Note: Descriptions are shown in the official language in which they were submitted.
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
PERSONALIZED CELLS, TISSUES, AND ORGANS FOR TRANSPLANTATION
FROM A HUMANIZED, BESPOKE, DESIGNATED-PATHOGEN FREE, (NON-
HUMAN) DONOR AND METHODS AND PRODUCTS RELATING TO SAME
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001]
This application claims priority of U.S. provisional patent application No.
62/975611,
filed Feb. 12, 2020, U.S. provisional patent application No. 62/964397, filed
Jan. 22, 2020, PCT
application No. PCT/U52019/054833, filed Oct. 4, 2019, U.S. provisional patent
application No.
62/848272, filed May 15, 2019, U.S. and provisional patent application No.
62/823455, filed Mar.
25, 2019, the disclosures of all of which are incorporated herein by reference
in their entireties.
BACKGROUND OF THE INVENTION
[0002]
According to the United Network for Organ Sharing ("UNOS"), every ten minutes,
someone is added to the national transplant waiting list, and nearly 20 people
die each day waiting
for a transplant. As of March 2020, there were about 112,385 people in need of
a lifesaving organ
transplant in the United States, with only about 19,000 donors identified and
about 39,000
transplants performed in 2019 (data from the United Network for Organ Sharing
(UNOS)). The
need for specific organs in the United States is as follows:
Organ Candidates in Need
Kidney 102,730
Liver 12,926
Pancreas 879
Kidney / Pancreas 1,820
Heart 3,702
Lung 1,283
Heart/Lung 52
Intestine 238
Total 124,630
[0003]
Over the past 5 years, from 2014 through 2019, an average of about 6,400
candidates
died each year while on the waiting list and without receiving an organ
transplant. About the same
number were not able to receive a long-awaited transplant because they were
too sick to receive a
1
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
transplant for the requisite surgery. While the rate of divergence between
available donors and
unmet need of recipients has been improved marginally, this disparity has
continued to present day
and remains considerable; the supply remains disastrously inadequate. Of
course, the patients in
need are awaiting for organs from human donors, which would represent the
transplantation of
organs from one species to another (allotransplantation).
[0004]
Allotransplantation presents many significant multifaceted problems, involving
safety, logistical, ethical, legal, institutional, and cultural complications.
From a safety perspective,
allogeneic tissues from human donors carry significant infectious disease
risks. For example some
in the transplantation field have report that "[human] cytomegalovirus (CMV)
is the single most
important infectious agent affecting recipients of organ transplants, with at
least two-thirds of these
patients having CMV infection after transplantation." Denner J (2018)
Reduction of the survival
time of pig xenotransplants by porcine cytomegalovirus. Virology Journal,
15(1): 171; Rubin RH
(1990) Impact of cytomegalovirus infection on organ transplant recipients.
Reviews of Infectious
Diseases, 12 Suppl 7:S754-766.
[0005]
Regulations regarding tissue transplants include criteria for donor screening
and
testing for adventitious agents, as well as strict regulations that govern the
processing and
distribution of tissue grafts. The transmission of viruses has occurred as a
result of
allotransplantation. Exogenous retroviruses (Human T-cell leukemia virus type
1 (HTLV-1),
Human T-cell leukemia virus type 2 (HTLV-2), and Human immunodeficiency virus
(HIV) have
been transmitted by human tissues during organ and cell transplantation, as
have viruses such as
human cytomegalovirus, and even rabies. Due to technical and timing
constraints surrounding
organ viability and post-mortem screening, absolute testing is hindered, and
this risk cannot be
eliminated.
[0006]
Immunological disparities between recipient and donor prevent graft-survival
for
extended durations, without immunosuppressive regimens that pose their own set
of complications
and additional risks. When a patient receives an organ from a (non-self) donor
(living or deceased),
the recipient's immune system will recognize the transplant as foreign. This
recognition will cause
their immune system to mobilize and "reject" the organ unless concomitant
medications that
suppress the immune system's natural processes are utilized. The response to
an allogeneic skin
graft is a potent immune response involving engagement of both the innate and
adaptive immune
systems. Abbas AK, Lichtman AHH, Pillai S (2017) Cellular and Molecular
Immunology.
2
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[0007]
With regard to the use of immunosuppressants, immunosuppressive drugs prolong
survival of the transplanted graft in acute and chronic rejection schemas.
However, they leave
patients vulnerable to infections from even the most routine of pathogens and
require continued
use for life but expose the patient to an increased risk of infection, even
cancer.
immunosuppressant can blunt the natural immunological processes;
unfortunately, these
medications are often a lifelong requirement after organ transplantation and
increase recipient
susceptibility to otherwise routine pathogens. While these drugs allow
transplant recipients to
tolerate the presence of foreign organs, they also increase the risk of
infectious disease and
symptoms associated with a compromised immune system, as a broad array of
organisms may be
transmitted with human allografts." Fishman JA, Greenwald MA, Grossi PA (2012)
Transmission
of Infection With Human Allografts: Essential Considerations in Donor
Screening. Clinical
Infectious Diseases, 55(5):720-727.
[0008]
Logistically, numerous factors must be considered prior to a successful organ
donation and transplant procedure. Blood type and other medical factors must
be evaluated for
every donated organ, but further, each organ type presents unique
characteristics that also must be
weighed, such as post-mortem ischemia, immunological compatibility, patient
location, and
institutional capabilities.
[0009]
For these patients, and the millions not included in these statistics who also
would
benefit significantly from tissue transplants such as cornea or pancreatic
islet cells, some in the
field have confirmed that "allotransplantation will never prove to be a
sufficient source." Ekser B,
Cooper DKC, Tector AJ (2015) The Need for Xenotransplantation as a Source of
Organs and Cells
for Clinical Transplantation. International journal of surgery (London,
England), 23(0 0): 199-204.
[00010]
Despite such drawbacks, organ transplantation is unquestionably the preferred
therapy for most patients with end stage organ failure, in large part due to a
lack of viable
alternatives. However, the advent of organ transplantation as a successful
life-saving therapeutic
intervention, juxtaposed against the paucity of organs available to
transplant, unfortunately places
medical professionals in an ideologically vexing position of having to decide
who lives and who
dies. Ultimately, alternative and adjunct treatment options that would
minimize the severe
shortcomings of allotransplant materials while providing the same mechanism of
action that makes
them so effective would be of enormous benefit to patients worldwide.
3
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[00011] The urgent need for organs and other transplantation tissue
generally, including for
temporary therapies while more permanent organs or other tissue are located
and utilized, has led
to investigation into utilization of organs, cells and tissue from non-human
sources, including other
animals for temporary and/or permanent xenotransplantation.
[00012] Xenotransplantation, such as the transplantation of a non-human
animal organ into
a human recipient, has the potential to reduce the shortage of organs
available for transplant,
potentially helping thousands of people worldwide. Swine have been considered
a potential non-
human source of organs, tissue and/or cells for use in human
xenotransplantation given that their
size and physiology are compatible with humans. However, xenotransplantation
using standard,
unmodified pig tissue into a human or other primate is accompanied by
rejection of the
transplanted tissue.
[00013] Wild type swine organs would evoke rejection by the human immune
system upon
transplantation into a human where natural human antibodies target epitopes on
the swine cells,
causing rejection and failure of the transplanted organs, cells or tissue. The
rejection may be a
cellular rejection (lymphocyte mediated) or humoral (antibody mediated)
rejection including but
not limited to hyperacute rejection, an acute rejection, a chronic rejection,
may involve survival
limiting thrombocytopenia coagulopathy and an acute humoral xenograft reaction
(AHXR). Other
roadblocks with respect to swine to human xenotransplantation include risks of
cross-species
transmission of disease or parasites.
[00014] One cause of hyperacute rejection results from the expression of
alpha-1,3-
galactosyltransferase ("alpha-1,3-GT") in porcine cells, which causes the
synthesis of alpha-1,3-
galactose epitopes. Except for humans, apes and Old World monkeys, most
mammals carry
glycoproteins on their cell surfaces that contain galactose alpha 1,3-
galactose (see, e.g., Galili et
al., "Man, apes, and old world monkeys differ from other mammals in the
expression of a-
galactosyl epitopes on nucleated cells," I Biol. Chem. 263: 17755-17762
(1988). Humans, apes
and Old World monkeys have a naturally occurring anti-alpha gal antibody that
is produced and
binds to glycoproteins and glycolipids having galactose alpha-1,3 galactose
(see, e.g., Cooper et
al., "Genetically engineered pigs," Lancet 342:682-683 (1993)).
[00015] Accordingly, when natural type swine products are utilized in
xenotransplantation,
human antibodies will be invoked to confront the foreign alpha-1,3-galactose
epitopes, and
hyperacute rejection normally follows. Beyond alpha-1,3-GT, swine cells
express multiple genes
4
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
which are not found in human cells. These include, but are not limited to,
Neu5GC, and f31,4-N-
acetylgalactosaminyltransferase (B4GALNT2). Antibodies to the a-Gal, Neu5GC,
f31,4-N and
Sda-like antigens are present in human blood prior to implantation of xeno-
tissue, and are involved
in the intense and immediate antibody-mediated rejection of implanted tissue.
[00016] Additionally pig cells express Class I and Class II SLAs on
endothelial cells. The
SLA cross-reacting antibodies contribute to the intense and immediate
rejection of the implanted
porcine tissue. SLA antigens may also be involved with the recipient's T-cell
mediated immune
response. Porcine SLAs may include, but are not limited to, antigens encoded
by the SLA-1, SLA-
2, SLA-3, SLA-4, SLA-5, SLA-6, SLA-8, SLA-9, SLA-11 and SLA-12 loci. Porcine
Class II
SLAs include antigens encoded by the SLA-DQ and SLA-DR loci.
[00017] Many attempts have been made by others to modify swine to serve as
a source for
xenotransplantation products, however such attempts have not yielded a
successful swine model
to date. Such commercial, academic and other groups have focused on
interventions, gene
alterations, efforts to induce tolerance through chimerism, inclusion of
transgenes, concomitant
use of exogenous immunosuppressive medications aimed to reduce the recipients'
natural
immunologic response(s) and other approaches. These groups have sought to
create a "one size
fits all" source animal aiming to create one, standardized source animal for
all recipients.
[00018] Specifically, certain groups have focused on creating transgenic
swine free of
PERV and utilizing transgenic bone marrow for therapy (see, e,g., eGenesis,
Inc.
PCT/U52018/028539); creating transgenic swine utilizing stem cell scaffolding
(see, e,g, United
Therapeutics/Revivicor [US20190111180A1]); mixed chimerism and utilizing
transgenic bone
marrow for therapy to tolerize patient T-cells (see, e,g, Columbia University
[U520180070564A1]). These "downstream" approaches ¨ post recognition by the
human immune
system ¨ have not succeeded in producing swine that produce products suitable
for prolonged use
in xenotransplantation or that survive the above-referenced transgenic and
other alterations.
[00019] In contrast to the above-referenced approaches, the present
invention achieves a
"patient-specific" solution by modifying the genome of donor swine cells to
escape detection from
the human immune system in the first instance, avoiding the immune cascade
that follows when a
patient's T-cells and antibodies are primed to destroy foreign material. This
"upstream" approach
is achieved through, in one aspect, specific combinations of minimal genetic
alterations that render
the donor animal's cells, tissues, and organs tolerogenic when transplanted
into a human without
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
sacrificing the animal's immune function. The present invention therefore
addresses long-felt but
unmet need for translating the science of xenotransplantation into a clinical
reality.
[00020] This "upstream" approach is achieved through, in one aspect,
specific combinations
of minimal genetic alterations that render the donor animal's cells, tissues,
and organs tolerogenic
when transplanted into a human without sacrificing the animal's immune
function. The present
invention therefore addresses long-felt but unmet need for translating the
science of
xenotransplantation into a clinical reality.
SUMMARY OF THE INVENTION
[00021] In one aspect, the present disclosure includes a biological system
for generating and
preserving a repository of personalized, humanized transplantable cells,
tissues, and organs for
transplantation, wherein the biological system is biologically active and
metabolically active, the
biological system comprising genetically reprogrammed cells, tissues, and
organs in a non-human
animal for transplantation into a human recipient. For example, the non-human
animal is a
genetically reprogrammed swine for xenotransplantation of cells, tissue,
and/or an organ isolated
from the genetically reprogrammed swine, the genetically reprogrammed swine
comprising a
nuclear genome that has been reprogrammed to replace a plurality of
nucleotides in a plurality of
exon regions of a major histocompatibility complex of a wild-type swine with a
plurality of
synthesized nucleotides from a human captured reference sequence. In one
aspect, cells of said
genetically reprogrammed swine do not present one or more surface glycan
epitopes selected from
alpha-Gal, Neu5Gc, and SDa. Further, genes encoding alpha-1,3
galactosyltransferase, cytidine
monophosphate-N-acetylneuraminic acid hydroxylase (CMAH), and f31,4-N-
acetylgalactosaminyltransferase are altered such that the genetically
reprogrammed swine lacks
functional expression of surface glycan epitopes encoded by those genes. In
some aspects, the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of: i) at least one of the wild-type swine's SLA-1, SLA-2, and SLA-3
with nucleotides
from an orthologous exon region of HLA-A, HLA-B, and HLA-C, respectively, of
the human
captured reference sequence; and ii) at least one the wild-type swine's SLA-6,
SLA-7, and SLA-8
with nucleotides from an orthologous exon region of HLA-E, HLA-F, and HLA-G,
respectively,
of the human captured reference sequence; and iii) at least one of the wild-
type swine's SLA-DR
and SLA-DQ with nucleotides from an orthologous exon region of HLA-DR and HLA-
DQ,
6
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
respectively, of the human captured reference sequence. In some aspects, the
reprogrammed
genome comprises at least one of A-C:
A) wherein the reprogrammed swine nuclear genome comprises site-directed
mutagenic substitutions of nucleotides at exon regions of the wild-type
swine's f32-
microglobulin with nucleotides from orthologous exons of a known human 02-
microglobulin
from the human captured reference sequence;
B) wherein the reprogrammed swine nuclear genome comprises a polynucleotide
that
encodes a polypeptide that is a humanized beta 2 microglobulin (hB2M)
polypeptide
sequence that is at least 95% identical to the amino acid sequence of beta 2
microglobulin
glycoprotein expressed by the human captured reference genome;
C) wherein the reprogrammed swine nuclear genome has been reprogrammed such
that, at the swine's endogenous 02-microglobulin locus, the nuclear genome has
been
reprogrammed to comprise a nucleotide sequence encoding 02-microglobulin
polypeptide of
the human recipient. Further, in some aspects, the reprogrammed swine nuclear
genome has
been reprogrammed such that the genetically reprogrammed swine lacks
functional
expression of the wild-type swine's endogenous 02-microglobulin polypeptides.
Further,
the reprogramming does not introduce any frameshifts or frame disruptions.
[00022] In other aspects, the present disclosure includes a method of
preparing a genetically
reprogrammed swine comprising a nuclear genome that lacks functional
expression of surface
glycan epitopes selected from alpha-Gal, Neu5Gc, and SD a and is genetically
reprogrammed to
express a humanized phenotype of a human captured reference sequence
comprising:
a. obtaining a porcine fetal fibroblast cell, a porcine zygote, a porcine
Induced
Pluripotent Stem Cells (IPSC), or a porcine germ-line cell;
b. genetically altering said cell in a) to lack functional alpha-1,3
galactosyltransferase,
cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMAH), and f31,4-
N-acetylgalactosaminyltransferase;
c. genetically reprogramming said cell in b) using clustered regularly
interspaced
short palindromic repeats (CRISPR)/Cas for site-directed mutagenic
substitutions
of nucleotides at exon regions of: i) at least one of the wild-type swine's
SLA-1,
SLA-2, and SLA-3 with nucleotides from an orthologous exon region of HLA-A,
HLA-B, and HLA-C, respectively, of the human captured reference sequence; and
7
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
ii) at least one the wild-type swine's SLA-6, SLA-7, and SLA-8 with
nucleotides
from an orthologous exon region of HLA-E, HLA-F, and HLA-G, respectively, of
the human captured reference sequence; and iii) at least one of the wild-type
swine's SLA-DR and SLA-DQ with nucleotides from an orthologous exon region
of HLA-DR and HLA-DQ, respectively, of the human captured reference
sequence,
wherein intron regions of the wild-type swine's genome are not reprogrammed,
and
wherein the reprogrammed genome comprises at least one of A-C:
A) wherein the reprogrammed swine nuclear genome comprises site-directed
mutagenic substitutions of nucleotides at exon regions of the wild-type
swine's f32-
microglobulin with nucleotides from orthologous exons of a known human 02-
microglobulin
from the human captured reference sequence;
B) wherein the reprogrammed swine nuclear genome comprises a polynucleotide
that
encodes a polypeptide that is a humanized beta 2 microglobulin (hB2M)
polypeptide
sequence that is at least 95% identical to beta 2 microglobulin expressed by
the human
captured reference genome;
C) wherein the reprogrammed swine nuclear genome has been reprogrammed such
that the genetically reprogrammed swine lacks functional expression of the
wild-type
swine's endogenous 02-microglobulin polypeptides, wherein the reprogrammed
swine
nuclear genome has been reprogrammed such that, at the swine's endogenous f32-
microglobulin locus, the nuclear genome has been reprogrammed to comprise a
nucleotide
sequence encoding 02-microglobulin polypeptide of the human recipient,
wherein said reprogramming does not introduce any frameshifts or frame
disruptions,
d. generating an embryo from the genetically reprogrammed cell in c); and
e. transferring the embryo into a surrogate pig and growing the transferred
embryo
in the surrogate pig.
[00023] In another aspect, the present disclosure includes a method of
producing a donor
swine tissue or organ for xenotransplantation, wherein cells of said donor
swine tissue or organ
are genetically reprogrammed to be characterized by a recipient-specific
surface phenotype
comprising:
8
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
a. obtaining a biological sample containing DNA from a prospective human
transplant
recipient;
b. performing whole genome sequencing of the biological sample to obtain a
human
capture reference sequence;
c. comparing the human capture reference sequence with the wild-type genome of
the
donor swine at loci (i)-(v):
(i) exon regions encoding at least one of SLA-1, SLA-2, and SLA-3;
(ii) exon regions encoding at least one of SLA-6, SLA-7, and SLA-8;
(iii) exon regions encoding at least one of SLA-DR and SLA-DQ;
(iv) one or more exons encoding beta 2 microglobulin (B2M);
(v) exon regions of SLA-MIC-2 gene and a gene encoding at least one of PD-
L1, CTLA-4, EPCR, TBM, and TFPI,
d. creating synthetic donor swine nucleotide sequences of 10 to 350 basepairs
in length
for one or more of said loci (i)-(v), wherein said synthetic donor swine
nucleotide
sequences are at least 95% identical to the human capture reference sequence
at
orthologous loci (vi)-(x) corresponding to swine loci (i)-(vi), respectively:
(vi) exon regions encoding at least one of HLA-A, HLA-B, and HLA-C;
(vii) exon regions encoding at least one of HLA-E, HLA-F, and HLA-G;
(viii) exon regions encoding at least one of HLA-DR and HLA-DQ;
(ix) one or more exons encoding human beta 2 microglobulin (hB2M);
(x) exon regions encoding at least one of MIC-A, MIC-B, PD-L1, CTLA-4, EPCR,
TBM, and TFPI from the human capture reference sequence,
e. replacing nucleotide sequences in (i)-(v) with said synthetic donor swine
nucleotide
sequences; and
f. obtaining the swine tissue or organ for xenotransplantation from a
genetically
reprogrammed swine having said synthetic donor swine nucleotide sequences.
[00024] In another aspect, the present disclosure includes a method of
screening for off
target edits or genome alterations in the genetically reprogrammed swine
comprising a nuclear
genome of the present disclosure including:
9
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
a. performing whole genome sequencing on a biological sample containing DNA
from a
donor swine before performing genetic reprogramming of the donor swine nuclear
genome, thereby obtaining a first whole genome sequence;
b. after reprogramming of the donor swine nuclear genome, performing whole
genome
sequencing to obtain a second whole genome sequence;
c. aligning the first whole genome sequence and the second whole genome
sequence to
obtain a sequence alignment;
d. analyzing the sequence alignment to identify any mismatches to the swine's
genome at
off-target sites.
[00025] In another aspect, the present disclosure includes a synthetic
nucleotide sequence
having wild-type swine intron regions from a wild-type swine MHC Class Ia, and
reprogrammed
at exon regions encoding the wild-type swine's SLA-3 with codons of HLA-C from
a human
capture reference sequence that encode amino acids that are not conserved
between the SLA-3 and
the HLA-C from the human capture reference sequence. In some aspects, the wild-
type swine's
SLA-1 and SLA-2 each comprise a stop codon.
[00026] In another aspect, the present disclosure includes a synthetic
nucleotide sequence
having wild-type swine intron regions from a wild-type swine MHC Class lb, and
reprogrammed
at exon regions encoding the wild-type swine's SLA-6, SLA-7, and SLA-8 with
codons of HLA-
E, HLA-F, and HLA-G, respectively, from a human capture reference sequence
that encode amino
acids that are not conserved between the SLA-6, SLA-7, and SLA-8 and the HLA-
E, HLA-F, and
HLA-G, respectively, from the human capture reference sequence.
[00027] In another aspect, the present disclosure includes a synthetic
nucleotide sequence
having wild-type swine intron regions from a wild-type swine MEW Class II, and
reprogrammed
at exon regions encoding the wild-type swine's SLA-DQ with codons of HLA-DQ,
respectively,
from a human capture reference sequence that encode amino acids that are not
conserved between
the SLA-DQ and the HLA-DQ, respectively, from the human capture reference
sequence, and
wherein the wild-type swine's SLA-DR comprises a stop codon.
[00028] In another aspect, the present disclosure includes a synthetic
nucleotide sequence
having wild-type swine intron regions from a wild-type swine beta-2-
microglobulin and
reprogrammed at exon regions encoding the wild-type swine's beta-2-
microglobulin with codons
of beta-2-microglobulin from a human capture reference sequence that encode
amino acids that
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
are not conserved between the wild-type swine's beta-2-microglobulin and the
beta-2-
microglobulin from the human capture reference sequence, wherein the synthetic
nucleotide
sequence comprises at least one stop codon in an exon region such that the
synthetic nucleotide
sequence lacks functional expression of the wild-type swine's 02-microglobulin
polypeptides.
[00029] In another aspect, the present disclosure includes a synthetic
nucleotide sequence
having wild-type swine intron regions from a wild-type swine MIC-2, and
reprogrammed at exon
regions of the wild-type swine's MIC-2 with codons of MIC-A or MIC-B from a
human capture
reference sequence that encode amino acids that are not conserved between the
MIC-2 and the
MIC-A or the MIC-B from the human capture reference sequence.
[00030] In another aspect, the present disclosure includes a synthetic
nucleotide sequence
having wild-type swine intron regions from a wild-type swine CTLA-4, and
reprogrammed at exon
regions encoding the wild-type swine's CTLA-4 with codons of CTLA-4 from a
human capture
reference sequence that encode amino acids that are not conserved between the
wild-type swine's
CTLA-4 and the CTLA-4 from the human capture reference sequence.
[00031] In another aspect, the present disclosure includes a synthetic
nucleotide sequence
having wild-type swine intron regions from a wild-type swine PD-Li and
reprogrammed at exon
regions encoding the wild-type swine's PD-Li with codons of PD-Li from a human
capture
reference sequence that encode amino acids that are not conserved between the
wild-type swine's
PD-Li and the PD-Li from the human capture reference sequence.
[00032] In another aspect, the present disclosure includes a synthetic
nucleotide sequence
having wild-type swine intron regions from a wild-type swine EPCR and
reprogrammed at exon
regions encoding the wild-type swine's EPCR with codons of EPCR from a human
capture
reference sequence that encode amino acids that are not conserved between the
wild-type swine's
EPCR and the EPCR from the human capture reference sequence.
[00033] In another aspect, the present disclosure includes a synthetic
nucleotide sequence
having wild-type swine intron regions from a wild-type swine TBM and
reprogrammed at exon
regions encoding the wild-type swine's TBM with codons of TBM from a human
capture reference
sequence that encode amino acids that are not conserved between the wild-type
swine's TBM and
the TBM from the human capture reference sequence.
[00034] In another aspect, the present disclosure includes a synthetic
nucleotide sequence
having wild-type swine intron regions from a wild-type swine TFPI and
reprogrammed at exon
11
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
regions encoding the wild-type swine's TFPI with codons of TFPI from a human
capture reference
sequence that encode amino acids that are not conserved between the wild-type
swine's TFPI and
the TFPI from the human capture reference sequence.
[00035] In contrast to the above-referenced approaches, the present
invention achieves a
"patient-specific" solution by modifying the genome of donor swine cells to
escape detection from
the human immune system in the first instance, avoiding the immune cascade
that follows when a
patient's T-cells and antibodies are primed to destroy foreign material. This
"upstream" approach
is achieved through, in one aspect, minimal, modifications to the swine genome
involving distinct
combinations of disruptions (such as knocking out a1,3-galactosyltransferase
(aGal), cytidine
monophosphate-N-acetylneuraminic acid hy droxyl as e (CMAH) and/or I 1-4 N-
acetylgalactosaminyltransferase such that the donor swine cells do not express
such on its cell
surfaces), regulation of expression of certain genes (for example, CTLA-4 and
PD-1), and
replacement of specific sections of the swine genome with synthetically
engineered sections based
upon recipient human capture sequences (for example, in certain SLA sequences
to regulate the
swine's expression of, for example, MHC-I and MHC-II). The present invention
therefore
addresses long-felt but unmet need for translating the science of
xenotransplantation into a clinical
reality.
[00036] Such modifications result in the reduce the extent of, the
causative, immunological
disparities and associated, deleterious immune processes that result from the
recognition of "non-
self', by selectively altering the extracellular antigens of the donor to
increase the likelihood of
acceptance of the transplant.
[00037] In certain aspects, the present disclosure centralizes
(predicates) the creation of
hypoimmunogenic and/or tolerogenic cells, tissues, and organs that does not
necessitate the
transplant recipients' prevalent and deleterious use of exogenous
immunosuppressive drugs (or
prolonged immunosuppressive regimens) following the transplant procedure to
prolong the life-
saving graft. This approach is countervailing to the existing and previous
dogmatic approaches;
instead of accepting that innate and immovable disparity between donor and
recipient, and thus
focusing on interventions, gene alterations, and/or concomitant exogenous
immunosuppressive
medications used as a method of reducing/eliminating/negatively-altering the
recipients' naturally
resulting immunologic response, shifting (if not reversing) the focus of the
otherwise area of
fundamental scientific dogma.
12
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[00038] In certain other aspects, the present disclosure provides
genetically modified, non-
transgenic swine that are minimally altered. For example, in the present
invention, certain distinct
sequences appearing on the donor swine SLA comprising native base pairs are
removed and
replaced with a synthetic sequence comprising the same number of base pairs
but reprogrammed
based on the recipient's human capture sequence. This minimal alteration keeps
other aspects of
the native swine genome in place and does not disturb, for example, introns
and other codons
naturally existing in the swine genome.
[00039] In certain other aspects, the present invention provides swine
with such and other
modifications, created in a designated pathogen environment in accordance with
the processes and
methods provided herein.
[00040] In certain other aspects the products derived from such swine for
xenotransplantation are minimally manipulated, viable, live cell, and capable
of making an organic
union with the transplant recipient, including, but not limited to, inducing
vascularization and/or
collagen generation in the transplant recipient.
[00041] In certain other aspects products derived from such source animals
are preserved,
including, but not limited to, through cryopreservation, in a manner that
maintains viability and
live cell characteristics of such products.
[00042] In certain other aspects, such products are for homologous use,
i.e., the repair,
reconstruction, replacement or supplementation of a recipient's organ, cell
and/or tissue with a
corresponding organ, cell and/or tissue that performs the same basic function
or functions as the
donor (e.g., swine skin is used as a transplant for human skin, swine kidney
is used as a transplant
for human kidney, swine liver is used as a transplant for human liver, swine
nerve is used as a
transplant for human nerve and so forth).
[00043] In certain other aspects, the present invention that the
utilization of such products
in xenotransplantation be performed with or without the need to use
immunosuppressant drugs or
therapies which inhibit or interfere with normal immune function.
BRIEF DESCRIPTION OF THE DRAWINGS
[00044] The accompanying drawings, which are incorporated herein and form
part of the
disclosure, help illustrate various aspects of the present invention and,
together with the
description, further serve to describe the invention to enable a person
skilled in the pertinent art to
13
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
make and use the aspects disclosed herein. In the drawings, like reference
numbers indicate
identical or functionally similar elements.
[00045] FIG. 1 illustrates an image of human trophoblast and trophoblast
cells.
[00046] FIG. 2 schematically illustrates a T Cell Receptor (TCR) binding
MHC Class I and
a peptide.
[00047] FIG. 3 schematically illustrates HLA Class I on the surface of a
cell.
[00048] FIG. 4 schematically illustrates a Cytotoxic T Cell (CD8+) -
Target Cell Interaction.
[00049] FIG. 5 schematically illustrates a Cytotoxic T Cell (CD4+) -
Target Cell Interaction.
[00050] FIG. 6 schematically illustrates codominant expression of HLA
genes and the
position of HLA genes on human chromosome 6.
[00051] FIG. 7 is a table listing numbers of serological antigens,
proteins, and alleles for
human MHC Class I and Class II isotypes.
[00052] FIG. 8 schematically illustrates HLA Class I and Class II on the
surface of a cell.
[00053] FIG. 9 shows the structure of MHC Class 1(A) and Class II proteins
(B). The two
globular domains furthest from the plasma membrane that form the peptide
binding region (PBR)
are shaded in blue. The two Ig-like domains, including the 02-microglobulin,
are shaded in grey.
[00054] FIG. 10 shows the HLA genomic loci map.
[00055] FIG. 11 schematically illustrates Human MHC Class I and Class II
isotypes.
[00056] FIG. 12 shows the schematic molecular organization of the HLA
Class I genes.
Exons are represented by the rectangles and introns by lines.
[00057] FIG. 13 shows the schematic molecular organization of the HLA
Class II genes.
Exons are represented by the rectangles and introns by lines.
[00058] FIG. 14 showing composite genetic alteration design for
"humanization" of
extracellular porcine cell expression
[00059] FIG. 15 shows comparative genomic organization of the human and
swine major
histocompatibility complex (MHC) Class I region. The human leukocyte antigen
(HLA) Class I
map is adapted from Ref [17] and the swine leukocyte antigen (SLA) Class I map
is based only
on one fully sequenced haplotype (Hp-1.1, H01) [4]. Note that not all the
genes are shown here
and the scale is approximate. The number and location of expressed SLA Class I
genes may vary
between haplotypes.
14
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[00060] FIG. 16 shows comparative genomic organization of the human and
swine major
histocompatibility complex (MHC) Class II region. The human leukocyte antigen
(HLA) Class II
map is adapted from Ref [17] and the swine leukocyte antigen (SLA) Class II
map is based only
on one fully sequenced haplotype (H01) [4]. Note that not all the genes are
shown here and the
scale is approximate. *The number and location of expressed HLA-DRB genes and
pseudogenes
may vary between haplotypes.
[00061] FIG. 17 shows a physical map of the SLA complex. Black boxes: loci
containing
MHC-related sequences. White boxes: loci without MHC-related sequences. From
the long arm
to the short arm of the chromosome, the order of the regions is Class 11 (11),
Class III (III) and
Class 1(1).
[00062] FIG. 18 shows the schematic molecular organization of the SLA
genes. Exons are
represented by the gray ovals and introns by lines. Gene length is approximate
to that found for
the Hp-1.1 genome sequence.
[00063] FIG. 19 shows a side-by-side genomic analysis of the peptide
sequences.
[00064] FIGS. 20 shows the location and the length al (exon 2) of SLA-DQA
and f31(exon
2) of SLA-DQB1.
[00065] FIG. 21 shows a spreadsheet detailing nucleotide sequences of
exons and introns
of SLA-DQA and SLA-DQB1.
[00066] FIG. 22 shows SLA-DQ betal domain of sus scrofa (wild boar).
[00067] FIG. 23 illustrates nomenclature of HLA alleles. Each HLA allele
name has a
unique number corresponding to up to four sets of digits separated by colons.
The length of the
allele designation is dependent on the sequence of the allele and that of its
nearest relative. All
alleles receive at least a four digit name, which corresponds to the first two
sets of digits, longer
names are only assigned when necessary. The digits before the first colon
describe the type, which
often corresponds to the serological antigen carried by an allotype. The next
set of digits are used
to list the subtypes, numbers being assigned in the order in which DNA
sequences have been
determined. Alleles whose numbers differ in the two sets of digits must differ
in one or more
nucleotide substitutions that change the amino acid sequence of the encoded
protein. Alleles that
differ only by synonymous nucleotide substitutions (also called silent or non-
coding substitutions)
within the coding sequence are distinguished by the use of the third set of
digits. Alleles that only
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
differ by sequence polymorphisms in the introns, or in the 5' or 3'
untranslated regions that flank
the exons and introns, are distinguished by the use of the fourth set of
digits.
[00068] FIG. 24 shows the length of exons and introns in HLA-DQA
[00069] FIG. 25A shows nucleotide sequence library between recipient
specific HLA-DQA
and HLA-DQA acquired from database, FIG. 25B shows Nucleotide Sequence Library
identifying
complete divergence between HLA vs SLA(DQ-A, Exon 2), FIG. 25C shows Human
Capture
Reference Sequence for DQ-Al for Three Patients, FIG. 25D shows Human Capture
Reference
Sequence for DQ-Bl for Three Patients, FIG. 25E shows Human Capture Reference
Sequence for
DR-A for Three Patients, FIG. 25F shows Human Capture Reference Sequence for
DQR-Bl for
Three Patients.
[00070] FIG. 26A shows example of Human Capture Reference Sequence(DQ-A1)
for
Three Patients, FIG. 26B shows example of Human Capture Reference Sequence(DQ-
B1) for
Three Patients, FIG. 26C shows example of Human Capture Reference Sequence(DR-
A) for Three
Patients, FIG. 26D shows example of Human Capture Reference Sequence(DR-B1)
for Three
Patients.
[00071] FIG. 27 shows the wild-type human beta-2 rnicroglobulin protein
and schematic
molecular organization of the human B2M gene and swine B2M gene.
[00072] FIG. 28 shows comparison of amino acid sequences of exon 2 of
human B2M vs
exon 2 of swine B2M
[00073] FIG. 29 shows Phenotyping analysis of porcine alveolar macrophages
(PAM). Cells
were cultured in medium alone (control), or were activated for 72 hours with
100 ng/mL IFN-y or
loaded 30 pg/mL KLH for 24 hours. The cells were stained for SLA-DQ and marker
is detected
using anti mouse APC-conjugated polyclonal IgG secondary antibody. Data is
presented as
histograms of count (y axis) versus fluorescence intensity in log scale (x
axis). Percentage of
positive and negative cells for SLA-DQ for activated cells are shown on
histograms.
[00074] FIG. 30 shows SI values for BrdU ELISA. Proliferation response of
three human
CD4+ T cells (A) and PBMCs (B) to untreated and IFN-y activated PAM cells
(15K) after seven
days incubation.
[00075] FIG. 31 shows a schematic depiction of a humanized porcine cell
according to the
present disclosure
16
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[00076] FIG. 32 shows a graph wherein 1 x 105 purified human CD8+ T cells
(A) or human
PBMC (B) were stimulated with increasing numbers of irradiated (30 Gy) porcine
PBMC from
four-fold knockout pig 10261 or a wild-type pig. Proliferation was measured
after 5 d + 16 h by
3H-thymidine incorporation. Data represent mean cpm SEM of triplicate
cultures obtained with
cells from one human blood donor in a single experiment. Similar response
patterns were observed
using responder cells from a second blood donor and stimulator cells from four-
fold knockout pig
10262. Proliferation of human CD8+ T cells decreased after stimulation with
four-fold knockout
porcine PBMC. (Fischer, et al.,2019)
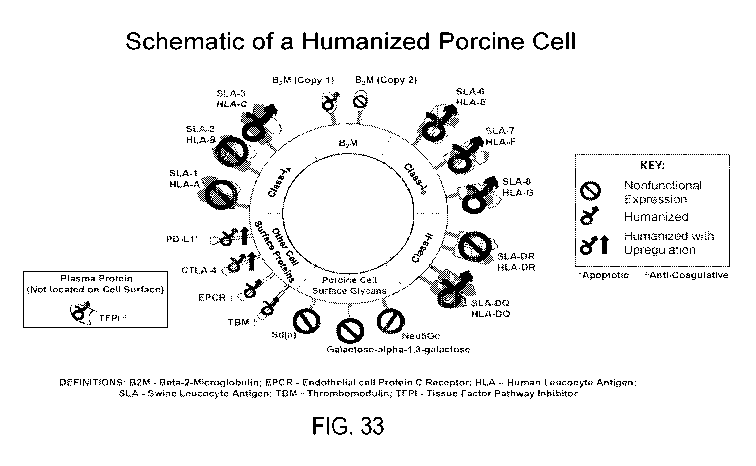
[00077] FIG. 33 shows schematic depiction of a humanized porcine cell
according to the
present disclosure.
[00078] FIG. 34 shows graph of proliferation of human plasma donors run on
3 separate
days with WT 128-11 and Gal T-KO B-174 PBMCs
[00079] FIG. 35 shows NK cytotoxicity of two donors (upper panel: KH;
lower panel: MS)
against 13 271 cells transfected with HLA-E/A2 (left column) and HLA-E/B7
(right column)
compared to the lysis of untransfected 13 271 cells. Results are depicted as
percentage of specific
lysis and were obtained at four different E:T ratios. Data are representative
of three independent
experiments. Open triangles represent HLA-E-transfected 13 271 cells, filled
diamonds represent
un-transfected 13 271 cells. (Forte, et al.,2005)
[00080] FIG. 36 shows graph of % cytotoxicity for each concentration
(dilution) of plasma,
and the results plotted in Prism. Based on the cytotoxicity curve, the
required dilution for 50% kill
(IC50) was determined.
[00081]
[00082] FIG. 37 illustrates a source animal facility and corresponding
designated pathogen
free facilities, animals, and herds in accordance with the present invention.
[00083] FIG. 38 illustrates an extracorporeal liver filter and circuit in
accordance with the
present invention.
[00084] FIG. 39 illustrates a combination skin product in accordance with
the present
invention.
[00085] FIG. 40A depicts POD-15. H&E, H&E, high power image depicts tissue
viability
with surface and follicular epithelial necrosis. FIG. 40B depicts POD-22 H&E,
high power image
demonstrating residual autograft (asterisks) with good overall viability. No
surface epithelium and
17
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
some surface necrosis noted, along with extensive fibrosis with infiltration
into the autograft
(arrows).
[00086] FIG. 41 depicts longitudinal progression of porcine split-
thickness skin graft used
as a temporary wound closure in treatment of full-thickness wound defects in a
non-human primate
recipient. Left: POD-0, xenotransplantation product at Wound Site 2. Right:
POD-30, same
xenotransplantation product at Wound Site 2.
[00087] FIG. 42 shows POD-30 histological images for: Top, Center: H&E,
Low power
image of wound site depicts complete epithelial coverage. Dotted line
surrounds the residual
xenotransplantation product.
[00088] FIG. 43A graphs the total serum IgM ELISA ( g/mL) for all four
subjects (2001,
2002, 2101, 2102) during the course of the study. FIG. 43B graphs the total
serum IgG ELISA
( g/mL) for all four subjects (2001, 2002, 2101, 2102) during the course of
the study.
[00089] FIG. 44A graphs systemic concentrations of soluble CD4OL as
measured by
Luminex 23-plex at POD-0, POD-7, POD-14, POD-21, and POD-30. FIG. 44B graphs
systemic
concentrations of TGF-alpha as measured by Luminex 23-plex at POD-0, POD-7,
POD-14, POD-
21, and POD-30. FIG. 44C graphs systemic concentrations of IL-12/23 (p40) as
measured by
Luminex 23-plex at POD-0, POD-7, POD-14, POD-21, and POD-30.
[00090] FIG. 45 illustrates a method for preparing a skin product in
accordance with the
present invention.
[00091] FIG. 46 shows a cryovial used to store a xenotransplantation
product.
[00092] FIG. 47 shows a shipping process of a xenotransplantation product.
[00093] FIG. 48 shows a secondary closure or container system for storing
a
xenotransplantation product at temperatures below ambient temperature,
including, but not limited
to, -150 degrees Celsius and other temperatures.
[00094] FIG. 49A depicts porcine split-thickness skin grafts at wound
sites 1, 2, 3, and 4,
respectively from left to right at POD-12. FIG. 49B depicts porcine split-
thickness skin grafts at
wound site 4 at POD-12 (left) and POD-14 (right).
[00095] FIG. 50A graphs MTT reduction assays fresh vs. cryopreserved (7
years) in porcine
tissue samples showing no statistical difference. FIG. 50B graphs MTT
reduction assays heat
deactivated vs. cryopreserved (7 years) in porcine tissue samples showing a
statistically significant
different in quantity of formazan produced.
18
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[00096] FIG. 51A-G shows images of a xenotransplantation product of the
present
disclosure for treatment of severe and extensive partial and full thickness
burns in a human patient.
[00097] FIG. 52 shows a graph of proliferative response of human
lymphocytes responder
peripheral blood mononuclear cells (PBMC) in the presence of mitomycin C
treated porcine
stimulator cells.
[00098] FIG. 53 shows anti-xenogeneic IgM (A) and IgG (B) antibody binding
data relative
to Median Fluorescence Intensities (MFI) for Xeno-001-00-1 patient sample at
multiple time
points, Pre, Day 7, Day 16, and Day 30. The data is shown for the plasma
samples tested at 1:2
dilutions.
DETAILED DESCRIPTION OF THE INVENTION
[00099] While aspects of the subject matter of the present disclosure may
be embodied in a
variety of forms, the following description is merely intended to disclose
some of these forms as
specific examples of the subject matter encompassed by the present disclosure.
Accordingly, the
subject matter of this disclosure is not intended to be limited to the forms
or aspects so described.
[000100] Unless otherwise defined, all technical and scientific terms used
herein have the
same meaning as commonly understood by one of ordinary skill in the art to
which this invention
belongs. Methods and materials are described herein for use in the present
invention; other,
suitable methods and materials known in the art can also be used. The
materials, methods, and
examples are illustrative only and not intended to be limiting. All
publications, patent applications,
patents, sequences, database entries, and other references mentioned herein
are incorporated by
reference in their entirety. Other features and advantages of the invention
will be apparent from
the following detailed description and figures, and from the claims.
[000101] "Best alignment" or "optimum alignment" means the alignment for
which the
identity percentage determined as described below is the highest. Comparisons
of sequences
between two nucleic acid sequences are traditionally made by comparing these
sequences after
aligning them optimally, the said comparison being made by segment or by
"comparison window"
to identify and compare local regions for similar sequences. For the
comparison, sequences may
be optimally aligned manually, or by using alignment software, e.g.,Smith and
Waterman local
homology algorithm (1981), the Neddleman and Wunsch local homology algorithm
(1970), the
Pearson and Lipman similarity search method (1988), and computer software
using these
algorithms (GAP, BESTFIT, BLAST P, BLAST N, FASTA and TFASTA in the Wisconsin
19
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Genetics Software Package, Genetics Computer Group, 575 Science Dr., Madison,
Wis.). In some
aspects, the optimum alignment is obtained using the BLAST program with the
BLOSUM 62
matrix or software having similar functionality. The "identity percentage"
between two sequences
of nucleic acids or amino acids is determined by comparing these two optimally
aligned sequences,
the sequence of nucleic acids or amino acids to be compared possibly including
additions or
deletions from the reference sequence for optimal alignment between these two
sequences. The
identity percentage is calculated by determining the number of positions for
which the nucleotide
or the amino acid residue is identical between the two sequences, by dividing
this number of
identical positions by the total number of compared positions and multiplying
the result obtained
by 100 to obtain the identity percentage between these two sequences.
[000102]
"Conservative," and its grammatical equivalents as used herein include a
conservative amino acid substitution, including substitution of an amino acid
residue by another
amino acid residue having a side chain R group with similar chemical
properties (e.g., charge or
hydrophobicity). Conservative amino acid substitutions may be achieved by
modifying a
nucleotide sequence so as to introduce a nucleotide change that will encode
the conservative
substitution. In general, a conservative amino acid substitution will not
substantially change the
functional properties of interest of a protein, for example, the ability of
MHC Ito present a peptide
of interest. Examples of groups of amino acids that have side chains with
similar chemical
properties include aliphatic side chains such as glycine, alanine, valine,
leucine, and isoleucine;
aliphatic-hydroxyl side chains such as serine and threonine; amide-containing
side chains such as
asparagine and glutamine; aromatic side chains such as phenylalanine,
tyrosine, and tryptophan;
basic side chains such as lysine, arginine, and histidine; acidic side chains
such as aspartic acid
and glutamic acid; and, sulfur-containing side chains such as cysteine and
methionine.
Conservative amino acids substitution groups include, for example,
valine/leucine/isoleucine,
phenylalanine/tyrosine, lysine/arginine,
alanine/valine, glutamate/aspartate, and
asparagine/glutamine. One skilled in the art would understand that in addition
to the nucleic acid
residues encoding a human or humanized MHC I polypeptide and/or (32
microglobulin described
herein, due to the degeneracy of the genetic code, other nucleic acid
sequences may encode the
polypeptide(s) of the invention. Therefore, in addition to a genetically
modified non-human animal
that comprises in its genome a nucleotide sequence encoding MHC I and/or (32
microglobulin
polypeptide(s) with conservative amino acid substitutions, a non- human animal
whose genome
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
comprises a nucleotide sequence(s) that differs from that described herein due
to the degeneracy
of the genetic code is also provided.
[000103] "Conserved" and its grammatical equivalents as used herein include
nucleotides or
amino acid residues of a polynucleotide sequence or amino acid sequence,
respectively, that are
those that occur unaltered in the same position of two or more related
sequences being compared.
Nucleotides or amino acids that are relatively conserved are those that are
conserved amongst more
related sequences than nucleotides or amino acids appearing elsewhere in the
sequences. Herein,
two or more sequences are said to be "completely conserved" if they are 100%
identical to one
another. In some embodiments, two or more sequences are said to be "highly
conserved" if they
are at least 70% identical, at least 80% identical, at least 90% identical, or
at least 95% identical,
but less than 100% identical, to one another. In some embodiments, two or more
sequences are
said to be "conserved" if they are at least 30% identical, at least 40%
identical, at least 50%
identical, at least 60% identical, at least 70% identical, at least 80%
identical, at least 90% identical,
or at least 95% identical, but less than 100% identical, to one another. In
some embodiments, two
or more sequences are said to be "conserved" if they are about 30% identical,
about 40% identical,
about 50% identical, about 60% identical, about 70% identical, about 80%
identical, about 90%
identical, about 95% identical, about 98% identical, or about 99% identical to
one another.
[000104] "Designated pathogen free," and its grammatical equivalents as
used herein
include reference to animals, animal herds, animal products derived therefrom,
and/or animal
facilities that are free of one or more specified pathogens. Preferably, such
"designated pathogen
free" animals, animal herds, animal products derived therefrom, and/or animal
facilities are
maintained using well-defined routines of testing for such designated
pathogens, utilizing proper
standard operating procedures (SOPs) and practices of herd husbandry and
veterinary care to
assure the absence and/or destruction of such designated pathogens, including
routines, testing,
procedures, husbandry, and veterinary care disclosed and described herein. It
will be further
understood that as used herein the term "free," and like terms when used in
connection with
"pathogen free" are meant to indicate that the subject pathogens are not
present, not alive, not
active, or otherwise not detectable by standard or other testing methods for
the subject pathogens.
[000105] "Alter," "altering," "altered" and grammatical equivalents as used
herein include
any and/or all modifications to a gene including, but not limited to,
deleting, inserting, silencing,
modifying, reprogramming, disrupting, mutating, rearranging, increasing
expression, knocking-in,
21
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
knocking out, and/or any or all other such modifications or any combination
thereof
[000106] "Endogenous loci" and its grammatical equivalents as used herein
include the
natural genetic loci found in the animal to be transformed into the donor
animal.
[000107] "Functional," e.g., in reference to a functional polypeptide, and
its grammatical
equivalents as used herein include a polypeptide that retains at least one
biological activity
normally associated with the native protein. For example, in some embodiments
of the invention,
a replacement at an endogenous locus (e.g., replacement at an endogenous non-
human MHC I,
MHC II, and/or (32 microglobulin locus) results in a locus that fails to
express a functional
endogenous polypeptide. Likewise, the term "functional" as used herein in
reference to functional
extracellular domain of a protein, can refer to an extracellular domain that
retains its functionality,
e.g., in the case of MHC I, ability to bind an antigen, ability to bind a T
cell co-receptor, etc. In
some embodiments of the invention, a replacement at the endogenous MHC locus
results in a locus
that fails to express an extracellular domain (e.g., a functional
extracellular domain) of an
endogenous MHC while expressing an extracellular domain (e.g., a functional
extracellular
domain) of a human MHC.
[000108] "Genetic or molecular marker," and their grammatical equivalents
as used herein
include polymorphic locus, i.e. a polymorphic nucleotide (a so-called single
nucleotide
polymorphism or SNP) or a polymorphic DNA sequence at a specific locus. A
marker refers to a
measurable, genetic characteristic with a fixed position in the genome, which
is normally inherited
in a Mendelian fashion, and which can be used for mapping of a trait of
interest. Thus, a genetic
marker may be a short DNA sequence, such as a sequence surrounding a single
base-pair change,
i.e. a single nucleotide polymorphism or SNP, or a long DNA sequence, such as
microsatellites or
Simple Sequence Repeats (SSRs). The nature of the marker is dependent on the
molecular analysis
used and can be detected at the DNA, RNA or protein level. Genetic mapping can
be performed
using molecular markers such as, but not limited to, RFLP (restriction
fragment length
polymorphisms; Botstein et al. (1980), Am J Hum Genet. 32:314-331; Tanksley et
al. (1989),
Bio/Technology 7:257-263), RAPD [random amplified polymorphic DNA; Williams et
al. (1990),
NAR 18:6531-6535], AFLP [Amplified Fragment Length Polymorphism; Vos et al.
(1995) NAR 23:4407-4414], SSRs or microsatellites [Tautz et al. (1989), NAR
17:6463-6471].
Appropriate primers or probes are dictated by the mapping method used.
[000109] "Improving" and its grammatical equivalents as used herein include
any
22
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
improvement recognized by one of skill in the art. For example, improving
transplantation can
mean lessening hyperacute rejection, which can encompass a decrease,
lessening, or diminishing
of an undesirable effect or symptom. In some aspects, a clinically relevant
improvement is
achieved.
[000110] "Locus" (loci plural) or "site" and their grammatical equivalents
as used herein
include a specific place or places on a chromosome where, for example, a gene,
a genetic marker
or a QTL is found.
[000111] "Minimally altered" and its grammatical equivalents as used herein
include
alteration of a donor animal genome including removing and replacing certain
distinct sequences
of native base pairs appearing on the donor animal's genome and replacing each
such sequence
with a synthetic sequence comprising the same number of base pairs, with no
net change to the
number of base pairs in the donor animal's genome, while not disturbing other
aspects of the donor
animal's native genome including, for example, introns and other codons
naturally existing in the
donor animal genome. For example, in the case of a swine as donor animal, a
minimally altered
swine can include specific alterations removing or deactivating certain SLA
exons to regulate the
donor swine cell's extracellular expression or non-expression of MEW Class II,
Ia, and/or Ib;
reprogramming certain native, naturally occurring swine cell SLA exons to
regulate the swine
cell's extracellular expression or non-expression of MEW Class II; conserving
or otherwise not
removing swine introns existing in or in the vicinity of the otherwise
engineered sequences;
increasing the expression of swine CTLA4 and PD-1; and removing or
deactivating alpha-1,3
galactosyltransferase, cytidine monophosphate-N-acetylneuraminic acid
hydroxylase, and 01,4-
N-acetylgalactosaminyltransferase.
[000112] "Minimally manipulated" and its grammatical equivalents as used
herein include
treatment of source animals, biological products derived from those source
animals, and other
biological products with minimal physical alteration of the related cells,
organs or tissues such that
such animals and products are substantially in their natural state.
[000113] "Ortholog," "orthologous," and their grammatical equivalents as
used herein
include a polynucleotide from one species that corresponds to a polynucleotide
in another species,
which has the same function as the gene or protein or QTL, but is (usually)
diverged in sequence
from the time point on when the species harboring the genes or quantitative
trait loci diverged (i.e.
the genes or quantitative trait loci evolved from a common ancestor by
speciation).
23
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000114] "Quantitative trait locus (QTL)" and its grammatical equivalents
as used herein
include a stretch of DNA (such as a chromosome arm, a chromosome region, a
nucleotide
sequence, a gene, and the like) that is closely linked to a gene that
underlies the trait in question.
"QTL mapping" involves the creation of a map of the genome using genetic or
molecular markers,
like AFLP, RAPD, RFLP, SNP, SSR, and the like, visible polymorphisms and
allozymes, and
determining the degree of association of a specific region on the genome to
the inheritance of the
trait of interest. As the markers do not necessarily involve genes, QTL
mapping results involve the
degree of association of a stretch of DNA with a trait rather than pointing
directly at the gene
responsible for that trait. Different statistical methods are used to
ascertain whether the degree of
association is significant or not. A molecular marker is said to be "linked"
to a gene or locus, if
the marker and the gene or locus have a greater association in inheritance
than would be expected
from independent assortment, i.e. the marker and the locus co-segregate in a
segregating
population and are located on the same chromosome. "Linkage" refers to the
genetic distance of
the marker to the locus or gene (or two loci or two markers to each other).
The closer the linkage,
the smaller the likelihood of a recombination event taking place, which
separates the marker from
the gene or locus. Genetic distance (map distance) is calculated from
recombination frequencies
and is expressed in centiMorgans (cM) [Kosambi (1944), Ann. Eugenet. 12:172-
175].
[000115] "Capture sequence" or "reference sequence" and their grammatical
equivalents as
used herein include a nucleic acid or amino acid sequence that has been
obtained, sequenced or
otherwise become known from a sample, animal (including humans), or
population. For example,
a capture sequence from a human patient is a "human patient capture sequence."
A capture
sequence from a particular human population is a "human population-specific
human capture
sequence." And a capture sequence from a human allele group is an "allele-
group-specific human
capture sequence."
[000116] "Humanized" and its grammatical equivalents as used herein include
embodiments
wherein all or a portion of an endogenous non-human gene or allele is replaced
by a corresponding
portion of an orthologous human gene or allele. For example, in some
embodiments, the term
"humanized" refers to the complete replacement of the coding region (e.g., the
exons) of the
endogenous non-human MHC gene or allele or fragment thereof with the
corresponding capture
sequence of the human MHC gene or allele or fragment thereof, while the
endogenous non-coding
region(s) (such as, but not limited to, the promoter, the 5' and/or 3'
untranslated region(s), enhancer
24
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
elements, etc.) of the non-human animal is not replaced.
[000117] "Personalized" or "individualized," and their grammatical
equivalents as used
herein, include a gene, allele, genome, proteome, cell, cell surface, tissue,
or organ from a non-
human animal which is adapted to the needs or special circumstances of an
individual human
recipient or a specific human recipient subpopulation.
[000118] "Reprogram," "reprogrammed," including in reference to
"immunogenomic
reprogramming," and their grammatical equivalents as used herein, refer to the
replacement or
substitution of endogenous nucleotides in the donor animal with orthologous
nucleotides based on
a separate reference sequence, wherein frameshift mutations are not introduced
by such
reprogramming. In addition, reprogramming results in no net loss or net gain
in the total number
of nucleotides in the donor animal genome, or results in a net loss or net
gain in the total number
of nucleotides in the donor animal genome that is equal to no more than 1%, no
more than 2%, no
more than 3%, no more than 4%, no more than 5%, no more than 6%, no more than
7%, no more
than 8%, no more than 9%, no more than 10%, no more than 12%, no more than
15%, or no more
than 20% of the number of nucleotides in the separate reference sequence. In
one example of
"reprogramming," an endogenous non-human nucleotide, codon, gene or fragment
thereof is
replaced with a corresponding synthetic nucleotide, codon, gene or fragment
thereof based on a
human capture sequence, through which the total number of base pairs in the
donor animal
sequence is equal to the total number of base pairs of the human capture
sequence.
[000119] "Tolerogenic" and its grammatical equivalents as used herein
include
characteristics of an organ, cell, tissue, or other biological product that
are tolerated by the reduced
response by the recipient's immune system upon transplantation.
[000120] "Transgenic" and its grammatical equivalents as used herein,
include donor animal
genomes that have been modified to introduce non-native genes from a different
species into the
donor animal's genome at a non-orthologous, non-endogenous location such that
the homologous,
endogenous version of the gene (if any) is retained in whole or in part.
"Transgene," "transgenic,"
and grammatical equivalents as used herein do not include reprogrammed
genomes, knock-outs or
other modifications as described and claimed herein. By way of example,
"transgenic" swine
include those having or expressing hCD46 ("human membrane cofactor protein,"
or "MCP"),
hCD55 ("human decay-accelerating factor," "DAF"), human B2M (beta-2-
microglobulin), and/or
other human genes, achieved by insertion of human gene sequences at a non-
orthologous, non-
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
endogenous location in the swine genome without the replacement of the
endogenous versions of
those genes.
Immunogenomic Reprogrammed Swine
[000121] As disclosed herein, tolerogenic non-human animal cells, tissues
and organs for
several human Class I and/or Class II WIC molecules are provided.
[000122] The human immune response system is a highly complex and efficient
defense
system against invading organisms. T-cells are the primary effector cells
involved in the cellular
response. Just as antibodies have been developed as therapeutics, (TCRs), the
receptors on the
surface of the T-cells, which give them their specificity, have unique
advantages as a platform for
developing therapeutics. While antibodies are limited to recognition of
pathogens in the blood and
extracellular spaces or to protein targets on the cell surface, TCRs recognize
antigens displayed by
WIC molecules on the surfaces of cells (including antigens derived from
intracellular proteins).
Depending on the subtype of T-cells that recognize displayed antigen and
become activated, TCRs
and T-cells harboring TCRs participate in controlling various immune
responses. For instance,
helper T-cells are involved in regulation of the humoral immune response
through induction of
differentiation of B cells into antibody secreting cells. In addition,
activated helper T-cells initiate
cell-mediated immune responses by cytotoxic T-cells. Thus, TCRs specifically
recognize targets
that are not normally seen by antibodies and also trigger the T-cells that
bear them to initiate wide
variety of immune responses.
[000123] It will be understood, that T-cell recognizes an antigen presented
on the surfaces of
cells by means of the TCRs expressed on their cell surface. TCRs are disulfide-
linked
heterodimers, most consisting of a and 0 chain glycoproteins. T-cells use
recombination
mechanisms to generate diversity in their receptor molecules similar to those
mechanisms for
generating antibody diversity operating in B cells (Janeway and Travers,
Immunobiology 1997).
Similar to the immunoglobulin genes, TCR genes are composed of segments that
rearrange during
development of T-cells. TCR polypeptides consist of variable, constant,
transmembrane and
cytoplasmic regions. While the transmembrane region anchors the protein and
the intracellular
region participates in signaling when the receptor is occupied, the variable
region is responsible
for specific recognition of an antigen and the constant region supports the
variable region-binding
surface. The TCR a chain contains variable regions encoded by variable (V) and
joining (J)
segments only, while the 0 chain contains additional diversity (D) segments.
26
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000124] Major histocompatibility complex Class I (MHCI) and Class II
(MHCII) molecules
display peptides on antigen-presenting cell surfaces for subsequent T-cell
recognition. See FIG. 2.
Within the human population, allelic variation among the classical MHCI and II
gene products is
the basis for differential peptide binding, thymic repertoire bias and
allograft rejection. MHC
molecules are cell-surface glycoproteins that are central to the process of
adaptive immunity,
functioning to capture and display peptides on the surface of antigen-
presenting cells (APCs).
MHC Class I (MHCI) molecules are expressed on most cells, bind endogenously
derived peptides
with sizes ranging from eight to ten amino acid residues and are recognized by
CD8 cytotoxic T-
lymphocytes (CTL). See FIG. 3 and FIG. 4. On the other hand, MHC Class II
(MHCII) are present
only on specialized APCs, bind exogenously derived peptides with sizes varying
from 9 to 22
residues, and are recognized by CD4 helper T-cells. See FIG. 5. These
differences indicate that
MHCI and MHCII molecules engage two distinct arms of the T-cell-mediated
immune response,
the former targeting invasive pathogens such as viruses for destruction by CD8
CTLs, and the
latter inducing cytokine-based inflammatory mediators to stimulate CD4 helper
T-cell activities
including B-cell activation, maturation and antibody production. In some
aspects, the biological
product of the present disclosure is not recognized by CD8+ T cells, do not
bind anti-HLA
antibodies, and are resistant to NK-mediated lysis.
[000125] The human leukocyte antigen (HLA) system or complex is a gene
complex
encoding the major histocompatibility complex (MHC) proteins in humans. These
cell-surface
proteins are responsible for the regulation of the immune system in humans.
The HLA gene
complex resides on a 3 Mbp stretch within chromosome 6p21. See FIG. 6. HLA
genes are highly
polymorphic, which means that they have many different alleles, allowing them
to fine-tune the
adaptive immune system. See FIG. 7. The proteins encoded by certain genes are
also known as
antigens, as a result of their historic discovery as factors in organ
transplants. Different classes
have different functions. See FIG. 8 and FIG. 9.
[000126] The HLA segment is divided into three regions (from centromere to
telomere),
Class II, Class III and Class I. See FIG. 10. Classical Class I and Class II
HLA genes are contained
in the Class I and Class II regions, respectively, whereas the Class III locus
bears genes encoding
proteins involved in the immune system but not structurally related to MHC
molecules. The
classical HLA Class I molecules are of three types, HLA-A, HLA-B and HLA-C.
Only the a chains
of these mature HLA Class I molecules are encoded within the Class I HLA locus
by the respective
27
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
HLA-A, HLA-B and HLA-C genes. See Fig. 11. In contrast, the beta-2
microglobulin f32m chain
encoded by the f32m gene is located on chromosome 15. The classical HLA Class
II molecules are
also of three types (HLA-DP, HLA-DQ and HLA-DR), with both the a and f3 chains
of each
encoded by a pair of adjacent loci. In addition to these classical HLA Class I
and HLA Class II
genes, the human MHC locus includes a long array of HLA pseudogenes as well as
genes encoding
non-classical MHCI and MHCII molecules. HLA-pseudogenes are an indication that
gene
duplication is the main driving force for HLA evolution, whereas non-classical
MHCI and MHCII
molecules often serve a restricted function within the immune system quite
distinct from that of
antigen presentation to af3 TCRs.
[000127] Aside from the genes encoding the antigen-presenting proteins,
there are a large
number of other genes, many involved in immune function, located on the HLA
complex.
Diversity of HLAs in the human population is one aspect of disease defense,
and, as a result, the
chance of two unrelated individuals with identical HLA molecules on all loci
is extremely low.
HLA genes have historically been identified as a result of the ability to
successfully transplant
organs between HLA-similar individuals.
[000128] Class I MHC molecules are expressed on all nucleated cells,
including tumor cells.
They are expressed specifically on T and B lymphocytes, macrophages, dendritic
cells and
neutrophils, among other cells, and function to display peptide fragments
(typically 8-10 amino
acids in length) on the surface to CD8+ cytotoxic T lymphocytes (CTLs). CTLs
are specialized to
kill any cell that bears an MHC I-bound peptide recognized by its own membrane-
bound TCR.
When a cell displays peptides derived from cellular proteins not normally
present (e.g., of viral,
tumor, or other non-self origin), such peptides are recognized by CTLs, which
become activated
and kill the cell displaying the peptide.
[000129] As shown in FIG. 12, MHC Class I protein comprises an
extracellular domain
(which comprises three domains: al, az and a3), a transmembrane domain, and a
cytoplasmic tail.
The al and az domains form the peptide-binding cleft, while the a3 interacts
with 02-microglobulin.
Class I molecules consist of two chains: a polymorphic a-chain (sometimes
referred to as heavy
chain) and a smaller chain called 02-microglobulin (also known as light
chain), which is generally
not polymorphic. These two chains form a non-covalent heterodimer on the cell
surface. The cc-
chain contains three domains (al, a2 and a3). As illustrated in FIG. 12, Exon
1 of the a-chain gene
encodes the leader sequence, exons 2 and 3 encode the al and a2 domains, exon
4 encodes the a3
28
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
domain, exon 5 encodes the transmembrane domain, and exons 6 and 7 encode the
cytoplasmic
tail. The a-chain forms a peptide-binding cleft involving the al and a2
domains (which resemble
Ig-like domains) followed by the a3 domain, which is similar to (32-
microglobulin.
[000130] (32 microglobulin is a non-glycosylated 12 kDa protein; one of its
functions is to
stabilize the MEW Class I a-chain. Unlike the a-chain, the (32 microglobulin
does not span the
membrane. The human (32 microglobulin locus is on chromosome 15 and consists
of 4 exons and
3 introns. Circulating forms of 132 microglobulin are present in serum, urine,
and other body fluids;
non-covalently MHC I-associated 132 microglobulin can be exchanged with
circulating 132
microglobulin under physiological conditions.
[000131] As shown in FIG. 13, MHC Class II protein comprises an
extracellular domain
(which comprises three domains: al, az , 131, and (31), a transmembrane
domain, and a cytoplasmic
tail. The al and 131 domains form the peptide-binding cleft, while the al and
131 interacts with the
transmembrane domain.
[000132] In addition to the aforementioned antigens, the Class I antigens
include other
antigens, termed non-classical Class I antigens, in particular the antigens
HLA-E, HLA-F and
HLA-G; this latter, in particular, is expressed by the extravillous
trophoblasts of the normal human
placenta in addition to HLA-C.
Cell Phenotype
[000133] Referring generally to FIG. 1, Dr. Peter Medawar profoundly said
"the success of
human pregnancy, where the fetus resides comfortably within the maternal
uterus for 9 months,
defies the precepts of immunology." Paraphrasing, he observed that the most
common, successful
transplant on earth is pregnancy.
[000134] The trophoblast expression of cell surface markers is well
characterized, and by
replicating such phenotype in the porcine cell where appropriate and necessary
to retain, critical
and desired cellular function can be obtained. According to literature,
extravillous trophoblast cells
express HLA Class Ia molecule (HLA-C) and all of HLA Class lb molecules.
Compared to HLA-
E and HLA-G, both of which are highly expressed on extravillous trophoblast
cells, HLA-C and
HLA-F are weakly expressed. See, e.g., Djurisic et at., "HLA Class lb
Molecules and Immune
Cells in Pregnancy and Preeclampsia," Frontiers in Immunology, Vol 5, Art. 652
(2014). In
addition to MHC molecules, PD-Li is upregulated in trophoblastic cells in
normal pregnancy,
particularly in syncytiotrophoblast cells. HLA Class II molecules are not
present on trophoblasts,
29
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
which may facilitate survival and detection of the embryo in the presence of
maternal
lymphocytes. See, e.g., 1/eras et at., "PD-Li Expression in Human Placentas
and gestational
Trophoblastic Diseases," Int. I Gynecol. Pathol. 36(2): 146-153 (2017).
[000135] The present invention provides a method of creating a tolerogenic
xenotransplantation swine cell that mimics the extracellular configuration of
a human trophoblast.
This method includes, but is not limited to, removing or deactivating certain
SLA exons to regulate
the swine cell's extracellular expression or non-expression of MHC Class II,
Ia, and/or Ib;
reprogramming certain native, naturally occurring swine cell SLA exons to
regulate the swine
cell's extracellular expression or non-expression of MHC Class II; conserving
or otherwise not
removing swine introns existing in or in the vicinity of the otherwise
engineered sequences;
increasing the expression of swine CTLA4 and PD-1; and removing or
deactivating alpha-1,3
galactosyltransferase, cytidine monophosphate-N-acetylneuraminic acid
hydroxylase, and 01,4-
N-acetylgalactosaminyltransferase. Such removal, reprogramming, and
modification to cause such
increase of expression, and other engineered aspects of a swine genome, to
create a tolerogenic
xenotransplantation swine cell that mimics the extracellular configuration of
a human trophoblast,
is described as follows.
[000136] The former and current attempts to this unmet clinical need has
precisely followed
the classic medical dogma of "one-size fits all". We refer to this as the
"downstream" approach -
which must contend with addressing all of the natural immune processes in
sequence. Instead of
adopting this limited view, the present invention takes a "patient-specific"
solution to dramatically
improve clinical outcome measures. The latter, our approach, we term the
"upstream" approach -
one which represents the culmination of unfilled scientific effort into a
coordinated translational
effort. The central theorem of our approach is countervailing to the existing
and previous dogmatic
approaches. The "downstream" approach accepts the innate and immovable
disparity between
donor and recipient, and focuses on interventions, gene alterations, and/or
concomitant exogenous
immunosuppressive medications used as a method of
reducing/eliminating/negatively-altering the
recipients' naturally resulting immunologic response. In contrast, we
intentionally choose to
reverse the focus of the otherwise area of fundamental scientific dogma.
Rather than accept the
immunological incompatibilities between the donor and recipient, specifically
(but not limited to)
those mismatches of the Major Histocompatibility Complex(es), we alter these
catalytic antigens
at the source, thereby eliminating all of the precipitating mechanisms that
are the causative
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
effectors of cell, tissue, and organ rejection between donor and recipient.
This approach applies
beyond the field of xenotransplantation including, but not limited to, the
fields of genetics,
obstetrics, infectious disease, oncology, agriculture, animal husbandry, food
industry and other
areas.
[000137] The present disclosure embodies the above modification in creating
a non-
transgenic genetically reprogrammed swine for xenotransplantation, wherein the
MEW surface
characterization of the swine mimic that of the recipient's trophoblast,
wherein the immune
response from the xenotransplantation is significantly reduced. The human
extravillous
trophoblast cells express HLA-C, HLA-E, HLA-F, and HLA-G, but not HLA-A, HLA-
B, HLA-
DQ and HLA-DR. As such, the current embodiment combines the unique MEW surface
characterization of human trophoblast with site-directed mutagenic
substitutions to minimize or
remove the immune response associated with xenotransplantation while
minimizing off target
effects on the native donor swine's SLANIFIC gene.
[000138] The human immune response system is a highly complex and efficient
defense
system against invading organisms. T-cells are the primary effector cells
involved in the cellular
response. Just as antibodies have been developed as therapeutics, (TCRs), the
receptors on the
surface of the T-cells, which give them their specificity, have unique
advantages as a platform for
developing therapeutics. While antibodies are limited to recognition of
pathogens in the blood and
extracellular spaces or to protein targets on the cell surface, TCRs recognize
antigens displayed by
MEW molecules on the surfaces of cells (including antigens derived from
intracellular proteins).
Depending on the subtype of T-cells that recognize displayed antigen and
become activated, TCRs
and T-cells harboring TCRs participate in controlling various immune
responses. For instance,
helper T-cells are involved in regulation of the humoral immune response
through induction of
differentiation of B cells into antibody secreting cells. In addition,
activated helper T-cells initiate
cell-mediated immune responses by cytotoxic T-cells. Thus, TCRs specifically
recognize targets
that are not normally seen by antibodies and also trigger the T-cells that
bear them to initiate wide
variety of immune responses.
[000139] As shown in FIG. 2, a T-cell recognizes an antigen presented on
the surfaces of
cells by means of the TCRs expressed on their cell surface. TCRs are disulfide-
linked
heterodimers, most consisting of a and 0 chain glycoproteins. T-cells use
recombination
mechanisms to generate diversity in their receptor molecules similar to those
mechanisms for
31
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
generating antibody diversity operating in B cells (Janeway and Travers,
Immunobiology 1997).
Similar to the immunoglobulin genes, TCR genes are composed of segments that
rearrange during
development of T-cells. TCR polypeptides consist of variable, constant,
transmembrane and
cytoplasmic regions. While the transmembrane region anchors the protein and
the intracellular
region participates in signaling when the receptor is occupied, the variable
region is responsible
for specific recognition of an antigen and the constant region supports the
variable region-binding
surface. The TCR a chain contains variable regions encoded by variable (V) and
joining (J)
segments only, while the I chain contains additional diversity (D) segments.
[000140] A TCR recognizes a peptide antigen presented on the surfaces of
antigen presenting
cells in the context of self- Major Histocompatibility Complex(MHC) molecules.
Two different
types of MHC molecules recognized by TCRs are involved in antigen
presentation, the Class I
MHC and class II MHC molecules. Mature T-cell subsets are defined by the co-
receptor molecules
they express. These co-receptors act in conjunction with TCRs in the
recognition of the MHC-
antigen complex and activation of the T-cell. Mature helper T-cells recognize
antigen in the
context of MHC Class II molecules and are distinguished by having the co-
receptor CD4.
Cytotoxic T-cells recognize antigen in the context of MHC Class I determinants
and are
distinguished by having the CD8 co-receptor.
[000141] In the human, MHC molecules are referred to as HLA, an acronym for
human
leukocyte antigens, and are encoded by the chromosome 6p21.3- located HLA
region.8,9 The HLA
segment is divided into three regions (from centromere to telomere), Class II,
Class III and Class
I. See FIG. 10. Classical Class I and Class II HLA genes are contained in the
Class I and Class II
regions, respectively, whereas the Class III locus bears genes encoding
proteins involved in the
immune system but not structurally related to MHC molecules. The classical HLA
Class I
molecules are of three types, HLA-A, HLA-B and HLA-C. Only the a chains of
these mature HLA
Class I molecules are encoded within the Class I HLA locus by the respective
HLA-A, HLA-B
and HLA-C genes. See FIG. 11. In contrast, the beta-2 microglobulin (32m chain
encoded by the
(32m gene is located on chromosome 15. The classical HLA Class II molecules
are also of three
types (HLA-DP, HLA-DQ and HLA-DR), with both the a and I chains of each
encoded by a pair
of adjacent loci. In addition to these classical HLA Class I and HLA Class II
genes, the human
MHC locus includes a long array of HLA pseudogenes as well as genes encoding
non-classical
MHCI and MHCII molecules. HLA-pseudogenes are an indication that gene
duplication is the
32
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
main driving force for HLA evolution, whereas non-classical MHCI and MHCII
molecules often
serve a restricted function within the immune system quite distinct from that
of antigen
presentation to c43 TCRs.
[000142] Human leukocyte antigen (HLA) genes show incredible sequence
diversity in the
human population. For example, there are >4,000 known alleles for the HLA-B
gene alone. The
genetic diversity in HLA genes in which different alleles have different
efficiencies for presenting
different antigens is believed to be a result of evolution conferring better
population-level
resistance against the wide range of different pathogens to which humans are
exposed. This genetic
diversity also presents problems during xenotransplantation where the
recipient's immune
response is the most important factor dictating the outcome of engraftment and
survival after
transplantation.
[000143] In humans, the classical Class I genes, termed HLA-A, HLA-B and
HLA-C, consist
of two chains: a polymorphic a-chain (sometimes referred to as heavy chain)
and a smaller chain
called (32-microglobulin (also known as light chain), which is generally not
polymorphic. These
two chains form a non-covalent heterodimer on the cell surface. As shown in
FIG. 12, the a-chain
contains three domains (al, a2 and a3). Exon 1 of the a-chain gene encodes the
leader sequence,
exons 2 and 3 encode the al and a2 domains, exon 4 encodes the a3 domain, exon
5 encodes the
transmembrane domain, and exons 6 and 7 encode the cytoplasmic tail. The a-
chain forms a
peptide-binding cleft involving the al and a2 domains (which resemble Ig-like
domains) followed
by the a3 domain, which is similar to (32-microglobulin.
[000144] (32 microglobulin is a non-glycosylated 12 kDa protein; one of its
functions is to
stabilize the MHC Class I a-chain. Unlike the a-chain, the (32 microglobulin
does not span the
membrane. The human (32 microglobulin locus is on chromosome 15 and consists
of 4 exons and
3 introns. (32-microglobulin-bound protein complexes undertake key roles in
various immune
system pathways, including the neonatal Fc receptor (FcRn), cluster of
differentiation 1 (CD1)
protein, non-classical major histocompatibility complex (MHC), and well-known
MHC Class I
molecules.
[000145] Class I MHC molecules are expressed on all nucleated cells,
including tumor cells.
They are expressed specifically on T and B lymphocytes, macrophages, dendritic
cells and
neutrophils, among other cells, and function to display peptide fragments
(typically 8-10 amino
acids in length) on the surface to CD8+ cytotoxic T lymphocytes (CTLs). CTLs
are specialized to
33
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
kill any cell that bears an MHC I-bound peptide recognized by its own membrane-
bound TCR.
When a cell displays peptides derived from cellular proteins not normally
present (e.g., of viral,
tumor, or other non-self origin), such peptides are recognized by CTLs, which
become activated
and kill the cell displaying the peptide.
[000146] MHC loci exhibit the highest polymorphism in the genome. All Class
I and II MHC
genes can present peptide fragments, but each gene expresses a protein with
different binding
characteristics, reflecting polymorphisms and allelic variants. Any given
individual has a unique
range of peptide fragments that can be presented on the cell surface to B and
T cells in the course
of an immune response.
[000147] In addition to the aforementioned antigens, the Class I antigens
include other
antigens, termed non-classical Class I antigens, in particular the antigens
HLA-E, HLA-F and
HLA-G; this latter, in particular, is expressed by the extravillous
trophoblasts of the normal human
placenta in addition to HLA-C.
[000148] MHC Class II protein comprises an extracellular domain (which
comprises three
domains: al, a2 , (31, and (31), a transmembrane domain, and a cytoplasmic
tail as shown in FIG.
13. The a2 and 132 domains form the peptide-binding cleft, while the al and
131 interacts with the
transmembrane domain.
[000149] With respect to the MHC-I proteins, the current disclosure either
inactivate, or
where necessary to retain the function of the "find and replace" orthologous
SLA proteins with
HLA analogs that would result in minimal immune recognition. In some aspects,
silencing the
genes which encode and are responsible for the expression of SLA-1 removes the
highly-
problematic and polymorphic HLA-A analog. Similarly, inactivation or complete
removal of genes
associated with SLA-2 would reduce the burden imposed by mismatched HLA-B
proteins. This
would, at the cell surface interface, appear to the human recipient's T cells
as a HLA-A and HLA-
B negative cell. With respect to the last of the classical MHC Class I
proteins, HLA-C, site-directed
mutagenesis of genes that encode for SLA-3 using a reference HLA-C sequence
would mimic an
allo-transplant with such a disparity. Given the "less- polymorphic" nature of
HLA-C, as compared
to HLA-A and HLA-B, this would be further improved by the replacement of SLA-3
with a
reference replacement sequence based on the subclass of HLA-C that is
naturally prevalent in
nature, and also invoking mechanisms that would allow for the minimal but
requisite level of
34
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
expression that would afford functionality and non-interruption of the
numerous known and also
those unknown MHC-I dependent processes.
[000150] With respect to the MHC-I proteins, the current disclosure either
inactivate, or
where necessary to retain the function of the "find and replace" orthologous
SLA proteins with
HLA analogs that would result in minimal immune recognition. In some aspects,
silencing the
genes which encode and are responsible for the expression of SLA-1 removes the
highly-
problematic and polymorphic HLA-A analog. Similarly, inactivation or complete
removal of genes
associated with SLA-2 would reduce the burden imposed by mismatched HLA-B
proteins. This
would, at the cell surface interface, appear to the human recipient's T cells
as a HLA-A and HLA-
B negative cell. With respect to the last of the classical MHC Class I
proteins, HLA-C, site-directed
mutagenesis of genes that encode for SLA-3 using a reference HLA-C sequence
would mimic an
allo-transplant with such a disparity. Given the "less- polymorphic" nature of
HLA-C, as compared
to HLA-A and HLA-B, this would be further improved by the replacement of SLA-3
with a
reference replacement sequence based on the subclass of HLA-C that is
naturally prevalent in
nature, and also invoking mechanisms that would allow for the minimal but
requisite level of
expression that would afford functionality and non-interruption of the
numerous known and also
those unknown MHC-I dependent processes.
[000151] Furthermore, the expression of non-classical MHC proteins - those
included in the
I-b category, which include HLA-E, F, and G are vitally important to both the
survival of the fetus
and synergistic existence of the trophoblast(s). Fortunately, these are
significantly less
polymorphic than the "classical" MHC-Ia variety. Without expression of these,
heightened
upregulation of cell lysis is a direct result of NK cell recognition and
activation is observed. In an
identical manner as described to the MHC-Ia components, the orthologous SLA
proteins with HLA
analogs are either inactivated, or where necessary, to "find and replace(d)"
FIG. 14 shows specific
alterations that are included in the present disclosure.
[000152] HLA-G can be a potent immuno-inhibitory and tolerogenic molecule.
HLA-G
expression in a human fetus can enable the human fetus to elude the maternal
immune response.
Neither stimulatory functions nor responses to allogeneic HLA-G have been
reported to date.
HLA-G can be a non-classical HLA Class I molecule. It can differ from
classical MHC Class I
molecules by its genetic diversity, expression, structure, and function. HLA-G
can be characterized
by a low allelic polymorphism. Expression of HLA-G can be restricted to
trophoblast cells, adult
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
thymic medulla, and stem cells. The sequence of the HLA-G gene (HLA-6.0 gene)
has been
described by GERAGHTY et al., (Proc. Natl. Acad. Sci. USA, 1987, 84, 9145-
9149): it comprises
4,396 base pairs and exhibits an intron/exon organization which is homologous
to that of the HLA-
A, HLA-B and HLA-C genes. More precisely, this gene comprises 8 exons and an
untranslated,
3'UT, end, with the following respective correspondence: exon 1: signal
sequence, exon 2: al
domain, exon 3: a2 domain, exon 4: a3 domain, exon 5: transmembrane region,
exon 6:
cytoplasmic domain I, exon 7: cytoplasmic domain II, exon 8: cytoplasmic
domain III and 3'
untranslated region (GERAGHTY et al., mentioned above, ELLIS et al., J.
Immunol., 1990, 144,
731-735). However, the HLA-G gene differs from the other Class I genes in that
the in-frame
translation termination codon is located at the second codon of exon 6; as a
result, the cytoplasmic
region of the protein encoded by this gene HLA-6.0 is considerably shorter
than that of the
cytoplasmic regions of the HLA-A, HLA-B and HLA-C proteins.
[000153] Natural killer (NK) cell-mediated immunity, comprising
cytotoxicity and cytokine
secretion, plays a major role in biological resistance to a number of
autologous and allogeneic
cells. The common mechanism of target cell recognition appears to be the lack
or modification of
self MHC Class 1-peptide complexes on the cell surface, which can lead to the
elimination of
virally infected cells, tumor cells and major histocompatibility MHC-
incompatible grafted cells.
KIR's, members of the Ig superfamily which are expressed on NK cells, have
recently been
discovered and cloned. KIR's are specific for polymorphic MHC Class I
molecules and generate a
negative signal upon ligand binding which leads to target cell protection from
NK cell-mediated
cytotoxicity in most systems. In order to prevent NK cell autoimmunity, i.e.,
the lysis of normal
autologous cells, it is believed that every given NK cell of an individual
expresses at least on KIR
recognizing at least one of the autologous HLA-A, B, C, or G alleles.
[000154] According to the present disclosure, in the context of swine-to-
human
xenotransplantation, each human recipient will have a major histocompatibility
complex (MHC)
(Class I, Class II and/or Class III) that is unique to that individual and
will not match the MHC of
the donor swine. Accordingly, when a donor swine graft is introduced to the
recipient, the swine
MHC molecules themselves act as antigens, provoking an immune response from
the recipient,
leading to transplant rejection.
[000155] According to this aspect of the present disclosure (i.e.,
reprogramming the
SLA/MHC to express specifically selected human MHC alleles), when applied to
swine cells,
36
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
tissues, and organs for purposes of xenotransplantation will decrease
rejection as compared to
cells, tissues, and organs derived from a wild-type swine or otherwise
genetically modified swine
that lacks this reprogramming, e.g., transgenic swine or swine with non-
specific or different
genetic modifications.
[000156] With the previous modifications incorporated, insertion or
activation of additional
extracellular ligands that would create a protective, localized immune
response as seen with the
maternal-fetal symbiosis, would be an additional step to minimize deleterious
cellular-mediated
immunological functions that may remain as a result of minor-antigen
disparities. Therefore,
porcine ligands for SLA-MIC2 is orthologously reprogrammed with human
counterparts, MICA.
Human Major Histocompatibility Complex Class I Chain-Related gene A (MICA) is
a cell surface
glycoprotein expressed on endothelial cells, dendritic cells, fibroblasts,
epithelial cells, and many
tumours. It is located on the short arm of human chromosome 6 and consists of
7 exons, 5 of which
encodes the transmembrane region of the MICA molecule. MICA protein at normal
states has a
low level of expression in epithelial tissues but is upregulated in response
to various stimuli of
cellular stress. MICA is classified as a non-classical MHC Class I gene ,and
functions as a ligand
recognized by the activating receptor NKG2D that is expressed on the surface
of NK cells and
CD8+ T cells (atlasgeneticsoncology.org/Genes/MICAID41364ch6p21.html).
[000157] In addition, porcine ligands for PD-L1, CTLA-4, and others are
overexpressed
and/or otherwise orthologously reprogrammed with human counterparts. PD-Li is
a
transmembrane protein that has major role in suppressing the adaptive immune
system in
pregnancy, allografts, and autoimmune diseases. It is encoded by the CD274
gene in human and
is located in chromosome 9. PD-Li binds to PD-1, a receptor found on activated
T cells, B cells,
and myeloid cells, to modulate activation or inhibition. Particularly, the
binding of PD-Li to
receptor PD-1 on T cells inhibits activation of IL-2 production and T cell
proliferation. CTLA4 is
a protein receptor that also functions as an immune checkpoint that
downregulates immune
responses. It is encoded by the CTLA4 gene and is located in chromosome 2 in
human. It is
constitutively expressed on regulatory T cells but are upregulated in
activated T cells. Gene
expression for CTLA-4 and PD-Li is increased, for example, based on
reprogramming promoters
thereof. There is a relationship between genotype and CTLA-4 or PD-Li
expression. For
example, individuals carrying thymine at position -318 of the CTLA4 promoter
(T(-318)) and
homozygous for adenine at position 49 in exon 1 showed significantly increased
expression both
37
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
of cell-surface CTLA-4 after cellular stimulation and of CTLA-4 mRNA in non-
stimulated cells
in Ligers A, et al. CTLA-4 gene expression is influenced by promoter and exon
1 polymorphisms,
Genes Immun. 2001 May;2(3):145-52, which is incorporated herein by reference
in its entirety for
all purposes. A similar upregulation can be achieved to overexpress PD-Li
using a PD-Li
promoter reprogramming.
[000158] Further, anti-coagulant porcine ligands for Endothelial protein C
receptor (EPCR),
Thrombomodulin (TBM), Tissue Factor Pathway Inhibitor(TFPI), and others are
orthologously
reprogrammed with human counterparts, as shown in FIG. 14. Endothelial protein
C receptor is
endothelial cell-specific transmembrane glycoprotein encoded by PROCR gene
that is located in
chromosome 20 in human. It enhances activation of Protein C, an anti-coagulant
serine protease,
and has crucial role in activated protein C mediated cytoprotive signaling.
Thrombomodulin is an
integral membrane glycoprotein present on surface of endothelial cells. It is
encoded by THBD
gene that is located in chromosome 20 in human. In addition to functioning as
cofactor in the
thrombin-induced activation of protein C in the anticoagulant pathway, it also
functions in
regulating C3b inactivation. Tissue Factor Pathway Inhibitor (TFPI) is a
glycoprotein that
functions as natural anticoagulant by inhibiting Factor Xa. It encoded by TFPI
gene located in
chromosome 2 in human and the protein structure consists of three tandemly
linked Kunitz
domains. In human, two major isoforms of TFPi exists, TFPIa and TFPIP. TFPIa
consists of three
inhibitory domains (K1, K2, and K3) and a positively charged C terminus while
TFPIP consists of
two inhibitory domains (K1 and K2) and C terminus. While K1 and K2 domains are
known to
bind and inhibit Factor VII and Factor Xa, respectively, the inhibitory
function of K3 is unknown.
In certain aspects, the present disclosure centralizes (predicates) the
creation of hypoimmunogenic
and/or tolerogenic cells, tissues, and organs that does not necessitate the
transplant recipients'
prevalent and deleterious use of exogenous immunosuppressive drugs (or
prolonged
immunosuppressive regimens) following the transplant procedure to prolong the
life-saving organ.
[000159] The table provided in FIG. 14 shows conceptual design that exhibit
summation of
various edits to create tolerogenic xenotransplantation swine cell that mimics
the extracellular
configuration of a human trophoblast. As exhibited in the FIG. 14, SLA-1, a
swine gene
orthologous to HLA-A, is silenced to mimic trophoblast, as HLA-A is not
expressed on
trophoblast. As further exhibited in the FIG. 14, SLA-8, a swine gene
orthologous to HLA-G, is
humanized through replacement with "human-capture" reference sequence, as HLA-
G is
38
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
expressed in trophoblast and has crucial role in maternal fetal tolerance,
given its interaction with
NK cells.
[000160] It is therefore understood that multiple source animals, with an
array of biological
properties including, but not limited to, genome modification and/or other
genetically engineered
properties, can be utilized to reduce immunogenicity and/or immunological
rejection (e.g., acute,
hyperacute, and chronic rejections) in humans resulting from
xenotransplantation. In certain
aspects, the present disclosure can be used to reduce or avoid thrombotic
microangiopathy by
transplanting the biological product of the present disclosure into a human
patient. In certain
aspects, the present disclosure can be used to reduce or avoid glomerulopathy
by transplanting the
biological product of the present disclosure into a human patient. It will be
further understood that
the listing of source animals set forth herein is not limiting, and the
present invention encompasses
any other type of source animal with one or more modifications (genetic or
otherwise) that serve(s)
to reduce immunogenicity and/or immunological rejection, singularly or in
combination.
Bioinformatic Sequence Analysis Comparing Identities of Conserved and Non-
Conserved
Nucleotides between Human versus Swine Genomes at Various Immunologically
Critical Loci
[000161] To reprogram the MHC disparities between the Swine Leukocyte
Antigen (SLA)
and the Human Leukocyte Antigen (HLA), the present disclosure includes using
highly conserved
MHC-loci between these two species, e.g., numerous genes that correspond in
function. The MHC
Class Ia, HLA-A, HLA-B, and HLA-C have an analogous partner in the swine (the
SLA 1, 2 and
3 respectively). In MHC Class II there are also numerous matches to be
utilized during
immunogenomic reprogramming according to the present disclosure.
[000162] As illustrated in FIG. 15, MHC genes are categorized into three
classes; Class I,
Class II, and Class III, all of which are encoded on human chromosome 6. The
MHC genes are
among the most polymorphic genes of the swine and human genomes, MHC
polymorphisms are
presumed to be important in providing evolutionary advantage; changes in
sequence can result in
differences in peptide binding that allow for better presentation of pathogens
to cytotoxic T cells.
[000163] The known human HLA/MHC or an individual recipient's sequenced
HLA/MHC
sequence(s) may be utilized as a template to reprogram with precise
substitution the swine
leukocyte antigen (SLA)/MHC sequence to match, e.g., to have 80%, 85%, 90%,
95%, 98%, 99%,
or 100% sequence homology to a known human HLA/MHC sequence or the human
recipient's
HLA/MHC sequence. Upon identifying a known human recipient HLA/MHC sequence to
be used
39
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
or performing genetic sequencing of a human recipient to obtain HLA/MHC
sequences, 3
reprogramming may be performed to SLA/MHC sequences in cells of the swine
based on desired
HLA/MHC sequences. For example, several targeting guide RNA (gRNA) sequences
are
administered to the swine of the present disclosure to reprogram SLA/MHC
sequences in cells of
the swine with the template HLA/MHC sequences of the human recipient.
[000164] The term "MHC I complex" or the like, as used herein, includes the
complex
between the WIC I a chain polypeptide and the 02-microglobulin polypeptide.
The term "MHC I
polypeptide" or the like, as used herein, includes the MHC I a chain
polypeptide alone. Typically,
the terms "human MHC" and "HLA" can be used interchangeably.
[000165] For purposes of modifying donor SLANIFIC to match recipient
HLA/MHC,
comparative genomic organization of the human and swine histocompatibility
complex has been
mapped as illustrated in FIG. 16 and FIG. 17. For example, such SLA to HLA
mapping can be
found in: Lunney, J., "Molecular genetics of the swine major
histocompatibility complex, the SLA
complex," Developmental and Comparative Immunology 33: 362-374 (2009)
("Lunney"), the
entire disclosure of which is incorporated herein by reference. Further, by
comparing the loci of
HLA and schematic molecular organization of various HLA genes, as illustrated
in FIG. 12 and
FIG. 13, with the loci of SLA and schematic molecular organization of various
SLA genes, as
show in FIG. 17 and FIG. 18, it is readily discernible that the placement and
number of exons in
extracellular and transmembrane domain is common between HLA MHC and SLA MHC.
Accordingly, a person of ordinary skill in the art effectively and efficiently
genetically reprogram
swine cells in view of the present disclosure and using the mapping of Lunney
et al. as a reference
tool.
[000166] The donor swine's SLANIFIC gene is used as a reference template in
creating the
replacement template. In implementing the present disclosure, the swine's
SLANIFIC gene may
be obtained through online archives or database such as Ensembl
(http://vega.archive.ensembl.org/index.html). As illustrated in FIG. 19, FIG.
20, FIG. 21, and FIG.
22, the exact location of the SLA-DQA and SLA-DQB1 gene, the length of the
respective gene
(exon and intron), and the exact nucleotide sequences of SLA-DQA and SLA-DQB1
are mapped.
In an alternative aspect of the present disclosure, the donor swine's SLANIFIC
gene may be
sequenced. In an alternative aspect of the present disclosure, the swine's
whole genome may be
sequenced. In one aspect, the sequenced SLA/MHC gene of the donor swine that
can be used as a
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
reference template include but are not limited to SLA-3, SLA-6, SLA-7, SLA-8,
SLA-DQa, SLA-
DQb, and beta-2 microglobulin. In another aspect, the sequenced SLA/MHC gene
of the donor
swine that can be used as a base template include but are not limited exon
regions of SLA-3, SLA-
6, SLA-7, SLA-8, SLA-DQa, SLA-DQb, and beta-2 microglobulin. In some aspects,
other SLAs
are unaltered and intron regions of the reprogrammed SLA regions are
unaltered, thereby
producing a minimally altered reprogrammed swine genome that provides cells,
tissues and organs
that are tolerogenic when transplanted into a human.
[000167] In accordance with one aspect the present invention, a donor swine
is provided with
a genome that is biologically engineered to express a specific set of known
human HLA molecules.
Such HLA sequences can be obtained, e.g., from the IPD-IMGT/HLA database
(available at
ebi.ac.uk/ipd/imgt/h1a/) and the international ImMunoGeneTics information
system (available
at imgt.org). Nomenclature for such genes is illustrated in FIG. 23. For
example, HLA-A1, B8,
DR17 is the most common HLA haplotype among Caucasians, with a frequency of
5%. Thus, the
disclosed method can be performed using the known MHC/HLA sequence information
in
combination with the disclosures provided herein. The HLA sequences are
obtainable through
online archives or database such as Ensembl
(vega.archive.ensembl.org/index.html). As illustrated
in FIG. 24, the exact location of the HLA-DQA1 gene, the length of the
respective gene(exon and
intron), and the exact nucleotide sequences of HLA-DQA1 could be obtained.
[000168] In some aspects, the recipient's human leukocyte antigen (HLA)
genes and MHC
(Class I, II and/or III), are identified and mapped. It will be understood
that ascertaining the human
recipient's HLA/MHC sequence can be done in any number of ways known in the
art. For example,
HLA/MHC genes are usually typed with targeted sequencing methods: either long-
read
sequencing or long-insert short-read sequencing. Conventionally, HLA types
have been
determined at 2-digit resolution (e.g., A*01), which approximates the
serological antigen
groupings. More recently, sequence specific oligonucleotide probes (S SOP)
method has been used
for HLA typing at 4-digit resolution (e.g., A*01:01), which can distinguish
amino acid differences.
Currently, targeted DNA sequencing for HLA typing is the most popular approach
for HLA typing
over other conventional methods. Since the sequence-based approach directly
determines both
coding and non-coding regions, it can achieve HLA typing at 6-digit (e.g.,
A*01:01:01) and 8-
digit (e.g., A*01:01:01:01) resolution, respectively. HLA typing at the
highest resolution is
desirable to distinguish existing HLA alleles from new alleles or null alleles
from clinical
41
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
perspective. Such sequencing techniques are described in, for example, Elsner
HA, Blasczyk R:
(2004) Immunogenetics of HLA null alleles: implications for blood stem cell
transplantation.
Tissue antigens. 64 (6): 687-695; Erlich RL, et al (2011) Next-generation
sequencing for HLA
typing of Class I loci. BMC genomics. 12: 42-10.1186/1471-2164-12-42; Szolek
A, et al. (2014)
OptiType: Precision HLA typing from next-generation sequencing data.
Bioinformatics 30:3310-
3316; Nariai N, et al. (2015) HLA-VBSeq: Accurate HLA typing at full
resolution from whole-
genome sequencing data. BMC Genomics 16:S7; Dilthey AT, et al. (2016) High-
accuracy HLA
type inference from whole-genome sequencing data using population reference
graphs. PLoS
Comput Biol 12:e1005151; Xie C., et al. (2017) Fast and accurate HLA typing
from short-read
next-generation sequence data with xHLA 114 (30) 8059-8064, each of which is
incorporated
herein in its entirety by reference.
[000169] A complete disruption of MHC Class I expression on xenograft has
shown to have
detrimental effects on the viability of the animal. In a study, SLA Class I
expression on porcine
cells were abrogated by targeting exon 2 of the porcine beta-2-microglobulin
gene. The genomic
sequencing of the produced piglets showed modification at the B2M locus
leading to a frameshift,
a premature stop codon, and ultimately a functional knockout. However, the
piglets of the study
did not survive for more than 4 weeks due to unexpected disease processes,
revealing that such
disruptive genetic modification may have a negative impact on the viability of
the animals. Sake
HJ, Frenszel A, Lucas-Hahn A, et al. Possible detrimental effects of beta-2-
microglobulin
knockout in pigs. Xenotransplantation. 2019;26:e12525.
[000170] In one aspect, a replacement template is created for site-directed
mutagenic
substitutions of nucleotides of the donor swine's SLA/MHC wherein the
reprogramming
introduces non-transgenic, minimally-required alteration that does not result
in any frameshifts or
frame disruptions in specific exon regions of the native donor swine's
SLA/MHC. The nucleotide
sequence(s) of the replacement template is identified by: a) obtaining a
biological sample
containing DNA from a transplant recipient, b) sequencing MHC Class I and II
genes in the
transplant recipient's sample, c) comparing the nucleotide sequence of the
recipient with that of
the donor swine at various loci, and d) creating a replacement template for
one or more of said
loci, wherein said nucleotide sequence of the replacement template are at
least 95% identical to
the transplant recipient's nucleotide sequences, as further described below.
42
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000171] The spreadsheet in FIG. 25A and FIG. 25B, shows human capture
reference
sequence of exons of DQ-Ai and DQ-Bi, respectively, of three individual
recipients. As mentioned
above, known human HLA/MHC or an individual recipient's sequenced HLA/MHC
sequence(s)
may be utilized as a template to reprogram with precise substitution the swine
leukocyte antigen
(SLA)/MHC sequence to match, e.g., to have 90%, 95%, 98%, 99%, or 100%
sequence homology
to a known human HLA/MHC sequence or the human recipient's HLA/MHC sequence.
As shown
in FIG. 25C, the known human HLA-DQA acquired through online database and
individual
recipients' sequenced HLA-DQA, can be compared in a nucleotide Sequence
Library. FIG. 26D
shows comparison of exon 2 region of the swine's SLA-DQA acquired through
online database
and the known and sequenced recipient's HLA-DQA1. Both exon 2 region of SLA-
DQA and
HLA-DQA1 contain 249 nucleotides. As illustrated in FIG. 25D, it can be
observed that 11% of
the aligned 249 nucleotides between exon 2 regions of SLA-DQA1 and HLA-DQA1
are
completely divergent. Therefore, this disclosure disclose method of
identifying the non-conserved
nucleotide sequences at a specific exons of human and swine MHC complex.
Furthermore, by
using a human capture reference template, known or sequenced, a site-directed
mutagenesis can
be performed wherein the specific non-conserved nucleotide sequence between
the specific exon
regions of the SLA gene and the known or recipient's HLA gene are replaced
without causing any
frameshift. The site-directed mutagenesis of the SLA-DQA1 and SLA-DQB1 gene is
shown in
FIG. 26A and FIG. 26B, wherein the nucleotide sequences of the exon 2 region
of the recipient
specific HLA-DQA1 and HLA-DQB1 are used to create a human capture replacement
sequence.
Therefore, the use of synthetic replacement template specific to the exon
regions of the MHC gene,
leads to a non-transgenic, minimally altered genome that does not result in
any frameshifts or
frame disruptions in the native donor swine's SLA/MHC gene.
[000172] As mentioned above, disruptive genetic modification that causes
frameshifts may
have a negative impact on the viability of the animals. Therefore, the present
invention discloses
method of inhibiting expression of MHC proteins without causing frameshift in
the MHC gene.
The spreadsheet in FIG. 25E and FIG. 25F shows human capture reference
sequence of exons of
DR-A and DR-Bi, respectively, of three individual recipients. As shown in FIG.
26C and FIG.26D,
by replacing the initial three nucleotide sequences of the leader exon 1 to a
STOP codon, the
expression of DR molecule can be inhibited without causing frameshift.
Specifically, for HLA-
DRA and DRB 1, the initial three sequences of exon 1, ATG, is replaced with
stop codon, TAA.
43
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Therefore, by using synthetic replacement template, wherein stop codon is
placed in the beginning
of exon 1, the invention provides method of inhibiting expression of desired
MHC molecule,
wherein the non-transgenic, minimally alteration of genome does not result in
any frameshifts or
frame disruptions in the native donor swine's SLA/MHC gene.
[000173] Further, the beta-2-microglobulin protein which comprises the
heterodimer
structure of each of the MHC-I proteins is species-specific. Based on the pig
genome assembly
S SC10.2, a segmental duplication of ¨45.5 kb, encoding the entire B2M
protein, was identified in
pig chromosome 1, wherein functional duplication of the B2M gene identified
with a completely
identical coding sequence between two copies in pigs. The phylogenetic
analysis of B2M
duplication in ten mammalian species, confirming the presence of B2M
duplication in
cetartioldactyls, like cattle, sheep, goats, pigs and whales, but non-
cetartiodactyl species, like mice,
cats, dogs, horses, and humans. The density of long interspersed nuclear
element (LINE) at the
edges of duplicated blocks (39 to 66%) was found to be 2 to 3-fold higher than
the average
(20.12%) of the pig genome, suggesting its role in the duplication event. The
B2M mRNA
expression level in pigs was 12.71 and 7.57 times (2-AACt values) higher than
humans and mice,
respectively. The identification of partially remaining duplicated B2M
sequences in the genomes
of only cetartiodactyls indicates that the event was lineage specific. B2M
duplication could be
beneficial to the immune system of pigs by increasing the availability of MHC
class I light chain
protein, B2M, to complex with the proteins encoded by the relatively large
number of MHC class
I heavy chain genes in pigs. As shown in FIG. 27, B2M molecule with respect to
MHC Class I
molecule can be observed. Further as stated above and shown in FIG. 27, swine
has duplication of
B2M gene while human has one. Thus, in one embodiment of the present
disclosure, the first copy
of the swine B2M gene is reprogrammed through site-directed mutagenesis, as
previously
disclosed. As shown in FIG. 28, the amino acid sequences of exon 2 of the
swine B2M is compared
with that of the human, wherein the non-conserved regions are identified. In
addition, the
expression of the second copy of the swine B2M gene is inhibited by use of
STOP codon, as
previously disclosed. Thus, in one embodiment of the present disclosure
includes a genetic
modification, wherein the fist copy of the swine B2M gene is reprogrammed
through site-directed
mutagenesis and second duplicated B2M gene is not expressed, wherein the
reprogramming does
not result in frameshift of B2M gene.
Selection and Characterization of Pilot Cell Porcine Line for
44
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Humanization by Genetic Modification
[000174] Primary macrophages and other antigen presenting cells (APC) are
useful for
studying immune response, however, the long term use of primary cells is
limited by the cells'
short life span. In addition, primary cells can only be genetically engineered
and evaluated one
time before the cells reach senescence. In the pig model, investigators
frequently have used porcine
aortic endothelial cells (PAECs) for these type of studies. An immortalized
cell line that has the
desired characteristics (expression of MHC Class I and II molecules and
CD80/86) of a
macrophage or representative APC would be ideal to conduct multiple
modifications of the
genome and address impact on immunological reactivity using the same genetic
background. The
ability to generate a viable immortalized pig cell line has been limited to
fibroblasts and epithelial
cell lines which are not relevant for the study of the immune response in
xenotransplantation.
[000175] An immortalized porcine alveolar macrophage (PAM) line was
developed from
Landrace strain of pig [Weingartl 2002] and is commercially available through
ATCC [ 3D4/21,
ATCC CRL-2843]. The cell line showed some percentage of non-specific esterase
and
phagocytosis which was dependent upon conditions of the medium. Cells could be
grown as
anchorage dependent or in colonies under serum free conditions.
Myeloid/monocyte markers (e.g.
CD14) were detected. Desired characteristics of an immortalized cell line was
MHC Class I and
II. MHC Class I was shown to be broadly expressed on all cells, however, MHC
Class II, DR and
DQ, expression of 3D4/21 cells was initially reported as being low, 18% and
4%. PAEC have been
shown to be activated and DR expression could be upregulated with exposure to
IFN-gamma.
3D4/21 cells were exposed to IFN-gamma and Class II expression increased DR:
29.68% to
42.27% and DQ: 2.28% to 57.36% after 24 hours of exposure to IFN-gamma. In
addition, CD80/86
are expressed on the cell surface, these glycoproteins are essential for the
second signal of T cell
activation and proliferation. PAM cells, 34D/21, have the desired
characteristics of a porcine APC
in which genetic changes in genes associated with the MHC can be documented
using an
immortalized cell line and the resulting changes in the phenotype can be
assessed using flow
cytometry to address expression or lack of expression of the glycoproteins of
interest and cellular
immune responses, Mixed Lymphocyte Response (MLR).
[000176] To test for cellular immune response, a one way MLR is set up in
which one set of
cells is identified as the stimulator cells, these are donor cells or
unmodified or modified PAM
cells, and the other set of cells is the responder cells, these are cells from
the recipient (these could
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
be from recipient's who share a similar expression of MHC molecules are the
modified PAM cells.
The stimulator cells are treated with an agent to prevent the cells from
proliferating and this could
be either radiation or incubation with mitomycin C which covalently crosslinks
DNA, inhibiting
DNA synthesis and cell proliferation. Hence, the stimulator cells do not
proliferate in culture
however, the responder cells proliferate in response to interaction at the MHC
Class I and II and it
is this proliferation that is measured in a MLR. A cell culture containing
both stimulator and
responder cells is prepared and incubated for 5-7 days and proliferation/
activation is measured.
Proliferation can be measured by the amount of radioactive thymidine [31-1Tdr]
or BrdU [analog of
thymidine] that is incorporated into the DNA upon proliferation at the end of
5 or 7 days.
[000177] Combinations of the MLR. Responders cells can be either PBMC, CD4+
T cells,
CD8+ T cells or other subpopulations of T cells. PBMC represent all the immune
cells that are
present in the recipient and the measured response reflects the ability of the
responders to mount
an immune response to the stimulator cells, [unmodified or modified PAM
cells]. The measured
proliferation consists of both CD4+ and CD8+ T cells which interact with MHC
Class II and I,
respectively. Using only CD4+ T cells against the unmodified or modified PAM
cells is to measure
the response to MHC Class II glycoproteins, DR and DQ. To observe a specific
response to DQ,
human antigen presenting cells (APCs) are absent from the culture such that
the cellular response
is not the result of pig antigens presented by the APCs. In parallel,
responder CD8+ T cells will be
used to assess an immune response to MHC Class I glycoproteins, SLA 1 AND 2.
This type of
analysis removes the contribution to the immune response from responder APCs
as found in
PBMC. Comparative data will demonstrate the contribution of these respective
glycoproteins to
the immune response of the genetically defined responder and reflects on the
genetic modifications
made to the PAM cells.
[000178] Flow cytometry, phenotypic analysis of the genetically modified
PAM cells. The
cell phenotype of genetically modified cells, e.g., cells from a genetically
modified animal or cells
made ex vivo, are analyzed to measure the changes in expression of the
glycoproteins encoded by
the genes that were modified. Cells are incubated with an antibody with a
fluorescent label that
binds to the glycoprotein of interest and labeled cells are analyzed using
flow cytometry. The
analysis has been performed on unmodified PAM cells to identify the expression
of MHC Class I,
Class II (DR and DQ) and CD80/86. Changes in modified PAM cells will be
referenced to this
database. Flow cytometry will also be used to characterize the expression of
glycoproteins
46
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
encoded by genes for SLA 3, 6, 7, and 8 as the genes in the PAM cells are
modified with recipient
specific sequences related to HLA C, E, F, and G.
[000179] In addition, this type of analysis is also used to ensure the
glycoprotein encoded by
a gene that is knock-out is not expressed. This technique can also be used to
sort out genetically
modified cells from a pool of cells with mixed phenotypes.
[000180] Complement Dependent Cytotoxicity (CDC) assays may be performed to
determine if anti-HLA antibodies recognize the cells from the biological
product of the present
disclosure. Assay plates prepared by adding a specific human serum containing
previously
characterized anti-HLA antibodies (or control serum) can be used. IFN-y
treated donor cells are
resuspended and added to the assay plates, incubated with a source of
complement, e.g., rabbit
serum. After at least 1 hour of incubation at room temperature, acridine
orange/ethidium bromide
solution is added. Percent cytotoxicity is determined by counting dead and
live cells visualized on
a fluorescent microscope, subtracting spontaneous lysis values obtained in the
absence of anti-
HLA antibodies, and scoring with a scale.
[000181] NK cell reactivity, modulation to decrease cytotoxicity. Potential
mechanisms of
activation, recognition,and elimination of target cells by NK cells, alone or
in combination, induce
the release of the content of their lytic granules (perforin, granzyme, and
cytolysin). As an
example, NK cells recognize the lack of self-major histocompatibility complex
(MHC) Class I
molecules on target cells by inhibitory NK cell receptors leading to direct NK
cytotoxicity. This
is the case for xenotransplantation. NK cells are regulated by HLA C that is
recognized by
inhibitory NK cell inhibitory killer cell immunoglobulin-like receptors
(KIRs),
KIR2DL2/2DL3,KIR2DL1, and KIR3DL1. NK cells inhibitory receptor,
immunoglobulin-like
transcript 2 (ILT2) interacts with MHC Class I and CD94-NKG2A recognizing HLA-
E. HLA F
and G have similar roles on the trophoblast. The cytolytic activity of
recipient NK cells to an
unmodified PAM cell can be measured in vitro in which human NK cells are added
to an adherent
monolayer of unmodified PAM cells and cultured for 4 hours. Cell lysis is
measured by release of
radioactive Cr51 or a chromophore measured by flow cytometry. PAM cells with
modified SLA 3,
6, 7 or 8 to mirror HLA C, HLA E, HLA G or HLA F, respectively, can be
assessed using this
cytotoxicity assay.
[000182] For knock in cells, the desired sequences are knocked into the
cell genome through
insertion of genomic material using, e.g., homology-directed repair (HDR). To
optimize
47
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
expression of Class II molecules, the cells are incubated in porcine
interferon gamma (IFN-y) for
72 hours which stimulates expression. Expression is then measured by flow
cytometry using target
specific antibodies. Flow cytometry may include anti-HLA-C, HLA-E, HLA-G, or
other HLA
antibodies, or pan anti-HLA Class I or Class II antibodies. According to the
present disclosure,
cell surface HLA expression after knock-in is confirmed.
[000183] A study was conducted identify the impact of the stimulation by
IFN-y and IFN-y
+ LPS on the phenotype of the porcine alveolar macrophages (PAM) purchased
from ATCC
(3D4/21 cells cat # CRL-2843TM) by flow cytometry.
[000184] PAM cells were thawed in RPMI-1640/10% FBS and cultured for two
days in three
different culture plates. On Day 3, for macrophage activation culture medium
was replaced with
RPMI-1640/20% FBS medium containing 100 ng/mL IFN-y (Plate 1) and 100 ng/mL
IFN-y plus
ng/mL LPS (Plate 2). Untreated cells in RPMI-1640/20% FBS were used as control
(Plate 3).
Following 24 hours incubation, adherent cells were detached from the plate
using TrypLE
treatment. Cells were resuspended in FACS buffer (1X PBS pH=7.4, 2 mM EDTA,
0.5% BSA).
Cell count and viability were determined by trypan blue exclusion method. A
total of 1 x 105 cells
were stained with mouse anti pig SLA Class I, SLA Class II DR, SLA Class II DQ
antibodies for
30 min and APC-conjugated CD152(CTLA-4)-mulg fusion protein (binds to porcine
CD80/CD86)
for 45 min at 4 C. Cells were washed two times using FACS buffer and antibody
stained cells
resuspended in 100 IAL FACS buffer containing anti mouse APC-conjugated
polyclonal IgG
secondary antibody. Followed by incubation for 30 min at 4 C. Cells were
washed two times using
FACS buffer. All cells were resuspended in 200 IAL FACS buffer. Samples were
acquired in
Novacyte flow cytometry and data was analyzed using NovoExpress.
[000185] Analysis procedure is based on NovoExpress flow cytometry analysis
software.
Any equivalent software can be used for the data analysis. Depending on the
software used analysis
presentation maybe slightly different. Gates maybe named differently and %
values might be
slightly different.
[000186] As shown in FIG. 29, untreated PAM cells result 99.98%, 29.68%,
and 2.28% SLA
Class I, SLA Class II DR and DQ molecules expression respectively. These cells
were 4.81%
CD80/86+. 24 hours of culturing cells in the presence of IFN-y increased all
SLA molecule
expression (99.99% SLA Class I+ with increased median fluorescence intensity,
42.27% DR+,
48
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
57.36% DQ+) and CD80/86 levels (47.38%). IFN-y containing cells with LPS
resulted similar
levels of SLA molecules and CD80/86 expression compared to cells only treated
with IFN-y.
[000187] PAM cells were treated with porcine IFN-y for 24 hours and stained
with primary
MAbs and fluorescein conjugated secondary antibody and APC conjugated CD152
which has a
high affinity for co-stimulatory molecules CD80 (B7-1) and CD86 (B7-2). Upon
treatment with
IFN-y, the cells displayed increased SLA and CD80/86 costimulatory molecules
expression
compared to unstimulated PAM cells. While unstimulated cells were 99.98% SLA
Class I+,
29.68% DR+ 2.28 DQ+ and 4.81% CD80/86+, IFN-y stimulated cells were 99.99% SLA
Class I+,
42.27% DR+, 57.36% DQ+, 47.38% CD80/86 +. IFN-y containing cells with LPS
resulted similar
levels of SLA molecules and CD80/86 expression compared to cells only treated
with IFN-y.
[000188] In basal conditions, macrophages express low levels of SLA Class
II and CD80/86
costimulatory molecules. IFN-y and IFN-y-LPS treatment for 24 hours induces
the expression of
SLA Class II and CD80/86 costimulatory molecules as well as SLA Class I
molecules. Extended
incubations would perhaps increase the expression of these molecules further.
[000189] Further, a study was conducted to evaluate the immune
proliferative responsiveness
of human PBMCs (Peripheral Blood Mononuclear Cells), CD8 and CD4 positive T
cells when
they are co-cultured with porcine alveolar macrophages (PAM) cells. Human
donor PBMCs or
their CD4+ T cells were co-cultured with untreated, IFN-y activated and KLH
loaded PAM cells
for seven days. As shown in FIG. 30A and FIG. 30B, one-way allogeneic and
autologous MLR
experiments were performed using the cells isolated from Donor #11, #50, and
#57 as positive and
negative controls respectively. Background controls were performed for
Mitomycin C (X) treated
and untreated PAM cells, and each human donor cells. Proliferative response is
determined
utilizing a bromo-deoxy uridine (BrdU) ELISA assay. On Day 6, BrdU addition
was completed.
On Day 7 media was collected for cytokine (IFN-y and IL-2) analysis and
proliferative responses
were determined. Cells were observed under the Olympus CK40 microscopy at 200X
magnification on Day 7 of co-culturing.
[000190] As shown in FIG. 31, 72 hours of culturing PAM cells in the
presence of IFN-y
increased SLA Class II DQ molecule expression from 2.55% to 95.82%. KLH loaded
PAM cells
resulted expression of similar level of SLA Class II DQ molecules with
untreated cells. All the
allogeneic controls had a positive proliferative response over baseline values
and mitomycin C
treated PBMCs and PAM cells had a decreased proliferative response compared to
baseline values.
49
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
As shown in FIG. 32A and FIG. 32B, Human PBMCs and CD4+ proliferative
responses resulted
in allogeneic responses that were higher than the xenogeneic responses with
PAM cells. The
proliferative responses of three different human CD4+ T cells displayed
similar xenogeneic
responses with PAM cells SI (Stimulation Indexes) values being between 15 and
18.08. The
proliferative responses were highest in xenogeneic cultures from PBMC Donor
#57 (Rw/PAMX, PAM-
IFNyX, KLHx=3 . 12, 2.75, and 3.79).
Gene Editing Schema to Create Multiple, Independent, Single-Variable Humanized
Pilot Porcine
Cell Lines by CRISPR-Cas9 Genetic Modification
[000191] The genetic modification can be made utilizing known genome
editing techniques,
such as zinc-finger nucleases (ZFNs), transcription activator-like effector
nucleases (TALENs),
adeno-associated virus (AAV)-mediated gene editing, and clustered regular
interspaced
palindromic repeat Cas9 (CRISPR-Cas9). These programmable nucleases enable the
targeted
generation of DNA double-stranded breaks (DSB), which promote the upregulation
of cellular
repair mechanisms, resulting in either the error-prone process of non-
homologous end joining
(NHEJ) or homology-directed repair (HDR), the latter of which is used to
integrate exogenous
donor DNA templates. CRISPR-Cas9 may also be used to perform precise
modifications of genetic
material. For example, the genetic modification via CRISPR-Cas9 can be
performed in a manner
described in Kelton, W. et. al., "Reprogramming MHC specificity by CRISPR-Cas9-
assisted
cassette exchange," Nature, Scientific Reports, 7:45775 (2017) ("Kelton"), the
entire disclosure of
which is incorporated herein by reference. Accordingly, the present disclosure
includes
reprogramming using CRISPR-Cas9 to mediate rapid and scarless exchange of
entire alleles, e.g.,
MHC, HLA, SLA, etc.
[000192] According to the present disclosure, CRISPR-Cas9 is used to
mediate rapid and
scarless exchange of entire MHC alleles at specific native locus in swine
cells. Multiplex targeting
of Cas9 with two gRNAs is used to introduce single or double-stranded breaks
flanking the MHC
allele, enabling replacement with the template HLA/MHC sequence (provided as a
single or
double-stranded DNA template).
[000193] In some aspects, the expression of polymorphic protein motifs of
the donor animal's
MHC can be further modified by knock-out methods known in the art. For
example, knocking out
one or more genes may include deleting one or more genes from a genome of a
non-human animal.
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Knocking out may also include removing all or a part of a gene sequence from a
non-human
animal. It is also contemplated that knocking out can include replacing all or
a part of a gene in a
genome of a non-human animal with one or more nucleotides. Knocking out one or
more genes
can also include substituting a sequence in one or more genes thereby
disrupting expression of the
one or more genes. Knocking out one or more genes can also include replacing a
sequence in one
or more genes thereby disrupting expression of the one or more genes without
frameshifts or frame
disruptions in the native donor swine's SLA/MHC gene. For example, replacing a
sequence can
generate a stop codon in the beginning of one or more genes, which can result
in a nonfunctional
transcript or protein. For example, if a stop codon is created within one or
more genes, the resulting
transcription and/or protein can be disrupted, silenced and rendered
nonfunctional.
[000194] In another aspect, the present invention utilizes alteration by
nucleotide
replacement of STOP codon at exon regions of the wild-type swine's SLA-DR to
avoid
provocation of natural cellular mediated immune response (CD8+ T Cell) by the
recipient,
including making cells that lack functional expression of SLA-DR, SLA-1, SLA-
2. For example,
the present invention utilizes TAA . In other embodiments, the invention
utilizes TAG. In other
embodiments, the invention utilizes TGA.
[000195] In one aspect, the present invention utilizes insertion or
creation (by nucleotide
replacement) of STOP codon at exons regions of the wild-type swine's second,
identical
duplication B2-microglobulin gene to reduce the B2-microglobulin mRNA
expression level in
pigs. It will be understood that B2-microglobulin is a predominant immunogen,
specifically a non-
gal xeno-antigen.
[000196] In one aspect, the recipient's HLA/MHC gene is sequenced and
template
HLA/MHC sequences are prepared based on the recipient's HLA/MHC genes. In
another aspect,
a known human HLA/MHC genotype from a World Health Organization (WHO) database
may be
used for genetic reprogramming of swine of the present disclosure.
[000197] CRISPR-Cas9 plasmids are prepared, e.g., using polymerase chain
reaction and the
recipient's HLA/MHC sequences are cloned into the plasmids as templates.
CRISPR cleavage
sites at the SLA/MHC locus in the swine cells are identified and gRNA
sequences targeting the
cleavage sites and are cloned into one or more CRISPR-Cas9 plasmids. CRISPR-
Cas9 plasmids
are then administered into the swine cells and CRIPSR/Cas9 cleavage is
performed at the MHC
locus of the swine cells.
51
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000198] The SLA/MHC locus in the swine cells are precisely replaced with
one or more
template HLA/MHC sequences matching the known human HLA/MHC sequences or the
recipient's sequenced HLA/MHC genes. Cells of the swine are sequenced after
performing the
SLA/MHC reprogramming steps in order to determine if the SLA/MHC sequences in
the swine
cells have been successfully reprogrammed. One or more cells, tissues, and/or
organs from the
HLA/MHC sequence-reprogrammed swine are transplanted into a human recipient.
[000199] The modification to the donor SLA/MHC to match recipient HLA/MHC
causes
expression of specific MHC molecules in the new swine cells that are
identical, or virtually
identical, to the MHC molecules of a known human genotype or the specific
human recipient. In
one aspect, the present disclosure involves making modifications limited to
only specific portions
of specific SLA regions of the swine's genome to retain an effective immune
profile in the swine
while biological products are tolerogenic when transplanted into human
recipients such that use of
immunosuppressants can be reduced or avoided. In contrast to aspects of the
present disclosure,
xenotransplantation studies of the prior art required immunosuppressant use to
resist rejection. In
one aspect, the swine genome is reprogrammed to disrupt, silence, cause
nonfunctional expression
of swine genes corresponding to HLA-A, HLA-B, and DR, and to reprogram via
substitution of
HLA-C, HLA-E, HLA-F, and/or HLA-G. In some aspects, the swine genome is
reprogrammed to
knock-out swine genes corresponding to HLA-A, HLA-B, HLA-C, HLA-F, DQ, and DR,
and to
knock-in HLA-C, HLA-E, HLA-G. In some aspects, the swine genome is
reprogrammed to knock-
out swine genes corresponding to HLA-A, HLA-B, HLA-C, HLA-F, DQ, and DR, and
to knock-
in HLA-C, HLA-E, HLA-G, HLA-F, and DQ. In one aspect, the swine genome is
reprogrammed
to knock-out SLA-1; SLA-6,7,8; SLA-MIC2; and SLA-DQA; SLA-DQB1; SLA-DQB2, and
to
knock-in HLA-C; HLA-E; HLA-G; and HLA-DQ. In certain aspects, HLA-C expression
is
reduced in the reprogrammed swine genome. By reprogramming the swine cells to
be invisible to
a human's immune system, this reprogramming thereby minimizes or even
eliminates an immune
response that would have otherwise occurred based on swine MHC molecules
otherwise expressed
from the donor swine cells.
[000200] Various cellular marker combinations in swine cells are made and
tested to prepare
biologically reprogrammed swine cells for acceptance by a human patient's body
for various uses.
For these tests, Porcine Aorta Endothelial Cells, fibroblast, or a transformed
porcine macrophage
cell line available from ATCC (3D4/21) are used.
52
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000201] The knockout only and knockout plus knock in cell pools are
generated by
designing and synthesizing a guide RNA for the target gene. Each guide RNA is
composed of two
components, a CRISPR RNA (crRNA) and a trans-activating RNA (tracrRNA). These
components
may be linked to form a continuous molecule called a single guide RNA (sgRNA)
or annealed to
form a two-piece guide (cr:tracrRNA).
[000202] CRISPR components (gRNA and Cas9) can be delivered to cells in
DNA, RNA, or
ribonucleoprotein (RNP) complex formats. The DNA format involves cloning gRNA
and Cas9
sequences into a plasmid, which is then introduced into cells. If permanent
expression of gRNA
and/or Cas9 is desired, then the DNA can be inserted into the host cell's
genome using a lentivirus.
Guide RNAs can be produced either enzymatically (via in vitro transcription)
or synthetically.
Synthetic RNAs are typically more pure than IVT-derived RNAs and can be
chemically modified
to resist degradation. Cas9 can also be delivered as RNA. The
ribonucleoproteins (RNP) format
consists of gRNA and Cas9 protein. The RNPs are pre-complexed together and
then introduced
into cells. This format is easy to use and has been shown to be highly
effective in many cell types.
[000203] After designing and generating the guide RNA, the CRISPR
components are
introduced into cells via one of several possible transfection methods, such
as lipofection,
electroporation, nucleofection, or microinjection. After a guide RNA and Cas9
are introduced into
a cell culture, they produce a DSB at the target site within some of the
cells. The NHEJ pathway
then repairs the break, potentially inserting or deleting nucleotides (indels)
in the process. Because
NHEJ may repair the target site on each chromosome differently, each cell may
have a different
set of indels or a combination of indels and unedited sequences.
[000204] For knock in cells, the desired sequences are knocked into the
cell genome through
insertion of genomic material using, e.g., homology-directed repair (HDR).
[000205] It will be further understood that disruptions and modifications
to the genomes of
source animals provided herein can be performed by several methods including,
but not limited to,
through the use of clustered regularly interspaced short palindromic repeats
("CRISPR"), which
can be utilized to create animals having specifically tailored genomes. See,
e.g., Niu et al.,
"Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas-9,"
Science 357:1303-
1307 (22 September 2017). Such genome modification can include, but not be
limited to, any of
the genetic modifications disclosed herein, and/or any other tailored genome
modifications
53
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
designed to reduce the bioburden and immunogenicity of products derived from
such source
animals to minimize immunological rejection.
[000206] CRISPR/CRISPR-associated protein (Cas), originally known as a
microbial
adaptive immune system, has been adapted for mammalian gene editing recently.
The
CRISPR/Cas system is based on an adaptive immune mechanism in bacteria and
archaea to defend
the invasion of foreign genetic elements through DNA or RNA interference.
Through mammalian
codon optimization, CRISPR/Cas has been adapted for precise DNA/RNA targeting
and is highly
efficient in mammalian cells and embryos. The most commonly used and
intensively characterized
CRISPR/Cas system for genome editing is the type II CRISPR system from
Streptococcus
pyogenes; this system uses a combination of Cas9 nuclease and a short guide
RNA (gRNA) to
target specific DNA sequences for cleavage. A 20-nucleotide gRNA complementary
to the target
DNA that lies immediately 5' of a PAM sequence (e.g., NGG) directs Cas9 to the
target DNA and
mediates cleavage of double-stranded DNA to form a DSB. Thus, CRISPR/Cas9 can
achieve gene
targeting in any N20-NGG site.
[000207] Thus, also encompassed by the invention is a genetically modified
non-human
animal whose genome comprises a nucleotide sequence encoding a human or
humanized MHC I
polypeptide and/or (32 microglobulin polypeptide, wherein the polypeptide(s)
comprises
conservative amino acid substitutions of the amino acid sequence(s) described
herein.
[000208] One skilled in the art would understand that in addition to the
nucleic acid residues
encoding a human or humanized MEW I polypeptide and/or (32 microglobulin
described herein,
due to the degeneracy of the genetic code, other nucleic acids may encode the
polypeptide(s) of
the invention. Therefore, in addition to a genetically modified non-human
animal that comprises
in its genome a nucleotide sequence encoding MHC I and/or (32 microglobulin
polypeptide(s) with
conservative amino acid substitutions, a non-human animal whose genome
comprises a nucleotide
sequence(s) that differs from that described herein due to the degeneracy of
the genetic code is
also provided.
[000209] In an additional or alternative approach, the present disclosure
includes
reprogramming, or leveraging the inhibitory and co-stimulatory effects of the
MHC-I (Class B)
molecules. Specifically, the present disclosure includes a process that "finds
and replaces"
portions of the donor animal genome corresponding to portions of the HLA gene,
e.g., to
overexpress HLA-G where possible, retaining and overexpressing portions
corresponding to HLA-
54
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
E, and/or "finding and replacing" portions corresponding to HLA-F. As used
herein, the term
"find and replace" includes identification of the
homologous/analogous/orthologous conserved
genetic region and replacement of the section or sections with the
corresponding human
components through gene editing techniques.
[000210] Another aspect includes finding and replacing the beta-2
microglobulin protein
which is expressed in HLA -A, -B, -C, -E, -F, and -G.
Homologous/analogous/orthologous
conserved cytokine mediating complement inhibiting or otherwise
immunomodulatory cell
markers, or surface proteins, that would enhance the overall immune tolerance
at donor-recipient
cellular interface.
[000211] In an additional or alternative approach, the present invention
utilizes
immunogenomic reprogramming to reduce or eliminate MHC-I (Class A) components
to avoid
provocation of natural cellular mediated immune response by the recipient. In
another aspect,
exon regions in the donor animal (e.g., swine) genome corresponding to exon
regions of HLA-A
and HLA-B are disrupted, silenced or otherwise nonfunctionally expressed on
the donor animal.
In another aspect, exon regions in the donor animal (e.g., swine) genome
corresponding to exon
regions of HLA-A and HLA-B are disrupted, silenced or otherwise
nonfunctionally expressed in
the genome of the donor animal and exon regions in the donor animal (e.g.,
swine) genome
corresponding to exon regions of HLA-C may be modulated, e.g., reduced. In one
aspect, the
present disclosure includes silencing, knocking out, or causing the minimal
expression of source
animal's orthologous HLA-C (as compared to how such would be expressed without
such
immunogenomic reprogramming).
[000212] Further, the beta-2-microglobulin protein which comprises the
heterodimer
structure of each of the MHC-I proteins is species-specific. Thus, in one
embodiment of the present
disclosure, it is reprogrammed. In contrast to its counterparts, the genetic
instructions encoding for
this prevalent, building-block protein is not located in the MHC-gene loci.
Thus, in one
embodiment of the present disclosure includes a genetic modification in
addition to those specific
for the respective targets as described herein.
[000213] FIG. 33 is a schematic depiction of a humanized porcine cell
according to the
present disclosure. As shown therein, the present disclosure involves
reprogramming exons
encoding specific polypeptides or glycoproteins, reprogramming and
upregulating specific
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
polypeptides or glycoproteins, and reprogramming the nuclear genome to have
nonfunctional
expression of specific polypeptides or glycoproteins, all of which are
described in detail herein.
Characterization of Humanized Pilot Porcine Cell Lines and In Vitro Evaluation
of Resultant
Impact to Immunological
[000214] Genetically modified cells, e.g., cells from a genetically
modified animal or cells
made ex vivo, can be analyzed and sorted. In some cases, genetically modified
cells can be
analyzed and sorted by flow cytometry, e.g., fluorescence-activated cell
sorting. For example,
genetically modified cells expressing a gene of interest can be detected and
purified from other
cells using flow cytometry based on a label (e.g., a fluorescent label)
recognizing the polypeptide
encoded by the gene. In this application, the surface expression of SLA-1, SLA-
2, SLA-3, SLA-
6, SLA-7, SLA-8, SLA-DR and SLA-DQ on unmodified PAM cells is established
using labeled
antibodies directed to epitopes on those glycoproteins. In the case of
specific gene knock outs (e.g.
SLA-1, SLA-2 and SLA-DR), analysis by flow cytometry is used to demonstrate
the lack of
expression of these glycoproteins even after incubation of the cells with
interferon gamma. Genes
for SLA-3, SLA-6, SLA-7, SLA-8, and SLA-DQ will be modified such that
glycoproteins
expressed on the cell surface will reflect HLA-C, HLA-E, HLA-F, HLA-G and HLA-
DQ
glycoproteins, respectively. Hence a different set of antibodies specific for
the HLA epitopes will
be used to detect expression of the glycoproteins encoded by the modified
genes and antibodies
directed to the SLA related glycoproteins will not bind to the cell surface of
the modified PAM
cells.
[000215] When knocking out surface sugar glycan epitopes, a cell line that
does not express
the sugar moieties is obtained, so there is no binding of natural preformed
antibodies found in
human serum. This is detected using flow cytometry and human serum and a
labeled goat anti
human IgG or IgM antibody; or specific antibodies directed against sugars; or
labeled sugar
specific isolectins. The result is no binding of the antibodies (isolectins)
to the final cell line.
Positive control is the original cell line (WT) without genetic modifications.
In addition, a
molecular analysis demonstrates changes in those genes.
[000216] For knock in cells, the desired sequences are knocked into the
cell genome through
insertion of genomic material using, e.g., homology-directed repair (HDR). To
optimize
expression of Class II molecules, the cells are incubated in porcine
interferon gamma (IFN-y) for
up to 72 hours which stimulates expression. Expression is then measured by
flow cytometry using
56
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
target specific antibodies. Flow cytometry may include anti-HLA-C, HLA-E, HLA-
G, or other
HLA antibodies, or pan anti-HLA Class I or Class II antibodies. According to
the present
disclosure, cell surface HLA expression after knock-in is confirmed.
[000217] The immune response of the modified swine cells are evaluated
through Mixed
Lymphocyte Reaction (MLR) study. Responders cells can be either PBMC, CD4+ T
cells, CD8+
T cells or other subpopulations of T cells. PBMC represent all the immune
cells that are present in
the recipient and the measured response reflects the ability of the responders
to mount an immune
response to the stimulator cells, for example, a comparison of unmodified PAM
cells and modified
PAM cells. Alternatively, PAECs or fibroblasts may be used. The measured
proliferation consists
of both CD4+ and CD8+ T cells which interact with MHC Class II and I,
respectively. Using only
CD4+ T cells against the unmodified or modified PAM cells measures the
response to MHC Class
II glycoproteins, DR and DQ. For example, in an MLR where SLA DR is knocked
out in the PAM
cells, the CD4+ T cell proliferative response will be decreased; or when SLA-
DQ gene is modified
by using a sequence from a "recipient" [the responder] the proliferative
response will be decreased
since in this case the responder recognizes the DQ glycoprotein as self,
whereas, in the DR knock-
out, DR was absent and thus a signal could not be generated.
[000218] Responder CD8+ T cells were used to assess an immune response to
MHC Class I
glycoproteins, SLA-1 and SLA-2. 1 x 105 purified human CD8+ T cells (A) or
human PBMC (B)
were stimulated with increasing numbers of irradiated (30 Gy) porcine PBMC
from four-fold
knockout pig 10261 or a wild-type pig. Proliferation was measured after 5 d +
16 h by 3H-
thymidine incorporation. Data represent mean cpm SEM of triplicate cultures
obtained with cells
from one human blood donor in a single experiment. Similar response patterns
were observed
using responder cells from a second blood donor and stimulator cells from four-
fold knockout pig
10262. Proliferation of human CD8+ T cells decreased after stimulation with
four-fold knockout
porcine PBMC. (Fischer, et al., Viable pigs after simultaneous inactivation of
porcine MHC Class
I and three xenoreactive antigen genes GGTA1, CMAH and B4GALNT2,
Xenotransplantation,
2019). Modified knock out PAM cells not expressing SLA-1 and SLA-2 will not
generate a CD8+
T cell response. This is in contrast with a response using PBMC as the
responders. See FIG. 34.
[000219] Complement Dependent Cytotoxicity (CDC) assays may be performed to
determine if anti-HLA antibodies recognize the cells from the biological
product of the present
disclosure. Assay plates prepared by adding a specific human plasma containing
previously
57
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
characterized anti-HLA antibodies (or control plasma) can be used. Plasma is
serially diluted
starting at 1:50 to 1:36450 in HB SS media with calcium and magnesium,
incubated with modified
or unmodified PAM cells for 30 minutes at 4 C followed by incubation with
freshly reconstituted
baby rabbit complement for 1 hour at 37 C. The cells were then stained with
Fluorescein Diacetate
(FDA) and Propidium Iodide (PI) for 15 minutes and analyzed by flow cytometry.
Appropriate
compensation controls were run for each assay. Cells were acquired and
analyzed on an ACEA
NovoCyte Flow Cytometer. PAM cells can also be treated with interferon gamma
to increase
surface expression of MEW molecules.
[000220] Cell populations were determined for the following conditions:
a. Dead Cells: PI+, FDA-
b. Damaged Cells: PI+, FDA+
c. Live Cells: PI-, FDA+
[000221] Appropriate calculations were performed to determine %
cytotoxicity for each
concentration (dilution) of plasma, and the results plotted in Prism. Based on
the cytotoxicity
curve, the required dilution for 50% kill (IC50) was determined. This is
illustrated using human
plasma against WT or GalTKO porcine PBMC in FIG. 36A and FIG. 36B, where
reduced
cytotoxicity was identified against cells lacking a 1,3-galactose on the
glycoproteins.
[000222] NK cytotoxicity against unmodified and modified PAM cells where
genes for SLA
3, SLA 6, SLA 7, and SLA 8 are modified such that glycoproteins expressed on
the cell surface
will reflect HLA C, HLA E, HLA F, and HLA G glycoproteins, respectively. The
cytotoxic activity
of freshly isolated and IL-2-activated human NK cells was tested in 4-hr 51Cr
release assays in
serum-free AIM-V medium. Labeled unmodified and modified PAM cells are
cultured in
triplicate with serial 2-fold dilutions of NK cells four E:T ratios ranging
from 40:1 to 5:1. After
incubation for 4 hr at 37 C, the assays are stopped, 51Cr release is analyzed
on a gamma counter,
and the percentage of specific lysis was calculated. NK cells from a specific
genetically matched
"recipient" will have reduced killing of modified PAM cells compared to
unmodified PAM cells.
The protection provided by HLA E in transfected PAEC cells against NK cells is
illustrated in Fig.
34.
[000223] HLA E expression on porcine lymphoblastoid cells inhibits
xenogeneic human NK
cytotoxicity. NK cytotoxicity of 2 donors, KH and MS, against 13271-E/A2 or
13271-E/B7 (solid
diamonds) transfected with HLA E/A2 or HLA E/B7, respectively or untransfected
13271 cells
58
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
(open triangle). To optimize expression of Class II molecules, the cells are
incubated in porcine
interferon gamma (IFN-y) for 72 hours which stimulates expression. Expression
is then measured
by flow cytometry using target specific antibodies. Flow cytometry may include
anti-HLA-C,
HLA-E, HLA-G, or other HLA antibodies, or pan anti-HLA Class I or Class II
antibodies.
According to the present disclosure, cell surface HLA expression after knock-
in is confirmed.
Multiple, Simultaneous Genetic Modifications in a Single Pilot Porcine Cell
Line to Achieve
Relative Humanized Phenotype and Consequential Reduction of CD8+, CD4+, and
Natural
Killer Cell Immune Reactivity as a Direct Result of Multiple CRISPRCas9
Genetic Modification
Schema
[000224] In some aspects, genetic modifications in a porcine cell line to
insert the
modifications listed in table listed in FIG. 33. In some aspects, in addition
to the genetic
modifications listed in FIG. 33, the three predominant swine cell surface
glycans (alpha-Gal,
Neu5Gc, and Sda) are not expressed in order to reduce the hyperacute rejection
phenomenon and
the deleterious activation of antibody-mediated immune pathways, namely
activation of the
complement cascade. With this step, creation of an allogeneic-"like" cell with
respect to non-MHC
cell markers is grossly achieved.
[000225] Genetically modified cells, e.g., cells from a genetically
modified animal or cells
made ex vivo, are analyzed and sorted. In some cases, genetically modified
cells can be analyzed
and sorted by flow cytometry, e.g., fluorescence-activated cell sorting. For
example, genetically
modified cells expressing a gene of interest can be detected and purified from
other cells using
flow cytometry based on a label (e.g., a fluorescent label) recognizing the
polypeptide encoded by
the gene. The gene of interest may be as small as a few hundred pairs of cDNA
bases, or as large
as about a hundred thousand pairs of bases of a genic locus comprising the
exonic-intron encoding
sequence and regulation sequences necessary to obtain an expression controlled
in space and time.
Preferably, the size of the recombined DNA segment is between 25 kb and longer
than 500 kb. In
any case, recombined DNA segments can be smaller than 25 kb and longer than
500 kb.
[000226] It will be further understood that causing the donor swine cells,
tissues, and organs
to express a known human MHC genotype or the recipient's MHC specifically as
described herein,
combined with the elimination in the donor swine cells of alpha-1,3-
galactosytransferase, Neu5Gc,
and 01,4-N-acetylgalactosaminyltransferase (B4GALNT2) (e.g., "single
knockout," "double
knockout," or "triple knockout"), presents a swine whose cells will have a
decreased
59
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
immunological rejection as compared to a triple knockout swine that lacks the
specific SLA/MHC
reprogramming of the present disclosure.
[000227] The immune response of the modified swine cells are evaluated
through Mixed
Lymphocyte Reaction (MLR) study. The impact of the modification or non-
expression of MHC
Ia polypeptides on the immune response are measured through the immune
response of CD8+ T
Cells. The impact of the modification of MHC Ib polypeptides on the immune
response are
measured through the immune response of NK Cells. The impact of the
modification or non-
expression of MHC II polypeptides on the immune response are measured through
the immune
response of CD4+ T Cells. The MLR study, herein, not only measures the
efficacy of the site-
directed mutagenic substitution, but also evaluates and identifies the impact
of individual
modifications, individually and as a whole, as measurements are taken
iteratively as additional
site-directed mutagenic substitutions are made.
[000228] For knock in cells, the desired sequences are knocked into the
cell genome through
insertion of genomic material using, e.g., homology-directed repair (HDR). To
optimize
expression of Class II molecules, the cells are incubated in porcine
interferon gamma (IFN-y) for
72 hours which stimulates expression. Expression is then measured by flow
cytometry using target
specific antibodies. Flow cytometry may include anti-HLA-C, HLA-E, HLA-G, or
other HLA
antibodies, or pan anti-HLA Class I or Class II antibodies. According to the
present disclosure,
cell surface HLA expression after knock-in is confirmed.
[000229] Complement Dependent Cytotoxicity (CDC) assays may be performed to
determine if anti-HLA antibodies recognize the cells from the biological
product of the present
disclosure. Assay plates prepared by adding a specific human serum containing
previously
characterized anti-HLA antibodies (or control serum) can be used. IFN-y
treated donor cells are
resuspended and added to the assay plates, incubated with a source of
complement, e.g., rabbit
serum. After at least 1 hour of incubation at room temperature, acridine
orange/ethidium bromide
solution is added. Percent cytotoxicity is determined by counting dead and
live cells visualized on
a fluorescent microscope, subtracting spontaneous lysis values obtained in the
absence of anti-
HLA antibodies, and scoring with a scale.
[000230] When knocking out or otherwise silencing surface sugar glycans, a
cell line that
does not express the sugar moieties is obtained, so there is no binding of
natural preformed
antibodies found in human serum. This is detected using flow cytometry and
human serum and a
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
labeled goat anti human IgG or IgM antibody; or specific antibodies directed
against sugars. The
result is no binding of the antibodies to the final cell line. Positive
control is the original cell line
(WT) without genetic modifications. In addition, a molecular analysis
demonstrates changes in
those genes.
[000231] In knocking out or otherwise silencing expression of SLA Class I
molecules using
CRISPR technologies, the resulting cell line lacks the above sugar moieties as
well as SLA Class
I expression. Analysis by flow cytometry and molecular gene are performed to
demonstrate no
surface expression and changes made at the gene level. Cellular reactivity is
assessed using a mixed
lymphocyte reaction (MLR) with human PBMCs and the irradiated cell line. In
comparison to the
WT line, there is a reduction in the T cell proliferation, predominantly in
the CD8+ T cells.
[000232] Reactivity against expression of SLA Class II molecules, DR and DQ
is also
minimized or eliminated (there is no porcine DP). Analysis is performed at the
molecular level,
cell surface expression, and in vitro reactivity with human PBMC. There is a
significant downward
modulation of reactivity against the resulting cell line.
[000233] To test for cellular reactivity, all cells are incubated with
porcine IFN-y for 72 hours
then human CD4+ T cells are added to porcine cell lines and cultured for 7
days. The readout is a
form of activation/proliferation depending on the resources available.
[000234] To observe a specific response to DQ, human antigen presenting
cells (APCs) are
absent from the culture such that the cellular response is not the result of
pig antigens presented
by the APCs.
Creation of a Humanized, "Bespoke", Designated-Pathogen Free, (Non-Human)
Donor of Cells,
Tissues, and Organs for Transplantation
[000235] Others have attempted to develop homozygous transgenic pigs, which
is a slow
process, requiring as long as three years using traditional methods of
homologous recombination
in fetal fibroblasts followed by somatic cell nuclear transfer (SCNT), and
then breeding of
heterozygous transgenic animals to yield a homozygous transgenic pig. The
attempts at developing
those transgenic pigs for xenotransplantation has been hampered by the lack of
pluripotent stem
cells, relying instead on the fetal fibroblast as the cell upon which genetic
engineering was carried
out. For instance, the production of the first live pigs lacking any
functional expression of a(1,3)
galactosyltransferase (GTKO) was first reported in 2000. In contrast to such
prior attempts, the
present disclosure provides a faster and fundamentally different process for
making non-transgenic
61
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
reprogrammed swine as disclosed herein. In some aspects, porcine fetal
fibroblast cells are
reprogrammed using gene editing, e.g., by using CRISPR/Cas for precise
reprogramming and
transferring a nucleus of the genetically modified porcine fetal fibroblast
cell to a porcine
enucleated oocyte to generate an embryo; and d) transferring the embryo into a
surrogate pig and
growing the transferred embryo to the genetically modified pig in the
surrogate pig.
[000236] Upon confirmation of study results, genetically reprogrammed pigs
are bred so that
several populations of pigs are bred, each population having one of the
desirable human cellular
modifications determined from the above assays. The pigs' cellular activity
after full growth is
studied to determine if the pig expresses the desired traits to avoid
rejection of the pigs' cells and
tissues after xenotransplantation. Thereafter, further genetically
reprogrammed pigs are bred
having more than one of the desirable human cellular modifications to obtain
pigs expressing cells
and tissues that will not be rejected by the human patient's body after
xenotransplantation.
[000237] The generation of an induced pluripotent stem cell (iPSC) from
pigs offers an
opportunity beyond the use of primary cells from fetal fibroblasts. The
ability of iPSC to proliferate
almost indefinitely, which contrasts with the limited number of cell divisions
that primary somatic
cells can undergo before they senesce, likely means that the iPSC will
tolerate the multiple
selection steps needed to accommodate directed changes in several genes,
especially for gene
knock-outs and knock-ins, before nuclear transfer. Another advantage of iPSC
over somatic cells
is that it has been predicted that cloning efficiency should be inversely
correlated with
differentiation state and associated epigenetic state. The PAM cells presented
in this disclosure are
a transformed cell line but the genetic engineering schema can be transferred
to porcine iPSC. The
specific genetically modified iPSC line would then be used for somatic cell
nuclear transfer
(SCNT), transferring a nucleus of the genetically modified porcine fetal
fibroblast cell to a porcine
enucleated oocyte to generate an embryo; and transferring the embryo into a
surrogate pig and
growing the transferred embryo to the genetically modified pig in the
surrogate pig. This has the
advantage in that the transferred nucleus contains the specific genome, hence
the piglets do not
need to go through breeding to obtain a homozygous offspring. The genotype and
phenotype of
the piglets are identical to the iPSC.
[000238] Specific populations of gene modified iPSC can be cryopreserved as
a specific cell
line and used as required for development of pigs needed for that genetic
background. Thawed
iPSCs are cultured and nucleus is transferred into enucleated oocytes to
generate
62
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
blastocysts/embryos for implantation into surrogate pig. This creates a viable
bank of genetically
modified iPSC for generation of pigs required for patient specific tissue,
organ, or cell
transplantation.
[000239] Restated, the former/previous approach to this unmet clinical need
has precisely
followed the classic medical dogma of "one-size fits all". Instead of
following this limited
approach, we pragmatically demonstrate the ability to harness present
technological advances and
fundamental principles to achieve a "patient-specific" solution which
dramatically improves
clinical outcome measures. The former, we refer as the "downstream" approach -
which must
contend with addressing all of the natural immune processes in sequence. The
latter, our approach,
we optimistically term the "upstream" approach - one which represents the
culmination of unfilled
scientific effort into a coordinated translational effort.
[000240] In another aspect, disclosed herein is a method for making a
genetically modified
animal described in the application, comprising: a) obtaining a cell with
reduced expression of one
or more of a component of a MEW I-specific enhanceosome, a transporter of a
MHC I-binding
peptide, and/or C3; b) generating an embryo from the cell; and c) growing the
embryo into the
genetically modified animal. In some cases, the cell is a zygote.
[000241] In certain aspects, HLA/MEIC sequence-reprogrammed swine are bred
for at least
one generation, or at least two generations, before their use as a source for
live tissues, organs
and/or cells used in xenotransplantation. In certain aspects, the CRISPR/Cas9
components can also
be utilized to inactivate genes responsible for PERV activity, e.g., the pol
gene, thereby
simultaneously completely eliminating PERV from the swine donors.
[000242] In certain aspects, the present disclosure includes embryogenesis
and live birth of
SLA-free and HLA-expressing biologically reprogrammed swine. In certain
aspects, the present
disclosure includes breeding SLA-free and HLA-expressing biologically
reprogrammed swine to
create SLA-free and HLA-expressing progeny. In certain aspects, the
CRISPR/Cas9 components
are injected into swine zygotes by intracytoplasmic microinjection of porcine
zygotes. In certain
aspects, the CRISPR/Cas9 components are injected into swine prior to selective
breeding of the
CRISPR/Cas9 genetically modified swine. In certain aspects, the CRISPR/Cas9
components are
injected into donor swine prior to harvesting cells, tissues, zygotes, and/or
organs from the swine.
In certain aspects, the CRISPR/Cas9 components include all necessary
components for controlled
63
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
gene editing including self-inactivation utilizing governing gRNA molecules as
described in U.S.
Pat. No. 9,834,791 (Zhang), which is incorporated herein by reference in its
entirety.
[000243] Upon confirmation of study results, genetically reprogrammed pigs
are bred so that
several populations of pigs are bred, each population having one of the
desirable human cellular
modifications determined from the above assays. The pigs' cellular activity
after full growth is
studied to determine if the pig expresses the desired traits to avoid
rejection of the pigs' cells and
tissues after xenotransplantation. Thereafter, further genetically
reprogrammed pigs are bred
having more than one of the desirable human cellular modifications to obtain
pigs expressing cells
and tissues that will not be rejected by the human patient's body after
xenotransplantation.
[000244] Any of the above protocols or similar variants thereof can be
described in various
documentation associated with a medical product. This documentation can
include, without
limitation, protocols, statistical analysis plans, investigator brochures,
clinical guidelines,
medication guides, risk evaluation and mediation programs, prescribing
information and other
documentation that may be associated with a pharmaceutical product. It is
specifically
contemplated that such documentation may be physically packaged with cells,
tissues, reagents,
devices, and/or genetic material as a kit, as may be beneficial or as set
forth by regulatory
authorities.
[000245] In another aspect, disclosed herein is a method for making a
genetically modified
animal described in the application, comprising: a) obtaining a cell with
reduced expression of one
or more of a component of a MHC I-specific enhanceosome, a transporter of a
MHC I-binding
peptide, and/or C3; b) generating an embryo from the cell; and c) growing the
embryo into the
genetically modified animal. In some cases, the cell is a zygote.
[000246] Muscle and skin tissue samples taken from each of these pigs were
dissected and
cultured to grow out the fibroblast cells. The cells were then harvested and
used for somatic cell
nuclear transfer (SCNT) to produce clones. Multiple fetuses (up to 8) were
harvested on day 30.
Fetuses were separately dissected and plated on 150 mm dishes to grow out the
fetal fibroblast
cells. Throughout culture, fetus cell lines were kept separate and labeled
with the fetus number on
each tube or culture vessel. When confluent, cells were harvested and frozen
at about 1 million
cells/mL in FBS with 10% DMSO for liquid nitrogen cryo-storage.
[000247] Added from different example: In certain aspects, the CRISPR/Cas9
components
are injected into swine oocytes, ova, zygotes, or blastocytes prior to
transfer into foster mothers.
64
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Creation of, Procurement of Personalized, Tolerogenic Cells, Tissues, and
Organs Donor of
Cells, Tissues, and Organs for Transplantation from Humanized, "Bespoke",
Designated-
Pathogen Free, (Non-Human) Donor
Source Animal Facility ("SAF")
[000248] Referring to FIG. 37, a barrier source animal location, including,
but not limited to,
a Source Animal Facility ("SAF") 100, that can be used for the housing,
propagation, maintenance,
care and utilization of a closed colony swine, including a closed colony that
is designated pathogen
free ("DPF") ("DPF Closed Colony") 102, is shown. As contained herein, the SAF
has positive
pressure, biocontainment characteristics is operated under specific isolation-
barrier conditions.
[000249] As described herein, the DPF Closed Colony 102 is comprised of
source animals
maintained and propagated for harvesting various biological products for use
in human
xenotransplantation and other therapies, wherein such products have reduced
bioburden and
demonstrate reduced immunogenicity resulting from xenotransplantation and
other therapeutic
procedures. In some aspects, xenotransplantation products of the present
disclosure are less
immunogenic than a xenotransplantation product made from conventional Gal-T
knockout swine,
from conventional triple knockout swine, from transgenic swine, from wild-type
animals, and/or
allograft. For example, as shown in Examples 1 and 2, biological products made
according to the
present disclosure provided unexpectedly high clinical benefit when using a
single knockout pig
as the donor animal in that, despite the presence of Neu5Gc and porcine
B4GALNT2, the
biological product made according to the present disclosure had less
immunogenicity than
allograft, vascularized, and was resistant to rejection for the entire
duration of the study period.
[000250] As further described herein, the SAF 100 and each of its
accompanying areas (e.g.,
rooms, suites or other areas) can be utilized to house and maintain source
animals from which
biological products are harvested and/or processed. The SAF 100 and its areas
are designed to
minimize and eliminate the potential for contamination of the harvested and/or
processed
biological products and cross-contamination between such products.
[000251] Within the SAF 100, in some aspects, utilized animal areas are
ventilated. For
example, animal areas are ventilated with high efficiency particulate air
(HEPA)-filtered fresh air
from the roof of the building, for example, having at least 10-15 air changes
per hour. Additionally,
one or more laminar flow hoods (e.g., Class II Type A2 Laminar Airflow
Biosafety Cabinets) are
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
utilized in the SAF rooms, including in a xenotransplantation drug processing
suite to providing
additional ventilation to minimize or eliminate cross contamination.
[000252] In some aspects, utilized areas are also temperature controlled
and monitored. For
example, the areas are heated and cooled to maintain temperature within the
range specified by,
for example, the Guide for the Care and Use of Laboratory Animals. Utilized
animal holding rooms
are also alarmed and centrally monitored for high or low temperatures, and
staff are notified
immediately if temperatures are beyond required temperature.
[000253] In some aspects, the SAF 100 has multiple levels of containment
for the source
animals. For example, source animals are contained in a primary level of
containment consisting
of pens and cages which are secured by stainless steel latches. With respect
to secondary level of
containment, functionally designated areas (e.g., rooms, suites or other
areas) can have latched
inner doors, and an ante-room with card-controlled access to a hallway. A
tertiary level of
containment can include outside perimeter fencing.
[000254] The entire SAF is located within a single building. Primary
entrance is through a
single door via programmable identification (ID) card. All other external
doors are alarmed, remain
locked, and are for emergency use only.
[000255] Security is also a consideration to ensure security of the SAF 100
in general, and to
control individuals entering the SAF 100 to minimize the risk of outside
contaminants entering the
SAF 100 and reaching the source animals. Therefore, in one aspect, the primary
entrance to the
SAF 100 is through a single door 116 via programmable identification (ID) card
118. All other
external doors 120 are alarmed, remain locked, and are for emergency use only.
[000256] It will be understood that the SAF 100 and its features as
disclosed herein are set
out as examples, and it will be further understood that other facilities with
various features can
also be utilized to perform the methods and produce the products disclosed
herein.
[000257] In some aspects, the SAF 100 animal program is licensed and/or
accredited and
overseen, evaluated and operated by a team of highly experienced, professional
staff. For example,
the program is registered and/or accredited with the USDA Animal and Plant
Health Inspection
Service (as a licensed animal research facility), National Institute of Health
(NIH) Office of
Laboratory Animal Welfare (OLAW) (confirming compliance with Public Health and
Safety
(PHS) regulations, Association for Assessment and Accreditation of Laboratory
Animal Care
66
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
(AAALAC) (with veterinary care of the source animals housed at the SAF under
the direction of
an attending veterinarian), and other federal, state and local regulatory
authorities.
[000258] In some aspects, to ensure the welfare of the source animals, SAF
personnel, and
caretakers of source animals adhere to procedures for animal husbandry, tissue
harvesting, and
termination of animals that are approved by an appropriate Institutional
Animal Care and Use
Committee, in accordance with the Animal Welfare Act (7 U.S.C. 2131, et seq.),
accredited by the
AAALAC, and in compliance of the standards as set forth in the Guide for the
Care and Use of
Laboratory Animals.
[000259] In some aspects, caretakers have extensive training and experience
in handling and
caring for the source animals being managed in accordance with the present
invention. For
example, each caretaker undergoes a documented training program covering the
standard
operating procedures governing handling and care of these source animals, and
be skilled in
making daily health assessments and insuring prompt care is directed to any
animal in need. In
addition, the caretakers can be trained in scrubbing and gowning procedures
prior to entry into the
isolation areas (e.g., rooms, suites or other areas) as described herein, and
under a medical
surveillance program to ensure staff health and the health of the source
animals.
[000260] To minimize and eliminate contamination risk to the SAF, any
personnel or visitors
entering the SAF wear personnel protective equipment or change into facility
dedicated clothing
and footwear before entry into any containment areas. Visitors who wish to
enter animal areas
must not have had any contact with live swine for at least 24 hours preceding
the visit or must
shower at the facility prior to entry.
[000261] It will be understood that the approaches and procedures set forth
herein are
examples as to how to ensure contamination does not reach the source animals
within SAF 100. It
will be further understood that a multitude of approaches can also be utilized
to achieve a
designated pathogen free environment for source animals.
Source Animals
[000262] In some aspects, as described herein, swine can be utilized as
source animals. As
used herein, unless otherwise specified, the terms "swine," "pig" and
"porcine" are generic terms
referring to the same type of animal without regard to gender, size, or breed.
It will be understood
that any number of source animals could be utilized in accordance with the
present invention,
including, but not limited to, pigs, non-human primates, monkeys, sheep,
goats, mice, cattle, deer,
67
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
horses, dogs, cats, rats, mules, and any other mammals. Source animals could
also include any
other animals including, but not limited to, birds, fish, reptiles, and
amphibians.
[000263] It will be further understood that any animal serving as a source
animal hereunder,
including swine, regardless of how such swine may be configured, engineered,
or otherwise altered
and/or maintained, may be created, bred, propagated and/or maintained in
accordance with the
present disclosure to create and maintain animals and resulting biological
products to be used in
or in preparation or pursuit of clinical xenotransplantation.
[000264] For example, the present disclosure includes non-human animals,
e.g., swine,
having certain combinations of specific genetic characteristics, breeding
characteristics and
pathogen-free profile. Such animals may include, as described above and
herein, immunogenomic
reprogrammed swine having a biologically reprogrammed genome such that it does
not express
one or more extracellular surface glycan epitopes, e.g., genes encoding alpha-
1,3
galactosyltransferase, cytidine monophosphate-N-acetylneuraminic acid
hydroxylase (CMAH),
and 01,4-N-acetylgalactosaminyltransferase are disrupted such that surface
glycan epitopes
encoded by said genes are not expressed, as well as other modifications to the
swine's SLA to
express MHC-I or MHC-II, and regulation of PD-1 and CTLA4, as described above
and herein.
Resulting from the process described herein, the swine is free of at least the
following zoonotic
pathogens:
(i) Ascaris species, cryptosporidium species, Echinococcus, Strongyloids
sterocolis,
and Toxoplasma gondii in fecal matter;
(ii) Leptospira species, Mycoplasma hyopneumoniae, porcine reproductive and
respiratory syndrome virus (PRRSV), pseudorabies, transmissible
gastroenteritis virus
(TGE) / Procine Respiratory Coronavirus, Toxoplasma Gondii in antibody titers;
(iii) Porcine Influenza;
(iv) the following bacterial pathogens as determined by bacterial culture:
Bordetella
bronchisceptica, Coagulase-positive staphylococci, Coagulase-negative
staphylococci,
Livestock-associated methicillin resistant Staphylococcus aureus (LA MRSA),
Microphyton and Trichophyton spp.;
(v) Porcine cytomegalovirus; and
(vi) Brucella suis;
68
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
is raised and maintained according to a bioburden-reducing procedure, the
procedure comprising
maintaining the swine in an isolated closed herd, wherein all other animals in
the isolated closed
herd are confirmed to be free of said zoonotic pathogens; wherein the swine is
isolated from contact
with any non-human animals and animal housing facilities outside of the
isolated closed herd.
[000265] As indicated previously, in some aspects, the swine source animals
may have a
combination of one or more genetic modifications including "knockout" and/or
"knock-in" swine
having one or more characteristics of swine disclosed in U.S. Patent No.
7,795,493 ("Phelps"), the
entire disclosure of which is incorporated herein by reference. Such swine
lack active (and/or have
disrupted) a-(1,3) galactosyl epitopes responsible for hyperacute rejection in
humans upon
transplantation. Multiple methods of production of knockout/knock-in swine are
disclosed in
Phelps including: the inactivation of one or both alleles of the alpha-1,3-GT
gene by one or more
point mutations (for example by a T-to-G point mutation at the second base of
exon 9) and/or
genetic targeting events as disclosed at col. 9, line 6 to col. 10, line13;
col. 21, line 53 to co1.28,
line 47; and col. 31, line 48 to col. 38, line 22 of Phelps, incorporated
herein by reference. The
creation of such swine through the described methods, and/or the utilization
of such swine and
progeny following creation, can be employed in the practice of the present
invention, including,
but not limited to, utilizing organs, tissue and/or cells derived from such
swine.
[000266] Similarly, in other aspects, the swine source animals include
"knockout" and
"knock-in" swine having one or more characteristics of swine disclosed in U.S.
Patent No.
7,547,816 ("Day"), the entire disclosure of which is incorporated herein by
reference. Such swine
also lack active (and/or have disrupted) a-(1,3) galactosyl epitopes
responsible for hyper-acute
rejection in humans upon transplantation. Multiple methods of production of
knockout/knock-in
swine are disclosed in Day including: enucleating an oocyte, fusing the oocyte
with a porcine cell
having a non-functional alpha-1,3-GT gene, followed by implantation into a
surrogate mother, as
described more fully at col. 4, line 61 to col. 18, line 55 of Day,
incorporated herein by reference.
The creation of such swine through the described methods, and/or the
utilization of such swine
and progeny following creation, can be employed in the practice of the present
invention,
including, but not limited to, utilizing organs, tissue and/or cells derived
from such swine.
[000267] Similarly, in other aspects, the swine source animals include GGTA
Null
("knockouts" and "knock-ins") swine having one or more characteristics of
swine disclosed in
U.S. Patent No. 7,547,522 ("Hawley"), the entire disclosure of which is
incorporated herein by
69
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
reference. Such swine also lack active (and/or have disrupted) a-(1,3)
galactosyl epitopes
responsible for hyper-acute rejection in humans upon transplantation. As
disclosed in Hawley,
production of knockout/knock-in swine includes utilizing homologous
recombination techniques,
and enucleating oocytes followed by fusion with a cell having a non-functional
alpha-1,3-GT gene
and implantation into a surrogate mother (as disclosed more fully at col. 6,
line 1 to col. 14, line
31). The creation of such swine through the described methods, and/or the
utilization of such swine
and progeny following creation, can be employed in the practice of the present
invention,
including, but not limited to, utilizing organs, tissue and/or cells derived
from such swine.
[000268] In yet other aspects, the swine source animals include swine and
swine that lack
active (and/or have disrupted) a-(1,3) galactosyl epitopes having one or more
characteristics of
swine as described in U.S. Patent No. 9,883,939 ("Yamada"), the entire
disclosure of which is
incorporated by reference herein. In certain aspects, the swine source animals
for use or
modification in accordance with the present disclosure include the swine
having one or more
characteristics of swine described in U.S. 2018/0184630 (Tector, III), the
disclosure of which is
incorporated by reference herein in its entirety. The creation of such swine
through the described
methods, and/or the utilization of such swine and progeny following creation,
can be employed in
the practice of the present invention, including, but not limited to,
utilizing organs, tissue and/or
cells derived from such swine.
[000269] In yet other aspects, swine source animals include the swine
having one or more
characteristics of swine disclosed in U.S. Patent Nos. 8,106,251 (Ayares),
6,469,229 (Sachs),
7,141,716 (Sachs), each of the disclosures of which are incorporated by
reference herein. The
creation of such swine through the described methods, and/or the utilization
of such swine and
progeny following creation, can be employed in the practice of the present
invention, including,
but not limited to, utilizing organs, tissue and/or cells derived from such
swine.
[000270] In some aspects, the swine can originate from one or more highly
inbred herds of
pigs (whether genetically modified or not (i.e., wild-type)) with a co-
efficient of inbreeding of 0.50
or greater. A higher coefficient of inbreeding indicates the products derived
from the source
animals may have more consistent biological properties for use in pig-to-human
xenotransplantation (e.g., a coefficient of inbreeding of 0.80 or greater in
one aspect). Coefficients
of inbreeding for animals are disclosed in Mezrich et al., "Histocompatible
Miniature Swine: An
Inbred Large-Animal Model," Transplantation, 75(6):904-907 (2003). An example
of a highly
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
inbred herd of swine includes miniature swine descendant from the miniature
swine disclosed in
Sachs, et al., "Transplantation in Miniature Swine. I. Fixation of the Major
Histocompatibility
Complex," Transplantation 22:559 (1976), which is a highly inbred line
possessing reasonable
size matches particularly for organs eventually utilized for clinical
transplantation. The creation of
such swine through the described methods, and/or the utilization of such swine
and progeny
following creation, can be employed in the practice of the present invention,
including, but not
limited to, utilizing organs, tissue and/or cells derived from such swine.
[000271] Source animals can also include animals swine that lack active
(and/or have
disrupted) alpha-1,3- galactosyltransferase, Neu5Gc, and 01,4-N-
acetylgalactosaminyltransferase
as described in U.S. Patent Publication No. US2017/0311579 (Tector), the
entire disclosure of
which is incorporated herein by reference. The creation of such swine through
the described
methods, and/or the utilization of such swine and progeny following creation,
can be employed in
the practice of the present invention, including, but not limited to,
utilizing organs, tissue and/or
cells derived from such swine.
[000272] It is therefore understood that multiple source animals, with an
array of biological
properties including, but not limited to, genome modification and/or other
genetically engineered
properties, can be utilized to reduce immunogenicity and/or immunological
rejection (e.g., acute,
hyperacute, and chronic rejections) in humans resulting from
xenotransplantation. In certain
aspects, the present disclosure can be used to reduce or avoid thrombotic
microangiopathy by
transplanting the biological product of the present disclosure into a human
patient. In certain
aspects, the present disclosure can be used to reduce or avoid glomerulopathy
by transplanting the
biological product of the present disclosure into a human patient. It will be
further understood that
the listing of source animals set forth herein is not limiting, and the
present invention encompasses
any other type of source animal with one or more modifications (genetic or
otherwise) that serve(s)
to reduce immunogenicity and/or immunological rejection, singularly or in
combination.
[000273] In some embodiments, preterm swine fetuses and neonatal piglets
are derived as
offspring from DPF Closed Colony, a-1,3-galactosyltransferase [Gal-T] knockout
pigs, as shown
and described herein in accordance with the present invention.
[000274] Such preterm swine fetuses and neonatal piglets are utilized as a
source for cells,
tissues and organs for xenotransplantation therapies, including, but not
limited to, in regenerative
or direct transplantation therapies. It will be understood that such cells,
tissues and organs can be
71
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
utilized as fresh or following cryopreservation in accordance with the present
invention (e.g.,
cryopreservation in the range of -80 C).
[000275] In one aspect, mesenchymal cells, pluripotent cells, stem cells
and/or other cells
that have not differentiated are harvested from such preterm swine fetuses and
utilized for
regenerative therapies and other therapies as described herein, whereas such
undifferentiated cells
can be found in high proportion in swine fetuses as well as in neonatal
piglets. Since these cells
are derived from fetuses earlier along the gestation period, they are less
differentiated and more
pliable which offers greater potential for regenerative therapies.
Furthermore, since these cells may
be derived from DPF Closed Colony, a-1,3-galactosyltransferase [Gal-T]
knockout pigs, as shown
and described herein, they do not possess aggravating immunogenic, pathogenic
and/or other
aggravating factors causing rejection by the human immune system, and the
cells will persist and
differentiate inside a human recipient offering regain of function of growth
of model tissue using
these genetic and cellular building blocks.
[000276] By way of example, such cells may be utilized to generate an array
of organs and/or
tissues, through regenerative cell-therapy methods known in the art (e.g.,
through utilization of
biological scaffolds), for xenotransplantation including, but not limited to,
skin, kidneys, liver,
brain, adrenal glands, anus, bladder, blood, blood vessels, bones, brain,
brain, cartilage, ears,
esophagus, eye, glands, gums, hair, heart, hypothalamus, intestines, large
intestine, ligaments, lips,
lungs, lymph, lymph nodes and lymph vessels, mammary glands, mouth, nails,
nose, ovaries,
oviducts, pancreas, penis, pharynx, pituitary, pylorus, rectum, salivary
glands, seminal vesicles,
skeletal muscles, skin, small intestine, smooth muscles, spinal cord, spleen,
stomach, suprarenal
capsule, teeth, tendons, testes, thymus gland, thyroid gland, tongue, tonsils,
trachea, ureters,
urethra, uterus, uterus, and vagina, areolar, blood, adenoid, bone, brown
adipose, cancellous,
cartaginous, cartilage, cavernous, chondroid, chromaffin, connective tissue,
dartoic, elastic,
epithelial, epithelium, fatty, fibrohyaline, fibrous, Gamgee, Gelatinous,
Granulation, gut-
associated lymphoid, Haller's vascular, hard hemopoietic, indifferent,
interstitial, investing, islet,
lymphatic, lymphoid, mesenchymal, mesonephric, mucous connective, multilocular
adipose,
muscle, myeloid, nasion soft, nephrogenic, nerve, nodal, osseous, osteogenic,
osteoid, periapical,
reticular, retiform, rubber, skeletal muscle, smooth muscle, and subcutaneous
tissue.
72
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000277] Accordingly, preterm swine fetuses and neonatal piglets may be
utilized as a source
of tissue, cells and organs in accordance with the present invention based on
their characteristics
as compared to adult swine.
Closed Colonies
General Closed Colony
[000278] Referring now to FIG. 37, in one aspect, animals are secured from
the outside to
consider as candidates to add to the General Closed Colony 128 that is housed
within the SAF 100
to help propagate the DPF Closed Colony 102 also housed within the SAF 100 in
a separate
isolation area 152. Transportation of the animals secured from the outside to
the SAF is controlled
to mitigate exposure to potential infectious agents. Such mitigation
techniques include, but are not
limited to, using a sterilized HEPA filtered cage during transport using a van
cleaned with
chlorhexidine and containing no other animals.
[000279] Candidate animals are initially quarantined to check health status
and suitability for
intake into the General Closed Colony 128. For example, in some aspects,
animals coming from
the outside are first housed in a quarantine intake area 130 within the SAF
and accompanied by a
complete health record (including, but not limited to, date of birth,
vaccinations, infections, and
antibiotic history), pedigree, and results of genetic tests. These animals
reside in the quarantine
intake area 130 for at least seven (7) days as the accompanying records are
evaluated and other
health screening measures are taken, including screening for some infectious
agents.
[000280] In some aspects, animals with poor health, questionable medical
status, or are not
able to be treated for such medical issues, will not be accepted into the
General Closed Colony 128
and/or will otherwise be culled from the quarantine area 130. Examples of
acceptance criteria
include, but are not limited to: (a) source animals are not born with any
congenital defect that was
unanticipated from the herd and that could have impacted the quality of health
of the animal; (b)
source animals have received all vaccinations according to age and the
vaccinations were killed
agents; (c) any infections that occurred in the source animal's lifetime have
been reviewed as well
as the clinical intervention, and it was determined that the infection and any
treatment (if
applicable) did not impact the quality of the health of the animal; (d)
results of the surveillance
testing has been reviewed and it has been verified that the source animal has
been tested within
the last 3 months (with all source animals tested at sacrifice and all tests
must be negative); (e) if
73
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
the animal was injured in any way which required medical attention, a review
has been conducted
and it has been confirmed that the impact of the injury and the medical
intervention (if applicable)
had no impact on the health of the animal; and/or (f) PERV tests have been
performed and results
recorded.
[000281] In some aspects, animals that pass this screening process and
timetable are moved
out of the quarantine intake area 130 and into a general holding area 132
within the SAF 100 to
join or create an existing or newly formed General Closed Colony 128. It will
be understood that
the general holding area 132 is kept under closed colony conditions
substantially similar to the
conditions applied to the DPF Closed Colony 102 in the DPF Isolation Area 152.
[000282] It will be further understood that, excluding their offspring,
candidate animals
secured from the outside will never become members of the DPF Closed Colony.
Piglets from the
General Closed Colony 128 animals will be utilized to create and/or propagate
the DPF Closed
Colony as further described herein.
DPF Closed Colony
Pregnant Sows and DPF Piglets
[000283] In one aspect, pregnant sows 134 (or gilts) are obtained from the
outside or from
the General Closed Colony 128 to produce piglets to create and/or add to the
DPF Closed Colony
102 herd. For example, in one aspect, sows 134 are placed in a sow quarantine
area 136 within the
SAF until the time to give birth, in this aspect via Cesarean section in order
to avoid exposing the
piglet to potential pathogens, including Porcine Cytomegalovirus (pCMV).
Contraction of pCMV
in piglets can occur when the piglets travel through the vagina of the sow
during natural birth. The
piglets, by virtue of their birthing through Cesarean section as described
herein, prevents such
contraction and the piglets produced through the methods described herein are
pCMV-free.
[000284] Prior to the Cesarean section procedure, for example the morning
of the procedure,
an operating room 138 within the SAF 100 prepared according to standard
operating room
protocols in a sterile environment with 2 sides: Side A 140 for the Cesarean
section of the sow,
and Side B 142 to receive the piglets 144 that are candidates to either found
or add to the DPF
Closed Colony.
[000285] The sow 134 is brought into the operating room 138 for captive
bolt euthanasia.
Immediately following this, the sow 134 is placed in the left lateral
decubitus position and the
abdomen and torso are prepped widely with chlorhexidine and draped in a
sterile fashion. A flank
74
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
incision is expeditiously made and the abdominal muscles are split in order to
gain access into the
peritoneum. The uterus is exteriorized, incised and the piglets 144 are
removed after doubly
clamping and dividing the umbilical cord. Immediate execution of the surgical
procedures
following captive bolt euthanasia is critical to the survival of the piglets
144.
[000286] Infection controls for the piglets 144 are implemented at birth.
The piglets 144 are
placed in a warmed 1% chlorhexidine (or other sterilization agent, such as
betadine) in sterile
saline bath solution and then passed over to piglet handlers to a
resuscitation area 148 for
resuscitation, rewarming and gavage feeding of the first dose of colostrum.
The sow's 134 carcass
is closed by staff with suture and disposed of following appropriate
procedures.
[000287] The piglets 144 are subsequently quarantined in a separate sterile
piglet quarantine
room 150 then transferred to a designated pathogen free isolation area ("DPF
Isolation Area") 152
to either create or join the DPF Closed Colony 102. It will be understood that
the DPF Isolation
Area 152 can be of any size suitable to manage and maintain the DPF Closed
Colony to the extent
needed for breeding, rearing, birthing, harvesting, and overall management as
described herein.
[000288] In one aspect, the DPF Isolation Area 152 that supports the DPF
Closed Colony is
a restricted access, positive-pressure barrier isolation suite, approximately
500ft2, with an animal
husbandry capacity to support at least 9 animals (up to 20 kg each), inside
the larger SAF 100. It
will be understood that the DPF Isolation Area 152 can be significantly larger
than this, and can
include multiple areas (including, but not limited to, multiple rooms and
suites), depending on the
need of the number of source animals and demand for products, in accordance
with the products
and methods as described herein.
[000289] In some aspects, tracking of piglets is performed and piglets are
handled under
designated pathogen free conditions in the DPF Isolation Area 152. For
example, handling of
piglets is performed wearing personal protective equipment ("PPE") in the DPF
Isolation Area
152, including face mask, gloves, shoe covers, and hair bonnet. The animals
are handled by clean
personnel, personnel who have not entered any animal room or facility where
other swine are
housed. For tracking, piglets are ear notched 3 days after birth and ear
tagged with hand-labeled
plastic ear tags at weaning (usually 3-5 weeks).
[000290] It will be understood that some piglets are raised in the DPF
Closed Colony 102 in
the DPF Isolation Area 152 as a source for xenotransplantation products, and
some piglets in the
DPF Closed Colony 102 are allowed to mature and be used to propagate the
General Closed
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Colony 128. In the event of propagation of the General Closed Colony 128, the
matured animal is
removed from the DPF Isolation Area 152 and added to the General Closed Colony
128 for
breeding. Since the DPF Isolation Area 152 is controlled to be DPF, once these
or any other
animals leave DPF Isolation Area 152, those animals never return to the DPF
Isolation Area 152.
[000291] Precautions are taken to prevent the exposure of any animals
within the DPF Closed
Colony 102 to contamination (for example, blood, blood products or tissues
obtained from animals
outside the DPF Closed Colony 102). If any animals within the DPF Closed
Colony 102 are
inadvertently exposed to blood, blood products, or tissues obtained from
animals outside the DPF
Closed Colony 102, those animals are removed from the DPF Closed Colony 102
and will never
return to the DPF Closed Colony 102. Aseptic techniques and sterile equipment
for all parenteral
interventions are used, and routine procedures such as vaccinations, treatment
with drugs or
biologics, phlebotomy, and biopsies are performed. The DPF Isolation Area 152
is restricted by
card access only to specially authorized and trained staff
[000292] In another aspect of the invention, in some aspects, newborn
piglets are handled
and hand-reared by trained and gowned staff in the DPF Isolation Area 152 to
ensure their health
and that they are maintained as designated pathogen free.
Propagation
[000293] The DPF Closed Colony 102 can be propagated in multiple ways. For
example, as
described herein, sows 134 may be taken from the outside or General Closed
Colony 128,
quarantined, and have their piglets 144 delivered via Cesarean section, with
the piglets
resuscitated, sterilized, quarantined, and placed into the DPF Isolation Area
152. Newborn piglets
may be maintained at 26-30 C or 80-85 F. In some aspects, heat lamps are used
to keep animals
warm. Newborn piglets are initially housed in sterilized medium crates in the
SAF with sterile
towels/drapes on the bottom.
[000294] The DPF Closed Colony 102 may also be propagated in other ways.
For example,
in one aspect, the DPF Closed Colony 102 is propagated through natural
intercourse amongst the
animals in the DPF Closed Colony 102 occurring entirely within the DPF
Isolation Area 152. It
will be understood that pregnancies may also occur in the DPF Closed Colony
102 within the DPF
Isolation Area 152 as a result of artificial insemination or other breeding
techniques that do not
involve natural intercourse.
76
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000295] In such aspects, pregnant sows 154 (or gilts) in the DPF Closed
Colony 102 within
the DPF Isolation Area 152 carry the entire pregnancy and piglets are
delivered through live
vaginal birth and Caesarian section is not necessary. Importantly, the piglets
resulting from natural
intercourse and live vaginal birth within the DPF Isolation Area 152 are
designated pathogen free,
including no infection by pCMV.
[000296] Following the live vaginal birth, piglets are immediately taken
away from the sow
to prevent the sows from harming the piglets. The piglets are then hand-reared
from birth by
humans within the DPF Isolation Area 152 in the methods as described herein.
[000297] In the case of mating in the DPF Closed Colony 102 or General
Closed Colony 128,
the breeding of swine disclosed herein is typically homozygous to homozygous
breeding. Females
are given hormones two weeks before gestation then throughout pregnancy.
Furthermore, as with
the DPF Closed Colony 102, the General Closed Colony 128 may also be
propagated through
natural intercourse amongst the animals in the General Closed Colony 128, and
may also occur as
a result of artificial insemination or other assisted reproductive
technologies (ARTs) that do not
involve natural intercourse.
[000298] Various techniques have been developed and refined to obtain a
large number of
offspring from genetically superior animals or obtain offspring from infertile
(or subfertile)
animals. These techniques include: artificial insemination, cryopreservation
(freezing) of gametes
or embryos, induction of multiple ovulations, embryo transfer, in vitro
fertilization, sex
determination of sperm or embryos, nuclear transfer, cloning, etc.
[000299] Artificial insemination (Al) has been used to obtain offspring
from genetically
superior males for more than 200 years. Improvements in methods to
cryopreserve (freeze) and
store semen have made Al accessible to more livestock producers. In the same
manner as
cryopreservation of semen, embryo freezing allowed for the global
commercialization of animals
with high genetic qualities.
[000300] Multiple ovulation and embryo transfer: Development of embryo
transfer
technology allows producers to obtain multiple progeny from genetically
superior females.
Depending on the species, fertilized embryos can be recovered from females
(also called embryo
donors) of superior genetic merit by surgical or nonsurgical techniques. The
genetically superior
embryos are then transferred to females (also called embryo recipients) of
lesser genetic merit. In
cattle and horses, efficient techniques recover fertilized embryos without
surgery, but only one or
77
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
sometimes two embryos are produced during each normal reproductive cycle. In
swine and sheep,
embryos must be recovered by surgical techniques. To increase the number of
embryos that can
be recovered from genetically superior females, the embryo donor is treated
with a hormone
regimen to induce multiple ovulations, or superovulation.
[000301] In vitro Fertilization: As an alternative to collecting embryos
from donor animals,
methods have been developed recently to produce embryos in vitro (in the
laboratory). The
methods are also called in vitro embryo production. Immature oocytes (female
eggs) can be
obtained from ovaries of infertile or aged females, or from regular embryo
donors (described
above). Ovum (egg) pick up is a nonsurgical technique that uses ultrasound and
a guided needle
to aspirate immature oocytes from the ovaries. Once the immature oocytes have
been removed
from the ovary, they are matured, fertilized, and cultured in vitro for up to
seven days until they
develop to a stage that is suitable for transfer or freezing.
[000302] Since the mid 1980s, technology has been developed to transfer the
nucleus from
either a blastomere (cells from early, and presumably undifferentiated
cleavage stage embryos) or
a somatic cell (fibroblast, skin, heart, nerve, or other body cell) to an
enucleated oocyte
(unfertilized female egg cell with the nucleus removed). This "nuclear
transfer" produces multiple
copies of animals that are themselves nearly identical copies of other animals
(transgenic animals,
genetically superior animals, or animals that produce high quantities of milk
or have some other
desirable trait, etc.). This process is also referred to as cloning. To date,
somatic cell nuclear
transfer has been used to clone cattle, sheep, pigs, goats, horses, mules,
cats, rabbits, rats, and mice.
[000303] The technique involves culturing somatic cells from an appropriate
tissue
(fibroblasts) from the animal to be cloned. Nuclei from the cultured somatic
cells are then
microinjected into an enucleated oocyte obtained from another individual of
the same or a closely
related species. Through a process that is not yet understood, the nucleus
from the somatic cell is
reprogrammed to a pattern of gene expression suitable for directing normal
development of the
embryo. After further culture and development in vitro, the embryos are
transferred to a recipient
female and ultimately result in the birth of live offspring. The success rate
for propagating animals
by nuclear transfer is often less than 10 percent and depends on many factors,
including the species,
source of the recipient ova, cell type of the donor nuclei, treatment of donor
cells prior to nuclear
transfer, the techniques used for nuclear transfer, etc.
78
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000304] Most commonly used ARTs rely on fertilization as a first step.
This joining of egg
and sperm is accompanied by the recombination of the genetic material from the
sire and dam, and
is often referred to as "shuffling the genetic deck." It will be understood
that these breeding
techniques can be used either within the DPF Closed Colony, as a breeding step
within the DPF
Isolation Area 152, or could be used as a breeding step for females in the
General Closed Colony
and/or from the outside.
[000305] In the case of utilization of ART to impregnate females in the
General Closed
Colony, and/or a female from the outside, the birthing of piglets from such
females can be as
described herein, i.e., sows 134 may be taken from the outside or General
Closed Colony 128,
quarantined, and have their piglets 144 delivered via Cesarean section, with
the piglets
resuscitated, sterilized, quarantined, and placed into the DPF Isolation Area
152.
Maintenance of Closed Colonies
[000306] Designated pathogens may include any number of pathogens,
including, but not
limited to, viruses, bacteria, fungi, protozoa, parasites, and/or prions
(and/or other pathogens
associated with transmissible spongiform encephalopathies (TSEs)). Designated
pathogens could
include, but not be limited to, any and all zoonotic viruses and viruses from
the following families:
adenoviridae, anelloviridae, astroviridae, calicivirdae, circoviridae,
coronaviridae, parvoviridae,
picornaviridae, and reoviridae.
[000307] Designated pathogens could also include, but not be limited to,
adenovirus,
arbovirus, arterivirus, bovine viral diarrhea virus, calicivirus, cardiovirus,
circovirus 2, circovirus
1, coronavirus, encephalomyocarditus virus, eperytherozoon, haemophilus suis,
herpes and
herpes-related viruses, iridovirus, kobuvirus, leptospirillum, listeria,
mycobacterium TB,
mycoplasma, orthomyxovirus, papovirus, parainfluenza virus 3, paramyxovirus,
parvovirus,
pasavirus-1, pestivirus, picobirnavirus (PBV), picornavirus, porcine
circovirus-like (po-circo-like)
virus, porcine astrovirus, porcine bacovirus, porcine bocavirus-2, porcine
bocavirus-4, porcine
enterovirus-9, porcine epidemic diarrhea virus (PEDV), porcine polio virus,
porcine lymphotropic
herpes virus (PLHV), porcine stool associated circular virus (PoSCV),
posavirus-1, pox virus,
rabies-related viruses, reovirus, rhabdovirus, rickettsia, sapelovirus,
sapovirus, staphylococcus
hyicus, staphylococcus intermedius, staphylococcus epidermidis, coagulase-
negative
staphylococci, suipoxvirus, swine influenza, teschen, torovirus, torque teno
sus virus-2 (TTSuV-
2), transmissible gastroenteritus virus, vesicular stomatitis virus, and/or
any and/or all other
79
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
viruses, bacteria, fungi, protozoa, parasites, and/or prions (and/or other
pathogens associated with
TSEs). In some aspects, particularly in swine herds, testing for TSEs is not
performed because
TSEs are not reported in natural conditions in swine. In other aspects,
testing for TSEs is performed
as part of the methods of the present disclosure.
[000308] There are huge numbers of pathogens that could possibly be tested
for in animal
herds, and there is no regulatory guidance or standard, or understanding in
the field as to what
specific group of pathogens should be tested for in donor animals, and which
specific group of
pathogens should be removed from donor animal populations in order to ensure
safe and effective
xenotransplantation. In other words, before the present disclosure, there was
no finite number of
identified, predictable pathogens to be tested for and excluded.
[000309] Importantly, the present disclosure provides a specific group of
pathogens
identified by the present inventors that are critical to exclude for safe and
effective
xenotransplantation, as set forth in the following Table 1.
TABLE 1
Test Pathogen
Parasite Fecal Float Ascaris species
Cryptosporidium species
Echinococcus
Strongyloids sterocolis
Toxoplasma gondii
Brucella BAPA (buffered Brucella suis
acidified plate agglutination
test)
Lepto6 Screen Leptospira species
M Hyo Mycoplasma Hyopneumoniae
PRRS x3 ELISA Porcine Reproductive and Respiratory Syndrome
Virus (PRRSV)
PRVgb Test Pseudorabies
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
T GE/PRC V Test Porcine Respiratory Coronavirus
Toxoplasmosis ELISA Toxoplasma Gondii
Porcine Cytomegalovirus Porcine CMV
PCR
Porcine Influenza PCR Porcine Influenza A
Nasal swab Bordetella bronchiseptica
Skin culture Coagulase-positive staphylococci
Skin culture Coagulase-negative staphylococci
Skin culture Livestock-associated methicillin
resistant Staphylococcus aureus (LA MRSA)
Skin culture Microphyton and Trichophyton spp.
Porcine Endogenous Porcine Endogenous Retrovirus (PERV) C (PERV C)
Retrovirus RT-PCR Assay
[000310] In certain aspects, a product of the present disclosure is sourced
from animals
having antibody titer levels below the level of detection for a plurality of
or all of the pathogens
discussed in the present disclosure. In certain aspects, subjects transplanted
with a product of the
present disclosure are tested and found to have antibody titer levels below
the level of detection
for a plurality of or all of the pathogens discussed in the present
disclosure.
[000311] In some aspects, the present disclosure includes a method of
testing for a specific
group of pathogens consisting of no more than 18-35, e.g., 35, 34, 33, 32, 31,
30, 29, 28, 27, 26,
25, 24, 23, 22, 21, 20, 19, or 18 pathogens, the specific group of pathogens
including each of the
pathogens identified in Table 1. In some aspects, the present disclosure
includes creating,
maintaining and using donor animals that are free of the 18-35, e.g., 35, 34,
33, 32, 31, 30, 29, 28,
27, 26, 25, 24, 23, 22, 21, 20, 19, or 18 pathogens, the specific group of
pathogens including each
of the pathogens identified in Table 1.
[000312] As described herein, piglets born via live vaginal birth within
the DPF Closed
Colony 102 are not infected with pCMV, but are nonetheless tested for pCMV on
a continuous
basis. Testing for Porcine Cytomegalovirus (pCMV) and Porcine Endogenous
Retrovirus (PERV),
should be routine and continuous for screening and maintenance as described
herein, and should
81
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
occur routinely and continuously for the DPF Closed Colony. In some aspects of
the present
invention, the source animals described herein are positive for PERV A and B
only, and some are
positive for PERV A, B, and C. In other aspects, the source animals are free
of PERV A, B and/or
C (through utilization of CRISPR and other techniques).
[000313] With respect to PERV, it is understood that most, if not all,
swine are known to be
positive for PERV A and B. While PERV is recognized, the risk of transmission
of PERV from
treatment with swine derived tissue is expected to be rare. To date eight PERV
mRNAs are
expressed in all porcine tissues and in all breeds of swine and preclinical
and clinical
xenotransplantation studies of humans exposed to pig cells, tissues, and
organs including
pancreatic islets have failed to demonstrate transmission of PERV. See, e.g.,
Morozov VA,
Wynyard S, Matsumoto S, Abalovich A, Denner J, Elliott R, "No PERV
transmission during a
clinical trial of pig islet cell transplantation," Virus Res 2017;227:34-40.
In the unlikely event that
a human infection should occur, PERV is susceptible in vitro to nucleoside and
non-nucleoside
reverse transcriptase inhibitors in common clinical use. See, e.g., Wilhelm M,
Fishman JA,
Pontikis R, Aubertin AM, Wilhelm FX, "Susceptibility of recombinant porcine
endogenous
retrovirus reverse transcriptase to nucleoside and non-nucleoside inhibitors,"
Cellular &
Molecular Life Sciences 2002;59:2184-90; Schuurman, H., "Regulatory aspects of
clinical
xenotransplantation," Int. J. Surg., 23, (2015), pp. 312-321. Experimental
data using the
xenotransplantation product of the present disclosure indicated that PERV
genetic material was
not detected in the recipient's organs and that porcine DNA and cells did not
migrate into the
circulation of the recipient from the xenotransplanted organ.
[000314] The DPF Closed Colony 102 is maintained to ensure that the animals
remain
designated pathogen free and that appropriate standards of animal care and
well-being are applied
at all levels of the SAF 100 (i.e., breeding, maintenance, propagation). No
animal is permitted
into the DPF Closed Colony if it or a parent has tested positive for any of
the pathogens in Table
1. For example, continuous testing for pathogens and other biological markers
occurs including
the numerous pathogens identified herein (including, but not limited to, pCMV
and other
pathogens). Environmental and blood samples are collected as necessary for
genotyping and
testing for pathogens. Test result(s) obtained for pathogens or other health
concerns are evaluated
by the facility veterinarian who may recommend follow-up testing and
observations, and
quarantine of the facility or areas (e.g., rooms, suites or other areas)
within a facility as needed.
82
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Careful documentation of any antimicrobial agents used during routine care of
the source animals
should be maintained, and exclusive use of killed vaccines used. Examples of
antimicrobial agents
include cefazolin, bacitracin, neomycin, and polymyxin.
[000315] In some aspects, routine health surveillance and screening for
pathogens (e.g.,
adventitious agents) of source animals is performed every 3 months. Samples of
serum, nasal
swabs, and stool for each animal in the General and DPF Closed Colonies are
obtained and
provided for analytical tests for detection of such pathogens every 3 months.
Source animal
samples of serum, nasal swabs, and stool for testing are obtained immediately
after euthanasia via
captive bolt and evaluated as disclosed herein including one or more of:
conducting a sterility
assay and confirming that aerobic and anaerobic bacteria do not grow in the
sterility assay;
conducting a mycoplasma assay and confirming that mycoplasma colonies do not
grow in the
mycoplasma assay; conducting an endotoxin assay and confirming that the
biological product is
free of endotoxins in the endotoxin assay, conducting the MTT-reduction assay
and confirming
that the product has at least 50% cell viability in the MTT-reduction assay;
conducting flow
cytometry and confirming that the product does not have galactosyl-a-1,3-
galactose epitopes as
determined by the flow cytometry; conducting pathogen-detection assays
specific for 18 to 35
pathogens and confirming that the product is free of Ascaris species,
cryptosporidium species,
Echinococcus, Strongyloids sterocolis, Toxoplasma gondii, Brucella suis,
Leptospira species,
mycoplasma hyopneumoniae, porcine reproductive and respiratory syndrome,
pseudorabies,
staphylococcus species, Microphyton species, Trichophyton species, porcine
influenza, porcine
cytomegalovirus, arterivirus, coronavirus, Bordetella bronchiseptica, and
Livestock-
associated methicillin-resistant Staphylococcus aureus.
[000316] In some aspects, all swine undergo routine health monitoring,
which includes
documentation of all illnesses, medical care, procedures, drugs administered,
vaccinations,
physical examinations, any treatments received, and general health assessments
and observations
each day at time of feeding with a visual health inspection indicating the
animal is able to stand,
move freely and appears clinically normal, as well as observations relating to
the animal's
appearance, activity and appetite, recording on the Animal Husbandry Log any
deficiencies. In
some aspects, animals are vaccinated against Mycoplasma Hyopneumoniae,
Hemophilus Parasuis,
Streptococcus Suis, Pasteurella Multocida, Bordatella Bronchiseptica and
Erysipelothrix
Rhusiopathiae. All swine six months or older may be vaccinated against
Erysipelothrix
83
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Rhusi op athi ae, Leptospira (Cani col a-Gripp otyphos a-Hardj o-
Icterohaemorrhagiae-Pomona),
Influenza and Parvovirus. Repeat vaccination may be performed, e.g., every six
months.
[000317] In some aspects, health monitoring will normally be performed as
part of daily
husbandry procedures for cleaning and feeding to minimize entry into swine
holding areas (e.g.,
rooms, suites or other areas). Prior to entering, personnel must wear personal
protective equipment
(PPE) and ensure that their footwear is free from gross contamination (e.g.
visible dirt or mud).
They will then don disposable shoe/boot covers prior to entry. Personnel in
contact with any
animals not housed in the designated pathogen free facility will change PPE if
contaminated. All
implements (shovel, other necessary tools) will undergo chlorhexidine
immersion of no less than
2 minutes if exogenous to vivarium and judged necessary. Solid waste and
soiled bedding is
removed. Animal holding areas are sanitized with diluted Quat-PV or bleach a
minimum of once
every two weeks.
[000318] In some aspects, bedding is replaced daily using irradiated
bedding wood shavings.
The replacement amount is an approximate equal amount to that which was
removed. All bedding
is completely replaced on a weekly basis at a minimum. Daily activities
including health status
checks, cleaning and water levels are documented in the Animal Husbandry log.
Appropriately
labeled trash and biological waste is picked up by staff daily and
incinerated.
[000319] With regard to piglet, newborns are handled and cared for by
trained and gowned
staff in an isolation suite. All supplies, room and crates are sanitized prior
to housing of the piglets.
Sterile drapes and towels are used to line the bottom of the crates. Room
temperature is controlled
to 80-85 F. Animals crates are maintained at 85950 F through the use of heat
lamps. Piglets are
maintained in the crates through the first 2 weeks after which time piglets
are housed on the floor
with irradiated wood shavings. Crates are cleaned daily and shavings are
removed and replenished
daily. Piglets are initially fed fresh-made, sterile colostrum (Bovine
Colostrum IgG formulated for
swine, Sterling Nursemate ASAP or equivalent) using a feeding tube every 1 to
2 hours until piglet
is self-feeding from feeder. During the early days, the piglet is weighed
twice a day and well-being
is checked and recorded twice a day. Starting at day 14, piglets are fed 3
times per day with a Milk
Replacer (Ralco Birthright or equivalent) that is further supplemented with
irradiated piglet grain
(antibiotic free creep feed, Blue Seal 813 or equivalent). The amount each
piglet eats at each
feeding is recorded. Vaccinations, genotyping, ear notching, and needle teeth
trimming are
performed within the first 7 days after birth of the piglet. In some aspects,
vaccines use killed
84
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
agents. Piglets are vaccinated against Mycoplasma Hyopneumoniae, Hemophilus
Parasuis,
Streptococcus Suis, Pasteurella Multocida, Bordatella Bronchiseptica and
Erysipelothrix
Rhusiopathiae at day 7 after birth, with a booster vaccination at 28 days of
age. In one aspect,
vaccines are killed agents. All swine six months or older are vaccinated
against Erysipelothrix
Rhusiopathiae, Leptospira (Canicola-Grippotyphosa- Hardjo-Icterohaemorrhagiae-
Pomona),
Influenza and Parvovirus. Repeat vaccination is performed every six months.
[000320] The source animals for the xenotransplantation product are
maintained in a positive
pressure, biocontainment establishment, under specific isolation-barrier
conditions governed by
standard operation procedures adopted by the managers of the given program,
and receive
specialized care, under controlled conditions in order to mitigate
adventitious agents. To ensure
the welfare of the closed colony of source animals intended for
xenotransplantation use, the SAF,
personnel, and the caretakers of source animals adhere to procedures for
animal husbandry, tissue
harvesting, and sacrifice of animals. The source animals are housed in a
positive pressure,
biocontainment establishment, under specific isolation- barrier conditions.
[000321] In some aspects, food and bedding are delivered to a loading dock,
transported, and
stored in a specific feed room off of the clean cage wash area accessible only
to staff in the inner
hallway. All bedding and feed are sterilized by irradiation and double bagged
to insure sterility.
Feed used for the piglets and more mature animals is defined grain feed by a
specific manufacturer.
It does not contain any cattle protein. Water supply is provided either by use
of the facility sterile
system or purchased sterile water which is dispensed into sterile pans.
Records for storage and
delivery of feed, water, and other consumables are maintained, and include
manufacturer, batch
numbers, and other pertinent information, per protocol.
[000322] In some aspects, animal records are maintained to describe the
feed provided to
source animals for at least two generations before their use as a source for
live tissues, organs
and/or cells used in xenotransplantation. This includes source, vendor, and
the type of feed used
(including its contents). Use of feed that has been derived from animals is
prohibited. Source
animals are not provided feeds containing animal proteins or other cattle
materials that are
prohibited by the FDA feed ban as expanded in 2008 as source animals (21 CFR
589.2000) or
feeds containing significant drug contamination or pesticide or herbicide
residues for source
animals (21 CFR 589.2001).
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000323] In some aspect, purified water is provided in sufficient quality
to prevent
unnecessary exposure of animals to infectious pathogens, drugs, pesticides,
herbicides, and
fertilizers. Newborn animals are provided colostrum specifically qualified for
herd qualification.
In some aspects, Bovine Colostrum IgG formulated for swine, Sterling Nursemate
ASAP or
equivalent is used to feed newborn animals.
Biological Products Derived from DPF Closed Colony
Biological Products
[000324] As described herein, biological products for xenotransplantation
are derived from
source animals produced and maintained in accordance with the present
invention, including from
the DPF Closed Colony 102 as described herein. Such biological products
include, but are not
limited to, liver, kidney, skin, lung, heart, pancreas, intestine, nerve and
other organs, cells and/or
tissues.
[000325] The present disclosure provides a continuous manufacturing process
for a
xenotransplantation product that has reduced immunogenicity, reduced
antigenicity, increased
viability, increased mitochondrial activity, a specifically required pathogen
profile, and
unexpectedly long shelf-life in xenotransplantation tissues subject to
cryopreservation. The
continuous manufacturing process is surprisingly and unexpectedly effective in
avoiding
hyperacute rejection, delayed xenograft rejection, acute cellular rejection,
chronic rejection, cross-
species transmission of diseases, cross-species transmission of parasites,
cross-species
transmission of bacteria, cross-species transmission of fungi, and cross-
species transmission of
viruses. The continuous manufacturing process is surprisingly and unexpectedly
effective in
creating a closed herd in which the donor animals survive normally without
detectable pathological
changes.
[000326] Harvesting of such biological products occurs in a single,
continuous, and self-
contained, segregated manufacturing event that begins with the sacrifice of
the source animal
through completion of the production of the final product. The animal is
euthanized via captive
bolt euthanasia, may be moved, if necessary, in a sterile, non-porous bag, to
an operating room
where the procedure to harvest biological product from the source animal will
occur. All members
of the operating team should be in full sterile surgical gear, e.g., dressed
in sterile dress to maintain
designated pathogen free conditions prior to receiving the source animal and
in some instanced be
double-gloved to minimize contamination, and surgical areas and tools are
sterilized. The source
86
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
animal is removed from the bag and container in an aseptic fashion. The source
animal is scrubbed
by operating staff, e.g., for at least 1-10 minutes with antiseptic, e.g.,
Chlorhexidine, brushes over
the entire area of the animal where the operation will occur, periodically
pouring Chlorhexidine
over the area to ensure coverage. Surgical area(s) of the animal are scrubbed
with opened Betadine
brushes and sterile water rinse over the entire area of the animal where the
operation will occur
for, e.g., 1-10 minutes. For surgery, operators will be dressed in sterile
dress in accordance with
program and other standards to maintain designated pathogen free conditions.
All organs, cells or
tissue from the source animal that will be used for xenotransplantation is
harvested within 15 hours
of the animal being sacrificed.
[000327] Biological products can also include, but are not limited to,
those disclosed herein
(e.g., in the specific examples), as well as any and all other tissues,
organs, and/or purified or
substantially pure cells and cell lines harvested from the source animals. In
some aspects, tissues
that are utilized for xenotransplantation as described herein include, but are
not limited to, areolar,
blood, adenoid, bone, brown adipose, cancellous, cartaginous, cartilage,
cavernous, chondroid,
chromaffin, connective tissue, dartoic, elastic, epithelial, Epithelium,
fatty, fibrohyaline, fibrous,
Gamgee, Gelatinous, Granulation, gut-associated lymphoid, Haller's vascular,
hard hemopoietic,
indifferent, interstitial, investing, islet, lymphatic, lymphoid, mesenchymal,
mesonephric, mucous
connective, multilocular adipose, muscle, myeloid, nasion soft, nephrogenic,
nerve, nodal,
osseous, osteogenic, osteoid, periapical, reticular, retiform, rubber,
skeletal muscle, smooth
muscle, and subcutaneous tissue. In some aspects, organs that are utilized for
xenotransplantation
as described herein include, but are not limited to, skin, kidneys, liver,
brain, adrenal glands, anus,
bladder, blood, blood vessels, bones, cartilage, cornea, ears, esophagus, eye,
glands, gums, hair,
heart, hypothalamus, intestines, large intestine, ligaments, lips, lungs,
lymph, lymph nodes and
lymph vessels, mammary glands, mouth, nails, nose, ovaries, oviducts,
pancreas, penis, pharynx,
pituitary, pylorus, rectum, salivary glands, seminal vesicles, skeletal
muscles, skin, small intestine,
smooth muscles, spinal cord, spleen, stomach, suprarenal capsule, teeth,
tendons, testes, thymus
gland, thyroid gland, tongue, tonsils, trachea, ureters, urethra, uterus, and
vagina.
[000328] In some aspects, purified or substantially pure cells and cell
lines that are utilized
for xenotransplantation as describe herein include, but are not limited to,
blood cells, blood
precursor cells, cardiac muscle cells, chondrocytes, cumulus cells,
endothelial cells, epidermal
cells, epithelial cells, fibroblast cells, granulosa cells, hematopoietic
cells, Islets of Langerhans
87
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
cells, keratinocytes, lymphocytes (B and T), macrophages, melanocytes,
monocytes, mononuclear
cells, neural cells, other muscle cells, pancreatic alpha-1 cells, pancreatic
alpha-2 cells, pancreatic
beta cells, pancreatic insulin secreting cells, adipocytes, epithelial cells,
aortic endothelial cells,
aortic smooth muscle cells, astrocytes, basophils, bone cells, bone precursor
cells, cardiac
myocytes, chondrocytes, eosinophils, erythrocytes, fibroblasts, glial cells,
hepatocytes,
keratinocytes, Kupffer cells, liver stellate cells, lymphocytes, microvascular
endothelial cells,
monocytes, neuronal stem cells, neurons, neutrophils, pancreatic islet cells,
parathyroid cells,
parotid cells, platelets, primordial stem cells., Schwann cells, smooth muscle
cells, thyroid cells,
tumor cells, umbilical vein endothelial cells, adrenal cells, antigen
presenting cells, B cells, bladder
cells, cervical cells, cone cells, egg cells, epithelial cells, germ cells,
hair cells, heart cells, kidney
cells, leydig cells, lutein cells, macrophages, memory cells, muscle cells,
ovarian cells, pacemaker
cells, peritubular cells, pituitary cells, plasma cells, prostate cells, red
blood cells, retinal cells, rod
cells, Sertoli cells, somatic cells, sperm cells, spleen cells, T cells,
testicular cells, uterine cells,
vaginal epithelial cells, white blood cells, ciliated cells, columnar
epithelial cells, dopaminergic
cells, dopaminergic cells, embryonic stem cells, endometrial cells,
fibroblasts fetal fibroblasts.,
follicle cells, goblet cells, keratinized epithelial cells, lung cells,
mammary cells, mucous cells,
non-keratinized epithelial cells, osteoblasts, osteoclasts, osteocytes, and
squamous epithelial cells.
[000329] An organ is a group of related cells that combine together to
perform one or more
specific functions within the body. Biologically, skin is the body's largest
and fastest¨growing
organ, and is classified as the primary component of the integumentary system,
one of the ten
macro-organ systems found in "advanced" animals. Skin fulfills several
critical roles including
regulating temperature, providing a dynamic barrier to the external world, and
serving as a conduit
to support an immense network of sensory receptors. The skin performs several
functions that are
vital to the survival and health of the body. The skin heals to prevent the
loss of blood after wounds,
regulates body temperature by dissipating heat and as a layer against cold,
absorption, secretion,
thermal-regulation, sensory detection and orientation, and barrier protection.
In fact, not only has
success in transplantation of skin been recognized to correlate to
transplantation of other organs,
but skin transplants appear to be more sensitive to rejection than other
organs, e.g., immune
privileged organs such as liver, and skin transplants have even been suggested
for use as "sentinel
transplants," i.e., use of skin grafts in a human recipient as early
predictors of rejection of
transplanted solid organs in the same recipient. For example, as reported in
Ali et al. Transplant
88
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Proc. 2016 Oct;48(8):2565-2570, evidence provided by experience with abdominal
wall
transplantation in some intestinal and multivisceral transplant recipients
suggest that rejection may
manifest in the skin component before emergence in the intestinal allograft,
providing a "lead
time" during which treatment of rejection of the abdominal wall could prevent
the emergence of
intestinal rejection.
[000330] Further, United States Code Title 42, Section 274 and Section 301,
explicitly list
skin in its formal definition of human organs, i.e., "Human organ,' as covered
by section 301 of
the National Organ Transplant Act, as amended, means the human (including
fetal) kidney, liver,
heart, lung, pancreas, bone marrow and other hematopoietic stem/progenitor
cells without regard
to the method of their collection, cornea, eye, bone skin, and intestine,
including the esophagus,
stomach, small and/or large intestine, or any portion of the gastrointestinal
tract." Similarly, the
Human Organ Transplant Ordinance (HOTO), an internationally ratified ordinance
to prevent
organ trading and protect donor and recipient rights to self¨determination.
This global legislation
lists skin - and whole segments of the integumentary system - formally as an
organ, and more
broadly defines an organ as "any part of the human body consisting of a
structured arrangement of
tissues which, if wholly removed, cannot be regenerated by the body...."
Following, the formal
medical definition of a transplant is: "the removal of tissue from one part of
the body or from one
individual and its implantation or insertion in another especially by
surgery." The HOTO defines
a transplant as "the transfer of an organ from one person to another during a
transplant operation,
regardless of permanence."
[000331] With regard to skin, grafts typically consist of decellularized
and/or reconstituted
sheets of homogenized dermis that are used to achieve temporary, superficial
wound coverage.
Such grafts do not retain the original tissue structure nor the metabolically
active, otherwise
naturally present cells, and thus do not become vascularized; no capillary
ingrowth or vessel-to-
vessel connections are made. Consequently, immune rejection is not a concern -
the skin graft
becomes "ejected" rather than rejected by the growth of a complete host
epithelium underneath
the graft. Thus, while the term graft can be correctly applied to such
solutions, the primary qualities
that differentiate a transplant from a graft are that of heightened
complexity, organization, and
inclusion of one or more types of tissue. In the present case, a skin
transplant is fundamentally
differentiated from grafts known in the prior art. For example, a skin
xenotransplant is comprised
of live cells that perform the same function as the patient's original skin
before eventually
89
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
experiencing immune-mediated rejected. Thus, in this context, a skin
xenotransplant according to
the present disclosure is an organ transplant rather than a graft.
[000332] In terms of harvesting a biological product from the swine,
wherein the harvesting
comprises euthanizing the swine and aseptically removing the biological
product from the swine;
processing said biological product comprising sterilization after harvesting
using a sterilization
process that does not reduce cell viability to less than 50% cell viability in
a 344,5-
dimethylthiazol-2-y1)-2,5-diphenyltetrazolium bromide (MTT)-reduction assay
and does not
reduce mitochondrial activity to less than 50% mitochondrial activity; and
storing the biological
product in a sterile container; and the non-human animal is a non-transgenic
genetically
reprogrammed swine for xenotransplantation of cells, tissue, and/or an organ
isolated from the
non-transgenic genetically reprogrammed swine, the non-transgenic genetically
reprogrammed
swine comprising a nuclear genome that has been reprogrammed to replace a
plurality of
nucleotides in a plurality of exon regions of a major histocompatibility
complex of a wild-type
swine with nucleotides from orthologous exon regions of a known human major
histocompatibility
complex sequence from a human capture sequence, wherein said reprogramming
does not
introduce any frameshifts or frame disruptions. Further specific aspects,
details and examples are
provided in the following disclosures and claims and any and all combinations
of those aspects,
details and examples constitute aspects of the present disclosure.
[000333] In other aspect, Xenogeneic kidneys are derived from a genetically
engineered,
reprogrammed and designated pathogen free swine is produced in accordance with
the present
invention and transplanted into a non-human primate and a human. It is
expected that survival of
at least fourteen months is observed in each of the non-human primate and the
human. In some
aspects, it is expected that survival of at least 24 months is observed in
each of the non-human
primate and the human. In some aspects, it is expected that survival of at
least 36 months is
observed in each of the non-human primate and the human. In some aspects, it
is expected that
survival of at least 48 months is observed in each of the non-human primate
and the human. In
some aspects, it is expected that survival of at least 60 months is observed
in each of the non-
human primate and the human.
[000334] In another aspect, Xenogeneic lungs are derived from a genetically
engineered,
reprogrammed and designated pathogen free swine produced in accordance with
the present
invention and transplanted into a non-human primate and a human. It is
expected that survival of
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
at least 30 days is observed in each of the non-human primate and the human.
In some aspects, it
is expected that survival of at least 3 months is observed in each of the non-
human primate and the
human. In some aspects, it is expected that survival of at least 6 months is
observed in each of the
non-human primate and the human. In some aspects, it is expected that survival
of at least 12
months is observed in each of the non-human primate and the human. In some
aspects, it is
expected that survival of at least 24 months is observed in each of the non-
human primate and the
human. In some aspects, it is expected that survival of at least 36 months is
observed in each of
the non-human primate and the human. In some aspects, it is expected that
survival of at least 48
months is observed in each of the non-human primate and the human. In some
aspects, it is
expected that survival of at least 60 months is observed in each of the non-
human primate and the
human.
[000335] In another aspect, Xenogeneic hearts are derived from a
genetically engineered,
reprogrammed and designated pathogen free swine produced in accordance with
the present
invention and transplanted into a non-human primate and a human. It is
expected that survival of
at least 20 months is observed in each of the non-human primate and the human.
In some aspects,
it is expected that survival of at least 24 months is observed in each of the
non-human primate and
the human. In some aspects, it is expected that survival of at least 36 months
is observed in each
of the non-human primate and the human. In some aspects, it is expected that
survival of at least
48 months is observed in each of the non-human primate and the human. In some
aspects, it is
expected that survival of at least 60 months is observed in each of the non-
human primate and the
human.
[000336] In another aspect, Xenogeneic nerve tissues are derived from a
genetically
engineered, reprogrammed and designated pathogen free swine produced in
accordance with the
present invention and transplanted into a non-human primate and a human. It is
expected that
survival of at least 75 days is observed in each of the non-human primate and
the human. In some
aspects, it is expected that survival of at least 3 months is observed in each
of the non-human
primate and the human. In some aspects, it is expected that survival of at
least 6 months is observed
in each of the non-human primate and the human. In some aspects, it is
expected that survival of
at least 12 months is observed in each of the non-human primate and the human.
In some aspects,
it is expected that survival of at least 24 months is observed in each of the
non-human primate and
the human. In some aspects, it is expected that survival of at least 36 months
is observed in each
91
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
of the non-human primate and the human. In some aspects, it is expected that
survival of at least
48 months is observed in each of the non-human primate and the human. In some
aspects, it is
expected that survival of at least 60 months is observed in each of the non-
human primate and the
human. zx
[000337] In another aspect, Xenogeneic livers are derived from a
genetically engineered,
reprogrammed and designated pathogen free swine produced in accordance with
the present
invention and transplanted into a non-human primate and a human. It is
expected that survival of
at least 60 days is observed in each of the non-human primate and the human.
In some aspects, it
is expected that survival of at least 3 months is observed in each of the non-
human primate and the
human. In some aspects, it is expected that survival of at least 6 months is
observed in each of the
non-human primate and the human. In some aspects, it is expected that survival
of at least 12
months is observed in each of the non-human primate and the human. In some
aspects, it is
expected that survival of at least 24 months is observed in each of the non-
human primate and the
human. In some aspects, it is expected that survival of at least 36 months is
observed in each of
the non-human primate and the human. In some aspects, it is expected that
survival of at least 48
months is observed in each of the non-human primate and the human. In some
aspects, it is
expected that survival of at least 60 months is observed in each of the non-
human primate and the
human.
[000338] In some embodiments, use of pig livers produced in accordance with
the present
invention to serve as extracorporeal filters for humans are disclosed. In a
study by Levy, et al.,
"Liver allotransplantation after extracorporeal hepatic support with
transgenic (hCD55/hCD59)
porcine livers: Clinical results and lack of pig-to- human transmission of the
porcine endogenous
retrovirus," Transplantation, 69(2):272-280 (2000) ("Levy"), the entire
contents of which are
incorporated herein by reference, whole organ extracorporeal perfusion of a
genetically modified
transgenic porcine liver was proposed to sustain patients awaiting human liver
transplantation for
fulminant hepatic failure. The pig livers used were reported to be transgenic
for human CD55
(decay-accelerating factor) and human CD59, however, the livers failed to
suppress marked
increase of [alpha]-gal antibodies.
[000339] In accordance with the present invention, in one aspect, a liver
derived from a
genetically reprogrammed source animal in accordance with the present
invention is utilized for
extracorporeal perfusion as a temporary filter for a human patient until a
patient receives a human
92
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
transplant. It will be understood that pigs with additional genetic
modifications may also be
utilized, including pigs genetically reprogrammed for any number of traits
disclosed elsewhere
herein.
[000340] In one aspect, as shown in FIG. 38, an extracorporeal circuit
utilizes an oxygenator
(e.g., Minimax Plus hollow fiber oxygenator), a pump (e.g., Bio-Medicus model
540 Bio-
Console with a BP50 Pediatric Bio Pump centrifugal pump), and a warmer (Bio-
Medicus
model 370 BioCalTM Temperature Controller). The circuit also utilizes a roller
pump (e.g., Sarns
model 7000; Sams, Ann Arbor, MI) to supplement for lack of gravity return to
the patient. Bridges
and clamps are utilized to isolate both the perfused liver and the patient.
[000341] In an operating area within the DPF Isolation Area, a source
animal is placed under
a general anesthetic (ketamine, xylazine, enflurane) or euthanized by captive
bolt. A hepatectomy
is then performed on the source animal in designated pathogen free conditions.
[000342] The livers can be preserved in any number of ways known in the art
prior to use as
an extracorporeal filter, including, but not limited to, as disclosed in Levy
(e.g., "a 4 C lactated
Ringer's/albumin solution and cannulated in the portal vein (28F Research
Medical, model SPC-
641-28) and the inferior vena cava (36F Research Medical, model SPC-641-36)").
[000343] The common bile duct can be intubated in any number of ways,
including, but not
limited to, as set forth in Levy (e.g., "with an intravenous extension tube
(Extension Set 30, Abbott
Hospitals, Inc., Chicago, IL) to allow subsequent quantification of bile
production.")
[000344] The liver product derived from the source animal can be packaged
and transported
to the location of the procedure in accordance with current practice with
human donor livers.
[000345] The procedure to utilize the liver filtration product can be
performed, for example,
by percutaneously cannulating a patient's internal jugular vein for venous
return with a 12F
pediatric arterial cannula (e.g., Medtronic DLP, Grand Rapids, MI) and
percutaneously
cannulating a patient's femoral vein for venous outflow with a 19F femoral
artery cannula (e.g.,
Medtronic Bio-Medicus, Eden Prairie, MN). These cannulas are connected to a
bypass circuit,
having a centrifugal pump (e.g., Bio-Medicus), a heat exchanger (Medtronic Bio-
Medicus), an
oxygenator (e.g., Medtronic Cardiopulmonary, Anaheim, CA), and a roller pump
(e.g., Sarns)
incorporated therein.
[000346] This circuit is primed with crystalloids and run for a period of
time (e.g., 20
minutes) before the liver obtained from the genetically reprogrammed source
animal is
93
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
incorporated at a stabilized flow rate of 800 ml/min, maintained in a
crystalloid bath occasionally
supplemented with warm solution.
[000347] In other aspect, Xenogeneic pancreases are derived from a
genetically engineered,
reprogrammed and designated pathogen free swine is produced in accordance with
the present
invention Xenogeneic pancreas derived from a genetically reprogrammed swine
produced in
accordance with the present invention is transplanted into a non-human primate
and a human. It is
expected that survival of at least 20 months is observed in each of the non-
human primate and the
human. In some aspects, it is expected that survival of at least 24 months is
observed in each of
the non-human primate and the human. In some aspects, it is expected that
survival of at least 36
months is observed in each of the non-human primate and the human. In some
aspects, it is
expected that survival of at least 48 months is observed in each of the non-
human primate and the
human. In some aspects, it is expected that survival of at least 60 months is
observed in each of
the non-human primate and the human.
[000348] While the subject matter of this disclosure has been described and
shown in
considerable detail with reference to certain illustrative aspects, including
various combinations
and sub-combinations of features, those skilled in the art will readily
appreciate other aspects and
variations and modifications thereof as encompassed within the scope of the
present disclosure.
Moreover, the descriptions of such aspects, combinations, and sub-combinations
is not intended
to convey that the claimed subject matter requires features or combinations of
features other than
those expressly recited in the claims. Accordingly, the scope of this
disclosure is intended to
include all modifications and variations encompassed within the spirit and
scope of the following
appended claims.
[000349] In other aspect, Xenogeneic dermal combination product derived
from a genetically
engineered, reprogrammed and designated pathogen free swine is produced in
accordance with the
present invention.
[000350] Some skin transplantation products for the treatment of burns and
other ailments
utilize cultured epidermal autografts (see, e.g., products produced by Vericel
Corporation under
the Epicel brand name). Such epidermal autografts can be utilized for
patients with burns
(including severe burns) and result in reduced or no rejection in the
transplanted epidermal material
since the material is derived from the patient's own skin.
94
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000351] However, such products are limited to the epidermis only, and do
not include the
dermis portion of the skin. Referring to FIG. 39, it will be understood that
the dermis (which
typically accounts for 95% of the thickness of the skin) performs
significantly different functions
than the epidermis (which is the outer portion of the skin that typically
accounts for 5% of the
thickness of the skin).
[000352] Since epidermal autografts alone lack the ability to perform the
critical functions of
the dermis, such products are used in combination with a viable dermis. In
some injuries, the
wound bed includes remaining portions of the patient's own dermis, which is
the ideal dermis to
utilize in a procedure grafting cultured epidermal autografts onto a patient.
However, in some cases
the burn is more severe, and the patient's own dermis no longer exists or is
no longer viable. In
those instances, a different dermis is required since an epidermal autograft
alone will not suffice.
[000353] In one aspect, a full thickness skin graft wound dressing
consisting of dermal tissue
derived from designated pathogen free a-1,3-galactosyltransferase [Gal-T]
knockout swine in
accordance with the present invention is used in conjunction or combination
with cultured
epidermal autografts. One treatment process utilizing this combination is as
follows.
[000354] A patient with severe burn wounds is taken to an operating room
within 48-72 hours
of injury. A biopsy is taken as soon as possible after the patient undergoes
care, and the epidermis
skin cells are isolated and grown separately according to the known procedures
for creating
cultured epidermal autografts (see, e.g., products produced by Vericel
Corporation under the
Epicel brand name).
[000355] Depending on how much of the patient's body is damaged, epidermal
autografts
are taken from healthy areas to treat burned areas and/or to later create an
epidermal autograft
mesh used in the grafting process.
[000356] Areas of severe burns are treated with the skin products described
herein, e.g., skin
products derived from a designated pathogen free a-1,3-galactosyltransferase
[Gal-T] knockout
swine produced in accordance with the present invention. Such treatments
comprise temporary
wound coverage until sufficient autografts are utilized to treat the patient
long-term.
[000357] Prior to application of the epidermal autografts, significant
debridement of wound
bed is required to ensure an adequate substrate. To confirm a wound bed is
ready for an epidermal
autograft, apply the skin products described herein, e.g., skin products
derived from a designated
pathogen free a-1,3-galactosyltransferase [Gal-T] knockout swine produced in
accordance with
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
the present invention to confirm adherence. Once adherence is confirmed, the
temporary wound
coverage product is removed, and in some aspects, the wound bed is covered
with a meshed
autograft, and one or more cultured epidermal autograft products are placed on
top to close the
gaps in the autograft mesh.
[000358] The debridement may include mechanical debridement, chemical
debridement,
enzymatic debridement, or a combination thereof. Mechanical debridement may
include surgical
excision, e.g., tangential excision to remove thin layers of dermis until
healthy tissue is visualized,
or fascial excision to remove the full thickness of dermis down to the
underlying fascia. Tangential
excision allows less viable tissue to be removed with the necrotic tissue, but
typically results in
higher blood loss, is a larger physiologic stressor than fascial excision, and
is more likely to result
in "incomplete" debridement, with some devitalized tissue remaining in place.
In fascial excision,
blood loss and operative time are minimized, but often a large amount of
healthy tissue is removed
with the burned tissue. Debriding agents may include agents capable of
cleaning a burn wound by
removing foreign material and dead tissue. Many such agents are known. In
enzymatic
debridement, collagenases or other proteolytic enzymes are employed that break
down proteins of
the extracellular matrix, allowing devitalized tissue to be wiped away without
the need for surgery
while preferably leaving healthy tissue substantially intact. Enzymatic
debridement involves the
application of proteolytic and optionally other exogenous enzymes to a wound
surface to break
down necrotic tissue. Enzymatic debridement may be a relatively slow process,
carried out over a
period of a number of weeks in combination with other topical preparations,
soakings and repeated
dressings. Alternately, rapid enzymatic debridement can be accomplished using
multi-enzyme
products, for example, those extracted from the stem of the pineapple plant,
as disclosed for
example in WO 98/053850 and WO 2006/0006167, and as provided in the product
marketed under
the trade name Debraseg. A procedure for enzymatic debridement generally
utilizes an enzyme
such as bromelain derivatives, debridase, collagenase, papain derivatives,
streptokinase, sutilains,
fibrinolysin, deoxyribonuclease, krill derivatives, trypsin or combinations
thereof. Autolytic
debridement relies on enhancing the natural process of selective liquefaction,
separation and
digestion of necrotic tissue and eschar from healthy tissue that occurs in
wounds due to
macrophage and endogenous proteolytic activity. This is achieved by the use of
occlusive, semi-
occlusive or moist interactive dressings.Enzymatic debridement agents include
a bromelain
enriched enzyme product, other collagenases, or other enzyme products capable
of clearing
96
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
devitalized tissue or wound debris. NexoBridTM (MediWound Ltd.) is one such
bromelain enriched
product that specifically targets heat-denatured collagen for degradation,
resulting in partial-
thickness and full-thickness wounds requiring a wound coverage or dressing
product. Such
products and methods are described in U.S. Patent Nos. 8,540,983; 8,119,124;
7,128,719;
7,794,709; 8,624,077; and U52009/0010910A1, each of which is incorporated by
reference herein.
[000359] In some aspects, the wound bed may include or be a chronic wound
or an acute
wound. Chronic wounds include but are not limited to venous leg ulcers,
pressure ulcers, and
diabetic foot ulcers. Acute wounds include but are not limited to burns,
traumatic injuries,
amputation wounds, skin graft donor sites, bite wounds, frostbite wounds,
dermabrasions, and
surgical wounds.
[000360] In the cases where there is no dermis, skin products derived from
a designated
pathogen free a-1,3-galactosyltransferase [Gal-T] knockout swine produced in
accordance with
the present invention are utilized. The epidermis is removed from such
products (e.g., before
dermis harvesting on the pig with a VERSAJETTm Hydrosurgery system), so that
just the dermis
remains. Then, the subject swine dermis is placed on the patient's
subcutaneous tissue, serving as
a substrate for the cultured epidermal autograft process described above.
Product Characteristics, Testing and Therapeutic Uses
[000361] In some aspects, the xenotransplantation products described and
disclosed herein
are temporary, i.e., their use in patients for xenotransplantation is non-
permanent, utilized
primarily for the treatment of acute ailments and injuries, able to be
utilized for longer periods of
time as compared to products that are not produced in accordance with the
present invention. It
will be understood that some of the aspects of the products described and
disclosed herein may
also be permanent or more permanent, with transplanted organs, tissues and/or
cells being accepted
by human recipients over much longer periods of time without adverse
rejection.
[000362] In other aspects, the xenotransplantation products described and
disclosed herein
are viable, live cell (e.g., vital, biologically active) products; distinct
from synthetic or other tissue-
based products comprised of terminally sterilized, non-viable cells which are
incapable of
completing the vascularization process. Further, in some aspects, the product
of the present
disclosure is not devitalized, or "fixed" with glutaraldehydes or radiation
treatment.
97
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000363]
In yet other aspects, the xenotransplantation products described and disclosed
herein are minimally manipulated (e.g., without physical alteration of the
related cells, organs or
tissues) such that such products are substantially in their natural state.
[000364]
In certain aspects, the xenotransplantation products described and disclosed
herein
are obtained from a non-human animal, e.g., a non-transgenic genetically
reprogrammed swine,
including cells, tissue, and/or an organ isolated from the non-transgenic
genetically reprogrammed
swine, the non-transgenic genetically reprogrammed swine comprising a nuclear
genome that has
been reprogrammed to replace a plurality of nucleotides in a plurality of exon
regions of a major
histocompatibility complex of a wild-type swine with nucleotides from
orthologous exon regions
of a known human major histocompatibility complex sequence from a human
capture sequence,
and wherein cells of said genetically reprogrammed swine do not present one or
more surface
glycan epitopes, wherein said reprogramming does not introduce any frameshifts
or frame
disruptions.
For example, genes encoding alpha-1,3 galactosyltransferase, cytidine
monophosphate-N-acetylneuraminic acid hydroxylase (CMAH), and (31,4-N-
acetylgalactosaminyltransferase are disrupted such that surface glycan
epitopes encoded by said
genes are not expressed. Further specific aspects, details and examples are
provided in the
following disclosures and claims and any and all combinations of those
aspects, details and
examples constitute aspects of the present disclosure.
[000365]
In yet other aspects, the xenotransplantation products described and disclosed
herein are capable of making an organic union with the human recipient,
including, but not limited
to, being compatible with vascularization, collagen growth (e.g., in regard to
skin), and/or other
interactions from the transplant recipient inducing graft adherence, organic
union, or other
temporary or permanent acceptance by the recipient.
[000366]
In yet other aspects, the xenotransplantation products described and disclosed
herein are utilized in xenotransplantation without the need to use, or at
least reduction of use, of
immunosuppressant drugs or other immunosuppressant therapies to achieve
desired therapeutic
results.
[000367]
In other aspects, some of the xenotransplantation products described and
disclosed
herein (e.g., skin) are stored by cryopreservation, stored fresh (without
freezing), or stored via
other methods to preserve such products consistent with this invention.
Storage involves using
conditions and processes that preserve cell and tissue viability.
98
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000368] In some aspects, storage may involve storing organs, tissues, or
cells, in any
combination of a sterile isotonic solution (e.g., sterile saline with or
without antibiotics), on ice, in
a cryopreservation fluid, cryopreserved at a temperature of around -40 C or
around -80 C, and
other methods known in the field. Such storage can occur in a primary
containment system and
secondary containment system.
[000369] In yet other aspects, the xenotransplantation products described
and disclosed
herein are for homologous use, i.e., the repair, reconstruction, replacement
or supplementation of
a recipient's organ, cell and/or tissue with a corresponding organ, cell
and/or tissue that performs
the same basic function or functions as the donor (e.g., swine kidney is used
as a transplant for
human kidney, swine liver is used as a transplant for human liver, swine skin
is used as a transplant
for human skin, swine nerve is used as a transplant for human nerve and so
forth).
[000370] In yet other aspects, the xenotransplantation products described
and disclosed
herein have a low bioburden, minimizing pathogens, antibodies, genetic
markers, and other
characteristics that may serve to increase the product's bioburden and the
human body's
immunological rejection of the product upon xenotransplantation. This may
include the innate
immune system, through PRRs TLRs, detecting PAMPs and rejecting the subject
xenotransplantation product.
[000371] It will be understood that the aspects disclosed and described
herein can be applied
in any number of combinations to create an array or different aspects
comprising one or more of
the features and/or aspects of the aspects encompassed by the present
invention.
[000372] It will be understood that there are numerous therapeutic
applications for products
derived from DPF Closed Colony in accordance with the present invention. For
example, such
products may be utilized to treat acute and/or chronic disease, disorders, or
injuries to organ, cells
or tissue, and any and all other ailments that can utilize the products
disclosed herein. Such
treatments and/or therapies can include utilizing such products to repair,
reconstruct, replace or
supplement (in some aspects on a temporary basis and in other aspects a
permanent basis), a human
recipient's corresponding organ, cell and/or tissue that performs the same
basic function or
functions as the donor.
[000373] Specific treatment applications include, but are not limited to,
lung transplants, liver
transplants, kidney transplants, pancreas transplants, heart transplants,
nerve transplants and other
full or partial transplants. With regard to skin, treatment applications also
include, but are not
99
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
limited to, treatment of burn wounds, diabetic ulcerations, venous
ulcerations, chronic skin
conditions, and other skin ailments, injuries and/or conditions (including,
but not limited to, severe
and extensive, deep partial and full thickness injuries, ailments and/or
conditions) (see, e.g.,
Example 2 herein); use in adult and pediatric patients who have deep dermal or
full thickness burns
comprising a total body surface area greater than or equal to 30%, optionally
in conjunction with
split-thickness autografts, or alone in patients for whom split-thickness
autografts may not be an
option due to the severity and extent of their wounds/burns; treatment of
liver failure, wounds,
ailments, injuries and/or conditions with liver products derived in accordance
with the present
invention; treatment of peripheral nerve damage, and other nerve ailments,
injuries and/or
conditions; and cell and other therapies utilizing materials harvested from
the DPF Closed Colony,
including the therapeutic uses disclosed in U.S. Patent No. 7,795,493
("Phelps"), including cell
therapies and/or infusion for certain disorders (as disclosed in col. 30, line
1 to col. 31, line 9) and
treatment or certain disorders or pathologies (as disclosed in col. 31, lines
10 to 42), the disclosure
of which is incorporated by reference herein.
[000374] It will be understood that the specific recitation of therapies
herein in no way limits
the types of therapeutic applications for the products disclosed and described
herein, which
encompass acute and/or chronic disease, disorders, injuries to the following
organs, tissues and/or
cells: skin, kidneys, liver, brain, adrenal glands, anus, bladder, blood,
blood vessels, bones, brain,
brain, cartilage, ears, esophagus, eye, glands, gums, hair, heart,
hypothalamus, intestines, large
intestine, ligaments, lips, lungs, lymph, lymph nodes and lymph vessels,
mammary glands, mouth,
nails, nose, ovaries, oviducts, pancreas, penis, pharynx, pituitary, pylorus,
rectum, salivary glands,
seminal vesicles, skeletal muscles, skin, small intestine, smooth muscles,
spinal cord, spleen,
stomach, suprarenal capsule, teeth, tendons, testes, thymus gland, thyroid
gland, tongue, tonsils,
trachea, ureters, urethra, uterus, uterus, vagina, areolar, blood, adenoid,
bone, brown adipose,
cancellous, cartaginous, cartilage, cavernous, chondroid, chromaffin,
connective tissue, dartoic,
elastic, epithelial, Epithelium, fatty, fibrohyaline, fibrous, Gamgee,
Gelatinous, Granulation, gut-
associated lymphoid, Haller's vascular, hard hemopoietic, indifferent,
interstitial, investing, islet,
lymphatic, lymphoid, mesenchymal, mesonephric, mucous connective, multilocular
adipose,
muscle, myeloid, nasion soft, nephrogenic, nerve, nodal, osseous, osteogenic,
osteoid, periapical,
reticular, retiform, rubber, skeletal muscle, smooth muscle, and subcutaneous
tissue; blood cells,
blood precursor cells, cardiac muscle cells, chondrocytes, cumulus cells,
endothelial cells,
100
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
epidermal cells, epithelial cells, fibroblast cells, granulosa cells,
hematopoietic cells, Islets of
Langerhans cells, keratinocytes, lymphocytes (B and T), macrophages,
melanocytes, monocytes,
mononuclear cells, neural cells, other muscle cells, pancreatic alpha-1 cells,
pancreatic alpha-2
cells, pancreatic beta cells, pancreatic insulin secreting cells, adipocytes,
epithelial cells, aortic
endothelial cells, aortic smooth muscle cells, astrocytes, basophils, bone
cells, bone precursor
cells, cardiac myocytes, chondrocytes, eosinophils, erythrocytes, fibroblasts,
glial cells,
hepatocytes, keratinocytes, Kupffer cells, liver stellate cells, lymphocytes,
microvascular
endothelial cells, monocytes, neuronal stem cells, neurons, neutrophils,
pancreatic islet cells,
parathyroid cells, parotid cells, platelets, primordial stem cells, Schwann
cells, smooth muscle
cells, thyroid cells, tumor cells, umbilical vein endothelial cells, adrenal
cells, antigen presenting
cells, B cells, bladder cells, cervical cells, cone cells, egg cells,
epithelial cells, germ cells, hair
cells, heart cells, kidney cells, leydig cells, lutein cells, macrophages,
memory cells, muscle cells,
ovarian cells, pacemaker cells, peritubular cells, pituitary cells, plasma
cells, prostate cells, red
blood cells, retinal cells, rod cells, Sertoli cells, somatic cells, sperm
cells, spleen cells, T cells,
testicular cells, uterine cells, vaginal epithelial cells, white blood cells,
ciliated cells, columnar
epithelial cells, dopaminergic cells, dopaminergic cells, embryonic stem
cells, endometrial cells,
fibroblasts fetal fibroblasts., follicle cells, goblet cells, keratinized
epithelial cells, lung cells,
mammary cells, mucous cells, non-keratinized epithelial cells, osteoblasts,
osteoclasts, osteocytes,
and squamous epithelial cells. This listing is in no way meant to limit the
array of therapeutic uses
to treat acute and/or chronic disease, disorders, injuries, organ or tissue
failures, and any and all
other ailments that can utilize the products disclosed herein.
[000375] With respect to the treatment of burns, including but not limited
to e.g., second-
and third-degree burns, in some aspects, skin products derived in accordance
with the present
invention are used to treat human patients with severe and extensive deep
partial and/or full
thickness burn wounds. Such products contain terminally-differentiated cell
types that are not
expanded ex vivo prior to use and do not migrate from the site of application
during intended
duration of treatment. Therefore, potential for tumorigenicity is negligible.
[000376] Such products adhere to the wound bed and provides a barrier
function in the
immediate post-burn period. Such products have non-terminally sterilized,
viable cells, allowing
for vascularization of the graft tissue with the recipient. In some aspects,
the epidermis remains
fully intact, and dermal components are maintained without change to
structural morphology or
101
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
organization of the various cells and tissues. This physiologic mechanism
supports the prolonged
survival of the graft material, and provides at least a temporary barrier
function with significant
clinical impact on par with, or better than, allograft. In some aspects, if
clinical signs of infection,
e.g., pain, edema, erythema, warmth, drainage, odor or unexplained fever, are
present or
developing, the product of the present disclosure is not applied until the
clinical signs of the
infection are reduced or eliminated for a predetermined period of time, e.g.,
1, 2, 3, 4, 5, 6, or 7
days, 1, 2, 3, or 4 weeks, or if the subject has tested negative for the
infection. In some aspects, the
wound is cleaned, confirmed to be well-vascularized and nonexuding. If a
dermal substitute such
as cadaver allograft is also being used, the epidermal layer is removed from
engrafted allograft
prior to the application of the product without removing the engrafted dermis.
The epidermal layer
may be removed with a dermatome or other instrument according to standard
operating procedures
of the facility.
[000377] Grafts conventionally used in clinical practice consist of
decellularized and/or
reconstituted sheets of homogenized dermis that are used to achieve temporary,
superficial wound
coverage. Such conventional grafts do not retain the original tissue structure
nor the metabolically
active, otherwise naturally present cells, and thus do not become
vascularized; no capillary
ingrowth or vessel-to-vessel connections are made. In contrast, skin products
described herein are
fundamentally differentiated from such grafts because the product of the
present disclosure
includes live cells that perform the same function as the patient's original
skin, i.e., the product
acts as an organ transplant. Skin performs additional, critical roles related
to homeostasis,
temperature regulation, fluid exchange, and infection prevention. The absence
of a sufficient
amount of skin can compromise the ability to perform these functions leading
to high incidences
of mortality and morbidity from infections and fluid loss. Skin transplants
have been reliably used
with notable clinical benefit to prevent these outcomes in patients with
significant wounds;
regardless of whether the graft is temporary or permanent. Thus, unlike other
proposed transplants,
use of immunosuppressive drugs would be reduced or not be necessary. In fact,
such regimens
would be contraindicated in burn patients whose injuries already exhibit some
level of comprised
immune function. Thus, the xenotransplantation product of the present
disclosure should not be
confused with traditional "xenograft" products consisting of econstituted,
homogenized wild-type
porcine dermis fashioned into sheets or meshed, such as EZ-DermTm or
MediSkinTM. Such
porcine xenografts do not vascularize and are primarily only useful for
temporary coverage of
102
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
superficial burns. In stark contrast, the xenotransplantation product of the
present disclosure
contains metabolically active, minimally manipulated cells in identical
conformations and
unchanged morphologies as the source tissue.
[000378] In some aspects, the present disclosure includes using
xenotransplanted donor skin
as a test for prediction of rejection of other organs from the same animal
donor. Techniques for
performing such predictive tests using human donor skin have previously been
described, e.g., in
Moraes et al., Transplantation. 1989;48(6):951-2; Starzl, et al., Clinical and
Developmental
Immunology, vol. 2013, Article ID 402980, 1-9; Roberto et al., Shackman et
al., Lancet. 1975;
2(7934):521-4, the disclosures of which are incorporated herein by reference
in their entireties for
all purposes. Moraes reported that the crossmatch procedure was highly
accurate in predicting
early kidney transplant rejection. Shackman reported that the fate of skin
grafts taken from live
human prospective kidney donors correlates well with the outcome of kidney
transplantation from
the same donors. According to the present disclosure, in one aspect, the
present disclosure includes
a method of using a xenotransplanted skin sample in a human patient in order
to determine whether
there is a risk of rejection of other organs xenotransplanted from the same
animal donor in the
human patient.
[000379] The skin grafting methods described herein can be used to treat
any injury for which
skin grafts are useful, e.g., for coverage of partial thickness and full
thickness wounds including
but not limited to burns, e.g. partial thickness or excised full thickness
burn wounds; avulsed skin
e.g. on an extremity; diabetic wounds, e.g., non-healing diabetic foot wounds,
venous stasis ulcers.
[000380] In some aspects, the xenotransplantation product of the present
disclosure has
pharmacokinetic and pharmacodynamics properties that meet regulatory
requirements.
Characterization of such properties requires a unique approach with respect to
classical meanings
of drug absorption, distribution, metabolism, and excretion. "Absorption" of
the
xenotransplantation product for the purposes of consideration of
pharmacokinetics, may be
described by the vascularization process the xenotransplantation product
experiences. For
example, shortly after surgery, skin xenotransplantation products may present
as warm, soft, and
pink, whereas wild-type or traditional xenografts appear as non-vascularized
"white grafts." In
some aspects, the distribution of the transplant is limited to the site of
transplant as confirmed by
DNA PCR testing to demonstrate the presence or absence of pig cells in
peripheral blood beyond
the transplantation site.
103
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000381] In other aspects, the cells of the biological products produced in
accordance with
the present invention do not migrate following xenotransplantation into the
recipient, including
into the circulation of the recipient. This includes that PERV or PERV-
infected porcine cells do
not migrate into the recipient. Confirmation that such cells do not migrate
into the recipient can
be performed in a number of ways, including via DNA-PCR analysis of peripheral
blood
mononuclear cells (PBMCs) and samples from the transplantation site and of
highly perfused
organs (e.g., liver, lung, kidney and spleen) to determine and otherwise
demonstrate that
migrations of porcine cells (DNA) or porcine retroviral (RNA) components in
the peripheral blood
did not occur in the recipient.
[000382] Moreover, bioavailability and mechanism of action of the
xenotransplantation
product is not affected by size. The distribution of the xenotransplantation
product is limited to the
site of the administration. For example, in the case of a skin transplant, the
debrided wound bed
initially created by the trauma or burn injury is the site of administration.
The present disclosure
includes testing to detect distribution of cells from the xenotransplantation
product in the
peripheral blood, wound beds, spleen and/or kidney beyond the site of
administration. In certain
aspects, such testing will demonstrate an absence of cells from the
xenotransplantation product in
the peripheral blood, wound beds, spleen and/or kidney beyond the site of
administration. Such
testing may include DNA PCR testing for various cellular markers present in
the type of animal
from which the product is obtained, e.g., PERV, swine MHC, and other swine DNA
sequences. In
certain aspects, cells and nucleic acids from the xenotransplantation product
remain limited to the
site of administration.
[000383] The metabolism of the xenotransplantation product, traditionally
defined as the
metabolic breakdown of the drug by living organisms, typically via specialized
enzymes or
enzymatic systems, may be congruent with the aforementioned natural host
rejection phenomenon,
which occurs in the absence of exogenous immunosuppressive drugs. Via the same
formulation
and identical route of administration as intended for future human use, such
xenotransplantation
products undergo a delayed, immune rejection course similar to allograft
comparators for clinically
useful durations.
[000384] In similar fashion, excretion of the xenotransplantation product
could be modeled
and experientially monitored by the clinical "sloughing" phenomenon as a
result of necrotic
104
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
ischemia of the transplant, due to antibody-mediated vascular injury,
ultimately leading to the
death of the tissue.
[000385] The demonstrated efficacy of the xenotransplantation product of
the present
disclosure, along with safety, availability, storage, shelf-life, and
distribution, provide significant
advantages over current standards of care.
[000386] In some aspects, the "dosage" of the xenotransplantation product
of the present
disclosure is expressed as percentage of viable cells in the product per unit
area of transplantation.
As such, in some aspects, the xenotransplantation product of the present
disclosure can be
considered as analogous to the active pharmaceutical ingredient in a
pharmaceutical drug product.
[000387] Survival of the xenogeneic cells, tissues, or organs of the
present disclosure is
increased by avoiding: (a) infiltration of immune or inflammatory cells into
the
xenotransplantation product or alteration of such cells in other relevant
compartments, such as the
blood and cerebrospinal fluid; (b) fibrotic encapsulation of the
xenotransplantation product, e.g.,
resulting in impaired function or xenotransplantation product loss; (c)
xenotransplantation product
necrosis; (d) graft versus host disease (GVHD); and (e) in vivo function and
durability of
encapsulation or barriers intended to diminish rejection or inflammatory
responses.
[000388] Blood samples from piglets are obtained and tested for phenotype,
lack of
expression of alpha galactose on the cell surface of blood cells using FITC-
1134 labeling and flow
cytometry. At this stage of development, all progeny will be genotyped at
birth. A PCR assay has
been established to determine if a pig has a wild type galactose-a1,3galactose
transferase gene
(Gal-T) or if it is heterozygous or homozygous for the Gal-T knockout (Gal-T-
KO) using DNA
isolated from ear notches or PBMC. Genomic DNA is isolated from PBMC (or skin
tissues) using
DNeasy Kit following the Qiagen DNeasy kit directions. PCR is performed on
genomic DNA and
control template DNA, Wild type Gal-T (+/+) Heterozygote Gal-T-KO (+/-) and
Homozygous
Gal-T-KO (-/-).
[000389] Punch biopsies of skin grafts are co-cultured with subconfluent
target cells, human
293 (kidney epithelium) and porcine ST-IOWA cell lines maintained in culture
medium
(Dulbecco' s modified Eagle's medium supplemented with 10% fetal bovine serum
and glutamine,
penicillin, and streptomycin) in a 75-cm2 flask. The biopsies are kept in
contact with the target
cells for 5 days, after which the culture medium and remaining tissue are
removed and the target
cell co-cultures are maintained by subculturing as necessary. PERV infection
of target cells is
105
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
determined by the presence of reverse transcriptase (RT) activity in the
culture supernatants.
Transmission assays are maintained for a minimum of 60 days before being
considered negative.
[000390]
Product characterization to measure safety, identity, purity and potency is
performed. Safety tests include bacterial and fungal sterility, mycoplasma,
and viral agents. The
present disclosure includes cryopreserving and archiving for further testing,
as needed, samples of
all final xenotransplantation products (i.e., cells or tissues or biopsies of
organs), whether fresh or
from culture ex vivo. In some cases, for example if the xenotransplantation
product is a whole
intact organ, a relevant surrogate sample (e.g., adjacent tissues or contra-
lateral organ) is archived.
[000391]
With regard to skin, storage and cryopreservation of porcine skin has not been
fully
characterized, especially with regards to viability, as most porcine
xenografts are intentionally
devitalized, or "fixed" with glutaraldehydes or radiation treatment. Such
information is necessary
to support the use of vital porcine skin grafts - or porcine skin transplants -
as a temporary and
clinically advantageous option.
[000392]
In procedures in which the xenotransplantation product is transplanted
immediately
after removal from the source animal, such as xenotransplantation of whole
organs, results of
testing of the xenotransplantation product may not be available before its
clinical use. In such
cases, testing of the source animal, itself, may be all the testing that is
possible before the
procedure. Testing of samples taken from such xenotransplantation products or
appropriate
relevant biological surrogates, e.g., adjacent tissues or contra-lateral
organs, may be performed
according to the present disclosure. Microbiological examination methods may
include aspects set
forth in the following Table 2:
TABLE 2
TEST DETAILS GROWTH
SUITABILITY OF
PROMOTION
COUNTING METHOD IN
THE PRESENCE OF
PRODUCT
Microorgani Preparation Total Total Total Aerobic
Total
sm of Test Aerobic Yeasts and Microbial
Yeasts and
Strain Microbial Molds Count
Molds
Count Count Count
106
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Staphylococcu Soybean- Soybean- Soybean-Casein
s aureus such Casein Digest Casein Digest
as ATCC Agar or Digest Agar Agar/MPN
6538, NCIMB Soybean- and Soy- Soybean- Casein
95 1 8, CIP Casein Digest bean- Casein Digest Broth
4.83, or Broth Digest Broth 100 cfu
NBRC 3O0350 100 cfu 300350
13276 18- 24 hours 300350 3 days
3 days
Pseudomonas Soybean- Soybean- Soybean-Casein
aeruginosa Casein Digest Casein Digest
such as ATCC Agar or Digest Agar Agar/MPN
9027, NCIMB Soybean- and Soy- Soyb ean- Casein
8626, CIP Casein Digest bean- Casein Digest Broth
82.118, Broth Digest Broth 100 cfu
or NBRC 1 30 -35 100 cfu 30 -35
3275 18- 24 hours 30 -35 3 days
3days
Bacillus Soybean- Soybean- Soybean-Casein
subtilis such Casein Digest Casein Digest
as ATCC Agar or Digest Agar Agar/MPN
6633, NCIMB Soybean- and Soy- Soybean- Casein
8054, CIP Casein Digest bean- Casein Digest Broth
52.62, or Broth Digest Broth 100 cfu
NBRC 3134 3O0350 100 cfu 30 -35
18- 24 hours 30 -35 3 days
3days
Candida Sabouraud Soybean- Sabouraud Soybean- Casein Sabouraud
albicans such Dextrose Agar Casein Dextrose 1 Digest Agar Dextrose
as ATCC or Sabouraud Digest Agar 00 cfu 100 cfu Agar
10231, NCPF Dextrose 100 cfu 20 -25 30 -35 100 cfu
3179, Broth 20 - 25 30 - 35 5 days 5days 20 - 25
IP 48.72, or 2-3 days 5 days MPN: not 5days
NBRC 1594 applicable
Aspergillus Sabouraud Soybean ¨ Sabouraud Soybean - Casein
Sabouraud
brasiiiensis Dextrose Casein Di- Dextrose Digest Agar Dextrose
such as Agar or gest Agar 100 cfu 100 cfu Agar
ATCC16404, Potato¨ 100 cfu 20 -25 30 -35 100 cfu
IMI 149007, Dextrose 30 - 35 5 days 5days 20 - 25
IP 1431.83, or Agar 20 - 25 < 5 days MPN: not 5days
NBRC 9455 5-7 days, or applicable
until good
107
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
sporulation is
achieved
[000393] The present disclosure includes using Buffered Sodium
Chloride¨Peptone Solution
pH 7.0 or Phosphate Buffer Solution pH 7.2 to make test suspensions; to
suspend A. brasiliensis
spores, 0.05% of polysorbate 80 may be added to the buffer. The present
disclosure includes using
the suspensions within 2 hours, or within 24 hours if stored between 2 C and 8
C. As an alternative
to preparing and then diluting a fresh suspension of vegetative cells of A.
brasiliensis or B. subtilis,
a stable spore suspension is prepared and then an appropriate volume of the
spore suspension is
used for test inoculation. The stable spore suspension may be maintained at 2
to 8 for a validated
period of time. To verify testing conditions, a negative control is performed
using the chosen
diluent in place of the test preparation. There must be no growth of
microorganisms. A negative
control is also performed when testing the products as described under Testing
of Products. A
failed negative control requires an investigation. Microbiological Examination
may be performed
according to USP 61, USP 63, USP 71, USP 85 EP section 2.6.13 Microbial
Examination of Non-
sterile Products (Test for Specified Microorganisms), each of which is
incorporated herein by
reference in its entirety.
[000394] With regard to testing for porcine cytomegalovirus (PCMV), source
animals are
screened for PCMV on a quarterly basis. However, caesarian derived piglets,
which are then
consistently raised in the closed colony are not infected with PCMV. Analysis
for PCMV was
conducted during the studies in Example 1 herein and no PCMV was detected in
the punch biopsies
using the following PCR method. These results were consistent to the PCR
results from nasal
swabs. Quantitative Real-Time PCR is utilized for PCMV testing. Target DNA
sequences were
quantified by real-time PCR using a Stratagene Mx3005P. Sequence-specific
primers and TaqMan
probe were generated for each gene target. Each 25uL PCR reaction included
target DNA, 800nM
primers 200nMTaqMan probe, 20 nM Rox reference and lx Brilliant III Ultra Fast
Master Mix.
The PCR cycling conditions were as follows: 1 cycle at 95 C for 5 min followed
by 50 cycles of
denaturation at 95 C for 10 seconds, and annealing-extension at 60 C for 30
seconds with data
collection following each extension. Serial dilutions of gel-extracted
amplicon cloned into
108
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Invitrogen TOPO plasmid served as quantifying standards. Target DNA is
detected with a linear
dynamic range of 10 to 106 copies. For quantification of PCMV DNA, 300 ng of
xenograft pig
kidney DNA was run in a TaqMan PCR in triplicate. Primers and probes specific
for PCMV DNA
polymerase gene have been shown to have no cross-reactivity with PLHV-1.
Utilization of
cesarean-derived swine as source animals, combined with animal husbandry of
the resulting closed
colony and maintenance of the barrier-isolation conditions is attributed the
animals being PCMV
free. With regard to skin, the inventors noted that the safety and efficacy
results achieved in
Example 1 using single knockout swine (as opposed to triple knockout or even
further genetically
modified swine) were quite surprising given the comparable performance to
allograft.
[000395] In some aspects, the analytical procedures used to test the
xenotransplantation
product can also include:
a. USP<71> Sterility. Samples are transferred to Tryptic Soy Broth (TSB) or
Fluid
Thioglycollate Medium (FTM) as appropriate. For Bacteriostasis and
fungistasis, TSB samples are
spiked with an inoculum of < 100 Colony Forming Units (CFUs) of 24 -hour
cultures of Bactillus
subtilis, Candida albicans, and with <100 spores of Aspergilius braseiliensis.
The FTM samples
will be spiked with an inoculum of <100 CFU's of 24-hour cultures of
Staphyloccocus aureus,
Pseudomonas aeruginosa, and Clostridium sporogenes. If growth is not observed,
the product is
found to be bacteriostatic or fungistatic and fails the USP <71> Sterility
Test.
b. Aerobic and Anaerobic Bacteriological Cultures. Samples are transferred to
Tryptic
Soy Broth (TSB) or Fluid Thioglycollate Medium (FTM) as appropriate. Vessels
will be incubated
to allow for potential growth. If no evidence of microbial growth is found,
the product will be
judged to comply with the test for sterility as described by USP<71>.
c. Mycoplasma Assay USP <63>. Fresh samples will be added to 100mL of
Mycoplasma Hayflick broth and incubated at 37 C for up to 21 days. The sample
is subcultured
after 2-4 days, 7-10 days, 14 days, and 21 days. The plates are then incubated
at 37 C for up to
14 days and checked for the presence of Mycoplasma colonies. If none are
detected, the product
is found to be in compliance with USP<63> and is mycoplasma free.
d. Endotoxin USP<85>. Three samples from the same lot will be tested for the
Inhibition/Enhancement of the Limulus amoebocyte lysate (LAL) test. Samples
will be extracted
with 40mL of WFI per sample at 37 C for 1 hour. Samples will then be tested in
the LAL Kinetic
109
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Chromogenic Test with a standard curve ranging from 5-50EU/mL at a validated
dilution. Assays
will be performed in compliance with USP<85>.
e. MTT Assay for Cell Viability. The metabolic activity of the drug product
is tested
relative to control tissue samples using a biochemical assay for [3-4,5
dimethylthiazol-2-y1]-2,5
diphenyltetrazolium bromide (MTT) metabolism. Positive and negative control
samples of fresh
xenotransplantation product tissue (positive control) or heat inactivated
discs of
xenotransplantation product tissue (negative control) or the test article of
Xenotransplantation
product are placed in amber microcentrifuge tubes containing MTT solution (0.3
m g/mL in
DMEM, 0.5 mL). The discs are treated with MTT formazan and incubated for 180
15 minutes
at 37 C and an atmosphere of 5% CO 2 in air. The reaction is terminated by
removal of the discs
and the formazan is extracted by incubation at either ambient temperature for
< 24 hours or
refrigerated at 4 C for < 72 hours. Samples are protected from light during
this time. Aliquots are
taken after the extraction is complete and the absorbance at 550 nm (with a
reference wavelength
of 630 nm) is measured and compared to a standard curve.
f. IB4 Assay for Extracellular Glycan Epitope. The absence of the
galactosyl-a-1,3-
galactose (Alpha-Gal) epitope on cells will be determined using fluorescence
activated flow
cytometry. White blood cells in whole blood are stained with a fluorochrome
labeled isolectin-B4
(FITC-I-B4) and comparisons are made against blood obtained from wild type
positive controls
and the Gal-T-KO source animal twice. First, all source animals are tested at
birth. Second, the
same test will be performed from whole blood collected at sacrifice of the
source animal and tested
for stability of the gene knockout, and the negative phenotype for Alpha-Gal.
The isolectin binds
to the epitope on cells from the wild type pig but no binding occurs on the
cells from the Gal-T-
KO pigs. The assay serves to confirm alpha-gal epitope is not present in the
genetically engineered
source animal. Spontaneous re-activation of the gene, and re-expression of the
Alpha-Gal moiety
post sacrifice is highly improbable and unreasonable to expect; its inclusion
would only deteriorate
the efficacy of the xenotransplantation product causing it to resemble wild-
type porcine tissue and
hyperacutely reject as previously demonstrated.
g. PERV Viral Assay. PERV pol quantitation lOuL of a 1:625 dilution of the RT
reaction was amplified in a 50 cycle PERV polymerase quantitative TaqMan PCR
in triplicate
using a Stratagene MX300P real-time thermocycler (Agilent Technologies). 1 OuL
of a 1:25
dilution of the "No RT enzyme" control RT reaction was similarly treated. PCR
conditions
110
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
included PERV pol forward and reverse primers at 800nM final concentration and
PERV pol probe
at 200nM final concentration. Brilliant III Ultra Fast master mix (600880
Agilent Technologies)
was used supplemented to 20 nM with ROX reporter dye (600880 A gilent
Technologies) and 0.04
U nits/i.tL UNG nuclease (N8080096, Life Technologies). Cycling conditions
included 1 cycle of
minutes at 50 C followed by one cycle of 10 minutes at 95 C and 50 cycles of
10 seconds at
950 C followed by 30 seconds at 60 C with data collected at the end of each
cycle. Absolute
copies of PERV pol, and of porcine MHC-I and porcine GAPDH nucleic acids were
measured per
nanogram of input cDNA. Punch biopsies of thawed as described herein and
washed
xenotransplantation product are tested for the presence of PERV DNA and RNA.
h. Histology and Morphology. Samples of the xenotransplantation product,
following
the described manufacturing process, are sampled for examination for cell
morphology and
organization. Verification under microscope via visible examination to ensure
correct cell
morphology and organization of xenotransplantation product tissues and absent
for abnormal cell
infiltrate populations.
i. Release Assay Sampling Methodology. Once all units of the final
xenotransplantation product lot have been created, units are independently,
randomly selected for
use in manufacturing release assays for the required acceptance criteria.
These units will be marked
for lot release to the various laboratory contractors, and the various
analytical tests will be
performed per the required cGMP conditions.
[000396] Similarly, prior to validation for human clinical use, all final
xenotransplantation
product must meet acceptance criteria for selecting a donor pig for material
including (i) reviewing
the medical record for a defined pedigree, (ii) reviewing the medical record
for the test results for
alpha-1,3-galactose by Flowmetrics, (iii) reviewing the medical record for a
history of full
vaccinations; (iv) reviewing the medical record for the surveillance tests
performed over the
lifetime of the pig; (v) adventitious agent screening of source animal; (vi)
reviewing the medical
record for infections over the lifetime of the pig; and (vi) reviewing the
medical record for any skin
abnormalities noted in the animal's history.
[000397] The final xenotransplantation product control strategy and
analytical testing is
conducted at the conclusion of the manufacturing process prior to release for
clinical use. Results
of the required analytical tests will be documented via a xenotransplantation
product drug product
111
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Certificate of Analysis (COA) that is maintained with a master batch record
pertaining to each lot
of xenotransplantation product drug product.
[000398] The following Table 3 is a list of the assays and results of the
battery of tests
performed on the xenotransplantation product materials.
TABLE 3
Test Test Method Sample Result
Material
Tested
Sterility Testing Tissue Culture 3mm Punch No growth detected
Biopsy of
Aerobic Bacteria Xenotranspl
Anaerobic Bacteria antation
product
(Post Thaw)
Fungi
Acid fast cultures
Specific bacterial
screen
Mycological Screen Mycoplasma Assay 3mm Punch No growth
Biopsy of detected after 28
Xenotranspl days
antation
product
(Post Thaw)
Bacteriostasis & USP<71> Xenotranspl Bacteriostatic, no
Fungistasis Gibraltar Laboratory antation growth of specific
product indicator organism
(Post Thaw)
Endotoxin Test USP<85> LAL, Xenotranspl <0.2 EU/unit
Kinetic antation
Chromogenic Test product
(Post Thaw)
Endogenous Viral Testing RT-qPCR 3mm Punch Presence of
(PERV) Co-culture Assay Biopsy of PERV A, B,
Xenotranspl
MGH antation confirmed
Infectious product
Disease (Post Thaw)
112
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Fishman
Laboratory
Viability Testing MTT and 3mm Punch Greater than 70%
Phenyl Acetate Biopsy Mitochondrial
Assays of Activity remaining
Xenotranspl following freeze-
thaw
antation cycle, confirmed by
product both assays
(Post Thaw)
Identity Histology, 3mm Punch No abnormalities
Hematoxylin and Biopsy noted.
Cell Morphology Eosin Staining of Cell morphology
Xenotranspl and organization
antation consistent with skin
product graft
No presence of
Alpha- GAL detected
(Post Thaw)
Confirmation
of absence of Flow Cytometry, Whole Blood, 2
Alpha-GAL isolectin-B4 (FITC- ml, obtained
(Gal-T- I- B4) from source
Knockout animal, at
confirmation) sacrifice.
[000399] In another aspect it will be understood that there includes an
adventitious agent
control strategy developed based on the source animal, including the species,
strain, geographic
origin, type of tissue, and proposed indication. Analytical Tests are
conducted for adventitious
agents, to include bacteria, fungi, mycoplasma, and viral microorganisms,
including as follows:
113
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
j. Bacteriological Free Status ¨ The bacteriological screen is conducted to
confirm
the drug product is free of potential biological agents of concern Humans.
Both Aerobic and
Anaerobic screens are conducted to ensure sterility. Samples are thawed as
described herein and
transferred to Tryptic Soy Broth (TSB) or Fluid Thioglycollate Medium (FTM) as
appropriate.
Vessels will be incubated to allow for potential growth. If no evidence of
microbial growth is
found, the product will be judged to comply with the test for sterility.
k. Mycological (Fungal) Free Status ¨ The mycological screen is conducted to
confirm the Drug Product is free of potential fungal agents of concern.
Samples are thawed as
described herein. After thawing, samples are transferred to a soybean-casein
digest agar. Vessels
will be incubated to allow for potential growth. If no evidence of fungal
growth is found, the
product will be judged to comply with the test for sterility per USP<71>.
1. Mycoplasma Free Status - The mycoplasma screen is conducted to
confirm the drug
product is free of mycoplasma. Samples are thawed as described herein and
added to 100mL of
Mycoplasma broth and incubated at 37 C for up to 21 days. The sample is sub-
cultured after 2-4
days, 7-10 days, 14 days, and 21 days. The plates are then incubated at 37 C
for up to 14 days and
checked for the presence of Mycoplasma colonies. If none are detected, the
product is found to be
in compliance with USP<63> and is mycoplasma free.
m. Endotoxin Free Status ¨ The endotoxin free status is conducted to confirm
the drug
product is free of endotoxins and related agents of concern. Three samples
from the same lot will
be tested for the Inhibition/Enhancement of the Limulus amoebocyte lysate
(LAL) test. Samples
will be thawed as described herein and extracted with 40mL of WFI per sample
at 37 C for 1 hour.
Samples will then be tested in the LAL Kinetic Chromogenic Test with a
standard curve ranging
from 5-50EU/mL at a validated dilution. Assays will be performed in compliance
with USP<85>.
n. Viral Assays Conducted ¨ The viral assays are conducted to confirm the
source
animal is free of potential viral agents of concern, confirmation of
endogenous viruses (see below).
This includes co-culturing and RT-PCR testing for specific latent endogenous
viruses including
PERV. In vivo assays are also conducted on the animal source to monitor animal
health and
freedom from viral infection as key aspects of the lot release criteria. Due
to the endemic nature
of PERV in porcine tissue, this qualifies as a positive result that does not
preclude the use of such
tissue. However, the virus is identified and characterized in lot release to
provide information for
monitoring the recipient of the xenotransplantation product.
114
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
o. Cell Viability Assay ¨ The MTT assay is conducted to confirm the
biologically
active status of cells in the xenotransplantation product. Evidence of
viability is provided through
surrogate markers of mitochondrial activity as compared to positive (fresh,
not cryopreserved) and
negative (heat- denatured) controls. The activity of the cells is required for
the xenotransplantation
product to afford the intended clinical function. This is required as a lot
release criteria, and is
currently established that tissue viability should not be less than 50% of the
metabolic activity
demonstrated by the fresh tissue control comparator.
p. Histology and Morphology - Verification under microscope via visible
examination
of Hematoxylin and Eosin (H&E) section staining of the epidermal and dermal
layers, to ensure
correct cell morphology and organization of the xenotransplantation product
tissues and cell
infiltrate populations. This is conducted to confirm the appropriate
physiologic appearance and
identity of cells present in the xenotransplantation product. The
xenotransplantation product is
composed of minimally manipulated porcine dermal and epidermal tissue layers.
This is required
as a lot release criteria. Evidence of the following cell layers (from most
superficial to deepest), in
the epidermal layer are verified:
i. Stratum Corneum
ii. Stratum Granulosum
iii. Stratum Spinosum
iv. Stratum Basale
Evidence of the following cellular structures in the dermal layer are
verified:
v. Blood vessels, evidence of vasculature
vi. Nerves
vii. Various glands
viii. Hair follicles
ix. Collagen
[000400] The genetically engineered source animals do not contain any
foreign, introduced
DNA into the genome; the gene modification employed is exclusively a knock-out
of a single gene
that was responsible for encoding for an enzyme that causes ubiquitous
expression of a cell-surface
antigen. It will be understood that the xenotransplantation product in one or
more aspects do not
incorporate transgene technologies, such as CD-46 or CD-55 transgenic
constructs.
115
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000401] An endotoxin free status is conducted to confirm the drug product
is free of
endotoxins and related agents of concern. Protocols for the assurance of
Endotoxin free status are
as follows: Three samples from the same lot are tested for
Inhibition/Enhancement of the Limulus
amoebocyte lysate (LAL) test. Samples are thawed, extracted, and tested in the
LAL Kinetic
Chromogenic Test with a standard curve ranging from 5-50EU/mL at a validated
dilution in
compliance with USP<85>.
[000402] The MTT assay is conducted to confirm the biologically active
status of cells in the
product. Evidence of viability is provided through surrogate markers of
mitochondrial activity as
compared to positive (fresh, not cryopreserved) and negative (heat-denatured)
controls. The
activity of the cells is required for the product to afford the intended
clinical function and the
viability parameters for one aspect ranging from 50% to 100% mitochondrial
activity.
[000403] Verification under microscope via visible examination of
Hematoxylin and Eosin
(H&E) section staining of the epidermal and dermal layers, to ensure correct
cell morphology and
organization of the xenotransplantation product tissues and cell infiltrate
populations. This is
conducted to confirm the appropriate physiologic appearance and identity of
cells present in the
product.
[000404] For skin xenotransplantation products, evidence of the following
cell layers (from
most superficial to deepest), in the epidermal layer are verified: Stratum
Corneum; Stratum
Granulosum; Stratum Spinosum; Stratum Basale. Evidence of the following
cellular structures in
the dermal layer are verified: Blood vessels, evidence of vasculature; Nerves;
Various glands; Hair
follicles; Collagen.
[000405] The xenotransplantation product may be further processed to ensure
that it remains
free of aerobic and anaerobic bacteria, fungi, viruses, and mycoplasma. Under
sterile conditions
in a laminar flow hood in a drug product processing suite using applicable
aseptic techniques,
immediately after, within 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 seconds, within 10
seconds to 1 minute, within
1 minute to 1 hour, within 1 hour to 15 hours, or within 15 hours to 24 hours
following harvest,
the xenotransplantation product is sterilized, e.g., using one or more of UV
irradiation or an anti-
microbial/anti-fungal. In one aspect, the product may be placed into an anti-
microbial/anti-fungal
bath ("antipathogen bath"). The antipathogen bath may include: one or more
anti-bacterial agents,
e.g., ampicillin, ceftazidime, neomycin, streptomycin, chloramphenicol,
cephalosporin, penicillin,
tetracycline, vancomyocin, and the like; one or more anti-fungal agents, e.g.,
amphotericin-B,
116
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
azoles, imidazoles, triazoles, thiazoles, candicidin, hamycin, natamycin,
nystatin, rimocidin,
allylamines, echinocandins, and the like; and/or one or more anti-viral
agents. The anti-pathogen
bath may include a carrier or medium as a diluent, e.g., RPMI-1640 medium. In
some aspects, the
anti-pathogen bath may include at least 2 anti-bacterial agents. In some
aspects, the anti-pathogen
bath may include at least 2 anti-bacterial agents and at least one anti-fungal
agent. In some aspects,
the anti-pathogen bath may include at least four agents. In some aspects, the
anti-pathogen bath
may include no more than 4, 5, 6, 7, 8, 9, or 10 agents. In some aspects, the
anti-pathogen bath
may include any combination of the foregoing.
[000406] The product may be sterilized using UV light sterilization. For
example, the
product is placed under the UV lamp for a desired period of time, e.g., 0.5,
1, 1,5, 2, 3, 4, 5, 6,
minutes or more, then turned over to the other side, and put under the UV lamp
for the same or a
different period of time on opposite side. The time period for exposing a
given sample to the UV
may be varied based on the specific biological agents or the types of
biological agents to be
sterilized, e.g., as shown in the following Table 11 below. For example, the
product may be
sterilized using a UV lamp having a UV-C intensity of at least 100 uW/cm2 for
at least 2 minutes
and up to 15, 12, 10, 8, 6, 5, 4, 3, or 2.5 minutes, and turned over such that
its opposite surface is
exposed to the UV lamp for at least 2 minutes and up to 15, 12, 10, 8, 6, 5,
4, 3, or 2.5 minutes to
obtain a UV-treated product; a UV-C dosage of at least 100,000 uW sec/cm2 and
up to 800,000,
700,000, 600,000, 500,000, 400,000, 300,000 or 200,000 uW sec/cm2; a UV-C
dosage of at least
200,000 uW sec/cm2 and up to 800,000, 700,000, 600,000, 500,000, 400,000, or
300,000 uW
sec/cm2; a UV lamp having a UV-C intensity of at least 100 uW/cm2 for at least
2 minutes and up
to 15, 12, 10, 8, 6, 5, 4, 3, or 2.5 minutes.
[000407] Product processing occurs in a single, continuous, and self-
contained, segregated
manufacturing event that begins with the sacrifice of the source animal
through completion of the
production of the final product. The animal is euthanized via captive bolt
euthanasia, may be
moved, if necessary, in a sterile, non-porous bag, to an operating room where
the procedure to
harvest biological product from the source animal will occur. All members of
the operating team
should be in full sterile surgical gear, e.g., dressed in sterile dress to
maintain designated pathogen
free conditions prior to receiving the source animal and in some instanced be
double-gloved to
minimize contamination, and surgical areas and tools are sterilized. The
source animal is removed
from the bag and container in an aseptic fashion. The source animal is
scrubbed by operating staff,
117
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
e.g., for at least 1-10 minutes with antiseptic, e.g., Chlorhexidine, brushes
over the entire area of
the animal where the operation will occur, periodically pouring Chlorhexidine
over the area to
ensure coverage. Surgical area(s) of the animal are scrubbed with opened
Betadine brushes and
sterile water rinse over the entire area of the animal where the operation
will occur for, e.g., 1-10
minutes.
[000408] In one aspect, with regard to skin, a full thickness skin graft
wound dressing
consisting of dermal tissue derived from a swine in accordance with the
present invention is used
in conjunction or combination with cultured epidermal autografts to produce a
product according
to the present disclosure and that can be used in methods of the present
disclosure. Prior to
application of the epidermal autografts, significant debridement of wound bed
is required to ensure
an adequate substrate. To confirm a wound bed is ready for an epidermal
autograft, apply the skin
products described herein, e.g., biological skin products derived from animals
of the present
disclosure to confirm adherence. Once adherence is confirmed, the temporary
wound coverage
product is removed, and in some aspects, the wound bed is covered with a
meshed autograft, and
one or more cultured epidermal autograft products are placed on top to close
the gaps in the
autograft mesh.
[000409] The debridement may include mechanical debridement, chemical
debridement,
enzymatic debridement, or a combination thereof. Mechanical debridement may
include surgical
excision, e.g., tangential excision to remove thin layers of dermis until
healthy tissue is visualized,
or fascial excision to remove the full thickness of dermis down to the
underlying fascia. Tangential
excision allows less viable tissue to be removed with the necrotic tissue, but
typically results in
higher blood loss, is a larger physiologic stressor than fascial excision, and
is more likely to result
in "incomplete" debridement, with some devitalized tissue remaining in place.
In fascial excision,
blood loss and operative time are minimized, but often a large amount of
healthy tissue is removed
with the burned tissue. Debriding agents may include agents capable of
cleaning a burn wound by
removing foreign material and dead tissue. Many such agents are known. In
enzymatic
debridement, collagenases or other proteolytic enzymes are employed that break
down proteins of
the extracellular matrix, allowing devitalized tissue to be wiped away without
the need for surgery
while preferably leaving healthy tissue substantially intact. Enzymatic
debridement involves the
application of proteolytic and optionally other exogenous enzymes to a wound
surface to break
down necrotic tissue. Enzymatic debridement may be a relatively slow process,
carried out over a
118
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
period of a number of weeks in combination with other topical preparations,
soakings and repeated
dressings. Alternately, rapid enzymatic debridement can be accomplished using
multi-enzyme
products, for example, those extracted from the stem of the pineapple plant,
as disclosed for
example in WO 98/053850 and WO 2006/0006167, and as provided in the product
marketed under
the trade name Debraseg. A procedure for enzymatic debridement generally
utilizes an enzyme
such as bromelain derivatives, debridase, collagenase, papain derivatives,
streptokinase, sutilains,
fibrinolysin, deoxyribonuclease, krill derivatives, trypsin or combinations
thereof. Autolytic
debridement relies on enhancing the natural process of selective liquefaction,
separation and
digestion of necrotic tissue and eschar from healthy tissue that occurs in
wounds due to
macrophage and endogenous proteolytic activity. This is achieved by the use of
occlusive, semi-
occlusive or moist interactive dressings. Enzymatic debridement agents include
a bromelain
enriched enzyme product, other collagenases, or other enzyme products capable
of clearing
devitalized tissue or wound debris. NexoBridTM (MediWound Ltd.) is one such
bromelain enriched
product that specifically targets heat-denatured collagen for degradation,
resulting in partial-
thickness and full-thickness wounds requiring a wound coverage or dressing
product. Such
products and methods are described in U.S. Patent Nos. 8,540,983; 8,119,124;
7,128,719;
7,794,709; 8,624,077; and U52009/0010910A1, each of which is incorporated by
reference herein.
[000410] In some aspects, the wound bed may include or be a chronic wound
or an acute
wound. Chronic wounds include but are not limited to venous leg ulcers,
pressure ulcers, and
diabetic foot ulcers. Acute wounds include but are not limited to burns,
traumatic injuries,
amputation wounds, skin graft donor sites, bite wounds, frostbite wounds,
dermabrasions, and
surgical wounds.
[000411] In the cases where there is no dermis, biological products
produced in accordance
with the present invention are utilized. The epidermis is removed from such
products (e.g., before
dermis harvesting on the pig with a VERSAJETTm Hydrosurgery system), so that
just the dermis
remains. Then, the subject biological product is placed on the patient's
subcutaneous tissue,
serving as a substrate for the cultured epidermal autograft process described
herein.
[000412] In one aspect, a liver derived in accordance with the present
disclosure is utilized
for extracorporeal perfusion as a temporary filter for a human patient until a
patient receives a
human transplant. In an operating area within the DPF Isolation Area, a source
animal is placed
under a general anesthetic (ketamine, xylazine, enflurane) or euthanized by
captive bolt. A
119
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
hepatectomy is then performed on the source animal in designated pathogen free
conditions. The
liver product derived from the source animal can be packaged and transported
to the location of
the procedure in accordance with current practice with human donor livers. The
procedure to
utilize the liver filtration product can be performed, for example, by
percutaneously cannulating a
human patient's internal jugular vein for venous return with an arterial
cannula and percutaneously
cannulating a patient's femoral vein for venous outflow with an artery
cannula. These cannulas are
connected to a bypass circuit, having a centrifugal pump, a heat exchanger, an
oxygenator, and a
roller pump incorporated therein. This circuit is primed with crystalloids and
run for a period of
time (e.g., 10-30 minutes) before the liver from an animal according to the
present disclosure is
incorporated at a stabilized flow rate, e.g., 600-1000 ml/min, maintained in a
crystalloid bath
occasionally supplemented with warm solution, e.g., 30-40 C.
[000413] It will be understood that, in the context of swine-to-human
xenotransplantation,
each human recipient will have a major histocompatibility complex (MHC) (Class
I, Class II
and/or Class III) that is unique to that individual and will not match the MHC
of the donor swine.
Accordingly, it will be understood that when a donor swine graft is introduced
to the recipient, the
swine MHC molecules themselves act as non-gal xeno-antigens, provoking an
immune response
from the recipient, leading to transplant rejection.
[000414] Human leukocyte antigen (HLA) genes show incredible sequence
diversity in the
human population. For example, there are >4,000 known alleles for the HLA-B
gene alone. The
genetic diversity in HLA genes in which different alleles have different
efficiencies for presenting
different antigens is believed to be a result of evolution conferring better
population-level
resistance against the wide range of different pathogens to which humans are
exposed. This genetic
diversity also presents problems during xenotransplantation where the
recipient's immune
response is the most important factor dictating the outcome of engraftment and
survival after
transplantation.
[000415] In accordance with one aspect the present invention, a donor swine
is provided with
a genome that is biologically engineered to express a specific set of known
human HLA molecules.
Such HLA sequences are available, e.g., in the IPD-IMGT/HLA database
(available at
ebi.ac.uk/ipd/imgt/h1a/) and the international ImMunoGeneTics information
system (available
at imgt.org). For example, HLA-Al, B8, DR17 is the most common HLA haplotype
among
120
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Caucasians, with a frequency of 5%. Thus, the disclosed method can be
performed using the known
MHC/HLA sequence information in combination with the disclosures provided
herein.
[000416] In some aspects, the recipient's human leukocyte antigen (HLA)
genes and MHC
(Class I, II and/or III), are identified and mapped. It will be understood
that ascertaining the human
recipient's HLA/MHC sequence can be done in any number of ways known in the
art. For example,
HLA/MHC genes are usually typed with targeted sequencing methods: either long-
read
sequencing or long-insert short-read sequencing. Conventionally, HLA types
have been
determined at 2-digit resolution (e.g., A*01), which approximates the
serological antigen
groupings. More recently, sequence specific oligonucleotide probes (S SOP)
method has been used
for HLA typing at 4-digit resolution (e.g., A*01:01), which can distinguish
amino acid differences.
Currently, targeted DNA sequencing for HLA typing is the most popular approach
for HLA typing
over other conventional methods. Since the sequence-based approach directly
determines both
coding and non-coding regions, it can achieve HLA typing at 6-digit (e.g.,
A*01:01:01) and 8-
digit (e.g., A*01:01:01:01) resolution, respectively. HLA typing at the
highest resolution is
desirable to distinguish existing HLA alleles from new alleles or null alleles
from clinical
perspective. Such sequencing techniques are described in, for example, Elsner
HA, Blasczyk R:
(2004) Immunogenetics of HLA null alleles: implications for blood stem cell
transplantation.
Tissue antigens. 64 (6): 687-695; Erlich RL, et al (2011) Next-generation
sequencing for HLA
typing of Class I loci. BMC genomics. 12: 42-10.1186/1471-2164-12-42; Szolek
A, et al.
(2014) OptiType: Precision HLA typing from next-generation sequencing data.
Bioinformatics 30:3310-3316; Nariai N, et al. (2015) HLA-VB Seq: Accurate HLA
typing at full
resolution from whole-genome sequencing data. BMC Genomics 16:S7; Dilthey AT,
et
al. (2016) High-accuracy HLA type inference from whole-genome sequencing data
using
population reference graphs. PLoS Comput Biol 12:e1005151; Xie C., et al.
(2017) Fast and
accurate HLA typing from short-read next-generation sequence data with xHLA
114 (30) 8059-
8064, each of which is incorporated herein in its entirety by reference.
[000417] The known human HLA/MHC or an individual recipient's sequenced
HLA/MHC
sequence(s) may be utilized as a template to modify the swine leukocyte
antigen (SLA)/MHC
sequence to match, e.g., to have 80%, 85%, 90%, 95%, 98%, 99%, or 100%
sequence homology
to a known human HLA/MHC sequence or the human recipient's HLA/MHC sequence.
Upon
identifying a known human recipient HLA/MHC sequence to be used or performing
genetic
121
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
sequencing of a human recipient to obtain HLA/MHC sequences, biological
reprogramming may
be performed to SLA/MHC sequences in cells of the swine based on desired
HLA/MHC
sequences. For example, several targeting guide RNA (gRNA) sequences are
administered to the
swine of the present disclosure to reprogram SLA/MHC sequences in cells of the
swine with the
template HLA/MHC sequences of the human recipient.
[000418] CRISPR-Cas9 is used to mediate rapid and scarless exchange of
entire MHC alleles
at specific native locus in swine cells. Multiplex targeting of Cas9 with two
gRNAs is used to
introduce single or double-stranded breaks flanking the MHC allele, enabling
replacement with
the template HLA/MHC sequence (provided as a single or double-stranded DNA
template). In
certain aspects, the CRISPR/Cas9 components are injected into swine oocytes,
ova, zygotes, or
blastocytes prior to transfer into foster mothers.
[000419] In certain aspects, the present disclosure includes embryogenesis
and live birth of
SLA-free and HLA-expressing biologically reprogrammed swine. In certain
aspects, the present
disclosure includes breeding SLA-free and HLA-expressing biologically
reprogrammed swine to
create SLA-free and HLA-expressing progeny. In certain aspects, the
CRISPR/Cas9 components
are injected into swine zygotes by intracytoplasmic microinjection of porcine
zygotes. In certain
aspects, the CRISPR/Cas9 components are injected into swine prior to selective
breeding of the
CRISPR/Cas9 genetically modified swine. In certain aspects, the CRISPR/Cas9
components are
injected into donor swine prior to harvesting cells, tissues, zygotes, and/or
organs from the swine.
In certain aspects, the CRISPR/Cas9 components include all necessary
components for controlled
gene editing including self-inactivation utilizing governing gRNA molecules as
described in U.S.
Pat. No. 9,834,791 (Zhang), which is incorporated herein by reference in its
entirety.
[000420] The genetic modification can be made utilizing known genome
editing techniques,
such as zinc-finger nucleases (ZFNs), transcription activator-like effector
nucleases (TALENs),
adeno-associated virus (AAV)-mediated gene editing, and clustered regular
interspaced
palindromic repeat Cas9 (CRISPR-Cas9). These programmable nucleases enable the
targeted
generation of DNA double-stranded breaks (DSB), which promote the upregulation
of cellular
repair mechanisms, resulting in either the error-prone process of non-
homologous end joining
(NHEJ) or homology-directed repair (HDR), the latter of which can be used to
integrate exogenous
donor DNA templates. CRISPR-Cas9 may also be used to remove viral infections
in cells. For
example, the genetic modification via CRISPR-Cas9 can be performed in a manner
described in
122
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Kelton, W. et. al., "Reprogramming MHC specificity by CRISPR-Cas9-assisted
cassette
exchange," Nature, Scientific Reports, 7:45775 (2017) ("Kelton"), the entire
disclosure of which
is incorporated herein by reference. Accordingly, the present disclosure
includes reprogramming
using CRISPR-Cas9 to mediate rapid and scarless exchange of entire alleles,
e.g., MHC, HLA,
SLA, etc.
[000421] In one aspect, the recipient's HLA/MHC gene is sequenced and
template
HLA/MHC sequences are prepared based on the recipient's HLA/MHC genes. In
another aspect,
a known human HLA/MHC genotype from a WHO database may be used for genetic
reprogramming of swine of the present disclosure. CRISPR-Cas9 plasmids are
prepared, e.g.,
using polymerase chain reaction and the recipient's HLA/MHC sequences are
cloned into the
plasmids as templates. CRISPR cleavage sites at the SLA/MHC locus in the swine
cells are
identified and gRNA sequences targeting the cleavage sites and are cloned into
one or more
CRISPR-Cas9 plasmids. CRISPR-Cas9 plasmids are then administered into the
swine cells and
CRIPSR/Cas9 cleavage is performed at the MHC locus of the swine cells.
[000422] The SLA/MHC locus in the swine cells are replaced with one or more
template
HLA/MHC sequences matching the known human HLA/MHC sequences or the
recipient's
sequenced HLA/MHC genes. Cells of the swine are sequenced after performing the
SLA/MHC
reprogramming steps in order to determine if the HLA/MHC sequences in the
swine cells have
been successfully reprogrammed. One or more cells, tissues, and/or organs from
the HLA/MHC
sequence-reprogrammed swine are transplanted into a human recipient.
[000423] In certain aspects, HLA/MHC sequence-reprogrammed swine are bred
for at least
one generation, or at least two generations, before their use as a source for
live tissues, organs
and/or cells used in xenotransplantation. In certain aspects, the CRISPR/Cas9
components can also
be utilized to inactivate genes responsible for PERV activity, e.g., the poi
gene, thereby
simultaneously completely eliminating PERV from the swine donors.
[000424] For purposes of modifying donor SLA/MHC to match recipient
HLA/MHC,
comparative genomic organization of the human and swine histocompatibility
complex has been
mapped. For example, such SLA to HLA mapping can be found in: Lunney, J.,
"Molecular
genetics of the swine major histocompatibility complex, the SLA complex,"
Developmental and
Comparative Immunology 33: 362-374 (2009) ("Lunney"), the entire disclosure of
which is
incorporated herein by reference. Accordingly, a person of ordinary skill in
the art effectively and
123
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
efficiently genetically reprogram swine cells in view of the present
disclosure and using the
mapping of Lunney et al. as a reference tool.
[000425] The modification to the donor SLA/MHC to match recipient HLA/MHC
causes
expression of specific MHC molecules from the swine cells that are identical,
or virtually identical,
to the MHC molecules of a known human genotype or the specific human
recipient. In one aspect,
the present disclosure involves making modifications limited to only specific
portions of specific
SLA regions of the swine's genome to retain an effective immune profile in the
swine while
biological products are hypoimmunogenic when transplanted into human
recipients such that use
of immunosuppressants can be reduced or avoided. In contrast to aspects of the
present disclosure,
xenotransplantation studies of the prior art required immunosuppressant use to
resist rejection. In
one aspect, the swine genome is reprogrammed to knock-out swine genes
corresponding to HLA-
A, HLA-B, HLA-C, and DR, and to knock-in HLA-C, HLA-E, HLA-G. In some aspects,
the swine
genome is reprogrammed to knock-out swine genes corresponding to HLA-A, HLA-B,
HLA-C,
HLA-F, DQ, and DR, and to knock-in HLA-C, HLA-E, HLA-G. In some aspects, the
swine
genome is reprogrammed to knock-out swine genes corresponding to HLA-A, HLA-B,
HLA-C,
HLA-F, DQ, and DR, and to knock-in HLA-C, HLA-E, HLA-G, HLA-F, and DQ. In one
aspect,
the swine genome is reprogrammed to knock-out SLA-11; SLA-6,7,8; SLA-MIC2; and
SLA-
DQA; SLA-DQB1; SLA-DQB2, and to knock-in HLA-C; HLA-E; HLA-G; and HLA-DQ. In
certain aspects, HLA-C expression is reduced in the reprogrammed swine genome.
By
reprogramming the swine cells to be invisible to a human's immune system, this
reprogramming
thereby minimizes or even eliminates an immune response that would have
otherwise occurred
based on swine MHC molecules otherwise expressed from the donor swine cells.
[000426] It will therefore be understood that this aspect (i.e.,
reprogramming the SLA/MHC
to express specifically selected human MHC alleles), when applied to swine
cells, tissues, and
organs for purposes of xenotransplantation will decrease rejection as compared
to cells, tissues,
and organs derived from a wild-type swine or otherwise genetically modified
swine that lacks this
reprogramming, e.g., transgenic swine or swine with non-specific or different
genetic
modifications.
[000427] It will be further understood that causing the donor swine cells,
tissues, and organs
to express a known human MHC genotype or the recipient's MHC specifically as
described herein,
combined with the elimination in the donor swine cells of alpha-1,3-
galactosytransferase, Neu5Gc,
124
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
and f31,4-N-acetylgalactosaminyltransferase (B4GALNT2) (e.g., "single
knockout," "double
knockout," or "triple knockout"), presents a swine whose cells will have a
decreased
immunological rejection as compared to a triple knockout swine that lacks the
specific SLA/MEIC
reprogramming of the present disclosure.
[000428] Cryopreservation and storage according to the present disclosure
includes
preparing biological product according to the present disclosures, placing in
a container, adding
freeze media to the container and sealing. For example. 15% dimethyl sulfoxide
(DMSO)
cryoprotective media is combined with fetal porcine serum (FPS) or donor serum
(if FPS is
unavailable) in a 1:1 ratio, filtered (0.45 micron), and chilled to 4 C prior
to use. The containers
are subsequently frozen in a controlled rate, phase freezer at a rate of 1 C
per minute to -40 C, then
rapidly cooled to a temperature -80 C. DMSO displaces intracellular fluid
during the freezing
process. Cryoprotective media, e.g., CryoStor is used in an amount of about 40-
80%, or 50-70%
based on maximum internal volume of the cryovial (10m1) less the volume of the
xenotransplantation product. In order to thaw the cryopreserved biological
product for surgical
use, sealed vials were placed in ¨37 C water baths for approximately 0.5 to 2
minutes, at which
point the container is opened and the product was removed using sterile
technique. Subsequently,
products undergo three, 1-minute serial washes, e.g., in saline with gentle
agitation, in order to
dilute and systematically remove ambient, residual DMSO and prevent loss of
cell viability. The
product may then be used surgically.
[000429] It will be understood that the xenotransplantation product may be
processed, stored,
transported, and/or otherwise handled using materials, containers and
processes to ensure
preserved sterility and prevent damage thereto. In some aspects, a sterile non-
adhesive material
may be used to protect the xenotransplantation product, e.g., to support the
xenotransplantation
product and prevent adhesive of the product to surfaces and/or to prevent self-
adhesion of the
xenotransplantation product during manipulation, storage, or transport.
Unintentional adhesion of
the xenotransplantation product may disrupt the integrity of the
xenotransplantation product and
potentially reduce its therapeutic viability. Inclusion of the sterile non-
adhesive material provides
protection and/or physical support and prevents adhesion. In some aspects, the
sterile non-
adhesive material is not biologically or chemically active and does not
directly impact the
metabolic activity or efficacy of the xenotransplantation product itself
125
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000430] Aspects of the present disclosure are further described by the
following non-
limiting list of items:
Item 1. A biological system for generating and preserving a repository of
personalized,
humanized transplantable cells, tissues, and organs for transplantation,
wherein the biological
system is biologically active and metabolically active, the biological system
comprising
genetically reprogrammed cells, tissues, and organs in a non-human animal for
transplantation
into a human recipient,
wherein the non-human animal is a genetically reprogrammed swine for
xenotransplantation of cells, tissue, and/or an organ isolated from the
genetically reprogrammed
swine, the genetically reprogrammed swine comprising a nuclear genome that has
been
reprogrammed to replace a plurality of nucleotides in a plurality of exon
regions of a major
histocompatibility complex of a wild-type swine with a plurality of
synthesized nucleotides from
a human captured reference sequence, and
wherein cells of said genetically reprogrammed swine do not present one or
more surface
glycan epitopes selected from alpha-Gal, Neu5Gc, and SDa,
and
wherein genes encoding alpha-1,3 galactosyltransferase, cytidine monophosphate-
N-
acetylneuraminic acid hydroxylase (CMAH), and 01,4-N-
acetylgalactosaminyltransferase are
altered such that the genetically reprogrammed swine lacks functional
expression of surface
glycan epitopes encoded by said genes,
wherein the reprogrammed genome comprises site-directed mutagenic
substitutions of
nucleotides at exon regions of: i) at least one of the wild-type swine's SLA-
1, SLA-2, and SLA-3
with nucleotides from an orthologous exon region of HLA-A, HLA-B, and HLA-C,
respectively,
of the human captured reference sequence; and ii) at least one the wild-type
swine's SLA-6,
SLA-7, and SLA-8 with nucleotides from an orthologous exon region of HLA-E,
HLA-F, and
HLA-G, respectively, of the human captured reference sequence; and iii) at
least one of the wild-
type swine's SLA-DR and SLA-DQ with nucleotides from an orthologous exon
region of HLA-
DR and HLA-DQ, respectively, of the human captured reference sequence,
wherein the reprogrammed genome comprises at least one of A-C:
A) wherein the reprogrammed swine nuclear genome comprises site-directed
mutagenic
substitutions of nucleotides at exon regions of the wild-type swine's 02-
microglobulin with
126
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
nucleotides from orthologous exons of a known human 02-microglobulin from the
human
captured reference sequence;
B) wherein the reprogrammed swine nuclear genome comprises a polynucleotide
that
encodes a polypeptide that is a humanized beta 2 microglobulin (hB2M)
polypeptide sequence
that is at least 95% identical to the amino acid sequence of beta 2
microglobulin glycoprotein
expressed by the human captured reference genome;
C) wherein the reprogrammed swine nuclear genome has been reprogrammed such
that, at
the swine's endogenous 02-microglobulin locus, the nuclear genome has been
reprogrammed to
comprise a nucleotide sequence encoding 02-microglobulin polypeptide of the
human recipient,
wherein the reprogrammed swine nuclear genome has been reprogrammed such that
the
genetically reprogrammed swine lacks functional expression of the wild-type
swine's
endogenous 02-microglobulin polypeptides, and
wherein said reprogramming does not introduce any frameshifts or frame
disruptions.
Item 2. The biological system of item 1, wherein the genetically reprogrammed
swine is
non-transgenic.
Item 3. The biological system of item 1 or item 2, wherein intron regions of
the wild-type
swine's genome are not reprogrammed.
Item 4. The biological system of any one of or combination of items 1-3,
wherein said
genetically reprogrammed swine is free of at least the following pathogens:
Ascaris species,
cryptosporidium species, Echinococcus, Strongyloids sterocolis, Toxoplasma
gondii, Brucella
suis, Leptospira species, mycoplasma hyopneumoniae, porcine reproductive and
respiratory
syndrome, pseudorabies, staphylococcus species, Microphyton species,
Trichophyton species,
porcine influenza, porcine cytomegalovirus, arterivirus, coronavirus,
Bordetella bronchiseptica,
and Livestock-associated methicillin-resistant Staphylococcus aureus.
Item 5. The biological system of any one of or combination of items 1-4,
wherein said
genetically reprogrammed swine is maintained according to a bioburden-reducing
procedure,
said procedure comprising maintaining the swine in an isolated closed herd,
wherein all other
animals in the isolated closed herd are confirmed to be free of said
pathogens, and wherein the
swine is isolated from contact with any non-human animals and animal housing
facilities outside
of the isolated closed herd.
127
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Item 6. The biological system of any one of or combination of items 1-4,
wherein the
wild-type swine genome comprises reprogrammed nucleotides at SLA-MIC-2 gene
and at exon
regions encoding SLA-3, SLA-6, SLA-7, SLA-8, SLA-DQ, CTLA-4, PD-L1, EPCR, TBM,
TFPI, and beta-2-microglobulin using the human capture reference sequence,
wherein the human
cell, tissue, or organ lacks functional expression of swine beta-2-
microglobulin, SLA-1, SLA-2,
and SLA-DR.
Item 7. The biological system of any one of or combination of items 1-5,
wherein the
wild-type swine genome comprises reprogrammed nucleotides at one or more of a
CTLA-4
promoter and a PD-Li promoter, wherein the one or more of the CTLA-4 promoter
and the PD-
Li promoter are reprogrammed to increase expression of one or both of
reprogrammed CTLA-4
and reprogrammed PD-Li compared to the wild-type swine's endogenous expression
of CTLA-4
and PD-Li.
Item 8. The biological system of any one of or combination of items 1-6,
wherein a total
number of the synthesized nucleotides is equal to a total number of the
replaced nucleotides,
such that there is no net loss or net gain in number of nucleotides after
reprogramming the
genome of the wild-type swine with the synthesized nucleotides.
Item 9. The biological system of any one of or combination of items 1-7,
wherein the
reprogramming with the plurality of synthesized nucleotides do not include
replacement of
nucleotides in codon regions that encode amino acids that are conserved
between the wild-type
swine MHC sequence and the human captured reference sequence
Item 10. The biological system of any one of or combination of items 1-8,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at the
major histocompatibility complex of the wild-type swine with orthologous
nucleotides from the
human captured reference sequence.
Item 11. The biological system of any one of or combination of items 1-9,
wherein site-
directed mutagenic substitutions are made in germ-line cells used to produce
the non-human
animal.
Item 12. The biological system of any one of or combination of items 1-10,
wherein the
human captured reference sequence is a human patient capture sequence, a human
population-
specific human capture sequence, or an allele-group-specific human capture
sequence.
128
CA 03133638 2021-09-14
WO 2020/198397
PCT/US2020/024780
Item 13. The biological system of any one of or combination of items 1-11,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-1 with nucleotides from an orthologous
exon region of a
HLA-A captured reference sequence.
Item 14. The biological system of any one of or combination of items 1-12,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-2 with nucleotides from an orthologous
exon region of a
HLA-B captured reference sequence.
Item 14. The biological system of any one of or combination of items 1-13,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-3 with nucleotides from an orthologous
exon region of a
HLA-C captured reference sequence.
Item 15. The biological system of any one of or combination of items 1-14,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-6 with nucleotides from an orthologous
exon region of a
HLA-E captured reference sequence.
Item 16. The biological system of any one of or combination of items 1-15,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-7 with nucleotides from an orthologous
exon region of a
HLA-F captured reference sequence.
Item 17. The biological system of any one of or combination of items 1-16,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-8 with nucleotides from an orthologous
exon region of a
HLA-G captured reference sequence.
Item 18. The biological system of any one of or combination of items 1-17,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's MHC class I chain-related 2 (MIC-2).
Item 19. The biological system of any one of or combination of items 1-18,
wherein the
reprogrammed genome lacks functional expression of SLA-1, SLA-2, SLA-DR, or a
combination thereof.
129
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Item 20. The biological system of any one of or combination of items 1-19,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-DQA from an orthologous exon region of a
HLA-DQA1
captured reference sequence.
Item 21. The biological system of any one of or combination of items 1-20,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-DQB from an orthologous exon region of a
HLA-DQB1
captured reference sequence.
Item 22. The biological system of any one of or combination of items 1-21,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-DRA and SLA-DRB lwith nucleotides from
orthologous
exon regions of HLA-DRA1 and HLA-DRBlof the human captured reference sequence,
or
wherein the reprogrammed genome lacks functional expression of SLA-DRA and SLA-
DRB1.
Item 23. The biological system of any one of or combination of items 1-22,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-DQA and SLA-DQB1 with nucleotides from
orthologous
exon regions of HLA-DQA1 and HLA-DQB1 of the human captured reference
sequence.
Item 24. The biological system of any one of or combination of items 1-23,
wherein the
site-directed mutagenic substitutions of nucleotides are at codons that are
not conserved between
the wild-type swine's nuclear genome and the known human sequence.
Item 25. The biological system of any one of or combination of items 1-24,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's B2-microglobulin with nucleotides from
orthologous exons of a
known human B2-microglobulin.
Item 26. The biological system of any one of or combination of items 1-25,
wherein the
reprogrammed swine nuclear genome comprises a polynucleotide that encodes a
polypeptide that
is a humanized beta 2 microglobulin (hB2M) polypeptide sequence that is at
least 95% identical
to the amino acid sequence of beta 2 microglobulin glycoprotein expressed by
the human
captured reference genome;
130
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Item 27. The biological system of any one of or combination of items 1-26,
wherein said
nuclear genome has been reprogrammed such that the genetically reprogrammed
swine lacks
functional expression of the wild-type swine's endogenous 02-microglobulin
polypeptides.
Item 28. The biological system of any one of or combination of items 1-27,
wherein said
nuclear genome has been reprogrammed such that, at the swine's endogenous 02-
microglobulin
locus, the nuclear genome has been reprogrammed to comprise a nucleotide
sequence encoding
132-microglobulin polypeptide of the human captured reference sequence.
Item 29. The biological system of any one of or combination of items 1-28,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of SLA-3, SLA-6, SLA-7, SLA-8, and MIC-2.
Item 30. The biological system of any one of or combination of items 1-29,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of SLA-DQ and MIC-2.
Item 31. The biological system of any one of or combination of items 1-30,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at SLA-3,
SLA-6, SLA-7, SLA-8, SLA-DQ, and MIC-2.
Item 32. The biological system of any one of or combination of items 1-31,
wherein the
reprogrammed genome lacks functional expression of SLA-DR, SLA-1, and/or SLA-
2.
Item 33. The biological system of any one of or combination of items 1-32,
wherein the
nuclear genome is reprogrammed using scarless exchange of the exon regions,
wherein there are
no frameshifts, insertion mutations, deletion mutations, missense mutations,
and nonsense
mutations.
Item 34. The biological system of any one of or combination of items 1-33,
wherein the
nuclear genome is reprogrammed without introduction of any net insertions,
deletions,
truncations, or other genetic alterations that would cause a disruption of
protein expression via
frame shift, nonsense, or missense mutations.
Item 35. The biological system of any one of or combination of items 1-34,
wherein
nucleotides in intron regions of the nuclear genome are not altered.
Item 36. The biological system of any one of or combination of items 1-35,
wherein said
nuclear genome is reprogrammed to be homozygous at the reprogrammed exon
regions.
131
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Item 37. The biological system of any one of or combination of items 1-36,
wherein said
nuclear genome is reprogrammed such that extracellular, phenotypic surface
expression of
polypeptide is tolerogenic in a human recipient.
Item 38. The biological system of any one of or combination of items 1-37,
wherein said
nuclear genome is reprogrammed such that expression of cytotoxic T-lymphocyte-
associated
protein 4 (CTLA-4) is increased by reprogramming a CTLA-4 promoter sequence.
Item 39. The biological system of any one of or combination of items 1-38,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type CTLA-4 with nucleotides from orthologous exons of a
human captured
reference sequence CTLA-4.
Item 40. The biological system of any one of or combination of items 1-39,
wherein the
reprogrammed nuclear genome comprises a polynucleotide that encodes a protein
that is a
humanized CTLA-4 polypeptide sequence that is at least 95% identical to CTLA-4
expressed by
the human captured reference genome.
Item 41. The biological system of any one of or combination of items 1-40,
wherein said
nuclear genome is reprogrammed such that expression of Programmed death-ligand
1(PD-L1) is
increased by reprogramming a PD-Li promoter sequence.
Item 42. The biological system of any one of or combination of items 1-41,
wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type PD-Li with nucleotides from orthologous exons of a
known human PD-
Ll.
Item 43. The biological system of any one of or combination of items 1-42,
wherein the
reprogrammed nuclear genome comprises a polynucleotide that encodes a protein
that is a
humanized PD-Llpolypeptide sequence that is at least 95% identical to PD-Li
expressed by the
human captured reference genome.
Item 44. A genetically reprogrammed, biologically active and metabolically
active non-
human cell, tissue, or organ obtained from the biological system of any one of
or combination of
items 1-43.
Item 45. The genetically reprogrammed, biologically active and metabolically
active non-
human cell, tissue, or organ of item 44, wherein the genetically reprogrammed,
biologically
132
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
active and metabolically active non-human cell is a stem cell, an embryonic
stem cell, a
pluripotent stem cell, or a differentiated stem cell.
Item 46. The genetically reprogrammed, biologically active and metabolically
active non-
human cell, tissue, or organ of item 45, wherein the stem cell is a
hematopoietic stem cell.
Item 47. The genetically reprogrammed, biologically active and metabolically
active non-
human cell, tissue, or organ of item 44, wherein the genetically reprogrammed,
biologically
active and metabolically active non-human tissue is a nerve, cartilage, or
skin.
Item 48. The genetically reprogrammed, biologically active and metabolically
active non-
human cell, tissue, or organ of item 44, wherein the genetically reprogrammed,
biologically
active and metabolically active non-human organ is a solid organ.
Item 49. A method of preparing a genetically reprogrammed swine comprising a
nuclear
genome that lacks functional expression of surface glycan epitopes selected
from alpha-Gal,
Neu5Gc, and SD a and is genetically reprogrammed to express a humanized
phenotype of a
human captured reference sequence comprising:
a. obtaining a porcine fetal fibroblast cell, a porcine zygote, a porcine
Induced Pluripotent
Stem Cells (IPSC), or a porcine germ-line cell;
b. genetically altering said cell in a) to lack functional alpha-1,3
galactosyltransferase,
cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMAH), and 01,4-N-
acetylgalactosaminyltransferase;
c. genetically reprogramming said cell in b) using clustered regularly
interspaced short
palindromic repeats (CRISPR)/Cas for site-directed mutagenic substitutions of
nucleotides at exon regions of: i) at least one of the wild-type swine's SLA-
1, SLA-2, and
SLA-3 with nucleotides from an orthologous exon region of HLA-A, HLA-B, and
HLA-
C, respectively, of the human captured reference sequence; and ii) at least
one the wild-
type swine's SLA-6, SLA-7, and SLA-8 with nucleotides from an orthologous exon
region of HLA-E, HLA-F, and HLA-G, respectively, of the human captured
reference
sequence; and iii) at least one of the wild-type swine's SLA-DR and SLA-DQ
with
nucleotides from an orthologous exon region of HLA-DR and HLA-DQ,
respectively, of
the human captured reference sequence,
wherein intron regions of the wild-type swine's genome are not reprogrammed,
and
wherein the reprogrammed genome comprises at least one of A-C:
133
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
A) wherein the reprogrammed swine nuclear genome comprises site-directed
mutagenic
substitutions of nucleotides at exon regions of the wild-type swine's 02-
microglobulin with
nucleotides from orthologous exons of a known human 02-microglobulin from the
human
captured reference sequence;
B) wherein the reprogrammed swine nuclear genome comprises a polynucleotide
that
encodes a polypeptide that is a humanized beta 2 microglobulin (hB2M)
polypeptide sequence
that is at least 95% identical to beta 2 microglobulin expressed by the human
captured reference
genome;
C) wherein the reprogrammed swine nuclear genome has been reprogrammed such
that the
genetically reprogrammed swine lacks functional expression of the wild-type
swine's
endogenous 02-microglobulin polypeptides, wherein the reprogrammed swine
nuclear genome
has been reprogrammed such that, at the swine's endogenous 02-microglobulin
locus, the nuclear
genome has been reprogrammed to comprise a nucleotide sequence encoding 02-
microglobulin
polypeptide of the human recipient,
wherein said reprogramming does not introduce any frameshifts or frame
disruptions,
d. generating an embryo from the genetically reprogrammed cell in c); and
e. transferring the embryo into a surrogate pig and growing the transferred
embryo in the
surrogate pig.
Item 50. The method of item 49, wherein step (a) further comprises replacing a
plurality
of nucleotides in a plurality of exon regions of a major histocompatibility
complex of a wild-type
swine with nucleotides from orthologous exon regions of a major
histocompatibility complex
sequence from the human captured reference sequence, wherein said replacing
does not
introduce any frameshifts or frame disruptions.
Item 51. The method of any one of or combination of items 49-50, wherein said
replacing
comprises performing site-directed mutagenic substitutions of nucleotides at
the major
histocompatibility complex of the wild-type swine with orthologous nucleotides
from the known
human major histocompatibility complex sequence.
Item 52. The method of any one of or combination of items 49-51, wherein the
human
captured reference sequence is a human patient capture sequence, a human
population-specific
human capture sequence, or an allele-group-specific human capture sequence.
134
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Item 53. The method of any one of or combination of items 49-52, wherein the
orthologous exon regions are at one or more polymorphic glycoproteins of the
wild-type swine's
major histocompatibility complex.
Item 54. The method of any one of or combination of items 49-53, further
comprising:
impregnating the surrogate pig with the embryo, gestating the embryo, and
delivering a piglet
from the surrogate pig through Cesarean section,
confirming that said piglet is free of at least the following zoonotic
pathogens:
(i) Ascaris species, cryptosporidium species, Echinococcus, Strongyloids
sterocolis, and
Toxoplasma gondii in fecal matter;
(ii) Leptospira species, Mycoplasma hyopneumoniae, porcine reproductive and
respiratory
syndrome virus (PRRSV), pseudorabies, transmissible gastroenteritis virus
(TGE) / Porcine
Respiratory Coronavirus, and Toxoplasma Gondii by determining antibody titers;
(iii) Porcine Influenza;
(iv) the following bacterial pathogens as determined by bacterial culture:
Bordetella
bronchisceptica, Coagulase-positive staphylococci, Coagulase-negative
staphylococci,
Livestock-associated methicillin resistant Staphylococcus aureus (LA MRSA),
Microphyton and
Trichophyton spp.;
(v) Porcine cytomegalovirus; and
(vi) Brucella suis; and
maintaining the piglet according to a bioburden-reducing procedure, said
procedure comprising
maintaining the piglet in an isolated closed herd, wherein all other animals
in the isolated closed
herd are confirmed to be free of said zoonotic pathogens, wherein the piglet
is isolated from
contact with any non-human animals and animal housing facilities outside of
the isolated closed
herd.
Item 55. The method of any one of or combination of items 49-54, wherein the
wild-type
swine genome comprises reprogrammed nucleotides at SLA-MIC-2 gene and at exon
regions
encoding SLA-3, SLA-6, SLA-7, SLA-8, SLA-DQ, CTLA-4, PD-L1, EPCR, TBM, TFPI,
and
beta-2-microglobulin using the human capture reference sequence, wherein the
human cell,
tissue, or organ lacks functional expression of swine beta-2-microglobulin,
SLA-DR, SLA-1, and
SLA-2.
135
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Item 56. The method of any one of or combination of items 49-55, wherein the
wild-type
swine genome comprises reprogrammed nucleotides at one or more of a CTLA-4
promoter and a
PD-Li promoter, wherein the one or more of the CTLA-4 promoter and the PD-Li
promoter are
reprogrammed to increase expression of one or both of reprogrammed CTLA-4 and
reprogrammed PD-Li compared to the wild-type swine's endogenous expression of
CTLA-4 and
PD-Li.
Item 57. The method of any one of or combination of items 49-56, wherein a
total
number of the synthesized nucleotides is equal to a total number of the
replaced nucleotides,
such that there is no net loss or net gain in number of nucleotides after
reprogramming the
genome of the wild-type swine with the synthesized nucleotides.
Item 58. The method of any one of or combination of items 49-57, wherein the
reprogramming with the plurality of synthesized nucleotides do not include
replacement of
nucleotides in codon regions that encode amino acids that are conserved
between the wild-type
swine MHC sequence and the human captured reference sequence
Item 59. The method of any one of or combination of items 49-58, wherein the
reprogrammed
genome comprises site-directed mutagenic substitutions of nucleotides at the
major
histocompatibility complex of the wild-type swine with orthologous nucleotides
from the human
captured reference sequence.
Item 60. The method of any one of or combination of items 49-59, wherein site-
directed
mutagenic substitutions are made in germ-line cells used to produce the non-
human animal.
Item 61. The method of any one of or combination of items 49-60, wherein the
human
captured reference sequence is a human patient capture sequence, a human
population-specific
human capture sequence, or an allele-group-specific human capture sequence.
Item 62. The method of any one of or combination of items 49-61, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-1 with nucleotides from an orthologous
exon region of a
HLA-A captured reference sequence.
Item 63. The method of any one of or combination of items 49-62, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-2 with nucleotides from an orthologous
exon region of a
HLA-B captured reference sequence.
136
CA 03133638 2021-09-14
WO 2020/198397
PCT/US2020/024780
Item 64. The method of any one of or combination of items 49-63, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-3 with nucleotides from an orthologous
exon region of a
HLA-C captured reference sequence.
Item 65. The method of any one of or combination of items 49-64, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-6 with nucleotides from an orthologous
exon region of a
HLA-E captured reference sequence.
Item 66. The method of any one of or combination of items 49-65, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-7 with nucleotides from an orthologous
exon region of a
HLA-F captured reference sequence.
Item 67. The method of any one of or combination of items 49-66, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-8 with nucleotides from an orthologous
exon region of a
HLA-G captured reference sequence.
Item 68. The method of any one of or combination of items 49-67, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's MHC class I chain-related 2 (MIC-2).
Item 69. The method of any one of or combination of items 49-68, wherein the
reprogrammed genome lacks functional expression of SLA-1, SLA-2, SLA-DR, or a
combination thereof.
Item 70. The method of any one of or combination of items 49-69, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-DQA from an orthologous exon region of a
HLA-DQA1
captured reference sequence.
Item 71. The method of any one of or combination of items 49-70, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-DQB from an orthologous exon region of a
HLA-DQB1
captured reference sequence.
137
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Item 72. The method of any one of or combination of items 49-71, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-DRA and SLA-DRB lwith nucleotides from
orthologous
exon regions of HLA-DRA1 and HLA-DRBlof the human captured reference sequence.
Item 73. The method of any one of or combination of items 49-72, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's SLA-DQA and SLA-DQB1 with nucleotides from
orthologous
exon regions of HLA-DQA1 and HLA-DQB1 of the human captured reference
sequence.
Item 74. The method of any one of or combination of items 49-73, wherein the
site-
directed mutagenic substitutions of nucleotides are at codons that are not
conserved between the
wild-type swine's nuclear genome and the known human sequence.
Item 75. The method of any one of or combination of items 49-74, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type swine's B2-microglobulin with nucleotides from
orthologous exons of a
known human B2-microglobulin.
Item 76. The method of any one of or combination of items 49-75, wherein the
reprogrammed swine nuclear genome comprises a polynucleotide that encodes a
polypeptide that
is a humanized beta 2 microglobulin (hB2M) polypeptide sequence that is at
least 95% identical
to the amino acid sequence of beta 2 microglobulin glycoprotein expressed by
the human
captured reference genome;
Item 77. The method of any one of or combination of items 49-76, wherein said
nuclear
genome has been reprogrammed such that the genetically reprogrammed swine
lacks functional
expression of the wild-type swine's endogenous 02-microglobulin polypeptides.
Item 78. The method of any one of or combination of items 49-77, wherein said
nuclear
genome has been reprogrammed such that, at the swine's endogenous 02-
microglobulin locus,
the nuclear genome has been reprogrammed to comprise a nucleotide sequence
encoding f32-
microglobulin polypeptide of the human captured reference sequence.
Item 79. The method of any one of or combination of items 49-78, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of SLA-3, SLA-6, SLA-7, SLA-8, and MIC-2.
138
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Item 80. The method of any one of or combination of items 49-79, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of SLA-DQ, and MIC-2.
Item 81. The method of any one of or combination of items 49-80, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at SLA-3,
SLA-6, SLA-7, SLA-8, SLA-DQ, and MIC-2.
Item 82. The method of any one of or combination of items 49-81, wherein the
reprogrammed genome lacks functional expression of SLA-DR, SLA-1, and/or SLA-
2.
Item 83. The method of any one of or combination of items 49-82, wherein the
nuclear
genome is reprogrammed using scarless exchange of the exon regions, wherein
there are no
frameshifts, insertion mutations, deletion mutations, missense mutations, and
nonsense
mutations.
Item 84. The method of any one of or combination of items 49-83, wherein the
nuclear
genome is reprogrammed without introduction of any net insertions, deletions,
truncations, or
other genetic alterations that would cause a disruption of protein expression
via frame shift,
nonsense, or missense mutations.
Item 85. The method of any one of or combination of items 49-84, wherein
nucleotides
in intron regions of the nuclear genome are not altered.
Item 86. The method of any one of or combination of items 49-85, wherein said
nuclear
genome is reprogrammed to be homozygous at the reprogrammed exon regions.
Item 87. The method of any one of or combination of items 49-86, wherein said
nuclear
genome is reprogrammed such that extracellular, phenotypic surface expression
of polypeptide is
tolerogenic in a human recipient.
Item 88. The method of any one of or combination of items 49-87, wherein said
nuclear
genome is reprogrammed such that expression of cytotoxic T-lymphocyte-
associated protein 4
(CTLA-4) is increased by reprogramming a CTLA-4 promoter sequence.
Item 89. The method of any one of or combination of items 49-88, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type CTLA-4 with nucleotides from orthologous exons of a
human captured
reference sequence CTLA-4.
139
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Item 90. The method of any one of or combination of items 49-89, wherein the
reprogrammed nuclear genome comprises a polynucleotide that encodes a protein
that is a
humanized CTLA-4 polypeptide sequence that is at least 95% identical to CTLA-4
expressed by
the human captured reference genome.
Item 91. The method of any one of or combination of items 49-90, wherein said
nuclear genome
is reprogrammed such that expression of Programmed death-ligand 1(PD-L1) is
increased by
reprogramming a PD-Li promoter sequence.
Item 92. The method of any one of or combination of items 49-91, wherein the
reprogrammed genome comprises site-directed mutagenic substitutions of
nucleotides at exon
regions of the wild-type PD-Li with nucleotides from orthologous exons of a
known human PD-
Ll.
Item 93. The method of any one of or combination of items 49-92, wherein the
reprogrammed nuclear genome comprises a polynucleotide that encodes a protein
that is a
humanized PD-Llpolypeptide sequence that is at least 95% identical to PD-Li
expressed by the
human captured reference genome.
Item 94. A method of inducing at least partial immunological tolerance in a
recipient
human to a xenotransplanted cell, tissue, or organ, the method comprising:
producing or obtaining non-human cell, tissue, or organ obtained from the
biological system of
any one of or combination of items 1-48, wherein the wild-type swine genome
comprises
reprogrammed nucleotides at SLA-MIC-2 gene and at exon regions of one or more
encoding the
wild-type swine's MHC Class Ia, MHC class lb, MHC Class II, and beta-2-
microglobulin using
the human capture reference sequence and wherein the human cell, tissue, or
organ lacks
functional expression of swine beta-2-microglobulin; and
implanting the non-human cell, tissue, or organ into the recipient human.
Item 95. A method of reducing Natural Killer cell-mediated rejection of a
xenograft
comprising: producing or obtaining non-human cell, tissue, or organ obtained
from the biological
system of any one of or combination of items 1-48, wherein the wild-type swine
genome
comprises reprogrammed nucleotides at SLA-MIC-2 gene and at exon regions
encoding one or
more of the wild-type swine's MHC Class Ia, MHC class Ib, MHC Class II, and
beta-2-
microglobulin using the human capture reference sequence and wherein the human
cell, tissue,
or organ lacks functional expression of swine beta-2-microglobulin, and
wherein the wild-type
140
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
swine genome comprises reprogrammed nucleotides at exon regions encoding one
or more of the
wild-type swine's CTLA-4 and PD-Li; and implanting the non-human cell, tissue,
or organ into
the recipient human.
Item 96. A method of reducing Cytotoxic T-cell Lymphocyte cell-mediated
rejection of a
xenograft comprising:
producing or obtaining non-human cell, tissue, or organ obtained from the
biological
system of any one of or combination of items 1-48, wherein the wild-type swine
genome
comprises reprogrammed nucleotides at SLA-MIC-2 gene and at exon regions
encoding one or
more of the wild-type swine's MHC Class Ia, MHC class Ib, MHC Class II, and
beta-2-
microglobulin using the human capture reference sequence and wherein the human
cell, tissue,
or organ lacks functional expression of swine beta-2-microglobulin, and
wherein the wild-type
swine genome comprises reprogrammed nucleotides at exon regions encoding one
or more of the
wild-type swine's CTLA-4 and PD-Li; and
implanting the non-human cell, tissue, or organ into the recipient human.
Item 97. A method of preventing or reducing coagulation and/or thrombotic
ischemia in
a recipient human to a xenotransplanted cell, tissue, or organ, the method
comprising:
producing or obtaining non-human cell, tissue, or organ obtained from the
biological system of
any one of or combination of items 1-48, wherein the wild-type swine genome
comprises
reprogrammed nucleotides at SLA-MIC-2 gene and at exon regions encoding one or
more of the
wild-type swine's MHC Class Ia, MHC class lb, MHC Class II, and beta-2-
microglobulin using
the human capture reference sequence, wherein the human cell, tissue, or organ
lacks functional
expression of swine beta-2-microglobulin, and wherein the wild-type swine
genome comprises
reprogrammed nucleotides at exon regions encoding one or more of the wild-type
swine's
endothelial protein C receptor (EPCR), thrombomodulin (TBM), and tissue factor
pathway
inhibitor (TFPI); and
implanting the non-human cell, tissue, or organ into the recipient human.
Item 98. A method of reducing MHC Class Ia-mediated rejection of a xenograft
comprising:
producing or obtaining non-human cell, tissue, or organ obtained from the
biological
system of any one of or combination of items 1-48, wherein the wild-type swine
genome
comprises reprogrammed nucleotides at SLA-MIC-2 gene and at exon regions
encoding SLA-3
141
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
and one or more of the wild-type swine's MHC class lb, MHC Class II, and beta-
2-
microglobulin using the human capture reference sequence, wherein the human
cell, tissue, or
organ lacks functional expression of swine beta-2-microglobulin, SLA-1, and
SLA-2; and
implanting the non-human cell, tissue, or organ into the recipient human.
Item 99. A method of reducing MHC Class Ib-mediated rejection of a xenograft
comprising:
producing or obtaining non-human cell, tissue, or organ obtained from the
biological
system of any one of or combination of items 1-48, wherein the wild-type swine
genome
comprises reprogrammed nucleotides at SLA-MIC-2 gene and at exon regions
encoding SLA-6,
SLA-7, and SLA-8, and one or more of the wild-type swine's MHC class Ia, MHC
Class II, and
beta-2-microglobulin using the human capture reference sequence, wherein the
human cell,
tissue, or organ lacks functional expression of swine beta-2-microglobulin;
and
implanting the non-human cell, tissue, or organ into the recipient human.
Item 100. A method of reducing MHC Class II-mediated rejection of a xenograft
comprising:
producing or obtaining non-human cell, tissue, or organ obtained from the
biological
system of any one of or combination of items 1-48, wherein the wild-type swine
genome
comprises reprogrammed nucleotides at SLA-MIC-2 gene and at exon regions
encoding at least
one of SLA-DR and SLA-DQ, and one or more of the wild-type swine's MHC class
Ia, MHC
Class Ib, and beta-2-microglobulin using the human capture reference sequence,
wherein the
human cell, tissue, or organ lacks functional expression of swine beta-2-
microglobulin; and
implanting the non-human cell, tissue, or organ into the recipient human.
Item 101. A method of inhibiting apoptotic cell-mediated rejection of a
xenograft
comprising:
producing or obtaining non-human cell, tissue, or organ obtained from the
biological
system of any one of or combination of items 1-48, wherein the wild-type swine
genome
comprises reprogrammed nucleotides at SLA-MIC-2 gene and at exon regions
encoding one or
more of the wild-type swine's MHC Class Ia, MHC class Ib, MHC Class II, and
beta-2-
microglobulin using the human capture reference sequence and wherein the human
cell, tissue,
or organ lacks functional expression of swine beta-2-microglobulin, and
wherein the wild-type
142
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
swine genome comprises reprogrammed nucleotides at exon regions encoding one
or more of the
wild-type swine's CTLA-4 and PD-Li; and
implanting the non-human cell, tissue, or organ into the recipient human.
Item 102. A method of producing a donor swine tissue or organ for
xenotransplantation,
wherein cells of said donor swine tissue or organ are genetically reprogrammed
to be
characterized by a recipient-specific surface phenotype comprising:
obtaining a biological sample containing DNA from a prospective human
transplant recipient;
performing whole genome sequencing of the biological sample to obtain a human
capture
reference sequence;
comparing the human capture reference sequence with the wild-type genome of
the donor swine
at loci (i)-(v):
(i) exon regions encoding at least one of SLA-1, SLA-2, and SLA-3;
(ii) exon regions encoding at least one of SLA-6, SLA-7, and SLA-8;
(iii)
exon regions encoding at least one of SLA-DR and SLA-DQ;
(iv) one or more exons encoding beta 2 microglobulin (B2M);
(v) exon regions of SLA-MIC-2 gene and a gene encoding at least one of PD-L1,
CTLA-4,
EPCR, TBM, and TFPI,
creating synthetic donor swine nucleotide sequences of 10 to 350 basepairs in
length for one or
more of said loci (i)-(v), wherein said synthetic donor swine nucleotide
sequences are at least
95% identical to the human capture reference sequence at orthologous loci (vi)-
(x) corresponding
to swine loci (i)-(vi), respectively:
(vi) exon regions encoding at least one of HLA-A, HLA-B, and HLA-C;
(vii) exon regions encoding at least one of HLA-E, HLA-F, and HLA-G;
(viii) exon regions encoding at least one of HLA-DR and HLA-DQ;
(ix) one or more exons encoding human beta 2 microglobulin (hB2M);
(x) exon regions encoding at least one of MIC-A, MIC-B, PD-L1, CTLA-4, EPCR,
TBM, and
TFPI from the human capture reference sequence,
replacing nucleotide sequences in (i)-(v) with said synthetic donor swine
nucleotide sequences;
and
obtaining the swine tissue or organ for xenotransplantation from a genetically
reprogrammed
swine having said synthetic donor swine nucleotide sequences.
143
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Item 103. The method of item 102, further comprising confirming that the
genetically
reprogrammed swine having said synthetic donor swine nucleotide sequences is
free of at least
the following zoonotic pathogens:
(i) Ascaris species, cryptosporidium species, Echinococcus, Strongyloids
sterocolis, and
Toxoplasma gondii in fecal matter;
(ii) Leptospira species, Mycoplasma hyopneumoniae, porcine reproductive and
respiratory
syndrome virus (PRRSV), pseudorabies, transmissible gastroenteritis virus
(TGE) / Porcine
Respiratory Coronavirus, and Toxoplasma Gondii by determining antibody titers;
(iii) Porcine Influenza;
(iv) the following bacterial pathogens as determined by bacterial culture:
Bordetella
bronchisceptica, Coagulase-positive staphylococci, Coagulase-negative
staphylococci,
Livestock-associated methicillin resistant Staphylococcus aureus (LA MRSA),
Microphyton and
Trichophyton spp.;
(v) Porcine cytomegalovirus; and
(vi) Brucella suis.
Item 104. The method of any one of or combination of items 102-103, further
comprising maintaining the genetically reprogrammed swine according to a
bioburden-reducing
procedure, said procedure comprising maintaining the genetically reprogrammed
swine in an
isolated closed herd, wherein all other animals in the isolated closed herd
are confirmed to be
free of said zoonotic pathogens, wherein the genetically reprogrammed swine is
isolated from
contact with any non-human animals and animal housing facilities outside of
the isolated closed
herd.
Item 105. The method of any one of or combination of items 102-104, further
comprising harvesting a biological product from said swine, wherein said
harvesting comprises
euthanizing the swine and aseptically removing the biological product from the
swine.
Item 106. The method of any one of or combination of items 102-105, further
comprising processing said biological product comprising sterilization after
harvesting using a
sterilization process that does not reduce cell viability to less than 50%
cell viability as
determined by a 3-(4,5-dimethylthiazol-2-y1)-2,5-diphenyltetrazolium bromide
(MTT)-reduction
assay.
144
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Item 107. The method of any one of or combination of items 102-106, further
comprising storing said biological product in a sterile container under
storage conditions that
preserve cell viability.
Item 108. A method of screening for off target edits or genome alterations in
the genetically
reprogrammed swine comprising a nuclear genome of any one of or combination of
items 1-49,
comprising:
performing whole genome sequencing on a biological sample containing DNA from
a
donor swine before performing genetic reprogramming of the donor swine nuclear
genome,
thereby obtaining a first whole genome sequence;
after reprogramming of the donor swine nuclear genome, performing whole genome
sequencing
to obtain a second whole genome sequence;
aligning the first whole genome sequence and the second whole genome sequence
to obtain a
sequence alignment;
analyzing the sequence alignment to identify any mismatches to the swine's
genome at off-target
sites.
Item 109. A synthetic nucleotide sequence having wild-type swine intron
regions from a
wild-type swine MHC Class Ia, and reprogrammed at exon regions encoding the
wild-type
swine's SLA-3 with codons of HLA-C from a human capture reference sequence
that encode
amino acids that are not conserved between the SLA-3 and the HLA-C from the
human capture
reference sequence.
Item 110. The synthetic nucleotide sequence of item 109, wherein the wild-type
swine's
SLA-1 and SLA-2 each comprise a stop codon.
Item 111. A synthetic nucleotide sequence having wild-type swine intron
regions from a
wild-type swine MHC Class lb, and reprogrammed at exon regions encoding the
wild-type
swine's SLA-6, SLA-7, and SLA-8 with codons of HLA-E, HLA-F, and HLA-G,
respectively,
from a human capture reference sequence that encode amino acids that are not
conserved
between the SLA-6, SLA-7, and SLA-8 and the HLA-E, HLA-F, and HLA-G,
respectively, from
the human capture reference sequence.
Item 112. A synthetic nucleotide sequence having the synthetic nucleotide
sequences of
both items 109 and 111 or both items 110 and 111.
145
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Item 113. A synthetic nucleotide sequence having wild-type swine intron
regions from a
wild-type swine MHC Class II, and reprogrammed at exon regions encoding the
wild-type
swine's SLA-DQ with codons of HLA-DQ, respectively, from a human capture
reference
sequence that encode amino acids that are not conserved between the SLA-DQ and
the HLA-
DQ, respectively, from the human capture reference sequence, and wherein the
wild-type
swine's SLA-DR comprises a stop codon.
Item 114. A synthetic nucleotide sequence having the synthetic nucleotide
sequences of:
both items 109 and 113; both items 110 and 113; or both items 112 and 113.
Item 115. A synthetic nucleotide sequence having wild-type swine intron
regions from a
wild-type swine beta-2-microglobulin and reprogrammed at exon regions encoding
the wild-type
swine's beta-2-microglobulin with codons of beta-2-microglobulin from a human
capture
reference sequence that encode amino acids that are not conserved between the
wild-type
swine's beta-2-microglobulin and the beta-2-microglobulin from the human
capture reference
sequence, wherein the synthetic nucleotide sequence comprises at least one
stop codon in an
exon region such that the synthetic nucleotide sequence lacks functional
expression of the wild-
type swine's 02-microglobulin polypeptides.
Item 116. A synthetic nucleotide sequence having wild-type swine intron
regions from a
wild-type swine MIC-2, and reprogrammed at exon regions of the wild-type
swine's MIC-2 with
codons of MIC-A or MIC-B from a human capture reference sequence that encode
amino acids
that are not conserved between the MIC-2 and the MIC-A or the MIC-B from the
human capture
reference sequence.
Item 117. A synthetic nucleotide sequence having wild-type swine intron
regions from a
wild-type swine CTLA-4, and reprogrammed at exon regions encoding the wild-
type swine's
CTLA-4 with codons of CTLA-4 from a human capture reference sequence that
encode amino
acids that are not conserved between the wild-type swine's CTLA-4 and the CTLA-
4 from the
human capture reference sequence.
Item 118. A synthetic nucleotide sequence having wild-type swine intron
regions from a
wild-type swine PD-Li and reprogrammed at exon regions encoding the wild-type
swine's PD-
Li with codons of PD-Li from a human capture reference sequence that encode
amino acids that
146
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
are not conserved between the wild-type swine's PD-Li and the PD-Li from the
human capture
reference sequence.
Item 119. A synthetic nucleotide sequence having wild-type swine intron
regions from a
wild-type swine EPCR and reprogrammed at exon regions encoding the wild-type
swine's EPCR
with codons of EPCR from a human capture reference sequence that encode amino
acids that are
not conserved between the wild-type swine's EPCR and the EPCR from the human
capture
reference sequence.
Item 120. A synthetic nucleotide sequence having wild-type swine intron
regions from a
wild-type swine TBM and reprogrammed at exon regions encoding the wild-type
swine's TBM
with codons of TBM from a human capture reference sequence that encode amino
acids that are
not conserved between the wild-type swine's TBM and the TBM from the human
capture
reference sequence.
Item 121. A synthetic nucleotide sequence having wild-type swine intron
regions from a
wild-type swine TFPI and reprogrammed at exon regions encoding the wild-type
swine's TFPI
with codons of TFPI from a human capture reference sequence that encode amino
acids that are
not conserved between the wild-type swine's TFPI and the TFPI from the human
capture
reference sequence.
[000431] The present invention is described in further detail in the
following examples which
are provided to be illustrative only, and are not intended to limit the scope
of the invention.
EXAMPLE 1
DPF CLOSED COLONY SKIN GRAFT (MONKEY STUDIES)
[000432] It has been discovered that skin grafts derived from a DPF Closed
Colony, a-1,3-
galactosyltransferase [Gal-T] knockout pigs produced in accordance with the
present invention
exhibit significantly longer rejection times than skin grafts derived from a-
1,3 -
galactosyltransferase [Gal-T] knockout pigs but that were not derived from DPF
Closed Colony
pigs.
[000433] Numerous prior studies evaluating rejection time of a-1,3-
galactosyltransferase
[Gal-T] knockout pigs (not derived from a DPF Closed Colony) on monkeys show
rejection times
in the range of 11-13 days. See, e.g., Albritton et al., Lack of Cross-
Sensitization Between alpha-
1, 3-Galactosyltransferase Knockout Porcine and Allogeneic Skin Grafts Permits
Serial Grafting,
Transplantation & Volume 97, Number 12, June 27, 2014, (Gal-T-KO skin grafts
on recipient
147
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
baboons fully rejected by 12 or 13 days); Barone et al., "Genetically modified
porcine split-
thickness skin grafts as an alternative to allograft for provision of
temporary wound coverage:
preliminary characterization," Burns 41 (2015) 565-574 (Gal-T-KO skin grafts
on recipient
baboons fully rejected by 11 days); and Weiner et al., Prolonged survival of
Gal-T-KO swine skin
on baboons, Xenotransplantation, 2010, 17(2): 147-152 (Gal-T-KO xenogeneic
split-thickness
skin grafts on baboons fully rejected by 11 days).
[000434] The subject invention has been shown in nonclinical studies to
perform on par and
surprisingly better than its allograft comparators, without the inherent
disadvantage of inconsistent
quality and unreliable and limited availability. That is, surprisingly, at
least Study No. 1 shows
skin grafts derived from a DPF Closed Colony, a-1,3-galactosyltransferase [Gal-
T] knockout pigs
produced in accordance with the present invention performed better than
allograft.
[000435] Two recent studies (Study No. 1 and Study No. 2 set out below) by
applicant
demonstrate that skin grafts derived from DPF Closed Colony, a-1,3-
galactosyltransferase [Gal-
l] knockout pigs produced in accordance with the present invention on monkeys
show
significantly higher rejection times, in Study 2 longer than 30 days. The
genetically engineered
source animals in this example did not contain any foreign, introduced DNA
into the genome; the
gene modification employed was exclusively a knock-out of a single gene that
was responsible for
encoding for an enzyme that causes ubiquitous expression of a cell-surface
antigen. The
xenotransplantation product in this example does not incorporate transgene
technologies, such as
CD-46 or CD-55 transgenic constructs.
Study No. 1
[000436] This study evaluated DPF Closed Colony, a-1,3-
galactosyltransferase [Gal-T]
knockout porcine xenotransplantation product material compared to allografts
as temporary wound
grafts prior to autograft placement in cynomolgus monkeys (Macaca fascicularis
) in an
experimental model of full thickness skin lesions.
[000437] Primary end points included screening for porcine endogenous
retrovirus (PERV)
in the grafts and the recipient as well as evaluation of the
xenotransplantation product and allograft
rejection and their potential effects on ultimate autograft take. Secondary
end points included
microbiologic and histopathologic analysis of kidney, spleen, liver, lung,
grafts, and wound bed
tissues collected at necropsy.
148
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000438] Four (4) cynomolgus monkeys were enrolled in this study. Four (4),
full thickness
wound beds measuring approximately 2-3 cm x 2-3 cm were created on the dorsal
region of each
animal on Day 0.
[000439] Initially, wounds were treated with either Xenogeneic skin
(xenotransplantation
product), a split-thickness Gal-T-transgenic porcine xenotransplantation
product material, or
Allogenic skin (allograft), a split-thickness allograft material, on Day 0.
[000440] On Day 15 of the study, the xenotransplantation product and
allografts were
removed and replaced with split-thickness autologous skin grafts (autografts),
after which the
animals were survived to Day 22 of study (with the exception of moribund
sacrifice Animals 1001
and 1004).
[000441] Microscopic evaluation of full thickness wound beds in a
cynomolgus monkey
model treated with xenotransplantation product or allograft and removed on Day
12 or 15 (FIG.
40A) and survived up to Day 22 (FIG. 40B) demonstrated no evidence of acute
tissue rejection
with either the xenotransplantation product or allograft comparable to
slightly better performance
overall with the xenotransplantation product test article when compared to the
allograft test article,
and average to good autograft performance following pretreatment with either
xenotransplantation
product or allograft test articles. The significant survival times of the
xenotransplantation product
prompted a follow-on study (Study No. 2).
Study No. 2
[000442] The objective of this study was to evaluate the safety and
immunogenicity of DPF
Closed Colony, a-1,3-gal actosyltransferase [Gal-T] knockout porcine
xenotransplantation product
material in cynomolgus monkeys (macaca fascicularis).
[000443] Primary end points included screening for porcine endogenous
retrovirus (PERV)
pre- and post-graft placement and evaluation of the xenotransplantation
product rejection.
[000444] Four (4) cynomolgus monkeys were enrolled in this study. Two (2) 9
cm2 full
thickness wound beds were created on the dorsal region of each animal created
on Day 0.
[000445] Wounds were treated with split-thickness Gal-T-knockout porcine
xenograft
material consisting of dermal and epidermal tissue layers.
[000446] FIG. 41 shows the longitudinal progression of porcine split-
thickness skin graft
used as a temporary wound closure in treatment of full-thickness wound defects
in a non-human
primate recipient. Left: POD-0, xenograft at Wound Site 2. Right: POD-30, same
xenograft at
149
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Wound Site 2. FIG. 42 shows POD-30 histological images for: Top, Center: H&E,
Low power
image of wound site depicts complete epithelial coverage. Dotted line
surrounds the residual
xenograft tissue. Bottom, Left: H&E, Higher power image of the large inset
box. To the right and
below the dotted line is the dermal component of the xenograft, with the
xenograft dermal matrix
indicated by an open arrow. To the left of the dotted line is the host dermis
(black arrow) and the
host dermal matrix. Mild inflammation is present and interpreted to be in
response to the xenograft
test article. Bottom, Right: H&E, higher power image of the small inset box.
The dotted line
roughly demonstrates the junction between the xenograft test article (below
dotted line) and new
collagen tissue (above dotted line), with intact epithelium at the top of the
image. Mild
inflammation in response to the xenograft (open arrows) is observed.
[000447] FIG. 43A graphs the total serum IgM ELISA ( g/mL) for all four
subjects (2001,
2002, 2101, 2102) during the course of the study. FIG. 43B graphs the total
serum IgG ELISA
( g/mL) for all four subjects (2001, 2002, 2101, 2102) during the course of
the study. In some
aspects, subjects transplanted with the product of the present disclosure will
have serum IgM and
IgG levels of less than 20,000 [tg/m1 each. In some aspects, subjects
transplanted with the product
of the present disclosure will have serum IgM and/or IgG levels below or less
than 10%, 5%, 3%,
or 1% higher than serum IgM and IgG levels measured prior to transplantation.
In some aspects,
the claimed method may demonstrate an immunoreactivity incidence rate of less
than 5%, 3%, or
1% of subjects transplanted with the product of the present disclosure.
[000448] FIG. 44A graphs systemic concentrations of soluble CD4OL as
measured by
Luminex 23-plex at POD-0, POD-7, POD-14, POD-21, and POD-30. FIG. 44B graphs
systemic
concentrations of TGF-alpha as measured by Luminex 23-plex at POD-0, POD-7,
POD-14, POD-
21, and POD-30. FIG. 44C graphs systemic concentrations of IL-12/23 (p40) as
measured by
Luminex 23-plex at POD-0, POD-7, POD-14, POD-21, and POD-30.
[000449] Animals were terminated at 30 or 31 Days, wound sites were
collected and fixed in
10% neutral buffered formalin (NBF) or Modified Davidson's Solution for the
testis and
epididymis. It should be noted that while the animals were terminated at 30 or
31 days due to the
study design and for comparison purposes, the xenotransplantation product of
the present
disclosure is capable of resisting rejection for longer than the study period
used in this example.
150
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000450]
Microscopic evaluation of full thickness wound beds in a cynomolgus monkey
model treated with xenograft and terminated on Day 30 or 31 demonstrated good
filling of the
wound defect with host and xenograft tissue.
[000451]
Screening for porcine endogenous retroviruses (PERV) and porcine
cytomegalovirus (PCMV) was performed separately at specified post-operative
intervals via
specialized (porcine specific) polymerase chain reaction (PCR) and reverse
transcriptase PCR
(RT-PCR) testing of samples. The porcine xenografts, lysed PBMCS of the
recipient, recipient
wound bed, and highly perfused organs from the recipients at necropsy were
evaluated for presence
of porcine cell migration. All tests were performed in triplicate with
internal controls for DNA and
RNA, as well as assay performance. Microbiologic (bacterial, fungal, viral)
assays and
histopathologic analysis of kidney, spleen, liver, lung, xenografts,
allografts, wound bed tissues
collected at necropsy, and analysis of peripheral blood were performed to test
for xenograft-related
immunogenic biomarkers. DNA PCR was performed to test for porcine cell
migration in PBMCs
from the cynomolgus monkey model treated with the product of the present
disclosure for the
following samples: (A.)
(3) full-thickness (xenograft) wound beds, (B) (3) full-thickness
(allograft) wound beds; (C) (2) spleen samples; and (D)(2) kidney samples.
There was no evidence
of cell migration or zoonotic transmission systemically to the host. The
presence of PERV is
attributed to the residual pig cells in the wound bed, as verified with
porcine MHC controls. Our
results suggest that porcine DNA and cells did not migrate into the
circulation of the graft
recipients from the grafts, and likewise PERV or PERV-infected porcine cells
did not migrate past
the wound bed.
[000452]
The following Table 4 shows the analysis for porcine cell migration and
transmission:
TABLE 4
PSK17-01 Sample Analysis PCMV PERV MHC CCR5
(swine) (Control)
Item No. PBMC @ End-of-Study
Subject # (EoS Date)
1 NHP-1001 (POD-I S)
151
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
2 NHP-1002 (POD-22) Neg (-) Neg (-) Neg (-) Pos (+)
3 NHP-1003 (POD-22) Neg (-) Neg (-) Neg (-) Pos (+)
4 NHP-1004 (POD-12) Neg (-) Neg (-) Neg (-) Pos (+)
Wound Bed @ End-of-Study
Subject # (Test Article) (EoS
Date)
NHP-1001 (Xenograft) (POD- IS) * * * *
6 NHP-1001 (Allograft) (POD-I S) * * * *
7 NHP-1002 (Xenograft) (POD-22) Neg (-) Neg (-) Neg (-) Pos (+)
8 NHP-1002 (Allograft) (POD-22) Neg (-) Neg (-) Neg (-) Pos (+)
9 NHP-1003 (Xenograft) (POD -22) Neg ( -) Neg (-) Neg (-) Pos (+)
NHP-1003 (Allograft) (POD-22) Neg ( -) Neg (-) Neg (-) Pos (+)
11 NHP-1004 (Xenograft) (POD-12) Neg (-) Pos (+)(A) Neg (-) Pos ( + )
12 NHP-1004 (Allograft) (POD-12) Neg (-) Neg (-) Neg (-) Pos (+)
Spleen @ End-of-Study
13 NHP-1001 Neg (-) Neg (-) Neg (-) Pos (+)
14 NHP-1004 Neg (-) Neg (-) Neg (-) Pos (+)
Kidney @ End-of-Study
NHP-1001 Neg (-) Neg (-) Neg (-) Pos (+)
16 NHP-1004 Neg (-) Neg (-) Neg (-) Pos (+)
Key for Table 4:
Neg (-) = Negative
Pos (+) = Positive
*= Test Not Performed or Sample Not Acceptable, due to unrelated, study design-
related
logistical or preservation issue
Pos (+)A The wound bed for NHP 1004 (PERV positive) underwent co-culture
studies to
ascertain whether the detected virus present at the interface between graft
and recipient
(host) could infect permissive human cells.
Co-culture of the xenograft and recipient wound bed cells with permissive
human cells for
PERV infection and replication did not demonstrate productive infection in the
target cells
(HEK293), after a 23-day culture.
152
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Table 5. Banff Grades and Pathologic Component Scores' of Skin Xenotransplants
at POD-30
Surgeon Banff 2 pa el e 3 .4 5
6 7 8
Animal Graft pc V C
cav
assessment Grade
100% re-
2001 1 III 3 3 3 1 0 1 0
epithelialized
100% re-
2001 2 III-IV 3 3 3 2 0 1 0
epithelialized
30% re-
2002 1 III-IV 3 3 3 3 0 0 0
epithelialized
2002 2 30%re-
III-IV 3 3 3 3 0 0 0
epithelialized
40% re-
2101 1 II 3 3 2 3 0 0 0
epithelialized
40% re-
2101 2 III-IV 3 3 3 2 0 0 0
epithelialized
20% re-
2102 1 III-IV 3 3 3 3 0 0 0
epithelialized
20% re-
2102 2 II-III 3 3 3 3 0 0 0
epithelialized
1. Pathologic Component Scores developed by Rosales, et al. 47
2. pc = perivascular cells ¨ number of cells surrounding dermal vessels
(venules, capillaries, and arterioles) in deep and superficial
dermis; scored on the most involved vessels; pc3 >50 cells/vessel
3. pa = perivascular dermal infiltrate area ¨percent area occupied by the most
involved dermal vessels at 40x magnification;
pa3 >75%
4. ei = epidermal infiltrate ¨ total number of mononuclear cells per four 20x
fields; ei3 = transepidermal infiltrate, ei2 >20 cells
5. e = epidermal injury and necrosis ¨ presence of keratinocyte apoptosis and
necrosis; e3 = sloughed, e2 = focal necrosis,
el = apoptosis
6. v = endarteritis ¨ mononuclear cells underneath arterial endothelium;
scored on the most involved artery; v0 = none
7. c = capillaritis ¨ maximum number of cells per capillary cross section;
scored on most involved capillaries; cl = 2-4/capillary,
c0 = 0-1/capillary
8. cav = chronic allograft vasculopathy ¨ intimal thickening with luminal
reduction; scored as percent luminal reduction;
cavO = none
[000453] The general appearance for all xenotransplants for the course of
the study was pink,
warm to the touch and adherent to the wound bed. Epidermolysis (mild to
moderate) was first
noted by the surgeon on POD-14 but the dermis was adherent. Assessment at POD-
21 revealed
that the wound bed was re-granulating and there were signs of re-
epithelialization, such that by
POD-30, 20% to 100% of the wound had been re-epithelialized. (Table 5) During
the clinical
course of these skin xenotransplants, there was no sloughing of the
xenotransplant tissue and
exposure of the wound bed.
[000454] Hematoxylin and eosin (H&E) - prepared sections containing
residual skin
xenotransplants and the underlying wound beds, obtained at POD-30, were
microscopically
evaluated by a blinded pathologist.H&E staining showed minimal to moderate
inflammatory
response. There was ulceration of the epithelia in four out of eight treated
sites. The response at
the wound sites was characterized by filling of the wound defect with a mature
dermal collagen
153
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
network surrounded by a variable layer of new collagen. This mature collagen
network was distinct
in appearance from the host dermis bordering the wound site, and was
interpreted to be the
xenotransplant dermis. The skin xenotransplants were assessed using a
systematic pathologic
component scoring and Banff classification 47.The Banff classification is
useful in categorizing
xenotransplant rejection, and it is complemented by the component score
approach, providing a
more comprehensive array of clinical thresholds for the diagnosis of
rejection.The results of this
assessment and Banff Grades for POD-30 are shown in Table 5. The Banff 2007
Working
Classification for Composite Tissue Allografts is based on the level of
epidermal apoptosis,
epidermal infiltrates, and perivascular/dermal infiltrates 48. The Banff
Grades ranged from II
(moderate) to IV (necrotizing acute rejection) with most showing Grade III
(severe).
Table 6. Changes in Serum Cytokines and Chemokines after Xenograft
Transplantation (pg/mL)
Cytokine/ POD-0 POD-7 POD-14 POD-21 POD-30
Chemokine
sCD40L 1900 7900T 77001: 86001: 85001:
1000 3100 3100 4000 5200
IL-lra 7.6 50T 28T 66T 24T
2.8 44 11 83 13
IL-2 29 42T 37T 41T 30
11 18 11 9 12
IL-6 0.31 7.3t 4.1t 8.5t 3.3t
0.6 8.3 2.6 6.3 2.7
IL-8 2500 4200T 3700T 3900T 2500
1300 3200 2600 2300 2100
IL-12/23 (p40) 0.6 1.8 26T 16T 6.7t
1.0 2.7 22 11 7.7
IL-15 3.1 6.0t 7.1t 5.0t 6.0t
1.9 2.0 1.3 1.3 1.5
MCP-1 360 710T 420T 460T 310
150 540 110 110 120
TGF-cc 4.5 22T 16T 5.2 9.9
4.6 11 11 3.6 8.9
POD = Postoperative day
154
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Values are means (n=4) SD
Significant datapoints (p<0.05) compared to POD 0, student t-test
Values include data at the upper level of detection (12,000 pg/mL)
[000455] As an evaluation of cell-mediated immune response, a total of 23
inflammatory and
anti-inflammatory cytokines characteristic of initial wound healing processes
or those anticipated
in an immunological response to xenogeneic cells were measured. Twelve of the
23
cytokines/chemokines assayed were consistently below the level of detection
throughout the entire
study period: TNF-a, IFN-y, TGF-(3, G-CSF, GM-CSF, IL-1- (3, IL-4, IL-5, IL-
10, IL-13, IL-17,
IL-18, and MIP-1-a. VEGF exceeded the level of detection at only three
individual timepoints,
and levels of MIP-1-beta were discernable only once (data not presented). Nine
cytokines/chemokines detected over the period of the study are listed in Table
6. All
cytokines/chemokines shown in the table were observed to increase above
background at POD-7,
the first day of sampling. IL-2, IL-8, MCP-1 and TGF-a peaked at POD-7 and
decreased over time.
IL-15 and IL-12/23 (p40) peaked at POD-14, while sCD40L, IL-lra and IL-6 had
an elevated peak
at POD-21. In general, all of the factors showed a return to normal by POD-30
with the exception
of sCD40L, which remained elevated at POD-30. Of interest, levels of IL-12/23
(p40) were nearly
absent until conspicuously elevated on POD-14, gradually reducing in
concentration over the
remainder of the study.
Table 7. Post-Transplant Changes in Binding of Recipient Serum IgM and IgG to
PBMC1 Targets from GalT-K02
Swine Donors
Pre/Post IgNI5 IgG6
Recipient
Transplant3
rMFI4 Fold Change rMFI4 Fold Change
Pre 8.51 0.0 16.28 0.0
2001
Post 45.72 4.4 1089.85 65.9
Pre 5.07 0.0 28.29 0.0
2002
Post 30.01 4.9 840.64 28.7
Pre 7.92 0.0 16.03 0.0
2101
Post 22.48 1.8 730.83 44.6
Pre 6.47 0.0 5.19 0.0
2102
Post 15.49 1.4 372 . 88 70.8
1. PBMC= peripheral blood mononuclear cell
2. Ga1T-K0= alpha-1,3 galactosyltransferase knockout
3. Pre-transplant=POD-0; Post-transplant=POD-30
4. rMFI = relative Mean Fluorescent Intensity
5. IgM = immunoglobulin M
6. IgG = immunoglobulin G
155
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000456] To assess the production of antibody to xenogeneic after skin
transplant, binding of
recipient serum IgM and IgG to peripheral blood mononuclear cell (PBMC)
targets from GalT-
KO donors was measured by flow cytometry. Serum IgM and IgG antibody levels
were analyzed
at pre-transplant and at POD-30. In Table 7, the relative mean fluorescent
intensity (MFI) and fold
increase in binding are summarized for each recipient. An increase in anti-
xenogeneic IgM and
IgG was detected in all animals. Between pre-transplant (POD-0) and post-
transplant (POD-30),
IgM anti-porcine antibodies increased between 1.4 to 4.9 fold and IgG anti-
porcine antibodies
increased between 28.7 to 70.8 fold. These results demonstrate a humoral
response to non-Gal
xenoantigens.
Table 8. Data for Postoperative Analysis of Wound Beds (Wound Site 1 and 2)
PERV
Animal ID Wound Site copies/500ng Micro- QC
(SD) chimerismt
W1 <LOW'
2001
W2 1495.6 ( 521)
W1 1518.8( 21)
2002
W2 <LOD
W1 527.1 ( 134)
2101
W2 137.8 ( 16)
W1 <LOD
2102
W2 <LOD
SD = Standard Deviation, LOD = Limit of Detection, QC= Quality Control
Porcine microchimerism cannot be accurately quantified due to mixture of cells
present in wound bed extraction
All QC gave a positive Ct , indicating no inhibition
[000457] Naïve skin xenotransplants were analyzed for PERV copy number and
as expected,
each cell contained copies of PERV A (32 1), B (9 0.1) and C (16 0.1). Sera
from the four
recipients were evaluated for the presence of circulating PERV; all samples
were found to be
negative for PERV poi and below the limit of detection. PBMC samples from each
of the four
recipients were also tested for PERV and for microchimerism (i.e., the
presence of circulating pig
cells) and were also found negative, at all time points. Tissues taken at the
end of the study (POD-
156
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
30) were evaluated for PERV expression and again were found negative. Wound
beds from animal
2102 were negative for the presence of PERV and for microchimerism. (Table 8)
For the other
animals, either one wound site or both were positive. This is not surprising
due to the direct contact
of the wound bed with the xenograft. It is expected that some porcine cells
not associated with the
graft may have sloughed off or been left behind in the process of removal at
the end of the study.
This is confirmed by the positive values achieved for the microchimerism assay
attributing the
PERV signal to porcine cell contamination. Altogether, these results provided
no evidence of
PERV transmission, consistent with previous studies.
EXAMPLE 2
[000458] The following example provides a description of a process of
harvesting and
processing skin from a genetically reprogrammed swine produced in accordance
with the present
invention, with the skin to be used as a xenogeneic skin product for human
transplantation. In
some of these aspects, the xenotransplantation product consists of split
thickness grafts consisting
of dermal and epidermal tissue layers containing vital, non-terminally
sterilized porcine cells
derived from specialized, genetically reprogrammed, Designated Pathogen Free
(DPF), source
animals.
[000459] In one aspect, the genetically reprogrammed source animal is any
genetically
reprogrammed animal described in the present disclosure. In one non-limiting
aspect, the
genetically engineered source animals in this example do not contain any
foreign, introduced DNA
into the genome; the gene modification includes a knock-out of a single gene
that was responsible
for encoding for an enzyme that causes ubiquitous expression of a cell-surface
antigen. The
xenotransplantation product in this example does not incorporate transgene
technologies, such as
CD-46 or CD-55 transgenic constructs.
[000460] The process and techniques disclosed herein are but examples, and
do not limit the
scope of the invention. It will be fully understood that while this example is
directed to
xenotransplantation skin products, several of the steps in the following
process and aspects of the
overall approach can be applied to other organs or tissues, including, but not
limited to, kidney,
lung, liver, pancreas, nerve, heart, intestine, and other organs or tissue. It
will be further understood
that modifications to the processes and methods disclosed in this example
(including additions or
omissions of one or more process or method steps) can be made in relation to
the harvesting and
processing of other organs or tissue besides skin. This understanding is based
in part on the fact
157
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
that other organs and tissue will have different physical characteristics and
so harvesting and
processing steps for such other organs or tissue will be different from this
example in certain
practical ways (e.g., a kidney, heart, liver, lung, or other whole organ will
not be cut to size and
packaged in a cryovial supported by nylon mesh). Nonetheless, it will be
further understood that
additions or omissions of one or more process or method steps as applied to
each such organ or
tissue may be made to this example utilizing approaches known in the art
(e.g., a harvested kidney,
heart, liver, lung, or other whole organ will, in some aspects, be placed in
an antipathogen bath or
exposed to UV light as described herein for the removal of pathogens following
harvest, and placed
in one or more closure systems. For example, such one or more closure systems
could include, but
not be limited to, a first closure system (e.g., utilizing an inert material
for initial closure to
surround the organ to prevent the organ from coming into contact with or
adhering to other
materials proximate to the organ) and/or a second closure system (e.g., a
sterile and secure outer
container that contains the organ and first closure system (if a first closure
system is utilized)).
Such organs within such closure system(s) are configured to be transported to
a clinical site as
whole organs, stored, protected and transported in temperatures, sterility,
and other conditions to
maintain sterility and cell viability for transplantation as described herein
at the clinical site.
Animal Preparation
[000461] Skin product processing occurs in a single, continuous, and self-
contained,
segregated manufacturing event that begins with the sacrifice of the source
animal through
completion of the production of the final product.
[000462] Xenogeneic skin grafts derived from the genetically reprogrammed
source animal
is received, with the swine being recently euthanized via captive bolt
euthanasia in another section
of the DPF Isolation Area. The source animal is contained in a sterile, non-
porous bag that is
contained within a plastic container which is delivered into the DPF Isolation
Area and placed in
an operating room where the procedure to harvest skin from the source animal
will occur. All
members of the operating team should be in full sterile surgical gear dressed
in sterile dress to
maintain designated pathogen free conditions prior to receiving the source
animal and in some
instanced be double-gloved to minimize contamination.
[000463] The operating area is prepared with materials required for
harvesting skin from the
source animal prior to decontamination (e.g., 24 hours prior with chlorine
dioxide gas treatment)
and prior to the procedure. Dermatome (electronic skin harvesting device,
e.g., Amalgatome by
158
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Exsurco) power supply, and extension cord are sterilized and placed in the
operating area prior to
the operation. Any materials not in the room during the chlorine dioxide gas
treatment (and
therefore non-sterile) will be sprayed with 70% ethanol or isopropanol prior
to entering the room.
[000464] The source animal is removed from the bag and container in an
aseptic fashion, for
example, a human lifting the source animal from the bag and container using
sterilized gloves
and/or sterilized device to aid lifting and minimize contamination. The source
animal is scrubbed
by operating staff for at least 2 minutes with Chlorhexidine brushes over the
entire area of the
animal where the operation will occur, periodically pouring Chlorhexidine over
the area to ensure
coverage.
[000465] The source animal is placed on its right lateral flank and dorsum
towards the
operating table leaving the left lateral flank and dorsum exposed. The exposed
surface is scrubbed
to the extreme visible surgical borders, and constrained by sterile drapes
secured with towel
clamps. The source animal is then scrubbed with opened Betadine brushes and
sterile water rinse
over the entire area of the animal where the operation will occur for
approximately 2 minutes.
[000466] This Chlorhexidine and Betadine mixture will sit on the source
animal for
approximately 2 minutes, and staff (dressed in sterile dress to maintain
designated pathogen free
conditions) will then rinse and dry the source animal with sterile water and
sterile gauze. The
source animal's hair is removed so as to not impact the membrane or introduce
another element
that would degrade the cells. Hair removal is done using sterilized clippers
and/or straight razor in
the designated pathogen free environment immediately post-mortem with a clean
blade utilizing a
chlorhexidine lather. Staff will use the clippers and/or straight razor
(lubricated in a sterile bath)
to remove any remaining hair on the operating site, taking care to not
puncture the skin. This
procedure will be repeated (scrubbing to shaving) by turning the source animal
onto the left lateral
flank so as to expose the right side. The source animal will be rinsed with
sterile water and dried
with sterile towels and sprayed with 70% ethanol. The source animal will be
inspected visually by
the surgeon to ensure proper coverage of scrubbing. After the sterile scrub
and final shaving, the
source animal is ready for skin harvest.
Skin Harvesting
[000467] Operators will be dressed in sterile dress in accordance with
program and other
standards to maintain designated pathogen free conditions. All tissue from the
source animal that
will be used for xenotransplantation is harvested within 15 hours of the
animal being sacrificed.
159
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000468] In one aspect, the source animal is laid on its side on an
operating table. In this
aspect, harvesting is done utilizing a dermatome circular blade, (for example
and Amalgatomeg
SD). As the staff secures the animal in place, the surgeon determines the most
appropriate width
(e.g., 1, 2, 3, or 4 inches) and uses the circular dermatome to remove strips
of split thickness skin
grafts at a chosen thickness (e.g., 0.50 mm, 0.55 mm, 0.62 mm).
[000469] By way of further example, the thickness of the skin grafts could
range from 0.01
mm to 4 mm, depending on the therapeutic needs at issue. It will also be
understood that in some
aspects a full thickness graft may also be utilized harvested with alternative
harvesting and grafting
procedures known in the art. Graft sizes can range from 1 cm2 to 1000 cm2 (or
approximately 1
ft2). It will be understood that larger graft sizes are also possible
depending on the application and
harvesting technique utilized and size of the source animal. It will be
understood that for all
aspects, other depths could be utilized as well, depending on the application
and needs of the task
at hand for therapeutic and/or other purposes.
[000470] In another aspect, skin harvesting involves surgically removing a
skin flap from the
animal first, then the skin flap is placed dermis-side down onto a harvest
board (e.g., a solid board
made of metal, plastic or other appropriate material) set upon on the
operating table. In this aspect,
sterile padding material is added beneath the skin flap and on top of the
harvest board, to allow
appropriate give for proper dermatome device function. The skin flap is then
affixed to the harvest
board firmly with steel clamps. Curved towel clamps are utilized on the side
of the skin flap
opposite the clamps until the skin is firm and taut. The surgeon will choose
the most appropriate
thickness on the dermatome and adjust per harvest conditions. The surgeon will
use the dermatome
on the secured skin flap, with an assistant maintaining tension along the
dermatome progress. A
second assistant may also provide assistance with skin flap tension, and may
use rat tooth forceps
to pull the graft product emerging from the dermatome.
[000471] Grafts are trimmed to desired sizes. By way of example, sizes can
be: 5 cm x 5 cm,
with a total surface area of 25 cm2 and uniform thickness of approximately
0.55 mm; 5 cm x 15
cm, with a total surface area of 75 cm2 and uniform thickness of approximately
0.55 mm; 8 cm x
7.5 cm, with a total surface area of 60 cm2 and uniform thickness of
approximately 0.55 mm; 8 cm
x 15 cm with a total surface area of 120 cm2 and uniform thickness of
approximately 0.55 mm. It
will be further understood that customizable sizes (i.e., width, thickness and
length) can be created
160
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
depending on patient needs, including larger sheets of skin can be harvested
for use in
xenotransplantation procedures.
[000472] The xenotransplantation product is further processed to be free of
aerobic and
anaerobic bacteria, fungus, and mycoplasma. Under sterile conditions in a
laminar flow hood in a
drug product processing suite using applicable aseptic techniques, immediately
after, within 1, 2,
3, 4, 5, 6, 7, 8, 9, 10 seconds, within 10 seconds to 1 minute, within 1
minute to 1 hour, within 1
hour to 15 hours, or within 15 hours to 24 hours following harvest, the
xenotransplantation product
is placed into an anti-microbial/anti-fungal bath ("antipathogen bath"). With
regard to a skin
product, this can occur after the skin product is trimmed to the proper dose
size and shape (e.g.,
trimmed to squares, rectangles, or others shapes of desired size(s))
[000473] The antipathogen bath includes ampicillin, ceftazidime,
vancomyocin,
amphotericin-B placed in a sterile container and the xenotransplantation
products are diluted as
outlined in the following Table 5 and added to RPMI-1640 medium as outlined in
the following
Table 6. In one aspect, about 10 mL of medium is removed from the bottle
before adding the above
items.
TABLE 5
Drug Vial Diluent Vol Diluent Approx. Vol Approx.
Mg available concentration
Ceftazidime 1000 10.0 mL Sterile water 10.8 mL 100 mg/mL
Ampicillin 2000 10.0 mL Sterile water 11 mL 180 mg/mL
Vancomycin 500 10.0 mL Sterile water 50 ug/ mL
Amphotericin B 50 5.0 mL Sterile water 10 mg/mL
TABLE 6
Drug Final Concentrations mg/500 mL Media Volume (mL) added
to
500 mL RPM! 1640
Ceftazidime 500-2500 mg/L 250-1250 mg 2.5-10
Ampicillin 500-2500 mg/L 250-1250 mg 1-6
Vancomycin 25-125 mg/L 10-75 mg 0.25-2
161
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Amphotericin B 40-200 mg/L 20-100 mg 2-10
Total volume added 5.75-28
[000474] It will be understood that while this example is directed to
xenotransplantation skin
products, other organs, including, but not limited to, kidney, lung, heart,
liver, pancreas, and other
organs can be bathed in the antipathogen bath in accordance with the present
invention. The
amounts of combination of drugs and other chemicals, and duration of exposure
to such
antipathogen bath, are performed to minimize the affect such exposure has on
cell viability and
mitochondrial activity to achieve both the desired antipathogen result and
minimal manipulation
of the xenotransplantation products in accordance with the present invention.
[000475] As an alternative, or in addition to, removing pathogens via the
antipathogen bath,
the products are made designated pathogen free by a process and system
utilizing ultraviolet light.
In this aspect, the operator is dressed in sterile dress in accordance with
institutional standards to
maintain designated pathogen free conditions. The operator wears eye
protection safety glasses for
ultraviolet light and lasers.
[000476] An ultraviolet laser lamp is set up in a laminar flow hood. Each
of the four corners
of the lamp is placed on two container lids that are stacked on top of each
other, i.e., four pairs of
lids are used to support the lamp, or other supporting items, able to position
the lamp in a temporary
or fixed position above the working surface of the hood. The distance from the
lamp bulbs (2 bulb
tubes total) to the floor of the hood is approximately 1.5 inches. The entire
interior of the hood is
sprayed with alcohol, e.g., ethanol or isopropanol. The lamp is turned on and
the operator performs
a calculation of time for desired exposure based on lamp specifications,
number of bulbs, and
distance between the bulbs and the xenotransplantation product.
[000477] The operator pours two baths (one chlorhexidine and one alcohol)
into two separate
bowls and places the two bowls under the hood.
[000478] A package of new sterilized cryovials is placed under the hood.
Cryovial caps are
unscrewed and placed into the chlorhexidine bath. Each cryovial (without cap)
is then turned
upside down and plunged open ended into the chlorhexidine bath, for one minute
each and then
set upright to air dry. Thereafter, the exterior of each cryovial is wiped
with chlorhexidine and
alcohol utilizing sterile gauze. The cryovial caps are removed from the
chlorhexidine bath and
162
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
placed on sterile gauze. The open ends of each vial were plunged into alcohol
bath for 1 minute
each and then set aside to air dry.
[000479] Xenotransplantation products recently obtained from the
harvest/procurement
phase in the surgical room are transferred into the product processing room,
via a one-way entrance
into the laminar flow hood. Anything entering the sterile field is wiped down
with 70% ethanol
prior to transfer to the operator. The operator will have access to all
required materials in the
laminar flow hood: xenotransplantation product (in sterile container),
cryovials, 10mL syringes
and needles, phase freezer holding rack, and pre-cut nylon mesh. Only one size
of the products is
processed at a time to ensure proper control to final vials. The operator is
seated at the laminar
flow hood in compliance with sterile, aseptic techniques.
[000480] When using UV light sterilization, the product is placed under the
UV lamp for a
desired period of time, e.g., 2 minutes or more, then turned over to the other
side, and put under
the UV lamp for the same period of time, e.g., 2 minutes or more on opposite
side. The time period
for exposing a given sample to the UV is varied based on the specific
biological agents or the types
of biological agents to be sterilized, e.g., as shown in the following Table
7:
TABLE 7
Biological Agent Type of UV-C
Dosage (uW sec/cm2) Sterilization time
Biological Agent for 90% sterilization (sec)*
Penicillium spp. Fungus 224,000 1800
Aspergillus flavus Fungus 34,900 300
Aspergillus niger Fungus 31,500 250
Yeast Fungus 4000 30
Influenza A Virus 1900 15
HIV-1 Virus 28,000 220
Vaccinia Virus 1500 10
Escherichia coli Bacteria 2000 20
Staphylococcus Bacteria 6600 50
aureus
Bacillus subtilis Bacteria 6800 50
Mycoplasma spp. Bacteria 8400 70
Pseudomonas Bacteria 2200 20
aeruginosa
*Using a UV-C intensity of 125 uW/cm2
163
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000481] With regard to other whole organs, product yield will typically
depend on how
many of each such whole organ a given source animal may have (e.g., one liver,
two lungs, two
kidneys, one heart, one pancreas and so forth).
[000482] It will also be understood that while this example is directed to
xenotransplantation
skin products, other organs, including, but not limited to, kidney, heart,
lung, liver, pancreas, and
other organs can be exposed to ultraviolet light and made designated pathogen
free in accordance
with the present invention. The UV exposure dosages, intensity, and duration
of exposure to such
ultraviolet light, are performed to minimize the affect such exposure has on
cell viability and
mitochondrial activity to achieve both the desired antipathogen result and
minimal manipulation
of the xenotransplantation products in accordance with the present invention.
Manufacturing Process
Generally
[000483] Through the continuous manufacturing event, source animals are
processed into
aseptic xenotransplantation products. Several items are involved in the
manufacture of the product
relating to the source animals, including, but not limited to:
a) care and husbandry of the source animals (including, as described
herein, providing
certain vaccinations, carefully maintaining and analyzing pedigree records,
performing proper animal husbandry, and maintaining the animals in isolation
barrier conditions);
b) product manufacturing (including, as described herein, processing the
source
animals into the subject product from euthanizing to harvest);
c) analytical testing of the source animals (including, as described herein,
screening
for adventitious agents including parasitology, bacteriology, and virology
assays);
d) analytical testing of the source animals (including, as described herein,
confirming
the source animal is an alpha-1,3-galactotransferase knockout or has other
characteristics that are desired for a given application); and
e) analytical testing of the source animals (including, as described
herein, viral assay
for Endogenous Viruses (PERV)).
[000484] Several items are also involved in the manufacture and release
testing of the
resulting products, including, but not limited to:
164
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
a) product manufacturing (including, as described herein, processing the drug
product,
storing the drug product, and releasing the drug product);
b) analytical testing of the drug product (including, as described herein,
viability
testing (via, e.g., MTT assay)),
c) sterility testing (including, as described herein, aerobic bacteria
culture, anaerobic
bacteria culture, fungal culture, mycoplasma assay, endotoxin test, USP
<71>)),
d) adventitious agent testing (including, as described herein, PCR Assay for
e.g.,
Endogenous Viruses (PERV)); and
e) analytical testing of the drug product (including, as described herein,
histology).
[000485] For skin, the quantity of product yield from each animal can vary
depending on the
size of each animal. By way of example, some animals could yield between 3,000
and 6,000 cm2
in product. In one aspect, a single batch of skin product is harvested from a
single source animal
in a continuous process. A batch description of the xenotransplantation
product is provided in
Table 8 and batch formula for the xenotransplantation product is provided in
Table 9.
TABLE 8
Batch Size
Product (strength) Lot Size
Xenotransplantation product Drug Product, 200 Units (180-220)
Dosage Strength 1(7.5 grams, 25 cm2) (1.5 kgs per lot) (1.35 kg to 1.65kg)
Xenotransplantation product Drug Product, 67 units (60-75)
Dosage Strength 2 (22.5 grams, 75 cm2) (1.5 kgs per lot) (1.35 kg to
1.65kg)
TABLE 9
Batch Formula
Component Nominal Amount per Vial Nominal Amount per Lot
Xenotransplantation
25cm2 200 Units
product Drug Substance
Dosage Strength 1
CryoStor 7m1 1.4L
Nylon Mesh 60 cm2 1200 cm2
Total Batch Size 7.5 grams 1.5 kgs
Component Nominal Amount per Vial Nominal Amount per Lot
165
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Xenotransplantation 75cm2 67 Units
product Drug Substance
Dosage Strength 1
CryoStor 5m1 350 ml
Nylon Mesh 180 cm2 3600 cm2
Total Batch Size 22.5 grams 1.5 kgs
[000486] Prior lot testing is performed under good laboratory practice
("GLP") conditions to
ensure process sterility is maintained consistently. Assurance of sterility of
the final product is
determined prior to material release and clinical use. Prior to validation for
human clinical use, all
xenotransplantation products will meet certain acceptance criteria, including
as described herein.
The final drug product control strategy and analytical testing is conducted at
the conclusion of the
manufacturing process prior to release for clinical use. Results of the
required analytical tests will
be documented via a drug product certificate of analysis (COA) that is
maintained with a master
batch record pertaining to each lot of xenotransplantation products.
[000487] Source animal sample archives are generated and maintained through
procurement
of tissue samples of lung, liver, spleen, spinal cord, brain, kidney, and
skin. These tissues are
collected for source animal tissues for testing, archive, and stored for
potential future testing.
Archived samples of source animal tissue and bodily fluids should be stored at
minus (-) 70 degrees
Celsius or lower, as appropriate for preserving the sample. In other aspects,
fixed samples can be
maintained at room temperature. Appropriate tissue samples should be collected
for formalin
fixation and paraffin-embedding and for cryopreservation from source animals
at the time the live
cells, tissues, or organs are procured. Cryopreservation should be at least
ten 0.5 cc aliquots of
citrated-or EDTA-anticoagulated plasma; five aliquots of viable leukocytes
(1x107/aliquot, for
subsequent isolation of nucleic acids and proteins or for use as a source of
viable cells for co-
culture or other tissue culture assays.
Product Processing Following Harvesting
[000488] The previously harvested and minimally manipulated
xenotransplantation skin
product (here the skin integrity being minimally manipulated dermal and
epidermal tissue layers
with standard cellular morphology and organization) enters the separate,
adjacent room with
positive pressure above that of the surgical suite, designated as the Class
10,000 (ISO-7) product
processing room.
166
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000489] The operating room will be setup per operating preparation
procedures and the
operating personnel will be dressed in Tyvex suits for fume hood work. If
requested, an assistant
will also be dressed in a Tyvex suit. Gowning and Dressing is done with
aseptic techniques. Gloves
and sleeves will be sprayed with alcohol if needed. The ABSL-2 laminar flow
hood, having been
prior sterilized via gaseous chlorine dioxide sterilization process, will be
sprayed with alcohol, e.g,
70% ethanol, and the laminar flow exhaust will be initiated. Utilizing aseptic
techniques,
previously sterilized via autoclave, surgical instrument, cryovials, cryotray,
flasks, syringes,
needles, additional containers, and all processing equipment will be placed
within the laminar flow
hood. Exterior packaging is sprayed with alcohol prior to being transferred to
the operator.
[000490] As described herein, prior to operation, nylon mesh graft backing
should be cut into
squares of appropriate size for the dosage levels, sealed in an autoclavable
pouch, and sterilized
via steam. Exterior of pouch will then be sterilized with 70% ethanol and
placed in the fume hood.
Exterior package of 10mL Cryovials will be decontaminated with 70% ethanol and
placed into the
fume hood. Sterile, autoclaved surgical instrument package should be sprayed
with 70% ethanol
and transferred to the operator.
[000491] Sterile syringes and needles should be sprayed with 70% ethanol
and transferred to
the operator. Graft tissue recently harvest form the porcine donor will be
transferred to the hood.
Anything entering the sterile field is wiped down with 70% ethanol prior to
transfer to the operator.
Operator will have access to all required materials in the fume hood: Grafts
(in sterile container),
Cryovials, 10mL syringes and needles, Phase Freezer holding rack, and cut
Nylon mesh. Operator
should be seated at the fume hood with in compliance with sterile, aseptic
technique.
[000492] Referring to FIG. 45, each cryovial will be sterilized and labeled
in advance to
reduce processing time and unnecessary material exposure to DMSO prior to
cryopreservation.
Pans containing each xenotransplantation product and the RPMI 1640 Tissue
Culture Media at
room temperature with antibiotics (e.g., antipathogen bath) is placed under
the laminar flow hood.
The products had been bathing in the anti-pathogen bath for not less than 30
minutes to sterilize
the xenotransplantation product.
[000493] In one aspect, when using UV light sterilization, the cryovials
are sterilized using
the UV lamp as described above. After the product is inserted into each vial,
each new cap is placed
on each new vial and screwed on securely. Each vial is placed under the lamp
and periodically
rolled for desired even exposure to light on the exterior of the vial. The
vials are placed inside a
167
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
glass jar that has an interior that has been previously sterilized and the
exterior is sterilized by the
operator with alcohol and chlorhexidine, including threads and caps. Vials are
wiped down with
alcohol and are placed into glass jars. The exteriors of the glass jars are
drenched with alcohol
outside of the hood. Under the hood, the operator bathes the glass jar lids
and plunges the open
ends of the jars into alcohol and wipes the exterior of the jars with alcohol
(and optionally
chlorhexidine) including threads of the jar. The vials are wiped with alcohol
utilizing gauze and
placed inside each glass jar with an instrument. The lids of the glass jars
are then secured and the
jars are handed to the assistant. Frequently and on a periodic basis
throughout these processes, the
assistant sprays the operator's gloves and arms with alcohol.
[000494] In this example, the xenotransplantation skin product, which was
cut to form in the
surgical suite with sterile scissors and was trimmed with 10-blade scalpel,
will be re-measured
with a sterile, stainless steel ruler to verify technical specifications and
dimensions have been met.
The xenotransplantation skin product is visually inspected to ensure no rips,
tears, observable
defects, or excessive or insufficient thickness are present.
[000495] Under the laminar flow hood the operator will use forceps to take
a single
xenotransplantation skin product from the antipathogen bath and place it upon
a piece of nylon
mesh that has been previously cut to fit the cryovial, centered on the nylon
mesh, with the dermis
side in contact with the mesh (e.g., dermis side down), taking 1 minute for
each product
(understanding the time could be less or more, and up to 5 minutes for each
product). It will be
understood that the sterile nylon mesh packaging component is utilized, among
other things, to
support the xenotransplantation product and prevent self-adhesion of the
xenotransplantation
product when rolled.
[000496] It will be further understood that the sterile nylon mesh
packaging component can
be of any dimension that would allow the xenotransplantation product to be
placed onto the sterile
nylon mesh packaging component and fit within the two dimensional surface area
(i.e., the length
and width not including the thickness) of the sterile nylon mesh packaging
component (e.g., the
two dimensional area dimension of the xenotransplantation product would be
less than the two
dimensional area dimension of the sterile nylon mesh packaging component).
[000497] It will be further understood that the dimensions of the sterile
nylon mesh packaging
component would be sized in accordance with the xenotransplantation product
size and dosage.
For example, the sterile nylon mesh packaging component is 8 cm x 7.5 cm (60
cm2) to fit a 5 cm
168
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
x 5 cm xenotransplantation skin product (25 cm2) (7.5 grams) utilizing 7 ml of
cryoprotective
media when placed in the cryovial. It will be even further understood that the
dimensions of the
sterile nylon mesh packaging component is 8 cm x 22.5 cm (180 cm2) to fit a 5
cm x 15 cm
xenotransplantation skin product (75 cm2) (22.5 grams) utilizing 5 ml of
cryoprotective media
when placed in the cryovial.
[000498] Unintentional adhesion of epidermal or dermal regions of the
xenotransplantation
skin product during packaging may disrupt the integrity of the
xenotransplantation skin product
and potentially reduce its therapeutic viability. Inclusion of the sterile
nylon-mesh packaging
component is intended to provide internal physical support to and prevent self-
adhesion. The
sterile nylon-mesh packaging component is not biologically or chemically
active and does not
directly impact the metabolic activity or efficacy of the xenotransplantation
skin product itself.
[000499] During the course of numerous experiments, including the monkey
studies
described in Example 1 herein, use of this sterile nylon-mesh packaging
component has never been
observed to cause an adverse, undesired reaction with the xenotransplantation
product, or degrade
and contaminate the final xenotransplantation product causing adverse
reactions or outcomes to
the recipient. The sterile, nylon-mesh packaging component is not used in the
grafting procedure.
Following cryopreservation and thawing, and prior to use of the
xenotransplantation product, it is
discarded. Thus, selection of the specific material and associated
specifications were carefully
chosen for the given application. Medifab 100-Micron Nylon Mesh (Part # 03-
100/32-Medifab) is
manufactured per cGMP standards, and was selected because of its physical
characteristics and
certified acceptability for human, clinical use.
[000500] Under the laminar flow hood, the operator will then tightly roll
this combination of
xenotransplantation product and nylon mesh packaging component and place the
combination
within a cryovial (e.g., 10 ml vial) taking 1 minute for each product
(understanding the time could
be less or more, and up to 5 minutes for each product). In this aspect, the
mesh material is rolled
to ensure that the vertical height of the cylinder is 8 cm and uniformly fits
within the 10 ml cryovial
(e.g., 10 cm length and 17 mm diameter) and once completed, can be secured
with a threaded seal
cap. The mesh material is oriented such that the protective mesh material is
on the exterior of the
xenotransplantation product, and that once the rolled is complete there is no
exposed or visible
xenotransplantation material and it is fully encased in the protective insert.
The intrinsic tensile
and material properties of the sterile nylon-mesh packaging component are
homogenous, and the
169
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
inelasticity or stiffness of the material causes it to expand to fill the
volume of the cryovial. Thus,
regardless of the initial "roll-density", the material will uniformly loosen
and is therefore
standardized.
[000501] Under the laminar flow hood the operator will then use a sterile
syringe to draw up
enough sterile cryoprotective media (e.g., 5-7 ml of the media with 5%
dimethyl sulfoxide
(DMSO) (Cryostor CS5, BioLife Solutions)) to fill the cryovial until the skin
product roll is fully
immersed, ensuring that the combination of xenotransplantation skin material,
mesh backing, and
cryoprotectant media is flush with the 10 ml fill line, taking 1 minute for
each product
(understanding the time could be less or more, and up to 5 minutes for each
product).
[000502] Under the laminar flow hood, the operator will seal the cryovial
with the threaded
cap. The identity of the contents and label information are confirmed by the
operator. Labels are
prepopulated and applied to the exterior of the cryovials containing the
product in advance of the
product processing.
[000503] It will be understood that the preparation of the
xenotransplantation products and
packaging components described herein could be in the form of therapeutic
dosages. For example,
the xenotransplantation drug product consists of:
q. Xenotransplantation split-thickness skin Drug Substance
r. Primary Container Closure System which includes
i. Primary Packaging Component: a sterile, clear, polypropylene 10 ml
cryovial with threaded seal-cap
ii. Sterile nylon-mesh packaging component
Cryoprotective media packaging component
The indicated dosage of Xenotransplantation product is 300mg of vital,
metabolically active,
porcine xenotransplantation drug substance per cm2, with a constant thickness
of 0.55 mm.
Example formulations include:
s. Dosage Strength 1: a 25 cm2 split thickness skin graft, with uniform
thickness of
0.55mm, which weighs approximately 7.5 grams.
t. Dosage Strength 2: a 75 cm2 split thickness skin graft, with uniform
thickness of
0.55mm, which weights approximately 22.5 grams.
[000504] An example xenotransplantation drug product primary packaging
component is a
sterile, clear, polypropylene 10 ml cryovial with threaded seal-cap. For
example, the Simport
170
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Cryovial, T310 (10-ml) is manufactured by Simport Scientific. This product is
composed of
medical grade resin that is BPA free, Heavy Metal Free, and LATEX Free and
meets USP Class
VI limits.
[000505] A nylon-mesh packaging component is utilized during the
xenotransplantation drug
product manufacturing process. The prepared xenotransplantation drug product
is placed on sterile
nylon-mesh packaging component (e.g., Medifab 100-Micron Nylon Mesh) that has
been
previously trimmed to the following dimensions:
u. Dosage Strength 1: 7.5 cm in width by 8 cm in height; total area of 60 cm2
v. Dosage Strength 2: 22.5 cm in width by 8 cm in height; total area of 180
cm2
[000506] A cryoprotective media packaging component is also utilized during
the drug
manufacturing process. The xenotransplantation drug product is immersed in the
following
volumes of cryoprotective media packaging component prior to cryopreservation:
w. Dosage Strength 1: 7 ml of Cryostor C55 (containing 5% DMSO).
x. Dosage Strength 2: 5 ml of Cryostor C55 (containing 5% DMSO).
[000507] With regard to the assurance of saturation of cryoprotective
media, the indicated
amount of CryoStor C55 media (per Dosage Strength) is applied via 10 ml
syringe with the
cryovial (such as a type of cryovial shown in FIG. 46) in the vertical
position, under a laminar flow
hood (IS0-5, FED STD 209E Class 100 conditions) Cryomedia fills the voided
space(s), and
gravity ensures that the fill-process begins from the base of the vertically
oriented cryovial towards
the fill line at the apex. Volume is added until it reaches the manufacturers
demarcated 10 ml fill
line. Filling the vial in this manner also facilitates the removal of air
bubbles. Once complete, the
threaded cap is sealed. Visual and physical verification of saturation and
fill is accomplished,
ensuring that contents the xenotransplantation product are unable to shift
internally.
Cry opreservation
[000508] Product materials will be placed in the appropriate freezer rack
containing cryovials
with product as described above, and placed in a certified, Q-A control rate-
phase freezer. Using
a certified, Q-A control rate-phase freezer, the entire product is
cryopreserved via one standardized
control-rate freezing process:
y. Starting at 4 C, internal chamber and sample temperature probe will
lower at a rate
of 1 Celsius per minute until a temperature of -40 C is achieved.
171
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
z. Once temperature of -40 C has been reached in a controlled rate, control-
rate
freezer sample temperature probe should lower rapidly from -40 C to -80 C.
aa. Material is then transferred to a GLP certified, -80 C freezer until use.
Taking 40 minutes per batch time from room temperature to -80 C
(understanding the time could
be less or more, and up to 2 hours). In some aspects, penetrative
cryoprotectants such as DMSO,
may be used to protect morphology and tissue structure, and retain metabolic
activity levels
comparable to that of fresh skin. In some aspects, cryopreservation may
alternatively or
additionally include one or more of glycerol, gentamicin, Nystatin, L-
glutamine, and other
processing solutions. In some aspects, 0-lactam antibiotics are not used.
[000509] Inclusion of the cryoprotective-media packaging component is
intended to support
cell survival during the freeze-thaw cycle required for the
xenotransplantation product. Failure to
include the cryoprotective media packaging component of xenotransplantation
product during
packaging may disrupt the integrity of the xenotransplantation product or
impede the
cryopreservation process, and may potentially reduce the xenotransplantation
product viability
below acceptance criteria. Cryopreservation of the xenotransplantation product
without inclusion
a cryoprotective media results in destruction of biologically active cells
contained in the
xenotransplantation product. Rapid formation of ice crystals and disruption of
cellular membranes
and mitochondrial organelle barriers occurs during the freezing process, and
the dimethyl-
sulfoxide ingredient acts to displace intracellular fluid. Thus, the
cryoprotective media reduces the
formation of such ice crystals and rapid, disruptive increase in total
cellular volume that would
negatively impact the cellular viability and, thus, the efficacy of the Drug
Product.
[000510] During the course of a number of experiments, including the monkey
studies in
Example 1 herein, use of this cryoprotective-media packaging component has
never been observed
to cause an adverse, undesired reaction with the xenotransplantation product,
or degrade and
contaminate the final xenotransplantation product causing adverse reactions or
outcomes to the
recipient. Thus, selection of the specific material and associated
specifications were chosen to meet
appropriate standards necessary of a xenotransplantation product intended for
human, clinical use.
This including identifying a cryoprotective media with minimal, subclinical
levels of DMSO, one
that would satisfactorily perform without the need for inclusion of an
additional
xenotransplantation material (porcine serum) in the formulation. The
cryoprotective media-
packaging component is not used in the grafting procedure. Upon thawing, and
prior to use of the
172
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
xenotransplantation for therapeutic uses including as a drug product, it is
discarded. CryoStor CS5
is manufactured per cGMP standards and was selected because of its certified
acceptability for
human, clinical use.
Shipping to Clinical Site
[000511] Shipping the product to the clinical site should be done to
maintain the
xenotransplantation skin product material at -80 C storage condition. One
example shipping
container is the EXP-6 Standard Dry Vapor Shipper having an extensive, having
the following
specifications:
= Dynamic Holding Time 10 Days
= Holding Temperature -150 C or Colder
= Core Technology Dry Vapor Liquid Nitrogen
= Specimen Chamber 2.8" (71 mm) Diameter
= 11.5" (292 mm) Depth
= Weight Dry 9.7 lbs / 4.4 kg
= Charged 18.3 lbs / 8.3 kg
= Domestic Dimensional 21.07 lbs / 9.56 kg
= International Dimensional 24.87 lbs / 11.28 kg
= Outer Box 12" x 12" x 22"
= (305 x 305 x 559mm)
Aspects of the shipping process are also shown in FIG. 47 including, but not
limited to, (1)
cryopreservation storage; (2) xenotransplantation product in cryovial and
media as described
herein while in cryopreservation storage; (3) cryovial placed in dry vapor
shipping container (or
secondary closure system); (4) container and vial shipped via courier; (5)
xenotransplantation
product controlled and monitored at delivery location (can last at least 10
days at minus (-) 150
degrees Celsius or colder); (6) xenotransplantation product in cryovial and
media as described
herein removed from container/ secondary closure system; (7)
xenotransplantation product in
cryovial and media as described herein placed in freezer at location being
stored at -80 C.
173
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Clinical Site Preparation
[000512] In one aspect, the drug product arrives at the clinical site as a
cryopreserved
xenotransplantation product. Prior to use, the xenotransplantation product
must be thawed in a
37 C water bath, removed from the vial and washed in a series of 3 sterile
0.9% saline baths at
room temperature.
[000513] For the thawing process, sterile equipment and aseptic techniques
are used:
a) Prepare 200mL of normal saline into each of three 500mL sterile, surgical
bowls.
b) Place the unopened cryovial with the skin product in water bath having a
temperature of about25 C. In some embodiments, the temperature is about 37 C.
c) In the bath, swirl gently for approximately 5 minutes or until tissue is
mobile within
the cryovial, taking care to minimize unnecessary exposure time the
xenotransplantation skin product tissue is suspended in the thawed DMSO as
much
as possible.
d) Open the cryovial and use sterile forceps to quickly remove tissue and mesh
to
transfer into a bowl of normal saline.
e) Using sterile forceps, ensure tissue is fully submerged in saline for 15
seconds,
agitating by swirling gently to maximize coverage. The underlying, supportive
mesh material should be separated from the skin xenotransplantation skin
product
material. Use a second pair of sterile forceps to separate if necessary. Mesh
can be
left in the bowl, or discarded.
f) Using sterile forceps, transfer the skin into a second bowl wash. Submerge
fully
and gently swirl for 15 seconds; this is a serial dilution or "rinse".
g) Repeat the previous step, using sterile forceps to transfer the skin into a
third wash
of normal saline. Submerge fully and gently swirl for about 15 seconds.
h) The entire duration of the rinse process should be completed within 60
seconds to
minimize unnecessary exposure time the product is suspended in thawed DMSO in
order to maximize product efficacy.
i) Tissue is now thawed, rinsed, and ready for application. Leave in normal
saline
until use, not to exceed 2 hours at about 25 C.
[000514] After the complete, thaw and rinse process is complete, the
xenotransplantation
product is ready for placement on the wound site. Serial washes in saline,
once thawed provide
174
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
ample dilutive solvent to remove the residual cryoprotectant (5% DMSO
solution, CryoStor C S5)
and replace the intracellular fluid levels to normal homeostatic conditions.
Such dilution and use
of a cryoprotective media containing a sub-clinical level of DMSO ensures that
any minimal,
residual DMSO remaining on the xenotransplantation skin product material post-
thaw would be
non-appreciable and would be highly unlikely to be clinically significant.
This process also ensures
retention of the maximum amount of metabolically active cells, and thereby
maximizing the
efficacy of the xenotransplantation product.
[000515] Example of Thawing. Following is one example of a thawing
procedure for a
xenotransplantation product_Thawing can occur in a BioSafety Cabinet with
operator in sterile
gloves as follows: (i) prepare 200mL of Normal saline into each of three 500mL
surgical bowls;
(ii) prepare the water bath by wiping it clean with chlorhexidine then
spraying it down with 70%
ethanol; (iii) after the ethanol has dried add sterile water solution into the
water bath and heat to
37 C +1_ 2 C;
(iv) the xenotransplantation drug product is in a double bag, leave it
unopened and
place it into the 37 C water bath; (v) swirl gently for approximately 5
minutes or until the tissue is
mobile within the cryovial; (vi) minimize the time the tissue spends in thawed
DMSO as much as
possible; (vii) spray the outside bags with ethanol and remove the vial from
the outer bags and
spray the xenotransplantation drug product cryovials with 70% ethanol before
placing into
Biosafety Cabinet; (viii) unscrew the cryovial and use forceps to quickly
remove tissue and mesh
to transfer into a bowl of normal saline; (ix) use forceps to ensure tissue is
fully submerged in
saline for 60 seconds, agitating by swirling gently to maximize coverage; (x)
the mesh should be
separated from the skin, using a second pair of forceps to separate if
necessary; (xi) the mesh can
be left in the bowl, or discarded; (xii) using forceps transfer the skin into
the second bowl wash;
(xiii) submerge fully and gently swirl for 60 seconds; (xiv) using forceps
transfer the skin into the
third bowl wash and submerge fully and gently swirl for 60 seconds. Tissue is
now thawed and
ready for application. Keep it moist with sterile saline in a sterile pan.
[000516] The process of rolling the inert, nylon mesh backing and the
xenotransplantation
skin product results in uniform "roll-density" of the xenotransplantation
product. All mesh
materials are cut to uniform dimensions, according to the prescribed
dimensions for the given
application, and are obtained from the same material lot, thus affording
uniform material properties
for all units of the skin product manufactured within a specific lot.
175
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000517] The intrinsic tensile and material properties of the nylon mesh
insert are
homogenous, and the inelasticity or stiffness of the material causes it to
expand to fill the volume
of the primary container closure system (cryovial). Thus, regardless of the
initial "roll-density",
the material will uniformly loosen and is therefore standardized.
[000518] The indicated amount of CryoStor CS5 media (per Dosage Strength)
is applied via
10ml-syringe with the cryovial in the vertical position, under Class 100, IS05
conditions within
an ABSL-2 laminar flow hood.
[000519] Cryomedia fills the voided space(s), and gravity ensures that the
fill-process begins
from the base of the vertically oriented cryovial towards the fill line at the
apex. Volume is added
until it reaches the manufacturers demarcated 10m1 fill line. Filling the vial
in this manner also
facilitates the removal of air bubbles.
[000520] Once complete, the threaded cap is sealed. Visual and physical
assurance of
saturation and fill is accomplished by the shaking the skin product ensuring
that contents are unable
to shift internally. Aspects of the cryovial are also shown in FIG. 46, with
aspects that can include,
among other things, 10 ml volume, size of 17 mm x 84 mm, vertical ribs
facilitating cap removal,
silicone washer, cap and tube made of the same polypropylene material with the
same coefficient
of expansion ensuring seal at all temperatures, 1 and 1/4 turn thread design,
thick wall, large white
marking area, and round bottom allowing for ease of emptying contents.
[000521] Aspects of the secondary closure system is shown in FIG. 48, with
aspects that can
include, among other things, Tyvek ¨ 1073B medical grade construction, 5
inches wide x 12" high,
storage ability of 15 cames or 2 cryovial boxes, holding temperature of -150
degrees Celsius or
colder, utilization of dry vapor liquid nitrogen, IATA rated 10 days of
dynamic holding time under
normal shipping conditions, specimen chamber diameter of 2.8 inches (71 mm),
specimen chamber
depth of 11.5 inches (292 mm), dry weight of 9.7 lbs / 4.4 kg, charged weight
of 18.3 lbs. / 8.3 kg,
domestic dimensional weight of 21.07 lbs. / 9.56 kg, international dimensional
weight of 24.87
lbs. / 11.28 kg, outer box dimensions of 12" x 12" x 22."
[000522] No additional or external impurities in the product are
anticipated to be present
since processing involves only the minimal mechanical manipulation of the
product, and no other
chemical or biological agents are introduced during this closed process.
Acceptance criteria testing
required for use of the source animals for the product manufacturing process
is conducted as
176
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
described herein and documented via the Drug Product COA. The final product is
evaluated for
viral adventitious agents as described herein.
[000523] In terms of shelf life, continuous storage of the
xenotransplantation product as
described support a shelf life long-term stability (cell-viability) of up to
at least 7 years (in one
embodiment is a shelf life of 6 months) when stored continuously at -80 C. The
shelf-life duration
of continued cryopreservation of the xenotransplantation product with of at
least 7 years. Table 10
shows stability time points that the xenotransplantation product will be
tested.
TABLE 10
Stability Study Time Points
Assay Time points (Months)
0 12 24 36 60
Histology A
Sterility A
Endotoxin A
Viability A
A = initial product release testing
B = stability testing for Xenotransplantation product
[000524] In accordance with one aspect, following in Table 11 are items
that can be utilized
in a certificate of analysis and release.
TABLE 11
Test Results
Test Method Acceptance Criteria Results
Clear, colorless to slightly
Appearance Visual Inspection yellow liquid with no
visible Conforms
particulates
TM5110 7.5 to 7.7
pH USP <791> 7.6
Cell viability is 75% to
Metabolic 200% of cells preserved in 87
Activity Assay TMS100 the internal standard at Day
1 recovery following
preservation.
177
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Kinetic Chromogenic
Endotoxin USP <85> s 0.5 EU/ml Conforms
Membrane Filtration
Sterility USP <71> Sterile Conforms
TMS111 FT-IR Conforms to CryoStor CS S
Identification Reference Standard Conforms
TM 5112
Osmolality USP <785> 1360-1390 mOsm/ kgH20 1388
Specific Gravity TM5114 1.055- 1.063 1.059
Gas Chromatography
DMSO Content (FID) 4.0%-7.0% 5.0
EXAMPLE 3
[000525] Porcine skin shares fundamental properties with human skin and
represents a
potential alternative to human cadaver skin grafts for temporary coverage of
severe burns. The
impact of extended cryopreservation on porcine grafts on graft viability,
graft take, and barrier
function was examined in a study using a model of MHC matched and mismatched
MHC Class II
skin transplants.
[000526] Cellular viability was assessed using formazan-MTT and the
biological properties
of the grafts, were assessed by grafting on swine recipients. To complement
the in vivo clinical
assessments, histologic, and morphologic analyses, a series of MTT-reduction
assays were
performed to evaluate the residual viability of porcine grafts after
cryopreservation and long-term
storage. Mitochondria reduce MTT into a formazan metabolite, which can be
observed as purple
hue. Harnessing this phenomenon, an analysis of changes in optical density
values measured by a
spectrophotometer, or an interpolation of the quantities of formazan produced
against standard
curves, can provide differential assessments of cellular viability, between
experimental samples
and positive and negative controls. There were 2 cohorts of 2 animals each
(total, N=4) based upon
the MHC match and each swine received 4 grafts: one autograft and three
allografts of identical
MHC-profiles. Grafts were clinically assessed for graft-take, adherence, and
time to graft rejection.
Rejection was also assessed histologically via the Banff grading scale.
[000527] Direct comparisons between otherwise equivalent materials yield
meaningful,
differential times of survival, based solely on duration of storage, holding
all other factors constant.
Side-by-side, in vivo evaluations are performed between equivalent grafts,
preserved in identical
178
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
fashion, stored for periods of 15 minutes versus 7 years. Clinical gross
assessments and
photographs, paired with independent histological assessments, determine
whether any
appreciable differences in graft survival exist relative to the length of time
in the frozen state. In
tandem, separate in vitro assessments of graft viability, quantified by MTT-
reduction assays,
characterize the metabolic activity of cells post-cryopreservation and various
storage terms.
Further, independent histomorphological analysis, using standard histological
(H&E) staining,
provides evidence as to whether these processes cause observable changes to
the graft material at
a structural level. This study advantageously used materials that had been
stored, uninterrupted,
for such a time, along with the associated surgical records and standardized
institutional protocols.
Further, processing methods and protocols between the comparative groups were
standardized,
and identically applied, with respect to cryopreservation and thawing
protocols, reagents, and
methods employed. Combined, this allowed for isolated, side-by-side evaluation
of duration of
storage, and alleviated the need to model or extrapolate findings, or
otherwise use normative
predictive methods. Furthermore, the use of MHC-matched and Class II
mismatched donor-
recipient pairs in this model of allogeneic skin transplantation served as
internal controls to both
confirm the identity of the tissues obtained seven years earlier, and the
veracity of the surgical
notes and documentation. Further, equivalent behavior exhibited by the
allografts also
demonstrates that the antigenicity of the grafts was not altered as a result
of the duration of storage.
[000528] There were no technical failures; all grafts adhered to their
respective wound beds
and re-vascularized. In cohort 1 (MHC-matched donor-recipient pair), all
grafts remained
adherent, and appeared uniformly healthy at postoperative day (POD) 12 (FIG.
49A), but at POD-
14, signs of necrosis, progressive erythema and loss of adherence were
observed (FIG. 49B).
Clinical assessment of the 6 grafts in cohort 1 showed rejection at POD-14 to
18. In cohort 2, MHC
Class II mismatched, allogeneic grafts appeared comparable to autografts
through POD-4.
However, by POD-8, all allogeneic grafts demonstrated mild erythema,
consistent with rejection
and were considered fully rejected by POD-10. No statistically significant
difference in the
duration, quality of adherence, or cellular viability among the fresh,
recently preserved, and long
term preserved skin grafts were observed. The cryopreserved materials were,
statistically speaking,
more alive than dead, and this finding was empirically witnessed in vivo, as
all 7-year grafts
demonstrated adherence to the wound bed and prolonged survivability. Such
survivability would
not have been exhibited by non-vital allografts. Without limiting the
invention, it will be
179
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
understood that the time period for cryopreservation for the present invention
may, for example,
include any length of time up to about 7 years.
[000529] Materials and Methods:
[000530] The study was conducted in accordance an IACUC approved protocol
(2005N000279, Amendment 69) at the Center for Transplantation Sciences, and in
compliance
with the U.S. Department of Agriculture's (USDA) Animal Welfare Act (9 CFR
Parts 1, 2 and 3),
the Guide for the Care and Use of Laboratory Animals, and all state, local
laws and regulations.
Study protocols, surgical procedures, and animal care guidelines were
independently reviewed and
monitored by a standing IACUC committee.
[000531] A total of eight swine were enrolled in this experiment, and all
were members of
the Sachs-NIH, inbred miniature swine colony. At the time of surgery, all
swine were between 10
and 20-kg in total body weight and between 2 and 4 months of age.
Immunosuppression regimen(s)
were not administered at any time during this experiment. Animals 24074 and
24075 were assigned
to Cohort 1 and represented a MHC-matched donor-recipient pair. Animals 24043
and 24070 were
assigned to Cohort 2 and represented a mis-match of MHC Class II donor-
recipient pair.
Separately, for the in vitro, MTT series of analyses, five, additional wild-
type Gottingen miniature
swine provided tissues for positive and negative controls.
[000532] Swine donors were anesthetized with I.M. 2 mg/kg telazol
(tiletamine HC1 and
zolazepam HC1, Zoetis Inc., Kalamazoo, MI) and brought to the operating room
for orotracheal
intubation. Anesthesia was maintained using 2% isoflurane and oxygen. Skin
surfaces were
disinfected before surgery with chlorhexidine acetate (NolvasanR Surgical
Scrub, Fort Dodge
Animal Health, Fort Dodge, IA) and povidone-iodine, 10% (Betadine Solution,
Purdue Products,
L.P., Stamford, CT). The animals were then draped, leaving the right side of
the dorsum exposed.
Split-thickness skin grafts, measuring approximately 25 cm2 (surface area)
were harvested
between the scapula and inferior margin of the lowermost rib from each animal
using an air-driven
Zimmer dermatome (Medfix Solution, Inc., Tucson, AZ) with the depth set to
0.056-cm (0.022
inches).
[000533] Following skin graft harvest, grafts intended for cryopreservation
and storage for
limited duration grafts underwent a standardized institutional protocol and
were maintained at -
80 C for 15 minutes prior to thawing. Long-term cryopreserved grafts had been
continuously
stored at -80 C for a period of more than 7 years. All grafts, previously
sized to approximately 25
180
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
cm2, were placed on a sterile nylon mesh backing for structural support and
rolled for placement
into a threaded seal cryovial under a laminar flow hood. Once all grafts were
prepared,
approximately 5-mL of freeze media was added to the vial and sealed. The
protocol required freeze
media prepared by combining 15% dimethyl sulfoxide (DMSO) cryoprotective media
(Lonza
BioWhittaker) with fetal porcine serum (FPS) or donor serum (if FPS is
unavailable) in a 1:1 ratio,
filtering (0.45 micron), and chilling to 4 C prior to use. The vials were
subsequently frozen in a
controlled rate, phase freezer at a rate of 1 C per minute to -40 C, then
rapidly cooled to a
temperature -80 C, at which they remained for 15 minutes for those test
articles in the control
group subjected to limited storage duration, or for a period of more than 7
years in the case of the
those experimental grafts in the test group exposed to extended duration of
cryopreservation.
DMSO displaces intracellular fluid during the freezing process. Cryoprotective
media, e.g.,
CryoStor is used in an amount of about 40-80%, or 50-70% based on maximum
internal volume
of the cryovial (10m1) less the volume of the xenotransplantation product.
[000534] In order to thaw the grafts for surgical use, sealed vials were
placed in 37 C water
baths for approximately 1 minute, at which point the vial was opened and the
frozen graft was
removed using sterile technique. Subsequently, grafts underwent 3, 1-minute
serial washes in
normal saline with gentle agitation, in order to dilute and systematically
remove ambient, residual
DMSO and prevent loss of cell viability. Grafts were then taken to the
surgical field in normal
saline at 25 C for engraftment.
[000535] Two separate, but identical, surgical events were performed in
succession. The
entire surgical plan included a total of four (n=4) donor-recipient swine,
employing two animals
per each of the two experimental cohorts (Cohort 1 and Cohort 2), paired
intentionally based on
SLA-configurations as described previously. In total, four technical controls
and twelve (n=12)
experimental grafts were engrafted and subsequently observed.
[000536] Each animal received four deep-partial defects along the animal's
right dorsum, in
a linear (caudal to cranial) orientation, ordered from 1 to 4, respectively.
Deep-partial wound
defects were surgically introduced via additional passes with the dermatome
after the initial split
thickness graft harvest. The resulting wound beds were uniform, free of
visible debris, and
demonstrated independent, punctate bleeding. These defects were interrupted,
and not made in a
single continuous pass with the dermatome. Instead, care was given to create
four, isolated but
equivalent wounds with regards to overall size, depth, and anatomical
location.
181
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000537] Following thawing, but prior to engraftment, all split-thickness
skin grafts were
fenestrated using a 15 (size) blade to prevent seroma or hematoma formation.
Graft test articles
were independently placed on the prepared wound bed and uniformly sutured in
place using simple
interrupted, 3-0 nylon sutures, applied in a graft-to-wound bed manner.
Approximately 16 points
of fixation were introduced per graft, spaced evenly around the graft, with
the resulting knot
located on the wound border, not the graft article. This technique ensured
that minimal, but
adequate, residual tension was present and uniform, which is necessary for
optimal graft-to-wound
adherence, minimization of hematomas, and optimal graft survivability.
[000538] At Wound Site 1 (most caudal), a split-thickness autograft was
placed, serving as a
technical control. This autograft test article was harvested during the wound
bed creation,
subsequently underwent the same freeze-thaw process concomitantly with all
experimental grafts,
and was held in an identical, cryopreserved state for the same duration as the
control grafts
identified for a limited duration (15 minutes at -80 C). At Wound Site 2, a
split-thickness allograft
from its respective cohort pair-mate was sutured into place. This graft
represented test articles
exposed to cryopreservation for a limited duration (15 minutes at -80 C). At
Wound Site 3, a split-
thickness allograft from the wild-type donor, which represented a split-
thickness graft, with
identical SLA matching as those at Wound Site 2 that had experienced
"extended" storage in the
cryopreserved state (more than 7 years at -80 C). At Wound Site 4 (most
cranial), a split-thickness
allograft from a genetically engineered knockout donor, which represented a
split-thickness graft,
with identical SLA matching as those grafts at Wound Site 2, sourced from the
genetically
engineered donor animal, that had also experienced "extended" exposure in the
cryopreserved state
(-80 C) for more than 7 years.
[000539] Overlying pressure dressings, consisting of Xeroform petrolatum
gauze
(Medtronic), TelfaTm non-adhesive dressing (Covidien, Minneapolis, MN), and
sterile gauze were
maintained in place and dry with multiple, overlapping sheets of TegadermTm
(3M, St. Paul, MN).
Recipients were then dressed with cotton jackets to reduce interference with
the grafts. Graft
dressings were removed on POD-2 and changed daily thereafter. Total
postoperative follow up
was 20 days. Animals were monitored for signs of pain including vocalization,
tachypnea, loss of
appetite, and changes in attitude, behavior, and mobility. Transdermal
fentanyl patches were
applied for post-operative analgesia. All sutures were removed by POD-7.
182
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000540] To validate the assay method and establish boundary conditions
specific to test
articles of split thickness skin porcine skin, two independent assay series
were performed on fresh
(n=5, 5) and heat denatured samples (n=5, 5). The (geometric) average formazan
produced on
fresh samples was 0.221 0.022-mg/mL and 0.300 0.035-mg/mL, respectively.
In contrast, the
(geometric) average formazan produced by heat-denatured samples was 0.094
0.020-mg/mL and
0.105 0.009-mg/mL, respectively. These differences were statistically
significant in both cases
(p<0.05).
[000541] All four porcine recipients tolerated the surgical procedure and
recovered fully
without incident. All sixteen (n=16) grafts re-vascularized without evidence
of technical
complication, and uniformly exhibited adherence to the underlying wound bed
(i.e. "good take").
Over the course of the post-operative observational period, no grafts were
lost due to mechanical
disturbance or exhibited any clinical signs of wound infection. All four (n=4)
autografts at Wound
Site 1 healed permanently and were indistinguishable from surrounding tissues
at the study end-
point, acting as a technical control for the skin grafting, cryopreservation
and thawing technique.
[000542] In Cohort 1, all six (n=6) allogeneic grafts demonstrated
equivalent adherence to
the underlying wound bed and uniformly exhibited clinical signs consistent
with vascularization
and perfusion on postoperative days (POD) 2 and 4. Notable, however, was the
contrast (loss) of
color exhibited by the allografts that had been cryopreserved for an extended
duration. All four of
these grafts appeared paler as compared to the autograft and allografts at
Wound Site 2. This
appearance fully resolved in all grafts, in both Animals, by POD-6. All six
(n=6) allografts
exhibited mild sloughing of the superficial epidermis by POD-8, but grafts
remained viable,
adherent, and appeared otherwise healthy at inspection on POD-12. In Animal
24074, grafts at
Wound Sites 2 and 3 showed initial signs of necrosis, progressive erythema,
and loss of adherence
by POD-14, and presented increasing signs of immune-mediated rejection, until
final rejection at
POD-18. However, the allograft at Wound Site 4 (most-cranial) did not
similarly persist; instead,
on POD-14 this graft was significantly darker and exhibited signs of complete
necrosis and was
clinically assessed to be fully rejected at this time. The rapid loss of the
graft 4, from viability at
POD-12 to complete avulsion by POD-14, dissimilar and distinct from Wound Site
2 and Wound
Site 3, was notable. For grafts on Animal 24075, all grafts were rejected on
POD-14.
[000543] In Cohort 2, animals presented similarly to those in Cohort 1
through POD-4, and
equivalently to each other. Overall, clinical signs were comparable in
progression to the minor-
183
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
mismatched grafts in Cohort 1, but at an accelerated pace. The grafts that had
experienced extended
cryopreservation appeared paler at POD-2 and POD-4 than the grafts that had
not experienced
cryopreservation, and all grafts showed increased evidence of perfusion and
vascularization by
POD-6. By POD-8, all three allogeneic grafts in Animal 24043, showed clear
signs of rejection
and were considered fully rejected. In Animal 24070, all three allogeneic
grafts showed clear signs
of rejection and were considered fully rejected by POD-10. However, all
allogeneic grafts survived
at the same rate, irrespective of the genetics or length of storage.
[000544] With respect to grafts subjected to limited or extended durations
of
cryopreservation, 100% of allograft comparators at Wound Sites 2 and 3 (n=4 of
4) were identical
with respect to clinical assessment of duration of graft survival. Comparison
of Wound Sites 2 and
4 were coincident (n=3), with the exception of the allograft at Wound Site 4,
Animal 24074, which
survived until POD-14 (n=1), determined to be clinically and rejected four
days prior to its
counterparts.
[000545] Overall, histological assessments closely mirrored the clinical
assessments.
Following surgery, all grafts, including autografts, exhibited early signs of
acute inflammation
during initial observations on POD-2 and 4, that later resolved with time. All
allografts in Cohort
2, as compared to those in Cohort 1, uniformly exhibited accelerated
progression towards immune-
mediated rejection.
[000546] Ultimately, all six (n=6) allogeneic grafts in Cohort 1, and three
allogeneic grafts
(n=3) from Animal 24043 in Cohort 2, independently demonstrated histological
and microscopic
signs of rejection coterminous with the independent gross clinical
assessments. The single
exception were the three allografts engrafted on Animal 24070, where each
graft received Banff
scores of 4 (of 4) on POD-10, but were not deemed officially rejected until
POD-12, one
assessment period (2 days) later than the corresponding clinical designation
assigned at POD-10.
[000547] With respect to grafts subjected to limited or extended durations
of
cryopreservation, 100% of allograft comparators at Wound Sites 2 and 3 (n=4 of
4) were identical
with respect to histological assessment of duration of graft survival.
Comparison of Wound Sites
2 and 4 were coincident (n=3), with the exception of the allograft at Wound
Site 4, Animal 24074,
which survived 14 days post-operatively (n=1), determined to be histologically
rejected four days
prior to its counterparts.
184
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000548] Neither the MTT nor the neutral red staining technique, as applied
on either testing
occasion, were deemed effective for histological and microscopic evaluation,
however the standard
hemotoxylin and eosin staining demonstrated observable tissue destruction of
the heat denatured
specimens.
[000549] Overall, using a linear, mixed effect model with random intercept,
the mean
survival of grafts at Wound Site 3 was 0.00 (95% CI: -1.10, 1.10 days) less
than allografts at
Wound Site 2. The mean survival of grafts at Wound Site 4 was 2.00 (95% CI:
1.10, 3.10 days)
less than allografts at Wound Site 2. Histological assessment finds on average
0.5 days more
survival than grafts assessed grossly, but this is not statistically
distinguishable (p =0.28). Seven of
the eight experimental grafts fared equivalently to their comparators. The in
vivo experiments
showed no statistical difference between grafts subjected to short versus long-
term storage. With
the exception of the graft at Wound Site 4 on Animal 24074, which was assessed
as fully rejected
four days earlier than its comparators, graft performance and survivability
were indistinguishable
between the two groups.
[000550] As noted in previous publications, cryopreserved grafts appeared
notably paler
during the early imbibition and vascularization periods. This contrast was
starkly evident for grafts
at Wound Sites 3 and 4 in all animals. Ultimately, grafts fully resolved and
adhered to the
underlying wound bed to an equivalent degree.
[000551] Demonstrated viability was evidenced uniformly across the three,
independent
evaluation methods. The statistical analysis of the MTT-assay shows there was
no significant
difference between cryopreserved and fresh specimens (FIG. 50A), but
significant differences
were observed between fresh and cryopreserved specimens versus heat-denatured
ones (FIG. 50B).
This suggests broadly that the cryopreserved materials were, statistically
speaking, more alive than
dead. This outcome is substantiated in the in vivo outcomes in which all 7-
year grafts demonstrated
adherence to the wound bed and prolonged survivability, which would not be
exhibited by non-
vital grafts.
[000552] Regarding the MTT-reduction assays, substantial variability
existed between
absolute values resulting from such assays, from specimen to specimen and from
cohort-to-cohort.
Indeed, absolute values of formazan production were actually higher than those
obtained from
non-cryopreserved samples; it is unlikely that freezing enhanced cellular
activity.
185
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000553] Pig skin can be cryopreserved for years, e.g., 1, 3, 5, 7 or more
years and retain cell
viability and that the genetic modification, Gal-T-KO, did not impact
metabolic stability when
compared to wild type pig skin processed and stored using the same procedures.
[000554] Furthermore, the use of MHC-matched and Class II mismatched donor-
recipient
pairs in this model of allogeneic skin transplantation served as internal
controls to compare the
effect of long term cryopreservation (7 years) on the survival of allogeneic
skin grafts. The cell
viability data after long term cryopreservation is supported by the survival
of the skin in vivo. This
also demonstrated that the genetic differences (wild type versus Gal-T-KO) of
the grafts did not
impact the survival of the grafts.
[000555] The hypothesis was that graft take, and overall survival, would be
inversely
proportional to the length of storage duration. In other words, it was
expected that the longer the
graft had been frozen, the less likely it would survive and mimic the
comparator grafts preserved
for shorter durations. Surprisingly, these studies revealed that the porcine
tissue can be
cryopreserved for significant durations, 7 years in the case of the present
disclosure, and retain
adequate cell viability. Moreover, the genetic modification (Gal-T-KO) did not
impact metabolic
activity, when compared to wild type skin processed identically. Lastly, the
results confirm that
the MTT-reduction assay can reliably provide an accurate, useful diagnostic
method, and
applicable to the assessment of porcine skin graft viability.
[000556] The promising results of this study indicate that it may be
feasible to cryopreserve
and store porcine skin for logistically relevant durations, and our findings
are consistent with
current industry practices and the multi-year "shelf life" guidance that the
American Association
for Tissue Banks has established for human cadaveric tissues.
[000557] Further, these data indicate that scalable, clinically useful
methods of preserving
and storing porcine xenotransplantation products with adequate viability are
disclosed, and that
vital porcine xenotransplantation products that can be effectively stored and
distributed.
EXAMPLE 4
Product Processing
Generally
[000558] A xenotransplantation product of the present disclosure was
processed according
to the following procedures.
Personnel
186
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
[000559] The operator was dressed in sterile dress in accordance with
institutional standards
to maintain designated pathogen free conditions. The operator wore eye
protection safety glasses
for ultraviolet light and lasers.
Preparation of Laminar Flow Hood and Product Processing
[000560] An ultraviolet laser lamp (Model #) was set up in a laminar flow
hood. Each of the
four corners of the lamp was placed on two container lids that were stacked on
top of each other,
i.e., four pairs of lids were used to support the lamp. The distance from the
lamp bulbs (2 bulb
tubes total) to the floor of the hood was approximately 1.5 inches. The entire
interior of the hood
was sprayed with alcohol, e.g., ethanol or isopropanol. The lamp was turned on
and the operator
performed a calculation of time for desired exposure based on lamp
specifications, number of
bulbs, and distance between the bulbs and the xenotransplantation product.
[000561] The operator poured two baths (one chlorhexidine and one alcohol)
into two
separate bowls and placed the two bowls under the hood.
[000562] A package of new sterilized vials was placed under the hood. Vial
caps were
unscrewed and placed into the chlorhexidine bath. Each vial (without cap) was
then turned upside
down and plunged open ended into the chlorhexidine bath, for one minute each
and then set upright
to air dry. Thereafter, the exterior of each vial was wiped with chlorhexidine
and alcohol utilizing
sterile gauze. The vial caps were removed from the chlorhexidine bath and
placed on sterile gauze.
The open ends of each vial were plunged into alcohol bath for 1 minute each
and then set aside to
air dry.
[000563] A xenotransplantation product "#46 product" (5x15 cm) having a
mesh backing
prepared according to Example 2 was removed from its original vial and the
operator placed
original vial into an empty bowl. Operator placed the #46 product on the paper
side of an opened
sterilized instrument package. The operator unrolled the #46 product and
placed it under the lamp
for 2 minutes, then turned it over to the other side, removed the mesh
backing, and put it under the
lamp for 2 minutes on opposite side, while still on the same paper. The time
period for exposing a
given sample to the UV light can be varied based on the specific biological
agents or the types of
biological agents to be sterilized, e.g., as shown in the following Table 12:
TABLE 12
187
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
Biological Agent Type of UV-C Dosage (uW
Sterilization time
Biological sec/cm2) for 90% (sec)*
Agent sterilization
Penicillium spp. Fungus 224,000 1800
Aspergillus flavus Fungus 34,900 300
Aspergillus niger Fungus 31,500 250
Yeast Fungus 4000 30
Influenza A Virus 1900 15
HIV-1 Virus 28,000 220
Vaccinia Virus 1500 10
Escherichia coli Bacteria 2000 20
Staphylococcus Bacteria 6600 50
aureus
Bacillus subtilis Bacteria 6800 50
Mycoplasma spp. Bacteria 8400 70
Pseudomonas Bacteria 2200 20
aeruginosa
*Using a UV-C intensity of 125 uW/cm2
[000564] Then the "#46 product was removed and cut in half. Each half was
rolled by hand
and placed into a new vial sterilized as explained above. Each new cap was
placed on each new
vial and screwed on securely. Each vial was placed under the lamp and
periodically rolled for
desired even exposure to light on the exterior of the vial. The vials were
placed inside a glass jar
that had an interior that had been previously sterilized and the exterior was
sterilized by the
operator with alcohol and chlorhexidine, including threads and caps.
[000565] A similar process was performed for the following
xenotransplantation products,
except instead of being placed on sterile paper prior to entry under the lamp,
the mesh was not
removed from the products and the products were placed under the lamp skin
side up for 2 minutes,
then the products were folded over so a first half of the bottom portion of
each product faced the
lamp for 2 minutes, then the second half of each product was folded over so
that the other half of
188
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
the bottom of each product faced the lamp for 2 minutes. Some of the products
were cut into
smaller sections and exposed to light, some for periods for longer than 2
minutes, but never less
than 2 minutes.
[000566] Products #40 (5x15 cm), #63 (10x15 cm), #69 (10x15 cm), and #25,
underwent the
above processes and products #69 and #25 were rolled exclusively using
instruments and the
operator did not directly handle those products. As with #46, after operator
securely screwed the
cap on each vial, each vial was placed under the lamp and rolled for even
exposure to light emitted
from lamp. Vials were later removed from under the lamp and wiped down with
alcohol prior to
being placed into glass jars.
[000567] Four glass jars were utilized to store each of the sets of vials.
Prior to being handed
to the operator, the assistant drenched the exteriors of the glass jars with
alcohol via a spray bottle.
The assistant handed the glass jars to the operator by holding the bottom of
each jar and handing
to operator outside of hood. After receiving the glass jars from assistant,
under the hood, the
operator bathed the glass jar lids and plunged the open ends of the jars into
alcohol and wiped the
exterior of the jars with alcohol including threads of the jar.
[000568] The vials were wiped with alcohol utilizing gauze and placed
inside each glass jar
with an instrument. The lids of the glass jars were then secured and the jars
were handed to the
assistant. Frequently and on a periodic basis throughout these processes the
assistant sprayed the
operator's gloves and arms with alcohol.
[000569] Thereafter, the products were placed into the phase freezer at the
conclusion of the
procedures.
EXAMPLE 5
[000570] In a human evaluation of a xenotransplantation product of the
present disclosure
for treatment of severe and extensive partial and full thickness burns in a
human patient, the
following results were obtained:
[000571] The patient presented with a mixed depth, flame-induced burn
injury, resulting in
a 14% Total Body Surface Area (TBSA) defect to the (anatomic) right, upper
torso ¨ specifically,
bordered: from the right lateral axilla (superior border) to the sixth right
lateral rib (inferior border)
as shown in Fig. 51A.
[000572] The surgeon temporarily grafted part of the affected wound area
with Human
Deceased Donor (HDD) allograft and the xenotransplant product of the present
disclosure. The
189
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
remaining regions of the wound area were covered with a negative pressure
wound therapy
(NPWT). The patient received approximately 150 cm2 of HDD allograft, meshed to
a 1:1.5 ratio,
and 25 cm2 of xenotransplant product of the present disclosure meshed to a 1:1
ratio during
surgery, which is specifically shown in FIG. 51B
[000573] Both temporary wound closure dressings were placed adjacently, but
not in direct
contact, and were secured with staples on the perimeter of the tissue(s),
overlaid with NPWT.
[000574] Upon clinical visual inspection of the first wound dressing change
on POD-5, the
HDD allograft and xenotransplant product of the present disclosure were both
observed to be fully
adherent to the underlying wound bed and were indistinguishable as shown in
FIG. 51C.
[000575] The patient experienced no adverse events and no serious adverse
events were
observed or reported.
[000576] In accordance with the regular clinical standard of care, both HDD
allograft and the
xenotransplant product of the present disclosure were removed at the first
wound dressing change.
Following mechanical removal, the underlying wound beds were equally perfused
(with visible
punctate bleeding) and otherwise appeared equivalent as shown in FIG. 51D.
[000577] A POD-5 close-up image of the wound bed for the xenotransplant
product of the
present disclosure adjacent to wound bed for HDD allograft is shown in FIG.
51E.
[000578] On POD-5 following removal, per clinical standard of care, the
entire affected area
received definitive wound closure via engraftment with a self (auto)graft
(autologous split-
thickness skin graft), obtained from the patient as shown in FIG. 51F.
[000579] Per protocol, blood samples for infectious disease, immunological
response, and
long-term evaluation were obtained, as well as pre-operative, pen-operative,
and post-operative
photographs.
[000580] On POD-14 (from the first operation), clinical observations at the
wound dressing
change demonstrated no discernible differences in the wound healing rate or
quality at any location
as shown in FIG. 51G.
[000581] Per protocol, blood samples for infectious disease, immunological
response, and
long-term evaluation were obtained, as well as pre-operative, pen-operative,
and post-operative
photographs.
[000582] Testing for detection of PERV by quantitative RT-PCR was performed
on baseline
blood samples (25 mL), first dressing change (21 mL), and two week blood
samples (23 mL). The
190
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
results were as follows:
[000583] PERV was not detected by qPCR in either RNA or DNA isolated from
PBMC and
RNA isolated from plasma. Evidence of porcine cells as determined by qPCR
directed to the
porcine mtC0II gene was not found in RNA isolated from the PBMC.
Source Cq PERV pol Cq porcine mtC0II PERV Porcine cells
DNA-PBMC <LOD <LOD Negative Negative
RNA-PBMC <LOD <LOD Negative Negative
RNA-plasma <LOD <LOD Negative Negative
[000584] Further, a study is conducted to assess the proliferative response
of human
lymphocytes responder peripheral blood mononuclear cells (PBMC) in the
presence of mitomycin
C treated porcine stimulator cells (alpha-galactosyltransferase knock out (KO)
pig B173) over
time. PBMC samples were obtained from patients enrolled in Sponsor Study XT-
001, both before
and after the transplantation of porcine skin grafts. The porcine skin grafts
were obtained from
genetically modified alpha-galactosyltransferase knock out (KO) pigs.
[000585] Patient PBMC samples were previously prepared by Ficoll gradient
centrifugation
and cryopreserved. Whole blood from the skin donor pig (B173) was previously
shipped to Xeno
Diagnostics (XD) and PBMCs isolated by Ficoll gradient centrifugation and
cryopreserved. The
day prior to setting up the MLR, samples were thawed at 37 C, washed, and
rested overnight in
10%FBS/RPMI. Porcine PBMCs were mitomycin C treated (stimulators) and mixed
with an equal
number of test human PBMCs (responders). The MLR was incubated for seven days
with BrdU
added on day six. On day seven, a BrdU ELISA was performed and proliferation
measured.
[000586] As shown in FIG. 52, PBMC obtained from skin graft Patient XT-001
generated
positive xenogeneic MLR PBMC mixed lymphocyte responses (MLR) when cocultured
with
alpha-Gal KO pig 173 PBMC (same source as skin graft). The xenogeneic
proliferative responses
were highest in cultures from sampling Days Sep 4 and Sep 19. In contrast, the
xenogeneic
proliferative response from sampling Day Oct 3 was reduced and near autologous
MLR response
levels. Overall, the xenogeneic response with KO pig 173 in all time periods
tested was less than
the human IRE 11 allogeneic comparator.
[000587] Furthermore, a study is conducted to measure the levels of human
plasma anti-
porcine IgM and IgG binding to porcine peripheral blood mononuclear cells
(PBMCs) obtained
191
CA 03133638 2021-09-14
WO 2020/198397 PCT/US2020/024780
from alpha-galactosyltransferase knock out (KO) pigs over time. Plasma samples
are obtained
from patients enrolled in Sponsor Study XT-001, both before and after the
transplantation of
porcine skin grafts. The porcine skin grafts were obtained from genetically
modified alpha-
galactosyltransferase knock out (KO) pigs.
[000588] In the study, the plasma samples were decomplemented in a 56 C dry
heat bath for
30 minutes. The samples were cooled and serially diluted in FACS
binding/washing media. The
diluted plasma samples were then incubated with KO porcine PBMCs followed by
incubation with
secondary antibody (PE-Goat anti human IgG and FITC-Goat anti human IgM).
Appropriate
compensation, Fluorescence Minus One (FMO), and Limit of Blank (LOB) controls
were run in
the same assay. Cells were acquired and analyzed on an ACEA NovoCyte Flow
Cytometer.
Binding of anti-porcine IgM and IgG was assessed using Median Fluorescence
Intensity (MFI)
and relative MFI obtained as follows: Relative MFI = Actual MFI value / LOB
(MFI obtained
using secondary antibody only in the absence of plasma).
[000589] The human plasma IgM and IgG binding was measured at four time
points
including pre-grafting and post grafting (Day 7, Day 16, Day 30). All actual
test samples at 1:2,
and 1:10 dilutions showed MFI values higher than LOB values. As shown in FIG.
53, an increase
in anti-xenogeneic IgM and IgG levels was obtained above pre-existing levels
on Day 16 and Day
30 as shown by an increase in relative median fluorescence intensities. The
average post-assay cell
viability value determined by 7AAD was 92.82%. Cells were only gated on ALIVE
cells to
determine IgM and IgG binding to porcine PBMCs.
192