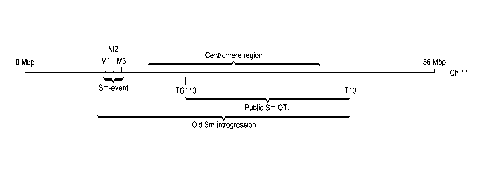Note: Descriptions are shown in the official language in which they were submitted.
TOMATO PLANTS WITH IMPROVED DISEASE RESISTANCE
FIELD OF THE INVENTION
[0001] The present invention relates to the field of plant breeding and more
specifically to
methods and compositions for producing tomato plants exhibiting improved
disease resistance
without linked deleterious traits.
BACKGROUND
[0002] Disease resistance is an important trait in agriculture, particularly
for the production of
food crops. Although disease resistance alleles have been identified in
tomato, efforts to introduce
these alleles into cultivated lines have been hindered by a lack of specific
markers linked to the
alleles, as well as the presence of deleterious alleles genetically linked to
disease resistance alleles
that lead to an unacceptable reduction in yield, fruit size, and fruit
quality. The use of marker-
assisted selection (MAS) in plant breeding has made it possible to select
plants based on genetic
markers linked to traits of interest. However, accurate markers for
identifying or tracking desirable
traits in plants are frequently unavailable even if a gene associated with the
trait has been
characterized. These difficulties are further complicated by factors such as
polygenic or
quantitative inheritance, epistasis, and an often incomplete understanding of
the genetic
background underlying expression of a desired phenotype. In the absence of
accurate and validated
markers for use in MAS, it may not be feasible to produce new plant lines
exhibiting certain disease
resistance phenotypes and acceptable yield, fruit size, and fruit quality.
SUMMARY
[0003] In one aspect, the invention provides a Solanum lycopersicum plant
comprising a
recombinant chromosomal segment on chromosome 11, wherein said chromosomal
segment
comprises a Stemphylium resistance allele from Solanum pimpinellifilium
conferring increased
resistance to Stemphylium to said plant compared to a plant not comprising
said allele, and wherein
the chromosomal segment lacks a deleterious allele genetically linked to said
Stemphylium
resistance allele that confers small fruit size when present. In some
embodiments, said
Stemphylium resistance allele is located within a chromosomal segment flanked
by marker locus
M1 (SEQ ID NO:1) and marker locus M3 (SEQ ID NO:3) on chromosome 11 in said
plant. In
certain embodiments, said introgressed Stemphylium resistance allele is within
a chromosomal
1
Date Recue/Date Received 2021-08-25
segment on chromosome 11 comprising a marker locus selected from the group
consisting of
marker locus M2 (SEQ ID NO:2), marker locus M4 (SEQ ID NO:4), marker locus M5
(SEQ ID
NO:9), marker locus M6 (SEQ ID NO:10), marker locus M7 (SEQ ID NO:15), marker
locus M8
(SEQ ID NO:20), marker locus M9 (SEQ ID NO:25), and marker locus M10 (SEQ ID
NO:30). In
further embodiments, said plant comprises a Solanum lycopersicum allele at
marker locus M1
(SEQ ID NO:1) and an Solanum pimpinellifilium allele at marker locus M2 (SEQ
ID NO:2). In
yet further embodiments, said plant further comprises a Solanum lycopersicum
allele at marker
locus M3 (SEQ ID NO:3).
[0004] In another aspect, cells, seed, and plant parts comprising a
recombinant chromosomal
segment on chromosome 11, wherein said chromosomal segment comprises a
Stemphylium
resistance allele from Solanum pimpinellifilium conferring increased
resistance to Stemphylium to
said plant compared to a plant not comprising said allele, and wherein the
chromosomal segment
lacks a deleterious allele genetically linked to said Stemphylium resistance
allele that confers small
fruit size when present are provided. In certain embodiments, a representative
sample of seed
comprising said chromosomal segment has been deposited under NCMA Accession
No.
202103011. Cells, seeds, and plant parts comprising said chromosomal segment
are further
provided.
[0005] In yet another aspect, the invention provides a Solanum lycopersicum
plant comprising a
recombinant chromosomal segment on chromosome 11, wherein said chromosomal
segment
comprises a Stemphylium resistance allele from Solanum pimpinellifilium
conferring increased
resistance to Stemphylium to said plant compared to a plant not comprising
said allele, and wherein
the chromosomal segment lacks a deleterious allele genetically linked to said
Stemphylium
resistance allele that confers small fruit size when present, and wherein said
recombinant
chromosomal segment further comprises a Tomato Brown Rugose Fruit Virus
(TBRFV) resistance
allele. In some embodiments, said TBRFV resistance allele is located within a
chromosomal
segment flanked by marker locus M1 (SEQ ID NO:1) and marker locus M3 (SEQ ID
NO:3) on
chromosome 11 in said plant. In other embodiments, said plant is homozygous
for said TBRFV
resistance allele.
[0006] In a further aspect, the invention provides a recombinant DNA segment
comprising a
Stemphylium resistance allele from Solanum pimpinellifilium that confers to a
Solanum
lycopersicum plant increased resistance to Stemphylium and lacking a
deleterious allele genetically
2
Date Recue/Date Received 2021-08-25
linked thereto that confers small fruit size. In some embodiments, said
recombinant DNA segment
comprises a marker locus selected from the group consisting of marker locus M2
(SEQ ID NO:2),
marker locus M4 (SEQ ID NO:4), marker locus M5 (SEQ ID NO:9), marker locus M6
(SEQ ID
NO:10), marker locus M7 (SEQ ID NO:15), marker locus M8 (SEQ ID NO:20), marker
locus M9
(SEQ ID NO:25), and marker locus M10 (SEQ ID NO:30). In further embodiments,
said
recombinant DNA segment is further defined as comprised within a plant, plant
part, plant cell, or
seed. In yet further embodiments, said DNA segment confers to said plant
increased resistance to
Stemphylium.
[0007] In another aspect, methods are provided for producing a plant
exhibiting resistance to
Stemphylium, comprising: a) crossing the plant of claim 1 with itself or with
a second tomato plant
of a different genotype to produce one or more progeny plants; and b)
selecting a progeny plant
comprising said Stemphylium resistance allele. In some embodiments, selecting
said progeny plant
comprises detecting a marker locus genetically linked to said Stemphylium
resistance allele. In
other embodiments, selecting said progeny plant comprises detecting a marker
locus within or
genetically linked to a chromosomal segment flanked in the genome of said
plant by marker locus
M1 (SEQ ID NO:1) and marker locus M3 (SEQ ID NO:3) on chromosome 11. In
further
embodiments, selecting a progeny plant comprises detecting at least one
polymorphism at a locus
selected from the group consisting of marker locus M1 (SEQ ID NO:1), marker
locus M2 (SEQ
ID NO:2), marker locus M3 (SEQ ID NO:3), marker locus M4 (SEQ ID NO:4), marker
locus M5
(SEQ ID NO:9), marker locus M6 (SEQ ID NO:10), marker locus M7 (SEQ ID NO:15),
marker
locus M8 (SEQ ID NO:20), marker locus M9 (SEQ ID NO:25), and marker locus M10
(SEQ ID
NO:30). In yet further embodiments, selecting a progeny plant comprises
detecting: a) a
polymorphism at marker locus M1 (SEQ ID NO:1) and a marker locus selected from
the group
consisting of marker locus M2 (SEQ ID NO:2), marker locus M4 (SEQ ID NO:4),
marker locus
M5 (SEQ ID NO:9), marker locus M6 (SEQ ID NO:10), marker locus M7 (SEQ ID
NO:15),
marker locus M8 (SEQ ID NO:20), marker locus M9 (SEQ ID NO:25), and marker
locus M10
(SEQ ID NO:30); or b) a polymorphism at marker locus M3 (SEQ ID NO:3) and a
marker locus
selected from the group consisting of marker locus M2 (SEQ ID NO:2), marker
locus M4 (SEQ
ID NO:4), marker locus M5 (SEQ ID NO:9), marker locus M6 (SEQ ID NO:10),
marker locus M7
(SEQ ID NO:15), marker locus M8 (SEQ ID NO:20), marker locus M9 (SEQ ID
NO:25), and
marker locus M10 (SEQ ID NO:30). In certain embodiments, selecting a progeny
plant comprises
3
Date Recue/Date Received 2021-08-25
detecting: a) a recurrent parent allele at marker locus M1 (SEQ ID NO:1); and
b) a donor allele at
a marker locus selected from the group consisting of marker locus M2 (SEQ ID
NO:2), marker
locus M4 (SEQ ID NO:4), marker locus M5 (SEQ ID NO:9), marker locus M6 (SEQ ID
NO:10),
marker locus M7 (SEQ ID NO:15), marker locus M8 (SEQ ID NO:20), marker locus
M9 (SEQ ID
NO:25), and marker locus M10 (SEQ ID NO:30). In other embodiments, selecting a
progeny plant
further comprises detecting a recurrent parent allele at marker M3 (SEQ ID
NO:3). In some
embodiments, said progeny plant is an F2-F6 progeny plant or producing said
progeny plant
comprises backcrossing.
[0008] In further aspects, methods are provided for producing a tomato plant
exhibiting resistance
to Stemphylium, comprising introgressing into a plant a Stemphylium resistance
allele from
Solanum pimpinellifilium within a recombinant chromosomal segment flanked in
the genome of
said plant by: marker locus M1 (SEQ ID NO:1) and marker locus M3 (SEQ ID NO:3)
on
chromosome 11; wherein said introgressed Stemphylium resistance allele confers
to said plant
increased resistance to Stemphylium compared to a plant not comprising said
allele, and wherein
said recombinant chromosomal segment lacks a deleterious allele genetically
linked to said
Stemphylium resistance allele that confers a small fruit size trait to said
plant when present. In
certain embodiments, said introgressed Stemphylium resistance allele is within
a recombinant
chromosomal segment on chromosome 11 comprising a marker locus selected from
the group
consisting of marker locus M2 (SEQ ID NO:2), marker locus M4 (SEQ ID NO:4),
marker locus
M5 (SEQ ID NO:9), marker locus M6 (SEQ ID NO:10), marker locus M7 (SEQ ID
NO:15),
marker locus M8 (SEQ ID NO:20), marker locus M9 (SEQ ID NO:25), and marker
locus M10
(SEQ ID NO:30). In some embodiments, said recombinant chromosomal segment is
defined by:
a) a non-introgressed allele at marker locus M1 (SEQ ID NO:1); b) an
introgressed allele at a
marker locus selected from the group consisting of marker locus M2 (SEQ ID
NO:2), marker locus
M4 (SEQ ID NO:4), marker locus M5 (SEQ ID NO:9), marker locus M6 (SEQ ID
NO:10), marker
locus M7 (SEQ ID NO:15), marker locus M8 (SEQ ID NO:20), marker locus M9 (SEQ
ID NO:25),
and marker locus M10 (SEQ ID NO:30); and c) a non-introgressed allele at
marker locus M3 (SEQ
ID NO:3). Said introgressing may comprise backcrossing, marker-assisted
selection, or assaying
for Stemphylium resistance. Tomato plants obtainable by the methods disclosed
herein are further
provided.
4
Date Recue/Date Received 2021-08-25
[0009] In yet a further aspect, methods are provided for selecting a tomato
plant exhibiting
resistance to Stemphylium, comprising: a) crossing the tomato plant of claim 1
with itself or with
a second tomato plant of a different genotype to produce one or more progeny
plants; and b)
selecting a progeny plant comprising said Stemphylium resistance allele. In
certain embodiments,
selecting said progeny plant comprises detecting a marker locus genetically
linked to said
Stemphylium resistance allele. In some embodiments, selecting said progeny
plant comprises
detecting a marker locus within or genetically linked to a chromosomal segment
flanked in the
genome of said plant marker locus M1 (SEQ ID NO:1) and marker locus M3 (SEQ ID
NO:3) on
chromosome 11. In further embodiments, selecting a progeny plant comprises
detecting at least
one polymorphism at a locus selected from the group consisting of marker locus
M1 (SEQ ID
NO:1), marker locus M2 (SEQ ID NO:2), marker locus M3 (SEQ ID NO:3), marker
locus M4
(SEQ ID NO:4), marker locus M5 (SEQ ID NO:9), marker locus M6 (SEQ ID NO:10),
marker
locus M7 (SEQ ID NO:15), marker locus M8 (SEQ ID NO:20), marker locus M9 (SEQ
ID NO:25),
and marker locus M10 (SEQ ID NO:30). In yet further embodiments, said progeny
plant is an F2-
F6 progeny plant, or producing said progeny plant comprises backcrossing.
BRIEF DESCRIPTION OF THE DRAWINGS
[0010] FIG. 1: Shows a schematic overview of different Stemphylium
introgressions on the
physical map of chromosome 11.
DETAILED DESCRIPTION
[0011] Gray leaf spot disease in tomato occurs worldwide and is caused by four
fungal species of
Stemphylium: S. solani, S. floridanum, S botyosum, and S. vesicarum. These
fungi cause gray
lesions on the foliage of plants and, in cases of severe disease pressure,
complete defoliation. Gray
leaf spot disease can be managed through application of fungicides or the use
of Stemphylium
resistant tomato cultivars. Stemphylium resistance is conferred by the Sm
gene, which originates
from wild tomato accession Solanum pimpinellifilium PI 79532 and confers
resistance to all four
Stemphylium species that cause gray leaf spot disease. The Sm gene was found
to be located
between markers T10 (isozyme marker) and TG110 (RFLP marker) on tomato
chromosome 11
and found to act in an incompletely dominant manner. However, PCR-based high-
throughput
makers that could be used for marker-assisted breeding have not been reported.
Date Recue/Date Received 2021-08-25
[0012] To date the Stemphylium resistance conferred by the Sm gene has been
associated with
commercially unacceptable reductions in yield and small fruit size. This has
hampered the ability
to use the resistance in a commercially relevant manner. The current inventors
were surprisingly
able to remove the deleterious yield and small fruit size phenotype from the
Stemphylium
resistance conferred by the Sm gene, allowing it to be used in a commercial
setting. This was
surprisingly possible despite the Sm gene being closely associated with the
centromeric region of
chromosome 11, which has a very low rate of recombination. Despite this, the
current inventors
were able to generate very small reduced recombinant segments with a
centromeric region that
originates from S. lycopersicum rather than S. pimpinellifilium, and lacking
associated deleterious
traits.
[0013] The present inventors also found that the Sm locus is located outside
the region flanked by
markers T10 and TG110, based on the physical map of tomato chromosome 11 (FIG.
1). TG110
was found to be located at 20.4 Mbp of tomato chromosome 11 of the public
tomato genome map
version SL2.50 (The Tomato Genome Consortium; Nature 485:635-641, 2012;
publically
available on the internet through solgenomics.net). T10, whose sequence was
found as Genbank
accession number X14041.1, was found to be located at 52.1 Mbp of tomato
chromosome 11 of
the public tomato genome map version SL2.50. The present inventors
surprisingly found that the
Sm locus is located between 8.9 and 9.8 Mbp of tomato chromosome 11 and were
able to identify
novel genetic markers for identifying and tracking Sm during plant breeding
and allowing removal
of genomic regions that confer undesirable traits, such as small fruit size.
Having discovered that
the Stemphylium resistance alleles and the small fruit size alleles can be
removed using the novel
markers provided herein, the inventors created for the first time a
recombinant event that can be
used by breeders to readily transfer the Stemphylium resistance alleles
without the reduced yield
and small fruit size trait to other tomato plants.
[0014] The present inventors have discovered for the first time that Ml, a SNP
marker with a
[C/A] change at 8,894,829 bp on chromosome 11 of the public tomato genome map
version
SL2.50, M2, a SNP marker with a [G/A] change at 9,591,834 bp of chromosome 11
of the public
tomato genome map version SL2.50, and M3, a SNP marker with a [T/A] change at
9,826,973 bp
of chromosome 11 of the public tomato genome map version SL2.50 can be used to
identify this
region, wherein M1 and M3 are flanking markers. The public genome of tomato is
available at
for example www.solgenomics.net, and one skilled in the art would understand
that the marker
6
Date Recue/Date Received 2021-08-25
sequences provided for the first time in the instant application could be
located on any version (or
later version) of the public genome. One aspect of the invention therefore
provides plants
comprising Solanum lycopersicum DNA at marker locus M1 (SEQ ID NO:1), and
introgressed
donor DNA at marker locus M2 (SEQ ID NO:2). In further embodiments, the
invention provides
plants comprising Solanum lycopersicum parent DNA at marker locus M3 (SEQ ID
NO:3), and
introgressed donor DNA at marker locus M2 (SEQ ID NO:2).
[0015] In certain embodiments, tomato plants are provided comprising an
introgressed allele on
chromosome 11, wherein said introgressed allele confers to said plant
increased resistance to
Stemphylium compared to a plant not comprising said allele. In further
embodiments, said plant
lacks a further allele, genetically linked to said introgressed allele, that
confers small fruit size
when present.
[0016] Further provided herein are reduced recombinant introgressions
comprising a genomic
interval between marker locus M1 (SEQ ID NO:1) and marker locus M3 (SEQ ID
NO:3) on
chromosome 11, wherein said reduced genomic interval lacks deleterious small
fruit size alleles
associated with larger Stemphylium resistance introgressions.
[0017] In other embodiments, the invention provides plants comprising one or
more of the novel
recombinant introgressions provided herein. These novel introgressions provide
robust resistance
to Stemphylium, while avoiding the reduction in performance characteristics
associated with
conventional introgressions of the Sm gene. Methods of producing the plants
described herein are
further provided. In certain embodiments, the invention provides tomato line
FDR-I15-0403V
comprising an exemplary reduced introgression described herein, a sample of
the seed of which
has been deposited under NCMA Accession No. 202103011.
[0018] The invention further provides novel trait-linked markers which can be
used to produce
plants comprising novel recombinant introgressions on chromosome 11 conferring
Stemphylium
resistance as described herein. In particular embodiments, the invention
provides the markers
shown in Table 1.
[0019] Methods of producing plants comprising the reduced recombinant
introgressions described
herein are further provided. In some examples, donor DNA from a resistant
donor parent is
introgressed into a cultivated plant line (the recurrent parent line). M1 (SEQ
ID NO:1) is used to
select the allele of the recurrent parent, and M2 (SEQ ID NO:2) is used to
select the allele of the
resistance donor parent resulting in a reduced genomic interval lacking
deleterious traits associated
7
Date Recue/Date Received 2021-08-25
with larger Stemphylium resistance introgressions. In further embodiments, M3
(SEQ ID NO:3)
is further used to select the allele of the recurrent parent resulting in a
further reduced genomic
interval conferring Stemphylium resistance.
[0020] In certain embodiments, the invention provides methods of producing or
selecting a tomato
plant exhibiting resistance to Stemphylium comprising: a) crossing a tomato
plant provided herein
with itself or with a second tomato plant of a different genotype to produce
one or more progeny
plants; and b) selecting a progeny plant comprising said first introgressed
allele or said second
introgressed allele. In some embodiments, methods of the invention comprise
selecting a progeny
plant by detecting at least one polymorphism at a locus selected from the
group consisting of
marker locus M1 (SEQ ID NO:1), M2 (SEQ ID NO:2), and M3 (SEQ ID NO:3).
[0021] Because genetically diverse plant lines can be difficult to cross, the
introgression of
Stemphylium resistance alleles into cultivated lines using conventional
breeding methods could
require prohibitively large segregating populations for progeny screens with
an uncertain outcome.
Marker-assisted selection (MAS) is therefore essential for the effective
introgression of
Stemphylium resistance alleles into elite cultivars. However, previously known
markers for
Stemphylium resistance have failed to discriminate between donor DNA
conferring disease
resistance and donor DNA conferring deleterious traits. This has been further
complicated by the
previous inability to resolve the specific regions associated with disease
resistance. For the first
time, the present invention enables effective MAS by providing improved and
validated markers
for detecting genotypes associated with disease resistance without the need to
grow large
populations of plants to maturity in order to observe the phenotype.
[0022] Further provided herein is use of the genetic region on chromosome 11
that confers
resistance to Stemphylium, wherein the region also confers resistance against
leaf and fruit
symptoms caused by the Tomato Brown Rugose Fruit Virus (TBRFV). Tomato Brown
Rugose
Fruit Virus is a rapidly spreading tobamovirus that causes spots to form on
the leaves of tomato
plants and developing fruit. The brown and black necrotic spots on the fruit
render the fruit
unmarketable. This virus was originally discovered in 2016 in Jordan but has
been reported to have
spread to important tomato growing regions, such as Israel, Turkey, the
Netherlands, Mexico, and
western USA.
[0023] In certain embodiments, SNP markers Ml, M2, and M3, which are described
above, can
also be used to select for the TBRFV resistance trait. Additional markers that
can be used to select
8
Date Recue/Date Received 2021-08-25
for the TBRFV resistance trait are M4, a SNP marker with a [T/G] change at
8,891,489 bp on
chromosome 11 of the public tomato genome map version SL2.50; M5, a SNP marker
with a [C/TI
change at 9,355,794 bp on chromosome 11 of the public tomato genome map
version SL2.50; M6,
a SNP marker with a [A/T] change at 9,401,319 bp on chromosome 11 of the
public tomato
genome map version SL2.50; M7, a SNP marker with a [G/T] change at 9,406,414
bp on
chromosome 11 of the public tomato genome map version SL2.50; M8, a SNP marker
with a [A/T]
change at 9,421,426 bp on chromosome 11 of the public tomato genome map
version SL2.50; M9,
a SNP marker with a [T/C] change at 9,470,789 bp on chromosome 11 of the
public tomato genome
map version SL2.50; and M10, a SNP marker with a [A/G] change at 9,756,371 bp
on chromosome
11 of the public tomato genome map version SL2.50. Markers M4, M5, M6, M7, M8,
M9, and
M10 can also be used to select for the Stemphylium resistance allele described
herein.
[0024] In certain embodiments markers M1 and M3 are used to select the allele
of the recurrent
parent line and any of markers M2, M4, M5, M6, M7, M8, M9, or M10 is used to
select for the
donor parent line.
I.
Genomic Regions, Alleles, and Polymorphisms Associated With Stemphylium
Resistance in Tomato Plants
[0025] The invention provides novel introgressions of one or more alleles
associated with
Stemphylium disease resistance without the detrimental small fruit size trait
in tomato plants,
together with polymorphic nucleic acids and linked markers for tracking the
introgressions during
plant breeding.
[0026] Tomato lines exhibiting Stemphylium resistance are known in the art and
may be used
together with the novel trait-linked markers provided herein in accordance
with certain
embodiments of the invention. For example, the wild tomato accession Solanum
pimpinellifolium
PI 79532, which also carries the designation LA2348, can be used as a source
for Stemphylium
resistance. This line is available at the U.S. National Plant Germplasm System
and the Tomato
Genetic Resource Center in Davis, California, USA. Using the improved genetic
markers and
assays of the invention, the present inventors were able to successfully
identify novel reduced
introgressions from S. pimpinellifolium that confer Stemphylium resistance to
the plant with fewer
deleterious traits when introgressed into a cultivated line. In certain
embodiments, the invention
provides tomato plants comprising donor DNA between marker loci M1 (SEQ ID
NO:1) and M3
(SEQ ID NO:3) on chromosome 11.
9
Date Recue/Date Received 2021-08-25
[0027] The novel introgressions provided herein confer robust resistance to
Stemphylium, while
avoiding the reduction in yield and small fruit size seen with conventional
introgressions. The
invention therefore represents a significant advance in the art.
II. Introgression of Genomic Regions Associated with Stemphylium Resistance
[0028] Marker-assisted introgression involves the transfer of a chromosomal
region defined by
one or more markers from a first genetic background to a second. Offspring of
a cross that contain
the introgressed genomic region can be identified by the combination of
markers characteristic of
the desired introgressed genomic region from a first genetic background and
both linked and
unlinked markers characteristic of the second genetic background.
[0029] The present invention provides novel accurate markers for identifying
and tracking
introgression of one or more of the genomic regions disclosed herein from a
Stemphylium resistant
plant into a cultivated line. The invention further provides markers for
identifying and tracking
the novel introgressions disclosed herein during plant breeding, including the
markers set forth in
Table 1.
[0030] Markers within or linked to any of the genomic intervals of the present
invention may be
useful in a variety of breeding efforts that include introgression of genomic
regions associated with
disease resistance into a desired genetic background. For example, a marker
within 40 cM, 20 cM,
15 cM, 10 cM, 5cM, 2 cM, or 1 cM of a marker associated with disease
resistance described herein
can be used for marker-assisted introgression of genomic regions associated
with a disease
resistant phenotype.
[0031] Tomato plants comprising one or more introgressed regions associated
with a desired
phenotype wherein at least 10%, 25%, 50%, 75%, 90%, or 99% of the remaining
genomic
sequences carry markers characteristic of the recurrent parent germplasm are
also provided.
Tomato plants comprising an introgressed region comprising regions closely
linked to or adjacent
to the genomic regions and markers provided herein and associated with a
disease resistance
phenotype are also provided.
III. Development of Disease Resistant Tomato Varieties
[0032] For most breeding objectives, commercial breeders work with germplasm
that is
"cultivated," "cultivated type," or "elite." These cultivated lines may be
used as recurrent parents
or as a source of recurrent parent alleles during breeding. Cultivated or
elite germplasm is easier
Date Recue/Date Received 2021-08-25
to breed because it generally performs well when evaluated for horticultural
performance. Many
cultivated tomato types have been developed and are known in the art as being
agronomically elite
and appropriate for commercial cultivation. However, the performance advantage
a cultivated
germplasm provides can be offset by a lack of allelic diversity. Breeders
generally accept this
tradeoff because progress is faster when working with cultivated material than
when breeding with
genetically diverse sources.
[0033] In contrast, when cultivated germplasm is crossed with non-cultivated
germplasm, a
breeder can gain access to novel alleles from the non-cultivated type. Non-
cultivated germplasm
may be used as a source of donor alleles during breeding. However, this
approach generally
presents significant difficulties due to fertility problems associated with
crosses between diverse
lines, and genetically linked deleterious alleles from the non-cultivated
parent. For example, non-
cultivated tomato types can provide alleles associated with disease
resistance. However, these non-
cultivated types may have poor horticultural qualities such as poor quality,
poor architecture, low
yield, or small fruit size.
[0034] The process of introgressing desirable resistance genes from non-
cultivated lines into elite
cultivated lines while avoiding problems with genetically linked deleterious
alleles or low
heritability is a long and often arduous process. In deploying alleles derived
from wild relatives it
is often desirable to introduce a minimal or truncated introgression that
provides the desired trait
but lacks detrimental effects. To aid introgression reliable marker assays are
preferable to
phenotypic screens. Success is furthered by simplifying genetics for key
attributes to allow focus
on genetic gain for quantitative traits such as disease resistance. Moreover,
the process of
introgressing genomic regions from non-cultivated lines can be greatly
facilitated by the
availability of accurate markers for MAS.
[0035] One of skill in the art would therefore understand that the alleles,
polymorphisms, and
markers provided by the invention allow the tracking and introduction of any
of the genomic
regions identified herein into any genetic background. In addition, the
genomic regions associated
with disease resistance disclosed herein can be introgressed from one genotype
to another and
tracked using MAS. Thus, the inventors' discovery of accurate markers
associated with disease
resistance will facilitate the development of tomato plants having beneficial
phenotypes. For
example, seed can be genotyped using the markers of the present invention to
select for plants
11
Date Recue/Date Received 2021-08-25
comprising desired genomic regions associated with disease resistance.
Moreover, MAS allows
identification of plants homozygous or heterozygous for a desired
introgression.
[0036] Inter-species crosses can also result in suppressed recombination and
plants with low
fertility or fecundity. For example, suppressed recombination has been
observed for the tomato
nematode resistance gene Mi, the Mla and Mk genes in barley, the Yr 1 7 and
Lr20 genes in wheat,
the Run] gene in grapevine, and the Rma gene in peanut. Meiotic recombination
is essential for
classical breeding because it enables the transfer of favorable alleles across
genetic backgrounds,
the removal of deleterious genomic fragments, and pyramiding traits that are
genetically tightly
linked. Therefore suppressed recombination forces breeders to enlarge
segregating populations for
progeny screens in order to arrive at the desired genetic combination.
[0037] Phenotypic evaluation of large populations is time-consuming, resource-
intensive and not
reproducible in every environment. Marker-assisted selection offers a feasible
alternative.
Molecular assays designed to detect unique polymorphisms, such as SNPs, are
versatile. However,
they may fail to discriminate alleles within and among tomato species in a
single assay. Structural
rearrangements of chromosomes such as deletions impair hybridization and
extension of
synthetically labeled oligonucleotides. In the case of duplication events,
multiple copies are
amplified in a single reaction without distinction. The development and
validation of accurate and
highly predictive markers are therefore essential for successful MAS breeding
programs.
IV. Marker Assisted Breeding and Genetic Engineering Techniques
[0038] Genetic markers that can be used in the practice of the present
invention include, but are
not limited to, restriction fragment length polymorphisms (RFLPs), amplified
fragment length
polymorphisms (AFLPs), simple sequence repeats (SSRs), simple sequence length
polymorphisms
(SSLPs), single nucleotide polymorphisms (SNPs), insertion/deletion
polymorphisms (Indels),
variable number tandem repeats (VNTRs), and random amplified polymorphic DNA
(RAPD),
isozymes, and other markers known to those skilled in the art. Marker
discovery and development
in crop plants provides the initial framework for applications to marker-
assisted breeding activities
(U.S. Patent Pub. Nos.: 2005/0204780, 2005/0216545, 2005/0218305, and
2006/00504538). The
resulting "genetic map" is the representation of the relative position of
characterized loci
(polymorphic nucleic acid markers or any other locus for which alleles can be
identified) to each
other.
12
Date Recue/Date Received 2021-08-25
[0039] Polymorphisms comprising as little as a single nucleotide change can be
assayed in a
number of ways. For example, detection can be made by electrophoretic
techniques including a
single strand conformational polymorphism (Orita, et al. (1989) Genomics,
8(2), 271-278),
denaturing gradient gel electrophoresis (Myers (1985) EPO 0273085), or
cleavage fragment length
polymorphisms (Life Technologies, Inc., Gaithersburg, MD), but the widespread
availability of
DNA sequencing often makes it easier to simply sequence amplified products
directly. Once the
polymorphic sequence difference is known, rapid assays can be designed for
progeny testing,
typically involving some version of PCR amplification of specific alleles
(PASA; Sommer, et al.
(1992) Biotechniques 12(1), 82-87), or PCR amplification of multiple specific
alleles (PAMSA;
Dutton and Sommer (1991) Biotechniques,11(6), 700-7002).
[0040] Polymorphic markers serve as useful tools for assaying plants for
determining the degree
of identity of lines or varieties (U.S. Patent No. 6,207,367). These markers
form the basis for
determining associations with phenotypes and can be used to drive genetic
gain. In certain
embodiments of methods of the invention, polymorphic nucleic acids can be used
to detect in a
tomato plant a genotype associated with disease resistance, identify a tomato
plant with a genotype
associated with disease resistance, and to select a tomato plant with a
genotype associated with
disease resistance. In certain embodiments of methods of the invention,
polymorphic nucleic acids
can be used to produce a tomato plant that comprises in its genome an
introgressed locus associated
with disease resistance. In certain embodiments of the invention, polymorphic
nucleic acids can
be used to breed progeny tomato plants comprising a locus or loci associated
with disease
resistance.
[0041] Genetic markers may include "dominant" or "codominant" markers.
"Codominant"
markers reveal the presence of two or more alleles (two per diploid
individual). "Dominant"
markers reveal the presence of only a single allele. Markers are preferably
inherited in codominant
fashion so that the presence of both alleles at a diploid locus, or multiple
alleles in triploid or
tetraploid loci, are readily detectable, and they are free of environmental
variation, i.e., their
heritability is 1. A marker genotype typically comprises two marker alleles at
each locus in a
diploid organism. The marker allelic composition of each locus can be either
homozygous or
heterozygous. Homozygosity is a condition where both alleles at a locus are
characterized by the
same nucleotide sequence. Heterozygosity refers to a condition where the two
alleles at a locus are
different.
13
Date Recue/Date Received 2021-08-25
[0042] Nucleic acid-based analyses for determining the presence or absence of
the genetic
polymorphism (i.e. for genotyping) can be used in breeding programs for
identification, selection,
introgression, and the like. A wide variety of genetic markers for the
analysis of genetic
polymorphisms are available and known to those of skill in the art. The
analysis may be used to
select for genes, portions of genes, QTL, alleles, or genomic regions that
comprise or are linked to
a genetic marker that is linked to or associated with disease resistance in
tomato plants.
[0043] As used herein, nucleic acid analysis methods include, but are not
limited to, PCR-based
detection methods (for example, TaqMan assays), microarray methods, mass
spectrometry-based
methods and/or nucleic acid sequencing methods, including whole genome
sequencing. In certain
embodiments, the detection of polymorphic sites in a sample of DNA, RNA, or
cDNA may be
facilitated through the use of nucleic acid amplification methods. Such
methods specifically
increase the concentration of polynucleotides that span the polymorphic site,
or include that site
and sequences located either distal or proximal to it. Such amplified
molecules can be readily
detected by gel electrophoresis, fluorescence detection methods, or other
means.
[0044] One method of achieving such amplification employs the polymerase chain
reaction (PCR)
(Mullis et al. (1986) Cold Spring Harbor Symp. Quant. Biol. 51:263-273;
European Patent 50,424;
European Patent 84,796; European Patent 258,017; European Patent 237,362;
European Patent
201,184; U.S. Patent 4,683,202; U.S. Patent 4,582,788; and U.S. Patent
4,683,194), using primer
pairs that are capable of hybridizing to the proximal sequences that define a
polymorphism in its
double-stranded form. Methods for typing DNA based on mass spectrometry can
also be used.
Such methods are disclosed in US Patents 6,613,509 and 6,503,710, and
references found therein.
[0045] Polymorphisms in DNA sequences can be detected or typed by a variety of
effective
methods well known in the art including, but not limited to, those disclosed
in U.S. Patent Nos.
5,468,613, 5,217,863; 5,210,015; 5,876,930; 6,030,787; 6,004,744; 6,013,431;
5,595,890;
5,762,876; 5,945,283; 5,468,613; 6,090,558; 5,800,944; 5,616,464; 7,312,039;
7,238,476;
7,297,485; 7,282,355; 7,270,981 and 7,250,252 all of which are incorporated
herein by reference
in their entirety. However, the compositions and methods of the present
invention can be used in
conjunction with any polymorphism typing method to detect polymorphisms in
genomic DNA
samples. These genomic DNA samples used include but are not limited to,
genomic DNA isolated
directly from a plant, cloned genomic DNA, or amplified genomic DNA.
14
Date Recue/Date Received 2021-08-25
[0046] For instance, polymorphisms in DNA sequences can be detected by
hybridization to allele-
specific oligonucleotide (ASO) probes as disclosed in U.S. Patent Nos.
5,468,613 and 5,217,863.
U.S. Patent No. 5,468,613 discloses allele specific oligonucleotide
hybridizations where single or
multiple nucleotide variations in nucleic acid sequence can be detected in
nucleic acids by a
process in which the sequence containing the nucleotide variation is
amplified, spotted on a
membrane and treated with a labeled sequence-specific oligonucleotide probe.
[0047] Target nucleic acid sequence can also be detected by probe ligation
methods, for example
as disclosed in U.S. Patent No. 5,800,944 where sequence of interest is
amplified and hybridized
to probes followed by ligation to detect a labeled part of the probe.
[0048] Microarrays can also be used for polymorphism detection, wherein
oligonucleotide probe
sets are assembled in an overlapping fashion to represent a single sequence
such that a difference
in the target sequence at one point would result in partial probe
hybridization (Borevitz et al.,
Genome Res. 13:513-523 (2003); Cui et al., Bioinformatics 21:3852-3858 (2005).
On any one
microarray, it is expected there will be a plurality of target sequences,
which may represent genes
and/or noncoding regions wherein each target sequence is represented by a
series of overlapping
oligonucleotides, rather than by a single probe. This platform provides for
high throughput
screening of a plurality of polymorphisms. Typing of target sequences by
microarray-based
methods is described in US Patents 6,799,122; 6,913,879; and 6,996,476.
[0049] Other methods for detecting SNPs and Indels include single base
extension (SBE)
methods. Examples of SBE methods include, but are not limited, to those
disclosed in U.S. Patent
Nos. 6,004,744; 6,013,431; 5,595,890; 5,762,876; and 5,945,283.
[0050] In another method for detecting polymorphisms, SNPs and Indels can be
detected by
methods disclosed in U.S. Patent Nos. 5,210,015; 5,876,930; and 6,030,787 in
which an
oligonucleotide probe having a 5' fluorescent reporter dye and a 3' quencher
dye covalently linked
to the 5' and 3' ends of the probe. When the probe is intact, the proximity of
the reporter dye to
the quencher dye results in the suppression of the reporter dye fluorescence,
e.g. by Forster-type
energy transfer. During PCR, forward and reverse primers hybridize to a
specific sequence of the
target DNA flanking a polymorphism while the hybridization probe hybridizes to
polymorphism-
containing sequence within the amplified PCR product. In the subsequent PCR
cycle DNA
polymerase with 5' 4 3' exonuclease activity cleaves the probe and separates
the reporter dye
from the quencher dye resulting in increased fluorescence of the reporter.
Date Recue/Date Received 2021-08-25
[0051] In another embodiment, a locus or loci of interest can be directly
sequenced using nucleic
acid sequencing technologies. Methods for nucleic acid sequencing are known in
the art and
include technologies provided by 454 Life Sciences (Branford, CT), Agencourt
Bioscience
(Beverly, MA), Applied Biosystems (Foster City, CA), LI-COR Biosciences
(Lincoln, NE),
NimbleGen Systems (Madison, WI), Illumina (San Diego, CA), and VisiGen
Biotechnologies
(Houston, TX). Such nucleic acid sequencing technologies comprise formats such
as parallel bead
arrays, sequencing by ligation, capillary electrophoresis, electronic
microchips, "biochips,"
microarrays, parallel microchips, and single-molecule arrays.
[0052] Some embodiments include methods for treating tomato, tomato plant
parts, or the soil or
substrate in which tomato plants are grown or intended to be grown with an
active compound or a
combination of active compounds. In some embodiments, the tomato plants are
suspected of being
or becoming infected with a disease, or the methods are for protecting or
treating plants from
fungal and bacterial infections. In some embodiments the disease is a fungal
infection, and the
embodiments include methods for protecting from a fungal disease. In further
embodiments, the
tomato plant comprises a recombinant chromosomal segment on chromosome 11 that
comprises a
Stemphylium resistance allele. In some embodiments, said chromosomal segment
lacks a
deleterious allele that confers a small fruit size trait to said plant when
present. In further
embodiments, the treatment increases tomato yield. In some embodiments, the
active compound
or combination of active compounds comprises a fungicidal active ingredient.
In certain
embodiments, the active compound is selected from the following groups: (1)
inhibitors of the
ergosterol synthesis, (2) inhibitors of the respiratory chain at complex I or
II, (3) inhibitors of the
respiratory chain at complex III, (4) inhibitors of the mitosis and cell
division, (5) compounds
capable of having a multisite action, (6) compounds capable of inducing a host
defense, (7) inhibitors
of the amino acid and/or protein biosynthesis, (8) inhibitors of the ATP
production, (9) inhibitors of
the cell wall synthesis, (10) inhibitors of the lipid and membrane synthesis,
(11) inhibitors of the
melanine biosynthesis, (12) inhibitors of the nucleic acid synthesis, (13)
inhibitors of the signal
transduction, (14) compounds capable of acting as uncoupler, and (15) other
fungicides. Examples
of such active compounds, their synthesis, and analysis are provided in
European Patent
Application EP3335559A1.
[0053] In some embodiments, inhibitors of the ergosterol synthesis are
selected from the group
consisting of (1.001) cyproconazole, (1.002) difenoconazole, (1.003)
epoxiconazole, (1.004)
16
Date Recue/Date Received 2021-08-25
fenhexamid, (1.005) fenpropidin, (1.006) fenpropimorph, (1.007) fenpyrazamine,
(1.008)
fluquinconazole, (1.009) flutriafol, (1.010) imazalil, (1.011) imazalil
sulfate, (1.012) ipconazole,
(1.013) metconazole, (1.014) myclobutanil, (1.015) paclobutrazol, (1.016)
prochloraz, (1.017)
propiconazole, (1.018) prothioconazole, (1.019) pyrisoxazole, (1.020)
spiroxamine, (1.021)
tebuconazole, (1.022) tetraconazole, (1.023) triadimenol, (1.024) tridemorph,
(1.025)
triticonazole, (1.026) (1R,2S,5S)-5-(4-chlorobenzy1)-2-(chloromethyl)-2-methyl-
1-(111-1,2,4-
triazol-1-ylmethyl)cyclopentanol, (1.027) (1 S,2R,5R)-5-(4-chlorobenzy1)-2-
(chloromethyl)-2-
methy1-1-(1H-1,2,4-tri az ol-1-ylmethyl)-cyclopentanol, (1.028) (2R)-2-(1-
chlorocyclopropy1)-4-
[(1R)-2,2-dichlorocyclopropy1]-1-(1H-1,2,4-triazol-1-y1)butan-2-ol,
(1.029) (2R)-2-(1-
chlorocyclopropy1)-4-[(1S)-2,2-dichlorocyclopropy1]-1-(1H-1,2,4-triazol-1-
y1)butan-2-ol, (1.030)
(2R)-244-(4-chlorophenoxy)-2-(trifluoromethyl)pheny1]-1-(1H-1,2,4-triazol-1-
y1)propan-2-ol,
(1.031)
(2 S)-2-(1-chl orocyclopropy1)-4- [(1R)-2,2-dichlorocyclopropyl] -1-(1H-1,2,4-
tri azol-1-
yObutan-2-ol, (1.032) (2 S)-2-(1 -chlorocyclopropy1)-4- [(1 S)-2,2-dichl
orocyclopropy1]-1-(1H-
1,2,4-tri azol-1-yl)butan-2-ol, (1.033) (2 S)-244-(4-chlorophenoxy)-2-
(trifluoromethyl)pheny1]-1-
(1H-1,2,4-tri azol-1-yl)propan-2-ol,
(1.034) (R)-[3 -(4-chloro-2-fluoropheny1)-5-(2,4-
difluoropheny1)-1,2-oxazol-4-yl] (pyridin-3 -yl)m ethanol,
(1.035) (S)43-(4-chloro-2-
fluoropheny1)-5-(2,4-difluoropheny1)-1,2-oxazol-4-y1](pyridin-3-y1)methanol,
(1.036) [3 -(4-
chloro-2-fluoropheny1)-5 -(2,4-difluoropheny1)-1,2-oxazol-4-yl] (pyridin-3 -
yl)methanol, (1.037)
1-( {(2R,4 S)-2- [2-chloro-4-(4-chlorophenoxy)phenyl] -4-methy1-1,3 -di oxolan-
2-yllmethyl)-1H-
1,2,4-tri azole,
(1.038) 1-( {(2 S,4 S)-2- [2-chloro-4-(4-chlorophenoxy)phenyl] -4-methy1-
1,3 -
dioxolan-2-y1 1 methyl)-1H-1,2,4-triazole,
(1.039) 1- { [3 -(2-chloropheny1)-2-(2,4-
difluorophenyl)oxiran-2-yl]methyll-1H-1,2,4-tri azol-5-y1 thiocyanate, (1.040)
1- {[rel(2R,3R)-3-
(2-chloropheny1)-2-(2,4-difluorophenyl)oxiran-2-yl]methy11-1H-1,2,4-triazol-5-
y1 thiocyanate,
(1.041) 1- {[rel(2R,3S)-3-(2-chloropheny1)-2-(2,4-difluorophenyl)oxiran-2-
yl]methy11-1H-1,2,4-
triazol-5-y1 thiocyanate,
(1.042) 2- [(2R,4R,5R)-1-(2,4-dichloropheny1)-5-hydroxy-2,6,6-
trimethylheptan-4-y1]-2,4-dihydro-3H-1,2,4-tri azol e-3 -thi one, (1.043) 2-
[(2R,4R,5 S)-1-(2,4-
dichloropheny1)-5-hydroxy-2,6,6-trimethylheptan-4-yl] -2,4-dihydro-3H-i,2,4-
tri azole-3 -thi one,
(1.044)
2- [(2R,4 S,5R)-1-(2,4-dichloropheny1)-5-hydroxy-2,6,6-trimethylheptan-4-yl] -
2,4-
dihydro-3H-1,2,4-tri azole-3 -thi one, (1.045) 2- [(2R,4 S,5 S)-1-(2,4-dichl
oropheny1)-5-hydroxy-
2,6,6-trimethylheptan-4-y1]-2,4-dihydro-3H-1,2,4-triazole-3 -thione, (1.046) 2-
[(2S,4R,5R)-1-
(2,4-dichloropheny1)-5-hydroxy-2,6,6-trimethylheptan-4-y1]-2,4-dihydro-3H-
1,2,4-tri azol e-3 -
17
Date Recue/Date Received 2021-08-25
thi one, (1.047) 2- [(2 S,4R,5 S)-1-(2,4-dichloropheny1)-5-hydroxy-2,6,6-
trimethylheptan-4-yl] -2,4-
dihydro-311-1,2,4-tri azole-3 -thi one, (1.048) 2- [(2 S,4 S,5R)-1-(2,4-dichl
oropheny1)-5-hydroxy-
2,6,6-trimethylheptan-4-y1]-2,4-dihydro-311-1,2,4-triazole-3 -thione, (1.049)
2-[(2S,4S,5 S)-1-(2,4-
dichloropheny1)-5-hydroxy-2,6,6-trimethylheptan-4-yl] -2,4-dihydro-311-1,2,4-
tri azole-3 -thi one,
(1.050) 2- [1-(2,4-dichloropheny1)-5-hydroxy-2,6,6-trimethylheptan-4-yl] -2,4-
dihydro-311-1,2,4-
tri azole-3 -thi one, (1.051) 2- [2-chloro-4-(2,4-dichlorophenoxy)phenyl] -1-
(1H-1,2,4-tri azol-1-
yl)propan-2-ol, (1.052) 242-chloro-4-(4-chlorophenoxy)pheny1]-1-(1H-1,2,4-
triazol-1-y1)butan-
2-ol, (1.053) 2- [4-(4-chlorophenoxy)-2-(trifluoromethyl)phenyl] -1-(1H-1,2,4-
tri azol-1-yl)butan-
2-ol, (1.054) 244-(4-chlorophenoxy)-2-(trifluoromethyl)pheny1]-1-(1H-1,2,4-
triazol-1-y1)pentan-
2-ol, (1.055) 244-(4-chlorophenoxy)-2-(trifluoromethyl)pheny1]-1-(1H-1,2,4-
triazol-1-y1)propan-
2-ol, (1.056) 2- { [3 -(2-chloropheny1)-2-(2,4-difluorophenyl)oxiran-2-
yl]methyl } -2,4-dihydro-3H-
1,2,4-tri azole-3 -thi one, (1.057) 2- { [rel(2R,3R)-3 -(2-chloropheny1)-2-
(2,4-difluorophenyl)oxiran-
2-yl]methyl } -2,4-dihydro-3H-1,2,4-triazole-3-thione, (1.058) 2- { [rel(2R,3
S)-3 -(2-chloropheny1)-
2-(2,4-difluorophenyl)oxiran-2-yl]methyl } -2,4-dihydro-3H-1,2,4-tri azol e-3 -
thi one, (1.059) 544-
chlorobenzy1)-2-(chloromethyl)-2-methyl-1-(1H-1,2,4-tri azol-1-
ylmethyl)cyclopentanol, (1.060)
5-(allylsulfany1)-1- { [3 -(2-chloropheny1)-2-(2,4-difluorophenyl)oxiran-2-
yl]methyl } -1H-1,2,4-
tri azole, (1.061)
5-(allylsulfany1)-1- { [rel(2R,3R)-3-(2-chloropheny1)-2-(2,4-
difluorophenyl)oxiran-2-yl]methyl } -1H-1,2,4-triazole, (1.062) 5-
(allylsulfany1)-1- { [rel(2R,3 S)-3 -
(2-chloropheny1)-2-(2,4-difluorophenyl)oxiran-2-yl]methyl } -1H-1,2,4-
triazole, (1.063) N'-(2,5-
dimethy1-4- { [3 -(1,1,2,2-tetrafluoroethoxy)phenyl] sulfanyl } pheny1)-N-
ethyl-N-
methylimidoformamide, (1.064)
N'-(2,5-dimethy1-4- { [3 -(2,2,2-
trifluoroethoxy)phenyl]sulfanyl } phenyl)-N-ethyl-N-methylimidoformamide,
(1.065) N'-(2,5-
dimethy1-4- { [3 -(2,2,3,3 -tetrafluoropropoxy)phenyl]sulfanyl } pheny1)-N-
ethyl-N-
methylimidoformamide, (1.066)
N'-(2,5-dimethy1-4- { [3-
(pentafluoroethoxy)pheny1]su1fany1 } phenyl)-N-ethyl-N-methylimidoformamide,
(1.067) N'-(2,5-
dimethy1-4- {3 - [(1,1,2,2-tetrafluoroethyl)sulfanyl]phenoxy } pheny1)-N-ethyl-
N-
methylimidoformamide, (1.068)
N'-(2,5-dimethy1-4- {3 - [(2,2,2-
trifluoroethyl)sulfanyl]phenoxy } phenyl)-N-ethyl-N-methylimidoformamide,
(1.069) N'-(2,5-
dimethy1-4- {3 - [(2,2,3,3 -tetrafluoropropyl)sulfanyl]phenoxy } pheny1)-N-
ethyl-N-
methylimidoformamide, (1.070)
N'-(2,5-dimethy1-4- {3 -
[(pentafluoroethyl)sulfanyl]phenoxy } phenyl)-N-ethyl-N-methylimidoformamide,
(1.071) N'-
18
Date Recue/Date Received 2021-08-25
(2,5-dimethy1-4-phenoxypheny1)-N-ethyl-N-methylimidoformamide,
(1.072) N'-(4-{[3-
(di fluorom ethoxy)phenyl] sulfanyl } -2,5-dim ethylpheny1)-N-ethyl-N-m ethyl
imi doform ami de,
(1.073)
N'-(4- {3 - [(di fluorom ethyl)sulfanyl]phenoxy } -2,5-dim ethylpheny1)-N-
ethyl-N-
methylimidoformamide, (1.074)
N'- [5-bromo-6-(2,3 -dihydro-111-inden-2-yloxy)-2-
m ethylpyri din-3 -yl] -N-ethyl-N-m ethylimi doform ami de, (1.075) N'- {4-
[(4,5 -di chl oro-1,3 -thi azol-
2-y0oxy]-2,5-dimethylphenyl } -N-ethyl-N-methylimidoformamide, (1.076) N'- {5-
bromo-6-
[(1R)-1 -(3,5-di fluorophenyl)ethoxy]-2-methylpyri din-3 -yl } -N-ethyl-N-m
ethylimi doform ami de,
(1.077) N'- {5-brom o-6- [(1 S)-1 -(3,5-di fluoroph enyl)ethoxy] -2-m
ethylpyri din-3 -yl } -N-ethyl-N-
methylimidoformamide, (1.078)
N'- {5-bromo-6-[(cis-4-isopropylcyclohexyl)oxy]-2-
m ethylpyri din-3 -yl} -N-ethyl-N-m ethylimi doform ami de,
(1.079) N'- {5-bromo-6-[(trans-4-
isopropylcyclohexyl)oxy]-2-methylpyridin-3-yll -N-ethyl-N-
methylimidoformamide, (1.080) N'-
{5-brom 0-641 -(3,5-di fluorophenyl)ethoxy] -2-m ethylpyri din-3 -yl } -N-
ethyl-N-
methylimidoformamide, (1.081) Mefentrifluconazole, and (1.082)
Ipfentrifluconazole.
[0054] In some embodiments, inhibitors of the respiratory chain at complex I
or II are selected
from the group consisting of (2.001) benzovindiflupyr, (2.002) bixafen,
(2.003) boscalid, (2.004)
carboxin, (2.005) fluopyram, (2.006) flutolanil, (2.007) fluxapyroxad, (2.008)
furametpyr, (2.009)
Isofetamid, (2.010) isopyrazam (anti-epimeric enantiomer 1R,4S,9S), (2.011)
isopyrazam (anti-
epimeric enantiomer 1S,4R,9R), (2.012) isopyrazam (anti-epimeric racemate
1RS,4SR,9SR),
(2.013) isopyrazam (mixture of syn-epimeric racemate 1RS,4SR,9RS and anti-
epimeric racemate
1RS,4SR,9SR), (2.014) isopyrazam (syn-epimeric enantiomer 1R,4 S,9R), (2.015)
isopyrazam
(syn-epimeric enantiomer 1S,4R,9S), (2.016) isopyrazam (syn-epimeric racemate
1RS,4SR,9RS),
(2.017) penflufen, (2.018) penthiopyrad, (2.019) pydiflumetofen, (2.020)
Pyraziflumid, (2.021)
sedaxane, (2.022) 1,3 -dimethyl-N-(1,1,3 -trimethy1-2,3 -dihydro-111-inden-4-
y1)-111-pyrazole-4-
c arb oxami de, (2.023) 1,3 -dim ethyl-N- [(3R)-1,1,3 -trim ethyl-2,3 -dihydro-
111-inden-4-y1]-1H-
pyrazol e-4-c arb oxami de, (2.024) 1,3 -dim ethyl-N-[(3 S)-1,1,3 -trim ethyl-
2,3 -dihydro-1H-inden-4-
y1]-1H-pyrazole-4-carboxamide, (2.025)
1 -m ethy1-3 -(tri fluorom ethyl)-N- [2'-
(tri fluorom ethyl)bipheny 1-2-y1]-1H-pyrazol e-4-c arb oxami de,
(2.026) 2-fluoro-6-
(tri fluorom ethyl)-N-(1,1,3 -trim ethyl-2,3 -dihydro-1H-inden-4-yl)b enz ami
de, (2.027) 3 -
(di fluorom ethyl)-1 -m ethyl-N-(1,1,3 -trim ethyl-2,3 -dihydro-1H-inden-4-y1)-
1H-pyrazol e-4-
c arb oxami de, (2.028) 3 -(di fluorom ethyl)-1 -m ethyl-N-[(3R)-1,1,3 -trim
ethyl-2,3 -dihydro-1H-
inden-4-y1]-1H-pyrazol e-4-c arb oxami de, (2.029) 3 -(di fluorom ethyl)-1 -m
ethyl-N- [(3 S)-1,1,3 -
19
Date Recue/Date Received 2021-08-25
trimethy1-2,3-dihydro-1H-inden-4-y1]-1H-pyrazole-4-carboxamide, (2.030) 3 -
(difluoromethyl)-
N-(7-fluoro-1,1,3 -trimethy1-2,3 -dihydro-1H-inden-4-y1)-1-methy1-1H-pyrazole-
4-carb oxamide,
(2.031)
3 -(difluoromethyl)-N-[(3R)-7-fluoro-1,1,3 -trimethy1-2,3 -dihydro-1H-inden-4-
yl] -1 -
methy1-1H-pyrazole-4-carb oxami de,
(2.032) 3 -(difluoromethyl)-N-[(3 S)-7-fluoro-1,1,3 -
trimethy1-2,3 -dihydro-1H-inden-4-yl] -1-methy1-1H-pyrazole-4-carb oxamide,
(2.033) 5,8-
difluoro-N-[2-(2-fluoro-4- { [4-(trifluoromethyl)pyridin-2-
yl]oxylphenyl)ethyl]quinazolin-4-
amine, (2.034) N-(2-cy clopenty1-5-fluorobenzy1)-N-cyclopropyl-3 -
(difluoromethyl)-5-fluoro-1-
methy1-1H-pyrazole-4-carb oxami de, (2.035) N-(2-tert-butyl-5-methylbenzy1)-N-
cyclopropyl-3 -
(difluoromethyl)-5-fluoro-l-methyl-1H-pyrazole-4-carb ox amide, (2.036) N-(2-
tert-butylbenzy1)-
N-cyclopropy1-3-(difluoromethyl)-5-fluoro-1-methyl-1H-pyrazole-4-carboxamide,
(2.037) N-(5 -
chloro-2-ethylbenzy1)-N-cyclopropy1-3 -(difluoromethyl)-5-fluoro-1 -methy1-1H-
pyrazole-4-
carb oxamide, (2.038) N-(5-chloro-24 sopropylbenzy1)-N-cyclopropy1-3 -
(difluoromethyl)-5-
fluoro-l-methy1-1H-pyrazole-4-carb ox amide, (2.039) N-[(1R,4 S)-9-
(dichloromethylene)-1,2,3,4-
tetrahydro-1,4-methanonaphthalen-5-yl] -3 -(difluoromethyl)-1-methy1-1H-
pyrazole-4-
carb oxamide, (2.040)
N- [(1 S,4R)-9-(dichloromethylene)-1,2,3,4-tetrahydro-1,4-
methanonaphthalen-5 -yl] -3 -(difluoromethyl)-1-methy1-1H-pyrazole-4-carb
oxamide, (2.041) N-
[1-(2,4-dichloropheny1)-1-methoxypropan-2-yl] -3 -(difluoromethyl)-1-m ethy1-
1H-pyrazole-4-
carb oxamide, (2.042) N-[2-chloro-6-(trifluoromethyl)benzy1]-N-cyclopropy1-3-
(difluoromethyl)-
5-fluoro-l-methyl-1H-pyrazole-4-carboxamide, (2.043)
N43-chloro-2-fluoro-6-
(trifluoromethyl)benzyl]-N-cyclopropyl-3-(difluoromethyl)-5-fluoro-1-methyl-1H-
pyrazole-4-
carboxamide, (2.044) N-[5-chloro-2-(trifluoromethyl)benzy1]-N-cyclopropy1-3-
(difluoromethyl)-
5-fluoro-l-methyl-1H-pyrazole-4-carboxamide, (2.045) N-cyclopropy1-3-
(difluoromethyl)-5-
fluoro-1-methyl-N45-methyl-2-(trifluoromethyl)benzyl]-1H-pyrazole-4-
carboxamide, (2.046) N-
cyclopropy1-3 -(difluoromethyl)-5-fluoro-N-(2-fluoro-64 sopropylbenzy1)-1-
methy1-1H-pyrazole-
4-carb oxamide, (2.047)
N-cyclopropy1-3-(difluoromethyl)-5-fluoro-N-(2-isopropy1-5-
methylbenzy1)-1-methyl-1H-pyrazole-4-carboxamide, (2.048) N-cyclopropy1-3 -
(difluoromethyl)-
5-fluoro-N-(24 sopropylb enzy1)-1-m ethy1-1H-pyrazole-4-carb othi amide,
(2.049) N-cyclopropyl-
3 -(difluoromethyl)-5-fluoro-N-(24 sopropylbenzy1)-1-methy1-1H-pyrazol e-4-
carb oxamide,
(2.050)
N-cyclopropy1-3 -(difluoromethyl)-5-fluoro-N-(5-fluoro-24 sopropylbenzy1)-1-
methyl-
1H-pyrazole-4-carb ox amide,
(2.051) N-cyclopropy1-3-(difluoromethyl)-N-(2-ethyl-4,5-
dimethylbenzy1)-5-fluoro-1-methyl-1H-pyrazole-4-carboxamide,
(2.052) N-cyclopropy1-3 -
Date Recue/Date Received 2021-08-25
(di fluorom ethyl)-N-(2-ethy1-5-fluorob enzy1)-5-fluoro-1 -m ethy1-1H-pyrazol
e-4-c arb oxami de,
(2.053) N-cyc lopropy1-3 -(difluorom ethyl)-N-(2-ethy1-5-m ethylb enzy1)-5-
fluoro-1 -m ethyl-1H-
pyrazole-4-carboxamide, (2.054)
N-cyclopropyl-N-(2-cyclopropy1-5-fluorobenzy1)-3-
(difluorom ethyl)-5-fluoro-1 -m ethy1-1H-pyrazol e-4-c arb ox amide, (2.055) N-
cyclopropyl-N-(2-
cyclopropy1-5-methylbenzy1)-3-(difluoromethyl)-5-fluoro-l-methyl-1H-pyrazole-4-
carboxamide, and (2.056) N-cyclopropyl-N-(2-cyclopropylbenzy1)-3-
(difluoromethyl)-5-fluoro-
1-m ethy1-1H-pyrazole-4-c arb oxamide.
[0055] In some embodiments, inhibitors of the respiratory chain at complex III
are selected from
the group consisting of (3.001) ametoctradin, (3.002) amisulbrom, (3.003)
azoxystrobin, (3.004)
coumethoxystrobin, (3.005) coumoxystrobin, (3.006) cyazofamid, (3.007)
dimoxystrobin, (3.008)
enoxastrobin, (3.009) famoxadone, (3.010) fenami done, (3.011)
flufenoxystrobin, (3.012)
fluoxastrobin, (3.013) kresoxim-methyl, (3.014) metominostrobin, (3.015)
orysastrobin, (3.016)
picoxystrobin, (3.017) pyraclostrobin, (3.018) pyrametostrobin, (3.019)
pyraoxystrobin, (3.020)
trifloxystrobin, (3.021)
(2E)-2- {2-[( { [(1E)-1-(3- {[(E)-1-fluoro-2-
phenylvinyl]oxy } phenyl)ethylidene] amino} oxy)methyl]phenyl } -2-(m
ethoxyimino)-N-
methylacetamide, (3.022)
(2E,3Z)-5- { [1 -(4-chloropheny1)-1H-pyrazol-3 -yl] oxy } -2-
(methoxyimino)-N,3 -dimethylpent-3 -enamide, (3.023)
(2R)-2- {2- [(2,5-
dim ethylphenoxy)m ethyl]phenyl } -2-m ethoxy-N-m ethylac etamide, (3.024)
(2S)-2- {2- [(2,5-
dim ethylphenoxy)m ethyl]phenyl } -2-m ethoxy-N-m ethylac etami de, (3.025) (3
S,6S,7R,8R)-8-
benzy1-3 - [( {3 -[(isobutyryloxy)methoxy]-4-methoxypyridin-2-y1 }
carbonyl)amino]-6-methy1-4,9-
di oxo-i,5-di oxonan-7-y1 2-methylpropanoate, (3.026)
2-{2-[(2,5-
dimethylphenoxy)methyl]phenyl 1 -2-methoxy-N-methylacetamide, (3.027) N-(3 -
ethy1-3,5,5-
trimethylcyclohexyl)-3 -formamido-2-hydroxybenzamide, (3.028) (2E,3Z)-5- { [1 -
(4-chloro-2-
fluoropheny1)-1H-pyrazol-3 -yl] oxy} -2-(m ethoxyimino)-N,3 -dim ethylpent-3 -
enami de, and
(3.029) methyl {5- [3 -(2,4-dim ethylpheny1)-1H-pyrazol-1 -yl] -2-m ethylb
enzylIc arb am ate.
[0056] In some embodiments, inhibitors of the mitosis and cell division are
selected from the
group consisting of (4.001) carbendazim, (4.002) diethofencarb, (4.003)
ethaboxam, (4.004)
fluopicolide, (4.005) pencycuron, (4.006) thiabendazole, (4.007) thiophanate-
methyl, (4.008)
zoxami de, (4.009) 3 -chl oro-4-(2,6-di fluoropheny1)-6-m ethy1-5-phenylpyri
dazine, (4.010) 3 -
chl oro-5-(4-chl oropheny1)-4-(2,6-di fluoropheny1)-6-m ethylpyri dazine,
(4.011) 3 -chl oro-5-(6-
chl oropyri din-3 -y1)-6-m ethy1-4-(2,4,6-tri fluorophenyl)pyri dazine,
(4.012) 4-(2-brom o-4-
21
Date Recue/Date Received 2021-08-25
fluoropheny1)-N-(2,6-di fluoropheny1)-1,3 -dim ethy1-111-pyrazol-5-amine,
(4.013) 4-(2-brom o-4-
fluoropheny1)-N-(2-brom o-6-fluoropheny1)-1,3 -di m ethy1-111-pyrazol-5-amine,
(4.014) 4-(2 -
brom o-4-fluoroph eny1)-N-(2-bromoph eny1)-1,3 -dim ethy1-111-pyrazol-5-amine,
(4.015) 4-(2-
brom o-4-fluoroph eny1)-N-(2-chl oro-6-fluoropheny1)-1,3 -dim ethy1-111-
pyrazol-5-amine, (4.016)
4-(2-brom o-4-fluoropheny1)-N-(2-chl oropheny1)-1,3 -dim ethy1-111-pyrazol-5-
amine, (4.017) 4 -(2-
brom o-4-fluoroph eny1)-N-(2-fluoropheny1)-1,3 -dim ethyl -111-pyrazol -5-
amine, (4.018) 4-(2-
chl oro-4-fluoropheny1)-N-(2,6-di fluoropheny1)-1,3 -dim ethy1-111-pyrazol-5-
amine, (4.019) 4-(2-
chl oro-4-fluoropheny1)-N-(2-chl oro-6-fluoropheny1)-1,3 -dim ethy1-111-
pyrazol-5-amine, (4.020)
4-(2-chloro-4-fluoropheny1)-N-(2-chl oropheny1)-1,3 -dim ethy1-111-pyrazol-5-
ami ne, (4.021) 4-(2-
chl oro-4-fluoropheny1)-N-(2-fluoropheny1)-1,3 -dim ethy1-111-pyrazol -5-
amine, (4.022) 4-(4-
chl oropheny1)-5 -(2,6-di fluoropheny1)-3,6-dim ethy 1pyri dazin e,
(4.023) N-(2-bromo-6-
fluoropheny1)-4-(2-chloro-4-fluoropheny1)-1,3 -dim ethy1-111-pyrazol-5-am ine,
(4.024) N-(2-
brom opheny1)-4-(2-chl oro-4-fluoropheny1)-1,3 -dim ethy1-111-pyrazol-5-am
ine, and (4.025) N-(4-
chl oro-2,6-di fluoropheny1)-4-(2-chl oro-4-fluoropheny1)-1,3 -dim ethy1-111-
pyrazol-5-amine.
[0057] In some embodiments, compounds capable of having a multisite action are
selected from
the group consisting of (5.001) bordeaux mixture, (5.002) captafol, (5.003)
captan, (5.004)
chlorothalonil, (5.005) copper hydroxide, (5.006) copper naphthenate, (5.007)
copper oxide,
(5.008) copper oxychloride, (5.009) copper(2+) sulfate, (5.010) dithianon,
(5.011) dodine, (5.012)
folpet, (5.013) mancozeb, (5.014) maneb, (5.015) metiram, (5.016) metiram
zinc, (5.017) oxine-
copper, (5.018) propineb, (5.019) sulfur and sulfur preparations including
calcium polysulfide,
(5.020) thiram, (5.021) zineb, (5.022) ziram, and (5.023) 6-ethy1-5,7-dioxo-
6,7-dihydro-511-
pyrrolo[3',4':5,6][1,4]dithiino[2,3-c][1,2]thiazole-3-carbonitrile.
[0058] In some embodiments, compounds capable of inducing a host defense are
selected from
the group consisting of (6.001) acibenzolar-S-methyl, (6.002) isotianil,
(6.003) probenazole, and
(6.004) tiadinil.
[0059] In some embodiments, inhibitors of the amino acid and/or protein
biosynthesis are selected
from the group consisting of (7.001) cyprodinil, (7.002) kasugamycin, (7.003)
kasugamycin
hydrochloride hydrate, (7.004) oxytetracycline, (7.005) pyrimethanil, and
(7.006) 3-(5-fluoro-
3,3,4,4-tetram ethy1-3,4-dihydroi soquinolin-1 -yl)quinol one.
[0060] In some embodiments, inhibitor of the ATP production is selected from
the group
consisting of (8.001) silthiofam.
22
Date Recue/Date Received 2021-08-25
[0061] In some embodiments, inhibitors of the cell wall synthesis are selected
from the group
consisting of (9.001) benthiavalicarb, (9.002) dimethomorph, (9.003) flumorph,
(9.004)
iprovalicarb, (9.005) mandipropamid, (9.006) pyrimorph, (9.007) valifenalate,
(9.008) (2E)-3-(4-
tert-butylpheny1)-3 -(2-chl oropyri din-4-y1)-1 -(m orpholin-4-yl)prop-2-en-1 -
one, and (9.009) (2Z)-
3 -(4-tert-butylpheny1)-3 -(2-chl oropyri din-4-y1)-1 -(m orpholin-4-yl)prop-2
-en-1 -one.
[0062] In some embodiments, inhibitors of the lipid and membrane synthesis are
selected from
the group consisting of (10.001) propamocarb, (10.002) propamocarb
hydrochloride, and (10.003)
tolclofos-methyl.
[0063] In some embodiments, inhibitors of the melanine biosynthesis are
selected from the group
consisting of (11.001) tricyclazole, and (11.002) 2,2,2-trifluoroethyl {3-
methy1-1-[(4-
methylbenzoyl)amino]butan-2-y1 1 carbamate.
[0064] In some embodiments, inhibitors of the nucleic acid synthesis are
selected from the group
consisting of (12.001) benalaxyl, (12.002) benalaxyl-M (kiralaxyl), (12.003)
metalaxyl, and
(12.004) metalaxyl-M (mefenoxam).
[0065] In some embodiments, inhibitors of the signal transduction are selected
from the group
consisting of (13.001) fludioxonil, (13.002) iprodione, (13.003) procymidone,
(13.004)
proquinazid, (13.005) quinoxyfen, and (13.006) vinclozolin.
[0066] In some embodiments, compounds capable of acting as uncoupler are
selected from the
group consisting of (14.001) fluazinam, and (14.002) meptyldinocap.
[0067] In some embodiments, other fungicides are selected from the group
consisting of (15.001)
abscisic acid, (15.002) benthiazole, (15.003) bethoxazin, (15.004) capsimycin,
(15.005) carvone,
(15.006) chinomethionat, (15.007) cufraneb, (15.008) cyflufenamid, (15.009)
cymoxanil, (15.010)
cyprosulfamide, (15.011) flutianil, (15.012) fosetyl-aluminium, (15.013)
fosetyl-calcium, (15.014)
fosetyl-sodium, (15.015) methyl isothiocyanate, (15.016) metrafenone, (15.017)
mildiomycin,
(15.018) natamycin, (15.019) nickel dimethyldithiocarbamate, (15.020)
nitrothal-isopropyl,
(15.021) oxamocarb, (15.022) oxathiapiprolin, (15.023) oxyfenthiin, (15.024)
pentachlorophenol
and salts, (15.025) phosphorous acid and its salts, (15.026) propamocarb-
fosetylate, (15.027)
pyriofenone (chlazafenone), (15.028) tebufloquin, (15.029) tecloftalam,
(15.030) tolnifanide,
(15.031)
1-(4- {4- [(5R)-5-(2,6-di fluoropheny1)-4,5-dihydro-1,2-ox azol-3 -yl] -1,3 -
thi azol-2-
yl 1 piperidin-l-y1)-245-methy1-3-(trifluoromethyl)-111-pyrazol-1-yl]ethanone,
(15.032) 1-(4- {4 -
[(5 S)-5-(2,6-di fluoropheny1)-4,5-dihydro-1,2-oxazol-3 -yl] -1,3 -thi azol-2-
yllpiperi din-1 -y1)-2- [5-
23
Date Recue/Date Received 2021-08-25
methyl-3-(trifluoromethyl)-1H-pyrazol-1-yl]ethanone,
(15.033) 2-(6-benzylpyridin-2-
yl)quinazoline, (15.034) 2,6-dimethy1-1H,5H-[1,4]dithiino[2,3-c:5,6-
cldipyrrole-1,3,5,7(2H,6H)-
tetrone, (15.035)
2- [3,5-bi s(difluoromethyl)-1H-pyrazol-1-y1]-1-[4-(4- {5- [2-(prop-2-yn-1-
yloxy)pheny1]-4,5-dihydro-1,2-oxazol-3 -yll -1,3 -thiazol-2-yl)piperidin-l-yl]
ethanone, (15.036) 2-
[3,5-bi s(difluoromethyl)-1H-pyrazol-1-yl] -1- [4-(4- {5- [2-chloro-6-(prop-2-
yn-1-yloxy)phenyl] -
4,5-dihydro-1,2-oxazol-3-y1 1 -1,3-thiazol-2-yOpiperidin- 1 -yl]ethanone,
(15.037) 2-[3,5-
bis(difluoromethyl)-1H-pyrazol-1-y1]-1-[4-(4- {5- [2-fluoro-6-(prop-2-yn-l-
yloxy)phenyl] -4,5-
dihydro-1,2-oxazol-3 -yll -1,3 -thi azol-2-yl)piperidin-l-yl] ethanone,
(15.038) 2- [6-(3 -fluoro-4-
methoxypheny1)-5-methylpyridin-2-yl]quinazoline,
(15.039) 2- {(5R)-3-[2-(1-{ [3,5-
bis(difluoromethyl)-1H-pyrazol-1 -yl]acetyl 1 piperidin-4-y1)-1,3-thiazol-4-
y1]-4,5-dihydro-1,2-
oxazol-5-y1 1 -3 -chlorophenyl methanesulfonate,
(15.040) 2- {(5S)-3-[2-(1-{ [3,5-
bis(difluoromethyl)-1H-pyrazol-1-yl]acetyl 1 piperidin-4-y1)-1,3-thiazol-4-y1]-
4,5-dihydro-1,2-
oxazol-5-y1 1 -3 -chlorophenyl methanesulfonate, (15.041) 2- {247,8-difluoro-2-
methylquinolin-3-
y0oxy]-6-fluorophenyllpropan-2-ol, (15.042) 2- {2-fluoro-6- [(8-fluoro-2-
methylquinolin-3 -
yl)oxy]phenyl 1 propan-2-ol,
(15.043) 2434241- { [3,5-bis(difluoromethyl)-1H-pyrazol-1-
yl] acetyl 1 piperidin-4-y1)-1,3 -thiazol-4-y1]-4,5-dihydro-1,2-oxazol-5-y1 1 -
3 -chlorophenyl
methanesulfonate, (15.044)
2- {34241- { [3,5-bi s(difluoromethyl)-1H-pyrazol-1-
yl] acetyl 1 piperidin-4-y1)-1,3 -thiazol-4-y1]-4,5-dihydro-1,2-oxazol-5-y1 1
phenyl
methanesulfonate, (15.045) 2-phenylphenol and salts, (15.046) 3 -(4,4,5-
trifluoro-3,3 -dimethyl-
3,4-dihydroisoquinolin-1-yl)quinoline,
(15.047) 3 -(4,4-difluoro-3,3 -dimethy1-3,4-
dihydroisoquinolin- 1 -yl)quinoline, (15.048) 4-amino-5-fluoropyrimidin-2-ol
(tautomeric form: 4-
amino-5-fluoropyrimidin-2(1H)-one), (15.049) 4-oxo-4-[(2-
phenylethyl)amino]butanoic acid,
(15.050) 5-amino-1,3,4-thiadiazole-2-thiol, (15.051) 5-chloro-N'-phenyl-N'-
(prop-2-yn-1-
yl)thiophene-2-sulfonohydrazide, (15.052) 5-fluoro-2-[(4-
fluorobenzyl)oxy]pyrimidin-4-amine,
(15.053) 5-fluoro-2-[(4-methylbenzyl)oxy]pyrimidin-4-amine, (15.054) 9-fluoro-
2,2-dimethy1-5-
(quinolin-3 -y1)-2,3 -dihydro-1,4-benzoxazepine, (15.055) but-3 -yn-l-yl {64(
{[(Z)-(1-methy1-1H-
tetrazol-5-y1)(phenyOmethylene]aminol oxy)methyl]pyridin-2-y1 1 carbamate,
(15.056) ethyl (2Z)-
3 -amino-2-cyano-3 -phenylacrylate, (15.057) phenazine- 1-carboxylic acid,
(15.058) propyl 3,4,5-
trihydroxybenzoate, (15.059) quinolin-8-ol, (15.060) quinolin-8-ol sulfate
(2:1), (15.061) tert-
butyl
{64( { [(1-methyl-1H-tetrazol-5-y1)(phenyOmethylene] amino} oxy)methyl]pyridin-
2-
24
Date Recue/Date Received 2021-08-25
yl 1 carbamate, and (15.062) 5-fluoro-4-imino-3 -methyl-1- [(4-m
ethylphenyl)sulfony1]-3,4-
dihydropyrimi din-2(1H)-one.
[0068] In certain embodiments, the active compound or combination of active
compounds is
selected from:
(1.001) cyproconazole, (1.002) difenoconazole, (1.003) epoxiconazole, (1.004)
fenhexamid, (1.010) imazalil, (1.012) ipconazole, (1.013) metconazole, (1.017)
propiconazole, (1.018) prothioconazole, (1.020) spiroxamine, (1.021)
tebuconazole,
(1.026) (1R,2S,5S)-5-(4-chlorobenzy1)-2-(chloromethyl)-2-methyl-1-(1H-1,2,4-
triazol-1-
ylmethyl)cyclopentanol, (1.027) (1 S,2R,5R)-5-(4-chl orob enzy1)-2-(chl orom
ethyl)-2-
methy1-1-(1H-1,2,4-tri azol-1-ylmethyl)cyclopentanol,
(1.059) 5-(4-chlorobenzy1)-2-
(chloromethyl)-2-methyl-1-(1H-1,2,4-triazol-1-ylmethyl)cyclopentanol,
(1.081)
Mefentrifluconazole, and (1.082) Ipfentrifluconazole,
(2.001) benzovindiflupyr, (2.002) bixafen, (2.003) boscalid, (2.005)
fluopyram, (2.007)
fluxapyroxad, (2.009) Isofetamid, (2.010) isopyrazam (anti-epimeric enantiomer
1R,4S,9S), (2.011) isopyrazam (anti-epimeric enantiomer 1S,4R,9R), (2.012)
isopyrazam
(anti-epimeric racemate 1RS,4SR,9SR), (2.013) isopyrazam (mixture of syn-
epimeric
racemate 1RS,4SR,9RS and anti-epimeric racemate 1RS,4SR,9SR), (2.014)
isopyrazam
(syn-epimeric enantiomer 1R,4S,9R), (2.015) isopyrazam (syn-epimeric
enantiomer
1 S,4R,9 S), (2.016) isopyrazam (syn-epimeric racemate 1RS,4SR,9RS), (2.017)
penflufen,
(2.018) penthiopyrad, (2.019) pydiflumetofen, (2.021) sedaxane, (2.027) 3 -
(di fluoromethyl)-1 -m ethyl-N-(1,1,3 -trim ethyl-2,3 -dihydro-1H-inden-4-y1)-
1H-pyrazol e-
4-c arb oxami de, (2.030) 3 -(di fluorom ethyl)-N-(7-fluoro-1,1,3 -trim ethyl-
2,3 -dihydro-1H-
inden-4-y1)-1 -m ethy1-1H-pyrazol e-4-c arb ox ami de,
(2.038) N-(5-chloro-2-
isopropylbenzy1)-N-cyclopropy1-3-(difluoromethyl)-5-fluoro-l-methyl-1H-
pyrazole-4-
carboxamide,
(3.003) azoxystrobin, (3.007) dimoxystrobin, (3.012) fluoxastrobin, (3.013)
kresoxim-
methyl, (3.016) picoxystrobin, (3.017) pyraclostrobin, (3.020)
trifloxystrobin, (3.025)
(3 S,6 S,7R,8R)-8-b enzy1-3 - [( {3 - [(i s obutyryl oxy)m ethoxy] -4-m
ethoxypyri di n-2-
yl 1 carbonyl)amino]-6-methyl-4,9-dioxo-1,5-dioxonan-7-y1 2-methylpropanoate,
(3.026)
2- {2- [(2,5 -dim ethylphenoxy)m ethyl]phenyll -2-m ethoxy-N-m ethyl ac etami
de,
Date Recue/Date Received 2021-08-25
(4.005) pencycuron, (4.007) thiophanate-methyl, (4.012) 4-(2-bromo-4-
fluoropheny1)-N-
(2,6-di fluoropheny1)-1,3 -dim ethy1-1H-pyrazol-5-amine,
(4.015) 4-(2-brom o-4-
fluoropheny1)-N-(2-chl oro-6-fluoropheny1)-1,3 -dim ethy1-1H-pyrazol-5-am ine,
(4.025) N-
(4-chl oro-2,6-di fluoroph eny1)-4-(2-chl oro-4-fluoropheny1)-1,3 -dim ethy1-
1H-pyrazol-5-
amine,
(5.003) captan, (5.004) chlorothalonil, (5.011) dodine, (5.012) folpet,
(5.013) mancozeb,
(5.015) metiram, (5.018) propineb,
(6.002) isotianil,
(7.001) cyprodinil, (7.005) pyrimethanil,
(12.003) metalaxyl, (12.004) metalaxyl-M (mefenoxam),
(13.001) fludioxonil, (13.002) iprodione, (13.004) proquinazid, (13.005)
quinoxyfen,
(14.001) fluazinam, (14.002) meptyldinocap,
(15.008) cyflufenamid, (15.010) cyprosulfamide, (15.011) flutianil, (15.012)
fosetyl-
aluminium, (15.016) metrafenone, (15.027) pyriofenone (chlazafenone), and
(15.047) 3-
(4,4-difluoro-3,3 -dimethy1-3,4-dihydroi soquinolin-1 -yl)quinolone, (15.048)
4-amino-5-
fluoropyrimidin-2-ol (tautomeric form: 4-amino-5-fluoropyrimidin-2(1H)-one),
(15.052)
5-fluoro-2-[(4-fluorobenzyl)oxy]pyrimidin-4-amine, (15.053)
5-fluoro-2-[(4-
methylbenzyl)oxy]pyrimidin-4-amine, (15.062)
5-fluoro-4-imino-3 -methyl-1- [(4-
methylphenyl)sulfony1]-3,4-dihydropyrimidin-2(1H)-one.
[0069] In certain embodiments, the active compound or combination of active
compounds is
selected from:
(1.002) difenoconazole, (1.010) imazalil, (1.012) ipconazole, (1.018)
prothioconazole,
(1.020) spiroxamine, (1.021) tebuconazole, (1.026) (1R,2S,5S)-5-(4-
chlorobenzy1)-2-
(chloromethyl)-2-methyl-1-(1H-1,2,4-triazol-1-ylmethyl)cyclopentanol,
(1.027)
(1 S,2R,5R)-5-(4-chlorob enzy1)-2-(chlorom ethyl)-2-m ethy1-1 -(1H-1,2,4-tri
azol-1 -
ylm ethyl)cyclopentanol, (1.059) 5-(4-chlorobenzy1)-2-(chloromethyl)-2-methyl-
1-(1H-
1,2,4-triazol-1-ylmethyl)cyclopentanol, (1.081) Mefentrifluconazole, and
(1.082)
Ipfentrifluconazole,
(2.001) benzovindiflupyr, (2.002) bixafen, (2.005) fluopyram, (2.007)
fluxapyroxad,
(2.017) penflufen, (2.018) penthiopyrad, (2.019) pydiflumetofen, (2.021)
sedaxane,
(2.027) 3 -(di fluorom ethyl)-1 -methyl-N-(1,1,3 -trim ethyl-2,3 -dihydro-1H-
inden-4-y1)-1H-
26
Date Recue/Date Received 2021-08-25
pyrazol e-4-c arb oxami de, (2.030) 3 -(difluorom ethyl)-N-(7-fluoro-1,1,3 -
trim ethyl-2,3 -
dihydro-1H-inden-4-y1)-1 -m ethy1-1H-pyrazol e-4-c arb oxami de, (2.038) N-(5-
chloro-2-
isopropylbenzy1)-N-cyclopropy1-3-(difluoromethyl)-5-fluoro-l-methyl-1H-
pyrazole-4-
carboxamide,
(3.003) azoxystrobin, (3.012) fluoxastrobin, (3.016) picoxystrobin, (3.017)
pyraclostrobin,
(3.020) trifloxystrobin, (3.025) (3S,6S,7R,8R)-8-benzy1-3-[({3-
[(isobutyryloxy)methoxy]-
4-methoxypyridin-2-yll carbonyl)amino]-6-methyl-4,9-dioxo-1,5-dioxonan-7-y1
2-
methylpropanoate, (3.026) 2- {2-[(2,5-dimethylphenoxy)methyl]phenyll -2-
methoxy-N-
methylacetamide,
(4.005) pencycuron, (4.007) thiophanate-methyl, (4.012) 4-(2-bromo-4-
fluoropheny1)-N-
(2,6-di fluoropheny1)-1,3 -dim ethy1-1H-pyrazol-5-amine, (4.015)
4-(2-brom o-4-
fluoropheny1)-N-(2-chl oro-6-fluoropheny1)-1,3 -dim ethy1-1H-pyrazol-5-am ine,
(4.025) N-
(4-chl oro-2,6-di fluoroph eny1)-4-(2-chl oro-4-fluoropheny1)-1,3 -dim ethy1-
1H-pyrazol-5-
amine,
(5.004) chlorothalonil, (5.011) dodine, (5.012) folpet, (5.013) mancozeb,
(5.018) propineb,
(6.002) isotianil,
(7.005) pyrimethanil,
(12.003) metalaxyl, (12.004) metalaxyl-M (mefenoxam),
(13.001) fludioxonil, (13.004) proquinazid,
(14.001) fluazinam, (14.002) meptyldinocap,
(15.008) cyflufenamid, (15.027) pyriofenone (chlazafenone), (15.047) 3-(4,4-
difluoro-
3,3 -dimethy1-3,4-dihydroi soquinolin-l-yl)quinolone, (15.048)
4-amino-5-
fluoropyrimidin-2-ol (tautomeric form: 4-amino-5-fluoropyrimidin-2(1H)-one),
(15.052)
5-fluoro-2-[(4-fluorobenzyl)oxy]pyrimidin-4-amine, (15.053)
5-fluoro-2-[(4-
methylbenzyl)oxy]pyrimidin-4-amine, (15.062)
5-fluoro-4-imino-3-methy1-1-[(4-
methylphenyl)sulfonyl]-3,4-dihydropyrimidin-2(1H)-one.
[0070] In certain embodiments, the active compound or combination of active
compounds is
selected from:
(1.012) ipconazole, (1.018) prothioconazole, (1.020) spiroxamine, (1.021)
tebuconazole,
(2.002) bixafen, (2.005) fluopyram, (2.017) penflufen, (2.027) 3-
(difluoromethyl)-1-
methyl-N-(1,1,3 -trimethy1-2,3 -dihydro-1H-inden-4-y1)-1H-pyrazole-4-c arb
oxamide,
27
Date Recue/Date Received 2021-08-25
(2.038)
N-(5-chloro-2-isopropylbenzy1)-N-cyclopropy1-3-(difluoromethyl)-5-fluoro-1-
methyl-1H-pyrazole-4-carboxamide,
(3.020) trifloxystrobin, (3.025) (3S,6S,7R,8R)-8-benzy1-3-[({3-
[(isobutyryloxy)methoxy]-
4-methoxypyridin-2-yll carbonyl)amino]-6-methyl-4,9-dioxo-1,5-dioxonan-7-y1
2-
methylpropanoate,
(4.005) pencycuron,
(5.004) chlorothalonil, (5.013) mancozeb, (5.018) propineb,
(12.003) metalaxyl, (12.004) metalaxyl-M (mefenoxam),
(13.001) fludioxonil, (13.004) proquinazid,
(15.008) cyflufenamid, and (15.047)
3-(4,4-difluoro-3,3-dimethy1-3,4-
dihydroisoquinolin-1-yl)quinoline.
[0071] In certain embodiments, the active compound or combination of active
compounds are
selected from the group (G1) consisting of the following mixtures: (I.01) +
(1.001), (I.01) +
(1.002), (I.01)+ (1.003), (I.01)+ (1.004), (I.01)+ (1.005), (I.01)+ (1.006),
(I.01)+ (1.007), (I.01)
+ (1.008), (I.01) + (1.009), (I.01) + (1.010), (I.01) + (1.011), (I.01) +
(1.012), (I.01) + (1.013),
(I.01) + (1.014), (I.01) + (1.015), (I.01) + (1.016), (I.01) + (1.017), (I.01)
+ (1.018), (I.01) +
(1.019), (I.01)+ (1.020), (I.01)+ (1.021), (I.01)+ (1.022), (I.01)+ (1.023),
(I.01)+ (1.024), (I.01)
+ (1.025), (I.01) + (1.026), (I.01) + (1.027), (I.01) + (1.028), (I.01) +
(1.029), (I.01) + (1.030),
(I.01) + (1.031), (I.01) + (1.032), (I.01) + (1.033), (I.01) + (1.034), (I.01)
+ (1.035), (I.01) +
(1.036), (I.01)+ (1.037), (I.01)+ (1.038), (I.01)+ (1.039), (I.01)+ (1.040),
(I.01)+ (1.041), (I.01)
+ (1.042), (I.01) + (1.043), (I.01) + (1.044), (I.01) + (1.045), (I.01) +
(1.046), (I.01) + (1.047),
(I.01) + (1.048), (I.01) + (1.049), (I.01) + (1.050), (I.01) + (1.051), (I.01)
+ (1.052), (I.01) +
(1.053), (I.01)+ (1.054), (I.01)+ (1.055), (I.01)+ (1.056), (I.01)+ (1.057),
(I.01)+ (1.058), (I.01)
+ (1.059), (I.01) + (1.060), (I.01) + (1.061), (I.01) + (1.062), (I.01) +
(1.063), (I.01) + (1.064),
(I.01) + (1.065), (I.01) + (1.066), (I.01) + (1.067), (I.01) + (1.068), (I.01)
+ (1.069), (I.01) +
(1.070), (I.01)+ (1.071), (I.01)+ (1.072), (I.01)+ (1.073), (I.01)+ (1.074),
(I.01)+ (1.075), (I.01)
+ (1.076), (I.01) + (1.077), (I.01) + (1.078), (I.01) + (1.079), (I.01) +
(1.080), (I.01) + (1.081),
(I.01) + (1.082), (I.01) + (2.001), (I.01) + (2.002), (I.01) + (2.003), (I.01)
+ (2.004), (I.01) +
(2.005), (I.01)+ (2.006), (I.01)+ (2.007), (I.01)+ (2.008), (I.01)+ (2.009),
(I.01)+ (2.010), (I.01)
+ (2.011), (I.01) + (2.012), (I.01) + (2.013), (I.01) + (2.014), (I.01) +
(2.015), (I.01) + (2.016),
(I.01) + (2.017), (I.01) + (2.018), (I.01) + (2.019), (I.01) + (2.020), (I.01)
+ (2.021), (I.01) +
28
Date Recue/Date Received 2021-08-25
(2.022), (I.01) + (2.023), (I.01) + (2.024), (I.01) + (2.025), (I.01) +
(2.026), (I.01) + (2.027), (I.01)
+ (2.028), (I.01) + (2.029), (I.01) + (2.030), (I.01) + (2.031), (I.01) +
(2.032), (I.01) + (2.033),
(I.01) + (2.034), (I.01) + (2.035), (I.01) + (2.036), (I.01) + (2.037), (I.01)
+ (2.038), (I.01) +
(2.039), (I.01) + (2.040), (I.01) + (2.041), (I.01) + (2.042), (I.01) +
(2.043), (I.01) + (2.044), (I.01)
+ (2.045), (I.01) + (2.046), (I.01) + (2.047), (I.01) + (2.048), (I.01) +
(2.049), (I.01) + (2.050),
(I.01) + (2.051), (I.01) + (2.052), (I.01) + (2.053), (I.01) + (2.054), (I.01)
+ (2.055), (I.01) +
(2.056), (I.01) + (3.001), (I.01) + (3.002), (I.01) + (3.003), (I.01) +
(3.004), (I.01) + (3.005), (I.01)
+ (3.006), (I.01) + (3.007), (I.01) + (3.008), (I.01) + (3.009), (I.01) +
(3.010), (I.01) + (3.011),
(I.01) + (3.012), (I.01) + (3.013), (I.01) + (3.014), (I.01) + (3.015), (I.01)
+ (3.016), (I.01) +
(3.017), (I.01) + (3.018), (I.01) + (3.019), (I.01) + (3.020), (I.01) +
(3.021), (I.01) + (3.022), (I.01)
+ (3.023), (I.01) + (3.024), (I.01) + (3.025), (I.01) + (3.026), (I.01) +
(3.027), (I.01) + (3.028),
(I.01) + (3.029), (I.01) + (4.001), (I.01) + (4.002), (I.01) + (4.003), (I.01)
+ (4.004), (I.01) +
(4.005), (I.01) + (4.006), (I.01) + (4.007), (I.01) + (4.008), (I.01) +
(4.009), (I.01) + (4.010), (I.01)
+ (4.011), (I.01) + (4.012), (I.01) + (4.013), (I.01) + (4.014), (I.01) +
(4.015), (I.01) + (4.016),
(I.01) + (4.017), (I.01) + (4.018), (I.01) + (4.019), (I.01) + (4.020), (I.01)
+ (4.021), (I.01) +
(4.022), (I.01) + (4.023), (I.01) + (4.024), (I.01) + (4.025), (I.01) +
(5.001), (I.01) + (5.002), (I.01)
+ (5.003), (I.01) + (5.004), (I.01) + (5.005), (I.01) + (5.006), (I.01) +
(5.007), (I.01) + (5.008),
(I.01) + (5.009), (I.01) + (5.010), (I.01) + (5.011), (I.01) + (5.012), (I.01)
+ (5.013), (I.01) +
(5.014), (I.01) + (5.015), (I.01) + (5.016), (I.01) + (5.017), (I.01) +
(5.018), (I.01) + (5.019), (I.01)
+ (5.020), (I.01) + (5.021), (I.01) + (5.022), (I.01) + (5.023), (I.01) +
(6.001), (I.01) + (6.002),
(I.01) + (6.003), (I.01) + (6.004), (I.01) + (7.001), (I.01) + (7.002), (I.01)
+ (7.003), (I.01) +
(7.004), (I.01) + (7.005), (I.01) + (7.006), (I.01) + (8.001), (I.01) +
(9.001), (I.01) + (9.002), (I.01)
+ (9.003), (I.01) + (9.004), (I.01) + (9.005), (I.01) + (9.006), (I.01) +
(9.007), (I.01) + (9.008),
(I.01) + (9.009), (I.01) + (10.001), (I.01) + (10.002), (I.01) + (10.003),
(I.01) + (11.001), (I.01) +
(11.002), (I.01)+ (12.001), (I.01) + (12.002), (I.01)+ (12.003), (I.01)+
(12.004), (I.01)+ (13.001),
(I.01) + (13.002), (I.01) + (13.003), (I.01) + (13.004), (I.01) + (13.005),
(I.01) + (13.006), (I.01) +
(14.001), (I.01)+ (14.002), (I.01) + (15.001), (I.01)+ (15.002), (I.01)+
(15.003), (I.01)+ (15.004),
(I.01) + (15.005), (I.01) + (15.006), (I.01) + (15.007), (I.01) + (15.008),
(I.01) + (15.009), (I.01) +
(15.010), (I.01)+ (15.011), (I.01) + (15.012), (I.01)+ (15.013), (I.01)+
(15.014), (I.01)+ (15.015),
(I.01) + (15.016), (I.01) + (15.017), (I.01) + (15.018), (I.01) + (15.019),
(I.01) + (15.020), (I.01) +
(15.021), (I.01)+ (15.022), (I.01) + (15.023), (I.01)+ (15.024), (I.01)+
(15.025), (I.01)+ (15.026),
29
Date Recue/Date Received 2021-08-25
(I.01) + (15.027), (I.01) + (15.028), (I.01) + (15.029), (I.01) + (15.030),
(I.01) + (15.031), (I.01) +
(15.032), (I.01)+ (15.033), (I.01)+ (15.034), (I.01)+ (15.035), (I.01)+
(15.036), (I.01)+ (15.037),
(I.01) + (15.038), (I.01) + (15.039), (I.01) + (15.040), (I.01) + (15.041),
(I.01) + (15.042), (I.01) +
(15.043), (I.01)+ (15.044), (I.01)+ (15.045), (I.01)+ (15.046), (I.01)+
(15.047), (I.01)+ (15.048),
(I.01) + (15.049), (I.01) + (15.050), (I.01) + (15.051), (I.01) + (15.052),
(I.01) + (15.053), (I.01) +
(15.054), (I.01)+ (15.055), (I.01)+ (15.056), (I.01)+ (15.057), (I.01)+
(15.058), (I.01)+ (15.059),
(I.01) + (15.060), (I.01) + (15.061), and (I.01) + (15.062).
[0072] In certain embodiments, the active compound or combination of active
compounds are
selected from the group (G2) consisting of the following mixtures: (1.59) +
(1.001), (1.59) +
(1.002), (1.59) + (1.003), (1.59) + (1.004), (1.59) + (1.005), (1.59) +
(1.006), (1.59) + (1.007), (1.59)
+ (1.008), (1.59) + (1.009), (1.59) + (1.010), (1.59) + (1.011), (1.59) +
(1.012), (1.59) + (1.013),
(1.59) + (1.014), (1.59) + (1.015), (1.59) + (1.016), (1.59) + (1.017), (1.59)
+ (1.018), (1.59) +
(1.019), (1.59) + (1.020), (1.59) + (1.021), (1.59) + (1.022), (1.59) +
(1.023), (1.59) + (1.024), (1.59)
+ (1.025), (1.59) + (1.026), (1.59) + (1.027), (1.59) + (1.028), (1.59) +
(1.029), (1.59) + (1.030),
(1.59) + (1.031), (1.59) + (1.032), (1.59) + (1.033), (1.59) + (1.034), (1.59)
+ (1.035), (1.59) +
(1.036), (1.59) + (1.037), (1.59) + (1.038), (1.59) + (1.039), (1.59) +
(1.040), (1.59) + (1.041), (1.59)
+ (1.042), (1.59) + (1.043), (1.59) + (1.044), (1.59) + (1.045), (1.59) +
(1.046), (1.59) + (1.047),
(1.59) + (1.048), (1.59) + (1.049), (1.59) + (1.050), (1.59) + (1.051), (1.59)
+ (1.052), (1.59) +
(1.053), (1.59) + (1.054), (1.59) + (1.055), (1.59) + (1.056), (1.59) +
(1.057), (1.59) + (1.058), (1.59)
+ (1.059), (1.59) + (1.060), (1.59) + (1.061), (1.59) + (1.062), (1.59) +
(1.063), (1.59) + (1.064),
(1.59) + (1.065), (1.59) + (1.066), (1.59) + (1.067), (1.59) + (1.068), (1.59)
+ (1.069), (1.59) +
(1.070), (1.59) + (1.071), (1.59) + (1.072), (1.59) + (1.073), (1.59) +
(1.074), (1.59) + (1.075), (1.59)
+ (1.076), (1.59) + (1.077), (1.59) + (1.078), (1.59) + (1.079), (1.59) +
(1.080), (1.59) + (1.081),
(1.59) + (1.082), (1.59) + (2.001), (1.59) + (2.002), (1.59) + (2.003), (1.59)
+ (2.004), (1.59) +
(2.005), (1.59) + (2.006), (1.59) + (2.007), (1.59) + (2.008), (1.59) +
(2.009), (1.59) + (2.010), (1.59)
+ (2.011), (1.59) + (2.012), (1.59) + (2.013), (1.59) + (2.014), (1.59) +
(2.015), (1.59) + (2.016),
(1.59) + (2.017), (1.59) + (2.018), (1.59) + (2.019), (1.59) + (2.020), (1.59)
+ (2.021), (1.59) +
(2.022), (1.59) + (2.023), (1.59) + (2.024), (1.59) + (2.025), (1.59) +
(2.026), (1.59) + (2.027), (1.59)
+ (2.028), (1.59) + (2.029), (1.59) + (2.030), (1.59) + (2.031), (1.59) +
(2.032), (1.59) + (2.033),
(1.59) + (2.034), (1.59) + (2.035), (1.59) + (2.036), (1.59) + (2.037), (1.59)
+ (2.038), (1.59) +
(2.039), (1.59) + (2.040), (1.59) + (2.041), (1.59) + (2.042), (1.59) +
(2.043), (1.59) + (2.044), (1.59)
Date Recue/Date Received 2021-08-25
+ (2.045), (1.59) + (2.046), (1.59) + (2.047), (1.59) + (2.048), (1.59) +
(2.049), (1.59) + (2.050),
(1.59) + (2.051), (1.59) + (2.052), (1.59) + (2.053), (1.59) + (2.054), (1.59)
+ (2.055), (1.59) +
(2.056), (1.59) + (3.001), (1.59) + (3.002), (1.59) + (3.003), (1.59) +
(3.004), (1.59) + (3.005), (1.59)
+ (3.006), (1.59) + (3.007), (1.59) + (3.008), (1.59) + (3.009), (1.59) +
(3.010), (1.59) + (3.011),
(1.59) + (3.012), (1.59) + (3.013), (1.59) + (3.014), (1.59) + (3.015), (1.59)
+ (3.016), (1.59) +
(3.017), (1.59) + (3.018), (1.59) + (3.019), (1.59) + (3.020), (1.59) +
(3.021), (1.59) + (3.022), (1.59)
+ (3.023), (1.59) + (3.024), (1.59) + (3.025), (1.59) + (3.026), (1.59) +
(3.027), (1.59) + (3.028),
(1.59) + (3.029), (1.59) + (4.001), (1.59) + (4.002), (1.59) + (4.003), (1.59)
+ (4.004), (1.59) +
(4.005), (1.59) + (4.006), (1.59) + (4.007), (1.59) + (4.008), (1.59) +
(4.009), (1.59) + (4.010), (1.59)
+ (4.011), (1.59) + (4.012), (1.59) + (4.013), (1.59) + (4.014), (1.59) +
(4.015), (1.59) + (4.016),
(1.59) + (4.017), (1.59) + (4.018), (1.59) + (4.019), (1.59) + (4.020), (1.59)
+ (4.021), (1.59) +
(4.022), (1.59) + (4.023), (1.59) + (4.024), (1.59) + (4.025), (1.59) +
(5.001), (1.59) + (5.002), (1.59)
+ (5.003), (1.59) + (5.004), (1.59) + (5.005), (1.59) + (5.006), (1.59) +
(5.007), (1.59) + (5.008),
(1.59) + (5.009), (1.59) + (5.010), (1.59) + (5.011), (1.59) + (5.012), (1.59)
+ (5.013), (1.59) +
(5.014), (1.59) + (5.015), (1.59) + (5.016), (1.59) + (5.017), (1.59) +
(5.018), (1.59) + (5.019), (1.59)
+ (5.020), (1.59) + (5.021), (1.59) + (5.022), (1.59) + (5.023), (1.59) +
(6.001), (1.59) + (6.002),
(1.59) + (6.003), (1.59) + (6.004), (1.59) + (7.001), (1.59) + (7.002), (1.59)
+ (7.003), (1.59) +
(7.004), (1.59) + (7.005), (1.59) + (7.006), (1.59) + (8.001), (1.59) +
(9.001), (1.59) + (9.002), (1.59)
+ (9.003), (1.59) + (9.004), (1.59) + (9.005), (1.59) + (9.006), (1.59) +
(9.007), (1.59) + (9.008),
(1.59) + (9.009), (1.59) + (10.001), (1.59) + (10.002), (1.59) + (10.003),
(1.59) + (11.001), (1.59) +
(11.002), (1.59) + (12.001), (1.59) + (12.002), (1.59) + (12.003), (1.59) +
(12.004), (1.59) + (13.001),
(1.59) + (13.002), (1.59) + (13.003), (1.59) + (13.004), (1.59) + (13.005),
(1.59) + (13.006), (1.59) +
(14.001), (1.59) + (14.002), (1.59) + (15.001), (1.59) + (15.002), (1.59) +
(15.003), (1.59) + (15.004),
(1.59) + (15.005), (1.59) + (15.006), (1.59) + (15.007), (1.59) + (15.008),
(1.59) + (15.009), (1.59) +
(15.010), (1.59) + (15.011), (1.59) + (15.012), (1.59) + (15.013), (1.59) +
(15.014), (1.59) + (15.015),
(1.59) + (15.016), (1.59) + (15.017), (1.59) + (15.018), (1.59) + (15.019),
(1.59) + (15.020), (1.59) +
(15.021), (1.59) + (15.022), (1.59) + (15.023), (1.59) + (15.024), (1.59) +
(15.025), (1.59) + (15.026),
(1.59) + (15.027), (1.59) + (15.028), (1.59) + (15.029), (1.59) + (15.030),
(1.59) + (15.031), (1.59) +
(15.032), (1.59) + (15.033), (1.59) + (15.034), (1.59) + (15.035), (1.59) +
(15.036), (1.59) + (15.037),
(1.59) + (15.038), (1.59) + (15.039), (1.59) + (15.040), (1.59) + (15.041),
(1.59) + (15.042), (1.59) +
(15.043), (1.59) + (15.044), (1.59) + (15.045), (1.59) + (15.046), (1.59) +
(15.047), (1.59) + (15.048),
31
Date Recue/Date Received 2021-08-25
(1.59) + (15.049), (1.59) + (15.050), (1.59) + (15.051), (1.59) + (15.052),
(1.59) + (15.053), (1.59) +
(15.054), (1.59) + (15.055), (1.59) + (15.056), (1.59) + (15.057), (1.59) +
(15.058), (1.59) + (15.059),
(1.59) + (15.060), (1.59) + (15.061), and (1.59) + (15.062).
[0073] In certain embodiments, the active compound or combination of active
compounds are
selected from the group (G3) consisting of the following mixtures: (I.81) +
(1.001), (I.81) +
(1.002), (I.81)+ (1.003), (I.81)+ (1.004), (I.81) + (1.005), (I.81)+ (1.006),
(I.81)+ (1.007), (I.81)
+ (1.008), (I.81) + (1.009), (I.81) + (1.010), (I.81) + (1.011), (I.81) +
(1.012), (I.81) + (1.013),
(I.81) + (1.014), (I.81) + (1.015), (I.81) + (1.016), (I.81) + (1.017), (I.81)
+ (1.018), (I.81) +
(1.019), (I.81)+ (1.020), (I.81)+ (1.021), (I.81) + (1.022), (I.81)+ (1.023),
(I.81)+ (1.024), (I.81)
+ (1.025), (I.81) + (1.026), (I.81) + (1.027), (I.81) + (1.028), (I.81) +
(1.029), (I.81) + (1.030),
(I.81) + (1.031), (I.81) + (1.032), (I.81) + (1.033), (I.81) + (1.034), (I.81)
+ (1.035), (I.81) +
(1.036), (I.81)+ (1.037), (I.81)+ (1.038), (I.81) + (1.039), (I.81)+ (1.040),
(I.81)+ (1.041), (I.81)
+ (1.042), (I.81) + (1.043), (I.81) + (1.044), (I.81) + (1.045), (I.81) +
(1.046), (I.81) + (1.047),
(I.81) + (1.048), (I.81) + (1.049), (I.81) + (1.050), (I.81) + (1.051), (I.81)
+ (1.052), (I.81) +
(1.053), (I.81)+ (1.054), (I.81)+ (1.055), (I.81) + (1.056), (I.81)+ (1.057),
(I.81)+ (1.058), (I.81)
+ (1.059), (I.81) + (1.060), (I.81) + (1.061), (I.81) + (1.062), (I.81) +
(1.063), (I.81) + (1.064),
(I.81) + (1.065), (I.81) + (1.066), (I.81) + (1.067), (I.81) + (1.068), (I.81)
+ (1.069), (I.81) +
(1.070), (I.81)+ (1.071), (I.81)+ (1.072), (I.81) + (1.073), (I.81)+ (1.074),
(I.81)+ (1.075), (I.81)
+ (1.076), (I.81) + (1.077), (I.81) + (1.078), (I.81) + (1.079), (I.81) +
(1.080), (I.81) + (1.081),
(I.81) + (1.082), (I.81) + (2.001), (I.81) + (2.002), (I.81) + (2.003), (I.81)
+ (2.004), (I.81) +
(2.005), (I.81)+ (2.006), (I.81)+ (2.007), (I.81) + (2.008), (I.81)+ (2.009),
(I.81)+ (2.010), (I.81)
+ (2.011), (I.81) + (2.012), (I.81) + (2.013), (I.81) + (2.014), (I.81) +
(2.015), (I.81) + (2.016),
(I.81) + (2.017), (I.81) + (2.018), (I.81) + (2.019), (I.81) + (2.020), (I.81)
+ (2.021), (I.81) +
(2.022), (I.81)+ (2.023), (I.81)+ (2.024), (I.81) + (2.025), (I.81)+ (2.026),
(I.81)+ (2.027), (I.81)
+ (2.028), (I.81) + (2.029), (I.81) + (2.030), (I.81) + (2.031), (I.81) +
(2.032), (I.81) + (2.033),
(I.81) + (2.034), (I.81) + (2.035), (I.81) + (2.036), (I.81) + (2.037), (I.81)
+ (2.038), (I.81) +
(2.039), (I.81)+ (2.040), (I.81)+ (2.041), (I.81) + (2.042), (I.81)+ (2.043),
(I.81)+ (2.044), (I.81)
+ (2.045), (I.81) + (2.046), (I.81) + (2.047), (I.81) + (2.048), (I.81) +
(2.049), (I.81) + (2.050),
(I.81) + (2.051), (I.81) + (2.052), (I.81) + (2.053), (I.81) + (2.054), (I.81)
+ (2.055), (I.81) +
(2.056), (I.81)+ (3.001), (I.81)+ (3.002), (I.81) + (3.003), (I.81)+ (3.004),
(I.81)+ (3.005), (I.81)
+ (3.006), (I.81) + (3.007), (I.81) + (3.008), (I.81) + (3.009), (I.81) +
(3.010), (I.81) + (3.011),
32
Date Recue/Date Received 2021-08-25
(I.81) + (3.012), (I.81) + (3.013), (I.81) + (3.014), (I.81) + (3.015), (I.81)
+ (3.016), (I.81) +
(3.017), (I.81) + (3.018), (I.81) + (3.019), (I.81) + (3.020), (I.81) +
(3.021), (I.81) + (3.022), (I.81)
+ (3.023), (I.81) + (3.024), (I.81) + (3.025), (I.81) + (3.026), (I.81) +
(3.027), (I.81) + (3.028),
(I.81) + (3.029), (I.81) + (4.001), (I.81) + (4.002), (I.81) + (4.003), (I.81)
+ (4.004), (I.81) +
(4.005), (I.81) + (4.006), (I.81) + (4.007), (I.81) + (4.008), (I.81) +
(4.009), (I.81) + (4.010), (I.81)
+ (4.011), (I.81) + (4.012), (I.81) + (4.013), (I.81) + (4.014), (I.81) +
(4.015), (I.81) + (4.016),
(I.81) + (4.017), (I.81) + (4.018), (I.81) + (4.019), (I.81) + (4.020), (I.81)
+ (4.021), (I.81) +
(4.022), (I.81) + (4.023), (I.81) + (4.024), (I.81) + (4.025), (I.81) +
(5.001), (I.81) + (5.002), (I.81)
+ (5.003), (I.81) + (5.004), (I.81) + (5.005), (I.81) + (5.006), (I.81) +
(5.007), (I.81) + (5.008),
(I.81) + (5.009), (I.81) + (5.010), (I.81) + (5.011), (I.81) + (5.012), (I.81)
+ (5.013), (I.81) +
(5.014), (I.81) + (5.015), (I.81) + (5.016), (I.81) + (5.017), (I.81) +
(5.018), (I.81) + (5.019), (I.81)
+ (5.020), (I.81) + (5.021), (I.81) + (5.022), (I.81) + (5.023), (I.81) +
(6.001), (I.81) + (6.002),
(I.81) + (6.003), (I.81) + (6.004), (I.81) + (7.001), (I.81) + (7.002), (I.81)
+ (7.003), (I.81) +
(7.004), (I.81) + (7.005), (I.81) + (7.006), (I.81) + (8.001), (I.81) +
(9.001), (I.81) + (9.002), (I.81)
+ (9.003), (I.81) + (9.004), (I.81) + (9.005), (I.81) + (9.006), (I.81) +
(9.007), (I.81) + (9.008),
(I.81) + (9.009), (I.81) + (10.001), (I.81) + (10.002), (I.81) + (10.003),
(I.81) + (11.001), (I.81) +
(11.002), (I.81) + (12.001), (I.81) + (12.002), (I.81) + (12.003), (I.81)+
(12.004), (I.81) + (13.001),
(I.81) + (13.002), (I.81) + (13.003), (I.81) + (13.004), (I.81) + (13.005),
(I.81) + (13.006), (I.81) +
(14.001), (I.81) + (14.002), (I.81) + (15.001), (I.81) + (15.002), (I.81)+
(15.003), (I.81) + (15.004),
(I.81) + (15.005), (I.81) + (15.006), (I.81) + (15.007), (I.81) + (15.008),
(I.81) + (15.009), (I.81) +
(15.010), (1.81) + (15.011), (1.81) + (15.012), (1.81) + (15.013), (1.81) +
(15.014), (1.81) + (15.015),
(I.81) + (15.016), (I.81) + (15.017), (I.81) + (15.018), (I.81) + (15.019),
(I.81) + (15.020), (I.81) +
(15.021), (1.81) + (15.022), (1.81) + (15.023), (1.81) + (15.024), (1.81) +
(15.025), (1.81) + (15.026),
(I.81) + (15.027), (I.81) + (15.028), (I.81) + (15.029), (I.81) + (15.030),
(I.81) + (15.031), (I.81) +
(15.032), (1.81) + (15.033), (1.81) + (15.034), (1.81) + (15.035), (1.81) +
(15.036), (1.81) + (15.037),
(I.81) + (15.038), (I.81) + (15.039), (I.81) + (15.040), (I.81) + (15.041),
(I.81) + (15.042), (I.81) +
(15.043), (1.81) + (15.044), (1.81) + (15.045), (1.81) + (15.046), (1.81) +
(15.047), (1.81) + (15.048),
(I.81) + (15.049), (I.81) + (15.050), (I.81) + (15.051), (I.81) + (15.052),
(I.81) + (15.053), (I.81) +
(15.054), (1.81) + (15.055), (1.81) + (15.056), (1.81) + (15.057), (1.81) +
(15.058), (1.81) + (15.059),
(I.81) + (15.060), (I.81) + (15.061), and (I.81) + (15.062).
33
Date Recue/Date Received 2021-08-25
[0074] In certain embodiments, the active compound or combination of active
compounds are
selected from the group (G4) consisting of the following mixtures: (I.91) +
(1.001), (I.91) +
(1.002), (I.91)+ (1.003), (I.91)+ (1.004), (I.91)+ (1.005), (I.91)+ (1.006),
(I.91)+ (1.007), (I.91)
+ (1.008), (I.91) + (1.009), (I.91) + (1.010), (I.91) + (1.011), (I.91) +
(1.012), (I.91) + (1.013),
(I.91) + (1.014), (I.91) + (1.015), (I.91) + (1.016), (I.91) + (1.017), (I.91)
+ (1.018), (I.91) +
(1.019), (I.91)+ (1.020), (I.91)+ (1.021), (I.91)+ (1.022), (I.91)+ (1.023),
(I.91)+ (1.024), (I.91)
+ (1.025), (I.91) + (1.026), (I.91) + (1.027), (I.91) + (1.028), (I.91) +
(1.029), (I.91) + (1.030),
(I.91) + (1.031), (I.91) + (1.032), (I.91) + (1.033), (I.91) + (1.034), (I.91)
+ (1.035), (I.91) +
(1.036), (I.91)+ (1.037), (I.91)+ (1.038), (I.91)+ (1.039), (I.91)+ (1.040),
(I.91)+ (1.041), (I.91)
+ (1.042), (I.91) + (1.043), (I.91) + (1.044), (I.91) + (1.045), (I.91) +
(1.046), (I.91) + (1.047),
(I.91) + (1.048), (I.91) + (1.049), (I.91) + (1.050), (I.91) + (1.051), (I.91)
+ (1.052), (I.91) +
(1.053), (I.91)+ (1.054), (I.91)+ (1.055), (I.91)+ (1.056), (I.91)+ (1.057),
(I.91)+ (1.058), (I.91)
+ (1.059), (I.91) + (1.060), (I.91) + (1.061), (I.91) + (1.062), (I.91) +
(1.063), (I.91) + (1.064),
(I.91) + (1.065), (I.91) + (1.066), (I.91) + (1.067), (I.91) + (1.068), (I.91)
+ (1.069), (I.91) +
(1.070), (I.91)+ (1.071), (I.91)+ (1.072), (I.91)+ (1.073), (I.91)+ (1.074),
(I.91)+ (1.075), (I.91)
+ (1.076), (I.91) + (1.077), (I.91) + (1.078), (I.91) + (1.079), (I.91) +
(1.080), (I.91) + (1.081),
(I.91) + (1.082), (I.91) + (2.001), (I.91) + (2.002), (I.91) + (2.003), (I.91)
+ (2.004), (I.91) +
(2.005), (I.91)+ (2.006), (I.91)+ (2.007), (I.91)+ (2.008), (I.91)+ (2.009),
(I.91)+ (2.010), (I.91)
+ (2.011), (I.91) + (2.012), (I.91) + (2.013), (I.91) + (2.014), (I.91) +
(2.015), (I.91) + (2.016),
(I.91) + (2.017), (I.91) + (2.018), (I.91) + (2.019), (I.91) + (2.020), (I.91)
+ (2.021), (I.91) +
(2.022), (I.91)+ (2.023), (I.91)+ (2.024), (I.91)+ (2.025), (I.91)+ (2.026),
(I.91)+ (2.027), (I.91)
+ (2.028), (I.91) + (2.029), (I.91) + (2.030), (I.91) + (2.031), (I.91) +
(2.032), (I.91) + (2.033),
(I.91) + (2.034), (I.91) + (2.035), (I.91) + (2.036), (I.91) + (2.037), (I.91)
+ (2.038), (I.91) +
(2.039), (I.91)+ (2.040), (I.91)+ (2.041), (I.91)+ (2.042), (I.91)+ (2.043),
(I.91)+ (2.044), (I.91)
+ (2.045), (I.91) + (2.046), (I.91) + (2.047), (I.91) + (2.048), (I.91) +
(2.049), (I.91) + (2.050),
(I.91) + (2.051), (I.91) + (2.052), (I.91) + (2.053), (I.91) + (2.054), (I.91)
+ (2.055), (I.91) +
(2.056), (I.91)+ (3.001), (I.91)+ (3.002), (I.91)+ (3.003), (I.91)+ (3.004),
(I.91)+ (3.005), (I.91)
+ (3.006), (I.91) + (3.007), (I.91) + (3.008), (I.91) + (3.009), (I.91) +
(3.010), (I.91) + (3.011),
(I.91) + (3.012), (I.91) + (3.013), (I.91) + (3.014), (I.91) + (3.015), (I.91)
+ (3.016), (I.91) +
(3.017), (I.91)+ (3.018), (I.91)+ (3.019), (I.91)+ (3.020), (I.91)+ (3.021),
(I.91)+ (3.022), (I.91)
+ (3.023), (I.91) + (3.024), (I.91) + (3.025), (I.91) + (3.026), (I.91) +
(3.027), (I.91) + (3.028),
34
Date Recue/Date Received 2021-08-25
(I.91) + (3.029), (I.91) + (4.001), (I.91) + (4.002), (I.91) + (4.003), (I.91)
+ (4.004), (I.91) +
(4.005), (I.91) + (4.006), (I.91) + (4.007), (I.91) + (4.008), (I.91) +
(4.009), (I.91) + (4.010), (I.91)
+ (4.011), (I.91) + (4.012), (I.91) + (4.013), (I.91) + (4.014), (I.91) +
(4.015), (I.91) + (4.016),
(I.91) + (4.017), (I.91) + (4.018), (I.91) + (4.019), (I.91) + (4.020), (I.91)
+ (4.021), (I.91) +
(4.022), (I.91) + (4.023), (I.91) + (4.024), (I.91) + (4.025), (I.91) +
(5.001), (I.91) + (5.002), (I.91)
+ (5.003), (I.91) + (5.004), (I.91) + (5.005), (I.91) + (5.006), (I.91) +
(5.007), (I.91) + (5.008),
(I.91) + (5.009), (I.91) + (5.010), (I.91) + (5.011), (I.91) + (5.012), (I.91)
+ (5.013), (I.91) +
(5.014), (I.91) + (5.015), (I.91) + (5.016), (I.91) + (5.017), (I.91) +
(5.018), (I.91) + (5.019), (I.91)
+ (5.020), (I.91) + (5.021), (I.91) + (5.022), (I.91) + (5.023), (I.91) +
(6.001), (I.91) + (6.002),
(I.91) + (6.003), (I.91) + (6.004), (I.91) + (7.001), (I.91) + (7.002), (I.91)
+ (7.003), (I.91) +
(7.004), (I.91) + (7.005), (I.91) + (7.006), (I.91) + (8.001), (I.91) +
(9.001), (I.91) + (9.002), (I.91)
+ (9.003), (I.91) + (9.004), (I.91) + (9.005), (I.91) + (9.006), (I.91) +
(9.007), (I.91) + (9.008),
(I.91) + (9.009), (I.91) + (10.001), (I.91) + (10.002), (I.91) + (10.003),
(I.91) + (11.001), (I.91) +
(11.002), (I.91) + (12.001), (I.91) + (12.002), (I.91) + (12.003), (I.91)+
(12.004), (I.91) + (13.001),
(I.91) + (13.002), (I.91) + (13.003), (I.91) + (13.004), (I.91) + (13.005),
(I.91) + (13.006), (I.91) +
(14.001), (I.91) + (14.002), (I.91) + (15.001), (I.91) + (15.002), (I.91)+
(15.003), (I.91) + (15.004),
(I.91) + (15.005), (I.91) + (15.006), (I.91) + (15.007), (I.91) + (15.008),
(I.91) + (15.009), (I.91) +
(15.010), (1.91) + (15.011), (1.91) + (15.012), (1.91) + (15.013), (1.91) +
(15.014), (1.91) + (15.015),
(I.91) + (15.016), (I.91) + (15.017), (I.91) + (15.018), (I.91) + (15.019),
(I.91) + (15.020), (I.91) +
(15.021), (1.91) + (15.022), (1.91) + (15.023), (1.91) + (15.024), (1.91) +
(15.025), (1.91) + (15.026),
(I.91) + (15.027), (I.91) + (15.028), (I.91) + (15.029), (I.91) + (15.030),
(I.91) + (15.031), (I.91) +
(15.032), (1.91) + (15.033), (1.91) + (15.034), (1.91) + (15.035), (1.91) +
(15.036), (1.91) + (15.037),
(I.91) + (15.038), (I.91) + (15.039), (I.91) + (15.040), (I.91) + (15.041),
(I.91) + (15.042), (I.91) +
(15.043), (1.91) + (15.044), (1.91) + (15.045), (1.91) + (15.046), (1.91) +
(15.047), (1.91) + (15.048),
(I.91) + (15.049), (I.91) + (15.050), (I.91) + (15.051), (I.91) + (15.052),
(I.91) + (15.053), (I.91) +
(15.054), (1.91) + (15.055), (1.91) + (15.056), (1.91) + (15.057), (1.91) +
(15.058), (1.91) + (15.059),
(I.91) + (15.060), (I.91) + (15.061), and (I.91) + (15.062).
[0075] In certain embodiments, the active compound or combination of active
compounds are
selected from the mixtures belonging to group (G1) or (G2).
[0076] In certain embodiments, the active compound or combination of active
compounds are
selected from the group (G1 -A) consisting of the following mixtures: (1.01) +
(1.012), (1.01) +
Date Recue/Date Received 2021-08-25
(1.018), (I.01) + (1.020), (I.01) + (1.021), (I.01) + (2.002), (I.01) +
(2.005), (I.01) + (2.017), (I.01)
+ (2.027), (I.01) + (2.038), (I.01) + (3.020), (I.01) + (3.025), (I.01) +
(4.005), (I.01) + (5.004),
(I.01) + (5.013), (I.01) + (5.018), (I.01) + (12.003), (I.01) + (12.004),
(I.01) + (13.001), (I.01) +
(13.004), (I.01) + (15.008), (I.01) + (15.047).
[0077] In certain embodiments, the active compound or combination of active
compounds are
selected from the group (G2-A) consisting of the following mixtures: (1.59) +
(1.012), (1.59) +
(1.018), (1.59) + (1.020), (1.59) + (1.021), (1.59) + (2.002), (1.59) +
(2.005), (1.59) + (2.017), (1.59)
+ (2.027), (1.59) + (2.038), (1.59) + (3.020), (1.59) + (3.025), (1.59) +
(4.005), (1.59) + (5.004),
(1.59) + (5.013), (1.59) + (5.018), (1.59) + (12.003), (1.59) + (12.004),
(1.59) + (13.001), (1.59) +
(13.004), (1.59) + (15.008), (1.59) + (15.047).
[0078] In certain embodiments, the active compound or combination of active
compounds are
selected from the group (G3 -A) consisting of the following mixtures: (I.81) +
(1.012), (I.81) +
(1.018), (I.81) + (1.020), (I.81) + (1.021), (I.81) + (2.002), (I.81) +
(2.005), (I.81) + (2.017), (I.81)
+ (2.027), (I.81) + (2.038), (I.81) + (3.020), (I.81) + (3.025), (I.81) +
(4.005), (I.81) + (5.004),
(I.81) + (5.013), (I.81) + (5.018), (I.81) + (12.003), (I.81) + (12.004),
(I.81) + (13.001), (I.81) +
(13.004), (I.81) + (15.008), (I.81) + (15.047).
[0079] In certain embodiments, the active compound or combination of active
compounds are
selected from the group (G4-A) consisting of the following mixtures: (I.91) +
(1.012), (I.91) +
(1.018), (I.91) + (1.020), (I.91) + (1.021), (I.91) + (2.002), (I.91) +
(2.005), (I.91) + (2.017), (I.91)
+ (2.027), (I.91) + (2.038), (I.91) + (3.020), (I.91) + (3.025), (I.91) +
(4.005), (I.91) + (5.004),
(I.91) + (5.013), (I.91) + (5.018), (I.91) + (12.003), (I.91) + (12.004),
(I.91) + (13.001), (I.91) +
(13.004), (I.91) + (15.008), (I.91) + (15.047).
[0080] In certain embodiments, the active compound or combination of active
compounds are
selected from the mixtures belonging to group (G1-A) or (G2-A).
[0081] In certain embodiments, the active compound or combination of active
compounds can be
present in a broad range of effective weight ratio of A:B, for example in a
range of 100:1 to 1:100,
preferably in a weight ratio of 50:1 to 1:50, most preferably in a weight
ratio of 20:1 to 1:20. Further
ratios of A:B which can be used according to the present invention with
increasing preference in the
order given are: 95:1 to 1:95, 90:1 to 1:90, 85:1 to 1:85, 80:1 to 1:80, 75:1
to 1:75, 70:1 to 1:70, 65:1
to 1:65, 60:1 to 1:60, 55:1 to 1:55, 45:1 to 1:45, 40:1 to 1:40, 35:1 to 1:35,
30:1 to 1:30, 25:1 to 1:25,
15:1 to 1:15, 10:1 to 1:10, 5:1 to 1:5, 4:1 to 1:4,3:1 to 1:3, 2:1 to 1:2.
36
Date Recue/Date Received 2021-08-25
[0082] Where a compound (A) or a compound (B) can be present in isomeric forms
and/or
tautomeric forms, such a compound is understood hereinabove and hereinbelow
also to include,
where applicable, corresponding isomeric and/or tautomeric forms or mixtures
thereof, even when
these are not specifically mentioned in each case.
[0083] Various genetic engineering technologies have been developed and may be
used by those
of skill in the art to introduce traits in plants. In certain aspects of the
claimed invention, traits are
introduced into tomato plants via altering or introducing a single genetic
locus or transgene into
the genome of a variety or progenitor thereof. Methods of genetic engineering
to modify, delete,
or insert genes and polynucleotides into the genomic DNA of plants are well-
known in the art.
[0084] In specific embodiments of the invention, improved tomato lines can be
created through
the site-specific modification of a plant genome. Methods of genetic
engineering include, for
example, utilizing sequence-specific nucleases such as zinc-finger nucleases
(see, for example,
U.S. Pat. Appl. Pub. No. 2011-0203012); engineered or native meganucleases;
TALE-
endonucleases (see, for example, U.S. Pat. Nos. 8,586,363 and 9,181,535); and
RNA-guided
endonucleases, such as those of the CRISPR/Cas systems (see, for example,
U.S. Pat. Nos. 8,697,359 and 8,771,945 and U.S. Pat. Appl. Pub. No. 2014-
0068797). One
embodiment of the invention thus relates to utilizing a nuclease or any
associated protein to carry
out genome modification. This nuclease could be provided heterologously within
donor template
DNA for templated-genomic editing or in a separate molecule or vector. A
recombinant DNA
construct may also comprise a sequence encoding one or more guide RNAs to
direct the nuclease
to the site within the plant genome to be modified. Further methods for
altering or introducing a
single genetic locus include, for example, utilizing single-stranded
oligonucleotides to introduce
base pair modifications in a tomato plant genome (see, for example Sauer et
al., Plant Physiol,
170(4):1917-1928, 2016).
[0085] Methods for site-directed alteration or introduction of a single
genetic locus are well-
known in the art and include those that utilize sequence-specific nucleases,
such as the
aforementioned, or complexes of proteins and guide-RNA that cut genomic DNA to
produce a
double-strand break (DSB) or nick at a genetic locus. As is well-understood in
the art, during the
process of repairing the DSB or nick introduced by the nuclease enzyme, a
donor template,
transgene, or expression cassette polynucleotide may become integrated into
the genome at the
site of the DSB or nick. The presence of homology arms in the DNA to be
integrated may promote
37
Date Recue/Date Received 2021-08-25
the adoption and targeting of the insertion sequence into the plant genome
during the repair process
through homologous recombination or non-homologous end joining (NHEJ).
[0086] In another embodiment of the invention, genetic transformation may be
used to insert a
selected transgene into a plant of the invention or may, alternatively, be
used for the preparation
of transgenes which can be introduced by backcrossing. Methods for the
transformation of plants
that are well-known to those of skill in the art and applicable to many crop
species include, but are
not limited to, electroporation, microprojectile bombardment, Agrobacterium-
mediated
transformation, and direct DNA uptake by protoplasts.
[0087] To effect transformation by electroporation, one may employ either
friable tissues, such
as a suspension culture of cells or embryogenic callus or alternatively one
may transform immature
embryos or other organized tissue directly. In this technique, one would
partially degrade the cell
walls of the chosen cells by exposing them to pectin-degrading enzymes
(pectolyases) or
mechanically wound tissues in a controlled manner.
[0088] An efficient method for delivering transforming DNA segments to plant
cells is
microprojectile bombardment. In this method, particles are coated with nucleic
acids and delivered
into cells by a propelling force. Exemplary particles include those comprised
of tungsten,
platinum, and preferably, gold. For the bombardment, cells in suspension are
concentrated on
filters or solid culture medium. Alternatively, immature embryos or other
target cells may be
arranged on solid culture medium. The cells to be bombarded are positioned at
an appropriate
distance below the macroprojectile stopping plate.
[0089] An illustrative embodiment of a method for delivering DNA into plant
cells by
acceleration is the Biolistics Particle Delivery System, which can be used to
propel particles coated
with DNA or cells through a screen, such as a stainless steel or Nytex screen,
onto a surface
covered with target cells. The screen disperses the particles so that they are
not delivered to the
recipient cells in large aggregates. Microprojectile bombardment techniques
are widely
applicable, and may be used to transform virtually any plant species.
[0090] Agrobacterium-mediated transfer is another widely applicable system for
introducing gene
loci into plant cells. An advantage of the technique is that DNA can be
introduced into whole
plant tissues, thereby bypassing the need for regeneration of an intact plant
from a protoplast.
Modern Agrobacterium transformation vectors are capable of replication in E.
coil as well as
Agrobacterium, allowing for convenient manipulations (Klee et al., Nat.
Biotechnol., 3(7):637-
38
Date Recue/Date Received 2021-08-25
642, 1985). Moreover, recent technological advances in vectors for
Agrobacterium-mediated gene
transfer have improved the arrangement of genes and restriction sites in the
vectors to facilitate the
construction of vectors capable of expressing various polypeptide coding
genes. The vectors
described have convenient multi-linker regions flanked by a promoter and a
polyadenylation site
for direct expression of inserted polypeptide coding genes. Additionally,
Agrobacterium
containing both armed and disarmed Ti genes can be used for transformation.
[0091] In those plant strains where Agrobacterium-mediated transformation is
efficient, it is the
method of choice because of the facile and defined nature of the gene locus
transfer. The use of
Agrobacterium-mediated plant integrating vectors to introduce DNA into plant
cells is well known
in the art (Fraley et al., Nat. Biotechnol ., 3:629-635, 1985; U.S. Patent No.
5,563,055).
[0092] Transformation of plant protoplasts also can be achieved using methods
based on calcium
phosphate precipitation, polyethylene glycol treatment, electroporation, and
combinations of these
treatments (see, for example, Potrykus et al., MoL Gen. Genet., 199:183-188,
1985; Omirulleh et
aL, Plant MoL Biol., 21(3):415-428, 1993; Fromm et al.,Nature, 312:791-793,
1986; Uchimiya et
al., MoL Gen. Genet., 204:204, 1986; Marcotte et al., Nature, 335:454, 1988).
Transformation of
plants and expression of foreign genetic elements is exemplified in Choi et
al. (Plant Cell Rep.,
13:344-348, 1994), and Ellul et al. (Theor. AppL Genet., 107:462-469, 2003).
V. Definitions
[0093] The following definitions are provided to better define the present
invention and to guide
those of ordinary skill in the art in the practice of the present invention.
Unless otherwise noted,
terms are to be understood according to conventional usage by those of
ordinary skill in the
relevant art.
[0094] As used herein, the term "plant" includes plant cells, plant
protoplasts, plant cells of tissue
culture from which tomato plants can be regenerated, plant calli, plant clumps
and plant cells that
are intact in plants or parts of plants such as pollen, flowers, seeds,
leaves, stems, and the like.
[0095] As used herein, the term "population" means a genetically heterogeneous
collection of
plants that share a common parental derivation.
[0096] As used herein, the terms "variety" and "cultivar" mean a group of
similar plants that by
their genetic pedigrees and performance can be identified from other varieties
within the same
species.
39
Date Recue/Date Received 2021-08-25
[0097] As used herein, an "allele" refers to one of two or more alternative
forms of a genomic
sequence at a given locus on a chromosome.
[0098] A "quantitative trait locus" (QTL) is a chromosomal location that
encodes for at least a
first allele that affects the expressivity of a phenotype.
[0099] As used herein, a "marker" means a detectable characteristic that can
be used to
discriminate between organisms. Examples of such characteristics include, but
are not limited to,
genetic markers, biochemical markers, metabolites, morphological
characteristics, and agronomic
characteristics.
[00100] As used herein, the term "phenotype" means the detectable
characteristics of a cell or
organism that can be influenced by gene expression.
[00101] As used herein, the term "genotype" means the specific allelic makeup
of a plant.
[00102] As used herein, "elite" or "cultivated" variety means any variety that
has resulted from
breeding and selection for superior agronomic performance. An "elite plant"
refers to a plant
belonging to an elite variety. Numerous elite varieties are available and
known to those of skill in
the art of tomato breeding. An "elite population" is an assoi _______________
intent of elite individuals or varieties
that can be used to represent the state of the art in terms of agronomically
superior genotypes of a
given crop species, such as tomato. Similarly, an "elite germplasm" or elite
strain of germplasm is
an agronomically superior germplasm.
[00103] As used herein, the term "introgressed," when used in reference to a
genetic locus, refers
to a genetic locus that has been introduced into a new genetic background,
such as through
backcrossing. Introgression of a genetic locus can be achieved through plant
breeding methods
and/or by molecular genetic methods. Such molecular genetic methods include,
but are not limited
to, various plant transformation techniques and/or methods that provide for
homologous
recombination, non-homologous recombination, site-specific recombination,
and/or genomic
modifications that provide for locus substitution or locus conversion.
[00104] As used herein, the terms "recombinant" or "recombined" in the context
of a
chromosomal segment refer to recombinant DNA sequences comprising one or more
genetic loci
in a configuration in which they are not found in nature, for example as a
result of a recombination
event between homologous chromosomes during meiosis.
Date Recue/Date Received 2021-08-25
[00105] As used herein, the term "linked," when used in the context of nucleic
acid markers and/or
genomic regions, means that the markers and/or genomic regions are located on
the same linkage
group or chromosome such that they tend to segregate together at meiosis.
[00106] As used herein, "tolerance locus" means a locus associated with
tolerance or resistance
to disease. For instance, a tolerance locus according to the present invention
may, in one
embodiment, control tolerance or susceptibility to Stemphylium.
[00107] As used herein, "tolerance" or "improved tolerance" in a plant refers
to the ability of the
plant to perform well, for example by maintaining yield, under disease
conditions. Tolerance may
also refer to the ability of a plant to maintain a plant vigor phenotype under
disease conditions.
Tolerance is a relative term, indicating that a "tolerant" plant is more able
to maintain performance
compared to a different (less tolerant) plant (e.g. a different plant variety)
grown in similar disease
conditions. One of skill will appreciate that plant tolerance to disease
conditions varies widely,
and can represent a spectrum of more-tolerant or less-tolerant phenotypes.
However, by simple
observation, one of skill can generally determine the relative tolerance of
different plants, plant
varieties, or plant families under disease conditions, and furthermore, will
also recognize the
phenotypic gradations of "tolerance."
[00108] As used herein "resistance" or "improved resistance" in a plant to
disease conditions is
an indication that the plant is more able to reduce disease burden than a non-
resistant or less
resistant plant. Resistance is a relative term, indicating that a "resistant"
plant is more able to reduce
disease burden compared to a different (less resistant) plant (e.g., a
different plant variety) grown
in similar disease conditions. One of skill will appreciate that plant
resistance to disease conditions
varies widely, and can represent a spectrum of more-resistant or less-
resistant phenotypes.
However, by simple observation, one of skill can generally determine the
relative resistance of
different plants, plant varieties, or plant families under disease conditions,
and furthermore, will
also recognize the phenotypic gradations of "resistant."
[00109] As used herein, "resistance allele" means the nucleic acid sequence
associated with
tolerance or resistance to disease.
[00110] The term "about" is used to indicate that a value includes the
standard deviation of error
for the device or method being employed to determine the value. The use of the
term "or" in the
claims is used to mean "and/or" unless explicitly indicated to refer to
alternatives only or the
alternatives are mutually exclusive, although the disclosure supports a
definition that refers to only
41
Date Recue/Date Received 2021-08-25
alternatives and to "and/or." When used in conjunction with the word
"comprising" or other open
language in the claims, the words "a" and "an" denote "one or more," unless
specifically noted.
The terms "comprise," "have" and "include" are open-ended linking verbs. Any
forms or tenses
of one or more of these verbs, such as "comprises," "comprising," "has,"
"having," "includes" and
"including," are also open-ended. For example, any method that "comprises,"
"has" or "includes"
one or more steps is not limited to possessing only those one or more steps
and also covers other
unlisted steps. Similarly, any plant that "comprises," "has" or "includes" one
or more traits is not
limited to possessing only those one or more traits and covers other unlisted
traits.
VI. Deposit Information
[00111] A deposit was made of at least 2500 seeds of tomato line FDR-I15-
0403V, which
comprises the reduced introgression described herein. The deposit was made
with the Provasoli-
Guillard National Center for Marine Algae & Microbiota (NCMA), at the Bigelow
Laboratory for
Ocean Science, 60 Bigelow Drive, East Boothbay, Me, 04544. The deposit is
assigned NCMA
Accession No. 202103011, and the date of deposit was March 11, 2021. Access to
the deposit will
be available during the pendency of the application to persons entitled
thereto upon request. The
deposit will be maintained in the NCMA Depository, which is a public
depository, for a period of
30 years, or 5 years after the most recent request, or for the enforceable
life of the patent, whichever
is longer, and will be replaced if nonviable during that period. Applicant
does not waive any
infringement of their rights granted under this patent or any other form of
variety protection,
including the Plant Variety Protection Act (7 U.S.C. 2321 et seq.).
Examples
Example 1. Mapping of the Stemphylium Resistance Locus
[00112] Two mapping populations were developed from an internal donor line
containing the
Stemphylium resistance locus derived from line PI 79532. In the F2 generation
of the mapping
populations segregating for the Stemphylium resistance locus, it was
determined that the resistance
is most likely to be completely dominant with a 3:1 segregation ratio. In a
preliminary mapping
study, it was determined that the Stemphylium resistance locus was located
within a 16 cM region
on chromosome 11. The interstitial markers within this region were tested for
accuracy across a
broad germplasm panel but were not found to be fully predictive of disease
resistance in all tested
42
Date Recue/Date Received 2021-08-25
lines. When the tomato genome sequence became available, we learned that this
16 cM region
spanned the centromere and was equivalent to ¨38.8Mbp of genomic sequence and
this precluded
our ability to accurately discern the actual position of the QTL in a
biparental population.
Accordingly, a modified association mapping approach was taken to identify
more predictive
markers across the diversity of the germplasm. In this approach, inbred lines
that were
phenotypically confirmed for Stemphylium resistance were cross-referenced with
lines that were
fingerprinted. 147 internal inbred lines were found to have a known
Stemphylium resistance
phenotype and that were fingerprinted. The lines were subsequently sorted in
two phenotypic
groups: resistant or susceptible. To ease the marker analysis, markers
monomorphic between the
two phenotypic groups were discarded and lines for which >20% missing data
were removed from
the analysis. For all the remaining markers, a Chi-square test statistic was
calculated and used to
determine if a marker was significantly associated with the disease phenotype
(i.e., whether
resistant class predominately had one homozygous genotype and the susceptible
class had the other
homozygous genotype for biallelic SNP assay). Using this approach, marker Ml,
a SNP marker
with a [C/A] change at 8,894,829 bp on chromosome 11 of the public tomato
genome map version
SL2.50, was found as significantly associated with Stemphylium resistance.
[00113] Additional markers that are associated with Stemphylium resistance
were identified using
a modified association mapping approach. A 50 Mbp region on chromosome 11 was
sequence
captured and resultant SNPs were cataloged. To identify potentially causative
SNPs, pairwise
comparisons were made between SNPs detected in resistant and susceptible
lines. Of all loci that
were polymorphic between resistant and susceptible genotypes in these
comparisons, only 76
tightly clustered SNPs on chromosome 11 were useful for consistently
discriminating resistant and
susceptible individuals in response to Stemphylium. This collection of loci
encompasses 1.8 cM
on chromosome 11 and corresponds to the interval of 8,894,829 to 9,826,973 bp
of the public
tomato genome map version SL2.50. This region confers S. pimpinellifilium-
derived resistance to
Stemphylium and contains markers Ml, M2, a SNP marker with a [G/A] change at
9,591,834 bp
of chromosome 11 of the public tomato genome map version SL2.50, and M3, a SNP
marker with
a [T/A] change at 9,826,973 bp of chromosome 11 of the public tomato genome
map version
SL2.50.
43
Date Recue/Date Received 2021-08-25
Table I. Markers for identifying and tracking the Stemphylium resistance
locus.
Marker Genetic Public
Marker SNP
Position
Marker Sequence Map Public Position
SNP
Size in
Marker
Name (SEQ ID Position Chr. of SNP 5L2.50
Change
(bp)
(bp)
NO.) (cM) (bp)
M1 1 34.35 11 8,894,829 1213
529 [C/A]
M2 2 35.7 11 9,591,834 199
99 [G/A]
M3 3 36.15 11 9,826,973 1968
787 [T/A]
44
Date Recue/Date Received 2021-08-25
Example 2. Breeding Event Creation
[00114] To aid breeding efforts, a breeding event donor was developed for the
Stemphylium
resistance allele without the detrimental small fruit size allele that could
be used across different
breeding programs. The mean fruit weight for each of the lines from the BC3F3
population was
compared to the fruit weights of the Stemphylium resistance donor line and the
recurrent parent
lines. As shown in Table 2, the line "BC3F3 Line 2" had mean fruit weight
similar to that of the
recurrent parent lines. Genetic analysis showed that this line also contained
the smallest
introgression and was subsequently used to develop the Stemphylium resistance
event donor. A
single BC3F5 line was selected as the event donor and finished as tomato line
FDR-I15-0403V,
and a sample of seed of this line has been deposited under NCIMB Accession No.
202103011.
The event was found to be 0.5 cM in size, while the distance between flanking
markers, M1 and
M3, is 1.8 cM. Markers Ml, M2, and M3 can be used to select for the reduced
introgression
conferring Stemphylium resistance. When introgressing the breeding event,
especially when
replacing an existing Stemphylium introgression, it is important to use marker
M2 to track the
resistance locus, which comprises an allele from S. pimpinellifilium, while
the flanking markers
M1 and M3 are used to track the recurrent parent allele.
Table 2. Comparison of mean fruit weights of BC3F3 introgression lines, the
original
Stemphylium resistance donor line, and the recurrent parent lines.
Pedigree Mean Fruit Weight (kg)
BC3F3 Line 1 0.15
BC3F3 Line 2 0.20
BC3F3 Line 3 0.16
BC3F3 Line 4 0.17
BC3F3 Line 5 0.16
BC3F3 Line 6 0.18
BC3F3 Line 7 0.17
BC3F3 Line 8 0.19
Large Sm Introgression Donor 0.12
Recurrent Parent 1 0.14
Recurrent Parent 2 0.19
Recurrent Parent 3 0.19
Date Recue/Date Received 2021-08-25
Example 3. Identification of TBRFV-Resistant Tomato Plants
[00115] A panel of 60 elite lines was tested for their performance against
Tomato Brown Rugose
Fruit Virus (TBRFV) infection. The following protocol was used to determine
resistance. To
prepare the inoculum for the experiment, a leaf infected with TBRFV showing
clear mosaic
symptoms was crushed with a small amount of water. The remaining leaf slurry
was diluted to a
concentration of 20 g leaf/100 mL water. A small amount of carborundum powder
was added with
a maximum of 1 tsp/L, however any other abrasive powder that is used in rub
inoculation may be
used. The inoculum suspension was kept on ice and in the dark until testing.
The experiment
contained control plants consisting of a resistant plant, e.g. FDR-I15-0403V,
and a susceptible
plant. The plants were grown in a randomized complete block design. Plants
were inoculated with
TBRFV 4 weeks after transplanting when the 2nd truss is flowering. Prior to
inoculation, the side
shoots were trimmed so that only the main stem remained. A portion of the
inoculum was placed
on two fully expanded leaves between the 1st and 2nd truss. The leaves with
inoculum were then
gently rubbed to introduce the virus into the leaf. TBRFV infection was first
evaluated one month
after infection and again two months after infection, when the plants had six
trusses. During the
evaluations, leaf, fruit spot, and fruit necrosis symptoms were measured on a
1-9 scale, where a 1
indicated a complete absence of symptoms and a 9 indicated complete
susceptibility. The
experiment was deemed successful if 90% of the controls performed as expected
for one of the
symptoms.
[00116] The level of fruit symptoms and leaf symptoms was determined
separately. From this
analysis it was found that one proprietary elite inbred line provided high
levels of resistance against
both fruit and leaf symptoms. This line was further used to map the genetic
region conferring the
resistance against TBRFV.
[00117] In a further screen, 156 elite tomato lines relevant for the market
segments in Jordan,
Israel, Turkey, and Mexico were screened for resistance against TBRFV to find
potential further
resistance donors and shorten the breeding process by working with material
that is adapted to the
relevant market. In this screen, a strong correlation between high resistance
against TBRFV and
the presence of a Stemphylium resistance introgression on chromosome 11 was
observed. This
relationship is confirmed by the genetic experiments of the TBRFV locus in the
following
example.
46
Date Recue/Date Received 2021-08-25
Example 4. Mapping TBRFV Resistance in Tomato
[00118] The proprietary elite inbred line identified as showing high levels of
resistance against
both fruit and leaf symptoms in Example 3 was used for further mapping. This
line was crossed
to two different susceptible tomato lines and the F2 progeny of each of these
crosses was genotyped
and phenotyped based on TBRFV resistance. In the subsequent QTL analysis for
both populations,
a QTL region conferring TBRFV resistance was found on chromosome 11. In
addition, it was
found that the resistance is recessive.
[00119] Further fine-mapping was done by selecting F3 lines that segregated
for the QTL interval
on chromosome 11. From the subsequent F4 generation, 35 lines were selected
with a fixed
recombination event in the QTL region. These lines were phenotyped for TBRFV
resistance and
genotyped with markers across the earlier found QTL region on chromosome 11.
This set of
markers included markers Ml, M2, and M3. In the subsequent QTL analysis, it
was found that the
region conferring TBRFV resistance overlapped with the Stemphylium resistance
locus on
chromosome 11. Specifically, the QTL for TBRFV resistance was mapped between
marker loci
M1 and M3, where marker M2 could be used as a trait linked marker. To confirm
that the
Stemphylium resistance breeding event described in Example 2 above also
conferred resistance to
TBRFV, the event donor FDR-I15-0403V was tested for TBRFV resistance. It was
found that this
donor, which contains a different genetic background than the mapping
population, also is highly
resistant to TBRFV. Additional markers in the region between marker loci M1
and M3 were
developed for further fine-mapping and selection of the trait (Table 3).
Additional markers that
can be used to select for the TBRFV resistance trait are: M4, a SNP marker
with a [T/G] change
at 8,891,489 bp on chromosome 11 of the public tomato genome map version
5L2.50; M5, a SNP
marker with a [C/TI change at 9,355,794 bp on chromosome 11 of the public
tomato genome map
version 5L2.50; M6, a SNP marker with a [A/T] change at 9,401,319 bp on
chromosome 11 of
the public tomato genome map version 5L2.50; M7, a SNP marker with a [G/T]
change at
9,406,414 bp on chromosome 11 of the public tomato genome map version 5L2.50;
M8, a SNP
marker with a [A/T] change at 9,421,426 bp on chromosome 11 of the public
tomato genome map
version 5L2.50; M9, a SNP marker with a [T/C] change at 9,470,789 bp on
chromosome 11 of
the public tomato genome map version 5L2.50; and M10, a SNP marker with a
[A/G] change at
9,756,371 bp on chromosome 11 of the public tomato genome map version 5L2.50.
Markers M4,
47
Date Recue/Date Received 2021-08-25
M5, M6, M7, M8, M9, and M10 may also be used to select for the reduced
introgression conferring
Stemphylium resistance.
48
Date Recue/Date Received 2021-08-25
Table 3. List of markers and favorable alleles at each marker for tracking
resistance QTLs.
SNP
Marker Fwd Rev
Genetic Public Marker position
Probe 1 Probe 2
Marker SNP Favorable sequence
primer primer
Chr. Position position size in
(SEQ (SEQ
name change allele (SEQ ID
(SEQ (SEQ
(cM) SNP (bp) (bp) marker
ID NO) ID NO)
NO) ID
NO) ID NO)
(bp)
M1 11 34.35 8,894,829 1213 529 [A/C] A 1
M4 11 34.64 8,891,489 101 51 [T/G] G 4
5 6 7 8
M5 11 35.25 9,355,794 201 101 [C/TI T 9
M6 11 35.35 9,401,319 201 101 [A/T] T 10
11 12 13 14
M7 11 35.36 9,406,414 201 101 [G/T] T 15
16 17 18 19
M8 11 35.39 9,421,426 201 101 [A/T] T 20
21 22 23 24
M9 11 35.48 9,470,789 187 101 [T/C] C 25
26 27 28 29
49
Date Recue/Date Received 2021-08-25
M2 11 35.7 9,591,834 199 99 [G/A] A 2
M10 11 36.01 9,756,371 184 101 [A/G] G
30 31 32 33 34
M3 11 36.15 9,826,973 1968 787 [A/T] A 3
Date Recue/Date Received 2021-08-25