabilizer when the ratio of
the first
reporter protein signal to second reporter protein signal in the presence of
the test
compound is increased as compared to the ratio of first reporter protein
signal to second
reporter protein signal in the absence of the test compound. The test compound
is
identified as a destabilizer when the ratio of first reporter protein signal
to second
reporter protein signal in the presence of the test compound is decreased as
compared to
the ratio of first reporter protein signal to second reporter protein signal
in the absence of
the test compound.
In some embodiments, the open reading frame is fused to the nucleotide
sequence
encoding a second reporter protein. In such embodiments, ratios of second
reporter
protein signal to first reporter protein signal are determined in presence and
absence of
the compound. The test compound is identified as a stabilizer when the ratio
of the
second reporter protein signal to first reporter protein signal in the
presence of the test
compound is increased as compared to the ratio of second reporter protein
signal to first
reporter protein signal in the absence of the test compound. The test compound
is
identified as a destabilizer when the ratio of second reporter protein signal
to first
reporter protein signal in the presence of the test compound is decreased as
compared to
the ratio of second reporter protein signal to first reporter protein signal
in the absence of
the test compound.
The present invention is further illustrated by the following Example, which
in no
way should be construed as further limiting. The entire contents of all of the
references
(including literature references, issued patents, published patent
applications, and
co-pending patent applications) cited throughout this application are hereby
expressly
incorporated by reference.
EXAMPLES
Example 1
pIRIGF constructs express in 293FT and HELA cells (FIG. 1A-C) and pUG-
FIRP constructs express in U-2 OS cells (FIG. 1D). Several different versions
of
mammalian and lentiviral plasmid constructs were tested for their ability to
generate
14
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
cells (e.g. 293FT, HELA, or U-2 OS cells) expressing tagged target proteins
(e.g., firefly
or NanoLuc tag) and co-expressing a reporter luciferase (e.g., Renilla or
Firefly).
293FT and HELA cells were transfected with IKZF1-firefly, IKZF3-firefly and
MYC-firefly fusion proteins and selected using puromycin and geneticin
respectively.
These pools were very unstable and lost signals in 10 to 30 days and generally
had very
small responses to IMiD's (FIG. 2A-C). Therefore, individual clones were
isolated using
a limited cloning strategy in 96-well plates. Surviving cells were isolated as
colonies,
further expanded and tested for luciferase signals and response to IMiDs.
Clone 2B4
was identified as a strong responder to lenalidomide. HELA cells expressed
very low
levels of luciferase making isolation of HELA clones very difficult. Detection
by
western blots of firefly, IKZF1 and myc confirmed expression of the fusion
protein and
relative expression correlated with firefly luciferase signals (FIG. 2D).
Cell line clones (IKZF1-2B4, IKZF1-2B11, myc-1C3 and myc-5F2) expressing
the indicated firefly fusion protein were evaluated in the dual-glo assay for
reproducibility. Potency of IMiD's and relative reduction in firefly
luciferase signals
confirmed the expected responses and generated data with Z' values sufficient
for
screening (FIG. 3A-D).
Pilot screen results for IKZF1 2B4 cells - Active compounds (Prestwick
collection and NCI collection are shown in FIG. 4A and 4B.
Pilot screen results for MYC 5F2 cells - Active compounds NCI collection are
shown in Fig. 5.
The hits tested on IKZF1 2B4 and MYC 5F2 cell lines were confirmed (FIG. 6A-
C). Summary retest data from commercial compounds and from DTP compounds is
shown in FIG. 6D-E.
FIG. 7 shows confirmation data using Western blots. IKZF1 2B4 cell line
examples (FIG. 7A-B), MYC 5F2 example (FIG. 7C)
FIG 8 shows further evaluations of HSP90 inhibitors. FIG. 8A demonstrates
testing of HSP 90 inhibitors CCT018159 and geldanamycin on cells transiently
transfected with the MYC-firefly fusion protein. FIG. 8B shows testing of HSP
90
inhibitors CCT018159 and geldanamycin on 293FT cells stably expressing MYC-
firefly
fusion protein. FIG. 8C shows testing of several HSP-90 inhibitors at various
doses on 5
different cell lines stably expressing the MYC-firefly fusions protein. FIG.
8D
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
compares the HSP90 inhibitor BIIB021 and pomalidomide on 293FT cells
transiently
expressing IKZF1-firefly fusion protein.
Example 2: Comparing activity in IKZF1 vs. MYC cell lines
Screening campaign at HMS screening facility ICCB
Cherry pick retests for IKZF1 were screened at ICCB (FIG. 9). Of the 44,460
compounds, 0.6% of the compounds that were screened had a hit rate based on
greater
than 35% decrease in Fluc/Rluc compared to plate average. 0.3% of the
compounds were
cherry picked. 81% (108/133) of the hits repeated with greater than 25%
decrease in
Fluc/Rluc compared to DMSO control. Approximately 90% (97/108) were hit in
both
cell lines. There was an 11 >25% difference in activity for IKZF1 vs. MYC. All
of the
above results were moderate or weak hits.
The results of the ICCB cherry picks IKZF1 vs. MYC selectivity comparison
show that the majority of hits in the IKZF1 cell line screen were also active
in the
counter screen assay suggesting a nonspecific mechanism (FIG. 10). Five
compounds
were inactive in the counter screen, but still reduced IKZF1 luciferase signal
more than
35%, showing some selectivity.
Dose response for HSP90 inhibitors
Two HSP90 inhibitors, BIIB021 (FIG. 11A) and PF-04929113 (FIG. 11B), show
similar activity in the 293FT IKZF1 cell line compared to the counter screen
293FT cell
line expressing MYC-firefly:renilla and U2OS cell line expressing MYC-
firefly:renilla
suggesting a mechanism nonselective for IKZF1. Knockdown of protein levels
were
confirmed by western blot indicating that the luciferase reporter system is
accurately
reflecting fusion protein reduction.
Stability time course for 7 MYC-luciferase fusion cell lines
Seven cell lines were used to measure the half life of the luciferases after
blocking all protein synthesis with cyclohexamide. The decay observed for both
fused
luciferases (MYC-firefly and MYC-nanoluc; FIG. 12B) were compared to the decay
observed for the unfused luciferases (renilla and firefly; FIG. 12A) using
both the MYC-
firefly:renilla and MYC-nanoluc:firefly cell lines 293T, U205 and the MYC-
firefly:renilla cell lines A549, H1299 and LS174T. As expected, the half-life
of untagged
16
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
firefly (approximately 4 hours) is shorter than the untagged renilla
(approximately 12
hours) since it contains a PEST domain. MYC nanoluc half life of approximately
2 hours
is longer than MYC-firefly half-life of less than 1 hour and closer to the
half-life of
untagged firefly. The balanced half-life of MYC-nanoluc and untagged firefly
should
reduce the number of artefact hits.
Seven MYC-luciferase reporter cell lines were used to measure changes in MYC-
luciferase expression after blocking the proteasome with MG132. The expression
of the
unfussed renilla and firefly were unchanged for about 6 hours but decreased
after 18
hours to a variable extent among cell lines (FIG. 13B). All cell lines showed
at least a
50% increase in MYC-luciferase fusion protein, but with different time course.
The
MYC-nanoluc demonstrated about 4-fold increase in luciferase signals
suggesting a
larger portion of the these fusion proteins are degraded by the proteasome
(FIG. 13A).
Seven MYC-luciferase reporter cell lines were used to measure changes in MYC-
luciferase expression after inhibition of ubiquitin dependent proteolysis with
the
neddylation inhibitor MLN-4924 (FIG. 14A-B). The expression of the unfussed
renilla
and firefly were minimally affected except for the 293T MYC-firefly:renilla
cell line. All
cell lines showed at least a 50% increase in MYC-luciferase fusion protein,
typically
peaking after 6 hours of treatment. These results demonstrate that ubiquitin
dependent
proteolysis is at least partially responsible for the stability of the MYC-
fusion proteins
in all 7 cell lines.
A549 and H1299 cell lines expressing MYC-firefly after siMYC knockdown
siRNA was used to knockdown the MYC-firefly luciferase fusion protein in the
A549 and H1299 cell lines using 48 hour treatment with siRNA directed to MRC
mRNA. The reduction in fusion protein, as observed by western blotting with
MYC and
firefly directed antibodies (FIG. 15A), is comparted to the decrease in
luciferase signals
(FIG. 15B). MYC antibody also detects the decrease in endogenous MYC. A
prominent
MYC-NICK band is observed in the A549 cells.
siGENOME siRNA Library
Figure 16A-D shows screening results from a commercial library of siRNA's
directed to the family of DUB enzymes with A549, H1299, and HEK293T cells
expressing MYC-firefly and U205 cells expressing MYC-nanoluc.
17
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
References
1. R. Martiniani, V. Di Loreto, C. Di Sano, A. Lombardo, A. M. Liberati,
Biological
activity of lenalidomide and its underlying therapeutic effects in multiple
myeloma. Adv Hematol 2012, 842945 (2012).
2. E. Terpos, N. Kanellias, D. Christoulas, E. Kastritis, M. A. Dimopoulos,
Pomalidomide: a novel drug to treat relapsed and refractory multiple myeloma.
OncoTargets and therapy 6, 531 (2013).
3. Y. X. Zhu, K. M. Kortuem, A. K. Stewart, Molecular mechanism of action
of
immune-modulatory drugs thalidomide, lenalidomide and pomalidomide in
multiple myeloma. Leukemia & lymphoma 54, 683 (Apr, 2013).
4. T. Ito et al., Identification of a primary target of thalidomide
teratogenicity.
Science 327, 1345 (Mar 12, 2010).
5. A. Lopez-Girona et al., Cereblon is a direct protein target for
immunomodulatory
and antiproliferative activities of lenalidomide and pomalidomide. Leukemia
26,
2326 (Nov, 2012).
6. L. H. Zhang et al., Lenalidomide efficacy in activated B-cell-like
subtype diffuse
large B-cell lymphoma is dependent upon IRF4 and cereblon expression. Br J
Haematol 160, 487 (Feb, 2013).
7. Y. Yang et al., Exploiting synthetic lethality for the therapy of ABC
diffuse large
B cell lymphoma. Cancer Cell 21, 723 (Jun 12, 2012).
8. Y. X. Zhu et al., Cereblon expression is required for the antimyeloma
activity of
lenalidomide and pomalidomide. Blood 118, 4771 (Nov 3, 2011).
9. A. Broyl et al., High cereblon expression is associated with better
survival in
patients with newly diagnosed multiple myeloma treated with thalidomide
maintenance. Blood 121, 624 (Jan 24, 2013).
10. D. Heintel et al., High expression of cereblon (CRBN) is associated
with
improved clinical response in patients with multiple myeloma treated with
lenalidomide and dexamethasone. Br J Haematol 161, 695 (Jun, 2013).
11. G. J. Zhang et al., Bioluminescent imaging of Cdk2 inhibition in vivo.
Nat Med
10, 643 (Jun, 2004).
18
CA 02964161 2017-04-07
WO 2016/057903
PCT/US2015/054914
12. M. Safran et al., Mouse model for noninvasive imaging of HIF prolyl
hydroxylase activity: assessment of an oral agent that stimulates
erythropoietin
production. Proc Natl Acad Sci U S A 103, 105 (Jan 3, 2006).
13. H. C. Yen, Q. Xu, D. M. Chou, Z. Zhao, S. J. Elledge, Global protein
stability
profiling in mammalian cells. Science 322, 918 (Nov 7, 2008).
14. T. A. Soucy et al., An inhibitor of NEDD8-activating enzyme as a new
approach
to treat cancer. Nature 458, 732 (Apr 9, 2009).
15. M. Ohh et al., An intact NEDD8 pathway is required for Cullin-dependent
ubiquitylation in mammalian cells. EMBO reports 3, 177 (Feb, 2002).
16. L. Cong et al., Multiplex genome engineering using CRISPR/Cas systems.
Science 339, 819 (Feb 15, 2013).
17. P. Mali et al., RNA-guided human genome engineering via Cas9. Science
339,
823 (Feb 15, 2013).
18. M. Merkenschlager, Ikaros in immune receptor signaling, lymphocyte
differentiation, and function. FEBS Lett 584, 4910 (Dec 15, 2010).
19. E. C. Thompson et al., Ikaros DNA-binding proteins as integral
components of B
cell developmental-stage-specific regulatory circuits. Immunity 26, 335 (Mar,
2007).
20. I. Ferreiros-Vidal et al., Genome-wide identification of Ikaros targets
elucidates
its contribution to mouse B-cell lineage specification and pre-B-cell
differentiation. Blood 121, 1769 (Mar 7, 2013).
21. S. Monticelli, F. Sallusto, Negative regulators take center stage.
Nature
immunology 13, 719 (Aug, 2012).
22. L. A. Garraway, W. R. Sellers, Lineage dependency and lineage-survival
oncogenes in human cancer. Nat Rev Cancer 6, 593 (Aug, 2006).
23. S. P. Shah et al., Mutation of FOXL2 in granulosa-cell tumors of the
ovary. N
Engl J Med 360, 2719 (Jun 25, 2009).
24. P. Kastner et al., Function of Ikaros as a tumor suppressor in B cell
acute
lymphoblastic leukemia. American journal of blood research 3, 1 (2013).
25. F. J. Quintana et al., Aiolos promotes TH17 differentiation by directly
silencing
112 expression. Nature immunology 13, 770 (Aug, 2012).
26. N. Avitahl et al., Ikaros sets thresholds for T cell activation and
regulates
chromosome propagation. Immunity 10, 333 (Mar, 1999).
19
CA 02964161 2017-04-07
WO 2016/057903 PCT/US2015/054914
27. J. Laubach, P. Richardson, K. Anderson, Multiple myeloma. Annu Rev Med
62,
249 (2011).
28. P. G. Richardson et al., Multicenter, phase I, dose-escalation trial of
lenalidomide
plus bortezomib for relapsed and relapsed/refractory multiple myeloma. J Clin
Oncol 27, 5713 (Dec 1, 2009).
29. G. R. Crabtree, S. L. Schreiber, Three-part inventions: intracellular
signaling and
induced proximity. Trends Biochem Sci 21, 418 (Nov, 1996).
30. M. R. Campanero, E. K. Flemington, Regulation of E2F through ubiquitin-
proteasome-dependent degradation: stabilization by the pRB tumor suppressor
protein. Proc Natl Acad Sci U S A 94, 2221 (Mar 18, 1997).
31. D. Wiederschain et al., Single-vector inducible lentiviral RNAi system
for
oncology target validation. Cell Cycle 8, 498 (Feb 1, 2009).
32. C. J. Ott et al., BET bromodomain inhibition targets both c-Myc and
IL7R in
high-risk acute lymphoblastic leukemia. Blood 120, 2843 (Oct 4, 2012).
33. K. Kondo, J. Klco, E. Nakamura, M. Lechpammer, W. G. Kaelin, Inhibition
of
HIF is necessary for tumor suppression by the von Hippel-Lindau protein.
Cancer
Cell 1, 237 (Apr, 2002).
34. J. Loven et al., Selective inhibition of tumor oncogenes by disruption
of super-
enhancers. Cell 153, 320 (Apr 11,2013).
We claim: