Note: Descriptions are shown in the official language in which they were submitted.
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
PEPTIDE INHIBITORS OF INTERLEUKIN-23 RECEPTOR AND THEIR USE
TO TREAT INFLAMMATORY DISEASES
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This application claims priority to and is a continuation-in-part of
U.S. Application No.
14/800,627 filed on July 15, 2015, and also claims priority to International
Patent Application
PCT/US2015/040658 filed on July 15, 2015, U.S. Provisional Application No.
62/264,820 filed
on December 8, 2015, and U.S. Provisional Application No. 62/281,123 filed on
January 20,
2016, all of which are incorporated by reference herein in theirentireties.
SEQUENCE LISTING
[0002] The instant application contains a Sequence Listing which has been
submitted
electronically in ASCII format and is hereby incorporated by reference in its
entirety. Said
ASCII copy, created on July 15, 2016, is named PRTH-002-03WO SL.txt and is 504
KB in size
FIELD OF THE INVENTION
[0003] The present invention relates to novel peptide inhibitors of the
interleukin-23 receptor,
and their use to treat or prevent a variety of diseases and disorders,
including inflammatory
bowel disease, Crohn's disease and psoriasis.
BACKGROUND
[0004] The interleukin-23 (IL-23) cytokine has been implicated as playing a
crucial role in the
pathogenesis of autoimmune inflammation and related diseases and disorders,
such as multiple
sclerosis, asthma, rheumatoid arthritis, psoriasis, and inflammatory bowel
diseases (IBDs), e.g.,
ulcerative colitis and Crohn's disease. Studies in acute and chronic mouse
models of IBD
revealed a primary role of IL-23R and downstream effector cytokines in disease
pathogenesis.
IL-23R is expressed on various adaptive and innate immune cells including Th17
cells,
T cells, natural killer (NK) cells, dendritic cells, macrophages, and innate
lymphoid cells,
which are found abundantly in the intestine. At the intestine mucosal surface,
the gene expression
and protein levels of IL-23R are found to be elevated in IBD patients. It is
believed that IL-23
1
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
mediates this effect by promoting the development of a pathogenic CD4+ T cell
population that
produces IL-6, IL-17, and tumor necrosis factor (TNF).
100051 Production of IL-23 is enriched in the intestine, where it is believed
to play a key role in
regulating the balance between tolerance and immunity through T-cell-dependent
and T-cell-
independent pathways of intestinal inflammation through effects on T-helper 1
(Thl) and Th17-
associated cytokines, as well as restraining regulatory T-cell responses in
the gut, favoring
inflammation. In addition, polymorphisms in the IL-23 receptor (IL-23R) have
been associated
with susceptibility to IBDs, further establishing the critical role of the IL-
23 pathway in intestinal
homeostasis.
[0006] Psoriasis, a chronic skin disease affecting about 2%-3% of the general
population has
been shown to be mediated by the body's T cell inflammatory response
mechanisms. 11-23 has
one of several interleukins implicated as a key player in the pathogenesis of
psoriasis,
purportedly by maintaining chronic autoimmune inflammation via the induction
of interleukin-
17, regulation of T memory cells, and activation of macrophages. Expression of
IL-23 and IL-
23R has been shown to be increased in tissues of patients with psoriasis, and
antibodies that
neutralize IL-23 showed IL-23-dependent inhibition of psoriasis development in
animal models
of psoriasis.
[0007] IL-23 is a heterodimer composed of a unique p19 subunit and the p40
subunit of IL-12,
which is a cytokine involved in the development of interferon-y (IFN-y)-
producing T helper 1
(TH1) cells. Although IL-23 and IL-12 both contain the p40 subunit, they have
different
phenotypic properties. For example, animals deficient in IL-12 are susceptible
to inflammatory
autoimmune diseases, whereas IL-23 deficient animals are resistant, presumably
due to a reduced
number of CD4+ T cells producing IL-6, IL-17, and TNF in the CNS of IL-23-
deficient animals.
IL-23 binds to IL-23R, which is a heterodimeric receptor composed of IL-12R31
and IL-23R
subunits. Binding of IL-23 to IL-23R activates the Jak-stat signaling
molecules, Jak2, Tyk2, and
Statl, Stat 3, Stat 4, and Stat 5, although Stat4 activation is substantially
weaker and different
DNA-binding Stat complexes form in response to IL-23 as compared with IL-12.
IL-23R
associates constitutively with Jak2 and in a ligand-dependent manner with
Stat3. In contrast to
2
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
IL-12, which acts mainly on naive CD4(+) T cells, IL-23 preferentially acts on
memory CD4(+)
T cells.
[0008] Efforts have been made to identify therapeutic moieties that inhibit
the IL-23 pathway,
for use in treating IL-23-related diseases and disorders. A number of
antibodies that bind to IL-
23 or IL-23R have been identified, including ustekinumab, a humanized antibody
that binds IL-
23, which has been approved for the treatment of psoriasis. More recently,
polypeptide
inhibitors that bind to IL-23R and inhibit the binding of IL-23 to IL-23R have
been identified
(see, e.g., US Patent Application Publication No. US2013/0029907). Clinical
trials in Crohn's
Disease or psoriasis with ustekinumab and briakinumab (which target the common
p40 subunit)
and tildrakizumab, guselkumab, MEDI2070, and BI-655066 (which target the
unique p19
subunit of IL-23) highlight the potential of IL-23 signaling blockade in
treatment of human
inflammatory diseases. While these findings are promising, challenges remain
with respect to
identifying stable and selective agents that preferentially target the IL-23
pathway in the
intestine, which can be used for the treatment of intestinal inflammation,
such as intestinal bowel
diseases, including Crohn's disease, ulcerative colitis and related disorders.
[0009] Clearly, there remains a need in the art for new therapeutics targeting
the IL-23 pathway,
which may be used to treat and prevent IL-23-asociated diseases, including
those associated with
autoimmune inflammation in the intestinal tract. In addition, compounds and
methods for
specific targeting of IL-23R from the luminal side of the gut may provide
therapeutic benefit to
IBD patients suffering from local inflammation of the intestinal tissue. The
present invention
addresses these needs by providing novel peptide inhibitors that bind IL-23R
to inhibit IL-23
binding and signaling and which are suitable for oral administration.
BRIEF SUMMARY OF THE INVENTION
[0010] The present invention provides inter alia novel peptide inhibitors of
IL-23R and related
methods of use.
In a first aspect, the present invention provides a peptide inhibitor of an
interleukin-23 receptor,
or a pharmaceutically acceptable salt or solvate thereof, wherein the peptide
inhibitor comprises
an amino acid sequence of Formula (Xa): X1-X2-X3-X4-X5-X6-X7-X8-X9-X10-X11-X12-
X13-X14-X15-X16-X17-X18-X19-X20 (Xa),
3
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
wherein:
Xl, X2 and X3 are any amino acid or absent
X4 is any amino acid or chemical moiety capable of forming a bond with X9;
X5, X6, X7 and X8 are any amino acid;
X9 is any amino acid or chemical moiety capable of forming a bond with X4;
X10, X11, X12, X13, X14 and X15 are any amino acid; and
X16, X17, X18, X19 and X20 are any amino acid or absent;
wherein the peptide inhibitor is cyclized via a bond between X4 and X9, and
wherein the peptide inhibitor inhibits the binding of an interleukin-23 (IL-
23) to an IL-23
receptor.
[0011] In certain embodiments of Xa:
[0012] X1 is absent; X2 is absent; X3 is absent; X4 is Cys, Abu or Pen; X5 is
Ala, E -MeOrn, E-
MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn, N-MeGln, N-MeArg, Orn, Gln, Arg,
Ser or Thr;
X6 is Asp or Thr; X7 is Trp or 6-Chloro-Trp; X8 is Glu, Gln or Val; X9 is Cys,
Abu or Pen; X10
is 2-Nal, a Phe analog, Tyr, or a Tyr analog; X11 is 1-Nal, 2-Nal, Phe(3,4-
dimethoxy), 5-
HydroxyTrp, Phe(3,4-C12), Trp or Tyr(3-tBu); X12 is 3-Pal, Acpc, Acbc, Acvc,
Achc, Agp, Aib,
a-DiethylGly, a-MeLys, a-MeLys(Ac), a-MeLeu, a-MeOrn, a-MeSer, a-MeVal, Cav,
Cha,
Cit, Cpa, D-Asn, Glu, His, hLeu, hArg, Lys, Leu, Octgly, Orn, 4-amino-4-
carboxy-piperidine,
Arg, Ser, Thr or MP; X13 is Cit, Asp, Dab, Dap, Phe, His, Dap(Peg2-Ac),
Dap(pyroglutaric
acid), Glu, HomoArg, Lys, Lys(Ac), Lys(Benzoic acid), Lys(glutaric acid),
Lys(IVA), Lys(Peg4-
isoGlu-Palm), Lys(pyroglutaric acid), Lys(succinic acid), Asn, Orn, Gln, Arg,
Thr or Val; X14 is
Asp, Dab(Ac), Dap(Ac), Phe, His, Lys(Ac), Met, Asn(isobutyl), Gln, Arg, Tyr or
Asp(1,4-
diaminobutane); and X15 is Ala, f3Ala, Glu, Gly, Asn, Gln, Arg or Ser.
[0013] In certain embodiments of Xa: X1 is absent; X2 is absent; X3 is absent;
X4 is Cys, Abu
or Pen; X5 is Ala, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn,
Orn, Gln, Arg,
4
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Ser or Thr; X6 is Asp or Thr; X7 is Trp or 6-Chloro-Trp; X8 is Gin or Val; X9
is Cys, Abu or
Pen; X10 is 2-Nal, a Phe analog, Tyr, or a Tyr analog; X11 is 1-Nal, 2-Nal,
Phe(3,4-dimethoxy),
5-HydroxyTrp, Phe(3,4-C12), Trp or Tyr(3-tBu); X12 is 3-Pal, Acpc, Acbc, Acvc,
Achc, Agp,
Aib, a-DiethylGly, a-MeLys, a-MeLys(Ac), a-MeLeu, a-MeOrn, a-MeSer, a-MeVal,
Cav,
Cha, Cit, Cpa, D-Asn, His, hLeu, hArg, Lys, Leu, Octgly, Orn, 4-amino-4-
carboxy-piperidine, or
THP; X13 is Cit, Asp, Dab, Dap, Phe, His, Dap(Peg2-Ac), Dap(pyroglutaric
acid), Glu, hArg,
Lys, Lys(Ac), Lys(Benzoic acid), Lys(glutaric acid), Lys(IVA), Lys(Peg4-isoGlu-
Palm),
Lys(pyroglutaric acid), Lys-(succinic acid), Asn, Orn,G1n, Arg, Thr or Val;
X14 is Dab(Ac),
Dap(Ac), Phe, His, Lys(Ac), Met, Asn, Gin, Arg, or Tyr; and X15 is Ala,
betaAla, Gly, Asn,
Gin, or Ser.
[0014] In certain embodiments of Xa: X1 is absent; X2 is absent; X3 is absent;
X4 is Cys, Abu
or Pen; X5 is Dap, Dap(Ac), Gly, Lys, Gin, Arg, Ser,Thr or Asn; X6 is Thr; X7
is Trp or 6-
Chloro-Trp; X8 is Gin; X9 is Cys, Abu or Pen; X10 is 2-Nal, a Phe analog, Tyr,
or a Tyr analog;
X11 is 1-Nal, 2-Nal, Phe(3,4-dimethoxy), Phe(3,4-C12), or Trp; X12 is Acpc,
Acbc, Acvc, Achc,
Aib, a-DiethylGly, a-MeLys, a-MeLys(Ac), a-MeLeu, a-MeOrn, a-MeSer, a-MeVal,
Cha,
Cit, hLeu, Lys, Leu, Arg or THP; X13 is Cit, Asp, Dap, Dap(Peg2-Ac),
Dap(pyroglutaric acid),
Glu, hArg, Lys, Lys(Ac), Lys(Benzoic acid), Lys(glutaric acid), Lys(IVA),
Lys(Peg4-isoGlu-
Palm), Lys(pyroglutaric acid), Lys-(succinic acid), Asn, Orn,G1n, Arg, or Val;
X14 is Dab(Ac),
Dap(Ac), His, Lys(Ac), Asn, Gin, or Tyr; and X15 is Ala, betaAla, Gly, Asn,
Gin, or Ser.
[0015] In certain embodiments of Xa: X1 is absent; X2 is absent; X3 is absent;
X4 is Cys, Abu
or Pen; X5 is Dap, Dap(Ac), Gin, Ser, Thr or Asn; X6 is Thr; X7 is Trp; X8 is
Gin; X9 is Cys,
Abu or Pen; X10 is a Phe analog, Tyr, or a Tyr analog; X11 is 2-Nal or Trp;
X12 is Acpc, Acbc,
Acvc, Achc, Aib, a-DiethylGly, a-MeLys, a-MeLys(Ac), a-MeLeu, a-MeOrn, a-
MeSer, a-
MeVal, hLeu, Leu, or THP; X13 is Cit, Asp, Glu, Lys, Lys(Ac), Asn, or Gin; X14
is Dab(Ac),
Asn, or His; and X15 is Ala, betaAla, Gly, Asn, or Gin.
[0016] In certain embodiments of Xa: X4 is Cys, Pen, hCys, D-Pen, D-Cys, D-
hCys, Met, Glu,
Asp, Lys, Orn, Dap, Dab, D-Dap, D-Dab, D-Asp, D-Glu, D-Lys, Sec, 2-
chloromethylbenzoic
acid, mercapto-propanoic acid, mercapto-butyric acid, 2-chloro-acetic acid, 3-
choropropanoic
acid, 4-chlorobutyric acid, 3-chloroisobutyric acid, Abu, f3-azido-Ala-OH,
propargylglycine, 2-
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
(3 '-butenyl)glycine, 2-allylglycine, 2-(3 '-butenyl)glycine,
2- (4'-pentenyl)glycine, 2-(5'-
hexenyl)glycine, or Abu; X7 is Trp, Glu, Gly, Ile, Asn, Pro, Arg, Thr or
OctGly, or a
corresponding a-methyl amino acid form of any of the foregoing; X9 is Cys,
Pen, hCys, D-Pen,
D-Cys, D-hCys, Glu, Lys, Orn, Dap, Dab, D-Dap, D-Dab, D-Asp, D-Glu, D-Lys,
Asp, Leu, Val,
Phe, or Ser, Sec, Abu, f3-azido-Ala-OH, propargylglycine, 2-2-allylglycine, 2-
(3'-
butenyl)glycine, 2-(4'-pentenyl)glycine, Ala, hCys, Abu, Met, MeCys, (D)Tyr or
2-(5'-
hexenyl)glycine; X10 is Tyr, Phe(4-0Me), 1-Nal, 2-Na!, Aic, a-MePhe, Bip,
(D)Cys, Cha,
DMT, (D)Tyr, Glu, His, hPhe(3,4-dimethoxy), hTyr, N-Me-Tyr, Trp, Phe(4-CONH2),
Phe(4-
phenoxy), Thr, Tic, Tyr(3-tBu), Phe(4-tBu), Phe(4-CN), Phe(4-Br), Phe(4-NH2),
Phe(4-F),
Phe(3,5-F2), Phe(4-CH2CO2H), Phe(penta-F), Phe(3,4-C12), Phe(4-CF3), Phe(4-
0CH3), Bip, Cha,
4-PyridylAlanine, f3hTyr, OctGly, Phe(4-N3), Phe(4-Br), Phe[4-(2-aminoethoxy)]
or Phe, a Phe
analog, a Tyr analog, or a corresponding a-methyl amino acid form of any of
the foregoing; X11
is 2-Na!, 1-Nal, 2,4-dimethylPhe, Bip, Phe(3,4-C12), Phe (3,4-F2), Phe(4-
CO2H), f3hPhe(4-F),
Me-Trp, 4-phenylcyclohexyl, Phe(4-CF3), a-MePhe, f3hNal, f3hPhe, f3hTyr,
f3hTrp, Nva(5-
phenyl), Phe, His, hPhe, Tic, Tqa, Trp, Tyr, Phe(4-0Me), Phe(4-Me), Trp(2,5,7-
tri-tert-Butyl),
Phe(4-0ally1), Tyr(3-tBu), Phe(4-tBu), Phe(4-guanidino, Phe(4-0Bz1), Octgly,
Glu(Bz1), 4-
Phenylbenzylalanine, Phe[4-(2-aminoethoxy)], 5-Hydroxy-Trp, 6-Chloro-Trp, N-
MeTrp,
1,2,3,4-tetrahydro-norharman, Phe(4-CONH2), Phe(3,4-Dimethoxy), Phe(2,3 -C12),
Phe(2,3-F2),
Phe(4-F), 4-phenylcyclohexylalanine, Bip, or a corresponding a-methyl amino
acid form of any
of the foregoing; X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit,
hLeu, 3-Pal, t-butyl-
Ala, 4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acvc, Acbc, Agp, Aib, a-
DiethylGly,
MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Aibõ D-Ala, (D)Asn,
(D)Asp,
(D)Leu, (D)Phe, (D)Tyr, Aib, a-MeLeu,a-MeOrn, R-
Aib, f3-Ala, f3hAla, PhArg, PhLeu, f3hVal, f3-spiro-pip, Glu, hArg, Ile, Lys,
N-MeLeu, N-MeArg,
Ogl, Orn, Pro, Gin, Ser, Thr, Tie, t-butyl-Gly, or a corresponding a-methyl
amino acid form of
any of the foregoing; X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Arg, Orn,
Val, f3hAla, Lys(Ac),
(D)Asn, (D)Leu, (D)Phe, (D)Thr, Ala, a-MeLeu, Aib, f3-Ala, f3-Glu, PhLeu,
f3hVal, f3-spiro-pip,
Cha, Chg, Asp, Dab, Dap, a-DiethylGly, hLeu, Asn, Ogl, Pro, Gin, Ser, f3-spiro-
pip, Thr, Tba,
Tie or Aib, Cit, hArg, Lys, Asn, Orn, Gin or a corresponding a-methyl amino
acid form of any
of the foregoing; X14 is Phe, Tyr, Glu, Gly, His, Lys, Leu, Met, Asn, Pro,
Gin, Arg, Ser, Thr,
6
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Ticf3hPhe, Arg, Lys(Ac), His; Dap(Ac), Dab(Ac), Asp or a corresponding a-
methyl amino acid
form of any of the foregoing; X15 is Gly, Ser, Thr, Gin, Ala, (D)Ala, (D)Asn,
(D)Asp, (D)Leu,
(D)Phe, (D)Thr, Aea, Asp, Asn, Glu, Phe, Gly, Lys, Leu, Pro, Arg, f3-Ala,
Sarc, or a
corresponding a-methyl amino acid form of any of the foregoing; X16 is Asp,
Glu, Ala, AEA,
AEP, f3hAla, Gaba, Gly, Ser, Pro, Asn, Thr or absent, or a corresponding a-
methyl amino acid
form of any of the foregoing; and X17 is Leu, Lys, Arg, Glu, Ser, Gly, Gin or
absent, or a
corresponding a-methyl amino acid form of any of the foregoing.
[0017] In certain embodiments of peptide inhibitors of Xa, the bond is a
disulfide bond, a
thioether bond, a lactam bond, a triazole ring, a selenoether bond, a
diselenide bond, or an olefin
bond.
[0018] In particular embodiments of peptide inhibitors of Xa, X4 is Cys and X9
is Cys, and the
bond is a disulfide bond. In particular embodiments, X4 is Pen and X9 is Pen,
and the bond is a
disulfide bond. In certain embodiments: X7 is Trp; X10 is Phe, Tyr, a Phe
analog, or a Tyr
analog; X11 is Trp, 1-Nal or 2-Nal; and X12 is Aib, a-Me-Lys, a-Me-Leu, Achc,
Acvc, Acpc,
Acbc or THP. In certain embodiments: X7 is Trp; X10 is Phe, Tyr, a Phe analog,
or a Tyr
analog; X11 is Trp, 1-Nal or 2-Nal; and X12 is Aib, a-Me-Lys or a-Me-Leu. In
particular
embodiments, the peptide inhibitor comprises any of the following the amino
acid sequences:
Pen-Q-T-W-Q-Pen-[Phe(4-0Me)]-[2-Nal]-[a-Me-Lys]-E-N-G; Pen-N-T-W-Q-[Pen]-
[Phe[4-(2-
aminoethoxy)]-[2-Nall-[Aib]-[Lys(Ac)]-N-N; Pen-Q-T-W-Q-[Pen]-[Phe[4-(2-
aminoethoxy)]-[2-
Nall-[a-MeLeu]-[Lys(Ac)]-N-N; or Pen-Q-T-W-Q-[Pen]-[Phe(4-CONH2)]-[2-Nall-[a-
MeLys]-
[Lys(Ac)]-N-N, wherein the peptide inhibitor comprises a disulfide bond
between the two Pen
amino acids.
[0019] In particular embodiments of peptide inhibitors of Xa, X4 is an amino
acid, aliphatic
acid, alicyclic acid or modified 2- methyl aromatic acid having a carbon side
chain capable of
forming a thioether bind with X9; X9 is a sulfur-containing amino acid capable
of forming a
thioether bond with X4, and the bond between X4 and X9 is a thioether bond. In
certain
embodiments, X4 is Abu, 2-chloromethylbenzoic acid, mercapto-propanoic acid,
mercapto-
butyric acid, 2-chloro-acetic acid, 3-chloro-propanoic acid, 4-chloro-butyric
acid, 3-chloro-
isobutyric acid; and X9 is Abu, Cys, Pen, hCys, D-Pen, D-Cys, or D-hCys.In
certain
7
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
embodiments, X4 is Abu; and X9 is Cys. In certain embodiments, X7 is Trp; X10
is Phe, Tyr, a
Phe analog, or a Tyr analog; X11 is Trp, 1-Nal or 2-Nal; and X12 is a-Me-Lys,
a-Me-Leu,
Me-Ser, a-Me-Val, Achc, Acvc, Acpc, Acbc, or [4-amino-4-carboxy-
tetrahydropyran]. In
certain embodiments, X7 is Trp; X10 is Phe, Tyr, a Phe analog, or a Tyr
analog; X11 is Trp, 1-
Nal or 2-Nal; and X12 is a-Me-Lys or [4-amino-4-carboxy-tetrahydropyran]. In
particular
embodiments, the peptide inhibitor comprises any of the following amino acid
sequences: [Abu]-
Q-T-W-Q-C-[Phe(4-0Me)]-[2-Nal]-[a-MeLys]-E-N-G;
[Abu]-Q-T-W-Q-C-[Phe(4-(2-
aminoethoxy))]-W-[a-MeLys]-E-N-G; or [Abu]-Q-T-W-Q-C-[Phe[4-(2-aminoethoxy)]]-
[2-
Nal]-[4-amino-4-carboxy-tetrahydropyran]-E-N-N, wherein the peptide inhibitor
comprises a
thioether bond between the Abu and the C.
[0020] In certain embodiments of peptide inhibitors of Xa: X4 is Pen, Cys or
hCys; X5 is any
amino acid; X6 is any amino acid; X7 is Trp, Bip, Gln, His, Glu(Bz1), 4-
Phenylbenzylalanine,
Tic, Phe[4-(2-aminoethoxy)], Phe(3,4-C12), Phe(4-0Me), 5-Hydroxy-Trp, 6-Chloro-
Trp, N-
MeTrp, a-Me-Trp, 1,2,3,4 -tetrahydro-norharman, Phe(4-CO2H), Phe(4-CONH2),
Phe(3,4-
Dimethoxy), Phe(4-CF3), Phe(4-tBu), f33-diPheAla, Glu, Gly, Ile, Asn, Pro,
Arg, Thr or Octgly,
or a corresponding a-methyl amino acid form of any of the foregoing; X8 is any
amino acid; X9
is Pen, Cys or hCys; X10 is 1-Nal, 2-Nal, Aic, Bip, (D)Cys, Cha, DMT, (D)Tyr,
Glu, Phe, His,
Trp, Thr, Tic, Tyr, 4-pyridylAla, Octgly, a Phe analog or a Tyr analog
(optionally, Phe(3,4-F2),
Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)], Phe[4-(2-(acetyl-aminoethoxy)],
Phe(4-Br),
Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-guanidino), Phe(4-Me), Phe(4-NH2),
Phe(4-N3),
Phe(4-0Me), or Phe(4-0Bz1)), or a corresponding a-methyl amino acid form of
any of the
foregoing; X11 is 2-Nal, 1-Nal, 2,4-dimethylPhe, Bip, Phe(3,4-C12), Phe (3,4-
F2), Phe(4-CO2H),
f3hPhe(4-F), a-Me-Trp, 4-phenylcyclohexyl, Phe(4-CF3), a-MePhe, f3hNal,
f3hPhe, f3hTyr,
f3hTrp, Nva(5-phenyl), Phe, His, hPhe, Tic, Tqa, Trp, Tyr, Phe(4-0Me), Phe(4-
Me), Trp(2,5,7-
tri-tert-Butyl), Phe(4-0ally1), Tyr(3-tBu), Phe(4-tBu), Phe(4-guanidino, Phe(4-
0Bz1), Octgly,
Glu(Bz1), 4-Phenylbenzylalanine, Phe[4-(2-aminoethoxy)], 5-Hydroxy-Trp, 6-
Chloro-Trp, N-
MeTrp, 1,2,3,4-tetrahydro-norharman, Phe(4-CONH2), Phe(3,4-0Me2) Phe(2,3-C12),
Phe(2,3-
F2), Phe(4-F), 4-phenylcyclohexylalanine or Bip, or a corresponding a-methyl
amino acid form
of any of the foregoing; X12 is a-MeLys, a-MeOrn, a-MeLeu, a-MeVal, 4-amino-4-
carboxy-
tetrahydropyran, Achc, Acpc, Acbc, Acvc, MeLeu, Aib, (D)Ala, (D)Asn, (D)Leu,
(D)Asp,
8
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
(D)Phe, (D)Thr, 3-Pal, Aib, f3-Ala, f3hGlu, f3hAla, PhLeu, f3hVal, f3-spiro-
pip, Cha, Chg, Asp,
Dab, Dap, a-DiethylGly, Glu, Phe, hLeu, hArg, hLeu, Ile, Lys, Leu, Asn, N-
MeLeu, N-MeArg,
Ogl, Orn, Pro, Gln, Arg, Ser, Thr or Tle, or a corresponding a-methyl amino
acid form of any of
the foregoing; X13 is Lys(Ac), (D)Asn, (D)Leu, (D)Thr, (D)Phe, Ala, Aib, a-
MeLeu, f3-Ala,
f3hGlu, f3hAla, PhLeu, f3hVal, f3-spiro-pip, Cha, Chg, Asp, Lys, Arg, Orn,
Dab, Dap, a-
DiethylGly, Glu, Phe, hLeu, Lys, Leu, Asn, Ogl, Pro, Gln, Asp, Arg, Ser, spiro-
pip, Thr, Tba,
Tlc, Val or Tyr, or a corresponding a-methyl amino acid form of any of the
foregoing; X14 is
Asn, Glu, Phe, Gly, His, Lys, Leu, Met, Asn, Pro, Gln, Arg, Ser, Thr, Tic or
Tyr, Lys(Ac), Orn
or a corresponding a-methyl amino acid form of any of the foregoing; X15 is
Gly, (D)Ala,
(D)Asn, (D)Asp, Asn, (D)Leu, (D)Phe, (D)Thr, Ala, AEA, Asp, Glu, Phe, Gly,
Lys, Leu, Pro,
Gln, Arg or Ser, f3-Ala, Arg or a corresponding a-methyl amino acid form of
any of the
foregoing; X16 is absent, Gly, Ala, Asp, Ser, Pro, Asn or Thr, or a
corresponding a-methyl
amino acid form of any of the foregoing; X17 is absent, Glu, Ser, Gly or Gln,
or a corresponding
a-methyl amino acid form of any of the foregoing; X18 is absent or any amino
acid; X19 is
absent or any amino acid; and X20 is absent or any amino acid. In particular
embodiments, the
bond between X4 and X9 is a disulfide bond. In certain embodiments, Xl, X2,
and X3 are
absent.In certain embodiments, X17, X19 and X20 are absent. In certain
embodiments, one or
both of X4 or X9 is Pen. In certain embodiments, both X4 and X9 are Pen. In
particular
embodiments, X18 is (D)-Lys. In certain embodiments, the peptide inhibitors
comprise one or
more, two or more, three or more, or four of the following: X5 is Arg, Asn,
Gln, Dap, Orn; X6 is
Thr or Ser; X7 is Trp, 2-Nal, 1-Nal, Phe(4-0Ally1), Tyr(3-tBu), Phe(4-tBu),
Phe(4-guanidino),
Phe(Bz1) or Phe(4-Me), 5-Hydroxy-Trp, 6-Chloro-Trp, N-MeTrp, a-MeTrp or
1,2,3,4 -
tetrahydro-norharman; and X8 is Gln, Val, Phe, Glu, Lys. In certain
embodiments, the peptide
inhibitors comprise one or more, two or more, three or more, four or more,
five or more, six or
more, or seven of the following: X10 is Tyr, Phe(4-0Bz1), Phe(4-0Me), Phe(4-
CONH2),
Phe(3,4-C12), Phe(4-tBu), Phe(4-NH2), Phe(4-Br), Phe(4-CN), Phe(4-CO2H), Phe(4-
(2aminoethoxy)) or Phe(4-guanadino); X11 is Trp, 2-Nal, 1-Nal, Phe(4-0Ally1),
Tyr(3-tBu),
Phe(4-tBu), Phe(4-guanidino), Phe(Bz1) or Phe(4-Me), 5-Hydroxy-Trp, 6-Chloro-
Trp, N-MeTrp,
a-MeTrp or 1,2,3,4 -tetrahydro-norharman; X12 is Arg, a-MeLys a-MeLeu, Aib or
a-MeOrn;
X13 is Lys, Glu or Lys(Ac); X14 is Phe or Asn; X15 is Gly, Sr or Ala; and X16
is absent or
9
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
AEA. In certain embodiments, X4 and X9 are Pen; X5 is Gin; X6 is Thr; X7 is
Trp; X8 is Gin;
X10 is Tyr, Phe(4-0Me) or 2-Nal; X11 is Trp, 2-Nal or 1-Nal; X12 is Arg,
aMeLys or a-
MeOrn; X13 is Lys, Glu or Lys(Ac); X14 is Phe or Asn; X15 is Gly; and X16 is
absent. In
certain embodiments, one or more of Xl, X2 and X3 are absent; and one or more,
two or more,
three or more, or four of X17, X18, X19 and X20 are absent.
[0021] In certain embodiments of peptide inhibitors of Xa: X4 is Abu, Pen, or
Cys; X7 is Trp,
Bip, Gin, His, Glu(Bz1), 4-Phenylbenzylalanine, Tic, Phe[4-(2-aminoethoxy)],
Phe(3,4-C12),
Phe(4-0Me), 5-Hydroxy-Trp, 6-Chloro-Trp, N-MeTrp, a-MeTrp, 1,2,3,4 -tetrahydro-
norharman, Phe(4-CO2H), Phe(4-CONH2), Phe(3,4-Dimethoxy), Phe(4-CF3), f33-
diPheAla,
Phe(4-tBu), Glu, Gly, Ile, Asn, Pro, Arg, Thr or Octgly, or a corresponding a-
methyl amino acid
form of any of the foregoing; X9 is Abu, Pen, or Cys; X10 is 1-Nal, 2-Nal,
Aic, Bip, (D)Cys,
Cha, DMT, (D)Tyr, Glu, Phe, His, Trp, Thr, Tic, Tyr, 4-pyridylAla, Octgly a
Phe analog or a
Tyr analog, or a corresponding a-methyl amino acid form of any of the
foregoing; X11 is 2-Nal,
1-Nal, 2,4-dimethylPhe, Bip, 4-phenylcyclohexyl, Glu(Bz1), 4-
Phenylbenzylalanine, Tic, Phe[4-
(2-aminoethoxy)], Phe(3,4-C12), Phe(3,4-F2), f3hPhe(4-F), Phe(4-0Me), 5-
Hydroxy-Trp, 6-
Chloro-Trp, N-MeTrp, a-MeTrp, 1,2,3,4 -tetrahydro-norharman, Phe(4-CO2H),
Phe(4-
CONH2), Phe(3,4-Dimethoxy), Phe(4-CF3), Phe(2,3-C12), Phe(2,3-F2),Phe(4-F), 4-
phenylcyclohexylalanine, a-MePhe, f3hNal, f3hPhe, f3hTyr, f3hTrp, Bip, Nva(5-
phenyl), Phe, His,
hPhe, Tqa, Trp, Tyr, Phe(4-Me), Trp(2,5,7-tri-tertButyl), Phe(4-0Ally1), Tyr(3-
tBu), Phe(4-tBu),
Phe(4-guanidino), Phe(4-0Bz1), or Octgly, or a corresponding a-methyl amino
acid form of any
of the foregoing; X12 is a-MeLys, a-MeOrn, a-MeLeu, MeLeu, Aib, (D)Ala,
(D)Asn, (D)Leu,
(D)Asp, (D)Phe, (D)Thr, 3-Pal, Aib, f3-Ala, f3hGlu, f3hAla, PhLeu, f3hVal, f3-
spiro-pip, Cha, Chg,
Asp, Dab, Dap, a-DiethylGly, Glu, Phe, hLeu, hArg, hLeu, Ile, Lys, Leu, Asn, N-
MeLeu, N-
MeArg, Ogl, Orn, Pro, Gin, Arg, Ser, Thr or Tie, or a corresponding a-methyl
amino acid form
of any of the foregoing; X13 is Lys(Ac), (D)Asn, (D)Leu, (D)Thr, (D)Phe, Ala,
Aib, a-MeLeu,
f3Ala, f3hGlu, f3hAla, PhLeu, f3hVal, f3-spiro-pip, Cha, Chg, Asp, Arg, Orn,
Dab, Dap, a-
DiethylGly, Glu, Phe, hLeu, Lys, Leu, Asn, Ogl, Pro, Gin, Asp, Arg, Ser, spiro-
pip, Thr, Tba,
Tic, Val or Tyr, or a corresponding a-methyl amino acid form of any of the
foregoing; X14 is
Asn, Glu, Phe, Gly, His, Lys, Leu, Met, Asn, Pro, Gin, Arg, Ser, Thr, Tic or
Tyr, or a
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
corresponding a-methyl amino acid form of any of the foregoing; X15 is Gly,
(D)Ala, (D)Asn,
(D)Asp, Asn, (D)Leu, (D)Phe, (D)Thr, Ala, AEA, Asp, Glu, Phe, Gly, Lys, Leu,
Pro, Gln, Arg or
Ser, or a corresponding a-methyl amino acid form of any of the foregoing, or
X15 is Gly,
(D)Ala, (D)Asn, (D)Asp, Asn, (D)Leu, (D)Phe, (D)Thr, Ala, Asn, Ser, AEA, Asp,
Glu, Phe, Gly,
Lys, Leu, Pro, Gln, Arg or Ser, or a corresponding a-methyl amino acid form of
any of the
foregoing; X16 is absent, Gly, Ala, Asp, Ser, Pro, Asn or Thr, or a
corresponding a-methyl
amino acid form of any of the foregoing; and X17 is absent, Glu, Ser, Gly or
Gln, or a
corresponding a-methyl amino acid form of any of the foregoing. In particular
embodiments, the
peptide inhibitor is cyclized via an intramolecular bond between X4 and X9.In
certain
embodiments, one or more of Xl, X2, and X3 are absent. In certain embodiments,
one or more
of X17, X19 and X20 are absent.In certain embodiments, one of X4 or X9 is Abu,
and the other
of X4 or X9 is not Abu.In certain embodiments, the peptide inhibitors comprise
one or more, two
or more, three or more, or four of the following: X5 is Arg, Gln, Dap or Orn;
X6 is Thr or Ser;
X7 is Trp, 2-Nal, 1-Nal, Phe(4-0Ally1), Tyr(3-tBu), Phe(4-tBu), Phe(4-
guanidino), Phe(4-0Bz1),
Phe(4-Me), 5-Hydroxy-Trp, 6-Chloro-Trp, N-MeTrp, or
a-MeTrp, 1,2,3,4 -tetrahydro-
norharman; and X8 is Gln, Val, Phe, Glu or Lys. In certain embodiments, the
peptide inhibitors
comprise one or more, two or more, three or more, four or more, five or more,
six or more, or
seven of the following: X10 is Tyr, Phe(4-0Bz1), Phe(4-0Me), Phe(4-CONH2),
Phe(3,4-C12),
Phe(4-tBu), Phe(4-NH2), Phe(4-Br), Phe(4-CN), Phe(4-CO2H), Phe(4-
(2aminoethoxy)) or Phe(4-
guanadino); X11 is Trp, 2-Nal, 1-Nal, Phe(4-0Ally1), Tyr(3-tBu), Phe(4-tBu),
Phe(4-guanidino),
Phe(Bz1) or Phe(4-Me), 5-Hydroxy-Trp, 6-Chloro-Trp, N-MeTrp, a-MeTrp or
1,2,3,4 -
tetrahydro-norharman; X12 is Arg, hLeu, (D)Asn, Aib, a-MeLys, a-MeLeu or a-
MeOrn; X13 is
Lys, Glu or Lys(Ac); X14 is Phe or Asn; X15 is Gly, Ser or Ala, or X15 is Asn,
Gly, Ser, f3Ala
or Ala; and X16 is absent or AEA.
[0022] In particular embodiments of any of the peptide inhibitors, X4 and X9
are Pen. In
particular embodiments, X4 and X9 form a disulfide bond.
[0023] In particular embodiments, X4 is Abu and X9 is Cys. In particular
embodiments, X4 and
X9 form a thioether bond.
11
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
In particular embodiments, the peptide inhibitor comprises an amino acid
sequence of any one of
SEQ ID NOS: 365-370, 857-1029. In particular embodiments, the peptide
inhibitor is cyclized
via a bond between X4 and X9, and the peptide inhibitor inhibits the binding
of an interleukin-23
(IL-23) to an IL-23 receptor.
[0024] In certain embodiments of peptide inhibitors of Xa, the peptide
inhibitor comprises an
amino acid sequence set forth in any of Formulas (V), (Va), (Vb), (Vc), (Vd),
(Ve), (Vf), (Vg)
and (Vh).
100251 In certain embodiments of peptide inhibitors of Xa, the peptide
inhibitor comprises any of
the following amino acid sequences:
[Palm] - [isoGlu] - [PEG4] - [Pen] -NTWQ- [Pen] - [Phe [4- (2-aminoethoxy)] -
[2-Nal] - [Aib] - [Ly s (Ac)] -
E12;
Ac- [Pen] -NTWQ- [Pen]- [Phe [4- (2-amino ethoxy)]- [2-Nal] - [Aib] - [Lys
(PEG4 - s oGlu-Palm)] -NN-
NH2;
Ac- [Pen]-QTWQ - [Pen] -Phe(4-CONH2)- [2-Nal] - [a -MeLy s (Ac)] - [Lys(Ac)] -
NN-NH2;
[Octany1]-[IsoGlu]-[PEG4]-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-
[Aib]-
[Lys(Ac)]-NN-NH2;
[0 ctanyl] - [PEG4] - [Pen] -NTWQ- [Pen] - [Phe [4 -(2 -aminoethoxy)] - [2-
Nal] - [Aib] - [Ly s(Ac)]-NN-
NH2;
[Palm] - [PEG4]- [Pen] -NTWQ- [Pen] - [Phe [4 -(2 -aminoethoxy)] - [2-Nal] -
[Aib] - [Lys (Ac)] -NN-
NH2;
Ac- [Pen] -NTWQ- [Pen]- [Phe[4-(2-aminoethoxY)] - [2-Nal] - [Ail* [Lys (PEG4-0
ctanyl)] -NN-NH2;
Ac- [Pen] -NTWQ- [Pen]- [Phe [442-amino ethoxY)] - [2-Nal] - [Aib] - [Lys
(PEG4 -Palm)] -NN-NH2;
Ac- [Pen] -NTWQ- [Pen] - [Phe [4 -(2-amino ethoxy)-(PEG4 -Palm)] - [2-Nal] -
[Aib] - [Ly s (Ac)]NN-
NH2;
12
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)-(PEG4-Laury1)]-[2-Nal]-[Aib]-
[Lys(Ac)]-NN-
NH2;
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-[a-MeLys(PEG4-Palm)-[Lys(Ac)]-NN-NH2;
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-[a-MeLys(PEG4-Laury1)]-[Lys(Ac)]-NN-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)-(PEG4-IsoGlu-Palm)]-[2-Nal]-[Aib]-
[Lys(Ac)]-NN-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)-(PEG4-IsoGLu-Laury1)]-[2-Nal]-[Aib]-
[Lys(Ac)]-NN-NH2;
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-[a-MeLys(PEG4-IsoGlu-Palm)]-[Lys(Ac)]-
NN-NH2;
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-[a-MeLys(PEG4-IsoGlu-Laury1)]-
[Lys(Ac)]-
NN-NH2;
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-[a-MeLys(IVA)]-[Lys(Ac)]-NN-NH2;
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-[a-MeLys(Biotin)]-[Lys(Ac)]-NN-NH2;
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-[a-MeLys(Octany1)]-[Lys(A0]-NN-NH2;
Ac-[Pen]-[Lys(IVA)]-TWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-
NN-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Na1]-[Aib]-[Lys(Ac)]-[Lys(IVA)]-
N-NH2;
Ac-[Pen]-[Lys(Biotin)]-TWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aib]-
[Lys(Ac)]-NN-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aib]-[Lys(Ac)]-
[Lys(Biotin)]-N-
NH2;
Ac-[Pen]-[Lys(Octany1)]-TWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aib]-
[Lys(Ac)]-NN-
NH2;
13
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aib]-[Lys(Ac)]-
[Lys(octany1)]-N-
NH2;
Ac-[Pen]-[Lys(Palm)]-TWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]--[A113]-
[Lys(Ac)]-NN-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-Lys(Palm)]-
N-NH2;
Ac-[Pen]-[Lys(PEG8)]-TWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-
NN-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-
[Lys(PEG8)]-N-NH2;
Ac-[Pen]-K(Pegll-Palm)TWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aib]-
[Lys(Ac)]-NN-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]--[Aib]-[Lys(Ac)]-
[Lys(Pegll-palm)]-
N-NH2;
Ac-[Pen]-[Cit]-TW-[Cit]-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]--[Aib]-[Lys(Ac)]-
NN-NH2;
Ac-[Pen]-[Lys(Ac)]-TW-[Cit]-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aib]-
[Lys(Ac)]-NN-
NH2;
Ac-[Pen]-NT-[Phe(3,4-0CH3)2]-Q-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aib]-
[Lys(Ac)]-
NN-NH2;
Ac-[Pen]-NT-[Phe(2,4-CH3)2]-Q-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aib]-
[Lys(Ac)]-NN-
NH2;
Ac-[Pen]-NT-[Phe(3-CH3)]-Q-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aib]-
[Lys(Ac)]-NN-
NH2;
Ac-[Pen]-NT-[Phe(4-CH3)]-Q-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aib]-
[Lys(Ac)]-NN-
NH2;
Ac[(D)Arg]-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal] - [Aib]-[Lys(Ac)]-
N-[j3Ala]-
NH2;
14
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Ac-[(D)Tyr]-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aib]-[Lys(Ac)]-N-
[f3Ala]-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(A0]-QN-NE12;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-[Lys(Ac)]-
N-NE12;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-N-
[Lys(Ac)]-NE12;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(A0]-QQ-NE12;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aib]-[Lys(Ac)]-Q-[f3Ala]-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-N-[Cit]-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[A1b]-[1-Ys(A0]-[Cit]-
NNH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-[Cit]-Q-
NE12;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-[Cit]-
[Lys(Ac)]-NE12;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-[Lys(Ac)]-
[Cit]-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-QN-[13Ala]-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-E-[Cit]-Q-NE12;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[A113]-[Cit]-N-[Cit]-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)] - [2-Nall-[Aib]-[Cn]-Q-[Cit]-M12;
Ac-[Pen]-[Cit]-TWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-NN-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-NN-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-P-Nall-[A113]-QNN-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-ENQ-NH2;
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Ac-[Pen]-GPWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[A113]-[Lys(Ac)]-NN-NH2;
Ac-[Pen]-PGWQ-[Pen]-[Phe[4-(2-aminoethcxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-NN-NH2;
Ac-[Pen]-NTWN-[Pen]-[Phe[4-(2-aminoethcxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-NN-NH2;
Ac-[Pen]-NSWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[A113]-[Lys(Ac)]-NN-NH2;
Ac-[Pen]-N-[Aib]-WQ-[Pen]-[Phe[4-(2-aminoethcxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-NN-
NH2;
Ac-[Pen]-NTW-[Aib]-[Pen]-[Phe[4-(2-aminoethoxY)] - [2-NalHAibl-[Lys(Ac)]N-
[Aib]-NH2;
Ac-[Pen]-QTW- [Ly s(Ac)] - [Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-
[Lys(Ac)]-NN-NH2;
Ac-[Pen]-[Lys(Ac)]-TWQ-[Pen]-[Phe[4-(2-aminoethcxY)]-[2-Nal]--[A113]-
[Lys(Ac)]NNNH2;
Ac-[Pen]-QVWQ-[Pen]-[Phe[4-(2-aminoethcxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-NN-NH2;
Ac-[Pen]-NT-[2-Nal]-Q-[Pen]-[Phe[4-(2-aminoethcxY)]-[2-Nal]-[A113]-[Lys(Ac)]-
NN-NH2;
Ac-[Pen]-NT-[1-Nal]-Q-[Pen]-[Phe[4-(2-aminoethcxY)]-[2-Nal]-[A113]-[Lys(Ac)]-
NN-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[a-MeLeu]-[Lys(Ac)]-NN-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[a-MeLys]-[Lys(Ac)]-NN-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(Ac)]-NN-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[a-MeLeu]-[Lys(Ac)]-N-
[f3Ala]-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[a-MeLys]-[Lys(Ac)]-N-
[f3Ala]-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(Ac)]-N-[f3Ala]-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethcxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-1_,N-NH2;
16
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-GN-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-SN-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-[Aib]-N-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(A0]-FN-NH2;
Ac-[Pen]-NTW-[Cit]-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-NN-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-[Tic]-
[(3Ala]-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-[nLeu]-
[[3Ala]-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-G-[(3Ala]-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-R-[f3Ala]-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]¨[A113]-[Lys(Ac)]-W-[PAla]-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]--[Aib]-[Lys(Ac)]-S-[(3Ala]-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]--[Aib]-[Lys(Ac)]-L-[f3Ala]-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]--[Ab]-[Lys(Ac)]-[MB]-
[(3Ala]-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]--[Aib]-[Lys(Ac)]-[N-MeAla]-
[f3Ala]-
NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-[2-
Nal*H3Alal-NH2;
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxY)]-[2-Nal]-[Aib]-[Lys(Ac)]-F-[f3Ala]-
NH2;
Ac-[(D)Arg]-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[ 4-
amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]NNNH2;
Biotin-[PEG4]-cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-
tetrahydropyran]-ENN-NH2;
17
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)l-NN-NH2;
Ac-[(D)Arg]-cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-
tetrahydropyran]-[Lys(Ac)l-NN-NH2;
Ac-[(D)Arg]-cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-
tetrahydropyran]-[Lys(Ac)l-NN-NH2;
Ac-E-[(D)Arg]-cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-
tetrahydropyranFENN-NH2;
Ac-RD)Asp]-[(D)Arg]-cyclo[[Abu]-QTWQCHPhe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-
4-
carboxy-tetrahydropyran]-ENN-NH2;
Ac-R-[(D)Arg]-cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-
tetrahydropyranFENN-NH2;
inoethoxy)]-[2-Nall-[4-amino-4-carboxy-tetrahydropyran]-ENN-NH2;
Ac-F-[(D)Arg]-cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-
tetrahydropyranFENN-NH2;
Ac-RD)Phe]-[(D)Arg]-cyclo[[Abu]-QTWQCHPhe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-
4-
carboxy-tetrahydropyran]-ENN-NH2;
Ac42-Nall-RD)Arg]-cyclo[[AbuFQTWQCHPhe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-tetrahydropyran]-ENN-NH2;
Ac-T-[(D)Arg]-cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-
tetrahydropyranFENN-NH2;
Ac-L-[(D)Arg]-cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-
tetrahydropyranFENN-NH2;
18
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Ac-[(D)Gln]-[(D)Arg]-cycloMbuFQTWQCHPhe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-tetrahydropyran]-ENN-NH2;
Ac-[(D)Asn]-[(D)Arg]-cyclo[[Abu]-QTWQC] - [Phe[4-(2-aminoethoxy)]-[2-Nall-[4-
amino-4-
carboxy-tetrahydropyran]-ENN-NH2;
Ac-cyclo[[Abu] -QTWQC]- [Phe[4-(2-aminoethoxy)-(PEG4-Alexa488)]-[2-Nall-[4-
amino-4-
carboxy-tetrahydropyran]-ENN-NH2;
[Alexa488]-[PEG4]-cyclo[[Abu] -QTWQC] - [Phe[4-(2-aminoethoxy)]-42-Nall-[4-
amino-4-
carboxy-tetrahydropyran]-ENN-NH2;
[Alexa647]-[PEG4]-cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-
4-
carboxy-tetrahydropyran]-ENN-NH2;
[Alexa-647]-[PEG4]-[(D)Arg]-cyclo[[Abu]-QTWQC] - [Phe[4-(2-aminoethoxy)]-[2-
Nall-[4-
amino-4-carboxy-tetrahydropyran]-[Lys(A0]-NN-NH2;
[Alexa647]- [PEG12]- [(D)Arg]-cyclo[ [Abu]-QTWQC] -[Phe[4-(2-aminoethoxy)]- [2-
Nall - [4-
amino-4-carboxy-tetrahydropyran]- [Lys(Ac)]-NN-NH2; and
[Alexa488]-[PEG4]-[(D)Arg]-cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-
[4-
amino-4-carboxy-tetrahydropyran]- [Lys(Ac)]-NN-NH2,
[0026] wherein the peptide inhibitor is cyclized via a disulfide bond between
two Pen residues or
by a thioether bond between Abu and a Cys residue, and wherein the peptide
inhibitor inhibits
the binding of an interleukin-23 (IL-23) to an IL-23 receptor.
[0027] In particular embodiments, any of the peptide inhibitors described
herein comprise one or
more half-life extension moiety and/or one or more linker moiety conjugated to
the peptide
inhibitor. In particular embodiments, the half-life extension moiety is
conjugated to the peptide
inhibitor via one or more linker moieties.
[0028] In certain embodiments, any of the peptide inhibitors described
hereinfurther comprise a
conjugated chemical substituent. In particular embodiments, the conjugated
chemical substituent
19
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
is a lipophilic substituent or a polymeric moiety, e.g., Ac, Palm, gamaGlu-
Palm, isoGlu-Palm,
PEG2-Ac, PEG4-isoGlu-Palm, (PEG)5-Palm, succinic acid, glutaric acid,
pyroglutaric acid,
benzoic acid, IVA, octanoic acid, 1,4 diaminobutane, isobutyl, or biotin. In
certain embodiments,
the conjugated chemical substituent is a polyethylene glycol with a molecular
mass of 400 Da to
40,000 Da.
[0029] In another aspect, the present invention includes peptide inhibitors
comprising the
structure of Formula I:
R1-X-R2 (I)
or a pharmaceutically acceptable salt or solvate thereof, wherein
Rl is a bond, hydrogen, a C1-C6 alkyl, a C6-C12 aryl, a C6-C12 aryl, a C 1 -C6
alkyl, a C 1 -C20
alkanoyl, and including PEGylated versions alone or as spacers of any of the
foregoing;
R2 is a bond, OH or NH2; and
X is any of the peptide sequences described herein, e.g., Xa, I, Ia-It, II, ha-
lid, III, Illa-Ille, IV,
IVa-IVb, V, or Va-Vh.
[0030] In a related aspect, the present invention includes a peptide dimer
inhibitor of an
interleukin-23 receptor, wherein the peptide dimer inhibitor comprises two
peptide monomer
subunits connected via one or more linker moieties, wherein each peptide
monomer subunit has a
sequence or structure set forth herein. In certain embodiments, one or both
peptide monomer
subunit is cyclized via an intramolecular bond between X4 and X9. In certain
embodiments, one
or both intramolecular bond is a disulfide bond, a thioether bond, a lactam
bond, a selenoether,
diselenide, or an olefin bond. In certain embodiments, the linker is any of
those shown in Table 2
or described herein. In certain embodiments, the linker moiety is a diethylene
glycol linker, an
iminodiacetic acid (IDA) linker, a P-Ala-iminodiaceticacid (f3-Ala-IDA)
linker, or a PEG linker.
In particular embodiments, the N-terminus of each peptide monomer subunit is
connected by the
linker moiety. In particular embodiments, the C-terminus of each peptide
monomer subunit is
connected by the linker moiety. In certain embodiments, the linker connects an
internal amino
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
acid residue of at least one of the peptide monomer subunits to the N-
terminus, C-terminus, or an
internal amino acid residue of the other peptide monomer subunit.
[0031] In a further related aspect, the present invention includes a
polynucleotide comprising a
sequence encoding a peptide inhibitor of the present invention or one or both
peptide monomer
subunit of a peptide dimer inhibitor of the present invention. The present
invention also includes
a vector comprising the polynucleotide.
[0032] In another aspect, the present invention includes a pharmaceutical
composition
comprising a peptide inhibitor or a peptide dimer inhibitor of the present
invention, and a
pharmaceutically acceptable carrier, excipient, or diluent. In particular
embodiments, the
pharmaceutical composition comprises an enteric coating. In certain
embodiments, the enteric
coating protects and releases the pharmaceutical composition within a
subject's lower
gastrointestinal system.
[0033] In another aspect, the present invention includes a method for treating
or preventing a
disease associated with IL-23 signalling, including but not limited to an
Inflammatory Bowel
Disease (IBD), ulcerative colitis, Crohn's disease, Celiac disease
(nontropical Sprue),
enteropathy associated with seronegative arthropathies, microscopic colitis,
collagenous colitis,
eosinophilic gastroenteritis, colitis associated with radio- or chemo-therapy,
colitis associated
with disorders of innate immunity as in leukocyte adhesion deficiency-1,
chronic granulomatous
disease, glycogen storage disease type 1 b, Hermansky-Pudlak syndrome, Chediak-
Higashi
syndrome, and Wiskott-Aldrich Syndrome, pouchitis resulting after
proctocolectomy and
ileoanal anastomosis, gastrointestinal cancer, pancreatitis, insulin-dependent
diabetes mellitus,
mastitis, cholecystitis, cholangitis, pericholangitis, chronic bronchitis,
chronic sinusitis, asthma,
psoriasis, or graft versus host disease in a subject, comprising providing to
the subject an
effective amount of a peptide inhibitor or pharmaceutical composition of the
present invention.
In certain embodiments, the inflammatory bowel disease is ulcerative colitis
or Crohn's disease.
In particular embodimnts, the peptide inhibitor or the peptide dimer inhibitor
inhibits binding of
an interleukin-23 (IL-23) to the interleukin-23 receptor (IL-23R). In certain
embodiments, the
pharmaceutical composition is provided to the subject by an oral, intravenous,
peritoneal,
intradermal, subcutaneous, intramuscular, intrathecal, inhalation,
vaporization, nebulization,
21
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
sublingual, buccal, parenteral, rectal, intraocular, inhalation, vaginal, or
topical route of
administration. In particular embodiments, the pharmaceutical composition is
provided orally
for treating Inflammatory Bowel Disease (IBD), ulcerative colitis, Crohn's
disease. In certain
embodiments, the pharmaceutical composition is provided to the subject
topically, parenterally,
intravenously, subcutaneously, peritonealy, or intravenously for treating
psoriasis.
BRIEF DESCRIPTION OF THE DRAWINGS
[0034] FIG. 1 Figure 1 provides an example of a rat IL-23 dose-response curve
as measured by
levels of IL-17A in the rat splenoctye assay.
[0035] Figure 2 is a graph showing IL-12-dependent production of IFNy from
human PBMCs
treated with the indicated amounts of Compound A or Compound B.
[0036] Figure 3 shows results for DAI values from Day 7. Statistical analysis
for significance
was determined using Student's t-test (GraphPad Prism). Differences were noted
as signficant
*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
[0037] Figure 4 shows an alignment of the amino acid sequences of human IL23R,
mouse IL-
23R, rat IL23R, chimp IL-23R, dog IL-23R and cow IL-23R, with highly conserved
amino acid
residues shaded. The region of mouse IL-23R lacking in the other IL-23R
species shown is
shown, and a region of IL23R that may be bound by certain peptide inhibitors
of the present
invention is indicated by a dashed line.
[0038] Figure 5 is a table outlining the study design for TNBS induced colitis
in rats.
[0039] Figures 6A-6D are graphs showing colon weight to length (Figure 6A),
colon wall
thickness (Table 6B, colon macroscopic score (Table 6C) or myeloperoxidase
(MPO) abundance
in proximal colon extracts quantified by ELISA, following sham treatment,
vehicle treatment, or
treatment with the indicated amounts of anti-IL23p19 antibody or Compound C.
Values are
shown as mean +SD. Statistical significance assessed by one-way ANOVA: *<0.05;
**<0.01;
***p<0.001; ****p<0.0001; ns, not significant.
22
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[0040] Figure 7 provides micrographs of colon lesions found in animals
following sham
treatment (upper left panel), vehicle treatment (upper right panel) showing
transmural
inflammation, presence of necrotic tissue, and mucosa devoid of crypts, anti-
IL23p19 antibody
(lower left panel), or 160 mg/kg/d Compound C (lower right panel) showing
restriction of
lesions to the mucosa.
[0041] Figures 8A-8E are graphs showing inflammation (Figure 8A), mucosal
necrosis (Figure
8B), grand loss (Figure 8C), colon wall thickness (Figure 8D) and histological
score (Figure 8E)
following vehicle treatment, treatment with anti-IL23p19 antibody, or
treatment with the
indicated amount of Compound C
[0042] Figure 9 shows the concentration of Compound C in the plasma and
proximal colon
determined one hour post last PO dose (left panel), and fold above IC75 of its
activity as
determined by the rat splenocyte assay (middle panel) and the rat IL23R ELISA
assay (right
panel).
[0043] Figure 10 provides a schematic diagram depicting the structure of
certain peptide
inhibitors and illustrating representative types of bonds between X4 and X9.
[0044] FIGS. 11A-11E show pharmacokinetic data for the IL-23R peptide
inhibitor Peptide 993
(SEQ ID NO: 993). FIG. 11A shows the concentration of Peptide 993 in serum
(nIVI) measured
at different time points up to 24 hours after oral administration of Peptide
993. FIGS. 11B-11D
show the concentration of Peptide 993 (in nIVI) in samples taken from the
Peyer's Patch (FIG.
11B), small intestine (FIG. 11C), and the colon (FIG. 11D). The dashed line
indicates 350 mIVI.
Fig 11E shows the amount of Peptide 993 detected in feces 24 hours after oral
administration (%
dose).
[0045] FIGS. 12A-12D summarize experiments comparing systemic treatments with
prodnisolone or anti-IL-23p19 neutralizing antibody with treatment with
Peptide 993 by oral
administration in the TNBS model of acute colitis. FIG. 12A shows the change
in body weight
(percentage) from day 0 to day 7 from sham, vehicle, and Peptide 993 treated
rats. FIG. 12B
shows the ratio of colon weight to colon length in mg/cm of colons harvested
from rats at day 7.
FIG. 12C shows the colon macroscopic score of colons harvested from rats at
day 7. FIG. 12D.
23
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
shows the sum of histopathology scores for colons taken from sham, vehicle,
and Peptide 993
treated rats. For all experiments, statistical comparisons between groups were
performed with a
1-Way ANOVA followed by a post hoc test: * p< 0.05; ** p< 0.01; *** p< 0.001;
**** p<
0.0001; ns, not significant.
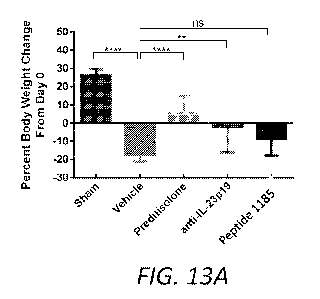
[0046] FIGS. 13A-13C summarizes experiments comparing systemic treatments with
prodnisolone or anti-IL-23p19 neutralizing antibody with treatment with
Peptide 1185 by oral
administration in the TNBS model of acute colitis. FIG. 13A shows the change
in body weight
(percentage) from day 0 to day 7 from sham, vehicle, and Peptide 1185 treated
rats. FIG. 13B
shows the ratio of colon weight to colon length in mg/cm of colons harvested
from rats at day 7.
FIG. 13C shows the colon macroscopic score of colons harvested from rats at
day 7. For all
experiments, statistical comparisons between groups were performed with a 1-
Way ANOVA
followed by a post hoc test: * p< 0.05; ** p< 0.01; *** p< 0.001; **** p<
0.0001; ns, not
significant.
[0047] FIGS. 14A-14D summarizes experiments comparing systemic treatments with
prodnisolone or anti-IL-23p19 neutralizing antibody with treatment with
Peptide 980 by oral
administration in the TNBS model of acute colitis. FIG. 14A shows the change
in body weight
(percentage) from day 0 to day 7 from sham, vehicle, and Peptide 980 treated
rats. FIG. 14B
shows the ratio of colon weight to colon length in mg/cm of colons harvested
from rats at day 7.
FIG. 13C shows the colon macroscopic score of colons harvested from rats at
day 7. FIG. 14D.
shows the sum of histopathology scores for colons taken from sham, vehicle,
and Peptide 980
treated rats. For all experiments, statistical comparisons between groups were
performed with a
1-Way ANOVA followed by a post hoc test: * p< 0.05; ** p< 0.01; *** p< 0.001;
**** p<
0.0001; ns, not significant.
[0048] FIGS. 15A-15E show levels of disease and IL-23 directed biomarkers
measured in
colons from rats in the sham (not TNBS-exposed) experimental group, or TNBS-
exposed
experimental groups that received treatment with vehicle or Peptide 993. Data
is shown for
MPO (FIG. 15A), IL-6 (FIG. 15B), IL-1 beta (FIG. 15C), IL-22 (FIG. 15D), and
IL-17A
(FIG.15E). For all experiments, statistical comparisons between groups were
performed with a
24
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1-Way ANOVA followed by a post hoc test: * p< 0.05; ** p< 0.01; *** p< 0.001;
**** p<
0.0001; ns, not significant.
[0049] FIGS. 16A-16B show levels of disease and IL-23 directed biomarkers
measured in
colons from rats in the sham (not TNBS-exposed) experimental group, or TNBS-
exposed
experimental groups that received treatment with vehicle or Peptide 980. Data
is shown for
MPO (FIG. 16A) and IL-22 (FIG. 16B). For all experiments, statistical
comparisons between
groups were performed with a 1-Way ANOVA followed by a post hoc test: * p<
0.05; ** p<
0.01; *** p< 0.001; **** p< 0.0001; ns, not significant
[0050] FIGS. 17A-17D show Schild analysis of inhibitor peptides. FIG. 17A
shows a graph
depicting the % Emax response as a function of increasing concentrations of IL-
23 in the
presence of Peptide 993 in concentrations of 0 nM (closed circles), 0.3 nM
(closed squares), 1
nM (triangles), 3 nM (inverted triangles), 10 nM (diamonds), 30 nM (open
circles), or 100 nM
(open squares). Properties of the curves are listed below the graph. FIG. 17B
depicts results
from the same set of experiments, and shows a graph displaying Log(dose
ratio') as a function
of Peptide 993 concentration (M) on a logarithmic scale. Properties of the
resulting linear
function are displayed below the graph. FIG. 17C shows a graph depicting the %
Emax response
as a function of increasing concentrations of IL-23 in the presence of the
peptide of SEQ ID NO:
1169 in concentrations of 0 nM (closed circles), 0.3 nM (closed squares), 1 nM
(triangles), 3 nM
(inverted triangles), 10 nM (diamonds), 30 nM (open circles), or 100 nM (open
squares).
Properties of the curves are listed below the graph. FIG. 17D shows a graph
depicting the %
Emax response as a function of increasing concentrations of IL-23 in the
presence of the peptide
of SEQ ID NO: 1211 in concentrations of 0 nM (closed circles), 0.3 nM (closed
squares), 1 nM
(triangles), 3 nM (inverted triangles), 10 nM (diamonds), 30 nM (open
circles), or 100 nM (open
squares). Properties of the curves are listed below the graph.
[0051] FIGS. 18A-18B show pharmacokinetic data for the IL-23R peptide
inhibitor Peptide
1185. FIG. 18A shows the concentration of Peptide 1185 in serum and in samples
taken from
small intestine and the colon. Fig 18B shows the amount of Peptide 1185
detected in urine and
feces 24 hours after oral administration (% dose).
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[0052] FIGS. 19A and 19B show pharmacokinetic data for the IL-23R peptide
inhibitor Peptide
980. FIG. 19A shows the concentration of Peptide 980 in serum and in samples
taken from
small intestine and the colon. Fig 19B shows the amount of Peptide 980
detected in urine and
feces 24 hours after oral administration (% dose).
DETAILED DESCRIPTION OF THE INVENTION
[0053] Unless otherwise defined herein, scientific and technical terms used in
this application
shall have the meanings that are commonly understood by those of ordinary
skill in the art.
Generally, nomenclature used in connection with, and techniques of, chemistry,
molecular
biology, cell and cancer biology, immunology, microbiology, pharmacology, and
protein and
nucleic acid chemistry, described herein, are those well-known and commonly
used in the art.
[0054] As used herein, the following terms have the meanings ascribed to them
unless specified
otherwise.
[0055] Throughout this specification, the word "comprise" or variations such
as "comprises"
or "comprising" will be understood to imply the inclusion of a stated integer
(or components)
or group of integers (or components), but not the exclusion of any other
integer (or
components) or group of integers (or components).
[0056] The singular forms "a," "an," and "the" include the plurals unless the
context clearly
dictates otherwise.
[0057] The term "including" is used to mean "including but not limited to."
"Including" and
"including but not limited to" are used interchangeably.
[0058] The terms "patient," "subject," and "individual" may be used
interchangeably and refer
to either a human or a non-human animal. These terms include mammals such as
humans,
primates, livestock animals (e.g., bovines, porcines), companion animals
(e.g., canines, felines)
and rodents (e.g., mice and rats).
[0059] The term "peptide," as used herein, refers broadly to a sequence of two
or more amino
acids joined together by peptide bonds. It should be understood that this term
does not connote a
26
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
specific length of a polymer of amino acids, nor is it intended to imply or
distinguish whether the
polypeptide is produced using recombinant techniques, chemical or enzymatic
synthesis, or is
naturally occurring.
[0060] The recitations "sequence identity", "percent identity", "percent
homology", or, for
example, comprising a "sequence 50% identical to," as used herein, refer to
the extent that
sequences are identical on a nucleotide-by-nucleotide basis or an amino acid-
by-amino acid basis
over a window of comparison. Thus, a "percentage of sequence identity" may be
calculated by
comparing two optimally aligned sequences over the window of comparison,
determining the
number of positions at which the identical nucleic acid base (e.g., A, T, C,
G, I) or the identical
amino acid residue (e.g., Ala, Pro, Ser, Thr, Gly, Val, Leu, Ile, Phe, Tyr,
Trp, Lys, Arg, His, Asp,
Glu, Asn, Gln, Cys and Met) occurs in both sequences to yield the number of
matched positions,
dividing the number of matched positions by the total number of positions in
the window of
comparison (i.e., the window size), and multiplying the result by 100 to yield
the percentage of
sequence identity.
[0061] Calculations of sequence similarity or sequence identity between
sequences (the terms
are used interchangeably herein) can be performed as follows. To determine the
percent identity
of two amino acid sequences, or of two nucleic acid sequences, the sequences
can be aligned for
optimal comparison purposes (e.g., gaps can be introduced in one or both of a
first and a second
amino acid or nucleic acid sequence for optimal alignment and non-homologous
sequences can
be disregarded for comparison purposes). In certain embodiments, the length of
a reference
sequence aligned for comparison purposes is at least 30%, preferably at least
40%, more
preferably at least 50%, 60%, and even more preferably at least 70%, 80%, 90%,
100% of the
length of the reference sequence. The amino acid residues or nucleotides at
corresponding amino
acid positions or nucleotide positions are then compared. When a position in
the first sequence is
occupied by the same amino acid residue or nucleotide as the corresponding
position in the
second sequence, then the molecules are identical at that position.
[0062] The percent identity between the two sequences is a function of the
number of identical
positions shared by the sequences, taking into account the number of gaps, and
the length of each
gap, which need to be introduced for optimal alignment of the two sequences.
27
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[0063] The comparison of sequences and determination of percent identity
between two
sequences can be accomplished using a mathematical algorithm. In some
embodiments, the
percent identity between two amino acid sequences is determined using the
Needleman and
Wunsch, (1970, J. Mol. Biol. 48: 444-453) algorithm which has been
incorporated into the GAP
program in the GCG software package, using either a Blossum 62 matrix or a
PAM250 matrix,
and a gap weight of 16, 14, 12, 10, 8, 6, or 4 and a length weight of 1, 2, 3,
4, 5, or 6. In yet
another preferred embodiment, the percent identity between two nucleotide
sequences is
determined using the GAP program in the GCG software package, using an
NVVSgapdna.CMP
matrix and a gap weight of 40, 50, 60, 70, or 80 and a length weight of 1, 2,
3, 4, 5, or 6. Another
exemplary set of parameters includes a Blossum 62 scoring matrix with a gap
penalty of 12, a
gap extend penalty of 4, and a frameshift gap penalty of 5. The percent
identity between two
amino acid or nucleotide sequences can also be determined using the algorithm
of E. Meyers and
W. Miller (1989, Cabios, 4: 11-17) which has been incorporated into the ALIGN
program
(version 2.0), using a PAM120 weight residue table, a gap length penalty of 12
and a gap penalty
of 4.
[0064] The peptide sequences described herein can be used as a "query
sequence" to perform a
search against public databases to, for example, identify other family members
or related
sequences. Such searches can be performed using the NBLAST and )(BLAST
programs (version
2.0) of Altschul, et al., (1990, J. Mol. Biol, 215: 403-10). BLAST nucleotide
searches can be
performed with the NBLAST program, score = 100, wordlength = 12 to obtain
nucleotide
sequences homologous to nucleic acid molecules of the invention. BLAST protein
searches can
be performed with the )(BLAST program, score = 50, wordlength = 3 to obtain
amino acid
sequences homologous to protein molecules of the invention. To obtain gapped
alignments for
comparison purposes, Gapped BLAST can be utilized as described in Altschul et
al. (Nucleic
Acids Res. 25:3389-3402, 1997). When utilizing BLAST and Gapped BLAST
programs, the
default parameters of the respective programs (e.g., )(BLAST and NBLAST) can
be used.
[0065] The term "conservative substitution" as used herein denotes that one or
more amino acids
are replaced by another, biologically similar residue. Examples include
substitution of amino
acid residues with similar characteristics, e.g., small amino acids, acidic
amino acids, polar
amino acids, basic amino acids, hydrophobic amino acids and aromatic amino
acids. See, for
28
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
example, the table below. In some embodiments of the invention, one or more
Met residues are
substituted with norleucine (Nle) which is a bioisostere for Met, but which,
as opposed to Met, is
not readily oxidized. Another example of a conservative substitution with a
residue normally not
found in endogenous, mammalian peptides and proteins is the conservative
substitution of Arg or
Lys with, for example, ornithine, canavanine, aminoethylcysteine or another
basic amino acid.
In some embodiments, one or more cysteines of a peptide analogue of the
invention may be
substituted with another residue, such as a serine. For further information
concerning
phenotypically silent substitutions in peptides and proteins, see, for
example, Bowie et.al.
Science 247, 1306-1310, 1990. In the scheme below, conservative substitutions
of amino acids
are grouped by physicochemical properties. I: neutral, hydrophilic, II: acids
and amides, III:
basic, IV: hydrophobic, V: aromatic, bulky amino acids.
I II III IV V
AN H M F
S DR L
TEK I
P Q V
[0066] In the scheme below, conservative substitutions of amino acids are
grouped by
physicochemical properties. VI: neutral or hydrophobic, VII: acidic, VIII:
basic, IX: polar, X:
aromatic.
VI VII VIII IX X
A E H M F
L D R S Y
T W
V
29
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[0067] The term "amino acid" or "any amino acid" as used here refers to any
and all amino
acids, including naturally occurring amino acids (e.g., a-amino acids),
unnatural amino acids,
modified amino acids, and non-natural amino acids. It includes both D- and L-
amino acids.
Natural amino acids include those found in nature, such as, e.g., the 23 amino
acids that combine
into peptide chains to form the building-blocks of a vast array of proteins.
These are primarily L
stereoisomers, although a few D-amino acids occur in bacterial envelopes and
some antibiotics.
The 20 "standard," natural amino acids are listed in the above tables. The
"non-standard,"
natural amino acids are pyrrolysine (found in methanogenic organisms and other
eukaryotes),
selenocysteine (present in many noneukaryotes as well as most eukaryotes), and
N-
formylmethionine (encoded by the start codon AUG in bacteria, mitochondria and
chloroplasts).
"Unnatural" or "non-natural" amino acids are non-proteinogenic amino acids
(i.e., those not
naturally encoded or found in the genetic code) that either occur naturally or
are chemically
synthesized. Over 140 unnatural amino acids are known and thousands of more
combinations are
possible. Examples of "unnatural" amino acids include f3-amino acids (f33 and
f32), homo-amino
acids, proline and pyruvic acid derivatives, 3-substituted alanine
derivatives, glycine derivatives,
ring-substituted phenylalanine and tyrosine derivatives, linear core amino
acids, diamino acids,
D-amino acids, alpha-methyl amino acids and N-methyl amino acids. Unnatural or
non-natural
amino acids also include modified amino acids. "Modified" amino acids include
amino acids
(e.g., natural amino acids) that have been chemically modified to include a
group, groups, or
chemical moiety not naturally present on the amino acid. According to certain
embodiments, a
peptide inhibitor comprises an intramolecular bond between two amino acid
residues present in
the peptide inhibitor. It is understood that the amino acid residues that form
the bond will be
altered somewhat when bonded to each other as compared to when not bonded to
each other.
Reference to a particular amino acid is meant to encompass that amino acid in
both its unbonded
and bonded state. For example, the amino acid residue homoSerine (hSer) or
homoSerine(C1) in
its unbonded form may take the form of 2-aminobutyric acid (Abu) when
participating in an
intramolecular bond according to the present invention. The present invention
inclues both
peptide inhibitors containing cross-links between X4 and X9, as well as the
peptide inhibitors
that do not contain cross-links between X4 and X9, e.g., before cross-link
formation. As such,
the names hSer and Abu are intended to indicate the same amino acids and are
used
interchangeably.
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[0068] For the most part, the names of naturally occurring and non-naturally
occurring
aminoacyl residues used herein follow the naming conventions suggested by the
IUPAC
Commission on the Nomenclature of Organic Chemistry and the IUPAC-IUB
Commission on
Biochemical Nomenclature as set out in "Nomenclature of a-Amino Acids
(Recommendations,
1974)" Biochemistry, 14(2), (1975). To the extent that the names and
abbreviations of amino
acids and aminoacyl residues employed in this specification and appended
claims differ from
those suggestions, they will be made clear to the reader. Some abbreviations
useful in describing
the invention are defined below in the following Table 1A.
[0069] Table 1A. Abbreviations of Non-Natural Amino Acids and Chemical
Moieties (for
amino acid derivatives, all L unless stated)
Abbreviation Definition
Ac- Acetyl
Hy Hydrogen (Free N-terminal)
Dap L-Diaminopropionic acid
Dab L-Diaminobutyric acid
Orn L-Ornathine
Pen L-Penicillamine
Sarc Sarcosine
Cit L-Citrulline
Cav L-Cavanine
Phe-(4-Guanidino) 4-Guanidine-L-Phenylalanine
N-MeArg N-Methyl-L-Arginine
N-MeTrp N-Methyl-L-Tryptophan
N-MeGln N-Methyl-L-Glutamine
31
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
N-MeAla N-Methyl-L-Alanine
N-MeLys N-Methyl-Lysine
N-MeAsn N-Methyl-L-Asparagine
6-ChloroTrp 6-Chloro-L-Tryptophan
5-HydroxyTrp 5-Hydroxy-L-Tryptophan
1,2,3,4-tetrahydro-norharman L-1,2,3,4-tetrahydro-norharman
2-Na!
L-2-Napthylalanine
(also referred to as 2-Nap)
1-Na!
L-1-Napthylalanine
(also referred to as 1-Nap)
Phe(4-0Me) 4-Methoxy-L-phenylalanine
Abu 2-Aminobutyric acid
B ip L-4,4' -B ipheny la lan in e
f3Ala beta-Alanine
PhTyr beta homo-L-Tyrosine
PhTrp beta homo-L-Trptophan
f3hAla beta homo-L-Alanine
phLeu, beta homo-L-Leucine
PhVal beta homo-L-Valine
Aib 2-aminoisobutyric acid
Azt L-azetidine-2-carboxylic acid
(3S)-1,2,3,4-Tetrahydroisoquinoline-7-hydroxy-3-carboxylic
Tic
Acid
Phe(4-0Me) 4-methoxy-L-phenylalanine
32
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
N-Me-Lys N-Methyl-L-Lysine
N-Me-Lys(Ac) N-E-Acetyl-D-lysine
CONH2 Carboxamide
COOH Acid
3-Pal L-3-Pyridylalanine
Phe(4-F) 4-Fluoro-L-Phenylalanine
DMT 2,6-DimethylTyrosine
Phe(4-0Me) 4-Methoxyphenylalanine
hLeu L-homoLeucine
hArg L-homoArginine
a-MeLys alpha-methyl-L-Lysine
a-MeOrn alpha-methyl-L-Ornathine
a-MeLeu alpha-methyl-L-Leucine
a-MeTrp alpha-methyl-L-Tryptophan
a-MePhe alpha-methyl-L-Phenylalanine
a-MeTyr alpha-methyl-L-Tyrosine
a¨DiethylGly a-DiethylGlycine
Lys(Ac) N-c-acetyl-L-Lysine
DTT Dithiothreotol
Nle L-Norleucine
fihTrp L-0-homoTrypophan
f3hPhe L-P-homophenylalanine
f3hPro L-P-homoproline
33
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Phe(4-CF3) 4-Trifluoromethyl-L-Phenylalanine
P-Glu L-P-Glutamic acid
f3hG1u L-0-homog1utamic acid
2-2-Indane 2-Aminoindane-2-carboxylic acid
1-1-Indane 1-Aminoindane-1-carboxylic acid
hCha L-homocyclohexylalanine
Cyclobutyl L-cyclobutylalanine
f3hPhe L-P-homo-phenylalanine
Gla Gama-Carboxy-L-Glutamic acid
Cpa Cyclopentyl-L-alanine
Cha Cyclohexyl-L-alanine
Octgly L-Octylglycine
t-butyl-Ala 3-(tert-buty1)-L-Ala-OH
t-butyl-Gly tert-butyl-glycine
AEP 3-(2-aminoethoxy)propanoic acid
AEA (2-aminoethoxy)acetic acid
Phe(4-Phenoxy)] 4-Phenoxy-L-phenylalanine
Phe(4-0Bz1) O-Benzyl-L-tyrosine
Phe(4-CONH2) 4-Carbamoyl-L-phenylalanine
Phe(4-CO2H) 4-Carboxy-L-phenylalanine
Phe(3,4-C12) 3,4 dichloro-L-phenylalanine
Tyr(3-t-Bu) 3-t-butyl-L-tyrosine
34
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Phe(t-Bu) t-butyl-L-phenylalanine
N H2
0
1401
Phe[4-(2-aminoethoxy)]
co2H
NH2
4-(2-aminoethoxy)-L-phenylalanine
Phe(4-CN) 4-cyano-L-phenylalanine
Phe(4-Br) 4-bromo-L-phenylalanine
Phe(4-NH2) 4-amino-L-phenylalanine
Phe(4-Me) 4-methyl-L-phenylalanine
4-Pyridylalanine 4-L-Pyridylalanine
H
N
4-amino-4-carboxy-piperidine
X
H2N co2H
4-amino-4-carboxy-piperidine
hPhe(3,4-dimethoxy) 3,4-dimethoxy-L-homophenylalanine
Phe(2,4-Me2) 2,4-dimethyl-L-phenylalanine
Phe(3,5-F2) 3,5-difluoro-L-phenylalanine
Phe(penta-F) pentafluoro-L-phenylalanine
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
2,5,7-tert butyl Trp 2,5,7-Tris-tert-butyl-L-tryptophan
CO2H
Tic
NH
L-1,2,3,4,-tetrahydro-isoquinoline-3-carboxylic acid
Phe(4-0Ally1) 0-Allyl-L-Tyrosine
Phe(4-N3) 4-azidophenylalanine
Achc
q
H2N co2H
1-aminocyclohexanecarboxylic acid
Acvc
R
H2N co2H
1-aminocyclopentanecarboxylic acid
Acbc
H2N co2H
1-aminocyclobutanecarboxylic acid
Acpc
7
H2N co2H
36
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1-aminocyclopropylcarboxylic acid
4-amino-4-carboxy-
tetrahydropyran
(also referred as THP) H2N co2H
4-amino-4-carboxy-tetrahydropyran
[0070] Throughout the present specification, unless naturally occurring amino
acids are referred
to by their full name (e.g. alanine, arginine, etc.), they are designated by
their conventional three-
letter or single-letter abbreviations (e.g. Ala or A for alanine, Arg or R for
arginine, etc.).
Unless otherwise indicated, three-letter and single-letter abbreviations of
amino acids refer to the
L-isomeric form of the amino acid in question. The term "L-amino acid," as
used herein, refers
to the "L" isomeric form of a peptide, and conversely the term "D-amino acid"
refers to the "D"
isomeric form of a peptide (e.g., Dasp, (D)Asp or D-Asp; Dphe, (D)Phe or D-
Phe). Amino acid
residues in the D isomeric form can be substituted for any L-amino acid
residue, as long as the
desired function is retained by the peptide. D-amino acids may be indicated as
customary in
lower case when referred to using single-letter abbreviations.
[0071] In the case of less common or non-naturally occurring amino acids,
unless they are
referred to by their full name (e.g. sarcosine, ornithine, etc.), frequently
employed three- or four-
character codes are employed for residues thereof, including, Sar or Sarc
(sarcosine, i.e. N-
methylglycine), Aib (a-aminoisobutyric acid), Dab (2,4-diaminobutanoic acid),
Dapa (2,3-
diaminopropanoic acid), y-Glu (y-glutamic acid), Gaba (y-aminobutanoic acid),
3-Pro
(pyrrolidine-3-carboxylic acid), and 8Ado (8-amino-3,6-dioxaoctanoic acid),
Abu (2-amino
butyric acid), f3hPro (f3-homoproline), PhPhe (P-homophenylalanine) and Bip
(3,r3
diphenylalanine), and Ida (Iminodiacetic acid).
[0072] As is clear to the skilled artisan, the peptide sequences disclosed
herein are shown
proceeding from left to right, with the left end of the sequence being the N-
terminus of the
37
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
peptide and the right end of the sequence being the C-terminus of the peptide.
Among sequences
disclosed herein are sequences incorporating a "Hy-" moiety at the amino
terminus (N-terminus)
of the sequence, and either an "-OH" moiety or an "-NH2" moiety at the carboxy
terminus (C-
terminus) of the sequence. In such cases, and unless otherwise indicated, a
"Hy-" moiety at the
N-terminus of the sequence in question indicates a hydrogen atom,
corresponding to the presence
of a free primary or secondary amino group at the N-terminus, while an "-OH"
or an "¨NH2"
moiety at the C-terminus of the sequence indicates a hydroxy group or an amino
group,
corresponding to the presence of an amido (CONH2) group at the C-terminus,
respectively. In
each sequence of the invention, a C-terminal "¨OH" moiety may be substituted
for a C-terminal
"¨NH2" moiety, and vice-versa.
[0073] The term "DRP," as used herein, refers to disulfide rich peptides.
[0074] The term "dimer," as used herein, refers broadly to a peptide
comprising two or more
monomer subunits. Certain dimers comprise two DRPs. Dimers of the present
invention include
homodimers and heterodimers. A monomer subunit of a dimer may be linked at its
C- or N-
terminus, or it may be linked via internal amino acid residues. Each monomer
subunit of a dimer
may be linked through the same site, or each may be linked through a different
site (e.g., C-
terminus, N-terminus, or internal site).
[0075] The term "NH2," as used herein, can refer to a free amino group present
at the amino
terminus of a polypeptide. The term "OH," as used herein, can refer to a free
carboxy group
present at the carboxy terminus of a peptide. Further, the term "Ac," as used
herein, refers to
Acetyl protection through acylation of the C- or N-terminus of a polypeptide.
In certain peptides
shown herein, the NH2 locates at the C-terminus of the peptide indicates an
amino group.
[0076] The term "carboxy," as used herein, refers to ¨CO2H.
[0077] The term "isostere replacement," as used herein, refers to any amino
acid or other analog
moiety having chemical and/or structural properties similar to a specified
amino acid.
[0078] The term "cyclized," as used herein, refers to a reaction in which one
part of a
polypeptide molecule becomes linked to another part of the polypeptide
molecule to form a
closed ring, such as by forming a disulfide bridge or other similar bond.
38
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[0079] The term "subunit," as used herein, refers to one of a pair of
polypeptide monomers that
are joined to form a dimer peptide composition.
[0080] The term "linker moiety," as used herein, refers broadly to a chemical
structure that is
capable of linking or joining together two peptide monomer subunits to form a
dimer.
[0081] The term "pharmaceutically acceptable salt," as used herein, represents
salts or
zwitterionic forms of the peptides or compounds of the present invention which
are water or oil-
soluble or dispersible, which are suitable for treatment of diseases without
undue toxicity,
irritation, and allergic response; which are commensurate with a reasonable
benefit/risk ratio,
and which are effective for their intended use. The salts can be prepared
during the final isolation
and purification of the compounds or separately by reacting an amino group
with a suitable acid.
Representative acid addition salts include acetate, adipate, alginate,
citrate, aspartate, benzoate,
benzenesulfonate, bisulfate, butyrate, camphorate, camphorsulfonate,
digluconate,
glycerophosphate, hemisulfate, heptanoate, hexanoate, formate, fumarate,
hydrochloride,
hydrobromide, hydroiodide, 2 -hydroxyethansulfonate (isethionate), lactate,
maleate,
mesitylenesulfonate, methanesulfonate,
naphthylenesulfonate, nicotinate, 2-
naphthalenesulfonate, oxalate, pamoate, pectinate, persulfate, 3 -
phenylproprionate, picrate,
pivalate, propionate, succinate, tartrate, trichloroacetate, trifluoroacetate,
phosphate, glutamate,
bicarbonate, para-toluenesulfonate, and undecanoate. Also, amino groups in the
compounds of
the present invention can be quaternized with methyl, ethyl, propyl, and butyl
chlorides,
bromides, and iodides; dimethyl, diethyl, dibutyl, and diamyl sulfates; decyl,
lauryl, myristyl,
and steryl chlorides, bromides, and iodides; and benzyl and phenethyl
bromides. Examples of
acids which can be employed to form therapeutically acceptable addition salts
include inorganic
acids such as hydrochloric, hydrobromic, sulfuric, and phosphoric, and organic
acids such as
oxalic, maleic, succinic, and citric. A pharmaceutically acceptable salt may
suitably be a salt
chosen, e.g., among acid addition salts and basic salts. Examples of acid
addition salts include
chloride salts, citrate salts and acetate salts. Examples of basic salts
include salts where the
cation is selected among alkali metal cations, such as sodium or potassium
ions, alkaline earth
metal cations, such as calcium or magnesium ions, as well as substituted
ammonium ions, such
as ions of the type N(R1)(R2)(R3)(R4)+, where R1, R2, R3 and R4 independently
will typically
designate hydrogen, optionally substituted C1-6-alkyl or optionally
substituted C2-6-alkenyl.
39
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Examples of relevant C1-6-alkyl groups include methyl, ethyl, 1-propyl and 2-
propyl groups.
Examples of C2-6-alkenyl groups of possible relevance include ethenyl, 1-
propenyl and 2-
propenyl. Other examples of pharmaceutically acceptable salts are described in
"Remington's
Pharmaceutical Sciences", 17th edition, Alfonso R. Gennaro (Ed.), Mark
Publishing Company,
Easton, PA, USA, 1985 (and more recent editions thereof), in the
"Encyclopaedia of
Pharmaceutical Technology", 3rd edition, James Swarbrick (Ed.), Informa
Healthcare USA
(Inc.), NY, USA, 2007, and in J. Pharm. Sci. 66: 2 (1977). Also, for a review
on suitable salts,
see Handbook of Pharmaceutical Salts: Properties, Selection, and Use by Stahl
and Wermuth
(Wiley-VCH, 2002). Other suitable base salts are formed from bases which form
non-toxic salts.
Representative examples include the aluminum, arginine, benzathine, calcium,
choline,
diethylamine, diolamine, glycine, lysine, magnesium, meglumine, olamine,
potassium, sodium,
tromethamine, and zinc salts. Hemisalts of acids and bases may also be formed,
e.g.,
hemisulphate and hemicalcium salts.
[0082] The term "N(alpha)Methylation", as used herein, describes the
methylation of the alpha
amine of an amino acid, also generally termed as an N-methylation.
[0083] The term "sym methylation" or "Arg-Me-sym", as used herein, describes
the
symmetrical methylation of the two nitrogens of the guanidine group of
arginine. Further, the
term "asym methylation" or "Arg-Me-asym" describes the methylation of a single
nitrogen of
the guanidine group of arginine.
[0084] The term "acylating organic compounds", as used herein refers to
various compounds
with carboxylic acid functionality that are used to acylate the N-terminus of
an amino acid or a
monomer or dimer, e.g., a monomer subunit prior to forming a C-terminal dimer.
Non-limiting
examples of acylating organic compounds include cyclopropylacetic acid, 4-
Fluorobenzoic acid,
4-fluorophenylacetic acid, 3-Phenylpropionic acid, Succinic acid, Glutaric
acid, Cyclopentane
carboxylic acid, 3,3,3-trifluoropropeonic acid, 3-Fluoromethylbutyric acid,
Tetrahedro-2H-
Pyran-4-carboxylic acid.
[0085] The term "alkyl" includes a straight chain or branched, noncyclic or
cyclic, saturated
aliphatic hydrocarbon containing from 1 to 24 carbon atoms. Representative
saturated straight
chain alkyls include, but are not limited to, methyl, ethyl, n-propyl, n-
butyl, n-pentyl, n-hexyl,
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
and the like, while saturated branched alkyls include, without limitation,
isopropyl, sec-butyl,
isobutyl, tert-butyl, isopentyl, and the like. Representative saturated cyclic
alkyls include, but
are not limited to, cyclopropyl, cyclobutyl, cyclopentyl, cyclohexyl, and the
like, while
unsaturated cyclic alkyls include, without limitation, cyclopentenyl,
cyclohexenyl, and the like.
[0086] The term "mammal" refers to any mammalian species such as a human,
mouse, rat, dog,
cat, hamster, guinea pig, rabbit, livestock, and the like.
[0087] As used herein, a "therapeutically effective amount" of the peptide
inhibitor of the
invention is meant to describe a sufficient amount of the peptide inhibitor to
treat an IL-23/IL-
23R-related disease, including but not limited to any of the diseases and
disorders described
herein (for example, to reduce inflammation associated with IBD). In
particular embodiments,
the therapeutically effective amount will achieve a desired benefit/risk ratio
applicable to any
medical treatment.
[0088] An "analog" of an amino acid, e.g., a "Phe analog" or a "Tyr analog"
means an analog of
the referenced amino acid. A variety of amino acid analogs are known and
available in the art,
including Phe and Tyr analogs. In certain embodiments, an amino acid analog,
e.g., a Phe analog
or a Tyr analog comprises one, two, three, four or five substitutions as
compared to Phe or Tyr,
respectively. In certain embodiments, the substitutions are present in the
side chains of the
amino acids. In certain embodiments, a Phe analog has the structure Phe(R2),
wherein R2 is a
Hy, OH, CH3, CO2H, CONH2, CONH2OCH2CH2NH2, t-Bu, OCH2CH2NH2, phenoxy, OCH3,
0Allyl, Br, Cl, F, NH2, N3, or guanadino. In certain embodiments, R2 is
CONH2OCH2CH2NH2,
OCH3, CONH2, OCH3 or CO2H. Examples of Phe analogs include, but are not
limited to: hPhe,
Phe(4-0Me), a-Me-Phe, hPhe(3,4-dimethoxy), Phe(4-CONH2), Phe(4-phenoxy), Phe(4-
guanadino), Phe(4-tBu), Phe(4-CN), Phe(4-Br), Phe(4-0Bz1), Phe(4-NH2), BhPhe(4-
F), Phe(4-
F), Phe(3,5 DiF), Phe(CH2CO2H), Phe(penta-F), Phe(3,4-C12), Phe (3,4-F2),
Phe(4-CF3), RR-
diPheAla, Phe(4-N3), Phe[4-(2-aminoethoxy)], 4-Phenylbenzylalanine, Phe(4-
CONH2), Phe(3,4-
Dimethoxy), Phe(4-CF3), Phe(2,3-C12), and Phe(2,3-F2). Examples of Tyr analogs
include, but
are not limited to: hTyr, N-Me-Tyr, Tyr(3-tBu), Tyr(4-N3) and f3hTyr.
41
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Peptide Inhibitors of IL-23R
[0089] Genome-wide association studies (GWAS) have demonstrated significant
association of
the IL-23 receptor (IL-23R) gene with inflammatory bowel disease (IBD),
suggesting that
perturbation of IL-23 signaling could be relevant to the pathogenesis of the
disease. The present
invention provides compositions and methods to modulate the IL-23 pathway
through selective
antagonism of IL-23R by oral treatment with peptides that are stable and
restricted to the
gastrointestinal (GI) tissue. Novel inhibitory peptides that are uniquely
resistant to
oxidative/reductive conditions and proteolytic degradation in a variety of
assays that mimic the
various compartments of the GI environment were identified. Functionally,
these peptides
potently neutralize IL-23-mediated signaling in a transformed human cell line
and in human
primary cells. The binding of IL-23R is selective, since the peptides do not
block the interaction
between IL-6 to IL-6R or antagonize the IL-12 signaling pathway. Furthermore,
these orally
delivered peptides are efficacious in attenuating colitis in a 2,4,6-
trinitrobenzenesulfonic acid
(TNBS)-induced acute rat model of IBD, as shown by a significant reduction in
the ratio of colon
weight to length, colon macroscopic score, neutrophil infiltration, and
histopathology
comparable to that of the control anti-IL-23p19 mAb.
[0090] The present invention relates generally to peptides that have IL-23R
antagonist activity,
including both peptide monomers and peptide dimers. In certain embodiments,
this invention
demonstrates a new paradigm for treatment of IBD and other diseases and
disorders by oral
delivery of antagonists of IL-23. IBD represents a local inflammation of the
intestinal tissue;
therefore, advantageous therapeutic agents would act from the luminal side of
the intestine,
yielding high drug concentrations in diseased tissue, minimizing systemic
availability and
resulting in improved efficacy and safety when compared to systemic
approaches. Oral
administration of the compounds of the present invention is expected to
maximize drug levels in
diseased intestinal tissues while limiting drug concentrations in circulation,
thereby providing
efficacious, safe, and durable delivery for life-long treatment of IBD and
other diseases and
disorders.
[0091] In certain embodiments, the present invention relates to various
peptides, or peptide
dimers comprising hetero- or homo-monomer subunits, that form cyclized
structures through
disulfide or other bonds. In certain embodiments, the disulfide or other bonds
are intramolecular
42
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
bonds. The cyclized structure of the peptide monomer inhibitors and the
monomer subunits of
the peptide dimer inhibitors has been shown to increase potency and
selectivity of the peptide
inhibitors. In certain embodiments, a peptide dimer inhibitor may include one
or more
intermolecular bonds linking the two monomer peptide subunits within the
peptide dimer
inhibitor, e.g., an intermolecular bridge between two cysteine residues, one
in each peptide
monomer subunit.
[0092] The present invention provides peptide inhibitors that bind to IL-23R,
which may be
monomers or dimers. In particular embodiments, the peptide inhibitors inhibit
the binding of IL-
23 to IL-23R. In certain embodiments, the IL-23R is human IL-23R, and the IL-
23 is human IL-
23. In certain embodiments, a peptide inhibitor of the present invention
reduces IL-23 binding to
IL-23R by at least 20%, at least 30%, at least 40%, at least 50%, at least
60%, at least 70%, at
least 80%, or at least 90% as compared to a negative control peptide. Methods
of determining
binding are known in the art and include ELISA assays, as described in the
accompanying
Examples.
[0093] In certain embodiments, a peptide inhibitor of the present invention
has an IC50 of > 1
mM, < 1 mM, 500 nM to 1000 nM, < 500 nIVI, <250 nIVI, < 100 nIVI, < 50 nM, <25
nIVI, < 10
<5 nM, <2 nM, < 1 nM, or < 5 mM, e.g., for inhibiting binding of IL-23 to IL-
23R (e.g.,
human IL-23 and human IL-23R). Methods of determining activity are known in
the art and
include any of those described in the accompanying Examples.
[0094] In certain embodiments, a peptide inhibitor of the present invention
has increased
stability, increased gastrointestinal stability, or increased stability in
stimulated intestinal fluid
(SIF) or simulated gastric fluid (SGF), and/or under redox conditions (e.g.,
DTT) as compared to
a control peptide. In certain embodiments, a control peptide is an unrelated
peptide of the same
or similar length. In particular embodiments, a control peptide is a peptide
having the identical
or a highly related amino acid sequence (e.g., > 90% sequence identity) as the
peptide inhibitor.
In particular embodiments, a control peptide is a peptide having the identical
or a highly related
amino acid sequence (e.g., > 90% sequence identity) as the peptide inhibitor,
but which does not
have a cyclized structure, e.g., through an intramolecular bond between two
amino acid residues
within the control peptide, or which is not dimerized, or which does not
comprise a conjugate for
43
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
stabilization. In particular embodiments, the only difference between the
peptide inhibitor and
the control peptide is that the peptide inhibitor comprises one or more amino
acid substitutions
that introduce one or more amino acid residues into the peptide inhibitor,
wherein the introduced
amino residue(s) forms an intrasulfide disulfide or thioether bond with
another amino acid
residue in the peptide inhibitor. One example of a control for a peptide dimer
inhibitor is a
monomer having the same sequence as one of the monomer subunits present in the
peptide dimer
inhibitor. One example of a control for a peptide inhibitor comprising a
conjugate is a peptide
having the same sequence but not including the conjugated moiety. In certain
embodiments, a
control peptide is a peptide (e.g., a naturally-occurring peptide)
corresponding to a region of IL-
23 that binds to IL-23R.
[0095] Methods of determining the stablity of a peptide are known in the art.
In certain
embodiments, the stability of a peptide inhibitor is determined using an SIF
assay, e.g., as
described in Example 3. In certain embodiments, the stability of a peptide
inhibitor is
determined using an SGF assay, e.g., as described in Example 3. In particular
embodiments, a
peptide inhibitor has a half-life (e.g., in SIF or SGF or DTT) under a given
set of conditions (e.g.,
temperature) of greater than 1 minute, greater than 10 minutes, greater than
20 minutes, greater
than 30 minutes, greater than 60 minutes, greater than 90 minutes, greater
than 120 minutes,
greater than 3 hours, or greater than four hours when exposed to SIF or SGF or
DTT. In certain
embodiments, the temperature is about 25 C, about 4 C, or about 37 C, and
the pH is a
physiological pH, or a pH about 7.4.
[0096] In some embodiments, the half-life is measured in vitro using any
suitable method known
in the art, e.g., in some embodiments, the stability of a peptide of the
present invention is
determined by incubating the peptide with pre-warmed human serum (Sigma) at 37
C. Samples
are taken at various time points, typically up to 24 hours, and the stability
of the sample is
analyzed by separating the peptide or peptide dimer from the serum proteins
and then analyzing
for the presence of the peptide or peptide dimer of interest using LC-MS.
[0097] In some embodiments, a peptide inhibitor of the present invention
exhibits improved
solubility or improved aggregation characteristics as compared to a control
peptide. Solubility
may be determined via any suitable method known in the art. In some
embodiments, suitable
44
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
methods known in the art for determining solubility include incubating
peptides in various
buffers (Acetate pH4.0, Acetate pH5.0, Phos/Citrate pH5.0, Phos Citrate pH6.0,
Phos pH 6.0,
Phos pH 7.0, Phos pH7.5, Strong PBS pH 7.5, Tris pH7.5, Tris pH 8.0, Glycine
pH 9.0, Water,
Acetic acid (pH 5.0 and other known in the art) and testing for aggregation or
solubility using
standard techniques. These include, but are not limited to, visual
precipitation, dynamic light
scattering, Circular Dichroism and fluorescent dyes to measure surface
hydrophobicity, and
detect aggregation or fibrillation, for example. In some embodiments, improved
solubility
means the peptide is more soluble in a given liquid than is a control peptide.
In some
embodiments, improved aggregation means the peptide has less aggregation in a
given liquid
under a given set of conditions than a control peptide.
[0098] In certain embodiments advantageous for achieving high compound
concentrations in
intestinal tissues when delivered orally, peptide inhibitors of the present
invention are stable in
the gastrointestinal (GI) environment. Proteolytic metabolism in the GI tract
is driven by
enzymes (including pepsins, trypsin, chymotrypsin, elastase, aminopeptidases,
and
carboxypeptidase A/B) that are secreted from the pancreas into the lumen or
are produced as
brush border enzymes. Proteases typically cleave peptides and proteins that
are in an extended
conformation. In the reducing environment of intestinal fluids, disulfide
bonds may be broken,
resulting in a linear peptide and rapid proteolysis. This luminal redox
environment is largely
determined by the Cys/CySS redox cycle. In enterocytes, relevant activities
include numerous
digestive enzymes such as CYP450 and UDP-glucuronsyl-transferase. Finally,
bacteria, present
in the large intestine at concentration ranging from 1010 to 1012 CFU/ml,
constitute another
metabolic barrier. In certain embodiments, the peptide inhibitors are stable
to various pHs that
range from strongly acidic in the stomach (pH 1.5-1.9), trending towards basic
in the small
intestine (pH 6-7.5), and then weakly acidic in the colon (pH 5-7). Such
peptide inhibitors are
stable during their transit through the various GI compartments, a process
that has been
estimated to take 3-4 h in the intestine and 6-48 h in the colon.
[0099] In some embodiments, the peptide inhibitors of the present invention
have less
degradation, e.g., over a period of time (i.e., more degradation stability),
e.g., greater than or
about 10% less, greater than or about 20% less, greater than or about 30%
less, greater than or
about 40 less, or greater than or about 50% less degradation than a control
peptide. In some
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
embodiments, degradation stability is determined via any suitable method known
in the art. In
some embodiments, the degradation is enzymatic degradation. For
example, in certain
embodiments, the peptide inhibitors have reduced susceptibility to degradation
by trypsin,
chhrmotrypsin or elastase. In some embodiments, suitable methods known in the
art for
determining degradation stability include the method described in Hawe et al.,
J Pharm Sci,
VOL. 101, No. 3, 2012, p 895-913, incorporated herein in its entirety. Such
methods are in some
embodiments used to select potent peptide sequences with enhanced shelf lifes.
In particular
embodiments, peptide stability is determined using a SIF assay or SGF assay as
described herein.
1001001In certain embodiments, peptide inhibitors of the present invention
inhibit or reduce IL-
23-mediated inflammation. In related embodiments, peptide inhibibitors of the
present invention
inhibit or reduce IL-23-mediated secretion of one or more cytokines, e.g., by
binding to IL-23R
on the cell surface, thus inhibiting IL-23 binding to the cell. In particular
embodiments, peptide
inhibitors of the present invention inhibit or reduce IL-23-mediated
activation of Jak2, Tyk2,
Statl, Stat3, Stat4, or Stat5. Methods of determining inhibition of cytokine
secretion and
inhibition of signaling molecules are known in the art. For example, inhibiton
of IL-23/IL-23R
signaling may be determined by measuring inhibition of phospho-Stat3 levels in
cell lysates, as
decribed in the accompanying Examples, including Example 2.
1001011In certain embodiments, peptide inhibitors of the present invention
inhibit or reduce IL-
23-mediated inflammation. In related embodiments, peptide inhibibitors of the
present invention
inhibit or reduce IL-23-mediated secretion of one or more cytokines, e.g., by
binding to IL-23R
on the cell surface, thus inhibiting IL-23 binding to the cell. In particular
embodiments, peptide
inhibitors of the present invention inhibit or reduce IL-23-mediated
activation of Jak2, Tyk2,
Statl, Stat3, Stat4, or Stat5. Methods of determining inhibition of cytokine
secretion and
inhibition of signaling molecules are known in the art. For example, inhibiton
of IL-23/IL-23R
signaling may be determined by measuring inhibition of phospho-Stat3 levels in
cell lysates, as
decribed in the accompanying Examples, including Example 2.
1001021In certain embodiments, peptide inhibitors have increased redox
stability as compared to
a control peptide. A variety of assays that may be used to determine redox
stability are known
46
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
and available in the art. Any of these may be used to determine the redox
stability of peptide
inhibitors of the present invention.
1001031In certain embodiments, the present invention provides various peptide
inhibitors that
bind or associate with the IL-23R, in vitro or in vivo, to disrupt or block
binding between IL-23
and IL-23R. In certain embodiments, the peptide inhibitors bind and/or inhibit
human IL-23R.
In certain embodiments, the peptide inhibitors bind and/or inhibit both human
and rodent IL-
23R. In certain embodiments, the peptide inhibitors bind and/or inhibit both
human and rat IL-
23R. In particular embodiments, the peptide inhibitors inhibit rat IL-23R at
least 50%, at least
60%, at least 70%, at least 80%, at least 90%, or at least 95% as well as they
bind or inhibit
human IL-23R, e.g., as determined by an assay described herein. In certain
embodiments, the
peptide inhibitors preferentially bind and/or inhibit human and/or rat IL-23R
as compared to
mouse IL-23R. In particular embodiments, the peptide inhibitors preferentially
bind to rat IL-
23R as compared to mouse IL-23R. In particular embodiments, the peptide
inhibitors
preferentially bind to human IL-23R as compared to mouse IL-23R. In certain
embodiments,
binding of a peptide inhibitor to mouse IL-23R is less than 75%, less than
50%, less than 40%,
less than 30%, less than 20%, or less than 10% of binding of the same peptide
inhibitor to human
IL-23R and/or rat IL-23R. In certain embodiments of peptide inhibitors that
preferentially bind
and/or inhibit human IL-23R and/or rat IL-23R as compared to mouse IL-23R, the
peptide
inhibitor binds to a region of IL-23R that is disrupted by the presence of
additional amino acids
present in mouse IL-23R but not human IL-23R or rat IL-23. In one embodiment,
the additional
amino acids present in the mouse IL-23R are in the region corresponding to
about amino acid
residue 315 to about amino acid residue 340 of the mouse IL23R protein, e.g.,
amino acid region
NVVQPWSSPFVHQTSQETGKR (see, e.g., Figure 4). In particular embodiments, the
peptide
inhibitors bind to a region of human IL-23R from about amino acid 230 to about
amino acid
residue 370.
1001041In certain embodiments, peptide inhibitors show GI-restricted
localization following oral
administration. In particular embodiments, greater than 50%, greater than 60%,
greater than
70%, greater than 80%, or greater than 90% of orally administered peptide
inhibitor is localized
to gastrointestinal organs and tissues. In particular embodiments, blood
plasma levels of orally
administered peptide inhibitor are less than 20%, less than 10%, less than 5%,
less than 2%, less
47
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
than 1% or less than 0.5% the levels of peptide inhibitor found in the small
intestine mucosa,
colon mucosa, or proximal colon.
1001051The various peptide inhibitors of the invention may be constructed
solely of natural
amino acids. Alternatively, the peptide inhibitors may include non-natural
amino acids
including, but not limited to, modified amino acids. In certain embodiments,
modified amino
acids include natural amino acids that have been chemically modified to
include a group, groups,
or chemical moiety not naturally present on the amino acid. The peptide
inhibitors of the
invention may additionally include one or more D-amino acids. Still further,
the peptide
inhibitors of the invention may include amino acid analogs.
1001061In certain embodiments, peptide inhibitors of the present invention
include one or more
modified or unnatural amino acids. For example, in certain embodiments, a
peptide inhibitor
includes one or more of Dab, Dap, Pen, Sarc, Cit, Cav, hLeu, 2-Nal, D-1-Nal, D-
2-Nal, Phe(4-
0Me), f3hTrp, a-MePhe, a-MeTyr, a-MeTrp, f3-HPhe, Phe(4-CF3), 2-2-Indane, 1-1-
Indane,
Cyclobutyl, f3-hPhe, Gla, Phe(4-NH2), hPhe, 1-Nal, Nle, homoamino acids, D-
amino acids, 4,4'-
Biphenylalanine (Bip), cyclobutyl-Ala, hCha, f3hPhe, f3G1u, Phe(4-Guanidino),
Phe[4-(2-
aminoethoxy)], Phe[4-(2-acetylaminoethoxy)], Phe(4-CONH2), Phe(4-Me),
Tyr(Bz1), or
Tyr(Me), Phe(3,4-diF2), Phe(3,4-C12), Phe(3-Me), Phe[4-(2-aminoethoxy)], Phe[4-
(2-
acetylaminoethoxy)], Phe(Br), Phe(4-CONH2), Phe(C1), Phe(4-CN), Phe(4-
guadino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Tyr, Tyr(Bz1), or Tyr(Me), Phe(3,4-dimethoxy), 5-
HydroxyTrP,
Phe(3,4-C12), Tyr(3-tBu), and various N-methylated amino acids and alpha-
methyl amino acids.
In some embodiments of the present invention, a peptide inhibitor includes one
or more non-
natural amino acids shown in Table 1A. One having skill in the art will
appreciate that other
modified or unnatural amino acids, and various other substitutions of natural
amino acids with
modified or unnatural amino acids, may be made to achieve similar desired
results, and such
substitutions are within the teaching and spirit of the present invention. In
certain embodiments,
peptide inhibitors of the present invention include any of those described
herein, including but
not limited to any of those comprising an amino acid sequence or peptide
inhibitor structure
shown in any one of the tables herein, the accompanying sequence listing or
the accompanying
figures, wherein one or more residues is substituted with a modified or
unnatural amino acid.
48
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1001071 The present invention also includes any of the peptide inhibitors
described herein in
either a free or a salt form. Thus, embodiments of any of the peptide
inhibitors described herein
(and related methods of use thereof) include a pharmaceutically acceptable
salt of the peptide
inhibitor.
1001081 The present invention also includes variants of any of the peptide
inhibitors described
herein, including but not limited to any of those comprising a sequence shown
in any one of the
tables herein, the accompanying sequence listing or the accompanying figures,
wherein one or
more L-amino acid residue is substituted with the D isomeric form of the amino
acid residue,
e.g., an L-Ala is substituted with a D-Ala.
1001091In particular embodiments of the peptide inhibitors described herein,
they comprise one
or more unnatural or non-natural amino acid residue.
1001101 The present invention also includes any of the peptide monomer
inhibitors described
herein linked to a linker moiety, including any of the specific linker
moieties described herein. In
particular embodiments, a linker is attached to an N-terminal or C-terminal
amino acid, while in
other embodiments, a linker is attached to an internal amino acid. In
particular embodiments, a
linker is attached to two internal amino acids, e.g., an internal amino acid
in each of two
monomer subunits that form a dimer. In some embodiments of the present
invention, a peptide
inhibitor is attached to one or more linker moieties shown.
1001111 The present invention also includes peptides and peptide dimers
comprising a peptide
having at least 90%, at least 95%, at least 98%, or at least 99% sequence
identity to the peptide
sequence of a peptide inhibitor described herein. In particular embodiments,
peptide inhibitors of
the present invention comprise a core peptide sequence and one or more N-
terminal and/or C-
terminal modification (e.g., Ac and NH2) and/or one or more conjugated linker
moiety and/or
half-life extension moiety. As used herein, the core peptide sequence is the
amino acid sequence
of the peptide absent such modifications and conjugates. For example, for the
peptide inhibitor:
[Palm] - [isoGlu] - [PEG4] - [Pen] -NTWQ- [Pen] - [Phe [4- (2-aminoethoxy)] -
[2-Nal] - [Aib] - [Ly s (Ac)] -
NN-NH2, the core peptide sequence is: [Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-
[2-Nal]-
[Aib]-[Lys(Ac)]-NN.
49
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1001121In certain embodiments, a peptide inhibitor or a monomer subunit of a
peptide inhibitor
of the present invention comprises, consists essentially of, or consists of 7
to 35 amino acid
residues, 8 to 35 amino acid residues, 9 to 35 amino acid residues, 10 to 35
amino acid residues,
7 to 25 amino acid residues, 8 to 25 amino acid residues, 9 to 25 amino acid
residues, 10 to 25
amino acid residues, 7 to 20 amino acid residues, 8 to 20 amino acid residues,
9 to 20 amino acid
residues, 10 to 20 amino acid residues, 7 to 18 amino acid residues, 8 to 18
amino acid residues,
9 to 18 amino acid residues, or 10 to 18 amino acid residues, and, optionally,
one or more
additional non-amino acid moieties, such as a conjugated chemical moiety,
e.g., a PEG or linker
moiety. In particular embodiments, a peptide inhibitor of the present
invention (or a monomer
subunit thereof), including but not limited to those of any embodiments of
Formula X, Formula I,
Formula II, Formula III, Formula IV, or Formula V is greater than 10, greater
than 12, greater
than 15, greater than 20, greater than 25, greater than 30 or greater than 35
amino acids, e.g., 35
to 50 amino acids. In certain embodiments, a peptide inhibitor (or a monomer
subunit thereof) is
less than 50, less than 35, less than 30, less than 25, less than 20, less
than 15, less than 12, or
less than 10 amino acids. In particular embodiments, a monomer subunit of a
peptide inhibitor
(or a peptide monomer inhibitor) comprises or consists of 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, or 35
amino acid residues. In
particular embodiments, a monomer subunit of a peptide inhibitor of the
present invention
comprises or consists of 10 to 18 amino acid residues and, optionally, one or
more additional
non-amino acid moieties, such as a conjugated chemical moiety, e.g., a PEG or
linker moiety. In
various embodiments, the monomer subunit comprises or consists of 7 to 35
amino acid residues,
7 to 20 amino acid residues, 8 to 20 amino acid residues, 9 to 20 amino acid
residues, 10 to 20
amino acid residues, 8 to 18 amino acid residues, 8 to 19 amino acid residues,
8 to 18 amino acid
residues, 9 to 18 amino acid residues, or 10 to 18 amino acid residues. In
particular
embodiments of any of the various Formulas described herein, X comprises or
consists of 7 to 35
amino acid residues, 8 to 35 amino acid residues, 9 to 35 amino acid residues,
10 to 35 amino
acid residues, 7 to 25 amino acid residues, 8 to 25 amino acid residues, 9 to
25 amino acid
residues, 10 to 25 amino acid residues, 7 to 18 amino acid residues, 8 to 18
amino acid residues,
9 to 18 amino acid residues, or 10 to 18 amino acid residues.
[00113] Certain illustrative peptide inhibitors described herein comprise 12
or more amino acid
residues. However, the present invention also includes peptide inhibitors
comprising a fragment
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
of any of the peptide sequences described herein, including peptide inhibitors
having 7, 8, 9, 10,
or 11 amino acid residues. For example, peptide inhibitors of the present
invention include
peptides comprising or consisting of X4-X9, X4-X10, X4-X11, X4-X12, X4-X13, X4-
X14, X4-
X15, or X4-X16. In particular embodiments, the present invention includes
peptide inhibitors
having any of the sequences described herein, including but not limited to,
those shown in any of
the formulas described herein, the sequence listing, or any of the tables
provided herein, wherein
one or more of X10, X11, X12, X13, X14, X15, or X16 is absent. In particular
embodiments,
one or more of X13, X14, X15 or X16 is absent.
[00114] In particular embodiments of the present invention, the peptide
inhibitors, or X regions
thereof, are not present within an antibody. In particular embodiments, the
peptide inhibitors, or
X regions thereof, are not present within a VH or VL region of an antibody.
1001151In particular embodiments of the peptide inhibitors described herein,
they comprise one
or more unnatural or non-natural amino acid residue.
1001161In particular embodiments, peptide inhibitors of the present invention
are cyclized via a
cyclic amide bond, a disulfide bond, or a thioether bond. In particular
embodiments, the bond is
an intramolecular bond between two amino acid residues within the peptide
inhibitor or a
monomer subunit thereof.
Peptide Inhibitors
[00117] Peptide inhibitors of the present invention include peptides having
any of the amino acid
sequences described herein, compounds having any of the structures described
herein, including
compounds comprising any of the peptide sequences described herein, and dimers
of any of such
peptides and compounds. Peptide inihibitors on the present invention include
both peptides not
having and those having a bond between X4 and X9, e.g., before and after a
cross-link is
introduced between X4 and X9. Illustrative peptides of the invention comprise
an amino acid
sequence or structure described in any of the accompanying tables, Examples,
figures and
sequence listing.
Si
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1001181In certain embodiments, the present invention includes a peptide
inhibitor of an
interleukin-23 receptor, or a pharmaceutically acceptable salt or solvate
thereof, wherein the
peptide inhibitor comprises an amino acid sequence of Formula (Xa):
X1 -X2-X3-X4-X5-X6-X7-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Xa)
wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is any amino acid or chemical moiety capable of forming a bond with X9;
X5 is any amino acid;
X6 is any amino acid;
X7 is any amino acid;
X8 is any amino acid;
X9 is any amino acid or chemical moiety capable of forming a bond with X4;
X10 is any amino acid;
X11 is any amino acid;
X12 is any amino acid;
X13 is any amino acid;
X14 is any amino acid;
X15 is any amino acid,
X16 is any amino acid or absent;
52
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
1001191wherein X4 and X9 are capable of forming a bond with each other. In
particular
embodiments, the bond is a disulfide bond, a thioether bond, a lactam bond, a
triazole ring, a
selenoether bond, a diselenide bond, or an olefin bond. In particular
embodiments, the bond is a
disulfide bond or a thioether bond. In certain embodiments, the peptide
inhibitor is cyclized via
the bond between X4 and X9. In certain embodiments, the peptide inhibitor
inhibits the binding
of an interleukin-23 (IL-23) to an IL-23 receptor. In particular embodiments,
when X4 is not an
amino acid, then X1 , X2, and X3 are absent. In certain embodiments, X1 is a D-
amino acid or
absent. In certain embodiments, X2 is a D-amino acid or absent. In certain
embodiments, X3 is a
D-amino acid or absent. In certain embodiments, X16 is a D-amino acid or
absent. In certain
embodiments, X17 is a D-amino acid or absent. In certain embodiments, X18 is a
D-amino acid
or absent. In certain embodiments, X19 is a D-amino acid or absent. In certain
embodiments,
X20 is a D-amino acid or absent.
[00120] In one embodiment of the peptide inhibitor of Formula Xa,
X1 is absent;
X2 is absent;
X3 is Glu, D-Glu, Arg, (D)Arg, Phe, (D)Phe, 2-Nal, Thr, Leu, (D)Gln or absent;
X4 is Cys, Abu or Pen;
X5 is Ala, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn, N-MeGln, N-
MeArg,
Orn, Gln, Arg, Ser or Thr;
X6 is Asp or Thr;
53
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X7 is Trp or 6-Chloro-Trp;
X8 is Glu, Gin or Val;
X9 is Cys, Abu or Pen;
X10 is 2-Nal, a Phe analog, Tyr, or a Tyr analog, wherein in particular
embodiments, X10 is 2-
Nal, Phe(3,4-diF2), Phe(3,4-C12), Phe(3-Me), Phe[4-(2-aminoethoxy)], Phe[4-(2-
acetylaminoethoxy)], Phe(Br), Phe(4-CONH2), Phe(C1), Phe(4-CN), Phe(4-
guadino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Tyr, Tyr(Bz1), or Tyr(Me);
X11 is 1-Nal, 2-Nal, Phe(3,4-dimethoxy), 5-HydroxyTrp, Phe(3,4-C12), Trp or
Tyr(3-tBu);
X12 is 3-Pal, Acpc, Acbc, Acvc, Achc, Agp, Aib, a-DiethylGly, a-MeLys, a-
MeLys(Ac), a-
MeLeu, a- a-MeOrn, a-MeSer, a-MeVal, Cav, Cha, Cit, Cpa, D-Asn, Glu, His,
hLeu, hArg,
Lys, Leu, Octgly, Orn, piperidine, Arg, Ser, Thr or THP;
X13 is Cit, Asp, Dab, Dap, Phe, His, Dap(Peg2-Ac), Dap(pyroglutaric acid),
Glu, hArg, Lys,
Lys(Ac), Lys(Benzoic acid), Lys(glutaric acid), Lys(IVA), Lys(Peg4-isoGlu-
Palm),
Lys(pyroglutaric acid), Lys-succinic acid, Asn, Orn,G1n, Arg, Thr or Val;
X14 is Asp, Dab(Ac), Dap(Ac), Phe, His, Lys(Ac), Met, Asn(isobutyl), Gin, Arg,
Tyr or
Asp(1,4-diaminobutane);
X15 is Ala, betaAla, Glu, Gly, Asn, Gin, Arg or Ser,
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
54
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
10012111N certain embodiments, X3 is absent. In particular embodiments, X16,
X17, X18, X19
and X20 are absent. In particular embodiments, X4 and X9 are Cys, and X4 and
X9 are linked
via a disulfide bond. In particular embodiments, X4 is Abu and X9 is Pen, and
X4 and X9 are
linked via a thioether bond. In particular embodiments, X4 is Abu and X9 is
Cys, and X4 and X9
are linked via a thioether bond.
[00122] In another embodiment of the peptide inhibitor of Formula Xa,
X1 is absent;
X2 is absent;
X3 is Glu, D-Glu, Arg, (D)Arg, Phe, (D)Phe, 2-Nal, Thr, Leu, (D)Gln or absent;
X4 is Cys, Abu or Pen;
X5 is Ala, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn, Orn, Gln,
Arg, Ser or
Thr;
X6 is Asp or Thr;
X7 is Trp or 6-Chloro-Trp;
X8 is Gln or Val;
X9 is Cys, Abu or Pen;
X10 is 2-Nal, a Phe analog, Tyr, or a Tyr analog, wherein in particular
embodiments, X10 is 2-
Nal, Phe(3,4-diF2), Phe(3-Me), Phe[4-(2-aminoethoxy)], Phe[4-(2-
acetylaminoethoxy)], Phe(Br),
Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-guadino), Phe(4-Me), Phe(4-NH2),
Phe(4-N3),
Tyr, Tyr(Bz1), or Tyr(Me);
X11 is 1-Nal, 2-Nal, Phe(3,4-dimethoxy), 5-HydroxyTrp, Phe(3,4-C12), Trp or
Tyr(3-tBu);
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X12 is 3-Pal, Acpc, Acbc, Acvc, Achc, Agp, Aib, a-DiethylGly, a-MeLys, a-
MeLys(Ac), a-
MeLeu, a-MeOrn, a-MeSer, a-MeVal, Cav, Cha, Cit, Cpa, D-Asn, His, hLeu, hArg,
Lys, Leu,
Octgly, Orn, 4-amino-4-carboxy-piperidine, or MP;
X13 is Cit, Asp, Dab, Dap, Phe, His, Dap(Peg2-Ac), Dap(pyroglutaric acid),
Glu, hArg, Lys,
Lys(Ac), Lys(Benzoic acid), Lys(glutaric acid), Lys(IVA), Lys(Peg4-isoGlu-
Palm),
Lys(pyroglutaric acid), Lys-succinic acid, Asn, Orn,G1n, Arg, Thr or Val;
X14 is Dab(Ac), Dap(Ac), Phe, His, Lys(Ac), Met, Asn, Gln, Arg, or Tyr;
X15 is Ala, f3Ala, Gly, Asn, Gln, or Ser,
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
1001231In some embodiments, X3 is absent. In particular embodiments, X16, X17,
X18, X19
and X20 are absent. In particular embodiments, X4 and X9 are Cys, and X4 and
X9 are linked
via a disulfide bond. In particular embodiments, X4 is Abu and X9 is Pen, and
X4 and X9 are
linked via a thioether bond. In particular embodiments, X4 is Abu and X9 is
Cys, and X4 and X9
are linked via a thioether bond.
[00124] In another embodiment of the peptide inhibitor of Formula Xa,
X1 is absent;
X2 is absent;
X3 is Glu, D-Glu, Arg, (D)Arg, Phe, (D)Phe, 2-Nal, Thr, Leu, (D)Gln or absent;
X4 is Cys, Abu or Pen;
56
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X5 is Dap, Dap(Ac), Gly, Lys, Gin, Arg, Ser,Thr or Asn;
X6 is Thr;
X7 is Trp or 6-Chloro-Trp;
X8 is Gin;
X9 is Cys, Abu or Pen;
X10 is 2-Nal, a Phe analog, Tyr, or a Tyr analog, wherein in particular
embodiments, X10 is 2-
Nal, Phe(3-Me), Phe[4-(2-aminoethoxy)], Phe[4-(2-acetylaminoethoxy)], Phe(4-
CONH2), Phe(4-
Me), Phe(4-NH2), Tyr, Tyr(Bz1), or Tyr(Me);
X11 is 1-Nal, 2-Nal, Phe(3,4-dimethoxy), Phe(3,4-C12), or Trp;
X12 is Acpc, Acbc, Acvc, Achc, Aib, a-DiethylGly, a-MeLys, a-MeLys(Ac), a-
MeLeu, a-
MeOrn, a-MeSer, a-MeVal, Cha, Cit, hLeu, Lys, Leu, Arg or THP;
X13 is Cit, Asp, Dap, Dap(Peg2-Ac), Dap(pyroglutaric acid), Glu, hArg, Lys,
Lys(Ac),
Lys(Benzoic acid), Lys(glutaric acid), Lys(IVA), Lys(Peg4-isoGlu-Palm),
Lys(pyroglutaric
acid), Lys(succinic acid), Asn, Orn,G1n, Arg, or Val;
X14 is Dab(Ac), Dap(Ac), His, Lys(Ac), Asn, Gin, or Tyr;
X15 is Ala, betaAla, Gly, Asn, Gin, or Ser,
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
57
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1001251In some embodiments, X3 is absent. In particular embodiments, X16, X17,
X18, X19
and X20 are absent. In particular embodiments, X4 and X9 are Cys, and X4 and
X9 are linked
via a disulfide bond. In particular embodiments, X4 is Abu and X9 is Pen, and
X4 and X9 are
linked via a thioether bond. In particular embodiments, X4 is Abu and X9 is
Cys, and X4 and X9
are linked via a thioether bond.
[00126] In another embodiment of the peptide inhibitor of Formula Xa,
X1 is absent;
X2 is absent;
X3 is Glu, D-Glu, Arg, (D)Arg, Phe, (D)Phe, 2-Nal, Thr, Leu, (D)Gln or absent;
X4 is Cys, Abu or Pen;
X5 is Dap, Dap(Ac), Gln, Ser, Thr or Asn;
X6 is Thr;
X7 is Trp;
X8 is Gln;
X9 is Cys, Abu or Pen;
X10 is a Phe analog, Tyr, or a Tyr analog, wherein in particular embodiments,
X10 is Phe[4-(2-
aminoethoxy)], Phe[4-(2-acetylaminoethoxy)], Phe(4-CONH2), Phe(4-Me), Tyr,
Tyr(Bz1), or
Tyr(Me);
X11 is 2-Nal or Trp;
X12 is Acpc, Acbc, Acvc, Achc, Aib, a-DiethylGly, a-MeLys, a-MeLys(Ac), a-
MeLeu, a-
MeOrn, a-MeSer, a-MeVal, hLeu, Leu, or THP;
X13 is Cit, Asp, Glu, Lys, Lys(Ac), Asn, or Gln;
58
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X14 is Dab(Ac), Asn, or His;
X15 is Ala, betaAla, Gly, Asn, or Gln;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
1001271In some embodiments, X3 is absent. In particular embodiments, X16, X17,
X18, X19
and X20 are absent. In particular embodiments, X4 and X9 are Cys, and X4 and
X9 are linked
via a disulfide bond. In particular embodiments, X4 is Abu and X9 is Pen, and
X4 and X9 are
linked via a thioether bond. In particular embodiments, X4 is Abu and X9 is
Cys, and X4 and X9
are linked via a thioether bond.
1001281In particular embodiments, the peptide inhibitor comprises the amino
acid sequence set
forth in any of the various formula described herein, e.g., Ia-It, Illa-
Ille, or IV.
1001291In certain embodiments, the present invention includes a peptide
inhibitor of an
interleukin-23 receptor, wherein the peptide inhibitor has the structure of
Formula I:
R1-X-R2 (I)
[00130] or a pharmaceutically acceptable salt or solvate thereof,
100131] wherein 1Z1 is a bond, hydrogen, an Cl -C6 alkyl, a C6-C12 aryl, a C6-
C12 aryl Cl -C6
alkyl, a Cl -C20 alkanoyl, and including PEGylated versions alone or as
spacers of any of the
foregoing;
1001321R2 is a bond, OH or NH2; and
59
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1001331X is an amino acid sequence, e.g., an amino acid comprising 7 to 35
amino acid residues.
In certain embodiments, R2 is OH or NH2.
[00134] In certain embodiments, X comprises a sequence of Formula Xa.
[00135] In particular embodiments of formula (I), X comprises the sequence of
Formula Ia:
X1 -X2-X3-X4-X5-X6-W-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Ia)
wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Met, Glu, Asp, Lys, Orn, Dap, Dab,
D-Dap, D-
Dab, D-Asp, D-Glu, D-Lys, Sec, 2-chloromethylbenzoic acid, mercapto-propanoic
acid,
mercapto-butyric acid, 2-chloro-acetic acid, 3-choro-propanoic acid, 4-chloro-
butyric acid, 3-
chloro-isobutyric acid, Abu, f3-azido-Ala-OH, propargylglycine, 2-(3'-
butenyl)glycine, 2-
allylglycine, 2-(3'-butenyl)glycine, 2-(4'-pentenyl)glycine, 2-(5'-
hexenyl)glycine or absent;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, D-Ala, D-Arg, D-Glu, D-
Phe, D-Leu, D-Thr,
D-Serõ a-MeOrn, a-MeSer, CitDap, Dab, Dap (Ac), Gly, Lys, Asn, N-Me-Gln, N-Me-
Arg, Orn
or Gln,
X6 is Asp, Thr, Asn, Phe, D-Asp, D-Thr, D-Asn, or D-Phe;
X8 is Val, Gln, Glu, or Lys;
X9 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Glu, Lys, Orn, Dap, Dab, D-Dap, D-
Dab, D-Asp,
D-Glu, D-Lys, Asp, Leu, Val, Phe, Ser, Sec, Abu, f3-azido-Ala-OH,
propargylglycine, 2-2-
allylglycine, 2-(3'-butenyl)glycine, 2-(4'-pentenyl)glycine, or 2-(5'-
hexenyl)glycine;
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X10 is Tyr, Phe, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-(acetyl-
aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr;
X11 is Trp, 1-Na!, 2-Na!, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu)
X12 is His, Phe, Arg, N-Me-His, or Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-
butyl-Ala, t-butyl-
Gly 4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-
DiethylGly, a-
MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha, Cit, Cpa,
(D)Asn, Glu,
hArg, or Lys;
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gln;
X14 is Phe, Tyr f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac), or Asp;
X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala, Glu, Arg or Asn;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
In particular embodiments of Ia: X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib,
Sarc, D-Ala, D-
Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib or D-Sarc; X10 is Tyr or Phe;
X11 is Trp, 1-
Na! or 2-Na!; X12 is His, Phe, Arg, N-Me-His, or Val, Cav, Cpa, Leu, Cit,
hLeu, 3-Pal, t-butyl-
Ala or t-butyl-Gly; X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, or
Aib; X14 is Phe,
Tyr or f3hPhe; X15 is Gly, Ser, Thr, Gln, Ala or Sarc; X16 is Asp, Glu, Ala,
AEA, AEP, f3hAla,
Gaba, or absent; and X17 is Leu, Lys, Arg, or absent.
[00136] In particular embodiments, X4 is present.
61
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1001371 In certain embodiments, the peptide inhibitor is cyclized.
1001381 In certain embodiments, the peptide inhibitor is linear or not
cyclized.
1001391In certain embodiments, the peptide inhibitor is cyclized, or contains
an intramolecular
bond, between X4 and X9.
1001401 In certain embodiments of Formula I, X comprises the sequence of
Formula Ib:
X1 -X2-X3-X4-X5-X6-W-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Ib),
[00141] wherein:
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Glu, Asp, Lys, Orn, Dap, Dab, D-
Dap, D-Dab, D-
Asp, D-Glu, D-Lys, Sec, 2-chloromethylbenzoic acid, mercapto-propanoic acid,
mercapto-
butyric acid, 2-chloro-acetic acid, 3-choro-propanoic acid, 4-chloro-butyric
acid, 3-chloro-
isobutyric acid, Abu, f3-azido-Ala-OH, propargylglycine, 2-(3'-
butenyl)glycine, 2-2-allylglycine,
2-(3'-butenyl)glycine, 2-(4'-pentenyl)glycine, 2-(5'-hexenyl)glycine, or
absent;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, D-Ala, D-Arg, D-Glu, D-
Phe, D-Leu, D-Thr,
D-Ser, a-MeOrn, a-MeSer, CitDap, Dab, Dap (Ac), Gly, Lys, Asn, N-Me-Gln, N-Me-
Arg, Orn
or Gln;
X6 is Asp, Thr, Asn, Phe, D-Asp, D-Thr, D-Asn, or D-Phe;
X8 is Val, Gln, Glu, or Lys;
X9 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Glu, Lys, Orn, Dap, Dab, D-Dap, D-
Dab, D-Asp,
D-Glu, D-Lys, Asp, Sec, Abu, f3-azido-Ala-OH, propargylglycine, 2-
allylglycine, 2-(3'-
butenyl)glycine, 2-(4'-pentenyl)glycine, or 2-(5'-hexenyl)glycine;
62
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X10 is Tyr, Phe, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-(acetyl-
aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr
X11 is Trp, 1-Na!, 2-Na!, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12), Tyr(3-t-
Bu)
X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-
Ala, t-butyl-Gly4-
amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-
DiethylGly, a-MeLys,
a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha, Cit, Cpa, (D)Asn, Glu,
hArg, or
Lys
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAlaAib, Lys(Ac), Cit, Asp,
Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gln
X14 is Phe, Tyr, or f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac) or
Asp;
X15 is Gly, Ser, Thr, Gln, Ala, or Sarc, f3-Ala, Glu, Arg or Asn;
X16 is any amino acid or absent;
X17 is any amino acid, or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
1001421In particular embodiments of Ib: X5 is Ala, Arg, Glu, Phe, Leu, Thr,
Ser, Aib, Sarc, D-
Ala, D-Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib or D-Sarc; X10 is Tyr or
Phe; X11 is
Trp, 1-Na! or 2-Na!; X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit,
hLeu, 3-Pal, t-
butyl-Ala or t-butyl-Gly; X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val,
f3hAla or Aib; X14 is
Phe, Tyr or f3hPhe; X15 is Gly, Ser, Thr, Gln, Ala, or Sarc; X16 is Asp, Glu,
Ala, AEA, AEP,
f3hAla, Gaba, or absent; and X17 is Leu, Lys, Arg, or absent.
[00143] In particular embodiments, X4 is present.
63
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[00144] In certain embodiments, the peptide inhibitor is cyclized.
1001451ln certain embodiments, the peptide inhibitor is linear or not
cyclized.
1001461In certain embodiments, the peptide inhibitor is cyclized, or contains
an intramolecular
bond, between X4 and X9.
1001471 In certain embodiments of Formula I, X comprises the sequence of
Formula Ic:
X1 -X2 -X3 -X4 -X5-X6-W-X8-X9-Y-X11 -X12-X13 -X14-X15-X16-X17-X18-X19-X20
(Ic)
[00148] wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Met, Glu, Asp, Lys, Orn, Dap, Dab,
D-Dap, D-
Dab, D-Asp, D-Glu, D-Lys, Sec, 2-chloromethylbenzoic acid, mercapto-propanoic
acid,
mercapto-butyric acid, 2-chloro-acetic acid, 3-choro-propanoic acid, 4-chloro-
butyric acid, 3-
chloro-isobutyric acid, Abu, f3-azido-Ala-OH, propargylglycine, 2-
allylglycine, 2-(3'-
butenyl)glycine, 2-(4'-pentenyl)glycine, 2-(5'-hexenyl)glycine, or absent;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, D-Ala, D-Arg, D-Glu, D-
Phe, D-Leu, D-Thr,
D-Ser, a-MeOrn, a-MeSer, CitDap, Dab, Dap (Ac), Gly, Lys, Asn, N-Me-Gln, N-Me-
Arg, Orn
or Gln
X6 is Asp, Thr, Asn, Phe, D-Asp, D-Thr, D-Asn, or D-Phe;
X8 is Val, Gln, Glu, or Lys;
64
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X9 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Glu, Lys, Orn, Dap, Dab, D-Dap, D-
Dab, D-Asp,
D-Glu, D-Lys, Asp, Sec, Abu, f3-azido-Ala-OH, propargylglycine, 2-
allylglycine, 2-(3'-
butenyl)glycine, 2-(4'-pentenyl)glycine, or 2-(5'-hexenyl)glycine;
X11 is Trp, 1-Na!, 2-Na! Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu)
X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-
Ala t-butyl-Gly;
4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-
DiethylGly,
MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha, Cit, Cpa,
(D)Asn, Glu,
hArg, or Lys
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, or Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gln
X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac) or Asp;
X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala, Glu, Arg or Asn;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
1001491In particular embodiments of Ic, X5 is Ala, Arg, Glu, Phe, Leu, Thr,
Ser, Aib, Sarc, D-
Ala, D-Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib, or D-Sarc; X11 is Trp, 1-
Na!, or 2-Na!;
X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-
Ala or t-butyl-
Gly; X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, or Aib; X14 is
Phe, Tyr, or f3hPhe;
X15 is Gly, Ser, Thr, Gln, Ala, or Sarc; X16 is Asp, Glu, Ala, AEA, AEP,
f3hAla, Gaba, or
absent; and X17 is Leu, Lys, Arg, or absent.
1001501ln particular embodiments, X4 is present.
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1001511 In certain embodiments, the peptide inhibitor is cyclized.
[00152] In certain embodiments, the peptide inhibitor is linear or not
cyclized.
1001531In certain embodiments, the peptide inhibitor is cyclized, or contains
an intramolecular
bond, between X4 and X9.
1001541 In certain embodiments of Formlua I, X comprises the sequence of
Formula Id:
X1 -X2-X3 -C-X5-X6-W-X8-C-X10-X11 -X12 -X13 -X14 -X15-X16-X17-X18-X19-X20
(Id)
[00155] wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, D-Ala, D-Arg, D-Glu, D-
Phe, D-Leu, D-Thr,
D-Ser, a-MeOrn, a-MeSer, CitDap, Dab, Dap (Ac), Gly, Lys, Asn, N-Me-Gln, N-Me-
Arg, Orn
or Gln;
X6 is Asp, Thr, Asn, Phe, D-Asp, D-Thr, D-Asn, or D-Phe;
X8 is Val, Gln, Glu, or Lys;
X10 is Tyr Phe, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-(acetyl-
aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr
X11 is Trp, 1-Nal, 2-Nal, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12), Tyr(3-t-
Bu)
X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-
Ala, t-butyl-Gly,
4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-
DiethylGly, a-
66
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha, Cit, Cpa,
(D)Asn, Glu,
hArg, or Lys
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gin
X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His, Dap(Ac), Dab(Ac) or Asp;
X15 is Gly, Ser, Thr, Gin, Ala, Sarc, f3-Ala, Glu, Arg or Asn;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
[00156] wherein X4 and X9 are optionally linked by a intramolecular disulphide
bridge.
1001571In certain embodiments of Id: X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser,
Aib, Sarc, D-Ala,
D-Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib, or D-Sarc; X10 is Tyr or Phe;
X11 is Trp,
1-Na!, or 2-Na!; X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit,
hLeu, 3-Pal, t-butyl-
Ala, or t-butyl-Gly; X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla,
or Aib; X14 is Phe,
Tyr, or f3hPhe; X15 is Gly, Ser, Thr, Gin, Ala, or Sarc; X16 is Asp, Glu, Ala,
AEA, AEP, f3hAla,
Gaba, or absent; and X17 is Leu, Lys, Arg, or absent.
[00158] In certain embodiments of Formula I, X comprises the sequence of
Formula le:
X1 -X2-X3-X4-X5-X6-W-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20
(le)
[00159] wherein
67
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is Pen, hCys, D-Pen, D-Cys, or D-hCys;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, D-Ala, D-Arg, D-Glu, D-
Phe, D-Leu, D-Thr,
D-Ser, a-MeOrn, a-MeSer, CitDap, Dab, Dap (Ac), Gly, Lys, Asn, N-Me-Gln, N-Me-
Arg, Orn
or Gln;
X6 is Asp, Thr, Asn, Phe, D-Asp, D-Thr, D-Asn, or D-Phe;
X8 is Val, Gln, Glu, or Lys;
X9 is Pen, hCys, D-Pen, D-Cys, D-hCys;
X10 is Tyr, Phe Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-(acetyl-
aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr;
X11 is Trp, 1-Nal, 2-Nal, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu);
X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-
Ala, t-butyl-Gly,
4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-
DiethylGly,
MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha, Cit, Cpa,
(D)Asn, Glu,
hArg, or Lys;
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gln;
X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac) or Asp;
X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala, Glu, Arg or Asn;
X16 is any amino acid or absent;
68
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
[00160] wherein X4 and X9 are optionally linked by a intramolecular disulphide
bridge.
1001611In certain embodiments of le: X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser,
Aib, Sarc, D-Ala,
D-Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib, or D-Sarc; X10 is Tyr or Phe;
X11 is Trp,
1-Nal, or 2-Nal; X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit,
hLeu, 3-Pal, t-butyl-
Ala, or t-butyl-Gly; X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla,
or Aib; X14 is Phe,
Tyr, or f3hPhe; X15 is Gly, Ser, Thr, Gln, Ala, or Sarc; X16 is Asp, Glu, Ala,
AEA, AEP, f3hAla,
Gaba, or absent; and X17 is Leu, Lys, Arg, or absent.
[00162] In particular embodiments, X4 is present.
1001631 In certain embodiments, the peptide inhibitor is cyclized.
1001641 In certain embodiments, the peptide inhibitor is linear or not
cyclized.
1001651In certain embodiments, the peptide inhibitor is cyclized, or contains
an intramolecular
bond, between X4 and X9.
1001661 In particular embodiments, X4 and X9 and both Pen.
1001671 In certain embodiments of Formula I, X comprises the sequence of
Formula If:
X1 -X2-X3-X4-X5-X6-W-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20
(If)
[00168] wherein
X1 is any amino acid or absent;
69
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is Glu, Lys, Orn, Dap, Dab, D-Dap, D-Dab, D-Asp, D-Glu, D-Lys, or Asp;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, D-Ala, D-Arg, D-Glu, D-
Phe, D-Leu, D-Thr,
D-Ser, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap (Ac), Gly, Lys, Asn, N-Me-Gln, N-
Me-Arg, Orn
or Gln;
X6 is Asp, Thr, Asn, Phe, D-Asp, D-Thr, D-Asn, or D-Phe;
X8 is Val, Gln, Glu, or Lys;
X9 is Glu, Lys, Orn, Dap, Dab, D-Dap, D-Dab, D-Asp, D-Glu, D-Lys, or Asp;
X10 is Tyr, Phe, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-(acetyl-
aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino), Phe(4-
Me), Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr;
X11 is Trp, 1-Nal, 2-Nal, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu);
X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-
Ala, or t-butyl-
Gly, 4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-
DiethylGly,
MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha, Cit, Cpa,
(D)Asn, Glu,
hArg, or Lys;
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gln;
X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac) or Asp;
X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala, Glu, Arg or Asn;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
[00169] wherein X4 and X9 are optionally cyclized through an intramolecular
bond.
1001701In certain embodiments of If: X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser,
Aib, Sarc, D-Ala,
D-Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib, or D-Sarc; X6 is Asp, Thr,
Asn, Phe, D-
Asp, D-Thr, D-Asn, or D-Phe; X8 is Val, Gln, Glu, or Lys; X9 is Glu, Lys, Orn,
Dap, Dab, D-
Dap, D-Dab, D-Asp, D-Glu, D-Lys, or Asp; X10 is Tyr or Phe;; X11 is Trp, 1-
Nal, or 2-Nal;
X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-
Ala, or t-butyl-
Gly; ;X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, or Aib; X14 is
Phe, Tyr, or f3hPhe;
X15 is Gly, Ser, Thr, Gln, Ala, or Sarc; X16 is Asp, Glu, Ala, AEA, AEP,
f3hAla, Gaba, or
absent; and X17 is Leu, Lys, Arg, or absent.
10017111n certain embodiments, the intramolecular bond is a lactam bond.
1001721 In certain embodiments of Formula I, X comprises the sequence of
Formula Ig:
X1 -X2-X3 -X4 -X5-X6-W-X8-X9-X10-X11 -X12-X13 -X14 -X15-X16-X17-X18-X19-X20
(Ig)
[00173] wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is f3-azido-Ala-OH, or propargylglycine;
71
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, D-Ala, D-Arg, D-Glu, D-
Phe, D-Leu, D-Thr,
D-Ser, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn, N-MeGln, N-
MeArg, Orn
or Gln;
X6 is Asp, Thr, Asn, Phe, D-Asp, D-Thr, D-Asn, or D-Phe;
X8 is Val, Gln, Glu, or Lys;
X9 is f3-azido-Ala-OH or propargylglycine, ;
X10 is Tyr, Phe, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-(acetyl-
aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr;
X11 is Trp, 1-Nal, 2-Nal, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu)
X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-
Ala, or t-butyl-
Gly, 4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-
DiethylGly,
MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha, Cit, Cpa,
(D)Asn, Glu,
hArg, or Lys;
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gln;
X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac) or Asp;
X15 is Gly, Ser, Thr, Gln, Ala, or Sarc, f3-Ala, Glu, Arg or Asn;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
72
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[00174] wherein X4 and X9 are optionally cyclized through an intramolecular
triazole ring.
1001751In particular embodiments of Ig: X5 is Ala, Arg, Glu, Phe, Leu, Thr,
Ser, Aib, Sarc, D-
Ala, D-Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib, or D-Sarc; X6 is Asp,
Thr, Asn, Phe,
D-Asp, D-Thr, D-Asn, or D-Phe; X8 is Val, Gln, Glu, or Lys; X9 is f3-azido-Ala-
OH or
propargylglycine; X10 is Tyr or Phe; X11 is Trp, 1-Nal, or 2-Nal; X12 is His,
Phe, Arg, N-Me-
His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-Ala, or t-butyl-Gly; X13 is
Thr, Sarc, Glu,
Phe, Arg, Leu, Lys, Val, f3hAla, or Aib; X14 is Phe, Tyr, or f3hPhe; X15 is
Gly, Ser, Thr, Gln,
Ala, or Sarc; X16 is Asp, Glu, Ala, AEA, AEP, f3hAla, Gaba, or absent; and X17
is Leu, Lys,
Arg, or absent.
[00176] In certain embodiments of Formula I, X comprises the sequence of
Formula Ih:
X1 -X2-X3-C-X5-X6-W-X8-C-Y-X11-H-X13-F-X15-X16-X17-X18-X19-X20
(Ih)
[00177] wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is 2-allylglycine, 2-(3'-butenyl)glycine, 2-(4'-pentenyl)glycine, or 2-(5'-
hexenyl)glycine;
X5 is Ala, Arg, Sarc, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn,
N-MeGln, N-
MeArg, Orn or Gln;
X6 is Asp, Thr, or Asn;
X8 is Val, Gln, or Glu;
X9 is 2-allylglycine, 2-(3'-butenyl)glycine, 2-(4'-pentenyl)glycine, or 2-(5'-
hexenyl)glycine;
X11 is Trp, 1-Nal, 2-Nal, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu)
73
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, f3hAla, Val, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gin;
X15 is Gly, Ser, Thr, Gin, Ala, Sarc, f3-Ala, Glu, Arg or Asn;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
[00178] wherein X4 and X9 are optionally cyclized via an intramolecular ring
closing methasis to
give the corresponding olefins.
1001791In particular embodiments of Ih: X5 is Ala, Arg, or Sarc; X6 is Asp,
Thr, or Asn; X11 is
Trp, 1-Nal, or 2-Nal; X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, f3hAla, Val,
or Aib; X15 is Gly,
Ser, Thr, Gin, Ala, or Sarc; X16 is Asp, Glu, Ala, AEA, AEP, f3hAla, Gaba, or
absent; and X17
is Leu, Lys, Arg, or absent.
[00180] In certain embodiments of Formula I, X comprises the sequence of
Formula Ii:
X1 -X2-X3-X4-X5-X6-W-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Ii),
[00181] wherein:
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
74
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X4 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, 2-chloromethylbenzoic acid,
mercapto-propanoic
acid, mercapto-butyric acid, 2-chloro-acetic acid, 3-choro-propanoic acid, 4-
chloro-butyric acid,
or 3-chloro-isobutyric acid;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, D-Ala, D-Arg, D-Glu, D-
Phe, D-Leu, D-Thr,
D-Ser, D-Aib, or D-Sarc, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys,
Asn, N-
MeGln, N-MeArg, Orn or Gln;
X6 is Asp, Thr, Asn, Phe, D-Asp, D-Thr, D-Asn, or D-Phe;
X8 is Val, Gln, Glu, or Lys;
X9 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, or Abu;
X10 is Tyr, Phe, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-(acetyl-
aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr;
X11 is Trp, 1-Nal, 2-Nal, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu)
X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-
Ala, t-butyl-Gly,
4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-
DiethylGly,
MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha, Cit, Cpa,
(D)Asn, Glu,
hArg, or Lys;
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gln;
X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac) or Asp;
X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala, Glu, Arg or Asn;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
[00182] wherein X4 and X9 are optionally cyclized via an intramolecular
thioether bond.
1001831In particular embodiments of Ii: X5 is Ala, Arg, Glu, Phe, Leu, Thr,
Ser, Aib, Sarc, D-
Ala, D-Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib, or D-Sarc; X6 is Asp,
Thr, Asn, Phe,
D-Asp, D-Thr, D-Asn, or D-Phe; X10 is Tyr or Phe; X11 is Trp, 1-Nal, or 2-Nal;
X12 is His,
Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-Ala, or t-
butyl-Gly; X13 is
Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, or Aib; X14 is Phe, Tyr, or
f3hPhe; X15 is Gly,
Ser, Thr, Gln, Ala, or Sarc; X16 is Asp, Glu, Ala, AEA, AEP, f3hAla, Gaba, or
absent; and X17
is Leu, Lys, Arg, or absent.
1001841 In certain embodiments of Formula I, X comprises the sequence of
Formula Ij:
X1 -X2-X3-X4-X5-X6-W-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Ij),
1001851 wherein:
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is Sec, 2-chloromethylbenzoic acid, 3-choro-propanoic acid, 4-chloro-
butyric acid, 3-chloro-
isobutyric acid, or Abu;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, D-Ala, D-Arg, D-Glu, D-
Phe, D-Leu, D-Thr,
D-Ser, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn, N-MeGln, N-
MeArg, Orn
or Gln;
X6 is Asp, Thr, Asn, Phe, D-Asp, D-Thr, D-Asn, or D-Phe;
76
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X8 is Val, Gin, Glu, or Lys;
X9 is Sec or Abu;
X10 is Tyr, Phe, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-
aminoethoxy), Phe[4-(2-(acetyl-aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-
C1), Phe(4-
CN), Phe(4-guanidino), Phe(4-Me), Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-
0Bz1) or Tyr;
X11 is Trp, 1-Na!, 2-Nal, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu);
X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-
Ala, t-butyl-Gly,
4-amino-4-carboxy-tetrahydropyran, Achc, Acpc, Acbc, Agp, Aib, a-DiethylGly, a-
MeLys, a-
MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha, Cit, Cpa, (D)Asn, Glu,
hArg, or
Lys;
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gin;
X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac) or Asp;
X15 is Gly, Ser, Thr, Gin, Ala, Sarc, f3-Ala, Glu, Arg or Asn;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
[00186] wherein X4 and X9 are optionally cyclized via an intramolecular
thioseleno or diselenide
bond.
1001871In particular embodiments of Ij: X5 is Ala, Arg, Glu, Phe, Leu, Thr,
Ser, Aib, Sarc, D-
Ala, D-Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib, or D-Sarc; X10 is Tyr or
Phe; X11 is
77
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Trp, 1-Na!, or 2-Na!; X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit,
hLeu, 3-Pal, t-
butyl-Ala, or t-butyl-Gly; X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val,
f3hAla, or Aib; X14 is
Phe, Tyr, or f3hPhe; X15 is Gly, Ser, Thr, Gln, Ala, or Sarc; X16 is Asp, Glu,
Ala, AEA, AEP,
f3hAla, Gaba, or absent; and X17 is Leu, Lys, Arg, or absent.
[00188] In certain embodiments of Formula I, X comprises the sequence of
Formula Ik:
X1 -X2-X3-X4-X5-X6-W-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Ik),
[00189] wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Met, Glu, Asp, Lys, Orn, Dap, Dab,
D-Dap, D-
Dab, D-Asp, D-Glu, D-Lys or absent;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, D-Ala, D-Arg, D-Glu, D-
Phe, D-Leu, D-Thr,
D-Ser, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn, N-MeGln, N-
MeArg, Orn
or Gln;
X6 is Asp, Thr, Asn, Phe, D-Asp, D-Thr, D-Asn, or D-Phe;
X8 is Val, Gln, Glu, or Lys;
X9 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Glu, Lys, Orn, Dap, Dab, D-Dap, D-
Dab, D-Asp,
D-Glu, D-Lys, Asp, Leu, Val, Phe, or Ser;
X10 is Tyr, Phe, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-(acetyl-
aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr;
X11 is Trp, 1-Na!, 2-Na!, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu);
78
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X12 is His, Phe, Arg, N-Me-His, Val, D-His, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-
butyl-Ala, t-
butyl-Gly,4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib,
a-
DiethylGly, a-MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha,
Cit, Cpa,
(D)Asn, Glu, hArg, or Lys;
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gln;
X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac) or Asp
or absent;
X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala, Glu, Arg or Asn or absent;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
1001901In particular embodiments of Ik: X5 is Ala, Arg, Glu, Phe, Leu, Thr,
Ser, Aib, Sarc, D-
Ala, D-Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib, or D-Sarc; X10 is Tyr or
Phe; X11 is
Trp, 1-Nal, or 2-Na!; X12 is His, Phe, Arg, N-Me-His, Val, D-His, Cav, Cpa,
Leu, Cit, hLeu, 3-
Pal, t-butyl-Ala, or t-butyl-Gly; X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys,
Val, f3hAla, Aib or
absent; X14 is Phe, Tyr, PhPhe or absent; X15 is Gly, Ser, Thr, Gln, Ala, Sarc
or absent; X16 is
Asp, Glu, Ala, AEA, AEP, f3hAla, Gaba, Leu, or absent; and X17 is Leu, Lys,
Arg, or absent.
In certain embodiments of Formula I, X comprises or consists of the sequence
of Formula Ii:
X1 -X2-X3-X4-X5-X6-W-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (I1),
[00191] wherein
X1 is any amino acid or absent;
79
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Met, Glu, Asp, Lys, Orn, Dap, Dab,
D-Dap, D-
Dab, D-Asp, D-Glu, D-Lys or absent;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, a-MeOrn, a-MeSer, Cit,
Dap, Dab, Dap(Ac),
Gly, Lys, Asn, N-MeGln, N-MeArg, Orn or Gln;
X6 is Asp, Thr, Asn, or Phe;
X8 is Val, Gln, Glu, or Lys;
X9 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Glu, Lys, Orn, Dap, Dab, D-Dap, D-
Dab, D-Asp,
D-Glu, D-Lys, Asp, Leu, Val, Phe, or Ser;
X10 is Tyr, Phe, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-(acetyl-
aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr;
X11 is Trp, 1-Nal, 2-Nalõ Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu);
X12 is His, Phe, Arg, N-Me-His, Val, D-His, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-
butyl-Ala, t-
butyl-Gly, 4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib,
a-
DiethylGly, a-MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha,
Cit, Cpa,
(D)Asn, Glu, hArg, or Lys;
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Val, f3hAla, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gln or absent;
X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac) or Asp
or absent;
X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala, Glu, Arg or Asn or absent;
X16 is any amino acid or absent;
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
1001921In particular embodiments of Ii: X5 is Ala, Arg, Glu, Phe, Leu, Thr,
Ser, Aib, or Sarc;
X10 is Tyr or Phe; X11 is Trp, 1-Nal, or 2-Nal; X12 is His, Phe, Arg, N-Me-
His, Val, D-His,
Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-Ala, or t-butyl-Gly; X13 is Thr,
Sarc, Glu, Phe, Arg,
Leu, Lys, Val, f3hAla, Aib or absent; X14 is Phe, Tyr, PhPhe or absent; X15 is
Gly, Ser, Thr,
Gln, Ala, Sarc or absent; X16 is Asp, Glu, Ala, AEA, AEP, f3hAla, Gaba, Leu,
or absent; and
X17 is Leu, Lys, Arg, or absent.
10019311n certain embodiments, X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys,
f3hAla, Aib, Lys(Ac),
Cit, Asp, Dab, Dap, Glu, hArg, Lys, Asn, Orn, or Gln. In certain embodiments,
X13 is Thr, Sarc,
Glu, Phe, Arg, Leu, Lys, f3hAla, or Aib.
1001941In certain embodiments, X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln,
Lys(Ac), His; Dap(Ac),
Dab(Ac) or Asp. In certain embodiments, X14 is Phe, Tyr, or f3hPhe.
1001951In certain embodiments, X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala,
Glu, Arg or Asn. In
certain embodiments, X15 is Gly, Ser, Thr, Gln, Ala, or Sarc.
[00196] In certain embodiments, X12 is alpha amino acid, e.g., 4-amino-4-
carboxy-
tetrahydropyran, Achc Acpc, Acbc, Aib, a-MeGly(diethyl), a-MeLys, a-MeLys(Ac),
a-Me-
Leu, a-MeOrn, a-MeSer, a-MeVal.
[00197] In certain embodiments, X13 is present.
[00198] In certain embodiments, X13 and X14 are present.
[00199] In certain embodiments, X13, X14 and X15 are present.
[00200] In particular embodiments, X4 is present.
81
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1002011 In certain embodiments, the peptide inhibitor is cyclized.
[00202] In certain embodiments, the peptide inhibitor is linear or not
cyclized.
1002031In certain embodiments, the peptide inhibitor is cyclized, or contains
an intramolecular
bond, between X4 and X9.
1002041In certain embodiments of the peptide inhibitor of Formula I, X
comprises or consists of
the sequence of Formula Im:
X1 -X2-X3 -X4-X5-X6-W-X8-X9-Y-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Im),
[00205] wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Met, Glu, Asp, Lys, Orn, Dap, Dab,
D-Dap, D-
Dab, D-Asp, D-Glu, D-Lys or absent;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, a-MeOrn, a-MeSer, Cit,
Dap, Dab, Dap(Ac),
Gly, Lys, Asn, N-MeGln, N-MeArg, Orn, or Gln;
X6 is Asp, Thr, Asn, or Phe;
X8 is Val, Gln, Glu, or Lys;
X9 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Glu, Lys, Orn, Dap, Dab, D-Dap, D-
Dab, D-Asp,
D-Glu, D-Lys, Asp, Leu, Val, Phe, or Ser;
X11 is Trp, 1-Nal, 2-Nal, Phe(3,4-0Me2); 5-Hydroxy-Trp, Phe(3,4-C12), or Tyr(3-
t-Bu);
X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-
Ala, or t-butyl-
Gly, 4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-
DiethylGly, a-
82
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha, Cit, Cpa,
(D)Asn, Glu,
hArg, or Lys;
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, f3hAla, Val, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gin or absent;
X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac), or Asp
or absent;
X15 is Gly, Ser, Thr, Gin, Ala, Sarc, f3-Ala, Glu, Arg or Asn or absent;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
1002061In certain embodiments of Im: X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser,
Aib, or Sarc; X11
is Trp, 1-Na!, or 2-Nal; X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu,
Cit, hLeu, 3-Pal, t-
butyl-Ala, or t-butyl-Gly; X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, f3hAla,
Val, Aib or absent;
X14 is Phe, Tyr, PhPhe or absent; X15 is Gly, Ser, Thr, Gin, Ala, Sarc or
absent; X16 is Asp,
Glu, Ala, AEA, AEP, f3hAla, Gaba, or absent; and X17 is Leu, Lys, Arg, or
absent.
1002071In certain embodiments, X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys,
f3hAla, or Aib. In
certain embodiments, X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, f3hAla, Aib,
Lys(Ac), Cit, Asp,
Dab, Dap, Glu, hArg, Lys, Asn, Orn, or Gin.
1002081 In certain embodiments, X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln,
Lys(Ac), His; Dap(Ac),
Dab(Ac) or Asp. In certain embodiments, X14 is Phe, Tyr, or f3hPhe.
1002091in certain embodiments, X15 is Gly, Ser, Thr, Gin, Ala, or Sarc, f3-
Ala, Glu, Arg or Asn.
In certain embodiments, X15 is Gly, Ser, Thr, Gin, Ala, or Sarc.
83
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1002101 In certain embodiments, X12 is alpha amino acid, e.g., 4-amino-4-
carboxy-
tetrahydropyran, Achc Acpc, Acbc, Aib, a-MeGly(diethyl), a-MeLys, a-MeLys(Ac),
a-Me-
Leu, a-MeOrn, a-MeSer, a-MeVal.
[00211]In certain embodiments, X13 is present.
1002121 In certain embodiments, X13 and X14 are present.
[00213]In certain embodiments, X13, X14, and X15 are present.
1002141 In particular embodiments, X4 is present.
1002151In certain embodiments, the peptide inhibitor is cyclized.
1002161 In certain embodiments, the peptide inhibitor is linear or not
cyclized.
1002171In certain embodiments, the peptide inhibitor is cyclized, or contains
an intramolecular
bond, between X4 and X9.
1002181 In certain embodiments of the peptide inhibitor of Formula I, X
comprises or consists of
the sequence of Formula In:
X1-X2-X3-C-X5-X6-W-X8-C-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (In)
[00219] wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, a-MeOrn, a-MeSer, Cit,
Dap, Dab, Dap(Ac),
Gly, Lys, Asn, N-MeGln, N-MeArg, Orn or Gln;
X6 is Asp, Thr, Asn, or Phe;
X8 is Val, Gln, Glu, or Lys;
84
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X10 is Tyr Phe, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-(acetyl-
aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr;
X11 is Trp, 1-Na!, 2-Na!, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu);
X12 is His, Phe, Arg, N-Me-His, Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-
Ala, or t-butyl-
Gly, 4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-
DiethylGly, a-
MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha, Cit, Cpa,
(D)Asn, Glu,
hArg, or Lys;
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, f3hAla, Val, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gln or absent;
X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac) or Asp
or absent;
X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala, Glu, Arg or Asn or absent;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
[00220] wherein the Cys at position X4 and and the Cys at position X9 are
optionally linked by a
disulphide bridge.
1002211In certain embodiments of In: X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser,
Aib, Sarc, a-
MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn, N-MeGln, N-MeArg, Orn
or Gln;
X10 is Tyr, Phe, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-(acetyl-
aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr; X11 is Trp, 1-Na!, 2-
Na!, Phe(3,4-
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
OMe2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-Bu); X12 is His, Phe, Arg, N-Me-
His, Val, Cav,
Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-Ala, or t-butyl-Gly, 4-amino-4-carboxy-
tetrahydropyran,
Achc Acpc, Acbc, Acvc, Agp, Aib, a-DiethylGly, a-MeLys, a-MeLys(Ac), a-Me-Leu,
a-
MeOrn, a-MeSer, a-MeVal, Cha, Cit, Cpa, (D)Asn, Glu, hArg, or Lys; X13 is Thr,
Sarc, Glu,
Phe, Arg, Leu, Lys, f3hAla, Val, Aib, Lys(Ac), Cit, Asp, Dab, Dap, Glu, hArg,
Lys, Asn, Orn, or
Gln or absent; X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac),
Dab(Ac) or Asp
or absent; X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala, Glu, Arg or Asn or
absent; X16 is Asp,
Glu, Ala, AEA, AEP, f3hAla, Gaba, or absent; and X17 is Leu, Lys, Arg, or
absent.
1002221In certain embodiments, X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys,
f3hAla, Aib, Lys(Ac),
Cit, Asp, Dab, Dap, Glu, hArg, Lys, Asn, Orn, or Gln. In certain embodiments,
X13 is Thr, Sarc,
Glu, Phe, Arg, Leu, Lys, f3hAla, or Aib.
1002231In certain embodiments, X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln,
Lys(Ac), His; Dap(Ac),
Dab(Ac) or Asp. In certain embodiments, X14 is Phe, Tyr, or f3hPhe.
1002241In certain embodiments, X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala,
Glu, Arg or Asn. In
certain embodiments, X15 is Gly, Ser, Thr, Gln, Ala, or Sarc.
1002251In certain embodiments, X12 is an alpha amino acid, e.g., 4-amino-4-
carboxy-
tetrahydropyran, Achc Acpc, Acbc, Acvc, Aib, a-DiethylGly, a-MeLys, a-
MeLys(Ac), a-Me-
Leu, a-MeOrn, a-MeSer, a-MeVal.
[00226] In certain embodiments, X13 is present.
[00227] In certain embodiments, X13 and X14 are present.
[00228] In certain embodiments, X13, X14 and X15 are present.
1002291In certain embodiments of the peptide inhibitor of Formula I, X
comprises or consists of
the sequence of Formula lo:
X1 -X2-X3-C-X5-X6-W-X8-C-Y-X11-H-X13-X14-X15-X16-X17-X18-X19-X20 (lo)
[00230] wherein
86
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, a-MeOrn, a-MeSer, Cit,
Dap, Dab, Dap(Ac),
Gly, Lys, Asn, N-MeGln, N-MeArg, Orn or Gln;
X6 is Asp, Thr, Asn, or Phe;
X8 is Val, Gln, Glu, or Lys;
X11 is Trp, 1-Nal, 2-Nal, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu);
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, f3hAla, Val, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, or Gln or absent;
X14 is Phe, Tyr, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac) or Asp or
absent;
X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala, Glu, Arg or Asn or absent;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
[00231] wherein the Cys at position X4 and and the Cys at position X9 are
optionally linked by a
disulphide bridge.
1002321In certain embodiments of To: X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser,
Aib, Sarc, a-
MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn, N-MeGln, N-MeArg, Orn
or Gln;
X11 is Trp, 1-Nal, 2-Nal, Phe(3,4-0Me2), 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-
t-Bu); X13 is
87
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Thr, Sarc, Glu, Phe, Arg, Leu, Lys, f3hAla, Val, Aib, Lys(Ac), Cit, Asp, Dab,
Dap, Glu, hArg,
Lys, Asn, Orn, Gin or absent; X14 is Phe, Tyr, Asn, Arg, Qln, Lys(Ac), His;
Dap(Ac), Dab(Ac)
Asp or absent; X15 is Gly, Ser, Thr, Gin, Ala, Sarc, f3-Ala, Glu, Arg or Asn
or absent; X16 is
Asp, Glu, Glu, Ala, AEA, AEP, f3hAla, Gaba, or absent; and X17 is Leu, Lys,
Arg, or absent.
1002331In certain embodiments, X12 is an alpha amino acid, e.g., 4-amino-4-
carboxy-
tetrahydropyran, Achc Acpc, Acbc, Acvc, Aib, a-DiethylGly, a-MeLys, a-
MeLys(Ac), a-Me-
Leu, a-MeOrn, a-MeSer, a-MeVal.
[00234] In certain embodiments, X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys,
f3hAla, Aib, Lys(Ac),
Cit, Asp, Dab, Dap, Glu, hArg, Lys, Asn, Orn, or Gin. In certain embodiments,
X13 is Thr, Sarc,
Glu, Phe, Arg, Leu, Lys, f3hAla or Aib.
1002351In certain embodiments, X14 is Phe, Tyr, Asn, Arg, Qln, Lys(Ac), His;
Dap(Ac),
Dab(Ac) or Asp. In certain embodiments, X14 is Phe or Tyr.
1002361In certain embodiments, X15 is Gly, Ser, Thr, Gin, Ala, Sarc, f3-Ala,
Glu, Arg or Asn. In
certain embodiments, X15 is Gly, Ser, Thr, Gin, Ala or Sarc.
[00237] In certain embodiments, X13 is present.
[00238] In certain embodiments, X13 and X14 are present.
[00239] In certain embodiments, X13, X14 and X15 are present.
1002401In certain embodiments of the peptide inhibitor of Formula I, X
comprises or consists of
the sequence of Formula Ip:
X1 -X2-X3-C-X5-X6-W-X8-C-Y-X11-H-X13-F-X15-X16-X17-X18-X19-X20 (Ip)
[00241] wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
88
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X3 is any amino acid or absent;
X5 is Ala, Arg, Sarc, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn,
N-MeGln, N-
MeArg, Orn or Gln;
X6 is Asp, Thr, or Asn;
X8 is Val, Gln, or Glu;
X11 is Trp, 1-Nal, 2-Nal, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu);
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, f3hAla, Val, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, Gln or absent;
X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala, Glu, Arg Asn or absent;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
[00242] wherein the Cys at position X4 and and the Cys at position X9 are
optionally linked by a
disulphide bridge.
1002431In certain embodimetns of Ip: X5 is Ala, Arg, or Sarc; X11 is Trp, 1-
Nal, or 2-Nal; X13
is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, f3hAla, Val, Aib or absent; X15 is Gly,
Ser, Thr, Gln, Ala,
Sarc or absent; X16 is Asp, Glu, Ala, AEA, AEP, f3hAla, Gaba, or absent; and
X17 is Leu, Lys,
Arg, or absent.
[00244] In certain embodiments, X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys,
f3hAla, Aib, Lys(Ac),
Cit, Asp, Dab, Dap, Glu, hArg, Lys, Asn, Orn, or Gln. In certain embodiments,
X13 is Thr, Sarc,
Glu, Phe, Arg, Leu, Lys, f3hAla or Aib.
89
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1002451In certain embodiments, X15 is Gly, Ser, Thr, Gin, Ala or Sarc, f3-Ala,
Glu, Arg or Asn.
In certain embodiments, X15 is Gly, Ser, Thr, Gin, Ala or Sarc.
[00246] In certain embodiments, X13 is present.
[00247] In certain embodiments, X13 and X14 are present.
[00248] In certain embodiments, X13, X14 and X15 are present.
1002491In certain embodiments of the peptide inhibitor of Formula I, X
comprises or consists of
the sequence of Formula Iq:
X1 -X2-X3-C-X5-X6-W-X8-C-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Iq),
wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, D-Ala, D-Arg, D-Glu, D-
Phe, D-Leu, D-Thr,
D-Ser, D-Aib, D-Sarc, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn,
N-MeGln,
N-MeArg, Orn or Gin;
X6 is Asp, Thr, Asn, Phe, D-Asp, D-Thr, D-Asn, or D-Phe;
X8 is Val, Gin, Glu, or Lys;
X10 is Tyr, Phe, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-aminoethoxy)],
Phe[4-(2-(acetyl-
aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino), Phe(4-Me),
Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr;
X11 is Trp, 1-Nal, 2-Nalõ Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-
Bu);
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X12 is His, Phe, Arg, N-Me-His, Val, or D-His, Cav, Cpa, Leu, Cit, hLeu, 3-
Pal, t-butyl-Ala, t-
butyl-Gly, 4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib,
a-
DiethylGly, a-MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal, Cha,
Cit, Cpa,
(D)Asn, Glu, hArg, or Lys;
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, f3hAla, Val, Aib, Lys(Ac), Cit,
Asp, Dab, Dap, Glu,
hArg, Lys, Asn, Orn, Gln or absent;
X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac), Asp or
absent;
X15 is Gly, Ser, Thr, Gln, Ala, Sarc, f3-Ala, Glu, Arg, Asn or absent;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
1002501 wherein the Cys at position X4 and and the Cys at position X9 are
optionally linked.
1002511In certain embodiments of Iq: X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser,
Aib, Sarc, D-Ala,
D-Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib, or D-Sarc; X10 is Tyr or Phe;
X11 is Trp,
1-Nal, or 2-Na!; X12 is His, Phe, Arg, N-Me-His, Val, or D-His, Cav, Cpa, Leu,
Cit, hLeu, 3-Pal,
t-butyl-Ala, or t-butyl-Gly; X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys,
f3hAla, Val, Aib or absent;
X14 is Phe, Tyr, PhPhe or absent; X15 is Gly, Ser, Thr, Gln, Ala, Sarc or
absent; X16 is Asp,
Glu, Ala, AEA, AEP, f3hAla, Gaba, Leu, or absent; and X17 is Leu, Lys, Arg, or
absent.
[00252] In certain embodiments, X12 is alpha amino acid, e.g., 4-amino-4-
carboxy-
tetrahydropyran, Achc Acpc, Acbc, Acvc, Aib, a-DiethylGly, a-MeLys, a-
MeLys(Ac), a-Me-
Leu, a-MeOrn, a-MeSer, a-MeVal.
91
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[00253] In certain embodiments, X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys,
f3hAla, Aib, Lys(Ac),
Cit, Asp, Dab, Dap, Glu, hArg, Lys, Asn, Orn, or Gin. In certain embodiments,
X13 is Thr, Sarc,
Glu, Phe, Arg, Leu, Lys, f3hAla or Aib.
1002541In certain embodiments, X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln,
Lys(Ac), His; Dap(Ac),
Dab(Ac), or Asp. In certain embodiments, X14 is Phe, Tyr or f3hPhe.
1002551In certain embodiments, X15 is Gly, Ser, Thr, Gin, Ala, Sarc, f3-Ala,
Glu, Arg or Asn. In
certain embodiments, X15 is Gly, Ser, Thr, Gin, Ala or Sarc.
[00256] In certain embodiments, X13 is present.
[00257] In certain embodiments, X13 and X14 are present.
[00258] In certain embodiments, X13, X14 and X15 are present.
[00259] In certain embodiments, Iq comprises or consists of the sequence of
Formula Iq':
X1 -X2-X3-C-X5-X6-W-X8-C-X10-X11-X12-X13-X14-X15 (Iq'),
wherein X1-X14 have the definition provided for Iq, and
wherein the Cys at position X4 and and the Cys at position X9 are optionally
linked.
1002601In certain embodiments of Iq': X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser,
Aib, Sarc, D-Ala,
D-Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib, or D-Sarc; X10 is Tyr or Phe;
X11 is Trp,
1-Nal, or 2-Nal; X12 is His, Phe, Arg, N-Me-His, Val, or D-His, Cav, Cpa, Leu,
Cit, hLeu, 3-Pal,
t-butyl-Ala, or t-butyl-Gly; X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys,
f3hAla, Val, Aib or absent;
X14 is Phe, Tyr, PhPhe or absent; X15 is Gly, Ser, Thr, Gin, Ala, Sarc or
absent; X16 is Asp,
Glu, Ala, AEA, AEP, f3hAla, Gaba, Leu, or absent; and X17 is Leu, Lys, Arg, or
absent.
[00261] In certain embodiments, X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys,
f3hAla, Aib, Lys(Ac),
Cit, Asp, Dab, Dap, Glu, hArg, Lys, Asn, Orn, or Gin. In certain embodiments,
X13 is Thr, Sarc,
Glu, Phe, Arg, Leu, Lys, f3hAla or Aib.
92
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1002621In certain embodiments, X14 is Phe, Tyr, f3hPhe, Asn, Arg, Qln,
Lys(Ac), His; Dap(Ac),
Dab(Ac) or Asp. In certain embodiments, X14 is Phe, Tyr or f3hPhe.
1002631In certain embodiments, X15 is Gly, Ser, Thr, Gin, Ala or Sarc, f3-Ala,
Glu, Arg or Asn.
In certain embodiments, X14 is Phe, Tyr or f3hPhe.
1002641 In certain embodiments, X13 is present.
1002651 In certain embodiments, X13 and X14 are present.
1002661 In certain embodiments, X13, X14 and X15 are present.
1002671In certain embodiments of the peptide inhibitor of Formula I, X
comprises or consists of
the sequence of Formula Ir:
X1 -X2-X3-X4-X5-X6-X7-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Ir)
wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X4 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Met, Glu, Asp, Lys, Orn, Dap, Dab,
D-Dap, D-
Dab, D-Asp, D-Glu, D-Lys, Sec, 2-chloromethylbenzoic acid, mercapto-propanoic
acid,
mercapto-butyric acid, 2-chloro-acetic acid, 3-choro-propanoic acid, 4-chloro-
butyric acid, 3-
chloro-isobutyric acid, Abu, f3-azido-Ala-OH, propargylglycine, 2-(3'-
butenyl)glycine, 2-
allylglycine, 2-(3'-butenyl)glycine, 2-(4'-pentenyl)glycine, 2-(5'-
hexenyl)glycine, Abu or absent;
X5 is any amino acid;
X6 is any amino acid;
X7 is Trp, Glu, Gly, Ile, Asn, Pro, Arg, Thr or OctGly, or a corresponding a-
methyl amino acid
form of any of the foregoing;
93
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X8 is any amino acid;
X9 is Cys, Pen, hCys, D-Pen, D-Cys, D-hCys, Glu, Lys, Orn, Dap, Dab, D-Dap, D-
Dab, D-Asp,
D-Glu, D-Lys, Asp, Leu, Val, Phe, or Ser, Sec, Abu, f3-azido-Ala-OH,
propargylglycine, 2-2-
allylglycine, 2-(3'-butenyl)glycine, 2-(4'-pentenyl)glycine, Ala, hCys, Abu,
Met, MeCys, (D)Tyr
or 2-(5'-hexenyl)glycine;
X10 is Tyr, Phe(4-0Me), 1-Nal, 2-Na!, Aic, a-MePhe, Bip, (D)Cys, Cha, DMT,
(D)Tyr, Glu,
His, hPhe(3,4-dimethoxy), hTyr, N-Me-Tyr, Trp, Phe(4-CONH2), Phe(4-phenoxy),
Thr, Tic,
Tyr(3-tBu), Phe(4-tBu), Phe(4-CN), Phe(4-Br), Phe(4-NH2), Phe(4-F), Phe(3,5-
F2), Phe(4-
CH2CO2H), Phe(penta-F), Phe(3,4-C12), Phe(4-CF3), Bip, Cha, 4-PyridylAlanine,
f3hTyr, OctGly,
Phe(4-N3), Phe(4-Br), Phe[4-(2-aminoethoxy)] or Phe, a Phe analog, a Tyr
analog, or a
corresponding a-methyl amino acid form of any of the foregoing;
X11 is 2-Na!, 1-Nal, 2,4-dimethylPhe, Bip, Phe(3,4-C12), Phe (3,4-F2), Phe(4-
CO2H), f3hPhe(4-
F), a-Me-Trp, 4-phenylcyclohexyl, Phe(4-CF3)õ Phe(3,4-0Me2), a-MePhe, f3hNal,
f3hPhe,
f3hTyr, f3hTrp, Nva(5-phenyl), Phe, His, hPhe, Tic, Tqa, Trp, Tyr, Phe(4-0Me),
Phe(4-Me),
Trp(2,5,7-tri-tert-Butyl), Phe(4-0ally1), Tyr(3-tBu), Phe(4-tBu), Phe(4-
guanidino, Phe(4-0Bz1),
Octgly, Glu(Bz1), 4-Phenylbenzylalanine, Phe[4-(2-aminoethoxy)], 5-Hydroxy-
Trp, 6-Chloro-
Trp, N-MeTrp, 1,2,3,4-tetrahydro-norharman, Phe(4-CONH2), Phe(3,4-Dimethoxy),
Phe(2,3-
C12), Phe(2,3-F2), Phe(4-F), 4-phenylcyclohexylalanine or Bip, or a
corresponding a-methyl
amino acid form of any of the foregoing;
X12 is His, Phe, Arg, N-Me-His, or Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-
butyl-Ala, a-MeLys,
D-Ala, (D)Asn, (D)Asp, (D)Leu, (D)Phe, (D)Tyr, Aib, a-MeLeu, a-MeOrn, (3-
Aib, f3-Ala, f3hAla, PhArg, PhLeu, f3hVal, f3-spiro-pip, Glu, hArg, Ile, Lys,
N-MeLeu, N-MeArg,
Ogl, Orn, Pro, Gln, Ser, Thr, Tle or t-butyl-Gly, 4-amino-4-carboxy-
tetrahydropyran, Achc
Acpc, Acbc, Acvc, Agp, Aib, a-DiethylGly, a-MeLys, a-MeLys(Ac), a-Me-Leu, a-
MeOrn,
MeSer, a-MeVal, Cha, Cit, Cpa, (D)Asn, Glu, hArg, or Lys or a corresponding a-
methyl amino
acid form of any of the foregoing;
94
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Asn, Cit, Lys, Arg, Orn, Val, f3hAla,
Lys(Ac), (D)Asn,
(D)Leu, (D)Phe, (D)Thr, Ala, a-MeLeu, Aib, f3-Ala, f3-Glu, PhLeu, f3hVal, f3-
spiro-pip, Cha,
Chg, Asp, Dab, Dap, a-DiethylGly, hLeu, Asn, Ogl, Pro, Gln, Ser, f3-spiro-pip,
Thr, Tba, Tle or
Aib, or a corresponding a-methyl amino acid form of any of the foregoing;
X14 is Phe, Tyr, Glu, Gly, His, Lys, Leu, Met, Asn, Lys(Ac), Dap(Ac), Asp,
Pro, Gln, Arg, Ser,
Thr, Tic or f3hPhe, or a corresponding a-methyl amino acid form of any of the
foregoing;
X15 is Gly, Ser, Thr, Gln, Ala, (D)Ala, (D)Asn, (D)Asp, (D)Leu, (D)Phe,
(D)Thr, Aea, Asp,
Asn, Glu, Phe, Gly, Lys, Leu, Pro, Arg, f3-Ala, or Sarc, or a corresponding a-
methyl amino acid
form of any of the foregoing;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
[00268] In particular embodiments, the peptide is cyclized via X4 and X9.
1002691 In particular embodiments, X3 is Glu, D-Glu, Arg, (D)Arg, Phe, (D)Phe,
2-Na!, Thr, Leu,
(D)Gln.
1002701In certain embodiments of Ir: X11 is 2-Na!, 1-Nal, 2,4-dimethylPhe,
Bip, Phe(3,4-C12),
Phe (3,4-F2), Phe(4-CO2H), f3hPhe(4-F), a-Me-Trp, 4-phenylcyclohexyl, Phe(4-
CF3), a-MePhe,
f3hNal, f3hPhe, f3hTyr, f3hTrp, Nva(5-phenyl), Phe, His, hPhe, Tic, Tqa, Trp,
Tyr, Phe(4-0Me),
Phe(4-Me), Trp(2,5,7-tri-tert-Butyl), Phe(4-0ally1), Tyr(3-tBu), Phe(4-tBu),
Phe(4-guanidino,
Phe(4-0Bz1), Octgly, Glu(Bz1), 4-Phenylbenzylalanine, Phe[4-(2-aminoethoxy)],
5-Hydroxy-
Trp, 6-Chloro-Trp, N-MeTrp, 1,2,3,4-tetrahydro-norharman, Phe(4-CONH2),
Phe(3,4-
Dimethoxy), Phe(2,3-C12), Phe(2,3-F2), Phe(4-F), 4-phenylcyclohexylalanine or
Bip, or a
corresponding a-methyl amino acid form of any of the foregoing; X12 is His,
Phe, Arg, N-Me-
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
His, or Val, Cav, Cpa, Leu, Cit, hLeu, 3-Pal, t-butyl-Ala, a-MeLys, D-Ala,
(D)Asn, (D)Asp,
(D)Leu, (D)Phe, (D)Tyr, Aib, a-MeLeu, a-MeOrn, (3-Aib, (3-Ala, (3hAla, (3hArg,
(3hLeu, (3hVal,
(3-spiro-pip, Glu, hArg, Ile, Lys, N-MeLeu, N-MeArg, Ogl, Orn, Pro, Gln, Ser,
Thr, Tle or t-
butyl-Gly, or a corresponding a-methyl amino acid form of any of the
foregoing; X13 is Thr,
Sarc, Glu, Phe, Arg, Leu, Lys, Arg, Orn, Val, (3hAla, Lys(Ac), (D)Asn, (D)Leu,
(D)Phe, (D)Thr,
Ala, a-MeLeu, Aib, (3-Ala, (3-Glu, (3hLeu, (3hVal, (3-spiro-pip, Cha, Chg,
Asp, Dab, Dap, a-
DiethylGly, hLeu, Asn, Ogl, Pro, Gln, Ser, (3-spiro-pip, Thr, Tba, Tle or Aib,
or a corresponding
a-methyl amino acid form of any of the foregoing; X14 is Phe, Tyr, Glu, Gly,
His, Lys, Leu,
Met, Asn, Pro, Gln, Arg, Ser, Thr, Tic or (3hPhe, or a corresponding a-methyl
amino acid form
of any of the foregoing; X15 is Gly, Ser, Thr, Gln, Ala, (D)Ala, (D)Asn,
(D)Asp, (D)Leu,
(D)Phe, (D)Thr, Aea, Asp, Asn, Glu, Phe, Gly, Lys, Leu, Pro, Arg or Sarc, or a
corresponding a-
methyl amino acid form of any of the foregoing; X16 is Asp, Glu, Ala, AEA,
AEP, (3hAla, Gaba,
Gly, Ser, Pro, Asn, Thr or absent, or a corresponding a-methyl amino acid form
of any of the
foregoing; and X17 is Leu, Lys, Arg, Glu, Ser, Gly, Gln or absent, or a
corresponding a-methyl
amino acid form of any of the foregoing.
[00271] In certain embodiments, both X4 and X9 are Pen. In particular
embodiments, X4 and X9
are cyclized via a disulfide bond.
1002721In certain embodiments, X4 is Abu and X9 is Cys. In certain
embodiments, X4 and X9
are cyclized via a thioether bond.
[00273] In particular embodiments, X5 is Ala, Arg, Glu, Phe, Leu, Thr, Ser,
Aib, Sarc, D-Ala, D-
Arg, D-Glu, D-Phe, D-Leu, D-Thr, D-Ser, D-Aib, Cys, Cit, Asp, Dab, Dap, Gly,
His, hCys, Lys,
Met, Asn, N-Me-Ala, N-Me-Asn, N-Me-Lys, N-Me-Gln, Orn, Pro, Pen, Gln, Val, aMe-
Lys,
aMe-Orn, or D-Sarc, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn, N-
MeGln, N-
MeArg, or Gln. In certain embodiments, X5 is Gln or Asn. In particular
embodiments, X5 is Ala,
Arg, Glu, Phe, Leu, Thr, Ser, Aib, Sarc, D-Ala, D-Arg, D-Glu, D-Phe, D-Leu, D-
Thr, D-Ser, D-
Aib, Cys, Cit, Asp, Dab, Dap, Gly, His, hCys, Lys, Met, Asn, N-Me-Ala, N-Me-
Asn, N-Me-Lys,
N-Me-Gln, N-Me-Arg, Orn, Pro, Pen, Gln, Val, aMe-Lys, aMe-Orn, or D-Sarc.In
certain
embodiments, X5 is Gln.
96
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1002741In particular embodiments, X6 is Asp, Thr, Asn, Phe, D-Asp, D-Thr, D-
Asn, Glu, Arg,
Ser or D-Phe. In particular embodiments, X6 is Thr.
1002751 In particular embodiments, X7 is Trp.
1002761In particular embodiments, X8 is Val, Gln, Glu, Phe, Asn, Pro, Arg,
Thr, Trp or Lys. In
particular embodiments, X8 is Gln.
1002771 In particular embodiments, X1 , X2 and X3 are absent.
1002781 In certain embodiments, X11 is a Trp analog.
1002791In particular embodiments, X10 is a Phe analog. In particular
embodiments, X10 is
Phe(4-0Me), Phe(4-CONH2), or Phe[4-(2-aminoethoxy)] (also referred to herein
as Phe[4-
2ae)]). In particular embodiments, X10 is Phe(4-0Me) or Phe[4-(2-aminoethoxy)]
(also referred
to herein as Phe[4-2ae)]).
1002801 In particular embodiments, X11 is 2-Nal or 1-Nal. In certain
embodiments, X11 is 2-Nal.
1002811In certain embodiments, X12 is a-MeLys, 4-amino-4-carboxy-
tetrahydropyran, Achc
Acpc, Acbc, Acvc, Agp, Aib, a-DiethylGly, a-MeLys, a-MeLys(Ac), a-Me-Leu, a-
MeOrn, a-
MeSer, or a-MeVal. In certain embodiments, X12 is a-MeLys.
1002821 In certain embodiments, X13 is Glu or Lys(Ac). In certain embodiments,
X13 is Glu.
[00283] In certain embodiments, X14 is Asn.
1002841 In certain embodiments, X15 is Gly or Asn. In certain embodiments, X15
is Gly.
1002851 In certain embodiments, one or more, two or more, three or more, or
four or more of X16,
X17, X18, X19 and X20 are absent. In particular embodiments, X16, X17, X18,
X19 and X20
are absent.
1002861In particular embodiments of Ir, X4 and X9 are Cys, X7 is Trp, and X18
is [(D)Lys]. In
particular embodiments of Ir, X4 and X9 are Cys, X7 is Trp, X10 is Tyr, and
X18 is [(D)Lys]. In
particular embodiments of Ir, X4 and X9 are Cys, X7 is Trp, Xi, X2 and X3 are
absent, X17 is
97
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
absent, X18 is [(D)Lys], and X19 and X20 are absent. In particular embodiments
of Ir, X4 and
X9 are Cys, X7 and X11 are Trp, X10 is Tyr, and X18 is [(D)Lys. In certain
embodiments, Xl,
X2, and X3 are absent; and in certain embodiments, X17 is absent.
1002871In particular embodiments of Ir, X4 and X9 are Pen, and X12 is a-MeLys.
In particular
embodiments of Ir, X4 and X9 are Pen, X12 is a-MeLys, and X16, X17, X18, X19
and X20 are
absent. In particular embodiments of Ir, X4 and X9 are Pen, X12 is a-MeLys, 4-
amino-4-
carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-DiethylGly, a-
MeLys(Ac), a-
Me-Leu, a-MeOrn, a-MeSer, a-MeVal, X16, X17, X18, X19 and X20 are absent, and
X7 is
Trp. In particular embodiments of Ir, X4 and X9 are Pen, X12 is a-MeLys, X16,
X17, X18, X19
and X20 are absent, and X7 is Trp. In particular embodiments of Ir, X4 and X9
are Pen, X7 is
Trp, and X12 is a-MeLys. In certain embodiments, X1 , X2, and X3 are absent.
In particular
embodiments, there is a disulfide bond between X4 and X9.
1002881In particular embodiments of Ir, X4 is Abu, X9 is Cys, and X12 is 4-
amino-4-carboxy-
tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-DiethylGly, a-MeLys, or a-
MeLys(Ac),
a-Me-Leu, a-MeOrn, a-MeSer, or a-MeVal. In particular embodiments of Ir, X4 is
Abu, X9 is
Cys, and X12 is a-MeLys. In particular embodiments of Ir, X4 is Abu, X9 is
Cys, X12 is a-
MeLys, 4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-
DiethylGly,
a-MeLys, or a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, or a-MeVal and X16, X17,
X18,
X19 and X20 are absent. In particular embodiments of Ir, X4 is Abu, X9 is Cys,
X12 is a-
MeLys, and X16, X17, X18, X19 and X20 are absent. In particular embodiments of
Ir, X4 is
Abu, X9 is Cys, X12 is a-MeLys, 4-amino-4-carboxy-tetrahydropyran, Achc Acpc,
Acbc, Acvc,
Agp, Aib, a-DiethylGly, a-MeLys, or a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer,
or a-
MeVal, X16, X17, X18, X19 and X20 are absent, and X7 is Trp. In particular
embodiments of Ir,
X4 is Abu, X9 is Cys, X12 is a-MeLys, X16, X17, X18, X19 and X20 are absent,
and X7 is Trp.
In particular embodiments of Ir, X4 is Abu, X9 is Cys, X7 is Trp, and X12 is a-
MeLys. In
certain embodiments, X1 , X2, and X3 are absent. In particular embodiments,
there is a thioether
bond between X4 and X9.
98
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1002891In certain embodiments of the peptide inhibitor of Formula I, X
comprises or consists of
the sequence of Formula Is:
X1 -X2-X3-C-X5-X6-W-X8-C-X10-X11-X12-X13-X14-G-X16-X17-X18-X19-X20 (Is)
wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
X5 is any amino acid;
X6 is any amino acid;
X8 is any amino acid;
X10 is Tyr, 1-Nal 2-Nal, Phe(3,4-F2), Phe(3,4-C12), F(3-Me), Phe[4-(2-
aminoethoxy)], Phe[4-(2-
(acetyl-aminoethoxy)], Phe(4-Br), Phe(4-CONH2), Phe(4-C1), Phe(4-CN), Phe(4-
guanidino),
Phe(4-Me), Phe(4-NH2), Phe(4-N3), Phe(4-0Me), Phe(4-0Bz1) or Tyr;
X11 is Trp 1-Nal, Phe(3,4-0Me2) 5-Hydroxy-Trp, Phe(3,4-C12) or Tyr(3-t-Bu);
X12 is Arg, Lys, His, hArg, Cit, Orn, 1-Nal, D-Ala, D-Leu, D-Phe, D-Asn, D-
Asp, Agp, Leu,
PhLeu, Aib, f3hAla, f3hVal, PhArg, hLeu, Dap, 4-amino-4-carboxy-
tetrahydropyran, Achc Acpc,
Acbc, Acvc, Agp, Aib, a-DiethylGly, a-MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-
MeSer,
a-MeVal, Cha, Cit, Cpa, (D)Asn, Glu, hArg, or Lys;
X13 is Cha, Ogl, Aib, Leu, Val, Dab, Glu, Lys, PhLeu, f3hAla, PhVal f3G1u,
Lys(Ac), Cit, Asp,
Dab, Dap, Glu, hArg, Lys, Asn, Orn, Lys(Ac), or Gln;
X14 is Phe, Tic, Asn Tyr, Asn, Arg, Qln, Lys(Ac), His; Dap(Ac), Dab(Ac) or
Asp;
X16 is any amino acid;
99
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X17 is absent;
X18 is D-Lys;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
1002901In particular embodiments of Is: X10 is Tyr, 1-Na! or 2-Na!; X11 is Trp
or 1-Na!; X12 is
Arg, Lys, His, hArg, Cit, Orn, 1-Na!, D-Ala, D-Leu, D-Phe, D-Asn, D-Asp, Agp,
Leu, PhLeu,
Aib, f3hAla, f3hVal, PhArg, hLeu or Dap; X13 is Cha, Ogl, Aib, Leu, Val, Dab,
Glu, Lys, PhLeu,
f3hAla, PhVal or PGLu; X14 is Phe, Tic, Asn or Tyr; and X16 is AEA, Ala or
f3Ala.
1002911In particular embodiments, X5 is Glu, Arg, Ala, N-Me-Arg, N-Me-Ala, N-
Me-Gln, Orn,
N-Me-Asn, N-Me-Lys, Ser, Gln, Orn, Asn or Dap. In particular embodiments, X5
is Glu, Arg,
Ala, N-Me-Arg, N-Me-Ala, N-Me-Gln, Orn, N-Me-Asn, N-Me-Lys, Ser, Asn or Dap.
[00292] In particular embodiments, X6 is Asp or Thr.
[00293] In particular embodiments, X8 is Gln or Val.
1002941 In particular embodiments, the peptide of Is is cyclized via a
disulfide bond between X4
and X9.
1002951In certain embodiments of the peptide inhibitor of Formula I, X
comprises or consists of
the sequence of Formula It:
X1 -X2-X3-C-X5-X6-W-X8-C-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (It)
wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any amino acid or absent;
100
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X5 is any amino acid;
X6 is any amino acid;
X8 is any amino acid;
X10 is Tyr, 1-Na!, 2-Na!, Phe[4-(2-aminoethoxy)], Phe(4-CONH2), Phe(4-0Me);
X11 is Trp, 1-Na!, 2-Na!, Bipõ Phe(3,4-0Me2) 5-Hydroxy-Trp,;
X12 is Arg, His, 3-Pal, Leu, Thr, Gln, Asn, Glu, Ile, Phe, Ser, Lys, hLeu, a-
MeLeu, D-Leu, D-
Asn, h-Leu, 4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp,
Aib, a-
DiethylGly, a-MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer or a-MeVal;
X13 is Thr, Glu, Tyr, Lys, Gln, Asn, Lys, Lys (Ac), Asp, Arg, Ala, Ser, Leu;
X14 is Phe, Tyr, Asn, Gly, Ser, Met, Arg, His, Lys, Leu or Gln;
X15 is Gly, Ser, Arg, Leu, Asp, Ala, f3-Ala, Glu, Arg or Asn;
X16 is absent or any amino acid;
X17 is absent or any amino acid;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent.
1002961In certain embodiments of It: X10 is Tyr, 1-Na! or 2-Na!; X11 is Trp, 1-
Na!, 2-Na! or
Bip; X12 is Arg, His, 3-Pal, Leu, Thr, Gln, Asn, Glu, Ile, Phe, Ser, Lys,
hLeu, a-MeLeu, D-Leu,
D-Asn, or h-Leu; X13 is Thr, Glu, Tyr, Lys, Gln, Asn, Lys, Asp, Arg, Ala, Ser,
Leu; X15 is Gly,
Ser, Arg, Leu, Asp or Ala; X16 is absent or Asn, Glu, Phe, Ala, Gly, Pro, Asp,
Gln, Ser, Thr, D-
Glu or Lys; and X17 is absent or Pro, Arg, Glu, Asp, Ser, Gly or Gln.
10029711n particular embodiments, X5 is Ser, Asp, Asn, Gln, Ala, Met, Arg, His
or Gly. In
particular embodiments, X5 is Ser, Asp, Gln, Ala, Met, Arg, His or Gly.
101
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1002981 In particular embodiments, X6 is any Asp, Ser or Thr.
1002991 In particular embodiments, X8 is Gln, Glu or Thr.
[00300] In particular embodiments, the peptide of It is cyclized via a
disulfide bond between X4
and X9.
1003011In a further embodiment, the present invention includes a peptide
inhibitor of an
interleukin-23 receptor, or a pharmaceutically acceptable salt or solvate
thereof, wherein the
peptide inhibitor comprises an amino acid sequence of Formula (Va):
X1 -X2 -X3 -X4-X5-X6-X7-X8-X9-X10-X11 -X12 -X13 -X14-X15-X16-X17-X18-X19-X20
(Va)
wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any D-amino acid or absent;
X4 is Cys, hCys, Pen, hPen, Abu, Ser, hSer or chemical moiety capable of
forming a bond with
X9;
X5 is Ala, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn, N-MeGln, N-
MeArg,
Orn, Gln, Arg, Ser or Thr;
X6 is Thr, Ser, Asp, Ile or any amino acid;
X7 is Trp, 6-Chloro-Trp, 1-Nap or 2-Nap;
X8 is Glu, Gln, Asn, Lys(Ac), Cit, Cav, Lys(N-c-(N-a-Palmitoyl-L-y-glutamy1)),
or Lys(N-c-
Palmitoyl;
X9 is Cys, hCys, Pen, hPen Abu, or any amino acid or chemical moiety capable
of forming a
bond with X4;
102
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X10 is 2-Na!, a Phe analog, Tyr, or a Tyr analog;
X11 is 1-Na!, 2-Na!, Phe(3,4-dimethoxy), 5-HydroxyTrp, Phe(3,4-C12), Trp or
Tyr(3-tBu);
X12 is Aib, 4-amino-4-carboxy-tetrahydropyran, any alpha-methylamino acid,
alpha-ethyl-
amino acid, Achc, Acvc, Acbc Acpc, 4-amino-4-carboxy-piperidine, 3-Pal, Agp, a-
DiethylGly,
a-MeLys, a-MeLys(Ac), a-MeLeu, a- a-MeOrn, a-MeSer, a-MeVal, Cav, Cha, Cit,
Cpa, D-
Asn, Glu, His, hLeu, hArg, Lys, Leu, Octgly, Orn, piperidine, Arg, Ser, Thr or
THP;
X13 is Lys(Ac), Gln, Cit, Glu, or any amino acid;
X14 is Asn, Gln, Lys(Ac), Cit, Cav, Lys(N-c-(N-a-Palmitoyl-L-y-glutamy1)),
Lys(N-c-
Palmitoy1), or any amino acid;
X15 is f3-Ala, Asn, Gly, Gln, Ala, Ser, Aib or Cit;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
wherein X4 and X9 are capable of forming a bond with each other. In particular
embodiments,
the bond is a ether, disulfide bond or a thioether bond. In certain
embodiments, the peptide
inhibitor is cyclized via the bond between X4 and X9.
1003021 In certain embodiments, X1 is a D-amino acid or absent. In certain
embodiments, X2 is a
D-amino acid or absent.
1003031In certain embodiments, X16 is a D-amino acid or absent. In certain
embodiments, X17
is a D-amino acid or absent. In certain embodiments, X18 is a D-amino acid or
absent. In
103
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
certain embodiments, X19 is a D-amino acid or absent. In certain embodiments,
X20 is a D-
amino acid or absent.
10030411n particular embodiments of Formula (Ia), X10 is 2-Nal, Phe(3,4-diF2),
Phe(3,4-C12),
Phe(3-Me), Phe[4-(2-aminoethoxy)], Phe[4-(2-acetylaminoethoxy)], Phe(Br),
Phe(4-CONH2),
Phe(C1), Phe(4-CN), Phe(4-guadino), Phe(4-Me), Phe(4-NH2), Phe(4-N3), Tyr,
Tyr(Bz1), or
Tyr(Me). In certain embodiments of Formula (Ia), X10 is Phe(4-ZR), Phe(3-ZR),
oe Phe(2-ZR),
where R= CH2(CH2).Y and n=1-25, Z=NH, 0, CO, CONH, or CH2, and Y=NH2, CO2H,
OH, or
CH3.
1003051ln a further related embodiments, the present invention includes a
peptide inhibitor of an
interleukin-23 receptor, or a pharmaceutically acceptable salt or solvate
thereof, wherein the
peptide inhibitor comprises an amino acid sequence of Formula (Vb):
X1 -X2-X3-X4-X5-X6-X7-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Vb)
wherein
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is D-Arg, D-Phe, any D amino acid or absent;
X4 is Cys, hCys, Pen, hPen, Abu, or a chemical moiety capable of forming a
bond with X9;
X5 is Gln, Asn, Lys(Ac), Cit, Cav, Lys(N-6-(N-a-Palmitoyl-L-y-glutamy1)), or
Lys(N-c-
Palmitoyl);
X6 is Thr, Ser, Asp, Ile or any amino acid;
X7 is Trp, 1-Nap or 2-Nap;
X8 is Gln, Asn, Lys(Ac), Cit, Cav, Lys(N-6-(N-a-Palmitoyl-L-y-glutamy1)), or
Lys(N-c-
Palmitoyl;
104
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X9 is Cys, hCys, Pen, hPen, Abu, any amino acid or a chemical moiety capable
of forming a
bond with X4;
X10 is Phe[4-(2-aminoethoxy)], Phe[4-(2-acetylaminoethoxy)], Phe(4-CONH2),
Phe(4-guadino),
Phe(4-NH2), Tyr(Me) or Phe(4-ZR), where R= CH2(CH2).Y; n=1-25; Z= 0, CO, NH,
CONH, or
CH2; and Y=NH2, CO2H, OH, or CH3,
X11 is 2-Nal or Trp;
X12 is Aib, 4-amino-4-carboxy-tetrahydropyran, a-DiethylGly, a-MeLys, a-
MeLys(Ac), a-
MeLeu, a-MeOrn, a-MeSer, a-MeVal, acid, Achc, Acvc, Acbc Acpc, or 4-amino-4-
carboxy-
piperidine;
X13 is Lys(Ac), Gln, Cit, Glu, or any amino acid;
X14 is Asn, Gln, Lys(Ac), Cit, Cav, Lys(N-6-(N-a-Palmitoyl-L-y-glutamy1)),
Lys(N-c-
Palmitoyl), or any amino acid;
X15 is f3-Ala, Asn, Gln, Ala, Ser, or Aib;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
X20 is any amino acid or absent,
wherein X4 and X9 are capable of forming a bond with each other. In particular
embodiments,
the bond is a disulfide bond or a thioether bond. In certain embodiments, the
peptide inhibitor is
cyclized via the bond between X4 and X9.
[00306] In certain embodiments, X1 is a D-amino acid or absent. In certain
embodiments, X2 is a
D-amino acid or absent.
105
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1003071In certain embodiments, X16 is a D-amino acid or absent. In certain
embodiments, X17
is a D-amino acid or absent. In certain embodiments, X18 is a D-amino acid or
absent. In
certain embodiments, X19 is a D-amino acid or absent. In certain embodiments,
X20 is a D-
amino acid or absent.
1003081In another related embodiment, the present invention includes a peptide
inhibitor of an
interleukin-23 receptor, or a pharmaceutically acceptable salt or solvate
thereof, wherein the
peptide inhibitor comprises an amino acid sequence of Formula (Vc):
X1 -X2-X3-X4-X5-X6-X7-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Vc)
wherein
X1 is absent;
X2 is absent;
X3 is D-Arg or absent;
X4 is Cys, Pen, Abu, or a chemical moiety capable of forming a bond with X9;
X5 is Gln, Asn, Lys(Ac), Cit, or Cav;
X6 is Thr or Ser;
X7 is Trp, 1-Nap or 2-Nap;
X8 is Gln, Asn, Lys(Ac), Cit, or Cav;
X9 is Cys, hCys, Pen, hPen, Abu, or any amino acid or chemical moiety capable
of forming a
bond with X4;
X10 is Phe[4-(2-aminoethoxy)], Phe(4-CONH2) or Phe(4-0R) where R= CH2(CH2)õY;
n=1-25;
and Y=NH2, CO2H, OH, or CH3;
X11 is Trp or 2-Nal;
106
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X12 is Aib, 4-amino-4-carboxy-tetrahydropyran, a-MeLys, a-MeLys(Ac), a-MeLeu,
Achc,
Acvc, Acbc or Acpc;
X13 is Lys(Ac) or Glu;
X14 is Asn, Gin, Lys(Ac), Lys(N-c-(N-a-Palmitoyl-L-y-glutamy1)), or Lys(N-c-
Palmitoy1);
X15 is Gly, f3-Ala, Asn, Gin, Ala, Ser, or Aib;
X16 is absent;
X17 is absent;
X18 is absent;
X19 is absent; and
X20 is absent,
wherein X4 and X9 are capable of forming a bond with each other. In particular
embodiments,
the bond is a disulfide bond or a thioether bond. In certain embodiments, the
peptide inhibitor is
cyclized via the bond between X4 and X9.
[00309] In certain embodiments, X1 is a D-amino acid or absent. In certain
embodiments, X2 is a
D-amino acid or absent.
1003101In certain embodiments, X16 is a D-amino acid or absent. In certain
embodiments, X17
is a D-amino acid or absent. In certain embodiments, X18 is a D-amino acid or
absent. In
certain embodiments, X19 is a D-amino acid or absent. In certain embodiments,
X20 is a D-
amino acid or absent.
1003111In another related embodiment, the present invention includes a peptide
inhibitor of an
interleukin-23 receptor, or a pharmaceutically acceptable salt or solvate
thereof, wherein the
peptide inhibitor comprises an amino acid sequence of Formula (Vd):
X1 -X2-X3-X4-X5-X6-X7-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Vd)
107
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
wherein
X1 is absent;
X2 is absent;
X3 is absent;
X4 is Pen or Abu;
X5 is Gin or Asn;
X6 is Thr or Ser;
X7 is Trp;
X8 is Gin or Asn;
X9 is Pen or Cys;
X10 is Phe[4-(2-aminoethoxy)] or Phe(4-CONH2);
X11 is Trp or 2-Nal;
X12 is Aib, 4-amino-4-carboxy-tetrahydropyran, a-MeLys, a-MeLeu, or Achc;
X13 is Lys(Ac) or Glu;
X14 is Asn, Gin or Lys(Ac);
X15 is Gly, Ala, Ser, f3-Ala, Asn, or Gin;
X16 is absent;
X17 is absent;
X18 is absent;
X19 is absent; and
108
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X20 is absent,
wherein X4 and X9 are capable of forming a bond with each other. In particular
embodiments,
the bond is a disulfide bond or a thioether bond. In certain embodiments, the
peptide inhibitor is
cyclized via the bond between X4 and X9.
[00312] Any of the peptide inhibitors of the present invention (e.g., any of
those of Formula I
(e.g., Ix, Ia-It) may be further defined, e.g., as described below. It is
understood that each of the
further defining features described herein may be applied to any peptide
inhibitors where the
amino acids designated at particular positions allow the presence of the
further defining feature.
[00313] In certain embodiments, any of the Phe[4-(2-aminoethoxy)] residues
present in a peptide
inhibitor may be substituted by Phe[4-(2-acetylaminoethoxy)].
1003141 In certain embodiments, X1 -X20 are any of the amino acids shown in
the corresponding
position relative to the cyclized Pen-Pen or cyclized Abu-Cys residues of the
illustrative peptide
inhibitors set forth in Tables 2-5.
1003151 In certain embodiments, any of the peptides inhibitors described
herein, including but not
limited to those of Formulas (X), (Va), (Vb), Vc), (Vd), (Ve), (Vf), (Vg) or
(Vh), further
comprises a linker or spacer moiety between any two amino acid residues of the
peptide. In
particular embodiments, the linker or spacer moiety is a PEG moiety.
[00316] In certain embodiments, the peptide inhibitor is cyclized by a
disulphide bridge.
1003171In certain embodiments, X10 is Tyr, Phe[4-(2-aminoethoxy)], Phe(4-
CONH2) or Phe(4-
0Me). In certain embodiments, X10 is Tyr.
1003181In certain embodiments, X11 is 2-Nal, Trp, or 5-Hydroxy-Trp. In certain
embodiments,
X11 is Trp.
1003191In certain embodiments, X10 is Tyr or Phe[4-(2-aminoethoxy)], and X11
is Trp or 2-Nal.
In certain embodiments, X10 is Tyr and X11 is Trp.
109
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[00320] In particular embodiments, X4 and X9 are both Cys.
[00321] In particular embodiments, X4 is Cys, Pen, hCys, or absent.
[00322] In particular embodiments, X7 and X11 are not both W.
[00323] In particular embodiments, X7 and X11 are both W.
1003241In particular embodiments, X7 and X11 are both W, X10 is Y, and X4 and
X9 are both
Cys.
1003251In particular embodiments, X15 is Gly, Asn, f3-ala or Ser. In
particular embodiments,
X15 is Gly or Ser.
[00326] In particular embodiments, X16 is AEA or AEP.
1003271In particular embodiments, X10 is Tyr, Phe or Phe[4-(2-aminoethoxy). In
particular
embodiments, X10 is Tyr or Phe.
[00328] In particular embodiments, X11 is Trp or 2-Nal. In particular
embodiments, X11 is Trp.
[00329] In particular embodiments, Xi, X2 and X3 are absent.
[00330] In particular embodiments, X18, X19 and X20 are absent.
[00331] In particular embodiments, Xi, X2, X3, X18, X19 and X20 are absent.
[00332] In particular embodiments, one or more of Xi, X2 or X3 are present.
1003331In particular embodiments of any of Ix, la-Ir, one of Xi, X2 and X3 is
present and the
other two are absent. In one embodiment, the Xi, X2 or X3 present is Ala.
1003341In certain embodiments, X3 is present. In particular embodiments, X3 is
Glu, (D)Glu,
Arg, (D)Arg, Phe, (D)Phe, 2-Nal, Thr, Leu, (D)G1n. In certain embodiments, X3
is (D)Arg or
(D)Phe. In particular embodiments, X1 and X2 are absent and X3 is present.
110
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1003351 In particular embodiments, two of Xl, X2 and X3 are present and the
other one is absent.
In certain embodiments, the two present consist of SG, NK, DA, PE, QV or DR.
1003361 In particular embodiments, Xl, X2 and X3 are present. In certain
embodiments, Xl, X2
and X3 consist of ADQ, KEN, VQE, GEE, DGF, NAB, ERN, RVG, KAN, or YED.
1003371In certain embodiments, the peptide comprises an AEP residue. In
particular
embodiments, any of X15, X16, X17, X18, X19 or X20 is AEP.
1003381In certain embodiments of any of the peptide inhibitors or peptide
monomer subunits,
X13 is Thr, Sarc, Glu, Phe, Arg, Leu, Lys, Lys(Ac), f3hAla, or Aib. In certain
embodiments of
any of the peptide inhibitors or peptide monomer subunits, X13 is Thr, Sarc,
Glu, Phe, Arg, Leu,
Lys, f3hAla, or Aib. In certain embodiments, X14 is Phe, Asn, Tyr, or f3hPhe.
In certain
embodiments, X14 is Phe, Tyr, or f3hPhe. In certain embodiments, X15 is Gly,
Asn Ser, Thr,
Gln, Ala, or Sarc. In certain embodiments, X15 is Gly, Ser, Thr, Gln, Ala, or
Sarc. In certain
embodiments, X12 is alpha amino acid, e.g., 4-amino-4-carboxy-tetrahydropyran,
Achc Acpc,
Acbc, Acvc, Aib, a-DiethylGly, a-MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-
MeSer, or a-
MeVal.
1003391 In certain embodiments, X13 is present.
1003401 In certain embodiments, X13 and 14 are present.
[00341]In certain embodiments, X13, X14 and X15 are present.
1003421In particular embodiments of any one of Ia-It, one or more of X16-X20
are present. In
particular embodiments, two or more or three or more of X16-X20 are present.
In particular
embodiments, X18 is [(D)Lys]. In particular embodiments, X17 is absent, and
X18 is [(D)Lys].
In certain embodiments wherein X4 and X9 are optionally Cys, X4 and X9 are
Cys, X7 is Trp,
and X18 is [(D)Lys]. In particular embodiments wherein X4 and X9 are
optionally Cys, X4 and
X9 are Cys, X7 is Trp, X10 is Tyr or Phe[4-(2-aminoethoxy)], and X18 is
[(D)Lys]. In particular
embodiments wherein X4 and X9 are optionally Cys, X4 and X9 are Cys, X7 is
Trp, X10 is Tyr,
and X18 is [(D)Lys]. In particular embodiments wherein X4 and X9 are
optionally Cys, X4 and
X9 are Cys, X7 is Trp, Xl, X2 and X3 are absent, X17 is absent, X18 is
[(D)Lys], and X19 and
111
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X20 are absent. In particular embodiments of Ir, X4 and X9 are Cys, X7 and X11
are Trp, X10
is Tyr, and X18 is [(D)Lys. In certain embodiments, Xi, X2, and X3 are absent;
and in certain
embodiments, X17 is absent.
1003431In certain embodiments, any of the peptide inhibitors (or monomer
subunits) described
herein is cyclized. In particular embodiments, the peptide inhibitor is
cyclized via a bond
between two or more internal amino acids of the peptide inhibitor. In
particular embodiments,
cyclized peptide inhibitors are not cyclized via a bond between the N-terminal
and C-terminal
amino acids of the peptide inhibitor. In certain embodiments, one of the amino
acid residues
participating in the intramolecular bond cyclizing the peptide in the amino
terminal amino acid
residue. In certain embodiments, any of the peptide inhibitors in cyclized via
a peptide bond
between its N-terminal amino acid and its C-terminal amino acid.
[00344] In certain embodiments of any of the peptide inhibitors, or one or
both monomer subunits
thereof, the peptide inhibitor (or one or both monomer subunit thereof) is
cyclized via an
intramolecular bond between X4 and X9 or by a triazole ring. In particular
embodiments, the
intramolecular bond is any disulfide bond, a thioether bond, a lactam bond, a
triazole, a
selenoether bond, a diselendide bond, or an olefin bond.
1003451 In one embodiment, X4 and X9 of the peptide inhibitor (or one or both
monomer subunits
thereof) are Cys, Pen, hCys, D-Pen, D-Cys or D-hCys, and the intramolecular
bond is a disulfide
bond. In certain embodiments, both X4 and X9 are Cys, or both X4 and X9 are
Pen, and the
intramolecular bond is a disulfide bond.
1003461 In one embodiment, X4 and X9 of the peptide inhibitor (or one or both
monomer subunits
thereof) are Glu, Asp, Lys, Orn, Dap, Dab, D-Dap, D-Dab, D-Asp, D-Glu or D-
Lys, and the
intramolecular bond is a lactam bond.
1003471In one embodiment, X4 is Abu, 2-chloromethylbenzoic acid, mercapto-
propanoic acid,
mercapto-butyric acid, 2-chloro-acetic acid, 3-choro-propanoic acid, 4-chloro-
butyric acid, or 3-
chloro-isobutyric acid; X9 is Abu, Cys, Pen, hCys, D-Pen, D-Cys or D-hCys; and
the
intramolecular bond is a thioether bond. In certain embodiments, X4 is Abu and
X9 is Pen, and
the intramolecular bond is a thioether bond. In particular embodiments, X4 is
a 2-methylbenzoyl
112
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
moiety capable of forming a thioether bond with X9, and X9 is selected from
Cys, N-Me-Cys,
D-Cys, hCys, Pen, and D-Pen. In particular embodiments, X4 is Abu and X9 is
Cys, and the
intramolecular bond is a thioether bond. In particular instances, a peptide
monomer, dimer, or
subunit thereof of any of the Formulas and peptides described herein, X4 is
selected from the
group consisting of modified Ser, modified hSer (e.g., Homo-Ser-C1), a
suitable isostere, and
corresponding D-amino acids. In other instances, X4 is an aliphatic acid
having from one to four
carbons and forming a thioether bond with X9. In some instances, X4 is a five-
or six-membered
alicyclic acid having a modified 2-methyl group that forms a thioether bond
with X9. In some
embodiments, X4 is a 2-methylbenzoyl moiety. In certain embodiments, X4 is
selected from
Cys, hCys, Pen, and a 2-methylbenzoyl moiety. In certain embodiments, X4 is
selected from the
group consisting of a modified Ser, a modified hSer, a suitable isostere, and
corresponding D-
amino acids. In one embodiment, X4 is a hSerC1 (before the thioether bond is
formed with X9
whereby the Cl is removed) or a hSer precursor (e.g., homoSer(0-TBDMS). In
other instances,
X4 is an aliphatic acid having from one to four carbons and forming a
thioether bond with X9.
In some instances, X4 is a five- or six-membered alicyclic acid having a
modified 2-methyl
group that forms a thioether bond with X9. In some instances, X4 is a 2-
methylbenzoyl moiety.
In certain embodiments wherein X4 is not an amino acid but is a chemical
moiety that binds to
X9, Xl, X2, and X3 are absent, and X4 is conjugated to or bound to X5. In some
embodiments,
the amino acid directly carboxyl to X9 is an aromatic amino acid. In certain
embodiments, X4 is
an amino acid, while in other embodiments, X4 is another chemical moiety
capable of binding to
X9, e.g., to form a thioether bond. In particular embodiments, X4 is another
chemical moiety
selected from any of the non-amino acid moieties described herein for X4. In
particular
embodiments wherein X4 is another chemical moiety, Xl, X2 and X3 are absent,
and the another
chemical moiety is bound to or conjugated to X5. In certain embodiments, X4 is
defined as a
chemical moiety including a group such as a chloride, e.g., in 2-
chloromethylbenzoic acid, 2-
chloro-acetic acid, 3-choropropanoic acid, 4-chlorobutyric acid, 3-
chloroisobutyric acid.
However, the skilled artisan will appreciate that once the peptide has
undergone ring closing
cyclization to form a thioether bond between X4 and X9, the chloride group is
no longer present.
The description of chemical moieties at X4 that include a reactant group such
as chloride thus
means both the group with the chloride and also the group without the
chloride, i.e., after
formation of the bond with X9. The present invention also includes peptides
comprising the same
113
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
structure as shown in any of the other formulas or tables described herein,
but where the
thioether bond is in the reverse orientation. In such embodiments of the
invention, it may
generally be considered that the amino acid residues or other chemical
moieties shown at X4 are
instead present at X9, and the amino acid residues shown at X9 are instead
present at X4, i.e., the
amino acid residue comprising the sulfur of the resulting thioether bond is
located at X4 instead
of X9, and the amino acid residue or other moiety having a carbon side chain
capable of forming
a thioether bond with X4 is located at X9. In this reverse orientation,
however, the amino acid or
chemical moiety at position X9 is one that comprises a free amine. For
example, in particular
embodiments, the amino acid at X9 is a protected homoserine, such as, e.g.,
homoserine
(OTBDMS). Thus, in particular reverse orientation embodiments of peptide
inhibitors of any of
the formulas described herein, X9 is an amino acid residue having a side chain
with one or two
carbons, and forming a thioether bond with X4, and X4 is selected from the
group consisting of
Cys, N-Me-Cys, D-Cys, HCys, Pen, and D-Pen. Specific examples of amino acid
residues and
other chemical moieties present at corresponding positions of other formulas
and tables are
described herein.
[00348] One of skill in the art will appreciate that certain amino acids and
other chemical moieties
are modified when bound to another molecule. For example, an amino acid side
chain may be
modified when it forms an intramolecular bridge with another amino acid side
chain, e.g., one or
more hydrogen may be removed or replaced by the bond. In addition, when hSer-
C1 binds to an
amino acid such as Cys or Pen via a thioether bond, the Cl moiety is released.
Accordingly, as
used herein, reference to an amino acid or modified amino acid, such as hSer-
C1, present in a
peptide dimer of the present invention (e.g., at position X4 or position X9)
is meant to include
the form of such amino acid or modified amino acid present in the peptide both
before and after
forming the intramolecular bond.
1003491In certain embodiments, the peptide inhibitor of the peptide inhibitor
(or one or both
monomer subunits thereof) is cyclized through a triazole ring. In certain
embodiments, the
peptide inhibitor of the peptide inhibitor (or one or both monomer subunits
thereof) is linear or
not cyclized. In certain embodiments of any of the peptide inhibitors
described herein, including
both monomer peptide inhibitors and dimer peptide inhibitors, one (or both)
peptide monomer
subunits comprise or consist of a cyclized peptide having a structure or
sequence set forth in any
114
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
of Ix, Ia, Ib, Ic, Id, Ie, If, Ig, Ih, Ti, Ij, Ik, Ti, Im, In, To, Ip, Iq,
Iq', Tr, Is or It, IIa-IId, Illa-Ille, Iva,
or IVb.
1003501In certain embodiments of any of the peptide inhibitors or monomer
subunits, X7 and
X11 are both W.
1003511In certain embodiments of any of the peptide inhibitors or monomer
subunits, X7 and
X11 are not both W. In particular embodiments, X7 is W and X11 is not W.
[00352] In certain embodiments of any of the peptide inhibitors or monomer
subunits, X4 and X9
are amino acid residues capable of forming an intramolecular bond between each
other that is a
thioether bond, a lactam bond, a triazole, a selenoether, a diselenide bond,
or an olefin bond.
1003531In certain embodiments, X7 and X11 are both W, X10 is Y, Phe[4-(2-
aminoethoxy) or
Phe(CONH2), and X4 and X9 are amino acid residues capable of forming an
intramolecular bond
between each other that is a thioether bond, a lactam bond, a triazole, a
selenoether, a diselenide
bond, or an olefin bond. In certain embodiments, X7 and X11 are both W, X10 is
Y, and X4 and
X9 are amino acid residues capable of forming an intramolecular bond between
each other that is
a thioether bond, a lactam bond, a triazole, a selenoether, a diselenide bond,
or an olefin bond.
[00354] In certain embodiments, X7 and X11 are both W, X10 is Y, and X4 and X9
are both C.
1003551In certain embodiments, X4 and X9 are each Cys, Pen, hCys, D-Pen, D-Cys
or D-hCys,
and the intramolecular bond is a disulfide bond.
1003561In certain embodiments, X4 and X9 are each Glu, Asp, Lys, Orn, Dap,
Dab, D-Dap, D-
Dab, D-Asp, D-Glu or D-Lys, and the intramolecular bond is a lactam bond.
1003571In certain embodiments, X4 and X9 are each f3-azido-Ala-OH or
propargylglycine, and
the peptide inhibitor (or monomer subunit) is cyclized through a triazole
ring.
1003581In certain embodiments, X4 and X9 are each 2-allylglycine, 2-(3'-
butenyl)glycine, 2-(4'-
pentenyl)glycine, or 2-(5'-hexenyl)glycinem and the peptide inhibitor (or
monomer subunit) is
cyclized via ring closing methasis to give the corresponding olefin / "stapled
peptide."
115
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1003591In certain embodiments, X4 is 2-chloromethylbenzoic acid, mercapto-
propanoic acid,
mercapto-butyric acid, 2-chloro-acetic acid, 3-choro-propanoic acid, 4-chloro-
butyric acid, 3-
chloro-isobutyric acid, or hSer(C1); X9 is hSer(C1), Cys, Pen, hCys, D-Pen, D-
Cys or D-hCys;
and the intramolecular bond is a thioether bond. In certain embodiments, X4 is
2-
chloromethylbenzoic acid or hSer(C1); X9 is Cys or Pen, and the intramolecular
bond is a
thioether bond. In certain embodiments, X4 is Abu, and X9 is Cys or Pen.
[00360] In certain embodiments, X4 is 2-chloromethylbenzoic acid, 2-chloro-
acetic acid, 3-choro-
propanoic acid, 4-chloro-butyric acid, 3-chloro-isobutyric acid, Abu or Sec;
X9 is Abu or Sec;
and the intramolecular bond is a selenoether bond.
10036111n certain embodiments, the intramolecular bond between X4 and X9 is a
diselenide
bond.
[00362] In certain embodiments of any of the peptide inhibitors described
herein that contain two
amino acid residues, e.g., cysteine residues, joined by an intramolecular
bond, e.g., disulphide
bond, the two amino acid residues participating in the intramolecular bond are
not both located at
either the N-terminal or C-terminal position of the peptide inhibitor. In
certain embodiments,
neither of the two amino acid residues, e.g., cysteines, particpating in the
intramolecular bond is
located at the N-terminal or C-terminal position of the peptide inhibitor. In
other words, in
certain embodiments, at least one, or both, of the two amino acid residues,
e.g., cysteines,
participating in the intramolecular bond are internal amino acid residues of
the peptide inhibitor.
In certain embodiments, neither of the two amino acid resiudes, e.g.,
cysteines, participating in
the intramolecular bond is located at the C-terminal position of the peptide
inhibitor. At certain
embodiment, the two amino acid residues participating in the intramolecular
bond are Cys, Pen,
hCys, D-Pen, D-Cys or D-hCys residues. In certain embodiments, the two amino
acid residues
participating in the intramolecular bond are located at X4 and X9. In one
embodiment, there is a
disulfide bond between the amino acid resiudes, e.g., cysteines or Pen
residues, at X4 and X9. In
particular embodiments, both X4 and X9 are Pen. In certain embodiments, one or
both peptide
monomer subunits in the peptide inhibitor is cyclized via a disulfide bond
between two Pen
residues, e.g., at positions X4 and X9.
116
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1003631In particular embodiments of any of the peptide inhibitors described
herein, one or both
peptide monomer subunits present in the peptide inhibitor, whether it is a
monomer or a dimer, is
cyclic or cyclized, e.g., by an intramolecular bond, such as a disulfide bond,
between two
cysteine residues present in the peptide monomer or peptide monomer subunit.
In certain
embodiments, a peptide inhibitor comprises two or more cysteine residues. In
some
embodiments, the peptide inhibitor is cyclized via an intramolecular disulfide
bond between the
two cysteine residues. In particular embodiments of peptide inhibitors having
any of the
Formulas described herein, the two cysteines occur at positions X4 and X9. In
other
embodiments, one or both peptide monomer subunits in the peptide inhibitor is
cyclized via a
disulfide bond between two Pen residues, e.g., at positions X4 and X9.
1003641In some embodiments, a peptide inhibitor has a structure of any of the
Formulas
described herein (e.g., Formula I) and comprises a disulfide bond, e.g., an
intramolecular
disulfide bond, or a thioether bond. Illustrative examples of such peptide
inhibitors are shown in
Tables 3A-3H and 4A, 4B, 9, 11 or 15. Such disulfide bonded peptides may have
a particular
advantage in that the disulfide bonds enhance structural stability and can
improve biological
activity of many bioactive peptides. However, in certain situations, these
bonds are labile to
reducing agents. One of skill in the art will appreciate that disulfide is
amenable to simple
isosteric replacement. Illustrative examples of such replacements include, but
are not limited to,
thioethers, dithioethers, selenoethers, diselenides, triazoles, lacatams,
alkane and alkene groups.
Accordingly, in certain embodiments of any of the peptide inhibitors described
herein, one, two
or more cysteine residues are substitued, e.g., with a thioether, dithioether,
selenoether,
diselenide, triazoles, lacatam, alkane or alkene group, including but not
limited to any of those
shown below or described herein. In particular embodiments, two of these
substituted groups
form a bond (e.g., an intramolecular bond), thus cyclizing the peptide
inhibitor or one or both
monomer subunits thereof.
1003651ln certain embodiments, a peptide inhibitor of the present invention
comprises or consists
of an amino acid sequence shown herein, e.g., in any one of Tables 3A-3H, 4A,
4B, 5A-5C, 6, 7,
8, 9, 10, 11, 12, 13, 14 or 15. In certain embodiments, a peptide inhibitor of
the present invention
has a structure shown herein, e.g., in any one of Tables 3A-3H, 4A, 4B, 5A-5C,
6, 7, 8, 9, 10, 11,
12, 13, 14 or 15.
117
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1003661 In certain embodiments, the present invention includes a peptide
inhibitor that comprises
a core consensus sequence selected from one of the following (shown in N-
terminal to C-
terminal direction):
X1X2X2WX2X1X2W;
X1X2X2WX2X1X2 (1-Nal);
X1X2X2WX2X1X2 (2-Nal);
X1X2X2WX2X1YVV;
X iX2X2WX2X1Y(1 -Nal);
X1X2X2WX2X1Y(2-Nal);
X1X2X2WX2X1X2X2;
X1X2X2WX2X1X2X2X2X2X2-[(D)Lys];
X1X2X2WX2X1X3X2;
X1X2X2WX2X1X3 (1-Nal); and
X iX2X2WX2X iX3(2-Nal) .
[00367] wherein W is tryptophan, Y is tyrosine, each the two X1 residues are
amino acids or other
chemical moieties capable of forming an intramolecular bond with each other;
each X2 is
independently selected from all amino acids, which include, e.g., natural
amino acids, L-amino
acids, D-amino acids, non-natural amino acids, and unnatural amino acids; and
X3 is any amino
acid. In particular embodiments, X3 is Phe, a Phe analog (e.g., Phe[4-(2-
aminoethoxy)] or
Phe(4-CONH2)), Tyr, or a Tyr analog (e.g., Tyr(Me)). In particular
embodiments, each X1 is
selected from Cys, Pen and Abu. In particular embodiments, each X1 is Cys. In
certain
embodiments, each X1 is Pen. In certain embodiments, one X1 is Cys and the
other X1 is Abu.
In particular embodiments, the N-terminal X1 is Abu and the C-terminal X1 is
Cys. In particular
embodiments, the N-terminal X1 is Cys and the C-terminal X1 is Abu. In
particular
embodiments, the residues between the two X1 residues are Gln, Thr, Trp and
Gln. In particular
embodiments, each X1 is selected from Cys, Pen and Abu; and X3 is Phe, a Phe
analog (e.g.,
Phe[4-(2-aminoethoxy)] or Phe(4-carbomide)), Tyr, or a Tyr analog (e.g.,
Tyr(Me)). In
particular embodiments, X3 is a Phe analog.
118
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1003681In certain embodiments, peptide inhibitors of the present invention
comprises any of the
following consensus sequences, wherein X1 , X2, X3, X4, X5, X6, X7, X8, X9,
X10, X11, X12,
X13, X14 and X15 are defined as shown in any of the various Formula or peptide
inhibitors
described herein:
X1 -X2-X3-Pen-X5-X6-W-X8-Pen-X10-X11-X12-X13-X14-X15;
Pen-X5-X6-W-Q-Pen;
Pen-X5-X6-W-X8-Pen;
Pen-X5-X6-W-X8-Pen-[Phe(4-CONH2)];
Pen-X5-X6-W-X8-Pen-[Phe[4-(2-aminoethoxy)]];
X1 -X2-X3-Abu-X5-X6-W-X8-C-X9-X10-X11-X12-X13-X14-X15;
Abu-X5-X6-W-Q-C;
Abu-X5-X6-W-X8-C;
Abu-X5-X6-W-X8-C-[Phe(4-CONH2)]; or
Abu-X5-X6-W-X8-C-[Phe[4-(2-aminoethoxy)]].
1003691In certain embodiments of any of the peptide inhibitors or monomer
subunits, X7 and
X11 are both W. In certain embodiments of any of the peptide inhibitors, X7
and X11 are both
W, and X10 is Y. In certain embodiments, X7 and X11 are both W and X10 is
Phe[4-(2-
aminoethoxy)] or Phe(4-0Me) .
1003701In certain embodiments of any of the peptide inhibitors or monomer
subunits, X7 and
X11 are not both W.
1003711In certain embodiments of peptide inhibitors of Formula I, X4 and X9
are each Pen, and
the intramolecular bond is a disulfide bond.
119
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[00372] In certain embodiments, a peptide inhibitor of the present invention
comprises or consists
of an amino acid sequence shown in any one of the tables, sequence listing or
the accompanying
figures herein.
[00373] In certain embodiments of any of the peptide inhibitors described
herein that contain two
amino acid residues, e.g., Pen residues, joined by an intramolecular bond,
e.g., disulphide bond,
one or both of the two amino acid residues participating in the intramolecular
bond are not
located at either the N-terminal or C-terminal position of the peptide
inhibitor. In certain
embodiments, neither of the two amino acid residues, e.g., Pen, particpating
in the intramolecular
bond is located at the N-terminal or C-terminal position of the peptide
inhibitor. In other words,
in certain embodiments, at least one, or both, of the two amino acid residues,
e.g., Pens,
participating in the intramolecular bond are internal amino acid residues of
the peptide inhibitor.
In certain embodiments, neither of the two amino acid residues, e.g., Pens,
participating in the
intramolecular bond is located at the C-terminal position of the peptide
inhibitor.
1003741In some embodiments, wherein a peptide of the invention is conjugated
to an acidic
compound such as, e.g., isovaleric acid, isobutyric acid, valeric acid, and
the like, the presence of
such a conjugation is referenced in the acid form. So, for example, but not to
be limited in any
way, instead of indicating a conjugation of isovaleric acid to a peptide by
referencing isovaleroyl
(e.g., is ovaleroyl- [Pen]-QTWQ [Pen] - [Phe(4-0Me)]- [2-Nal] - [a-MeLys]-
[Lys(Ac)]-NG-NH2 in
some embodiments, the present application references such a conjugation as
isovaleric acid-
[Pen]- Q TWQ [Pen] - [Phe(4-0Me)] - [2-Nal] - [a-MeLys] - [Lys(Ac)]-NG-NH2.
1003751 The present invention further includes peptide inhibitors that
selectively bind to an
epitope or binding domain present within amino acid residues 230 ¨ 349 of the
human IL23R
protein. In particular embodiments, the peptide inhibitor binds human IL23R
and not mouse IL-
23R. In certain embodiments, the peptide inhibitor also binds to rat IL-23R.
[00376] In certain embodiments of peptide inhibitors of Formula I, X4 is Abu;
X9 is Cys, Pen,
homocys, and the intramolecular bond is a thioether bond.
1003771In certain embodiments, peptide inhibitors do not include compounds,
disclosed in PCT
Application No. PCT/U52014/030352 or PCT Application No. PCT/U52015/038370.
120
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Illustrative Peptide Inhibitors Comprising Pen-Pen Disulfide Bonds
1003781In certain embodiments, the present invention includes a peptide
inhibitor of an
interleukin-23 receptor, wherein the peptide inhibitor has the structure of
Formula II:
R1-X-R2 (II)
[00379] or a pharmaceutically acceptable salt or solvate thereof,
wherein Rl is a bond, hydrogen, a C1-C6 alkyl, a C6-C12 aryl, a C6-C12 aryl, a
C1-C6 alkyl, a
C1-C20 alkanoyl, an alkylsulphonate, an acid, y-Glu or pG1u, appended to the N-
terminus, and
including PEGylated versions (e.g., 200 Da to 60,000 Da), alone or as a spacer
of any of the
foregoing;
[00380]R2 is a bond, OH or NH2; and
1003811X is an amino acid sequence of 8 to 20 amino acids or 8 to 35 amino
acids.
1003821In particular embodiments of peptide inhibitor of Formula II, X
comprises or consists of
the sequence of Formula Ha:
X1 -X2-X3-X4-X5-X6-X7-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Ha)
wherein
X1 is absent or any amino acid;
X2 is absent or any amino acid;
X3 is absent or any amino acid;
X4 is Pen, Cys or homo-Cys;
X5 is any amino acid;
X6 is any amino acid;
X7 is Trp, Bip, Gln, His, Glu(Bz1), 4-Phenylbenzylalanine, Tic, Phe[4-(2-
aminoethoxy)],
Phe(3,4-C12), Phe(4-0Me), 5-Hydroxy-Trp, 6-Chloro-Trp, N-MeTrp, a-Me-Trp,
1,2,3,4 -
tetrahydro-norharman, Phe(4-CO2H), Phe(4-CONH2), Phe(3,4-Dimethoxy), Phe(4-
CF3),
Phe(4-tBu), f3f3-diPheAla, Glu, Gly, Ile, Asn, Pro, Arg, Thr or Octgly, or a
corresponding a-
methyl amino acid form of any of the foregoing;
X8 is any amino acid;
121
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X9 is Pen, Cys or hCys;
X10 is 1-Na!, 2-Na!, Aic, Bip, (D)Cys, Cha, DMT, (D)Tyr, Glu, Phe, His, Trp,
Thr, Tic, Tyr, 4-
pyridylAla, Octgly, a Phe analog or a Tyr analog (optionally, Phe(3,4-F2),
Phe(3,4-C12), F(3-Me),
Phe[4-(2-aminoethoxy)], Phe[4-(2-(acetyl-aminoethoxy)], Phe(4-Br), Phe(4-
CONH2), Phe(4-C1),
Phe(4-CN), Phe(4-guanidino), Phe(4-Me), Phe(4-NH2), Phe(4-N3), Phe(4-0Me), or
Phe(4-
OBz1)), or a corresponding a-methyl amino acid form of any of the foregoing;
X11 is 2-Na!, 1-Na!, 2,4-dimethylPhe, Bip, Phe(3,4-C12), Phe (3,4-F2), Phe(4-
CO2H), f3hPhe(4-
F), a-Me-Trp, 4-phenylcyclohexyl, Phe(4-CF3), a-MePhe, f3hNal, f3hPhe, f3hTyr,
f3hTrp, Nva(5-
phenyl), Phe, His, hPhe, Tic, Tqa, Trp, Tyr, Phe(4-0Me), Phe(4-Me), Trp(2,5,7-
tri-tert-Butyl),
Phe(4-0ally1), Tyr(3-tBu), Phe(4-tBu), Phe(4-guanidino, Phe(4-0Bz1), Octgly,
Glu(Bz1), 4-
Phenylbenzylalanine, Phe[4-(2-aminoethoxy)], 5-Hydroxy-Trp, 6-Chloro-Trp, N-
MeTrp,
1,2,3,4-tetrahydro-norharman, Phe(4-CONH2), Phe(3,4-0Me2) Phe(2,3-C12),
Phe(2,3-F2), Phe(4-
F), 4-phenylcyclohexylalanine or Bip, or a corresponding a-methyl amino acid
form of any of
the foregoing;
X12 is a-MeLys, a-MeOrn, a-MeLeu, a-MeVal, 4-amino-4-carboxy-tetrahydropyran,
Achc
Acpc, Acbc, Acvc, MeLeu, Aib, (D)Ala, (D)Asn, (D)Leu, (D)Asp, (D)Phe, (D)Thr,
3-Pal, Aib,
f3-Ala, f3hGlu, f3hAla, PhLeu, f3hVal, f3-spiro-pip, Cha, Chg, Asp, Dab, Dap,
a-DiethylGly, Glu,
Phe, hLeu, hArg, hLeu, Ile, Lys, Leu, Asn, N-MeLeu, N-MeArg, Ogl, Orn, Pro,
Gln, Arg, Ser,
Thr or Tle, or a corresponding a-methyl amino acid form of any of the
foregoing;
X13 is Lys(Ac), (D)Asn, (D)Leu, (D)Thr, (D)Phe, Ala, Aib, a-MeLeu, f3-Ala,
f3hGlu, f3hAla,
PhLeu, f3hVal, f3-spiro-pip, Cha, Chg, Asp, Lys, Arg, Orn, Dab, Dap, a-
DiethylGly, Glu, Phe,
hLeu, Lys, Leu, Asn, Ogl, Pro, Gln, Asp, Arg, Ser, spiro-pip, Thr, Tba, Tlc,
Val or Tyr, or a
corresponding a-methyl amino acid form of any of the foregoing;
X14 is Asn, Glu, Phe, Gly, His, Lys, Leu, Met, Asn, Pro, Gln, Arg, Ser, Thr,
Tic or Tyr,
Lys(Ac), Orn or a corresponding a-methyl amino acid form of any of the
foregoing;
X15 is Gly, (D)Ala, (D)Asn, (D)Asp, Asn, (D)Leu, (D)Phe, (D)Thr, Ala, AEA,
Asp, Glu, Phe,
Gly, Lys, Leu, Pro, Gln, Arg or Ser, f3-Ala, Arg or a corresponding a-methyl
amino acid form of
any of the foregoing;
X16 is absent, Gly, Ala, Asp, Ser, Pro, Asn or Thr, or a corresponding a-
methyl amino acid form
of any of the foregoing;
122
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X17 is absent, Glu, Ser, Gly or Gin, or a corresponding a-methyl amino acid
form of any of the
foregoing;
X18 is absent or any amino acid;
X19 is absent or any amino acid; and
X20 is absent or any amino acid.
1003831In certain embodiments of Ha: X10 is 1-Nal, 2-Nal, Aic, Bip, (D)Cys,
Cha, DMT,
(D)Tyr, Glu, Phe, His, Trp, Thr, Tic, Tyr, 4-pyridylAla, Octgly, a Phe analog
or a Tyr analog, or
a corresponding a-methyl amino acid form of any of the foregoing; X11 is 2-
Nal, 1-Nal, 2,4-
dimethylPhe, Bip, Phe(3,4-C12), Phe (3,4-F2), Phe(4-CO2H), f3hPhe(4-F), a-Me-
Trp, 4-
phenylcyclohexyl, Phe(4-CF3), a-MePhe, f3hNal, f3hPhe, f3hTyr, f3hTrp, Nva(5-
phenyl), Phe,
His, hPhe, Tic, Tqa, Trp, Tyr, Phe(4-0Me), Phe(4-Me), Trp(2,5,7-tri-tert-
Butyl), Phe(4-0ally1),
Tyr(3-tBu), Phe(4-tBu), Phe(4-guanidino, Phe(4-0Bz1), Octgly, Glu(Bz1), 4-
Phenylbenzylalanine, Phe[4-(2-aminoethoxy)], 5-Hydroxy-Trp, 6-Chloro-Trp, N-
MeTrp,
1,2,3,4-tetrahydro-norharman, Phe(4-CONH2), Phe(3,4-Dimethoxy), Phe(2,3-C12),
Phe(2,3-F2),
Phe(4-F), 4-phenylcyclohexylalanine or Bip, or a corresponding a-methyl amino
acid form of
any of the foregoing; X12 is a-MeLys, a-MeOrn, a-MeLeu, MeLeu, Aib, (D)Ala,
(D)Asn,
(D)Leu, (D)Asp, (D)Phe, (D)Thr, 3-Pal, Aib, f3-Ala, f3hGlu, f3hAla, PhLeu,
f3hVal, f3-spiro-pip,
Cha, Chg, Asp, Dab, Dap, a-DiethylGly, Glu, Phe, hLeu, hArg, hLeu, Ile, Lys,
Leu, Asn, N-
MeLeu, N-MeArg, Ogl, Orn, Pro, Gin, Arg, Ser, Thr or Tie, or a corresponding a-
methyl amino
acid form of any of the foregoing; X13 is Lys(Ac), (D)Asn, (D)Leu, (D)Thr,
(D)Phe, Ala,
Aib, a-MeLeu, f3-Ala, f3hGlu, f3hAla, PhLeu, f3hVal, f3-spiro-pip, Cha, Chg,
Asp, Lys, Arg, Orn,
Dab, Dap, a-DiethylGly, Glu, Phe, hLeu, Lys, Leu, Asn, Ogl, Pro, Gin, Asp,
Arg, Ser, spiro-pip,
Thr, Tba, Tic, Val or Tyr, or a corresponding a-methyl amino acid form of any
of the foregoing;
X14 is Asn, Glu, Phe, Gly, His, Lys, Leu, Met, Asn, Pro, Gin, Arg, Ser, Thr,
Tic or Tyr, or a
corresponding a-methyl amino acid form of any of the foregoing; and X15 is
Gly, (D)Ala,
(D)Asn, (D)Asp, Asn, (D)Leu, (D)Phe, (D)Thr, Ala, AEA, Asp, Glu, Phe, Gly,
Lys, Leu, Pro,
Gin, Arg or Ser, or a corresponding a-methyl amino acid form of any of the
foregoing.
123
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1003841In certain embodiments, X3 is present. In particular embodiments, X3 is
Glu,(D)Glu,
Arg, (D)Arg, Phe, (D)Phe, 2-Na!, Thr, Leu, (D)Gln. In certain embodiments, X3
is (D)Arg or
(D)Phe. In particular embodiments, X1 and X2 are absent and X3 is present.
1003851In certain embodiments, X5 is Gln, Ala, Cit, Asp, Dab, Dap, Cit Glu,
Phe, Gly, His,
hCys, Lys, Leu, Met, Asn, N-Me-Ala, N-Me-Asn, N-Me-Lys, a¨Me-Lys, a¨Me-Orn, N-
Me-
Gln, N-Me-Arg, a-MeSer, Orn, Pro, Arg, Ser, Thr, or Val. In certain
embodiments, X5 is Gln,
Ala, Cit, Asp, Dab, Dap, Glu, Phe, Gly, His, hCys, Lys, Leu, Met, Asn, N-Me-
Ala, N-Me-Asn,
N-Me-Lys, aMe-Lys, aMe-Orn, N-Me-Gln, N-Me-Arg, Orn, Pro, Arg, Ser, Thr, or
Val. In
certain embodiments, X5 is Gln or Asn.
[00386] In certain embodiments, X6 is Thr, Asp, Glu, Phe, Asn, Pro, Arg, or
Ser.
[00387] In certain embodiments, X7 is Trp.
[00388] In certain embodiments, X8 is Gln, Glu, Phe, Lys, Asn, Pro, Arg, Val,
Thr, or Trp.
[00389] In certain embodiments, X10 is a Tyr analog or a Phe analog. In
particular embodiments,
X10 is a Phe analog.
1003901In certain embodiments wherein X10 is a Phe analog, X10 is selected
from hPhe, Phe(4-
0Me), a-Me-Phe, hPhe(3,4-dimethoxy), Phe(4-CONH2), Phe(4-phenoxy), Phe(4-
guanadino),
Phe(4-tBu), Phe(4-CN), Phe(4-Br), Phe(4-0Bz1), Phe(4-NH2), Phe(4-F), Phe(3,5
DiF),
Phe(CH2CO2H), Phe(penta-F), Phe(3,4-C12), Phe(4-CF3), f33-diPheAla, Phe(4-N3)
and Phe[4-(2-
aminoethoxy)]. In particular embodiments, X10 is Phe(4-0Me) or Phe[4-(2-
aminoethoxy)]. In
particular embodiments, X10 is Phe(4-0Me), Phe(4-CONH2) or Phe[4-(2-
aminoethoxy)]. In
certain embodiments where X10 wherein X10 is a Phe analog, X10 is selected
from hPhe, Phe(4-
0Me), a-Me-Phe, hPhe(3,4-dimethoxy), Phe(4-CONH2), Phe(4-phenoxy), Phe(4-
guanadino),
Phe(4-tBu), Phe(4-CN), Phe(4-Br), Phe(4-0Bz1), Phe(4-NH2), Phe(4-F), Phe(3,5
DiF),
Phe(CH2CO2H), Phe(penta-F), Phe(3,4-C12), Phe(4-CF3), f33-diPheAla, Phe(4-N3)
and Phe[4-(2-
aminoethoxy)]. In particular embodiments, X10 is Phe(4-0Me).
10039111n certain embodiments where X10 is a Tyr analog, X10 is selected from
hTyr, a-MeTyr,
N-Me-Tyr, Tyr(3-tBu), Phe(4-CONH2), Phe[4-(2-aminoethoxy)], and bhTyr. In
certain
124
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
embodiments where X10 is a Tyr analog, X10 is selected from hTyr, a-MeTyr, N-
Me-Tyr,
Tyr(3-tBu), and bhTyr.
1003921In certain embodiments, X10 is Tyr, Phe(4-0Me), Phe[4-(2-aminoethoxy)],
Phe(4-
CONH2), or 2-Nal. In certain embodiments, X10 is Phe(4-0Me) or Phe[4-(2-
aminoethoxy)]. In
certain embodiments, X10 is not Tyr.
In certain embodiments, X11 is a Trp analog. In particular embodiments, X11 is
2-Nal or 1-Nal.
In certain embodiments, X11 is 2-Nal.
[00393] In certain embodiments, X12 is Aib, a-MeLys or a¨MeLeu.
[00394] In particular embodiments of a peptide inhibitor of Formula II, one or
both of X4 or X9 is
Pen. In particular embodiments, both X4 and X9 are Pen.
1003951In certain embodiments, the peptide inhibitor of Formula II is
cyclized. In particular
embodiments, the peptide inhibitor of Formula II is cyclized via an
intramolecular bond between
X4 and X9. In particular embodiments, the intramolecular bond is a disulfide
bond. In particular
embodiments, X4 and X9 are both Pen.
1003961In certain embodiments, the peptide inhibitor of Formula II is linear
or not cyclized. In
particular embodiments of the linear peptide inhibitor of Formula I, X4 and/or
X9 are any amino
acid.
[00397] In particular embodiments of a peptide inhibitor of Formula II, one or
more, two or more,
or all three of Xi, X2, and X3 are absent. In certain embodiments, X1 is
absent. In certain
embodiments, X1 and X2 are absent. In certain embodiments, Xi, X2 and X3 are
absent.
[00398] In particular embodiments of a peptide inhibitor of Formula II, one or
more, two or more,
three or more, four or more, or all of X16, X17, X18, X19 and X20 are absent.
In particular
embodiments of a peptide inhibitor of Formula I, one or more, two or more,
three or more, or all
of X17, X18, X19 and X20 are absent. In certain embodiments, one or more, two
or more, or all
three of X17, X19 and X20 are absent. In certain embodiments, one or more of
Xi, X2 and X3
125
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
are absent; and one or more, two or more, three or more, or four of X17, X18,
X19 and X20 are
absent.
[00399] In particular embodiments of a peptide inhibitor of Formula II, X18 is
(D)-Lys. In certain
embodiments, X18 is (D)-Lys and X17 is absent.
1004001In particular embodiments of a peptide inhibitor of Formula II, the
peptide inhibitor
comprises one or more, two or more, three or more, or four of the following
features: X5 is Asn,
Arg or Gln; X6 is Thr; X7 is Trp; and X8 is Gln. In particular embodiments of
a peptide inhibitor
of Formula I, X4 is Pen; X5 is Gln, Asn or Arg; X6 is Thr; X7 is Trp, 5-
hydroxy-Trp, 6-chloro-
Trp, N-MeTrp, alpha-Me-Trp, or 1,2,3,4-tetrahydro-norharman; X8 is Gln; and X9
is Pen. In
particular embodiments, X5 is Gln. In certain embodiments, Xl, X2 and X3 are
absent. In
particular embodiments, both X4 and X9 are Pen.
1004011In particular embodiments of a peptide inhibitor of Formula II, the
peptide inhibitor
comprises one or more, two or more, three or more, four or more, five or more,
six or more, or
seven of the following features: X10 is Tyr, a Phe analog, a Tyr analog or 2-
Nal; X11 is Trp, 5-
hydroxy-Trp, 6-chloro-Trp, N-MeTrp, alpha-Me-Trp, 1,2,3,4-tetrahydro-
norharman, 2-Nal or 1-
Nal; X12 is Aib, a-MeLys, a-MeOrn and a-MeLeu; X13 is Lys, Glu or Lys(Ac); X14
is Phe or
Asn; X15 is Gly, Ser or Ala; and X16 is absent or AEA. In certain embodiments,
X10 is Tyr,
Phe(4-0Me), Phe[4-(2-aminoethoxy)], Phe(CONH2), or 2-Nal. In certain
embodiments, X11 is
2-Nal or 1-Nal. In certain embodiments, X10 is not Tyr. In certain
embodiments, Xl, X2 and X3
are absent. In particular embodiments, both X4 and X9 are Pen.
1004021In particular embodiments of a peptide inhibitor of Formula II, the
peptide inhibitor
comprises one or more, two or more, three or more, four or more, five or more,
six or more,
seven or more, eight or more, nine or more, ten or more, or eleven of the
following features: X5
is Arg or Gln; X6 is Thr; X7 is Trp; X8 is Gln; X10 is a Phe analog; X11 is
Trp, 2-Nal or 1-Nal;
X12 is Aib, a-MeLys or a-MeOrn; X13 is Lys, Glu or Lys(Ac); X14 is Asn; X15 is
Gly, Ser or
Ala; and X16 is absent or AEA. In certain embodiments, X10 is Phe(4-0Me) or
Phe[4-(2-
aminoethoxy)]. In certain embodiments, X11 is 2-Nal or 1-Nal. In certain
embodiments, Xl, X2
and X3 are absent. In particular embodiments, both X4 and X9 are Pen.
126
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[00403] In particular embodiments of a peptide inhibitor of Formula II, the
peptide is cyclized via
X4 and X9; X4 and X9 are Pen; X5 is Gln; X6 is Thr; X7 is Trp; X8 is Gln; X10
is Tyr, a Phe
analog or 2-Nal; X11 is Trp, 2-Nal or 1-Nal; X12 is Arg, a¨MeLys, a-MeOrn, or
a-MeLeu;
X13 is Lys, Glu or Lys(Ac); X14 is Phe or Asn; X15 is Gly, Ser or Ala; and X16
is absent. In
certain embodiments, X10 is Tyr, Phe(4-0Me), Phe[4-(2-aminoethoxy)], Phe(4-
0Me) or 2-Nal.
In certain embodiments, X10 is Phe(4-0Me). In certain embodiments, X10 is not
Tyr. In certain
embodiments, X11 is 2-Nal or 1-Nal. In certain embodiments, X1 , X2 and X3 are
absent.
1004041 In particular embodiments of a peptide inhibitor of Formula II, the
peptide is cyclized via
X4 and X9; X4 and X9 are Pen; X5 is Gln; X6 is Thr; X7 is Trp; X8 is Gln; X10
is Tyr, Phe(4-
OMe) or 2-Nal; X11 is Trp, 2-Nal or 1-Nal; X12 is Arg, a-MeLys or a-MeOrn; X13
is Lys, Glu
or Lys(Ac); X14 is Phe or Asn; X15 is Gly; and X16 is absent. In certain
embodiments, X10 is
Phe(4-0Me). In certain embodiments, X11 is 2-Nal or 1-Nal. In certain
embodiments, Xi, X2
and X3 are absent.
1004051 In particular embodiments of a peptide inhibitor of Formula II, the
peptide is cyclized via
X4 and X9; X4 and X9 are Pen; X5 is Gln; X6 is Thr; X7 is Trp; X8 is Gln; X10
is Phe(4-0Me)
or Phe[4-(2-aminoethoxy)]; X11 is Trp, 2-Nal or 1-Nal; X12 is a¨MeLys, a-
MeOrn, or a-
MeLeu; X13 is Lys, Glu or Lys(Ac); X14 is Asn; X15 is Gly, Ser or Ala; and X16
is absent. In
certain embodiments, X10 is Phe(4-0Me). In certain embodiments, X11 is 2-Nal
or 1-Nal. In
certain embodiments, Xi, X2 and X3 are absent.
[00406] In particular embodiments of a peptide inhibitor of Formula II, X10 is
not Tyr.
1004071In certain embodiments, the present invention includes a peptide,
optionally 8 to 35, 8 to
20, 8 to 16 or 8 to 12 amino acids in length, optionally cyclized, comprising
or consisting of
having a core sequence of Formula IIb:
Pen-Xaa5-Xaa6-Trp-Xaa8-Pen-Xaa10- [(2-Nal)] (II13)
[00408] wherein Xaa5, Xaa6 and Xaa8 are any amino acid residue; and Xaal 0 is
a Phe analogue,
wherein the peptide inhibits binding of IL-23 to IL-23R. In particular
embodiments, X10 is a
Phe analog selected from a¨Me-Phe, Phe(4-0Me), Phe(4-0Bz1), Phe(4-0Me), Phe(4-
CONH2),
127
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Phe(3,4-C12), Phe(4-tBu), Phe(4-NH2), Phe(4-Br), Phe(4-CN), Phe(4-CO2H), Phe[4-
(2-
aminoethoxy)] or Phe(4-guanadino). In particular embodiments, Xaal 0 is Phe(4-
0Me) or
Phe[4-(2-aminoethoxy)]. In one embodiment, Xaal 0 is Phe(4-0Me). In certain
embodiments,
the peptide is cyclized via an intramolecular bond between Pen at Xaa4 and Pen
at Xaa9. In
particular embodiments, the peptide is a peptide inhibitor of Formula II, and
wherein in certain
embodiments, Xl, X2 and X3 are absent. In particular embodiments, the peptide
inhibits the
binding of IL-23 to IL-23R. In certain embodiments, a peptide of Formula IIb
further comprises
an amino acid bound to the N-terminal Pen residue. In particular embodiments,
the bound amino
acid is Glu,(D)Glu, Arg, (D)Arg, Phe, (D)Phe, 2-Nal, Thr, Leu, or (D)G1n. In
certain
embodiments, it is is (D)Arg or (D)Phe.
1004091In certain embodiments, the present invention includes a peptide,
optionally 8 to 35, 8 to
20, 8 to 16, or 8 to 12 amino acids in length, optionally cyclized, comprising
or consisting of a
core sequence of Formula IIc:
Pen-Xaa5-Xaa6-Trp-Xaa8-Pen-Xaa10- [(2-Nal)] (IIc)
1004101 wherein Xaa5, Xaa6 and Xaa8 are any amino acid residue; and Xaal 0 is
Tyr, a Phe
analog, a-Me-Tyr, a-Me-Trp or 2-Nal, wherein the peptide inhibits binding of
IL-23 to IL-23R.
In certain embodiments, X10 is Tyr, Phe(4-0Me), Phe[4-(2-aminoethoxy)], a-Me-
Tyr, a¨Me-
Phe, a-Me-Trp or 2-Nal. In certain embodiments, Xaal 0 is Tyr, Phe(4-0Me),
Phe(CONH2),
Phe[4-(2-aminoethoxy)] or 2-Nal. In certain embodiments, Xaal 0 is Tyr, Phe(4-
0Me), Phe[4-(2-
aminoethoxy)] or 2-Nal. In particular embodiments, Xaal 0 is Phe(4-0Me) or
Phe[4-(2-
aminoethoxy)]. In one embodiment, Xaal 0 is Phe[4-(2-aminoethoxy)] or
Phe(CONH2). In
particular embodiments, Xaal 0 is Phe(4-0Me) or Phe[4-(2-aminoethoxy)]. In one
embodiment,
Xaal 0 is Phe[4-(2-aminoethoxy)]. In certain embodiments, Xaal 0 is not Tyr.
In certain
embodiments, the peptide is cyclized via an intramolecular bond between Pen at
Xaa4 and Pen at
Xaa9. In particular embodiments, the peptide is a peptide inhibitor of Formula
II, and wherein in
certain embodiments, Xl, X2 and X3 are absent. In particular embodiments, the
peptide inhibits
the binding of IL-23 to IL-23R. In certain embodiments, a peptide of Formula
IIc further
comprises an amino acid bound to the N-terminal Pen residue. In particular
embodiments, the
128
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
bound amino acid is Glu,(D)Glu, Arg, (D)Arg, Phe, (D)Phe, 2-Na!, Thr, Leu, or
(D)G1n. In
certain embodiments, it is is (D)Arg or (D)Phe.
10041111n certain embodiments, the present invention includes a peptide,
optionally 8 to 35, 8 to
20, 8 to 16 or 8 to 12 amino acids in length, optionally cyclized, comprising
or consisting of a
core sequence of Formula lid:
Pen-Xaa5-Xaa6-Trp-Xaa8-Pen- Phe [4 -(2 -aminoethoxy)] - [2-Na!] (lid)
[00412] wherein Xaa5, Xaa6 and Xaa8 are any amino acid residue. In certain
embodiments, the
peptide comprises a disulfide bond between Xaa4 and Xaa9. In certain
embodiments, the peptide
is a peptide inhibitor of Formula I, and wherein in certain embodiments, X1 ,
X2 and X3 are
absent. In particular embodiments, the peptide inhibits the binding of IL-23
to IL-23R. In certain
embodiments, a peptide of Formula lid further comprises an amino acid bound to
the N-terminal
Pen residue. In particular embodiments, the bound amino acid is Glu,(D)Glu,
Arg, (D)Arg, Phe,
(D)Phe, 2-Na!, Thr, Leu, or (D)G1n. In certain embodiments, it is is (D)Arg or
(D)Phe.
[00413] In particular embodiments of a peptide inhibitor of Formula II, the
peptide inhibitor has a
structure shown in any of Tables 2, 3, 4A, 4B, 8, 11 or 15 or comprises an
amino acid sequence
set forth in Tables 2, 3, 4A, 4B, 8, 11 or 15.
Table 2. Illustrative Di-Pen Inhibitors
[Palm]-[isoGluHPEG4HPenl-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-
NalHAibHLys(Ac)]-NN-N H2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(PEG4-isoGlu-Palm)]-
NN-NH2
Ac-[Per]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-[a-MeLys(Ac)HLys(Ac)]-NN-NH2
[Octanyl]-[lsoGluHPEG4HPerd-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-
NalHAibHLys(Ac)]-NN-NH2
[Octany1]-[PEG4HPerd-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(Ac)]-NN-
NH2
[Palm]-[PEG4HPenl-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(Ac)]-NN-N
H2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(PEG4-Octany1)]-NN-
NH2
129
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-NalHAibHLys(PEG4-Palm)]-NN-N
H2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)-(PEG4-Palm)H2-NalHAibHLys(Ac)1NN-
NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)-(PEG4-Lauryl)H2-NalHAibHLys(Ac)]-NN-
NH2
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal[a-MeLys(PEG4-Palm)-[Lys(Ac)]-NN-N H2
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal[a-MeLys(PEG4-Lauryl)HLys(Ac)]-NN-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)-(PEG4-lsoGlu-Palm)H2-
NalHAibHLys(Ac)]-NN-NH2
Ac-[Pen]-NTWQ-[Pen]- [Phe[4-(2-arninoethoxy)-(PEG4-IsoGLu-Lauryl)H2-
NalHAibHLys(Ac)]-NN-NH2
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-[a-MeLys(PEG4-IsoGlu-Palm)HLys(Ac)]-
NN-NH2
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-a-Me-K(PEG4-IsoGlu-Lauryl)HLys(Ac)]-
NN-NH2
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-[a-MeLys(IVA)HLys(Ac)]-NN-NH2
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-[a-MeLys(Biotin)HLys(Ac)]-NN-NH2
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-[a-MeLys(Octanyl)HLys(Ac)]-NN-N H2
Ac-[Pen]Lys(IVA)]-TWQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-Nal]-[AibHLys(Ac)]-NN-N
H2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(Ac)HLys(IVA)1-N-N
H2
Ac-[Pen]- [Lys(Biotin)l-TWQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-NalHAibHLys(Ac)]-
NN-NH2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-NalHAibHLys(Ac)HLys(Biotin)]-N-
N H2
Ac-[Pen]-[Lys(octany1)]-TWQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-NalHAibHLys(Ac)]-
NN-NH2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-NalHAibHLys(Ac)HLys(octany1)]-
N-NH2
Ac-[Pen]-[Lys(Palm)]-TWQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-Nall--[Aib]-
[Lys(Ac)]-NN-N H2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-NalHAibHLys(Ac)]-Lys(Palm)]-N-
NH2
Ac-[Pen]- [Lys(PEG8)1-TWQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-Nal]--[Aib]-
[Lys(Ac)]-NN-NH2
130
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]--[Aib]-
[Lys(Ac)HLys(PEG8)]-N-NH2
Ac-[Pen]-K(Pegll-Palm)TWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(Ac)]-NN-
NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall--[Aib]-[Lys(Ac)HLys(Pegll-
palm)]-N-NH2
Ac-[Pen]-[Citl-TW-[Cit]-[PenHPhe[4-(2-aminoethoxy)]-[2-Nal]--[Aib]-[Lys(Ac)]-
NN-NH2
Ac-[Pen]-[Lys(Ac)]-TW-[Cit]-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-
[AibHLys(Ac)]-NN-NH2
Ac-[Pen]-NT-[Phe(3,4-0CH3)2]-Q-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-
NalHAibHLys(Ac)]-NN-NH2
Ac-[Pen]-NT-[Phe(2,4-CH3)2]-Q-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-
[AibHLys(Ac)]-NN-NH2
Ac-[Pen]-NT-[Phe(3-CH3)]-Q-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall--[Aib]-
[Lys(Ac)]-NN-NH2
Ac-[Penl-NT-[Phe(4-CH3)]-Q-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(Ac)]-
NN-NH2
Ac[(D)Arg]-[Penl-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall--[Aib]-[Lys(Ac)]-N-
[13Ala]-NH2
Ac-[(D)Tyr]-[Pen]-NTWQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-NalHAibHLys(Ac)]-N-
[13Ala]-NH2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(Ac)]-QN-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibHLys(Ac)HLys(Ac)]-N-N H2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2_aminoethoxy)H2-NalHAibHLys(Ac)]-N-[Lys(Ac)]-NH2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibHLys(Ac)]-QQ-NH2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2_aminoethoxy)H2-NalHAibHLys(Ac)1-Q-U3Alal-NH2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-NalHAibHLys(Ac)]-N-[Cit]-NH2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-NalHAibHLys(Ac)HCitl-NNH2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(Ac)HCitl-Q-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(Ac)HCitHLys(Ac)]-N
H2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Na IHAibl-[Lys(Ac)]-[Lys(Ac)]-
[Citl-N H2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-QN-[13Alal-NH2
131
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-E-[Cit]-Q-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-CitNCitNH2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHCitl-Q-[Cit]-NH2
Ac-[Pen]-[Citl-TWQ-[Pen]-[Phe[4-(2-arninoethoxy)]-[2-NalHAibHLys(Ac)]-NN-NH2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(Ac)]-NN-NH2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-CINN-N H2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-ENCI-NH2
Ac-[Pen]-GPWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[AibHLys(Ac)]-NN-N H2
Ac-[Pen]-PGWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[AibHLys(Ac)]-NN-N H2
Ac-[Pen]-NTVVN-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(Ac)]-NN-NH2
Ac-[Pen]-NSWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibHLys(Ac)]-NN-NH2
Ac-[Pen]-N-[Aib]-WQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[AibHLys(Ac)]-NN-NH2
Ac-[Pen]-NTVV-[Aib]-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-NalHAibHLys(Ac)]N-[Aib]-
NH2
Ac-[Pen]-QTW-[Lys(Ac)]-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[AibHLys(Ac)]-NN-
NH2
Ac-[Pen]-[Lys(Ac)]-TWQ-[Pen]-[Phe[4-(2-arninoethoxy)]-[2-Nall--[Aib]-
[Lys(Ac)]NNNH2
Ac-[Pen]-QVWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[AibHLys(Ac)]-NN-N H2
Ac-[Penl-NT-[2-Nal]-Q-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]- [AibHLys(Ac)]-NN-
NH2
Ac-[Penl-NT-[1-Nal]-Q-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]- [AibHLys(Ac)]-NN-
NH2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-NalHa-MeLeuHLys(Ac)]-NN-N H2
Ac-[Pen]-NTVVQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-NalHa-MeLysHLys(Ac)]-NN-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-NN-
NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHa-MeLeuHLys(Ac)]-N-U3Alal-
NH2
132
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHa-MeLysHLys(Ac)]-N-U3Alal-
NH2
Ac-[Perd-NTWQ-[PenHPhe[4-(2-aminoethoxy)H2-Nall-[4-amino-4-carboxy-
tetrahydropyranHLys(AcH-N-
[13Ala]-NH2
Ac-[Per]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)1-[2-Nal]-[AibHLys(Ac)]-LN-NH2
Ac-[Per]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)1-[2-Nal]-[AibHLys(Ac)]-GN-N H2
Ac-[Per]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[AibHLys(Ac)]-SN-N H2
Ac-[Per]-NTVVQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-Nal]-[AibHLys(Ac)HAibl-N-NH2
Ac-[Per]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)1-[2-Nal]-[AibHLys(Ac)]-FN-N H2
Ac-[Per]-NTW-[Cit]-[PenHPhe[4-(2-aminoethoxy)]-[2-NalHAibHLys(Ac)]-NN-NH2
Ac-[Per]-NTVVQ-[Pen]-[Phe[4-(2_aminoethoxy)]-[2-Nal]-[AibHLys(Ac)HTicH13Alal-
NH2
Ac-[Perd-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibHLys(Ac)HnLeu]-[13Alal-NH2
Ac-[Perd-NTVVQ-[PenHPhe[4-(2_aminoethoxy)1-[2-NalHAibHLys(Ac)]-G-[13Alal-NH2
Ac-[Perd-NTVVQ-[PenHPhe[4-(2_aminoethoxy)1-[2-NalHAibHLys(Ac)]-R-[13Alal-N H2
Ac-[Perd-NTVVQ-[PenHPhe[4-(2-aminoethoxy)1-[2-Nall--[AibHLys(Ac)]-W-[13Alal-
NH2
Ac-[Perd-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-Nall--[AibHLys(Ac)]-S-[13Alal-NH2
Ac-[Perd-NTVVQ-[PenHPhe[4-(2_aminoethoxy)1-[2-Nall--[AibHLys(Ac)]-L-[13Alal-
NH2
Ac-[Perd-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-Nall--[AibHLys(Ac)HAIBHI3Alal-NH2
Ac-[Perd-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-Nall--[AibHLys(Ac)HN-MeAlal-
U3Alal-NH2
Ac-[Perd-NTVVQ-[PenHPhe[4-(2_aminoethoxy)1-[2-NalHAibHLys(Ac)H2-NapH13Alal-NH2
Ac-[Perd-NTVVQ-[PenHPhe[4-(2_aminoethoxy)1-[2-NalHAibHLys(Ac)]-F-[13Alal-NH2
Ac-[(D)ArgHPerd-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-Nall-[ 4-arnino-4-carboxy-
tetrahydropyrard-
[Lys(AcH-NN-NH2
Table 3. Illustrative Peptides Containing the Ac-[Pen]-)0(WX-[Pen]-)00(X Motif
and
Analogues Thereof
133
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NN-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNE-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNF-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNK-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNN-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNW-NH
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNG-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNT-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNPK-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNPG-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNEP-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGK-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNPT-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNKGF-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGW-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGO-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGGG-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNKKK-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNEEE-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNFFF-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNTTT-N H2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGGGR-NH2
134
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGGGF-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGGGE-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGGGQ-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGGGT-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]- [Lys(Ac)]-
NNGGGGR-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGGGGF-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGGGGE-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGGGGQ-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNGGGGT-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNRRRRR-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNFFFFF-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNEEEEE-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNQQQQQ-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NNTTTTT-NH2
Ac-GGG-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(Ac)]-NN-NH2
Ac-RRR-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(Ac)]-NN-NH2
Ac-FFF-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(Ac)]-NN-NH2
Ac-EEE-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(Ac)]-NN-NH2
Ac-QQQ-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(Ac)]-NN-NH2
Ac-TTT-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(Ac)]-NN-NH2
Ac-RG-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NN-NH2
Ac-FG-[Perd-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyranHLys(AcH-
NN-NH2
135
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Ac-EG-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NN-NH2
Ac-C1G-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(AcH-NN-NH2
Ac-TG-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Palm)]-
NN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(isoGlu-
Palm)]-NN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(PEG11-
Palm)]-NN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ahx-
Palm)]-NN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(isoGlu-
Ahx-Palm)]-NN-NH2
[Palm]-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(AcH-NN-N H2
[Palm-isoGlu]-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-tetrahydropyran]-
[Lys(AcH-NN-NH2
[Palm-PEG11]-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-tetrahydropyran]-
[Lys(AcH-NN-NH2
[Palm-Ahx]-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(AcH-NN-NH2
[Palm-Ahx-isoGlu]-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-
tetrahydropyran]-[Lys(Ac)]-NN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NN-Lys[Palml-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NN-Lys[isoGlu-Palm]-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NN-Lys[PEG11-Palm]-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NN-Lys[Ahx-Palm]-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-
NN-Lys[isoGlu-Ahx-Palm]-NH2
Illustrative Peptide Inhibitors Comprising Thioether Bonds
10041411n certain embodiments, the present invention includes a peptide
inhibitor of an
interleukin-23 receptor, wherein the peptide inhibitor has the structure of
Formula III:
136
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
R1-X-R2 (III)
[00415] or a pharmaceutically acceptable salt or solvate thereof,
[00416] wherein 1Z1 is a bond, hydrogen, a C1-C6 alkyl, a C6-C12 aryl, a C6-
C12 aryl, a C1-C6
alkyl, a C1-C20 alkanoyl, an alkylsulphonate, an acid, y-Glu or pG1u, appended
to the N-
terminus, and including PEGylated versions (e.g., 200 Da to 60,000 Da), alone
or as a spacer of
any of the foregoing;
1004171R2 is a bond, OH or NH2; and
1004181X is an amino acid sequence of 8 to 20 amino acids or 8 to 35 amino
acids,
[00419] In particular embodiments of peptide inhibitors of Formula III, X
comprises or consists of
the sequence of Formula Ma:
X1 -X2-X3-X4-X5-X6-X7-X8-X9-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20 (Ma)
wherein
X1 is absent or any amino acid;
X2 is absent or any amino acid;
X3 is absent or any amino acid;
X4 is Abu, Pen, or Cys;
X5 is any amino acid;
X6 is any amino acid;
X7 is Trp, Bip, Gln, His, Glu(Bz1), 4-Phenylbenzylalanine, Tic, Phe[4-(2-
aminoethoxy)],
Phe(3,4-C12), Phe(4-0Me), 5-Hydroxy-Trp, 6-Chloro-Trp, N-MeTrp, a-MeTrp,
1,2,3,4 -
tetrahydro-norharman, Phe(4-CO2H), Phe(4-CONH2), Phe(3,4-(OCH3)2), Phe(4-CF3),
f3f3-
diPheAla, Phe(4-tBu), Glu, Gly, Ile, Asn, Pro, Arg, Thr or Octgly, or a
corresponding a-methyl
amino acid form of any of the foregoing;
X8 is any amino acid;
X9 is Abu, Pen, or Cys;
X10 is 1-Nal, 2-Nal, Aic, Bip, (D)Cys, Cha, DMT, (D)Tyr, Glu, Phe, His, Trp,
Thr, Tic, Tyr, 4-
pyridylAla, Octgly a Phe analog or a Tyr analog (optionally, Phe(3,4-F2),
Phe(3,4-C12), F(3-Me),
137
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Phe[4-(2-aminoethoxy)], Phe[4-(2-(acetyl-aminoethoxy)], Phe(4-Br), Phe(4-
CONH2), Phe(4-C1),
Phe(4-CN), Phe(4-guanidino), Phe(4-Me), Phe(4-NH2), Phe(4-N3), Phe(4-0Me),
Phe(4-0Bz1)),
or a corresponding a-methyl amino acid form of any of the foregoing;
X11 is 2-Na!, 1-Na!, 2,4-dimethylPhe, Bip, 4-phenylcyclohexyl, Glu(Bz1), 4-
Phenylbenzylalanine, Tic, Phe[4-(2-aminoethoxy)], Phe(3,4-C12), Phe(3,4-F2),
f3hPhe(4-F),
Phe(4-0Me), 5-Hydroxy-Trp, 6-Chloro-Trp, N-MeTrp, a-MeTrp, 1,2,3,4 -tetrahydro-
norharman, Phe(4-CO2H), Phe(4-CONH2), Phe(3,4-Dimethoxy), Phe(4-CF3), Phe(2,3-
C12),
Phe(3,4-C12), Phe(2,3-F2),Phe(4-F), 4-phenylcyclohexylalanine, a-MePhe,
f3hNal, f3hPhe, f3hTyr,
f3hTrp, Bip, Nva(5-phenyl), Phe, His, hPhe, Tqa, Trp, Tyr, Phe(4-Me),
Trp(2,5,7-tri-tertButyl),
Phe(4-0Ally1), Tyr(3-tBu), Phe(4-tBu), Phe(4-guanidino), Phe(4-0Bz1), or
Octgly, or a
corresponding a-methyl amino acid form of any of the foregoing;
X12 is a-MeLys, a-MeOrn, a-MeLeu, MeLeu, Aib, (D)Ala, (D)Asn, (D)Leu, (D)Asp,
(D)Phe,
(D)Thr, 3-Pal, Aib, f3-Ala, f3hGlu, f3hAla, PhLeu, f3hVal, f3-spiro-pip, Cha,
Chg, Asp, Dab, Dap,
a-DiethylGly, Glu, Phe, hLeu, hArg, hLeu, Ile, Lys, Leu, Asn, N-MeLeu, N-
MeArg, Ogl, Orn,
Pro, Gln, Arg, Ser, Thr or Tle, amino-4-carboxy-tetrahydropyran (THP), Achc
Acpc, Acbc,
Acvc, Aib, or a corresponding a-methyl amino acid form of any of the
foregoing;
X13 is Lys Lys(Ac), (D)Asn, (D)Leu, (D)Thr, (D)Phe, Ala, Aib, a-MeLeu, f3Ala,
f3hGlu, f3hAla,
PhLeu, f3hVal, f3-spiro-pip, Cha, Chg, Asp, Arg, Orn, Dab, Dap, a-DiethylGly,
Glu, Phe, hLeu,
Lys, Leu, Asn, Ogl, Pro, Gln, Asp, Arg, Ser, spiro-pip, Thr, Tba, Tlc, Val or
Tyr, or a
corresponding a-methyl amino acid form of any of the foregoing;
X14 is Asn, Glu, Phe, Gly, His, Lys, Lys (Ac), Leu, Met, Asn, Pro, Gln, Arg,
Ser, Thr, Tic, Asp
or Tyr, or a corresponding a-methyl amino acid form of any of the foregoing;
X15 is Gly, (D)Ala, (D)Asn, (D)Asp, Asn, (D)Leu, (D)Phe, (D)Thr, Ala, AEA,
Asp, Glu, Phe,
Gly, Lys, Leu, Pro, Gln, Arg, f3-Ala, or Ser, or a corresponding a-methyl
amino acid form of
any of the foregoing;
X16 is absent, Gly, Ala, Asp, Ser, Pro, Asn or Thr, or a corresponding a-
methyl amino acid form
of any of the foregoing;
X17 is absent, Glu, Ser, Gly or Gln, or a corresponding a-methyl amino acid
form of any of the
foregoing;
X18 is absent or any amino acid;
138
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X19 is absent or any amino acid; and
X20 is absent or any amino acid.
1004201In certain embdoiments, X14 is Asn, Glu, Phe, Gly, His, Lys, Lys (Ac),
Leu, Met, Asn,
Pro, Gin, Arg, Ser, Thr, Tic, or Tyr, or a corresponding a-methyl amino acid
form of any of the
foregoing
[00421]In certain embodiments of Ma: X7 is Trp, Bip, Gin, His, Glu(Bz1), 4-
Phenylbenzylalanine, Tic, Phe[4-(2-aminoethoxy)], Phe(3,4-C12), Phe(4-0Me), 5-
Hydroxy-Trp,
6-Chloro-Trp, N-MeTrp, a-MeTrp, 1,2,3,4 -tetrahydro-norharman, Phe(4-CO2H),
Phe(4-
CONH2), Phe(3,4-Dimethoxy), Phe(4-CF3), f33-diPheAla, Phe(4-tBu), Glu, Gly,
Ile, Asn, Pro,
Arg, Thr or Octgly, or a corresponding a-methyl amino acid form of any of the
foregoing; X10
is 1-Nal, 2-Nal, Aic, Bip, (D)Cys, Cha, DMT, (D)Tyr, Glu, Phe, His, Trp, Thr,
Tic, Tyr, 4-
pyridylAla, Octgly a Phe analog or a Tyr analog, or a corresponding a-methyl
amino acid form
of any of the foregoing; X11 is 2-Nal, 1-Nal, 2,4-dimethylPhe, Bip, 4-
phenylcyclohexyl,
Glu(Bz1), 4-Phenylbenzylalanine, Tic, Phe[4-(2-aminoethoxy)], Phe(3,4-C12),
Phe(3,4-F2),
f3hPhe(4-F), Phe(4-0Me), 5-Hydroxy-Trp, 6-Chloro-Trp, N-MeTrp, a-MeTrp,
1,2,3,4 -
tetrahydro-norharman, Phe(4-CO2H), Phe(4-CONH2), Phe(3,4-Dimethoxy), Phe(4-
CF3),
Phe(2,3-C12), Phe(2,3-F2),Phe(4-F), 4-phenylcyclohexylalanine, a-MePhe,
f3hNal, f3hPhe, f3hTyr,
f3hTrp, Bip, Nva(5-phenyl), Phe, His, hPhe, Tqa, Trp, Tyr, Phe(4-Me),
Trp(2,5,7-tri-tertButyl),
Phe(4-0Ally1), Tyr(3-tBu), Phe(4-tBu), Phe(4-guanidino), Phe(4-0Bz1), or
Octgly, or a
corresponding a-methyl amino acid form of any of the foregoing; X12 is a-
MeLys, a-MeOrn,
a-MeLeu, MeLeu, Aib, (D)Ala, (D)Asn, (D)Leu, (D)Asp, (D)Phe, (D)Thr, 3-Pal,
Aib, f3-Ala,
f3hGlu, f3hAla, PhLeu, f3hVal, f3-spiro-pip, Cha, Chg, Asp, Dab, Dap, a-
DiethylGly, Glu, Phe,
hLeu, hArg, hLeu, Ile, Lys, Leu, Asn, N-MeLeu, N-MeArg, Ogl, Orn, Pro, Gin,
Arg, Ser, Thr or
Tie, or a corresponding a-methyl amino acid form of any of the foregoing; X13
is Lys(Ac),
(D)Asn, (D)Leu, (D)Thr, (D)Phe, Ala, Aib, a-MeLeu, f3Ala, f3hGlu, f3hAla,
PhLeu, f3hVal, (3-
spiro-pip, Cha, Chg, Asp, Arg, Orn, Dab, Dap, a-DiethylGly, Glu, Phe, hLeu,
Lys, Leu, Asn,
Ogl, Pro, Gin, Asp, Arg, Ser, spiro-pip, Thr, Tba, Tic, Val or Tyr, or a
corresponding a-methyl
amino acid form of any of the foregoing; X14 is Asn, Glu, Phe, Gly, His, Lys,
Leu, Met, Asn,
Pro, Gin, Arg, Ser, Thr, Tic or Tyr, or a corresponding a-methyl amino acid
form of any of the
139
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
foregoing; and X15 is Gly, (D)Ala, (D)Asn, (D)Asp, Asn, (D)Leu, (D)Phe,
(D)Thr, Ala, AEA,
Asp, Glu, Phe, Gly, Lys, Leu, Pro, Gln, Arg or Ser, or a corresponding a-
methyl amino acid
form of any of the foregoing.
[00422] In certain embodiments, X3 is present. In particular embodiments, X3
is Glu,(D)Glu,
Arg, (D)Arg, Phe, (D)Phe, 2-Nal, Thr, Leu, or (D)Gln. In certain embodiments,
it is (D)Arg or
(D)Phe.
1004231 In particular embodiments, X5 is Gln, Ala, Cys, Cit, Asp, Dab, Dap,
Glu, Phe, Gly, His,
hCys, Lys, Leu, Met, Asn, N-Me-Ala, N-M-Asn, N-Me-Lys, N-Me-Gln, N-Me-Arg,
Orn, Pro,
Pen, Gln, Arg, Ser, Thr, or Val.
1004241 In particular emodiments, X6 is Thr, Asp, Glu, Phe, Asn, Pro, Arg,
Ser, or Thr.
1004251 In particular embodiments, X8 is Gln, Glu, Phe, Lys, Asn, Pro, Arg,
Val, Thr, or Trp.
1004261 In certain embodiments, X10 is a Tyr analog or a Phe analog. In
particular embodiments,
X10 is Phe(4-0Me), Phe(CONH2) or Phe[4-(2-aminoethoxy)]. In certain
embodiments, X10 is a
Tyr analog or a Phe analog. In particular embodiments, X10 is Phe(4-0Me) or
Phe[4-(2-
aminoethoxy)].
1004271In certain embodiments where X10 is a the Phe analog, X10 is selected
from hPhe,
Phe(4-0Me), a-MePhe, hPhe(3,4-dimethoxy), Phe(4-CONH2), Phe(4-0-Bz1)), Phe(4-
guanadino), Phe(4-tBu), Phe(4-CN), Phe(4-Br), Phe(4-NH2), Phe(4-F), Phe(3,5
DiF),
Phe(CH2CO2H), Phe(penta-F), Phe(3,4-C12), Phe(4-CF3), f33-diPheAla, Phe(4-N3)
and Phe[4-(2-
aminoethoxy)]. In particular embodiments, X10 is Phe[4-(2-aminoethoxy)] or
Phe(CONH2). In
particular embodiments, X10 is Phe[4-(2-aminoethoxy)].
1004281In certain embodiments where X10 is a Tyr analog, X10 is selected from
hTyr, N-Me-
Tyr, Tyr(3-tBu), Phe(4-0Me) and bhTyr. In particular embodiments, X10 is Phe(4-
0Me).
1004291In particular embodiments, X10 is Tyr, Phe(4-0Me), Phe(4-0Bz1), Phe(4-
0Me), Phe(4-
CONH2), Phe(3,4-C12), Phe(4-tBu), Phe(4-NH2), Phe(4-Br), Phe(4-CN), Phe(4-
carboxy), Phe[4-
(2aminoethoxy)] or Phe(4-guanadino). In particular embodiments, X10 is not
Tyr.
140
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1004301In certain embodiments, X11 is Trp or a Trp analog. In particular
embodiments, X11 is
2-Nal or 1-Nal.
1004311In particular embodiments, the peptide inhibitor of Formula III is
cyclized. In certain
embodiments, the peptide inhibitor is cyclized via an intramolecular bond
between X4 and X9.
In certain embodiments, the intramolecular bond is a thioether bond.
1004321In certain embodiments, the peptide inhibitor of Formula III is linear
or not cyclized. In
particular embodoiments of the linear peptide inhibitor of Formula III, X4
and/or X9 are any
amino acid.
1004331In particular embodiments of a peptide inhibitor of Formula III, one or
more, two or
more, or all three of Xi, X2, and X3 are absent. In certain embodiments, X1 is
absent. In certain
embodiments, X1 and X2 are absent. In certain embodiments, Xi, X2 and X3 are
absent.
1004341In particular embodiments of a peptide inhibitor of Formula III, one or
more, two or
more, three or more, four or more, or all of X16, X17, X18, X19 and X20 are
absent. In
particular embodiments of a peptide inhibitor of Formula III, one or more, two
or more, three or
more, or all of X17, X18, X19 and X20 are absent. In certain embodiments, one
or more, two or
more, or all three of X17, X19 and X20 are absent. In certain embodiments, one
or more of Xi,
X2 and X3 are absent; and one or more, two or more, three or more, or four of
X17, X18, X19
and X20 are absent.
1004351 In particular embodiments of a peptide inhibitor of Formula III, one
of X4 or X9 is Abu,
and the other of X4 or X9 is not Abu. In certain embodiments, X4 is Abu and X9
is Cys.
1004361 In particular embodiments, a peptide inhibitor of Formula III
comprises one or more, two
or more, three or more, or four of the following features: X5 is Arg or Gln;
X6 is Thr; X7 is Trp;
and X8 is Gln. In particular embodiments, X5 is Gln, X6 is Thr, X7 is Trp, and
X8 is Gln. In
certain embodiments, X5 is Gln. In certain embodiments, Xi, X2 and X3 are
absent. In certain
embodiments, X4 is Abu and X9 is Cys.
1004371 In particular embodiments, a peptide inhibitor of Formula III
comprises one or more, two
or more, three or more, four or more, five or more, six or more, or seven of
the following
141
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
features: X10 is Tyr or a Phe analog; X11 is Trp, 2-Nal, 1-Nal, Phe(4-0-
Ally1), Tyr(3-tBu),
Phe(4-tBu), Phe(4-guanidino), Phe(4-0Bz1) or Phe(4-Me); X12 is Arg, hLeu,
(D)Asn, or any
alpha methyl amino acids including, Aib, a-MeLys, a-MeLeu or a-MeOrn; X13 is
Lys, Glu or
Lys(Ac); X14 is Phe or Asn; X15 is f3-Ala, Gln, Gly, Ser, Ala; and X16 is
absent or AEA. In
particular embodiments, a peptide inhibitor of Formula III comprises one or
more, two or more,
three or more, four or more, five or more, six or more, or seven of the
following features: X10 is
Tyr or a Phe analog; X11 is Trp, 2-Nal, 1-Nal, Phe(4-0-Ally1), Tyr(3-tBu),
Phe(4-tBu), Phe(4-
guanidino), Phe(4-0Bz1) or Phe(4-Me); X12 is Arg, hLeu, (D)Asn, or any alpha
methyl amino
acids including, Aib, a-MeLys, a-MeLeu or a-MeOrn; X13 is Lys, Glu or Lys(Ac);
X14 is Phe
or Asn; X15 is Gly, Ser, Ala; and X16 is absent or AEA. In certain
embodiments, the Phe analog
is Phe(4-0Bz1), Phe(4-0Me), Phe(4-CONH2), Phe(3,4-C12), Phe(4-tBu), Phe(4-
NH2), Phe(4-Br),
Phe(4-CN), Phe(4-carboxy), Phe[4-(2aminoethoxy)] or Phe(4-guanadino). In
certain
embodiments, X11 is 2-Nal or 1-Nal. In certain embodiments, X1 , X2 and X3 are
absent. In
certain embodiments, X4 is Abu and X9 is Cys.
[00438] In particular embodiments, a peptide inhibitor of Formula III
comprises one or more, two
or more, three or more, four or more, five or more, six or more, or seven of
the following
features: X10 is Tyr or a Phe analog; X11 is Trp, 2-Nal, 1-Nal, Phe(4-0-
Ally1), Tyr(3-tBu),
Phe(4-tBu), Phe(4-guanidino), Phe(4-0Bz1) or Phe(4-Me); X12 is Arg, hLeu,
(D)Asn, 4-amino-
4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-DiethylGly, a-
MeLys, a-
MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-MeVal,; X13 is Lys, Glu or Lys(Ac);
X14 is Phe
or Asn; X15 is Gly; and X16 is absent or AEA. In certain embodiments, the Phe
analog is
Phe(4-0Bz1), Phe(4-0Me), Phe(4-CONH2), Phe(3,4-C12), Phe(4-tBu), Phe(4-NH2),
Phe(4-Br),
Phe(4-CN), Phe(4-carboxy), Phe(4-(2aminoethoxy)) or Phe(4-guanadino). In
certain
embodiments, X11 is 2-Nal or 1-Nal. In certain embodiments, Xi, X2 and X3 are
absent. In
certain embodiments, X4 is Abu and X9 is Cys.
[00439] In particular embodiments, a peptide inhibitor of Formula III
comprises one or more, two
or more, three or more, four or more, five or more, six or more, seven or
more, eight or more,
nine or more, ten or more, or eleven of the following features: X5 is Arg or
Gln; X6 is Thr; X7
is Trp; X8 is Gln; X10 is a Phe analog; X11 is Trp, 2-Nal, 1-Nal, Phe(4-0-
Ally1), Tyr(3-tBu),
Phe(4-tBu), Phe(4-guanidino), Phe(Bz1) or Phe(4-Me); X12 is Aib, a-MeLys, a-
MeLeu, 4-
142
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Agp, Aib, a-
DiethylGly, a-MeLys,
a-MeLys(Ac), a-Me-Leu, a-MeSer, a-MeVal,a-MeOrn; X13 is Lys, Glu or Lys(Ac);
X14 is
Phe or Asn; X15 is f3-ala, Gly, Ser, Ala; and X16 is absent or AEA. In
particular embodiments, a
peptide inhibitor of Formula III comprises one or more, two or more, three or
more, four or
more, five or more, six or more, seven or more, eight or more, nine or more,
ten or more, or
eleven of the following features: X5 is Arg or Gln; X6 is Thr; X7 is Trp; X8
is Gln; X10 is a
Phe analog; X11 is Trp, 2-Nal, 1-Nal, Phe(4-0-Ally1), Tyr(3-tBu), Phe(4-tBu),
Phe(4-
guanidino), Phe(Bz1) or Phe(4-Me); X12 is Aib, a-MeLys, a-MeLeu or a-MeOrn;
X13 is Lys,
Glu or Lys(Ac); X14 is Phe or Asn; X15 is Gly, Ser, Ala; and X16 is absent or
AEA. In certain
embodiments, the Phe analog is Phe(4-0Bz1), Phe(4-0Me), Phe[4-(2aminoethoxy)],
Phe(4-
CONH2), Phe(3,4-C12), Phe(4-tBu), Phe(4-NH2), Phe(4-Br), Phe(4-CN), Phe(4-
CO2H), or Phe(4-
guanadino). In certain embodiments, X11 is 2-Nal or 1-Nal. In certain
embodiments, X1 , X2
and X3 are absent. In certain embodiments, X4 is Abu and X9 is Cys.
[00440] In particular embodiments, a peptide inhibitor of Formula III
comprises one or more, two
or more, three or more, four or more, five or more, six or more, seven or
more, eight or more,
nine or more, ten or more, or eleven of the following features: X5 is Arg or
Gln; X6 isThr; X7 is
Trp; X8 is Gln; X10 is Tyr or a Phe analog; X11 is Trp, 2-Nal, 1-Nal, Phe(4-0-
Ally1), Tyr(3-
tBu), Phe(4-tBu), Phe(4-guanidino), Phe(Bz1) or Phe(4-Me); X12 is Arg, hLeu,
(D)Asn, 4-
amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, Aib, a-DiethylGly, a-
MeLys, a-
MeLys(Ac), a-Me-Leu, a-MeSer, a-MeVal,; X13 is Lys, Glu or Lys(Ac); X14 is Phe
or Asn;
X15 is f3-Ala, Asn or Gly; and X16 is absent or AEA. In particular
embodiments, a peptide
inhibitor of Formula III comprises one or more, two or more, three or more,
four or more, five or
more, six or more, seven or more, eight or more, nine or more, ten or more, or
eleven of the
following features: X5 is Arg or Gln; X6 isThr; X7 is Trp; X8 is Gln; X10 is
Tyr or a Phe
analog; X11 is Trp, 2-Nal, 1-Nal, Phe(4-0-Ally1), Tyr(3-tBu), Phe(4-tBu),
Phe(4-guanidino),
Phe(Bz1) or Phe(4-Me); X12 is Arg, hLeu, (D)Asn, a-MeLys, a-MeLeu or a-MeOrn,
Aib; X13
is Lys, Glu or Lys(Ac); X14 is Phe or Asn; X15 is Gly; and X16 is absent or
AEA. In certain
embodiments, the Phe analog is Phe(4-0Bz1), Phe(40Me), Phe(4-CONH2), Phe(3,4-
C12), Phe(4-
tBu), Phe(4-NH2), Phe(4-Br), Phe(4-CN), Phe(4-CO2H), Phe(4-(2-aminoethoxy)) or
Phe(4-
143
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
guanidino). In certain embodiments, X11 is 2-Na! or 1-Nal. In certain
embodiments, Xl, X2 and
X3 are absent. n certain embodiments, X4 is Abu and X9 is Cys.
1004411In certain embodiments, the present invention includes a peptide of 8
to 20, 8 to 16 or 8
to 12 amino acids, optionally cyclized, comprising or consisting of a core
sequence of Formula
Xaa4-Xaa5-Xaa6-Trp-Xaa8-Xaa9-Xaa 1 0-Xaa11 (Tub)
1004421wherein Xaa4 and Xaa9 are each independently selected from Abu and Cys,
wherein
Xaa4 and Xaa9 are not both the same; Xaa5, Xaa6 and Xaa8 are any amino acid
residue; Xaal 0
is Tyr, a Phe analog or 2-Na!, and Xaall is 2-Na! or Trp, wherein the peptide
inhibits binding of
IL-23 to IL-23R. In particular embodiments, Xaal 0 is Phe(4-0Me), 2-Na!, or
Phe[4-(2-
aminoethoxy)]. In one embodiment, Xaal0 is Phe(4-0Me). In one embodiment, Xaa7
is Phe[4-
(2-aminoethoxy)]. In one embodiment, Xaal 1 is 2-Nal. In certain embodiments,
the peptide is
cyclized via Xaa4 and Xaa9. In particular embodiments, the Phe analog is Phe[4-
(2aminoethoxy)] or Phe(4-0Me). In certain embodiments, Xaa4 is Abu and Xaa9 is
Cys, and the
peptide is cyclized via Xaa4 and Xaa9. In particular embodiments, the peptide
is a peptide
inhibitor of Formula III, and wherein in certain embodiments, Xl, X2 and X3
are absent. In
particular embodiments, the peptide inhibits the binding of IL-23 to IL-23R.
In certain
embodiments, a peptide of Formula Illb comprises a Glu,(D)Glu, Arg, (D)Arg,
Phe, (D)Phe, 2-
Na!, Thr, Leu, or (D)Gln bound to Xaa4. In certain embodiments, it is (D)Arg
or (D)Phe.
1004431In certain embodiments, the present invention includes a peptide of 8
to 20, 8 to 16 or 8
to 12 amino acids, optionally cyclized, comprising or consisting of a core
sequence of Formula
IIIc:
Abu-Xaa5-Xaa6-Trp-Xaa8-Cys-[Phe(4-0Me)]-(2-Nal) (IIIc)
1004441wherein Xaa5, Xaa6 and Xaa8 are any amino acid residue; and wherein the
peptide
inhibits binding of IL-23 to IL-23R. In certain embodiments, the peptide is
cyclized via Abu at
Xaa4 and Cys at Xaa9. In certain embodiments, the peptide is a peptide
inhibitor of Formula III,
and wherein in certain embodiments, Xl, X2 and X3 are absent. In particular
embodiments, the
peptide inhibits the binding of IL-23 to IL-23R. In certain embodiments, a
peptide of Formula
144
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Mc comprises a Glu, (D)Glu, Arg, (D)Arg, Phe, (D)Phe, 2-Na!, Thr, Leu, or
(D)Gln bound to
Abu. In certain embodiments, it is (D)Arg or (D)Phe.
1004451 In certain embodiments, the present invention includes a peptide of 8
to 20, 8 to 16 or 8
to 12 amino acids, optionally cyclized, comprising or consisting of a core
sequence of Formula
IIId:
Abu-Xaa5 -Xaa6 - Trp-Xaa8 -Cy s-Xaal0 - Trp (IIId)
[00446] wherein Xaa5, Xaa6 and Xaa8 are any amino acid residue; Xaal 0 is a
modified Phe; and
wherein the peptide inhibits binding of IL-23 to IL-23R. In particular
embodiments, the
modified Phe is Phe(4-tBu), Phe(4-guanidino), Phe[4-(2-aminoethoxy)], Phe(4-
CO2H), Phe(4-
CN), Phe(4-Br), Phe(4-NH2), PHe(CONH2) or Phe(4-Me). In particular
embodiments, the
modified Phe is Phe(4-tBu), Phe(4-guanidino), Phe[4-(2-aminoethoxy)], Phe(4-
CO2H), Phe(4-
CN), Phe(4-Br), Phe(4-NH2), or Phe(4-Me). In one embodiment, Xaal 0 is Phe[4-
(2-
aminoethoxy)] or Phe(4-0Me). In one embodiment, Xaal 0 is Phe[4-(2-
aminoethoxy)]. In certain
embodiments, the peptide is cyclized via Abu at Xaa4 and Cys at Xaa9. In
certain embodiments,
the peptide is a peptide inhibitor of Formula III, and wherein in certain
embodiments, Xl, X2
and X3 are absent. In particular embodiments, the peptide inhibits the binding
of IL-23 to IL-
23R. In certain embodiments, a peptide of Formula IIId comprises a Glu,
(D)Glu, Arg, (D)Arg,
Phe, (D)Phe, 2-Na!, Thr, Leu, or (D)Gln bound to Abu. In certain embodiments,
it is (D)Arg or
(D)Phe.
1004471 In certain embodiments, the present invention includes a peptide,
optionally 8 to 20, 8 to
16 or 8 to 12 amino acids, optionally cyclized, comprising or consisting of a
core sequence of
Formula Me:
Abu-Xaa5 -Xaa6 - Trp-Xaa8 -Cy s- Phe [4 -(2-aminoethoxy)] - [2-Na!] (Me)
[00448] wherein Xaa5, Xaa6 and Xaa8 are any amino acid residue. In certain
embodiments, the
peptide is cyclized via Abu at Xaa4 and Cys at Xaa9. In certain embodiments,
the peptide is a
peptide inhibitor of Formula III, and wherein in certain embodiments, Xl, X2
and X3 are absent.
In particular embodiments, the peptide inhibits the binding of IL-23 to IL-
23R. In certain
145
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
embodiments, a peptide of Formula Illb comprises a Glu, (D)Glu, Arg, (D)Arg,
Phe, (D)Phe, 2-
Nal, Thr, Leu, or (D)Gln bound to Abu. In certain embodiments, it is (D)Arg or
(D)Phe.
1004491In one embodiment, Xaa5 and Xaa8 is Gln. In one embodiment, Xaa6 is
Thr. In certain
embodiments, the peptide is cyclized via Abu at Xaa4 and Cys at Xaa9.
[00450] In particular embodiments of a peptide inhibitor of Formula III, the
peptide inhibitor has
a structure shown in any of Tables 5A-5C or comprises an amino acid sequence
set forth in
Tables 5A-5C.
1004511In certain aspects, the present invention provides a peptide inhibitor
of an interleukin-23
receptor, or a pharmaceutically acceptable salt or solvate thereof, wherein
the peptide inhibitor
comprises an amino acid sequence of Formula (Vf):
X1 -X2-X3-Abu-X5-X6-X7-X8-Cys-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20
(VI),
wherein:
X1 is absent;
X2 is absent or X2 is D-Asp, E, R, D-Arg, F, D-Phe, 2-Nal, T, L, D-Gln, or D-
Asn;
X3 is D-Arg;
X5 is N, Q, Cit, Lys, or a Lys conjugate (e.g., Lys(IVA), Lys(biotin),
Lys(octanyl), Lys(Palm),
Lys(PEG), Lys(PEG8), Lys(PEG11-Palm), Lys(Ac));
X6 is T, S or V;
X7 is W, 1-Nal, or 2-Nal;
X8 is Q, Cit, N, Aib or Lys(Ac);
X10 is Phe[4-(2-aminoethoxy)], Phe[4-(2-acetylaminoethoxy)] or Phe(4-CONH2);
X11 is 2-Nal;
146
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X12 is 4-amino-4-carboxy-tetrahydropyran, Aib, aMeLeu, aMeLys, or an aMeLys
conjugate
(e.g., aMeLys(Ac),
aMeLys(PEG4-Palm), aMeLys(PEG4-
Laury1), aMeLys(PEG4IsoGluPalm), aMeLys(PEG4IsoGluLaury1), aMeLys(IVA),
aMeLys(bi
otin), or aMeLys(octany1));
X13 is Q, E, Cit or a Lys conjugate (e.g., Lys(Ac), Lys(PEG4-isoGlu-Palm),
Lys(PEG4-octanyl),
Lys(PEG4-Palm), Lys(biotin), Lys(octanyl), Lys(Palm), Pys(PEG8), or Lys(PEG11-
Palm));
X14 is N, Cit, Q, L, G, S, Aib, F, 2-Nap, N-Me-Ala, R, W, nLeu, Tic or a Lys
conjugate (e.g.,
Lys(Ac));
X15 is N, Cit, Q, f3Ala, Lys(Ac) or Aib; and
X16, X17, X18, X19 and X20 are absent.
In particular embodiments, X2 is D-Asp, E, R, D-Arg, F, D-Phe, 2-Nal, T, L, D-
Gln, or
D-Asn
1004521In certain aspects, the present invention provides a peptide inhibitor
of an interleukin-23
receptor, or a pharmaceutically acceptable salt or solvate thereof, wherein
the peptide inhibitor
comprises an amino acid sequence of Formula (Vh):
X1 -X2-X3-Abu-X5-X6-X7-X8-Cys-X10-X11-X12-X13-X14-X15-X16-X17-X18-X19-X20
(Vh),
wherein:
X1 is any amino acid or absent;
X2 is any amino acid or absent;
X3 is any D-amino acid or absent;
X4 is Cys, hCys, Pen, hPen, Abu, Ser, hSer or chemical moiety capable of
forming a bond with
X9;
147
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
X5 is Ala, a-MeOrn, a-MeSer, Cit, Dap, Dab, Dap(Ac), Gly, Lys, Asn, N-MeGln, N-
MeArg,
Orn, Gln, Arg, Ser, Glu or Thr;
X6 is Thr, Ser, Asp, Ile or any amino acid;
X7 is Trp, 6-Chloro-Trp, 1-Nap or 2-Nap;
X8 is Glu, Gln, Asn, Lys(Ac), Cit, Cav, Lys(N-c-(N-a-Palmitoyl-L-y-glutamy1)),
or Lys(N-c-
Palmitoyl;
X9 is Cys, hCys, Pen, hPen Abu, or any amino acid or chemical moiety capable
of forming a
bond with X4;
X10 is 2-Nal, a Phe analog, Tyr, or a Tyr analog;
X11 is 1-Nal, 2-Nal, Phe(3,4-dimethoxy), 5-HydroxyTrp, Phe(3,4-C12), Trp or
Tyr(3-tBu);
X12 is Aib, 4-amino-4-carboxy-tetrahydropyran, any alpha-methylamino acid,
alpha-ethyl-
amino acid, Achc, Acvc, Acbc Acpc, 4-amino-4-carboxy-piperidine, 3-Pal, Agp, -
DiethylGly,
a-MeLys, a-MeLys(Ac), a-MeLeu, a-MeOrn, a-MeSer, a-MeVal, Cav, Cha, Cit, Cpa,
D-Asn,
Glu, His, hLeu, hArg, Lys, Leu, Octgly, Orn, piperidine, Arg, Ser, Thr or THP;
X13 is Lys(Ac), Gln, Cit, Glu, or any amino acid;
X14 is Asn, Gln, Lys(Ac), Cit, Cav, Lys(N-c-(N-a-Palmitoyl-L-y-glutamy1)),
Lys(N-c-
Palmitoy1), Asp, or any amino acid;
X15 is f3-Ala, Asn, Gly, Gln, Ala, Ser, Aib, Asp or Cit;
X16 is any amino acid or absent;
X17 is any amino acid or absent;
X18 is any amino acid or absent;
X19 is any amino acid or absent; and
148
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X20 is any amino acid or absent.
1004531In certain embodiments of any of the peptide inhibitors described
herein, including but
not limited to those of Formula (If) and (Ih), the peptide inhibitor is
cyclized via a bond, e.g., a
thioether bond, between X4 and X9. In certain embodiments, the peptide
inhibitor inhibits the
binding of an interleukin-23 (IL-23) to an IL-23 receptor.
1004541In certain embodiments, X1 , X2 and X3 are absent. In certain
embodiments, X1 and X2
are absent. In certain embodiments, X1 is a D-amino acid or absent. In certain
embodiments, X2
is a D-amino acid or absent.
1004551In certain embdoiments, X5 is Ala, a-MeOrn, a-MeSer, Cit, Dap, Dab,
Dap(Ac), Gly,
Lys, Asn, N-MeGln, N-MeArg, Orn, Gln, Arg, Ser, or Thr;
1004561 In certain embodiments, X5 is N, X6 is T, X7 is W, or X8 is Q. In
certain embodiments,
X5 is N, X6 is T, X7 is W, and X8 is Q.
1004571In certain embodiments, X5 is Q, X6 is T, X7 is W, or X8 is Q. In
certain embodiments,
X5 is Q, X6 is T, X7 is W, and X8 is Q.
1004581 In certain embodiments, X5 is N, X6 is T, X7 is W, and X8 is Cit.
1004591 In certain embodiments, X10 is Phe[4-(2-aminoethoxy)].
1004601In certain embodiments, X12 is 4-amino-4-carboxy-tetrahydropyran, Aib,
aMeLeu, or
aMeLys. In certain embodiments, X12 is 4-amino-4-carboxy-tetrahydropyran.
[00461]In certain embodiments, X13 is E or Lys(Ac). In certain embodiments,
X13 is Lys(Ac).
1004621In certain embodiments, X14 is Asn, Gln, Lys(Ac), Cit, Cav, Lys(N-6-(N-
a-Palmitoyl-L-
y-glutamy1)), Lys(N-E-Palmitoy1), or any amino acid;
1004631 In certain embdoiments, X15 is f3-Ala, Asn, Gly, Gln, Ala, Ser, Aib,
or Cit.
1004641 In certain embodiments, X14 is N.
149
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1004651 In certain embodiments, X15 is N.
1004661In certain embodiments, X16 is a D-amino acid or absent. In certain
embodiments, X17
is a D-amino acid or absent. In certain embodiments, X18 is a D-amino acid or
absent. In
certain embodiments, X19 is a D-amino acid or absent. In certain embodiments,
X20 is a D-
amino acid or absent.
1004671In certain embodiments, X2 is absent; X3 is absent; X5 is Q, X6 is T,
X7 is W, and X8 is
Q; X10 is Phe[4-(2-aminoethoxy)]; X12 is 4-amino-4-carboxy-tetrahydropyran,
Aib, aMeLeu,
or aMeLys; X13 is E or Lys(Ac); X14 is N; and X15 is N. In certain
embodiments, X12 is 4-
amino-4-carboxy-tetrahydropyran and X13 is Lys(Ac).
[00468] In certain embodiments, any of the amino acids of the peptide
inhibitor are connected by
a linker moiety, e.g., a PEG.
1004691 In certain embodiments, the N-terminus of the peptide inhibitor
comprises an Ac group.
[00470] In certain embodiments, the C-terminus of the peptide inhibitor
comprises an NH2 group.
1004711 In certain embodiments, the present invention includes a peptide
comprising or consisting
of an amino acid sequence shown in any of the Table 4s or Table 5s, or a
peptide inhibitor
comprising or consisting of a structure shown in any of the Table 4s or Table
5s (or a
pharmaceutically acceptable salt thereof). In particular embodiments, the
peptide does not
include the conjugated moieties but does include the Abu residue. In
particular embodiments,
the peptide or inhibitor comprises a thioether bond between the two Abu and
Cys residues, or
between the two outermost amino acids within the brackets folloing the term
"cyclo", which
indicated the presence of a cyclic structure. In particular embodiments, the
inhibitor is an acetate
salt. The peptide sequence of illustrative inhibitors is shown in Tables 4 and
5 from N-term to
C-term, with conjugated moieties, and N-terminal Ac and/or C-terminal NH2
groups indicated.
The cyclic structure is indicated by "Cyclo" as illustrated in Table 5,
indicating the presence of a
thioether bond between the bracketed Abu at X4 and Cys at X9.
Table 4. Illustrative Thioether Peptide Inhibitors
Biotin-[PEG11-cyclo[[Abul-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-[4-amino-4-
carboxy-tetrahydropyran]-
ENN-NH2
150
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyranHLys(Ac)]-NN-
NH2
Ac-[(D)Arg]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-tetrahydropyrar]-
[Lys(Ac)]-NN-NFI2
Ac-[(D)Arg]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-tetrahydropyrar]-
[Lys(Ac)]-NN-NFI2
Ac-E-[(D)Argl-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-[4-amino-4-carboxy-
tetrahydropyrard-
ENN-NH2
Ac-[(D)Asp]-[(D)Argl-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-
4-carboxy-
tetrahydropyran]-ENN-NH2
Ac-R-[(D)Argl-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-[4-amino-4-carboxy-
tetrahydropyrard-
ENN-NH2
Ac-[(D)Arg]-[(D)Argl-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-
amino-4-carboxy-
tetrahydropyran]-ENN-NH2
Ac-F-[(D)Arg]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-tetrahydropyrar]-
ENN-NH2
Ac-[(D)Phe]-[(D)Arg]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-
amino-4-carboxy-
tetrahydropyran]-ENN-N H2
Ac-[2-Nal]-[(D)Arg]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-
amino-4-carboxy-
tetrahydropyran]-ENN-NH2
Ac-T-[(D)Argl-cyclo[[Abul-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-[4-amino-4-
carboxy-tetrahydropyrard-
ENN-NH2
Ac-L-[(D)Argl-cyclo[[Abul-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-[4-amino-4-
carboxy-tetrahydropyrard-
ENN-NH2
Ac-[(D)G1n]-[(D)Arg]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-
amino-4-carboxy-
tetrahydropyran]-ENN-NH2
Ac-[(D)Asn]-[(D)Arg]-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-
4-carboxy-
tetrahydropyran]-ENN-NH2
Ac-cycloRAbul-QTWOC1-[Phe[4-(2-aminoethoxy)-(PEG4-Alexa488)]-[2-Nal]-[4-amino-
4-carboxy-
tetrahydropyran]-ENN-NH2
[A1exa488]-[PEG4]-cycloRAbul-QTWOC1-[Phe[4-(2-aminoethoxy)] [2 Nal] [4 amino-4-
carboxy-
tetrahydropyran]-ENN-NH2
[Alexa647]-[PEG4]-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-
tetrahydropyran]-ENN-NH2
[Alexa-647]-[PEG4]-[(D)Arg]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-
[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-NN-N H2
[A1exa647]-[PEG12]-[(D)Arg]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-
[4-amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-NN-N H2
[Alexa488]-[PEG4]-[(D)Arg]-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nal]-[4-
amino-4-carboxy-
tetrahydropyran]-[Lys(Ac)]-NN-N H2
Table 5A. Illustrative Thioether Peptide Inhibitors
151
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
_____________ S----
Ac,
N N--[Phe(4-0Me)]-[2-Nal]-XXXX-N H2
H 0 H
0
Ac-Cyclo-[[Abu]-XXWXC]-[Phe(4-0Me)[2-Na11-XXXX-NH2
Sequence
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-ENN-
NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-ENNE-
NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-ENNF-
NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-ENNK-
NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-ENNN-
NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-
ENNW-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-ENNT-
NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-ENNG-
NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-
ENNPK-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-
ENNPG-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-
ENNEP-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-
ENNGK-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-
ENNPT-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-
ENNGF-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-
ENNGW-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-
ENNGO-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-
ENNGGG-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyrar]-
ENNKKK-NH2
152
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNEEE-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNFFF-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNTTT-N H2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNGGGR-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNGGGF-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNGGGE-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNGGGQ-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNGGGT-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNGGGGR-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNGGGGF-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNGGGGE-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNGGGGQ-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNGGGGT-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNRRRRR-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNGFFFFF-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNEEEEE-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNQCICICICI-N H2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENNTTTTT-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-ENN-
NH2
Ac-GGG-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENN-N H2
Ac-RRR-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENN-N H2
Ac-FFF-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENN-N H2
153
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-EEE-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENN-N H2
Ac-000-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENN-N H2
Ac-TTT-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENN-N H2
Ac-RG-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENN-N H2
Ac-FG-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENN-N H2
Ac-EG-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENN-N H2
Ac-C1G-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
ENN-N H2
Ac-TG-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENN-N H2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(Palm)]-NN-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(PEG11-Palm)]-NN-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(isoGlu-Palm)]-NN-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(Ahx-Palm)]-NN-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(isoGlu-Ahx-Palm)]-NN-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(isoGlu-Ahx-Palm)]-NN-NH2
[Palm]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-
ENN-N H2
[Palm-isoGlu]-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-
tetrahydropyranl-ENN-N H2
[Palm-PEG11]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-
carboxy-
tetrahydropyran]-ENN-N H2
[Palm-Ahx]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-
tetrahydropyran]-ENN-N H2
[Palm-Ahx-isoGlu]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-
4-carboxy-
tetrahydropyran]-ENN-N H2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-ENN-
Lys[Palml-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-ENN-
Lys[Pegll-Palm]-NH2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-amino-4-carboxy-
tetrahydropyran]-ENN-
Lys[isoGlu-Palm]-NH2
154
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-cyclo[[Abu]-QTVVOC]-[Phe[4-(2-aminoethoxy)]-[2-NalH4-amino-4-carboxy-
tetrahydropyranl-ENN-
Lys[Ahx-Palml-NH2
Ac-cyclo[[Abu]-QTVVOC]-[Phe[4-(2-aminoethoxy)]-[2-NalH4-amino-4-carboxy-
tetrahydropyranl-ENN-
Lys[isoGlu-Ahx-Palml-NH2
Table 5B. Illustrative Thioether Peptides
Ac-[D-Arg]- Cyclo4Abu-QTWOC1-[Phe(4-2ae)H2-NaIHTHH-ENN-NH2
Ac-[D-Arg]- Cyclo4Abu-QTWOC1-[Phe(4-2ae)H2-NaIHTHP11-END-NH2
Ac-[D-Arg]- Cyclo4Abu-QTWOC1-[Phe(4-2ae)H2-NaIHTHH-EDN-NH2
Ac-[D-Arg]- Cyclo4Abu-QTWEC1-[Phe(4-2ae)H2-NaIHTHH-ENN-NH2
Ac-[D-Arg]- Cyclo4Abu-ETWOC1-[Phe(4-2ae)H2-NaIHTHP1-ENN-NH2
Ac-[D-Arg]- Cyclo4Abu-QTWOC1-[Phe(4-2ae)H2-NaIHTHH-EDD-N H2
Ac-[D-Arg]- Cyclo4Abu-QTWEC1-[Phe(4-2ae)H2-NaIHTHH-END-NH2
Ac-[D-Arg]- Cyclo4Abu-ETWCIC1-[Phe(4-2ae)H2-Nall-[Tetrahydropyran-A1-END-N H2
Ac-[D-Arg]- Cyclo4Abu-QTWEC1-[Phe(4-2ae)H2-NaIHTHH-EDN-NH2
Ac-[D-Arg]- Cyclo4Abu-ETWCIC1-[Phe(4-2ae)H2-Nall-[Tetrahydropyran-A1-EDN-N H2
Ac-[D-Arg]- Cyclo4Abu-ETWEC1-[Phe(4-2ae)H2-NaIHTHP1-ENN-NH2
Ac-[D-Arg]- Cyclo4Abu-QTWOC1-[Phe(4-2ae)H2-NaIHTHH-ENN-NH2
Ac-[D-Arg]- Cyclo4Abu-QTWOC1-[Phe(4-2ae)H2-NaIHTHH-END-NH2
Ac-[D-Arg]- Cyclo4Abu-QTWCIC1-[Phe(4-2ae)H2-Nall-[Tetrahydropyran-A1-EDN-N H2
Ac-[D-Arg]- Cyclo4Abu-ETWEC1-[Phe(4-2ae)H2-NaIHTHP1-ENN-NH2
Ac-[D-Arg]- Cyclo4Abu-ETWOC1-[Phe(4-2ae)H2-NaIHTetrahydropan-Al-ENN-OH
Ilustrative Peptide Inhibitors Containing Cyclic Amides
155
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
10047211n certain embodiments, the present invention includes a peptide
inhibitor of an
interleukin-23 receptor, wherein the peptide inhibitor has the structure of
Formula IV:
R1-X-R2 (IV)
[00473] or a pharmaceutically acceptable salt or solvate thereof,
[00474] wherein 1Z1 is a bond, hydrogen, an C1-C6 alkyl, a C6-C12 aryl, a C6-
C12 aryl C1-C6
alkyl, a Cl -C20 alkanoyl, and including PEGylated versions alone or as
spacers of any of the
foregoing;
[00475] R2 is a bond, OH or NH2; and
1004761X is an amino acid sequence of 8 to 20 amino acids, comprising or
consisting of the
sequence of Formula IVa:
X1 -X2-X3 -X4 -X5-X6-W-X8-X9-X10-X11 -X12 -X13 -X14-X15-X16-X17-X18-X19-X20
(IVO
[00477] wherein
X1 is absent or any amino acid;
X2 is absent or any amino acid;
X3 is absent or any amino acid;
X4 is Dap, Dab, Glu, Asp, (D)-Asp or (D)-Dab;
X5 is Gln, Ala, Cys, Cit, Asp, Dab, Dap, Glu, Phe, Gly, His, hCys, Lys, Leu,
Met, Asn, N-Me-
Ala, N-M-Asn, N-Me-Lys, N-Me-Gln, N-Me-Arg, Orn, Pro, Pen, Gln, Arg, Ser, Thr,
or Val;
X6 is Thr, Asp, Glu, Phe, Asn, Pro, Arg, Ser, or Thr;
X7 is Trp, Glu, Gly, Ile, Asn, Pro, Arg, Thr or OctGly;
X8 is Gln, Glu, Phe, Lys, Asnõ Pro, Arg, Thr, or Trp;
X9 is Dap, Dab, Glu, Asp, (D)-Asp or (D)-Dab;
156
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
X10 is Tyr(OMe)Phe(4-0Me), 1-Na!, 2-Na!, Aic, a-MePhe, Bip, (D)Cys, Cha, DMT,
(D)Tyr),
Glu, Phe, His, hPhe(3,4-dimethoxy), hTyr, N-Me-Tyr, Trp, Phe(4-CONH2), Phe(4-
phenoxy),
Thr, Tic, Tyr, Tyr(3-tBu), Phe(4-tBu), Phe(4-CN), Phe(4-Br), Phe(4-NH2), Phe(4-
F), Phe(3,5-
F2),Phe(penta-F), Phe(3,4-C12), Phe(4-CF3), Bip, Cha, 4-pyridylalanine,
f3hTyr, OctGly, Phe(4-
N3), Phe(4-Br) or Phe[4-(2-aminoethoxy)];
X11 is 2-Na!, 1-Na!, 2,4-dimethylPhe, Bip, Phe(3,4-C12), Phe(3,5-F2), Phe(4-
CONH2), Phe(4-F),
4-phenylcyclohexylalanine, Phe(4-CF3), a-MePhe, f3hPhe, f3hTyr, f3hTrp, BIP,
Nva(5-phenyl),
Phe, His, hPhe, Tic, Tqa, Trp, Tyr, Phe(4-0Me), Phe(4-Me), Trp(2,5,7-tri-
tertButyl), Phe(4-
0Ally1), Tyr(3-tBu), Phe(4-tBu), Phe(4-guanidino), Tyr(Bz1), or OctGly;
X12 is a-MeLys, a-MeOrn, a-MeLeu, Aib, (D)Ala, (D)Asn, (D)Leu, (D)Asp, (D)Phe,
(D)Thr,
3-Pal, Aib, f3-Ala, f3-Glu, f3hAla, PhLeu, f3hVal, f3-spiro-pip, Cha, Chg,
Asp, Dab, Dap, a-
DiethylGly, Glu, Phe, hLeu, hArg, hLeu, Ile, Lys, Leu, Asn, N-MeLeu, N-MeArg,
Ogl, Orn, Pro,
Gln, Arg, Ser, Thr Tle, 4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc,
Acvc, a-
DiethylGly, a-MeLys, a-MeLys(Ac), a-Me-Leu, a-MeSer, a-MeVal,;
X13 is Lys(Ac), (D)Asn, (D)Leu, (D)Thr, (D)Phe, Ala, Aib, a-MeLeu, Aib, f3-
Ala, f3-Glu,
f3hAla,f3hLeu, f3hVal, f3-spiro-pip, Cha, Chg, Asp, Dab, Dap, a-DiethylGly,
Glu, Phe, hLeu, Lys,
Lys(Ac), Leu, Asn, Ogl, Pro, Gln, Arg, Ser, f3-spiro-pip, Thr, Tba, Tlc, Val
or Tyr;
X14 is Asn, Glu, Phe, Gly, His, Lys, Leu, Met, Asn, Pro, Gln, Arg, Ser, Thr,
Tic or Tyr;
X15 is f3-ala, Asn, Gly, (D)Ala, (D)Asn, (D)Asp, (D)Leu, (D)Phe, (D)Thr, Ala,
AEA, Asp, Glu,
Phe, Gly, Lys, Leu, Pro, Gln, Arg or Ser;
X16 is absent, Gly, Ala, Asp, Ser, Pro, Asn or Thr;
X17 is absent, Glu, Ser, Gly or Gln;
X18 is absent or any amino acid;
X19 is absent or any amino acid; and
X20 is absent or any amino acid.
157
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
10047811n certain embodiments of IVa: X12 is a-MeLys, a-MeOrn, a-MeLeu, Aib,
(D)Ala,
(D)Asn, (D)Leu, (D)Asp, (D)Phe, (D)Thr, 3-Pal, Aib, f3-Ala, f3-Glu, f3hAla,
PhLeu, f3hVal, (3-
spiro-pip, Cha, Chg, Asp, Dab, Dap, a-DiethylGly, Glu, Phe, hLeu, hArg, hLeu,
Ile, Lys, Leu,
Asn, N-MeLeu, N-MeArg, Ogl, Orn, Pro, Gln, Arg, Ser, Thr or Tle; X13 is
Lys(Ac), (D)Asn,
(D)Leu, (D)Thr, (D)Phe, Ala, Aib, a-MeLeu, Aib, f3-Ala, f3-Glu, f3hAla,f3hLeu,
f3hVal, f3-spiro-
pip, Cha, Chg, Asp, Dab, Dap, a-DiethylGly, Glu, Phe, hLeuLys, Leu, Asn, Ogl,
Pro, Gln, Arg,
Ser, f3-spiro-pip, Thr, Tba, Tlc, Val or Tyr; X14 is Asn, Glu, Phe, Gly, His,
Lys, Leu, Met, Asn,
Pro, Gln, Arg, Ser, Thr, Tic or Tyr; and X15 is Gly, (D)Ala, (D)Asn, (D)Asp,
(D)Leu, (D)Phe,
(D)Thr, Ala, AEA, Asp, Glu, Phe, Gly, Lys, Leu, Pro, Gln, Arg or Ser.
1004791In particular embodiments of a peptide inhibitor of Formula (IV): X5 is
Cys, Cit, Asp,
Dab, Dap, Gly, His, hCys, Lys, Met, Asn, N-Me-Ala, N-Me-Asn, N-Me-Lys, N-Me-
Gln, N-Me-
Arg, Orn, Pro, Pen, Gln, Val; X6 is Glu, Arg, Ser; X7 is Trp, Glu, Gly, Ile,
Asn, Pro, Arg, Thr or
OctGly; X8 is Phe, Asn, Pro, Arg, Thr, Trp; X10 is Phe(4-0Me), 1-Nal, 2-Nal,
Aic, a-MePhe,
Bip, (D)Cys, Cha, DMT, (D)Tyr, Glu, His, hPhe(3,4-dimethoxy), hTyr, N-Me-Tyr,
Trp, Phe(4-
CONH2), Phe-(4-phenoxy), Thr, Tic, Tyr(3-tBu), Phe(4-tBu), Phe(4-CN), Phe(4-
Br), Phe(4-
NH2), Phe(4-F), Phe(3,5-F2), PheCH2CO2H, Phe(penta-F), Phe(3,4-C12), Phe(4-
CF3), Bip, Cha,
4-PyridylAlanine, f3hTyr, OctgGly, Tyr(4-N3), Phe(4-Br), Phe[4-(2-
aminoethoxy)]; X11 is 2-
Nal, 1-Nal, 2,4-dimethylPhe, Bip, Phe(3,4-C12), Phe(3,5-F2), Phe(4-CONH2),
Phe(4-F),4-
phenylcyclohexyl, Phe(4-CF3), a-MePhe, Nal, f3hPhe, f3hTyr, f3hTrp, BIP, Nva(5-
phenyl), Phe,
His, hPhe, Tic, Tqa, Tyr, Phe(4-0Me), Phe(4-Me), Tyr(2,5,7-tri-tert-Butyl),
Phe(4-0Ally1),
Phe(3-tBu), Phe(4-tBu), Phe(4-guanidino), Tyr(Bz1), OctGly; X12 is a-Me-Lys, D-
Ala, (D)Asn,
(D)Asp, (D)Leu, (D)Phe, (D)Tyr, Aib, a-MeLeu, a-MeOrn, Aib, f3-Ala, f3hAla,
PhArg, PhLeu,
f3hVal, f3-spiro-pip, Glu, hArg, Ile, Lys, N-MeLeu, N-MeArg, Ogl, Orn, Pro,
Gln, Ser, Thr, Tle,
4-amino-4-carboxy-tetrahydropyran, Achc Acpc, Acbc, Acvc, a-DiethylGly, a-
MeLys(Ac)õ a-
MeSer, a-MeVal; X13 is Lys, Lys(Ac), (D)Asn, (D)Leu, (D)Phe, (D)Thr, Ala, a-
MeLeu, Aib, (3-
Ala, f3-Glu, PhLeu, f3hVal, f3-spiro-pip, Cha, Chg, Asp, Dab, Dap, a-
DiethylGly, hLeu, Asn,
Ogl, Pro, Gln, Ser, Thr, Tba, Tle; X14 is Glu, Gly, His, Lys, Leu, Met, Asn,
Pro, Gln, Arg, Ser,
Thr, Tic; X15 is (D)Ala, (D)Asn, (D)Asp, (D)Leu, (D)Phe, (D)Thr, Aea, Asp,
Glu, Phe, Gly,
Lys, Leu, Pro, Asn, Arg or f3-Ala; X16 is Gly, Ser, Pro, Asn, Thr; or X17 is
Glu, Ser, Gly, Gln.
158
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1004801In particular embodiments of a peptide inhibitor of Formula (IV): X5 is
Cys, Cit, Asp,
Dab, Dap, Gly, His, hCys, Lys, Met, Asn, N-Me-Ala, N-Me-Asn, N-Me-Lys, N-Me-
Gln, N-Me-
Arg, Orn, Pro, Pen, Gln, Val; X6 is Glu, Arg, Ser; X7 is Trp, Glu, Gly, Ile,
Asn, Pro, Arg, Thr or
OctGly; X8 is Phe, Asn, Pro, Arg, Thr, Trp; X10 is Phe(4-0Me), 1-Nal, 2-Nal,
Aic, a-MePhe,
Bip, (D)Cys, Cha, DMT, (D)Tyr, Glu, His, hPhe(3,4-dimethoxy), hTyr, N-Me-Tyr,
Trp, Phe(4-
CONH2), Phe-(4-phenoxy), Thr, Tic, Tyr(3-tBu), Phe(4-tBu), Phe(4-CN), Phe(4-
Br), Phe(4-
NH2), Phe(4-F), Phe(3,5-F2), PheCH2CO2H, Phe(penta-F), Phe(3,4-C12), Phe(4-
CF3), Bip, Cha,
4-PyridylAlanine, (3hTyr, OctgGly, Tyr(4-N3), Phe(4-Br), Phe[4-(2-
aminoethoxy)]; X11 is 2-
Nal, 1-Nal, 2,4-dimethylPhe, Bip, Phe(3,4-C12), Phe(3,5-F2), Phe(4-CONH2),
Phe(4-F),4-
phenylcyclohexyl, Phe(4-CF3), a-MePhe, Nal, (3hPhe, (3hTyr, (3hTrp, BIP, Nva(5-
phenyl), Phe,
His, hPhe, Tic, Tqa, Tyr, Phe(4-0Me), Phe(4-Me), Tyr(2,5,7-tri-tert-Butyl),
Phe(4-0Ally1),
Phe(3-tBu), Phe(4-tBu), Phe(4-guanidino), Tyr(Bz1), OctGly; X12 is a-Me-Lys, D-
Ala, (D)Asn,
(D)Asp, (D)Leu, (D)Phe, (D)Tyr, Aib, a-MeLeu, a-MeOrn, Aib, (3-Ala, (3hAla,
(3hArg, (3hLeu,
(3hVal, (3-spiro-pip, Glu, hArg, Ile, Lys, N-MeLeu, N-MeArg, Ogl, Orn, Pro,
Gln, Ser, Thr, Tle;
X13 is Lys(Ac), (D)Asn, (D)Leu, (D)Phe, (D)Thr, Ala, a-MeLeu, Aib, (3-Ala, (3-
Glu, (3hLeu,
(3hVal, (3-spiro-pip, Cha, Chg, Asp, Dab, Dap, a-DiethylGly, hLeu, Asn, Ogl,
Pro, Gln, Ser, Thr,
Tba, Tle; X14 is Glu, Gly, His, Lys, Leu, Met, Asn, Pro, Gln, Arg, Ser, Thr,
Tic; X15 is (D)Ala,
(D)Asn, (D)Asp, (D)Leu, (D)Phe, (D)Thr, Aea, Asp, Glu, Phe, Gly, Lys, Leu,
Pro, Arg; X16 is
Gly, Ser, Pro, Asn, Thr; or X17 is Glu, Ser, Gly, Gln.
1004811 In certain embodiments, the peptide inhibitor is cyclized. In
particular embodiments, the
peptide is cyclized through an intramolecular bond between X4 and X9. In
particular
embodiments, the intramolecular bond is an amide bond.
1004821 In certain embodiments, the peptide inhibitor is linear or not
cyclized.
1004831In particular embodiments of a peptide inhibitor of Formula IV, one or
more, two or
more, or all three of Xl, X2, and X3 are absent.
1004841In certain embodiments, X3 is Glu, (D)Glu, Arg, (D)Arg, Phe, (D)Phe, 2-
Nal, Thr, Leu,
or (D)Gln. In certain embodiments, X3 is (D)Arg or (D)Phe.
159
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1004851In particular embodiments of a peptide inhibitor of Formula IV, one or
more, two or
more, or all three of X17, X19 and X20 are absent.
1004861In particular embodiments of a peptide inhibitor of Formula IV, X4 is
Dap, Dab, or
(D)Dab, and X9 is Glu, (D)Asp, or Asp. In particular embodiments of a peptide
inhibitor of
Formula I, X4 is Glu, (D)Asp or Asp, and X9 is Dab, Dap or (D)Dab.
1004871In particular embodiments of a peptide inhibitor of Formula IV, X18 is
(D)-Lys. In
certain embodiments, X17 is absent and X18 is (D)-Lys.
1004881In particular embodiments of a peptide inhibitor of Formula IV, the
peptide inhibitor
includes one or more, two or more, three or more, or all four of the following
features: X5 is Gln;
X6 isThr; X7 is Trp; and X8 is Gln.
1004891In particular embodiments of a peptide inhibitor of Formula IV, the
peptide inhibitor
includes one or more, two or more, three or more, four or more, five or more,
six or more, or
seven of the following features: X10 is Tyr, Phe[4-(2-aminoethoxy)], Phe(4-
CONH2) or Phe(4-
0Me); X11 is 2-Nal or Trp; X12 is 4-amino-4-carboxy-tetrahydropyran, Achc
Acpc, Acbc,
Acvc, Aib, a-DiethylGly, a-MeLys, a-MeLys(Ac), a-Me-Leu, a-MeOrn, a-MeSer, a-
MeVal,
or Arg; X13 is Glu or Lys(Ac); X14 is Asn; X15 is Gly, Asn, or f3-Ala; and X16
is AEA. In
particular embodiments of a peptide inhibitor of Formula IV, the peptide
inhibitor includes one
or more, two or more, three or more, four or more, five or more, six or more,
or seven of the
following features: X10 is Tyr; X11 is Trp; X12 is Arg; X13 is Glu; X14 is
Asn; X15 is Gly; and
X16 is AEA.
1004901In particular embodiments of a peptide inhibitor of Formula IV, the
peptide inhibitor
includes one or more, two or more, three or more, four or more, five or more,
six or more, seven
or more, eight or more, nine or more ten or more or all of the following
features: X5 is Gln; X6
isThr; X7 is Trp; X8 is Gln; X10 is Tyr; X11 is Trp; X12 is Arg; X13 is Glu or
Lys(Ac); X14 is
Asn; X15 is Gly; and X16 is AEA. In particular embodiments of a peptide
inhibitor of Formula
IV, the peptide inhibitor includes one or more, two or more, three or more,
four or more, five or
more, six or more, seven or more, eight or more, nine or more ten or more or
all of the following
160
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
features: X5 is Gin; X6 isThr; X7 is Trp; X8 is Gin; X10 is Tyr; X11 is Trp;
X12 is Arg; X13 is
Glu; X14 is Asn; X15 is Gly; and X16 is AEA.
1004911In certain embodiments of a peptide inhibitor of Formula IV, the
peptide is cyclized via
X4 and X9; X5, X6, X7 and X8 are Gin, Thr, Trp, and Gin, respectively; and
X10, X11, X12,
X13, X14, X15, and X16 are Tyr, Trp, Arg, Glu, Asn, Gly, and AEA,
respectively.
1004921In certain embodiments, the present invention includes a peptide of 8
to 20 amino acids,
optionally cyclized, comprising or consisting of having a core sequence
comprising:
Xaa4-Xaa5-Xaa6-Trp-Xaa8-Xaa9-[Phe(4-0Me)]-[ 2-Nail (Formula IVb)
[00493] wherein Xaa4 and Xaa9 are each independently selected from Dap, Dab,
Glu, Asp, (D)-
Asp and(D)-Dab, wherein Xaa4 and Xaa9 are capable of forming an intramolecular
bond, e.g., a
cyclic amide; and Xaa5, Xaa6 and Xaa8 are any amino acid residue, wherein the
peptide inhibits
binding of IL-23 to IL-23R. In particular embodiments, the peptide inhibitor
is a peptide
inhibitor of Formula IV. In particular embodiments, the peptide inhibits the
binding of IL-23 to
IL-23R
1004941In certain embodiments, of a peptide inhibitor of Formula IV, the
peptide inhibitor has a
structure shown in Table 7 or comprises an amino acid sequence set forth in
Table 7.
1004951Certain illustrative peptide inhibitors of the present invention are
also shown in any of
Formulas (Va), (Vb), (Vc), (Vd), (Ve), (Vf), (Vg) and (Vh), and in Tables 2-5,
which provide the
amino acid sequence of selected peptide inhibitors. These peptide inhibitors
are acetate salts.
Optional Characteristics of Peptide Inhibitors
1004961Any of the peptide inhibitors of the present invention may be further
defined, e.g., as
described below. It is understood that each of the further defining features
described herein may
be applied to any peptide inhibitors where the amino acids designated at
particular positions
allow the presence of the further defining feature.
1004971In certain embodiments of any of the peptide inhibitors described
herein, the peptide
inhibitor is cyclized.
161
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1004981In certain embodiments of any of the peptide inhibitors described
herein, the peptide
inhibitor or monomer subunit thereof is linear or not cyclized. In certain
embodiments where the
peptide is linear or not cyclized, X4 and X9 can be any amino acid.
[00499] In certain embodiments, the peptide inhibitor is cyclized, e.g.,
through X4 and X9.
1005001 In various embodiments, Rl is a bond, hydrogen, a C1-C6 alkyl, a C6-
C12 aryl, a C6-C12
aryl Cl -C6 alkyl, or a Cl -C20 alkanoyl, and including PEGylated versions
alone or as spacers of
any of the foregoing, e.g., acetyl. It is understood that the Rl may replace
or be present in
addition to the typical amine group located at the amino terminus of a
peptide. It is further
understood that Rl may be absent. In certain embodiments, the peptide
inhibitor comprises an N-
terminus selected from hydrogen, a C1-C6 alkyl, a C6-C12 aryl, a C6-C12 aryl
C1-C6 alkyl, or a
C1-C20 alkanoyl, and including PEGylated versions alone or as spacers of any
of the foregoing,
e.g., acetyl. In particular embodiments of any of the peptide inhibitors
described herein, Rl or the
N-terminal moiety is hydrogen. In certain embodiments, Rl is a bond, e.g., a
covalent bond.
1005011In certain embodiments of any of the peptide inhibitors having any of
the various
Formulas set forth herein, Rl or the N-terminal moiety is selected from
methyl, acetyl, formyl,
benzoyl, trifluoroacetyl, isovaleryl, isobutyryl, octanyl, and the conjugated
amides of lauric acid,
hexadecanoic acid, and y-Glu-hexadecanoic acid. In one embodiment, Rl or the N-
terminal
moiety is pG1u. In certain embodiments, Rl is hydrogen. In particular
embodiments, Rl is acetyl,
whereby the peptide inhibitor is acylated at its N-terminus, e.g., to cap or
protect an N-terminal
amino acid residue, e.g., an N-terminal Pen or Abu residue.
1005021In certain embodiments of any of the peptide inhibitors described
herein, Rl or the N-
terminal moiety is an acid. In certain embodiments, Rl or the N-terminal
moiety is an acid
selected from acetic acid, formic acid, benzoic acid, trifluoroacetic acid,
isovaleric acid,
isobutyric acid, octanoic acid, lauric acid, hexadecanoic acid, 4-
Biphenylacetic acid, 4-
fluorophenylacetic acid, gallic acid, pyroglutamic acid, cyclopentanepropionic
acid, glycolic
acid, oxalic acid, pyruvic acid, lactic acid, malonic acid, succinic acid,
malic acid, maleic acid,
fumaric acid, tartaric acid, citric acid, palmitic acid, benzoic acid, 3-(4-
hydroxybenzoyl) benzoic
acid, cinnamic acid, mandelic acid, 4-methylbicyclo(2.2.2)-oct-2-ene-1 -
carboxylic acid,
glucoheptonic acid, 3-phenylpropionic acid, trimethylacetic acid, tertiary
butylacetic acid, lauryl
162
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
sulfuric acid, gluconic acid, glutamic acid, hydroxynaphthoic acid, salicylic
acid, stearic acid,
muconic acid, an alkylsulfonic acid and an arylsulfonic acid.
1005031In particular embodiments, Rl or the N-terminal moiety is an
alkylsulfonic acid selected
from methanesulfonic acid, ethanesulfonic acid, 1,2-ethane-disulfonic acid,
and 2-
hydroxyethanesulfonic acid.
1005041In particular embodiments, Rl or the N-terminal moiety is an
arylsulfonic acid selected
from benzenesulfonic acid, 4- chlorobenzenesulfonic acid, 2-
naphthalenesulfonic acid, 4-
toluenesulfonic acid, and camphorsulfonic acid.
1005051In some embodiments, wherein a peptide of the present invention
comprises a
conjugation to an acidic compound such as, e.g., isovaleric acid, isobutyric
acid, valeric acid, and
the like, the presence of such a conjugation is referenced in the acid form.
So, for example, but
not to be limited in any way, instead of indicating a conjugation of
isovaleric acid to a peptide by
referencing i s oval eroyl (e. g. , isovaleroyl- [Pen] - Q TWQ [Pen]- [Phe(4-
0Me)]- [2-Nal] - [a-MeLys]-
[Lys(Ac)]-NG-NH2, in some embodiments, the present application references such
a conjugation
as isovaleric acid-[Pen]-QTWQ[Pen]-[Phe(4-0Me)]-[2-Nal]-[a-MeLys]-[Lys(Ac)]-NG-
NH2.
Reference to the conjugation in its acid form is intended to encompass the
form present in the
peptide inhibitor.
1005061In certain embodiments, the peptide inhibitor comprises a C-terminus
(e.g., R2 or the C-
termial moiety) selected from a bond, OH or NH2. In certain embodiments, R2 is
a bond. In
various embodiments of any of the peptide inhibitors having any of the various
Formulas set
forth herein, R2 or the C-terminal moiety is OH or NH2. It is understood that
the R2 or the C-
terminal moiety may replace or be present in addition to the carboxyl group
typically located at
the carboxy terminus of a peptide. It is further understood that R2 may be
absent.
1005071In particular embodiments of any of the peptide inhibitors having any
of the various
Formulae set forth herein, X comprises or consists of 7 to 35 amino acid
residues, 8 to 35 amino
acid residues, 9 to 35 amino acid residues, 10 to 35 amino acid residues, 7 to
25 amino acid
residues, 8 to 25 amino acid residues, 9 to 25 amino acid residues, 10 to 25
amino acid residues,
7 to 20 amino acid residues, 8 to 20 amino acid residues, 9 to 20 amino acid
residues, 7 to 18
163
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
amino acid residues, 8 to 18 amino acid residues, 9 to 18 amino acid residues,
or 10 to 18 amino
acid residues.
[00508] In certain embodiments of any of the Formulae set forth herein, X
either or both does not
comprise or does not consist of an amino acid sequence set forth in US Patent
Application
Publication No. US2013/0029907. In certain embodiments of any of the Formulae
set forth
herein, X either or both does not comprise or does not consist of an amino
acid sequence set
forth in US Patent Application Publication No. US2013/0172272.
1005091In certain embodiments of any of the peptide inhibitors described
herein, the peptide
inhibitor, or each monomer subunit thereof, comprises or consists of at least
3, at least 4 at least
5, at least 6, or at least 7 amino acid residues carboxy terminal of the X9
amino acid residue. In
particular embodiments of any of the peptide inhibitors described herein, the
peptide inhibitor
comprises 3 to 11,3 to 10,3 to 9,3 to 8,3 to 7,3 to 6,3 to 5,3 to 4, 3, 4, 5,
6, 7, 8, 9, 10, or 11
amino acid residues carboxy terminal of the X9 amino acid residue.
1005101In certain embodiments of any of the peptide inhibitors described
herein, the peptide
inhibitor, or each monomer subunit thereof, comprises or consists of 4 amino
acid residues
between X4 and X9. In one embodiment, both X4 and X9 are cysteines.
[00511]In certain embodiments, a peptide inhibitor of any of the Formulae
described herein
comprises the amino acid residues or moieties indicated as X4-X15. In
particular embodiments,
the peptide inhibitor does not include X1 -X3 or X16-X20. In certain
embodiments, the peptide
inhibitors include an N-terminal extension of one to three amino acid residues
corresponding to
any of X1 -X3. In particular embodiments, any one or more of X1 , X2 and X3,
when present, are
a D-amino acid. In certain embodiments, the peptide inhibitors include an C-
terminal extension
of one to five amino acid residues corresponding to any of X16-X20. In
particular embodiments,
any one or more of X16, X17, X18, X19 and X20, when present, are a D-amino
acid.
Illustrative amino acid residues that may be present in the N-terminal and/or
C-terminal
extensions are shown in Tables 3 and 5. These tables each show a first peptide
inhibitor, with
derivates thereof comprising N-terminal extensions, C-terminal extensions,
and/or conjugated
moieties. The present invention includes derivatives of any fo the peptide
inhibitors described
herein comprising one or more such N-terminal extension, C-terminal extension,
and/or
164
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
conjugated moiety. In certain embodiments, any of the amino acid residues
shown in the
extended positions in Tables 3 and 5 may be present in any combination in a
peptide inhibitor of
the present invention. In particular embodiments, the N-terminal and/or C-
terminal extensions
are associated with an increased half-life, e.g., upon administration to a
subject.
1005121In certain embodiments of any of the peptide inhibitors described
herein, the peptide
inhibitor, or each monomer subunit thereof, comprises the amino acid sequence
motif, W-X-X-
Y-W, e.g., at positions X7-X11. In certain embodiments, the peptide inhibitor,
or each monomer
subunit thereof, comprises the amino acid sequence motif, C-X-X-W-X-C-Y-W,
e.g., at
positions X4-X11. In certain embodiments, the peptide inhibitor, or each
monomer subunit
thereof, comprises the amino acid sequence motif, Pen-X-X-W-X-Pen-Y-W, e.g.,
at positions
X4-X11. In certain embodiments of any of the peptide inhibitors described
herein, the peptide
inhibitor, or both monomer subunit thereof, does not comprise the amino acid
sequence motif,
W-X-X-Y-W, e.g., at positions X7-X11, where X is any amino acid.
1005131In certain embodiments of any of the Formula or peptide inhibitors
described herein, the
peptide inhibitor comprises one or more amino acid residues N-terminal to X4.
In particular
embodiments, X3 is present. In certain embodiments, X3 is Glu, (D)Glu, Arg,
(D)Arg, Phe,
(D)Phe, 2-Nal, Thr, Leu, or (D)G1n. In certain embodiments, X3 is (D)Arg or
(D)Phe.
1005141In particular embodiments of any of the Formula or peptide inhibitors
described herein,
the peptide inhibitor comprises an amino acid at X2. In particular
embodiments, X2 is Glu,
(D)Asp, Arg, (D)Arg, Phe, (D)Phe, 2-Nal, Thr, Leu, (D)G1n, or (D)Asn. In
certain embodiments,
X2 and X3 are present. In particular embodiments, X2 is Glu, (D)Asp, Arg,
(D)Arg, Phe,
(D)Phe, 2-Nal, Thr, Leu, (D)G1n, or (D)As, and X3 is (D)Arg.
1005151In certain embodiments, a peptide inhibitor of the present invention,
or one or both
monomer subunits thereof, comprises, optionally at its C-terminus, one of the
following amino
acid sequences:
ENG;
ENN;
[4-amino-4-carboxy-tetrahydropyran] -ENN;
165
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[Lys(Ac)]-NN;
[a-MeLys]-ENG;
[a-MeLys]- [Lys(Ac)]-NN;
[a-MeLeu]- [Lys(Ac)]-NN
[a-MeLeu]-ENG;
[a-MeOrn]-[Lys(Ac)]-NG;
[a-MeLeu]-ENG;
Aib-[Lys(Ac)]-NG;
Aib-[Lys(Ac)]-NN;
NG-[AEA](D)-Lys];
[Dapa]-NG-[AEA] - [(D)-Lys];
[Orn]-NG-[AEA]-[(D)-Lys];
[a-MeLys]-ENN;
[4-amino-4-carboxy-tetrahydropyran]- [Lys(Ac)]-NN;
[Achc]-[Lys(Ac)]-NN; or
[Acpc]-[Lys(Ac)]-NN.
[00516] In particular embodiments, one of these amino acid sequences
constitutes the terminal C-
terminal amino acids of the peptide. In particular embodiment, these amino
acid sequences
correspond to X13-X15 or X12-X15 or X14-X16 or X13-X17.
1005171In certain embodiments, a peptide inhibitor of the present invention,
or one or both
monomer subunits thereof, comprises, optionally at its C-terminus, one of the
following amino
acid sequences:
WQCY42-NalHa-MeLys];
WQC-[Phe(4-0Me)]-[2-Nal]-[a-MeLys];
166
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
WQC- [Phe(4-0Me)]- [2-Nall- [Al b];
WQ-[Pen]-[Phe(4-0Me)]- [2-Nall- [a-MeLys];
W-Xaa8-C-Phe[4-(2-aminoethoxy)] 42-Nall;
W-Xaa8-C-Phe[4-(2-aminoethoxy)] ;
W-Xaa8-C-Phe[4-(2-aminoethoxy)]; or
1005181 W-Xaa8-C-[Phe(4-0CH3)]. In particular embodiments, one of these amino
acid
sequences constitutes the terminal C-terminal amino acids of the peptide. In
particular
embodiment, these amino acid sequences correspond to X7 to X12 or X7 to X11 or
X7 to X10.
1005191In certain embodiments of any of the peptide inhibitors described
herein, including both
peptide monomer inhibitors and monomer subunits of peptide dimer inhibitors,
the peptide
monomer inhibitor or monomer subunit is cyclized via a peptide bond between
its N-terminal
amino acid residue and its C-terminal amino acid residue. In particular
embodiments, the
peptide inhibitor (or monomer subunit thereof) comprises both an
intramolecular bond between
X4 and X9 and a peptide bond between its N-terminal amino acid residue and its
C-terminal
amino acid residue. In certain embodiments, the intramolecular bond is any of
those described
herein, e.g., a disulfide bond or a thioether bond.
[00520] In certain embodiments, the present invention includes a peptide
inhibitor that comprises
a core consensus sequence selected from one of the following (shown in N-
terminal to C-
terminal direction):
X1 -X2 -X3 -Pen-X5-X6-W-X8-Pen-X10-X11 -X12 -X13 -X14-X15;
Pen-X5-X6-W-Q-Pen;
Pen-X5-X6-W-X8-Pen;
Pen-X5 -X6-W-X8 -Pen- [Phe(4-CONH2)]; and
Pen-X5-X6-W-X8-Pen- [Phe [4- (2-aminoethoxy)] ,
167
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
wherein the Pen residues arejoined by an intramolecular bond, e.g., disulphide
bond. Xl, X2,
X3, X5, X6, X8, X10, X11, X12, X13, X14, and X15 may be any amino acid. In
some
embodiment X5 is Arg, Asn, Gln, Dap, Orn; X6 is Thr or Ser; and X8 is Gln,
Val, Phe, Glu, Lys.
In particular embodiments, Xl, X2, X3, X5, X6, X8, X10, X11, X12, X13, X14,
and X15 are
defined as described in any of the various Formulas and peptide inhibitors
described herein.
1005211 In certain embodiments, the present invention includes a peptide
inhibitor that comprises
a core consensus sequence selected from one of the following (shown in N-
terminal to C-
terminal direction):
X1 -X2-X3-Abu-X5-X6-W-X8-C-X9-X10-X11-X12-X13-X14-X15;
Abu-X5-X6-W-Q-C;
Abu-X5-X6-W-X8-C;
Abu-X5-X6-W-X8-C-[Phe(4-CONH2)]; and
Abu-X5-X6-W-X8-C-[Phe[4-(2-aminoethoxy)]],
where Abu and C are linked through a intra moleculer thiother bond. Xl, X2,
X3, X5, X6, X8,
X10, X11, X12, X13, X14, and X15 may be any amino acid. In some embodiment X5
is Arg,
Asn, Gln, Dap, Orn; X6 is Thr or Ser; and X8 is Gln, Val, Phe, Glu, Lys. In
particular
embodiments, Xl, X2, X3, X5, X6, X8, X10, X11, X12, X13, X14, and X15 are
defined as
described in any of the various Formulas and peptide inhibitors described
herein.
1005221In certain embodiments, any of the peptide inhibitors described herein
may be further
cyclized via a peptide bond between its N-terminal amino acid residue and its
C-terminal amino
acid residue. In particular embodiments, the peptide inhibitor comprises a
peptide bond between
X3 or X4 and any one of X9, X10, X11, X12, X13, X14, X15, X16, X17, X18, X19
or X20. In
particular embodiments, peptide inhibitors of the present invention comprise a
peptide bond
between their N-terminal and C-terminal amino acid residues, and they also
comprise an
intramolecular bond between X4 and X9. In certain embodiments, the
intramolecular bond is a
disulfide bond, a thioether bond, a lactam bond or any of the other bonds
described herein.
168
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Peptide Dimers
1005231In certain embodiments, the present invention includes dimers of the
monomer peptide
inhibitors described herein, including dimers of any of the monomer peptide
inhibitors described
herein or in the accompanyingtables, figures or sequences listing. These
dimers fall within the
scope of the general term "peptide inhibitors" as used herein. Illustrative
dimers of the present
invention are also shown in the accompanying tables, which indicate the
dimerized monomer
subnits in brackets followed by the linker. Unless otherwise indicated, the
subunits are linked
via their C-termini. The term "dimer," as in a peptide dimer, refers to
compounds in which two
peptide monomer subinits are linked. A peptide dimer inhibitor of the present
invention may
comprise two identical monomer subunits, resulting in a homodimer, or two non-
identical
monomer subunits, resulting in a heterodimer. A cysteine dimer comprises two
peptide
monomer subunits linked through a disulfide bond between a cysteine residue in
one monomer
subunit and a cysteine residue in the other monomer subunit.
1005241In some embodiments, the peptide inhibitors of the present invention
may be active in a
dimer conformation, in particular when free cysteine residues are present in
the peptide. In
certain embodiments, this occurs either as a synthesized dimer or, in
particular, when a free
cysteine monomer peptide is present and under oxidizing conditions, dimerizes.
In some
embodiments, the dimer is a homodimer. In other embodiments, the dimer is a
heterodimer.
1005251In certain embodiments, a peptide dimer inhibitor of the present
invention is a peptide
dimer comprising two peptide inhibitors of the invention, including but not
limited to a
homodimer or heterdimer comprising any of the peptide sequences shown herein,
e.g., in Tables
3A-3H, 4A, 4B, 5A-5C, 6, 7, 8, 9, 10, 11, 12, 13, 14 or 15.
[00526] Certain amino acid sequences listed in Tables 3A-3H, 4A, 4B, 5A-5C, 6,
7, 8, 9, 10, 11,
12, 13, 14 or 15 are shown using one letter codes for amino acids. Where only
the monomer
peptide inhibitor sequences are shown; however it is understood that, in
certain embodiments,
these monomer peptide inhibitors, i.e., monomer subunits, are dimerized to
form peptide dimer
inhibitors, in accordance with the present teaching and as shown generally,
e.g., in Tables 3A-
3H, 4A, 4B, 5A-5C, 6, 7, 8, 9, 10, 11, 12, 13, 14 or 15.
169
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[00527] In certain embodiments, monomer subunits of the present invention may
be dimerized by
a suitable linking moiety, e.g., a disulphide bridge between two cysteine
residues, one in each
peptide monomer subunit, or by another suitable linker moiety, including but
not limited to those
defined herein. Some of the monomer subunits are shown having C- and N-termini
that both
comprise free amine. Thus, to produce a peptide dimer inhibitor, the monomer
subunit may be
modified to eliminate either the C- or N-terminal free amine, thereby
permitting dimerization at
the remaining free amine. Further, in some instances, a terminal end of one or
more monomer
subunits is acylated with an acylating organic compound selected from the
group consisting of:
Trifluoropentyl, Acetyl, Octonyl, Butyl, Pentyl, Hexyl, Palmityl,
Trifluoromethyl butyric,
cy cl op entane carboxylic, cyclopropylacetic, 4-fluorobenzoic, 4-fluorophenyl
acetic, 3 -
Phenylpropionic, tetrahedro-2H-pyran-4carboxylic, succinic acid, and glutaric
acid. In some
instances, monomer subunits comprise both a free carboxy terminal and a free
amino terminal,
whereby a user may selectively modify the subunit to achieve dimerization at a
desired terminus.
One having skill in the art therefore, will appreciate that the monomer
subunits of the instant
invention may be selectively modified to achieve a single, specific amine for
a desired
dimerization.
1005281It is further understood that the C-terminal residues of the monomer
subunits disclosed
herein are optionally amides. Further, it is understood that, in certain
embodiments, dimerization
at the C-terminus is facilitated by using a suitable amino acid with a side
chain having amine
functionality, as is generally understood in the art. Regarding the N-terminal
residues, it is
generally understood that dimerization may be achieved through the free amine
of the terminal
residue, or may be achieved by using a suitable amino acid side chain having a
free amine, as is
generally understood in the art.
1005291 The linker moieties connecting monomer subunits may include any
structure, length,
and/or size that is compatible with the teachings herein. In at least one
embodiment, a linker
moiety is selected from the non-limiting group consisting of cysteine, lysine,
DIG, PEG4, PEG4-
biotin, PEG13, PEG25, PEG1K, PEG2K, PEG3.4K, PEG4K, PEG5K, IDA, ADA, Boc-IDA,
Glutaric acid, Isophthalic acid, 1,3-phenylenediacetic acid, 1,4-
phenylenediacetic acid, 1,2-
phenylenediacetic acid, Triazine, Boc-Triazine, IDA-biotin, PEG4-Biotin, AADA,
suitable
aliphatics, aromatics, heteroaromatics, and polyethylene glycol based linkers
having a molecular
170
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
weight from approximately 400Da to approximately 40,000Da. Non-limiting
examples of
suitable linker moieties are provided in Table 2A.
Table 2A. Illustrative Linker Moieties
Abbrivation Discription Structure
DIG DIGlycolic acid,
0
0
Bifunctional PEG linker with 4 PolyEthylene
PEG4
Glycol units a
Bifunctional PEG linker with 13 PolyEthylene
PEG13
Glycol units
Bifunctional PEG linker with 25 PolyEthylene
PEG25
Glycol units
Bifunctional PEG linker with PolyEthylene
PEG1K
Glycol Mol wt of 1000Da
Bifunctional PEG linker with PolyEthylene
PEG2K
Glycol Mol wt of 2000Da
Bifunctional PEG linker with PolyEthylene
PEG3.4K
Glycol Mol wt of 3400Da
Bifunctional PEG linker with PolyEthylene
PEG5K
Glycol Mol wt of 5000Da
DIG DIGlycolic acid
171
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
0
0
(3-A1a-IDA (3-A1a-Iminodiacetic acid
0
Y-
Boc- (3 -Ala-Iminodiacetic acid
Ala-IDA
0
Ac-(3 -Ala -
Ac- (3 -Ala-Iminodiacetic acid
IDA
0
Palmityl- (3 -Ala-Iminodiacetic acid
Palm 0
)=3
0
GTA Glutaric acid
PMA Pemilic acid
172
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
oo
AZA Azelaic acid
DDA Dodecanedioic acid
0
IPA Isopthalic aicd
0
1,3-PDA 1,3- Phenylenediacetic acid
0 0 0 0
0
1,4-PDA 1,4- Phenylenediacetic acid 0
1,2-PDA 1,2 - Phenylenediacetic acid
0
N=<
Triazine Amino propyl Triazine di-acid ti¨c 114
N¨)1_0
173
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1 1 0
Ni)-
Boc- \ N=(
Boc-Triazine di-acid N4 14
Triazine 14 4
0
Amino diacetic acid , 0
ADA (which may also referred to as Iminodiacetic 0/4J.L0
acid)
n-Acetyl amino acetic acid
AADA (which may also referred to as N-acetyl )0jL
0 0
Iminodiacetic acid)
,-,
PEG4- PEG4-Biotin (Product number 10199,
Biotin QuantaBioDesign)
'l .NC\r"\-1H-\''',.---\;,--ve'c,-%"./11,:,=,
---'N
IDA-Biotin N-Biotin- (3 -Ala-Iminodiacetic acid
e )
c,..
OH
174
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
H2N
OH
Lys Lysine H2N
0
10053011n some embodiments, a peptide dimer inhibitor is dimerized via a
linker moiety. In
some embodiments, a peptide dimer inhibitor is dimerized via an intermolecular
disulfide bond
formed between two cysteine residues, one in each monomer subunit. In some
embodiments, a
peptide dimer inhibitor is dimerized via both a linker moiety and an
intermolecular disulfide
bond formed between two cysteine residues. In some embodiments, the
intramolecular bond is a
thioether, lactam, triazole, selenoether, diselenide or olefin, instead of the
disulfide bond.
[00531] An illustrative diageam of one embodiments of a dimer is shown below:
o
o NH2 NH2
it o
o./ 0 0 0 0 .N
0
N N N
H 4HNH2 0
0 H.,....1-1OjciH H
N NH
HNN N [Nlj= 1\1,..,NH2
N
H H H H
0 0 0 0 NH2
0
-
NH /K 1101
o NH2
111 NH -1)
HN,Ire
S S 0
0
0
01)
0
NH
0 NH2 NH2
o/HH
0HO o o o o JNH2 0
OH
I-I...... H H
HN\/cN N ENIA N
N N N N N [gi r\h(rNH2
H H
0 NH2 H
H
0 0 0 0 0 NH2
-
/ IL 0
NH
00 WI
HN
S S
0
Compound D .
175
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[00532] One having skill in the art will appreciate that the linker (e.g., C-
and N-terminal linker)
moieties disclosed herein are non-limiting examples of suitable, and that the
present invention
may include any suitable linker moiety. Thus, some embodiments of the present
invention
comprises a homo- or heterodimer peptide inhibitor comprised of two monomer
subunits
selected from the peptides shown in any of tables herein or comprising or
consisting of a
sequence presented in any of tables herein, wherein the C- or N-termini of the
respective
monomer subunits (or internal amino acid residues) are linked by any suitable
linker moiety to
provide a dimer peptide inhibitor having IL-23R inhibitory activity. In
certain embodiments, a
linker binds to the N- or C-terminus of one monomer subunit and an internal
amino acid residue
of the other monomer subunit making up the dimer. In certain embodiments, a
linker binds to an
internal amino acid residue of one monomer subunit and an internal amino acid
residue of the
other monomer subunit making up the dimer. In further embodiments, a linker
binds to the N-or
C-terminus of both subunits.
1005331In particular embodiments, a peptide inhibitor of the present invention
comprise two or
more polypeptide sequences of monomer peptide inhibitors described herein.
1005341In one embodiment, a peptide dimer inhibitor of the present invention
comprises two
peptide monomer subunits connected via one or more linker moieties, wherein
each peptide
monomer subunit comprises or consists of 7 to 35 amino acid residues, 8 to 35
amino acid
residues, 9 to 35 amino acid residues, 10 to 35 amino acid residues, 7 to 25
amino acid residues,
8 to 25 amino acid residues, 9 to 25 amino acid residues, 10 to 25 amino acid
residues, 7 to 20
amino acid residues, 8 to 20 amino acid residues, 9 to 20 amino acid residues,
7 to 18 amino acid
residues, 8 to 18 amino acid residues, 9 to 18 amino acid residues, or 10 to
18 amino acid
residues and comprises the sequence of Formula Ia, as described herein.
1005351In particular embodiments, one or both of the monomer subunits comprise
the sequence
of any one of Formula Formula X, Formula I, II, III, IV or V as described
herein.
1005361In certain embodiments, a peptide dimer inhibitor comprises two peptide
monomer
subunits connected via one or more linker moieties, wherein each peptide
monomer subunit is 8-
20 amino acids in length and comprises a sequence of any one of the formulas
describd herein,
e.g., Formula X, Formula I, II, III, IV or V. In certain embodiments, a
peptide dimer inhibitor
176
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
comprises two peptide monomer subunits connected via one or more linker
moieties, wherein
each peptide monomer subunit is 8-20 amino acids in length and comprises a
sequence of any
one of Formula X, Formula I, II, III, IV or V.
[00537] In certain embodiments, a peptide dimer inhibitor has the structure of
Formula VI:
(R1-X-R2)2-L (VI)
[00538] or a pharmaceutically acceptable salt or solvate thereof,
[00539] wherein each Rl is independently absent, a bond (e.g., a covalent
bond), or R1 is selected
from hydrogen, a C1-C6 alkyl, a C6-C12 aryl, a C6-C12 aryl C1-C6 alkyl, a C 1 -
C20 alkanoyl,
and including PEGylated versions alone or as spacers of any of the foregoing;
[00540] each R2 is independently absent, a bond (e.g., a covalent bond), or
selected from OH or
NH2;
1005411L is a linker moiety; and
[00542] each X is an independently selected peptide monomer subunit comprising
or consisting of
7 to 35 amino acid residues, 8 to 35 amino acid residues, 9 to 35 amino acid
residues, 10 to 35
amino acid residues, 7 to 25 amino acid residues, 8 to 25 amino acid residues,
9 to 25 amino acid
residues, 10 to 25 amino acid residues, 7 to 20 amino acid residues, 8 to 20
amino acid residues,
9 to 20 amino acid residues, 7 to 18 amino acid residues, 8 to 18 amino acid
residues, 9 to 18
amino acid residues, or 10 to 18 amino acid residues amino acids in length,
each comprising or
consisting of the sequence of Formula Ia, as described herein. In particular
embodiments, each
peptide monomer subunit comprises or consists of a sequence of Formula Ix, Ia,
Ib, Ic, Id, Ie, If,
Ig, Ih, Ti, Ij, Ik, Tl, Im, In, To, Ip, Iq, Iq', Is, It, IIa, IIb, IIc, IId,
Ma, Mb, IIIc, IIId, Me, IVa, IVb,
or Va-Vh as described herein.
1005431In certain embodiments, one or both peptide monomer subunit of a
peptide dimer
inhibitor is cyclized, e.g., via an intramolecular bond between X4 and X9. In
certain
embodiments wherein both peptide monomer subunits are cyclized, the
intramolecular bond may
be the same or different between the two peptide monomer subinits. In certain
embodiments, one
177
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
or both intramolecular bond is a disulfide bond, a thioether bond, a lactam
bond, a selenoether,
diselenide, or an olefin bond.
1005441In one embodiment, X4 and X9 of the one or both cyclized peptide
monomer subunit is
independently selected from Cys, Pen, hCys, D-Pen, D-Cys and D-hCys, and the
intramolecular
bond is a disulfide bond.
1005451In one embodiment, X4 and X9 of the one or both cyclized peptide
monomer subunit is
independently selected from Glu, Asp, Lys, Orn, Dap, Dab, D-Dap, D-Dab, D-Asp,
D-Glu and
D-Lys, and the intramolecular bond is a lactam bond.
1005461 In one embodiment, X4 and X9 of the one or both cyclized peptide
monomer subunit are
each independently selected from f3-azido-Ala-OH, propargylglycine, and the
peptide dimer
inhibitor is cyclized through a triazole ring. In one embodiment, X4 and X9 of
the one or both
cyclized peptide monomer subunit are each independently selected from 2-
allylglycine, 2-(3'-
butenyl)glycine, 2-(4'-pentenyl)glycine, 2-(5'-hexenyl)glycine, and the
peptide dimer inhibitor is
cyclized vi a ring closing methasis to give the corresponding olefins /
'stapled peptides'.
10054711n one embodiment, X4 of one or both cyclized peptide monomer subunit
is 2-
chloromethylbenzoic acid, mercapto-propanoic acid, mercapto-butyric acid, 2-
chloro-acetic acid,
3-choro-propanoic acid, 4-chloro-butyric acid, 3-chloro-isobutyric acid, or
hSer(C1), X9 of one
or both cyclized peptide monomer subunit is hSer(C1), Cys, Pen, hCys, D-Pen, D-
Cys or D-hCys,
and the intramolecular bond is a thioether bond.
10054811n one embodiment, X4 of one or both cyclized peptide monomer subunit
is 2-
chloromethylbenzoic acid, 2-chloro-acetic acid, 3-choro-propanoic acid, 4-
chloro-butyric acid, 3-
chloro-isobutyric acid, hSer(C1), or Sec, X9 of one or both cyclized peptide
monomer subunit is
hSer(C1) or Sec, and the intramolecular bond is a selenoether bond.
1005491 In certain embodiments, one or both intramolecular bond is a
diselenide bond.
1005501In certain embodiments, one or both peptide monomer subunits is linear
or not cyclized.
178
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1005511In particular embodiments, of the peptide dimer inhibitors, each X7 and
each X11 are
both W. In certain embodiments, each X7 and each X11 are both W, each X10 is
Y, and each
X4 and X9 are both C. In certain embodiments, each X7 and each X11 are both W,
each X10 is
Y, and each X4 and X9 are amino acids capable of forming an intramolecular
bond that is a
thioether bond, a lactam bond, a triazole, a selenoether, a diselenide bond,
or an olefin bond.
1005521In certain embodiments of the peptide dimer inhibitors, one or both
peptide monomer
subunit has a structure shown herein, e.g., in Tables 3A-3I, or comprises an
amino acid sequence
shown herein, e.g., as set forth in Tables 3A-3I, or wherein the peptide dimer
inhibitor has a
structure shown herein, e.g., in Table 3F, or comprises an amino acid sequence
shown herein,
e.g., as set forth in Table 3F.
1005531In particular embodiments, each Rl is independently a bond (e.g., a
covalent bond), or
selected from hydrogen, a C 1 -C6 alkyl, a C6-C12 aryl, a C6-C12 aryl C 1 -C6
alkyl, a C1-C20
alkanoyl, and including PEGylated versions alone or as spacers of any of the
foregoing. In
particular embodimetns, the N-terminus of each subunit includes a moiety
selected from
hydrogen, a C1-C6 alkyl, a C6-C12 aryl, a C6-C12 aryl C 1 -C6 alkyl, a C1-C20
alkanoyl, and
including PEGylated versions alone or as spacers of any of the foregoing.
10055411n certain embodiments of any of the peptide inhibitors having any of
the various
Formulae set forth herein, each R1 (or N-terminal moiety) is selected from
methyl, acetyl,
formyl, benzoyl, trifluoroacetyl, isovaleryl, isobutyryl, octanyl, and the
conjugated amides of
lauric acid, hexadecanoic acid, and y-Glu-hexadecanoic acid.
1005551ln particular embodiments, each R2 (or C-terminal moiety) is
independently a bond (e.g.,
a covalent bond), or selected from OH or NH2.
1005561In particular embodiments of any of the peptide inhibitors having any
of the various
Formulae set forth herein, each X comprises or consists of 7 to 35 amino acid
residues, 8 to 35
amino acid residues, 9 to 35 amino acid residues, 10 to 35 amino acid
residues, 7 to 25 amino
acid residues, 8 to 25 amino acid residues, 9 to 25 amino acid residues, 10 to
25 amino acid
residues, 7 to 18 amino acid residues, 8 to 18 amino acid residues, 9 to 18
amino acid residues,
or 10 to 18 amino acid residues.
179
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1005571In particular embodiments, one or both X comprises or consists of the
sequence of any
one of the formulas described herein. In certain embodiments of any of the
peptide inhibitors,
including dimers, or Formulae set forth herein, an X does not comprise or
consist of an amino
acid sequence set forth in US Patent Application Publication No.
US2013/0029907. In certain
embodiments of any of the peptide inhibitors, including dimers, or formulas
set forth herein, an
X does not comprise or consist of an amino acid sequence set forth in US
Patent Application
Publication No. US2013/0172272.
1005581 In particular embodiments of peptide inhibitors of the present
invention (both monomers
and dimers) comprising Cys at position X4 and Cys at position X9, the Cys at
position X4 and
and the Cys at position X9 are linked by a disulphide bridge.
1005591In particular embodiments of peptide inhibitors of the present
invention, each X7 and
each X11 are not both W.
1005601In particular embodiments of peptide inhibitors of the present
invention, each X7 and
each X11 are both W.
1005611In particular embodiments of peptide inhibitors of the present
invention, each X7 and
each X11 are both W, X10 is Y, and X4 and X9 are both C.
1005621In certain embodiments, at least two cysteine residues of the peptide
dimer inhibitor are
linked by a disulphide bridge, either intramolecular or intermolecular.
10056311n particular embodiments of either or both monomer subunit (e.g., Ix,
Ia-It where
permissible) present in a peptide dimer inhibitor, X4 and X9 are both Cys.
10056411n particular embodiments of either or both monomer subunit (e.g., Ix,
Ia-It where
permissible) present in a peptide dimer inhibitor, X7 and X11 are both W.
10056511n particular embodiments of either or both monomer subunit (e.g., Ia-
It where
permissible) present in a peptide dimer inhibitor, X7 and X11 are both W, X10
is Y, and X4 and
X9 are both Cys.
180
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
10056611n particular embodiments of either or both monomer subunit (e.g., Ia-
It where
permissible) present in a peptide dimer inhibitor, X15 is Gly or Ser.
10056711n particular embodiments of either or both monomer subunit (e.g., Ia-
It where
permissible) present in a peptide dimer inhibitor, X16 is AEA or AEP.
10056811n particular embodiments of either or both monomer subunit (e.g., Ia-
It where
permissible) present in a peptide dimer inhibitor, X10 is Tyr or Phe, or an
analog of Tyr or Phe.
10056911n particular embodiments of either or both monomer subunit (e.g., Ia-
It where
permissible) present in a peptide dimer inhibitor, X11 is Trp.
1005701In particular embodiments of any of the peptide dimer inhibitors
described herein, either
or both 1Z1 is hydrogen.
[00571] In particular embodiments of peptide dimer inhibitors of the present
invention, the linker
moiety (L) is any of the linkers described herein or shown in Table 2A or 2B.
In certain
embodiments, L is a lysine linker, a diethylene glycol linker, an
iminodiacetic acid (IDA) linker,
a P-Ala-iminodiaceticacid (f3-Ala-IDA) linker, or a PEG linker.
1005721In various embodiments of any of the peptide dimer inhibitors, each of
the peptide
monomer subunits is attached to a linker moiety via its N-terminus, C-
terminus, or an internal
amino acid residue.
1005731In certain embodiments of any of the peptide dimer inhibitors, the N-
terminus of each
peptide monomer subunit is connected by a linker moiety.
1005741In certain embodiments of any of the peptide dimer inhibitors, the C-
terminus of each
peptide monomer subunit is connected by a linker moiety.
1005751In certain embodiments of any of the peptide dimer inhibitors, each
peptide monomer
subunit is connected by a linker moiety attached to an internal amino acid.
181
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1005761In certain embodiements of peptide dimer inhibitors, the linker moiety
is a diethylene
glycol linker, an iminodiacetic acid (IDA) linker, a f3-Ala-iminodiaceticacid
(j3-Ala-IDA) linker,
or a PEG linker.
1005771In certain embodiments of the peptide dimer inhibitors, one or both
peptide monomer
subunit has a structure shown in any of the tables in the Examples or
comprises an amino acid
sequence set forth in any of the tables in the Examples.
1005781In certain embodiments of any of the peptide inhibitors, including
dimers, or Formulae
set forth herein, an X does not comprise or consist of an amino acid sequence
set forth in US
Patent Application Publication No. US2013/0029907. In certain embodiments of
any of the
peptide inhibitors, including dimers, or Formulas set forth herein, an X does
not comprise or
consist of an amino acid sequence set forth in US Patent Application
Publication No.
US2013/0172272.
1005791In particular embodiments of peptide inhibitors of the present
invention, each X7 and
each X11 are both W, X10 is Y, and X4 and X9 are both Pen.
In certain embodiments, at least two cysteine residues of the peptide dimer
inhibitor are linked
by a disulphide bridge, either intramolecular or intermolecular.
Peptide Inhibitor Conjugates and Biopolymers
1005801In certain embodiments, peptide inhibitors of the present invention,
including both
monomers and dimers, comprise one or more conjugated chemical substituents,
such as
lipophilic substituents and polymeric moieties, which may be referred to
herein as half-life
extension moieties. Without wishing to be bound by any particular theory, it
is believed that the
lipophilic substituent binds to albumin in the bloodstream, thereby shielding
the peptide inhibitor
from enzymatic degradation, and thus enhancing its half-life. In addition, it
is believed that
polymeric moieties enhance half-life and reduce clearance in the bloodstream.
1005811In additional embodiments, any of the peptide inhibitors, e.g. petides
of Formulas (Va)-
(Vh), further comprise a linker moiety attached to an amino acid residue
present in the inhibitor,
e.g., a linker moiety may be bound to a side chain of any amino acid of the
peptide inhibitor, to
182
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
the N-terminal amino acid of the peptide inhibitor, or to the C-terminal amino
acid of the peptide
inhibitor.
1005821In additional embodiments, any of the peptide inhibitors e.g. petides
of Formulas (Va)-
(Vh), further comprise half-life extension moiety attached to an amino acid
residue present in the
inhibitor, e.g., a half-life extension moiety may be bound to a side chain of
any amino acid of the
peptide inhibitor, to the N-terminal amino acid of the peptide inhibitor, or
to the C-terminal
amino acid of the peptide inhibitor.
1005831In additional embodiments, any of the peptide inhibitors e.g. petides
of Formulas (Va)-
(Vh), further comprise half-life extension moiety attached to a linker moiety
that is attached to an
amino acid residue present in the inhibitor, e.g., a half-life extension
moiety may be bound to a
linker moiety that is bound to a side chain of any amino acid of the peptide
inhibitor, to the N-
terminal amino acid of the peptide inhibitor, or to the C-terminal amino acid
of the peptide
inhibitor.
1005841In particular embodiments, an IL23R analogue comprises a half-life
extension moiety
having the structure shown below, wherein n=0 to 24 or n=14 to 24:
n=0 to 24
X kiy2:
X=CH3, CO2H, NH2, OH
0
1005851In certain embodiments, a IL23R analogue of the present invention
comprises a half-life
extension moiety shown in Table 7.
Table 7. Illustrative Half-Life Extension Moieties
Half-Life Extension Moietys
0
Cl SS.
C12 (Lauric acid)
183
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
0
C2 SSN
C14 (Mysteric acid)
0
C3
Sg{
C16 (Palm or Palmitic acid)
0
C4 S'S
C18 (Stearic acid)
0
C5 -SS:
C20
0
0
C6 rPrN
OH C12 diacid
0
C7 HO sr:
C14 diacid
0
0
C8 HO
3-53.
0 C16 diacid
0
C9 HO/
C18 diacid
0
0
C10 HO
54
C20 diacid
0
184
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
10058611n certain embodiments, a half-life extension moiety is bound directly
to a peptide
inhibitor, while in other embodiments, a half-life extension moiety is bound
to the peptide
inhibitor via a linker moiety, e.g., any of those depicted in Tables 6 or 8.
[00587] Table 8. Illustrative Linker Moieties
Linker Moiety
0
Li
CO2H
IsoGlu
N H2
124 N
L2
Dapa
N3-(
L3
Ahx
Lipidic based linkers:
L4
in N
n=1 to 24
0
N )211
L5
0
PEG1
0
L6
PEG2
1 85
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
H
.5*SjK
/ oo NIX
L7
0 n=11
PEG11 (40 atoms) also known as PEG12
''..s.. sJ.,..,..........,.... 0 ..,........$, A
N
\ n H
L8
0
n=1 to 25
PEG based linkers
s-CS5.0(3 A
L9 N
H
OEG
0
0
H
N-Z31
L10 H
CO2H 0
IsoGlu-Ahx
CO2H o
L11 H H
0 0
IsoGlu-OEG-OEG
0
H
Nni...,....../=i.N o.,..õ.õ,-
,...so.õ.....o.õ........õ,,,,o.....o.õ,....õ..-...Nc-
L12
CO2H o
1soGlu-PEG5
0
H
L13
n=1-25
CO2H 0
IsoGlu-PEGn
186
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
0 0
L14 ;22''NNOC):31-
H H
pAla-PEG2
9 9
N 1.
L15 H \ H
n=11
pAla-PEG11 (40 atoms)
1005881In particular embodiments, a peptide inhibitor of the present invention
comprises any of
the linker moieties shown in Table 8 and any of the half-life extension
moieties shown in Table
7, including any of the following combinations shown in Table 9a.
Table 9a. Illustrative Combinations of Linkers and Half-Life Extension
Moieties in Peptide
Inhibitors
Linker Half-Life Linker Half-Life Linker Half-Life
Extension Extension Extension
Moiety Moiety Moiety
Li Cl Li C2 Li C3
L2 Cl L2 C2 L2 C3
L3 Cl L3 C2 L3 C3
L4 Cl L4 C2 L4 C3
L5 Cl L5 C2 L5 C3
L6 Cl L6 C2 L6 C3
L7 Cl L7 C2 L7 C3
L8 Cl L8 C2 L8 C3
L9 Cl L9 C2 L9 C3
L10 Cl L10 C2 L10 C3
L11 Cl L11 C2 L11 C3
L12 Cl L12 C2 L12 C3
L13 Cl L13 C2 L13 C3
L14 Cl L14 C2 L14 C3
L15 Cl L15 C2 L15 C3
Linker Half-Life Linker Half-Life Linker Half-Life
Extension Extension Extension
187
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Moiety Moiety Moiety
Li C4 Li C5 Li C6
L2 C4 L2 C5 L2 C6
L3 C4 L3 C5 L3 C6
L4 C4 L4 C5 L4 C6
L5 C4 L5 C5 L5 C6
L6 C4 L6 C5 L6 C6
L7 C4 L7 C5 L7 C6
L8 C4 L8 C5 L8 C6
L9 C4 L9 C5 L9 C6
L10 C4 L10 C5 L10 C6
Ll 1 C4 L11 C5 Ll 1 C6
L12 C4 L12 C5 L12 C6
L13 C4 L13 C5 L13 C6
L14 C4 L14 C5 L14 C6
L15 C4 L15 C5 L15 C6
Linker Half-Life Linker Half-Life Linker Half-Life
Extension Extension Extension
Moiety Moiety Moiety
Li C7 Li C8 Li C9
L2 C7 L2 C8 L2 C9
L3 C7 L3 C8 L3 C9
L4 C7 L4 C8 L4 C9
L5 C7 L5 C8 L5 C9
L6 C7 L6 C8 L6 C9
L7 C7 L7 C8 L7 C9
L8 C7 L8 C8 L8 C9
L9 C7 L9 C8 L9 C9
L10 C7 L10 C8 L10 C9
Ll 1 C7 L11 C8 Ll 1 C9
L12 C7 L12 C8 L12 C9
L13 C7 L13 C8 L13 C9
L14 C7 L14 C8 L14 C9
L15 C7 L15 C8 L15 C9
Linker Half-Life Linker Half-Life Linker Half-Life
Extension Extension Extension
Moiety Moiety Moiety
Li C10 L6 C10 L11 C10
L2 C10 L7 C10 L12 C10
L3 C10 L8 C10 L13 C10
L4 C10 L9 C10 L14 C10
L5 C10 L10 C10 L15 C10
188
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
10058911n some embodiments there may be multiple linkers present between the
peptide the
conjugated moiety, e.g., half-life extension moiety, e.g., as depicted in
Table 9b.
Table 9b. Illustrative Combinations of Linkers and Half-Life Extension
Moieties in Peptide
Inhibitors
Linker Half-Life Extension Linker Half-Life Extension
Moiety Moiety
L1-L2 C10 L1-L2 C8
L2-L5-L3 C 1 0 L2-L5-L3 C8
L3-L8 C 1 0 L3-L8 C8
L1-L2-L3 C 1 0 L1-L2-L3 C8
L5-L3-L3-L3 C10 L5-L3-L3-L3 C8
1005901Illustrative examples of peptide inhibitors of the present invention,
including those
having a conjugates linker and/or half-life extension moiety are shown below.
All amino acids
are L amino acids unless otherwise stated. The present invention also includes
salt forms of any
of these peptide inhibitors, including, but not limited to, acetate salts
thereof.
NH2
I-1
oNH2 o
. NH
0 o N H2
z o o 0 cr 0 hl 40 0
,.
H H
kil JL H
(N))LN N N
N N NH2
H H H
0 NH 0 40 X 10 0
S H2P1 NH ) 1 HO 0 0
OH IN
0 o
HN 0
Example 1: cyclo[[Abt]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-
tetrahydropyran]-ENN-NH2
189
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
NH2
L--1
NH NH2
o%..... NH2
*
0
V 0 0
0 0 I.:
0
H JL H
N H
NH JL
VY N h,yr N
N NH2
H H H
0 NH 0 0 HO 0
0 H 0
H2N¨
s-
NH ) 0
0 0
H
OH CD) NINH
\
,0
0
D
H2N 0 NH
H
H2N N 0)
HN
Example 1a: Ac-[(D)-Arg]-cyclo[[Abu]-QTWQC] - [Phe[4-(2-aminoethoxy)]-[2-Nal]-
[4-amino-
4-carboxy-tetrahydropyran]-ENN-NH2
NH2
0) NH2
= Oj
NH NH2
140 o
Z o o o o 0
N
H
FN H H
Her N H rN LFNI4
NH2
N
0 NH 0 0 0 0 NH2
Ht N
y t
NH _\sI 0
r
OH H WI NHAc
0 Ir'NH
H2N 0
0
0
Example 2: Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-
tetrahydropyran]-[Lys(Ac)]-NN-NH2
190
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
half-Life
Extension Moiety
Linker )
NH ___________________________________
H
lik NH 0 NH2 0
NH2
, 4111 (0
0 0 0
_...,y H H N y
Nyks N NrFij(N4
N N NH2
H H H H
0 NH 0 0 0 0
S H2N
NH .) 0
410 HO 0 0
OH kil
0 Ir*NH
...,.,
0 o
H2N 0
Example 3: cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)-(Linker-Half-Life
Extension
Moiety)]-[2-Nall-[4-amino-4-carboxy-tetrahydropyran]-ENN-NE12
191
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Half-Life
Extension Moiety
__________________________________________ =
Linker
..= 1
NH _______________________________________
H
* 0 NH2 0
NH NH2
Z0111
0 00 0 0 40 0
H)....1., H 2iii.....TK
N N
H H
N N
N N NH2
H ir N H H H
0 NH Os 0
0 -...1. 0
S H2N
0
H 0 0
.....j NH ill
OH i\11
...,..
0 yCiNo 0
\......,H D
H2N 0
NH
H2N H y N,......._.....õ, 0),......
HN
Example 3a: Ac-[(D)-Arg]cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)-(Linker-Half-
Life
Extension Moiety)]-[2-Nal]-[4-amino-4-carboxy-tetrahydropyran]-ENN-NE-12
NH2
Ll
NH 0.,....., NH2 0
41 0 NH2
7 0 0 0 1\pr rj......e.... 40
0
HH r 2
......\...r, N).õ1.. N
N N N NH
H H H H
0 NH 0 0 0 0
S H2N
.....-IX NH ...) 0
OH 0 kil (''NH
....õ
0 o WI Hi IME
IIalf-Life
Extension Moiety
H2N o
Example 4: cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-
tetrahydropyrari]-[Lys(Linker-Half-Life Extension Moiety)]-NN-NE-12
192
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
NH2
4
0%.....,. NH2 0
Ll
. NH 111)
NH2
Z
0 0 0
Nr N
H H H
N .........).1. N N H2
0....., N H 0 ,..= 0 0 ......(11...' 0
S
H2N-(
0
OH kl WI HI OEM
0 N H
..,...c
Half-Life
D Extension Moiety
H2N 0 NH
H
H2N y, N .................... )..,......
0
N H
Example 4a: Ac-[(D)-Arg]-cyclo[[Abu] -QTWQC] - [Phe[4-(2-aminoethoxy)]-[2-Nal]-
[4-amino-
4-carboxy-tetrahydropyran]-[Lys(Linker-Half-Life Extension Moiety)]-NN-NE-12
Half-Life
Extension Moiety
NH2 Cen
L-1 I
NH
0)õ, NH2 0 NH2 NH
V 411 0
0 0 0 0 .0 0 4
H H
kli NH2
N N NPIr \11......N N
H H H H H
0 NH 0 0 0 0 0
S H2N
X NH ill HO o 0
OH 0 PI ye
..õ
NH
0 o)
H2N 0
Example 5: cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-
tetrahydropyran]-ENN-[Lys(Linker-Half-Life Extension Moiety)]-1\1E12
193
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1 la11-1 de
NH2 EM ,
* NH :)_NH2 01-10
NH2 NH
0
' 0
H
H
N
N x.11,N N .,...,r1 f42 r....1A, N ,.....IN H2
N N
H H 0 H 0 H 0
0 NH 0 0
S
0H2N
ki
NH illl HO
OH 0 l 0 0
yeN H
....
0 \
\ ")
H2N 0
.........NH
H
H2NyN....,.......... 0),....,
HN
Example 5a: Ac[(D)-Arg]-cyclo[[Abu] -QTWQC] -[Phe[4-(2-aminoethoxy)] - [2-
NalH4-amino-
4-carboxy-tetrahydropyran]-ENN-[Lys(Linker-Half-Life Extension Moiety)]-NH2
NH2
LI
* 0,NH2 0
NH NH2
)
0
/ VL
0 0 = 0 0 0 0
H H rH H N))LN r\ N N
N Nr NH2
H H H
0 NH 0 0 0 HH20 N
S
*0
NH 40) HO 0
OH NI
_______________________ . , ______ .
INC) \H r Linker,1 Half-1i fe
,Extension moiety,
H2N o
Example 6: [Half-Life Extension Moiety-Linker]-[cyclo[[Abu]-QTWQC]-[Phe[4-
(2-
aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-tetrahydropyran]-ENN-NH2
194
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
NH2
0...õ N H2 1.---1
0
4Ik NH NH2
0
0 0 0 0
H
H H
,...T., N ....õ....,1, N
7rTILN NH2
H H H H
0,.............õ..NH 0 0
kl 0 0 0
2N
y.......NH .40 HOO
OH oõ),........õ..
y...(INH
0 \O
D
H2N 0 NH
H I
H2N ________________________________________ =
N r Linker Half-Lith
Extension Moiety,
NH
Example 6a: [Half-Life Extension Moiety-Linked-[(D)-Arg]-[cyclo[[Abt]-QTWQC]-
[Phe[4-
(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-tetrahydropyran]-ENN-NH2
NH2
* NH
0.õ......, NH2 0 0 Oj NH2
V 0
n
0 0 0 "......L0 0
H H H
N
N
N hi )( N
.="....k 0 'ICIL hl .....-y NiT)LF1 N H2
FIN -----y- H
0 NH 0 0 0
S H2N
_\s1
1lLi 0
'II NH
W
H
OH ())N NH
Ir NHAc
H2N 1,===== 0
0 Linker Half-Life N
Extension Moiety ,
o
Example 7: [Half-Life Extension Moiety-Linked-[Pen]-NTWQ-[Pen]-[Phe[4-
(aminoethoxy)]-
[2-Nal]-[4-amino-4-carboxy-tetrahydropyran]-[Lys(Ac)]-NN-NH2
195
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Half-Life
Extension Moiety
NH2 EME
0%...,NH2 0 0) NH NH2 NH
r CD,
../' 0 0 0 0 0 0
N....cr. H H Hj LN41
N N
ri4 NH2
.11....'N N
H H
0........., NH 0 0 0 0 NH2 0
S
I
rNH s III 0
OH ..,,,,IVI 411 NHAc
0 --i---NH
H2N 0
0
Example 8: Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-
tetrahydropyran]-[Lys(Ac)]-NN-[Lys(Linker-Half-Life Extension Moiety)]-NE-12
Half-Life
Extension Moiety
MIE
NH
41 0 0.........) NH 0 NH2 NH2
0 0
,
40H 0
.a..., H 0 Nqr s...(11,....
N N N
N H. N N NH2
H H H H
.....:;xNH 0 0 0 0 NH2
S
I
Ilit 0
wil r----
NHAc
OH o.....õN
NH
H2N 0 o)
0
Example 9: Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(aminoethoxy)-(Linker-Half-Life
Extension
Moiety)]-[2-Nal]-[4-amino-4-carboxy-tetrahydropyran]-[Lys(Ac)]-NN-NE12
196
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
NH2
NH
0)
NH2
a a a a
40 0
M*LN
LHN NH2
NL
0 NH 0 0 0 0 NH2
S
=
si
101 0
Half-Life
OH o N HN Linker
Extension Moiety)
H2N 0 oA
Example 10: Ac- [Pen] -NTWQ- [Pen]- [Phe[4-(aminoethoxy)] - [2-Na!]- [4-
amino-4-carboxy-
tetrahydropyran]-[Lys(Linker-Half-Life Extension moiety )]-NN-NE12.
10059111n certain embodiments, the half-life of a peptide inhibitor of the
invention that includes a
conjugated chemical substituent, i.e., a half-life extension moiety, is at
least 100%, at least
120%, at least 150%, at least 200%, at least 250%, at least 300%, at least
400%, or at least 500%
of the half-life of the same peptide inhibitor but without the conjugated
chemical substituent. In
certain embodiments, the lipophilic substituents and/or polypermic moieties
enhance the
permeability of the peptide inhibitor through the epithelium and/or its
retention in the lamina
propria. In certain embodiments, the permeability through the epithelium
and/or the retention in
the lamina propria of a peptide inhibitor of the invention that includes a
conjugated chemical
substituent is at 100%, at least 120%, at least 150%, at least 200%, at least
250%, at least 300%,
at least 400%, or at least 500% of the half-life of the same peptide inhibitor
but without the
conjugated chemical substituent.
1005921 In one embodiment, a side chain of one or more amino acid residues
(e.g., Lys residues)
in a peptide inhibitor of the invention is conjugated (e.g., covalently
attached) to a lipophilic
substituent. The lipophilic substituent may be covalently bonded to an atom in
the amino acid
side chain, or alternatively may be conjugated to the amino acid side chain
via one or more
spacers. The spacer, when present, may provide spacing between the peptide
analogue and the
lipophilic substituent. In particular embodiments, the peptide inhibitor
comprises any of the
conjugated moieties shown in Tables 2-5.
197
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1005931In certain embodiments, the lipophilic substituent may comprise a
hydrocarbon chain
having from 4 to 30 C atoms, for example at least 8 or 12 C atoms, and
preferably 24 C atoms or
fewer, or 20 C atoms or fewer. The hydrocarbon chain may be linear or branched
and may be
saturated or unsaturated. In certain embodiments, the hydrocarbon chain is
substituted with a
moiety which forms part of the attachment to the amino acid side chain or the
spacer, for
example an acyl group, a sulfonyl group, an N atom, an 0 atom or an S atom. In
some
embodiments, the hydrocarbon chain is substituted with an acyl group, and
accordingly the
hydrocarbon chain may form part of an alkanoyl group, for example palmitoyl,
caproyl, lauroyl,
myristoyl or stearoyl.
1005941A lipophilic substituent may be conjugated to any amino acid side chain
in a peptide
inhibitor of the invention. In certain embodiment, the amino acid side chain
includes a carboxy,
hydroxyl, thiol, amide or amine group, for forming an ester, a sulphonyl
ester, a thioester, an
amide or a sulphonamide with the spacer or lipophilic substituent. For
example, the lipophilic
substituent may be conjugated to Asn, Asp, Glu, Gln, His, Lys, Arg, Ser, Thr,
Tyr, Trp, Cys or
Dbu, Dpr or Orn. In certain embodiments, the lipophilic substituent is
conjugated to Lys. An
amino acid shown as Lys in any of the formula provided herein may be replaced
by, e.g., Dbu,
Dpr or Orn where a lipophilic substituent is added.
[00595] In certain embodiments, the peptide inhibitors of the present
invention may be modified,
e.g., to enhance stability, increase permeability, or enhance drug like
characteristics, through
conjugation of a chemical moiety to one or more amino acid side chain within
the peptide. For
example, the N(epsilon) of lysine N(epsilon), the (3¨carboxyl of aspartic, or
the y¨carboxyl of
glutamic acid may be appropriately functionalized. Thus, to produce the
modified peptide, an
amino acid within the peptide may be appropriately modified. Further, in some
instances, the
side chain is acylated with an acylating organic compound selected from the
group consisting of:
Trifluoropentyl, Acetyl, Octonyl, Butyl, Pentyl, Hexyl, Palmityl,
Trifluoromethyl butyric,
cyclopentane carboxylic, cyclopropylacetic, 4-fluorobenzoic, 4-fluorophenyl
acetic, 3-
Phenylpropionic, tetrahedro-2H-pyran-4carboxylic, succinic acid glutaric acid
or bile acids. One
having skill is the art will appreciate that a series of conjugates can be
linked, e.g., for example
PEG4, isoglu and combinations thereof. One having skill is the art will
appreciate that an amino
198
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
acid with the peptide can be isosterically replaced, for example, Lys may be
replaced for Dap,
Dab, a-MeLys orOrn. Examples of modified residues within a peptide are shown
in Table 1B.
Table 1B. Examples of modified Lysine, Asp and Asn within the peptide
0 0
HN). H1\1)
) ) 14
H2N OH OH
H2N
0 0
Ne-Lys(Ac) Ne-Lys(Palm)
0
0
EN1 ifH
HN ) H N) - [\11 HO2C 0 14 II
) 0 14
/ CO2H
/
H2N rOH
H2N (OH
0
Ne-Lys-gamaGlu-Palm o Ne-Lys-isoGiu-Palm
0
H 0 CO2H
)
HNI)LV0l-----LNK 2 0
i 4 0 H 14
H2N4r0H
H2N rOH
Ne-Lys(PEG2-Ac) Ne-Lys(PEG4-isoGIu-PaIrn)
0 0
0 0
HNO INI
HN )-H.r0H
-N-
) 5 0 14
) 0
H2N H2N
ThrOH ThrOH
0 0
Ne-Lys(PEG)5-Palm Ne-Lys(succinic acid)
199
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
0 0 0
HN).*LON HN)Lr EN-11
tO
ThrOH ThrOH
H2N H2N
0 0
Ne-Lys(glutaric acid) Ne-Lys(Pyroglutaric acid)
o 11
HN HN
H2N.r0H
H2NOH
0 0
N8-Lys(Benzoic acid) Ne-Lys(IVA)
0
HN
HN
0
H2N.r0H
H2NrOH
0
0
Asp(1,4 diaminobutane)
Ne-Lys(octanoic acid)
0
HN)õ
0
HN-7¨tHH
H2NrOH
o 0
H2N rOH
Asn(isobutyl) 0
Ne-Lys(Biotin)
1005961In further embodiments of the present invention, alternatively or
additionally, a side-
chain of one or more amino acid residues in a peptide inhibitor of the
invention is conjugated to a
polymeric moiety, for example, in order to increase solubility and/or half-
life in vivo (e.g. in
200
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
plasma) and/or bioavailability. Such modifications are also known to reduce
clearance (e.g.
renal clearance) of therapeutic proteins and peptides.
1005971As used herein, "Polyethylene glycol" or "PEG" is a polyether compound
of general
formula H-(0-CH2-CH2)n-OH. PEGS are also known as polyethylene oxides (PE0s)
or
polyoxyethylenes (POEs), depending on their molecular weight PEO, PEE, or POG,
as used
herein, refers to an oligomer or polymer of ethylene oxide. The three names
are chemically
synonymous, but PEG has tended to refer to oligomers and polymers with a
molecular mass
below 20,000 Da, PEO to polymers with a molecular mass above 20,000 Da, and
POE to a
polymer of any molecular mass. PEG and PEO are liquids or low-melting solids,
depending on
their molecular weights. Throughout this disclosure, the 3 names are used
indistinguishably.
PEGS are prepared by polymerization of ethylene oxide and are commercially
available over a
wide range of molecular weights from 300 Da to 10,000,000 Da. While PEG and
PEO with
different molecular weights find use in different applications, and have
different physical
properties (e.g. viscosity) due to chain length effects, their chemical
properties are nearly
identical. The polymeric moiety is preferably water-soluble (amphiphilic or
hydrophilic), non-
toxic, and pharmaceutically inert. Suitable polymeric moieties include
polyethylene glycols
(PEG), homo- or co-polymers of PEG, a monomethyl-substituted polymer of PEG
(mPEG), or
polyoxyethylene glycerol (POG). See, for example, Int. J. Hematology 68:1
(1998);
Bioconjugate Chem. 6:150 (1995); and Crit. Rev. Therap. Drug Carrier Sys.
9:249 (1992). Also
encompassed are PEGS that are prepared for purpose of half life extension, for
example, mono-
activated, alkoxy-terminated polyalkylene oxides (P0A's) such as mono-methoxy-
terminated
polyethyelene glycols (mPEG's); bis activated polyethylene oxides (glycols) or
other PEG
derivatives are also contemplated. Suitable polymers will vary substantially
by weights ranging
from about 200 Da to about 40,000 Da or from about 200 Da to about 60,000 Da
are usually
selected for the purposes of the present invention. In certain embodiments,
PEGs having
molecular weights from 200 to 2,000 or from 200 to 500 are used. Different
forms of PEG may
also be used, depending on the initiator used for the polymerization process ¨
a common
common initiator is a monofunctional methyl ether PEG, or methoxypoly(ethylene
glycol),
abbreviated mPEG.
201
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[00598]Lower-molecular-weight PEGS are also available as pure oligomers,
referred to as
monodisperse, uniform, or discrete. These are used in certain embodiments of
the present
invention.
[00599] PEGS are also available with different geometries: branched PEGS have
three to ten PEG
chains emanating from a central core group; star PEGs have 10 to 100 PEG
chains emanating
from a central core group; and comb PEGS have multiple PEG chains normally
grafted onto a
polymer backbone. PEGS can also be linear. The numbers that are often included
in the names
of PEGS indicate their average molecular weights (e.g. a PEG with n = 9 would
have an average
molecular weight of approximately 400 daltons, and would be labeled PEG 400.
[00600] As used herein, "PEGylation" is the act of covalently coupling a PEG
structure to the
peptide inhibitor of the invention, which is then referred to as a "PEGylated
peptide inhibitor".
In certain embodiments, the PEG of the PEGylated side chain is a PEG with a
molecular weight
from about 200 to about 40,000. In some embodiments, a spacer of a peptide of
formula I,
formula I', or formula I" is PEGylated. In certain embodiments, the PEG of a
PEGylated spacer
is PEG3, PEG4, PEGS, PEG6, PEG7, PEG8, PEG9, PEG10, or PEG11. In certain
embodiments,
the PEG of a PEGylated spacer is PEG3 or PEG8.
[00601] Other suitable polymeric moieties include poly-amino acids such as
poly-lysine, poly-
aspartic acid and poly-glutamic acid (see for example Gombotz, et al. (1995),
Bioconjugate
Chem., vol. 6: 332-351; Hudecz, et al. (1992), Bioconjugate Chem., vol. 3, 49-
57 and Tsukada,
et al. (1984), J. Natl. Cancer Inst., vol. 73, : 721-729. The polymeric moiety
may be straight-
chain or branched. In some embodiments, it has a molecular weight of 500-
40,000 Da, for
example 500-10,000 Da, 1000-5000 Da, 10,000-20,000 Da, or 20,000-40,000 Da.
1006021In some embodiments, a peptide inhibitor of the invention may comprise
two or more
such polymeric moieties, in which case the total molecular weight of all such
moieties will
generally fall within the ranges provided above.
1006031In some embodiments, the polymeric moiety is coupled (by covalent
linkage) to an
amino, carboxyl or thiol group of an amino acid side chain. Certain examples
are the thiol group
202
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
of Cys residues and the epsilon amino group of Lys residues, and the carboxyl
groups of Asp and
Glu residues may also be involved.
[00604] The skilled worker will be well aware of suitable techniques which can
be used to
perform the coupling reaction. For example, a PEG moiety bearing a methoxy
group can be
coupled to a Cys thiol group by a maleimido linkage using reagents
commercially available from
Nektar Therapeutics AL. See also WO 2008/101017, and the references cited
above, for details
of suitable chemistry. A maleimide-functionalised PEG may also be conjugated
to the side-chain
sulfhydryl group of a Cys residue.
[00605] As used herein, disulfide bond oxidation can occur within a single
step or is a two step
process. As used herein, for a single oxidation step, the trityl protecting
group is often employed
during assembly, allowing deprotection during cleavage, followed by solution
oxidation. When a
second disulfide bond is required, one has the option of native or selective
oxidation. For
selective oxidation requiring orthogonal protecting groups, Acm and Trityl is
used as the
protecting groups for cysteine. Cleavage results in the removal of one
protecting pair of cysteine
allowing oxidation of this pair. The second oxidative deprotection step of the
cysteine protected
Acm group is then performed. For native oxidation, the trityl protecting group
is used for all
cysteines, allowing for natural folding of the peptide. A skilled worker will
be well aware of
suitable techniques which can be used to perform the oxidation step.
[00606] Several chemical moieties, including poly(ethylene)glycol, react with
functional groups
present in the twenty naturally occurring amino acids, such as, for example,
the epsilon amino
group in lysine amino acid residues, the thiol present in cysteine amino acid
residues, or other
nucleophilic amino acid side chains. When multiple naturally occurring amino
acids react in a
peptide inhibitor, these non-specific chemical reactions result in a final
peptide inhibitor that
contains many isomers of peptides conjugated to one or more
poly(ethylene)glycol strands at
different locations within the peptide inhibitor.
[00607] One advantage of certain embodiments of the present invention includes
the ability to add
one or more chemical moiety (such as PEG) by incorporating one or more non-
natural amino
acid(s) that possess unique functional groups that react with an activated PEG
by way of
chemistry that is unreactive with the naturally occurring amino acids present
in the peptide
203
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
inhibitor. For example, azide and alkyne groups are unreactive with all
naturally occurring
functional groups in a protein. Thus, a non-natural amino acid may be
incorporated in one or
more specific sites in a peptide inhibitor where PEG or another modification
is desired without
the undesirable non-specific reactions. In certain embodiments, the particular
chemistry involved
in the reaction results in a stable, covalent link between the PEG strand and
the peptide inhibitor.
In addition, such reactions may be performed in mild aqueous conditions that
are not damaging
to most peptides. In certain embodiments, the non-natural amino acid residue
is AHA.
[00608] Chemical moieties attached to natural amino acids are limited in
number and scope. By
contrast, chemical moieties attached to non-natural amino acids can utilize a
significantly greater
spectrum of useful chemistries by which to attach the chemical moiety to the
target molecule.
Essentially any target molecule, including any protein (or portion thereof)
that includes a non-
natural amino acid, e.g., a non-natural amino acid containing a reactive site
or side chain where a
chemical moiety may attach, such as an aldehyde- or keto-derivatized amino
acid, can serve as a
substrate for attaching a chemical moiety.
1006091 Numerous chemical moieties may be joined or linked to a particular
molecule through
various known methods in the art. A variety of such methods are described in
U.S. Patent No.
8,568,706. As an illustrative example, azide moieties may be useful in
conjugating chemical
moieties such as PEG or others described herein. The azide moiety serves as a
reactive functional
group, and is absent in most naturally occurring compounds (thus it is
unreactive with the native
amino acids of naturally occurring compounds). Azides also undergo a selective
ligation with a
limited number of reaction partners, and azides are small and can be
introduced to biological
samples without altering the molecular size of significantly. One reaction
that allows
incorporation or introduction of azides to molecules is the copper-mediated
Huisgen [3+2]
cycloaddition of an azide. This reaction can be used for the selective
PEGylation of peptide
inhibitors. (Tornoe et al., J. Org. Chem. 67: 3057, 2002; Rostovtsev et al.,
Angew. Chem., Int.
Ed. 41: 596, 2002; and Wang et al., J. Am. Chem. Soc. 125: 3192, 2003, Speers
et al., J. Am.
Chem. Soc., 2003, 125, 4686).
204
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Illustrative Peptide Inhibitors and Peptide Dimer Inhibitors, and Methods of
Making the Same
[00610] The present invention thus provides various peptide inhibitors which
bind or associate
with IL-23, to disrupt or block binding between IL-23 and IL-23R.
[006111Illustrative peptide inhibitors and peptide dimer inhibitors of the
present invention are
shown in Tables 3A-3H, 4A, 4B, 5A-5C, 6, 7, 8, 9, 10, 11, 12, 13, 14 or 15
provides the amino
acid sequence of selected monomer peptide inhibitors and peptide dimer
inhibitors, and indicates
the linker moiety present in the peptide dimer inhibitors. According to the
protocols discussed
herein, a number of the peptide inhibitors and peptide dimer inhibitors shown
in the
accompanying tables were synthesized and cyclyzed. Tables E3A-E3H, E4A, E4B,
ESA-ESC,
E6, E7, E8, E9, El 0, Ell, E12, E13, E14 or El 5 provide the IC50 values for
selected monomer
peptide inhibitors and peptide dimer inhibitors in inhibiting IL-23 binding to
the IL-23R, or in
inhibiting IL-23 signaling as determined by measuring changes in phospho-STAT3
levels, as
described in the accompanying Examples. Illustrative peptide inhibitors of the
present invention
are shown in Formulas (V), and in Tables 2-5, which provide the amino acid
sequence of
selected peptide inhibitors. These peptide inhibitors are acetate salts.
[00612] The peptide inhibitors of the present invention may be synthesized by
many techniques
that are known to those skilled in the art. In certain embodiments, monomer
subunits are
synthesized, purified, and dimerized using the techniques described in the
accompanying
Examples. In certain embodiments, the present invention provides a method of
producing a
peptide inhibitor (or monomer subunit thereof) of the present invention,
comprising chemically
synthesizing a peptide comprising, consisting of, or consisting essentially of
a peptide having an
amino acid sequence described herein, including but not limited to any of the
amino acid
sequences set forth in any of Formulas I, II, III, IV, V or VI or tables
herein. In other
embodiments, the peptide is recombinantly synthesized, instead of being
chemically synthesized.
In certain embodiments, the peptide inhibitor is a dimer, and the method
comprises synthezing
both monomer subunits of the peptide dimer inhibitor and then dimerizing the
two monomer
subunits to produce the peptide dimer inhibitor. In various embodiments,
dimerization is
accomplished via any of the various methods described herein. In particular
embodiments,
methods of producing a peptide inhibitor (or monomer subunit thereof) further
comprise
cyclizing the peptide inhibitor (or monomer subunit thereof) after its
synthesis. In particular
205
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
embodiments, cyclization is accomplished via any of the various methods
described herein. In
certain embodiments, the present invention provides a method of producing a
peptide inhibitor
(or monomer subunit thereof) of the present invention, comprising introducing
an intramolecular
bond, e.g., a disulfide, an amide, or a thioether bond between two amino acids
residues within a
peptide comprising, consisting of, or consisting essentially of a peptide
having an amino acid
sequence described herein, including but not limited to any of the amino acid
sequences set forth
in any of Formulas I, II, III, IV, V or VI, or the accompanying Examples,
Tables, or Sequence
Listing.
1006131In related embodiments, the present invention includes polynucleotides
that encode a
polypeptide having a sequence set forth in any one of Formulas I, II, III, IV,
V or VI, or the
accompanying Examples, Tables, or Sequence Listing.
1006141 In addition, the present invention includes vectors, e.g., expression
vectors, comprising a
polynucleotide of the present invention.
Methods of Treatment
1006151In certain embodiments, the present invention includes methods of
inhibiting IL-23
binding to an IL-23R on a cell, comprising contacting the IL-23 with a peptide
inhibitor of the
present invention. In certain embodiments, the cell is a mammalian cell. In
particular
embodiments, the method is performed in vitro or in vivo. Inhibition of
binding may be
determined by a variety of routine experimental methods and assays known in
the art.
1006161In certain embodiments, the present invention includes methods of
inhibiting IL-23
signaling by a cell, comprising contacting the IL-23 with a peptide inhibitor
of the present
invention. In certain embodiments, the cell is a mammalian cell. In particular
embodiments, the
method is performed in vitro or in vivo. In particular embodiments, the
inhibition of IL-23
signalling may be determined by measuring changes in phospho-STAT3 levels in
the cell.
1006171In some embodiments, the present invention provides methods for
treating a subject
afflicted with a condition or indication associated with IL-21 or IL-23R
(e.g., activation of the
IL-23/IL-23R signaling pathway), wherein the method comprises administering to
the subject a
peptide inhibitor of the present invention. In one embodiment, a method is
provided for treating
206
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
a subject afflicted with a condition or indication characterized by
inappropriate, deregulated, or
increased IL-23 or IL-23R activity or signaling, comprising administering to
the individual a
peptide inhibitor of the present invention in an amount sufficient to inhibit
(partially or fully)
binding of IL-23 to IL-23R in the subject. In particular embodiments, the
inhibition of IL-23
binding to IL-23R occurs in particular organs or tissues of the subject, e.g.,
the stomach, small
intestine, large intestine/colon, intestinal mucosa, lamina propria, Peyer's
Patches, mesenteric
lymph nodes, or lymphatic ducts.
1006181In some embodiments, methods of the present invention comprise
providing a peptide
inhibitor of the present invention to a subject in need thereof. In particular
embodiments, the
subject in need thereof has been diagnosed with or has been determined to be
at risk of
developing a disease or disorder associated with IL-23/IL-23R. In particular
embodiments, the
subject is a mammal.
1006191In certain embodiments, the disease or disorder is autoimmune
inflammation and related
diseases and disorders, such as multiple sclerosis, asthma, rheumatoid
arthritis, inflammatory
bowel diseases (IBDs), juvenile IBD, adolescent IBD, Crohn's disease,
sarcoidosis, Systemic
Lupus Erythematosus, ankylosing spondylitis (axial spondyloarthritis),
psoriatic arthritis, or
psoriasis. In particular embodiments, the disease or disorder is psoriasis
(e.g., plaque psoriasis,
guttate psoriasis, inverse psoriasis, pustular psoriasis, Palmo-Plantar
Pustulosis, psoriasis
vulgaris, or erythrodermic psoriasis), atopic dermatitis, acne ectopica,
ulcerative colitis, Crohn's
disease, Celiac disease (nontropical Sprue), enteropathy associated with
seronegative
arthropathies, microscopic colitis, collagenous colitis, eosinophilic
gastroenteritis/esophagitis,
colitis associated with radio- or chemo-therapy, colitis associated with
disorders of innate
immunity as in leukocyte adhesion deficiency-1, chronic granulomatous disease,
glycogen
storage disease type lb, Hermansky-Pudlak syndrome, Chediak-Higashi syndrome,
Wiskott-
Aldrich Syndrome, pouchitis resulting after proctocolectomy and ileoanal
anastomosis,
gastrointestinal cancer, pancreatitis, insulin-dependent diabetes mellitus,
mastitis, cholecystitis,
cholangitis, primary biliary cirrhosis, viral-associated enteropathy,
pericholangitis, chronic
bronchitis, chronic sinusitis, asthma, uveitis, or graft versus host disease.
207
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1006201In certain related embodiments, the present invention provides a method
of selectively
inhibiting IL-23 or IL-23R signaling (or the binding of IL-23 to IL-23R) in a
subject in need
thereof, comprising providing to the subject a peptide inhibitor of the
present invention. In
particular embodiments, the present invention includes a method of selectively
inhibiting IL-23
or IL-23R signaling (or the binding of IL-23 to IL-23R) in the GI tract of a
subject in need
thereof, comprising providing to the subject a peptide inhibitor of the
present invention by oral
administration. In particular embodiments, exposure of the administered
peptide inhibitor in GI
tissues (e.g., small intestine or colon) is at least 10-fold, at least 20-
fold, at least 50-fold, or at
least 100-fold greater than the exposure in the blood. In particular
embodiments, the present
invention includes a method of selectively inhibiting IL23 or IL23R signaling
(or the binding of
IL23 to IL23R) in the GI tract of a subject in need thereof, comprising
providing to the subject a
peptide inhibitor, wherein the peptide inhibitor does not block the
interaction between IL-6 and
IL-6R or antagonize the IL-12 signaling pathway. In a further related
embodiment, the present
invention includes a method of inhibiting GI inflammation and/or neutrophil
infiltration to the
GI, comprising providing to a subject in need thereof a peptide inhibitor of
the present
invention.In some embodiments, methods of the present invention comprise
providing a peptide
inhibitor of the present invention (i.e., a first therapeutic agent) to a
subject in need thereof in
combination with a second therapeutic agent. In certain embodiments, the
second therapeutic
agent is provided to the subject before and/or simultaneously with and/or
after the peptide
inhibitor is administered to the subject. In particular embodiments, the
second therapeutic agent
is an anti-inflammatory agent. In certain embodiments, the second therapeutic
agent is a non-
steroidal anti-inflammatory drug, steroid, or immune modulating agent. In
another embodiment,
the method comprises administering to the subject a third therapeutic agent.
In certain
embodiments, the second therapeutic agent is an antibody that binds IL-23 or
IL-23R.
1006211In particular embodiments, the peptide inhibitor, or the pharmaceutical
composition
comprising a peptide inhibitor, is suspended in a sustained-release matrix. A
sustained-release
matrix, as used herein, is a matrix made of materials, usually polymers, which
are degradable by
enzymatic or acid-base hydrolysis or by dissolution. Once inserted into the
body, the matrix is
acted upon by enzymes and body fluids. A sustained-release matrix desirably is
chosen from
biocompatible materials such as liposomes, polylactides (polylactic acid),
polyglycolide
(polymer of glycolic acid), polylactide co-glycolide (copolymers of lactic
acid and glycolic acid)
208
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
polyanhydrides, poly(ortho)esters, polypeptides, hyaluronic acid, collagen,
chondroitin sulfate,
carboxylic acids, fatty acids, phospholipids, polysaccharides, nucleic acids,
polyamino acids,
amino acids such as phenylalanine, tyrosine, isoleucine, polynucleotides,
polyvinyl propylene,
polyvinylpyrrolidone and silicone. One embodiment of a biodegradable matrix is
a matrix of one
of either polylactide, polyglycolide, or polylactide co-glycolide (co-polymers
of lactic acid and
glycolic acid).
1006221In certain embodiments, the present invention includes pharmacetical
compositions
comprising one or more peptide inhibitors of the present invention and a
pharmaceutically
acceptable carrier, diluent or excipient. A pharmaceutically acceptable
carrier, diluent or
excipient refers to a non-toxic solid, semi-solid or liquid filler, diluent,
encapsulating material or
formulation auxiliary of any type. Prevention of the action of microorganisms
may be ensured by
the inclusion of various antibacterial and antifungal agents, for example,
paraben, chlorobutanol,
phenol sorbic acid, and the like. It may also be desirable to include isotonic
agents such as
sugars, sodium chloride, and the like.
1006231In certain embodiments, the compositions are administered orally,
parenterally,
intracisternally, intravaginally, intraperitoneally, intrarectally, topically
(as by powders,
ointments, drops, suppository, or transdermal patch), by inhalation (such as
intranasal spray),
ocularly (such as intraocularly) or buccally. The term "parenteral" as used
herein refers to modes
of administration which include intravenous, intramuscular, intraperitoneal,
intrasternal,
subcutaneous, intradermal and intraarticular injection and infusion.
Accordingly, in certain
embodiments, the compositions are formulated for delivery by any of these
routes of
administration.
1006241In certain embodiments, pharmaceutical compositions for parenteral
injection comprise
pharmaceutically acceptable sterile aqueous or nonaqueous solutions,
dispersions, suspensions or
emulsions, or sterile powders, for reconstitution into sterile injectable
solutions or dispersions
just prior to use. Examples of suitable aqueous and nonaqueous carriers,
diluents, solvents or
vehicles include water, ethanol, polyols (such as glycerol, propylene glycol,
polyethylene glycol,
and the like), carboxymethylcellulose and suitable mixtures thereof, f3-
cyclodextrin, vegetable
oils (such as olive oil), and injectable organic esters such as ethyl oleate.
Proper fluidity may be
209
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
maintained, for example, by the use of coating materials such as lecithin, by
the maintenance of
the required particle size in the case of dispersions, and by the use of
surfactants. Thes
compositions may also contain adjuvants such as preservative, wetting agents,
emulsifying
agents, and dispersing agents. Prolonged absorption of an injectable
pharmaceutical form may be
brought about by the inclusion of agents which delay absorption, such as
aluminum monostearate
and gelatin.
1006251Injectable depot forms include those made by forming microencapsule
matrices of the
peptide inhibitor in one or more biodegradable polymers such as polylactide-
polyglycolide,
poly(orthoesters), poly(anhydrides), and (poly)glycols, such as PEG. Depending
upon the ratio
of peptide to polymer and the nature of the particular polymer employed, the
rate of release of
the peptide inhibitor can be controlled. Depot injectable formulations are
also prepared by
entrapping the peptide inhibitor in liposomes or microemulsions compatible
with body tissues.
[00626] The injectable formulations may be sterilized, for example, by
filtration through a
bacterial-retaining filter, or by incorporating sterilizing agents in the form
of sterile solid
compositions which can be dissolved or dispersed in sterile water or other
sterile injectable
medium just prior to use.
[00627] Topical administration includes administration to the skin or mucosa,
including surfaces
of the lung and eye. Compositions for topical lung administration, including
those for inhalation
and intranasal, may involve solutions and suspensions in aqueous and non-
aqueous formulations
and can be prepared as a dry powder which may be pressurized or non-
pressurized. In non-
pressurized powder compositions, the active ingredientmay be finely divided
form may be used
in admixture with a larger-sized pharmaceutically acceptable inert carrier
comprising particles
having a size, for example, of up to 100 micrometers in diameter. Suitable
inert carriers include
sugars such as lactose.
[00628] Alternatively, the composition may be pressurized and contain a
compressed gas, such as
nitrogen or a liquefied gas propellant. The liquefied propellant medium and
indeed the total
composition may bey such that the active ingredient does not dissolve therein
to any substantial
extent. The pressurized composition may also contain a surface active agent,
such as a liquid or
210
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
solid non-ionic surface active agent or may be a solid anionic surface active
agent. It is preferred
to use the solid anionic surface active agent in the form of a sodium salt.
[00629] A further form of topical administration is to the eye. A peptide
inhibitor of the invention
may be delivered in a pharmaceutically acceptable ophthalmic vehicle, such
that the peptide
inhibitor is maintained in contact with the ocular surface for a sufficient
time period to allow the
peptide inhibitor to penetrate the corneal and internal regions of the eye, as
for example the
anterior chamber, posterior chamber, vitreous body, aqueous humor, vitreous
humor, cornea,
iris/ciliary, lens, choroid/retina and sclera. The pharmaceutically acceptable
ophthalmic vehicle
may, for example, be an ointment, vegetable oil or an encapsulating material.
Alternatively, the
peptide inhibitors of the invention may be injected directly into the vitreous
and aqueous
humour.
[00630] Compositions for rectal or vaginal administration include
suppositories which may be
prepared by mixing the peptide inhibitorss of this invention with suitable non-
irritating
excipients or carriers such as cocoa butter, polyethylene glycol or a
suppository wax, which are
solid at room temperature but liquid at body temperature and, therefore, melt
in the rectum or
vaginal cavity and release the active compound.
[00631] Peptide inhibitors of the present invention may also be administered
in liposomes or other
lipid-based carriers. As is known in the art, liposomes are generally derived
from phospholipids
or other lipid substances. Liposomes are formed by mono- or multi-lamellar
hydrated liquid
crystals that are dispersed in an aqueous medium. Any non-toxic,
physiologically acceptable and
metabolizable lipid capable of forming liposomes can be used. The present
compositions in
liposome form can contain, in addition to a peptide inhibitor of the present
invention, stabilizers,
preservatives, excipients, and the like. In certain embodiments, the lipids
comprise
phospholipids, including the phosphatidyl cholines (lecithins) and serines,
both natural and
synthetic. Methods to form liposomes are known in the art.
[00632Wharmaceutical compositions to be used in the invention suitable for
parenteral
administration may comprise sterile aqueous solutions and/or suspensions of
the peptide
inhibitos made isotonic with the blood of the recipient, generally using
sodium chloride,
glycerin, glucose, mannitol, sorbitol, and the like.
211
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1006331In some aspects, the invention provides a pharmaceutical composition
for oral delivery.
Compositions and peptide inhibitors of the instant invention may be prepared
for oral
administration according to any of the methods, techniques, and/or delivery
vehicles described
herein. Further, one having skill in the art will appreciate that the peptide
inhibitors of the
instant invention may be modified or integrated into a system or delivery
vehicle that is not
disclosed herein, yet is well known in the art and compatible for use in oral
delivery of peptides.
1006341In certain embodiments, formulations for oral administration may
comprise adjuvants
(e.g. resorcinols and/or nonionic surfactants such as polyoxyethylene oleyl
ether and n-
hexadecylpolyethylene ether) to artificially increase the permeability of the
intestinal walls,
and/or enzymatic inhibitors (e.g. pancreatic trypsin inhibitors,
diisopropylfluorophosphate (DFF)
or trasylol) to inhibit enzymatic degradation. In certain embodiments, the
peptide inhibitor of a
solid-type dosage form for oral administration can be mixed with at least one
additive, such as
sucrose, lactose, cellulose, mannitol, trehalose, raffinose, maltitol,
dextran, starches, agar,
alginates, chitins, chitosans, pectins, gum tragacanth, gum arabic, gelatin,
collagen, casein,
albumin, synthetic or semisynthetic polymer, or glyceride. These dosage forms
can also contain
other type(s) of additives, e.g., inactive diluting agent, lubricant such as
magnesium stearate,
paraben, preserving agent such as sorbic acid, ascorbic acid, alpha-
tocopherol, antioxidants such
as cysteine, disintegrators, binders, thickeners, buffering agents, pH
adjusting agents, sweetening
agents, flavoring agents or perfuming agents.
1006351In particular embodiments, oral dosage forms or unit doses compatible
for use with the
peptide inhibitors of the present invention may include a mixture of peptide
inhibitor and
nondrug components or excipients, as well as other non-reusable materials that
may be
considered either as an ingredient or packaging. Oral compositions may include
at least one of a
liquid, a solid, and a semi-solid dosage forms. In some embodiments, an oral
dosage form is
provided comprising an effective amount of peptide inhibitor, wherein the
dosage form
comprises at least one of a pill, a tablet, a capsule, a gel, a paste, a
drink, a syrup, ointment, and
suppository. In some instances, an oral dosage form is provided that is
designed and configured
to achieve delayed release of the peptide inhibitor in the subject's small
intestine and/or colon.
212
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1006361 In one embodiment, an oral pharmaceutical composition comprising a
peptide inhibitor of
the present invention comprises an enteric coating that is designed to delay
release of the peptide
inhibitor in the small intestine. In at least some embodiments, a
pharmaceutical composition is
provided which comprises a peptide inhibitor of the present invention and a
protease inhibitor,
such as aprotinin, in a delayed release pharmaceutical formulation. In
some instances,
pharmaceutical compositions of the instant invention comprise an enteric coat
that is soluble in
gastric juice at a pH of about 5.0 or higher. In at least one embodiment, a
pharmaceutical
composition is provided comprising an enteric coating comprising a polymer
having dissociable
carboxylic groups, such as derivatives of cellulose, including
hydroxypropylmethyl cellulose
phthalate, cellulose acetate phthalate and cellulose acetate trimellitate and
similar derivatives of
cellulose and other carbohydrate polymers.
1006371In one embodiment, a pharmaceutical composition comprising a peptide
inhibitor of the
present invention is provided in an enteric coating, the enteric coating being
designed to protect
and release the pharmaceutical composition in a controlled manner within the
subject's lower
gastrointestinal system, and to avoid systemic side effects. In addition to
enteric coatings, the
peptide inhibitors of the instant invention may be encapsulated, coated,
engaged or otherwise
associated within any compatible oral drug delivery system or component. For
example, in some
embodiments a peptide inhibitor of the present invention is provided in a
lipid carrier system
comprising at least one of polymeric hydrogels, nanoparticles, microspheres,
micelles, and other
lipid systems.
[00638] To overcome peptide degradation in the small intestine, some
embodiments of the present
invention comprise a hydrogel polymer carrier system in which a peptide
inhibitor of the present
invention is contained, whereby the hydrogel polymer protects the peptide
inhibitor from
proteolysis in the small intestine and/or colon. The peptide inhibitors of the
present invention
may further be formulated for compatible use with a carrier system that is
designed to increase
the dissolution kinetics and enhance intestinal absorption of the peptide.
These methods include
the use of liposomes, micelles and nanoparticles to increase GI tract
permeation of peptides.
[00639] Various bioresponsive systems may also be combined with one or more
peptide inhibitor
of the present invention to provide a pharmaceutical agent for oral delivery.
In some
213
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
embodiments, a peptide inhibitor of the instant invention is used in
combination with a
bioresponsive system, such as hydrogels and mucoadhesive polymers with
hydrogen bonding
groups (e.g., PEG, poly(methacrylic) acid [PMAA], cellulose, Eudragit ,
chitosan and alginate)
to provide a therapeutic agent for oral administration. Other embodiments
include a method for
optimizing or prolonging drug residence time for a peptide inhibitor disclosed
herein, wherein
the surface of the peptide inhibitor surface is modified to comprise
mucoadhesive properties
through hydrogen bonds, polymers with linked mucins or/and hydrophobic
interactions. These
modified peptide molecules may demonstrate increase drug residence time within
the subject, in
accordance with a desired feature of the invention. Moreover, targeted
mucoadhesive systems
may specifically bind to receptors at the enterocytes and M-cell surfaces,
thereby further
increasing the uptake of particles containing the peptide inhibitor.
1006401 Other embodiments comprise a method for oral delivery of a peptide
inhibitor of the
present invention, wherein the peptide inhibitor is provided to a subject in
combination with
permeation enhancers that promote the transport of the peptides across the
intestinal mucosa by
increasing paracellular or transcellular permeation. Various permeation
enhancers and methods
for the oral delivery of therapeutic agents is described in Brayden, D.J.,
Mrsny, R.J., 2011. Oral
peptide delivery: prioritizing the leading technologies. Ther. Delivery 2
(12), 1567-1573.
1006411In certain embodiments, pharmaceutical compositions and formulations of
the present
invention comprises a peptide inhibitor of the present invention and one or
more permeation
enhancer. Examples of absorption enhancers may include Bile salts, fatty
acids, surfactants
(anionic, cationic, and nonanionic) chelators, Zonular OT, esters,
cyclodextrin, dextran sulfate,
azone, crown ethers, EDTA, sucrose esters, and phosphotidyl choline, for
example. Although
absorption enhancers are not typically carriers by themselves, they are also
widely associated
with other carriers to improve oral bioavailability by transporting of
peptides and proteins across
the intestinal mucosa. Such substances can be added to the formulation as
excipients or
incorporated to form non specific interactions with the intended peptide
inhibitor.
[00642]Dietary components and/or other naturally occurring substances affirmed
as enhancing
tight junction permeation and as Generally Recognized As Safe (GRAS) include,
e.g.,
asglycerides, acylcarnitines, bile salts, and medium chain fatty acids. Sodium
salts of medium
214
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
chain fatty acids (MCFAS) were also suggested to be permeation enhancers. The
most
extensively studied MCFAS is sodium caprate, a salt of capric acid, which
comprises 2-3% of
the fatty acids in the milk fat fraction. To date, sodium caprate is mainly
used as an excipient in a
suppository formulation (DoktacillinTM) for improving rectal ampicillin
absorption. The
permeation properties of another dietary MCFAS, sodium caprylate (8-carbon),
were shown in
vitro to be lower when compared to sodium caprate. Sodium caprylate and a
peptidic drug were
formulated in an admixture with other excipients in oil to generate an oily
suspension (OS) that
enhanced permeability (Tuvia, S. et al., Pharmaceutical Research, Vol. 31, No.
8, pp. 2010-2021
(2014).
[00643]For example, in one embodiment, a permeation enhancer is combined with
a peptide
inhibitor, wherein the permeation enhancer comprises at least one of a medium-
chain fatty acid,
a long-chain fatty acid, a bile salt, an amphiphilic surfactant, and a
chelating agent. In certain
embodiments, medium-chain fatty acid salts promote absorption by increasing
paracellular
permeability of the intestinal epithelium. In one embodiment, a permeation
enhancer comprising
sodium N4hydroxybenzoyl)amino] caprylate is used to form a weak noncovalent
association
with the peptide inhibitor of the instant invention, wherein the permeation
enhancer favors
membrane transport and further dissociation once reaching the blood
circulation. In another
embodiment, a peptide inhibitor of the present invention is conjugated to
oligoarginine, thereby
increasing cellular penetration of the peptide into various cell types.
Further, in at least one
embodiment a noncovalent bond is provided between a peptide inhibibitor of the
present
invention and a permeation enhancer selected from the group consisting of a
cyclodextrin (CD)
and a dendrimers, wherein the permeation enhancer reduces peptide aggregation
and increasing
stability and solubility for the peptide inhibitor molecule.
1006441In certain embodiments, a pharmaceutical composition or formulation
comprises a
peptide inhibitor of the present invention and a transient permeability
enhancers (TPEs).
Permeation enhancers and TPEs may be used to increase orally bioavailability
or the peptide
inhibitor. One example of a TPE that may be used is an oily suspension
formulation that
disperses a powder containing sodioum caprylate and a therapeutic agent
(Tuvia, S. et al.,
Pharmaceutical Research, Vol. 31, No. 8, pp. 2010-2021 (2014).
215
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1006451In certain embodiments, pharmaceutical composition and formulations may
include a
peptide inhibitor of the present invention and one or more absorption
enhancers, enzyme
inhibitors, or mucoso adhesive polymers.
1006461In particular embodiments, peptide inhibors of the present invention
are formulated in a
formulation vehicle, such as, e.g., emulsions, liposomes, microsphere or
nanoparticles.
[00647] Other embodiments of the invention provide a method for treating a
subject with a
peptide inhibitor of the present invention having an increased half-life. In
one aspect, the present
invention provides a peptide inhibitor having a half-life of at least several
hours to one day in
vitro or in vivo (e.g., when administered to a human subject) sufficient for
daily (q.d.) or twice
daily (b.i.d.) dosing of a therapeutically effective amount. In another
embodiment, the peptide
inhibitor has a half-life of three days or longer sufficient for weekly (q.w.)
dosing of a
therapeutically effective amount. Further, in another embodiment, the peptide
inhibitor has a
half-life of eight days or longer sufficient for bi-weekly (b.i.w.) or monthly
dosing of a
therapeutically effective amount. In another embodiment, the peptide inhibitor
is derivatized or
modified such that is has a longer half-life as compared to the underivatized
or unmodified
peptide inhibitor. In another embodiment, the peptide inhibitor contains one
or more chemical
modifications to increase serum half-life.
[00648] When used in at least one of the treatments or delivery systems
described herein, a
peptide inhibitor of the present invention may be employed in pure form or,
where such forms
exist, in pharmaceutically acceptable salt form.
[00649] The total daily usage of the peptide inhibitors and compositions of
the present invention
can be decided by the attending physician within the scope of sound medical
judgment. The
specific therapeutically effective dose level for any particular subject will
depend upon a variety
of factors including: a) the disorder being treated and the severity of the
disorder; b) activity of
the specific compound employed; c) the specific composition employed, the age,
body weight,
general health, sex and diet of the patient; d) the time of administration,
route of administration,
and rate of excretion of the specific peptide inhibitor employed; e) the
duration of the treatment;
f) drugs used in combination or coincidental with the specific peptide
inhibitor employed, and
like factors well known in the medical arts.
216
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1006501 In particlar embodiments, the total daily dose of the peptide
inhibitors of the invention to
be administered to a human or other mammal host in single or divided doses may
be in amounts,
for example, from 0.0001 to 300 mg/kg body weight daily or 1 to 300 mg/kg body
weight daily.
Non-invasive Detection of Intestinal Inflammation
1006511The peptide inhibitors of the invention may be used for detection,
assessment and
diagnosis of intestinal inflammation by microPET imaging, wherein the peptide
inhibitor is
labeled with a chelating group or a detectable label, as part of a a non-
invasive diagnostic
procedure. In one embodiment, a peptide inhibitor is conjugated with a
bifunctional chelator. In
another embodiment, a peptide inhibitor is radiolabeled. The labeled peptide
inhibitor is then
administered to a subject orally or rectally. In one embodiment, the labeled
peptide inhibitor is
included in drinking water. Following uptake of the peptide inhibitor,
microPET imaging may
be used to visualize inflammation throughout the subject's bowels and
digestive track.
Identification of Peptide Inhibitors that Inhibit IL-23 Signalling
1006521 As described herein, in certain embodiments, peptide inhibitors of the
present invention
preferentially bind to human IL-23R and/or rat IL-23R as compared to mouse IL-
23R. Mouse
IL-23R contains additional amino acids as compared to human IL-23R or rat IL-
23R in the
region corresponding to about amino acid residue 315 to about amino acid
residue 340 of the
mouse IL23R protein, e.g., amino acid region NVVQPWSSPFVHQTSQETGKR (see, e.g.,
Figure
4). In particular embodiments, the peptide inhibitors bind to a region of
human IL-23R from
about amino acid 230 to about amino acid residue 370.
1006531 The present invention provides a new method to identify an inhibitor
(e.g., a peptide
inhibitor) of IL-23R, based on identifying an agent (e.g., a peptide) that
preferentially binds to
human IL-23R or rat IL-23R as compared to mouse IL-23R. In certain
embodiments, the
method comprises: (a) determining an amount of binding of a candidate agent to
a human IL-
23R polypeptide or a rat IL-23R polypeptide; (b) determining an amount of
binding of the
candidate agent to the mouse IL-23R polypeptide; and (c) comparing the
determined amount of
binding to the human IL-23R polypeptide or the rat IL-23R polypeptide to the
determined
amount of binding to the mouse IL-23R polypeptide, wherein if the determined
amount of
217
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
binding to the human IL-23R polypeptide or the rat IL-23R polypeptide is
greater than the
amount of binding to the mouse IL-23R polypeptide, the candidate compound is
an inhibitor of
IL-23R. In particular embodiments, the candidate compound is identified as an
inhibitor of IL-
23R if the determined amount of binding to the human IL-23R polypeptide or the
rat IL-23R
polypeptide is at least 1.5-fold, at least 2-fold, at least 3-fold, at least 4-
fold, at least 5-fold, at
least 10-fold, at least 20-fold, at least 30-fold, at least 40-fold, at least
50-fold, at least 100-fold,
at least 200-fold, at least 500-fold, or at least 100-fold the determined
amount of binding to the
mouse IL-23R polypeptide. In particular embodiments, the candidate compound is
a peptide. In
particular embodiments, the peptide is a peptide of one of the formulas
described herein. In
particular embodiments, the human IL-23 polypeptide or rat IL-23R polypeptide
comprises or
consists of the full length human IL-23R or rat IL-23R protein, respectively.
In other
embodiments, the human IL-23R polypeptide is a fragment of the full length
human IL-23R
protein, comprising 8 or more amino acid residues within the region of human
IL-23R from
about amino acid residue 230 to about amino acid residue 370. In other
embodiments, the rat IL-
23R polypeptide is a fragment of the full length rat IL-23R protein,
comprising 8 or more amino
acid residues within the region of rat IL-23R from about amino acid residue
245 to about amino
acid residue 385.
1006541In another embodiment, the present invention provides a new method to
identify an
inhibitor (e.g., a peptide inhibitor) of IL-23R, based on identifying an agent
that binds to a region
of human IL-23R or rat IL-23 that is disrupted in mouse IL-23R by the presence
of additional
amino acids from about amino acid residues 315 to about amino acid residue 340
of the mouse
IL23R protein, e.g., amino acid region NWQPWSSPFVHQTSQETGKR (see, e.g., Figure
4). In
certain embodiments, the method comprises: (a) determining an amount of
binding of a
candidate agent to a fragment of human IL-23R polypeptide that falls within
about amino acid
residue 230 to about amino acid residue 370, or to a fragment of rat IL-23R
polypeptide that falls
within about amino acid residue 245 to about amino acid residue 385; (b)
determining an amount
of binding of the candidate agent to a negative control (e.g., a negative
control peptide unrelated
to human IL-23R or rat-IL-23R); and (c) comparing the determined amount of
binding to the
fragment of human IL-23R polypeptide or the fragment of rat IL-23R polypeptide
to the
determined amount of binding to the negative control, wherein if the
determined amount of
binding to the human IL-23R polypeptide fragment or the rat IL-23R polypeptide
fragment is
218
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
greater than the amount of binding to the negative control, the candidate
compound is an
inhibitor of IL-23R. In particular embodiments, the candidate compound is
identified as an
inhibitor of IL-23R if the determined amount of binding to the human IL-23R
polypeptide
fragment or the rat IL-23R polypeptide fragment is at least 1.5-fold, at least
2-fold, at least 3-
fold, at least 4-fold, at least 5-fold, at least 10-fold, at least 20-fold, at
least 30-fold, at least 40-
fold, at least 50-fold, at least 100-fold, at least 200-fold, at least 500-
fold, or at least 100-fold the
determined amount of binding to the negative control. In particular
embodiments, the candidate
compound is a peptide. In particular embodiments, the peptide is a peptide of
one of the
formulas described herein. In particular embodiments, the fragment of human IL-
23R includes
at least 8, at least 12, at least 20, at least 50, or at least 100, or all
amino acid residues within the
region of human IL-23R from about amino acid residue 230 to about amino acid
residue 370. In
other embodiments, the fragment of rat IL-23R polypeptide includes at least 8,
at least 12, at
least 20, at least 50, or at least 100, or all amino acid residues within the
region of rat IL-23R
from about amino acid residue 245 to about amino acid residue 385.
1006551 Methods of determining binding of a candidate compound to an IL-23
polypeptide are
known in the art and include but are not limited to in vitro and cell-based
binding assays,
including those described herein. For example, a labeled candidate compound
may be incubated
with a recombinantly produced IL-23R polypeptide or negative control bound to
a solid support
under conditions and for a time sufficient to allow binding, and then binding
determined by
measuring the amount of label associated with the bound IL-23R polypeptide.
Non-invasive Detection of Intestinal Inflammation
1006561 The peptide inhibitors of the invention may be used for detection,
assessment and
diagnosis of intestinal inflammation by microPET imaging, wherein the peptide
inhibitor is
labeled with a chelating group or a detectable label, as part of a a non-
invasive diagnostic
procedure. In one embodiment, a peptide inhibitor is conjugated with a
bifunctional chelator. In
another embodiment, a peptide inhibitor is radiolabeled. The labeled peptide
inhibitor is then
administered to a subject orally or rectally. In one embodiment, the labeled
peptide inhibitor is
included in drinking water. Following uptake of the peptide inhibitor,
microPET imaging may
be used to visualize inflammation throughout the subject's bowels and
digestive track.
219
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Animal Models of IBD
[00657] The present invention includes models of animal disease, including
inflammatory
diseases and disorders, such as inflammatory bowel diseases, e.g., Crohn's
disease and colitis.
As described in the accompanying Examples, several animal models of
inflammatory diseases
and disorders were developed.
1006581In one embodiment, the present invention includes a method of assessing
the ability of a
candidate compound to inhibit or reduce an inflammatory disease disorder,
comprising:
1006591(a) providing to a rat an amount of dextran sulfate sodium (DSS)
sufficient to induce
IBD;
1006601(b) providing to the rat an amount of a candidate compound; and
1006611(c) measuring an amount of IBD symptoms present in the rat after being
provided with
the DSS and the candidate compound;
[00662] wherein if the amount of IBD symptoms measured in (c) are
significantly lower than the
amount measured in a control rat provided with the amount of DSS and either an
amount of a
control compound or no peptide (e.g., vehicle control), the candidate compound
inhibits or
reduces the inflammatory disease or disorder.
[00663] In certain embodiments, the rat is provided with DSS for about 5 to 12
days, e.g., about 9
days. In particular embodiments, the rat is provided with DSS by providing to
the rat ad lib
exposure to drinking water containing DSS, e.g., about 1% to about 10% DSS,
about 2% to
about 5% DSS, or about 3% DSS. In particular embodiments, the rat is provided
with the test
compound at about 5 mg/kg to about 100 mg/kg, or about 10 mg/kg to about 50
mg/kg, or about
20 mg/kg or about 30 mg/kg. In particular embodiments, the rat is provided
with test compound
orally, e.g., in drinking water. In certain embodiments, the DSS assay is
performed as described
in the accompanying Examples.
1006641In another embodiment, the present invention includes a method of
assessing the ability
of a candidate compound to inhibit or reduce an inflammatory disease disorder,
comprising:
220
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1006651(a) providing to a rat an amount of 2,4,6-Trinitrobenzenesu1fonic acid
(TNBS) sufficient
to induce IBD;
1006661(b) providing to the rat an amount of a candidate compound; and
1006671(c) measuring an amount of IBD symptoms present in the rat after being
provided with
the TNBS and the candidate compound;
[00668] wherein if the amount of IBD symptoms measured in (c) are
significantly lower than the
amount measured in a control rat provided with the amount of TNBS and either
an amount of a
control compound or no peptide (e.g., vehicle control), the candidate compound
inhibits or
reduces the inflammatory disease or disorder.
1006691In certain embodiments, the animals are provided with about 10mg/kg to
about 200
mg/kg TNBS, e.g., about 10 mg/kg, about 20 mg/kg, about 30 mg/kg, about 40
mg/kg, about 50
mg/kg, about 60 mg/kg, about 70 mg/kg, about 80 mg/kg, about 90 mg/kg, about
100 mg/kg,
about 120 mg/kg, about 150 mg/kg or about 200 mg/kg of TNBS. In certain
embodiemnts, the
TNBS is in alcohol, e.g., in 45%-50% ethanol. In particular embodiments, the
TNBS is
administered intrarectally. In particular embodiments, the rat is provided
with the test compound
at about 5 mg/kg to about 100 mg/kg, or about 10 mg/kg to about 50 mg/kg, or
about 20 mg/kg
or about 30 mg/kg. In particular embodiments, the rat is provided with the
test compound orally,
e.g., in drinking water. In certain embodiments, the TNBS assay is performed
as described in the
accompanying Examples.
[00670] In particular embodiments IBD symptoms are measured immediately
following provision
of the DSS or TNBS and candidate compound (or test compound or no compound),
or later, e.g.,
at about 3 days, 5 days, or 9 days following initial provision of DSS or TNBS
and candidate
compound (or test compound or no compound). In particular embodiments, the IBD
symptoms
measured include one or more of percent body weight loss, stool consistency, a
quantitative
hemoccult score, and ratio of colon weight:colon length. In certain
embodiments, the IBD
symptoms are measured using a disease activity index (DAI) score and/or ratio
of colon
weight: colon length, wherein the DAI score consists of ratings from three
parameters, including
221
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
percent body weight loss, stool consistency, and a quantitative hemoccult
score, and can achieve
a maximum of three units.
[00671]In certain embodiments, a neutralizing anti-IL-23p19 antibody is used
as a comparator or
positive control.
1006721In certain embodiments, to assess the extent of the inflammatory
response, animals are
observed, e.g., daily, for clinical signs which included percent body weight
loss and signs of
loose stools or diarrhea. Following a time period after inoculation of with
DSS or TNBS (e.g., 5
days, 6, days, or seven days), rats are sacrificed and their entire colon
length and colon weight
from cecum to rectum recorded. The severity of colitis may be evaluated by a
pathologist
blinded to the identity of treatments. In addition to the colon wall
thickness, the gross colon
damage may be assessed based on a 0-4 scale according to Table 19 below, and
histopathological
scores were determined based on below parameters (Tables 20 and 21).
1006731 In certain embodiments, IBD symptoms are measured in three groups of
rats, each with at
least 3 animals, e.g., six animals each, wherein the three groups include:
vehicle, DSS or TNBS,
and DSS or TNBS with a positive control (e.g., sulfasalazine administered at
100 mg/kg PO,
QD).
EXAMPLES
EXAMPLE 1
SYNTHESIS OF PEPTIDE MONOMERS
1006741Peptide monomers of the present invention were synthesized using the
Merrifield solid
phase synthesis techniques on Protein Technology's Symphony multiple channel
synthesizer.
The peptides were assembled using EIBTU (0-Benzotriazole-N,N,N',N'-tetramethyl-
uronium-
hexafluoro-phosphate), Diisopropylethylamine(DIEA) coupling conditions. For
some amino acid
couplings PyA0P(7-Azabenzotriazo1-1-y1oxy)tripyrro1idinophosponium
hexafiuorophosphate)
and DIEA conditions were used. Rink Amide 1\41311A resin (100-200 mesh, 0.57
mmol/a) was
used for peptide with C-tenninal amides and pre-loaded Wang Resin with N-a-
Frnoc protected
amino acid was used for peptide with C-terminal acids. The coupling reagents
(I-IBTU and
DIEA premixed) were prepared at 100mmol concentration. Similarly amino acids
solutions were
222
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
prepared at 100 mmol concentration. Peptide inhibitors of the present
invention were identified
based on medical chemistry optimization and/or phage display and screened to
identify those
having superior binding and/or inhibitory properties.
Assembly
[00675] The peptides were assembled using standard Symphony protocols. The
peptide sequences
were assembled as follows: Resin (250 mg, 0.14 mmol) in each reaction vial was
washed twice
with 4m1 of DMF followed by treatment with 2.5ml of 20% 4-methyl piperidine
(Fmoc de-
protection) for 10min. The resin was then filtered and washed two times with
DMF (4m1) and
re-treated with N-methyl piperifine for additional 30 minute. The resin was
again washed three
times with DMF (4 ml) followed by addition 2.5ml of amino acid and 2.5ml of
HBTU-DIEA
mixture. After 45min of frequent agitations, the resin was filtered and washed
three timed with
DMF (4 ml each). For a typical peptide of the present invention, double
couplings were
performed. After completing the coupling reaction, the resin was washed three
times with DMF
(4 ml each) before proceeding to the next amino acid coupling.
Ring Closing Metathesis to form Olefins
[00676] The resin (100 umol) was washed with 2 ml of DCM (3 x 1 min) and then
with 2 ml of
DCE (3 x 1 min) before being treated with a solution of 2 ml of a 6 mM
solution of Grubbs' first-
generation catalyst in DCE (4.94 mg m1-1; 20 mol% with regard to the resin
substitution). The
solution was refluxed overnight (12 h) under nitrogenbefore being drained. The
resin was
washed three times with DMF (4 ml each); DCM (4 ml) before being dried and
cleavaed.
Cleavage
[00677] Following completion of the peptide assembly, the peptide was cleaved
from the resin by
treatment with cleavage reagent, such as reagent K (82.5% trigluoroacetic
acid, 5% water, 5%
thioanisole, 5% phenol, 2.5% 1,2-ethanedithiol). The cleavage reagent was able
to successfully
cleave the peptide from the resin, as well as all remaining side chain
protecting groups.
[00678] The cleaved peptides were precipitated in cold diethyl ether followed
by two washings
with ethyl ether. The filtrate was poured off and a second aliquot of cold
ether was added, and
the procedure repeated. The crude peptide was dissolved in a solution of
acetonitrile:water (7:3
223
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
with 1% TFA) and filtered. The quality of linear peptide was then verified
using electrospray
ionization mass spectrometry (ESI-MS) (Micromass/Waters ZQ) before being
purified.
Disulfide Bond Formation via Oxidation
[00679] The peptide containing the free thiol (for example diPen) was
assembled on a Rink
Amide-MBHA resin following general Fmoc-SPPS procedure. The peptide was
cleaved from the
resin by treatment with cleavage reagent 90% trifluoroacetic acid, 5% water,
2.5% 1,2-
ethanedithiol, 2.5% tri-isopropylsilane). The cleaved peptides were
precipitated in cold diethyl
ether followed by two washings with ethyl ether. The filtrate was poured off
and a second
aliquot of cold ether was added, and the procedure repeated. The crude peptide
was dissolved in
a solution of acetonitrile:water (7:3 with 1% TFA) and filtered giving the
wanted unoxidized
peptide crude peptide
[00680] The crude, cleaved peptide with X4 and X9 possessing either Cys, Pen,
hCys, (D)Pen,
(D)Cys or (D)hCys, was dissolved in 20m1 of water: acetonitrile. Saturated
Iodine in acetic acid
was then added drop wise with stirring until yellow color persisted. The
solution was stirred for
15 minutes, and the reaction was monitored with analytic HPLC and LCMS. When
the reaction
was completed, solid ascorbic acid was added until the solution became clear.
The solvent
mixture was then purified by first being diluted with water and then loaded
onto a reverse phase
HPLC machine (Luna C18 support, 10u, 100A, Mobile phase A: water containing
0.1% TFA,
mobile phase B: Acetonitrile (ACN) containing 0.1% TFA, gradient began with 5%
B, and
changed to 50% B over 60 minutes at a flow rate of 15m1/min). Fractions
containing pure
product were then freeze-dried on a lyophilyzer.
Lactam Bond Formation
[00681] 100mg of crude, cleaved peptide (approx. 0.12mmol) is dissolved in
100m1 of anhydrous
dichloromethane. HOBt (1-Hydroxybenzotriazole hydrate) (0.24mmol, 2
equivalents) is added
followed by DIEA (N,N-Diisopropylethylamine) (1.2mmol, 10equivalents) and TBTU
(0-
(Benzotriazol-1-y1)-N,N,N',N' -tetramethyluronium tetrafluoroborate)(0.24
mmol, 2 equivalents).
The mixture is stirred overnight and followed the reaction by HPLC. When the
reaction is
completed, dichloromethane is evaporated and diluted with water and
Acetonitrile and then
loaded onto a reverse phase HPLC machine (Luna C18 support, 10u, 100A, Mobile
phase A:
224
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
water containing 0.1% TFA, mobile phase B: Acetonitrile (ACN) containing 0.1%
TFA, gradient
begins with 5% B, and is changed to 50% B over 60 minutes at a flow rate of
15m1/min).
Fractions containing pure product are then freeze-dried on a lyophilyzer.
Triazole bond formation
[00682] The purified peptide containing the relevant amino acids alkyne and
azide was stirred at
room temperature in a phosphate / Me0H (2 :1) at pH 7.4 (1 mg per 2 ml). CuSO4
.5 H20 (10
equiv.), and sodium ascorbate (10 equiv.) was added and the mixture was
agitated in at room
temperature for 36 h. Me0H was removed and the solution was acidified to pH 3
with 1%TFA
water mix. The solution was then filtered before being loaded onto EIPLC for
peptide
purification.
Thioether Bond Formation
[00683] The peptide containing the free thiol (eg Cys) and hSer(OTBDMS) was
assembled on a
Rink Amide-MBHA resin following general Fmoc-SPPS procedure. Chlorination was
carried
out by treating the resin with PPh3 (10 equiv.) and C13CCN (10 equiv.) in DCM
for 2 h. The
peptide was cleaved from the resin by treatment with cleavage reagent 90%
trifluoroacetic acid,
5% water, 2.5% 1,2-ethanedithiol, 2.5% tri-isopropylsilane). The cleaved
peptides were
precipitated in cold diethyl ether followed by two washings with ethyl ether.
The filtrate was
poured off and a second aliquot of cold ether was added, and the procedure
repeated. The crude
peptide was dissolved in a solution of acetonitrile:water (7:3 with 1% TFA)
and filtered giving
the wanted uncyclized crude peptide
[00684] The crude peptide possessing a free thiol (eg, Cys, Pen, hCys, (D)Pen,
(D)Cys or
(D)hCys) and the alkyl halide (hSer(C1)) at either the X4 and X9 position or
X9 and X4 position
was dissolved in 0.1 M TRIS buffer pH 8.5. Cyclization was allowed to take
place overnight at
RT. The solvent mixture was then purified by first being diluted two-fold with
water and then
loaded onto a reverse phase EIPLC machine (Luna C18 support, 10u, 100A, Mobile
phase A:
water containing 0.1% TFA, mobile phase B: Acetonitrile (ACN) containing 0.1%
TFA, gradient
began with 5% B, and changed to 50% B over 60 minutes at a flow rate of
15m1/min). Fractions
containing pure product were then freeze-dried on a lyophilyzer.
S elenoether Bond Formation
225
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[00685] Crude peptide containing the thiol protected -Selenium amino acid and
the alkyl halide at
X4 and X9 was dissolved in 0.1 M sodium phosphate buffer pH 5.5 containing DTT
(40 equ.).
Cyclization was allowed to take place over 24 h at RT. The solution was then
diluted two-fold
with water, and the final cyclized peptide was purified using RP-HPLC,
affording the
selenoether.
Diselenide Bond Formation
1006861Diselenide precursor was dissolved in a solution of 0.1 M phosphate
buffer pH 6.0 and
isopropanolcontaining DTT (40 equiv), and the reaction mixture was incubated
at 37 C. After
20h, additional DTT (10 equiv) was added to the reaction. After a total of
32h, the cyclization
reaction was then diluted with twofold water, and the final cyclized peptide
was purified using
RP-HPLC, affording the diselenide.
Purification
[00687] Analytical reverse-phase, high performance liquid chromatography
(HPLC) was
performed on a Gemini C18 column (4.6 mm x 250 mm) (Phenomenex). Semi-
Preparative
reverse phase HPLC was performed on a Gemini 10 pm C18 column (22 mm x 250 mm)
(Phenomenex) or Jupiter 10 pm, 300 A C18 column (21.2 mm x 250 mm)
(Phenomenex).
Separations were achieved using linear gradients of buffer B in A (Mobile
phase A: water
containing 0.15% TFA, mobile phase B: Acetonitrile (ACN) containing 0.1% TFA),
at a flow
rate of 1 mL/min (analytical) and 15 mL/min (preparative). Separations were
achieved using
linear gradients of buffer B in A (Mobile phase A: water containing 0.15% TFA,
mobile phase
B: Acetonitrile (ACN) containing 0.1% TFA), at a flow rate of 1 mL/min
(analytical) and
15mL/min (preparative).
Linker Activation and Dimerization
1006881Peptid monomer subunits were linked to form peptide dimer inhibitors as
described
below.
[00689] Small Scale DIG Linker Activation Procedure: 5mL of NMP was added to a
glass vial
containing IDA diacid (304.2 mg, 1 mmol), N-hydroxysuccinimide (NHS, 253.2 mg,
2.2 eq.
2.2mmol) and a stirring bar. The mixture was stirred at room temperature to
completely dissolve
the solid starting materials. N, N'-Dicyclohexylcarbodiimide (DCC, 453.9mg,
2.2 eq., 2.2
226
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
mmol) was then added to the mixture. Precipitation appeared within 10 min and
the reaction
mixture was further stirred at room temperature overnight. The reaction
mixture was then
filtered to remove the precipitated dicyclohexylurea (DCU). The activated
linker was kept in a
closed vial prior to use for dimerization. The nominal concentration of the
activated linker was
approximately 0.20 M.
[00690]For dimerization using PEG linkers, there is no pre-activation step
involved.
Commercially available pre-activated bi-functional PEG linkers were used.
[00691]Dimerization Procedure: 2mL of anhydrous DMF was added to a vial
containing peptide
monomer (0.1 mmol). The pH of the peptide was the adjusted to 8-9 with DIEA.
Activated
linker (IDA or PEG13, PEG 25) (0.48eq relative to monomer, 0.048 mmol) was
then added to
the monomer solution. The reaction mixture was stirred at room temperature for
one hour.
Completion of the dimerization reaction was monitored using analytical HPLC.
The time for
completion of dimerization reaction varied depending upon the linker. After
completion of
reaction, the peptide was precipitated in cold ether and centrifuged. The
supernatant ether layer
was discarded. The precipitation step was repeated twice. The crude dimer was
then purified
using reverse phase HPLC (Luna C18 support, 10u, 100A, Mobile phase A: water
containing
0.1% TFA, mobile phase B: Acetonitrile (ACN) containing 0.1% TFA, gradient of
15%B and
change to 45%B over 60min, flow rate 15m1/min). Fractions containing pure
product were then
freeze-dried on a lyophilyzer.
EXAMPLE 2
CHARACTERIZATION OF PEPTIDE INHIBITION OF BINDING OF INTERLEUKIN-23 TO THE
INTERLEUKIN-23 RECEPTOR
1006921 Peptide optimization was performed to identify peptide inhibitors of
IL-23 signalling that
were active at low concentrations (e.g., IC50 <10 nM) while exhibiting
gastrointestinal (GI)
stability. Certain peptides were tested to identify peptides that inhibit the
binding of IL-23 to
human IL-23R and inhibit IL-23/IL-23R functional activity, as described below.
Peptides tested
included peptides containing a variety of different cyclization chemistries,
including, e.g., cyclic
amides (side chain cyclizations), peptides containing a disulfide linkage,
e.g., between two Pen
residues, and peptides containing a thioether linkage. Peptide inhibitors of
the present invention
227
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
include but are not limited to peptides having any of the structures depicted
herein. In addition,
peptide inhibitors of the present invention include those having the same
amino acid sequence of
the peptides or structures described herein, without being required to have
the same or any N- or
C-terminal "capping" groups, such as, e.g., Ac or NH2.
[00693] Assays performed to determine peptide activity are described below,
and the results of
these assays is provided in Tables E3A-E3H, E4A and E4B, E5A-E5C, E6, E7, and
E8. Human
ELISA indicates the 1L23-IL23R competitive binding assay described below, Rat
ELISA
indicates the rat IL-23R competitive binding ELISA assay described below, and
pStat3HTRF
indicates the DB cells IL-23R pSTAT3 cell assay described below. The peptides
depicted in
Tables E3B-E3E are cyclized via a disulfide bridge formed between two cysteine
residues in
these peptides. The peptides depicted in Table E3F are dimerized via a linker
moiety or through
internal cysteine moieties, as indicated. The peptides depicted in Tables E4A
and E4B are
cyclized via the two Pen residues present in each of these peptides. The
peptides depicted in
Table E5A are cyclized via a thioether bond between the indicated amino acid
residues. Table
E5B provides an illustrative structure depicting thioether cyclization, which
is indicated in the
table by the term "Cyclo," with the cyclic region bracketed immediately
following. The
monomer subunits of the peptide dimers shown in Table E5C are cyclized as
indicated by the
term "Cyclo" and linked to each other via the indicated linker. The peptides
shown in Table E6
are cyclized via ring closing metathesis of the indicated residues. Table E7
provides two
illustrative structures depicting side chain cyclizations via cyclic amides,
and the peptides in this
table are cyclized as indicated following the term "Cyclo." Table E8 depicts
peptides cyclized
via a cysteine residue and a Pen residue.
[00694] Peptide inhibitors of the present invention include both the cyclized
form of the peptides
shown herein, as well as the non-cyclized forms. For certain peptides, the
residue Abu is present
where indicated, whereas in other embodiments related to the non-cyclized
form, the Abu may
be referred to as a hSer(C1) or homoSer residue.
1L23-IL23R Competitive Binding ELISA
[00695] An Immulon 4I-113X plate was coated with 50 ng/well of IL23R huFC
and
incubated overnight at 4 C. The wells were washed four times with PBST,
blocked with PBS
228
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
containing 3% Skim Milk for 1 hour at room temperature, and washed again four
times with
PBS T. Serial dilutions of test peptides and IL-23 at a final concentration of
2 nIVI diluted in
Assay Buffer (PBS containing 1% Skim Milk) were added to each well, and
incubated for 2
hours at room temperature. After the wells were washed, bound IL-23 was
detected by
incubation with 50 ng/well of goat anti-p40 polyclonal antibodies (R&D Systems
#AF309)
diluted in Assay Buffer for 1 hour at room temperature. The wells were again
washed four times
with PBST. The secondary antibodies, EIRP conjugated donkey anti-goat IgG
(Jackson
ImmunoResearch Laboratories #705-035-147) diluted 1:5000 in Assay Buffer was
then added,
and incubated for 30 minutes at room temperature. The plate was finally washed
as above.
Signals were visualized with TMB One Component EIRP Membrane Substrate,
quenched with 2
M sulfuric acid and read spectrophotometrically at 450 nm. IC50 values for
various test peptides
determined from these data are shown in Tables E3A-E3H, E4A and 4EB, ESA-ESC,
E6, E7,
and E8.
Rat IL-23R Competitive Binding ELISA
100696] An assay plate was coated with 300 ng/well of Rat IL-23R huFC and
incubated
overnight at 4 C. The wells were washed, blocked, and washed again. Serial
dilutions of test
peptides and IL-23 at a final concentration of 7 nIVI were added to each well,
and incubated for 2
hours at room temperature. After the wells were washed, bound IL-23 was
detected with goat
anti-p40 polyclonal antibodies, followed by an EIRP conjugated donkey anti-
goat IgG. Signals
were visualized with TMB One Component EIRP Membrane Substrate and quenched
with 2 M
sulfuric acid. IC50 values for various test peptides determined from these
data are shown in
Tables E3G, E3H, E4A, E4B, E5B, ESC and E8.
DB Cells IL23R pSTAT3 Cell Assay
1006971 IL-23 plays a central role in supporting and maintaining Th17
differentiation in vivo.
This process is thought to mediated primarily through the Signal Transducer
and Activator of
Transcription 3 (STAT3), with phosphorylation of STAT3 (to yield pSTAT3)
leading to
upregulation of RORC and pro-inflammatory IL-17. This cell assay examines the
levels of
pSTAT3 in IL-23R-expressing DB cells when stimulated with IL-23 in the
presence of test
compounds. DB cells (ATCC #CRL-2289), cultured in RPMI-1640 medium (ATCC #30-
2001)
229
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
supplemented with 10% FBS and 1% Glutamine, were seeded at 5 X 10E5 cells/well
in a 96 well
tissue culture plate. Serial dilutions of test peptides and IL-23 at a final
concentration of 0.5 nIVI
were added to each well, and incubated for 30 minutes at 37 C in a 5% CO2
humidified
incubator. Changes in phospho-STAT3 levels in the cell lysates were detected
using the Cisbio
HTRF pSTAT3 Cellular Assay Kit, according to manufacturer's Two Plate Assay
protocol. IC50
values determined from these data are shown in Tables E3E, E3G, E3H, E4A, E4B,
E5B, ESC,
and E8 as absolute values or within ranges. Where not shown, data was not
determined.
Table E3A. Illustrative Non-cyclic Peptides and Activities
ELBA
=.
=
SEQ I L23R /
NO Sequenceii
11,23
.
in ii Moles
=(11050) ........
1 Ac-[Ailp] - [Ail3FTWQDYVVLY4AibFR-NEI. 2 >100,000
2 Ac-CAMTWQDYVVLYGRC-NH2 7200
3 Ac-[Aib]-[Aib]-TWQDYVVLYGR-NH2 >100,000
4 Ac-AMTWQDYVVLYGRK-NH2 4100
Ac-CAMTWQDYVVLYGRCK-NH2 8500
6 Ac-KAMTWQDYWLYGR-NH2 5600
7 Ac-KCAMTWQDYWLYGRC-NH2 10600
8 Ac-AMTWAibDYVVLYGR-NH2 >37,500
9 Ac-AMTWQDYVVLYGR-NH2 6100
Cyclo-[AMTWQDYVVLYGR] Not active
11 Hy-AATWQDYWLYGR-OH 7785
12 Hy-AMAWQDYVVLYGR-OH 24225
13 Hy-AMTAQDYVVLYGR-OH N/A
14 Hy-AMTWADYVVLYGR-OH 6248
Hy-AMTWQAYVVLYGR-OH 9589
Table E3B. Illustrative Peptides Containing the C)000(C Motif with IC50 >1 uM
in IL23-
IL23R Competitive Binding ELISA
SEQ
ID NO. Sequence
=
87 Hy-CSDWECYVVHIFG-NH2
88 Hy-CETWECYVVHSFS-NH2
89 Hy-CQSWECYVVHYYG-NH2
90 Hy-CSDWRCYWHVFG-NH2
91 Hy-CHTWVCYVVEIEFS-NH2
92 Hy-CTDWVCYVVEIEYS-NH2
93 Hy-CQTWVCYVVHTYG-NH2
230
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ.
ID NO. õ Sequence
94 Hy-CGNVVECYVVHVYG-NH2
95 Hy-CKDWKCYVVHIYG-NH2
96 Hy-CRTWVCYVVHVFG-NH2
97 Hy-CAD-[1-Nal]-VCYVVHTFG-NH2
98 Hy-CAD-[2-Nall-VCYVVHTFG-NH2
99 Hy-CAD-[1-BIP]-VCYVVHTFG-NH2
100 Hy-CAD-[Tic]-VCYVVHTFG-NH2
101 Hy-CAD-[[3hW]-VCYVVHTFG-NH2
102 Hy-CADWVCY-[1-BIP]-HTFG-NH2
103 Hy-CADWVCY-[Tic]-HTFG-NH2
104 Hy-CADWVCY-[f3hW]-HTFG-NH2
105 Hy-CADWVCYAHTFG-NH2
106 Hy-ACDWVCYVVHTFG-NH2
107 Hy-ACDWCCYVVCTFG-NH2
108 Hy-AADWCAYVVCTFG-NH2
109 Hy-CADWCCYVVCTFG-NH2
110 Hy-CADWCCYVVCTFG-NH2
111 Hy-CADWCCYVVCTFG-NH2
112 Hy-CADWVCYVVHTF-NH2
113 Hy-CADWVCYVVHT-NH2
114 Hy-CADWVCYVV-NH2
115 Hy-[(3-Ala]-SCADWVCYVVHTFG-OH
116 Ac-[(D)Lys]-SCADWVCYVVHTFG-OH
117 Ac-[(D)Lys]-[(3-Ala]-CADWVCYVVHTFG-OH
118 Hy-[AEA]-CADWVCYVVHTFG-OH
119 Ac-[(D)Lys]-CADWVCYVVHTFG-OH
120 Ac-CKDWVCYVVHTFG-OH
121 Ac-CADWKCYVVHTFG-OH
122 Ac-CADWVCYVVKTFG-OH
123 Ac-CADWVCYVVEIKFG-OH
124 Ac-CADWVCYVVHTKG-OH
125 Ac-CADWVCYWHTF-[(D)Lys]-0H
126 Ac-CADWVCYVVHTFG-NH2
127 Hy-CADWVCY-[1-Nal]-HTF-OH
128 Hy-CADWVCY-[1-Nall-HT-[N-Me-Phe]-NH2
129 Hy-CADWVCY-[1-Nal]-H-[Sarc]-F-OH
130 Hy-CADWVCY-[1-Nall-[N-Me-His]-TF-OH
131 Hy-CADWVCYVVHTFGK-OH
132 Hy-C-[Sarc]-DWVCY-[1-Nall-HTF-OH
133 Hy-CAD-[N-Me-Trp]-VCY-[1-Nal]-HTF-OH
134 Hy-CADW-[Sarc]-CY-[1-Nal]-HTF-OH
135 Hy-CADWVCY-[1-Nal]-HT-[(D)Phe]-0H
136 Hy-CADWVCY-[1-Nal]-HTF-[Sarc]-0H
231
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ.
ID NO. Sequence
137 Ac-CATWVCYWHTFG-NH2
138 Ac-CADWECYWHTFG-NH2
139 Ac-CADWVCYVVEIRCGWWGC-NH2
140 Ac-CADWVCY-[1-Nall-H-RD)AlaFFG-NH2
141 Ac-CADWVCY-[1-Nall-H-[Aib]-FG-NH2
142 Ac-CADWVCY-[1-Nall-H-[b-Ala]-FG-NH2
143 Ac-CADWVCY-[1-Nall-FTFG-NH2
144 Ac-CADWVCY-[1-Nall-RD)Ala]-TFG-NH2
145 Ac-CADWVCY-[1-Nall-H-[Aib]-[(D)Phe]-G-NH2
146 Ac-CADWVCY-[1-Nall-HTF-[Aib]-NH2
147 Ac-CADWVCY41-Nall4N-Me-HisH(D)AlaFF-[Aib]-NH2
148 Ac-CADWVCY-[1-Nall-H-[AEP]-G-NH2
149 Ac-CADWVCYVV-[N-MeHis]-TFG-[AEA]-[(D)Lys]-NH2
150 Ac-CADWVCY-[Aic]-HTFG-[AEA]-[(D)Lys]-NH2
151 Ac-CADWVCY-[Bip]-HTFG-[AEA]-[(D)Lys]-NH2
152 Ac-CQTWQCYVV-[N-MeArg]-ENG-[AEA]- [(D)-Lys] -NH2
153 Ac-CQTWQCYWR-[N-MeArg]-NG-[AEA]- [(D)-Lys] -NH2
154 Ac-CQTWQCYVVR-[N-MeLys]-NG-[AEA1-[(D)-Lys] -NH2
155 Ac-CQTWQCYVVR-[Sarc]-NG-[AEA]- [(D)-Lys]-NH2
156 Ac-CQTWQCYVVR-RD)Glu]-NG-[AEA]-[(D)-Lys]-NH2
157 Ac-CQTWQCYVV- [(D)Arg]-ENG-[AEA]-[(D)-Lys]-NH2
158 Ac-CQTWQCYVV-RD)Arg]-[(D)Glu]-NG-[AEA]-[(D)Lys]-NH2
159 Ac-CQTWQCYW-[N-MeGlu]-NG-[AEA]-[(D)-Lys]
160 Ac-CADWVC-NH2
161 Ac-CRDWQCYVV -[N-MeArg]-KFG-[AEP]-[(D)-Lys] -NH2
162 Ac-CRDWQCYVVR-RD)Lys]-FG-[AEP]-[(D)-Lys] -NH2
163 Ac-CRDWQCYVV-RD)Arg]-KFG-[AEP]-[(D)-Lys] -NH2
164 Ac-CRDWQCYVV-RD)Arg]-[(D)Lys]-FG-[AEPH(D)-Lys] -NH2
165 Ac-CQTWQCYVV-[N-MeArg]-ENG-[AEA]-[(D)-Lys] -NH2
Table E3C. Illustrative Peptides Containing the C)000(C Motif with IC50 of
500nM to
1000nM in 1L23-IL23R Competitive Binding ELISA
SEQ
ID NO. Sequence
166 Hy-CTDWKCYVVHEFG-NH2
167 Hy-CRTWTCYVVHVYG-NH2
168 Hy-CPNVVECYVVEIRFG-NH2
169 Hy-CADWVCYVVHTFG-NH2
170 Hy-CADWIVICYVVHEYG-NH2
171 Hy-CTTWKCYVVHQYG-NH2
172 Hy-CSNVVECYVVEIHYG-NH2
173 Hy-CSDWVCYVVHVYG-NH2
174 Hy-CDTWKCYVVEIRQS-NH2
232
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
175 Hy-CADWVCY-[1-Nal]-HTFG-NH2
176 Hy-CADWVCY-[2-Nal]-HTFG-NH2
177 Hy-CADWVCYVVHTFG-NH2
178 Ac-CADWVCYVVHTFG-[(D)Lys]-0H
179 Ac-CADWVCYVVHTFGAP-[(D)Lys]-0H
180 Ac-CTDWKCYVVHTFG-NH2
181 Ac-CRDWVCYWHTFG-NH2
182 Ac-CADWVCYVVEIEFG-NH2
183 Ac-CADWVCYVVEIFHQLRDA-NH,
184 Ac-CADWVCYVVEIEHSERVG-NH2
185 Ac-CADWVCYVVHNHSEGSG-NH2
186 Ac-CADWVCYVVEIRSTGGQH-NH2
187 Ac- [(D)Ly s] -CRDWQ CY- [1 -Na!] -HTH- [ S arc]- [AEP] - [(D)Arg] -
NH2
188 Ac-TQFDCRTWECYVVHTFG-NH2
189 Ac-GGVECNDWQCYVVHTFG-NH2
190 Ac-REGTCSTWKCYVVHTFG-NH2
191 Ac-DTPRCRTWECYVVHTFG-NH2
192 Ac-GGGECENVVECYVVHTFG-NH2
193 Ac-GDHKCSSWECYVVHTFG-NH2
194 Ac-GSVHCMTWECYWHTFG-NH2
195 Ac-CADWVCY-[1-Nal]-VTFG-NH2
196 Ac-CADWVCYVV-[(D)His]-TFG-[AEA]-[(D)Lys]-NH2
Table E3D. Illustrative Peptides Containing the C)000(C Motif with IC50 <500nM
in IL23-
IL23R Competitive Binding ELISA
SEQ
Sequence
ID NO.
"
197 Hy-CRDWQCYVVEIKFG-NH2
198 Hy-CSNVVVCYVVHTYG-NH2
199 Ac-CADWVCYVVHTFG-[(3-Ala]-[(D)Lys]-0H
200 Ac-CADWVCYVVHTFG-[AEA]-[(D)Lys]-0H
201 Ac-CADWVCYVVHTFG-OH
202 Ac-CADWVCYVVHTFG-[AEP]-(D)Arg]-0H
203 Ac-CADWVCYWHTFG-[AEP]-K-OH
204 Ac-CADWVCYVVHTFG-[Gaba]-[(D)Lys]-0H
205 Ac-CADWVCYWHTFG-[Hexanoic]-[(D)Lys]-0H
206 Ac-CADWVCYVVHTFG-[(PEG)2-[(D)-Lys]-0H
207 Ac-CADWVCYWHTFGP-[(D)Lys]-0H
208 Ac-CADWVCYWHTFG-[Azt]-[(D)-Lys]-0H
209 Ac-CADWVCYVVHTFGA-[(D)Lys]-0H
210 Ac-CADWVCYVVHTFGAP-[(D)Lys]-0H
211 Ac-CADWVCYVVHTFGA[Azt]-[(D)Lys]-0H
212 Ac-CADWVCYWHTFGAA[(D)Lys]-0H
213 Ac-CRDWQCYVVEIKFG-[AEP]-[(D)Lys]-0H
233
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ.
ID NO. Sequence
214 Ac-CATWQCYVVEIEYG-NH2
215 Ac-CKTWTCYVVEIEFG-NH2
216 Ac-CTTWTCYWHQYG-NH2
217 Ac-CRTWECYVVEIEFG-NH2
218 Ac-CRTWQCYWHEYG-NH2
219 Ac-CQTWQCYVVRENG-NH2
220 Ac-CRTWECYVVEIEYG-NH2
221 Ac-CTTWECYVVEIEYG-NH2
222 Ac-CRTWECYVVEIEQS-NH2
223 Ac-CTTWECYWHQFG-NH2
224 Ac-CTTWECYVVEIEFG-NH2
225 Ac-CQTWECYVVEILYG-NH2
226 Ac-CEDWKCYVVEIKYG-NH2
227 Ac-CTDWVCYWHTFG-NH2
228 Ac-CADWVCYVVHTYG-NH2
229 Ac-CADWVCYVVEIRHADRVK-NH2
230 Ac-CADWVCYVVHTFGER-NH2
231 Ac-CADWVCYVVHTHGER-NH2
232 Ac-DTPRCRTWECYWHTFG-NH2
233 Ac-CQTWVCYVVRENG- [AEA]- [(D)-Lys] -NH2
234 Ac-CQTWQCYVVRENG-[AEA]-[(D)-Lys]-NH2
235 Ac-CQTWQCYVVRTNG- [AEA]- [(D)-Lys] -NH2
236 Ac-CQTWQCYVVRKNG-[AEA]- [(D)-Lys] -NH2
237 Ac-CQTWQCYVVRRNG-[AEA] [(D)-Lys] -NH2
238 Ac-CQTWQCYVVR-[Dapa]-NG- [AEA]- [(D)-Lys] -NH2
239 Ac-CQTWQCYWR-[Orn]-NG- [AEA]- [(D)-Lys] -NH2
240 Ac-CRTWQCYVVRKFG-[AEA] [(D)-Lys] -NH2
241 Ac-CQTWQCYVVRENG-[AEA]-[(D)Arg]-NH2
242 Ac-CQTWQCYVVRENG-[AEA]-[(D)-Lys]-NH2
243 Ac-CQDWQCYVVRENG-[AEA]- [(D)-Lys] -NH2
244 Ac-CQTWQCYVVRENG-[AEA]-[(D)-Lys]-NH2
245 Ac-CQTWQCYVVRTNG-[AEA]-[(D)-Lys]-NH2
246 Ac-CQTWVCYVVRENG-[AEA]-[(D)-Lys]-NH2
247 Ac-CQTWQCYWRKNG- [AEA]- [(D)-Lys]-NH2
248 Ac-CQTWQCYW-[Cav]-ENG-NH2
249 Ac-CQTWQCYW-[Cpa]-ENG-NH2
250 Ac-CQTWQCYVVLENG-NH2
251 Ac-CQTWQCYVV[-hLeu]-ENG-NH2
252 Ac-CQTWQCYWR-[K-Ac]-NG-NH2
253 Hy-CRTWQCYVVRKFG-NH2
Table E3E. IC50 of Illustrative Peptides Containing the C)000(C Motif with
Activities
234
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
ELISA
ii SEQ ,. pStat3
I-ITRF in ii
ii ID NO. quencoi
1L23 in00
n Moles i
.:
= n NI
169 Hy-CADWVCYWHTFG-NH2 ****
****
178 Ac-CADWVCYVVHTFG-[(D)Lys]-0H **** ****
210 Ac-CADWVCYVVHTFGAP-[(D)Lys]-0H **** ND
211 Ac-CADWVCYWHTFGA[Azt]-[(D)Lys]-0H **** ND
180 Ac-CTDWKCYWHTFG-NH2 **** ****
Ac-CADWVCYW-RD)His]-TFG-[AEA]-[(D)Lys]-
196 **** ****
NH2
281 DIG dimererisation through N-termina Lysine ***** *****
(Ac-KMTWQDYVVLYGR-NH2)2
284 DIG dimererisation through C-terminal Lysine *****
*****
(Ac-AMTWQDYVVLYGK-NH2)2
*=<10nIVI; **=10-25 nM *** = 25-100 nM, **** = 100-1000 nM, *****=1000-10,000
nM.
Table E3F. IC50 of Illustrative Peptide Dimers
Human
SEQ ID 'iii ::::::
......
: :: ..
.. ii ii IL23R/IL23
itinlier NI oictSti ::::::
:: : Seci u en cOii NO. ::iii : ::
:.:.:.
...... E LISA
== =
oxidized dimer through
277 (Hy-FPTWEWYWCNRD-NH2)2 *****
the cysteine
oxidized dimer through
278 (Hy-ALTWEFYWLCRE-NH2)2 >10,000
the cysteine
291 DIG through Lysine (Hy-113A1a1SCADWVCYWHTFG-OH)2DIG
>10,000
(Ac-RD)Lys] -SCADWVCYWHTFG-OH)
292 DIG through Lysine
>10,000
2DIG
(Ac-(D)Lys-H3Alal -CADWVCYWHTFG-
293 DIG through Lysine
>10,000
OH)2DIG
294 DIG through Lysine (Hy-AEA-CADWVCYWHTFG-OH)2DIG
>10,000
(Ac-RD)Lys] -CADWVCYWHTFG-OH)
295 DIG through Lysine
>10,000
2DIG
296 DIG through Lysine (Ac-CKDWVCYWHTFG-OH)2DIG
>10,000
297 DIG through Lysine (Ac-CADWKCYWHTFG-OH)2DIG
>10,000
298 DIG through Lysine (Ac-CADWVCYWKTFG-OH)2DIG
299 DIG through Lysine (Ac-CADWVCYWHKFG-OH)2DIG
>10,000
300 DIG through Lysine (Ac-CADWVCYWHTKG-OH)2DIG *****
301 DIG through Lysine (Ac-CADWVCYWHTFDK-OH)2DIG
>10,000
302 DIG through Lysine (Ac-CADWVCYWHTFGDK)2DIG *****
303 DIG through Lysine
(Ac-CADWVCYWHTFG- H3-Alal - RD)Lys] -
304 DIG through Lysine ***
OH)2DIG
(Ac-CADWVCYWHTF G- [AEA] - [(D)Ly s] -
***
OH)2DIG
DIG through C terminal
305 (Hy-CADWVCYWHTFGK-OH)2DIG *****
Lysine
306 PEG25 through Lysine (Hy- 113Alal-SCADWVCYWHTFG-OH)
235
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Humaii
SEQ ID :: ... iiii
I L23R/I L23
i)l_,er ink Moietf iiiiii iiiSequenc0ii
NO. g :.ELISA
2PEG25
307 PEG25 through Lysine (Ac-RD)Lys1-SCADWVCYWHTFG-OH)2
308 PEG25 through Lysine
(Ac-(D)Lys)-H3A1a1-CADWVCYWHTFG-
OH)2
309 PEG25 through Lysine (Hy-{AEA1-CADWVCYWHTFG-OH)2
310 PEG25 through
Lysine (Ac-[(D)Lys1-CADWVCYWHTFG-OH)2
311 PEG25 through Lysine (Ac-CKDWVCYWHTFG-OH)2
312 PEG25 through Lysine (Ac-CADWKCYWHTFG-OH)2
313 PEG25 through Lysine (Ac-CADWVCYWKTFG-OH)2
314 PEG25 through Lysine (Ac-CADWVCYWHKFG-OH)2
315 PEG25 through Lysine (Ac-CADWVCYWHTKG-OH)2
316 PEG25 through Lysine (Ac-CADWVCYWHTF-RD)Lys1-0H)2
317 PEG25 through
Lysine (Ac-CADWVCYWHTFG-[(D)Lys1-0H)2
(Ac-CADWVCYWHTFG-[bAla1-[(D)Lys1-
318 PEG25 through Lysine
OH)2
319 PEG25 through Lysine
(Ac-CADWVCYWHTFG4AEA1-[(D)Lys1-
OH)2
PEG25 through C-
320 (Hy-CADWVCYWHTFGK-OH)2
terminal Lysine
*=<10nIVI; **=10-25 nM *** = 25-100 nIVI, **** = 100-1000 nIVI, *****=1000-
10,000 nIVI.
Table E3G. IC50 of Illustrative Peptides Containing the 00(WX0000(X-[(D)Lys]
Motif
_
: Human .::
Rat :" pStat3 :
.....
:H ELISA .E.L.I.SA
HTRE
ill SEQ I
ID NO Sequence ii:: IL23/ :.. IL23/ (nNI)
. .:iii
ii IL23R IL23R
=
.:
... .....
.. ...
16 Ac-CQDWQCYWR-[Cha]-FG-[AEA1-[(D)Lys1-NH2 113
17 Ac-CQTWQCYWR-[0g11-FG-[AEA1-[(D)Lys1-NH2 206
18 Ac-CQTWQCYWK4Dap1-FG-[AEA1-[(D)Lys1-NH2 32
19 Ac-CQTWQCYWH4Dap1-FG-[AEA1-[(D)Lys1-NH2 49 59
20 Ac-CQTWQCYWRLFG-[AEA1-[(D)Lys1-NH2 51 47
21 Ac-CQTWQCYW-[hArg]-[Dap1-FG-[AEA1-[(D)Lys1-NH2 56
22 Ac-CQTWQCYW-[Cit]-[Dap]-FG-[AEA1-[(D)Lys1-NH2 25
23 Ac-CQTWQCYWRVFG-[AEA1-[(D)Lys1-NH2 39 62 14
24 Ac-CQTWQCYWR-[Dap]-[Tic]-G-[AEA1-[(D)Lys1-NH2 892 65 12
25 Ac-CQTWQCY4Tic14Orn1-KFG4AEA14(D)Lys1-NH2 >30000
26 Ac-CQTWQCYWR-[Dab]-FG-[AEA1-[(D)Lys1-NH2 37
27 Ac-CQTWQCYW-[Orn1-{Dap1-FG-[AEA1-[(D)Lysl-NH2 79 276 37
28 Ac-CQTWQCYWHENGA-[(D)Lys1-NH2 220
29 Ac-CRTWQCYWRENGA4(D)Lys1-NH2 102 86 17
30 Ac-CRTWQCYWREYGA4(D)Lys1-NH2 78 80 8
31 Ac-C-[N-MeAla1-DWVCYWHTFG-[AEA1-[(D)Lys1-NH2 183
236
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Human Rat 1. pStat3
ELISA ELISA HTRF
SEQ
ID NO Sequence IL23/ IL23/ (nNI)
..,iii
IL23R IL23R
(nNI) (n111)
32 Ac-CADWVCYWRKFG- [13Alal- [(D)Ly s] -NH2 57 33(1) 13
33 Ac-CADWVCYW- [(D)Lys] -NH2 52 29
34 Ac-CADWVCYW-[Cit]-[Tlel-FG-H3-Alal-RD)Lysl-NH2 518
35 Ac-CADWVCYW- [Cit]- [Tba] -FG- [13-A1al - [(D)Lys] -NH2 153
36 Ac-CADWVCYW-[Cit]-[Chal-FG-[(3-Alal-RD)LYs1-NH2 223
37 Ac-CADWVCY41-Na1l - [Cit] -VFG- [13-Alal - [(D)Lys] -NH2 79
22
38 Ac-CADWVCYW- [(D)Lys] -NH2 124
39 Ac-CADWVCYW- [Cit]- [Chg] -FG- [13-A1al - [(D)Lys] -NH2 >30000
40 Ac-CADWVCYW- [Cit] - [13A1al -FG- [(D)Lys] -NH2 2584
41 Ac-CADWVCYW- [Tle] - [Tle] -FG- [13-A1al - [(D)Lys] -NH2 ¨30000
42 Ac-CADWVCYW- [Tle] -KFG- [13-A1al - [(D)Lys] -NH2 199
43 Ac-CQTWQCYW- [(D)Ala] -VFG- [AEA] - [(D)Lys] -NH2 232
44 Ac-CQTWQCYW- [13Alal -VFG- [AEAl- [(D)Lys] -NH2 2207
45 Ac-CQTWQCYW- [(D)Leu] -VFG- [AEAH(D)Lys] -NH2 188
46 Ac-CQTWQCYW-RD)Phel -VFG- [AEAl- [(D)Lys] -NH2 848
47 Ac-CQTWQCYW- [(D)Asn] -VFG-[AEA] - [(D)Lys] -NH2 61
48 Ac-CQTWQCYW-RD)Thr] -VFG-[AEA] -[(D)Lys] -NH2 3662
49 Ac-CQTWQCYW- [(D)Asp] -VFG-[AEA] - [(D)Lys] -NH2 129
50 Ac-CQTWQCYW- [Cit] - [(D)Leu] -FG- [AEA] - [(D)Lys] -NH2 709
51 Ac-CQTWQCYW- [Cit] - [(D)Phel -FG- [AEAH(D)Lys] -NH2 1304
52 Ac-CQTWQCYW- [Citl- [(D)Asn] -FG-[AEAl- [(D)Lys] -NH2 269
53 Ac-CQTWQCYW- [Citl- [(D)Thr] -FG- [AEAl- [(D)Lys] -NH2 1214
54 Ac-CQTWQCYW- [Agp] -VNG-[AEA] -[(D)Lys] -NH2 241
55 Ac-CQTWQCY- [cc-MeTrp] -RVNG- [AEA] - [(D)Lys] -NH2 ¨6000
56 Ac-CQTWQCY- [cc¨MeTrp] - [Cit]- [hLeul -NG- [AEA] -
¨6000
[(D)Lys] -NH2
57 Ac-CQTWQCYW- [Cit]-VNG- [AEAl- [(D)Lys] -NH2 73
58 Ac-CQTWQCYW- [Agp]- [Dap] -NG- [AEA] - [(D)Lys]-NH2 38
59 Ac-CQTWQCYW-[Cit] -VF- [(D)Ala] - [AEAH(D)Lys] -NH2 397
60 Ac-CQ TWQCYW-[Cit] -VF- [(D)Leu] -[AEA] -[(D)Ly s] -NH2 444
61 Ac-CQTWQCYW-[Cit] -VF- [(D)Phel- [AEA] - [(D)Lys] -NH2 784
62 Ac-CQTWQCYW- [Cit] -VF- [(D)Asn]- [AEA] - [(D)Lys] -NH2 93
63 Ac-CQTWQCYW-[Cit] -VF- [(D)Thr] - [AEAH(D)Lys] -NH2 518
64 Ac-CQTWQCYW- [Cit] -VF- [(D)Asp] - [AEAH(D)Lys] -NH2 551
65 Ac-C- [N-MeArg] -TWQCYWRVFG-[AEA] - [(D)Lys] -NH2 149
192 107
66 Ac-C- [N-MeQ1n] -TWQCYWRVFG-[AEA] - [(D)Lys] -NH2 69 85
101
67 Ac-C- [Cit]-TWQCYWRVFG- [AEA] -[(D)Ly sl-NH2 50 76
107
68 Ac-CADWVCYW- [Ornl- [Dap]-FG- [AEA] - [(D)Lysl-NH2 382
69 Ac-CADWVCY41 -Nal] -[Ornl- [Dal:11-FG- [AEAl- [(D)Lys] -
302
NH2
70 Ac-CADWVCY- [(D)Trp] -[Orn] -[Dap] -FG- [AEA] - >30000
237
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Human Rat 1. pStat3
ELISA ELISA HTRF
SEQ
ID NO Sequence IL23/ IL23/ (nNI)
.
IL23R IL23R
(nNI) (n111)
RD)Lys] -NH2
71 Ac-CADWVCY- [hPhel- [Ornl- [Dap] -FG- [AEA]- [(D)Lys]-
-30000
NH2
72 Ac-CADWVCY- [Bipl- [Ornl- [Dap] -FG- [AEA] -[(D)Lys] -
>30000
NH2
73 Ac-CADWVCY- [Phe(3,5-F2)]- [Orn]-[Dap]-FG- [AEA] -
¨6000
[(D)Lys] -NH2
74 Ac-CADWVCY- [Phe(CONH2)]- [Orn] - [Dap]-FG- [AEA] -
¨6000
[(D)Lys] -NH2
75 Ac-CADWVCY- [Phe(4-CF3)]- [Orn] - [Dal:11-FG- [AEA] -
>1000
[(D)Lys] -NH2
76 Ac-CADWVCY- [Phe(2,4-Me2)]- [Orn] - [Dap] -FG- [AEA] -
1525
[(D)Lys] -NH2
77 Ac-CMTWQCYWLYGR- [AEAl- [(D)Lys] -NH2 398
77 Hy-CMTWQCYWLYGR- [AEA] - [(D)Lysl-NH2 >30000
78 Ac-CADWVCY- [13hTrp] - [Oral- [Dap]-FG- [AEA] - [(D)Lys]-
6000
NH2
79 Ac-CADWVCYW- [Orn] - [c(-MeLeul -FG- [AEA] -[(D)Ly s] -
¨6000
NH2
80 Ac-CADWVCYW- [Orn] - [13-spira1-pip] -FG-[AEA] -
579
[(D)Lys] -NH2
81 Ac-CADWVCY44-Pheny1cy1cohexy1a1aninel- [Orn] - [Dap] -
>3000
FG- [AEAH(D)Lys] -NH2
82 Ac-CADWVCYW-[Orn]-[Aib] -FG- [AEAH(D)Lys] -NH2 1085
83 Ac-CADWVCYW-[Orn]-[DiethylGlyl-FG- [AEA] -
¨6000
[(D)Lys] -NH2
84 Ac-CADWVCY- [c(-MePhe(4-F)l- [Ornl- [Dap] -FG-[AEA] -
>30000
[(D)Lys] -NH2
85 Ac-CQTWQCY- [13hPhel -RVNG- [AEA] - [(D)Lys] -NH2 >30000
86 Ac-CQTWQCY- [13(1-Nal)] -RVNG-[AEA] -[(D)Lys] -NH2 >30000
321 Ac-CQTWQCY- [13hTyrl-RVNG- [AEA] - [(D)Lysl-NH2 >30000
322 Ac-CQTWQCY-H3hPhe(4-F) -RVNG-[AEAH(D)Lysl-NH2 >30000
323 Ac-CQTWQCY-H3Nya(5-Pheny1)]-RVNG-[AEAl-
>30000
[(D)Lys] -NH2
324 Ac-CQTWQCY- [Phe(3,4-C12)l-RVNG- [AEA] - [(D)Lys] -
>30000
NH2
325 Ac-CQTWQCY- [Tqal-RVNG- [AEA] - [(D)Ly s] -NH2 >30000
326 Ac-CQTWQCYWR-H3hLeul -NG- [AEA] - [(D)Lys] -NH2 224
327 Ac-CQTWQCYWR- [Aib] -NG- [AEA] - [(D)Lys] -NH2 1065
328 Ac-CQTWQCYWR-H3hAlal -NG- [AEA] -[(D)Lys] -NH2 457
329 Ac-CQTWQCYWR-H3hVall -NG- [AEA] -[(D)Lys] -NH2 328
330 Ac-CQTWQCYWR-H3-spira1-pip] -NG- [AEA] - [(D)Lysl-
405
NH2
238
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Human ' Rat 1. pStat3
ELISA ELISA HTRF
ID NO Sequence iii. IL23/ :. IL23/
(nNI)
. .,iii
IL23R IL23R i
..........................................iiii...... (nNI)
......p........ (n111) ......
............................................ii
331 Ac-CQTWQCYWR-[13Glul-NG-[AEA1-[(D)Lysl-NH2 250
332 Ac-CQTWQCYW-[13hLeul-VNG-[AEA1-[(D)Lysl-NH2 311
333 Ac-CQTWQCYW-H3Aibl-VNG-[AEA1-[(D)Lysl-NH2 2903
334 Ac-CQTWQCYW-H3hAlal-VNG-[AEA1-[(D)Lysl-NH2 355
335 Ac-CQTWQCYW-H3hVall-VNG-[AEAl-RD)Lysl-NH2 501
336 Ac-CQTWQCYW-H3-spiral-pipl-VNG-[AEA1-[(D)Lysl-
>6000
NH2
337 Ac-CQTWQCYW-H3hArgl-VNG-[AEA1-[(D)Lysl-NH2 922
338 Ac-MRTWQ-[MeCysl-YWRKFG-[AEA1-[(D)Lysl-NH2 4251
339 Ac-ACDWVCYWRKFG-[AEA1-[(D)Lysl-NH2 630
340 Ac-SRTWQSYWRKFG-{AEA1-[(D)Lysl-NH2 2816
341 Ac-CDWVCYWRKFG-[AEA1-[(D)Lysl-NH2 664
342 Ac-ARTWQ-[MeCysl-YWRKFG-[AEA1-[(D)Lysl-NH2 7571
343 Ac-ARTWQAYWRKFG-[AEA1-[(D)Lysl-NH2 3194
344 Ac-CQTWQCYW-[hLeul-EN-[AEAl-RD)Lysl-NH2 132
345 Ac-CQTWQCYWOLcul-ENG-[AEA1-[(D)Lysl-NH2 222
346 Ac-CSTWECYWRVYG-[AEAl-RD)Lysl-NH2 47
347 Ac-C4Ornl-TWQCYWRVFG4AEA1-[(D)Lysl-NH2 22 69 95
348 Ac-CQTWQCYW-[Orn1-[Dapl-FG-[AEA1-[(D)Lysl-NH2 96
349 Ac-C-[N-MeAsnl-TWQCYWRVFG-[AEAl-RD)Lysl-NH2 148
350 Ac-C-[N-MeLysl-TWQCYWRVFG-[AEA1-[(D)Lysl-NH2 80
351 Ac-C4Dabl-TWQCYWRVFG-[AEAl-RD)Lysl-NH2 23 51 99
352 Ac-CQTWQCYY4Orn14Dapl-FG4AEA14(D)Lysl-NH2 710
353 Ac-CSTWQCYW4Orn14Dapl-YG4AEA1-[(D)Lysl-NH2 371
354 Ac-CSTWECYW-[Cit]-[Dap]-YG-[AEA1-[(D)Lysl-NH2 74
355 Ac-CQTWQCFF-[Orn1-{Dapl-FG-[AEA1-[(D)Lysl-NH2 4274
356 Ac-CPTWQCYWRVFG-[AEA1-[(D)Lysl-NH2 422
347 Ac-CSTWECYW4Orn14Dabl-YG4AEA1-[(D)Lysl-NH2 338
358 Ac-CSTWECYWRVFG-[AEA1-[(D)Lysl-NH2 48
359 Ac-CLTWQCYWRVFG-[AEA1-[(D)Lysl-NH2 134
360 Ac-CQTWQCYF-[Orn1-{Dapl-FG-[AEA1-[(D)Lysl-NH2 1885
461 Ac-CNTWQCYWRVFG-[AEA14(D)Lysl-NH2 21 79 96
362 Ac-C-[Dapl-TWQCYWRVFG-[AEAl-RD)Lysl-NH2 31
100
363 Ac-C-[N-Me-Alal-TWQCYWRVFG-[AEAl-RD)Lysl-NH2 139
364 Ac-CKTWQCYWRVFG-[AEA1-[(D)Lysl-NH2 40
365 Ac-CQDWQCYWR-[Cha]-FG-[AEAl-RD)Lysl-NH2 113
366 Ac-CQTWQCYWR-[0g11-FG-[AEA1-[(D)Lysl-NH2 206
367 Ac-CQTWQCYWK4Dapl-FG-[AEA1-[(D)Lysl-NH2 32
368 Ac-CQTWQCYWH4Dapl-FG4AEA1-[(D)Lysl-NH2 49 59
369 Ac-CQTWQCYWRLFG-[AEA1-[(D)Lysl-NH2 51 47
370 Ac-CQTWQCYW-[hArg]-[Dap]-FG-[AEA1-[(D)Lysl-NH2 56
239
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Table E3H. IC50 of Illustrative Peptides Containing the 00(WX0000( Motif
Human .iii Rat
A pStat3
SE ID 11,23/ iii I L23/
HTRF
Q iiiii
Sequence iiii
IL23R iii 11,23R (nM)
ELISA iii ELISA
...............................................................................
.......................................................................iiii....
.. (nM) ...... iii...... (nM) ......iii
............................................iii
371 Ac-CSTWECYWRTFG-NH2 252
372 Ac-CDSWECYWRTYG-NH2 366
373 Ac-CSTWECYWHTYG-NH2 181 286 97
374 Ac-CKTWTCYWHTYG-NH2 381
375 Ac-CRTWECYWHEYS-NH2 416
376 Ac-CRTWTCYWHEYG-NH2 434
377 Ac-CFTWQCYWHEYS-NH2 515
378 Ac-CQTWQCYW43-Pall-ENG-NH2 56 20
101
379 Ac-CQTWQC-NH2 >30000
380 Ac-CRTWQC-NH2 >30000
381 Ac-CADWVCY-NH2 >30000
382 Ac-CADWVCYW-NH2 >30000
383 Ac-CADWVCYWH-NH2 ¨30000
384 Ac-CADWVCYWHT-NH2 4795
385 Ac-CADWVCYWHTF-NH2 3277
386 Ac-CMTWQCYWLYGR-NH2 613
387 Ac-CRTWQCYWHEFG-NH2
388 Ac-CRTWECYWHTFG-NH2
389 Ac-CQTWQCYWHEFG-NH2
390 Ac-CRTWQCYWQQFGGE-NH2 81
391 Ac-CRSWQCYWLNFGPD-NH2 101
392 Ac-CRTWQCYWLKMGDS-NH2 39
393 Ac-CQTWQCYWIKRDQG-NH2 67
394 Ac-CSTWQCYWLKHGGE-NH2 19 24 2
395 Ac-CSTWECYWSQRADQ-NH2 240
396 Ac-CQTWECYWRTFGPS-NH2 58
397 Ac-CRTWQCYWQEKGTD-NH2 118
398 Ac-CQTWQCYWLDSLGD-NH2 93
399 Ac-CRTWQCYWTKFGSEP-NH2 87 57
340 Ac-CRSWQCYWNKFGADD-NH2 142
341 Ac-CHTWQCYWLNFGDEE-NH2 323
342 Ac-CRTWQCYWLNFGNEQ-NH2 127
343 Ac-CRTWQCYWSEFGTGE-NH2 180 778
103
344 Ac-CRTWQCYWLRLGDEG-NH2 352 483
181
345 Ac-CHTWQCYWSTLGPEA-NH2 222
346 Ac-CSTWQCYWSKQSGGS-NH2 133 204 89
347 Ac-CHTWQCYWLNNGTSQ-NH2 113
348 Ac-CHTWQCYWRANDGRD-NH2 210
349 Ac-SGCRTWQCYWHEFG-NH2 390
350 Ac-NKCRTWQCYWHEYG-NH2 112
240
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
V Human Rat
pStat3
SEQ ID 1L23/ IL23/ HTRF
NO Sequence IL23R 11,23R (nM)
.
ELISA ELISA
(nM) (nM)
351 Ac-SGCRTWECYWHEYG-NH2 257
352 Ac-DACRTWECYWHKFG-NH2 165
353 Ac-PECRTWECYWHKFG-NH2 197
354 Ac-QVCQTWECYWREFG-NH2 145
355 Ac-DRCVTWECYWREFG-NH2 217
356 Ac-ADQCRTWQCYWHEFG-NH2 228
357 Ac-KENCRTWECYWREFG-NH2 148
358 Ac-VQECSTWQCYWRTFG-NH2 138
359 Ac-GEECSTWQCYWRKFG-NH2 53 24
360 Ac-DGSCRTWQCYWHQFG-NH2 240
361 Ac-NADCHSWECYWREFG-NH2 872
362 Ac-ERNCSTWECYWRAFG-NH2 855
363 Ac-RVGCSTWECYWREFG-NH2 417
364 Ac-KANCRTWQCYWRKFE-NH2 412
365 Ac-YEDCRTWQCYWENFG-NH2 280
366 Ac-CQTWQCYWRNFGDS-NH2
367 Ac-CQTWQCYWRNFESG-NH2
368 Ac-CQDWQCYWREFGPG-NH2
369 Ac-CQDWQCYWRSFGPQ-NH2
370 Ac-CQTWQCYWRTLGPS-NH2
371 Ac-CRTWQCYWQNFG-NH2 235
372 Ac-CGTWQCYWRTFGPS-NH2 76
373 Ac-CSTWQCYWHKFGNE-NH2 182
374 Ac-CRTWECYWRTYGPS-NH2 116
375 Ac-CRTWQCYWWENSQM-NH2 99
376 Ac-CQTWQCYWREFGGG-NH2 165
377 Ac-CQTWQCYWRTHGDR-NH2 83
378 Ac-CRDWQCYWLSRP-NH2 330
379 Ac-CQTWQCYW-[K(Palm)l-ENG-NH2 4880
380 Ac-CQTWQCYW4K(PEG8)1-ENG-NH2 153
381 Ac-CQTWQCYW4hLetil-EQG-NH2 128
382 Ac-CQTWQABUC-RD)Tyr1-WOLeul-ENG-NH2 >30000
383 Ac-CQTWQC-[(N-MeTyil-W-[hLeul-ENG-NH2 >30000
384 Ac-CQTWQC-[Tic-OH1-W4hLeul-ENG-NH2 >30000
385 Ac-CQTWQCEW-[hLeul-ENG-NH2 >30000
386 Ac-CQTWQCTW-[hLeul-ENG-NH2 >30000
387 Ac-CQTWQC4Chal-W4hLetil-ENG-NH2 ¨6000
388 Ac-CQTWQCYW-[cc-MeLeul-ENG-NH2 22 27 5
389 Ac-CQTWQCYW-RD)Leul-ENG-NH2 319
390 Ac-CQTWQCYW4hLeul-ENG-[(D)Lysl-NH2 121
391 Ac-CQTWQCYW-[hLeul-ENG-OH 317
392 Ac-CQTWQCYW4hLetil-ENE-NH2 222 1002
310
393 Ac-CQTWQCYWOLeul-ENR-NH2 93
241
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Human .ii Rat A pStat3
SEQ ID I L23/ iii IL23/ HTRF
iiiii
NO Sequence iiii IL23R
iii 11,23R (nM)
. iiiii
ELISA ELISA
...............................................................................
.......................................iiii...... (nM) ...... iii......
(nM) ....iii ............................................iii
394 Ac-CQTWQCYW4hLeul-ENF-NH2 82 182 69
395 Ac-CQTWQCYW4hLeul-ENP-NH2 253 114 31
396 Ac-CQTWQCYW-[hLeul-ENQ-NH2 347
397 Ac-CQTWQCYW-[hLeul-ENL-NH2 45
398 Ac-CQTWQCYW4hLeul-EEG-NH2 135 53 16
399 Ac-CQTWQCYW-[hLeul-ERG-NH2 647
400 Ac-CQTWQCYW4hLeul-EPG-NH2 108 140 27
401 Ac-CQTWQCYW-[hLeul-ELG-NH2 158
402 Ac-CQTWQCYW-[hLeul-ETG-NH2 818
403 Ac-CQTWQCYW4hLeul-FNG-NH2 395
404 Ac-CQTWQCYW4hLeul-PNG-NH2 4828
405 Ac-CQTWQCYW4hLeul-NNG-NH2 89 26
406 Ac-CQTWQCYW4hLeul-LNG-NH2 78
407 Ac-CQTWQCYW4hLeul-TNG-NH2 109
408 Ac-CQTWQCYWFENG-NH2 185
409 Ac-CQTWQCYWPENG-NH2 >30000
410 Ac-CQTWQCYWQENG-NH2 173
411 Ac-CQTWQCYWTENG-NH2 114
412 Ac-CQTWQCYWEENG-NH2 147
413 Ac-CQTWFCYW4hLeul-ENG-NH2 1412
414 Ac-CQTWPCYW-[hLeul-ENG-NH2 2735
415 Ac-CQTWNCYW-[hLeul-ENG-NH2 1849
416 Ac-CQTWRCYW-[hLeul-ENG-NH2 278
417 Ac-CQTWTCYW4hLeul-ENG-NH2 114
418 Ac-CQTWECYW-[hLeul-ENG-NH2 164
419 Ac-CQTGQCYW4hLeul-ENG-NH2 >10,000
420 Ac-CQTPQCYWOLeul-ENG-NH2 >10,000
421 Ac-CQTNQCYW4hLeul-ENG-NH2 >10,000
422 Ac-CQTRQCYW4hLeul-ENG-NH2 >10,000
423 Ac-CQTTQCYW4hLeul-ENG-NH2 >10,000
424 Ac-CQTEQCYW4hLeul-ENG-NH2 >10,000
425 Ac-CQFWQCYW4hLeul-ENG-NH2 1152
426 Ac-CQPWQCYW4hLeul-ENG-NH2 >10,000
427 Ac-CQNWQCYW-[hLeul-ENG-NH2 336
428 Ac-CQRWQCYW-[hLeul-ENG-NH2 469
429 Ac-CQEWQCYW-[hLeul-ENG-NH2 773
450 Ac-CFTWQCYWOLeul-ENG-NH2 205
451 Ac-CPTWQCYWOLeul-ENG-NH2 27412
452 Ac-CNTWQCYW4hLeul-ENG-NH2 61
453 Ac-CGTWQCYW4hLeul-ENG-NH2 167
454 Ac-CTTWQCYW4hLeul-ENG-NH2 59 28 10
455 Ac-CETWQCYW-[hLeul-ENG-NH2 101
456 Ac-CQTWQCYW-[N-MeLeul-ENG-NH2 >6000
242
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
============'=================
==================t-
n p Human Rat
,:. pStat3
SEQ ID I L23/ iii IL23/
HTRF
iiiii
NO Sequence iiii IL23R iii 11,23R (nM)
. iiiii
ELISA iii ELISA
...............................................................................
.....................iiii........ (nM) ...... iii...... (nM)
......iii ............................................iii
457 Ac-CQTWQCYW- [cc-MeOrn] -ENG-NH2 46 64 12
458 Ac-CQTWQCYW- [cc-MeOrn] -ENG-NH2 28 31 7
459 Ac-CQTWQC-[cc-MePhel-W-[hLeul-ENG-NH2 ¨30000
460 Ac-CQTWQCYW4Aibl-ENG-NH2 31 34 12
461 Ac-CQTWQC-[hTyr]-W-[hLeul-ENG-NH2 ¨6000
462 Ac-CQTWQC-[Bipl-W-[hLeul-ENG-NH2 237
463 Ac-CQTWQCYW40g11-ENG-NH2 66 163 76
464 Ac-CQTWQCYW- [hLeul - [Lys(Ac) ] -NG-NH2 19 32 3
465 Ac-CQTWQCYW4hLeul-ENGG-NH2 61 140 24
466 Ac-CQTWQCYW-[hLeul-ENGP-NH2 97
467 Ac-CQTWQCYW-[hLeul-ENGE-NH2 180
468 Ac-CQTWQCYWOLeul-ENG-(D)Glu-NH2 183
469 Ac-CQTWQCY-[cc-MePhel-[hLeul-ENG-NH2 ¨30000
470 Ac-CQTWQCYW-[hLeul-ENGP-NH2 239
471 Ac-CQTWQCYW-[hLeul-ENGG-NH2 362
472 Ac-CQTWQCYW- [hLeul -ENGL -NH2 174
473 Ac-CQTWQCYW4hLeul-ENGF-NH2 131
474 Ac-CQTWQCYW-[hLeul-ENGE-NH2 129
475 Ac-CQTWQCYW-[hLeul-ENGN-NH2 66 23
476 Ac-CQTWQCYW- [hLeul -ENGT -NH2 160
477 Ac-CQTWQCYW4hLeul-ENGR-NH2 >10,000
>1000
478 Ac-PCQTWQCYW-[hLeul-ENG-NH2 97
479 Ac-LCQTWQCYW4hLeul-ENG-NH2 61 26 21
480 Ac-FCQTWQCYW4hLeul-ENG-NH2 56 25 16
481 Ac-ECQTWQCYW4hLeul-ENG-NH2
482 Ac-NCQTWQCYW4hLeul-ENG-NH2
483 Ac-RCQTWQCYW4hLeul-ENG-NH2
484 Ac-CQTWQCY- [2-Na1l - [hLeul -ENG-NH2
485 Ac-CQTWQCY41-Nall4hLeul-ENG-NH2 18 37 6
486 Ac-CQTWQC42-Nall-W4hLeul-ENG-NH2 48 73 11
487 Ac-CQ TWQC- [1 -Nal] - [2-Nall -[hLetil -ENG-NH2 78 125 17
488 Ac-CQ TWQC- [2-Nall - [1-Nall -[hLetil -ENG-NH2 117
489 Ac-CQTWQC-[Aic]-W-[hLeul-ENG-NH2 126
490 Ac-CQTWQCHW-[hLeul-ENG-NH2 ¨6000
491 Ac-CQTWQCYH- [hL eh] -ENG-NH2 398
492 Ac-CQ TWQC- [Tyr(OMe)] -W- [hLeul -ENG-NH2 ¨30000
493 Ac-CQTWQCY-[134:11-[hLeul-ENG-NH2 42 51 11
494 Ac-CQTWQCY- [Tyr(OMe)] - [hLeul -ENG-NH2 998
495 Ac-CQTWQCHH- [hL eh] -ENG-NH2 148
496 Ac-CQTWQCY- [cc¨MeTrp] - [hLeul -EQG-NH2 >30000
497 Ac-CQTW-RK(PEG8)1-CYWLENG-NH2 212
498 Ac-CQTWQCYWZ-LNG-NH2 800
499 Ac-CQTW- [K(PEG8)] CYW- [K(PEG8)1-ENG-NH2 753
243
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
: Human Rat
::. pStat3
... .....
...
i
SE ID 11,23/ ii IL23/
HTRF
Q iiiii :::
Sequence iii IL23R iii 11,23R :: (nM)
ELISA iii ELISA
..
.==
.....
::
..
.==
:: ...
= ::
500 Ac-CQTW- [K(Palm)1-CYWLENG-NH2 ¨30000
501 Ac-CQTWQCYW-[Orn1-[K(Palm)1-NG-NH2 >6000
502 Ac-Gly-[(D)Asn]-(D)Glu-(D)Leu-(D)Trp-(D)Tyr-(D)Cys-
>30000
(D)G1n-(D)Trp-(D)Thr-(D)G1n-(D)Cys-NH2
503 Ac-CQTWQCYW-[(Orn)14K(Peg8)1-NG-NH2 169
504 Ac-CRTWQCYWHEFG-NH2 166
505 Ac-CRTWECYWHTFG-NH2 333
506 Ac-CQTWQCYWHEFG-NH2 169
507 Ac-CQTWQCYWRNFGDS-NH2 96
508 Ac-CQTWQCYWRNFESG-NH2 315
509 Ac-CQDWQCYWREFGPG-NH2 82
510 Ac-CQDWQCYWRSFGPQ-NH2 117
511 Ac-CQTWQCYWRTLGPSNH2 66
512 Ac-CQTWQCYW-[(D)Prol-ENG-NH2 >30000
513 Ac-CQTWQCYWELNG-NH2 79
514 Ac-CQTWECYWELNG-NH2 154
515 Ac-CQTWQCYR1-Nall-[cc-MeLeul-ENG-NH2 22 67 13
516 Ac-CQTWQCY41-Nall4RDAsnl-ENG-NH2 145 98
517 Ac-CQTWQCYWLE-[K(Palm)I-G-NH2 >6000
518 Ac-CQTWQCYWLEN-[K(Palm)1-NH2 2800
519 Ac-CSTWECYWRTFG-NH2 252
520 Ac-CDSWECYWRTYG-NH2 366
521 Ac-CSTWECYWHTYG-NH2 181 286 97
Table E4A. IC50 of Illustrative examples of dimers of Peptides Containing the
Ac-[Pen]-
)0(WX-[Pen]-X)00( Motif and analogues
SEQ ID =
Human Rat
1)St8t3
NO SequencOi ELISA ELISA HTRF
. .
[Ac- [Penl-QTWQ-[Pen]-[Phe(4-0Me)]-[2-Nall -[cc-MeLys]-
522 ** *
ENG-NH212DIG
[Ac- [Penl-QTWQ- [Pei+ [Phe(4-0Me)1- [2-Nall - [ec-MeLysl-
523 * **
ENG-NH212 PEG25
[Ac- [Penl-QTWQ- [Penl- [Phe [4-(2-aminoethoxy)142-Nall - [ec-
524 ** **
Me-Leul-QNN-NH212 DIG
[Ac- [Penl-QTWQ- [Penl- [Phe [4-(2-aminoethoxy)142-Nall - [ec-
525 * **
Me-Leul-QNN-NH212 PEG25
[Ac- [Penl-QTWQ- [Penl- [Phe [4-(2-aminoethoxy)1- [2-Nall -
526 *** ***
[Aibl4Lys(Ac)1-NQ-NH212 DIG
[Ac- [Penl-QTWQ- [Penl- [Phe [4-(2-aminoethoxy)1- [2-Nall -
527 ** ***
[Aibl4Lys(Ac)1-NQ-NH212 PEG25
244
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
H um:tit-1¨ Rat¨ p Stat3.-
SEQ ID
Sequ en c0
ELISA ELISA HTRF
NO.
(n M) (n M)
(n M)
528 [Ac-[Pen]-QTWQ-[Pen1-[Phe(4-0Me)]-[2-Nal]-[a-MeVall-
[Lys(Ac)1-NN4D)Lys112 DIG
529 [Ac- [Penl-QTWQ [Pei+ [Phe P-(2-acety1aminoethoxy)1- [2-
Nall- [cc-MeVa1l-[Lys(Ac)1-NN4D)Lys112 DIG
[Ac- [Penl-QTWQ [Pei+ [Phe P-(2-acetylaminoethoxy)1-
530 **
Nailcc-MeVall-KNN-NH212 DIG
531 [Ac- [Pen] -QTWQ [Pei+ [Phe [4 -(2 -acetylaminoethoxy)] - [2- ***
Nall-K-[Lys(Ac)1-NN-NH212DIG
532 [Ac- Wen] -QTWQ- [Pei+ [Phe (4-0Me)] - [2-Nall - [cc-MeLys] - **
[Lys(Ac)1-NN-NH212 DIG
533 [Ac- [cc-MeLysl-Wenl-QTWQ- [Pei+ [Phe(4-CONH2)1- 2-Nail - ****
[cc-MeVall-[Lys(Ac)1-NN-NH212 DIG
534 [Ac-[Penl-QTWQ-[Pen1-[Phe(4-CONH2)1-P-Nail-k-MeLysl- **
[Lys(Ac)1-NN-NH212 DIG
535 [Ac-[Penl-NTWQ-[Pen1-[Phe(4-CONH2)1-P-Nail-[Aibl-KNN-
**
NH212 DIG
536 [Ac-[Penl-NTWQ-[Pen1-[Phe(4-CONH2)1-P-Nail-P-amino-4-
carboxy-tetrahydropyranl-KNN-NH212 DIG
537 [Ac-IPenl-NTWQ-[Pen1-[Phe(4-CONH2)1-P-Nall-[Achcl-
KNN-NH212 DIG
538[Pen] -NTWQ- [Pen] - [Phe(4-CONH2)1- -Nail - [Acvc] - **
KNN-NH212 DIG
539 [Ac-[Penl-NTWQ-[Pen1-[Phe(4-CONH2)1-P-Nail-k-MeLeul-
KNN-NH212 DIG
540 [Ac- [Penl-NTWQ- [Penl- [Phe(4-0Me)1- [2-Nall- [Aibl-KNN-
NH212 DIG
541 [Ac- [Penl-NTWQ- [Penl- [Phe(4-0Me)1- [2-Nall- 4-amino-4-
carboxy-tetrahydropyranl-KNN-NH212DIG
542 [Ac- [Penl-NTWQ- [Penl- [Phe(4-0Me)1- [2-Nall- [Achcl-KNN-
NH212 DIG
543 [Ac-[Penl-NTWQ-[Pen1-[Phe(4-0Me)]-[2-Nall-[Acycl-KNN-
NH212 DIG
[Ac- [Penl-NTWQ- [Pei+ [Phe(4-0Me)1- [2-Nall - [cc-MeLet+
544
KNN-NH212 DIG
545 [Ac-[Penl-QTWQ-[Pen1-[Phe(4-CONH2)1-P-Nail-k-MeLysl-
[Lys(Ac)1-NN-NH212 IDA
546 [Ac-[Penl-QTWQ-[Pen1-[Phe(4-CONH2)1-P-Nail-k-MeLys1-
[Lys(Ac)1-NN-NH212 [IDA-13Alal
*=<10nIVI; **=10-25 nM*** = 25-100 nJVI,**** = 100-1000 nIVI, *****=1000-
10,000
245
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Table E4B. IC50 of Illustrative Peptides Containing the Ac-[Pen]-X(WX-[Pen]-
)000( Motif
and Analogues
...............................................................................
.............................................................i.
SE" D Human.¨ Rat ......
i)Stat.3.:
1
I .,
SequencOi ELISA ELISA HTRF
NO.
...............................................................................
...............................................................................
...............................................................................
...............................................................................
.........................................t..... -- (nM) -- ......õõ.... --
(nM) -- (nM)....
547 Ac-[Pen] -RTWQ- [Pen] -YWRKFG- [AEAl- RD)-Lys] -NH2 **** ****
***
548 Ac-A- [Pen] -DWV- [Pen] -YWRKF G- [AEA] - [(D)-Lysl-NH2 >30000
549 Ac- [[Pen] -QTWQ- [Pen] -YW-[hLetil -ENG-NH2 ****
550 Ac- [Pen] -QTWQ- [Pen] -YW [N-MeArg] -ENG-NH2 >30000
551 Ac- [Pen] -QTWQ- [Penl-YW- [hLeul -ENG-NH2 ****
552 Ac- [Pen] -QTWQ- [Penl-YW- [N-MeArg] -ENG-NH2 >30000
Ac-A- [Pen] -DWV- [Pen] -YW- [Ornl- [Dap] -F G- [AEA] - RD)-Lys] -
>30000
553
NH2
554 Ac- [Pen] -QTWQ- [Pen] -YW- [cc-MeLeul -ENG-NH2 *** **** **
555 Ac- [Pen] -QTWQ- [Penl-YW- [(D)Asn] -ENG-NH2 *****
556 Ac- [Pen] -QTWQ- [Pen] -Y- [2-Nall - [cc-MeLy s] -ENG-NH2 ***
**** *
Ac- [Pen]-QTWQ- [Penl- [Phe(4-0Me)] 42-Nall - [cc-MeLys] -ENG- ***
557 **** **
NH2
558 Ac- [Pen] -OTWQ- [Pen] -12-Na11-12-Nall - [cc-MeLy s] -ENG-NH2 ****
**** **
559 Ac- [Pen] -OTWO- [Pen] -Y42-Nall - [cc-MeOrn] -ENG-NH2 *** ****
**
560 Ac- [Pen] -OTWO- [Penl-YW- [oc-MeOrn] -ENG-NH2 **** **** ***
561 Ac- [Pen] -OTWO- [Pen] -Y41-Nall - [cc-MeOrn] -ENG-NH2 **** ****
***
Ac- [Pen] -QTWQ- [Penl- [Phe(4-0Me)] 42-Nall - [cc-MeOrn] -
562 **** ***
[Lys(Ac) ] -NG-NH2
563 Ac- [Pen] -QTWQ- [Pen] -YW- [cc-MeLys] - [Lys(Ac) ] -NG-NH2 ****
***
Ac- [Pen] -QTWQ- [Penl- [Phe-(4-0Me)]-W- [cc-MeLy s] -
564 *** *** **
[Lys(Ac) ] -NG-NH2
Ac- [Pen] -QTWQ- [Penl- [Phe(4-0Me)] 42-Nall - [cc-MeLys] -
565 *** *** *
[Lys(Ac) ] -NG-NH2
246
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Hurnan...''... Rat pStat.3.'
SEQ ID
SequencOi ELISA ELISA HTRF
NO.
(nM) (nM)
Ac-[Pen]-QTWQ-[Pen]-[Phe(4-0Me)]-[1-Nal]-[ct-MeLysl-
566 *** **** ***
[Lys(Ac)l-NG-NH2
Ac-[Penl-QTWQ-[Pen1-[BIP1-[2-Nall-[ec-MeLysl-[Lys(Ac)]-
567 >10,000
NG-NH2
Ac-[Penl-QTWQ-[Penl-Phe(3,4-C12)42-Nall-[ec-MeLysl-
568 ****
[Lys(Ac)l-NG-NH2
Ac-[Penl-QTWQ-[Pen1-[Phe(3,5-F2)]-[2-Nall-[ec-MeLysl-
569 ****
[Lys(Ac)l-NG-NH2
Ac-[Penl-QTWQ-[Pen1-[Phe(4-NH2)142-Nall-[ec-MeLysl-
570 ****
[Lys(Ac)l-NG-NH2
571 Ac-[Penl-QTWQ-[Pen1-[2-Nall-[cc-MeLys1-[Lys(Ac)]-NG-NH2 >10000
572 Ac-[Penl-QTWQ[Pen1-[Phe(3,4-C12)]-[2-Nall-[cc-MeOrnl-ENG- ****
NH2
Ac-[Penl-QTWQ[Pen1-[Phe(4-CN)142-Nall-k-MeOrnl-ENG-
573 ****
NH2
Ac-[Penl-QTWQ[Pen1-[Phe(3,5-F2)]-[2-Nall-[ec-MeOrnl-ENG-
574 ****
NH2
Ac-[Penl-QTWQ[Pen1-[Phe(4-CH2CO2M-P-Nall-[cc-MeOrnl-
575
ENG-NH2
576 Ac-[Penl-QTWQ[Pen1-[Phe(4-CH2C0E12)1-P-Nall-[cc-MeOrnl-
ENG-NH2
Ac-[Penl-QTWQ[Pen1-[Phe(penta-F)]-[2-Nall-[cc-MeOrnl-ENG-
577
NH2
578
Ac-[Penl-QTWQ[Pen1-[Phe(4-CF3)]-[2-Nall-k-MeLysl-ENG-
NH2
Ac-[Penl-QTWQ[Pen1-[Phe[4-(2-aminoethoxy)]-[2-Nall-[ec-
579
MeLysl-ENG-NH2
580 Ac-[Penl-QTWQ[Pen1-[Phe[4-(2-aminoethoxy)]-[2-Nall-[ec-
MeLysl-ENG-NH2
Ac-[Penl-QTWQ-[Pen1-[Phe(4-0Me)142-Nall-k-MeLysl-
581 ****
K(iyDde)-NG-NH2
succinic acid-[Penl-QTWQ[Pen1-[Phe(4-0Me)]-[2-Nall-k-
582 *** ***
MeLys1-[Lys(Ac)]-NG-NH2
glutaric acid-[Penl-QTWQ[Pen1-[Phe(4-0Me)142-Nall-[ec-
583 *** *** **
MeLys1-[Lys(Ac)]-NG-NH2
4-methylmorpholine-2,6-dione-[Penl-QTWQ[Pen1-[Phe(4-
584 *** *** **
OMe)l-p-Nall-k-MeLys1-[Lys(Ac)]-NG-NH2
pyroglutamic acid-[Penl-QTWQ[Pen1-[Phe(4-0Me)142-Nall-[ec- ***
585 ***
MeLys1-[Lys(Ac)]-NG-NH2
247
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Human¨ Rat pStat.3.'
SEQ ID
SequencOi ELISA ELISA HTRF
NO.
(nNI) (01)
isovaleric acid-[Pen[-QTWQ[Pen]-[Phe(4-0Me)142-Nall-ret-
586 *** ***
**
MeLysl-[Lys(Ac)l-NG-NH2
587 gallic acid-[Penl-QTWQ[Pen]-[Phe(4-0Me)]-[2-Nall-[cc- *****
MeLysl-[Lys(Ac)l-NG-NH2
octanoic acid-[Penl-QTWQ[Pen]-[Phe(4-0Me)]-[2-Nall-[cc-
588 ****
MeLysl--[Lys(Ac)l-NG-NH2
589 4-Biphenylacetic acid-[Penl-QTWQ[Pen]-[Phe(4-0Me)l-[2-Nall- ****
-[cc-MeLysl-[Lys(Ac)l-NG-NH2
4-fluorophenylacetic acid-[Penl-QTWQ-[Pen]-[Phe(4-0Me)]-[2- ***
590 ****
Nall-[cc-MeLysl-[Lys(Ac)l-NG-NH2
591 Hy-[Penl-ADWV-[Penl-YWHTFG-NH2 >6000
Ac-[Penl-GTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[cc-
592 **
MeLysl-ENG-NH2
Ac-[Penl-TTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[cc-
593 **
MeLysl-ENG-NH2
Ac-[Penl-STWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[cc-
594 **
MeLysl-ENG-NH2
Ac-[Pen]-[Dapl-TWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-
595 ***
11cc-MeLysl-ENG-NH2
Ac- [Pen] - [cc-MeOrn] -TWQ [Pen] - [Phe [4-(2-amino ethoxy)] -112-
596 ****
Nall-[cc-MeLysl-ENG-NH2
Ac-[Penl-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[cc-
597
MeLysl-ENG-NH2
Ac-[Penl-QTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[cc-
598 ***
MeLysl-[Lys(Ac)l-NG-NH2
Ac-[Penl-QTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall4a-
599 **
MeLysl-[Lys(Ac)l-NN-NH2
Ac-[Penl-QTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[cc-
600 **
MeLysl-ENG-NH2
Ac-[Penl-QTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[cc-
601 ***
MeLysl-ENA-NH2
602 Ac-[Penl-QTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[cc-
MeLeul-[Lys(Ac)l-NN-NH2
603 Ac-[Penl-QTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[cc-
MeLeul-QNN-NH2
604 Ac-[Penl-QTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[Aibl-
ENN-NH2
605 Ac-[Penl-QTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-Aib-
11Lys(Ac)l-NN-NH2
248
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Hum Rat pStat.3.'
SEQ ID
Sequel] cOi ELISA ELISA HTRF
NO.
(n NI) (n M)
606 Ac- [Pert] -QTWQ-[Pen] -[Phe [4-(2-aminocthoxy)] -[Aibl -
[Ly s(Ac)] -NQ-NH2
Ac- [Pert] -Dap (Ac)TWQ- [Pen] - [Phe [4-(2-acetylaminoethoxy)] -
607 **
[2-Na1l- [c(-MeLys(Ac)] -ENG-NH2
Ac- [Pen] - [c(-MeOrn(Ac)l-TWQ- [Pen] - [Phe [4-(2-
608 ****
acetylaminoethoxY)1-12-Nall- [c(-MeLy s(Ac)] -ENG-NH2
609 Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-acety1aminoethoxy)]
[c(-MeLy s(Ac)] - [Ly s(Ac)l-NG-NH2
610 Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-acety1aminoethoxy)]
[c(-MeLy s(Ac)] - [Ly s(Ac)l-NN-NH2
Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-acetylaminoethoxy)]
611 **
[cc-MeLy s(Ac)] -ENG-NH2
Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-acetylaminoethoxy)]
612 **
[c(-MeLy s(Ac)] -ENA-NH2
613 Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-acetylaminoethoxy)]
[c(-MeLeul-[Lys(Ac)]-NN-NH2
614 Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-acetylaminoethoxy)]
[c(-MeLeul-QNN-NH2
Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-acetylaminoethoxy)]
615 **
[Aibl-ENN-NH2
616 Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-acetylaminoethoxy)]
[Aibl - [Ly s(Ac)] -NN-NH2
617 Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-acetylaminoethoxy)]
[Aibl - [Ly s(Ac)] -NQ-NH2
618 Ac- [Pen] -QTWQ- [Penl- [Phe(4-0Me)142-Nall-[Aibl-ENN-NH2
Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-aminoethoxy)] -
619 **
[hLeul-ENA-NH2
620 Ac- [Pen] -TTWQ- [Pen] -[Phc [4-(2-aminoethoxy)] - [2-Nall- [Aib] -
[Ly s(Ac)] -NN-NH2
621 Ac- [Pen] -QTWQ- [Pen] - [Phe(4-0Me)] - [2-Nall - [Aib] - [Lys(Ac)l-
NA-NH2
622 Ac- [Pen] -TTWQ- [Pen] -[Phc [4-(2-aminoethoxy)] - [2-Nall- [Aib] -
[Ly s(Ac)] -NQ-NH2
623 Ac- [Pen] -QTWQ- [Pen] - [Phe(4-0Me)] - [2-Nall - [Aib] - [Lys(Ac)l-
NQ-NH2
624 Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-aminoethoxy)] - [2-Nall -[Aibl -
[Ly s(Ac)] -NA-NH2
625 Ac- [Pen] -QTWQ- [Pen] - [Phe(4-0Me)] - [2-Nall - [Aib] - [Lys(Ac)l-
NN-NH2
249
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Hum Rat
pStat.3.'
SEQ ID
Sequel] cOi ELISA ELISA HTRF
NO.
(n NI) (n M)
626 Ac- [Pen] -QTWQ-[Pen] -[Phe [4-(2-aminoethoxy)] -[2-Nall -
RILeul 4Lys(Ac)1-N-H3Alal -NH2
627 Ac- [Pen] -QTWQ- [Pen] - [Phe (4 -0Me)1- [2-Nall - [hLeu] -
[Lys(Ac)1-N-H3Alal -NH2
628 Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-am ino ethoxy)] - [2-Nall -[Aib] -
[Lys(Ac)1-N-H3Alal -NH2
629 Ac- [Pen] -QTWQ- [Pen] - [Phe (4-0Me)] - [2-Nall - [Aib] - [Lys(Ac)l-
N-13Alal -NH2
630 Ac- [Pen] -NTWQ- [Pen]- [Phe(4-0Me)142-Nall- [Aib] -ENN-NH2
631 Ac- [Pen] -NTWQ- [Pen] - [Phe [4-(2-aminoethoxy)] -
RiLeul -ENA-NH2
632 Ac- [Pen] -NTWQ- [Pen] - [Phe [4-(2-am ino ethoxy)] - [2-Nall -[Aib] -
[Ly s (Ac)] -NN-NH2
633 Ac- [Pen] -NTWQ- [Pen] - [Phe (4-0Me)] - [2-Nall - [Aib] - [Lys(Ac)l-
N-H3Ala] -NH2
634 Ac- [Pen] -NTWQ- [Pen] - [Phe [4-(2-am ino ethoxy)] - [2-Nall -[Aib] -
[Ly s (Ac)] -NQ -NH2
635 Ac- [Pen] -NTWQ- [Pen] - [Phe (4-0Me)] - [2-Nall - [Aib] - [Lys(Ac)l-
NA-NH2
636 Ac- [Pen] -NTWQ- [Pen] - [Phe [4-(2-am ino ethoxy)] - [2-Nall -[Aib] -
[Ly s (Ac)] -NA-NH2
637 Ac- [Pen] -NTWQ- [Pen] - [Phe (4-0Me)] - [2-Nall - [Aib] - [Lys(Ac)l-
NN-NH2
638 Ac- [Pen] -NTWQ- [Pen] - [Phe (4-0Me)] - [2-Nall - [Aib] - [Lys(Ac)l-
NQ-NH2
Ac- [Pen] -NTWQ- [Pen] - [Phe [4-(2-am ino ethoxy)] - [2-Nall -[Aib] -
639 **
[Lys(Ac)1-N-H3Alal -NH2
640 Ac- [Pen] -NTWQ- [Pen] - [Phe (4 -0Me)1- [2-Nall - [hLeu] -
[Lys(Ac)1-N-H3Alal -NH2
641 Ac- [Pen] -NTWQ- [Pen] - [Phe [4-(2-aminoethoxy)] -
RiLeul4Lys(Ac)1-N- H3Ala] -NH2
642 Ac-E- [Pen] -QTWQ- [Penl- [Phe [4-(2-aminoethoxy)1- [2 -Nal] -
[Aib] - [Ly s (Ac)] -NN-NH2
643 Ac-(D)Asp- [Pen] -QTWQ - [Pen] - [Phe [4 -(2 -am ino ethoxy)] -[2-
Nall- [Aibl-[Lys(Ac)1-NN-NH2
644 Ac-R- [Pen] -Q TWQ -Wen] -[Phe [4 -(2 -am ino ethoxy)] - [2-Nall -
[Aib] - [Ly s (Ac)] -NN-NH2
645 Ac-(D)Arg -Wen] -QTWQ- [Pen]- [Phe [4-(2-aminoethoxy)] - [2 -
Nall- [Aibl-[Lys(Ac)1-NN-NH2
250
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Hu man¨ Rat i)Stat3.:
SEQ ID
Sequel) cOi
ELISA ELISA HTRF
NO.
(n M) (n
(n M )
646 Ac-Phe-[Perd -QTWQ- [Pen1-[Phe [4-(2-aminoethoxy)] 42-Nail -
[Aib] - [Ly s (Ac)] -NN-NH2
647 Ac-(D)Phe- [Pen] -Q TWQ- [Pen] - [Phe [4-(2-am inoethoxy)] -112-
Nail - [Aib] - [Ly s (Ac)] -NN-Nt12
648 Ac- 112-N al] - [Pen] -QTWQ- [Pen]- [Phe [4-(2-aminoethoxy)1- 112-
Nail - [Aib] - [Ly s (Ac)] -NN-Nt12
649 Ac-T- [Pen] -QTWQ- [Penl- [Phe [4-(2-am inoethoxy)1- 112-Nail-
[Aib] - [Ly s (Ac)] -NN-NH2
650 Ac-L- [Pen] -QTWQ- [Penl- [Phe [4-(2-am inoethoxy)1- 112-Nail-
[Aib] - [Ly s (Ac)] -NN-NH2
651
Ac-(D)G1n- [Pen] -Q TWQ- [Pen] - [Phe [4-(2-am inoethoxy)] -112-
Nail - [Aib] - [Ly s (Ac)] -NN-Nt12
652
Ac- [(D)Asn] -[Pen] -QTWQ- [Pen] - [Phe [4-(2-aminoethoxy)] -112-
Nail - [Aib] - [Ly s (Ac)] -NN-NH2
653 Ac- [Pen] -QTWQ- [Pen]- [Phe(4-0Me)] -[c(-MeVall -
[Ly s (Ac)] -NN- [(D)Lys] -NH2
654 Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-acetylaminoethoxy)] -
[c(-MeV al] -KNN-Nt12
Ac- [Pen] -QTWQ- [Pen] - [Phe [4-(2-acetylaminoethoxy)] -
655 ** *
K- [Ly s (Ac)] -NN-NH2
666 Ac- [Pen] -QTWQ- [Pen]- [Phe(4-0Me)] - [c(-MeLys] -
[Ly s (Ac)] -NN-NH2
Ac- [(D)Ly s] - [Pen] -QTWQ- [Pen] - [Phe(4-C ONH2)] - [cc-
667 *****
MeVal] - [Lys (Ac)] -NN-Nt12
Ac- [Pen] -QTWQ- [Pen] - [Phe (4-C ONH2)] - [c(-MeLy s] -
668 * **
[Ly s (Ac)] -NN-NH2
Ac- [Pen] -QTWQ- [Pen] - [Phe (4-C ONH2)] -[c(-MeVall -
669 **
[Ly s (Ac)] -NN-NH2
Ac- [Pen] -QTWQ [Per+ [Phe (4-C ONH2)1-1Phe (3,4-0Me2)14c(-
670 **
MeVal] - [Lys (Ac)] -NN-Nt12
671
Ac-RD)Phe] -[Pen] -NTWQ [Pen] -[Phe [4-(2-aminoethoxy)] -112-
Nail - [Aib] - [Ly s (Ac)] -NN-Nt12
672
Ac- [(D)Phe] - [Pen] -NTWQ - [Pen]- [Phe [4-(2-aminoethoxy)] 42-
Nail - [4-amino-4-c arboxy-tetrahydropyran] -[Ly s (Ac)] -NN-Nt12
673
Ac-RD)Phe] -[Pen] -NTWQ [Pen] -[Phe [4-(2-aminoethoxy)] -112-
Nail- [Ache] - [Ly s (Ac)] -NN-Nt12
674
Ac- [(D)Phe] - [Pen] -NTWQ - [Pen]- [Phe [4-(2-aminoethoxy)] -112-
Nail - [4-amino-4-c arboxy-tetrahydropyran] - [Cit] -NN-Nt12
675
Ac- [(D)Phe] - [Pen] -NTWQ - [Pen]- [Phe [4-(2-aminoethoxy)] -112-
Nail - [Ache] - [Cit] -NN-NH2
251
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
SEQ ID
SequencOi
ELISA ELISA HTRF
NO.
676
Ac-[(D)Phe]-[Penl-NTWQ-[Pen1-[Phe[4-(2-aminoethoxy)]-[2-
Nall- [Aib] -{Lys(Ac) ] -N- H3A1a] -NH2
677
Ac- RD)Phe] -{Pen] -NTWQ- [Pen] - [Phe(4-0Me)] - [2-Na1l -114-
am ino-4-c arb oxy-tetrahydropyran] - [Lys (Ac)] -NN-NH2
678 Ac- RD)Phe] -{Pen] -NTWQ- [Pen] - 11Phe(4-0Me)1- 112-Nall- [Achc] -
[Lys(Ac) ] -NN-NH2
679
Ac- RD)Phe] -{Pen] -NTWQ- [Pen] - 11Phe(4-0Me)1- 112-Nall -114-
am ino-4-c arb oxy-tetrahydropyran] - rit] -NN-NH2
680 Ac- RD)Phe] -{Pen] -NTWQ- [Pen] - 11Phe(4-0Me)1- 112-Nall- [Achc] -
rit] -NN-NH2
681 Ac- RD)Phe] -{Pen] -NTWQ- [Pen] - 11Phe(4-0Me)1- 112-Nall- [Achc] -
EN-NH2
682 Ac- [Pen] -NTWQ Wei+ [Phe (4-C ONH2)1- 112-Nall - [Aib] -
[Ly s (Ac)] -NN-NH2
683
Ac- [Pen] -NTWQ Wei+ [Phe (4-C ONH2)1- 112-Nall- 114-amino-4-
carboxy-tetrahydropyran] -Ly s (Ac)] -NN-NH2
684 Ac- [Pen] -NTWQ Wei+ [Phe (4-C ONH2)1- 112-Nall- [Achc] -
[Ly s (Ac)] -NN-NH2
685 Ac- [Pen] -NTWQ Wei+ [Phe (4-C ONH2)1- 112-Nall- [Acpc] -
[Ly s (Ac)] -NN-NH2
686
Ac- [Pen] -NTWQ Wei+ [Phe (4-C ONH2)1- 112-Nall - [cc-MeL eu] -
[Ly s (Ac)] -NN-NH2
687 Ac- [Pen] -NTWQ [Pen] 4Phe (4-0Me)l- [2-Nall- [Aib] -[Ly s (Ac)] -
NN-NH2
688
Ac- [Pen] -NTWQ [Pen] 4Phe (4-0Me)] - [2-Nall- [4-am ino-4-
carboxy-tetrahydropyran] - [Ly s (AO] -NN-NH2
689 Ac- [Pen] -NTWQ [Pen] 4Phe (4-0Me)l- [2-Nall- [Achc] -[Ly s (Ac)] -
NN-NH2
670 Ac- [Pen] -NTWQ [Pen] 4Phe (4-0Me)l- [2-Nall- [Acpc] -[Ly s (Ac)] -
NN-NH2
671 Ac- [Pen] -NTWQ [Pen] 4Phe(4-0Me)] - [2-Nall- [cc-MeLeu] -
[Ly s (Ac)] -NN-NH2
*=<10nIVI; **=10-25 nM*** = 25-100 nJVI,**** = 100-1000 nIVI, *****=1000-
10,000
Table E5A. IC50 of Illustrative Peptide Inhibitors (Thioethers)
SEQ Human
ID iii$equence/StructurC ELISA
NO. (nNI)
0
[N-MeAla]-DWVCYWHTF G-[AEA]-[(D)Lys] -NH2
672
¨6000
252
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
.SEQ Human
ID iiiSeq en ce/Stru ctu reiii HASA ..
NO. .
0
[N-MeAla]-DWV-[Pen]-YWHTFG-[AEA]-[(D)Lys] -NH2
673
>30000
0
[N-MeAla]-DWV-(D)Pen]-YWHTFG-[AEA]-[(D)Lys] -NH2
674
>30000
0
[N-MeAlal-DWV-[hCysl-YWHTFG-[AEA1-[(D)Lys] -NH2
675
¨6000
0
ADWVCYWHTFG-[AEA]-[(D)Lys] -NH2
676
¨3000
0
ADWV_-_[PYWHTFG-[AEA1-[(D)Lys] -NH2
677
>30000
0
ADWV-[(D)Pen]-YWHTFG-[AEA1-[(D)Lys] -NH2
678
>30000
0
ADWV-[hCys]-YWHTFG-[AEA]-[(D)Lys] -NH2
679
100 ¨6000
0
680 tj....?DWQCYWRENG-
[AEA](D)Lys] -NH2
>6000
0
681 <1¨ QDWQCYWRENG-[AEA]-
[(D)Lys] -NH2
¨30000
0
682 QDWQCYWRENG-[AEA]-[(D)Lys] -NH2
¨6000
0
QDWQCYWRENG-[AEA]-[(D)Lys] -NH2
683
¨6000
0
QDWQCYWRENG-[AEA]-[(D)Lys] -NH2
684
¨30000
253
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ = Human
ID iiiSequence/Structureiii ELISA
NO. . ( iNI)
0
QDWQCYWRENG- [AEA]-[(D)Lys] -NH2
685
>6000
0
686 QDWQ-[hCys]-YWRENG-[AEA]-[(D)Lys] -NH2
>6000
0
687 QDWQ-[hCys]-YWRENG- [AEA] -[(D)Lys] -NH2
>6000
0
QDWQ- [hCys]-YWRENG-[AEA]-[(D)Lys] -NH2
688
S >6000
0
QDWQ- [hCys]-YWRENG- [AEA]- [(D)Lys] -NH2
689
S ¨30000
0
QDWQCYWRENG-[AEA]-[(D)Lys] -NH,
690
s >30000
0
QDWQ-[hCysl-CYWRENG-[AEA]-[(D)Lys] -NH2
691
>30000
s¨j
Table E5B. IC50 of Illustrative Peptide Inhibitors (Thioethers)
Ac,
N----[Phe(4-0Me)]-[2-Nal]-XXXX-NH 2
0
0
Ac-Cyc10-[[Abui-XXWXCHPhe(4-0Mc)]42-Na1]-XXX-NH 2
SEQ Human = Rat pStat3."
ID Sequence ELISA .ELISA H.TRF
NO ,4
692
Ac-Cyclo-[[Abu]RTWQC]-YVVRKFG-
*** **** ***
[AEA]-[(D)Lys]-NH2
254
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
N.Q. (nm) .... .(1m)
693
Ac-Cyclo-[CRTWQ-[Abu]]-YVVRKFG- **** **** ***
[AEA]-[(D)Lys]- NH2
694
Ac-Cyclo-[[Abu]-QTWQC]-YVVRENG- **** **** ***
[AEA]-[(D)Lys]- NH2
Ac-Cyclo-[[Abu]-RTWQ-[Pen]]-
695 *****
YVVRKFG-[AEA]-[(D)Lys]- NH2
Ac- Cyclo-[[Pen]-RTWQ-[Abu]]-
696 ****
YVVRKFG-[AEA]-[(D) Lys]-NH2
Ac-Cyclo-[[(D)Cys]-RTWQ- [Abu]]-
697 ****
YVVRKFG-[AEA]-[(D)-Lys]- NH2
698 Ac-Cyclo-[[Abu]-QTWQC]-YVV-[Orn]- ****
[Dap]-NG-[AEA]-[(D) Lys]- NH2
Ac-Cyclo-[[Abu]-QTWQC]-YW-[hLeu]- ***
699 **
ENG- NH2
700 Ac-Cyclo-[Abu]-QTWQ-(D)Cys]]-YVV- *****
[hLeu]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQ- [Pen]]-YVV-
701 *****
[hLeu]-ENG-NH2
702
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
*** ****
OMe)]-[2-Nall-[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQCFYVV- [a-
703 ** ***
MeLeu]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQCFY-[2-Nal]-
704 ** **
[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
705 ** **
OMe)]-[2-Nall-[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
706 ** ***
OMe)]-[2-Nall-[a-MeOrn]-ENG-NH2
707
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
*** ***
OMe)]-W-[a-MeOrn]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
708 ** ***
OMe)]-[2-Nall-[a-MeLys]-ENG-NH2
255
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ" Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO.
Ac-Cy cl o- [ [Abu] -Q TWQ C] - [Phe(4-
709 ** ** **
OMe)]-W-[a-MeLys]- [Ly s(Ac)] -NG-NH2
Ac-Cy cl o- [ [Abu] -Q TWQ C] - [Phe(4-
710 ** * ** **
OMe)]-W-[a-MeLys]-ENG-NH2
Ac-Cy cl o- [ [Abu] -Q TWQ C] - [Phe(4-
711 OMe)] - [1 -Nal] - [a-MeLys] - [Lys(Ac)]- ** * * ** **
NG-NH2
Ac-Cy cl o- [ [Abu] -Q TWQ C] - [Phe(4-
712 OMe)] - [2-Nal] - [a-MeLys] - [Lys(Ac)]- **
NG-NH2
Ac-Cyclo-[[Abu]-QTWQCFYVV- [a-
713 ** * * ** **
MeOrn]- [Ly s(Ac)] -NG-NH2
Ac-Cy cl o- [ [Abu] -Q TWQ C] - [Phe(4-
714 OMe)] - [2-Nal] - [(D)Asn] - [Lys(Ac)] -NG- *** **** ***
NH2
Ac-Cy cl o- [ [Abu] -Q TWQ C] - [Phe(4-
715 Phenoxy)]- [2-Nail- [a-MeLysHLys(Ac)]- ****
NG-NH2
Ac-Cyclo- [ [Abu] -QTWQC] - [hPhe(3 ,4-
716 dirnethoxy)] - [2-Nail- [a-MeLys]- *****
[Lys(Ac)]-NG-NH2
717 Ac-Cyclo- [ [Abu] -Q TWQC] - [DMT] - [2- * * * * *
Nail - [a-MeLy s] - [Lys(Ac)]-NG-NH2
Ac-Cy cl o- [ [Abu] -Q TWQ C] - [Phe(4-
718 CONH2)]-[2-Nal[a-MeLys]- * **
[Lys(Ac)]NG-NH2
Ac-Cyclo- [ [Abu] -Q TWQC] -Phe(3 ,4-
719 C12) [2-Nal]-[a-MeLysHLys(Ac)]NG- **** ***
NH2
720 Ac-Cyclo- [ [Abu] -QTWQ- [Pen]] - [Phe(4- * * * * * * * * *
* *
OMe)] - [2-Nall -[a-MeLys]-ENG-NH2
Ac-Cyclo- [ [Abu] -QTWQ- [Pen]] - [Phe(4-
721 OMe)]-[2-Nal[a-MeLys]- [Lys(Ac)]NG- *** **** ***
NH2
Ac-Cy cl o- [ [Pen] -Q TWQ- [Abu] ] - [Phe(4-
722 OMe)] - [2-Nall -[a-MeLys]- [Ly s(Ac)]NG-
NH2
Ac-Cy cl o- [ [Abu] -Q TWQ C] - [Phe(4-
723 >10,000
OMe)] - [Trp(2,5,7-tri-tert-B utyl)] - [cc-
256
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO.
MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
724 OMe)]- [Phe(4-0ally1)] - [a-MeLysFENG- ****
NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
725 OMe)]-[Tyr(3-tBu)]-[a-MeLys]-ENG- *** **** **
NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
726 OMe)]-[Phe(4-tBu)]-[a-MeLys]-ENG- *****
NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
727 OMe)]-[Phe(4-guanidino)]-[a-MeLys]- ****
ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
728 ****
OMe)]-[Phe(Bz1)]-[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQCHTyr(3-tBu)]-
729 >10,000
W-[a-MeLys]-ENG-NH2
780 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-tBu)]- *****
W-[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
781 *** *** ***
guanidino)]-W-[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-
782 ** **
aminoethoxy)1-W-[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
783 ****
CO2H)]-W-[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
784 *** *** **
phenoxy)1-W-[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe(4-CNA *** -
785 ***
W-[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe(4-BrA *** -
786 *** ***
W-[a-MeLys]-ENG-NH2
787
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
*** ***
NH2)]-W-[a-MeLys]-ENG-NH2
257
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ" Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO.
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
788 ****
OMe)]-Phe(4-Me)-[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
789 *** *** **
OMe)]-[1-Nall-[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
790 OMe)]-[2-Nal]-[a-MeOrn]-[Lys(Ac)]- ** **
NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[2-Nall-[2-
791 *** ****
Nall-[a-MeOrn]-[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Bip]-[2-
792 ****
Nall-[a-MeLys]-[Lys(Ac)l-NG-NH2
793 Ac-Cyclo-[[Abu]-QTWQC]-Cha-[2-Nall- *****
[a-MeLys]-[Lys(Ac)l-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[2-Nall-[2-
794 *** *** **
Nall-[a-MeLys]-[Lys(Ac)l-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[4-
795 Pyridylalanine]-[2-NalHa-MeLys]- ****
[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[(3-
796 homoTyr1[2-Nail[a-MeLys]-[Lys(Ac)]- ¨10000
NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
797 CONH2)]-[2-Nall-[a-MeLys]-[Lys(Ac)]- ** **
NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[2-Nall-[2-
798 *** ***
Nall-[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT42-Nall-QC]-
799 [Phe(4-0Me)]-[2-Nall-[a-MeLys]- ****
[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QT-[1-Nall-QC]-
800 [Phe(4-0Me)]-[2-Nall-[a-MeLys]- ****
[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QTYQCHPhe(4-
801 OMe)]-[2-Nall-[a-MeLys]-[Lys(Ac)]- ¨10000
NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
802 OMe)]-[2-Nall-[a-MeLys]-[Lys(Ac)]-
NG-NH2
258
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ" Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO.
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
803 OMe)]-[2-Nal]-[a-MeLys]-[Lys(Ac)]- ***
NGGE-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
804 OMe)]-[2-Nal]-[a-MeLys]-[Lys(Ac)]-
NGAE-NH2
Ac-Cyclo-[[Abu]-STWQC]-[Phe(4-
805 OMe)]-[2-Nal]-[a-MeLys]-[Lys(Ac)]-
NGGE-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
806 OMe)]-W-[a-MeLys]-[Lys(Ac)]-NGGE-
NH2
807
Ac-Cyclo-[[Abu]-QTWQC]-Y-[2-Nal]-
[a-MeLys]-[Lys(Ac)]-NGGE-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
808 OMe)]-[2-Nal[a-MeLys]-[Lys(Ac)]-NS- ** **
NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
809 OMe)]-[2-Nal]-[a-MeLys]-[Lys(Ac)]- **
NA-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
810 ** ***
OMe)]-[2-Nall-[Aib]-[Lys(A0]-NG-NH2
811 Ac-Cyclo-[[Abu]-QTWQC]-[Phe-4-N3]- *** *** **
[2-Nall-[a-MeLys]-[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
812 OMe)]-[2-Nal]-[a-MeLys]-[Lys(Ac)]- *** ***
QG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
813 OMe)]-[2-Nal]-[a-MeLys]-[Lys(Ac)]-
[Cit]-G-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
814 *** ***
OMe)]-[2-Nall-[a-MeLys]-VNG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
815 ****
OMe)]-[2-Na1]-[Orti]-[Lys(A0]-NG- NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
816 ****
OMe)]-[2-Nall-[0m]-[Dap]-NG-NH2
Ac-Cyclo-[[Abu]-NTWQC]-[Phe(4-
817 OMe)]-[2-Nal]-[a-MeLys]-[Lys(Ac)]- ** ***
NG-NH2
259
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
ir-SEQ Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO. .1
Ac-Cyclo-RAbu]-QT- [Bip]-QC]- [Phe(4-
818 OMe)] - [2-Nal] - [a-MeLys] - [Lys(Ac)]- ¨10000
NG-NH2
Ac-Cy clo- [ [Abu] -Q TWQC] - [Phe(4-
819 *** ***
OMe)] - [2-Nall - [Cha] - [Lys(Ac)] -NG-NH2
Ac-Cy clo- [ [Abu] -Q TWQC] - [Phe(4-
820 ***
OMe)] - [2-Nall - [Chg]- [Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QT-[Octgly]-QC]-
821 [Phe(4-OMe)] - [2-Nal] - [a-MeLy s]- >10000
[Lys(Ac)] -NG-NH2
Ac-Cyclo-[[Abu]-QTWQCHOctgly]- [2-
822 ¨10000
Nail - [a-MeLy s] - [Lys(Ac)]-NG-NH2
Ac-Cy clo- [ [Abu] -Q TWQC] - [Phe(4-
823 OMe)]-[Octgly]- [a-MeLys]-[Lys(Ac)]- ¨10000
NG-NH2
Ac-Cy clo- [ [Abu] -Q TWQC] - [Phe(4-
824 OMe)] - [2-Nal] - [a-MeLys] - [Lys(Ac)]- *** ***
NGE-NH2
Ac-Cy clo- [ [Abu] -Q TWQC] - [Phe(4-
825 OMe)] - [2-Nal] - [a-MeLys] - [Lys(Ac)]- **
NAE-NH2
Ac-Cy clo- [ [Abu] -S TWQC] - [Phe(4-
826 OMe)] - [2-Nal] - [a-MeLys] - [Lys(Ac)]- *** *** ***
NGE-NH2
Ac-Cy clo- [ [Abu] -Q TWQC] - [Phe(4-
827 OMe)]-W-[a-MeLys]- [Lys(Ac)]-NGE- ****
NH2
Ac-Cyclo-[[Abu]-QTWQCFY- [2-Nall-
828 ***
[a-MeLy s] - [Lys(Ac)] -NGE-NH2
Ac-Cyclo- [[Abu]-Q TWQC]- [Phe[4-(2-
829 aminoethoxy)]]-[2-Nal[a-MeLysFENG- *
NH2
Ac-Cyclo- [[Abu]-Q TQQC]- [Phe[4-(2-
830 aminoethoxy)]]- [2-Nall 4a-MeLysFENG- >3000
NH2
Ac-Cyclo- [[Abu]-Q THQC]- [Phe[4-(2-
831 aminoethoxy)]]- [2-Nall 4a-MeLysFENG- >3000
NH2
260
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ" Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO. .1
Ac-Cyclo-[[Abu]-QT-[hPhe]-QCHPhet4-
832 (2-aminoethoxy)]]-[2-Nall-[a-MeLys]- >3000
ENG-NH2
Ac-Cyclo-[[Abu]-QT-[Glu(Bz1)]-QC]-
833 [Phe[4-(2-aminoethoxy)]]-[2-NalHa- >3000
MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT-[Bip]-QC]-[Phe[4-
834 (2-aminoethoxy)]]-[2-Nall-[a-MeLys]- >3000
ENG-NH2
Ac-Cyclo-[[Abu]-QT-[Tic]-QC]-[Phe[4-
835 (2-aminoethoxy)]]-[2-Nall-[a-MeLys]- >3000
ENG-NH2
Ac-Cyclo-[[Abu]-QT-[Phe[4-(2-
aminoethoxy)]]-QC]-[Phe[4-(2-
836 >3000
aminoethoxy)]]-[2-NalHa-MeLysFENG-
NH2
Ac-Cyclo-[[Abu]-QT-[Phe(3,4-C12)]-
837 QCF[Phe[4-(2-aminoethoxy)]]-[2-Nall- >3000
[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT-[Phe(4-0Me)]-QC]-
838 [Phe[4-(2-aminoethoxy)]]-[2-Nall-[a- >3000
MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT-[Orn(Benzyl)]-QC]-
839 [Phe[4-(2-aminoethoxy)]]-[2-Nall-[a- >3000
MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT-
840
[Orn(Benzaldehyde)]-QC]-[Phe[4-(2-
aminoethoxy)]]-[2-NalHa-MeLysFENG-
NH2
Ac-Cyclo-[[Abu]-QTWQC]-
841 [PheOCH2CH2NHAc]-[2-Nall-[a-
MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
842 aminoethoxy)]]-[2-NalHa-MeLeuFENG-
NH2
Ac-Cyclo-[[Abu]-QT-[5-hydroxyTrp]-
843 QCF[Phe[4-(2-aminoethoxy)]]-[2-Nall- ¨3000
[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT-[6-chloroTrp]-QC]-
844 [Phe[4-(2-aminoethoxy)]]-[2-Nail[a- ** **
MeLys]-ENG-NH2
261
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO. .1
Ac-Cyclo-[[Abu]-QT-[N-MeTrp]-0q-
845 [Phe[4-(2-aminoethoxy)]]-[2-Nall-[a- >3000
MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT-[1,2,3,4-tetrahydro-
norharman]-QC]-[Phe[4-(2-
846 ****
aminoethoxy)]]-[2-Nall4a-MeLysFENG-
NH2
Ac-Cyclo-[[Abu]-QT-[Phe(4-CO2H)]-
847 QCF[Phe[4-(2-aminoethoxy)]]-[2-Nall- >3000
[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT-[Ph(4-CONH2)]-
848 QCF[Phe[4-(2-aminoethoxy)]]-[2-Nall- >3000
[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT-[Ph(4-CONH2)]-
849 QCF[Phe[4-(2-aminoethoxy)]]-[2-Nall- >3000
[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT-[Phe(3,4-0Me)]-
850 QCF[Phe[4-(2-aminoethoxy)]]-[2-Nall- ¨3000
[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT4a-MePhe]-QC]-
851 [Phe[4-(2-aminoethoxy)]]-[2-Nail[a- ****
MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT-[Phe(4-CF3)]-QC]-
852 [Phe[4-(2-aminoethoxy)]]-[2-Naila- ¨3000
MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT-[Phe(4-tBu)]-QC]-
853 [Phe[4-(2-aminoethoxy)]]-[2-NalHa- >3000
MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QT-[Phe(2,4-Me2)]-
854 QCF[Phe[4-(2-aminoethoxy)]]-[2-Nall- ****
[a-MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
855 aminoethoxy)]]-[2-Nail[a-MeLys]- **
DNG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
856 aminoethoxy)]]-[2-Nall4a-MeLys]-
QNG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
857 aminoethoxy)]]-[2-Nall4a-MeLys]-
[Lys(Benzoic acid)]-NG-NH2
262
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ" Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO. .1
Ac-Cyclo-[[Abu]-QTWOC]-[Phe[4.-(2-
858 aminoethoxy)]]-[2-Nal[a-MeLys]- **
[Lys(succinic acid)]-NG-NE12
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
859
aminoethoxy)]]-[2-NalHa-MeLys]-
[Lys(glutaric acid)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
860
aminoethoxy)]]-[2-NalHa-MeLys]-
[Lys(pyroglutamic acid)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
861
aminoethoxy)]]-[2-Nal[a-MeLys]- **
[Lys(isovaleric acid)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
862 aminoethoxy)]]-[2-Nal] - [a-MeLys]- -- ¨3000
[Lys(Palm)]-NG-NE12
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
863 aminoethoxy)]]-[2-Nal] - [a-MeLys]-
Lys [(PEG1)]-NG-NE12
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
864 aminoethoxy)]]-[2-Nal] - [a-MeLys]-
[Lys(PEG2)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
865 aminoethoxy)]]-[2-Nal] - [a-MeLys]-
[Dap(Benzoic acid)]-NG-NE12
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
866 aminoethoxy)]]-[2-Nal] - [a-MeLys]-
[Dap(succinic acid)]-NG-NE12
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
867 aminoethoxy)]]-[2-Nal] - [a-MeLys]-
[Dap(glutaric acid)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
868 aminoethoxy)]]-[2-Nal] - [a-MeLys]- **
[Dap(pyroglutamic acid)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
869 aminoethoxy)]]-[2-Nal] - [a-MeLys]-
Dap(IVA)NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
870 aminoethoxy)]]-[2-Nal] - [a-MeLys]-
[Dap(PEG1)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
871 aminoethoxy)]]-[2-Nal] - [a-MeLys]-
[Dap(PEG2)]-NG-NH2
263
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ" Hamad' Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO. .1
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-
872 aminoethoxy)]]-[2-Nal] - [a-MeLys]- ** **
[Dap(PEG2-Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
873 aminoethoxy)]]42-Nall4a-MeLys]-
[Lys(Ac)]--NG-[AEA]-[(D)Lys]-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
874 aminoethoxy)]]42-Nall4a-MeLys]-
[Lys(A0]-NG-RD)14s1-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
875 aminoethoxy)]]42-Nall4a-MeLys]-
[Lys(Ac)]-NG-[AEA]-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
876 aminoethoxy)]]42-Nall4AibHLys(Ac)]- **
QG-N2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
877 **
aminoethoxY)]]-[2-Nall-[Aib]-QNG-NH2
878 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
aminoethoxY)]]-[2-Nall-[AibFENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
879 aminoethoxy)]]-1-Nal[Aib]-[Lys(Ac)]- *** **
NG-NH
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
880 aminoethoxy)]]42-Nall4AibHLys(Ac)]- **
NA-NH
881 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
aminoethoxY)]]-[2-Nall-[Aib]-KNG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
882 aminoethoxy)]]4Phe(4-CO2H)14a- ****
MeLys]-[Lys(A0]-NG-NH2
Ac-Cyclo-[[Abu]-[Dap]-TWQC]-[Phe[4-
883 (2-aminoethoxy)]]-[Phe(4-Phenoxy)]-[cc- ****
MeLys]-[Lys(A0]-NG-NH2
Ac-Cyclo-[[Abu]-DapTWQC]-[Phe[4-(2-
aminoethoxy)]]-[Phe[4-(2-
884 ****
aminoethoxy)]]-[a-MeLys]-[Lys(Ac)]-
NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
885 aminoethoxy)]Ha-MeLysHLys(Ac)]- >3000
NG-NH
264
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ" Hamad' Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO. .1
Ac-Cyclo-[[Abu]-DabTWQC]-[Phe[4-(2-
886 aminoethoxy)]]-[hPhe] - [a-MeLys]- >1000
[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-DapTWQC]-[Phe[4-(2-
887 aminoethoxy)]]-[Glu(Bz1)] - [a-MeLys]- >3000
[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
888 aminoethoxy)]]-W-[a-Me-Orn]-ENG- **
NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
889 aminoethoxy)]]-W-[a-MeLys]-[Lys(Ac)]- *
NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
890 aminoethoxy)]]-W-[a-Me-Orn]- ** **
[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
891 aminoethoxy)]]-[2-Nall4a-Me-Orn]-
[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
892 aminoethoxy)]]-[2-Nal]-[a-MeLys]-
[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
893 aminoethoxy)]]-[2-Nal]- [OrnHLys(Ac)]- ** **
NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
894 ***
aminoethoxY)]i-W-[Orn]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
895 ****
aminoethoxy)]]-W-[Orn]-[Dap]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
896 aminoethoxy)]]-W-[Orn]-[Dap(Ac)]-NG- ****
NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
897 aminoethoxy)]]-[2-Nal]- [Orn]-[Dap]-NG- *** ***
NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
898 aminoethoxy)]]-[2-Nall-[Orn]-[Dap(Ac)]- ***
NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
899 **
aminoethoxY)]i-W-[hLeu]-ENG-NH2
265
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ" Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO. .1
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[442:-
900 (acetyl-aminoethoxy)] ] - [2-Nal] - [a-
MeLys(Ac)] - [Lys(Ac)] -NG-NH2
Ac-Cyclo- [[Abu]-Q TWQC]- [Phe[4-(2-
901 aminoethoxy)] ] -W- [a-Me-Leu] -ENG-
NH2
Succicinyl-Cyclo- [[Abu]-QTWQC]-
902 [Phe[4-(2-aminoethoxy)]]-[2-Nall4a-
MeLys] - [Lys(Ac)] -NG-NH2
Ac-Cyclo- [[Abu]-Q TWQC]- [Phe[4-(2-
903 aminoethoxy)]]-W- [a-MeLys]- [Lys(Ac)]- *****
[Dap] -G-NH2
Ac-Cyclo- [[Abu]-Q TWQC]- [Phe[4-(2-
904 aminoethoxy)]]-W- [a-MeLys]- [Lys(Ac)]- ***
[6-amino-1,4-diazepane-2,5-dione] -NH2
Ac-Cyclo- [[Abu]-Q TWQC]- [Phe[4-(2-
905 aminoethoxy)] ] -W-Chg- [Lys(Ac)] -NG- ***
NH2
Ac-Cyclo- [[Abu]-Q TWQC]- [Phe[4-(2-
906 (acetyl-aminoethoxy)] ] - [2-Nall - [a-
MeLys(Ac)]-ENG-NH2
Ac-Cyclo- [[Abu]-Q TWQC]- [Phe[4-(2-
907 aminoethoxy)]]- [Phe(4-CONH2)]-[a- ****
MeLys] - [Lys(Ac)] -NG-NH2
Ac-Cyclo- [[Abu]-Q TWQC]- [Phe[4-(2-
908 aminoethoxy)] ] - [Phe(3,4-0Me2] - [a- ** ***
MeLys] - [Lys(Ac)] -NG-NH2
Ac-Cyclo- [[Abu]- [Dap]-TWQC]- [Phe[4-
909 (2-aminoethoxy)]]-[Tic]-[a-MeLys]- >3000
[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-DapTWQC]- [Phe[4-(2-
910 aminoethoxy)]]- [Phe(3,4-C12)]- [a- ***
MeLys] [Lys(Ac)] -NG-NH2
Ac-Cyclo- [[Abu]-Q TWQC]- [Phe[4-(2-
911 aminoethoxy)]]-[2-Nal]- [a-MeLys]-ENQ- *
NH2
Ac-Cyclo- [[Abu]-Q TWQC]- [Phe[4-(2-
912 aminoethoxy)]]- [2-Nall 4a-MeLysFENN- *
NH2
Ac-Cyclo- [[Abu]-TTWQC]- [Phe[4-(2-
913 aminoethoxy)]]- [2-Nall 4a-MeLysFENG- *
NH2
266
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ" Hamad' Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO. .1
Ac-Cyclo-[[Abu]-QTWOCHPhe[4-(2-
914 aminoethoxy)]]-[2-NalHa-Me-
Gly(Ethyl)] Lys(Ac)l-NG-NH2
Ac-Cyclo-[[Abu]-QTWOCHPhe[4-(2-
915 aminoethoxy)]]-[2-NalHa-MeVal]-
[Lys(Ac)l-NG-NH2
Ac-Cyclo-[[Abu]-QTWOCHPhe[4-(2-
916 aminoethoxy)]]-[2-NalHa-MeSer]-
[Lys(Ac)l-NG-NH2
Ac-Cyclo-[[Abu]-QTDapQC]-[Phe[4-(2-
917 aminoethoxy)]]-[2-NalHa-MeLys]- >3000
[Lys(Ac)l-NG-NH2
Ac-Cyclo-[[Abu]-QTWOCHPhe[4-(2-
918 aminoethoxy)]]-[Dap[a-MeLys]- >3000
[Lys(Ac)l-NG-NH2
Ac-Cyclo-[[Abu]-QTRQC]-[Phe[4-(2-
919 aminoethoxy)]]-[2-NalHa-MeLys]- >3000
[Lys(Ac)l-NG-NH2
Ac-Cyclo-[[Abu]-QTWOCHPhe[4-(2-
920 aminoethoxy)fi-R4a-MeLysHLys(Ac)]- >3000
NG-NH2
Ac-Cyclo-[[Abu]-QTDapQC]-[Phe[4-(2-
921 aminoethoxy)]]-[Dap]-[a-MeLys]- >3000
[Lys(Ac)l-NG-NH2
Ac-Cyclo-[[Abu]-QTDOCHPhe[4-(2-
922 aminoethoxy)]]-[2-NalHa-MeLys]- >3000
[Lys(Ac)l-NG-NH2
Ac-Cyclo-[[Abu]-QTWOCHPhe[4-(2-
923 aminoethoxy)fi-D4a-MeLysHLys(Ac)]- >3000
NG-NH2
Ac-Cyclo-[[Abu]-QTDOCHPhe[4-(2-
924 aminoethoxy)fi-D4a-MeLysHLys(Ac)]- >3000
NG-NH2
Ac-(D)Lys-[Cyclo-[[Abu]-QTWQQ-
925 [Phe(4-0Me)]-[2-Nall-[a-MeLeu]-ENG- **
NH2
Ac-Cyclo-[[Abu]-QTWOCHPhe[4-(2-
926 aminoethoxy)]]-[2-Nall-[a-MeLysFRNG- **
NH2
Ac-Cyclo-[[Abu]-QTWOCHPhe[4-(2-
927 aminoethoxy)]]-[2-NalHa-MeLys]-
[Orn]-NG-NH2
267
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ" Hamad' Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO. .1
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
928 arninoethoxy)]]-[2-NalHa-MeLys]-
KNG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
929 arninoethoxy)]]-[2-NalHa-MeLys]-
hRNG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
930 arninoethoxy)]]-[2-Nall-[hLeu]-
[Lys(Ac)]-N-[f3Ala]-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
931 arninoethoxy)]]-[2-Nall-[CitHDap]-NG- **
NH2
Ac-Cyclo-[[Abu]-[a-Me-Orn]-TWQC]-
932 [Phe[4-(2-aminoethoxy)]]-[2-Nail[a- *** **
MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
933 arninoethoxy)]]-[2-NalHa-MeLys]-
NNG-NH2
Ac-Cyclo-[[Abu]-STWQC]-[Phe[4-(2-
934 arninoethoxy)]]-[2-NalHa-MeLys]- ****
KNGGE-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
935 (acetyl-arninoethoxy)]]-[2-Nall-[a-
MeLys(A01-ENQ-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
936 (acetyl-aminoethoxy)]]-[2-Nall-[cc-
MeLys(Ac)]-ENN-NH2
Ac-Cyclo-[[Abu]-TWQC]-[Phe[4-(2-
937 aminoethoxy)]]-[2-NalHa-MeLysFENG-
NH2
938 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-Me)]- *
[2-Nall-[a-MeLys]-[Lys(A01-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(3-Me)]- **
939
[2-Nall-[a-MeLys]-[Lys(A01-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[hTyr]-[2-
940 *****
Nall-[a-MeLys]-[Lys(Ac)l-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
941 arninoethoxy)]Ha-MeTrpHa-MeLys]- ¨3000
[Lys(Ac)]-NG-NH2
268
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ" Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO. .1
Ac-Cyclo-RAbul-ra-MeSed-TWQ6-
942 [Phe[4-(2-aminoethoxy)]]-[2-Nal]-[a- *** **
MeLys]-[Lys(A0]-NG-NH2
Ac-Cyclo-[[Abu]-Q4a-MeSed-WQC]-
943 [Phe[4-(2-aminoethoxy)]]-[2-Nall-[a- >3000
MeLys]-[Lys(A0]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
944 aminoethoxy)]]-[a-MePhe]- [a-MeLys]- >3000
[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-
945 **
aminoethoxY)]i-W-[AiN-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
946 aminoethoxy)]]-[2-Nall-[AiN4Lys(Ac)]-
NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
947 aminoethoxy)]]-[2-Nall-[AiN-E- **
[Dap(Ac)]-G-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
948 aminoethoxy)]]-[2-Nal]-[AiN-E-
[Dab(Ac)]-G-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
949 aminoethoxy)]]-[2-Nall-[AiN-E- **
[Lys(A0]-G-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-
950 **
aminoethoxY)]I-W-[AiN-ENN-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
951 aminoethoxy)]]-[2-Nal]-[a-MeLeu]-ENN- *
NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
952 aminoethoxy)]]-[Phe(3,4-0Me2)]-[AiN- *** **
ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
953 aminoethoxy)]]-[Phe(3,4-C12)]-[AiN- ***
ENN-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
954 aminoethoxy)]]-[2-NalHa-MeLeu)-[Cit]- *
NN-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
955 aminoethoxy)]]-[2-Nal]-[a-MeLeu]-
[Lys(Ac)]-NN-NH2
269
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO.
I
956 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-Me)]- **
[2-Nall-[AiN-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(3,4-
957 *** **
F2)1-[2-NalHAN-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe(3-
958 ****
CONH2)]-[2-Nall-[AiN-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQCF[Phe(2,4-
959 ****
C12)]-[2-Nall-[AilA-ENG-NH2
960 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(3-Me)]- **
[2-Nall-[AiN-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe(4-C1)]-
961 **
[2-Nall-[AiN-ENG-NH2
962 Ac-Cyclo-[[Abu]-QTWQCHPhe(4-F)l- ****
[2-Nall-[AiN-ENG-NH2
963 Ac-Cyclo-[[Abtil-QTWQCF[Phe(2,4-C12, *****
4-0Bz)]-[2-Nall-[AiN-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-
964 OMe)]-[2-Nall-[a-MeLeu]-ENG- *** **
[(D)Lys] -NH2
Ac-E-Cyclo-[[Abti]-QTWQC]-[Phe[4-(2-
965 aminoethoxy)]]-[2-Nail[a-MeLysFENN- *
NH2
Ac-(D)Glu-[Cyclo-[[Abu]-QTWQC]-
966 [Phe[4-(2-aminoethoxy)]]-[2-NalHa-
MeLys]-ENN-NH2
Ac-Arg-Cyclo-[[Abu]-QTWQC]-[Phe[4-
967 (2-aminoethoxy)]]-[2-Nall-[a-MeLys]-
ENN-NH2
Ac-RD)Arg]-Cyclo-[[Abu]-QTWQC]-
968 [Phe[4-(2-aminoethoxy)]]-[2-NalHa-
MeLys]-ENN-NH2
Ac-F-Cyclo-[[Abu]-QTWQC]]-[Phe[4-(2-
969 aminoethoxy)]]-[2-Nail[a-MeLysFENN- *
NH2
Ac-[(D)Phe]-Cyclo-[[Abti]-QTWQC]-
970 [Phe[4-(2-aminoethoxy)]]-[2-NalHa-
MeLys]-ENN-NH2
270
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ" Hamad Rat pStat..3.4
ID Sequence ELISA ELISA HTRF
NO. .1
Ac-[2-Nal]-Cyclo-RAbuFQTWQC. ]-
971 [Phe[4-(2-anainoethoxy)]]-[2-Nail[a- **
MeLys]-ENN-NH2
Ac-T-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
972 aminoethoxy)]]-[2-Nall[a-MeLysFENN- *
NH2
Ac-Leu-Cyclo-[[Abu]-QTWQC]-[Phe[4-
973 (2-anainoethoxy)]]-[2-Nall-[a-MeLys]-
ENN-NH2
Ac-[(D)Gln]-Cyclo-[[Abu]-QTWQC]-
974 [Phe[4-(2-anainoethoxy)]]-[2-NalHa-
MeLys]-ENN-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
975 anainoethoxy)]]-[2-Nall-[AcpcFENN- **
NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
976 aminoethoxy)]]-[2-Nall-[AcbcFENN-
NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
977 aminoethoxy)]]-[2-Nall-[Achc]- ENN-
NH2
978 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
aminoethoxY)]]-[2-Nall-[Acvc]-ENN-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
979 aminoethoxy)]]-[2-Nall44-amino-4-
carboxy-piperidineFENN-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
980 aminoethoxy)]]-[2-Nall44-amino-4-
carboxy-tetrahydropyran]-ENN-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
981 aminoethoxy)]]-[2-Nall-[a-MeLeu]-
[Lys(Ac)]-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
982 aminoethoxy)]]-[2-Nall4a-MeLeuFENG-
NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
983 aminoethoxy)]]-[2-Nall-[a-MeLeu]-
QNG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
984 aminoethoxy)]]-[2-Nall-[a-MeLeuFQN-
[f3Ala]-NH2
271
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
SEQ Human Rat pStat3
ID Sequence ELISA ELISA HTRF
NO. == (nNI
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
***
985 aminoethoxy)]]-[2-Nal]-[a-MeLeu]-
QDG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-
****
986 cycloaPhe[4-(2-aminoethoxy)]]-[2-Nall-
[a-MeLeu]-QD]-G-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
987 aminoethoxy)]]-[2-Nall-[Aib] -QN-[j3Ala]-
NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
988 aminoethoxy)]]-[1,2,3,4-tetrahydro¨
norharman]-[A113]-QNG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2- **
989 aminoethoxy)]]-[5-hydroxyTrp]-[Aib]-
QNG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
990 aminoethoxy)]]-[2-Nal] -[a-MeLys]- ***
[Lys(Ac)]-[Asn(isobutyl)]-G-N-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
aminoethoxy)]]-[2-Nall-[a-MeLys]-
991 ***
[Lys(Ac)]-[Asp(1,4-diaminoethane)]-G-
NH2
Ac-(D)Phe-Cyclo-[[Abu]-QTWQC]-
992
[Phe[4-(2-aminoethoxy)]]-[2-Nall-[4-
amino-4-carboxy-tetrahydropyran]-ENN-
NH2
Ac-[(D)Arg]-Cyclo-[[Abu]-QTWQC]-
993
[Phe[4-(2-aminoethoxy)]]-[2-Nall-[4-
amino-4-carboxy-tetrahydropyran]-ENN-
NH2
*=<10nIVI; **=10-25 nM *** = 25-100 nJVI,**** = 100-1000 nM, *****=>1000
Table ESC. IC50 of Illustrative Thioether Peptide Dimers Synthesized
Rat
SEQ= = Human pStat3ID '
Linker ELIS
Moiety Sequenceii [LISA HTRF
== A
NO. (iiM) (nM)
272
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
[Ac-[(D)Lys]-Cyclo-[[Abu]-
DIG through QTWQC]-[Phe(4-0Me)]- [2-
***
994
(D)Lys Nal]-[a-MeLeu]-ENG-NH2]2
DIG
DIG through
[Ac-Cyclo-[[Abu]-QTWQC]-
Phe[4-(2-
995 [Phe[4-(2-aminoethoxy)]]- [2- *** **
aminoethox
[Aib]-QNG-NH2]2 DIG
[Ac-Cyclo-[[Abu]-QTWQC]-
DIG through
996 [Phe(4-0Me)]-[2-Nal]- [a- **
a-MeLys
MeLys]-ENG-NH2]2 DIG
[Ac-[(D)Lys]-Cyclo-[[Abu]-
PEG25
QTWQC]-[Phe(4-0Me)]- [2- **
997 through a-
Nall4a-MeLysFENG-NH2]2
MeLys
PEG25
[Ac-Cyclo-[[Abu]-QTWQC]-
DIG through
998 [Phe(4-0Bz1)]-W-[a-MeLys]-
(D)Lys
ENG-NH2]2 DIG
PEG25 [Ac-Cyclo-[[Abu]-QTWQC]-
999 through Y(Bz1)-W-[a-MeLys]-ENG-
(D)Lys NH2]2PEG25
Alexa488-
[D-Arg]-Cyclo-[[Abu]-
PEG4
QTWQC]-[Phe(4-2ae)]-[2-
through [D-
Nail -
Arg]
*=<10nIVI; **=10-25 nM *** = 25-100 nJVI,**** = 100-1000 nM, *****=>1000
Table E5D. Illustrative Thioether Peptides
SEQ ID Human
NO. IL23R /
Sequence IL23
ELISA
(nM)
993 Ac-[D-Arg]- Cyclo-[[Abu]-QTWQC]-[Phe(4-2ae)]-[2-
Nall-[THP1-ENN-NH2
Ac-[D-Arg]- Cyclo-[[Abu]-QTWQC]-[Phe(4-2ae)]-[2-
**
Nall-[THP1]-END-NH2
Ac-[D-Arg]- Cyclo-[[Abu]-QTWQC]-[Phe(4-2ae)]-[2-
***
Nall-[THP1-EDN-NH2
Ac-[D-Arg]- Cyclo-[[Abu]-QTWEC]-[Phe(4-2ae)]-[2-
**
Nall-[THP]-ENN-NH2
Ac-[D-Arg]- Cyclo-[[Abu]-ETWQC]-[Phe(4-2ae)]-[2-
**
Nall-[THP]-ENN-NH2
273
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-[D-Arg]- Cyclo-[[Abu]-QTWQC]-[Phe(4-2ae)]-[2-
***
Nall-[THP1-EDD-NH2
Ac-[D-Arg]- Cyclo-[[Abu]-QTWEC]-[Phe(4-2ae)]-[2-
**
Nall-[THP1-END-NH2
Ac-[D-Arg]- Cyclo-[[Abu]-ETWQC]-[Phe(4-2ae)]-[2-
**
NaI]-[Tetrahydropyran-A]-END-NH2
Ac-[D-Arg]- Cyclo-[[Abu]-QTWEC]-[Phe(4-2ae)]-[2-
***
Nall-[THP1-EDN-NH2
Ac-[D-Arg]- Cyclo-[[Abu]-ETWQC1-[Phe(4-2ae)1-[2-
***
Nall-[THP1-EDN-NH2
Ac-[D-Arg]- Cyclo-[Abu-ETWEC1-[Phe(4-2ae)]-[2-Nal]-
**
[THP]-ENN-NH2
Ac-[D-Arg]- Cyclo-[[Abu]-QTWQC1-[Phe(4-2ae)1-[2-
**
Nall-[THP1-ENN-OH
Ac-[D-Arg]- Cyclo-[[Abu]-QTWQC1-[Phe(4-2ae)1-[2-
***
Nall-[THP1-END-OH
Ac-[D-Arg]- Cyclo-[[Abu]-QTWQC1-[Phe(4-2ae)1-[2-
***
Nall-[THP]-EDN-OH
Ac-[D-Arg]- Cyclo-[[Abu]-QTWEC1-[Phe(4-2ae)1-[2-
**
Nall-[THP1-ENN-OH
Ac-[D-Arg]- Cyclo-[[Abu1ETWQC1-[Phe(4-2ae)1-[2-
**
Nall-[THP]-ENN-OH
* =< 1 nIVI; ** = 1 nM - 10 nM; *** = 10 nM¨ 100 nM
Table E6. IC50 of Peptide Inhibitors (Ring Closing Metathesis)
Human
SE'' ID
NO ii5eq u en ce/Stru ctu rgi EL I SA
.
(ii M)
0 H 0
1000 NH2 --))L YWHTFG-NH2 ¨
20000
Ac-NH 0
1001 ,ADWV YWHTFG-NH2 ¨30000
Ac-NH 0 H
RTWQ N
1002 YWRKFG-[AEA1-[(D)Lys] -NH2 *****
2 ¨ 3
274
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
0 H
Ac-NH
1003 'õ..R_TWQ-- NO)L
YWRKFG-[AEA]-[(D)Lys] -NH2 *****
( )
3 _
2
O ()))LH0
Ac-NH
Q
RTW ' N
1004 YWRKFG-[AEA]-[(D)Lys] -NH2 *****
3
0 H
Ac-NH
1005 RTWQ --.))L YWRKFG-
[AEA]-[(D)Lys] -NH2 ****
(
3 _
Ac-NH 0 H
1006 .....RTWQ;NO)L YWRKFG-[AEA]-[(D)Lys] -NH2 ****
( )
2 - 2
O H
Ac-NH
1007 RTWQ "...) YWRKFG- [AEA] -
[(D)Lys] -NH2 ****
(
O H
Ac-NH
1008 - RTWQ ' N YWRKF G- [AEA] -
[(D)Lys] -NH2 ****
)2
*=<10nIVI; **=10-25 nM *** = 25-100 nIVI, **** = 100-1000 nIVI, *****=1000-
10,000 nIVI.
Table E7. IC50 of Illustrative Peptides Containing Cyclic amides (side chain
cyclizations)
o
0
H
(
t 4....i._ 1
1 -Y7.2 ) 1.2 1 I 1.2
NThrYWXXXX-AEA-(D)Lys-N H2 Ac......Ni XXWX , N ,y(WXXXX-AEA-(D)Lys-
N H 2
H 6 H H 0 H
0
0
'
SE' ID
Uhlman
:
NO Sequence
. õii
...............................................................................
..............................................................................t
.. ELnIS'I;(..;
(N
1009 Ac-Cyclo-[[Dap]-QTWQE]-YWRENG-[AEA]-[(D)Lys]-NH2 ¨6000
1010 Ac-Cyclo-[EQTWQ-[Dab]]-YVVRENG-[AEA]-[(D)Lys]-NH2 >6000
1011 Ac-Cyclo-[EQTWQ-[Dap]]-YVVRENG-[AEA]-[(D)Lys]-NH2 ¨6000
1012 Ac-Cyclo-[[Dab]QTWQE]-YVVRENG-[AEA]-[(D)Lys]-NH2
¨30000
1013 Ac-Cyclo-[[Dap]-QTWQ-[(D)Asp]-YVVRENG-[AEA]-[(D)Lys]-NH2 >30000
1014 Ac-Cyclo-[[Dap]-QTWQD]-YVVRENG-[AEA]-[(D)Lys]-NH2
>30000
275
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
1015 Ac-Cyclo-[[DQTWQ-[Dab]]-YWRENG-[AEA]-[(D)Lys]-NH2
¨6000
1016 Ac-Cyclo-[[Dab]QTWQD]-YVVRENG-[AEA]-[(D)Lys]-NH2
>6000
1017 Ac-Cyclo-[[(D)Dab]-QTWQ-[(D)Asp]]-YVVRENG-[AEA]-[(D)Lys]-
6000
NH2
1018 Ac-Cy cl o- [ [(D)Asp]-Q TWQ- [(D)D ab] ] -YVVRENG- [AEA] - [(D)Ly
s] - _1400
NH2
1019 Ac-Cyclo-[[(D)Asp]-QTWQ-[(D)Dap]]-YVVRENG-[AEA]-[(D)Lys]-
30000
NH2
Table E8. IC50 of Illustrative Peptides Containing the Ac-[Pen]-XXWX)0000(
Motif
and Ac-)00(WX-[Pen]-)000( analogues
SEQ Human........Rat
pStat.31
ID SetittenM ELISA ELISA HTRF
NO.
( nM) ( nM)
(nM)
................
1020 Ac-[Penl-ADWVCYWHTFG-NH2 *****
1021 Ac-CADWV-[Penl-YWHTFG-NH2 *****
1022 Ac4(D)Penl-ADWVCYWHTFG4AEAl-RD)-Lysl-NH2 **** ***** ****
1023 Ac-CADWV4(D)Penl-YWHTFG4AEA]-[(D)-Lysl-NH2 >30000 ***** ****
1024 Ac-{Penl-RTWQCYWRKFG-{AEAI-RD)-Lysl-NH2 **** **** ****
1025 Ac-ACDWV-[Penl-YWRKFG-[AEAl-RD)-Lysl-NH2 *****
1026 Ac-A-[Penl-DWVCYWRKFG-[AEAl-RD)-Lysl-NH2 ****
1027 Ac-A-[hCys]-DWV-[Penl-YWRKFG-[AEAl-RD)-Lysl-NH2 ¨30000
1028 Ac-CQTWQ-[Penl-YW-[cc-MeLeul-ENG-NH2 **** ****
1029 Ac-CQTWQ-[Penl-YW-RD)Asnl-ENG-NH2 *****
*=<10nIVI; **=10-25 nM *** = 25-100 nJVI,**** = 100-1000 nIVI, *****=1000-
10,000
[00698] SAR analysis of the activities of the peptide inhibitors tested
indicated that the C)000(C
disulphide is associated with high activity. The two Trp residues and the Phe
residue are also
associated with high activity, but it is recognized that these amino acids can
be readily
exchanged with similar homologs (e.g., 1-Nal substitued for Trp and/or Phe
substituted for Tyr).
In addition, the data suggested that the presence of one or more basic
residues at the C-terminus
276
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
is associated with high activity. Also, His-9 can be replaced by Arg or
another homolog and
maintain or improve activity. The schematic below provides one illustrative
consensus sequence
showing certain residues associated with high activity.
1 2 3 4 5 6 7 8 9 10 11 12 13 14
Cys-Xxx-Xxx-Trp-Xxx-Cys-Tyr-Trp-His-Xxx-Phe-Xxx-Xxx-(D)Lys-OH
EXAMPLE 3
STABILITY OF PEPTIDE INHIBITORS IN SIMULATED INTESTINAL FLUID (SIF), SIMULATED
GASTRIC
FLUID (SGF) AND REDOX CONDITIONS
[00699] Studies were carried out in simulated intestinal fluid (SIF) and
simulated gastric fluid
(SGF) to evaluate gastric stability of the peptide inhibitors of the present
invention. In addition,
studies were carried out to assess redox stability of the peptide inhibiotrs
of the present
invention.
1007001SIF was prepared by adding 6.8 g of monobasic potassium phosphate and
10.0 g of
pancreatin to 1.0 L of water. After dissolution, the pH was adjusted to 6.8
using NaOH. DMSO
stocks (2 mM) were first prepared for the test compounds. Aliquots of the DMSO
solutions were
dosed into 6 individual tubes, each containing 0.5 mL of SIF, which is pre-
warmed to 37 C. The
final test compound concentration was 20 p.M. The vials were kept in a
benchtop Thermomixer
for the duration of the experiment. At each timepoint (0, 5, 10, 20, 40, 60,
or 360 minutes or 24
hours), 1.0 mL of acetonitrile containing 1% formic acid was added to one vial
to terminate the
reaction. Samples were stored at 4 C until the end of the experiment. After
the final timepoint is
sampled, the tubes were mixed and then centrifuged at 3,000 rpm for 10
minutes. Aliquots of the
supernatant were removed, diluted 1:1 into distilled water containing internal
standard, and
analyzed by LCMSNIS. Percent remaining at each timepoint was calculated based
on the peak
area response ratio of test to compound to internal standard. Time 0 was set
to 100%, and all
later timepoints were calculated relative to time 0. Half-lives were
calculated by fitting to a first-
order exponential decay equation using Graphpad. Stablity in SIF assays is
shown in Tables E9
and E10.
277
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
[00701] SGF was prepared by adding 20 mg NaC1, 32 mg porcine pepsin (MP
Biochemicals,
catalog 02102599), and 70p1 HC1 to 10m1 water (final pH=2). Aliquots of SGF
(0.5ml each)
were pre-warmed at 37 C. To start the reaction, 1 1 of peptide stock solution
(10mM in DMSO)
was added to 0.5ml SGF and thoroughly mixed such that the final peptide
concentration was
20p.M. The reactions were incubated at 37 C with gentle shaking. At each time
point (0, 15, 30,
60 min) 500 aliquots were removed and added to 200 ul acetonitrile containing
0.1% formic
acid to quench the reaction. Samples are stored at 4 C until the end of the
experiment and
centrifuged at 10,000 rpm for 5 minutes. Aliquots of the supernatant were
removed, diluted 1:1
into distilled water containing internal standard, and analyzed by LCMSNIS.
Percent remaining
at each timepoint was calculated based on the peak area response ratio of test
to compound to
internal standard. Time 0 was set to 100%, and all later timepoints were
calculated relative to
time 0. Half-lives were calculated by fitting to a first-order exponential
decay equation using
GraphPad. Stability in SGF assays in shown in Tables E9 and El O.
Table E9. Stability of Illustrative Peptides containing the Ac-[Pen]-)0(WX-
[Pen]-)000( Motif
and Analogues in Simulated Intestinal Fluid (SIF) and Simulated Gastric Fluid
(SGF)
SEQ SGF' S
IF
ID NO: t I /2 t
I /2
iiiSeAl nen*
=
=== ( m in )
(ruin )
.=
549 Ac- [[Pen] -QTWQ- [Pen] -YW- [hLeul -ENG-NH2 *****
1030 Ac- [Pen] -QTWQ- [Pen] -YWN-Me-RENG-NH2
****
551 Ac- [Pen] -QTWQ- [Pen] -YW- [hL eu] -ENG-NH2 *****
552 Ac- [Pen] -QTWQ- [Pen] -YW- [N-MeArg] -ENG-NH2
***
554 Ac- [Pen] -QTWQ-[Pen] -YW- [cc-MeLeul-ENG-NH2 **
1028 Ac-CQ TWQ- [Pen] -YW- [cc-MeL eu] -ENG-NH2
*****
555 Ac- [Pen] -QTWQ- [Pen] -YW- [(D)Asn] -ENG-NH2 **
1029 Ac-CQTWQ-[Penl-YW-RD)Asnl-ENG-NH2
*****
556 Ac- [Pen] -QTWQ- [Pen] -[cc-MeLy s] -ENG-NH2 **
557 Ac- [Pen] -QTWQ- [Pen] - [Phe(4-0M01- [2-Na11- [cc-MeLy s] -ENG-NH2
*** **
558 Ac- [Pen] -QTWQ- [Peril -12-Nall -12-Nall - [cc-MeLy s] -ENG-NH2 **
278
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ '" SG F' S
IF
ID NO: ti!' ti/'
i e(luenlif
=
(min)
(min)
559 Ac413enl-QTWQ-1Penl-Y-12-Nall-1cc-MeOrn1-ENG-NH2 **
560 Ac-113enl-QTWQ-1Penl-YW-10t-MeOrn1-ENG-NH2 **
561 Ac-113enl-QTWQ-1Penl-Y-11-Nall-1cc-MeOrn1-ENG-NH2 **
1031 Ac-113en] -QTWQ-1Pen1-11Phe(4 -OW)] (OW)] -12-Nal] -1cc-MeOrn1-
11Lys(Ac)1-NG-NH2
563 Ac-113enl-QTWQ-1Penl-YW-1cc-MeLys1-1Lys(Ac)1-NG-NH2
1032 Ac-113enl-QTWQ-1Penl- 11Phe(4-0Me)1-W-1cc-MeLys1-1Lys(Ac)1-NG-NH2
565 Ac-113enl-QTWQ-1Pen1-1Phe(4-0Me)]-12-Nall-1cc-MeLys1-1Lys(Ac)1-NG-
NH2
566 Ac-113enl-QTWQ-1Pen1-1Phe(4-0Me)]-11-Nall-1cc-MeLys1-1Lys(Ac)1-NG-
NH2
1033 succinic anhydride- [Pen] -Q TWQ [Pen] - [Phe (4-0Me)] -12-Nall - [cc-
MeLys] - **
11Lys(Ac)1-NG-NH2
585 pyroglutamic acid-113enl-QTWQ[Pen1-1Phe(4-0Me)]-12-Nall-1cc-MeLysl-
**
11Lys(Ac)1-NG-NH2
1034 Ac-113enl-QTWQ-1Pen1-1Phe14-(2-aminoethoxy)]-12-Nall-1cc-MeLysl-
[Lys(Ac)1-NN-NH2
601 Ac-113enl-QTWQ-1Pen1-1Phe14-(2-aminoethoxy)]-12-Nall-1cc-MeLysl-
ENA-NH2
602 Ac-113enl-QTWQ-1Pen1-1Phe14-(2-aminoethoxy)]-12-Nall-1cc-MeLeul- **
***
11Lys(Ac)1-NN-NH2
603 Ac-113enl-QTWQ-1Pen1-1Phe14-(2-aminoethoxy)]-12-Nall-1cc-MeLeul-
QNN-NH2
604 Ac-113enl-QTWQ-1Pen1-1Phe14-(2-aminoethoxy)]-12.-Nall-1Aibl-ENN-
NH2
605 Ac-113enl-QTWQ-1Pen1-1Phe14-(2-aminoethoxy)]-12-Nall-Aib-1Lys(Ac)l-
** ***
NN-NH2
606 Ac-113enl-QTWQ-1Pen1-1Phe14-(2-aminoethoxy)]-12-Nall-1Aibl-
[Lys(Ac)1-NQ-NH2
607 Ac-113enl-Dap(Ac)TWQ-1Pen1-1Phe14-(2-acetylaminoethoxy)]-12-Nall-1cc-
279
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ " SG F'
S I F
ID NO: t1/2
t1/2
i equenlif
MeLys(Ac)1-ENG-NH2
608 Ac-[Pen1-[cc-MeOrn(Ac)1-TWQ-[Pen1-[Phe[4-(2-acetylaminoethoxy)1-p-
**
****
Nall-[cc-MeLys(Ac)-ENG-NH2
609 Ac-[Penl-QTWQ-[Pen1-[Phe[4-(2-acetylaminoethoxy)]-[2-Nall-[cc-
MeLys(Ac)-[Lys(Ac)-NG-NH2
610 Ac-[Penl-QTWQ-[Pen1-[Phe[4-(2-acetylaminoethoxy)]-[2-Nall-[cc-
MeLys(Ac)-[Lys(Ac)1-NN-NH2
611 Ac-[Penl-QTWQ-[Pen1-[Phe[4-(2-acetylaminoethoxy)]-[2-Nall-[cc- **
MeLys(Ac)1-ENG-NH2
612 Ac-[Penl-QTWQ-[Pen1-[Phe[4-(2-acetylaminoethoxy)]-[2-Nall-[cc-
MeLys(Ac)1-ENA-NH2
613 Ac-[Penl-QTWQ-[Pen1-[Phe[4-(2-acetylaminoethoxy)]-[2-Nall-[cc-
MeLeul-[Lys(Ac)l-NN-NH2
614 Ac-[Penl-QTWQ-[Pen1-[Phe[4-(2-acetylaminoethoxy)]-[2-Nall-[cc-
MeLeul-QNN-NH2
615 Ac-[Penl-QTWQ-[Pen1-[Phe[4-(2-acetylaminoethoxy)]-[2-Nall-[Aibl-
ENN-NH2
616 Ac-[Penl-QTWQ-[Pen1-[Phe[4-(2-acetylaminoethoxy)]-[2-Nall-[Aibl-
[Lys(Ac)1-NN-NH2
617 Ac-[Penl-QTWQ-[Pen1-[Phe[4-(2-acetylaminoethoxy)]-[2-Nall-[Aibl-
[Lys(Ac)1-NQ-NH2
522 11Ac-[Penl-QTWQ-11Pen1-11Phe(4-0Me)]-112-Nan-11cc-MeLysl-ENG-Nt1212
****
DIG
618 Ac-[Penl-QTWQ-[Pen1-[Phe(4-0Me)]-[2-Nall-[Aibl-ENN-NH2 ***
619 Ac-[Penl-QTWQ-[Pen1-[Phe[4-(2-aminoethoxy)]-[2-Nall-[hLeul-ENA-
***** *****
NH2
620 Ac-[Penl-TTWQ-[Pen1-[Phe[4-(2-aminoethoxy)]-[2-Nall-[Aibl-
[Lys(Ac)1-NN-NH2
625 Ac-[Penl-QTWQ-[Pen1-[Phe(4-0Me)]-[2-Nan-[Aibl-[Lys(Ac)l-NN-NH2 **
628 Ac-[Penl-QTWQ-[Pen1-[Phe[4-(2-aminoethoxy)]-[2-Nall-[Aibl-
[Lys(Ac)1-N-H3Alal-NH2
280
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
SEQ SG F' S
I F
ID NO: t1/2
t1/2
i e(luenlif
630 Ac- [Pen] -NTWQ-Pen] - [Phe(4-0Me)] - [2-Na1l -[Aibl-ENN-NH2 ***
631 Ac- [Pen] -NTWQ- [Pen] - [Phe [4-(2-aminoethoxy)] - [2-Nall -[hLeul -
ENA-
***** ****
NH2
632 Ac- [Pen] -NTWQ- [Pen] -[Phe [4-(2-am inoethoxy)] - [2-Na1l -[Aibl -
[Ly s (Ac)] -NN-NH2
633 Ac- [Pen] -NTWQ-[Pen] - [Phe(4-0Me)] - [2-Na1l - [Aib] - [Lys(Ac) ] -
**
NH2
634 Ac- [Pen] -NTWQ- [Pen] -[Phe [4-(2-am inoethoxy)] - [2-Na1l -[Aibl -
[Lys(Ac)] -NQ-Nt12
636 Ac- [Pen] -NTWQ- [Pen] -[Phe [4-(2-am inoethoxy)] - [2-Na1l -[Aibl -
[Ly s (Ac)] -NA-NH2
637 Ac- [Pen] -NTWQ-[Pen] - [Phe(4-0Me)] - [2-Na1] - [Aib]- [Ly s(Ac)l-NN-
Nt12
638 Ac- [Pen] -NTWQ-[Pen] - [Phe(4-0Me)] - [2-Na1] - [Aib]- [Ly s(Ac)l-NQ-
Nt12
639 Ac- [Pen] -NTWQ- [Pen] -[Phe [4-(2-am inoethoxy)] - [2-Na1l -[Aibl -
[Lys(Ac) ] -N- [PAW -NH2
640 Ac- [Pen] -NTWQ-[Pen] - [Phe(4-0Me)] -[hLeul - [Lys (Ac)] -N-
***** *****
[13A1al -NH2
641 Ac- [Pen] -NTWQ- [Pen] -[Phe [4-(2-am inoethoxy)] - [2-Nall -[hLeul -
***** *****
[Lys(Ac) ] -N- [PAW -NH2
669 Ac- [Pen] -QTWQ- [Penl- [Phe(4-CONH2)] - [2-Na1l -[cc-MeV al] -
[Lys(Ac) ] - **
NN-NH2
534 [Ac-[Pen] -QTWQ- [Penl- [Phe(4-CONH2)] - [2-Nall- [cc-MeLy sl- [Lys
(Ac)] - **
NN-NH212 DIG
1035 Ac-[(D)Phel - [Pen] -NTWQ- [Pen] - [Phe [4-(2-aminoethoxy)] [Aib]-
[Ly s (Ac)] -NN-NH2
676 Ac-[(D)Phel - [Pen] -NTWQ- [Pen] - [Phe [4-(2-aminoethoxy)] -[Aib]-
**
[Lys(Ac) ] -N- [PAla] -NH2
682 Ac- [Pen] -NTWQ [Pen] - [Phe(4-CONH2)] - [2-Na1l - [AIN- [Lys(Ac) ] -NN-
** ****
NH2
683 Ac- [Pen] -NTWQ [Pen] - [Phe(4-CONH2)] - [2-N al] 44-am ino-4-c arboxy-
**
tetrahydropyranl-Ly s(Ac)] -NN-NH2
281
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
ii SEQ '" iii "
SG F' iii SIF
ii ID NO: .:: iii il t1/2
iii t1/2
iietluen114ii
.:..:
(m in ) 1::: (m in )
..
684 Ac-[Penl-NTWQ[Pen]-[Phe(4-CONH2)]-[2-Nall-[Achc]- [Lys(Ac)[-NN- *
*
NH2
1036 Ac-[Penl-NTWQ[Pen]-[Phe(4-CONH2)]-[2-Nall-[Acycl- [Lys(Ac)]-NN- *
*
NH2
686 Ac-[Penl-NTWQ[Pen]-[Phe(4-CONH2)]-[2-Nall-k-MeLeul-[Lys(Ac)]- *
*
NN-NH2
688 Ac-[Penl-NTWQ[Pen]-[Phe(4-0Me)]-[2-Nall-[4-amino-4-carboxy- *
*
tetrahydropyranl-[Lys(Ac)l-NN-Nt12
689 Ac-[Penl-NTWQ[Pen]-[Phe(4-0Me)]-[2-Nall-[Achc]-[Lys(Ac)i-NN-Nt12 *
**
1037 Ac-[Penl-NTWQ[Pen]-[Phe(4-0Me)]-[2-Nall-[Acycl-[Lys(Ac)l-NN-Nt12 **
*
671 Ac-[Penl-NTWQ[Pen]-[Phe(4-0Me)]-[2-Nall-[a-MeLeul-[Lys(Ac)]-NN- *
*
NH2
535 [Ac4Penl-NTWQ-[Pen]-[Phe(4-CONH2)1-12-Nall-[Aibl-KNN-NH2l2DIG **
**
536 [Ac4Penl-NTWQ4Pen]4Phe(4-CONH2)]42-Nall44-amino-4-carboxy- *
*
tetrahydropyranl-KNN-NH2l2 DIG
537 [Ac4Penl-NTWQ-[Pen]-[Phe(4-CONH2)142-Nall4Achcl-KNN-NH2l2 **
***
DIG
539 [Ac-[Penl-NTWQ-[Pen]-[Phe(4-CONH2)]-[2-Nall-k-MeLeul-KNN-
** **
NH2l2 DIG
the matrix used is 100 fold dilution of standard SIF concentration
*=>360min; **=180-360minn; ***=120-180min; ****=<60-120min; *****=<60min
Table E10. Stability of Illustrative Peptides Containing Thioethers Motif and
Analogues Within
Simulated Intestinal Fluid (SIF) and Simulated Gastric Fluid (SGF)
ir SEQ ID SIP
SG1
...:. ...
ii 4NO;:: iiiii iiequen0iii
iii ii 11/2 iii 11/2
-
(min) (min)
.= .....
...
692 Ac-Cyclo-r[Abu]RTWQC1-YWRKFG-[AEA]-[(D)Lys]-NH2 *****
694 Ac-Cyclo-[[Abu]-QTWQC]-YVVRENG-[AEA]-[(D)Lys]- NH2 *****
699 Ac-Cyclo-[[Abu]-QTWQC]-YW-[hLeu]-ENG- NH2 ***** ND
700 Ac-Cyclo-[Abu]-QTWQ-(D)Cys]]-YW-[hLeu]-ENG-NH2 ****
282
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
ipS'EQifl
SIP"ninSGPri
NO Siqutnce 11/2 11/2
(min) (min)
701 Ac-Cy cl o- [ [Abu]- Q TWQ - [Pen] ] -YW- [hLeu] -ENG-NH2 *****
703 Ac-Cyclo-[[Abu]-QTWQC]-YW- [a-MeLeu]-ENG-NH2 *****
704 Ac-Cyclo- [[Abu]-QTWQC]-Y- [2-Na!]- a-MeLysFENG-NH2 *****
702 Ac-Cyclo- [[Abu]-QTWQC]-[Phe(4-0Me)]- [2-Nal]-[a-
*** *****
MeLys]-ENG-NH2
706 Ac-Cyclo- [[Abu]-QTWQC]-[Phe(4-0Me)]- [2-Nal]-[a-
*** *****
MeOrnFENG-NH2
707 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-W-[a-MeOrn]- **
*****
ENG-NH2
702 Ac-Cyclo- [[Abu]-QTWQC]-[Phe(4-0Me)]- [2-Na!]-[a- **
*****
MeLys]-ENG-NH2
709 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-W-[a-MeLys]-
*****
[Lys(Ac)]-NG-NH2
710 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-W-[a-MeLys]-
*****
ENG-NH2
711 Ac-Cyclo- [[Abu]-QTWQC]-[Phe(4-0Me)]- [1-Nal]-[a-
*****
MeLys]-[Lys(Ac)]-NG-NH2
712 Ac-Cyclo- [[Abu]-QTWQC]-[Phe(4-0Me)]- [2-Nal]-[a-
** *****
MeLy s] - [Ly s(Ac)] -NG-NH2
713 Ac-Cyclo-[[Abu]-QTWQC]-YVV- [a-MeOrn]-[Lys(Ac)]-NG-
**
NH2
714 Ac-Cy cl o- [ [Abu] -Q TWQ C] - [Phe(4-0Me)] - [2-Na!]- [(D)Asn]-
[Lys(Ac)]-NG-NH2
715 Ac-Cyclo- [ [Abu] -QTWQC]- [Phe(4-Phenoxy)]-[2-Nal] - [a-
MeLy s] - [Ly s(Ac)] -NG-NH2
716 Ac-Cyclo- [[Abu]-QTWQC]-[hPhe(3,4-dimethoxy)]- [2-Na!] - [a-
MeLys]-[Lys(Ac)]-NG-NH2
717 Ac-Cyclo- [ [Abu] -QTWQC]-[DMT]-[2-NalHa-MeLys]-
[Lys(Ac)]-NG-NH2
718 Ac-Cyclo- [[Abu]-QTWQC]-[Phe(4-CONH2)]- [2-Nal]-[a-
*** *****
MeLys]-[Lys(A0]NG-NH2
719 Ac-Cyclo- [ [Abu] - QTWQC] -Phe(3,4-C12) [2-Na!]- [a-MeLys]- ***
[Lys(Ac)]NG-NH2
720 Ac-Cyclo- [ [Abu]- QTWQ - [Pen]] - [Phe(4-0Me)] - [2-Nal]-[a- **
***
MeLys]-ENG-NH2
721 Ac-Cyclo- [ [Abu]- QTWQ - [Pen]] - [Phe(4-0Me)] - [2-Nal]-[a- **
***
MeLys]-[Lys(Ac)]NG-NH2
782 Ac-Cyclo- [ [Abu] -Q TWQC] - [Phe[4-(2-aminoethoxy)] -W- [a-
***
MeLys]-ENG-NH2
283
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
NO Siqutnce 11/2 11/2
(min) (min)
790 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-[2-Nal]-[a-
*** *****
MeOrn]-[Lys(A0]-NG-NH2
791 Ac-Cyclo-[[Abu]-QTWQC]-[2-Nal]-[2-Nal]-[a-MeOrn]- *** ND
[Lys(Ac)]-NG-NH2
794 Ac-Cyclo-[[Abu]-QTWQC]-[2-Nal]-[2-Nal]-[a-MeLys]- ** ND
[Lys(Ac)]-NG-NH2
797 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-CONH2)]-[2-Nal]-[a- **** *****
MeLys]-[Lys(A0]-NG-NH2
798 Ac-Cyclo-[[Abu]-QTWQC]-[2-Nall-[2-NalHa-MeLysFENG- .
ND
NH2
810 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-[2-Nall-[Aib]-
[Lys(Ac)]-NG-NH2
815 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-[2-Nall-[Orn]-
[Lys(Ac)]-NG- NH2
820 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-[2-Nall-[Chg]-
***
[Lys(Ac)]-NG-NH2
822 Ac-Cyclo-[[Abu]-QTWQC]-[Octgly]-[2-Nal]-[a-MeLys]- *****
[Lys(Ac)]-NG-NH2
823 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-[Octgly]-[a- ****
MeLys]-[Lys(A01-NG-NH2
823 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-[Octgly]-[a- *****
MeLys]-[Lys(A0]-NG-NH2
829 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- *
[a-MeLys]-ENG-NH2
857 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- .
[a-MeLys]-[Lys(Benzoic acid)]NG-NH2
861 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- *
[a-MeLys]-[Lys(isovaleric acid)]-NG-NH2
876 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- *.
*****
[Aib]-[Lys(A0]-QG-NH2
877 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- *.
**
[Aib]-QNG-NH2
878 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- ***.
***
[Aib]-ENG-NH2
879 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-1-
Nal[Aib]-[Lys(Ac)]-NG-NH2
880 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- ****
*****
[Aib]-[Lys(Ac)]-NA-NH2
891 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- .
**
[a-Me-Orn]-[Lys(A01-NG-NH2
892 Ac-Cyclo-[[Abu]-QTWQC] - [Phe[4-(2-aminoethoxy)]]-[2-Nall- *
284
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
NO Siqutnce 11/2 11/2
(min)
(min).
[a-MeLys]-[Lys(Ac)]-NG-M-12
893 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- ***.
[Orn]-[Lys(Ac)]-NG-NH2
894 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-W-
*****
[Orn]-ENG-NH2
895 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-W-
*****
[Orn]-[Dap]-NG-NH2
896 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-W-
*****
[Orn]-[Dap(Ac)]-NG-NH2
897 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- *****
[Orn]-[Dap]-NG-NH2
898 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- *****
[Orn]-[Dap(Ac)]-NG-NH2
899 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-W-
***** *****
[hLeu]-ENG-NH2
900 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-(acetyl-aminoethoxy)]]-
** *****
[2-Nall-[a-MeLys(Ac)]-[Lys(Ac)]-NG-NH2
901 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-W-[a- **
Me-Leu]-ENG-NH2
902 Succicinyl-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]- **
[2-Nall-[a-MeLys]-[Lys(Ac)]-NG-NH2
906 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-(acetyl-aminoethoxy)]]- ****
*****
[2-Nall-[a-MeLys(Ac)]-ENG-NH2
820 Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-[2-Nall-[Chg]- **
[Lys(Ac)]-NG-NH2
911 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- .
**
[a-MeLys]-ENQ-NH2
912 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- .
**
[a-MeLys]-ENN-NH2
913 Ac-Cyclo-[[Abu]-TTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- .
**
[a-MeLys]-ENG-NH2
914 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- ****
*****
[a-Me-Gly(Ethyl)] Lys(Ac)l-NG-NH2
915 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- ***
****
[a-MeVa1]-[Lys(A0]-NG-NH2
916 Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]- *****
***
[a-MeSer]-[Lys(Ac)]-NG-NH2
925 Ac-(D)Lys-[Cyclo-[[Abu]-QTWQC]]-[Phe(4-0Me)]-[2-Nal]-
**** *****
[a-MeLeu]-ENG-NH2
1039 [Ac-[(D)Lys]-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-[2-Nall- ****
*****
[a-MeLeu]-ENG-NH2]2 DIG: dimerization through (D)Lys
285
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
NO Siqutnce 11/2 11/2
(min) (min)
930 Ac-Cyclo-r[AbuFQTWQC]-[Phe[4-(2-aminoethoxy)]142-Nall- *****
ND
[hLeu]-[Lys(Ac)]-N-[f3Ala]-NH2
933 Ac-Cyclo-[[AbuFQTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall- .
**
[a-MeLys]-NNG-NH2
946 Ac-Cyclo-[[AbuFQTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall- ****
****
[Aib]-[Lys(Ac)]-NG-NH2
955 Ac-Cyclo-[[AbuFQTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall- ***
*****
[a-MeLeu]-[Lys(A01-NN-NH2
1040 [Ac-Cyclo-[[Abu]-QTWQCFY(Bz1)-W4a-MeLysFENG-
** *****
NH212;PEG25 through [a-MeLys]
965 Ac-E-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2- ***
Nall-[a-MeLys]-ENN-NH2
966 Ac-(D)Glu-[Cyclo-[[Abu]-QTWQC]-[Phe[4-(2- ****
aminoethoxY)lF[2-Nall-[a-MeLys]-ENN-NH2
967 Ac-Arg-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2- *****
***
Nall-[a-MeLys]-ENN-NH2
1041 Ac-[(D)Arg-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2- **
aminoethoxy)]]-[2-Nall-[a-MeLys]-ENN-NH2
969 Ac-F-Cyclo-[[Abu]-QTWQC]]-[Phe[4-(2-aminoethoxy)]]-[2-
**** ***
Nall-[a-MeLys]-ENN-NH2
970 Ac-[(D)Phe]-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
** ***
aminoethoxy)]]-[2-Nall-[a-MeLys]-ENN-NH2
972 Ac-T-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2- ***
Nall-[a-MeLys]-ENN-NH2
973 Ac-Leu-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-
***
Nall-[a-MeLys]-ENN-NH2
1042 Ac-RD)Q1n]-Cyclo-[[Abu]-QTWQCHPhe[4-(2- ***
aminoethoxy)]]-[2-Nall-[a-MeLys]-ENN-NH2
975 Ac-Cyclo-[[AbuFQTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall- *****
[Acpc]-ENN-NH2
976 Ac-Cyclo-[[AbuFQTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall- ***** ****
[Acbc]-ENN-NH2
1043 Ac-Cyclo-[[AbuFQTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall- .
[Acpc]- ENN-NH2
978 Ac-Cyclo-[[AbuFQTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall- *.
[Acyc]-ENN-NH2
979 Ac-Cyclo-[[AbuFQTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall- *
[4-amino-4-carboxy-piperidineFENN-NH2
972 Ac-T-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-
Nall-[a-MeLys]-ENN-NH2
286
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
the matrix used is 100 fold dilution of standard SIF concentration
*=>360min; **=180-360minn; ***=120-180min; ****=<60-120min; *****=<60min
[00702] For each peptide tested, the DTT stability assay was conducted by
adding 5p1 of a 10mM
peptide stock solution in DMSO to lml of 100mM Tris-C1, pH 7.5 (final peptide
concentration is
50pM). At time 0 min, Sul of a freshly thawed 100mM DTT solution was added to
the
incubation tube containing the peptide, such that the final DTT concentration
was 0.5mM. The
reactions were incubated at room temperature. At different time points up to
120 minutes (20
min, 40 min, 80 min, 120 min), 500 aliquots were removed, and the reaction was
quenched by
adding 10p1 of 5M acetic acid. To measure disappearance of the parent peptide,
the quenched
samples (300) were analyzed by reverse phase EIPLC and UV absorbance at 220nm.
The
fraction oxidized remaining was graphed versus time, and half-lives were
calculated by fitting to
a first-order exponential decay equation using Excel. The results of these
studies are shown in
Table El 1. The peptides having half-life >120 min are all considered stable.
Table El 1. Stability of Illustrative Peptides in DTT Assay
DTT
Sequence Stability
(min)
Ac-CRTWECYWHEFG-NH2 <10
Ac-CQTWQCYW-[hLeu]-ENG-NH2
Ac-CADWVWCYVVHTFGA-[Azt]-[(D)Lys]-NH2
Ac-Cyclo-[[Abu]-RTWQC]-YVVRKFG-[AEA]-[(D)Lys]-NH2 >120
Ac-Pen] -QTWQ- [Pen] -YW- [hLetil -ENG-Nt12 >120
Ac-{Pen] -QTWQ- [Pen] -YW- [c(-MeLeul -ENG-Nt12 >120
Ac-Cyclo- [ [Abu] -Q TWQ - [Phe(4-0Me)]- [2-Nal]- [a-MeLys]-
>120
ENG-NH2
Ac-{Pen] -QTWQ- [Phe(4 -Ome)142-Nall - [cc-MeLysl-ENG-Nt12 >120
Ac-Cyclo- [ [Abu] -QTWQ C] - [Phe [4-(2-aminoethoxy)] -W- [a-
MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC] - [Phe[4-(2-aminoethoxy)]]-[2-Nal]-[4-
>120
amino-4-carboxy-tetrahydropyran]-ENN-NH2
[Ac-[Pen] -QTWQ- [Phe(4-CONH2)] - -Nall- [c(-MeLy s] - [Lys(Ac) ] -
>120
NN-NH212 DIG
*=10-120 min
EXAMPLE 4
287
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
CROSS-REACTIVITY OF PEPTIDE INHIBITORS
1007031 The amino acids of the extracellular domain of the human IL-23R are
95%, 77% and
70% identical to the cyno IL-23R, rat IL-23R and mouse IL-23R, respectively.
Interestingly, the
mouse receptor contains an insertion of 21 residues that are absent in human,
mouse, chimp, dog
and cow receptor. These additional amino acids are located in a region where
human IL-23R is
thought to bind to IL-23.
[00704] To identify peptide inhibitors that cross-reacted with species other
than human IL-23R,
the ability of certain peptide inhibitors to inhibit human IL-23R, cyno IL-
23R, rat IL-23R and
mouse IL-23R by ELISA assay. In line with the observation regarding the
sequence differences
between human IL-23R and mouse IL-23R, the peptide antagonists tested showed a
lack of or
very weak inhibitory activities in the mouse IL-23R ELISA (see Table E12). In
contrast, the
antagonists tested to date displayed comparable potency towards the rat
receptor and slightly less
activity towards the cyno receptor.
[00705] Various bioassays performed to determine the potency, cross reactivity
and the selectivity
of IL-23R antagonists are described below.
Assays for Selectivity of specific IL-23R Antagonists
Human IL-12Rf31 ELI SA
[00706] An assay plate was coated with 100 ng/well of human IL-12Rf31 huFC and
incubated
overnight at 4 C. The wells were washed, blocked, and washed again. Serial
dilutions of test
peptides and IL-23 at a final concentration of 2.5 nIVI were added to each
well, and incubated for
2 hours at room temperature. After the wells were washed, bound IL-23 was
detected with goat
anti-p40 polyclonal antibodies, followed by an EIRP conjugated donkey anti-
goat IgG. Signals
were visualized with TMB One Component EIRP Membrane Substrate and quenched
with 2 M
sulfuric acid.
Mouse IL-23R Competitive Binding ELISA
[00707] An assay plate was coated with 50 ng/well of Mouse IL-23R huFC and
incubated
overnight at 4 C. The wells were washed, blocked, and washed again. Serial
dilutions of test
288
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
peptides and IL-23 at a final concentration of 4 nM were added to each well,
and incubated for 2
hours at room temperature. After the wells were washed, bound IL-23 was
detected with goat
anti-p40 polyclonal antibodies, followed by an HIRP conjugated donkey anti-
goat IgG. Signals
were visualized with TMB One Component HIRP Membrane Substrate and quenched
with 2 M
sulfuric acid.
Rat IL-23R Competitive Binding ELISA
[00708]An assay plate was coated with 300 ng/well of Rat IL-23R huFC and
incubated
overnight at 4 C. The wells were washed, blocked, and washed again. Serial
dilutions of test
peptides and IL-23 at a final concentration of 7 nM were added to each well,
and incubated for 2
hours at room temperature. After the wells were washed, bound IL-23 was
detected with goat
anti-p40 polyclonal antibodies, followed by an HIRP conjugated donkey anti-
goat IgG. Signals
were visualized with TMB One Component HIRP Membrane Substrate and quenched
with 2 M
sulfuric acid.
Cyno IL-23R Competitive Binding ELISA
1007091 An assay plate was coated with 50 ng/well of Cyno IL-23R huFC and
incubated
overnight at 4 C. The wells were washed, blocked, and washed again. Serial
dilutions of test
peptides and IL-23 at a final concentration of 2 nM were added to each well,
and incubated for 2
hours at room temperature. After the wells were washed, bound IL-23 was
detected with goat
anti-p40 polyclonal antibodies, followed by an HIRP conjugated donkey anti-
goat IgG. Signals
were visualized with TMB One Component HIRP Membrane Substrate and quenched
with 2 M
sulfuric acid.
Table E12. Cross-Reactivity of Illustrative Peptide Inhibitors
Human IL-23R Rodent and Cyno IL-23R
Cross
Activity (nM) Reactivity (nM)
Cmpd.
ELISA ELISA ELISA
Number Cell Assay ELISA cyno
hulL23R mouse IL23R rat IL23R
IL23
pSTAT3 HTRF IL23 IL23 IL23R IL23
22
197 ++ ND ++ ND
169 ++ ++ ++
289
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
198 +++ +++ ND +++ +++
213 +++ +++ ND +++ ND
219 +++ +++ ND +++ ND
230 +++ +++ ND +++ ND
+++ indicates 0-250 nIVI
++ indicates 251-1000 nIVI
+ indicates 1001-10,000 nM
- indicates > 25,000 nIVI
EXAMPLE 5
NK CELL ASSAY
1007101Natural killer (NK) cells, purified from human peripheral blood of
healthy donors by
negative selection (Miltenyi Biotech, Cat # 130-092-657), were cultured in
complete media
(RPMI 1640 containing 10% FBS, L-glutamine and penicillin-streptomycin) in the
presence of
IL-2 (RnD, Cat # 202-IL-010/CF) at 25 ng/mL. After 7 days, cells were
centrifuged, and
resuspended in complete media at 1E6 cells/mL. Recombinant IL-23 at
predetermined EC50 to
EC75 and IL-18 (RnD, Cat # B003-5) at 10 ng/mL were mixed with varying
concentrations of
peptides, and added to NK cells seeded at 1E5 cells per well. After 20 to 24
hours, IFNy in the
supernatant was quantified using Quantikine ELISA (RnD, Cat # DIF50).
Table E13. IC50 of Illustrative Peptide Inhibitors in Primary Cell Line (NK
Cell Assay)
NK cell assay
Sequence
(nM)
Ac-Cyclo-[[Abu]-QTWQC]-Y42-NalHa-MeLysFENG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-[2-Nal]-[a-MeLys]-
ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC] - [Phe[4-(2-aminoethoxy)]-W4a-
MeLys]-ENG-NH2
Ac-Cyclo-[[Abu]-QTWQC] - [Phe[4-(2-aminoethoxy)]]-[2-Nal]-
[a-MeLys]-[Lys(isovaleric acid)]-NG-NE12
Ac-Cyclo-[[Abu]-QTWQC] - [Phe[4-(2-aminoethoxy)]]-[2-Nal]-
[Ai13]-QNG-NE12
Ac-Cyclo-[[Abu]-QTWQC] - [Phe[4-(2-aminoethoxy)]]-[2-Nal]-
[Aib]-[Lys(Ac)]-NA-NH2
290
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Ac-Cyclo- [[Abu]-QTWQC]-[Phe[4-(2-(acetyl-aminoethoxy)]]-
[2-Nall-[a-MeLys(Ac)]-[Lys(Ac)1-NG-NH2
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[Phe(3,4-
OMe21-[a-MeLys]-[Lys(Ac)1-NG-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall-
[a-MeLys]-ENQ-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall-
[a-MeLys]-ENN-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall-
[a-MeVal]-[Lys(Ac)1-NG-NH2
[Ac-Cyclo-[[Abu]-QTWQC]-[Phe(4-0Me)]-[2-Nall-[a-MeLys]-
ENG-NH2]2; DIG through a-MeLys
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall-
[a-MeLeu)-[Cit]-NN-NH2
Ac-[(D)Phe]-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
aminoethoxy)]]- [a-MeLys]-ENN-NH2
Ac-T-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-
Nall-[a-MeLysFENN-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall-
[Acbc]-ENN-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall-
[Acpc]- ENN-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall-
[AchcFENN-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall-
[4-amino-4-carboxy-tetrahydropyran]-ENN-NH2
Ac-Cyclo-[[Abu]-QTWQCHPhe[4-(2-aminoethoxy)]]-[2-Nall-
[a-MeLeu]-QN-[f3Ala]-NH2
Ac-(D)Phe-Cyclo-[ [Abu]-QTWQC]- [Phe[4-(2-aminoethoxy)]]-
[2-Nall-[4-amino-4-carboxy-tetrahydropyran]-ENN-NH2
Ac-[(D)Arg]-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-
aminoethoxy)]]-[2-Nall- [4-amino-4-carboxy-tetrahydropyran]-
ENN-NH2
*=<25nIVI
Table 14. IC50 of Illustrative Peptides Containing the Ac-[Pen]-XXWX-[Pen]-
)000( Motif and
analogues (NK cell assay)
291
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
NK Cell
Sequence assay
(nM)
Ac-[Pen] -QTWQ- [Pen] -[Phe [4-(2-am ino ethoxy)] - [2-Nall - [cc-MeLeul -
[Lys(Ac) ] -NN-NH2
Ac-[Pen] -NTWQ- [Pen] -[Phe [4-(2-amino ethoxy)] - [2-Nall - [Aib]- [Lys(Ac)]-
NN-NH2
Ac-[Pen] -NTWQ- [Pen] -[Phe [4-(2-amino ethoxy)] - [2-Nall - [Aib]- [Lys(Ac)]-
N- [13Alal -NH2
Ac-[Pen] -QTWQ- [Pen] -[Phe(4-0Me)] - [cc-MeLy s] - [Lys
(Ac)] -NN-
NH2
Ac- [Pen] -QTWQ- [Pen] -[Phe(4-CONH2)] - [2-Nall -[cc-MeLysl- [Lys(Ac)]-
NN-NH2
Ac- [Pen] -QTWQ- [Pen] -[Phe(4-CONH2)] - [2-Nall -[cc-MeV al] -[Lys(Ac)] -
**
NN-NH2
[Ac- [Pen] -QTWQ [Pen] - [Phe [4-(2-acetylamino ethoxy)] - [2-Nall - [cc-
MeVal] -KNN-NH212 DIG
[Ac- [Pen] -QTWQ [Pen] - [Phe[4-(2-acetylaminoethoxy)l- 112-Nall -K-
[Lys(Ac)] -NN-NH212 DIG
[Ac- [Pen] -QTWQ- [Pen] - [Phe(4-0Me)] - [2-Nall- [cc-MeLy s] - [Lys(Ac)]-NN-
NH212 DIG
11Ac-11Pen1-01W0-11Pen1-11Phe(4-CONH2)1-I2-Nall-kx-MeLysl-[Lys(Ac)l-
NN-NH212 DIG
Ac- [(D)Phel -[Pen] -NTWQ [Pen] - [Phe(4-0Me)] - 112-Nall - [4-amino-4-
carboxy-tetrahydropyran] - [Cit] -NN-Nt12
Ac- [(D)Phel -[Pen] -NTWQ [Pen] - [Phe(4-0Me)] - - 11Achcl-ENN-Nt12
Ac-[Pen] -NTWQ [Pen] - [Phe(CONH2)] - 112-Nail -[Aibl - 11Lys(Ac)l-NN-NH2
[Ac- [Pen] -NTWQ- [Pei+ 11Phe(4-CONH2)1- 11Aibl-KNN-NH212 DIG
*=<10nIVI; **=10-25nM
EXAMPLE 6
BIOASSAY CHARACTERIZATION OF PEPTIDE INHIBITORS
[00711] The potency, cross reactivity, and selectivity of certain peptide
inhibitors was determined
using various bioassays developed for this purpose and described below.
Rat Splenocyte Assay
292
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
[00712] A new assay developed was the rat splenocyte assay. This assay
examined the levels of
IL-17A in activated rat splenocytes following stimulation with IL-23 in the
presence of test
compound.
[00713] Briefly, splenocytes freshly isolated from rat were seeded in 96-well
tissue culture plates
in complete medium containing IL-1 ft Serial dilutions of test compounds were
distributed to
each well along with rat IL-23 at a final concentration that is between EC50
to EC80 values;
plates then were incubated for 3 days at 37 C in a 5% CO2 humidified
incubator. Changes in IL-
17A levels in the supernatants were detected using an ELISA.
Rat Colitis Model: 9 Days of 3% DSS-Containing Drinking Water
[00714] There is a body of evidence in the literature supporting the
pathogenic role of IL-23/IL-
23R signaling in animal models of colitis. For the IL-23 ligand, this
requirement has been shown
in multiple models, including an IL-104- spontaneous colitis model, a
Helicobacter hepaticus-
driven colitis model, the anti-CD40 innate colitis model, and the chronic
CD45RBh1gh CD4+ T-
cell transfer model. For the IL-23 receptor, the requirement for colitis
development has been
shown in the acute models of colitis induced by DSS or by anti-CD40, as well
as the chronic
CD45RBhigh CD4+ T-cell transfer model. Since certain peptide inhibitors of the
present invention
do not cross react with the IL-23 receptor from mouse but do recognize that
from the rat, a rat
model of IBD relevant to the IL-23 pathway was developed.
1007151In this model, colitis was induced in SD rats by 9 days of ad lib
exposure to drinking
water containing 3% DSS. The disease activity index (DAI) score and ratio of
colon
weight: colon length were compared between three study groups (n=6
rats/group): vehicle, 3%
DSS, and 3% DSS with positive control (sulfasalazine administered at 100 mg/kg
PO, QD). The
DAI score consisted of ratings from three parameters, including percent body
weight loss, stool
consistency, and a quantitative hemoccult score, and could achieve a maximum
value of 3 units.
DSS-exposed animals displayed significantly elevated DAI score (compared to
vehicle control)
from Day 4 onward, with DAI values peaking at approximately 2.5 by the end of
the study (Day
9). Treatment of the DSS-exposed rats with the positive control
(sulfasalazine) attenuated the
disease score (compared to DSS alone) from Day 5. The differences observed in
the terminal
293
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
ratio of colon weight:colon length also were significant for DSS-induced
disease animals with
and without sulfasalazine treatment.
Ex Vivo Activity and Stability
[00716] Two peptides (Compound A and Compound B) were selected for use in
further biological
studies (shown below). One contained a thioether linkage and the other
contained a Pen-Pen
disulfide bond. The activity, selectivity and ex vivo stability profiles of
the two compounds are
provided herein.
100717] Assays for selectivity of peptide inhibitors included a human IL-12Rb
1 ELISA and
measurement of the production of IL-12 in PHA activated human PBMC, which are
described
briefly below.
Human IL-12Rf31 ELIS A
[00718] An assay plate was coated with 100 ng/well of human IL-12Rbl huFC and
incubated
overnight at 4 C. The wells were washed, blocked, and washed again. Serial
dilutions of test
peptides and IL-23 at a final concentration of 2.5 nIVI were added to each
well, and incubated for
2 hours at room temperature. After the wells were washed, bound IL-23 was
detected with goat
anti-p40 polyclonal antibodies, followed by an EIRP conjugated donkey anti-
goat IgG. Signals
were visualized with TMB One Component EIRP Membrane Substrate and quenched
with 2 M
sulfuric acid. Data from these assays is provided herein.
Production of IFNy by IL-12 in PHA Activated Human PBMC
[00719] This assay examined the ability of IL-23R antagonists to neutralize
production of IFNy
proteins in IL-12-stimulated human PBMCs. IL-23R peptide inhibitors specific
to the IL-23/IL-
23R pathway are not expected to alter the levels of IFNy produced. Compound A
and
Compound B were tested in this assay, and a graph showing that they do not
alter the levels of
IFNy produced at most concentrations tested is provided in Figure 2.
294
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
is 0
0 ______________________
0
ANQ T W Q-N))1 N __ N __ N EN G-NH2
0 0 0 0
104.
Compound A
0
NQ
0 _________
0
T W N N ____ N ____ EN G-NH2
0 0 0 0
104.
Compound B
In Vivo Activity
[00720] Acute colitis was induced by feeding female Sprague Dawley rats with
3% (wt/vol) DSS
dissolved in drinking water. For nine days starting at the same day as DSS,
Compounds A or B
was administered orally three times per day at 20 mg/kg or 30 mg/kg. Compounds
A was also
administered intraperitoneally three times per day at 30 mg/kg. A neutralizing
anti-IL-23p19
antibody was used as a comparator, and was administered intraperitoneally at 4
mg/kg on the
same day and fifth day after starting DSS. To quantify colitis with clinical
activity, disease
activity index (DAI) was determined daily for each animal as an average of
three parameters:
body weight change (scale 0-3), stool consistency (scale 0-3) and hemoccult
blood (scale 0-3), as
shown in Table E15. At necropsy, the entire colon was removed from the cecum
to the rectum.
The colon was measured for length, flushed with PBS to remove feces, weighed,
and opened
295
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
longitudinally to determine macroscopic score. The visible damage of the colon
was scored on a
scale from 0-3, as shown in Table E16.
100721] Table E17 shows that at Day 7, treatment with Compound A and B
significantly
improved DAI scores compared to vehicle treated group. FIG. 1 shows results
for DAI values
from Day 7. In addition, a significant reduction was also observed in the
colon weight to colon
length ratios, and colon macroscopic scores (FIG. 3). The reduction in
inflammation observed
with orally delivered peptides was similar to the effects observed from
neutralizing anti-IL23p19
monoclonal antibody. Statistical analysis for significance was compared to the
vehicle treated
group and was determined using Student's T-test (GraphPad Prism). Differences
were noted as
signficant *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Table El 5. Scoring of the Disease Activity Index
Score Percent Body Weight Change Stool Consistency Hemoccult Score
0 None Normal Normal
1 1 to 7 Semi solid Guaiac+
2 8 to 15 Loose Bleeding+
3 > 15 Diarrhea Bleeding++
Table E16. Scoring of Gross Morphologic Damage of the Colon
Score Gross morphology
0 Normal
1 Erythemia
2 Erythemia, slight edema, small erosions
3 Two are more
bleeding ulcers, inflammation, moderate adhesions
4 Severe ulceration, stenosis with dilations, severe adhesions
Table E17. Disease activity index scores and the individual parameters scores
at Day 7, colon
weight to length ratios and colon macroscopic scores at Day 9.
Day 7 Day 9 Necropsy
296
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Colon
Colon
Percent Body Weight Stool Hemoccult Weight/Length
Macrosopic
Change Consistency Score DAI (g/cm)
Score
Group Mean SD Mean SD Mean SD Mean SD Mean SD
Mean SD
No DSS 11.00 2.08**** 0 0**** 0 0**** 0 0**** 75.51 7.03***
ND ND
3% DSS, 2.00 0.58 1.50 0.50 1.72 0.45
124.36 17.11 1.00 1.00
Vehicle -6.39 1.11
Anti-
1.00 0.58* 0.50 0.5* 0.67 0.43** 99.96 16.19* 0.00 0**
IL23p19
mAb -0.05 1.92****
Compound 1.17 0.90 0.50 0.5* 0.56 0.46** 98.38 6.91*
0.00 0**
A, PO 3.18 2.09****
Compound 0.83 0.69* 0.67 0.47* 0.61 0.3*** 97.36 9.32*
0.00 0**
B, PO 0.13 1.24****
Compound
1.17 0.69 0.83 0.69 0.83 0.54* 104.32 12.45 0.33 0.47
A, IP -0.50 1.88***
EXAMPLE 7
IN VITRO ASSAYS AND SURFACE PLASMON RESONANCE (SPR) ANALYSIS
1007221 In vitro assays and SPR were performed to further characterize an
illustrative compound,
Compound C:
40/ N1-12
0
Q T W Q-N N NJ
0
____________________________________________________ N ____ EN G-NH2
0 0 0 0
110 N
Ac-Cyclo-[[Abti]-QTWQC]-[Phe[4-(2-aminoethoxy)]-W-[a-MeLys]-ENG-NI-12
Compound C.
297
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
[00723] Assays described in previous examples were performed to demonstrate
that Compound C
is a potent, selective and competitive inhibitor of IL-23R, showing potent
inhibition of IL-23-
dependent upregulation of phosphor-STAT3 (pSTAT3) in human DB cells and IFNy
production
in human peripheral blood natural killer (PB NK) cells. In adition, Compound C
was selective,
showing little inhibition in a cell free ELISA for human IL6R, or in IL-12-
dependent production
of IFNg in PBMC. Data is shown below in Table El 8A. Compound C also cross-
reacted with
cynomolgus IL-23R (IC50 7 nM) and rat IL-23R (IC50 17 nM), and inhibited IL-23-
dependent
IL-17A production in rat splenocytes (IC50 130 nM) (data not shown).
Table El 8A. In vitro Characterization of Compound C
IC50 KB
pSTAT3
IFNy/PB NK IFNy/IL-12 IL-
/DB IL-6/IL-6R IL-23R
Primary Cell PBMC Cell
12Rf31
Cell ELISA Surface
Assay Assay Surface
Assay
Compound
4 nM 27nM >100 uM >100 uM 2.4nM None
[00724] Compound C exposure was also restricted to the GI following oral
administration to rats
does PO at 20 mg/kg, with AUC values of 355 ug.h/g for small intestine mucosa;
77 ug.h/g for
colon mucosa; and 0.3 ug.h/mL for plasma, with a 40% recovery in feces.
[00725] Compound C was also stable in a variety of GI fluids and reducing
environment, having a
SIF half-life of > 24 h; a SGF half-life of > 24 h; a human intestinal fluid
half-life of > 24 h, and
a half-life of > 2 h in a DTT assay.
[00726] SPR experiments were carried out using a Biacore 2000 instrument and
T100 optical
biosensors equipped with Biacore CM4 and Xantec HC1500m sensor chips.
Recombinant
human IL-23R huFC (RnD), or recombinant human IL-12Rf31 huFC (RnD) or a
mixture of the
two receptor subunits were captured on an anti-human IgG surface. Recombinant
human IL-23
298
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
(Humanzyme) or Compound C was used as the analyte. SPR sensorgrams were fitted
to a one
to one interaction model, giving rise to a rough estimate of the association
rate constant (km),
dissociation rate constant (koff) and dissociation constant (KD) of the
complexes, as shown in
Table El 1. The data show that Compound C does not bind to IL-12Rf31, and
binds to IL-23R
and the mixed surface of IL-12Rf31 and IL-23R with similar potency, at 2.42nM
and 2.56nM,
respectively. This affinity for IL-23R is comparable to that from IL-23. In
contrast, the affinity
of IL-23 to the mixed surface is approximately 14x faster than that from
Compound C.
Table E18B. Binding characteristics of IL-23 and Compound C for IL-12Rf31, IL-
23R or mixed
IL-12Rf31 and IL-23R as determined by SPR.
IL-23 Compound C
Surface ka(M1 sec-1) kd (sec-i) KD (um) ka(M1 sec1)
kd (sec-i) KD (nM)
IL-12Rbl huFC 5.01E+05 4.38E-04 0.87 does not bind up to
16.7 uM
IL-23R huFC 7.82E+05 0.00132 1.69 1.37E+07 0.033
2.42
IL-12Rbl huFC/IL-
23R huFC 6.31E+05 1.15E-04 0.18 1.59E+07 0.041
2.56
EXAMPLE 8
EFFICACY OF IL-23R ANTAGONISTS IN TNBS INDUCED COLITIS IN RAT
1007271To further evaluate the efficacy of IL-23R antagonists in an animal
model of disease,
acute colitis was induced by providing 7-week-old female Sprague-Dawley rats
with 60 mg/kg
2,4, 6-Trin itro benzenes Mfonic acid (TNBS) in 45%-50% ethanol (TNBS/ethanol)
administered
intrarectally at Day 0. Compound C (described in Example 7) was administered
orally three
times a day at 20 mg/kg or 6.7 mg/kg and was provided in drinking water at 0.6
mg/mL or 0.2
mg/mL, respectively, for 8 days starting approximately 24 hours (Day -1) prior
TNBS
inoculation. A neutralizing anti-IL-23p19 antibody was used as a comparator,
and was
administered intraperioneally at 4 mg/kg on Day -1 and again on Day 3. All
animals received
orally PBS (pH 7.4) vehicle which was used to formulate Compound C. The study
design in
shown in Figure 5.
1007281 To assess the extent of the inflammatory response, animals were
observed daily for
clinical signs which included percent body weight loss and signs of loose
stools or diarrhea. Six
days after inoculation of TNBS, rats were sacrificed and the entire colon
length and colon weight
299
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
from cecum to rectum from each animal were recorded. The severity of colitis
was evaluated by
a pathologist blinded to the identity of treatments. In addition to the colon
wall thickness, the
gross colon damage was scored on a 0-4 scale according to Table E19 below, and
histopathological scores were determined based on below parameters (Tables E20
and E21).
Table El 9. Definitions for colon macroscopic scores
Score Colon Gross Morphology
0 Normal
1 Erythema
2 Erythema, slight edema, small erosions
3 Two or more bleeding ulcers, inflammation, moderate adhesions
4 Severe ulceration, stenosis with dilations, severe adhesions
Table E20. Definitions for histopathology
Parameter Definition
Extent and severity of inflammatory cells infiltration, localized and/or
diffuse
involving full thickness of the colon section (transmural). Inflammatory cells
include polymorpho-nuclear leukocytes (neutrophils), mononuclear cells
Inflammation (macrophages + lymphopcytes), fibroplasia and neovascularization.
Necrosis in the mucosa with loss of surface epithelium, hemorrhage and
cellular
Mucosal debris; measured as the length of the lesion on the total length
of the colon
Necrosis section to determine % area affected
Gland Loss % crypt epithelial degeneration with or without superficial
mucosal erosion
Colon the average thickness of the colon measured transmurally (full
thickness) from
Thickness the mucosal surface to the serosa
Table E21. Scoring criteria
Score Inflammation
0 Normal tissue, no inflammation
Very minimal localized infiltrates in the superficial mucosa affecting <2% of
the
0.5 colon section
Minimal degree of multifocal infiltrates in the mucosa affecting approximately
2
1 - 10% of the colon section
Mild degree of multifocal infiltrates in the mucosa, submucosa, outer muscle
2 band, and serosa affecting approximately 11 - 25% of the colon
section
Moderate degree of multifocal infiltrates in the mucosa submucosa, outer
3 muscle band and serosa affecting approximately 26 - 50% of the
colon section
Marked degree of multifocal to diffuse infiltrates in the mucosa submucosa,
4 outer muscle band and serosa affecting approximately 51- 75% of
the colon
300
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
section
Sever degree of multifocal to diffuse infiltrates in the mucosa submucosa,
outer
muscle band and serosa affecting approximately >75% of the colon section
Score Mucosal Necrosis
0 No Necrosis
0.5 Very minimal and localized region affecting <2% of the total colon
section
1 Minimal focal to multifocal regions affecting 2 - 10% of the total
colon section
2 Mild focal to multifocal regions affecting 11 - 25% of the total
colon section
Moderate focal to multifocal regions affecting 26 - 50% of the total colon
3 section
4 Marked focal to multifocal regions affecting 51 - 75% of the total
colon section
5 Severe focal to multifocal regions affecting >75% of the total
colon section
Score Gland Loss
0 No loss, normal crypt epithelium and mucosa
0.5 Very minimal loss not exceeding 1 - 2 regions of mucosa/gland
affected
1 Minima1,1-10% regions of mucosa/gland affected
2 Mild, 11-25% regions of mucosa/gland affected
3 Moderate, 26-50% regions of mucosa/gland affected
4 Marked, 51-75% regions of mucosa/gland affected
5 Severe, >75% regions of mucosa/gland affected
Score Colon Thickness
0 Normal = <350 microns or less
0.5 Very Minimal = 351-400 microns
1 Minimal = 400- 500 microns
2 Mild = 501 - 600 microns
3 Moderate = 601 - 700 microns
4 Marked = 701 - 800 microns
5 Severe = >801 microns
1007291 Compared to the sham group, rats challenged with TNBS suffered acute
weight loss,
displayed increased incidence of loose stools, and increased colon weight to
length ratio. These
data were confirmed by the macroscopic examination of colon which revealed
mild colonic
injury characterized by erythema, edema and small erosions. Treatment with
Compound C
attenuated these changes as compared to the TNBS colitis group. At the high
dose, Compound C
was significantly effective in reducing the colon weight to length ratio,
diminishing the thickness
of the colon walls, and more importantly, improving the colon gross pathology
scores to normal
301
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
in 70% of the animals. Statistical significances were observed at the low dose
in all above
indications except colon wall thickness although a trend was evident. The
reduction in
inflammation observed with orally delivered Compound C was similar to the
effect observed
from the neutralizing anti-IL-23p19 monoclonal antibody (Figure 6).
[00730] Histological examination of H&E stained distal colons show that the
majority of the
lesions observed from the vehicle group are transmural, characterized by
necrosis with
inflammatory cells transversing the entire thickness of the colon, presence of
necrotic tissue
debris on the lumen surface, and mucosa devoid of crypts. The animals treated
with Compound
C generally showed localized lesions limited in the mucosa and submucosa
regions, with colon
tissues showed potential signs of healing at sites of necrosis. Specifically,
the animals treated
with 160 mg/kg/d Compound C showed a significant reduction in inflammation,
mucosal
necrosis and colon wall thickness leading to a significant reduction in the
overall histological
score, comparable to that from the anti-IL-23p19 antibody control (FIG. 7).
[00731] Concentration analysis of samples collected 1 hour post the last PO
dose show that the
plasma concentrations of Compound C detected from all animals are <=2X below
the IC75 of
the compounds as determined in the rat splenocyte/IL-17A cell based assay or
the rat IL-23R
ELISA, suggesting that the efficacies observed from oral treatment are most
likely due to its
local activity at the colon (see FIG. 8). Collectively, these data highlights
the protective effect of
an IL-23R antagonist in the development of TNBS colitis.
[00732] These studies demonstrate that peptides of the present invention are
potent, selective and
orally efficacious IL-23R peptide antagonists that are promising therapeutics
for the treatment of
IBD and other disorders. As shown herein, the present invention provides
petpides that are:
potent blockers of IL-23/IL-23R signaling in a human cell line and in human
primary cells;
selective for IL-23R, and do not inhibit binding to IL-6R or signaling through
IL-12R; cross-
reactive towards rat and cynomolgus but not mouse homologs, enabling in vivo
studies in these
species; resistant to proteolytic and reducing environments of the GI,
resulting in high drug
levels in the intestinal tissues and limited drug concentrations in the
circulation, offering
potential safety advantages over systemically delivered therapeutics; and
effective and
302
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
comparable to an anti-IL23p19 monoclonal antibody in attenuating colitis in a
TNBS-induced rat
colitis model, most like through GI-restricted activities.
EXAMPLE 9
IN VITRO CHARACTERIZATION OF AN ILLUSTRATIVE IL-23R ANTAGONISTS
[00733] To evaluate the properties of efficacy the IL-23R peptide antagonist
of SEQ ID NOS:
980 (Peptide 980), 993 (Peptide 993), and 1185 (Peptide 1185) experiments to
determine
potency, selectivity, and stability of the peptides were performed as
described above. The IC50
values of the peptides as measured by quantitative ELISA for IL-23/IL-23R
competitive binding
assays (performed as described above in Example 2) for human (Hu), cynomolgus
monkey
(Cyno), and rat (Rat) IL-23 and IL-23R binding are shown in Table E22. The
potencies of the
peptides were also evaluated by IL-23R activity assays as described above. The
IC50 values of
the peptides as determined by the reduction of phospho-STAT3 (pSTAT3) levels
in human DB
cells exposed to IL-23 (Hu DB Cell (pSTAT3); performed as described above in
Example 2); by
the reduction of IFNy produced by human NK cells exposed to IL-23 (Hu NK cell;
performed as
described above n Example 5); and by the reduction of IL-17A produced by
activated
splenocytes exposed to IL-23 (Rat(spleen); performed as described above in
Example 6).
Selectivity was evaluated by measuring the IC50 of the peptides for inhibiting
human IL-23/IL-
12R betal interactions (see Example 4) or human IL-6/IL-6R interactions (see
Example 6). The
stabilities of the peptides were determined by measuring the half-life of the
peptides exposed to
simulated intestinal fluid (SIF), simulated gastric fluid (SGF), or human
intestinal fluid (SIF).
Table E22. Properties of illustrative IL-23R antagonists Peptide
303
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=.:1::=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:
=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:11.:::=:=:=:=:=:=:=:=:=:=:=:=:=:=:=
:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:
=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=*=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=
:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=p:=:=:=:=:=:=
:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:=:
=. EL1SA all ass* Seleen% it). Stabilil
.
.
.
.
.
.
.
.
.
.
.
.
.
(n.1\1) 011\4) (EL ISA, 111\1) 0 1 2. hr)
=
=.
.
.................................... .............. ............
.......... ........
Hu DB Cell Hu NK Rat IL-23/ IL-6/
iL
Peptide Hu C' to Rat , SIF SGF HIP (pSTAT.1) cell
(Spleen) IL-12R11 IL-6R
..
' ' I === ===
I I
Peptide
2.0 2.0 2.0 0.6 2.2 18.3 >100,000 >100,000 12 6 24
993
Peptide 1
1.3 3 0.6 2.8 11.5 >100,000 >100,000 11 10
980
Peptide 2
3 3 2 3.9 35 >100,000 >100,000 33 12 24
1185
The structure of Peptide 993 is shown below:
kit*
et
e.'
Ci=--9.
Ni=4
.,:ei ,...: ,
e, N. =-1
N.. e 0
L
0 I ''' q 1 I . 0 1 0 0
_,.11õ , N ii, A. I, it.õ xii JJ. ,:. A, 3 ,Il 1,,
, .,," 4õ. - lc ,,,....,,, .y, N. , .<.= .
14... .. N.,...,,,,,, 4NN:z.
1: N 4 14 i I
' 41
..-,:f: . .e
I 1%,
I. N
0.*-= N'r'''' y t¨
is. .i.:
re-- .
: .
...,..
, .
ligit 0 hir
HztisrAs'W
Ac-[(D)-Arg]-cyclo[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-
tetrahydropyran]-ENN-N1-12 (SEQ ID NO: 993)
The structure of Peptide 1185 is shown below:
304
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
f r
,,lot,
41. ,,,,.:9-,
tõ,,--0 mt.,
.,..,1
.=., õA'
i'-'7 $7.ff=
tt.tti Akb
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]- [2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-
[Lys(Ac)]-NN-NH2 (SEQ ID NO: 1185)
The structure of Peptide 980 is shown below:
r
r
..),,..õ
I.....'.. NH ,.,..,t,N.y,40
14.1
. ,
.1
D
" - 1 I
tit i4 õ...,
9 .0 0
gi i ( Iii
. ,....1,,, a=C .N. , N.,,Arte .--N-,,,õ"4,1r4- x-R---,-õ,,,k, ,--
e-N--,..A"
...¨ci,.
,,.
,sp
.-s=
$
1
111:1*, N ).. fik#1'
0
NN'ee' . hill I lia- 0
L?)
i
Ac-Cyclo-[[Abu]-QTWQC]-[Phe[4-(2-aminoethoxy)]]-[2-Nal]-[4-amino-4-carboxy-
tetrahydropyran]-ENN-NH2 (SEQ ID NO: 980)
[00734] The results summarized in Table E22 indicate that the peptides
potently and selectively
inhibit IL-23/IL-23R binding as compared to IL-23/IL-12R betal or IL-6/IL-6R
interactions.
This inhibition was confirmed with cell culture assays that measure the IL-23R-
dependent such
305
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
as STAT3 phosphorylation, IFNy production, and IL-17A production. The peptides
were also
determined to have a high degree of stability when exposed to simulated
intestinal fluid,
simulated gastric fluid, or human intestinal fluid.
1007351 Data from the Human DB Cell (pSTAT3) experiment were used for S child
analysis (see
FIG. 17). For Schild analysis, concentrations of 0.3nM, 1nM, 3nM, 1 OnM, 30nM,
100nM of
Peptide 993 were tested. The Schild slope was determined to be 1.068,
indicating that Peptide
993 behaves as a simple competitive antagonist. S child analysis was also
performed on a peptide
with the structure of SEQ ID NO: 1169, which is highly similar to the
stuctures of Peptide 1185.
The Schild slope was determined to be 0.91, indicating that peptides with
similar structure,
including Petide 1185, are likely behave as a simple competitive antagonist.
Schild analysis was
also performed on the peptide of SEQ ID NO: 1211, which has a structure
similar to to Peptide
980. The Schild slope was determined to be 0.76. However, when the slop was
fixed to 1, the
R2 value was 0.975. These data suggest that peptides with similar structure,
including Petide
1185, could behave as a simple competitive antagonist.
EXAMPLE 10
IN VIVO PHARMACOKINETICS OF ILLUSTRATIVE IL-23R ANTAGONISTS
[00736] Pharmacokinetic properties of example peptides were measured in vivo.
Sprague Dawley
rats were administered Peptide 993 at 10 mg/kg P.O.
1007371A single oral dose of Peptide 933 was administered to normal female
Sprague-Dawley
rats (N=3 rats per time point) either with or without a drinking water dose
that was provided ad
libitum (see FIG. 11). Following the oral dose, the exposure of Peptide 993
was determined in
the plasma at 0.25, 0.5, 1, 3, 6, 8, and 24 hours post-dose. The levels of
Peptide 993 were also
determined in small intestine, colon, small intestine mucosa, colon mucosa,
small intestine
mucus, Peyer's patch and Mesenteric Lymph Node (MLN) at 1, 3, 6, 8 and 24
hours post-dose.
Urine and feces were collected at 6 and 24 hours to determine the excretion of
Peptide 993.
Plasma, feces, and tissue samples were stored at ¨80 10 C prior to
analysis. For plasma, 50
[IL of the sample was used to extract compound by means of protein
precipitation using a quench
solution (MeOH:ACN w/ 0.1% formic acid, 50:50 volume) with internal standard.
For feces,
samples were homogenized with 0.1% Formic acid in water (water:feces ratio of
4:1) prior to
306
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
extraction. 50 [IL of fecal homogenate was used to extract compound by means
of protein
precipitation using a quench solution (MeOH:ACN w/ 0.1% formic acid, 50:50
volume) with
internal standard. For tissues such as colon or small intestine, samples were
homogenized with
0.1% Formic acid in water (water:tissue ratio of 3:1) prior to extraction. For
tissue such as
Peyer's Patch and Mesenteric Lymph Nodes, samples were homogenized with 0.1%
Formic acid
in water (water:tissue ratio of 20:1) prior to extraction. 50 [IL of tissue
homogenate was used to
extract compound by means of protein precipitation using a quench solution
(MeOH:ACN w/
0.1% formic acid, 50:50 volume) with internal standard. The precipitated
protein was removed
by filter plate and the collected supernatant was dried and reconstituted.
Processed samples were
analyzed on an AB/MDS Sciex API 4000 mass spectrometer. Positive ions were
monitored in
the multiple reaction-monitoring (MRM) mode. Quantitation was by peak area
ratio.
[00738] No detectable levels of Peptide 993 were observed in rat plasma
between 0 and 24 hours
following administration (see FIG. 11A). In contrast, detectable levels of
Peptide 993 were
present in the Peyer's Patch and small intestine for at least six hours (see
FIGS. 11B and 11C),
and for at least 8 hours in the colon post-dose administration (see FIG 11D).
Levels greater than
5% of the total administered dose of Peptide 993 were detected in the rat
feces at 24 hours
following administration, further indicating that Peptide 993 has a high
degree of oral stability.
Taken together, these results demonstrate that Peptide 993 is an orally
stabile GI restricted
peptide, that demonstrates a high GI content and limited systemic distribution
following oral
administration.
[00739] Sprague Dawley rats were administered Peptide 1185 at 10 mg/kg P.O. A
single oral dose
of Peptide 1185 was administered to normal female Sprague-Dawley rats (N=3
rats per time
point). Following the oral dose, the exposure of Peptide 1185 was determined
in the plasma in
samples taken up to 8 hours post-dose. Urine and feces were collected to hours
to determine the
excretion of Peptide 1185 (FIG 18).
[00740] Sprague Dawley rats were administered Peptide 980 at 10 mg/kg P.O. A
single oral dose
of Peptide 980 was administered to normal female Sprague-Dawley rats (N=3 rats
per time
point). Following the oral dose, the exposure of Peptide 980 was determined in
the plasma in
307
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
samples taken up to 8 hours post-dose. Urine and feces were collected to hours
to determine the
excretion of Peptide 980 (FIG 19).
EXAMPLE 11
SAFTEY PROFILE OF ILLUSTRATIVE IL-23R ANTAGONISTS
1007411 Saftey profiles of illustrative peptide inhibitors were characterized.
Peptide 993 and
Peptide 1185 were evaluated in a safety panel examining binding to a panel of
44 targets. The
targets included G protein coupled receptors (GPCRs), transporters, e.g.,
dopamine transporter
(DAT), and ion channels. For all targets, these peptides displayed no
activity, as defined by a
change (inhibitory or stimulatory) in the target's activity of greater than
25%. The targets tested
in the safety panel are listed in Table E24. For each target, Peptide 993 and
Peptide 1185 was
determined to be inactive at concentrations of up to 10 M. The safety profile
of Peptide 980
was evaluated by testing compounds selected from Table E24. For all compounds
tested,
Peptide 980 showed only moderate activity in the acetylcholinesterase assay
(33%), and
otherwise did not display any activity, as defined by by a change in the
target's activity of greater
than 25%.
Table E24. Peptide 993 and Peptide 1185 are Inactive in a Safety Panel
Panel of 44 targets
A2A M3 5-HT3
2+
alphaiA delta2 (DOP) Ca channel (L, dihydropyridine site)
alpha2A kappa (KOP) hERG (membrane preparation)
beta mu (MOP) K channel
beta2 5-HT1A Na channel (site 2)
CB 5-HT1B AR
CB2 5-HT2A GR
CCK (CCKA) 5-HT
2B Lck kinase
1
Via COX
1
D2S dopamine transporter (DAT) COX2
norepinephrine transporter
ETA (NET) acetylcholinesterase
308
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Panel of 44 targets
5-HT transporter (SERT) MAO-A
H2 BZD (central) PDE3A
1 NMDA PDE4D2
N neuronal a4132
2
EXAMPLE 12
EFFECTS OF AN ILLUSTRATIVE PEPTIDE INHIBITOR IN A RAT MODEL OF ACUTE COLITIS
[00742] To evaluate the efficacy of the illustrative peptide inhibitor Peptide
1185 in an animal
model of disease, acute colitis was induced by providing 7-week-old female
Sprague-Dawley
rats with 64 mg/kg TNBS in 50% ethanol administered intrarectally at Day 0.
Peptide 1185 was
administered orally 37mg/kg/day (combined PO and in drinking water), PO BID,
day -1 to day 7.
A sham group that was not exposed to TNBS and a TNBS treated group that
received treatment
with vehicle were used as control groups. For comparators, neutralizing anti-
IL-23p19 antibody
was administered intraperioneally at 4 mg/kg on Day -1 and again on Day 3, and
prednisolone
was administered at 10 mg/day P.O. All animals received orally water as the
vehicle which was
used to formulate Peptide 993.
[00743] As described above, animals were observed daily for clinical signs
which included
percent body weight loss and signs of loose stools or diarrhea. Six days after
inoculation of
TNBS, rats were sacrificed and the entire colon length and colon weight from
each animal were
recorded. The severity of colitis was evaluated by a pathologist. In addition
to the colon wall
thickness, the gross colon damage was scored on a 0-5 scale according to Table
E29, and
histopathological scores were determined based on the parameters listed in
Table E30.
[00744] Treatment with Peptide 1185 significantly reduced some of the disease
parameters that
were observed in the TNBS rat model of acute colitis. While rats in the sham
group continued to
gain weight over the course of the study, rats exposed to TNBS and treated
with vehicle
experienced weight loss. Oral treatment with prednisolone or systemic
treatment with anti-IL-
23p19 prevented the weight loss in TNBS exposed rats. Treatment with orally
administered
309
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
Peptide 1185 did not significantly prevent the weight loss in TNBS challenged
rats (see FIG. 13).
A significant reduction was also observed in the colon weight to colon length
ratios following
treatment with prednisolone or anti-IL-23p19 as compared to treatment with
vehicle. Oral
administration of Peptide1185 resulted in similar reductions of colon weight
to colon length
ratios and colon macroscopic scores in TNBS exposed rats. Higher colon
macroscopic scores
indicated a higher degree of colon pathology. The colon macroscopic score was
determined by
adding the scores for adhesion, strictures, ulcer, and colon wall thickness,
all of which were
significantly reduced by treatment with prednisolone, anti-IL-23p19, or
Peptide 1185, as
compared to vehicle treated controls. These data demonstrate that oral
administration of Peptide
1185 has comparable efficacy to systemic administration of anti-IL-23p19
monoclonal antibody.
1007451 The pathological features of tissue sections from colon taken from
rats in the sham,
vehicle, anti-IL-23p19, and Peptide 1185 groups were examined. Mucosal
inflammation,
transmural inflammation, gland loss, and erosion were scored according to the
criteria listed in
Table E29 For all of these features, treatment with anti-IL-23p19 or
Prednisolone reduced the
histopathology scores associated with TNBS exposure. Treatment with Peptide
1185 did not
significantly reduce the histopathology scores.
EXAMPLE 13
EFFECTS OF AN ILLUSTRATIVE PEPTIDE INHIBITOR IN A RAT MODEL OF ACUTE COLITIS
1007461 To evaluate the efficacy of the illustrative IL-23R peptide inhibitor
Peptide 993 in an
animal model of disease, acute colitis was induced by providing 7-week-old
female Sprague-
Dawley rats with 64 mg/kg TNBS in 50% ethanol administered intrarectally at
Day 0. Peptide
993 was administered orally two times a day at 10 mg/kg, for a total of 42
mg/kg per day, for 8
days starting approximately 24 hours (Day -1) prior TNBS inoculation. A sham
group that was
not exposed to TNBS and a TNBS treated group that received treatment with
vehicle were used
as control groups. All animals received orally water as the vehicle which was
used to formulate
Peptide 993.
1007471 As described above, animals were observed daily for clinical signs
which included
percent body weight loss and signs of loose stools or diarrhea. Six days after
inoculation of
310
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
TNBS, rats were sacrificed and the entire colon length and colon weight from
each animal were
recorded. The severity of colitis was evaluated by a pathologist. In addition
to the colon wall
thickness, the gross colon damage was scored on a 0-5 scale according to Table
E29, and
histopathological scores were determined based on the parameters listed in
Table E30.
1007481 Treatment with Peptide 993 significantly reduced all disease
parameters that were
observed in the TNBS rat model of acute colitis. While rats in the sham group
continued to gain
weight over the course of the study, rats exposed to TNBS and treated with
vehicle experienced
weight loss. Treatment with orally administered Peptide 993 also prevented the
weight loss in
TNBS challenged rats (see FIG. 12). In addition, a significant reduction was
also observed in the
colon weight to colon length ratios following treatment with oral
administration of Peptide 993.
Higher colon macroscopic scores indicated a higher degree of colon pathology
and was
significantly reduced by treatment with Peptide 993, as compared to vehicle
treated controls.
Oral administration of Peptide 993 has comparable efficacy to systemic
administration of anti-
IL-23p19 monoclonal antibody, which served as a positive control.
Histopathological scores
were significantly reduced in colons from Peptide 993 treated rats as compared
to vehicle group.
EXAMPLE 14
EFFECTS OF AN ILLUSTRATIVE PEPTIDE INHIBITOR IN A RAT MODEL OF ACUTE COLITIS
1007491 To evaluate the efficacy of the illustrative IL-23R peptide inhibitor
Peptide 980 in an
animal model of disease, acute colitis was induced by providing 7-week-old
female Sprague-
Dawley rats with 64 mg/kg Ti\TBS in 50% ethanol administered intrarectally at
Day 0. Peptide
980 was administered orally 37mg/kg/day (combined PO and in drinking water),
PO BID, day -1
to day 7. A sham group that was not exposed to TNBS and a TNBS treated group
that received
treatment with vehicle were used as control groups. All animals received
orally PBS as the
vehicle which was used to formulate Peptide 980.
1007501 As described above, animals were observed daily for clinical signs
which included
percent body weight loss and signs of loose stools or diarrhea. Six days after
inoculation of
TNBS, rats were sacrificed and the entire colon length and colon weight from
each animal were
recorded. The severity of colitis was evaluated by a pathologist. In addition
to the colon wall
thickness, the gross colon damage was scored on a 0-5 scale according to Table
E29, and
311
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
histopathological scores were determined based on the parameters listed in
Table E30.
Treatment with Peptide 980 significantly reduced all disease parameters that
were observed in
the TNBS rat model of acute colitis.
1007511Treatment with orally administered Peptide 980 prevented the weight
loss in TNBS
challenged rats (see FIG. 14). In addition, a significant reduction was also
observed in the colon
weight to colon length ratios following treatment with oral administration of
Peptide 980.
Higher colon macroscopic score indicates a higher degree of colon pathology,
and was
significantly reduced by treatment with Peptide 980, as compared to vehicle
treated controls (see
FIG. 14).
1007521Pathological features of tissue sections from colon taken from rats in
the sham, vehicle,
and Peptide 980 treated groups were examined.
Mucosal inflammation, transmural
inflammation, gland loss, and erosion (see FIG. 14D) were scored according to
the criteria listed
in Table E30. The sum of the histopathology score was significantly reduced by
Peptide 980
treatment as compared to vehicle.
EXAMPLE 15
LEVELS OF BIOMARKERS FOLLOWING TREATMENT WITH PEPTIDE INHIBITORS IN
A RAT MODEL OF ACUTE COLITIS
1007531 Levels of inflammatory markers were examined in the colons. The distal
colon tissue
samples, designated for protein expression analysis, were flash frozen after
collection. For
protein extraction, the samples were thawed, weighed and homogenized in the
extraction buffer
(PBS pH 7.2 supplemented with Protease Inhibitors, 3x volume:weight). The
homogenates were
centrifuged at 13krpm at 4 C for 15 minutes, a total of two times to remove
the debris. The
supernatants were saved in multiple aliquots in -80 C and subsequently used
for protein
expression analysis on ELISA. The total protein in each sample was quantified
using BCA assay.
MPO, IL-1(3, IL-6, IL-17A and IL-22 protein expression in the distal colon
samples were
analyzed using commercially available rat ELISA kits.
[00754] Treatment with Peptide 993 reduced levels of inflammatory markers
present in the colon.
The disease defining (MPO, IL-6 and IL- 1 beta) and IL-23 directed biomarkers
(IL-22, and IL-
312
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
17A) were reduced by treatment with Peptide 993 as compared to vehicle treated
controls (see
FIG. 15). These data demonstrate that administration of Peptide 993 in amounts
that can reduce
pathology in vivo also decrease levels of biomarkers present in the colon that
are associated with
IL-23R activity. Treatment with Peptide 980 reduced levels MPO and IL-22 as
compared to
vehicle treated controls (see FIG. 16). Treatment with Peptide 1185 at the
dose tested did not
significantly reduce levels of MPO, IL-22, or IL-17A.
Table E29. Colon Macroscopic Score
Colonic Score Parameter r¨ihe alues for each amma was
surnmed to obtain the colonic,:scoreltnaxinium...value..T7
Adhesions:
none =0
minimal = 1
involving several bowel loops = 2
Strictures:
none =0
minimal = 1
mild = 2
severe, proximal dilatation = 3
Ulcers:
none
focal hyperemia, no ulcers = 1
ulcer without significant inflammation (hyperemia and bowel wall
thickening = 2
ulceration of 1- <3 cm = 3
ulceration 3- <6 cm = 4
ulceration > 6 cm = 5
Wall thickness:
Wall thickness:
less than 1 mm = 0
1-3 mm= 1
> 3 mm = 2
Table E30a. Histopathology - Mucosal/Submucosal Inflammation Score
Mucosal/Submucosal Inflammation Score
The extent of macrophage, lymphocyte, neutrophil and other
inflammatory infiltrateswas assigned severity scores according
to the following criteria:
0= Normal
313
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
1=Minimal, larger focal area with MNIC and neutrophils or
minimal diffuse,no separation of glands, may be mostly in areas
of submucosal edema or mesentery
2=Mild, diffuse mild, or multifocal affecting 11-25% of mucosa
with minor focal ormultifocal gland separation, no separation in
most areas
3=Moderate, 26-50% of mucosa affected with minimal to mild
focal ormultifocal separation of glands by inflammatory cell
infiltrate, milder in remaining areasof mucosa with some areas
having no gland separation by inflammation
4=Marked, 51-75% of mucosa affected with mild to moderate
separation of glandsby inflammatory cell infiltrate, minimal to
mild in remaining areas of mucosa but allglands have some
separation by infiltrate
5=Severe, 76-100% of mucosa affected with moderate to
marked areas of gland separation by inflammatory cell infiltrate,
mild to moderate in remaining areas of mucosa
Table E30b.
Mucosa! Thickness Score
Mucosal thickness was scored in the colon in a non-tangential
area of the section that best represented the overall mucosa!
thickness. This parameter is indicative of gland elongation and
mucosa! hyperplasia. A hyperplasia score was determined as
follows.
- =No mucosa present
0= Normal
1= Minimal, 5-10% thicker than control mucosa
2=Mild, 11-25% thicker than control mucosa
3=Moderate, 26-50% thicker than control mucosa
4=Marked, 51-75% thicker than control mucosa
Table E30c.
Transmural Inflammation
Presence of inflammatory cell infiltrates within the tunica
muscularis mucosa and increased fibroblasts/fibrocytes with
perpendicularly arranged blood vessels (granulation tissue),
possibly extending to the serosa.
0= Normal
1=Minimal, 5-10% infiltration
2=Mild, 11-25% infiltration
3=Moderate, 26-50% infiltration
314
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
4=Marked, 51-75% infiltration
5=Severe, infiltrates reach the serosa and mesentery
Table E30d.
Gland Loss Score
Crypt epithelial and remaining gland epithelial loss was scored
based on the approximate percent
of the mucosa that was affected as follows:
0=None
1=Minimal, 1-10% of the mucosa affected
2=Mild, 11-25% of the mucosa affected
3=Moderate, 26-50% of the mucosa affected
4= Marked, 51-75% of the mucosa affected
5=Severe, 76-100% of the mucosa affected
Table E30e.
Erosion Score
The loss of surface epithelium was scored based on the
approximate percent of the mucosa thatwas affected as follows.
0=None
1=Minimal, 1-10% of the mucosa affected
2=Mild, 11-25% of the mucosa affected
3=Moderate, 26-50% of the mucosa affected
4= Marked, 51-75% of the mucosa affected
5=Severe, 76-100% of the mucosa affected
Table E3 Of.
Histopathology Sum
A sum of inflammation, gland loss, erosion, and transmural
inflammation scores was calculated.
EXAMPLE 16
CHARACTERIZATION OF ADDITIONAL PEPTIDE INHIBITION OF BINDING OF INTERLEUKIN-23
TO THE INTERLEUKIN-23 RECEPTOR
315
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
1007551 Peptide optimization was performed to identify additional peptide
inhibitors of IL-23
signalling that were active at low concentrations (e.g., IC50 <10 nM) while
exhibiting
gastrointestinal (GI) stability. Certain peptides were tested to identify
peptides that inhibit the
binding of IL-23 to human IL-23R and inhibit IL-23/IL-23R functional activity,
as described
below. Peptides tested included peptides containing a variety of different
cyclization
chemistries, including, e.g., peptides containing a disulfide linkage, e.g.,
between two Pen
residues, and peptides containing a thioether linkage. Peptide inhibitors of
the present invention
include but are not limited to peptides having any of the structures depicted
herein. In addition,
peptide inhibitors of the present invention include those having the same
amino acid sequence of
the peptides or structures described herein, without being required to have
the same or any N- or
C-terminal "capping" groups, such as, e.g., Ac or NH2.
[00756] Assays performed to determine peptide activity were performed as
described in Example
2 above. Human ELISA indicates the 1L23-IL23R competitive binding assay, Rat
ELISA
indicates the rat IL-23R competitive binding ELISA assay, and pStat3HTRF
indicates the DB
cells IL-23R pSTAT3 cell assay. The peptides depicted in Table E31 are
cyclized via a disulfide
bridge formed between two residues in these peptides. The peptides depicted in
Table E32 are
cyclized via a thioether bond between the indicated amino acid residues. Table
E32 provides an
illustrative structure depicting thioether cyclization, which may also be
indicated in the table by
the term "Cyclo," with the cyclic region bracketed immediately following.
Table E31. Illustrative Peptides Containing the Ac-[Pen]-)0(WX-[Pen]-)000(
Motif and
Analogues
SE Q Human Rat
pStat3
ID Sequence
ELISA ELISA HTRF
NO:
(nM) (nM) (nM)
[PalmHisoGluHPEG4]-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-
1115 **
[2-Nall-[Aib]-[Lys(Ac)]-NN-NFI2
1116 Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
[Lys(PEG4-isoGlu-Palm)]-NN-NH2
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-NalHa-MeLys(Acil-
1117 **
[Lys(Ac)]-NN-N H2
[OctanylHlsoGluHPEG4HPerd-NTWQ-[PenHPhe[4-(2-
1118 **
arninoethoxy)]-[2-NalHAibHLys(Ac)]-NN-NH2
[Octany1HPEG4HPerd-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-
1119 **
NalHAilDHLys(Ac)]-NN-N H2
316
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
[PalmHPEG4]-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-
1120 *
[AilD]-[Lys(Ac)l-NN-NFI2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1121 ** *
[Lys(PEG4-OctanyI)]-NN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1122 *
[Lys(PEG4-Palm)]-NN-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)-(PEG4-Palm)]-[2-
1123 ***
Nal]-[Aibl-[Lys(Ac)]NN-NFI2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)-(PEG4-Laury1)]-[2-
1124 **
Nal]-[Aib]-[Lys(Ac)]-NN-N H2
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-NalHa-[a-Palm)-
1125 *
[Lys(Ac)]-NN-N H2
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-NalHa-[a-
1126 *
LauryI)]-[Lys(Ac)]-NN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)-(PEG4-IsoGlu-
1127 ***
Palm)]-[2-Nal]-[Aibl-[Lys(Ac)]-NN-NFI2
1128
Ac-[Pen]-NTWQ-[Pen]- [Phe[4-(2-aminoethoxy)-(PEG4-lsoGLu-
***
Laury1)]-[2-Nal]-[AibHLys(Ac)]-NN-N H2
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-NalHa-[a-
1129 ** *
IsoGlu-Palm)]-[Lys(Ac)]-NN-N H2
Ac-[Per]-QTWQ-[Pen]-Phe(4-CONH2)-[2-Nal]-a-Me-K(PEG4-IsoGlu-
1130 *
LauryI)]-[Lys(Ac)]-NN-NH2
Ac-[Pen]-QTWQ-[Pen]-Phe(4-CONH2)-[2-NalHa-[a-
1131 ** *
[Lys(Ac)]-NN-N H2
Ac-[Per]-QTWQ-[Pen]-phe(4-CONH2)-[2-NalHa-MeLys(Biotinil-
1132 * ** *
[Lys(Ac)]-NN-N H2
Ac-[Pen]-QTWQ-[Pen]-phe(4-CONH2)-[2-NalHa-MeLys(Octanylil-
1133 ** *
[Lys(Ac)]-NN-N H2
Ac-[Pen]-[Lys(IVA)]-TWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-
1134 ** *
[AilD]-[Lys(Ac)l-NN-NFI2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1135 *
[Lys(Ac)]-[Lys(IVA)]-N-NFI2
Ac-[PenHLys(Biotin)]-TWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-
1136 ** *
[AilD]-[Lys(Ac)l-NN-NFI2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1137 **
[Lys(Ac)]-[Lys(Biotin)]-N-NH2
Ac-[Pen]-TWQ-[PenHPhe[4-(2-aminoethoxy)]-[2-
1138 **
Nal]-[Aib]-[Lys(Ac)]-NN-N H2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1139 **
[Lys(AO]-[Lys(octany1)1-N-N H2
Ac-[PenHLys(Palm)]-TWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]--
1140 >1000
[AilD]-[Lys(Ac)l-NN-NFI2
317
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1141 >1000
[Lys(Ac)]-Lys(Palm)l-N-NH2
Ac-[PenHLys(PEG8)1-TWQ-[Per]-[Phe[4-(2-arninoethoxy)H2-Nall--
1142 **
[AilD]-[Lys(Ac)]-NN-NFI2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall--[Aib]-
1143 **
[Lys(Ac)]-[Lys(PEG8)1-N-NFI2
Ac-[Per]-K(Peg11-Palm)TWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-
1144 **
NalHAilDHLys(Ac)]-NN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall--[Aib]-
1145 **
[Lys(Ac)]-[Lys(Peg11-palm)]-N-N H2
Ac-[Pen]-TW-[CitHPenHPhe[4-(2-[Pen]-Nall--
1146 * *
[AilD]-[Lys(Ac)]-NN-NFI2
Ac-[Pen]-[Lys(Ac)]-TW-[Cit]-[Perd-[Phe[4-(2-aminoethoxy)]-[2-Nall-
1147 ** *
[AilD]-[Lys(Ac)]-NN-NFI2
Ac-[Per]-NT-[Phe(3,4-0CF13)2]-Q-[PenHPhe[4-(2-aminoethoxy)H2-
1148 ¨1000
NalHAilDHLys(Ac)]-NN-N H2
Ac-[Pen]-NT-[Phe(2,4-CF13)2]-Q-[Pen]-(2-aminoethoxy)H2-
1149 ***
NalHAilDHLys(Ac)]-NN-N H2
1150
Ac-[Per]-NT-[Phe(3-CH3)]-Q-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall--
***
[AilD]-[Lys(Ac)]-NN-NFI2
Ac-[Per]-NT-[Phe(4-CH3)]-Q-[PenHPhe[4-(2-aminoethoxy)H2-Nall-
1151 ***
[AilD]-[Lys(Ac)]-NN-NFI2
Ac[(D)Arg]-[Perd-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall--
1152 * * *
[AilD]-[Lys(Ac)]-N-[13Ala]-NH2
Ac-[(D)Tyr]-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-
1153 * * *
[AilD]-[Lys(Ac)]-N-[13Ala]-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1154 *
[Lys(Ac)]-QN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1155 *
[Lys(Ac)]-[Lys(Ac)]-N-NE12
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-
1156 *
[Lys(Ac)]-N-[Lys(Ac)]-NFI2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-
1157 ** *
[Lys(Ac)]-QQ-N H2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1158 ** *
[Lys(A0]-Q-[PAIa]-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1159 *
[Lys(Ac)]-N-[Cit]-NFI2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1160 ** *
[Lys(Ac)]-[Cit]-NNFI2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1161 *
[Lys(Ac)]-[ati-Q-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1162 **
[Lys(Ac)]-[Cit]-[Lys(Ac)]-NFI2
318
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-
1163 **
[Lys(Ac)]-[Lys(Ac)]-[Cit]-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-QN-
1164 * * *
[13Ala]-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-E-[Cit]-
1165 *
0-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-
1166 * *
CitNCitNH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibHCitl-
1167 *
Q-[Cit]-NH2
Ac-[PenHCitl-TWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-
1168 * *
[Lys(Ac)]-NN-N H2
1169
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
[Lys(Ac)]-NN-N H2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-QNN-
1170 * *
NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-ENQ-
1171 ** *
NH2
Ac-[Per]-GPWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-
1172 >1000
[Lys(Ac)]-NN-N H2
1173
Ac-[Per]-PGWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
-1000
[Lys(Ac)]-NN-N H2
Ac-[Per]-NTWN-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-
1174 **
[Lys(Ac)]-NN-N H2
Ac-[Per]-NSWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-
1175 *
[Lys(Ac)]-NN-N H2
Ac-[Per]-N-[Aib]-WQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-
1176 >1000
[Lys(Ac)]-NN-N H2
Ac-[Per]-NTW-[Aib]-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-
1177 **
[Lys(Ac)]N-[Aib]-NH2
Ac-[Per]-QTW-[Lys(Ac)]-[Pen]-[Phe[4-(2-aminoethoxy)H2-Nall-
1178 * *
[AilD]-[Lys(Ac)l-NN-NH2
Ac-[Pen]-TWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-Nall--
1179 *
[AilD]- [Lys(Ac)]NNNH2
Ac-[Per]-QVWQ-[PenHPhe[4-(2-aminoethoxy)H2-NalHAibl-
1180 *
[Lys(Ac)]-NN-N H2
Ac-[Per]-NT-[2-Nal]-Q-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-
1181 **
[Lys(Ac)]-NN-N H2
Ac-[Per]-NT-[1-Nal]-Q-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHAibl-
1182 **
[Lys(Ac)]-NN-N H2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-NalHa-MeLeul-
1183 * * *
[Lys(Ac)]-NN-N H2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)H2-[2-MeLys]-[a
1184 * * *
[Lys(Ac)]-NN-N H2
319
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
1185 * * *
carboxy-tetrahydropyran]-[Lys(Ac)]-NN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHa-MeLeul-
1186 * * *
[Lys(Ac)]-N-[13Ala]-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHa-MeLysl-
1187 * * *
[Lys(Ac)]-N-[13Ala]-NH2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
1188 * * *
carboxy-tetrahydropyran]-[Lys(Ac)]-N-U3Alal-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1189 *
[Lys(Ac)]-LN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1190 *
[Lys(Ac)]-GN-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1191 ** *
[Lys(Ac)]-SN-N H2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1192 *
[Lys(Ac)]-[Aib]-N-N H2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1193 *
[Lys(Ac)]-FN-N H2
Ac-[Per]-NTW-[Cit]-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1194 * * *
[Lys(Ac)]-NN-N H2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1195 ***
[Lys(Ac)]-[Tic]PAlal-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1196 *
[Lys(Ac)]-[nLeu]-[13Ala]-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1197 *
[Lys(Ac)]-G-[13Ala]-N H2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1198 *
[Lys(Ac)]-R-[13Ala]-N H2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]--[Aib]-
1199 **
[Lys(Ac)]-W-U3Alal-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]--[Aib]-
1200 *
[Lys(Ac)]-5-[13Ala]-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]--[Aib]-
1201 *
[Lys(Ac)]-L-[13Ala]-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]--[Aib]-
1202 *
[Lys(Ac)[AIBHI3Alal-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]--[Aib]-
1203 ***
[Lys(Ac)]-[N-MeAla]-U3Alal-NH2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1204 ** *
[Lys(Ac)][2-NapHPAlal-N H2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1205 ** *
[Lys(Ac)]-F-[13Ala]-NH2
Ac-[(D)Arg]-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[ 4-
1206 *
amino-4-carboxy-tetrahydropyran]-[Lys(Ac)]NN-NH2
320
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
Table E32. IC50 of Illustrative Peptide Inhibitors (Thioethers)
......õ-- S--õ..,...
Ac, r....XXWX,N
N '\i[Phe(4-0Me)]-[2-Nal]-XXXX-NH2
H 0 H
0
Ac-Cyclo- [[Abu]-XXWXC]4Phe(4-0Me)]- [2-Nal]-XXX-1\TH 2
SEQ Human Rat pStat3
ID Sequence ELISA ELISA HTRF
NO: (nM) (nM) (nM)
Biotin-[PEG4]-cyclo[[Abu]-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-
1207 TBC * *
[4-amino-4-carboxy-tetrahydropyran]-ENN-NH2
Ac-cyclo[[Abu]-QTWOCHPhe[4-(2-aminoethoxy)H2-NalH4-amino-
1208 TBC * *
4-carboxy-tetrahydropyranHLys(Ac)l-NN-NH2
Ac-[(D)Ard-cyclo[[Abu]-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-
1209 TBC * *
[4-arnino-4-carboxy-tetrahydropyran]-[Lys(Ac)]-NN-NH2
Ac-[(D)Ard-cyclo[[Abu]-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-
1210 * * *
[4-arnino-4-carboxy-tetrahydropyran]-[Lys(Ac)]-NN-NH2
1211
Ac-E-[(D)Arg]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-
[4-amino-4-carboxy-tetrahydropyran]-ENN-NH2
1212
Ac-[(D)Asp]-[(D)Argl-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-
[2-Nal]-[4-amino-4-carboxy-tetrahydropyrard-ENN-NH2
Ac-R-[(D)Arg]-cyclo[[Abu]-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-
1213 * * *
[4-amino-4-carboxy-tetrahydropyran]-ENN-NH2
Ac-[(D)ArgH(D)Argl-cyclo[[Abu]-QTWOCHPhe[4-(2-aminoethoxy)]-
1214 TBC * *
[2-Nal]-[4-amino-4-carboxy-tetrahydropyrard-ENN-NH2
Ac-F-[(D)Arg]-cyclo[[Abu]-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-
1215 *
[4-amino-4-carboxy-tetrahydropyran]-ENN-NH2
1216
Ac-[(D)Phe]-[(D)Arg]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-
[2-Nal]-[4-amino-4-carboxy-tetrahydropyrard-ENN-NH2
Ac-[2-Nal]-[(D)Arg]-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-
1217 *
[2-Nal]-[4-amino-4-carboxy-tetrahydropyrard-ENN-NH2
Ac-T-[(D)Arg]-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-
1218 *
[4-amino-4-carboxy-tetrahydropyran]-ENN-NH2
Ac-L-[(D)Arg]-cyclo[[Abu]-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-
1219 *
[4-amino-4-carboxy-tetrahydropyran]-ENN-NH2
Ac-[(D)G1n]-[(D)Arg]-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-
1220 TBC * *
[2-Nal]-[4-amino-4-carboxy-tetrahydropyrard-ENN-NH2
Ac-[(D)Asn]-[(D)Arg]-cyclo[[Abu]-QTWOCHPhe[4-(2-aminoethoxy)]-
1221 * *
[2-Nal]-[4-amino-4-carboxy-tetrahydropyrard-ENN-NH2
321
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
1222 Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)-(PEG4-A1exa488)]-
[2-Nal]-[4-amino-4-carboxy-tetrahydropyrard-ENN-NH2
1223 [A1exa488]-[PEG41-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]--
[2-Nal]-[4-amino-4-carboxy-tetrahydropyrard-ENN-NH2
[Alexa647HPEG41-cyclo[[Abu]-QTWOCHPhe [4-(2-aminoethoxy)H2-[2
1224 TBC
NalH4-amino-4-carboxy-tetrahydropyranl-ENN-NH2
[Alexa-647]-[PEG4H(D)Argl-cycloRAbul-QTWOCHPhe[4-(2-
1225 aminoethoxy)]-[2-NalH4-amino-4-carboxy-tetrahydropyrard-
[Lys(Ac)]-NN-NFI2
[A1exa647]-[PEG121-[(D)Arg]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-
1226 aminoethoxy)]-[2-NalH4-amino-4-carboxy-tetrahydropyrard-
[Lys(Ac)]-NN-NFI2
[A1exa488]-[PEG4H(D)Argl-cycloRAbul-QTWOCHPhe [4-(2-
1227 aminoethoxy)]-[2-NalH4-amino-4-carboxy-tetrahydropyrard-
[Lys(Ac)]-NN-NFI2
* < 1 0 nIVI; ** > 10 and < 100 nM; *** > 100 and < 1,000 nIVI
EXAMPLE 17
STABILITY OF ADDITIONAL PEPTIDE INHIBITORS IN SIMULATED INTESTINAL FLUID
(SIF),
SIMULATED GASTRIC FLUID (SGF) AND REDOX CONDITIONS
[00757] Studies were carried out in simulated intestinal fluid (SIF) and
simulated gastric fluid
(SGF) to evaluate gastric stability of additional peptide inhibitors of the
present invention. In
addition, studies were carried out to assess redox stability of the additional
peptide inhibitors of
the present invention.
Table E33. Thioethers and Dipens
SIF SGF
SEQ ID t1/2 t1/2
Sequence
NO:
(min) (min)
Biotin-[PEE1]-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-[4-
1228 >90 >180
a mino-4-carboxy-tetra hydropyranl-ENN-NH2
Ac-cyclo[[Abu]-QTWOCH Phe[4-(2-aminoethoxy)H2-NalH4-amino-4-
1229 >180 >180
carboxy-tetrahydropyranHLys(Ac)]-NN-N H2
Ac-[(D)Arg]-cyclo[[Abu]-QTWOCHPhe[4-(2-aminoethoxy)H2-Nall-[4-
1230 >180 <180
a mino-4-carboxy-tetra hydropyran]-[Lys(Ac)]-NN-N H2
1231 Ac-[(D)Arg]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-Nall-[4-
>180, >180
a mino-4-carboxy-tetra hydropyran]-[Lys(Ac)]-NN-N H2 >180 >180
322
CA 02991984 2018-01-09
WO 2017/011820
PCT/US2016/042680
[Octany1]-[PEG4]-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-
1232 >180 >180
[AilD]-[Lys(Ac)1-NN-NFI2
[PalmHPEG4]-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nall-
1233 >180 >10
[AilD]-[Lys(Ac)1-NN-NFI2
Ac-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)-(PEG4-Alexa488)]-[2-
1234 >180 >90
Nal]-[4-amino-4-carboxy-tetrahydropyranl-ENN-NH2
[Alexa488]-[PEG4]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)] [2
1235 > 180 > 180
Nal]-[4-amino-4-carboxy-tetrahydropyranl-ENN-NH2
[A1exa647]-[PEG4]-cyclo[[Abu]-QTWOC]-[Phe[4-(2-aminoethoxy)]-[2-
1236 >180 >180
Nal]-[4-amino-4-carboxy-tetrahydropyranl-ENN-NH2
Ac[(D)Arg]-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]--[Aib]-
1237 >180 >180
[Lys(Ac)]-N-[13Alal-NFI2
Ac-[(D)Tyr]-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-
1238 >180 >180
[Lys(Ac)]-N-[13Alal-NFI2
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aibl-QN-WAlal-
1239 Stable >180
NH2
>180
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[a-MeLeul- >180,
1240 >180
[Lys(Ac)]-NN-N H2 >180
>180,
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4- >180,
1241 >180
carboxy-tetrahydropyran]-[Lys(AcH-NN-NH2 >180
Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[a-MeLeul- Stable
1242 >180
[Lys(Ac)]-N-[13Alal-NFI2
Ac-[Pen]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4- >180
>180
1243
carboxy-tetrahydropyran]-[Lys(Ac)]-N-[Ala]-NFI2
Ac-[Pen]-NTW-[Cit]-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aibl- >180
>180
1244
[Lys(Ac)]-NN-N H2
Ac-[(D)Arg]-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4- > 180
> 180
1245
AMINO-4-CARBOXY-TETRAHYDROPYRAN]-[Lys(Ac)]NNNH2
[A1exa488]-[PEG4]-[(D)Arg]-cycloRAbul-QTWOCHPhe[4-(2-
> 180 >180
1246 arninoethoxy)]-[2-Nall-[4-arnino-4-carboxy-tetrahydropyran]-[Lys(AcH-
NN-NH2
>90 = less than or equal to 180 min and greater than 90 min; >45 min = less
than or equal to 90
min and greater than 45 min; >10 = less than or equal to 45 min and greater
than 10 min; <10 =
less than 10 min.
EXAMPLE 18
NK CELL AS SAY
1007581 Natural killer (NK) cells, purified from human peripheral blood of
healthy donors by
negative selection (Miltenyi Biotech, Cat # 130-092-657), were cultured in
complete media
(RPMI 1640 containing 10% FBS, L-glutamine and penicillin-streptomycin) in the
presence of
323
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
IL-2 (RnD, Cat # 202-IL-010/CF) at 25 ng/mL. After 7 days, cells were
centrifuged, and
resuspended in complete media at 1E6 cells/mL. Recombinant IL-23 at
predetermined EC50 to
EC75 and IL-18 (RnD, Cat # B003-5) at 10 ng/mL were mixed with varying
concentrations of
peptides, and added to NK cells seeded at 1E5 cells per well. After 20 to 24
hours, IFNy in the
supernatant was quantified using Quantikine ELISA (RnD, Cat # DIF50). Results
are shown in
Table E34. Multiple results shown for a single peptide are from separate
assays.
Table E34. Primary Cell Assay (Thioethers and Dipens)
SEQ ID NK cell
NO: Sequence Assay
(nM)
Ac-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-carboxy-
1247 * * *
tetrahydropyran]-[Lys(Ac)]-NN-NH2
1248 Ac-[(D)Arg]-cycloRAbul-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-
4- * ** **
carboxy-tetrahydropyran]-[Lys(Ac)]-NN-NH2
[Alexa647]-[PEG4]-cyclo[[Abu]-QTWOCHPhe[4-(2-aminoethoxy)]-[2-Nal]-[4-
1249 **, *
amino-4-carboxy-tetrahydropyran]-ENN-NH2
Ac[(D)Arg]-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]--[Aib]-
1250 **
[Lys(Ac)]-N-[13Ala]-NH2
1251 Ac-[Per]-NTVVQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aibl-QN-
[13Ala]-N H2 **
Ac-[Per]-QTW-[Lys(Ac)]-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-NalHAibl-[Lys(Ac)]-
1252 **
NN-NH2
1253 Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[a-
MeLeuHLys(Ac)]-
NN-NH2
1254 Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-
tetrahydropyran]-[Lys(Ac)]-NN-NH2
1255 Ac-[Per]-NTWQ-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[4-amino-4-
carboxy-
tetrahydropyran]-[Lys(Ac)]-N-U3Alal-NH2
1256 Ac-[Per]-NTW-[Cit]-[Pen]-[Phe[4-(2-aminoethoxy)]-[2-Nal]-[Aibl-
[Lys(Ac)]-NN-
NH2
1007591A11 of the above U.S. patents, U.S. patent application publications,
U.S. patent
applications, foreign patents, foreign patent applications and non-patent
publications referred to
in this specification and/or listed in the Application Data Sheet, are
incorporated herein by
reference, in their entirety.
324
CA 02991984 2018-01-09
WO 2017/011820 PCT/US2016/042680
1007611From the foregoing it will be appreciated that, although specific
embodiments of the
invention have been described herein for purposes of illustration, various
modifications may be
made without deviating from the spirit and scope of the invention.
Accordingly, the invention is
not limited except as by the appended claims.
325