Note: Descriptions are shown in the official language in which they were submitted.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
1
METHODS AND COMPOSITIONS FOR OCULAR CELL THERAPY
I. SEQUENCE LISTING
The instant application contains a Sequence Listing which has been submitted
electronically
in ASCII format and is hereby incorporated by reference in its entirety. Said
ASCII copy,
created on September 17, 2019, is named
PAT058298_sequence_listing_2019_5T25.bd
and is 224 KB in size.
II. FIELD
The present invention relates to methods of generating an expanded population
of
genetically modified ocular cells, for example limbal stem cells (LSCs) or
corneal endothelial
cells (CECs), wherein the cells are expanded involving the use of a LATS
inhibitor and the
expression of B2M in the cells has been reduced or eliminated. The present
invention also
relates to a population of such modified cells, preparations, uses and methods
of therapy
comprising said cells.
III. BACKGROUND
Organ regeneration and/or healing is an issue crucial to treat many serious
health issues.
For example in the eye, it is known that corneal blindness is the third
leading cause of
blindness worldwide. Approximately half of all the cornea transplants
worldwide are
performed for treatment of corneal endothelial dysfunction.
The cornea is a transparent tissue comprising different layers: corneal
epithelium, Bowman's
membrane, stroma, Descemet's Membrane and endothelium. The corneal endothelium
also
comprises a monolayer of human corneal endothelial cells and helps maintain
corneal
transparency via its barrier and ionic pump functions. It plays a crucial role
in maintaining the
balance of fluid, nutrients and salts between the corneal stroma and the
aqueous humor. To
maintain transparency, endothelial cell density must be maintained, however
endothelial cell
density can be significantly decreased as a result of trauma, disease or
endothelial
dystrophies. The density of the cells also decreases with aging. Human corneal
endothelium
has a limited propensity to proliferate in vivo. If the density of cells falls
too low, the barrier
function may be compromised. Loss of endothelial barrier function results in
corneal edema
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
2
and loss of visual acuity. The clinical condition of bullous keratopathy may
be one resulting
complication.
Currently the only treatment for blindness caused by corneal endothelial
dysfunction is
corneal transplantation. Although corneal transplantation is one of the most
common forms
of organ transplantation, the availability of donor corneas required is
extremely limited. A
2012-2013 global survey quantified the considerable shortage of corneal graft
tissue, finding
that only one cornea is available for every 70 needed (Gain at el., (2016)
Global Survey of
Corneal Transplantation and Eye Banking. JAMA Ophthalmol. 134:167-173).
New therapeutic approaches to supply corneal endothelial cells for the
treatment of corneal
endothelial dysfunction are thus greatly needed.
The corneal epithelium also needs to be maintained in the eye. The corneal
epithelium is
composed of a layer of basal cells and multiple layers of a non-keratinized,
stratified,
squamous epithelium. It is essential in maintaining the clarity and the
regular refractive
surface of the cornea. It acts as a transparent, renewable protective layer
over the corneal
stroma and is replenished by a stem cell population located in the limbus. In
limbal stem cell
deficiency, a condition in which limbal stem cells are diseased or absent, a
decrease in the
number of healthy limbal stem cells results in a decreased capacity for
corneal epithelium
renewal.
Limbal stem cell deficiency may arise as a result of injuries from chemical or
thermal burns,
ultraviolet and ionizing radiation, or even as a result of contact lens wear;
genetic disorders
like aniridia, and immune disorders such as Stevens Johnson syndrome and
ocular
cicatricial pemphigoid. Loss of limbal stem cells can be partial or total; and
may be unilateral
or bilateral. Symptoms of limbal stem cell deficiency include pain,
photophobia, non healing
painful corneal epithelial defects, corneal neovascularization, replacement of
the corneal
epithelium by conjunctival epithelium, loss of corneal transparency and
decreased vision
that can eventually lead to blindness.
A product for use in treating limbal stem cell deficiency was granted a
conditional marketing
authorisation in the European Union in 2015 (under the name Holoclar0), making
it the first
Advanced Therapy Medicinal Product (ATMP) containing stem cells in Europe.
Holoclar is
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
3
an ex vivo expanded preparation of autologous human corneal epithelial cells
containing
stem cells. A biopsy of healthy limbal tissue is taken from the patient,
expanded ex vivo and
frozen until surgery. For administration to the patient, the thawed cells are
grown on a
membrane comprising fibrin and then surgically implanted onto the eye of the
patient. The
therapy is intended for use in adults with moderate to severe limbal stem cell
deficiency due
to physical or chemical ocular burns (Rama P, Matuska S, Paganoni G, Spinelli
A, De Luca
M, Pellegrini G. (2010). Limbal stem-cell therapy and long-term corneal
regeneration. N Engl
J Med. 363:147-155). However the method is limited in that it is for
autologous use only, and
there must be enough surviving limbus in one eye to allow a minimum of one to
two square
millimeters of undamaged tissue to be extracted from the patient. There is
also the risk that
for each specific patient the culture of his/her cells may not be successful
and the patient
cannot receive this treatment. Furthermore feeder cells of murine origin are
used to prepare
the Holoclar cell preparation which introduces potential safety concerns due
to the risk of
disease transmission and potential immunogenicity into the preparation for use
in humans.
Moreover, the Holoclar cell preparation only contains approximately 5% of
limbal stem cells,
as identified by p63a1pha staining.
New therapeutic approaches to supply limbal stem cells for the treatment of
limbal stem cell
deficiency are thus greatly needed.
IV. SUMMARY
The inventions described herein relate to compositions and methods for ocular
cell therapy,
for example, ocular cells modified at specific target sequences in their
genome, including as
modified by introduction of CRISPR systems (e.g., S. pyogenes Cas9 CRISPR
systems)
that include gRNA molecules which target said target sequences. For example,
the present
disclosure relates to gRNA molecules, CRISPR systems, ocular cells, and
methods using
genome edited cells, e.g., modified limbal stem cells, for treating ocular
diseases.
The present invention provides a modified limbal stem cell, which has reduced
or eliminated
expression of beta-2-microglobulin (B2M) relative to an unmodified limbal stem
cell.
The present invention further provides a population of modified limbal stem
cells, which have
reduced or eliminated expression of B2M relative to an unmodified limbal stem
cell.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
4
In one aspect, a modified limbal stem cell includes an insertion or deletion
of a base pair,
e.g., more than one base pair, at or near B2M relative to an unmodified limbal
stem cell. In
another aspect, the invention provides a population of cells including the
modified limbal
stem cell, wherein in at least about 30% of the cells, at least one said
insertion or deletion is
a frameshift mutation, e.g., as measured by next-generation sequencing (NGS).
In certain aspects, the invention provides a modified limbal stem cell, which
has reduced or
eliminated expression of beta-2-microglobulin (B2M) relative to an unmodified
limbal stem
cell, wherein the B2M expression is reduced or eliminated by a CRISPR system
(e.g., S.
pyogenes Cas9 CRISPR system) comprising a gRNA molecule comprising a targeting
domain complementary to a target sequence in the B2M gene.
In other aspects, the invention provides a modified limbal stem cell, which
has reduced or
eliminated expression of beta-2-microglobulin (B2M) relative to an unmodified
limbal stem
cell, wherein the B2M expression is reduced or eliminated by a CRISPR system
(e.g., S.
pyogenes Cas9 CRISPR system) comprising a nucleic acid molecule encoding a
gRNA
molecule comprising a targeting domain complementary to a target sequence in
the B2M
gene.
In certain aspects, the invention provides a modified limbal stem cell, which
has reduced or
eliminated expression of beta-2-microglobulin (B2M) relative to an unmodified
limbal stem
cell, wherein the B2M expression is reduced or eliminated by a CRISPR system
(e.g., S.
pyogenes Cas9 CRISPR system) comprising a gRNA molecule comprising a targeting
domain complementary to a target sequence in the B2M gene, wherein the
modified limbal
stem cell was exposed to (e.g., was cultured in media comprising) a LATS
inhibitor.
In other aspects, the invention provides a modified limbal stem cell, which
has reduced or
eliminated expression of beta-2-microglobulin (B2M) relative to an unmodified
limbal stem
cell, wherein the B2M expression is reduced or eliminated by a CRISPR system
(e.g., S.
pyogenes Cas9 CRISPR system) comprising a nucleic acid molecule encoding a
gRNA
molecule comprising a targeting domain complementary to a target sequence in
the B2M
gene, wherein the modified limbal stem cell was exposed to a LATS inhibitor.
The present invention also provides a modified corneal endothelial cell, which
has reduced
or eliminated expression of B2M relative to an unmodified corneal endothelial
cell.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
The present invention further provides a population of modified corneal
endothelial cells,
which have reduced or eliminated expression of B2M relative to an unmodified
corneal
endothelial cell.
5
In one aspect, a modified corneal endothelial cell includes an insertion or
deletion of a base
pair, e.g., more than one base pair, at or near B2M relative to an unmodified
corneal
endothelial cell. In another aspect, the invention provides a population of
cells including the
modified corneal endothelial, wherein in at least about 30% of the cells, at
least one said
insertion or deletion is a frameshift mutation, e.g., as measured by next-
generation
sequencing (NGS).
The invention further provides methods of treating a patient suffering from an
ocular disease
comprising: providing a population of limbal stem cells, wherein the
population of limbal
stem cells has been cultured in the presence of a LATS inhibitor; introducing
into the
population of limbal stem cells a CRISPR system (e.g., S. pyogenes Cas9 CRISPR
system)
comprising a gRNA molecule comprising a targeting domain complementary to a
target
sequence in the B2M gene; and administering the population of cells to a
patient in need
thereof.
The invention also provides methods of preparing a population of modified
limbal stem cells
for ocular cell therapy comprising: modifying a population of limbal stem
cells by reducing or
eliminating expression of B2M comprising introducing into the limbal stem
cells a gRNA
molecule with a targeting domain comprising the sequence of any one of SEQ ID
NOs: 23-
105 or SEQ ID NOs: 108-119 or SEQ ID NOs: 134-140, wherein the limbal stem
cells have
optionally been cultured in the presence of a LATS inhibitor; and further
expanding the
modified limbal stem cells in cell culture media comprising a LATS inhibitor.
In certain aspects, a LATS inhibitor useful in a method of the invention is a
compound of
Formula Al
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
6
,R2
R5
xi
x2y
N
A
k I
R3 Al
or a salt thereof.
Non-limiting embodiments of the present disclosure are described in the
following
embodiments:
1. A modified limbal stem cell, which has reduced or eliminated expression
of beta-2-
microglobulin (B2M) relative to an unmodified limbal stem cell, wherein the
B2M
expression is reduced or eliminated by a CRISPR system comprising a gRNA
molecule comprising a targeting domain complementary to a target sequence in
the
B2M gene.
2. A modified limbal stem cell, which has reduced or eliminated expression
of beta-2-
microglobulin (B2M) relative to an unmodified limbal stem cell, wherein the
B2M
expression is reduced or eliminated by a CRISPR system comprising a nucleic
acid
molecule encoding a gRNA molecule comprising a targeting domain complementary
to a target sequence in the B2M gene.
3. The modified limbal stem cell of embodiment 1 or 2, wherein the modified
limbal stem
cell was cultured in media comprising a large tumor suppressor kinase ("LATS")
inhibitor,
optionally wherein the LATS inhibitor is a compound of Formula Al
,R2
Rb
X1
N
X2C-0
A
R3
Al
or a salt thereof, wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
7
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
_11i4
N N NH *"-N
N
\
NH ¨NH and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, Ci
6a1ky1, hydroxyC1_6alkyl, C1_6haloalkyl, R , -NH2, C1_6alkylamino, and di-(C1_
6a1ky1)amino;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
8
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6a1ky1 that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form a
4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional
heteroatoms independently selected from N, 0, and S, wherein the 4- to 6-
membered
heterocycloalkyl formed by R1 and R2 taken together with the nitrogen atom to
which both
are bound is unsubstituted or substituted by 1 to 3 substituents independently
selected from
halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein
the 3-to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises
1 to 2 oxygen atoms as chain members and is unsubstituted or substituted by R
.
4. The modified limbal stem cell according to embodiment 3, wherein the
compound is
selected from: N-methyl-2-(pyridin-4-y1)-N-(1,1,1-trifluoropropan-2-
yOpyrido[3,4-
d]pyrimidin-4-amine; 2-methyl-1-(2-methyl-2-{[2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-4-
yl]amino}propoxy)propan-2-ol; 2,4-dimethy1-4-{[2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-4-
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
9
yl]amino}pentan-2-ol; N-tert-butyl-2-(pyrimidin-4-y1)-1,7-naphthyridin-4-
amine; 2-
(pyridin-4-yI)-N-[1-(trifluoromethyl)cyclobutyl]pyrido[3,4-d]pyrimidin-4-
amine; N-propy1-
2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-amine; N-(propan-2-y1)-2-(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; 3-(pyridin-4-y1)-N-(1-
(trifluoromethyl)cyclopropy1)-
2,6-naphthyridin-1-amine; 2-(3-methy1-1H-pyrazol-4-y1)-N-(1-
methylcyclopropyl)pyrido[3,4-d]pyrimidin-4-amine; 2-methy1-2-{[2-(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-yl]amino}propan-1-ol; 2-(pyridin-4-y1)-4-(3-
(trifluoromethyDpiperazin-1-yOpyrido[3,4-d]pyrimidine; N-cyclopenty1-2-
(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; N-propy1-2-(3-(trifluoromethyl)-1H-pyrazol-4-
yl)pyrido[3,4-d]pyrimidin-4-amine; N-(2-methylcyclopenty1)-2-(pyridin-4-
yhpyrido[3,4-
d]pyrimidin-4-amine; 2-(3-chloropyridin-4-y1)-N-(1,1,1-trifluoro-2-
methylpropan-2-
yOpyrido[3,4-d]pyrimidin-4-amine; 2-(2-methy1-2-{[2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-
4-yl]amino}propoxy)ethan-1-ol; N-(1-methylcyclopropy1)-7-(pyridin-4-
yOisoquinolin-5-
amine; (1S,2S)-2-{[2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-
yl]amino}cyclopentan-1-ol;
N-methy1-2-(pyridin-4-y1)-N-[(2S)-1,1,1-trifluoropropan-2-yl]pyrido[3,4-
d]pyrimidin-4-
amine; N-methyl-N-(propan-2-y1)-2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-amine;
N-
(propan-2-y1)-2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-amine; 3-(pyridin-4-y1)-
N-(1-
(trifluoromethyl)cyclopropy1)-2,6-naphthyridin-1-amine and N-methy1-2-(pyridin-
4-y1)-N-
[(2 R)-1,1 ,1-trifluoropropan-2-yl]pyrido[3,4-d]pyrimidin-4-amine.
5. The modified limbal stem cell according to embodiment 3, wherein the
compound is
selected from: 3-(pyridin-4-y1)-N-(1-(trifluoromethyl)cyclopropy1)-2,6-
naphthyridin-1-
amine; N-(1-methylcyclopropy1)-7-(pyridin-4-yOisoquinolin-5-amine; 2-(pyridin-
4-y1)-4-
(3-(trifluoromethyl)piperazin-1-yOpyrido[3,4-d]pyrimidine; N-(tert-buty1)-2-
(pyridin-4-y1)-
1,7-naphthyridin-4-amine; and N-methy1-2-(pyridin-4-y1)-N-[(2S)-1,1,1-
trifluoropropan-
2-yl]pyrido[3,4-d]pyrimidin-4-amine.
6. The modified limbal stem cell according to embodiment 3, wherein the
compound is
selected from: 3-(pyridin-4-y1)-N-(1-(trifluoromethyl)cyclopropy1)-2,6-
naphthyridin-1-
amine; N-(1-methylcyclopropy1)-7-(pyridin-4-yOisoquinolin-5-amine; and 2-
(pyridin-4-
y1)-4-(3-(trifluoromethyl)piperazin-1-yOpyrido[3,4-d]pyrimidine.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
7. The modified limbal stem cell according to embodiment 3, wherein the
compound is
selected from: N-(tert-butyl)-2-(pyridin-4-y1)-1,7-naphthyridin-4-amine; and N-
methyl-2-
(pyridin-4-y1)-N-[(2S)-1,1,1-trifluoropropan-2-yl]pyrido[3,4-d]pyrimidin-4-
amine.
5 8. The modified limbal stem cell according to embodiment 3, wherein
the compound is N-
(tert-butyl)-2-(pyridin-4-y1)-1,7-naphthyridin-4-amine.
9. The modified limbal stem cell according to any one of embodiments 3 to
8, wherein the
compound is present in a concentration of 3 to 10 micromolar.
10. The modified limbal stem cell of embodiment any one of embodiments 1-9,
wherein
the targeting domain of the gRNA molecule is complementary to a sequence
within a
genomic region selected from: chr15:44711469-44711494, chr15:44711472-
44711497, chr15:44711483-44711508, chr15:44711486-44711511, chr15:44711487-
44711512, chr15:44711512-44711537, chr15:44711513-44711538, chr15:44711534-
44711559, chr15:44711568-44711593, chr15:44711573-44711598, chr15:44711576-
44711601, chr15:44711466-44711491, chr15:44711522-44711547, chr15:44711544-
44711569, chr15:44711559-44711584, chr15:44711565-44711590, chr15:44711599-
44711624, chr15:44711611-44711636, chr15:44715412-44715437, chr15:44715440-
44715465, chr15:44715473-44715498, chr15:44715474-44715499, chr15:44715515-
44715540, chr15:44715535-44715560, chr15:44715562-44715587, chr15:44715567-
44715592, chr15:44715672-44715697, chr15:44715673-44715698, chr15:44715674-
44715699, chr15:44715410-44715435, chr15:44715411-44715436, chr15:44715419-
44715444, chr15:44715430-44715455, chr15:44715457-44715482, chr15:44715483-
44715508, chr15:44715511-44715536, chr15:44715515-44715540, chr15:44715629-
44715654, chr15:44715630-44715655, chr15:44715631-44715656, chr15:44715632-
44715657, chr15:44715653-44715678, chr15:44715657-44715682, chr15:44715666-
44715691, chr15:44715685-44715710, chr15:44715686-44715711, chr15:44716326-
44716351, chr15:44716329-44716354, chr15:44716313-44716338, chr15:44717599-
44717624, chr15:44717604-44717629, chr15:44717681-44717706, chr15:44717682-
44717707, chr15:44717702-44717727, chr15:44717764-44717789, chr15:44717776-
44717801, chr15:44717786-44717811, chr15:44717789-44717814, chr15:44717790-
44717815, chr15:44717794-44717819, chr15:44717805-44717830, chr15:44717808-
44717833, chr15:44717809-44717834, chr15:44717810-44717835, chr15:44717846-
44717871, chr15:44717945-44717970, chr15:44717946-44717971, chr15:44717947-
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
11
44717972, chr15:44717948-44717973, chr15:44717973-44717998, chr15:44717981-
44718006, chr15:44718056-44718081, chr15:44718061-44718086, chr15:44718067-
44718092, chr15:44718076-44718101, chr15:44717589-44717614, chr15:44717620-
44717645, chr15:44717642-44717667, chr15:44717771-44717796, chr15:44717800-
44717825, chr15:44717859-44717884, chr15:44717947-44717972, chr15:44718119-
44718144, chr15:44711563-44711585, chr15:44715428-44715450, chr15:44715509-
44715531, chr15:44715513-44715535, chr15:44715417-44715439, chr15:44711540-
44711562, chr15:44711574-44711596, chr15:44711597-44711619, chr15:44715446-
44715468, chr15:44715651-44715673, chr15:44713812-44713834, chr15:44711579-
44711601, chr15:44711542-44711564, chr15:44711557-44711579, chr15:44711609-
44711631, chr15:44715678-44715700, chr15:44715683-44715705, chr15:44715684-
44715706, chr15:44715480-44715502.
11. The modified limbal stem cell of embodiment 10, wherein the targeting
domain of the
gRNA molecule is complementary to a sequence within a genomic region selected
from: chr15:44715513-44715535, chr15:44711542-44711564, chr15:44711563-
44711585, chr15:44715683-44715705, chr15:44711597-44711619, or
chr15:44715446-44715468.
12. The modified limbal stem cell of embodiment 10, wherein the targeting
domain of the
gRNA molecule is complementary to a sequence within a genomic region
chr15:44711563-44711585.
13. The modified limbal stem cell of any one of embodiments 1-9, wherein
the targeting
domain of the gRNA molecule to B2M comprises a targeting domain comprising the
sequence of any one of SEQ ID NOs: 23-105 or 108-119 or 134-140.
14. The modified limbal stem cell of embodiment 13, wherein the targeting
domain of the
gRNA molecule to B2M comprises a targeting domain comprising the sequence of
any
one of SEQ ID NOs: 108, 111, 115, 116, 134 or 138.
15. The modified limbal stem cell of embodiment 13, wherein the targeting
domain of the
gRNA molecule to B2M comprises a targeting domain comprising the sequence of
SEQ ID NO: 108.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
12
16. The modified limbal stem cell of embodiment 13, wherein the targeting
domain of the
gRNA molecule to B2M comprises a targeting domain comprising the sequence of
SEQ ID NO: 115.
17. The modified limbal stem cell of embodiment 13, wherein the targeting
domain of the
gRNA molecule to B2M comprises a targeting domain comprising the sequence of
SEQ ID NO: 116.
18. The modified limbal stem cell of any one of embodiments 1-9, wherein
the gRNA
comprises the sequence of any one of SEQ ID NO: 120, 160-177.
19. The modified limbal stem cell of embodiment 18, wherein the gRNA
comprises the
sequence of any one of SEQ ID NO: 120, 162, 166, 167, 171, and 175.
20. The modified limbal stem cell of embodiment 18, wherein the gRNA comprises
the
sequence of SEQ ID NO: 120.
21. The modified limbal stem cell of embodiment 18, wherein the gRNA
comprises the
sequence of SEQ ID NO: 166.
22. The modified limbal stem cell of embodiment 18, wherein the gRNA
comprises the
sequence of SEQ ID NO: 167.
23. The modified limbal stem cell of embodiments 1-22, wherein the CRISPR
system is an
S. pyogenes Cas9 CRISPR system.
24. The modified limbal stem cell of embodiment 23, wherein the CRISPR
system
comprises a Cas9 molecule comprising SEQ ID NO: 106 or 107 or any of SEQ ID
NO:
124 to 134.
25. The modified limbal stem cell of embodiment 23, wherein the CRISPR
system
comprises a Cas9 molecule comprising SEQ ID NO: 106 or 107.
26. A modified limbal stem cell comprising a genome in which the b2
microglobulin
(B2M) gene on chromosome 15 has been edited
(a) to delete a contiguous stretch of genomic DNA comprising the sequence of
any
one of SEQ ID NOs: 141 to 159, thereby eliminating surface expression of MHC
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
13
Class I molecules in the cell, or
(b) to form an indel at or near the target sequence complementary to the
targeting
domain of the gRNA molecule comprising the sequence of any one of SEQ ID NOs:
23-105 or 108-119 or 134-140, thereby eliminating surface expression of MHC
Class
I molecules in the cell.
27. The modified limbal stem cell of embodiment 26 comprising a genome in
which the
b2 microglobulin (B2M) gene on chromosome 15 has been edited:
(a) to delete a contiguous stretch of genomic DNA comprising the sequence of
any
one of SEQ ID NOs: 141, 148 or 149, thereby eliminating surface expression, of
MHC Class I molecules in the cell, or
(b) to form an indel at or near the target sequence complementary to the
targeting
domain of the gRNA molecule domain comprising the sequence of any one of SEQ
ID NOs: 108, 111, 115, 116, 134 or 138, thereby eliminating surface expression
of
MHC Class I molecules in the cell.
28. The modified limbal stem cell of embodiment 26 comprising a genome in
which the
b2 microglobulin (B2M) gene on chromosome 15 has been:
(a) edited to delete a contiguous stretch of genomic DNA comprising the
sequence of
SEQ ID NOs: 141, thereby eliminating surface expression, of MHC Class I
molecules
in the cell, or
(b) to form an indel at or near the target sequence complementary to the
targeting
domain of the gRNA molecule domain comprising the sequence of any one of SEQ
ID NOs: 108, thereby eliminating surface expression of MHC Class I molecules
in the
cell.
29. A modified limbal stem cell comprising a genome in which the b2
microglobulin (B2M)
gene on chromosome 15 has been edited:
(a) to delete a contiguous stretch of genomic DNA region selected from any one
of:
chr15:44711469-44711494, chr15:44711472-44711497, chr15:44711483-44711508,
chr15:44711486-44711511, chr15:44711487-44711512, chr15:44711512-44711537,
chr15:44711513-44711538, chr15:44711534-44711559, chr15:44711568-44711593,
chr15:44711573-44711598, chr15:44711576-44711601, chr15:44711466-44711491,
chr15:44711522-44711547, chr15:44711544-44711569, chr15:44711559-44711584,
chr15:44711565-44711590, chr15:44711599-44711624, chr15:44711611-44711636,
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
14
chr15:44715412-44715437, chr15:44715440-44715465, chr15:44715473-44715498,
chr15:44715474-44715499, chr15:44715515-44715540, chr15:44715535-44715560,
chr15:44715562-44715587, chr15:44715567-44715592, chr15:44715672-44715697,
chr15:44715673-44715698, chr15:44715674-44715699, chr15:44715410-44715435,
chr15:44715411-44715436, chr15:44715419-44715444, chr15:44715430-44715455,
chr15:44715457-44715482, chr15:44715483-44715508, chr15:44715511-44715536,
chr15:44715515-44715540, chr15:44715629-44715654, chr15:44715630-44715655,
chr15:44715631-44715656, chr15:44715632-44715657, chr15:44715653-44715678,
chr15:44715657-44715682, chr15:44715666-44715691, chr15:44715685-44715710,
chr15:44715686-44715711, chr15:44716326-44716351, chr15:44716329-44716354,
chr15:44716313-44716338, chr15:44717599-44717624, chr15:44717604-44717629,
chr15:44717681-44717706, chr15:44717682-44717707, chr15:44717702-44717727,
chr15:44717764-44717789, chr15:44717776-44717801, chr15:44717786-44717811,
chr15:44717789-44717814, chr15:44717790-44717815, chr15:44717794-44717819,
chr15:44717805-44717830, chr15:44717808-44717833, chr15:44717809-44717834,
chr15:44717810-44717835, chr15:44717846-44717871, chr15:44717945-44717970,
chr15:44717946-44717971, chr15:44717947-44717972, chr15:44717948-44717973,
chr15:44717973-44717998, chr15:44717981-44718006, chr15:44718056-44718081,
chr15:44718061-44718086, chr15:44718067-44718092, chr15:44718076-44718101,
chr15:44717589-44717614, chr15:44717620-44717645, chr15:44717642-44717667,
chr15:44717771-44717796, chr15:44717800-44717825, chr15:44717859-44717884,
chr15:44717947-44717972, chr15:44718119-44718144, chr15:44711563-44711585,
chr15:44715428-44715450, chr15:44715509-44715531, chr15:44715513-44715535,
chr15:44715417-44715439, chr15:44711540-44711562, chr15:44711574-44711596,
chr15:44711597-44711619, chr15:44715446-44715468, chr15:44715651-44715673,
chr15:44713812-44713834, chr15:44711579-44711601, chr15:44711542-44711564,
chr15:44711557-44711579, chr15:44711609-44711631, chr15:44715678-44715700,
chr15:44715683-44715705, chr15:44715684-44715706, chr15:44715480-44715502,
thereby eliminating surface expression of MHC Class I molecules in the cell,
or
(b) to form an indel at or near the genomic DNA region selected from any one
of:
chr15:44711469-44711494, chr15:44711472-44711497, chr15:44711483-44711508,
chr15:44711486-44711511, chr15:44711487-44711512, chr15:44711512-44711537,
chr15:44711513-44711538, chr15:44711534-44711559, chr15:44711568-44711593,
chr15:44711573-44711598, chr15:44711576-44711601, chr15:44711466-44711491,
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
chr15:44711522-44711547, chr15:44711544-44711569, chr15:44711559-44711584,
chr15:44711565-44711590, chr15:44711599-44711624, chr15:44711611-44711636,
chr15:44715412-44715437, chr15:44715440-44715465, chr15:44715473-44715498,
chr15:44715474-44715499, chr15:44715515-44715540, chr15:44715535-44715560,
5 chr15:44715562-44715587, chr15:44715567-44715592, chr15:44715672-
44715697,
chr15:44715673-44715698, chr15:44715674-44715699, chr15:44715410-44715435,
chr15:44715411-44715436, chr15:44715419-44715444, chr15:44715430-44715455,
chr15:44715457-44715482, chr15:44715483-44715508, chr15:44715511-44715536,
chr15:44715515-44715540, chr15:44715629-44715654, chr15:44715630-44715655,
10 chr15:44715631-44715656, chr15:44715632-44715657, chr15:44715653-
44715678,
chr15:44715657-44715682, chr15:44715666-44715691, chr15:44715685-44715710,
chr15:44715686-44715711, chr15:44716326-44716351, chr15:44716329-44716354,
chr15:44716313-44716338, chr15:44717599-44717624, chr15:44717604-44717629,
chr15:44717681-44717706, chr15:44717682-44717707, chr15:44717702-44717727,
15 chr15:44717764-44717789, chr15:44717776-44717801, chr15:44717786-
44717811,
chr15:44717789-44717814, chr15:44717790-44717815, chr15:44717794-44717819,
chr15:44717805-44717830, chr15:44717808-44717833, chr15:44717809-44717834,
chr15:44717810-44717835, chr15:44717846-44717871, chr15:44717945-44717970,
chr15:44717946-44717971, chr15:44717947-44717972, chr15:44717948-44717973,
chr15:44717973-44717998, chr15:44717981-44718006, chr15:44718056-44718081,
chr15:44718061-44718086, chr15:44718067-44718092, chr15:44718076-44718101,
chr15:44717589-44717614, chr15:44717620-44717645, chr15:44717642-44717667,
chr15:44717771-44717796, chr15:44717800-44717825, chr15:44717859-44717884,
chr15:44717947-44717972, chr15:44718119-44718144, chr15:44711563-44711585,
chr15:44715428-44715450, chr15:44715509-44715531, chr15:44715513-44715535,
chr15:44715417-44715439, chr15:44711540-44711562, chr15:44711574-44711596,
chr15:44711597-44711619, chr15:44715446-44715468, chr15:44715651-44715673,
chr15:44713812-44713834, chr15:44711579-44711601, chr15:44711542-44711564,
chr15:44711557-44711579, chr15:44711609-44711631, chr15:44715678-44715700,
chr15:44715683-44715705, chr15:44715684-44715706, chr15:44715480-44715502,
thereby eliminating surface expression, of MHC Class I molecules in the cell.
30. The modified limbal stem cell of embodiment 29 comprising a genome in
which the b2
microglobulin (B2M) gene on chromosome 15 has been edited:
(a) to delete a contiguous stretch of genomic DNA region selected from:
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
16
chr15:44715513-44715535, chr15:44711542-44711564, chr15:44711563-44711585,
chrl 5:44715683-44715705, chrl 5:44711597-44711619, or chrl 5:44715446-
44715468, or
(b) to form an indel at or near the genomic DNA region selected from any one
of:
chr15:44715513-44715535, chr15:44711542-44711564, chr15:44711563-44711585,
chr15:44715683-44715705, chr15:44711597-44711619, or chr15:44715446-
44715468.
31. The modified limbal stem cell of embodiment 28 comprising a genome in
which the b2
microglobulin (B2M) gene on chromosome 15 has been edited
(a) to delete a contiguous stretch of genomic DNA region chr15:44711563-
44711585,
thereby eliminating surface expression of MHC Class I molecules in the cell,
or:
(b) to form an indel at or near the genomic DNA region, thereby eliminating
surface
expression of MHC Class I molecules in the cell.
32. The modified limbal stem cell of any one of the previous embodiments,
wherein the
modified limbal stem cell comprises an indel formed at or near the target
sequence
complementary to the targeting domain of the gRNA molecule.
33. The modified limbal stem cell of any one of embodiments 26(b), 27(b),
28(b), 29(b),
30(b) ir 31(b) or 32, wherein wherein the indel comprises a deletion of 10 or
greater
than 10 nucleotides, optionally 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24,
25, 26, 27, 28, 29, 30, 31,32, 33, 34, or 35 nucleotides.
34. The modified limbal stem cell any one of embodiments 26 to 33, wherein the
modified
limbal stem cell was cultured in media comprising a large tumor suppressor
kinase ("LATS")
inhibitor, optionally wherein the LATS inhibitor is a compound of Formula Al
R1 R2
R5
X1
N
A
R3
Al
or a salt thereof, wherein
X1 and X2 are each independently CH or N;
Ring A is
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
17
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
NH
N N and N
* NH 1
,
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_10cycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, Ci
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
18
6alkyl, hydroxyC1_6alkyl, C16haloalkyl, R , -NH2, C1_6alkylamino, and di-(C1_
6alkyl)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9-or 10-membered fused bicyclic heteroaryl comprising, as ring member,
Ito 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form a
4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional
.. heteroatoms independently selected from N, 0, and S, wherein the 4- to 6-
membered
heterocycloalkyl formed by R1 and R2 taken together with the nitrogen atom to
which both
are bound is unsubstituted or substituted by 1 to 3 substituents independently
selected from
halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein
the 3-to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
19
35. The modified limbal stem cell according to embodiment 34, wherein the
compound is
selected from: N-methy1-2-(pyridin-4-y1)-N-(1,1,1-trifluoropropan-2-
yOpyrido[3,4-
d]pyrimidin-4-amine; 2-methy1-1-(2-methy1-2-{[2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-4-
yl]amino}propoxy)propan-2-ol; 2,4-dimethy1-4-{[2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-4-
yl]amino}pentan-2-ol; N-tert-butyl-2-(pyrimidin-4-y1)-1,7-naphthyridin-4-
amine; 2-
(pyridin-4-y1)-N-[I -(trifluoromethyl)cyclobutyl]pyrido[3,4-d]pyrimidin-4-
amine; N-propy1-
2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-amine; N-(propan-2-y1)-2-(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; 3-(pyridin-4-y1)-N-(1-
(trifluoromethyl)cyclopropy1)-
2,6-naphthyridin-1-amine; 2-(3-methyl-1H-pyrazol-4-y1)-N-(1 -
methylcyclopropyl)pyrido[3,4-d]pyrimidin-4-amine; 2-methy1-2-{[2-(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-yl]amino}propan-1-ol; 2-(pyridin-4-y1)-4-(3-
(trifluoromethyDpiperazin-1-yOpyrido[3,4-d]pyrimidine; N-cyclopenty1-2-
(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; N-propy1-2-(3-(trifluoromethyl)-1H-pyrazol-4-
y1)pyrido[3,4-d]pyrimidin-4-amine; N-(2-methylcyclopenty1)-2-(pyridin-4-
yhpyrido[3,4-
d]pyrimidin-4-amine; 2-(3-chloropyridin-4-y1)-N-(1,1,1-trifluoro-2-
methylpropan-2-
yOpyrido[3,4-d]pyrimidin-4-amine; 2-(2-methy1-2-{[2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-
4-yl]amino}propoxy)ethan-1-ol; N-(1-methylcyclopropy1)-7-(pyridin-4-
yOisoquinolin-5-
amine; (1S,2S)-2-{[2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-
yl]amino}cyclopentan-1-ol;
N-methyl-2-(pyridin-4-y1)-N-[(2S)-1 ,1 ,1-trifluoropropan-2-yl]pyrido[3,4-
d]pyrimidin-4-
amine; N-methyl-N-(propan-2-y1)-2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-amine;
N-
(propan-2-y1)-2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-amine; 3-(pyridin-4-y1)-
N-(1-
(trifluoromethyl)cyclopropy1)-2,6-naphthyridin-1-amine and N-methy1-2-(pyridin-
4-y1)-N-
[(2R)-1 ,1,1-trifluoropropan-2-yl]pyrido[3,4-d]pyrimidin-4-amine.
36. The modified limbal stem cell according to embodiment 34, wherein the
compound is
selected from: 3-(pyridin-4-y1)-N-(1-(trifluoromethyl)cyclopropy1)-2,6-
naphthyridin-1-
amine; N-(1-methylcyclopropy1)-7-(pyridin-4-yOisoquinolin-5-amine; 2-(pyridin-
4-y1)-4-
(3-(trifluoromethyl)piperazin-1-yOpyrido[3,4-d]pyrimidine; N-(tert-buty1)-2-
(pyridin-4-y1)-
1,7-naphthyridin-4-amine; and N-methy1-2-(pyridin-4-y1)-N-[(2S)-1,1,1-
trifluoropropan-
2-yl]pyrido[3,4-d]pyrimidin-4-amine.
37. The modified limbal stem cell according to embodiment 34, wherein the
compound is
selected from: 3-(pyridin-4-y1)-N-(1-(trifluoromethyl)cyclopropy1)-2,6-
naphthyridin-1-
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
amine; N-(1-methylcyclopropy1)-7-(pyridin-4-yOisoquinolin-5-amine; and 2-
(pyridin-4-
y1)-4-(3-(trifluoromethyl)piperazin-1-yhpyrido[3,4-d]pyrimidine.
38. The modified limbal stem cell according to embodiment 34, wherein the
compound is
5 selected from: N-(tert-butyl)-2-(pyridin-4-y1)-1,7-naphthyridin-4-amine;
and N-methy1-2-
(pyridin-4-y1)-N-[(2S)-1,1,1-trifluoropropan-2-yl]pyrido[3,4-d]pyrimidin-4-
amine.
39. The modified limbal stem cell according to embodiment 34, wherein the
compound is
N-(tert-butyl)-2-(pyridin-4-y1)-1,7-naphthyridin-4-amine.
40. The modified limbal stem cell according to any one of embodiments 34 to
39, wherein
the compound is present in a concentration of 3 to 10 micromolar.
41. The modified limbal stem cell of any of embodiments 1-40, wherein the
cell is
autologous with respect to a patient to be administered said cell.
42. The modified limbal stem cell of any of embodiments 1-40, wherein the
cell is
allogeneic with respect to a patient to be administered said cell.
43. A method of preparing a modified limbal stem cell or a population of
modified limbal
stem cells for ocular cell therapy comprising,
a) modifying a limbal stem cell or a population of limbal stem cells by
reducing or
eliminating expression of B2M comprising introducing into the limbal stem cell
or the
population of limbal stem cells a CRISPR system comprising a gRNA molecule
with
a targeting domain
(i) comprising the sequence of any one of SEQ ID NOs: 23-105 or 108-119, or
134
to 140, or
(ii) complementary to a sequence within a genomic region selected from:
chr15:44711469-44711494, chr15:44711472-44711497, chr15:44711483-
44711508, chr15:44711486-44711511, chr15:44711487-44711512,
chr15:44711512-44711537, chr15:44711513-44711538, chr15:44711534-
44711559, chr15:44711568-44711593, chr15:44711573-44711598,
chr15:44711576-44711601, chr15:44711466-44711491, chr15:44711522-
44711547, chr15:44711544-44711569, chr15:44711559-44711584,
chr15:44711565-44711590, chr15:44711599-44711624, chr15:44711611-
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
21
44711636, chr15:44715412-44715437, chr15:44715440-44715465,
chr15:44715473-44715498, chr15:44715474-44715499, chr15:44715515-
44715540, chr15:44715535-44715560, chr15:44715562-44715587,
chr15:44715567-44715592, chr15:44715672-44715697, chr15:44715673-
44715698, chr15:44715674-44715699, chr15:44715410-44715435,
chr15:44715411-44715436, chr15:44715419-44715444, chr15:44715430-
44715455, chr15:44715457-44715482, chr15:44715483-44715508,
chr15:44715511-44715536, chr15:44715515-44715540, chr15:44715629-
44715654, chr15:44715630-44715655, chr15:44715631-44715656,
chr15:44715632-44715657, chr15:44715653-44715678, chr15:44715657-
44715682, chr15:44715666-44715691, chr15:44715685-44715710,
chr15:44715686-44715711, chr15:44716326-44716351, chr15:44716329-
44716354, chr15:44716313-44716338, chr15:44717599-44717624,
chr15:44717604-44717629, chr15:44717681-44717706, chr15:44717682-
44717707, chr15:44717702-44717727, chr15:44717764-44717789,
chr15:44717776-44717801, chr15:44717786-44717811, chr15:44717789-
44717814, chr15:44717790-44717815, chr15:44717794-44717819,
chr15:44717805-44717830, chr15:44717808-44717833, chr15:44717809-
44717834, chr15:44717810-44717835, chr15:44717846-44717871,
chr15:44717945-44717970, chr15:44717946-44717971, chr15:44717947-
44717972, chr15:44717948-44717973, chr15:44717973-44717998,
chr15:44717981-44718006, chr15:44718056-44718081, chr15:44718061-
44718086, chr15:44718067-44718092, chr15:44718076-44718101,
chr15:44717589-44717614, chr15:44717620-44717645, chr15:44717642-
44717667, chr15:44717771-44717796, chr15:44717800-44717825,
chr15:44717859-44717884, chr15:44717947-44717972, chr15:44718119-
44718144, chr15:44711563-44711585, chr15:44715428-44715450,
chr15:44715509-44715531, chr15:44715513-44715535, chr15:44715417-
44715439, chr15:44711540-44711562, chr15:44711574-44711596,
chr15:44711597-44711619, chr15:44715446-44715468, chr15:44715651-
44715673, chr15:44713812-44713834, chr15:44711579-44711601,
chr15:44711542-44711564, chr15:44711557-44711579, chr15:44711609-
44711631, chr15:44715678-44715700, chr15:44715683-44715705,
chr15:44715684-44715706, chr15:44715480-44715502,
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
22
wherein the limbal stem cell or the population of limbal stem cells have
optionally
been cultured in the presence of a LATS inhibitor; and
b) further expanding the modified limbal stem cell or thepopulation of
modified limbal
stem cells in cell culture media comprising a LATS inhibitor; and
c) optionally, enriching the population of limbal stem cells with the limbal
stem cells
having reduced or eliminated expression of B2M by fluorescene activated cell
sorting (FACS) or magnetic activated cell sorting (MACS).
44. The method of embodiment 43, wherein the LATS inhibitor is a compound
of Formula
Al
R1 R2
R5 'N'
Xi
N X2-L'~rs-N\,,
R3
Al
or a salt thereof, wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the remainder
of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
N NH
NH
/QIN
N
*
= N
\ NH , -\\
-NH L-
--N " , and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
23
R2 is selected from
(a) C1_8alkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OR , wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, Ci
6a1ky1, hydroxyC1_6alkyl, C1_6haloalkyl, R , -NH2, C1_6alkylamino, and di-(C1_
6a1ky1)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
24
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form a
4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional
heteroatoms independently selected from N, 0, and S, wherein the 4- to 6-
membered
heterocycloalkyl formed by R1 and R2 taken together with the nitrogen atom to
which both
are bound is unsubstituted or substituted by 1 to 3 substituents independently
selected from
halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein
the 3-to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises
1 to 2 oxygen atoms as chain members and is unsubstituted or substituted by R
.
45. The method according to embodiment 44, wherein the compound is
selected from: N-
methy1-2-(pyridin-4-y1)-N-(1,1,1-trifluoropropan-2-yOpyrido[3,4-d]pyrimidin-4-
amine; 2-
methy1-1-(2-methy1-2-{[2-(pyridin-4-y1)pyrido[3,4-d]pyrimidin-4-
yl]amino}propoxy)propan-2-ol; 2,4-dimethy1-4-{[2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-4-
yl]amino}pentan-2-ol; N-tert-butyl-2-(pyrimidin-4-y1)-1,7-naphthyridin-4-
amine; 2-
(pyridin-4-y1)-N-[I -(trifluoromethyl)cyclobutyl]pyrido[3,4-d]pyrimidin-4-
amine; N-propy1-
2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-amine; N-(propan-2-y1)-2-(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; 3-(pyridin-4-y1)-N-(1-
(trifluoromethyl)cyclopropy1)-
2,6-naphthyridin-1-amine; 2-(3-methy1-1H-pyrazol-4-y1)-N-(1-
methylcyclopropyl)pyrido[3,4-d]pyrimidin-4-amine; 2-methy1-2-{[2-(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-yl]amino}propan-1-ol; 2-(pyridin-4-y1)-4-(3-
(trifluoromethyDpiperazin-1-yOpyrido[3,4-d]pyrimidine; N-cyclopenty1-2-
(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; N-propy1-2-(3-(trifluoromethyl)-1H-pyrazol-4-
yl)pyrido[3,4-d]pyrimidin-4-amine; N-(2-methylcyclopenty1)-2-(pyridin-4-
yhpyrido[3,4-
d]pyrimidin-4-amine; 2-(3-chloropyridin-4-y1)-N-(1,1,1-trifluoro-2-
methylpropan-2-
yOpyrido[3,4-d]pyrimidin-4-amine; 2-(2-methy1-2-{[2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-
4-yl]amino}propoxy)ethan-1-ol; N-(1-methylcyclopropy1)-7-(pyridin-4-
yOisoquinolin-5-
amine; (1S,25)-2-{[2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-
yl]amino}cyclopentan-1-ol;
N-methyl-2-(pyridin-4-y1)-N-[(25)-1 ,1 ,1-trifluoropropan-2-yl]pyrido[3,4-
d]pyrimidin-4-
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
amine; N-methyl-N-(propan-2-y1)-2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-amine;
N-
(propan-2-y1)-2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-amine; 3-(pyridin-4-y1)-
N-(1-
(trifluoromethyl)cyclopropy1)-2,6-naphthyridin-1-amine and N-methy1-2-(pyridin-
4-y1)-N-
[(2 R)-1,1 ,1-trifluoropropan-2-yl]pyrido[3,4-d]pyrimidin-4-amine.
5
46. The method according to embodiment 44, wherein the compound is selected
from 3-
(pyridin-4-y1)-N-(1-(trifluoromethyl)cyclopropy1)-2,6-naphthyridin-1-amine; N-
(1-
methylcyclopropy1)-7-(pyridin-4-yOisoquinolin-5-amine; 2-(pyridin-4-y1)-4-(3-
(trifluoromethyDpiperazin-1-yOpyrido[3,4-d]pyrimidine; N-(tert-buty1)-2-
(pyridin-4-y1)-
10 1,7-naphthyridin-4-amine; and N-methy1-2-(pyridin-4-y1)-N-[(2S)-1,1,1-
trifluoropropan-
2-yl]pyrido[3,4-d]pyrimidin-4-amine.
47. The method according to embodiment 44, wherein the compound is selected
from 3-
(pyridin-4-y1)-N-(1-(trifluoromethyl)cyclopropy1)-2,6-naphthyridin-1-amine; N-
(1-
15 methylcyclopropy1)-7-(pyridin-4-yOisoquinolin-5-amine; and 2-(pyridin-
4-y1)-4-(3-
(trifluoromethyDpiperazin-1-yOpyrido[3,4-d]pyrimidine.
48. The method according to embodiment 44, wherein the compound is selected
from: N-
(tert-buty1)-2-(pyridin-4-y1)-1,7-naphthyridin-4-amine; and N-methy1-2-
(pyridin-4-y1)-N-
20 [(2S)-1,1,1-trifluoropropan-2-yl]pyrido[3,4-d]pyrimidin-4-amine.
49. The method according to embodiment 44, wherein the compound is N-(tert-
buty1)-2-
(pyridin-4-y1)-1,7-naphthyridin-4-amine.
25 50. The method according to any one of embodiments 44 to 49, wherein the
compound is
present in a concentration of 3 to 10 micromolar.
51. The method of any one of embodiments 43-50, wherein the CRISPR system
is an S.
pyo genes Cas9 CRISPR system.
52. The method of embodiment 51, wherein the CRISPR system comprises a Cas
9
molecule comprising SEQ ID NO: 106 or 107or 107 or any of SEQ ID NO: 124 to
134.
53. The method of embodiment 51, wherein the CRISPR system comprises a Cas
9
molecule comprising SEQ ID NO: 106 or 107or 107.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
26
54. A cell population comprising the modified limbal stem cell of any one
of embodiments
1 to 42 or the modified limbal stem cell obtained by the method of any one of
embodiments 43-53.
55. The cell population of embodiment 54, wherein the modified limbal stem
cell comprises
an indel formed at or near the target sequence complementary to the targeting
domain
of the gRNA molecule domain.
56. The cell population of embodiment 55, wherein the indel comprises a
deletion of 10 or
greater than 10 nucleotides, optionally 11, 12, 13, 14, 15, 16, 17, 18, 19,
20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31,32, 33, 34, or 35 nucleotides.
57. The cell population of embodiment 55 or 56, wherein the indel is formed
in at least
about 40%, e.g., at least about 50%, e.g., at least about 60%, e.g., at least
about 70%,
e.g., at least about 80%, e.g., at least about 90%, e.g., at least about 95%,
e.g., at
least about 96%, e.g., at least about 97%, e.g., at least about 98%, e.g., at
least about
99%, of the cells of the cell population, e.g., as detectible by next
generation
sequencing and/or a nucleotide insertional assay..
58. The cell population of any one of embodiments 55 to 57, wherein an off-
target indel is
detected in no more than about 5%, e.g., no more than about 1%, e.g., no more
than
about 0.1%, e.g., no more than about 0.01%, of the cells of the cell
population, e.g., as
detectible by next generation sequencing and/or a nucleotide insertional
assay.
59. A composition comprising the modified limbal stem cell of any one of
embodiments 1
to 42 or the modified limbal stem cell obtained by the method of any one of
embodiments 43-53 or the cell population of any one of embodiments 54-58 or
the
population of modified limbal stem cells obtained by the method of any one of
embodiments 43-53.
60. The composition of embodiment 54, wherein the modified limbal stem cell
comprises
an indel formed at or near the target sequence complementary to the targeting
domain
of the gRNA molecule domain.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
27
61. The composition of embodiment 55, wherein the indel comprises a deletion
of 10 or
greater than 10 nucleotides, optionally 11, 12, 13, 14, 15, 16, 17, 18, 19,
20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31,32, 33, 34, or 35 nucleotides.
62. The composition of embodiment 55 or 56, wherein the indel is formed in at
least about
40%, e.g., at least about 50%, e.g., at least about 60%, e.g., at least about
70%, e.g.,
at least about 80%, e.g., at least about 90%, e.g., at least about 95%, e.g.,
at least
about 96%, e.g., at least about 97%, e.g., at least about 98%, e.g., at least
about 99%,
of the cells of the population.
63. The composition of any one of embodiments 55 to 57, wherein an off-
target indel is
detected in no more than about 5%, e.g., no more than about 1%, e.g., no more
than
about 0.1%, e.g., no more than about 0.01%, of the cells of the population of
cells e.g.,
as detectible by next generation sequencing and/or a nucleotide insertional
assay.
64. The modified limbal stem cell of any one of embodiments 1 to 42 or the
cell population
of any one of embodiments 54 to 58 or the composition of any one of
embodiments 59
to 63 for use in treatment of an an ocular disease.
65. The modified limbal stem cell or the cell population or the composition
for use
according to embodiment 64, wherein the ocular disease is limbal stem cell
deficiency.
66. The modified limbal stem cell or the cell population or the composition
for use
according to embodiment 65, wherein the ocular disease is unilateral limbal
stem cell
deficiency.
67. The modified limbal stem cell or the cell population or the composition
for use
according to embodiment 65, wherein the ocular disease is bilateral limbal
stem cell
deficiency.
68. The modified limbal stem cell or the cell population or the composition
for use
according to any one of embodiments 59 to 62, wherein the cell is autologous
with
respect to a patient to be administered said cell.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
28
69. The modified limbal stem cell or the cell population or the
composition for use
according to any one of embodiments 59 to 62, wherein the cell is allogeneic
with
respect to a patient to be administered said cell.
70. A method of treating a patient suffering from an ocular disease comprising
the step of
administering to the patient in need thereof the modified limbal stem cell of
any one of
embodiments 1-42 or the cell population of any one of embodiments 54 to 58 or
the
composition of any one of embodiments 59 to 63.
71. The method of embodiment 70, wherein the ocular disease is limbal stem
cell
deficiency.
72. The method of embodiment 71, wherein the ocular disease is unilateral
limbal stem
cell deficiency.
73. The method of embodiment 71, wherein the ocular disease is bilateral
limbal stem cell
deficiency.
74. The method of any one of embodiments 71 to 73, wherein the cell is
autologous with
respect to a patient to be administered said cell.
75. The method of any one of embodiments 71 to 73, wherein the cell is
allogeneic with
respect to a patient to be administered said cell.
76. Use of the modified limbal stem cell of any one of embodiments 1 to 42
or the cell
population of any one of embodiments 54 to 58 or the composition of any one of
embodiments 59 to 63 for the treatment of an ocular disease.
77. Use of embodiment 76, wherein the ocular disease is limbal stem cell
deficiency.
Other features and advantages of the present invention will be apparent from
the following
detailed description and claims.
V. BRIEF DESCRIPTION OF THE FIGURES
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
29
Figure 1: LATS inhibitors (compound ex. 3 and ex. 4) induce YAP
dephosphorylation in
LSCs within one hour of treatment as shown by Western blot.
Figure 2: Immunolabelling of p63-alpha in limbal stem cell cultures indicates
that the LSC
population can be expanded when it is maintained in medium comprising the LATS
inhibitors
(compound ex. 3 and ex. 4). Figure 2A: In the presence of growth medium and
DMSO, only
a few isolated cells attach to the culture dish and survive up to 6 days. Most
cells expressed
the human nuclear marker, but few expressed p63a1pha. Figures 2B and 2C: In
contrast, in
the presence of LATS inhibitors: compound example no. 3 and example no. 4, the
cells
.. formed colonies and expressed p63a1pha. This result indicated that the LATS
inhibitors
promote the expansion of the population of cells with the p63a1pha-positive
phenotype.
Figure 20: Passaging cells and culturing them in the presence of LATS
inhibitor compound
for two weeks enabled cell population expansion and the formation of confluent
cultures
expressing p63a1pha.
Figure 3: FACS analyses show CRISPR-mediated deletion of B2M with sgRNA SEQ ID
NO: 120 and subsequent elimination of HLA A, B and C occurred in about 70
percent of the
LSCs.
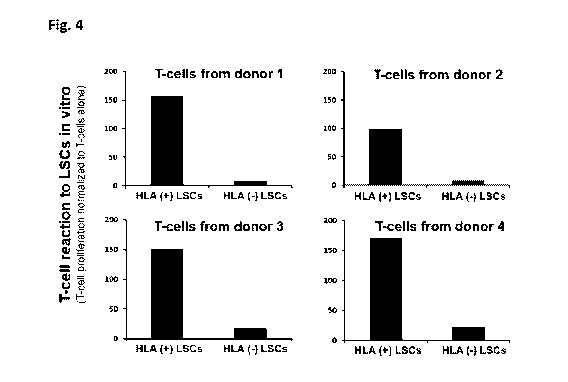
Figure 4: Graphs showing the results of gene edited LSCs (CRISPR-mediated
deletion of
B2M with sgRNA SEQ ID NO: 120) co-cultured with CD8+ T-cells from 4 different
donors.
Figure 5: Efficiency of B2M deletion. Figure 5 shows FACS data detecting B2M
surface
protein on gene edited limbal stem cells, which were CRISPR-edited with sgRNA
CR00442,
CR000446, CR000455, 1-CR004366, 4-CR004366, 6-HEYJA000001, 8-HEYJA000004, and
9-HEYJA000005 represented in Table 6. All sgRNAs show a B2M surface protein
knockout
of between 27-62%.
Figure 6: Efficiency of HLA A,B,C elimination. Figure 6 shows FACS data
detecting HLA-
ABC surface protein on gene edited limbal stem cells, which were CRISPR-edited
with
sgRNA CR00442, CR000446, CR000455, 1-CR004366, 4-CR004366, 6-HEYJA000001, 8-
HEYJA000004, and 9-HEYJA000005 represented in Table 6. All sgRNAs show an HLA-
ABC surface protein elimination of between 28-60%.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
Figure 7: MACS-mediated selection of B2M-negative LSCs. Figure 7 shows FACS
data
detecting B2M surface protein on gene edited limbal stem cells, which were
MACS treated
after nucleofection to obtain a B2M negative LSC culture. All sgRNAs tested
(CR00442,
CR000446, CR000455, 1-CR004366, 4-CR004366, 6-HEYJA000001, 8-HEYJA000004, and
5 9-HEYJA000005, represented in Table 6) show a pure (-99 t0100%) B2M
negative LSC
culture.
Figure 8: MACS-mediated selection of HLA A,B,C-negative LSCs. Figure 8 shows
FACS
data detecting HLA-ABC surface protein on gene edited limbal stem cells, which
were
10 MACS treated after nucleofection to obtain a B2M/HLA-ABC negative LSC
culture. All
sgRNAs tested (CR00442, CR000446, CR000455, 1-CR004366, 4-CR004366, 6-
HEYJA000001, 8-HEYJA000004, and 9-HEYJA000005, represented in Table 6) show a
pure (-99 t0100%) HLA-ABC negative LSC culture.
15 VI. DETAILED DESCRIPTION
LATS
LATS is the abbreviated name of the large tumor suppressor kinase. LATS as
used herein
20 refers to LATS1 and/or LATS2. LATS1 as used herein refers to the large
tumor suppressor
kinase 1 and LATS2 refers to the large tumor suppressor kinase 2. LATS1 and
LATS2 both
have serine/threonine protein kinase activity. LATS1 and LATS2 have been given
the
Human Genome Organisation (HUGO) Gene Nomenclature Committee identifiers: HGNC
ID
6514 and HGNC ID 6515 respectively. LATS1 is sometimes also referred to in the
art as
25 WARTS or wts, and LATS2 is sometimes referred to in the art as KPM.
Representative
LATS sequences, include, but are not limited to, the protein sequences
available from the
National Center for Biotechnology Information protein database with the
accession numbers
NP_004681.1 (LATS1) and NP_001257448.1 (LATS1) and NP_055387.2 (LATS 2), as
shown below.
LATS1: NP 004681.1 (Serine/threonine¨protein kinase LATS1 isoform 1, homo
sapiens)(SEQ ID NO: 1:)
1 mkrsekpegy rqmrpktfpa snytvssrqm lqeiresIrn Iskpsdaaka ehnmskmste
61 dprqvrnppk fgthhkalqe irnsllpfan etnssrstse vnpqmlqdlq aagfdedmvi
121 qalqktnnrs ieaaiefisk msyqdprreq maaaaarpin asmkpgnvqq svnrkqswkg
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
31
181 skeslvpqrh gpplgesvay hsespnsqtd vgrplsgsgi safvqahpsn gqrvnppppp
241 qvrsvtpppp prgqtppprg ttppppswep nsqtkrysgn meyvisrisp vppgawqegy
301 ppppintspm nppnqgqrgi ssvpvgrqpi imqssskfnf psgrpgmqng tgqtdfmihq
361 nvvpagtvnr qppppyplta angqspsalq tggsaapssy tngsipqsmm vpnrnshnme
421 lynisvpglq tnwpqsssap aqsspssghe iptwqpnipv rsnsfnnplg nrashsansq
481 psattvtait papiqqpvks mrvIkpelqt alapthpswi pqpiqtvqps pfpegtasnv
541 tvmppvaeap nyqgppppyp khllhqnpsv ppyesiskps kedqpslpke deseksyenv
601 dsgdkekkqi ttspitvrkn kkdeerresr iqsyspqafk ffmeqhvenv lkshqqr1hr
661 kkqlenemmr vglsqdaqdq mrkmlcqkes nyinkrakm dksmfvkikt Igigafgevc
721 larkvdtkal yatktlrkkd vlIrnqvahv kaerdilaea dnewvvrlyy sfqdkdnlyf
781 vmdyipggdm msllirmgif peslarfyia eltcavesvh kmgfihrdik pdnilidrdg
841 hikltdfglc tgfrwthdsk yyqsgdhprq dsmdfsnewg dpsscrcgdr Ikplerraar
901 qhqrclahsl vgtpnyiape vlIrtgytql cdwwsvgvil femlvgqppf laqtpletqm
961 kvinwqtslh ippqaklspe asdliiklcr gpedrIgkng adeikahpff ktidfssdlr
1021 qqsasyipki thptdtsnfd pvdpdklwsd dneeenvndt Ingwykngkh pehafyeftf
1081 rrffddngyp ynypkpieye yinsqgseqq sdeddqntgs eiknrdlvyv
LATS1: serine/threonine-protein kinase LATS1 isoform 2 [Homo sapiens]
NCB! Reference Sequence: NP_001257448.1 (SEQ ID NO: 2:)
1 mkrsekpegy rqmrpktfpa snytvssrqm lqeiresIrn Iskpsdaaka ehnmskmste
61 dprqvrnppk fgthhkalqe irnsllpfan etnssrstse vnpqmlqdlq aagfdedmvi
121 qalqktnnrs ieaaiefisk msyqdprreq maaaaarpin asmkpgnvqq svnrkqswkg
181 skeslvpqrh gpplgesvay hsespnsqtd vgrplsgsgi safvqahpsn gqrvnppppp
241 qvrsvtpppp prgqtppprg ttppppswep nsqtkrysgn meyvisrisp vppgawqegy
301 ppppintspm nppnqgqrgi ssvpvgrqpi imqssskfnf psgrpgmqng tgqtdfmihq
361 nvvpagtvnr qppppyplta angqspsalq tggsaapssy tngsipqsmm vpnrnshnme
421 lynisvpglq tnwpqsssap aqsspssghe iptwqpnipv rsnsfnnplg nrashsansq
481 psattvtait papiqqpvks mrvIkpelqt alapthpswi pqpiqtvqps pfpegtasnv
541 tvmppvaeap nyqgppppyp khllhqnpsv ppyesiskps kedqpslpke deseksyenv
601 dsgdkekkqi ttspitvrkn kkdeerresr iqsyspqafk ffmeqhvenv lkshqqr1hr
661 kkqlenemmr vkpfkmsifi InhlfawcIf
LATS 2: NP_055387.2 serine/threonine-protein kinase LATS2 [Homo sapiens].
((SEQ ID
NO: 3:)
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
32
1 mrpktfpatt ysgnsrqrlq eireglkqps kssvqglpag pnsdtsldak vlgskdatrq
61 qqqmratpkf gpyqkalrei rysllpfane sgtsaaaevn rqmlqelvna gcdqemagra
121 Ikqtgsrsie aaleyiskmg yldprneqiv rvikqtspgk glmptpvtrr psfegtgdsf
181 asyhqlsgtp yegpsfgadg ptaleemprp yvdylfpgvg phgpghqhqh ppkgygasve
241 aagahfplqg ahygrphllv pgeplgygvq rspsfqsktp petggyaslp tkgqggppga
301 glafpppaag lyvphphhkq agpaahqlhv Igsrsqvfas dsppqslltp smslnvdly
361 elgstsvqqw paatlarrds lqkpgleapp rahvafrpdc pvpsrtnsfn shqprpgppg
421 kaepslpapn tvtavtaahi Ihpyksvrvl rpepqtavgp shpawvpapa papapapapa
481 aegldakeeh alalggagaf pldveyggpd rrcppppypk hIlIrskseq ydldslcagm
541 eqslragpne peggdksrks akgdkggkdk kqiqtspvpv rknsrdeekr esriksyspy
601 afkffmeqhv enviktyqqk vnrrlqleqe makaglceae qeqmrkilyq kesnynnkr
661 akmdksmfvk iktlgigafg evclackvdt halyamktlr kkdvinrnqv ahvkaerdil
721 aeadnewvvk lyysfqdkds lyfvmdyipg gdmmsllirm evfpehlarf yiaeltlaie
781 svhkmgfihr dikpdnilid Idghikltdf glctgfrwth nskyyqkgsh vrqdsmepsd
841 lwddvsncrc gdrIktleqr arkqhqrcla hslvgtpnyi apevlIrkgy tqlcdwwsvg
901 vilfemlvgq ppflaptpte tqlkvinwen tlhipaqvkl speardlitk IccsadhrIg
961 rngaddlkah pffsaidfss dirkqpapyv ptishpmdts nfdpvdeesp wndasegstk
1021 awdtltspnn khpehafyef tfrrffddng ypfrcpkpsg aeasqaessd lessdlvdqt
1081 egcqpvyv
LATS is thought to negatively regulate YAP1 activity. "YAP1" refers to the yes-
associated
protein 1, also known as YAP or YAP65, which is a protein that acts as a
transcriptional
regulator of genes involved in cell proliferation. LATS kinases are
serine/threonine protein
kinases that have been shown to directly phosphorylate YAP which results in
its cytoplasmic
retention and inactivation. Without phosphorylation by LATS, YAP translocates
into the
nucleus, forming a complex with a DNA binding protein, TEAD, and results in
downstream
gene expression. (Barry ER & Camargo FD (2013) The Hippo superhighway:
signaling
crossroads converging on the Hippo/Yap pathway in stem cells and development.
Current
opinion in cell biology 25(2):247-253.; Mo JS, Park HW, & Guan KL (2014) The
Hippo
signaling pathway in stem cell biology and cancer. EMBO reports 15(6):642-656;
Pan D
(2010) The hippo signaling pathway in development and cancer. Developmental
cell
19(4):491-505.)
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
33
The Hippo/YAP pathway is involved in numerous cell types and tissues in
mammalian
systems, including various cancers. In particular, the Hippo pathway is
evidently involved in
the intestine, stomach and esophagus, pancreas, salivary gland, skin, mammary
gland,
ovary, prostate, brain and nervous system, bone, chrondrocytes, adipose cells,
myocytes, T
lymphocytes, B lymphocytes, myeloid cells, kidney, and lung. See Nishio et
al., 2017,
Genes to Cells 22:6-31.
LATS1 and LATS2 Inhibition
Compounds of Formula Al or subformulae thereof (e.g., Formula A2), in free
form or in salt
form are potent inhibitors of LATS1 and/or LATS2.
In a preferred embodiment, the compounds of Formula A2 or subformulae thereof,
in free
form or in salt form are potent inhibitors of LATS1 and LATS2.
LATS Inhibitors
The invention therefore relates to a compound of Formula A2:
R1 R2
R5 'NI
X1
N
A
R3 2
or a salt, or stereoisomer thereof, wherein
XI is CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
a 9-membered fused bicyclic heteroaryl that is selected from
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
34
401
N N N
.1
NH NH mN , and ,
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
and
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) C1_8alkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R6;
(viii) -NR7aR7b, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R6;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R6;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, Ci
6a1ky1, hydroxyC1_6alkyl, C1_6haloalkyl, R , -NH2, C1_6alkylamino, and di-(C1_
6a1ky1)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
5 (b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
10 6a1ky1 that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
15 C(0)R ;
or, provided that when X1 is CH, R1 and R2 can be taken together with the
nitrogen atom to
which both are bound to form a 4- to 6-membered heterocycloalkyl that can
include, as
ring members, 1 to 2 additional heteroatoms independently selected from N, 0,
and S,
wherein the 4- to 6-membered heterocycloalkyl formed by R1 and R2 taken
together
20 with the nitrogen atom to which both are bound is unsubstituted or
substituted by 1 to 3
substituents independently selected from halogen, C1_6alkyl, C1_6haloalkyl,
and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
25 comprises 1 to 2 oxygen atoms as chain members and is unsubstituted
or substituted
by R ;
with the proviso that:
(1) when X1 is N, ring A is 4-primidinyl or 3-fluoro-4-primidinyl, R1 is H
or methyl, R3 is H or
Cl and R5 is H; then R2 is not C2_4alkyl that is substituted with a
substituent selected
30 from ¨NH2, C1_6alkylamino or t-butyl-carbamoyl-amino and that is
optionally further
substituted with unsubstituted phenyl; and
(2) when X1 is N, ring A is indazol-5-yl, R1, R3 and R5 are H; then R2 is not
Caalkyl that is
substituted with ¨N H2.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
36
Unless specified otherwise, the term "compounds of the present invention"
refers to
compounds of Formula Al or subformulae thereof (e.g., Formula A2), or salts
thereof, as
well as all stereoisomers (including diastereoisomers and enantiomers),
rotamers, tautomers
and isotopically labeled compounds (including deuterium substitutions), as
well as inherently
formed moieties.
Various (enumerated) embodiments of the invention are described herein. It
will be
recognized that features specified in each embodiment may be combined with
other
specified features to provide further embodiments of the present invention.
When an
.. embodiment is described as being "according to" a previous embodiment, the
previous
embodiment includes sub-embodiments thereof, for example such that when
Embodiment
is described as being "according to" embodiments 1 to 19, embodiments 1 to 19
includes
embodiments 19 and 19A.
15 Embodiment I. A method of cell population expansion,
comprising the step of a)
culturing a population of cells comprising limbal stem cells in the presence
of a LATS
inhibitor to generate an expanded population of cells comprising limbal stem
cells,
wherein the limbal stem cells have reduced or eliminated expression of B2M by
a
CRISPR system (e.g., S. pyogenes Cas9 CRISPR system), for example, a CRISPR
20 system comprising a gRNA selected from those described in Table 1 or
Table 4 or Table
6.
Embodiment 2. A method of cell population expansion, comprising the
step of a)
culturing a population of cells comprising corneal endothelial cells in the
presence of a
LATS inhibitor to generate an expanded population of cells comprising corneal
endothelial cells, wherein the corneal endothelial cells have reduced or
eliminated
expression of B2M by a CRISPR system, for example, a CRISPR system (e.g., S.
pyo genes Cas9 CRISPR system) comprising a gRNA selected from those described
in
Table 1 or Table 4 or Table 6.
Embodiment 3. The method of cell population expansion according to Embodiment
1 or
Embodiment 2, wherein the LATS inhibitor is a compound of Formula Al
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
37
R2
R5
Xi
x2y
N
A
R3 k I
Al
or a salt thereof, wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the remainder
of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
N,, ir"\
-1!))
N
NH /91
\¨NH NH and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) C1_8alkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
38
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R6;
(viii) -NR7aR7b, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R6;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, C1_
6a1ky1, hydroxyC1_6alkyl, C1_6haloalkyl, R , -NH2, C1_6alkylamino, and di-(C1_
6a1ky1)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6a1ky1 that is unsubstituted or substituted by R or ¨C(0)R6; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R6;
or R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form a
4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional
heteroatoms independently selected from N, 0, and S, wherein the 4- to 6-
membered
heterocycloalkyl formed by R1 and R2 taken together with the nitrogen atom to
which
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
39
both are bound is unsubstituted or substituted by 1 to 3 substituents
independently
selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3-to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises Ito
2 oxygen atoms as chain members and is unsubstituted or substituted by R ..
Embodiment 4. A method of cell population expansion, comprising the step
of a) culturing
a seeding population of cells comprising limbal stem cells in the presence of
a compound of
Formula A1,
R1 R2
R5
X1
N
A
R3
Al
or a salt thereof to generate an expanded population of cells comprising
limbal stem cells,
wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
*
N N
*
' N
\ NH , ¨NHILz-zikir4H NH
N , and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
5 (i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
10 (vi) C1_6haloalkyl;
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
15 (ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, Ci
6a1ky1, hydroxyC1_6alkyl, C1_6haloalkyl, R , -NH2, C1_6alkylamino, and di-(C1_
20 6a1ky1)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
25 (xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
30 (b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
41
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form a
4-to 6-membered heterocycloalkyl that can include, as ring members, Ito 2
additional
heteroatoms independently selected from N, 0, and S, wherein the 4- to 6-
membered
heterocycloalkyl formed by R1 and R2 taken together with the nitrogen atom to
which
both are bound is unsubstituted or substituted by 1 to 3 substituents
independently
selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
Embodiment 5. A method of cell population expansion, comprising the step
of a) culturing
a seeding population of cells comprising corneal endothelial cells in the
presence of a
compound of Formula Al,
,R2
R5 N
'" X1
N -
X2 17-
R3
Al
or a salt thereof to generate an expanded population of cells comprising
corneal endothelial
cells, wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
42
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
N,
NH
N and N
* NH
¨NH
N
, ,
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, Ci
6a1ky1, hydroxyC1_6alkyl, C16haloalkyl, R , -NH2, C1_6alkylamino, and di-(C1_
6a1ky1)amino;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
43
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6a1ky1 that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form a
4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional
heteroatoms independently selected from N, 0, and S, wherein the 4- to 6-
membered
heterocycloalkyl formed by R1 and R2 taken together with the nitrogen atom to
which
both are bound is unsubstituted or substituted by 1 to 3 substituents
independently
selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
Embodiment 6. The method of cell population expansion according to
Embodiment 3
to Embodiment 5, wherein the compound is selected from: N-methy1-2-(pyridin-4-
y1)-
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
44
N-(1,1,1-trifluoropropan-2-yOpyrido[3,4-d]pyrimidin-4-amine; 2-methy1-1-(2-
methy1-2-
{[2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-yl]amino}propoxy)propan-2-ol; 2,4-
dimethy1-
4-{[2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-yl]amino}pentan-2-ol; N-tert-buty1-
2-
(pyrimidin-4-y1)-1 ,7-naphthyridin-4-amine; 2-(pyridin-4-yI)-N-[1-
(trifluoromethyl)cyclobutyl]pyrido[3,4-d]pyrimidin-4-amine; N-propy1-2-
(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; N-(propan-2-y1)-2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-4-amine; 3-(pyridin-4-y1)-N-(1-(trifluoromethyl)cyclopropy1)-2,6-
naphthyridin-1 -amine; 2-(3-methy1-1H-pyrazol-4-y1)-N-(1-
methylcyclopropyl)pyrido[3,4-d]pyrimidin-4-amine; 2-methy1-2-{[2-(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-yl]amino}propan-1-ol; 2-(pyridin-4-y1)-4-(3-
(trifluoromethyl)piperazin-1-yOpyrido[3,4-d]pyrimidine; N-cyclopenty1-2-
(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; N-propy1-2-(3-(trifluoromethyl)-1H-pyrazol-4-
yOpyrido[3,4-d]pyrimidin-4-amine; N-(2-methylcyclopenty1)-2-(pyridin-4-
yOpyrido[3,4-
d]pyrimidin-4-amine; 2-(3-chloropyridin-4-yI)-N-(1,1,1-trifluoro-2-
methylpropan-2-
yl)pyrido[3,4-d]pyrimidin-4-amine; 2-(2-methy1-2-{[2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-4-yl]amino}propoxy)ethan-1-ol; N-(1-methylcyclopropy1)-7-(pyridin-
4-
yOisoquinolin-5-amine; (1S,2S)-2-{[2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-
yl]amino}cyclopentan-1-ol; N-methyl-2-(pyridin-4-y1)-N-[(2S)-1 ,1,1-
trifluoropropan-2-
yl]pyrido[3,4-d]pyrimidin-4-amine; N-methyl-N-(propan-2-yI)-2-(pyridin-4-
yl)pyrido[3,4-d]pyrimidin-4-amine; N-(propan-2-y1)-2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-4-amine; 3-(pyridin-4-y1)-N-(1-(trifluoromethyl)cyclopropy1)-2,6-
naphthyridin-1-amine and N-methyl-2-(pyridin-4-y1)-N-[(2R)-1 ,1,1-
trifluoropropan-2-
yl]pyrido[3,4-d]pyrimidin-4-amine.
Embodiment 7. The method of cell population expansion according to
Embodiment 3
to Embodiment 5, wherein the compound is selected from 3-(pyridin-4-y1)-N-(1-
(trifluoromethyl)cyclopropy1)-2,6-naphthyridin-1-amine; N-(1-
methylcyclopropy1)-7-
(pyridin-4-yOisoquinolin-5-amine; 2-(pyridin-4-y1)-4-(3-
(trifluoromethyDpiperazin-1-
yOpyrido[3,4-d]pyrimidine; N-(tert-butyl)-2-(pyridin-4-y1)-1,7-naphthyridin-4-
amine;
and N-methyl-2-(pyridin-4-y1)-N-[(2S)-1 ,1,1-trifluoropropan-2-yl]pyrido[3,4-
d]pyrimidin-4-amine.
Embodiment 8. The method of cell population expansion according to
Embodiment 3
to Embodiment 5, wherein the compound is selected from 3-(pyridin-4-yI)-N-(1-
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
(trifluoromethyl)cyclopropyI)-2,6-naphthyridin-1-amine; N-(1-
methylcyclopropy1)-7-
(pyridin-4-yOisoquinolin-5-amine; and 2-(pyridin-4-y1)-4-(3-
(trifluoromethyDpiperazin-
1-yOpyrido[3,4-d]pyrimidine.
5 Embodiment 9. The
method of cell population expansion according to Embodiment 3
to Embodiment 5, wherein the compound is selected from: N-(tert-buty1)-2-
(pyridin-4-
y1)-1,7-naphthyridin-4-amine; and N-methy1-2-(pyridin-4-y1)-N-[(2S)-1,1,1-
trifluoropropan-2-yl]pyrido[3,4-d]pyrimidin-4-amine.
10
Embodiment 10. The method of cell population expansion according to Embodiment
3
to Embodiment 5, wherein the compound is selected is N-(tert-buty1)-2-(pyridin-
4-y1)-
1,7-naphthyridin-4-amine.
Embodiment 11. The method of cell population expansion according to Embodiment
3
15 to
Embodiment 5, wherein said compound is present in a concentration of 0.5 to
100
micromolar, preferably 0.5 to 25 micromolar, more preferably 1 to 20
micromolar,
particularly preferably of about 3 to 10 micromolar.
Embodiment 12. The method of cell population expansion according to Embodiment
3
20 to
Embodiment 5, wherein in step a) the compound is present for one to two weeks
and subsequently step b) is performed wherein the cells are cultured for a
period in
growth medium without supplementation with said compound, preferably wherein
the
period is one to two weeks.
25
Embodiment 13. The method of cell population expansion according to Embodiment
1
to Embodiment 5, wherein the method produces greater than 10 fold expansion of
the seeded amount of cells.
Embodiment 14. The method of cell population expansion according to Embodiment
1
30 to Embodiment 5, wherein the method produces 15 fold to 600 fold,
preferably 20
fold to 550 fold expansion of the seeded amount of cells.
Embodiment 15. The method of cell population expansion according to Embodiment
1
or Embodiment 2, wherein the LATS inhibitor inhibits LATS1 and LATS2.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
46
Embodiment 16. The method of cell population expansion according to any one of
Embodiment 2 to Embodiment 3 or Embodiment 5 to Embodiment 15, wherein said
method further comprises genetically modifying said corneal endothelial cells.
Embodiment 17. The method of cell population expansion according to any one of
Embodiment 1 or Embodiment 4 or Embodiment 6 to Embodiment 15, wherein said
method further comprises genetically modifying said limbal stem cells.
Embodiment 18. The method of cell population expansion according to Embodiment
16
or Embodiment 17, wherein said genetically modifying comprises reducing or
eliminating the expression and/or function of a gene associated with
facilitating a
host versus graft immune response.
Embodiment 19. The method of cell population expansion according to any one of
Embodiment 16 to Embodiment 18, wherein said genetically modifying comprises
introducing into said cell a gene editing system which specifically targets a
gene
associated with facilitating a host versus graft immune response.
Embodiment 20. The method of cell population expansion according to Embodiment
19, wherein said gene editing system is a CRISPR gene editing system.
Embodiment 21. The method of cell population expansion according to any one of
Embodiment 16 to Embodiment 20, wherein said gene is B2M.
Embodiment 22. The method of cell population expansion according to any one of
Embodiment 1 to Embodiment 21, which comprises the further step after
generation
of an expanded population of cells of rinsing those cells to substantially
remove the
compound.
Embodiment 23. A cell population obtainable by the method of any one of
Embodiment
1 to Embodiment 22.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
47
Embodiment 24. A cell population obtained by the method of any one of
Embodiment 1
to Embodiment 22.
Embodiment 25. A cell population comprising corneal endothelial cells or the
cell
population of Embodiment 23 or Embodiment 24, wherein one or more of said
cells
comprises a non-naturally occurring insertion or deletion of one or more
nucleic acid
residues of a gene associated with facilitating a host vs graft immune
response,
wherein insertion and/or deletion results in reduced or eliminated expression
or
function of said gene.
Embodiment 26. The cell population according to Embodiment 25, wherein said
gene is
B2M.
Embodiment 27. A composition comprising the cell population according to
Embodiment 25 or Embodiment 26.
Embodiment 28. A method of culturing cells comprising culturing a population
of cells
comprising corneal endothelial cells in the presence of a LATS inhibitor.
Embodiment 29. The method of culturing cells according to Embodiment 28,
wherein
the LATS inhibitor is a compound of Formula Al,
R1 R2
R5
N
"`' X1
X211)110
R3
Al
or a salt thereof, wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
48
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
."` ,
sN, NH N
,NH
.NH NH
, and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or Ci_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R6;
(viii) -NR7aR713, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R6;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, Ci
6a1ky1, hydroxyC1_6alkyl, C1_6haloalkyl, R , -NH2, C1_6alkylamino, and di-(C1_
6a1ky1)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
49
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form a
4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional
heteroatoms independently selected from N, 0, and S, wherein the 4- to 6-
membered
heterocycloalkyl formed by R1 and R2 taken together with the nitrogen atom to
which
both are bound is unsubstituted or substituted by 1 to 3 substituents
independently
selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
Embodiment 30. A method of culturing cells comprising culturing a population
of cells
comprising corneal endothelial cells in the presence of a compound of Formula
Al,
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
R2
R5
Xi
x2y
N
A
R3 k I
Al
or a salt thereof, wherein
X1 and X2 are each independently CH or N;
Ring A is
5 (a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
10 heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
N,, ir"\
-1!))
N
NH /91
\¨NH NH and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
15 from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
20 (a) C1_8alkyl that is unsubstituted or substituted by 1 to 3
substituents independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
25 (iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
51
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R6;
(viii) -NR7aR7b, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R6;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, C1_
6a1ky1, hydroxyC1_6alkyl, C1_6haloalkyl, R , -NH2, C1_6alkylamino, and di-(C1_
6a1ky1)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6a1ky1 that is unsubstituted or substituted by R or ¨C(0)R6; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R6;
or R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form a
4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional
heteroatoms independently selected from N, 0, and S, wherein the 4- to 6-
membered
heterocycloalkyl formed by R1 and R2 taken together with the nitrogen atom to
which
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
52
both are bound is unsubstituted or substituted by 1 to 3 substituents
independently
selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
Embodiment 31. A method of culturing cells comprising culturing a population
of cells
comprising limbal stem cells in the presence of a LATS inhibitor.
Embodiment 32. The method of culturing cells according to Embodiment 31,
wherein
the LATS inhibitor is a compound of Formula Al,
,R2
R5 N
X1
N X210
R3 A
Al
or a salt thereof, wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
,C1 NH
N N N
NH \
NH
, and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
53
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, C1_
6a1ky1, hydroxyC1_6alkyl, C16haloalkyl, R , -NH2, C1_6alkylamino, and di-(C1_
6a1ky1)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
54
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6a1ky1 that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form a
4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional
heteroatoms independently selected from N, 0, and S, wherein the 4- to 6-
membered
heterocycloalkyl formed by R1 and R2 taken together with the nitrogen atom to
which
both are bound is unsubstituted or substituted by 1 to 3 substituents
independently
selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
Embodiment 33. A method of culturing cells comprising culturing a population
of cells
comprising limbal stem cells in the presence of a compound of Formula Al,
Ri R2
R5 sl\l'
X1
N
X211y*Th\,,
R3
A
Al
or a salt thereof, wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
5 (b) a 9-membered fused bicyclic heteroaryl that is selected from
_11i4
N N NH *"-N
N
\
NH ¨NH and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
10 C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
15 selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
20 (v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
25 hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by
¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, Ci
30 6a1ky1, hydroxyC1_6alkyl, C1_6haloalkyl, R , -NH2, C1_6alkylamino,
and di-(C1_
6a1ky1)amino;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
56
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6a1ky1 that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form a
4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional
heteroatoms independently selected from N, 0, and S, wherein the 4- to 6-
membered
heterocycloalkyl formed by R1 and R2 taken together with the nitrogen atom to
which
both are bound is unsubstituted or substituted by 1 to 3 substituents
independently
selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
Embodiment 34. . Use of a compound of the Formula Al, or a salt thereof,
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
57
,R2
R5
Xi
x2y
N
A
R3 k I
Al
in a method of generating an expanded population of limbal stem cells,
preferably ex vivo,
wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
JL
N N N
NH
\ -NH , NH N N
, and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
58
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R6;
(viii) -NR7aR7b, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R6;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, C1_
6a1ky1, hydroxyC1_6alkyl, C1_6haloalkyl, R , -NH2, C1_6alkylamino, and di-(C1_
6a1ky1)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6a1ky1 that is unsubstituted or substituted by R or ¨C(0)R6; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R6;
or R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form a
4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional
heteroatoms independently selected from N, 0, and S, wherein the 4- to 6-
membered
heterocycloalkyl formed by R1 and R2 taken together with the nitrogen atom to
which
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
59
both are bound is unsubstituted or substituted by 1 to 3 substituents
independently
selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
Embodiment 35. Use of a compound of the Formula Al, or a salt thereof,
R1 R2
R5 sN'
X1
N
X2--CO
R3
Al
in a method of generating an expanded corneal endothelial cell population,
preferably ex
vivo, wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the remainder
of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
N *
N
, and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
R2 is selected from
(a) C1_8alkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
5 (ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
10 (vii) -OR , wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or C1_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
15 (x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen, Ci
6a1ky1, hydroxyC1_6alkyl, C1_6haloalkyl, R , -NH2, C1_6alkylamino, and di-(C1_
6a1ky1)amino;
20 (xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted
by 1 to 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl,
C1_6alkylamino, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
25 (xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring
members, 1 to 4
heteroatoms independently selected from N and 0; and
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
30 (c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
61
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R :
or R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form a
4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional
heteroatoms independently selected from N, 0, and S, wherein the 4- to 6-
membered
heterocycloalkyl formed by R1 and R2 taken together with the nitrogen atom to
which
both are bound is unsubstituted or substituted by 1 to 3 substituents
independently
selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
Embodiment 36. Use of a compound of the Formula Al or a salt thereof,
according to
Embodiment 34 or Embodiment 35, wherein the compound is of the formula
selected
from Formulae Ito IV:
R1 R2 R:: ,R2
R5 R5 N
N
r:4.'`'^r)CA) N
R3 R3
R1 ,R2 R1 R2
R5 N R5 'NI'
N N
( 9
R3 C-;) IV. Ill,
and
Embodiment 37. Use of a compound of Formula Al or a salt thereof, according to
Embodiment 34 or Embodiment 35, wherein the compound is selected from 3-
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
62
(pyridin-4-y1)-N-(1-(trifluoromethyl)cyclopropy1)-2,6-naphthyridin-1-amine; N-
(1-
methylcyclopropy1)-7-(pyridin-4-yOisoquinolin-5-amine; and 2-(pyridin-4-y1)-4-
(3-
(trifluoromethyDpiperazin-1-yOpyrido[3,4-d]pyrimidine.
Embodiment 38. Use of a compound of the Formula Al, or a salt thereof,
according to
Embodiment 34 or Embodiment 35 , wherein the compound is selected from: N-
methy1-2-(pyridin-4-y1)-N-(1,1,1-trifluoropropan-2-yOpyrido[3,4-d]pyrimidin-4-
amine;
2-methy1-1-(2-methy1-2-{[2-(pyridin-4-y1)pyrido[3,4-d]pyrimidin-4-
yl]amino}propoxy)propan-2-ol; 2,4-dimethy1-4-{[2-(pyridin-4-yl)pyrido[3,4-
d]pyrimidin-
4-yl]amino}pentan-2-ol; N-tert-butyl-2-(pyrimidin-4-y1)-1,7-naphthyridin-4-
amine; 2-
(pyridin-4-y1)-N-[I -(trifluoromethyl)cyclobutyl]pyrido[3,4-d]pyrimidin-4-
amine; N-
propy1-2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-amine; N-(propan-2-y1)-2-
(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; 3-(pyridin-4-y1)-N-(1-
(trifluoromethyl)cyclopropy1)-
2,6-naphthyridin-1-amine; 2-(3-methy1-1H-pyrazol-4-y1)-N-(1-
methylcyclopropyl)pyrido[3,4-d]pyrimidin-4-amine; 2-methy1-2-{[2-(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-yl]amino}propan-1-ol; 2-(pyridin-4-y1)-4-(3-
(trifluoromethyl)piperazin-1-yOpyrido[3,4-d]pyrimidine; N-cyclopenty1-2-
(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; N-propy1-2-(3-(trifluoromethyl)-1H-pyrazol-4-
yOpyrido[3,4-d]pyrimidin-4-amine; N-(2-methylcyclopenty1)-2-(pyridin-4-
yOpyrido[3,4-
d]pyrimidin-4-amine; 2-(3-chloropyridin-4-y1)-N-(1,1,1-trifluoro-2-
methylpropan-2-
yOpyrido[3,4-d]pyrimidin-4-amine; 2-(2-methy1-2-{[2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-4-yl]amino}propoxy)ethan-1-ol; N-(1-methylcyclopropy1)-7-(pyridin-
4-
yOisoquinolin-5-amine; (1S,2S)-2-{[2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-
yl]amino}cyclopentan-1-ol; N-methyl-2-(pyridin-4-y1)-N-[(2S)-1 ,1,1-
trifluoropropan-2-
yl]pyrido[3,4-d]pyrimidin-4-amine; N-methyl-N-(propan-2-y1)-2-(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; N-(propan-2-y1)-2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-4-amine; 3-(pyridin-4-y1)-N-(1-(trifluoromethyl)cyclopropy1)-2,6-
naphthyridin-1-amine and N-methyl-2-(pyridin-4-y1)-N-[(2R)-1 ,1,1-
trifluoropropan-2-
yl]pyrido[3,4-d]pyrimidin-4-amine.
Embodiment 39. Use of a compound of the Formula Al, or a salt thereof,
according to
Embodiment 34 or Embodiment 35 , wherein the compound is selected from: N-
(tert-
buty1)-2-(pyridin-4-y1)-1,7-naphthyridin-4-amine; and N-methy1-2-(pyridin-4-
y1)-N-
[(2S)-1,1,1-trifluoropropan-2-yl]pyrido[3,4-d]pyrimidin-4-amine.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
63
Embodiment 40. Use of a compound of the Formula Al, or a salt thereof,
according to
Embodiment 34 or Embodiment 35 , wherein the compound is N-(tert-buty1)-2-
(pyridin-4-y1)-1,7-naphthyridin-4-amine.
Embodiment 41. A method of treatment of an ocular disease or disorder
comprising
administering to a subject in need thereof a modified cell population, wherein
the cell
population has been grown in the presence of a compound of Formula Al, or a
salt
thereof,
R1 R2
R5 sN'
NO
R3
Al
wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
N
1
NH
NH N and
*¨N /.:1".N
,
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
64
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OR , wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or-C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or Ci_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted or
substituted by 1 to 2 substituents independently selected from halogen,
C1_6alkyl,
hydroxyC1_6alkyl, C1_6haloalkyl, R , -N H2, C1_6alkylamino, and di-
(C1_6alkyl)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted by 1 to
2 substituents independently selected from hydroxyl, halogen, C1_6alkyl,
C1_6alkylamino,
and di-(C1_6alkyhamino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9-or 10-membered fused bicyclic heteroaryl comprising, as ring member,
Ito 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or, R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form
5 a 4- to 6-membered heterocycloalkyl that can include, as ring members, 1
to 2
additional heteroatoms independently selected from N, 0, and S, wherein the 4-
to 6-
membered heterocycloalkyl formed by R1 and R2 taken together with the nitrogen
atom
to which both are bound is unsubstituted or substituted by 1 to 3 substituents
independently selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
10 R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
Embodiment 42. A method of treatment of an ocular disease or disorder
comprising
administering to a subject in need thereof a modified limbal stem cell
population,
wherein said population has been grown in the presence of a compound of
Formula
Al, or a salt thereof,
R1 R2
R5
x2- --ED
R3
Al
wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
66
1 NH
N N
NH
t--AH NH N
, and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R6;
(viii) -NR7aR713, wherein R7a is hydrogen or C1_6alkyl, and R713 is
selected from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R6;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted
or substituted by 1 to 2 substituents independently selected from halogen,
C1_6alkyl,
hydroxyC1_6alkyl, C16haloalkyl, R , -N H2, C1_6alkylamino, and di-
(C1_6alkyl)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted by
1 to 2 substituents independently selected from hydroxyl, halogen, C1_6alkyl,
Ci
6a1ky1amin0, and di-(C1_6alkyl)amino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring
members, 1 to 4
heteroatoms independently selected from N and 0; and
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
67
(xv) 9- or 10-membered fused bicyclic heteroaryl comprising, as
ring member, 1 to
2 heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or, R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form
a 4-to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional heteroatoms independently selected from N, 0, and S, wherein the 4-
to 6-
membered heterocycloalkyl formed by R1 and R2 taken together with the nitrogen
atom
to which both are bound is unsubstituted or substituted by 1 to 3 substituents
independently selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
Embodiment 43. A method of treatment of an ocular disease or disorder
comprising
administering to a subject in need thereof a modified corneal endothelial cell
population, wherein the population has been grown in the presence of a
compound
of Formula Al, or a salt thereof,
R1 R2
R5
X
N
X2*C0
A
R3
Al
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
68
wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
N N NH
,NE1 I
\
IN N T ¨NH -NH , and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or Ci_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
69
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted or
substituted by 1 to 2 substituents independently selected from halogen,
C1_6alkyl,
hydroxyC1_6alkyl, C1_6haloalkyl, R , -N H2, C1_6alkylamino, and di-
(C1_6alkyl)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted by 1 to
2 substituents independently selected from hydroxyl, halogen, C1_6alkyl,
C1_6alkylamino,
and di-(C1_6alkyhamino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9-or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or, R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form
a 4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional heteroatoms independently selected from N, 0, and S, wherein the 4-
to 6-
membered heterocycloalkyl formed by R1 and R2 taken together with the nitrogen
atom
to which both are bound is unsubstituted or substituted by 1 to 3 substituents
independently selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
Embodiment 44. A method of treatment of an ocular disease or disorder
according to
Embodiment 41 to Embodiment 43 , wherein the compound is of the formula
selected from Formulae Ito IV:
R1 R2 R1 R2
R5 N R5 'N'
N
N N N
R3 R3 Cr-)
5
R1 R2 R1 R2
R5 'N' Rb 'N'
N
N N 0
,-
A
R3 R3
III, and IV.
Embodiment 45. A method of treatment of an ocular disease or disorder
according to
10 Embodiment 41 to Embodiment 43 ,wherein the compound is selected
from 3-
(pyridin-4-y1)-N-(1-(trifluoromethyl)cyclopropy1)-2,6-naphthyridin-1-amine; N-
(1-
methylcyclopropy1)-7-(pyridin-4-yOisoquinolin-5-amine; 2-(pyridin-4-y1)-4-(3-
(trifluoromethyDpiperazin-1-yOpyrido[3,4-d]pyrimidine; N-(tert-buty1)-2-
(pyridin-4-y1)-
1,7-naphthyridin-4-amine; and N-methy1-2-(pyridin-4-y1)-N-[(2S)-1,1,1-
trifluoropropan-
15 2-yl]pyrido[3,4-d]pyrimidin-4-amine.
Embodiment 46. A method of treatment of an ocular disease or disorder
according to
Embodiment 41 to Embodiment 43 , wherein the compound is selected from: N-
methy1-2-(pyridin-4-y1)-N-(1,1,1-trifluoropropan-2-yOpyrido[3,4-d]pyrimidin-4-
amine;
20 2-methy1-1-(2-methy1-2-{[2-(pyridin-4-y1)pyrido[3,4-d]pyrimidin-4-
yl]amino}propoxy)propan-2-ol; 2,4-dimethy1-4-{[2-(pyridin-4-yl)pyrido[3,4-
d]pyrimidin-
4-yl]amino}pentan-2-ol; N-tert-butyl-2-(pyrimidin-4-y1)-1,7-naphthyridin-4-
amine; 2-
(pyridin-4-y1)-N-[I -(trifluoromethyl)cyclobutyl]pyrido[3,4-d]pyrimidin-4-
amine; N-
propy1-2-(pyridin-4-yOpyrido[3,4-d]pyrimidin-4-amine; N-(propan-2-yI)-2-
(pyridin-4-
25 yOpyrido[3,4-d]pyrimidin-4-amine; 3-(pyridin-4-y1)-N-(1-
(trifluoromethyl)cyclopropy1)-
2,6-naphthyridin-1-amine; 2-(3-methy1-1H-pyrazol-4-y1)-N-(1-
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
71
methylcyclopropyl)pyrido[3,4-d]pyrimidin-4-amine; 2-methy1-2-{[2-(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-yl]amino}propan-1-ol; 2-(pyridin-4-y1)-4-(3-
(trifluoromethyl)piperazin-1-yOpyrido[3,4-d]pyrimidine; N-cyclopenty1-2-
(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; N-propy1-2-(3-(trifluoromethyl)-1H-pyrazol-4-
yl)pyrido[3,4-d]pyrimidin-4-amine; N-(2-methylcyclopenty1)-2-(pyridin-4-
yOpyrido[3,4-
d]pyrimidin-4-amine; 2-(3-chloropyridin-4-y1)-N-(1,1,1-trifluoro-2-
methylpropan-2-
yOpyrido[3,4-d]pyrimidin-4-amine; 2-(2-methy1-2-{[2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-4-yl]amino}propoxy)ethan-1-ol; N-(1-methylcyclopropy1)-7-(pyridin-
4-
yOisoquinolin-5-amine; (1S,2S)-2-{[2-(pyridin-4-yl)pyrido[3,4-d]pyrimidin-4-
yl]amino}cyclopentan-1-ol; N-methy1-2-(pyridin-4-y1)-N-[(2S)-1,1,1-
trifluoropropan-2-
yl]pyrido[3,4-d]pyrimidin-4-amine; N-methyl-N-(propan-2-y1)-2-(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-amine; N-(propan-2-y1)-2-(pyridin-4-yOpyrido[3,4-
d]pyrimidin-4-amine; 3-(pyridin-4-y1)-N-(1-(trifluoromethyl)cyclopropy1)-2,6-
naphthyridin-1-amine and N-methy1-2-(pyridin-4-y1)-N-[(2R)-1,1,1-
trifluoropropan-2-
yl]pyrido[3,4-d]pyrimidin-4-amine.
Embodiment 47. A method of treatment of an ocular disease or disorder
according to
Embodiment 41 to Embodiment 43 , wherein the compound is selected from: N-
(tert-
buty1)-2-(pyridin-4-y1)-1,7-naphthyridin-4-amine; and N-methy1-2-(pyridin-4-
y1)-N-
[(2S)-1,1,1-trifluoropropan-2-yl]pyrido[3,4-d]pyrimidin-4-amine.
Embodiment 48. A method of treatment of an ocular disease or disorder
according to
Embodiment 41 to Embodiment 43, wherein the compound is N-(tert-buty1)-2-
(pyridin-4-y1)-1,7-naphthyridin-4-amine.
Embodiment 49. A method of promoting cell proliferation of modified limbal
stem cells
or modified corneal endothelial cells, the method comprising culturing the
modified
limbal stem cells or modified corneal endothelial cells in a cell
proliferation medium
comprising a compound of Formula Al, or a salt thereof,
R1 R2
N
R3
Al
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
72
wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
N N NH
,NE1 I
\
IN N T ¨NH -NH , and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OR , wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or Ci_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
73
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted or
substituted by 1 to 2 substituents independently selected from halogen,
C1_6alkyl,
hydroxyC1_6alkyl, C1_6haloalkyl, R , -N H2, C1_6alkylamino, and di-
(C1_6alkyl)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted by 1 to
2 substituents independently selected from hydroxyl, halogen, C1_6alkyl,
C1_6alkylamino,
and di-(C1_6alkyhamino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9-or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or, R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form
a 4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional heteroatoms independently selected from N, 0, and S, wherein the 4-
to 6-
membered heterocycloalkyl formed by R1 and R2 taken together with the nitrogen
atom
to which both are bound is unsubstituted or substituted by 1 to 3 substituents
independently selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3- to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted
by R .
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
74
Embodiment 50. A cell preparation comprising a LATS inhibitor and modified
corneal
endothelial cells.
Embodiment 51. The cell preparation according to Embodiment 50, wherein the
LATS
inhibitor is a compound of Formula Al,
R1 R2
R5
X.1
N eCip
R3 'µ\_A
Al
.. wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
N,
*
I N N NH erIN
NH I 61
NH -NH , and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
5 (iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OR , wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
10 substituted by R or-C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or Ci_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
15 (xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are
each unsubstituted or
substituted by 1 to 2 substituents independently selected from halogen,
C1_6alkyl,
hydroxyC1_6alkyl, C1_6haloalkyl, R , -N H2, C1_6alkylamino, and di-
(C1_6alkyl)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted by 1 to
20 2 substituents independently selected from hydroxyl, halogen,
C1_6alkyl, C1_6alkylamino,
and di-(C1_6alkyhamino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
25 (xv) 9-or 10-membered fused bicyclic heteroaryl comprising, as ring
member, Ito 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
30 (d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
76
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or, R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form
a 4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional heteroatoms independently selected from N, 0, and S, wherein the 4-
to 6-
membered heterocycloalkyl formed by R1 and R2 taken together with the nitrogen
atom
to which both are bound is unsubstituted or substituted by 1 to 3 substituents
independently selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3-to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises Ito
2 oxygen atoms as chain members and is unsubstituted or substituted by R .
Embodiment 52. A cell preparation comprising a compound of Formula Al,
R1 R2
R5 sl\l'
N'
N
X2-kri)
R3
Al
and modified corneal endothelial cells, wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
77
N N
\ N
I -1'1A
-NH .,
,N1-1
L'"'N , and ,
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OR , wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or Ci_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_10cycloalkyl that are each
unsubstituted or
substituted by 1 to 2 substituents independently selected from halogen,
C1_6alkyl,
hydroxyC1_6alkyl, C16haloalkyl, R , -N H2, C1_6alkylamino, and di-
(C1_6alkyl)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted by 1 to
2 substituents independently selected from hydroxyl, halogen, C1_6alkyl,
C1_6alkylamino,
and di-(C1_6alkyhamino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
78
(xv) 9-or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or, R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form
a 4-to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional heteroatoms independently selected from N, 0, and S, wherein the 4-
to 6-
membered heterocycloalkyl formed by R1 and R2 taken together with the nitrogen
atom
to which both are bound is unsubstituted or substituted by 1 to 3 substituents
independently selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein
the 3-to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted by
R .
Embodiment 53. A cell preparation comprising a LATS inhibitor and modified
limbal
stem cells.
Embodiment 54. The cell preparation according to Embodiment 53, wherein the
LATS
inhibitor is a compound of Formula Al,
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
79
RI R2
R5 'N'
Xi
N
R3
Al
wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the remainder
of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
17'N)
1-1 H
N
¨NH -NH, and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) C1_8alkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
(vii) -OR , wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or Ci_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
5 .. (ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted or
substituted by 1 to 2 substituents independently selected from halogen,
C1_6alkyl,
hydroxyC1_6alkyl, C16haloalkyl, R , -N H2, C1_6alkylamino, and di-
(C1_6alkyl)amino;
10 (xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted by 1 to
2 substituents independently selected from hydroxyl, halogen, C1_6alkyl,
C1_6alkylamino,
and di-(C1_6alkyhamino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
15 (xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring
members, 1 to 4
heteroatoms independently selected from N and 0; and
(xv) 9-or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
20 (c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
25 (e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
30 or, R1 and R2 can be taken together with the nitrogen atom to which both
are bound to form
a 4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional heteroatoms independently selected from N, 0, and S, wherein the 4-
to 6-
membered heterocycloalkyl formed by R1 and R2 taken together with the nitrogen
atom
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
81
to which both are bound is unsubstituted or substituted by 1 to 3 substituents
independently selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein the
3-to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises Ito
2 oxygen atoms as chain members and is unsubstituted or substituted by R .
Embodiment 55. A cell preparation comprising a compound of Formula Al,
R1 R2
R5 sN'
.õx
N
A
Al
and modified limbal stem cells, wherein
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the remainder
of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
NH
, and
I *¨Nfr
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or C1_6alkoxy;
R1 is hydrogen or C1_6alkyl;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
82
R2 is selected from
(a) C1_8alkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OR , wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R ;
(viii) -NR7aR7b, wherein R7a is hydrogen or Ci_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by ¨C(0)R ;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted or
substituted by 1 to 2 substituents independently selected from halogen,
C1_6alkyl,
hydroxyC1_6alkyl, C1_6haloalkyl, R , -N H2, C1_6alkylamino, and di-
(C1_6alkyl)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
independently selected from N, 0 and S and that is unsubstituted or
substituted by 1 to
2 substituents independently selected from hydroxyl, halogen, C1_6alkyl,
C1_6alkylamino,
and di-(C1_6alkyhamino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
heteroatoms independently selected from N and 0; and
(xv) 9-or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
83
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
selected from N, 0 and S and that is unsubstituted or substituted by 1 to 2
substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or, R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form
a 4- to 6-membered heterocycloalkyl that can include, as ring members, 1 to 2
additional heteroatoms independently selected from N, 0, and S, wherein the 4-
to 6-
membered heterocycloalkyl formed by R1 and R2 taken together with the nitrogen
atom
to which both are bound is unsubstituted or substituted by 1 to 3 substituents
independently selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein
the 3-to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted by
R .
Embodiment 56. The cell preparation according to any one of Embodiment 50 to
Embodiment 55, which further comprises a growth medium, wherein the growth
medium is selected from the group consisting of Dulbecco's Modified Eagle's
Medium supplemented with Fetal Bovine Serum, human endothelial serum free
medium with human serum, X-VIV015 medium and mesenchymal stem cell-
conditioned medium; preferably X-VIV015 medium.
Embodiment 57. A method for expanding a population of modified cells ex vivo
which
comprises contacting the cells with a compound of Formula Al,
R1 R2
N
'J
Al
wherein
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
84
X1 and X2 are each independently CH or N;
Ring A is
(a) a 5- or 6-membered monocyclic heteroaryl that is linked to the
remainder of the
molecule through a carbon ring member and comprises, as ring member, 1 to 4
heteroatoms that are independently selected from N, 0 and S, provided that at
least
one of the heteroatom ring member is an unsubstituted nitrogen (-N=)
positioned at the
3- or the 4-position relative to the linking carbon ring member of the 5-
membered
heteroaryl or at the para ring position of the 6-membered heteroaryl; or
(b) a 9-membered fused bicyclic heteroaryl that is selected from
NH
N N
NH I *-Na'N
N
NH -NH , , and
wherein "*" represents the point of attachment of ring A to the remainder of
the molecule;
wherein ring A is unsubstituted or substituted by 1 to 2 substituents
independently selected
from halogen, cyano, C16alkyl, C1_6haloalkyl, -NH2, C1_6alkylamino, di-
(C1_6alkyl)amino,
C3_6cycloalkyl, and phenylsulfonyl;
R is hydroxyl or Ci_6alkoxy;
R1 is hydrogen or C1_6alkyl;
R2 is selected from
(a) Ci_salkyl that is unsubstituted or substituted by 1 to 3 substituents
independently
selected from
(i) halogen;
(ii) cyano;
(iii) oxo;
(iv) C2alkenyl;
(v) C2alkynyl;
(vi) C1_6haloalkyl;
(vii) -OW, wherein R6 is selected from hydrogen, C1_6alkyl that is
unsubstituted or
substituted by R or -C(0)R6;
(viii) -NR7aR7b, wherein R7a is hydrogen or Ci_6alkyl, and R713 is selected
from
hydrogen, -C(0)R , C1_6alkyl that is unsubstituted or substituted by -C(0)R6;
(ix) -C(0)R8, wherein R8 is R or -NH-Ci_6alkyl-C(0)R ;
(x) -S(0)2C1_6alkyl;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
(xi) monocyclic C3_6cycloalkyl or polycyclic C7_1ocycloalkyl that are each
unsubstituted or
substituted by 1 to 2 substituents independently selected from halogen,
C1_6alkyl,
hydroxyC1_6alkyl, C16haloalkyl, R , -N H2, C1_6alkylamino, and di-
(C1_6alkyl)amino;
(xii) 6-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
5 independently selected from N, 0 and S and that is unsubstituted or
substituted by 1 to
2 substituents independently selected from hydroxyl, halogen, C1_6alkyl,
C1_6alkylamino,
and di-(C1_6alkyhamino;
(xiii) phenyl that is unsubstituted or substituted by halogen;
(xiv) 5- or 6-membered monocyclic heteroaryl comprising, as ring members, 1 to
4
10 heteroatoms independently selected from N and 0; and
(xv) 9-or 10-membered fused bicyclic heteroaryl comprising, as ring member, 1
to 2
heteroatoms independently selected from N and 0;
(b) -S(0)2C1_6alkyl;
(c) phenyl that is unsubstituted or substituted by 1 to 2 substituents
independently
15 selected from halogen, C1_6alkyl and R ;
(d) C3_6cycloalkyl that is unsubstituted or substituted by 1 to 2
substituents independently
selected from C1_6haloalkyl, R , C1_6alkylamino, di-(C1_6alkyl)amino, -C(0)R ,
and C1-
6alkyl that is unsubstituted or substituted by R or ¨C(0)R ; and
(e) 4-membered heterocycloalkyl comprising, as ring members, 1 to 2
heteroatoms
20 selected from N, 0 and S and that is unsubstituted or substituted by 1
to 2 substituents
independently selected from C1_6haloalkyl, R , C1_6alkylamino,
6a1ky1)amino, -C(0)R , and C1_6alkyl that is unsubstituted or substituted by R
or ¨
C(0)R ;
or, R1 and R2 can be taken together with the nitrogen atom to which both are
bound to form
25 a 4- to 6-membered heterocycloalkyl that can include, as ring members, 1
to 2
additional heteroatoms independently selected from N, 0, and S, wherein the 4-
to 6-
membered heterocycloalkyl formed by R1 and R2 taken together with the nitrogen
atom
to which both are bound is unsubstituted or substituted by 1 to 3 substituents
independently selected from halogen, C1_6alkyl, C1_6haloalkyl, and R ;
30 R3 is selected from hydrogen, halogen and C1_6alkyl; and
R5 is selected from hydrogen, halogen and ¨NH-(3- to 8-membered heteroalkyl),
wherein
the 3-to 8-membered heteroC3_8alkyl of the ¨NH-(3- to 8-membered heteroalkyl)
comprises 1 to 2 oxygen atoms as chain members and is unsubstituted or
substituted by
R .
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
86
Embodiment 58. The method according to Embodiment 57, wherein said modified
cells
are gene edited cells.
Embodiment 59. Cells obtained by the method according to any one of Embodiment
57
to 58.
In one embodiment, said compound of the present invention is present in a
concentration of
about 0.5 to about 100 micromolar, preferably of about 0.5 to about 25
micromolar, more
preferably of about 1 to about 20 micromolar, particularly preferably of about
3 to about 10
micromolar. In one embodiment, said compound of the present invention is
present in a
concentration of 0.5 to 100 micromolar, preferably 0.5 to 25 micromolar, more
preferably 1
to 20 micromolar, particularly preferably of 3 to 10 micromolar. In a specific
embodiment,
said compound of the present invention is present in a concentration of 3 to
10 micromolar.
In another embodiment, the present invention relates to a method of treatment
of an ocular
disease or disorder comprising administering to a subject in need thereof a
cell population
(e.g., cell population comprising modified limbal stem cell with reduced or
eliminated
expression of B2M by a CRISPR system), wherein the population has been grown
in the
presence of an agent capable of inhibiting the activity of LATS1 and LATS2
kinases;
thereby inducing YAP translocation and driving downstream gene expression for
cell
proliferation. In a further embodiment, the agent is a compound of Formula Al
or
subformulae thereof (e.g., Formula A2), or pharmaceutically acceptable salt
thereof.
In another embodiment, the present invention relates to a method of treatment
of an ocular
disease or disorder comprising administering to a subject in need thereof a
limbal stem cell
population (e.g., cell population comprising modified limbal stem cell with
reduced or
eliminated expression of B2M by a CRISPR system), wherein the population has
been
grown in the presence of an agent capable of inhibiting the activity of LATS1
and LATS2
kinases; thereby inducing YAP translocation and driving downstream gene
expression for
cell proliferation. In a further embodiment, the agent is a compound of
Formula Al or
subformulae thereof (e.g., Formula A2), or pharmaceutically acceptable salt
thereof.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
87
In another embodiment, the present invention relates to a method of treatment
of an ocular
disease or disorder comprising administering to a subject in need thereof a
corneal
endothelial cell population (e.g., cell population comprising modified corneal
endothelial
cells with reduced or eliminated expression of B2M by a CRISPR system),
wherein the
population has been grown in the presence of an agent capable of inhibiting
the activity of
LATS1 and LATS2 kinases; thereby inducing YAP translocation and driving
downstream
gene expression for cell proliferation. In a further embodiment, the agent is
a compound of
Formula Al or subformulae thereof (e.g., Formula A2), or a pharmaceutically
acceptable
salt thereof.
In another embodiment, the present invention relates to a method of promoting
ocular
wound healing comprising administering to an eye of a subject a
therapeutically effective
amount of a cell population (e.g., cell population comprising modified cells
with reduced or
eliminated expression of B2M by a CRISPR system) obtainable or obtained by the
method
of cell population expansion according to the invention. In one embodiment,
the ocular
wound is a corneal wound. In other embodiments, the ocular wound is an injury
or surgical
wound.
DEFINITIONS
The general terms used hereinbefore and hereinafter preferably have within the
context of
this invention the following meanings, unless otherwise indicated, where more
general terms
wherever used may, independently of each other, be replaced by more specific
definitions
or remain, thus defining more detailed embodiments of the invention.
All methods described herein can be performed in any suitable order unless
otherwise
indicated herein or otherwise clearly contradicted by context. The use of any
and all
examples, or exemplary language (e.g. "such as") provided herein is intended
merely to
better illuminate the invention and does not pose a limitation on the scope of
the invention
otherwise claimed.
As used herein, the terms "a," "an," "the" and similar terms used in the
context of the present
invention (especially in the context of the claims) are to be construed to
cover both the
singular and plural unless otherwise indicated herein or clearly contradicted
by the context.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
88
As used herein, the term "Ci_salkyl" refers to a straight or branched
hydrocarbon chain
radical consisting solely of carbon and hydrogen atoms, containing no
unsaturation, having
from one to eight carbon atoms, and which is attached to the rest of the
molecule by a
single bond. The term "C1_4alkyl" is to be construed accordingly. As used
herein, the term "n-
alkyl" refers to straight chain (un-branced) alkyl radical as defined herein.
Examples of C1_
salkyl include, but are not limited to, methyl, ethyl, n-propyl, 1-methylethyl
(iso-propyl), n-
butyl, n-pentyl, 1,1-dimethylethyl (t-butyl), -C(CH3)2CH2CH(CH3)2 and -
C(CH3)2CH3.
As used herein, the term "C2_6alkenyl" refers to a straight or branched
hydrocarbon chain
radical group consisting solely of carbon and hydrogen atoms, containing at
least one
double bond, having from two to six carbon atoms, which is attached to the
rest of the
molecule by a single bond. As used herein, the term "C2_4alkenyl" is to be
construed
accordingly. Examples of C2_6alkenyl include, but are not limited to, ethenyl,
prop-1-enyl,
but-1-enyl, pent-1-enyl, pent-4-enyl and penta-1,4-dienyl.
As used herein, the term "alkylene" refers to a divalent alkyl group. For
example, as used
herein, the term "C1-6a1ky1ene" or "Ci to C6 alkylene" refers to a divalent,
straight, or
branched aliphatic group containing 1 to 6 carbon atoms. Examples of alkylene
include, but
are not limited to methylene (-CH2-), ethylene (-CH2CH2-), n-propylene (-
CH2CH2CH2-), iso-
propylene (-CH(CH3)CH2-), n-butylene, sec-butylene, iso-butylene, tert-
butylene, n-
pentylene, isopentylene, neopentylene and n-hexylene.
As used herein, the term "C2_6alkynyl" refers to a straight or branched
hydrocarbon chain
radical group consisting solely of carbon and hydrogen atoms, containing at
least one triple
bond, having from two to six carbon atoms, and which is attached to the rest
of the molecule
by a single bond. As used herein, the term "C2_4alkynyl" is to be construed
accordingly.
Examples of C2_6alkynyl include, but are not limited to, ethynyl, prop-1-ynyl,
but-1-ynyl, pent-
1-ynyl, pent-4-ynyl and penta-1,4-diynyl.
As used herein, the term "C1_6alkoxy" refers to a radical of the formula -0Ra,
where Ra is a
C1_6alkyl radical as generally defined above. As used herein, the term "Ci-
6alkoxy" or "Ci to
C6 alkoxy" is intended to include C1, C2, C3, C4, C5, and C6 alkoxy groups
(that is Ito 6
carbons in the alkyl chain). Examples of C1_6alkoxy include, but are not
limited to, methoxy,
ethoxy, propoxy, isopropoxy, butoxy, isobutoxy, pentoxy, and hexoxy.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
89
As used herein, the term "C1_6alkylamino" refers to a radical of the formula -
NH-Ra, where Ra
is a Ci_aalkyl radical as defined above.
As used herein, the term "di-(Ci_6alkyl)amino" refers to a radical of the
formula -N(Ra)-Ra,
where each Ra is a C1_4alkyl radical, which may be the same or different, as
defined above.
As used herein, the term "cyano" means the radical
As used herein, the term "cycloalkyl" refers to nonaromatic carbocyclic ring
that is a fully
hydrogenated ring, including mono-, bi- or poly-cyclic ring systems.
"C3_1ocycloalkyl" or "C3
to Cio cycloalkyl" is intended to include C3, C4, C5, C6, C7, C8, Cg and C10
cycloalkyl groups
that is 3 to 10 carbon ring members). Example cycloalkyl groups include, but
are not limited
to, cyclopropyl, cyclobutyl, cyclopentyl, cyclohexyl and norbornyl.
As used herein, the term "fused ring"refers to a multi-ring assembly wherein
the rings
comprising the ring assembly are so linked that the ring atoms that are common
to two rings
are directly bound to each other. The fused ring assemblies may be saturated,
partially
saturated, aromatics, carbocyclics, heterocyclics, and the like. Non-exclusive
examples of
common fused rings include decalin, naphthalene, anthracene, phenanthrene,
indole,
benzofuran, purine, quinoline, and the like.
As used herein, the term "halogen" refers to bromo, chloro, fluoro or iodo;
preferably fluoro,
chloro or bromo.
As used herein, the term "haloalkyl" is intended to include both branched and
straight-chain
saturated alkyl groups as defined above having the specified number of carbon
atoms,
substituted with one or more halogens. For example, "C1-6ha10a1ky1" or "C1 to
C6 haloalkyl" is
intended to include C1, C2, C3, C4, C5, and C6 alkyl chain. Examples of
haloalkyl include, but
are not limited to, fluoromethyl, difluoromethyl, trifluoromethyl,
trichloromethyl,
pentafluoroethyl, pentachloroethyl, 2,2,2-trifluoroethyl, 1,3-dibromopropan-2-
yl, 3-bromo-2-
fluoropropyl and 1,4,4-trifluorobutan-2-yl, heptafluoropropyl, and
heptachloropropyl.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
As used herein, the term "heteroalkyrrefers to an alkyl, as defined herein,
where one or
more of the carbon atoms within the alkyl chain are replaced by heteroatoms
independently
selected from N, 0 and S. In Cx_yhetereoalkyl or x- to y-membered heteroalkyl,
as used
herein, x-y describe the number of chain atoms (carbon and heteroatoms) on the
5 heteroalkyl. For example C3_8heteroalkyl refers to an alkyl chain with 3
to 8 chain atoms.
Unless otherwise indicated, the atom linking the radical to the remainder of
the molecule
must be a carbon. Representative example of 3- to 8-membered heteroalkyl
include, but are
not limited to -(CH2)0CH3, -(CH2)20CH(CH3)2, -(CH2)2-0-(CH2)2-0H and -(CH2)2-
(0-
(CH2)2)2-0H.
As used herein, the term "heteroaryl" refers to aromatic moieties containing
at least one
heteroatom (e.g., oxygen, sulfur, nitrogen or combinations thereof) within a 5-
to 10-
membered aromatic ring system. Examples of heteroaryl include, but are not
limited to
pyrrolyl, pyridyl, pyrazolyl, indolyl, indazolyl, thienyl, furanyl,
benzofuranyl, oxazolyl,
isoxazolyl, imidazolyl, triazolyl, tetrazolyl, triazinyl, pyrimidinyl,
pyrazinyl, thiazolyl, purinyl,
benzimidazolyl, quinolinyl, isoquinolinyl, quinoxalinyl, benzopyranyl,
benzothiophenyl,
benzoimidazolyl, benzoxazolyl and 1H-benzo[d][1,2,3]triazolyl. The
heteroaromatic moiety
may consist of a single or fused ring system. A typical single heteroaryl ring
is a 5- to 6-
membered ring containing one to four heteroatoms independently selected from
N, 0 and S
and a typical fused heteroaryl ring system is a 9- to 10-membered ring system
containing
one to four heteroatoms independently selected from N, 0 and S. The fused
heteroaryl ring
system may consist of two heteroaryl rings fused together or a heteroaryl
fused to an aryl
(e.g., phenyl).
As used herein, the term "heteroatoms" refers to nitrogen (N), oxygen (0) or
sulfur (S)
atoms. Unless otherwise indicated, any heteroatom with unsatisfied valences is
assumed to
have hydrogen atoms sufficient to satisfy the valences, and when the
heteroatom is sulfur, it
can be unoxidized (S) or oxidized to 5(0) or S(0)2.
As used herein, the term "hydroxyl" or "hydroxy"refers to the radical ¨OH.
As used herein, the term "heterocycloalkyl" means cycloalkyl, as defined in
this application,
provided that one or more of the ring carbons indicated, are replaced by a
moiety selected
from -0-, -N=, -NH-, -S-, -5(0)- and -S(0)2-. Examples of 3 to 8 membered
heterocycloalkyl
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
91
include, but are not limited to, oxiranyl, aziridinyl, azetidinyl,
imidazolidinyl, pyrazolidinyl,
tetrahydrofuranyl, tetrahydrothienyl, tetrahydrothienyl 1,1-dioxide,
oxazolidinyl, thiazolidinyl,
pyrrolidinyl, pyrrolidiny1-2-one, morpholinyl, piperazinyl, piperidinyl,
pyrazolidinyl,
hexahydropyrimidinyl, 1,4-dioxa-8-aza-spiro[4.5]dec-8-yl, thiomorpholinyl,
sulfanomorpholinyl, sulfonomorpholinyl and octahydropyrrolo[3,2-b]pyrrolyl.
As used herein, the term "oxo"refers to the divalent radical =0.
As used herein, the term "substituted" means that at least one hydrogen atom
is replaced
with a non-hydrogen group, provided that normal valencies are maintained and
that the
substitution results in a stable compound. When a substituent is oxo =0),
then two
hydrogens on the atom are replaced. In cases wherein there are nitrogen atoms
(e.g.,
amines) present in compounds of the present invention, these may be converted
to
N-oxides by treatment with an oxidizing agent (e.g., mCPBA and/or hydrogen
peroxides) to
.. afford other compounds of the present invention.
As used herein, the term "unsubstituted nitrogen" refers to a nitrogen ring
atom that has no
capacity for substitution due to its linkage to its adjacent ring atoms by a
double bond and a
single bond (-N=). For example, the nitrogen at the para position of the 4-
pyridyl
.. is an "unsubstituted" nitrogen, andthe nitrogen at the 4-position, in
reference to the linking
NH
C-ring atom, of 1H-pyrazol-4-yl, N , is an "unsubstituted" nitrogen.
As a person of ordinary skill in the art would be able to understand, for
example, a ketone (-
CH-C(=0)-) group in a molecule may tautomerize to its enol form(-C=C(OH)-).
Thus, the
invention is intended to cover all possible tautomers even when a structure
depicts only one
of them.
As used herein, "and "x-k" are symbols denoting the point of attachment
of X, to
other part of the molecule.
When any variable occurs more than one time in any constituent or formula for
a compound
of the present invention, its definition at each occurrence is independent of
its definition at
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
92
every other occurrence. Thus, for example, if a group is shown to be
substituted with 0-3 R
groups, then said group may be unsubstituted or substituted with up to three R
groups, and
at each occurrence R is selected independently from the definition of R.
Unless specified otherwise, the term "compound of the present invention" or
"compounds of
the present invention" refers to compounds of Formula Al and subformulae
thereof (e.g.,
Formula A2), as well as isomers, such as stereoisomers (including
diastereoisomers,
enantiomers and racemates), geometrical isomers, conformational isomers
(including
rotamers and astropisomers), tautomers, isotopically labeled compounds
(including
deuterium substitutions), and inherently formed moieties (e.g., polymorphs,
solvates and/or
hydrates). When a moiety is present that is capable of forming a salt, then
salts are included
as well, in particular pharmaceutically acceptable salts.
It will be recognized by those skilled in the art that the compounds of the
present invention
may contain chiral centers and as such may exist in different isomeric forms.
As used
herein, the term "isomers" refers to different compounds of the present
invention that have
the same molecular formula but differ in arrangement and configuration of the
atoms.
As used herein, the term "enantiomers" are a pair of stereoisomers that are
non-
superimposable mirror images of each other. A 1:1 mixture of a pair of
enantiomers is a
"racemic" mixture. As used herein, the term is used to designate a racemic
mixture where
appropriate. When designating the stereochemistry for the compounds of the
present
invention, a single stereoisomer with known relative and absolute
configuration of the two
chiral centers is designated using the conventional RS system (e.g., (1S,2S));
a single
stereoisomer with known relative configuration but unknown absolute
configuration is
designated with stars (e.g., (1R*,2R*)); and a racemate with two letters (e.g,
(1RS,2RS) as a
racemic mixture of (1R,2R) and (1S,2S); (1RS,2SR) as a racemic mixture of
(1R,2S) and
(1S,2R)). As used herein, the term "diastereoisomers" are stereoisomers that
have at least
two asymmetric atoms, but which are not mirror-images of each other. The
absolute
stereochemistry is specified according to the Cahn-Ingold-Prelog R-S system.
When a
compound of the present invention is a pure enantiomer, the stereochemistry at
each chiral
carbon may be specified by either R or S. Resolved compounds of the present
invention
whose absolute configuration is unknown can be designated (+) or (¨) depending
on the
direction (dextro- or levorotatory) which they rotate plane polarized light at
the wavelength of
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
93
the sodium D line. Alternatively, the resolved compounds of the present
invention can be
defined by the respective retention times for the corresponding
enantiomers/diastereomers
via chiral HPLC.
Certain of the compounds of the present invention described herein contain one
or more
asymmetric centers or axes and may thus give rise to enantiomers,
diastereomers, and
other stereoisomeric forms that may be defined, in terms of absolute
stereochemistry, as
(R)- or (S)-.
Geometric isomers may occur when a compound of the present invention contains
a double
bond or some other feature that gives the molecule a certain amount of
structural rigidity. If
the compound contains a double bond, the substituent may be E or Z
configuration. If the
compound contains a disubstituted cycloalkyl, the cycloalkyl substituent may
have a cis- or
trans-configuration.
As used herein, the term "conformational isomers" or "conformers" are isomers
that can
differ by rotations about one or more bonds. Rotamers are conformers that
differ by rotation
about only a single bond.
As used herein, the term "atropisomer" refers to a structural isomer based on
axial or planar
chirality resulting from restricted rotation in the molecule.
Unless specified otherwise, the compounds of the present invention are meant
to include all
such possible isomers, including racemic mixtures, optically pure forms and
intermediate
mixtures. Optically active (R)- and (S)- isomers may be prepared using chiral
synthons or
chiral reagents, or resolved using conventional techniques (e.g., separated on
chiral SFC or
HPLC chromatography columns, such as CHIRALPAKO and CHIRALCELO available from
DAICEL Corp. using the appropriate solvent or mixture of solvents to achieve
good
separation).
The compounds of the present invention can be isolated in optically active or
racemic forms.
Optically active forms may be prepared by resolution of racemic forms or by
synthesis from
optically active starting materials. All processes used to prepare compounds
of the present
invention and intermediates made therein are considered to be part of the
present invention.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
94
When enantiomeric or diastereomeric products are prepared, they may be
separated by
conventional methods, for example, by chromatography or fractional
crystallization.
As used herein, the term "LATS" is the abbreviated name of the large tumor
suppressor
protein kinase. As used herein, the term "LATS" refers to LATS1 and/or LATS2.
As used
herein, the term "LATS1" refers to the large tumor suppressor kinase 1 and the
term
"LATS2" refers to the large tumor suppressor kinase 2. LATS1 and LATS2 both
have
serine/threonine protein kinase activity.
As used herein, the term "YAP1" refers to the yes-associated protein 1, also
known as YAP
or YAP65, which is a protein that acts as a transcriptional regulator of genes
involved in cell
proliferation.
As used herein, the term "MST1/2" refers to mammalian sterile 20¨like kinase -
1 and -2.
The term "effective amount" or "therapeutically effective amount" are used
interchangeably
herein, and refer to an amount of a compound, formulation, material, or
composition, as
described herein effective to achieve a particular biological result.
As used herein, the term "a therapeutically effective amount" of a compound of
the present
invention refers to an amount of the compound of the present invention that
will elicit the
biological or medical response of a subject, for example, reduction or
inhibition of an
enzyme or a protein activity, or ameliorate symptoms, alleviate conditions,
slow or delay
disease progression, or prevent a disease, etc. In one non-limiting
embodiment, the term "a
therapeutically effective amount" as used herein refers to the amount of the
LATS
compound of the present invention that, when administered to a subject, is
effective to (1) at
least partially alleviate, inhibit, prevent and/or ameliorate a condition, or
a disorder or a
disease (i) mediated by LATS activity, or (ii) characterized by activity
(normal or abnormal)
of LATS; or (2) reduce or inhibit the activity of LATS; or (3) reduce or
inhibit the expression
of LATS. In another non-limiting embodiment, the term "a therapeutically
effective amount"
as used herein refers to the amount of the compound of the present invention
that, when
administered to a cell, or a tissue, or a non-cellular biological material, or
a medium, is
effective to at least partially reducing or inhibiting the activity of LATS;
or at least partially
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
reducing or inhibiting the expression of LATS.
Further, as used herein, the term "a therapeutically effective amount" of a
modified limbal
stem cell of the present invention refers to an amount of the cells of the
present invention
5 that will elicit the biological or medical response of a subject, for
example, ameliorate
symptoms, alleviate conditions, slow or delay disease progression, inhibit or
prevent a
disease, in particular ocular disease, in particular limbal stem cell
deficiency.
As used herein, the term "subject" includes human and non-human animals. Non-
human
10 animals include vertebrates, e.g., mammals and non-mammals, such as non-
human
primates, sheep, cats, horses, cows, chickens, dog, mouse, rat, goat, rabbit,
and pig.
Preferably, the subject is human. Except when noted, the terms "patient" or
"subject" are
used herein interchangeably.
15 As used herein, the term "IC50" refers to the molar concentration of an
inhibitor that produces
50% of the inhibition effect.
As used herein, the term "treat", "treating" or "treatment" of any disease or
disorder refers to
alleviating or ameliorating the disease or disorder (i.e., slowing or
arresting the development
20 of the disease or at least one of the clinical symptoms thereof); or
alleviating or ameliorating
at least one physical parameter or biomarker associated with the disease or
disorder,
including those which may not be discernible to the patient.
As used herein, the term "prevent", "preventing" or "prevention" of any
disease or disorder
25 refers to the prophylactic treatment of the disease or disorder; or
delaying the onset or
progression of the disease or disorder.
As used herein, a subject is "in need of" a treatment if such subject would
benefit
biologically, medically or in quality of life from such treatment.
Depending on the process conditions, the compounds of the present invention
are obtained
either in free (neutral) or salt form. Both the free form and salt form, and
particularly
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
96
"pharmaceutically acceptable salts" of these compounds, are within the scope
of the
invention.
As used herein, the terms "salt" or "salts" refers to an acid addition or base
addition salt of a
compound of the present invention. "Salts" include, in particular,
"pharmaceutically
acceptable salts". As used herein, the term "pharmaceutically acceptable
salts" refers to
salts that retain the biological effectiveness and properties of the compounds
of the present
invention and, which typically are not biologically or otherwise undesirable.
In many cases,
the compounds of the present invention are capable of forming acid and/or base
salts by
virtue of the presence of amino and/or carboxyl groups or groups similar
thereto.Pharmaceutically acceptable acid addition salts can be formed with
inorganic acids
and organic acids.
Inorganic acids from which salts can be derived include, for example,
hydrochloric acid,
hydrobromic acid, sulfuric acid, nitric acid, phosphoric acid, and the like.
Organic acids from which salts can be derived include, for example, acetic
acid, propionic
acid, glycolic acid, oxalic acid, maleic acid, malonic acid, succinic acid,
fumaric acid, tartaric
acid, citric acid, benzoic acid, mandelic acid, methanesulfonic acid,
ethanesulfonic acid,
toluenesulfonic acid, sulfosalicylic acid, and the like.
Pharmaceutically acceptable base addition salts can be formed with inorganic
and organic
bases.
Inorganic bases from which salts can be derived include, for example, ammonium
salts and
metals from columns Ito XII of the periodic table. In certain embodiments, the
salts are
derived from sodium, potassium, ammonium, calcium, magnesium, iron, silver,
zinc, and
copper; particularly suitable salts include ammonium, potassium, sodium,
calcium and
magnesium salts.
Organic bases from which salts can be derived include, for example, primary,
secondary,
and tertiary amines, substituted amines including naturally occurring
substituted amines,
cyclic amines, basic ion exchange resins, and the like. Certain organic amines
include
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
97
isopropylamine, benzathine, cholinate, diethanolamine, diethylamine, lysine,
meglumine,
piperazine and tromethamine.
In another aspect, the present invention provides compounds of Formula Al or
subformulae
thereof (e.g., Formula A2) in acetate, ascorbate, adipate, aspartate,
benzoate, besylate,
bromide/hydrobromide, bicarbonate/carbonate, bisulfate/sulfate,
camphorsulfonate, caprate,
chloride/hydrochloride, chlortheophyllonate, citrate, ethandisulfonate,
fumarate, gluceptate,
gluconate, glucuronate, glutamate, glutarate, glycolate, hippurate,
hydroiodide/iodide,
isethionate, lactate, lactobionate, laurylsulfate, malate, maleate, malonate,
mandelate,
mesylate, methylsulphate, mucate, naphthoate, napsylate, nicotinate, nitrate,
octadecanoate, oleate, oxalate, palmitate, pamoate, phosphate/hydrogen
phosphate/dihydrogen phosphate, polygalacturonate, propionate, sebacate,
stearate,
succinate, sulfosalicylate, sulfate, tartrate, tosylate trifenatate,
trifluoroacetate or xinafoate
salt form.
Any formula given herein is also intended to represent unlabeled forms as well
as
isotopically labeled forms of the compounds. Isotopically labeled compounds of
the present
invention have structures depicted by the formulas given herein except that
one or more
atoms are replaced by an atom having a selected atomic mass or mass number.
Isotopes
that can be incorporated into compounds of the present invention include, for
example,
isotopes of hydrogen.
Further, incorporation of certain isotopes, particularly deuterium (i.e., 2H
or D), may afford
certain therapeutic advantages resulting from greater metabolic stability, for
example
increased in vivo half-life or reduced dosage requirements or an improvement
in therapeutic
index or tolerability. It is understood that deuterium in this context is
regarded as a
substituent of a compound of Formula Al or subformulae thereof (e.g., Formula
A2). The
concentration of deuterium may be defined by the isotopic enrichment factor.
As used
herein, the term "isotopic enrichment factor" means the ratio between the
isotopic
abundance and the natural abundance of a specified isotope. If a substituent
in a compound
of the present invention is denoted as being deuterium, such compound has an
isotopic
enrichment factor for each designated deuterium atom of at least 3500 (52.5%
deuterium
incorporation at each designated deuterium atom), at least 4000 (60% deuterium
incorporation), at least 4500 (67.5% deuterium incorporation), at least 5000
(75% deuterium
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
98
incorporation), at least 5500 (82.5% deuterium incorporation), at least 6000
(90% deuterium
incorporation), at least 6333.3 (95% deuterium incorporation), at least 6466.7
(97%
deuterium incorporation), at least 6600 (99% deuterium incorporation), or at
least 6633.3
(99.5% deuterium incorporation). It should be understood that the term
"isotopic enrichment
factor" as used herein can be applied to any isotope in the same manner as
described for
deuterium.
Other examples of isotopes that can be incorporated into compounds of the
present
invention include isotopes of hydrogen, carbon, nitrogen, oxygen, phosphorous,
fluorine,
and chlorine, such as 3H, 11C, 13C, 14C, 15N, 18F 31p, 321D, 355, 36C1, 1231,
1241, and 1251,
respectively. Accordingly it should be understood that the invention includes
compounds
that incorporate one or more of any of the aforementioned isotopes, including
for example,
radioactive isotopes, such as 3H and 14C, or those into which non-radioactive
isotopes, such
as 2H and 13C are present. Such isotopically labelled compounds are useful in
metabolic
.. studies (with 14C), reaction kinetic studies (with, for example 2H or 3H),
detection or imaging
techniques, such as positron emission tomography (PET) or single-photon
emission
computed tomography (SPECT) including drug or substrate tissue distribution
assays, or in
radioactive treatment of patients. In particular, an 15F or labeled compound
may be
particularly desirable for PET or SPECT studies. Isotopically-labeled
compounds of the
present invention can generally be prepared by conventional techniques known
to those
skilled in the art or by processes analogous to those described in the
accompanying
Examples and Preparations using an appropriate isotopically-labeled reagents
in place of
the non-labeled reagent previously employed.
Any asymmetric atom (e.g., carbon or the like) of the compound(s) of the
present invention
can be present in racemic or enantiomerically enriched, for example the (R)-,
(S)- or (R,S)-
configuration. In certain embodiments, each asymmetric atom has at least 50 %
enantiomeric excess, at least 60 % enantiomeric excess, at least 70 %
enantiomeric
excess, at least 80 % enantiomeric excess, at least 90 % enantiomeric excess,
at least 95
__ % enantiomeric excess, or at least 99 % enantiomeric excess in the (R)- or
(S)-
configuration. Substituents at atoms with unsaturated double bonds may, if
possible, be
present in cis- (Z)- or trans- (E)- form.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
99
Accordingly, as used herein a compound of the present invention can be in the
form of one
of the possible stereoisomers, rotamers, atropisomers, tautomers or mixtures
thereof, for
example, as substantially pure geometric (cis or trans) stereoisomers,
diastereomers,
optical isomers (antipodes), racemates or mixtures thereof.
Any resulting mixtures of stereoisomers of the compounds of the present
invention can be
separated on the basis of the physicochemical differences of the constituents,
into the pure
or substantially pure geometric or optical isomers, diastereomers, racemates,
for example,
by chromatography and/or fractional crystallization.
Any resulting racemates of final compounds of the present invention or
intermediates
thereof can be resolved into the optical antipodes by known methods, e.g., by
separation of
the diastereomeric salts thereof, obtained with an optically active acid or
base, and
liberating the optically active acidic or basic compound. In particular, a
basic moiety may
thus be employed to resolve the compounds of the present invention into their
optical
antipodes, e.g., by fractional crystallization of a salt formed with an
optically active acid, e.g.,
tartaric acid, dibenzoyl tartaric acid, diacetyl tartaric acid, di-0,0'-p-
toluoyl tartaric acid,
mandelic acid, malic acid or camphor-10-sulfonic acid. Racemic products can
also be
resolved by chiral chromatography, e.g., high pressure liquid chromatography
(H PLC) using
a chiral adsorbent.
As used herein, the term "percentage of sequence identity" is determined by
comparing two
optimally aligned sequences over a comparison window, wherein the portion of
the
polynucleotide sequence in the comparison window may comprise additions or
deletions
(i.e., gaps) as compared to the reference sequence (e.g., a polypeptide of the
invention),
which does not comprise additions or deletions, for optimal alignment of the
two sequences.
The percentage is calculated by determining the number of positions at which
the identical
nucleic acid base or amino acid residue occurs in both sequences to yield the
number of
matched positions, dividing the number of matched positions by the total
number of
positions in the window of comparison and multiplying the result by 100 to
yield the
percentage of sequence identity.
As used herein, the terms "identical" or percent "identity," in the context of
two or more
nucleic acids or polypeptide sequences, refer to two or more sequences or
subsequences
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
100
that are the same sequences. Two sequences are "substantially identical" if
two sequences
have a specified percentage of amino acid residues or nucleotides that are the
same (i.e., at
least 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98% or 99% sequence identity over a
specified region, or, when not specified, over the entire sequence of a
reference sequence),
when compared and aligned for maximum correspondence over a comparison window,
or
designated region as measured using one of the following sequence comparison
algorithms
or by manual alignment and visual inspection. The invention provides
polypeptides or
polynucleotides that are substantially identical to the polypeptides or
polynucleotides,
respectively, exemplified herein.
As used herein, the term "isolated" means altered or removed from the natural
state. For
example, a nucleic acid or a peptide or cell naturally present in a living
animal is not
"isolated," but the same nucleic acid or peptide or cell partially or
completely separated from
the coexisting materials of its natural state is "isolated."
As used herein, the term "nucleic acid" or "polynucleotide" refers to
deoxyribonucleic acids
(DNA) or ribonucleic acids (RNA) and polymers thereof in either single- or
double-stranded
form. Unless specifically limited, the term encompasses nucleic acids
containing known
analogues of natural nucleotides that have similar binding properties as the
reference
nucleic acid and are metabolized in a manner similar to naturally occurring
nucleotides.
Unless otherwise indicated, a particular nucleic acid sequence also implicitly
encompasses
conservatively modified variants thereof (e.g., degenerate codon
substitutions), alleles,
orthologs, SNPs, and complementary sequences as well as the sequence
explicitly
indicated. Specifically, degenerate codon substitutions may be achieved by
generating
.. sequences in which the third position of one or more selected (or all)
codons is substituted
with mixed-base and/or deoxyinosine residues (Batzer et al., Nucleic Acid Res.
19:5081
(1991); Ohtsuka et al., J. Biol. Chem. 260:2605-2608 (1985); and Rossolini et
al., Mol. Cell.
Probes 8:91-98 (1994)).
.. As used herein, the term "cell population" or "population of cells"
comprises cells that
proliferate in the presence of a LATS1 and/or LATS2 inhibitor in vivo or ex
vivo. In such
cells, Hippo signaling typically suppresses cell growth, but will proliferate
when the pathway
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
101
is disrupted by LATS inhibition. In certain embodiments, a cell population
useful in a method,
preparation, medium, agent, or kit of the invention comprises cells from
tissues described
above or cells described or provided herein. Such cells include, but are not
limited to ocular
cells (e.g., limbal stem cells, corneal endothelial cells), epithelial cells
(e.g., from skin),
neural stem cells, mesenchymal stem cells, basal stem cells of the lungs,
embryonic stem
cells, adult stem cells, induced pluripotent stem cells and liver progenitor
cells.
Pharmacology and Utility
In one embodiment, the present invention relates to ex vivo cell therapies
involvining
expansion of cells using small molecule LATS kinase inhibitors, said cells
being modified as
described herein.
Ex vivo cell therapies generally involve expansion of a cell population
isolated from a patient
or healthy donor to be transplanted to a patient to establish a transient or
stable graft of the
expanded cells. Ex vivo cell therapies can be used to deliver a gene or
biotherapeutic
molecule to a patient, wherein gene transfer or expression of the
biotherapeutic molecule is
achieved in the isolated cells. Non-limiting examples of ex vivo cell
therapies include, but are
not limited to, stem cell transplantation (e.g., hematopoietic stem cell
transplantation,
autologous stem cell transplantation, or cord blood stem cell
transplantation), tissue
regeneration, cellular immunotherapy, and gene therapy. See, e.g., Naldini,
2011, Nature
Reviews Genetics volume 12, pages 301-315.
Ex vivo procedures are well known in the art and are discussed more fully
below. Briefly,
cells are isolated from a mammal (e.g., a human) and genetically modified
(i.e., transduced
or transfected in vitro) with a gRNA molecule of the invention. The modified
cell can be
administered to a mammalian recipient to provide a therapeutic benefit. The
mammalian
recipient may be a human and the cell can be autologous with respect to the
recipient.
Alternatively, the cells can be allogeneic with respect to the recipient.
The term "autologous" refers to any material derived from the same individual
into whom it is
introduced.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
102
The term "allogeneic" refers to any material derived from a different animal
of the same
species as the individual to whom the material is introduced. Two or more
individuals are
said to be allogeneic to one another when the genes at one or more loci are
not identical. In
some aspects, allogeneic material from individuals of the same species may be
sufficiently
unlike genetically to interact antigenically.
Pharmaceutical Composition and Administration
Pharmaceutical compositions of the present invention may comprise a cell
(e.g., a modified
cell, such as LSC or CEC, with reduced or eliminated expression of B2M by a
CRISPR
system), e.g., a plurality of cells, as described herein, in combination with
one or more
pharmaceutically or physiologically acceptable carriers, diluents or
excipients. Such
compositions may comprise buffers such as neutral buffered saline, phosphate
buffered
saline and the like; carbohydrates such as glucose, mannose, sucrose or
dextrans,
mannitol; proteins; polypeptides or amino acids such as glycine; antioxidants;
chelating
agents such as EDTA or glutathione; adjuvants (e.g., aluminum hydroxide); and
preservatives.
In one embodiment, the pharmaceutical compositions of the present invention
are
cryopreserved compositions. The cryopreserved compositions comprise a cell
(e.g., a
modified cell, such as LSC or CEC, with reduced or eliminated expression of
B2M by a
CRISPR system), e.g., a plurality of cells) and a cryoprotectant. The term
"cryoprotectant",
as used herein, refers to chemical compounds which are added to biological
samples in
order to minimize the deleterious effects of cryopreservation procedures. In
one
embodiment, the cryopreserved compositions comprise a cell (e.g., a modified
cell, such as
LSC or CEC, with reduced or eliminated expression of B2M by a CRISPR system),
e.g., a
plurality of cells) and a cryoprotectant selected from the list of glycerol,
DMSO
(dimethylsulfoxide) polyvinylpyrrolidone, hydroxyethyl starch, propylene
glycol, acetamide,
monosaccharides, algae-derived polysaccharides, and sugar alcohols, or a
combination
thereof. In a more specific embodiment, the cryopreserved compositions
comprise a cell
(e.g., a modified cell, such as LSC or CEC, with reduced or eliminated
expression of B2M by
a CRISPR system), e.g., a plurality of cells) and DMSO concentration of 0.5%
to 10%, e.g.,
1%_ 10%, 2%_7%, 3%_6%, 4% _ 5%, preferably 5%. DMSO acts as a cryoprotecting
agent
against formation of water crystals within and outside the cells, which could
lead to cell
.. damage during cryopreservation steps. In a further embodiment, the
cryopreserved
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
103
compositions further comprise a suitable buffer, for example CryoStor CS5
buffer (BioLife
Solutions).
Compositions of the present invention are in one aspect formulated for
intravenous
administration. Composition of the present invention are in one aspect
formulated for topical
administration, in particular for topical eye administartion.
Pharmaceutical compositions of the present invention may be administered in a
manner
appropriate to the disease to be treated (or prevented). The quantity and
frequency of
administration will be determined by such factors as the condition of the
patient, and the
type and severity of the patient's disease, although appropriate dosages may
be determined
by clinical trials.
In one embodiment, the pharmaceutical composition is substantially free of,
e.g., there are
no detectable levels of a contaminant, e.g., selected from the group
consisting of endotoxin,
mycoplasma, replication competent lentivirus (RCL), p24, VSV-G nucleic acid,
HIV gag,
residual anti-CD3/anti-CD28 coated beads, mouse antibodies, pooled human
serum, bovine
serum albumin, bovine serum, culture media components, vector packaging cell
or plasmid
components, a bacterium and a fungus. In one embodiment, the bacterium is at
least one
selected from the group consisting of Alcaligenes faecalis, Candida albicans,
Escherichia
coli, Haemophilus influenza, Neisseria meningitides, Pseudomonas aeruginosa,
Staphylococcus aureus, Streptococcus pneumonia, and Streptococcus pyogenes
group A.
In another aspect, in embodiments of the invention relating to in vivo use,
the present
invention provides a pharmaceutical composition comprising a modified limbal
stem cell of
the present invention, or a cell population obtainable or obtained by the
method of cell
population expansion according to the invention, and a pharmaceutically
acceptable carrier.
In a further embodiment, the composition comprises at least two
pharmaceutically
acceptable carriers, such as those described herein.
In certain instances, it may be advantageous to administer the cell population
(e.g., a cell
population comprising modified cells, such as LSCs or CECs, with reduced or
eliminated
expression of B2M by a CRISPR system, e.g., S. pyo genes Cas9 CRISPR system)
obtainable or obtained by the method of cell population expansion according to
the invention
in combination with at least one additional pharmaceutical (or therapeutic)
agent, such as an
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
104
immunosuppressant for example corticosteroids, cyclosporine, tacrolimus, and
combinations
of immunosuppressants. In particular, compositions will either be formulated
together as a
combination therapeutic or administered separately.
PREPARATION OF LATS INHIBITOR COMPOUNDS
The LATS inhibitor compounds useful in methods of the present invention can be
prepared
in a number of ways known to one skilled in the art of organic synthesis in
view of the
methods, reaction schemes and examples provided herein. Such compounds of the
present
invention can be synthesized using the methods described in U.S. Patent
Application No.
15/963,816, filed April 26, 2018, and International Application No.
PCT/162018/052919 (WO
2018/198077), filed April 26, 2018, which are incorporated herein in itheir
entirety.
For example, LATS inhibitor compounds can be synthesized using the methods
described
below, together with synthetic methods known in the art of synthetic organic
chemistry, or by
.. variations thereon as appreciated by those skilled in the art. Preferred
methods include, but
are not limited to, those described below. The reactions are performed in a
solvent or
solvent mixture appropriate to the reagents and materials employed and
suitable for the
transformations being effected. It will be understood by those skilled in the
art of organic
synthesis that the functionality present on the molecule should be consistent
with the
.. transformations proposed. This will sometimes require a judgment to modify
the order of the
synthetic steps or to select one particular process scheme over another in
order to obtain a
desired compound of the present invention.
The starting materials are generally available from commercial sources such as
Aldrich
.. Chemicals (Milwaukee, Wis.) or are readily prepared using methods well
known to those
skilled in the art (e.g., prepared by methods generally described in Louis F.
Fieser and Mary
Fieser, Reagents for Organic Synthesis, v. 1-19, Wiley, New York (1967-1999
ed.), Larock,
R.C., Comprehensive Organic Transformations, 2nd-ed., Wiley-VCH Weinheim,
Germany
(1999), or Beilsteins Handbuch der organischen Chemie, 4, Aufl. ed. Springer-
Verlag,
Berlin, including supplements (also available via the Beilstein online
database).
For illustrative purposes, the reaction schemes depicted below provide
potential routes for
synthesizing the compounds of the present invention as well as key
intermediates. For a
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
105
more detailed description of the individual reaction steps, see the Examples
section below.
Those skilled in the art will appreciate that other synthetic routes may be
used to synthesize
the inventive compounds. Although specific starting materials and reagents are
depicted in
the schemes and discussed below, other starting materials and reagents can be
easily
substituted to provide a variety of derivatives and/or reaction conditions. In
addition, many of
the compounds prepared by the methods described below can be further modified
in light of
this disclosure using conventional chemistry well known to those skilled in
the art.
In the preparation of compounds of the present invention, protection of remote
functionality
of intermediates may be necessary. The need for such protection will vary
depending on the
nature of the remote functionality and the conditions of the preparation
methods. The need
for such protection is readily determined by one skilled in the art. For a
general description
of protecting groups and their use, see Greene, T.W. et al., Protecting Groups
in Organic
Synthesis, 4th Ed., Wiley (2007). Protecting groups incorporated in making of
the
compounds of the present invention, such as the trityl protecting group, may
be shown as
one regioisomer but may also exist as a mixture of regioisomers.
Abbreviations
Abbreviations as used herein, are defined as follows: "lx" for once, "2x" for
twice, "3x" for
thrice, " C" for degrees Celsius, "aq" for aqueous, "Col" for column, "eq" for
equivalent or
equivalents, "g" for gram or grams, "mg" for milligram or milligrams, "nm" for
nanometer or
nanometers, "L" for liter or liters, "mL" or "ml" for milliliter or
milliliters, "ul", "uL", "p1", or "pL"
for microliter or microliters, "nL" or "nl" for nanoliter or nanoliters, " "N"
for normal, "uM" or
"pM" micromolar, "nM" for nanomolar, "mol" for mole or moles, "mmol" for
millimole or
millimoles, "min" for minute or minutes, "h" or "hrs" for hour or hours, "RT"
for room
temperature, ON for overnight, "atm" for atmosphere, "psi" for pounds per
square inch,
"conc." for concentrate, "aq" for aqueous, "sat" or "sat'd" for saturated, "WM
for molecular
weight, "mw" or "pwave" for microwave, "mp" for melting point, "Wt" for
weight, "MS" or
"Mass Spec" for mass spectrometry, "ESI" for electrospray ionization mass
spectroscopy,
"HR" for high resolution, "HRMS" for high resolution mass spectrometry, "LCMS"
for liquid
chromatography mass spectrometry, "HPLC" for high pressure liquid
chromatography, "RP
HPLC" for reverse phase HPLC, "TLC" or "tic" for thin layer chromatography,
"NMR" for
nuclear magnetic resonance spectroscopy, "n0e" for nuclear Overhauser effect
spectroscopy, "1H" for proton, "6 " for delta, "s" for singlet, "d" for
doublet, "t" for triplet, "q"
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
106
for quartet, "m" for multiplet, "br" for broad, "Hz" for hertz, "cc" for
"enantiomeric excess"
and "a", "8", "R","r", "S", "s", "E", and "Z" are stereochemical designations
familiar to one
skilled in the art.
The following abbreviations used herein below have the corresponding meanings:
AC Active Control
AIBN azobisisobutyronitrile
ATP adenosine triphosphate
Bn benzyl
Boc tert-butoxy carbonyl
Boc20 di-tert-butyl dicarbonate
BSA bovine serum albumin
Bu butyl
Cs2CO3 cesium carbonate anhydrous
CHCI3 chloroform
DAST diethylaminosulfurtrifluoride
DBU 2,3,4,6,7,8,9,10-octahydropyrimido[1,2-a]azepine
DCM dichloromethane
DMAP 4-dimethylaminopyridine
DMEM Dulbecco's modified Eagle's medium
DMF dimethylformamide
DMSO dimethylsulfoxide
DPPA diphenylphosphoryl azide
DTT dithiolthreitol
EA ethyl acetate
EDTA ethylenediaminetetraacetic acid
Equiv. equivalence
Et ethyl
Et20 diethyl ether
Et0H ethanol
Et0Ac ethyl acetate
FBS fetal bovine serum
HATU 2-(7-Aza-1H-benzotriazole-1-yI)-1,1,3,3-
tetramethyluronium
hexafluorophosphate
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
107
HCI hydrochloric acid
HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic
acid
HPMC (hydroxypropyl)methyl cellulose
HTRF homogeneous time resolved fluorescence
i-Bu isobutyl
i-Pr isopropyl
KOAc potassium acetate
LiAIH4 lithium aluminium hydride
LATS large tumor suppressor
LSC limbal stem cell
LSCD limbal stem cell deficiency
Me methyl
mCPBA 3-chloroperoxybenzoic acid
MeCN acetonitrile
Mn02 manganese dioxide
N2 nitrogen
NaBH.4 sodium borohydride
NaHCO3 sodium bicarbonate
Na2SO4 sodium sulfate
NBS N-Bromosuccinimide
NC Neutral Control
PBS phosphate buffered saline
PFA paraformaldehyde
Ph phenyl
PPh3 triphenylphosphine
Ph3P=0 triphenylphosphine oxide
pYAP phospho-YAP
Rf retention factor
RT room temperature ( C)
Ser serine
t-Bu or But tert-butyl
T3P Propane phosphonic acid anhydride
TEA triethylamine
TFA trifluoroacetic acid
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
108
THF tetrahydrofuran
UVA Ultraviolet A
YAP Yes associated protein (NCB! Gene ID: 10413;
official symbol:
(YAP I)
I. General Synthetic Routes
Compounds of Formulae Ito VI can be prepared as illustrated in the General
Schemes Ito
III and in greater details in Schemes Ito 6 below.
General Scheme I for the preparation of compounds of Formula I or ll
L 0 cyclization and L CI
amination and R2 R1
L N
chlorination coupling
N
NH2 N
N A
GSla GS1b M GS1c
M, L = halide, alky, -ON, etc
Y = OH, NH2
further functionalization
R2õ ,R1
R5 N
X
N
N A
R3
Formula 1111
The bicyclic dichloride GS1b could be commercially available when X = C or
could be
prepared from aminoisonicotinic acid/amide GS1a through cyclization and
chlorination. The
dichloride of GS1b could be aminated and coupled with the appropriate agents
to form
GS1c, which further functionalized to yield Formula I or Formula II through
any necessary
functionalization, such as but not limited to protection and de-protection
steps, reduction,
hydrolysis, alkylation, amination, coupling, etc
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
109
General Scheme ll for the preparation of compounds of Formula Ill
cyclization and N L Cl
alkylation 1 '''=== N chlorination
- "" N
__________________________ r
11 E-1 Ei 11
-------------------------------------------------- ;.-
N õ.=
N ,--" .,,'
A
M KA 0 A m
M, L = halide, alky, -ON, etc
amination
,-R 2õR1
R" N L R2.;N,R1
further funotionalization
-.. ----------------------------------------------
NI ---,,,(1.,1,,,A
R3 111 M
General Scheme Ill for the preparation of compounds of Formula IV
L L L Br
coupling brornination
i 1101's'
N.,..--. .,.' N ,-,-
= Br _____ N A , A
M M M
M, L = halide, alky, -ON, etc
amination
'-'
,-R2NõRi
L R2. R1
further functionalization
--.,
-. _______________________________________________
N,--
R3 IV A
Po
Scheme 1.
Compounds of Formula V can be prepared as illustrated in Scheme 1 below. Step
C could
include amination and any necessary functionalization, such as but not limited
to protection
and de-protection steps, reduction, hydrolysis, alkylation, etc.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
1 1 0
Scheme 1
0
01-1 CI
0
Cr- I -Ph
N
NaHS03, I Step B I
N N
p-Ts01-1 lb lc
la
Step A
amine
Step C
DIEA
KF
R2N
V
Scheme 2.
Alternatively, compounds of Formula V can be prepared as illustrated in Scheme
2. Step C
could include amination and any necessary functionalization, such as but not
limited to
protection and de-protection steps, reduction, hydrolysis, alkylation, etc.
Further
functionalization of mono-chloride intermediate 2d by but not limited to metal
mediated
coupling, amination, alkylation etc. and necessary protection and de-
protection steps, leads
to compounds of Formula V.
Scheme 2
0 0 QCI
H2N NH2 POCI3
NH
Step A N N0 Step B N
N CI
NH2
2a 2c
2b
amine
DIEA
Step C KE
R2;,
1 ,1 Further
functionalization
'NAN
NNA
V 2d
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
1 1 1
Scheme 3.
Compounds of Formula I where R5 is hydrogen can be prepared as illustrated in
Scheme 3.
Step C could include amination and any necessary functionalization, such as
but not limited
to protection and de-protection steps, reduction, hydrolysis, alkylation, etc.
Further
functionalization of mono-chloride intermediate 3d by but not limited to metal
mediated
coupling, amination, alkylation etc. and necessary protection and de-
protection steps, leads
to compounds of Formula (I) where R5 is hydrogen.
Scheme 3
OH 11 CI
0
CI- Ph
N
ryli-N"2 N I I
N
N NaHS03, Step B
N
CI p-Ts0H CI
CI 3b 3c
3a Step A
amine
Step C
DIEA
KF
R. R1 R2,N,R1
Further functionalization
R3N CI N
3d
where R5 is H
Scheme 4.
Compounds of Formula I, where R3 and R5 are both hydrogen, can be prepared as
illustrated in Scheme 4. Step C could include amination and any necessary
functionalization, such as but not limited to protection and de-protection
steps, reduction,
hydrolysis, alkylation, etc. leads to compounds of Formula I where R3 and R5
are both
hydrogen.
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
112
Scheme 4
0 OH CI
-..._ )1N- Chlorination by
(...=^' OH NC,,
A r---%'-'''''N
1 SOCl2 or POCI3 1
---------------------------------------------------------- --"' N ,,--
---------------------------- , N,õ,,,,,N-5=01,,A N,,....5.--,,' NE1 N A
2
Step A Step B
4a` 4b" 4c"
amine
Step C DIEA
KF
t
R2;, ,R1
'N
-- - N
il
N A
i
where R3 and R5 are H
Scheme 5.
Compounds of Formula I, where R3 is hydrogen, can be prepared as illustrated
in Scheme
5. Step D could include amination and any necessary functionalization, such as
but not
limited to protection and de-protection steps, reduction, hydrolysis,
alkylation, etc. Further
functionalization of mono- chloride intermediate 5d by, but not limited to,
metal mediated
coupling, amination, alkylation etc. and necessary protection and de-
protection steps, leads
to compounds of Formula I where R3 is hydrogen,
Scheme 5
NH
Br j' , .,--k,..----,,,, Br 0 NH Br OH
ii .
H2N 1 N-1 I I
,,,,, ' CO2H =,,,,)1"- -'1',õ,-'-`-1
rr.'\"=-r`s1
11
H ,
N., ,(),F 't N,,,,,,,,,;--.., .õ- N
N.,,7---,,,et,,,,,,,,-;õ\.,,
Step A F Step B 1 1
5a 5b 5c
0
ii
Step C
R2 R1 R2 Ri . õ 5 ' amine CI
CI
RNJ' CI N Further functionalization
DIEA
KF rj''''-=''')"I
N
y'z''N
1 I
NN .,-,,,-.-õ, ,,;-,'t -. ---------------------------------
'''" N '''-'1 - --....,'"-N--""*""..C-1 Step D N
1 1
5d
where R3 is H
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
113
Scheme 6.
Compounds of Formula VI can be prepared from commercially available dichloride
6a' (2,4-
dichloro-1,7-naphthyridine, Aquila Pharmatech) as illustrated in Scheme 6.
Step A could
include metal mediated coupling and any necessary functionalization, such as
but not
limited to protection and de-protection steps, cyclization, reduction,
hydrolysis, alkylation,
etc. Step B could include amination and any necessary functionalization, such
as but not
limited to protection and de-protection steps, reduction, hydrolysis,
alkylation, etc.
Scheme 6
CI CI R2, ,R1
amoe
Coupling DIEA
KF
N
N CI Step A Step B
N A
6a 6b'
Preparation of Exemplified Examples
The following Examples have been prepared, isolated and characterized using
the methods
disclosed herein. The following examples demonstrate a partial scope of the
invention and
are not meant to be limiting of the scope of the invention.
Unless specified otherwise, starting materials are generally available from a
non-excluding
commercial sources such as TCI Fine Chemicals (Japan), Shanghai Chemhere Co.,
Ltd.(Shanghai, China), Aurora Fine Chemicals LLC (San Diego, CA), FCH Group
(Ukraine),
Aldrich Chemicals Co. (Milwaukee, Wis.), Lancaster Synthesis, Inc. (Windham,
N.H.), Acros
Organics (Fairlawn, N.J.), Maybridge Chemical Company, Ltd. (Cornwall,
England), Tyger
Scientific (Princeton, N.J.), AstraZeneca Pharmaceuticals (London, England),
Chembridge
Corporation (USA), Matrix Scientific (USA), Conier Chem & Pharm Co., Ltd
(China),
Enamine Ltd (Ukraine), Combi-Blocks, Inc. (San Diego, USA), Oakwood Products,
Inc.
(USA), Apollo Scientific Ltd. (UK), Allichem LLC. (USA) and Ukrorgsyntez Ltd
(Latvia).
LCMS Methods Employed in Characterization of Examples
Analytical LC/MS is carried out on Agilent systems using ChemStation software.
The
systems consist of:
= Agilent G1312 Binary Pump
= Agilent G1367 Well Plate Autosampler
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
114
= Agilent G1316 Thermostated Column Compartment
= Agilent G1315 Diode Array Detector
= Agilent 6140/6150 Mass Spectrometer
= SOFTA Evaporative Light Scattering Detector
Typical method conditions are as follows:
= Flow Rate: 0.9mL/min
= Column: 1.8 micrometres 2.1x50mm Waters Acquity HSS T3 C18 column
= Mobile Phase A: Water+0.05% TFA
= Mobile Phase B: Acetonitrile+0.035% TFA
= Run Time: 2.25 minutes
= The system runs a gradient from 10% B to 90% B in 1.35 minutes. A 0.6
minute wash at
100% B follows the gradient. The remaining duration of the method returns the
system
to initial conditions.
= Typical mass spectrometer Scan range is 100 to 1000 amu.
NMR Employed in Characterization of Examples
Proton spectra are recorded on a Bruker AVANCE ll 400 MHz with 5 mm QNP
Cryoprobe or
a Bruker AVANCE III 500 MHz with 5 mm QNP probe unless otherwise noted.
Chemical
shifts are reported in ppm relative to dimethyl sulfoxide (6 2.50), chloroform
(6 7.26),
methanol (6 3.34), or dichloromethane (6 5.32). A small amount of the dry
sample (2-5 mg)
is dissolved in an appropriate deuterated solvent (1 mL).
Reagents and materials
Solvents and reagents were purchased from suppliers and used without any
further
purification. Basic ion exchange resin cartridges PoraPakTM Rxn CX 20cc (2g)
were
purchased from Waters. Phase separator cartridges (Isolute Phase Separator)
were
purchased from Biotage. !solute absorbant (Isolute HM-N) was purchased from
Biotage.
ISCO Methods Employed in Purification of Examples
ISCO flash chromatography is carried on Teledyne COMBIFLASHO system with
prepacked
silica RediSepO column.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
115
Preparative HPLC Methods Employed in Purification of Examples
Preparative HPLC is carried out on Waters Autoprep systems using MassLynx and
Fraction Lynx software. The systems consist of:
= Waters 2767 Autosampler/Fraction Collector
= Waters 2525 Binary Pump
= Waters 515 Makeup pump
= Waters 2487 Dual Wavelength UV Detector
= Waters ZQ Mass Spectrometer
Typical method conditions are as follows:
= Flow Rate: 100mL/min
= Column: 10 micrometres 19x50mm Waters Atlantis T3 C18 column
= Injection Volume: 0-1000 microlitres
= Mobile Phase A: Water+0.05% TFA
= Mobile Phase B: Acetonitrile+0.035% TFA
= Run Time: 4.25 minutes
The system runs a gradient from x% B to y% B as appropriate for the examples
in 3 minutes
following a 0.25 minute hold at initial conditions. A 0.5 minute wash at 100%6
follows the
gradient. The remaining duration of the method returns the system to initial
conditions.
Fraction collection is triggered by mass detection through Fraction Lynx
software.
Chiral Preparative HPLC Methods Employed in Purification of Examples
SFC chiral screening is carried out on a Thar Instruments Prep Investigator
system coupled
to a Waters ZQ mass spectrometer. The Thar Prep Investigator system consists
of:
= Leap HTC PAL autosampler
= Thar Fluid Delivery Module (0 to 10 mL/min)
= Thar SFC 10 position column oven
= Waters 2996 PDA
= Jasco CD-2095 Chiral Detector
= Thar Automated Back Pressure Regulator.
All of the Thar components are part of the SuperPure Discovery Series line.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
116
The system flows at 2mL/min (4mL/min for the Whelk0-1 column) and is kept at
30 degrees
C. The system back pressure is set to 125 bar. Each sample is screened through
a battery
of six 3 micrometre columns:
= 3 micrometre 4.6x50 mm ChiralPak AD
= 3 micrometre 4.6x50 mm ChiralCel OD
= 3 micrometre 4.6x50 mm ChiralCel OJ
= 3 micrometre 4.6x250 mm Whelk 0-1
= 3 micrometre 4.6x50 mm ChiralPak AS
= 3 micrometre 4.6x50 mm Lux-Cellulose-2
The system runs a gradient from 5% co-solvent to 50% co-solvent in 5 minutes
followed by
a 0.5 minute hold at 50% co-solvent, a switch back to 5% co-solvent and a 0.25
minute hold
at initial conditions. In between each gradient there is a 4 minute
equilibration method the
flows at 5% co-solvent through the next column to be screened. The typical
solvents
screened are Me0H, Me0H+20mM NH3, Me0H+0.5%DEA, IPA, and IPA+20mM NH3.
Once separation is detected using one of the gradient methods an isocratic
method will be
developed and, if necessary, scaled up for purification on the Thar Prep80
system.
Example 1: N-methyl-2-(pyridin-4-yI)-N-(1,1,1-trifluoropropan-2-yl)pyrido[3,4-
d]pyrimidin-4-amine
N
N--N
N
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
117
Q o
9 cl
____________ 1-0-j1-NH, - r Poc13 ,,,,. ,,,
''''. NH N
,
1 Jr 1 ______________ ) 11
Step 1 N ,,--'= N0 Step 2 N ,,,,, N-,-,,-
Ci
N1-12
H
2a 2b 2c
CF3
D1EA
H KF
,
Step 3
CF3 CF3 1) Suzuki CF3
NI " OH
HObtl
li
24) Chiral seperation
1
48b .',. N
48a ''''. k Step 4 47
Step 1: A mixture of urea (40.00 g, 666.00 mmol) and 3-aminoisonicotinic acid
(2a,
18.40 g, 133.20 mmol) was heated at 210 C for 1 hr (NOTE: no solvent was
used). NaOH
(2N, 320 mL) was added, and the mixture was stirred at 90 C for lh. The solid
was
collected by filtration, and washed with water. The crude product thus
obtained was
suspended in HOAc (400 mL), and stirred at 100 C for lh. The mixture was
cooled to RT,
filtered, and the solid was washed with a large amount of water, and then
dried under the
vacuum to give pyrido[3,4-d]pyrimidine-2,4(1H,3H)-dione (2b, 17.00 g, 78%
yield) without
further purification. LCMS (m/z [M+H]F): 164Ø
Step 2: To a mixture of pyrido[3,4-d]pyrimidine-2,4(1H,3H)-dione (2b, 20.00 g,
122.60 mmol) and P0CI3 (328.03 g, 2.14 mol) in toluene (200 mL) was added DIEA
(31.69
g, 245.20 mmol) dropwise and this reaction mixture stirred at 25 C overnight
(18hr) to give
suspension.
The solvent and P0CI3 was removed under vacuum, diluted with DCM (50 mL),
neutralized with DIEA to pH=7 at -20 C and concentrated again, the residue
was purified by
column (20-50% EA/PE) to give 2,4-dichloropyrido[3,4-d]pyrimidine (2c, 20.00g,
99.99
mmol, 82% yield) as a yellow solid. 1H NMR (400 MHz, CHLOROFORM-d) 6 9.52 (s,
1 H),
8.92 (d, J=5.6 Hz, 1 H), 8.04 (d, J=5.6 Hz, 1 H). LCMS (m/z [M+H]F): 200Ø
Step 3: In a 20mL vial 2,4-dichloropyrido[3,4-d]pyrimidine (600 mg, 3.0 mmol)
was
stirred in DMSO (0.7mL) at room temperature and degassed with N2. DIEA (1 mL,
6 mmol)
was added and stirred for 5 minutes then KF (174 mg, 3 mmol). This mixture was
stirred at
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
118
room temperature for 15 minutes then racemic 1,1,1-trifluoro-N-methylpropan-2-
amine (419
mg, 3.3 mmol) was added and degassed then stirred at 60 C for 4 hours. The
reaction was
then concentrated and purified by flash chromatography on a COMBIFLASHO system
(ISCO) using 0-10% Me0H/DCM to afford 2-chloro-N-methyl-N-(1,1,1-
trifluoropropan-2-
.. yOpyrido[3,4-d]pyrimidin-4-amine (680 mg, 74%). 1H NMR (500 MHz, Acetone-
d6) 6 9.09
(d, J = 0.9 Hz, 1H), 8.59 (d, J = 5.9 Hz, 1H), 8.22 (dd, J = 5.9, 0.9 Hz, 1H),
5.93 (dddd, J =
15.3, 8.3, 7.0, 1.2 Hz, 1H), 3.61 (q, J = 1.0 Hz, 3H), 1.63 (d, J = 7.0 Hz,
3H). LCMS (m/z
[M+H]F): 291.7.
Step 4: In a 20 mL microwave reactor was added PalladiumTetrakis (99 mg, 0.086
mmol), potassium carbonate (2.15 mL, 4.3 mmol), and 2 chloro-N-methyl-N-(1,1,1-
trifluoropropan-2-yOpyrido[3,4-d]pyrimidin-4-amine (500 mg, 1.72 mmol) and
pyridin-4-
ylboronic acid (233 mg, 1.89 mmol) in acetonitrile (8 mL) to give an yellow
suspension. The
reaction mixture was stirred at 130 C for 30 min under microwave. The crude
mixture was
diluted with DCM, H20, separated and extracted with DCM x3. Combined the
organic layers
and dried Na2SO4, filtered and concentrated. The residue was purified by flash
chromatography on a COMBIFLASHO system (ISCO) using 0-10% Me0H/DCM to give
Example 1, the racemic product, then followed by chiral HPLC (21x250mm OJ-H
column
with 85% CO2 as phase A and 15% Me0H as phase B, flow rate 2mL/min, 30 C, 3.5
min
elution time) to separate the enantiomers to afford Examples la and lb.
Example la: N-methyl-2-(pyridin-4-y1)-N-[(25)-1,1,1-trifluoropropan-2-
yl]pyrido[3,4-
d]pyrimidin-4-amine
F*F
N
N
NN
N
1H NMR (500 MHz, DMSO-d6) 6 9.33 (d, J = 0.8 Hz, 1H), 8.86 - 8.75 (m, 2H),
8.63
(d, J = 5.9 Hz, 1H), 8.38 - 8.30 (m, 2H), 8.20 (dd, J = 6.0, 0.9 Hz, 1H), 6.11
(qt, J = 8.5, 7.4
Hz, 1H), 3.50 (d, J = 1.1 Hz, 3H), 1.61 (d, J = 7.0 Hz, 3H). LCMS (m/z [M+H]):
334.1. Chiral
HPLC TR = 1.73 min. Absolute stereochemistry was confirmed by X-ray crystal
structure.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
119
Example lb: N-methyl-2-(pyridin-4-y1)-N-[(2R)-1,1,1-trifluoropropan-2-
yl]pyrido[3,4-
d]pyrimidin-4-amine
F
N
NN
N
1H NMR (500 MHz, DMSO-d6) 6 9.33 (d, J = 0.8 Hz, 1H), 8.86 - 8.75 (m, 2H),
8.63
(d, J = 5.9 Hz, 1H), 8.38 - 8.30 (m, 2H), 8.20 (dd, J = 6.0, 0.9 Hz, 1H), 6.11
(qt, J = 8.5, 7.4
Hz, 1H), 3.50 (d, J = 1.1 Hz, 3H), 1.61 (d, J = 7.0 Hz, 3H). LCMS (m/z [M+H]):
334.1. Chiral
HPLC TR = 1.25 min. Absolute stereochemistry was confirmed by X-ray crystal
structure.
Example 2: N-(tert-butyl)-2-(pyridin-4-y1)-1,7-naphthyridin-4-amine:
NH
Nc
suzuki
9H ci
ci HO DIEA
ii
¨
H2N KF
Step Step 2
1
N
N
Step 1: In a 20 mL microwave reactor was added PalladiumTetrakis (58.1 mg,
0.050
mmol), potassium carbonate (1.256 mL, 2.51 mmol), and 2,4-dichloro-1,7-
naphthyridine
(200 mg, 1.005 mmol) and pyridin-4-ylboronic acid (130 mg, 1.055 mmol) in
Acetonitrile
(Volume: 2 mL) to give an orange suspension. The reaction mixture was stirred
at 120 C for
60 min under microwave. The crude mixture was diluted with DCM, H20, separated
and
extracted with DCM x3. Combined the organic layers and dried Na2SO4, filtered
and
concentrated. The residue was purified by flash chromatography on a
COMBIFLASHO
system (ISCO) using 0-10% Me0H/DCM to give the product (62%). 1H NMR (400 MHz,
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
120
DMSO-d6) 6 9.58 (d, J = 0.9 Hz, 1H), 8.85 - 8.78 (m, 4H), 8.32 - 8.29 (m, 2H),
8.11 (dd, J =
5.8, 0.9 Hz, 1H). LCMS [M+H] = 242.
Step 2: In a 40m1 vial was added potassium fluoride (11.54 mg, 0.199 mmol), 4-
chloro-2-(pyridin-4-yI)-1,7-naphthyridine (40 mg, 0.166 mmol), and 2-
methylpropan-2-amine
(0.035 mL, 0.331 mmol) in DMSO (Volume: 2 mL) to give a yellow suspension. The
reaction
mixture was stirred at 130 C for 24 hrs. Solvent was evaporated under air
flow. The residue
was purified by flash chromatography on a COMBIFLASHO system (ISCO) using 0-
10%
Me0H/DCM to give the product (82%). 1H NMR (400 MHz, DMSO-d6) 6 9.22 (d, J =
0.7
Hz, 1H), 8.78 - 8.72 (m, 2H), 8.48 (d, J = 5.8 Hz, 1H), 8.30 (dd, J = 6.0, 0.9
Hz, 1H), 8.15 -
8.06 (m, 2H), 7.28 (s, 1H), 6.73 (s, 1H), 1.56 (s, 9H). LCMS [M+H] = 279.2.
Example 3: 2,4-dimethy1-44[2-(pyridin-4-y1)pyrido[3,4-d]pyrimidin-4-
yl]amino}pentan-
2-01
j<DH
HN
N
N =*.N
111
1H NMR (400 MHz, Acetone-d6) 59.57 (s, 1H), 9.15 (d, J = 0.9 Hz, 1H), 8.82 -
8.72
(m, 2H), 8.56 (d, J = 5.6 Hz, 1H), 8.44 - 8.37 (m, 2H), 7.69 (dd, J = 5.6, 0.9
Hz, 1H), 2.08 (s,
2H), 1.87 (s, 6H), 1.48 (d, J = 0.8 Hz, 6H). LCMS (m/z [M+H]): 338.2.
Example 4: 2-(3-methyl-1H-pyrazol-4-y1)-N-(1-methylcyclopropyl)pyrido[3,4-
cl]pyrimidin-4-amine
Fir;>1
N
NLN
1H NMR (500 MHz, Methanol-d4) 59.01 (s, 1H), 8.41 (d, J = 5.7 Hz, 1H), 8.26
(s,
1H), 7.91 (dd, J = 5.7, 0.9 Hz, 1H), 2.83 (s, 3H), 1.60 (s, 3H), 1.05 - 0.94
(m, 2H), 0.91 -
0.82 (m, 2H). LCMS (m/z [M+H]+): 281.1.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
121
Startinq material to prepare an expanded population of cells:
Autologous method
The seeding population of cells for use in the method of cell population
expansion to obtain
an expanded population of cells may be obtained from a recipient
himself/herself. In patients
where tissue, organ, or cell deficiency is partial, for example healthy cells
are present, the
seeding population of cells may be obtained from non-affected tissue or organ
or cell
source. For example, in the case of unilateral ocular cell deficiency, the
seeding population
may be obtained from a biopsy on the non-affected eye. It may also be obtained
from
healthy tissue remaining in an organ that is partially damaged.
Allogenic method
In a preferred embodiment, the seeding population of cells for use in the
method of cell
population expansion to obtain an expanded population of cells may be obtained
from cells
originally derived from donor tissue (e.g., human, rabbit, monkey etc.,
preferably human).
For example a source of human tissue is from cadaveric donors or tissues from
living
donors, including living relatives.
From autologous or allogenic tissue derived as described above under
autologous and
allogenic methods which has been removed from the body, the cells may, for
example, be
extracted and prepared as follows: The desired area may be dissected, for
example, using
scalpels and the cells then dissociated (e.g. using collagenase, dispase,
trypsin, accutase or
TripLE; for example 1 mg/ml collagenase at 37 C), until cell detachment
becomes apparent
by microscopic observation (e.g., using a Zeiss Axiovert inverted microscope)
from 45
minutes to 3 hours.
Suitably, the cells, e.g., LSCs or CECs, isolated from several corneas or from
different
donors may be pooled for further processing, such as cell population expansion
and B2M-
gene-editting.
For use in the cell population expansion method according to the invention the
isolated cells
are then added to medium, for example by pipetting, as described below in the
section "Cell
population expansion".
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
122
In a preferred embodiment according to the invention, an assessment of the
quality of
cellular material harvested from the donor is performed. For example,
approximately 24
hours after harvesting the cells and beginning culturing in medium (growth or
cell
.. proliferation medium, as described below), a visual assessment under
brightfield microscope
to look for floating cells present (as an indicator of dead cells) may be
performed. Ideally this
assessment is to show that there is approximately less than 10% as floating
cells for the
material to be suitable for use to generate an expanded population of cells
according to the
invention.
The number of cells suitable for use in the method of cell population
expansion according to
the invention is not limited, but as an example for illustrative purposes, the
seeding cell
population suitable for use in the method of cell population expansion
according to the
invention may comprise approximately 1000 cells.
If it is desired to measure the cell numbers in the seeding cell population,
this may be done
for example by manual or automated cell counting using a light microscope,
immunohistochemistry or FACS according to standard protocols well known in the
art.
Ex-vivo ocular cell population expansion and use in therapy
Described below in more detail is a description of the methodology relating to
expansion of
ocular cell populations (preparation of starting material, followed by cell
population
expansion phase, storage of cells) as applied to ocular cells with the
specific examples of
limbal stem cells and corneal endothelial cells.
Starting material to prepare an expanded population of limbal stem cells:
Corneal epithelial and limbal cells
Autolodous method
The seeding population of cells for use in the method of cell population
expansion to obtain
an expanded population of limbal stem cells may be obtained from the recipient
himself/herself. In patients where limbal stem cell deficiency is partial, the
seeding
population of cells may be obtained from non-affected parts of the limbus. For
example, in
the case of unilateral limbal stem cell deficiency, the seeding population may
be obtained
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
123
from a biopsy on the non-affected eye. It may also be obtained from healthy
tissue
remaining in a limbus that is partially damaged.
Allogenic method
In a preferred embodiment, the seeding population of cells for use in the
method of cell
population expansion to obtain an expanded population of limbal stem cells may
be obtained
from cells originally derived from donor mammalian corneal tissue (e.g.,
human, rabbit,
monkey etc., preferably human).
For example a source of human corneal tissue is from cadaveric donors (for
example
sourced through eye banks) or tissues from living donors, including living
relatives. A range
of donor limbal tissue is suitable for use according to the invention. In a
preferred
embodiment limbal tissue is obtained from living relatives or donors with a
compatible HLA
profile.
The tissue that is used to obtain the LSCs may, for example, be a ring of
limbal tissue of
approximately 4mm in width and 1 mm in height.
From the corneal tissue as described above under autologous and allogenic
methods which
has been removed from the body, the LSCs may, for example, be extracted and
prepared as
follows: The limbal epithelial area may be dissected, for example, using
scalpels and the
cells then dissociated (e.g., using collagenase, dispase, trypsin, accutase or
TripLE; for
example 1 mg/ml collagenase at 37 C), until cell detachment becomes apparent
by
microscopic observation (e.g., using a Zeiss Axiovert inverted microscope)
from 45 minutes
to 3 hours.
Suitably, the cells, e.g., LSCs or CECs, isolated from several corneas or from
different
donors may be pooled for further processing, such as cell population expansion
and B2M-
gene-editting.
For use in the cell population expansion method according to the invention the
isolated cells
are then added to medium, for example by pipetting, as described below in the
section "Cell
population expansion".
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
124
In a preferred embodiment according to the invention, an assessment of the
quality of
cellular material harvested from the donor cornea is performed. For example,
approximately
24 hours after harvesting the cells and beginning culturing in medium (growth
or cell
proliferation medium, as described below), a visual assessment under
brightfield microscope
to look for floating cells present (as an indicator of dead cells) may be
performed. Ideally this
assessment is to show that there is approximately less than 10% as floating
cells for the
material to be suitable for use to generate an expanded population of cells
according to the
invention.
The number of cells suitable for use in the method of cell population
expansion according to
the invention is not limited, but as an example for illustrative purposes, the
seeding cell
population suitable for use in the method of cell population expansion
according to the
invention may comprise approximately 1,000 limbal stem cells.
If it is desired to measure the cell numbers in the seeding cell population,
this may be done
for example by manual or automated cell counting using a light microscope,
immunohistochemistry or FACS according to standard protocols well known in the
art.
Starting material to prepare an expanded population of corneal endothelial
cells
The seeding population of corneal endothelial cells (CECs) for use in the
method of cell
population expansion may be obtained from cells originally derived from
mammalian corneal
tissue (e.g., human, rabbit, monkey etc., preferably human). For example, a
source of
human corneal tissue is from cadaveric human donors (which may be sourced
through eye
banks).
The age of the donors can range, for example, from infancy to 70 years of age.
Preferably
also suitable donors are those who have no history of corneal disease or
trauma. In one
.. embodiment according to the invention, preferred donor corneas are those
where the
corneal endothelial cell count is above 2 000 cells/mm2 (area). In a more
preferred
embodiment according to the invention the corneal endothelial cell count is 2
000 to 3 500
cells/mm2 (area). This is measured for example by examining the cornea of the
donor
material under a direct light microscope or a specular microscope as per
standard Eye Bank
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
125
techniques known in the art for evaluation of donor tissue before
transplantation to patients
(see Tran et al (2016) Comparison of Endothelial Cell Measurements by Two Eye
Bank
Specular Micorscopes; International Journal of Eye Banking; vol 4., no 2; 1-8,
which is
herein incorporated by reference).
The surface of cornea that is used to obtain the CECs is not limited, but may,
for example,
be an area of approx. 8-10mm in diameter.
The CECs may, for example, be extracted and prepared as follows from the donor
corneal
tissue: The corneal endothelial cell layer and Descemet's membrane (DM) are
scored, for
example with a surgical-grade reverse Sinsky endothelial stripper. The DM-
endothelial cell
layer is peeled off the corneal stroma and cells are dissociated from the DM,
for example
using 1 mg/ml collagenase at 37 C until cell detachment becomes apparent by
microscopic
observation (e.g. using a Zeiss Axiovert inverted microscope) (from 45 minutes
to 3 hours).
As the DM only carries corneal endothelial cells in the cornea, the cell
population isolated in
this manner is a population of CECs, which is suitable for use as a seeding
population of
cells according to the invention.
For use in the method of cell population expansion according to the invention
the isolated
corneal endothelial cells may be added to medium as described below in the
section "Cell
population expansion".
In a preferred embodiment according to the invention, an assessment of the
quality of
cellular material harvested from the donor cornea is performed. For example,
approximately
24 hours after harvesting the cells and beginning culturing in medium (growth
or cell
proliferation medium, as described below), a visual assessment under
brightfield microscope
to look for floating cells present (as an indicator of dead cells) may be
performed. Ideally this
assessment is to show that there is approximately less than 10% as floating
cells for the
material to be suitable for use to generate an expanded population of cells
according to the
invention.
The starting number of cells suitable for use in the method of cell population
expansion
according to the invention is not limited, but as an example for illustrative
purposes, the
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
126
seeding population of corneal endothelial cells suitable for use in the method
of cell
population expansion according to the invention may be 100 000 to 275 000
cells.
If it is desired to measure the cell numbers in the seeding cell population,
this may be done
for example by taking an aliquot and performing immunocytochemistry (e.g., to
count nuclei
stained with Sytox Orange) or by live cell imaging under brightfield
microscope to count the
number of cells.
The Sytox Orange assay may be performed according to standard protocols known
in the
art. In brief, after cells have attached to the cell culture dish (typically
24h after cell plating),
the cells are fixed in paraformaldehyde. The cells are then permeabilized
(e.g., using a
solution of 0.3% Triton X-100) and they are then labeled in a solution of
Sytox Orange (e.g.,
using 0.5 micromolar of Sytox Orange in PBS). The number of nuclei stained
with Sytox
Orange per surface area are then counted under a Zeiss epifluorescence
microscope.
Cell population expansion
In one embodiment of the invention, a population of cells comprising cells
from a patient or a
donor, can be grown in medium in a culture container known in the art, such as
plates, multi-
well plates, and cell culture flasks. For example, a culture dish may be used
which is non-
coated or coated with collagen, synthemax, gelatin or fibronectin. A preferred
example of a
suitable culture container is a non-coated plate. Standard culturing
containers and
equipment such as bioreactors known in the art for industrial use may also be
used.
The term "culture medium", "cell culture medium", "cell medium" or "medium" is
used to
describe (i) a cellular growth medium in which cells are grown, for example,
stem cells,
progenitor cells, or differentiated cells or (ii) a cell proliferation medium
in which cells are
prolifirated, for example, stem cells, progenitor cells, or differentiated
cells.
The medium used may be a growth medium or a cell proliferation medium. In
general, a
growth medium is a culture medium supporting the growth and maintenance of a
population
of cells. Those of skill in art can readily determine an appropriate growth
medium for a
particular type of cell population. Suitable growth mediums are known in the
art for stem cell
culture or epithelial cell culture are for example: DMEM (Dulbecco's Modified
Eagle's
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
127
Medium) supplemented with FBS (Fetal Bovine Serum) (Invitrogen), human
endothelial SF
(serum free) medium (Invitrogen) supplemented with human serum, X-VIV015
medium
(Lonza), or DMEM/F12 (Thermo Fischer Scientific) (optionally supplemented with
calcium
chloride). These may be additionally supplemented with growth factors (e.g.
bFGF), and/or
antibiotics such as penicillin and streptomycin.
Alternatively, isolated cells may be added first to a cell proliferation
medium according to the
invention. The cell proliferation medium as defined herein comprises a growth
medium and a
LATS inhibitor according to the invention.
In certain embodiments, a cell proliferation medium of the invention comprises
a growth
medium and a LATS inhibitor according to the invention. The LATS inhibitor is
preferably
selected from the group comprising compounds according to Formula Al or
subformulae
thereof (e.g., Formula A2) and as further described under the section "LATS
inhibitors".
In a preferred embodiment the LATS inhibitors according to Formula Al or
subformulae
thereof (e.g., Formula A2) are added at a concentration of about 0.5 to 100
micromolar,
preferably about 0.5 to 25 micromolar, more preferably about 1 to 20
micromolar. In a further
embodiment the LATS inhibitors according to Formula Al or subformulae thereof
(e.g.,
Formula A2) are added at a concentration of 0.5 to 100 micromolar, preferably
0.5 to 25
micromolar, more preferably 1 to 20 micromolar. In a specific embodiment the
LATS
inhibitors according to Formula Al or subformulae thereof (e.g., Formula A2)
are added at a
concentration of about 3 to 10 micromolar. In a more specific embodiment the
LATS
inhibitors according to Formula Al or subformulae thereof (e.g., Formula A2)
are added at a
concentration of 3 to 10 micromolar.
In one embodiment, the stock solution of the compound according to Formula Al
or
subformulae thereof (e.g., Formula A2) may be prepared by dissolving the
compound
powder to a stock concentration of 1 mM to 100 mM in DMSO, e.g., 1 mM to 50
mM, 5 mM
to 20 mM, 10 mM to 20mM, in particularly 10mM. In one embodiment, the stock
solution of
the compound according to Formula Al or subformulae thereof (e.g., Formula A2)
may be
prepared by dissolving the compound powder to a stock concentration of 10mM in
DMSO.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
128
In one aspect of the invention the LATS inhibitor according to the invention
inhibits LATS1
and/or LATS2 activity in the cell population. In a preferred embodiment the
LATS inhibitor
inhibits LATS1 and LATS2.
In one embodiment, a cell proliferation medium of the invention optionally
further comprises
a rho-associated protein kinase (ROCK) inhibitor. The addition of a ROCK
inhibitor was
found to prevent cell death and promote attachment of cells in suspensions,
especially when
culturing stem cells. The ROCK inhibitor are known in the art and in one
example, selected
from (R)-(+)-trans-4-(1-aminoethyl)-N-(4-Pyridyl)cyclohexanecarboxamide
dihydrochloride
monohydrate ((1R,40-44(R)-1-aminoethyl)-N-(pyridin-4-y0cyclohexanecarboxamide;
Y-
27632; Sigma-Aldrich), 5-(1,4-diazepan-1-ylsulfonyl) isoquinoline (fasudil or
HA 1077;
Cayman Chemical), H-1152, H-1152P, (S)-(+)-2-Methyl-1-[(4-methyl-5-
isoquinolinyl)sulfonyl]homopiperazine, 2HCI, ROCK Inhibitor, Dimethylfasudil
(diMF, H-
11 52P), N-(4-Pyridy1)-N'-(2,4,6-trichlorophenyOurea, Y-39983, Wf-536, SNJ-
1656, and (5)-
+)-2-methyl-1-[(4-methyl-5-isoquinolinyOsulfonyl]-hexahydro-1H-1,4-diazepine
dihydrochloride (H-1152; Tocris Bioscience), and its derivatives and analogs.
Additional
ROCK inhibitors include imidazole-containing benzodiazepines and analogs (see,
e.g., WO
97/30992). Others include those described in International Application
Publication Nos.: WO
01/56988; WO 02/100833; WO 03/059913; WO 02/076976; WO 04/029045; WO
03/064397;
WO 04/039796; WO 05/003101; WO 02/085909; WO 03/082808; WO 03/080610; WO
04/112719; WO 03/062225; and WO 03/062227, for example. In some of these
cases,
motifs in the inhibitors include an indazole core; a 2-
aminopyridine/pyrimidine core; a 9-
deazaguanine derivative; benzamide-comprising; aminofurazan-comprising; and/or
a
combination thereof. Rock inhibitors also include negative regulators of ROCK
activation
such as small GTP-binding proteins (e.g., Gem, RhoE, and Rad), which can
attenuate
ROCK activity. In specific embodiments of the disclosure, ROCK1 is targeted
instead of
ROCK2, for example, WO 03/080610 relates to imidazopyridine derivatives as
kinase
inhibitors, such as ROCK inhibitors, and methods for inhibiting the effects of
ROCK1 and/or
ROCK2. The disclosures of the applications cited above are incorporated herein
by
reference. The Rho inhibitor can also act downstream by interaction with ROCK
(Rho-
activated kinase) leading to an inhibition of Rho. Such inhibitors are
described in U.S. Pat.
No. 6,642,263 (the disclosures of which are incorporated by reference herein
in their
entirety). Other Rho inhibitors that may be used are described in U.S. Pat.
Nos. 6,642,263,
and 6,451,825. Such inhibitors can be identified using conventional cell
screening assays,
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
129
e.g., described in U.S. Pat. No. 6,620,591 (all of which are herein
incorporated by reference
in their entirety).
In a preferred embodiment, the ROCK inhibitor used in the cell proliferation
medium of the
present invention is (R)-(+)-trans-4-(1-aminoethyl)-N-(4-
Pyridyl)cyclohexanecarboxamide
dihydrochloride monohydrate ((1R,40-44(R)-1-aminoethyl)-N-(pyridin-4-
y0cyclohexanecarboxamide; Y-27632; Sigma-Aldrich; described in Nature 1997,
vol. 389,
pp. 990-994; JP4851003, JP11130751; JP2770497; U55478838; U56218410, all of
which
are herein incorporated by reference in their entirety).
In one embodiment, said ROCK inhibitor, in particular Y-27632, is present in a
concentration
of about 0.5 to about 100 micromolar, preferably of about 0.5 to about 25
micromolar, more
preferably of about 1 to about 20 micromolar, particularly preferably of about
10 micromolar.
In one embodiment, said compound of the present invention is present in a
concentration of
0.5 to 100 micromolar, preferably 0.5 to 25 micromolar, more preferably 1 to
20 micromolar,
particularly preferably 10 micromolar. In a specific embodiment, said ROCK
inhibitor, in
particular Y-27632, is present in a concentration of 10 micromolar.
In a specific embodiment, a cell proliferation medium of the invention
comprises DMEM/F12
(1:1), 5-20% human serum or fetal bovine serum or a serum substitute, 1-2 mM
calcium
chloride, 1 micromolar to 20 micromolar LATS inhibitor, and optionally, 1
micromolar to 20
micromolar ROCK inhibitor. In a more specific embodiment, a cell proliferation
medium of
the invention comprises DMEM/F12 (1:1), 10-20% human serum or fetal bovine
serum or a
serum substitute, e.g., 10% human serum or fetal bovine serum or a serum
substitute, 1-2
mM calcium chloride, 3 micromolar to 10 micromolar LATS inhibitor, and
optionally, 10
micromolar ROCK inhibitor.
The cells may go through a round or rounds of addition of fresh growth medium
and/or cell
proliferation medium. The cells do not need to be passaged in order for fresh
medium to be
added, but passaging cells is also a way to add fresh medium.
A series of mediums may be also used, in various combinations of orders: for
example a cell
proliferation medium, followed by addition of a growth medium (which is not
supplemented
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
130
with LATS inhibitors according to the invention, and may be different to the
growth medium
used as the base for the cell proliferation medium).
The cell population expansion phase according to the invention occurs during
the period the
cells are exposed to the cell proliferation medium.
Standard temperature conditions known in the art for culturing cells may be
used, for
example preferably about 30 C to 40 C. Particularly preferably cell growth,
as well as the
cell population expansion phase is carried out at about 37 C. A conventional
cell incubator
with 5-10% CO2 levels may be used. Preferably the cells are exposed to 5% CO2.
The cells may be passaged during the culturing in the growth or cell
proliferation medium as
necessary. Cells may be passaged when they are sub-confluent or confluent.
Preferably the
cells are passaged when they reach approximately 90%-100% confluency, although
lower
percentage confluency levels may also be performed. The passaging of cells is
done
according to standard protocols known in the art. For example, in brief cells
are passaged by
treating cultures with Accutase (e.g., for 10 minutes), rinsing the cell
suspension by
centrifugation and plating cells in fresh growth medium or cell proliferation
medium as
desired. Cell splitting ratios range, for example, from 1:2 to 1:5.
For the cell population expansion phase of the method of cell population
expansion
according to the invention, the expansion of the seeding cell population in
the cell
proliferation medium may be performed until the required amount of cellular
material is
obtained.
The cells may be exposed to the cell proliferation medium for a range of time
periods in
order to expand the cell population.
In a preferred embodiment the seeding cell population is exposed to the LATS
inhibitors
according to the invention (such as those compounds according to Formula Al or
subformulae thereof (e.g., Formula A2)) directly after cell isolation from the
patient or donor
tissue and maintained for the entire time that cell proliferation is required,
for example 12 to
16 days.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
131
In one embodiment according to the invention, a gene editing technique may
optionally be
performed to genetically modify cells and/or to express a biotherapeutic
compound. For
example, the cells may be modified to reduce or eliminate the expression
and/or function of
an immune response mediating gene, which may otherwise contribute to immune
rejection
when the cell population is delivered to the patient. The application of gene
editing
techniques in the method of cell population expansion according to the
invention is optional,
and the administration to the patient of topical immunosuppressants and/or
anti-
inflammatory agents (as described further under the section Immunosuppressant
and Anti-
inflammatory agent) may instead be used if desired to mitigate issues with
immunorejection
of the transplanted material in the patient.
According to one aspect of the invention, genetically modifying comprises
reducing or
eliminating the expression and/or function of a gene associated with
facilitating a host
versus graft immune response. In a preferred embodiment, genetically modifying
comprises
introducing into an isolated stem cell or stem cell population a gene editing
system which
specifically targets a gene associated with facilitating a host versus graft
immune response.
In a specific embodiment, said gene editing system is CRISPR (CRISPR:
clustered regularly
interspaced short palindromic repeats, also known as CRISPR/Cas systems).
The gene editing technique may be performed at different points, such as for
example (1) on
tissue, before cell isolation or (2) at the time of cell isolation or (3)
during the cell population
expansion phase in vitro (when the cells are exposed to a LATS inhibitor
according to the
invention in vitro) or (4) in vitro at the end of the cell population
expansion phase (after the
cells are exposed to a LATS inhibitor according to the invention in vitro). In
one embodiment,
CRISPR is used after two weeks of in vitro expansion of the cell population in
the presence
of the LATS inhibitor according to the invention.
The gene editing techniques suitable for use in the method of cell population
expansion are
further described under the section "reduction of immunorejection".
In the method of cell population expansion according to the invention the LATS
inhibitors,
which are preferably compounds, produce greater than 2 fold expansion of the
seeded
population of cells.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
132
In one aspect of the method of cell population expansion according to the
invention the
compounds according to Formula Al or subformulae thereof (e.g., Formula A2)
produce
greater than 30 fold expansion of the seeded population of isolated cells
(i.e., cells obtained
from a patient or a donor). In a specific embodiment of the method of cell
population
expansion according to the invention, the LATS inhibitors according to Formula
Al or
subformulae thereof produce 100 fold to 2200 fold expansion of the seeded
population of
isolated cells. In a more specific embodiment of the method of cell population
expansion
according to the invention, the LATS inhibitors according to Formula Al or
subformulae
thereof (e.g., Formula A2) produce 600 fold to 2200 fold expansion of the
seeded population
of isolated cells. The fold expansion factor achieved by the method of cell
population
expansion according to the invention may be achieved in one or more passages
of the cells.
In another aspect of the invention the fold expansion factor achieved by the
method of cell
population expansion according to the invention may be achieved after exposure
to the
compound according to Formula Al or subformulae thereof (e.g., Formula A2) for
about 12
to 16 days, preferably about 14 days. In one mebodiment, the expanded seeded
population
of isolated LSCs according to the invention comprises at least 40% of
undifferentiated LSCs,
e.g., at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at
least 70%, at
least 75%, at least 80%, at least 85%, at least 90%, at least 95% of
undifferentiated LSCs.
In a specific embodiment, the expanded seeded population of isolated LSCs
according to
.. the invention comprises at least 60% of undifferentiated LSCs. In a more
specific
embodiment, the expanded seeded population of isolated LSCs according to the
invention
comprises at least 80% of undifferentiated LSCs. In a preferred embodiment,
the expanded
seeded population of isolated LSCs according to the invention comprises at
least 90% of
undifferentiated LSCs.
If it is desired to measure the cell number or expansion of the cell
population, this may be
done for example by taking an aliquot and performing immunocytochemistry
(e.g., to count
nuclei stained with Sytox Orange) or by live cell imaging under brightfield
microscope to
count the number of cells or by performing real-time quantitative live-cell
analysis of cell
confluence at various time points during the cell population expansion phase
of the method
according to the invention.
The Sytox Orange assay may be performed according to standard protocols known
in the
art. In brief, after cells have attached to the cell culture dish (typically
24h after cell plating),
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
133
the cells are fixed in paraformaldehyde. The cells are then permeabilized
(e.g. using a
solution of 0.3% Triton X-100) and they are then labeled in a solution of
Sytox Orange (e.g.,
using 0.5 micromolar of Sytox Orange in PBS). The number of nuclei stained
with Sytox
Orange per surface area are then counted under a Zeiss epifluorescence
microscope.
The cell population expanded by the method of cell population expansion
according to the
invention may be added to a solution and then stored, for example in a
preservation or
cryopreservation solution (such as those described below), or added directly
to a
composition suitable for delivery to a patient. The preservation,
cryopreservation solution or
composition suitable for ocular delivery may optionally comprise a LATS
inhibitor according
to the invention.
In a more preferred embodiment according to the invention, the cell population
preparation
which is delivered to a patient comprises very low to negligible levels of a
LATS inhibitor
compound. Thus in a specific embodiment, the method of cell population
expansion
according to the invention comprises the further step of rinsing to
substantially remove the
compound of the present invention (such as the compound according to Formula
Al or
subformulae thereof (e.g., Formula A2)). This may involve rinsing the cells
after the cell
population expansion phase according to the invention. To rinse the cells, the
cells are
detached from the culture dish (e.g., by treating with Accutase), the detached
cells are then
centrifuged, and a cell suspension is made in PBS or growth medium according
to the
invention. This step may be performed multiple times, e.g., one to ten times,
to rinse out the
cells. Finally the cells may be resuspended in a preservation solution,
cryopreservation
solution, a composition suitable for ocular delivery, growth medium or
combinations thereof
as desired.
The expanded population of cells prepared by the method of cell population
expansion and
rinsed of cell proliferation medium comprising a LATS inhibitor according the
invention may
be transferred to a composition suitable for delivery to a patient, such as
for example a
localising agent. Optionally the cell population is stored for a period before
addition to a
localising agent suitable for delivery to a patient. In a preferred
embodiment, the expanded
cell population may first be added to a solution suitable for preservation or
cryopreservation,
which preferably does not comprise a LATS inhibitor, and the cell population
stored
(optionally with freezing) before addition to a localising agent suitable for
delivery to a
patient, which also preferably does not comprise a LATS inhibitor.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
134
Typical solutions suitable for cryopreservation, glycerol, dimethyl sulfoxide,
propylene glycol
or acetamide may be used in the cryopreservation solution of the present
invention. The
cryopreserved preparation of cells is typically kept at -20 C or -80 C. In
one embodiment,
the cryopreserved compositions comprise a cell (e.g., a modified cell, such as
LSC or CEC,
with reduced or eliminated expression of B2M by a CRISPR system), e.g., a
plurality of
cells) and a cryoprotectant selected from the list of glycerol, DMSO
(dimethylsulfoxide)
polyvinylpyrrolidone, hydroxyethyl starch, propylene glycol, acetamide,
monosaccharides,
algae-derived polysaccharides, and sugar alcohols, or a combination thereof.
In a more
specific embodiment, the cryopreserved compositions comprise a cell (e.g., a
modified cell,
such as LSC or CEC, with reduced or eliminated expression of B2M by a CRISPR
system),
e.g., a plurality of cells) and DMSO concentration of 0.5% to 10%, e.g., 1%-
10%, 2%-7%,
3%-6%, 4% - 5%, preferably 5%. DMSO acts as a cryoprotecting agent against
formation of
water crystals within and outside the cells, which could lead to cell damage
during
cryopreservation steps. In a further embodiment, the cryopreserved
compositions further
comprise a suitable buffer, for example CryoStor CS5 buffer (BioLife
Solutions).
Cell population expansion: To prepare an expanded population of limbal stem
cells
In one embodiment of the invention, a population of cells comprising corneal
epithelial and
limbal cells, including limbal stem cells, for example obtained as described
in the section
"Starting material to prepare an expanded population of limbal stem cells:
Corneal epithelial
and limbal cells", can be grown in medium in a culture container known in the
art, such as
plates, multi-well plates, and cell culture flasks. For example, a culture
dish may be used
which is non-coated or coated with collagen, synthemax, gelatin or
fibronectin. A preferred
example of a suitable culture container is a non-coated plate. Standard
culturing containers
and equipment such as bioreactors known in the art for industrial use may also
be used.
The medium used may be a growth medium or a cell proliferation medium. A
growth
medium is defined herein as a culture medium supporting the growth and
maintenance of a
population of cells. Suitable growth mediums are known in the art for stem
cell culture or
epithelial cell culture are for example: DMEM (Dulbecco's Modified Eagle's
Medium)
supplemented with FBS (Fetal Bovine Serum) (Invitrogen), human endothelial SF
(serum
free) medium (Invitrogen) supplemented with human serum, X-VIV015 medium
(Lonza), or
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
135
DMEM/F12 (Thermo Fischer Scientific) (optionally supplemented with calcium
chloride).
These may be additionally supplemented with growth factors (e.g., bFGF),
and/or antibiotics
such as penicillin and streptomycin. A preferred growth medium according to
the invention is
X-VIV015 medium (which is not additionally supplemented with growth factors).
Alternatively, the isolated cells may be added first to a cell proliferation
medium according to
the invention. The cell proliferation medium as defined herein comprises a
growth medium
and a LATS inhibitor according to the invention. In the cell proliferation
medium according to
the invention the growth medium component is selected from the group
consisting of DMEM
(Dulbecco's Modified Eagle's Medium) supplemented with FBS (Fetal Bovine
Serum)
(Invitrogen), human endothelial SF (serum free) medium (Invitrogen)
supplemented with
human serum, X-VIV015 medium (Lonza or DMEM/F12 (Thermo Fischer Scientific)
(optionally supplemented with calcium chloride). These may be additionally
supplemented
with growth factors (e.g., bFGF), and/or antibiotics such as penicillin and
streptomycin.
A preferred cell proliferation medium according to the invention is X-VIV015
medium
(Lonza) with a LATS inhibitor according to the invention. This cell
proliferation medium has
the advantage that it does not need additional growth factors or feeder cells
to facilitate the
proliferation of the LSCs. X-VIVO medium comprises inter alia pharmaceutical
grade human
albumin, recombinant human insulin, and pasteurized human transferrin.
Optionally
antibiotics may be added to X-VIV015 medium. In a preferred embodiment, X-
VIV015
medium is used without the addition of antibiotics.
Suitably, in a specific embodiment, a cell proliferation medium according to
the invention is
DMEM/F12 medium supplemented with serum albumin, e.g., human serum or fetal
bovone
serum or a serum substitute, and further comprising a LATS inhibitor according
to the
invention. Optionally antibiotics may be added to DMEM/F12 medium. In a
preferred
embodiment, DMEM/F12 medium is used without the addition of antibiotics.
The cell proliferation medium comprises a growth medium and a LATS inhibitor
according to
the invention. The LATS inhibitor is preferably selected from the group
comprising
compounds according to Formula Al or subformulae thereof (e.g., Formula A2)
and as
further described under the section "LATS inhibitors".
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
136
In a preferred embodiment the LATS inhibitors according to Formula Al or
subformulae
thereof (e.g., Formula A2) are added at a concentration of about 0.5 to 100
micromolar,
preferably about 0.5 to 25 micromolar, more preferably about 1 to 20
micromolar. In a
preferred embodiment the LATS inhibitors according to Formula Al or
subformulae thereof
(e.g., Formula A2) are added at a concentration of 0.5 to 100 micromolar,
preferably 0.5 to
25 micromolar, more preferably 1 to 20 micromolar. In a specific embodiment
the LATS
inhibitors according to Formula Al or subformulae thereof (e.g., Formula A2)
are added at a
concentration of about 3 to 10 micromolar. In a more specific embodiment the
LATS
inhibitors according to Formula Al or subformulae thereof (e.g., Formula A2)
are added at a
concentration of 3 to 10 micromolar.
In one embodiment, the stock solution of the compound according to Formula Al
or
subformulae thereof (e.g., Formula A2) may be prepared by dissolving the
compound
powder to a stock concentration of 10mM in DMSO. In one embodiment, the stock
solution
of the compound according to Formula Al or subformulae thereof (e.g., Formula
A2) may be
prepared by dissolving the compound powder to a stock concentration of 1 mM to
100 mM
in DMSO, e.g., 1 mM to 50 mM, 5 mM to 20 mM, 10 mM to 20mM, in particularly
10mM.
In one aspect of the invention the LATS inhibitor according to the invention
inhibits LATS1
and/or LATS2 activity in the limbal cells. In a preferred embodiment the LATS
inhibitor
inhibits LATS1 and LATS2.
In one embodiment, a cell proliferation medium of the invention optionally
further comprises
a rho-associated protein kinase (ROCK) inhibitor. The addition of a ROCK
inhibitor was
found to prevent cell death and promote attachment of cells in suspensions,
especially when
culturing stem cells. The ROCK inhibitor are known in the art and in one
example, selected
from (R)-(+)-trans-4-(1-aminoethyl)-N-(4-Pyridyl)cyclohexanecarboxamide
dihydrochloride
monohydrate ((lR,40-4-((R)-1-aminoethyl)-N-(pyridin-4-
y0cyclohexanecarboxamide; Y-
27632; Sigma-Aldrich), 5-(1,4-diazepan-l-ylsulfonyl) isoquinoline (fasudil or
HA 1077;
Cayman Chemical), H-1152, H-1152P, (S)-(+)-2-Methyl-1-[(4-methyl-5-
isoquinolinyl)sulfonyl]homopiperazine, 2HCI, ROCK Inhibitor, Dimethylfasudil
(diMF, H-
11 52P), N-(4-Pyridy1)-N'-(2,4,6-trichlorophenyOurea, Y-39983, Wf-536, SNJ-
1656, and (5)-
-0-2-methyl-I -[(4-methyl-5-isoquinolinyOsulfonyl]-hexahydro-1 H-1,4-diazepine
dihydrochloride (H-1152; Tocris Bioscience), and its derivatives and analogs.
Additional
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
137
ROCK inhibitors include imidazole-containing benzodiazepines and analogs (see,
e.g., WO
97/30992). Others include those described in International Application
Publication Nos.: WO
01/56988; WO 02/100833; WO 03/059913; WO 02/076976; WO 04/029045; WO
03/064397;
WO 04/039796; WO 05/003101; WO 02/085909; WO 03/082808; WO 03/080610; WO
04/112719; WO 03/062225; and WO 03/062227, for example. In some of these
cases,
motifs in the inhibitors include an indazole core; a 2-
aminopyridine/pyrimidine core; a 9-
deazaguanine derivative; benzamide-comprising; aminofurazan-comprising; and/or
a
combination thereof. Rock inhibitors also include negative regulators of ROCK
activation
such as small GTP-binding proteins (e.g., Gem, RhoE, and Rad), which can
attenuate
ROCK activity. In specific embodiments of the disclosure, ROCK1 is targeted
instead of
ROCK2, for example, WO 03/080610 relates to imidazopyridine derivatives as
kinase
inhibitors, such as ROCK inhibitors, and methods for inhibiting the effects of
ROCK1 and/or
ROCK2. The disclosures of the applications cited above are incorporated herein
by
reference. The Rho inhibitor can also act downstream by interaction with ROCK
(Rho-
activated kinase) leading to an inhibition of Rho. Such inhibitors are
described in U.S. Pat.
No. 6,642,263 (the disclosures of which are incorporated by reference herein
in their
entirety). Other Rho inhibitors that may be used are described in U.S. Pat.
Nos. 6,642,263,
and 6,451,825. Such inhibitors can be identified using conventional cell
screening assays,
e.g., described in U.S. Pat. No. 6,620,591 (all of which are herein
incorporated by reference
in their entirety).
In a preferred embodiment, the ROCK inhibitor used in the cell proliferation
medium of the
present invention is (R)-(+)-trans-4-(1-aminoethyl)-N-(4-
Pyridyl)cyclohexanecarboxamide
dihydrochloride monohydrate ((1R,40-44(R)-1-aminoethyl)-N-(pyridin-4-
yl)cyclohexanecarboxamide; Y-27632; Sigma-Aldrich; described in Nature 1997,
vol. 389,
pp. 990-994; JP4851003, JP11130751; JP2770497; U55478838; U56218410, all of
which
are herein incorporated by reference in their entirety).
In one embodiment, said ROCK inhibitor, in particular Y-27632, is present in a
concentration
of about 0.5 to about 100 micromolar, preferably of about 0.5 to about 25
micromolar, more
preferably of about 1 to about 20 micromolar, particularly preferably of about
10 micromolar.
In one embodiment, said compound of the present invention is present in a
concentration of
0.5 to 100 micromolar, preferably 0.5 to 25 micromolar, more preferably 1 to
20 micromolar,
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
138
particularly preferably 10 micromolar. In a specific embodiment, said ROCK
inhibitor, in
particular Y-27632, is present in a concentration of 10 micromolar.
In a specific embodiment, a cell proliferation medium of the invention
comprises DMEM/F12
(1:1), 5-20% human serum or fetal bovine serum or a serum substitute, 1-2 mM
calcium
chloride, 1 micromolar to 20 micromolar LATS inhibitor, and optionally, 1
micromolar to 20
micromolar ROCK inhibitor. In a more specific embodiment, a cell proliferation
medium of
the invention comprises DMEM/F12 (1:1), 10-20% human serum or fetal bovine
serum or a
serum substitute, e.g., 10% human serum or fetal bovine serum or a serum
substitute, 1-2
mM calcium chloride, 3 micromolar to 10 micromolar LATS inhibitor, and
optionally, 10
micromolar ROCK inhibitor.
The cells may go through a round or rounds of addition of fresh growth medium
and/or cell
proliferation medium. The cells do not need to be passaged in order for fresh
medium to be
added, but passaging cells is also a way to add fresh medium.
A series of mediums may be also used, in various combinations of orders: for
example a cell
proliferation medium, followed by addition of a growth medium (which is not
supplemented
with LATS inhibitors according to the invention, and may be different to the
growth medium
used as the base for the cell proliferation medium).
The cell population expansion phase according to the invention occurs during
the period the
cells are exposed to the cell proliferation medium.
Standard temperature conditions known in the art for culturing cells may be
used, for
example preferably about 30 C to 40 C. Particularly preferably cell growth,
as well as the
cell population expansion phase is carried out at about 37 C. A conventional
cell incubator
with 5-10% CO2 levels may be used. Preferably the cells are exposed to 5% CO2.
The cells may be passaged during the culturing in the growth or cell
proliferation medium as
necessary. Cells may be passaged when they are sub-confluent or confluent.
Preferably the
cells are passaged when they reach approximately 90%-100% confluency, although
lower
percentage confluency levels may also be performed. The passaging of cells is
done
according to standard protocols known in the art. For example, in brief cells
are passaged by
treating cultures with Accutase (e.g., for 10 minutes), rinsing the cell
suspension by
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
139
centrifugation and plating cells in fresh growth medium or cell proliferation
medium as
desired. Cell splitting ratios range, for example, from 1:2 to 1:5.
For the cell population expansion phase of the method of cell population
expansion
according to the invention, the expansion of the seeding cell population in
the cell
proliferation medium may be performed until the required amount of cellular
material is
obtained.
The cells may be exposed to the cell proliferation medium for a range of time
periods in
order to expand the cell population. For example this may include the entire
time that the
LSCs are kept in culture, or for the first week after LSC isolation or for 24
hours after
dissection of the limbus from the cornea.
In a preferred embodiment the seeding cell population is exposed to the LATS
inhibitors
according to the invention (such as those compounds according to Formula Al or
subformulae thereof (e.g., Formula A2)) directly after cell isolation from the
cornea and
maintained for the entire time that LSC proliferation is required, for example
12 to 16 days.
In one embodiment according to the invention, a gene editing technique may
optionally be
performed to genetically modify cells, to reduce or eliminate the expression
and/or function
of an immune response mediating gene which may otherwise contribute to immune
rejection
when the cell population is delivered to the patient. The application of gene
editing
techniques in the method of cell population expansion according to the
invention is optional,
and the administration to the patient of topical immunosuppressants and/or
anti-
inflammatory agents (as described further under the section Immunosuppressant
and Anti-
inflammatory agent) may instead be used if desired to mitigate issues with
immunorejection
of the transplanted material in the patient.
According to one aspect of the invention, genetically modifying comprises
reducing or
eliminating the expression and/or function of a gene associated with
facilitating a host
versus graft immune response. In a preferred embodiment, genetically modifying
comprises
introducing into a limbal stem cell a gene editing system which specifically
targets a gene
associated with facilitating a host versus graft immune response. In a
specific embodiment,
said gene editing system is CRISPR (CRISPR: clustered regularly interspaced
short
palindromic repeats, also known as CRISPR/Cas systems).
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
140
The gene editing technique may be performed at different points, such as for
example (1) on
limbal epithelial tissue, before LSC isolation or (2) at the time of cell
isolation or (3) during
the cell population expansion phase in vitro (when the cells are exposed to a
LATS inhibitor
according to the invention in vitro) or (4) in vitro at the end of the cell
population expansion
phase (after the cells are exposed to a LATS inhibitor according to the
invention in vitro). In
a one embodiment CRISPR is used after two weeks of in vitro expansion of the
cell
population in the presence of the LATS inhibitor according to the invention.
The gene editing techniques suitable for use in the method of cell population
expansion are
further described under the section "reduction of immunorejection".
In the method of cell population expansion according to the invention the LATS
inhibitors,
which are preferably compounds, produce greater than 2 fold expansion of the
seeded
population of cells.
In one aspect of the method of cell population expansion according to the
invention the
compounds according to Formula Al or subformulae thereof (e.g., Formula A2)
produce
greater than 30 fold expansion of the seeded population of limbal cells. In a
specific
embodiment of the method of cell population expansion according to the
invention, the LATS
inhibitors according to Formula Al or subformulae thereof (e.g., Formula A2)
produce 100
fold to 2200 fold expansion of the seeded population of limbal cells. In a
more specific
embodiment of the method of cell population expansion according to the
invention, the LATS
inhibitors according to Formula Al or subformulae thereof (e.g., Formula A2)
produce 600
fold to 2200 fold expansion of the seeded population of limbal cells. The fold
expansion
factor achieved by the method of cell population expansion according to the
invention may
be achieved in one or more passages of the cells. In another aspect of the
invention the fold
expansion factor achieved by the method of cell population expansion according
to the
invention may be achieved after exposure to the compound according to Formula
Al or
subformulae thereof (e.g., Formula A2) for about 12 to 16 days, preferably
about 14 days.
In one aspect of the method of cell population expansion according to the
invention, the
LATS inhibitors according to Formula Al or subformulae thereof (e.g., Formula
A2) produce
a cell population with more than 6% of p63a1pha positive cells compared to the
total amount
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
141
of cells. In a specific embodiment of the method of cell population expansion
according to
the invention, the LATS inhibitors according to Formula Al or subformulae
thereof (e.g.,
Formula A2) produce a cell population with more than 20% of p63a1pha positive
cells
compared to the total amount of cells. In another specific embodiment of the
method of cell
population expansion according to the invention, the LATS inhibitors according
to Formula
Al or subformulae thereof (e.g., Formula A2) produce a cell population with
more than 70%
of p63a1pha positive cells compared to the total amount of cells. In yet
another specific
embodiment of the method of cell population expansion according to the
invention the LATS
inhibitors according to Formula Al or subformulae thereof (e.g., Formula A2)
produce a cell
population with more than 95% of p63a1pha positive cells compared to the total
amount of
cells. The increase in the percentage of p63a1pha positive cells achieved by
the method of
cell population expansion according to the invention may be achieved in one or
more
passages of the cells. In another aspect of the invention the increase in the
percentage of
p63a1pha positive cells achieved by the method of cell population expansion
according to
the invention may be achieved after exposure to the compound according to
Formula Al or
subformulae thereof (e.g., Formula A2) for about 12 to 16 days, preferably
about 14 days.
If it is desired to measure the cell number or expansion of the cell
population, this may be
done for example by taking an aliquot and performing immunocytochemistry (e.g.
to count
nuclei stained with Sytox Orange) or by live cell imaging under brightfield
microscope to
count the number of cells or by performing real-time quantitative live-cell
analysis of cell
confluence at various time points during the cell population expansion phase
of the method
according to the invention.
The Sytox Orange assay may be performed according to standard protocols known
in the
art. In brief, after cells have attached to the cell culture dish (typically
24h after cell plating),
the cells are fixed in paraformaldehyde. The cells are then permeabilized
(e.g., using a
solution of 0.3% Triton X-100) and they are then labeled in a solution of
Sytox Orange (e.g.,
using 0.5 micromolar of Sytox Orange in PBS). The number of nuclei stained
with Sytox
Orange per surface area are then counted under a Zeiss epifluorescence
microscope.
Suitably, according to the invention the LSCs obtainable or obtained by the
method of cell
population expansion can be isolated from the other cells in the culture using
a variety of
methods known to those of skill in the art such as immunolabeling and
fluorescence sorting,
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
142
for example solid phase adsorption, fluorescence-activated cell sorting
(FACS), magnetic-
affinity cell sorting (MACS), and the like. In certain embodiments, the LSCs
are isolated
through sorting, for example immunofluorescence sorting of certain cell-
surface markers.
Two preferred methods of sorting well known to those of skill in the art are
MACS and
FACS. The LSCs markers suitable for said cell-sorting are p63a1pha, ABCB5,
ABCG2, and
CIEBP5.
Thus, in one aspect, the present invention relates to a method of preparing a
modified limbal
stem cell or a population of modified limbal stem cells for ocular cell
therapy comprising,
a) modifying a limbal stem cell or a population of limbal stem cells by
reducing or
eliminating expression of B2M comprising introducing into the limbal stem cell
or the
population of limbal stem cells a CRISPR system comprising a gRNA molecule
with a
targeting domain
(i) comprising the sequence of any one of SEQ ID NOs: 23-105 or 108-119, or
134 to
140, or
(ii) complementary to a sequence within a genomic region selected from:
chr15:44711469-44711494, chr15:44711472-44711497, chr15:44711483-44711508,
chr15:44711486-44711511, chr15:44711487-44711512, chr15:44711512-44711537,
chr15:44711513-44711538, chr15:44711534-44711559, chr15:44711568-44711593,
chr15:44711573-44711598, chr15:44711576-44711601, chr15:44711466-44711491,
chr15:44711522-44711547, chr15:44711544-44711569, chr15:44711559-44711584,
chr15:44711565-44711590, chr15:44711599-44711624, chr15:44711611-44711636,
chr15:44715412-44715437, chr15:44715440-44715465, chr15:44715473-44715498,
chr15:44715474-44715499, chr15:44715515-44715540, chr15:44715535-44715560,
chr15:44715562-44715587, chr15:44715567-44715592, chr15:44715672-44715697,
chr15:44715673-44715698, chr15:44715674-44715699, chr15:44715410-44715435,
chr15:44715411-44715436, chr15:44715419-44715444, chr15:44715430-44715455,
chr15:44715457-44715482, chr15:44715483-44715508, chr15:44715511-44715536,
chr15:44715515-44715540, chr15:44715629-44715654, chr15:44715630-44715655,
chr15:44715631-44715656, chr15:44715632-44715657, chr15:44715653-44715678,
chr15:44715657-44715682, chr15:44715666-44715691, chr15:44715685-44715710,
chr15:44715686-44715711, chr15:44716326-44716351, chr15:44716329-44716354,
chr15:44716313-44716338, chr15:44717599-44717624, chr15:44717604-44717629,
chr15:44717681-44717706, chr15:44717682-44717707, chr15:44717702-44717727,
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
143
chr15:44717764-44717789, chr15:44717776-44717801, chr15:44717786-44717811,
chr15:44717789-44717814, chr15:44717790-44717815, chr15:44717794-44717819,
chr15:44717805-44717830, chr15:44717808-44717833, chr15:44717809-44717834,
chr15:44717810-44717835, chr15:44717846-44717871, chr15:44717945-44717970,
chr15:44717946-44717971, chr15:44717947-44717972, chr15:44717948-44717973,
chr15:44717973-44717998, chr15:44717981-44718006, chr15:44718056-44718081,
chr15:44718061-44718086, chr15:44718067-44718092, chr15:44718076-44718101,
chr15:44717589-44717614, chr15:44717620-44717645, chr15:44717642-44717667,
chr15:44717771-44717796, chr15:44717800-44717825, chr15:44717859-44717884,
chr15:44717947-44717972, chr15:44718119-44718144, chr15:44711563-44711585,
chr15:44715428-44715450, chr15:44715509-44715531, chr15:44715513-44715535,
chr15:44715417-44715439, chr15:44711540-44711562, chr15:44711574-44711596,
chr15:44711597-44711619, chr15:44715446-44715468, chr15:44715651-44715673,
chr15:44713812-44713834, chr15:44711579-44711601, chr15:44711542-44711564,
chr15:44711557-44711579, chr15:44711609-44711631, chr15:44715678-44715700,
chr15:44715683-44715705, chr15:44715684-44715706, chr15:44715480-44715502,
wherein the limbal stem cell or the population of limbal stem cells have
optionally been
cultured in the presence of a LATS inhibitor; and
b) further expanding the modified limbal stem cell or the population of
limbal stem cells
in cell culture media comprising a LATS inhibitor, and, optionally, ROCK
inhibitor; and
c) optionally, enriching the population of limbal stem cells with the
undifferentiated
limbal stem cells having expression of LSCs bionarkers, such as p63a1pha,
ABCB5, ABCG2,
and C/EBP5, by fluorescene activated cell sorting (FACS) or magnetic activated
cell sorting
(MACS), and
d) optionally, enriching the population of limbal stem cells with the limbal
stem cells having
reduced or eliminated expression of B2M by fluorescene activated cell sorting
(FACS) or
magnetic activated cell sorting (MACS).
In one aspect, the present invention relates to a cell population comprising
the modified LSC
of the present invention or the modified LSC obtained by the method of the
present
invention.
In one embodiment, the cell population of the present invention comprises the
modified
limbal stem cell of the present invention or the modified limbal stem cell
obtained by the
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
144
method of the present invention, wherein the modified limbal stem cell
comprises an indel
formed at or near the target sequence complementary to the targeting domain of
the qRNA
molecule domain. In one embodiment, the indel comprises a deletion of 10 or
greater than
nucleotides, optionally 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27,
5 28, 29, 30, 31,32, 33, 34, or 35 nucleotides. In a further embodiment,
the indel is formed in
at least about 40%, e.g., at least about 50%, e.g., at least about 60%, e.g.,
at least about
70%, e.g., at least about 80%, e.g., at least about 90%, e.g., at least about
95%, e.g., at
least about 96%, e.g., at least about 97%, e.g., at least about 98%, e.g., at
least about 99%,
of the cells of the cell population, e.g., as detectible by next generation
sequencing and/or a
10 nucleotide insertional assay.
In one embodiment, the cell population of the present invention comprises the
modified
limbal stem cell of the present invention or the modified limbal stem cell
obtained by the
method of the present invention, wherein the modified limbal stem cell
comprises an indel
formed at or near the target sequence complementary to the targeting domain of
the gRNA
molecule domain, and wherein an off-target indel is detected in no more than
about 5%,
e.g., no more than about 1%, e.g., no more than about 0.1%, e.g., no more than
about
0.01%, of the cells of the cell population, e.g., as detectible by next
generation sequencing
and/or a nucleotide insertional assay.
In one aspect according to the invention the LSC population obtainable or
obtained by the
method of cell population expansion according to the invention preferably
shows at least one
of the following characteristics. More preferably, it shows two or more, more
preferably all, of
the following characteristics.
(1) The cell preparation is positive for p63a1pha cells. The expression of
p63a1pha may be
estimated by standard techniques known in the art, such as for example
immunohistochemistry and quantitative RT-PCR.
(2) The cell preparation comprises more than 6% p63a1pha positive cells.
Preferably the cell
preparation comprises more than 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80% or 90%
p63a1pha positive cells. In a preferred embodiment the cell preparation
comprises more than
95% p63a1pha positive cells. The percentage of p63a1pha cells may be measured
by
immunohistochemistry or FACS.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
145
(3) The cells express one or more of ABCB5, ABCG2, and C/EBP5. The expression
of
ABCB5, ABCG2, and C/EBP5 may be estimated by standard techniques known in the
art,
such as for example immunohistochemistry and quantitative RT-PCR.
(4) The cells can differentiate into corneal epithelium cells as observed by
keratin-12
expression. These characteristics can be observed by immunohistochemistry or
FACS.
(5) The cell preparation comprises more than 50% B2M and /or HLA-ABC negative
cells.
Preferably the cell preparation comprises more than 50%, 55%, 60%, 65%, 70%,
75%, 80%,
85%, 90%, 95% or 99% B2M and /or HLA-ABC negative cells. In a preferred
embodiment
the cell preparation comprises more than 95% B2M and /or HLA-ABC negative
cells. The
percentage of B2M and /or HLA-ABC negative cells may be measured by
immunohistochemistry or FACS or MACS.
In a preferred embodiment, the cell preparation comprises more than 95%
p63a1pha positive
cells and more than 95% B2M and /or HLA-ABC negative cells.
The cell population expanded by the method of cell population expansion
according to the
invention may be added to a solution and then stored, for example in a
preservation or
cryopreservation solution (such as those described below), or added directly
to a
composition suitable for ocular delivery. The preservation, cryopreservation
solution or
composition suitable for ocular delivery may optionally comprise a LATS
inhibitor according
to the invention.
In a more preferred embodiment according to the invention, the cell population
preparation
which is delivered to the eye comprises very low (e.g., low trace level) to
neglible levels of a
LATS inhibitor compound. Thus in a specific embodiment, the method of cell
population
expansion according to the invention comprises the further step of rinsing to
substantially
remove the compound of the present invention (such as the compound according
to
Formula Al or subformulae thereof). This may involve rinsing the cells after
the cell
population expansion phase according to the invention. To rinse the cells, the
cells are
detached from the culture dish (e.g. by treating with Accutase), the detached
cells are then
centrifuged, and a cell suspension is made in PBS or growth medium according
to the
invention. This step may be performed multiple times, e.g., one to ten times,
to rinse out the
cells. Finally the cells may be resuspended in a preservation solution,
cryopreservation
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
146
solution, a composition suitable for ocular delivery, growth medium or
combinations thereof
as desired.
The expanded population of cells prepared by the method of cell population
expansion and
.. rinsed of cell proliferation medium comprising a LATS inhibitor according
the invention may
be transferred to a composition suitable for ocular delivery, such as for
example a localising
agent. Optionally the cell population is stored for a period before addition
to a localising
agent suitable for ocular delivery. In a preferred embodiment, the expanded
cell population
may first be added to a solution suitable for preservation or
cryopreservation, which
preferably does not comprise a LATS inhibitor, and the cell population stored
(optionally with
freezing) before addition to a localising agent suitable for ocular delivery,
which also
preferably does not comprise a LATS inhibitor.
Typical solutions suitable for preservation of LSCs are Optisol or PBS or
CryoStor C55
buffer (BioLife Solutions), preferably Optisol. Optisol is a corneal storage
medium
comprising chondroitin sulfate and dextran to enhance corneal dehydration
during storage
(see for example Kaufman et al., (1991) Optisol corneal storage medium; Arch
Ophthalmol
Jun; 109(6): 864-8). For cryopreservation, glycerol, dimethyl sulfoxide,
propylene glycol or
acetamide may be used in the cryopreservation solution of the present
invention. The
cryopreserved preparation of cells is typically kept at -20 C or -80 C. In
one embodiment,
the cryopreserved compositions comprise a cell (e.g., a modified cell, such as
LSC or CEC,
with reduced or eliminated expression of B2M by a CRISPR system), e.g., a
plurality of
cells) and a cryoprotectant selected from the list of glycerol, DMSO
(dimethylsulfoxide)
polyvinylpyrrolidone, hydroxyethyl starch, propylene glycol, acetamide,
monosaccharides,
algae-derived polysaccharides, and sugar alcohols, or a combination thereof.
In a more
specific embodiment, the cryopreserved compositions comprise a cell (e.g., a
modified cell,
such as LSC or CEC, with reduced or eliminated expression of B2M by a CRISPR
system),
e.g., a plurality of cells) and DMSO concentration of 0.5% to 10%, e.g., 1%-
10%, 2%-7%,
3%-6%, 4% - 5%, preferably 5%. DMSO acts as a cryoprotecting agent against
formation of
water crystals within and outside the cells, which could lead to cell damage
during
cryopreservation steps. In a further embodiment, the cryopreserved
compositions further
comprise a suitable buffer, for example CryoStor C55 buffer (BioLife
Solutions).
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
147
In one aspect the invention relates to a preserved or cryopreserved
preparation of limbal
stem cells obtainable by the method of cell population expansion according to
the invention.
In an alternative aspect the invention relates to a fresh cell preparation
where limbal stem
cells obtainable by the method of cell population expansion according to the
invention are in
suspension in PBS and/or growth medium or combined with a localising agent.
The fresh
cell preparation is typically kept at about 15 to 37 C. Standard cell
cultures containers
known in the art may be used to store the cells, such as a vial or a flask.
In a preferred embodiment according to the invention, before use in the eye, a
cryopreserved preparation of cells is thawed (for example by incubating at a
temperature of
about 37 C in an incubator or waterbath). Preferably 10 volumes of PBS or
growth medium
may be added to rinse off the cells from the cryopreservant solution. Cells
may then be
rinsed by centrifugation, and a cell suspension may be made in PBS and/or
growth medium,
before combination with a localising agent for ocular delivery, which also
preferably does not
comprise a LATS inhibitor.
In one aspect of the invention the expanded population of cells prepared by
the method of
cell population expansion, are prepared as a suspension (for example in PBS
and/or growth
medium, such as for example X-VIVO medium or DMEM/F12) and combined with a
localising agent suitable for ocular delivery, (such as a biomatrix like GelMA
or fibrin glue). In
a specific embodiment of the method of treatment according to the invention,
this
combination of cells, PBS and/or growth medium, and biomatrix is delivered to
the eye via a
carrier (such as a contact lens). In yet another specific embodiment this
combination of cells,
PBS and/or growth medium, and biomatrix comprises at most only trace levels of
a LATS
inhibitor.
The term "trace levels" as used herein means less than 5% w/v (e.g., no more
than 5% w/v,
4% w/v, 3% w/v, 2% w/v, or 1% w/v), and preferably less than 0.01% w/v (e.g.,
no more than
0.01% w/v, 0.009% w/v, 0.008% w/v, 0.007% w/v, 0.006% w/v, 0.005% w/v, 0.004%
w/v,
0.003% w/v, 0.002% w/v, or 0.001% w/v), which can be measured, for example
using high-
resolution chromatography as described in the Examples herein. In certain
embodiments,
trace levels of a LATS inhibitor compound of the invention are the levels of
residual
compounds present after one or more wash steps, which collectively are below
the cellular
potency of such compounds, and accordingly they do not induce biological
effect in vivo.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
148
Accordingly, residual levels of compounds are below the amount expected to
have a
biological effect on cell population expansion in cell culture or in a subject
(e.g., after
transplantation of an expanded cell population to the subject). Trace levels
can be
measured, for example, as the wash-off efficiency, which can be calculated as
follows:
Wash-off efficiency = 100 - (average concentration in post-wash pellet x
pellet volume x
molecule weight) / (compound concentration x culture media volume x molecule
weight). As
used herein, "rinsing to substantially remove" a LATS inhibitor compound of
the invention
from cells refers to steps for establishing trace levels of the LATS inhibitor
compound.
Alternatively, the cells may be cultured and the cell population proliferation
phase may occur
in cell proliferation medium on a localising agent suitable for cell delivery
to the ocular
surface (for example fibrin, collagen).
In one aspect, the present invention relates to a composition comprising the
modified limbal
stem cell of the present invention or the modified limbal stem cell obtained
by the method of
the present invention or the cell population of the present invention or the
population of
modified limbal stem cells obtained by the method of the present invention.
Suitably, the
modified limbal stem cell of the composition comprises an indel formed at or
near the target
sequence complementary to the targeting domain of the gRNA molecule domain.
Suitably,
the indel comprises a deletion of 10 or greater than 10 nucleotides,
optionally 11, 12, 13, 14,
15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31,32, 33, 34,
or 35
nucleotides. Suitably, the indel is formed in at least about 40%, e.g., at
least about 50%,
e.g., at least about 60%, e.g., at least about 70%, e.g., at least about 80%,
e.g., at least
about 90%, e.g., at least about 95%, e.g., at least about 96%, e.g., at least
about 97%, e.g.,
at least about 98%, e.g., at least about 99%, of the cells of the population.
In one
embodiment, an off-target indel is detected in no more than about 5%, e.g., no
more than
about 1%, e.g., no more than about 0.1%, e.g., no more than about 0.01%, of
the cells of the
population of cells e.g., as detectible by next generation sequencing and/or a
nucleotide
insertional assay.
Cell population expansion: To prepare an expanded population of corneal
endothelial
cells
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
149
In a preferred embodiment of the invention, corneal endothelial cells, for
example isolated
and obtainable as described in the section "Starting material to prepare an
expanded
population of corneal endothelial cells", can be grown in medium in a culture
container
known in the art, such as plates, multi-well plates, and cell culture flasks.
For example, a
culture dish may be used which is non-coated or coated with collagen,
synthemax, gelatin or
fibronectin. A preferred example of a suitable culture container is a non-
coated plate.
Standard culturing containers and equipment such as bioreactors known in the
art for
industrial use may also be used.
The medium used may be a growth medium or a cell proliferation medium. A
growth
medium is defined herein as a culture medium supporting the growth and
maintenance of a
population of cells. Suitable growth mediums are known in the art for corneal
endothelial cell
culture are for example: DMEM (Dulbecco's Modified Eagle's Medium)
supplemented with
FBS (Fetal Bovine Serum) (Invitrogen), human endothelial SF (serum free)
medium
(Invitrogen) supplemented with human serum, X-VIV015 medium (Lonza) or
mesenchymal
stem cell-conditioned medium. These may be additionally supplemented with
growth factors
(e.g., bFGF), and/or antibiotics such as penicillin and streptomycin. A
preferred growth
medium according to the invention is X-VIV015 medium (which is not
additionally
supplemented with growth factors).
Alternatively, the isolated cells may be added first to a cell proliferation
medium according to
the invention. The cell proliferation medium as defined herein comprises a
growth medium
and a LATS inhibitor according to the invention. In the cell proliferation
medium according to
the invention the growth medium component is selected from the group
consisting of DMEM
(Dulbecco's Modified Eagle's Medium) supplemented with FBS (Fetal Bovine
Serum)
(Invitrogen), human endothelial SF (serum free) medium (Invitrogen)
supplemented with
human serum, X-VIV015 medium (Lonza) or mesenchymal stem cell-conditioned
medium.
These may be additionally supplemented with growth factors (e.g., bFGF),
and/or antibiotics
such as penicillin and streptomycin.
A preferred cell proliferation medium according to the invention is X-VIV015
medium
(Lonza) with a LATS inhibitor according to the invention. This cell
proliferation medium has
the advantage that it does not need additional growth factors or feeder cells
to facilitate the
proliferation of the CECs. X-VIVO medium comprises inter alia pharmaceutical
grade human
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
150
albumin, recombinant human insulin, and pasteurised human transferrin.
Optionally
antibiotics may be added to X-VIV015 medium. In a preferred embodiment, X-
VIV015
medium is used without the addition of antibiotics.
The cell proliferation medium comprises a growth medium and a LATS inhibitor
according to
the invention. The LATS inhibitor is preferably selected from the group
comprising
compounds according to Formula Al or subformulae thereof (e.g., Formula A2)
and as
further described under the section "LATS Inhibitors".
In a preferred embodiment the LATS inhibitors according to Formula Al or
subformulae
thereof (e.g., Formula A2) are added at a concentration of about 0.5 to 100
micromolar,
preferably about 0.5 to 25 micromolar, more preferably about 1 to 20
micromolar. In a
preferred embodiment the LATS inhibitors according to Formula Al or
subformulae thereof
(e.g., Formula A2) are added at a concentration of 0.5 to 100 micromolar,
preferably 0.5 to
.. 25 micromolar, more preferably 1 to 20 micromolar. In a specific embodiment
the LATS
inhibitors according to Formula Al or subformulae thereof (e.g., Formula A2)
are added at a
concentration of about 3 to 10 micromolar. In a more specific embodiment the
LATS
inhibitors according to Formula Al or subformulae thereof (e.g., Formula A2)
are added at a
concentration of 3 to 10 micromolar.
In one embodiment, the stock solution of the compound according to Formula Al
or
subformulae thereof (e.g., Formula A2) may be prepared by dissolving the
compound
powder to a stock concentration of 10mM in DMSO. In one embodiment, the stock
solution
of the compound according to Formula Al or subformulae thereof (e.g., Formula
A2) may be
prepared by dissolving the compound powder to a stock concentration of 1 mM to
100 mM
in DMSO, e.g., 1 mM to 50 mM, 5 mM to 20 mM, 10 mM to 20mM, in particularly
10mM.
In one aspect of the invention the LATS inhibitor according to the invention
inhibits LATS1
and/or LATS2 activity in the corneal endothelial cells. In a preferred
embodiment the LATS
inhibitor inhibits LATS1 and LATS2.
In one embodiment, a cell proliferation medium of the invention optionally
further comprises
a rho-associated protein kinase (ROCK) inhibitor. The addition of a ROCK
inhibitor was
found to prevent cell death and promote attachment of cells in suspensions,
especially when
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
151
culturing stem cells. In a preferred embodiment, the ROCK inhibitor used in
the cell
proliferation medium of the present invention is (R)-(+)-trans-4-(1-
aminoethyl)-N-(4-
Pyridyl)cyclohexanecarboxamide dihydrochloride monohydrate ((1R,40-44(R)-1-
aminoethyl)-N-(pyridin-4-yhcyclohexanecarboxamide; Y-27632; Sigma-Aldrich;
described in
Nature 1997, vol. 389, pp. 990-994; JP4851003, JP11130751; JP2770497;
U55478838;
U56218410, all of which are herein incorporated by reference in their
entirety).
In one embodiment, said ROCK inhibitor, in particular Y-27632, is present in a
concentration
of about 0.5 to about 100 micromolar, preferably of about 0.5 to about 25
micromolar, more
preferably of about 1 to about 20 micromolar, particularly preferably of about
10 micromolar.
In one embodiment, said compound of the present invention is present in a
concentration of
0.5 to 100 micromolar, preferably 0.5 to 25 micromolar, more preferably 1 to
20 micromolar,
particularly preferably 10 micromolar. In a specific embodiment, said ROCK
inhibitor, in
particular Y-27632, is present in a concentration of 10 micromolar.
In a specific embodiment, a cell proliferation medium of the invention
comprises DMEM/F12
(1:1), 5-20% human serum or fetal bovine serum or a serum substitute, 1-2 mM
calcium
chloride, 1 micromolar to 20 micromolar LATS inhibitor, and optionally, 1
micromolar to 20
micromolar ROCK inhibitor. In a more specific embodiment, a cell proliferation
medium of
.. the invention comprises DMEM/F12 (1:1), 10-20% human serum or fetal bovine
serum or a
serum substitute, e.g., 10% human serum or fetal bovine serum or a serum
substitute, 1-2
mM calcium chloride, 3 micromolar to 10 micromolar LATS inhibitor, and
optionally, 10
micromolar ROCK inhibitor.
The cells may go through a round or rounds of addition of fresh growth medium
and/or cell
proliferation medium. The cells do not need to be passaged in order for fresh
medium to be
added, but passaging cells is also a way to add fresh medium.
A series of mediums may be also used, in various combinations of orders: for
example a cell
proliferation medium, followed by addition of a growth medium (which is not
supplemented
with LATS inhibitors according to the invention, and may be different to the
growth medium
used as the base for the cell proliferation medium).
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
152
The cell population expansion phase according to the invention occurs during
the period the
cells are exposed to the cell proliferation medium.
Standard temperature conditions known in the art for culturing cells may be
used, for
example preferably about 30'C to 40C. Particularly preferably cell growth, as
well as the
cell population expansion phase is carried out at about 37C. A conventional
cell incubator
with 5-10% CO2 levels may be used. Preferably the cells are exposed to 5% CO2.
The cells may be passaged during the culturing in the growth or cell
proliferation medium as
necessary. Cells may be passaged when they are sub-confluent or confluent.
Preferably the
cells are passaged when they reach approximately 90%-100% confluency, although
lower
percentage confluency levels may also be performed. The passaging of cells is
done
according to standard protocols known in the art. For example, in brief the
cells are
detached from the culture container, for example using collagenase. The cells
are then
centrifuged and rinsed in PBS or the cell growth medium according to the
invention and
plated in fresh growth or cell proliferation medium as desired at a dilution
of, for example,
1:2 to 1:4.
For the cell population expansion phase of the method of cell population
expansion
according to the invention, the expansion of the seeding cell population in
the cell
proliferation medium may be performed until the required amount of cellular
material is
obtained.
The cells may be exposed to the cell proliferation medium for a range of time
periods in
order to expand the cell population. For example, this may include the entire
time that the
CECs are kept in culture, or only for the first one to two weeks after CEC
isolation or only for
24 hours after dissection of the cornea.
In a preferred embodiment, the corneal endothelial cells are exposed to the
LATS inhibitors
according to the invention (such as those compounds according to Formula Al or
subformulae thereof (e.g., Formula A2)) directly after cell isolation from the
cornea, and
maintained for the entire time that CEC proliferation is required, for example
one to two
weeks.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
153
In a more preferred embodiment of the invention, after the cell population
expansion phase
in vitro (i.e., after the cells are exposed to a LATS inhibitor according to
the invention fora
period of time to expand the population of cells), the method of cell
population expansion
according to the invention comprises a further step wherein the cells may be
grown for a
period of time (e.g., two weeks) in growth medium without supplementation of a
LATS
inhibitor, to enable a mature corneal endothelium to form. A mature corneal
endothelium is
defined herein as a monolayer of CECs with hexagonal morphology, ZO-1-positive
tight
junctions and expression of Na/K ATPase. In a preferred embodiment the cells
are not
passaged while the mature corneal endothelium is formed.
In one embodiment according to the invention, a gene editing technique may
optionally be
performed to genetically modify cells, to reduce or eliminate the expression
and/or function
of an immune response mediating gene which may otherwise contribute to immune
rejection
when the cell population is delivered to the patient. The application of gene
editing
techniques in the method of cell population expansion according to the
invention is optional,
and the administration to the patient of topical immunosuppressants and/or
anti-
inflammatory agents (as described further under the section Immunosuppressant
and Anti-
inflammatory agent) may instead be used if desired to mitigate issues with
immunorejection
of the transplanted material in the patient.
According to one aspect of the invention, for the scenario that a gene editing
technique is
used, genetically modifying comprises reducing or eliminating the expression
and/or function
of a gene associated with facilitating a host versus graft immune response. In
a preferred
embodiment, genetically modifying comprises introducing into a corneal
endothelial cell a
gene editing system which specifically targets a gene associated with
facilitating a host
versus graft immune response. In a specific embodiment, said gene editing
system is
CRISPR (CRISPR: clustered regularly interspaced short palindromic repeats,
also known as
CRISPR/Cas systems).
A gene editing technique, if it is to be used, may be performed at different
points, such as for
example (1) on corneal tissue, before CEC isolation or (2) at the time of cell
isolation or (3)
during the cell population expansion phase in vitro (when the cells are
exposed to a LATS
inhibitor according to the invention in vitro) or (4) in vitro at the end of
the cell population
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
154
expansion phase (after the cells are exposed to a LATS inhibitor according to
the invention
in vitro).
The gene editing techniques suitable for use in the method of cell population
expansion are
further described under the section "reduction of immunorejection".
In the method of cell population expansion according to the invention the LATS
inhibitors,
which are preferably compounds, produce greater than 2 fold expansion of the
seeded
population of cells.
In one aspect of the method of cell population expansion according to the
invention the
compounds according to Formula Al or subformulae thereof (e.g., Formula A2)
produce
greater than 10 fold expansion of the seeded population of corneal endothelial
cells. In a
specific embodiment of the method of cell population expansion according to
the invention,
the LATS inhibitors according to Formula Al or subformulae thereof (e.g.,
Formula A2)
produce 15 fold to 600 fold expansion of the seeded population of corneal
endothelial cells.
In a more specific embodiment of the method of cell population expansion
according to the
invention, the LATS inhibitors according to Formula Al or subformulae thereof
(e.g.,
Formula A2) produce 20 fold to 550 fold expansion of the seeded population of
corneal
endothelial cells. The fold expansion factor achieved by the method of cell
population
expansion according to the invention may be achieved in one or more passages
of the cells.
In another aspect of the invention the fold expansion factor achieved by the
method of cell
population expansion according to the invention may be achieved after exposure
to the
compound according to Formula Al or subformulae thereof (e.g., Formula A2) for
one to two
weeks, preferably after about 10 days.
If it is desired to measure the cell number or expansion of the cell
population, this may be
done for example by taking an aliquot and performing immunocytochemistry
(e.g., to count
nuclei stained with Sytox Orange) or by live cell imaging under brightfield
microscope to
count the number of cells or by performing real-time quantitative live-cell
analysis of cell
confluence at various time points during the cell population expansion phase
of the method
according to the invention.
Suitably, according to the invention the CECs obtainable or obtained by the
method of cell
population expansion can be isolated from the other cells in the culture using
a variety of
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
155
methods known to those of skill in the art such as immunolabeling and
fluorescence sorting,
for example solid phase adsorption, fluorescence-activated cell sorting
(FACS), magnetic-
affinity cell sorting (MACS), and the like. In certain embodiments, the CECs
are isolated
through sorting, for example immunofluorescence sorting of certain cell-
surface markers.
Two preferred methods of sorting well known to those of skill in the art are
MACS and
FACS. The CECs markers suitable for said cell-sorting are Na/K ATPase, 8a2,
AQP1 and
SLC4A11.
Thus, in one aspect, the present invention relates to a method of preparing a
modified CEC
or a population of modified CECs for ocular cell therapy comprising,
a) modifying a CEC or a population of CECs by reducing or eliminating
expression of
B2M comprising introducing into the CE or the population of CECs a CRISPR
system
comprising a gRNA molecule with a targeting domain
(i) comprising the sequence of any one of SEQ ID NOs: 23-105 or 108-119, or
134 to
140, or
(ii) complementary to a sequence within a genomic region selected from:
chr15:44711469-44711494, chr15:44711472-44711497, chr15:44711483-44711508,
chr15:44711486-44711511, chr15:44711487-44711512, chr15:44711512-44711537,
chr15:44711513-44711538, chr15:44711534-44711559, chr15:44711568-44711593,
chr15:44711573-44711598, chr15:44711576-44711601, chr15:44711466-44711491,
chr15:44711522-44711547, chr15:44711544-44711569, chr15:44711559-44711584,
chr15:44711565-44711590, chr15:44711599-44711624, chr15:44711611-44711636,
chr15:44715412-44715437, chr15:44715440-44715465, chr15:44715473-44715498,
chr15:44715474-44715499, chr15:44715515-44715540, chr15:44715535-44715560,
chr15:44715562-44715587, chr15:44715567-44715592, chr15:44715672-44715697,
chr15:44715673-44715698, chr15:44715674-44715699, chr15:44715410-44715435,
chr15:44715411-44715436, chr15:44715419-44715444, chr15:44715430-44715455,
chr15:44715457-44715482, chr15:44715483-44715508, chr15:44715511-44715536,
chr15:44715515-44715540, chr15:44715629-44715654, chr15:44715630-44715655,
chr15:44715631-44715656, chr15:44715632-44715657, chr15:44715653-44715678,
chr15:44715657-44715682, chr15:44715666-44715691, chr15:44715685-44715710,
chr15:44715686-44715711, chr15:44716326-44716351, chr15:44716329-44716354,
chr15:44716313-44716338, chr15:44717599-44717624, chr15:44717604-44717629,
chr15:44717681-44717706, chr15:44717682-44717707, chr15:44717702-44717727,
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
156
chr15:44717764-44717789, chr15:44717776-44717801, chr15:44717786-44717811,
chr15:44717789-44717814, chr15:44717790-44717815, chr15:44717794-44717819,
chr15:44717805-44717830, chr15:44717808-44717833, chr15:44717809-44717834,
chr15:44717810-44717835, chr15:44717846-44717871, chr15:44717945-44717970,
chr15:44717946-44717971, chr15:44717947-44717972, chr15:44717948-44717973,
chr15:44717973-44717998, chr15:44717981-44718006, chr15:44718056-44718081,
chr15:44718061-44718086, chr15:44718067-44718092, chr15:44718076-44718101,
chr15:44717589-44717614, chr15:44717620-44717645, chr15:44717642-44717667,
chr15:44717771-44717796, chr15:44717800-44717825, chr15:44717859-44717884,
chr15:44717947-44717972, chr15:44718119-44718144, chr15:44711563-44711585,
chr15:44715428-44715450, chr15:44715509-44715531, chr15:44715513-44715535,
chr15:44715417-44715439, chr15:44711540-44711562, chr15:44711574-44711596,
chr15:44711597-44711619, chr15:44715446-44715468, chr15:44715651-44715673,
chr15:44713812-44713834, chr15:44711579-44711601, chr15:44711542-44711564,
chr15:44711557-44711579, chr15:44711609-44711631, chr15:44715678-44715700,
chr15:44715683-44715705, chr15:44715684-44715706, chr15:44715480-44715502,
wherein the CEC or the population of CECs have optionally been cultured in the
presence of
a LATS inhibitor; and
b) further expanding the modified CEC or the population of CECs in cell
culture media
comprising a LATS inhibitor, and, optionally, ROCK inhibitor; and
c) optionally, enriching the population of CECs with the undiffirentiated
CECs having
expression of CECs biomarkers, such as Na/K ATPase, 8a2, AQP1 and SLC4A11, by
fluorescene activated cell sorting (FACS) or magnetic activated cell sorting
(MACS), and
d) optionally, enriching the population of CECs with the CECs having reduced
or eliminated
expression of B2M by fluorescene activated cell sorting (FACS) or magnetic
activated cell
sorting (MACS).
In one aspect, the present invention relates to a cell population comprising
the modified
CEC of the present invention or the modified CEC obtained by the method of the
present
invention.
In one embodiment, the cell population of the present invention comprises the
modified CEC
of the present invention or the modified CEC obtained by the method of the
present
invention, wherein the modified CEC comprises an indel formed at or near the
target
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
157
sequence complementary to the targeting domain of the qRNA molecule domain. In
one
embodiment, the indel comprises a deletion of 10 or greater than 10
nucleotides, optionally
11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29,
30, 31,32, 33, 34, or
35 nucleotides. In a further embodiment, the indel is formed in at least about
40%, e.g., at
least about 50%, e.g., at least about 60%, e.g., at least about 70%, e.g., at
least about 80%,
e.g., at least about 90%, e.g., at least about 95%, e.g., at least about 96%,
e.g., at least
about 97%, e.g., at least about 98%, e.g., at least about 99%, of the cells of
the cell
population, e.g., as detectible by next generation sequencing and/or a
nucleotide insertional
assay.
In one embodiment, the cell population of the present invention comprises the
modified CEC
of the present invention or the modified CEC obtained by the method of the
present
invention, wherein the modified CEC comprises an indel formed at or near the
target
sequence complementary to the targeting domain of the gRNA molecule domain,
and
wherein an off-target indel is detected in no more than about 5%, e.g., no
more than about
1%, e.g., no more than about 0.1%, e.g., no more than about 0.01%, of the
cells of the cell
population, e.g., as detectible by next generation sequencing and/or a
nucleotide insertional
assay.
In one aspect according to the invention the CEC population obtainable or
obtained by the
method of cell population expansion according to the invention preferably
shows at least one
of the following characteristics. More preferably, it shows two or more,
particularly preferably
all, of the following characteristics.
(1) The cells express Na/K ATPase. The expression of Na/K ATPase may be
estimated by
standard techniques known in the art, such as for example
immunohistochemistry,
quantitative RT-PCR or by FACS analysis.
(2) The cells express one or more of Collagen 8a2, AQP1 (aquaporin 1) and
SLC4A11
(Soiute Camer Fundy 4 Member 11). Preferably the relative expression levels
are higher
than cells which do not typically express collagen 8a2, AQP1 and SLC4A11, such
as, for
example, in dermal fibroblasts. The expression of Collagen 8a2, AQP1 or
SLC4A11 may be
estimated by standard techniques known in the art, such as for example
immunohistochemistry, quantitative RT-PCR or by FACS analysis.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
158
(3) The cells do not express (or at most express relatively low levels of)
RPE65 (a marker of
retinal pigmented epithelium) and/or CD31 (a marker of vascular endothelium).
The relative
expression levels are similar to cells which do not typically express RPE65,
CD31, such as
in dermal fibroblasts. The expression of RPE65 and CD31 may be estimated by
standard
techniques known in the art, such as for example quantitative RT-PCR,
immunohistochemistry or FACS analysis.
(4) The cells express relatively low levels of CD73. The relative expression
levels are lower
than cells which have undergone endothelial to mesenchymal transition. The
expression of
CD73 may be estimated by standard techniques known in the art, such as for
example
FACS analysis or immunohistochemistry.
(5) The cell preparation comprises more than 50% B2M and /or HLA-ABC negative
cells.
Preferably the cell preparation comprises more than 50%, 55%, 60%, 65%, 70%,
75%, 80%,
85%, 90%, 95% or 99% B2M and /or HLA-ABC negative cells. In a preferred
embodiment
the cell preparation comprises more than 95% B2M and /or HLA-ABC negative
cells. The
percentage of B2M and /or HLA-ABC negative cells may be measured by
immunohistochemistry or FACS or MACS.
In a preferred embodiment, the cell preparation comprises more than 95% Na/K
ATPase,
8a2, AQP1 or SLC4A11 positive cells and more than 95% B2M and /or HLA-ABC
negative
cells.
In another aspect according to the invention, when in a layer, for example
when cultured on
a plate, the CEC population obtainable by the method of cell population
expansion
according to the invention preferably shows at least one of the following
characteristics.
More preferably, it shows two or more, particularly preferably all, of the
following
characteristics:
(1) The cells are able to form a single layer structure. This is one of the
characteristics of the
corneal endothelial cell layer in the body. This may be observed by nuclear
staining (e.g.,
with nuclear dye such as Sytox, Hoechst) followed by examination by
microscopy.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
159
(2) The cells are able to form tight junctions. This may be checked by a
standard technique
known in the art, immunofluorescence staining of tight-junction marker Zonula
Occludens-1
(ZO-1).
(3) The cells are able to be regularly arranged in the cell layer. This may be
checked by a
standard technique known in the art, immunofluorescence staining of tight-
junction marker
Zonula Occludens-1 (ZO-1). In the healthy corneal endothelial cell layer in
the body, the
cells constituting the layer are regularly arrayed, due to which corneal
endothelial cells are
considered to maintain normal function and high transparency and the cornea is
considered
to appropriately exhibit water control function.
The cell population expanded by the method of cell population expansion
according to the
invention may be added to a solution and then stored, for example in a
preservation or
cryopreservation solution (such as those described below), or added directly
to a
composition suitable for ocular delivery. The preservation, cryopreservation
solution or
composition suitable for ocular delivery may optionally comprise a LATS
inhibitor according
to the invention.
In a more preferred embodiment according to the invention, the cell population
preparation
which is delivered to the eye comprises very low to negligible levels of a
LATS inhibitor
compound. Thus in a specific embodiment, the method of cell population
expansion
according to the invention comprises the further step of rinsing to
substantially remove the
compound of the present invention (such as the compound according to Formula
Al or
subformulae thereof (e.g., Formula A2)). This may involve rinsing the cells
after the cell
population expansion phase according to the invention (directly after the cell
population
expansion phase and/or after the cells have been cultured to form a mature
corneal
endothelium in growth medium which has not been supplemented by a LATS
inhibitor). To
rinse the cells, the cells are centrifuged, and a cell suspension is made in
PBS or growth
medium according to the invention. This step may be performed multiple times,
e.g. one to
ten times, to rinse out the cells. Finally the cells may be resuspended in a
preservation
solution, cryopreservation solution, a composition suitable for ocular
delivery, growth
medium or combinations thereof as desired.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
160
The expanded population of cells prepared by the method of cell population
expansion and
rinsed of cell proliferation medium comprising a LATS inhibitor according the
invention may
be transferred to a composition suitable for ocular delivery, such as for
example a localising
agent. Optionally the cell population is stored for a period before addition
to a localising
agent suitable for ocular delivery. In a preferred embodiment, the expanded
cell population
may first be added to a solution suitable for preservation or
cryopreservation, which
preferably does not comprise a LATS inhibitor, and the cell population stored
(optionally with
freezing) before addition to a localising agent suitable for ocular delivery,
which also
preferably does not comprise a LATS inhibitor.
Typical solutions for suitable for preservation of CECs are Optisol or PBS,
preferably
Optisol. Optisol is a corneal storage medium comprising chondroitin sulfate
and dextran to
enhance corneal dehydration during storage (see for example Kaufman et al.,
(1991) Optisol
corneal storage medium; Arch Ophthalmol Jun; 109(6): 864-8). For
cryopreservation,
glycerol, dimethyl sulfoxide, propylene glycol or acetamide may be used in the
cryopreservation solution of the present invention. The cryopreserved
preparation of cells is
typically kept at -20 C or -80 C.
In one aspect the invention relates to a preserved or cryopreserved
preparation of corneal
endothelial cells obtainable by the method of cell population expansion
according to the
invention. In an alternative aspect the invention relates to a fresh cell
preparation where
corneal endothelial cells obtainable by the method of cell population
expansion according to
the invention are in suspension in PBS and/or growth medium or combined with a
localising
agent. The fresh cell preparation is typically kept at about 37 C. Standard
cell cultures
containers known in the art may be used to store the cells, such as a vial or
a flask.
In a preferred embodiment according to the invention, before use in the eye, a
cryopreserved preparation of cells is thawed (for example by incubating at a
temperature of
about 37 C in an incubator or waterbath). Preferably 10 volumes of PBS or
growth medium
may be added to rinse off the cells from the cryopreservant solution. Cells
may then be
rinsed by centrifugation, and a cell suspension may be made in PBS and/or
growth medium,
before combination with a localising agent for ocular delivery, which also
preferably does not
comprise a LATS inhibitor.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
161
In one aspect of the invention the expanded population of cells prepared by
the method of
cell population expansion, (preferably also including the step of growth in
medium without
supplementation with LATS inhibitor to form a mature corneal endothelium), are
prepared as
a suspension (for example in PBS and/or growth medium, such as for example X-
VIVO
medium) and combined with a localising agent suitable for ocular delivery,
(such as a
biomatrix like GelMA or fibrin glue). In a specific embodiment of the method
of treatment
according to the invention, this combination of cells, PBS and/or growth
medium, and
biomatrix is delivered as a suspension to the eye. In yet another specific
embodiment this
combination of cells, PBS and/or growth medium, and biomatrix comprises at
most only
trace levels of a LATS inhibitor.
Alternatively, the cells may be cultured and the cell population proliferation
phase may occur
in cell proliferation medium on a localising agent suitable for cell delivery
to the ocular
surface.
In an embodiment of the invention the cell population expanded according to
the invention
may be isolated as a contiguous cell sheet for delivery to the cornea, using
methods known
in the art (for examples, see Kim et al, JSM Biotechnol. Bioeng., 2016,
p.1047). Cell sheets
may be mechanically supported on a material or materials for delivery to the
cornea.
In one aspect, the present invention relates to a composition comprising the
modified CEC
of the present invention or the modified CEC obtained by the method of the
present
invention or the cell population of the present invention or the population of
modified CEC
obtained by the method of the present invention. Suitably, the modified CEC of
the
composition comprises an indel formed at or near the target sequence
complementary to the
targeting domain of the gRNA molecule domain. Suitably, the indel comprises a
deletion of
10 or greater than 10 nucleotides, optionally 11, 12, 13, 14, 15, 16, 17, 18,
19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31,32, 33, 34, or 35 nucleotides. Suitably,
the indel is formed
in at least about 40%, e.g., at least about 50%, e.g., at least about 60%,
e.g., at least about
70%, e.g., at least about 80%, e.g., at least about 90%, e.g., at least about
95%, e.g., at
least about 96%, e.g., at least about 97%, e.g., at least about 98%, e.g., at
least about 99%,
of the cells of the population. In one embodiment, an off-target indel is
detected in no more
than about 5%, e.g., no more than about 1%, e.g., no more than about 0.1%,
e.g., no more
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
162
than about 0.01%, of the cells of the population of cells e.g., as detectible
by next generation
sequencing and/or a nucleotide insertional assay.
Reduction of Immunorejection
Upon transplantation, allogeneic limbal stem cells or corneal endothelial
cells are at risk of
rejection by the recipient's immune system. Immunosuppression regimens can be
used to
reduce the risk of immunorejection of transplanted cells, such as LSCs or
CECs.
Suitable systemic immunosuppressant agents used in recipients of allogeneic
LSCs or
CECs include tacrolimus, mycophenolate mofetil, prednisone and prophylactic
valganciclovir
and trimethoprim/sulfamethoxazole. (See: Holland EJ, Mogilishetty G, Skeens
HM, Hair DB,
Neff KD, Biber JM, Chan CC (2012) Systemic immunosuppression in ocular surface
stem
cell transplantation: results of a 10-year experience. Cornea. 2012
Jun;31(6):655-61).
As the methods of cell population expansion according the present invention
provide high
expansion capabilities of a population of cells, optionally gene-editing
technologies may be
used to remove drivers of immunorejection or add genes that reduce the
recipient's immune
response.
In one aspect of the invention gene editing is carried out on a cell
population "ex vivo". In
another aspect of the invention gene-editing technologies may optionally be
used to reduce
or eliminate the expression of a gene associated with facilitating a host
versus graft immune
response. In a preferred embodiment the gene is selected from the group
consisting of:
B2M, HLA-A, HLA-B and HLA-C. In a specific embodiment the gene is B2M. B2M is
beta 2
microglobulin and is a component of the class I major histocompatibility
complex (MHC). It
has the HUGO Gene Nomenclature Committee (HGNC) identifier 914. HLA-A is major
histocompatibility complex, class I, A (HGNC ID 4931). HLA-B is major
histocompatibility
complex, class I, B (HGNC ID 4932). HLA-C is major histocompatibility complex,
class I, C
(HGNC ID 4933).
In a preferred embodiment, the gene editing method used in a method of the
invention is
CRISPR (CRISPR: clustered regularly interspaced short palindromic repeats,
also known as
CRISPR/Cas systems). In one aspect of the invention, the gene editing is
carried out on a
cell population "ex vivo".
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
163
CRISPR Gene Editing Systems
"CRISPR" as used herein refers to a set of clustered regularly interspaced
short palindromic
repeats, or a system comprising such a set of repeats. "Cas," as used herein,
refers to a
CRISPR-associated protein. The diverse CRISPR-Cas systems can be divided into
two
classes according to the configuration of their effector modules: class 1
CRISPR systems
utilize several Cas proteins and the crRNA to form an effector complex,
whereas class 2
CRISPR systems employ a large single-component Cas protein in conjunction with
crRNAs
to mediate interference. One example of class 2 CRISPR-Cas system employs Cpf1
(CRISPR from Prevotella and Francisella 1). See, e.g., Zetsche et al., Cell
163:759-771
(2015), the content of which is herein incorporated by reference in its
entirety. The term
"Cpf1" as used herein includes all orthologs, and variants that can be used in
a CRISPR
system.
The terms "CRISPR system", "Cas system" or "CRISPR/Cas system" refer to a set
of
molecules comprising an RNA-guided nuclease or other effector molecule and a
gRNA
molecule that together are necessary and sufficient to direct and effect
modification of
nucleic acid at a target sequence by the RNA-guided nuclease or other effector
molecule. In
one embodiment, a CRISPR system comprises a gRNA and a Cas protein, e.g., a
Cas9
protein. Such systems comprising a Cas9 or modified Cas9 molecule are referred
to herein
as "Cas9 systems" or "CRISPR/Cas9 systems". In one example, the gRNA molecule
and
Cas molecule may be complexed, to form a ribonuclear protein (RNP) complex.
Naturally-occurring CRISPR systems are found in approximately 40% of sequenced
eubacteria genomes and 90% of sequenced archaea. Grissa et al. (2007) BMC
Bioinformatics 8: 172. This system is a type of prokaryotic immune system that
confers
resistance to foreign genetic elements such as plasmids and phages and
provides a form of
acquired immunity. Barrangou etal. (2007) Science 315: 1709-1712; Marragini
etal. (2008)
Science 322: 1843-1845.
The CRISPR system has been modified for use in gene editing (silencing,
enhancing or
changing specific genes) in eukaryotes such as mice, primates and humans.
Wiedenheft et
al. (2012) Nature 482: 331-8. This is accomplished by, for example,
introducing into the
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
164
eukaryotic cell one or more vectors encoding a specifically engineered guide
RNA (gRNA)
(e.g., a gRNA comprising sequence complementary to sequence of a eukaryotic
genome)
and one or more appropriate RNA-guided nucleases, e.g., Cas proteins. The RNA
guided
nuclease forms a complex with the gRNA, which is then directed to the target
DNA site by
hybridization of the gRNA's sequence to complementary sequence of a eukaryotic
genome,
where the RNA-guided nuclease then induces a double or single-strand break in
the DNA.
Insertion or deletion of nucleotides at or near the strand break creates the
modified genome.
As these naturally occur in many different types of bacteria, the exact
arrangements of the
CRISPR and structure, function and number of Cas genes and their product
differ somewhat
from species to species. Haft et al. (2005) PLoS Comput. Biol. 1: e60; Kunin
et al. (2007)
Genome Biol. 8: R61; Mojica etal. (2005) J. MoL EvoL 60: 174-182; Bolotin
etal. (2005)
MicrobioL 151: 2551-2561; Pourcel et al. (2005) MicrobioL 151: 653-663; and
Stern etal.
(2010) Trends. Genet. 28: 335-340. For example, the Cse (Cas subtype, E. coli)
proteins
(e.g., CasA) form a functional complex, Cascade, that processes CRISPR RNA
transcripts
into spacer-repeat units that Cascade retains. Brouns et al. (2008) Science
321: 960-964. In
other prokaryotes, Cas6 processes the CRISPR transcript. The CRISPR-based
phage
inactivation in E. coli requires Cascade and Cas3, but not Cas1 or Cas2. The
Cmr (Cas
RAMP module) proteins in Pyrococcus furiosus and other prokaryotes form a
functional
complex with small CRISPR RNAs that recognizes and cleaves complementary
target
RNAs.
A simpler CRISPR system relies on the protein Cas9, which is a nuclease with
two active
cutting sites, one for each strand of the double helix. Combining Cas9 and
modified CRISPR
locus RNA can be used in a system for gene editing. Pennisi (2013) Science
341: 833-836.
Cas9
In some embodiments, the RNA-guided nuclease is a Cas molecule, e.g., a Cas9
molecule.
The terms "Cas9" or "Cas9 molecule" refer to an enzyme from bacterial Type II
CRISPR/Cas
system responsible for DNA cleavage. Cas9 also includes wild-type protein as
well as
functional and nonfunctinal mutants thereof. The "Cas9 molecule," can interact
with a gRNA
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
165
molecule (e.g., sequence of a domain of a tracr, also known as tracrRNA or
trans activating
CRISPR RNA) and, in concert with the gRNA molecule, localize (e.g., target or
home) to a
site which comprises a target sequence and PAM (protospacer adjacent motif)
sequence.
According to the present invention, Cas9 molecules used in the methods and
compositions
described herein can be from, derived from, or otherwise based on, the Cas9
proteins of a
variety of species. For example, Cas9 molecules of, derived from, or based on,
e.g., S.
pyogenes, S. thermophilus, Staphylococcus aureus and/or Neisseria meningitidis
Cas9
molecules, can be used in the systems, methods and compositions described
herein.
Additional Cas9 species include: Acidovorax avenae, Actinobacillus
pleuropneumoniae,
Actinobacillus succinogenes, Actinobacillus suis, Actinomyces sp., cycliphilus
denitrificans,
Aminomonas paucivorans, Bacillus cereus, Bacillus smithii, Bacillus
thuringiensis,
Bacteroides sp., Blastopirellula marina, Bradyrhiz obium sp., Brevibacillus
latemsporus,
Campylobacter coli, Campylobacter jejuni, Campylobacter lad, Candidatus
Puniceispirillum,
Clostridiu cellulolyticum, Clostridium perfringens, Corynebacterium accolens,
Corynebacterium diphtheria, Corynebacterium matruchotii, Dinoroseobacter
sliibae,
Eubacterium dolichum, gamma proteobacterium, Gluconacetobacler diazotrophicus,
Haemophilus parainfluenzae, Haemophilus sputorum, Helicobacter canadensis,
Helicobacter cinaedi, Helicobacter mustelae, Ilyobacler polytropus, Kingella
kingae,
Lactobacillus crispatus, Listeria ivanovii, Listeria monocytogenes,
Listeriaceae bacterium,
Methylocystis sp., Methylosinus trichosporium, Mobiluncus mulieris, Neisseria
bacilliformis,
Neisseria cinerea, Neisseria flavescens, Neisseria lactamica. Neisseria sp.,
Neisseria
wadsworthii, Nitrosomonas sp., Parvibaculum lavamentivorans, Pasteurella
multocida,
Phascolarctobacterium succinatutens, Ralstonia syzygii, Rhodopseudomonas
palustris,
Rhodovulum sp., Simonsiella muelleri, Sphingomonas sp., Sporolactobacillus
vineae,
Staphylococcus lugdunensis, Streptococcus sp., Subdoligranulum sp., Tislrella
mobilis,
Treponema sp., or Verminephrobacter eiseniae.
In some embodiments, the ability of an active Cas9 molecule to interact with
and cleave a
target nucleic acid is PAM sequence dependent. A PAM (protospacer adjacent
motif)
sequence is a sequence in the target nucleic acid. It is typically short, for
example 2 to 7
base pairs long. In an embodiment, cleavage of the target nucleic acid occurs
upstream
from the PAM sequence. Active Cas9 molecules from different bacterial species
can
recognize different sequence motifs (e.g., PAM sequences). In an embodiment,
an active
Cas9 molecule of S. pyogenes recognizes the sequence motif NGG and directs
cleavage of
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
166
a target nucleic acid sequence 1 to 10, e.g., 3 to 5, base pairs upstream from
that sequence.
See, e.g., Mali el al, SCIENCE 2013; 339(6121): 823- 826. In an embodiment, an
active
Cas9 molecule of S. thermophilus recognizes the sequence motif NGGNG (SEQ ID
NO: 4)
and NNAG AAW (SEQ ID NO: 5) (W = A or T and N is any nucleobase) and directs
cleavage of a core target nucleic acid sequence 1 to 10, e.g., 3 to 5, base
pairs upstream
from these sequences. See, e.g., Horvath et al., SCIENCE 2010; 327(5962): 167-
170, and
Deveau et al, J BACTERIOL 2008; 190(4): 1390- 1400. In an embodiment, an
active Cas9
molecule of S. mutans recognizes the sequence motif NGG or NAAR (R - A or G)
and
directs cleavage of a core target nucleic acid sequence 1 to 10, e.g., 3 to 5
base pairs,
upstream from this sequence. See, e.g., Deveau et al., J BACTERIOL 2008;
190(4): 1390-
1400.
In an embodiment, an active Cas9 molecule of S. aureus recognizes the sequence
motif
NNGRR (SEQ ID NO: 6) (R = A or G) and directs cleavage of a target nucleic
acid sequence
1 to 10, e.g., 3 to 5, base pairs upstream from that sequence. See, e.g., Ran
F. et al.,
NATURE, vol. 520, 2015, pp. 186-191. In an embodiment, an active Cas9 molecule
of N.
meningitidis recognizes the sequence motif NNNNGATT (SEQ ID NO: 7) and directs
cleavage of a target nucleic acid sequence 1 to 10, e.g., 3 to 5, base pairs
upstream from
that sequence. See, e.g., Hou et al., PNAS EARLY EDITION 2013, 1 -6. The
ability of a
Cas9 molecule to recognize a PAM sequence can be determined, e.g., using a
transformation assay described in Jinek et al , SCIENCE 2012, 337:816.
Exemplary naturally occurring Cas9 molecules are described in Chylinski et al,
RNA Biology
2013; 10:5, 727-737. Such Cas9 molecules include Cas9 molecules of a cluster 1
bacterial
family, cluster 2 bacterial family, cluster 3 bacterial family, cluster 4
bacterial family, cluster 5
bacterial family, cluster 6 bacterial family, a cluster 7 bacterial family, a
cluster 8 bacterial
family, a cluster 9 bacterial family, a cluster 10 bacterial family, a cluster
11 bacterial family,
a cluster 12 bacterial family, a cluster 13 bacterial family, a cluster 14
bacterial family, a
cluster 15 bacterial family, a cluster 16 bacterial family, a cluster 17
bacterial family, a
cluster 18 bacterial family, a cluster 19 bacterial family, a cluster 20
bacterial family, a
cluster 21 bacterial family, a cluster 22 bacterial family, a cluster 23
bacterial family, a
cluster 24 bacterial family, a cluster 25 bacterial family, a cluster 26
bacterial family, a
cluster 27 bacterial family, a cluster 28 bacterial family, a cluster 29
bacterial family, a
cluster 30 bacterial family, a cluster 31 bacterial family, a cluster 32
bacterial family, a
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
167
cluster 33 bacterial family, a cluster 34 bacterial family, a cluster 35
bacterial family, a
cluster 36 bacterial family, a cluster 37 bacterial family, a cluster 38
bacterial family, a
cluster 39 bacterial family, a cluster 40 bacterial family, a cluster 41
bacterial family, a
cluster 42 bacterial family, a cluster 43 bacterial family, a cluster 44
bacterial family, a
cluster 45 bacterial family, a cluster 46 bacterial family, a cluster 47
bacterial family, a
cluster 48 bacterial family,. a cluster 49 bacterial family, a cluster 50
bacterial family, a
cluster 51 bacterial family, a cluster 52 bacterial family, a cluster 53
bacterial family, a
cluster 54 bacterial family, a cluster 55 bacterial family, a cluster 56
bacterial family, a
cluster 57 bacterial family, a cluster 58 bacterial family, a cluster 59
bacterial family, a
cluster 60 bacterial family, a cluster 61 bacterial family, a cluster 62
bacterial family, a
cluster 63 bacterial family, a cluster 64 bacterial family, a cluster 65
bacterial family, a
cluster 66 bacterial family, a cluster 67 bacterial family, a cluster 68
bacterial family, a
cluster 69 bacterial family, a cluster 70 bacterial family, a cluster 71
bacterial family, a
cluster 72 bacterial family, a cluster 73 bacterial family, a cluster 74
bacterial family, a
cluster 75 bacterial family, a cluster 76 bacterial family, a cluster 77
bacterial family, or a
cluster 78 bacterial family.
Exemplary naturally occurring Cas9 molecules include a Cas9 molecule of a
cluster 1
bacterial family. Examples include a Cas9 molecule of: S. pyogenes (e.g.,
strain SF370,
MGAS 10270, MGAS 10750, MGA52096, MGAS315, MGAS5005, MGAS6180, MGA59429,
NZ131 and SSI- 1), S. thermophilus (e.g., strain LMD-9), S. pseudoporcinus
(e.g., strain
SPIN 20026), S. mutans (e.g., strain UA 159, NN2025), S. macacae (e.g., strain
NCTC1
1558), S. gallolylicus (e.g., strain UCN34, ATCC BAA-2069), S. equines (e.g.,
strain ATCC
9812, MGCS 124), S. dysdalactiae (e.g., strain GGS 124), S. bovis (e.g.,
strain ATCC
700338), S. cmginosus (e.g.; strain F021 1 ), S. agalactia* (e.g., strain
NEM316, A909),
Listeria monocytogenes (e.g., strain F6854), Listeria innocua (L. innocua,
e.g., strain Clip
11262), EtUerococcus italicus (e.g., strain DSM 15952), or Enterococcus
faecium (e.g., strain
1,23 ,408). Additional exemplary Cas9 molecules are a Cas9 molecule of
Neisseria
meningitidis (Hou etal. PNAS Early Edition 2013, 1-6) and a S. aureus Cas9
molecule.
In an embodiment, a Cas9 molecule, e.g., an active Cas9 molecule comprises an
amino
acid sequence: having at least 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 96%,
97%,
98%, or 99% homology with; differs at no more than 1%, 2%, 5%, 10%, 15%, 20%,
30%, or
40% of the amino acid residues when compared with; differs by at least 1, 2,
5, 10 or 20
amino acids but by no more than 100, 80, 70, 60, 50, 40 or 30 amino acids
from; or is
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
168
identical to; any Cas9 molecule sequence described herein or a naturally
occurring Cas9
molecule sequence, e.g., a Cas9 molecule from a species listed herein or
described in
Chylinski et al. , RNA Biology 2013, 10:5, 121-T,1 Hou et al. PNAS Early
Edition 2013, 1-6.
In an embodiment, a Cas9 molecule comprises an amino acid sequence having at
least
60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% homology with;
differs at no more than 1%, 2%, 5%, 10%, 15%, 20%, 30%, or 40% of the amino
acid
residues when compared with; differs by at least 1, 2, 5, 10 or 20 amino acids
but by no
more than 100, 80, 70, 60, 50, 40 or 30 amino acids from; or is identical to;
S. pyogenes
Cas9 (UniProt Q99ZW2). In one embodiment, a Cas9 molecule comprises an amino
acid
sequence having 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99%
homology with; differs at no more than 1%, 2%, 5%, 10%, 15%, 20%, 30%, or 40%
of the
amino acid residues when compared with; differs by at least 1, 2, 5, 10 or 20
amino acids
but by no more than 100, 80, 70, 60, 50, 40 or 30 amino acids from; or is
identical to; S.
.. pyogenes Cas9:
Met Asp Lys Lys Tyr Ser Ile Gly Leu Asp Ile Gly Thr Asn Ser Val
1 5 10 15
Gly Trp Ala Val Ile Thr Asp Glu Tyr Lys Val Pro Ser Lys Lys Phe
20 25 30
Lys Val Leu Gly Asn Thr Asp Arg His Ser Ile Lys Lys Asn Leu Ile
35 40 45
Gly Ala Leu Leu Phe Asp Ser Gly Glu Thr Ala Glu Ala Thr Arg Leu
50 55 60
Lys Arg Thr Ala Arg Arg Arg Tyr Thr Arg Arg Lys Asn Arg Ile Cys
65 70 75 80
Tyr Leu Gin Glu Ile Phe Ser Asn Glu Met Ala Lys Val Asp Asp Ser
85 90 95
Phe Phe His Arg Leu Glu Glu Ser Phe Leu Val Glu Glu Asp Lys Lys
100 105 110
His Glu Arg His Pro Ile Phe Gly Asn Ile Val Asp Glu Val Ala Tyr
115 120 125
His Glu Lys Tyr Pro Thr Ile Tyr His Leu Arg Lys Lys Leu Val Asp
130 135 140
Ser Thr Asp Lys Ala Asp Leu Arg Leu Ile Tyr Leu Ala Leu Ala His
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
169
145 150 155 160
Met Ile Lys Phe Arg Gly His Phe Leu Ile Glu Gly Asp Leu Asn Pro
165 170 175
Asp Asn Ser Asp Val Asp Lys Leu Phe Ile Gin Leu Val Gin Thr Tyr
180 185 190
Asn Gin Leu Phe Glu Glu Asn Pro Ile Asn Ala Ser Gly Val Asp Ala
195 200 205
Lys Ala Ile Leu Ser Ala Arg Leu Ser Lys Ser Arg Arg Leu Glu Asn
210 215 220
Leu Ile Ala Gin Leu Pro Gly Glu Lys Lys Asn Gly Leu Phe Gly Asn
225 230 235 240
Leu Ile Ala Leu Ser Leu Gly Leu Thr Pro Asn Phe Lys Ser Asn Phe
245 250 255
Asp Leu Ala Glu Asp Ala Lys Leu Gin Leu Ser Lys Asp Thr Tyr Asp
260 265 270
Asp Asp Leu Asp Asn Leu Leu Ala Gin Ile Gly Asp Gin Tyr Ala Asp
275 280 285
Leu Phe Leu Ala Ala Lys Asn Leu Ser Asp Ala Ile Leu Leu Ser Asp
290 295 300
Ile Leu Arg Val Asn Thr Glu Ile Thr Lys Ala Pro Leu Ser Ala Ser
305 310 315 320
Met Ile Lys Arg Tyr Asp Glu His His Gin Asp Leu Thr Leu Leu Lys
325 330 335
Ala Leu Val Arg Gin Gin Leu Pro Glu Lys Tyr Lys Glu Ile Phe Phe
340 345 350
Asp Gin Ser Lys Asn Gly Tyr Ala Gly Tyr Ile Asp Gly Gly Ala Ser
355 360 365
Gin Glu Glu Phe Tyr Lys Phe Ile Lys Pro Ile Leu Glu Lys Met Asp
370 375 380
Gly Thr Glu Glu Leu Leu Val Lys Leu Asn Arg Glu Asp Leu Leu Arg
385 390 395 400
Lys Gin Arg Thr Phe Asp Asn Gly Ser Ile Pro His Gin Ile His Leu
405 410 415
Gly Glu Leu His Ala Ile Leu Arg Arg Gin Glu Asp Phe Tyr Pro Phe
420 425 430
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
170
Leu Lys Asp Asn Arg Glu Lys Ile Glu Lys Ile Leu Thr Phe Arg Ile
435 440 445
Pro Tyr Tyr Val Gly Pro Leu Ala Arg Gly Asn Ser Arg Phe Ala Trp
450 455 460
Met Thr Arg Lys Ser Glu Glu Thr Ile Thr Pro Trp Asn Phe Glu Glu
465 470 475 480
Val Val Asp Lys Gly Ala Ser Ala Gin Ser Phe Ile Glu Arg Met Thr
485 490 495
Asn Phe Asp Lys Asn Leu Pro Asn Glu Lys Val Leu Pro Lys His Ser
500 505 510
Leu Leu Tyr Glu Tyr Phe Thr Val Tyr Asn Glu Leu Thr Lys Val Lys
515 520 525
Tyr Val Thr Glu Gly Met Arg Lys Pro Ala Phe Leu Ser Gly Glu Gin
530 535 540
Lys Lys Ala Ile Val Asp Leu Leu Phe Lys Thr Asn Arg Lys Val Thr
545 550 555 560
Val Lys Gin Leu Lys Glu Asp Tyr Phe Lys Lys Ile Glu Cys Phe Asp
565 570 575
Ser Val Glu Ile Ser Gly Val Glu Asp Arg Phe Asn Ala Ser Leu Gly
580 585 590
Thr Tyr His Asp Leu Leu Lys Ile Ile Lys Asp Lys Asp Phe Leu Asp
595 600 605
Asn Glu Glu Asn Glu Asp Ile Leu Glu Asp Ile Val Leu Thr Leu Thr
610 615 620
Leu Phe Glu Asp Arg Glu Met Ile Glu Glu Arg Leu Lys Thr Tyr Ala
625 630 635 640
His Leu Phe Asp Asp Lys Val Met Lys Gin Leu Lys Arg Arg Arg Tyr
645 650 655
Thr Gly Trp Gly Arg Leu Ser Arg Lys Leu Ile Asn Gly Ile Arg Asp
660 665 670
Lys Gin Ser Gly Lys Thr Ile Leu Asp Phe Leu Lys Ser Asp Gly Phe
675 680 685
Ala Asn Arg Asn Phe Met Gin Leu Ile His Asp Asp Ser Leu Thr Phe
690 695 700
Lys Glu Asp Ile Gin Lys Ala Gin Val Ser Gly Gin Gly Asp Ser Leu
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
171
705 710 715 720
His Glu His Ile Ala Asn Leu Ala Gly Ser Pro Ala Ile Lys Lys Gly
725 730 735
Ile Leu Gin Thr Val Lys Val Val Asp Glu Leu Val Lys Val Met Gly
740 745 750
Arg His Lys Pro Glu Asn Ile Val Ile Glu Met Ala Arg Glu Asn Gin
755 760 765
Thr Thr Gin Lys Gly Gin Lys Asn Ser Arg Glu Arg Met Lys Arg Ile
770 775 780
Glu Glu Gly Ile Lys Glu Leu Gly Ser Gin Ile Leu Lys Glu His Pro
785 790 795 800
Val Glu Asn Thr Gin Leu Gin Asn Glu Lys Leu Tyr Leu Tyr Tyr Leu
805 810 815
Gin Asn Gly Arg Asp Met Tyr Val Asp Gin Glu Leu Asp Ile Asn Arg
820 825 830
Leu Ser Asp Tyr Asp Val Asp His Ile Val Pro Gin Ser Phe Leu Lys
835 840 845
Asp Asp Ser Ile Asp Asn Lys Val Leu Thr Arg Ser Asp Lys Asn Arg
850 855 860
Gly Lys Ser Asp Asn Val Pro Ser Glu Glu Val Val Lys Lys Met Lys
865 870 875 880
Asn Tyr Trp Arg Gin Leu Leu Asn Ala Lys Leu Ile Thr Gin Arg Lys
885 890 895
Phe Asp Asn Leu Thr Lys Ala Glu Arg Gly Gly Leu Ser Glu Leu Asp
900 905 910
Lys Ala Gly Phe Ile Lys Arg Gin Leu Val Glu Thr Arg Gin Ile Thr
915 920 925
Lys His Val Ala Gin Ile Leu Asp Ser Arg Met Asn Thr Lys Tyr Asp
930 935 940
Glu Asn Asp Lys Leu Ile Arg Glu Val Lys Val Ile Thr Leu Lys Ser
945 950 955 960
Lys Leu Val Ser Asp Phe Arg Lys Asp Phe Gin Phe Tyr Lys Val Arg
965 970 975
Glu Ile Asn Asn Tyr His His Ala His Asp Ala Tyr Leu Asn Ala Val
980 985 990
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
172
Val Gly Thr Ala Leu Ile Lys Lys Tyr Pro Lys Leu Glu Ser Glu Phe
995 1000 1005
Val Tyr Gly Asp Tyr Lys Val Tyr Asp Val Arg Lys Met Ile Ala Lys
1010 1015 1020
Ser Glu Gln Glu Ile Gly Lys Ala Thr Ala Lys Tyr Phe Phe Tyr Ser
1025 1030 1035 1040
Asn Ile Met Asn Phe Phe Lys Thr Glu Ile Thr Leu Ala Asn Gly Glu
1045 1050 1055
Ile Arg Lys Arg Pro Leu Ile Glu Thr Asn Gly Glu Thr Gly Glu Ile
1060 1065 1070
Val Trp Asp Lys Gly Arg Asp Phe Ala Thr Val Arg Lys Val Leu Ser
1075 1080 1085
Met Pro Gln Val Asn Ile Val Lys Lys Thr Glu Val Gln Thr Gly Gly
1090 1095 1100
Phe Ser Lys Glu Ser Ile Leu Pro Lys Arg Asn Ser Asp Lys Leu Ile
1105 1110 1115 1120
Ala Arg Lys Lys Asp Trp Asp Pro Lys Lys Tyr Gly Gly Phe Asp Ser
1125 1130 1135
Pro Thr Val Ala Tyr Ser Val Leu Val Val Ala Lys Val Glu Lys Gly
1140 1145 1150
Lys Ser Lys Lys Leu Lys Ser Val Lys Glu Leu Leu Gly Ile Thr Ile
1155 1160 1165
Met Glu Arg Ser Ser Phe Glu Lys Asn Pro Ile Asp Phe Leu Glu Ala
1170 1175 1180
Lys Gly Tyr Lys Glu Val Lys Lys Asp Leu Ile Ile Lys Leu Pro Lys
1185 1190 1195 1200
Tyr Ser Leu Phe Glu Leu Glu Asn Gly Arg Lys Arg Met Leu Ala Ser
1205 1210 1215
Ala Gly Glu Leu Gln Lys Gly Asn Glu Leu Ala Leu Pro Ser Lys Tyr
1220 1225 1230
Val Asn Phe Leu Tyr Leu Ala Ser His Tyr Glu Lys Leu Lys Gly Ser
1235 1240 1245
Pro Glu Asp Asn Glu Gln Lys Gln Leu Phe Val Glu Gln His Lys His
1250 1255 1260
Tyr Leu Asp Glu Ile Ile Glu Gln Ile Ser Glu Phe Ser Lys Arg Val
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
173
1265 1270 1275 1280
Ile Leu Ala Asp Ala Asn Leu Asp Lys Val Leu Ser Ala Tyr Asn Lys
1285 1290 1295
His Arg Asp Lys Pro Ile Arg Glu Gin Ala Glu Asn Ile Ile His Leu
1300 1305 1310
Phe Thr Leu Thr Asn Leu Gly Ala Pro Ala Ala Phe Lys Tyr Phe Asp
1315 1320 1325
Thr Thr Ile Asp Arg Lys Arg Tyr Thr Ser Thr Lys Glu Val Leu Asp
1330 1335 1340
Ala Thr Leu Ile His Gin Ser Ile Thr Gly Leu Tyr Glu Thr Arg Ile
1345 1350 1355 1360
Asp Leu Ser Gin Leu Gly Gly Asp
1365
(SEQ ID NO: 123).
In certain embodiments, the Cas9 molecule is a S. pyogenes Cas9 variant, such
as a variant
described in Slaymaker et al., Science Express, available online December 1,
2015 at
Science DOI: 10.1126/science.aad5227; Kleinstiver et al., Nature, 529, 2016,
pp. 490-495,
available online January 6, 2016 at doi:10.1038/nature16526; or
U52016/0102324, the
contents of which are incorporated herein in their entirety.
In some embodiments, the Cas9 molecule is a S. pyogenes Cas9 variant of SEQ ID
NO:
123 that includes one or more mutations to positively charged amino acids
(e.g., lysine,
arginine or histidine) that introduce an uncharged or nonpolar amino acid,
e.g., alanine, at
said position. In embodiments, the mutation is to one or more positively
charged amino
acids in the nt-groove of Cas9. In embodiments, the Cas9 molecule is a S.
pyogenes Cas9
variant of SEQ ID NO: 123 that includes a mutation at position 855 of SEQ ID
NO: 123, for
example a mutation to an uncharged amino acid, e.g., alanine, at position 855
of SEQ ID
NO: 123. In embodiments, the Cas9 molecule has a mutation only at position 855
of SEQ ID
NO: 123, relative to SEQ ID NO: 123, e.g., to an uncharged amino acid, e.g.,
alanine. In
embodiments, the Cas9 molecule is a S. pyogenes Cas9 variant of SEQ ID NO: 123
that
includes a mutationat position 810, a mutation at position 1003, and/or a
mutation at position
1060 of SEQ ID NO: 123, for example a mutation to alanine at position 810,
position 1003,
and/or position 1060 of SEQ ID NO: 123. In embodiments, the Cas9 molecule has
a
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
174
mutation only at position 810, position 1003, and position 1060 of SEQ ID NO:
123, relative
to SEQ ID NO: 123, e.g., where each mutation is to an uncharged amino acid,
for example,
alanine. In embodiments, the Cas9 molecule is a S. pyogenes Cas9 variant of
SEQ ID NO:
123 that includes a mutationat position 848, a mutation at position 1003,
and/or a mutation
at position 1060 of SEQ ID NO: 123, for example a mutation to alanine at
position 848,
position 1003, and/or position 1060 of SEQ ID NO: 123. In embodiments, the
Cas9 molecule
has a mutation only at position 848, position 1003, and position 1060 of SEQ
ID NO: 123,
relative to SEQ ID NO: 123, e.g., where each mutation is to an uncharged amino
acid, for
example, alanine. In embodiments, the Cas9 molecule is a Cas9 molecule as
described in
.. Slaymaker et al., Science Express, available online December 1, 2015 at
Science DOI:
10.1126/science.aad5227.
In embodiments, the Cas9 molecule is a S. pyogenes Cas9 variant of SEQ ID NO:
123 that
includes one or more mutations. In embodiments, the Cas9 variant comprises a
mutation at
position 80 of SEQ ID NO: 123, e.g., includes a leucine at position 80 of SEQ
ID NO: 123
(i.e., comprises or consists of SEQ ID NO: 123 with a C8OL mutation). In
embodiments, the
Cas9 variant comprises a mutation at position 574 of SEQ ID NO: 123, e.g.,
includes a
glutamic acid at position 574 of SEQ ID NO: 123 (i.e., comprises or consists
of SEQ ID NO:
123 with a C574E mutation). In embodiments, the Cas9 variant comprises a
mutation at
position 80 and a mutation at position 574 of SEQ ID NO: 123, e.g., includes a
leucine at
position 80 of SEQ ID NO: 123, and a glutamic acid at position 574 of SEQ ID
NO: 123 (i.e.,
comprises or consists of SEQ ID NO: 123 with a C8OL mutation and a C574E
mutation).
Without being bound by theory, it is believed that such mutations improve the
solution
properties of the Cas9 molecule.
In embodiments, the Cas9 molecule is a S. pyogenes Cas9 variant of SEQ ID NO:
123 that
includes one or more mutations. In embodiments, the Cas9 variant comprises a
mutation at
position 147 of SEQ ID NO: 123, e.g., includes a tyrosine at position 147 of
SEQ ID NO: 123
(i.e., comprises or consists of SEQ ID NO: 123 with a D147Y mutation). In
embodiments,
the Cas9 variant comprises a mutation at position 411 of SEQ ID NO: 123, e.g.,
includes a
threonine at position 411 of SEQ ID NO: 123 (i.e., comprises or consists of
SEQ ID NO: 123
with a P411T mutation). In embodiments, the Cas9 variant comprises a mutation
at position
147 and a mutation at position 411 of SEQ ID NO: 123, e.g., includes a
tyrosine at position
147 of SEQ ID NO: 123, and a threonine at position 411 of SEQ ID NO: 123
(i.e., comprises
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
175
or consists of SEQ ID NO: 123 with a D147Y mutation and a P41 IT mutation).
Without
being bound by theory, it is believed that such mutations improve the
targeting efficiency of
the Cas9 molecule, e.g., in yeast.
In embodiments, the Cas9 molecule is a S. pyogenes Cas9 variant of SEQ ID NO:
123 that
includes one or more mutations. In embodiments, the Cas9 variant comprises a
mutation at
position 1135 of SEQ ID NO: 123, e.g., includes a glutamic acid at position
1135 of SEQ ID
NO: 123 (i.e., comprises or consists of SEQ ID NO: 123 with a D1135E
mutation). Without
being bound by theory, it is believed that such mutations improve the
selectivity of the Cas9
molecule for the NGG PAM sequence versus the NAG PAM sequence.
In embodiments, the Cas9 molecule is a S. pyogenes Cas9 variant of SEQ ID NO:
123 that
includes one or more mutations that introduce an uncharged or nonpolar amino
acid, e.g.,
alanine, at certain positions. In embodiments, the Cas9 molecule is a S.
pyogenes Cas9
variant of SEQ ID NO: 123 that includes a mutation at position 497, a mutation
at position
661, a mutation at position 695 and/or a mutation at position 926 of SEQ ID
NO: 123, for
example a mutation to alanine at position 497, position 661, position 695
and/or position 926
of SEQ ID NO: 123. In embodiments, the Cas9 molecule has a mutation only at
position
497, position 661, position 695, and position 926 of SEQ ID NO: 123, relative
to SEQ ID NO:
123, e.g., where each mutation is to an uncharged amino acid, for example,
alanine.
Without being bound by theory, it is believed that such mutations reduce the
cutting by the
Cas9 molecule at off-target sites
It will be understood that the mutations described herein to the Cas9 molecule
may be
combined, and may be combined with any of the fusions or other modifications
described
herein, and the Cas9 molecule may be tested in any of the assays described
herein.
Various types of Cas molecules can be used herein. In some embodiments, Cas
molecules
of Type ll Cas systems are used. In other embodiments, Cas molecules of other
Cas
systems are used. For example, Type I or Type III Cas molecules may be used.
Exemplary
Cas molecules (and Cas systems) are described, e.g., in Haft et al., PLoS
COMPUTATIONAL BIOLOGY 2005, 1(6): e60 and Makarova et al., NATURE REVIEW
MICROBIOLOGY 2011, 9:467-477, the contents of both references are incorporated
herein
by reference in their entirety.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
176
In an embodiment, a Cas or Cas9 molecule used in the methods disclosed herein
comprises
one or more of the following activities: a nickase activity; a double stranded
cleavage activity
(e.g., an endonuclease and/or exonuclease activity); a helicase activity; or
the ability,
together with a gRNA molecule, to localize to a target nucleic acid.
In some embodiments, the Cas9 molecule, e.g., a Cas9 of S. pyogenes, may
additionally
comprise one or more amino acid sequences that confer additional activity. In
some
aspects, the Cas9 molecule may comprise one or more nuclear localization
sequences
(NLSs), such as at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more NLSs.
Typically, an NLS
consists of one or more short sequences of positively charged lysines or
arginines exposed
on the protein surface, but other types of NLS are known. Non-limiting
examples of NLSs
include an NLS sequence comprising or derived from: the NLS of the SV40 virus
large T-
antigen, having the amino acid sequence PKKKRKV (SEQ ID NO: 8). Other suitable
NLS
sequences are known in the art (e.g., Sorokin, Biochemistry (Moscow) (2007)
72:13, 1439-
1457; Lange J Biol Chem. (2007) 282:8, 5101-5). In any of the aforementioned
embodiments, the Cas9 molecule may additionally (or alternatively) comprise a
tag, e.g., a
His tag, e.g., a His(6) tag (His His His His His His, SEQ ID NO: 121) or
His(8) tag (His His
His His His His His His, SEQ ID NO: 122) e.g., at the N terminus or the C
terminus.
In specific aspects, provided herein are modified human cells, such as LSCs or
CECs, with
reduced or eliminated expression of B2M by a CRISPR system, e.g., S. pyogenes
Cas9
CRISPR system, wherein the modified cells have been transduce to express a
Cas9
suitable for gene editing. In a particular aspect, provided herein are
modified human cells,
such as LSCs or CECs, with reduced or eliminated expression of B2M by a CRISPR
system, wherein the modified cells express a Cas9 suitable for gene editing.
In some embodiments, a Cas9 molecule comprises an amino sequence having at
least
about 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% homology
with; differs at no more than 1%, 2%, 5%, 10%, 15%, 20%, 30%, or 40% of the
amino acid
residues when compared with; differs by at least 1, 2, 5, 10 or 20 amino acids
but by no
more than 100, 80, 70, 60, 50, 40 or 30 amino acids from; or is identical to
to a Cas9
sequence provided herein, e.g., SEQ ID NO: 123, SEQ ID NO: 106, SEQ ID NO:
107, SEQ
ID NO: 124, SEQ ID NO: 125, SEQ ID NO: 126, SEQ ID NO: 127, SEQ ID NO: 128,
SEQ ID
NO: 129, SEQ ID NO: 130, SEQ ID NO: 131, SEQ ID NO: 132, or SEQ ID NO: 133. In
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
177
specific embodiments, a Cas9 molecule comprises an amino sequence selected
from SEQ
ID NO: 123, SEQ ID NO: 106, SEQ ID NO: 107, SEQ ID NO: 124, SEQ ID NO: 125,
SEQ ID
NO: 126, SEQ ID NO: 127, SEQ ID NO: 128, SEQ ID NO: 129, SEQ ID NO: 130, SEQ
ID
NO: 131, SEQ ID NO: 132, and SEQ ID NO: 133.
In certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of iProt 20109496 (SEQ ID NO: 106):
MARKKKRKVDKKYSIGLDIGTNSVGWAVITDEYKVPSKKFKVLGNTDRHSIKKNLIG
ALLFDSGETAEATRLKRTARRRYTRRKNRILYLQEIFSNEMAKVDDSFEHRLEESFL
VEEDKKH ERH PI FGN IVDEVAYHEKYPTIYHLRKKLVDSTDKADLRLIYLALAH MIKFR
GHFLIEGDLNPDNSDVDKLFIQLVQTYNQLFEENRNASGVDAKAILSARLSKSRRLE
NLIAQLEGEKKNGLFGINLIALSLGLTPNEKSNFDLAEDAKLQLSKDTYDDDLDNLLA
QIGDQYADLFLAAKNLSDAILLSDILRVNTEITKAPLSASMIKRYDEHHQDLTLLKALV
RQQLPEKYKEIFFDQSKNGYAGYIDGGASQEEFYKFIKPILEKNIDGTEELLVKLNRE
DLLRKQRTFDNGSIPHQIHLGELHAILRRQEDFYPFLKDNREKIEKILTFRIPYYVGPL
ARGNSRFAWMTRKSEETITPWNFEEVVDKGASAQSFIERIVITNFDKNLPNEKVLPK
HSLLYEYFTVYNELTKVKYVTEGMRKPAFLSGEQKKAIVDLLFKTNRKVTVKQLKED
YFKKIEEEDSVE1SGVEDRFNASLGTYHDLLKIIKDKDFLDNEENEDILEDIVLTLTLFE
DR EMIEERLKTYAH LFDDKVrVIKQLKRRRYTGVVGRLSRKLINGI RDKQSGKTILDFLK
SDGFANRNFMQUHDDSLTFKEDIQKAQVSGQGDSLHEHIANLAGSPAIKKGILQTV
KVVDELVKVMGRHKPENIVIEMARENQTTQKGQKNSRERMKRIEEGIKELGSORKE
HPVENTQLQNEKLYLYYLOAGRDMYVDQELDINRLSDYDVDHIVPQSFLKDDSIDN
KVLTRSDKNRGKSDNVPSEEVVKKMKNYWRQLLNAKUTORKEDNLTKAERGGLS
E LDKAG FIKRQ LVETRQITKHVAQILDSR NI NT KYDEN DKLI REVKVITLKSKLVSDFR
KDFQFYKVREINNYHHAHDAYLNAVVGTALIKKYFKLESEFVYGDYKVYDVRKMIAK
SEQEIGKATAKYFEYSNIMNEFKTEITLANGEIRKRPLIETNGETGEIVWDKGRDFAT
VRKVLSMPQVNIVKKTEVQTGGFSKES1LPKRNSDKLIARKKDVVDPKKYGGEDSPT
VAYSVLVVAKVEKGKSKKLKSVKELLGITIM ERSSFEKN PI DFLEAKGYKEVKKDLII K
LPKYSLFELENGRKRrVILASAGELQKGNELALPSKYVNFLYLASHYEKLKGSPEDNE
QKQLFVEQHKHYLDEIIEQISEFSKRVILADANLDKVLSAYNKHRDKPIREQAENIIHL
FTLTNLGAPAAFKYFDTTIDRKRYTSTKEVLDATLIHQSITGLYETRIDLSQLGGDSRA
DHHHHHH
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
178
In certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of as shown in the Examples herein as SEQ ID NO:
107.In
certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of iProt105026 (also referred to as iProt106154,
iProt106331,
iProt106545, and PID426303, depending on the preparation of the protein) (SEQ
ID NO:
107):
MAPKKKRKVD KKYSIGLDIG TNSVGWAVIT DEYKVPSKKF KVLGNTDRHS IKKNLIGALL
FDSGETAEAT RLKRTARRRY TRRKNRICYL QEIFSNEMAK VDDSFFHRLE ESFLVEEDKK
HERHPIFGNI VDEVAYHEKY PTIYHLRKKL VDSTDKADLR LIYLALAHMI KFRGHFLIEG
DLNPDNSDVD KLFIQLVQTY NQLFEENPIN ASGVDAKAIL SARLSKSRRL ENLIAQLPGE
KKNGLFGNLI ALSLGLTPNF KSNFDLAEDA KLQLSKDTYD DDLDNLLAQI GDQYADLFLA
AKNLSDAILL SDILRVNTEI TKAPLSASMI KRYDEHHQDL TLLKALVRQQ LPEKYKEIFF
DQSKNGYAGY IDGGASQEEF YKFIKPILEK MDGTEELLVK LNREDLLRKQ RTFDNGSIPH
QIHLGELHAI LRRQEDFYPF LKDNREKIEK ILTFRIPYYV GPLARGNSRF AWMTRKSEET
ITPWNFEEVV DKGASAQSFI ERMTNFDKNL PNEKVLPKHS LLYEYFTVYN ELTKVKYVTE
GMRKPAFLSG EQKKAIVDLL FKTNRKVTVK QLKEDYFKKI ECFDSVEISG VEDRFNASLG
TYHDLLKIIK DKDFLDNEEN EDILEDIVLT LTLFEDREMI EERLKTYAHL FDDKVMKQLK
RRRYTGWGRL SRKLINGIRD KQSGKTILDF LKSDGFANRN FMQLIHDDSL TFKEDIQKAQ
VSGQGDSLHE HIANLAGSPA IKKGILQTVK VVDELVKVMG RHKPENIVIE MARENQTTQK
GQKNSRERMK RIEEGIKELG SQILKEHPVE NTQLQNEKLY LYYLQNGRDM YVDQELDINR
LSDYDVDHIV PQSFLKDDSI DNKVLTRSDK NRGKSDNVPS EEVVKKMKNY WRQLLNAKLI
TQRKFDNLTK AERGGLSELD KAGFIKRQLV ETRQITKHVA QILDSRMNTK YDENDKLIRE
VKVITLKSKL VSDFRKDFQF YKVREINNYH HAHDAYLNAV VGTALIKKYP KLESEFVYGD
YKVYDVRKMI AKSEQEIGKA TAKYFFYSNI MNFFKTEITL ANGEIRKRPL IETNGETGEI
VWDKGRDFAT VRKVLSMPQV NIVKKTEVQT GGFSKESILP KRNSDKLIAR KKDWDPKKYG
GFDSPTVAYS VLVVAKVEKG KSKKLKSVKE LLGITIMERS SFEKNPIDFL EAKGYKEVKK
DLIIKLPKYS LFELENGRKR MLASAGELQK GNELALPSKY VNFLYLASHY EKLKGSPEDN
EQKQLFVEQH KHYLDEIIEQ ISEFSKRVIL ADANLDKVLS AYNKHRDKPI REQAENIIHL
FTLTNLGAPA AFKYFDTTID RKRYTSTKEV LDATLIHQSI TGLYETRIDL SQLGGDSRAD
PKKKRKVHHH HHH
In certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of iProt106518 (SEQ ID NO: 124):
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
179
MAPKKKRKVD KKYSIGLDIG TNSVGWAVIT DEYKVPSKKF KVLGNTDRHS IKKNLIGALL
FDSGETAEAT RLKRTARRRY TRRKNRILYL QEIFSNEMAK VDDSFFHRLE ESFLVEEDKK
HERHPIFGNI VDEVAYHEKY PTIYHLRKKL VDSTDKADLR LIYLALAHMI KFRGHFLIEG
DLNPDNSDVD KLFIQLVQTY NQLFEENPIN ASGVDAKAIL SARLSKSRRL ENLIAQLPGE
KKNGLFGNLI ALSLGLTPNF KSNFDLAEDA KLQLSKDTYD DDLDNLLAQI GDQYADLFLA
AKNLSDAILL SDILRVNTEI TKAPLSASMI KRYDEHHQDL TLLKALVRQQ LPEKYKEIFF
DQSKNGYAGY IDGGASQEEF YKFIKPILEK MDGTEELLVK LNREDLLRKQ RTFDNGSIPH
QIHLGELHAI LRRQEDFYPF LKDNREKIEK ILTFRIPYYV GPLARGNSRF AWMTRKSEET
ITPWNFEEVV DKGASAQSFI ERMTNFDKNL PNEKVLPKHS LLYEYFTVYN ELTKVKYVTE
GMRKPAFLSG EQKKAIVDLL FKTNRKVTVK QLKEDYFKKI EEFDSVEISG VEDRFNASLG
TYHDLLKIIK DKDFLDNEEN EDILEDIVLT LTLFEDREMI EERLKTYAHL FDDKVMKQLK
RRRYTGWGRL SRKLINGIRD KQSGKTILDF LKSDGFANRN FMQLIHDDSL TFKEDIQKAQ
VSGQGDSLHE HIANLAGSPA IKKGILQTVK VVDELVKVMG RHKPENIVIE MARENQTTQK
GQKNSRERMK RIEEGIKELG SQILKEHPVE NTQLQNEKLY LYYLQNGRDM YVDQELDINR
LSDYDVDHIV PQSFLKDDSI DNKVLTRSDK NRGKSDNVPS EEVVKKMKNY WRQLLNAKLI
TQRKFDNLTK AERGGLSELD KAGFIKRQLV ETRQITKHVA QILDSRMNTK YDENDKLIRE
VKVITLKSKL VSDFRKDFQF YKVREINNYH HAHDAYLNAV VGTALIKKYP KLESEFVYGD
YKVYDVRKMI AKSEQEIGKA TAKYFFYSNI MNFFKTEITL ANGEIRKRPL IETNGETGEI
VWDKGRDFAT VRKVLSMPQV NIVKKTEVQT GGFSKESILP KRNSDKLIAR KKDWDPKKYG
GFDSPTVAYS VLVVAKVEKG KSKKLKSVKE LLGITIMERS SFEKNPIDFL EAKGYKEVKK
DLIIKLPKYS LFELENGRKR MLASAGELQK GNELALPSKY VNFLYLASHY EKLKGSPEDN
EQKQLFVEQH KHYLDEIIEQ ISEFSKRVIL ADANLDKVLS AYNKHRDKPI REQAENIIHL
FTLTNLGAPA AFKYFDTTID RKRYTSTKEV LDATLIHQSI TGLYETRIDL SQLGGDSRAD
PKKKRKVHHH HHH
In certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of iProt106519 (SEQ ID NO: 125):
MGSSHHHHHH HHENLYFQGS MDKKYSIGLD IGTNSVGWAV ITDEYKVPSK KFKVLGNTDR
HSIKKNLIGA LLFDSGETAE ATRLKRTARR RYTRRKNRIC YLQEIFSNEM AKVDDSFFHR
LEESFLVEED KKHERHPIFG NIVDEVAYHE KYPTIYHLRK KLVDSTDKAD LRLIYLALAH
MIKFRGHFLI EGDLNPDNSD VDKLFIQLVQ TYNQLFEENP INASGVDAKA ILSARLSKSR
RLENLIAQLP GEKKNGLFGN LIALSLGLTP NFKSNFDLAE DAKLQLSKDT YDDDLDNLLA
QIGDQYADLF LAAKNLSDAI LLSDILRVNT EITKAPLSAS MIKRYDEHHQ DLTLLKALVR
QQLPEKYKEI FFDQSKNGYA GYIDGGASQE EFYKFIKPIL EKMDGTEELL VKLNREDLLR
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
180
KQRTFDNGSI PHQIHLGELH AILRRQEDFY PFLKDNREKI EKILTFRIPY YVGPLARGNS
RFAWMTRKSE ETITPWNFEE VVDKGASAQS FIERMTNFDK NLPNEKVLPK HSLLYEYFTV
YNELTKVKYV TEGMRKPAFL SGEQKKAIVD LLFKTNRKVT VKQLKEDYFK KIECFDSVEI
SGVEDRFNAS LGTYHDLLKI IKDKDFLDNE ENEDILEDIV LTLTLFEDRE MIEERLKTYA
HLFDDKVMKQ LKRRRYTGWG RLSRKLINGI RDKQSGKTIL DFLKSDGFAN RNFMQLIHDD
SLTFKEDIQK AQVSGQGDSL HEHIANLAGS PAIKKGILQT VKVVDELVKV MGRHKPENIV
IEMARENQTT QKGQKNSRER MKRIEEGIKE LGSQILKEHP VENTQLQNEK LYLYYLQNGR
DMYVDQELDI NRLSDYDVDH IVPQSFLKDD SIDNKVLTRS DKNRGKSDNV PSEEVVKKMK
NYWRQLLNAK LITQRKFDNL TKAERGGLSE LDKAGFIKRQ LVETRQITKH VAQILDSRMN
TKYDENDKLI REVKVITLKS KLVSDFRKDF QFYKVREINN YHHAHDAYLN AVVGTALIKK
YPKLESEFVY GDYKVYDVRK MIAKSEQEIG KATAKYFFYS NIMNFFKTEI TLANGEIRKR
PLIETNGETG EIVWDKGRDF ATVRKVLSMP QVNIVKKTEV QTGGFSKESI LPKRNSDKLI
ARKKDWDPKK YGGFDSPTVA YSVLVVAKVE KGKSKKLKSV KELLGITIME RSSFEKNPID
FLEAKGYKEV KKDLIIKLPK YSLFELENGR KRMLASAGEL QKGNELALPS KYVNFLYLAS
HYEKLKGSPE DNEQKQLFVE QHKHYLDEII EQISEFSKRV ILADANLDKV LSAYNKHRDK
PIREQAENII HLFTLTNLGA PAAFKYFDTT IDRKRYTSTK EVLDATLIHQ SITGLYETRI
DLSQLGGDGG GSPKKKRKV
In certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of iProt106520 (SEQ ID NO: 126):
MAHHHHHHGG SPKKKRKVDK KYSIGLDIGT NSVGWAVITD EYKVPSKKFK VLGNTDRHSI
KKNLIGALLF DSGETAEATR LKRTARRRYT RRKNRICYLQ EIFSNEMAKV DDSFFHRLEE
SFLVEEDKKH ERHPIFGNIV DEVAYHEKYP TIYHLRKKLV DSTDKADLRL IYLALAHMIK
FRGHFLIEGD LNPDNSDVDK LFIQLVQTYN QLFEENPINA SGVDAKAILS ARLSKSRRLE
NLIAQLPGEK KNGLFGNLIA LSLGLTPNFK SNFDLAEDAK LQLSKDTYDD DLDNLLAQIG
DQYADLFLAA KNLSDAILLS DILRVNTEIT KAPLSASMIK RYDEHHQDLT LLKALVRQQL
PEKYKEIFFD QSKNGYAGYI DGGASQEEFY KFIKPILEKM DGTEELLVKL NREDLLRKQR
TFDNGSIPHQ IHLGELHAIL RRQEDFYPFL KDNREKIEKI LTFRIPYYVG PLARGNSRFA
WMTRKSEETI TPWNFEEVVD KGASAQSFIE RMTNFDKNLP NEKVLPKHSL LYEYFTVYNE
LTKVKYVTEG MRKPAFLSGE QKKAIVDLLF KTNRKVTVKQ LKEDYFKKIE CFDSVEISGV
EDRFNASLGT YHDLLKIIKD KDFLDNEENE DILEDIVLTL TLFEDREMIE ERLKTYAHLF
DDKVMKQLKR RRYTGWGRLS RKLINGIRDK QSGKTILDFL KSDGFANRNF MQLIHDDSLT
FKEDIQKAQV SGQGDSLHEH IANLAGSPAI KKGILQTVKV VDELVKVMGR HKPENIVIEM
ARENQTTQKG QKNSRERMKR IEEGIKELGS QILKEHPVEN TQLQNEKLYL YYLQNGRDMY
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
181
VDQELDINRL SDYDVDHIVP QSFLKDDSID NKVLTRSDKN RGKSDNVPSE EVVKKMKNYW
RQLLNAKLIT QRKFDNLTKA ERGGLSELDK AGFIKRQLVE TRQITKHVAQ ILDSRMNTKY
DENDKLIREV KVITLKSKLV SDFRKDFQFY KVREINNYHH AHDAYLNAVV GTALIKKYPK
LESEFVYGDY KVYDVRKMIA KSEQEIGKAT AKYFFYSNIM NFFKTEITLA NGEIRKRPLI
ETNGETGEIV WDKGRDFATV RKVLSMPQVN IVKKTEVQTG GFSKESILPK RNSDKLIARK
KDWDPKKYGG FDSPTVAYSV LVVAKVEKGK SKKLKSVKEL LGITIMERSS FEKNPIDFLE
AKGYKEVKKD LIIKLPKYSL FELENGRKRM LASAGELQKG NELALPSKYV NFLYLASHYE
KLKGSPEDNE QKQLFVEQHK HYLDEIIEQI SEFSKRVILA DANLDKVLSA YNKHRDKPIR
EQAENIIHLF TLTNLGAPAA FKYFDTTIDR KRYTSTKEVL DATLIHQSIT GLYETRIDLS
QLGGDSRADP KKKRKV
In certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of iProt106521 (SEQ ID NO: 127):
MAPKKKRKVD KKYSIGLDIG TNSVGWAVIT DEYKVPSKKF KVLGNTDRHS IKKNLIGALL
FDSGETAEAT RLKRTARRRY TRRKNRICYL QEIFSNEMAK VDDSFFHRLE ESFLVEEDKK
HERHPIFGNI VDEVAYHEKY PTIYHLRKKL VDSTDKADLR LIYLALAHMI KFRGHFLIEG
DLNPDNSDVD KLFIQLVQTY NQLFEENPIN ASGVDAKAIL SARLSKSRRL ENLIAQLPGE
KKNGLFGNLI ALSLGLTPNF KSNFDLAEDA KLQLSKDTYD DDLDNLLAQI GDQYADLFLA
AKNLSDAILL SDILRVNTEI TKAPLSASMI KRYDEHHQDL TLLKALVRQQ LPEKYKEIFF
DQSKNGYAGY IDGGASQEEF YKFIKPILEK MDGTEELLVK LNREDLLRKQ RTFDNGSIPH
QIHLGELHAI LRRQEDFYPF LKDNREKIEK ILTFRIPYYV GPLARGNSRF AWMTRKSEET
ITPWNFEEVV DKGASAQSFI ERMTNFDKNL PNEKVLPKHS LLYEYFTVYN ELTKVKYVTE
GMRKPAFLSG EQKKAIVDLL FKTNRKVTVK QLKEDYFKKI ECFDSVEISG VEDRFNASLG
TYHDLLKIIK DKDFLDNEEN EDILEDIVLT LTLFEDREMI EERLKTYAHL FDDKVMKQLK
RRRYTGWGRL SRKLINGIRD KQSGKTILDF LKSDGFANRN FMQLIHDDSL TFKEDIQKAQ
VSGQGDSLHE HIANLAGSPA IKKGILQTVK VVDELVKVMG RHKPENIVIE MARENQTTQK
GQKNSRERMK RIEEGIKELG SQILKEHPVE NTQLQNEKLY LYYLQNGRDM YVDQELDINR
LSDYDVDHIV PQSFLKDDSI DNKVLTRSDK NRGKSDNVPS EEVVKKMKNY WRQLLNAKLI
TQRKFDNLTK AERGGLSELD KAGFIKRQLV ETRQITKHVA QILDSRMNTK YDENDKLIRE
VKVITLKSKL VSDFRKDFQF YKVREINNYH HAHDAYLNAV VGTALIKKYP KLESEFVYGD
YKVYDVRKMI AKSEQEIGKA TAKYFFYSNI MNFFKTEITL ANGEIRKRPL IETNGETGEI
VWDKGRDFAT VRKVLSMPQV NIVKKTEVQT GGFSKESILP KRNSDKLIAR KKDWDPKKYG
GFDSPTVAYS VLVVAKVEKG KSKKLKSVKE LLGITIMERS SFEKNPIDFL EAKGYKEVKK
DLIIKLPKYS LFELENGRKR MLASAGELQK GNELALPSKY VNFLYLASHY EKLKGSPEDN
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
182
EQKQLFVEQH KHYLDEIIEQ ISEFSKRVIL ADANLDKVLS AYNKHRDKPI REQAENIIHL
FTLTNLGAPA AFKYFDTTID RKRYTSTKEV LDATLIHQSI TGLYETRIDL SQLGGDSRAD
HHHHHH
In certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of iProt106522 (SEQ ID NO: 128):
MAHHHHHHGG SDKKYSIGLD IGTNSVGWAV ITDEYKVPSK KFKVLGNTDR HSIKKNLIGA
LLFDSGETAE ATRLKRTARR RYTRRKNRIC YLQEIFSNEM AKVDDSFFHR LEESFLVEED
KKHERHPIFG NIVDEVAYHE KYPTIYHLRK KLVDSTDKAD LRLIYLALAH MIKFRGHFLI
EGDLNPDNSD VDKLFIQLVQ TYNQLFEENP INASGVDAKA ILSARLSKSR RLENLIAQLP
GEKKNGLFGN LIALSLGLTP NFKSNFDLAE DAKLQLSKDT YDDDLDNLLA QIGDQYADLF
LAAKNLSDAI LLSDILRVNT EITKAPLSAS MIKRYDEHHQ DLTLLKALVR QQLPEKYKEI
FFDQSKNGYA GYIDGGASQE EFYKFIKPIL EKMDGTEELL VKLNREDLLR KQRTFDNGSI
PHQIHLGELH AILRRQEDFY PFLKDNREKI EKILTFRIPY YVGPLARGNS RFAWMTRKSE
ETITPWNFEE VVDKGASAQS FIERMTNFDK NLPNEKVLPK HSLLYEYFTV YNELTKVKYV
TEGMRKPAFL SGEQKKAIVD LLFKTNRKVT VKQLKEDYFK KIECFDSVEI SGVEDRFNAS
LGTYHDLLKI IKDKDFLDNE ENEDILEDIV LTLTLFEDRE MIEERLKTYA HLFDDKVMKQ
LKRRRYTGWG RLSRKLINGI RDKQSGKTIL DFLKSDGFAN RNFMQLIHDD SLTFKEDIQK
AQVSGQGDSL HEHIANLAGS PAIKKGILQT VKVVDELVKV MGRHKPENIV IEMARENQTT
QKGQKNSRER MKRIEEGIKE LGSQILKEHP VENTQLQNEK LYLYYLQNGR DMYVDQELDI
NRLSDYDVDH IVPQSFLKDD SIDNKVLTRS DKNRGKSDNV PSEEVVKKMK NYWRQLLNAK
LITQRKFDNL TKAERGGLSE LDKAGFIKRQ LVETRQITKH VAQILDSRMN TKYDENDKLI
REVKVITLKS KLVSDFRKDF QFYKVREINN YHHAHDAYLN AVVGTALIKK YPKLESEFVY
GDYKVYDVRK MIAKSEQEIG KATAKYFFYS NIMNFFKTEI TLANGEIRKR PLIETNGETG
EIVWDKGRDF ATVRKVLSMP QVNIVKKTEV QTGGFSKESI LPKRNSDKLI ARKKDWDPKK
YGGFDSPTVA YSVLVVAKVE KGKSKKLKSV KELLGITIME RSSFEKNPID FLEAKGYKEV
KKDLIIKLPK YSLFELENGR KRMLASAGEL QKGNELALPS KYVNFLYLAS HYEKLKGSPE
DNEQKQLFVE QHKHYLDEII EQISEFSKRV ILADANLDKV LSAYNKHRDK PIREQAENII
HLFTLTNLGA PAAFKYFDTT IDRKRYTSTK EVLDATLIHQ SITGLYETRI DLSQLGGDSR
ADPKKKRKV
In certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of iProt106658 (SEQ ID NO: 129):
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
183
MGSSHHHHHH HHENLYFQGS MDKKYSIGLD IGTNSVGWAV ITDEYKVPSK KFKVLGNTDR
HSIKKNLIGA LLFDSGETAE ATRLKRTARR RYTRRKNRIC YLQEIFSNEM AKVDDSFFHR
LEESFLVEED KKHERHPIFG NIVDEVAYHE KYPTIYHLRK KLVDSTDKAD LRLIYLALAH
MIKFRGHFLI EGDLNPDNSD VDKLFIQLVQ TYNQLFEENP INASGVDAKA ILSARLSKSR
RLENLIAQLP GEKKNGLFGN LIALSLGLTP NFKSNFDLAE DAKLQLSKDT YDDDLDNLLA
QIGDQYADLF LAAKNLSDAI LLSDILRVNT EITKAPLSAS MIKRYDEHHQ DLTLLKALVR
QQLPEKYKEI FFDQSKNGYA GYIDGGASQE EFYKFIKPIL EKMDGTEELL VKLNREDLLR
KQRTFDNGSI PHQIHLGELH AILRRQEDFY PFLKDNREKI EKILTFRIPY YVGPLARGNS
RFAWMTRKSE ETITPWNFEE VVDKGASAQS FIERMTNFDK NLPNEKVLPK HSLLYEYFTV
YNELTKVKYV TEGMRKPAFL SGEQKKAIVD LLFKTNRKVT VKQLKEDYFK KIECFDSVEI
SGVEDRFNAS LGTYHDLLKI IKDKDFLDNE ENEDILEDIV LTLTLFEDRE MIEERLKTYA
HLFDDKVMKQ LKRRRYTGWG RLSRKLINGI RDKQSGKTIL DFLKSDGFAN RNFMQLIHDD
SLTFKEDIQK AQVSGQGDSL HEHIANLAGS PAIKKGILQT VKVVDELVKV MGRHKPENIV
IEMARENQTT QKGQKNSRER MKRIEEGIKE LGSQILKEHP VENTQLQNEK LYLYYLQNGR
DMYVDQELDI NRLSDYDVDH IVPQSFLKDD SIDNKVLTRS DKNRGKSDNV PSEEVVKKMK
NYWRQLLNAK LITQRKFDNL TKAERGGLSE LDKAGFIKRQ LVETRQITKH VAQILDSRMN
TKYDENDKLI REVKVITLKS KLVSDFRKDF QFYKVREINN YHHAHDAYLN AVVGTALIKK
YPKLESEFVY GDYKVYDVRK MIAKSEQEIG KATAKYFFYS NIMNFFKTEI TLANGEIRKR
PLIETNGETG EIVWDKGRDF ATVRKVLSMP QVNIVKKTEV QTGGFSKESI LPKRNSDKLI
ARKKDWDPKK YGGFDSPTVA YSVLVVAKVE KGKSKKLKSV KELLGITIME RSSFEKNPID
FLEAKGYKEV KKDLIIKLPK YSLFELENGR KRMLASAGEL QKGNELALPS KYVNFLYLAS
HYEKLKGSPE DNEQKQLFVE QHKHYLDEII EQISEFSKRV ILADANLDKV LSAYNKHRDK
PIREQAENII HLFTLTNLGA PAAFKYFDTT IDRKRYTSTK EVLDATLIHQ SITGLYETRI
DLSQLGGDGG GSPKKKRKV
In certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of iProt106745 (SEQ ID NO: 130):
MAPKKKRKVD KKYSIGLDIG TNSVGWAVIT DEYKVPSKKF KVLGNTDRHS IKKNLIGALL
FDSGETAEAT RLKRTARRRY TRRKNRICYL QEIFSNEMAK VDDSFFHRLE ESFLVEEDKK
HERHPIFGNI VDEVAYHEKY PTIYHLRKKL VDSTDKADLR LIYLALAHMI KFRGHFLIEG
DLNPDNSDVD KLFIQLVQTY NQLFEENPIN ASGVDAKAIL SARLSKSRRL ENLIAQLPGE
KKNGLFGNLI ALSLGLTPNF KSNFDLAEDA KLQLSKDTYD DDLDNLLAQI GDQYADLFLA
AKNLSDAILL SDILRVNTEI TKAPLSASMI KRYDEHHQDL TLLKALVRQQ LPEKYKEIFF
DQSKNGYAGY IDGGASQEEF YKFIKPILEK MDGTEELLVK LNREDLLRKQ RTFDNGSIPH
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
184
QIHLGELHAI LRRQEDFYPF LKDNREKIEK ILTFRIPYYV GPLARGNSRF AWMTRKSEET
ITPWNFEEVV DKGASAQSFI ERMTNFDKNL PNEKVLPKHS LLYEYFTVYN ELTKVKYVTE
GMRKPAFLSG EQKKAIVDLL FKTNRKVTVK QLKEDYFKKI ECFDSVEISG VEDRFNASLG
TYHDLLKIIK DKDFLDNEEN EDILEDIVLT LTLFEDREMI EERLKTYAHL FDDKVMKQLK
RRRYTGWGRL SRKLINGIRD KQSGKTILDF LKSDGFANRN FMQLIHDDSL TFKEDIQKAQ
VSGQGDSLHE HIANLAGSPA IKKGILQTVK VVDELVKVMG RHKPENIVIE MARENQTTQK
GQKNSRERMK RIEEGIKELG SQILKEHPVE NTQLQNEKLY LYYLQNGRDM YVDQELDINR
LSDYDVDHIV PQSFLKDDSI DNAVLTRSDK NRGKSDNVPS EEVVKKMKNY WRQLLNAKLI
TQRKFDNLTK AERGGLSELD KAGFIKRQLV ETRQITKHVA QILDSRMNTK YDENDKLIRE
VKVITLKSKL VSDFRKDFQF YKVREINNYH HAHDAYLNAV VGTALIKKYP KLESEFVYGD
YKVYDVRKMI AKSEQEIGKA TAKYFFYSNI MNFFKTEITL ANGEIRKRPL IETNGETGEI
VWDKGRDFAT VRKVLSMPQV NIVKKTEVQT GGFSKESILP KRNSDKLIAR KKDWDPKKYG
GFDSPTVAYS VLVVAKVEKG KSKKLKSVKE LLGITIMERS SFEKNPIDFL EAKGYKEVKK
DLIIKLPKYS LFELENGRKR MLASAGELQK GNELALPSKY VNFLYLASHY EKLKGSPEDN
EQKQLFVEQH KHYLDEIIEQ ISEFSKRVIL ADANLDKVLS AYNKHRDKPI REQAENIIHL
FTLTNLGAPA AFKYFDTTID RKRYTSTKEV LDATLIHQSI TGLYETRIDL SQLGGDSRAD
PKKKRKVHHH HHH
In certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of iProt106746 (SEQ ID NO: 131):
MAPKKKRKVD KKYSIGLDIG TNSVGWAVIT DEYKVPSKKF KVLGNTDRHS IKKNLIGALL
FDSGETAEAT RLKRTARRRY TRRKNRICYL QEIFSNEMAK VDDSFFHRLE ESFLVEEDKK
HERHPIFGNI VDEVAYHEKY PTIYHLRKKL VDSTDKADLR LIYLALAHMI KFRGHFLIEG
DLNPDNSDVD KLFIQLVQTY NQLFEENPIN ASGVDAKAIL SARLSKSRRL ENLIAQLPGE
KKNGLFGNLI ALSLGLTPNF KSNFDLAEDA KLQLSKDTYD DDLDNLLAQI GDQYADLFLA
AKNLSDAILL SDILRVNTEI TKAPLSASMI KRYDEHHQDL TLLKALVRQQ LPEKYKEIFF
DQSKNGYAGY IDGGASQEEF YKFIKPILEK MDGTEELLVK LNREDLLRKQ RTFDNGSIPH
QIHLGELHAI LRRQEDFYPF LKDNREKIEK ILTFRIPYYV GPLARGNSRF AWMTRKSEET
ITPWNFEEVV DKGASAQSFI ERMTNFDKNL PNEKVLPKHS LLYEYFTVYN ELTKVKYVTE
GMRKPAFLSG EQKKAIVDLL FKTNRKVTVK QLKEDYFKKI ECFDSVEISG VEDRFNASLG
TYHDLLKIIK DKDFLDNEEN EDILEDIVLT LTLFEDREMI EERLKTYAHL FDDKVMKQLK
RRRYTGWGRL SRKLINGIRD KQSGKTILDF LKSDGFANRN FMQLIHDDSL TFKEDIQKAQ
VSGQGDSLHE HIANLAGSPA IKKGILQTVK VVDELVKVMG RHKPENIVIE MARENQTTQK
GQKNSRERMK RIEEGIKELG SQILKEHPVE NTQLQNEALY LYYLQNGRDM YVDQELDINR
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
185
LSDYDVDHIV PQSFLKDDSI DNKVLTRSDK NRGKSDNVPS EEVVKKMKNY WRQLLNAKLI
TQRKFDNLTK AERGGLSELD KAGFIKRQLV ETRQITKHVA QILDSRMNTK YDENDKLIRE
VKVITLKSKL VSDFRKDFQF YKVREINNYH HAHDAYLNAV VGTALIKKYP ALESEFVYGD
YKVYDVRKMI AKSEQEIGKA TAKYFFYSNI MNFFKTEITL ANGEIRKAPL IETNGETGEI
VWDKGRDFAT VRKVLSMPQV NIVKKTEVQT GGFSKESILP KRNSDKLIAR KKDWDPKKYG
GFDSPTVAYS VLVVAKVEKG KSKKLKSVKE LLGITIMERS SFEKNPIDFL EAKGYKEVKK
DLIIKLPKYS LFELENGRKR MLASAGELQK GNELALPSKY VNFLYLASHY EKLKGSPEDN
EQKQLFVEQH KHYLDEIIEQ ISEFSKRVIL ADANLDKVLS AYNKHRDKPI REQAENIIHL
FTLTNLGAPA AFKYFDTTID RKRYTSTKEV LDATLIHQSI TGLYETRIDL SQLGGDSRAD
PKKKRKVHHH HHH
In certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of iProt106747 (SEQ ID NO: 132):
MAPKKKRKVD KKYSIGLDIG TNSVGWAVIT DEYKVPSKKF KVLGNTDRHS IKKNLIGALL
FDSGETAEAT RLKRTARRRY TRRKNRICYL QEIFSNEMAK VDDSFFHRLE ESFLVEEDKK
HERHPIFGNI VDEVAYHEKY PTIYHLRKKL VDSTDKADLR LIYLALAHMI KFRGHFLIEG
DLNPDNSDVD KLFIQLVQTY NQLFEENPIN ASGVDAKAIL SARLSKSRRL ENLIAQLPGE
KKNGLFGNLI ALSLGLTPNF KSNFDLAEDA KLQLSKDTYD DDLDNLLAQI GDQYADLFLA
AKNLSDAILL SDILRVNTEI TKAPLSASMI KRYDEHHQDL TLLKALVRQQ LPEKYKEIFF
DQSKNGYAGY IDGGASQEEF YKFIKPILEK MDGTEELLVK LNREDLLRKQ RTFDNGSIPH
QIHLGELHAI LRRQEDFYPF LKDNREKIEK ILTFRIPYYV GPLARGNSRF AWMTRKSEET
ITPWNFEEVV DKGASAQSFI ERMTNFDKNL PNEKVLPKHS LLYEYFTVYN ELTKVKYVTE
GMRKPAFLSG EQKKAIVDLL FKTNRKVTVK QLKEDYFKKI ECFDSVEISG VEDRFNASLG
TYHDLLKIIK DKDFLDNEEN EDILEDIVLT LTLFEDREMI EERLKTYAHL FDDKVMKQLK
RRRYTGWGRL SRKLINGIRD KQSGKTILDF LKSDGFANRN FMQLIHDDSL TFKEDIQKAQ
VSGQGDSLHE HIANLAGSPA IKKGILQTVK VVDELVKVMG RHKPENIVIE MARENQTTQK
GQKNSRERMK RIEEGIKELG SQILKEHPVE NTQLQNEKLY LYYLQNGRDM YVDQELDINR
LSDYDVDHIV PQSFLADDSI DNKVLTRSDK NRGKSDNVPS EEVVKKMKNY WRQLLNAKLI
TQRKFDNLTK AERGGLSELD KAGFIKRQLV ETRQITKHVA QILDSRMNTK YDENDKLIRE
VKVITLKSKL VSDFRKDFQF YKVREINNYH HAHDAYLNAV VGTALIKKYP ALESEFVYGD
YKVYDVRKMI AKSEQEIGKA TAKYFFYSNI MNFFKTEITL ANGEIRKAPL IETNGETGEI
VWDKGRDFAT VRKVLSMPQV NIVKKTEVQT GGFSKESILP KRNSDKLIAR KKDWDPKKYG
GFDSPTVAYS VLVVAKVEKG KSKKLKSVKE LLGITIMERS SFEKNPIDFL EAKGYKEVKK
DLIIKLPKYS LFELENGRKR MLASAGELQK GNELALPSKY VNFLYLASHY EKLKGSPEDN
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
186
EQKQLFVEQH KHYLDEIIEQ ISEFSKRVIL ADANLDKVLS AYNKHRDKPI REQAENIIHL
FTLTNLGAPA AFKYFDTTID RKRYTSTKEV LDATLIHQSI TGLYETRIDL SQLGGDSRAD
PKKKRKVHHH HHH
In certain embodiments, a Cas9 protein used in a method or composition of the
present
invention has the sequence of iProt106884 (SEQ ID NO: 133):
MAPKKKRKVD KKYSIGLDIG TNSVGWAVIT DEYKVPSKKF KVLGNTDRHS IKKNLIGALL
FDSGETAEAT RLKRTARRRY TRRKNRICYL QEIFSNEMAK VDDSFFHRLE ESFLVEEDKK
HERHPIFGNI VDEVAYHEKY PTIYHLRKKL VDSTDKADLR LIYLALAHMI KFRGHFLIEG
DLNPDNSDVD KLFIQLVQTY NQLFEENPIN ASGVDAKAIL SARLSKSRRL ENLIAQLPGE
KKNGLFGNLI ALSLGLTPNF KSNFDLAEDA KLQLSKDTYD DDLDNLLAQI GDQYADLFLA
AKNLSDAILL SDILRVNTEI TKAPLSASMI KRYDEHHQDL TLLKALVRQQ LPEKYKEIFF
DQSKNGYAGY IDGGASQEEF YKFIKPILEK MDGTEELLVK LNREDLLRKQ RTFDNGSIPH
QIHLGELHAI LRRQEDFYPF LKDNREKIEK ILTFRIPYYV GPLARGNSRF AWMTRKSEET
ITPWNFEEVV DKGASAQSFI ERMTAFDKNL PNEKVLPKHS LLYEYFTVYN ELTKVKYVTE
GMRKPAFLSG EQKKAIVDLL FKTNRKVTVK QLKEDYFKKI ECFDSVEISG VEDRFNASLG
TYHDLLKIIK DKDFLDNEEN EDILEDIVLT LTLFEDREMI EERLKTYAHL FDDKVMKQLK
RRRYTGWGAL SRKLINGIRD KQSGKTILDF LKSDGFANRN FMALIHDDSL TFKEDIQKAQ
VSGQGDSLHE HIANLAGSPA IKKGILQTVK VVDELVKVMG RHKPENIVIE MARENQTTQK
GQKNSRERMK RIEEGIKELG SQILKEHPVE NTQLQNEKLY LYYLQNGRDM YVDQELDINR
LSDYDVDHIV PQSFLKDDSI DNKVLTRSDK NRGKSDNVPS EEVVKKMKNY WRQLLNAKLI
TQRKFDNLTK AERGGLSELD KAGFIKRQLV ETRAITKHVA QILDSRMNTK YDENDKLIRE
VKVITLKSKL VSDFRKDFQF YKVREINNYH HAHDAYLNAV VGTALIKKYP KLESEFVYGD
YKVYDVRKMI AKSEQEIGKA TAKYFFYSNI MNFFKTEITL ANGEIRKRPL IETNGETGEI
VWDKGRDFAT VRKVLSMPQV NIVKKTEVQT GGFSKESILP KRNSDKLIAR KKDWDPKKYG
GFDSPTVAYS VLVVAKVEKG KSKKLKSVKE LLGITIMERS SFEKNPIDFL EAKGYKEVKK
DLIIKLPKYS LFELENGRKR MLASAGELQK GNELALPSKY VNFLYLASHY EKLKGSPEDN
EQKQLFVEQH KHYLDEIIEQ ISEFSKRVIL ADANLDKVLS AYNKHRDKPI REQAENIIHL
FTLTNLGAPA AFKYFDTTID RKRYTSTKEV LDATLIHQSI TGLYETRIDL SQLGGDSRAD
PKKKRKVHHH HHH
In a preferred embodoiment, the CRISPR system used in the present invention
comprises a
Cas9 molecule comprising SEQ ID NO: 106 or 107or 107.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
187
Thus, engineered CRISPR gene editing systems, e.g., for gene editing in
eukaryotic cells,
typically involve (1) a guide RNA molecule (gRNA) comprising a targeting
domain (which is
capable of hybridizing to the genomic DNA target sequence), and a sequence
which is
capable of binding to a Cas, e.g., Cas9 enzyme, and (2) a Cas, e.g., Cas9,
protein. The
sequence which is capable of binding to a Cas protein may comprise a domain
referred to
as a tracr domain or tracrRNA. The targeting domain and the sequence which is
capable of
binding to a Cas, e.g., Cas9 enzyme, may be disposed on the same (sometimes
referred to
as a single gRNA, chimeric gRNA or sgRNA) or different molecules (sometimes
referred to
as a dual gRNA or dgRNA). If disposed on different molecules, each includes a
hybridization
domain which allows the molecules to associate, e.g., through hybridization.
dRNA
The terms "guide RNA", "guide RNA molecule", "gRNA molecule" or "gRNA" are
used
interchangeably, and refer to a set of nucleic acid molecules that promote the
specific
directing of a RNA-guided nuclease or other effector molecule (typically in
complex with the
gRNA molecule) to a target sequence. In some embodiments, said directing is
accomplished
through hybridization of a portion of the gRNA to DNA (e.g., through the gRNA
targeting
domain), and by binding of a portion of the gRNA molecule to the RNA-guided
nuclease or
other effector molecule (e.g., through at least the gRNA tracr). In
embodiments, a gRNA
molecule consists of a single contiguous polynucleotide molecule, referred to
herein as a
"single guide RNA" or "sgRNA" and the like. In other embodiments, a gRNA
molecule
consists of a plurality, usually two, polynucleotide molecules, which are
themselves capable
of association, usually through hybridization, referred to herein as a "dual
guide RNA" or
"dgRNA" and the like. gRNA molecules are described in more detail below, but
generally
include a targeting domain and a tracr. In embodiments the targeting domain
and tracr are
disposed on a single polynucleotide. In other embodiments, the targeting
domain and tracr
are disposed on separate polynucleotides.
The term "targeting domain" as the term is used in connection with a gRNA, is
the portion of
the gRNA molecule that recognizes, e.g., is complementary to, a target
sequence, e.g., a
target sequence within the nucleic acid of a cell, e.g., within a gene.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
188
The term "crRNA" as the term is used in connection with a gRNA molecule, is a
portion of
the gRNA molecule that comprises a targeting domain and a region that
interacts with a
tracr to form a flagpole region.
The term "flagpole" as used herein in connection with a gRNA molecule, refers
to the portion
of the gRNA where the crRNA and the tracr bind to, or hybridize to, one
another.
The term "tracr" as used herein in connection with a gRNA molecule, refers to
the portion of
the gRNA that binds to a nuclease or other effector molecule. In embodiements,
the tracr
comprises nucleic acid sequence that binds specifically to Cas9. In
embodiments, the tracr
comprises nucleic acid sequence that forms part of the flagpole.
The term "target sequence" refers to a sequence of nucleic acids
complimentary, for
example fully complementary, to a gRNA targeting domain. In embodiments, the
target
sequence is disposed on genomic DNA. In an embodiment the target sequence is
adjacent
to (either on the same strand or on the complementary strand of DNA) a
protospacer
adjacent motif (PAM) sequence recognized by a protein having nuclease or other
effector
activity, e.g., a PAM sequence recognized by Cas9. The target sequence refers
herein to a
target sequence of beta-2-microglobulin or B2M.
The term "complementary" as used in connection with nucleic acid, refers to
the pairing of
bases, A with T or U, and G with C. The term complementary refers to nucleic
acid
molecules that are completely complementary, that is, form A to T or U pairs
and G to C
pairs across the entire reference sequence, as well as molecules that are at
least 80%,
85%, 90%, 95%, 99% complementary.
"Beta-2-microglobulin" or "B2M", also known as IMD43, is a component of MHC
class I
molecules. B2M is a serum protein found in association with the major
histocompatibility
complex (MHC) class I heavy chain on the surface of nearly all nucleated
cells. The protein
has a predominantly beta-pleated sheet structure that can form amyloid fibrils
in some
pathological conditions. The encoded antimicrobial protein displays
antibacterial activity in
amniotic fluid. A mutation in this gene has been shown to result in
hypercatabolic
hypoproteinemia (NCBI: Gene ID: 567).
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
189
The term "a target sequence in the B2M gene" or "a target polynucleotide
sequence in the
B2M gene" refers to a contigious sequence within the B2M polynucleotide
sequence (NCBI:
Gene ID: 567). The B2M polynucleotide sequence encodes B2M protein, a serum
protein
found in association with the major histocompatibility complex (MHC) class I
heavy chain on
the surface of nearly all nucleated cells. The B2M gene has 4 exons which span
approximately 8 kb.
In some embodiments, the target polynucleotide sequence is a variant of B2M.
in some
embodiments, the target polynucleotide sequence is a homolog of B2M, In some
embodiments, the target polynucleotide sequence is an ortholog of B2M.
The term "genomic DNA of B2M" refers to the B2M polynucleotide sequence (NCBI:
Gene
ID: 567).
gRNA molecule formats are known in the art. An exemplary gRNA molecule, e.g.,
dgRNA
molecule, as disclosed herein comprises, e.g., consists of, a first nucleic
acid having the
sequence:
5'nnnnnnnnnnnnnnnnnnnnGUUUUAGAGCUAUGCUGUUUUG 3' (SEQ ID NO: 9),
where the "n"s refer to the residues of the targeting domain, e.g., as
described herein, and
may consist of 15-25 nucleotides, e.g., consists of 20 nucleotides;
and a second nucleic acid sequence having the exemplary sequence:
5'AACUUACCAAGGAACAGCAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUG
AAAAAGUGGCACCGAGUCGGUGC 3', optionally with 1, 2, 3, 4, 5, 6, or 7 (e.g., 4 or
7,
e.g., 7) additional U nucleotides at the 3' end (SEQ ID NO: 10).
The second nucleic acid molecule may alternatively consist of a fragment of
the sequence
above, wherein such fragment is capable of hybridizing to the first nucleic
acid. An example
of such second nucleic acid molecule is:
5'AACAGCAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCAC
CGAGUCGGUGC 3', optionally with 1, 2, 3, 4, 5, 6, or 7 (e.g., 4 or 7, e.g., 7)
additional U
nucleotides at the 3' end (SEQ ID NO:11).
Another exemplary gRNA molecule, e.g., a sgRNA molecule, as disclosed herein
comprises,
e.g., consists of a first nucleic acid having the sequence:
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
190
5'nnnnnnnnnnnnnnnnnnnGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCC
GUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGC 3'(SEQ ID NO:12), where the "n"s
refer to the residues of the targeting domain, e.g., as described herein, and
may consist of
15-25 nucleotides, e.g., consist of 20 nucleotides, optionally with 1, 2, 3,
4, 5, 6, or 7 (e.g., 4
or 7, e.g., 4) additional U nucleotides at the 3' end.
Additional components and/or elements of CRISPR gene editing systems known in
the art,
e.g., are described in U.S. Publication No.2014/0068797, W02015/048577, and
Cong
(2013) Science 339: 819-823, the contents of which are hereby incorporated by
reference in
__ their entirety. Such systems can be generated which inhibit a target gene,
by, for example,
engineering a CRISPR gene editing system to include a gRNA molecule comprising
a
targeting domain that hybridizes to a sequence of the target gene. In
embodiments, the
gRNA comprises a targeting domain which is fully complementarity to 15-25
nucleotides,
e.g., 20 nucleotides, of a target gene. In embodiments, the 15-25 nucleotides,
e.g., 20
nucleotides, of the target gene, are disposed immediately 5' to a protospacer
adjacent motif
(PAM) sequence recognized by the RNA-guided nuclease, e.g., Cas protein, of
the CRISPR
gene editing system (e.g., where the system comprises a S. pyogenes Cas9
protein, the
PAM sequence comprises NGG, where N can be any of A, T, G or C).
In some embodiments, the gRNA molecule and RNA-guided nuclease, e.g., Cas
protein, of
the CRISPR gene editing system can be complexed to form a RNP
(ribonucleoprotein)
complex. Such RNP complexes may be used in the methods described herein. In
other
embodiments, nucleic acid encoding one or more components of the CRISPR gene
editing
system may be used in the methods described herein.
In some embodiments, foreign DNA can be introduced into the cell along with
the CRISPR
gene editing system, e.g., DNA encoding a desired transgene, with or without a
promoter
active in the target cell type. Depending on the sequences of the foreign DNA
and target
sequence of the genome, this process can be used to integrate the foreign DNA
into the
genome, at or near the site targeted by the CRISPR gene editing system. For
example, 3'
and 5' sequences flanking the transgene may be included in the foreign DNA
which are
homologous to the gene sequence 3' and 5' (respectively) of the site in the
genome cut by
the gene editing system. Such foreign DNA molecule can be referred to
"template DNA."
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
191
In an embodiment, the CRISPR gene editing system of the present invention
comprises
Cas9, e.g., S. pyogenes Cas9, and a gRNA comprising a targeting domain which
hybridizes
to a sequence of a gene of interest. In an embodiment, the gRNA and Cas9 are
complexed
to form a RNP(ribonucleoprotein). In an embodiment, the CRISPR gene editing
system
comprises nucleic acid encoding a gRNA and nucleic acid encoding a Cas
protein, e.g.,
Cas9, e.g., S. pyogenes Cas9. In an embodiment, the CRISPR gene editing system
comprises a gRNA and nucleic acid encoding a Cas protein, e.g., Cas9, e.g., S.
pyogenes
Cas9.
In some embodiments, inducible control over Cas9, sgRNA expression can be
utilized to
optimize efficiency while reducing the frequency of off-target effects thereby
increasing
safety. Examples include, but are not limited to, transcriptional and post-
transcriptional
switches listed as follows; doxycycline inducible transcription Loew et al.
(2010) BMC
Biotechnol. 10:81, Shield1 inducible protein stabilization Banaszynski et al.
(2016) Cell 126:
995-1004, Tamoxifen induced protein activation Davis et al. (2015) Nat. Chem.
Biol. 11:
316-318, Rapamycin or optogenetic induced activation or dimerization of split
Cas9 Zetsche
(2015) Nature Biotechnol. 33(2): 139-142, Nihongaki et al. (2015) Nature
Biotechnol. 33(7):
755-760, Polstein and Gersbach (2015) Nat. Chem. Biol. 11: 198-200, and SMASh
tag drug
inducible degradation Chung et al. (2015) Nat. Chem. Biol. 11:713-720.
In general, the CRISPR-Cas or CRISPR system refers collectively to transcripts
and other
elements involved in the expression of or directing the activity of CRISPR-
associated ("Cas")
genes, including sequences encoding a Cas gene, a tracr (trans-activating
CRISPR)
sequence (e.g., tracrRNA or an active partial tracrRNA), a tracr-mate sequence
(encompassing a "direct repeat" and a tracrRNA-processed partial direct repeat
in the
context of an endogenous CRISPR system), a guide sequence (also referred to as
a
"spacer" in the context of an endogenous CRISPR system), or "RNA(s)" as that
term is
herein used (e.g., RNA(s) to guide Cas9, e.g., CRISPR RNA and transactivating
(tracr) RNA
or a single guide RNA (sgRNA) (chimeric RNA)) or other sequences and
transcripts from a
CRISPR locus. In general, a CRISPR system is characterized by elements that
promote the
formation of a CRISPR complex at the site of a target sequence (also referred
to as a
protospacer in the context of an endogenous CRISPR system). In the context of
formation of
a CRISPR complex, "target sequence" refers to a sequence to which a guide
sequence is
designed to have complementarity, where hybridization between a target
sequence and a
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
192
guide sequence promotes the formation of a CRISPR complex. A target sequence
may
comprise any polynucleotide, such as DNA or RNA polynucleotides. In some
embodiments,
a target sequence is located in the nucleus or cytoplasm of a cell. In some
embodiments it
may be preferred in a CRISPR complex that the tracr sequence has one or more
hairpins
and is 30 or more nucleotides in length, 40 or more nucleotides in length, or
50 or more
nucleotides in length; the guide sequence is between 10 to 30 nucleotides in
length, the
CRISPR/Cas enzyme is a Type ll Cas9 enzyme. In embodiments of the invention
the terms
guide sequence and guide RNA ("gRNA") are used interchangeably. In general, a
guide
sequence is any polynucleotide sequence having sufficient complementarity with
a target
polynucleotide sequence to hybridize with the target sequence and direct
sequence-specific
binding of a CRISPR complex to the target sequence. In some embodiments, the
degree of
complementarity between a guide sequence and its corresponding target
sequence, when
optimally aligned using a suitable alignment algorithm, is about or more than
about 50%,
60%, 75%, 80%, 85%, 90%, 95%, 97.5%, 99%, or more. Optimal alignment may be
determined with the use of any suitable algorithm for aligning sequences, non-
limiting
example of which include the Smith- Waterman algorithm, the Needleman-Wunsch
algorithm, algorithms based on the Burrows-Wheeler Transform (e.g. the Burrows
Wheeler
Aligner), ClustalW, Clustal X, BLAT, Novoalign (Novocraft Technologies); ELAND
(Illumine,
San Diego, CA), and SOAP. In some embodiments, a guide sequence is about or
more than
about 5, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 , 22, 23, 24, 25, 26,
27, 28, 29, 30, 35,
40, 45, 50, 75, or more nucleotides in length. In some embodiments, a guide
sequence is
less than about 75, 50, 45, 40, 35, 30, 25, 20, 15, 12, or fewer nucleotides
in length.
Preferably the guide sequence is 10 - 30 nucleotides long. The ability of a
guide sequence
to direct sequence- specific binding of a CRISPR complex to a target sequence
may be
assessed by any suitable assay. For example, the components of a CRISPR system
sufficient to form a CRISPR complex, including the guide sequence to be
tested, may be
provided to a host cell having the corresponding target sequence, such as by
transfection
with vectors encoding the components of the CRISPR sequence, followed by an
assessment of preferential cleavage within the target sequence, such as by
Surveyor assay.
Similarly, cleavage of a target polynucleotide sequence may be evaluated in a
test tube by
providing the target sequence, components of a CRISPR complex, including the
guide
sequence to be tested and a control guide sequence different from the test
guide sequence,
and comparing binding or rate of cleavage at the target sequence between the
test and
control guide sequence reactions. Other assays are possible, and will occur to
those skilled
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
193
in the art. A guide sequence may be selected to target any target sequence. In
some
embodiments, the target sequence is a sequence within a genome of a cell.
Exemplary
target sequences include those that are unique in the target genome. For
example, for the
S. pyogenes Cas9, a unique target sequence in a genome may include a Cas9
target site of
the form MM M MMMNNNNNNNNNNNNXGG (SEQ ID NO: 13), where NNN NNN NN XGG
(SEC) ID NO: 179) (N is A, G, T, or C; and X can be anything) has a single
occurrence in
the genome. A unique target sequence in a genome may include an S. pyogenes
Cas9
target site of the form MMM MMMMMNNNNNNNNNNNXGG (SEQ ID NO: 14), where N N N
N XGG (N is A, G, T, or C; and X can be anything) has a single occurrence in
the genome.
For the S. thermophilus CRISPRI Cas9, a unique target sequence in a genome may
include
a Cas9 target site of the form MMMMMMMMNN N N NN XXAGAAW (SEQ ID NO: 15),
where NNN NN N )0(AGAAW (SEQ ID NO: 180) (N is A, G, T, or C; X can be
anything; and
W is A or T) has a single occurrence in the genome. A unique target sequence
in a genome
may include an S. thermophilus CRISPRI Cas9 target site of the form MMMMMM MN
N
NNN NNXXAGAAW (SEQ ID NO: 16), where NNNNNNNNNNNXXAGAAW (SEQ ID NO:
181) (N is A, G, T, or C; X can be anything; and W is A or T) has a single
occurrence in the
genome. For the S. pyogenes Cas9, a unique target sequence in a genome may
include a
Cas9 target site of the form MMMMMMMMNNNN NNNNNNXGGXG (SEQ ID NO: 17),
where NNNNNNNNNNNNXGGXG (SEC) ID NO: 182) (N is A, G, T, or C; and X can be
anything) has a single occurrence in the genome. A unique target sequence in a
genome
may include an S. pyogenes Cas9 target site of the form
MMMMMMMMMNNNNNNNNNNNXGGXG (SEQ ID NO: 183) where
NNNNNNNNNNNXGGXG (SEQ ID NO: 18), (N is A, G, T, or C; and X can be anything)
has
a single occurrence in the genome. In each of these sequences, N is any
nucleobase and
.. "M "may be A, G, T, or C, and need not be considered in identifying a
sequence as unique.
In some embodiments, a guide sequence is selected to reduce the degree
secondary
structure within the guide sequence. In some embodiments, about or less than
about 75%,
50%, 40%, 30%, 25%, 20%, 15%, 10%, 5%, 1%, or fewer of the nucleotides of the
guide
sequence participate in self-complementary base pairing when optimally folded.
Optimal
.. folding may be determined by any suitable polynucleotide folding algorithm.
Some programs
are based on calculating the minimal Gibbs free energy. An example of one such
algorithm
is mFold, as described by Zuker and Stiegler (Nucleic Acids Res. 9(1981), 133-
148).
Another example folding algorithm is the online webserver RNAfold, developed
at Institute
for Theoretical Chemistry at the University of Vienna, using the centroid
structure prediction
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
194
algorithm (see e.g. A.R. Gruber et al., 2008, Cell 106(1): 23-24; and PA Carr
and GM
Church, 2009, Nature Biotechnology 27(12): 1151-62).
Methods for Designing gRNA Molecules
Methods for selecting, designing, and validating targeting domains for use in
the gRNAs
described herein are provided. Exemplary targeting domains for incorporation
into gRNAs
are also provided herein.
Methods for selection and validation of target sequences as well as off-target
analyses have
been described (see, e.g., Mali 2013 ; Hsu 2013; Fu 2014; Heigwer 2014; Bae
2014; and
Xiao 2014). For example, target sequences can be chosen by identifying the PAM
sequence
fora Cas9 molecule (for example, relevant PAM e.g., NGG PAM for S. pyogenes,
NNNNGATT (SEQ ID NO: 19), or NNNNGCTT PAM (SEQ ID NO: 20), for N.
meningitides,
and NNGRRT (SEQ ID NO: 21), or NNGRRV PAM (SEQ ID NO: 22), for S. aureus), and
identifying the adjacent sequence as the target sequence for a CRISPR system
(e.g., S.
pyogenes Cas9 CRISPR system) using that Cas9 molecule. A software tool can be
used to
further refine the choice of potential targeting domains corresponding to a
users target
sequence, e.g., to minimize total off-target activity across the genome.
Candidate targeting
domains and gRNAs comprising those targeting domains can be functionally
evaluated by
using methods known in the art and/or as set forth herein.
As a non-limiting example, targeting domains for use in gRNAs for use with S.
pyogenes, N.
meningiitidis and S. aureus Cas9s are identified using a DNA sequence
searching algorithm.
17-mer, 18-mer, 19-mer, 20-mer, 21-mer, 22-mer, 23-mer, and/or 24-mer
targeting domains
are designed for each Cas9. With respect to S. pyogenes Cas9, preferably, the
targeting
domain is a 20-mer. gRNA design is carried out using a custom gRNA design
software
based on the public tool cas-offinder (Bae 2014). This software scores guides
after
calculating their genome-wide off-target propensity.
Provided in the table below (i.e., Table 1, Table 4) are targeting domains for
gRNA
molecules for use in the compositions and methods of the present invention in
altering
expression of or altering the B2M gene.
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
195
In specific embodiments, cells described herein, such as LSCs and CECs, have
reduced or
eliminated expression of B2M by a CRISPR system (e.g., S. pyogenes Cas9 CRISPR
system) comprising a gRNA selected from those described in Table 1 or Table 2
or Table 4
or Table 6. Use of CRISPR and gRNA molecules targeting the B2M gene are also
described, for example, in Mandal et al., 2014, Cell Stem Cell, 15:643-652;
International
Patent Application Publication Nos. W016073955, W017093969, W016011080,
W016183041, W017106537, W02017143210, W02017212072, and W02018064594.
Table 1: gRNA Targeting Domains for Exemplary Allogeneic Ocular Cell Targets
Target r
Region a SEQ
(e.g., n Genomic Location ID
ii:Target Exon) d (hg38) Guide RNA Sequence
chr15:44711469- UGGCUGGGCACGCGUU
B2M Exon 1 + 44711494 UAAUAUAAG 23
chr15:44711472- CUGGGCACGCGUUUAAU
B2M Exon 1 + 44711497 AUAAGUGG 24
chr15:44711483- UUUAAUAUAAGUGGAGG
B2M Exon 1 + 44711508 CGUCGCGC 25
chr15:44711486- AAUAUAAGUGGAGGCGU
B2M Exon 1 + 44711511 CGCGCUGG 26
chr15:44711487- AUAUAAGUGGAGGCGUC
B2M Exon 1 + 44711512 GCGCUGGC 27
chr15:44711512- GGGCAUUCCUGAAGCUG
B2M Exon 1 + 44711537 ACAGCAUU 28
chr15:44711513- GGCAUUCCUGAAGCUGA
B2M Exon 1 + 44711538 CAGCAUUC 29
chr15:44711534- AUUCGGGCCGAGAUGUC
B2M Exon 1 + 44711559 UCGCUCCG 30
chr15:44711568- CUGUGCUCGCGCUACUC
B2M Exon 1 + 44711593 UCUCUUUC 31
chr15:44711573- CUCGCGCUACUCUCUCU
B2M Exon 1 + 44711598 UUCUGGCC 32
chr15:44711576- GCGCUACUCUCUCUUUC
B2M Exon 1 + 44711601 UGGCCUGG 33
chr15:44711466- AUAUUAAACGCGUGCCC
B2M Exon 1 - 44711491 AGCCAAUC 34
chr15:44711522- UCUCGGCCCGAAUGCUG
B2M Exon 1 - 44711547 UCAGCUUC 35
chr15:44711544- GCUAAGGCCACGGAGCG
B2M Exon 1 - 44711569 AGACAUCU 36
chr15:44711559- AGUAGCGCGAGCACAGC
B2M Exon 1 - 44711584 UAAGGCCA 37
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
196
chr15:44711565- AGAGAGAGUAGCGCGAG
B2M Exon 1 - 44711590 CACAGCUA 38
chr15:44711599- GAGAGACUCACGCUGGA
B2M Exon 1 - 44711624 UAGCCUCC 39
chr15:44711611- GCGGGAGGGUAGGAGA
B2M Exon 1 - 44711636 GACUCACGC 40
chr15:44715412- UAUUCCUCAGGUACUCC
B2M Exon 2 + 44715437 AAAGAUUC 41
chr15:44715440- UUUACUCACGUCAUCCA
B2M Exon 2 + 44715465 GCAGAGAA 42
chr15:44715473- CAAAUUUCCUGAAUUGC
B2M Exon 2 + 44715498 UAUGUGUC 43
chr15:44715474- AAAUUUCCUGAAUUGCU
B2M Exon 2 + 44715499 AUGUGUCU 44
chr15:44715515- ACAUUGAAGUUGACUUA
B2M Exon 2 + 44715540 CUGAAGAA 45
chr15:44715535- AAGAAUGGAGAGAGAAU
B2M Exon 2 + 44715560 UGAAAAAG 46
chr15:44715562- GAGCAUUCAGACUUGUC
B2M Exon 2 + 44715587 UUUCAGCA 47
chr15:44715567- UUCAGACUUGUCUUUCA
B2M Exon 2 + 44715592 GCAAGGAC 48
chr15:44715672- UUUGUCACAGCCCAAGA
B2M Exon 2 + 44715697 UAGUUAAG 49
chr15:44715673- UUGUCACAGCCCAAGAU
B2M Exon 2 + 44715698 AGUUAAGU 50
chr15:44715674- UGUCACAGCCCAAGAUA
B2M Exon 2 + 44715699 GUUAAGUG 51
chr15:44715410- AUCUUUGGAGUACCUGA
B2M Exon 2 - 44715435 GGAAUAUC 52
chr15:44715411- AAUCUUUGGAGUACCUG
B2M Exon 2 - 44715436 AGGAAUAU 53
chr15:44715419- UAAACCUGAAUCUUUGG
B2M Exon 2 - 44715444 AGUACCUG 54
chr15:44715430- GAUGACGUGAGUAAACC
B2M Exon 2 - 44715455 UGAAUCUU 55
chr15:44715457- GGAAAUUUGACUUUCCA
B2M Exon 2 - 44715482 UUCUCUGC 56
chr15:44715483- AUGAAACCCAGACACAU
B2M Exon 2 - 44715508 AGCAAUUC 57
chr15:44715511- UCAGUAAGUCAACUUCA
B2M Exon 2 - 44715536 AUGUCGGA 58
chr15:44715515- UUCUUCAGUAAGUCAAC
B2M Exon 2 - 44715540 UUCAAUGU 59
chr15:44715629- CAGGCAUACUCAUCUUU
B2M Exon 2 - 44715654 UUCAGUGG 60
chr15:44715630- GCAGGCAUACUCAUCUU
B2M Exon 2 - 44715655 UUUCAGUG 61
chr15:44715631- GGCAGGCAUACUCAUCU
B2M Exon 2 - 44715656 UUUUCAGU 62
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
197
chr15:44715632- CGGCAGGCAUACUCAUC
B2M Exon 2 - 44715657 UUUUUCAG 63
chr15:44715653- GACAAAGUCACAUGGUU
B2M Exon 2 - 44715678 CACACGGC 64
chr15:44715657- CUGUGACAAAGUCACAU
B2M Exon 2 - 44715682 GGUUCACA 65
chr15:44715666- UAUCUUGGGCUGUGACA
B2M Exon 2 - 44715691 AAGUCACA 66
chr15:44715685- AAGACUUACCCCACUUA
B2M Exon 2 - 44715710 ACUAUCUU 67
chr15:44715686- UAAGACUUACCCCACUU
B2M Exon 2 - 44715711 AACUAUCU 68
chr15:44716326- AGAUCGAGACAUGUAAG
B2M Exon 3 + 44716351 CAGCAUCA 69
chr15:44716329- UCGAGACAUGUAAGCAG
B2M Exon 3 + 44716354 CAUCAUGG 70
chr15:44716313- AUG UCUCGAUCUAUGAA
B2M Exon 3 - 44716338 AAAGACAG 71
chr15:44717599- UUUUCAGGUUUGAAGAU
B2M Exon 4 + 44717624 GCCGCAUU 72
chr15:44717604- AGGUUUGAAGAUGCCGC
B2M Exon 4 + 44717629 AUUUGGAU 73
chr15:44717681- CACUUACACUUUAUGCA
B2M Exon 4 + 44717706 CAAAAUG U 74
chr15:44717682- ACUUACACUUUAUGCAC
B2M Exon 4 + 44717707 AAAAUGUA 75
chr15:44717702- AUGUAGGGUUAUAAUAA
B2M Exon 4 + 44717727 UGUUAACA 76
chr15:44717764- GUCUCCAUGUUUGAUGU
B2M Exon 4 + 44717789 AUCUGAGC 77
chr15:44717776- GAUGUAUCUGAGCAGGU
B2M Exon 4 + 44717801 UGCUCCAC 78
chr15:44717786- AGCAGGUUGCUCCACAG
B2M Exon 4 + 44717811 GUAGCUCU 79
chr15:44717789- AGGUUGCUCCACAGGUA
B2M Exon 4 + 44717814 GCUCUAGG 80
chr15:44717790- GGUUGCUCCACAGGUAG
B2M Exon 4 + 44717815 CUCUAGGA 81
chr15:44717794- GCUCCACAGGUAGCUCU
B2M Exon 4 + 44717819 AGGAGGGC 82
chr15:44717805- AGCUCUAGGAGGGCUG
B2M Exon 4 + 44717830 GCAACUUAG 83
chr15:44717808- UCUAGGAGGGCUGGCAA
B2M Exon 4 + 44717833 CU UAGAGG 84
chr15:44717809- CUAGGAGGGCUGGCAAC
B2M Exon 4 + 44717834 UUAGAGGU 85
chr15:44717810- UAGGAGGGCUGGCAACU
B2M Exon 4 + 44717835 UAGAGGUG 86
chr15:44717846- AUUCUCUUAUCCAACAU
B2M Exon 4 + 44717871 CAACAUCU 87
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
198
chr15:44717945- CAAUUUACAUACUCUGC
B2M Exon 4 + 44717970 UUAGAAU U 88
chr15:44717946- AAUUUACAUACUCUGCU
B2M Exon 4 + 44717971 UAGAAUU U 89
chr15:44717947- AUUUACAUACUCUGCUU
B2M Exon 4 + 44717972 AGAAUUUG 90
chr15:44717948- UUUACAUACUCUGCUUA
B2M Exon 4 + 44717973 GAAUUUGG 91
chr15:44717973- GGGAAAAUUUAGAAAUA
B2M Exon 4 + 44717998 UAAUUGAC 92
chr15:44717981- UUAGAAAUAUAAUUGAC
B2M Exon 4 + 44718006 AGGAUUAU 93
chr15:44718056- UACUUCUUAUACAUUUG
B2M Exon 4 + 44718081 AUAAAGUA 94
chr15:44718061- CU UAUACAU UUGAUAAA
B2M Exon 4 + 44718086 GUAAGGCA 95
chr15:44718067- CAUUUGAUAAAGUAAGG
B2M Exon 4 + 44718092 CAUGGUUG 96
chr15:44718076- AAGUAAGGCAUGGUUGU
B2M Exon 4 + 44718101 GGUUAAUC 97
chr15:44717589- CUUCAAACCUGAAAAGA
B2M Exon 4 - 44717614 AAAGAAAA 98
chr15:44717620- AUUUGGAAUUCAUCCAA
B2M Exon 4 - 44717645 UCCAAAUG 99
chr15:44717642- UAUUAAAAAGCAAGCAA
B2M Exon 4 - 44717667 GCAGAAUU 100
chr15:44717771- GCAACCUGCUCAGAUAC
B2M Exon 4 - 44717796 AUCAAACA 101
chr15:44717800- UUGCCAGCCCUCCUAGA
B2M Exon 4 - 44717825 GCUACCUG 102
chr15:44717859- UCAAAUCUGACCAAGAU
B2M Exon 4 - 44717884 GUUGAUGU 103
chr15:44717947- CAAAUUCUAAGCAGAGU
B2M Exon 4 - 44717972 AUGUAAAU 104
chr15:44718119- CAAGUUUUAUGAUUUAU
B2M Exon 4 - 44718144 UUAACUUG 105
In specific embodiments, modified cells described herein, such as LSCs or
CECs, have
reduced or eliminated expression of B2M by a CRISPR system (e.g., S. pyogenes
Cas9
CRISPR system) comprising a gRNA selected from those described in Table 1 or
Table 4 or
Table 6 in the Examples, wherein such modified cells comprise gene editing of
B2M within
Exon 1.
In specific embodiments, modified cells described herein, such as LSCs or
CECs, have
reduced or eliminated expression of B2M by a CRISPR system (e.g., S. pyogenes
Cas9
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
199
CRISPR system) comprising a gRNA selected from those described in Table 1 or
Table 4 or
Table 6, wherein such modified cells comprise gene editing of B2M within Exon
2.
In specific embodiments, modified cells described herein, such as LSCs or
CECs, have
reduced or eliminated expression of B2M by a CRISPR system (e.g., S. pyogenes
Cas9
CRISPR system) comprising a gRNA selected from those described in Table 1 or
Table 4 or
Table 6, wherein such modified cells comprise gene editing of B2M within Exon
3.
In specific embodiments, modified cells described herein, such as LSCs or
CECs, have
reduced or eliminated expression of B2M by a CRISPR system (e.g., S. pyogenes
Cas9
CRISPR system) comprising a gRNA selected from those described in Table 1 or
Table 4 or
Table 6, wherein such modified cells comprise gene editing of B2M within Exon
4.
In specific embodiments, modified cells described herein, such as LSCs or
CECs, have
reduced or eliminated expression of B2M by a CRISPR system (e.g., S. pyogenes
Cas9
CRISPR system) comprising a gRNA selected from those described in Table 1 or
Table 4,
wherein such modified cells comprise gene editing of B2M within a genomic
location (e.g.,
chr15:44711469-44711494) selected from those described in Table 1 or Table 4.
In some
embodiments, the targeting domain of the gRNA molecule used in the present
invention is
complementary to a sequence within a genomic region selected from:
chr15:44711469-
44711494, chr15:44711472-44711497, chr15:44711483-44711508, chr15:44711486-
44711511, chr15:44711487-44711512, chr15:44711512-44711537, chr15:44711513-
44711538, chr15:44711534-4471 1559, chr15:44711568-44711593, chr15:44711573-
44711598, chr15:44711576-44711601, chr15:44711466-44711491, chr15:44711522-
44711547, chr15:44711544-44711569, chr15:44711559-44711584, chr15:44711565-
44711590, chr15:44711599-44711624, chr15:44711611-44711636, chr15:44715412-
44715437, chr15:44715440-44715465, chr15:44715473-44715498, chr15:44715474-
44715499, chr15:44715515-44715540, chr15:44715535-44715560, chr15:44715562-
44715587, chr15:44715567-44715592, chr15:44715672-44715697, chr15:44715673-
44715698, chr15:44715674-44715699, chr15:44715410-44715435, chr15:44715411-
44715436, chr15:44715419-44715444, chr15:44715430-44715455, chr15:44715457-
44715482, chr15:44715483-44715508, chr15:44715511-44715536, chr15:44715515-
44715540, chr15:44715629-44715654, chr15:44715630-44715655, chr15:44715631-
44715656, chr15:44715632-44715657, chr15:44715653-44715678, chr15:44715657-
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
200
44715682, chr15:44715666-44715691, chr15:44715685-44715710, chr15:44715686-
44715711, chr15:44716326-44716351, chr15:44716329-44716354, chr15:44716313-
44716338, chr15:44717599-44717624, chr15:44717604-44717629, chr15:44717681-
44717706, chr15:44717682-44717707, chr15:44717702-44717727, chr15:44717764-
44717789, chr15:44717776-44717801, chr15:44717786-44717811, chr15:44717789-
44717814, chr15:44717790-44717815, chr15:44717794-44717819, chr15:44717805-
44717830, chr15:44717808-44717833, chr15:44717809-44717834, chr15:44717810-
44717835, chr15:44717846-44717871, chr15:44717945-44717970, chr15:44717946-
44717971, chr15:44717947-44717972, chr15:44717948-44717973, chr15:44717973-
44717998, chr15:44717981-44718006, chr15:44718056-44718081, chr15:44718061-
44718086, chr15:44718067-44718092, chr15:44718076-44718101, chr15:44717589-
44717614, chr15:44717620-44717645, chr15:44717642-44717667, chr15:44717771-
44717796, chr15:44717800-44717825, chr15:44717859-44717884, chr15:44717947-
44717972, chr15:44718119-44718144, chr15:44711563-44711585, chr15:44715428-
44715450, chr15:44715509-44715531, chr15:44715513-44715535, chr15:44715417-
44715439, chr15:44711540-44711562, chr15:44711574-44711596, chr15:44711597-
44711619, chr15:44715446-44715468, chr15:44715651-44715673, chr15:44713812-
44713834, chr15:44711579-44711601, chr15:44711542-44711564, chr15:44711557-
44711579, chr15:44711609-44711631, chr15:44715678-44715700, chr15:44715683-
44715705, chr15:44715684-44715706, chr15:44715480-44715502. In a specific
embodiment, the targeting domain of the gRNA molecule is complementary to a
sequence
within a genomic region selected from: chr15:44715513-44715535, chr15:44711542-
44711564, chr15:44711563-44711585, chr15:44715683-44715705, chr15:44711597-
44711619, or chr15:44715446-44715468. In one embodiment, the targeting domain
of the
gRNA molecule is complementary to a sequence within a genomic region
chr15:44711597-
44711619. In another embodiment, the targeting domain of the gRNA molecule is
complementary to a sequence within a genomic region chr15:44715446-44715468.
In a
preferred embodiment, the targeting domain of the gRNA molecule is
complementary to a
sequence within a genomic region chr15:44711563-44711585.
In particular embodiments, modified cells described herein, such as LSCs or
CECs, have
reduced or eliminated expression of B2M by a CRISPR system (e.g., S. pyogenes
Cas9
CRISPR system) comprising a gRNA targeting domain sequence selected from those
described in Table 1 or Table 4. In one embodiment, the targeting domain of
the gRNA
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
201
molecule to B2M comprises a targeting domain comprising the sequence of any
one of SEQ
ID NOs: 23-105 or 108-119 or 134-140. In a specific embodiment, the targeting
domain of
the gRNA molecule to B2M comprises a targeting domain comprising the sequence
of any
one of SEQ ID NOs: 108, 111, 115, 116, 134 or 138. In a preferred embodiment,
the
targeting domain of the gRNA molecule to B2M comprises a targeting domain
comprising
the sequence of SEQ ID NO: 108. In another embodiment, the targeting domain of
the
gRNA molecule to B2M comprises a targeting domain comprising the sequence of
SEQ ID
NO: 115. In another embodiment, the targeting domain of the gRNA molecule to
B2M
comprises a targeting domain comprising the sequence of SEQ ID NO: 116.
In some embodiments, modified cells described herein, such as LSCs or CECs,
have
reduced or eliminated expression of B2M by a CRISPR system (e.g., S. pyogenes
Cas9
CRISPR system) comprising a gRNA targeting a sequence complementary to any of
the
sequences selected from those described in Table 5. In some embodiments,
modified cells
described herein, such as LSCs or CECs, have reduced or eliminated expression
of B2M by
a CRISPR system (e.g., S. pyogenes Cas9 CRISPR system) comprising a gRNA
targeting a
sequence complementary to any of the sequences selected from SEQ ID NOs: 141
to 159.
In particular embodiment, modified cells described herein, such as LSCs or
CECs, have
reduced or eliminated expression of B2M by a CRISPR system (e.g., S. pyogenes
Cas9
CRISPR system) comprising a gRNA, wherein the gRNA comprises the sequence of
any
one of SEQ ID NO: 120, 160-177. In a specific embodiment, modified cells
described herein,
such as LSCs or CECs, have reduced or eliminated expression of B2M by a CRISPR
system (e.g., S. pyogenes Cas9 CRISPR system) comprising a gRNA, wherein the
gRNA
comprises the sequence of any one of SEQ ID NO: 120, 162, 166, 167, 171, and
175. In a
preferred embodiment, the gRNA comprises the sequence of SEQ ID NO: 120. In
another
embodiment, the gRNA comprises the sequence of SEQ ID NO: 166 or 167.
In particular embodiments, modified cells described herein, such as LSCs or
CECs, have
reduced or eliminated expression of B2M by a CRISPR system (e.g., S. pyogenes
Cas9
CRISPR system) comprising a gRNA comprising one, two, three, four, five, six,
seven or
eight nucleotide modifications (e.g., addition, substitution, or deletion)
relative to a gRNA
sequence described in Table 1 or Table 4 or Table 6.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
202
In one aspect, the present invention relates to a modified LSC or CEC
comprising a genome
in which the b2 microglobulin (B2M) gene on chromosome 15 has been edited to
delete a
contiguous stretch of genomic DNA comprising the sequence of any one of SEQ ID
NOs:
141 to 159, thereby eliminating surface expression of MHC Class I molecules in
the cell. In
one embodiment, the modified LSC or CEC of the present invention comprises a
genome in
which the b2 microglobulin (B2M) gene on chromosome 15 has been edited to
delete a
contiguous stretch of genomic DNA comprising the sequence of any one of SEQ ID
NOs:
141, 144, 148, 149, 153 or 157, thereby eliminating surface expression of MHC
Class I
molecules in the cell. In a more specific embodiment, the modified LSC or CEC
of the
.. present invention comprises a genome in which the b2 microglobulin (B2M)
gene on
chromosome 15 has been edited to delete a contiguous stretch of genomic DNA
comprising
the sequence of any one of SEQ ID NOs: 141, 148 or 149, thereby eliminating
surface
expression of MHC Class I molecules in the cell. In a preferred embodiment,
the modified
LSC or CEC comprises a genome in which the b2 microglobulin (B2M) gene on
chromosome 15 has been edited to delete a contiguous stretch of genomic DNA
comprising
the sequence of SEQ ID NOs: 141, thereby eliminating surface expression of MHC
Class I
molecules in the cell.
In one aspect, the present invention relates to a modified LSC or CEC
comprising a genome
in which the b2 microglobulin (B2M) gene on chromosome 15 has been edited to
delete a
contiguous stretch of genomic DNA region selected from any one of:
chr15:44711469-
44711494, chr15:44711472-44711497, chr15:44711483-44711508, chr15:44711486-
44711511, chr15:44711487-44711512, chr15:44711512-44711537, chr15:44711513-
44711538, chr15:44711534-44711559, chr15:44711568-44711593, chr15:44711573-
44711598, chr15:44711576-44711601, chr15:44711466-44711491, chr15:44711522-
44711547, chr15:44711544-44711569, chr15:44711559-44711584, chr15:44711565-
44711590, chr15:44711599-44711624, chr15:44711611-44711636, chr15:44715412-
44715437, chr15:44715440-44715465, chr15:44715473-44715498, chr15:44715474-
44715499, chr15:44715515-44715540, chr15:44715535-44715560, chr15:44715562-
44715587, chr15:44715567-44715592, chr15:44715672-44715697, chr15:44715673-
44715698, chr15:44715674-44715699, chr15:44715410-44715435, chr15:44715411-
44715436, chr15:44715419-44715444, chr15:44715430-44715455, chr15:44715457-
44715482, chr15:44715483-44715508, chr15:44715511-44715536, chr15:44715515-
44715540, chr15:44715629-44715654, chr15:44715630-44715655, chr15:44715631-
44715656, chr15:44715632-44715657, chr15:44715653-44715678, chr15:44715657-
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
203
44715682, chr15:44715666-44715691, chr15:44715685-44715710, chr15:44715686-
44715711, chr15:44716326-44716351, chr15:44716329-44716354, chr15:44716313-
44716338, chr15:44717599-44717624, chr15:44717604-44717629, chr15:44717681-
44717706, chr15:44717682-44717707, chr15:44717702-44717727, chr15:44717764-
44717789, chr15:44717776-44717801, chr15:44717786-44717811, chr15:44717789-
44717814, chr15:44717790-44717815, chr15:44717794-44717819, chr15:44717805-
44717830, chr15:44717808-44717833, chr15:44717809-44717834, chr15:44717810-
44717835, chr15:44717846-44717871, chr15:44717945-44717970, chr15:44717946-
44717971, chr15:44717947-44717972, chr15:44717948-44717973, chr15:44717973-
44717998, chr15:44717981-44718006, chr15:44718056-44718081, chr15:44718061-
44718086, chr15:44718067-44718092, chr15:44718076-44718101, chr15:44717589-
44717614, chr15:44717620-44717645, chr15:44717642-44717667, chr15:44717771-
44717796, chr15:44717800-44717825, chr15:44717859-44717884, chr15:44717947-
44717972, chr15:44718119-44718144, chr15:44711563-44711585, chr15:44715428-
44715450, chr15:44715509-44715531, chr15:44715513-44715535, chr15:44715417-
44715439, chr15:44711540-44711562, chr15:44711574-44711596, chr15:44711597-
44711619, chr15:44715446-44715468, chr15:44715651-44715673, chr15:44713812-
44713834, chr15:44711579-44711601, chr15:44711542-44711564, chr15:44711557-
44711579, chr15:44711609-44711631, chr15:44715678-44715700, chr15:44715683-
44715705, chr15:44715684-44715706, chr15:44715480-44715502, thereby
eliminating
surface expression of MHC Class I molecules in the cell. In one embodiment,
the modified
LSC or CEC of the present invention comprises a genome in which the b2
microglobulin
(B2M) gene on chromosome 15 has been edited to delete a contiguous stretch of
genomic
DNA region selected from: chr15:44715513-44715535, chr15:44711542-44711564,
chr15:44711563-44711585, chr15:44715683-44715705, chr15:44711597-44711619, or
chr15:44715446-44715468. In a specific embodiment, the modified LSC or CEC of
the
present invention comprises a genome in which the b2 microglobulin (B2M) gene
on
chromosome 15 has been edited to delete a contiguous stretch of genomic DNA
region
selected from: chr15:44711563-44711585, chr15:44711597-44711619, or
chr15:44715446-
44715468, thereby eliminating surface expression of MHC Class I molecules in
the cell. In a
preferred embodiment, the modified LSC or CEC of the present invention
comprises a
genome in which the b2 microglobulin (B2M) gene on chromosome 15 has been
edited to
delete a contiguous stretch of genomic DNA region chr15:44711563-44711585,
thereby
eliminating surface expression of MHC Class I molecules in the cell.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
204
In one aspect, the present invention relates to a modified LSC or CEC
comprising a genome
in which the b2 microglobulin (B2M) gene on chromosome 15 has been edited to
to form an
indel at or near the target sequence complementary to the targeting domain of
the gRNA
molecule.
An "indel," as the term is used herein, refers to a nucleic acid comprising
one or more
insertions of nucleotides, one or more deletions of nucleotides, or a
combination of
insertions and delections of nucleotides, relative to a reference nucleic
acid, that results after
being exposed to a composition comprising a gRNA molecule, for example a
CRISPR
system. Indels can be determined by sequencing nucleic acid after being
exposed to a
composition comprising a gRNA molecule, for example, by NGS. With respect to
the site of
an indel, an indel is said to be "at or near" a reference site (e.g., a site
complementary to a
targeting domain of a gRNA molecule) if it comprises at least one insertion or
deletion within
about 10, 9 , 8, 7, 6, 5, 4, 3, 2, or 1 nucleotide(s) of the reference site,
or is overlapping with
part or all of said refrence site (e.g., comprises at least one insertion or
deletion overlapping
with, or within 10,9, 8, 7,6, 5,4, 3,2, or 1 nucelotides of a site
complementary to the
targeting domain of a gRNA molecule, e.g., a gRNA molecule described herein).
.. An "indel pattern," as the term is used herein, refers to a set of indels
that results after
exposure to a composition comprising a gRNA molecule. In an embodiment, the
indel
pattern consists of the top three indels, by frequency of appearance. In an
embodiment, the
indel pattern consists of the top five indels, by frequency of appearance. In
an embodiment,
the indel pattern consists of the indels which are present at greater than
about 5% frequency
relative to all sequencing reads. In an embodiment, the indel pattern consists
of the indels
which are present at greater than about 10% frequency relative to to total
number of indel
sequencing reads (i.e., those reads that do not consist of the unmodified
reference nucleic
acid sequence). In an embodiment, the indel pattern includes of any 3 of the
top five most
frequently observed indels. The indel pattern may be determined, for example,
by
sequencing cells of a population of cells which were exposed to the gRNA
molecule.
In one aspect, the present invention provides a modified LSC or CEC comprising
a genome
in which the b2 microglobulin (B2M) gene on chromosome 15 has been edited to
form an
indel at or near the target sequence complementary to the targeting domain of
the gRNA
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
205
molecule comprising the sequence of any one of SEQ ID NOs: 23-105 or 108-119
or 134-
140, thereby eliminating surface expression of MHC Class I molecules in the
cell. In one
embodiment, the modified LSC or CEC of the present invention comprises a
genome in
which the b2 microglobulin (B2M) gene on chromosome 15 has been edited to form
an indel
at or near the target sequence complementary to the targeting domain of the
gRNA
molecule comprising the sequence of any one of SEQ ID NOs: 108, 111, 115, 116,
134 or
138, thereby eliminating surface expression of MHC Class I molecules in the
cell. In a more
specific embodiment, the modified LSC or CEC of the present invention
comprises a
genome in which the b2 microglobulin (B2M) gene on chromosome 15 has been
edited to
form an indel at or near the target sequence complementary to the targeting
domain of the
gRNA molecule comprising the sequence of any one of SEQ ID NOs: 108, 115, or
116,
thereby eliminating surface expression of MHC Class I molecules in the cell.
In a preferred
embodiment, the modified LSC or CEC comprises a genome in which the b2
microglobulin
(B2M) gene on chromosome 15 has been edited to form an indel at or near the
target
sequence complementary to the targeting domain of the gRNA molecule comprising
the
sequence SEQ ID NOs: 108, thereby eliminating surface expression of MHC Class
I
molecules in the cell.
In one aspect, the present invention provides a modified LSC or CEC comprising
a genome
in which the b2 microglobulin (B2M) gene on chromosome 15 has been edited to
form an
indel at or near the genomic DNA region selected from any one of:
chr15:44711469-
44711494, chr15:44711472-44711497, chr15:44711483-44711508, chr15:44711486-
44711511, chr15:44711487-44711512, chr15:44711512-44711537, chr15:44711513-
44711538, chr15:44711534-44711559, chr15:44711568-44711593, chr15:44711573-
44711598, chr15:44711576-44711601, chr15:44711466-44711491, chr15:44711522-
44711547, chr15:44711544-44711569, chr15:44711559-44711584, chr15:44711565-
44711590, chr15:44711599-44711624, chr15:44711611-44711636, chr15:44715412-
44715437, chr15:44715440-44715465, chr15:44715473-44715498, chr15:44715474-
44715499, chr15:44715515-44715540, chr15:44715535-44715560, chr15:44715562-
44715587, chr15:44715567-44715592, chr15:44715672-44715697, chr15:44715673-
44715698, chr15:44715674-44715699, chr15:44715410-44715435, chr15:44715411-
44715436, chr15:44715419-44715444, chr15:44715430-44715455, chr15:44715457-
44715482, chr15:44715483-44715508, chr15:44715511-44715536, chr15:44715515-
44715540, chr15:44715629-44715654, chr15:44715630-44715655, chr15:44715631-
44715656, chr15:44715632-44715657, chr15:44715653-44715678, chr15:44715657-
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
206
44715682, chr15:44715666-44715691, chr15:44715685-44715710, chr15:44715686-
44715711, chr15:44716326-44716351, chr15:44716329-44716354, chr15:44716313-
44716338, chr15:44717599-44717624, chr15:44717604-44717629, chr15:44717681-
44717706, chr15:44717682-44717707, chr15:44717702-44717727, chr15:44717764-
44717789, chr15:44717776-44717801, chr15:44717786-44717811, chr15:44717789-
44717814, chr15:44717790-44717815, chr15:44717794-44717819, chr15:44717805-
44717830, chr15:44717808-44717833, chr15:44717809-44717834, chr15:44717810-
44717835, chr15:44717846-44717871, chr15:44717945-44717970, chr15:44717946-
44717971, chr15:44717947-44717972, chr15:44717948-44717973, chr15:44717973-
44717998, chr15:44717981-44718006, chr15:44718056-44718081, chr15:44718061-
44718086, chr15:44718067-44718092, chr15:44718076-44718101, chr15:44717589-
44717614, chr15:44717620-44717645, chr15:44717642-44717667, chr15:44717771-
44717796, chr15:44717800-44717825, chr15:44717859-44717884, chr15:44717947-
44717972, chr15:44718119-44718144, chr15:44711563-44711585, chr15:44715428-
44715450, chr15:44715509-44715531, chr15:44715513-44715535, chr15:44715417-
44715439, chr15:44711540-44711562, chr15:44711574-44711596, chr15:44711597-
44711619, chr15:44715446-44715468, chr15:44715651-44715673, chr15:44713812-
44713834, chr15:44711579-44711601, chr15:44711542-44711564, chr15:44711557-
44711579, chr15:44711609-44711631, chr15:44715678-44715700, chr15:44715683-
44715705, chr15:44715684-44715706, chr15:44715480-44715502, thereby
eliminating
surface expression of MHC Class I molecules in the cell. In one embodiment,
the modified
LSC or CEC of the present invention comprises a genome in which the b2
microglobulin
(B2M) gene on chromosome 15 has been edited to form an indel at or near the
genomic
DNA region selected from any one of: chr15:44715513-44715535, chr15:44711542-
44711564, chr15:44711563-44711585, chr15:44715683-44715705, chr15:44711597-
44711619, or chr15:44715446-44715468. In a specific embodiment, the modified
LSC or
CEC of the present invention comprises a genome in which the b2 microglobulin
(B2M)
gene on chromosome 15 has been edited to form an indel at or near the genomic
DNA
region selected from any one of: chr15:44711563-44711585, chr15:44711597-
44711619, or
chr15:44715446-44715468, thereby eliminating surface expression of MHC Class I
molecules in the cell. In a preferred embodiment, the modified LSC or CEC of
the present
invention comprises a genome in which the b2 microglobulin (B2M) gene on
chromosome
15 has been edited to form an indel at or near the genomic DNA region
chr15:44711563-
44711585, thereby eliminating surface expression of MHC Class I molecules in
the cell.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
207
In some embodiment, the formed indel comprises a deletion of 10 or greater
than 10
nucleotides, optionally 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28,
29, 30, 31,32, 33, 34, or 35 nucleotides.
In some embodiments, the indel is formed at or near the target sequence
complementary to
the targeting domain of the gRNA molecule in at least about 40%, e.g., at
least about 50%,
e.g., at least about 60%, e.g., at least about 70%, e.g., at least about 80%,
e.g., at least
about 90%, e.g., at least about 95%, e.g., at least about 96%, e.g., at least
about 97%, e.g.,
at least about 98%, e.g., at least about 99%, of the cells of the population.
In some embodiments, the indel comprising a deletion of 10 or greater than 10
nucleotides
is detected in at least about 5%, optionally at least about 10%, 15%, 20%,
25%, 30% or
more, of the cells of the population.
In some embodiments, the indel is as measured by next generation sequencing
(NGS).
In one embodiment, the present invention provides a modified LSC or CEC
comprising a
genome in which the b2 microglobulin (B2M) gene on chromosome 15 has been
edited to
form an indel at or near the target sequence, and wherein no off-target indels
are formed in
said modified LSC or CEC, e.g., as detectable by next generation sequencing
and/or a
nucleotide insertional assay. In one mebodiment, the present invention
provides a
population of modified LSCs or CECs comprising a genome in which the b2
microglobulin
(B2M) gene on chromosome 15 has been edited to form an indel at or near the
target
sequence, and wherein an off-target indel is detected in no more than about
5%, e.g., no
more than about 1%, e.g., no more than about 0.1%, e.g., no more than about
0.01%, of the
cells of the population of the modifoed LSCs or CECs, e.g., as detectible by
next generation
sequencing and/or a nucleotide insertional assay.
An "off-target indel", as the term used herein, refers to an indel at or near
a site other than
the target sequence of the targeting domain of the gRNA molecule. Such sites
may
comprise, for example, 1, 2, 3, 4, 5 or more mismatch nucleotides relative to
the sequence
of the targeting domain of the gRNA. In exemplary embodiments, such sites are
detected
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
208
using targeted sequencing of in silico predicted off-target sites, or by an
insertional method
known in the art.
In some embodiments, the modified LSC or CEC of the present invention is
autologous with
respect to a patient to be administered said cell. In other embodiments, the
modified LSC or
CEC of the present invention is allogeneic with respect to a patient to be
administered said
cell.
Functional Analysis of Candidate Molecules
Candidate Cas9 molecules, candidate gRNA molecules, candidate Cas9
molecule/gRNA
molecule complexes, can be evaluated by art-known methods or as described
herein. For
example, exemplary methods for evaluating the endonuclease activity of Cas9
molecule
have been described previously (Jinek 2012). Each technique described herein
may be
.. used alone or in combination with one or more techniques to evaluate the
candidate
molecule. The techniques disclosed herein may be used for a variety of methods
including,
without limitation, methods of determining the stability of a Cas9
molecule/gRNA molecule
complex, methods of determining a condition that promotes a stable Cas9
molecule/gRNA
molecule complex, methods of screening for a stable Cas9 molecule/gRNA
molecule
complex, methods of identifying an optimal gRNA to form a stable Cas9
molecule/gRNA
molecule complex, and methods of selecting a Cas9/gRNA complex for
administration to a
subject.
Binding and Cleavage Assay: Testing the endonuclease activity of Cas9 molecule
The
ability of a Cas9 molecule/gRNA molecule complex to bind to and cleave a
target nucleic
acid can be evaluated in a plasmid cleavage assay. In this assay, synthetic or
in vitro-
transcribed gRNA molecule is pre-annealed prior to the reaction by heating to
95 C and
slowly cooling down to room temperature. Native or restriction digest-
linearized plasmid
DNA (300 ng (-8 nM)) is incubated for 60 min at 37 C with purified Cas9
protein molecule
(50-500 nM) and gRNA (50-500 nM, 1: 1) in a Cas9 plasmid cleavage buffer (20
mM
HEPES pH 7.5, 150 mM KC1, 0.5 mM DTT, 0.1 mM EDTA) with or without 10 mM
MgCl2.
The reactions are stopped with 5X DNA loading buffer (30% glycerol, 1.2% SDS,
250 mM
EDTA), resolved by a 0.8 or 1% agarose gel electrophoresis and visualized by
ethidium
bromide staining. The resulting cleavage products indicate whether the Cas9
molecule
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
209
cleaves both DNA strands, or only one of the two strands. For example, linear
DNA products
indicate the cleavage of both DNA strands. Nicked open circular products
indicate that only
one of the two strands is cleaved.
Alternatively, the ability of a Cas9 molecule/gRNA molecule complex to bind to
and cleave a
target nucleic acid can be evaluated in an oligonucleotide DNA cleavage assay.
In this
assay, DNA oligonucleotides (10 pmol) are radiolabeled by incubating with 5
units T4
polynucleotide kinase and -3-6 pmol (-20-40 mCi) [y-32P]-ATP in IX T4
polynucleotide
kinase reaction buffer at 37 C for 30 min, in a 50 microlitre reaction. After
heat inactivation
(65 C for 20 min), reactions are purified through a column to remove
unincorporated label.
Duplex localising agents (100 nM) are generated by annealing labeled
oligonucleotides with
equimolar amounts of unlabeled complementary oligonucleotide at 95 C for 3
min, followed
by slow cooling to room temperature. For cleavage assays, gRNA molecules are
annealed
by heating to 95 C for 30 s, followed by slow cooling to room temperature.
Cas9 (500 nM
final concentration) is pre-incubated with the annealed gRNA molecules (500
nM) in
cleavage assay buffer (20 mM HEPES pH 7.5, 100 mM KCI, 5 mM MgC12, 1 mM DTT,
5%
glycerol) in a total volume of 9 microlitre. Reactions are initiated by the
addition of 1
microlitre target DNA (10 nM) and incubated for 1 h at 37 C. Reactions are
quenched by
the addition of 20 microlitre of loading dye (5 mM EDTA, 0.025% SDS, 5%
glycerol in
formamide) and heated to 95 C for 5 min. Cleavage products are resolved on 12%
denaturing polyacrylamide gels containing 7 M urea and visualized by
phosphorimaging.
The resulting cleavage products indicate that whether the complementary
strand, the non-
complementary strand, or both, are cleaved.
One or both of these assays can be used to evaluate the suitability of a
candidate gRNA
molecule or candidate Cas9 molecule.
Indel Detection and Identification. Targeted genome modifications can also be
detected by
either Sanger or deep sequencing. For the former, genomic DNA from the
modified region
can be amplified with either primers flanking the target sequence of the gRNA.
Amp!icons
can be subcloned into a plasmid such as pUC19 for transformation, and
individual colonies
should be sequenced to reveal the clonal genotype.
Alternatively, deep sequencing is suitable for sampling a large number of
samples or target
sites. NGS primers are designed for shorter amplicons, typically in the 100-
200-bp size
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
210
range. For the detection of indels, it is important to design primers situated
at least 50 bp
from the Cas9 target site to allow for the detection of longer indels.
Amp!icons may be
assessed using commercially-available instruments, for example, the Illumine
system.
Detailed descriptions of NGS optimization and troubleshooting can be found in
the Illumine
user manual.
Ocular administration of the expanded cell population
In one aspect of the invention the expanded cell population obtainable by the
methods
.. according to the invention as described above is delivered to the eye. The
delivery is
performed under aseptic conditions.
In one embodiment relating to use for limbal stem cell therapy after a 360
limbal peritomy
the fibrovascular corneal pannus may be carefully removed from the surface.
In one aspect of the invention, the cell population is combined with a
localising agent
suitable for ocular delivery (as described further below) and delivered to the
eye. In a
preferred embodiment the cells and localising agent suitable for ocular
delivery are
combined and administered to the eye via a carrier such as for example a
therapeutic
contact lens or amniotic membrane. In an alternative embodiment the cells and
localising
agent suitable for use in the eye, such as a light curable biomatrix, like
GelMA, are delivered
to the eye via bioprinting.
In one embodiment, the invention provides a method of transplanting a
population of cells
comprising limbal stem cells or corneal endothelial cells onto the cornea of a
subject, the
method comprising expanding a population of cells comprising limbal stem cells
or corneal
endothelial cells by culturing said population with cell proliferation medium
comprising a
LATS inhibitor according to the invention, rinsing the expanded population of
cells to
substantially remove the LATS inhibitor, and administering said cells onto the
cornea of said
subject. Preferably said cells are combined with a biomatrix prior to said
administration. In a
specific embodiment said cells are combined with a biomatrix which is GelMA
prior to said
administration. In a more specific embodiment said corneal endothelial cells
are combined
with a biomatrix which is bioprinted onto the ocular surface. Particularly
preferably said
limbal stem cells or corneal endothelial cells are combined with a biomatrix
which is GelMA
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
211
and bioprinted onto the ocular surface by polymerising the GelMA by a light
triggered
reaction. In another embodiment said cells are combined with (1) thrombin and
fibrinogen or
(2) fibrin glue prior to said administration.
In another embodiment, the invention provides a method of transplanting a
population of
cells to the eye of a subject, comprising combining the cells with a biomatrix
to form a
cell/biomatrix mixture, injecting the mixture into the eye of the subject or
applying the mixture
onto the surface of the eye of the subject, and bioprinting the cells in or on
the eye by
guiding and fixing the cells, such as on the cornea, using a light source,
such as an
Ultraviolet A or white light source. In certain embodiments, the light source
produces light of
a wavelength that is at least 350 nm. In certain embodiments, the light source
produces light
in the 350 nm to 420 nm range. For example, an LED light source can be used to
produce a
light having a wavelength of 365 nm or 405 nm, or any other wavelength above
350 nm, or a
mercury lamp with a bandpass filter can be used to produce a light having a
wavelength of
365 nm. In another embodiment, the light source produces visible, white light
having a
wavelength, for example, in the 400 nm to 700 nm range. In certain
embodiments, the cells
are ocular cells, such as corneal cells (e.g., corneal endothelial cells),
lens cells, trabecular
mesh cells, or cells found in the anterior chamber. In a particular
embodiment, the cells are
corneal endothelial cells. Certain embodiments of such method include:
Embodiment x1. A method of transplanting a population of isolated cells to the
eye of
a subject, comprising combining the cells with a biomatrix to form a
cell/biomatrix
mixture, injecting the mixture into the eye of the subject, (e.g., into the
anterior
chamber) and bioprinting the cells in the eye by guiding and fixing the cells
in the eye
using a light source.
Embodiment x2. The method of Embodiment x1, wherein the isolated cells are
combined with a biomatrix which is GelMA and bioprinted onto the cornea by
polymerising the GelMA by a light triggered reaction.
Embodiment x3. The method of Embodiment x1 or Embodiment x2, wherein the light
source produces a light having a wavelength in the 350 nm to 700 nm range.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
212
Embodiment x4. The method of any one of Embodiments x1 to x3, wherein the
wavelength is 350 nm to 420 nm.
Embodiment x5. The method of any one of Embodiments x1 to x4, wherein the
wavelength is 365 nm.
Embodiment x6. The method of any one of Embodiments x1 to x5, wherein the
isolated cells are corneal endothelial cells.
Embodiment x7. A method of transplanting a population of isolated cells to the
eye of
a subject, comprising combining the cells with a biomatrix to form a
cell/biomatrix
mixture, applying the mixture onto the eye of the subject, and bioprinting the
cells on
the eye by guiding and fixing the cells on the eye using a light source.
Embodiment x8. The method of Embodiment x7, wherein the isolated cells are
combined with a biomatrix which is GelMA and bioprinted onto the ocular
surface by
polymerising the GelMA by a light triggered reaction.
Embodiment x9. The method of Embodiment x7 or Embodiment x8, wherein the light
source produces a light having a wavelength in the 350 nm to 700 nm range.
Embodiment x10. The method of any one of Embodiments x7 to x9, wherein the
wavelength is 350 nm to 420 nm.
Embodiment x11. The method of any one of Embodiments x7 to x10, wherein the
wavelength is 365 nm.
Embodiment x12. The method of any one of Embodiments x7 to x11, wherein the
isolated cells are limbal stem cells.
In an alternative embodiment the expanded cell population obtainable by the
methods
according to the invention as described above may be delivered directly via a
therapeutic
contact lens to the eye, without use of a localising agent suitable for ocular
delivery (such as
GelMA or fibrin glue).
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
213
Localising agent suitable for ocular delivery
In an embodiment of the invention the cell preparation may be delivered to the
eye via a
localising agent suitable for ocular use. The cells may be embedded within the
localising
agent or adhered to the surface of the localising agent, or both.
The type of localising agent is not limited as long as it is able to carry
LSCs or CECs and is
suitable for use in the eye. In a preferred embodiment, the localising agent
is degradable
and biocompatible. Where CECs are delivered, preferably the localising agent
can facilitate
CEC attachment to the cornea after surgical delivery to the surface of the
eye.
In a preferred embodiment the cells are only combined with the localising
agent after cell
population expansion. In a particularly preferred embodiment the expanded cell
population is
combined with the localising agent suitable for ocular delivery after rinsing
the cell
population to substantially remove the presence of the LATs inhibitor
according to the
invention. In one embodiment, the LSCs or CECs and localising agent are
combined and
stored in a form suitable for ocular use. In another embodiment, the LSCs or
CECs and
localising agent are stored separately and combined immediately prior to
ocular use.
The localising agent is preferably selected from the list consisting of
fibrin, collagen, gelatin,
cellulose, amniotic membrane, fibrin glue, a combination of thrombin and
fibrinogen,
polyethylene (glycol) diacrylate (PEGDA), GelMA, (which is methacrylamide
modified
gelatin, and is also known as gelatin methacrylate), localising agents
comprising a polymer,
cross-linked polymer, or hydrogel comprising one or more of hyaluronic acid,
polyethylene
glycol, polypropylene glycol, polyethylene oxide, polypropylene oxide,
poloxamer, polyvinyl
alcohol, polyacrylic acid, polymethacrylic acid, polyvinyl pyrrolidone,
poly(lactide-co-
glycolide), alginate, gelatin, collagen, fibrinogen, cellulose,
methylcellulose,
hydroxyethylcellulose, hydroxypropyl cellulose, hydroxypropylmethylcellulose,
hydroxypropyl-guar, gellan gum, guar gum, xanthan gum and
carboxymethylcellulose, as
well as derivatives thereof, co-polymers thereof, and combinations thereof.
In a more preferred embodiment the localising agent is selected from the list
consisting of
fibrin, collagen, gelatin, amniotic membrane, fibrin glue, a combination of
thrombin and
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
214
fibrinogen, polyethylene (glycol) diacrylate (PEGDA), GelMA, localising agents
comprising a
polymer, cross-linked polymer, or hydrogel comprising one or more of
hyaluronic acid,
polyethylene glycol, polypropylene glycol, polyethylene oxide, polypropylene
oxide,
poloxamer, polyacrylic acid, poly(lactide-co-glycolide), alginate, gelatin,
collagen, fibrinogen,
hydroxypropylmethylcellulose and hydroxypropyl-guar, as well as derivatives
thereof, co-
polymers thereof, and combinations thereof.
In a preferred embodiment the expanded cell population according to the
invention may be
delivered to a recipient via a localising agent which is a biomatrix. In a
more preferred
embodiment the localising agent is a light curable, degradable biomatrix.
Preferably this is
able to be injected into the eye. A specific example of a biomatrix is GelMA,
which is
methacrylamide modified gelatin, and is also known as gelatin methacrylate.
GelMA may be prepared according to standard protocols known in the art (Van
Den Bulcke
et al., Biomacromolecules, 2000, p.31-38; Yue et al., Biomaterials, 2015,
p.254-271). For
example, gelatin from porcine skin (gel strength 300 g Bloom, Type A) is
dissolved in PBS
without calcium and magnesium (Dulbeccos PBS), and methacrylic anhydride may
be
added with strong agitation into the gelatin solution to reach the desired
concentration (e.g.,
8% (vol/vol). The mixture may be stirred before and after adding further DPBS.
The diluted
mixture may be purified via dialysis against Milli-Q water using dialysis
tubing to remove
methacrylic acid. The purified samples may optionally be lyophilized and the
solid stored at -
80 C, -20 C, or 4 C until further use.
A GelMA stock solution is prepared by dissolving lyophilized GelMA in a
formulation suitable
for ocular use comprising pharmaceutically acceptable excipients. To prepare a
GelMA
stock solution, lyophilized GelMA may be dissolved in DPBS. After the GelMA is
fully
dissolved, a photoinitiator (for example such as lithium phenyl-2,4,6-
trimethylbenzoylphosphinate) may be introduced into the GelMA solution. To
adjust the pH
to neutral, NaOH may be added to the solution before filtering using 0.22
micrometre sterile
membranes. The final filtrate may be separated into aliquots and stored at 4
C until further
use.
In one aspect according to the invention, the cells are encapsulated within
the biomatrix
using a photoinitiator to polymerise the biomatrix, which is preferably GelMa.
Suitable
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
215
photoinitiator agents are Irgacure 2959, lithium phenyl-2,4,6-
trimethylbenzoylphosphinate,
sodium phenyl-2,4,6-trimethylbenzoylphosphinate, lithium bis(2,4,6-
trimethylbenzoyl)phosphinate, sodium bis(2,4,6-trimethylbenzoyl)phosphinate,
Dipheny1(2,4,6-trimethylbenzoyOphosphine oxide, eosin Y, riboflavin phosphate,
camphorquinone, Quantacure BPQ, Irgacure 819, Irgacure 1850, and Darocure
1173. In a
preferred embodiment, the photoiniator is lithium phenyl-2,4,6-
trimethylbenzoylphosphinate,
sodium phenyl-2,4,6-trimethylbenzoylphosphinate, riboflavin phosphate. In
another
embodiment the photoinitiator is lithium phenyl-2,4,6-
trimethylbenzoylphosphinate.
.. Prior to polymerization, the light curable biomatrix is combined with a
suitable photoinitiator
in a formulation suitable for ocular use comprising pharmaceutically
acceptable excipients in
suitable containers known in the art such as vials. The photoinitiator may be
combined with
the biomatrix prior to mixing with cells; alternatively the photoinitiator may
be combined with
the biomatrix after mixing with cells; alternatively the photoiniator may be
added to the cells
first, then combined with the biomatrix. The concentration of biomatrix and
photoinitiator is
dependent on the specific biomatrix and specific photoinitiator used, but is
chosen to provide
polymerization within a convenient light exposure duration, typically less
than about 5
minutes; preferably less than about 2 minutes; more preferably less than about
one minute.
In one embodiment the photoinitiator is lithium phenyl-2,4,6-
trimethylbenzoylphosphinate
and its concentration in the formulation for cell delivery to the eye is about
0.01% w/v to
about 0.15% w/v. In another aspect the lithium phenyl-2,4,6-
trimethylbenzoylphosphinate
concentration in the formulation for cell delivery to the eye is about 0.05%
w/v or about
0.075% w/v. LAP may be synthesized using published procedure (Biomaterials
2009, 30,
6702-6707) and is also available from TCI (Prod. # L0290) and Biobots
(BioKey).
The cells may be added to the GelMA in suitable containers known in the art
such as vials or
tubes. The cells may for example be added by pipetting into the GelMA and
mixing by gentle
pipetting up and down. In one embodiment the GelMA concentration in the
composition
suitable for ocular delivery is about 10 to about 200 mg/mL, or about 25 to
about 150
mg/mL, or about 25 to about 75 mg/mL. In a preferred embodiment the GelMA
concentration
in the composition suitable for ocular delivery is about 25 mg/mL, about 50
mg/mL or about
75 mg/mL.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
216
To polymerise the light curable biomatrix, the biomatrix, photoinitiator, and
cells are exposed
to a light source for a preferred duration, as described above. The wavelength
of light used
for polymerization will depend on the photochemical properties of the specific
photoinitiator
used. For example, photoinitation of polymerization for Irgacure 2959 will
occur with light of
wavelength between 300-370nm; photoinitation of polymerization for lithium
phenyl-2,4,6-
trimethylbenzoylphosphinate will occur with light of wavelength between 300-
420nm;
photoinitation of polymerization for riboflavin-5'-phosphate will occur with
light of wavelength
between 300-500nm. The light source used may emit a range of wavelengths, like
that
achieved with incandescent lamps, gas discharge lamps, or metal vapor lamps;
alternatively, the light source used may emit a narrow range of wavelengths,
like that
achieved with optical filters or with an light emitting diode (LED).
Preferably, the light source
used does not emit light with wavelength less than 315nm to avoid the damaging
effects of
UV irradiation on cells. In one embodiment, the light source is a white light
source with a
spectral range of 415-700nm. In another embodiment the light source is a LED
light source
with spectral range of about 365 5nm, about 375 5nm, about 385 5nm, about 395
5nm,
about 405 5nm, about 415 5nm, about 425 5nm, about 435 5nm, about 445 5nm,
about
455 5nm, or about 465 5nm. The intensity of light is chosen to minimize
phototoxicity and
provide polymerization within a convenient light exposure duration, typically
less than about
5 minutes; preferably less than about 2 minutes; more preferably less than
about one
minute. One indication of polymerization is an increase in solution viscosity.
Another
indication of polymerization is the onset of gelation.
The polymerization of the biomatrix may occur on the ocular surface via
bioprinting
techniques, or alternatively on a carrier that is then transplanted to the
ocular surface.
.. Optionally the polymerization of the biomatrix may occur on the cornea
surface in the
anterior chamber, or alternatively on a carrier that is then transplanted to
the cornea surface
in the anterior chamber.
Carrier
The cells (e.g., modified LSCs) and localising agent suitable for ocular
delivery are
preferably delivered via a carrier such as a contact lens or amniotic
membrane.
Contact lenses suitable for use according to the invention (e.g., for use with
modified LSCs)
are preferably those which conform to the patient's corneal curvature and are
able to be well
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
217
tolerated by the patient in clinical practice for continuous use as bandage
contact lenses for
several days.
Examples of suitable types of contact lens according to the invention are
consistent with
what has been extensively validated in clinical use for long-term bandage
contact lens use
with Boston keratoprosthesis type 1 (which can be also used in patients with
limbal stem cell
deficiency) and described in : Thomas, Merina M.D.; Shorter, Ellen 0.D.;
Joslin, Charlotte E.
0.D., Ph.D.; McMahon, Timothy J. 0.D.; Cortina, M. Soledad M.D.Contact Lens
Use in
Patients With Boston Keratoprosthesis Type 1: Fitting, Management, and
Complications.
Eye Contact Lens. 2015 Nov;41(6):334-40.
A contact lens can be of any appropriate material known in the art or later
developed, and
can be a soft lens, a hard lens, or a hybrid lens, preferably a soft lens,
more preferably a
conventional hydrogel contact lens or a silicone hydrogel (SiHy) contact lens.
A "conventional hydrogel contact lens" refers to a contact lens comprising a
hydrogel bulk
(core) material which is a water-insoluble, crosslinked polymeric material, is
theoretically free
of silicone, and can contain at least 10% by weight of water within its
polymer matrix when
fully hydrated. A conventional hydrogel contact lens typically is obtained by
copolymerization
of a conventional hydrogel lens formulation (i.e., polymerizable composition)
comprising
silicone-free, hydrophilic polymerizable components known to a person skilled
in the art.
Examples of conventional hydrogel lens formulation for making commercial
hydrogel contact
lenses include, without limitation, alfafilcon A, acofilcon A, deltafilcon A,
etafilcon A, focofilcon
A, helfilcon A, helfilcon B, hilafilcon B, hioxifilcon A, hioxifilcon B,
hioxifilcon D, methafilcon A,
methafilcon B, nelfilcon A, nesofilcon A, ocufilcon A, ocufilcon B, ocufilcon
C, ocufilcon D,
omafilcon A, phemfilcon A, polymacon, samfilcon A, telfilcon A, tetrafilcon A,
and vifilcon A.
A "SiHy contact lens" refers to a contact lens comprising a silicone hydrogel
bulk (core)
material which is a water-insoluble, crosslinked polymeric material containing
silicone and can
contains at least 10% by weight of water within its polymer matrix when fully
hydrated. A
silicone hydrogel contact lens typically is obtained by copolymerization of a
silicone hydrogel
lens formulation comprising at least silicone-containing polymerizable
component and
hydrophilic polymerizable components known to a person skilled in the art.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
218
Examples of SiHy lens formulation for making commercial SiHy contact lenses
include,
without limitation, asmofilcon A, balafilcon A, comfilcon A, delefilcon A,
efrofilcon A, enfilcon
A, fanfilcon A, galyfilcon A, lotrafilcon A, lotrafilcon B, narafilcon A,
narafilcon B, senofilcon A,
senofilcon B, senofilcon C, smafilcon A, somofilcon A, and stenfilcon A.
In a preferred embodiment the carrier is a contact lens selected from the
group consisting of
Balafilcon A, Lotrafilcon A, Lotrafilcon B, Senofilcon A and methafilcon A.
In a particularly preferred embodiment the carrier is a contact lens, which is
Lotrafilcon B.
The carrier may be held in place on the ocular surface using fibrin glue or
sutures to prevent
eye movements from dislodging the construct.
The carrier combined with biomatrix and cells may be left on the eye for a
range of times in
order to deliver the cells, for example a few days to one week, preferably one
week.
Other delivery methods:
In an alternative embodiment, the LSCs may be delivered as a cell suspension
to the ocular
surface (without a localising agent such as a biomatrix and with/or without a
carrier such as
a contact lens). Compounds and excipients known in the art to improve tissue
adhesion
such as mucoadhesive agents, viscosity enhancers, or reverse thermal gelators
may be
included in the formulation.
.. Bioprintind step
The population of ocular cells, e.g., corneal endothelial cells, obtainable
according to the
method of cell population expansion according to the invention may be grafted
to the eye of
a subject, e.g., to the cornea of a subject.
The cell population according to the invention may be delivered via a
localising agent
suitable for ocular use which is a light curable, degradable biomatrix such as
GelMA. The
following methods describe procedures for controlling the delivery to the
inner wall of the
cornea.
.. Method 1. Bubble depression method
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
219
The dysfunctional endothelial cells may first be detached from the inner wall
of the cornea
by peeling/scraping or in a controlled manner using photodisruption with a
femtosecond
laser. A small bolus of the cell-laden biomatrix is then injected near the
interior surface of the
cornea. This may be done manually using a standard syringe or custom
applicator. It can
also be controlled through a surgical system (e.g., constellation) or syringe
pumps. A gas
bubble is then injected beneath the bolus. The gas bubble squeezes the bolus
against the
posterior cornea, creating a thin coating. The entire gel is then cured using
a using a UV or
near UV light source, or any other spectral band needed to cure the biomatrix.
Alternatively,
the dysfunctional tissue may be left, and the biomatrix cured over top of it.
The light source
can be focused into different sizes using other optical focusing methods to
control the curing
area. The remaining uncured area can be flushed out using
irrigating/aspirating canula.
Method 2. Subtractive method using femtosecond laser
The dysfunctional endothelial cells may first be detached from the inner wall
of the cornea
by peeling/scraping or in a controlled manner using photodisruption with a
femtosecond
laser. Alternatively, they may be left in place. The cell-laden biomatrix is
then injected onto
the interior surface of the cornea covering the void where tissue was removed
or over the
dysfunctional tissue. This may be done manually using a standard syringe or
custom
applicator. It can also be controlled through a surgical system (e.g.
constellation) or syringe
pumps. The biomatrix is then cured using a using a UV or near UV light source,
or any other
spectral band needed to cure the biomatrix. The femtosecond laser is then used
to detach
excess material, controlling the thickness and area to a desired distribution.
The excess
material is then removed with forceps through a corneal incision.
.. Method 3. Stain mask and absorption based thickness control
A biocompatible stain (Trypan Blue, Brilliant Blue, etc.) is firstly used to
dye the inner surface
of the cornea. The dysfunctional endothelial cells are then detached from the
inner wall of
the cornea by peeling/scraping. The cell-laden biomatrix containing the
biocompatible stain
is then injected onto the interior surface of the cornea covering the void
where tissue was
removed. The biomatrix is then cured using a using a UV or near UV light
source, or any
other spectral band needed to cure the biomatrix. The stain in the corneal
tissue increases
the light absorption acting as a mask to control the area of the cured
biomatrix. Similarly, the
stain in the biomatrix increases the absorption of light thereby controlling
the depth/thickness
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
220
of the cured material. Uncured gel material is then flushed from the anterior
chamber using
an irrigating/aspirating cannula.
Method 4. Dry anterior chamber application
The dysfunctional endothelial cells may first be detached from the inner wall
of the cornea
by peeling/scraping or in a controlled manner using photodisruption with a
femtosecond
laser. Alternatively it may be left in place. The anterior chamber of the
anterior segment is
then drained of aqueous and replaced with gas (e.g. air). The cell-laden
biomatrix is then
applied to interior surface of the cornea in small controlled droplets
(allowing surface tension
to disperse the drops), or painted using a brush or soft tip cannula.
Hyaluronic acid may be
applied to the biomatrix to alter its viscous properties and enable better
control over
dispensing/application. The entire biomatrix is then cured using a using a UV
or near UV
light source, or any other spectral band needed to cure the biomatrix.
Finally, the anterior
chamber is then filled again with balanced salt solution.
Method 5. Naturally Buoyant Formulation
The dysfunctional endothelial cells may first be detached from the inner wall
of the cornea
by peeling/scraping or in a controlled manner using photodisruption with a
femtosecond
laser. A small bolus of the cell-laden biomatrix is then injected near the
interior surface of the
cornea. The biomatrix is formulated to be naturally buoyant relative to
aqueous humor or
aerated to achieve the same effect. This causes the biomatrix to naturally
rise to posterior
cornea, creating a thin coating. The entire biomatrix is then cured using a
using a UV or near
UV light source, or any other spectral band needed to cure the biomatrix.
Alternatively, the
dysfunctional tissue may be left, and the biomatrix cured over top of it. The
UV light source
can be focused into different sizes using optical focusing methods to control
the curing area.
The remaining uncured area can be flushed out using aspiration canula.
Other delivery methods
In an alternative embodiment an expanded cell population, such as CECs as
described
herein, may be delivered as a cell suspension (without a localising agent such
as a light
curable, degradable biomatrix) and left to attach by gravity by having the
patient look down
for 3 hours. Compounds and excipients known in the art to improve tissue
adhesion such as
adhesive agents, viscosity enhancers, or reverse thermal gelators may be
included in the
formulation.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
221
In yet another alternative embodiment an expanded cell population, such as
CECs as
described herein can also be delivered by using magnetic beads. A suspension
of
CECs/beads in a medium suitable for ocular delivery is prepared and this is
then injected
into the eye. Cell attachment is promoted by a magnet applied to the eye.
(Magnetic field-
guided cell delivery with nanoparticle-loaded human corneal endothelial cells.
Moysidis SN,
Alvarez-Delfin K, Peschansky VJ, Salero E, Weisman AD, Bartakova A, Raffa GA,
Merkhofer RM Jr, Kador KE, Kunzevitzky NJ, Goldberg JL.Nanomedicine. 2015
Apr;11(3):499-509. doi: 10.1016/j.nano.2014.12.002.)
Therapeutic uses
The modified ocular cell or the ocular cell population according to the
present disclosure
(e.g., LSC, CEC, LSC population or CEC populiation) may be used in a method of
treatment
or prophylaxis of an ocular disease or disorder comprising administering to a
subject in need
thereof of a therapeutically effective amount of a cell population comprising
ocular cells
(e.g., LSCs or CECs).
The limbal stem cell population according to the invention (e.g., LSCs with
reduced or
eliminated expression of B2M by a CRISPR system, e.g., S. pyogenes Cas9 CRISPR
system) may be used in a method of treatment or prophylaxis of an ocular
disease or
disorder comprising administering to a subject in need thereof of a
therapeutically effective
amount of a cell population comprising limbal stem cells. Preferably the
ocular disease or
disorder is associated with limbal stem cell deficiency.
Limbal stem cell deficiency may arise as a result of several diverse
conditions including but
not limited to:
- direct stem cell damage from chemical or thermal burns or radiation
injury;
- congenital conditions such as aniridia, sclerocornea, multiple endocrine
neoplasia;
- autoimmune disorders such as Stevens Johnson syndrome or ocular cicatricial
pemphigoid or collagen vascular diseases;
- chronic non-auto-immune inflammatory disorders such as contact lens use,
dry eye
disease, rosacea, staph marginal, keratitis (bacterial, fungal & viral),
pterygia or
neoplasm;
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
222
- iatrogenic, such as after multiple eye surgeries, excision of pterygia or
neoplasm,
cryotherapy;
- as a result of medication toxicity such as preservatives (thimerosal,
benzalkonium),
topical anesthetics, pilocarpine, beta blockers, mitomycin, 5-fluorouracil,
silver
nitrate, and oral medications causing Stevens Johnson syndrome.
(See: Dry Eye: a practical guide to ocular surface disorders and stem cell
surgery. SLACK
2006- Rzany B, Mockenhaupt M, Baur S et al. J. Clin. Epidemiol. 49,769-773
(1996)).
The most commonly encountered causes of limbal stem cell deficiency in
clinical practice
are chemical burns, aniridia, Stevens Johnson Syndrome and contact lens use.
More preferably the ocular disease or disorder is limbal stem cell deficiency
which arises
due an injury or disease or disorder selected from the group consisting of
chemical burns,
thermal burns, radiation injury, aniridia, sclerocornea, multiple endocrine
neoplasia, Stevens
Johnson syndrome, ocular cicatricial pemphigoid, collagen vascular diseases,
chronic non-
auto-immune inflammatory disorders arising from contact lens use, dry eye
disease,
rosacea, staph marginal, keratitis (including bacterial, fungal & viral
keratitis), pterygia or
neoplasm, limbal stem cell deficiency arising after multiple eye surgeries or
excision of
pterygia or neoplasm or cryotherapy; and limbal stem cell deficiency arising
as a result of
medication toxicity from a medication such as a medication selected from the
group
consisting of preservatives (e.g., thimerosal, benzalkonium), topical
anaesthetics,
pilocarpine, beta blockers, mitomycin, 5-fluorouracil, silver nitrate, and
oral medications
causing Stevens Johnson syndrome.
.. In a specific embodiment, the present invention provides a method of
treating limbal stem
cell deficiency by administering to a subject in need thereof an effective
amount of a limbal
stem cell population (e.g., limbal stem cell population with reduced or
eliminated expression
of B2M by a CRISPR system, e.g., S. pyogenes Cas9 CRISPR system) obtainable by
the
method of cell population expansion according to the invention.
In a more specific embodiment, the present invention provides a method of
treating limbal
stem cell deficiency which arises due an injury or disorder selected from the
group
consisting of chemical burns, thermal burns, radiation injury, aniridia,
sclerocornea, multiple
endocrine neoplasia, Stevens Johnson syndrome, ocular cicatricial pemphigoid,
collagen
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
223
vascular diseases, chronic non-auto-immune inflammatory disorders arising from
contact
lens use, dry eye disease, rosacea, staph marginal, keratitis (including
bacterial, fungal &
viral keratitis), pterygia or neoplasm, limbal stem cell deficiency arising
after multiple eye
surgeries, or excision of pterygia or neoplasm or cryotherapy; and limbal stem
cell deficiency
arising as a result of medication toxicity from a medication selected from the
group
consisting of preservatives (thimerosal, benzalkonium), topical anesthetics,
pilocarpine, beta
blockers, mitomycin, 5-fluorouracil, silver nitrate, and oral medications
causing Stevens
Johnson syndrome by administering to a subject in need thereof a
therapeutically effective
amount of a limbal stem cell population (e.g., limbal stem cell population
with reduced or
eliminated expression of B2M by a CRISPR system, e.g., S. pyogenes Cas9 CRISPR
system) obtainable by the method of cell population expansion according to the
invention.
In yet a more specific embodiment, the present invention provides a method of
treating
limbal stem cell deficiency which arises due an injury or disease or disorder
selected from
the group consisting of chemical burns, aniridia, Stevens Johnson Syndrome and
contact
lens use by administering to a subject in need thereof a thereapeutically
effective amount of
a limbal stem cell population (e.g., limbal stem cell population with reduced
or eliminated
expression of B2M by a CRISPR system, e.g., S. pyogenes Cas9 CRISPR system)
obtainable by the method of cell population expansion according to the
invention.
When an adult is a recipient (transplant recipient), in a particular
embodiment, greater than
1,000 p63a1pha expressing cells may be administered to a patient in the
methods of
treamtent according to the invention. In a particular embodiment, 1,000 to
100,000 p63a1pha
expressing cells may be administered to a patient in the methods of treatment
according to
the invention.
The corneal endothelial cell population (e.g., corneal endothelial cell
population with reduced
or eliminated expression of B2M by a CRISPR system, e.g., S. pyogenes Cas9
CRISPR
system) according to the invention may be used in a method of treatment or
prophylaxis of
an ocular disease or disorder comprising administering to a subject in need
thereof of a
therapeutically effective amount of a cell population comprising corneal
endothelial cells.
Preferably the ocular disease or disorder is associated with decreased corneal
endothelial
cell density. In a preferred embodiment the ocular disease or disorder is
corneal endothelial
dysfunction.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
224
More preferably the ocular disease or disorder is corneal endothelial
dysfunction which is
selected from the group consisting of Fuchs endothelial corneal dystrophy,
bullous
keratopathy (including pseudophakic bullous keratopathy and aphakic bullous
keratopathy),
corneal transplant failure, posterior polymorphous corneal dystrophy,
congenital hereditary
endothelial dystrophy, X-linked endothelial corneal dystrophy, aniridia, and
corneal
endothelitis. In a specific embodiment the ocular disease or disorder is
selected from the
group consisting of Fuchs endothelial corneal dystrophy, bullous keratopathy
(including
pseudophakic bullous keratopathy and aphakic bullous keratopathy) and corneal
transplant
failure.
In a specific embodiment, the present invention provides a method of treating
corneal
endothelial dysfunction by administering to a subject in need thereof an
effective amount of
a corneal endothelial cell population (e.g., corneal endothelial cell
population with reduced or
eliminated expression of B2M by a CRISPR system, e.g., S. pyogenes Cas9 CRISPR
system) obtainable by the method of cell population expansion according to the
invention.
In a more specific embodiment, the present invention provides a method of
treating corneal
endothelial dysfunction which is selected from the group consisting of Fuchs
endothelial
corneal dystrophy, bullous keratopathy (including pseudophakic bullous
keratopathy and
aphakic bullous keratopathy), corneal transplant failure, posterior
polymorphous corneal
dystrophy, congenital hereditary endothelial dystrophy, X-linked endothelial
corneal
dystrophy, aniridia, and corneal endothelitis by administering to a subject in
need thereof an
effective amount of a corneal endothelial cell population (e.g., corneal
endothelial cell
population with reduced or eliminated expression of B2M by a CRISPR system,
e.g., S.
pyogenes Cas9 CRISPR system) obtainable by the method of cell population
expansion
according to the invention.
In yet a more specific embodiment, the present invention provides a method of
treating
corneal endothelial dysfunction selected from the group consisting of Fuchs
endothelial
corneal dystrophy, bullous keratopathy (including pseudophakic bullous
keratopathy and
aphakic bullous keratopathy) and corneal transplant failure by administering
to a subject in
need thereof an effective amount of a corneal endothelial cell population
(e.g., corneal
endothelial cell population with reduced or eliminated expression of B2M by a
CRISPR
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
225
system, e.g., S. pyogenes Cas9 CRISPR system) obtainable by the method of cell
population expansion according to the invention.
When an adult is a recipient (transplant recipient), in particular aspects,
the corneal
endothelial cell population (e.g., corneal endothelial cell population with
reduced or
eliminated expression of B2M by a CRISPR system, e.g., S. pyogenes Cas9 CRISPR
system) for use in the method of treatment according to the invention
preferably has a final
cell density in the eye of about at least 500 cells/mm2 (area), preferably
1,000 to 3,500
cells/mm2 (area), more preferably 2,000 to about 4,000 cells/mm2 (area).
In certain embodiments, a patient's vision is improved by a method of
treatment provided
herein. Visual acuity tests are well known in the art, including, for example
the Snellen and
Sloan acuity tests, and Early Treatment Diabetic Retinopathy Study (ETDRS)
acuity test. An
improvement in vision can be measured, for example, using a best corrected
visual acuity
(BCVA) measurement. In certain embodiments, the BCVA of a patient treated as
provided
herein improves by at least 1, 2, 3, 4, 5 or more lines as measured by ETDRS
letters
following treatment with a modified cell or cell population or composition of
the invention as
provided herein.
EXAMPLES
The following examples are provided to further illustrate the invention but
not to limit its
scope. Other variants of the invention will be readily apparent to one of
ordinary skill in the
art and are encompassed by the appended claims.
Unless defined otherwise, the technical and scientific terms used herein have
the same
meaning as that usually understood by a specialist familiar with the field to
which the
disclosure belongs.
Example 1: Human limbal epithelial cell isolation
Research-consented cadaveric human corneas were obtained from eye banks.
Limbal rims
were dissected and partially dissociated in a 1.2 mg/ml dispase solution for 2
hours at 37 C
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
226
followed by 10 minutes in TrypLE (Life Technologies). Pieces of limbal crypts
were then
carefully cut out of the partially dissociated limbal rims and rinsed by
centrifugation). Cells
obtained in this manner were used in the Examples below.
Example 2: Exposure of cells to LATS inhibitors and measurement of
intracellular YAP distribution
Cells obtained as described in Example 1 were plated in glass-bottom black
wall 24-well
dishes in limbal epithelium cell culture medium (DMEM F12 supplemented with
10% human
serum and 1.3 mM calcium chloride) supplemented with LATS inhibitor compound
example
no. 4 or 3 at a concentration of 10 micromolar or supplemented in DMSO as a
negative
control. Cells were cultured under these conditions for 24 hours at 37 C in 5%
CO2.
To measure the effect of the LATS inhibitors on the downstream target YAP,
intracellular
YAP distribution was analyzed by immunohistochemistry. Cell cultures were
fixed with 4%
PFA for 20 minutes, permeabilized and blocked in a blocking solution of 0.3%
Triton X-100
(Sigma-Aldrich) and 3% donkey serum in PBS for 30 minutes. Cells were then
labeled with
primary antibody in the blocking solution for 12 hours at 4 C. Primary
antibody used was
anti-YAP from Santa Cruz Biotechnology. Samples were washed in PBS three times
and
donkey-raised secondary antibody Alexa Fluor 488 (Molecular Probes) at 1:500
dilution
were applied for 30 minutes at room temperature. Negative control was omitted
primary
antibody (data not shown). Fluorescence was observed using a Zeiss LSM 880
confocal
microscope.
Only weak YAP immunostaining was observed in the nucleus of LSCs cultured
without the
LATS inhibitors (DMSO control). YAP immunostaining was stronger in the nucleus
of LSCs
exposed to the LATS inhibitor compound 2-(3-methyl-1H-pyrazol-4-y1)-N-(1-
methylcyclopropyl)pyrido[3,4-d]pyrimidin-4-amine or 2,4-dimethy1-4-{[2-
(pyridin-4-
yOpyrido[3,4-d]pyrimidin-4-yl]amino}pentan-2-ol prepared as described in U.S.
Patent
.. Application No. 15/963,816 and International Application No.
PCT/162018/052919 (WO
2018/198077), filed April 26, 2018 (data not shown).
Example 3 : Exposure of cells to LATS inhibitors and measurement of YAP
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
227
phosphorylation
Cells obtained as described in Example 1 were detached from the culture dish
with
Accutase for 10 minutes at 37 C, cell suspensions were rinsed by
centrifugation and plated
in DMEM F12 supplemented with 10% human serum and 1.3 mM calcium chloride in 6-
well
plates (Corning) and cultured without LATS inhibitor compounds for 2-4 days.
The medium was then replaced by fresh limbal epithelium cell culture medium
(DMEM F12
supplemented with 10% human serum and 1.3 mM calcium chloride) supplemented
with
LATS inhibitor compound example no. 4 or 3 at a concentration of 10 micromolar
or
supplemented in DMSO as a negative control. Cells were cultured under these
conditions
for 1 hour at 37 C in 5% CO2.
To measure the effect of the LATS inhibitors on the downstream target YAP, the
YAP
phosphorylation levels were measured by western blot as follows. The cell
pellets were
obtained by trypsin dissociation and centrifugation and washed with PBS. The
pellets were
lysed with 30 microlitres of RIPA lysis buffer containing protease inhibitor
cocktail (Life
Technologies) for 30 minutes, with vortexing every 10 minutes. The cell debris
were then
pelleted at 4 C for 15 minutes at 14k rpm and the protein lysate was
collected. Protein
concentration was quantified using a micro BCA kit (Pierce). Fifteen
micrograms of total
protein was loaded in each well of 4-20% TGX gels (BioRad) and Western
blotting was
performed according to the manufacturer's instructions. Membranes were probed
with
phospho-YAP (5er127) (CST, 1:500) or total Yap (Abnova, 1:500) antibody and
actin
(Abcam) labelling was used as loading control. Membranes were stained with HRP-
conjugated secondary antibodies, rinsed and imaged using a ChemiDoc system
(Biorad)
according to the manufacturer's instructions.
Western blot analysis (see Figure 1) showed that both compound example no. 4
and 3
caused a reduction in YAP phosphorylation levels in human LSCs.These results
suggest
that the LATS inhibitor compound example no. 4 and 3 can activate YAP
signaling in human
LSCs.
Example 4: Human limbal stem cell population expansion and
immunohistochemical observation of cellular phenotype
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
228
Cells obtained as described in Example 1 were plated in 24-well plates
(Corning) in limbal
epithelium cell culture medium (DMEM F12 supplemented with 10% human serum and
1.3
mM calcium chloride) supplemented with LATS inhibitor compound example no. 4
or 3 at a
concentration of 10 micromolar or supplemented in DMSO as a negative control.
Cells were
first cultured at 37 C in 5% CO2 for 6 days after isolation without passaging
(Figures 2A, 2B
and 2C).
To evaluate the ability of the compounds to enable LSC expansion after two
passages,
LSCs were passaged and cultured for two weeks in the presence of compound
example 3 to
enable expansion (Figure 2D). Limbal stem cells (LSCs) were passaged by
treating cultures
with Accutase for 10 minutes at 37 C, rinsing the cell suspension by
centrifugation and
plating cells in fresh LSC culture medium supplemented with LATS inhibitor
compound
example 3.
In order to observe that the expanded cell population expressed p63a1pha, this
was
measured by immunohistochemistry as follows. Cell cultures were fixed with 4%
PFA for 20
minutes, permeabilized and blocked in a blocking solution of 0.3% Triton X-100
(Sigma-
Aldrich) and 3% donkey serum in PBS for 30 minutes. Cells were then labeled
with primary
antibody in the blocking solution for 12 hours at 4 C. Primary antibody used
was p63a1pha
from Cell Signalling. Samples were washed in PBS three times and donkey-raised
secondary antibody Alexa Fluor 488 (Molecular Probes) at 1:500 dilution were
applied for 30
minutes at room temperature. Cells were counter-stained with a human nuclear
antigen
antibody (Millipore) at a 1:500 dilution in order to label all cells in the
culture and confirm
their human identity. Negative control was omitted primary antibody (data not
shown).
Fluorescence was observed using a Zeiss LSM 880 confocal microscope.
Figure 2A shows that in the presence of growth medium and DMSO, only a few
isolated
cells attach to the culture dish and survive up to 6 days. Most cells
expressed the human
nuclear marker, but few expressed p63a1pha. In contrast, in the presence of
LATS inhibitors
compound example no. 4 (Figure 2B) and compound example no. 3 (Figure 2C), the
cells
formed colonies and expressed p63a1pha. This result indicated that the LATS
inhibitors
promote the expansion of the population of cells with the p63a1pha-positive
phenotype.
Figure 2D: Passaging cells and culturing them in the presence of LATS
inhibitor compound
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
229
example no. 3 for two weeks enabled cell population expansion and the
formation of
confluent cultures expressing p63a1pha.
Example 5: Human limbal stem cell population expansion and measurement
thereof
Cells obtained as described in Example 1 were plated in 48-well plates
(Corning) in
XVIV015 medium (Lonza) supplemented with LATS inhibitors (as listed in Table 2
and 3
below) at a concentration of 10 micromolar or supplemented in DMSO as a
negative control.
Cells were cultured at 37 C in 5% CO2.
For each compound, two sets of cultures were generated. A first set of
cultures was fixed in
4% PFA for 20 minutes at room temperature after cells isolated from the cornea
had
attached to the cell culture dish (typically 24h after cell plating). A second
set of cultures was
fixed in 4% PFA for 20 minutes at room temperature after being cultured for
two passages.
Cells were passaged when they reached 90-100% confluence.
In order to observe that the expanded cell population expressed p63a1pha, this
was
measured by immunohistochemistry as follows. The fixed cell cultures were
permeabilized
and blocked in a blocking solution of 0.3% Triton X-100 (Sigma-Aldrich) and 3%
donkey
serum in PBS for 30 minutes. Cells were then labeled with primary antibody in
the blocking
solution for 12 hours at 4 C. Primary antibody used was p63a1pha from Cell
Signalling.
Samples were washed in PBS three times and donkey-raised secondary antibody
Alexa
Fluor 488 (Molecular Probes) at 1:500 dilution were applied for 30 minutes at
room
temperature. Cell nuclei were then labeled in a solution of 0.5 micromolar of
Sytox Orange
(ThermoFisher) in PBS for 5 minutes at room temperature.
To evaluate the percentage of p63a1pha-positive cells, the number of cells
labeled by the
anti-p63a1pha antibody was counted and the total number of cells was
determined by
counting the number of nuclei stained by Sytox Orange. The proportion of
p63a1pha-positive
cells was then determined by calculating the percentage of Sytox-orange-
positive nuclei that
also expressed p63a1pha.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
230
To evaluate cell expansion ratios, nuclei were counted using a Zeiss LSM 880
confocal
microscope. The expansion factor was then determined by calculating the ratio
of the
expanded population of cells to population of seeded cells.
Results in the Tables below indicate that the LATS inhibitors enabled cell
population
expansion. In the presence of the LATS inhibitors, 57 to 97 percent of the
cells express the
p63a1pha-positive phenotype.
Table 2
Expansion Compound Example
Expansio
Compound factor No. n factor
N-methyl-2-(pyridin-4-y1)-N-(1,1,1- 2-(pyridin-4-
yI)-4-(3-
trifluoropropan-2-yl)pyrido[3,4-
(trifluoromethyl)piperazi
d]pyrimidin-4-amine n-1-
yl)pyrido[3,4-
2137 d]pyrimidine
1051
2-methyl-1-(2-methy1-2-{[2-(pyridin- N-
cyclopenty1-2-(pyridin-
4-yl)pyrido[3,4-d]pyrimidin-4- 4-
yl)pyrido[3,4-
yl]amino}propoxy)propan-2-ol 2087 d]pyrimidin-
4-amine 1048
N-propy1-2-(3-
2,4-dimethy1-4-{[2-(pyridin-4-
(trifluoromethyl)-1H-
yl)pyrido[3,4-d]pyrimidin-4- pyrazol-4-
yOpyrido[3,4-
yl]amino}pentan-2-ol (Ex. 3) 2029 d]pyrimidin-
4-amine 991
N-(2-methylcyclopentyI)-
2-(pyridin-4-
N-(tert-buty1)-2-(pyridin-4-y1)-1,7-
yl)pyrido[3,4-
naphthyridin-4-amine 1717 d]pyrimidin-
4-amine 976
2-(3-ch loropyridin-4-yI)-
N-(1,1,1-trifluoro-2-
2-(pyridin-4-yI)-N-[1- methylpropan-
2-
(trifluoromethyl)cyclobutyl]pyrido[3,
yl)pyrido[3,4-
4-d]pyrimidin-4-amine 1712 d]pyrimidin-
4-amine 961
2-(2-methy1-2-{[2-
(pyridin-4-yl)pyrido[3,4-
d]pyrimidin-4-
N-propy1-2-(pyridin-4-yl)pyrido[3,4-
yl]amino}propoxy)ethan-
d]pyrimidin-4-amine 1423 1-01
705
N-(1-methylcyclopropy1)-
N-(propan-2-y1)-2-(pyridin-4- 7-(pyridin-4-
yl)pyrido[3,4-d]pyrimidin-4-amine 1275
yl)isoquinolin-5-amine 681
(1S,2S)-2-{[2-(pyridin-4-
yl)pyrido[3,4-
3-(pyridin-4-yI)-N-(1- d]pyrimidin-
4-
(trifluoromethyl)cyclopropyI)-2,6-
yl]amino}cyclopentan-1-
naphthyridin-1-amine 1241 ol 39
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
231
Expansion Compound Example Expansio
Compound factor No. n factor
243-methy1-1H-pyrazol-4-y1)-N41-
methylcyclopropyl)pyrido[3,4-
d]pyrimidin-4-amine (Ex. 4) 1205 DMSO 35
2-methy1-2-{[2-(pyridin-4-
yl)pyrido[3,4-d]pyrimidin-4-
yl]amino}propan-1-ol 1160
Table 3
Percentage Percentag
of p63a- e of p63a-
positive Compound Example positive
Compound cells No. cells
N-methyl-2-(pyridin-4-y1)-N-(1,1,1- 2-(pyridin-4-
yI)-4-(3-
trifluoropropan-2-yl)pyrido[3,4-
(trifluoromethyDpiperazi
d]pyrimidin-4-amine n-1-
yl)pyrido[3,4-
97 d]pyrimidine 90
2-methy1-142-methy1-24[2-
(pyridin-4-yOpyrido[3,4- N-cyclopenty1-2-
d]pyrimidin-4- (pyridin-4-
yl)pyrido[3,4-
yl]amino}propoxy)propan-2-ol 95 d]pyrimidin-4-
amine 87
N-propy1-2-(3-
2 ,4-dimethy1-4-{[2-(pyridin-4-
(trifluoromethyl)-1H-
yl)pyrido[3,4-d]pyrimidin-4- pyrazol-4-
yOpyrido[3,4-
yl]amino}pentan-2-ol (Ex. 3) 92 d]pyrimidin-4-
amine 86
N-(2-
methylcyclopentyI)-2-
N-(tert-buty1)-2-(pyridin-4-y1)-1,7- (pyridin-4-
yl)pyrido[3,4-
naphthyridin-4-amine 93 d]pyrimidin-4-
amine 86
2-(3-chloropyridin-4-yI)-
N-(1 ,1 ,1 -trifluoro-2-
2-(pyridin-4-y1)-N-[I - methylpropan-2-
(trifluoromethyl)cyclobutyl]pyrido[3 yl)pyrido[3,4-
,4-d]pyrimidin-4-amine 95 d]pyrimidin-4-
amine 87
242-methy1-24[2-
(pyridin-4-yl)pyrido[3,4-
d]pyrimidin-4-
N-propy1-2-(pyridin-4-
yl]amino}propoxy)ethan
yl)pyrido[3,4-d]pyrimidin-4-amine 93 -1-ol 86
N-(1-
methylcyclopropyI)-7-
N4propan-2-y1)-2-(pyridin-4- (pyridin-4-
yl)pyrido[3,4-d]pyrimidin-4-amine 93 yl)isoquinolin-
5-amine 80
(1S,2S)-2-{[2-(pyridin-
4-yl)pyrido[3,4-
3-(pyridin-4-yI)-N-(1- d]pyrimidin-4-
(trifluoromethyl)cyclopropyI)-2,6-
yl]amino}cyclopentan-
naphthyridin-1-amine 95 1-01 6
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
232
Percentage
Percentag
of p63a- e
of p63a-
positive Compound Example positive
Compound cells No. cells
2-(3-methyl-1H-pyrazol-4-y1)-N-(1-
methylcyclopropyl)pyrido[3,4-
d]pyrimidin-4-amine (Ex. 4) 89 DMSO 3
2-methyl-2-{[2-(pyridin-4-
yl)pyrido[3,4-d]pyrimidin-4-
yl]amino}propan-1-ol 89
Example 6: Reducing Immune Rejection by CRISPR/Cas9-mediated deletion of the
beta-2-microglobulin gene in HEK293
In the example below, HLA class I expression was eliminated from the HEK293
surface by
CRISPR-mediated deletion of the beta-2-microglobulin gene.
Guide RNA (gRNA) targeting B2M were obtained from Dharmacon (Layfette, CO)
(sequences 1-5 in Table 4). Seven additional gRNA were also designed (6-12 in
Table 4).
Table 5 shows the PAM sequence for each gRNA ID, the target sequence location,
the
sequence of B2M gene that corresponds to the gRNA targeting domain and is
complementary to the target sequence in the B2M gene. Table 6 represents
sequences of
sgRNAs. These gRNAs (SEQ ID NO 108-119) were tested for the ability to reduce
or
eliminate expression of B2M in HEK293 cells using a lipofection approach as
follows
Table 4
SEQ ID Sequence of the targeting domain
gRNA ID
NO: of the gRNA
1-CR004366 108 GAGUAGCGCGAGCACAGCUA
2-CR004366 109 CGUGAGUAAACCUGAAUCUU
3-CR004366 110 AAGUCAACUUCAAUGUCGGA
4-CR004366 111 CAGUAAGUCAACUUCAAUGU
5-CR004366 112 CUGAAUCUUUGGAGUACCUG
6-HEYJA000001 113 GGCCGAGAUGUCUCGCUCCG
7-HEYJA000003 114 CUCGCGCUACUCUCUCUUUC
8-HEYJA000004 118 ACUCACGCUGGAUAGCCUCC
9-HEYJA000005 116 UCACGUCAUCCAGCAGAGAA
10-HEYJA000007 117 AGUCACAUGGUUCACACGGC
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
233
11-HEYJA000008 118 CCACCUCUUGAUGGGGCUAG
12-H EYJA000009 119 GCUACUCUCUCUUUCUGGCC
CR000442 134 GGCCACGGAGCGAGACAUCU
CR000443 135 CGCGAGCACAGCUAAGGCCA
CR000446 136 AGGGUAGGAGAGACUCACGC
CR000453 137 CACAGCCCAAGAUAGUUAAG
CR000455 138 UUACCCCACUUAACUAUCUU
CR000456 139 CUUACCCCACUUAACUAUCU
CR006979 140 UCCUGAAUUGCUAUGUGUCU
Table 5
SEQ ID NO
of sequence
of B2M
Target Sequence of B2M gene that
gRNA ID PAM Sequence corresponds to
gRNA targeting gene that
Location domain
correspond
s to gRNA
targeting
domain
chr15:44711563 GAGTAGCGCGAGCACAGCT 141
1-CR004366 AGG
-44711585 A
chr15:44715428 CGTGAGTAAACCTGAATCTT 142
2-CR004366 TGG
-44715450
chr15:44715509 AAGTCAACTTCAATGTCGGA 143
3-CR004366 TGG
-44715531
chr15:44715513 CAGTAAGTCAACTTCAATGT 144
4-CR004366 CGG
-44715535
chr15:44715417 CTGAATCTTTGGAGTACCTG 145
5-CR004366 AGG
-44715439
6- TGG
chr15:44711540 GGCCGAGATGTCTCGCTCC 146
HEYJA000001 -44711562 G
7- TGG
chr15:44711574 CTCGCGCTACTCTCTCTTTC 147
HEYJA000003 -44711596
8- AGG
chr15:44711597 ACTCACGCTGGATAGCCTC 148
HEYJA000004 -44711619 C
9- TGG
chr15:44715446 TCACGTCATCCAGCAGAGA 149
HEYJA000005 -44715468 A
10- AAG
chr15:44715651 AGTCACATGGTTCACACGG 150
HEYJA000007 -44715673 C
11- TAG
chr15:44713812 CCACCTCTTGATGGGGCTA 151
HEYJA000008 -44713834 G
12- TGG
chr15:44711579 GCTACTCTCTCTTTCTGGCC 152
HEYJA000009 -44711601
CR000442 CGG chr15:44711542
GGCCACGGAGCGAGACATC 153
-44711564 T
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
234
CR000443 CGG chr15:44711557 CGCGAGCACAGCTAAGGCC 154
-44711579 A
CR000446 TGG chr15:44711609 AGGGTAGGAGAGACTCACG 155
-44711631 C
CR000453 TGG chr15:44715678 CACAGCCCAAGATAGTTAA 156
-44715700 G
CR000455 GGG chr15:44715683 TTACCCCACTTAACTATCTT 157
-44715705
CR000456 TGG chr15:44715684 CTTACCCCACTTAACTATCT 158
-44715706
CR006979 GGG chr15:44715480 TCCTGAATTGCTATGTGTCT 159
-44715502
PAM=Protospacer adjacent Motif; gRNAs 1-5 from Dharmacon
Table 6
gRNA ID SEQ ID NO: Sequence of the sgRNA
GAGUAGCGCGAGCACAGCUAGUUUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
1-CR004366 120
CCGUUAUCAACUUGAAAAAGUGGCACCGA
GUCGGUGCUUUU
CGUGAGUAAACCUGAAUCUUGUUUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
2-CR004366 160
CCGUUAUCAACUUGAAAAAGUGGCACCGA
GUCGGUGCUUUU
AAGUCAACUUCAAUGUCGGAGUUUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
3-CR004366 161
CCGUUAUCAACUUGAAAAAGUGGCACCGA
GUCGGUGCUUUU
CAGUAAGUCAACUUCAAUGUGUUUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
4-CR004366 162
CCGUUAUCAACUUGAAAAAGUGGCACCGA
GUCGGUGCUUUU
CUGAAUCUUUGGAGUACCUGGUUUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
5-CR004366 163
CCGUUAUCAACUUGAAAAAGUGGCACCGA
GUCGGUGCUUUU
GGCCGAGAUGUCUCGCUCCGGUUUUAGA
GCUAGAAAUAGCAAGUUAAAAUAAGGCUA
6-HEYJA000001 164
GUCCGUUAUCAACUUGAAAAAGUGGCACC
GAGUCGGUGCUUUU
CUCGCGCUACUCUCUCUUUCGUUUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
7-HEYJA000003 165
CCGUUAUCAACUUGAAAAAGUGGCACCGA
GUCGGUGCUUUU
ACUCACGCUGGAUAGCCUCCGUUUUAGAG
8-HEYJA000004 166
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
235
CCGUUAUCAACU UGAAAAAGUGGCACCGA
GUCGGUGCUUUU
UCACGUCAUCCAGCAGAGAAGU UUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
9-H EYJA000005 167
CCGUUAUCAACU UGAAAAAGUGGCACCGA
GUCGGUGCUUUU
AG UCACAUGGU UCACACGGCGUUUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
10-H EYJA000007 168
CCGUUAUCAACU UGAAAAAGUGGCACCGA
GUCGGUGCUUUU
CCACCUCU UGAUGGGGCUAGGUUU UAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
11-H EYJA000008 169
CCGUUAUCAACU UGAAAAAGUGGCACCGA
GUCGGUGCUUUU
GCUACUCUCUCUUUCUGGCCGUUUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
12-H EYJA000009 170
CCGUUAUCAACU UGAAAAAGUGGCACCGA
GUCGGUGCUUUU
GGCCACGGAGCGAGACAUCUGUUUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
CR000442 171
CCGUUAUCAACU UGAAAAAGUGGCACCGA
GUCGGUGCUUUU
CGCGAGCACAGCUAAGGCCAGUUUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
CR000443 172
CCGUUAUCAACU UGAAAAAGUGGCACCGA
GUCGGUGCUUUU
AGGGUAGGAGAGACUCACGCGUUUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
CR000446 173
CCGUUAUCAACU UGAAAAAGUGGCACCGA
GUCGGUGCUUUU
CACAGCCCAAGAUAGUUAAGGUUUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
CR000453 174
CCGUUAUCAACU UGAAAAAGUGGCACCGA
GUCGGUGCUUUU
UUACCCCACUUAACUAUCUUGU UUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
CR000455 175
CCGUUAUCAACU UGAAAAAGUGGCACCGA
GUCGGUGCUUUU
CU UACCCCACU UAACUAUCUGU UUUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
CR000456 176
CCGUUAUCAACU UGAAAAAGUGGCACCGA
GUCGGUGCUUUU
UCCUGAAU UGC UAUGUGUCUGU U UUAGAG
CUAGAAAUAGCAAGUUAAAAUAAGGCUAGU
CR006979 177
CCGUUAUCAACU UGAAAAAGUGGCACCGA
GUCGGUGCUUUU
Lipofection:
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
236
One day before transfection, 500'000 HEK293cells (ATCC, Manassas, VA) were
plated in a
35mm dish and grown in DMEM/10%FBS. The following day, cells were transfected
with a
mixture of tracrRNA-gRNA-Cas9 mRNA. A stock of 10 micromolar was prepared
resuspending 20 nanomoles of gRNA and 20 nanomoles tracrRNA in 2000 microlitre
10
millimolar Tris buffer pH7.4 each. Additionally, Cas9 mRNA was pre-diluted
1:10 adding 10
microlitre of 1 microgram/microlitre Cas9 mRNA to 90 microlitre 10 millimolar
Tris buffer
pH7.4.
To obtain the mixture for the size of a 35mm petri dish, 12.5 microlitre of 10
micromolar
tracrRNA (Dharmacon, Cat # U-002000-20), 12.5 microlitre of 10 micromolar gRNA
targeting human B2M (Table 4; SEQ ID NOs: 108-119), 50 microlitre of 0.1
microgram/microlitre Cas9 mRNA (Dharmacon, Cat #CAS11195), and 15 microlitre
of
DharmaFECT Duo Transfection Reagent (Dharmacon, Cat #T-2010-02) were combined
and
incubated for 20 minutes at room temperature. The mixture was added drop wise
to the
culture dish in 2.5m1 DMEM/10%FBS medium. The transfection reagent alone
represented
the transfection negative control.
After 6 h incubation in 5% CO2 at 37 C medium was replaced with fresh
DMEM/10%FBS
medium. After 72h in a 5% CO2 incubator cells were prepared for FACS analysis.
FACS analysis: HEK293 cells were treated with Accutase (ThermoFisher, Cat #
A1110501)
for 20 minutes in 5% CO2 at 37 C. The reaction was stopped by using cell
culture medium
containing 10% Serum and transferred to a falcon tube fora centrifugation step
(1000 rpm, 5
minutes). After aspirating the medium cells were resuspended in 200 microlitre
FACS buffer
(PBS/10%FBS).
To analyze the expression of B2M and HLA-ABC, 5 microlitre APC mouse anti-
human p 2 -
microglobulin antibody (Biolegend, Cat #316312) and 20 microlitre PE mouse
anti-human
HLA-ABC antibody (BD Bioscience, Cat # 560168) were added to the cell
suspension and
incubated for 30 minutes on ice. Cells were washed 3 times after antibody
labelling with
FACS buffer and resuspended in 500 microlitre in FACS buffer.
Each sample was transferred to one well of a round bottom 96 well plate and
analyzed on a
BD LSRFortessa X-20 device. FACS data were analyzed using BD FACSDiva
software.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
237
The results are shown in Table 7 below.
Table 7
B2M knockout
gRNA ID efficiency in
HEK293 [%)
1-CR004366 70
2-CR004366 33
3-CR004366 12
4-CR004366 48
5-CR004366 33
6-HEYJA000001 47
7-HEYJA000003 37
8-HEYJA000004 65
9-HEYJA000005 39
10-HEYJA000007 13
11-HEYJA000008 3
12-HEYJA000009 17
Example 7 : Reducing Immune Rejection by CRISPR/Cas9-mediated deletion of the
beta-2-microglobulin gene in LSCs
In the example below, HLA class I expression was eliminated from the LSC
surface by
CRISPR-mediated deletion of the beta-2-microglobulin gene.
The sgRNA ID SEQ NO 120 was tested for the ability to reduce or eliminate
expression of
B2M in LSCs using a nucleofection approach as follows.
Nucleofection:
LSCs at passage 0 were trypsinized with TryLETmExpress Enzym (ThermoFisher,
Cat
#12605010) for 15min in 5%CO2 at 37 C. After scraping the cells, the reaction
was stopped
by using cell culture medium containing 10% Serum and transferred to a falcon
tube. After
counting cells using Vi-cell 200'000 cells were prepared per reaction by
transferring 200'000
cells in single tubes and centrifuged at 1000rpm for 5min.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
238
The supernatant was aspirated using manually pipetting to avoid cell loss and
the cells were
resuspended in the Stem cell nucleofector solution 11 (Lonza, Cat #VPH-5022).
Resuspend
cells in nucleofector solution immediately before adding the Cas9
protein:sgRNA mixture. A
stock of 100pM (3.23pg/p1) was prepared resuspending 5.1 nanomoles of single-
guideRNA
(sgRNA) in 51p1 10mM Tris buffer pH7.4. To obtain the nucleofection mixture,
8pg high
concentrated (5pg/p1) Cas9 protein (shown below) (volume = 1.6p1) was mixed
with a
16.2pg sgRNA combined with a sequence targeting 1-CR004366 sequence from Table
4
(shown below, SEQ ID NO: 120) (volume = 5p1) and incubated for 20 min at room
temperature to form a Cas9 protein-sgRNA complex. A molar ratio of 1:10
(50pm01 Cas9
protein: 500pm01 sgRNA) was used.
Cas9 Protein (SEQ ID NO: 107)
MAPKKKRKVD KKYSIGLDIG TNSVGWAVIT DEYKVPSKKF KVLGNTDRHS
IKKNLIGALL FDSGETAEAT RLKRTARRRY TRRKNRICYL QEIFSNEMAK
VDDSFFHRLE ESFLVEEDKK HERHPIFGNI VDEVAYHEKY PTIYHLRKKL
VDSTDKADLR LIYLALAHMI KFRGHFLIEG DLNPDNSDVD KLFIQLVQTY
NQLFEENPIN ASGVDAKAIL SARLSKSRRL ENLIAQLPGE KKNGLFGNLI
ALSLGLTPNF KSNFDLAEDA KLQLSKDTYD DDLDNLLAQI GDQYADLFLA
AKNLSDAILL SDILRVNTEI TKAPLSASMI KRYDEHHQDL TLLKALVRQQ
LPEKYKEIFF DQSKNGYAGY IDGGASQEEF YKFIKPILEK MDGTEELLVK
LNREDLLRKQ RTFDNGSIPH QIHLGELHAI LRRQEDFYPF LKDNREKIEK
ILTFRIPYYV GPLARGNSRF AWMTRKSEET ITPWNFEEVV DKGASAQSFI
ERMTNFDKNL PNEKVLPKHS LLYEYFTVYN ELTKVKYVTE GMRKPAFLSG
EQKKAIVDLL FKTNRKVTVK QLKEDYFKKI ECFDSVEISG VEDRFNASLG
TYHDLLKIIK DKDFLDNEEN EDILEDIVLT LTLFEDREMI EERLKTYAHL
FDDKVMKQLK RRRYTGWGRL SRKLINGIRD KQSGKTILDF LKSDGFANRN
FMQLIHDDSL TFKEDIQKAQ VSGQGDSLHE HIANLAGSPA IKKGILQTVK
VVDELVKVMG RHKPENIVIE MARENQTTQK GQKNSRERMK RIEEGIKELG
SQILKEHPVE NTQLQNEKLY LYYLQNGRDM YVDQELDINR LSDYDVDHIV
PQSFLKDDSI DNKVLTRSDK NRGKSDNVPS EEVVKKMKNY WRQLLNAKLI
TQRKFDNLTK AERGGLSELD KAGFIKRQLV ETRQITKHVA QILDSRMNTK
YDENDKLIRE VKVITLKSKL VSDFRKDFQF YKVREINNYH HAHDAYLNAV
VGTALIKKYP KLESEFVYGD YKVYDVRKMI AKSEQEIGKA TAKYFFYSNI
MNFFKTEITL ANGEIRKRPL IETNGETGEI VWDKGRDFAT VRKVLSMPQV
NIVKKTEVQT GGFSKESILP KRNSDKLIAR KKDWDPKKYG GFDSPTVAYS
VLVVAKVEKG KSKKLKSVKE LLGITIMERS SFEKNPIDFL EAKGYKEVKK
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
239
DLIIKLPKYS LFELENGRKR MLASAGELQK GNELALPSKY VNFLYLASHY
EKLKGSPEDN EQKQLFVEQH KHYLDEIIEQ ISEFSKRVIL ADANLDKVLS
AYNKHRDKPI REQAENIIHL FTLTNLGAPA AFKYFDTTID RKRYTSTKEV
LDATLIHQSI TGLYETRIDL SQLGGDSRAD PKKKRKVHHH HHH
sgRNA (SEQ ID NO:120)
GAGUAG C GC GAGCACAGCUAGUUUUAGAGCUAGAAAUAG CAAGUUAAAAUAAG GCUAGU C C GUUAU CAA
CUUGAAAAAGUGGCACCGAGUCGGUGCUUUU
The Cas9 protein-sgRNA complex was added to the cell suspension and
transferred to the
electroporation cuvette immediately. Cells were transfected using the
nucelofector device
(Lonza, Amaxa Nucleofector II) and program A023. After nucleofection cells
were
transferred from cuvette to one well of a 48 well synthemax coated plate
containing pre-
warmed LSC medium including 3pM LATS inhibitor and 10pM Rockinhibitor Y-27632
(Nature 1997, vol. 389, pp. 990-994). Incubate LSCs in a 5% CO2 incubator for
around 5
days until cells are 90% confluent.
FACS analysis:
LSCs were treated with TryLETmExpress Enzym (ThermoFisher, Cat #12605010) for
15
minutes in 5% CO2 at 37 C. After scraping the cells, the reaction was stopped
by using cell
culture medium containing 10% Serum and transferred to a falcon tube for a
centrifugation
step (1000 rpm, 5 minutes). After aspirating the medium cells were resuspended
in 200p1
FACS buffer (PBS/10%FBS).
To analyze the expression of B2M and HLA-ABC, 5pIAPC mouse anti-human p 2 -
microglobulin antibody (Biolegend, Cat #316312) and 20p1 PE mouse anti-human
HLA-ABC
antibody (BD Bioscience, Cat # 560168) were added to the cell suspension and
incubated
for 30 minutes on ice.
The same amount and incubation time of isotype control was used for each color
(5p1 of
Biolegend APC Mouse IgG1, K lsotype Ctrl (FC) Antibody #316311 and 20p1 of BD
Biosciences PE mouse IgG1, K lsotype Ctrl #555749) to set up the negative
control gate
later in FACS. Cells were washed 3 times after antibody labelling with FACS
buffer and
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
240
resuspended in 500p1 in FACS buffer (depending on the cell number). Before
FACS sorting,
cells were filtered through a 70pm filter and stored on ice until sorting.
In order to prevent cells sticking to the wall, collection tubes were filled
with FACS buffer for
30 minutes before the sort and aspirated before adding the collection medium.
Cells were
sorted on a BD FACSAria 11 instrument into prepared collection tubes, using
human serum
enriched LSC medium including compound. FACS data were analyzed using BD
FACSDiva
software and FlowJo software.
The results confirmed about 70% of the cells CRISPR-edited with sgRNA SEQ ID
NO: 120
did not express B2M and eliminate HLA I expression on cell surface of limbal
stem cells
(Figure 3).
LSC/T-cell reaction assay:
An LSC/T-cell assay was performed in flat bottom 96we11 synthemax coated
plates in
duplicates and incubated in 5% CO2 at 37 C for 10 days. RPMI-1640 supplemented
with
HEPES (100pM), non-essential aas (10x), sodium pyruvate (10mM), 2-
Mercapthoethanol
(10x), 10% FBS and 1% Penicillin-Streptomycin (Gibco by Life Technologies) was
used as
medium for co-culture. Alternatively, RPMI-1640 supplemented with HEPES
(10mM), non-
essential aas (1x), sodium pyruvate (1mM), 2-Mercapthoethanol (1x), 10% FBS
and 1%
Penicillin-Streptomycin (Gibco by Life Technologies) was used as medium for co-
culture.
One day before co-culture, LSCs (stimulator cells) were passed and cultured to
a confluency
of around 70% (30'000 - 50'000 cells) and cultured with LSC medium including
compound.
On day two, peripheral blood mononuclear cells (PBMC) were separated using
EDTA blood
with the Ficoll-Paque method (GE Healthcare Life Sciences, cat #17-1440-03).
After PBMC
isolation, the CD8+ T-cell isolation Kit (Miltenyi Biotec, Cat #130-096-495)
was used to
separate CD8+ cells from all other cell populations. The cell suspension with
1-10x10^6
CD8+ cells were stained with 1pM CellTrace Violet (Invitrogen, Cat #C34557)
and were
incubated for 20 minutes at 37 C in the dark. After incubation 2m1 ice cold
heat inactivated
FBS was added to each 5m1 cell suspension and cells were incubated for
additional 5
minutes at 37 C. After 3 washing steps with culture medium, stained CD8+ cells
were
diluted to a final concentration of 100'000 cells per well and 100 pl of CD8+
cell dilution was
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
241
added to each well containing LSCs after washing off LSC medium. For positive
control,
stained CD8+ cells were incubated in a pre-coated 10pg/m1 anti-human CD3+
(eBioscience,
Cat #16-0037-85) well including diluted 3pg/mlanti-human CD28 (eBioscience,
Cat # 16-
0289-85). One separate duplicate with stained CD8+ cells with medium only was
used as
negative control.
After 10 days, CD8+ cells were transferred to a U-bottomed 96-well plate and
washed 3
times using autoMAC rinsing solution (Miltenyi Biotec, Cat #130-091-222)
including MACS
BSA stock solution (Miltenyi Biotec, Cat #130-091-376). Cells were measured on
a BD
LSRFortessa X-20. FACS data were analyzed using BD FACSDiva software and
FlowJo
software.
Fig. 4 shows gene edited limbal stem cells (LSCs) co-cultured with CD8+ T-
cells from 4
different donors. In all 4 donors T-cell immunresponse was almost completely
eliminated co-
cultured with B2M/HLA-Class I negative LSCs, which were CRISPR-edited with
sgRNA SEQ
ID NO: 120.
Example 8: Screening for efficiency of sgRNAs in reducing or eliminating
expression
of B2M in LSCs and elimination of HLA I expression on cell surface of limbal
stem
cells
Limbal Stem Cell Isolation and Culture:
Cells obtained as described in Example 1 were plated in a 10 cm synthemax
coated petri
dish in limbal epithelium cell culture medium (DMEM F12 supplemented with 10%
human
serum and 1.3 mM calcium chloride) supplemented with 3pM LATS inhibitor
compound and
10pM Rock inhibitor Y-27632 (Nature 1997, vol. 389, pp. 990-994). Cells were
cultured
under these conditions for 24-48 hours at 37 C in 5% CO2.
LSCs were nuclofected with selected gRNAs (Table 6) followed by FACS analysis
/ MACS
separation.
Nucleofection approach for sgRNA screening (SEQ ID NO 120 and 160-177)-in LSCs
was
performed as follows: LSCs at passage 3 were trypsinized with TryLETMExpress
Enzym
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
242
(ThermoFisher, Cat #12605010) for 15min in 5%CO2 at 37 C. After scraping the
cells, the
reaction was stopped by using cell culture medium containing 10% Serum and
transferred to
a falcon tube. After counting cells using Vi-cell 300'000 cells were prepared
per reaction by
transferring 300'000 cells in single tubes and centrifuged at 1000 rpm for
5min. The
supernatant was aspirated using manually pipetting to avoid cell loss and the
cells were
resuspended in the Stem cell nucleofector solution 11 (Lonza, Cat #VPH-5022).
Resuspend
cells in nucleofector solution immediately before adding the Cas9 RNP:sgRNA
mixture. To
obtain the nucleofection mixture, 5pg high concentrated (5pg/p1) Cas9 protein
of SEQ ID
NO: 106 (volume = 0.78p1) was mixed with 19.5pg sgRNA of Table 6 (volume =
12.2p1) and
incubated for 20 min at room temperature. A molar ratio of ¨1:20 (31.5pm01
Cas9 RNP:
605pm01 sgRNA) was used. The Cas9 protein-guideRNA complex was added to the
cell
suspension and transferred to the electroporation cuvette immediately. Cells
were
transfected using the nucelofector device (Lonza, Amaxa Nucleofector II) and
program
A023. After nucleofection cells were transferred from cuvette to one well of a
24 well
synthemax coated plate containing pre-warmed LSC medium including 3pM LATS
compound and 10pM Rock inhibitor Y-27632 (Nature 1997, vol. 389, pp. 990-994).
Incubate
LSCs in a 5% CO2 incubator for around 3 days until cells are 90% confluent.
FACS analysis:
LSCs were treated with TryLETmExpress Enzym (ThermoFisher, Cat #12605010) for
15
minutes in 5% CO2 at 37 C. After scraping the cells, the reaction was stopped
by using cell
culture medium containing 10% Serum and transferred to a falcon tube for a
centrifugation
step (1000 rpm, 5 minutes). After aspirating the medium cells were resuspended
in 200p1
FACS buffer (PBS/10%FBS).
To analyze the expression of B2M and HLA-ABC, 5pIAPC mouse anti-human p 2 -
microglobulin antibody (Biolegend, Cat #316312) and 20p1 PE mouse anti-human
HLA-ABC
antibody (BD Bioscience, Cat # 560168) were added to the cell suspension and
incubated
for 30 minutes on ice.
The same amount and incubation time of isotype control was used for each color
(5plof
Biolegend APC Mouse IgG1, K lsotype Ctrl (FC) Antibody #316311 and 20p1 of BD
Biosciences PE mouse IgG1, K lsotype Ctrl #555749) to set up the negative
control gate
CA 03116512 2021-04-14
WO 2020/084580
PCT/IB2019/059162
243
later in FACS. Cells were washed 3 times after antibody labelling with FACS
buffer and
resuspended in 200plin FACS buffer (depending on the cell number).
FACS data were analyzed using BD FACSDiva software and FlowJo software.
The results of B2M knockout efficiency in LSCs after nucleofection are shown
in Table 8
below.
Table 8.
B2M knockout
efficiency in
gRNA ID LSCs after
nucleofection
[%)
1-CR004366 55
2-CR004366 2
3-CR004366 2
4-CR004366 22
5-CR004366 2
6-HEYJA000001 33
7-HEYJA000003 8
8-HEYJA000004 22
9-HEYJA000005 12
10-HEYJA000007 1
11-HEYJA000008 1
12-HEYJA000009 1
CR000442 62
CR000443 25
CR000446 27
CR000453 5
CR000455 24
CR000456 3
CR006979 2
Example 9: Efficiency of sgRNAs in reducing or eliminating expression of B2M
in
LSCs and elimination of HLA I expression on cell surface of limbal stem cells
FACS and MACS of B2M-negative LSCs
Limbal Stem Cell Isolation and Culture as performed in Example 8.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
244
Nucleofection for sgRNA selected for on/off target analysis:
LSCs at passage 3 were trypsinized with TryLETmExpress Enzym (ThermoFisher,
Cat
#12605010) for 15min in 5%CO2 at 37 C. After scraping the cells, the reaction
was stopped
by using cell culture medium containing 10% Serum and transferred to a falcon
tube. After
counting cells using Vi-cell 1'000'000 cells were prepared per reaction by
transferring
1'000'000 cells in single tubes and centrifuged at 1000 rpm for 5min.
The supernatant was aspirated using manually pipetting to avoid cell loss and
the cells were
resuspended in Stem cell nucleofector solution 11 (Lonza, Cat #VPH-5022).
Resuspend cells
in nucleofector solution immediately before adding the Cas9 RNP:sgRNA mixture.
To obtain the nucleofection mixture, 10pg high concentrated (5pg/p1) Cas9
protein (volume
= 1.56p1; SEQ ID NO: 106) was mixed with 40.2pg sgRNA (volume = 25p1;
sequences of
sgRNAs are presented in Table 6: SEQ ID NO 120, 162, 164, 166, 167, 171, 173,
175) and
incubated for 20 min at room temperature. A molar ratio of 1:20 (62.5pm01 Cas9
RNP:
1250pm01 sgRNA) was used.
The Cas9 protein-guideRNA complex was added to the cell suspension and
transferred to
the electroporation cuvette immediately. Cells were transfected using the
nucelofector
device (Lonza, Amaxa Nucleofector II) and program A023. After nucleofection
cells were
transferred from cuvette to one well of a 12 well synthemax coated plate
containing pre-
warmed LSC medium including 3pM LATS compound and 10pM Rockinhibitor Y-27632
(Nature 1997, vol. 389, pp. 990-994). Incubate LSCs in a 5% CO2 incubator for
around 3
days until cells are 90% confluent.
FACS:
LSCs were treated with TryLETMExpress Enzym (ThermoFisher, Cat #12605010) for
15
minutes in 5% CO2 at 37 C. After scraping the cells, the reaction was stopped
by using cell
culture medium containing 10% Serum and transferred to a falcon tube for a
centrifugation
step (1000 rpm, 5 minutes). After aspirating the medium cells were resuspended
in 200p1
FACS buffer (PBS/10%FBS).
To analyze the expression of B2M and HLA-ABC, 2.5pIAPC mouse anti-human p 2 -
microglobulin antibody (Biolegend, Cat #316312) and 10p1 PE mouse anti-human
HLA-ABC
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
245
antibody (BD Bioscience, Cat # 560168) were added to the cell suspension and
incubated
for 30 minutes on ice.
The same amount and incubation time of isotype control was used for each color
(2.5p1 of
Biolegend APC Mouse IgG1, K lsotype Ctrl (FC) Antibody #316311 and 10p1 of BD
Biosciences PE mouse IgG1, K lsotype Ctrl #555749) to set up the negative
control gate
later in FACS. Cells were washed 3 times after antibody labelling with FACS
buffer and
resuspended in 300p1 in FACS buffer. A small aliquot of labelled LSCs (-15'000
LSCs) were
analyzed by FACS to confirm B2M knockout after nucleofection. FACS data were
analyzed
using BD FACSDiva software and FlowJo software.
To obtain a purified B2M negative LSC culture, the second and bigger portion
of antibody
labelled LSCs were sorted using MACS to separete B2M negative from B2M
positives.
The results of B2M knockout efficiency in LSCs after nucleofection are shown
on Figure 5.
Efficiency of elimination of HLA I expression on cell surface of limbal stem
cells after
nucleofection are shown on Figure 6.
MACS:
To obtain a purified B2M negative LSC culture, the second and bigger portion
of antibody
labelled LSCs were sorted using MACS to separate B2M negative from B2M
positives.
After labelling LSCs with B2M and HLA-ABC antibodies as decribed above, the
reaction was
stopped by adding 2m1 MACS buffer (Miltenyi Biotec, #130-091-222) and
centrifuged at
1000rpm for 5 minutes. For each step MACS buffer was always supplemented with
3pM
LATS inhibitor compound, 10pM Rock inhibitor Y-27632 (Nature 1997, vol. 389,
pp. 990-
994) and BSA (Miltenyi Biotec, #130-091-376).
After aspirating the supernatant, cells were resuspended in 80p1 MACS buffer
and 10p1 of
anti-APC micorbeads (Miltenyi Biotec, #130-090-855) and 10p1 anti-PE
microbeads (Miltenyi
Biotec, #130-048-801) were added to the cell suspension. Antibody labelled
LSCs including
magnetic beads were incubated for 15 minutes in the refrigerator in the dark.
After
incubation cells were washed by adding 2m1 of MACS buffer and centrifudged for
5 minutes
at 1000rpm. 500p1 of MACS buffer was added after aspiration of supernatant.
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
246
To prepare the LS columns (Miltenyi Biotec, #130-042-401) for separation of
B2M negative
from B2M positive LSCs, LS columns were placed on the magnet device (Miltenyi
Biotec,
Quadro magnet) and washed with 3m1 MACS buffer. The flow through was
discarded.
Cell supsension was applied on top of column and flow through was collected in
a separate
15m1 falcon tube to collect B2M negative LSCs. Once all of the cell suspension
was in the
flow through fraction, 3m1 MACS buffer was applied on the column. This step
was repeated
3 times by adding new MACS buffer when the column reservoir was empty. B2M
negative
LSC fraction was centrifuged for 5 minutes at 1000rpm. After aspirating the
supernatant,
B2M negative LSCs were resuspended in LSC media including 3pM LATS inhibitor
compound and 3pM Rock inhibitor Y-27632 (Nature 1997, vol. 389, pp. 990-994)
and plated
on 1 well of a 48 synthemax coated plate. After 8-21days (depending on cell
expansion)
LSCs were treated with TryLETmExpress Enzym (ThermoFisher, Cat #12605010) for
15
minutes in 5% CO2 at 37 C and a small aliquot of B2M negative was prepared for
FACS to
confirm purity of B2M negative LSC culture (Figures7 and 8) and the second and
bigger
fraction was prepared for On/off-target analysis.
Figures 7 and 8 show FACS data detecting B2M and HLA-ABC surface protein on
gene
edited limbal stem cells, which were MACS treated after nucleofection to
obtain a B2M/HLA-
ABC negative LSC culture. All sgRNAs tested showed a pure (-99-100%) B2M/HLA-
ABC
negative LSC culture.
Example 10: Characterization of gRNA specificity and analysis of CRISPR/Cas9-
mediated off-target editing events
A biochemical method (See, e.g., Cameron et al., Nature Methods. 6, 600-606;
2017) was
used to determine potential off-target genomic sites cleaved by Cas9 and
selected B2M
guides. Guides showing B2M indel activity were tested for potential off-target
genomic
cleavage sites with this assay. In this experiment, 11 sgRNAs targeting human
B2M were
screened using genomic DNA purified from male human peripheral blood
mononuclear cells
(PBMCs) alongside a control guide with a known off-target profile. The number
of potential
off-target sites detected using a guide concentration of 64 nM in the
biochemical assay are
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
247
shown in Table 10. As a result of this analysis, several gRNAs were selected
for analysis of
potential off-target activity in limbal stem cells.
Off-target activity detection in limbal stem cells:
Potential CRISPR/Cas9-mediated cutting sites identified above were evaluated
using
targeted PCR and NGS in genome-edited expanded LSCs.
Selected sgRNAs (SEQ ID NO: 120, 162, 166, 167, 171, and 175) were further
analyzed by
amplicon sequencing in edited and non-edited cells. Primers flanking the
potential off-target
sites for each guide were used to detect indels by NGS analysis in edited LSCs
and non-
edited peripheral blood mononuclear cells. Sites that had either (1) a
difference in mean
indel percentage between edited and unedited cells greater than 0.5%; or (2) a
p value less
than 0.05 between edited and unedited indel were further analyzed. NGS
sequence reads
for such sites were assessed for characteristic indel patterns near the
putative Cas9 cut site.
Based on the results, we could assess the specificity of gRNAs and their
suitability for
therapeutic applications.
Results:
The gRNA on-target and off-target results are shown below. All sgRNAs of Table
10 were
analyzed by biochemical assay, with select results further analyzed by
amplicon
sequencing. NGS results showed B2M sgRNAs (SEQ ID NO: 120, 162, 166, 167, 171,
and
175) can achieve ¨99% indels in purified LSC populations. In the NGS results,
no predicted
sites tested positive for off-target activity with any of the sgRNAs (SEQ ID
NO: 120, 162,
166, 167, 171, and 175). For SEQ ID NO: 120,64 out of 69 off-target loci were
sequenced,
and zero validated indels for off-target activity in LSCs were identified. For
SEQ ID NO: 162,
88 out of 92 off-target loci were sequenced, and zero validated indels for off-
target activity in
LSCs were identified. For SEQ ID NO: 166,60 out of 62 off-target loci were
sequenced, and
zero validated indels for off-target activity in LSCs were identified. For SEQ
ID NO: 167, 35
out of 35 off-target loci were sequenced, and zero validated indels for off-
target activity in
LSCs were identified. For SEQ ID NO: 171, 28 out of 29 off-target loci were
sequenced, and
zero validated indels for off-target activity in LSCs were identified. For SEQ
ID NO: 175, 46
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
248
out of 48 off-target loci were sequenced, and zero validated indels for off-
target activity in
LSCs were identified.
Table 10. Off-Target Analysis
gRNA ID (sgRNA Target Sequence of the off- B2M
Number of sites
SEQ ID NO) targeting domain of target
purified positive for off-
the gRNA (SEQ ID Site_ rhAmpSeq target activity
in
NO) Count indel LSCs
6-HEYJA000001 B2M GGCCGAGAUGUCU 8 ND ND
(SEQ ID NO: 164) CGCUCCG
(SEQ ID NO: 113)
CR000442 B2M GGCCACGGAGCGA 29 99.10 0.26% 0 of 28
(SEQ ID NO: 171) GACAUCU
(SEQ ID NO:134)
1-CR004366 B2M GAGUAGCGCGAGCA 69 99.60 0.44% 0 of 64
(SEQ ID NO: 120) CAGCUA
(SEQ ID NO: 108)
CR000443 B2M CGCGAGCACAGCUA 117 ND ND
(SEQ ID NO: 172) AGGCCA
(SEQ ID NO: 135)
8-HEYJA000004 B2M ACUCACGCUGGAUA 62 99.95 0.07% 0 of 60
(SEQ ID NO: 166) GCCUCC
(SEQ ID NO: 115)
CR000446 B2M AGGGUAGGAGAGAC 98 ND ND
(SEQ ID NO: 173) UCACGC
(SEQ ID NO: 136)
4-CR004366 B2M CAGUAAGUCAACUU 92 99.85 0.21% 0 of 88
(SEQ ID NO: 162) CAAUGU
(SEQ ID NO: 111)
CR000453 B2M CACAGCCCAAGAUA 82 ND ND
(SEQ ID NO: 174) GUUAAG
(SEQ ID NO: 137)
CR000455 B2M UUACCCCACUUAAC 48 99.33 0.32% 0 of 46
(SEQ ID NO: 175) UAUCUU
(SEQ ID NO: 138)
CR000456 B2M CUUACCCCACUUAA 66 ND ND
(SEQ ID NO: 176) CUAUCU
(SEQ ID NO: 139)
9-HEYJA000005 B2M UCACGUCAUCCAGC 35 99.80 0.26% 0 of 35
(SEQ ID NO: 167) AGAGAA
(SEQ ID NO: 116)
Control VEGF GACCCCCUCCACCC 756 ND ND
A CGCCUC (SEQ ID
NO: 178)
ND: no data
Unless indicated otherwise, all methods, steps, techniques and manipulations
that are not
specifically described in detail can be performed and have been performed in a
manner
known per se, as will be clear to the skilled person. Reference is for example
again made to
CA 03116512 2021-04-14
WO 2020/084580 PCT/IB2019/059162
249
the standard handbooks and the general background art mentioned herein and to
the further
references cited therein. Unless indicated otherwise, each of the references
cited herein is
incorporated in its entirety by reference.
.. Claims to the invention are non-limiting and are provided below. Although
particular
embodiments and claims have been disclosed herein in detail, this has been
done by way of
example for purposes of illustration only, and is not intended to be limiting
with respect to the
scope of the appended claims, or the scope of subject matter of claims of any
corresponding
future application. In particular, it is contemplated by the inventors that
various substitutions,
alterations, and modifications may be made to the disclosure without departing
from the
spirit and scope of the disclosure as defined by the claims. The choice of
nucleic acid
starting material, clone of interest, or library type is believed to be a
matter of routine for a
person of ordinary skill in the art with knowledge of the embodiments
described herein.
Other embodiments, advantages, and modifications are considered to be within
the scope of
the following claims. Those skilled in the art will recognize or be able to
ascertain, using no
more than routine experimentation, many equivalents of the specific
embodiments of the
invention described herein.