Note: Descriptions are shown in the official language in which they were submitted.
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
IL-15-BASED FUSIONS TO IL-12 AND IL-18
RELATED APPLICATIONS
This application claims the benefit U.S. provisional application 62/467,623
filed
March 6, 2017, which is incorporated herein by reference in its entirety.
FIELD OF THE INVENTION
This invention relates generally to the field of multimeric fusion molecules.
BACKGROUND OF THE INVENTION
Prior to the invention described herein, there was a pressing need to develop
new
strategies to augment immune responses and provide therapeutic benefit to
patients with
neoplasia or infectious diseases.
SUMMARY OF THE INVENTION
The invention is based, at least in part, on the surprising discovery that
multi-specific
interleukin-15 (IL-15)-based fusion protein complexes enhance the stimulation
of immune
cells and promote their activity against disease cells, thereby resulting in
reduction or
prevention of disease. These IL-15-based fusion protein complexes may also
show increased
binding to disease and target antigens. Provided herein are multi-specific IL-
15-based fusion
protein complexes comprising IL-12 and IL-18 binding domains (FIG. 1A, 1B).
Specifically,
described herein are fusion protein complexes comprising an IL-15N72D:IL-
15RaSu-Ig Fc
scaffold fused to IL-12 and/or IL-18 binding domains. As described in detail
below, when
characterized using human immune cells, these fusion protein complexes exhibit
binding and
biological activity of each of the IL-15, IL-12 and IL-18 cytokines.
Additionally, these
fusion protein complexes induce cytokine-induced memory-like (CIML) natural
killer (NK)
cells with elevated activation markers, increased cytotoxicity against tumor
cells and
enhanced production of IFN-y.
As such, the fusion protein complex as a single molecule binds to and signals
via
multiple cytokine receptors on NK cells to provide the responses previously
observed only
with a combination of multiple individual cytokines. Additionally, these
fusion protein
complexes comprise the Fc region of Ig molecules, which can form a dimer to
provide a
soluble multi-polypeptide complex, bind Protein A for the purpose of
purification and
interact with Fey receptors on NK cells and macrophages, thereby providing
advantages to
1
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
the fusion protein complex that are not present in the combination of
individual cytokines.
Mammalian cell expression-based methods for making these fusion protein
complexes
suitable for large scale production of clinical grade material are described
herein. Additional
methods for making and using CIML NK cells induced by the fusion protein
complex of the
invention are also provided.
Accordingly, provided is an isolated soluble fusion protein complex comprising
at
least two soluble proteins. For example, the first protein comprises an IL-15
polypeptide,
e.g., a variant IL-15 polypeptide comprising an N72D mutation (IL-15N72D). The
second
protein comprises a soluble IL-15 receptor alpha sushi-binding domain (IL-
15RaSu) fused to
an immunoglobulin Fc domain (IL-15RaSu/Fc). A third component of the isolated
soluble
fusion protein complex comprises a binding domain of IL-12, wherein the IL-12
binding
domain is fused to the either the IL-15N72D or the IL-15RaSu/Fc protein. A
forth
component of the isolated soluble fusion protein complex comprises a binding
domain of IL-
18, wherein the IL-18 binding domain is fused to the either the IL-15N72D or
the IL-
15RaSu/Fc protein. In some cases, the IL-12 and/or IL-18 binding domains are
fused to both
the IL-15N72D and IL-15RaSu/Fc proteins. In other cases, either the IL-12 or
IL-18 binding
domain is fused to the IL-15N72D or the IL-15RaSu/Fc proteins and another
binding domain
is fused to the other protein. In other cases, the complex comprises an IL-18
binding domain
fused to the IL-15N72D:IL-15RaSu-Ig Fc scaffold without IL-12 or an IL-18
binding domain
fused to the IL-15N72D:IL-15RaSu-Ig Fc scaffold without IL-12. The fusions may
be made
at the N- or C-terminus of the proteins. The IL-12 protein may comprise a
heterodimer of the
p40 and p35 IL-12 subunits. Alternatively, the IL-12 protein may comprise a
single-chain
format in which the p40 and p35 subunits are linked by a flexible polypeptide
linker. The
single-chain IL-12 may comprise either the C-terminus of p40 linked to the N-
terminus of
p35 or the C-terminus of p35 linked to the N-terminus of p40. An exemplary
fusion protein
complex comprises an IL-18 polypeptide covalently linked to an IL-15N72D and a
single-
chain IL-12 polypeptide covalently linked to an IL-15RaSu/Fc fusion protein.
Alternatively,
the fusion protein complex comprises a single-chain IL-12 polypeptide
covalently linked to
an IL-15N72D and an IL-18 polypeptide covalently linked to an IL-15RaSu/Fc
fusion protein
(FIG. 1A, 1B).
Exemplary first proteins comprise the amino acid sequences set forth in SEQ ID
NO:
2 and SEQ ID NO: 6. Exemplary second proteins comprise the amino acid
sequences set
forth in SEQ ID NO: 4 and SEQ ID NO: 8. Exemplary nucleic acid sequences
encoding the
first protein comprise the sequences set forth in SEQ ID NO: 1 and SEQ ID NO:
5.
2
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
Exemplary nucleic acid sequences encoding the second protein comprise the
sequences set
forth in SEQ ID NO: 3 and SEQ ID NO: 7. In one aspect, the nucleic acid
sequence(s)
further comprises a promoter, translation initiation signal, and leader
sequence operably
linked to the sequence encoding the fusion protein. Also provided are DNA
vectors
comprising the nucleic acid sequences described herein. For example, the
nucleic acid
sequence is in a vector for replication, expression, or both.
Also provided is a soluble fusion protein complex comprising a first soluble
fusion
protein complex covalently linked to a second soluble fusion protein complex.
For example,
the soluble fusion protein complexes of the invention are multimerized, e.g.,
dimerized,
trimerized, or otherwise multimerized (e.g., 4 complexes, 5 complexes, etc.).
For example,
the multimers are homomultimers or heteromultimers. The soluble fusion protein
complexes
are joined by covalent bonds, e.g., disulfide bonds, chemical cross-linking
agents. In some
cases, one soluble fusion protein is covalently linked to another soluble
fusion protein by a
disulfide bond linking the Fc domain of the first soluble protein to the Fc
domain of the
second soluble protein.
The Fc domain or functional fragment thereof includes an Fc domain selected
from
the group consisting of IgG Fc domain, human IgG1 Fc domain, human IgG2 Fc
domain,
human IgG3 Fc domain, human IgG4 Fc domain, IgA Fc domain, IgD Fc domain, IgE
Fc
domain, and IgM Fc domain; mouse IgG2A domain, or any combination thereof
Optionally,
the Fc domain includes an amino acid change that results in an Fc domain with
altered
complement or Fc receptor binding properties or altered dimerization or
glycosylation
profiles. Amino acid changes to produce an Fc domain with altered complement
or Fc
receptor binding properties or altered dimerization or glycosylation profiles
are known in the
art. For example, a substitution of leucine residues at positions 234 and 235
of the IgG1 CH2
(numbering based on antibody consensus sequence) (i.e., ... PELL GG ...) with
alanine
residues (i.e., ... PEAAGG ...) results in a loss of Fc gamma receptor
binding, whereas
the substitution of the lysine residue at position 322 of the IgG1 CH2
(numbering based on
antibody consensus sequence) (i.e., ... KCKSL ...) with an alanine residue
(i.e., K C A
S L ...) results in a loss of complement activation. In some examples, such
mutations are
combined.
In some aspects, the IL-12 or IL-18 binding domains is covalently linked to an
IL-15
polypeptide (or functional fragment thereof) by a polypeptide linker sequence.
Similarly, the
IL-12 or IL-18 binding domain is covalently linked to an IL-15Ra polypeptide
(or functional
fragment thereof) by a polypeptide linker sequence. Optionally, the IL-15Ra
polypeptide (or
3
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
functional fragment thereof) is covalently linked to the Fc domain (or
functional fragment
thereof) by a polypeptide linker sequence. Each polypeptide linker sequence
can be selected
independently. Optionally, the polypeptide linker sequences are the same.
Alternatively,
they are different.
Optionally, the soluble fusion protein complexes of the invention are provided
wherein at least one of the soluble fusion proteins comprise one or more
binding domain or
detectable labels. Such binding domains may comprise antibodies, soluble T
cell receptors,
ligands, soluble receptor domains or functional fragments thereof IL-15-based
fusion
protein complexes comprising such binding domains have been previously
described in U.S.
Patent No. 8,492,118, incorporated herein by reference. Detectable labels
include, but are not
limited to, biotin, streptavidin, an enzyme or catalytically active fragment
thereof, a
radionuclide, a nanoparticle, a paramagnetic metal ion, or a fluorescent,
phosphorescent, or
chemiluminescent molecule, or any combination thereof
The invention provides methods for making the soluble fusion protein complexes
of
the invention. The method includes the steps of: a) introducing into a first
host cell a DNA
vector with appropriate control sequences encoding the first protein, b)
culturing the first host
cell in media under conditions sufficient to express the first protein in the
cell or the media, c)
purifying the first protein from the host cells or media, d) introducing into
a second host cell a
DNA vector with appropriate control sequences encoding the second protein, e)
culturing the
second host cell in media under conditions sufficient to express the second
protein in the cell
or the media, 0 purifying the second protein from the host cells or media, and
g) mixing the
first and second proteins under conditions sufficient to allow binding between
IL-15 domain
of a first protein and the soluble IL-15Ra domain of a second protein to form
the soluble
fusion protein complex.
In some cases, the method further includes mixing the first and second protein
under
conditions sufficient to allow formation of a disulfide bond between the
polypeptides
expressed from the expression vectors.
Alternatively, methods for making soluble fusion protein complexes of the
invention
are carried out by a) introducing into a host cell a DNA vector with
appropriate control
sequences encoding the first protein and a DNA vector with appropriate control
sequences
encoding the second protein, b) culturing the host cell in media under
conditions sufficient to
express the proteins in the cell or the media and allow association between IL-
15 domain of a
first protein and the soluble IL-15Ra domain of a second protein to form the
soluble fusion
4
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
protein complex, and c) purifying the soluble fusion protein complex from the
host cells or
media.
In one aspect, the method further includes mixing the first and second protein
under
conditions sufficient to allow formation of a disulfide bond between the
polypeptides
expressed from the expression vectors.
Also provided are methods for making soluble fusion protein complexes
comprising
a) introducing into a host cell a DNA vector with appropriate control
sequences encoding the
first and second proteins, b) culturing the host cell in media under
conditions sufficient to
express the proteins in the cell or the media and allow association between IL-
15 domain of a
first protein and the soluble IL-15Ra domain of a second protein to form the
soluble fusion
protein complex, and to allow formation of a disulfide bond between the
polypeptides, and
c) purifying the soluble fusion protein complex from the host cells or media.
Optionally, the method further includes mixing the first and second protein
under
conditions sufficient to allow formation of a disulfide bond between the
polypeptides
expressed from the expression vectors.
In some cases, the method further includes purification of the fusion protein
complex
by Protein A affinity chromatography, size exclusion chromatography, ion
exchange
chromatography and/or other standard methods (including viral inactivation
and/or filtration)
sufficient to generate a sufficiently pure fusion protein complex suitable for
use as a clinical
reagent or therapeutic.
In certain aspects of the soluble fusion protein complexes of the invention,
the IL-15
polypeptide is an IL-15 variant having a different amino acid sequence than
native IL-15
polypeptide. The human IL-15 polypeptide is referred to herein as huIL-15, hIL-
15, huIL15,
hIL15, IL-15 wild type (wt), and variants thereof are referred to using the
native amino acid,
its position in the mature sequence and the variant amino acid. For example,
huIL15N72D
refers to human IL-15 comprising a substitution of N to D at position 72. In
one aspect, the
IL-15 variant functions as an IL-15 agonist as demonstrated, e.g., by
increased binding
activity for the IL-15R3yC receptors compared to the native IL-15 polypeptide.
Alternatively, the IL-15 variant functions as an IL-15 antagonist as
demonstrated by e.g.,
decreased binding activity for the IL-15R3yC receptors compared to the native
IL-15
polypeptide.
Methods of enhancing immune function are carried out by a) contacting a
plurality of
cells with a soluble fusion protein complex of the invention, wherein the
plurality of cells
further include immune cells comprising the IL-15R chains recognized by the IL-
15 domain,
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
the IL-12R chains recognized by the IL-12 domain and/or the IL-18R chains
recognized by
the IL-18 domain, and b) activating the immune cells via signaling of the IL-
15R, IL-12R
and/or IL-18R. In one aspect, the method of enhancing immune function further
includes
activation the immune cells via signaling of a combination of at least two or
all of the IL-
15R, IL-12R and IL-18R by the soluble fusion protein complex. Exemplary
methods for
enhancing immune function include activation of NK cells via signaling of the
IL-15R, IL-
12R and IL-18R by the soluble fusion protein complex. Such methods include
activation of
NK cells resulting in increased activation markers (i.e., CD25, CD69),
elevated cytotoxicity
against diseased cells or increased production of IFN-y. In some aspects,
methods include
induction of CIML NK cells by the soluble fusion protein complex of the
invention.
Methods for killing a target cell are carried out by a) contacting a plurality
of cells
with a soluble fusion protein complex of the invention, wherein the plurality
of cells further
include immune cells comprising the IL-15R chains recognized by the IL-15
domain, the IL-
12R chains recognized by the IL-12 domain and/or the IL-18R chains recognized
by the IL-
18 domain, and the target disease cells, b) activating the immune cells via
signaling of the IL-
15R, IL-12R and/or IL-18R, and c) killing the target disease cells by the
activated immune
cells. In one aspect, the method includes activation the immune cells via
signaling of a
combination of at least two or all of the IL-15R, IL-12R and IL-18R by the
soluble fusion
protein complex. Exemplary methods include activation of NK cells, in
particular CIML NK
cells, via signaling of the IL-15R, IL-12R and IL-18R by the soluble fusion
protein complex.
Such methods include activation of NK cells resulting in activation markers
(i.e., CD25,
CD69), elevated cytotoxicity against target cells.
The invention also provides methods for preventing or treating disease in a
patient,
the method including the steps of: a) mixing immune cells comprising the IL-
15R chains
recognized by the IL-15 domain, the IL-12R chains recognized by the IL-12
domain and/or
the IL-18R chains recognized by the IL-18 domain with a soluble fusion protein
complex of
the invention, b) activating the immune cells via signaling of the IL-15R, IL-
12R and/or IL-
18R, c) administering (or adoptively transfer) to the patient the activated
immune cells, and
d) damaging or killing the disease cells via the activated immune cells
sufficient to prevent or
treat the disease in the patient. In one aspect, the method includes
activation the immune
cells via signaling of a combination of at least two or all of the IL-15R, IL-
12R and IL-18R
by the soluble fusion protein complex. Exemplary methods include activation of
NK cells, in
particular, CIML NK cells, via signaling of the IL-15R, IL-12R and IL-18R by
the soluble
fusion protein complex. Other aspects of the method include use of
immortalized immune
6
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
cells, such as NK-92, aNK, haNK or taNK cells, which may be irradiated prior
to transfer. In
some embodiments of the invention, the patient is pretreated or preconditioned
to facilitate
engraftment or survival of the adoptively transferred cells. Examples of
preconditioning
include treatment with cyclophosphamide and fludarabine. Additionally, the
patient may be
treated with agents that promote activation, survival, or persistence of the
adoptively
transferred cells pre- and/or post-cell transfer. Examples of such treatment
include use of IL-
2, IL-15, ALT-803 or other immunostimulatory agents. Other therapeutic
approaches of
known in the field of adoptive cell therapy (i.e., including but not limited
to allogeneic,
autologous, haploidentical, DLI, stem cell, CAR T, NK92-based and CAR NK
therapies)
may also be used in the methods herein.
Also provided are methods for preventing or treating disease in a patient, the
method
including the steps of: a) administering to the patient a soluble fusion
protein complex of the
invention, b) activating the immune cells in the patient via signaling of the
IL-15R, IL-12R
and/or IL-18R, and c) damaging or killing the disease cells via the activated
immune cells
sufficient to prevent or treat the disease in the patient.
Administration of the fusion protein complexes of the invention induces an
immune
response in a subject. For example, administration of the fusion protein
complexes of the
invention induces an immune response against cells associated with neoplasia
or infectious
disease. In one aspect, the fusion protein complex of the invention increases
immune cell
proliferation, activation markers, cytotoxicity against target cells, and/or
production of pro
inflammatory cytokines.
The invention provides methods of stimulating immune responses in a mammal by
administering to the mammal an effective amount of the soluble fusion protein
complex of
the invention. The invention also provides methods of suppressing immune
responses in a
mammal by administering to the mammal an effective amount of the soluble
fusion protein
complex of any one of the invention.
Methods for treating a neoplasia or infectious disease in a subject in need
thereof are
carried out by administering to a subject an effective amount of activated
immune cells or a
pharmaceutical composition comprising a soluble fusion protein complex
described herein.
For example, methods for treating solid or hematological malignancies in a
subject in need
thereof are carried out by administering to a subject an effective amount of
CIML NK cells
activated ex vivo by the soluble fusion protein complex of the invention,
thereby treating the
malignancy. Exemplary soluble fusion protein complexes comprise the amino acid
7
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
sequences set forth in SEQ ID NO: 2 and SEQ ID NO: 6 and in SEQ ID NO: 4 and
SEQ ID
NO: 8.
Suitable neoplasias for treatment with the methods described herein include a
glioblastoma, prostate cancer, acute myeloid leukemia, B-cell neoplasm,
multiple myeloma,
B-cell lymphoma, B cell non-Hodgkin's lymphoma, Hodgkin's lymphoma, chronic
lymphocytic leukemia, acute myeloid leukemia, cutaneous T-cell lymphoma, T-
cell
lymphoma, a solid tumor, urothelial/bladder carcinoma, melanoma, lung cancer,
renal cell
carcinoma, breast cancer, gastric and esophageal cancer, head and neck cancer,
prostate
cancer, pancreatic cancer, colorectal cancer, ovarian cancer, non-small cell
lung carcinoma,
and squamous cell head and neck carcinoma.
An exemplary infection for treatment using the methods described herein
include
infections with human immunodeficiency virus (HIV) or cytomegalovirus (CMV).
The
methods described herein are also useful to treat bacterial infections (e.g.,
gram positive or
gram negative bacteria) (See, e.g., Oleksiewicz et al. 2012. Arch Biochem
Biophys. 526:124-
31, incorporated herein by reference).
Cell therapies of the invention comprise administration of an effective amount
of
activated immune cells. For example, an effective amount of activated NK cells
is between
1 x 104 cells/kg and 1 x 1010 cells/kg, e.g., 1 x 104, 1 x 105, 1 x 106, 1 x
107, 1 x 108, 1 x 109,
and 1 x 1010 cells/kg, or such amounts that can be isolated by leukapheresis.
Alternatively,
activated immune cells are administered as a fixed dose or based on body
surface area (i.e.,
per m2). Cells can be administered after ex vivo activation or cryogenically
preserved and
administered after thawing (and washing as needed).
The pharmaceutical composition comprising a fusion protein complex is
administered
in an effective amount. For example, an effective amount of the pharmaceutical
composition
is between about 1 g/kg and 100 g/kg, e.g., 1, 5, 10, 15, 20, 25, 30, 35,
40, 45, 50, 55, 60,
65, 70, 75, 80, 85, 90, 95, or 100 g/kg. Alternatively, the fusion protein
complex is
administered as a fixed dose or based on body surface area (i.e., per m2).
The adoptively transferred immune cells or pharmaceutical composition
comprising
the fusion protein complex is administered at least one time per month, e.g.,
twice per month,
once per week, twice per week, once per day, twice per day, every 8 hours,
every 4 hours,
every 2 hours, or every hour. Suitable modes of administration for the
adoptively transferred
immune cells include systemic administration, intravenous administration, or
local
administration. Suitable modes of administration for the pharmaceutical
composition include
systemic administration, intravenous administration, local administration,
subcutaneous
8
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
administration, intramuscular administration, intratumoral administration,
inhalation, and
intraperitoneal administration.
In an aspect, the present disclosure provides an isolated soluble fusion
protein
complex comprising at least two soluble proteins, where the first soluble
protein comprises an
interleukin-15 (IL-15) polypeptide domain and the second soluble protein
comprises a
soluble IL-15 receptor alpha sushi-binding domain (IL-15RaSu) fused to an
immunoglobulin
Fc domain, where one of the first or second soluble protein further comprises
an IL-18
binding domain or functional fragment thereof, where one of the first or
second soluble
protein further comprises an IL-12 binding domain or functional fragment
thereof and
wherein the IL-15 domain of the first soluble protein binds to the IL-15RaSu
domain of the
second soluble protein to form a soluble fusion protein complex.
In an embodiment, the IL-15 polypeptide is an IL-15 variant comprising an N72D
mutation (IL-15N72D).
In an embodiment, the IL-12 binding domain comprises the p40 and p35 subunits
of
IL-12. In an embodiment, the p40 and p35 subunits of IL-12 are linked by a
flexible
polypeptide linker into a single-chain format.
In an embodiment, the first soluble protein comprises the amino acid sequence
set
forth in one of SEQ ID NOs: 2 or 6.
In an embodiment, the second soluble protein comprises the amino acid sequence
set
forth in one of SEQ ID NOs: 4 or 8.
In an embodiment, a first soluble fusion protein complex may be covalently
linked to
a second soluble fusion protein complex.
In an embodiment, the first soluble fusion protein complex is covalently
linked to the
second soluble fusion protein complex by a disulfide bond linking the Fc
domain of the first
soluble fusion protein complex to the Fc domain of the second soluble fusion
protein
complex.
In an embodiment, the first or second soluble protein further comprises a
binding
domain that recognizes a disease antigen.
In an embodiment, the first or second soluble protein further comprises a
binding
domain that recognizes an immune checkpoint or signaling molecule.
In an embodiment, the disease antigen is associated with neoplasia or
infectious
disease.
In an embodiment, the first soluble protein is encoded by the sequence set
forth in one
of SEQ ID NOs: 1 or S. In an embodiment, the nucleic acid sequence further
comprises a
9
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
promoter, translation initiation signal, and leader sequence operably linked
to the sequence
encoding the soluble protein.
In an embodiment, the second soluble protein may be encoded by the nucleic
acid
sequence set forth in one of SEQ ID NOs: 3 or 7. In an embodiment, the nucleic
acid
sequence further comprises a promoter, translation initiation signal, and
leader sequence
operably linked to the sequence encoding the soluble protein.
In an embodiment, a DNA vector may comprise any of the above enumerated
nucleic
acid sequences.
In an embodiment, a method for enhancing immune function, the method
comprising:
a) contacting a plurality of cells with any of the above soluble fusion
protein complexes,
where the plurality of cells further comprises immune cells comprising the IL-
15R chains
recognized by the IL-15 domain, the IL-12R chains recognized by the IL-12
domain and/or
the IL-18R chains recognized by the IL-18 domain, and b) activating the immune
cells via
signaling of the IL-15R, IL-12R and/or IL-18R.
In an aspect, the present disclosure provides a method for killing a target
cell,
comprising: a) contacting a plurality of cells with any of the above soluble
fusion protein
complexes, where the plurality of cells further include immune cells
comprising the IL-15R
chains recognized by the IL-15 domain, the IL-12R chains recognized by the IL-
12 domain
and/or the IL-18R chains recognized by the IL-18 domain, and the target
disease cells, b)
activating the immune cells via signaling of the IL-15R, IL-12R and/or IL-18R,
and c)
killing the target disease cells by the activated immune cells.
In an embodiment, the target cells are tumor cells or infected cells.
In an aspect, the present disclosure provides a method of enhancing immune
responses in a subject, comprising: a) contacting a plurality of cells with
any of the above
soluble fusion protein complexes, where the plurality of cells further include
immune cells
comprising the IL-15R chains recognized by the IL-15 domain, the IL-12R chains
recognized
by the IL-12 domain and/or the IL-18R chains recognized by the IL-18 domain,
b) activating
the immune cells via signaling of the IL-15R, IL-12R and/or IL-18R, c)
administering (or
adoptively transfer) to the patient the activated immune cells; and d)
enhancing immune
responses in the patient.
In an aspect, the present disclosure provides a method of preventing or
treating
disease in a patient, comprising: a) contacting a plurality of cells with a
soluble fusion protein
complex, wherein the plurality of cells further include immune cells
comprising the IL-15R
chains recognized by the IL-15 domain, the IL-12R chains recognized by the IL-
12 domain
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
and/or the IL-18R chains recognized by the IL-18 domain, b) activating the
immune cells via
signaling of the IL-15R, IL-12R and/or IL-18R, c) administering (or adoptively
transfer) an
effective amount of the activated immune cells to the patient, and d) damaging
or killing the
disease cells via the activated immune cells sufficient to prevent or treat
the disease in the
patient.
In an embodiment, the disease is a neoplasia or infectious disease.
In an aspect, the present disclosure provides a method of enhancing immune
responses in a subject comprising administering to the subject an effective
amount of any of
the above soluble fusion protein complexes.
In an aspect, the present disclosure provides a method for treating a
neoplasia or
infectious disease in a subject in need thereof comprising administering to
said subject an
effective amount of a pharmaceutical composition comprising any of the above
soluble fusion
protein complexes, thereby treating said neoplasia or infectious disease.
In an embodiment, the neoplasia is selected from the group consisting of a
glioblastoma, prostate cancer, hematological cancer, B-cell neoplasms,
multiple myeloma, B-
cell lymphoma, B cell non-Hodgkin lymphoma, Hodgkin's lymphoma, chronic
lymphocytic
leukemia, acute myeloid leukemia, cutaneous T-cell lymphoma, T-cell lymphoma,
a solid
tumor, urothelial/bladder carcinoma, melanoma, lung cancer, renal cell
carcinoma, breast
cancer, gastric and esophageal cancer, prostate cancer, pancreatic cancer,
colorectal cancer,
ovarian cancer, non-small cell lung carcinoma, and squamous cell head and neck
carcinoma.
In an embodiment, the immune cells are NK cells or cytokine induced memory
like
(CIML) NK cells.
In an embodiment, the effective amounts of the activated immune cells are
between 1
x 104 cells/kg and 1 x 1010 cells/kg.
In an embodiment, the immune cells are administered at least one time per
week.
In an embodiment, the effective amount is between about 1 and 100 fig/kg said
fusion
protein complex.
In an embodiment, the fusion protein complex is administered at least one time
per
week.
In an embodiment, the fusion protein complex increases immune cell
proliferation,
activation markers, cytotoxicity against target cells, and/or production of
pro inflammatory
cytokines, including IFN-y.
11
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
Preferably, the fusion protein complex increases serum levels of interferon
gamma
(IFN-y), and/or stimulates CD4+ and CD8+ T cells and NK cells to kill diseased
cells or tumor
cells in a subject.
Unless defined otherwise, all technical and scientific terms used herein have
the
meaning commonly understood by a person skilled in the art to which this
invention belongs.
The following references provide one of skill with a general definition of
many of the terms
used in this invention: Singleton et al., Dictionary of Microbiology and
Molecular Biology
(2nd ed. 1994); The Cambridge Dictionary of Science and Technology (Walker
ed., 1988);
The Glossary of Genetics, 5th Ed., R. Rieger et al. (eds.), Springer Verlag
(1991); and Hale &
Marham, The Harper Collins Dictionary of Biology (1991). As used herein, the
following
terms have the meanings ascribed to them below, unless specified otherwise.
By "agent" is meant a peptide, nucleic acid molecule, or small compound.
By "TxM" is meant a fusion protein complex comprising an IL-15N72D:IL-
15RaSu/Fc scaffold linked to a binding domain (FIG. 1A, 1B). An exemplary TxM
is an IL-
15N72D:IL-15RaSu fusion protein complex comprising fusions to IL-12 and IL-18
cytokines.
By "ameliorate" is meant decrease, suppress, attenuate, diminish, arrest, or
stabilize
the development or progression of a disease.
By "analog" is meant a molecule that is not identical, but has analogous
functional or
structural features. For example, a polypeptide analog retains the biological
activity of a
corresponding naturally-occurring polypeptide, while having certain
biochemical
modifications that enhance the analog's function relative to a naturally
occurring polypeptide.
Such biochemical modifications could increase the analog's protease
resistance, membrane
permeability, or half-life, without altering, for example, ligand binding. An
analog may
include an unnatural amino acid.
The invention includes antibodies or fragments of such antibodies, so long as
they
exhibit the desired biological activity. Also included in the invention are
chimeric antibodies,
such as humanized antibodies. Generally, a humanized antibody has one or more
amino acid
residues introduced into it from a source that is non-human. Humanization can
be performed,
for example, using methods described in the art, by substituting at least a
portion of a rodent
complementarity-determining region for the corresponding regions of a human
antibody.
The term "antibody" or "immunoglobulin" is intended to encompass both
polyclonal
and monoclonal antibodies. The preferred antibody is a monoclonal antibody
reactive with
the antigen. The term "antibody" is also intended to encompass mixtures of
more than one
12
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
antibody reactive with the antigen (e.g., a cocktail of different types of
monoclonal antibodies
reactive with the antigen). The term "antibody" is further intended to
encompass whole
antibodies, biologically functional fragments thereof, single-chain
antibodies, and genetically
altered antibodies such as chimeric antibodies comprising portions from more
than one
species, bifunctional antibodies, antibody conjugates, humanized and human
antibodies.
Biologically functional antibody fragments, which can also be used, are those
peptide
fragments derived from an antibody that are sufficient for binding to the
antigen. "Antibody"
as used herein is meant to include the entire antibody as well as any antibody
fragments (e.g.
F(ab1)2, Fab', Fab, Fv) capable of binding the epitope, antigen or antigenic
fragment of
interest.
By "binding to" a molecule is meant having a physicochemical affinity for that
molecule.
The term "binding domain" is intended to encompass an antibody, single chain
antibody, Fab, Fv, T-cell receptor binding domain, ligand binding domain,
receptor binding
domain, or other antigen-specific polypeptides known in the art.
As used herein, the term "biologically active polypeptide" or "effector
molecule" is
meant an amino acid sequence such as a protein, polypeptide or peptide; a
sugar or
polysaccharide; a lipid or a glycolipid, glycoprotein, or lipoprotein that can
produce the
desired effects as discussed herein. Effector molecules also include chemical
agents. Also
contemplated are effector molecule nucleic acids encoding a biologically
active or effector
protein, polypeptide, or peptide. Thus, suitable molecules include regulatory
factors,
enzymes, antibodies, or drugs as well as DNA, RNA, and oligonucleotides. The
biologically
active polypeptides or effector molecule can be naturally-occurring or it can
be synthesized
from known components, e.g., by recombinant or chemical synthesis and can
include
heterologous components. A biologically active polypeptide or effector
molecule is generally
between about 0.1 to 100 KD or greater up to about 1000 KD, preferably between
about 0.1,
0.2, 0.5, 1, 2, 5, 10, 20, 30 and 50 KD as judged by standard molecule sizing
techniques such
as centrifugation or SDS-polyacrylamide gel electrophoresis. Desired effects
of the invention
include, but are not limited to, for example, forming a fusion protein complex
of the
invention with increased binding activity, killing a target cell, e.g. either
to induce cell
proliferation or cell death, initiate an immune response, in preventing or
treating a disease, or
to act as a detection molecule for diagnostic purposes. For such detection, an
assay could be
used, for example an assay that includes sequential steps of culturing cells
to proliferate
13
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
same, and contacting the cells with a fusion protein complex of the invention
and then
evaluating whether the fusion protein complex inhibits further development of
the cells.
Covalently linking the effector molecule to the fusion protein complexes of
the
invention in accordance with the invention provides a number of significant
advantages.
Fusion protein complexes of the invention can be produced that contain a
single effector
molecule, including a peptide of known structure. Additionally, a wide variety
of effector
molecules can be produced in similar DNA vectors. That is, a library of
different effector
molecules can be linked to the fusion protein complexes for recognition of
infected or
diseased cells. Further, for therapeutic applications, rather than
administration of the fusion
protein complex of the invention to a subject, a DNA expression vector coding
for the fusion
protein complex can be administered for in vivo expression of the fusion
protein complex.
Such an approach avoids costly purification steps typically associated with
preparation of
recombinant proteins and avoids the complexities of antigen uptake and
processing
associated with conventional approaches.
As noted, components of the fusion proteins disclosed herein, e.g., effector
molecule
such as cytokines, chemokines, growth factors, protein toxins, immunoglobulin
domains or
other bioactive molecules and any peptide linkers, can be organized in nearly
any fashion
provided that the fusion protein has the function for which it was intended.
In particular,
each component of the fusion protein can be spaced from another component by
at least one
suitable peptide linker sequence if desired. Additionally, the fusion proteins
may include
tags, e.g., to facilitate modification, identification and/or purification of
the fusion protein.
More specific fusion proteins are in the Examples described below.
"Detect" refers to identifying the presence, absence or amount of the analyte
to be
detected.
By "disease" is meant any condition or disorder that damages or interferes
with the
normal function of a cell, tissue, or organ. Examples of diseases include
neoplasias and viral
infections.
By the terms "effective amount" and "therapeutically effective amount" of a
formulation or formulation component is meant a sufficient amount of the
formulation or
component, alone or in a combination, to provide the desired effect. For
example, by "an
effective amount" is meant an amount of a compound, alone or in a combination,
required to
ameliorate the symptoms of a disease relative to an untreated patient. The
effective amount
of active compound(s) used to practice the present invention for therapeutic
treatment of a
disease varies depending upon the manner of administration, the age, body
weight, and
14
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
general health of the subject. Ultimately, the attending physician or
veterinarian will decide
the appropriate amount and dosage regimen. Such amount is referred to as an
"effective"
amount.
By "fragment" is meant a portion of a polypeptide or nucleic acid molecule.
This
portion contains, preferably, at least 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%,
or 90% of
the entire length of the reference nucleic acid molecule or polypeptide. For
example, a
fragment may contain 10, 20, 30, 40, 50, 60, 70, 80, 90, or 100, 200, 300,
400, 500, 600, 700,
800, 900, or 1000 nucleotides or amino acids. However, the invention also
comprises
polypeptides and nucleic acid fragments, so long as they exhibit the desired
biological
activity of the full length polypeptides and nucleic acid, respectively. A
nucleic acid
fragment of almost any length is employed. For example, illustrative
polynucleotide
segments with total lengths of about 10,000, about 5,000, about 3,000, about
2,000, about
1,000, about 500, about 200, about 100, about 50 base pairs in length
(including all
intermediate lengths) are included in many implementations of this invention.
Similarly, a
polypeptide fragment of almost any length is employed. For example,
illustrative polypeptide
segments with total lengths of about 10,000, about 5,000, about 3,000, about
2,000, about
1,000, about 5,000, about 1,000, about 500, about 200, about 100, or about 50
amino acids in
length (including all intermediate lengths) are included in many
implementations of this
invention.
The terms "isolated", "purified", or "biologically pure" refer to material
that is free to
varying degrees from components which normally accompany it as found in its
native state.
"Isolate" denotes a degree of separation from original source or surroundings.
"Purify"
denotes a degree of separation that is higher than isolation.
A "purified" or "biologically pure" protein is sufficiently free of other
materials such
that any impurities do not materially affect the biological properties of the
protein or cause
other adverse consequences. That is, a nucleic acid or peptide of this
invention is purified if it
is substantially free of cellular material, viral material, or culture medium
when produced by
recombinant DNA techniques, or chemical precursors or other chemicals when
chemically
synthesized. Purity and homogeneity are typically determined using analytical
chemistry
techniques, for example, polyacrylamide gel electrophoresis or high
performance liquid
chromatography. The term "purified" can denote that a nucleic acid or protein
gives rise to
essentially one band in an electrophoretic gel. For a protein that can be
subjected to
modifications, for example, phosphorylation or glycosylation, different
modifications may
give rise to different isolated proteins, which can be separately purified.
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
Similarly, by "substantially pure" is meant a nucleotide or polypeptide that
has been
separated from the components that naturally accompany it. Typically, the
nucleotides and
polypeptides are substantially pure when they are at least 60%, 70%, 80%, 90%,
95%, or
even 99%, by weight, free from the proteins and naturally-occurring organic
molecules with
they are naturally associated.
By "isolated nucleic acid" is meant a nucleic acid that is free of the genes
which flank
it in the naturally-occurring genome of the organism from which the nucleic
acid is derived.
The term covers, for example: (a) a DNA which is part of a naturally occurring
genomic
DNA molecule, but is not flanked by both of the nucleic acid sequences that
flank that part of
the molecule in the genome of the organism in which it naturally occurs; (b) a
nucleic acid
incorporated into a vector or into the genomic DNA of a prokaryote or
eukaryote in a
manner, such that the resulting molecule is not identical to any naturally
occurring vector or
genomic DNA; (c) a separate molecule such as a cDNA, a genomic fragment, a
fragment
produced by polymerase chain reaction (PCR), or a restriction fragment; and
(d) a
recombinant nucleotide sequence that is part of a hybrid gene, i.e., a gene
encoding a fusion
protein. Isolated nucleic acid molecules according to the present invention
further include
molecules produced synthetically, as well as any nucleic acids that have been
altered
chemically and/or that have modified backbones. For example, the isolated
nucleic acid is a
purified cDNA or RNA polynucleotide. Isolated nucleic acid molecules also
include
messenger ribonucleic acid (mRNA) molecules.
By an "isolated polypeptide" is meant a polypeptide of the invention that has
been
separated from components that naturally accompany it. Typically, the
polypeptide is
isolated when it is at least 60%, by weight, free from the proteins and
naturally-occurring
organic molecules with which it is naturally associated. Preferably, the
preparation is at least
75%, more preferably at least 90%, and most preferably at least 99%, by
weight, a
polypeptide of the invention. An isolated polypeptide of the invention may be
obtained, for
example, by extraction from a natural source, by expression of a recombinant
nucleic acid
encoding such a polypeptide; or by chemically synthesizing the protein. Purity
can be
measured by any appropriate method, for example, column chromatography,
polyacrylamide
gel electrophoresis, or by HPLC analysis.
By "marker" is meant any protein or polynucleotide having an alteration in
expression
level or activity that is associated with a disease or disorder.
By "neoplasia" is meant a disease or disorder characterized by excess
proliferation or
reduced apoptosis. Illustrative neoplasms for which the invention can be used
include, but
16
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
are not limited to leukemias (e.g., acute leukemia, acute lymphocytic
leukemia, acute
myelocytic leukemia, acute myeloblastic leukemia, acute promyelocytic
leukemia, acute
myelomonocytic leukemia, acute monocytic leukemia, acute erythroleukemia,
chronic
leukemia, chronic myelocytic leukemia, chronic lymphocytic leukemia),
polycythemia vera,
lymphoma (Hodgkin's disease, non-Hodgkin's disease), Waldenstrom's
macroglobulinemia,
heavy chain disease, and solid tumors such as sarcomas and carcinomas (e.g.,
fibrosarcoma,
myxosarcoma, liposarcoma, chondrosarcoma, osteogenic sarcoma, chordoma,
angiosarcoma,
endotheliosarcoma, lymphangiosarcoma, lymphangioendotheliosarcoma, synovioma,
mesothelioma, Ewing's tumor, leiomyosarcoma, rhabdomyosarcoma, colon
carcinoma,
pancreatic cancer, breast cancer, ovarian cancer, prostate cancer, squamous
cell carcinoma,
basal cell carcinoma, adenocarcinoma, sweat gland carcinoma, sebaceous gland
carcinoma,
papillary carcinoma, papillary adenocarcinomas, cystadenocarcinoma, medullary
carcinoma,
bronchogenic carcinoma, renal cell carcinoma, hepatoma, nile duct carcinoma,
choriocarcinoma, seminoma, embryonal carcinoma, Wilm's tumor, cervical cancer,
uterine
cancer, testicular cancer, lung carcinoma, small cell lung carcinoma, bladder
carcinoma,
epithelial carcinoma, glioma, glioblastoma multiforme, astrocytoma,
medulloblastoma,
craniopharyngioma, ependymoma, pinealoma, hemangioblastoma, acoustic neuroma,
oligodenroglioma, schwannoma, meningioma, melanoma, neuroblastoma, and
retinoblastoma). In particular embodiments, the neoplasia is multiple myeloma,
beta-cell
lymphoma, urothelial/bladder carcinoma or melanoma. As used herein,
"obtaining" as in
"obtaining an agent" includes synthesizing, purchasing, or otherwise acquiring
the agent.
By "reduces" is meant a negative alteration of at least 5%, 10%, 25%, 50%,
75%, or
100%.
By "reference" is meant a standard or control condition.
A "reference sequence" is a defined sequence used as a basis for sequence
comparison. A reference sequence may be a subset of or the entirety of a
specified sequence;
for example, a segment of a full-length cDNA or gene sequence, or the complete
cDNA or
gene sequence. For polypeptides, the length of the reference polypeptide
sequence will
generally be at least about 16 amino acids, preferably at least about 20 amino
acids, more
preferably at least about 25 amino acids, and even more preferably about 35
amino acids,
about 50 amino acids, or about 100 amino acids. For nucleic acids, the length
of the
reference nucleic acid sequence will generally be at least about 50
nucleotides, preferably at
least about 60 nucleotides, more preferably at least about 75 nucleotides, and
even more
17
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
preferably about 100 nucleotides or about 300 nucleotides or any integer
thereabout or
therebetween.
By "specifically binds" is meant a compound or antibody that recognizes and
binds a
polypeptide of the invention, but which does not substantially recognize and
bind other
molecules in a sample, for example, a biological sample, which naturally
includes a
polypeptide of the invention.
Nucleic acid molecules useful in the methods of the invention include any
nucleic
acid molecule that encodes a polypeptide of the invention or a fragment
thereof Such
nucleic acid molecules need not be 100% identical with an endogenous nucleic
acid
sequence, but will typically exhibit substantial identity. Polynucleotides
having "substantial
identity" to an endogenous sequence are typically capable of hybridizing with
at least one
strand of a double-stranded nucleic acid molecule. Nucleic acid molecules
useful in the
methods of the invention include any nucleic acid molecule that encodes a
polypeptide of the
invention or a fragment thereof Such nucleic acid molecules need not be 100%
identical
with an endogenous nucleic acid sequence, but will typically exhibit
substantial identity.
Polynucleotides having "substantial identity" to an endogenous sequence are
typically
capable of hybridizing with at least one strand of a double-stranded nucleic
acid molecule.
By "hybridize" is meant pair to form a double-stranded molecule between
complementary
polynucleotide sequences (e.g., a gene described herein), or portions thereof,
under various
conditions of stringency. (See, e.g., Wahl, G. M. and S. L. Berger (1987)
Methods Enzymol.
152:399; Kimmel, A. R. (1987) Methods Enzymol. 152:507).
For example, stringent salt concentration will ordinarily be less than about
750 mM
NaCl and 75 mM trisodium citrate, preferably less than about 500 mM NaCl and
50 mM
trisodium citrate, and more preferably less than about 250 mM NaCl and 25 mM
trisodium
citrate. Low stringency hybridization can be obtained in the absence of
organic solvent, e.g.,
formamide, while high stringency hybridization can be obtained in the presence
of at least
about 35% formamide, and more preferably at least about 50% formamide.
Stringent
temperature conditions will ordinarily include temperatures of at least about
30 C, more
preferably of at least about 37 C, and most preferably of at least about 42
C. Varying
additional parameters, such as hybridization time, the concentration of
detergent, e.g., sodium
dodecyl sulfate (SDS), and the inclusion or exclusion of carrier DNA, are well
known to
those skilled in the art. Various levels of stringency are accomplished by
combining these
various conditions as needed. In a preferred: embodiment, hybridization will
occur at 30 C
in 750 mM NaCl, 75 mM trisodium citrate, and 1% SDS. In a more preferred
embodiment,
18
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
hybridization will occur at 37 C in 500 mM NaCl, 50 mM trisodium citrate, 1%
SDS, 35%
formamide, and 100 lig /ml denatured salmon sperm DNA (ssDNA). In a most
preferred
embodiment, hybridization will occur at 42 C in 250 mM NaCl, 25 mM trisodium
citrate,
1% SDS, 50% formamide, and 200 ug/m1 ssDNA. Useful variations on these
conditions will
be readily apparent to those skilled in the art.
For most applications, washing steps that follow hybridization will also vary
in
stringency. Wash stringency conditions can be defined by salt concentration
and by
temperature. As above, wash stringency can be increased by decreasing salt
concentration or
by increasing temperature. For example, stringent salt concentration for the
wash steps will
preferably be less than about 30 mM NaCl and 3 mM trisodium citrate, and most
preferably
less than about 15 mM NaCl and 1.5 mM trisodium citrate. Stringent temperature
conditions
for the wash steps will ordinarily include a temperature of at least about 25
C, more
preferably of at least about 42 C, and even more preferably of at least about
68 C. In a
preferred embodiment, wash steps will occur at 25 C in 30 mM NaCl, 3 mM
trisodium
citrate, and 0.1% SDS. In a more preferred embodiment, wash steps will occur
at 42 C in 15
mM NaCl, 1.5 mM trisodium citrate, and 0.1% SDS. In a more preferred
embodiment, wash
steps will occur at 68 C in 15 mM NaCl, 1.5 mM trisodium citrate, and 0.1%
SDS.
Additional variations on these conditions will be readily apparent to those
skilled in the art.
Hybridization techniques are well known to those skilled in the art and are
described, for
example, in Benton and Davis (Science 196:180, 1977); Grunstein and Hogness
(Proc. Natl.
Acad. Sci., USA 72:3961, 1975); Ausubel et al. (Current Protocols in Molecular
Biology,
Wiley Interscience, New York, 2001); Berger and Kimmel (Guide to Molecular
Cloning
Techniques, 1987, Academic Press, New York); and Sambrook et al., Molecular
Cloning: A
Laboratory Manual, Cold Spring Harbor Laboratory Press, New York.
By "substantially identical" is meant a polypeptide or nucleic acid molecule
exhibiting at least 50% identity to a reference amino acid sequence (for
example, any one of
the amino acid sequences described herein) or nucleic acid sequence (for
example, any one of
the nucleic acid sequences described herein). Preferably, such a sequence is
at least 60%,
more preferably 80% or 85%, and more preferably 90%, 95% or even 99% identical
at the
amino acid level or nucleic acid to the sequence used for comparison.
Sequence identity is typically measured using sequence analysis software (for
example, Sequencher, Gene Codes Corporation, 775 Technology Drive, Ann Arbor,
MI;
Vector NTI, Life Technologies, 3175 Staley Rd. Grand Island, NY). Such
software matches
identical or similar sequences by assigning degrees of homology to various
substitutions,
19
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
deletions, and/or other modifications. Conservative substitutions typically
include
substitutions within the following groups: glycine, alanine; valine,
isoleucine, leucine;
aspartic acid, glutamic acid, asparagine, glutamine; serine, threonine;
lysine, arginine; and
phenylalanine, tyrosine. In an exemplary approach to determining the degree of
identity, a
BLAST program may be used, with a probability score between e' and e-th
indicating a
closely related sequence.
By "subject" is meant a mammal, including, but not limited to, a human or non-
human mammal, such as a bovine, equine, canine, ovine, or feline. The subject
is preferably
a mammal in need of such treatment, e.g., a subject that has been diagnosed
with B cell
lymphoma or a predisposition thereto. The mammal is any mammal, e.g., a human,
a
primate, a mouse, a rat, a dog, a cat, a horse, as well as livestock or
animals grown for food
consumption, e.g., cattle, sheep, pigs, chickens, and goats. In a preferred
embodiment, the
mammal is a human.
Ranges provided herein are understood to be shorthand for all of the values
within the
range. For example, a range of 1 to 50 is understood to include any number,
combination of
numbers, or sub-range from the group consisting 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 13, 14, 15,
16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34,
35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48, 49, or 50.
The terms "treating" and "treatment" as used herein refer to the
administration of an
agent or formulation to a clinically symptomatic individual afflicted with an
adverse
condition, disorder, or disease, so as to affect a reduction in severity
and/or frequency of
symptoms, eliminate the symptoms and/or their underlying cause, and/or
facilitate
improvement or remediation of damage. It will be appreciated that, although
not precluded,
treating a disorder or condition does not require that the disorder, condition
or symptoms
associated therewith be completely eliminated. Agents or formulations used in
treatment
may comprise cells or tissues.
Treatment of patients with neoplasia may include any of the following:
Adjuvant
therapy (also called adjunct therapy or adjunctive therapy) to destroy
residual tumor cells that
may be present after the known tumor is removed by the initial therapy (e.g.
surgery), thereby
preventing possible cancer reoccurrence; neoadjuvant therapy given prior to
the surgical
procedure to shrink the cancer; induction therapy to cause a remission,
typically for acute
leukemia; consolidation therapy (also called intensification therapy) given
once a remission is
achieved to sustain the remission; maintenance therapy given in lower or less
frequent doses
to assist in prolonging a remission; first line therapy (also called standard
therapy); second (or
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
3rd, 4th, etc.) line therapy (also called salvage therapy) is given if a
disease has not responded
or reoccurred after first line therapy; and palliative therapy (also called
supportive therapy) to
address symptom management without expecting to significantly reduce the
cancer.
The terms "preventing" and "prevention" refer to the administration of an
agent or
composition to a clinically asymptomatic individual who is susceptible or
predisposed to a
particular adverse condition, disorder, or disease, and thus relates to the
prevention of the
occurrence of symptoms and/or their underlying cause.
Unless specifically stated or obvious from context, as used herein, the term
"or" is
understood to be inclusive. Unless specifically stated or obvious from
context, as used
herein, the terms "a", "an", and "the" are understood to be singular or
plural.
Unless specifically stated or obvious from context, as used herein, the term
"about" is
understood as within a range of normal tolerance in the art, for example
within 2 standard
deviations of the mean. About can be understood as within 10%, 9%, 8%, 7%, 6%,
5%, 4%,
3%, 2%, 1%, 0.5%, 0.1%, 0.05%, or 0.01% of the stated value. Unless otherwise
clear from
context, all numerical values provided herein are modified by the term about.
The recitation of a listing of chemical groups in any definition of a variable
herein
includes definitions of that variable as any single group or combination of
listed groups. The
recitation of an embodiment for a variable or aspect herein includes that
embodiment as any
single embodiment or in combination with any other embodiments or portions
thereof
Any compositions or methods provided herein can be combined with one or more
of
any of the other compositions and methods provided herein.
The transitional term "comprising," which is synonymous with "including,"
"containing," or "characterized by," is inclusive or open-ended and does not
exclude
additional, unrecited elements or method steps. By contrast, the transitional
phrase
"consisting of' excludes any element, step, or ingredient not specified in the
claim. The
transitional phrase "consisting essentially of' limits the scope of a claim to
the specified
materials or steps "and those that do not materially affect the basic and
novel
characteristic(s)" of the claimed invention.
Other features and advantages of the invention will be apparent from the
following
description of the preferred embodiments thereof, and from the claims. Unless
otherwise
defined, all technical and scientific terms used herein have the same meaning
as commonly
understood by one of ordinary skill in the art to which this invention
belongs. Although
methods and materials similar or equivalent to those described herein can be
used in the
practice or testing of the present invention, suitable methods and materials
are described
21
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
below. All published foreign patents and patent applications cited herein are
incorporated
herein by reference.
Genbank and NCBI submissions indicated by accession number cited herein are
incorporated herein by reference. All other published references, documents,
manuscripts and
scientific literature cited herein are incorporated herein by reference. In
the case of conflict,
the present specification, including definitions, will control. In addition,
the materials,
methods, and examples are illustrative only and not intended to be limiting.
BRIEF DESCRIPTION OF THE DRAWINGS
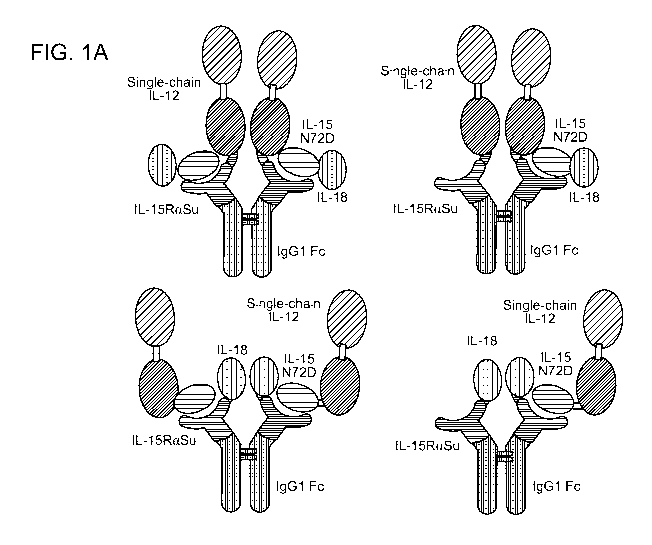
FIG. 1A is a schematic diagram illustrating different TxM fusion protein
complexes
comprising the IL-15N72D:IL-15RaSu/Fc scaffold fused to IL-12 and IL-18
binding
domains. In some cases, the dimeric IL-15RaSu/Fc fusion protein complexes
comprise one
or two IL-15N72D fusion proteins. FIG. 1B is a schematic diagram illustrating
different
TxM fusion protein complexes comprising the IL-15N72D:IL-15RaSu/Fc scaffold
fused to
IL-18 binding domains.
FIG. 2A is a line graph showing the chromatographic profile of hIL18/IL12/TxM
protein-containing cell culture supernatant following binding and elution on a
Protein A
resin. FIG. 2B is a line graph showing the chromatographic profile of Protein
A-purified
hIL18/IL12/TxM protein following elution on a preparative size exclusion
column. FIG. 2C
is a line graph showing the chromatographic profile of Protein A/SEC-purified
hIL18/IL12/TxM protein following elution on an analytical size exclusion
column,
demonstrating separation of monomeric multiprotein hIL18/IL12/TxM fusion
protein
complexes from protein aggregates.
FIG. 3 is a photograph showing a sodium dodecyl sulfate polyacrylamide gel (4
¨
12%) electrophoresis (SDS-PAGE) analysis of the hIL18/IL12/TxM fusion protein
complex
following disulfide bond reduction. Left lane: See Blue Plus2 marker, right
lane: Protein A-
purified hIL18/IL12/TxM.
FIG. 4A is a line graph showing the binding activity of the hIL18/IL12/TxM
fusion
protein complex to antibodies specific to human IL-15 and human IgG. FIG. 4B
is a line
graph showing the binding activity of the hIL12/IL18/TxM fusion protein
complex to
antibodies specific to human IL-15 and human IgG. FIG. 4C is a line graph
showing the
binding activity of the two headed IL18/TxM fusion protein complex to
antibodies specific to
22
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
human IL-15 and human IL-18. Controls include anti-CD20 TxM (2B8T2M), ALT-803
and
hIL12/IL18/TxM depending on the assay format.
FIG. 5 is a line graph illustrating the proliferation of IL-15-dependent 32D13
cells
mediated by hIL18/IL12/TxM fusion protein complex compared to ALT-803.
FIG. 6 is a line graph further illustrating the proliferation of IL-15-
dependent 32E43
cells mediated by hIL18/IL12/TxM fusion protein complex compared to ALT-803.
FIG. 7 is a line graph further illustrating the activation of IL-18-sensitive
HEK18
reporter cells mediated by hIL18/IL12/TxM fusion protein complex compared to
IL-18.
FIG. 8 is a line graph further illustrating the activation of IL-12-sensitive
HEK12
reporter cells mediated by hIL18/IL12/TxM fusion protein complex compared to
IL-12.
FIG. 9A and 9B are line graphs showing IL-12 biological activity of
hIL18/IL12/TxM
(FIG. 9A) or a combination of recombinant IL-12, IL-18 and ALT-803
(rIL12+rIL18+ALT-
803) (FIG. 9B) (red lines) compared to media control (black lines) in
stimulating
phosphorylation of STAT4 in aNK cells. FIG. 9C and 9D are line graphs showing
IL-18
biological activity of hIL18/IL12/TxM (A) or a combination of recombinant IL-
12, IL-18 and
ALT-803 (rIL12+rIL18+ALT-803) (B) (red lines) compared to media control (black
lines) in
stimulating phosphorylation of p38 MAPK in purified human NK cells. FIG. 9E
and 9F are
line graphs showing IL-15 biological activity of hIL18/IL12/TxM (A) or a
combination of
recombinant IL-12, IL-18 and ALT-803 (rIL12+rIL18+ALT-803) (B) (red lines)
compared to
media control (black lines) in stimulating phosphorylation of STAT5 in aNK
cells.
FIG. 10A is a bar chart illustrating the combined cytokine immunostimulatory
activity
of hIL18/IL12/TxM fusion protein complex compared to cytokines alone or in
combination
to induce IFN-y production by aNK cells. FIG. 10B is a line graph illustrating
the cytokine
immunostimulatory activity of two headed IL18/TxM fusion protein complex
compared to
ALT-803 or hIL18/IL12/TxM to induce IFN-y production by aNK cells.
FIG. 11A and 11B are line graphs showing biological activity of hIL18/IL12/TxM
(FIG. 11A) or a combination of recombinant IL-12, IL-18 and ALT-803
(rIL12+rIL18+ALT-
803) (FIG. 11B) (red lines) compared to media control (black lines) for
inducing CD25 by
purified human NK cells. FIG. 11C and 11D are line graphs showing biological
activity of
hIL18/IL12/TxM (FIG. 11A) or a combination of recombinant IL-12, IL-18 and ALT-
803
(rIL12+rIL18+ALT-803) (FIG. 11B) (red lines) compared to media control (black
lines) for
inducing CD69 by purified human NK cells. FIG. 11E and 11F are line graphs
showing
biological activity of hIL18/IL12/TxM (A) or a combination of recombinant IL-
12, IL-18 and
23
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
ALT-803 (rIL12+rIL18+ALT-803) (B) (red lines) compared to media control (black
lines)
for inducing intracellular IFN-y by purified human NK cells.
FIG. 12A is a line graph illustrating the induction of the activation marker
CD25 on
the surface of human NK cells mediated by hIL18/IL12/TxM fusion protein
complex
compared to IL-18+IL-12+ALT-803. FIG. 12B is a line graph illustrating the
induction of
intracellular IFN-y in human NK cells mediated by hIL18/IL12/TxM fusion
protein complex
compared to IL-18+IL-12+ALT-803.
FIG. 13A is a bar chart illustrating maintenance of CD25 on the surface of
human
CIML NK cells induced by priming with hIL18/IL12/TxM fusion protein complex
(compared to ALT-803) followed by resting in ALT-803. FIG. 13B is a bar chart
illustrating
enhanced levels of intracellular IFN-y in human CIML NK cells induced by
priming with
hIL18/IL12/TxM fusion protein complex (compared to ALT-803) followed by
resting in
ALT-803 and restimulation with IL-12+ALT-803 or 1(562 leukemia targets.
FIG. 14A show contour plots illustrating proliferation (CTV dilution) and IFN-
y
expression in human CIML NK cells induced by priming with hIL18/IL12/TxM
fusion
protein complex, individual cytokines or IL-18+IL-12+ALT-803 followed by
resting in IL-15
and restimulation with IL-12+ALT-803 compared to no restimulation. FIG. 14B
show
contour plots illustrating proliferation (CTV dilution) and IFN-y expression
in human CIML
NK cells induced by priming with hIL18/IL12/TxM fusion protein complex,
individual
cytokines or IL-18+IL-12+ALT-803 followed by resting in ALT-803 and
restimulation with
IL-12+ALT-803 compared to no restimulation.
FIG. 15 show histogram plots illustrating proliferation (CTV dilution) in
human
CIML NK cells induced by priming with hIL18/IL12/TxM fusion protein complex,
individual cytokines or IL-18+IL-12+ALT-803 followed by resting in IL-15 or
ALT-803 and
restimulation with IL-12+ALT-803.
FIG. 16 is a bar chart illustrating the cytotoxicity of human NK cells against
MDA-
MB-231 human breast cancer cells induced by hIL18/IL12/TxM fusion protein
complex or
ALT-803 (IL-15N72D:IL-15Ra/Fc complex).
FIG. 17A is a bar graph illustrating the induction of intracellular granzyme B
in
human NK cells mediated by hIL18/IL12/TxM fusion protein complex compared to
ALT-
803 or no treatment. FIG. 17B is a bar graph illustrating direct cytotoxicity
(vehicle bars) or
antibody dependent cellular cytotoxicity (aTF Ab bars) of human NK cells
against Tissue
Factor-positive SW1990 human pancreatic adenocarcinoma cells following priming
with
hIL18/IL12/TxM fusion protein complex compared to media alone or ALT-803. FIG.
17C is
24
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
a bar graph illustrating increased expression of IFN-y by human NK cells
incubated with
SW1990 human pancreatic adenocarcinoma cells with anti-TF Ab or media alone
(vehicle)
following priming with hIL18/IL12/TxM fusion protein complex compared to media
alone or
ALT-803.
FIG. 18A is a bar graph showing changes in spleen weights following
administration
of hIL18/IL12/TxM (20 mg/kg) vs. PBS in C57BL/6 mice. FIG. 18B is a bar graph
showing
changes in the percentage of CD8 T cells and NK cells in the spleen of C57BL/6
mice
following administration of hIL18/IL12/TxM (20 mg/kg) compared to PBS
controls. FIG.
18C is a bar graph showing changes in the absolute CD8 T cell and NK cell
counts in the
blood of C57BL/6 mice following administration of hIL18/IL12/TxM (20 mg/kg)
compared
to PBS controls. FIG. 18D is a bar graph showing changes in the percentage of
CD8 T cells
and NK cells in the blood of C57BL/6 mice following administration of
hIL18/IL12/TxM (20
mg/kg) compared to PBS controls.
DETAILED DESCRIPTION
Therapies employing natural killer (NK) cells and T cells have emerged as
potential
treatments for cancer and viral infections due to the ability of these cells
to kill diseased cells
and release pro-inflammatory cytokines (See, e.g., Fehniger TA and Cooper MA.
Trends
Immunol. 2016; 37:877-888; and Cerwenka A and Lanier LL. Nat Rev Immunol. 2016
16:112-23). Of particular interest are cytokine-induced memory-like (CIML)
natural killer
(NK) cells, which exhibit long-lasting non-antigen-specific NK cell effector
function. These
cells can be induced ex vivo following overnight stimulation of purified NK
cells with
saturating amounts of interleukin-12 (IL-12, 10 ng/ml), IL-15 (50 ng/ml), and
IL-18 (50
ng/ml). These primed NK cells exhibit memory like properties such as 1)
enhanced
proliferation, 2) expression of IL-2 receptor a (IL-2Ra, CD25), perforin,
granzymes, and
other activation markers, and 3) increased interferon-y (IFN-y) production
following re-
stimulation.
Initial therapeutic evaluation of CIML NK cells in a first-in-human phase 1
clinical
trial utilized ex vivo IL-12/IL-15/IL-18 stimulation of allogeneic
haploidentical NK cells
followed by adoptive transfer of the CIML NK cells into patients with relapsed
or refractory
acute myeloid leukemia (AML) who had been preconditioned with cyclophosphamide
and
fludarabine. Following transfer, patients received low dose IL-2 to support
the cells in vivo.
These transferred, primed NK cells peaked in frequency between 7 and 14 days
after
infusion, comprising greater than 90% of all NK cells in the blood 7 days
after transfer. Of
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
the nine evaluable patients at the time of publication, four had a complete
remission, in
addition to one patient having a morphologic leukemia free state, suggesting
promising
therapeutic activity mediated by the adoptively transferred CIML NK cells
(See, Romee, R,
et al. Sci Transl Med. 2016; 8:357ra123, incorporated herein by reference).
Prior to the invention described herein, optimal methods for generating CIML
NK
cells were not fully elucidated. Prior to the invention described herein,
strategies employed
recombinant human IL-12 (produced in insect cells), human IL-18 (produced in
E. coil), and
human IL-15 (produced in E. coil), which differ in glycosylation and
potentially other post-
transcriptional modifications compared to mammalian cell-produced cytokines.
The
recombinant cytokines may also have different purity and stability and are not
generally
available as clinical grade material. Additionally, each cytokine is expected
to have unique
receptor binding, internalization and recycling properties.
Accordingly, described herein are multi-specific IL-15-based fusion protein
complexes comprising IL-12 and IL-18 binding domains (FIG. 1A, 1B).
Specifically,
described herein are fusion protein complexes comprising an IL-15N72D:IL-
15RaSu-Ig Fc
scaffold fused to IL-12 and IL-18 binding domains. When characterized using
human
immune cells, these fusion protein complexes exhibit binding and biological
activity of each
of the IL-15, IL-12 and IL-18 cytokines. Additionally, these fusion protein
complexes act to
induce CIML NK cells with elevated activation markers, increased cytotoxicity
against tumor
cells and enhanced production of IFN-y. Thus, the fusion protein complex as a
single
molecule binds to and signals via multiple cytokine receptors on NK cells to
provide the
synergistic responses previously only observed with a combination of multiple
individual
cytokines. Additionally, these fusion protein complexes comprise the Fc region
of Ig
molecules, which can form a dimer to provide a soluble multi-polypeptide
complex, bind
Protein A for the purpose of purification and interact with Fcy receptors on
NK cells and
macrophages, thus providing advantages to the fusion protein complex that are
not present in
the combination of individual cytokines. Mammalian cell expression-based
methods for
making these fusion protein complexes suitable for large scale production of
clinical grade
material are described herein. Additional methods for making and using CIML NK
cells
induced by the fusion protein complex of the invention are also provided.
Interleukin-15
Interleukin-15 (IL-15) is an important cytokine for the development,
proliferation, and
activation of effector NK cells and CD8+ memory T cells. IL-15 binds to the IL-
15 receptor
26
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
a (IL-15Ra) and is presented in trans to the IL-2/IL-15 receptor (3 - common y
chain (IL-
15Rf3y,) complex on effector cells. IL-15 and IL-2 share binding to the IL-
15Rf3y,, and signal
through STAT3 and STAT5 pathways. However, unlike IL-2, IL-15 does not support
maintenance of CD4+CD25+FoxP3+ regulatory T (Treg) cells or induce cell death
of activated
CD8+ T cells, effects that may have limited the therapeutic activity of IL-2
against multiple
myeloma. Additionally, IL-15 is the only cytokine known to provide anti-
apoptotic signaling
to effector CD8+ T cells. IL-15, either administered alone or as a complex
with the IL-15Ra,
exhibits potent anti-tumor activities against well-established solid tumors in
experimental
animal models and, thus, has been identified as one of the most promising
immunotherapeutic drugs that could potentially cure cancer.
To facilitate clinical development of an IL-15-based cancer therapeutic, an IL-
15
mutant (IL-15N72D) with increased biological activity compared to IL-15 was
identified
(Zhu et al., J Immunol, 183: 3598-3607, 2009). The pharmacokinetics and
biological activity
of this IL-15 super-agonist (IL-15N72D) was further improved by the creation
of IL-
15N72D:IL-15Ra/Fc fusion protein complex (ALT-803), such that the super
agonist complex
has at least 25-times the activity of the native cytokine in vivo (Han et al.,
Cytokine, 56: 804-
810, 2011).
IL-15:IL-15Ra protein complex
As described above, an IL-15:IL-15Ra fusion protein complex can refer to a
complex
having IL-15 non-covalently bound to the soluble IL-15Ra domain of the native
IL-15Ra. In
some cases, the soluble IL-15Ra is covalently linked to a biologically active
polypeptide
and/or to an IgG Fc domain. The IL-15 can be either IL-15 or IL-15 covalently
linked to a
second biologically active polypeptide. The crystal structure of the IL-15:IL-
15Ra protein
complex is shown in Chirifu et al., 2007 Nat Immunol 8, 1001-1007,
incorporated herein by
reference.
In various embodiments of the above aspects or any other aspect of the
invention
delineated herein, the IL-15Ra fusion protein comprises soluble IL-15Ra, e.g.,
IL-15Ra
covalently linked to a biologically active polypeptide (e.g., the heavy chain
constant domain of
IgG, an Fc domain of the heavy chain constant domain of IgG, or a cytokine).
In other
embodiments of the invention of the above aspects, IL-15 comprises IL-15,
e.g., IL-15
covalently linked to a second biologically active polypeptide, e.g., a
cytokine. In other
embodiments, purifying the IL-15:IL-15Ra fusion protein complex from the host
cell or media
involves capturing the IL-15:IL-15Ra fusion protein complex on an affinity
reagent that
27
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
specifically binds the IL-15:IL-15Ra fusion protein complex. In other
embodiments, the IL-
15Ra fusion protein contains an IL-15Ra/Fc fusion protein and the affinity
reagent specifically
binds the Fc domain. In other embodiments, the affinity reagent is Protein A
or Protein G. In
other embodiments, the affinity reagent is an antibody. In other embodiments,
purifying the
IL-15:IL-15Ra fusion protein complex from the host cell or media comprises ion
exchange
chromatography. In other embodiments, purifying the IL-15:IL-15Ra fusion
protein complex
from the host cell or media comprises size exclusion chromatography.
In other embodiments, the IL-15Ra comprises IL-15RaSushi (IL-15RaSu). In other
embodiments, the IL-15 is a variant IL-15 (e.g., IL-15N72D). In other
embodiments, the IL-
15 binding sites of the IL-15:IL-15Ra fusion protein complex are fully
occupied. In other
embodiments, both IL-15 binding sites of the IL-15:IL-15RaSu/Fc fusion protein
complex are
fully occupied. In other embodiments, the IL-15:IL-15Ra fusion protein complex
is purified
based on the fusion protein complex charge or size properties. In other
embodiments, the fully
occupied IL-15N72D:IL-15RaSu/Fc fusion protein complex is purified by anion
exchange
chromatography based on the fusion protein complex charge properties. In other
embodiments,
the fully occupied IL-15N72D:IL-15RaSu/Fc fusion protein complex is purified
using a
quaternary amine-based resin with binding conditions employing low ionic
strength neutral
pH buffers and elution conditions employing buffers of increasing ionic
strength.
In certain embodiments, a soluble fusion protein complex comprises a first and
second
soluble protein, wherein: the first soluble protein comprises an interleukin-
15 (IL-15)
polypeptide domain linked to an IL-12 or IL-18 binding domain or functional
fragment
thereof; the second soluble protein comprises a soluble IL-15 receptor alpha
sushi-binding
domain (IL-15RaSu) fused to an immunoglobulin Fc domain, wherein the IL-15RaSu
domain is linked to an IL-12 or IL-18 binding domain or functional fragment
thereof; and, the
IL-15 polypeptide domain of the first soluble protein binds to the IL-15RaSu
domain of the
second soluble protein to form a soluble fusion protein complex.
In certain embodiments, an isolated soluble fusion protein complex comprises
an
interleukin-15 (IL-15) polypeptide domain linked to an IL-12 and/or IL-18
binding domain or
functional fragment thereof In certain embodiments, the IL-15 polypeptide
domain is an IL-
15 variant comprising an N72D mutation (IL-15N72D).
In certain embodiments, an isolated soluble fusion protein complex comprising
a
soluble IL-15 receptor alpha sushi-binding domain (IL-15RaSu) fused to an
immunoglobulin
Fc domain, wherein the IL-15RaSu domain is linked to an IL-12 and/or IL-18
binding
domain or functional fragment thereof
28
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
The isolated protein fusion complex can be a "two headed" fusion protein
complex.
These complexes can vary in their combination of IL-15, IL-15RaSu/Fc,
interleukins,
comprising, for example, IL-18/IL-15RaSu/Fc and IL-15N72D fusion proteins or
IL-
15RaSu/Fc and IL-18/IL-15N72D fusion proteins (FIG. 1B). Similarly, these
fusion protein
complexes comprise IL-12/IL-15RaSu/Fc and IL-15N72D fusion proteins or IL-
15RaSu/Fc
and IL-12/IL-15N72D fusion proteins. The combinations can be varied, as well
as the types
of molecules which include variants, mutants, homologs, analogs, modified
molecules and
the like.
Accordingly, in certain embodiments, an isolated soluble fusion protein
complex
comprises a first and second soluble protein, wherein the first soluble
protein comprises an
interleukin-15 receptor alpha sushi-binding domain (IL-15RaSu) fused to an
immunoglobulin
Fc domain, wherein the IL-15RaSu domain is fused to an IL-18 binding domain or
functional
fragment thereof; the second soluble protein comprises an interleukin-15 (IL-
15) polypeptide
domain fused to an IL-18 domain; wherein the IL-15 polypeptide domain of the
first soluble
protein binds to the IL-15RaSu domain of the second soluble protein to form a
soluble fusion
protein complex.
In other embodiments, an isolated soluble fusion protein complex comprises a
first
and second soluble protein, wherein the first soluble protein comprises an
interleukin-15
receptor alpha sushi-binding domain (IL-15RaSu) fused to an immunoglobulin Fc
domain,
wherein the IL-15RaSu domain is fused to an IL-12 binding domain or functional
fragment
thereof; the second soluble protein comprises an interleukin-15 (IL-15)
polypeptide domain
fused to an IL-12 domain; wherein the IL-15 polypeptide domain of the first
soluble protein
binds to the IL-15RaSu domain of the second soluble protein to form a soluble
fusion protein
complex.
In certain embodiments, an isolated soluble fusion protein comprises an
interleukin-
15 polypeptide domain, a first and second soluble protein wherein the first
soluble protein
comprises an interleukin-15 receptor alpha sushi-binding domain (IL-15RaSu)
fused to an
immunoglobulin Fc domain, wherein the IL-15RaSu domain is linked to an IL-12
and/or IL-
18 binding domain or functional fragment thereof and a second soluble protein
comprising an
interleukin-15 receptor alpha sushi-binding domain (IL-15RaSu) fused to an
immunoglobulin
Fc domain, wherein the IL-15RaSu domain is linked to an IL-12 and/or IL-18
binding
domain or functional fragment thereof, and, wherein the IL-15 polypeptide
domain binds to
the IL-15RaSu domain of the first and/or second soluble protein to form a
soluble fusion
protein complex.
29
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
In another embodiment, an isolated soluble fusion protein comprises an
interleukin-15
receptor alpha sushi-binding domain (IL-15RaSu) fused to an immunoglobulin Fc
domain, a
first and second soluble protein wherein the first soluble protein comprises
an interleukin-15
polypeptide domain linked to an IL-12 and/or IL-18 binding domain or
functional fragment
thereof and a second soluble protein comprising an interleukin-15 polypeptide
domain linked
to an IL-12 and/or IL-18 binding domain or functional fragment thereof, and,
wherein the IL-
15 polypeptide domain of the first and/or second soluble protein binds to the
IL-15RaSu
domain to form a soluble fusion protein complex.
In certain embodiments of the soluble fusion protein complexes of the
invention, the
IL-15 polypeptide is an IL-15 variant having a different amino acid sequence
than native IL-
15 polypeptide. The human IL-15 polypeptide is referred to herein as huIL-15,
hIL-15,
huIL15, hIL15, IL-15 wild type (wt) and variants thereof are referred to using
the native
amino acid, its position in the mature sequence and the variant amino acid.
For example,
huIL15N72D refers to human IL-15 comprising a substitution of N to D at
position 72. In
certain embodiments, the IL-15 variant functions as an IL-15 agonist as
demonstrated, e.g.,
by increased binding activity for the IL-15R3yC receptors compared to the
native IL-15
polypeptide. In certain embodiments, the IL-15 variant functions as an IL-15
antagonist as
demonstrated by e.g., decreased binding activity for the IL-15R3yC receptors
compared to
the native IL-15 polypeptide. In certain embodiments, the IL-15 variant has
increased
binding affinity or a decreased binding activity for the IL-15R3yC receptors
compared to the
native IL-15 polypeptide. In certain embodiments, the sequence of the IL-15
variant has at
least one (i.e., 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) amino acid change
compared to the native
IL-15 sequence. The amino acid change can include one or more of an amino acid
substitution or deletion in the domain of IL-15 that interacts with IL-15R0
and/or IL-15RyC.
In certain embodiments, the amino acid change is one or more amino acid
substitutions or
deletions at position 8, 61, 65, 72, 92, 101, 108, or 111 of the mature human
IL-15 sequence.
For example, the amino acid change is the substitution of D to N or A at
position 8, D to A at
position 61, N to A at position 65, N to R at position 72 or Q to A at
position 108 of the
mature human IL-15 sequence, or any combination of these substitutions. In
certain
embodiments, the amino acid change is the substitution of N to D at position
72 of the mature
human IL-15 sequence.
ALT-803
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
ALT-803 comprises an IL-15 mutant with increased ability to bind IL-2Rf3y and
enhanced biological activity (U.S. Patent No. 8,507, 222, incorporated herein
by reference).
This super-agonist mutant of IL-15 was described in a publication (Zu et al.,
2009 J Immunol,
183: 3598-3607, incorporated herein by reference). This IL-15 super-agonist in
combination
with a soluble IL-15a receptor fusion protein (IL-15RaSu/Fc) results in a
fusion protein
complex with highly potent IL-15 activity in vitro and in vivo (Han et al.,
2011, Cytokine, 56:
804-810; Xu, et al., 2013 Cancer Res. 73:3075-86, Wong, et al., 2013,
OncoImmunology
2:e26442). The IL-15 super agonist complex comprises an IL-15 mutant (IL-
15N72D) bound
to an IL-15 receptor a/IgG1 Fc fusion protein (IL-15N72D:IL-15RaSu/Fc) is
referred to as
"ALT-803."
Pharmacokinetic analysis indicated that the fusion protein complex has a half-
life of
25 hours following i.v. administration in mice. ALT-803 exhibits impressive
anti-tumor
activity against aggressive solid and hematological tumor models in
immunocompetent mice.
It can be administered as a monotherapy using a twice weekly or weekly i.v.
dose regimen or
as combinatorial therapy with an antibody. The ALT-803 anti-tumor response is
also
durable. Tumor-bearing mice that were cured after ALT-803 treatment were also
highly
resistant to re-challenge with the same tumor cells indicating that ALT-803
induces effective
immunological memory responses against the re-introduced tumor cells.
The sequence for ALT-803 (IL-15N72D associated with a dimeric IL-15RaSu/Fc
fusion protein) comprises SEQ ID NO: 9:
IL-15N72D protein sequence (with leader peptide)
METDTLLLWVLLLWVPGSTG-
[Leader peptide]
NWVNVISDLKKIEDLIQSMHIDATLYTESDVHPSCKVTAMKCFLLELQVISLESGDAS
IHDTVENLIILANDSLSSNGNVTESGCKECEELEEKNIKEFLQSFVHIVQMFINTS
[IL-15N72D]
IL-15RaSu/Fc protein sequence (with leader peptide)
MDRLTSSFLLLIVPAYVLS-
[Leader peptide]
ITCPPPMSVEHADIWVKSYSLYSRERYICNSGFKRKAGTSSLTECVLNKATNVAHWT
TPSLKCIR-
[IL-1 5RaStil
31
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
EPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEV
KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKA
LPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNG
QPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSL
SLSPGK
[IgG1 CH2-CH3 (Fc domain)]
IL-12
IL-12 is a member of a cytokine family consists of IL-12, IL-23, IL-27 and IL-
35,
which have diverse functions and play a role in both pro- and anti-
inflammatory responses.
IL-12 is typically expressed by activated antigen presenting cells (APCs). IL-
12 promotes
Thl differentiation and IFN-y production by T cells, and plays a role in
induction of anti-
tumor responses. As described herein, IL-12 in combination with IL-15 and IL-
18 is capable
of inducing CIML NK cells.
IL-12 is a disulfide-linked heterodimer consisting an a subunit (p35) and a13
subunit
(p40) in which the a subunit consists of a four-helix bundle long-chain
cytokine and the 13
subunit are homologous to non-signaling receptors of the IL-6 family. Crystal
structure and
mutagenesis analyses of IL-12 have defined amino acid residues at the p35/p40
interface
important for subunit interactions (Yoon, et al. .2000, EMBO J. 9, 3530-354).
For example, a
key arginine residue in p35 (R189) interacts with an aspartic acid in p40
(D290), such that the
R189 side chain is buried in a hydrophilic pocket on p40. Additionally,
conformation
changes in p40 may be important in optimizing these interactions. Based on
this information,
IL-12 variants containing amino acid changes could be generated that exhibit
improved
subunit interactions. Moreover, single-chain forms of IL-12 can be generated
consisting of
the p35 subunit linked to the p40 subunit by a flexible linker, either through
the C-terminus
of p35 linked to the N-terminus of p40 or vice versa. Such variants could be
incorporated
into the fusion protein complex of the invention to optimize expression,
subunit interactions
and/or stability of the IL-12 binding domain. Similarly, the IL-12 genes and
expression
constructs could be modified (i.e., codon optimization, removal of secondary
structures) to
improve gene expression, translation, post translational modification and/or
secretion.
The actions of IL-12 are mediated by binding to a transmembrane receptor
comprised
of two subunits (IL-12R(31 and IL-12R(32). Each subunit of the receptor is
composed of an
extracellular ligand-binding domain, a transmembrane domain and a cytosolic
domain that
mediates binding of Janus-family tyrosine kinases. IL-12 binding is believed
to result in
heterodimerization of (31 and 132 and the generation of a high-affinity
receptor complex
capable of signal transduction. In this model, dimerization of the receptor
leads to
32
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
juxtaposition of the cytosolic domains and the subsequent tyrosine
phosphorylation and
activation of the receptor-associated Janus-family kinases, Jak2 and Tyk-2.
These activated
kinases, in turn, tyrosine phosphorylate and activate several members of the
signal transducer
and activator of transcription (STAT) family (STAT-1, -3 and -4). The STATs
translocate to
the nucleus to activate transcription of several immune-responsive genes,
including IFN-y.
Although the crystal structure of the IL-12:IL-12R complex has not yet been
determined, IL-
12 variants with increased receptor binding/signaling activity can be isolated
by standard
screening assays (Leong et al. 2003, PNAS 100:1163-1168). Fragments of the IL-
12
heterodimer, including just the p35 subunit, may exhibit biological activity.
IL-12 variants
could also be isolated that modify IL-12/IL-12R surface residence time,
turnover and/or
recycling. Moreover, IL-12 variants could be incorporated into the fusion
protein complex of
the invention to optimize and/or balance the combined cytokine activities to
induce immune
cell responses, particularly CIML NK cell activity.
IL-18
Interleukin 18 (IL-18) is a pleiotropic IL-1 superfamily cytokine involved in
the
regulation of innate and acquired immune response. In the milieu of IL-12 or
IL-15, IL-18 is
a potent inducer of IFN-y in NK cells and CD4 T helper (Th) 1 lymphocytes.
However, IL-18
also modulates Th2 and Th17 cell responses, as well as the activity of CD8
cytotoxic cells
and neutrophils, in a host microenvironment-dependent manner. The biological
activity of IL-
18 is mediated by its binding to the heterodimeric IL-18Ra/r3 complexes
expressed on T cells,
NK cells, macrophages, neutrophils, and endothelial cells which induces
downstream signals
leading to the activation of NF-KB. In addition, the activity of IL-18 can be
modulated by the
levels of the high-affinity, constitutively expressed, and circulating IL-18
binding protein (IL-
18BP), which competes with cell surface receptors for IL-18 and neutralizes IL-
18 activity.
Variants of IL-18 (e.g., with amino acid mutations/deletions) that decrease
interactions with
IL-18BP and/or increase binding/signaling of the IL-18Ra/r3 complexes may be
useful in
enhancing IL-18 activity. Identification of such variants can be made through
by standard
screening assays (Kim et al. 2001, PNAS 98:3304-3309). Fragments of the IL-18
may
exhibit biological activity. IL-18 variants could also be isolated that modify
IL-18/IL-18R
surface residence time, turnover and/or recycling. Such IL-18 variants could
be incorporated
into the fusion protein complex of the invention to optimize IL-18 activity
and/or balance the
combined cytokine activities to induce immune cell responses, particularly
CIML NK cell
activity. In addition, IL-18 variants could be incorporated into the fusion
protein complex of
33
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
the invention to optimize expression and/or stability of the IL-18 binding
domain. Similarly,
the IL-18 genes and expression constructs could be modified (i.e., codon
optimization,
removal of secondary structures) to improve gene expression, translation, post
translational
modification and/or secretion.
Antigen-specific Binding Domains
Antigen-specific binding domains consist of polypeptides that specifically
bind to
targets on diseased cells. Alternatively, these domains may bind to targets on
other cells that
support the diseased state, such as targets on stromal cells that support
tumor growth or
targets on immune cells that support disease-mediated immunosuppression.
Antigen-specific
binding domains include antibodies, single chain antibodies, Fabs, Fv, T-cell
receptor binding
domains, ligand binding domains, receptor binding domains, domain antibodies,
single
domain antibodies, minibodies, nanobodies, peptibodies, or various other
antibody mimics
(such as affimers, affitins, alphabodies, atrimers, CTLA4-based molecules,
adnectins,
anticalins, Kunitz domain-based proteins, avimers, knottins, fynomers,
darpins, affibodies,
affilins, monobodies and armadillo repeat protein-based proteins (Weidle, UH,
et al. 2013.
Cancer Genomics & Proteomics 10: 155-168)) known in the art.
In certain embodiments, the antigen for the antigen-specific binding domain
comprises a cell surface receptor or ligand. In a further embodiment, the
antigen comprises a
CD antigen, cytokine or chemokine receptor or ligand, growth factor receptor
or ligand,
tissue factor, cell adhesion molecule, MHC/MHC-like molecules, Fc receptor,
Toll-like
receptor, NK receptor, TCR, BCR, positive/negative co-stimulatory receptor or
ligand, death
receptor or ligand, tumor associated antigen, or virus encoded antigen.
Preferably, the antigen-specific binding domain is capable of binding to an
antigen on
a tumor cell. Tumor-specific binding domain may be derived from antibodies
approved for
treatment of patients with cancer include rittlximab, ofatumumab, and
obinutuzumab (anti-
CD20 Abs); trastuzumab and pertuzumab (anti-HER2 Abs); cetuximab and
panitumumab
(anti-EGFR Abs); and alemtuzumab (anti-CD52 Ab). Similarly, binding domains
from
approved antibody-effector molecule conjugates specific to CD20 (90Y-labeled
ibritumomab
tiuxetan,
tositumomab), HER2 (ado-trastuzumab emtansine), CD30 (brentuximab
vedotin) and CD33 (gemtuzumab ozogamicin) (Sliwkowski MX, Mellman I. 2013
Science
341:1192) could be used.
Additionally, preferred binding domains of the invention may include various
other
tumor-specific antibody domains known in the art. The antibodies and their
respective
34
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
targets for treatment of cancer include but are not limited to nivolumab (anti-
PD-1 Ab), TA99
(anti-gp75), 3F8 (anti-GD2), 8H9 (anti-B7-H3), abagovomab (anti-CA-125
(imitation)),
adecatumumab (anti-EpCAM), afutuzumab (anti-CD20), alacizumab pegol (anti-
VEGFR2),
altumomab pentetate (anti-CEA), amatuximab (anti-mesothelin), AME-133 (anti-
CD20),
anatumomab mafenatox (anti-TAG-72), apolizumab (anti-HLA-DR), arcitumomab
(anti-
CEA), bavituximab (anti-phosphatidylserine), bectumomab (anti-CD22), belimumab
(anti-
BAFF), besilesomab (anti-CEA-related antigen), bevacizumab (anti-VEGF-A),
bivatuzumab
mertansine (anti-CD44 v6), blinatumomab (anti-CD19), BMS-663513 (anti-CD137),
brentuximab vedotin (anti-CD30 (TNFRSF8)), cantuzumab mertansine (anti-mucin
CanAg),
cantuzumab ravtansine (anti-MUC1), capromab pendetide (anti-prostatic
carcinoma cells),
carlumab (anti-MCP-1), catumaxomab (anti-EpCAM, CD3), cBR96-doxorubicin
immunoconjugate (anti-Lewis-Y antigen), CC49 (anti-TAG-72), cedelizumab (anti-
CD4),
Ch.14.18 (anti-GD2), ch-TNT (anti-DNA associated antigens), citatuzumab
bogatox (anti-
EpCAM), cixutumumab (anti-IGF-1 receptor), clivatuzumab tetraxetan (anti-
MUC1),
conatumumab (anti-TRAIL-R2), CP-870893 (anti-CD40), dacetuzumab (anti-CD40),
daclizumab (anti-CD25), dalotuzumab (anti-insulin-like growth factor I
receptor),
daratumumab (anti-CD38 (cyclic ADP ribose hydrolase)), demcizumab (anti-DLL4),
detumomab (anti-B-lymphoma cell), drozitumab (anti-DRS), duligotumab (anti-
HER3),
dusigitumab (anti-ILGF2), ecromeximab (anti-GD3 ganglioside), edrecolomab
(anti-
EpCAM), elotuzumab (anti-SLAMF7), elsilimomab (anti-IL-6), enavatuzumab (anti-
TWEAK receptor), enoticumab (anti-DLL4), ensituximab (anti-SAC), epitumomab
citircetan
(anti-episialin), epratuzumab (anti-CD22), ertumaxomab (anti-HER2/neu, CD3),
etaracizumab (anti-integrin av(33), faralimomab (anti-Interferon receptor),
farletuzumab (anti-
folate receptor 1), FBTA05 (anti-CD20), ficlatuzumab (anti-HGF), figitumumab
(anti-IGF-1
receptor), flanvotumab (anti-TYRP1(glycoprotein 75)), fresolimumab (anti-TGF
(3),
futuximab (anti-EGFR), galiximab (anti-CD80), ganitumab (anti-IGF-I),
gemtuzumab
ozogamicin (anti-CD33), girentuximab (anti-carbonic anhydrase 9 (CA-IX)),
glembatumumab vedotin (anti-GPNMB), guselkumab (anti-IL13), ibalizumab (anti-
CD4),
ibritumomab thrcetan (anti-CD20), icrucumab (anti-VEGFR-1), igovomab (anti-CA-
125),
IMAB362 (anti-CLDN18.2), IMC-CS4 (anti-CSF1R), IMC-TR1 (TGFORID, imgatuzumab
(anti-EGFR), inclacumab (anti-selectin P), indatuximab ravtansine (anti-SDC1),
inotuzumab
ozogamicin (anti-CD22), intetumumab (anti-CD51), ipilimumab (anti-CD152),
iratumumab
(anti-CD30 (TNFRSF8)), KM3065 (anti-CD20), KW-0761 (anti-CD194), LY2875358
(anti-
MET) labetuzumab (anti-CEA), lambrolizumab (anti-PDCD1), lexatumumab (anti-
TRAIL-
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
R2), lintuzumab (anti-CD33), lirilumab (anti-KIR2D), lorvotuzumab mertansine
(anti-CD56),
lucatumumab (anti-CD40), lumiliximab (anti-CD23 (IgE receptor)), mapatumumab
(anti-
TRAIL-R1), margettvdmab (anti-ch4D5), matuzumab (anti-EGFR), mavrilimumab
(anti-
GMCSF receptor a-chain), milatuzumab (anti-CD74), minretumomab (anti-TAG-72),
mitumomab (anti-GD3 ganglioside), mogamulizumab (anti-CCR4), moxetumomab
pasudotox (anti-CD22), nacolomab tafenatox (anti-C242 antigen), naptumomab
estafenatox
(anti-5T4), narnatumab (anti-RON), necitumumab (anti-EGFR), nesvacumab (anti-
angiopoietin 2), nimotuzumab (anti-EGFR), nivolumab (anti-IgG4), nofetumomab
merpentan, ocrelizumab (anti-CD20), ocaratuzumab (anti-CD20), olaratumab (anti-
PDGF-R
a), onartuzumab (anti-c-MET), ontuxizumab (anti-TEM1), oportuzumab monatox
(anti-
EpCAM), oregovomab (anti-CA-125), otlertuzumab (anti-CD37), pankomab (anti-
tumor
specific glycosylation of MUC1), parsatuzumab (anti-EGFL7), pascolizumab (anti-
IL-4),
patritumab (anti-HER3), pemtumomab (anti-MUC1), pertuzumab (anti-HER2/neu),
pidilizumab (anti-PD-1), pinatuzumab vedotin (anti-CD22), pintumomab (anti-
adenocarcinoma antigen), polatuzumab vedotin (anti-CD79B), pritumumab (anti-
vimentin),
PRO131921 (anti-CD20), quilizumab (anti-IGHE), racotumomab (anti-N-
glycolylneuraminic
acid), radretumab (anti-fibronectin extra domain-B), ramucirumab (anti-
VEGFR2),
rilotumumab (anti-HGF), robatumumab (anti-IGF-1 receptor), roledumab (anti-
RHD),
rovelizumab (anti-CD ii & CD18), samalizumab (anti-CD200), satumomab pendetide
(anti-
TAG-72), seribantumab (anti-ERBB3), SGN-CD19A (anti-CD19), SGN-CD33A (anti-
CD33), sibrotuzumab (anti-FAP), silttlximab (anti-IL-6), solitomab (anti-
EpCAM),
sontuzumab (anti-episialin), tabalumab (anti-BAFF), tacatuzumab tetraxetan
(anti-alpha-
fetoprotein), taplitumomab paptox (anti-CD19), telimomab aritox, tenatumomab
(anti-
tenascin C), teneliximab (anti-CD40), teprotumumab (anti-CD221), TGN1412 (anti-
CD28),
ticilimumab (anti-CTLA-4), tigatuzumab (anti-TRAIL-R2), TNX-650 (anti-IL-13),
tositumomab (anti-0520), tovetumab (anti-CD140a), TRBS07 (anti-GD2),
tregalizumab
(anti-CD4), tremelimumab (anti-CTLA-4), TRU-016 (anti-CD37), tucotuzumab
celmoleukin
(anti-EpCAM), ublituximab (anti-CD20), urelumab (anti-4-1BB), vantictumab
(anti-Frizzled
receptor), vapaliximab (anti-A0C3 (VAP-1)), vatelizumab (anti-ITGA2),
veltuzumab (anti-
CD20), vesencumab (anti-NRP1), visilizumab (anti-CD3), volociximab (anti-
integrin a5r31),
vorsetuzumab mafodotin (anti-CD70), votumumab (anti-tumor antigen CTAA16.88),
zalutumumab (anti-EGFR), zanolimumab (anti-CD4), zatuximab (anti-HER1),
ziralimumab
(anti-CD147 (basigin)), RG7636 (anti-ETBR), RG7458 (anti-MUC16), RG7599 (anti-
NaPi2b), MPDL3280A (anti-PD-L1), RG7450 (anti-STEAP1), and GDC-0199 (anti-Bc1-
2).
36
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
Other antibody domains or tumor target binding proteins useful in the
invention (e.g.
TCR domains) include, but are not limited to, those that bind the following
antigens (note, the
cancer indications indicated represent non-limiting examples): aminopeptidase
N (CD13),
annexin Al, B7-H3 (CD276, various cancers), CA125 (ovarian cancers), CA15-3
(carcinomas), CA19-9 (carcinomas), L6 (carcinomas), Lewis Y (carcinomas),
Lewis X
(carcinomas), alpha fetoprotein (carcinomas), CA242 (colorectal cancers),
placental alkaline
phosphatase (carcinomas), prostate specific antigen (prostate), prostatic acid
phosphatase
(prostate), epidermal growth factor (carcinomas), CD2 (Hodgkin's disease, NHL
lymphoma,
multiple myeloma), CD3 epsilon (T cell lymphoma, lung, breast, gastric,
ovarian cancers,
autoimmune diseases, malignant ascites), CD19 (B cell malignancies), CD20 (non-
Hodgkin's
lymphoma, B-cell neoplasms, autoimmune diseases), CD21 (B-cell lymphoma), CD22
(leukemia, lymphoma, multiple myeloma, SLE), CD30 (Hodgkin's lymphoma), CD33
(leukemia, autoimmune diseases), CD38 (multiple myeloma), CD40 (lymphoma,
multiple
myeloma, leukemia (CLL)), CD51 (metastatic melanoma, sarcoma), CD52
(leukemia), CD56
(small cell lung cancers, ovarian cancer, Merkel cell carcinoma, and the
liquid tumor,
multiple myeloma), CD66e (carcinomas), CD70 (metastatic renal cell carcinoma
and non-
Hodgkin lymphoma), CD74 (multiple myeloma), CD80 (lymphoma), CD98
(carcinomas),
CD123 (leukemia), mucin (carcinomas), CD221 (solid tumors), CD227 (breast,
ovarian
cancers), CD262 (NSCLC and other cancers), CD309 (ovarian cancers), CD326
(solid
tumors), CEACAM3 (colorectal, gastric cancers), CEACAM5 (CEA, CD66e) (breast,
colorectal and lung cancers), DLL4 (A-like-4), EGFR (various cancers), CTLA4
(melanoma),
CXCR4 (CD 184, heme-oncology, solid tumors), Endoglin (CD 105, solid tumors),
EPCAM
(epithelial cell adhesion molecule, bladder, head, neck, colon, NHL prostate,
and ovarian
cancers), ERBB2 (lung, breast, prostate cancers), FCGR1 (autoimmune diseases),
FOLR
(folate receptor, ovarian cancers), FGFR (carcinomas), GD2 ganglioside
(carcinomas), G-28
(a cell surface antigen glycolipid, melanoma), GD3 idiotype (carcinomas), heat
shock
proteins (carcinomas), HER1 (lung, stomach cancers), HER2 (breast, lung and
ovarian
cancers), HLA-DR10 (NHL), HLA-DRB (NHL, B cell leukemia), human chorionic
gonadotropin (carcinomas), IGF1R (solid tumors, blood cancers), IL-2 receptor
(T-cell
leukemia and lymphomas), IL-6R (multiple myeloma, RA, Castleman's disease, IL6
dependent tumors), integrins (av(33, a5(31, a6(34, al l(33, a5135, av135, for
various cancers),
MAGE-1 (carcinomas), MAGE-2 (carcinomas), MAGE-3 (carcinomas), MAGE 4
(carcinomas), anti-transferrin receptor (carcinomas), p97 (melanoma), MS4A1
(membrane-
spanning 4-domains subfamily A member 1, Non-Hodgkin's B cell lymphoma,
leukemia),
37
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
MUC I (breast, ovarian, cervix, bronchus and gastrointestinal cancer), MUC16
(CA125)
(ovarian cancers), CEA (colorectal cancer), gp100 (melanoma), MARTI
(melanoma), MPG
(melanoma), MS4A1 (membrane-spanning 4-domains subfamily A, small cell lung
cancers,
NHL), nucleolin, Neu oncogene product (carcinomas), P21 (carcinomas), nectin-4
(carcinomas), paratope of anti-(N- glycolylneuraminic acid, breast, melanoma
cancers),
PLAP-like testicular alkaline phosphatase (ovarian, testicular cancers), PSMA
(prostate
tumors), PSA (prostate), ROB04, TAG 72 (tumour associated glycoprotein 72,
AML, gastric,
colorectal, ovarian cancers), T cell transmembrane protein (cancers), Tie
(CD202b), tissue
factor, TNFRSF1OB (tumor necrosis factor receptor superfamily member 10B,
carcinomas),
TNFRSF13B (tumor necrosis factor receptor superfamily member 13B, multiple
myeloma,
NHL, other cancers, RA and SLE), TPBG (trophoblast glycoprotein, renal cell
carcinoma),
TRAIL-R1 (tumor necrosis apoptosis inducing ligand receptor 1, lymphoma, NHL,
colorectal, lung cancers), VCAM-1 (CD106, Melanoma), VEGF, VEGF-A, VEGF-2
(CD309) (various cancers). Some other tumor associated antigen targets have
been reviewed
(Gerber, et al, mAbs 2009 1:247-253; Novellino et al, Cancer Immunol
Immunother. 2005
54:187-207, Franke, et al, Cancer Biother Radiopharm. 2000, 15:459-76, Guo, et
al., Adv
Cancer Res. 2013; 119: 421-475, Parmiani et al. J Immunol. 2007 178:1975-9).
Examples of
these antigens include Cluster of Differentiations (CD4, CD5, CD6, CD7, CD8,
CD9, CD10,
CD1 la, CD1 lb, CD1 lc, CD12w, CD14, CD15, CD16, CDw17, CD18, CD21, CD23,
CD24,
CD25, CD26, CD27, CD28, CD29, CD31, CD32, CD34, CD35, CD36, CD37, CD41, CD42,
CD43, CD44, CD45, CD46, CD47, CD48, CD49b, CD49c, CD53, CD54, CD55, CD58,
CD59, CD61, CD62E, CD62L, CD62P, CD63, CD68, CD69, CD71, CD72, CD79, CD81,
CD82, CD83, CD86, CD87, CD88, CD89, CD90, CD91, CD95, CD96, CD100, CD103,
CD105, CD106, CD109, CD117, CD120, CD127, CD133, CD134, CD135, CD138, CD141,
CD142, CD143, CD144, CD147, CD151, CD152, CD154, CD156, CD158, CD163, CD166,
.CD168, CD184, CDw186, CD195, CD202 (a, b), CD209, CD235a, CD271, CD303,
CD304),
annexin Al, nucleolin, endoglin (CD105), ROB04, amino-peptidase N, -like-4
(DLL4),
VEGFR-2 (CD309), CXCR4 (CD184), Tie2, B7-H3, WT1, MUC1, LMP2, HPV E6 E7,
EGFRvIII, HER-2/neu, idiotype, MAGE A3, p53 nonmutant, NY-ES0-1, GD2, CEA,
MelanA/MART1, Ras mutant, gp100, p53 mutant, proteinase3 (PR1), bcr-abl,
tyrosinase,
survivin, hTERT, sarcoma translocation breakpoints, EphA2, PAP, ML-IAP, AFP,
EpCAM,
ERG (TMPRSS2 ETS fusion gene), NA17, PAX3, ALK, androgen receptor, cyclin B 1,
polysialic acid, MYCN, RhoC, TRP-2, GD3, fucosyl GM1, mesothelin, PSCA, MAGE
Al,
sLe(a), CYPIB I, PLAC1, GM3, BORIS, Tn, GloboH, ETV6-AML, NY-BR-1, RGS5,
38
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
SART3, STn, carbonic anhydrase IX, PAX5, 0Y-TES1, sperm protein 17, LCK,
HMWMAA, AKAP-4, SSX2, XAGE 1, B7H3, legumain, Tie 2, Page4, VEGFR2, MAD-
CT-1, FAP, PDGFR-0, MAD-CT-2, and Fos-related antigen 1.
Additionally, preferred binding domains of the invention include those
specific to
antigens and epitope targets associated with infected cells that are known in
the art. Such
targets include but are not limited those derived from the following
infectious agents are of
interest: HIV virus (particularly antigens derived from the HIV envelope spike
and/or gp120
and gp41 epitopes), Human papilloma virus (HPV), Mycobacterium tuberculosis,
Streptococcus agalactiae, methicillin-resistant Staphylococcus aureus ,
Legionella
pneumophilia, Streptococcus pyogenes, Escherichia coli, Neisseria gonorrhoeae,
Neisseria
meningitidis, Pneumococcus , Cryptococcus neoformans , Histoplasma capsulatum,
-
influenzae B, Treponema pallidum, Lyme disease spirochetes, Pseudomonas
aeruginosa,
Mycobacterium leprae, Brucella abortus, rabies virus, influenza virus,
cytomegalovirus,
herpes simplex virus I, herpes simplex virus II, human serum parvo-like virus,
respiratory
syncytial virus, varicella-zoster virus, hepatitis B virus, hepatitis C virus,
measles virus,
adenovirus, human T-cell leukemia viruses, Epstein-Barr virus, murine leukemia
virus,
mumps virus, vesicular stomatitis virus, sindbis virus, lymphocytic
choriomeningitis virus,
wart virus, blue tongue virus, Sendai virus, feline leukemia virus, reovirus,
polio virus,
simian virus 40, mouse mammary tumor virus, dengue virus, rubella virus, West
Nile virus,
Plasmodium fakiparum, Plasmodium vivax, Toxoplasma gondii, Trypanosoma
rangeli,
Trypanosoma cruzi, Trypanosoma rhodesiensei, Trypanosoma brucei, Schistosoma
mansoni,
Schistosoma japonicum, Babesia bovis, Elmeria tenella, Onchocerca volvulus ,
Leishmania
tropica, Trichinella spiralis, Theileria parva, Taenia hydatigena, Taenia
ovis, Taenia
saginata, Echinococcus granulosus ,Mesocestoides corti, Mycoplasma arthritidis
, M
hyorhinis, M orale, M arginini, Acholeplasma laidlawii, M salivarium and M
pneumoniae.
Immune Checkpoint Inhibitor and Immune Agonist Domains
In other embodiments, the binding domain is specific to an immune checkpoint
or
signaling molecule or its ligand and acts as an inhibitor of immune checkpoint
suppressive
activity or as an agonist of immune stimulatory activity. Such immune
checkpoint and
signaling molecules and ligands include PD-1, PD-L1, PD-L2, CTLA-4, CD28,
CD80,
CD86, B7-H3, B7-H4, B7-H5, ICOS-L, ICOS, BTLA, CD137L, CD137, HVEM, KIR, 4-
1BB, OX4OL, CD70, CD27, CD47, CIS, 0X40, GITR, IDO, TIM3, GAL9, VISTA, CD155,
TIGIT, LIGHT, LAIR-1, Siglecs and A2aR (Pardoll DM. 2012. Nature Rev Cancer
12:252-
39
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
264, Thaventhiran T, et al. 2012. J Clin Cell Immunol S12:004). Additionally,
preferred
antibody domains of the invention may include ipilimumab and/or tremelimumab
(anti-
CTLA4), nivolumab, pembrolizumab, pidilizumab, TSR-042, ANB011, AMP-514 and
AMP-
224 (a ligand-Fc fusion) (anti-PD1), atezolizumab (MPDL3280A), avelumab
(MSB0010718C), durvalumab (MEDI4736), MEDI0680, and BMS-9365569 (anti-PDL1),
MEDI6469 (anti-0X40 agonist), BMS-986016, IMP701, IMP731, IMP321 (anti-LAG3)
and
GITRligand.
T-Cell Receptors (TCRs)
T-cells are a subgroup of cells which together with other immune cell types
(polymorphonuclear cells, eosinophils, basophils, mast cells, B-cells, NK
cells), constitute the
cellular component of the immune system. Under physiological conditions, T-
cells function
in immune surveillance and in the elimination of foreign antigen. However,
under
pathological conditions, there is compelling evidence that T-cells play a
major role in the
causation and propagation of disease. In these disorders, breakdown of T-cell
immunological
tolerance, either central or peripheral is a fundamental process in the
causation of
autoimmune disease.
The TCR complex is composed of at least seven transmembrane proteins. The
disulfide-linked (a13 or y.5) heterodimer forms the monotypic antigen
recognition unit, while
the invariant chains of CD3, consisting of E, y, 8, C, and 11 chains, are
responsible for coupling
the ligand binding to signaling pathways that result in T-cell activation and
the elaboration of
the cellular immune responses. Despite the gene diversity of the TCR chains,
two structural
features are common to all known subunits. First, they are transmembrane
proteins with a
single transmembrane spanning domain--presumably alpha-helical. Second, all
TCR chains
have the unusual feature of possessing a charged amino acid within the
predicted
transmembrane domain. The invariant chains have a single negative charge,
conserved
between the mouse and human, and the variant chains possess one (TCR-) or two
(TCR-a)
positive charges. The transmembrane sequence of TCR-a is highly conserved in a
number of
species and thus phylogenetically may serve an important functional role. The
octapeptide
sequence containing the hydrophilic amino acids arginine and lysine is
identical between the
species.
A T-cell response is modulated by antigen binding to a TCR. One type of TCR is
a
membrane bound heterodimer consisting of an a and p chain resembling an
immunoglobulin
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
variable (V) and constant (C) region. The TCR a chain includes a covalently
linked V-a and
C-a chain, whereas the p chain includes a V-f3 chain covalently linked to a C-
f3 chain. The
V-a and V-f3 chains form a pocket or cleft that can bind a superantigen or
antigen in the
context of a major histocompatibility complex (MHC) (known in humans as an HLA
complex). See, Davis Ann. Rev. of Immunology 3: 537 (1985); Fundamental
Immunology 3rd
Ed., W. Paul Ed. Rsen Press LTD. New York (1993).
The extracellular domains of the TCR chains (af3 or y6) can also engineered as
fusions to heterologous transmembrane domains for expression on the cell
surface. Such
TCRs may include fusions to CD3, CD28, CD8, 4-1BB and/or chimeric activation
receptor
(CAR) transmembrane or activation domains. TCRs can also be the soluble
proteins
comprising one or more of the antigen binding domains of af3 or y6 chains.
Such TCRs may
include the TCR variable domains or function fragments thereof with or without
the TCR
constant domains. Soluble TCRs may be heterodimeric or single-chain molecules.
Fc Domain
Fusion protein complexes of the invention may contain an Fc domain. For
example,
hIL-18/IL12/TxM comprises an IL-18/IL-15N72D:IL-12/IL-15RaSu/Fc fusion protein
complex. Fusion proteins that combine the Fc regions of IgG with the domains
of another
protein, such as various cytokines and soluble receptors have been reported
(see, for example,
Capon et al., Nature, 337:525-531, 1989; Chamow et al., Trends Biotechnol.,
14:52-60,
1996); U.S. Pat. Nos. 5,116,964 and 5,541,087). The prototype fusion protein
is a
homodimeric protein linked through cysteine residues in the hinge region of
IgG Fc, resulting
in a molecule similar to an IgG molecule without the heavy chain variable and
CH1 domains
and light chains. The dimeric nature of fusion proteins comprising the Fc
domain may be
advantageous in providing higher order interactions (i.e. bivalent or
bispecific binding) with
other molecules. Due to the structural homology, Fc fusion proteins exhibit an
in vivo
pharmacokinetic profile comparable to that of human IgG with a similar
isotype.
Immunoglobulins of the IgG class are among the most abundant proteins in human
blood,
and their circulation half-lives can reach as long as 21 days. To extend the
circulating half-
life of IL-15 or an IL-15 fusion protein and/or to increase its biological
activity, fusion
protein complexes containing the IL-15 domain non-covalently bound to IL-15Ra
covalently
linked to the Fc portion of the human heavy chain IgG protein are described
herein.
41
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
The term "Fc" refers to the fragment crystallizable region which is the
constant region
of an antibody that interacts with cell surface receptors called Fc receptors
and some proteins
of the complement system. Such an "Fe" is in dimeric form. The original
immunoglobulin
source of the native Fc is preferably of human origin and may be any of the
immunoglobulins, although IgG1 and IgG2 are preferred. Native Fc's are made up
of
monomeric polypeptides that may be linked into dimeric or multimeric forms by
covalent
(i.e., disulfide bonds) and non-covalent association. The number of
intermolecular disulfide
bonds between monomeric subunits of native Fc molecules ranges from 1 to 4
depending on
class (e.g., IgG, IgA, IgE) or subclass (e.g., IgGl, IgG2, IgG3, IgAl, IgGA2).
One example
of a native Fc is a disulfide-bonded dimer resulting from papain digestion of
an IgG (see
Ellison et al. (1982), Nucleic Acids Res. 10: 4071-9). The term "native Fc" as
used herein is
generic to the monomeric, dimeric, and multimeric forms. Fc domains containing
binding
sites for Protein A, Protein G, various Fc receptors and complement proteins.
In some
embodiments, Fc domain of the fusion protein complex is capable of interacting
with Fc
receptors to mediate antibody-dependent cell-mediated cytotoxicity (ADCC)
and/or antibody
dependent cellular phagocytosis (ADCP). In other applications, the fusion
protein complex
comprises an Fc domain (e.g., IgG4 Fc) that is incapable of effectively
mediating ADCC or
ADCP.
In some embodiments, the term "Fc variant" refers to a molecule or sequence
that is
modified from a native Fc, but still comprises a binding site for the salvage
receptor, FcRn.
International applications WO 97/34631 and WO 96/32478 describe exemplary Fc
variants,
as well as interaction with the salvage receptor, and are hereby incorporated
by reference.
Thus, the term "Fc variant" comprises a molecule or sequence that is humanized
from a non-
human native Fc. Furthermore, a native Fc comprises sites that may be removed
because
they provide structural features or biological activity that are not required
for the fusion
molecules of the present invention. Thus, in certain embodiments, the term "Fc
variant"
comprises a molecule or sequence that alters one or more native Fc sites or
residues that
affect or are involved in (1) disulfide bond formation, (2) incompatibility
with a selected host
cell (3) N-terminal heterogeneity upon expression in a selected host cell, (4)
glycosylation,
(5) interaction with complement, (6) binding to an Fc receptor other than a
salvage receptor,
(7) antibody-dependent cellular cytotoxicity (ADCC) or (8) antibody-dependent
cellular
phagocytosis (ADCP). Such alterations can increase or decrease any one or more
of these Fc
properties. Fc variants are described in further detail hereinafter.
42
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
The term "Fc domain" encompasses native Fc and Fc variant molecules and
sequences as defined above. As with Fc variants and native Fc's, the term "Fc
domain"
includes molecules in monomeric or multimeric form, whether digested from
whole antibody
or produced by recombinant gene expression or by other means.
Linkers
In some cases, the fusion protein complexes of the invention also include a
flexible
linker sequence interposed between the IL-15 or IL-15Ra domains and the IL-12
and/or IL-
18 binding domain or the IL-12 subunits. The linker sequence should allow
effective
positioning of the polypeptide with respect to the IL-15 or IL-15Ra domains to
allow
functional activity of both domains. Alternatively, the linker should allow
formation of a
functional IL-12 binding domain.
In certain cases, the soluble fusion protein complex has a linker wherein the
first
polypeptide is covalently linked to IL-15 (or functional fragment thereof) by
polypeptide
linker sequence. In other aspects, the soluble fusion protein complex as
described herein has
a linker wherein the second polypeptide is covalently linked to IL-15Ra
polypeptide (or
functional fragment thereof) by polypeptide linker sequence.
The linker sequence is preferably encoded by a nucleotide sequence resulting
in a
peptide that can effectively position the binding groove of a TCR molecule for
recognition of
a presenting antigen or the binding domain of an antibody molecule for
recognition of an
antigen. As used herein, the phrase "effective positioning of the biologically
active
polypeptide with respect to the IL-15 or IL-15Ra domains", or other similar
phrase, is
intended to mean the biologically active polypeptide linked to the IL-15 or IL-
15Ra domains
is positioned so that the IL-15 or IL-15Ra domains are capable of interacting
with each other
to form a protein complex. For example, the IL-15 or IL-15Ra domains are
effectively
positioned to allow interactions with immune cells to initiate or inhibit an
immune reaction,
or to inhibit or stimulate cell development.
The fusion protein complexes of the invention preferably also include a
flexible linker
sequence interposed between the IL-15 or IL-15Ra domains and the
immunoglobulin Fc
domain. The linker sequence should allow effective positioning of the Fc
domain,
biologically active polypeptide and IL-15 or IL-15Ra domains to allow
functional activity of
each domain. For example, the Fc domains are effectively positioned to allow
proper fusion
protein complex formation and/or interactions with Fc receptors on immune
cells or proteins
43
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
of the complement system to stimulate Fc-mediated effects including
opsonization, cell lysis,
degranulation of mast cells, basophils, and eosinophils, and other Fc receptor-
dependent
processes; activation of the complement pathway; and enhanced in vivo half-
life of the fusion
protein complex.
Linker sequences can also be used to link two or more polypeptides of the
biologically active polypeptide to generate a single-chain molecule with the
desired
functional activity.
Preferably, the linker sequence comprises from about 7 to 20 amino acids, more
preferably from about 10 to 20 amino acids. The linker sequence is preferably
flexible so as
not hold the biologically active polypeptide or effector molecule in a single
undesired
conformation. The linker sequence can be used, e.g., to space the recognition
site from the
fused molecule. Specifically, the peptide linker sequence can be positioned
between the
biologically active polypeptide and the effector molecule, e.g., to chemically
cross-link same
and to provide molecular flexibility. The linker preferably predominantly
comprises amino
acids with small side chains, such as glycine, alanine and serine, to provide
for flexibility.
Preferably, about 80 or 90 percent or greater of the linker sequence comprises
glycine,
alanine or serine residues, particularly glycine and serine residues.
Different linker sequences could be used including any of a number of flexible
linker
designs that have been used successfully to join antibody variable regions
together (see,
Whitlow, M. et al., (1991) Methods: A Companion to Methods in Enzymology, 2:97-
105).
Adoptive cell therapy
Adoptive cell therapy (ACT) (including allogeneic and autologous hematopoietic
stem cell transplantation (HSCT) and recombinant cell (i.e., CAR T) therapies)
is the
treatment of choice for many malignant disorders (for reviews of HSCT and
adoptive cell
therapy approaches, see, Rager & Porter, Ther Adv Hematol (2011) 2(6) 409-428;
Roddie &
Peggs, Expert Opin. Biol. Ther. (2011) 11(4):473-487; Wang et al. Int. J.
Cancer: (2015)136,
1751-1768; and Chang, Y.J. and X.J. Huang, Blood Rev, 2013.27(1): 55-62). Such
adoptive
cell therapies include, but are not limited to, allogeneic and autologous
hematopoietic stem
cell transplantation, donor leukocyte (or lymphocyte) infusion (DLI), adoptive
transfer of
tumor infiltrating lymphocytes, or adoptive transfer of T cells or NK cells
(including
recombinant cells, i.e., CAR T, CAR NK, gene-edited T cells or NK cells, see
Hu et al. Acta
Pharmacologica Sinica (2018) 39: 167-176, Irving et al. Front Immunol. (2017)
8: 267.).
Beyond the necessity for donor-derived cells to reconstitute hematopoiesis
after radiation and
44
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
chemotherapy, immunologic reconstitution from transferred cells is important
for the
elimination of residual tumor cells. The efficacy of ACT as a curative option
for
malignancies is influenced by a number of factors including the origin,
composition and
phenotype (lymphocyte subset, activation status) of the donor cells, the
underlying disease,
the pre-transplant conditioning regimen and post-transplant immune support
(i.e., IL-2
therapy) and the graft-versus-tumor (GVT) effect mediated by donor cells
within the graft.
Additionally, these factors must be balanced against transplant-related
mortality, typically
arising from the conditioning regimen and/or excessive immune activity of
donor cells within
the host (i.e., graft-versus-host disease, cytokine release syndrome, etc.).
Approaches utilizing adoptive NK cell therapy have become of significant
interest. In
patients receiving autologous HSCT, blood NK cell numbers recover very early
after the
transplant and the levels of NK cells correlate with a positive outcome (Rueff
et al., 2014,
Biol. Blood Marrow Transplant. 20, 896-899). Although therapeutic strategies
with
autologous NK cell transfer have had limited success due to a number of
factors, adoptive
transfer of ex vivo-activated allogeneic (or haplo-identical) NK cells has
emerged as a
promising immunotherapeutic strategy for cancer (Guillerey et al. 2016. Nature
Immunol. 17:
1025-1036). The activity of these cells is less likely to be suppressed by
self-MHC molecules
compared to autologous NK cells. A number of studies have shown that adoptive
therapy
with haploidentical NK cells to exploit alloreactivity against tumor cells is
safe and can
mediate significant clinical activity in AML patients. Taking these findings
further, recent
studies have focused on optimizing ex vivo activation/expansion methods for NK
cells or NK
precursors (i.e., stem cells) and pre-transplant conditioning and post-
transplant immune
support strategies; use of NK cell lines or recombinant tumor-targeting NK
cells; evaluation
of combination therapies with other agents such as therapeutic Ab,
immunomodulatory
agents (lenalidomide), and anti-MR and checkpoint Abs. In each case, these
strategies could
be complemented by the fusion protein complex of the invention, which has the
capacity to
augment NK cell proliferation and activation. As indicated herein, ex vivo
incubation of NK
cells with the fusion protein complex of the invention result in induction of
CIML NK cell
exhibiting elevated activation markers, increased cytotoxicity against tumor
cells and
enhanced production of IFN-y. Additionally, the fusion protein complex of the
invention is
capable of activating human NK cell lines. Moreover, methods are provided for
augmenting
immune responses and treating neoplasia and infection disease by direct
administration of the
fusion protein complex of the invention or administration of immune cells
activated by the
fusion protein complex of the invention.
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
Pharmaceutical Therapeutics
The invention provides pharmaceutical compositions comprising fusion protein
complexes for use as a therapeutic. In one aspect, fusion protein complex of
the invention is
administered systemically, for example, formulated in a pharmaceutically-
acceptable buffer
such as physiological saline. Preferable routes of administration include, for
example,
instillation into the bladder, subcutaneous, intravenous, intraperitoneal,
intramuscular,
intratumoral or intradermal injections that provide continuous, sustained or
effective levels of
the composition in the patient. Treatment of human patients or other animals
is carried out
using a therapeutically effective amount of a therapeutic identified herein in
a
physiologically-acceptable carrier. Suitable carriers and their formulation
are described, for
example, in Remington's Pharmaceutical Sciences by E. W. Martin. The amount of
the
therapeutic agent to be administered varies depending upon the manner of
administration, the
age and body weight of the patient, and with the clinical symptoms of the
neoplasia.
Generally, amounts will be in the range of those used for other agents used in
the treatment of
other diseases associated with neoplasia or infectious diseases, although in
certain instances
lower amounts will be needed because of the increased specificity of the
compound. A
compound is administered at a dosage that enhances an immune response of a
subject, or that
reduces the proliferation, survival, or invasiveness of a neoplastic or,
infected cell as
determined by a method known to one skilled in the art.
Formulation of Pharmaceutical Compositions
The administration of the fusion protein complex of the invention for the
treatment of
a neoplasia or infectious disease is by any suitable means that results in a
concentration of the
therapeutic that, combined with other components, is effective in
ameliorating, reducing, or
stabilizing said neoplasia or infectious disease. The fusion protein complex
of the invention
may be contained in any appropriate amount in any suitable carrier substance,
and is
generally present in an amount of 1-95% by weight of the total weight of the
composition.
The composition may be provided in a dosage form that is suitable for
parenteral (e.g.,
subcutaneous, intravenous, intramuscular, intravesicular, intratumoral or
intraperitoneal)
administration route. For example, the pharmaceutical compositions are
formulated
according to conventional pharmaceutical practice (see, e.g., Remington: The
Science and
Practice of Pharmacy (20th ed.), ed. A. R. Gennaro, Lippincott Williams &
Wilkins, 2000
46
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
and Encyclopedia of Pharmaceutical Technology, eds. J. Swarbrick and J. C.
Boylan, 1988-
1999, Marcel Dekker, New York).
Human dosage amounts are initially determined by extrapolating from the amount
of
compound used in mice or non-human primates, as a skilled artisan recognizes
it is routine in
the art to modify the dosage for humans compared to animal models. For
example, the
dosage may vary from between about 1 lag compound/kg body weight to about 5000
mg
compound/kg body weight; or from about 5 mg/kg body weight to about 4,000
mg/kg body
weight or from about 10 mg/kg body weight to about 3,000 mg/kg body weight; or
from
about 50 mg/kg body weight to about 2000 mg/kg body weight; or from about 100
mg/kg
body weight to about 1000 mg/kg body weight; or from about 150 mg/kg body
weight to
about 500 mg/kg body weight. For example, the dose is about 1, 5, 10, 25, 50,
75, 100, 150,
200, 250, 300, 350, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900,
950, 1,000, 1,050,
1,100, 1,150, 1,200, 1,250, 1,300, 1,350, 1,400, 1,450, 1,500, 1,600, 1,700,
1,800, 1,900,
2,000, 2,500, 3,000, 3,500, 4,000, 4,500, or 5,000 mg/kg body weight.
Alternatively, doses
are in the range of about 5 mg compound/Kg body weight to about 20 mg
compound/kg body
weight. In another example, the doses are about 8, 10, 12, 14, 16 or 18 mg/kg
body weight.
Preferably, the fusion protein complex is administered at 0.5 mg/kg-about 10
mg/kg (e.g.,
0.5, 1, 3, 5, 10 mg/kg). Of course, this dosage amount may be adjusted upward
or downward,
as is routinely done in such treatment protocols, depending on the results of
the initial clinical
trials and the needs of a particular patient.
Pharmaceutical compositions are formulated with appropriate excipients into a
pharmaceutical composition that, upon administration, releases the therapeutic
in a controlled
manner. Examples include single or multiple unit tablet or capsule
compositions, oil
solutions, suspensions, emulsions, microcapsules, microspheres, molecular
complexes,
nanoparticles, patches, and liposomes. Preferably, the fusion protein complex
is formulated
in an excipient suitable for parenteral administration.
Parenteral Compositions
The pharmaceutical composition comprising a fusion protein complex of the
invention are administered parenterally by injection, infusion or implantation
(subcutaneous,
intravenous, intramuscular, intratumoral, intravesicular, intraperitoneal) in
dosage forms,
formulations, or via suitable delivery devices or implants containing
conventional, non-toxic
pharmaceutically acceptable carriers and adjuvants. The formulation and
preparation of such
47
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
compositions are well known to those skilled in the art of pharmaceutical
formulation.
Formulations can be found in Remington: The Science and Practice of Pharmacy,
supra.
Compositions comprising a fusion protein complex of the invention for
parenteral use
are provided in unit dosage forms (e.g., in single-dose ampoules).
Alternatively, the
composition is provided in vials containing several doses and in which a
suitable preservative
may be added (see below). The composition is in the form of a solution, a
suspension, an
emulsion, an infusion device, or a delivery device for implantation, or is
presented as a dry
powder to be reconstituted with water or another suitable vehicle before use.
Apart from the
active agent that reduces or ameliorates a neoplasia or infectious disease,
the composition
includes suitable parenterally acceptable carriers and/or excipients. The
active therapeutic
agent(s) may be incorporated into microspheres, microcapsules, nanoparticles,
liposomes for
controlled release. Furthermore, the composition may include suspending,
solubilizing,
stabilizing, pH-adjusting agents, tonicity adjusting agents, and/or
dispersing, agents.
As indicated above, the pharmaceutical compositions comprising a fusion
protein
complex of the invention may be in a form suitable for sterile injection. To
prepare such a
composition, the suitable active therapeutic(s) are dissolved or suspended in
a parenterally
acceptable liquid vehicle. Among acceptable vehicles and solvents that may be
employed are
water, water adjusted to a suitable pH by addition of an appropriate amount of
hydrochloric
acid, sodium hydroxide or a suitable buffer, 1,3-butanediol, Ringer's
solution, and isotonic
sodium chloride solution and dextrose solution. The aqueous formulation may
also contain
one or more preservatives (e.g., methyl, ethyl or n-propyl p-hydroxybenzoate).
In cases
where one of the compounds is only sparingly or slightly soluble in water, a
dissolution
enhancing or solubilizing agent can be added, or the solvent may include 10-
60% w/w of
propylene glycol.
The present invention provides methods of treating neoplasia or infectious
diseases or
symptoms thereof which comprise administering a therapeutically effective
amount of a
pharmaceutical composition comprising a compound of the formulae herein to a
subject (e.g.,
a mammal such as a human). Thus, one embodiment is a method of treating a
subject
suffering from or susceptible to a neoplasia or infectious disease or symptom
thereof The
method includes the step of administering to the mammal a therapeutic amount
of an amount
of a compound herein sufficient to treat the disease or disorder or symptom
thereof, under
conditions such that the disease or disorder is treated.
The methods herein include administering to the subject (including a subject
identified as in need of such treatment) an effective amount of a compound
described herein,
48
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
or a composition described herein to produce such effect. Identifying a
subject in need of
such treatment can be in the judgment of a subject or a health care
professional and can be
subjective (e.g. opinion) or objective (e.g. measurable by a test or
diagnostic method).
The therapeutic methods of the invention (which include prophylactic
treatment) in
general comprise administration of a therapeutically effective amount of the
compounds
herein, such as a compound of the formulae herein to a subject (e.g., animal,
human) in need
thereof, including a mammal, particularly a human. Such treatment will be
suitably
administered to subjects, particularly humans, suffering from, having,
susceptible to, or at
risk for a neoplasia, infectious disease, disorder, or symptom thereof
Determination of those
subjects "at risk" can be made by any objective or subjective determination by
a diagnostic
test or opinion of a subject or health care provider (e.g., genetic test,
enzyme or protein
marker, Marker (as defined herein), family history, and the like). The fusion
protein
complexes of the invention may be used in the treatment of any other disorders
in which an
increase in an immune response is desired.
The invention also provides a method of monitoring treatment progress. The
method
includes the step of determining a level of diagnostic marker (Marker) (e.g.,
any target
delineated herein modulated by a compound herein, a protein or indicator
thereof, etc.) or
diagnostic measurement (e.g., screen, assay) in a subject suffering from or
susceptible to a
disorder or symptoms thereof associated with neoplasia in which the subject
has been
administered a therapeutic amount of a compound herein sufficient to treat the
disease or
symptoms thereof The level of Marker determined in the method can be compared
to known
levels of Marker in either healthy normal controls or in other afflicted
patients to establish the
subject's disease status. In some cases, a second level of Marker in the
subject is determined
at a time point later than the determination of the first level, and the two
levels are compared
to monitor the course of disease or the efficacy of the therapy. In certain
aspects, a pre-
treatment level of Marker in the subject is determined prior to beginning
treatment according
to this invention; this pre-treatment level of Marker can then be compared to
the level of
Marker in the subject after the treatment commences, to determine the efficacy
of the
treatment.
Combination Therapies
Optionally, the fusion protein complex of the invention or immune cells
treated with the
fusion protein complex of the invention are administered in combination with
any other
standard therapy; such methods are known to the skilled artisan and described
in Remington's
49
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
Pharmaceutical Sciences by E. W. Martin. If desired, the fusion protein
complex of the
invention or immune cells treated with the fusion protein complex of the
invention are
administered in combination with any conventional anti-neoplastic therapy,
including but not
limited to, immunotherapy, adoptive cell therapy, vaccines, therapeutic and
checkpoint
inhibitor antibodies, targeted therapy, surgery, radiation therapy, or
chemotherapy.
Kits or Pharmaceutical Systems
Pharmaceutical compositions comprising the fusion protein complex of the
invention
or immune cells treated with the fusion protein complex of the invention may
be assembled
into kits or pharmaceutical systems for use in ameliorating a neoplasia or
infectious disease.
Kits or pharmaceutical systems according to this aspect of the invention
comprise a carrier
means, such as a box, carton, tube, having in close confinement therein one or
more container
means, such as vials, tubes, ampoules, bottles and the like. The kits or
pharmaceutical
systems of the invention may also comprise associated instructions for using
the fusion
protein complex of the invention. In one embodiment, the kit includes
appropriate containers
such as bags, bottles, tubes, to allow ex vivo treatment of immune cells using
the fusion
protein complex of the invention and/or administration of such cells to a
patient. Kits may
also include medical devices comprising the fusion protein complex of the
invention.
Recombinant Protein Expression
In general, preparation of the fusion protein complexes of the invention
(e.g.,
components of a TxM complex) can be accomplished by procedures disclosed
herein and by
recognized recombinant DNA techniques.
In general, recombinant polypeptides are produced by transformation of a
suitable
host cell with all or part of a polypeptide-encoding nucleic acid molecule or
fragment thereof
in a suitable expression vehicle. Those skilled in the field of molecular
biology will
understand that any of a wide variety of expression systems may be used to
provide the
recombinant protein. The precise host cell used is not critical to the
invention. A
recombinant polypeptide may be produced in virtually any eukaryotic host
(e.g.,
Saccharomyces cerevisiae, insect cells, e.g., Sf21 cells, or mammalian cells,
e.g., NIH 3T3,
HeLa, or preferably COS cells). Such cells are available from a wide range of
sources (e.g.,
the American Type Culture Collection, Rockland, Md.; also, see, e.g., Ausubel
et al., Current
Protocol in Molecular Biology, New York: John Wiley and Sons, 1997). The
method of
transfection and the choice of expression vehicle will depend on the host
system selected.
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
Transformation methods are described, e.g., in Ausubel et al. (supra);
expression vehicles
may be chosen from those provided, e.g., in Cloning Vectors: A Laboratory
Manual (P. H.
Pouwels et al., 1985, Supp. 1987).
A variety of expression systems exist for the production of recombinant
polypeptides.
Expression vectors useful for producing such polypeptides include, without
limitation,
chromosomal, episomal, and virus-derived vectors, e.g., vectors derived from
bacterial
plasmids, from bacteriophage, from transposons, from yeast episomes, from
insertion
elements, from yeast chromosomal elements, from viruses such as baculoviruses,
papova
viruses, such as SV40, vaccinia viruses, adenoviruses, fowl pox viruses,
pseudorabies viruses
and retroviruses, and vectors derived from combinations thereof
Once the recombinant polypeptide is expressed, it is isolated, e.g., using
affinity
chromatography. In one example, an antibody (e.g., produced as described
herein) raised
against the polypeptide may be attached to a column and used to isolate the
recombinant
polypeptide. Lysis and fractionation of polypeptide-harboring cells prior to
affinity
chromatography may be performed by standard methods (see, e.g., Ausubel et
al., supra).
Once isolated, the recombinant protein can, if desired, be further purified,
e.g., by high
performance liquid chromatography (see, e.g., Fisher, Laboratory Techniques in
Biochemistry and Molecular Biology, eds., Work and Burdon, Elsevier, 1980).
As used herein, biologically active polypeptides or effector molecules of the
invention
may include factors such as cytokines, chemokines, growth factors, protein
toxins,
immunoglobulin domains or other bioactive proteins such as enzymes. Also,
biologically
active polypeptides may include conjugates to other compounds such as non-
protein toxins,
cytotoxic agents, chemotherapeutic agents, detectable labels, radioactive
materials and such.
Cytokines of the invention are defined by any factor produced by cells that
affect
other cells and are responsible for any of a number of multiple effects of
cellular immunity.
Examples of cytokines include but are not limited to the IL-2 family,
interferon (IFN), IL-10,
IL-12, IL-18, IL-1, IL-17, TGF and TNF cytokine families, and to IL-1 through
IL-35, IFN-
a, IFN-p, IFNy, TGF-p, TNF-a, and TNFP.
In an aspect of the invention, the first protein comprises a first
biologically active
polypeptide covalently linked to interleukin-15 (IL-15) domain or a functional
fragment
thereof IL-15 is a cytokine that affects T-cell activation and proliferation.
IL-15 activity in
affecting immune cell activation and proliferation is similar in some respects
to IL-2,
51
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
although fundamental differences have been well characterized (Waldmann, T A,
2006,
Nature Rev. Immunol. 6:595-601).
In another aspect of the invention, the first protein comprises an interleukin-
15 (IL-
15) domain that is an IL-15 variant (also referred to herein as IL-15 mutant).
The IL-15
variant preferably comprises a different amino acid sequence that the native
(or wild type) IL-
15 protein. The IL-15 variant preferably binds the IL-15Ra polypeptide and
functions as an
IL-15 agonist or antagonist. Preferably, IL-15 variants with agonist activity
have super
agonist activity. The IL-15 variant can function as an IL-15 agonist or
antagonist
independent of its association with IL-15Ra. IL-15 agonists are exemplified by
comparable
or increased biological activity compared to wild type IL-15. IL-15
antagonists are
exemplified by decreased biological activity compared to wild type IL-15 or by
the ability to
inhibit IL-15-mediated responses. In some examples, the IL-15 variant binds
with increased
or decreased activity to the IL-15RPyC receptors. In some cases, the sequence
of the IL-15
variant has at least one amino acid change, e.g. substitution or deletion,
compared to the
native IL-15 sequence, such changes resulting in IL-15 agonist or antagonist
activity.
Preferably, the amino acid substitutions/deletions are in the domains of IL-15
that interact
with IL-15R P and/or yC. More preferably, the amino acid
substitutions/deletions do not
affect binding to the IL-15Ra polypeptide or the ability to produce the IL-15
variant.
Suitable amino acid substitutions/deletions to generate IL-15 variants can be
identified based
on putative or known IL-15 structures, comparisons of IL-15 with homologous
molecules
such as IL-2 with known structure, through rational or random mutagenesis and
functional
assays, as provided herein, or other empirical methods. Additionally, suitable
amino acid
substitutions can be conservative or non-conservative changes and insertions
of additional
amino acids. Preferably, IL-15 variants of the invention contain one or more
than one amino
acid substitutions/deletions at position 6, 8, 10, 61, 65, 72, 92, 101, 104,
105, 108, 109, 111,
or 112 of the mature human IL-15 sequence; particularly, D8N ("D8" refers to
the amino acid
and residue position in the native mature human IL-15 sequence and "N" refers
to the
substituted amino acid residue at that position in the IL-15 variant), I6S,
D8A, D61A, N65A,
N72R, V104P or Q108A substitutions result in IL-15 variants with antagonist
activity and
N72D substitutions result in IL-15 variants with agonist activity.
Chemokines, similar to cytokines, are defined as any chemical factor or
molecule
which when exposed to other cells are responsible for any of a number of
multiple effects of
cellular immunity. Suitable chemokines may include but are not limited to the
CXC, CC, C,
52
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
and CX3C chemokine families and to CCL-1 through CCL-28, CXC-1 through CXC-17,
XCL-1, XCL-2, CX3CL1, MIP-lb, IL-8, MCP-1, and Rantes.
Growth factors include any molecules which when exposed to a particular cell
induce
proliferation and/or differentiation of the affected cell. Growth factors
include proteins and
chemical molecules, some of which include: GM-CSF, G-CSF, human growth factor
and
stem cell growth factor. Additional growth factors may also be suitable for
uses described
herein.
Toxins or cytotoxic agents include any substance that has a lethal effect or
an
inhibitory effect on growth when exposed to cells. More specifically, the
effector molecule
can be a cell toxin of, e.g., plant or bacterial origin such as, e.g.,
diphtheria toxin (DT), shiga
toxin, abrin, cholera toxin, ricin, saporin, pseudomonas exotoxin (PE),
pokeweed antiviral
protein, or gelonin. Biologically active fragments of such toxins are well
known in the art and
include, e.g., DT A chain and ricin A chain. Additionally, the toxin can be an
agent active at
the cell surface such as, e.g., phospholipase enzymes (e.g., phospholipase C).
Further, the effector molecule can be a chemotherapeutic drug such as, e.g.,
vindesine,
vincristine, vinblastin, methotrexate, adriamycin, bleomycin, or cisplatin.
Additionally, the effector molecule can be a detectably-labeled molecule
suitable for
diagnostic or imaging studies. Such labels include biotin or
streptavidin/avidin, a detectable
nanoparticles or crystal, an enzyme or catalytically active fragment thereof,
a fluorescent
label such as green fluorescent protein, FITC, phycoerythrin, cychome, texas
red or quantum
dots; a radionuclide e.g., iodine-131, yttrium-90, rhenium-188 or bismuth-212;
a
phosphorescent or chemiluminescent molecules or a label detectable by PET,
ultrasound or
MRI such as Gd-- or paramagnetic metal ion-based contrast agents. See e.g.,
Moskaug, et al.
Biol. Chem. 264, 15709 (1989); Pastan, I. et al. Cell 47, 641, 1986; Pastan et
al.,
Recombinant Toxins as Novel Therapeutic Agents, Ann. Rev. Biochem. 61, 331,
(1992);
"Chimeric Toxins" Olsnes and Phil, Pharmac. Ther., 25, 355 (1982); published
PCT
application no. WO 94/29350; published PCT application no. WO 94/04689;
published PCT
application no. W02005046449 and U.S. Pat. No. 5,620,939 for disclosure
relating to
making and using proteins comprising effectors or tags.
The IL-15 and IL-15Ra polypeptides of the invention suitably correspond in
amino
acid sequence to naturally occurring IL-15 and IL-15Ra molecules, e.g. IL-15
and IL-15Ra
molecules of a human, mouse or other rodent, or other mammal. Sequences of
these
polypeptides and encoding nucleic acids are known in the literature, including
human
interleukin 15 (IL15) mRNA - GenBank: U14407.1 (incorporated herein by
reference), Mus
53
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
musculus interleukin 15 (IL15) mRNA - GenBank: U14332.1 (incorporated herein
by
reference), human interleukin-15 receptor alpha chain precursor (IL15RA) mRNA -
GenBank: U31628.1 (incorporated herein by reference), Mus musculus interleukin
15
receptor, alpha chain - GenBank: BC095982.1 (incorporated herein by
reference).
In some settings, it can be useful to make the protein fusion or conjugate
complexes
of the present invention polyvalent, e.g., to increase the valency of the sc-
antibody. In
particular, interactions between the IL-15 and IL-15Ra domains of the fusion
protein
complex provide a means of generating polyvalent complexes. In addition, the
polyvalent
fusion protein can be made by covalently or non-covalently linking together
between one and
four proteins (the same or different) by using e.g., standard biotin-
streptavidin labeling
techniques, or by conjugation to suitable solid supports such as latex beads.
Chemically
cross-linked proteins (for example cross-linked to dendrimers) are also
suitable polyvalent
species. For example, the protein can be modified by including sequences
encoding tag
sequences that can be modified such as the biotinylation BirA tag or amino
acid residues with
chemically reactive side chains such as Cys or His. Such amino acid tags or
chemically
reactive amino acids may be positioned in a variety of positions in the fusion
protein,
preferably distal to the active site of the biologically active polypeptide or
effector molecule.
For example, the C-terminus of a soluble fusion protein can be covalently
linked to a tag or
other fused protein which includes such a reactive amino acid(s). Suitable
side chains can be
included to chemically link two or more fusion proteins to a suitable
dendrimer or other
nanoparticle to give a multivalent molecule. Dendrimers are synthetic chemical
polymers that
can have any one of a number of different functional groups of their surface
(D. Tomalia,
Aldrichimica Acta, 26:91:101(1993)). Exemplary dendrimers for use in
accordance with the
present invention include e.g. E9 starburst polyamine dendrimer and E9 combust
polyamine
dendrimer, which can link cysteine residues. Exemplary nanoparticles include
liposomes,
core-shell particles or PLGA-based particles.
In another aspect, one or both of the polypeptides of the fusion protein
complex
comprises an immunoglobulin domain. Alternatively, the protein binding domain-
IL-15
fusion protein can be further linked to an immunoglobulin domain. The
preferred
immunoglobulin domains comprise regions that allow interaction with other
immunoglobulin
domains to form multichain proteins as provided above. For example, the
immunoglobulin
heavy chain regions, such as the IgG1 CH2-013, are capable of stably
interacting to create the
Fc region. Preferred immunoglobulin domains including Fc domains also comprise
regions
with effector functions, including Fc receptor or complement protein binding
activity, and/or
54
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
with glycosylation sites. In some aspects, the immunoglobulin domains of the
fusion protein
complex contain mutations that reduce or augment Fc receptor or complement
binding
activity or glycosylation or dimerization, thereby affecting the biological
activity of the
resulting protein. For example, immunoglobulin domains containing mutations
that reduce
binding to Fc receptors could be used to generate fusion protein complex of
the invention
with lower binding activity to Fc receptor-bearing cells, which may be
advantageous for
reagents designed to recognize or detect specific antigens.
Nucleic Acids and Vectors
The invention further provides nucleic acid sequences and particularly DNA
sequences that encode the present fusion proteins (e.g., components of TxM).
Preferably, the
DNA sequence is carried by a vector suited for extrachromosomal replication
such as a
phage, virus, plasmid, phagemid, cosmid, YAC, or episome. In particular, a DNA
vector that
encodes a desired fusion protein can be used to facilitate preparative methods
described
herein and to obtain significant quantities of the fusion protein. The DNA
sequence can be
inserted into an appropriate expression vector, i.e., a vector that contains
the necessary
elements for the transcription and translation of the inserted protein-coding
sequence. A
variety of host-vector systems may be utilized to express the protein-coding
sequence. These
include mammalian cell systems infected with virus (e.g., vaccinia virus,
adenovirus, etc.);
insect cell systems infected with virus (e.g., baculovirus); microorganisms
such as yeast
containing yeast vectors, or bacteria transformed with bacteriophage DNA,
plasmid DNA or
cosmid DNA. Depending on the host-vector system utilized, any one of a number
of suitable
transcription and translation elements may be used. See, Sambrook et al.,
supra and Ausubel
et al. supra.
Included in the invention are methods for making a soluble fusion protein
complex,
the method comprising introducing into a host cell a DNA vector as described
herein
encoding the first and second proteins, culturing the host cell in media under
conditions
sufficient to express the fusion proteins in the cell or the media and allow
association
between IL-15 domain of a first protein and the soluble IL-15Ra domain of a
second protein
to form the soluble fusion protein complex, purifying the soluble fusion
protein complex
from the host cells or media.
In general, a preferred DNA vector according to the invention comprises a
nucleotide
sequence linked by phosphodiester bonds comprising, in a 5' to 3' direction a
first cloning site
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
for introduction of a first nucleotide sequence encoding a biologically active
polypeptide,
operatively linked to a sequence encoding an effector molecule.
The fusion protein components encoded by the DNA vector can be provided in a
cassette format. By the term "cassette" is meant that each component can be
readily
substituted for another component by standard recombinant methods. In
particular, a DNA
vector configured in a cassette format is particularly desirable when the
encoded fusion
protein complex is to be used against pathogens that may have or have capacity
to develop
serotypes.
To make the vector coding for a fusion protein complex, the sequence coding
for the
biologically active polypeptide is linked to a sequence coding for the
effector peptide by use
of suitable ligases. DNA coding for the presenting peptide can be obtained by
isolating DNA
from natural sources such as from a suitable cell line or by known synthetic
methods, e.g. the
phosphate triester method. See, e.g., Oligonucleotide Synthesis, IRL Press (M.
J. Gait, ed.,
1984). Synthetic oligonucleotides also may be prepared using commercially
available
automated oligonucleotide synthesizers. Once isolated, the gene coding for the
biologically
active polypeptide can be amplified by the polymerase chain reaction (PCR) or
other means
known in the art. Suitable PCR primers to amplify the biologically active
polypeptide gene
may add restriction sites to the PCR product. The PCR product preferably
includes splice
sites for the effector peptide and leader sequences necessary for proper
expression and
secretion of the biologically active polypeptide -effector fusion complex. The
PCR product
also preferably includes a sequence coding for the linker sequence, or a
restriction enzyme
site for ligation of such a sequence.
The fusion proteins described herein are preferably produced by standard
recombinant
DNA techniques. For example, once a DNA molecule encoding the biologically
active
polypeptide is isolated, sequence can be ligated to another DNA molecule
encoding the
effector polypeptide. The nucleotide sequence coding for a biologically active
polypeptide
may be directly joined to a DNA sequence coding for the effector peptide or,
more typically,
a DNA sequence coding for the linker sequence as discussed herein may be
interposed
between the sequence coding for the biologically active polypeptide and the
sequence coding
for the effector peptide and joined using suitable ligases. The resultant
hybrid DNA molecule
can be expressed in a suitable host cell to produce the fusion protein
complex. The DNA
molecules are ligated to each other in a 5' to 3' orientation such that, after
ligation, the
translational frame of the encoded polypeptides is not altered (i.e., the DNA
molecules are
56
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
ligated to each other in-frame). The resulting DNA molecules encode an in-
frame fusion
protein.
Other nucleotide sequences also can be included in the gene construct. For
example,
a promoter sequence, which controls expression of the sequence coding for the
biologically
active polypeptide fused to the effector peptide, or a leader sequence, which
directs the fusion
protein to the cell surface or the culture medium, can be included in the
construct or present
in the expression vector into which the construct is inserted. An
immunoglobulin or CMV
promoter is particularly preferred.
In obtaining variant biologically active polypeptide, IL-15, IL-15Ra or Fc
domain
coding sequences, those of ordinary skill in the art will recognize that the
polypeptides may
be modified by certain amino acid substitutions, additions, deletions, and
post-translational
modifications, without loss or reduction of biological activity. In
particular, it is well-known
that conservative amino acid substitutions, that is, substitution of one amino
acid for another
amino acid of similar size, charge, polarity and conformation, are unlikely to
significantly
alter protein function. The 20 standard amino acids that are the constituents
of proteins can
be broadly categorized into four groups of conservative amino acids as
follows: the nonpolar
(hydrophobic) group includes alanine, isoleucine, leucine, methionine,
phenylalanine,
proline, tryptophan and valine; the polar (uncharged, neutral) group includes
asparagine,
cysteine, glutamine, glycine, serine, threonine and tyrosine; the positively
charged (basic)
group contains arginine, histidine and lysine; and the negatively charged
(acidic) group
contains aspartic acid and glutamic acid. Substitution in a protein of one
amino acid for
another within the same group is unlikely to have an adverse effect on the
biological activity
of the protein. In other instance, modifications to amino acid positions can
be made to reduce
or enhance the biological activity of the protein. Such changes can be
introduced randomly
or via site-specific mutations based on known or presumed structural or
functional properties
of targeted residue(s). Following expression of the variant protein, the
changes in the
biological activity due to the modification can be readily assessed using
binding or functional
assays.
Homology between nucleotide sequences can be determined by DNA hybridization
analysis, wherein the stability of the double-stranded DNA hybrid is dependent
on the extent
of base pairing that occurs. Conditions of high temperature and/or low salt
content reduce the
stability of the hybrid, and can be varied to prevent annealing of sequences
having less than a
selected degree of homology. For instance, for sequences with about 55% G-C
content,
hybridization and wash conditions of 40-50 C, 6 x SSC (sodium chloride/sodium
citrate
57
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
buffer) and 0.1% SDS (sodium dodecyl sulfate) indicate about 60-70% homology,
hybridization and wash conditions of 50-65 C, 1 x SSC and 0.1% SDS indicate
about 82-97%
homology, and hybridization and wash conditions of 52 C, 0.1 x SSC and 0.1%
SDS indicate
about 99-100% homology. A wide range of computer programs for comparing
nucleotide
and amino acid sequences (and measuring the degree of homology) are also
available, and a
list providing sources of both commercially available and free software is
found in Ausubel et
al. (1999). Readily available sequence comparison and multiple sequence
alignment
algorithms are, respectively, the Basic Local Alignment Search Tool (BLAST)
(Altschul et
al., 1997) and ClustalW programs. BLAST is available on the world wide web at
ncbi.nlm.nih.gov and a version of ClustalW is available at 2.ebi.ac.uk.
The components of the fusion protein can be organized in nearly any order
provided
each is capable of performing its intended function. For example, in one
embodiment, the
biologically active polypeptide is situated at the C or N terminal end of the
effector molecule.
Preferred effector molecules of the invention will have sizes conducive to the
function
for which those domains are intended. The effector molecules of the invention
can be made
and fused to the biologically active polypeptide by a variety of methods
including well-
known chemical cross-linking methods. See, e.g., Means, G. E. and Feeney, R.
E. (1974) in
Chemical Modification of Proteins, Holden-Day. See also, S. S. Wong (1991) in
Chemistry of
Protein Conjugation and Cross-Linking, CRC Press. However it is generally
preferred to use
recombinant manipulations to make the in-frame fusion protein.
As noted, a fusion molecule or a conjugate molecule in accord with the
invention can
be organized in several ways. In an exemplary configuration, the C-terminus of
the
biologically active polypeptide is operatively linked to the N-terminus of the
effector
molecule. That linkage can be achieved by recombinant methods if desired.
However, in
another configuration, the N-terminus of the biologically active polypeptide
is linked to the
C-terminus of the effector molecule.
Alternatively, or in addition, one or more additional effector molecules can
be
inserted into the biologically active polypeptide or conjugate complexes as
needed.
Vectors and Expression
A number of strategies can be employed to express the components of fusion
protein
complex of the invention (e.g., TxM). For example, a construct encoding one or
more
components of fusion protein complex of the invention can be incorporated into
a suitable
vector using restriction enzymes to make cuts in the vector for insertion of
the construct
58
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
followed by ligation. The vector containing the gene construct is then
introduced into a
suitable host for expression of the fusion protein. See, generally, Sambrook
et al., supra.
Selection of suitable vectors can be made empirically based on factors
relating to the cloning
protocol. For example, the vector should be compatible with, and have the
proper replicon
for the host that is being employed. The vector must be able to accommodate
the DNA
sequence coding for the fusion protein complex that is to be expressed.
Suitable host cells
include eukaryotic and prokaryotic cells, preferably those cells that can be
easily transformed
and exhibit rapid growth in culture medium. Specifically, preferred hosts
cells include
prokaryotes such as E. coil, Bacillus subtillus, etc. and eukaryotes such as
animal cells and
yeast strains, e.g., S. cerevisiae. Mammalian cells are generally preferred,
particularly J558,
NSO, 5P2-0 or CHO. Other suitable hosts include, e.g., insect cells such as
Sf9.
Conventional culturing conditions are employed. See, Sambrook, supra. Stable
transformed
or transfected cell lines can then be selected. Cells expressing a fusion
protein complex of
the invention can be determined by known procedures. For example, expression
of a fusion
protein complex linked to an immunoglobulin can be determined by an ELISA
specific for
the linked immunoglobulin and/or by immunoblotting. Other methods for
detecting
expression of fusion proteins comprising biologically active polypeptides
linked to IL-15 or
IL-15Ra domains are disclosed in the Examples.
As mentioned generally above, a host cell can be used for preparative purposes
to
propagate nucleic acid encoding a desired fusion protein. Thus, a host cell
can include a
prokaryotic or eukaryotic cell in which production of the fusion protein is
specifically
intended. Thus host cells specifically include yeast, fly, worm, plant, frog,
mammalian cells
and organs that are capable of propagating nucleic acid encoding the fusion.
Non-limiting
examples of mammalian cell lines which can be used include CHO dhfr-cells
(Urlaub and
Chasm, Proc. Natl. Acad. Sci. USA, 77:4216 (1980)), 293 cells (Graham et al.,
J Gen. Virol.,
36:59 (1977)) or myeloma cells like 5P2 or NSO (Galfre and Milstein, Meth.
Enzymol.,
73(B):3 (1981)).
Host cells capable of propagating nucleic acid encoding a desired fusion
protein
complexes encompass non-mammalian eukaryotic cells as well, including insect
(e.g., Sp.
frugiperda), yeast (e.g., S. cerevisiae, S. pombe, P. pastoris., K lactis, H
polymorpha; as
generally reviewed by Fleer, R., Current Opinion in Biotechnology, 3(5):486496
(1992)),
fungal and plant cells. Also contemplated are certain prokaryotes such as E.
coil and
Bacillus.
59
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
Nucleic acid encoding a desired fusion protein can be introduced into a host
cell by
standard techniques for transfecting cells. The term "transfecting" or
"transfection" is
intended to encompass all conventional techniques for introducing nucleic acid
into host
cells, including calcium phosphate co-precipitation, DEAE-dextran-mediated
transfection,
lipofection, electroporation, microinjection, viral transduction and/or
integration. Suitable
methods for transfecting host cells can be found in Sambrook et al. supra, and
other
laboratory textbooks.
Various promoters (transcriptional initiation regulatory region) may be used
according to the invention. The selection of the appropriate promoter is
dependent upon the
proposed expression host. Promoters from heterologous sources may be used as
long as they
are functional in the chosen host.
Promoter selection is also dependent upon the desired efficiency and level of
peptide
or protein production. Inducible promoters such as tac are often employed in
order to
dramatically increase the level of protein expression in E coil.
Overexpression of proteins
may be harmful to the host cells. Consequently, host cell growth may be
limited. The use of
inducible promoter systems allows the host cells to be cultivated to
acceptable densities prior
to induction of gene expression, thereby facilitating higher product yields.
Various signal sequences may be used according to the invention. A signal
sequence
which is homologous to the biologically active polypeptide coding sequence may
be used.
Alternatively, a signal sequence which has been selected or designed for
efficient secretion
and processing in the expression host may also be used. For example, suitable
signal
sequence/host cell pairs include the B. subtilis sacB signal sequence for
secretion in B.
subtilis, and the Saccharomyces cerevisiae a-mating factor or P. pastoris acid
phosphatase
phoI signal sequences for P. pastoris secretion. The signal sequence may be
joined directly
through the sequence encoding the signal peptidase cleavage site to the
protein coding
sequence, or through a short nucleotide bridge consisting of usually fewer
than ten codons,
where the bridge ensures correct reading frame of the downstream TCR sequence.
Elements for enhancing transcription and translation have been identified for
eukaryotic protein expression systems. For example, positioning the
cauliflower mosaic
virus (CaMV) promoter 1,000 bp on either side of a heterologous promoter may
elevate
transcriptional levels by 10- to 400-fold in plant cells. The expression
construct should also
include the appropriate translational initiation sequences. Modification of
the expression
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
construct to include a Kozak consensus sequence for proper translational
initiation may
increase the level of translation by 10 fold.
A selective marker is often employed, which may be part of the expression
construct
or separate from it (e.g., carried by the expression vector), so that the
marker may integrate at
a site different from the gene of interest. Examples include markers that
confer resistance to
antibiotics (e.g., bla confers resistance to ampicillin for E. coil host
cells, nptII confers
kanamycin resistance to a wide variety of prokaryotic and eukaryotic cells) or
that permit the
host to grow on minimal medium (e.g., HIS4 enables P. pastoris or His-S
cerevisiae to grow
in the absence of histidine). The selectable marker has its own
transcriptional and
translational initiation and termination regulatory regions to allow for
independent expression
of the marker. If antibiotic resistance is employed as a marker, the
concentration of the
antibiotic for selection will vary depending upon the antibiotic, generally
ranging from 10 to
600 jig of the antibiotic/mL of medium.
The expression construct is assembled by employing known recombinant DNA
techniques (Sambrook et al., 1989; Ausubel et al., 1999). Restriction enzyme
digestion and
ligation are the basic steps employed to join two fragments of DNA. The ends
of the DNA
fragment may require modification prior to ligation, and this may be
accomplished by filling
in overhangs, deleting terminal portions of the fragment(s) with nucleases
(e.g., ExoIII), site
directed mutagenesis, or by adding new base pairs by PCR. Polylinkers and
adaptors may be
employed to facilitate joining of selected fragments. The expression construct
is typically
assembled in stages employing rounds of restriction, ligation, and
transformation of E. coil.
Numerous cloning vectors suitable for construction of the expression construct
are known in
the art (XZAP and pBLUESCRIPT SK-1, Stratagene, La Jolla, CA, pET, Novagen
Inc.,
Madison, WI, cited in Ausubel et al., 1999) and the particular choice is not
critical to the
invention. The selection of cloning vector will be influenced by the gene
transfer system
selected for introduction of the expression construct into the host cell. At
the end of each
stage, the resulting construct may be analyzed by restriction, DNA sequence,
hybridization
and PCR analyses.
The expression construct may be transformed into the host as the cloning
vector
construct, either linear or circular, or may be removed from the cloning
vector and used as is
or introduced onto a delivery vector. The delivery vector facilitates the
introduction and
maintenance of the expression construct in the selected host cell type. The
expression
construct is introduced into the host cells by any of a number of known gene
transfer systems
61
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
(e.g., natural competence, chemically mediated transformation, protoplast
transformation,
electroporation, biolistic transformation, transfection, or conjugation)
(Ausubel et al., 1999;
Sambrook et al., 1989). The gene transfer system selected depends upon the
host cells and
vector systems used.
For instance, the expression construct can be introduced into S. cerevisiae
cells by
protoplast transformation or electroporation. Electroporation of S. cerevisiae
is readily
accomplished, and yields transformation efficiencies comparable to spheroplast
transformation.
The present invention further provides a production process for isolating a
fusion
protein of interest. In the process, a host cell (e.g., a yeast, fungus,
insect, bacterial or animal
cell), into which has been introduced a nucleic acid encoding the protein of
the interest
operatively linked to a regulatory sequence, is grown at production scale in a
culture medium
to stimulate transcription of the nucleotides sequence encoding the fusion
protein of interest.
Subsequently, the fusion protein of interest is isolated from harvested host
cells or from the
culture medium. Standard protein purification techniques can be used to
isolate the protein of
interest from the medium or from the harvested cells. In particular, the
purification
techniques can be used to express and purify a desired fusion protein on a
large-scale (i.e. in
at least milligram quantities) from a variety of implementations including
roller bottles,
spinner flasks, tissue culture plates, bioreactor, or a fermentor.
An expressed protein fusion complex can be isolated and purified by known
methods.
Typically the culture medium is centrifuged or filtered and then the
supernatant is purified by
affinity or immunoaffinity chromatography, e.g. Protein-A or Protein-G
affinity
chromatography or an immunoaffinity protocol comprising use of monoclonal
antibodies that
bind the expressed fusion protein complex. The fusion proteins of the present
invention can
be separated and purified by appropriate combination of known techniques.
These methods
include, for example, methods utilizing solubility such as salt precipitation
and solvent
precipitation, methods utilizing the difference in molecular weight such as
dialysis, ultra-
filtration, gel-filtration, and SDS-polyacrylamide gel electrophoresis,
methods utilizing a
difference in electrical charge such as ion-exchange column chromatography,
methods
utilizing specific affinity such as affinity chromatography, methods utilizing
a difference in
hydrophobicity such as reverse-phase high performance liquid chromatography
and methods
utilizing a difference in isoelectric point, such as isoelectric focusing
electrophoresis, metal
affinity columns such as Ni-NTA. See generally Sambrook et al. and Ausubel et
al. supra for
disclosure relating to these methods.
62
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
It is preferred that the fusion proteins of the present invention be
substantially pure.
That is, the fusion proteins have been isolated from cell substituents that
naturally accompany
it so that the fusion proteins are present preferably in at least 80% or 90%
to 95%
homogeneity (w/w). Fusion proteins having at least 98 to 99% homogeneity (w/w)
are most
preferred for many pharmaceutical, clinical and research applications. Once
substantially
purified the fusion protein should be substantially free of contaminants for
therapeutic
applications. Once purified partially or to substantial purity, the soluble
fusion proteins can
be used therapeutically, or in performing in vitro or in vivo assays as
disclosed herein.
Substantial purity can be determined by a variety of standard techniques such
as
chromatography and gel electrophoresis.
The present fusion protein complexes are suitable for in vitro or in vivo use
with a
variety of cells that are cancerous or are infected or that may become
infected by one or more
diseases.
Human interleukin-15 (huIL-15) is trans-presented to immune effector cells by
the
human IL-15 receptor a chain (huIL-15Ra) expressed on antigen presenting
cells. IL-15Ra
binds huIL-15 with high affinity (38 pM) primarily through the extracellular
sushi domain
(huIL-15RaSu). As described herein, the huIL-15 and huIL-15RaSu domains can be
used as
a scaffold to construct multi-domain fusion protein complexes.
IgG domains, particularly the Fc fragment, have been used successfully as
dimeric
scaffolds for a number of therapeutic molecules including approved biologic
drugs. For
example, etanercept is a dimer of soluble human p75 tumor necrosis factor-a
(TNF-a)
receptor (sTNFR) linked to the Fc domain of human IgGl. This dimerization
allows
etanercept to be up to 1,000 times more potent at inhibiting TNF-a activity
than the
monomeric sTNFR and provides the fusion with a five-fold longer serum half-
life than the
monomeric form. As a result, etanercept is effective at neutralization of the
pro-
inflammatory activity of TNF-a in vivo and improving patient outcomes for a
number of
different autoimmune indications.
In addition to its dimerization activity, the Fc fragment also provides
cytotoxic
effector functions through the complement activation and interaction with Fcy
receptors
displayed on natural killer (NK) cells, neutrophils, phagocytes and dendritic
cells. In the
context of anti-cancer therapeutic antibodies and other antibody domain-Fc
fusion proteins,
these activities likely play an important role in efficacy observed in animal
tumor models and
in cancer patients. However these cytotoxic effector responses may not be
sufficient in a
number of therapeutic applications. Thus, there has been considerable interest
in improving
63
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
and expanding on the effector activity of the Fc domain and developing other
means of
recruiting cytolytic immune responses, including T cell activity, to the
disease site via
targeted therapeutic molecules. IgG domains have been used as a scaffold to
form bispecific
antibodies to improve the quality and quantity of products generated by the
traditional
hybridoma fusion technology. Although these methods bypass the shortcomings of
other
scaffolds, it has been difficult to produce bispecific antibodies in mammalian
cells at levels
sufficient to support clinical development and use.
In an effort to develop human-derived immunostimulatory multimeric scaffold,
human IL-15 (huIL-15) and IL-15 receptor domains were used. huIL-15 is a
member of the
small four a-helix bundle family of cytokines that associates with the huIL-15
receptor a-
chain (huIL-15Ra) with a high binding affinity (equilibrium dissociation
constant (KD) ¨1 0-
11 M.\
)The resulting complex is then trans-presented to the human IL-2/15 receptor
13/common y chain (huIL-15R(3yC) complexes displayed on the surface of T cells
and NK
cells. This cytokine/receptor interaction results in expansion and activation
of effector T cells
and NK cells, which play an important role in eradicating virally infected and
malignant cells.
Normally, huIL-15 and huIL-15Ra are co-produced in dendritic cells to form
complexes
intracellularly that are subsequently secreted and displayed as heterodimeric
molecules on
cell surfaces. Thus, the characteristics of huIL-15 and huIL-15Ra interactions
suggest that
these inter chain binding domains could serve as a human-derived
immunostimulatory
scaffold to make soluble dimeric molecules capable of target-specific binding.
The practice of the present invention employs, unless otherwise indicated,
conventional techniques of molecular biology (including recombinant
techniques),
microbiology, cell biology, biochemistry and immunology, which are well within
the purview
of the skilled artisan. Such techniques are explained fully in the literature,
such as,
"Molecular Cloning: A Laboratory Manual", second edition (Sambrook, 1989);
"Oligonucleotide Synthesis" (Gait, 1984); "Animal Cell Culture" (Freshney,
1987);
"Methods in Enzymology" "Handbook of Experimental Immunology" (Weir, 1996);
"Gene
Transfer Vectors for Mammalian Cells" (Miller and Cabs, 1987); "Current
Protocols in
Molecular Biology" (Ausubel, 1987); "PCR: The Polymerase Chain Reaction",
(Mullis,
1994); "Current Protocols in Immunology" (Coligan, 1991). These techniques are
applicable
to the production of the polynucleotides and polypeptides of the invention,
and, as such, may
be considered in making and practicing the invention. Particularly useful
techniques for
particular embodiments will be discussed in the sections that follow.
64
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
The following examples are put forth so as to provide those of ordinary skill
in the art
with a complete disclosure and description of how to make and use the assay,
screening, and
therapeutic methods of the invention, and are not intended to limit the scope
of what the
inventors regard as their invention.
EXAMPLES
Example 1: Generation and Characterization of Fusion Protein Complexes
Comprising IL-
15, IL-12 and IL-18 domains
An important therapeutic approach for treating cancer or infectious disease
relies on
augmenting immune cell activity against the diseased cells. This strategy
includes
stimulating immune cells ex vivo followed by adoptive transfer and/or directly
increasing
immune cell levels or activity in vivo in the patient. Immune cells involved
in these
approaches may be those of the innate (i.e., NK cells) or adaptive (i.e., T
cells) immune
system.
One approach for augmenting immune activity is to provide immunostimulatory
cytokines to the immune cells. Such cytokines are known in the art and can be
used alone or
in combination with other cytokines or agents. As described in detail below,
fusion protein
complexes comprising an IL-15N72D:IL-15RaSu/Fc scaffold fused to IL-12 and/or
IL-18
binding domains were generated (FIG. 1A and FIG.1B). These fusion protein
complexes
have advantages in binding to NK cells and signaling cell responses via each
of the cytokine
receptors. The Fc region of Ig molecules forms a dimer to provide a soluble
multi-
polypeptide complex, can bind Protein A for the purpose of purification and
can interact with
Fcy receptors on NK cells and macrophages, thus providing advantages to the
fusion protein
complex that are not present in the combination of individual cytokines.
Additionally,
interactions between the IL-15N72D and IL-15RaSu domains provides a means to
link the
IL-15N72D, IL-12 and IL-18 (and possibly other protein domains or agents) into
a single
immunostimulatory fusion protein complex.
Specifically, constructs were made linking IL-12 and/or IL-18 domains to the
IL-
15N72D and IL-15RaSu/Fc chains. In the case of IL-12, the mature cytokine
consists of two
polypeptide subunits (p40 and p35) that can be linked via a flexible linker
sequence to
generate an active single-chain form. In some cases, either IL-12 or IL-18
polypeptide is
linked to the N-terminus of the IL-15N72D and/or IL-15RaSu/Fc chains. In other
cases, the
IL-12 or IL-18 polypeptide is linked to the N-terminus of IL-15N72D and/or IL-
15RaSu/Fc
chains. Specific fusion protein complexes comprising an IL-15N72D:IL-15RaSu/Fc
scaffold
fused to IL-12 and/or IL-18 binding domains are described below.
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
1) A fusion protein complex was generated comprising IL-12/IL-15RaSu/Fc and
IL-18/IL-15N72D fusion proteins. The human IL-12 subunit sequences and human
IL-18
sequence were obtained from the UniProt website and DNA for these sequences
was
synthesized by Genewiz. Specifically, constructs were made linking the IL-12
subunit beta
(p40) to IL-12 subunit alpha (p35) to generate a single chain version of IL-12
and then
directly linking the IL-12 sequence to the IL-15RaSu/Fc chain. The synthesized
IL-12
sequence was linked to the N-terminal coding region of IL-15RaSu/Fc via
overlapping PCR.
The nucleic acid and protein sequences of a construct comprising IL-12 linked
to the N-
terminus of IL-15RaSu/Fc are shown below.
The nucleic acid sequence of the IL-12/IL-15RaSu/Fc construct (including
signal
peptide sequence) is as follows (SEQ ID NO: 1):
(Signal peptide)
atgaagtgggtgaccttcatcagcctgctgttcctgttctccagcgcctactcc
(Human IL-12 subunit beta (p40))
atctgggagctgaagaaagacgtgtatgtcgtggagctggactggtatcctgacgcccccggcgagatggtggtgctga
catgcgac
acccctgaggaggatggcatcacatggaccctggaccaaagcagcgaggtgctgggctccggaaagaccctgaccatcc
aggtga
aggagttcggcgacgccggccagtatacctgccataagggaggcgaggtgctgtcccactccctgctcctgctgcacaa
gaaggaa
gatggcatctggagcaccgatattctgaaggaccagaaggagcccaagaacaaaacctbctgcggtgcgaggccaagaa
ttattcc
ggcaggttcacctgctggtggctgaccacaatctccaccgacctgaccttcagcgtcaagagctccaggggatcctccg
atcctcagg
gcgtgacctgtggagctgccaccctgtccgctgagagggtgaggggcgacaacaaggagtacgagtactccgtcgagtg
tcagga
ggactccgcctgccctgctgccgaagagagcctgcctatcgaagtcatggtggacgccgtgcacaagctgaagtatgag
aactacac
cagcagcttcttcatccgggacattatcaagcctgatccccctaagaacctgcagctcaagcccctgaagaattcccgg
caagtcgag
gtgtcctgggagtaccccgacacctggtccacccctcactcctattttagcctgaccttctgcgtgcaggtgcagggca
agagcaaga
gggagaagaaagaccgggtgttcaccgacaagaccagcgctaccgtgatctgtcggaagaacgcttccatttccgtgcg
ggctcag
gacaggtattactcctcctcctggtccgagtgggctagcgtcccctgcagc
(Linker)
ggaggtggcggatccggaggtggaggttctggtggaggtgggagt
(Human IL-12 subunit alpha (p35))
aggaacctgcccgtggctacacccgaccctggaatgttcccctgtctccaccacagccaaaacctcctgcgggccgtgt
ccaacatg
ctgcaaaaggctcggcagacactggagttctacccctgcaccagcgaggagatcgaccatgaggacatcacaaaggaca
agacaa
gcaccgtggaggcttgcctccccctggaactgaccaagaatgagtcctgcctcaacagccgggagacatccttcatcac
caatggctc
ctgtctggcttcccggaagacaagcttcatgatggccctgtgcctgtccagcatctatgaggacctgaagatgtaccag
gtcgagtttaa
gaccatgaacgccaagctgctgatggaccccaagcggcaaatcttcctggaccagaacatgctggctgtgatcgacgag
ctgatgca
66
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
ggctctgaacttcaacagcgagaccgtgccccagaagtcctccctggaggagcctgatttttacaagaccaaaatcaag
ctctgcatcc
tcctgcacgccttccggatcagggccgtgaccatcgatcgggtgatgtcctacctgaatgcttcc
(Human IL-15R a sushi domain)
atcacgtgtcctcctcctatgtccgtggaacacgcagacatctgggtcaagagctacagcttgtactccagggagcggt
acatttgtaac
tctggittcaagcgtaaagccggcacgtccagcctgacggagtgcgtgttgaacaaggccacgaatgtcgcccactgga
caaccccc
agtctcaaatgcattagA
(Human IgG1 CH2-CH3 (Fc) domain)
gagccgaaatcttgtgacaaaactcacacatgcccaccgtgcccagcacctgaactcctggggggaccgtcagtcttcc
tcttccccc
caaaacccaaggacaccctcatgatctcccggacccctgaggtcacatgcgtggtggtggacgtgagccacgaagaccc
tgaggtc
aagttcaactggtacgtggacggcgtggaggtgcataatgccaagacaaagccgcgggaggagcagtacaacagcacgt
accgtg
tggtcagcgtcctcaccgtcctgcaccaggactggctgaatggcaaggagtacaagtgcaaggtctccaacaaagccct
cccagccc
ccatcgagaaaaccatctccaaagccaaagggcagccccgagaaccacaggtgtacaccctgcccccatcccgggatga
gctgac
caagaaccaggtcagcctgacctgcctggtcaaaggcttctatcccagcgacatcgccgtggagtgggagagcaatggg
cagccgg
agaacaactacaagaccacgcctcccgtgctggactccgacggctccttcttcctctacagcaagctcaccgtggacaa
gagcaggt
ggcagcaggggaacgtcttctcatgctccgtgatgcatgaggctctgcacaaccactacacgcagaagagcctctccct
gtctcctgg
taaa
The amino acid sequence of the IL-12/IL-15RaSu/Fc fusion protein (including
signal
peptide sequence) is as follows (SEQ ID NO: 2):
(Signal peptide)
MKWVTFISLLFLF S SAYS
(Human IL-12 subunit beta (p40))
IWELKKDVYVV ELDWYP DAP GEMVVLTCDTPEEDGITWTLDQ S S EVL GS GKTL TI Q
VKEFGDAGQYTCHKGGEVLSHSLLLLHKKEDGIWSTDILKDQKEPKNKTFLRCEAK
NY S GRF T CWWL TTI S TDLTF SVKS SRGS SDP QGVTCGAATL S AERVRGDNKEYEY S V
ECQED S ACP AAEES LPIEVMVDAVHKLKYENYTS SF FIRDIIKPDPP KNL QLKPLKNSR
QVEVSWEYPDTWSTPHSYFSLTFCVQVQGKSKREKKDRVFTDKTSATVICRKNASIS
VRAQDRYYS S SW SEWASVP C S
(Linker)
GGGGSGGGGSGGGGS
(Human IL-12 subunit alpha (p35))
RNLPVATPDPGMFPCLHHSQNLLRAV SNMLQKARQTLEFYPCTSEEIDHEDITKDKT
STVEACLPLELTKNESCLNSRETSFITNGSCLASRKTSFMMALCLSSIYEDLKMYQVE
67
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
FKTMNAKLLMDPKRQIFLDQNMLAVIDELMQALNFNSETVPQKS SLEEPDFYKTKIK
LCILLHAFRIRAVTIDRVMSYLNAS
(Human IL-15R a sushi domain)
ITCPP PM S VEHADIWVKSY S LY S RERYI CN S GF KRKAGT S SLTECVLNKATNVAHWT
TPSLKCIR
(Human IgG1 CH2-CH3 (Fc) domain)
EPKSCDKTHTCPPCPAPELLGGP SVFLFPPKPKDTLMISRTPEVTCVVVDV SHEDPEV
KFNWYVDGVEVHNAKTKPREEQYN STYRVV SVLTVLHQDWLNGKEYKC KV SNKA
LPAPIEKTISKAKGQPREPQVYTLPP SRDELTKNQVSLTCLVKGFYP SDIAVEWESNG
QPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVF SC SVMHEALHNHYTQKSL
SLSPGK
In some cases, the leader peptide is cleaved from the intact polypeptide to
generate
the mature form that may be soluble or secreted.
Constructs were also made linking the synthesized IL-18 sequence to the N-
terminus
coding region of IL-15N72D via overlapping PCR. The nucleic acid and protein
sequences
of a construct comprising IL-18 linked to the N-terminus of IL-15N72D are
shown below.
The nucleic acid sequence of the IL-18/IL-15N72D construct (including leader
sequence) is as follows (SEQ ID NO: 3):
(Signal peptide)
atgaagtgggtgaccttcatcagcctgctgttcctgttctccagcgcctactcc
(Human IL-18)
tacttcggcaagctggagtccaagctgtccgtgatcaggaacctgaacgaccaggtgctgttcatcgaccagggcaaca
ggcccctg
ttcgaggacatgaccgactccgactgcagggacaacgcccctaggaccatcttcatcatctccatgtataaggacagcc
agcccagg
ggaatggccgtgaccatctccgtgaagtgcgagaagatctccaccctgtcctgcgagaacaagatcatctccttcaagg
agatgaacc
cccccgacaacatcaaggacaccaagtccgacatcatcttcttccagcggtccgtgcccggacacgacaacaagatgca
gttcgagt
cctcctcctacgagggctactttctggcctgtgagaaggagagggacctcttcaagctcatcctgaagaaggaggacga
gctgggcg
acaggtccatcatgttcaccgtgcagaacgaggac
(Human IL-15N72D)
aactgggttaacgtaataagtgatttgaaaaaaattgaagatcttattcaatctatgcatattgatgctactttatata
cggaaagtgatgttc
accccagttgcaaagtaacagcaatgaagtgctttctcttggagttacaagttatttcacttgagtccggagatgcaag
tattcatgataca
gtagaaaatctgatcatcctagcaaacgacagtttgtcttctaatgggaatgtaacagaatctggatgcaaagaatgtg
aggaactggag
gaaaaaaatattaaagaat I tgcagagttttgtacatattgtccaaatgttcatcaacacttct
68
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
The amino acid sequence of the IL-18/IL-15N72D fusion protein (including
leader
sequence) is as follows (SEQ ID NO: 4):
(Signal peptide)
MKWVTFISLLFLFS SAYS
(Human IL-18)
YFGKLESKLSVIRNLNDQVLFIDQGNRPLFEDMTDSDCRDNAPRTIFIISMYKDSQPR
GMAVTISVKCEKISTLSCENKIISFKEMNPPDNIKDTKSDIIFFQRSVPGHDNKMQFESS
SYEGYFLACEKERDLFKLILKKEDELGDRSIMFTVQNED
(Human IL-15N72D)
NWVNVISDLKKIEDLIQSMHIDATLYTESDVHPSCKVTAMKCFLLELQVISLESGDAS
IHDTVENLIILANDSLSSNGNVTESGCKECEELEEKNIKEFLQSFVHIVQMFINTS
In some cases, the leader peptide is cleaved from the intact polypeptide to
generate
the mature form that may be soluble or secreted.
The IL-12/IL-15RaSu/Fc and IL-18/IL-15N72D constructs were cloned into
expression vectors as described previously (U.S. Patent No. 8,507,222, at
Example 1,
incorporated herein by reference), and the expression vectors were transfected
into CHO
cells. Co-expression of the two constructs in CHO cells allowed for formation
and secretion
of a soluble IL-18/IL-15N72D:IL-12/IL-15RaSu/Fc fusion protein complex
(referred to as
hIL18/IL12/TxM). The hIL18/IL12/TxM protein was purified from CHO cell culture
supernatant by Protein A affinity chromatography and size exclusion
chromatography
resulting in soluble (non-aggregated) fusion protein complexes consisting of
IL-12/IL-
15RaSu/Fc dimers and IL-18/IL-15N72D fusion proteins (FIG. 2A-2C).
Reduced SDS-PAGE analysis of the Protein A-purified IL-18/IL-15N72D:IL-12/IL-
15RaSu/Fc fusion protein complexes is shown in FIG. 3. Bands corresponding to
the soluble
IL-12/IL-15RaSu/Fc and IL-18/IL-15N72D proteins at ¨90 kDa and ¨30 kDa,
respectively,
were observed (FIG. 3).
2) For a second approach, a similar fusion protein complex was generated
comprising
IL-18/IL-15RaSu/Fc and IL-12/IL-15N72D fusion proteins. Specifically,
constructs were
made by attaching IL-18 directly to the IL-15RaSu/Fc chain. The synthesized IL-
18 is linked
to the N-terminal coding region of IL-15RaSu/Fc via overlapping PCR. The
nucleic acid and
69
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
protein sequences of a construct comprising the IL-18 linked to the N-terminus
of IL-
15RaSu/Fc are shown below.
The nucleic acid sequence of the IL-18/IL-15RaSu/Fc construct (including
signal
peptide sequence) is as follows (SEQ ID NO: 5):
(Signal peptide)
atgaagtgggtgaccttcatcagcctgctgttcctgttctccagcgcctactcc
(Human IL-18)
tacttcggcaagctggagtccaagctgtccgtgatcaggaacctgaacgaccaggtgctgttcatcgaccagggcaaca
ggcccctg
ttcgaggacatgaccgactccgactgcagggacaacgcccctaggaccatcttcatcatctccatgtataaggacagcc
agcccagg
ggaatggccgtgaccatctccgtgaagtgcgagaagatctccaccctgtcctgcgagaacaagatcatctccttcaagg
agatgaacc
cccccgacaacatcaaggacaccaagtccgacatcatcttcttccagcggtccgtgcccggacacgacaacaagatgca
gttcgagt
cctcctcctacgagggctactttctggcctgtgagaaggagagggacctcttcaagctcatcctgaagaaggaggacga
gctgggcg
acaggtccatcatgttcaccgtgcagaacgaggac
(Human IL-15R a sushi domain)
atcacgtgtcctcctcctatgtccgtggaacacgcagacatctgggtcaagagctacagcttgtactccagggagcggt
acatttgtaac
tctggtttcaagcgtaaagccggcacgtccagcctgacggagtgcgtgttgaacaaggccacgaatgtcgcccactgga
caaccccc
agtctcaaatgcattaga
(Human IgG1 CH2-CH3 (Fc) domain)
gagccgaaatcttgtgacaaaactcacacatgcccaccgtgcccagcacctgaactcctggggggaccgtcagtcttcc
tcttccccc
caaaacccaaggacaccctcatgatctcccggacccctgaggtcacatgcgtggtggtggacgtgagccacgaagaccc
tgaggtc
aagttcaactggtacgtggacggcgtggaggtgcataatgccaagacaaagccgcgggaggagcagtacaacagcacgt
accgtg
tggtcagcgtcctcaccgtcctgcaccaggactggctgaatggcaaggagtacaagtgcaaggtctccaacaaagccct
cccagccc
ccatcgagaaaaccatctccaaagccaaagggcagccccgagaaccacaggtgtacaccctgcccccatcccgggatga
gctgac
caagaaccaggtcagcctgacctgcctggtcaaaggcttctatcccagcgacatcgccgtggagtgggagagcaatggg
cagccgg
agaacaactacaagaccacgcctcccgtgctggactccgacggctccttcttcctctacagcaagctcaccgtggacaa
gagcaggt
ggcagcaggggaacgtcttctcatgctccgtgatgcatgaggctctgcacaaccactacacgcagaagagcctctccct
gtctcctgg
taaa
The amino acid sequence of the IL-18/IL-15RaSu/Fc fusion protein (including
signal
peptide sequence) is as follows (SEQ ID NO: 6):
(Signal peptide)
MKWVTFISLLFLF S SAYS
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
(Human IL-18)
YFGKLESKLSVIRNLNDQVLFIDQGNRPLFEDMTDSDCRDNAPRTIFIISMYKDSQPR
GMAVTISVKCEKISTL SCENKIISFKEMNPPDNIKDTKSDIIFFQRSVP GHDNKMQFES S
SYEGYFLACEKERDLFKLILKKEDELGDRSIMFTVQNED
(Human IL-15R a sushi domain)
ITCPP PM S VEHADIWVKSY S LY S RERYI CN S GF KRKAGT S SLTECVLNKATNVAHWT
TPSLKCIR
(Human IgG1 CH2-CH3 (Fc) domain)
EPKSCDKTHTCPPCPAPELLGGP SVFLFPPKPKDTLMISRTPEVTCVVVDV SHEDPEV
KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKA
LPAPIEKTISKAKGQPREPQVYTLPP SRDELTKNQVSLTCLVKGFYP SDIAVEWESNG
QPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVF SC SVMHEALHNHYTQKSL
SLSPGK
In some cases, the leader peptide is cleaved from the intact polypeptide to
generate
the mature form that may be soluble or secreted.
Constructs were also made linking the synthesized IL-12 sequence to the N-
terminus
coding region of IL-15N72D via overlapping PCR. As describe above, a single-
chain version
of IL-12 (p40-linker-p35) was used. The nucleic acid sequence of the IL-12/IL-
15N72D
construct (including leader sequence) is as follows (SEQ ID NO: 7):
(Signal peptide)
atgaagtgggtgaccttcatcagcctgctgttcctgttctccagcgcctactcc
(Human IL-12 subunit beta (p40))
atctgggagctgaagaaagacgtgtatgtcgtggagctggactggtatcctgacgcccccggcgagatggtggtgctga
catgcgac
acccctgaggaggatggcatcacatggaccctggaccaaagcagcgaggtgctgggctccggaaagaccctgaccatcc
aggtga
aggagttcggcgacgccggccagtatacctgccataagggaggcgaggtgctgtcccactccctgctcctgctgcacaa
gaaggaa
gatggcatctggagcaccgatattctgaaggaccagaaggagcccaagaacaaaacctttctgcggtgcgaggccaaga
attattcc
ggcaggttcacctgctggtggctgaccacaatctccaccgacctgaccttcagcgtcaagagctccaggggatcctccg
atcctcagg
gcgtgacctgtggagctgccaccctgtccgctgagagggtgaggggcgacaacaaggagtacgagtactccgtcgagtg
tcagga
ggactccgcctgccctgctgccgaagagagcctgcctatcgaagtcatggtggacgccgtgcacaagctgaagtatgag
aactacac
cagcagcttcttcatccgggacattatcaagcctgatccccctaagaacctgcagctcaagcccctgaagaattcccgg
caagtcgag
gtgtcctgggagtaccccgacacctggtccacccctcactcctattttagcctgaccttctgcgtgcaggtgcagggca
agagcaaga
71
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
gggagaagaaagaccgggtgttcaccgacaagaccagcgctaccgtgatctgtcggaagaacgcttccatttccgtgcg
ggctcag
gacaggtattactcctcctcctggtccgagtgggctagcgtcccctgcagc
(Linker)
ggaggtggcggatccggaggtggaggttctggtggaggtgggagt
(Human IL-12 subunit alpha (p35))
aggaacctgcccgtggctacacccgaccctggaatgttcccctgtctccaccacagccaaaacctcctgcgggccgtgt
ccaacatg
ctgcaaaaggctcggcagacactggagttctacccctgcaccagcgaggagatcgaccatgaggacatcacaaaggaca
agacaa
gcaccgtggaggcttgcctccccctggaactgaccaagaatgagtcctgcctcaacagccgggagacatccttcatcac
caatggctc
ctgtctggcttcccggaagacaagcttcatgatggccctgtgcctgtccagcatctatgaggacctgaagatgtaccag
gtcgagtttaa
gaccatgaacgccaagctgctgatggaccccaagcggcaaatcttcctggaccagaacatgctggctgtgatcgacgag
ctgatgca
ggctctgaacttcaacagcgagaccgtgccccagaagtcctccctggaggagcctgatttttacaagaccaaaatcaag
ctctgcatcc
tcctgcacgccttccggatcagggccgtgaccatcgatcgggtgatgtcctacctgaatgcttcc
(Human IL-15N72D)
aactgggttaacgtaataagtgatttgaaaaaaattgaagatcttattcaatctatgcatattgatgctactttatata
cggaaagtgatgttc
accccagttgcaaagtaacagcaatgaagtgctttctcttggagttacaagttatttcacttgagtccggagatgcaag
tattcatgataca
gtagaaaatctgatcatcctagcaaacgacagtttgtcttctaatgggaatgtaacagaatctggatgcaaagaatgtg
aggaactggag
gaaaaaaatattaaagaatttttgcagagttttgtacatattgtccaaatgttcatcaacacttct
The amino acid sequence of the IL-12/IL-15N72D fusion protein (including
leader
sequence) is as follows (SEQ ID NO: 8):
(Signal peptide)
MKWVTFISLLFLFS SAYS
(Human IL-12 subunit beta (p40))
IWELKKDVYVV ELDWYP DAP GEMVVLTCDTPEEDGITWTLDQS S EVL GS GKTL TI Q
VKEFGDAGQYTCHKGGEVLSHSLLLLHKKEDGIWSTDILKDQKEPKNKTFLRCEAK
NY S GRF T CWWL TTI S TDLT F S VKS SRGS S DP Q GV T C GAATL S AERVRGDNKEYEY S
V
ECQEDSACPAAEESLPIEVMVDAVHKLKYENYTS SF FIRDIIKPDPPKNL QLKPLKNSR
QVEVSWEYPDTWSTPHSYFSLTFCVQVQGKSKREKKDRVFTDKTSATVICRKNASIS
VRAQDRYYS S SW SEWASVP C S
(Linker)
GGGGSGGGGSGGGGS
(Human IL-12 subunit alpha (p35))
72
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
RNLPVATPDPGMFPCLHHSQNLLRAVSNMLQKARQTLEFYPCTSEEIDHEDITKDKT
STVEACLPLELTKNESCLNSRETSFITNGSCLASRKTSFMMALCLSSIYEDLKMYQVE
FKTMNAKLLMDPKRQIFLDQNMLAVIDELMQALNFNSETVPQKSSLEEPDFYKTKIK
LCILLHAFRIRAVTIDRVMSYLNAS
(Human IL-15N72D)
NWVNVISDLKKIEDLIQSMHIDATLYTESDVHPSCKVTAMKCFLLELQVISLESGDAS
IHDTVENLIILANDSLSSNGNVTESGCKECEELEEKNIKEFLQSFVHIVQMFINTS
In some cases, the leader peptide is cleaved from the intact polypeptide to
generate
the mature form that may be soluble or secreted.
The IL-18/IL-15RaSu/Fc and IL-12/IL-15N72D constructs were cloned into
expression vectors as described previously (U.S. Patent No. 8,507,222, at
Example 1,
incorporated herein by reference), and the expression vectors were transfected
into CHO
cells. Co-expression of the two constructs in CHO cells allowed for formation
and secretion
of the soluble IL-12/IL-15N72D:IL-18/IL-15RaSu/Fc fusion protein complex
(referred to as
hIL12/IL18/TxM), which can be purified by Protein A affinity and other
chromatography
methods.
3) Similar fusion protein complexes could be generated comprising IL-18/IL-
15RaSu/Fc and IL-18/IL-15N72D fusion proteins or comprising IL-12/IL-15RaSu/Fc
and
IL-12/IL-15N72D fusion proteins. "Two headed" fusion protein complexes could
be
generated comprising IL-18/IL-15RaSu/Fc and IL-15N72D fusion proteins or IL-
15RaSu/Fc
and IL-18/IL-15N72D fusion proteins (FIG. 1B). Similarly, "two headed" fusion
protein
complexes could be generated comprising IL-12/IL-15RaSu/Fc and IL-15N72D
fusion
proteins or IL-15RaSu/Fc and IL-12/IL-15N72D fusion proteins. Such complexes
were
generated as described above.
Example 2: In Vitro Characterization of the Activities of hIL18/IL12/TxM and
hIL12/IL18/TxM Fusion Protein Complexes
ELISA-based methods confirmed the formation of the hIL18/IL12/TxM and
hIL12/IL18/TxM fusion protein complexes. In FIG. 4A, the IL-18/IL-15N72D:IL-
12/IL-
15RaSu/Fc fusion protein complexes in the culture supernatant from transfected
CHO cells
were detected using a huIgGl/IL15-specific ELISA with a capture antibody, anti-
human IL-
15 antibody (MAB647, R&D Systems) and a detection antibody, horseradish
peroxidase
conjugated. This is compared to a similar antibody TxM fusion protein complex
(2B8T2M)
73
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
with a known concentration. The signal from the hIL18/IL12/TxM fusion protein
complex
can be compared to that of the 2B8 T2M control to estimate the fusion protein
concentration.
Similar results were obtained from hIL12/IL18/TxM fusion protein complexes
(FIG. 4B).
For the purified "two-headed" IL-18/TxM complex, an ELISA with anti-IL-18 Ab
capture
and anti-IL-15 Ab detection verified formation of the fusion proteins complex
(FIG. 4C). The
results from these assays demonstrate that soluble IL-18/IL-15N72D, IL-12/IL-
15RaSu/Fc,
IL-12/IL-15N72D and IL-18/IL-15RaSu/Fc proteins can be produced in CHO cells
and the
hIL18/IL12/TxM and hIL12/IL18/TxM fusion protein complexes can form and be
secreted
into the culture media.
To assess the IL-15 immunostimulatory activity of the hIL18/IL12/TxM fusion
protein complexes, proliferation of IL-15-dependent 32D13 cells, a mouse
hematopoietic cell
line, was assessed. Increasing levels of hIL18/IL12/TxM were added to 32D13
cells (104
cell/well) in 200 p.L RPMI:10% FBS media and cells were incubated for 2 days
at 37 C.
WST-1 proliferation reagent (10 4/well) then was added. After 4 hours,
absorbance was
measured at 450 nm to determine cell proliferation based on cleavage of WST-1
to a soluble
formazan dye by metabolically active cells. The bioactivity of the IL-
15N72D:IL-15RaSu/Fc
complex (ALT-803) was assessed as a positive control. As shown in FIG. 5,
hIL18/IL12/TxM was able to promote cell proliferation of 32D13 cells, thereby
demonstrating
IL-15 activity. The activity of hIL18/IL12/TxM was reduced compared to that of
ALT-803,
possibly due to the linkage of IL-18 to the IL-15N72D domain.
To further assess the IL-15 activity of hIL18/IL12/TxM, increasing
concentrations of
hIL18/IL12/TxM were added to 32D13 cells (104 cells/well) in 200 p.L IMDM:10%
FBS
media and incubated for 3 days at 37 C. PrestoBlue cell viability reagent (20
p.L/well) then
was added. After 4 hours, absorbance was measured at 570 nm (with a 600 nm
reference
wavelength for normalization) to determine cell proliferation based on
reduction of
PrestoBlue, a resazurin-based solution, by metabolically active cells. The
half maximal
effective concentration (EC50) of IL-15 bioactivity for hIL18/IL12/TxM was
then determined
based on the relationship between absorbance and protein concentration. The
bioactivity of
ALT-803 was assessed as a positive control. As shown in FIG. 6, hIL18/IL12/TxM
was able
to promote cell proliferation of 32D13 cells, thereby demonstrating IL-15
activity. The
activity of hIL18/IL12/TxM was reduced compared to that of ALT-803, possibly
due to the
linkage of IL-18 to IL-15N72D.
To assess the IL-18 activity of hIL18/IL12/TxM, activation of IL-18 reporter
HEK-
Blue IL-18 (HEK18) cells was assessed. Increasing concentrations of
hIL18/IL12/TxM were
74
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
added to HEK18 cells (5x104 cells/well) in 200 4 IMDM:10% FBS HEK-Blue media
and
incubated for 20-22 hours at 37 C. Culture supernatant (20 4/well) was then
added to
QUANTI-Blue reagent (180 4/well). After 20 hours, absorbance was measured at
650 nm to
determine cell activation based on reduction of QUANTI-Blue, a secreted
embryonic alkaline
phosphatase (SEAP) detection reagent. The half maximal effective concentration
(EC50) of
IL-18 bioactivity of hIL18/IL12/TxM was then determined based on the
relationship between
absorbance and protein concentration. The bioactivity of recombinant IL-18 was
assessed as
a positive control. As shown in FIG. 7, hIL18/IL12/TxM was able to activate
HEK18 cells,
thereby demonstrating IL-18 activity. The activity of hIL18/IL12/TxM was
reduced
compared to that of recombinant IL-18, possibly due to the linkage of IL-18 to
IL-15N72D.
To assess the IL-12 activity of hIL18/IL12/TxM, activation of IL-12 reporter
HEK-
Blue IL-12 (HEK12) cells was assessed. Increasing concentrations of
hIL18/IL12/TxM were
added to HEK12 cells (5x104 cells/well) in 200 4 IMDM:10% FBS HEK-Blue media
and
incubated for 20-22 hours at 37 C. Culture supernatant (20 4/well) was then
added to
QUANTI-Blue reagent (180 4/well). After 20 hours, absorbance was measured at
650 nm to
determine cell activation based on reduction of QUANTI-Blue, a secreted
embryonic alkaline
phosphatase (SEAP) detection reagent. The half maximal effective concentration
(EC50) of
IL-12 bioactivity of hIL18/IL12/TxM was then determined based on the
relationship between
absorbance and protein concentration. The bioactivity of recombinant IL-12 was
assessed as
a positive control. As shown in FIG. 8, hIL18/IL12/TxM was able to activate
HEK12 cells,
similar to recombinant IL-12, thereby demonstrating IL-12 activity.
In order to further demonstrate the individual activity of each cytokine (IL-
12, IL-18,
and IL-15), flow cytometry-based intracellular phosphoprotein assays were
developed
utilizing proteins that are uniquely phosphorylated in response to receptor
signaling by each
cytokine (IL-12: STAT4, IL-18: p38 MAPK, and IL-15: STAT5). Following short
term
stimulation (5-15 minutes) of NK92 (aNK) cells or purified human NK cells
(>95% CD56+)
with 1 ug/m1hIL18/IL12/TxM resulted in similar responses to that seen with the
optimal
combinations of recombinant IL-12 (long/m1), IL-18 (50 ng/ml) and ALT-803 (50
ng/ml IL-
15 activity) (FIG. 9A-F). These results demonstrate that each of the cytokine
domains of the
hIL18/IL12/TxM fusion protein complex retains its specific immunostimulatory
biological
activity.
It is known that the combination of IL-12, IL-18 and IL-15 activity is more
effective
at inducing IFN-y production by NK cells than any of these cytokines alone. In
order to
evaluate the combined cytokine activity of the hIL18/IL12/TxM complex, aNK
cells were
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
incubated with hIL18/IL12/TxM complex (50 nM), combinations of IL-12 (0.5 nM),
IL-18 (3
nM), and ALT-803 (10 nM), or each cytokine alone. After 2 days, IFN-y levels
in the culture
supernatants were determined with ELISA methods. As shown in FIG. 10A, IL-12,
IL-18
and ALT-803 alone had little effect on aNK cells whereas combinations of IL-
12+ALT-803
and IL-18+ALT-803 induced low level production of IFN-y by aNK cells. However,
hIL18/IL12/TxM fusion protein complex alone and combinations of IL-12+IL-18
and IL-
12+IL-18+ALT-803 exhibited high level production of IFN-y by aNK cells. These
results
verify that hIL18/IL12/TxM fusion protein complex exhibit the expected
immunostimulatory
activity of the combined IL-12, IL-18 and IL-15 cytokines. Similar studies
with the "two
headed" IL-18/TxM complex demonstrated the ability of this complex to induce
IFN-y by
aNK cells but to a lesser degree than hIL18/IL12/TxM fusion protein complex
(FIG. 10B).
Example 3: Induction of Cytokine Induced Memory Like NK Cells by
hIL18/IL12/TxM
Fusion Protein Complexes
Previous studies have shown that cytokine induced memory like NK cells can be
induced ex vivo following overnight stimulation of purified NK cells with
saturating amounts
of IL-12 (10 ng/ml), IL-15 (50 ng/ml), and IL-18 (50 ng/ml). These cells
exhibit memory-like
properties such as 1) enhanced proliferation, 2) expression of IL-2 receptor a
(IL-2Ra, CD25)
and other activation markers, and 3) increased IFN-y production. To evaluate
the ability of
hIL18/IL12/TxM to promote generation of cytokine induced memory like NK cells,
purified
human NK cells (>95% CD56+) (5x106 cells/nil) were stimulated for 18 hours
with 1 pg/ml
hIL18/IL12/TxM or the optimal combination of recombinant IL-12 (10 ng/ml), IL-
18 (50
ng/ml), and ALT-803 (50 ng/ml IL-15 activity). Induction of cytokine induced
memory like
cells was assessed as increased cell-surface CD25 and CD69 (stimulation
marker) expression
and intracellular IFN-y levels as determined by antibody-staining and flow
cytometric
methods. The results indicated that hIL18/IL12/TxM fusion protein complex was
capable of
inducing CD25, CD69 and intracellular IFN-y to a similar extent as the optimal
combination
of IL-12, IL-18 and IL15 following overnight incubation with human NK cells
(Fig. 11A ¨
Fig. 11F). Thus, overnight incubation with hIL18/IL12/TxM fusion protein
complexes can
generate cytokine induced memory like NK cells.
Previous studies have shown that cytokine-induced memory-like NK cells can be
induced ex vivo following overnight stimulation of purified NK cells with
saturating amounts
of IL-12 (10 ng/ml), IL-15 (50 ng/ml), and IL-18 (50 ng/ml). These cells
exhibit memory-like
76
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
properties such as 1) enhanced proliferation, 2) expression of IL-2 receptor a
(IL-2Ra,
CD25), 3) increased IFNy production, and 4) augmented cytotoxicity mediated by
perforin
and granzymes. To evaluate the ability of hIL18/IL12/TxM to promote generation
of
cytokine-induced memory-like NK cells, purified human NK cells (>95% CD56+,
5x106
cells/m1) were stimulated for 12-18 hours with increasing concentrations of
hIL18/IL12/TxM
or the optimal combination of recombinant IL-12 (10 ng/ml), IL-18 (50 ng/ml),
and ALT-803
(50 ng/ml IL-15, 3.88 nM). Induction of a pre-activated cytokine-induced
memory-like cell
phenotype was assessed as increased cell surface CD25 expression and
intracellular IFNy
levels as determined by antibody staining and flow cytometric methods. As
shown in FIG.
12A and 12B, hIL18/IL12/TxM was capable of inducing CD25 and intracellular
IFNy to a
similar extent as the optimal combination of IL-12, IL-18, and ALT-803
following overnight
incubation with human NK cells.
In order to demonstrate generation of cytokine induced memory like NK cells by
hIL18/IL12/TxM, primary human NK cells (2x106/m1) were primed for 16 hours as
above
with hIL18/IL12/TxM (38.8 nM), washed, and rested in low dose ALT-803 (77.6
pM,
equivalent to 1 ng/ml IL-15) for 6 days, to allow the primed NK cells to
differentiate into
cytokine induced memory like NK cells. Maintenance of CD25 expression and
enhanced
IFN-y production following 6 hour re-stimulation with cytokines (IL-12 (10
ng/ml) and ALT-
803 (50 ng/ml IL-15 equivalent)) or leukemia targets (K562 cells, 5:1 ratio),
in the presence
of brefeldin A and monensin, were assessed as correlates for generation of
cytokine induced
memory like NK cells. In all cases, priming with hIL18/IL12/TxM, compared to
low dose IL-
15 (77.6 pM ALT-803) as a control, resulted in enhanced levels of CD25 (FIG.
13A) and
increased expression of IFN-y following re-stimulation with both cytokines and
leukemia
targets (FIG. 13B).
Similar studies were conducted to further compare the effects of short-term
priming
with hIL18/IL12/TxM or different cytokine combinations on human NK cell that
were
subsequently rested in low dose ALT-803 or IL-15 and restimulated with IL-12
and IL-15.
For these studies, proliferation and IFN-y production of the CIML NK cells was
assessed as a
measure of immune activation. As shown in FIG. 14A and B, human NK cells were
labeled
with CellTrace Violet and primed as above with media alone, IL-12 (0.5 nM), IL-
18 (3 nM),
ALT-803 (10 nM), ALT-803 (10 nM) + IL-18 (3 nM), ALT-803 (10 nM) + IL-18 (3
nM) +
IL-12 (0.5 nM), or hIL18/IL12/TxM (10 nM). Following priming, the cells were
washed and
maintained in media containing lng/mL IL-15 (FIG. 14A) or 75pM ALT-803 (FIG.
14B)
followed by no re-stimulation or restimulation with lOng/mL IL-12 + 5Ong/mL IL-
15. The
77
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
proliferation of the CIML NK cells was determined by dilution of the CellTrace
Violet label
and intracellular IFN-y expression was determined by intracellular staining
and flow
cytometry. Compared to no priming or priming with individual cytokines, ALT-
803+IL-18,
or ALT-803+IL-18+IL-12, NK cells expressed higher levels of intracellular IFN-
y following
priming with hIL18/IL12/TxM and subsequent resting in IL-15 or ALT-803 then
restimulation with IL-12 + IL-15. Specifically, >83% of the CIML NK cells
generated by
priming with hIL18/IL12/TxM were found to express IFN-y following
restimulation
compared to ¨74% of the NK cells primed with the standard hIL18+IL12+ALT-803
combination and 50%-60% of the NK cells primed with individual cytokines. CIML
NK
cells primed with hIL18/IL12/TxM also showed greater proliferation as measured
by
CellTrace Violet dilution than NK cells primed with hIL18+IL12+ALT-803 or the
individual
cytokines (FIG. 15). There results confirm that short-term priming of human NK
cells with
hIL18/IL12/TxM can result in an CIML NK cell (i.e., increased proliferation
and immune
activation (CD25, IFN-y expression)) equivalent or better than priming with
hIL18+IL12+IL-
15.
Additionally, the effect of hIL18/IL12/TxM fusion protein complexes on
cytotoxicity
of human NK cells against human tumor cells was investigated. Human breast
cells (MDA-
MB-231) (Celltrace violet labelled) were incubated with purified human NK
cells (2
independent donors; NK1 and NK2) (E:T ratio; 1:1) in the presence of
hIL18/IL12/TxM
complex (10 nM) or ALT-803 (10 nM) as a control. After 2 days, the percentage
of dead
tumor cells (Violet + PIP) was assessed by flow cytometry following staining
with propidium
iodide (PI). As shown in FIG. 16, hIL18/IL12/TxM induced significantly more
effective
human NK cell cytotoxicity against breast tumor cells than ALT-803. These
results are
consistent with the ability of IL-12, IL-18 and IL-15 combination treatment to
enhance anti-
tumor NK cell activity.
The hIL18/IL12/TxM fusion protein complexes were also able to augment
expression
of granzyme B in human NK cells, as compared to ALT-803 or no treatment (FIG.
17A).
Furthermore, these hIL18/IL12/TxM activated NK cells were also more effective
in direct or
antibody-mediated cytotoxicity assays against a human tumor target (FIG. 17B),
including
enhanced production of IFNy (FIG. 17C). Thus, overnight incubation with
hIL18/IL12/TxM
can generate a phenotype associated with cytokine-induced memory-like NK
cells.
78
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
Example 4: Antitumor Activities of Immune Cells Stimulated by hIL18/IL12/TxM
Fusion
Protein Complexes
The ability of hIL18/IL12/TxM fusion protein complexes to induce cytokine
induced
memory like NK cells with in vivo antitumor activity will be assessed. Splenic
NK cells will
be isolated from mice by standard methods and stimulated at 5x106 cells/ml for
18 hours with 1
pg/ml hIL18/IL12/TxM, a combination of recombinant IL-12 (10 ng/ml), IL-18 (50
ng/ml),
and ALT-803 (50 ng/ml IL-15 activity) or ALT-803 alone (50 ng/ml IL-15
activity). The
cells will then be washed and adoptively transferred (1x106 cells/mouse) i.v.
into C57BL/6
mice that bear subcutaneous RMA-S lymphoma and received 5 Gy of total body
radiation 3
hours prior to cell transfer. Survival of mice will be monitored. Tumor-
bearing mice treated
with IL-12+IL-18+ALT-803-activated NK cells (CIML NK cells) are expected to
survival
longer that mice treated with ALT-803-activated NK cells (Ni, J, et al. J.
Exp. Med. 2012
209:2351-2365). Prolonged survival of tumor-bearing mice receiving
hIL18/IL12/TxM-
activated NK cells will provide evidence that hIL18/IL12/TxM can serve as an
ex-vivo agent
to augment in vivo antitumor activity of immune cells.
Similarly, purified human NK cells will be stimulated at 5x106 cells/ml for 18
hours
with 1 pg/ml hIL18/IL12/TxM, a combination of recombinant IL-12 (10 ng/ml), IL-
18 (50
ng/ml), and ALT-803 (50 ng/ml IL-15 activity) or ALT-803 alone (50 ng/ml IL-15
activity).
The cells will then be washed and adoptively transferred (1x106 cells/mouse)
i.v. into NSG
mice that bear K562 leukemia cells and received low-dose rhIL-2 post cell
transfer. Survival
of mice will be monitored. Tumor-bearing mice treated with IL-12+IL-18+ALT-803-
activated NK cells (CIML NK cells) are expected to survival longer that mice
treated with
ALT-803-activated NK cells (Romee, R, et al. Sci Transl Med. 2016;
8:357ra123).
Prolonged survival of tumor-bearing mice receiving hIL18/IL12/TxM-activated
human NK
cells will provide evidence that hIL18/IL12/TxM can serve as an ex-vivo agent
to augment in
vivo antitumor activity of immune cells.
For treatment of patients with malignancies such as relapsed or refractory
acute
myeloid leukemia (AML) (Romee, R, et al. Sci Transl Med. 2016; 8:357ra123),
patients will
be treated with preconditioned with cyclophosphamide and fludarabine and then
treated with
CIML NK cells which were generated from allogeneic haploidentical NK cells
incubated ex
vivo with hIL18/IL12/TxM or hIL12/IL18/TxM for 16 to 24 hours. Following cell
transfer,
patients may receive low dose IL-2 to support the cells in vivo. Antitumor
responses
(objective responses, progression free survival, overall survival, time to
relapse, etc.) will be
assessed and will provide evidence that hIL18/IL12/TxM or hIL12/IL18/TxM can
serve as an
79
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
ex-vivo agent to augment antitumor activity of human immune cells in patients
with
malignancies. Similar studies will be carried out in patients with other
hematologic or solid
tumor or infectious diseases.
In each of these studies, the persistence and functionality of the NK cells
can be
evaluated post transfer. For example, PBMCs could be isolated from patients 7
to 14 days
post transfer and the percentage of Ki67-positive (proliferation marker) donor
NK cells can
be determined by flow cytometry. The NK cells can also be restimulated with
tumor cells
and the levels of IFN-y production can be assessed by flow cytometry. The
results of these
studies will indicate if pre-transfer treatment of NK cells ex vivo with
hIL18/IL12/TxM or
hIL12/IL18/TxM augments their subsequent immune responses in vivo.
Example 5: Immunostimulatory Effects of hIL18/IL12/TxM Fusion Protein
Complexes
Following Administration to Mice
As indicated above hIL18/IL12/TxM fusion protein complexes were highly
effective
at stimulating proliferation and responses of immune cells in vitro. To assess
the activity of
these complexes in vivo, female C57BL/6 mice were injected intraperitoneally
with 20 mg/kg
hIL18/IL12/TxM or PBS as a control. After 3 days, the mice were sacrificed and
blood and
spleen samples were taken to determine changes in immune cell subsets as
measured by flow
cytometry following staining with antibodies to CD8 T cells (CD8), CD4 T cells
(CD4), B
cells (CD19) and NK cells (NKp46). As shown in FIG. 18A, hIL18/IL12/TxM
treatment
resulted in a 2.5-fold increase in the weight of the spleens compared to PBS
control treated
mice. However, there were no signs of clinical toxicity with 20 mg/kg
hIL18/IL12/TxM
treatment. Administration of hIL18/IL12/TxM also resulted in a greater than 2-
fold increase
in the percentage of CD8 T cells and a 5.5-fold increase in the percentage of
NK cells in the
spleens of treated mice compared to the PBS controls (FIG. 18B). Additionally,
absolute cell
counts in the blood increased 6.4-fold for CD8 T cells and 23-fold for NK
cells and blood cell
percentages increased 3.9-fold for CD8 T cells and 13-fold for NK cells
following treatment
of mice with hIL18/IL12/TxM compared to PBS (FIG. 18C and FIG. 18D). The
results of
this study clearly indicate that administration of hIL18/IL12/TxM provided an
immunostimulatory effect to immune cells, particularly CD8 T cells and NK
cells, in mice
without causing overt toxicity.
OTHER EMBODIMENTS
While the invention has been described in conjunction with the detailed
description
thereof, the foregoing description is intended to illustrate and not limit the
scope of the
CA 03055318 2019-09-04
WO 2018/165208
PCT/US2018/021220
invention, which is defined by the scope of the appended claims. Other
aspects, advantages,
and modifications are within the scope of the following claims.
The patent and scientific literature referred to herein establishes the
knowledge that is
available to those with skill in the art. All United States patents and
published or unpublished
United States patent applications cited herein are incorporated by reference.
All published
foreign patents and patent applications cited herein are hereby incorporated
by reference.
Genbank and NCBI submissions indicated by accession number cited herein are
hereby
incorporated by reference. All other published references, documents,
manuscripts and
scientific literature cited herein are hereby incorporated by reference.
While this invention has been particularly shown and described with references
to
preferred embodiments thereof, it will be understood by those skilled in the
art that various
changes in form and details may be made therein without departing from the
scope of the
invention encompassed by the appended claims.
81