Note: Descriptions are shown in the official language in which they were submitted.
1
Description
Title of Invention: ANTI-C-MET ANTIBODY AND USE THEREOF
Technical Field
The present invention relates to an antibody or an antigen binding fragment
thereof,
specifically binding to a human hepatocyte growth factor receptor (c-Met), and
a
composition for preventing or treating cancer comprising the same.
[2]
Background Art
[3] Receptor tyrosine kinases (RTK) act as a vital modulator in cell
growth, differentiation,
neovascularization, tissue recovery, etc. Besides such general physiological
processes, an
abnormal expression of a certain RTK is associated with the development and
progression
of many kinds of cancer. Thus, such RTK has been considered as a promising
drug target
for cancer treatment.
[4] A hepatocyte growth factor receptor (HGFR; c-Met), which is a kind of
the RTK, is a
receptor on the surface of cells with regard to hepatocyte growth factor known
as a scatter
factor (HGF/SF) (Laird AD et al., Expert. Opin. Investig. Drugs 12: 51-64
(2003)). An
abnormal c-Met activation by HGF, which is one of the representative oncogenic
mechanisms, is known to be associated with tumor proliferation, apoptosis
inhibition,
neovascularization, invasion, metastasis and the like (Bottaro DP et al.,
Science 251: 802-
804 (1991), Day RM et al., Oncogene 18: 3399-3406 (1999)). And also, it is
reported that
the abnormal c-Met activation by c-Met mutation and amplification is
associated with
various cancers such as lung cancer, colon cancer, head and neck cancer,
stomach cancer,
breast cancer, etc., and is also involved in an increase in tumor
aggressiveness and its
unfavorable prognosis (Lefebvre J et al., FASEB J 26: 1387-1399 (2012), Liu X
etal.,
Trends Mol Med 16: 37-45 (2010), Smolen GA etal., Proc Natl Acad Sci USA 103:
2316-
2321 (2006), Foveau B et al., Mol Biol Cell 20: 2495-2507 (2009)).
[5]
[6] Thus, c-Met has drawn much attention as a target antigen for treating
such various
cancers and various approaches have been made to inhibit the expression and
activity
of c-Met. As a c-Met-specific small molecule tyrosine kinase inhibitor, which
has been
known so far, there are Tivantinib (ArQule), INC280 (Novatis), AMG337 (Amgen),
etc.
And, Rilotumumab (Amgen), Ficlatuzumab (AVEP Pharmaceuticals), HuL2G7 (Galaxy
Biotech), etc., have been developed as an HGF-specific monoclonal antibody,
which is
a ligand of c-Met. Also, as an antagonist monoclonal antibody, which targets
Date Regue/Date Received 2023-01-10
2
c-Met, there are Onartuzurnab (WO 2006/015371) in clinical phase 111 of
development by
Genentech, Emibetuzumab (WO 2010/059654) in clinical phase II by Lilly, SA1T-
301 (US
2014154251) in clinical phase I of development, ABT-700 (Wang J et al., BMC
Cancer. 16:
105-118(2016)), etc. Onartuzumab is a monovalent antagonistic antibody derived
from a
bivalent monoclonal antibody (5D5), which acts on c-Met as an agent (Mark
Merchant, et
al., Proc Natl Acad Sci U S A. 110(32): E2987¨E299 (2013)). As such, various
drugs have
been developed with regard to c-Met, but c-Met is associated with the
occurrence and
progression of various cancers as described above, thus it is constantly
driving a
continuous demand for developing a new therapeutic agent capable of treating
cancer by
targeting c-Met.
[7]
Disclosure of Invention
Technical Problem
[8] The present inventors have developed a novel anti-c-Met antibody
binding to c-Met
with a high affinity and have also identified that such anti-c-Met antibody, a
chimera
thereof and humanized and affinity-optimized antibodies remarkably inhibit a
proliferation
of tumor cells and have an excellent anticancer effect, thus having completed
the present
invention.
[9]
Solution to Problem
[10] One objective of the present invention is to provide an antibody or an
antigen binding
fragment thereof that specifically binds to a hepatocyte growth factor
receptor (c-Met).
[11] Another objective of the present invention is to provide a nucleic
acid molecule
encoding the antibody or the antigen binding fragment thereof, an expression
vector
comprising the nucleic acid molecule, a host cell having the expression vector
introduced
therein, a method for producing an antibody or an antigen binding fragment
thereof using
the host cell.
[12] Yet another objective of the present invention is to provide a
composition for detecting
c-Met comprising the antibody or the antigen binding fragment thereof, a kit
for detection
comprising the same, and a method for detecting a c-Met antigen using the
same.
[13] Still yet another objective of the present invention is to provide a
composition for
preventing or treating cancer comprising the antibody or the antigen binding
fragment
thereof.
[I 3a] Another objective of the present invention is to provide a
composition for preventing or
treating cancer, comprising the antibody or the antigen binding fragment as
defined herein
and a pharmaceutically accepted excipient.
CA 3061704 2019-11-26
2a
[13b] Another objective of the present invention is to provide a
composition for the
preparation of a medicament for preventing or treating cancer, comprising the
antibody or
the antigen binding fragment as defined herein and a pharmaceutically accepted
excipient.
[13c] Another objective of the present invention is to provide a use of the
antibody or the
antigen binding fragment as defined herein for preventing or treating cancer.
[13d] Another objective of the present invention is to provide a use of the
antibody or the
antigen binding fragment as defined herein for the preparation of a medicament
for
preventing or treating cancer.
Advantageous Effects of Invention
[14] The antibody or the antigen binding fragment thereof of the
present invention that
specifically binds to a hepatocyte growth factor receptor (c-Met), has a novel
sequence,
Date Recue/Date Received 2022-02-11
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
3
and shows an excellent cancer cell proliferation inhibitory activity and a
remarkably
excellent anticancer activity even by a little amount thereof, thus
effectively preventing
or treating the disease such as cancer.
[15]
Brief Description of Drawings
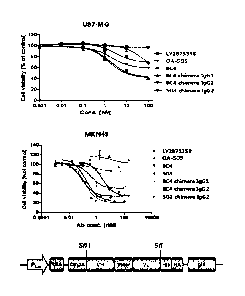
[16] FIG. 1 shows results of an in vitro test on tumor cell proliferation
inhibitory activity
of hybridoma c-Met antibody of the present invention.
[17] FIG. 2 shows a schematic diagram of a vector for expressing a separate
transcriptome
for seFv display.
[18] FIG. 3 shows results of analyzing a tumor cell proliferation
inhibitory activity by
hu8C4 affinity-optimized antibody of the present invention,
[19] FIG. 4 shows results of analyzing a tumor cell proliferation
inhibitory activity by a
bispecific antibody of the present invention.
[20] FIG. 5 shows results of analyzing a tumor cell proliferation
inhibitory activity by a
bispecific antibody of the present invention.
[21] FIG. 6 shows results of comparing a tumor cell proliferation
inhibitory activity
between the bispecific antibody of the present invention and a combined
therapy in U-
87 MG (gilioblatoma), NCI-H292 (NSCLC), NCI-H1648 (NSCLC) and NCI-H596
(NSCLC) cell lines.
[22] FIG. 7 shows results of comparing a tumor cell proliferation
inhibitory activity
between the bispecific antibody of the present invention and a combined
therapy in
LS174T (colon), BT20 (TN BC) and KP4 (pancreatic) cell lines.
[23] FIG. 8 shows results of comparing a tumor cell proliferation
inhibitory activity
between the bispecific antibody of the present invention and a combined
therapy in
HCC827 (NSCLC) and NCI-H596 (NSCLC) cell lines.
[24] FIG. 9 shows results of measuring a binding capacity of the anti-c-Met
antibody and
the bispecific antibody of the present invention with regard to various kinds
of c-Met
and EGFR antigens by an ELISA method.
[25] FIG. 10 shows results of measuring an effect of decreasing a receptor
level by the
bispecific antibody of the present invention in an NCI-H820 (NSCLC) cell line.
[26] FIG. 11 shows results of measuring an inhibition of c-Met and EGFR
phospho-
rylation by the anti-c-Met antibody and the bispecific antibody of the present
invention
in an NCI-H820 (NSCLC) cell line.
[27] FIG. 12 shows results of measuring an anticancer effect of the
bispecific antibody of
the present invention in a U-87 MG (glioblastoma) cell xenograft model.
[28] FIG. 13 shows results of measuring an anticancer effect of the
bispecific antibody of
the present invention in an NCI-H820 (NSCLC) cell xenograft model.
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
4
[29] FIG. 14 shows results of analyzing a tumor cell proliferation
inhibitory activity by
treating the anti-c-Met antibody of the present invention and the anti-HER2
antibody
by a combined therapy in an NCI-H21.70 (NSCLC) cell line.
[30] FIG. 15 shows results of measuring an anticancer effect of a combined
therapy with
the anti-c-Met antibody of the present invention and the anti-HER2 antibody in
an
NCI-H2170 (NSCLC) cell xenograft model.
[31] FIG. 16 shows results of measuring an anticancer effect of the
bispecific antibody of
the present invention in an NCI-H596 (NSCLC) cell xenograft model.
[32] FIG. 17 shows results of measuring an anticancer effect of the
bispecific antibody of
the present invention in an EBC-1 (NSCLC) cell xenograft model.
[33] FIG. 18 shows results of indicating an amount of c-Met on the surface
of cells,
measured after treating an HCC827 cell line with a bispecific antibody (Im8C4
x
Vectibix scFv), etc.
[34] FIG. 19 shows results of indicating an amount of EGFR on the surface
of cells,
measured after treating an HCC827 cell line with a bispecific antibody (hu8C4
x
'Vectibix scFv), etc.
[35] FIG. 20 shows results of indicating an epitope of a bispecific
antibody, analyzed by a
hydrogen-deuterium exchange mass spectrometry (HDX-MS), in a tertiary
structure.
[36]
Best Mode for Carrying out the Invention
1371 Hereinafter, the present invention will be described in more detail as
follows.
Meanwhile, each description and embodiment disclosed in the present invention
may
be applied to other descriptions and embodiments respectively as well. In
other words,
all the combinations of various elements disclosed in the present invention
are within
the scope of the present invention. Also, the scope of the present invention
may not be
restricted by the detailed descriptions below.
[38]
[39] To achieve the objectives above, one aspect of the present invention
provides an
antibody or an antigen binding fragment thereof that specifically binds to a
hepatocyte
growth factor receptor (c-Met).
[40] The antibody or the antigen binding fragment thereof of the present
invention,
specifically binding to c-Met, binds to c-Met with a high affinity to inhibit
an ex-
pression or activity thereof, thus showing an excellent tumor cell
proliferation in-
hibitory activity, such that the antibody alone or with conventional
pharmaceutically
acceptable carriers, other anticancer drugs, anticancer adjuvants, etc. may be
valuably
used as an anticancer composition for preventing or treating cancer.
[41] In the present invention, the term "antibody" means a protein molecule
serving as a
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
receptor for specifically recognizing an antigen, comprising an immunoglobulin
molecule immunologically having reactivity with a certain antigen, wherein
examples
thereof may comprise a monoclonal antibody, a polyclonal antibody, a full-
length
antibody and antibody fragments all. Also, the term may comprise a bivalent or
bispecific molecule (e.g., a bispecific antibody), a diabody, a triabody or a
tetrabody.
[42] In the present invention, the term "monoclonal antibody" refers to an
antibody
molecule of a single molecule composition obtained from substantially the same
antibody population, wherein such monoclonal antibody shows a single binding
specificity and affinity for a certain epitope. In the present invention, the
term "full-
length antibody" has a structure with two full-length light chains and two
full-length
heavy chains, wherein each of light chains is linked to a heavy chain by a
disulfide
bond. A constant region of the heavy chain has gamma (y), mu (11), alpha (a),
delta (8)
and epsilon (s) types, and also has gammal (y1), gamma2 (y2), gamma3 (y3),
gamma4
(y4), alpha] (al) and a1pha2 (a2) as a subclass. A constant region of the
light chain
has kappa (K) and lambda (X) types. IgG comprises IgGl, IgG2, IgG3 and IgG4 as
a
subtype.
[43] In the present invention, the terms "fragment," "antibody fragment"
and "antigen
binding fragment" refer to any fragments of the antibody of the present
invention
having an antigen binding function of the antibody, wherein such terms are
used inter-
changeably with each other, Exemplary antigen binding fragments comprise Fab,
Fab',
F(abl)2, Fv and the like, but not limited thereto.
[44] The Fab has a structure with a variable region of light and heavy
chains, a constant
region of light chain and a first constant region of heavy chain (CHI domain),
and also
has one antigen binding site. An antigen binding fragment of an antibody
molecule or
an antibody fragment means a fragment having an antigen binding function, and
Fab' is
different from Fab in that the former has a hinge region having one or more
cysteine
residue in C terminus of a heavy chain CH1 domain. F(ab')2 antibody is created
in such
a way that a cysteine residue of a hinge region of Fab' forms a disulfide
bond. Fv is a
minimal antibody fragment having only a heavy chain variable region and a
light chain
variable region, wherein a recombinant technology for creating Fv fragments is
disclosed in PCT International Patent Publication Applications WO 88/10649, WO
88/106630, WO 88/07085, WO 88/07086, WO 88/09344 and the like. Two-chain Fv is
formed in such a way that a heavy chain variable region and a light chain
variable
region are linked to each other by a non-covalent bond, while single-chain Fv
is
formed in such a way that a heavy chain variable region and a single chain
variable
region are generally linked with each other either by a covalent bond through
a peptide
linker or directly linked in C-terminus, thus forming a structure like a dimer
as shown
in the two-chain Fv. Such antibody fragment may be obtained by using a protein
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
6
hydrolase (for example, Fab may be obtained by performing a restriction
digestion of a
whole antibody by papain and F(ab1)2 fragment may be obtained by performing a
digestion of the same by pepsin) or may be produced by a gene recombination
technology, but not limited thereto.
[45]
[46] Particularly in the present invention, it may be provided that the
antibody specifically
binding to c-Met is:
[47] (a) an antibody comprising a light chain variable region comprising a
light chain
CDR1 represented by SEQ ID NO: 1; a light chain CDR2 represented by SEQ ID NO:
2; a light chain CDR3 represented by SEQ ID NO: 3, and a heavy chain variable
region comprising a heavy chain CDR1 represented by SEQ ID NO: 7; a heavy
chain
CDR2 represented by SEQ ID NO: 8; and a heavy chain CDR3 represented by SEQ ID
NO: 9;
[48] (h) an antibody comprising a light chain variable region comprising a
light chain
CDRI represented by SEQ ID NO: 4; a light chain CDR2 represented by SEQ ID NO:
5; a light chain CDR3 represented by SEQ ID NO: 6, and a heavy chain variable
region comprising a heavy chain CDR1 represented by SEQ ID NO: 10; a heavy
chain
CDR2 represented by SEQ ID NO: 11; and a heavy chain CDR3 represented by SEQ
ID NO: 12; or
[49] (c) affinity-optimized antibodies thereof.
[50] In the present invention, the term "heavy chain" may comprise both a
full-length
heavy chain and a fragment thereof comprising a variable region domain VH with
an
amino acid sequence having a variable region sequence enough to give
specificity to
an antigen, as well as three constant region domains CHI, CH2 and CH3. Also,
in the
present invention, the term "light chain" may comprise both a full-length
light chain
and a fragment thereof comprising a variable region domain VI with an amino
acid
sequence having a variable region sequence enough to give specificity to an
antigen, as
well as a constant region domain CL.
[51] In the present invention, the antibody may comprise both a mouse
antibody produced
from a mouse, and a mutant thereof, wherein a part of an amino acid sequence
of a
parent antibody is substituted, added and/or deleted to improve the affinity,
immunity,
etc., of the antibody. The mutant may comprise a chimeric antibody, a
humanized
antibody, an affinity-optimized antibody, etc., as an example, but not limited
thereto.
In the present invention, the mutant comprehensively refers to an antibody,
wherein a
part of a CDR amino acid sequence of a parent antibody is mutated
(substituted, added
or deleted) on condition of having the same CDR as that of the parent antibody
or
targeting the same epitope as that of the parent antibody. Such mutant may be
appro-
priately adjusted by those skilled in the art to improve the affinity,
immunity and the
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
7
like of an antibody within the scope of maintaining a binding capacity for the
same
epitope.
[52] In other words, the antibody or the antigen binding fragment thereof
of the present
invention may comprise a sequence of anti-e-Met antibody described herein as
well as
biological equivalents thereof, within the scope of specifically recognizing c-
Met. For
example, an additional change may be made in an amino acid sequence of the
antibody, in order to further improve the binding affinity and/or other
biological char-
acteristics of the antibody. Such change comprises, for example, the deletion,
insertion
and/or substitution of an amino acid sequence residue of the antibody. Such
amino acid
mutation is made based on relative similarity of amino acid side chain
substituent, e.g.,
hydrophobicity, hydrophilicity, charge, size, etc. By analyzing the size,
shape and type
of amino acid side chain substituent, it can be seen that arginine, lysine and
histidine
are all positive charge residues; alanine, glycine and serine have a similar
size; and
phenylalanine, tryptophan and tyrosine have a similar shape. Thus, based on
such con-
siderations, it can be seen that arginine, lysine and histidine; alanine,
glycine and
serine; and phenylalanine, tryptophan and tyrosine are biologically functional
equivalents.
[53] In the present invention, the term "chimeric antibody" is an antibody
formed in such
a way that a variable region of a mouse antibody is recombined with a constant
region
of a human antibody, which results in a greatly improved immune reaction in
comparison with a mouse antibody.
[54] In the present invention, the term "humanized antibody" means an
antibody formed
in such a way that a protein sequence of an antibody derived from other
species than
human is modified to be similar to that of an antibody mutant naturally
produced from
human. For example, the humanized antibody may be prepared by preparing a
humanized variable region through a recombination of CDR derived from a mouse
with FR derived from a human antibody and then by recombining the same with a
constant region of a preferred human antibody. However, a simple CDR grafting
only
results in a low affinity of the humanized antibody, so several key FR amino
acid
residues, which are considered to possibly influence a three-dimensional
structure of
CDR, may develop an affinity with those of mouse antibody, thus reaching the
same
level as the affinity of an original mouse antibody.
[55] In the present invention, the term "affinity-optimized antibody,"
which is a mutant
formed in such a way that a part of CDR sequence of a certain antibody is
substituted,
added or deleted, means an antibody with a better binding affinity to an
antigen while
binding to the same antigen epitope as that of the certain antibody.
Particularly, the
affinity-optimized antibody of the present invention refers to a mutant
antibody binds
to the same epitope as that of: (a) an antibody comprising a light chain
variable region
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
8
comprising a light chain CDR1 represented by SEQ ID NO; 1; a light chain CDR2
rep-
resented by SEQ ID NO: 2; a light chain CDR3 represented by SEQ ID NO: 3, and
a
heavy chain variable region comprising a heavy chain CDR1 represented by SEQ
ID
NO: 7; a heavy chain CDR2 represented by SEQ ID NO: 8; a heavy chain CDR3 rep-
resented by SEQ ID NO: 9; or (b) an antibody comprising a light chain variable
region
comprising a light chain CDR1 represented by SEQ ID NO: 4; a light chain CDR2
rep-
resented by SEQ ID NO: 5; a light chain CDR3 represented by SEQ ID NO: 6, and
a
heavy chain variable region comprising a heavy chain CDR1 represented by SEQ
ID
NO: 10; a heavy chain CD12 represented by SEQ ID NO: 11; a heavy chain CDR3
represented by SEQ ID NO: 12. A person of ordinary skill in the art may
prepare the
affinity-optimized antibody by using a known technology based on certain light
chain
and heavy chain CDR sequences. For example, the affinity-optimized antibody of
the
present invention may be prepared through a phage display. In the present
invention,
the term "phage display" refers to a technology, which displays a mutant
polypeptide
as a fusion protein with at least a part of coat protein on a phage, for
example, on the
surface of fibrous phage particles. The usefulness of the phage display lies
in the fact
that it targets a large library of randomized protein mutants, thus promptly
and ef-
ficiently classifying sequences binding to a target antigen with a high
affinity.
Displaying a library of peptides and proteins on the phage has been used for
screening
millions of polypeptides in order to see a polypeptide with a specific binding
charac-
teristic.
[56]
[57] In one exemplary embodiment of the present invention, it may be
provided that the
antibody is an antibody comprising: (a) a light chain variable region
represented by
SEQ ID NO: 13 and a heavy chain variable region represented by SEQ ID NO: 15;
or
(b) a light chain variable region represented by SEQ ID NO: 14 and a heavy
chain
variable region represented by SEQ ID NO: 16. As an example, it may be
provided
that the antibody is an antibody comprising: (a) a light chain variable region
coded by
a nucleotide represented by SEQ ID NO: 17 and a heavy chain variable region
coded
by a nucleotide represented by SEQ ID NO: 19; or (b) a light chain variable
region
coded by a nucleotide represented by SEQ ID NO: 18 and a heavy chain variable
region coded by a nucleotide represented by SEQ ID NO: 20, but not limited
thereto.
[58] According to one specific embodiment of the present invention, a
hybridoma cell
group was obtained from a mouse, wherein a human c-Met Sema domain/Fe fusion
protein is an antigen, from which anti-c-Met antibody specifically binding to
c-Met
was selected by screening with an EL1SA analysis method using c-Met/His fusion
protein as an antigen. The selected antibody and the chimeric antibody thereof
have a
tumor cell proliferation inhibitory activity, which is equal to or more
excellent than
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
9
even commercially available known LY2875358 and 0A-5D5 (Table 3 and FIG. 1),
thus being very valuably used in prevention or treatment of cancer.
[59]
[60] In another exemplary embodiment of the present invention, it may be
provided that
the antibody comprises:
[61] (a) a light chain variable region represented by SEQ ID NO: 21 and a
heavy chain
variable region represented by SEQ ID NO: 23; (b) a light chain variable
region rep-
resented by SEQ ID NO: 22 and a heavy chain variable region represented by SEQ
ID
NO: 24; (c) a light chain variable region represented by SEQ ID NO: 29 and a
heavy
chain variable region represented by SEQ ID NO: 31; or (d) a light chain
variable
region represented by SEQ ID NO: 30 and a heavy chain variable region
represented
by SEQ ID NO: 32. As an example, it may be provided that the antibody is an
antibody
comprising: (a) a light chain variable region coded by a nucleotide
represented by SEQ
ID NO: 25 and a heavy chain variable region coded by a nucleotide represented
by
SEQ ID NO: 27; (b) a light chain variable region coded by a nucleotide
represented by
SEQ ID NO: 26 and a heavy chain variable region coded by a nucleotide
represented
by SEQ ID NO: 28; (c) a light chain variable region coded by a nucleotide
represented
by SEQ ID NO: 33 and a heavy chain variable region coded by a nucleotide rep-
resented by SEQ ID NO: 35; or (d) a light chain variable region coded by a
nucleotide
represented by SEQ ID NO: 34 and a heavy chain variable region coded by a nu-
cleotide represented by SEQ ID NO: 36, but not limited thereto. Also, it may
be
provided that the antibody comprises a hinge region represented by one of SEQ
ID
NO: 37 to SEQ ID NO: 44.
[62] In one specific embodiment of the present invention, a humanized
antibody
comprising CDR of the antibody obtained through a phage display selection was
prepared, and it was identified that such antibody showed an anticancer
activity, which
was similar to that of the chimera antibody of the present invention (Examples
2 and
3). Also, in another specific embodiment of the present invention, a tumor
cell pro-
liferation inhibitory activity of the antibody was evaluated according to a
hinge region
sequence, and it was identified that a proliferation of most tumor cells was
effectively
inhibited, even with a somewhat difference in the activity depending on the
difference
of hinge sequence (Table 7).
[63]
[64] In yet another exemplary embodiment of the present invention, but not
limited
thereto, it may be provided that an affinity-optimized antibody for the
humanized
antibody is an antibody, wherein one or more amino acid sequence is
substituted from
an antibody comprising: a light chain variable region comprising a light chain
CDR1
represented by SEQ ID NO: 1; a light chain CDR2 represented by SEQ ID NO: 2; a
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
light chain CDR3 represented by SEQ ID NO: 3, and a heavy chain variable
region
comprising a heavy chain CDR1 represented by SEQ ID NO: 7; a heavy chain CDR2
represented by SEQ ID NO: 8; a heavy chain CDR3 represented by SEQ ID NO: 9,
and wherein, (i) Gina 1st position of the light chain CDR] is substituted with
A, E, K,
L, N, R, S. V or W; A in a 2nd position thereof is substituted with C, G, I,
P, S, T or V;
S in a 3rd position thereof is substituted with G, M, N, P, Q, R, S or T; E in
a 4th
position thereof is substituted with A, D, F, G, H, K, M, Q, R, S, T or V; N
in a 5th
position thereof is substituted with A, D, E, G, K, L, P. Q, R, S, T or V; I
in a 6th
position thereof is substituted with A, F, L, M, Q, R, S. T or V; Y in a 7th
position
thereof is substituted with F, H, R or V; or G in a 8th position thereof is
substituted
with D, F, H, M, N, R, S, T or V; (ii) G in a 1st position of the light chain
CDR2 is
substituted with D, F, H, K, P. Q, S, V or Y; T in a 3rd position thereof is
substituted
with Q; or N in a 4th position thereof is substituted with G; (iii) Q in a 1st
position of
the light chain CDR3 is substituted with E, G, I, M or N; N in a 2nd position
thereof is
substituted with A, D, E, H, L, Q, S or T; V in a 3rd position thereof is
substituted with
1, L, M, N, Q, S or T; L in a 4th position thereof is substituted with F, H,
I, M, R, S, V,
W or Y; S in a 51h position thereof is substituted with C, D, E, F, G, H, K,
L, N, Q, R,
T, V or Y; S in a 6th position thereof is substituted with D, E, F, G, H, I.
L, M, N, P, Q,
R, T, V or Y; P in a 7th position thereof is substituted with A, D, E, G, N,
Q, S or V; Y
in an 8th position thereof is substituted with E, F, L, M or Q; or T in a 9th
position
thereof is substituted with D, F, G, 1, L, N, S. V. W or Y; (iv) D in a 1st
position of the
heavy chain CDRI is substituted with G or Q; Yin a 2nd position thereof is
substituted
with Q; or I in a 4th position thereof is substituted with A or Q; (v) F in a
3rd position
of the heavy chain CDR2 is substituted with D, E, W or Y; G in a 5th position
thereof
is substituted with D, H or Y; S in a 6th position thereof is substituted with
F, P, W or
Y; G in a 7th position thereof is substituted with A, F, L, N or T; N in an
8th position
thereof is substituted with F, P. S. T or Y; T in a 9th position thereof is
substituted with
A, D, E, F, G, H, L, P. S or V; H in a 10th position thereof is substituted
with A, D, F,
M, R, S, T, V, W or Y; F in an llth position thereof is substituted with G, H,
I, L, M,
N, P, Q, V or Y; S in a 12th position thereof is substituted with A, D, G, H,
I, L, P, T
or V; A in a 13th position thereof is substituted with D, E, F, G, H, I, K, L,
M, P. R, S,
T, V or Y; R in a 14th position thereof is substituted with A, E, G, H, L, N,
P, Q, S, W
or Y; F in a 15th position thereof is substituted with D, E, G, L, M, P, R, S,
V or W; K
in a 16th position thereof is substituted with A, E, F, G, H, L, R, S, T, V or
Y; or G in a
17th position thereof is substituted with E, F, H, L, M, N, P, Q, R, S, T, V
or W; or (vi)
G in a 1st position of the heavy chain CDR3 is substituted with E, F, H, N, Q,
V or W;
D in a 2nd position thereof is substituted with E; Y in a 3rd position thereof
is sub-
stituted with L, Q, T or V; G in a 4th position thereof is substituted with W;
F in a 5th
CA 03061704 2019-10-28
WO 2018/221969
PCT/KR2018/006182
11
position thereof is substituted with L or Y; L in a 6th position thereof is
substituted
with Q, S or Y; or Y in a 7th position thereof is substituted with C, L, M, N
or Q.
Herein, it may be provided that the light chain CDR1 comprises 0 to 5
substitutions,
the light chain CDR2 comprises 0 to 1 substitution, the light chain CDR3
comprises 0
to 7 substitutions, the heavy chain CDR1 comprises 0 to 1 substitution, the
heavy chain
CDR2 comprises 0 to 11 substitutions, and the heavy chain CDR3 comprises 0 to
6
substitutions.
[65] Particularly, in still yet another exemplary embodiment of the
present invention, it
may be provided that the affinity-optimized antibody comprises a light chain
variable
region comprising a light chain CDR1 represented by any one of SEQ ID NO: 1
and
SEQ ID NO: 229 to SEQ ID NO: 268; a light chain CDR2 represented by any one of
SEQ ID NO: 2, SEQ ID NO: 182 to SEQ ID NO: 190, SEQ ID NO: 227 and SEQ ID
NO: 228; a light chain CDR3 represented by any one of SEQ ID NO: 3, SEQ ID NO:
142 to SEQ ID NO: 181, SEQ ID NO: 191 to SEQ TD NO: 226 and SEQ ID NO: 269
to SEQ ID NO: 301; and a heavy chain variable region comprising a heavy chain
CDR1 represented by any one of SEQ ID NO: 7 and SEQ ID NO: 108 to SEQ ID NO:
112; a heavy chain CDR2 represented by any one of SEQ ID NO: 8, SEQ ID NO: 54
to SEQ ID NO: 63, SEQ ID NO: 72 to SEQ ID NO: 107 and SEQ ID NO: 118 to SEQ
ID NO: 141; a heavy chain CDR3 represented by any one of SEQ ID NO: 9, SEQ ID
NO: 64 to SEQ ID NO: 71 and SEQ ID NO: 113 to SEQ ID NO: 117, more par-
ticularly, comprising a light chain variable region represented by any one of
SEQ ID
NO: 21 and SEQ ID NO: 306 to SEQ ID NO: 311, and a heavy chain variable region
represented by any one of SEQ ID NO: 23 and SEQ ID NO: 302 to SEQ ID NO: 305,
and much more particularly comprising: (a) a light chain variable region
represented
by SEQ ID NO: 21 and a heavy chain variable region represented by SEQ ID NO:
302;
(b) a light chain variable region represented by SEQ ID NO: 21 and a heavy
chain
variable region represented by SEQ ID NO: 305; (c) a light chain variable
region rep-
resented by SEQ ID NO: 310 and a heavy chain variable region represented by
SEQ
ID NO: 23; (d) a light chain variable region represented by SEQ ID NO: 308 and
a
heavy chain variable region represented by SEQ ID NO: 305; (e) a light chain
variable
region represented by SEQ ID NO: 306 and a heavy chain variable region
represented
by SEQ ID NO: 303; (f) a light chain variable region represented by SEQ ID NO:
307
and a heavy chain variable region represented by SEQ ID NO: 304; (g) a light
chain
variable region represented by SEQ ID NO: 308 and a heavy chain variable
region rep-
resented by SEQ ID NO: 304; (h) a light chain variable region represented by
SEQ ID
NO: 309 and a heavy chain variable region represented by SEQ ID NO: 304; (i) a
light
chain variable region represented by SEQ ID NO: 311 and a heavy chain variable
region represented by SEQ ID NO: 304; or (j) a light chain variable region
represented
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
12
by SEQ ID NO: 306 and a heavy chain variable region represented by SEQ ID NO:
302, but not limited thereto.
[66] In one specific embodiment of the present invention, a competitive
selection method
was used to select an antibody with a more improved affinity than the
humanized
antibody, thus obtaining a number of affinity-optimized antibodies (Tables 8
to 10 and
12). The affinity-optimized antibody has a tumor cell proliferation inhibitory
effect that
is 4.3 to 28.5 times more excellent than the humanized body (Table 11, 13 and
FIG. 3).
[67]
[68] In the present invention, it may be provided that the antibody is an
antibody or an
antigen binding fragment thereof specifically further binding to an epidermal
growth
factor receptor (EGFR) in addition to specifically binding to c-Met.
[69] It is known that the EGFR, one of ErbB tyrosine kinases, is abnormally
activated in
many epidermal cell tumors comprising non-small-cell lung carcinoma, causes
cell
proliferation, invasion, metastasis and angiogenesis, and increases cell
survival.
Gefitinib (Iressa), elotinib (Tarceva) and osimertinib (Tagrisso), which are
EGFR
tyrosine kinase inhibitors, are used as a representative lung cancer
therapeutic agent;
and cetuximab (Erbitux) and panitumumab (Vectibix), which are EGFR target an-
tibodies, are used as a colon cancer therapeutic agent (Yewale C et al.,
Biomaterials.
2013 34(34):8690-707 (2013), Deric L. Wheeler et al., Nature Reviews Clinical
Oncology 7, 493-507 (2010)).
[70] Such EGFR target therapeutic agents cause resistance one year before
and after
treatment, wherein c-Met amplification, mutation and HGF-induced activation
are
known as a key mechanism of resistance (Simona Corso Cancer Discovery 3:978-
992
(2013), Curtis R Chong et al., Nature Medicine 19, 1389-1400 (2013)). Also, it
is
reported that EGFR and c-Met are simultaneously expressed in various tumor
cells,
wherein, upon inhibiting EGFR, c-Met becomes activated, thus promptly
developing
the resistance of EGFR TKI (Engelman, J.A., et al., Science, 316:1039-43
(2007)),
[71] Based on such mechanism, a single treatment with a c-Met target drug
alone and a
combined treatment with an EGFR target drug have been now in a clinical trial,
but
their efficacy has not been verified yet as a therapeutic agent and there is a
need for de-
veloping a therapeutic agent for c-Met-related cancerous tumors, known as a
key cause
of resistance. Accordingly, the present inventors have prepared c-Met/EGFR
bispecific
antibody based on the antibody described above. The bispecific antibody not
only ef-
fectively inhibits a proliferation of tumor cells, which are resistant to
existing EGFR
therapeutic agents, but also shows an excellent proliferation inhibitory
activity against
tumor cells, thus being valuably used in treatment of diseases such as c-Met-
mediated
cancers through various mechanisms.
[72] It may be provided that the bispecific antibody is formed in such a
way that an
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
13
antibody or an antigen binding fragment thereof specifically binding to EGFR
is linked
to one light chain or heavy chain terminus of c-Met specific antibody, for
example,
being linked to a heavy chain C-terminus, but not limited thereto.
[73] It may be provided that the binding fragment specifically binding to
EGFR is Fab,
Fab', F(a13')2 or Fv.
[74] In one exemplary embodiment of the present invention, it may be
provided that the
Fv is a scFv fragment, wherein the scFv fragment is linked by a connector
capable of
linking the scFv fragment to one light chain or heavy chain terminus of c-Met
antibody. In one exemplary embodiment of the present invention, an antibody
specifically binding to EGFR is further prepared by linking with a connector
rep-
resented by SEQ ID NO: 312.
[75] It may be provided that the EGFR scFv fragment is an EGFR scFv capable
of
specifically binding to EGFR, known in the art, wherein, for example, there
are
Erbitux, Vectibix, Portrazza, TheraCIM or the like, but not limited thereto.
[76] In one exemplary embodiment of the present invention, it may be
provided that the
EGFR scFv is an Erbitux or Vectibix scFv fragment, particularly the EGFR scFv
comprises an amino acid sequence represented by SEQ ID NO: 313 or SEQ ID NO:
314, wherein the Vectibix scFv comprises an amino acid sequence represented by
SEQ
ID NO: 315, but not limited thereto.
[77] According to one specific embodiment of the present invention, as a
result of
identifying a tumor cell proliferation inhibitory activity of the bispecific
antibody, it
was identified that the antibody had a more excellent tumor activity
inhibitory efficacy
than a hu8C4 optimized antibody (Tables 16 and 17, and FIGS. 4, 5, 16 and 17).
In
particular, it was identified that the antibody of the present invention had
an excellent
cell proliferation inhibitory effect on even NCI-H292 and NCI-H1648 cell
lines, in
which c-Met and EGFR are normally expressed (Tables 17 and 19 and FIG. 6).
Based
on such results, it can be seen that an anticancer effect of the antibody of
the present
invention is not particularly limited by an abnormality of c-Met expression or
a
presence or absence of c-Met mutation, etc.
[78] Furthermore, it was identified that the bispecific antibody of the
present invention
had a more excellent tumor cell proliferation inhibitory capacity than a
combined
therapy of two antibodies (Tables 18 to 21 and FIGS. 6 to 8). Also, as a
result of
identifying an effect of the bispecific antibody of the present invention on
the activity
of antigens and signal transduction materials, it was identified that the
bispecific
antibody of the present invention had a more excellent signal transduction
inhibitory
efficacy than an antibody alone (Fla 11).
[79] It may be provided that the antibody or the antigen binding fragment
thereof of the
present invention binds to an epitope region represented by an amino acid
sequence
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
14
selected from the group represented by SEQ ID NO: 331, SEQ ID NO: 332, SEQ ID
NO: 333 and/or SEQ ID NO: 334. An affinity-optimized antibody prepared based
on a
certain antibody (reference antibody) is characterized by having a high
homology with
the light chain and heavy chain CDR sequences of a variable region with regard
to the
reference antibody, thus binding to the same epitope region as the reference
antibody,
such that such affinity-optimized antibody can share all the biological
characteristics
such as a pharmaceutical mechanism and a pharmaceutical efficacy caused by a
binding site, specificity and antibody and exhibit a more excellent effect on
binding
affinity than the reference antibody.
[80] The epitope region respectively means, for example, YVSKPGAQL (SEQ ID
NO:
331) in 321th to 329th positions, IGASLNDDI (SEQ ID NO: 332) in 3331h to 341th
positions, PIKYVND (SEQ ID NO: 333) in 366th to 372th positions, and
QVVVSRSGPST (SEQ ID NO: 334) in 464th to 474th positions from N-terminus of a
reference c-Met antigen (SEQ ID NO: 335), wherein c-Met antigen sequence with
the
antibody or the antigen binding fragment thereof of the present invention
binding
thereto comprises a partial mutation (substitution, addition or deletion) or a
binding
antigen exists in a form of a c-Met fragment, precursor or subtype, thus its
binding
sites or sequences may somewhat vary accordingly. Nevertheless, a person of
ordinary
skill in the art may clearly specify a position and a sequence, to which the
antigen or
the antigen binding fragment thereof of the present invention binds based on
an epitope
sequence information of a reference c-Met antigen.
[81] In one specific embodiment of the present invention, it was identified
that the
bispecific antibody hu8C4 x Vectibix scEv of the present invention binds to 4
epitope
regions of Y321 -L329 (SEQ ID NO: 331), T333 -1341 (SEQ ID NO: 332), P366 -
D372 (SEQ ID NO: 333), and Q464 - S474 (SEQ ID NO: 334) of a human c-Met sema
domain 13 chain (Table 28).
[82] The "antibody or antigen binding fragment thereof specifically binding
to c-Met" of
the present invention means the one binding to a human c-Met by KD 1 X lfi'M
or
less. It may be provided that the antibody or the antigen binding fragment
thereof binds
to human c-Met, for example, by KD 5 X 101 M or less, KD 1 X 10-8 M or less,
KD 5 X
9 M or less, or KD 1 X 10 9 M or less, but not limited thereto.
[83] In one specific embodiment of the present invention, it was directly
identified that
the antibody or the antigen binding fragments thereof of the present invention
had a
high binding affinity to c-Met antigen by identifying a binding affinity of
hu8C4,
hu8C4 AH71 and hu8C4 x Vectibix seFv to c-Met ECD, thus identifying KD values
of
3.173 X 10-w, 9.993 X 104' and 2.78 X 104'), respectively (Table 22). It was
identified
that the antibody or the antigen binding fragment thereof of the present
invention had a
cross-reactivity to a c-Met antigen of a cynomolgus monkey, which is an ape
(Table
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
22), but did not bind to other animal-derived antigens (e.g., rodents) (FIG.
9). Also, it
was identified that the antibody or the antigen binding fragment thereof of
the present
invention did not bind to other receptors on the surface of cells than c-Met
(Table 24).
Thus, it can be seen from the results above that the antibody or the antigen
binding
fragment thereof of the present invention showed a binding specificity to c-
Met antigen
of humans and monkeys.
[84] As used herein, the term "binding constant (IQ" means a binding ratio
of a certain
antibody-antigen interaction, and the term "dissociation constant (Koff)"
means a dis-
sociation ratio of a certain antibody-antigen interaction. Also, in the
present invention,
the term "affinity to antigen (Ku)" is the one that a ratio of Kaf : K. (i.e.,
Koff / K.) is
indicated as a molar concentration (M). It may be provided that a KD value for
an
antibody is measured by using a method widely established in the art. For
example, as
a method for measuring a Kt, value of an antibody, it may be provided by a
surface
plasmon resonance analysis using a BiocoreTM system, but not limited thereto.
[85] Another aspect of the present invention provides a method for
producing a nucleic
acid molecule for coding the antibody or the antigen binding fragment thereof,
an ex-
pression vector comprising the nucleic acid molecule, a host cell having the
expression
vector introduced therein, an antibody using the host cell or an antigen
binding
fragment thereof.
[86] The antibody and the antigen binding fragment thereof are such as that
described
above.
[87] As used herein, the term "nucleic acid molecule" has a meaning that
compre-
hensively comprises DNA and RNA molecules, wherein a nucleotide, a basic con-
stituent unit in the nucleic acid molecule, comprises not only a natural
nucleotide, but
also an analogue, in which a sugar or base portion is modified (Scheit,
Nucleotide
Analogs, John Wiley, New York (1980); Uhlman and Peyman, Chemical Reviews,
(1990) 90:543-584). A sequence of a nucleic acid molecule for coding the heavy
chain
and light chain variable regions of the present invention may be modified,
wherein the
modification comprises an addition, deletion, or non-conservative or
conservative sub-
stitution of nucleotide.
[88] It is understood that the nucleic acid molecule of the present
invention also comprises
a nucleotide sequence representing a substantial identity with the
aforementioned nu-
cleotide sequence. In the present invention, in case of aligning the
aforementioned nu-
cleotide sequence of the present invention with any other sequences in the
most corre-
sponding way and analyzing the aligned sequences by an algorithm
conventionally
used in the art, the substantial identity means a nucleotide sequence that
represents a
minimal 80% homology, particularly a minimal 90% homology, more particularly a
minimal 95% homology.
16
[89] As used herein, the term "vector," which is a means for expressing a
target gene in a host
cell, comprises a plasmid vector; a cosmid vector; and virus vector such as a
bacteriophage
vector, an adenovirus vector, a retrovirus vector and an adeno-related virus,
particularly a
plasmid vector, but not limited thereto.
[90] In the vector of the present invention, it may be provided that a
nucleic acid molecule for
coding a light chain variable region and a nucleic acid molecule for coding a
heavy chain
variable region are operatively linked with a promoter.
[91] In the present invention, the term "operatively linked" means a
functional binding between a
nucleic acid expression regulatory sequence (e.g., a promoter, a signal
sequence, or an array in a
transcriptional regulatory factor binding site) and other nucleic acid
sequence, thus the regulatory
sequence controls a transcription and/or decoding of the other nucleic acid
sequence.
[92] The recombinant vector system of the present invention may be built
through various
methods known in the art. For example, such detailed methods are disclosed in
Sambrook et
al., Molecular Cloning, A Laboratory Manual, Cold Spring Harbor Laboratory
Press (2001).
[93] The vector of the present invention may be typically built as a vector
for cloning or a
vector for expression. Also, the vector of the present invention may be built
in such a way
that a prokaryotic cell or an eukaryotic cell is a host.
[94] For example, if the vector of the present invention is an expression
vector and the
prokaryotic cell is a host, it is general to comprise powerful promotors
capable of carrying
out transcription (e.g., tac promotor, lac promotor, lacUV5 promotor, 1pp
promotor, pLX
promotor, pRX promotor, rac5 promotor, amp promotor, recA promotor, SP6
promotor, trp
promotor, T7 promotor and the like), a ribosome binding site for starting
decoding and
transcription/decoding termination sequence. If E. coli (e.g., HB101, BL21,
DH5a, etc.) is
used as a host cell, promotor and operator portions of E. coli tryptophan
biosynthetic pathway
(Yanofsky, C., J. Bacteriol., (1984) 158:1018-1024), and a leftward promotor
of phage
(pLk promotor, Herskowitz, I. and Hagen, D., Ann, Rev. Genet., (1980) 14:399-
445) may be
used as a regulatory portion. If Bacillus sp. is used as a host cell, a
promotor of toxin protein
gene of Bacillus thuringiensis (Appl. Environ. Microbiol. (1998) 64:3932-3938;
Mol. Gen.
Genet. (1996) 250:734-741) or any promotors expressible in Bacillus sp. may be
used as a
regulatory portion.
[95] Meanwhile, the recombinant vector of the present invention may be
prepared by
manipulating plastnid (e.g., pCL, pSC101, pGV1106, pACYC177, ColE1, pKT230,
pME290,
pBR322, pUC8/9, pUC6, pBD9, pHC79, pIJ61, pLAFR1, pHV14, pGEX series, pET
series,
pUC19 and the like), phage (e.g., kgt4=XB, k-Charon, XAzl, M13
Date Recue/Date Received 2021-02-17
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
17
and the like) or virus (e.g., SV40, etc.) often used in the art.
[96] Meanwhile, if the vector of the present invention is an expression
vector and an eu-
karyotic cell is a host, promotors derived from a genome of mammal cells
(e.g., metal-
lothionein promotor, (3-actin promotor, human hemoglobin promotor and human
muscle creatin promotor) or promotors derived from mammal virus (e.g.,
adenoviral
late promotor, vaccinia virus 7.5K promotor, SV40 promotor, cytomegalovirus
(CMV)
promotor, tk promotor of HSV, mouse breast tumor virus (MMTV) promotor, LTR
promotor of HIV, promotor of Moloney virus, promotor of Epstein-barr virus
(EBV)
and promotor of Rous sarcoma virus (RSV)) may be used, wherein they generally
have
a polyadenylation sequence as a transcription termination sequence.
Particularly, the
recombinant vector of the present invention comprises a CMV promotor.
[97] The recombinant vector of the present invention may be fused with
other sequences
in order to facilitate refining of an antibody expressed therefrom. As
examples of fused
sequences, there are glutathione S-Ira]sferase (Pharmacia, USA), maltose
binding
protein (NEB, USA), FLAG (JET, USA), 6x His (hexahistidine; Quiagen, USA) and
the like. Also, a protein expressed by the vector of the present invention is
an antibody,
thus the expressed antibody may be easily purified through a protein A column,
etc.,
without an additional sequence for refining.
[98] Meanwhile, the recombinant vector of the present invention comprises
an antibiotic
resistance gene conventionally used in the art as a selected marker, wherein
it may
comprise, for example, resistance genes to ampicillin, gentamicin,
carbenicillin, chlo-
ramphenicol, streptomycin, kanamycin, geneticin, neomycin and tetracycline.
[99] As a vector for expressing the antibody of the present invention,
there may be both a
vector system, in which a light chain and a heavy chain are simultaneously
expiessed
in one vector, and a system, in which a light chain and a heavy chain are
respectively
expressed in a separate vector. In the latter case, two vectors may be
introduced into a
host cell, for example, through co-transformation or targeted transformation.
The co-
transformation is a method for selecting cells that express both light and
heavy chains
after simultaneously introducing each vector DNA for coding light and heavy
chains
into a host cell. The targeted transformation is a method for selecting a cell
transformed with a vector comprising a light (or heavy) chain and transforming
a
selected cell again with a vector comprising a heavy (or light) chain to
finally select a
cell that expresses both light and heavy chains.
[100] As long as they are capable of stably and continuously cloning and
expressing the
vector of the present invention, any host cells known in the art may be used,
wherein
such host cells may comprise Bacillus sp. strains such as Escherichia coli,
Bacillus
subtilis and Bacillus thuringiensis and prokaryotic host cells such as
Streptomyces,
Pseudomonas (e.g., Pseudomonas putida), Proteus mirabilis or Staphylococcus
(e.g.,
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
18
Staphylococcus carnosus), but not limited thereto.
[101] As suitable enkaryotic host cells of the vector, there may be mycetes
such as As-
pergillus species, yeasts such as Pichia pastoris, Saccharomyces cerevisiae,
Schizosac-
charomyces and Neurospora erassa, other lower eukaryotic cells, cells of
higher eu-
karyotes such as insect-derived cells, and cells derived from plants or
mammals.
[102] Particularly, host cells may be COS7 cells (monkey kidney cells), NSO
cells, SP2/0,
Chinese hamster ovary (CHO) cells, W138, baby hamster kidney (BHK) cells,
MDCK,
myeloma cell lines, HuT 78 cells or 293 cells, more particularly CHO cells,
but not
limited thereto.
[103] In the present invention, "transformation" and/or "transfection" into
host cells may be
performed by selecting a suitable standard technology according to host cells
as known
in the art, comprising any methods for introducing nucleic acid into
organisms, cells,
tissues or organs. The methods comprise electroporation, plasmogamy, calcium
phosphate (CaPO4) precipitation, calcium chloride (CaCl2) precipitation,
agitation
using silicon carbide fiber, agrobacteria-mediated transformation, PEG,
dextran
sulfate, lipofectamine, drying/suppression-mediated transformation and the
like, but
not limited thereto.
[104] In the present invention, the method for producing an antibody or an
antigen binding
fragment thereof using a host cell may particularly comprise steps of: (a)
culturing a
host cell transformed with a recombinant vector of the present invention; and
(b) ex-
pressing an anti-c-Met antibody or an antigen binding fragment thereof in the
host cell.
[105] In preparing the antibody above, culturing of a transformed host cell
may be
performed in an appropriate medium and under culturing conditions known in the
art.
Such culturing process may be easily adjusted according to a selected strain
by those
skilled in the art. Such culturing method is disclosed in various documents
(e.g., James
M. Lee, Biochemical Engineering, Prentice-Hall International Editions, 138-
176). Cell
culture is divided into suspension culture and attachment culture according to
a cell
growth type, and batch culture, fed-batch culture and continuous culture
according to a
culture method. A medium used in culture has to appropriately satisfy
requirements of
a certain strain,
[106] In culturing of animal cells, the medium comprises various carbon
sources, nitrogen
sources and microelement ingredients. Examples of usable carbon sources may
comprise carbohydrates such as glucose, sucrose, lactose, fructose, maltose,
starch and
cellulose; fats such as soybean oil, sunflower oil, castor oil and coconut
oil; fat acids
such as palmitic acid, stearic acid and linoleic acid; alcohols such as
glycerol and
ethanol; and organic acids such as acetic acid, wherein such carbon sources
may be
used alone or in combination.
[107] Nitrogen sources, which may be used in the present invention, may
comprise, for
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
19
example, organic nitrogen sources such as peptone, yeast extract, meat juice,
malt
extract, corn steep liquor (CSL) and soybean-wheat, and inorganic nitrogen
sources
such as urea, ammonium sulfate, ammonium chloride, ammonium phosphate,
ammonium carbonate and ammonium nitrate, wherein such nitrogen sources may be
used alone or in combination. As a phosphorus source, the medium may comprise
potassium dihydrogen phosphate, dipotassium hydrogen phosphate and sodium-
containing salt corresponding thereto. Also, the medium may comprise metallic
salts
such as magnesium sulphate or iron sulfate. Besides, the medium may comprise
amino
acids, vitamins, appropriate precursors and the like.
[108] During culture, compounds such as ammonium hydroxide, potassium
hydroxide,
ammonia, phosphoric acid and sulfuric acid arc added to a culture product in
an ap-
propriate way to adjust a pH of the culture product. Also, during culture,
bubble
formation may be suppressed by using a defoaming agent such as fatty acid
polyglycol
ester. Also, oxygen or oxygen-containing gas (e.g., air) is injected into a
culture
product in order to maintain an aerobic state of the culture product. A
temperature of
the culture product is normally 20 C to 45 C, preferably 25 C to 40 C.
[109] The production method may further comprise a step of: (c) collecting
an anti-c-Met
antibody or an antigen binding fragment thereof expressed in the host cell. An
antibody
obtained by culturing the transformed host cell may be used in a non-purified
state, or
further used in a purified state with high purity by using various
conventional methods,
for example, dialysis, salt precipitation, chromatography and the like. Out of
those
methods, a method for using chromatography is most often used, wherein a type
and
order of column may be selected from ion-exchange chromatography, size
exclusion
chromatography, affinity chromatography, etc., according to antibody
characteristics,
culture method, etc.
11101
[111] Another aspect of the present invention provides a composition for
detecting c-Met,
comprising the antibody or the antigen binding fragment thereof, a kit for
detection
comprising the same, and a method for detecting c-Met antibody using the same.
[112] The composition for detecting c-Met and the kit comprising the same
form an
antigen-antibody complex in such a way that an antibody specifically binding
to c-Met
or an antigen binding fragment thereof comes into contact with a specimen
sample,
thus effectively detecting c-Met.
[113] As used herein, the term "antigen-antibody complex" means a conjugate
between c-
Met and an antibody for recognizing the same, in order to identify a tumor or
a cancer
cell of expressing c-Met in a sample.
[114] A method for quantifying c-Met antigen using a composition for
detecting c-Met and
using a kit comprising the same may be performed by identifying a formation of
an
CA 03061704 2019-10-28
WO 2018/221969
PCT/KR2018/006182
antigen-antibody complex, wherein identifying of the formation of an antigen-
antibody
complex may be performed by enzyme immunoassay (ELISA), western blotting, im-
munofluorescence, immunohistochemistry staining, flow cytometry, immunocyto-
chemistry, radioir.n.m.un.oassay
i.m.m.unopreci.pi.tation assay, im.m.unodiffusion
assay, complement fixation assay, a protein chip, etc., but not limited
thereto. The
ELISA comprises various ELISA methods such as a direct ELISA using a labeled
antibody for recognizing an antigen attached to a solid support; an indirect
ELISA
using a labeled -secondary antibody for recognizing a capture antibody in a
complex of
an antibody for recognizing an antigen attached to a solid support; a direct
sandwich
ELISA using another labeled antibody for recognizing an antigen in a complex
of an
antibody and an antigen attached to a solid support; an indirect sandwich
ELISA using
a labeled secondary antibody for reacting with another antibody for
recognizing an
antigen in a complex of an antibody and an antigen attached to a solid support
and then
recognizing such antibody, etc.
[115] As a label for qualitatively or quantitatively making a formation of
an antigen-
antibody complex measurable, there are an enzyme, a fluorescent material, a
ligand, a
luminous material, a microparticle, a redox molecule, radio isotope and the
like, but
not necessarily limited thereto. As the enzymes, there are p-glucuronidase, p-
D-glucosida.se, II-D-galactosidase, urea.se, peroxidase, alkaline phosphatase,
acetyl-
cholinesterase, glucose oxidase, hexokinase and GDPase, RNase, glucose oxidase
and.
luciferase, phosphofructokinase, phosphoenolpyruvate carboxylase, aspartate
amino-
transferase, phosphoenolpyruvate decarboxylase,13-lactamase, etc., but not
limited
thereto.
[116]
[117] Another aspect of the present invention provides a composition for
preventing or
treating cancer comprising the antibody or the antigen binding fragment
thereof of the
present invention.
[118] Yet another aspect of the present invention provides a method for
preventing or
treating cancer, comprising a step of administering a composition comprising
the
antibody or the antigen binding fragment thereof of the present invention to
an in-
dividual being in danger of developing cancer or having the same.
[119] Still yet another aspect of the present invention provides a use of
cancer treatment
and a use of preparing an anticancer drug, with regard to a composition
comprising the
antibody or the antigen binding fragment thereof of the present invention.
[120] The antibody and the antigen binding fragment thereof are such as
that described
above.
[121] The antibody or the antigen binding fragment thereof of the present
invention is
capable of binding to c-Met alone or a combination of c-Met and EGFR with high
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
21
affinity to inhibit a growth of cancer cells, such that the antibody alone or
in com-
bination with conventional pharmaceutically acceptable carriers can be used in
treatment, prevention and diagnosis of hyperproliferative diseases such as
cancer.
[122] In the present invention, the term "prevention" means all the acts,
which prevent or
delay diseases such as cancer, etc., from occurrence or recurrence by an
administration
of the composition of the present invention, and the term "treatment" means an
in-
hibition of development of diseases such as cancer, reduction of cancer, or
removal of
cancer.
[123] It may be provided that cancer, a disease applied to the composition
of the present
invention, is particularly lung cancer, stomach cancer, colon cancer, rectal
cancer,
triple negative breast cancer (TNBC), glioblastoma, pancreatic cancer, head
and neck
cancer, breast cancer, ovarian cancer, renal cancer, bladder cancer, prostate
cancer,
solenoma, salivary gland tumor or thyroid cancer, more particularly lung
cancer,
stomach cancer, colon cancer, rectal cancer, triple negative breast cancer
(TNBC),
glioblastoma, pancreatic cancer, head and neck cancer, breast cancer, and much
more
particularly lung cancer, stomach cancer, colon cancer, rectal cancer, triple
negative
breast cancer (TNBC), glioblastoma, pancreatic cancer, head and neck cancer,
but not
limited thereto. In the present invention, it may be provided that cancer is
the one
caused by, in particular, c-Met overexpression, amplification, mutation or
activation,
but not limited thereto. In other words, a composition comprising the antibody
or the
binding fragment thereof of the present invention has an inhibitory effect on
pro-
liferation of all the cancerous tumors irrespective of abnormal expression or
mutation
of c-Met, such that a pharmaceutical use of the present invention is not
limited by an
expression aspect or presence or absence of mutation of c-Met.
[124] The composition may be a form of a pharmaceutical composition, a
quasi-drug com-
position and a composition for health food.
[125] The composition of the present invention for preventing or treating
cancer may
further comprise a pharmaceutically acceptable carrier. The pharmaceutically
ac-
ceptable carrier is the one conventionally used in preparing a formulation,
comprising
lactose, dextrose, sucrose, sorbitol, mannitol, starch, acacia rubber, calcium
phosphate,
alginate, gelatin, calcium silicate, microcrystalline cellulose,
polyvinylpyrrolidone,
cellulose, water, syrup, methylcellulose, methyl hydroxybenzoate, propyl
hydroxy-
benzoate, talc, magnesium stearate, mineral oil and the like, but not limited
thereto.
Besides the ingredients, the composition of the present invention for
preventing or
treating cancer may further comprise lubricant, humectant, sweetening agent,
flavoring
agent, emulsifier, suspending agent, preservative, etc. Suitable
pharmaceutically ac-
ceptable carriers and preparations are described in detail in Remington's
Pharma-
ceutical Sciences (19th ed., 1995).
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
22
[126] The composition of the present invention may be administered orally
or parenterally
wherein a parenteral administration may be performed by intravenous infusion,
sub-
cutaneous infusion, intramuscular injection, intraperitoneal injection,
endothelial ad-
ministration, local administration, intranasal administration, intrapulmonary
admin-
istration, rectal administration and the like. During an oral administration,
protein or
peptide is digested, so an oral composition may be formulated in such a way
that its
active drug is coated or protected from decomposition in stomach. A
composition of
the present invention may be administered by a predetermined device through
which
an active substance may be moved into a target cell.
[127] A suitable dosage of the composition of the present invention for
preventing or
treating cancer varies depending on such factors as a formulation method, an
admin-
istration type, a patient' age, weight, gender, morbid condition, food,
administration
time, administration path, excretion speed and response sensitivity, wherein
an
ordinary skilled doctor may easily determine and prescribe an effective dose
for a
desired treatment or prevention. According to one exemplary embodiment of the
present invention, a daily dose of the pharmaceutical composition of the
present
invention may amount to 0.001-100 mg/kg or more. In the present
specifications, the
term "pharmaceutical effective dose" means an amount enough to treat, prevent
and
diagnose diseases such as cancer.
[128] The composition of the present invention for preventing or treating
cancer may be
formulated into a preparation by using pharmaceutically acceptable carriers
and/or ex-
pedients according to a method, which may be easily performed by those skilled
in the
art, to which the present invention pertains, such that such composition can
be
prepared in a mono-dose form or prepared by being inserted into a multi-dose
container. At this time, a dosage form may be in a form of solution in oil or
aqueous
medium, suspension or emulsion, or in a form of extract, powder, suppository,
powdered drug, granule, tablet or capsule, and may further comprise a
dispersing agent
or a stabilizer.
[129] The composition of the present invention may be administered as an
individual
therapeutic agent or administered in combination with other therapeutic
agents, and
may be administered sequentially or simultaneously with conventional
therapeutic
agents.
[130] The antibody or the antigen binding fragment thereof of the present
invention may be
used in treatment of cancer in such a way that it is injected in vivo in a
form of an
antibody-therapeutic agent (functional molecule) and a bispecific antibody-
therapeutic
agent (functional molecule) conjugate, which are such as that described above.
Ap-
propriate and desirable various conditions for targeting a drug to a specific
target site
are reported in documents, for example, Trouet et al., Plenum Press, New York
and
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
23
London, (1982) 19-30.
[131] According to one specific embodiment of the present invention, as a
result of
identifying an antitumor activity of the composition of the present invention
for
preventing or treating cancer in a xenograft mouse model, it was identified
that its
tumor activity inhibitory efficacy was remarkably excellent compared to the
control
group (FIGS. 12 and 13).
[132] c-Met, targeted by an antibody or an antigen binding fragment thereof
included in the
composition of the present invention is a molecule expressed on the surface of
cancer
cells, thus it may be used in the prevention, treatment and diagnosis of c-Met
related
cancer in such a way that a functional molecule further is bound to the
antibody of the
present invention or is administered in combination therewith. The functional
molecule
may comprise a chemical substance, radioactive nuclide, immunotherapeutic
agent,
cytokine, chemokine, toxin, biotic agent, enzyme inhibitor and the like.
[133] The functional molecule capable of coupling with the antibody or the
fragment
thereof of the present invention results in antibody drug-conjugates (ADC) may
be a
chemical substance, cytokine or chemokine, but not limited thereto. The
chemical
substance may be, for example, an anticancer drug, particularly, acivicin,
aclarubicin,
acodazole, acronycine, adozelesin, alanosine, aldesleukin, allopurinol sodium,
al-
tretamine, aminoglutethimide, amonafide, ampligen, amsacrine, androgens,
anguidine,
aphidicolin glycinate, asaley, asparaginase, 5-azacytidine, azathioprine,
bacillus
calmettc-guerin (BCG), Baker's antifol, beta-2-dioxythioguanosinc, bisantrenc
HC1,
bleomycin sulfate, bulsufan, buthionine sulfoximine, BWA773U82, BW502U83/HC1,
BW 7U85 mesylate, ceracemide, carbetimer, carboplatin, carmustine,
chlorambucil,
chloroquinoxalin-sulfonamide, chlorozotocin, chromomycin A3, cisplatin,
cladribine,
corticosteroid, corynebacterium pary um, CPT-11, crisnatol, cyclocytidine, cy-
clophosphamide, cytarabinc, cytembcna, dabis male ate, decarbazine,
dactinomycin,
daunorubicin HC1, deazauridine, dexrazoxane, dianhydro galactitol, diaziquone,
dibro-
modulcitol, didemnin B, diethyldithio carbamate, diglycoaldehyde, dihydro-
5-azacytidine, doxorubicin, echinomycin, dedatrexate, edelfosine, eflomithine,
Elliot's
solution, elsamitrucin, epirubicin, esorubicin, estramustine phosphate,
estrogen,
etanidazole, ethiophos, etoposide, fadrazole, fazarabine, fenretinide,
filgrastim, fi-
nasteride, flavone acetic acid, floxuridine, fludarabine phosphate, 5'-
fluorouracil,
Fluosolml, flutamide, gallium nitrate, gemcitabine, goserelin acetate,
hepsulfam, hex-
amethylene bisacetamide, homoharringtonine, hydrazine sulfate,
4-hydroxyandrostenedione, hydroxyurea, idarubicin HC1, ifosfamide, 4-
ipomeanole,
iproplatin, isotretinoin, leucovorin calcium, leuprolide acetate, levamisol,
liposomal
daunorubicin, liposome trapping doxorubicin, lomustine, lonidamine,
maytansine,
mechlorethamine hydrochloride, melphalan, menogaril, merbarone, 6-
mercaptopurine,
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
24
mesna, methanol extract of bacillus calmette-guerin, methotrexate, N-
methylformamide, mifepristone, mitoguazone, mitomyein-C, mitotane,
mitoxantrone
hydrochloride, monocyte/macrophage colony-stimulating factor, nabi.lone,
nafoxidine,
neocarzinostatin, octreotide acetate, ormaplatin, oxaliplatin, paclitaxel,
pala, pen-
tostatin, piperazinedione, pipobroman, pirarubicin, piritrexim, piroxantrone
hy-
drochloride, PIXY-321, plicamycin, porfimer sodium, prednimustine,
procarbazine,
progestins, pyrazofurin, razo.xane, sargramostim, semustine, spirogermanium,
spiromustine, streptonigrin, streptozocin, sulofenur, suramin sodium,
tamoxifen,
taxorere, tegafur, teniposide, terephthalamidine, teroxirone, thioguanine,
thiotepa,
thymidine injection, tiazofurin, topotecan, toremifene, tretinoin,
trifluoperazine hy-
drochloride, trifluridine, trimetexate, tumor necrosis factor (TNF), uracil
mustard,
vinblastin sulfate, vincristine sulfate, vindesine, vinorelbine, vinzolidine,
Yoshi 864,
zorubicin, cytosine arabinoside, etoposide, melphalan, taxotere, taxol and
mixtures
thereof, but not limited thereto.
[134]
Mode for the Invention
[135] Hereinafter, the present invention will be described in more detail
through Examples.
The following Examples are provided only for the purpose of illustrating the
present
invention in more detail. Thus, according to the purpose of the present
invention, it is
apparent to those skilled in the art that the Examples are not construed to
limit the
scope of the present invention.
[136]
[137] Example 1. Preparation of hybridoma cell for producing c-Met specific
antibody
and identification of tumor cell proliferation inhibitory activity thereof
[138]
[139] (1) Preparation and selection of hybridoma cell line for producing
monoclonal
antibody to c-Met protein
[140] A human c-Met Sema domain/Fe fusion protein (self-produced) was
intraperitoneally
injected as an antigen into a mouse, in order to obtain an immunized mouse
needed for
developing a hybridoma cell line through animal immunization. Screening was
performed through an ELISA analysis method using a human c-Met/His fusion
protein
as an antigen, in order to select a hybridoma cell specifically responding to
c-Met
protein only out of a hybridoma cell group.
[141]
[142] (2) c-Met antibody
[143] Light chain and heavy chain CDR amino acid sequences of a mouse
antibody
obtained from a selected hybridoma cell line are shown in Tables 1 and 2
respectively.
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
[144]
[145] [Table 1]
[146] Hybridoma light chain CDR
Antibody CDR 1 SEQ DDN0s1 CDR 2 SEQ ID NOs CDR 3
SEQ P) NOs
8C4 GASENIVGALN 1 ' GATNLAD 2 QNVILSSPYT 3
563 SATSSVRYMY 4 DTSNLAS 5 QQWSSYPRT 6
[147] [Table 2]
[148] Hybridoma heavy chain CDR
Antibody CDR 1 SEQ ID NOs CDR 2 SEQ ID NOs CDR 3 SEQ ID NOs
8C4 DYY1N 7 EIFPGSGNTHFSARFKG 8 GDYGFLY 9
5G3 DYTLH 10 YINPYSGYTNYNQUIP 11 GINDY 12
[149]
[150] (3) In vitro tumor cell proliferation inhibitory activity of
hybridoma C-Met
antibody
[151] With regard to a c-Met specific mouse antibody obtained film a
hybridoma cell line
as well as a chimera antibody prepared by fusing the antibody with human heavy
chain
and light chain constant regions, a tumor cell proliferation inhibitory
activity was
tested in a human glioblastoma cell line U-87 MG and a human stomach cancer
cell
line MKN45.
[152]
[153] Particularly, the U-87 MG cells (ATCC, #HTB14) were diluted in a
culture medium
EMEM (ATCC, #30-2003) containing 10% (v/v) FBS, 100 U / 500 ml penicillin and
100 fig / 500 ml streptomycin (Invitrogen, #15140-122), after which resulting
cells
were added by 100 pi into each well of a 96-well plate at a concentration of
2.5 X 103
cells, such that the plate was cultured under 37 C, 95% RH and 5% (v/v) CO2
conditions for 18 - 24 hours. The cell culture medium was removed from each
well,
after which an EMEM medium containing 2% (v/v) FBS was added by 100 pi into
each well, and an antibody prepared at 2X of a final concentration (100 nM)
was con-
tinuously diluted at a ratio of 1/10, such that resulting cells were added by
100 id, into
each well at six concentrations (i.e., 200 nM, 20 nM, 2 nM, 200 pM, 20 pM and
2 pM)
for each antibody. Then, the plate was cultured for 5 days under 37 C, 95% RH
and
5% (v/v) CO2 conditions, after which resulting cells were fixed with 10% TCA
(Trichloroacetic acid; Sigma, #T0699) solution on a final day. The resulting
fixed cells
were dyed for 25 minutes in such a way that 80 ge of 0.4% SRB (sulforhodamine
B)
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
26
solution was added into each well, after which resulting cells were washed 5
times
with 1% acetic acid solution. Then, 150 ,u.f of 10 rriM Tris solution was
inserted into
each well of a dried plate to dissolve SRB dye, after which its optical
density was
measured at a wavelength of 540nm by using a microplate reader.
[154] Also, MKN45 (#JCRB0254) cell lines were diluted in an RPMI-1640
medium
(Gibco, #A10491) containing 10% (v/v) FBS, after which the resulting cell
lines were
divided by 2.5 X 103 into each well of a 96-well plate, such that the
resulting plate was
cultured overnight under 37 C, 5% CO2 conditions. Then, the medium of each
well of
the plate was replaced with 100 jii of an RPMI-1640 medium containing 1% (v/v)
PBS, after which a test antibody was sequentially diluted at a ratio of 1/10
(i.e., 100
nM, 10 nM, 1 nM, 100 pM, 10 pM and 1 pM) to reach 1 pM at a final
concentration of
100 nM, such that the resulting antibody was added by 100 itt into each well.
Then,
the plate was cultured for 5 days under 37 C, 5% CO2 conditions, after which
the
medium was removed therefrom, such that a TCA solution was inserted by 200 jd
into
each well to fix cells. As shown in the test on the U87 MG cell, the cells of
the plate
were dyed according to a conventional SRB colorimetric assay method, after
which an
optical density of each well was measured at a wavelength of 540 nm by using a
mi-
croplate reader. Results of the U87 MG and MICN45 cell lines are shown in
Table 3
and FIG. 1.
[155]
[156] [Table 3]
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
27
[157] Results of in vitro test on tumor cell proliferation inhibitory
activity of hybridoma c-Met
antibody
MKN45
U-87 MG
(Gastric cancer, c-Met
(GB, HGF autocrine)
amplified)
1050 (nM)
IC50 (nM)
LY2875358
> 100 0.34
(Eli Lilly)
OA-505
>100 >100
(Genentech)
hybridoma 8C4 17.5 9.78
hybridoma 5G3 > 100 0.32
8C4 chimera 4401 32.4 > 100
804 chimera IgG2 > 100 12.92
5G3 chimera IgG2 > 100 0.41
[158] As seen in Table 3 and FIG. 1 above, the anti-c-Met 8C4, 563
antibodies and
chimera antibodies thereof of the present invention all have a tumor cell
proliferation
inhibitory activity, which is equal to or more excellent than the known c-Met
an-
tibodies LY2875358 and 0A-5D5 (control group). Thus, the 8C4, 5G3 antibodies
and
mutants thereof such as chimera antibodies, humanized antibodies and affinity-
optimized antibodies to antigen of the present invention may be very valuably
used in
preventing or treating c-Met related cancer.
[159] Specific consensus sequences for light chain and heavy chain variable
regions of the
8C4, 5G3 antibodies of the present invention are shown in the following Table
4.
[160]
[161] [Table 4]
CA 03061704 2010-10-20
WO 2018/221969 PCT/KR2018/006182
28
[1621 Consensus SEQ ID NOs for light chain and heavy chain variable regions
of 8C4, 5G3
antibodies
Consensus amino acids sequence Consensus nucleotides sequence
light chain heavy chain light chain heavy
chain
. . gaggttcagctgcagca
gatattctgatgaccca
gtctggagetgagctgg
gtctccagatcactgt
cgaggcceggggettca
ctgcatctgtgggagaa
gtgaagctgtcctgcaa
actgtcaccatcacatg
ggcttctggctacacct
tggagcaagtgagaata
tcagtgactactatata
macggtgctttaaat
DILQSPASLSASVGE EVQLQQSGAELARPGAS ggtatcagcgaaaaca aactgggtgaagcaggg
MT
gactggacagggccttg
TVTITCGASENIYGALN VIISCKASGYIFSDYYI rggaaaatacctcagc
agtggattggagagatt
WYQRKQGKSPQLLIYGA NWVKQGTGQGLEWIGEI tcctgatctatggtgca
tttcctggaagtggaaa
INLADGESRFSGSGSG FPGSWITIFSARFKGKA kccaacttggcagatgg
BC4 lactcacttcagtgcga
RQFSLKITSLHPDDVAT ILTADKSSSTAYMQLSS Icatgtcatcgaggttca
ggttcawgggcaaggcc
YYCQNVLSSPYTF(X;GT LTSIDSAVYFCAGGDYG gtggcagtgggtctggt
acactgactgcagacaa
KLEIK (SEQ ID NO:FLYWGROLVITSA lagacagttactctcaa
atcctccagcacagcct
13) (SEQ ID NO: 15) atcactagcctgcatc
acatgcagctcagcagc
tgacgatgttgcaacg
ctgacatctacggactc
lattactgtcaaaatgt
tgcagtctatttctgtg
gctaagtagtccgtaca
ccangtgactacggg
cgttcggaggggggacc
tttctttactggggccg
aageggaaatcaaa
. agggactctggtcactg
(SEQ ID NO: 17)
,
1 tctctgca (SEQ ID
'
1 __ =
=
. _ ..._
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
29
,
[163] 0: 19)
.............. ... .. ... ..
Icagggccagctgcagca
caaattgttacaccca gtaggggctgaaagg
= tct ccagcaat catgt caagacctggggcctca
ctgcataccaggggag tgaagatgtcagcaa
aaggtcaccalgacag gataggctacacct
cagtgccacctcaagtg ttactgactacacgctg
..
tacgttacatgtactgg Cactgggtaaaacagag
IVLIQSPAIMSASPGE . GQLQQSGAELARPGAS t accagcagaagccagg cctggacagggt ctgg
VFMTCSATSSVRYMYW 1 SCKASGYTFTDYTL a t cct cceccagactec .a a tggat tggat acat t
b
' 'QQKPGSSPRI,I, I YDTS HWYKQRPGQGLEW I GY I 1 ga 1. t t a Lgacacattc 'at CCL
tacagtatta
LASGVPGRFSGSGSGT NPYSGYTNYNQKFKDKA aacctggatctggagt tactaattacaatcaga
PG3
SLTISRLEAEDAATY ILTADKSSSTAYMQLSG ccuggtcgcttcagcg 'oattcaaggacaaggcc
CQQWSSYPRTFGGGTK LTSEDSAVFYCARGHMD Tcagtgggtagggacc acattgactgcagacaa
EIK (SEQ ID NO: WGQGTSVTVSS (S .itctaactactcacaat atectccagcacagect
,
14) ID NO: 16) cagccgaaggaggag 'acatgcaaagagcggc
aagatgagccacttat agacatagaagacte
t actgccagcagtggag tgcagtct Mat tgtg
iagtLacccacggacgt Caagaggacatatggac
I cggt ggaggc a c caag t aciggggt caaggaac
ctggaaat caaa (SEQc t cagt caccgt c t cct
:
:
Ill NO: 18) 'ca (SEQ ID NO:
20)
. ________________________________________________ .J.... __
[164] Example 2. Preparation of humanized antibody of 8C4 antibody and
identi-
fication of in vitro tumor cell proliferation inhibitory activity thereot
[165]
[166] As one example, the mouse antibody 8C4 was humanized and an in vitro
tumor cell
proliferation inhibitory activity thereof was identified, in order to further
identify an
effect of an antibody prepared in the present invention,
[167]
[168] For a humanized design of 8C4 antibody heavy chains, a human germline
gene
having a high homology with a gene in a heavy chain variable region of a mouse
antibody 8C4 was analyzed first through Ig Blast
(http://www.ncbi.nlm.nih.gov/igblast/). In result, it was identified that
IGHV3-23 had
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
48% homology with the 8C4 antibody in an amino acid level, and also identified
that
IGHV3-11 had 46% homology with the 8C4 antibody in an amino acid level.
[169] The CDR-H1, CDR-H2 and CDR-H3 of the mouse antibody 8C4 was defined
by
Kabat numbering, and hu8C4-1 was prepared in such a way that the CDR portion
of
the mouse antibody 8C4 was represented by be introduced into a framework of
IGHV3-23. At this time, no. 48 (V¨>I), no. 49 (S¨>G), no. 71 (R¨>A), no. 73
(N¨+K),
no. 78 (L¨+A) and no. 94 (K-0G) amino acids were back-mutated into an original
amino acid sequence of the mouse antibody 8C4 to finally build a heavy chain
of
hu8C4-1. In case of hu8C4-2, the CDR portion of the mouse antibody 8C4 was rep-
resented by be introduced into a framework of IGHV3-11, and no. 48 (V¨>I), no.
49
(S¨>G), no. 71 (R¨>A), no, 73 (N¨>K), no. 78 (L¨>A) and no. 94 (R¨>G) amino
acids
were back-mutated into an original amino acid sequence of the mouse antibody
8C4 to
finally build a heavy chain of hu8C4-2.
[170] Even in case of a light chain of 13C4 antibody, for a humanized
design, a human
germline gene having a high homology with a gene in a light chain variable
region of
the mouse antibody 8C4 was analyzed through Ig Blast
(http://www.ncbi.nlm.nih.gov/igblast/). In result, it was identified that
IGKV1-27 had
65.3% homology with the 8C4 antibody in an amino acid level, and that IGKV1-33
had 64.2% homology with the 8C4 antibody in an amino acid level.
[171] The CDR-L1, CDR-L2 and CDR-L3 of the mouse antibody 8C4 were defined
by
Kabat numbering, and the CRD portion of thc mouse antibody 8C4 was represented
by
be introduced into a framework of IGKV1-33 and a framework of IGKV1-27, thus
preparing hu8C4-1 and hu8C4-2 respectively. At this time, amino acid no. 69
(T¨>R)
of both and hu8C4-2 were back-mutated into an original amino acid sequence of
the
mouse antibody 8C4.
1172,J
[173] The 8C4 humanized antibody was expressed in a 293T cell by using a
pCLS05
vector (Korea Patent Registration No. 10-1420274). With regard to such
obtained
humanized antibodies in a form of IgGl, it was identified whether or not they
had a
tumor cell proliferation inhibitory activity in U-87 MG, a human glioblastoma
cell line,
by the same method as shown in Example 1 above,
[174] In result, it was identified that the IC50 values of hu8C4-1 and
hu8C4-2 amounted to
30 nM and 24.6 nM respectively, thus indicating a similar level of anticancer
activity
to that of a chimera 8C4 antibody (IC50, 32.4 nM).
[175] Specific consensus sequences for light chain and heavy chain variable
regions of the
hu8C4-1 and hu8C4-2 humanized antibodies are shown in Table 5.
[176]
[177] [Table 5]
Ch 03061704 2010-10-20
WO 2018/221969
PCT/ICR2018/006182
31
[178] Consensus SEQ ID NOs for light chain and heavy chain variable regions
of hu8C4-1 and hu8C4-
2 humanized antibodies
Consensus mei no acids sequence 1 Consensus nue 1 eot ides sequence
light chain heavy chain light chain heavy chain
a tat ccagat gacc gaggt tcagt tagtg
agtct cccagcagt gaat ccggaggagga
tttccgcttctgtg ctggtgcagcctggt
kgtgatagggtgacg ggaagt t tgaggctg
taact tgcggagca tcatgcgcagccagt
gtgagaatatttac ggctacaccttcagt
g i
tgctttaaattgg gactactatataaac
DIQIITC/SPSSLSASV EVQLVESGGGLIMPG t [ accagcagaagcct tgggt aagacaggct
GDRVTITCGASENIY GSLRLSCAASGYTFS g gaaagctccaaag cccggaaaagggctg
GALNWYQQ1(PGICAPK D'YYINIIVRQAPGKGL Ictgct gatctatggt gagtggattggagag
LLIYGATNLADGVPS EVIIGEIFPGSGNTIF gcaaccaacttggca at t t tt cct ggaagt
hu8C4-1
RFSGSGSGRDFIFTI SARFKGRAILSADILS ga tggcgt ccc t agc gga a a t act cacttc
I
SSLQPEDIA'fYYCQN ENTAYLQMNSLRAEDg ttcagcggcagt agtgcgaggttcaag
VLSSPYTFGQGIIIVE TAVYYCAGGDYGFLYgg
aagcggcagagac ggccgagccaccctc
1K (SEQ ID NO : WGQGTLVTVSS It tcact
ttcacaatc tcagcagacaaaagc
21) (SEQ ID NO:
23) tcctccctgcaaccc aagaataccgcct at
gaggacat tgcaacc ctgcagatgaatagc
t act a t tgtcaaaat ct t cgcgcagaagat
gtgctaagtagtccg ac t gccgtgt at t ac
t acacgt t tggccag tgtgccgggggtgac
gaaccaaggt tgaa tacgggtttctttac
ttaaa (SEQ ID ggggacagggcacc
0: 25)
ttggtgacagtctct
Ch 03061704 2010-10-20
WO 2018/221969 PCT/ICR2018/006182
32
[179] tct (SEQ ID
NO:
27)
caggt tcagt tagtg
gacat ccagatgacc gaat ccggaggagga
cagtct ccatcct cc ctggtgaagcctggt
ctgtctgcatctgt a ggaagt ttgaggctg
ggagacagagtcacc t cat gcgcagccagt
at cact tgcggagca ggct acacct tcagt
agtgagaatatttac gactactatataaac
ggtgct t taaat tgg tggat cagacaggct
DIQPITRSPSSISASVQVQLVESGGGLVKPG t at cagcagaaacca cccggaaaagggctg
GDRVTI TCGASEN I Y GSLRLSCAASGYNS gggaaagt t cc t aag gagtggat t ggagag
GALNWYQRKPOORK DYYINWIRRAPGKGL ct cctgatctatggt at t t t tcctggaagt
LLIYGATNLADGVPS EWIGEIFPGSGNTIF gcaaccaact tggca ggaaatactcact tc
hu8C4-2
RFSGSGSGRDFTLT I SARFKGRATISADKA gat ggggt ccc at ct agt gcgaggt t caag
SSUIPEDVATYYCRN KNSAYLQIINSLRAED cggt t cagtggcag t ggccgagccacc at c
VLSSPYTFGQGTKVE TAVYYCAGGDYGFLY ggatctgggcgagat tcagcagacaaagcg
IK (SEQ ID NO: WGRGILVTVSS tt cact
ctcaccatc aagaatagcgcct at
22) (SEQ ID NO:
24) agcagcctgcagcct ctgcagatgaatagc
gaagatgt tgcaact ct t cgcgcagaagat
t at tactgtcaaaat actgccgtgt at t ac
gtgct aagtagtccg tgtgccgggggtgac
t acacgt t tggccag tacgggt t t ct t tac
ggaaccaaggt tgaa tggggacagggcacc
at taaa (SEQ IDttggtgacagtctct
NO: 26) tct (SEQ ID
NO:
28)
[180] Example 3. Preparation of humanized antibody of 5G3 antibody and
identi-
fication of in vitro tumor cell proliferation inhibitory activity thereof
[181]
[182] Then, the mouse antibody 5G3 of the present invention was humanized
to identify an
in vitro tumor cell proliferation inhibitory activity themof.
[183]
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
33
[184] Particularly, for a heavy chain design of hu5G3-1, a human germline
gene having a
highest homology with a gene in a heavy chain variable region of the mouse
antibody
5G3 was analyzed first through Ig Blast
(http://www.ncbi.nlm.nih.gov/igblast/). In
result, it was identified that IGHV1-46 had 67.3% homology with the 5G3
antibody in
an amino acid level. The CDR-H1, CDR-H2 and CDR-H3 of the mouse antibody 5G3
were defined by Kabat numbering, and the CRD portion of the mouse antibody 5G3
was represented by be introduced into a framework of IGHV1-46. At this time,
amino
acid no. 48 (M¨>I), no. 69 (M¨)L), no. 71 (R¨*A), no. 73 (T¨>K) and no. 78 (V-
0A)
were back-mutated into an original amino acid sequence of the mouse antibody
5G3.
By doing so, a heavy chain of hu5G3-1 was built.
[185] For a light chain of hu5G3-1, CDR-grafting was performed in IGKV3-20
gene
having 63.5% homology with the 5G3 antibody, and amino acid no. 43 (AS), no.
60
(D¨>A) and no. 71 (F¨>N) were back-mutated to build a light chain of hu5G3-1.
[186] Also, to design a heavy chain of hu5G3-2, the CDR-H1, CDR-H2 and CDR-
H3 of
the mouse antibody 5G3 defined by Kabat numbering were introduced by using VH3
subtype, which was conventionally known to be most stable. At this time, amino
acid
no. 67 (F¨>A), no. 69 (I¨>L), no. 73 (T¨>K), no. 90 (Y¨>F) and no. 94 (T¨>R)
were
back-mutated into an original amino acid sequence of the mouse antibody 5G3.
By
doing so, a heavy chain of hu5G3-2 was built.
[187] For a light chain of hu5G3-2, CDR-grafting was performed in IGVK ifi
gene, which
was known to stably form a structure with VH3 subtype, and back-mutation was
not
performed.
[188]
[189] The 5G3 humanized antibody was expressed in a 293T cell by using a
pCLS05
vector (Korea Patent Registration No. 10-1420274). With regard to such
obtained
humanized antibodies in a form of 1gG2, it was identified whether or not they
had a
tumor cell proliferation inhibitory activity in MKN45, a human stomach cancer
cell
line, by the same method as shown in Example 1 above.
[190] In result, it was identified that the IC50 values of hu5G3-1 and
hu5G3-2 amounted to
0.52 nM and 0.5 nM respectively, thus indicating a similar level of anticancer
activity
to that of a chimera 5G3 antibody (IC50. 0.41 nM).
[191]
[192] Consensus sequences for light chain and heavy chain variable regions
of the hu5G3-1
and hu5G3-2 humanized antibodies are shown in Table 6.
[193]
[194] [Table 6]
CA 03061704 2019-10-28
WO 2018/221969
PCT/KR2018/006182
34
[195] Consensus
SEQ ID NOs for light chain and heavy chain variable regions of hu5G3 - 1 and
hu5G3-2 humanized antibodies
Consensus amino acids sequence Consensus nucleotides sequence
light chain heavy chain light chain heavy chain
gaaat tgtgt tgaca caggtgcagctggtg
cagtctccagccacc cagtctggggctgag
ctgtct t tgtctcca gtgaagaagcctggg
ggggaaagagccacc gcctcagtgaaggt t
ct ct cc t gcagt gcc tcctgcaaggcatct
acct caagtgt acgt ggatacacct t cacc
t acatgtactggtac gactacacgctgcac
E I VLTQSPATLSLSP QVQLVQSGABIG(PG cagcagaaacctggc tg,ggtgcgacaggcc
GERAILSCSATSSYR ASYKVS1ASGY1FT cagt ct cccaggct c cctggacaagggct t
YMYRYQQI(PGQSPRL DYTLIDYYRQAPGQII, ctcatctatgacaca gagtggat aggatac
LI YDTSNLASGIPAR EWIGYINPYSGYTNY t ccaacctggct t ct at t aa t cc t t acagt
hu5G3 -1
FSGSGSGIDNILTISNOTKDRVTLTADKSggcatcccagcaaggggttatactaattac
RLEPEDFAVYYCQQ11 TSTAYMELSSLRSED ttcagtggcagtggg aatcagaaat tcaag
SSYWIYGGGTIKVEI TAVYYCARGHNDYWG t ctgggacagacaac gacagagt cacct tg
K (SEQ ID NO: QGTLVTYSS (SEQactctcaccatcagc accgcagacaaatcc
29) ID NO: 31)
agactggagcctgaa acgagcacagcctac
gattttgcagtt tat atggagctgagcagc
t actgtcagcagtgg ctgagatctgaggac
agtagt tacccacgg acggccgtgt at tac
acgt tcggcggaggg tgtgctagaggacat
accaaggtggagatc a tggact ac tggggc
aaa (SEQ ID NO: caaggaaccctggtc
33) accgt ct cc t c a
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
[196] (SEQ ID NO: 35)
1 gaagt ccaacttgtg
gacat cc agatgac t
gagt caggaggcggg
cagagt ccctct tct
ctcgtgcagccaggc
ctgtctgcctcagtg
ggatcattgcgactt
ggagat cgggt c ac a
tcttgtgctgcctca
atcacatgttcagca
gggt acacct tcact
I acaagct cagtgcga
gat t atacct tgcat
tacatgtattggtac
tgswt t cgccaagca
I OITQSPSSLSASV EVQLVESGGGLVQPG cagcagaagccaggc
cccggtaagggtct c
GDRVTITCSATSSVR GSLRLSCAASGYTET aaagccccaaagctg
ga a t gggt aggatac
YMYWYQQKPGKARKL DYTLHINVRQARGKGL ct gat ct atgacac a
at t aat ccat acagc
L I YDTSNLASGVPSR PA iliY INPYSGYTNY t ctaat ctggccagc
ggc t acaccaactac
hu5G3-2 FSGSGSG1 WILT' S NQKEKDRATLSADICS ggcgt cc cat ct cgc
aaccagaaattcaaa
'LQPEDF'ATYYCQQW KNTAYLQMNSLRAED t t ct caggctccgga
gacagggctaccct t
SYPRTFGQGTKVE I TAIFYCARGINDYWG agcggt actgat t t t
agtgccgacaagtct
(SEQ ID NO:QGTLVTVSS (SEQgccctgactatttct
aagaacaccgcctac
0) ID NO: 32) tcettgcagcctgag
cttcagatgaactcc
gacttczcaacctat
cttagagccgaggat
t at tgccagcagtgg
actgctgtgttttat
t ct agct accctcgc
tgcgctaggggt cat
acattcggccaggga
atggactactgggga
accaaggt cgaaat t
caggggaccttggtg
aaa (SEQ ID NO:
actgtgtcttcc
34)
(SEQ ID NO: 36)
[197] Example 4. Preparation of hinge mutant and testing of tumor cell
proliferation
inhibitory activity thereof
[198]
[199] Then, a lest on tumor cell proliferation inhibitory activity was
performed according
to a hinge sequence of human IgG1 heavy chain constant region.
[200]
[201] First of all, a hinge of the human IgG1 heavy chain constant region
had an amino
acid sequence of "EPKSCDKTHTCPPCP (SEQ ID NO: 37)," which was substituted to
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
36
obtain a hinge region mutant having an amino acid sequence of SEQ. ID NO: 38
to
SEQ ID NO: 44.
[202] The resulting mutants were respectively cloned into a vector
comprising the heavy
chain variable region of h.u8C4-1, hu8C4-2 hum.anized antibodies prepared i.n.
Example
2 above. An in vitro tumor cell proliferation inhibitory activity according to
a hinge
sequence was identified in U-87 MG by the same method as shown in Example 1
above.
[203] Also, an effect of the 8C4 humanized antibody was analyzed as follows
with regard
to non-small cell lung cancer cell line NCI.-H1993 (ATCC, #CRL-5909). The NCI-
H1993 cell lines were diluted in an RPMI-1640 medium (Gibco, #A10491)
containing
10% (v/v) FBS, after which resulting cell lines were divided by 3.0 X 103 into
each
well of a 96-well plate, such that the resulting plate was cultured overnight
under 37 C,
5% CO2 conditions. After that, the medium of each well of the plate was
replaced with
100 pl of an RI MI-1640 medium containing 2% (v/v) FBS, after which a test
antibody
was sequentially diluted at a ratio of 1/10 (i.e., 100 nM, 10 nM, 1 nM, 100
pM, 10 pM
and 1 pM) to reach 0.001 nM at a final concentration of 100 nM, such that the
resulting
antibody was added by 100 Id into each well. Then, the plate was cultured for
5 days
under 37 C, 5% CO2 conditions, after which the medium was removed therefrom,
such
that a TCA solution (Sigma, #T0699) was inserted by 200 a into each well to
fix the
cells. Also, the cells of the plate were dyed according to a conventional SRB
col-
orimetric assay method, after which an optical density of each well was
measured at a
wavelength of 540 nm by using a microplate reader.
[204] Results of hu8C4-1 in U-87 MG and NCI-H1993 (ATCC, #CRL-5909) are
shown in
Table 7.
[205]
[206] [Table 7]
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
37
[207] Hinge region mutant sequences and results of in vitro test on tumor
cell proliferation inhibitory
activity
NCI-111993
U-87 MG
SEQ SEQ (NSCLC, c-
(GBM, HGF
ID Amino acids ID Nucleotides Met
autocrine)
NOs NOs amplified)
(IC,50 nM)
(IC50 nil)
EPLSCDKTIITCPP gagcccaaatcttgtgacaaaactcacacatgcc
37 45 12.6 > 100
CP caccgtgccca
gagcgaaaatgttgtgtcgagtgcccaccgtgcc
38 ERKCCVECPPCP 146 31.0 0.30
ca
39 ECCVECPPCP 47 gagtgagtgtegagtgcccaccgtgccca 57.3 > 100
40 ERKCCCPPCP 48 gagcgaaaatgttgagcccaccgtgecca 37.6 0.23
41 ECCCPPCP 49 gagtgttgttgcccaccgtgccca 25.3 > 100
42 EKCCVECPPCP 50 gagaaatgagIgtcgagtgcccaccgtgccca 31.4 0.48
43 ERKCCVCPPCP 51 gagcgaaaatgttgtgtctgcccaccgtgccca 30.8 0.47
44 EKCCVCPPCP 52 gagaaatgttegtctgcccaccgtgccca 75.9 0.38
[208] As seen in Table 7, there is some difference in the tumor cell
proliferation inhibitory
activity of the ]u8C4 antibody according to a difference of hinge sequence,
but it was
identified that such antibody effectively inhibited a proliferation of most
tumor cells.
Accordingly, hereinafter an IgGi humanized antibody representatively having a
hinge
region of SEQ ID NO: 38 in hu8C4-1 was named as hu8C4, and an affinity-
optimized
antibody thereto was prepared to identify an effect thereof.
[209]
[210] Example 5. Preparation of affinitv-optimized antibody of hu8C4 and
identi-
fication of in vitro tumor cell proliferation inhibitory activity thereof
[211]
[212] To prepare an affinity-optimized antibody of hu8C4, a phage-displayed
scFv library
was first prepared by using a phagemid vector displayed in a combined form of
scFv
and pill, wherein a schematic structure of the vector is illustrated in FIG.
2. The
phagemid vector comprises a scFv fragment of an antibody under a control of an
IPTG-inductive lac promotor, wherein a linker sequence used was GGGGS GGGGS
GGGGS (SEQ. No. 53).
[213] Then, a mutation-inducing oligonucleotide having an NNK codon was
used to
introduce variety into the heavy chain and light chain CDR domain of hu8C4. Ac-
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
38
cordingly, a hu8C4 scFv library with a fusion of His, HA and pIII was
prepared, after
which a human c-Met specific antibody was selected from the prepared antibody
library.
[214] Particularly, a competitive selection method was used to select an.
antibody with an
improved affinity. A human c-Met antigen was bound according to the
manufacturer
guidelines in Dynabeads M-280 (Thermo Fisher Scientific, 11205D). A bead with
an
antigen binding thereto was blocked for 2 hours by a superblock T:ris buffered
saline
(TBS, Pierce), Also recombinant phage grew overnight at 37 C, and then
recombinant
phage was centrifuged and a phage of its supernatant was blocked with
superblock
TBS, 0.05% Tween 20 for 2 hours. Then, the bead was washed with PBS containing
0.05% Twin 20. A blocked phage solution was added into the washed bead, after
which the resulting bead was incubated in a rotator for 2 hours for phage
binding, such
that the resulting bead was washed with PBS containing 0.05% Twin 20. Then, a
human c-Met antigen was added into PBS 1 ml containing 0.05% Twin 20, after
which
the resulting antigen was incubated in a rotator for 24 hours (Rouet R et al.
(2012) Nat
Protoc. 7:364-373). After that, the phage binding to the bead was eluted with
100 mM
triethanolamine for 5 minutes, after which an eluent was neutralized with 0.5
M Tris/
Cl (pH 7.2). An eluted phage neutralization liquid was infected with E. coil
TG1.
[215] An individual clone selected through the experiment grew in a 96-well
format of
2xYT broth 200 jug with added carbenicillin and ampicillin, after which a
culture su-
pernatant thereof was directly used for 'ELISA to select a phage-displayed say
binding
to a plate coated with target protein. Amino acid sequences of light chain and
heavy
chain CDR regions of a detected antibody are shown in Tables 8 and 9, and the
repre-
sentative amino acid sequences of light chain and heavy chain variable regions
of an
affinity-optimized antibody are shown in Table 10.
[216]
[217] [Table 8]
Ch 03061704 2010-10-29
WO 2018/221969 PCT/KR2018/006182
39
[218] List of heavy chain CDR sequences
List of heavy chain CDR sequences .
CDR1 SEQ ID NOs CDR2 SEQ ID Wis CDR3
SEQ ID NOs
AH01 DYYIN 7 EIDPGSGNTHFSARFKG 54 GDYGFLY 9
A302 DYYIN 7 EIEPGSGNTHFSARFIG 55 GDYGFLY 9
I003 DYYIN 7 ETWPGSCNTHFSARFIG 56 GDYGFLY __ 9
_. _
A104 DYYIN 7 EIYP5SG1T1FSARF1cG ¨ 57 GDYGFLY 9
A305 DYYIN 7 EIFPGWGNTHFSARFKG 58 GDYGFLY 9
AH06 DYYIN 7 EIFPGYGNTHESARFKG 59 , GDYGFLY 9
AH07 DYYIN 7 EIFPGSGYTHFSARFKG 60 GDYGFLY 9
A1108 DYYIN 7 EIFPGSGNTWFSARFKG 61 GDYGFLY 9
A939 DYYIN 7 EIFPGSWYFSARFKG 92 GDYGFLY __ 9
AH12 DYYIN 7 EIFPGWGINFSARFKG 63 GDYGFLY 9
A813 DYYIN 7 EIFPGSGNTHFSARFKG 8 QDYGFLY 64
AH14 DYYIN 7 EIFPGSMIFSARFKG 8 ELIGFLY 66
AH15 DYYIN 7 EIFPGSGNTHFSARFKG 8 IMPLY es
AH16 DYYIN 7 EIFPGSGNTHFSARFIG 8 NDYGFLY 67
AH17 DYYIN 7 EIFPGSGMFSARFIG 8 VELGFLY 68
AH18 DYYIN 7 EIFPGSGNIHFSARFKG 8 , FE1MYL 69
AH19 DYYIN , 7 EIFPGSGNTHFSARFKG 8 GEYGYQN 70
AH20 DYYIN 7 EIFPGSGNTHFSARFKG 8 WEYGLSN 71
AH21 DYYIN 7 ElFPWrSDHFSARFKG 72 GDYGFLY 9
AH22 DYYIN 7 EIFPGSGNTHFSAWNGT 73 GDYGFLY 9
A823 DYYIN 7 EIFPGSGNESVSFRFKG 74 GDYGFLY 9
A1124 DYYIN 7 EIFPGSGNSAVISRFKG 75 , GDYGFLY 9
AH25 DYYIN 7 EIERGSGMTVVIRFIG 76 GDYGFLY 9
AH26 DYYIN 7 ETFPGSCALSMHGRING 77 GDYGFLY 9
A1127 DYYIN 7 EIFPGW11111PVPRFKG 78 GDYGFLY 9
AH28 DYYIN 7 EIFPGSGNPFLT1RFIG 79 GDYGFLY 9
AH29 DYYIN 7 EIFPGSGNSHP/SRFIG 80 GDYGFLY 9
AH30 DYYIN 7 EIFPGSGNLSGIRSFKG 81 GDYGFLY 9
AH31 DYYIN 7 EIFPGSDIFFHGKRFKG 82 GDYGFLY 9
AH32 DYYIN 7 EIFPGSGNPRLGARFKG 83 GDYGFLY 9
AH33 DYYIN 7 EIFPGSGNVSQVERFEG 84 GDYGFLY 9
AH34 , DYYIN 7 EIFPGSGNFHGASRFKG 85 GDYGFLY 9
AH35 , DYYIN 7 EIFPGSGNVVGGYRFKG 86 GDYGFLY 9
AH36 DYYIN 7 EIFPGSGtIPMYDERFIG 87 GDYGFLY 9
A1137 DYYIN 7 EIFPGSGNADLT1RFIG 88 GDYGFLY 9
AII38 DYYIN 7 EFPGSGETNLYRKG 89 GDYGFLY 9
A839 DYYIN 7 EIFPGSGNLDIPPRFKG 90 GDYGFLY , 9
AH40 DYYIN 7 EIFPGSGNTRFSSAPLP 91 GDYGFLY 9
A1141 DYYIN 7 EIFPGSGNTHFSSEFYS 92 GDYGFLY 9
A1142 DYYIN 7 EIFPGSGNINFSMSESF 93 GDYGFLY 9
A1143 DYYIN 7 EIFPGSG.1111FSOGSRN 94 GDYGFLY 9
A944 DYYIN 7 RTFINAGNUFSSSIISR 9S CDVGFIY 0
AH45 DYYIN 7 EIFPGSGNTHFSRSYSG 96 GDYGFLY 9
AH46 DYYIN 7 EIFPGSGNTHFSGLSEV 97 GDYGFLY 9
AH47 DYYIN 7 EIFPGSMTHFSHYWAS 98 GDYGFLY 9
A1148 DYYIN 7 EIFPGSGNTHFSIGLIQ 99 GDYGFLY 9
AH49 DYYIN 7 EIFPGSGMIFSRHRLH 100 GDYGFLY 9
A1160 DYYIN 7 EIFPGSWIMFSVITSM 101 GDYGFLY 9
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
[219]
List of heavy chain CDR sequences (continue)
CDR1 SEQ ID N')..; CDR2 SEQ ID NOs CDR3 SEQ ID NOs
A151 DYYIN 7 EiFPGSGRTHFSLQDYL 102 GDYGFLY 9
A1152 DYYIN 7 EIFPGSGNTHFSDGVSS 103 GDYGFLY 9
AH53 DYYIN 7 EIFPGSGICHFSWGSE 104 GDYGFLY 9
A854 DYYIN 7 EIFPGSGNTHFSGNM 105 GDYGFLY 9 ,
AH55 DYYIN _ 7 EIFPGSGWTHFSRSPTP 106
GDYGFLY ' 9
A1156 DYYIN 7 EIFPGSGNTHFSLRNFP 107 GDYGFLY , 9
AH57 DYYAN 108 EIFPGSGNTHFSARFKG 8 GDYGFLY 9
A1158 GYYIN 109 EIFPGSGNTHFSARFKG 8 GDYGFLY 9
AH59 QYYIN 110 EIFPGSGNTHFSARFKG 8 GDYGFLY 9
A1160 DQYIN 111 EIFPGSGITIIIFSARFKG 8 GDYGFLY 9
A1161 MAO 112 E1FPGSGNTHFSARFK6 8 GDYGFLY 9
AH62 DYYIN 7 EIFPGSGNTHFSARFKG 8 GDVGFLY 113
A1163 DYYIN 7 EIFPGSGNTHFSARFKG 8 GDYGFQY
114.
A1164 DYYIN 7 EIFPGSGNIHFSARFKG 8 GDYGFIQ . 115
AH66 DYYIN 7 EI FPGSGNTHFSARFKG 8 GDOLLC 116 .
A1166 DYYIN 7 EIFPGSGNTHFSARFKG 8 WDYGFLY 117
A1167 DYYIN 7 EIFPDSAPSHFSARFKG 118 GDYGFLY _ 9
A368 DYYIN 7 EIFIIDIPPHFSARFKG 119 GDYGFLY . 9
A469 DYYIN 7 EIFPGPFTPHFSARFKG 120 GDYGFLY 9
AH70 DYYIN 7 EIFPGSNFGHFSARFKG 121 GDYGFLY 9
AH71 DYYIN 7 EIFPGIGHTHFSARFKG 58 QDYGFLY 64
A5172 DYYIN 7 EIFINAIGNYHFSRSPTP 122 GDYGFLY 9
A1173 DYYIN 7 EIFPGEGNSHWSRFKG 123 GDYGFLY 9
AH74 DYYIN 7 EIFPGYGNTHFSARFKG 59 QDYGFLY 64
A175 DYYIN 7 EIFPGYGNTYFSARFKG 124 GDYGFLY 9
A876 DYYIN 7 EIFPGYGNTHFSEPTF 125 GDYGFLY 9
AH77 DYYIN 7 EIFPGYGNSHVYSRFKG 126 GDYGFLY 9
AH78 DYYIN 7 EIFPGSGNTYFSARFKLi 62 QD\GFLY 64
AH79 DYYIN 7 EIFPGSGNTYFSRSPIP 127 GDYGFLY ! 9
AH80 DYYIN 7 EIFPGSGNSHVVSRFKG BO QDYGFLY 64
AH81 DYYIN 7 EIFPGSGNSHVVRSPIP 128 GDYGFLY 9
A182 DYYIN 7 EIRPGSGNSHYVERT9 128 GDYGFLY 9
AH83 DYYIN 7 ET FPGYIGNTYFSARFKG 63 QDYGFLY 64
AH84 DYYIN 7 EIFPGENTHFSRSPTh 122 QDYGFLY 64
A1185 DYYIN 7 EIFPGWGNSHITSRFKG 123 QDYGFLY 64
10(86 DYYIN 7 EIFPGYGNTYFSARFKG 124 QDYGFIX 64
10187 DYYIN 7 EIFPGYGNSHINSRFKG 126 QDYGFLY 64
A1188 DYYIN 7 EIFPGSGITTHFSRSFTP 106 QDYGFLY 64
AH89 DYYIN 7 EIFPGYGNTHFSRSPTP 125 QDYGFLY .
64
A1190 DYYIN 7 EIFPGSGNTYFSRSPTP 127 QDYGFLY .
64
AH91 DYY1N 7 EIFPGSGNSHYVRSPTP 128 QDYGFLY 64
A192 DYYIN 7 EIFPGSGNSHVVSSFTP 129 QDYGFLY 64
A193 DYYIN 7 EIFPDSAPSYFSARFKG 130 GDYGFLY 9
A5194 DYYIN 7 EIFPGPFTPYFSARFKG 131 GDYGFLY 9
AH95 DYYIN 7 EIFPGSNFGYFSRSPTP 132 GDYGFLY 9
A1196 DYYIN 7 EIPPDSAPSHVVSRFKG 133 GDYGFLY . 9
AH97 DYYIN 7 EIFPGPFTSHVVSRFKG 134 GDYGFLY , 9
AH98 DYYIN 7 EIFPGSNFSHINSRFKG 135 GDYGFLY 9
AH99 DYYIN 7 EIFPDSAPSHFSISPIP 136 GDYGFLY . 9
AHMO DYYIN 7 EIRINTFTPHFSRSPTP 137 GDYGFLY 9
CA 03061704 2010-10-20
WO 2018/221969 PCT/KR2018/006182
41
[220]
List of heavy chain CDR sequences (continue) .
CDR1 SEQ ID NOs CDR2 SW ID NOs CDR3 SEQ ID NOs
AH101 DYYIN 7 EIFPGSNFGHFSRSPTP 138
GDYGFLY 9 ,
41102 DYYIN 7 E1FPDSAPS11VVSSP7P 139 GDYGFLY
0
AH103 DYYIN 7 EIFPGPFTSHVVSSPTP , 140 GDYGFLY I.
9
A1104 DYYIN 7 EIFPGSNFSHVVSSPTP 141 GDYGFLY , 9
. A11105 QYYIN 110 __ EIFPDSAPSHFSARFKG 118 _ _GDYGFLY 9
AH106 QYYIN 110 E1FPGPFTPHFSARFKG 120 GDYGFLY --9
A11107 QYYIN 110 EIFPGSNFGHFSARFKG 121 GDYGFLY
9
A11108 DYYIN 7 EIFPDSAPSHFSARFIG 118 QDYGFLY
64
A11109 DYYIN 7 EIFPGPFTPIIFMRFKG 120 QDYGFLY
64
A11110 DYYIN 7 EIFPGSNFGHFSARFKG 121 QDYGFLY
64
A11111 DYYIN 7 EIFPDSAPSHFSARFKG 118 GDYGFQY 114
,
A1I12 DYYIN 7 EIFPGPFTPHFSARFKG 120 GDYGFQY 114
A11113 DYYIN 7 ElFPGSNFGHFSARFKG 121 GDYGFQY 114
A11114 DYYIN 7 ElFPDSAPSIFSARFKG 118 GDYGFLQ 115
A1115 DYYIN 7 EIFPGPFTPHFSARFIG 120 GDYGFLQ 115
AH116 DYYIN 7 EIFPGSNFGHFSARFKG 121 GDYGFLQ 115
A1117 DYYIN 7 EIFPGSGNTHFSNSESF 93 HDYGFLY
66
AH118 DYYIN 7 EIFFGSGNINFSLQDYL 102
HDYGFLY 66 ,
AH119 DYYIN 7 EIFPGSGNTHFSNQGSE 104 HDYGFLY
66
[221]
[222] [Table 9]
CA 03061704 2010-10-20
WO 2018/221969
PCT/KR2018/006182
42
[223]
List of light chain CDR sequences
CDR1 SEQ ID NUs CDR2 SEQ ID NOs CDR3 SEQ
ID NOs
ALOI GASFYIYGALN 1 GATNLAD 2 QNVWSSPYT 112
AL02 GASENIYGALN 1 GATNLAD 2 QNVLNSPYT . 143
A1.03 GASENIYGALN 1 GATNIAD 2 QNVIESPYT 144
AL04 GASENIVGALN 1 GATNLAD 2 QNVLKSPYr 145
AL05 GASENIYGALN 1 GATNLAD 9 QNVLYSPYT 146
A1.06 GA SDI I YGAIN 1 GATNLAD 9 QNVLSRPYr
147
AL07 GASENIYGALN 1 GATNLAD 2 QNVLSSPFX . 148
AL08 , GASENIYGALN 1 GATNLAD 2 QYVLSEPYI 149
AL11 GASENIYGALN 1 GATNLAD 2 QNVLESPET 150
AL12 GASENIYGALN 1 GAINILAD 2 QNVLSVPET 151
ALI3 GASEWIYGALN 1 GATNLAD 2 QNVLSLPEr 159
AL14 GASENIYGALN 1 GATNLAD 2 QNVLSIPEr 153
AL15 GASENIYGALN 1 GATNLAD 2 QNVLSMPET 154
AL16 GASENIYGALN 1 GATNLAD 2 QNILSSPEr 155
AL17 GASENIYGALN 1 GA1NLAD 2 QNLISSPEr 156
ALI8 GASENIYGALN 1 GATNLAD 9 QMMISSPET 137
AL19 , GASENIYGALN 1 GATNLAD 2 QNIISLPEr 158
AL20 GASENIYGALN 1 GATNLAD 2 QNIISIPEr 159 ,
AL21 GASENIYGALN 1 GATNLAD 2 QNSLSSPET 160
_
AL22 GASENIYGALN 1 GAINLAD 2 QNTLSSPET 161
AL23 GASENIYGALN 1 GATNLAD 2 QNVSSSPEr 162
AL24 GASTNIYGALN 1 GAINLAD 2 QNVISSPET 163
AL25 GASENIYGALN 1 GATNLAD 2 QNVFSSPET 164
AL26 GASENIYGALN 1 GATNLAD 2 QNVYSSPET 165
AL27 GASENIYGALN 1 GATNLAD 2 QNVRSSPEr 166
AL28 GASENIYGALN 1 GAiNLAD 2 QNLVSSVEf 161
AL29 GASENIYGALN 1 GATNLAD 2 QNLISSPEr _ 156
AL30 GASENIYGALN 1 GATNLAD 2 QNLMSSPEr 168
AL31 GASENIYGALN 1 GATNLAD 2 QNIMSSPET 169
AL32 GASENIYGALN 1 GATNLAD 2 QNVHSSPET 170
AL33 GASENIYGALN 1 GATNLAD 2 QNVMSSPEr 171
AL34 GASENIYGALN 1 GATNLAD 2 QNLLSSPEr 172
AL35 GASENIYGALN 1 GATNLAD 2 QSVLFSPFS 173
.
AL36 GASENIYGALN 1 GATNLAD 2 QQVLFFPET 174
AL37 GASENIYGALN 1 GATNLAD 2 QNLLSPSFY 175
AL38 GASENIYGALN 1 GATNLAD 2 QSVLFSPFT 176
AL39 GASENIYGALN 1 GATNLAD 2 QNILSSPLF 177
AL40 GASENIYGALN 1 GATNLAD 2 QNILHYSLIT 178 ,
AL41 GASENIYGALN 1 GATNLAD 2 QQVLFFPU. 179
AL12 GASENIYGALN 1 GAINLAD 2 QQVLDFVFY , 180
AL43 GASENIYGALN 1 GATtLAD 2 OVVSSPET 181
AL44 GASENIYGALN 1 DAINLAD 182 QMVLSSPYr 3
ALAS GASENIYGALN 1 FATNLAD 183 QNVLSSPYT 3
AL46 GASENIYGALN 1 HAMAD 184 QNVLSSPYT , 3
AL47 GASENIYGALN 1 KATNLAD 185 QNVLSSPYT 3
AL48 GASENIYGALN 1 PATNLAD 186 QNVLSSPYT 3
AL/19 GASENIYGALM 1 QATNLAD 187 QNVLSSPYT 3
ALSO GASENIYGALN 1 SATNLAD 188 QNVLSSPYT 3
CA 03061704 2019-10-28
WO 2018/221969
PCT/KR2018/00618 2
43
[224]
List of light chain CDR sequences (continue)
CORI SEQ ID NOs CDR2 SEQ ID NOs CDR3 SEQ ID
NOs
AL51 GASENITGALN 1 VATNLAD 189 QNVLSSPYT 3
AL52 GASFAITGALN 1 YATNLAD 190 QNVLSSPYT 3
AL53 GASENIYGALN 1 GATNLAD 2 ITVLSPPYT 191
AL54 GASENIMALN 1 GATNLAD 2 QNNINFPFN 192
AL55 GASENITGALN 1 GATNLAD 2 QHYLFLPYV 193
AL56 GASENINALN 1 GATNLAD 2 QAVLINAYT 194
AL57 GASENITGALN 1 GATNLAD 2 QNVLRVGYL 195
AL58 . GASENITGALN 1 GATILAD 2 QSYLRVGYL 196
AL59 GASENIYGALN 1 GATNLAD 2 QNIISSPYT 197
AL60 GASENIYGALN 1 GATNLAD 2 QQVLCESFL 198
AL61 GASENITGALN 1 GATNLAD 2 QNNISQSLL 199
AL62 i GASENITGALN 1 GATNLAD 2 QP1APSYL 200
AL63 GASENIYGALN 1 GATNLAD 2 QNLLFQPLS 201
AL64 GASENIYGALN 1 GATNLAD 2 QNYLFQPLV 202
AL65 GASCNITGALN 1 GATNLAD 2 CINQLDPSLF 203
AL66 GASENIYGALN 1 GATNLAD 2 NULESPYT 204
ALF/ 1 GASENITGALN 1 GATNLAD 2 QALUANT 20b
AL68 GASENIYGALN 1 GATNLAD 2 Q0LLESPYT 206
AL69 GASENIYGALN 1 GATNLAD 2 NLTLVSPYT 207
AL70 GASENIYGALN 1 GATNLAD 2 GNILDSPYT 208
AL71 GASPAITGALN 1 GATNLAD 2 MATLISPYT 209
AL72 GASENITGALN 1 GATNLAD 2 NNLLDSPYT 210
AL73 GASENIYGALN 1 GATNLAD 2 EFAUSFYT 211
AL74 GASENIYGALN 1 GATNLAD 2 QNILFVDYT , 212
AL75 GASENIYGALN 1 GATNLAD 2 QNVLHLNYT 213
AL76 GASENIYGALN 1 GATNLAD 2 QNVIATPYT 214
AL77 GASENITGALN 1 GATNLAD 2 QNILHPGYT 215 ,
AL78 GASENITGALN 1 GATNLAD 2 QNYLTRGYT 216
AL79 GASENITGALN 1 GATNLAD 2 IINILTSPYT 217
A180 GASENITGALN 1 GATNLAD 2 QNVLGGGQG 218
AL81 GASENITGALN 1 GATNLAD 2 QNYLEIPLI 219
82 GASENIYGALN 1 GATNLAD 2 QNVLDDPFD 220
AL83 GASEINIIGALN 1 GATNLAD 2 QNYLDFPLL . 221
ALM GASDRYGALN 1 GATNLAD 2 QNVLYPSLV 222
AL85 GASENINALN 1 GATNLAD 2 QNYLFDQQS 223
AL86 GASENIYGALN 1 GATNLAD 2 QNVLSNEET 224
AL87 GASENIYGALN 1 ranun 2 QNVLKHPYT 225
AL88 GASENTYGALN 1 GATNLAD 2 QNVLSFGMW 226
AL89 GASENIYGALN 1 GATGLAD 227 QNNISSPTT 3
AL90 GASFATYGALN 1 GAQNLAD 228 QNVISSIWT 3
AL91 GSSRSIYGALN 229 GATNLAD 2 QNVLSSPYT 3
AL92 RAGRSITGALN 230 GATNLAD 2 QNVLSSPYT 3
AL93 LGRRGIYGALN 231 GATNLAD 2 QNVLSSPYT 3
A194 EVQVGITGALN 232 GATNLAD 2 QNVLSSPYT 3
- 1
AL95 i RPSEKIYGALN 233 GATNLAD 2 QNVLSSPYT 3
AL96 RASAVIYGALN 234 GATNLAD 2 QNVLSSPYT 3
87 KTGOLITGALN 235 GATNLAD 2 QNVLSSPYT 3
AL98 SCRVPITGALN 236 GATNLAD 2 QNVLSSPYT 3
,
AL99 , VASRGITGALN 237 GATNLAD 2 QNVLSSPYT 3
AL100 RGROITGALN 238 GATNLAD 2 QN\l,SSPYT 3
CA 03061704 2019-10-28
WO 2018/221969
PCT/KR2018/006182
44
[225] List of tight chain CDR sequences (continue)
CDR1 SEQ ID NOs C0R2 SEQ ID NOs CDR3 SEQ
ID NOs
AL101 , AAPRGIYGALN 239 , GATNLAD . 9 QNVLSSPYT , 3
AL102 SAPFKIYGALN 240 GATNLAD 2 QNVLSSPYT
3
AL103 , LGMDDIYGALN , 211 GATNLAD , 2 QPIVLSSPYT 3
. . .
AL104 NVRRGIVGALN 242 GATNLAD 2 QNVLSSPYT
3
AL105 NTS6R1YGALN 243 GATNLAD 9 QNVLSSPYT
3
ALM LVSRPIYGALN 214 GATNLAD 9 QNVLSSPYT
3
!L107 WTNRPIYGALN 245 GAINLAD 2 QNVLSSPYT
3
AL108 , R1PSAIYGALN 246 GATNLAD 2 QNVLSSPYT 3
. , .
AL109 GATRGIYGALN 247 GATNLAD 2 QNVLSSPYT
3
AL110 EGGSPIYGALN 248 GAINLAD , 2 QNVLSSPYT
3
AL111 GASRGMFRALN 249 GATNLAD 9 QNVLSSPYT
3
AL112 GASGLVFSALN 250 , GATNLAD _ 2 QNVLSSPYT
3
AL113 GASRGTHMALN 251 GATNLAD , 2 QNVLSSPYT
3 ,
AL114. GASSRFHNALN 252 GATNLAD 2 QNVLSSPYT
3
AL115 GASRTAFTALN 253 GATNLAD 2 QNVLSSPYT
3
AL116 GASRSTFSALN 254 GATNLAD 9 QNVLSSPYT
3
AL117 GASGPMFDALN 255 GATNLAD 2 QNVLSSPYT
3
AL118 . GASHDLYGALN 256 GATNLAD 2 QNVLSSPYT 3
AL119 GASULFGALN 257 GATNLAD 2 OVLSSPYT , 3
.
P1190 , GASKAAFGALN 258 GATNLAD . 2 QNVLSSPYT 3
AL121 GASEGIVGALN 259 GATNLAD 2 QNVLSSPYT
3
AL122 GASHEIHVALN 260 GATNLAD 2 QNVLSSPYT
3
AL123 , GASRGVFGALN 261 GATNLAD 2 QNVLSSPYT 3
AL124 GASGRVRGALN 262 GATNLAD 2 QNVLSSPYT
3
AL125 GASTGSFSALN 263 GATNLAD 2 QNVLSSPYT
3
P1126 GASCNISFDALN 264 GATNLAD 9 QNVLSSPYT
3
AL127 GASMSYFALN 265 GATNLAD 9 QNVLSSPYT 3
AL128 , GASFIUSALN 266 GATNLAD 2 QNVLSSPYT 3
AL129 GASAPRHSALN 267 , GATNLAD , 2 QNVLSSPYT _
3
L1:30 GASMPLFHALN 268 , GATNLAD . 2 QNVLSSPYT ,
3
A1131 GASENIYCALN 1 GATNLAD 9 QNILSSPYT 269
A1122 _ GASENIKALN 1 . CA1ALAB . 2 IIVLSMAT , 270
,
AL193 GASFICYGALN 1 GATNLAD , 2 QNVLSEPET 271
.
AL134 , GASENIYGALN 1 GATNLAD 2 QNVLYSPET 272 ,
. .
AL135 GASEUVGALN 1 GATNLAD . 2 OVLEEPYT 273
ALl.:.96 GASENIYGALN 1 GATNLAD 9 QNVLELPFT 271
A1137 GASENIYCALN 1 GAINLAD 9 QNVLEMPET 275
AL128 . CASFIIIVGALN 1 . GAINLAD ___ 2 OILESPET 276
AL139 , GASENIYGALN , 1 GATNLAD 2 QNVIESPET 277 ,
AL140 GASENIYGALN 1 GATNLAD 2 QNVMESPET 278
AL141 GASENIYGALN 1 GAINLAD , 2 OLLESPET 279
AL142 GASENIYGALN 1 GAINLAD 9 QNVLYEPYT 280
11L143 GASENIYGALN 1 GATNLAD , 2 QNILSEPET 281
AL144 CASENIVCALN 1 GATNLAD 2 QNVISEPET , 282
AL145 GASENIYGALN 1. GAINLAD . 2 QNVMSEPET 283
AL146 GASDITYGALN 1 GATNLAD 2 QNLLSEPFT 284
AL117 GAS , ENIYCALN 1 GATNLAD 2 QSVLFEPFS
285
_
A1148 GASENIYGALN 1 . GAINLAD . 2 QSVLFEPFT 286
AL1.49 ' GASENIYGALN 1 GATNLAD 2 QNILYSPET 287
AL150 GASENIVGALN 1 GATNLAD 2 QNILSLPET 286
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
[226]
List of light chain CDR sequences
CDR1 SEQ fD ?Ds I CDR2 SE.) ID NOs CDR2
SEQ 1D NOs
ALIBI GASENIYGALN 1 GATNLAD 2 . OfILSMPET 289
. _
AL152 'GASENIYGALN 1 GATNLAD 2 QNVLYMPET 290
AL153 GASENIYGALN 1 GATNLAD 2 QNVISMPET 291
. .
AL154 GASENIYGALN 1 GATNLAD 2 QNVMSMPET 292
_
AL135 GASENIYGALN 1 GATNLAD 2 QNLLSMPET 293
AL156 GASENIYGALN . 1 GATNLAD 2 . QNIISSPET
294
AL157 GASENIYGALN . 1 GATNLAD , 2 , QNVLYLPET . 295
AL158 GASENIYGALN 1 GATNLAD 2 OVINSPET 296
. _
_ AL159 GASENIYGALN 1 GATNLAD 2 QNVMYSPET 297
ALISO GASENIYGALN 1 GATNLAD 2 QNLLYSPET 298
AL161 GASENIYGALN 1 , GATNLAD 2 QNVISLPET
299
AL162 , GASENIYGALN 1 GATNLAD 2 QNVMSLPET
300
AL163 GASENIYGALN . 1 GATNLAD 2 QNLIõ.512ET
301
AL164 RASAYIYGALN 234 GATGLAD 227 QNVLSSPYT 3
AL165 GASENIYGALN 1 GATGLAD . 227 QNVLESPYT
144
AL166 GASENIYGALN . 1 , GATGLAD 227 QNVLSPYT
149
AL167 GASENIYGALN _ 1 GATGLAD 227 QNVLSSPET
148
AL168 GASENIYGALN 1 GATGLAD 227 QNVLYSPYT
146
AL159 GASENIYGALN 1 GATGLAD 227 QNILSSPET
155
AL170 GASENIYGALN 1 GATGLAD 227 QNLLSSPET
172
AL171 GASENIYGALN 1 GATGLAD 227 QNVISSPET
163
AL172 GASENIYGALN . 1 GATGLAD 227 QNVMSSPET
171
AL173 GASENIYGALN , 1 , GATGLAD 227 QNVLSLPET
152
. AL174 GASENIYGALN 1 GATGLAD 227 QNVLSMPET
154
AL175 GASENIYGALN 1 GATGLAD 227 QSVLFSPFS
173
AL176 GASENIYGALN _ 1. ..GATGLAD , 227 QNLLFQPLS
.201
AL177 GASENIYGALN . 1 GATGLAD 227 QQVLFFPLL
179
_
AL178 GASENIYGALN , 1 GATGLAD 227 QSVLFSPFT
176
AL179 RASAYIYGALN 224 GATNLAD 2 QNVLESPYT 144
ALISO RASAYIYGALN 234 GATNLAD 2 QNVLSEPYT 149
ALIBI RASAYIYGALN . 234 GATNLAD 2 QNVLSSPET 148
. AL182 RASAY1YGALN . 234 , GATNLAD 2 QNVLYSPYT
146
AL183 RASAYIYGALN 234 GATNLAD 2 QNILSSPET 155
AL184 DASAVIYGALN 234 GATNLAD 2 QNLLSSPET 172
AL185 RASAYIYGALN _ 234 , GATNLAD 2 QNV1SSPET
163
AL186 RASAYIYGALN , 234 GATNLAD 2 QNVMSSPET 171
.
AL187 RASAYIYGALN _ 234 GATNLAD 2 QNVLSLPET 152
AL1B8 RASAYIYGALN 234 GATNLAD 2 QNVLSMPET 154
AL1S9 RASAYIYGALN 234 GATNLAD 2 QSVLFSPFS 173
AL190 RASAYIYGALN 234 , GATNLAD 2 QNLLFQPLS
201
AL191 RASAVIYGALN 234 GATNLAD 2 QQVLFFPLL 179
AL192 .. RASAYIYGALN _ 234 , GATNLAD . 2 , QSVLFSPFT
176
AL193 GASRSTFSALN 254 GATNLAD 2 QNVLSIPET 153
AL194 GASMPLFHALN . 268 GATNLAD 2 QNVLSIPLT 153
AL195 GASRSTFSALN . 254 , GATNLAD 2 QNVLEFPYT
273
AL196 GASMPLFHALN 268 GATNLAD 2 QNVLEEPYT 273
[227]
[228] [Table 10]
CA 03061704 2019-10-28
WO 2018/221969
PCT/KR2018/006182
46
[229] List of sequences of light chain and heavy chain variable regions of
affinity-optimized
antibody
Amino acids sequence SEQ ID NOs
EVQLVESGGGLVQPGGSLRLSCAASGYIFSDYYINWVRQAPGKGLEW
AH71 IGEIFPGWGNTHFSARFKGRATLSADKSKNIAYLQMNSLRADDIAVY 302
YCAGQDYGFLYWGQ611,17VSS
EVQLVESGGGLVQPGGSLRLSCAASGYTFSDYYINWVRQAPGKGLEW
A1172 IGEIFFGRGNTHFSEPTPRATLSOKSKNTAYLOINSLRAEDTAVY 303
YCAGGDYGFLYWGQGTLVTVSS
IEVQLVESGGGINPGG8LRLSCAASGYIFSDYYININRQAPGKGLEW'
A1173 IGEIFFGRGNSHVVSRFKGRATLSADKSKNTAYWENSLRAEDTAVY 304
YCAGGDYGFLYWGQGILVTVSS
EVQLVESGGGINPGGSLRLSCAASGYTFSDYYINWVRQAPGKGLEW
A1185 IGEIFPGWGNSHVVSRFKGRATLSADKSKNTAYLQMNSLRAEDTAVY 305
YCAGQDYGFLYWGQGILA/TVSS
DIQIIMSPSSISASVGDRVTITCGASMFIYHALNWYQQKPGKAPKLL
AL130 IYGATNLADGVPSRFSGSGSGRDFTFTISSLQPFDIATYYCQNVLSS 306
PYTFGQGTKVEIK
DIQMTQSPSSLSASVGDRVTITCGASENIVGALNWYQQKPGKAPKLL
AL135 IYGABLADGVPSRFSGSGSGRDFTFTISSLQPEDIATYYCQNVLEE 307
PYTFGQGINVEIK
bIQMTQSPSSLSASVGDRVTITCGASENIYCKINIYUPGKAPKLL
AL165 lYGATGLADGVPSRFSGSGSGRIFTFTISSLQPEDIAMCQNITLES 308
PYTFGQGTKVEIK
bIQPITQSPSSLSASVGDRVIITCGASENIYGALNlYMKPGKAPEL
AL166 309
IYGATQADCATSRFSGSGSGRDFTFTISSLOPEDIATYYMNVLSE
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
47
[230]
PYTFGQGTKVEIK
DIQMTQSPSSLSASVGDRVTITCGASMPIIIIALNWYQQKPGKAPKLL
AL194 IYGATNLADGYPSITSGSGSGRDFTFT1SSLQPED1ATYYCQNVLS1 310
PETITIQGTIOTEIK
DIQMTOSPSSLSASVGDRVTITOGASRSIT'SALNWYQQKPGKAPKLL
AL195 IYGATNLADG'VPSITSGSGSGEDFTFTISSLQPEDIATYYCQNVLEE 311
PYTFGQGTKVEIK
[231] Also, an in vitro test on proliferation inhibitory activity was
performed on U-87 MG
cell line by using a part of the affinity-optimized antibodies, wherein
results thereof are
shown in Table 11.
[232]
[233] [Table 11]
[234] in vitro tumor cell proliferation inhibitory activity by hu8C4 light
chain and heavy chain
affinity-optimized antibodies
U-87 MG (GBM, HGF autocrine)
CR11 proliferation inhibition assay, IC50
Antibodies (nM) IC50 Fold
affinity-optimized
hu8C4
antibodies
hu8C4 A1-171 " 11.3 95.5 8.5
hu8C4 A1172 10.9 95.5 8.8
hu8C4 AH73 10.9 95.5 8.8
.....
""-fii-i"8"1"1"-AT-18T 10.1 95,5 9.5
hu8C4 AL130 5.0 45.0 9.0
hu8C4 AL135 7.1 31.9 4.5
hu8C4 AL165 6.8 39.0 5.7
hu8C4 AL166 9.1 39.0 4.3
hu8C4 AL194 9.6 94.5 9.8
hu8C4 AL195 18.0 94.5 5.3
)
[235] As seen in Table 11, it was identified that 1050 of tumor cell
proliferation inhibitory
activity of a hu8C4 affinity-optimized antibody in a U-87 MG cell amounted to
5.0 -
18 nM, wherein efficacy thereof was increased 4.3 - 9.8 times more than a
parent
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
48
antibody hu8C4. The results above represent a test performed on a part of
antibodies
having an amino acid sequence presented in Tables 8 to 10, wherein an affinity
of the
parent hu8C4 antibody was optimized and all the antibodies were selected based
on an
antigen affinity through a selection process. Thus, it is expected that there
may be a
sufficiently equal effect even with regard to the rest of affinity-optimized
antibodies as
well as antibodies with a combination of presented heavy chain and light chain
variable region CDRs.
[236]
[237] For an additional experiment, 10 kinds of affinity-optimized antibody
were prepared
by combining the light chain and heavy chain variable regions. A specific
combination
of light chain and heavy chain sequences are shown in Table 12.
[238]
[239] [Table 12]
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
49
[240] List ofcombinedvariable region sequences ofaffinity-optimizedantibody
Heavy chain variable Light chain variable
region region
Light chain variable
hu8C4 Alf71 A1171(SEQ ID NO: 302) region of hu8C4 -1
antibody (SEQ ID NO: 21)1
Light chain variable
hu8C4 AH85 A1185(SEQ ID NO: 305) region of hu8C4 -1
antibody (SEQ ID NO: 21)
Heavy chain variable
hn8C4 AL194 region of hu8C4-1 antibody AL194(SEQ ID NO: 310)
(SEQ ID NO: 23)
hu8C4 A56 A1185(SEQ ID NO: 305) - AL165(SEQ ID NO: 308)
hu8C4 A62 A1172(SEQ ID NO: 303) AL130(SEQ ID NO: 306)
hu8C4 A71 AH73(SEQ ID NO: 304) AL135(SEQ ID NO: 307)
hu8C4 A72 'A1173SEQ'ID NO: 304) AL165(SEQ ID NO: 308)
hu8C4 A73 AH73(SEQ ID NO: 304) - AL166(SEQ ID NO: 309)
hu8C4 A76 AH73(SEQ ID NO: 304) AL195(SEQ ID NO: 311)
hu8C4 A78 All71(SEQ ID NO: 302) AL130(SEQ ID NO: 306)
[241] Then, a tumor cell proliferation inhibitory activity was evaluated by
the same method
as shown in Example 1 above, wherein results thereof are shown in Table 13 and
FIG.
3.
[242]
[243] [Table 13]
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
[244] In vitro tumor cell proliferation inhibitory activity by affinity-
optimized antibody
U-87 MG (GBM, HGF autocrine)
Cell proliferation inhibition assay,
Antibodies I C50 (nil) IC50 Fold
Affinity-optimized
hu8C4
antibody
hu8C4 AH71 3.6 49.0 18.6
hu8C4 AH85 3.2 49.0 15.2
hu8C4 AL194 5.3 49.0 9.2
hu8C4 A56 1.7 49.0 28.5
hu8C4 A62 1.8 49.0 27.6
hu8C4 A71 5.0 49.0 9.7
hu8C4 A72 3.6 49.0 13.8
hu8C4 A78 4.0 49.0 12.3
hu8C4 A76 4.3 49.0 11.3
hu8C4 A78 2.6 49.0 18.9
[245] As seen in Table 13 above, it was identified that hu8C4 as well as 10
kinds of key
antibody with a combination of light chain and heavy chain variable regions of
an
affinity-optimized antibody thereof showed a tumor cell proliferation
inhibitory
activity, too. In particular, IC50 of the 10 kinds of antibody amounted to 1.7
- 5.3 nM
and it was identified that they had a tumor cell proliferation inhibitory
effect, which
was 9.2 - 28.5 times more excellent than the parent antibody hu8C4.
[246]
[247] Example 6. Preparation of bispecific antibody and in vitro tumor cell
proliferation
inhibitory activity
[248]
[249] To prepare a bispecific antibody specifically binding to c-Met and
EGFR, Erbitux
and Vectibix scFv fragments, known to specifically bind to EGFR, were linked
re-
spectively to a heavy chain C-terminus of the c-Met antibody of the present
invention
by a GGGGSGGGGS (SEQ. No. 312) connector.
[250] To increase the stability of the scFv, a 44th residue of a heavy
chain and a 100th
residue of a light chain were substituted with cystine (Reiter Y. et al.,
Biochemistry
CA 03061704 2019-10-28
WO 2018/221969
PCT/K1R2018/006182
51
33(18):5451-5459 (1994)). Erbitux and Vectibix scFv sequences, amino acid
sequences of heavy chain of bispecific antibody and a combination of variable
regions
of bispecific antibody are shown in the following Tables 14 and 15.
[251]
[252] [Table 141
[253] List of amino acid sequences of EGFR antibody for preparing
bispecific antibody as well as
bispecific antibody
Am i no ac ids sequence SRO ID KOs
rRLSI
QVQLKQSGPGLVQ ?SC)SLS : "1::::VSGESLIWYGVHWVIKISPGKCIALGY I WEGGEDY MTPF
NKONSKSQVFEK141\ SI giNDIA I TYCARALITY DYEFAYW(WINTVSAGGGGSGG
Erbi tux scFv 3L 313
GGSGGGGSGGG(iSDILLIQSPV:LSVSPGERVSFSCRASQSIGTNIHWYQQRTNGSPRLLIKY
ASESISGIPSRFSGSGWBFILSINSVESEDIADYYCQQNNNUTTFGCGTILFLK
JILLIQSPVILSYSPGKI1VSFSCKASQS.GTHIHIVYWHINGSFKU,IKYAS:SISGIPSIN
GSGSGTDFTLSI NVESEDIADYYCQQN NNIVPITFGCGTKLELKC6(ZSGGGGSGGGGSGGCC
Erbi tux scFv :all 314
SWQLKQSGPGLVQP9(1SLS I TaVSGFSLTNYGVI IVAFFQSPGKCLEI/LGV 1 WSGGNTDI
FTSRLSI NKDKSKSQVFFKNNSIASNDTAITYCARALTYYDYEFAYFGQGTLVTVSA
QVQLQESGPGLIIPSEILSLTCTVTIGSVSSGDYMT I RQSPGKCLEFIG1IIYYSGNINYW
SI ISKLIISIDTSKI(OLKISSVTAADIAIT YCYRDK VIGAI.1)111tXX11111/TYSS6(41(1:116
Yee t i bix scFv 315
CCSGCCGSGGCCSDIQIIMSPSSL,SASVGDRVTITCQASQDISIs'YLVINQQVGKAPKLLITD
ASNLETGYPSRFSGSGSCIDFTFTISSOPEDIATVFC0JHF1HLP1AFGCG1WEIK
INQLVEFAXXIVQP(MLF 1 SCAAS(1141.1;DY Y INFYIK/APGKC11111 GE! P6FtGl1IH [TAR
FKGRATLSADKSK.VTAYLQ111+1SUMEDTAWYCAGGDYGFLYWOZGILVIVSMSTKGPSVFP
111PSSKSTWTAALGCLVIWYPEPVTVSINSGALTSGVH1FFAVIASSGLYSISSIT1,7VPS
SSLGTQTYICKVNIIKPSKTKVIIKVERKCCVECPPCPAPELLGGPSVFLFPPKPICDTL111SRT
PEVIUVVDVSHEDPEYKFMVDGVEVIINALTKPREEQYKSTYRWSVLIVIIIQDWIAGKEY
h tiCA a IrI:i lix KLIVSNKA1PAP I KKT1SK AK; 74'KENYYTLPEstMELTENVS XL1 .v
Kt; =NPSDI A vt/F.
316
scFv ILL heavy caain
SICQPENNYKTTPPVLDSDGSFFLYSKLTVDERVQQGINFSCSVIIIIFALlIN:IYTQESLSLSP
CACCIZSGGGGSQVQLKQSGPMVQPSUISITCTVSGFSLTNYCAKINRQPGICCLEWILYI
1193GNIDYNIPFTSRLSINKDNSKSQVUFKKNSLQSNDTAIYYCAR4L1YYDYCFAYKGQGIL
VIVSAGGGGSGGGGSGGGGSGGGGSDI LLNSPVILSVSPGERVSFSCRASQSIGTNIHWYM
RTM111SPRILIK1' ASESISCIPSRFSGS:NaT111,SIKSVESEDIADYYCCRINNTIFITEUX;
TKLELK
EVQLVF,SCCGLYQPGGSLRLSCAASGYIFSDYY I 11111/VRQAPGKGLENIGEI F?GliGtiTHFSiR
FKGRATLSADISINTAYLCANSIRAEDTAWYCAGQDYGFLYWGQGTLVITSSASTKGPSVFP
LAPSSKSIMGTIALGCLVADYFPFPVTn1114SGALTSGVirIFFAVIXISSGLYSISSVVINPe
.611,yrvICKINHEPSKTK VI1KKVE.RIMECPECPAPHAI;GESVFIFPFAFKITTLIJISRT
PEWCVVVDVSHEDPFIKPAYYDGVEVIINAKTKPREEMTYRVI1SVLIV:11QDENGKEY
huRC4 A1171 x Ftrhi tiix KCKVSNICAI PAP TEKTISK
AKCQPREPCPTLPPSRDFLTINQVSITTINE71/PIDI AVFIF
317
scFv It heavy ciain
SNOWENNYILTIPPVLDSDGSFFLYSKIDDIESRWQQGNVITSCRINEALIIHYTQKSLSGSP
GKGGGGSGGGGSQVQLKSGPGLVCIPSQSLSI1CTVSGFSLTNYCX111/VRQS.IGKCLEITLGYI
1410'rINNITFTSRLSINKIASKVEFKIINSIASNITTAIYYCARAI;IYY1)YEFAYWGQ611,
FIVSACAZGSGGGGSGGGGSGGOGSDI LLTOPVILSVSPGERVSFSCRASQSIGTNIHM
RINGSPELL I KY ASESISC I PSRFSGS:TSGTDFTLS INSVESEDIADYYMTINIVPTTFGOG
BULK
Ch 03061704 2010-10-20
WO 2018/221969
PCT/KR2018/006182
52
[254]
Amino acids sequence (continue) i nue ) SE)2 11)
N3s
EVQLVESGGGLYQPGGSLRLSCAASGYTESDYY INWVRQAPGEGLEFIGE FPGICATIFSES
11112,41 LSADKkFTAY Q\IN.' K )'F VY YMCA AlkLYWa/GTLYINSSASLI(GPS1/11)
LAPSSESTSGCTAALGCLVEDYFEEPVTVSWNSGALTSGlinEPAVLQSSGLYSLSSWINPS
SELGIQTY ICKVNIIKESKTKVDEKVFIZECCVECPPCPAPELLGGPSI'FLEPPEREITLAII SET
PEVIVIITIVSHEDPEVKFHWYVEGVEVIINA1EPREEQYNSTYRITSVLTVLHQD1LNGICEY
hu8C4 AH72 x Erbi t Inc ECKVSNIIALEAF
IF,KT1SKAKGQPREPQVYTLEPSEDELTENQVSLICLYEGFYPSDI AVERT 318
scEv IL heavy chain
SHGQPENNYETTPRIESDGSFELYSKUNDESEWNGNVESCSVMHEALIINHYTOBLSLSP
GEGGGSGGGGSQVQ1.11QSGEGLVQPSQSLSITCTVFGESEMGV111/VRQSPG1CCLEWLGV 1
ESCICTIµTDYNTPFTSRLSINKENSK5OFFENINSLQSNDTAIYYCARALTYYNEFAltiVGQGIL
YTVSAGGGGSGGOGMEGGSGGC4SDILLTQSPVILSVSPGERVSFSCRA5QSIGTNIEWCI
RINGSPR1LIKYASESISCIPSEFSCSCSGIEFrLSINSVESED IADYYCQQAINFTTFCCG
DELELK
EWLVESGGGLVQFGGSLRLSCAASCYTFSDYY INWVRQAPCKGLEVI GE I FPGAGNSHWER
FIGRATLSADESIMAYLOINSLILAEDTAVYKAGGDIGFLYWGQGTLYINSSASTKGPSVFP
LAPSSICSTSCOMALGUNIWYFEEPVINSWNSGALTSGVHTFPA7LQSSGLYSLSSWINPS
SSLGMTY 1 CEIFICKESKII(VDKIEVERECCYECPPCPAPELLGGPSVFLEPPEFEDIINI SET
PEVITVINDVSIIEDENCENVIVEGVEVENAETKPREFQYNSTYRITSYLTVIIIQDENGEBY
hu8C4 AH73 x Erbi tux KagSNLAIPAF
IEKTISKAKGQPREP(ANTLPFSRDELTINQVSLTCLVEGFYPSDIAVENE
319
scEv HL heavy chain 11(ICIPKIAYETI
KNIIISIXiSFFLYSKLIVDESICittnaftiFSCSAHEALHNHYTQl(SLS1.9
GEGGGGSGGCIGSQVQLEQSGEGLVQPSQSLSITCTVSTSLTHYGVINNRCISKECLEWLGVI
NSGMTDYNTIITSRLSINKENSMVFFKINUSNIYIAlYYCARALTYYDYEFAYWCQGTL
VIVSAGGIXEGG(XiS(21X;S(XXIGED I LINSPV I I SVSKER VSFSCRASOS I GIN I HVN(4)
RTNGSTELL1EYASESISGIPSEFSGSGSGTOFfLSINSVESKOIADYYQ(INNNWITTFGOG
MIRK
EVQLVESCGGINPGGSLELSCAASCYTFSDYY1NWVIIQAPGKGLE1110E1FPC110811VVER
FKGRATLSADESENTAYLQ)lNSLRAEDTAVYYCAGQDYGFLYWKIGTLITiSSASTI(GPSVFP
LAPSSKSTSGGIAALGCLVEDYEPEPVTISWNSGALTSGMEPAVLQSSGLYSLESVVTITS
SSLORY1ClaNHKEWIEVDKEVERECCVECPPCPAPELLGGESVFLEPPEID1114I SET
mot:yid VINSICKUPKVKliNwY VIXNEVHNAKTKPRRIOINSIV RV VSVI:r V I 1441114.11GKIni
1u8C1 10185 x Er bi tux ILICVSNILALPAF I EKT
ISKAKGQPREPQVYILPFSRDELTIDIQVSIICLYEGFYPSDI AVM 320
scEv HI. heavy chain
ENGQPENNYETTPPVLBDGSFPLYSELTVDESRWQQGNVESCSVMHEALHNHYNKSLSLY
GEGGGGSGGGGSQVQ1EQSGPGLIVIWSLSI'fCMYSLTNYGVI11/VRQSPGELEWLGVI
FSGGIMYPTIIITSRLSINKENSIWOFFIOINSLQSNDTAIYYCAILALTYYDYEFAIIVGQGTL
VIVSAGGGGSGGGGSGCCIGSGGOGSDILLTQSIN ILVSPGERVSFSCRASQSIG1141 IWO
RINGSPELLI KYASESISG I NEFSGSGSGTEFTLS I NSVESED I ADYYCQQ190111PTTEGCG
MILK
CA 03061704 2019-10-28
WO 2018/221969
PCT/KR2018/00618 2
53
[255]
no C. i ris equence (coif i nue) SFL) I)
NOs
ENLVESGGIGIAPGGSYL SC Ali SG Y sti I) V Y1 R(,):1?(;KGLEIFIGIII
FFGNGYFHINAll
FISMAILSADEKNTAYLOINSLILIEIY:Al liCAGGIAGH1.11(ZILVTVSSASTEFSVIT
LAPSSKSTSTMALUIVKDI FPEPVINSWNSGALTSGVHTFPAVLQSSGLYASSV VMS
SSLGTQTY I CNINHKPS1111VDKKVERIMVECPPCPAPELLGGPSVFLITAPILUTIA 1 SRI
PINTCVWDYSHEOPEWNWYVDGVEVIINAKTICPREEQYKSINOVSYLTVLIEVENGKElf
1118C4 x Er .Di tux IECKVSNKALPAP 1
EKT1SKAKGQPREPQVYTLPPRDELTKNQYSLICLIIKGFYPSDIAVER
327.
3c K.: LI heavy c ha i 91QPENKYIETPRil
INIY;SPFLISKLTVDKSM(041;SCSVNIEALHNHYAKSLSLSP
MGGGG SGGGGSD I LLTQSPVI L,SVSPGERVSFSCRASQSIGIN I HWYQ:ENGSPRLL I KYA
SESI SG I PSRFSGSCSGTDFTLSI NSVESEDIADYYCQQAWNWP rTICCGTICIELKGGGGSGG
GGSGOGGSGGGOSQVQLKSGPGIAPSQSLSITCVSGPSMYGVHWRQSPGliCLEWLGV
IVSGGNIDYNTPFTSRLSINKDNSMTFOINSLQSNCIATYYCARALTYYDYVAITGQGT
LVWSA
NACCHSGGGLV0PGGS.121.SAASGY7SDIY I NIVVRQAPGI(GLEW I GE I FPGIEITI ESAR
FKGRATLSADKaNTAYLCNNSLILEDTAVYKAGQDYGFLYWGQGTLVIVSSASTEFSVFP
LAPSMSTSGGTAALGCLVKDYRITY:raNSGALTSGIRTITAYLQSSGLYSLSSVVIVPS
SSLGTQTY I GIVNEKPSNEVDKKVERKCCVECPPCPAPELLGGPSVFLAFFFEITIS. I SRI
PENTCVWDYSIIEOPEVIOWYVDGVETNAKTKPRIMYNSTYRYISYLTVIIIIIIKNGKFX
hu8C 4 ran x Erb i tux KCKVSNKALPAP 1
EKTISKAKGQPREMYTI.PPSRDELTKNUSLICINEGFYPSDI AVER
322
scFv 13 heavy cha i n SiCLRENNYIETPPVILSOGSFFLYSKI.TVDKSRWQ(ANVFSC SYNE&
HNHYRIKSI.S1,51'
"2(GGCAISCAGGSD11.1.11QSPVI ISVSPGFRITSFSCRASQS1GIN11111YQQRTNGSPRI A ,1
313SISGIPSICFSGSGSGrDFILSINSVESED1ADYYCQQWWPTTFGCGTKIELEGGMG
a;SCrIGGSGGGGSOUNIQSCPG1,14.)PSQS1,S11ITSGFSLTNYOFIVVRQSPCJICI,FNILV
TIVSCIGNIDYNTPFTSRISI NKT)NSIMV1TINK111:MDTATYYCARALTYYDITTAYtiGQGT
LVTVSA
EL9LVESGGGLVQPCGaRLSCAAWYTFSDYY NrittgAFGICGLEIVIGlilFYGIEYTHFSRS
IIVRAILSADISKNTAYWNNSULIEDTAVYKAGLINWLYWGWILYPISSAStliGFSVFP
LAMISTS(1GTAAL(XLVIWYWEINTVStraiiILTSGV4ffFPAYLQSSGLYSLSSVVIVPS
SSLGTQTY ICNVNIAPSMIVDICINFIKCCVECPPCPAPELLGGPSFLFPPI(PIEDILM 1 SRI
PITTCVVVDVSIIEDPEVIEFNWYVDGVEVHNAKTKPREEQYNSTYRVI'SVIIVL101/LNGBEV
hti8( 4 A1172 x Erb i tux 1CCKVSNKA1PAP 1 EKTISHAKGQPRFPVITITI) SIZDELTKNQ
VSLICLVKGFYPSO 1 AVM
323
scFv 13 heavy chain 24QPENNYITTPRILDSDGSFFLYSELTVDKSIKQGNVFSCSVNIIEALI
INIKQKSLSLS1)
GIGGGGSGGGGSDILLIQSPVI LSVSPGCRITSPSCRASQSIGIN 1 1111YQQRT1GSPRLL KYA
SESI SG 1 PSRFSCMSGTDFT:SI NSVESEDIADYKUNNWPTIMCGTELELKGGGGG
GGSGGGGSGIGGGSQVQMSG7GLVQPSQSLSI1CIVSGFSLINYGIRMASPGIICLEHLGV
IIISGGNIDYNTPFTSRLSINKONSEQVFF111/1CLQSNDTAIYYC4RALTYYDYEFAYMQG7
LITVSA
CI1 03061704 2010-10-20
WO 2018/221969
PCT/ICR2018/006182
54
[256] Amino acids sequence (cant i nue) SD3 ID
NOs
EVQLVEqnEl VOPCGSLRLSCAASCYTFSDYY1NWVEQAPGKGLFS 1 CHI FPCithSHVVS1
FORA TLSABESKNTAYUZiNSLRAEDTAVYYC.ACGDYGFLYWGQGTLVTVSSASTEGPSVF?
UPSSKSCTPJI1 VKDYFPEPVTVSIIIMALTSGVINWAYLQS,SGLYSLSSVVT4TS
SSLGTOTYIGNNHI(PSNTKVDKINERINCVDCPPCPAPELLGGPSYFLFPPI(PKCIILN LSI?!
PENTCVVVINSIIRDPEVIROVVDGVEVHNAKTKPRFAQVICriRVVSVLTVLIQDWISGKEV
ha8C4 A1173 x Erbi tux KCEVSNICALPAP 1
EKTISKAKGQPREPQVYTLITSRDELTKNQVSLTCLYEGFYPSDIAVEF 324
v LH heavy chain SNGOPENNYKTIPPVLIEDGSFFLYSKL
IVDISRIVQQGNWSCSVNHEALHNHTFASLSL5?
aliiIGG3SCOCCSD 1LLY4SPV 1 LSVSPG FRVSFSCRASQSI GIN IHIYQQRTHCSPRLLIKY1
KIPSRFSGSGSODFILSINSVESEDIADYY03MNIVITFGCGTKLELIMIGGSGG
GGSGGGGSGCCGSQVCOGISCIPGLV3I'SQSLSITCTVSGFSLTNYGYINNIIQSPGI(CLEIYLGV
IrbGGNTDYNDFTSRLSINKDNSKSQVFFIGINSLOSNDTAIYYCARALTYYDYEFAYWGQGT
LVTVSA
EVQLVE93GOLVQPGGSLELSCAASGYTFSDYY INWVIZQAPGKGLET GE1 PPGICNSAVVSR
KGRATLSADKSKNTAYLOWSI.RAEDTAVYYCAOQDY(FLYWGQGTLVITSSASTIMPSVF?
LAPSSESTXIGTAALGCLVEDYFPEPTINSIVNXiALTSGVIITFPAUSSGLYSLSSVVIVPS
SSLMI)TY1C1iVNHIONY11(W)KKVIiRliCCVECPPCPP.PELUZPStilIFPPIIPKIY11111SIff
PEVTCVVVDVSINOPEVICFNIVYVDGVEVI1NAKTKPREEQYKSTYRVVVLTVLHQD1INGKEY
ha8C4 A1185 x Erbi t liCKVSNKALPAP
1FXTISKAKGQPREPOWTLPFSEDELTIMVSLICLVNGFYPSC MIFF
325
wFv LH heavy chain SNGQPENNYKTIPPM6DGSFFLYSKLTVDERNOCATSCSVNHEALHNHY1QESISLS?
GRGGCLSGGGGSD111.1T)SPVI LSVSPGERVSFSCRASOSIGTN IIIIVNORTNGSPRLL ID I
SET SST PSRFSGSGSGTOFTI,SiNSVESED1AOYYOICOMIIPTTFGCGTELELEGGGSGG
GGSGMGSGGGGSQVQLICQSGPGLVQPSQSLSITCINSGF5LTNYGAIIIVINSPGICCLEWLGV
1 IISCCNTINNIYI=TSKI S I NKOKSKS()V111(NNSI PSIII ill 1 YYCARALTYY DYITAYMX.0
LVTVSA
EVQLVF,SGGGINQPGGSLRLSCAASGYTFSDYY f M'IVRQAPCAGI El GE1 FPGSGKIIIFSAR
FKGRATISADI(SENTAYLONNSLRAEDTAVYYCAGGDYGFLYNGQGTLVITSSASTILGPSVF?
LAPSSKSTSGGDALGCLVEDYFPEPTASIIINSGAL'fSGVHIFPAYLOSGLYSLSSVVTVPS
SSLGT31YlaVNIEPS4TICVDKKVERECCVECPPCPA2ELLGGPSVFLFPPMILMI SET
FUTCVVYDVS1 TOPEVENWYVDGVEVIIHAK18PRECQYNSTIRVV5VLTVUORNGIU
hu8C4 x Vcc t i bi x KCKVSNICALPAP 1
EKTISLAKGQPREPQVULPPSEDELTINQVSLTCLVKGFYPSC AVEIP
326
scFv heavy chain
StIGQPENNYKUPPVLIEDGSFFLYSKLIVEI5RICQQGIVFSCSINIIBALIINIIY1X)1(SLSLS?
(il(GSOGGGSQVQ14110,SGRIVKPSIO .51:1MCGSVSSGDYWIT 1 liCISPGKCIAW 1
III ITSATNYNTSLKSRLTI SI DTSKTQFSLKISSIVADTAIITCYRDRVTGAFDIWGQGIII
VTVSSaKi(B(.:(LOS(WGIMGGS1)1()NIQSFSSLSP.SVGDRVIi1OQAS(201StelLAWYQQ
l(PGKAPIiLL 1 YDASALEICVPSRFSGSGSGTDFIFT SSLQPED 1 ATITCQHFDILPLAFCCG
nala
CA 03061704 2019-10-28
WO 2018/221969 PCT/K1R201
8/00618 2
[257]
Amino acids sequence (continue) i nue ) SEC 11) NC
s
SDYY! Nh QUO( GI ,F.lt I GE I FP( iWiiNTI- FSAI?
FICGRATLSAIXSKN1AYLQ1ANSLRAMTAVYYGIGQDYGFLYWGQG11NIVSSASTKGPSVFP
LAPSSKSTIVTAALGCLVIWYFPENTVSIVNSGALT%4111'FPAACtSSGLYSLSSV4TVPS
SSLGTCHT ICNVNHICPSEIVDIXVIIIIKCCVECPPCPAPFILGGPSVFLFPPITIEDILM SRT
PETICMOVSHEDPEVICIWVOGVEVFNAB1KPRIta)YNST\1INSVIIVLHQDENG111
14E4 AH71 x Vec t ihix KCI(VSNKA: PAP HKT 1
SKANQPRITQW11.PPSHDHINKNQVS1,111.V1(MPS1)1 Alf HIVE
327
scFv heavy chain
SNGQPENNYKTIPK116DG5FFLYSKUR'DICSRVANGWFSCSNNHEALHNHYTQKS1,SISP
II1GEGGGSGCGCEIVQ1ASGPGLVITSETLSLTCNSOGSVSSGDYYN1IIKSPGICCLEIFIG
YYSGYIWNPSIASRI,TISIDISKTQFSLlaSSMADTAIMIRDRVTGAFONGQG111
47VSSWAISGGGGSG(36(TSGWISD1(110WPSSLSASVGDRYf ITCQ,Aa)E1SNYLNINQ
KPONAPK I .:.1 YDAS41,KTGVPSKFFASGNMYPT I MAP:DI ATV Fa.)H141)HIY1 AFC XX;
11EVE11(
EVQLVESGCCLVQPGGSLRLSCAAXYTFSDYYININRO.AFGELETIGFIFFCIFUNITIFSRS
PTPRATLSADKSITTAYIAMNSLRAEDTAVYYCAGGDYGFLYWGQCILV1VSSASTKGPSVFP
LAPSSISTWGTAALGCLVI(DYFFEPVTVSIVNSGALTSGVIITFPAVIASSGLYSLSSVVIVPS
SSLGTQTYICNINWPSYTKVDKIEVCRECVCCPPCPAPELLGGPSVPLITPEPI1D11111 SRI
PliVICVVV)VSHEDPEVICFMTVIEVEVINAKTKPREFX/YNSTYRVVSNLTVLHQDWIAGKFS
hu8C4 :11172 x Vec t ibix
ICIVSNKA:PAPIEKTISKAKCQPEFQWILPFS?.DELTIKVSLTCLVI(GFYPSDI AVER
328
scFv heavy chain
SNGQPENNYHTIPPV'LDSOGSFFLYSKLTVDISSIMQGNVFSCSVERALHNHYTHICSLSLSP
GEGGGSGGGGSQVQLQESGPGLVI(PSETISLICIVSGGSVSSGDYYTNIKSPGICCLETIG
H I IN SliNTWNPSI AS121,1' I SI IVISKTQFSI AISSVIAADIA I YYCV111)RYI1A111) I
W(1.4(;111
VTVSSGGCGSGOGGSGGGGSGCZSi DlOUQSPSSLSASVGDRVTITOQA93DISMYLNITQQ
KPCICAPKLIYDASNLEIMPSRFSGSCSGTIFTFTISSLQPEDIATINCQHFDHLPLAFCCG
EVQLVESGGGLVQPGGSLRL5CAA%1ITSDYYI NFIVRQAPGKGLE111GEIFFPIGNSHVVSR
FIGRATLSADIGSKNTAYLQMNSLRAEDTAVYYCAGGDYGFLYWGQGTINIVSSASTIMPSVFP
LAFSSKSTSGGTAALGCLVICDYFPEPVINSWNSGALT%VHTFPAVLQ5SGLYSISSVVMS
SSLORYICNINIIKPUTICVD1H(VERKCCVFMCPAPHILGCIPSIRFPRP111111)41SRT
V elkiVh1-
1NAILIKYNZifile,VVSVLIVLHQUWLIkblinT
lafiC4 A1173 x Vec t i i x
KaCVSNKAIPAPIEKTISKAKGQPRITQWILMRDELTIDIQVSLTCLVEGFYPSDIAVEE
329
scFv heavy chain
SNGQPENNYKTIPPV1MGSFFLYSICLIVDKSR1/QQGNVFSCSVIIHEALHNHYTQICSLS1SP
GEGGGSGGGGSQVQLQESGPGLVICPSEMSLTUVSGGSVSSGDYYRTIVIRCSPGI(CLEVIIG
III YYSGhT)11(NPSLKSRLT SI DTSICTQFSLICISSVTAADTA I YYCVRDRV'TGAFDINCQG111
VITSSGGGGSOCCIGSCAGGSGGGGSDIOITQTSS,SASVGDRVTIXQASQDISNYLAYQQ
OGICAPKLAYDASALEMVPSITSGSGSGuartP II SWIPED IKIYFCQHFDILPLAFGCG
TKVE I K
[258] Amino az itls sequence (continue) i nue )
SR) It) NO8
EVQIXESCAGLVQPGGSLRLSCA.ASGYTFSDYYINWITQAPAGLIMIGEIFPGCNSHVVR
FKGRATISADVINTAYLWISI.RAEDTAVYKAGQDYGFINIVa)G71XFVSSASTKGPSVFP
LAPSSKS1114.LAALGCLVIDYF?EPVTVSPISGALTSGVIIMPAUMLYSLSSVVIVPS
SSLGTQTYICNVNHXPSITIVDEVERKCCVMPPCPAPELLGGPSIFIIPPKPICDTLII SIT
PEVICVVVDVSHEDPEVIFRYDGVEVIINAKIICPREEMSTYRWSVLTVLHINKNGKEY
hu3C4 A1185 x Vec t it ix
KCIVSNKALPAPIFKTISKAKCQ?REPQNYTIPPSRDELTRIQVSITCLVEFYPSDI AVER
330
scFv heavy jain
SNGQPENNYHTTPPVLDSDGSFF:,YSFITVDIERWQQGNVFSCSITHALINHYTQLSESLV
GICGCCGSGGGGSQVQLQESGPCLVI(PSEILSLICTVSCCSVSSGDYYITIVIRQSPOECLERG
HIYY SGNTNYNPSLKSRLTI SI DTSKTQFSLICISSVTAADTA 1 YYCIRDRVTGAFDINGQG111
VrYSSCGGGYJCGCSGCGGSGCCGSDIQIITQSPSSLSASVGDRVTITCQASWISNYLN1/YQ:1
ITGKAPPILLI YDASNLETGY? SRFSGSGSAUt. In ISSLEDIA1YFIHFDHLPLAFGCG
TUE I
[259] [Table 15]
CI 09061704 2010-10-20
WO 2018/221969
PCT/KR2018/006182
56
[260] List of combined variable region sequences of bispecific antibody
Heavy chain variable Light chain variable
region region
light chain variable
hu8C4 x Erbitux scFv HL
hu8C4 x Erbitux scFv HL region of hu8C4-1
(SEQ ID NO: 316)
antibody (SEQ ID NO: 21)
light chain variable
hu8C4 A1171 x Erbitux scFv hu8C4 AH71 x Erbitux scFv
region of hu8C4 -1
HL HL (SEQ ID NO: 317)
antibody (SEQ ID NO: 21)
light chain variable
hu8C4 AH85 x Erbitux scFv hu8C4 A1185 x Erbitux scFv
region of hu8C4 -1
HL HL (SEQ ID NO: 320)
antibody (SEQ ID NO: 21)
hu8C4 AL194 x Erbitux scFv hu8C4 x Erbitux scFv HL
AL194(SEQ ID NO: 310)
IL (SEQ ID NO: 316)
hu8C4 A56 x Erbitux scFv hu8C4 A1185 x Erbitux scFv
AL165(SEQ ID NO: 308)
HL HL (SEQ ID NO: SAO
hu8C4 A62 x Erbitux scFv hu8C4 AH72 x Erbitux scFv
AL130(SEQ ID NO: 306)
M, HL (SEQ ID NO: 318)
hu8C4 A71 x Erbitux scFv hu8C4 A1173 x Erbitux scFv
AL135(SEQ ID NO: 307)
IL HL (SEQ ID NO: 319)
hu8C4 A72 x Erbitux scFv hu8C4 AH73 x Erbitux scFv
AL165(SEQ ID NO: 308)
HL HL (SEQ ID NO: 319)
hu8C4 A73 x Erbitux scFv hu8C4 AH73 x Erbitux scFv
AL166(SEQ ID NO: 309)
HL HL (SEQ ID NO: 319)
hu8C4 A76 x-E-rbitux scFv hu8C4 AH73 x Erbitux scFv
AL195(SEQ ID NO: 311)
HL HL (SEQ ID NO: 319)
Ch 03061704 2010-10-20
WO 2018/221969
PCT/ICR2018/006182
57
U261]
hu8C4 A78 x Erbitux scFv hu8C4 A1171 x Erbitux scFv
4L130(SEQ ID NO: 306)
HL HL (SEQ ID NO: 317)
light chain variable
hu8C4 x Erbitux scFv LH
hu8C4 x Erbitux scFv LH region of hu8C4-1
(SEQ ID NO: 321)
antibody (SEQ ID NO: 21)
--light chain variable
hu8C4 A1171 x Erbitux scFv hu8C4 AH71 x Erbitux scFv
region of hu8C4 -1
111 LH (SEQ ID NO: 322)
antibody (SEQ ID NO: 21)
tight chain variable
hu8C4 A1185 x Erbitux scFv hu8C4 A1185 x Erbitux scFv
region of hu8C4 -1
LH LH (SEQ ID NO: 325)
antibody (SEQ ID NO: 21)
hu8C4 AL194 x Erbitux scFv hu8C4 x Erbitux scFv LH
4L194(SEQ ID NO: 310)
I.H (SEQ ID NO: 321)
hu8C4 A56 x Erbitux scFv hu8C4 A1185 x Erbitux scFv
AL165(SEQ ID NO: 308)
LH LH (SEQ ID NO: 325)
hu8C4 A62 x Erbitux scFv hu8C4 A1172 x Erbitux scFv-
AL130(SEQ ID NO: 306)
LH LH (SEQ ID NO: 323)
hu8C4 A71 x Erbitux scFv hu8C4 A1173 x Erbitux scFv
4L135(SEQ ID NO: 307)
LH LH (SEQ ID NO: 324)
hu8C4 A72 x Erbitux scFv hu8C4 AH73 x Erbitux scFv
AL165(SEQ ID NO: 308)
LH LH (SEQ ID NO: 324)
hu8C4 A73 x Erbitux scFv hu8C4 AH73 x Erbitux scFv
AL166(SEQ ID NO: 309)
UI LH (SEQ ID NO: 324)
hu8C4 A76 x Erbitux scFv hu8C4 A1173 x Erbitux scFv
AL195(SEQ ID NO: 311)
LH LH (SEQ ID NO: 324)
hu8C4 A78 x Erbitux scFv hu8C4 A1171 x Erbitux scFv AL130(SEQ ID NO: 306)
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
58
[262] LH LH (SEQ ID NO: 322)
light chain variable
hu8C4 x Vectibix scFv
hu8C4 x Vectibix scFv region of hu8C4-1
(SEQ ID NO: 326)
antibody (SEQ ID NO: 21)
light chain variable
huSC4 11171 x Vectibix
hu8C4 AH71 x Vectibix scFv region of hu8C4 -1
scFv (SEQ ID NO: 327)
antibody (SEQ ID NO: 21)
light chain variable
hu8C4 AH85 x Vectibix
hu8C4 A1185 x Vectibix scFv region of hu8C4 -1
scFv (SEQ ID NO: 330)
antibody (SEQ ID NO: 21)
hu8C4 AL194 x Vectibix hu8C4 x Vectibix self
AL194(SEQ ID NO: 310)
scFv (SEQ ID NO: 326)
hu8C4 A1185 x Vectibix
hu8C4 A56 x Vectibix scFAr AL165(SEQ ID NO: 308)
scFv (SEQ ID NO: 330)
hu8C4 A1172 x Vectibix
hu8C4 A62 x Vectibix 3cFY AL130(SEQ ID NO: 306)
scFv (SEQ ID NO: 328)
hu8C4 AH73 x Vectibix
hu8C4 A71 x Vectibix scFv AL135(SEQ ID NO: 307)
scFv (SEQ ID NO: 329)
hu8C4 A1173 x Vectibix
hu8C4 A72 x Vectibix scFv AL165(SEQ ID NO: 308)
scFv (SEQ ID NO: 329)
hu8C4 A1173 x Vectibix
hu8C4 A73 x Vectibix scFv AL166(SEQ ID NO: 309)
scFv (SEQ ID NO; 329)
hu8C4 AH73 x Vectibix
hu8C4 A76 x Vectibix scFv AL195(SEQ ID NO: 811)
scFv (SEQ ID NO: 329)
hu8C4 AH71 x Vectibix
hu8C4 A78 x Vectibix scFv AL130(SEQ ID NO: 306)
scFv (SEQ ID NO: 327)
[263] Then, an in vitro anticancer efficacy of a bispecific antibody
linking Erbitux and
Vectibix scFv fragments was evaluated in a U-87 MG tumor cell line by the same
method as shown in Example 1.
[264] Also, a tumor cell proliferation inhibitory activity was evaluated by
using NCI-
H1993, NCI-H292 and NCI-H820 lung cancer cell lines. Particularly, with regard
to an
NCI-H1993 (ATCC, #CRL-5909) cell line with c-Met gene overexpressed therein,
an
NCI-H292 (ATCC, #CRL-1848) cell line with EGFR and c-Met normally expressed
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
59
therein, and NCI-H820 (ATCC, #HTB-181) with threonine (T) mutated into me-
thionine (M) in EGFR amino acid no. 790, a tumor cell proliferation inhibitory
activity
was performed by the following method. Each cell line was diluted in an RPMI-
1640
medium (Gibco, #A10491) containing 10% (v/v) FBS, after which the resulting
cell
lines were divided by 2.0 X 103 into each well of a 96-well plate, such that
the
resulting plate was cultured overnight under 37 C, 5% CO2 conditions. Then,
each well
of the plate was replaced with 100 a of a serum-free medium, after which the
resulting plate was cultured under 37 C, 5% CO, conditions for 18 hours. After
that,
the medium was replaced with 100 ite of the RPMI- 1 640 medium containing 2%
(v/v)
PBS or HGF 50 ng/ml, after which a test antibody was sequentially diluted at a
ratio of
1/10 (i.e., 100 nM, 10 nM, 1 nM, 100 pM, 10 pM and 1 pM) to reach 0.001 nM at
a
final concentration of 100 nM, such that the resulting antibody was added by
100 ge
into each well. Subsequently, the plate was cultured for 5 days under 37 C, 5%
CO2
conditions, after which the medium was removed therefrom, such that a TCA
solution
was inserted by 200 fit into each well to fix cells. Also, the cells of the
plate were dyed
according to a conventional SRB colorimetrie assay method, after which an
optical
density of each well was measured at a wavelength of 540 nm by using a
microplate
reader.
[265] Results of proliferation inhibitory activity in each cell line above
are shown in Tables
16 and 17 and FIGS. 4 and 5.
[266]
[267] [Table 16]
Ch 09061704 2010.40.10
WO 2018/221969
PCT/KR2018/006182
[268] In vitro tumor cell proliferation inhibitory activity by bispecific
antibody
Cell proliferation inhibition assay, TC50
(nM)
Bispecific antibodies U-87 MG
--
NCI-H1993
(GBM, IIGF
(NSCLC, c -Met amplified)
autocrine)
hu8C4 x Vectibix scFv. 0.06 0.32
hu8C4 A1171 x Erbitux scFv
0.06 0.41
HL
hu8C4 A1185 x Erbitux scFv
0.06 0.48
HL
hu8C4 AL194 x Erbitux scFv
0.07 0.64
HL
hu8C4 A56 x Erbitux scFv
0.07 0.57
HL
hu8C4 A62 x Erbitux scFv
0.08 0.65
HL
hu8C4 A70 x Erbitux scFv
0.07 0.67
HL
hu8C4 A72 x Erbitux scFv
0.06 0.49
HL
hu8C4 A73 x Erbitux scFv
0.06 0.50
ML
hu8C4 A76 x Erbitux scFv
0.06 0.49
ML
bu8C4 A78 x Erbitux scFv
0.06 0.76
HL 1
[269] [Table 17]
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
61
[270] In vitro lung cancer cell line proliferation inhibitory activity by
bispecific antibody
Cell proliferation inhibition assay, IC50
(nM)
NCI-H820
Bispecific antibodies NCI-11292
(NSCLC EGFR 1790M,
(NSCLC)
c-Met amplified)
no HGF HGF 50ng/m1 no HGF HGF 50ng/m1
hu8C4 x Vect ibix scFv 0.70 0.24 > 100 4.2
hu8C4 Af171 x Erbitux seFv-
. 51 0.22 > 100 8.5
HL
Ini8C4 A1185 x Erbitux scFv
0.43 0.23 > 100 7.6
hu8C4 AL164 x Erbitux scFv
0.41 0.24 > 100 19.0
HL
hu8C4 A56 x Erbitux scFv
0.42 0.29 > 100 21.7
ilL
hu8C4 A62 x Erbitux scFv
0.74 0.28 > 100 40.2
lll
hu8C4 A10 x Erbitux say
0.74 0.23 > 100 40.9
hu8C4 A72 x Erbitux scFv
0.78 0.23 > 100 19.5
HL
hu8C4 A73 x Erbitux scFv.
0.87 0.26 > 100 38.4
HL
_
hu8C4 A76 x Erbitux scFv
0.73 0.21 > 100 10.3
Ills
[271] In result, there was no difference in efficacy between bispecific
antibodies prepared
from U-87 MG tumor cell line by the method and it was identified that an
activity in-
hibitory efficacy thereof was about 15 times more excellent than IC50 of hu8C4
optimized antibody. Also, as a result of evaluating a tumor cell proliferation
inhibitory
activity using NCI-H1993, NCI-H292 and NCI-H820 lung cancer cell lines, it was
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
62
identified that there was no difference in efficacy between bispecific
antibodies
prepared.
[272] Such the results suggest that the antibody of the present invention
has a proliferation
inhibitory effect on all the cancer types regardless of an overexpression or
mutation of
c-Met and EGFR, thus may be effectively used in these cancer types.
[273]
[274] Example 7. Comparative evaluation of in vitro tumor cell
proliferation in-
hibitory activity of bispecific antibody compared to combined therapy
[275]
[276] Eight types of cancer were used to compare a tumor cell proliferation
inhibitory
activity between a combined therapy of each antibody targeting c-Met and EGFR
re-
spectively and the bispecific antibody of the present invention.
[277] Particularly, a tumor cell proliferation inhibitory activity was
evaluated in a lung
cancer cell line NCI-H292 (ATCC, #CRL-1848), an HGF-autocrinal glioblastoma
cell
line U-87 MG (ATCC, #HTB-14), lung cancer cell lines NCI-H1648 (ATCC
#CRL-5882) and NCI-H596 (ATCC #HTB-178), HCC827 (ATCC, #CRL2868), a
colon cancer cell line LS174T (ATCC, #CL-188), a triple negative breast cancer
(TNBC) cell line BT20 (ATCC, #HTB-19) and a pancreatic cancer cell line KP4
(JCRB, #RCB1005). The NCI-H1648 cell line is characterized by a normal
expression
of EGFR and c-Met, the NCI-H596 cell line is characterized by a deletion of
some
sequence of exon no. 14 of MET acne, and the HCC827 cell line is characterized
by a
deletion of some sequence of exon no. 19 of EGFR gene. Also, the LS174T cell
line
has a KRAS mutation and the KP4 is characterized by autocrining HGF.
[278] The U-87 MG cell line was evaluated by a method of Example 1 and the
NCI-H292
cell line was evaluated by a method of Example 6. Also, the NCI-H1648, NCI-
11596
and HCC827 cell lines were diluted in an RPM1-1640 medium (Gibco, #A10491)
containing 10% (v/v) FBS, after which the resulting cell lines were divided by
2.0 X
103 in each well of a 96-well plate. The LS174T cell line was diluted in a
DMEM
medium (Gibco, #11995-065) containing 10% (v/v) FBS, after which the resulting
cell
lines were divided by 2.0 X 103, The BT20 cell line was diluted in an EMEM
medium
(ATCC, #30-2003) containing 10% (v/v) FBS, after which the resulting cell
lines were
divided by 3.0 X 103. And, the KP4 cell line was diluted in an RPMI-1640
medium
(Gibco, #A10491) containing 10% (v/v) FBS, after which the resulting cell
lines were
divided by 1.5 X 103, such that the resulting plate was cultured overnight
under 37 C,
5% CO2 conditions. Then, each well of the plate was replaced with 100 p1 of a
serum-
free medium, after which the resulting plate was cultured under 37 C, 5% CO2
conditions for 18 hours. After that, the medium was replaced with 100 gi of
the RPMI-
1640 medium containing 2% (v/v) FBS or HGF 50 ng/ml, after which a test
antibody
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
63
was sequentially diluted at a ratio of 1/10 (i.e., 100 nM, 10 nM, 1 nM, 100
pM, 10 pM
and 1 pM) to reach 1 pM at a final concentration of 100 nM, such that the
resulting
antibody was added by 100 Lte into each well. Then, the plate was incubated
for 5 days
under 37 C, 5% CO2 conditions, after which the medium was removed therefrom,
such
that a TCA solution was inserted by 200 id into each well to fix cells. Also,
the cells of
the plate were dyed according to a conventional SRB colorimetric assay method,
after
which an optical density of each well was measured at a wavelength of 540 nrn
by
using a microplate reader.
[279]
[280] Results of this Example are shown in Tables 18 to 21 and FIGS. 6 to
8.
[281]
[282] [Table 18]
[283] Comparative evaluation of in vitro tumor cell proliferation
inhibitory activity between
combined therapy and bispecific antibody in U-87 MG and NC1-11292 cell lines
Cell proliferation inhibition assay, IC5.3 (nM)
U-87 MG NCI-11292 (NSCLC)
Antibodies
(GBM, HGF
No HGF HGF 50 mg/m1
autocrine)
Vectibix >100 0.09 >100
hu8C4 83.9 >100 >100
hu8C4 + Vectibix
79.0 0.10 0.34
combined
hu8C4 x Vectibix scFv 0.4 0.15 0.12
C-EM1-MAb > 100 5.29 5.73
C-LA480 858,8
C-0A-51)5 171.9
C-AbF46 > 100
[284] [Table 19]
CA 03061704 2019-10-28
WO 2018/221969
PCT/K1R2018/006182
64
[285] Comparative evaluation of in vitro tumor cell proliferation
inhibitory activity between
combined therapy and bispecific antibody in NCI-H1648 and NCI-H596 cell lines
Cell proliferation inhibition assay. ICso (nM)
NCI -H1648 NCI-11596
Antibodies
(NSCLC) (NSCLC, c -Met mutated)
No HGF HGF 50ng/mi No HGF HGF 50ng/m1
Vectibix > 100 > 100 > 100 > 100
hu8C4 > 100 > 100 > 100 2.3
hu8C4 + Vectibix
>100 >100 >100 2.4
combined
hu8C4 x Vectibix scFv 15.4 29.5 > 100 0.4
[286] [Table 20]
1287] Comparative
evaluation of in vitro tumor cell proliferation inhibitory activity between
combined therapy and bispecific antibody in LS174T, BT20 and KP4 cell lines
Cell proliferation inhibition assay, Ka (nM)
LS174T BT20 KP4
Antibodies (Colon, KRAS G12V) (TNBC) (Pancreas)
HGF
HGF 5Ong/m1 HGF 50ng/m1
autocrine
Vectibix > 100 > 100 > 100
hu8C4 > 100 > 100 42.0
hu8C4 + Vectibix
34.4 > 100 36.4
combined
hu8C4 x Vectibix scFN, 33.4 - 100 27.0
C-EM1-MAb > 100 > 100
1288] [Table 21]
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
[289] Comparative evaluation of in vitro tumor cell proliferation
inhibitory activity between
combined therapy and bispecific antibody in HCC827 and NCI-H596 cell lines
Cell proliferation inhibition assay, IC5o
(ral)
NCI-H596
HCC827
Antibodies (NSCLC, c-Met
(NSCLC, EGFR mutated)
mutated)
HGF 50
No HGF HGF 50 ng/ml
ng/ml
Tarceva 2.96 > 100 > 100
Yectibix > 100 > 100 > 100
hu8C4 > 100 > 100 67.2
hu8C4 x Vectibix scFv > 100 > 100 0.8
LA480 >100 >100 >100
INC280 > 100 > 100 42.5
MD1214063 > 100 > 100 68.2
Xalkori 87.3
Tarceva + hu8C4 combined 3.24 3.09
Tarceva + hu8C4 x Vectibix scFv
2.35 2.42
combined
Tarceva + LA480 combined 3.24 4.78 -
Tarceva + ING280 combined 3.06 2.88
Tarceva + U91214063 combined 2.80 4.10
[290] In result, it was identified that a tumor cell proliferation
inhibitory capacity of the
bispecific antibody of the present invention was more excellent than that of
hu8C4,
Vectibix or a combined therapy of two antibodies in the 8 kinds of tumor cell
line all.
Also, it was identified that it had a remarkably excellent tumor cell
proliferation in-
hibitory capacity in U-87MG, NC1-H292, BT20 and 104 cell lines when compared
to
EMI-MAb (Janssen) used as a control bispecific antibody.
[291] Moreover, it was identified that both hu8C4 and hu8C4 x Vectibix scFv
had an
excellent tumor cell proliferation inhibitory capacity compared to a control
antibody,
when compared to LA480 (Lilly), 0A-5D5 (Genentech) and AbF46 (Samsung), which
were c-Met target antibodies in U-87MG cell lines.
[292] Also, Tarceva, an EGFR tyrosine kinase inhibitor in HCC827 cell line,
showed re-
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
66
sistance under HGF processing conditions, but it was identified that it showed
an
excellent tumor cell proliferation inhibitory capacity when being processed in
com-
bination with Tarceva, hu8C4, hu8C4 x Vectibix scFv or c-Met inhibitors under
such
conditions.
[293] Also, as a result of comparing various EGFR inhibitors and c-Met
inhibitors in NCI-
H596 cell line, it was identified that a tumor cell proliferation inhibitory
capacity of
hu8C4 x Vectibix scFv was excellent compared to EGFR or c-Met single target
drug.
[294]
[295] Example 8. Measurement of binding capacity to ECD (BIAcorel
[296]
[297] Then, to measure a binding capacity of the c-Met antibody of the
present invention to
an extracellular domain (ECD), binding of c-Met antibody and bispecific
antibody to
c-Met ECD and EGFR ECD was measured between human and cynomolgus monkey
by using BIAcore.
[298] Particularly, a human c-Met ECD (ACROBiosystems, MET-H5227), a
cynomolgus
monkey c-Met ECD (SiNo. Biological, 90304-CO8H), a human EGFR ECD strep
(ACROBiosystems, EGR-H5285) and a cynomolgus monkey EGFR ECD (SiNo. Bi-
ological, 90285-008B) were used.
[299] First of all, to capture an anti-c-Met antibody and a bispecific
antibody, an Pc-
specific anti-human IgG antibody (SouthernBiotech, 2047-01) was fixed to a CM5
sensor chip in the level of 10000 RU. The antibodies were diluted in HBS-EP
buffer
(0.01 M HEPES pH 7.4, 0.15 M NaC1, 3 mM EDTA and 0.005% (v/v) Surfactant P20)
at a concentration of 1 - 2 ig/ml, after which the resulting antibodies were
injected into
a CM5 chip with an anti-human Tg Fc fixed thereto at a flow rate of 30 fit/min
for 10 -
120 seconds, and then captured in a range of 150 - 200 RU. Each antigen was
used
after being diluted at 10, 5, 2.5, 1.25, 0.625, 0.3125 and 0.15625 nM, after
which the
resulting antigens were sequentially injected from a lower concentration.
Then, the
resulting antigens were injected at a flow rate of 30 tt(/min for 5 minutes to
carry out
binding, after which a running buffer was injected thereinto for 10 - 15
minutes to
carry out a dissociation. 15 id of 10 mM Glycine-HC1 (pH 1.5) was used to
revive the
chip. A binding and dissociation speed for each cycle was evaluated by using a
"1:1
Langmuir binding" model in BIAevaluation software version 4.1, and biacore
data are
summarized in Tables 22 and 23.
[300]
[301] [Table 22]
09061704 2010-10-20
WO 2018/221969 PCT/KR2018/006182
67
[302] Measurement of affinity to c-Met ECD
Binding Dissociation
Affinity to antigen
hu8C4 constant constant
11)
(4,, 1/Ms) (tat, 1/s)
Human c-Met 6.77 x 105 2.148 x 10-4 3.173 x
1(119
Cynomolgus monkey c-
7.467 x 105 3.447 x 10-4 4.616 x 10
Met
Binding Dissociation
Affinity to antigen
hu8C4 AH71 constant constant
N)
Oion, 1/Ms) (kou, 1/s)
Human c-Net 8.306 x 105 8.301 x 10-5 9.993 x
10.11
Cynomolgus monkey c-
Met
_
Binding Dissociation
Affinity to antigen
hu8C4 x Voctibix say constant constant
ao, M)
(kon, 1/Ms) (kadf, 1/s)
Human c-Met 7.339 x 1CP 2.041 x 10-4¨ 2.78 x
Cynomolgus monkey c-
7.77 x 105 3.37 x 10-4 4.338 x 10-m
Met
13031 [Table 231
[304] Measurement of affinity to E,GFR ECD
Binding Dissociation
Affinity to antigen
Vectihix constant constant
(Kn, N)
1/Ms) (coif, 1/s)
Human EGFR 5.278 x 105 1.5 x 10-4 2.841 x 10119
CYnomolgms monkey EGET' 9.37 x 105 1.963 x 10F4 2.095 x 1(110
Binding Dissociation
Affinity to antigen
hu8C4 x Vectibix scIN constant constant
N)
(kon, 1/Ms) (koff, 1/s)
Human ER 7.776 x 104 1.257 x 10-4 1.617 x 10-
9
Cynomolgus monkey EGFR 1.424 x 105 1.274 x 10-4 8.942 x
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
68
[305] The data were used to prove that the hu8C4, hu8C4 x Vectibix scFv
bispecific an-
tibodies of the present invention bind to c-Met ECD of human and cynomolgus
monkey with an excellent affinity.
[306]
[307] Example 9. Measurement of c-Met antibody binding capacity to c-Met
ECD.
EGFR ECD between various animal species (ELISA)
[308]
[309] Binding of c-Met antibody and bispecific antibody to c-Met ECD and
EGFR ECD
between mouse, cynomolgus monkey and human was measured by using ELISA.
[310] Particularly, mouse c-Met (SiNo. Biological Inc, 50622-MO8H),
cynomolgus
monkey c-Met (SiNo. Biological Inc, 90304-CO8H), human c-Met (R&D Systems,
358-MT), mouse EGFR (SiNo. Biological Inc, 51091-MO8H), cynomolgus monkey
EGFR (SiNo. Biological, 90285-008B) and human EGFR (Abcam, 155639) antigens
were all divided into a 96-well plate at a concentration of 2 /Wm], after
which the
resulting plate was reacted at 4 C overnight. After being blocked at room
temperature
for 1 hour, hu8C4 x Vectibix scFv bispecific antibody was sequentially diluted
at a
ratio of 1/5 from 100 nM to measure its binding capacity in 7 concentration
sections
(i.e., 100 nM, 20 nM, 4 nM, 800 pM, 160 pM, 32 pM and 6.4 pM).
[311] After binding the hu8C4 x Vectibix scFv bispecific antibody at room
temperature for
1 hour, anti-human IgG, F(ab')2 fragment specific-HRP conjugated antibody
(Jackson
Immunoresearch, 109-035-097) was diluted at a ratio of 1: 2500, after which
the
resulting antibody was reacted at room temperature for 1 hour. Color
development was
made by using TMB (Sigma, T4444) solution, wherein its value was measured at
an
optical density of 450 nm and its ELISA results are shown in FIG. 9.
[312] In result, it was identified that hu8C4 monospecific antibody and
hu8C4 x Vectibix
scFv bispecific antibody did not bind to a mouse c-Met and a mouse EGFR, but
bind to
monkey and human c-Mets and EGERs. Also, it was identified that a human IgG
antibody, used as a negative control group, did not bind at all. The results
above
suggest that the c-Met antibody of the present invention is specific only to
human and
monkey c-Mets and EGFRs.
[313]
[314] Example 10. Cross-reactivity of c-Met antibody to various receptors
on the
surface of cells
[315]
[316] Specificity of hu8C4 antibody specifically binding to c-Met according
to the present
invention as well as its cross-reactivity to other receptor tyrosine kinase
antigens were
analyzed by an indirect ELISA method, and 5 antigens of FGF R3, VEGFR R2, IGF
IR, PDGF R and RON were selected out of key receptor tyrosine kinases to
perform an
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
69
analysis.
[317] In this Example, human c-Met Fc chimera (R&D systems, 358-MT_CF),
human FGF
R3 (111c) Fc chimera (R&D systems, 766-1-R), human IGF-IR (R&D systems,
391-GR-050), human PDGF R1.3 Fc chimera (R&D systems, 385-PR_CF), human
VEGF R2 Fc chimera (R&D systems, 357-KD_CF) and human MSP R/Ron (R&D
systems, 1947-MS-050) were used as an antigen.
[318] Each antigen was diluted in 0.05 M carbonate-bicarbonate (Sigma,
C3041) buffer at
a concentration of 1 ,ug/ml, after which the resulting antigen was added into
each well
of a 96-well plate (Corning, #2592), such that the resulting plate was coated
at 4 C
overnight. The plate was washed once with TBS-T, after which TBS-T containing
4%
- skim milk was added by 200 a into each well of the resulting plate in order
to inhibit
a non-specific binding, such that the resulting plate was reacted at 37eC for
1 hour.
Then, the plate was washed once with TBS-T buffer, after which a primary
antibody
was sequentially diluted in TBS-T buffer containing 2% - skim milk from a
highest
concentration of 30 nM to 0.002 nM, such that the resulting antibody was added
by
100 a into each well, thus being reacted at 37 C for 2 hours. After being
washed three
times with TBS-T buffer, an anti-human kappa light chains-peroxidase (Sigma,
A7164) was diluted at a ratio of 1: 5000 as a secondary antibody, after which
the
resulting antibody was added by 100 g12, into each well, thus being reacted at
37 C for 1
hour. Then, after being washed three times with TBS-T buffer, TMB solution
(Sigma,
__________ was added by 100 a into each well to carry out an color developing
reaction,
after which 2 N ammonium sulfate solution was added by 50 p...e into each well
to stop
the reaction. An optical density was measured based on a value at a wavelength
of 450
nm by using a microplate reader and a reference wavelength of 570 nm was used.
A
degree of binding of an anti-c-Met antibody to each antigen was proportionate
to an
optical density signal value, wherein results thereof are shown in Table 24.
[319]
[320] [Table 24]
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
[321] Binding specificity of anti-c-Met antibody hu8C4 to various antigens
" hU8C4 binding (A4b-onfli - A5701.)
Ab.conc.
c-Met IGF-IR RON PDGFR VEGFR2 FGFR3
(nM)
30.000 -2.55 2.51 0.00 0.00. 0.0010.00 0.01 0.01 0.00 0.01 0.01 0.02 "
6.000 1.96 2.03 0.00 0.00- 0.00 0.00 -0.01 -0.01-0.01-0.01 0.00 0.01
1.200 1.81 1.74 0.00 0.00 0.00 0,00 -0.01 -0.01-0.01-0.01 0.00 0,01
0.240 1.48 1.54 0.00 0.00 0.00 0.00 -0.01 -0.01-0.02-0.02 -0.01 - 0.00
0.048 0,76 0.76 0.00 0.00 0.00 0.00 -0.01 -0.01 -0.02 -0.01 0.00 0.00
0.010 0.21 0.20 0.00 0.00 0.00 0.00 -0.01 -0.01 -0.01--0.01 0.00 0.00
0.002 0.05 0.05 0.00 0.00 0.00 0.00 -0.01 -0.01 -0.01 -0.01 0.00 0.00
Blank 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0000.00 0.00 0.00
[322] As seen in Table 24, the 1. u8C4 antibody of the present invention
preferentially binds
to c-Met, and it was identified that it did hardly bind to other antigens of
FGF R3,
VEGFR R2, IGF IR, PDGF R and RON.
[323]
[324] Example 11. In vitro internalization activity of c-Met antibody and c-
Met level
inhibitory activity of bispecific antibody
[325]
[326] It was identified that the c-Met antibody of the present invention
had an in vitro in-
ternalization activity in tumor cells as well as an effect on reducing a
receptor level by
a bispecific antibody capable of simultaneously binding to c-Met and EGFR.
[327] First of all, an antibody internalization occurs by a physiological
activity of a normal
receptor, wherein, when binding to a specific ligand, the receptor normally
expressed
outside cells becomes activated through a homo- or hetero- dimcrization and
causes a
receptor-mediated endocytosis. An antibody specific to a receptor of a cell
has a
capacity to induce such phenomenon and is internalized into the cell by
causing the en-
docytosis, thus inducing a decomposition of the receptor, reducing a degree of
ex-
pression thereof, and possibly inhibiting a signal transduction by a certain
receptor. An
amount of antibodies bound outside cells may be detected by using a
fluorescence-
activated cell sorting (FACS) device, thus finding an amount of antibodies
internalized
inside the cells. In case of binding antibodies by using an antibody with FITC
binding
to an anti-human kappa LC as a secondary antibody for a light chain of an
antibody to
be measured, it is possible to quantitatively measure an amount of antibodies,
which
are not internalized, but remain binding to a target receptor outside cells,
thus
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
71
identifying an amount of internalized antibodies accordingly. It is possible
to measure
a background signal by a non-specific binding of an antibody used in a test by
using a
human IgG antibody, non-specific to an antigen, thus measuring a fluorescent
signal by
an actual specific binding.
[328] In this Example, a MKN45 cell line (#JCRB0254), which was a stomach
cancer cell
line, was used to identify an in vitro internalization activity of c-Met
antibody inside
tumor cells. MKN45 expresses a c-Met receptor at a high level by amplification
of
MET gene, such that a phosphorylation of the c-Met receptor is induced in an
HGF-
nondependent way. A test was performed as follows to see if a c-Met receptor
is in-
ternalized into a cell by an anti-c-Met antibody hu8C4, thus reducing a level
of ex-
pression.
[329] First of all, MKN45 stomach cancer cell lines were divided by 5.0 x
105 into each
well of a 6-well plate containing an RPMI-1640 medium (2 ml) containing 10%
(v/v)
FBS, after which the plate was cultured under 37 C, RH 95% and 5% CO2
conditions
for 24 hours. An anti-c-Met antibody to be analyzed as well as an anti-IgG
antibody
(control group) were diluted to reach a final concentration of 100 nM, after
which the
resulting antibodies were reacted overnight. As a plate to be used as a non-
internalized
control group was treated as an anti-c-Met antibody and a human IgG antibody
(control group), after which the resulting plate was reacted at 4 C for 1
hour. Then,
cells of each well were collected with 1 ml of an enzyme-free cell
dissociation buffer
(Gibco, #13151), after which the collected cells were washed twice with a cold
PBS.
As a secondary antibody, anti-human kappa LC-FITC (LSBio #LS-C60539) was
diluted at a ratio of 1: 2000, after which the resulting antibody was added
thereinto,
thus being reacted at 4 C for 1 hour. Then, the cells were washed twice with
PBS, after
which the resulting cells were fixed with 100 ge of BD Cytofix (BD, #554655)
and
washed once with PBS, such that an FITC geo-mean (MR) value, a degree of flu-
orescent staining, was measured by using a BD FACS Canto II parenchymatous
cell
analyzer. An amount of antibodies bound outside cells was obtained by a
following
formula, wherein results thereof are shown in Table 25.
[330]
[331] Surface bound Ab(%) = RMF11370c expd MFI[IgG control]) I (MFIPT
contron MFILigG control j)j X
100
[332]
[333] [Table 25]
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
72
[334] Measurement of internalization of hu8C4 and OA-5D5 control antibodies
to MKN45
stomach cancer cell line
Antibody OA-5D5 hu8C4
FITC MFI [IgG
127 127
control]
FITC MEI [4 C
1763 1444
control]
FITC MFI [37 C exp.] 1724 858
Surface bound Ab(%) 98 56
[335] As seen in Table 25 above, it can be shown that 0A-5D5, an anti-c-Met
antibody
used as a control group, was hardly internalized into cells, while the hu8C4
antibody of
the present invention was internalized about 40% or more into cells in MKN45
stomach cancer cell line. That is, it is shown that the hu8C4 antibody
remarkably
reduces a level of expression of a c-Met receptor.
[336] Then, a test for measuring a receptor level on NC1-11820 lung cancer
cell line was
performed in order to identify an effect of reducing a receptor level by a
bispecific
antibody capable of simultaneously binding to c-Met receptor and EGFR
receptor. The
NCI-H820 cell line is a cell line suitable for measuring an effect of reducing
a receptor
level by an anti-c-Met x EGFR bispecific antibody, because a c-Met receptor
was
expressed in a level of about 83,000 SABC (specific antibody-binding capacity)
and an
EGFR receptor is expressed in a level of about 74,000 SABC.
[337]
[338] First of all, NCI-H820 cell lines were divided by 1.0 x 105 into each
well of a 6-well
plate with an RPMI-1640 medium (2 ml) containing 10% (v/v) FBS, after which
the
resulting plate was cultured overnight under 37 C, RH 95% and 5% CO2
conditions for
24 hours. Then, it was replaced with a serum-free medium, after which the
resulting
plate was cultured overnight under 37 C, RH 95% and 5% CO2 conditions for 24
hours. Then, an anti-c-Met antibody, an anti-c-Met x EGFR bispecific antibody,
an
anti-EGFR antibody and a human IgG antibody as a control group, which were to
be
analyzed, were diluted and treated in a medium containing 2% - FBS to reach a
final
concentration of 10 nM, after which the resulting antibodies were cultured for
5 days.
After that, cells of each well were collected with 1 ml of an enzyme-free cell
dis-
sociation buffer, after which the collected cells were washed twice with a
cold PBS.
Subsequently, goat F(abs)2 anti-mouse IgG-CSF (R&D Systems Cat.#F0103B) was
added by 10 0, into each well as a secondary antibody, thus being reacted at 4
C for 1
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
73
hour. Then, the cells were washed twice with PBS, after which the resulting
cells were
fixed with 100 ge of BD Cytofix (BD, #554655) and washed once with PBS, such
that
an F1TC geo-mean (MFI) value, a degree of fluorescent staining, was measured
by
using a BD FACS Canto II parenchymatous cell analyzer.
[339] In result, when treating an anti-c-Met antibody hu8C4, an EGFR
receptor was hardly
decreased, but a c-Met receptor was remarkably decreased to a level of 2%
(FIG. 10).
Also, an anti-EGFR antibody Vectibix reduced the EGFR receptor to a level of
about
83%, but a c-Met receptor was hardly decreased. By contrast, in case of ft-
eating the
hu8C4 x Vectibix hi specific antibody of the present invention simultaneously
binding
to c-Met and EGFR receptors, it was identified that the EGFR receptor was
decreased
to a level of about 21% and the c-Met receptor was decreased to a level of
about 4%,
respectively,
[340] Thus, it was identified that the hu8C4 x Vectibix bispecific antibody
of the present
invention remarkably reduced a level of expression of c-Met and EGFR receptors
si-
multaneously.
[341]
[342] Example 12. Identification of c-Met and EGFR in vitro signal
inhibitory activity of
bispecific antibody
[343]
[344] Then, an experiment using an NCI-H820 cell line was performed to
identify an effect
of the bispecific antibody of the present invention on the activity of antigen
and signal
transduction materials.
[345]
[346] First of all, NCI-H820 cell lines were divided into a 6-well plate at
a concentration of
x l0 cells per well, after which the resulting plate was cultured overnight
under
37 C, 5% CO2 conditions, such that it was replaced with a serum-free medium
and
cultured overnight again. An antibody was diluted and treated in a serum-free
medium
at a concentration of 100 nM, after which the resulting antibody was reacted
for 24
hours, such that HGF (G.ibco, PHG0254) and EGF (R&D Systems, 236-EG-200) were
treated at a concentration of 50 ng/ml and 10 ng/ml respectively 15 minutes
before
collecting cells. Then, the cells were dissolved in a dissolution buffer to
carry out a
collection of cells, after which a protein concentration was quantified by
using a Lowry
assay method. 20 fig of protein was loaded onto each well and run in SDS-PAGE,
after
which blotting was performed in a nitrocellulose membrane. After blocking the
membrane, all the primary antibodies were diluted and reacted at a ratio of 1:
1,000,
after which HRP-binding anti-rabbit antibody was diluted at a ratio of 1:
5,000 and
reacted as secondary cells. Then, the antibodies absorbed onto the membrane
were
reacted with enhanced chemiluminescence (ECL), after which the resulting
antibodies
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
74
were measured by using an LC-3000 device.
[347] In result, as seen in FIG. 11, when treating hu8C4 x Vectibix scFv
bispecific
antibody, the EGI-R phosphorylation, Erk phosphorylation and Akt
phosphorylation
were remarkably decreased more than treating hu8C4 or Vectibix antibody alone.
[348] Thus, the hu8C4 x Vectibix scFv bispecific antibody of the present
invention may
reduce an activity of receptor such as EGFR, Erk, Akt, etc., and downstream
signal
transduction substances in NCI-H820 cell line. In result, it is shown that the
antibody
of the present invention shows an efficacy through a signal transduction
inhibition,
[349]
[350] Example 13. Identification of tumor cell proliferation inhibitory
activity in U-87
MG xenograft mouse model
[351]
[352] An experiment was performed representatively by using hu8C4 IgG2 x
Vectibix
scFv in order to identify a tumor cell proliferation inhibitory activity by
the bispecific
antibody of the present invention in an HGF-dependent U-87 MG cell xenograft
model.
[353]
[354] First of all, human glioblastoma U-87 MG cell lines were cultured
under 37 C, 5%
CO2 conditions by using an EMEM (ATCCO 3O2003TM) medium containing L-
glutamine (300 mg/e), 25 mM HEPES, 25 mM NaHCO3, 10 % heat inactivated FBS
and the like. Then, U-87 MG cells were subcutaneously inoculated by 200 fri
into a
flank of a 6 to 8 week-old male athymic nude mouse (Harlan) at a concentration
of 1 x
107 per mouse. After identifying that a tumor volume formed in 25 days after
in-
oculation reached 60 - 130 mm3, a grouping was performed, after which a test
material
was intraperitoneally administered once a week for 4 weeks (total 5 times: 0,
7, 14, 21
and 28 days). The test material was administered 5 mg/kg, and a tumor volume
and a
mouse weight were measured twice a week. For data, a comparison between an
excipient control group and a test material-administered group was generally
verified
by using Student t-test, and a statistical method used was Origin Pro 8.5
program.
"Maximum inhibition %" indicates an inhibition % of tumor growth compared to a
solvent-treated control group.
[355] In result, a group administered with 3.5 mg/kg and 6.8 mg/kg of hu8C4
IgG2 x
Vectibix scFv had a maximum inhibition 96% for a tumor volume compared to a
solvent control group, and a group administered with 1.5 mg/kg thereof had a
maximum inhibition 80%, thus reducing a tumor volume to a significant level
from a
7th day after administration until the final day of the test (p <0.01) (FIG.
12). Also,
when compared to BsAB-01 as a positive control group, the bispecific antibody
of the
present invention reduced a tumor growth to a significant level (p <0.01).
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
[356] Thus, it was identified from results above that the bispecific
antibody of the present
invention remarkably reduced a tumor growth, thus having an excellent
antitumor
efficacy.
[357]
[358] Example 14. Identification of tumor cell proliferation inhibitory
activity in NCI-
H820 xe,nograft mouse model
[359]
[360] NCI-H820 cell line, which is a cell line with threonine (T) of EGFR
amino acid no.
790 mutated into methionine (M) and with a MET gene amplified, is known as a
resistant cell line of AZD9291 (osimertinib, tagrisso), which is a third
generation
EGFR TM (Darren A. E. Cross, ct al., Cancer Discov. 4(9): 1046-1061 (2014)).
An
evaluation was made in an NCI-H820 xenograft mouse model by representatively
using hu8C4 x Ve,ctibix scFv out of the bispecific antibodies of the present
invention,
in order to see a tumor cell proliferation inhibitory activity of the
bispecific antibody in
NCI-H820 cell line having resistance to such EGFR TKI.
[361] Particularly, a mouse used in this Example was a 6-week-old male
mouse (Jackson
Laboratory, STOCK Hgftm1.1 (HGF) Aveo Prkdcscid/J), wherein a mouse HGF gene
was removed therefrom and transformed to express a human HGF gene. The NCI-
H820 (ATCC, #HTB-181) cell line was inserted into a flask for cell culture
along with
an RPMI1640 medium containing 10% FBS, after which the resulting flask was
cultured under 37 C, 5% CO2 conditions. Then, the resulting cells were washed
with
PBS and 2.5% trypsin-EDTA (Gibco, 15090) was diluted 10 times, after which it
was
added thereinto to separate the cells. After that, a centrifugation (1,000
rpm, 5 min.)
was performed to get rid of supernatant and obtain a cell suspension in a new
medium.
Subsequently, a cell viability was identified by a microscope, after which the
resulting
cells were diluted in a serum-free medium at a concentration of 5.0 x
107cellstml, thus
preparing cell lines. The cell lines prepared were subcutaneously administered
into a
mouse by an amount of 0.1 ml/head. After administration, when a tumor size in
a
region with cell lines transplanted thereinto reached about 100 - 150 mm3,
cell lines
were distributed so that a tumor size of each group can be evenly dispersed
according
to a ranked tumor size. Then, oncogenesis was identified twice a week from a
7th day
after starting cell administration until 28th day after a day of grouping (day
of starting
an administration of test material) and after closing an administration of
test material,
after which a tumor's major axis and minor axis were measured by a calipers,
thus cal-
culating a tumor size (ab2/2 (a: a length of major axis, b: a length of minor
axis)). Sta-
tistical analysis was performed by Prism 5.03 (GraphPad Software Inc., San
Diego,
CA, USA). If a p value is less than 0.05, it was judged as statistically
significant.
[362] In result, in all the groups administered with hu8C4 x Vectibix scFv
from a 4th day
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
76
after starting an administration of test material until 28th day thereof, it
was shown that
a tumor proliferation inhibitory activity was significantly higher than a
solvent control
group (p<0.001), and it was also identified that a tumor inhibition ratio
amounted to
maximum100% (FIG. 13). On the other hand, AZD9291 (Selleckchem), used as a
positive control group, did not show a significant difference from the solvent
control
group.
[363]
[364] Example 15. Identification of in vitro tumor cell proliferation
inhibitory activity
by a combined administration of 5G3 c-Met antibody and HER2 antibody
[365]
[366] An in vitro test on cell proliferation inhibitory activity was
performed by NCI-H2170
cell line, in order to evaluate a tumor cell proliferation inhibitory activity
according to
a combination of the anti-c-Met antibody 5G3 of the present invention and anti-
HER2
antibody. NCI-H2170 cell line (ATCC #CRL-5928) is a non-small cell lung cancer
(NSCLC) tumor cell line, wherein, as a result of measuring its receptor level,
EGFR
was expressed in the level of about 2,700 specific antibody-binding capacity
(SABC),
while c-Met was expressed in the level of about 11,000 SABC.
[367] Particularly, NCI-H2170 cells were diluted in an RPIVII-1640 culture
medium
containing 10% (v/v) FBS, after which the resulting cells were added by 100 a
into a
plate at a concentration of 3.0 X 10 cells per well, such that the resulting
plate was
cultured under 37 C, 95% RH and 5% (v/v) CO2 conditions for 18 - 24 hours.
Then,
the cell culture medium of each well was removed therefrom, after which an
RPMI-
1640 medium containing 2% (v/v) FBS was added by 100 a into each well. After
that,
antibodies prepared at 2X of a final concentration (100 nM) were continuously
diluted
at a ratio of 1/10, such that the resulting antibodies were added by 100 pi
into each
well at six concentrations (i.e., 200 nM, 20 nM, 2 nM, 200 pM, 20 pM and 2 pM)
for
each antibody. The plate was cultured for 5 days under 37 C, 95% RH and 5%
(v/v)
CO, conditions, after which 20 ;A of WST-8 solution (CCK-8, Dojindo) was added
into each well on the final day to carry out color development for 1 - 2
hours, such that
an optical density was measured at a wavelength of 450 nm by a microplate
reader.
[368] Results of cell proliferation inhibitory activity are shown in Table
26 and FIG. 14.
[369]
[370] [Table 26]
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
77
[371] In vitro tumor cell proliferation inhibitory activity by a combined
therapy of anti-c-Met
antibody and anti-HER2 antibody
Cell proliferation inhibition assay, IC50 (DI)
Antibodies NCI-H2170 (NSCLC)
No HGF HGF 5g/ml
A091-F1 > 100 > 100
5G3 > 100 >100
A091-F1 + 5G3 combined > 100 11_22
[372] As seen in Table 26, it was identified that a combined treatment of
5G3 and A091
antibody (Korea Patent Registration No. 10-1515535) as an anti-HER2 antibody
had a
more excellent tumor cell proliferation inhibitory capacity than a single
treatment of
each antibody in NCI-H2170 tumor cell line.
[373]
[374] Example 16, Identification of in vivo tumor cell proliferation
inhibitory activity by
a combined administration of 5G3 c-Met antibody and 1IER2 antibody in an NO-
H2170 xenograft mouse model as a human lung cancer cell line
[375]
[376] An anticancer activity experiment was performed on an NCI-H2170
xenograft mouse
model as a lung cancer cell line, in order to see a combined efficacy of HER2
antibody
and c-Met antibody.
[377] Particularly, in this Example a tumor size of a mouse was measured by
the same
method as shown in Example 14 by using the same mouse as shown in Example 13
above. Results of evaluating an antitumor efficacy by a combination of A091
and 5G3
in an NCI-H2170 xenograft mouse model as a lung tumor cell are shown in FIG.
15.
[378] In result, in case of carrying out a single administration of A091
alone or a combined
administration of A091 and 5G3, a tumor volume was decreased to a significant
level
compared to a solvent control group from a 14th day after administration (p
<0.05).
Also, a group administered with a combination of A091 and 5G3 showed a
significant
decrease in a tumor volume compared to a group administered with A091 alone or
a
group administered with BsAB02 (US2010/0254988 Al) as a control bispecific
antibody (p < 0.01).
[379]
[380] Example 17. Identification of tumor cell proliferation inhibitory
activity in NCI-
H596 xenograft mouse model
[381]
[382] As NCI-H596 cell line was a lung cancer cell line with a mutation in
exon14 of c-
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
78
Met, an evaluation was made on an NCI-H596 xenograft mouse model, in order to
identify an anticancer effect of hu8C4 x Vectibix scFv.
[383] In this Example, a tumor size of a mouse was measured by using the
same mouse and
the same method as shown in Example 14 above.
[384] Results of evaluating an anticancer efficacy after administering
hu8C4 x Vectibix
scFv once or twice a week for total 4 weeks in an NCI-H596 xenograft model as
a lung
tumor cell are shown in FIG. 16.
[385] As a result of measuring a tumor size, a level of tumor size in a
group administered
with hu8C4 x Vectibix scFv 10 mg/kg twice a week showed a statistically
significant
difference compared to a control group from an 11th day after starting an
admin-
istration of test material until the end of an experiment, and levels of tumor
sizes in a
group administered with hu8C4 x Vectibix scFv 5 mg/kg twice a week and a group
ad-
ministered with hu8C4 x Vectibix scFv 10 mg/kg once a week were also
significantly
lower compared to a control group from an 18th day after starting an
administration of
test material. Also, a level of tumor size in a group administered with test
material had
a tendency of change in a dose-correlated way according to a test material
dose, and a
tumor size of a test group was lower compared to a control group even after a
final day
of administering a test material (Day 28).
[386]
[387] Example 18. Identification of tumor cell proliferation inhibitory
activity in EBC -
1 xenograft mouse model
[388]
[389] As EBC-1 was a lung cancer cell line with an amplification of c-Met
gene, an
evaluation was made on an EBC-1 xenograft mouse model, in order to identify an
an-
ticancer effect of hu8C4 x Vectibix scFv.
1390J A mouse used in this Example was a six-week-old female athymic nude
mouse
(Harlan). EBC-1 (JCRB, #JCRB0820) cell lines were inserted into a flask for
cell
culture together with an EMEM medium containing 10% FBS, after which the
resulting cell lines were cultured under 37 C, 5% CO2 conditions. Cell lines
were
prepared in such a way that the resulting cell lines were diluted in a serum-
free
medium at a concentration of 5.0 x 107 cells/ml, after which the cell lines
were subcu-
taneously administered into a mouse by an amount of 0.1 ml/head. When a tumor
size
in a region with cell lines transplanted thereinto reached about 100 - 150
mm3, hu8C4
x Vectibix scFv was administered once or twice a week for total 4 weeks, after
which a
tumor size of the mouse was measured by the same method as shown in Example
14.
[391] Results of evaluating an anticancer efficacy by hu8C4 x Vectibix scFv
in an EBC-1
xenograft model as a lung cancer cell are shown in FIG. 17.
[392] As a result of measuring a tumor size, a level of tumor size in a
group administered
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
79
with hu8C4 x Vectibix say 10 mg/kg twice a week showed a statistically
significant
difference compared to a control group from a 7th day after starting an
administration
of test material until a 56th day after starting an administration of test
material. A
group administered with hu8C4 x Vectibix scFv 5 mg/kg twice a week and a group
ad-
ministered with the same once a week showed a significant low level compared
to a
control group from an 18th day after starting an administration of test
material. Also, a
level of tumor size in a group administered with test material had a tendency
of change
in a dose-correlated way according to a test material dose, and a level of
tumor size in
a group administered with hu8C4 x Vectibix scFv 10 mg/kg twice a week during
an
observation period after a final day (Day 28) of administering a test material
was sig-
nificantly low compared to a control group until a 56th day after starting an
admin-
istration of test material. In particular, it was found that one individual in
a group ad-
ministered with hu8C4 x Vectibix scFv 10 mg/kg twice a week had a complete
response on an 18th day after starting an administration of test material.
[393]
[394] Example 19. Effect of reducing c-Met and EGFR on the surface of
cancer cells by
bispecific antibody
[395]
[396] An effect of reducing c-Met and EGFR on the surface of in vitro tumor
cells by the
bispecific antibody (hu8C4 x Vectibix scFv) of the present invention was
identified
and compared with an effect of the c-Met antibody (hu8C4) of the present
invention,
vectibix, c-Met/EGFR combination, and other antibodies.
[397] A receptor generally located on a cell membrane was internalized into
a cell when
binding to an antibody, thus an amount thereof located on the cell membrane
was
decreased. A decrease in the receptor on such cell membrane causes an
inhibition of
receptor activation and a decrease in a downstream signal thereof by a ligand
binding.
[398] In this Example, a lung adenocarcinoma cell line HCC827 was used to
observe a
decrease in c-Met and EGFR on a cell membrane. HCC827 has an EGFR E746-A750
deletion mutation and overexpresses c-Met. HCC827 was treated with the
bispecific
antibody (hu8C4 x Vectibix scFv) of the present invention and other
antibodies, after
which immunofluorescence staining was performed by an antibody specific to c-
Met
and EG1-R, such that the resulting cell line was analyzed with a fluorescence
activated
cell sorter, thus measuring an amount of c-Met and EGFR on the surface of
cells. A
detailed method is as follows.
[399] First of all, HCC827 cells (ATCOO CRL-2868TM) were divided by 3.0 x
105 into
each well of a 6-well plate containing an RPMI-1640 medium (2 ml) containing
10%
(v/v) FBS, after which the plate was cultured under 37 C, RH 95% and 5% CO2
conditions for 24 hours. The bispecific antibody (hu8C4 x Vectibix scFv) of
the
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
present invention, the c-Met antibody (hu8C4) of the present invention,
vectibix, a
mixture of the c-Met antibody (hu8C4) of the present invention and vectibix, C-
EM1
and LA480 were diluted to reach a final concentration of 100 nM, after which
the
resulting antibodies were treated and reacted for 18 hours. As a plate to be
used as a
non-decreasing control group with c-Met and EGFR, a human IgG antibody was
treated and reacted for 18 hours. Then, cells of each well were collected by
500 fd, of
an enzyme-free cell dissociation buffer (Gibco, #13151), after which cells
were
separated from the enzyme-free cell dissociation buffer by a centrifugal
separator, such
that the enzyme-free cell dissociation buffer was removed therefrom. For
immunofluo-
rescence staining, a goat-derived c-Met antibody (R&D systems, AF276), a goat-
derived EGFR antibody (R&D systems, AF231) or a non-specific goat-derived
antibody for measuring an amount of staining were mixed by 2 jig respectively
with
200 gi of a cold PBS containing 2% (v/v) FBS, after which the resulting
antibodies
were treated into each well, such that the resulting plate was reacted at 4 C
for 1 hour.
Then, the resulting plate was washed twice with a cold PBS containing 2% (v/v)
FBS.
ALEXA488 was bound as a secondary antibody, after which 1 j of a donkey-
derived
antibody (Thermo Fisher, A-11055) binding to a goat antibody was diluted with
200
fik of a cold PBS containing 2% (v/v) FBS, such that the resulting antibody
was used.
After being reacted with the secondary antibody at 4 C for 1 hour, the
resulting cells
were washed twice with a cold PBS containing 2% (v/v) FBS, after which the
resulting
cells were fixed by using 200 a of BD Cytofix(BD, #554655). After being washed
once with PBS, an ALEXA488 Geo-mean (MFI) value, a degree of fluorescent
staining, was measured by using a BD FACS Canto II fluorescence activated cell
sorter. An amount of c-Met and EGFR located on a cell membrane was indicated
as
geo mean fluorescence intensity (MFI) by a following formula. With regard to
values
obtained after repeatedly performing a test three times, an average and
standard
deviation thereof are shown in Table 27 and FIGS. 18 and 19.
[400]
[401] c-Met or EGER. surface amount = geo MFI[experimental groupl - geo
MFI[õõõF.,65, pat-derived
antibody]
[402]
[403] [Table 27]
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
81
[404] Amount of c-Met and EGFR on the surface of cells measured after
treating HCC827 cell line
with bispecific antibody (hu8C4 x Vectibix scFv), etc.
c-Met EGFR
Treated antibody Means Means
S.D. S.D.
(geo MFI) (geo WI)
human IgG 5653 1032 11494 3276
hu8C4 3436 892 11593 3448
Vectibix 5653 1309 10326 3256
hu8C4 Vectibix combined 3551 1047 10111 2932
hu8C4 x Vectibix scFv 1689 321 9930 3305
C-EM1 3665 878 11503 3715
C-LA480 3267 764 11655 4156
[405] As seen in Table 27 above, all the antibodies binding to c-Met
decreased c-Met on
the surface of cells by 40-70%, while antibodies binding to EGFR showed an in-
significant effect of decreasing by 10 - 15%. Further considering an effect of
reducing
c-Met, hu8C4, combination of hu8C4 + Vectibix, C-EM1 and C-LA480 decreased c-
Met on the surface of cells by about 40% or so, while hu8C4 x Vectibix scFv
decreased c-Met on the surface of cells by 70%, thus showing a more excellent
effect
of reducing c-Met on the surface of cells than other antibodies and a
combination of
antibodies.
[406] Results above show that the bispecific antibody (hu8C4 x Vectibix
scFv) of the
present invention remarkably decreases an amount of c-Met on the surface of
cells.
[407]
[408] Example 20. Epitope Mapping
[409] To figure out an epitope of the bispecific antibody (hu8C4 x Vectibix
scFv) of the
present invention on a human c-Met antigen, its analysis was commissioned to
the
molecule model design support team of the Osong Medical Innovation Foundation
(KBIO, Korea). The analysis was performed by hydrogen-deuterium exchange mass
spectrometry (HDX-MS).
[410] c-Met sema domain consists of two a/1i chains, thus identifying each
coverage for the
two chains. Due to a presence of a number of disulfide bonds in a sample, a
peptide
coverage was optimized by adjusting a quench holding time, a TCEP
concentration, a
pepsin concentration, etc. Finally, an experiment was performed under quench
buffer
conditions with 100 mM K.Phosphate, 125 mM TCEP, 0.5 M Guanidine-HC1 and pH
2.66.
CA 03061704 2019-10-28
WO 2018/221969 PCT/KR2018/006182
82
[411] Antigens and antibodies were prepared at a concentration of 3.3 mg/ml
and 65 mg/ml
respectively, and 37 pmol of cMET antigens and 36 pmol of antibodies were
bound 3
hours before the experiment. A deuterium labeling buffer was reacted for 0,
0.33, 10,
60 and 240 minutes. Labeling was stopped with a quench buffer in accordance
with
each labeling time and vortexing was performed, after which they were
immediately
frozen in liquid nitrogen, thus being stored at -80 C before the analysis. The
resulting
antigens and antibodies were loaded onto a pepsin column and analyzed with a
mass
spectrometer (MS).
[412] As a result of the analysis, it was identified that the bispecific
antibody (hu8C4 x
Vectibix scFv) of the present invention binds to a 3-dimensional form of
epitopes in 4
regions of Y321 - L329 (SEQ. No. 331), 1333 - 1341 (SEQ. No. 332), P366 - D372
(SEQ. No. 333), and Q464 - S474 (SEQ. No. 334) of a human c-Met sema domain
chain (Table 28). A labeling was performed on a tertiary structure of a human
c-Met
antigen (PDB No. 4K3J) by using a PyMOL program, wherein results thereof are
shown in FIG. 20.
[413]
[414] [Table 28]
[415] Amino acid sequence of epitope region
Epitope region Amino acids sequence SEQ ID NO
_
Y321-L329 YVSKPGAQL 331
1333-1341 IGASLNDDI 332
P366-D372 I PIKYVND 333
Q464-S474
QVVVSRSGPST 334
[416] From the results above, it can be seen that the mouse antibody,
humanized antibody,
affinity-optimized antibody or antigen binding fragments thereof of the
present
invention, specifically binding to c-Met, selectively act on c-Met, wherein
they show
an excellent cancer cell proliferation inhibitory activity as well as a
remarkably
excellent anticancer activity even by a little amount thereof, thus
effectively preventing
or treating cancer.
[417]
[418] While specific portions of the present invention have been described
in detail above,
it is apparent to those skilled in the art that such detailed descriptions are
set forth to il-
lustrate exemplary embodiments only, but are not construed to limit the scope
of the
present invention. Thus, it should be understood that the substantial scope of
the
present invention is defined by the accompanying claims and equivalents
thereto.
83
In some aspects, embodiments of the present invention as described herein
include the
following items:
Item 1. An antibody or an antigen binding fragment thereof that specifically
binds to a
hepatocyte growth factor receptor (c-Met), wherein the antibody or the antigen
binding fragment
thereof is:
(a) an antibody comprising a light chain variable region comprising a light
chain CDR1
represented by SEQ II) NO: 1; a light chain CDR2 represented by SEQ ID NO: 2;
a light chain CDR3
represented by SEQ NO: 3, and a heavy chain variable region comprising a heavy
chain CDR1
represented by SEQ ID NO: 7; a heavy chain CDR2 represented by SEQ ID NO: 8;
and a heavy chain
CDR3 represented by SEQ ID NO: 9;
(b) an antibody comprising a light chain variable region comprising a light
chain CDR1
represented by SEQ ID NO: 4; a light chain CDR2 represented by SEQ ID NO: 5; a
light chain
CDR3 represented by SEQ ID NO: 6, and a heavy chain variable region comprising
a heavy chain
CDR1 represented by SEQ ID NO: 10; a heavy chain CDR2 represented by SEQ ID
NO: 11; a
heavy chain CDR3 represented by SEQ ID NO: 12; or
(c) an affinity-optimized antibody thereof,
wherein the affinity-optimized antibody comprises:
(i) a light chain variable region represented by SEQ ID NO: 21 and a heavy
chain variable
region represented by SEQ ID NO: 302;
(ii) a light chain variable region represented by SEQ ID NO: 21 and a heavy
chain variable
region represented by SEQ ID NO: 305;
(iii) a light chain variable region represented by SEQ ID NO: 310 and a heavy
chain
variable region represented by SEQ ID NO: 23;
(iv) a light chain variable region represented by SEQ ID NO: 308 and a heavy
chain
variable region represented by SEQ ID NO: 305;
(v) a light chain variable region represented by SEQ ID NO: 306 and a heavy
chain
variable region represented by SEQ ID NO: 303;
(vi) a light chain variable region represented by SEQ ID NO: 307 and a heavy
chain
variable region represented by SEQ ID NO: 304;
(vii) a light chain variable region represented by SEQ ID NO: 308 and a heavy
chain
variable region represented by SEQ ID NO: 304;
(viii) a light chain variable region represented by SEQ ID NO: 309 and a heavy
chain
variable region represented by SEQ ID NO: 304;
(ix) a light chain variable region represented by SEQ ID NO: 311 and a heavy
chain
Date Regue/Date Received 2023-01-10
84
variable region represented by SEQ ID NO: 304; or
(x) a light chain variable region represented by SEQ ID NO: 306 and a heavy
chain
variable region represented by SEQ ID NO: 302.
Item 2. The antibody or the antigen binding fragment thereof according to item
1, wherein
the antibody comprises: (a) a light chain variable region represented by SEQ
ID NO: 13 and a
heavy chain variable region represented by SEQ ID NO: 15; or (b) a light chain
variable region
represented by SEQ ID NO: 14 and a heavy chain variable region represented by
SEQ ID NO: 16.
Item 3. The antibody or the antigen binding fragment thereof according to item
1, wherein
the antibody comprises:
(a) a light chain variable region represented by SEQ ID NO: 21 and a heavy
chain variable
region represented by SEQ ID NO: 23;
(b) a light chain variable region represented by SEQ ID NO: 22 and a heavy
chain variable
region represented by SEQ ID NO: 24;
(c) a light chain variable region represented by SEQ ID NO: 29 and a heavy
chain variable
region represented by SEQ ID NO: 31; or
(d) a light chain variable region represented by SEQ ID NO: 30 and a heavy
chain variable
region represented by SEQ ID NO: 32_
Item 4. The antibody or the antigen binding fragment thereof according to any
one of items
1 to 3, wherein the antibody comprises a hinge region represented by any one
of SEQ ID NO: 37
to SEQ ID NO: 44.
Item 5. The antibody or the antigen binding fragment thereof according to any
one of items
1 to 4, wherein the antibody further specifically binds to an epidermal growth
factor receptor
(EGFR).
Item 6. The antibody or the antigen binding fragment thereof according to item
5, wherein
the antibody that is an antibody or an antigen binding fragment thereof
binding to EGFR is linked
to one light chain or heavy chain terminus of c-Met specific antibody.
Item 7. The antibody or the antigen binding fragment thereof according to item
5, wherein
the antigen binding fragment binding to the EGFR is Fab, Fab', F(a1:02 or Fv.
Date Regue/Date Received 2023-01-10
85
Item 8. The antibody or the antigen binding fragment thereof according to item
7, wherein
the Fv is one or more scFv fragment selected from the group consisting of
ErbituxTm, Vectibix ,
PorirazzaTM and TheraCIMTM.
Item 9. The antibody or the antigen binding fragment thereof according to item
8, wherein
the ErbituxTM scFv comprises an amino acid sequence represented by SEQ ID NO:
313 or SEQ ID
NO: 314.
Item 10. The antibody or the antigen binding fragment thereof according to
item 8, wherein
the Vectibix scFv comprises an amino acid sequence represented by SEQ ID NO:
315.
Item 11. The antibody or the antigen binding fragment thereof according to
item 6 wherein
the antibody or the antigen binding fragment thereof is linked by a connector
represented by SEQ
ID NO: 312.
Item 12_ The antibody or the antigen binding fragment thereof according to
item 1, wherein
the antigen binding fragment is Fab, Fab', F(ab')2 or Fv.
Item 13_ A nucleic acid molecule encoding the antibody or the antigen binding
fragment
thereof as defined in any one of items 1 to 12.
Item 14_ An expression vector comprising the nucleic acid molecule of item 13.
Item 15. A host cell having the expression vector introduced therein of item
14.
Item 16. A method for producing an antibody or an antigen binding fragment
thereof,
comprising:
(a) a step of expressing an anti-c-Met antibody or an antigen binding fragment
thereof by
culturing the host cell of item 15; and
(b) a step of collecting an anti-c-Met antibody or an antigen binding fragment
thereof.
Item 17. A composition for detecting c-Met, comprising the antibody or the
antigen binding
fragment thereof of any one of items 1 to 12 and a pharmaceutically accepted
excipient.
Date Regue/Date Received 2023-01-10
86
Item 18. A kit for detecting c-Met, comprising the composition for detecting c-
Met of item
17, and instructions.
Item 19. A method for detecting a c-Met antigen, comprising a step of
contacting the
antibody or the antigen binding fragment thereof of any one of items 1 to 12
with a specimen
sample.
Item 20. A composition for treating cancer, comprising the antibody or the
antigen binding
fragment as defined in any one of items 1 to 12 and a pharmaceutically
accepted excipient, wherein
the cancer is caused by c-Met overexpression, amplification, mutation or
activation.
Item 21. A composition for the preparation of a medicament for treating
cancer, comprising
the antibody or the antigen binding fragment as defined in any one of items 1
to 12 and a
pharmaceutically accepted excipient, wherein the cancer is caused by c-Met
overexpression,
amplification, mutation or activation.
Item 22. The composition for treating cancer according to item 20 or the
composition for the
preparation of a medicament for treating cancer according to item 21, wherein
the antibody or the
antigen binding fragment thereof binds to c-Met to inhibit a receptor
activity.
Item 23. The composition for treating cancer according to item 20 or 22 or the
composition for
the preparation of a medicament for treating cancer according to item 21 or
22, wherein the antibody
or the antigen binding fragment thereof further binds to EGFR to inhibit the
receptor activity.
Item 24. The composition for treating cancer according to any one of items 20
and 22 to 23
or the composition for the preparation of a medicament for treating cancer
according to any one of
items 21 to 23, wherein the cancer is selected from the group consisting of
lung cancer, stomach
cancer, colon cancer, rectal cancer, triple negative breast cancer (TNBC),
glioblastoma, pancreatic
cancer, head and neck cancer, breast cancer, ovarian cancer, liver cancer,
renal cancer, bladder
cancer, prostate cancer, brain cancer, uterine cancer, solenoma, thyroid
cancer, acute myeloid
leukemia, chronic myeloid leukemia, myeloma, multiple myeloma, melanoma,
lymphoma and
adrenal cortex cancer.
Item 25. Use of the antibody or the antigen binding fragment as defined in any
one of items
Date Recue/Date Received 2023-06-01
87
1 to 12 for treating cancer, wherein the cancer is caused by c-Met
overexpression, amplification,
mutation or activation.
Item 26. Use of the antibody or the antigen binding fragment as defined in any
one of items
1 to 12 for the preparation of a medicament for treating cancer, wherein the
cancer is caused by c-
Met overexpression, amplification, mutation or activation.
Item 27_ The use according to item 25 or 26, wherein the antibody or the
antigen binding
fragment thereof binds to c-Met to inhibit a receptor activity.
Item 28_ The use according to item 27, wherein the antibody or the antigen
binding fragment
thereof further binds to EGFR to inhibit the receptor activity.
Item 29. The use according to any one of items 25 to 28, wherein the cancer is
selected from
the group consisting of lung cancer, stomach cancer, colon cancer, rectal
cancer, triple negative
breast cancer (TNBC), glioblastoma, pancreatic cancer, head and neck cancer,
breast cancer,
ovarian cancer, liver cancer, renal cancer, bladder cancer, prostate cancer,
brain cancer, uterine
cancer, solenoma, thyroid cancer, acute myeloid leukemia, chronic myeloid
leukemia, myeloma,
multiple myeloma, melanoma, lymphoma and adrenal cortex cancer.
Date Regue/Date Received 2023-01-10