Note: Descriptions are shown in the official language in which they were submitted.
CA 03113059 2021-03-16
WO 2020/061560
PCT/US2019/052396
SYNTHETIC BINDING AGENTS FOR LIMITING PERMEATION THROUGH
MUCUS
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This patent application claims priority to U.S. provisional patent
application no.
62/734,771, filed on September 21, 2018 (titled "SYNTHETIC BINDING AGENTS FOR
MUCOSAL TRAPPING"), herein incorporated by reference in its entirety.
INCORPORATION BY REFERENCE
[0002] All publications and patent applications mentioned in this
specification are herein
incorporated by reference in their entirety to the same extent as if each
individual publication
or patent application was specifically and individually indicated to be
incorporated by
reference.
STATEMENT AS TO FEDERALLY SPONSORED RESEARCH
[0003] This invention was made with Government support under Grant No.
R56HD095629 and U54HD096957 awarded by the National Institutes of Health. The
Government has certain rights in the invention.
REFERENCE TO SEQUENCE LISTING
[0004] The present application includes a listing of sequences. A Sequence
Listing in
electronic format is submitted with this utility application.
FIELD
[0005] The present disclosure generally relates to methods and compositions
for
enhancing agglutination of a target, facilitate enchaining of a target, and/or
muco-trapping of
a target to prevent conception (e.g., for contraception), and/or to prevent or
treat infection,
including viral, bacterial and/or fungal infections.
BACKGROUND
[0006] The mucosal barrier plays an important potential protective role as
a barrier to
prevent foreign matter from entering the body. The mucosal barrier may be
further enhanced
by local immunity that allows a robust immune system response to occur at
mucosal
membranes of the intestines, the urogenital tract and the respiratory system,
i.e., surfaces that
1
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
are in contact with the external environment. The mucosal immune system may
provide
protection against pathogens but maintains a tolerance towards non-harmful
commensal
microbes and benign environmental substances. Since the mucosal membranes are
the
primary contact point between a host and its environment, a large amount of
secondary
lymphoid tissue is found here. The mucosa-associated lymphoid tissue, or MALT,
provides a
critical element of the mucosal immune response. The mucosal immune system
provides
three main functions: serving as the body's first line defense from antigens
and infection,
preventing systemic immune responses to commensal bacteria and food antigens
(primarily
food proteins in the gut-associated lymphoid tissue, so-called oral
tolerance), and regulating
appropriate immune responses to pathogens encountered on a daily basis.
[00071 Unfortunately, the mucosal immune response may be inadequate, and it
is often
difficult to elicit the necessary immune response for sufficient duration.
This is exemplified
by the lack of effective vaccines against the majority of sexually transmitted
infections,
including HIV, Herpes, Chlamydia and Gonorrhea. Consequently, enhancements of
the
mucus barrier and the mucosal immune system by direct delivery of antibodies
have been
suggested as one method of treating or preventing infection. See, e.g., US
20150284451,
which describes the use of compositions to prevent pathogen infection by
applying antibodies
that may interact with mucus.
[0008] Although some antibodies have been shown to interact with mucins to
adhesively
crosslink individual antibody-coated pathogens to mucins and thereby
immobilizing them in
mucus (a process frequently referred to as muco-trapping), it would be
beneficial to provide
antibodies or antibody constructs having further improved ability to more
effectively prevent
foreign matter, including viruses and bacteria, from permeating through mucus
to reach target
cells. In addition, such improved constructs may be beneficially used as a
contraceptive, by
blocking or limiting passage of sperm to the egg within the female
reproductive tract.
Beyond crosslinking foreign entities to mucins, it is possible to further
enhance the potencies
of the antibodies by improving the agglutination and/or enchainment of foreign
entities
together in a manner that limits their effective permeation through mucus.
[00091 Nearly half of all pregnancies in the U.S. are unintended,
underscoring the critical
need for additional options for contraception. Non-hormonal contraceptives
would be of
particular use.
[000101 Described herein are methods and compositions (including compositions
of
engineered/synthetic binding agents) for enhancing agglutination, enchainment
and/or muco-
2
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
trapping of a target, i.e., reducing the fraction of target entities that
could permeate through
mucus, including pathogens and sperm.
SUMMARY OF THE DISCLOSURE
[00011] In general, described herein are synthetic binding agents for
enhancing
agglutination and/or muco-trapping of a target, and methods of enhancing
agglutination
and/or muco-trapping of a target using any of these synthetic binding agents.
The target
typically has one or more epitopes, and may be a virus, bacteria, fungus,
sperm or parasite.
The synthetic binding agents described herein are multimeric, having multiple
epitope-
binding regions. All of these epitope-binding regions may be immunoglobulin
fragment
antigen binding (Fab) regions or fragments, and may include a core of a human
or humanized
Immunoglobulin G (IgG) having Fab and Fe domains. All of the Fab
fragments/domains
(including those of the core humanized IgG) may be directed to the same
epitope to recognize
the foreign body. Thus, the synthetic binding agents for enhancing
agglutination and/or
muco-trapping described herein may include a human or humanized IgG that is
linked to one
or more additional Fab domains, wherein the one or more additional Fab domains
and the
parent IgG Fab domains all specifically bind to the target epitope with high
affinity, and may
reduce the mobility of the target in mucus to less than about 50% relative to
its native
mobility in mucus. The synthetic binding agents may be recombinant (e.g.,
engineered)
antibodies. Any of the synthetic binding agents described herein may be
further configured
(or selected) to enhance mucin crosslinking once bound to the target, but may
otherwise be
relatively free to diffuse through mucus (e.g., have a low affinity for
mucins). As used herein
the term "native mobility" refers to the mobility of the target (e.g., sperm,
virus, bacteria,
etc.) in the same environment (e.g., mucus, saline, etc.) in the absence of a
synthetic binding
agent or antibody.
[00012] Also described herein are also methods and compositions (including
compositions
of engineered/synthetic binding agents) that could provide bactericidal and/or
microbicidal
effect by more effectively clumping together pathogens that undergo cell
division, which
leads to a chain of bacteria and/or other pathogens, and potentially
inhibiting replication,
triggering cell death, e.g., forming aggregations (including in some
variations multi-pathogen
aggregations) and/or preventing the spread of the infection through either
agglutination or
enchained growth.
[00013] In particular, described herein are synthetic binding agents
configured as
recombinant antibodies that may be used as a contraceptive. A contraceptive
synthetic
3
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
binding agent may be referred to herein as a human contraceptive agent (HCA),
although
these methods may also be used for non-human (e.g., animal) contraception. For
example,
described herein are contraceptive methods and HCA compositions, including
recombinant
engineered antibodies (Ab) that can block sperm permeation through mucus. A
major effector
function for Ab in mucus is to arrest the forward motion of foreign entities
such as viruses
and highly motile bacteria, and block them from reaching target cells. This
function can be
accomplished in two ways. First, when concentrations of the foreign entity are
high such that
the foreign bodies would frequently collide, Ab can crosslink two or more
bodies together,
resulting not only in an increase in hydrodynamic diameter but also an
effective
neutralization of the net forward motion of swimming bodies. This process is
commonly
referred to as agglutination. Second, when concentrations of the foreign
entity are modest
such that collisions between foreign bodies are relatively infrequent, Ab can
immobilize by
directly crosslinking the foreign body to the mucin matrix present in mucus
via multiple Fe-
mucin bonds. This process, which is herein referred to as mucin-crosslinldng
or muco-
trapping, has remained largely unrecognized because the affinity between each
Ab molecule
and mucin was long thought to be much too weak to effectively bind individual
foreign
bodies to mucins. However, vaginally dosed antigen-specific IgG that are tuned
to possess
weak affinity to mucins can trap viruses in mucus by forming multiple weakly
adhesive
bonds between the virus and the mucin mesh (akin to a VELCRO 03 patch with
individually
weak hooks). Finally, for foreign bodies that can divide on its own (e.g.
bacteria, fungus,
etc.), aggregates of the bacteria may be formed by enchaining the daughter
cell of the
dividing bacteria with the mother cell i.e. enchained growth. The end result
is a clump of
foreign bodies (similar to what would be formed by agglutination) but without
requiring
independent and distinct foreign bodies from colliding with each other.
100014] Sperm concentration varies widely in the female reproductive tract,
with the
maximum concentration in semen immediately following ejaculation and lower
concentrations in more distal sites, such as the cervical canal.
[000151 The various HCA constructs described herein are configured to act by
blocking
sperm permeation through mucus and preventing sperm from reaching the egg and
may
therefore harness both agglutination and muco-trapping mechanisms. Polyvalent
Ig such as
sIgA and IgM are markedly more potent agglutinators than IgG (IgM is ¨1000-
fold more
potent at agglutination than IgG). Unfortunately, large scale manufacturing of
IgM or sIgA
remains exceptionally challenging, and both IgM and sIgA suffer from stability
issues. IgG
represents the predominant isotype of Ab under clinical development and has an
outstanding
4
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
track record of safety in humans. Thus, from a translational development
perspective, IgG
represents the most logical platform for developing more potent HCA. Greater
potency not
only translates to lower doses of HCA needed but also maximizes the potential
effectiveness
of HCA-based contraception.
[00016] To date, a multimeric Ab construct to create more potent HCA has not
been
developed. Described herein are engineered multimeric Ab that may radically
improve on
the current monomeric IgG1 -based HCA by more potently agglutinating sperm,
thus
achieving greater potency in blocking permeation through mucus.
[00017] For example, a synthetic binding agent for enhancing agglutination
and/or rnuco-
trapping of a target having an epitope may include: a human or humanized
Immunoglobulin
G (IgG) having a pair of Fab domains, wherein the human or humanized IgG is
linked to one
or more additional immurioglobulin fragment antigen binding (Fab) domains,
wherein the one
or more additional Fab domains and the IgG Fab domains all specifically bind
to the epitope
of the target, so that the synthetic binding agent binds to the target with
high affinity and
reduces the mobility of the target in mucus to less than about 50% relative to
its native
mobility in mucus (or in some variations, in water). The reduction of mobility
in mucus may
be due to enhanced agglutination by the synthetic binding agent (construct),
and/or due to
enhanced enchainment of targets that can divide. In any of the variations
described herein the
one or more additional Fab domains and the IgG Fab domains may bind different
epitopes on
the same target (e.g., pathogen).
[00018] Any number (preferably an even number) of additional Fab domains may
be
included. For example, the one or more additional Fab domains may comprise 2,
4, 6 or 8
additional Fab domains.
1000191 In some variations, the synthetic binding agent is a contraceptive
synthetic binding
agent (e.g., a contraceptive antibody), and the target is sperm and all of the
one or more Fab
domains and the IgG Fab domains specifically bind repeating poly-n-
acetyllactosaminyl
structures on sperm, an N-linked glyeosylated form of SEQ ID: 1 (e.g., an
amino acid
sequence comprising GQNDTSQTSSPS), where the glycans are poly-n-
acetyllactosamine.
This target is referred to herein as CD52g.
[00020] The additional Fab domains may each comprise: (i) a heavy chain (HC)
with a
variable region (VH) comprising complementarity determining region(s) (CDRs)
having the
amino acid sequence of: SEQ ID NO: 4 (e.g., an HC CDR sequence as SEQ ID NO:
4);
and/or (ii) a light chain (LC) with a variable region (VL) comprising
complementarity
determining the amino acid sequence of: SEQ ID NO: 7 (e.g., an LC CDR sequence
as SEQ
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
ID NO: 7). In some variations, the at least one additional Fab domain of the
synthetic
binding agent is linked to a Fab domain of the pair of Fab domains of the IgG;
alternatively
or additionally, the at least one additional Fab domain may be linked to an Fe
region of the
IgG. The IgG may comprise at least one Fc region that is a naturally occurring
sequence. As
described below, in some variations the Fc sequence may also be modified (for
instance, to
prolong systemic circulation and reduced interactions with other immune
cells).
[000211 The amino acid sequences of the additional Fab domains do not have to
be
identical to each other or to the IgG Fab domains, although they may all bind
to the same
antigen with approximately the same affinity. One of reasonable skill in the
art may apply
known methods to vary the sequence while retaining substantially all of the
binding affinity.
For example, conservative amino acid substitutions may be made (e.g., an
exchange between
two amino acids separated by a small physicochemical distance). For example,
in any of the
variations described herein, the additional Fab domains may comprise: (i) a
heavy chain (HC)
with a variable region (VH) comprising complementarity determining region(s)
(CDRs)
having an amino acid sequence that is between 100% and 75% (e.g., between 100%-
80%,
between 100%-85%, between 100%-90%, between 100%-95%, etc.) identical to the
amino
acid sequence of the IgG Fab domain (e.g., for an CD52g synthetic binding
peptide, having
an amino acid sequence that is between 100% and 75% (e.g., between 100%-80%,
between
100%-85%, between 100%-90%, between 100%-95%, etc.) identical to the amino
acid
sequence of SEQ ID NO: 4, e.g., an HC CDR sequence as SEQ ID NO: 4); and/or
(ii) a light
chain (LC) with a variable region (VL) comprising complementarity determining
the amino
acid sequence having an amino acid sequence that is between 100% and 75%
(e.g., between
100%-80%, between 100%-85%, between 100%-90%, between 100%-95%, etc.)
identical to
the amino acid sequence of the IgG Fab domain light chain VL (e.g., for a
CD52g synthetic
binding peptide, having an amino acid sequence that is between 100% and 75%
(e.g.,
between 100%-80%, between 100%-85%, between 100%-90%, between 100%-95%, etc.)
identical to the amino acid sequence of; SEQ ID NO: 7, e.g., an LC CDR
sequence as SEQ
ID NO: 7).
[00022] In some variations, the one or more additional Fab domains may be
linked to the
IgG via a flexible linker comprising an amino acid sequence comprising n
pentapeptide
repeats consisting of Glycine (G) and Serine (S), wherein n is between 2 and
15 (e.g.,
between 2 and 14, between 2 and 13, between 2 and 12, between 2 and 11,
between 2 and 10,
between 2 and 9, between 2 and 8, between 3 and 15, between 3 and 14, between
3 and 13,
between 3 and 12, between 3 and 11, between 3 and 10, between 3 and 9, between
3 and 8,
6
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
etc.). Other linkers, not limited to amino acid/peptide linkers, may be used,
for example,
non-amino acid polymers such as polynucleotide linkers and other synthetic
linkers, In
general, the linkers do not need to be identical. Linkers may be one set of
glycine serine
linkers that are used for connecting one Fab, but another set of linkers uses,
e.g., (EAAAK)3,
and/or yet another set of linker uses (Ala-Pro). (10 ¨ 34 aa) linker.
[00023] Also described herein are isolated nucleic acid molecules encoding any
of the
synthetic binding agents described herein. Also described herein are vectors
comprising any
of these nucleic acid molecules and/or an isolated host cell or a non-human
organism
transformed or transfected with the nucleic acid molecule. Also described
herein are
compositions comprising any of the synthetic binding agents and a
pharmaceutically
acceptable carrier.
[00024] Although many of the variations described herein are directed
specifically to
compositions (e.g., synthetic binding agents) and methods for contraception,
it should be
understood that any of these compositions and methods may be directed to the
treatment or
prevention of infection by a pathogen (e.g., virus, bacteria, fungi, etc.).
[00025] For example, described herein are methods of enhancing agglutination
and/or
muco-trapping of a target (e.g., sperm, virus, bacteria, fungi, etc.) having
an epitope, the
method comprising: administering a synthetic binding agent to a patient, the
synthetic
binding agent comprising a human or humanized Immunoglobulin G (IgG) having a
pair of
Fab domains, wherein the human or humanized IgG is linked to one or more
additional
immunoglobulin fragment antigen binding (Fab) domains, wherein the one or more
additional
Fab domains and the IgG Fab domains all specifically bind to the epitope of
the target, so that
the synthetic binding agent binds to the target with high affinity and
enhances agglutination
and/or muco-trapping of the target in the patient's mucus to reduce the
mobility of the target
to less than about 50% (e.g., less than about 40%, 30%, 20%, 10%, etc.) of its
native mobility
e.g., relative to its native mobility in mucus, or in some variations in
water. In some
variations, the synthetic binding agents described herein reduce the fraction
of progressively
motile sperm, e.g. >95% (90% or greater, 85% or greater, 80% or greater, 75%
or greater,
70% or greater, 65% or greater, 60% or greater, etc.) as compared to the
fraction of
progressively motile sperm in control. For example, >50% reduction in
progressively motile
sperm populations.
[00026] As mentioned above, any number (preferably an even number) of
additional Fab
domains may be included. For example, the one or more additional Fab domains
may
comprise 2, 4, 6 or 8 additional Fab domains.
7
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[00027] In particular, the target may be a sperm and all of the one or more
Fab domains
and the IgG Fab domains may specifically bind to an epitope of CD52g. The
mobility of the
sperm is slowed down by at least 50% (e.g., relative to its native mobility in
mucus), e.g., less
than about 40%, 30%, 20%, or 10% native mobility.
[00028] In general, administering comprises administering to the patient
vaginally. In
some variations (e.g., directed to respiratory or other infection or
infectious routes)
administering may comprise topical administration, such as, but not limited
to, administering
via inhalation (e.g., of an aerosol), oral, eye-drop, lavage, etc. In any of
these variations,
administering may comprise administering systemically to the patient,
including systemic
delivery for mucosal (e.g. vaginal, respiratory, gastrointestinal)
applications.
[00029] In some variations, administering comprises delivering from an
intravaginal ring
(IVR) or vaginal film. In some variations, administering comprises delivering
to lung
mucosa using a nebulizer.
[00030] Any appropriate amount of the synthetic binding agent may be
administered. For
example, administering may comprise delivering between 0.01 mg and 100 mg/day
of the
synthetic binding agent. For example, when using a contraceptive synthetic
binding agent,
administering may comprise administering an amount sufficient to agglutinate
the target to
enhance overall ability to limit sperm peimeation across mucus.
[00031] In some variations, the synthetic binding agents described herein are
synthetic
binding agents for inhibiting sperm mobility through mucus. For example a
synthetic binding
agents for inhibiting sperm mobility through mucus may include: a human or
humanized
Immunoglobulin G (IgG) having a pair of Fab domains, wherein the human or
humanized
IgG is linked to one or more additional immunoglobulin fragment antigen
binding (Fab)
domains, wherein the one or more additional Fab domains and the IgG Fab
domains all
specifically bind to an epitope of CD52g, so that the synthetic binding agent
reduces the
mobility of sperm in mucus to less than about 50% relative to its native
mobility in mucus
and/or reduces the fraction of progressively motile sperm by 50%. In some
variations, the
one or more additional Fab domains may comprise, for example, 2, 4, 6 or 8
additional Fab
domains. The epitope of CD52g may comprise an N-linked glycosylated form of
SEQ ID
NO: 1.
[00032] In any of the variations described herein, the additional Fab domains
may each
comprise: (i) a heavy chain (HC) with a variable region (VH) comprising
complementarity
determining regions (CDRs) having the amino acid sequence of: SEQ ID NO: 4;
and/or (ii) a
8
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
light chain (LC) with a variable region (VL) comprising complementarity
determining
regions (CDRs) having the amino acid sequence of: SEQ ID NO: 7.
[00033] The one or more additional Fab domains may be linked to a Fab domain
of the
pair of Fab domains of the IgG. For example, the at least one additional Fab
domain may be
linked to an Fe region of the IgG. The IgG may comprise at least one Fe region
that is a
naturally occurring sequence.
[00034] The one or more additional Fab domains may be linked to the IgG via a
flexible
linker comprising an amino acid sequence comprising n pentapeptide repeats
consisting of
Glycine (G) and Serine (S), wherein n is between 3 and 8.
[00035] Also described herein are isolated nucleic acid molecules encoding the
synthetic
binding agent, and/or a vector comprising the corresponding nucleic acid
molecule, and/or an
isolated host cell or a non-human organism transformed or transfeeted with the
nucleic acid
molecules. Any of the synthetic binding agents described herein may be part of
a
composition comprising the synthetic binding agent and a pharmaceutically
acceptable
carrier.
[00036] Any appropriate delivery device may be used with the compositions
and/or
synthetic binding agents described herein. For example, an intravaginal ring
(IVR) or vaginal
film may be used.
[00037] Described herein are methods of providing contraception in a female
subject.
These methods may include administering to a mucosa of a reproductive tract of
the subject
any of the synthetic binding agents (and particularly those that bind a sperm-
selective marker,
including CD52g, as described herein) in an amount effective to provide
contraception. For
example, described herein are methods of inhibiting the mobility of sperm in
the mucus of a
reproductive tract of a female subject that include contacting the mucus
(e.g., in the female
genital tract) with any of these synthetic binding agents (including via a
delivery device) in
an amount effective to inhibit the mobility of at least 80% of sperm present
in the mucus. In
some variations, the mobility of the sperm may be slowed down by at least 50%
relative to its
normal or native mobility in mucus and/or reduces the fraction of
progressively motile sperm
by 50%. The synthetic binding agent or composition may be delivered via
vaginal
administration (e.g., using an intravaginal ring (IVR)); alternatively or
additionally, the
synthetic binding agent or composition may be delivered via systemic
administration. An
IVR may be configured to release an effective amount for at least 15 days. In
some
variations, the composition may be delivered in a film that dissolves
intravaginally releasing
the synthetic binding agent.
9
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[00038] In general, the Fab fragments may be on the N- or C- terminal ends of
the core
IgG. For example, the additional Fab domain(s) may be inserted at the N-
terminus (i.e.
extending another Fab arm) or at the C-terminus (i.e. after the Fe domain). In
some
variations, the synthetic binding agent may include at least two additional
Fab domains (also
referred to herein as Fab fragments) before and/or after the human or
humanized IgG. In
particular, the synthetic binding agent for enhancing agglutination,
enchainment and/or
muco-trapping of a target having an epitope (including an epitope specific to
sperm and
therefore effective to trap sperm) may include a human or humanized Immuno
globulin G
(IgG) having a pair of Fab domains (and a pair of Fe domains), in which the
human or
humanized IgG is linked to four additional Fab domains, and the IgG Fab
domains as well as
the additional Fab domains all specifically bind to the epitope of the target,
so that the
synthetic binding agent binds to the target with high affinity and reduces the
mobility of the
target in mucus to less than about 50% relative to its native mobility in
mucus. In this
example, the synthetic binding agent includes additional Fabs on both sides of
the core IgG
(e.g., 2 on the N terminal and 2 on the C terminal ends, for a total of 6 Fabs
on the molecule).
[00039] In variations configured as a contraceptive, the potency of the
binding agent may
be determined in part by the minimum concentration of the synthetic binding
agent that is
able to effectively agglutinate the sperm and prevent it from freely swimming
(i.e. remain as
progressively motile sperm). In general, the synthetic binding agents having 6
or more total
Fab fragments have been found to have an order of magnitude better potency
(e.g.,
agglutination potency) compared to just the IgG (including the IgG
glycosylated to enhance
mucosal binding). Specifically, many of the synthetic binding agents
configured as
contraceptives described herein have been shown to reduce progressively motile
sperm (e.g.
by 95% vs. untreated control) to a similar extent as native IgG at greater
than I Ox lower
binding agent concentrations. The potency of the binding agent may also be
determined in
part by enhanced "muco-trapping", which refers to the synthetic binding agent
crosslinldng a
greater fraction of sperm to mucins compared to native IgG, or crosslinking a
similar fraction
of sperm to mucins as IgG at lower binding agent concentrations.
[000401 The human or humanized IgG forming the core of the synthetic binding
agents
described herein may include non-native Fe regions (e.g., Fe regions modified
to increase
stability/half-life in the body, Fe regions modified to decrease
immunoreactivity, etc.). For
example, the Fe region of the IgG portion of the synthetic binding agent may
be modified to
include one or more specific mutations whereby specific immune functions are
modified. For
instance, the Fe region may have enhanced FeRn affinity to extend circulation
kinetics. For
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
example, a mutation in human IgG (e.g., IgG1) of T250Q/M428L may increase
binding to
Fan, and increase the half-life, and/or a mutation of M252Y/S254T/T256E +
H433K/N434F
may increase binding to Fan and increase the half-life. In some variations the
synthetic
binding agent includes a modified Fe region having reduced FcR affinity which
may help
ensure that the Ab does not prime the immune system to develop antibodies
against sperm.
For example, one or more mutations in the human IgG (e.g., IgG1) that decrease
binding to
FcR (e.g., FeyR) may be included, such as E233P/L234V/L235A/G236 +
A327G/A330S/P331S, L234A/L235A/ P329G, and/or K322A.
[00041] For example, as described above, a method of enhancing agglutination
and/or
muco-trapping of a target having an epitope may include: administering a
synthetic binding
agent to a subject, the synthetic binding agent comprising a human or
humanized
Immunoglobulin G (IgG) having a pair of Fab domains, wherein the human or
humanized
IgG is linked to one or more additional immunoglobulin fragment antigen
binding (Fab)
domains, wherein the one or more additional Fab domains and the IgG Fab
domains all
specifically bind to the epitope of the target, so that the synthetic binding
agent binds to the
target with high affinity and enhances the ability for the subject's mucus to
limit permeability
of the target across mucus, as reflected by a reduction of the mobility of the
target to less than
about 50% relative to its native mobility in mucus, or the fraction of motile
target to less than
50% relative to untreated control. The one or more additional Fab domains
comprises 2, 4, 6
or 8 additional Fab domains.
[00042] In some variations, and particularly contraceptive methods, target may
be sperm.
In some variations, the antigen is CD52g. The mobility of the sperm in mucus
may be
slowed down by at least 50% compared to the native mobility of the sperm in
the mucus.
[09043] In some variations, the target may be a pathogen, e.g., all of the one
or more Fab
domains and the IgG Fab domains may specifically bind to a pathogen. For
example, the
pathogen may be one (or more) of: Acinetobacter baumannii; Bacteroides
fragilis;
Burkholderia cepacia; Clostridium difficile; Clostridium sordellii; Carbapenem-
resistant
Enterobacteriaceae; Enterococcus faecalis; Escherichia coli; Hepatitis A;
Hepatitis B;
Hepatitis C; human immunodeficiency virus HIV-1 and HIV-2 (HIV, AIDS);
Influenza;
Klebsiella pneumonia; Methicillin-resistant Staphylococcus aureus; Morgctnella
morganii;
Mycobacterium abscessus; Norovirus; Psuedomonas aeruginosa; Staphylococcus
aureus;
Stenotrophomonas rnaltophilia; Mycobacterium tuberculosis; Vancomyin-resistant
Staphylococcus aureus; Vancomycin-resistant Enterococci; Neisseria gonorrhoeae
(gonorrhea); Chlamydia trachomatis (chlamydia, lymphogranuloma venereum);
Treponema
11
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
pallidum (syphilis); Haernophilus ducreyi (chancroid); Klebsiella granulomatis
or
Calymmatobacterium granulomatis (donovanosis), Mycoplasma genitalium,
Ureaplasma
urealyticurn (mycoplasmas); HTLV-1 (T-lymphotrophic virus type 1); herpes
simplex virus
type 1 and type 2 (HSV-1 and HSV-2); Epstein-Barr virus; cytomegalovirus;
human
herpesvirus 6; varicella-zoster virus; human papillomaviruses (genital warts);
hepatitis A
virus, hepatitis B virus, hepatitis C virus (viral hepatitis); molluscum
contagiosum virus
(MCV); Trichomona vaginal is (trichomoniasis); and yeasts, such as Candida
albicans
(vulvovaginal candidiasis). In some variations, the pathogen includes a
fungus, such as
Aspergillus.
100044] Administering may comprise administering by any appropriate route, or
more than
one route. For example, administering may comprise administering to the
subject vaginally
(e.g., from an intravaginal ring, IVR). Administering may comprise
administering
systemically to the subject. Administering may comprise administering to the
subject as a
vaginal film. Administering may comprise administering from a nebulizer.
Administering
may comprise administering by inhalation. Administering may comprise an eye
drop.
Administering may comprise an oral capsule or pill. Administering may comprise
a mouth
wash. In some variations, administering comprises delivering between 0.01 mg
and 100
mg/day of the synthetic binding agent. Administering may comprise
administering in an
amount sufficient to agglutinate or form enchainment of the target while
maintaining or
enhancing muco-trapping, with the overall net effect of reducing the
permeability of the
target through mucus, and/or reducing the growth or presence of the target.
[00045] As mentioned above, the IgG Fab domains may have an amino acid
sequence that
is not identical to the one or more additional Fab domains. For example, the
IgG domains
may have an amino acid sequence that is between 100% (identical) and 75%, 80%,
85%,
90%, 95%, etc. In general, the IgG Fab and the additional Fab domains
recognize the same
antigen with approximately the same affinity, regardless of their sequence.
1000461 For example, described herein are methods of inhibiting fertilization
and/or
conception by agglutination and/or muco-trapping of sperm that may include:
administering a
synthetic binding agent to a subject, the synthetic binding agent comprising a
human or
humanized Immunoglobulin G (IgG) having a pair of Fab domains, wherein the
human or
humanized IgG is linked to one or more additional immunoglobulin fragment
antigen binding
(Fab) domains, wherein the one or more additional Fab domains and the IgG Fab
domains all
specifically bind to an epitope of sperm, so that the synthetic binding agent
binds to the
sperm with high affinity and enhances agglutination and/or muco-trapping of
the sperm in
12
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
mucus. The net effect is either a reduction in the fraction of progressively
motile sperm,
and/or a reduction in the mobility of motile sperm. As mentioned, the one or
more additional
Fab domains may comprise 2, 4, 6 or 8 additional Fab domains.
[00047] The mobility of the sperm in mucus may be slowed down by at least 50%
compared to the native mobility of the sperm in the mucus. In some variations,
the mobility
of the target (e.g,, sperm, pathogen, etc.) may be slowed by at least 40%
compared to the
native mobility of the target, slowed by at least 30% compared to the native
mobility, slowed
by at least 20% compared to the native mobility, slowed by at least 15%
compared to the
native mobility, etc.
[00048] In some variations, the synthetic binding agents described herein
reduce the
fraction of progressively motile sperm among all sperm, e.g. >95% (90% or
greater, 85% or
greater, 80% or greater, 75% or greater, 70% or greater, 65% or greater, 60%
or greater, etc.)
than reduction in fraction of progressively motile sperm vs. control. For
example, >50%
reduction in progressively motile sperm populations.
[00049] As mentioned, any appropriate administration route may be used. For
example,
administering may comprise administering to the subject via one or more of:
vaginally (e.g.,
from an intravaginal ring), topically, systemically, as a vaginal film, from a
nebulizer.
Administering may comprise administering in an amount sufficient to
agglutinate the target,
and/or muco-trapping of the target, with the overall effect of reducing target
permeation
through mucus.
[00050] Also described herein are methods of treating or preventing an
infection by a
pathogen, the method comprising administering a synthetic binding agent to a
subject, the
synthetic binding agent comprising a human or humanized Immunoglobulin G (IgG)
having a
pair of Fab domains, wherein the human or humanized IgG is linked to one or
more
additional immunoglobulin fragment antigen binding (Fab) domains, wherein the
one or more
additional Fab domains and the IgG Fab domains all specifically bind to a
single epitope of
the pathogen, so that the synthetic binding agent binds to the pathogen with
high affinity and
enhances agglutination of the target, inducing enchained growth of the target,
and/or muco-
trapping of the target in the subject's mucus. The one or more additional Fab
domains
comprises 2, 4, 6 or 8 additional Fab domains. The mobility of the pathogen in
mucus is
slowed down by at least 10% (at least 50%, at least 40%, at least 30%, at
least 20%, at least
15%, etc.) compared to the native mobility of the pathogen in the mucus,
[00051] The pathogen may be one or more of: influenza (including influenza A,
B, and C);
severe acute respiratory syndrome (SARS); respiratory syncytial virus (RSV);
parainfluenza;
13
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
adenovirus; human rhinovirus; coronavirus; and norovirus. The pathogen may be
one or
more of: Salmonella and Escherichia colt. The pathogen may be one or more of:
Neisseria
gonorrhoeae (gonorrhea); Chlarnydia trachornatis (chlarnydia, lymphogranuloma
venereum);
Treponema pallidum (syphilis); Haemophilus ducreyi (chancroid); Klebsiella
granulornatis or
Calymmatobaeterium granulomatis (donovanosis), Mycoplasma genital/urn,
Ureaplasma
urealyticum (mycoplasmas); human immunodeficiency virus HIV-1 and HIV-2 (HIV,
AIDS);
HTLV-1 (T-lymphotrophic virus type 1); herpes simplex virus type 1 and type 2
(HSV-1 and
HSV-2); Epstein-Barr virus; cytornegalovirus; human herpesvinis 6; varicella-
zoster virus;
human papillomavinises (genital warts); hepatitis A virus, hepatitis B virus,
hepatitis C virus
(viral hepatitis); molluscum contagiosum virus (MCV); Trichomona vaginalis
(triehomoniasis); and yeasts, such as Candida albicans (vulvovaginal
candidiasis).
[00052] Administering may be any of the types of delivery described herein,
including but
not limited to: administering systemically, orally, intramuscular injection,
intravascular
injection, subcutaneous injection, parenteral, inhalation (e.g., from a
nebulizer), topical, etc.
Administering comprises delivering between 0.01 mg and 100 mg/day of the
synthetic
binding agent. Administering may comprise administering in an amount
sufficient to
agglutinate the pathogen and/or preserving or further enhancing muco-trapping
of the
pathogen. In some variations administering may comprise administering in
sufficient amount
to cause enchained growth, linking dividing bacteria from the same mother
bacteria together
into a long chain, which has the effect of creating large clumps too large to
permeate through
mucus. In some variations administering may comprise administering in
sufficient amount to
cause enchained growth, linking dividing bacteria from the same mother
bacteria together
into a long chain, which has the effect of creating large clumps that limit
their spread
throughout the body and/or limit their growth rate,
[00053] As mentioned above, the IgG Fab domains may have an amino acid
sequence that
is not identical to the one or more additional Fab domains.
[00054] Any of the synthetic binding agents described herein for enhancing
agglutination,
enchained growth and/or muco-trapping of a target having an epitope may
include: a human
or humanized Imnnmoglobulin G (IgG) having a pair of Fab domains, wherein the
human or
humanized IgG is linked to one or more additional immunoglobulin fragment
antigen binding
(Fab) domains by a linker (e.g., an amino acid/peptide linker), wherein the
one or more
additional Fab domains and the IgG Fab domains may all specifically bind to
the epitope of
the target, so that the synthetic binding agent binds to the target with high
affinity and
reduces the mobility of the target in mucus (e.g., to less than about x%
relative to its native
14
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
mobility in mucus, such as less than about 15%, 20%, 30%, 40%, 50%, etc.). The
one or
more additional Fab domains may comprise 2, 4, 6 or 8 additional Fab domains.
[00055] The target may be sperm and all of the one or more Fab domains and the
IgG Fab
domains may specifically bind to and epitope of CD52g (e.g., a repeating poly-
n-
acetyllactosaminyl structures on sperm, an N-linked glycosylated form of SEQ
ID: 1). As
mentioned, the additional Fab domains of the synthetic binding agent targeting
CD52g may
each comprise: (i) a heavy chain (HC) with a variable region (VH) comprising
complementarity determining regions (CDRs) having an amino acid sequence that
is between
100% and 80% identical to the amino acid sequence of: SEQ ID NO: 4; and/or
(ii) a light
chain (LC) with a variable region (VI) comprising complementarity determining
regions
(CDRs) having an amino acid sequence that is between 100% and 80% identical to
the amino
acid sequence of: SEQ ID NO: 7.
[00056] For example, a synthetic binding agent for inhibiting sperm mobility
through
mucus, may include: a human or humanized Imrnunoglobulin G (IgG) having a pair
of Fab
domains, wherein the human or humanized IgG is linked to one or more
additional
immunoglobulin fragment antigen binding (Fab) domains, wherein the one or more
additional
Fab domains and the IgG Fab domains all specifically bind to an epitope of
CD52g, so that
the synthetic binding agent reduces the mobility of sperm in mucus to less
than about 50%
relative to its native mobility in mucus. The one or more additional Fab
domains may
comprise 2, 4, 6 or 8 additional Fab domains. The epitope of CD52g may be
repeating poly-
n-acetyllactosaminyl structures, an N-linked glycosylated form of SEQ ID NO:
1. The
additional Fab domains may each comprise: (i) a heavy chain (HC) with a
variable region
(VH) comprising complementarity determining regions (CDRs) having an amino
acid
sequence that is between 100% and 80% identical to the amino acid sequence of:
SEQ ID
NO: 4; and/or (ii) a light chain (LC) with a variable region (VI) comprising
complementarity
determining regions (CDRs) having an amino acid sequence that is between 100%
and 80%
identical to the amino acid sequence of: SEQ ID NO: 7.
[00057] Also described herein are specific examples of synthetic binding
agents that target
bacterial pathogens, such as, in one non-limiting example, Klebsiella
bacillus. For example,
the IgG may be directed to an antigen of Klebsiella, e.g., an example of which
is provided in
SEQ ID NO: 39 to SEQ ID NO: 45, and the additional Fab domains may each
comprise: (i) a
heavy chain (HC) with a variable region (VH) comprising complementarity
determining
regions (CDRs) having the amino acid sequences that is identical or similar to
the HC VH
region of the IgG (e.g., in relation to the example of SEQ ID NO: 39 to SEQ ID
NO: 45,
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SEQ ID NO: 41); and (ii) a light chain (LC) with a variable region (VI)
comprising
complementarity determining regions (CDRs) having the amino acid sequences
that is
identical or similar to that of the IgG (e.g., in relation to the example of
SEQ ID NO: 39 to
SEQ ID NO: 45, SEQ ID NO: 44). For example, an synthetic binding agent
directed to an
antigen of Kiebsiella may have additional Fab domains each comprise: (i) a
heavy chain (HC)
with a variable region (VH) comprising complementarity determining regions
(CDRs) having
an amino acid sequence that is between 100% and 80% identical to the amino
acid sequence
of: SEQ ID NO: 41; and/or (ii) a light chain (LC) with a variable region (VI)
comprising
complementarity determining regions (CDRs) having an amino acid sequence that
is between
100% and 80% identical to the amino acid sequence of: SEQ ID NO: 44.
[00058] For example, a synthetic binding agent for treating or preventing
infection by a
Klebsiella bacillus pathogen may include: a human or humanized Immunoglobulin
G (IgG)
having a pair of Fab domains, wherein the human or humanized IgG is linked to
one or more
additional immunoglobulin fragment antigen binding (Fab) domains, wherein the
one or more
additional Fab domains and the IgG Fab domains all specifically bind to an
epitope specific
to Klebsiella bacillus, so that the synthetic binding agent reduces the
mobility of Klebsiella
bacillus in mucus. The one or more additional Fab domains may comprise 2, 4, 6
or 8
additional Fab domains. The additional Fab domains may each comprise: (1) a
heavy chain
(HC) with a variable region (VH) comprising complementarity determining
regions (CDRs)
having an amino acid sequence that is between 100% and 80% identical to the
amino acid
sequence of: SEQ ID NO: 41; and/or (ii) a light chain (LC) with a variable
region (VL)
comprising complementarity determining regions (CDRs) having an amino acid
sequence
that is between 100% and 80% identical to the amino acid sequence of: SEQ ID
NO: 44.
1000591 Another non-limiting example of a synthetic binding agent that targets
a bacterial
pathogen are synthetic binding agents that target Salmonella bacillus. The IgG
portion of the
synthetic binding agent may be directed to an antigen of Salmonella (such as
described in
SEQ ID NO: 67-73), and the additional Fab domains (which are directed to the
same target
antigen) may have a similar or identical amino acid sequence as the Fab domain
of the IgG.
For example, the additional Fab domains may each comprise: (i) a heavy chain
(HC) with a
variable region (VH) comprising complementarity determining regions (CDRs)
having the
amino acid sequences of the IgG (e.g., SEQ ID NO: 69); and/or (ii) a light
chain (LC) with a
variable region (VI) comprising complementarity determining regions (CDRs)
having the
amino acid sequences of the IgG (e.g., SEQ ID NO: 72). In some variations, the
synthetic
binding agent directed to an antigen of Salmonella includes additional Fab
domains that each
16
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
comprise: (i) a heavy chain (HC) with a variable region (VH) comprising
complementarity
determining regions (CDRs) having an amino acid sequence that is between 100%
and 80%
identical to the amino acid sequence of: SEQ ID NO: 69; and/or (ii) a light
chain (LC) with a
variable region (VL) comprising complementarity determining regions (CDRs)
having an
amino acid sequence that is between 100% and 80% identical to the amino acid
sequence of:
SEQ ID NO: 72.
[000601 For example, a synthetic binding agent for treating or preventing
infection by a
Salmonella bacillus pathogen may include: a human or humanized Immunoglobulin
G (IgG)
having a pair of Fab domains, wherein the human or humanized IgG is linked to
one or more
additional immunoglobulin fragment antigen binding (Fab) domains, wherein the
one or more
additional Fab domains and the IgG Fab domains all specifically bind to an
epitope specific
to Salmonella bacillus, so that the synthetic binding agent reduces the
mobility of Salmonella
bacillus in mucus. The one or more additional Fab domains may comprise 2, 4, 6
or 8
additional Fab domains. The additional Fab domains may each comprise: (i) a
heavy chain
(HC) with a variable region (VH) comprising complementarity determining
regions (CDRs)
having an amino acid sequence that is between 100% and 80% identical to the
amino acid
sequence of: SEQ ID NO: 69; and/or (ii) a light chain (LC) with a variable
region (VL)
comprising complementarity determining regions (CDRs) having an amino acid
sequence
that is between 100% and 80% identical to the amino acid sequence of: SEQ ID
NO: 72.
[000611 Another non-limiting example of a synthetic binding agent that targets
a bacterial
pathogen are synthetic binding agents that target Neisseria gonorrhoeae. The
IgG portion of
the synthetic binding agent may be directed to an antigen of Neisseria
gonorrhoeae (such as
described in SEQ ID NO: 102-108), and the additional Fab domains (which are
directed to
the same target antigen) may have a similar or identical amino acid sequence
as the Fab
domain of the IgG. For example, the additional Fab domains may each comprise:
(i) a heavy
chain (HC) with a variable region (VH) comprising complementarity detellnining
regions
(CDRs) having the amino acid sequence that is similar or identical to the HC
VH of the IgG
(e.g., SEQ ID NO: 104); and/or (ii) a light chain (LC) with a variable region
(VL) comprising
complementarity determining regions (CDRs) having the amino acid sequence that
is similar
or identical to that of the LC VL of the IgG (e.g., SEQ ID NO: 107). For
example, a synthetic
binding agent directed against Neisseria gonorrhoeae may include an IgG
against an antigen
of Neisseria gonorrhoeae and additional Fab domains that each comprise: (i) a
heavy chain
(HC) with a variable region (VH) comprising complementarity determining
regions (CDRs)
having an amino acid sequence that is between 100% and 80% identical to the
amino acid
17
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
sequence of the IgG (e.g., SEQ ID NO: 104); and/or (ii) a light chain (LC)
with a variable
region (VI) comprising complementarity determining regions (CDRs) having an
amino acid
sequence that is between 100% and 80% identical to the amino acid sequence of
that of the
IgG (e.g., SEQ ID NO: 107).
[00062] For example, a synthetic binding agent for treating or preventing
infection by a
Neisseria gonorrhoeae may include: a human or humanized Immunoglobulin G (IgG)
having
a pair of Fab domains, wherein the human or humanized IgG is linked to one or
more
additional immunoglobulin fragment antigen binding (Fab) domains, wherein the
one or more
additional Fab domains and the IgG Fab domains all specifically bind to an
epitope specific
to Neisseria gonorrhoeae, so that the synthetic binding agent reduces the
mobility of
Neisseria gonorrhoeae in mucus. The one or more additional Fab domains may
comprise 2,
4, 6 or 8 additional Fab domains. The additional Fab domains may each
comprise: (i) a heavy
chain (HC) with a variable region (VH) comprising complementarity determining
regions
(CDRs) having an amino acid sequence that is between 100% and 80% identical to
the amino
acid sequence of: SEQ ID NO: 104; and/or (ii) a light chain (LC) with a
variable region (VI)
comprising complementarity determining regions (CDRs) having an amino acid
sequence
that is between 100% and 80% identical to the amino acid sequence of: SEQ ID
NO: 107.
100063] Any of the synthetic binding agents described herein may include at
least one
additional Fab domains is linked to a Fab domain of the pair of Fab domains of
the IgG;
alternatively or additionally, any of the synthetic binding agents described
herein may include
at least one addition additional Fab domain that is linked to an Fe region of
the IgG. The IgG
may comprise at least one Fe region that is a naturally occurring sequence.
The IgG may
comprise at least one Fe region comprising one or more mutations that enhance
or decrease
binding to Fe receptors.
[00064] The one or more additional Fab domains may be linked to the IgG via a
linker, as
described here, such as a flexible peptide linker comprising an amino acid
sequence
comprising n pentapeptide repeats consisting of Glycine (G) and Serine (S),
wherein n is
between 3 and 8
[00065] In general, the IgG Fab domains may have an amino acid sequence that
is not
identical to the one or more additional Fab domains, while still recognizing
the same antigen
with the same (or nearly equivalent) affinity.
[00066] Also described herein are isolated nucleic acid molecules encoding any
of these
synthetic binding agents, and/or a vector comprising such isolated nucleic
acid molecules. In
some variations, the nucleotide sequence encoding the additional IgG (e.g., HV
and/or LC)
18
CA 03113059 2021-03-16
WO 2020/061560
PCT/US2019/052396
may be different from the nucleotide sequence encoding the region of the IgG
having
corresponding binding affinity; the resulting amino acid sequence may be
identical or nearly
identical (e.g., having 75% homology or more, 80% homology or more, 85%
homology or
more, 90% homology or more, 95% homology or more, etc., including
corresponding
substitutions), Also described herein are isolated host cells or a non-human
organisms
transformed or transfected with these nucleic acid molecules.
[00067] Also described herein are compositions of any of these synthetic
binding agents
and a pharmaceutically acceptable carrier.
BRIEF DESCRIPTION OF THE DRAWINGS
[000681 The novel features of the invention are set forth with particularity
in the claims
that follow. A better understanding of the features and advantages of the
present invention
will be obtained by reference to the following detailed description that sets
forth illustrative
embodiments, in which the principles of the invention are utilized, and the
accompanying
drawings of which:
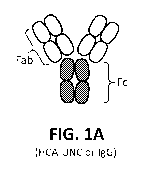
[00069] FIG. IA is a schematic example of a mAb directed against an epitope
that may
form a core of a synthetic binding agent (e.g., recombinant inAb) having
enhanced
agglutination and/or muco-trapping. In some variations the core antibody is
directed to an
antigen associated with sperm, and particularly human sperm. Alternatively,
other epitopes,
including viral or bacterial epitopes, may be used.
[00070] FIGS. 1B-1F illustrate examples of synthetic, engineered, constructs
against an
epitope of a target having enhanced agglutination and/or muco-trapping, using
the core
shown in FIG. IA. FIG. 18 is an example of a Fab-IgG variation (e.g.,
duplicate Fab
domain(s) linked to the amino ends of the core IgG). FIG. 1C is an example of
an IgG-Fab
variation (e.g., duplicate Fab domain(s) linked to the carboxyl ends of the
core IgG). FIG.
ID is an example of a Fab-IgG-Fab variation (e.g., duplicate Fab domains
linked to both the
carboxyl ends and the amino ends of the core IgG). FIG. lE is an example of a
Fab-IgG-Fab-
Fab variation (e.g., duplicate Fab domains linked to both the carboxyl ends
and the amino
ends of the core IgG). FIG. 1F is an example of a Fab-Fab-IgG-Fab-Fab
variation of a
synthetic binding agent as described herein (e.g., a 10-mer, having 4
additional pairs of Fab
domains, two linked to the carboxyl ends and two to the amino ends of the core
IgG).
[00071] FIG. 1G is a graphical table further illustrating structural
properties of the core
IgG and synthetic constructs shown in FIGS. 1A-1E.
19
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[00072] FIGS. 2A-2B schematically illustrate muco-trapping. In this example,
the target is
a virus; other targets may include bacteria and sperm. In FIG. 2A the
schematic shows how a
target (e.g., virus) may readily diffuse across native mucus (in the absence
of any virus-
specific Ab). FIG. 213 shows how an anti-viral Ab, and particularly one that
only weakly (in
an unbound state) interacts with mucins and therefore freely diffuses through
the mucus, may
trap the target in mucus by adhesive interactions. Arrows indicate the small
fraction of free
(not virus-bound) Ab that will interact with mucins at any given time.
[00073] FIG. 3A illustrates agglutination of human sperm.
[00074] FIG. 3B illustrates trapping of sperm in mucus (e.g., cervicovaginal
mucus
(CVM) and endocervical mucus (CM)).
[00075] FIG. 4A illustrates an SDS-PAGE analysis of one example synthetic
binding
agent having enhanced agglutination and/or muco-trapping potencies (referred
to as MM-
006) compared to unmodified IgG. In this example, the synthetic binding agent
is
configured as an Fab-IgG-Fab construct and is compared to HCA IgG).
[00076] FIG. 4B shows a color-matched SEC/MALS analysis of MM-006 from FIG.
4A.
The SEC curves (solid curves, right y-axis) shows homogenous expression
profile and MALS
data (thicker line, left y-axis) confirms desired molecular weight (MW).
[00077] FIGS. 5A and 5B illustrate the application of the construct (MM-006)
for FIG.
4A-4B to reduce the mobility/motility of sperm. FIG. 5A is a brightfield image
of sperm 30s
after exposure to MM-006. FIG. 5B shows the sperm agglutination potency of MM-
006 and
HCA relative to PBS. * indicates p <0.05; ** p< 0.01; *** p <0.001.
[00078] FIGS. 6A-6B illustrate examples of an apparatus for delivering a
construct (e.g., a
contraceptive synthetic binding agent having multiple Fab repeats). FIG. 6A
shows a
transparent view of a ring with four capsules inserted into ring cavities.
FIG. 6B shows
exploded and assembled views of sustained release capsules: coated antibody
pellet (center
region, top), closed end capsule piece (right end of capsule), and capsule cap
with release
window (left end of capsule).
[00079] FIG. 6C shows daily (top) and cumulative (bottom) release of human IgG
over at
least 28 days with different sustained release capsule formulations.
[00080] FIG. 7 shows a silver stained gel showing human IgG recovered from
capsules
remained intact even after up to 4 weeks of exposure to human CVM (replaced
every 5 days
to maintain the degradative environment). Pure IgG band and PBS are shown as
controls; all
incubations were at 37 C. Each band represents IgG recovered from an
individual capsule.
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
1000811 FIGS. 8A-8B show sperm agglutination in the presence of one of the
synthetic
binding agents having multiple Fab repeats described herein (configured as an
HCA). FIG.
8A shows a light microscopy image of agglutinated sperm with a round cell
(shown by
arrow). FIG. 813 is a fluorescent image of the same field; the construct (HCA)
in this example
was conjugated to Dylight633 (red) and labeled the entire length of all
spermatozoa.
CMFDA (Live CellTracker green) labeled seminal leukocyte was also positive for
HCA as
indicated by the co-labeling (white arrow).
[00082] FIG. 9 is a graph illustrating differential scanning calorimetry of an
exemplary
synthetic binding agent having multiple Fab repeats (e.g., Fab-IgG, bottom)
and an scFv-IgG
(top) construct. The scFv construct shows unfolding at a much lower
temperature than the
synthetic binding agent having multiple Fab repeats (Fab-IgG) due to the lack
of a CH1/CL
domain.
1000831 FIG. 10 shows an Analytical Size Exclusion Chromatography graph of the
core
alone (IgG, bottom), an scFv-IgG construct (middle), and an exemplary
synthetic binding
agent having multiple Fab repeats (Fab-IgG, top) after single-step protein A
purification.
Both the IgG and Fab-IgG construct showed a single sharp peak at their
expected molecular
weight. The scFv-IgG construct shows formation of high molecular-weight
aggregates.
1000841 FIG. 11A shows an SDS-PAGE analysis of multimeric synthetic binding
agent
having multiple Fab repeats. In this example the multimeric constructs are HCA
(e.g.,
contraceptive) synthetic binding agents having multiple Fab repeats, where all
of the Fabs are
directed against the same epitope of CD52g (e.g., all share the same targeting
sequence).
[00085] FIG. 11B is a color-matched SEC/MALS analysis of the different
multimeric
HCA constructs from FIG. 11A. SEC curves for each construct (solid curves,
right y-axis)
shows homogenous expression profile and MALS data (dotted lines, left y-axis)
confirms
desired molecular weight of each construct (also compared to the core IgG).
[00086] FIG. 12A shows a whole-sperm ELISA verifying that different multimeric
synthetic binding agents having multiple Fab repeats (configured as HCA
constructs) possess
functional Fab that binds sperm.
[00087] FIGS. 1213 and 12C show brightfield images of sperm 1 minute after
treatment
with PBS (FIG. 1213), or after 30 seconds after treatment with a multimeric
synthetic binding
agent having multiple Fab repeats (e.g., configured as a Fab-IgG-Fab HCA
construct) (FIG.
12C).
[00088] FIG. 13 shows the muco-trapping potency of a Fab-IgG, as reflected by
the
ensemble geometric average of effective diffusivity (Deff) of PEGylated
nanoparticles (PS-
21
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
PEG) in human CVM with no Ab, control IgG (Ctrl IgG) or a multimeric synthetic
binding
agent having multiple Fab repeats (e.g., an anti-PEG Fab-IgG construct),
compared to
uncoated nanoparticles (PS-COOH) in untreated CVM. Data is plotted for
distinct samples
(indicated with different color circles) with averages indicated by solid
lines. * indicates a
statistically significant difference (p <0.05).
[00089] FIGS. 14A and 14B show an amino acid sequence comparison between
portions
of the heavy (FIG. 14A; the VH portion of SEQ ID NO: 3 is compared to a VH
portion of a
germline sequence) and light (FIG. 14B; the VL portion of SEQ ID NO: 7 is
compared to a
VL portion of a germline sequence) chain sequences of a core IgG against an
epitope on
human sperm, CD52g, compared to a germline sequence (e.g., native IgG).
100090] FIGS. 15A-15D illustrate one example of the production and
characterization of
multimeric anti-sperm IgG antibodies. FIG. 15A schematically illustrates
examples of anti-
sperm IgG, Fab, Fe, Fab-IgG, and IgG-Fab. FIG. 15B is a gel showing non-
reducing IgG,
Fab-IgG, and IgG-Fab. FIG. 15C shows a reducing SDS-Page analysis comparing
IgG, Fab-
IgG, and IgG-Fab after expression in Expi293 cells and purification by protein
A/G
chromatography. FIG. 15D is a graph illustrating the purity and homogeneity of
the purified
multimeric antibodies (IgG compared to Fab-IgG and IgG-Fab) via analytical SEC-
MALS
analysis.
[00091] FIGS. 16A-16C illustrate the characterization of multimeric anti-sperm
IgG
antibodies. FIG. 16A shows the molar mass versus time of the IgG, Fab-IgG and
IgG-Fab
respectively as determined by SEC¨MALS. FIG. 16B shows the values of melting
temperature (Tm) and aggregating temperature (Tagg) of as determined by
nanoDSF by
measuring intrinsic fluorescence of a protein and changes in back-reflection
respectively.
FIG. 16C is a graph showing whole sperm ELISA analysis to assess the binding
potency of
indicated antibodies. Motavizumab (anti-RSV IgG1) is used as the isotype
control. ELISA
was performed in triplicates and repeated three times using 3 unique
specimens. Lines
indicate arithmetic mean concentration and standard error of mean.
[00092] FIGS, 17A-17B illustrate sperm agglutination potency of parent IgG,
Fab-IgG and
IgG-Fab using purified motile sperm (10x106progressively motile sperm per mL).
FIG. 17A
graphically illustrates measured sperm agglutination potency of the parent and
multimeric
anti-sperm IgGs by quantifying the percentage of sperm that remains
progressively motile
post Ab-treatment at different concentrations compared to pre-treatment
condition. FIG. 17B
shows the percentage of agglutination-escaped progressive sperm post-treatment
normalized
22
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
to the negative control for further comparison. Data represents 6 unique sperm
specimens.
Lines indicate arithmetic mean concentration and standard error of mean.
[00093] FIGS. I8A-18B show sperm agglutination kinetics of parent IgG, Fab-IgG
and
IgG-Fab using purified motile spenu (10x106progressively motile sperm per mL).
FIG. 18A
is a graph showing quantification of the time required to achieve 90%
agglutination of
progressive sperm compared to untreated control. The CASA analysis was
obtained every
30s post-treatment until 90s. FIG. 18B shows the rate of speim agglutination
of indicated
anti-sperm antibodies by quantifying the percentage of agglutinated sperm post
Ab-treatment
at three different time points compared to pre-treatment. Data represents 6
unique sperm
specimens. Lines indicate arithmetic mean concentration and standard error of
mean.
[000941 FIG. 19 illustrates the muco-trapping potency of parent IgG, Fab-IgG
and IgG-
Fab using pH-neutralized female CVM and purified motile sperm
(1x106progressively motile
sperm/mL). Muco-trapping potency of indicated antibodies was assessed by
performing real-
time video microscopy on fluorescently labelled sperm suspended in Ab-treated
(25ug/m1)
CVM. A neural network tracker customized with standardized sperm motility
parameters was
used in all recorded videos to quantify the percentage of progressively motile
sperm present
in the mucus specimen.
[00095] FIGS. 20A-20D illustrate production and characterization of multimeric
anti-
sperm IgG antibodies. FIG. 20A schematically illustrates anti-sperm Fab, Fe,
IgG, Fab-IgG-
Fab (FIF), Fab-IgG-Fab-Fab (FIFF) and Fab-Fab-IgG-Fab-Fab (FFIFF). In this
example, N-
terminal and C-terminal Fabis of FIF, FIFF and FFIFF contains fully intact
anti-sperm Fab/s
with VH, VL, CH1, and CL. FIG. 20B is a gel showing non-reducing FIF, FIFF and
FFIFF
(compared to IgG). FIG, 20C is a reducing SDS-Page analysis of the indicated
antibodies
(IgG, FIF, FIFF, and FFIFF) after expression in Expi293 cells and purification
by protein
AJG chromatography. FIG. 20D is a graph demonstrating the purity and
homogeneity of the
purified multimeric antibodies via analytical SEC-MALS analysis.
[00096] FIGS. 21A-21C illustrate additional characterization of multimeric
anti-sperm IgG
antibodies. FIG. 21A illustrates the molar mass versus time of the IgG, FIF,
FIFF and FFIFF
respectively as determined by SEC¨MALS. FIG. 21B graphically illustrates the
value of
melting temperature (Tm) and aggregating temperature (Tagg) of indicated
antibodies as
determined by nanoDSF by measuring intrinsic fluorescence of a protein and
changes in
back-reflection respectively. FIG. 21C is a graph showing a whole Sperm ELISA
analysis to
assess the binding potency of the indicated antibodies. Motavizumab (anti-RSV
IgG1) is used
23
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
as the isotype control. ELISA was performed in in triplicates and repeated
three times using 3
unique specimens. Lines indicate arithmetic mean concentration and standard
error of mean.
1000971 FIG. 22A-22D illustrates sperm agglutination potency of parent IgG and
multimeric constructs using purified motile sperm (10x106 progressively motile
sperm per
mL) and whole semen. FIG. 22A is a graph showing sperm agglutination potency
of the
parent IgG, FIF, FIFE and FFIFF by quantifying the percentage of sperm that
remains
progressively motile post Ab-treatment compared to pre-treatment condition
using purified
motile sperm (10x106 progressively motile sperm per mL). FIG. 22B shows the
percentage
of agglutination-escaped progressive sperm post Ab-treatment using purified
motile sperm
that was normalized to the negative control for further comparison. FIG. 22C
is a graph
showing measured sperm agglutination potency of the parent IgG and FFIFF by
quantifying
the percentage of sperm that remains progressively motile post Ab-treatment
compared to
pre-treatment condition using whole semen. FIG. 22D shows the percentage of
agglutination-escaped progressive sperm post Ab-treatment using whole semen
that was
normalized to the negative control for further comparison. Data represents 6
unique sperm
specimens. Lines indicate arithmetic mean concentration and standard error of
mean.
1000981 FIGS. 23A-23D shows sperm agglutination kinetics of parent IgG and
multimeric
constructs using purified motile sperm (10x106progressive1Ly motile sperm per
mL) and
whole semen. FIG. 23A illustrates agglutination kinetics of parent IgG, FIF,
FIFF and FFIFF
determined by quantifying the time required to achieve 90% agglutination of
progressive
sperm compared to untreated control using purified motile sperm
(10x106progressively
motile sperm per mL). CASA analysis was obtained every 30s post-treatment
until 90s. FIG.
23B illustrates the rate of sperm agglutination of parent IgG, FIF, FIFF and
FFIFF by
quantifying the percentage of agglutinated sperm post Ab-treatment at three
different time
points compared to pre-treatment using purified motile sperm (10x106
progressively motile
sperm per mL). FIG. 23C shows the agglutination kinetics of parent IgG and
FFIFF by
quantifying the time required to achieve 90% agglutination of progressive
sperm compared to
untreated control using whole semen. The CASA analysis was obtained every 30s
post-
treatment until 90s, FIG. 23D is a graph showing the rate of sperm
agglutination of parent
IgG and FFIFF by quantifying the percentage of agglutinated sperm post Ab-
treatment at
three different time points compared to pre-treatment using whole semen. Data
represents 6
unique sperm specimens. Lines indicate arithmetic mean concentration and
standard error of
mean.
24
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[00099] FIGS. 24A-24D show sperm agglutination kinetics of parent IgG and
FFIFF using
low and high concentration of purified motile sperm (2x106and
50x106progressive
sperm/mL). FIG. 24A shows the agglutination kinetics of IgG and FFIFF by
quantifying the
time required to achieve 90% agglutination of progressive sperm compared to
untreated
control. The CASA analysis was obtained every 30s post-treatment until 90s
using purified
motile spetni (2x106 progressive sperm/mL). FIG. 24B shows the rate of sperm
agglutination
of IgG and FFIFF by quantifying the percentage of agglutinated sperm post Ab-
treatment at
three different time points compared to pre-treatment using purified motile
sperm (2x106
progressive sperm/mL). FIG. 24C shows the agglutination kinetics of IgG and
FFIFF by
quantifying the time required to achieve 90% agglutination of progressive
sperm compared to
untreated control using purified motile sperm (50x106progressive sperm/mL).
The CASA
analysis was obtained every 30s post-treatment until 90s. FIG. 24D shows the
rate of sperm
agglutination of IgG and FFIFF by quantifying the percentage of agglutinated
sperm post Ab-
treatment at three different time points compared to pre-treatment using
purified motile sperm
(50x106 progressive sperm/mL). Data represents 6 unique sperm specimens. Lines
indicate
arithmetic mean concentration and standard error of mean.
[000100] FIG. 25 is a graph showing spew' agglutination potency of parent IgG,
FIF and
FFIFF using whole semen in sheep study. Agglutination potency of IgG, FIF and
FFIFF was
measured in vivo by instilling Abs into sheep vagina, followed by human semen
and
simulated intercourse. The sperm motility was assessed immediately in the
fluids from the
sheep vagina. Data represents 3 unique sheep studies for FIF and FFIFF, and 1
sheep study
for IgG at both 33 ug/ml and 333 ug/ml. Lines indicate arithmetic mean
concentration and
standard error of mean,
[000101] FIGS. 26 and 27A illustrate agglutination potency of exemplary films
(e.g.,
Nicotiana-produced films) of parent IgG and FIF using purified motile sperm
(10x106
progressive sperm/mL) and whole semen. FIG, 26 shows sperm agglutination
potency of the
parent IgG-Film and FIF-Film by quantifying the percentage of sperm that
remains
progressively motile post Ab-treatment compared to pre-treatment condition
using purified
motile sperm (10x106 progressively motile sperm per mL). FIG. 27A illustrates
the
percentage of agglutination-escaped progressive sperm post Ab-treatment using
purified
motile sperm; the data is normalized to the negative control.
[000102] FIG. 27B illustrates sperm agglutination potency of the parent IgG-
Film and FIF-
Film by quantifying the percentage of sperm that remains progressively motile
post Ab-
treatment compared to pre-treatment condition using whole semen.
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[000103] FIG. 27C shows the percentage of agglutination-escaped progressive
sperm post
Ab-treatment using whole semen, normalized to the negative control. Data
represents 6
unique sperm specimens. Lines indicate arithmetic mean concentration and
standard error of
mean.
[000104] FIGS. 28A-28D show agglutination kinetics of exemplary films (e.g.,
Nicotiana-
produced films) of parent IgG and FIF using purified motile sperm
(10x106progressively
motile sperrn/mL) and whole semen. FIG. 28A illustrates agglutination kinetics
of indicated
antibodies by quantifying the time required to achieve 90% agglutination of
progressive
sperm compared to untreated control using purified motile sperm (10x106
progressively
motile sperm/mL). The CASA analysis was obtained every 30s post-treatment
until 90s. FIG.
28B illustrates the rate of sperm agglutination of indicated anti-sperm
antibodies by
quantifying the percentage of agglutinated sperm post Ab-treatment at three
different time
points compared to pre-treatment using purified motile sperm
(10x106progressively motile
sperm/mL). FIG. 28C is a graph showing agglutination kinetics of indicated
antibodies by
quantifying the time required to achieve 90% agglutination of progressive
sperm compared to
untreated control using whole semen. The CASA analysis was obtained every 30s
post-
treatment until 90s. FIG. 28D shows the measured rate of sperm agglutination
of indicated
anti-sperm antibodies (quantifying the percentage of agglutinated sperm post
Ab-treatment at
three different time points compared to pre-treatment using whole semen). Data
represents 6
unique sperm specimens. Lines indicate arithmetic mean concentration and
standard error of
mean.
[000105] FIGS. 29A-29D illustrate agglutination kinetics exemplary films
(e.g., Nicotiana-
produced films) of parent IgG and FIF using low and high concentration of
purified motile
speun (2x106 and 50x106 progressively motile sperm/mL). FIG. 29A shows the
agglutination
kinetics of indicated antibodies based on quantification of the time required
to achieve 90%
agglutination of progressive sperm compared to untreated control using
purified motile sperm
(2x106 progressively motile speitn/mL). The CASA analysis was obtained every
30s post-
treatment until 90s. FIG. 29B shows the rate of sperm agglutination of
indicated anti-sperm
antibodies as determined by quantifying the percentage of agglutinated sperm
post Ab-
treatment at three different time points compared to pre-treatment using
purified motile sperm
(2x106 progressively motile sperm/mL). FIG. 29C shows the agglutination
kinetics of
indicated antibodies by quantifying the time required to achieve 90%
agglutination of
progressive sperm compared to untreated control using purified motile sperm
(50x106
progressively motile sperm/mL). The CASA analysis was obtained every 30s post-
treatment
26
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
until 90s. FIG. 29D shows the rate of sperm agglutination of indicated anti-
sperm antibodies
by quantifying the percentage of agglutinated sperm post Ab-treatment at three
different time
points compared to pre-treatment using purified motile sperm (50x106
progressively motile
speillilmL). Data represents 6 unique sperm specimens. Lines indicate
arithmetic mean
concentration and standard error of mean.
[000106] FIG. 30 illustrates the agglutination kinetics of exemplary films
(e.g., Nicotiana-
produced films) of parent IgG and FIF in an acidic environment using purified
motile sperm
(20x106progressively motile sperm/mL) and 24hr treatment with lactic acid. The
agglutination kinetics of lactic acid-treated antibodies were assessed by
quantifying the time
required to achieve 90% agglutination of progressive sperm compared to
untreated control.
The CASA analysis was obtained every 30s post-treatment until 90s. Data
represents 2
unique sperm specimens. Note: Lactic acid (LA) treated antibodies were
neutralized with
seminal plasma (SP) and then followed by dilution with SP or saline media i.e.
MI-IM.
[000107] FIGS. 31A-31D illustrate characterization of a Nicotiana-produced FIF-
Film (film
of Fab-IgG-Fab synthetic binding agent) and Expi293-produced FFIFF (e.g., Fab-
Fab-IgG-
Fab-Fab) synthetic binding agent post-nebulization. FIG. 31A shows a non-
reducing SDS-
Page analysis of the indicated antibodies before and after nebulization using
mesh-nebulizer,
and FIG. 31B shows a reducing SDS-Page analysis of the indicated antibodies
before and
after nebulization using mesh-nebulizer. In FIGS. 31C and 31D, whole-sperm
ELISA
analysis was used to assess the binding potency of the FIF-Film (shown in FIG.
31A) and
FFIFF post-nebulization (FIG. 31D). ELISA was performed in triplicates and
repeated twice
using the same donor specimens. Lines indicate arithmetic mean concentration
and standard
error of mean.
10001081 FIGS. 32A-32B illustrate the production and characterization of
multimeric anti-
RSV IgG antibodies. FIG. 32A shows a non-reducing gel and reducing SDS-Page
analysis of
a multimeric IgG against RSV (Motavizumab as parent IgG) after expression in
Expi293
cells and purification by protein A/G chromatography. FIG. 32B is an RSV ELISA
analysis
to assess the binding potency of the indicated antibodies. Synagis/Palivizumab
(anti-RSV
IgG1) is used as the positive control, ELISA was performed in triplicates.
DETAILED DESCRIPTION
[0001091 The methods and compositions (including the multimeric synthetic
binding agent
having multiple Fab repeats for enhancing agglutination, facilitating
enchained growth and/or
improving muco-trapping) described herein are based, in part, on the discovery
that foreign
27
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
bodies, including pathogens such as virus and bacteria, as well as sperm, may
be more
strongly trapped by mucus following binding with a multirneric antibody-based
constructs.
The constructs may be engineered to stop the penetration of target (e.g.,
pathogen, sperm,
etc.) through mucus by improving the agglutination potency, facilitating
enchained growth of
the pathogen and/or enabling muco-trapping, and may prevent and/or treat
infection, and/or
provide contraception.
10001101 The present invention is explained in greater detail below. This
description is not
intended to be a detailed catalog of all the different ways in which the
invention may be
implemented, or all the features that may be added to the instant invention.
For example,
features illustrated with respect to one embodiment may be incorporated into
other
embodiments, and features illustrated with respect to a particular embodiment
may be deleted
from that embodiment. In addition, numerous variations and additions to the
various
embodiments suggested herein will be apparent to those skilled in the art in
light of the
instant disclosure which do not depart from the instant invention. Hence, the
following
specification is intended to illustrate some particular embodiments of the
invention, and not
to exhaustively specify all permutations, combinations and variations thereof.
10001111 Unless the context indicates otherwise, it is specifically intended
that the various
features of the invention described herein can be used in any combination.
Moreover, the
present invention also contemplates that in some embodiments of the invention,
any feature
or combination of features set forth herein can be excluded or omitted, To
illustrate, if the
specification states that a complex comprises components A, B and C, it is
specifically
intended that any of A, B or C, or a combination thereof, can be omitted and
disclaimed
singularly or in any combination.
[0001121 Unless otherwise defined, all technical and scientific terms used
herein have the
same meaning as commonly understood by one of ordinary skill in the art to
which this
invention belongs. The terminology used in the description of the invention
herein is for the
purpose of describing particular embodiments only and is not intended to be
limiting of the
invention.
[000113] Except as otherwise indicated, standard methods known to those
skilled in the art
may be used for production of recombinant and synthetic polypeptides,
antibodies or antigen-
binding fragments thereof, manipulation of nucleic acid sequences, and
production of
transformed cells. Such techniques are known to those skilled in the art. See,
e.g.,
SAMBROOK et al., MOLECULAR CLONING: A LABORATORY MANUAL 2nd Ed.
(Cold Spring Harbor, N.Y., 1989); F. M. AUSUBEL et oL CURRENT PROTOCOLS IN
28
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
MOLECULAR BIOLOGY (Green Publishing Associates, Inc. and John Wiley & Sons,
Inc.,
New York).
[000114] All publications, patent applications, patents, nucleotide sequences,
amino acid
sequences and other references mentioned herein are incorporated by reference
in their
entirety.
[000115] As used in the description of the invention and the appended claims,
the singular
forms "a," "an" and "the" are intended to include the plural forms as well,
unless the context
clearly indicates otherwise.
[000116] As used herein, "and/or" refers to and encompasses any and all
possible
combinations of one or more of the associated listed items, as well as the
lack of
combinations when interpreted in the alternative ("or").
[000117] Moreover, the present invention also contemplates that in some
embodiments of
the invention, any feature or combination of features set forth herein can be
excluded or
omitted.
[000118] The term "about," as used herein when referring to a measurable value
such as an
amount of a compound or agent of this invention, dose, time, temperature, and
the like, is
meant to encompass variations of 10%, 5%, 1%, 0.5%, or even 0.1% of the
specified
amount.
[000119] Unless otherwise indicated, all numbers expressing quantities of
ingredients,
properties such as reaction conditions, and so forth used in the specification
and claims are to
be understood as being modified in all instances by the term "about".
Accordingly, unless
indicated to the contrary, the numerical parameters set forth in this
specification and claims
are approximations that can vary depending upon the desired properties sought
to be obtained
by the presently-disclosed subject matter,
[000120] As used herein, ranges can be expressed as from "about" one
particular value,
and/or to "about" another particular value. It is also understood that there
are a number of
values disclosed herein, and that each value is also herein disclosed as
"about" that particular
value in addition to the value itself. For example, if the value "10" is
disclosed, then "about
10" is also disclosed. It is also understood that each unit between two
particular units is also
disclosed. For example, if 10 and 15 are disclosed, then 11, 12, 13, and 14
are also disclosed.
[000121] The transitional phrase "consisting essentially of' means that the
scope of a claim
is to be interpreted to encompass the specified materials or steps recited in
the claim, and
those that do not materially affect the basic and novel characteristic(s) of
the claimed
invention.
29
CA 03113059 2021-03-16
WO 2020/061560
PCT/US2019/052396
[000122] As used herein, the term "poiypeptide" encompasses both peptides and
proteins,
unless indicated otherwise.
[000123] A "nucleic acid" or "nucleotide sequence" is a sequence of nucleotide
bases, and
may be RNA, DNA or DNA-RNA hybrid sequences (including both naturally
occurring and
non-naturally occurring nucleotide), but is preferably either single or double
stranded DNA
sequences.
[000124] As used herein, an "isolated" antibody means an antibody separated or
substantially free from at least some of the other components of the naturally
occurring
organism or virus, for example, the cell structural components or other
polypeptides or
nucleic acids commonly found associated with the antibody. The term also
encompasses
antibodies that have been prepared synthetically.
[000125] By the terms "treat," "treating," or "treatment of' (or grammatically
equivalent
terms) it is meant that the severity of the subject's condition is reduced or
at least partially
improved or ameliorated and/or that some alleviation, mitigation or decrease
in at least one
clinical symptom is achieved and/or there is a delay in the progression of the
condition.
[000126] As used herein, the terms "prevent," "prevents," or "prevention" and
"inhibit,"
"inhibits," or "inhibition" (and grammatical equivalents thereof) are not
meant to imply =
complete abolition of disease and encompasses any type of prophylactic
treatment that
reduces the incidence of the condition, delays the onset of the condition,
and/or reduces the
symptoms associated with the condition after onset.
[000127] An "effective," "prophylactically effective," or "therapeutically
effective" amount
as used herein is an amount that is sufficient to provide some improvement or
benefit to the
subject. Alternatively stated, an "effective," "prophylactically effective,"
or "therapeutically
effective" amount is an amount that will provide some delay, alleviation,
mitigation, or
decrease in at least one clinical symptom in the subject. Those skilled in the
art will
appreciate that the effects need not be complete or curative, as long as some
benefit is
provided to the subject.
[000128] As used herein, the term "trapping potency" refers to the ability of
an antibody
that specially binds to a target pathogen or sperm to inhibit movement of the
pathogen or
sperm through mucus. Trapping potency can be measured by methods known in the
art and as
disclosed herein. Trapping potency can be quantitated, e.g., as the amount of
antibody (e.g.,
concentration of antibody in mucus) needed to reduce the mobility of at least
50% (e.g., at
least 55%, at least 60%, at least 65%, at least 70%, at least 75%, etc.) of
the pathogen or
sperm within the mucus gel to at least one-half (e.g., one-quarter, one-tenth,
etc.) of its native
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
mobility in solution (e.g., saline) and/or in mucus. For sperm, trapping
potency can also be
quantitated, e.g., as the amount of antibody (e.g., concentration of antibody
in mucus) needed
to reduce the fraction of progressively motile sperm by at least 50% (e.g. at
least 55%, at least
60%, at least 65%, at least 70%, at least 75%, etc.) as determined by Computer
Assisted
Sperm Analysis (CASA). Mobility in mucus can be measured using techniques well
known
in the art and described herein. Alternatively, trapping potency can be
quantitated as the
reduction in percentage of pathogens or sperm that penetrate mucus.
[000129] The twin "enhances trapping potency" refers to enhancement compared
to the
core antibody (e.g., core IgG). Further, any of the multimeric synthetic
binding agents
having multiple Fab repeats described herein may be selected or further
configured to
enhance mucin-crosslinking by including a glycosylation pattern comprising the
biantennary
core glycan structure Manal-6(Mana I -3)Mani31 -4G1cNAcf11-4G1cNAc01 with
terminal N-
acetylglucosamine on each branch. This glycosylation pattern may be on the Fe
region of the
core Ab (e.g., the core IgG). Alternatively or additionally, a composition of
the synthetic
binding agent having multiple Fab repeats described herein may be selected or
configured
such that at least x% of the synthetic binding agent having multiple Fab
repeats have a
glycosylation pattern comprising the biantennary core glycan structure Manal-
6(Manal -
3)Manf3l -4GIcNAcf31-4G I cNAci31 with terminal N-acetylglucosamine on each
branch,
where x% is 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%,
85%,
90%, 95%, or substantially all). A composition in which, for example, greater
than 40% of
the synthetic binding agent having multiple Fab repeats described herein (to
enhance
agglutination potency) while also possessing an oligosaccharide that provides
increased
rnucin crosslinking, may be particularly beneficial for muco-trapping of a
target once bound
to the target, compared to core IgG as found in nature prior to any
modification and/or
selection.
[000130] As used herein, the term "bind specifically" or "specifically binds"
in reference to
an antibody of the presently-disclosed subject matter means that the antibody
of the invention
will bind with an epitope (including one or more epitopes) of a target
pathogen or sperm, but
does not substantially bind to other unrelated epitopes or molecules. In
certain embodiments,
the term refers to an antibody that exhibits at least about 60% binding, e.g.,
at least about
70%, 80%, 90%, or 95% binding, to the target epitope relative to binding to
other unrelated
epitopes or molecules.
[000131] The antibodies, compositions, and methods described herein may
include methods
for inhibiting and/or treating pathogen infection, eliminating pathogen from a
mucosal
31
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
surface, and providing contraception. In particular, the presently-disclosed
subject matter
relates to synthetic binding agents having multiple Fab repeats and
compositions of these that
are capable of facilitating aggregation and/or enchained growth of pathogens
and sperm,
trapping pathogens and sperm in mucus, thereby inhibiting transport of
pathogens or sperm
across or through mucus secretions, which may lead to the destruction and/or
natural
elimination of these pathogens and/or sperm.
[000132] Any of the synthetic binding agents having multiple Fab repeats
described herein
be directed to non-neutralizing epitopes of pathogens; in some variations, the
synthetic
binding agents having multiple Fab repeats described herein be directed to
neutralizing
epitopes of pathogens.
[000133] Antibodies are naturally found in mucus. The synthetic binding agent
having
multiple Fab repeats described herein may generally diffuse rapidly through
mucus, slowed
only slightly by weak, transient adhesive interactions with mucins within the
mucus. This
rapid diffusion allows the synthetic binding agent having multiple Fab repeats
to accumulate
rapidly on pathogen or sperm surfaces. When a plurality of synthetic binding
agents have
accumulated on the surface of a pathogen or sperm, the adhesive interactions
between the
plurality of antibodies and the mucus become sufficient to trap the bound
pathogen or sperm
in the mucus, thereby preventing infection/providing contraception. Moreover,
and somewhat
surprisingly, binding multiple pathogens using the same synthetic binding
agent having
multiple Fab repeats may more effectively trap the complex formed by the
multiple
pathogens/sperm and the synthetic binding agent, either by aggregation of
distinct
pathogens/sperm together or facilitating enchained growth of pathogens.
Pathogens or sperm
trapped in CVM cannot reach their target cells in the mucosal surface, and
will instead be
shed with post-coital discharge and/or inactivated by spontaneous thermal
degradation as
well as additional protective factors in mucus, such as defensins (Cole, Curr.
Top. Micro biol.
Immunol. 306:199 (2006); Doss et al., I Leukoc. Biol. 87:79 (2010)). As
disclosed herein,
this pathogen agglutination and/or trapping activity provides for protection
without
neutralization, and can effectively inhibit infection at sub-neutralization
doses and/or using
antibodies to non-neutralizing epitopes of a pathogen. The low-affinity
interactions that the
synthetic binding agent having multiple Fab repeats described herein may form
with mucins
are not only Pc-dependent, but may also influenced by antibody glycosylation.
[000134] Accordingly, the synthetic binding agent having multiple Fab repeats
described
herein may include an oligosaccharide at a glycosylation site, the
oligosaccharide comprising,
consisting essentially of, or consisting of a pattern correlating with
(providing) enhanced
32
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
trapping potency of the antibody in mucus, and wherein the antibody
specifically binds an
epitope of a target (e.g., pathogen or sperm). The unique glycosylation
pattern/unique
oligosaccharide component of the antibody may maximize trapping potency of the
synthetic
binding agent once a synthetic binding agent forms a complex with one or more
target (e.g.,
pathogen or sperm), without unduly hindering the ability of the unbound
synthetic binding
agent to diffuse readily through mucus to rapidly bind a target. In certain
embodiments, the
synthetic binding agent having multiple Fab repeats described herein is one
that exhibits a
mobility in mucus that is reduced no more than about 50%, e.g., no more than
about 40%,
30%, 20%, 10%, or 5%, relative to its native mobility in solution (e.g.,
mucus, saline or
water) and effectively traps a target pathogen or sperm in mucus once
complexed with one or
more targets (e.g., at least 50% of target slowed by at least on half). In
some embodiments,
the synthetic binding agent having multiple Fab repeats described herein
reduces the mobility
of at least 50% of the target, e.g., at least 50%, 60%, 70%, 80%, or 90% or
more of the target,
by at least 50% (e.g., 60%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, or
99%, etc.)
or more. In other embodiments, the synthetic binding agent having multiple Fab
repeats
described herein reduces the percentage of target (e.g., pathogens or sperm)
that can penetrate
mucus by at least 10%, e.g., at least 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%,
or more.
For example, the synthetic binding agent having multiple Fab repeats described
herein may
have a sufficient binding rate to an epitope of the target to trap the target
pathogen or sperm
in mucus within one hour (e.g., within 30 minutes or 15 minutes) at a
synthetic binding agent
concentration in the mucus of less than 10 mg/ml (e.g., less than 5 mg/ml,
less than 1 mg/ml,
less than 0.1 mg/ml, less than 50 uglnal, less than 30, less than 20, less
than 10, less than 5,
less than 2.5, less than 1, less than 0.5, less than 0.11.tg/ml, etc.).
[000135] In some embodiments, the synthetic binding agent having multiple Fab
repeats
described herein may include an oligosaccharide component that is bound to an
N-linked
glycosylation site in an Fe region of the synthetic binding agent (e.g., the
core IgG portion of
the synthetic binding agent). The N-linked glycosylation site can be an
asparagine residue on
the Fe region of the core, for example, the Asn 297 asparagine residue. The
amino acid
numbering is with respect to the standard amino acid structure of a human IgG
molecule.
[000136] The N-glycan structure may be GO/GOF form, or a pure GnGn form (e.g.,
with
terminal N-acetylglucosamine on each branch without terminal galactose or
sialic acid). In
some embodiments, the oligosaccharide component, i.e., the glycan, attached to
the antibody
comprises, consists essentially of, or consists of a core structure without
any fucose residue.
33
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
In other embodiments, the glycan does not contain any galactose residues. In
some
embodiments the glycan does not include galactose.
10001371 The synthetic binding agent having multiple Fab repeats described
herein may
include a mixture of synthetic binding agents having different oligosaccharide
components.
In some embodiments, the mixture comprises at least about 30% synthetic
binding agent
having multiple Fab repeats described herein having the GO/GOF core glycan
structure (e.g.,
with or without the fucose residue), e.g., at least about 40%, 50%, 60%, 70%,
80%, 90% or
more.
10001381 In some embodiments, the synthetic binding agent having multiple Fab
repeats
described herein are generated in a human cell line, e.g., a 293 cell line,
e.g., a 2931 cell line,
other mammalian cell lines (e.g. CHO), in plants (e.g. Nicotianct), or in
other microorganisms
(e.g. Trichoderma).
[0001391 The synthetic binding agent having multiple Fab repeats described
herein may be
useful for binding target to trap the target in mucus to inhibit infection or
impregnation by the
target. In variations in which the target is a pathogen, the synthetic binding
agent having
multiple Fab repeats described herein can be directed to any pathogen that can
infect a
subject through a mucus membrane. Pathogens can be in the categories of algae,
bacteria,
fungi, parasites (helminths, protozoa), viruses, and subviral agents. Target
pathogens further
include synthetic systems comprising an antigen having an epitope, for example
particles or
particulates (e.g., polystyrene beads) comprising attached proteins, e.g., as
might be used for
bioterrorism. Pathogens include those that cause sexually-transmitted diseases
(listed with
the diseases caused by such pathogens), including, without limitation,
Neisseria gonorrhoeae
(gonorrhea); Chlamydia trachomatis (ehlamydia, lymphogranuloma venereum);
Treponerna
pallidum (syphilis); Haernophilus ducreyi (chancroid); Klebsiella granulomatis
or
Calymmatobacterium granulomatis (donovanosis), Adycoplasma genital/urn,
Ureaplasma
2,trealytieum (mycoplasmas); human immunodeficiency virus HIV-1 and HIV-2
(HIV, AIDS);
HTLV-1 (T-Iymphotrophic virus type 1); herpes simplex virus type 1 and type 2
(HSV-1 and
HSV-2); Epstein-Barr virus; cytomegalovirus; human herpesvirus 6; varicella-
zoster virus;
human papillomaviruses (genital warts); hepatitis A virus, hepatitis B virus,
hepatitis C virus
(viral hepatitis); molluscum contagiosum virus (MCV); Triehomona vaginalis
(trichomoniasis); and yeasts, such as Candida albicans (vulvovaginal
candidiasis). The
antibodies and compositions may also be active against other diseases that are
transmitted by
contact with bodily fluids that may also be transmissible by sexual contact
and are capable of
being prevented by administration of the compositions according to this
invention.
34
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Accordingly, the phrase, "sexually transmitted diseases (STDs)," is to be
interpreted herein as
including any disease that is capable of being transmitted in the course of
sexual contact,
whether or not the genital organs are the site of the resulting pathology.
Pathogens also
include those that cause respiratory diseases, including, without limitation,
influenza
(including influenza A, B, and C); severe acute respiratory syndrome (SARS);
respiratory
syncytial virus (RSV); parainfluenza; adenovirus; human rhinovirus;
coronavirus; and
norovirus. Other pathogens include, without limitation, Salmonella and
Escherichia coil,
Other pathogens include Klebsiella bacillus.
[000140] Pathogens may include those that affect non-human animals, such as
livestock,
e.g., swine (e.g., porcine epidemic diarrhea virus (PEDV), transmissible
gastroenteritis virus
(TGEV), rotavinis, classical swine fever virus (CSFV), porcine circovirus type
2 (PCV2),
encephalomyocarditis virus (EMCV), porcine reproductive and respiratory
syndrome virus
(PRRSV), porcine parvovirus (PPV), pseudorabies virus (PRV), Japanese
encephalitis virus
(JEV), Brucella, Leptospira, Salmonella, and Lawsonia intracellularis,
Pasteurella
multocida, Brctchyspira hyodysenteriae, Mycoplasma hyopneumoniae), ruminants
(e.g.,
bovine virus diarrhoea virus (BVDV), border disease virus (BDV), bovine
papular stomatitis
virus (BPSV), pseudocowpox virus (PCPV), Pasteurella haemolytica, Pasteurella
multocida,
Haemophilus somnus, Haemophilus agnii, Moraxella bovis, Mycoplasma mycoides,
Theileria
annulctta, Mycobacterium avium paratuberculosis), ungulates (e.g., Brucella
abortus,
Mycobacterium bovis, Theikria parva, Rift Valley fever virus, foot-and-mouth
disease virus,
lumpy skin disease virus), horses (e.g., Rhodococcus equi, Salmonella
choleraesuis,
Pasteurella multocida, equine herpesvirus-1, equine herpesvirus-4, equine
influenza virus,
Streptococcus equi), poultry (e.g., fowl pox virus, Newcastle disease virus,
Marek's disease
virus, avian influenza virus, infectious bursal disease virus (IBDV), avian
infectious
bronchitis virus (IBV)), and the like.
[000141] The terms virus and viral pathogen are used interchangeably herein,
and further
refer to various strains of virus, e.g., influenza is inclusive of new strains
of influenza, which
would be readily identifiable to one of ordinary skill in the art. The terms
bacterium, bacteria,
and bacterial pathogen are used interchangeably herein, and further refer to
antibiotic-
resistant or multidrug resistant strains of bacterial pathogens. As used
herein when referring
to a bacterial pathogen, the term "antibiotic-resistant strain" or "multidrug
resistant strain"
refers to a bacterial pathogen that is capable of withstanding an effect of an
antibiotic or drug
used in the art to treat the bacterial pathogen (i.e., a non-resistant strain
of the bacterial
pathogen).
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
10001421 In some embodiments, it is contemplated that a synthetic binding
agent having
multiple Fab repeats described herein is capable of broadly binding to viruses
containing lipid
envelopes, which are not necessarily specific to one virus.
[000143] In variations when the synthetic binding agent having multiple Fab
repeats
described herein specifically binds a neutralizing epitope of the target
pathogen, a sub-
neutralization dose can be used. A sub-neutralization doses is a dose below
that which would
be needed to achieve effective neutralization. For example, in the case of
polyelonal anti-
HSV gG antibodies targeting HSV, as described hereinbelow, an effective
neutralization dose
is approximately 5 pg/ml. However, effective agglutination and/or trapping
using the
synthetic binding agent having multiple Fab repeats described herein can be
achieved at a
dose below 5 ughnl, and even below a dose of 1 ug/ml.
10001441 As will be recognized by one of skill in the art, doses appropriate
for agglutination
and/or trapping bacterial pathogens can be higher in some embodiments than the
doses
appropriate for trapping viral pathogens. It will further be recognized that
appropriate doses
may differ between pathogens, between mucosal surfaces, and also between
individuals. It
will also be recognized that different subjects and different mucosal surfaces
may have
different optimal glyean patterns and optimal antibody-mucin affinities,
contributing to
different optimal doses.
10001451 It is further proposed herein that synthetic binding agent having
multiple Fab
repeats described herein that selectively bind non-neutralizing epitopes of a
target pathogen
can be used to effectively trap the target pathogen in mucus. As such, in some
embodiments,
the synthetic binding agent having multiple Fab repeats specifically binds a
non-neutralizing
epitope, e.g., one or more non-neutralizing epitopes.
[000146] The presently disclosed subject matter further includes synthetic
binding agent
having multiple Fab repeats that selectively binds a conserved epitope of a
target. A benefit
of targeting a conserved epitope would be to preserve efficacy of the
synthetic binding agent
having multiple Fab repeats as against new strains of the pathogen. Targeting
such epitopes
has been avoided at times in the past because they were viewed as being
ineffective targets;
however, in view of the disclosure herein such epitopes can serve as effective
targets.
[000147] The synthetic binding agent having multiple Fab repeats described
herein may be
particularly useful for binding sperm to trap the sperm in mucus to inhibit
fertilization of an
egg by the sperm. Sperm specific antigens that can be used as antibody targets
are well
known in the art. See, e.g., U.S. Pat. Nos. 8,211,666, 8,137,918, 8,110,668,
8,012,932,
7,339,029, 7,230,073, and 7,125,550, each incorporated by reference in its
entirety. As will
36
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
be described herein one particular epitope region for human sperm may include
the N-linked
glycan of sperm CD52 glycoform. See also U.S. Pat. Nos. 5,227,160 and
6,355,235, herein
incorporated by reference in their entirety.
[000148] The low-affinity binding interactions that the synthetic binding
agent having
multiple Fab repeats described herein forms with mucins may be influenced by
glycosylation,
and may also be Fc-dependent. As such, the synthetic binding agent having
multiple Fab
repeats described herein may have a preserved and/or engineered Fe region in
the core IgG
region. Such synthetic binding agents may be one or more subclasses of IgG,
e.g., IgGi, IgG2,
IgG3, IgG4, or any combination thereof.
[000149] In some embodiments, the synthetic binding agent having multiple Fab
repeats
described herein has a sufficient binding rate and/or binding affinity to an
epitope of the
target to accumulate on the surface of the target at sufficient levels to trap
the target within
one hour after administration of the synthetic binding agent having multiple
Fab repeats
described herein at a concentration of less than about 10 mg/mL (e.g., less
than about 5
mg/mL, less than 2 mg/mL, less than about 1 mg/mL, less than about 03 mg/mL,
less than
about 50 tg/ml, less than about 40 ughnl, less than about 30 ug/ml, less than
about 20 ug/ml,
less than about 10 ug/ml, less than about 5 ug/ml, less than about 1 ug/ml,
less than about 0.5
ughril, less than about 0.1 ug/ml, etc.). The term "trap" in this instance
refers to reduction of
further movement through the mucus. In some embodiments, the target (e.g.,
pathogen or
sperm) may be trapped within about 30 minutes, e.g., about 25, 20, 15, 10, 5
or 1 minutes
after administration of the synthetic binding agent having multiple Fab
repeats described
herein. In some embodiments, the synthetic binding agent traps the target at a
synthetic
binding agent concentration of less than about 5 mg/ml, 2.5 mg/ml, 1 mg/ml,
100 ug/ml, 50
ug/ml, 10 ug/ml, 5 jig/ml, 4 jig/ml, 3 jig/ml, 2 ughnl, or 1 jig/ml.
[000150] The following discussion is presented as a general overview of the
techniques
available for the production of synthetic binding agent having multiple Fab
repeats; however,
one of skill in the art will recognize that many variations upon the following
methods are
known.
[000151] The term "antibody" or "antibodies" as used herein refers to all
types of
immunoglobulins, including IgG, IgM, IgA, IgD, and IgE. The antibody can be
monoclonal
or polyelonal and can be of any species of origin, including (for example)
mouse, rat, rabbit,
horse, goat, sheep, camel, or human, or can be a chimeric or humanized
antibody. See, e.g.,
Walker et al., Mokc. Immunol. 26:403 (1989). The antibodies can be recombinant
monoclonal antibodies produced according to the methods disclosed in U.S. Pat.
No.
37
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
4,474,893 or U.S. Pat. No. 4,816,567, the antibodies can also be chemically
constructed
according to the method disclosed in U.S. Pat. No. 4,676,980.
[000152] Antibody fragments included within the scope of the present invention
include, for
example, Fab, Fab', F(ab)2, and Fv fragments; domain antibodies, diabodies;
nanobodies;
vaccibodies, linear antibodies; single-chain antibody molecules, scFv; and
multispecific
antibodies formed from antibody fragments. Such fragments can be produced by
known
techniques. For example, F(ab')2 fragments can be produced by pepsin digestion
of the
antibody molecule, and Fab fragments can be generated by reducing the
disulfide bridges of
the F(ab1)2 fragments. Alternatively, Fab expression libraries can be
constructed to allow
rapid and easy identification of monoclonal Fab fragments with the desired
specificity (Huse
et al., Science 254:1275 (1989)). In some embodiments, the term "antibody
fragment" as used
herein may also include any protein construct that is capable of binding a
target.
[000153] Antibodies, including the core Ab forming part of the synthetic
binding agent
having multiple Fab repeats described herein may be humanized or camelized.
Humanized
forms of non-human (e.g., murine) antibodies are chimeric immunoglobulins,
immunoglobulin chains or fragments thereof (such as Fv, Fab, Fab', F(abr)2 or
other antigen-
binding subsequences of antibodies) which contain minimal sequence derived
from non-
human immunoglobulin. Humanized antibodies include human immunoglobulins
(recipient
antibody) in which residues from a complementarily determining region (CDR) of
the
recipient are replaced by residues from a CDR of a non-human species (donor
antibody) such
as mouse, rat or rabbit having the desired specificity, affinity and capacity.
In some instances,
Fv framework residues of the human immunoglobulin are replaced by
corresponding non-
human residues. Humanized antibodies may also comprise residues which are
found neither
in the recipient antibody nor in the imported CDR or framework sequences. In
general, the
humanized antibody will comprise substantially all of at least one, and
typically two, variable
domains, in which all or substantially all of the CDR regions correspond to
those of a non-
human immunoglobulin and all or substantially all of the framework (FR)
regions (i.e., the
sequences between the CDR regions) are those of a human immunoglobulin
consensus
sequence. The humanized antibody optimally also will comprise at least a
portion of an
immunoglobulin constant region (Fe), typically that of a human immunoglobulin
(Jones et
al., Nature 321:522 (1986); Riechmann et al., Nature, 332:323 (1988); and
Presta, Curr. Op.
Struct. Biol. 2:593 (1992)),
[000154] Methods for humanizing non-human antibodies are well known in the
art.
Generally, a humanized antibody has one or more amino acid residues introduced
into it from
38
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
a source which is non-human. These non-human amino acid residues are often
referred to as
"import" residues, which are typically taken from an "import" variable domain.
Humanization can essentially be performed following the method of Winter and
co-workers
(Jones et al., Nature 321:522 (1986); Riechmann et al., Nature 332;323 (1988);
Verhoeyen et
al., Science 239:1534 (1988)), by substituting rodent CDRs or CDR sequences
for the
corresponding sequences of a human antibody. Accordingly, such "humanized"
antibodies
are chimeric antibodies (U.S. Pat. No. 4,816,567), wherein substantially less
than an intact
human variable domain has been substituted by the corresponding sequence from
a non-
human species. In practice, humanized antibodies are typically human
antibodies in which
some CDR residues (e.g., all of the CDRs or a portion thereof) and possibly
some FR
residues are substituted by residues from analogous sites in rodent
antibodies.
[000155] Human antibodies and synthetic binding agent having multiple Fab
repeats based
on human or humanized IgG as described herein can also be produced using
various
techniques known in the art, including phage display libraries (Hoogenboom and
Winter,
Mol. Biol. 227:381 (1991); Marks et al., J. Mol. Biol. 222:581 (1991)). The
techniques of
Cole et al. and Boemer et al. are also available for the preparation of human
monoclonal
antibodies (Cole et al., Monoclonal Antibodies and Cancer Therapy, Alan R.
Liss, p. 77
(1985) and Boemer et al., J Immunol. 147:86 (1991)). Similarly, human
antibodies can be
made by introducing human irnmunoglobulin loci into transgenic animals, e.g.,
mice in which
the endogenous immunoglobulin genes have been partially or completely
inactivated. Upon
challenge, human antibody production is observed, which closely resembles that
seen in
humans in all respects, including gene rearrangement, assembly, and antibody
repertoire.
This approach is described, for example, in U.S. Pat. Nos. 5,545,807;
5,545,806; 5,569,825;
5,625,126; 5,633,425; 5,661,016, and in the following scientific publications:
Marks et al.,
Bio/Technology 10:779 (1992); Lonberg et al., Nature 368:856 (1994); Morrison,
Nature
368:812 (1994); Fishwild et al., Nature Biotechnol. 14:845 (1996); Neuberger,
Nature
Biotechnol. 14:826 (1996); Lonberg and Huszar, Intern. Rev, Itrununol. 13:65
(1995).
[000156] Immunogens (antigens) are used to produce antibodies specifically
reactive with
target polypeptides. Recombinant or synthetic polypeptides and peptides, e.g.,
of at least 5
(e.g., at least 7 or 10) amino acids in length, or greater, are the preferred
immunogens for the
production of monoclonal or polyclonal antibodies. In one embodiment, an
immunogenic
polypeptide conjugate is also included as an immunogen. The peptides are used
either in
pure, partially pure or impure form. Suitable polypeptides and epitopes for
target pathogens
and sperm are well known in the art. Polynucleotide and polypeptide sequences
are available
39
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
in public sequence databases such as GENBANKO/GENPEPT . Large numbers of
neutralizing and non-neutralizing antibodies that specifically bind to target
pathogens and
sperm have been described in the art and can be used as starting material to
prepare the
antibodies of the present invention. Alternatively, new antibodies can be
raised against target
pathogens and sperm using the techniques described herein and well known in
the art.
10001571 Recombinant polypeptides are expressed in eukaryotic or prokaryotic
cells and
purified using standard techniques. The polypeptide, or a synthetic version
thereof, is then
injected into an animal capable of producing antibodies. Either monoclonal or
polyclonal
antibodies can be generated for subsequent use in immunoassays to measure the
presence and
quantity of the polypeptide.
[0001581 Methods of producing polyclonal antibodies are known to those of
skill in the art.
In brief, an immunogen, e.g., a purified or synthetic peptide, a peptide
coupled to an
appropriate carrier (e.g., glutathione-S-transferase, keyhole limpet
hemanocyanin, etc.), or a
peptide incorporated into an immunization vector such as a recombinant
vaccinia virus is
optionally mixed with an adjuvant and animals are immunized with the mixture.
The animal's
immune response to the immunogen preparation is monitored by taking test
bleeds and
determining the titer of reactivity to the peptide of interest. When
appropriately high titers of
antibody to the immunogen are obtained, blood is collected from the animal and
antisera are
prepared. Further fractionation of the antisera to enrich for antibodies
reactive to the peptide
is performed where desired. Antibodies, including binding fragments and single
chain
recombinant versions thereof, against the polypeptides are raised by
immunizing animals,
e.g., using immunogenic conjugates comprising a polypeptide covalently
attached
(conjugated) to a carrier protein as described above. Typically, the immunogen
of interest is a
polypeptide of at least about 10 amino acids, in another embodiment the
polypeptide is at
least about 20 amino acids in length, and in another embodiment, the fragment
is at least
about 30 amino acids in length. For example, the polypeptide can comprise
amino acids acid
residues 1 through 200 from the N-terminal of the papillomavirus L2 protein.
The
immunogenic conjugates are typically prepared by coupling the polypeptide to a
carrier
protein (e.g., as a fusion protein) or, alternatively, they are recombinantly
expressed in an
immunization vector.
[0001591 Monoclonal antibodies are prepared from cells secreting the desired
antibody.
These antibodies are screened for binding to normal or modified peptides, or
screened for
agonistic or antagonistic activity. Specific monoclonal and polyclonal
antibodies will usually
bind with a KD of at least about 50 mM, e.g., at least about 1 mM, e.g., at
least about 0.1 mM
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
or better. In some instances, it is desirable to prepare monoclonal antibodies
from various
mammalian hosts, such as mice, rodents, primates, humans, etc. Description of
techniques for
preparing such monoclonal antibodies are found in Kohler and Milstein 1975
Nature
256:495-497. Summarized briefly, this method proceeds by injecting an animal
with an
immunogen, e.g., an immunogenic peptide either alone or optionally linked to a
carrier
protein. The animal is then sacrificed and cells taken from its spleen, which
are fused with
myeloma cells. The result is a hybrid cell or "hybridoma" that is capable of
reproducing in
vitro. The population of hybridomas is then screened to isolate individual
clones, each of
which secrete a single antibody species to the immunogen. In this manner, the
individual
antibody species obtained are the products of immortalized and cloned single B
cells from the
immune animal generated in response to a specific site recognized on the
immunogenic
substance.
[000160] Alternative methods of immortalization include transfoimation with
Epstein Barr
Virus, oncogenes, or retroviruses, or other methods known in the art. Colonies
arising from
single immortalized cells are screened for production of antibodies of the
desired specificity
and affinity for the antigen, and yield of the monoclonal antibodies produced
by such cells is
enhanced by various techniques, including injection into the peritoneal cavity
of a vertebrate
(preferably mammalian) host. The polypeptides and antibodies of the present
invention are
used with or without modification, and include chimeric antibodies such as
humanized
murine antibodies. Other suitable techniques involve selection of libraries of
recombinant
antibodies in phage or similar vectors. See, Huse et al. 1989 Science 246:1275-
1281; and
Ward et al. 1989 Nature 341:544-546.
[0001611 Antibodies specific to the target polypeptide can also be obtained by
phage
display techniques known in the art.
[000162] Synthetic binding agent having multiple Fab repeats as described
herein can be
labeled by joining, either covalently or noncovalently, a substance which
provides a
detectable signal. A wide variety of labels and conjugation techniques are
known and are
reported extensively in both the scientific and patent literature. Suitable
labels include
radionuclides, enzymes, substrates, cofactors, inhibitors, fluorescent
moieties,
chemiluminescent moieties, magnetic particles, and the like. Synthetic binding
agent having
multiple Fab repeats as described herein may be useful for detecting or
diagnosing the
presence of a target on which an antigen is found.
[000163] Method of making synthetic binding agent having multiple Fab repeats
as
described herein with a glycosylation pattern of interest can be achieved by
any method
41
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
known to those or skill in the art. For example, in some embodiments,
mammalian cells can
be used, such as, Chinese hamster ovary (CHO) cells, baby hamster kidney (BHK)
cells, and
NSO- and SP2/0-mouse myeloma cells, to produce antibodies having the desired
glycosylation pattern. In certain embodiments, human cell lines can be used,
e.g., 293 cells.
In some embodiments, non-mammalian cells can be used. The cell line can be
genetically
engineered to produce the antibodies with the desired oligosaccharide. Such
cell lines can
have altered expression, for example, of one or more enzymes affecting
glycosylation
patterns, e.g., glycosyltransferases. Glycosyltransferases include, without
limitation, a
galactosyltransferase, a fucosyltransferase, a glucosyltransferase, an N-
acetylgalactosaminyltransferase, an N-acetylglucosaminyltransferase, a
glucuronyltransferase, a sialyltransferase, a mannosyltransferase, a
glucuronic acid
transferase, a galacturonic acid transferase, an oligosaccharyltransferase, or
any combination
thereof. Specific examples include, without limitation,
oligosaccharyltransferase, UDP-N-
acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, GDP-
fucose
protein:0-fucosyltransferase 1, GDP-fucose protein:0-fucosyltransferase 2,
protein:0-
glucosyltransferase, UDP-N-acetylglueosamine:peptide N-
aeetylglucosaminyltransferase,
protein:0-mannosyltransferase,131,4 galactosyltransferase, and any combination
thereof.
Enzymes involved in glycosylation of proteins are well known in the art and
can be
manipulated using routine techniques. See, for example, U.S. Pat. Nos.
8,383,106, 8,367,374,
8,080,415, 8,025,879, 8,021,856, 7,906,329, and 7,846,434, each incorporated
herein by
reference in its entirety. In other embodiments, glycans can be synthesized in
specific
patterns and linked to the synthetic binding agent having multiple Fab repeats
described
herein. In further embodiments, synthetic binding agent having multiple Fab
repeats
described herein with mixed glycosylation patterns can be separated to isolate
antibodies with
the desired glycosylation pattern.
[000164] As would be recognized by one skilled in the art, the synthetic
binding agent
having multiple Fab repeats described herein can also be formed into suitable
compositions,
e.g., pharmaceutical compositions for administration to a subject in order to
act as a
contraceptive and/or to treat or prevent an infection caused by a target
pathogen or a disease
or disorder caused by infection by a target pathogen. A composition may
comprise, consist
essentially of, or consist of a synthetic binding agent having multiple Fab
repeats described
herein in a prophylactically or therapeutically effective amount and a
pharmaceutically-
acceptable carrier.
42
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[0001651 Pharmaceutical compositions containing the synthetic binding agent
having
multiple Fab repeats described herein can be formulated in combination with
any suitable
pharmaceutical vehicle, excipient or carrier that would commonly be used in
this art,
including such conventional materials for this purpose, e.g,, saline,
dextrose, water, glycerol,
ethanol, and combinations thereof As one skilled in this art would recognize,
the particular
vehicle, excipient or carrier used will vary depending on the subject and the
subjects
condition, and a variety of modes of administration would be suitable for the
compositions of
synthetic binding agent having multiple Fab repeats described herein. Suitable
methods of
administration of any pharmaceutical composition disclosed in this application
include, but
are not limited to, topical, oral, intranasal, buccal, inhalation, anal, and
vaginal
administration, wherein such administration achieves delivery of the antibody
to a mucus
membrane of interest.
[0001661 The composition can be any type of composition suitable for
delivering a
synthetic binding agent having multiple Fab repeats described herein to a
mucosa' surface
and can be in various forms known in the art, including solid, semisolid, or
liquid form or in
lotion form, either oil-in-water or water-in-oil emulsions, in aqueous gel
compositions.
Compositions include, without limitation, gel, paste, suppository, douche,
ovule, foam, film,
spray, ointment, pessary, capsule, tablet, jelly, cream, milk, dispersion,
liposomes,
powder/talc or other solid, suspension, solution, emulsion, microemulsion,
nanoemulsion,
liquid, aerosol, microcapsules, time-release capsules, controlled release
formulation,
sustained release formulation or bioadhesive gel (e.g., a mucoadhesive
thermogelling
composition) or in other forms embedded in a matrix for the slow or controlled
release of the
antibody to the surface onto which it has been applied or in contact.
10001671 If topical administration is desired, the composition may be
formulated as needed
in a suitable form, e.g., an ointment, cream, gel, lotion, drops (such as eye
drops and ear
drops), or solution (such as mouthwash). The composition may contain
conventional
additives, such as preservatives, solvents to promote penetration, and
emollients. Topical
formulations may also contain conventional carriers such as cream or ointment
bases,
ethanol, or oleyl alcohol. Other formulations for administration, including
intranasal
administration, etc., are contemplated for use in connection with the
presently-disclosed
subject matter. All formulations, devices, and methods known to one of skill
in the art which
are appropriate for delivering the synthetic binding agent having multiple Fab
repeats
described herein or a composition containing the synthetic binding agent
having multiple Fab
43
CA 03113059 2021-03-16
WO 2020/061560
PCT/US2019/052396
repeats described herein to one or more mucus membranes of a subject can be
used in
connection with the presently-disclosed subject matter.
[000168] Any of the compositions described herein may include mixtures of the
synthetic
binding agent having multiple Fab repeats described herein, including mixtures
having
different numbers of Fab repeats (e.g., some with 4 Fab repeats, some with 6
Fab repeats,
etc.).
[000169] The compositions used in the methods described herein may include
other agents
that do not negatively impact or otherwise affect the inhibitory and/or
contraceptive
effectiveness of the components of the composition, including antibodies,
antimicrobial
agents, and/or sperm-function inhibitors. For example, solid, liquid or a
mixture of solid and
liquid pharmaceutically acceptable carriers, diluents, vehicles, or excipients
may be
employed in the pharmaceutical compositions. Suitable physiologically
acceptable,
substantially inert carriers include water, a polyethylene glycol, mineral oil
or petrolatum,
propylene glycol, hydroxyethylcellulose, carboxymethyl cellulose, cellulosic
derivatives,
polycarboxylic acids, linked polyacrylic acids, such as carbopols; and other
polymers such as
poly(lysine), poly(glutamic acid), poly(maleic acid), polylactic acid),
thermal polyaspartate,
and aliphatic-aromatic resin; glycerin, starch, lactose, calcium sulphate
dihydrate, terra alba,
sucrose, talc, gelatin, pectin, acacia, magnesium stearate, stearic acid,
syrup, peanut oil, olive
oil, saline solution, and the like.
[000170] The pharmaceutical compositions described herein useful in the
methods of the
present invention may further include diluents, fillers, binding agents,
colorants, stabilizers,
perfumes, gelling agents, antioxidants, moisturizing agents, preservatives,
acids, and other
elements known to those skilled in the art. For example, suitable
preservatives are well
known in the art, and include, for example, methyl paraben, propyl paraben,
butyl paraben,
benzoic acid and benzyl alcohol.
[000171] For injection, the carrier may typically be a liquid, such as sterile
pyrogen-free
water, pyrogen-free phosphate-buffered saline solution, bacterio static water,
or Cremophor
EL (BASF, Parsippany, N.J.). For other methods of administration, the carrier
can be either
solid or liquid.
[000172] For oral administration, the synthetic binding agent having multiple
Fab repeats
described herein can be administered in solid dosage forms, such as capsules,
tablets, and
powders, or in liquid dosage forms, such as elixirs, syrups, and suspensions.
Compositions
can be encapsulated in gelatin capsules together with inactive ingredients and
powdered
carriers, such as glucose, lactose, sucrose, mannitol, starch, cellulose or
cellulose derivatives,
44
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
magnesium stearate, stearic acid, sodium saccharin, talcum, magnesium
carbonate and the
like. Examples of additional inactive ingredients that can be added to provide
desirable color,
taste, stability, buffering capacity, dispersion or other known desirable
features are red iron
oxide, silica gel, sodium lauryl sulfate, titanium dioxide, edible white ink
and the like.
Similar diluents can be used to make compressed tablets. Both tablets and
capsules can be
manufactured as sustained release products to provide for continuous release
of medication
over a period of hours. Compressed tablets can be sugar coated or film coated
to mask any
unpleasant taste and protect the tablet from the atmosphere, or enteric-coated
for selective
disintegration in the gastrointestinal tract. Liquid dosage forms for oral
administration can
contain coloring and flavoring to increase patient acceptance.
[000173] Compositions suitable for buccal (sub-lingual) administration include
tablets or
lozenges comprising the antibody in a flavored base, usually sucrose and
acacia or
tragacanth; and pastilles comprising the antibody in an inert base such as
gelatin and glycerin
or sucrose and acacia. The composition can comprise an orally dissolvable or
degradable
composition. Alternately, the composition can comprise a powder or an
aerosolized or
atomized solution or suspension comprising the antibody. Such powdered,
aerosolized, or
atomized compositions, when dispersed, preferably have an average particle or
droplet size in
the range from about 0.1 to about 200 nanometers
[000174] Compositions of the synthetic binding agent having multiple Fab
repeats
described herein that are suitable for parenteral administration comprise
sterile aqueous and
non-aqueous injection solutions of the synthetic binding agent having multiple
Fab repeats
described herein, which preparations are preferably isotonic with the blood of
the intended
recipient. These preparations can contain anti-oxidants, buffers,
bacteriostats and solutes
which render the composition isotonic with the blood of the intended
recipient. Aqueous and
non-aqueous sterile suspensions can include suspending agents and thickening
agents. The
compositions can be presented in unit/dose or multi-dose containers, for
example sealed
ampoules and vials, and can be stored in a freeze-dried (lyophilized)
condition requiring only
the addition of the sterile liquid carrier, for example, saline or water-for-
injection
immediately prior to use.
[000175] Extemporaneous injection solutions and suspensions can be prepared
from sterile
powders, granules and tablets of the kind previously described. For example,
in one aspect,
there is provided an injectable, stable, sterile composition comprising a
synthetic binding
agent having multiple Fab repeats described herein, in a unit dosage form in a
sealed
container. The synthetic binding agent having multiple Fab repeats described
herein may be
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
provided in the foini of a lyophilizate which is capable of being
reconstituted with a suitable
pharmaceutically acceptable carrier to form a liquid composition suitable for
injection thereof
into a subject.
[000176] Compositions suitable for rectal administration may be presented as
unit dose
suppositories. These can be prepared by admixing the synthetic binding agent
having
multiple Fab repeats described herein with one or more conventional solid
carriers, for
example, cocoa butter, and then shaping the resulting mixture.
10001771 The synthetic binding agent having multiple Fab repeats described
herein can
alternatively be formulated for nasal administration or otherwise administered
to the lungs of
a subject by any suitable means, e.g., administered by an aerosol suspension
of respirable
particles comprising the synthetic binding agent having multiple Fab repeats
described
herein, which the subject inhales. The respirable particles can be liquid or
solid. The term
"aerosol" includes any gas-borne suspended phase, which is capable of being
inhaled into the
bronchioles or nasal passages. Specifically, aerosol includes a gas-borne
suspension of
droplets, as can be produced in a metered dose inhaler or nebulizer, or in a
mist sprayer.
Aerosol also includes a dry powder composition suspended in air or other
carrier gas, which
can be delivered by insufflation from an inhaler device, for example. See
Ganderton & Jones,
Drug Delivery to the Respiratory Tract, Ellis Harwood (1987); Gonda (1990)
Critical
Reviews in Therapeutic Drug Carrier Systems 6:273-313; and Raebum et al., J.
Pharmacol.
Toxicol. Meth. 27:143 (1992). Aerosols of liquid particles comprising the
synthetic binding
agent having multiple Fab repeats described herein can be produced by any
suitable means,
such as with a pressure-driven aerosol nebulizer or an ultrasonic nebulizer,
as is known to
those of skill in the art. See, e.g., U.S. Pat. No. 4,501,729. Aerosols of
solid particles
comprising the synthetic binding agent having multiple Fab repeats described
herein can
likewise be produced with any solid particulate medicament aerosol generator,
by techniques
known in the phaanaceutical art.
[000178] Alternatively, one can administer the synthetic binding agent having
multiple Fab
repeats described herein in a local rather than systemic manner, for example,
in a depot or
sustained-release formulation.
1000179] The synthetic binding agent having multiple Fab repeats described
herein may be
coated or impregnated on a device (or a composition including the synthetic
binding agent
having multiple Fab repeats described herein may be coated or impregnated).
The device can
be for delivery of the synthetic binding agent having multiple Fab repeats
described herein
and compositions of the synthetic binding agent to a mucus membrane, e.g., to
the vagina or
46
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
uterus. In one embodiment, a device includes a solid support adapted to be
inserted into the
vagina. The support can be impregnated with or coated with a composition of
the synthetic
binding agent having multiple Fab repeats described herein. The release of
synthetic binding
agent from the devices may be controlled by the material composing these
devices, such as
silicone elastomers, ethylene vinyl acetate and polyurethane polymers.
Devices, such as
cervicovaginal and rectal devices, include, without limitation, a ring, rod,
applicator, sponge,
cervical cap, tampon, diaphragm, or intrauterine device. Applicators can be
those currently
used commercially to deliver spermicidal gels or anti-yeast compounds and
include, without
limitation, plunger-type applicators, pessaries, sprays, squeezable tubes,
vaginal rings,
cervical rings, sponges, and the like. All such means for delivery are
intended to be
encompassed by the present invention.
[0001801 As noted herein, synthetic binding agent having multiple Fab repeats
described
herein is capable of diffusing through mucus when it is unbound, to allow the
synthetic
binding agent having multiple Fab repeats to bind a target (e.g., pathogen or
sperm) at a
desirable rate. It is also desirable that, when synthetic binding agent having
multiple Fab
repeats described herein is bound to the target, the cumulative effect of the
antibody-mucin
interactions effectively traps the pathogen or sperm in the mucus and/or
agglutinates the
target. To facilitate this goal, in some embodiments, it can be desirable to
provide a
composition that includes more than one synthetic binding agent having
multiple Fab repeats
described herein, wherein each synthetic binding agent specifically binds a
different epitope
of the pathogen or sperm. Such a composition may provide the ability for an
increased
number of synthetic binding agents having multiple Fab repeats to become bound
to the
pathogen or sperm, thereby strengthening the antibody-mucin interactions that
serve to trap
the pathogen or sperm in the mucus.
[000181] In some embodiments, a composition includes a first synthetic binding
agent
having multiple Fab repeats described herein and a second synthetic binding
agent having
multiple Fab repeats described herein, wherein the first synthetic binding
agent specifically
binds a first epitope of the target and the second binding agent specifically
binds a second
epitope of the target, wherein the first epitope is distinct from the second
epitope. In certain
embodiments, the composition includes three or more different synthetic
binding agents
having multiple Fab repeats described herein, e.g., 3, 4, 5, 6, 7, 8, 9, 10,
or more different
synthetic binding agents having multiple Fab repeats described herein, wherein
each synthetic
binding agent specifically binds a different epitope of the target.
47
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[0001821 It is also desirable to provide a composition that can provide
treatment or
prevention of infection due to more than one target pathogen. In some
embodiments of the
presently-disclosed subject matter, a composition includes a first synthetic
binding agent
having multiple Fab repeats and a second synthetic binding agent having
multiple Fab
repeats, wherein the first synthetic binding agent specifically binds an
epitope of a first target
pathogen and the second synthetic binding agent specifically binds an epitope
of second
target pathogen. In certain embodiments, the composition includes three or
more different
synthetic binding agents having multiple Fab repeats, e.g., 3, 4, 5, 6, 7, 8,
9, 10, or more
different synthetic binding agents, wherein each synthetic binding agent
specifically binds an
epitope of a different target. As discussed above, in some variations the
target may be the
same, but the synthetic binding agents having multiple Fab repeats may have
different
numbers of Fab repeats,
[0001831 In other embodiments, a composition provides both contraception and
treatment
or prevention of infection by one or more target pathogens. In some
embodiments, a
composition includes a first synthetic binding agent having multiple Fab
repeats and a second
synthetic binding agent having multiple Fab repeats, wherein the first
synthetic binding agent
specifically binds an epitope of sperm and the second synthetic binding agent
specifically
binds an epitope of a target pathogen. In certain embodiments, the composition
includes three
or more different synthetic binding agents having multiple Fab repeats
described herein, e.g.,
3, 4, 5, 6, 7, 8, 9, 10, or more different synthetic binding agents having
multiple Fab repeats,
wherein one or more synthetic binding agents having multiple Fab repeats bind
different
epitopes of sperm and one or more synthetic binding agent having multiple Fab
repeats
specifically binds an epitope of a target pathogen or multiple target
pathogens.
[0001841 In some embodiments, the pharmaceutical composition can further
include an
additional active agent, e.g., a prophylactic or therapeutic agent. For
example, the additional
active agent can be an antimicrobial agent, as would be known to one of skill
in the art. The
antimicrobial agent may be active against algae, bacteria, fungi, parasites
(helminths,
protozoa), viruses, and subviral agents. Accordingly, the antimicrobial agent
may be an
antibacterial, antifungal, antiviral, antiparasitic, or antiprotozoal agent.
The antimicrobial
agent is preferably active against infectious diseases. Suitable antiviral
agents include, for
example, virus-inactivating agents such as nonionic, anionic and cationic
surfactants, and
C31 G (amine oxide and alkyl betaine), polybiguanides, docosanol,
acylcarnitine analogs,
octyl glycerol, and antimicrobial peptides such as magainins, gramicidins,
protegrins, and
retrocyclins. Mild surfactants, e.g., sorbitan monolaurate, may advantageously
be used as
48
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
antiviral agents in the compositions described herein. Other antiviral agents
that may
advantageously be utilized in the compositions described herein include
nucleotide or
nucleoside analogs, such as tenofovir, acyclovir, amantadine, didanosine,
foscamet,
ganciclovir, ribavirin, vidarabine, zaleitabine, and zidovudine. Further
antiviral agents that
may be used include non-nucleoside reverse transcriptase inhibitors, such as
UC-781
(thiocarboxanilide), pyridinones, TIBO, nevaripine, delavirdine, calanolide A,
capravirine
and efavirenz. From these reverse transcriptase inhibitors, agents and their
analogs that have
shown poor oral bioavailability are especially suitable for administration to
mucosal tissue, in
combination with antibodies and compositions of the invention, to prevent
sexual
transmission of HIV. Other antiviral agents that may be used are those in the
category of HIV
entry blockers, such as cyanovirin-N, cyclodextrins, carregeenans, sulfated or
sulfonated
polymers, mandelic acid condensation polymers, monoclonal antibodies,
chemokine receptor
antagonists such as TAK-779, SCH-C/D, and AMD-3100, and fusion inhibitors such
as T-20
and 1249.
10001851 Suitable antibacterial agents include antibiotics, such as
aminoglycosides,
cephalosporins, including first, second and third generation cephalosporins;
macrolides,
including erythromycins, penicillins, including natural penicillins,
penicillinase-resistant
penicillins, aminopenicillins, extended spectrum penicillins; sulfonamides,
tetracyclines,
fluoroquinolones, metronidazole and urinary tract antiseptics.
[000186] Suitable antifungal agents include amphotericin B, nystatin,
griseofulyin,
flucytosine, fluconazole, potassium iodide, intraconazole, clortrimazole,
miconazole,
ketoconazole, and tolnaftate. Suitable antiprotozoal agents include
antimalarial agents, such
as chloroquine, primaquine, pyrimethamine, quinine, fansidar, and mefloquine;
amebicides,
such as dioloxamide, emetine, iodoquinol, metronidazole, paromomycine and
quinacrine;
pentarnidine isethionate, atovaquone, and eflomithine.
[000187] In certain embodiments, the additional active agent can be a sperm-
function
inhibitor, e.g., an agent that has the ability to inhibit the function of
sperm, to otherwise
inhibit fertilization of an egg by sperm and/or to otherwise prevent
pregnancy, such as by
killing and/or functionally inactivating sperm or by other effects on the
activity of the sperm.
In some embodiments, the active agent may have at least dual functions, such
as acting as a
sperm-function inhibitor and as an antimicrobial agent.
[000188] Sperm-function inhibitors include, without limitation, surfactants,
including
nonionic surfactants, cationic surfactants, and anionic surfactants;
spermicides, such as
nonoxyno1-9 (a-(4-Nonylpheny1)-(o-hydroxynona(oxyethylene); other sperm-
inactivators
49
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
such as sulfated or sulfonated polymers such as polystyrene sulfonate,
mandelic acid
condensation polymers, cyclodextrins; antimicrobial peptides such as
gramicidins, magainins,
indolicidin, and melittin; and acid-buffering compositions, such as BufferGel
and AcidForm.
Nonionic surfactants include, for example, sorbitan mono laurate,
nonylphenoxypolyethoxy
ethanol, p-diisobutyphenoxypolyethoxy ethanol, polyoxyethylene (10) oleyl
ether and onyx-
ol. Suitable anionic surfactants include, without limitation, sodium alkyl
sulfonates and the
sodium alkylbenzene sulfonates. Cationic surfactants include, for example, the
quaternary
ammonium surfactants, such as cetyl pyrimidinium chloride and benzalkonium
chlorides.
Zwitterionic surfactants such as acylcamitine analogs and C3 1G are especially
suitable for
their mild skin and mucosal irritation properties.
[000189] The presently-disclosed subject matter further includes a kit
including the
synthetic binding agent having multiple Fab repeats described herein or a
composition
comprising the synthetic binding agent having multiple Fab repeats as
described herein; and
optionally a device for administering the synthetic binding agent or
composition. In some
embodiments, the kit can include multiple synthetic binding agents having
multiple Fab
repeats and/or compositions containing such synthetic binding agents. In some
embodiments,
each of the multiple synthetic binding agents provided in such a kit can
specifically bind to a
different epitope of the target, e.g., pathogen or sperm. In other
embodiments, each of the
multiple synthetic binding agents having multiple Fab repeats as described
herein provided in
such a kit can specifically bind to an epitope of a different target pathogen
or to an epitope of
sperm. In some embodiments, the kit can further include an additional active
agent, e.g.,
antimicrobial, such as an antibiotic, an antiviral, or other antimicrobial, or
a sperm-function
inhibitor as would be known to one of skill in the art.
SYNTHETIC BINDING AGENTS HAVING MULTIPLE FAB REPEATS
[000190] In general, the synthetic binding agent having multiple Fab repeats
described
herein may include a core IgG that is directed to an epitope of a target. The
synthetic binding
agent having multiple Fab repeats may be constructed by coupling multiple
additional copies
of the same (or a portion of the same) Fab domain of the IgG core. In some
embodiments,
the additional Fab may not be identical to the Fab domain of the IgG core but
still bind the
same epitope. The additional copies may be added to the amino and/or carboxyl
ends of the
IgG core. This is schematically illustrated in FIGS. 1A-1G. In addition, the
core IgG
includes an Fe region that may be glycosylated (or a composition including the
synthetic
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
binding agent having multiple Fab repeats may be selected to enrich for
glycosylation) in a
pattern that enhances muco-trapping, such as the GO glycosylation form.
[000191] For example, FIGS. 1A-1E illustrate different multimeric constructs
("synthetic
binding agent having multiple Fab repeats") that may be produced and
characterized, and the
agglutination and muco-trapping potencies measured. The one or more
constructs, e.g.,
having the greatest potency, may be combined and used.
[0001921 FIG. lA shows an example of a core 1gG including a pair of Fab and Fe
regions.
As shown in FIGS. 1B-1E, the same Fab regions may then be combined in pairs
(e.g., 2
additional, 4 additional, 6 additional, 8 additional, 10 additional, etc.) to
the core IgG to form
synthetic binding agents having multiple Fab repeats.
Contraceptive Synthetic Binding Agents Having Multiple Fab Repeats
[000193] There is a strong unmet demand for non-hormonal contraceptives: Per
the CDC,
roughly half of ¨30 million women between the ages of 20-35 in the U.S. seek
some fonn of
reversible contraceptive method (e.g. pills, IUDs, condoms, rings, etc.).
Socio-societal issues
no doubt contribute to the limited uptake and adherence of common
contraceptive methods;
nevertheless, numerous studies have also demonstrated the need for alternative
contraceptive
methods, particularly non-hormonal options. The vast majority of women start
with hormonal
contraceptives, which is readily available and highly effective. However, many
women are
naturally averse to exogenous hormones despite counselling. Over half of women
quit
(majority within 3-6 months) due to reasons including real and perceived side
effects
associated with hormonal contraceptives (such as weight gain, mood swings and
depression,
headaches and nausea), strongly underscoring the need for non-hormonal
contraception.
Also, both oral and IUD-based hormonal contraception frequently lead to
intermenstrual
"spotting" (light bleeding in weeks prior to the period). Although this may be
viewed as mere
inconvenience in western societies, many women/couples find it seriously
objectionable.
Spotting can significantly limit use of hormonal contraception among certain
populations,
since men contacting women's menstrual blood can be a serious taboo for
religious reasons
(e.g., Muslims and orthodox Jews).
[000194] The synthetic binding agent having multiple Fab repeats described
herein may be
non-hormonal contraceptives that can block sperm permeation through mucus. A
major
effector function for Ab in mucus is to arrest the forward motion of foreign
entities such as
viruses and highly motile bacteria, and block them from reaching target cells.
This function
can be accomplished in two ways. First, when concentrations of the foreign
entity are high
51
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
such that the foreign bodies would frequently collide, Ab can crosslink two or
more bodies
together, resulting not only in an increase in hydrodynamic diameter but more
importantly an
effective neutralization of the net forward motion of swimming bodies. This
process is
commonly referred to as agglutination (see, e.g., FIG. 3A). Second, when
concentrations of
the foreign entity are modest such that collisions between foreign bodies are
relatively
infrequent, Ab can immobilize by directly crosslinking the foreign body to the
mucin matrix
present in mucus via multiple Fc-mucin bonds (see, e.g., FIG. 3B). This
process, which is
referred to herein as muco-trapping, remained largely unrecognized because the
affinity
between each Ab molecule and mucin was long thought to be much too weak to
effectively
bind individual foreign bodies to mucins. Vaginally dosed antigen-specific IgG
can trap
viruses in mucus by forming multiple weakly adhesive bonds between the virus
and the
mucin mesh (akin to a Velcro patch with individually weak hooks). IgG-
mediated trapping
effectively reduced the flux of viruses arriving at target cells and directly
blocked vaginal
Herpes transmission in mice.
[000195] Sperm concentration varies widely in the female reproductive tract,
with the
maximum concentration in semen immediately following ejaculation and lower
concentrations in more distal sites, such as the cervical canal. The ideal
human contraceptive
Ab (RCA, i.e. an Ab molecule that can block sperm permeation through mucus and
prevent
sperm from reaching the egg) should therefore harness one or both
agglutination and trapping
in mucus. Polyvalent Ig such as sIgA and IgM are markedly more potent
agglutinators than
IgG (IgM is ¨1000-fold more potent at agglutination than IgG). Unfortunately,
large scale
manufacturing of IgM or sIgA remains exceptionally challenging, and IgG
represents the
predominant isotype of Ab under clinical development. However, it may be
beneficial to use
IgG.
[000196] Described herein are synthetic binding agents having multiple Fab
repeats with
greater agglutination potency as compared to current monomeric IgG 1-based HCA
(see, e.g.,
JPS638400A) by engineering multimeric HCA that can more potently agglutinate
sperm.
[000197] Thus, described herein are multimeric HCA constructs (e.g., synthetic
binding
agent having multiple Fab repeats) with enhanced agglutination potency. These
synthetic
binding agents may include a Fab from a human IgM that binds a unique antigen
restricted to
only sperm and cells in the male reproductive tract, CD52g, and appears to be
universal in all
men (see, e.g., Norton et al., Tissue Antigens 2002, 60:354-364, August 14,
2002). This Fab
may serve as the basis for the HCA molecule (e.g., the synthetic binding
agent). As
described above, different synthetic binding agents having multiple Fab
repeats constructs
52
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
may be formed, comprised of increasing valency of Fab domains relative to
traditional IgG,
while maintaining its native muco-trapping potency. The different synthetic
binding agents
may include: Fab-IgG, IgG-Fab, Fab-IgG-Fab, and Fab-IgG-Fab-Fab; the core IgG
may be
used as a control. See FIG. 1A-1G, discussed above. All of these synthetic
binding agent
constructs were tested for expression and activity, including (i) ELISA and
biolayer
interferometry for antigen affinity, (ii) gel electrophoresis and SEC to
assess incorporation of
additional Fab domains, and (iii) verification of the ability to purify
different HCA constructs
using conventional Protein A/G columns.
[0001981 Described herein are synthetic binding agents having multiple Fab
repeats that
may be formed as discussed above in relation to FIGS. 1A-1G. For example, in
FIG. 1A, the
core antibody may be an IgG form of the HCA-UNC antibody targeting the CD52g
glycan, as
described in greater detail below. In some variations the synthetic binding
agent having
multiple Fab repeats includes the core IgG and a pair of additional copies of
the Fab (copied
from the IgG core) attached to the amino terminal ends, as shown in FIG. 1B,
or the carboxyl
ends, as shown in FIG. IC. In some variations, additional Fab copies are
attached to either or
both the amino and/or carboxyl ends, as shown in FIG. 1D (additional Fab
copies at both
amino and carboxyl ends) and FIG. lE (two additional Fab copies at the amino
end and four
additional Fab copies at the carboxyl end). FIG. 1G shows examples of these
structures. The
attached sequence listing provides examples of sequences for each of these
five structures.
For example, SEQ ID NO: 9 is an exemplary listing of a full-length Fab-IgG
heavy chain
portion, while SEQ ID No: 13 is the corresponding full length Fab-IgG light
chain amino acid
sequence. Similarly, SEQ ID NO: 15 and SEQ ID NO: 19 illustrate an example of
amino
acid sequences of heavy chain and light chain, respectively, of an IgG-Fab
synthetic binding
agent having multiple Fab repeats. SEQ ID NO: 21 and SEQ ID NO: 25 illustrate
an
example of amino acid sequences of heavy chain and light chain, respectively,
of a Fab-IgG-
Fab synthetic binding agent having multiple Fab repeats. Further, SEQ ID NO:
27 and SEQ
ID NO: 31 illustrate an example of amino acid sequences of heavy chain and
light chain,
respectively, of a Fab-IgG-Fab-Fab synthetic binding agent having multiple Fab
repeats.
[000199] These various synthetic binding agents having multiple Fab repeats
were
examined against each other, as well as against the core IgG (e.g., as encoded
by the amino
acid sequences of SEQ ID NO: 3 and SEQ ID NO: 7). These synthetic binding
agents were
also examined against single-chain variable fragment (scFv) moieties or camel-
derived
nanobodies. scFv-based multimeric Ab constructs frequently suffer from low
stability,
heterogeneous expression, and decreased affinity and specificity stemming from
the removal
53
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
of the CH I/CL interface present in a full-length Fab (FIG. 9). In contrast,
the synthetic
binding agent having multiple Fab repeats exhibit enhanced agglutination
potential compared
to HCA-UNC target the same antigen. These synthetic binding agents having
multiple Fab
repeats have increased valency, span, and directional separation of the Fabs
(diametrically
opposed orientation). Because all Fabs bind the same target epitope and have
essentially the
same amino acids, the synthetic binding agents are not limited by incorrect
heavy and light
chain pairing that necessitates the use of sav-based designs typical for
engineering
multimeric bispecific antibodies that target two distinct epitopes. Instead,
these multimeric
Ab constructs (synthetic binding agent having multiple Fab repeats) link full
Fab domains to
IgG. These Fab-based HCA constructs will likely result in improved stability
and
manufacturability compared to scFv-based constructs, preserved binding
affinity to specific
epitopes, and preserved Fc-mucin affinity.
[000200] For example, initial studies with Fab-IgG (one form of synthetic
binding agent
having multiple Fab repeats) showed that these molecules can be expressed and
purified
using industry-standard techniques while avoiding the formation of aggregates
commonly
observed with scFv-based multimeric constructs, as shown in FIG. 10, showing
an analytical
size exclusion chromatography result for IgG (bottom), scFv-IgG (middle), and
Fab-IgG
(top) after single-step protein A purification. Both IgG and Fab-IgG formats
show a single
sharp peak at their expected molecular weight, while scFv-IgG shows formation
of high
molecular-weight aggregates.
[000201] Thus, a synthetic binding agent having multiple Fab repeats may be
used for IgG-
based FICA for contraception. There is at least 10-fold more IgG present in
CVM than IgA,
which suggests IgG is the optimal mAb for vaginal protection in humans.
[000202] Undiluted, physiological human genital secretions were used in an ex
vivo
investigation of sperm and STI trapping of the synthetic binding agents having
multiple Fab
repeats targeting a CD52g epitope, Trapping of sperm in fresh, minimally
perturbed ex vivo
samples of CVM and CM was observed to ensure that our observations reflect
physiological
conditions as closely as possible.
[000203] Synthetic binding agents having multiple Fab repeats configured as
multimeric
HCA constructs (e.g., Fab-IgG, IgG-Fab, Fab-IgG-Fab) were created using
standard cloning
methods. Briefly, genes encoding HCA VHNL domains and flexible linkers
(GSSSSx3
(SEQ ID NO: 32) were synthesized and cloned into an in-house HCA IgG1
mammalian
expression vector.
54
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[000204] Constructs were expressed by transient expression in 30mL cultures of
Expi293
cells, and the corresponding HCA constructs were purified using protein-A
affinity
chromatography. The purity was verified by SDS-PAGE (FIG. 11A; note: we
compared both
an in-house IgG HCA and an IgG HCA that was independently prepared by Mapp
Biopharmaceutical). We also performed size-exclusion chromatography/multi-
angle light
scattering (SEC/MALS) on each multimeric HCA construct to ensure homogenous
expression of the desired antibody construct (FIG. 11B). In all cases,
constructs expressed at
high yield with minimal (<10%) aggregation.
[0002051 Synthetic binding agent having multiple Fab repeats configured as
multimeric
HCA binds and agglutinates sperm. The multimeric HCA was shown to bind sperm.
We first
performed a whole-sperm ELISA assay using equimolar quantities of each
construct (1.5
nM), followed by detection using anti-Fe HRP secondary. Our pilot assay showed
that each
of the multimeric constructs tested (Fab-IgG, IgG-Fab, Fab-IgG-Fab) had
comparable if not
superior binding to sperm than the native IgG (FIG. I2A), demonstrating that
the new HCA
constructs (the synthetic binding agents having multiple Fab repeats directed
to CD52g) can
indeed bind sperm. To further verify that the HCA constructs can mediate
agglutination, we
mixed IgG-Fab HCA and PBS into semen and imaged the distribution of cells in
the semen
after 1 minute, and found that IgG-Fab HCA potently agglutinated cells (FIGS.
1213-12C).
FIG. 12B shows the PBS control, while FIG. 12C shows the agglutination due to
a synthetic
binding agent having multiple Fab repeats against CD52g (IgG-Fab) after I
minute, as
quantified by the fraction of progressively motile sperm.
[000206] Muco-trapping was shown with IgG-Fab constructs. The synthetic
binding agent
having multiple Fab domains retained its muco-trapping potency, as confirmed
by
microscopy studies using anti-HER2 x anti-PEG IgG-Fab, showing that the
construct can
immobilize ¨100 nm PEG-coated nanoparticles (PS-PEG) in human CVM. The anti-
PEG Fab
portion is functional and binds the antigen of interest (PEG), and the IgG-Fab
structure
retains adequate muco-affinity to trap virus-sized particles in mucus. This is
illustrated in
FIG. 13. PS-PEG in native CVM (no Ab) as well as in CVM treated with control
IgG-Fab
(anti-HER2 x anti-VSVG) both exhibited rapid diffusion only a few fold slower
than their
theoretical rates in pure water. In contrast, addition of PEG-binding IgG-Fab
to CVM
resulted in extensive trapping of PS-PEG, with the fraction of mobile
particles reduced from
71% to only 3%, comparable to muco-adhesive uncoated nanoparticles (PS-COOH).
The
degree of trapping was similar to the level we demonstrated previously with
native anti-PEG
IgG. These results underscore the potential of IgG-Fab and other synthetic
binding agents
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
with multiple Fabs to facilitate trapping of foreign bodies in mucus,
including virus, bacteria
and sperm.
10002071 Sperm trapping and agglutination observed in fresh, minimally
perturbed ex vivo
samples of CVM and CM provide evidence of physiological relevance.
[000208] As discussed herein, agglutination potential of the native IgG1 HCA
(e.g., IgG-
UNC) has been shown in various synthetic binding agents having multiple Fab
repeats
directed against an epitope of CD52g. HCA constructs with different
polyvalency (i.e,
number of Fab domains per molecule), as shown in FIGS. 1B-1F, were constructed
and
examined. Preliminary results suggest a greater agglutination potency will be
achieved by
increasing the number of CD52g-binding Fab domains along an IgG backbone, and
that we
may achieve agglutination potency comparable to native IgM molecules with our
Fab-IgG-
Fab-Fab and Fab-Fab-IgG-Fab-Fab constructs. The evaluation of both Fab-IgG and
IgG-Fab
formats, which differ only in the location of the appended Fab fragment,
provide additional
information regarding the importance of geometric orientation and sterics
(particularly
separation of Fabs, or "spanning" capability) in sperm agglutination and
trapping potency. By
using whole Fab domains rather than scFv, we expect the rnultimeric HCA
constructs to
remain stable, and that each Fab domain possess comparably high affinity to
the CD52g
antigen. We anticipate that HCA constructs with identical Fc-N-glycan profiles
as native IgG
molecules would possess comparable muco-trapping potency.
[0002091 A baseline IgG1 HCA construct has been used to incorporate additional
identical
Fab domains against CD52g at different locations along the heavy chain. The
heavy- and
light-chain gene sequences for IgG control antibodies and each of the Fab-
based multimeric
antibodies may be codon-optimized, synthesized, and cloned into mammalian
expression
vectors (Integrated DNA Technologies). For each format, Fab-components may be
separated
by a flexible peptide linker (e.g., a flexible linker comprising an amino acid
sequence
comprising n pentapeptide repeats consisting of Glycine (G) and Serine (S),
wherein n is
between 3 and 8 amino acids, such as 6 repeated units of GSSSS (SEQ ID NO:
33), GGGGS
(SEQ ID NO:34), etc.).
[000210] Upon verification of the cloning, small batches (30-60 mL) of each
HCA
construct will be expressed by transient transfection in Expi293 mammalian
cells. After three
days of cell growth, the various HCA constructs will be purified from culture
supernatant by
protein A affinity chromatography. Expression yield will be quantified using
absorbance at
280nrrt and BCA assay using human IgG as standard, and purified products will
be assessed
for purity using SDS-PAGE electrophoresis under both reducing and non-reducing
56
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
conditions. The correct assembly, thermal stability and binding kinetics for
each of the
multimeric HCA formats may be verified; for example, correct assembly may be
determined
by molecular weight evaluation by size-exclusion chromatography/multi-angle
light
scattering (Wyatt DAWN HELEOS II; see, e.g,, FIG. 10). Theinial stability (Tm)
may be
measured using differential scanning calorimetry (MicroCal VP-DSC; see FIG.
9).
Antibodies that express and purify with less than 95% correct assembled
product, or that are
destabilized >10 C from the parental sequence will not be further tested
without
optimization.
[000211] Whole-sperm ELISA may be used to quantify different HCA mAbs. First,
high-
affinity 96-well half-area plates (Thermo Scientific, Rockford, IL) may be
coated overnight at
4 C with 50uL per well of sperm at 106/m1 (measured using cell counter).
Plates are washed
three times with 0.05% Tween in PBS (PBS-T), blocked with 5% milk for at least
lhr, and
incubated for at least 2hr with serial dilutions of each HCA mAbs. Following
three PBS-T
washes, plates are incubated with F(abf)2 anti-human IgG Fe (Goat)-HRP
conjugate (709-
1317; Rockland, Gilbertsville, PA) for 1 hr. 1-Step Ultra TMB substrate
(Thermo Scientific,
Rockford, IL) is used to develop the HRP conjugated IgG for 15 min followed by
quenching
with 2N sulfuric acid. Absorbance is measured at 450 nm using a BioTek Synergy
2 plate
reader. Binding kinetics to sperm will also be evaluated by bio-layer
interferometry (Octet
Red384) by using anti-hIgG Pc Capture biosensors dipped into TritonX-100
treated sperm
lysates (which serve as source of HCA antigen). We anticipate the HCA will be
structurally
intact, stable and bind sperm.
10002121 The synthetic binding agents having multiple Fab repeats described
herein are
derived from fully human Ab from an immune infertile but otherwise healthy
woman (Isojim
et al.). The epitope may include the glycosylation structure; and may
specifically recognize a
poly-n-acetyllactosamine region (e.g., repeating poly-n-acetyllactosaminyl
structures) an
antibody such as H6-3C4 may bind to an internal stretch of N-
acetyllactosamines,
and unlike antibodies against blood group i, it is not affected by terminal
sialylation. This
glycoform (referred to as CD52g) is believed to be specific to male-derived
cells (e.g.,
spetni), Thus, a Fab may binds this CD52g (see, SEQ ID NO: 1) glycoprotein
that is unique
to the male genital tract and present on the surface of all sperm and other
cells in semen.
Although CD52g shares a short peptide backbone with leukocyte CD52, the HCA-
UNC used
as the core IgG does NOT bind CD 52, and only binds the unique form of CD52g
that is
produced and secreted only by epithelial cells lining the lumen of the
epididymis, vas
deferens and seminal vesicles. CD52g contains a glycosylphosphatidylinositol
(GPI) anchor,
57
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
and is transferred to the plasma membrane of sperm as they mature in the
epididymis. As
shown in FIGS. 8A-8B, HCA also co-agglutinates leukocytes in semen; these
cells are
potential HIV-infected "Trojan Horse leukocytes" that may act as motile
vectors for HIV
transmission, implying that HCA may also afford some protection against cell-
mediated
transmissions. A WHO-sponsored anti-sperm vaccine workshop has identified
CD52g as a
promising antifertility vaccine candidate due to its unique expression in the
male reproductive
tract, potent antigenicity, and its ability to induce infertility in otherwise
healthy individuals.
Importantly, this HCA target appears to be ubiquitous in men: we have tested
fresh semen
samples from 100 men (73% Caucasian, 26% African American, and 1% Asian), and
all
specimens had >90% of sperm agglutinated within seconds by the prototype FICA.
1000213] The synthetic binding agents having multiple Fab repeats described
herein may be
produced in CHO cells, in Nicotiana plants, and in Trichoderma (for the latter
two, they can
be produced in modified plants or yeast containing the human glycosylation
pathway, and are
capable of making fully human mAb, such as ZMapp in Nicotiana).
[000214] The total dose of synthetic binding agents having multiple Fab
repeats for
contraception (e.g., HCA) may be, e.g., ¨20-80 mg to maintain ¨400 pg/mL of
HCA in CVM
for 28 days. Improving agglutination potency by just 10-fold over, e.g., HCA-
UNC in the
synthetic binding agents having multiple Fab repeats described herein may
allow
substantially lower concentrations to be delivered.
[000215] These synthetic binding agents having multiple Fab repeats were then
tested to
measure sperm agglutination and trapping potency in vitro. Sperm agglutination
and trapping
potency for different synthetic binding agents having multiple Fab repeats
(referred to in this
example as HCA constructs) were tested. Fresh human cervicovaginal mucus (CVM)
and
mid-cycle endocervical mucus (CM) was used to measure the real-time mobility
for
thousands of individual sperm cells in mucus treated with different HCA
constructs to
determine the precise extent the mobility and mobile fraction of speimatozoa
in mucus is
reduced by agglutination and muco-trapping over time.
[000216] A sheep vagina model may be further used to evaluate the potency of
HCA in
reducing free motile sperm by agglutinating and/or trapping human sperm in
vaginal mucus.
The anatomy of the sheep vagina is similar to the human vagina, and is the
best available
animal model for preclinical assessment of vaginal products. To examine
potential in vivo
efficacy, agglutination and trapping of fresh human semen in the sheep vagina
may be
assessed at different times after dosing semen, such as 2 min after
deposition.
58
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[000217] Sperm must swim through mucus to reach the egg. In some infertile
women
without other known causes of infertility, Ab have been isolated that bind the
surface of
living sperm and block sperm from penetrating mucus, with IgM offering the
most potent
combination of agglutination and trapping among naturally occurring Ab (see,
e.g., Isojima et
al., "Establishment and characterization of a human hybridoma secreting
monoclonal
antibody with high titers of spetni immobilizing and agglutinating activities
against human
seminal plasma." J Reprod Immunol, 1987. 10(1): p. 67-78; and Tsuji et al.,
"Human sperm
carbohydrate antigens defined by an antisperm human monoclonal antibody
derived from an
infertile woman bearing antisperm antibodies in her serum." J Exp Med, 1988.
168(1): p.
343-56. PMCID: 2188971). In good agreement with human studies, animal studies
have also
shown that vaginal sperm-binding IgG, sIgA and IgM can provide contraception.
This natural
mechanism of infertility may be used to design a synthetic binding agent
having multiple Fab
repeats to enable non-hormonal contraception. A fully human mAb, termed HCA-
UNC (or
"HCA original"), which binds a highly validated and well characterized antigen
target
ubiquitously present only on the surface of sperm and cells in the male
reproductive tract was
used to form the core IgG of the synthetic binding agent having multiple Fab
repeats for use
as a contraceptive.
[000218] Multimeric HCA constructs (e.g., synthetic binding agents having
multiple Fab
repeats) were constructed having multiple Fab domains linked to a parent IgG
molecule, with
the overall goal of engineering an HCA that possesses IgM-like agglutination
potency, while
still amenable to commercial IgG purification process using, e.g., Protein
A/G, to enable a
potent, topical, non-hormonal contraceptive via an HCA that is cost effective
and sorely
needed by women around the world.
[000219] Antibodies can bind antigen on the sperm surface in the context of
immune
infertility. Immune infertility broadly refers to immune mechanisms that can
contribute to
infertility, and can be mediated by a variety of antibodies, including anti-
phospholipid, anti-
thyroid and anti-sperm antibodies (ASA). ASA refers to a broad spectrum of
antibodies that
can bind any sperm-associated antigens. The vast majority of naturally
occurring ASA bind
cytoplasmic antigens only accessible after sperm die, and are thus irrelevant
for
contraception. However, some Ab isolated from women who are immune infertile
can cause
infertility even without directly blocking sperm-egg interactions, including
the IgM molecule
isolated by Isojima from an infertile woman that serves as the basis for our
current HCA. The
HCA-UNC that may be used as the basis for the synthetic binding agent having
multiple Fab
repeats described herein binds an accessible surface antigen unique to sperm
and cells in the
59
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
male reproductive tract, and can prevent sperm from reaching the egg by
agglutinating and/or
immobilizing sperm in mucus. Indeed, it is partially because of the increased
barrier function
imparted by sperm-binding Ab in mucus that the motility of sperm in CM is
often measured
in clinical evaluation of infertility. Other ASAs may form the basis (e.g.,
core IgG) for other
HCAs using the principles described herein.
[000220] Vaginally delivered HCA is likely to provide highly effective and
safe
contraception. Sperm must swim through mucus to reach and fertilize the egg.
Not
surprisingly, poor sperm motility in cervical mucus is generally a good
correlate to infertility,
and sperm motility in mucus remains a gold standard test in diagnosing
infertility. By
extension, arresting sperm motility in mucus through antibodies that can
agglutinate and
immobilize individual sperm in mucus, by directly reducing the number of sperm
that reach
the egg, should provide an effective form of contraception. Indeed, such sperm-
binding Ab
have been isolated from the cervicovaginal secretions of infertile women.
Studies have shown
that these sperm-binding Ab (IgG, IgA or IgM) can trap vigorously motile sperm
in cervical
mucus without interfering with the sperm motility apparatus (Ab-coated sperm
will swim
freely in buffer), and trapped sperm shake in place for hours in mucus until
they die. This
"shaking phenomenon" was and continues to be a standard clinical diagnosis for
a cause of
infertility in humans, Local delivery of sperm-binding Ab is highly effective
in vivo,
reducing egg fertilization by at least 95% in a highly fertile rabbit model.
[000221] The female reproductive tract is coated with far smaller volumes of
mucus (-1-2
mL) than the volume of blood in circulation (-5,000 mL). Thus, by delivering
HCA locally,
contraceptive concentrations may be achieved with far lower amounts of HCA
than with
systemic delivery. Vaginally delivered mAb are poorly absorbed into the
systemic
circulation, further reducing the HCA amount needed to sustain contraceptive
levels in the
female reproductive tract.
[000222] HCA delivered into the vagina is highly unlikely to generate systemic
toxicity,
because: HCA is a fully human IgG; HCA is unlikely to be absorbed into the
systemic
circulation, the vagina is poorly responsive to immunization, and the target
antigen of HCA is
found exclusively in cells originating from the male reproductive tract, and
is not present in
females. The exceptionally limited systemic uptake could lead to a sufficient
safety profile
for HCA. Vaginal secretions possess very low complement activity, and have
exceedingly
few, if any, live leukocytes due to continuous acidification of the vagina to
pH ¨4 by lactic
acid from commensal Lactobacilli (leukocytes are effectively immobilized or
killed at pH <
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
6). Thus, HCA, especially if delivered at doses below total IgG present in
CVM, is unlikely
to trigger toxicity or inflammation in local vaginal tissues, yet remain
effective at vaginal pH.
[000223] Weak and transient mucin bonds with the synthetic binding agent
having multiple
Fab repeats may allow the synthetic binding agent to freely diffuse in mucus
most of the time
and rapidly accumulate on the pathogens. In turn, the array of bound Ab on
Ab/pathogen
complex can form a sufficient number of weak erosslinks with the mucin mesh to
trap
pathogen with permanent avidity. Interactions between IgG and mucins appear to
occur
through N-glycans on IgG-Fc. IgG can be harnessed to trap even highly motile
bacterial
pathogens and enable pathogen trapping in different mucus secretions,
including from the
airways as well as GI and female reproductive tracts, underscoring pathogen
trapping by IgG-
mucin affinity as a universal mucosal protective mechanism. Our multimeric HCA
constructs
retain the muco-trapping potencies relative to IgG and effectively immobilize
individual
sperm in mucus.
Release Device Example: intravaginal ring
[0002241 FIGS. 6A-6C describe a capsule-IVR (intravaginal ring) system that
may be used
with the synthetic binding agent having multiple Fab repeats described herein.
This delivery
apparatus makes use of conventional pill-processing to fabricate capsules that
can be
embedded in IVR and facilitate sustained release of the synthetic binding
agent. In FIG. 6A
the device is a ring that may be placed in the vagina (e.g., intrauterine) and
multiple time-
release capsules (FIG. 6B) may be loaded thereon. The capsules have been shown
to
maintain structural stability of the synthetic binding agent having multiple
Fab repeats for at
least 4 weeks when immersed in human CVM at 37 C (CVM replaced every 3-4
days). This
is illustrated in FIG. 6C showing both daily and cumulative release over one
month. The
release rates can be readily tuned over a wide range of release rates,
including as low as in the
0.1-0.3 mg/day range (Figure 4C; Formulation D) for 28 days or more. The
capsules can also
be formulated to provide greater release rates during the Days 2-6 window
(Formulation A &
B); it may be desirable to have greater dose of HCA delivered immediately
before the fertility
window. The synthetic binding agent having multiple Fab repeats configured as
HCA
described herein, when loaded into suitable IVR systems, will enable a
reliable and safe
contraceptive product that is not only non-hormonal but also economically
feasible, and does
not require daily or coitally-associated administration.
61
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
EXAMPLES: Sequences
[000225] The synthetic binding agents described herein typically include
multiple
additional copies of Fab regions, as described above. As describe above in
FIGS. 1A-1G,
these synthetic binding agents may be arranged in in a variety of different
configurations of a
core IgG (including Fe and Fab domains), in which the duplicated copies of the
Fab domains
(directed to the same target epitope as the Fab domains on the core IgG) are
attached at either
or both the NH2 and/or the COOH ends. Any Fab domain may be used, and may be
linked to
the amino or carboxyl ends via a flexible linker comprising an amino acid
sequence
comprising n pentapeptide repeats consisting of Glycine (G) and Serine (S),
wherein n is
between 3 and 8.
[000226] As an example, a synthetic binding agent may be directed to an N-
linked glycan
of an epitope specific to sperm, referred to as CD52 glycoform ("CD52g"). SEQ
ID NO: 1
shows one example of an amino acid sequence corresponding to CD52g (see, e.g.,
Diekman
et al., FASEB Journal, vol. 13: 1303-1313, August 1999). The variable domains
(heavy
and/or light) of any antibody directed against a protein including this
sequence may be used,
and configured as a synthetic binding agent as described herein. The exemplary
synthetic
binding agents described by SEQ ID NOS: 2-31 illustrate examples of such
antibodies
directed against a sperm-specific epitope. For example, a synthetic binding
agent that is
directed to an epitope specific to sperm, such as CD52g (e.g., an n-
glycosylated form of
CD52) includes both heavy chain and light chain. SEQ ID NO: 2 is an exemplary
DNA
sequence for a heavy chain domain of a core IgG directed to an epitope of
CD52g, and SEQ
ID NO: 3 is an example of an amino acid sequence for a heavy chain portion of
the IgG.
SEQ ID NO: 4 is an example of an amino acid sequence of a Fab fragment for a
heavy chain.
SEQ ID NO: 5 is an example of an amino acid sequence of an Fe fragment of a
heavy chain.
SEQ ID NO: 6 is an example of an exemplary DNA sequence for a light chain
domain of a
core IgG directed to an epitope of CD52g. SEQ ID NO: 7 is an example of an
amino acid
sequence of a light chain domain of a core IgG directed to an epitope of
CD52g.
[000227] SEQ ID NO: 8 to SEQ ID NO: 13 show exemplary DNA and amino acid
sequences for heavy and light chain portions of a synthetic binding agent
(e.g., recombinant
mAb) that may reduce sperm mobility in mucus having a structure similar to
that shown in
FIG. 1B (e.g., Fab-IgG). SEQ ID NO: 8 is an exemplary DNA sequence for a heavy
chain
domain of a Fab-IgG synthetic binding agent directed to an epitope of CD52g,
and SEQ ID
NO: 9 is an example of an amino acid sequence for a heavy chain portion of a
Fab-IgG. SEQ
62
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
ID NO: 10 is an example of an amino acid sequence of a Fab fragment for a
heavy chain of a
synthetic binding agent including a Fab extending from the N-terminal end of a
core IgG.
SEQ ID NO: 11 is an example of an amino acid sequence of an Fe fragment of a
heavy chain.
SEQ ID NO: 12 is an example of an exemplary DNA sequence for a light chain
domain of a
core IgG directed to an epitope of CD52g. SEQ ID NO: 13 is an example of an
amino acid
sequence of a light chain domain of a synthetic binding agent including a Fab
extending from
the N-terminal end of a core IgG directed to an epitope of CD52g.
[000228] SEQ ID NO: 14 to SEQ ID NO: 19 show exemplary DNA and amino acid
sequences for heavy and light chain portions of a synthetic binding agent
(e.g., recombinant
mAb) that may reduce sperm mobility in mucus having a structure similar to
that shown in
FIG. 1C (e.g., IgG-Fab). SEQ ID NO: 14 is an exemplary DNA sequence for a
heavy chain
domain of an IgG-Fab synthetic binding agent directed to an epitope of CD52g,
and SEQ ID
NO: 15 is an example of an amino acid sequence for a heavy chain portion of
such an IgG-
Fab. SEQ ID NO: 16 is an example of an amino acid sequence of a Fab fragment
for a heavy
chain of a synthetic binding agent including a Fab extending from the N-
terminal end of a
core IgG. SEQ ID NO: 17 is an example of an amino acid sequence of an Fe
fragment of a
heavy chain. SEQ ID NO: 18 is an example of an exemplary DNA sequence for a
light chain
domain directed to an epitope of CD52g. SEQ ID NO: 19 is an example of an
amino acid
sequence of a light chain domain of a synthetic binding agent directed to an
epitope of
CD52g.
[000229] SEQ ID NO: 20 to SEQ ID NO: 25 show exemplary DNA and amino acid
sequences for heavy and light chain portions of a synthetic binding agent
(e.g., recombinant
mAb) that may reduce sperm mobility in mucus having a structure similar to
that shown in
FIG. 1D (e.g., Fab-IgG-Fab). SEQ ID NO: 20 is an exemplary DNA sequence for a
heavy
chain domain of an Fab-IgG-Fab synthetic binding agent directed to an epitope
of CD52g,
and SEQ ID NO: 21 is an example of an amino acid sequence for a heavy chain
portion of
such a Fab-IgG-Fab. SEQ ID NO: 22 is an example of an amino acid sequence of a
Fab
fragment for a heavy chain of a synthetic binding agent directed to an epitope
of CD52g.
SEQ ID NO: 23 is an example of an amino acid sequence of an Fe fragment of a
heavy chain
of a synthetic binding agent directed to an epitope of CD52g. SEQ ID NO: 24 is
an example
of an exemplary DNA sequence for a light chain domain directed to an epitope
of CD52g.
SEQ ID NO: 25 is an example of an amino acid sequence of a light chain domain
of a
synthetic binding agent directed to an epitope of CD52g.
63
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[000230] SEQ ID NO: 26 to SEQ ID NO: 31 show exemplary DNA and amino acid
sequences for heavy and light chain portions of a synthetic binding agent
(e.g., recombinant
mAb) that may reduce sperm mobility in mucus having a structure similar to
that shown in
FIG. lE (e.g., Fab-IgG-Fab-Fab). SEQ ID NO: 26 is an exemplary DNA sequence
for a
heavy chain domain of an Fab-IgG-Fab-Fab synthetic binding agent directed to
an epitope of
CD52g, and SEQ ID NO: 27 is an example of an amino acid sequence for a heavy
chain
portion of such a Fab-IgG-Fab-Fab. SEQ ID NO: 28 is an example of an amino
acid
sequence of a Fab fragment for a heavy chain of a synthetic binding agent
directed to an
epitope of CD52g. SEQ ID NO: 29 is an example of an amino acid sequence of an
Fe
fragment of a heavy chain of a synthetic binding agent directed to an epitope
of CD52g. SEQ
ID NO: 30 is an example of an exemplary DNA sequence for a light chain domain
directed to
an epitope of CD52g. SEQ ID NO: 31 is an example of an amino acid sequence of
a light
chain domain of a synthetic binding agent directed to an epitope of CD52g.
[000231] FIGS. 14A and 14B illustrate a comparison between the amino acid
sequences of
the heavy (FIG. 14A) and light (FIG. 14B) chain sequences described in the
sequence listing,
as compared to a gerrnline sequence (e.g., native IgG). The notation of the
different
constructs are listed from NH2 to COOH ends, with IgG implying Fab-Fe.
[000232] SEQ ID NO: 32 to SEQ ID NO: 38 show an exemplary DNA and amino acid
sequences for heavy and light chain portions of a synthetic binding agent
(e.g., recombinant
mAb) that may reduce sperm mobility in mucus having a structure of Fab-Fab-IgG-
Fab-Fab.
SEQ ID NO: 32 is an exemplary DNA sequence for a heavy chain domain of a Fab-
Fab-IgG-
Fab-Fab synthetic binding agent directed to an epitope of CD52g, and SEQ ID
NO: 33 is an
example of an amino acid sequence for a heavy chain portion of such a Fab-Fab-
IgG-Fab-
Fab. SEQ ID NO: 34 is an example of a DNA sequence for a light chain of the
anti-CD53g
Fab-Fab-IgG-Fab-Fab synthetic protein. SEQ ID NO: 35 is an example of an amino
acid
sequence of the anti-CD52g Fab-Fab-IgG-Fab-Fab synthetic binding agent. SEQ ID
NO: 36
is an amino acid sequence of the Fab fragment of the Fab-Fab-IgG-Fab-Fab
(heavy chain)
portion, while SEQ ID NO: 37 is the amino acid sequence of anti-CD52g Fab
fragment of
Fab-Fab-IgG-Fab-Fab. SEQ ID NO: 38 is an example of an amino acid sequence of
an Fe
fragment of a heavy chain of a synthetic binding agent directed to an epitope
of CD52g,
including configured as a Fab-Fab-IgG-Fab-Fab.
[000233] In another example, a synthetic binding agent, which in particular
may reduce the
fraction of pathogen that can permeate through mucus and/or freely divide as
described
herein, may be directed against Klebsiella (e.g., having anti-Klebsiella
activity). For
64
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
example, a human or humanized IgG (mAb) that specifically recognizes an
epitope of
Klebsiella pneumonia 01 may be used. For example, the anti- Klebsiella mAb
illustrated by
SEQ ID NO: 39 to SEQ ID NO: 45 is directed against the D-galactan-II antigen
of Klebsiella
pneumonia; other epitopes or other anti-Klebsiella mAbs may be used instead.
For example,
SEQ ID NO: 39 is a polynucleotide (DNA) sequence of the heavy chain of an anti-
Klebsiella
IgG. SEQ ID NO: 40 is an amino acid sequence of an anti-Klebsiella heavy
chain. SEQ ID
NO: 43 is a polynucleotide (DNA) sequence of a light chain of the anti-
Klebsiella IgG; SEQ
ID NO: 44 is an amino acid sequence of the anti-Klebsiella light chain. SEQ ID
NO: 45 is an
amino acid sequence of a Fab fragment of this anti-Klebsiella IgG light chain,
while SEQ ID
NO: 41 is an amino acid sequence of a Fab fragment of an anti-Klebsiella heavy
chain and
SEQ ID NO: 42 is an amino acid sequence of an Fc fragment of the heavy chain
of this anti-
Kiebsielia antibody.
[0002341 An example of a synthetic binding agent specific to Klebsiella
constructed as
described herein as a Fab-IgG construct (similar to FIG. 1B) is described by
SEQ ID NO: 46
to SEQ ID NO: 52. SEQ ID NO: 46 is the DNA sequence of the anti-Klebsiella Fab-
IgG
heavy chain, while SEQ ID NO: 47 is an amino acid sequence of a heavy chain of
a Fab-IgG.
SEQ ID NO: 48 is an amino acid sequence of this anti-Klebsiella Fab fragment
of a Fab-IgG
heavy chain. SEQ ID NO: 49 is an amino acid sequence of the Fe fragment of an
IgG-Fab.
SEQ ID NO: 50 is a DNA sequence of the light chain of the Fab-IgG, and SEQ ID
NO: 51 is
an amino acid sequence of the light chain of the Fab-IgG. SEQ ID NO: 52 shows
an amino
acid sequence of a Fab fragment of Fab-IgG Light Chain.
10002351 Another example of a synthetic binding agent specific to Klebsiella,
constructed
as an IgG-Fab construct (similar to FIG. 1C) is described by SEQ ID NO: 53 to
SEQ ID NO:
59. SEQ ID NO: 53 is the DNA sequence of the anti-Klebsiella IgG-Fab heavy
chain, while
SEQ ID NO: 54 is an amino acid sequence of a heavy chain of a IgG-Fab. SEQ ID
NO: 55
is an amino acid sequence of an anti-Klebsiella Fab fragment of a IgG-Fab
heavy chain. SEQ
ID NO: 56 is an amino acid sequence of an Fe fragment of an IgG-Fab. SEQ ID
NO: 57 is a
DNA sequence of the light chain of this IgG-Fab synthetic binding agent, and
SEQ ID NO:
58 is an amino acid sequence of the light chain of the IgG-Fab. SEQ ID NO: 59
shows an
amino acid sequence of a Fab fragment of IgG-Fab Light Chain.
[0002361 An example of a synthetic binding agent specific to Klebsiella
constructed as
described herein as a Fab-IgG-Fab construct (similar to FIG. ID) is described
by SEQ ID
NO: 60 to SEQ ID NO: 66. SEQ ID NO: 60 is the DNA sequence of the anti-
Klebsiella Fab-
IgG-Fab heavy chain, and SEQ ID NO: 61 is an amino acid sequence of a heavy
chain of a
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Fab-IgG-Fab. SEQ ID NO: 62 is an amino acid sequence of this anti-Klebsiella
Fab
fragment of a Fab-IgG-Fab heavy chain. SEQ ID NO: 63 is an amino acid sequence
of the Fc
fragment of an Fab-IgG-Fab. SEQ ID NO: 64 is a DNA sequence of the light chain
of the
Fab-IgG-Fab, and SEQ ID NO: 65 is an amino acid sequence of the light chain of
the Fab-
IgG-Fab. SEQ ID NO: 66 shows an amino acid sequence of a Fab fragment of Fab-
IgG-Fab
Light Chain.
[000237] An example of a synthetic binding agent specific to Klebsiella
constructed as
described herein as a Fab-Fab-IgG-Fab-Fab construct (similar to FIG. 1F) is
described by
SEQ ID NO: 160 to SEQ ID NO: 166. SEQ ID NO: 160 is the DNA sequence of the
anti-
Klebsiella Fab-Fab-IgG-Fab-Fab heavy chain, and SEQ ID NO: 161 is an amino
acid
sequence of a heavy chain of a Fab-Fab-IgG-Fab-Fab, SEQ ID NO: 162 is an amino
acid
sequence of this anti-Klebsiella Fab fragment of a Fab-Fab-IgG-Fab-Fab heavy
chain. SEQ
ID NO: 163 is an amino acid sequence of the Fe fragment of a Fab-Fab-IgG-Fab-
Fab, SEQ
ID NO: 164 is a DNA sequence of the light chain of the Fab-Fab-IgG-Fab-Fab,
and SEQ ID
NO: 165 is an amino acid sequence of the light chain of the Fab-Fab-IgG-Fab-
Fab. SEQ ID
NO: 166 shows an amino acid sequence of a Fab fragment of Fab-Fab-IgG-Fab-Fab
Light
Chain.
[000238] In another example, a synthetic binding agent, which in particular
may reduce the
fraction of pathogen that can permeate through mucus and/or freely divide as
described
herein, may be directed against Salmonella (e.g., having anti- Salmonella
activity). For
example, a human or humanized IgG (mAb) that specifically recognizes an
epitope of
Salmonella may be used. For example, the anti- Salmonella inAb illustrated by
SEQ ID NO:
67 to SEQ ID NO: 73 is directed against an antigen of Salmonella. Any
appropriate epitope
or other anti- Salmonella mAbs may be used. For example, SEQ ID NO: 67 is a
polynucleotide (DNA) sequence of the heavy chain of an anti- Salmonella IgG.
SEQ ID NO:
68 is an amino acid sequence of an anti-Salmonella heavy chain. SEQ ID NO: 71
is a
polynucleotide (DNA) sequence of a light chain of the anti-Salmonella IgG; SEQ
ID NO: 72
is an amino acid sequence of the anti-Salmonella light chain. SEQ ID NO: 69 is
an amino
acid sequence of a Fab fragment of this anti-Salmonella IgG heavy chain, while
SEQ ID NO:
73 is an amino acid sequence of a Fab fragment of an anti-Salmonella light
chain and SEQ ID
NO: 70 is an amino acid sequence of an Fe fragment of the heavy chain of this
anti-Klebsiella
antibody.
[000239] An example of a Fab-IgG synthetic anti-Salmonella LP binding agent is
described
by SEQ ID NO: 74 to SEQ ID NO: 80, including the DNA sequence of a synthetic
Fab-IgG
66
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Heavy Chain in SEQ ID NO: 74 (the amino acid sequence of this heavy chain is
shown in
SEQ ID NO: 75). The amino acid residue of the Fab fragment of Fab-IgG Heavy
Chain is
provided in SEQ ID NO: 76 and the amino acid residues of the Fe fragment of
Fab-IgG is
provided in SEQ ID NO: 77. SEQ ID NO: 78 is a DNA sequence of Fab-IgG (Light
Chain)
portion, and the amino acid sequence is in SEQ ID NO: 79. SEQ ID NO: 80 lists
the amino
acid residues of the Fab fragment of Fab-IgG Light Chain.
[000240] An example of an IgG-Fab synthetic anti-Salmonella LPS binding agent
is
described by SEQ ID NO: 81 to SEQ ID NO: 87, including the DNA sequence of a
synthetic
Fab-IgG Heavy Chain in SEQ ID NO: 81 (the amino acid sequence of this heavy
chain is
shown in SEQ ID NO: 82). The amino acid residue of the Fab fragment of Fab-IgG
Heavy
Chain is provided in SEQ ID NO: 83 and the amino acid residues of the Fe
fragment of Fab-
IgG is provided in SEQ ID NO: 84. SEQ ID NO: 85 is a DNA sequence of Fab-IgG
(Light
Chain) portion, and the amino acid sequence is in SEQ ID NO: 86. SEQ ID NO: 87
lists the
amino acid residues of the Fab fragment of Fab-IgG Light Chain.
[000241] An example of a Fab-IgG-Fab synthetic anti-Salmonella LPS binding
agent is
described by SEQ ID NO: 88 to SEQ ID NO: 94, including the DNA sequence of a
synthetic
Fab-IgG Heavy Chain in SEQ ID NO: 88 (the amino acid sequence of this heavy
chain is
shown in SEQ ID NO: 89). The amino acid residue of the Fab fragment of Fab-IgG
Heavy
Chain is provided in SEQ ID NO: 90 and the amino acid residues of the Pc
fragment of Fab-
IgG is provided in SEQ ID NO: 91. SEQ ID NO: 92 is a DNA sequence of Fab-IgG
(Light
Chain) portion, and the amino acid sequence is in SEQ ID NO: 93. SEQ ID NO: 94
lists the
amino acid residues of the Fab fragment of Fab-IgG Light Chain.
[000242] An example of a Fab-Fab-IgG-Fab-Fab synthetic anti-Salmonella LPS
binding
agent is described by SEQ ID NO: 95 to SEQ ID NO: 101, including the DNA
sequence of a
synthetic Fab-IgG Heavy Chain in SEQ ID NO: 95 (the amino acid sequence of
this heavy
chain is shown in SEQ ID NO: 96). The amino acid residue of the Fab fragment
of Fab-IgG
Heavy Chain is provided in SEQ ID NO: 97 and the amino acid residues of the Fe
fragment
of Fab-IgG is provided in SEQ ID NO: 98. SEQ ID NO: 99 is a DNA sequence of
Fab-IgG
(Light Chain) portion, and the amino acid sequence is in SEQ ID NO: 100. SEQ
ID NO: 101
lists the amino acid residues of the Fab fragment of Fab-IgG Light Chain.
[000243] In another example, a synthetic binding agent, which in particular
may reduce the
fraction of pathogen that can permeate through mucus and/or freely divide as
described
herein, may be directed against Neisseria gonorrhoeae (e.g., having anti-
Gonorrhea
activity). For example, a human or humanized IgG (mAb) that specifically
recognizes an
67
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
epitope of Neisseria gonorrhoeae may be used. For example, the anti- Gonorrhea
mAb
(2C7) illustrated by SEQ ID NO: 102 to SEQ ID NO: 108 is directed against an
antigen of
Neisseria gonorrhoeae. Any appropriate epitope or other anti-gonorrhoeae mAbs
may be
used. For example, SEQ ID NO: 102 is a polynucleotide (DNA) sequence of the
heavy chain
of an anti-gonorrhoeae IgG. SEQ ID NO: 103 is an amino acid sequence of an
anti-
gonorrhoeae heavy chain. SEQ ID NO: 106 is a polynucleotide (DNA) sequence of
a light
chain of the anti-gonorrhoeae IgG; SEQ ID NO: 107 is an amino acid sequence of
the anti-
gonorrhoeae light chain. SEQ ID NO: 104 is an amino acid sequence of a Fab
fragment of
this anti-gonorrhoeae IgG heavy chain, while SEQ ID NO: 108 is an amino acid
sequence of
a Fab fragment of an anti-gonorrhoeae light chain and SEQ ID NO: 105 is an
amino acid
sequence of an Fe fragment of the heavy chain of this anti-gonorrhoeae
antibody.
10002441 An example of a Fab-IgG synthetic anti-gonorrhoeae (2C7) binding
agent is
described by SEQ ID NO: 109 to SEQ ID NO: 115, including the DNA sequence of a
synthetic Fab-IgG Heavy Chain in SEQ ID NO: 109 (the amino acid sequence of
this heavy
chain is shown in SEQ ID NO: 110). The amino acid residue of the Fab fragment
of Fab-IgG
Heavy Chain is provided in SEQ ID NO: 111 and the amino acid residues of the
Fe fragment
of Fab-IgG is provided in SEQ ID NO: 112. SEQ ID NO: 113 is a DNA sequence of
Fab-
IgG (Light Chain) portion, and the amino acid sequence is in SEQ ID NO: 114.
SEQ ID NO:
115 lists the amino acid residues of the Fab fragment of Fab-IgG Light Chain.
[000245] An example of an IgG-Fab synthetic anti-gonorrhoeae binding agent is
described
by SEQ ID NO: 116 to SEQ ID NO: 122, including the DNA sequence of a synthetic
IgG-
Fab Heavy Chain in SEQ ID NO: 116 (the amino acid sequence of this heavy chain
is shown
in SEQ ID NO: 117). The amino acid residue of the Fab fragment of IgG-Fab
Heavy Chain
is provided in SEQ ID NO: 118 and the amino acid residues of the Fe fragment
of IgG-Fab is
provided in SEQ ID NO: 119. SEQ ID NO: 120 is a DNA sequence of IgG-Fab (Light
Chain) portion, and the amino acid sequence is in SEQ ID NO: 121. SEQ ID NO:
122 lists
the amino acid residues of the Fab fragment of IgG-Fab Light Chain.
[000246] An example of a Fab-IgG-Fab synthetic anti-gonorrhoeae binding agent
is
described by SEQ ID NO: 123 to SEQ ID NO: 129, including the DNA sequence of a
synthetic Fab-IgG-Fab Heavy Chain in SEQ ID NO: 123 (the amino acid sequence
of this
heavy chain is shown in SEQ ID NO: 124). The amino acid residue of the Fab
fragment of
Fab-IgG-Fab Heavy Chain is provided in SEQ ID NO: 125 and the amino acid
residues of the
Fe fragment of Fab-IgG-Fab is provided in SEQ ID NO: 126. SEQ ID NO: 127 is a
DNA
sequence of Fab-IgG-Fab (Light Chain) portion, and the amino acid sequence is
in SEQ ID
68
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
NO: 128. SEQ ID NO: 129 lists the amino acid residues of the Fab fragment of
Fab-IgG-Fab
Light Chain.
[000247] An example of a Fab-Fab-IgG-Fab-Fab synthetic anti-gonorrhoeae
binding agent
is described by SEQ ID NO: 153 to SEQ ID NO: 159, including the DNA sequence
of a
synthetic Fab-Fab-IgG-Fab-Fab Heavy Chain in SEQ ID NO: 153 (the amino acid
sequence
of this heavy chain is shown in SEQ ID NO: 154). The amino acid residue of the
Fab
fragment of Fab-Fab-IgG-Fab-Fab Heavy Chain is provided in SEQ ID NO: 155 and
the
amino acid residues of the Fe fragment of Fab-Fab-IgG-Fab-Fab is provided in
SEQ ID NO:
156. SEQ ID NO: 157 is a DNA sequence of Fab-Fab-IgG-Fab-Fab (Light Chain)
portion,
and the amino acid sequence is in SEQ ID NO: 158. SEQ ID NO: 159 lists the
amino acid
residues of the Fab fragment of Fab-Fab-IgG-Fab-Fab Light Chain.
[000248] In another example, a synthetic binding agent, which in particular
may reduce the
fraction of pathogen that can permeate through mucus as described herein, may
be directed
against Respiratory Syncytial Virus (RSV). For example, a human or humanized
IgG (mAb)
that specifically recognizes an epitope of RSV may be used. For example, an
anti-RSV mAb
(modeled after published Motavizumab) is illustrated by SEQ ID NO: 132 to SEQ
ID NO:
138 and is directed against an antigen of RSV. Any appropriate epitope or
other anti-RSV
mAbs may be used, SEQ ID NO: 132 is a polynucleotide (DNA) sequence of the
heavy
chain of an anti-RSV IgG. SEQ ID NO: 133 is an amino acid sequence of an anti-
RSV heavy
chain. SEQ ID NO: 136 is a polynucleotide (DNA) sequence of a light chain of
the anti-RSV
IgG; SEQ ID NO: 137 is an amino acid sequence of the anti- RSV light chain.
SEQ ID NO:
134 is an amino acid sequence of a Fab fragment of this anti-RSV IgG heavy
chain, while
SEQ ID NO: 138 is an amino acid sequence of a Fab fragment of an anti-RSV
light chain and
SEQ ID NO: 135 is an amino acid sequence of an Fe fragment of the heavy chain
of this anti-
RSV antibody.
[000249] An example of a Fab-IgG synthetic anti-RSV binding agent is described
by SEQ
ID NO: 139 to SEQ ID NO: 145, including the DNA sequence of a synthetic Fab-
IgG Heavy
Chain in SEQ ID NO: 139 (the amino acid sequence of this heavy chain is shown
in SEQ ID
NO: 140). The amino acid residue of the Fab fragment of Fab-IgG Heavy Chain is
provided
in SEQ ID NO: 141 and the amino acid residues of the Fe fragment of Fab-IgG is
provided in
SEQ ID NO: 142. SEQ ID NO: 143 is a DNA sequence of Fab-IgG (Light Chain)
portion,
and the amino acid sequence is in SEQ ID NO: 144. SEQ ID NO: 145 lists the
amino acid
residues of the Fab fragment of Fab-IgG Light Chain.
69
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[000250] An example of an IgG-Fab synthetic anti-RSV binding agent is
described by SEQ
ID NO: 146 to SEQ ID NO: 152, including the DNA sequence of a synthetic Fab-
IgG Heavy
Chain in SEQ ID NO: 146 (the amino acid sequence of this heavy chain is shown
in SEQ ID
NO: 147). The amino acid residue of the Fab fragment of Fab-IgG Heavy Chain is
provided
in SEQ ID NO: 148 and the amino acid residues of the Fe fragment of Fab-IgG is
provided in
SEQ ID NO: 149. SEQ ID NO: 150 is a DNA sequence of Fab-IgG (Light Chain)
portion,
and the amino acid sequence is in SEQ ID NO: 151. SEQ ID NO: 152 lists the
amino acid
residues of the Fab fragment of Fab-IgG Light Chain.
[000251] Other synthetic binding agents (e.g., multimeric constructs) may be
directed
against Psuedomonas aeruginosa, Methicillin-resistant Staphylococcus aureus,
Acinetobacter
baumannii, and Clostridium difficile, Sequences for IgG mAbs against surface
antigens for
these (and other pathogens) are published and synthetic binding agents may be
formed as
described herein. Thus, although specific sequences of exemplary synthetic
binding agents
that may reduce the fraction of pathogen that can permeate through mucus
and/or freely
divide are described above, one of skill in the art, may understand that the
specification
generally teaches the method of making and using synthetic binding agents from
an IgG,
particularly IgGs directed against surface antigens.
EXAMPLES
[000252] The synthetic binding agents described herein are synthetic human or
humanized
Immunoglobulin G (IgG) having a pair of Fab domains to which additional Fab
domains
directed to the same antigen are linked by a flexible linker at either or both
the end(s) of the
Fab domains of the IgG and/or the Fe region of the IgG, in tandem. The
resulting synthetic
binding agent has been found to dramatically reduce the mobility of the target
(e.g., pathogen,
such as bacteria, virus, yeast, etc. and/or sperm, etc.) in mucus. The
synthetic binding agents
were found to be stable across a variety of delivery forms, including
nebulized forms, and can
be readily produced using the methods and techniques described herein.
[000253] For example, studies were performed to demonstrate that virtually any
starting
IgG (e.g., IgG1 mAb) having specific binding for an antigen (or antigen
region) of a target,
such as sperm or a pathogen (virus, bacteria, yeast, mold, etc.), a synthetic
binding agent as
described herein may be generated. In some variations the variable heavy chain
and light
chain, in some cases as well as constant heavy and light chain sequences, of a
starting IgG1
mAb were codon-optimized for Homo sapiens using the optimization tool, such as
that
provided by GeneArt (ThermoFisher Scientific). Codon-optimized sequences of
VH, CH1,
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
VL and CL may be used to design the gene fragments required to assemble the
synthetic
binding agents described herein (e.g., using software such as Benchling
software).
[0002541 For example, to assemble a Fab-IgG synthetic binding agent, a gene
fragment
comprised of VH-CH1-6xG4S-Linkers-VH was designed to be cloned into mammalian
expression vector comprised of CHI -CH2-CH3 DNA sequences. Similarly, to
assemble Fab-
Fab-IgG-Fab-Fab, the gene fragments comprised of VH-CH1-6xG4SLinkers-VH-CH1-
6xG4SLinkers-VH and 6xG4SLinkers-VH-CH1-6xG4SLinkers-VH-CH1 were designed to
be further cloned into an IgG1 expression vector. In some examples, to
minimize synthesis
problems that could occur due to repeated sequences, the DNA sequences for
repeated
fragments may be, e.g., manually, codon-optimized resulting in increased
variability of DNA
sequences and subsequent reduced complexity for gene synthesis. After codon-
optimization,
gene sequences may be further processed through a complexity-analyzing tool
provided (e.g.,
such as that provided by IDT (Integrated DNA Technologies) to obtain a
complexity score.
Gene fragments with complexity scores < 25 are known to be easily and
successfully
synthesized via GeneArt Gene synthesis.
10002551 Expression vectors encoding the synthetic binding agent may be
generated. For
example, an expression plasmid encoding the light chain, the gene fragment
consisting of VL
and CL (CA) DNA sequences may be synthesized using custom gene-synthesis
service (e.g.,
Integrated DNA Technologies) and cloned into an empty mammalian expression
vector
using, e.g., KpnI (5') and EcoRI (3') restriction sites. For the construction
of expression
plasmids encoding heavy chains (HC) for the synthetic binding agent, in some
examples four
cloning vectors comprising of VH-CHI-6xG4SLinkers-VH, VH-CH1-6xG4SLinkers-VH-
CH1-6xG4SLinkers-VH, 6xG4SLinkers-VH-CHI and 6xG4SLinkers-VH-CH1-
6xG4SLinkers-VH-CH1 DNA sequences were synthesized using GeneArt gene
synthesis
service (ThermoFisher Scientific). In some examples, for the construction of
expression
plasmid encoding HC for IgG, VH fragment was amplified from the cloning vector
comprising of VH-CHI-6xG4SLinkers-VH vector using forward primer, 5t-
TAAGCAGGTACCGCCACCATGAAGTG-3' (SEQ ID NO: 130), and reverse primer, 5'-
TGCTTAGCTAGCTGGAGAAACTGTC-3r (SEQ ID NO: 131), and then cloned into the
mammalian expression vector comprised of CH1-CH2-CH3 DNA sequences using KpnI
(5')
and Nhei (3') restriction sites. In some examples, for the construction of
expression plasmid
encoding HC for Fab-IgG, VH-CH1-6xG4SLinkers-VH fragment was cloned into the
same
mammalian expression vector using KpnI (5') and NheI (3') restriction sites.
For example,
71
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
the construction of an expression plasmid encoding HC for IgG-Fab may include
using a
6xG4SLinkers-VH-CH1 fragment that is cloned into the IgG mammalian expression
vector
using BamHI (5') and MIul (3') restriction sites. For the construction of
expression plasmid
encoding HC for Fab-IgG-Fab, VH-CH1-6xG4SLinkers-VH fragment may be first
cloned
into the mammalian expression vector using KpnI (5') and NheI (3') restriction
sites followed
by the cloning of 6xG4SLinkers-VH-CHI fragment using BamHI (5') and MluI (3')
restriction sites. For the construction of expression plasmid encoding HC for
Fab-IgG-Fab-
Fab, VH-CHI-6xG4SLinkers-VH fragment may be first cloned into the mammalian
expression vector using KpnI (5') and NheI (3') restriction sites followed by
the cloning of
6xG4SLinkers-VH-CHI-6xG4SLinkers-VH-CH1 fragment using BamHI (5') and MluI
(3')
restriction sites. For the construction of expression plasmid encoding HC for
Fab-Fab-IgG-
Fab-Fab, VH-CH1-6xG4SLinkers-VH-CHI-6xG4SLinkers-VH fragment may be first
cloned
into the mammalian expression vector using KpnI (5') and NheI (3') restriction
sites followed
by the cloning of 6xG4SLinkers-VH-CH1-6xG4SLinkers-VH-CH1 fragment using BamHI
(5') and MluI (3') restriction sites. For the ligation of all heavy chains as
well as a light chain
into the expression vectors, quick ligation kit (New England Bio labs,
Ipswich, MA) may be
used. All ligated DNA constructs may be transformed into chemically competent
TOP 10 E.
coli cells (Life Technologies) and plated on ampicillin plates for selection.
Bacterial colonies
may be picked, cultured, and the plasmids prepped (e.g., Qiagen MiniPrep Kit).
Correct
assembly of the constructs into the expression vector may be confirmed by
Sanger
sequencing (e.g., Eurofins Genomics).
10002561 In some of the experiments described herein, expression plasmids
encoding the
heavy chain (HC) and light chain (LC) for IgG, Fab-IgG, IgG-Fab, FIF, FIFF and
FFIFF
antibodies were scaled up by transforming the sequencing-confirmed expression
plasmids in
chemically competent TOP10 E. coli, inoculating the transformation mix into
100 mL Luria
broth in a 250 mL baffled flask and overnight shaking at 220 r.p.m at 37 C.
Midi-prep
plasmid purifications were done using NucleoBond Xtra Midi EF Kits (Macherey-
Nagel)
according to the manufacturer's protocols. Proteins were expressed in Expi293
cells using
ExpiFectamineTM 293 Transfection reagents and protocols provided by the
manufacturer
(ThermoFisher Scientific). For IgG, one HC and one LC plasmid were co-
transfected using a
1:1 ratio at 1 ug total DNA per 1 mL of culture. For both Fab-IgG and IgG-Fab,
one HC and
one LC plasmid were co-transfected using a 1:2 ratio at 1 tig total DNA per 1
mL culture. For
Fab-IgG-Fab, one HC and one LC plasmid were co-transfected using a 1:3 ratio
at 1 jig total
DNA per 1 mL culture. For Fab-IgG-Fab-Fab, one HC and one LC plasmid were co-
72
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
transfected using a 1:4 ratio at 1 ug total DNA per 1 mL culture. For Fab-Fab-
IgG-Fab-Fab,
one HC plasmid and one LC plasmid were co-transfected using a 1:5 ratio at 1
ug total DNA
per 1 rriL culture. Transfected cells were grown at 37 C in a 5% CO2 incubator
while
shaking at 125 r.p.m. for 5 days. Supernatants were harvested by
centrifugation at 5000g for
mm and passed through 0.22- m filters for purification using standard protein
A affinity
chromatography. Briefly, 30 mL of transfected supernatant was incubated with
400 [IL
PBS-washed PierceTM Protein A Plus Agarose Resin (ThermoFisher Scientific)
overnight
at 4 C. Next, the resin-supernatant solution was flown through the gravity
columns
followed by the washing of resin. Protein was eluted by adding 9001,LL of
PierceTM IgG
Elution Buffer (ThermoFisher Scientific) into PBS-washed resin and was
immediately
neutralized by adding 100111, of UltraPureTM 1 M Tris-HCI Buffer, pH 7.5
(ThermoFisher
Scientific). Eluted proteins were further dialyzed into PBS using Amiconel
Ultra
Centrifugal Filters (Millipore Sigma).
[000257] In some variations the synthetic binding agent may be delivered to a
mucosa, as
described herein, Delivery may be via topical delivery, including aerosol,
liquid, or gel
(including dissolvable gel). For example in some variations a film may be used
to deliver
the synthetic binding agent. In some cases, a vaginal film may be used.
[000258] To examine this in the context of a synthetic anti-sperm binding
agent, genes
containing the complete heavy chain and light chain sequences of IgG and Fab-
IgG-Fab were
cloned into plant expression vectors (TMV and PVX; Icon Genetics) followed by
transformation into Agrobacterium turnefaciens strain ICF320 (Icon Genetics).
Next, the
transformation mixture was infiltrated into the 4 wk old N. bentharniana
plants that were
genetically modified to produce highly homogenous mammalian N-glycans of the
GnGn
glycoform. Seven days later, anti-sperm antibodies were extracted from the
leaf tissue and
purified using protein A chromatography. To remove the endotoxins, purified
rnAbs were
passed through an Acrodisc Units with Mustang Q Membrane (Pall Life Sciences).
Endosafe
PTS (Charles River) was utilized to measure the endotoxin level, which was
found to less
than 150 EU/mg. A film, such as a Nicotiana-produced HCA film, may then be
made. In
some experiments, the HCA films were formulated as a 2-inch by 2-inch
polyvinyl alcohol
(PVA) film casts and dried from an aqueous wet blend. The aqueous wet blend
was
composed of approximately 38.5 mL formulated antibody concentrate (200 mg/mL
maltitol,
10.0 inM histidine, 0.05 mg/mL, polysorbate 20) mixed with an aqueous polymer
concentrate
(17.88 g PVA 8-88 dissolved in 53.6 g WFI). IgG-Film and FIF-Film (e.g., Fab-
IgG-Fab)
73
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
were composed with 20 mg and 10 mg of mAbs per film respectively. The films
were
dissolved in ultra-pure water to perform characterization studies followed by
sperm potency
and agglutination kinetics assays.
[0002591 The synthetic binding agents described herein may also be nebulized
for delivery
without significantly reducing their efficacy. For example, nebulization of
F1F (Fab-IgG-Fab)
constructs and FFIFF (Fab-Fab-IgG-Fab-Fab) constructs were examined. FIF and
FFTFF
antibodies were nebulized using a PART eRapid vibrating mesh nebulizer system.
The
nebulized solutions were collected into a 50 mL conical tube, and the
stability of the
nebulized antibodies was assessed using SDS-PAGE, and the affinity of the
antibody to its
antigen assessed using whole-sperm ELISA assay.
[0002601 In some of the examples described herein, biophysical
characterization of the
synthetic binding agents (e.g., Fab-IgG, IgG-Fab, Fab-IgG-Fab, Fab-Fab-IgG,
IgG-Fab-Fab,
Fab-Fab-IgG-Fab, Fab-IgG-Fab-Fab, Fab-Fab-IgG-Fab-Fab) were done using SDS-
PAGE,
SEC-MALS and nano-DSF. SDS-PAGE experiments were performed using 4-12% NuPage
Bis-Tris gels (ThermoFisher Scientific) in 1X NuPage MOPS buffer under both
reducing
and non-reducing conditions to confirm the correct assembly of all HCA protein
constructs.
For each protein sample, 1 lig of protein was diluted in 3.75 p.L LDS sample
buffer followed
by the addition of 11.25 pL nuclease-free water. Proteins were then denatured
at 70 C for 10
min in a thermocycler. Next, 0.3 uL of 0.5 M TCEP was added as a reducing
agent to the
denatured protein for reduced samples and incubated at room temperature for 5
min. Bio-Rad
Precision Protein Plus Unstained Standard and NovexTM Sharp Pre-stained
Protein Standard
were used as ladders. After loading the samples, the gel was run for 50 min at
a constant
voltage of 200 V and washed 3 times with Milli-Q water. Then, the protein
bands were
visualized by staining with Imperial Protein Stain (ThermoFisher Scientific)
for 1hr followed
by overnight de-staining with Milli-Q water.
[000261] SEC-MALS experiments were performed at room temperature using a GE
Superdex 200 10/300 column connected to an Agilent FPLC system, a Wyatt DAWN
HELEOS II multi-angle light-scattering instrument, and a Wyatt T-rEX
refractometer. The
flow rate was maintained at 0.5 mLimin. The column was equilibrated with IX
PBS, pH 7.4
containing 200 mg/liter of NaN3 prior to sample loading. 50-100ug of each
sample was
injected onto the column, and data were collected for 50 min, The MALS data
were collected
and analyzed using Wyatt ASTRA software (Ver. 6).
74
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[9002621 NanoDSF experiments were performed using a Nanotemper Prometheus
NT.48
system. Samples were diluted to 0.5 mg/mL in 1X PBS at pH 7.4 and loaded into
Prometheus
NT.48 capillaries. Thermal denaturation experiments were perfatined from 25 C
to 95 C at
the rate of 1 Chnin, measuring the intrinsic tryptophan fluorescence at 330 nm
and at 350
nm. The melting temperature (Tm) for each experiment was calculated
automatically by
Nanotemper PR. Themicontrol software by plotting ratiometric measurement of
the
fluorescent signal against increasing temperature. The aggregation temperature
(Tagg) for
each experiment was also calculated automatically by Nanotemper PR.
Thermcontrol
software via the detection of the back-reflection intensity of a light beam
that passes the
sample.
10002631 The anti-sperm synthetic binding agents described herein were
examined to
determine sperm agglutination potency as well as muco-trapping potency. Fresh
semen was
examined. For example, male subjects were asked to refrain from sexual
activity for 24 hrs
prior to semen collection. Semen was collected by masturbation into sterile 50
mL sample
cups and incubated for a minimum of 15 min post-ejaculation at room
temperature to allow
liquefaction. Semen volume was measured, and density gradient sperm separation
procedure
was used to extract motile sperm from liquefied ejaculates. Briefly, 1.5 mL of
liquefied
semen was carefully layered over 1.5 mL of Isolate (90% density gradient
medium, Irvine
Scientific) at room temperature, and centrifuged at 300g for 20 min. Following
centrifugation, the upper layer containing dead cells and seminal plasma was
carefully
removed without disturbing the motile sperm pellet in the lower layer. The
sperm pellet was
then washed twice with sperm washing medium (Irvine Scientific) by
centrifugation at 300g
for 10 min. Finally, the purified motile sperm pellet was resuspended in sperm
washing
medium, and an aliquot was taken for determination of sperm count and motility
using
computer-assisted sperm analysis (CASA). All semen samples used in the
functional assays
exceeded lower reference limits for sperm count (15 x 106 spermatozoa per mL)
and total
motility (40%) as indicated by WHO guidelines. A Hamilton-Thorne computer-
assisted
sperm analyzer, 12.3 version, was used for the sperm count and motility
analysis in all
experiments unless stated otherwise This device consists of a phase-contrast
microscope
(Olympus CX41), a camera, an image digitizer and a computer with a Hamilton-
Thorne
Ceros 12.3 software to save and analyze the procured data. For each analysis,
4.4 .L of
semen sample was inserted into MicroTool counting chamber slides (Cytonix).
Then, six
randomly selected microscopic fields, near the center of the slide, were
imaged and analyzed
for progressive and non-progressive motile sperm count. The parameters that
were assessed
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
by CASA for motility analysis were as follows: average pathway velocity (VAP:
the average
velocity of the spermatozoa through a smoothed cell path in urn/sec), the
straight-line
velocity (VSL: the average velocity measured in a straight line from the
beginning to the end
of track in um/sec), the curvilinear velocity (VCL: the average velocity
measured over the
actual point-to-point track of the cell in gm/sec), the lateral head amplitude
(ALH: amplitude
of lateral head displacement in um), the beat cross-frequency (BCF: frequency
of sperm head
crossing the sperm average path in Hz), the straightness (STR: the average
value of the ratio
VSLNAP in %), and the linearity (UN: the average value of the ratio VSLNCL in
%),
Progressive motile sperm were defined as having a minimum of 25 lam/sec VAP
and 80% of
SIR.
10002641 Whole Sperm ELISA was also performed. Briefly, half-area polystyrene
plates
(CLS3690, Corning) were coated with 2.0x105 sperm per well in 50 iL of NaHCO3
buffer
(pH 9.6). After overnight incubation at 4 C, the plates were centrifuged at
the speed of 300g
for 20min. The supernatant was discarded, and the plates were air-dried for 1
hr at 45 C. The
plates were washed once with 1XPBS. 1004 of 5% milk was incubated at room
temperature for 1 hr to prevent non-specific binding of an antibody to the
microwells. The
serial dilution of monoclonal antibodies in 1% milk were added to the
microwells and
incubated overnight at 4 C. Motavizumab, a mAb against respiratory syncytial
virus, was
constructed and expressed in the laboratory by accessing the published
sequence and used as
a negative control for this assay. After primary incubation, the plates were
washed three times
using PBS. Then, the secondary antibody, goat anti-human IgG F(abt)2 antibody
HRP-
conjugated (1:50,000 dilutions in 1% milk, 209-1304, Rockland Inc.) was added
to the wells
and incubated for 1 hr at room temperature. The washing procedure was repeated
and 50 uL
of the buffer containing substrate (1-Step Ultra TMB ELISA Substrate,
ThermoFisher
Scientific) was added to develop the colorimetric reaction for 15min. The
reaction was
quenched using 50 uL of 2N H2504, and the absorbance at 450 nm (signal) and
570 nm
(background) was measured using SpectraMax M2 Microplate Reader (Molecular
Devices).
Each experiment was done with samples in triplicates and repeated at least
twice as a
measure assay variability.
[000265] Sperm escape assays were also performed with purified motile sperm
and whole
semen. Purified motile sperm were diluted in sperm washing medium to a final
concentration
of 10x 106 progressively motile sperm per mL. Next, 40 uL aliquots of diluted
sperm or
whole semen were transferred to individual 0.2 mL PCR tubes, and sperm count,
and motility
76
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
was again perfoitned on each 40 ttL aliquot using CASA. This count serves as
the original
(untreated) concentration of sperm for evaluating the agglutination potencies
of respective
HCA constructs. Following CASA, 304 of sperm were added to 30 tL of HCA
constructs,
and gently mixed by pipetting. The tubes were then fixed at 45 angles in a
custom 3D
printed tube holder for 5 min at room temperature. Following this incubation
period, 4.4 !IL
was extracted from the top layer of the mixture with minimal perturbation of
the tube and
transferred to the CASA instrument to quantify the number of progressively
motile sperm.
The percentage of the progressively motile sperm that escaped agglutination
was computed
by dividing the sperm count obtained after treatment with HCA constructs by
the original
spelni count in each respective tube, correcting for the 2-fold dilution with
antibody. Each
experimental condition was evaluated in duplicates on each semen specimen, and
the average
from the two experiments was used in the analysis. At least 5 independent
experiments were
done per assay, each using a single semen donor.
10002661 Agglutination kinetics assays with purified motile sperm and whole
semen were
also performed to characterize the sperm ("anti-sperm") synthetic binding
agent. Purified
motile sperm were diluted in spew' washing medium to a final concentration of
10x 106
progressively motile sperm per mL or 50x 106 progressively motile sperm per mL
or 2x 106
progressively motile sperm per mL. Next, 4,44 of diluted sperm or whole semen
was added
to 4.4 viL of HCA constructs in 0.2 mL PCR tubes, and mixed by gently
pipetting up and
down three times over 3 s. A timer was started immediately by a second person
while 4.4 jiL
of the mixture was transferred to chamber slides with a depth of 20 1il\/1
(Cytonix, Beltsville,
MD), and video microscopy (Olympus CKX41) using a 10x objective lens focused
on the
center of chamber slide was captured up to 90 s at 60 frames/s. Progressive
sperm count was
measured by CASA every 30 s up to 90 s. The percentage of the agglutinated
sperm at each
time point was computed by normalizing the progressive sperm count obtained
after
treatment with HCA constructs to the progressive sperm count obtained after
treatment with
sperm washing media. Each experimental condition was evaluated in duplicates
on each
semen specimen, and the average from the two experiments used in the analysis.
At least 6
independent experiments were done per assay, each using a single semen donor.
[0002671 Acidic pH stability of IgG- and FIF-Film via agglutination kinetics
assay were
performed using IgG-Film and FIF-Film constructs that were incubated in 0.5%
Lactic Acid
(LA) or sperm washing medium (MUM; control) for 24 hrs at 37 C. HCA constructs
incubated in LA were neutralized using equal volume of seminal plasma (SP).
Next,
77
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
neutralized HCA were diluted further using either SP or MI-IM media. Purified
motile sperm
were diluted in sperm washing medium to a final concentration of 20x 106
progressively
motile sperm per mL. Next, 4.4 !IL of diluted sperm was added to 4.44 of HCA
constructs
(HCA-LA/SP or HCA-LA/MHM or HCA-MHM/MHM) in 0.2 mL PCR tubes, and mixed by
gently pipetting up and down three times over 3 s. A timer was started
immediately by a
second person while 4.4 j.iL of the mixture was transferred to chamber slides
with a depth of
20 tiM (Cytonix, Beltsville, MD), and video microscopy (Olympus CKX41) using a
10x
objective lens focused on the center of chamber slide was captured up to 90 s
at 60 frames/s.
Progressive sperm count was measured by CASA every 30 s up to 90 s. The
percentage of the
agglutinated sperm at each time point was computed by normalizing the
progressive sperm
count obtained after treatment with HCA constructs to the progressive sperm
count obtained
after treatment with sperm washing media.
[0002681 In some cases sperm were fluorescently labeled. Purified motile sperm
were
fluorescently labelled using Live/Dead Sperm Viability Kit (Invitrogen
Molecular Probes),
which stains live sperm with SYBR 14 dye, a membrane-permeant nucleic acid
stain, and
dead sperm with propidium iodide, a membrane impermeant nucleic acid stain.
For labelling
of imL of washed semen, final concentration of 200 nM and 12 JAM were
respectively used
for SYBR 14 and Propidium Iodide dye. Once labelled, the sperm solution was
washed twice
to remove unbound fluorophores by centrifuging at 300g for 10 min using Sperm
Washing
Media (Irvine Scientific). Next, the labelled motile sperm pellet was
resuspended in sperm
washing medium, and an aliquot was taken for determination of sperm count and
motility
using CASA.
[0002691 In some cases cervicovaginal mucus (CVM) was used. Briefly, undiluted
CVM
secretions, averaging 0.5 g per sample, were obtained from women of
reproductive age,
ranging from 20 to 32 years old (27.4 0.9 years, mean SD), by using a self-
sampling
menstrual collection device (Instead Softcup). Participants inserted the
device into the vagina
for at least 30 s, removed it, and placed it into a 50 mL centrifuge tube.
Samples were
centrifuged at 230g for 5 min to collect the secretions. Aliquots of CVM for
lactic acid and
Ab measurements (diluted 1:5 with lx PBS and stored at ¨80 C) and slides for
gram staining
were prepared immediately, and the remainder of the sample was stored at 4 C
until
microscopy, typically within a few hours.
[0002701 Multiple particle tracking of fluorescently labelled sperm in CVM was
used to
mimic the dilution and neutralization of CVM by alkaline seminal fluid. CVM
was first
diluted three-fold using sperm washing media and titrated to pH 6.8-7.1 using
small volumes
78
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
(-3% v/v) of 3 N NaOH, The pH was confirmed using a micro pH electrode
(Microelectrodes, Inc., Bedford, NH) calibrated to pH 4, 7 and 10 buffers.
Next, 4 pL of
HCA constructs or control (anti-RSV IgG1) was added to 60 pL of diluted and pH
adjusted
CVM and mixed well in a CultureWellTm chamber slides (invitrogen #C37000,
ThermoFisher
Scientific) followed by addition of 4 'IL of 7.5x105 per mL of fluorescently
labelled sperm.
Once mixed, sperm, antibody and CVM were incubated for 15 min at room
temperature.
Then, translational motions of the sperm were recorded using an electron
multiplying
charge-coupled-device camera (Evolve 512; Photometrics, Tucson, AZ) mounted on
an
inverted epifluorescence microscope (Axio0bserver Dl; Zeiss, Thomwood, NY)
equipped
with an Alpha Plan-Apo 20/0.4 objective, an environmental (temperature and
CO2) control
chamber, and light-emitting diode (LED) light source (Lumencor Light Engine
DAPI/GFP/543/623/690). 6 videos (512 x 512 pixels, 16-bit image depth) were
captured for
each antibody condition with MetaMorph imaging software (Molecular Devices,
Sunnyvale,
CA) at a temporal resolution of 66.7 ms and spatial resolution of 50 nm
(nominal pixel
resolution, 0.78pm/pixel) for 20s. Microscopy videos obtained for this
trapping were run
through a Neural Net Tracker to determine the percentage of progressively
motile sperm after
incubation with different concentrations of HCA constructs.
f0002711 In some experiments, HCA constructs were instilled to the sheep's
vagina
followed by human semen and simulated intercourse (-30s) with a vaginal
dilator. Two
minutes later, the fluids from the sheep vagina were recovered and immediately
assessed for
progressive sperm motility. The condition with multimeric HCA constructs was
repeated two
more times in the same sheep with at least 7 days in between experiments. For
each
experiment, semen samples were pooled from 3-4 donors.
[000272] In one example described herein, native IgG sequences and synthetic
anti-RSV
binding agents based on the anti-RSV IgG were generated and characterized. The
codon-
optimized sequences of VH, CH!, VL and CL were utilized to design the
sequences for anti-
RSV IgG, Fab-IgG and IgG-Fab antibodies. The complete sequence of IgG, IgG-Fab
and
Fab-IgG were ordered using GeneArt Gene Synthesis. To minimize synthesis
problems that
could occur due to repeated sequences in IgG-Fab and Fab-IgG, the DNA
sequences for
repeated fragments were manually codon-optimized resulting in increased
variability of DNA
sequences and subsequent reduced complexity for gene synthesis. After manual
codon-
optimization, gene sequences were further processed through the complexity-
analyzing tool
provided by IDT to obtain a complexity score. Gene fragments with complexity
scores <25
had been easily and successfully synthesized via GeneArt Gene synthesis.
79
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[000273] Plasmids encoding native anti-RSV mAbs and anti-RSV synthetic binding
agents
were generated as described above, The variable heavy (VH) and variable light
(VL) DNA
sequences for anti-RSV antibodies were obtained from the publication of
Motavizumab. For
the construction of expression plasmid encoding the light chain, the gene
fragment consisting
of VL and CL (Ck) DNA sequences was synthesized using GeneArt gene synthesis
service
and cloned into the empty mammalian expression vector (pAH) using Kpni (5')
and EcoRI
(3') restriction sites. For the construction of expression plasmids encoding
heavy chains (BC)
for IgG, Fab-IgG and IgG-Fab, the complete gene sequences of all Abs were
synthesized
using GeneArtea gene synthesis service (ThermoFisher Scientific) and cloned
into empty
mammalian expression vector (pAH) sequences using Kpni (5') and MluI (3')
restriction
sites. For the ligation of all heavy chains as well as a light chain into the
expression vectors,
quick ligation kit (New England Biolabs, Ipswich, MA) was used. All ligated
DNA
constructs were transformed into chemically competent TOP 10 E. coil cells
(Life
Technologies) and plated on ampicillin plates for selection. Bacterial
colonies were picked,
cultured, and the plasmids were prepped (Qiagen MiniPrep Kit). Correct
assembly of the
constructs into the expression vector were confirmed by Sanger sequencing
(Eurofins
Genomics).
[000274] The expression plasmids encoding the heavy chain (HC) and light chain
(LC) for
IgG, Fab-IgG and IgG- antibodies were scaled up by transforming the sequencing-
confirmed
expression plasmids in chemically competent TOP10 E. coli, inoculating the
transformation
mix into 100 mL Luria broth in a 250 mL baffled flask and overnight shaking at
220 r.p.m at
37 C. Midi-prep plasmid purifications were done using NucleoBond Xtra Midi EF
Kits
(Macherey-Nagel) according to the manufacturer's protocols. Proteins were
expressed in
Expi293 cells using ExpiFectamineTM 293 Transfection reagents and protocols
provided by
the manufacturer (ThermoFisher Scientific). For IgG, one HC and one LC plasmid
were co-
transfected using a 1:1 ratio at 1 pig total DNA per 1 mL of culture. For both
Fab-IgG and
IgG-Fab, one HC and one LC plasmid were co-transfected using a 1:2 ratio at 1
pig total DNA
per 1 mL culture. Transfected cells were grown at 37 C in a 5% CO2 incubator
while shaking
at 125 r.p.m. for 5 days. Supernatants were harvested by centrifugation at
5000g for 10 min
and passed through 0.22 pm filters for purification using standard protein A
affinity
chromatography. Briefly, 30 mL of transfected supernatant was incubated with
400 pL
PBS-washed PierceTM Protein A Plus Agarose Resin (ThermoFisher Scientific)
overnight
at 4 C. Next, the resin-supernatant solution was flown through the gravity
columns
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
followed by the washing of resin. Protein was eluted by adding 9001.iL of
PierceTM IgG
Elution Buffer (ThermoFisher Scientific) into PBS-washed resin and was
immediately
neutralized by adding 100 [IL of UltraPureTM 1 M Tris-HCI Buffer, pH 7.5
(ThermoFisher
Scientific). Eluted proteins were further dialyzed into PBS using Amicon Ultra
Centrifugal Filters (Millipore Sigma).
1000275] SDS-PAGE experiments were performed using 4-12% NuPage Bis-Tris gels
(ThermoFisher Scientific) in IX NuPage MOPS buffer under both reducing and non-
reducing conditions to confinu the correct assembly of all anti-RSV antibody
constructs. For
each protein sample, I lig of protein was diluted in 3.75111, LDS sample
buffer followed by
the addition of 11.25 111, nuclease-free water. Proteins were then denatured
at 70 C for 10
min in a thermocycler. Next, 0.3 IAL of 0.5 M TCEP was added as a reducing
agent to the
denatured protein for reduced samples and incubated at room temperature for 5
mm. Bio-Rad
Precision Protein Plus Unstained Standard was used as protein ladder. After
loading the
samples, the gel was run for 50 min at a constant voltage of 200 V and washed
3 times with
Milli-Q water. Then, the protein bands were visualized by staining with
Imperial Protein
Stain (ThermoFisher Scientific) for ihr followed by overnight de-staining with
Milli-Q water.
[000276] Anti-RSV synthetic binding agents were examined using ELISA. Briefly,
half-
area polystyrene plates (CLS3690, Corning) were coated with 50 iitL of 10
ug/mL of human
RSV (ATCC VR-1540P) per well using NaHCO3 buffer (pH 9.6). After overnight
incubation at 4 C9 the plates were incubated under the UV for 1 hr to
inactivate the virus. The
plates were washed twice with IXPBS, 100 uL of 5% milk was incubated at room
temperature for I hr to prevent non-specific binding of an antibody to the
microwells. The
serial dilution of monoclonal antibodies in I% milk were added to the
microwells and
incubated overnight at 4 C. Palivizumab (Synagis), an FDA-approved mAb against
RSV,
was used as a positive control for this assay. After primary incubation, the
plates were
washed three times using IXPBS. Then, the secondary antibody, goat anti-human
IgG F(a1:)2
antibody HRP-conjugated (1:50,000 dilutions in I% milk, 209-1304, Rockland
Inc.) was
added to the wells and incubated for 1 hr at room temperature. The washing
procedure was
repeated and 50 III, of the buffer containing substrate (1-Step Ultra TMB
ELISA Substrate,
ThermoFisher Scientific) was added to develop the colorimetric reaction for
15min. The
reaction was quenched using 50 IALof 2N H2SO4, and the absorbance at 450 nm
(signal) and
570 nm (background) was measured using SpectraMax M2 Microplate Reader
(Molecular
81
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Devices). Each experiment was done with samples in triplicates and repeated at
least twice as
a measure assay variability.
[0002771 In general, the anti-RSV synthetic binding agents were synthetized as
described
herein to form Fab-IgG, IgG-Fab, Fab-IgG-Fab, Fab-Fab-IgG, Fab-Fab-IgG-Fab,
IgG-Fab-
Fab, Fab-IgG-Fab-Fab, and Fab-Fab-IgG-Fab-Fab. Both the synthesis and
characterization
of these anti-RSV synthetic binding agents were similar to those described for
the other
synthetic binding agents described, resulting in improved muco-trapping. In
particular, as
compared to IgG, the synthetic binding agents described herein had a superior
muco-trapping
and agglutination effect on the target. In particular, motile targets, such as
sperm, bacteria,
and other pathogens, may show an increase in mucosal tracking as compared to
native
antibodies. Preliminary evidence also suggests that the synthetic binding
agents described
herein may specifically inhibit growth of bacteria. Surface antigens on
targets (e.g., virus)
were used in all of the examples described herein.
[0002781 For example, FIG. 15A illustrates native (IgG, top, including
component Fab and
Fe regions), and F1 (Fab-IgG) and IF (IgG-Fab) synthetic binding agents
(bottom). For
example, the diagrams shown in FIG. 15A may be of anti-sperm IgG, Fab-IgG, and
IgG-Fab;
the N-terminal Fab of Fab-IgG and C-terminal Fab of IgG-Fab may contain a
fully intact
anti-speirn Fab with VH, VL, Cu, and CL, as described above. In FIG. 1513 a
non-reducing
gel shows that the synthesized Fab-IgG and IgG-Fab were produced as expected,
and were
expressed with high efficiency. FIG. 15C shows a reducing SDS-Page analysis of
the
indicated antibodies after expression in Expi293 cells and purification by
protein A/G
chromatography. FIG. 15D is a demonstration of the purity and homogeneity of
the purified
multimeric antibodies via analytical SEC-MALS analysis.
[0002791 FIGS. 16A-16C further characterize the synthetic binding agents
described herein.
In this example, anti-sperm synthetic binding agents were compared to anti-
sperm IgG
antibodies, As shown in FIG. 16A, the molar mass versus time of the IgG, Fab-
IgG and IgG-
Fab respectively as determined by SEC¨MALS showed that the synthetic binding
agents
described herein resembled the native mAb from which they originated. This was
also
apparent in FIG. 16B. The value of melting temperature (Tm) and aggregating
temperature
(Tagg) of as determined by nanoDSF by measuring intrinsic fluorescence of a
protein and
changes in back-reflection respectively were nearly the same. In FIG. 16C,
whole Sperm
ELISA analysis was used to assess the binding potency of indicated antibodies.
Motavizumab
(e.g., anti-RSV IgG1) was used as the isotype control. ELISA was performed in
triplicates
82
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
and repeated three times using 3 unique specimens. Lines indicate arithmetic
mean
concentration and standard error of mean.
[000280] FIGS. 17A-17B show the effect of anti-sperm synthetic binding agents
as
described herein (e.g., with additional FAB regions linked by flexible linkers
to either or both
the native variable (Fab) or constant ends). In FIG. 17A, the sperm
agglutination potency of
parent IgG, Fab-IgG and IgG-Fab was examined using purified motile sperm
(10x106
progressively motile sperm per mL). The sperm agglutination potency of the
parent and
multimerie anti-sperm IgGs (e.g., the synthetic binding agents) were measured
by quantifying
the percentage of sperm that remains progressively motile post Ab-treatment at
different
concentrations compared to pre-treatment condition. In FIG. 17B, the
percentage of
agglutination-escaped progressive sperm post-treatment was normalized to the
negative
control for further comparison. Data represents 6 unique sperm specimens.
Lines indicate
arithmetic mean concentration and standard error of mean. There was little
appreciable
difference between the Fab-IgG and the IgG-Fab synthetic binding agents,
however a
substantial improvement was seen over the native IgG in both the relative
extent of
agglutination (i.e. fraction agglutinated), and the minimum concentrations of
Fab-IgG or IgG-
Fab needed to agglutinate a comparable fraction of sperm.
[000281] FIGS. 18A-18B show the effect of sperm agglutination kinetics for
parent IgG,
Fab-IgG and IgG-Fab using purified motile sperm (10x106 progressively motile
sperm per
mL). In FIG. 18A, the agglutination kinetics of indicated antibodies was
assayed by
quantifying the time required to agglutinate 90% of progressively motile sperm
compared to
untreated control. The CASA analysis was obtained every 30s post-treatment
until 90s. In
FIG. 1811 the rate of sperm agglutination of indicated anti-speim antibodies
was measured by
quantifying the percentage of agglutinated sperm post Ab-treatment at three
different time
points compared to pre-treatment. Data represents 6 unique sperm specimens.
Lines indicate
arithmetic mean concentration and standard error of mean. Again, the synthetic
binding
agents has far superior sperm-agglutination potential compared to IgG alone,
particularly at
lower concentrations (e.g., less than about 2 ug/ml).
[000282] FIG. 19 shows the muco-trapping potency of parent IgG, Fab-IgG and
IgG-Fab
using pH-neutralized female CVM and purified motile sperm (1.5x106
progressively motile
sperm/mL). The muco-trapping potency of indicated antibodies was assessed by
perfoiming
real-time video microscopy on fluorescently labelled sperm suspended in Ab-
treated
(25ug/m1) CVM. We employed neural network tracker customized with standardized
sperm
83
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
motility parameters in all recorded videos to quantify the percentage of
progressively motile
sperm.
[0002831 In general there is further improvement with additional Fab groups,
up to 10 Fab
groups. The 10-mer (e.g., Fab-Fab-IgG-Fab-Fab, having a total of 10 Fabs)
performed
slightly better than 6 mer (Fab-IgG-Fab, Fab-Fab-IgG, IgG-Fab-Fab) or 8-mer
(Fab-Fab-IgG-
Fab, Fab-IgG-Fab-Fab) and 4 mer (Fab-IgG, IgG-Fab). FIGS. 20A-20D show the
characterization of multimeric anti-sperm IgG antibodies. FIG. 20A is a
schematic diagrams
of anti-sperm IgG, Fab-IgG-Fab (FIF), Fab-IgG-Fab-Fab (FIFF) and Fab-Fab-IgG-
Fab-Fab
(FFIFF). N-terminal and C-terminal Fabs of FIF, FIFF and FFIFF contains fully
intact anti-
sperm Fabs with VH, VL, CH1, and CL. FIGS. 20B and 20C show non-reducing and
reducing SDS-Page analysis, respectively, of the indicated antibodies after
expression in
Expi293 cells and purification by protein A/G chromatography. FIG. 20D shows a
demonstration of the purity and homogeneity of the purified multimeric
antibodies via
analytical SEC-MALS analysis.
[0002841 FIGS. 21A-21C also illustrate that the synthetic binding agents
(e.g., multimeric
anti-sperm IgG antibodies) described herein have comparable properties as
compared to each
other and to the native IgG. In FIG. 21A, molar mass versus time of the IgG,
FIF, FIFF and
FFIFF respectively is determined by SEC--MALS. FIG. 21B shows the value of
melting
temperature (Tm) and aggregating temperature (Tagg) of indicated antibodies as
determined
by nanoDSF by measuring intrinsic fluorescence of a protein and changes in
back-reflection
respectively. In FIG. 21C, the whole Sperm ELISA analysis is used to assess
the binding
potency of the indicated antibodies. Motavizumab (anti-RSV IgG1) is used as
the isotype
control. ELISA was performed in in triplicates and repeated three times using
3 unique
specimens. Lines indicate arithmetic mean concentration and standard error of
mean.
[0002851 As mentioned, the synthetic binding agents, including the high-Fab
number (e.g.,
8 mer, 10 mer) were highly effective at enhancing sperm agglutination. FIG.
22A-22B show
sperm agglutination potency of parent IgG and multimeric constructs using
purified motile
sperm (10x106 progressively motile sperm per mL) and whole semen. In FIG. 22A,
sperm
agglutination potency of the parent IgG, FIF, FIFF and FFIFF was measured by
quantifying
the percentage of sperm that remains progressively motile post Ab-treatment
compared to
pre-treatment condition using purified motile sperm (10x106 progressively
motile sperm per
mL). In FIG. 22B, the percentage of agglutination-escaped progressive sperm
post Ab-
treatment using purified motile sperm was normalized to the negative control
for further
comparison. As mentioned, the 10 mer (Fab-Fab-IgG-Fab-Fab) was slightly better
than the
84
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
lower Fab constructs, particularly at lower concentrations, and faster times
to agglutination.
In FIG. 22C, the sperm agglutination potency of the parent IgG and FFIFF was
measured by
quantifying the percentage of sperm that remains progressively motile post Ab-
treatment
compared to pre-treatment condition using whole semen. In FIG. 22D, the
percentage of
agglutination-escaped progressive sperm post Ab-treatment using whole semen
was
normalized to the negative control for further comparison. Data represents 6
unique sperm
specimens. Lines indicate arithmetic mean concentration and standard error of
mean.
10002861 Sperm agglutination kinetics of parent IgG and multimeric constructs
using
purified motile spent (10x1 06 progressively motile sperm per mL) and whole
semen is also
illustrated in FIGS. 23A-23B. In FIG. 23A, the agglutination kinetics of
parent IgG, FIF,
FIFF and FFIFF was examined by quantifying the time required to achieve 90%
agglutination
of progressive speim compared to untreated control using purified motile sperm
(10x106
progressively motile sperm per mL). The CASA analysis was obtained every 30s
post-
treatment until 90s. In FIG, 23B the rate of sperm agglutination of parent
IgG, FIF, FIFF and
FFIFF was measured by quantifying the percentage of agglutinated sperm. post
Ab-treatment
at three different time points compared to pre-treatment using purified motile
sperm (10x106
progressively motile sperm per mL). The agglutination kinetics of parent IgG
and FFIFF was
assessed (in FIG. 23C) by quantifying the time required to achieve 90%
agglutination of
progressive sperm compared to untreated control using whole semen. The CASA
analysis
was obtained every 30s post-treatment until 90s. In FIG. 23D, the rate of
sperm agglutination
of parent IgG and FFIFF was measured by quantifying the percentage of
agglutinated sperm
post Ab-treatment at three different time points compared to pre-treatment
using whole
semen, Data represents 6 unique sperm specimens. Lines indicate arithmetic
mean
concentration and standard error of mean.
[000287] Sperm agglutination kinetics of parent IgG and FFIFF was examined
using low
and high concentration of purified motile sperm (2x106 and 50x106 progressive
sperm/mL),
as shown in FIGS. 24A-24B. In FIG, 24A, the agglutination kinetics of IgG and
FFIFF was
examined by quantifying the time required to achieve 90% agglutination of
progressive
sperm compared to untreated control. The CASA analysis was obtained every 30s
post-
treatment until 90s using purified motile sperm (2x106 progressive sperm/mL).
The rate of
sperm agglutination of IgG and FFIFF was measured by quantifying the
percentage of
agglutinated spenu post Ab-treatment at three different time points compared
to pre-
treatment using purified motile sperm (2x106 progressive sperm/mL), as shown
in FIG. 24B.
The agglutination kinetics of IgG and FFIFF were examined in FIG. 24C by
quantifying the
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
time required to achieve 90% agglutination of progressive sperm compared to
untreated
control using purified motile sperm (50x106 progressive sperm/mL). The CASA
analysis
was obtained every 30s post-treatment until 90s. In FIG. 24D, the rate of
sperm agglutination
of IgG and FFIFF was measured by quantifying the percentage of agglutinated
sperm post
Ab-treatment at three different time points compared to pre-treatment using
purified motile
sperm (50x106 progressive sperm/mL). Data represents 6 unique sperm specimens.
Lines
indicate arithmetic mean concentration and standard error of mean.
[000288] FIG. 25 shows the sperm agglutination potency of parent IgG, FIF and
FFIFF
using whole semen in a sheep study. The agglutination potency of IgG, FIF and
FFIFF was
measured in viva by instilling Abs into sheep vagina, followed by human semen
and
simulated intercourse. Sperm motility was examined immediately in the fluids
from the sheep
vagina. Data represents 3 unique sheep studies for FIF and FFIFF, and 1 sheep
study for IgG
at both 33 ug/ml and 333 ug/ml. Lines indicate arithmetic mean concentration
and standard
error of mean.
[000289] As mentioned above, films may also be used to deliver the synthetic
binding
agents described herein. For example, FIG. 26 and 27A-27C illustrate
agglutination potency
of a Nicotiana-produced film of parent IgG and FIF using purified motile sperm
(10x106
progressive sperm/mL) and whole semen. FIG. 26 shows measured sperm
agglutination
potency of the parent IgG-Film and FIF-Film by quantifying the percentage of
sperm that
remains progressively motile post Ab-treatment compared to pre-treatment
condition using
purified motile sperm (10x106 progressively motile sperm per mL). FIG. 27A
shows the
percentage of agglutination-escaped progressive sperm post Ab-treatment using
purified
motile sperm was normalized to the negative control for further comparison.
The sperm
agglutination potency of the parent IgG-Film and FIF-Film was measured (in
FIG. 27B) by
quantifying the percentage of sperm that remains progressively motile post Ab-
treatment
compared to pre-treatment condition using whole semen. In FIG. 27C, the
percentage of
agglutination-escaped progressive sperm post Ab-treatment using whole semen
was
normalized to the negative control for further comparison. Data represents 6
unique spent'
specimens. Lines indicate arithmetic mean concentration and standard error of
mean.
[000290] FIGS. 28A-28B illustrate agglutination kinetics of a Nieotiana-
produced films of
parent IgG and FIF using purified motile sperm (10x106 progressively motile
sperm/mL) and
whole semen. FIG. 28A shows the agglutination kinetics of indicated antibodies
by
quantifying the time required to achieve 90% agglutination of progressive
sperm compared to
untreated control using purified motile sperm (10x106 progressively motile
sperin/mL). The
86
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
CASA analysis was obtained every 30s post-treatment until 90s. The rate of
sperm
agglutination of indicated anti-sperm antibodies was assessed (in FIG. 288) by
quantifying
the percentage of agglutinated sperm post Ab-treatment at three different time
points
compared to pre-treatment using purified motile sperm (10x106 progressively
motile
sperm/mL).
[0002911 Agglutination kinetics of the Nicotiana-produced films of parent IgG
and FIF was
assessed using purified motile sperm (10x106 progressively motile sperm/mL)
and whole
semen, as shown in FIGS. 28C and 28D. The agglutination kinetics of indicated
antibodies
was assessed by quantifying the time required to achieve 90% agglutination of
progressive
sperm compared to untreated control using whole semen. The CASA analysis was
obtained
every 30s post-treatment until 90s. The rate of sperm agglutination of
indicated anti-sperm
antibodies was measured by quantifying the percentage of agglutinated sperm
post Ab-
treatment at three different time points compared to pre-treatment using whole
semen. Data
represents 6 unique sperm specimens. Lines indicate arithmetic mean
concentration and
standard error of mean.
[000292] FIGS. 29A-28B illustrate agglutination kinetics of the Nicotiana-
produced films
of parent IgG and FIF using low and high concentration of purified motile
sperm (2x106 and
50x106 progressively motile sperm/mL). The agglutination kinetics of indicated
antibodies
was assessed by quantifying the time required to achieve 90% agglutination of
progressive
sperm compared to untreated control using purified motile sperm (2x106
progressively motile
sperm/mL). The CASA analysis was obtained every 30s post-treatment until 90s.
We also
measured the rate of sperm agglutination of indicated anti-sperm antibodies by
quantifying
the percentage of agglutinated sperm post Ab-treatment at three different time
points
compared to pre-treatment using purified motile sperm (2x106 progressively
motile
spermimL) (FIG. 29B). The agglutination kinetics of indicated antibodies was
assessed by
quantifying the time required to achieve 90% agglutination of progressive
sperm compared to
untreated control using purified motile sperm (50x106 progressively motile
sperm/mL) as
shown in FIG. 29C. The CASA analysis was obtained every 30s post-treatment
until 90s. We
also measured the rate of sperm agglutination of indicated anti-sperm
antibodies by
quantifying the percentage of agglutinated sperm post Ab-treatment at three
different time
points compared to pre-treatment using purified motile sperm (50x106
progressively motile
sperm/mL) (FIG. 29D). Data represents 6 unique sperm specimens. Lines indicate
arithmetic
mean concentration and standard error of mean.
87
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
[0002931 FIG. 30 shows agglutination kinetics of the Nicotiana-produced films
of parent
IgG and FIF in the acidic environment using purified motile sperm (20x106
progressively
motile spermimL) and 24hr treatment with lactic acid. The agglutination
kinetics of lactic
acid-treated antibodies was assessed by quantifying the time required to
achieve 90%
agglutination of progressive sperm compared to untreated control. The CASA
analysis was
obtained every 30s post-treatment until 90s. Data represents 2 unique sperm
specimens. Note:
Lactic acid (LA) treated antibodies were neutralized with seminal plasma (SP)
and then
followed by dilution with SP or saline media i.e. MHM.
[000294] FIGS, 31A-31D show the characterization of Nicotiana-produced FIF-
Film and
Expi293-produced FFIFF antibodies post-nebulization. Non-reducing (FIG. 31A)
and
reducing (FIG. 31B) SDS-Page analysis of the indicated antibodies before and
after
nebulization using mesh-nebulizer. Whole Sperm ELISA analysis to assess the
binding
potency of (FIG. 31C) FIF-Film and (FIG. 31D) FFIFF post-nebulization. ELISA
was
performed in triplicates and repeated twice using the same donor specimens.
Lines indicate
arithmetic mean concentration and standard error of mean. Thus, nebulizing the
synthetic
binding agents described herein did not damage or disrupt them, and they may
be effectively
delivered by nebulization (e.g., by aerosolization).
[000295] FIGS. 32A-32B illustrate the production and characterization of
multimeric anti-
RSV IgG antibodies. FIG. 32A shows non-reducing and reducing SDS-Page
analysis,
respectively, of ant-RSV synthetic binding agents (e.g., formed from a
Motavizumab parent
IgG) after expression in Expi293 cells and purification by protein A/G.
chromatography. FIG.
32B shows RSV ELISA analysis to assess the binding potency of the indicated
antibodies.
Synagis/Palivizumab (anti-RSV IgG1) was used as the positive control. ELISA
was
performed in triplicates.
[000296] As compared to control, the anti-RSV synthetic binding agents (in
this case Fab-
IgG and IgG-Fab, however Fab-Fab-IgG-Fab-Fab and Fab-IgG-Fab may have similar
results
as described above), were superior to native IgG in binding the same target,
which will result
in substantially greater muco-trapping potency and therefore therapeutic
efficacy, particularly
at lower concentrations and faster times.
10002971 As mentioned above, in some variations a synthetic binding agent may
be made
with multiple scFvs or nanobodies in the same manner as described with Fabs.
Further, in
some variations multimeric antibodies similar to those described herein may be
made without
Fe regions.
88
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SEQUENCE LISTING
SEQ ID NO: I
Amino Acid sequence of CD52g
GQNDTSQTSSPS
SEQ ID NO: 2
Annotated DNA sequence of HCA-UNC (anti-CD52g) (Heavy Chain)
CAGGTTCAGCTGCAGCAATGGGGAGCCGGACTGCTGAAGCCTAGCGAGACACTGTCTCTGAC
CTGTGCCGTGTACGGCGGCAGOTTCAGOGGCTACTACTGGTCCTGGATCAGACAGCCTCCTG
GCAAAGGCCTGGAATGGATCGGCGAGATCAATCACAGCGGCAGCACCAACTACAACCCCAGC
CTGAGAAGCAGAGTGACCATCAGCGTGGACACCAGCAAGAACCAGTTCAGCCTGAAGCTGAG
AAGCGTGACAGCCGCCGATACCGCCGTGTACTATTGCGCCAGAGGCTTTATGGTCCGAGGCA
TCATGTGGAACTACTACTACATGGACGTGTGGGGCAAGGGCACCACCGTGACAGTTTCTCCA
GCTAGCACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGG
CACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGA
ACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTOCTACAGTCCTCAGGACTC
TACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTG
CAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTG
ACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTC
CTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGT
GGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGG
AGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTC
AGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTC
CAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAG
AACCACAGGTGTACACCCTGCCCCCATCCCGGGACGAGCTGACCAAGAACCAGGTCAGCCTG
ACCTGCCTGGTCAAAGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCA
GCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCT
ACAGCAAGCTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTOTTCTCATGCTCCGTG
ATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAA
SEQ ID NO: 3
Annotated Amino Acid sequence of HCA-UNC (anti-CD52g)(Heavy
Chain)
QVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRUPGKGLEWIGEINHSGSTNYNE'S
LRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYYMDVWGKGTTVTVSP
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVF
LFPPKPMDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVV
SVLTVLHOWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLIDEDSRDELTKNQVSL
TCLVKGFYPSDIAVEWESNGUENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSV
MHEALHNHYTQKSLSLSPGK
SEQ ID NO: 4
89
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Annotated Amino Acid sequence of Fab fragment of HCA-UNC
(anti-CD52g) (Heavy Chain)
QVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRQPPGKGLEWIGEINHSGSTNYNPS
LRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGEMVRGIMWNYYYMDVWGKGTTVTVSP
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 5
Annotated Amino Acid sequence of Fc fragment of HCA-UNC (anti-
CD52g) (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKOTLMISRTPEVTCVVVDVSHEDPEVKFNWYVOGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHOWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGUENNYKTTPPVLDSIDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 6
DNA sequence of HCA-UNC (anti-CD52g) (Light Chain)
AGGAGCGAGCTGACACAGGATCCAGTGGTGTCTGTGGCCCTGGGCCAGACAGTGCGGATTAC
TTGTCAGGGCGACAGCCTGAGAACCTACCACGCCTCTTGGTATCAGCAGAAGCCCAGACAGG
CCCCTGTGCTGGTCATCTACGACGAGAACAACAGACCCAGCGGCATCCCCGATAGATTCAGC
GGCAGCACATCTGGCAATACCGCCAGCCTGACAATCACTGGCGCCCAGGCTGAAGATGAGGC
CGACTACTACTGCAACAGCAGAGACAGCAGCGGCAACCGGCTGGTTTTTGGCGGAGGCACAA
AGCTGACAGTGCTGGGCCAACCTAAGGCCGCTCCTAGCGTGACACTGTTCCCTCCAAGCAGC
GAAGAACTGCAGGCCAACAAGGCCACACTCGTGTGCCTGATCAGCGACTTTTATCCTGGCGC
CGTGACCGTGGCCTGGAAGGCTGATAGTTCTCCTGTGAAGGCCGGCGTGGAAACCACCACAC
CTAGCAAGCAGAGCAACAACAAATACGCCGCCAGCAGCTACCTGAGCCTGACACCTGAGCAG
TGGAAGTCCCACAGATCCTACAGCTGCCAAGTGACCCACGAGGGCAGCACCGTGGAAAAAAC
AGTGGCCCCTACCGAGTGCAGC
SEQ ID NO: 7
Annotated Amino Acid sequence of HCA-UNC (anti-CD52g) (Light
Chain)
SSELTQDPVVSVALGQTVRITCQGDSLRTYHASWYQQKPRQAPVLVIYDENNRPSGIPDRES
GSTSGNTASLTITGAQAEDEADYYCNSRDSSGNRLVEGGGTKLTVLGQPKAAPSVTLFPPSS
EELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKONNKYAASSYLSLTPEQ
WKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 8
Annotated DNA sequence of Synthetic anti-CD52g Fab-IgG (Heavy
Chain)
CAGGTTCAGCTGCAGCAATGGGGAGCCGGACTGCTGAAGCCTAGCGAGACACTGTCTCTGAC
CTGTGCCGTGTACGGCGGCAGCTTCAGCGGCTACTACTGGTCCTGGATCAGACAGCCTCCTG
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
GCAAAGGCCTGGAATGGATCGGCGAGATCAATCACAGCGGCAGCACCAACTACAACCCCAGC
CTGAGAAGCAGAGTGACCATCAGCGTGGACACCAGCAAGAACCAGTTCAGCCTGAAGCTGAG
AAGCGTGACAGCCGCCGATACCGCCGTGTACTATTGCGCCAGAGGCTTTATGGTCCGAGGCA
TCATGTGGAACTACTACTACATGGACGTGTGGGGCAAGGGCACCACCGTGACAGTTTCTCCA
GCCTCTACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGG
CACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGA
ACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTC
TACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGOTTGGGCACCCAGACCTACATCTG
CAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTG
ACAAAACTCACACAGGTGGCGGAGGATCTGGCGGAGGGGGGAGCGGAGGGGGTGGGTCCGGC
GGGGGGGGTTCCGGGGGTGGTGGATCAGGCGGCGGAGGAAGTCAGGTTCAGCTGCAGCAATG
GGGAGCCGGACTGCTGAAGCCTAGCGAGACACTGTCTCTGACCTGTGCCGTGTACGGCGGCA
GCTTCAGCGGCTACTACTGGTCCTGGATCAGACAGCCTCCTGGCAAAGGCCTGGAATGGATC
GGCGAGATCAATCACAGCGGCAGCACCAACTACAACCCCAGCCTGAGAAGCAGAGTGACCAT
CAGCGTGGACACCAGCAAGAACCAGTTCAGCCTGAAGCTGAGAAGCGTGACAGCCGCCGATA
CCGCCGTGTACTATTGCGCCAGAGGCTTTATGGTCCGAGGCATCATGTGGAACTACTACTAC
ATGGACGTGTGGGGCAAGGGCACCACCGTGACAGTTTCTCCAGCTAGCACCAAGGGCCCATC
GGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCC
TGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGC
GGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGT
GACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCA
GCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACACATGCCCA
CCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAA
GGACACCCTCATGATCTCCCGGACCOCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACG
AAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAATGCCAAGACA
AAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCA
CCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCAGCCC
CCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCTG
CCCCCATCCCGGGACGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAAAGGCTT
CTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGA
CCACGCCTOCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCRAGCTCACCGTGGAC
AAGAGCAGGTGGCAGCAGGGGAACGTOTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAA
CCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAA
SEQ ID NO: 9
Annotated Amino Acid sequence of Synthetic anti-CD52q Fab-IqG
(Heavy Chain)
QVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRUPGKGLEWIGEINHSGSTNYNPS
LRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGEMVRGIMWNYYYMDVWGKGTTVTVSP
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGSG
GGGSGGGGSGGGGSQVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRQPPGKGLEWI
GEINHSGSTNYNPSLRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYY
MDVWGKGTTVTVSPASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTS
GVHTFPAVLOSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCP
PCPAPELLGGPSVFLFETKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVIDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGUENNYKTTFPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
91
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SEQ ID NO: 10
Annotated Amino Acid sequence of Synthetic anti-CD52g Fab
fragment of Fab-IgG (Heavy Chain)
QVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRUPGKGLEWIGEINHSGSTNYNPS
LRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYYMDVWGKGTTVTVSP
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLOSCL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 11
Annotated Amino Acid sequence of Synthetic anti-CD52g Fe
fragment of Fab-IgG (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPTEKTISKAKGQPREPTFYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 12
DNA sequence of Synthetic anti-CD52g Fab-IgG (Light Chain)
AGCAGCGAGCTGACACAGGATCCAGTGGTGTCTGTGGCCCTGGGCCAGACAGTGCGGATTAC
TTGTCAGGGCGACAGCCTGAGAACCTACCACGCCTCTTGGTATCAGCAGAAGCCCAGACAGG
CCCCTGTGCTGGTCATCTACGACGAGAACAACAGACCCAGOGGCATCCCCGATAGATTCAGC
GGCAGCACATCTGCCAATACCGCCAGCCTGACAATCACTGGCGCCCAGGCTGAAGATGAGGC
CGACTACTACTGCAACAGCAGAGACAGCAGOGGCAACCGGCTGGTTTTTGGCGGAGGCACAA
AGCTGACAGTGCTGGGCCAACCTAAGGCCGCTCCTAGCGTGACACTGTTCCCTCCAAGCAGC
GAAGAACTGCAGGCCAACAAGGCCACACTCGTGTGCCTGATCAGCGACTTTTATCCTGGCGC
CGTGACCGTGGCCTGGAAGGCTGATAGTTCTCCTGTGAAGGCCGGCGTGGAAACCACCACAC
CTAGCAAGCAGAGCAACAACAAATACGCCGCCAGCAGCTACCTGAGCCTGACACCTGAGCAG
TGGAAGTCCCACAGATCCTACAGCTGCCAAGTGACCCACGAGGGCAGCACCGTGGAAAAAAC
AGTGGCCCCTACCGAGTGCAGC
SEQ ID NO: 13
Annotated Amino Acid sequence of Synthetic anti-CD52g Fab-IgG
(Light Chain)
SSELTOPVVSVALGQTVRITCQGDSLRTYHASWYQQKPRQAPVLVIYDENNRTSGIPDRFS
GSTSGNTASLTITGAQAEDEADYYCNSRDSSGNRLVFGGGTKLTVLGQPKAAPSVTLFPPSS
EELQANKATINCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKUNNKYAASSYLSLTPEQ
WKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 14
Annotated DNA sequence of Synthetic anti-CD52g IgG-Fab (Heavy
Chain)
92
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
CAGGTTCAGCTGCAGCAATGGGGAGCCGGACTGCTGAAGCCTAGCGAGACACTGTCTCTGAC
CTGTGCCGTGTACGGCGGCAGCTTCAGOGGCTACTACTGGTCCTGGATCAGACAGCCTCCTG
GCAAAGGCCTGGAATGGATCGGCGAGATCAATCACAGCGGCAGCACCAACTACAACCCCAGC
CTGAGAAGCAGAGTGACCATCAGCGTGGACACCAGCAAGAACCAGTTCAGCCTGAAGCTGAG
AAGCGTGACAGCCGCCGATACCGCCGTGTACTATTGCGCCAGAGGCTTTATGGTCCGAGGCA
TCATGTGGAACTACTACTACATGGACGTGTGGGGCAAGGGCACCACCGTGACAGTTTCTCCA
GCTAGCACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGG
CACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGA
ACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTC
TACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTG
CAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTG
ACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTC
CTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGT
GGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGG
AGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTC
AGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTC
CAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAG
AACCACAGGTGTACACCCTGCCCCCATCCCGGGACGAGCTGACCAAGAACCAGGTCAGCCTG
ACCTGCCTGGTCAAAGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCA
GCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTOTTCCTCT
ACAGCAAGCTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTG
ATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAAGG
ATCCGGTGGCGGAGGATCTGGCGGAGGGGGGAGCGGAGGGGGTGGGTCCGGCGGGGGGGGTT
CCGGGGGTGGTGGATCAGGCGGCGGAGGAAGTCAGGTTCAGCTGCAGCAATGGGGAGCCGGA
CTGCTGAAGCCTAGCGAGACACTGTCTCTGACCTGTGCCGTGTACGGCGGCAGCTTCAGOGG
CTACTACTGGTCCTGGATCAGACAGCCTCCTGGCAAAGGCCTGGAATGGATCGGCGAGATCA
ATCACAGCGGCAGCACCAACTACAACCCCAGCCTGAGAAGGAGAGTGACCATCAGCGTGGAC
ACCAGCAAGAACCAGTTCAGCCTGAAGCTGAGAAGCGTGACAGCCGCCGATACCGCCGTGTA
CTATTGCGCCAGAGGCTTTATGGTCCGAGGCATCATGTGGAACTACTACTACATGGACGTGT
GGGGCAAGGGCACCACCGTGACAGTTTCTCCAGCCTCTACCAAGGGCCCATCGGTOTTCCCC
CTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGA
CTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACA
CCTTCCOGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCC
TCCAGCAGOTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAA
GGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACACA
SEQ ID NO: 15
Annotated Amino Acid sequence of Synthetic anti-CD52g IgG-Fab
(Heavy Chain)
OVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWTRUPGKGLEWIGEINHSGSTNYNPS
LRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYYMDVWGKGTTVTVSP
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVF
LFPPKPKDTLMISRTPEVTCVVVINSHEDPEVKFNWYVDGVEVANAKTKPREEONSTYRVV
SVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSL
TUNKGFYPSDIAVEWESNGUENNYKTTETVLDSIDGSFFLYSKLTVDKSRWQQGNVFSCSV
MHEALHNHYTQKSLSLSPGKGSGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSQVQLQQWGAG
LLKPSETLSLTCAVYGGSFSGYYWSWIRUPGKGLEWIGEINESGSTNYNPSLRSRVTISVD
TSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYYMDVWGKGTTVTVSPASTKGPSVFP
93
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
LAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVP
SSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 16
Annotated Amino Amid sequence of Synthetic anti-CD52g Fab
fragment of IgG-Fab (Heavy Chain)
QVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRUPGKGLEWIGEINHSGSTNYNPS
LRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGEMVRGIMWNYYYMDVWGKGTTVTVSP
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLOSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 17
Annotated Amino Acid sequence of Synthetic anti-CD52g Fc
fragment of IgG-Fab (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGUREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 18
DNA sequence of Synthetic anti-CD52g IgG-Fab (Light Chain)
AGCAGCGAGCTGACACAGGATCCAGTGGTGTCTGTGGCCCTGGGCCAGACAGTGCGGATTAC
TTGTCAGGGCGACAGCCTGAGAACCTACCACGCCTCTTGGTATCAGCAGAAGCCCAGACAGG
CCCCTGTGCTGGTCATCTACGACGAGAACAACAGACCCAGCGGCATCCCCGATAGATTCAGC
GGCAGCACATCTGGCAATACCGCCAGCCTGACAATCACTGGCGCCCAGGCTGAAGATGAGGC
CGACTACTACTGCAACAGGAGAGACAGCAGCGGCAACCGGCTGGTTTTTGGCGGAGGCACAA
AGCTGACAGTGCTGGGCCAACCTAAGGCCGCTCCTAGCGTGACACTGTTCCCTCCAAGGAGC
GAAGAACTGCAGGCCAACAAGGCCACACTCGTGTGCCTGATCAGCGACTTTTATCCTGGCGC
CGTGACCGTGGCCTGGAAGGCTGATAGTTCTCCTGTGAAGGCCGGCGTGGAAACCACCACAC
CTAGCAAGCAGAGCAACAACAAATACGCCGCCAGGAGCTACCTGAGCCTGACACCTGAGCAG
TGGAAGTCCCACAGATCCTACAGCTGCCAAGTGACCCACGAGGGCAGCACCGTGGAAAAAAC
AGTGGCCCCTACCGAGTGCAGC
SEQ ID NO: 19
Annotated Amino Acid sequence of Synthetic anti-CD52g IgG-Fab
(Light Chain)
SSELTQDPVVSVALGQTVRITCQGDSLRTYHASWYQQKPRQAPVLVIYDENNRPSGIPDRFS
GSTSGNTASLTITGAQAEDEADYYCNSRDSSGNRLVFGGGTKLTVLGQPKAAPSVTLFPPSS
EELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPEQ
WKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 20
94
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Annotated DNA sequence of Synthetic anti -CD52g Fab-IgG-Fab
(Heavy Chain)
CAGGTTCAGCTOCAGCAATGGGGAGCCGGACTGCTGAAGCCTAGCGAGACACTGTCTCTGAC
CTGTGCCGTGTACGGCGGCAGCTTCAGCGGCTACTACTGGTCCTGGATCAGACAGCCTCCTG
GCAAAGGCCTGGAATGGATCGGCGAGATCAATCACAGOGGCAGCACCAACTACAACCCCAGC
CTGAGAAGGAGAGTGACCATCAGCGTGGACACCAGCAAGAACCAGTTCAGCCTGAAGCTGAG
AAGCGTGACAGCCGCCGATACCGCCGTGTACTATTGCGCCAGAGGCTTTATGGTCCGAGGCA
TCATGTGGAACTACTACTACATGGACGTGTGGGGCAAGGGCACCACCGTGACAGTTTCTCCA
GCCTCTACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGG
CACAGOGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGA
ACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTC
TACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTG
CAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTG
ACAAAACTCACACAGGTGGCGGAGGATCTGGCGGAGGGGGGAGCGGAGGGGGTGGGTCCGGC
GGGGGGGGTTCCGGGGGTGGTGGATCAGGCGGCGGAGGAAGTCAGGTTCAGCTGCAGCAATG
GGGAGCCGGACTGCTGAAGCCTAGCGAGACACTGTCTCTGACCTGTGCCGTGTACGGCGGCA
GCTTCAGCGGCTACTACTGGTCCTGGATCAGACAGCCTCCTGGCAAAGGCCTGGAATGGATC
GGCGAGATCAATCACAGOGGCAGCACCAACTACAACCCCAGCCTGAGAAGCAGAGTGACCAT
CAGCGTGGACACCAGCAAGAACCAGTTCAGCCTGAAGCTGAGAAGCGTGACAGCCGCCGATA
CCGCCGTGTACTATTGCGCCAGAGGCTTTATGGTCCGAGGCATCATGTGGAACTACTACTAC
ATGGACGTGTGGGGCAAGGGCACCACCGTGACAGTTTCTCCAGCTAGCACCAAGGGCCCATC
GGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCC
TGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGC
GGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGT
GACCGTGCCCTCCAGCAGOTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCA
GCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACACATGCCCA
CCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCOCCAAAACCCAA
GGACACCCTCATGATCTCCOGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACG
AAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAATGCCAAGACA
AAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCA
CCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTOCCAGCCC
CCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCTG
CCCCCATCCOGGGACGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAAAGGCTT
CTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGA
CCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGAC
AAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAA
CCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAAGGATCCGGTGGCGGAGGATCTG
GOGGAGGGGGGAGCGGAGGGGGTGGGTCCGGCGGGGGGGGTTCCGGGGGTGGTGGATCAGGC
GGCGGAGGAAGTCAGGTTCAGCTGCAGCAATGGGGAGCCGGACTGCTGAAGCCTAGCGAGAC
ACTGTCTCTGACCTGTGCCGTGTACGGCGGCAGCTTCAGCGGCTACTACTGGTCCTGGATCA
GACAGCCTCCTGGCAAAGGCCTGGAATGGATCGGCGAGATCAATCACAGCGGCAGCACCAAC
TACAACCCCAGCCTGAGAAGCAGAGTGACCATCAGCGTGGACACCAGCAAGAACCAGTTCAG
CCTGAAGCTGAGAAGCGTGACAGCCGCCGATACCGCCGTGTACTATTGCGCCAGAGGCTTTA
TGGTCCGAGGCATCATGTGGAACTACTACTACATGGACGTGTGGGGCAAGGGCACCACCGTG
ACAGTTTCTCCAGCCTCTACCAAGGGCCCATCGGTCTTCCCOCTGGCACCCTCCTCCAAGAG
CACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGA
CGGTGTCGTGGAACTCAGGCGCCCTGACCAGOGGCGTGCACACCTTCCCGGCTGTCCTACAG
TCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCA
GACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGC
CCAAATCTTGTGACAAAACTCACACA
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SEQ ID NO: 21
Annotated Amino Acid sequence of Synthetic anti-CD52g Fab-IgG-
Fab (Heavy Chain)
QVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRUPGKGLEWIGEINHSGSTNYNPS
LRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYYMDVWCKGTTVTVSP
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLOSGL
YSLSSVVTVPSSSLGTQTYICNVNEKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGSG
GGGSGGGGSGGGGSQVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRQPPGKGLEWI
GEINHSGSTNYNPSLRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYY
MDVWGKGTTVTVSPASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTS
GVHTFPAVLUSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCP
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVIDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTIPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKGSGGGGSGGGGSGGGGSGGGGSGGGGSG
GGGSQVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRUPGKGLEWIGEINHSGSTN
YNPSLRSRVTISVDTSKNQFSLKLRSVIAADTAVYYCARGFMVRGIMWNYYYMDVWGKGTTV
TVSPASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQ
SSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 22
Annotated Amino Acid sequence of Synthetic anti-CD52g Fab
fragment of Fab-IgG-Fab (Heavy Chain)
QVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRQPPGKGLEWIGEINHSGSTNYNPS
LRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYYMDVWGKGTIVTVSP
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 23
Annotated Amino Acid sequence of Synthetic anti-CD52g Fc
fragment of Fab-IgG-Fab (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFELYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 24
DNA sequence of Synthetic anti-CD52g Fab-IgG-Fab (Light Chain)
AGCAGCGAGCTGACACAGGATCCAGTGGTGTCTGTGGCCCTGGGCCAGACAGTGCGGATTAC
TTGTCAGGGCGACAGCCTGAGAACCTACCACGCCTCTTGGTATCAGCAGAAGCCCAGACAGG
CCCCTGTGCTGGTCATCTACGACGAGAACAACAGACCCAGCGGCATCCCCGATAGATTCAGC
GGCAGCACATCTGGCAATACCGCCAGCCTGACAATCACTGGCGCCCAGGCTGAAGATGAGGC
96
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
CGACTACTACTGCAACAGCAGAGACAGCAGCGGCAACCGGCTGGTTTTTGGCGGAGGCACAA
AGCTGACAGTGCTGGGCCAACCTAAGGCCGCTCCTAGCGTGACACTGTTCCCTCCAAGCAGC
GAAGAACTGCAGGCCAACAAGGCCACACTCGTGTGCCTGATCAGCGACTTTTATCCTGGCGC
CGTGACCGTGGCCTGGAAGGCTGATAGTTCTCCTGTGAAGGCCGGCGTGGAAACCACCACAC
CTAGCAAGCAGAGCAACAACAAATACGCCGCCAGCAGCTACCTGAGCCTGACACCTGAGCAG
TGGAAGTCCCACAGATCCTACAGCTGCCAAGTGACCCACGAGGGCAGCACCGTGGAAAAAAC
AGTGGCCCCTACCGAGTGCAGC
SEQ ID NO: 25
Annotated Amino Acid sequence of Synthetic anti-CD52g Fab-IgG-
Fab (Light Chain)
SSELTQDPVVSVALGQTVRITCQGDSLRTYHASWYQQKPRQAPVLVIYDENNRPSGIPDRES
GSTSGNTASLTITGAQAEDEADYYCNSRDSSGNRLVEGGGTKLTVLGQPKAAPSVTLEPPSS
EELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPEQ
WKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 26
Annotated DNA sequence of Synthetic anti-CD52g Fab-IgG-Fab-Fab
(Heavy Chain)
CAGGTTCAGCTGCAGCAATGGGGAGCCGGACTGCTGAAGCCTAGCGAGACACTGTCTCTGAC
CTGTGCCGTGTACGGCGGCAGOTTCAGOGGCTACTACTGGTCCTGGATCAGACAGCCTCCTG
GCAAAGGCCTGGAATGGATCGGCGAGATCAATCACAGCGGCAGCACCAACTACAACCCCAGC
CTGAGAAGCAGACTGACCATCAGCGTGGACACCAGCAAGAACCAGTTCAGCCTGAAGCTGAG
AAGCGTGACAGCCGCCGATACCGCCGTGTACTATTGCGCCAGAGGCTTTATGGTCCGAGGCA
TCATGTGGAACTACTACTACATGGACGTGTGGGGCAAGGGCACCACCGTGACAGTTTCTCCA
GCCTCTACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGG
CACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGA
ACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTC
TACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGGAGCTTGGGCACCCAGACCTACATCTG
CAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTG
ACAAAACTCACACAGGTGGCGGAGGATCTGGCGGAGGGGGGAGCGGAGGGGGTGGGTCCGGC
GGGGGGGGTTCCGGGGGTGGTGGATCAGGCGGCGGAGGAAGTCAGGTTCAGCTGCAGCAATG
GGGAGCCGGACTGCTGAAGCCTAGCGAGACACTGTOTCTGACCTGTGCCGTGTACGGCGGCA
GCTTCAGCGGCTACTACTGGTOCTGGATCAGACAGCCTCCTGGCAAAGGCCTGGAATGGATC
GGCGAGATCAATCACAGCGGCAGCACCAACTACAACCCCAGCCTGAGAAGCAGAGTGACCAT
CAGCGTGGACACCAGCAAGAACCAGTTCAGCCTGAAGCTGAGAAGCGTGACAGCCGCCGATA
CCGCCGTGTACTATTGCGCCAGAGGCTTTATGGTCCGAGGCATCATGTGGAACTACTACTAC
ATGGACGTGTGGGGCAAGGGCACCACCGTGACAGTTTCTCCAGCTAGCACCAAGGGCCCATC
GGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCC
TGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGC
GGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGT
GACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCA
GCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACACATGCCCA
CCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAA
GGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACG
AAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAATGCCAAGACA
AAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACCGTOCTGCA
97
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
CCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCAGCCC
CCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCTG
CCCCCATCCCGGGACGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAAAGGCTT
CTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGA
CCACGCCTOCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGAC
AAGAGGAGGTGGCAGGAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAA
CCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAAGGATCCGGTGGCGGAGGCTCTG
GTGGAGGAGGCAGTGGCGGAGGCGGTTCTGGTGGTGGTGGCTCTGGTGGCGGCGGTTCAGGC
GGTGGCGGATCTCAAGTTCAGCTGCAGCAATGGGGAGCGGGCCTGCTGAAGCCTTCTGAGAC
ACTGTCTCTGACCTGCGCCGTGTACGGCGGCAGCTTTAGCGGCTACTACTGGTCCTGGATCA
GACAGCCTCCTGGCAAAGGCCTGGAATGGATCGGCGAGATCAATCACAGCGGCAGCACCAAC
TACAACCCCAGCCTGAGAAGGAGAGTGACCATCAGCGTGGACACCAGCAAGAACCAGTTCAG
CCTGAAGCTGOGGAGCGTGACAGCCGCTGATACAGCCGTGTACTACTGCGCCAGAGGCTTTA
TGGTCCGAGGCATCATGTGGAACTACTACTACATGGACGTGTGGGGCAAGGGCACCACCGTG
ACAGTTTCTCCAGCCAGCACAAAGGGCCCCAGCGTTTTCCCACTGGCTCCTAGGAGCAAGAG
CACATCTGGTGGAACAGCCGCTCTGGGCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGA
CCGTGTCCTGGAATTCTGGCGCTOTGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAA
AGCAGCGGCCTGTACTCTCTGAGGAGCGTGGTCACAGTGCCAAGCTCTAGCCTGGGCACCCA
GACCTACATCTGCAATGTGAACCACAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAAC
CCAAGAGCTGCGACAAGACCCATACAGGGGGCGGTGGAAGCGGAGGCGGGGGTAGCGGTGGT
GGCGGCAGCGGCGGAGGCGGATCAGGGGGCGGCGGAAGTGGTGGCGGTGGTTCTCAGGTTCA
ACTCCAACAGTGGGGCGOTGGACTGCTGAAACCTAGCGAAACCCTGAGCCTGACATGTGCTG
TGTATGGCGGCTCCTTCTCCGGCTACTATTGGAGCTGGATTCGGCAGCCACCAGGCAAGGGA
CTCGAGTGGATTGGAGAGATCAACCACTCCGGCTCCACCAATTACAATCCATCTCTGOGGTC
CCGCGTGACCATCTCCGTGGATACCTCTAAGAATCAGTTCTCACTGAAGCTGAGATCCGTGA
CCGCTGCCGACACTGCCGTGTATTATTGTGCCCGGGGATTCATGGTTCGAGGGATTATGTGG
AATTACTATTATATGGATGTCTGGGGAAAAGGGACGACCGTGACTGTGTCCCCTGCCTCTAC
AAAGGGAGCCTCCGTGTTTCCTCTGGCTCCCAGCTCTAAGTCTACCAGCGGAGGAACAGCTG
CCCTGGGATGTCTCGTGAAAGACTACTTCCCCGAACCAGTGACAGTCAGCTGGAACAGCGGA
GCCCTGACTTCTGGGGTGCACACATTCCCTGCCGTCCTGCAATCTTCTGGCCTGTACAGCCT
GTCCAGCGTCGTGACCGTTCCTTCTAGCTCTCTGGGAACACAGACATATATCTGTAATGTGA
ATCACAAACCCTCCAATACGAAGGTCGACAAAAAGGTCGAGCCTAAGTCCTGTGATAAGACC
CACACC
SEQ ID NO: 27
Annotated Amino Acid sequence of Synthetic anti-CD52g Fab-IgG-
Fab-Fab (Heavy Chain)
QVQLQNGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRQPPGKGLEWIGETNESGSTNYNPS
LRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYYMDVWGKGTTVTVSP
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGSG
GGGSGGGGSGGGGSQVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRQPPGKGLEWI
GEINHSGSTNYNPSLRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYY
MDVWGKGTTVTVSPASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTS
GVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCP
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVIDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHOWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGOPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVESCSVMHEALHNHYTQKSLSLSPGKGSGGGGSGGGGSGGGGSGGGGSGGGGSG
98
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
GGGSQVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYNSWIRUPGKGLEWIGEINHSGSTN
YNPSLRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYYMDVWGKGTTV
TVSPASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQ
SSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGG
GGSGGGGSGGGGSGGGGSQVQLQINGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRUPGKG
LEWIGEINHSGSTNYNPSLRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMW
NYYYMDVWGKGTTVTVSPASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSG
ALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKT
HT
SEQ ID NO; 28
Annotated Amino Acid sequence of Synthetic anti-CD52g Fab
fragment of Fab-IgG-Fab-Fab (Heavy Chain)
QVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRUPGKGLEWIGEINHSGSTNYNE'S
LRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYYMDVWGKGTTVTVSP
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLOSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 29
Annotated Amino Acid sequence of Synthetic anti-CD52g Fc
fragment of Fab-IgG-Fab-Fab (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 30
DNA sequence of Synthetic anti-CD52g Fab-IgG-Fab-Fab (Light
Chain)
AGCAGCGAGCTGACACAGGATCCAGTGGTGTCTGTGGCCCTGGGCCAGACAGTGCGGATTAC
TTGTCAGGGCGACAGCCTGAGAACCTACCACGCCTCTTGGTATCAGCAGAAGCCCAGACAGG
CCCCTGTGCTGGTCATCTACGACGAGAACAACAGACCCAGCGGCATCCCCGATAGATTCAGC
GGCAGCACATCTGGCAATACCGCCAGCCTGACAATCACTGGCGCCCAGGCTGAAGATGAGGC
CGACTACTACTGCAACAGCAGAGACAGGAGCGGCAACCGGCTGGTTTTTGGCGGAGGCACAA
AGCTGACAGTGCTGGGCCAACCTAAGGCCGCTCCTAGCGTGACACTGTTCCCTCCAAGCAGC
GAAGAACTGCAGGCCAACAAGGCCACACTCGTGTGCCTGATCAGCGACTTTTATCCTGGCGC
CGTGACCGTGGCCTGGAAGGCTGATAGTTCTCCTGTGAAGGCCGGCGTGGAAACCACCACAC
CTAGCAAGCAGAGCAACAACAAATACGCCGCCAGCAGCTACCTGAGCCTGACACCTGAGCAG
TGGAAGTCCCACAGATCCTACAGCTGCCAAGTGACCCACGAGGGCAGCACCGTGGAAAAAAC
AGTGGCCCCTACCGAGTGCAGC
SEQ ID NO: 31
Annotated Amino Acid sequence of Synthetic anti-CD52g Fab-IgG-
Fab-Fab (Light Chain)
99
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SSELTQDPVVSVALGQTVRITCQGDSLRTYHASWYWKPRQAPVLVIYDENNRPSGIPDRES
GSTSGNTASLTITGAQAEDEADYYCNSRDSSGNRLVFGGGTKLTVL
GUKAAPSVTLFPPSSEELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQS
NNKYAASSYLSLTPEQWKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 32
Annotated DNA sequence of Synthetic anti-CD52g Fab-Fab-IgG-
Fab-Fab (Heavy Chain)
CAAGTTCAGCTGCAGCAATGGGGAGCCGGCCTGCTGAAGCCTTCTGAGACACTGTCTCTGAC
CTGCGCCGTGTACGGCGGCAGCTTTAGCGGCTACTACTGGTCCTGGATCAGACAGCCTCCTG
GCAAAGGCCTGGAATGGATCGGCGAGATCAATCACAGCGGCAGCACCAACTACAACCCCAGC
CTGAGAAGCAGAGTGACCATCAGCGTGGACACCAGCAAGAACCAGTTCAGCCTGAAGCTGCG
GAGCGTGACAGCCGCTGATACAGCCGTGTACTACTGCGCCAGAGGCTTTATGGTCCGAGGCA
TCATGTGGAACTACTACTACATGGACGTGTGGGGCAAGGGCACCACCGTGACAGTTTCTCCA
GCCAGCACAAAGGGCCCCAGCGTTTTCCCACTGGCTCCTAGGAGCAAGAGCACATCTGGTGG
AACAGCCGCTCTGGGCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCCTGGA
ATTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGGAGCGGCCTG
TACTCTCTGAGCAGCGTGGTCACAGTGCCAAGCTCTAGCCTGGGCACCCAGACCTACATCTG
CAATGTGAACCACAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCG
ACAAGACCCATACAGGGGGCGGTGGAAGCGGAGGCGGGGGTAGCGGTGGTGGCGGCAGCGGC
GGAGGCGGATCAGGGGGCGGCGGAAGTGGTGGCGGTGGTTCTCAGGTTCAACTCCAACAGTG
GGGCGCTGGACTGCTGAAACCTAGCGAAACCCTGAGCCTGACATGTGCTGTGTATGGCGGCT
COTTCTCCGGCTACTATTGGAGCTGGATTCGGCAGCCACCAGGCAAGGGACTCGAGTGGATT
GGAGAGATCAACCACTCCGGCTCCACCAATTACAATCCATCTCTGCGGTCCCGCGTGACCAT
CTCCGTGGATACCTCTARGAATCAGTTCTCACTGAAGCTGAGATCCGTGACCGCTGCCGACA
CTGCCGTGTATTATTGTGCCCGGGGATTCATGGTTCGAGGGATTATGTGGAATTACTATTAT
ATGGATGTCTGGGGAAAAGGGACGACCGTGACTGTGTCCCCTGCCTCTACAAAGGGACCCTC
CGTGTTTCCTCTGGCTCCCAGCTCTAAGTCTACCAGCGGAGGAACAGCTGCCCTGGGATGTC
TCGTGAAAGACTACTTCCCCGAACCAGTGACAGTCAGCTGGAACAGCGGAGCCCTGACTTCT
GGGGTGCACACATTCCCTGCCGTOCTGCAATCTTCTGGCCTGTACAGCCTGTCCAGCGTCGT
GACCGTTCCTTCTAGCTCTCTGGGAACACAGACATATATCTGTAATGTCAATCACAAACCCT
CCAATACGAAGGTCGACAAAAAGGTCGAGCCTAAGTCCTGTGATAAGACCCACACCGGTGGC
GGAGGCTCTGGGGGAGGAGGCAGTGGCGGAGGCGGTTCTGGTGGTGGTGGCTCTGGTGGCGG
CGGTTCAGGCGGTGGCGGATCTCAGGTTCAGCTGCAGCAATGGGGAGCCGGACTGCTGAAGC
CTAGCGAGACACTGTOTCTGACCTGTGCCGTGTACGGCGGCAGCTTCAGCGGCTACTACTGG
TCCTGGATCAGACAGCCTCCTGGCAAAGGCCTGGAATGGATCGGCGAGATCAATCACAGCGG
CAGCACCAACTACAACCCCAGCCTGAGAAGGAGAGTGACCATCAGCGTGGACACCAGCAAGA
ACCAGTTCAGCCTGAAGCTGAGAAGCGTGACAGCCGCCGATACCGCCGTGTACTATTGCGCC
AGAGGCTTTATGGTCCGAGGCATCATGTGGAACTACTACTACATGGACGTGTGGGGCAAGGG
CACCACCGTTACAGTCTCACCTGCTAGCACCAAGGGCCCATCGGTCTTCCOCCTGGCACCCT
CCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCC
GAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGC
TGTCCTACAGTCCTCAGGACTCTACTCCCTCAGGAGCGTGGTGACCGTGCCCTCCAGCAGCT
TGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAG
AAAGTTGAGCCCAAATCTTGTGACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACT
CCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCC
GGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTC
AACTGGTACGTGGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGGAGTA
CAACAGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCA
100
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
AGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCC
AAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCTGCCCCCATCCCGGGACGAGCT
GACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAAAGGCTTCTATCCCAGCGACATCGCCG
TGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGAC
TCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGACAAGAGCAGGTGGCAGGAGGG
GAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCC
TCTCCCTGTCTCCGGGTAAAGGATCCGGTGGCGGAGGCTCTGGTGGAGGAGGCAGTGGCGGA
GGCGGTTCTGGTGGTGGTGGCTCTGGTGGCGGCGGTTCAGGCGGTGGCGGATCTCAAGTTCA
GCTGCAGCAATGGGGAGCCGGCCTGCTGAAGCCTTCTGAGACACTGTCTCTGACCTGCGCCG
TGTACGGCGGCAGCTTTAGCGGCTACTACTGGTCCTGGATCAGACAGCCTCCTGGCAAAGGC
CTGGAATGGATCGGCGAGATCAATCACAGCGGCAGCACCAACTACAACCCCAGCCTGAGAAG
CAGAGTGACCATCAGCGTGGACACCAGCAAGAACCAGTTCAGCCTGAAGCTGCGGAGCGTGA
CAGCCGCTGATACAGCCGTGTACTACTGCGCCAGAGGCTTTATGGTCCGAGGCATCATGTGG
AACTACTACTACATGGACGTGTGGGGCAAGGGCACCACCGTGACAGTTTCTCCAGCCAGCAC
AAAGGGCCCCAGCGTTTTCCCACTGGCTCCTAGCAGCAAGAGCACATCTGGTGGAACAGCCG
CTCTGGGCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCCTGGAATTCTGGC
GCTCTGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGCAGCGGCCTGTACTCTCT
GAGCAGCGTGGTCACAGTGCCAAGCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGA
ACCACAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACC
CATACAGGGGGCGGTGGAAGCGGAGGCGGGGGTAGCGGTGGTGGCGGCAGCGGCGGAGGCGG
ATCAGGGGGCGGCGGAAGTGGTGGCGGTGGTTCTCAGGTTCAACTCCAACAGTGGGGCGCTG
GACTGCTGAAACCTAGCGAAACCCTGAGCCTGACATGTGCTGTGTATGGCGGCTCCTTCTCC
GGCTACTATTGGAGCTGGATTOGGCAGCCACCAGGCAAGGGACTCGAGTGGATTGGAGAGAT
CAACCACTCCGGCTCCACCAATTACAATCCATCTCTGCGGTCCCGCGTGACCATCTCCGTGG
ATACCTCTAAGAATCAGTTCTCACTGAAGCTGAGATCCGTGACCGCTGCCGACACTGCCGTG
TATTATTGTGCCCGGGGATTCATGGTTCGAGGGATTATGTGGAATTACTATTATATGGATGT
CTGGGGAAAAGGGACGACCGTGACTGTGTCCCCTGCCTCTACAAAGGGACCCTCCGTGTTTC
CTCTGGCTCCCAGCTCTAAGTCTACCAGCGGAGGAACAGCTGCCCTGGGATGTCTCGTGAAA
GACTACTTCCCCGAACCAGTGACAGTCAGCTGGAACAGCGGAGCCCTGACTTCTGGGGTGCA
CACATTCCCTGCCGTCCTGCAATCTTCTGGCCTGTACAGCCTGTCCAGCGTCGTGACCGTTC
CTTCTAGCTCTCTGGGAACACAGACATATATCTGTAATGTCAATCACAAACCCTCCAATACG
AAGGTCGACAAAAAGGTCGAGCCTAAGTCCTGTGATAAGACCCACACC
SEQ ID NO: 33
Annotated Amino Acid sequence of Synthetic anti-CD52q Fab-Fab-
IgG-Fab-Fab (Heavy Chain)
QVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRUPGKGLEWIGEINHSGSTNYNPS
LRSRVTISVDTSKNUSLKLRSVTAADTAWYCARGFMVRGIMWNYYYMEWWGKGTTVTVSP
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGSG
GGGSGGGGSGGGGSQVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIRINTGKGLEWI
GEINHSGSTNYNPSLRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYY
MDVWGKGTTVTVSPASTKGPSVFPLAPSSKSTSGGTAALGUNKDYFETPVTVSWNSGALTS
GVHTFPAVLOSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVE2KSCDKTHTGG
GGSGGGGSGGGGSGGGGSGGGGSGGGGSQVUQQWGAGLLKPSETLSLTCAVYGGSFSGYYW
SWIRQPPGKGLEWIGEINHSGSTNYNPSLRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCA
RGFMVRGIMWNYYYMDVWGKGTTVTVSPASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFP
EPVTVSWNSGALTSGVHTFPAVLOSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDK
KVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPIOTLMISRTPEVTCVVVWSHEDETVKF
101
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
NWYVDGVEVHNAKTKPREEQYNS TYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAP I EKT I S
KAKGQPREPQVYTL P P S RDELTKNQVS LTCLVKG FY PS DIAVEWESNGQPENNYKTTPPVL D
SDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKGSGGGGSGGGGSGG
GGSGGGGSGGGGSGGGGSQVQLQQWGAGLI,KPSETLSLTCAVYGGSFSGYYWSWIROPGKG
LEWIGEINHSGSTNYNPSLRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMW
NYYYMDVWGKGTTVTVSPASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSG
ALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKT
HTGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSQVQLQQWGAGLLKPSETLSLTCAVYGGSFS
GYYWSWIRQPPGKGLEWIGEINHSGSTNYNPSLRSRVTISVDTSKNQFSLKLRSVTAADTAV
YYCARGFMVRGIMWNYYYMDVWGKGTTVTVSPASTKGPSVFPLAPSSKSTSGGTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNT
KVDKKVEPKSCDKTHT
SEQ ID NO: 34
Annotated DNA sequence of Synthetic anti-CD52g Fab-Fab-IgG-
Fab-Fab (Light Chain)
AGCAGCGAGCTGACACAGGATCCAGTGGTGTCTGTGGCCCTGGGCCAGACAGTGCGGATTAC
TTGTCAGGGCGACAGCCTGAGAACCTACCACGCCTCTTGGTATCAGCAGAAGCCCAGACAGG
CCCCTGTGCTGGTCATCTACGACGAGAACAACAGACCCAGCGGCATCCCCGATAGATTCAGC
GGCAGCACATCTGGCAATACCGCCAGCCTGACAATCACTGGCGCCCAGGCTGAAGATGAGGC
CGACTACTACTGCAACAGCAGAGACAGGAGCGGCAACCGGCTGGTTTTTGGCGGAGGCACAA
AGCTGACAGTGCTGGGCCAACCTAAGGCCGCTCCTAGCGTGACACTGTTCCCTCCAAGCAGC
GAAGAACTGCAGGCCAACAAGGCCACACTCGTGTGCCTGATCAGCGACTTTTATCCTGGCGC
CGTGACCGTGGCCTGGAAGGCTGATAGTTCTCCTGTGAAGGCCGGCGTGGAAACCACCACAC
CTAGCAAGCAGAGCAACAACAAATACGCCGCCAGCAGCTACCTGAGCCTGACACCTGAGCAG
TGGAAGTCCCACAGATCCTACAGCTGCCAAGTGACCCACGAGGGCAGCACCGTGGAAAAAAC
AGTGGCCCCTACCGAGTGCAGC
SEQ ID NO: 35
Annotated Amino Acid sequence of Synthetic anti-CD52g Fab-Fab-
IgG-Fab-Fab (Light Chain)
SSELTQDPVVSVALGUVRITCQGDSLRTYHASWYQQKPRQAPVLVIYDENNRPSGIPDRFS
GSTSGNTASLTITGAQAEDEADYYCNSRDSSGNRINFGGGTKLTVLGOKAAPSVTLFPPSS
EELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPEQ
WKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 36
Annotated Amino Acid sequence of Synthetic anti-CD52g Fab
fragment of Fab-Fab-IgG-Fab-Fab (Heavy Chain)
QVQLQQWGAGLLKPSETLSLTCAVYGGSFSGYYWSWIROPGKGLEWIGEINHSGSTNYNPS
LRSRVTISVDTSKNQFSLKLRSVTAADTAVYYCARGFMVRGIMWNYYYMDVWGKGTTVTVSP
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
102
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SEQ ID NO: 37
Annotated Amino Amid sequence of Synthetic anti-CD52g Fab
fragment ofFab-Fab-IgG-Fab-Fab (Light Chain)
SSELTQDPVVSVALGQTVRITCQGDSLRTYHASWYWKPRQAPVLVIYDENNRRSGIPDRFS
GSTSGNTASLTITGAQAEDEADYYCNSRDSSGNRLVFGGGTKLTVLGUKAAPSVTLFPPSS
EELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPEQ
WKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 38
Annotated Amino Acid sequence of Synthetic anti-CD52g Fc
fragment of Fab-Fab-IgG-Fab-Fab (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
XPREEQYNSTYRVVSVLTVLHOWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 39
Annotated DNA sequence of anti-klebsiella IgG (Heavy Chain)
GAGGTGCAGCTGGTTGAATCTGGCGGAGGACTGGTTCAGCCTGGCGGATCTCTGAGACTGTC
TTGCGCCGCCAGCTTTAGCCTGACAAGCTACGCCGTGCACATCCACTGGGTTCGACAGGCCC
CTGGCAAAGGCCTTGAATGGGTTGCCAGAGTGATCTGGGCTGGCGGCATCACCCACTACAAT
AGCGCCCTGATGAGCAGATACGCCGACAGCGTGAAGGGCAGATTCACCATCAGCGCCGACAC
CAGCAAGAACACCGCCTACCTGCAGATGAACAGCCTGAGAGCCGAGGACACCGCCGTGTACT
ATTGCGCCAGAGGCAACTGGGCCTTCGACTATTGGGGACAGGGCACCCTGGTCACCGTTAGC
TCTGCCTCTACAAAGGGCCCTAGTGTGTTCCCTCTGGCTOCCAGCAGCAAGTCTACATCTGG
CGGAACAGCCGCTCTGGGCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCCT
GGAATTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGCAGCGGC
CTGTACTCTCTGAGCAGCGTGGTCACAGTGCCTAGCTCTAGCCTGGGCACCCAGACCTACAT
CTGCAATGTGAACCACAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCT
GCGACAAGACCCACACCTGTCCTCCATGTCCTGCTCCAGAACTGCTCGGCGGACCTTCCGTG
TTCCTGTTTCCTCCAAAGCCTAAGGACACCCTGATGATCAGCAGAACCCCTGAAGTGACCTG
CGTGGTGGTGGATGTGTCCCACGAGGATCCCGAAGTGAAGTTCAATTGGTACGTGGACGGCG
TGGAAGTGCACAACGCCAAGACCAAGCCTAGAGAGGAACAGTACAACAGCACCTACAGAGTG
GTGTCCGTGCTGACCGTGCTGCACCAGGATTGGCTGAACGGCAAAGAGTACAAGTGCAAGGT
GTCCAACAAGGCCCTGCCTGCTCCTATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCTA
GGGAACCCCAGGTTTACACACTGCCTCCAAGCCGGGAAGAGATGACCAAGAACCAGGTGTCC
CTGACCTGCCTCGTGAAGGGCTTCTACCCTTCCGATATCGCCGTGGAATGGGAGAGCAATGG
CCAGCCTGAGAACAACTACAAGACAACCCCTCCTGTGCTGGACAGCGACGGCTCATTCTTCC
TGTACAGCAAGCTGACAGTGGACAAGTCCAGATGGCAGCAGGGCAACGTGTTCAGCTGCAGC
GTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTOCCTGAGCCTGTCTCCTGGCAA
A
SEQ ID NO: 40
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Annotated Amino Acid sequence of anti-klebsiella IgG (Heavy
Chain)
EVQLVESGGGLVQPGGSLRLSCAASFSLTSYAVHIHWVRQAPGKGLEWVARVIWAGGITHYN
SALMSRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGNWAFDYWGQGTLVTVS
SASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSG
LYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSV
FLFPPKPKIDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRV
VSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLE,PSREEMTKNQVS
LTCLVKGFYPSDIAVEWESNGUENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCS
VMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 41
Annotated Amino Acid sequence of Fab fragment of anti-
klebsiella (Heavy Chain)
EVQLVESGGGLVUGGSLRLSCAASFSLTSYAVHIHWVRQAPGKGLEWVARVIWAGGITHYN
SALMSRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGNWAFDYWGQGTLVTVS
SASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSG
LYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 42
Annotated Amino Acid sequence of Fc fragment of anti-
kiebsiella (Heavy Chain)
PCPAPELLGGPSVFLFETKPKDTLMISRTPEVTCVVVIDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 43
DNA sequence of anti-klebsiella (Light Chain)
GACATCCAGATGACACAGAGCCCTAGCAGCCTGTCTGCCAGCGTGGGAGACAGAGTGACCAT
CACCTGTAGAGCCAGCAGCGCCAGATCCAGCGTGTCCTATATTCACGTGGCCTGGTATCAGC
AGAAGCCCGGCAAAGCCOCTAAGCTGCTGATCTACGACACCAGCAAACTGGCCAGCTTCCTG
TACAGCGGCGTGCCCTCTAGATTCAGCGGCAGCAGATCTGGCACCGACTTCACCCTGACCAT
AAGCAGCCTGCAGCCTGAGGACTTCGCCACCTACTACTGCTTTCAAGGCAGCGGCTACCCCT
ACACCTTTGGCCAGGGAACAAAGGTGGAAATCAAGAGAACAGTGGCCGCTCCTAGCGTGTTC
ATCTTCCCACCTTCCGACGAGCAGCTGAAGTCTGGCACAGCCTCTGTCGTGTGCCTGCTGAA
CAACTTCTACCCCAGAGAAGCCAAGGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ATAGCCAAGAGAGCGTGACCGAGGAGGACAGCAAGGACTCTACCTACAGCCTGAGCAGCACC
CTGACACTGAGCAAGGCCGACTACGAGAAGCACAAAGTGTACGCCTGCGAAGTGACCCACCA
GGGCCTTTCTAGCCCTGTGACCAAGAGCTTCAACCGGGGCGAATGT
SEQ ID NO: 44
104
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Annotated Amino Acid sequence of anti-klebsiella (Light Chain)
DIQMTOPSSLSASVGDRVTITCRASSARSSVSYIHVAWYQQKPGKAPKLLIYDTSKLASFL
YSGVPSRFSGSRSGTDFTLTISSLUEDFATYYCFQGSGYPYTFGQGTKVEIKRTVAAPSVF
IFPPSDEQLKSGTASVVOLLNNFYPREAKVQWKVDNALUGNSQESVTENSKDSTYSLSST
LTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 45
Annotated Amino Acid sequence of Fab fragment of anti-
klebsiella IgG (Light Chain)
DIQMTUPSSLSASVGDRVTITCRASSARSSVSYTHVAWYWKPGKAPKLLIYDTSKLASFL
YSGVPSRFSGSRSGTDFTLTISSLQPEDFATYYCFQGSGYPYTFGQGTKVEIKRTVAAPSVF
IFPPSDEQLKSGTASVVCLIANNFYPREAKVQWKVDNALUGNSQESVTEUSKDSTYSLSST
LTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 46
Annotated DNA sequence of Synthetic anti-klebsiella Fab-IgG
(Heavy Chain)
GAGGTGCAGTTGGTTGAATCCGGTGGCGGGTTGGTCCAGCCTGGCGGGAGCCTTCGGCTTAG
TTGTGCCGCATCATTTAGCCTGACATCTTACGCTGTCCATATCCATTGGGTGCGGCAAGCGC
CTGGTAAGGGCCTGGAATGGGTGGCAAGGGTGATATGGGCAGGGGGTATTACGCATTACAAC
TCTGCATTGATGAGTCGGTACGCCGACAGCGTCAAAGGTCGGTTCACCATTTCTGCCGATAC
CTCTAAGAACACAGCCTACCTCCAGATGAACTCACTGCGAGCGGAGGACACTGCTGTGTACT
ATTGCGCCCGCGGCAATTGGGCATTTGACTACTGGGGGCAAGGTACACTCGTAACGGTCTCA
TCTGCCTCTACAAAGGGCCCTAGTGTGTTCCCTCTGGCTCCCAGGAGCAAGTCTACATCTGG
CGGAACAGCCGCTCTGGGCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCCT
GGAATTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGGAGCGGC
CTGTACTCTCTGAGCAGCGTGGTCACAGTGCCTAGCTCTAGCCTGGGCACCCAGACCTACAT
CTGCAATGTGAACCACAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCT
GCGACAAGACCCACACCGGTGGCGGAGGATCTGGTGGAGGAGGGAGCGGAGGGGGTGGGTCC
GGAGGGGGTGGTTCCGGAGGTGGTGGATCAGGTGGCGGAGGAAGTGAGGTGCAGCTGGTTGA
ATCTGGCGGAGGACTGGTTCAGCCTGGCGGATCTCTGAGACTGTCTTGCGCCGCCAGCTTTA
GCCTGACAAGCTACGCCGTGCACATCCACTGGGTTCGACAGGCCOCTGGCAAAGGCCTTGAA
TGGGTTGCCAGAGTGATCTGGGCTGGCGGCATCACCCACTACAATAGCGCCCTGATGAGGAG
ATACGCCGACAGCGTGAAGGGCAGATTCACCATCAGCGCCGACACCAGCAAGAACACCGCCT
ACCTGCAGATGAACAGCCTGAGAGCCGAGGACACCGCCGTGTACTATTGCGCCAGAGGCAAC
TGGGCCTTCGACTATTGGGGACAGGGCACCCTGGTCACCGTTAGCTCTGCCTCTACAAAGGG
CCCTAGTGTGTTCCCTCTGGCTCCCAGCAGCAAGTCTACATCTGGCGGAACAGCCGCTCTGG
GCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCCTGGAATTCTGGCGCTCTG
ACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGGAGCGGCCTGTACTCTCTGAGGAG
CGTGGTCACAGTGCCTAGCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGAACCACA
AGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCACACC
TGTCCTCCATGTCCTGCTCCAGAACTGCTCGGCGGACCTTCCGTGTTCCTGTTTCCTCCAAA
GCCTAAGGACACCCTGATGATCAGGAGAACCOCTGAAGTGACCTGCGTGGTGGTGGATGTGT
CCCACGAGGATCCCGAAGTGAAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCC
AAGACCAAGCCTAGAGAGGAACAGTACAACAGCACCTACAGAGTGGTGTCCGTGCTGACCGT
GCTGCACCAGGATTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGCCCTGC
105
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
CTGCTCCTATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCTAGGGAACCCCAGGTTTAC
ACACTGCCTCCAAGCCGGGAAGAGATGACCAAGAACCAGGTGTCCCTGACCTGCCTCGTGAA
GGGCTTCTACCCTTCCGATATCGCCGTGGAATGGGAGAGCAATGGCCAGCCTGAGAACAACT
ACAAGACAACCCCTCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTACAGCAAGCTGACA
GTGGACAAGTCCAGATGGCAGCAGGGCAACGTGTTCAGCTGCAGCGTGATGCACGAGGCCCT
GCACAACCACTACACCCAGAAGTCCCTGAGCCTGTCTCCTGGCAAA
SEQ ID NO: 47
Annotated Amino Acid sequence of Synthetic anti-klebsiella IgG
Fab-IgG (Heavy Chain)
EVQLVESGGGLVQPGGSLRLSCAASFSLTSYAVHIHWVRQAPGKGLEWVARVIWAGGITHYN
SALMSRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGNWAFDYWGQGTLVTVS
SASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSG
LYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGS
GGGGSGGGGSGGGGSEVQLVESGGGLVQPGGSLRLSCAASFSLTSYAVHIHWVRQAPGKGLE
WVARVIWAGGITHYNSALMSRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGN
WAFDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGAL
TSGVHTFPAVLOSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
CPPCPAPELLGGPSVFLFPPKPECTLMISRTPEVTCVVVDVSHEDPEVKFNWYVIDGVEVHNA
KTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVY
TLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLT
VDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 48
Annotated Amino Acid sequence of Synthetic anti-klebsiella Fab
fragment of Fab-IgG (Heavy Chain)
EVQLVESGGGLVQPGGSLRLSCAASFSLTSYAVHIHWVRQAPGKGLEWVARVIWAGGITHYN
SALMSRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGNWAFDYWGQGTLVTVS
SASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLOSG
LYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 49
Annotated Amino Acid sequence of Synthetic anti-klebsiella Fc
fragment of Fab-IgG (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEONSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNCUENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 50
DNA sequence of Synthetic anti-klebsiella Fab-IgG (Light
Chain)
GACATCCAGATGACACAGAGCCCTAGCAGCCTGTCTGCCAGCGTGGGAGACAGAGTGACCAT
CACCTGTAGAGCCAGCAGCGCCAGATCCAGCGTGTCCTATATTCACGTGGCCTGGTATCAGC
106
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
AGAAGCCCGGCAAAGCCOCTAAGCTGCTGATCTACGACACCAGCAAACTGGCCAGCTTCCTG
TACAGCGGCGTGCCCTCTAGATTCAGCGGCAGCAGATCTGGCACCGACTTCACCCTGACCAT
AAGCAGCCTGCAGCCTGAGGACTTCGCCACCTACTACTGCTTTCAAGGCAGCGGCTACCCCT
ACACCTTTGGCCAGGGAACAAAGGTGGAAATCAAGAGAACAGTGGCCGCTCCTAGCGTGTTC
ATCTTCCCACCTTCCGACGAGCAGCTGAAGTCTGGCACAGCCTCTGTCGTGTGCCTGCTGAA
CAACTTCTACCCCAGAGAAGCCAAGGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ATAGCCAAGAGAGCGTGACCGAGCAGGACAGCAAGGACTCTACCTACAGCCTGAGCAGCACC
CTGACACTGAGCAAGGCCGACTACGAGAAGCACAAAGTGTACGCCTGCGAAGTGACCCACCA
GGGCCTTTCTAGCCCTGTGACCAAGAGCTTCAACCGGGGCGAATGT
SEQ ID NO: 51
Annotated Amino Acid sequence of Synthetic anti-klebsiella
Fab-IgG (Light Chain)
DIQMTQSPSSLSASVGDRVTITCRASSARSSVSYTHVAWYQQKPGKAPKLLIYDTSKLASFL
YSGVPSRFSGSRSGTDFTLTISSLUEDFAMCFQGSGYPYTFGQGTKVEIKRTVAAPSVF
IFPPSDEQLKSGTASVVCLLNNEYPREAKVQWKVDNALOGNSQESVTEQDSKDSTYSLSST
LTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 52
Annotated Amino Acid sequence of Synthetic anti-klebsiella Fab
fragment of Fab-IgG (Light Chain)
DIQMTOPSSLSASVGDRVTITCRASSARSSVSYTHVAWYQQKPGKAPKLLTYDTSKLASEL
YSGVPSRFSGSRSGTDFTLTISSLUEDFATyyCFQGSGYPYTEGQGTKVEIKRTVAAPSVF
IFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSST
LTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 53
Annotated DNA sequence of Synthetic anti-klebsiella IgG-Fab
(Heavy Chain)
GAGGTGCAGCTGGTTGAATCTGGCGGAGGACTGGTTCAGCCTGGCGGATCTCTGAGACTGTC
TTGCGCCGCCAGCTTTAGCCTGACAAGCTACGCCGTGCACATCCACTGGGTTCGACAGGCCC
CTGGCAAAGGCCTTGAATGGGTTGOCAGAGTGATCTGGGCTGGCGGCATCACCCACTACAAT
AGCGCCCTGATGAGCAGATACGCCGACAGCGTGAAGGGCAGATTCACCATCAGCGCCGACAC
CAGCAAGAACACCGCCTACCTGCAGATGAACAGCCTGAGAGCCGAGGACACCGCCGTGTACT
ATTGCGCCAGAGGCAACTGGGCCTTCGACTATTGGGGACAGGGCACCCTGGTCACCGTTAGC
TCTGCCTCTACAAAGGGCCCTAGTGTGTTCCCTCTGGCTCCCAGCAGCAAGTCTACATCTGG
CGGAACAGCCGCTCTGGGCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCCT
GGAATTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGCAGCGGC
CTGTACTCTCTGAGCAGCGTGGTCACAGTGCCTAGCTCTAGCCTGGGCACCCAGACCTACAT
CTGCAATGTGAACCACAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCT
GCGACAAGACCCACACCTGTCCTCCATGTOCTGCTCCAGAACTGCTCGGCGGACCTTCCGTG
TTCCTGTTTCCTCCAAAGCCTAAGGACACCCTGATGATCAGCAGAACCCCTGAAGTGACCTG
CGTGGTGGTGGATGTGTCCCACGAGGATCCCGAAGTGAAGTTCAATTGGTACGTGGACGGCG
TGGAAGTGCACAACGCCAAGACCAAGCCTAGAGAGGAACAGTACAACAGCACCTACAGAGTG
GTGTCCGTGCTGACCGTGCTGCACCAGGATTGGCTGAACGGCAAAGAGTACAAGTGCAAGGT
GTCCAACAAGGCCCTGCCTGCTCCTATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCTA
107
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
GGGAACCCCAGGTTTACACACTGCCTCCAAGCCGGGAAGAGATGACCAAGAACCAGGTGTCC
CTGACCTGCCTCGTGAAGGGCTTCTACCCTTCCGATATCGCCGTGGAATGGGAGAGCAATGG
CCAGCCTGAGAACAACTACAAGACAACCOCTCCTGTGCTGGACAGCGACGGCTCATTCTTCC
TGTACAGCAAGCTGACAGTGGACAAGTCCAGATGGCAGCAGGGCAACGTGTTCAGCTGCAGC
GTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGAGCCTGTCTCCTGGCAA
AGGATCCGGTGGCGGAGGATCTGGTGGAGGAGGGAGCGGAGGGGGTGGGTCCGGAGGGGGTG
GTTCCGGAGGTGGTGGATCAGGTGGCGGAGGAAGTGAGGTGCAGTTGGTTGAATCCGGTGGC
GGGTTGGTCCAGCCTGGCGGGAGCCTTCGGCTTAGTTGTGCCGCATCATTTAGCCTGACATC
TTACGCTGTCCATATCCATTGGGTGCGGCAAGCGCCTGGTAAGGGCCTGGAATOGGTGGCAA
GGGTGATATGGGCAGGGGGTATTACGCATTACAACTCTGCATTGATGAGTCGGTACGCCGAC
AGCGTCAAAGGTCGGTTCACCATTTCTGCCGATACCTCTAAGAACACAGCCTACCTCCAGAT
GAACTCACTGCGAGCGGAGGACACTGCTGTGTACTATTGCGCCCGCGGCAATTGGGCATTTG
ACTACTGGGGGCAAGGTACACTCGTAACGGTCTCATCTGCCTCTACAAAGGGCCCTAGTGTG
TTCCCTCTGGCTCCCAGGAGCAAGTCTACATCTGGCGGAACAGCCGCTCTGGGCTGCCTGGT
CAAGGATTACTTTCCCGAGCCTGTGACCGTGTOCTGGAATTCTGGCGCTCTGACAAGCGGCG
TGCACACCTTTCCAGCTGTGCTGCAAAGCAGCGGCCTGTACTCTCTGAGCAGCGTGGTCACA
GTGCCTAGCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGAACCACAAGCCTAGCAA
CACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCACACC
SEQ ID NO: 54
Annotated Amino Amid sequence of Synthetic anti-klebsiella
IgG-Fab (Heavy Chain)
EVQLVESGGGLVUGGSLRLSCAASFSLTSYAVHIHWVKAPGKGLEWVARVIWAGGITHYN
SALMSRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGNWAFDYWGQGTLVTVS
SASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSG
LYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSV
FLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRV
VSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQ=LPPSREEMTKNQVS
LTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVESCS
VMHEALHNHYTQKSLSLSPGKGSGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSEVQLVESGG
GLVQPGGSLRLSCAASPSLTSYAVHIHWVRQAPGKGLEWVARVIWAGGITHYNSALMSRYAD
SVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGNWAFDYWGQGTLVTVSSASTKGPSV
FPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLOSGLYSLSSVVT
VPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 55
Annotated Amino Acid sequence of Synthetic anti-klebsiella Fab
fragment of IgG-Fab (Heavy Chain)
EVQLVESGGGLVUGGSLRLSCAASFSLTSYAVHIHWVRQAPGEGLEWVARVIWAGGITHYN
SALMSRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGNWAFDYWGQGTLVTVS
SASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSG
LYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 56
Annotated Amino Amid sequence of Synthetic anti-klebsiella Fa
fragment of IgG-Fab (Heavy Chain)
108
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGUENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 57
DNA sequence of Synthetic anti-klebsiella IgG-Fab (Light
Chain)
GACATCCAGATGACACAGAGCCCTAGGAGCCTGTCTGCCAGCGTGGGAGACAGAGTGACCAT
CACCTGTAGAGCCAGGAGCGCCAGATCCAGCGTGTCCTATATTCACGTGGCCTGGTATCAGC
AGAAGCCCGGCAAAGCCCCTAAGCTGCTGATCTACGACACCAGCAAACTGGCCAGCTTCCTG
TACAGCGGCGTGCCCTCTAGATTCAGCGGCAGGAGATCTGGCACCGACTTCACCCTGACCAT
AAGGAGCCTGCAGCCTGAGGACTTCGCCACCTACTACTGCTTTCAAGGCAGCGGCTACCCCT
ACACCTTTGGCCAGGGAACAAAGGTGGAAATCAAGAGAACAGTGGCCGCTCCTAGCGTGTTC
ATCTTCCCACCTTCCGACGAGCAGCTGAAGTCTGGCACAGCCTCTGTCGTGTGCCTGCTGAA
CAACTTCTACCCCAGAGAAGCCAAGGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ATAGCCAAGAGAGCGTGACCGAGCAGGACAGCAAGGACTCTACCTACAGCCTGAGGAGCACC
CTGACACTGAGCAAGGCCGACTACGAGAAGCACAAAGTGTACGCCTGCGAAGTGACCCACCA
GGGCCTTTCTAGCCCTGTGACCAAGAGCTTCAACCGGGGCGAATGT
SEQ ID NO: 58
Annotated Amino Acid sequence of Synthetic anti-klebsiella
IgG-Fab (Light Chain)
DIQMTOPSSLSASVGDRVTITCRASSARSSVSYTHVAWYWKPGKAPKLLTYDTSKLASFL
YSGVPSRFSGSRSGTDFTLTISSLUEDFATYYCFQGSGYPYTFGQGTKVEIKRTVAAPSVF
IFPPSDEQLKSGTASVVOLLNNEYPREAKVQWKVDNALUGNSQESVTENSKDSTYSLSST
LTLSKADYEKHKVYACEVTHQGLSSPVTKSENRGEC
SEQ ID NO: 59
Annotated Amino Acid sequence of Synthetic anti-klebsiella Fab
fragment of IgG-Fab (Light Chain)
DIQMTUPSSLSASVGDRVTITCRASSARSSVSYIHVAWYQQKPGKAPKLLTYDTSKLASFL
YSGVPSRFSGSRSGTDFTLTISSLQPEDFATYYCFQGSGYPYTEGQGTKVEIKRTVAAPSVF
IFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTENSKDSTYSLSST
LTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 60
Annotated DNA sequence of Synthetic anti-klebsiella Fab-IgG-
Fab (Heavy Chain)
GAGGTGCAGTTGGTTGAATCCGGTGGCGGGTTGGTCCAGCCTGGCGGGAGCCTTCGGCTTAG
TTGTGCCGCATCATTTAGCCTGACATCTTACGCTGTCCATATCCATTGGGTGCGGCAAGCGC
CTGGTAAGGGCCTGGAATGGGTGGCAAGGGTGATATGGGCAGGGGGTATTACGCATTACAAC
TCTGCATTGATGAGTCGGTACGCCGACAGCGTCAAAGGTCGGTTCACCATTTCTGCCGATAC
CTCTAAGAACACAGCCTACCTCCAGATGAACTCACTGCGAGCGGAGGACACTGCTGTGTACT
109
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
ATTGCGCCCGCGGCAATTGGGCATTTGACTACTGGGGGCAAGGTACACTCGTAACGGTCTCA
TCTGCCTCTACAAAGGGCCCTAGTGTGTTCCCTCTGGCTCCCAGCAGCAAGTCTACATCTGG
CGGAACAGCCGCTCTGGGCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCCT
GGAATTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGGAGCGGC
CTGTACTCTCTGAGGAGCGTGGTCACAGTGCCTAGCTCTAGCCTGGGCACCCAGACCTACAT
CTGCAATGTGAACCACAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCT
GCGACAAGACCCACACCGGTGGCGGAGGATCTGGTGGAGGAGGGAGCGGAGGGGGTGGGTCC
GGAGGGGGTGGTTCCGGAGGTGGTGGATCAGGTGGCGGAGGAAGTGAGGTGCAGCTGGTTGA
ATCTGGCGGAGGACTGGTTCAGCCTGGCGGATCTCTGAGACTGTCTTGCGCCGCCAGOTTTA
GCCTGACAAGCTACGCCGTGCACATCCACTGGGTTCGACAGGCCCCTGGCAAAGGCCTTGAA
TGGGTTGCCAGAGTGATCTGGGCTGGCGGCATCACCCACTACAATAGCGCCCTGATGAGGAG
ATACGCCGACAGCGTGAAGGGCAGATTCACCATCAGCGCCGACACCAGCAAGAACACCGCCT
ACCTGCAGATGAACAGCCTGAGAGCCGAGGACACCGCCGTGTACTATTGCGCCAGAGGCAAC
TGGGCCTTCGACTATTGGGGACAGGGCACCCTGGTCACCGTTAGCTCTGCCTCTACAAAGGG
CCCTAGTGTGTTCCCTCTGGCTCCCAGCAGCAAGTCTACATCTGGCGGAACAGCCGCTCTGG
GCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCCTGGAATTCTGGCGCTCTG
ACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGCAGCGGCCTGTACTCTCTGAGGAG
CGTGGTCACAGTGCCTAGCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGAACCACA
AGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCACACC
TGTOCTCCATGTCCTGCTCCAGRACTGCTCGGCGGACCTTCCGTGTTCCTGTTTCCTCCAAA
GCCTAAGGACACCCTGATGATCAGGAGAACCCCTGAAGTGACCTGCGTGGTGGTGGATGTGT
CCCACGAGGATCCCGAAGTGAAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCC
AAGACCAAGCCTAGAGAGGAACAGTACAACAGCACCTACAGAGTGGTGTCCGTGCTGACCGT
GCTGCACCAGGATTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGCCCTGC
CTGCTCCTATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCTAGGGAACCCCAGGTTTAC
ACACTGCCTCCAAGCCGGGAAGAGATGACCAAGAACCAGGTGTCCCTGACCTGCCTCGTGAA
GGGCTTCTACCCTTCCGATATCGCCGTGGAATGGGAGAGCAATGGCCAGCCTGAGAACAACT
ACAAGACAACCCCTCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTACAGCAAGCTGACA
GTGGACAAGTCCAGATGGCAGGAGGGCAACGTGTTCAGCTGCAGCGTGATGCACGAGGCCCT
GCACAACCACTACACCCAGAAGTCCCTGAGCCTGTCTCCTGGCAAAGGATCCGGTGGCGGAG
GATCTGGTGGAGGAGGGAGCGGAGGGGGTGGGTCCGGAGGGGGTGGTTCCGGAGGTGGTGGA
TCAGGTGGCGGAGGAAGTGAGGTGCAGTTGGTTGAATCCGGTGGCGGGTTGGTCCAGCCTGG
CGGGAGCCTTOGGCTTAGTTGTGCCGCATCATTTAGCCTGACATCTTACGCTGTCCATATCC
ATTGGGTGCGGCAAGCGCCTGGTAAGGGCCTGGAATGGGTGGCAAGGGTGATATGGGCAGGG
GGTATTACGCATTACAACTCTGCATTGATGAGTCGGTACGCCGACAGCGTCAAAGGTCGGTT
CACCATTTCTGCCGATACCTCTAAGAACACAGCCTACCTCCAGATGAACTCACTGCGAGCGG
AGGACACTGCTGTGTACTATTGCGCCCGCGGCAATTGGGCATTTGACTACTGGGGGCAAGGT
ACACTCGTAACGGTCTCATCTGCCTCTACAAAGGGCCCTAGTGTGTTCCCTCTGGCTCCCAG
CAGCAAGTCTACATCTGGCGGAACAGCCGCTCTGGGCTGCCTGGTCAAGGATTACTTTCCCG
AGCCTGTGACCGTGTCCTGGAATTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCT
GTGCTGCAAAGGAGCGGCCTGTACTCTCTGAGCAGCGTGGTCACAGTGCCTAGCTCTAGCCT
GGGCACCCAGACCTACATCTGCAATGTGAACCACAAGCCTAGCAACACCAAGGTGGACAAGA
AGGTGGAACCCAAGAGCTGCGACAAGACCCACACC
SEQ ID NO: 61
Annotated Amino Acid sequence of Synthetic anti-klebsiella
Fab-IgG-Fab (Heavy Chain)
EVQLVESGGGLVQPGGSLRLSCAASFSLTSYAVHIHWVRQAPGKGLEWVARVIWAGGITHYN
SALMSRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGNWAFDYWGQGTIXTVS
110
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSG
LYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGS
GGGGSGGGGSGGGGSEVQLVESGGGLVUGGSLRLSCAASFSLTSYAVHIHWVRQAPGKGLE
WVARVIWAGGITHYNSALMSRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGN
WAFDYWGQGTINTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGAL
TSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
CPPCPAPELLGGPSVFLFETKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNA
KTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVY
TLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGUENNYKTTPPVLDSDGSFFLYSKLT
VDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKGSGGGGSGGGGSGGGGSGGGGSGGGG
SGGGGSEVQLVESGGGLVQPGGSLRLSCAASFSLTSYAVHIHWVRQAPGKGLEWVARVIWAG
GITHYNSALMSRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGNWAFDYWGQG
TLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPA
VLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 62
Annotated Amino Acid sequence of Synthetic anti-klebsiella Fab
fragment of Fab-IgG-Fab (Heavy Chain)
EVQLVESGGGLVQPGGSLRLSCAASFSLTSYAVHIHWVRQAPGKGLEWVARVIWAGGITHYN
SALMSRYADSVKGRETISADTSKNTAYLONSLRAEDTAVYYCARGNWAFDYWGQGTLVTVS
SASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSG
LYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 63
Annotated Amino Acid sequence of Synthetic anti-klebsiella Fc
fragment of Fab-IgG-Fab (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEONSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 64
DNA sequence of Synthetic anti-kiebsiella Fab-IgG-Fab (Light
Chain)
GACATCCAGATGACACAGAGCCCTAGCAGCCTGTCTGCCAGCGTGGGAGACAGAGTGACCAT
CACCTGTAGAGCCAGCAGCGCCAGATCCAGCGTGTCCTATATTCACGTGGCCTGGTATCAGC
AGAAGCCOGGCAAAGCCCCTAAGCTGCTGATCTACGACACCAGCAAACTGGCCAGCTTCCTG
TACAGCGGCGTGCCCTCTAGATTCAGCGGCAGCAGATCTGGCACCGACTTCACCCTGACCAT
AAGCAGCCTGCAGCCTGAGGACTTCGCCACCTACTACTGCTTTCAAGGCAGCGGCTACCOCT
ACACCTTTGGCCAGGGAACAAAGGTGGAAATCAAGAGAACAGTGGCCGCTCCTAGCGTGTTC
ATCTTCCCACCTTCCGACGAGCAGCTGAAGTCTGGCACAGCCTCTGTCGTGTGCCTGCTGAA
CAACTTCTACCCCAGAGAAGCCAAGGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ATAGCCAAGAGAGCGTGACCGAGCAGGACAGCAAGGACTCTACCTACAGCCTGAGCAGCACC
CTGACACTGAGCAAGGCCGACTACGAGAAGCACAAAGTGTACGCCTGCGAAGTGACCCACCA
GGGCCTTTCTAGCCCTGTGACCAAGAGCTTCAACCGGGGCGAATGT
111
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SEQ ID NO: 65
Annotated Amino Acid sequence of Synthetic anti-klebsiella
Fab-IgG-Fab (Light Chain)
DIQMTQSPSSLSASVGDRVTITCRASSARSSVSYTHVAWYQQKPGKAPKLLIYDTSKLASFL
YSGVPSRFSGSRSGTDFTLTISSLUEDFATYYCFQGSGYPYTEGQGTKVEIKRTVAAPSVF
IFPPSDEQLKSGTASVVOLLNNEYPREAKVQWKVDNALUGNSQESVTEQDSKDSTYSLSST
LTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 66
Annotated Amino Acid sequence of Synthetic anti-klebsiella Fab
fragment of Fab-IgG-Fab (Light Chain)
DIQMTOPSSLSASVGDRVTITCRASSARSSVSYIHVAWYQQKPGKAPKLLIYDTSKLASFL
YSGVPSRFSGSRSGTDFTLTISSLUEDFATYYCFQGSGypyTFGQGTKVEIKRTVAAPSVF
IFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALUGNSQESVTEQDSKDSTYSLSST
LTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 67
Annotated DNA sequence of anti-Salmonella LPS IgG (Heavy
Chain)
GAAGTGAAGCTGGTGGAATCTGGCGGCGGACTGGTTCAACCTGGCGGATCTCTGTCTCTGAG
CTGTGCCGCCAGCGGCTTCACCTTCAGCGACTACTACATGACCTGGGTCCGACAGGCCCCTG
GAAAAGCTCCTGAATGGCTGGCCCTGATCCGGAACAAGAGAAACGGCGATACCGCCGAGTAC
AGCGCCTCTGTGAAGGGCAGATTCACCATCAGCCGGGACTACAGCCGGTCCATCCTGCACCT
TCAGATGAACGCCCTGAGAACCGAGGACTCCGCCACCTACTATTGCGTGCGACAAGGCAGAG
GCTACACCCTGGATTATTGGGGCCAGGGCACAAGCGTGACAGTGTCTAGCGCTAGCACCAAG
GGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCT
GGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCC
TGACCAGCGGCGTGCACACCTTCCOGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGC
AGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCA
CAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACA
CATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCOCCCA
AAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGT
GAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAATG
CCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACC
GTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCT
CCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGT
ACACCCTGCCCCCATCCCGGGACGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTC
AAAGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAA
CTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCA
CCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCT
CTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAA
SEQ ID NO: 68
112
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Annotated Amino Acid sequence of anti-Salmonella LPS IgG
(Heavy Chain)
EVKLVESGGGLVQPGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIRNKRNGDTAEY
SASVKGRFTISRDYSRSILHLQMNALRTEDSAMCVRQGRGYTLDYNGQGTSVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTIATCPPCPAPELLGGPSVFLFPP
KPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLT
VLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLV
KGFYPSDIAVEWESNGUENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEA
LHNHYTQKSLSLSPGK
SEQ ID NO: 69
Annotated Amino Acid sequence of Fab fragment of anti-
Salmonella LPS IgG (Heavy Chain)
EVKLVESGGGLVQPGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIRNKRNGDTAEY
SASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVRQGRGYTLDYWGQGTSVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 70
Annotated Amino Acid sequence of Fc fragment of anti-
Salmonella LPS (Heavy Chain)
CPAPELLGGPSVFLFPPKPKIDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVIDGVEVHNAKTK
PREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGUREPQVYTLP
PSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGOENNYKTTPPVLDSDGSFFLYSKLTVIDK
SRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 71
DNA sequence of anti-Salmonella LPS (Light Chain)
GACATCCAGATGAATCAGAGCCCCAGCAGCCTGTCTGCCAGCCTGGGAGATACCATCAGCAT
CACCTGTCGGGCCAGCCAGAACATCAACATCTGGCTGAGCTGGTATCAGCAGAAACCCGGCA
ACGTGCCCAAGCTGCTGATCTACAAGGCCAGCAATCTGCACACCGGCGTGCCCAGCAGATTT
TCTGGCTCTGGCAGOGGCACCGACTTCACCCTGATCATATCTAGCCTGCAGCCTGAGGATAT
CGCCACCTACTACTGCCTGCAAGGCCAGAGCTACCCCAGAACATTTGGCGGAGGCACCAAGC
TGGAAATCAAGACGGTGGCTGCACCATCTGTCTTCATCTTCCCGCCATCTGATGAGCAGTTG
AAATCTGGAACTGCCTCTGTTGTGTGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGT
ACAGTGGAAGGTGGATAACGCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGG
ACAGCAAGGACAGCACCTACAGCCTCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAG
AAACACAAAGTCTACGCCTGCGAAGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAG
CTTCAACAGGGGAGAGTGT
SEQ ID NO: 72
113
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Annotated Amino Acid sequence of anti-Salmonella LPS (Light
Chain)
DIONQSPSSLSASLGDTISITCRASQNINIWLSWYQQKPGNVPKLLIYKASNLHTGVPSRF
SGSGSGTDFTLIISSLQPEDIATYYCLQGQSYPRTFGGGTKLEIKTVAAPSVFIFPPSDEQL
KSGTASVVCLLNNFYPREAKVQWKVIDNALQSGNSQESVTEUSKIDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 73
Annotated Amino Acid sequence of Fab fragment of Synthetic
anti-Salmonella LPS IgG (Light Chain)
DIQMNOPSSLSASLGDTISITCRASQNINIWLSWYQQKKNVE,KLLIYKASNLHTGVPSRF
SGSGSGTDFTLIISSLUEDIATYYCLQGQSYPRTFGGGTKLEIKTVAAPSVFIF2PSDEQL
KSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEUSKDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 74
Annotated DNA sequence of Synthetic anti-Salmonella LPS Fab-
IgG (Heavy Chain)
GAAGTGAAGCTGGTGGAATCTGGCGGCGGACTGGTTCAACCTGGCGGATCTCTGTCTCTGAG
CTGTGCCGCCAGOGGCTTCACCTTCAGCGACTACTACATGACCTGGGTCCGACAGGCCCCTG
GAAAAGCTCCTGAATGGCTGGCCCTGATCCGGAACAAGAGAAACGGCGATACCGCCGAGTAC
AGCGCCICTGTGAAGGGCAGATTCACCATCAGCCGGGACTACAGCCCGTCCATCCTGCACCT
TCAGATGAACGCCCTGAGAACCGAGGACTCCGCCACCTACTATTGCGTGCGACAAGGCAGAG
GCTACACCCTGGATTATTGGGGCCAGGGCACAAGCGTGACAGTGTCTAGCGCCAGCACAAAG
GGCCCCTCTGTTTTTCCACTGGCTOCCAGCAGCAAGAGCACAAGCGGAGGAACAGCTGCCCT
GGGATGCCTCGTGAAGGACTACTTCCCTGAACCAGTGACCGTGTCCTGGAACTCTGGCGCTC
TGACTTCTGGGGTCCACACTTTCCCAGCTGTCCTGCAGTCTAGOGGACTGTACTCTCTGAGC
AGCGTGGTCACAGTGCCTAGCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGAACCA
CAAGCCTAGCAACACGAAGGTCGACAAAAAGGTGGAACCCAAGAGCTGCGACAAGACCCATA
CAGGCGGCGGAGGATCTGGCGGAGGTGGAAGCGGAGGCGGAGGAAGCGGTGGCGGCGGTAGT
GGCGGTGGTGGTTCAGGCGGTGGCGGATCTGAAGTGAAACTGGTTGAAAGCGGCGGAGGCCT
GGTTCAGCCAGGTGGAAGTCTCTCTCTGTOTTGTGCCGCCTCTGGCTTTACCTTCTCTGATT
ACTATATGACGTGGGTTCGCCAAGCTCCTGGCAAGGCACCAGAATGGCTCGCTCTGATTAGA
AACAAGCGGAATGGCGACACAGCCGAGTATTCCGCCAGCGTGAAAGGCCGGTTCACCATCTC
CAGAGACTACTCCCGCAGCATCCTGCATCTGCAAATGAATGCTCTGCGGACCGAGGACAGCG
CTACCTATTACTGCGTTAGGCAAGGCCGGGGATACACACTGGACTACTGGGGACAAGGCACC
TCCGTGACTGTGTCCTCCGCTAGCACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTC
CAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAAC
CGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGOGGCGTGCACACCTTCCCGGCTGTC
CTACAGTOCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGG
CACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAG
TTGAGCCCAAATCTTGTGACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTG
GGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCOGGAC
CCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACT
GGTACGTGGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAAC
114
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
AGCACGTACCGTGTGGTCAGCGTCCTOACCGTOCTGCACCAGGACTGGCTGAATGGCAAGGA
GTACAAGTGCAAGGTOTCCAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAG
CCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCTGCCCCCATCCCGGGACGAGCTGACC
AAGAACCAGGTCAGCCTGACCTGCCTGGTCAAAGGCTTCTATCCCAGCGACATCGCCGTGGA
GTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCG
ACGGCTCCTTCTTCCTCTACAGCAAGCTOACCGTGGACAAGAGCAGGTGGCAGCAGGGGAAC
GTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTC
CCTGTCTCCGGGTAAA
SEQ ID NO: 75
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
IgG Fab-IgG (Heavy Chain)
EVKLVESGGGLVUGGSLSLSCAASGFTFSDYYMTWVRQAPGRAPEWLALIRNKRNGDTAEY
SASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVRQGRGYTLDYWGQGTSVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLOSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGSGGGGS
GGGGSGGGGSEVKLVESGGGLVUGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIR
NKRNGDTAEYSASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVRQGRGYTLDYWGQGT
SVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAV
LUSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELL
GGPSVFLFPPKPKDTLMISRTPEVTOVVVDVSHEDPEVKFNWYVIDGVEVHNAKTKPREEQYN
STYRVVSVLTVLHQDWLNGKEYKCKVSNKALIDAPIEKTISKAKGUREPQVYTLFTSRDELT
KNQVSLTCLVKGFYPSDIAVEWESNGUENNYKTTP-PVLDSDGSFFLYSKLTVDKSRWQQGN
VFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 76
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fab fragment of Fab-IgG (Heavy Chain)
EVKLVESGGGLVQPGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIRNKRNGDTAEY
SASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVRQGRGYTLDYWGQGTSVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 77
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fc fragment of Fab-IgG (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKOKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGUENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVESCSVMHEALENHYTUSLSLSPGK
SEQ ID NO: 78
115
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
DNA sequence of Synthetic anti-Salmonella LPS Fab-IgG (Light
Chain)
GACATCCAGATGAATCAGAGCCCCAGCAGCCTGTCTGCCAGCCTGGGAGATACCATCAGCAT
CACCTGTCGGGCCAGCCAGAACATCAACATCTGGCTGAGCTGGTATCAGCAGAAACCCGGCA
ACGTGCCCAAGCTGCTGATCTACAAGGCCAGCAATCTGCACACCGGCGTGCCCAGCAGATTT
TCTGGCTCTGGCAGOGGCACCGACTTCACCCTGATCATATCTAGCCTGCAGCCTGAGGATAT
CGCCACCTACTACTGCCTGCAAGGCCAGAGCTACCCCAGAACATTTGGCGGAGGCACCAAGC
TGGAAATCAAGACGGTGGCTGCACCATCTGTCTTCATCTTCCCGCCATCTGATGAGCAGTTG
AAATCTGGAACTGCCTCTGTTGTGTGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGT
ACAGTGGAAGGTGGATAACGCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGG
ACAGCAAGGACAGCACCTACAGCCTCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAG
AAACACAAAGTCTACGCCTGCGAAGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAG
CTTCAACAGGGGAGAGTGT
SEQ ID NO: 79
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fab-IgG (Light Chain)
DIQMNOPSSLSASLGDTISITCRASQNINIWLSWYQQKPGNVPKLLIYKASNLHTGVPSRF
SGSGSGTDFTLIISSLUEDIATYYCLQGUYPRTFGGGTKLEIKTVAAPSVFIFPPSDEQL
KSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYE
KHKVYACEVTHOGLSSPVTKSFNRGEC
SEQ ID NO: 80
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fab fragment of Fab-IgG (Light Chain)
DIQMNQSPSSLSASLGDTISITCRASONINIWLSWYQQKPGNVPKIJLIYKASNLHTGVPSRF
SGSGSGTDFTLIISSLUEDIATYYCLQGUYPRTFGGGTKLEIKTVAAPSVFIFPPSDEQL
KSGTASVVCIINNFYPREAKVQWKVDNALOGNSQESVTENSKDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 81
Annotated DNA sequence of Synthetic anti-Salmonella LPS IgG-
Fab (Heavy Chain)
GAAGTGAAGCTGGTGGAATCTGGCGGCGGACTGGTTCAACCTGGCGGATCTCTGTOTCTGAG
CTGTGCCGCCAGCGGCTTCACCTTCAGCGACTACTACATGACCTGGGTCCGACAGGCCCCTG
GAAAAGCTCCTGAATGGCTGGCCCTGATCCGGAACAAGAGAAACGGCGATACCGCCGAGTAC
AGCGCCTCTGTGAAGGGCAGATTCACCATCAGCCGGGACTACAGCCGGTCCATCCTGCACCT
TCAGATGAACGCCCTGAGAACCGAGGACTCCGCCACCTACTATTGCGTGCGACAAGGCAGAG
GCTACACCCTGGATTATTGGGGCCAGGGCACAAGCGTGACAGTGTCTAGCGCTAGCACCAAG
GGCCCATCGGTCTTCCOCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCT
GGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCC
TGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGC
AGCGTGGTGACCGTGCCCTCCAGCAGOTTGGGCACCCAGACCTACATCTGCAACGTGAATCA
CAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACA
116
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
CATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCOCCCA
AAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGT
GAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAATG
CCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACC
GTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCT
CCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGT
ACACCCTGCCCCCATCCCGGGACGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTC
AAAGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAA
CTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCA
CCGTGGACAAGAGCAGGTGGCAGGAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCT
CTGCACAACCACTACACGCAGAAGAGCCTCTOCCTGTCTCCGGGTAAAGGATCCGGCGGAGG
CGGATCTGGTGGCGGAGGTAGTGGCGGCGGAGGTTCAGGTGGTGGTGGTAGCGGAGGTGGCG
GTTCTGGCGGTGGTGGATCTGAAGTGAAGCTGGTGGAATCTGGCGGAGGCCTGGTTCAACCT
GGCGGATCTCTGTOTCTGAGCTGTGCCGCCAGCGGCTTCACCTTCAGCGACTACTACATGAC
CTGGGTCCGACAGGCCOCTGGAAAAGCTCCTGAATGGCTGGCCCTGATCCGGAACAAGAGAA
ACGGCGATACCGCCGAGTACAGCGCCTCTGTGAAGGGCAGATTCACCATCAGCCGGGACTAC
AGCCGGTCCATCCTGCACCTTCAGATGAACGCCCTGAGAACCGAGGATAGCGCCACCTACTA
CTGCGTGCGACAAGGCAGAGGCTACACCCTGGATTATTGGGGCCAGGGCACAAGCGTGACAG
TGTCTAGCGCCTCTACAAAGGGCCCCAGCGTTTTCCCACTGGCTCCTAGGAGCAAGAGCACA
AGCGGAGGAACAGCCGCTCTGGGCTGTCTGGTCAAGGACTACTTTCCCGAGCCTGTGACCGT
GTCCTGGAATTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGCA
GCGGCCTGTACTCTCTGAGGAGCGTGGTCACAGTGCCAAGCTCTAGCCTGGGCACCCAGACC
TACATCTGCAATGTGAACCACAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAA
GAGCTGCGACAAGACCCACACC
SEQ ID NO: 82
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
IgG-Fab (Heavy Chain)
EVKLVESGGGLVQPGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIRNKRNGDTAEY
SASVKGRFTISRDYSRSILHLQMNALRTEDSAMCVRQGRGYTLDYWGQGTSVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPP
KPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLT
VLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLV
KGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEA
LHNHYTQKSLSLSPGKGSGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSEVKLVESGGGLVQP
GGSLSLSCAASGFTESDYYMTWVRQAPGKAPEWLALIRNKRNGDTAEYSASVXGRETISRDY
SRSILHLQMNALRTEDSATYYCVRQGRGYTLDYWGQGTSVTVSSASTKGPSVETLAPSSKST
SGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLOSGLYSLSSVVTVPSSSLGTQT
YICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 83
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fab fragment of IgG-Fab (Heavy Chain)
EVKLVESGGGLVQPGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIRNKRNGDTAEY
SASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVRQGRGYTLDYWGQGTSVTVSSASTK
117
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 84
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fc fragment of IgG-Fab (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEONSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGUREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 85
DNA sequence of Synthetic anti-Salmonella LPS IgG-Fab (Light
Chain)
GACATCCAGATGAATCAGAGCCCCAGCAGCCTGTCTGCCAGCCTGGGAGATACCATCAGCAT
CACCTGTCGGGCCAGCCAGAACATCAACATCTGGCTGAGCTGGTATCAGCAGAAACCCGGCA
ACGTGCCCAAGCTGCTGATCTACAAGGCCAGCAATCTGCACACCGGCGTGCCCAGCAGATTT
TCTGGCTCTGGCAGOGGCACCGACTTCACCCTGATCATATCTAGCCTGCAGCCTGAGGATAT
CGCCACCTACTACTGCCTGCAAGGCCAGAGCTACCCCAGAACATTTGGCGGAGGCACCAAGC
TGGAAATCAAGACGGTGGCTGCACCATCTGTOTTCATCTTCCCGCCATCTGATGAGCAGTTG
AAATCTGGAACTGCCTCTGTTGTGTGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGT
ACAGTGGAAGGTGGATAACGCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGG
ACAGCAAGGACAGCACCTACAGCCTCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAG
AAACACAAAGTCTACGCCTGCGAAGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAG
CTTCAACAGGGGAGAGTGT
SEQ ID NO: 86
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
IgG-Fab (Light Chain)
DIQMNQSPSSLSASLGDTISITCRASQNINIWLSWYQOPGNVPKLLIYKASNLHTGVPSRF
SGSGSGTDFTLITSSLUEDIATYYCLQGQSYPRTFGGGTKLEIKTVAAPSVFIFPPSDEQL
KSGTASVVCLLNNFYPREAKVQWKVDNALUGNSQESVTEUSKDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 87
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fab fragment of IgG-Fab (Light Chain)
DIQMNQSPSSLSASLGDTISITCRASQNINIWLSWYQQKPGNWKLLIYKASNLHTGVPSRF
SGSGSGTDFTLIISSLUEDIATYYCLQGQSYPRTFGGGTKLEIKTVAAPSVFIFPPSDEQL
KSGTASVVCLLNNFYPREAKVQWKVDNALUGNSQESVTEUSKDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
118
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
SEQ ID NO: 88
Annotated DNA sequence of Synthetic anti-Salmonella LPS Fab-
IgG-Fab (Heavy Chain)
GAAGTGAAGCTGGTGGAATCTGGCGGCGGACTGGTTCAACCTGGCGGATCTCTGTCTCTGAG
CTGTGCCGCCAGCGGCTTCACCTTCAGCGACTACTACATGACCTGGGTCCGACAGGCCCCTG
GAAAAGCTCCTGAATGGCTGGCCCTGATCCGGAACAAGAGAAACGGCGATACCGCCGAGTAC
AGCGCCTCTGTGAAGGGCAGATTCACCATCAGCCGGGACTACAGCCGGTCCATCCTGCACCT
TCAGATGAACGCCCTGAGAACCGAGGACTCCGCCACCTACTATTGCGTGCGACAAGGCAGAG
GCTACACCCTGGATTATTGGGGCCAGGGCACAAGCGTGACAGTGTOTAGCGCCAGCACAAAG
GGCCCCTCTGTTTTTCCACTGGCTCCCAGGAGCAAGAGCACAAGCGGAGGAACAGCTGCCCT
GGGATGCCTCGTGAAGGACTACTTCCCTGAACCAGTGACCGTGTCCTGGAACTCTGGCGCTC
TGACTTCTGGGGTCCACACTTTCCCAGCTGTCCTGCAGTCTAGCGGACTGTACTCTCTGAGC
AGCGTGGTCACAGTGCCTAGCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGAACCA
CAAGCCTAGCAACACGAAGGTCGACAAAAAGGTGGAACCCAAGAGCTGCGACAAGACCCATA
CAGGCGGCGGAGGATCTGGCGGAGGTGGAAGCGGAGGCGGAGGAAGCGGTGGCGGCGGTAGT
GGCGGTGGTGGTTCAGGCGGTGGCGGATCTGAAGTGAAACTGGTTGAAAGCGGCGGAGGCCT
GGTTCAGCCAGGTGGAAGTCTCTCTCTGTCTTGTGCCGCCTCTGGCTTTACCTTCTCTGATT
ACTATATGACGTGGGTTCGCCAAGCTCCTGGCAAGGCACCAGAATGGCTCGCTCTGATTAGA
AACAAGOGGAATGGCGACACAGCCGAGTATTCCGCCAGCGTGAAAGGCCGGTTCACCATCTC
CAGAGACTACTCCCGCAGCATCCTGCATCTGCAAATGAATGCTCTGOGGACCGAGGACAGCG
CTACCTATTACTGCGTTAGGCAAGGCCGGGGATACACACTGGACTACTGGGGACAAGGCACC
TCCGTGACTGTGTCCTCCGCTAGCACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTC
CAAGAGCACCTCTGGGGGCACAGOGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAAC
CGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTC
CTACAGTCCTCAGGACTCTACTCCCTCAGGAGCGTGGTGACCGTGCCCTCCAGGAGCTTGGG
CACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAG
TTGAGCCCAAATCTTGTGACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTG
GGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGAC
CCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACT
GGTACGTGGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAAC
AGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGA
GTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAG
CCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCTGCCCCCATCCCGGGACGAGCTGACC
AAGAACCAGGTCAGCCTGACCTGCCTGGTCAAAGGCTTCTATCCCAGCGACATCGCCGTGGA
GTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCG
ACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAAC
GTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTC
CCTGTCTCCGGGTAAAGGATCCGGCGGAGGCGGATCTGGTGGCGGAGGTAGTGGCGGCGGAG
GTTCAGGTGGTGGTGGTAGCGGAGGTGGCGGTTCTGGCGGTGGTGGATCTGAAGTGAAGCTG
GTGGAATCTGGCGGAGGCCTGGTTCAACCTGGCGGATCTCTGTCTCTGAGCTGTGCCGCCAG
CGGCTTCACCTTCAGCGACTACTACATGACCTGGGTCCGACAGGCCCCTGGAAAAGCTCCTG
AATGGCTGGCCCTGATCCGGAACAAGAGAAACGGCGATACCGCCGAGTACAGCGCCTCTGTG
AAGGGCAGATTCACCATCAGCCGGGACTACAGCCGGTCCATCCTGCACCTTCAGATGAACGC
CCTGAGAACCGAGGATAGCGCCACCTACTACTGCGTGCGACAAGGCAGAGGCTACACCCTGG
ATTATTGGGGCCAGGGCACAAGCGTGACAGTGTCTAGCGCCTCTACAAAGGGCCCCAGCGTT
TTCCCACTGGCTCCTAGGAGCAAGAGCACAAGCGGAGGAACAGCCGCTCTGGGCTGTCTGGT
CAAGGACTACTTTCCCGAGCCTGTGACCGTGTCCTGGAATTCTGGCGCTCTGACAAGCGGCG
TGCACACCTTTCCAGCTGTGCTGCAARGCAGCGGCCTGTACTCTCTGAGGAGCGTGGTCACA
119
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
GTGCCAAGCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGAACCACAAGCCTAGCAA
CACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCACACC
SEQ ID NO: 89
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fab-IgG-Fab (Heavy Chain)
EVKLVESGGGLVQPGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIRNKRNGDTAEY
SASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVROGRGYTLDYWGQGTSVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVIDKKVEPXSCDKTHTGGGGSGGGGSGGGGSGGGGS
GGGGSGGGGSEVKLVESGGGLVQPGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIR
NKRNGDTAEYSASVKGRFTISRDYSRSILHLQMNALRTEDSATTYCVRQGRGYTLDYWGQGT
SVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAV
LOSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELL
GGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYN
STYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGUREPQVYTLPPSRDELT
KNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTETVLDSDGSFFLYSKLTVDKSRWQQGN
VFSCSVMHEALHNHYTQKSLSLSPGKGSGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSEVKL
VESGGGLVOGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIRNKRNGOTAEYSASV
KGRFTISRDYSRSILHLONALRTEDSATYYCVRQGRGYTLDYWGQGTSVTVSSASTKGPSV
FPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVT
VPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 90
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fab fragment of Fab-IgG-Fab (Heavy Chain)
EVKLVESGGGLVUGGSLSLSCAASGFTFSDYYMTWVKAPGKAPEWLALIRNKRNGDTAEY
SASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVRWRGYTLDYWGQGTSVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLOSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCEIKTHT
SEQ ID NO: 91
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fc fragment of Fab-IgG-Fab (Heavy Chain)
PCPAPELLGGPSVFLFETKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGUENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVESCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 92
DNA sequence of Synthetic anti-Salmonella LPS Fab-IgG-Fab
(Light Chain)
120
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
GACATCCAGATGAATCAGAGCCCCAGCAGCCTGTCTGCCAGCCTGGGAGATACCATCAGCAT
CACCTGTCGGGCCAGCCAGAACATCAACATCTGGCTGAGCTGGTATCAGGAGAAACCCGGCA
ACGTGCCCAAGCTGCTGATCTACAAGGCCAGCAATCTGCACACCGGCGTGCCCAGCAGATTT
TCTGGCTCTGGCAGOGGCACCGACTTCACCCTGATCATATCTAGCCTGCAGCCTGAGGATAT
CGCCACCTACTACTGCCTGCAAGGCCAGAGCTACCCCAGAACATTTGGCGGAGGCACCAAGC
TGGAAATCAAGACGGTGGCTGCACCATCTGTOTTCATCTTCCCGCCATCTGATGAGCAGTTG
AAATCTGGAACTGCCTCTGTTGTGTGCCTGCTGAATAACTTCTATCCGAGAGAGGCCAAAGT
ACAGTGGAAGGTGGATAACGCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGGAGG
ACAGCAAGGACAGCACCTACAGCCTCAGGAGCACCCTGACGCTGAGCAAAGCAGACTACGAG
AAACACAAAGTCTACGCCTGCGAAGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAG
CTTCAACAGGGGAGAGTGT
SEQ ID NO: 93
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fab-IgG-Fab (Light Chain)
DIQMNQSPSSLSASLGIDTISITCRASQNINIWLSWYQQKPGNV2KLLTYKASNLHTGVPSRF
SGSGSGTDFTLIISSLUEDIATYYCLQGQSYPRTFGGGTKLEIKTVAAIDSVFIFPPSDEQL
KSGTASVVCLLNNFYPREAKVQWKVDNALOGNSQESVTEQDSKIDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 94
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fab fragment of Fab-IgG-Fab (Light Chain)
DIQMNQSPSSLSASLGDTISITCRASQNINIWLSWYQQKPGNVPKLLIYKASNLHTGVPSRF
SGSGSGTDFTLIISSLUEDIATYYCLQGQSYPRTFGGGTKLEIKTVAAPSVFIFPPSDEQL
KSGTASVVCLLNNFYPREAKVQWKVDNALUGNSQESVTENSKDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSENRGEC
SEQ ID NO: 95
Annotated DNA sequence of Synthetic anti-Salmonella LPS Fab-
Fab-IgG-Fab-Fab(Heavy Chain)
GAAGTGAAGCTGGTGGAAAGCGGCGGAGGACTGGTTCAACCTGGCGGATCTCTGAGCCTGTC
TTGTGCCGCCAGCGGCTTCACCTTCAGCGACTACTACATGACCTGGGTCCGACAGGCCCCTG
GAAAAGCTCCTGAATGGCTGGCCCTGATCCGGAACAAGAGAAACGGCGATACCGCCGAGTAC
AGCGCCTCTGTGAAGGGCAGATTCACCATCAGCCGGGACTACAGCCGGTCCATCCTGCACCT
TCAGATGAACGCCCTGAGAACCGAGGACTCCGCCACCTACTATTGCGTGCGACAAGGCAGAG
GCTACACCCTGGATTATTGGGGCCAGGGCACAAGCGTGACAGTGTCTAGCGCCAGCACAAAG
GGCCCCTCTGTTTTTCCACTGGCTOCCAGGAGCAAGAGCACAAGCGGAGGAACAGCTGCCCT
GGGATGCCTCGTGAAGGACTACTTCCCTGAACCAGTGACCGTGTCCTGGAACTCTGGCGCTC
TGACATCTGGGGTGCACACATTCCCTGCTGTGCTGCAGAGCAGCGGCCTGTATTCTCTGAGC
AGCGTGGTCACAGTGCCCAGCTCTAGTCTGGGCACCCAGACCTACATCTGCAATGTGAACCA
CAAGGCTAGCAACACGAAGGTCGACAAAAAGGTGGAACCCAAGAGCTGCGACAAGACCCATA
CAGGCGGAGGCGGATCTGGTGGTGGTGGATCTGGCGGTGGCGGTTCAGGTGGCGGCGGTAGC
GGAGGTGGTGGTAGTGGTGGTGGCGGCTCTGAAGTGAAACTCGTCGAATCTGGTGGCGGACT
GGTGCAGCCAGGTGGAAGTCTGTCTCTGAGCTGTGCCGCCTCTGGCTTTACCTTCTCTGATT
121
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
ACTATATGACGTGGGTTCGCCAAGCTCCTGGCAAGGCACCAGAATGGCTCGCTCTGATTAGA
AACAAGCGGAATGGCGACACAGCCGAGTATTCCGCCAGCGTGAAAGGCCGGTTCACCATCTC
CAGAGACTACTCCCGCAGCATCCTGCATCTGCAAATGAATGCTCTGCGGACCGAGGACAGCG
CTACCTATTACTGCGTTAGGCAAGGCCGGGGATACACACTGGACTACTGGGGACAAGGCACC
TCCGTGACTGTGTCCTCTGCCTCTACCAAGGGACCCAGCGTGTTCCCACTTGCACCTAGCAG
CAAGTCTACCAGCGGCGGAACTGCCGCTCTCGGATGCCTGGTCAAAGATTATTTCCCCGAAC
CTGTCACCGTCAGCTGGAATAGCGGAGCCCTTACCAGCGGAGTGCATACTTTCCCTGCCGTC
CTCCAGTCATCCGGGCTGTATAGTCTGTCCTCCGTGGTTACCGTGCCAAGCAGCTCTCTGGG
AACACAGACATATATCTGTAATGTCAATCACAAACCCTCCAACACAAAAGTGGACAAAAAAG
TCGAGCCGAAGTCCTGTGATAAGACACACACTGGCGGCGGAGGTTCTGGCGGAGGTGGAAGC
GGAGGCGGTGGCTCAGGCGGCGGTGGCAGTGGCGGAGGCGGTAGCGGCGGAGGCGGTTCTGA
AGTTAAGCTGGTTGAGTCCGGCGGTGGCCTTGTGCAGCCTGGTGGTTCTCTCTCTCTGTCCT
GTGCTGCCTCCGGATTCACCTTTTCCGATTATTACATGACATGGGTTCGACAAGCACCAGGG
AAAGCCCCAGAGTGGCTGGCACTCATCAGAAACAAACGCAACGGGGACACCGCCGAATACTC
TGCCAGTGTCAAAGGCAGGTTTACAATCAGCAGGGATTACTCTCGGAGCATTCTCCACCTCC
AAATGAACGCACTCCGCACAGAGGATAGCGCCACTTACTACTGTGTCCGGCAAGGACGGGGC
TATACCCTCGATTACTGGGGTCAAGGGACATCTGTGACCGTCAGTTCTGCTAGCACCAAGGG
CCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGG
GCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTG
ACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAG
CGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACA
AGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACACA
TGCCCACCGTGCCCAGCACCTGAACTICCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAA
ACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGA
GCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAATGCC
AAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACCGT
CCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTCC
CAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTAC
ACCCTGCCCCCATCCCGGGACGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAA
AGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACT
ACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACC
GTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCT
GCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAAGGATCCGGCGGAGGCG
GATCTGGCGGCGGAGGTAGTGGCGGCGGAGGTTCAGGTGGTGGTGGTAGCGGAGGTGGCGGT
TCTGGCGGTGGTGGAAGCGAAGTGAAGCTGGTGGAATCTGGCGGAGGCCTGGTTCAACCTGG
CGGATCTCTGTCTCTGAGCTGTGCCGCCAGCGGCTTCACCTTCAGCGACTACTACATGACCT
GGGTCCGACAGGCCCCTGGAAAAGCTCCTGAATGGCTGGCCCTGATCCGGAACAAGAGAAAC
GGCGATACCGCCGAGTACAGCGCCTCTGTGAAGGGCAGATTCACCATCAGCCGGGACTACAG
CCGGTCCATCCTGCAC=CAGATGAACGCCCTGAGAACCGAGGATAGCGCCACCTACTACT
GCGTGCGACAAGGCAGAGGCTACACCCTGGATTATTGGGGCCAGGGCACAAGCGTGACAGTG
TCTAGCGCCTCTACAAAGGGCCCCAGCGTTTTCCCACTGGCTCCTAGCAGCAAGAGCACAAG
CGGAGGAACAGCCGCTCTGGGCTGTCTGGTCAAGGACTACTTTCCCGAGCCTGTGACCGTGT
CCTGGAATTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGCAGC
GGCCTGTACTCTCTGAGCAGCGTGGTCACAGTGCCAAGCTCTAGCCTGGGCACCCAGACCTA
CATCTGCAATGTGAACCACAAGCCTAGCAACACCAAGGTGGACAAG.AAGGTGGAACCCAAGA
GCTGCGACAAGACCCATACAGGCGGTGGCGGAAGCGGAGGCGGTGGCTCAGGTGGCGGAGGT
TCTGGCGGAGGCGGCAGTGGTGGTGGTGGCAGTGGTGGCGGAGGATCTGAAGTCAAACTGGT
CGAAAGCGGAGGTGGACTGGTTCAGCCAGGTGGAAGCCTGTCTCTGTCTTGTGCCGCTTCCG
GCTTTACCTTCTCTGATTACTATATGACGTGGGTTCGCCAAGCTCCTGGCAAGGCACCAGAA
TGGCTCGCTCTGATTAGAAACAAGCGGAATGGCGACACAGCCGAGTATTCCGCCAGCGTGAA
AGGCCGGTTCACCATCTCCAGAGACTACTCCCGCAGCATCCTGCATCTGCAAATGAATGCTC
122
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
TGOGGACCGAGGACTCCGCCACATATTACTGTGTCAGACAAGGCOGGGGATACACACTCGAC
TACTGGGGACAGGGAACCTCCGTGACTGTGTCCTCTGCCAGCACAAAGGGGCCCTCCGTGTT
TCCTCTGGCTCCAAGCTCCAAGTCTACCAGCGGTGGAACTGCTGCCCTGGGATGCCTCGTGA
AGGATTACTTCCCAGAACCAGTGACAGTCAGCTGGAACAGCGGAGCCCTGACTTCTGGGGTG
CACACATTCCCTGCCGTCCTGCAATCTTCTGGCCTGTACAGCCTGTCCTCCGTCGTGACCGT
TCCTTCTAGCTCTCTGGGAACACAGACATATATCTGTAATGTCAATCACAAACCCTCCAATA
CGAAGGTCGACAAAAAGGTCGAGCCTAAGTCCTGTGATAAGACCCACACC
SEQ ID NO: 96
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fab-Fab-IgG-Fab-Fab (Heavy Chain)
EVKLVESGGGLVQPGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIRNKRNGDTAEY
SASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVRQGRGYTLDYNGQGTSVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGSGGGGS
GGGGSGGGGSEVKLVESGGGLVUGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIR
NKRNGDTAEYSASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVRWRGYTLDYWGQCT
SVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAV
LOSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGS
GGGGSGGGGSGGGGSGGGGSEVKLVESGGGLVUGGSLSLSCAASGFTFSDYYMTWVRQAPG
KAPEWLALIRNKRNGDTAEYSASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVRQGRG
YTLDYWGQGTSVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGAL
TSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKE'SNTKVDKKVEAKSCDKTHT
CPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNA
KTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGUREPQVY
TLPPSRDELTKNQVSLTCLVKGEYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLT
VDKSRWQQGNVFSCSVMHEALHNHYTOSLSLSPGKGSGGGGSGGGGSGGGGSGGGGSGGGG
SGGGGSEVKLVESGGGLVUGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIRNKRN
GDTAEYSASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVRQGRGYTLDYWGQGTSVTV
SSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSS
GLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGG
SGGGGSGGGGSGGGGSEVKLVESGGGLVUGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPE
WLALIRNKRNGDTAEYSASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVRQGRGYTLD
YWGQGTSVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGV
HTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEMSCDKTHT
SEQ ID NO: 97
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fab fragment of Fab-Fab-IgG-Fab-Fab (Heavy Chain)
EVKLVESGGGLVQPGGSLSLSCAASGFTFSDYYMTWVRQAPGKAPEWLALIRNKRNGDTAEY
SASVKGRFTISRDYSRSILHLQMNALRTEDSATYYCVRQGRGYTLDYWGQGTSVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 98
123
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fa fragment of Fab-Fab-IgG-Fab-Fab (Heavy Chain)
PCPAPELLGGPSVFLFETKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 99
DNA sequence of Synthetic anti-Salmonella LPS Fab-Fab-IgG-Fab-
Fab (Light Chain)
GACATCCAGATGAATCAGAGCCCCAGCAGCCTGTCTGCCAGCCTGGGAGATACCATCAGCAT
CACCTGTCGGGCCAGCCAGAACATCAACATCTGGCTGAGCTGGTATCAGCAGAAACCCGGCA
ACGTGCCCAAGCTGCTGATCTACAAGGCCAGCAATCTGCACACCGGCGTGCCCAGCAGATTT
TCTGGCTCTGGCAGCGGCACCGACTTCACCCTGATCATATCTAGCCTGCAGCCTGAGGATAT
CGCCACCTACTACTGCCTGCAAGGCCAGAGCTACCCCAGAACATTTGGCGGAGGCACCAAGC
TGGAAATCAAGACGGTGGCTGCACCATCTGTCTTCATCTTCCCGCCATCTGATGAGCAGTTG
AAATCTGGAACTGCCTCTGTTGTGTGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGT
ACAGTGGAAGGTGGATAACGCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGG
ACAGCAAGGACAGCACCTACAGCCTCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAG
AAACACAAAGTCTACGCCTGCGAAGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAG
CTTCAACAGGGGAGAGTGT
SEQ ID NO: 100
Annotated Amino Amid sequence of Synthetic anti-Salmonella LPS
Fab-Fab-IgG-Fab-Fab (Light Chain)
DIQMNOPSSLSASLGDTISITCRASOINIWLSWYQOPGNVPKLLIYKASNLHTGVPSRF
SGSGSGTEFTLIISSLQPEDIATYYCLQGQSYPRTFGGGTKLEIKTVAAPSVFIFPPSDEQL
KSGTASVVOLLNNFYPREAKVQWKVDNALOGNSQESVTEWSKDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 101
Annotated Amino Acid sequence of Synthetic anti-Salmonella LPS
Fab fragment of Fab-Fab-IgG-Fab-Fab (Light Chain)
DIQMNQSPSSLSASLGDTISITCRASQNINIWLSWYQQKPGNVPKLLIYKASNLHTGVPSRF
SGSGSGTDFTLIISSLQPEDIATYYCLQGQSYPRTFGGGTKLEIKTVAAPSVFIFPPSDEQL
KSGTASVVULNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKIDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 102
Annotated DNA sequence of anti-Gonorrhea (2C7) IgG (Heavy
Chain)
GAGGTTCAGCTGCAGCAGTCTGGACCTGAGCTGGTCAAGCCTGGCAGCAGCGTGAAGATCAG
124
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
CTGTAAAGGCAGCGGCTACACCTTCACCGACTACAACATGGAATGGGTCAAGCAGAGCCACG
GCAAGAGCCTGGAATGGATCGGCGTGATCAACCCCAACAACCGGTTCACCAGCTACAACCAG
AACTTCAGAGGCAAGGCCACACTGACCGTGGACAAGAGCAGCAGCACCGCCTACATGGATCT
GAGAAGCCTGACCAGCGACGACAGCGCCGTGTATTTTTGTGCCGGCAGCCGGIGGTATCAGT
ACGACTATTGGGGCCAGGGCACAACCCTGACCGTTAGCTCTGCTAGCACCAAGGGCCCATCG
GTCTTCCOCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCT
GGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCG
GCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGGAGCGTGGTG
ACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAG
CAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACACATGCCCAC
CGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTOTTCCTCTTCCCOCCAAAACCCAAG
GACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGA
AGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAATGCCAAGACAA
AGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTOCTCACCGTCCTGCAC
CAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCAGCCCC
CATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCTGC
CCCCATCCOGGGACGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAAAGGCTTC
TATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGAC
CACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGACA
AGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAAC
CACTACACGCAGAAGAGCCTCTOCCTGTOTCCGGGTAAA
SEQ ID NO: 103
Annotated Amino Acid sequence of anti-Gonorrhea (2C7) IgG
(Heavy Chain)
EVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFTSYNQ
NFRGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYQYDYWGQGTTLTVSSASTKGPS
VFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSGLYSLSSVV
TVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPK
DTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLH
QDWINGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGF
YPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHN
HYTQKSLSLSPGK
SEQ ID NO: 104
Annotated Amino Acid sequence of Fab fragment of anti-
Gonorrhea (2C7) (Heavy Chain)
EVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFTSYNQ
NERGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYQYDYWGQGTTLTVSSASTKGPS
VFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSGLYSLSSVV
TVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 105
Annotated Amino Acid sequence of Fc fragment of anti-Gonorrhea
(2C7) (Heavy Chain)
125
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
PCPAPELLGGPSVELFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHOWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGUENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 106
DNA sequence of anti-Gonorrhea (2C7) (Light Chain)
CAGGTGGTGGTCACACAAGAGAGCGCCCTGACAACAAGCCCTGGCGAGACAGTGACCCTGAC
CTGCAGATCTTCTACAGGCGCCGTGACCACCTCCAACTACGCCAATTGGGTGCAAGAGAAGC
CCGACCACCTGTTCACAGGCCTGATCGGCGGCATCAACAATAGAGCACCTGGCGTGCCAGCC
AGATTCAGOGGATCTCTGATCGGAGACAAGGCCGCACTGACAATCACAGGCGCCCAGACAGA
GGACGAGGCCATCTACTTTTGCGCCCTGTGGTACAGCAACCACTGGGTTTTCGGCGGAGGCA
CCAAGCTGACAGTTCTGGGCCAACCTAAGGCCGCTCCTAGCGTGACACTGTTCCCTCCAAGC
AGCGAAGAACTGCAGGCCAACAAGGCCACACTCGTGTGCCTGATCAGCGACTTTTATCCTGG
CGCCGTGACCGTGGCCTGGAAGGCTGATAGTTCTCCTGTGAAGGCCGGCGTGGAAACCACCA
CACCTAGCAAGCAGAGCAACAACAAATACGCCGCCAGCAGCTACCTGAGCCTGACACCTGAG
CAGTGGAAGTOCCACAGATCCTACAGCTGCCAAGTGACCCACGAGGGCAGCACCGTGGAAAA
AACAGTGGCCCCTACCGAGTGCAGC
SEQ ID NO: 107
Annotated Amino Acid sequence of anti-Gonorrhea (2C7) (Light
Chain)
QVVVTQESALTTSPGETVTLTCRSSTGAVTTSNYANWVQEKPDHLFTGLIGGINNRAPGVPA
RFSGSLIGDKAALTITGAQTEDEATYFCALWYSNHWVFGGGTKLTVLGUKAAPSVTLFPPS
SEELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPE
QWKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 108
Annotated Amino Acid sequence of Fab fragment of anti-
Gonorrhea (2C7) IgG (Light Chain)
QVVVTQESALTTSPGETVTLTCRSSTGAVTTSNYANWVQEKPDHLFTGLIGGINNRAPGVPA
RFSGSLIGDKAALTITGAQTEDEAIYFCALWYSNHWVFGGGTKLTVLGQPKAAPSVTLFPPS
SEELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPE
QWKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 109
Annotated DNA sequence of Synthetic anti-Gonorrhea (2C7) Fab-
IgG (Heavy Chain)
GAGGTACAACTGCAACAGAGTGGCCCCGAGCTTGTGAAGCCAGGGTCCAGCGTGAAGATTTC
TTGCAAGGGAAGTGGGTACACGTTCACGGACTACAACATGGAGTGGGTGAAACAAAGTCACG
GTAAATCCTTGGAGTGGATCGGAGTTATCAACCCAAACAACCGATTTACTAGCTACAACCAG
AATTTCAGGGGGAAGGCAACACTCACCGTCGACAAATCCTOTTCTACGGCATATATGGATCT
126
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
*
CCGCTCACTTACTAGCGAGGACTCTGCAGTCTATTTTTGCGCGGGGAGCCGATGGTATCAAT
ACGACTATTGGGGTCAAGGTACAACGCTTACTGTTAGCTCAGCTAGCACCAAGGGCCCATCG
GTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCT
GGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCG
GCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTG
ACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAG
CAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACACAGGTGGCG
GAGGATCTGGTGGAGGAGGGAGCGGAGGGGGTGGGTCCGGAGGGGGTGGTTCCGGAGGTGGT
GGATCAGGTGGCGGAGGAAGTGAGGTTCAGCTGCAGCAGTCTGGACCTGAGCTGGTCAAGCC
TGGCAGCAGCGTGAAGATCAGCTGTAAAGGCAGCGGCTACACCTTCACCGACTACAACATGG
AATGGGTCAAGCAGAGCCACGGCAAGAGCCTGGAATGGATCGGCGTGATCAACCCCAACAAC
CGGTTCACCAGCTACAACCAGAACTTCAGAGGCAAGGCCACACTGACCGTGGACAAGAGCAG
CAGCACCGCCTACATGGATCTGAGAAGCCTGACCAGCGAGGACAGCGCCGTGTATTTTTGTG
CCGGCAGCCGGTGGTATCAGTACGACTATTGGGGCCAGGGCACAACCCTGACCGTTAGCTCT
GCTAGCACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGG
CACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGA
ACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTC
TACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTG
CAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTG
ACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTC
CTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGT
GGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGG
AGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTC
AGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTC
CAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAG
AACCACAGGTGTACACCCTGCCCCCATCCCGGGACGAGCTGACCAAGAACCAGGTCAGCCTG
ACCTGCCTGGTCAAAGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCA
GCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCT
ACAGCAAGCTCACCGTGGACAAGAGGAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTG
ATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAA
SEQ ID NO: 110
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) Fab-IgG (Heavy Chain)
EVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFTSYNQ
NFRGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYODYWGQGTTLTVSSASTKGPS
VFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFRAVLQSSGLYSLSSVV
TVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGSGGGGSGGG
GSGGGGSEVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINLDNN
RFTSYNQNFRGKATLTVIDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYQYDYWGQGTTLTVSS
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVF
LFPPKPKDTLMISRTPEVTCVVVIDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVV
SVLTVLHOWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLETSRDELTKNQVSL
TCLVKGFYPSDIAVEWESNGUENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSV
MHEALHNHYTQKSLSLSPGK
SEQ ID NO: 111
127
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Annotated Amino Acid sequence of Synthetic anti -Gonorrhea
(2C7) Fab fragment of Fab-IgG (Heavy Chain)
EVOLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINMNRFTSYNQ
NFRGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYQYDYWGQGTTLTVSSASTKGPS
VFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVV
TVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 112
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) Fc fragment of Fab-IgG (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 113
DNA sequence of Synthetic anti-Gonorrhea (2C7) Fab-IgG (Light
Chain)
CAGGTGGTGGTCACACAAGAGAGCGCCCTGACAACAAGCCCTGGCGAGACAGTGACCCTGAC
CTGCAGATCTTCTACAGGCGCCGTGACCACCTCCAACTACGCCAATTGGGTGCAAGAGAAGC
CCGACCACCTGTTCACAGGCCTGATCGGCGGCATCAACAATAGAGCACCTGGCGTGCCAGCC
AGATTCAGCGGATCTCTGATCGGAGACAAGGCCGCACTGACARTCACAGGCGCCCAGACAGA
GGACGAGGCCATCTACTTTTGCGCCCTGTGGTACAGCAACCACTGGGTTTTCGGCGGAGGCA
CCAAGCTGACAGTTCTGGGCCAACCTAAGGCCGCTCCTAGCGTGACACTGTTCCCTCCAAGC
AGCGAAGAACTGCAGGCCAACAAGGCCACACTCGTGTGCCTGATCAGCGACTTTTATCCTGG
CGCCGTGACCGTGGCCTGGAAGGCTGATAGTTCTCCTGTGAAGGCCGGCGTGGAAACCACCA
CACCTAGCAAGCAGAGCAACAACAAATACGCCGCCAGCAGCTACCTGAGCCTGACACCTGAG
CAGTGGAAGTCCCACAGATCCTACAGCTGCCAAGTGACCCACGAGGGCAGCACCGTGGAAAA
AACAGTGGCCCCTACCGAGTGCAGC
SEQ ID NO: 114
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) Fab-IgG (Light Chain)
QVVVTQESALTTSPGETVTLTCRSSTGAVTTSNYANWVQEKPDHLFTGLIGGINNRAPGVPA
RFSGSLIGDKAALTITGAQTEDEAIYFCALWYSNHWVEGGGTKLTVLGUKAAPSVTLFPPS
SEELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPE
QWKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 115
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) Fab fragment of Fab-IgG (Light Chain)
128
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
QVVVTQESALTT S PGETVT LT CRSST GAVT T SNYANWVQEKP DHL FT GL I GG INNRA PGVPA
RFSGSLIGDKAALTITGAQTEDEAIYFCALWYSNHWVFGGGTKLTVLGUKAAPSVTLFPPS
SEELQANKATINCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPE
QWKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 116
Annotated DNA sequence of Synthetic anti-Gonorrhea (2C7) IgG-
Fab (Heavy Chain)
GAGGTTCAGCTGCAGCAGTCTGGACCTGAGCTGGTCAAGCCTGGCAGCAGCGTGAAGATCAG
CTGTAAAGGCAGCGGCTACACCTTCACCGACTACAACATGGAATGGGTCAAGCAGAGCCACG
GCAAGAGCCTGGAATGGATCGGCGTGATCAACCCCAACAACCGGTTCACCAGCTACAACCAG
AACTTCAGAGGCAAGGCCACACTGACCGTGGACAAGAGCAGCAGCACCGCCTACATGGATCT
GAGAAGCCTGACCAGCGAGGACAGCGCCGTGTATTTTTGTGCCGGCAGCCGGTGGTATCAGT
ACGACTATTGGGGCCAGGGCACAACCCTGACCGTTAGCTCTGCTAGCACCAAGGGCCCATCG
GTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCT
GGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCG
GCGTGCACACCTTCCOGGCTGTCCTACAGTCCTCAGGACTCTACTOCCTCAGCAGCGTGGTG
ACCGTGCCCTCCAGCAGOTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAG
CAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACACATGCCCAC
CGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTOTTCCCCCCAAAACCCAAG
GACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGA
AGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAATGCCAAGACAA
AGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCAC
CAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCAGCCCC
CATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCTGC
CCCCATCCCGGGACGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAAAGGCTTC
TATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATCGGCAGCCGGAGAACAACTACAAGAC
CACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGACA
AGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAAC
CACTACACGCAGAAGAGCCTCTOCCTGTCTCCGGGTAAAGGATCCGGTGGCGGAGGATCTGG
TGGAGGAGGGAGCGGAGGGGGTGGGTCCGGAGGGGGTGGTTCCGGAGGTGGTGGATCAGGTG
GCGGAGGAAGTGAGGTACAACTGCAACAGAGTGGCCCCGAGCTTGTGAAGCCAGGGTCCAGC
GTGAAGATTTCTTGCAAGGGAAGTGGGTACACGTTCACGGACTACAACATGGAGTGGGTGAA
ACAAAGTCACGGTAAATCCTTGGAGTGGATCGGAGTTATCAACCCAAACAACCGATTTACTA
GCTACAACCAGAATTTCAGGGGGAAGGCAACACTCACCGTCGACAAATCCTCTTCTACGGCA
TATATGGATCTCCGCTCACTTACTAGCGAGGACTCTGCAGTCTATTTTTGCGCGGGGAGCCG
ATGGTATCAATACGACTATTGGGGTCAAGGTACAACGCTTACTGTTAGCTCAGCTAGCACCA
AGGGCCCATCGGTCTTCCCCOTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGOGGCC
CTGGGCTOCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGC
CCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCA
GCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAAT
CACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCA
CACA
SEQ ID NO: 117
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) IgG-Fab (Heavy Chain)
129
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
EVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFTSYNQ
NFRGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYQYDYWGQGTTLTVSSASTKGPS
VFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSGLYSLSSVV
TVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPK
DTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLH
QDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGF
YPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHN
HYTQKSLSLSPGKGSGGGGSGGGGSGGGGSGGGGSGGCGSGGGGSEVQLQQSGPELVKPGSS
VKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFTSYNQNFRGKATLTVDKSSSTA
YMDLRSLTSEDSAVYFCACSRWYQYDYWGQGTTLTVSSASTKGPSVFPLAPSSKSTSGGTAA
LGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSGLYSLSSVVTVPSSSLGTQTYICNVN
HKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 118
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) Fab fragment of IgG-Fab (Heavy Chain)
EVQLQQSGPELVKPCSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFTSYNQ
NFRGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYQYDYWGQGTTLTVSSASTKGPS
VFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSGLYSLSSVV
TVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 119
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) Fc fragment of IgG-Fab (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNOSLTCLVKGFYPSDIAVEWESNCUENNYKTTPPVLDSDGSETLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 120
DNA sequence of Synthetic anti-Gonorrhea (2C7) IgG-Fab (Light
Chain)
CAGGTGGTGGTCACACAAGAGAGCGCCCTGACAACAAGCCCTGGCGAGACAGTGACCCTGAC
CTGCAGATCTTCTACAGGCGCCGTGACCACCTCCAACTACGCCAATTGGGTGCAAGAGAAGC
CCGACCACCTGTTCACAGGCCTGATCGGCGGCATCAACAATAGAGCACCTGGCCTGCCAGCC
AGATTCAGOGGATCTCTGATCGGAGACAAGGCCGCACTGACAATCACAGGCGCCCAGACAGA
GGACGAGGCCATCTACTTTTGCGCCCTCTGGTACAGCAACCACTGGGTTTTCGGCCGAGGCA
CCAAGCTGACAGTTCTGGGCCAACCTAAGGCCGCTCCTAGCGTGACACTGTTCCCTCCAAGC
AGCGAAGAACTGCAGGCCAACAAGGCCACACTCGTGTGCCTGATCAGCCACTTTTATCCTGG
CGCCGTGACCGTGGCCTGGAAGGCTGATAGTTCTCCTGTGAAGGCCGGCGTGGAAACCACCA
CACCTAGCAACCAGAGCAACAACAAATACGCCGCCAGCAGCTACCTGAGCCTGACACCTGAG
CAGTGGAAGTCCCACAGATCCTACAGCTGCCAAGTGACCCACGAGGGCAGCACCGTGGAAAA
AACAGTGGCCCCTACCGAGTGCAGC
SEQ ID NO: 121
130
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) IgG-Fab (Light Chain)
QVVVTQESALTTSPGETVTLTCRSSTGAVTTSNYANWVQEKPDHLFTGLIGGINNRAPGVPA
RFSGSLIGDKAALTITGAQTEDEAIYFCALWYSNHWVFGGGTKLTVLGUKAAPSVTLFPPS
SEELOANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPE
OWKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 122
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) Fab fragment of IgG-Fab (Light Chain)
OVVTQESALTTSPGETVTLTCRSSTGAVTTSNYANWVQEKPDHLFTGLIGGINNRAPGVPA
RFSGSLIGOKAALTITGAQTEDEAIYFCALWYSNHWVFGGGTKLTVLGQPKAAPSVTLFETS
SEELQANKATINCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKUNNKYAASSYLSLTET
ONKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 123
Annotated DNA sequence of Synthetic anti-Gonorrhea (2C7) Fab-
IgG-Fab (Heavy Chain)
GAGGTACAACTGCAACAGAGTGGCCCCGAGOTTGTGAAGCCAGGGTCCAGCGTGAAGATTTC
TTGCAAGGGAAGTGGGTACACGTTCACGGACTACAACATGGAGTGGGTGAAACAAAGTCACG
GTAAATCCTTGGAGTGGATCGGAGTTATCAACCCAAACAACCGATTTACTAGCTACAACCAG
AATTTCAGGGGGAAGGCAACACTCACCGTCGACAAATCCTCTTCTACGGCATATATGGATCT
CCGCTCACTTACTAGCGAGGACTCTGCAGTCTATTTTTGCGCGGGGAGCCGATGGTATCAAT
ACGACTATTGGGGTCAAGGTACAACGCTTACTGTTAGCTCAGCTAGCACCAAGGGCCCATCG
GTCTTCCOCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCT
GGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCG
GCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTG
ACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAG
CAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACACAGGTGGCG
GAGGATCTGGTGGAGGAGGGAGCGGAGGGGGTGGGTCCGGAGGGGGTGGTTCCGGAGGTGGT
GGATCAGGTGGCGGAGGAAGTGAGGTTCAGCTGCAGCAGTCTGGACCTGAGCTGGTCAAGCC
TGGCAGCAGCGTGAAGATCAGCTGTAAAGGCAGCGGCTACACCTTCACCGACTACAACATGG
AATGGGTCAAGCAGAGCCACGGCAAGAGCCTGGAATGGATCGGCGTGATCAACCCCAACAAC
CGGTTCACCAGCTACAACCAGAACTTCAGAGGCAAGGCCACACTGACCGTGGACAAGAGCAG
CAGCACCGCCTACATGGATCTGAGAAGCCTGACCAGCGAGGACAGCGCCGTGTATTTTTGTG
CCGGCAGCCGGTGOTATCAGTACGACTATTGGGGCCAGGGCACAACCCTGACCGTTAGCTCT
GCTAGCACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGG
CACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGA
ACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTC
TACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTG
CAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTG
ACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTC
CTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGT
GGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGG
AGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTC
AGCGTOCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTC
CAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAG
131
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
AACCACAGGTGTACACCCTGCCCCCATCCCGGGACGAGCTGACCAAGAACCAGGTCAGCCTG
ACCTGCCTGGTCAAAGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCA
GCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCT
ACAGCAAGCTCACCGTGGACAAGAGCAGGTGGCACCAGGGGAACGTCTTCTCATGCTCCGTG
ATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAAGG
ATCCGGTGGCGGAGGATCTGGTGGAGGAGGGAGCGGAGGGGGTGGGTCCGGAGGGGGTCGTT
CCGGAGGTGGTGGATCAGGTGGCGGAGGAAGTGAGGTACAACTGCAACAGAGTGGCCCCGAG
CTTGTGAAGCCAGGGTCCAGCGTGAAGATTTCTTGCAAGGGAAGTGGGTACACGTTCACGGA
CTACAACATGGAGTGGGTGAAACAAAGTCACGGTAAATCCTTGGAGTGGATCGGAGTTATCA
ACCCAAACAACCGATTTACTAGCTACAACCAGAATTTCAGGGGGAAGGCAACACTCACCGTC
GACAAATCCTCTTCTACGGCATATATGGATCTCCGCTCACTTACTAGCGAGGACTCTGCAGT
CTATTTTTGCGCGGGGAGCCGATGGTATCAATACGACTATTGGGGTCAAGGTACAACGCTTA
CTGTTAGCTCAGCTAGCACCAAGGGCCCATCGGTCTTCCOCCTGGCACCCTCCTCCAACACC
ACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGAC
GGTGTCGTGGAACTCAGGCGCCCTGACCAGOGGCGTGCACACCTTCCCGGCTGTCCTACAGT
CCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAG
ACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCC
CAAATCTTGTGACAAAACTCACACA
SEQ ID NO: 124
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) Fab-IgG-Fab (Heavy Chain)
EVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFTSYNQ
NFRGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYUDYWGQGTTLTVSSASTKGPS
VFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVV
TVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGCGSGGGGSGGG
GSGGGGSEVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHCKSLEWIGVINPNN
RFTSYNQNFRGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYQYDYWGQGTTLTVSS
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLUSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVF
LFETKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVV
SVLTVLHQDWINGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSL
TCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSV
MHEALHNHYTQKSLSLSPGKGSGGGGSGGGGSGGGGSGGGGSGGGGSGGGCSEVQLQQSGPE
LVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFTSYNQNFRGKATLTV
DKSSSTAYMDLRSLTSEDSAVYFCAGSRWYWDYWGQGTTLTVSSASTKGPSVFPLAPSSKS
TSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQ
TYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 125
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) Fab fragment of Fab-IgG-Fab (Heavy Chain)
EVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFTSYNQ
NFRGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYQYDYWGQGTTLTVSSASTKGPS
VFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVV
TVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
132
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SEQ ID NO: 126
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) Fc fragment of Fab-IgG-Fab (Heavy Chain)
PCPAPELLGGPSVFLEPPKPECTLMISRTPEVTCVVVINSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPTEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 127
DNA sequence of Synthetic anti-Gonorrhea (2C7) Fab-IgG-Fab
(Light Chain)
CAGGTGGTGGTCACACAAGAGAGCGCCCTGACAACAAGCCCTGGCGAGACAGTGACCCTGAC
CTGCAGATCTTCTACAGGCGCCGTGACCACCTCCAACTACGCCAATTGGGTGCAAGAGAAGC
CCGACCACCTGTTCACAGGCCTGATCGGCGGCATCAACAATAGAGCACCTGGCGTGCCAGCC
AGATTCAGOGGATCTCTGATCGGAGACAAGGCCGCACTGACAATCACAGGCGCCCAGACAGA
GGACGAGGCCATCTACTTTTGCGCCCTGTGGTACAGCAACCACTGGGTTTTCGGCGGAGGCA
CCAAGCTGACAGTTCTGGGCCAACCTAAGGCCGCTCCTAGCGTGACACTGTTCCCTCCAAGC
AGCGAAGAACTGCAGGCCAACAAGGCCACACTCGTGTGCCTGATCAGCGACTTTTATCCTGG
CGCCGTGACCGTGGCCTGGAAGGCTGATAGTTCTCCTGTGAAGGCCGGCGTGGAAACCACCA
CACCTAGCAAGCAGAGCAACAACAAATACGCCGCCAGCAGCTACCTGAGCCTGACACCTGAG
CAGTGGAAGTCCCACAGATCCTACAGCTGCCAAGTGACCCACCAGGGCAGCACCGTGGAAAA
AACAGTGGCCCCTACCGAGTGCAGC
SEQ ID NO: 128
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) Fab-IgG-Fab (Light Chain)
QVVVTQESALTTSPGETVTLTCRSSTGAVTTSNYANWVQEKPDHLFTGLIGGINNRAPGVPA
RFSGSLIGDKAALTITGAQTEDEAIYFCALWYSNHWVFGGGTKLTVLGUKAARSVTLFPPS
SEELQANKATLVCLISOFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPE
QWKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 129
Annotated Amino Acid sequence of Synthetic anti-Gonorrhea
(2C7) Fab fragment of Fab-IgG-Fab (Light Chain)
QVVVTQESALTTSPGETVTLTCRSSTGAVTTSNYANWVQEKPDHLFTGLIGGINNRAPGVPA
RFSGSLIGDKAALTITGAQTEDEATYFCALWYSNHWVEGGGTKLTVLGOKAAPSVTLFPPS
SEELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPE
QWKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 130
Forward primer
TAAGCAGGTACCGCCACCATGAAGTG
133
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SEQ ID NO: 131
Reverse primer
TGCTTAGCTAGCTGGAGAAACTGTC
SEQ ID NO: 132
Annotated DNA seq-uence of anti-RSV (Respiratory syncytia1
virus) IgG (Heavy Chain)
CAAGTGACCCTGAGAGAGTCTGGCCCCGCTCTGGTTAAGCCCACACAGACCCTGACACTGAC
CTGCACCTTCAGCGGCTTTAGCCTGTCTACAGCCGGCATGAGCGTOGGCTGGATTAGACAGC
CTCCTGGCAAAGCCCTGGAATGGCTGGCCGACATTTGGTGGGACGACAAGAAGCACTACAAC
CCCAGCCTGAAGGACCGGCTGACCATCAGCAAGGACACCAGCAAGAACCAGGTGGTGCTGAA
AGTGACCAACATGGACCCTGCCGACACCGCCACCTACTACTGCGCCAGAGACATGATCTTCA
ACTTCTACTTCGACGTGTGGGGCCAGGGCACCACCGTGACAGTTAGCTCTGCCTCTACAAAG
GGCCCCAGCGTGTTCCCTCTGGCTCCTAGCAGCAAGTCTACAAGCGGAGGAACAGCCGCTCT
GGGCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCCTGGAATTCTGGCGCTC
TGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGCAGCGGCCTGTACTCTCTGAGC
AGCGTGGTCACAGTGCCAAGCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGAACCA
CAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCACA
CCTGTCCTCCATGTCCTGCTCCAGAACTGCTCGGCGGACCTTCCGTGTTTCTGTTCCCTCCA
AAGCCTAAGGACACCCTGATGATCAGCAGAACCCCTGAAGTGACCTGCGTGGTGGTGGATGT
GTCCCACGAGGATCCCGAAGTGAAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACG
CCAAGACCAAGCCTAGAGAGGAACAGTACAACAGCACCTACAGAGTGGTGTCTGTGCTGACC
GTGCTGCACCAGGATTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGCCCT
GCCTGCTCCTATCGAGAAAACCATCTCCAAGGCCAAGGGCCAGCCTAGGGAACCCCAGGTTT
ACACACTGCCTCCAAGCAGGGACGAGCTGACCAAGAATCAGGTGTCCCTGACCTGCCTCGTG
AAGGGCTTCTACCOTTCCGATATCGCCGTGGAATGGGAGAGCAATGGCCAGCCTGAGAACAA
CTACAAGACAACCCCTCCTGTGCTGGACAGCGACGGCTCATTOTTCCTGTACAGCAAGCTGA
CAGTGGACAAGTCCAGATGGCAGCAGGGCAACGTGTTCAGCTGCAGCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGAGCCTGTCTCCAGGCAAG
SEQ ID NO: 133
Annotated Amino Acid sequence of anti-RSV (Respiratory
syncytial virus) IgG (Heavy Chain)
QVTLRESGPALVKPTQTLTLTCTFSGESLSTAGMSVGWIRUPGKALEWLADIWWDDKKHYN
PSLKDRLTISKDTSKNQVVLKVTNMDPADTATYYCARDMIFNFYFDVWGQGTTVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPP
KPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLT
VLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLV
KGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEA
LHNHYTQKSLSLSPGK
SEQ ID NO: 134
134
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Annotated Amino Acid sequence of Fab fragment of anti-RSV
(Respiratory syncytial virus) (Heavy Chain)
QVTLRESGPALVKPTQTLTLTCTFSGFSLSTAGMSVGWIRUPGKALEWLADIWWDDKKHYN
PSLKDRLTISKDTSKNQVVLKVTNMDPADTATYYCARDMIFNFYFDVWGQGTTVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 135
Annotated Amino Acid sequence of Fc fragment of anti-RSV
(Respiratory syncytial virus) (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHOWLNGKEYKCKVSNKALPAPIEKTISKAKGUREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSIDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 136
DNA sequence of anti-RSV (Respiratory syncytial virus) (Light
Chain)
GACATCCAGATGACACAGAGCCCCAGCACACTGTCTGCCAGCGTGGGAGACAGAGTGACCAT
CACATGTAGCGCCAGCAGCAGAGTGGGCTACATGCACTGGTATCAGCAGAAGCCTGGCAAGG
CCCCTAAGCTGCTGATCTACGACACAAGCAAGCTGGCCTCTGGCGTGCCCAGCAGATTTTCT
GGCTCTGGCAGCGGCACCGAGTTCACCCTGACCATCTCTAGCCTGCAGCCTGACGACTTCGC
CACCTACTACTGCTTTCAAGGCAGCGGCTACCCOTTCACCTTTGGCGGCGGAACAAAGGTGG
AAATCAAGCGGACAGTGGCCGCTCCTAGCGTGTTCATCTTTCCACCTAGCGACGAGCAGCTG
AAGTCTGGCACAGCCTCTGTCGTGTGCCTGCTCAACAACTTCTACCCCAGAGAAGCCAAGGT
GCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAATAGCCAAGAGAGCGTGACCGAGCAGG
ACAGCAAGGACTCTACCTACAGCCTGTCCTCCACACTGACCCTGAGCAAGGCCGACTACGAG
AAGCACAAAGTGTACGCCTGCGAAGTGACCCACCAGGGCCTTTCTAGCCCTGTGACCAAGAG
CTTCAACCGGGGCGAGTGC
SEQ ID NO: 137
Annotated Amino Acid sequence of anti-RSV (Respiratory
syncytial virus) (Light Chain)
DIQMTQSPSTLSASVGDRVTITCSASSRVGYMHWYQQKPGKAPKIALIYDTSKLASGVPSRFS
GSGSGTEFTLTISSLQPDDFATYYCFQGSGYPFTFGGGTKVEIKRTVAAPSVFIFPPSDEQL
KSGTASVVCLLNNFYPREAKVQWKVDNALOGNSQESVTEQDSKDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 138
Annotated Amino Acid sequence of Fab fragment of anti-RSV
(Respiratory syncytial virus) IgG Might Chain)
DIQMTOPSTLSASVGDRVTITCSASSRVGYMHWYQQKPGKAPKILIYDTSKLASGVPSRFS
GSGSGTEFTLTISSLUDDFATYYCFQGSGYPFTFGGGTKVEIKRTVAAPSVFIFPPSDEQL
135
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
KSGTASVVCLLNNFYPREAKVQWKVDNALUGNSQESVTEQDSKDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 139
Annotated DNA sequence of Synthetic anti-RSV (Respiratory
syncytial virus) Fab-IgG (Heavy Chain)
CAAGTGACCCTGAGAGAGTCTGGCCCCGCTCTGGTTAAGCCCACACAGACCCTGACACTGAC
CTGCACCTTCAGCGGCTTTAGCCTGTOTACAGCCGGCATGAGCGTOGGCTGGATTAGACAGC
CTCCTGGCAAAGCCCTGGAATGGCTGGCCGACATTTGGTGGGACGACAAGAAGCACTACAAC
CCCAGCCTGAAGGACCGGCTGACCATCAGCAAGGACACCAGCAAGAACCAGGTGGTGCTGAA
AGTGACCAACATGGACCCTGCCGACACCGCCACCTACTACTGCGCCAGAGACATGATCTTCA
ACTTCTACTTCGACGTGTGGGGCCAGGGCACCACCGTGACAGTTAGCTCTGCCTCTACAAAG
GGCCCCAGCGTGTTCCCTCTGGCTCCTAGCAGCAAGTOTACAAGCGGAGGAACAGCCGCTCT
GGGCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCCTGGAATTCTGGCGCTC
TGACAAGOGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGCAGCGGCCTGTACTCTCTGAGC
AGCGTGGTCACAGTGCCAAGCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGAACCA
CAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCATA
CAGGCGGCGGAGGATCTGGCGGAGGTGGAAGCGGAGGCGGAGGAAGCGGTGGCGGCGGTAGT
GGCGGTGGTGGTTCAGGTGGTGGTGGCTCTCAAGTCACACTGAGAGAAAGCGGCCCTGCTCT
CGTGAAGCCTACTCAGACACTCACCCTGACCTGTACATTCTCTGGCTTCAGCCTGAGCACCG
CCGGCATGTCTGTTGGATGGATCAGACAACCACCAGGCAAGGCTCTCGAGTGGCTCGCTGAT
ATTTGGTGGGATGATAAGAAACATTATAACCCATCTCTCAAGGACCGCCTCACAATCTCCAA
GGATACCTCCAAGAATCAGGTCGTCCTCAAAGTCACGAATATGGATCCCGCCGATACGGCCA
CATATTACTGTGCCCGGGATATGATCTTTAATTTCTATTTTGATGTCTGGGGCCAAGGGACA
ACCGTCACCGTGTCTAGCGCCAGCACAAAGGGACCCTCCGTGTTTCCACTGGCACCCAGCTC
TAAGAGCACCTCTGGTGGAACAGCTGCCCTGGGATGTCTCGTGAAAGACTACTTCCCCGAAC
CAGTGACAGTCAGCTGGAACAGCGGAGCCCTGACTTCTGGGGTGCACACATTCCCTGCCGTC
CTGCAATCTTCTGGCCTGTACAGCCTGTCCAGCGTCGTGACCGTTCCTTCTAGCTCTCTGGG
AACACAGACATATATCTGTAATGTCAATCACAAACCCTCCAATACGAAGGTCGACAAAAAGG
TCGAGCCTAAGTCCTGTGATAAGACCCACACCTGTCCTCCATGTCCTGCTCCAGAACTGCTC
GGCGGACCTTCTGTGTTTCTGTTCCCTCCAAAGCCTAAGGACACCCTGATGATCAGCAGAAC
CCCTGAAGTGACCTGCGTGGTGGTCGATGTGTCCCACGAGGATCCCGAAGTGAAGTTCAATT
GGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCTAGAGAGGAACAGTACAAC
AGCACCTACAGAGTGGTGTCTGTGCTGACCGTGCTGCACCAGGATTGGCTGAACGGCAAAGA
GTACAAGTGCAAGGTGTCCAACAAGGCCCTGCCTGCTCCTATCGAGAAAACCATCTCTAAGG
CCAAGGGCCAGCCTCGCGAACCCCAGGTTTACACACTTCCACCAAGCCGGGACGAGCTGACA
AAAAACCAGGTGTCCCTGACATGCCTCGTGAAGGGCTTCTACCCCTCCGATATCGCCGTGGA
ATGGGAGAGCAATGGCCAGCCTGAGAACAACTACAAGACCACACCTCCTGTGCTGGACAGCG
ACGGCTCATTCTTCCTGTACTCCAAGCTGACAGTGGACAAGTCCAGATGGCAGCAGGGCAAC
GTGTTCAGCTGCAGCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAAAGCCTGTC
TCTGAGCCCCGGCAAG
SEQ ID NO: 140
Annotated Amino Acid sequence of Synthetic anti-RSV
(Respiratory syncytial virus) IgG Fab-IgG (Heavy Chain)
QVTLRESGPALVKPTQTLTLTCTFSGFSLSTAGMSVGWIRQPPGKALEWLADIWWDDKKHYN
PSLKDRLTISKDTSKNQVVLKVTNMDPADTATYYCARDMIFNEYFDVWGQGTTVTVSSASTK
136
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGSGGGGS
GGGGSGGGGSQVTLRESGPALVKPTQTLTLTCTFSGFSLSTAGMSVGWIRQPPGKALEWLAD
IWWDDKKHYNPSLKDRLTISKDTSKNQVVLKVTNMDPADTATYYCARDMIFNFYFDVWGQGT
TVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAV
LQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCIWTHTCPPCPAPELL
GGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYN
STYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELT
KNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGN
VFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 141
Annotated Amino Acid sequence of Synthetic anti-RSV
(Respiratory syncytia1 virus) Fab fragment of Fab-IgG (Heavy
Chain)
QVTLRESGPALVKPTQTLTLTCTFSGFSLSTAGMSVGWIRQPPGKALEWLADIWWDDKKHYN
PSLKDRLTISKDTSKNQVVLKVTNMDPADTATYYCARDMIFNFYFDVWGQGTTVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 142
Annotated Amino Acid sequence of Synthetic anti-RSV
(Respiratory syncytia1 virus) Fc fragment of Fab-IgG (Heavy
Chain)
PCPAPELLGGPSVFLIPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHOWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 143
DNA sequence of Synthetic anti-RSV (Respiratory syncytial
virus) Fab-IgG (Light Chain)
GACATCCAGATGACACAGAGCCCCAGCACACTGTCTGCCAGCGTGGGAGACAGAGTGACCAT
CACATGTAGCGCCAGCAGCAGAGTGGGCTACATGCACTGGTATCAGCAGAAGCCTGGCAAGG
CCCCTAAGCTGCTGATCTACGACACAAGCAAGCTGGCCTCTGGCGTGCCCAGCAGATTTTCT
GGCTCTGGCAGCGGCACCGAGTTCACCCTGACCATCTCTAGCCTGCAGCCTGACGACTTCGC
CACCTACTACTGCTTTCAAGGCAGCGGCTACCCOTTCACCTTTGGCGGCGGAACAAAGGTGG
AAATCAAGCGGACAGTGGCCGCTCCTAGCGTGTTCATCTTTCCACCTAGCGACGAGCAGCTG
AAGTCTGGCACAGCCTCTGTCGTGTGCCTGCTCAACAACTTCTACCCCAGAGAAGCCAAGGT
GCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAATAGCCAAGAGAGCGTGACCGAGCAGG
ACAGCAAGGACTCTACCTACAGCCTGTOCTCCACACTGACCCTGAGCAAGGCCGACTACGAG
AAGCACAAAGTGTACGCCTGCGAAGTGACCCACCAGGGCCTTTCTAGCCCTGTGACCAAGAG
CTTCAACCGGGGCGAGTGC
137
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SEQ ID NO: 144
Annotated Amino Acid sequence of Synthetic anti-RSV
(Respiratory syncytial virus) Fab-IgG (Light Chain)
DIQMTOPSTLSASVGDRVTITCSASSRVGYMHWYQUPGKAPKLLMTSKLASGVPSRFS
GSGSGTEFTLTISSLUDDEATYYCFQGSGYPFTFGGGTKVEIKRTVAAPSVFIFPPSDEQL
KSGTASVVOLLNNFYPREAKVQWKVDNALUGNSQESVTEWSKDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 145
Annotated Amino Acid sequence of Synthetic anti-RSV
(Respiratory syncytial virus) Fab fragment of Fab-IgG (Light
Chain)
DIQMTQSPSTLSASVGDRVTITCSASSRVGYMHWYQQKPGKAPKLLITDTSKLASGVPSRFS
GSGSGTEFTLTISSLQPDDFATYYCFQGSGYPFTFGGGTKVEIKRTVAAPSVFIFPPSDEQL
KSGTASVVCLLNNFYPREAKVQWKVIDNALUGNSQESVTEUSKDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 146
Annotated DNA sequence of Synthetic anti-RSV (Respiratory
syncytial virus) IgG-Fab (Heavy Chain)
CAAGTGACCCTGAGAGAGTCTGGCCCCGCTCTGGTTAAGCCCACACAGACCCTGACACTGAC
CTGCACCTTCAGCGGCTTTAGCCTGTCTACAGCCGGCATGAGCGTCGGCTGGATTAGACAGC
CTCCTGGCAAAGCCCTGGAATGGCTGGCCGACATTTGGTGGGACGACAAGAAGCACTACAAC
CCCAGCCTGAAGGACCGGCTGACCATCAGCAAGGACACCAGCAAGAACCAGGTGGTGCTGAA
AGTGACCAACATGGACCCTGCCGACACCGCCACCTACTACTGCGCCAGAGACATGATCTTCA
ACTTCTACTTCGACGTGTGGGGCCAGGGCACCACCGTGACAGTTAGCTCTGCCTCTACAAAG
GGCCCCAGCGTGTTCCCTCTGGCTCCTAGCAGCAAGTCTACAAGCGGAGGAACAGCCGCTCT
GGGCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCCTGGAATTCTGGCGCTC
TGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGCAGCGGCCTGTACTCTCTGAGC
AGCGTGGTCACAGTGCCAAGCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGAACCA
CAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCACA
CCTGTOCTCCATGTOCTGCTCCAGAACTGCTCGGCGGACCTTCCGTGTTTCTGTTCCCTCCA
AAGCCTAAGGACACCCTGATGATCAGCAGAACCCCTGAAGTGACCTGCGTGGTGGTGGATGT
GTCCCACGAGGATCCCGAAGTGAAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACG
CCAAGACCAAGCCTAGAGAGGAACAGTACAACAGCACCTACAGAGTGGTGTCTGTGCTGACC
GTGCTGCACCAGGATTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGCCCT
GCCTGCTCCTATCGAGAAAACCATCTCCAAGGCCAAGGGCCAGCCTAGGGAACCCCAGGTTT
ACACACTGCCTCCAAGCAGGGACGAGCTGACCAAGAATCAGGTGTCCCTGACCTGCCTCGTG
AAGGGCTTCTACCCTTCCGATATCGCCGTGGAATGGGAGAGCAATGGCCAGCCTGAGAACAA
CTACAAGACAACCCCTCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTACAGCAAGCTGA
CAGTGGACAAGTCCAGATGGCAGCAGGGCAACGTGTTCAGCTGCAGCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGAGCCTGTCTCCAGGCAAAGGTGGCGGAGGATC
TGGCGGAGGTGGAAGCGGCGGAGGCGGTTCTGGTGGTGGCGGCTCTGGCGGCGGTGGTTCAG
GTGGCGGCGGTTCTCAAGTTACACTGAGAGAAAGCGGCCCAGCTCTCGTGAAGCCTACTCAG
ACACTCACCCTGACATGTACCTTCTCTGGCTTCAGCCTGAGCACCGCCGGCATGTCTGTTGG
138
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
ATGGATCAGACAACCACCAGGCAAGGCTCTCGAGTGGCTCGCTGATATTTGGTGGGATGATA
AGAAACATTATAACCCATCTCTCAAGGACCGCCTCACCATTTCCAAGGATACCTCCAAAAAT
CAGGTCGTGCTCAAAGTCACGAATATGGATCCCGCCGATACGGCCACATATTACTGTGCCCG
GGATATGATCTTTAATTTCTATTTTGATGTCTGGGGCCAAGGGACAACCGTCACCGTGTCTA
GCGCCAGCACAAAGGGACCCTCTGTGTTTCCACTGGCTCCCAGCTCTAAGAGCACCTCCGGT
GGAACAGCTGCCCTGGGATGTCTCGTGAAAGACTACTTCCCCGAACCAGTGACAGTCAGCTG
GAACAGCGGAGCCCTGACTAGTGGGGTGCACACATTCCCTGCCGTCCTGCAATCTAGOGGAC
TGTACAGCCTGTCCAGCGTCGTGACCGTGCCTTCTAGCTCTCTGGGAACACAGACATATATC
TGTAATGTCAATCACAAACCCTCCAATACGAAGGTCGACAAAAAGGTCGAGCCTAAGTCCTG
TGATAAGACGCACACA
SEQ ID NO: 147
Annotated Amino Acid sequence of Synthetic anti-RSV
(Respiratory syncytial virus) IgG-Fab (Heavy Chain)
QVTLRESGPALVKPTQTLTLTCTFSGFSLSTAGMSVGWIRQPPGKALEWLADIWWDDKKHYN
PSLKDRLTISKDTSKNQVVLKVTNMDPADTATYYCARDMIFNFYFDVWGQGTTVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVFPKSCDKTHTCPPCPAPELLGGPSVFLFPP
KPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLT
VLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLV
KGFYPSDIAVEWESNGUENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEA
LHNHYTQKSLSLSPGKGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSQVTLRESGPALVKPTQ
TLTLTCTFSGFSLSTAGMSVGWIRQPPGKALEWLADIWWDDKKHYNPSLKDRLTISKDTSKN
QVVLKVTNMDPADTATYYCARDMIFNFYFDVWGQGTTVTVSSASTKGPSVFPLAPSSKSTSG
GTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYI
CNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 148
Annotated Amino Acid sequence of Synthetic anti-RSV
(Respiratory syncytial virus) Fab fragment of IgG-Fab (Heavy
Chain)
QVTLRESGPALVKPTQTLTLTCTFSGFSLSTAGMSVGWIRUPGKALEWLADIWWDDKKHYN
PSLKDRLTISKDTSKNQVVLKVTNMDPADTATYYCARDMIFNFYFDVWGQGTTVTVSSASTK
GPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSCVHTFPAVLQSSGLYSLS
SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 149
Annotated Amino Acid sequence of Synthetic anti-RSV
(Respiratory syncytial virus) Fc fragment of IgG-Fab (Heavy
Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHOWLNGKEYKCKVSNKALPAPIEKTISKAKGUREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNWPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
139
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SEQ ID NO: 150
DNA sequence of Synthetic anti-RSV (Respiratory syncytial
virus) IgG-Fab (Light Chain)
GACATCCAGATGACACAGAGCCCCAGCACACTGTCTGCCAGCGTGGGAGACAGAGTGACCAT
CACATGTAGCGCCAGCAGCAGAGTGGGCTACATGCACTGGTATCAGCAGAAGCCTGGCAAGC
CCCCTAAGCTGCTGATCTACGACACAAGCAACCTGGCCTCTGGCGTGCCCAGCAGATTTTCT
GGCTCTGGCAGOGGCACCGACTTCACCCTGACCATCTCTAGCCTGCAGCCTGACGACTTCGC
CACCTACTACTGCTTTCAAGGCAGCGGCTACCCCTTCACCTTTGGCGCCGGAACAAACCTGG
AAATCAAGCGGACACTGGCCGCTCCTAGCGTGTTCATCTTTCCACCTAGCGACGAGCAGCTG
AAGTCTGGCACAGCCTCTGTCCTGTGCCTGCTCAACAACTTCTACCCCAGAGAAGCCAAGGT
GCAGTGGAAGGTGGACAACGCCCTGCAGAGCGCCAATAGCCAAGAGAGCGTGACCGAGCAGG
ACAGCAAGGACTCTACCTACAGCCTGTCCTCCACACTGACCCTGAGCAAGGCCGACTACGAG
AAGCACAAAGTGTACGCCTGCGAACTGACCCACCAGGGCCTTTCTACCCCTGTGACCAAGAG
CTTCAACCGGGGCGAGTGC
SEQ ID NO: 151
Annotated Amino Acid sequence of Synthetic anti-RSV
(Respiratory syncytial virus) IgG-Fab (Light Chain)
DIQMTQSPSTLSASVGDRVTITCSASSRVGYMHWYQQKPGKAPKLLIYETSKLASGVPSRFS
GSGSGTEFTLTISSLUDDFATYYCFQGSGYPFTFGGGTKVEIKRTVAAPSVFIFPPSDEQL
KSGTASVVOLLNNFYPREAKVQWKVDNALUGNSQESVTEQDSKOSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 152
Annotated Amino Acid sequence of Synthetic anti-RSV
(Respiratory syncytial virus) Fab fragment of IgG-Fab (Light
Chain)
DIQMTOPSTLSASVGDRVTITCSASSRVGYMHWYQOPGKAPKLLIYDTSKLASGVPSRES
GSGSGTEFTLTISSLUDDFATYYCFQGSGYPFTFGGGTKVEIKRTVAAPSVFIFPPSDEQL
KSCTASVVOLLNNFYPREAKVQWKVDNALUGNSQESVTENSKDSTYSLSSTLTLSKADYE
KHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 153
Annotated DNA sequence of anti-Gonorrhea (2C7) Fab-Fab-IgG-
Fab-Fab (Heavy Chain)
GAGGTTCAGCTGCAGCAGTCTGGACCTGAGCTGGTCAAGCCTGGCAGGAGCGTGAAGATCAG
CTGTAAAGGCAGCGGCTACACCTTCACCGACTACAACATGGAATGGCTCAAGCAGAGCCACG
GCAAGAGCCTGGAATGGATCGGCGTGATCAACCCCAACAACCGGTTCACCAGCTACAACCAG
AACTTCAGAGGCAAGGCCACACTGACCGTGGACAAGAGCAGCAGCACCGCCTACATGGATCT
GAGAAGCCTGACCAGCGAGGACAGCGCCGTGTATTTTTGTGCCGGCAGCCGGTGGTATCAGT
ACGACTATTGGGGCCAGGGCACAACCCTGACAGTGTCTAGCGCCTCTACAAAGGGCCCCAGC
GTTTTCCCACTGGCTCCTAGCAGCAAGAGCACATCTGGCGGAACAGCCGCTCTGGGCTGTCT
GGTCAAGGACTACTTTCCCGAGCCTGTGACCGTGTCCTGGAATTCTGGCGCTCTGACAAGCG
140
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
GCGTGCACACCTTTCCAGCTGTGCTGCAAAGCAGCGGCCTGTACTCTCTGAGCAGCGTGGTC
ACAGTGCCAAGCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGAACCACAAGCCTAG
CAACACCAAGGTCGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCATACAGGCGGCG
GAGGAAGCGGAGGCGGAGGATCTGGTGGTGGTGGATCTGGCGGTGGCGGAAGCGGCGGAGGC
GGTTCTGGTGGCGGAGGTAGTGAAGTGCAGCTTCAGCAGAGCGGCCCAGAACTCGTGAAGCC
TGGAAGCTCTGTGAAGATCTCCTGCAAAGGCTCTGGGTACACGTTTACGGATTACAATATGG
AATGGGTTAAGCAATCCCACGGGAAGTCCCTCGAGTGGATTGGAGTGATCAATCCGAACAAT
CGCTTTACCTCCTACAATCAAAACTTCCGGGGCAAAGCTACCCTGACTGTGGATAAGTCCAG
CTCCACAGCCTATATGGACCTGCGGTCCCTGACCTCTGAGGACTCCGCTGTGTACTTCTGCG
CCGGCTCCAGATGGTATCAATATGATTACTGGGGACAGGGAACCACTCTGACTGTGTCCTCC
GCCAGCACAAAGGGACCCTCCGTGTTTCCTCTGGCTCCCAGCTCTAAGTCTACCAGCGGAGG
AACAGCTGCCCTGGGATGCCTCGTGAAAGACTACTTCCCTGAACCAGTGACAGTCAGCTGGA
ACAGCGGAGCCCTGACTTCTGGGGTGCACACATTCCCTGCCGTCCTGCAATCTTCTGGCCTG
TACAGCCTGTCCAGCGTCGTGACCGTTCCTTCTAGCTCTCTGGGAACACAGACATATATCTG
TAATGTCAATCACAAACCCTCCAATACGAAGGTGGACAAAAAGGTCGAGCCTAAGTCCTGTG
ATAAGACCCACACAGGTGGTGGTGGTAGTGGTGGCGGCGGTTCAGGCGGAGGTGGTAGTGGC
GGAGGTGGCAGCGGTGGTGGCGGTAGCGGAGGTGGTGGAAGTGAAGTTCAACTGCAACAGTC
AGGCCCCGAGCTTGTGAAGCCAGGCTCCTCCGTGAAAATCAGTTGCAAAGGATCCGGGTATA
CCTTCACAGACTATAATATGGAATGGGTCAAACAGTCTCACGGCAAAAGTCTTGAGTGGATA
GGTGTCATTAACCCGAACAACAGATTCACCTCTTATAATCAAAATTTCAGAGGGAAAGCCAC
GCTCACAGTCGACAAGTCCTCCAGCACTG=ATATGGATCTCCGCAGCCTGACATCCGAGG
ATTCTGCCGTCTACTTCTGTGCAGGCAGTCGCTGGTATCAGTACGATTACTGGGGTCAAGGG
ACTACCCTGACCGTCAGCTCTGCTAGCACCAAGGGCCCATCGGTOTTCCCCCTGGCACCCTC
CTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCG
AACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCT
GTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTT
GGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGA
AAGTTGAGCCCAAATCTTGTGACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTC
CTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCC.AAGGACACCCTCATGATCTCCCG
GACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCA
ACTGGTACGTGGACGGCGTGGAGGTGC.ATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTAC
AACAGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAA
GGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCA
AAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCTGCCCCCATCCCGGGACGAGCTG
ACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAA_AGGCTTCTATCCCAGCGACATCGCCGT
GGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCC TCCCGTGCTGGACT
CCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGAC.AAGAGCAGGTGGCAGCAGGGG
AACGTOTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCCT
CTCCCTGTCTCCGGGTAAAGGATCCGGTGGTGGTGGTAGTGGTGGCGGCGGTTCAGGCGGAG
GTGGTAGTGGCGGAGGTGGCAGCGGTGGTGGCGGTAGCGGAGGTGGTGGAAGTGAGGTTCAG
CTGCAGCAGTCTGGACCTGAGCTGGTCAAGCCTGGCAGCAGCGTGAAGATCAGCTGTAAAGG
CAGCGGCTACACCTTCACCGACTACAACATGGAATGGGTCAAGCAGAGCCACGGCAAGAGCC
TGGAATGGATCGGCGTGATCAACCCCAACAACCGGTTCACCAGCTACAACCAGAACTTCAGA
GGCAAGGCCACACTGACCGTGGACAAGAGCAGCAGCACCGCCTACATGGATCTGAGAAGCCT
GACCAGCGAGGACAGCGCCGTGTATTTTTGTGCCGGCAGCCGGTGGTATCAGTACGACTATT
GGGGCCAGGGCACAACCCTGACAGTGTCTAGCGCCTCTACAAAGGGCCCCAGCGTTTTCCCA
CTGGCTCCTAGCAGCAAGAGCACATCTGGCGGAACAGCCGCTCTOGGCTGTCTGGTCAAGGA
CTACTTTCCCGAGCCTGTGACCGTGTCCTGGAATTCTGGCGCTCTGACAAGCGGCGTGCACA
CCTTTCCAGCTGTGCTGCAAAGCAGCGGCCTGTACTCTCTGAGCAGCGTGGTCACAGTGCCA
AGCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGAACCACAAGCCTAGCAACACCAA
GGTCGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCATACAGGCGGCGGAGGAAGCG
141
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
GAGGCGGAGGATCTGGTGGTGGTGGATCTGGCGGTGGCGGAAGCGGCGGAGGCGGTTCTGGT
GGCGGAGGTAGTGAAGTGCAGCTTCAGCAGAGCGGCCCACAACTCGTGAAGCCTGGAAGCTC
TGTGAAGATCTCCTGCAAAGGCTCTGGGTACACGTTTACGGATTACAATATGGAATGGCTTA
AGCAATCCCACGGGAAGTCCCTCGAGTGGATTGGAGTGATCAATCCGAACAATCGCTTTACC
TCCTACAATCAAAACTTCCGGGGCAAAGCTACCCTGACTGTGGATAAGTCCAGCTCCACAGC
CTATATGGACCTGCGGTCCCTGACCTCTGAGGACTCCGCTGTGTACTTCTGCGCCGGCTCCA
GATGGTATCAATATGATTACTGGGGACAGGGAACCACTCTGACTGTGTCCTCCGCCAGCACA
AAGGGACCCTCCGTGTTTCCTCTCGCTCCCAGCTCTAAGTCTACCAGCGGAGGAACAGCTGC
CCTGGGATGCCTCGTGAAAGACTACTTCCCTGAACCAGTGACAGTCAGCTGGAACAGCGGAG
CCCTGACTTCTGGGGTGCACACATTCCCTGCCGTCCTGCAATCTTCTGGCCTGTACAGCCTG
TCCAGCGTCGTGACCGTTCCTTCTAGCTCTCTGGGAACACAGACATATATCTGTAATGTCAA
TCACAAACCCTCCAATACGAAGGTGGACAAAAAGGTCGAGCCTAAGTCCTGTGATAAGACCC
ACACA
SEQ ID NO: 154
Annotated Amino Acid sequence of anti-Gonorrhea (2C7) Fab-Fab-
IgG-Fab-Fab (Heavy Chain)
EVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFTSYNQ
NFRGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYQYDYWGQGTTLTVSSASTKGPS
VFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVIJQSSGLYSLSSVV
TVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGSGGGGSGGG
GSGGGGSEVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNN
RFTSYNONFRCKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYWDYWGQGTTLTVSS
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGSG
GGGSGGGGSGGGGSEVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWI
GVINPNNRFTSYNQNFRGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYQYDYWGQG
TTLTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPA
VLOSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPEL
LGGPSVFLFETKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVIDGVEVHNAKTKPREEQY
NSTYRVVSVLTVLHQDWLNCKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDEL
TKNQVSLTCLVKGFYPSDIAVEWESNCQPENNYKTTPPVLDSDCSFFLYSKLTVDKSRWQQG
NVFSCSVMHEALHNHYTQKSLSLSPGKGSGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSEVQ
LQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFTSYNQNFR
GKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYUDYWGQGTTLTVSSASTKGPSVFP
LAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVP
SSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGSGGGGSGGGGSG
GGGSEVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFT
SYNQNFRGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYWDYWCQCTTLTVSSAST
KGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSL
SSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 155
Annotated Amino Acid sequence of Fab fragment of anti-
Gonorrhea (2C7) (Heavy Chain)
EVQLQQSGPELVKPGSSVKISCKGSGYTFTDYNMEWVKQSHGKSLEWIGVINPNNRFTSYNQ
NFRGKATLTVDKSSSTAYMDLRSLTSEDSAVYFCAGSRWYQYDYWGQGTTLTVSSASTKGPS
VFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVV
TVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
142
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
SEQ ID NO: 156
Annotated Amino Acid sequence of Fc fragment of anti-Gonorrhea
(2C7) (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 157
DNA sequence of anti-Gonorrhea (2C7) (Light Chain)
CAGGTGGTGGTCACACAAGAGAGCGCCCTGACAACAAGCCCTGGCGAGACAGTGACCCTGAC
CTGCAGATCTTCTACAGGCGCCGTGACCACCTCCAACTACGCCAATTGGGTGCAAGAGAAGC
CCGACCACCTGTTCACAGGCCTGATCGGCGGCATCAACAATAGAGCACCTGGCGTGCCAGCC
AGATTCAGCGGATCTCTGATCGGAGACAAGGCCGCACTGACAATCACAGGCGCCCAGACAGA
GGACGAGGCCATCTACTTTTGCGCCCTGTGGTACAGCAACCACTGGGTTTTCGGCGGAGGCA
CCAAGCTGACAGTTCTGGGCCAACCTAAGGCCGCTCCTAGCGTGACACTGTTCCCTCCAAGC
AGCGAAGAACTGCAGGCCAACAAGGCCACACTCGTGTGCCTGATCAGCGACTTTTATCCTGG
CGCCGTGACCGTGGCCTGGAAGGCTGATAGTTCTCCTGTGAAGGCCGGCGTGGAAACCACCA
CACCTAGCAAGGAGAGCAACAACAAATACGCCGCCAGCAGCTACCTGAGCCTGACACCTGAG
CAGTGGAAGTOCCACAGATCCTACAGCTGCCAAGTGACCCACGAGGGCAGCACCGTGGAAAA
AACAGTGGCCCCTACCGAGTGCAGC
SEQ ID NO: 158
Annotated Amino Acid sequence of anti-Gonorrhea (2C7) (Light
Chain)
QVVVTQESALTTSPGETVTLTCRSSTGAVTTSNYANWVQEMPDHLFTGLIGGINNRAPGVPA
RFSGSLIGDKAALTITGAQTEDEAIYFCALWYSNHWVEGGGTKLTVLGQPKAAPSVTLFPPS
SEELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPE
QWKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 159
Annotated Amino Acid sequence of Fab fragment of anti-
Gonorrhea (2C7) IgG (Light Chain)
QVVVTQESALTTSPGETVTLTCRSSTGAVTTSNYANWVQEKPDHLFTGLIGGINNRAPGVPA
RFSGSLIGDKAALTITGAQTEDEAIYFCALWYSNHWVFGGGTKLTVLGUKAAPSVTLEPPS
SEELQANKATLVCLISDFYPGAVTVAWKADSSPVKAGVETTTPSKQSNNKYAASSYLSLTPE
QWKSHRSYSCQVTHEGSTVEKTVAPTECS
SEQ ID NO: 160
Annotated DNA sequence of anti-klebsiella Fab-Fab-IgG-Fab-Fab
(Heavy Chain)
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
GAGGTGCAGCTGGTTGAATCTGGCGGAGGACTGGTTCAGCCTGGCGGATCTCTGAGACTGTC
TTGCGCCGCCAGCTTTAGCCTGACAAGCTACGCCGTGCACATCCACTGGGTTCGACAGGCCC
CTGGCAAAGGCCTTGAATGGGTTGCCAGAGTGATCTGGGCTGGCGGCATCACCCACTACAAT
AGCGCCCTGATGAGCAGATACGCCGACAGCGTGAAGGGCAGATTCACCATCAGCGCCGACAC
CAGCAAGAACACCGCCTACCTGCAGATGAACAGCCTGAGAGCCGAGGACACCGCCGTGTACT
ATTGCGCCAGAGGCAACTGGGCCTTCGACTATTGGGGACAGGGCACCCTGGTCACAGTGTCT
AGCGCCTCTACAAAGGGCCCCAGCGTTTTCCCACTGGCTCCTAGCAGCAAGAGCACATCTGG
TGGAACAGCCGCTCTGGGCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCCT
GGAATTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGCAGCGGC
CTGTACTCTCTGAGCAGCGTCGTGACAGTGCCAAGCAGCTCTCTGGGCACCCAGACCTACAT
CTGCAATGTGAACCACAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCT
GCGACAAGACCCATACAGGCGGAGGTGGAAGCGGAGGCGGAGGCTCTGGCGGCGGAGGAAGT
GGTGGCGGCGGTAGTGGCGGTGGTGGATCAGGTGGCGGAGGTTCTGAAGTCCAGCTGGTGGA
AAGTGGCGGCGGACTTGTGCAACCAGGTGGAAGTCTGAGGCTGAGCTGTGCCGCCTCTTTCA
GCCTGACCTCTTATGCCGTGCATATTCATTGGGTCCGCCAAGCTCCAGGCAAGGGCCTCGAG
TGGGTCGCACGAGTTATTTGGGCAGGCGGAATTACACACTATAACTCTGCCCTCATGTCCCG
CTACGCCGACTCTGTGAAAGGCCGGTTTACCATCTCCGCCGATACCTCCAAGAATACTGCCT
ATCTCCAAATGAACTCTCTGCGCGCCGAAGATACAGCCGTGTATTACTGTGCTCGCGGAAAT
TGGGCCTTTGATTACTGGGGCCAAGGCACACTGGTTACCGTCAGCTCTGCCAGCACAAAGGG
ACCCTCCGTGTTTCCTCTGGCTCCCAGCTCTAAGTCTACCAGCGGAGGAACAGCTGCCCTGG
GATGTCTCGTGAAAGACTACTTCCCCGAACCAGTGACAGTCAGCTGGAACAGCGGAGCCCTG
ACTTCTGGGGTGCACACATTCCCTGCCGTCCTGCAATCTTCTGGCCTGTACAGCCTGTCCAG
CGTGGTCACCGTTCCTAGCTCTAGCCTGGGAACACAGACATATATCTGTAATGTCAATCACA
AACCCTCCAATACGAAGGTCGACAAAAAGGTCGAGCCTAAGTCCTGTGATAAGACACACACT
GGCGGTGGCGGTTCAGGCGGAGGCGGAAGTGGCGGAGGCGGATCCGGCGGTGGTGGTAGTGG
TGGTGGCGGCAGCGGAGGCGGCGGATCTGAAGTTCAGCTTGTTGAGTCAGGTGGTGGCCTCG
TGCAACCTGGCGGAAGCCTTAGACTTTCCTGCGCCGCTTCATTCTCCCTGACCTCATACGCT
GTCCATATACACTGGGTCCGACAAGCACCCGGAAAAGGATTGGAGTGGGTTGCCCGGGTTAT
ATGGGCTGGTGGTATCACACATTATAACAGCGCTCTGATGTCTCGCTATGCCGATTCCGTCA
AGGGGCGCTTCACAATCTCTGCCGACACCTCTAAAAACACGGCTTACCTTCAAATGAATTCC
CTCCGCGCTGAGGATACCGCTGTCTACTACTGTGCACGCGGCAACTGGGCTTTCGACTACTG
GGGTCAAGGGACTCTCGTGACTGTGTCCTCTGCCTCTACAAAGGGCCCTAGTGTGTTCCCTC
TGGCTCCCAGCAGCAAGTCTACATCTGGCGGAACAGCCGCTCTGGGCTGCCTGGTCAAGGAT
TACTTTCCCGAGCCTGTGACCGTGTCCTGGAATTCTGGCGCTCTGACAAGCGGCGTGCACAC
CTTTCCAGCTGTGCTGCAAAGCAGCGGCCTGTACTCTCTGAGCAGCGTGGTCACAGTGCCTA
GCTCTAGCCTGGGCACCCAGACCTACATCTGCAATGTGAACCACAAGCCTAGCAACACCAAG
GTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCACACCTGTCCTCCATGTCCTGC
TCCAGAACTGCTCGGCGGACCTTCCGTGTTCCTGTTTCCTCCAAAGCCTAAGGACACCCTGA
TGATCAGCAGAACCCCTGAAGTGACCTGCGTGGTGGTGGATGTGTCCCACGAGGATCCCGAA
GTGAAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCTAGAGA
GGAACAGTACAACAGCACCTACAGAGTGGTGTCCGTGCTGACCGTGCTGCACCAGGATTGGC
TGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGCCCTGCCTGCTCCTATCGAGAAA
ACCATCAGCAAGGCCAAGGGCCAGCCTAGGGAACCCCAGGTTTACACACTGCCTCCAAGCCG
GGAAGAGATGACCAAGAACCAGGTGTCCCTGACCTGCCTCGTGAAGGGCTTCTACCCTTCCG
ATATCGCCGTGGAATGGGAGAGCAATGGCCAGCCTGAGAACAACTACAAGACAACCCCTCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGTACAGCAAGCTGACAGTGGACAAGTCCAGATG
GCAGCAGGGCAACGTGTTCAGCTGCAGCGTGATGCACGAGGCCCTGCACAACCACTACACCC
AGAAGTCCCTGAGCCTGTCTCCTGGCAAAGGATCCGGCGGTGGCGGTTCAGGCGGAGGCGGA
AGTGGCGGAGGCGGATCCGGCGGTGGTGGTAGTGGTGGTGGCGGCAGCGGAGGCGGCGGATC
TGAGGTGCAGCTGGTTGAATCTGGCGGAGGACTGGTTCAGCCTGGCGGATCTCTGAGACTGT
144
CA 03113059 2021-03-16
W02020/061560 PCT/US2019/052396
CTTGCGCCGCCAGCTTTAGCCTGACAAGCTACGCCGTGCACATCCACTGGGTTCGACAGGCC
CCTGGCAAAGGCCTTGAATGGGTTGCCAGAGTGATCTGGGCTGGCGGCATCACCCACTACAA
TAGCGCCCTGATGAGGAGATACGCCGACAGCGTGAAGGGCAGATTCACCATCAGCGCCGACA
CCAGCAAGAACACCGCCTACCTGCAGATGAACAGCCTGAGAGCCGAGGACACCGCCGTGTAC
TATTGCGCCAGAGGCAACTGGGCCTTCGACTATTGGGGACAGGGCACCCTGGTCACAGTGTC
TAGCGCCTCTACAAAGGGCCCCAGCGTTTTCCCACTGGCTCCTAGCAGCAAGAGCACATCTG
GTGGAACAGCCGCTCTGGGCTGCCTGGTCAAGGATTACTTTCCCGAGCCTGTGACCGTGTCC
TGGAATTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAAAGCAGCGG
CCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCAAGCAGCTCTCTGGGCACCCAGACCTACA
TCTGCAATGTGAACCACAAGCCTAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGC
TGCGACAAGACCCATACAGGCGGAGGTGGAAGCGGAGGCGGAGGCTCTGGCGGCGGAGGAAG
TGGTGGCGGCGGTAGTGGCGGTGGTGGATCAGGTGGCGGAGGTTCTGAAGTCCAGCTGGTGG
AAAGTGGCGGCGGACTTGTGCAACCAGGTGGAAGTCTGAGGCTGAGCTGTGCCGCCTCTTTC
AGCCTGACCTCTTATGCCGTGCATATTCATTGGGTCCGCCAAGCTCCAGGCAAGGGCCTCGA
GTGGGTCGCACGAGTTATTTGGGCAGGCGGAATTACACACTATAACTCTGCCCTCATGTCCC
GCTACGCCGACTCTGTGAAAGGCCGGTTTACCATCTCCGCCGATACCTCCAAGAATACTGCC
TATCTCCAAATGAACTCTCTGCGCGCCGAAGATACAGCCGTGTATTACTGTGCTCGCGGAAA
TTGGGCCTTTGATTACTGGGGCCAAGGCACACTGGTTACCGTCAGCTCTGCCAGCACAAAGG
GACCCTCCGTGTTTCCTCTGGCTOCCAGCTCTAAGTCTACCAGCGGAGGAACAGCTGCCCTG
GGATGTCTCGTGAAAGACTACTTCCCCGAACCAGTGACAGTCAGCTGGAACAGCGGAGCCCT
GACTTCTGGGGTGCACACATTCCCTGCCGTOCTGCAATCTTCTGGCCTGTACAGCCTGTCCA
GCGTGGTCACCGTTCCTAGCTCTAGCCTGGGAACACAGACATATATCTGTAATGTCAATCAC
AAACCCTCCAATACGAAGGTCGACAAAAAGGTCGAGCCTAAGTCCTGTGATAAGACACACAC
SEQ ID NO: 161
Annotated Amino Acid sequence of anti-klebsiella Fab-Fab-IgG-
Fab-Fab (Heavy Chain)
EVQLVESGGGLVQPGGSLRLSCAASFSLTSYAVHIHWVRQAPGKGLEWVARVIWAGGITHYN
SALMSRYADSVKGRFTISADTSKNTAYLWNSLRAEDTAVYYCARGNWAFDYWGQGTINTVS
SASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSG
LYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTGGGGSGGGGSGGGGS
GGGGSGGGGSGGGGSEVQLVESGGGINUGGSLRLSCAASFSLTSYAVHIHWVRQAPGKGLE
WVARVIWAGGITHYNSALMSRYADSVKGRFTISADTSKNTAYLWNSLRAEDTAVYYCARGN
WAFDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGAL
TSGVHTFPAVLOSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
GGGGSGGGGSGGGGSGGGGSGGGGSGGGGSEWLVESGGGLVQPGGSLRLSCAASFSLTSYA
VHIHWVRQAPGKGLEWVARVIWAGGITHYNSALMSRYADSVKGRFTISADTSKNTAYLQMNS
LRAEDTAVYYCARGNWAFDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKID
YEPEPVTVSWNSGALTSGVHTFPAVLQ,SSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTK
VIDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVEWSHEDPE
VKFNWYVDGVEVHNAKTKPREEONSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPTEK
TISKAKGQPREPQVYTLPPSREEMTKNWSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPP
VLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKGSGGGGSGGGG
SGGGGSGGGGSGGGGSGGGGSEVQLVESGGGLVQPGGSLRLSCAASFSLTSYAVHIHWVRQA
PGKGLEWVARVIWAGGITHYNSALMSRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVY
YCARGNWAFDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVS
WNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKS
CDKTHTGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSEVQLVESGGGINQPGGSLRLSCAASF
SLTSYAVHIHWVRQAPGKGLEWVARVIWAGGITHYNSALMSRYADSVKGRFTISADTSKNTA
145
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
YLQMNSLRAEDTAVYYCARGNWAFDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAAL
GCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNH
KPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 162
Annotated Amino Acid sequence of Fab fragment of anti-
klebsiella (Heavy Chain)
EVQLVESGGGLVUGGSLRLSCAASESLTSYAVHIHWVRQAPGKGLEWVARVIWAGGITHYN
SALMSRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGNWAFDYWGQGTLVTVS
SASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSG
LYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT
SEQ ID NO: 163
Annotated Amino Acid sequence of Fa fragment of anti-
klebsiella (Heavy Chain)
PCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 164
DNA sequence of anti-klebsiella (Light Chain)
GACATCCAGATGACACAGAGCCCTAGCAGCCTGTCTGCCAGCGTGGGAGACAGAGTGACCAT
CACCTGTAGAGCCAGCAGCGCCAGATCCAGCGTGTCCTATATTCACGTGGCCTGGTATCAGC
AGAAGCCCGGCAAAGCCCCTAAGCTGCTGATCTACGACACCAGCAAACTGGCCAGCTTCCTG
TACAGCGGCGTGCCCTCTAGATTCAGCGGCAGCAGATCTGGCACCGACTTCACCCTGACCAT
AAGCAGCCTGCAGCCTGAGGACTTCGCCACCTACTACTGCTTTCAAGGCAGCGGCTACCOCT
ACACCTTTGGCCAGGGAACAAAGGTGGAAATCAAGAGAACAGTGGCCGCTCCTAGCGTGTTC
ATCTTCCCACCTTCCGACGAGCAGCTGAAGTCTGGCACAGCCTCTGTCGTGTGCCTGCTGAA
CAACTTCTACCCCAGAGAAGCCAAGGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ATAGCCAAGAGAGCGTGACCGAGCAGGACAGCAAGGACTCTACCTACAGCCTGAGCAGCACC
CTGACACTGAGCAAGGCCGACTACGAGAAGCACAAAGTGTACGCCTGCGAAGTGACCCACCA
GGGCCTTTCTAGCCCTGTGACCAAGAGCTTCAACCGGGGCGAATGT
SEQ ID NO: 165
Annotated Amino Acid sequence of anti-klebsiella (Light Chain)
DIQMTQSPSSLSASVGDRVTITCRASSARSSVSYTHVAWYQQKPGKAPKLLIYDTSKLASFL
YSGVPSRFSGSRSGTOFTLTISSLUEDFATYYCFQGSGYPYTFGQGTKVEIKRTVAAPSVF
IFPPSDEQLKSGTASVVCLLNNEYPREAKVQWKVDNALQSGNSQESVTEUSKDSTYSLSST
LTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
SEQ ID NO: 166
146
CA 03113059 2021-03-16
WO 2020/061560 PCT/US2019/052396
Annotated Amino Acid sequence of Fab fragment of anti-
klebsiella Fab-Fab-IgG-Fab-Fab (Light Chain)
DIQMTQSPSSLSASVGDRVTITCRASSARSSVSYIHVAWYQQKPGKAPKLLIYDTSKLASFL
YSGVPSRFSGSRSGTDFTLTISSLQPEDFATYYCFQGSGYPYTEGQGTKVEIKRTVAAPSVF
IFPPSDEQLKSGTASVVCLLNNEYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSST
LTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
147