Note: Descriptions are shown in the official language in which they were submitted.
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
1
CRISPR-Based Compositions and Methods of Use
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application claims benefit of priority under 35 U.S.C. 119 to
U.S.
provisional patent applications bearing serial numbers 62/093,588 and
62/239,546, filed
December 18, 2014 and October 9, 2015, and entitled "CRISPR-BASED
COMPOSITIONS AND METHODS OF USE," the contents of which are herein
incorporated by reference in its entirety.
SEQUENCE LISTING
[0002] The instant application contains a Sequence Listing that has been
submitted
in ASCII format via EFS-Web and is hereby incorporated by reference in its
entirety.
The ASCII copy, created on December 18, 2015, is named IDT01-008-US 5T25.txt,
and
is 177,163 bytes in size.
FIELD OF THE INVENTION
[0003] This invention pertains to modified compositions for use in CRISPR
systems,
and their methods of use.
BACKGROUND OF THE INVENTION
[0004] The use of clustered regularly interspaced short palindromic repeats
(CRISPR) and associated Cas proteins (CRISPR-Cas system) for site-specific DNA
cleavage has shown great potential for a number of biological applications.
CRISPR is
used for genome editing; the genome-scale-specific targeting of
transcriptional
repressors (CRISPRi) and activators (CRISPRa) to endogenous genes; and other
applications of RNA-directed DNA targeting with Cas enzymes.
[0005] CRISPR-Cas systems are native to bacteria and Archaea to provide
adaptive
immunity against viruses and plasmids. There are three classes of CRISPR-Cas
systems
that could potentially be adapted for research and therapeutic reagents, but
Type-II
CRISPR systems have a desirable characteristic in utilizing a single CRISPR
associated
(Cas) nuclease (specifically Cas9) in a complex with the appropriate guide
RNAs ¨
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
2
either a 2-part RNA system similar to the natural complex in bacteria
comprising a
CRISPR-activating RNA:trans-activating crRNA (crRNA:tracrRNA) pair or an
artificial
chimeric single-guide-RNA (sgRNA) ¨ to mediate double-stranded cleavage of
target
DNA. In mammalian systems, these RNAs have been introduced by transfection of
DNA
cassettes containing RNA Pol III promoters (such as U6 or H1) driving RNA
transcription, viral vectors, and single-stranded RNA following in vitro
transcription (see
Xu, T., et al., Appl Environ Microbiol, 2014. 80(5): p. 1544-52).
[0006] In the CRISPR-Cas9 system, using, for example, the system present in
Streptococcus pyogenes as an example (S.py. or Spy), native crRNAs are about
42 bp
long, containing a 5'-region of about 20 bases complementary to a target
sequence (also
referred to as a protospacer sequence) and a 3' region typically about 22
bases long that
corresponds to a complementary region of the tracrRNA sequence. The native
tracrRNAs are about 85-90 bases long, having a 5'-region containing the region
complementary to the crRNA as well as about a 10-base region 5'-upstream. The
remaining 3' region of the tracrRNA includes secondary structures (herein
referred to as
the "tracrRNA 3'-tail").
[0007] Jinek et al. extensively investigated the portions of the crRNA and
tracrRNA
that are required for proper functioning of the CRISPR-Cas9 system (Science,
2012.
337(6096): p. 816-21). They devised a truncated crRNA:tracrRNA fragment that
could
still function in CRISPR-Cas9 wherein the crRNA was the wild type 42
nucleotides and
the tracrRNA was truncated to 75 nucleotides. They also developed an
embodiment
wherein the crRNA and tracrRNA are attached with a linker loop, forming a
single guide
RNA (sgRNA), which varies between 99-123 nucleotides in different embodiments.
The
configuration of the native 2-part crRNA:tracrRNA complex is shown in FIG. 1
and the
99 nucleotide embodiment of the artificial sgRNA single guide is shown in FIG.
2.
[0008] At least two groups have elucidated the crystal structure of
Streptococcus
pyogenes Cas9 (SpyCas9). In Jinek, M., et al., the structure did not show the
nuclease in
complex with either a guide RNA or target DNA. They carried out molecular
modeling
experiments to reveal predictive interactions between the protein in complex
with RNA
and DNA (Science, 2014. 343, p. 1215, DOI: 10.1126/science/1247997).
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
3
[0009] In Nishimasu, H., et al., the crystal structure of SpyCas9 is shown
in complex
with sgRNA and its target DNA at 2.5 angstrom resolution (Cell, 2014. 156(5):
p. 935-
49, incorporated herein in its entirety). The crystal structure identified two
lobes to the
Cas9 enzyme: a recognition lobe (REC) and a nuclease lobe (NUC). The
sgRNA:target
DNA heteroduplex (negatively charged) sits in the positively charged groove
between
the two lobes. The REC lobe, which shows no structural similarity with known
proteins
and therefore likely a Cas9-specific functional domain, interacts with the
portions of the
crRNA and tracrRNA that are complementary to each other.
[0010] Another group, Briner et al. (Mol Cell, 2014. 56(2): p. 333-9,
incorporated
herein in its entirety), identified and characterized the six conserved
modules within
native crRNA:tracrRNA duplexes and sgRNA.
[0011] The CRISPR-Cas9 system is utilized in genomic engineering as
follows: a
portion of the crRNA hybridizes to a target sequence, a portion of the
tracrRNA
hybridizes to a portion of the crRNA, and the Cas9 nuclease binds to the
entire construct
and directs cleavage. The Cas9 contains two domains homologous to
endonucleases
HNH and RuvC, wherein the HNH domain cleaves the DNA strand complementary to
the crRNA and the RuvC-like domain cleaves the noncomplementary strand. This
results
in a double-stranded break in the genomic DNA. When repaired by non-homologous
end
joining (NHEJ) the break is typically shifted by 1 or more bases, leading to
disruption of
the natural DNA sequence and in many cases leading to a frameshift mutation if
the
event occurs in the coding exon of a protein-encoding gene. The break by also
be
repaired by homology dependent recombination (HDR), which permits insertion of
new
genetic material via experimental manipulation into the cut site created by
Cas9
cleavage.
[0012] Some of the current methods for guide RNA delivery into mammalian
cells
include transfection of double-stranded DNA (dsDNA) containing RNA Pol III
promoters for endogenous transcription, viral delivery, transfection of RNAs
as in vitro
transcription (IVT) products, or microinjection of IVT products. There are
disadvantages
to each of these methods. Unmodified exogenous RNA introduced into mammalian
cells
is known to initiate the innate immune response via recognition by Toll-like
Receptors
(TLRs), RIG-I, OASI and others receptors that recognize pathogen-associated
molecular
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
4
patterns (PAMPs). However, in most published studies, RNA which has been in
vitro
transcribed (IVT) by a T7 RNA polymerase is delivered to the cells. This type
of RNA
payload has been shown to be a trigger for the innate immune response. The
alternative
delivery methods described above each have their own disadvantages as well.
For
example, dsDNA cassettes can lead to integration, guide RNA transcription
driven
endogenously by a RNA Pol II promoter can persist constitutively, and the
amount of
RNA transcribed is uncontrollable.
[0013] RNA is quickly degraded by nucleases present in serum and in cells.
Unmodified CRISPR RNA triggers (crRNAs, tracrRNAs, and sgRNAs) made by IVT
methods or chemical synthesis are quickly degraded during delivery or after
delivery to
mammalian cells. Greater activity would be realized if the RNA was chemically
modified to gain nuclease resistance. The most potent degradative activity
present in
serum and in cells is a 3'-exonuclease (Eder et al., Antisense Research and
Development
1:141-151, 1991). Thus "end blocking" a synthetic oligonucleotide often
improves
nuclease stability. Chemical modification of single-stranded anti sense
oligonucleotides
(AS0s) and double-stranded small interfering RNAs (siRNAs) has been well
studied and
successful approaches are in practice today (for reviews, see: Kurreck, Eur.
J. Biochem.,
270:1628-1644, 2003; Behlke, Oligonucleotides, 18:305-320, 2008; Lennox et
al., Gene
Therapy, 18:1111-1120, 2011). It is therefore desirable to devise chemical
modification
strategies for use with the RNA components of CRISPR/Cas. While the basic
toolbox of
chemical modifications available is well known to those with skill in the art,
the effects
that site-specific modification have on the interaction of a RNA species and
an effector
protein are not easily predicted and effective modification patterns usually
must be
empirically determined. In some cases, sequence of the RNA may influence the
effectiveness of a modification pattern, requiring adjustment of the
modification pattern
employed for different sequence contexts, making practical application of such
methods
more challenging.
[0014] There is therefore a need to modify the guide RNA to reduce its
toxicity to
cells and to extend lifespan and functionality in mammalian cells while still
performing
their intended purpose in the CRISPR-Cas system. The methods and compositions
of the
invention described herein provide RNA and modified RNA oligonucleotides for
use in a
CRISPR-Cas system. These and other advantages of the invention, as well as
additional
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
inventive features, will be apparent from the description of the invention
provided
herein.
BRIEF SUMMARY OF THE INVENTION
[0015] This invention pertains to modified compositions for use in CRISPR
systems,
and their methods of use. The compositions include modified internucleotide
linkages
and 2'-0-alkyl and 2'-0-fluoro modified RNA oligonucleotides to serve as the
guides
strands (crRNA:tracrRNA or sgRNA) for the CRISPR-Cas system. Compositions also
include end-modifications such as an inverted-dT base or other non-nucleotide
modifiers
that impeded exonuclease attack (such as the propanediol group (C3 spacer),
napthyl-azo
modifier, or others as are well known in the art).
[0016] In a first aspect, isolated tracrRNA including a length-modified
form of SEQ
ID NO.:18 is provided. The isolated tracrRNA displays activity in a Clustered
Regularly
Interspaced Short Palindromic Repeats (CRISPR)-CRISPR associated (Cas) (CRISPR-
Cas) endonuclease system.
[0017] In a second aspect, an isolated crRNA including a length-modified
form of
formula (I) is provided:
5'-X¨Z-3' (I),
wherein X represents sequences comprising a target-specific protospacer
domain comprising about 20 target-specific nucleotides, and Z represents
sequences
comprising a universal tracrRNA-binding domain comprising about 20
nucleotides. The
isolated crRNA displays activity in a Clustered Regularly Interspaced Short
Palindromic
Repeats (CRISPR)-CRISPR associated (Cas) (CRISPR-Cas) endonuclease system.
[0018] In a third aspect, an isolated tracrRNA including a chemically-
modified form
of one of SEQ ID NOs.:2, 18, 30-33 and 36-39 is provided. The isolated
tracrRNA
displays activity in a Clustered Regularly Interspaced Short Palindromic
Repeats
(CRISPR)-CRISPR associated (Cas) (CRISPR-Cas) endonuclease system.
[0019] In a fourth aspect, isolated crRNA including a chemically-modified
form of
formula (I) is provided:
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
6
5'-X¨Z-3' (I),
wherein X represents sequences comprising a target-specific protospacer
domain comprising from about 17 nucleotides to about 20 nucleotides, and Z
represents
sequences comprising a universal tracrRNA-binding domain comprising about 12
nucleotides to about 19 nucleotides. The isolated crRNA displays activity in a
Clustered
Regularly Interspaced Short Palindromic Repeats (CRISPR)-CRISPR associated
(Cas)
(CRISPR-Cas) endonuclease system.
BRIEF DESCRIPTION OF THE DRAWINGS
[0020] FIG. 1 is an illustration of a wild-type (WT) natural 2-part
crRNA:tracrRNA
complex with a 42 base unmodified crRNA (SEQ ID No. 46) and an 89 base
unmodified
tracrRNA (SEQ ID No. 18). Lowercase letters represent RNA.
[0021] FIG. 2 is an illustration of a 99 base artificial single-guide RNA
(SEQ ID
NO: 428) (sgRNA) that fuses the crRNA and tracrRNA elements into a single
sequence
through the addition of a new hairpin loop. Lowercase letters represent RNA.
[0022] FIG. 3 shows an alignment of the full-length and truncated tracrRNA
species
studied in Example 2. Sequences are RNA and are shown 5'-3'. Alignment is
based
upon the 89 base WT tracrRNA sequence at the top (SEQ ID No. 18). Internal
gaps
represent sites of internal truncation/deletion. Uppercase letters represent
RNA.
[0023] FIG. 4 shows an alignment of the full-length and truncated crRNA and
tracrRNA species studied in Example 3. Alignment is based upon the 42 base WT
crRNA (SEQ ID No. 46) and 89 base WT tracrRNA (SEQ ID No. 18) sequences at the
top of their respective groupings. The 20 base 5'-domain in the crRNAs is
sequence-
specific and targets human HPRT1. The 3'-domain in underlined and binds to a
region
towards the 5'-end of the tracrRNA. The 5'-domain in the tracrRNA is
underlined that
binds the 3'-end of the crRNA. Uppercase letters represent RNA.
[0024] FIG. 5 is an illustration of a truncated 2-part crRNA:tracrRNA
complex with
a 36 base crRNA (SEQ ID No. 48) and a 67 base tracrRNA (SEQ ID No. 2).
Lowercase
letters represent RNA.
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
7
[0025] FIG. 6 is a schematic showing structure of one embodiment of an
optimized
truncated and chemically-modified tracrRNA (SEQ ID No. 134). Length is 67
bases.
RNA is lower case and 2'0Me RNA is uppercase. Phosphorothioate (PS)
internucleotide linkages are indicated by "*". Residues which lead to
substantial loss of
function when converted from RNA to 2'0Me RNA are identified by large arrows
and
residues which lead to a moderate loss of function when converted from RNA to
2'0Me
RNA are identified by small arrows.
[0026] FIG. 7 is a schematic showing structure of one embodiment of an
optimized
truncated and chemically-modified crRNA (SEQ ID No. 239). Length is 36 bases.
RNA
is lower case and 2'0Me RNA is uppercase. Phosphorothioate (PS)
internucleotide
linkages are indicated by "*". Residues which lead to substantial loss of
function when
converted from RNA to 2'0Me RNA are identified by large arrows and residues
which
lead to a moderate loss of function when converted from RNA to 2'0Me RNA are
identified by small arrows. The 5'-end 20 base protospacer target-specific
guide domain
is indicated, which in this case is sequence specific to the human HPRT 1
gene. The 3'-
end 16 base tracrRNA binding domain is indicated.
[0027] FIG. 8 is a schematic showing structure of one embodiment of the
optimized
truncated/modified crRNA:tracrRNA complex as employed in Example 8. The crRNA
is positioned at the top with the 5'-protospacer domain 20 base underlined,
which in this
case is specific for target human HPRT 1 site 38285; the 3'-end is the 16 base
tracrRNA
binding domain. The tracrRNA is aligned below. RNA is lower case, 2'0Me RNA is
uppercase, and "*" indicates a phosphorothioate internucleotide linkage
modification.
This figure shows the complex formed by crRNA SEQ ID No. 178 and tracrRNA SEQ
ID No. 100.
[0028] FIG. 9 is a schematic showing structure of one embodiment of the
optimized
truncated/modified crRNA:tracrRNA complex that is highly modified. The crRNA
is
positioned at the top with the 5'-protospacer domain 20 base underlined, which
in this
case is specific for target human HPRT 1 site 38285; the 3'-end is the 16 base
tracrRNA
binding domain. The tracrRNA is aligned below. RNA is lower case, 2'0Me RNA is
uppercase, and "*" indicates a phosphorothioate internucleotide linkage
modification.
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
8
This figure shows the complex formed by crRNA SEQ ID No. 446 and tracrRNA SEQ
ID No. 134.
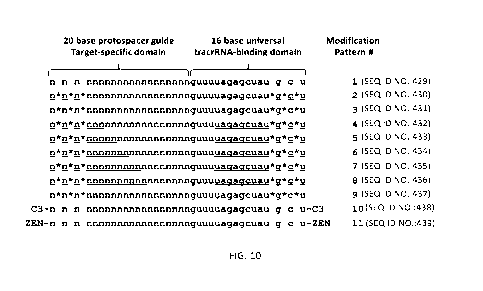
[0029] FIG. 10 is a schematic showing the crRNA modification patterns
employed in
Example 10. Oligonucleotide sequences (SEQ ID NOS 429-439, respectively, in
order
of appearance) are shown 5'-3'. Lowercase = RNA; Underlined = 2'-0-methyl RNA;
C3
= C3 spacer (propanediol modifier); * = phosphorothioate internucleotide
linkage; ZEN
= napthyl-azo modifier. The 5'-target specific protospacer domain is
indicated. Bases
are indicated by "N" in this domain as sequence is different for each target
site, although
the modification pattern employed remains constant. The 3'-universal tracrRNA
binding
domain is indicated. Modification patterns are numbered for reference between
Table 10
and FIG. 10.
[0030] FIG. 11 is a plot of the data in Table 10 showing the functional
gene editing
observed using the T7E1 assay in mammalian cells using crRNAs made with 11
different modification patterns tested at 12 different sites in the human
HPRT1 gene. All
crRNA variants were paired with an optimized, modified tracrRNA (SEQ ID No.
100).
[0031] FIG. 12 is a schematic showing structure of one embodiment of the
optimized
truncated/modified crRNA:tracrRNA complex that is highly modified using crRNA
Mod
Pattern 6 that is universal and can be applied in any sequence context. The
crRNA (SEQ
ID NO: 440) is positioned at the top with the 5'-protospacer domain 20 base
underlined
(N-bases); the 3'-end is the 16 base tracrRNA binding domain. The tracrRNA is
aligned
below (SEQ ID No. 134). RNA is lower case, 2'0Me RNA is uppercase, and "*"
indicates a phosphorothioate internucleotide linkage modification.
[0032] FIG. 13 shows a plot of RT-qPCR data from HEK-Cas9 cells transfected
with
different CRISPR gRNAs showing relative expression levels of IFIT1 and IFITM1,
2
genes involved in interferon signaling pathways.
DETAILED DESCRIPTION OF THE INVENTION
[0033] Aspects of this invention relate to modified compositions for use in
CRISPR
systems, and their methods of use.
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
9
[0034] The term "oligonucleotide," as used herein, refer to
polydeoxyribonucleotides
(containing 2-deoxy-D-ribose), polyribonucleotides (containing D-ribose), and
to any
other type of polynucleotide which is an N glycoside of a purine or pyrimidine
base (a
single nucleotide is also referred to as a "base" or "residue"). There is no
intended
distinction in length between the terms "nucleic acid", "oligonucleotide" and
"polynucleotide", and these terms can be used interchangeably. These terms
refer only
to the primary structure of the molecule. Thus, these terms include double-
and single-
stranded DNA, as well as double- and single-stranded RNA. For use in the
present
invention, an oligonucleotide also can comprise nucleotide analogs in which
the base,
sugar or phosphate backbone is modified as well as non-purine or non-
pyrimidine
nucleotide analogs. An oligonucleotide may comprise ribonucleotides,
deoxyribonucleotides, modified nucleotides (e.g., nucleotides with 2'
modifications,
synthetic base analogs, etc.) or combinations thereof.
[0035] Compositions of the present invention include any modification that
potentially reduces activation of the innate immune system. Modifications can
be placed
or substituted at a conventional phosphodiester linkage, at the ribose sugar,
or at the
nucleobase of RNA. Such compositions could include, for example, a modified
nucleotide such as 2'-0-methly-modified RNAs.
[0036] More broadly, the term "modified nucleotide" refers to a nucleotide
that has
one or more modifications to the nucleoside, the nucleobase, pentose ring, or
phosphate
group. For example, modified nucleotides exclude ribonucleotides containing
adenosine
monophosphate, guanosine monophosphate, uridine monophosphate, and cytidine
monophosphate and deoxyribonucleotides containing deoxyadenosine
monophosphate,
deoxyguanosine monophosphate, deoxythymidine monophosphate, and deoxycytidine
monophosphate. Modifications include those naturally occurring that result
from
modification by enzymes that modify nucleotides, such as methyltransferases.
Modified
nucleotides also include synthetic or non-naturally occurring nucleotides.
Modifications
also include base analogs and universal bases. Synthetic or non-naturally
occurring
modifications in nucleotides include those with 2' modifications, e.g., 2'-0-
alkyl
(including 2'-0-methyl), 2'-fluoro, 2'-methoxyethoxy, 2'-allyl, 2'-042-
(methylamino)-
2-oxoethyl], 4'-thio, bicyclic nucleic acids, 4'-CH2-0-2'-bridge, 4'-(CH2)2-0-
2'-
bridge, 2'-LNA, and 2'-0-(N-methylcarbamate) or those comprising base analogs.
Such
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
modified groups are described, e.g., in Eckstein et al., U.S. Pat. No.
5,672,695 and
Matulic-Adamic et al., U.S. Pat. No. 6,248,878.
[0037] The use of 2'-0-methyl has been documented in siRNA literature (See
Behlke, M.A., Oligonucleotides, 2008. 18(4): p. 305-19) as well as in mRNA
delivery
(see Sahin, U. et al., Nat Rev Drug Discov, 2014. 13(10): p. 759-80). Sahin et
al.,
describes modifications of mRNA therapeutics that extend beyond 2'-0Me
modification
and "non-immunogenic" mRNA.
[0038] The term "ribonucleotide" encompasses natural and synthetic,
unmodified
and modified ribonucleotides. Modifications include changes to the sugar
moiety, to the
base moiety and/or to the linkages between ribonucleotides in the
oligonucleotide.
[0039] The term "Cas9 protein" encompasses wild-type and mutant forms of
Cas9
having biochemical and biological activity when combined with a suitable guide
RNA
(for example sgRNA or dual crRNA:tracrRNA compositions) to form an active
CRISPR-Cas endonuclease system. This includes orthologs and Cas9 variants
having
different amino acid sequences from the Streptococcus pyogenese Cas9 employed
as
example in the present invention.
[0040] The term "length-modified," as that term modifies RNA, refers to a
shortened
or truncated form of a reference RNA lacking nucleotide sequences or an
elongated form
of a reference RNA including additional nucleotide sequences.
[0041] The term "chemically-modified," as that term modifies RNA, refers to
a form
of a reference RNA containing a chemically-modified nucleotide or a non-
nucleotide
chemical group covalently linked to the RNA. Chemically-modified RNA, as
described
herein, generally refers to synthetic RNA prepared using oligonucleotide
synthesis
procedures wherein modified nucleotides are incorporated during synthesis of
an RNA
oligonucleotide. However, chemically-modified RNA also includes synthetic RNA
oligonucleotides modified with suitable modifying agents post-synthesis.
[0042] Applicants have discovered novel crRNA and tracrRNA oligonucleotide
compositions that display robust activity in the Clustered Regularly
Interspaced Short
Palindromic Repeats (CRISPR)-CRISPR associated (Cas) (CRISPR-Cas) endonuclease
system. The oligonucleotide compositions include length-modified forms of
crRNA and
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
11
tracrRNA, as well as chemically-modified forms of crRNA and tracrRNA. The
length-
modified forms of crRNA and tracrRNA enable one to prepare active forms of
these
RNAs with cost-effective and efficient oligonucleotide synthesis protocols
routinely
available. The chemically-modified forms of crRNA and tracrRNA provide one
with
active agents tunable with certain specific properties, such as improved
stability in
cellular and in vivo contexts. The length-modified forms of crRNA and tracrRNA
can
also include modifications, thereby enabling access to a broad range of
compositions
having activity in CRISPR-Cas endonuclease system contexts. These
oligonucleotide
compositions and their properties in the CRISPR-Cas endonuclease system are
described
below.
[0043] Length-modified forms of crRNA and tracrRNA
[0044] FIG.1 depicts a representation of the wild-type S. pyogenes
crRNA:tracrRNA
complex, wherein an exemplary isolated crRNA (SEQ ID No. 46) is paired with an
isolated tracrRNA (SEQ ID No. 18). In a first aspect, an isolated tracrRNA
including a
length-modified form of SEQ ID NO.:18 is provided. The isolated tracrRNA
displays
activity in the CRISPR-Cas endonuclease system. In one respect, the isolated
tracrRNA
includes a length-modified form of SEQ ID NO.:18 nucleotide having deleted
sequence
information. In some embodiments, the length-modified form of SEQ ID NO.:18
includes shortened or truncated forms of SEQ ID NO.:18, wherein SEQ ID NO.:18
can
be shortened by 1 to 20 nucleotides at the 5'-end and by 1-10 nucleotides at
the 3'-end.
Such shortened or truncated forms of SEQ ID NO.:18 retain activity when paired
with a
functionally competent crRNA in the CRISPR-Cas endonuclease system. Where
shortening of the 5'-end of the tracrRNA is performed and extends into
sequence that
pairs with the 3'-end of the crRNA, improved activity may be obtained using
chemical
modification that enhance binding affinity in these domains. Where shortening
of the 3'-
end of the crRNA is performed and extends into sequence that pairs with the 5'-
end of
the tracrRNA, improved activity may be obtained using chemical modification
that
enhance binding affinity in these domains. Preferred examples of a length-
modified form
of SEQ ID NO.:18 having a shortened or truncated form include SEQ ID NOs:2, 30-
33
and 36-39. A highly preferred example of a length-modified form of SEQ ID
NO.:18
having a shortened or truncated form includes SEQ ID NO:2. For each of the
foregoing
exemplary length-modified forms of SEQ ID NO.:18 having a shortened or
truncated
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
12
form, SEQ ID NOs.:2, 30-33 and 36-69 can consist of chemically non-modified
nucleotides.
[0045] In a second aspect, an isolated crRNA comprising a length-modified
form of
formula (I) is provided:
5'-X--Z-3' (I),
wherein X represents sequences including a target-specific protospacer
domain, and Z represents sequences including a tracrRNA-binding domain.
[0046] The target-specific protospacer domain (X domain of formula (I))
typically
includes about twenty nucleotides having complementarity to a region of DNA
targeted
by the CRISPR-Cas endonuclease system. The tracrRNA-binding domain (the Z
domain
of formula (I)) typically includes about 20 nucleotides in most CRISPR
endonuclease
systems (in the native S.py. version, this domain is 22 nucleotides). The
isolated crRNA
displays activity in the CRISPR-Cas endonuclease system.
[0047] In one respect, the isolated crRNA includes a length-modified form
of
formula (I) having deleted sequence information. In some embodiments, the
length-
modified form of formula (I) includes shortened or truncated forms of formula
(I),
wherein formula (I) can be shortened by 1-8 nucleotides at the 3'-end of the Z
domain.
The length-modified form of formula (I) can be shortened at the 5-end of the X-
domain
to accommodate a target-specific protospacer domain having 17, 18, 19 or 20
nucleotides. Highly preferred examples of such length-modified form of formula
(I)
include target-specific protospacer domain having 19 or 20 nucleotides. The
exemplary
length-modified forms of formula (I) having a shortened or truncated form with
a target-
specific protospacer (X-domain) of 17-20 nucleotides in length and/or lacking
1-8
nucleotides at the 3'-end of the Z-domain can consist of chemically non-
modified
nucleotides.
[0048] Such shortened or truncated forms of formula (I) retain activity
when paired
with a competent tracrRNA in the CRISPR-Cas endonuclease system. Preferred
embodiments of isolated crRNA of formula (I) having a length-modified form of
formula
(I) can include chemically non-modified nucleotides and chemically modified
nucleotides.
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
13
[0049] Chemically-modified forms of crRNA and tracrRNA
[0050] In a third aspect, an isolated tracrRNA including a chemically-
modified
nucleotide or a non-nucleotide chemical modifier is provided. The isolated
tracrRNA
displays activity in the CRISPR-Cas endonuclease system. In one respect, the
isolated
tracrRNA includes a chemically-modified nucleotide having a modification
selected
from a group consisting of a ribose modification, an end-modifying group, and
internucleotide modifying linkages. Exemplary ribose modifications include 2'0-
alkyl
(e.g., 2'0Me), 2'F, bicyclic nucleic acid, and locked nucleic acid (LNA).
Exemplary
end-modifying groups include a propanediol (C3) spacer and napthyl-azo
modifier (N,N-
diethy1-4-(4-nitronaphthalen-1-ylazo)-phenylamine, or "ZEN"), and an inverted-
dT
residue. Exemplary internucleotide modifying linkages include phosphorothioate
modification. In one respect, the isolated tracrRNA having a chemically-
modified form
include SEQ ID NO. :46 and length-modified forms thereof, such as shortened or
truncated forms of SEQ ID NO. :46. Preferred shortened or truncated forms of
SEQ ID
NO. :46 having a chemically-modified nucleotide include SEQ ID NOs:2, 30-33
and 36-
39 having a chemically-modified nucleotide. Yet other examples of isolated
tracrRNA
having a chemically-modified nucleotide with robust activity in the CRISPR-Cas
endonuclease system are presented in the Examples.
[0051] In a fourth aspect, an isolated crRNA including a chemically-
modified
nucleotide is provided. The isolated crRNA displays activity in the CRISPR-Cas
endonuclease system. In one respect, the isolated crRNA includes a chemically-
modified
nucleotide having a modification selected from a group consisting of a ribose
modification, an end-modifying group, and internucleotide modifying linkage.
Exemplary ribose modifications include 2'0-alkyl (e.g., 2'0Me), 2'F, bicyclic
nucleic
acid, and locked nucleic acid (LNA). Exemplary end-modifying groups include a
propanediol (C3) spacer and napthyl-azo modifier (N,N-diethy1-4-(4-
nitronaphthalen-1-
ylazo)-phenylamine, or "ZEN"), and an inverted-dT residue. Exemplary
internucleotide
modifying linkages include phosphorothioate modification. In one respect, the
isolated
crRNA having a chemically-modified form include crRNA of formula (I) and
length-
modified forms thereof. Preferred shortened or truncated forms of crRNA of
formula (I)
having a chemically-modified nucleotide include SEQ ID NOs.:429-439. Highly
preferred examples of an isolated crRNA having a chemically-modified
nucleotide
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
14
include SEQ ID NOs.:434 and 435. These particular isolated crRNA species
represent
"universal" crRNAs having a chemically-modified nucleotide showing high
activity
when combined with a competent tracrRNA in the CRISPR-Cas endonuclease system.
Yet other examples of isolated crRNA having a chemically-modified nucleotide
with
robust activity in the CRISPR-Cas endonuclease system are presented in the
Examples.
[0052] The foregoing isolated, length-modified and chemically-modified of
crRNA
and tracrRNA preferably include chemical modifications at the 2'-OH groups
(for
example, 2'0Me, 2'F, bicyclic nucleic acid, locked nucleic acid, among others)
and end-
blocking modifications (for example, ZEN, C3 spacer, inverted-dT). Use of both
types of
general modifications provides isolated, length-modified and chemically-
modified of
crRNA and tracrRNA with biochemical stability and immunologic tolerance for
isolated,
length-modified and chemically-modified of crRNA and tracrRNA in biological
contexts.
[0053] The foregoing isolated, length-modified and chemically-modified of
crRNA
and tracrRNA can be mixed in different combinations to form active
crRNA:tracrRNA
as the guide RNA for Cas9. For example, an isolated, length-modified tracrRNA
can be
combined with an isolated chemically-modified crRNA to form an active
crRNA:tracrRNA as the guide RNA for Cas9. The Examples provide illustrations
of
different combinations of isolated, length-modified and chemically-modified of
crRNA
and tracrRNA resulting in active crRNA:tracrRNA as the guide RNA for Cas9.
[0054] The extent to which one needs particular chemically-modified
nucleotides
included in one (or both) of the isolated, length-modified and chemically-
modified
crRNA and tracrRNA depends upon the application for which the resultant active
crRNA:tracrRNA serves as the guide RNA for Cas9. In certain biochemical assays
of the
CRISPR-Cas endonuclease system, particularly where nucleases can be minimized
or
absent, one may not need extensively chemically-modified crRNA and tracrRNA to
effect robust activity of the resultant guide RNA for Cas9 of the CRISPR-Cas
endonuclease system. This is attributed to the fact that chemically-modified
nucleotides
that confer resistance to nucleases are not necessary when nucleases are
minimal or
absent. In certain biological (in vivo) contexts, wherein a mixture including
crRNA and
tracrRNA is delivered to cells inside carrier vehicles, such as liposome
nanoparticles, the
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
isolated length-modified and chemically-modified crRNA and tracrRNA may
require
less extensive chemically-modified nucleotides than mixtures of crRNA and
tracrRNA
delivered directly into the blood stream or injected into organ systems as
isolated,
"naked," RNA mixtures. The extent of chemical modification present in
chemically-
modified crRNA and tracrRNA can dictate the half-life of the relevant RNA
molecules
in vivo (that is, in the relevant biological context, such as, for example, in
the blood
stream or inside cells). Accordingly, the modification profile of chemically-
modified
crRNA and tracrRNA can be used to fine tune the biochemical and biological
activity of
the resultant crRNA:tracrRNA duplexes as a guide RNA for Cas9 in the CRISPR-
Cas
endonuclease system.
[0055] Although the prior art focuses on the structure of Cas9 as it
interacts with a
sgRNA, the disclosed design patterns described herein contemplate the
aforementioned
crRNA:tracrRNA dual RNA systems. A single strand guide RNA offers several
benefits,
such as simplicity of a therapeutic design. However, standard solid phase
phosphoramidite RNA synthesis shows diminishing yields for oligonucleotides as
length
increases and this problem becomes more apparent as length exceeds 60-70
bases. This
precludes robust, cost-effective synthesis of some tracrRNAs as well as the
chimeric
sgRNA, especially at larger scales needed for some commercial or therapeutic
applications. For this reason, the invention contemplates embodiments of not
only
sgRNA, but also alternate dual crRNA:tracrRNA as the guide RNA for Cas9.
However,
an isolated guide RNA having robust activity when combined with Cas9 in the
CRISPR-
Cas endonuclease system can be engineered by linkage or synthesis of
appropriate
crRNA and tracrRNA as an artificial, unimolecular sgRNA based upon the
isolated,
length-modified and chemically-modified forms of crRNA and tracrRNA provided
herein. Long single guides of this type may be obtained by direct synthesis or
by post-
synthetic chemical conjugation of shorter strands.
[0056] The design of length-modified and chemically-modified tracrRNA
compositions addresses the potential synthetic issues associated with tracrRNA
oligonucleotides that are >80 nucleotides in length. The coupling efficiency
of 2'-0Me-
modified RNA monomers (effectively containing a protecting group on the 2'-OH)
is
greater than RNA monomer coupling. Incorporating 2'-0Me modified RNAs provides
some advantages. First, it allows for longer oligonucleotides to be
synthesized as either
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
16
full 2'-0Me or RNA/2'-0Me mixed oligonucleotides. Secondly, the methods and
compositions of the invention lead to synthesis and transfection of
crRNA:tracrRNA that
can evade detection by the immune system. It is well known that exogenous,
unmodified
RNAs trigger an innate immune response in mammalian cells as well as whole
animals.
Using 2'0Me-modified oligonucleotides can confer RNA stability to nucleases (a
third
advantage) as well as reduce cell death and toxicity associated with
immunogenic
triggers. These advantages are not unique to 2'-0Me modification, per se, as
the other
disclosed modified nucleotides having different chemical moieties (for
example, 2'F,
other 2'0-alkyls, LNA, and other bicyclic nucleotides) can offer similar
benefits and
advantages in terms of conferring resistance to nucleases.
[0057] In another embodiment, the tracrRNA portion complementary to the
crRNA
contains at least one modified nucleotide, and in a further embodiment the
tracrRNA
portion complementary to the crRNA is comprised of more than 10% modified
residues,
and in a further embodiment the tracrRNA portion not complementary to the
crRNA is
comprised of more than 50% modified residues, and a further embodiment the
tracrRNA
portion not complementary to the crRNA is comprised of more than 90% modified
residues.
[0058] In another embodiment, the crRNA portion is unmodified and the
tracrRNA
portion is comprised of at least one modified nucleotide. In a further
embodiment the
crRNA portion is unmodified and the tracrRNA portion is comprised of more than
10%
modified bases.
[0059] In another embodiment, an isolated crRNA of formula (I) is designed
with
modifications that are empirically determined. As depicted in FIGs. 7 and 10,
the 12
nucleotides at the 3'-end of the Z domain (the tracrRNA-binding domain) and
the 10-12
nucleotides at the 5'-end of the X domain (within the protospacer domain)
represent
universal nucleotides amenable to substitution with chemically-modified
nucleotides,
wherein the resultant RNAs retain robust activity in the CRISPR-Cas
endonuclease
system. Yet other nucleotides within the 5'-end of the Z domain (the tracrRNA-
binding
domain) are intolerant to substitution with chemically-modified nucleotides
(FIG. 7). Yet
the ability of other sites within an isolated crRNA of formula (I) to accept
chemically-
modified nucleotides and retain activity in the CRISPR-Cas endonuclease system
is
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
17
largely determined empirically. The tracrRNA binding domain (Z domain) of the
crRNA is constant (i.e., sequence does not change as target site varies), so
the
modifications patterns described herein are universal to all crRNAs regardless
of target
site and can be broadly applied. The protospacer (X domain) of the crRNA
varies with
target, and the tolerance of some of the base positions within this domain to
chemical
modification vary with sequence context and, if maximal chemical modification
of a site
is desired, may benefit from empiric optimization. However, some of the
residues within
the target-specific protospacer (X) domain can be modified without
consideration to
sequence context. The 10-12 residues at the 5'-end of this domain can be
substituted
with 2'-modified residues with the expectation that full activity of the
modified crRNA
will be maintained. The remaining 8-10 bases towards the 3'-end of the
protospacer (X)
domain may tolerate modification or may not, depending on sequence context.
One
sequence context where 17 out of the 20 bases of the protospacer (X) domain
can be
modified while maintaining full activity are shown in FIG. 7. Sites were
modification
compromised activity are indicated.
[0060] The applications of Cas9-based tools are many and varied. They
include, but
are not limited to: plant gene editing, yeast gene editing, rapid generation
of
knockout/knockin animal lines, generating an animal model of disease state,
correcting a
disease state, inserting a reporter gene, and whole genome functional
screening.
[0061] The utility of the present invention is further expanded by
including mutant
versions of Cas enzymes, such as a DlOA and H840a double mutant of Cas9 as a
fusion
protein with transcriptional activators (CRISPRa) and repressors (CRISPRi)
(see Xu, T.,
et al., Appl Environ Microbiol, 2014. 80(5): p. 1544-52). The Cas9-sgRNA
complex also
can be used to target single-stranded mRNA as well (see O'Connell, M.R., et
al., Nature,
516:263, 2014). In the same way as targeting dsDNA, crRNA:tracrRNA can be used
with a PAMmer DNA oligonucleotide to direct Cas9 cleavage to the target mRNA
or use
it in the mRNA capture assay described by O'Connell.
[0062] By utilizing an approach to deliver synthetic RNA oligonucleotides
for
CRISPR/Cas9 applications, it is possible to 1) use mass spectroscopy to
confirm discrete
RNA sequences, 2) selectively insert 2'-0Me modified RNAs in well-tolerated
locations
to confer stability and avoid immunogenicity yet retain functional efficacy,
3)
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
18
specifically control the amount of RNA that is introduced into cells for a
controlled
transient effect, and 4) eliminate concern over introducing dsDNA that would
be
endogenously transcribed to RNA but could also become substrate in either
homology-
directed repair pathway or in non-homologous end joining resulting in an
integration
event. These integration events can lead to long term undesired expression of
crRNA or
tracrRNA elements. Further, integration can disrupt other genes in a random
and
unpredictable fashion, changing the genetic material of the cell in undesired
and
potentially deleterious ways. The present invention is therefore desirable as
a means to
introduce transient expression of elements of the CRISPR pathway in cells in a
way
which is transient and leaves no lasting evidence or change in the genome
outside of
whatever alteration is intended as directed by the crRNA guide.
[0063] CRISPR-Cas endonuclease systems
[0064] A competent CRISPR-Cas endonuclease system includes a
ribonucleoprotein
(RNP) complex formed with isolated Cas9 protein and isolated guide RNA
selected from
one of a dual crRNA:tracrRNA combination and a chimeric sgRNA. In some
embodiments, isolated length-modified and/or chemically-modified forms of
crRNA and
tracrRNA are combined with purified Cas9 protein (for example, SEQ ID NOs.:407-
410), an isolated mRNA encoding Cas9 protein (for example, SEQ ID NO.:413), or
a
gene encoding Cas9 protein (for example, SEQ ID NOs.: 411 and 412) in an
expression
vector. In certain assays, isolated length-modified and/or chemically-modified
forms of
crRNA and tracrRNA can be introduced into cell lines that stably express Cas9
protein
from an endogenous expression cassette encoding the Cas9 gene. In other
assays, a
mixture of length-modified and/or chemically-modified forms of crRNA and
tracrRNA
in combination with either Cas9 mRNA or Cas9 protein can be introduced into
cells.
EXAMPLE 1
[0065] This example illustrates functioning of chemically modified and
truncated
guide RNAs in an in vitro Cas9 DNA cleavage assay.
[0066] CrRNA and tracrRNA oligonucleotides were synthesized having various
chemical modifications and truncations relative to the WT sequences as
indicated (Table
1).
SUBSTITUTE SHEET (RULE 26)
(9Z '3'111N) JAMAS uuiisaris
bramnbnobnynobvfmnrambbnbormovovvoonvnynn 8 Vt
ra-mobribbonbvboovobbribvvvy
L9 17
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
DE
I t
bramnbnobnynobvfmnrambbnbormovovvoonvnynn
=ram
68 nnobribbon6vboovo.6.6n6vvvvvbrmovvonvnnboonbv 9
no.6.6vvnyvvynnbvvobvnvobvovvvvonnvoovv.6.6t-Ln6
I t
bramnbnobnynobvfmnrambbnbormovovvoonvnynn
ra-mobribbonbvboovobbribvvvy
L9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
V
I t
bramnbnobnynobvfmnrambbnbormovovvoonvnynn
ra-mobribbonbvboovobbribvvvy
L9 17
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
DZ
It
bramnbnobnynobvfmnrambbnbormovovvoonvnynn
=ram
68 nnobribbon6vboovo.6.6n6vvvvvbrmovvonvnnboonbv 9
no.6.6vvnyvvynnbvvobvnvobvovvvvonnvoovv.6.6t-Ln6
It'
bramnbnobnynobvfmnrambbnbormovovvoonvnynn
ra-mobribbonbvboovobbribvvvy
L9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
VZ
It'
bramnbnobnynobvfmnrambbnbormovovvoonvnynn
ra-mobribbonbvboovobbrib
L vvvy
9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
DI
S
nobnynobvfmnram.6.6nbormovovvoonvnynn
=ram
68
nnobribbon6vboovo.6.6n6vvvvvbrmovvonvnnboonbv
no.6.6vvnyvvynnbvvobvnvobvovvvvonnvoovv.6.ii i
6t-Ln6
S
nobnynobvfmnram.6.6nbormovovvoonvnynn
ra-mobribbonbvboovobbribvvvy
L9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
VI
S
nobnynobvfmnram.6.6nbormovovvoonvnynn
.uud
a3uanbas vNllnuil IN al
aunualp tOuarl
VMll
a3uanbas vtop3 Oas
.13C.11/.13
*SiTd VNIII3U.11 puu vtop3 snopun tinAt asuapnuopua
6su3 q vNa la&tul ullall jo aun1al3 jo sa!pno 1u3!tuatpopi Inn ui
61
Zr6990/SIOZSI1/13.1
16O01/910Z OM
ZT-90-LTOZ 890L6Z0 VD
(9Z '3'111N) JAMS uuiisaris
ra-mobribbonbvboovobbribvvvy
L9ZI
vbrmovvonvnnbo¨onbvno.6.6vvri.v¨vvvri.n6vvobvnvobv
NI
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn
ra-mobribbonbvboovobbribvvvy
L9 II
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
+++ DI
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn
ra-mobribbonbvboovobbribvvvy
L9 .17
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
39
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn 01
=ram
68 nnobribbon6vboovo.6.6n6vvvvvbrmovvonvnnboonbv 9
no.6.6vvnyvvynnbvvobvnvobvovvvvonnvoovv.6.6t-Ln6
EE9
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn 01
ra-mobribbonbvboovobbribvvvy
L9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
V9
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn 01
ra-mobribbonbvboovobbribvvvy
L9 .17
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
DS
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn 6
=ram
68 nnobribbon6vboovo.6.6n6vvvvvbrmovvonvnnboonbv 9
no.6.6vvnyvvynnbvvobvnvobvovvvvonnvoovv.6.6t-Ln6
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn 6
ra-mobribbonbvboovobbribvvvy
L9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
VS
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn 6
ra-mobribbonbvboovobbrib
L vvvy
9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
3.17
bramnbnobnynobvfmnrambbnbormovovvoonvnynn 8
=ram
68 nnobribbon6vboovo.6.6n6vvvvvbrmovvonvnnboonbv 9
no.6.6vvnyvvynnbvvobvnvobvovvvvonnvoovv.6.6t-Ln6
at
1 t
bramnbnobnynobvfmnrambbnbormovovvoonvnynn 8
ra-mobribbonbvboovobbribvvvy
L9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
J!ud
a3uanbas vNllnuil =oNI
aunualp tOual
VIµ111
a3uanbas vtop3 Oas
.13C.11/.13
OZ
Zr6990/SIOZSI1/13.1
I6001/910Z OM
ZT-90-LTOZ 890L6Z0 VD
(9Z '3'111N) JAMS uuiisaris
+++ SE
nobnynobvfmnram.6.6nbormovovvoonvnynn 91 991
ra-mobribbonbvboovobbribvvvy
L9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
+++
V9I
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn 91
ra-mobribbonbvboovobbribvvvy
L9 El
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
IS
SE
nobnynobvfmnram.6.6nbormovovvoonvnynnci
ra-mobribbonbvboovobbribvvvy
L9ZI
vbrmovvonvnnbo¨onbvno.6.6vvri.v¨vvvri.n6vvobvnvobv
SE
nobnynobvfmnram.6.6nbormovovvoonvnynnci
ra-mobribbonbvboovobbribvvvy
L9 ii
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
+++
OSI
SE
nobnynobvfmnram.6.6nbormovovvoonvnynnci
ra-mobribbonbvboovobbrib
L vvvy
9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
VSI
SE
nobnynobvfmnram.6.6nbormovovvoonvnynnci
ra-mobribbonbvboovobbribvvvy
L9 Et
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
SE nobnynobvfmnram.6.6nbormovovvoonvnynn t
ra-mobribbonbvboovobbribvvvy
L9ZI
vbrmovvonvnnbo¨onbvno.6.6vvri.v¨vvvri.n6vvobvnvobv
SE nobnynobvfmnram.6.6nbormovovvoonvnynn t
ra-mobribbonbvboovobbribvvvy
L9 ii
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
0171
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn 171
ra-mobribbonbvboovobbribvvvy
L9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
VtI
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn 171
ra-mobribbonbvboovobbribvvvy
L9 El
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
SE nobnynobvfmnram.6.6nbormovovvoonvnynn
.uud
a3uanbas vNllnuil ai
aunualp tOual
VMll
a3uanbas vtop3 Oas
.13C.11/.13
TZ
Zr6990/SIOZSI1/13.1
I6001/910Z OM
ZT-90-LTOZ 890L6Z0 VD
(9Z '3'111N) JAMS uuiisaris
ra-mobribbonbvboovobbribvvvy
L9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
vol
SE
nabnenabebennnnbbnbannaeoeepanenenn tZ
ra-mobribbonbvboovobbrib
L vvvy
9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
+++ V6
SE
nabnynabvfmnnnn.6.6nbannavavvaanvnynn EZ
ra-mobribbonbvboovobbrib
L vvvy
9
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
VL
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn ZZ
=ram
68 nnobribbon6vboovo.6.6n6vvvvvbrmovvonvnnbo¨onbv 1Z
no.6.6vvri.v¨vvvri.n6vvobvnvoEmovvvvonnvoovv.6.6nn.6
TZ
t
bramnbnobnynobvfmnrambbnbormovovvoonvnynn
nnrLrLrL
68 nnobribbon6vboovo.6.6n6vvvvvbrmovvonvnnboonbv OZ
no n nn n nn o.6.6t-Ln6
.6.6vvyvvybvvobvvobvovvvvovovv
+++ dZ
It
bramnbnobnynobvfmnrambbnbormovovvoonvnynn
=ram
68 nnobribbon6vboovo.6.6n6vvvvvbrmovvonvnnboonbv 61
no n nn n nn o.6.6t-Ln6
.6.6vvyvvybvvobvvobvovvvvovovv
+++ HZ
It
bramnbnobnynobvfmnrambbnbormovovvoonvnynn
=ram
68 nnobribbon6vboovo.6.6n6vvvvvbrmovvonvnnboonbv 81
no n nn n nn o.6.6t-Ln6
.6.6vvyvvybvvobvvobvovvvvovovv
+++ QZ
It
bramnbnobnynobvfmnrambbnbormovovvoonvnynn
ra-mobribbonbvboovobbribvvvy
L9LI
vEn¨ri.ovvonvnnboonbvno.6.6vvri.vvvvri.nbvvobvnvobv
+++ HI
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn
ra-mobribbonbvboovobbribvvvy
L9 El
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
+++
'191
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn 91
ra-mobribbonbvboovobbribvvvy
L9ZI
vbrmovvonvnnbo¨onbvno.6.6vvri.v¨vvvri.n6vvobvnvobv
+++
)19I
SE
nobnynobvfmnram.6.6nbormovovvoonvnynn 91
ra-mobribbonbvboovobbribvvvy
L9 II
vbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnvobv
.uud
a3uanbas vNllnuil ai
aunualp tOual
VMll
a3uanbas vtop3 Oas
.13C.11/.13
ZZ
Zr6990/SIOZSI1/13.1
I6001/910Z OM
ZT-90-LTOZ 890L6Z0 VD
CA 02970683 2017-06-12
WO 2016/100951 PCT/US2015/066942
23
cr/tracr SE Q crRNA Sequence
RNA
Length Cleavage
pair ID No. tracrRNA Sequence
7 uuauauccaacacuucgugguuuuagagcuaugcuguuuug 41
3D guuggaaccauucaaaacagcauagcaaguuaaaauaaggcu
18 aguccguuaucaacuugaaaaaguggcaccgagucggugcuu 89
uuuuu
8 uuauauccaacacuucgugguuuuagagcuaugcuguuuug 41
4D guuggaaccauucaaaacagcauagcaaguuaaaauaaggcu
18 aguccguuaucaacuugaaaaaguggcaccgagucggugcuu 89
uuuuu
25 uuauauccaacacuucgugguuuuagagcuaugcuguuuug 41
8D guuggaaccauucaaaacagcauagcaaguuaaaauaaggcu
18 aguccguuaucaacuugaaaaaguggcaccgagucggugcuu 89
uuuuu
26 uuauauccaacacuucgugguuuuagagcuaugcuguuuug 41
13D guuggaaccauucaaaacagcauagcaaguuaaaauaaggcu
18 aguccguuaucaacuugaaaaaguggcaccgagucggugcuu 89
uuuuu
26 uuauauccaacacuucgugguuuuagagcuaugcuguuuug 41
131 guuggaaccauucaaaacagcauagcaaguuaaaauaaggcu
21
aguccguuaucaacuugaaaaaguggcaccgagucggugcuu 89
uuuuu
Oligonucleotide sequences are shown 5'-3'. Lowercase = RNA, Underlined = 2'-
0-methyl RNA, Italics = 2'-fluoro RNA. Lengths of the RNA oligonucleotides
are indicated (bases). The relative efficiency of cleavage of the DNA target
by
recombinant Cas9 with each of the crRNA:tracrRNA pairs as visualized by
agarose gel electrophoresis is indicated with "+++" indicating complete
cleavage,
"++" and "+" indicating intermediate levels of cleavage, and "-" indicating no
cleavage.
[0067] The crRNAs contained a 19 base protospacer guide sequence matching a
site
in the human HPRT1 gene adjacent to a suitable `NGG" PAM site. A 938 base pair
region from the human HPRT1 gene was cloned into the pCR-Blunt vector (Life
Technologies). The plasmid was linearized by digestion with the restriction
endonuclease XmaI (New England BioLabs) prior to use in the Cas9 cleavage
assay.
Sequence of the HPRT1 target fragment is shown below. The target PAM site is
indicated in bold font and the protospacer guide sequence binding site is
underlined.
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
24
HPRT1 target sequence. SEQ ID No. 27.
GAAT GTT GT GATAAAAGGT GAT GCTCACCTCTCCCACACCCTTTTATAGTTTAGGGATT GTATTTCCAAGG
TTTCTAGACTGAGAGCCCTTTTCATCTTTGCTCATTGACACTCTGTACCCATTAATCCTCCTTATTAGCTC
CCCTTCAATGGACACATGGGTAGTCAGGGTGCAGGTCTCAGAACTGTCCTTCAGGTTCCAGGTGATCAACC
AAGTGCCTTGTCTGTAGTGTCAACTCATTGCTGCCCCTTCCTAGTAATCCCCATAATTTAGCTCTCCATTT
CATAGTCTTTCCTT GGGT GT GTTAAAAGT GACCAT GGTACACTCAGCACGGAT GAAAT GAAACAGT
GTTTA
GAAACGTCAGTCTTCTCTTTTGTAATGCCCTGTAGTCTCTCTGTATGTTATATGTCACATTTTGTAATTAA
CAGCTTGCTGGTGAAAAGGACCCCACGAAGTGTTGGATATAAGCCAGACTGTAAGTGAATTACTTTTTTTG
TCAATCATTTAACCATCTTTAACCTAAAAGAGTTTTAT GT GAAAT GGCTTATAATT GCTTAGAGAATATTT
GTAGAGAGGCACATTT GCCAGTATTAGATTTAAAAGT GAT GTTTTCTTTATCTAAAT GAT GAATTAT GATT
CTTTTTAGTTGTTGGATTTGAAATTCCAGACAAGTTTGTTGTAGGATATGCCCTTGACTATAATGAATACT
TCAGGGATTTGAATGTAAGTAATTGCTTCTTTTTCTCACTCATTTTTCAAAACACGCATAAAAATTTAGGA
AAGAGAATT GTTTTCTCCTTCCAGCACCTCATAATTT GAACAGACT GAT GGTTCCCATTAGTCACATAAAG
CT GTAGTCTAGTACAGACGTCCTTAGAACT GGAACCT GGCCAGGCTAGGGT GACACTTCTT GTT GGCT GAA
ATAGTTGAACAGCTT
[0068] The crRNA and tracrRNA pairs were tested for the ability to direct
degradation of a linearized plasmid DNA containing a cloned fragment of the
human
HPRT1 gene by recombinant Spy Cas9 (New England BioLabs). The crRNA:tracrRNA
were annealed in Duplex Buffer (30 mM HEPES pH 7.5, 100 mM potassium acetate)
at
150 nM concentration. Spy Cas9 (15 nM) was preincubated with the
crRNA:tracrRNA
for 10 min at 37 C at a 1:1 molar ratio. Subsequently, 3 nM of the linearized
target
plasmid was added and incubated at 37 C for 1 hour. The reaction products were
separated on a 1% agarose gel at 125 V for 1 hour. Bands were visualized by
GelRed
(Biotium) post-staining according to the manufacturer's protocol. The gel was
imaged on
a UV-transilluminator and results are summarized in Table 1 above.
[0069] Native wild-type (WT) CRISPR RNAs have a 19-20 base protospacer
domain
(guide, which binds to a target nucleic acid) at the 5'-end and a 22 base
domain at the 3'-
end that binds to the tracrRNA. Thus WT crRNAs are 41-42 bases long. The WT
tracrRNA is 89 bases long. We observed that a WT type crRNA:tracrRNA pair
supported full cleavage of the target DNA (cr/tracrRNA pair 2D). We
additionally
observed that a truncated version of the reagents with a 35 base crRNA (19
base
protospacer and 16 base tracrRNA binding domain) paired with a 67 base
tracrRNA
supported full cleavage of the target RNA (cr/tracrRNA pair 1A). The crRNA
tracrRNA
binding region was truncated 6 bases at the 3'-end (SEQ ID No. 1). The
tracrRNA was
truncated at both ends (SEQ ID No. 2). Pairwise combinations of the short
crRNA with
the long tracrRNA showed cleavage as well as the long crRNA with the short
tracrRNA
(pair 2A). These findings are significant as it permits use of shorter RNA
components to
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
direct Cas9 target recognition and cleavage. Shorter RNA oligonucleotides are
less
expensive and less difficult for chemical synthesis, requiring less
purification and giving
higher yields than longer RNA oligonucleotides.
[0070] Some elements of the native crRNA and tracrRNA (Fig. 1) were deleted
to
make a functional sgRNA (Fig. 2). However, the amount of duplex nucleic acid
binding
the crRNA to the tracrRNA in the sgRNA is limited to 11 base pairs, which is
typically
too short for duplex formation in biological salt conditions. The complex is
stable in
sgRNA format due to the unimolecular hairpin structure, however the same
sequences
split into 2 RNAs would be unstable. It was therefore unclear what length of
duplex
domain was needed to make a minimal yet functional 2-molecule (2-part) CRISPR
complex, or if this complex would function to direct target cleavage by Cas9.
The
present example demonstrates that having as little of 15 bases base paired
permits a
function 2-part crRNA:tracrRNA complex that is competent to direct Cas9
nuclease
activity against a target complementary to the crRNA protospacer domain (SEQ
ID Nos.
land 2).
[0071] Complete chemical modification of the crRNA with 2'0Me RNA was not
tolerated (pair 3A and pair 5A). Further, complete 2'0Me modification of the
22 base
tracrRNA binding domain of the crRNA did not support target cleavage (pair 4A,
pair
6A) and complete 2'0Me modification of the protospacer guide domain did not
support
cleavage (pair 7A). Complete chemical modification of the tracrRNA with 2'0Me
RNA
was also not tolerated (pair 1B, 1C and pair 2B, 2C).
[0072] Importantly, some highly 2'0Me-modified versions of both CRISPR RNA
species did support cleavage. Pair 1K shows high cleavage activity with a
tracrRNA
having 29 2'0Me residues at the 3'-end (SEQ ID No. 11). Pair 1L shows high
cleavage
activity with 9 2'0Me residues at the 5'-end and 29 2'0Me residues at the 3'-
end (SEQ
ID No. 13). Thus 38 out of 67 RNA residues in the short version of the
tracrRNA can be
converted to 2'0Me RNA (57%) with no loss of activity in an in vitro cleavage
assay.
[0073] Pair 14A demonstrates that 11 bases at the 3'-end of the crRNA (50%
of the
22 base tracrRNA binding domain) can be modified with 2'0Me RNA and support
target cleavage (SEQ ID No. 14). The modified crRNA retains full activity when
paired
with the modified tracrRNA (pair 14L, Seq ID Nos. 13 and 14). Modification of
11
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
26
bases towards the 5'-end of the crRNA (in the guide, protospacer domain, bases
2-12)
supports target cleavage (pair 15A) and this modification is also functional
when paired
with the modified tracrRNA (pair 15L, SEQ ID Nos. 13 and 15). The 2'0Me
modifications towards the 5'-end and 3'-end of the crRNA can be combined (SEQ
ID
No.16) such that 22 out of 35 residues are modified (63%) and still support
cleavage
(pair 16A), even when paired with the modified tracrRNA (pair 16L, SEQ ID Nos.
13
and 16).
[0074] The crRNA:tracrRNA pairs mentioned above all employed 2'0Me RNA as
the modifier. Additional studies showed that 2'F modification was also
tolerated by
Cas9 and enabled cleavage of a target DNA. Pair 9A employs a crRNA with 2'F
modification at all pyrimidine bases (SEQ ID No. 23) and this design supported
complete target cleavage. Likewise complete 2'F modification of the crRNA
supported
complete target cleavage (pair 10A, SEQ ID No. 24). Combined use of 2'0Me and
2'F
modifications may permit complete modification of both the crRNA and tracrRNA.
The
studies in the present example utilized in vitro biochemical analyses.
Performance may
vary in the context of mammalian gene editing where the sequences have to
function in
the milieu of a cell nucleus.
EXAMPLE 2
[0075] This example demonstrates functioning of truncated tracrRNAs to
direct
genome editing by the Spy Cas9 nuclease in mammalian cells.
[0076] Both functional Cas9 nuclease and the RNA triggers (a single sgRNA
or dual
crRNA:tracrRNA pair) must be present in the nucleus of mammalian cells for
CRISPR
genome editing to take place. Transfection of large plasmid vectors expressing
Cas9 is
inefficient and adds variability to experimental results. In order to
accurately assess the
impact that changes in length and chemical composition of the crRNA and
tracrRNA
have in mammalian cells in the absence of other variables, a cell line that
stably
expresses Spy Cas9 was constructed.
[0077] A HEK293 cell line having constitutive expression of SpyCas9 (human
codon-optimized) with stable vector integration and selection under G418 was
developed
as described below. Human optimized Spy Cas9 was ligated into a pcDNA3.1
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
27
expression vector (Life Technologies) and transfected into HEK293 cells using
Lipofectamine2000 (Life Technologies). The transfected cells were allowed to
grow for
2 days before being placed under selective pressure using Neomycin. After 7
days, cells
were plated to single colonies using limiting dilution techniques. Monoclonal
colonies
were screened for Cas9 activity and the clone having highest level of
expression was
used for future studies. A single copy integration event for Cas9 was
determined using
droplet digital PCR (ddPCR). Western blot using an anti-Cas9 antibody showed
low but
constant expression of Cas9 protein. This cell line is henceforth referred to
as "HEK-
Cas9".
[0078] The HEK-Cas9 cell line was employed in subsequent studies. In a
reverse
transfection format, anti-HPRT 1 crRNA:tracrRNA complexes were mixed with
Lipofectamine RNAiMAX (Life Technologies) and transfected into the HEK-Cas9
cells.
Transfections were done with 40,000 cells per well in 96 well plate format.
RNAs were
introduced at a final concentration of 30 nM in 0.75 11.1 of the lipid
reagent. Cells were
incubated at 37 C for 48 hours. Genomic DNA was isolated using QuickExtract
solution
(Epicentre). Genomic DNA was amplified with KAPA HiFi DNA Polymerase (Roche)
and primers targeting the HPRT region of interest (HPRT forward primer:
AAGAATGTTGTGATAAAAGGTGATGCT (SEQ ID No. 28); HPRT reverse primer:
ACACATCCATGGGACTTCTGCCTC (SEQ ID No. 29)). PCR products were melted
and re-annealed in NEB buffer 2 (New England BioLabs) to allow for
heteroduplex
formation followed by digestion with 2 units of T7 endonuclease 1 (T7EI; New
England
BioLabs) for 1 hour at 37 C. The digested products were visualized on a
Fragment
Analyzer (Advanced Analytics). Percent cleavage of targeted DNA was calculated
as
the average molar concentration of the cut products / (average molar
concentration of the
cut products + molar concentration of the uncut band) x 100.
[0079] TracrRNAs (Table 2) were synthesized having deletions at the 5'-end,
3'-end,
internal or combinations thereof. The tracrRNAs were complexed with unmodified
truncated anti-HPRT1 crRNA SEQ ID No. 1 (Table 1) which has a 19 base
protospacer
domain targeting HPRT 1 at the 5'-end and a 16 base tracrRNA binding domain at
the 3'-
end. The paired crRNA:tracrRNA RNA oligonucleotides were transfected into the
HEK-Cas9 cells and processed as described above. Relative gene editing
activities were
assessed by comparing cleavage rates in the HPRT 1 gene using the T7EI
mismatch
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
28
endonuclease cleavage assay with quantitative measurement of products done
using the
Fragment Analyzer. A representation of the wild-type S. pyogenes
crRNA:tracrRNA
complex is shown in FIG. 1 (which pairs crRNA SEQ ID No. 46 with tracrRNA SEQ
ID
No. 18). The relative location of deletions in the tracrRNA tested in this
example are
shown in sequence alignment format in FIG. 3.
Table 2: Effect of length truncations in the tracrRNA on efficiency of gene
editing
in mammalian cells by Cas9 endonuclease.
SEQ ID Cleavage
Truncation
tracrRNA Sequence 5'-3' Length
No. (%) positions
guuggaaccauucaaaacagcauagcaaguua
18 aaauaaggcuaguccguuaucaacuugaaaaa 38 89
guggcaccgagucggugcuuuuuuu
caaaacagcauagcaaguuaaaauaaggcuag
5' ¨ 12 bases
30 uccguuaucaacuugaaaaaguggcaccgagu 26 74
3' - 3 bases
cggugcuuuu
aacagcauagcaaguuaaaauaaggcuagucc
5' - 15 bases
31 guuaucaacuugaaaaaguggcaccgagucgg 32 70
ugcuuu 3 -4
bases
agcauagcaaguuaaaauaaggcuaguccguu
5' - 18 bases
2 aucaacuugaaaaaguggcaccgagucggugc 57 67
3' -4 bases
uuu
agcauagcaaguuaaaauaaggcuaguccguu
5' - 18 bases
32 aucaacuugaaaaaguggcaccgagucggugc 47 65
3' - 6 bases
33
cauagcaaguuaaaauaaggcuaguccguuau 27 63 5' -
20 bases
caacuugaaaaaguggcaccgagucggugcu 3' -
6 bases
5' - 18 bases
agcauagcaaguuaaaauaguuaucaacuuga
34 0 55 Int ¨ 10 bases
aaaaguggcaccgagucggugcu
3' -6 bases
5' - 18 bases
agcauagcaaguuaaaauaaacuugaaaaagu
35 0 49 Int ¨ 16 bases
ggcaccgagucggugcu
3' -6 bases
36
agcauagcaaguuaaaauaaggcuaguccguu 53 64 5' -
18 bases
aucaacuugaaaaaguggcaccgagucggugc 3' -
7 bases
37
agcauagcaaguuaaaauaaggcuaguccguu 56 63 5' -
18 bases
aucaacuugaaaaaguggcaccgagucggug 3' -
8 bases
38
agcauagcaaguuaaaauaaggcuaguccguu 56 62 5' -
18 bases
aucaacuugaaaaaguggcaccgagucggu 3' -
9 bases
39
agcauagcaaguuaaaauaaggcuaguccguu 53 61 5' -
18 bases
aucaacuugaaaaaguggcaccgagucgg 3' -
10 bases
agcauagcaaguuaaaauaaggcuaguccguu 5 58 5' -
18 bases
aucaacuugaaaaagugccgagucgg Int ¨
3 bases
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
29
SEQ ID Cleavage
Truncation
tracrRNA Sequence 5'-3' Length
No. (%) positions
3'- 10 bases
5' - 18 bases
agcauagcaaguuaaaauaaggcuaguccaac
41 0 59 Int ¨
6 bases
uugaaaaaguggcaccgagucggugcu
3' -6 bases
5' - 18 bases
agcauagcaaguuaaaauaaggcuaguccaac
42 0 55 Int ¨ 6
bases
uugaaaaaguggcaccgagucgg
3'- 10 bases
5' - 18 bases
agcauagcaaguuaaaauaaggcuaguccaac
43 0 52 Int ¨ 9
bases
uugaaaaagugccgagucgg
3'- 10 bases
44
agcauagcaaguuaaaauaaggcuaguccguu 0 49 5' -
18 bases
aucaacuugaaaaagug 3' -
22 bases
5' - 18 bases
agcauagcaaguuaaaauaaggcuaguccguu
45 0 52 Int ¨ 13
bases
aucagcaccgagucggugcu
3' -6 bases
5' - 18 bases
agcauagcaaguuaaaauaaggcuaguccguc
427 4 64 Int ¨ 3
bases
aacuugaaaaaguggcaccgagucggugcuuu
3' -4 bases
Oligonucleotide sequences are shown 5'-3'. Lowercase = RNA. Lengths of the RNA
oligonucleotides are indicated (bases). The number of RNA residues removed in
truncation studies at the 5'-end, 3'-end, and internal (int) are indicated.
The relative
functional activity of each species is indicated by the % cleavage in a T7EI
heteroduplex
assay.
[0080] This example demonstrates that for purposes of gene editing in
mammalian
cells that the tracrRNA can tolerate significant deletions from both the 5'-
end and 3'-end
and retain full functionality. Deletion of 18 bases from the 5'-end was well
tolerated.
Deletion of 20 bases from the 5'-end led to reduced activity, possibly due to
lower
affinity of binding of the crRNA. It is possible that this reduced length or
even shorter
might be functional if Tm-enhancing modifications were employed to stabilize
the short
duplex forming region. Deletion of up to 10 bases from the 3'-end was well
tolerated.
Additional deletions resulted in loss of activity. Internal deletions that
disrupted hairpin
elements or spacing between hairpin elements were not functional.
[0081] In summary, this example demonstrates that truncation of the
tracrRNA from
the 89 base length of the wild-type (WT, SEQ ID No. 18) to a 67 base length
(SEQ ID
No. 2) or to a 62 base length (SEQ ID No. 38), or to a 61 base length (SEQ ID
No. 39)
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
retained high functional activity. Use of shortened tracrRNAs of this kind
will be less
costly and easier to manufacture by chemical methods than the WT 89 base RNA.
Some
of the truncated species (SEQ ID No. 2, SEQ ID No. 38, and SEQ ID No. 39)
showed
increased functional activity over the 89 base WT tracrRNA. Therefore in
addition to
being less costly and easier to manufacture by chemical methods, the shortened
tracrRNAs of the present invention showed improved activity.
EXAMPLE 3
[0082] Examples 1 and 2 demonstrated that crRNA:tracrRNA complexes shorter
than the WT lengths of 42 and 89 bases, respectively, can show higher
functional activity
in mammalian gene editing. The present example shows further optimization of
the
lengths of these RNA species.
[0083] A series of crRNAs and tracrRNAs (Table 3) were synthesized having
different lengths as indicated. Truncations were made at the 3'-end of the
crRNA, the
5'-end of the tracrRNA, and/or the 3'-end of the tracrRNA. The crRNAs and
tracrRNA
were paired as indicated in Table 3. The crRNAs all employed a 20 base
protospacer
domain targeting HPRT1 at the 5'-end and variable length 3'-ends (tracrRNA
binding
domains). An alignment of the crRNA and tracrRNA sequences studied in this
example
is shown in FIG. 4 to make clear the positions of truncations relative to each
functional
domain.
[0084] The paired crRNA:tracrRNA RNA oligonucleotides were transfected into
the
HEK-Cas9 cells and processed as described in Example 2. Relative gene editing
activities were assessed by comparing cleavage rates in the HPRT1 gene using
the T7EI
mismatch endonuclease cleavage assay with quantitative measurement of products
done
using the Fragment Analyzer. Results are shown in Table 3. The relative
location of
deletions are shown in sequence alignment format in FIG. 4.
Table 3: Effect of length truncations in both the crRNA and tracrRNA on
efficiency
of gene editing in mammalian cells by Cas9 endonuclease.
cr/tracr crRNA Sequence
SEQ
Cleavage
RNA ID N Length
o. tracrRNA Sequence 0/0
pair
cuuauauccaacacuucgugguuuuagagcuaugcu
42/89 46 42 25
guuuug
SUBSTITUTE SHEET (RULE 26)
(9Z '3'111N) JAMS uuiisaris
ZE 9E nobnynobvbvnnnn.6.6nbonnovovvoonvnynno 817 OL/9E
nnnobn.6.6onEmboovo.6.6nbvvvvv.6nnovvon
OL IS
vnnboonfmno.6.6vvnyvvynnbvvobvnvobvovv
817 OL/6E
=Lb
6E Lt
nobnynobvbvnnnn.6.6nbonnovovvoonvnynno
nnnobn.6.6onEmboovo.6.6nbvvvvv.6nnovvon
OL IS
vnnboonfmno.6.6vvnyvvynnbvvobvnvobvovv
SS OL/Z17
.6nnnn.6
Z17 917
nobnynobvbvnnnn.6.6nbonnovovvoonvnynno
nn
17L
n.n.a6n.6.6onEmboovo.6.6nbvvvvv.6n.n.ovvon.vn.n. OS
OZ
boonfmn.Q.6.6vvn.vvvvn.n.6vvobvn.vobvovvvvo 17LItE
17
.6n.vn.obvfmn.n.n.n.6.6nbon.n.ovovvoon.vn.vn.n.o 617
nn
17L
n.n.a6n.6.6onEmboovo.6.6nbvvvvv.6n.n.ovvon.vn.n. OS
9Z
boonfmn.Q.6.6vvn.vvvvn.n.6vvobvn.vobvovvvvo 17L/9E
9
n.a6n.vn.obvfmn.n.n.n.6.6nbon.n.ovovvoon.vn.vn.n.o 817
nn
17L
n.n.a6n.6.6onEmboovo.6.6nbvvvvv.6n.n.ovvon.vn.n. OS
17
boonfmn.Q.6.6vvn.vvvvn.n.6vvobvn.vobvovvvvo 17L/6E
n.n.6
6E Lt
n.a6n.vn.obvfmn.n.n.n.6.6nbon.n.ovovvoon.vn.vn.n.o
nn
17L
n.n.a6n.6.6onEmboovo.6.6nbvvvvv.6n.n.ovvon.vn.n. OS
SE
boonfmn.Q.6.6vvn.vvvvn.n.6vvobvn.vobvovvvvo 17L/Z17
bn.n.n.n.6
Z17 917
n.a6n.vn.obvfmn.n.n.n.6.6nbon.n.ovovvoon.vn.vn.n.o
n.n.n.n.n.n.n.a6n.6.6onbv.6
68 00vo.6.6n6vvvvvbrmovvonvnnboonfmno.6.6vv 81
1Z
n.vvvvn.n.6vvobvn.vobvovvvvon.n.voovv.6.6n.n.6 68/17E
17
.6n.vn.obvfmn.n.n.n.6.6nbon.n.ovovvoon.vn.vn.n.o 617
n.n.n.n.n.n.n.a6n.6.6onbv.6
68 00vo.6.6n6vvvvvbrmovvonvnnboonfmno.6.6vv 81
8
n.vvvvn.n.6vvobvn.vobvovvvvon.n.voovv.6.6n.n.6 6819
9
n.a6n.vn.obvfmn.n.n.n.6.6nbon.n.ovovvoon.vn.vn.n.o 817
n.n.n.n.n.n.n.a6n.6.6onbv.6
68 00vo.6.6n6vvvvvbrmovvonvnnboonfmno.6.6vv 81
1
n.vvvvn.n.6vvobvn.vobvovvvvon.n.voovv.6.6n.n.6 6816E
n.n.6
6E Lt
n.a6n.vn.obvfmn.n.n.n.6.6nbon.n.ovovvoon.vn.vn.n.o
n.n.n.n.n.n.n.a6n.6.6onbv.6
68 00vo.6.6n6vvvvvbrmovvonvnnboonfmno.6.6vv 81
n.vvvvn.n.6vvobvn.vobvovvvvon.n.voovv.6.6n.n.6
%a3uanbas vNllnuil ai
tOual V1µ111
agunualp a3uanbas vtop3 Oas
.13C.11/.13
if
Zr6990/SIOZSI1/13.1 I6001/910Z OM
ZT-90-LTOZ 890L6Z0 VD
(9Z '3'111N) JAMS uuiisaris
nobribbonbyboovo.6.6n6vvvvvbrm
9 ES
ovvonynnboonbvnobbvvri.vvvvri.n6vvobvnvo
9
bralt-Ln6
Z.17 917
nobnynobvfmnram.6.6nbormovovvoonvnynno
nobribbonbyboovo.6.6n6vvvvvbrmov
S9 ZS
vonynnboonbvnobbvvri.vvvvri.n6vvobvnvobv
91 S9/17
17E bnynobvfmnram.6.6nbormovovvoonvnynno 617
nobribbonbyboovo.6.6n6vvvvvbrmov
S9 ZS
vonynnboonbvnobbvvri.vvvvri.n6vvobvnvobv
Lt S9/9
9 nobnynobvfmnram.6.6nbormovovvoonvnynno17
nobribbonbyboovo.6.6n6vvvvvbrmov
S9 ZS
vonynnboonbvnobbvvri.vvvvri.n6vvobvnvobv
917
nt-L6
6 Lt
nobnynobvfmnram.6.6nbormovovvoonvnynno
nobribbonbyboovo.6.6n6vvvvvbrmov
S9 ZS
vonynnboonbvnobbvvri.vvvvri.n6vvobvnvobv
OS S9/Zt
74 bralt-Ln6
917
nobnynobvfmnram.6.6nbormovovvoonvnynno
ra-mobribbonbvboovobbribvvvvvbrmov
L9
vonynnboonbvnobbvvri.vvvvri.n6vvobvnvobv
ft' L9/17
17E bnynobvfmnram.6.6nbormovovvoonvnynno 617
ra-mobribbonbvboovobbribvvvvvbrmov
L9
vonynnboonbvnobbvvri.vvvvri.n6vvobvnvobv
LS L9/9
9 nobnynobvfmnram.6.6nbormovovvoonvnynno 817
ra-mobribbonbvboovobbribvvvvvbrmov
L9
vonynnboonbvnobbvvri.vvvvri.n6vvobvnvobv
It' L9/6
nt-L6
6 Lt
nobnynobvfmnram.6.6nbormovovvoonvnynno
ra-mobribbonbvboovobbribvvvvvbrmov
L9
vonynnboonbvnobbvvri.vvvvri.n6vvobvnvobv
9 L9/t'
bralt-Ln6
Z.17 917
nobnynobvfmnram.6.6nbormovovvoonvnynno
ra-mobribbonbvbo ov obbribvvvvvbrmovv on
OL IS
vnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobvovv
6 OL/17
17E bnynobvfmnram.6.6nbormovovvoonvnynno 617
ra-mobribbonbvbo ov obbribvvvvvbrmovv on
OL IS
vnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobvovv
a3uanbas vNllnuil ai
tOualVMll
agunualp a3uanbas vtop3 Oas
.13C.11/.13
Z
Zr6990/SIOZSI1/13.1 I6001/910Z OM
ZT-90-LTOZ 890L6Z0 VD
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
33
cr/tracr crRNA Sequence
SEQ
Cleavage
RNA Length
pair ID No. tracrRNA Sequence
4'7
cuuauauccaacacuucgugguuuuagagcuaugcu
39
guu
39/63 13
3
cauagcaaguuaaaauaaggcuaguccguuaucaac
63
uugaaaaaguggcaccgagucggugcu
48 cuuauauccaacacuucgugguuuuagagcuaugcu 36
36/63 28
cauagcaaguuaaaauaaggcuaguccguuaucaac
53 63
uugaaaaaguggcaccgagucggugcu
49 cuuauauccaacacuucgugguuuuagagcuaug 34
34/63 33
cauagcaaguuaaaauaaggcuaguccguuaucaac
53 63
uugaaaaaguggcaccgagucggugcu
Oligonucleotide sequences are shown 5'-3'. Lowercase = RNA. Lengths of the
RNA oligonucleotides are indicated (bases). The relative functional activity
of
each crRNA:tracrRNA pair is indicated by the % cleavage in a T7EI
heteroduplex assay.
[0085] All of the compounds studied directed CRISPR/Cas editing at the
HPRT1
locus in HEK-Cas9 cells. Efficiency varied widely from 6% to 57%. The most
effective
crRNA+tracrRNA combination was the 36 base crRNA (SEQ ID No. 48) with the
67mer
tracrRNA (SEQ ID No. 2). A schematic representation of the truncated,
optimized
crRNA:tracrRNA complex is shown in FIG. 5. In this case the tracrRNA binding
domain of the crRNA was truncated to 16 bases from the WT 22 base sequence (3'-
end).
This hybridizes to the crRNA binding domain at the 5'-end of the tracrRNA. The
tracrRNA was truncated 18 bases at the 5'-end and 4 bases at the 3'-end to
product the
active 67 base product. For this pair, a blunt end is formed upon
hybridization of the 3'-
end of the crRNA with the 5'-end of the tracrRNA. Other versions also showed
high
activity, including the 42 base (WT) crRNA (SEQ ID No. 46) paired with the 70
base
tracrRNA (SEQ ID No. 51).
[0086] The shortest crRNA tested was 34 bases in length (SEQ ID No. 49)
and, in
general, showed lower activity than the longer variants. The shorter duplex
domain
formed between this variant and the tracrRNA has reduced binding affinity (Tm)
compared to the 36 base crRNA variant and that 34 base complex was less stable
at
37 C. Use of chemical modification that increase binding affinity (Tm), such
as 2'0Me
RNA, 2'F RNA, or LNA residues, will increase stability of this short duplex
domain and
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951 PCT/US2015/066942
34
will lead to improved activity, permitting use of very short crRNAs of this
design.
Extensive use of Tm-enhancing modifications will permit use of even shorter
tracrRNA
binding domains in the crRNA, such as 13 base, or 12 base, or 11 base, or 10
base, or 9
base, or 8 base or shorter, depending on the kind and number of modified
residues
employed.
EXAMPLE 4
[0087] Examples 1, 2, and 3 demonstrated that crRNA:tracrRNA complexes
shorter
than the WT lengths of 42 and 89 bases, respectively, can show higher
functional activity
in mammalian gene editing. In those examples, all truncations were made in the
universal domains of the RNAs. The present example tests the effects that
truncations
have on the target-specific protospacer domain of the guide crRNA.
[0088] A series of crRNAs (Table 4) were synthesized having protospacer
domain
lengths of 20, 19, 18, or 17 bases as indicated. Truncations were made at the
5'-end of
the crRNA, using a 16mer universal tracrRNA binding sequence at the 3'-end.
The
crRNAs were paired with an unmodified 67mer tracrRNA (SEQ ID No. 2). The
crRNAs
targeted 4 different sites in the same exon of the human HPRT1 gene.
[0089] The paired crRNA:tracrRNA RNA oligonucleotides were transfected into
the
HEK-Cas9 cells and processed as described in Example 2. Relative gene editing
activities were assessed by comparing cleavage rates in the HPRT1 gene using
the T7EI
mismatch endonuclease cleavage assay with quantitative measurement of products
done
using the Fragment Analyzer. Results are shown in Table 4.
Table 4: Effect of length truncations in the 5'-protospacer domain of the
crRNA on
efficiency of gene editing in mammalian cells by Cas9 endonuclease.
SEQ ID LengthCleavage HPRT1
crRNA Sequence 5'-3' Protospacer
No. (%) site
domain
cuuauauccaacacuucgugguuuuagagc
48 20 64
uaugcu
1 uuauauccaacacuucgugguuuuagagcu 19 62 38285
augcu
54 18 57
uauauccaacacuucgugguuuuagagcua
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
SEQ ID
LengthCleavage HPRT1
crRNA Sequence 5'-3 Protospacer
No.(%) site
domain
ugcu
55 auauccaacacuucgugguuuuagagcuau 17 42
gcu
aauuauggggauuacuaggaguuuuagagc
56 20 78
uaugcu
57 auuauggggauuacuaggaguuuuagagcu 19 81
augcu
38087
58 uuauggggauuacuaggaguuuuagagcua 18 82
ugcu
59 uauggggauuacuaggaguuuuagagcuau 17 82
gcu
auuucacauaaaacucuuuuguuuuagagc
60 20 52
uaugcu
61 uuucacauaaaacucuuuuguuuuagagcu 19 30
augcu
38358
62 uucacauaaaacucuuuuguuuuagagcua 18 12
ugcu
63 ucacauaaaacucuuuuguuuuagagcuau 17 0
gcu
uccauuucauagucuuuccuguuuuagagc
64 20 70
uaugcu
65 ccauuucauagucuuuccuguuuuagagcu 19 71
augcu
38094
66 cauuucauagucuuuccuguuuuagagcua 18 52
ugcu
67 auuucauagucuuuccuguuuuagagcuau 17 0
gcu
Oligonucleotide sequences are shown 5'-3'. Lowercase = RNA. The target-
specific protospacer domain is underlined and length is indicated (bases). The
relative functional activity of each species is indicated by the % cleavage in
a
T7EI heteroduplex assay.
[0090] Of the 4
sites studied, one (site 38087) showed high activity for all 4 crRNAs
with no changes seen as the protospacer domain was shortened. Site 38285
similar
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
36
efficacy for the 20 and 19 base protospacer crRNAs (SEQ ID Nos. 48 and 1), a
slight
decrease for the 18 base version (SEQ ID No. 54), and a large decrease for the
17 base
version (SEQ ID No. 55). Site 38094 showed similar efficacy for the 20 and 19
base
protospacer crRNAs (SEQ ID Nos. 64 and 65), a moderate decrease for the 18
base
version (SEQ ID No. 66), and no activity for the 17 base version (SEQ ID No.
67). Site
38358 showed good activity for the 20 base version (SEQ ID No. 60), lower
activity for
the 19 base version (SEQ ID No. 61), even lower activity for the 18 base
version (SEQ
ID No. 62) and no activity for the 17 base version (SEQ ID No. 63).
[0091] The use of shortened 17 base protospacer guide domains can lower the
occurrence of undesired off-target events compared to the wild-type 20 base
domain (Fu
et al., Nature Biotechnol., 32:279, 2014). We observe that on-target efficacy
varies in a
sequence-context specific fashion and that 20 base and 19 base protospacer
guide
domains are generally effective but that activity begins to decrease when 18
base
protospacer domains are used and activity significantly decreases when 17 base
protospacer domains are used. Therefore, to maintain desired on-target
efficiency, use of
20 and 19 base target-specific protospacer guide domains are employed herein.
Significant truncation of the protospacer guide domain in many cases lowers on-
target
cleavage of a DNA target by the Cas9 endonuclease. Use of chemical
modifications that
enhance Tm (increase binding affinity of the protospacer target-specific
domain of the
crRNA to the target DNA sequence) may permit use of shorter sequences such
that a 17
base protospacer guide may show similar on-target efficacy as an unmodified 20
base
protospacer guide domain.
EXAMPLE 5
[0092] This example demonstrates that truncated crRNA:tracrRNA complex show
improved gene editing activity at multiple sites. The prior examples studied
efficacy of
the truncated RNAs as triggers of CRISPR gene editing in mammalian cells at a
single
site in the human HRPT1 gene. Site/sequence specific effects may exist. The
present
example demonstrates improved performance of the truncated species of the
present
invention at 12 sites in an exon of the human HPRT1 gene.
[0093] A series of crRNAs (Table 5) were synthesized having a protospacer
domain
lengths of 20 bases specific to 12 sites in the human HPRT1 gene with a 16mer
universal
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951 PCT/US2015/066942
37
tracrRNA binding sequence at the 3'-end. The crRNAs were paired with an
unmodified
67mer tracrRNA (SEQ ID No. 2). The same 12 sites were studied using WT length
crRNA:tracrRNA complexes wherein the crRNA comprised a 20 base protospacer
guide
paired with a 22mer universal tracrRNA binding sequence at the 3'-end
complexed with
the WT 89mer tracrRNA (SEQ ID No. 18).
[0094] The paired crRNA:tracrRNA RNA oligonucleotides were transfected into
the
HEK-Cas9 cells and processed as described in Example 2. Relative gene editing
activities were assessed by comparing cleavage rates in the HPRT 1 gene using
the T7EI
mismatch endonuclease cleavage assay with quantitative measurement of products
done
using the Fragment Analyzer. Results are shown in Table 5.
Table 5: Effect of length truncations in both the crRNA and tracrRNA on
efficiency
of gene editing in mammalian cells by Cas9 endonuclease.
cr/tracr crRNA Sequence
SEQ Cleavage
RNA Length
pair ID No. tracrRNA Sequence
tic cauuucauagucuuuccuguuuuagagcuau
64 36
38094 gcu
short agcauagcaaguuaaaauaaggcuaguccguua 55
2 ucaacuugaaaaaguggcaccgagucggugcuu 67
tic cauuucauagucuuuccuguuuuagagcuau
68 42
38094 gcuguuuug
guuggaaccauucaaaacagcauagcaaguuaa 31
long
18 aauaaggcuaguccguuaucaacuugaaaaagu 89
ggcaccgagucggugcuuuuuuu
uuuuguaauuaacagcuugcguuuuagagcuau
69 36
38231 gcu
short agcauagcaaguuaaaauaaggcuaguccguua 7
2 ucaacuugaaaaaguggcaccgagucggugcuu 67
uuuuguaauuaacagcuugcguuuuagagcuau
70 42
38231 gcuguuuug
guuggaaccauucaaaacagcauagcaaguuaa 0
long
18 aauaaggcuaguccguuaucaacuugaaaaagu 89
ggcaccgagucggugcuuuuuuu
cuuagagaauauuuguagagguuuuagagcuau
71 36
38371 gcu
short agcauagcaaguuaaaauaaggcuaguccguua 57
2 ucaacuugaaaaaguggcaccgagucggugcuu 67
38371 72 cuuagagaauauuuguagagguuuuagagcuau
42 27
long gcuguuuug
SUBSTITUTE SHEET (RULE 26)
(9Z '3'111N) JAMS HillIIISELIS
.6nrambno6 Euoi
8 Z.17 917
nvnobvfmnram.6.6nbormovovvoonvnynno S8Z8E
n
L9
nnobribbon6vboovo.6.6n6vvvvvbrmovvon Z
I_Totis
8E
vnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobv
nob S8Z8E
9E 817
nvnobvfmnram.6.6nbormovovvoonvnynno
nramm-mobribbonEmboovo.6.6
68 nbvvvvvbrmovvonvnnboonbvno.6.6vvnyv 81
LE
Euoi
vvri.n6vvobvnvobvovvvvonnvoovv.6.6t-Ln6
.6nrambno6 EE18E
Z.17 6L
nvnobvfmnrawilm000vovovvrimmovon.6.6
n
L9
nnobribbon6vboovo.6.6n6vvvvvbrmovvon Z
I_Totis
ES
vnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobv
nob EEI8E
9E 8L
nvnobvfmnrawilm000vovovvrimmovon.6.6
nramm-mobribbonEmboovo.6.6
68 nbvvvvvbrmovvonvnnboonbvno.6.6vvnyv 81 Euoi
ES
vvri.n6vvobvnvobvovvvvonnvoovv.6.6t-Ln6
.6nrambnob L808E
Z.17 LL
nvnobvfmnrambv.6.6vnovralv.6.6.6.6nvnnyv
n
L9
nnobribbon6vboovo.6.6n6vvvvvbrmovvon Z
09 I_Totis vnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobv
9E
nob L808
9S
nvnobvfmnrambv.6.6vnovralv.6.6.6.6nvnnyv
nramm-mobribbonEmboovo.6.6
68 nbvvvvvbrmovvonvnnboonbvno.6.6vvnyv 81
Euoi
ZZ
vvri.n6vvobvnvobvovvvvonnvoovv.6.6t-Ln6
.6nrambnob 17LS8E
Z.17 9L
nvnobvfmnrambrmnyvvvvri.vobovovvvvo
n
L9
nnobribbon6vboovo.6.6n6vvvvvbrmovvon Z
I_Totis
8S
vnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobv
9E
nob 17LS8E
SL
nvnobvfmnrambrmnyvvvvri.vobovovvvvo
nramm-mobribbonEmboovo.6.6
68 nbvvvvvbrmovvonvnnboonbvno.6.6vvnyv 81 Euoi
L
vvri.n6vvobvnvobvovvvvonnvoovv.6.6t-Ln6
.6nrambnob 60S8E
Z.17 17L
nvnobvfmnrambormovnyvbnyvnynovbrm
n
L9
nnobribbon6vboovo.6.6n6vvvvvbrmovvon Z
9S
I_Totis
vnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobv
9E
nob 608E
EL
nvnobvfmnrambormovnyvbnyvnynovbrm
nramm-mobribbonEmboovo.6.6
68 nbvvvvvbrmovvonvnnboonbvno.6.6vvnyv 81
vvri.n6vvobvnvobvovvvvonnvoovv.6.6t-Ln6
% a3uanbas ifTNIp3U.I1 'ON ai
tOual VNII
agunualp a3uanbas vtop3 Oas
.13C.11/.13
8
Zr6990/SIOZSI1/13.1 I6001/910Z OM
ZT-90-LTOZ 890L6Z0 VD
(9Z '3'111N) JAMAS uniiisaris
./Cussu xaidnpaialaq
HLI u 1.1! aEuAuaio % alp Aq poluo!pu! s ud yl\111.10E.11:VNII.10 qoi
Jo ATIAnou puonounj aAnuial qj, (sasuci) paluo!pu! J1 samloaionuoEllo y!\111
wilJo sqlEual .y1\111= asualamol =,E-,c umogs spouanbas apRoaionuoEllo
nramm-mobribbonEmboovo.6.6
68 nbvvvvvbrmovvonvnnboonbvno.6.6vvnyv 81
Euo!
Z
vvri.n6vvobvnvobvovvvvonnvoovv.6.6t-Ln6
.6nrambnob L98
Z.17 98
nvnobvfmnrambnyvnovbribnynnt-Lobvovri.
rL
L9
nnobribbon6vboovo.6.6n6vvvvvbrmovvon
VOIIS
nob L98
9S8
nvnobvfmnrambnyvnovbribnynnt-Lobvovri.
nramm-mobribbonEmboovo.6.6
68 nbvvvvvbrmovvonvnnboonbvno.6.6vvnyv 81 Euo!
91
vvri.n6vvobvnvobvovvvvonnvoovv.6.6t-Ln6
.6nrambnob 998
Z.17 178
nvnobvfmnrambv.6.6nobn.6.6vbnynnyvvon
rL
L9
nnobribbon6vboovo.6.6n6vvvvvbrmovvon
potis
9Z
vnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobv
nob 998
9 8
nvnobvfmnrambv.6.6nobn.6.6vbnynnyvvon
nramm-mobribbonEmboovo.6.6
68 nbvvvvvbrmovvonvnnboonbvno.6.6vvnyv 81
Euo!
8
vvri.n6vvobvnvobvovvvvonnvoovv.6.6t-Ln6
.6nrambnob 8S8
Z.17 ZS
nvnobvfmnrambramnonovvvvnvovont-mv
rL
L9Z17
nnobribbon6vboovo.6.6n6vvvvvbrmovvon
VOIIS
nob 8S8
909
nvnobvfmnrambramnonovvvvnvovont-mv
nramm-mobribbonEmboovo.6.6
68 nbvvvvvbrmovvonvnnboonbvno.6.6vvnyv 81 Euo!
9
vvri.n6vvobvnvobvovvvvonnvoovv.6.6t-Ln6
.6nrambnob L8Z8
Z.17 18
nvnobvfmnrambbormovovvoonvnynno.6.6
rL
L9
nnobribbon6vboovo.6.6n6vvvvvbrmovvon
8 VOIIS
17
vnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobv
nob L8Z8
9 08
nvnobvfmnrambbormovovvoonvnynno.6.6
nramm-mobribbonEmboovo.6.6
68 nbvvvvvbrmovvonvnnboonbvno.6.6vvnyv 81
vvr!nbvvobvnvobvovvvvormvoovv.6.6t-m6
%a3uanbas VNIII3U.11 'ON (II
tOuarl
agunualp a3uanbas vtop3
IS
.13C4/.13
6
Zr6990/SIOZSI1/13.1
I6001/910Z OM
ZT-90-LTOZ 890L6Z0 VD
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
[0095] The relative efficiency of CRISPR mediated gene editing in the HEK-
Cas9
cells varied with sequence context. However, in all cases the shorter
optimized RNA
guides (36mer crRNA and 67mer tracrRNA) showed higher efficiency than the WT
RNAs (42mer crRNA and 89mer tracrRNA). Use of the shortened, optimized guide
RNAs of the present invention improve Cas9 cleavage of targeted DNAs relative
to the
WT RNAs, improving the gene editing rates.
EXAMPLE 6
[0096] Example 1 described chemical modification patterns that functioned
with
Cas9 in an in vitro biochemical target DNA cleavage assay. This example
demonstrates
functioning of chemically modified tracrRNAs to direct genome editing by the
Spy Cas9
nuclease in mammalian cells. Optimal modification patterns differ between in
vitro and
in vivo use.
[0097] A series of tracrRNAs (Table 6) were synthesized having a variety of
chemical modifications, including: the ribose modifications 2'0Me RNA and LNA;
the
end-modifying groups propanediol spacer and napthyl-azo modifier (N,N-diethy1-
4-(4-
nitronaphthalen-1-ylazo)-phenylamine, or "ZEN"); and select internucleotide
linkages
with phosphorothioate modifications. See: Lennox et al., Molecular Therapy
Nucleic
Acids 2:e117 2013 for structure of the napthyl-azo modified and use of the
napthyl-azo
modifier and propanediol modifier for use as end-groups to block exonuclease
attack.
The tracrRNAs listed in Table 6 were complexed with unmodified truncated anti-
HPRT 1
crRNA SEQ ID No. 1 (Table 1) which has a 19 base protospacer domain targeting
HPRT 1 at the 5'-end and a 16 base tracrRNA binding domain at the 3'-end. The
paired
crRNA:tracrRNA RNA oligonucleotides were transfected into the HEK-Cas9 cells
and
processed as described above. Relative gene editing activities were assessed
by
comparing cleavage rates in the HPRT 1 gene using the T7EI mismatch
endonuclease
cleavage assay with quantitative measurement of products done using the
Fragment
Analyzer.
SUBSTITUTE SHEET (RULE 26)
(9Z '3'111N) JAMAS uuiisaris
nobribbonbv.E=oovo
9
B 001
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvo**,
E - ntmobribbonEmbo ov ob
OZ 66
bribvvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobv- E
nAm.,¶-LobribbonEmboovo
8S 86
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvo*.6*E.
E - ntmobribbonEmbo ov ob
ES L6
bribvvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobv- E
nobribbonbyboo
Z9 96
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
nobribbonbyboo
Z9 S6
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
nobribbonbyboo
LS 176
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
nobribbonbyboo
LS 6
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
nobribbonbyboo
6S Z6
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
nobribbonbyboo
19 16
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
nobribbonbyboo
09 06
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
ra-mobribbonbvboo
179 68
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
ra-mobribbonbvboo
09 88
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
ra-mobribbonbvboo
19 L8
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
ra-mobribbonbvboo
179 1
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
ra-mobribbonbvboo
Z
obbribvvvvvbrmovv onynnbo-onbvno.6.6vvri.v-vvvri.n6vE. obvnv obv
ra-mobribbonbvboo
ZI LI
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
ra-mobribbonbvboo
9S II
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
ra-mobribbonbvboo
0 17
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
ra-mobribbonbvboo
S9
obbribvvvvvbrmovv onvnnbo onbvno.6.6vvri.vvvynnEmv obvnv obv
(s-,$) a3uanbas V101.13U.I1
agunualp Oas
=slia3 uulluunuutu
stuallud uo9u3Illpotu ap!loapnuollo v/01.13U.11 Jo uo9uz!uuldo :9 am",
It
Zr6990/SIOZSI1/13.1
I6001/910Z OM
ZT-90-LTOZ 890L6Z0 VD
(9Z '3'111N) JAMAS uuiisaris
= nobribbonbv.E=oovo
ii ZZI
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
,,*nobribbonbv.E=oovo
171 1Z I
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
OZ I
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
0 611
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
SI 811
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
09 Li I
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
09 911
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
171 Sit
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
19 1711
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
Z9
B '11
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo**,
= nobribbonbv.E=oovo
Z9 Z I I
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
19 I I I
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
19 Oil
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
ii 601
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
0 .6.6 801
n6vvvvvbrmovvonvnnboonbvn.6
o.6vvnyvvynnE=vvobvnvo*B*,
= nobribbonbv.E=oovo
8 LOI
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
-ri.,*.B,v,bonEmboovo
8S 901
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnE=vvobvnvo*B*,
Naz-ntmobribbonEmboovo.6.6
Z SO I
nbvvvvvbrmovvonvnnboonbvnobbvvnyvvvri.n6vvobvnvobv-Naz
Naz-ntmobribbonbvboo
SS 1701
vo.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnbvvobvnvobv
17S 01
bribvvvvvbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnv**E,v,
ra-mobribbonbvboo
6 ZO I
vo.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvnyvvynnbvvobvnvobv
ri.,¶-LAm.,*obri..6.6on6vboovo.6
SS ioi
bribvvvvvbrmovvonvnnboonbvno.6.6vvnyvvvri.n6vvobvnv*QA,6*E.
(0) =oN
(s-,$) a3uanbas V101.13U.11
agunuao Oas
zr
Zr6990/SIOZSI1/13.1
I6001/910Z OM
ZT-90-LTOZ 890L6Z0 VD
(9Z '3'111N) JAMAS uuiisaris
= nobribbonbv.E=oovo
ZSt
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvri.vvvv-nnbvvobvnvo*B*,
= nobribbonbv.E=oovo
0
.6.6n6vvvvvbrmovvonvn nbaanbenabbvvnyvvynnbvvobvnvo*B*,
= nobribbonbv.E=oovo
17 Z171
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6 eeneeeennE=vvobvnvo*B*,
= nobribbonbv.E=oovo
0 1171
.6.6n6vvvvvbrmovvonvn nb nb en abb eeneeeennE=vvobvnvo*B*,
= nobribbonbv.E=oovo
0 0171
.6.6n6vvvvvbrmovvonvazpooyoya,09.6vvnyvvvri.n6vvobvnvo*B*,
= nobribbonbv.E=oovo
0B 6E1
.6.6n6vvvvvbrmovvonvnnboonbvnoboyvvivvya,L6vvobvnvo**,
= nobribbonbv.E=oovo
0B 8E1
.6.6n6vvvvvbrmovvonvazooaloya,oppyvvivvya,L6vvobvnvo**,
-ri.,*.B,v,bonEmboovo
ElB LEI
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvo**,
-ri.,*.B,v,bonEmboovo
17S
B 9E1
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvo**,
= nobribbonbv.E=oovo
61B SEI
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvo**,
= nobribbonbv.E=oovo
8S17EI
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvo*B*,
nnnobribbon6vboovo.6.6n6vvvvvbri.no*v*E.,* Am.,* E. Aa-LAai.
0 EEI
*.6,* Am.,v, ,v,n,v,ri.,*.6 vobvnvobv
,lAu - n6.6 ttEreb ob
6S bribvvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobv- E ZEI
E - ribbon6vboova6
8S bribvvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobv- E Et
.1Ail I - ra-mobn.6.6onbv.600va6
0E1
bribvvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvobv- E
q.-F,*q.-FAalobri..6.6onbvboovo.6.6
LS 6ZI
nbvvvvvbrmovvonvnnboonbvnobbvvnyvvvri.n6vvobvnvo*.6+,*v+
= nobribbonbv.E=oovo
0 .6.6 8ZI
n6vvvvvbrmovvonvnnboonbvn.6
o.6vvri.vvvvri.n6vvobvnvo*B*,
= nobribbonbv.E=oovo
0 LZI
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvo*B*,
= nobribbonbv.E=oovo
179 9ZI
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvo*B*,
= nobribbonbv.E=oovo
0 SZI
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvo*B*,
= nobribbonbv.E=oovo
0 17ZI
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvo*B*,
= nobribbonbv.E=oovo
0B EZI
.6.6n6vvvvvbrmovvonvnnboonbvno.6.6vvri.vvvvri.n6vvobvnvo**,
(0) =oN
(s-,$) a3uanbas V101.13U.11
agunuao Oas
Zr6990/SIOZSI1/13.1
I6001/910Z OM
ZT-90-LTOZ 890L6Z0 VD
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
44
SEQ ID
Cleavage
tracrRNA Sequence (5'-3')
No. (%)
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
145 63
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
146 0
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
147 62
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
148 57
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
149 47
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
150 61
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
151 61
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
152 61
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
153 61
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
154 50
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
155 46
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
156 59
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
157 2
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
158 18
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
159 50
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
160 58
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
161 14
caccgagucggugcu*u*u
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
162 8
caccgagucggugcu*u*u
Oligonucleotide sequences are shown 5'-3'. Uppercase = DNA; Lowercase = RNA;
Underlined = 2'-0-methyl RNA; Italics = 2'-fluoro RNA; +a, +c, +t, +g = LNA;
C3 =
C3 spacer (propanediol modifier); * = phosphorothioate internucleotide
linkage; ZEN ¨
napthyl-azo modifier; Inv-dT = inverted-dT. The relative functional activity
of each
species is indicated by the % cleavage in a T7EI heteroduplex assay.
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
[0098] Modification is usually necessary for synthetic nucleic acids to
function well
in an intracellular environment due to the presence of exonucleases and
endonucleases
that degrade unmodified oligonucleotides. A wide range of modifications have
been
described that confer nuclease resistance to oligonucleotides. The precise
combination
and order of modifications employed that works well for a given application
can vary
with sequence context and the nature of the protein interactions required for
biological
function. Extensive prior work has been done relating to chemical modification
of
antisense oligonucleotides (which interact with RNase H1) and siRNAs (which
interact
with DICER, AG02, and other proteins). It is expected that chemical
modification will
improve function of the CRISPR crRNA:tracrRNA complex. However, it is not
possible
to predict what modifications and/or pattern of modifications will be
compatible with
functional complexation of the synthetic RNAs with Cas9. The present invention
defines minimal, moderate, and extensive chemical modification patterns for
the
tracrRNA that retain high levels of function to direct Cas9 mediated gene
editing in
mammalian cells.
[0099] The results in Table 6 demonstrate that extensive modification is
tolerated
throughout the 5' and 3' end domains of the tracrRNA. Modification of the
internal
domains of the tracrRNA showed reduced activity, likely due to altered
structure of the
folded RNA and/or blocking of protein contact points with the 2'-OH of key RNA
residues by the 2'0Me modification. For example, compound SEQ ID No. 100 has
39/67 residues modified with 2'0Me RNA (58%) and retains full activity
compared with
the unmodified sequence. SEQ ID No. 134 has 46/67 residues modified with 2'0Me
RNA (69%) and retains near full activity compared with the unmodified sequence
(FIG.
6). SEQ ID No. 134 is a truncated 67mer variant of the tracrRNA. Using SEQ ID
No.
134 as a model, modification of 11 sequential residues in the 5'-domain with
2'0Me
RNA was tolerated with no loss of activity. Modification of 35 sequential
residues in the
3'-domain with 2'0Me RNA was tolerated with not loss of activity. Of note, the
two
hairpin structures present in the 3'-domain are necessary for function as
deletion of
either of these features results in loss of activity (Example 2, FIG. 3), yet
both of these
domains can be completely modified with 2'0Me RNA without compromising
function.
Note that both SEQ ID Nos. 134 and 100 also have phosphorothioate (PS)
modified
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
46
internucleotide linkages at the 5'- and 3'-ends, which provides additional
protection
against exonuclease attack.
[00100] Specific residues were identified that led to large reductions or
complete loss
of activity when modified. Using the 67 base tracrRNA (for example, SEQ ID No.
134)
as reference, starting from the 5'-end of the sequence substitution of 2'0Me
RNA for the
natural RNA at residues U12, A15, G26, U27, G30, U31, and U32 led to
substantial loss
of activity (FIG. 6). Specific residues were also identified that led to
smaller yet
significant reductions in activity when modified. Using the 67 base tracrRNA
(for
example, SEQ ID No. 134) as reference, starting from the 5'-end of the
sequence
substitution of 2'0Me RNA for the natural RNA at residues U13, U18, C23, U24,
and
C28 led to reduced activity (FIG. 6). This study was performed using 2'0Me
RNA. Use
of other modifications, such as 2'F, LNA, DNA, etc. at these positions may be
better
tolerated. The central 21 residue domain of unmodified RNA in SEQ ID No. 134
was
modified with 2'-F RNA either completely (SEQ ID No. 141) or partially (SEQ ID
Nos.
142 and 143). These variants were not functional. The central 21 residue
domain of
unmodified RNA in SEQ ID No. 134 was modified with DNA either completely (SEQ
ID No. 138) or partially (SEQ ID Nos. 139 and 140). These variants were not
functional.
Modification of isolated residues in this domain may work, however large
continuous
blocks of modification in this domain reduce activity of the tracrRNA.
[00101] To further investigate which individual residues can be modified using
2'0Me RNA within the central domain of the tracrRNA, a single base
modification
2'0Me RNA 'walk' was done (SEQ ID Nos. 144-162). Within this series,
modification
as residues A14, A19, A20, G21, G22, A25, and C29 showed no loss of activity
and are
candidates for modification.
[00102] Antisense oligonucleotides are often made using complete PS
modification,
where every internucleotide linkage is phosphorothioate modified. This
extensive level
of modification is possible because the protein effector molecule RNase H1
(which
mediates ASO-directed mRNA degradation) tolerates the PS modification in the
ASO
when forming a functional substrate/enzyme complex. On the other hand, siRNAs
do
not tolerate full PS modification; extensive PS modification disrupts
productive
interaction with the effector protein AGO2 (which mediates siRNA-directed mRNA
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
47
degradation). Extensive PS modification of the tracrRNA in the internal RNA
loops
disrupts functional interaction with Cas9 (Seq ID No. 133; 29 PS
modifications).
Limited PS end-modification can be done with no loss of activity (SEQ ID Nos.
98 and
101; 2-3 PP linkages on each end). Less extensive PS modification may be
tolerated in
the central domain. In particular, RNase cleavage mapping (where incubation of
the
tracrRNA in a series of serum or cell extract dilutions are used to find the
sites that are
most sensitive to RNase attack) may be used to identify critical sites where
PS
modification of only one or a few linkages may stabilize the RNA without
disrupting
function.
[00103] There are applications where the PS modification contributes to
chemical
toxicity. In this case use of other methods to block exonuclease attack are
desirable.
Options include end-modifiers such as inverted-dT or abasic groups such as
dSpacer, C3
spacer (propanediol), ZEN (napthyl-azo modifier), and others. Placement of
such end-
modifying groups can eliminate the need for terminal PS internucleotide
linkages.
EXAMPLE 7
[00104] Example 1 described chemical modification patterns that functioned
with
Cas9 in an in vitro biochemical target DNA cleavage assay. This example
demonstrates
functioning of chemically modified crRNAs to direct genome editing by the Spy
Cas9
nuclease in mammalian cells. Optimal modification patterns differ between in
vitro and
in vivo use.
[00105] A series of crRNAs (Table 7) were synthesized having a variety of
chemical
modifications, including: the ribose modifications 2'0Me RNA, 2'F, and LNA;
the end-
modifying groups propanediol spacer and napthyl-azo modifier (N,N-diethy1-4-(4-
nitronaphthalen-1-ylazo)-phenylamine, or "ZEN"), and an inverted-dT residue;
and
select internucleotide linkages with phosphorothioate modifications. See:
Lennox et al.,
Molecular Therapy Nucleic Acids 2:e117 2013 for structure of the napthyl-azo
modified
and use of the napthyl-azo modifier and propanediol modifier for use as end-
groups to
block exonuclease attack. The crRNAs had either a 19 base protospacer domain
targeting HPRT1 at the 5'-end (SEQ ID Nos. 1, 9 10, 14-16, 22-24, 163-173) or
a 20
base protospacer domain targeting the same site (SEQ ID Nos. 48, 174-237) with
a 16
base tracrRNA binding domain at the 3'-end. The crRNAs listed in Table 7 were
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
48
complexed with unmodified truncated (67 base) tracrRNA SEQ ID No. 2 (Table 1)
or
chemically-modified truncated (67 base) tracrRNA SEQ ID No. 100 (Table 6). The
use
of two tracrRNAs enables determination if function of chemical modified crRNAs
varies
if paired with a modified tracrRNA. The paired crRNA:tracrRNA RNA
oligonucleotides
were transfected into the HEK-Cas9 cells and processed as described
previously.
Relative gene editing activities were assessed by comparing cleavage rates in
the HPRT1
gene using the T7EI mismatch endonuclease cleavage assay, with quantitative
measurement of products done using the Fragment Analyzer.
Table 7: Optimization of crRNA oligonucleotide modification patterns in
mammalian cells.
SEQ Cleavage % Cleavage %
ID
crRNA Sequence (5'-3') tracrRNA tracrRNA
No .
SEQ ID No 2 SEQ ID No. 100
1 uuauauccaacacuucgugguuuuagagcuaugcu
63 61
9 uuauauccaacacuucgugguuuuagagcuaugcu
1 0
10 uuauauccaacacuucgugguuuuagagcuaugcu 0 1
22 uuauauccaacacuucgugguuuuagagcuaugcu 1 1
23 uuauauccaacacuucgugguuuuagagcuaugcu
5 ND
24 uuauauccaacacuucgugguuuuagagcuaugcu
3 5
14 uuauauccaacacuucgugguuuuagagcuaugcu 63 26
15 uuauauccaacacuucgugguuuuagagcuaugcu 5 3
16 uuauauccaacacuucgugguuuuagagcuaugcu 5 5
163
C3 -uuauauccaacacuucgugguuuuagagcuau
65 49
gcu-C3
u*u*a*uauccaacacuucgugguuuuagagcuau
164 *g*c * 65 65
u
165 uuauauccaacacuucgugguuuuagagcuaugcu 0 3
166 uuauauccaacacuucgugguuuuagagcuaugcu 54 42
167 uuauauccaacacuucgugguuuuagagcuaugcu 49 58
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
49
SEQ Cleavage % Cleavage %
ID
crRNA Sequence (5'-3') tracrRNA tracrRNA
No.
SEQ ID No 2 SEQ ID No. 100
168 uuauauccaacacuucgugguuuuagagcuaugcu 64 60
169 uuauauccaacacuucgugguuuuagagcuaugcu 16 16
170 uuauauccaacacuucgugguuuuagagcuaugcu 3 3
171 uuauauccaacacuucgugguuuuagagcuaugcu 42 62
172 uuauauccaacacuucgugguuuuagagcuaugcu 4 13
173 uuauauccaacacuucgugguuuuagagcuaugcu 1 1
4 cuuauauccaacacuucgugguuuuagagcuaugc
8 61 60
u
cuuauauccaacacuucgugguuuuagagcuaugc
174 60 59
U
cuuauauccaacacuucgugguuuuagagcuaugc
175 62 60
U
cuuauauccaacacuucgugguuuuagagcuaugc
176 61 59
U
c*u*u*auauccaacacuucgugguuuuagagcua
177 60 59
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
1 '78 ¨ ¨ ¨ 61 59
u*g*c*u
C3 - cuuauauccaacacuucgugguuuuagagcua
179 61 58
ugcu
180
cuuauauccaacacuucgugguuuuagagcuaugc
57 59
u -C3
181
C3 - cuuauauccaacacuucgugguuuuagagcua
62 59
ugcu -C3
1 ZEN- cuuauauccaacacuucgugguuuuagagcu
82 64 62
augcu
cuuauauccaacacuucgugguuuuagagcuaugc
183 62 60
u- ZEN
ZEN- cuuauauccaacacuucgugguuuuagagcu
184 64 64
augcu- ZEN
ZEN- cuuauauccaacacuucgugguuuuagagcu
185 60 63
augcu- ZEN
u*u*a*uauccaacacuucgugguuuuagagcuau
186 64 62
*g*c *u
c*u*u*auauccaacacuucgugguuuuagagcua
18'7 65 65
u*g*c*u
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
SEQ Cleavage % Cleavage %
crRNA Sequence (5'-3') tracrRNA tracrRNA
ID No.
SEQ ID No 2 SEQ ID No. 100
c*u*u*auauccaacacuucgugguuuuagagcua
188 ¨ ¨ ¨ 0 0
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
189 ¨ ¨ ¨
63 64
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
64 62
190 _ _ _
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
64 63
191 _ _ _
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
192 _ _ _ 64 64
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
193 _ _ _ 64 65
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
194 _ _ _ 60 63
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
195 _ _ _ 63 62
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
196 _ _ _ _ 62 63
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
197 _ _ _ _ 61 64
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
198 ¨ ¨ ¨ 61 64
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
199 _ _ _ 63 68
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
200 _ _ _ 59 67
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
63 67
201 _ _ _
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
69
202 _ _ _
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
53 67
203 _ _ _
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
204 _ _ _ 54 67
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
205 _ _ _ 59 62
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
206 _ _ _ 58 61
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
207 _ _ _ 50 60
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
208 _ _ _
0 7
u*g*c*u
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
51
SEQ Cleavage % Cleavage %
ID
crRNA Sequence (5'-3') tracrRNA tracrRNA
No.
SEQ ID No 2 SEQ ID No. 100
2
09
c*u*u*auauccaacacuucgugguuuuagagcua _ _ _
0 0
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
210 _ _ _
u*g*c*u 0 0
c*u*u*auauccaacacuucgugguuuuagagcua
211 _ _ _
u*g*c*u 0 0
c*u*u*auauccaacacuucgugguuuuagagcua
212 _ _ _
u*g*c*u 56 68
c*u*u*auauccaacacuucgugguuuuagagcua
213 _ _ _ 41 64
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
214 _ _ _ 53 67
u*g*c*u
215
c*u*u*auauccaacacuucgugguuuuagagcua _ _ _
0 2
u*g*c*u
216
c*u*u*auauccaacacuucgugguuuuagagcua _ _ _
0 0
u*g*c*u
21
c*u*u*auauccaacacuucgugguuuuagagcua 7 _ _ _
0 0
u*g*c*u
21
8
c*u*u*auauccaacacuucgugguuuuagagcua _ _ _
0 0
u*g*c*u
2
c*u*u*auauccaacacuucgugguuuuagagcua 19 _ _ _
0 0
u*g*c*u
220
c*u*u*auauccaacacuucgugguuuuagagcua _ _ _
0 0
u*g*c*u
+c*+t*uauauccaacacuucgugguuuuagagcu
22158 61
aug*+c*+t
c*u*u*auauccaacacuucgugguuuuagagcua
222 _ _ _
u*g*c*u 31 54
c*u*u*auauccaacacuucgugguuuuagagcua
223 _ _ _
u*g*c*u 6 60
c*u*u*auauccaacacuucgugguuuuagagcua
224 _ _ _
u*g*c*u 27 57
225
c*u*u*auauccaacacuucgugguuuuagagcua _ _ _
0 2
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
6
22O _ _ _ 2 25
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
7
22 _ _ _ 3 31
u*g*c*u
8
c*u*u*auauccaacacuucgugguuuuagagcua
22 _ _ _ 4 35
u*g*c*u
22
c*u*u*auauccaacacuucgugguuuuagagcua 9 _ _ _
0 0
u*g*c*u
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
52
SEQ Cleavage % Cleavage %
crRNA Sequence (5'-3') tracrRNA tracrRNA
ID No.
SEQ ID No 2 SEQ ID No. 100
c*u*u*auauccaacacuucgugguuuuagagcua
230 _ _ _
0 0
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
2310 1
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
0 0
232 _ _ _
u*g*c *u
c*u*u*auauccaacacuucgugguuuuagagcua
33 67
233 _ _ _
u*g*c *u
c*u*u*auauccaacacuucgugguuuuagagcua
234 _ _ _ 24 66
u*g*c*u
C3 -cuuauauccaacacuucgugguuuuagagcua
235 56 65
ugcu - C 3
C3 -cuuauauccaacacuucgugguuuuagagcua
236 11 55
ugcu - C 3
C3 -cuuauauccaacacuucgugguuuuagagcua
237 62 65
ugcu- I nvT
c*u*u*auauccaacacuucgugguuuuagagcua
238 ¨ ¨ ¨ 17 67
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
239 _ _ _ 39 66
u*g*c*u
C3 -
240 cuuauauccaacacuucgugguuuuagagcuaugc 27 63
u-C3
C3 -
241 cuuauauccaacacuucgugguuuuagagcuaugc 14 46
u-C3
ZEN-
242 cuuauauccaacacuucgugguuuuagagcuaugc 41 67
u- ZEN
ZEN-
243 cuuauauccaacacuucgugguuuuagagcuaugc 23 24
u- ZEN
c*u*u*auauccaacacuucgugguuuuagagcua
244 _ _ _ _ ND 60
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
245 _ _ _ ND 65
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
246 _ _ _ ND 64
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
247 _ _ _ ND 64
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
248 _ _ _ ND 63
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
249 _ _ _ ND 53
u*g*c*u
c*u*u*auauccaacacUUCGUGGUUUuagagcua
250 _ _ _
u*g*c *u 0 2
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
53
SEQ Cleavage % Cleavage %
ID
crRNA Sequence (5'-3') tracrRNA tracrRNA
No.
SEQ ID No 2 SEQ ID No. 100
251
c*u*u*auauccaacacUUCGUGguuuuagagcua _ _ _
0 0
u*g*c*u
c*u*u*auauccaacacuucgugGUUUuagagcua
252 _ _ _
u*g*c*u 0 18
c*u*u*auauccaacac u ucgugguu uuagagcua
253 _ _ _
u*g*c*u 0 3
c*u*u*auauccaacac u ucgugguuuuagagcua
254 _ _ _ 5 0
u*g*c*u
c*u*u*auauccaacacuucguggu u uuagagcua 5 _ _ _
0 0
u*g*c*u
C3 -cuuauauccaacacuucgugguuuuagagcua
256 27 53
ugcu - C 3
C3 -cuuauauccaacacuucgugguuuuagagcua
257 10 50
ugcu - C 3
c*u*u*auauccaacacuucgugguuuuagagcua
258 - ¨ ¨ 29 47
u*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcua
259 _ _ _ 7 45
*u*g*c
2
c*u*u*auauccaacacuucgugguuuuagagcu* 60 _ _ _
0 4
a*u*g
c*u*u*auauccaacacuucgugguuuuagagc*u
2610 0
*a*u
c *u*u*auauccaacacuucgugguuuuagag*c *
2620 0
u*a
c*u*u*auauccaacacuucgugguuuuaga*g*c
263 0 0
*u
c*u*u*auauccaacacuucgugguuuuagagcua
264 _ _ _ 50 62
u*g*c*u
c*u*u*auauccaacacuucgugg*u*u*u*uaga
265 _ _ _ 45 59
gcuau*g*c*u
c*u*u*auaucca*a*c*a*c*u*u*c*g*u*g*g
266 26 36
uuuuagagcuau*g*c*u
c*u*u*auaucca*a*c*a*c*u*u*c*g*u*g*g
267 20 34
*u*u*u*uagagcuau*g*c*u
C3 -cuuauauccaacacuucgugguuuuagagcua
268 27 59
ugcu - C 3
C3 -cuuauauccaacacuucgugg*u*u*u*uaga
269 45 60
gcuaugcu - C 3
C3 -cuuauaucca*a*c*a*c*u*u*c*g*u*g*g
270 16 43
uuuuagagcuaugcu - C 3
C3 -cuuauaucca*a*c*a*c*u*u*c*g*u*g*g
271 22 45
*u*u*u*uagagcuaugcu - C 3
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
54
SEQ Cleavage % Cleavage %
ID
crRNA Sequence (5'-3') tracrRNA tracrRNA
No.
SEQ ID No 2 SEQ ID No. 100
cuuauauccaacacuucgugguuuuagagcuaugc
272 63 57
U
273 cuuauauccaacacuucgugguuuuagagcuaugc
59 60
cuuauauccaacacuucgugguuuuagagcuaugc
274 u 63 63
2'7 cuuauauccaacacuucgugguuuuagagcuaugc
64 62
cuuauauccaacacuucgugguuuuagagcuaugc
276 u 0 1
cuuauauccaacacuucgugguuuuagagcuaugc
277 u 5 16
78
cuuauauccaacacuucgugguuuuagagcuaugc
2 64 61
cuuauauccaacacuucgugguuuuagagcuaugc
279 64 63
U
cuuauauccaacacuucgugguuuuagagcuaugc
280 30 49
U
cuuauauccaacacuucgugguuuuagagcuaugc
281 56 60
cuuauauccaacacuucgugguuuuagagcuaugc
282 53 61
U
cuuauauccaacacuucgugguuuuagagcuaugc
283 0 3
cuuauauccaacacuucgugguuuuagagcuaugc
284 0 2
U
cuuauauccaacacuucgugguuuuagagcuaugc
285 2 8
cuuauauccaacacuucgugguuuuagagcuaugc
286 u 48 61
0
a*u*a*uccaacacuucgugguuuuagagcuau*g
2 0
87
u*a*u*ccaacacuucgugguuuuagagcuau*g*
288 0 0
c*u
289
+A*+T*a*uccaacacuucgugguuuuagagcuau
2 14
*g*c*u
+T*+A*u*ccaacacuucgugguuuuagagcuau*
2900 0
g*c*u
Oligonucleotide sequences are shown 5'-3'. Uppercase = DNA; Lowercase = RNA;
Underlined = 2'-0-methyl RNA; Italics = 2'-fluoro RNA; +a, +c, +t, +g = LNA;
C3 =
C3 spacer (propanediol modifier); * = phosphorothioate internucleotide
linkage; ZEN =
napthyl-azo modifier; InvT = inverted-dT. The relative functional activity of
each
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
species is indicated by the % cleavage in a T7EI heteroduplex assay when the
indicated
crRNA is paired with the indicated tracrRNA. ND = not determined.
[00106] Some kind of chemical modification is usually necessary for
synthetic nucleic
acids to function well in an intracellular environment due to the presence of
exonucleases and endonucleases that degrade unmodified oligonucleotides. A
wide
range of modifications have been described that confer nuclease resistance to
oligonucleotides. The precise combination and order of modifications employed
that
works well for a given application can vary with sequence context and the
nature of the
protein interactions required for biological function. Extensive prior work
has been done
relating to chemical modification of antisense oligonucleotides (which
interact with
RNase H1) and siRNAs (which interact with DICER, AG02, and other proteins). It
is
expected that chemical modification will improve function of the CRISPR
crRNA:tracrRNA complex. However, it is not possible to predict what
modifications
and/or pattern of modifications will be compatible with association of the
RNAs with
Cas9 in a functional way. The present invention defines minimal, moderate, and
extensive chemical modification patterns for the crRNA that retain high levels
of
function to direct Cas9 mediated gene editing in mammalian cells. The survey
in
Example 7 was performed targeting a single site in the human HPRT 1 gene. Note
that
modification patterns of the 20 base 5'-end protospacer guide domain of the
crRNA that
perform well may vary with sequence context. However, it is likely that
modification
patterns of the 3'-end tracrRNA binding domain that perform well as defined
herein will
be affected when the sequence of the adjacent protospacer domain changes when
different sites are targeted, so the 3'-domain modification patterns shown
here will be
"universal".
[00107] The results in Table 7 demonstrate that extensive modification is
tolerated
throughout the 5' and 3' ends of the crRNA. Modification of certain select
positions
within internal domains of the crRNA lead to reduced activity or totally
blocks activity,
likely due to altered structure of the folded RNA and/or blocking of protein
contact
points with the 2'-OH of key RNA residues by the 2'0Me modification. For
example,
compound SEQ ID No. 204 has 21/36 residues modified with 2'0Me RNA (58%) and
retains full activity compared with the unmodified sequence. Compound SEQ ID
No.
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
56
239 has 30/36 residues modified with 2'0Me RNA (83%) and retains full activity
compared with the unmodified sequence. Both of these compounds also have 3
phosphorothioate (PS) modified internucleotide linkages at the 5'- and 3'-
ends, which
provides additional protection against exonuclease attack. In contrast, SEQ ID
No. 165
has only 4/36 residues modified with 2'0Me RNA (11%) yet has totally lost
activity.
[00108] Large blocks of sequence were tolerant to 2'0Me modification at the 5'-
end
and 3'-end of the crRNA, however modification of certain residues in the
central portion
of the molecule led to inactivation. To further investigate which individual
residues can
be modified using 2'0Me RNA within the central domain of the crRNA, a single
base
modification 2'0Me RNA 'walk' was done (SEQ ID Nos. 272-286). Specific
residues
(positions within the crRNA) were identified that led to large reductions or
complete loss
of activity. Using the 36 base crRNA SEQ ID No. 239 as model and numbering
from
the 5'-end of the sequence, substitution of 2'0Me RNA for the natural RNA of
residues
U15 and U16 lead to substantial loss of activity and residue U19 led to a
moderate loss
of activity (FIG. 7). These 3 sites lie within the target-specific protospacer
guide
domain, so sequence varies with target (residues 15, 16, and 19, FIG. 7). It
is possible
that in certain sequence contexts that these sites will be tolerant to
modification. Within
the universal tracrRNA-binding domain (residues 21-36), substitution of 2'0Me
RNA
for the natural RNA of residues U22, U23, and U24 led to substantial loss of
activity.
Given that this domain does not change with sequence context, it is likely
that these sites
will not vary in modification tolerance as target sequence changes. Sequence-
specific
effects of modification in the 20-base target-specific protospacer guide
domain are
studies in greater detail in Example 10.
[00109] Antisense oligonucleotide are often made with complete PS
modification,
where every internucleotide linkage is phosphorothioate modified. This
extensive level
of modification is possible because the protein effector molecule RNase H1
tolerates the
PS modification in the ASO when forming a functional substrate/enzyme complex.
On
the other hand, siRNAs do not tolerate full PS modification; extensive PS
modification
disrupts productive interaction with the effector protein AG02. Limited PS end
modification of the crRNA can be done with no loss of activity (SEQ ID Nos.
177, 178,
239, etc., have 3 PS linkages on each end). End-modification is desirable as
this adds
additional protection from exonuclease attack. PS modification of select
internal sites
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
57
may also be tolerated and may provide additional protection from endonuclease
attack.
Using SEQ ID No. 264 as a base modification pattern, internal linkages were PS
modified in the tracrRNA-binding domain (SEQ ID No. 265), in the 3'-end of the
protospacer guide domain (seed region) (SEQ ID No. 266), or both regions (SEQ
ID No.
267). Increasing level of PS modification led to reduced functional activity,
with SEQ
ID No. 267 having ¨50% the activity of the less modified SEQ ID No. 264
variant. SEQ
ID No 267 has 21 out of 35 internucleotide linkages modified and will be
stable to
nuclease exposure. In cases where exposure to a high nuclease environment is
needed
(such as direct IV administration for research or therapeutic indications),
this highly
modified variant may actually show higher activity than the less modified
variants,
which will be degraded more quickly.
[00110] There are experimental settings where the PS modification contributes
to
chemical toxicity. In this case use of other methods to block exonuclease
attack are
desirable. The crRNA can have a C3 spacer (propanediol modifier) or a ZEN
(napthyl-
azo modifier) placed on either or both the 5'-end and 3'-end to block
exonuclease attack,
obviating the need the PS modification. This strategy can be employed to
eliminate the
PS-end block modification (See SEQ ID Nos. 179-186). This strategy can be used
to
reduce PS content of more highly modified crRNA variants. SEQ ID No. 271 has
the
internal protospacer domain and tracrRNA binding domain PS-modified in the
same
pattern as SEQ ID No. 267, yet employs only 15 PS internucleotide linkages
(instead of
21) and shows improved activity. Therefore combination of non-base end-blocks
with
internal PS modification may be used to increase nuclease stability while
maintaining
high activity.
EXAMPLE 8
[00111] The following example demonstrates improved potency of the modified
CRISPR crRNAs and tracrRNAs of the present invention. Examples 2-7 employed
transfection of crRNA:tracrRNA complexes into human HEK-Cas9 cells at 30 nM
concentration. Experimental testing had previously shown that this dose
represented the
upper shoulder of the dose response curve such that using higher doses of RNA
did not
improve gene editing efficiency but use of lower doses resulted lower gene
editing
efficiency. Those measurements were done using unmodified RNAs. The present
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
58
example re-examines the dose response of new optimized chemically modified
RNAs of
the present invention compared with unmodified RNAs and demonstrates that
chemical
modification (i.e., nuclease stabilization) results in more potent compounds
which can be
used at lower dose.
[00112] Example 5 demonstrated that the truncated guide RNAs of the present
invention performed superior to WT RNAs at 12 sites in the human HPRT1 gene.
Four
of these sites (38087, 38231, 38133, and 38285) were chosen for comparison of
unmodified vs. modified RNA in the present example. Unmodified crRNAs were
paired
with the unmodified tracrRNA (SEQ ID No. 2) at a 1:1 molar ratio. Unmodified
crRNAs were paired with the modified tracrRNA (SEQ ID No. 100) at a 1:1 molar
ratio.
Modified crRNAs were paired with the modified tracrRNA (SEQ ID No. 100) at a
1:1
molar ratio. Sequences are shown in Table 8. RNAs were transfected into HEK-
Cas9
cells as described previously at 30 nM, 10 nM, and 3 nM concentrations. Cells
were
incubated for 48 hours at 37 C, then were processed for DNA and studied for
evidence
of gene editing activity comparing cleavage rates at the HPRT1 locus in the
T7EI
mismatch endonuclease assay, with quantitative measurement of products done
using the
Fragment Analyzer as previously described. Results are shown in Table 8.
Table 8: Increased potency of modified vs. unmodified crRNA:tracrRNA complexes
to direct Cas9-mediated gene editing in mammalian cells.
cr/tracr30 nM 10 nM 3
nM
SEQ crRNA Sequence
RNA
Cleavage Cleavage Cleavage
pair ID No. tracrRNA Sequence
aauuauggggauuacuaggaguuuuagagcuaug
38087 56
Cu
Un-cr 80 76 35
agcauagcaaguuaaaauaaggcuaguccguuau
Un-tr 2 caacuugaaaaaguggcaccgagucggugcuuu
aauuauggggauuacuaggaguuuuagagcuaug
38087 56
Cu
Un-cr a*g*cauagcaaguuaaaauaaggcuaguccguu 83 76 50
Mod-tr 100 aucaacuugaaaaaguggcaccgagucggugcu*
u*u
a*a*u*uauggggauuacuaggaguuuuagagcu
38087 445 ¨au¨*g¨*c*u
Mod-cr a*g*cauagcaaguuaaaauaaggcuaguccguu 77 77 54
Mod-tr 100 aucaacuugaaaaaguggcaccgagucggugcu*
u*u
38231 69 uuuuguaauuaacagcuugcguuuuagagcuaug
31 4 0
Un-cr Cu
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
59
cr/tracr30 nM 10 nM 3
nM
SEQ crRNA Sequence
RNA
Cleavage Cleavage Cleavage
ID No.
pair tracrRNA Sequence
Un-tr agcauagcaaguuaaaauaaggcuaguccguuau
2 caacuugaaaaaguggcaccgagucggugcuuu
uuuuguaauuaacagcuugcguuuuagagcuaug
38231 69 Cu
Un-cr a*g*cauagcaaguuaaaauaaggcuaguccguu 45 14 1
Mod-tr 100 aucaacuugaaaaaguggcaccgagucggugcu*
u*u
u*u*u*uguaauuaacagcuugcguuuuagagcu
38231 446 ¨au¨*g*c*u
Mod-cr a*g*cauagcaaguuaaaauaaggcuaguccguu 48 25 4
Mod-tr 100 aucaacuugaaaaaguggcaccgagucggugcu*
u*u
ggucacuuuuaacacacccaguuuuagagcuaug
38133 78
Cu
Un-cr 73 61 27
agcauagcaaguuaaaauaaggcuaguccguuau
Un-tr 2 caacuugaaaaaguggcaccgagucggugcuuu
ggucacuuuuaacacacccaguuuuagagcuaug
38133 78
au.
Un-cr a*g*cauagcaaguuaaaauaaggcuaguccguu 74 61 37
Mod-tr 100 aucaacuugaaaaaguggcaccgagucggugcu*
u*u
g*g*u*cacuuuuaacacacccaguuuuagagcu
38133 447 au*g*c*u
Mod-cr a*g*cauagcaaguuaaaauaaggcuaguccguu 75 66 55
Mod-tr 100 aucaacuugaaaaaguggcaccgagucggugcu*
u*u
cuuauauccaacacuucgugguuuuagagcuaug
38285 48 Cu
Un-cr 66 16 2
agcauagcaaguuaaaauaaggcuaguccguuau
Un-tr 2 caacuugaaaaaguggcaccgagucggugcuuu
cuuauauccaacacuucgugguuuuagagcuaug
38285 48
au.
Un-cr a*g*cauagcaaguuaaaauaaggcuaguccguu 67 16 5
Mod-tr 100 aucaacuugaaaaaguggcaccgagucggugcu*
u*u
c*u*u*auauccaacacuucgugguuuuagagcu
38285 178 ¨au¨*g¨*c*u
Mod-cr a*g*cauagcaaguuaaaauaaggcuaguccguu 62 60 26
Mod-tr 100 aucaacuugaaaaaguggcaccgagucggugcu*
u*u
Oligonucleotide sequences are shown 5'-3'. Lowercase = RNA; Underlined = 2' -0-
methyl RNA; * = phosphorothioate internucleotide linkage. Unmodified crRNA =
Un-
cr. Unmodified tracrRNA = Un-tr. Modified crRNA = Mod-cr. Modified tracrRNA =
Mod-tr. The relative functional activity of each species is indicated by the %
cleavage in
a T7EI heteroduplex assay for each dose studied.
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
[00113] In general, modification of the crRNA and tracrRNA had a small impact
on
gene editing efficiency when the RNAs were transfected at high dose where the
RNAs
are present in excess. At lower doses, the modified reagents showed improved
potency
and, in some cases, markedly improved potency. The degree of improvement
varied
with site. The very potent site 38087 showed highly efficiency gene editing at
the 30 nM
and 10 nM doses with all crRNA/tracrRNA variants tested, but at the 3 nM use
of the
modified tracrRNA (with either of the crRNAs) showed improved activity. A low
potency site, such as 38231, showed improved gene editing efficiency even at
the highest
dose tested (30 nM) using the modified RNAs. Modification of the tracrRNA
alone
showed benefit, but the greatest benefit was realized when both the crRNA and
tracrRNA were modified. FIG. 8 shows a schematic of one effective modified
crRNA
(SEQ ID No. 178) paired with modified tracrRNA (SEQ ID No. 100), specific for
HPRT1 site 38285. FIG. 9 shows a schematic of a more highly modified pair that
is also
highly functional, crRNA (SEQ ID No. 239) paired with modified tracrRNA (SEQ
ID
No. 134), also specific for HPRT1 site 38285.
[00114] The present example employed transfection of the crRNA:tracrRNA
complex
into HEK-Cas9 cells, where Cas9 protein is constitutively expressed. Therefore
transfected RNAs can bind Cas9 protein immediately, minimizing risk of
degradation in
the cytoplasm by nucleases. It is anticipated that the benefit of chemical
modification of
the crRNA and/or tracrRNA will be greater in cases where the transfected RNAs
must
survive exposure to cellular nucleases while Cas9 protein is being made, as
occurs when
using protocols where Cas9 mRNA or a Cas9 expression vector is co-transfected
with
the targeting RNAs, such that Cas9 is not already expressed in the cells. The
benefits of
using highly modified RNAs will be greatest for in vivo applications (such as
medical
therapeutics) where the RNAs may be exposed to both nucleases present in serum
(following IV administration) and cellular cytoplasmic nucleases.
EXAMPLE 9
[00115] Examples 2-8 demonstrate activity of truncated and/or chemically-
modified
CRISPR crRNAs and/or tracrRNAs to trigger Cas9-mediated genome editing in
mammalian cells that constitutively express Cas9. The present example
demonstrates
that the truncated, modified RNA compositions of the present invention can
bind Cas9
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
61
protein and this complex can be transfected into human cells and further that
transfection
of the ribonuclear protein (RNP) complex is sufficient to trigger highly
efficient genome
editing.
[00116] Reagents specific for human HPRT1 site 38285 were employed in the
present
example. Unmodified crRNA was paired with unmodified tracrRNA at a 1:1 molar
ratio. Unmodified crRNA was paired with modified tracrRNA at a 1:1 molar
ratio.
Modified crRNA was paired with modified tracrRNA at a 1:1 molar ratio.
Sequences
are shown in Table 9. RNAs were transfected into unmodified HEK293 cells as
described above except that a 1:1 complex of Cas9 protein (Caribou
Biosciences) with
crRNA:tracrRNA were employed at 10 nM concentration using increased amounts of
RNAiMAX lipid transfection reagent (1.2 [AL, increased over the 0.75 [AL
amount used
per 100 [AL transfection in 96 well format for the 30 nM RNA-alone
transfections in
HEK-Cas9 cells). Cells were incubated for 48 hours at 37 C, then were
processed for
DNA and studied for evidence of gene editing activity comparing cleavage rates
at the
HPRT1 locus in the T7EI mismatch endonuclease assay, with quantitative
measurement
of products done using the Fragment Analyzer as previously described. Results
are
shown in Table 9.
Table 9: Increased potency of modified vs. unmodified crRNA:tracrRNA complexes
to direct Cas9-mediated gene editing in mammalian cells.
cr/tracr10 nM
SEQ crRNA Sequence
RNA Cleavage
pair ID No. tracrRNA Sequence
38285 48 cuuauauccaacacuucgugguuuuagagcuaugcu
Un-cr 42
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaa
Un-tr 2 aaaguggcaccgagucggugcuuu
38285 48 cuuauauccaacacuucgugguuuuagagcuaugcu
Un-cr 41
Modtr 100 a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuug
- ¨
aaaaaguggcaccgagucggugcu*u*u
38285 178 c*u*u*auauccaacacuucgugguuuuagagcuau*g*c*u
Mod-cr 54
Modtr 100 a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuug
- ¨
aaaaaguggcaccgagucggugcu*u*u
Oligonucleotide sequences are shown 5'-3'. Lowercase = RNA; Underlined = 2'-0-
methyl RNA; * = phosphorothioate internucleotide linkage. Unmodified crRNA =
Un-
cr. Unmodified tracrRNA = Un-tr. Modified crRNA = Mod-cr. Modified tracrRNA =
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
62
Mod-tr. The relative functional activity of each complex is indicated by the %
cleavage
in a T7EI heteroduplex assay for each dose studied.
[00117] All 3 CRISPR RNA complexes performed well in the RNP-transfection
protocol for mammalian genome editing. The unmodified crRNA + unmodified
tracrRNA pair (SEQ ID Nos. 48 and 2) and the unmodified crRNA + modified
tracrRNA
pair (SEQ ID Nos. 48 and 100) performed 2.5x better at 10 nM dose in the RNP
protocol
than in the HEK-Cas9 protocol, consistent with the less modified RNAs
suffering
degradation between transfection and eventual complexation with Cas9 protein
in the
cytoplasm or nucleus. Thus higher doses are needed for unmodified RNAs and in
some
settings it is likely that unmodified RNAs will fail to direct any genome
editing activity.
The modified crRNA + modified tracrRNA (SEQ ID NOs. 178 and 100), on the other
hand, worked with high efficiency in both protocols.
[00118] The modified, truncated CRISPR RNAs of the present invention work well
with direct Cas9 RNP transfection methods.
EXAMPLE 10
[00119] The chemical modification optimization studies performed in Examples 6
and
7 studied the activity of crRNAs having various modification patterns paired
with a
tracrRNA having various modification patterns. The tracrRNA is universal and
the same
sequence is employed at all target sites. It is expected that the performance
of various
modification patterns for the tracrRNA will be similar between different
target sites. The
crRNA, however, varies sequence between different target sites. In the
optimized
version tested in Examples 7 and 8, the 5'-20 bases of the crRNA are target-
specific (i.e.,
the "protospacer domain") and the 3'-16 bases are universal (i.e., "the
tracrRNA binding
domain"). Like the tracrRNA, it is expected that the performance of various
modification patterns in the universal 16 base 3'-domain of the crRNA will be
similar at
all target sites. However, it is possible that performance of different
modification
patterns may be influenced by the sequence context present in the 5'-20 base
target-
specific domain.
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
63
[00120] It is well established that effective modification patterns for
small interfering
RNAs (siRNAs) are affected by sequence context (Behlke, Oligonucleotides
18:305-320,
2008). For siRNAs, certain "limited modification" patterns can be applied to
all sites,
whereas for "heavy modification" it is not possible to predict which patterns
will be
functional for a given sequence and empiric testing is necessary. The present
example
studies the effect that sequence context has on the crRNA, testing different
modification
patterns within the 5'-20 base target-specific domain at different sites.
[00121] The modification studies in Examples 6 and 7 employed a single crRNA
PAM site in the human HPRT1 gene. The present study examines 12 sites in the
human
HPRT1 gene, including the site previously examined, comparing functional
performance
of different modification patterns and establishes a single modification
pattern that can
be employed with good results at all sites. See Example 5 for other studies
relating to
these 12 sites.
[00122] A series of crRNAs (Table 10) were synthesized having a protospacer
domain
lengths of 20 bases specific to 12 sites in the human HPRT1 gene with a 16mer
universal
tracrRNA binding sequence at the 3'-end. The crRNAs were made using a variety
of
chemical modifications, including: the ribose modifications 2'0Me RNA, the end-
modifying groups propanediol spacer and napthyl-azo modifier (N,N-diethy1-4-(4-
nitronaphthalen-1-ylazo)-phenylamine, or "ZEN"), an inverted-dT residue; and
select
internucleotide linkages with phosphorothioate modifications. A schematic
representation of the different modification patterns employed is shown in
FIG. 10.
[00123] The crRNAs were paired with a highly modified 67mer tracrRNA (SEQ ID
No. 100). The paired crRNA:tracrRNA RNA oligonucleotides were transfected into
the
HEK-Cas9 cells and processed as described in Example 2. Relative gene editing
activities were assessed by comparing cleavage rates in the HPRT1 gene using
the T7EI
mismatch endonuclease cleavage assay with quantitative measurement of products
done
using the Fragment Analyzer. Results are shown in Table 10 and in FIG. 11.
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
64
Table 10: Optimization of crRNA oligonucleotide modification patterns in
mammalian cells across 12 target sites.
HPRT1 SEQ M od
Cleavage %
P
Target ID attern crRNA Sequence (5'-3')
tracrRNA
site No. SEQ ID No. 100
38094 64 1 uccauuucauagucuuuccuguuuuagagcuaugcu 62
38231 69 1 uuuuguaauuaacagcuugcguuuuagagcuaugcu 35
38371 71 1 cuuagagaauauuuguagagguuuuagagcuaugcu 66
38509 73 1 uugacuauaaugaauacuucguuuuagagcuaugcu 71
38574 75 1 caaaacacgcauaaaaauuuguuuuagagcuaugcu 52
38087 56 1 aauuauggggauuacuaggaguuuuagagcuaugcu 72
38133 78 1 ggucacuuuuaacacacccaguuuuagagcuaugcu 65
38285 48 1 cuuauauccaacacuucgugguuuuagagcuaugcu 62
38287 80 1 ggcuuauauccaacacuucgguuuuagagcuaugcu 47
38358 60 1 auuucacauaaaacucuuuuguuuuagagcuaugcu 59
38636 83 1 ucaaauuaugaggugcuggaguuuuagagcuaugcu 27
38673 85 1 uacagcuuuaugugacuaauguuuuagagcuaugcu 49
u*c*c*auuucauagucuuuccuguuuuagagcuau
38094 291 271
*g*c*u
u*u*u*uguaauuaacagcuugcguuuuagagcuau
38231 292 2 _ _ _
*g*c*u 54
c*u*u*agagaauauuuguagagguuuuagagcuau
38371 293 265
*g*c*u
u*u*g*acuauaaugaauacuucguuuuagagcuau
38509 294 278
*g*c*u
c*a*a*aacacgcauaaaaauuuguuuuagagcuau
38574 295 2 _ _ _
*g*c*u 56
a*a*u*uauggggauuacuaggaguuuuagagcuau
38087 296 2 _ _ _
*g*c*u 76
g*g*u*cacuuuuaacacacccaguuuuagagcuau
38133 297 270
*g*c*u
c*u*u*auauccaacacuucgugguuuuagagcuau
38285 178 2 _ _ _
*g*c*u 65
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
HPRT1 SEQ Cleavage %
Mod
Target ID crRNA Sequence (5'-3')
tracrRNA
Pattern
site No. SEQ
ID No. 100
g*g*c*uuauauccaacacuucgguuuuagagcuau
38287 298 259
*g*c*u
38358 299 2 a*u*u*ucacauaaaacucuuuuguuuuagagcuau
*g*c*u 73
u*c*a*aauuaugaggugcuggaguuuuagagcuau
38636 300 229
*g*c*u
u*a*c *agcuuuaugugacuaauguuuuagagcuau
38673 301 260
*g*c*u
u*c*c*auuucauagucuuuccuguuuuagagcuau
38094 302 367
*g*c*u
u*u*u*uguaauuaacagcuugcguuuuagagcuau
38231 303 357
*g*c*u
c*u*u*agagaauauuuguagagguuuuagagcuau
38371 304 365
*g*c*u
u*u*g*acuauaaugaauacuucguuuuagagcuau
38509 305 379
*g*c*u
38574 306 3 c*a*a*aacacgcauaaaaauuuguuuuagagcuau
*g*c*u 52
38087 307 3 a*a*u*uauggggauuacuaggaguuuuagagcuau
*g*c*u 76
g*g*u*cacuuuuaacacacccaguuuuagagcuau
38133 308 366
*g*c*u
38285 309 3 c*u*u*auauccaacacuucgugguuuuagagcuau
*g*c*u 60
g*g*c*uuauauccaacacuucgguuuuagagcuau
38287 310 356
*g*c*u
38358 311 3 a*u*u*ucacauaaaacucuuuuguuuuagagcuau
*g*c*u 66
u*c*a*aauuaugaggugcuggaguuuuagagcuau
38636 312 324
*g*c*u
u*a*c *agcuuuaugugacuaauguuuuagagcuau
38673 313 3
51
*g*c*u
u*c*c*auuucauagucuuuccuguuuuagagcuau
38094 314 468
*g*c*u
u*u*u*uguaauuaacagcuugcguuuuagagcuau
38231 315 453
*g*c*u
c*u*u*agagaauauuuguagagguuuuagagcuau
38371 316 465
*g*c*u
u*u*g*acuauaaugaauacuucguuuuagagcuau
38509 317 476
*g*c*u
38574 318 4 c*a*a*aacacgcauaaaaauuuguuuuagagcuau
*g*c*u 51
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
66
HPRT1 SEQ
Cleavage %
Mod
Target ID crRNA Sequence (5'-3')
tracrRNA
Pattern
site No. SEQ
ID No. 100
38087 319 4 a*a*u*uauggggauuacuaggaguuuuagagcuau
_
*g*c*u 76
g*g*u*cacuuuuaacacacccaguuuuagagcuau
38133 320 470
*g*c*u
38285 321 4 c*u*u*auauccaacacuucgugguuuuagagcuau
*g*c*u 65
g*g*c*uuauauccaacacuucgguuuuagagcuau
38287 322 456
*g*c*u
38358 323 4 a*u*u*ucacauaaaacucuuuuguuuuagagcuau
*g*c*u 64
u*c*a*aauuaugaggugcuggaguuuuagagcuau
38636 324 423
*g*c*u
u*a*c*agcuuuaugugacuaauguuuuagagcuau
38673 325 448
*g*c*u
u*c*c*auuucauagucuuuccuguuuuagagcuau
38094 326 571
*g*c*u
u*u*u*uguaauuaacagcuugcguuuuagagcuau
38231 327 553
*g*c*u
c*u*u*agagaauauuuguagagguuuuagagcuau
38371 328 569
*g*c*u
u*u*g*acuauaaugaauacuucguuuuagagcuau
38509 329 577
*g*c*u
38574 330 5 c*a*a*aacacgcauaaaaauuuguuuuagagcuau
*g*c*u 51
38087 331 5 a*a*u*uauggggauuacuaggaguuuuagagcuau
*g*c*u 80
g*g*u*cacuuuuaacacacccaguuuuagagcuau
38133 332 570
*g*c*u
38285 333 5 c*u*u*auauccaacacuucgugguuuuagagcuau
*g*c*u 64
g*g*c*uuauauccaacacuucgguuuuagagcuau
38287 334 559
*g*c*u
38358 335 5 a*u*u*ucacauaaaacucuuuuguuuuagagcuau
*g*c*u 64
u*c*a*aauuaugaggugcuggaguuuuagagcuau
38636 336 525
*g*c*u
u*a*c*agcuuuaugugacuaauguuuuagagcuau
38673 337 556
*g*c*u
u*c*c*auuucauagucuuuccuguuuuagagcuau
38094 338 670
*g*c*u
u*u*u*uguaauuaacagcuugcguuuuagagcuau
38231 339 653
*g*c*u
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
67
HPRT1 SEQ
Cleavage %
Mod
Target ID crRNA Sequence (5'-3')
tracrRNA
Pattern
site No. SEQ
ID No. 100
c*u*u*agagaauauuuguagagguuuuagagcuau
38371 340 668
*g*c*u
u*u*g*acuauaaugaauacuucguuuuagagcuau
38509 341 672
*g*c*u
38574 342 6 c*a*a*aacacgcauaaaaauuuguuuuagagcuau
*g*c*u 51
38087 343 6 a*a*u*uauggggauuacuaggaguuuuagagcuau
_
*g*c*u 81
g*g*u*cacuuuuaacacacccaguuuuagagcuau
38133 344 671
*g*c*u
38285 345 6 c*u*u*auauccaacacuucgugguuuuagagcuau
*g*c*u 64
g*g*c*uuauauccaacacuucgguuuuagagcuau
38287 346 655
*g*c*u
38358 347 6 a*u*u*ucacauaaaacucuuuuguuuuagagcuau
*g*c*u 65
u*c*a*aauuaugaggugcuggaguuuuagagcuau
38636 348 624
*g*c*u
u*a*c*agcuuuaugugacuaauguuuuagagcuau
38673 349 655
*g*c*u
u*c*c*auuucauagucuuuccuguuuuagagcuau
38094 350 773
*g*c*u
u*u*u*uguaauuaacagcuugcguuuuagagcuau
38231 351 751
*g*c*u
c*u*u*agagaauauuuguagagguuuuagagcuau
38371 352 773
*g*c*u
u*u*g*acuauaaugaauacuucguuuuagagcuau
38509 353 778
*g*c*u
38574 354 7 c*a*a*aacacgcauaaaaauuuguuuuagagcuau
*g*c*u 50
38087 355 7 a*a*u*uauggggauuacuaggaguuuuagagcuau
*g*c*u 83
g*g*u*cacuuuuaacacacccaguuuuagagcuau
38133 356 763
*g*c*u
38285 357 7 c*u*u*auauccaacacuucgugguuuuagagcuau
*g*c*u 63
g*g*c*uuauauccaacacuucgguuuuagagcuau
38287 358 743
*g*c*u
38358 359 7 a*u*u*ucacauaaaacucuuuuguuuuagagcuau
*g*c*u 66
u*c*a*aauuaugaggugcuggaguuuuagagcuau
38636 360 728
*g*c*u
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
68
HPRT1 SEQ
Cleavage %
Mod
Target ID crRNA Sequence (5'-3')
tracrRNA
Pattern
site No. SEQ
ID No. 100
u*a*c*agcuuuaugugacuaauguuuuagagcuau
38673 361 761
*g*c*u
u*c*c*auuucauagucuuuccuguuuuagagcuau
38094 362 863
*g*c*u
u*u*u*uguaauuaacagcuugcguuuuagagcuau
38231 363 840
*g*c*u
c*u*u*agagaauauuuguagagguuuuagagcuau
38371 364 864
*g*c*u
u*u*g*acuauaaugaauacuucguuuuagagcuau
38509 365 867
*g*c*u
38574 366 8 c*a*a*aacacgcauaaaaauuuguuuuagagcuau
*g*c*u 18
38087 367 8 a*a*u*uauggggauuacuaggaguuuuagagcuau
*g*c*u 75
g*g*u*cacuuuuaacacacccaguuuuagagcuau
38133 368 848
*g*c*u
38285 369 8 c*u*u*auauccaacacuucgugguuuuagagcuau
*g*c*u 53
g*g*c*uuauauccaacacuucgguuuuagagcuau
38287 370 824
*g*c*u
38358 371 8 a*u*u*ucacauaaaacucuuuuguuuuagagcuau
*g*c*u 56
u*c*a*aauuaugaggugcuggaguuuuagagcuau
38636 372 822
*g*c*u
u*a*c*agcuuuaugugacuaauguuuuagagcuau
38673 373 850
*g*c*u
u*c*c*auuucauagucuuuccuguuuuagagcuau
38094 374 965
*g*c*u
u*u*u*uguaauuaacagcuugcguuuuagagcuau
38231 375 97
*g*c*u
c*u*u*agagaauauuuguagagguuuuagagcuau
38371 376 970
*g*c*u
u*u*g*acuauaaugaauacuucguuuuagagcuau
38509 377 957
*g*c*u
38574 378 9 c*a*a*aacacgcauaaaaauuuguuuuagagcuau
*g*c*u 8
38087 379 9 a*a*u*uauggggauuacuaggaguuuuagagcuau
*g*c*u 74
g*g*u*cacuuuuaacacacccaguuuuagagcuau
38133 380 938
*g*c*u
38285 222 9 c*u*u*auauccaacacuucgugguuuuagagcuau
*g*c*u 54
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
69
HPRT1 SEQ M od
Cleavage %
P
Target ID attern crRNA Sequence (5'-3')
tracrRNA
site No. SEQ
ID No. 100
g*g*c*uuauauccaacacuucgguuuuagagcuau
38287 381 932
*g*c*u
a*u*u*ucacauaaaacucuuuuguuuuagagcuau
38358 382 9 *g*c*u 58
u*c*a*aauuaugaggugcuggaguuuuagagcuau
38636 383 9 *g*c*u 19
u*a*c*agcuuuaugugacuaauguuuuagagcuau
38673 384 955
*g*c*u
C3-
38094 385 10 uccauuucauagucuuuccuguuuuagagcuaugcu 66
-C3
C3-
38231 386 10 uuuuguaauuaacagcuugcguuuuagagcuaugcu 54
-C3
C3-
38371 387 10 cuuagagaauauuuguagagguuuuagagcuaugcu 57
-C3
C3-
38509 388 10 uugacuauaaugaauacuucguuuuagagcuaugcu 75
-C3
C3-
38574 389 10 caaaacacgcauaaaaauuuguuuuagagcuaugcu 50
-C3
C3-
38087 390 10 aauuauggggauuacuaggaguuuuagagcuaugcu 71
-C3
C3-
38133 391 10 ggucacuuuuaacacacccaguuuuagagcuaugcu 68
-C3
C3-
38285 181 10 cuuauauccaacacuucgugguuuuagagcuaugcu 58
-C3
C3-
38287 392 10 ggcuuauauccaacacuucgguuuuagagcuaugcu 57
-C3
C3-
38358 393 10 auuucacauaaaacucuuuuguuuuagagcuaugcu 64
-C3
C3-
38636 394 10 ucaaauuaugaggugcuggaguuuuagagcuaugcu 22
-C3
C3-
38673 395 10 uacagcuuuaugugacuaauguuuuagagcuaugcu 50
-C3
ZEN-
38094 396 11 uccauuucauagucuuuccuguuuuagagcuaugcu 74
-ZEN
ZEN-
38231 397 11 uuuuguaauuaacagcuugcguuuuagagcuaugcu 44
-ZEN
ZEN-
38371 398 11 cuuagagaauauuuguagagguuuuagagcuaugcu 72
-ZEN
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
HPRT1 SEQ M od
Cleavage %
P
Target ID attern crRNA Sequence (5'-3')
tracrRNA
site No. SEQ
ID No. 100
ZEN-
38509 399 11 uugacuauaaugaauacuucguuuuagagcuaugcu 74
-ZEN
ZEN-
38574 400 11 caaaacacgcauaaaaauuuguuuuagagcuaugcu 57
-ZEN
ZEN-
38087 401 11 aauuauggggauuacuaggaguuuuagagcuaugcu 82
-ZEN
ZEN-
38133 402 11 ggucacuuuuaacacacccaguuuuagagcuaugcu 73
-ZEN
ZEN-
38285 184 11 cuuauauccaacacuucgugguuuuagagcuaugcu 60
-ZEN
ZEN-
38287 403 11 ggcuuauauccaacacuucgguuuuagagcuaugcu 62
-ZEN
ZEN-
38358 404 11 auuucacauaaaacucuuuuguuuuagagcuaugcu 69
-ZEN
ZEN-
38636 405 11 ucaaauuaugaggugcuggaguuuuagagcuaugcu 26
-ZEN
ZEN-
38673 406 11 uacagcuuuaugugacuaauguuuuagagcuaugcu 44
-ZEN
Oligonucleotide sequences are shown 5'-3'. Lowercase = RNA; Underlined = 2'-0-
methyl RNA; C3 = C3 spacer (propanediol modifier); * = phosphorothioate
internucleotide linkage; ZEN = napthyl-azo modifier. The relative functional
activity of
each species is indicated by the % cleavage in a T7EI heteroduplex assay when
the
indicated crRNA is paired with the indicated tracrRNA at each of 12 sites in
human
HRPT1.
[00124] The modified crRNAs employed a fixed modification pattern in the 16-
base
3'-end domain which is universal and binds the tracrRNA. Different
modification
pattern were tested/compared in the 5'-end domain that is target specific
(i.e., sequence
varies with target site). The test set comprised variants having 0, 3, 4, 6,
8, 10, 12, 13, or
14 contiguous 2'0Me RNA residues starting at the 5'-end and walking towards
the 3'-
end. The modification patterns avoided positions previously demonstrated to
reduce
functional performance of the crRNA (Example 7). Use of only non-base modifier
end
groups (C3 spacer or ZEN) were also tested (without additional modification).
When
functional activity is compared across all 12 sites in the survey, all sites
tested showed
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
71
full activity when 0-10 RNA residues at the 5'-end were replaced with 2'0Me
RNA
residues. Only 1/12 sites showed a slight reduction in activity with 12
residues modified,
however 3/12 sites showed a reduction in activity when 13 residues were
modified and
4/12 sites showed a reduction in activity when 14 residues were modified. The
end-
modifiers (C3, ZEN) showed full activity at all sites.
[00125] The highest level of crRNA modification that showed full activity at
all sites
tested included Mod Patterns 6 and 7 (FIG. 10). This represents 61% and 67% of
the
bases in the crRNA modified with 2'0Me RNA, respectively.
n*n*n*nnnnnnnnnnnnnnnnnguuuuagagcuau*g*c*u Mod Pattern 6
(SEQ ID NO.:434)
n*n*n*nnnnnnnnnnnnnnnnnguuuuagagcuau*g*c*u Mod Pattern 7
(SEQ ID NO. :435)
[00126] The data in the present example also demonstrates that individual
sites can be
modified at higher levels and retain potency. For example, 8 of the 12 sites
studied
showed full activity using Mod Pattern 8, which has 72% of the residues
modified.
Further, example 7 demonstrates that the crRNA targeting site 38285 in HPRT1
(SEQ ID
No. 239) has full activity and has 30/36 residues modified (83%, with only 6
unmodified
RNA residues remaining). A base modification pattern such as Mod Pattern 6 or
Mod
Pattern 7 can be used as a starting point for studies to empirically ascertain
the extent
that a particular sequence can be modified before activity is lost. FIG. 12
shows a
schematic where a Mod Pattern 6 crRNA is paired with a highly modified
tracrRNA,
SEQ ID No. 134.
EXAMPLE 11
[00127] The Examples herein employ the Cas9 endonuclease from Streptococcus
pyogenes. The native amino acid sequence of S.py. Cas9 (SpyCas9) is shown
below
(SEQ ID No. 407).
[00128] The native Cas9 DNA sequence was codon optimized for expression in
E.coli
bacteria and had elements added for mammalian nuclear localization (nuclease
localization signals) and aid protein purification (His-tag). The final amino-
acid
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
72
sequence of the recombinant protein is shown (SEQ ID No 408). The DNA sequence
employed to express the recombinant protein in E.coli is shown (SEQ ID No.
409).
[00129] The native Cas9 DNA sequence was codon optimized for expression in
human cells and had elements added for antibody recognition (V5 epitope) and
mammalian nuclear localization (nuclease localization signals, NLS) added. The
final
amino-acid sequence is shown (SEQ ID No. 410) and DNA sequence follows (SEQ ID
No 411).
[00130] The native S.py Cas9 DNA sequence codon was optimized for expression
in
human cells and assembled as a T7 RNA polymerase expression cassette (SEQ ID
No.
412). The sequence contains a T7 RNA polymerase promoter, a V5 epitope tag, a
nuclear localization signal, the codon optimized Cas9 sequence, a second
nuclear
localization signal, and the BGH (bovine growth hormone) gene 3'-UTR element
with a
polyadenylation signal. Sequence of mRNA made from this expression cassette is
shown (SEQ ID No. 413).
S.py. Cas9 amino acid sequence (SEQ ID No. 407).
MDKKYS I GLDI GINSVGWAVITDEYKVPSKKEKVLGNTDRHS I KKNL I GALL FDS
GETAEATRLKRTARRR
YTRRKNRI CYLQEI FSNEMAKVDDS FFHRLEES FLVEEDKKHERHP I FGNIVDEVAYHEKYPT I
YHLRKKL
VDS TDKADLRL I YLALAHMI KFRGHFL I EGDLNPDNS DVDKL FI QLVQTYNQL FEENP INAS
GVDAKAI LS
ARL S KS RRLENL IAQL P GEKKNGL FGNL IAL S LGLT PNEKSNEDLAEDAKLQL S
KDTYDDDLDNLLAQI GD
QYADL FLAAKNL S DAI LL S DI LRVNTEI TKAP L SASMI KRYDEHHQDLT LLKALVRQQL P
EKYKEI FFDQS
KNGYAGYI DGGASQEEFYKFI KP I LEKMDGTEELLVKLNREDLLRKQRT FDNGS I PHQIHLGELHILRRQE
DFYP FLKDNREKI EKI LT FRI PYYVGP LARGNS RFAWMTRKS EET I T PWNFEEVVDKGASAQS FI
ERMINE
DKNLPNEKVLPKHSLLYEYFTVYNELTKVKYVTEGMRKPAELSGEQKKAIVDLLEKTNRKVIVKQLKEDYF
KKI EC FDSVEI SGVEDRFNASLGTYHDLLKI I KDKDFLDNEENEDI LEDIVLTLT L FEDREMI
EERLKTYA
HL FDDKVMKQLKRRRYT GWGRL S RKL INGI RDKQS GKT I LDFLKS DGFANRNFMQL I HDDS LT
FKEDI QKA
QVSGQGDSLHEHIANLAGS PAIKKGILQTVKVVDELVKVMGRHKPENIVIEMARENQTTQKGQKNSRERMK
RI EEGI KELGSQI LKEHPVENTQLQNEKLYLYYLQNGRDMYVDQELDINRL S DYDVDHIVPQS FLKDDS ID
NKVLTRS DKNRGKS DNVP S EEVVKKMKNYWRQLLNAKL I TQRKEDNLIKAERGGL S ELDKAGFI
KRQLVET
RQI TKHVAQI LDS RMNTKYDENDKL I REVKVI T LKS KLVS
DERKDFQFYKVREINNYHHAHDAYLNAVVGT
AL I KKYPKLES EFVYGDYKVYDVRKMIAKS EQEI GKATAKYFFYSNIMNFFKTEI T LANGEI RKRP L I
ETN
GET GEIVWDKGRDFATVRKVL SMPQVNIVKKTEVQT GGFS KES I L PKRNS DKL
IARKKDWDPKKYGGFDS P
TVAYSVLVVAKVEKGKSKKLKSVKELLGITIMERS S FEKNP I DFLEAKGYKEVKKDL I I KL PKYS L
FELEN
GRKRMLASAGELQKGNELALPSKYVNFLYLASHYEKLKGS PEDNEQKQLFVEQHKHYLDEI I EQI S EFS KR
VI LADANLDKVL SAYNKHRDKP I REQAENI I HL FT LTNLGAPAAFKYFDTT I DRKRYT S
TKEVLDAT L I HQ
S I T GLYETRI DL SQLGGD
S.py Cas9 amino acid sequence expressed from DNA codon optimized for
expression in
E.coli containing 3 NLS sequences and a purification His-tag (SEQ ID No. 408).
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
73
MGS SAP KKKRKVGI HGVPAAMDKKYS I GLD I GTNSVGWAVITDEYKVP SKKFKVLGNTDRHS I KKNL
I GAL
L FD S GETAEAT RLKRTARRRYT RRKNRI CYLQE I FSNEMAKVDDS FFHRLEES FLVEEDKKHERHP I
FGNI
VDEVAYHEKYPT I YHLRKKLVD S T DKADLRL I YLALAHMI KFRGHFL I EGDLNP DNS DVDKL FI
QLVQTYN
QLFEENP INAS GVDAKAI L SARL S KS RRLENL IAQL P GEKKNGL FGNL IAL S LGLT
PNEKSNEDLAEDAKL
QL S KDTYDDDLDNLLAQ I GDQYADLFLAAKNLSDAI LL S D I LRVNT E I T KAP L SASMI
KRYDEHHQDLTLL
KALVRQQL P EKYKE I FFDQSKNGYAGYI DGGASQEEFYKFI KP I LEKMDGTEELLVKLNREDLLRKQRT
FD
NGS I PHQ I HLGELHAI LRRQEDFYP FLKDNREKI EKI LT FRI PYYVGPLARGNSRFAWMTRKSEET
IT PWN
FEEVVDKGASAQS FI ERMTNEDKNLPNEKVLPKHSLLYEYFTVYNELTKVKYVTEGMRKPAELSGEQKKAI
VDLL FKTNRKVTVKQLKEDYFKKI EC FD SVE I SGVEDRFNASLGTYHDLLKI I KDKD FLDNEENED I
LED I
VLT LT L FEDREMI EERLKTYAHLFDDKVMKQLKRRRYTGWGRLSRKLINGI RDKQSGKT I LD ELKS
DGFAN
RNFMQL I HDD S LT EKED I QKAQVS GQGD S LHEHIANLAGS PAI KKGI
LQTVKVVDELVKVMGRHKPENIVI
EMARENQTTQKGQKNSRERMKRI EEGI KELGS Q I LKEHPVENTQLQNEKLYLYYLQNGRDMYVDQELDINR
LSDYDVDHIVPQS FLKDDS I DNKVLT RS DKNRGKS DNVP S EEVVKKMKNYWRQLLNAKL I
TQRKFDNLT KA
ERGGLSELDKAGFI KRQLVET RQ I T KHVAQ I LD S RMNT KYDENDKL I REVKVI T LKS KLVS D
FRKD FQ FYK
VRE INNYHHAHDAYLNAVVGTAL I KKYP KLE S E FVYGDYKVYDVRKMIAKS EQE I
GKATAKYFFYSNIMNF
FKT E I T LANGE I RKRP L I ETNGET GE IVWDKGRD FATVRKVL SMPQVNIVKKT EVQT GGFS
KE S I LP KRNS
DKLIARKKDWDPKKYGGFDS PTVAYSVLVVAKVEKGKSKKLKSVKELLGIT IMERS S FEKNP I DFLEAKGY
KEVKKDL I I KLPKYSLFELENGRKRMLASAGELQKGNELALP SKYVNFLYLASHYEKLKGS PEDNEQKQLF
VEQHKHYLDE I I EQ I S E FS KRVI LADANLDKVLSAYNKHRDKP I REQAENI I HL FT
LTNLGAPAAFKYFDT
T I DRKRYT S T KEVLDAT L I HQ S I T GLYET RI
DLSQLGGDAAPKKKRKVDPKKKRKVAAALEHHHHHH
S.py Cas9 DNA sequence codon optimized for expression in E.coli containing 3
NLS
sequences and a purification His-tag (SEQ ID No. 409).
AT GGGCAGCAGC GC C C CAAAGAAGAAGC GGAAGGT C GGTAT C CAC GGAGT C C CAGCAGC CAT
GGACAAAAA
GTACT CTAT T GGC CT GGATAT C GGGAC CAACAGC GT C GGGT GGGCT GT TAT CAC C GAC
GAGTATAAAGTAC
CT T C GAAAAAGT T CAAAGT GCT GGGCAACAC C GAT C GC CAT T CAAT CAAAAAGAACT T GAT
T GGT GC GCT G
T T GT T T GACT C C GGGGAAAC C GC C GAGGC GACT C GC CT TAAAC GTACAGCAC GT C GC
C GGTACACT C GGC G
TAAGAAT C GCAT T T GCTAT T T GCAGGAAAT CT T TAGCAAC GAGAT GGCAAAAGT C GAT GACT
C GT T T T T C C
AC C GC CT C GAGGAAAGCT T T CT GGT GGAGGAAGACAAAAAGCAT GAGC GT CAC C C GAT CT
T C GGCAACAT T
GT C GAT GAAGTAGC GTAT CAT GAAAAATAC C CAAC CAT T TAC CACT TAC GCAAAAAGCT GGT
GGACAGCAC
T GACAAAGCT GAT T T GC GC CT TAT CTAT T TAGC C CT GGCACATAT GAT TAAGT T T C GT
GGT CACT T C CT GA
T C GAAGGAGACT TAAAT C C C GACAACAGT GAT GT T GATAAAT T GT T TAT T CAGCT T GT
C CAAACT TACAAT
CAACT GT T C GAGGAAAAC C C GAT CAAT GC CT C C GGT GT GGAT GCAAAAGC CAT T T
TAAGT GCAC GC CT TAG
CAAGT C C C GT C GCT TAGAAAAC CT TAT C GC GCAGCT GC C C GGC GAGAAAAAGAAT GGT T
T GT T T GGGAAC C
T TAT T GC CT T GAGCT TAGGC CT CAC C C C GAAT T T CAAAAGTAAT T T C GAT CT T
GCAGAAGAC GC CAAAT TA
CAACT GT C GAAGGATACT TAT GAT GAC GAT CT C GATAAT CT GT TAGC GCAGAT T GGT GAC
CAATAC GC C GA
T CT T T T T CT GGC GGCTAAAAAT CT GAGC GAC GC CAT CT T GCT T T C GGATAT T CT C
C GC GT TAACAC C GAAA
T CAC GAAAGC GC CT CT TAGT GC CAGCAT GAT TAAAC GT TAT GAT GAACAC CAC CAGGAC CT
GAC CT TACT C
AAAGC GT T GGT T C GC CAGCAACT GC CAGAGAAGTACAAAGAAAT CT T CT T T GAT CAGT
CAAAGAAT GGT TA
T GC C GGCTATAT T GAC GGGGGT GCAAGC CAAGAGGAAT T CTACAAAT T TAT CAAGC CTAT T
CT GGAGAAAA
T GGAT GGCAC C GAAGAGT TAT T GGT GAAGCT TAAC C GT GAAGAC CT C CT GC GGAAACAGC
GCACAT T C GAT
AAT GGT T C GAT C C CACAC CAAAT C CAT T T GGGGGAGT TACAC GCTAT T T T GC GT C
GC CAGGAAGACT T T TA
C C CT T T C CT GAAGGATAAC C GGGAGAAAAT T GAGAAGAT C CT TAC CT T T C GTAT T C
C GTAT TAC GTAGGC C
C CT TAGCAC GGGGTAATAGC C GT T T C GC GT GGAT GACAC GGAAGT C GGAAGAGAC GAT CAC
C C C GT GGAAC
T T C GAAGAGGTAGT C GACAAGGGC GCAT CAGC GCAGT CT T T TAT T GAAC GTAT GAC GAAT
T T C GATAAAAA
CT T GC C CAAT GAGAAGGT GCT T C C GAAACAT T C CT T GT TATAT GAATAT T T TACAGT
T TACAAC GAGCT GA
C CAAGGT TAAATAC GT GAC GGAAGGAAT GC GCAAGC C C GCT T T T CT TAGC GGT
GAGCAAAAAAAGGC GAT C
GT C GAC CT GT TAT T CAAAAC GAAT C GTAAG GT GACT GTAAAGCAACT CAAAGAAGAT TACT T
CAAAAAGAT
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
74
T GAGT GCTT C GACAGC GT C GAAAT CT CT GGGGTAGAGGAT C GGTTTAAC
GCAAGTTTAGGTACCTACCAT G
ACCTGCTTAAAAT CAT TAAGGATAAAGACTTCTTAGATAAT GAAGAGAACGAAGATATTCTCGAGGACAT C
GT CTT GAC GTTAACCTTATTT GAGGAT C GT GAAAT GATT GAGGAAC GCCT CAAAACTTAT
GCCCACCT GTT
C GAC GATAAGGT GAT GAAGCAGCT GAAAC GT C GGC GCTACACAGGAT GGGGCC GCTT GAGT C
GCAAACTTA
TTAAC GGAAT CC GT GACAAGCAAT CC GGCAAAAC GATT CT GGATTT CTT GAAGT C GGAC
GGATTT GCTAAT
C GCAACTT CAT GCAGTT GAT CCAT GAT GACT CCCT GACTTTTAAAGAGGATATT CAAAAGGC
GCAGGTTAG
T GGT CAAGGC GACAGCTTACAC GAACACAT C GCAAATTT GGCT GGTT C GCC GGC
CATTAAAAAGGGGAT CC
T CCAGACC GT GAAAGTT GTAGAT GAGCTT GTTAAGGT CAT GGGT C GT CATAAGCCC GAAAACAT C
GT GATT
GAAATGGCGCGGGAGAAT CAAACGACCCAGAAAGGACAAAAGAATAGCCGTGAACGGAT GAAGCGGATC GA
GGAAGGCATTAAAGAGCT GGGGT CT CAAAT CTT GAAGGAACACCCT GT GGAGAACACT CAGCT
CCAAAAT G
AAAAACTTTACCT GTACTATTT GCAGAAC GGAC GC GATAT GTAC GT GGACCAAGAGTT GGATATTAAT
C GG
CT GAGT GACTAC GAC GTT GAT CATAT C GT CCC GCAGAGCTT CCT CAAAGAC GATT CTATT
GACAATAAGGT
ACT GAC GC GCT CT GATAAAAACC GT GGTAAGT C GGACAAC GT GCCCT CC GAAGAGGTT GT
GAAAAAGAT GA
AAAATTATT GGC GCCAGCTTTTAAAC GC GAAGCT GAT CACACAAC GTAAATT C GATAATTT
GACCAAGGCT
GAAC GGGGT GGCCT GAGC GAGTTAGATAAGGCAGGATTTATTAAAC GCCAGTTAGT GGAGACT C GT
CAAAT
CAC CAAACAT GT C GC GCAGATTTT GGACAGCC GGAT GAACACCAAGTAC GAT GAAAAT GACAAACT
GAT CC
GT GAGGT GAAAGT CATTACT CT GAAGT CCAAATTAGTTAGT GATTT CC GGAAGGACTTT CAATT
CTACAAA
GT CC GT GAAATTAATAACTAT CAT CAC GCACAT GAC GC GTACCT GAAT GCAGT GGTT GGGACC
GCCCTTAT
CAAGAAATAT CCTAAGCT GGAGT C GGAGTTT GT CTAT GGC GACTATAAGGTATAC GAT GTT C
GCAAAAT GA
TT GC GAAAT CT GAGCAGGAGAT C GGTAAGGCAACC GCAAAATATTT CTTTTACT CAAACATTAT
GAATTT C
TTTAAGACAGAAAT CACT CT GGCCAAC GGGGAGATT C GCAAAC GT CC GTT GAT C GAAACAAAC
GGC GAGAC
TGGCGAAATTGTTTGGGACAAAGGGCGTGATTTCGCGACGGTGCGCAAGGTACTGAGCATGCCTCAAGTCA
ATATT GTTAAGAAAACC GAAGT GCAGAC GGGC GGGTTTT CCAAGGAAAGCAT CTTACCCAAAC GTAATT
CA
GATAAACTTATT GCAC GCAAAAAGGACT GGGAT CC GAAAAAGTAT GGAGGCTT C GACAGT CCAACC
GTAGC
CTACT CT GTT CT C GTT GTAGC GAAAGTAGAAAAGGGTAAAT CCAAGAAACT GAAAT CT GT
CAAGGAGTT GC
TT GGAAT CACCATTAT GGAGC GTAGCT CCTT C GAGAAGAACCC GATT GACTTT CT
GGAAGCCAAAGGATAT
AAAGAGGTCAAGAAAGATCTTATCATTAAGCTGCCTAAGTATTCACTCTTCGAGCTGGAAAATGGTCGTAA
AC GCAT GCT C GCTT CT GCC GGC GAGTT GCAGAAGGGCAAT GAATTAGCACTT CCAT CAAAGTAC
GTTAACT
T CCT GTATTT GGCCAGCCATTAC GAGAAACT GAAGGGGT CT CCAGAGGACAAC
GAACAGAAACAATTATTT
GTAGAGCAGCACAAGCATTAT CTT GAT GAAAT CATT GAGCAAATTT CC GAATT CAGTAAAC GC GTAAT
CCT
GGCC GAT GCAAACCT C GACAAGGT GCT GAGC GCTTACAATAAGCAT C GC GACAAACCTAT CC GT
GAGCAGG
CTGAAAATATCATTCACCTGTTCACATTAACGAACCTGGGCGCTCCGGCCGCTTTTAAATATTTCGACACG
ACAAT C GACC GTAAGC GCTATACCAGTAC GAAAGAAGT GTT GGAT GC GACCCTTATT CACCAGT
CAATTAC
AGGATTATAT GAGACCC GTAT C GACCTTAGCCAATTAGGT GGGGAT GC GGCCCC GAAGAAAAAAC
GCAAAG
T GGAT CC GAAGAAAAAAC GCAAAGT GGC GGCC GCACT C GAGCACCACCACCACCACCACT GA
S.py Cas9 amino acid sequence expressed from DNA codon optimized for
expression in
human cells containing a V5 epitope tag and 2 NLS sequences (SEQ ID No. 410).
MGKPI PNPLLGLDS TAPKKKRKVGI HGVPAADKKY S I GL DI GTNSVGWAVI TDEYKVP S KK FKVL
GNTDRHS IKKNL I GALL FDSGETAEAT RLKRTARRRY TRRKNRICYLQE I FSNEMAKVDDS FEHR
LEES FLVEEDKKHERHP I FGNIVDEVAYHEKYP T I YHLRKKLVDS TDKADL RL IYLALAHMIK FR
GHFL I EGDLNPDNSDVDKL FI QLVQTYNQL FEENP INAS GVDAKAIL SARL SKSRRLENLIAQLP
GEKKNGL FGNLIALSLGLT PN FKSN FDLAEDAKLQL S KDTYDDDL DNLLAQI GDQYADL FLAAKN
L SDAILL SDILRVNTEI TKAPLSASMIKRYDEHHQDLTLLKALVRQQLPEKYKEI FFDQSKNGYA
GYIDGGASQEE FYKFIKPILEKMDGTEELLVKLNREDLLRKQRT FDNGS I PHQIHLGELHAIL RR
QED FY P FLKDNREKI EKIL T FRI PYYVGPLARGNSRFAWMTRKSEET IT PWNFEEVVDKGASAQS
FI ERMTN FDKNL PNEKVL PKHSLLYEY FTVYNELT KVKYVT EGMRKPAFL S GEQKKAIVDLL FKT
NRKVTVKQLKEDY FKKI EC FDSVEI SGVEDRFNASLGTYHDLLKI IKDKDFLDNEENEDILEDIV
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
LTLTL FEDREMI EERLKTYAHL FDDKVMKQL KRRRYT GWGRL S RKL INGIRDKQS GKT I LD FL KS
DGFANRNFMQLIHDDSLT FKEDIQKAQVSGQGDSLHEHIANLAGS PAIKKGILQTVKVVDELVKV
MGRHKPENIVIEMARENQT TQKGQKNSRERMKRIEEGIKELGSQILKEHPVENTQLQNEKLYLYY
LQNGRDMYVDQELDINRLSDYDVDHIVPQS FLKDDS I DNKVLT RS DKNRGKSDNVP S EEVVKKMK
NYWRQLLNAKL I TQRKFDNLT KAERGGL S EL DKAGFI KRQLVETRQI TKHVAQIL DS RMNT KYDE
NDKL I REVKVI TL KS KLVS DERKDFQFYKVREINNYHHAHDAYLNAVVGTAL I KKYPKL ES E FVY
GDYKVYDVRKMIAKSEQEI GKATAKY F FY SNIMNF FKT E I T LANGEI RKRPL I ETNGET GE
IVWD
KGRDFATVRKVL SMPQVNIVKKT EVQT GGFS KE S I L PKRNS DKL IARKKDWDPKKYGGEDS PTVA
YSVLVVAKVEKGKSKKLKSVKELLGIT IMERSS FEKNPI DFLEAKGYKEVKKDL I IKLPKYSL FE
LENGRKRMLASAGELQKGNELALPSKYVNFLYLASHYEKLKGS PEDNEQKQL FVEQHKHYL DE I I
EQI SE FS KRVI LADANL DKVL SAYNKHRDKP IREQAENI IHLFTLTNLGAPAAFKYFDT T I DRKR
Y T S TKEVLDAT L I HQS I T GLYET RI DL SQLGGDSRADPKKKRKVE FHHTGLVDPS SVPSLSLNR
S.py Cas9 DNA sequence codon optimized for expression in human cells
containing a
V5 epitope tag and 2 NLS sequences (SEQ ID No. 411).
ATGGGCAAGCCCATCCCTAACCCCCTGTTGGGGCTGGACAGCACCGCTCCCAAAAAGAAAAGGAAGGTGGG
CATTCACGGCGTGCCTGCGGCCGACAAAAAGTACAGCATCGGCCTTGATATCGGCACCAATAGCGTGGGCT
GGGCCGTTATCACAGACGAATACAAGGTACCCAGCAAGAAGTTCAAGGTGCTGGGGAATACAGACAGGCAC
TCTATCAAGAAAAACCTTATCGGGGCTCTGCTGTTTGACTCAGGCGAGACCGCCGAGGCCACCAGGTTGAA
GAGGACCGCAAGGCGAAGGTACACCCGGAGGAAGAACAGGAT CT GCTAT CT GCAGGAGAT CTT CAGCAACG
AGAT GGCCAAGGT GGACGACAGCTT CTT CCACAGGCT GGAGGAGAGCTT CCTT GT
CGAGGAGGATAAGAAG
CACGAACGACACCCCATCTTCGGCAACATAGTCGACGAGGTCGCTTATCACGAGAAGTACCCCACCATCTA
CCACCTGCGAAAGAAATTGGTGGATAGCACCGATAAAGCCGACTTGCGACTTATCTACTTGGCTCTGGCGC
ACAT GATTAAGTT CAGGGGCCACTT CCT GAT CGAGGGCGACCTTAACCCCGACAACAGT GACGTAGACAAA
TT GTT CAT CCAGCTT GTACAGACCTATAACCAGCT GTT CGAGGAAAACCCTATTAACGCCAGCGGGGT GGA
TGCGAAGGCCATACTTAGCGCCAGGCTGAGCAAAAGCAGGCGCTTGGAGAACCTGATAGCCCAGCTGCCCG
GTGAAAAGAAGAACGGCCTCTTCGGTAATCTGATTGCCCTGAGCCTGGGCCTGACCCCCAACTTCAAGAGC
AACTT CGACCT GGCAGAAGAT GCCAAGCT GCAGTT GAGTAAGGACACCTAT GACGACGACTT GGACAAT
CT
GCTCGCCCAAATCGGCGACCAGTACGCTGACCTGTTCCTCGCCGCCAAGAACCTTTCTGACGCAATCCTGC
TTAGCGATATCCTTAGGGT GAACACAGAGAT CACCAAGGCCCCCCT GAGCGCCAGCAT GAT CAAGAGGTAC
GACGAGCACCAT CAGGACCT GACCCTT CT GAAGGCCCT GGT GAGGCAGCAACT GCCCGAGAAGTACAAGGA
GAT CTTTTT CGACCAGAGCAAGAACGGCTACGCCGGCTACAT CGACGGCGGAGCCAGCCAAGAGGAGTT CT
ACAAGTT CAT CAAGCCCAT CCT GGAGAAGAT GGAT GGCACCGAGGAGCT GCT GGT GAAGCT
GAACAGGGAA
GATTTGCTCCGGAAGCAGAGGACCTTTGACAACGGTAGCATCCCCCACCAGATCCACCTGGGCGAGCTGCA
CGCAATACTGAGGCGACAGGAGGATTTCTACCCCTTCCTCAAGGACAATAGGGAGAAAATCGAAAAGATTC
TGACCTTCAGGATCCCCTACTACGTGGGCCCTCTTGCCAGGGGCAACAGCCGATTCGCTTGGATGACAAGA
AAGAGCGAGGAGACCATCACCCCCTGGAACTTCGAGGAAGTGGTGGACAAAGGAGCAAGCGCGCAGTCTTT
CAT CGAACGGAT GACCAATTT CGACAAAAACCT GCCTAACGAGAAGGT GCT GCCCAAGCACAGCCT GCTTT
ACGAGTACTT CACCGT GTACAACGAGCT CACCAAGGT GAAATAT GT GACCGAGGGCAT GCGAAAACCCGCT
TT CCT GAGCGGCGAGCAGAAGAAGGCCAT CGT GGACCT GCT GTT CAAGACCAACAGGAAGGT GACCGT
GAA
GCAGCTGAAGGAGGACTACTTCAAGAAGATCGAGTGCTTTGATAGCGTGGAAATAAGCGGCGTGGAGGACA
GGTTCAACGCCAGCCTGGGCACCTACCACGACTTGTTGAAGATAATCAAAGACAAGGATTTCCTGGATAAT
GAGGAGAACGAGGATATACT CGAGGACAT CGT GCT GACTTT GACCCT GTTT GAGGACCGAGAGAT GATT
GA
AGAAAGGCT CAAAACCTACGCCCACCT GTT CGACGACAAAGT GAT GAAACAACT GAAGAGACGAAGATACA
CCGGCT GGGGCAGACT GT CCAGGAAGCT CAT CAACGGCATTAGGGACAAGCAGAGCGGCAAGACCAT CCT G
GATTTCCTGAAGTCCGACGGCTTCGCCAACCGAAACTTCATGCAGCTGATTCACGATGACAGCTTGACCTT
CAAGGAGGACATCCAGAAGGCCCAGGTTAGCGGCCAGGGCGACTCCCTGCACGAACATATTGCAAACCTGG
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
76
CAGGCT CCCCT GC GAT CAAGAAGGGCATACT GCAGACCGTTAAGGTT GT GGACGAATT GGT CAAGGT
CAT G
GGCAGGCACAAGCCCGAAAACATAGTTATAGAGATGGCCAGAGAGAACCAGACCACCCAAAAGGGCCAGAA
GAACAGCCGGGAGCGCATGAAAAGGATCGAGGAGGGTATCAAGGAACTCGGAAGCCAGATCCTCAAAGAGC
ACCCCGTGGAGAATACCCAGCTCCAGAACGAGAAGCTGTACCTGTACTACCTGCAGAACGGCAGGGACATG
TACGTTGACCAGGAGTTGGACATCAACAGGCTTTCAGACTATGACGTGGATCACATAGTGCCCCAGAGCTT
TCTTAAAGACGATAGCATCGACAACAAGGTCCTGACCCGCTCCGACAAAAACAGGGGCAAAAGCGACAACG
T GCCAAGCGAAGAGGT GGTTAAAAAGAT GAAGAACTACT GGAGGCAACT GCT CAACGCGAAATT GAT
CACC
CAGAGAAAGTT CGATAACCT GACCAAGGCCGAGAGGGGCGGACT CT CCGAACTT GACAAAGCGGGCTT CAT
AAAGAGGCAGCTGGTCGAGACCCGACAGATCACGAAGCACGTGGCCCAAATCCTCGACAGCAGAATGAATA
CCAAGTACGAT GAGAAT GACAAACT CAT CAGGGAAGT GAAAGT GATTACCCT GAAGAGCAAGTT GGT GT
CC
GACTTTCGCAAAGATTTCCAGTTCTACAAGGTGAGGGAGATCAACAACTACCACCATGCCCACGACGCATA
CCTGAACGCCGTGGTCGGCACCGCCCTGATTAAGAAGTATCCAAAGCTGGAGTCCGAATTTGTCTACGGCG
ACTACAAAGTTTAC GAT GT GAGGAAGAT GAT CGCTAAGAGCGAACAGGAGAT CGGCAAGGCCACCGCTAAG
TATTT CTT CTACAGCAACAT CAT GAACTTTTT CAAGACCGAGAT CACACTT GCCAACGGCGAAAT
CAGGAA
GAGGCCGCTTATCGAGACCAACGGTGAGACCGGCGAGATCGTGTGGGACAAGGGCAGGGACTTCGCCACCG
T GAGGAAAGT CCT GAGCAT GCCCCAGGT GAATATT GT GAAAAAAACT GAGGT
GCAGACAGGCGGCTTTAGC
AAGGAAT CCAT CCT GCCCAAGAGGAACAGCGACAAGCT GAT CGCCCGGAAGAAGGACT GGGACCCTAAGAA
GTATGGAGGCTTCGACAGCCCCACCGTAGCCTACAGCGTGCTGGTGGTCGCGAAGGTAGAGAAGGGGAAGA
GCAAGAAACT GAAGAGCGT GAAGGAGCT GCT CGGCATAACCAT CAT GGAGAGGT CCAGCTTT
GAGAAGAAC
CCCATT GACTTTTT GGAAGCCAAGGGCTACAAAGAGGT CAAAAAGGACCT GAT CAT CAAACT CCCCAAGTA
CTCCCTGTTTGAATTGGAGAACGGCAGAAAGAGGATGCTGGCGAGCGCTGGGGAACTGCAAAAGGGCAACG
AACTGGCGCTGCCCAGCAAGTACGTGAATTTTCTGTACCTGGCGTCCCACTACGAAAAGCTGAAAGGCAGC
CCCGAGGACAACGAGCAGAAGCAGCTGTTCGTGGAGCAGCACAAGCATTACCTGGACGAGATAATCGAGCA
AAT CAGCGAGTT CAGCAAGAGGGT GATT CT GGCCGACGCGAACCT GGATAAGGT CCT
CAGCGCCTACAACA
AGCACCGAGACAAACCCAT CAGGGAGCAGGCCGAGAATAT CATACACCT GTT CACCCT GACAAAT CT GGGC
GCACCTGCGGCATTCAAATACTTCGATACCACCATCGACAGGAAAAGGTACACTAGCACTAAGGAGGTGCT
GGATGCCACCTTGATCCACCAGTCCATTACCGGCCTGTATGAGACCAGGATCGACCTGAGCCAGCTTGGAG
GCGACTCTAGGGCGGACCCAAAAAAGAAAAGGAAGGTGGAATTCCACCACACTGGACTAGTGGATCCGAGC
T CGGTACCAAGCTTAAGTTTAAACCGCT GA
S.py Cas9 DNA sequence codon optimized for expression in human cells as a T7
RNA
polymerase expression cassette (SEQ ID No. 412). The sequence contains a T7
RNA
polymerase promoter, a V5 epitope tag, a nuclear localization signal, the
codon
optimized Cas9 sequence, a second nuclear localization signal, and the BGH
(bovine
growth hormone) gene 3'-UTR element with a polyadenylation signal.
TAATACGACTCACTATAGGGAGACCCAAGCTGGCTAGCGTTTAAACGGGCCCTCTAGACTCGAGCGGCCGC
CACCATGGGCAAGCCCATCCCTAACCCCCTGTTGGGGCTGGACAGCACCGCTCCCAAAAAGAAAAGGAAGG
TGGGCATTCACGGCGTGCCTGCGGCCGACAAAAAGTACAGCATCGGCCTTGATATCGGCACCAATAGCGTG
GGCTGGGCCGTTATCACAGACGAATACAAGGTACCCAGCAAGAAGTTCAAGGTGCTGGGGAATACAGACAG
GCACTCTATCAAGAAAAACCTTATCGGGGCTCTGCTGTTTGACTCAGGCGAGACCGCCGAGGCCACCAGGT
T GAAGAGGACCGCAAGGCGAAGGTACACCCGGAGGAAGAACAGGAT CT GCTAT CT GCAGGAGAT CTT CAGC
AACGAGAT GGCCAAGGT GGACGACAGCTT CTT CCACAGGCT GGAGGAGAGCTT CCTT GT
CGAGGAGGATAA
GAAGCACGAACGACACCCCATCTTCGGCAACATAGTCGACGAGGTCGCTTATCACGAGAAGTACCCCACCA
TCTACCACCTGCGAAAGAAATTGGTGGATAGCACCGATAAAGCCGACTTGCGACTTATCTACTTGGCTCTG
GCGCACAT GATTAAGTT CAGGGGCCACTT CCT GAT CGAGGGCGACCTTAACCCCGACAACAGT GACGTAGA
CAAATT GTT CAT CCAGCTT GTACAGACCTATAACCAGCT GTT CGAGGAAAACCCTATTAACGCCAGCGGGG
TGGATGCGAAGGCCATACTTAGCGCCAGGCTGAGCAAAAGCAGGCGCTTGGAGAACCTGATAGCCCAGCTG
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
77
CCCGGTGAAAAGAAGAACGGCCTCTTCGGTAATCTGATTGCCCTGAGCCTGGGCCTGACCCCCAACTTCAA
GAGCAACTTCGACCTGGCAGAAGATGCCAAGCTGCAGTTGAGTAAGGACACCTATGACGACGACTTGGACA
ATCTGCTCGCCCAAATCGGCGACCAGTACGCTGACCTGTTCCTCGCCGCCAAGAACCTTTCTGACGCAATC
CT GCTTAGCGATATCCTTAGGGT GAACACAGAGAT CACCAAGGCCCCCCT GAGCGCCAGCAT GAT CAAGAG
GTACGACGAGCACCAT CAGGACCT GACCCTT CT GAAGGCCCT GGT GAGGCAGCAACT GCCCGAGAAGTACA
AGGAGATCTTTTTCGACCAGAGCAAGAACGGCTACGCCGGCTACATCGACGGCGGAGCCAGCCAAGAGGAG
TT CTACAAGTT CAT CAAGCCCAT CCT GGAGAAGAT GGAT GGCACCGAGGAGCT GCT GGT GAAGCT
GAACAG
GGAAGATTTGCTCCGGAAGCAGAGGACCTTTGACAACGGTAGCATCCCCCACCAGATCCACCTGGGCGAGC
TGCACGCAATACTGAGGCGACAGGAGGATTTCTACCCCTTCCTCAAGGACAATAGGGAGAAAATCGAAAAG
ATTCTGACCTTCAGGATCCCCTACTACGTGGGCCCTCTTGCCAGGGGCAACAGCCGATTCGCTTGGATGAC
AAGAAAGAGCGAGGAGACCATCACCCCCTGGAACTTCGAGGAAGTGGTGGACAAAGGAGCAAGCGCGCAGT
CTTT CAT CGAACGGAT GACCAATTT CGACAAAAACCT GCCTAACGAGAAGGT GCT GCCCAAGCACAGCCT
G
CTTTACGAGTACTT CACCGT GTACAACGAGCT CAC CAAGGT GAAATAT GT GACCGAGGGCAT
GCGAAAACC
CGCTTTCCTGAGCGGCGAGCAGAAGAAGGCCATCGTGGACCTGCTGTTCAAGACCAACAGGAAGGTGACCG
TGAAGCAGCTGAAGGAGGACTACTTCAAGAAGATCGAGTGCTTTGATAGCGTGGAAATAAGCGGCGTGGAG
GACAGGTTCAACGCCAGCCTGGGCACCTACCACGACTTGTTGAAGATAATCAAAGACAAGGATTTCCTGGA
TAAT GAGGAGAACGAGGATATACT CGAGGACAT CGT GCT GACTTT GACCCT GTTT GAGGACCGAGAGAT
GA
TT GAAGAAAGGCT CAAAACCTACGCCCACCT GTT CGACGACAAAGT GAT GAAACAACT GAAGAGACGAAGA
TACACCGGCT GGGGCAGACT GT CCAGGAAGCT CAT CAACGGCATTAGGGACAAGCAGAGCGGCAAGACCAT
CCTGGATTTCCTGAAGTCCGACGGCTTCGCCAACCGAAACTTCATGCAGCTGATTCACGATGACAGCTTGA
CCTTCAAGGAGGACATCCAGAAGGCCCAGGTTAGCGGCCAGGGCGACTCCCTGCACGAACATATTGCAAAC
CTGGCAGGCTCCCCTGCGATCAAGAAGGGCATACTGCAGACCGTTAAGGTTGTGGACGAATTGGTCAAGGT
CAT GGGCAGGCACAAGCCCGAAAACATAGTTATAGAGAT GGCCAGAGAGAACCAGACCACCCAAAAGGGCC
AGAAGAACAGCCGGGAGCGCATGAAAAGGATCGAGGAGGGTATCAAGGAACTCGGAAGCCAGATCCTCAAA
GAGCACCCCGTGGAGAATACCCAGCTCCAGAACGAGAAGCTGTACCTGTACTACCTGCAGAACGGCAGGGA
CAT GTAC GTT GACCAGGAGTT GGACAT CAACAGGCTTT CAGACTAT GACGT GGAT CACATAGT
GCCCCAGA
GCTTTCTTAAAGACGATAGCATCGACAACAAGGTCCTGACCCGCTCCGACAAAAACAGGGGCAAAAGCGAC
AACGT GCCAAGCGAAGAGGT GGTTAAAAAGAT GAAGAACTACT GGAGGCAACT GCT CAACGCGAAATT GAT
CACCCAGAGAAAGTT CGATAACCT GACCAAGGCCGAGAGGGGCGGACT CT CCGAACTT GACAAAGCGGGCT
TCATAAAGAGGCAGCTGGTCGAGACCCGACAGATCACGAAGCACGTGGCCCAAATCCTCGACAGCAGAATG
AATACCAAGTACGAT GAGAAT GACAAACT CAT CAGGGAAGT GAAAGT GATTACCCT GAAGAGCAAGTT
GGT
GT CCGACTTT CGCAAAGATTT CCAGTT CTACAAGGT GAGGGAGAT CAACAACTACCACCAT GCCCACGACG
CATACCT GAACGCCGT GGT CGGCACCGCCCT GATTAAGAAGTAT CCAAAGCT GGAGT CCGAATTT GT
CTAC
GGCGACTACAAAGTTTACGAT GT GAGGAAGAT GAT CGCTAAGAGCGAACAGGAGAT CGGCAAGGCCACCGC
TAAGTATTT CTT CTACAGCAACAT CAT GAACTTTTT CAAGACCGAGAT CACACTT GCCAACGGCGAAAT
CA
GGAAGAGGCCGCTTAT CGAGACCAACGGT GAGACCGGCGAGAT CGT GT GGGACAAGGGCAGGGACTT CGCC
ACC GT GAGGAAAGT CCT GAGCAT GCCCCAGGT GAATATT GT GAAAAAAACT GAGGT
GCAGACAGGCGGCTT
TAGCAAGGAAT CCAT CCT GCCCAAGAGGAACAGCGACAAGCT GAT CGCCCGGAAGAAGGACT GGGACCCTA
AGAAGTATGGAGGCTTCGACAGCCCCACCGTAGCCTACAGCGTGCTGGTGGTCGCGAAGGTAGAGAAGGGG
AAGAGCAAGAAACT GAAGAGCGT GAAGGAGCT GCT CGGCATAACCAT CAT GGAGAGGT CCAGCTTT
GAGAA
GAACCCCATT GACTTTTT GGAAGCCAAGGGCTACAAAGAGGT CAAAAAGGACCT GAT CAT CAAACT CCCCA
AGTACTCCCTGTTTGAATTGGAGAACGGCAGAAAGAGGATGCTGGCGAGCGCTGGGGAACTGCAAAAGGGC
AACGAACT GGCGCT GCCCAGCAAGTACGT GAATTTT CT GTACCT GGCGT CCCACTACGAAAAGCT
GAAAGG
CAGCCCCGAGGACAACGAGCAGAAGCAGCTGTTCGTGGAGCAGCACAAGCATTACCTGGACGAGATAATCG
AGCAAAT CAGCGAGTT CAGCAAGAGGGT GATT CT GGCCGACGCGAACCT GGATAAGGT CCT
CAGCGCCTAC
AACAAGCACCGAGACAAACCCAT CAGGGAGCAGGCCGAGAATAT CATACACCT GTT CACCCT GACAAAT CT
GGGCGCACCTGCGGCATTCAAATACTTCGATACCACCATCGACAGGAAAAGGTACACTAGCACTAAGGAGG
TGCTGGATGCCACCTTGATCCACCAGTCCATTACCGGCCTGTATGAGACCAGGATCGACCTGAGCCAGCTT
GGAGGCGACT CTAGGGCGGACCCAAAAAAGAAAAGGAAGGT GGAATT CCACCACACT GGACTAGT GGAT CC
GAGCTCGGTACCAAGCTTAAGTTTAAACCGCTGATCAGCCTCGACTGTGCCTTCTAGTTGCCAGCCATCTG
TTGTTTGCCCCTCCCCCGTGCCTTCCTTGACCCTGGAAGGTGCCACTCCCACTGTCCTTTCCTAATAAAAT
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
78
GAGGAAAT T GCAT C GCAT T GT CT GAGTAGGT GT CAT T C TAT T CT GGGGGGT GGGGT
GGGGCAGGACAGCAA
GGGGGAGGAT T GGGAAGACAATAGCAGGCAT GC T GGGGAT GC GGT GGGCT C TAT GGC
S.py Cas9 mRNA (SEQ ID No. 413) as made from the expression cassette (SEQ ID
No.
412). The sequence contains a V5 epitope tag, a nuclear localization signal,
the codon
optimized Cas9 sequence, a second nuclear localization signal, and the BGH
(bovine
growth hormone) gene 3'-UTR element and poly-A tail.
GGGAGAC C CAAGCUGGCUAGC GUUUAAAC GGGC C CU CUAGACU C GAGC GGC C GC CAC
CAUGGGCAAGC C CA
UC C CUAAC C C C CU GUU GGGGCU GGACAGCAC C GCUC C CAAAAAGAAAAGGAAGGU GGGCAUU
CAC GGC GU G
C CU GC GGC C GACAAAAAGUACAGCAUC GGC CUUGAUAUC GGCAC CAAUAGC GU GGGCU GGGC C
GUUAU CAC
AGAC GAAUACAAGGUAC C CAGCAAGAAGUUCAAGGUGCUGGGGAAUACAGACAGGCACUCUAUCAAGAAAA
AC CUUAUC GGGGCU CU GCU GUUU GACU CAGGC GAGAC C GC C GAGGC CAC CAGGUUGAAGAGGAC
C GCAAGG
C GAAGGUACAC C C GGAGGAAGAACAGGAU CU GCUAU CU GCAGGAGAU CUU CAGCAAC GAGAUGGC
CAAGGU
GGAC GACAGCUUCUUC CACAGGCUGGAGGAGAGCUUC CUU GU C GAGGAGGAUAAGAAGCAC GAAC GACAC
C
C CAUCUUC GGCAACAUAGUC GAC GAGGUC GCUUAU CAC GAGAAGUAC C C CAC CAUCUAC CAC CU
GC GAAAG
AAAUUGGUGGAUAGCAC C GAUAAAGC C GACUU GC GACUUAU CUACUU GGCU CU GGC
GCACAUGAUUAAGUU
CAGGGGC CACUUC CU GAU C GAGGGC GAC CUUAAC C C C GACAACAGUGAC
GUAGACAAAUUGUUCAUC CAGC
UUGUACAGAC CUAUAAC CAGCUGUUC GAGGAAAAC C CUAUUAAC GC CAGC GGGGU GGAU GC GAAGGC
CAUA
CUUAGC GC CAGGCUGAGCAAAAGCAGGC GCUUGGAGAAC CU GAUAGC C CAGCU GC C C
GGUGAAAAGAAGAA
C GGC CU CUU C GGUAAU CU GAUU GC C CU GAGC CU GGGC CU GAC C C C
CAACUUCAAGAGCAACUUC GAC CU GG
CAGAAGAU GC CAAGCUGCAGUUGAGUAAGGACAC CUAUGAC GAC GACUU GGACAAU CU GCU C GC C
CAAAUC
GGC GAC CAGUAC GCUGAC CU GUU C CU C GC C GC CAAGAAC CUUU CU GAC GCAAUC CU
GCUUAGC GAUAUC CU
UAGGGU GAACACAGAGAU CAC CAAGGC CCCC CU GAGC GC CAGCAUGAUCAAGAGGUAC GAC GAGCAC
CAUC
AGGAC CU GAC C CUU CU GAAGGC C CU GGU GAGGCAGCAACU GC C C
GAGAAGUACAAGGAGAUCUUUUUC GAC
CAGAGCAAGAAC GGCUAC GC C GGCUACAUC GAC GGC GGAGC CAGC CAAGAGGAGUU CUACAAGUU
CAU CAA
GC C CAUC CU GGAGAAGAU GGAU GGCAC C GAGGAGCUGCUGGUGAAGCUGAACAGGGAAGAUUUGCUC C
GGA
AGCAGAGGAC CUUUGACAAC GGUAGCAUCCCC CAC CAGAUC CAC CU GGGC GAGCUGCAC
GCAAUACUGAGG
C GACAGGAGGAUUUCUAC C C CUUC CU CAAGGACAAUAGGGAGAAAAU C GAAAAGAUU CU GAC
CUUCAGGAU
C C C CUACUAC GU GGGC C CU CUU GC CAGGGGCAACAGC C GAUUC GCUUGGAUGACAAGAAAGAGC
GAGGAGA
C CAU CAC C C C CU GGAACUU C GAGGAAGUGGUGGACAAAGGAGCAAGC GC GCAGUCUUUCAUC GAAC
GGAUG
AC CAAUUUC GACAAAAAC CU GC CUAAC GAGAAGGU GCU GC C CAAGCACAGC CU GCUUUAC
GAGUACUU CAC
C GU GUACAAC GAGCU CAC CAAGGU GAAAUAU GU GAC C GAGGGCAU GC GAAAAC C C GCUUUC
CU GAGC GGC G
AGCAGAAGAAGGC CAUC GU GGAC CU GCU GUU CAAGAC CAACAGGAAGGUGAC C GU GAAGCAGCU
GAAGGAG
GACUACUUCAAGAAGAUC GAGUGCUUUGAUAGC GU GGAAAUAAGC GGC GU GGAGGACAGGUU CAAC GC
CAG
C CU GGGCAC CUAC CAC GACUUGUUGAAGAUAAUCAAAGACAAGGAUUUC CU GGAUAAU GAGGAGAAC
GAGG
AUAUACUC GAGGACAUC GU GCU GACUUU GAC C CU GUUU GAGGAC C
GAGAGAUGAUUGAAGAAAGGCUCAAA
AC CUAC GC C CAC CU GUU C GAC GACAAAGUGAUGAAACAACUGAAGAGAC GAAGAUACAC C
GGCUGGGGCAG
ACU GU C CAGGAAGCUCAUCAAC GGCAUUAGGGACAAGCAGAGC GGCAAGAC CAUC CU GGAUUU C CU
GAAGU
C C GAC GGCUUC GC CAAC C GAAACUU CAU GCAGCU GAUU CAC GAUGACAGCUUGAC
CUUCAAGGAGGACAUC
CAGAAGGC C CAGGUUAGC GGC CAGGGC GACUC C CU GCAC GAACAUAUUGCAAAC CU GGCAGGCU C
C C CU GC
GAUCAAGAAGGGCAUACUGCAGAC C GUUAAGGUU GU GGAC GAAUUGGUCAAGGUCAUGGGCAGGCACAAGC
C C GAAAACAUAGUUAUAGAGAUGGC CAGAGAGAAC CAGAC CAC C CAAAAGGGC CAGAAGAACAGC C
GGGAG
C GCAUGAAAAGGAUC GAGGAGGGUAUCAAGGAACUC GGAAGC CAGAUC CU CAAAGAGCAC C C C GU
GGAGAA
UAC C CAGCUC CAGAAC GAGAAGCUGUAC CU GUACUAC CU GCAGAAC GGCAGGGACAUGUAC GUUGAC
CAGG
AGUUGGACAUCAACAGGCUUUCAGACUAUGAC GU GGAU CACAUAGU GC C C CAGAGCUUUCUUAAAGAC
GAU
AGCAUC GACAACAAGGUC CU GAC C C GCUC C GACAAAAACAGGGGCAAAAGC GACAAC GU GC CAAGC
GAAGA
GGUGGUUAAAAAGAUGAAGAACUACUGGAGGCAACUGCUCAAC GC GAAAUU GAU CAC C CAGAGAAAGUUC G
AUAAC CU GAC CAAGGC C GAGAGGGGC GGACU CU C C GAACUUGACAAAGC
GGGCUUCAUAAAGAGGCAGCUG
GU C GAGAC C C GACAGAU CAC GAAGCAC GU GGC C CAAAUC CU C GACAGCAGAAUGAAUAC
CAAGUAC GAU GA
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
79
GAAUGACAAACUCAUCAGGGAAGUGAAAGUGAUUACCCUGAAGAGCAAGUUGGUGUCCGACUUUCGCAAAG
AUUUCCAGUUCUACAAGGUGAGGGAGAUCAACAACUACCACCAUGCCCACGACGCAUACCUGAACGCCGUG
GUCGGCACCGCCCUGAUUAAGAAGUAUCCAAAGCUGGAGUCCGAAUUUGUCUACGGCGACUACAAAGUUUA
CGAUGUGAGGAAGAUGAUCGCUAAGAGCGAACAGGAGAUCGGCAAGGCCACCGCUAAGUAUUUCUUCUACA
GCAACAUCAUGAACUUUUUCAAGACCGAGAUCACACUUGCCAACGGCGAAAUCAGGAAGAGGCCGCUUAUC
GAGACCAACGGUGAGACCGGCGAGAUCGUGUGGGACAAGGGCAGGGACUUCGCCACCGUGAGGAAAGUCCU
GAGCAUGCCCCAGGUGAAUAUUGUGAAAAAAACUGAGGUGCAGACAGGCGGCUUUAGCAAGGAAUCCAUCC
UGCCCAAGAGGAACAGCGACAAGCUGAUCGCCCGGAAGAAGGACUGGGACCCUAAGAAGUAUGGAGGCUUC
GACAGCCCCACCGUAGCCUACAGCGUGCUGGUGGUCGCGAAGGUAGAGAAGGGGAAGAGCAAGAAACUGAA
GAGCGUGAAGGAGCUGCUCGGCAUAACCAUCAUGGAGAGGUCCAGCUUUGAGAAGAACCCCAUUGACUUUU
UGGAAGCCAAGGGCUACAAAGAGGUCAAAAAGGACCUGAUCAUCAAACUCCCCAAGUACUCCCUGUUUGAA
UUGGAGAACGGCAGAAAGAGGAUGCUGGCGAGCGCUGGGGAACUGCAAAAGGGCAACGAACUGGCGCUGCC
CAGCAAGUACGUGAAUUUUCUGUACCUGGCGUCCCACUACGAAAAGCUGAAAGGCAGCCCCGAGGACAACG
AGCAGAAGCAGCUGUUCGUGGAGCAGCACAAGCAUUACCUGGACGAGAUAAUCGAGCAAAUCAGCGAGUUC
AGCAAGAGGGUGAUUCUGGCCGACGCGAACCUGGAUAAGGUCCUCAGCGCCUACAACAAGCACCGAGACAA
ACCCAUCAGGGAGCAGGCCGAGAAUAUCAUACACCUGUUCACCCUGACAAAUCUGGGCGCACCUGCGGCAU
UCAAAUACUUCGAUACCACCAUCGACAGGAAAAGGUACACUAGCACUAAGGAGGUGCUGGAUGCCACCUUG
AUCCACCAGUCCAUUACCGGCCUGUAUGAGACCAGGAUCGACCUGAGCCAGCUUGGAGGCGACUCUAGGGC
GGACCCAAAAAAGAAAAGGAAGGUGGAAUUCCACCACACUGGACUAGUGGAUCCGAGCUCGGUACCAAGCU
UAAGUUUAAACCGCUGAUCAGCCUCGACUGUGCCUUCUAGUUGCCAGCCAUCUGUUGUUUGCCCCUCCCCC
GUGCCUUCCUUGACCCUGGAAGGUGCCACUCCCACUGUCCUUUCCUAAUAAAAUGAGGAAAUUGCAUCGCA
UUGUCUGAGUAGGUGUCAUUCUAUUCUGGGGGGUGGGGUGGGGCAGGACAGCAAGGGGGAGGAUUGGGAAG
ACAAUAGCAGGCAUGCUGGGGAUGCGGUGGGCUCUAUGGC - pol yA
EXAMPLE 12
[00131] The following example demonstrates reduced stimulation of the innate
immune system in mammalian cells by the truncated chemically modified
crRNA:tracrRNA complexes of the present invention when compared with
unmodified
IVT sgRNAs.
[00132] Mammalian cells possess a variety of receptors intended to identify
and
respond to foreign RNAs as part of anti-viral immunity. This includes
receptors such as
TLR-3, TLR-7, TLR8, RIG-I, MDA5, OAS, PKR, and others. In broad terms, RNAs
that are short or contain chemical modifications present in mammalian cells
(such as
2'0Me RNA) evade detection or are less stimulatory than are long, unmodified
RNAs.
The present example compares the level of stimulation of 2 immune response
associated
genes (IFIT 1 and IFITM1) when mammalian REK293 cells are transfected with
truncated unmodified or truncated modified crRNA:tracrRNA complexes of the
present
invention with a commercial IVT sgRNA (Thermo Fisher Scientific, Waltham, MA).
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
[00133] CRISPR guide RNAs specific to human HPRT1 site 38285 were employed.
Sequences are shown in Table 11 below. The unmodified crRNA:tracrRNA complexes
(SEQ ID Nos. 48 and 2), the modified crRNA:tracrRNA complexes (SEQ ID Nos. 178
and 100) and the sgRNA (SEQ ID No. 414) were transfected into HEK-Cas9 cells
at 30
nM concentration as outlined in Example 2 above. RNA was prepared 24 hours
after
transfection using the 5V96 Total RNA Isolation Kit (Promega, Madison, WI).
cDNA
was synthesized using 150 ng total RNA with SuperScriptTm-II Reverse
Transcriptase
(Invitrogen, Carlsbad, CA) per the manufacturer's instructions using both
random
hexamer and oligo-dT priming. Transfection experiments were all performed a
minimum of three times.
[00134] Quantitative real-time PCR was performed using 10 ng cDNA per 10 tL
reaction with ImmolaseTM DNA Polymerase (Bioline, Randolph, MA), 200 nM
primers,
and 200 nM probe. Cycling conditions employed were: 95 C for 10 minutes
followed by
40 cycles of 2-step PCR with 95 C for 15 seconds and 60 C for 1 minute. PCR
and
fluorescence measurements were done using an ABI Prism' 7900 Sequence Detector
(Applied Biosystems Inc., Foster City, CA). All reactions were performed in
triplicate
using 2-color multiplexing. Expression data were normalized against an average
of two
internal control genes. Copy number standards were linearized cloned amplicons
for all
assays. Unknowns were extrapolated against standards to establish absolute
quantitative
measurements. Housekeeping internal control normalization assays were HPRT 1
(primers and probe SEQ ID Nos. 415-417) and SFRS9 (primers and probe SEQ ID
Nos.
418-420). Immune activation pathway assays were IFIT All (primers and probe
SEQ ID
Nos. 421-423) and IFIT 1 (primers and probe SEQ ID Nos. 424-426). The results
were
normalized using non-transfected cells as baseline and are shown in FIG. 13.
Table 11. Nucleic acid reagents employed in immune activation experiments in
Example 12.
SEQ
Reagent Sequence
ID No.
Unmodified
48 cuuauauccaacacuucgugguuuuagagcuaugcu
crRNA
2 Unmodified agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggca
tracrRNA ccgagucggugcuuu
Modified
178 c*u*u*auauccaacacuucgugguuuuagagcuau*g*c*u
crRNA
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO
2016/100951 PCT/US2015/066942
81
SEQ
Reagent Sequence
ID No.
100 Modified a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugg
tracrRNA caccgagucggugcu*u*u
ppp-gcuuauauccaacacuucgugguuuuagagcuagaaauagcaaguuaa
414 IVT sgRNA aauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucggugcuuu
uuuu
Hs HPRT
415 GACTTTGCTTTCCTTGGTCAG
F517
Hs HPRT
416 GGCTTATATCCAACACTTCGTGGG
R591
Hs HPRT
FAM-ATGGTCAAG ( ZEN) GTCGCAAGCTTGCTGGT- ZEN
P554
Hs SFRS9
418 TGTGCAGAAGGATGGAGT
F569
Hs SFRS9
419 CTGGTGCTTCTCTCAGGATA
R712
Hs SFRS9
HEX- TGGAATATG ( ZEN) CCCTGCGTAAACTGGA- ZEN
P644
Hs IFITM1
421 CTCTTCTTGAACTGGTGCTGTCTG
For
Hs IFITM1
422 CAGGATGAATCCAATGGTCATGAGG
Rev
1113 Hs IFITM1
FAM-AAGTGCCTG ( ZEN) AACATCTGGGCCCTGATT-ZEN
Probe FAM
Hs IFIT1
424 CCATTGTCTGGATTTAAGCGG
For
Hs IFIT1
425 GCCACAAAAAATCACAAGCCA
Rev
IV4 Hs IFIT1
HEX-TTTCTTTGC ( ZEN) TTCCCCTAAGGCAGGCTG-ZEN
Probe HEX
1 Compound I is an oligonucleotide having the formula SEQ ID NO: 417-(ZEN)-SEQ
ID NO: 441.
2 Compound II is an oligonucleotide having the formula SEQ ID NO: 420-(ZEN)-
SEQ ID NO: 442.
3 Compound III is an oligonucleotide having the formula SEQ ID NO: 423-(ZEN)-
SEQ ID NO: 443.
4 Compound IV is an oligonucleotide having the formula SEQ ID NO: 426-(ZEN)-
SEQ ID NO: 444.
Oligonucleotide sequences are shown 5'-3'. Uppercase = DNA; Lowercase = RNA;
Underlined = 2'-0-methyl RNA; * = phosphorothioate internucleotide linkage;
ppp =
triphosphate; ZEN = napthyl-azo modifier, dark quencher; FAM = 6-
carboxyfluorescein;
HEX = hexachlorofluorescein.
[00135] Treatment with the unmodified or chemically modified truncated
crRNA:tracrRNA complex did not lead to detectable increases in IFIT1 or IFITM1
expression over baseline. In contrast, treatment with the longer IVT sgRNA led
to a 45-
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
82
fold induction of IFITM1 and a 220-fold induction of IFIT1. Thus, significant
stimulation of the innate immune system occurred using the sgRNA that was
absent
using the short crRNA:tracrRNA complexes of the present invention.
EXAMPLE 13
[00136] The following example combines modification patterns identified in
Examples 6 and 7 as being particularly efficacious to demonstrate new highly
modified
crRNA and tracrRNA compositions that perform with high efficiency in mammalian
CRISPR genome editing applications.
[00137] A series of crRNAs and tracrRNAs (Table 12) were synthesized having
chemical modifications as indicated. The crRNAs employed a 20 base protospacer
domain targeting the same site in the human HPRT1 gene (38285) at the 5'-end
with a 16
base tracrRNA binding domain at the 3'-end. The tracrRNAs were synthesized
having
chemical modifications as indicated, using the 67 nucleotide or 62 nucleotide
truncated
versions of the tracrRNA sequence. The crRNAs and tracrRNAs listed in Table 12
were
paired as indicated and transfected into the HEK-Cas9 cells at 30 nM
concentration and
processed as described in previous Examples. Relative gene editing activities
were
assessed by comparing cleavage rates in the HPRT1 gene using the T7EI mismatch
endonuclease cleavage assay, with quantitative measurement of products done
using the
Fragment Analyzer.
[00138] Table 12: Activity of highly modified crRNA:tracrRNA complexes to
direct Cas9-mediated gene editing in mammalian cells.
cr/tracr
SEQ crRNA Sequence Cleavage
RNA
ID No.
pair tracrRNA Sequence
448 c*u*u*auauccaacacuucgugguuuuagagcuau*g*c*u
1 57
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaa
2 aaaguggcaccgagucggugcuuu
448 c*u*u* auauccaacacuucgugguuuuagagcuau* g* c*u
2 58
100
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuug
¨
aaaaaguggcaccgagucggugcu*u*u
3 48 cuuauauccaacacuucgugguuuuagagcuaugcu 58
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
83
cr/tracr
SEQ crRNA Sequence Cl
RNA eavage
ID No.
pair tracrRNA Sequence
a* g* cauagcaaguuaaaauaaggcuaguccguuaucaacuug
449 aaaaaguggcaccgagucggugcu*u*u
48 cuuauauccaacacuucgugguuuuagagcuaugcu
4 57
a* g* cau a gcaaguuaaaauaaggcuaguccguuaucaacuug
450 aaaaaguggcaccgagu cggu gc u* u*u
48 cuuauauccaacacuucgugguuuuagagcuaugcu
65
a*g*cauagcaaguuaaaauaaggcuaguccguuaucaacuug
451 ¨aaaaaguggcaccgagucg*g*u
[00139] Oligonucleotide sequences are shown 5'-3'. Lowercase = RNA;
Underlined = 2'-0-methyl RNA; Lowercase italic = 2'F RNA; * =
phosphorothioate internucleotide linkage. The relative functional activity of
each
complex is indicated by the % cleavage in a T7EI heteroduplex assay for each
dose studied.
[00140] The crRNA:tracrRNA pairs #1 and #2 show that a highly 2'F RNA modified
crRNA (SEQ ID No. 448, which has 22/36 residues modified, or 61%) is highly
functional when paired with either an unmodified tracrRNA (SEQ ID No. 2) or a
highly
2'0Me modified tracrRNA (SEQ ID No. 100). The crRNA:tracrRNA pairs #3 and #4
show that tracrRNA compositions having moderate (SEQ ID No. 450, with 19/67
residues modified, or 28%) or high (SEQ ID No. 449, with 46/67 residues
modified, or
69%) levels of 2'F RNA modification are highly functional. Information derived
from
Example 6 (in particular, the 2'0Me "walk", SEQ ID Nos. 144-162) was used to
identify
specific residues that can be modified within the internal domain of the
tracrRNA (see
Fig. 6). The crRNA:tracrRNA pair #5 demonstrates that an extremely highly
modified
tracrRNA, which in this case was a truncated 62 nucleotide design (SEQ ID No.
451,
having 51/62 residues modified with 2'0Me RNA, or 82%), has high potency in
triggering CRISPR genome editing in mammalian cells. Therefore, the original
89 RNA
nucleotide wild-type tracrRNA has been optimized herein to a form that has as
little as
11 RNA residues remaining (11/62), thereby significantly reducing risk of RNA-
based
activation of the mammalian innate immune system and reducing the nuclease-
susceptible RNA content of the tracrRNA to a minimal level.
SUBSTITUTE SHEET (RULE 26)
CA 02970683 2017-06-12
WO 2016/100951
PCT/US2015/066942
84
[00141] All references, including publications, patent applications, and
patents, cited
herein are hereby incorporated by reference to the same extent as if each
reference were
individually and specifically indicated to be incorporated by reference and
were set forth
in its entirety herein.
[00142] The use of the terms "a" and "an" and "the" and similar referents in
the
context of describing the invention (especially in the context of the
following claims) are
to be construed to cover both the singular and the plural, unless otherwise
indicated
herein or clearly contradicted by context. The terms "comprising," "having,"
"including," and "containing" are to be construed as open-ended terms (i.e.,
meaning
"including, but not limited to,") unless otherwise noted. Recitation of ranges
of values
herein are merely intended to serve as a shorthand method of referring
individually to
each separate value falling within the range, unless otherwise indicated
herein, and each
separate value is incorporated into the specification as if it were
individually recited
herein. All methods described herein can be performed in any suitable order
unless
otherwise indicated herein or otherwise clearly contradicted by context. The
use of any
and all examples, or exemplary language (e.g., "such as") provided herein, is
intended
merely to better illuminate the invention and does not pose a limitation on
the scope of
the invention unless otherwise claimed. No language in the specification
should be
construed as indicating any non-claimed element as essential to the practice
of the
invention.
[00143] Preferred embodiments of this invention are described herein,
including the
best mode known to the inventors for carrying out the invention. Variations of
those
preferred embodiments may become apparent to those of ordinary skill in the
art upon
reading the foregoing description. The inventors expect skilled artisans to
employ such
variations as appropriate, and the inventors intend for the invention to be
practiced
otherwise than as specifically described herein. Accordingly, this invention
includes all
modifications and equivalents of the subject matter recited in the claims
appended hereto
as permitted by applicable law. Moreover, any combination of the above-
described
elements in all possible variations thereof is encompassed by the invention
unless
otherwise indicated herein or otherwise clearly contradicted by context.
SUBSTITUTE SHEET (RULE 26)