Note: Descriptions are shown in the official language in which they were submitted.
WO 2020/210392
PCT/US2020/027320
TRISPECIFIC BINDING PROTEINS, METHODS, AND USES THEREOF
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This application claims priority to U.S. Provisional Application No.
62/831,572,
filed April 9, 2019, and EP Application No. EP19306311.2, filed October 8,
2019, the
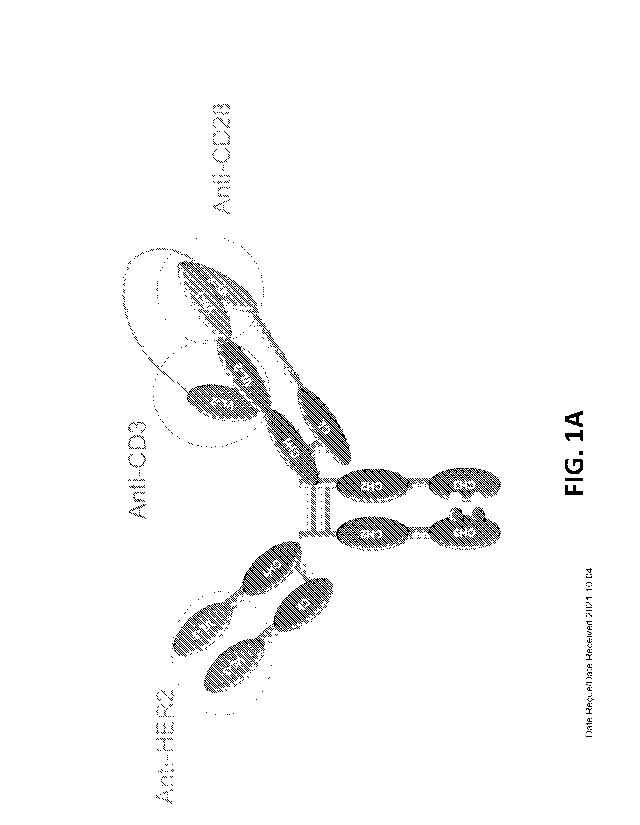
disclosures of each of which are incorporated herein by reference in their
entirety.
SUBMISSION OF SEQUENCE LISTING ON ASCII TEXT FILE
[0002] The content of the following submission on ASCII text file is
incorporated
herein by reference in its entirety: a computer readable form (CRF) of the
Sequence Listing
(file name: 183952032040SEQLIST.TXT, date recorded: April 6, 2020, size: 526
KB).
FIELD
[0003] The disclosure relates to trispecific and/or trivalent binding
proteins comprising
four polypeptide chains that form three antigen binding sites that
specifically bind one or
more target proteins, wherein a first pair of polypeptides forming the binding
protein
possess dual variable domains having a cross-over orientation. The disclosure
also relates
to methods for making trispecific and/or trivalent binding proteins and uses
of such binding
proteins.
BACKGROUND
[0004] Monoclonal antibody based biotherapeutics have become an important
avenue
for new drug development. Monoclonal antibody technology offers specific
targeting,
precise signaling delivery and/or payload to specific cell population, and
provides long
lasting biological effect through its Fc functions. Efforts in antibody
engineering have
allowed developing bispecific antibodies combining the specificities of two
monoclonal
antibodies for various biological applications, expanding the scope of
antibody drug
development. Newly discovered neutralizing antibodies with improved breadth
and
potency may provide more options for developing biotherapeutics to treat
complexed
diseases such as cancer, arthritis, and/or inflammatory disorders.
[0005] Immuno-oncology is a promising, emerging therapeutic approach to
disease
management in cancer. The immune system is the first line of defense against
cancer
development and progression. There is now large evidence that T cells are able
to control
1
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
tumor growth and prolong the survival of cancer patients in both early and
late stages of
disease. However, T cells specific for tumors can be limited in a number of
ways
preventing them from controlling the disease.
[0006] As part of the human adaptive immunity, T cell immunity plays
crucial role in
controlling viral infection and cancer, possibly eliminating infected cells
and malignant
cells which result in clearance of viral infection or cure of cancer. In
chronic infectious
diseases such as Herpes viral infection (HSV, CMV, EBV, etc.), HIV, and HBV,
viruses
establish their persistence in humans by various mechanisms including immune
suppression, T cell exhaustion, and latency establishment. Nevertheless, viral
infection
generally induces viral antigen specific immunity including antigen specific
CD8 T cells
that can readily recognize infected cells for controlling or killing through
cytokine release
or cytotoxic T cell (CTL) mediated killing processes.
[0007] Thus, viral antigen specific T cell activation and/or amplification
in vivo and/or
ex vivo may provide therapeutic strategies against chronic viral infections.
[0008] All references cited herein, including patent applications, patent
publications,
and UniProtKB/Swiss-Prot Accession numbers are herein incorporated by
reference in their
entirety, as if each individual reference were specifically and individually
indicated to be
incorporated by reference.
BRIEF SUMMARY
[0009] To meet these and other needs, provided herein are trispecific
binding proteins
(e.g., antibodies) that form three antigen binding sites. These binding
proteins can
specifically bind one, two, or three antigen targets or target proteins, such
as CD28, CD3,
and a tumor target protein. Some tumors express specific antigens. For
example, HER2
amplification and overexpression can be found in molecular subtypes of breast
cancer, and
also in gastric, ovarian, lung and prostate carcinomas. Optimal activation of
T cells requires
two factors: 1. Antigen recognition and 2. Co-stimulation. Using the
trispecific
HER2/CD28xCD3 trispecific binding proteins described herein, Signal 1 is
provided by an
agonist anti-CD3 binding site, and Signal 2 is provided by an agonist anti-
CD28 binding
site. The trispecific binding protein recruits T cells to the tumor via HER2,
CD38, or a
binding site recognizing another tumor target protein and activates the
engaged T cells via
anti-CD3 and ¨CD28. The resulting activation induces the killing potential of
the immune
cells against the nearby tumor cells. In addition, anti-CD3 binding sites are
described with
2
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
high affinity binding to human CD3 polypeptides and potential manufacturing
liabilities
(e.g., deamidation sites) removed.
[0010] Further provided herein are anti-CD38/CD28xCD3 trispecific
antibodies that
were developed and evaluated for their potential in activating T cells, and
subsequent
proliferation and/or amplification of antigen specific T cells. These
trispecific Abs can
effectively expand CD4 and CD8 effector and memory populations, including
antigen
specific CD8 T central memory and effector memory cells in vitro.
Specifically, in vitro
expansion of CMV, EBV, HIV-I, Influenza specific CD8 central memory and
effector
memory cells were demonstrated. The anti-CD38/CD28xCD3 trispecific antibodies
described herein exhibited novel properties by engaging CD3/CD28/CD38,
providing
signaling pathways to stimulate and expand T cells, which may offer an
effective strategy
treating chronic infectious diseases such as HSV, CMV, EBV, HIV-I, and HBV
infections.
[0011] To meet these and other needs, provided herein are binding proteins
that bind
CD38 polypeptides (e.g., human and cynomolgus monkey CD38 polypeptides),
including
monospecific, bispecific, or trispecific binding proteins with at least one
antigen binding
site that binds a CD38 polypeptide. Advantageously, these binding proteins
have the ability
to recruit T cells to the proximity of cancer cells, subsequently to activate
T cells and
promote the activated T cells killing of adjacent cancer cells through a
Granzyme/Perforin
mechanism, providing a different mode of action for anti-tumor activity from
anti-CD38
antibodies such as DARZALEX (daratumumab). Moreover, the ability to bind both
human and cynomolgus monkey CD38 polypeptides allows binding proteins to be
readily
tested in preclinical toxicological studies, e.g., to evaluate their safety
profiles for later
clinical use.
[0012] In some embodiments, provided herein are binding proteins comprising
four
polypeptide chains that form the three antigen binding sites, wherein a first
polypeptide
chain comprises a structure represented by the formula:
VL2-L1-VL1-L2-CL [I]
and a second polypeptide chain comprises a structure represented by the
formula:
VH1-L3-VH2-L4-CH1-hinge-CH2-CH3 [II]
and a third polypeptide chain comprises a structure represented by the
formula:
VH3-CH1-hinge-CH2-CH3 [III]
and a fourth polypeptide chain comprises a structure represented by the
formula:
VL3-CL [IV]
wherein:
3
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
VIA is a first immunoglobulin light chain variable domain;
VL2 is a second immunoglobulin light chain variable domain;
VL3 is a third immunoglobulin light chain variable domain;
VHi is a first immunoglobulin heavy chain variable domain;
VH2 is a second immunoglobulin heavy chain variable domain;
VH3 is a third immunoglobulin heavy chain variable domain;
CL is an immunoglobulin light chain constant domain;
CHi is an immunoglobulin CHi heavy chain constant domain;
CH2 is an immunoglobulin CH2 heavy chain constant domain;
CH3 is an immunoglobulin CH3 heavy chain constant domain;
hinge is an immunoglobulin hinge region connecting the CHi and CH2 domains;
and
Li, L2, L3 and L4 are amino acid linkers;
wherein the polypeptide of formula I and the polypeptide of formula II form a
cross-over
light chain-heavy chain pair; and
wherein VH1 and VLi form a first antigen binding site;
wherein VH2 and VL2 form a second antigen binding site that binds a CD3
polypeptide,
wherein the VH2 domain comprises a CDR-H1 sequence comprising the amino acid
sequence of GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the amino
acid sequence of IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising
the amino acid sequence of RGVYYALSPFDY (SEQ ID NO:57), and the VL2 domain
comprises a CDR-L1 sequence comprising the amino acid sequence of
QSLVHX1NX2X3TY, wherein Xi is E or Q, X2 is A or L, and X3 is Q, R, or F (SEQ
ID
NO:180), a CDR-L2 sequence comprising the amino acid sequence of KVS (SEQ ID
NO:64), and a CDR-L3 sequence comprising the amino acid sequence of GQGTQYPFT
(SEQ ID NO:65); and
wherein VH3 and VL3 form a third antigen binding site.
[0013] In
some embodiments, the first binding site binds a CD28 polypeptide. In some
embodiments, the VH1 domain comprises a CDR-H1 sequence comprising the amino
acid
sequence of GYTFTSYY (SEQ ID NO:49), a CDR-H2 sequence comprising the amino
acid sequence of IYPGNVNT (SEQ ID NO:50), and a CDR-H3 sequence comprising the
amino acid sequence of TRSHYGLDWNFDV (SEQ ID NO:51), and the VLi domain
comprises a CDR-L1 sequence comprising the amino acid sequence of QNIYVW (SEQ
ID
NO:52), a CDR-L2 sequence comprising the amino acid sequence of KAS (SEQ ID
NO:53), and a CDR-L3 sequence comprising the amino acid sequence of QQGQTYPY
4
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
(SEQ ID NO:54). In some embodiments, the VH1 domain comprises the amino acid
sequence of
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSTYPGN
VNTNYAQKFQGRATLTVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNEDV
WGKGTTVTVSS (SEQ ID NO:91), and/or the VLi domain comprises the amino acid
sequence of
DIQMTQSPSSLSASVGDRVTITCQASQNIYVWLNWYQQKPGKAPKWYKASNLHT
GVPSRF SGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTEGQGTKLEIK (SEQ ID
NO:92).
[0014] In some embodiments, the CDR-L1 sequence of the VL2 domain comprises
an
amino acid sequence selected from the group consisting of QSLVHQNAQTY (SEQ ID
NO:59), QSLVEIENLQTY (SEQ ID NO:60), QSLVHENLFTY (SEQ ID NO:61), and
QSLVHENLRTY (SEQ ID NO:62). In some embodiments, a binding protein of the
present disclosure comprises an antigen binding site comprising: an antibody
heavy chain
variable (VH) domain comprising a CDR-H1 sequence comprising the amino acid
sequence
of GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the amino acid
sequence of IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising the
amino acid sequence of RGVYYALSPFDY (SEQ ID NO:57); and/or an antibody light
chain variable (VL) domain comprising a CDR-L1 sequence comprising the amino
acid
sequence of QSLVHQNAQTY (SEQ ID NO:59), a CDR-L2 sequence comprising the
amino acid sequence of KVS (SEQ ID NO:64), and a CDR-L3 sequence comprising
the
amino acid sequence of GQGTQYPFT (SEQ ID NO:65). In some embodiments, a
binding
protein of the present disclosure comprises an antigen binding site
comprising: an antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the
amino acid sequence of IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence
comprising the amino acid sequence of RGVYYALSPFDY (SEQ ID NO:57); and/or an
antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the
amino acid sequence of QSLVHENLQTY (SEQ ID NO:60), a CDR-L2 sequence
comprising the amino acid sequence of KVS (SEQ ID NO:64), and a CDR-L3
sequence
comprising the amino acid sequence of GQGTQYPFT (SEQ ID NO:65). In some
embodiments, a binding protein of the present disclosure comprises an antigen
binding site
comprising: an antibody heavy chain variable (VH) domain comprising a CDR-H1
sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:55), a CDR-
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
H2 sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID NO:56),
and
a CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ ID
NO:57); and/or an antibody light chain variable (VL) domain comprising a CDR-
L1
sequence comprising the amino acid sequence of QSLVHENLFTY (SEQ ID NO:61), a
CDR-L2 sequence comprising the amino acid sequence of KVS (SEQ ID NO:64), and
a
CDR-L3 sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID
NO:65).
In some embodiments, a binding protein of the present disclosure comprises an
antigen
binding site comprising: an antibody heavy chain variable (VH) domain
comprising a CDR-
H1 sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:55), a
CDR-H2 sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID
NO:56), and a CDR-H3 sequence comprising the amino acid sequence of
RGVYYALSPFDY (SEQ ID NO:57); and/or an antibody light chain variable (VL)
domain
comprising a CDR-L1 sequence comprising the amino acid sequence of
QSLVEIENLRTY
(SEQ ID NO:62), a CDR-L2 sequence comprising the amino acid sequence of KVS
(SEQ
ID NO:64), and a CDR-L3 sequence comprising the amino acid sequence of
GQGTQYPFT
(SEQ ID NO:65). In some embodiments, the VH2 domain comprises the amino acid
sequence of
QVQLVESGGGVVQPGRSLRLSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKD
KSNSYATYYADSVKGRETISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPF
DYWGQGTLVTVSS (SEQ ID NO:93), and/or the VL2 domain comprises an amino acid
sequence selected from the group consisting of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVS
NRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIK
(SEQ ID NO:95),
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVS
NRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIK
(SEQ ID NO:96),
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLFTYLSWYLQKPGQSPQSLIYKVS
NRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIK
(SEQ ID NO:97), and
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLRTYLSWYLQKPGQSPQSLIYKVS
NRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIK
(SEQ ID NO:98). In some embodiments, the VH2 domain comprises the amino acid
sequence of
6
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
QVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMHWVRQAPGKQLEWVAQIKD
KSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPF
DYWGQGTLVTVSS (SEQ ID NO:93) or
QVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMHWVRQAPGKQLEWVAQIKD
KSNSYATYYASSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPF
DYWGQGTLVTVSS (SEQ ID NO:302), and/or the VL2 domain comprises an amino acid
sequence selected from the group consisting of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVS
NRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK
(SEQ ID NO:95),
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVS
NRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK
(SEQ ID NO:96),
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLFTYLSWYLQKPGQSPQSLIYKVS
NRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK
(SEQ ID NO:97), and
DIVMTQTPLSLSVTPGQPASISCK SSQSLVHENLRTYLSWYLQKPGQSPQSLIYKVS
NRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK
(SEQ ID NO:98). In some embodiments, a binding protein of the present
disclosure
comprises an antigen binding site comprising: an antibody heavy chain variable
(VH)
domain comprising the amino acid sequence of SEQ ID NO:93, and/or an antibody
light
chain variable (VL) domain comprising the amino acid sequence of SEQ ID NO:95.
In
some embodiments, a binding protein of the present disclosure comprises an
antigen
binding site comprising: an antibody heavy chain variable (VH) domain
comprising the
amino acid sequence of SEQ ID NO:302, and/or an antibody light chain variable
(VL)
domain comprising the amino acid sequence of SEQ ID NO:95. In some
embodiments, a
binding protein of the present disclosure comprises an antigen binding site
comprising: an
antibody heavy chain variable (VH) domain comprising the amino acid sequence
of SEQ
ID NO:93, and/or an antibody light chain variable (VL) domain comprising the
amino acid
sequence of SEQ ID NO:96. In some embodiments, a binding protein of the
present
disclosure comprises an antigen binding site comprising: an antibody heavy
chain variable
(VH) domain comprising the amino acid sequence of SEQ ID NO:93, and/or an
antibody
light chain variable (VL) domain comprising the amino acid sequence of SEQ ID
NO:97. In
some embodiments, a binding protein of the present disclosure comprises an
antigen
7
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
binding site comprising: an antibody heavy chain variable (VH) domain
comprising the
amino acid sequence of SEQ ID NO:93, and/or an antibody light chain variable
(VL)
domain comprising the amino acid sequence of SEQ ID NO:98.
[0015] In some embodiments, the third antigen binding site binds a tumor
target
protein. In some embodiments, the tumor target protein is a CD38 polypeptide
(e.g., a
human CD38 polypeptide). In some embodiments, the tumor target protein is a
HER2
polypeptide (e.g., a human HER2 polypeptide). In some embodiments, a tumor
target
protein of the present disclosure includes, without limitation, A2AR, APRIL,
ATPDase,
BAFF, BAFFR, BCMA, BlyS, BTK, BTLA, B7DC, B7H1, B7H4 (also known as VTCN1),
B7H5, B7H6, B7H7, B7RP1, B7-4, C3, C5, CCL2 (also known as MCP-1), CCL3 (also
known as MIP-1a), CCL4 (also known as MIP-1b), CCL5 (also known as RANTES),
CCL7 (also known as MCP-3), CCL8 (also known as mcp-2), CCL11 (also known as
eotaxin), CCL15 (also known as MIP-1d), CCL17 (also known as TARC), CCL19
(also
known as MIP-3b), CCL20 (also known as MIP-3a), CCL21 (also known as MIP-2),
CCL24 (also known as MPIF-2/eotaxin-2), CCL25 (also known as TECK), CCL26
(also
known as eotaxin-3), CCR3, CCR4, CD3, CD19, CD20, CD23 (also known as FCER2, a
receptor for IgE), CD24, CD27, CD28, CD38, CD39, CD40, CD70, CD80 (also known
as
B7-1), CD86 (also known as B7-2), CD122, CD137 (also known as 41BB), CD137L,
CD152 (also known as CTLA4), CD154 (also known as CD4OL), CD160, CD272, CD273
(also known as PDL2), CD274 (also known as PDL1), CD275 (also known as B7H2),
CD276 (also known as B7H3), CD278 (also known as ICOS), CD279 (also known as
PD-
1), CDH1 (also known as E-cadherin), chitinase, CLEC9, CLEC91, CRTH2, CSF-1
(also
known as M-CSF), CSF-2 (also known as GM-CSF), CSF-3 (also known as GCSF),
CX3CL1 (also known as SCYD1), CXCL12 (also known as SDF1), CXCL13, CXCR3,
DNGR-1, ectonucleoside triphosphate diphosphohydrolase 1, EGFR, ENTPD1,
FCER1A,
FCER1, FLAP, FOLH1, Gi24, GITR, GITRL, GM-CSF, Her2, HHLA2, HMGB1, HVEM,
ICOSLG, IDO, IFNa, IgE, IGF1R, IL2Rbeta, ILl, ILlA, IL1B, IL1F10, IL2, IL4,
IL4Ra,
IL5, IL5R, IL6, IL7, IL7Ra, IL8, IL9, IL9R, IL10, rhIL10, IL12, IL13, IL13Ra1,
IL13Ra2,
IL15, IL17, IL17Rb (also known as a receptor for IL25), IL18, IL22, IL23,
IL25, IL27,
IL33, IL35, ITGB4 (also known as b4 integrin), ITK, KIR, LAG3, LAMP1, leptin,
LPFS2,
MHC class II, MUC-1, NCR3LG1, NKG2D, NTPDase-1, 0X40, OX4OL, PD-1H, platelet
receptor, PROM1, S152, SISP1, SLC, SPG64, ST2 (also known as a receptor for
IL33),
STEAP2, Syk kinase, TACI, TDO, T14, TIGIT, TIM3, TLR, TLR2, TLR4, TLR5, TLR9,
TMEF1, TNFa, TNFRSF7, Tp55, TREM1, TSLP (also known as a co-receptor for
IL7Ra),
8
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
TSLPR, TWEAK, VEGF, VISTA, Vstm3, WUCAM, and XCR1 (also known as
GPR5/CCXCR1). In some embodiments, one or more of the above antigen targets
are
human antigen targets.
[0016] In some embodiments, the third antigen binding site binds a human
CD38
polypeptide. In some embodiments, the VH3 domain comprises a CDR-H1 sequence
comprising the amino acid sequence of GYTFTSYA (SEQ ID NO:13), a CDR-H2
sequence comprising the amino acid sequence of IYPGQGGT (SEQ ID NO:14), and a
CDR-H3 sequence comprising the amino acid sequence of ARTGGLRRAYFTY (SEQ ID
NO:15), and the VL3 domain comprises a CDR-L1 sequence comprising the amino
acid
sequence of QSVSSYGQGF (SEQ ID NO:16), a CDR-L2 sequence comprising the amino
acid sequence of GAS (SEQ ID NO:17), and a CDR-L3 sequence comprising the
amino
acid sequence of QQNKEDPWT (SEQ ID NO:18). In some embodiments, the VH3 domain
comprises a CDR-H1 sequence comprising the amino acid sequence of GYTLTEFS
(SEQ
ID NO:19), a CDR-H2 sequence comprising the amino acid sequence of FDPEDGET
(SEQ
ID NO:20), and a CDR-H3 sequence comprising the amino acid sequence of
TTGRFFDWF
(SEQ ID NO:21), and the VL3 domain comprises a CDR-L1 sequence comprising the
amino
acid sequence of QSVISRF (SEQ ID NO:22), a CDR-L2 sequence comprising the
amino
acid sequence of GAS (SEQ ID NO:23), and a CDR-L3 sequence comprising the
amino
acid sequence of QQDSNLPIT (SEQ ID NO:24). In some embodiments, the VH3 domain
comprises a CDR-H1 sequence comprising the amino acid sequence of GYAFTTYL
(SEQ
ID NO:25), a CDR-H2 sequence comprising the amino acid sequence of INPGSGST
(SEQ
ID NO:26), and a CDR-H3 sequence comprising the amino acid sequence of ARYAYGY
(SEQ ID NO:27), and the VL3 domain comprises a CDR-L1 sequence comprising the
amino
acid sequence of QNVGTA (SEQ ID NO:28), a CDR-L2 sequence comprising the amino
acid sequence of SAS (SEQ ID NO:29), and a CDR-L3 sequence comprising the
amino
acid sequence of QQYSTYPFT (SEQ m NO:30). In some embodiments, the VH3 domain
comprises a CDR-H1 sequence comprising the amino acid sequence of GYSFTNYA
(SEQ
ID NO:31), a CDR-H2 sequence comprising the amino acid sequence of ISPYYGDT
(SEQ
ID NO:32), and a CDR-H3 sequence comprising the amino acid sequence of
ARRFEGFYYSMDY (SEQ ID NO:33), and the VL3 domain comprises a CDR-L1
sequence comprising the amino acid sequence of QSLVHSNGNTY (SEQ ID NO:34), a
CDR-L2 sequence comprising the amino acid sequence of KVS (SEQ ID NO:35), and
a
CDR-L3 sequence comprising the amino acid sequence of SQSTHVPLT (SEQ ID
NO:36).
9
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
[0017] In some embodiments, the VH3 domain comprises a CDR-H1 sequence
comprising the amino acid sequence of GFTFSSYG (SEQ ID NO:37), a CDR-H2
sequence
comprising the amino acid sequence of IWYDGSNK (SEQ ID NO:38), and a CDR-H3
sequence comprising the amino acid sequence of ARDPGLRYFDGGMDV (SEQ ID
NO:39), and the VL3 domain comprises a CDR-L1 sequence comprising the amino
acid
sequence of QGISSY (SEQ ID NO:40), a CDR-L2 sequence comprising the amino acid
sequence of AAS (SEQ ID NO:41), and a CDR-L3 sequence comprising the amino
acid
sequence of QQLNSFPYT (SEQ ID NO:42). In some embodiments, the VH3 domain
comprises a CDR-H1 sequence comprising the amino acid sequence of GFTFSSYG
(SEQ
ID NO:43), a CDR-H2 sequence comprising the amino acid sequence of IWYDGSNK
(SEQ ID NO:44), and a CDR-H3 sequence comprising the amino acid sequence of
ARMFRGAFDY (SEQ ID NO:45), and the VL3 domain comprises a CDR-L1 sequence
comprising the amino acid sequence of QGIRND (SEQ ID NO:46), a CDR-L2 sequence
comprising the amino acid sequence of AAS (SEQ ID NO:47), and a CDR-L3
sequence
comprising the amino acid sequence of LQDYIYYPT (SEQ ID NO:48). In some
embodiments, the VH3 domain comprises the amino acid sequence of
QVQLVQSGAEVVKPGA SVKVSCK A SGYTFTSYAMHWVKEAPGQRLEWIGYIYPG
QGGTNYNQKFQGRATLTADTSASTAYMELSSLRSEDTAVYFCARTGGLRRAYFTY
WGQGTLVTVSS (SEQ ID NO:79), and/or the VL3 domain comprises the amino acid
sequence of
DIVLTQSPATLSLSPGERATISCRASQSVSSYGQGFMFIWYQQKPGQPPRLLIYGASS
RATGIPARFSGSGSGTDFTLTISPLEPEDFAVYYCQQNKEDPWTEGGGTKLEIK
(SEQ ID NO:80). In some embodiments, the VH3 domain comprises the amino acid
sequence of
QVQLVQSGAEVKKPGASVKVSCKVSGYTLTEF SIHWVRQAPGQGLEWMGGFDPE
DGETIYAQKFQGRVIMTEDTSTDTAYMEMNSLRSEDTAIYYCTTGRFEDWFWGQG
TLVTVSS (SEQ ID NO:81), and/or the VL3 domain comprises the amino acid
sequence of
EIILTQSPAILSLSPGERATLSCRASQSVISRFLSWYQVKPGLAPRLLIYGASTRATGIP
VRF SGSGSGTDF SLTISSLQPEDCAVYYCQQDSNLPITFGQGTRLEIK (SEQ ID
NO:82). In some embodiments, the VH3 domain comprises the amino acid sequence
of
QVQLVQSGAEVKKPGASVKVSCKASGYAFTTYLVEWIRQRPGQGLEWMGVINPG
SGSTNYAQKFQGRVTMTVDRSSTTAYMELSRLRSDDTAVYYCARYAYGYWGQG
TLVTVSS (SEQ ID NO:83), and/or the VL3 domain comprises the amino acid
sequence of
DIQMTQSPSSLSASVGDRVTITCRASQNVGTAVAWYQQKPGKSPKQUYSASNRYT
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
GVPSRFSGSGSGTDFTLTISSLQPEDLATYYCQQYSTYPFTFGQGTKLEIK (SEQ ID
NO:84). In some embodiments, the VH3 domain comprises the amino acid sequence
of
QVQLVESGGGVVQPGRSLRLSCAASGFTFSSYGMYWVRQAPGKGLEWVAVIWYD
GSNKYYADSVKGRFTISRDNSKNTLYLQMNSLRAEDTAVYHCARDPGLRYFDGG
MDVWGQGTTVTVSS (SEQ ID NO:87), and/or the VL3 domain comprises the amino acid
sequence of
DIQLTQSPSFLSASVGDRVTITCRASQGISSYLAWYQQKPGKAPKLLIFAASTLHSG
VPSRFSGSGSGTEFTLTISSLQPEDFATYYCQQLNSFPYTFGQGTKLEIK (SEQ ID
NO:88). In some embodiments, the VH3 domain comprises the amino acid sequence
of
QVQLVESGGGVVQPGRSLRLSCAASGFTFSSYGMHWVRQAPGKGLEWVAVIWYD
GSNKYYADSVKGRFTISGDNSKNTLYLQMNSLRAEDTAVYYCARMERGAFDYWG
QGTLVTVSS (SEQ ID NO:89), and/or the VL3 domain comprises the amino acid
sequence
of
AIQMTQSPSSLSASVGDRVTITCRASQGIRNDLGWYQQKPGKAPKLLIYAASSLQS
GVPSRFSGSGSGTDFTLTISGLQPEDSATYYCLQDYIYYPTFGQGTKVEIK (SEQ ID
NO:90). In some embodiments, the VH3 domain comprises the amino acid sequence
of
QVQLVQSGAEVKKPGASVKVSCKASGYSFTNYAVHWVRQAPGQGLEWMGVISPY
YGDTTYAQKFQGRVTMTVDKSSSTAYMELSRLRSDDTAVYYCARRFEGFYYSMD
YWGQGTLVTVSS (SEQ ID NO:85), and/or the VL3 domain comprises the amino acid
sequence of
DVVMTQSPLSLPVTLGQPASISCRPSQSLVHSNGNTYLNWYQQRPGQSPKWYKV
SKRFSGVPDRFSGSGSGTDFTLKISRVEAEDVGVYYCSQSTHVPLTFGGGTKVEIK
(SEQ ID NO:86).
[0018] In some embodiments, the first polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:156 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:156; the second polypeptide chain comprises
the
amino acid sequence of SEQ ID NO:157 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:157; the third polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:158 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:158; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:159 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:159. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:160 or an amino acid sequence that is at least 95% identical to the
amino acid
11
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
sequence of SEQ ID NO:160; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:161 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:161; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:162 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:162; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:163 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:163. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:164 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:164; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:165 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:165; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:166
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:166; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:167 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:167. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:168 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:168; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:169 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:169; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:170 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:170;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:171 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:171. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:172 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:172; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:173 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:173; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:174 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:174; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:175 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:175. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:176 or an
amino acid
12
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:176; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:177 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:177; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:178
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:178; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:179 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:179. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:181 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:181; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:182 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:182; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:183 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:183;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:184 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:184. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:185 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:185; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:186 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:186; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:187 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:187; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:188 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:188.
[0019] In some embodiments, the third antigen binding site binds a human
HER2
polypeptide. In some embodiments, the VH3 domain comprises a CDR-H1 sequence
comprising the amino acid sequence of GFNIKDTY (SEQ ID NO:1) or GFNIRDTY (SEQ
ID NO:2), a CDR-H2 sequence comprising the amino acid sequence of IYPTNGYT
(SEQ
ID NO:3), IYPTQGYT (SEQ ID NO:4), or IYP'TNAYT (SEQ ID NO:5), and a CDR-H3
sequence comprising the amino acid sequence of SRWGGDGFYAMDY (SEQ ID NO:6),
SRWGGEGFYAMDY (SEQ ID NO:7), or SRWGGSGFYAMDY (SEQ ID NO:8), and the VL3
domain comprises a CDR-L1 sequence comprising the amino acid sequence of
QDVNTA
(SEQ ID NO:9) or QDVQTA (SEQ ID NO:10), a CDR-L2 sequence comprising the amino
13
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
acid sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the
amino
acid sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, the VH3 domain
comprises a CDR-H1 sequence comprising the amino acid sequence of GFNIKDTY
(SEQ
ID NO:1), a CDR-H2 sequence comprising the amino acid sequence of IYPTNGYT
(SEQ
ID NO:3), and a CDR-H3 sequence comprising the amino acid sequence of
SRWGGDGFYAMDY (SEQ ID NO:6), and the VL3 domain comprises a CDR-L1
sequence comprising the amino acid sequence of QDVNTA (SEQ ID NO:9), a CDR-L2
sequence comprising the amino acid sequence of SAS (SEQ ID NO:11), and a CDR-
L3
sequence comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12). In some
embodiments, the VH3 domain comprises a CDR-H1 sequence comprising the amino
acid
sequence of GFNIRDTY (SEQ ID NO:2), a CDR-H2 sequence comprising the amino
acid
sequence of IYPTQGYT (SEQ ID NO:4), and a CDR-H3 sequence comprising the amino
acid sequence of SRWGGEGFYAMDY (SEQ ID NO:7), and the VL3 domain comprises a
CDR-L1 sequence comprising the amino acid sequence of QDVNTA (SEQ ID NO:9), a
CDR-L2 sequence comprising the amino acid sequence of SAS (SEQ ID NO:11), and
a
CDR-L3 sequence comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12).
In
some embodiments, the VH3 domain comprises a CDR-H1 sequence comprising the
amino
acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-H2 sequence comprising the
amino
acid sequence of IYPTNAYT (SEQ ID NO:5), and a CDR-H3 sequence comprising the
amino acid sequence of SRWGGSGFYAMDY (SEQ ID NO:8), and the VL3 domain
comprises a CDR-L1 sequence comprising the amino acid sequence of QDVNTA (SEQ
ID
NO:9), a CDR-L2 sequence comprising the amino acid sequence of SAS (SEQ ID
NO:11),
and a CDR-L3 sequence comprising the amino acid sequence of QQHYTTP (SEQ ID
NO:12). In some embodiments, the VH3 domain comprises a CDR-H1 sequence
comprising the amino acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-H2
sequence
comprising the amino acid sequence of IYPTQGYT (SEQ ID NO:4), and a CDR-H3
sequence comprising the amino acid sequence of SRWGGSGFYAMDY (SEQ ID NO:8),
and the VL3 domain comprises a CDR-L1 sequence comprising the amino acid
sequence of
QDVNTA (SEQ ID NO:9), a CDR-L2 sequence comprising the amino acid sequence of
SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the amino acid sequence
of
QQHYTTP (SEQ ID NO:12). In some embodiments, the VH3 domain comprises a CDR-
H1 sequence comprising the amino acid sequence of GFNIRDTY (SEQ ID NO:2), a
CDR-
H2 sequence comprising the amino acid sequence of IYPTNAYT (SEQ ID NO:5), and
a
CDR-H3 sequence comprising the amino acid sequence of SRWGGEGFYAMDY (SEQ ID
14
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
NO:7), and the VL3 domain comprises a CDR-L1 sequence comprising the amino
acid
sequence of QDVNTA (SEQ ID NO:9), a CDR-L2 sequence comprising the amino acid
sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the amino
acid
sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, the VH3 domain
comprises a CDR-H1 sequence comprising the amino acid sequence of GFNIKDTY
(SEQ
ID NO:1), a CDR-H2 sequence comprising the amino acid sequence of IYPTNGYT
(SEQ
ID NO:3), and a CDR-H3 sequence comprising the amino acid sequence of
SRWGGDGFYAMDY (SEQ ID NO:6), and the VL3 domain comprises a CDR-L1
sequence comprising the amino acid sequence of QDVQTA (SEQ ID NO:10), a CDR-L2
sequence comprising the amino acid sequence of SAS (SEQ ID NO:11), and a CDR-
L3
sequence comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12). In some
embodiments, the VH3 domain comprises the amino acid sequence of
EVQLVESGGGLVQPGGSLRLSCAASGFNIKDTYIHWVRQAPGKGLEWVARTYPTN
GYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGDGFYAMDY
WGQGTLVTVSS (SEQ ID NO:72),
EVQLVESGGGLVQPGGSLRLSCAASGFNIRDTYIHWVRQAPGKGLEWVARTYPTQ
GYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGEGFYAMDY
WGQGTLVTVSS (SEQ ID NO:73),
EVQLVESGGGLVQPGGSLRLSCAASGFNIRDTYIHWVRQAPGKGLEWVARTYPTQ
GYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGSGFYAMDY
WGQGTLVTVSS (SEQ ID NO:74),
EVQLVESGGGLVQPGGSLRLSCAASGFNIRDTYIHWVRQAPGKGLEWVARTYPTN
AYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGSGFYAMDY
WGQGTLVTVSS (SEQ ID NO:75), or
EVQLVESGGGLVQPGGSLRLSCAASGFNIRDTYIHWVRQAPGKGLEWVARTYPTN
AYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGEGFYAMDY
WGQGTLVTVSS (SEQ ID NO:76), and/or the VL3 domain comprises the amino acid
sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYS
GVPSRF SGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:77) or
DIQMTQSPSSLSASVGDRVTITCRASQDVQTAVAWYQQKPGKAPKWYSASFLYS
GVPSRF SGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:78). In some embodiments, the VH3 domain comprises the amino acid sequence
of
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
EVQLVESGGGLVQPGGSLRLSCAASGFNIKDTYIHWVRQAPGKGLEWVARIYPTN
GYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGDGFYAMDY
WGQGTLVTVSS (SEQ ID NO:72), and/or the VL3 domain comprises the amino acid
sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKLLIYSASFLYS
GVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:77). In some embodiments, the VH3 domain comprises the amino acid sequence
of
EVQLVESGGGLVQPGGSLRLSCAASGFNIRDTYIHWVRQAPGKGLEWVARTYPTQ
GYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGEGFYAMDY
WGQGTLVTVSS (SEQ ID NO:73), and/or the VL3 domain comprises the amino acid
sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKLLIYSASFLYS
GVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:77). In some embodiments, the VH3 domain comprises the amino acid sequence
of
EVQLVESGGGLVQPGGSLRLSCAASGFNIRDTYIHWVRQAPGKGLEWVARTYPTN
AYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGSGFYAMDY
WGQGTLVTVSS (SEQ ID NO:75), and/or the VL3 domain comprises the amino acid
sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKLLIYSASFLYS
GVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:77). In some embodiments, the VH3 domain comprises the amino acid sequence
of
EVQLVESGGGLVQPGGSLRLSCAASGFNIRDTYIHWVRQAPGKGLEWVARTYPTQ
GYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGSGFYAMDY
WGQGTLVTVSS (SEQ ID NO:74), and/or the VL3 domain comprises the amino acid
sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKLLIYSASFLYS
GVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:77). In some embodiments, the VH3 domain comprises the amino acid sequence
of
EVQLVESGGGLVQPGGSLRLSCAASGFNIRDTYIHWVRQAPGKGLEWVARTYPTN
AYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGEGFYAMDY
WGQGTLVTVSS (SEQ ID NO:76), and/or the VL3 domain comprises the amino acid
sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKLLIYSASFLYS
GVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
16
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
NO:77). In some embodiments, the VH3 domain comprises the amino acid sequence
of
EVQLVESGGGLVQPGGSLRL SCAASGFNIKDTYIHWVRQAPGKGLEWVARIYPTN
GYTRYADSVKGRF TISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGDGFYAMDY
WGQGTLVTVSS (SEQ ID NO:72), and/or the VL3 domain comprises the amino acid
sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVQTAVAWYQQKPGKAPKWYSASFLYS
GVPSRF SGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:78).
[0020] In some embodiments, the first polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:100 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:100; the second polypeptide chain comprises
the
amino acid sequence of SEQ ID NO:101 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:101; the third polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:102 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:102; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:103 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:103. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:104 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:104; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:105 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:105; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:106 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:106; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:107 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:107. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:112 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:112; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:113 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:113; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:114
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:114; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:115 or an amino acid sequence that is at least 95% identical to the
amino acid
17
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
sequence of SEQ ID NO:115. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:116 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:116; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:117 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:117; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:118 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:118;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:119 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:119. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:120 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:120; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:121 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:121; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:122 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:122; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:123 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:123. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:124 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:124; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:125 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:125; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:126
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:126; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:127 or an amino acid sequence that is at least 9 5 % identical to the
amino acid
sequence of SEQ ID NO:127. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:128 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:128; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:129 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:129; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:130 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:130;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:131 or an
amino acid
18
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:131. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:132 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:132; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:133 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:133; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:134 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:134; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:135 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:135. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:136 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:136; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:137 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:137; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:138
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:138; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:139 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:139. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:140 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:140; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:141 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:141; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:142 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:142;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:143 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:143. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:144 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:144; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:145 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:145; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:146 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:146; and the fourth polypeptide chain
comprises
19
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
the amino acid sequence of SEQ ID NO:147 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:147. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:148 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:148; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:149 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:149; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:150
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:150; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:151 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:151. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:152 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:152; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:153 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:153; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:154 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:154;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:155 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:155. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:286 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:286; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:287 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:287; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:288 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:288; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:289 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:289. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:290 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:290; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:291 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:291; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:292
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
ID NO:292; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:293 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:293. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:294 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:294; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:295 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:295; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:296 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:296;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:297 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:297. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:298 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:298; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:299 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:299; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:300 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:300; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:301 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:301.
[0021] In some embodiments that may be combined with any other embodiments
described herein, at least one of Li, L2, L3 or L4 is independently 0 amino
acids in length.
In some embodiments, Li, L2, L3 and L4 each independently are zero amino acids
in length
or comprise a sequence selected from the group consisting of GGGGSGGGGS (SEQ
ID
NO:69), GGGGSGGGGSGGGGS (SEQ ID NO: 70), S, RT, TKGPS (SEQ ID NO: 68),
GQPKAAP (SEQ ID NO: 67), and GGSGSSGSGG (SEQ ID NO: 71). In some
embodiments, Li, L2, L3 and L4 each independently comprise a sequence selected
from the
group consisting of GGGGSGGGGS (SEQ ID NO:69), GGGGSGGGGSGGGGS (SEQ ID
NO:70), S, RT, TKGPS (SEQ ID NO:68), GQPKAAP (SEQ ID NO: 67), and
GGSGSSGSGG (SEQ ID NO:71). In some embodiments, Li comprises the sequence
GQPKAAP (SEQ ID NO: 67), L2 comprises the sequence TKGPS (SEQ ID NO:68), L3
comprises the sequence S, and L4 comprises the sequence RT. In some
embodiments, at
least one of Li, L2, L3 or L4 comprises the sequence DKTHT (SEQ ID NO:66). In
some
embodiments, Li, L2, L3 and L4 comprise the sequence DKTHT (SEQ ID NO:66).
21
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
[0022] In some embodiments that may be combined with any other embodiments
described herein, the hinge-CH2-CH3 domains of the second and the third
polypeptide chains
are human IgG4 hinge-CH2-CH3 domains, and wherein the hinge-CH2-CH3 domains
each
comprise amino acid substitutions at positions corresponding to positions 234
and 235 of
human IgG4 according to EU Index, wherein the amino acid substitutions are
F234A and
L235A. In some embodiments, the hinge-CH2-CH3 domains of the second and the
third
polypeptide chains are human IgG4 hinge-CH2-CH3 domains, and wherein the hinge-
CH2-
CH3 domains each comprise amino acid substitutions at positions corresponding
to positions
233-236 of human IgG4 according to EU Index, wherein the amino acid
substitutions are
E233P, F234V, L235A, and a deletion at 236. In some embodiments, the hinge-CH2-
CH.3
domains of the second and the third polypeptide chains are human IgG4 hinge-
CH2-CH3
domains, and wherein the hinge-CH2-CH3 domains each comprise amino acid
substitutions at
positions corresponding to positions 228 and 409 of human IgG4 according to EU
Index,
wherein the amino acid substitutions are S228P and R409K. In some embodiments,
the
hinge-Cm-CH3 domains of the second and the third polypeptide chains are human
IgG1
hinge-Cm-CH3 domains, and wherein the hinge-CH2-CH3 domains each comprise
amino acid
substitutions at positions corresponding to positions 234, 235, and 329 of
human IgG1
according to EU Index, wherein the amino acid substitutions are L234A, L235A,
and
P329A. In some embodiments, the hinge-CH2-CH3 domains of the second and the
third
polypeptide chains are human IgG1 hinge-CH2-CH3 domains, and wherein the hinge-
CH2-
CH3 domains each comprise amino acid substitutions at positions corresponding
to positions
298, 299, and 300 of human IgG1 according to EU Index, wherein the amino acid
substitutions are S298N, T299A, and Y300S. In some embodiments, the hinge-CH2-
CH3
domain of the second polypeptide chain comprises amino acid substitutions at
positions
corresponding to positions 349, 366, 368, and 407 of human IgG1 or IgG4
according to EU
Index, wherein the amino acid substitutions are Y349C, T366S, L368A, and
Y407V; and
wherein the hinge-CH2-CH3 domain of the third polypeptide chain comprises
amino acid
substitutions at positions corresponding to positions 354 and 366 of human
IgG1 or IgG4
according to EU Index, wherein the amino acid substitutions are S354C and
T366W. In
some embodiments, the hinge-CH2-CH3 domain of the second polypeptide chain
comprises
amino acid substitutions at positions corresponding to positions 354 and 366
of human
IgG1 or IgG4 according to EU Index, wherein the amino acid substitutions are
S354C and
T366W; and wherein the hinge-CH2-CH3 domain of the third polypeptide chain
comprises
amino acid substitutions at positions corresponding to positions 349, 366,
368, and 407 of
22
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
human IgG1 or IgG4 according to EU Index, wherein the amino acid substitutions
are
Y349C, T366S, L368A, and Y407V.
[0023] In some embodiments, provided herein are isolated nucleic acid
molecules
comprising a nucleotide sequence encoding the binding protein of any one of
the above
embodiments. In some embodiments, provided herein are expression vectors
comprising
the nucleic acid molecule of any one of the above embodiments. In some
embodiments,
provided herein are isolated host cells comprising the nucleic acid molecule
of any one of
the above embodiments or the expression vector of any one of the above
embodiments. In
some embodiments, the host cell is a mammalian or insect cell.
[0024] In some embodiments, provided herein are pharmaceutical compositions
comprising the binding protein of any one of the above embodiments and a
pharmaceutically
acceptable carrier.
[0025] In some embodiments, provided herein are methods of preventing
and/or
treating cancer in a patient comprising administering to the patient a
therapeutically
effective amount of at least one binding protein or pharmaceutical composition
of any one
of the above embodiments. In some embodiments, provided herein is a binding
protein or
pharmaceutical composition according to any one of the above embodiments for
use in a
method of preventing and/or treating cancer in a patient, wherein the method
comprises
administering to the patient a therapeutically effective amount of the binding
protein or
pharmaceutical composition. In some embodiments, provided herein is a binding
protein or
pharmaceutical composition according to any one of the above embodiments for
use in
manufacturing a medicament for preventing and/or treating cancer in a patient.
[0026] In some embodiments, the at least one binding protein is co-
administered with a
chemotherapeutic agent. In some embodiments, the patient is a human.
[0027] In some embodiments, the third antigen binding site binds a human
CD38
polypeptide, and wherein cancer cells from the individual or patient express
CD38. In
some embodiments, the cancer is multiple myeloma. In some embodiments, the
cancer is
acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), chronic
lymphocytic
leukemia (CLL), or a B cell lymphoma. In some embodiments, prior to
administration of
the binding protein, the patient has been treated with daratumumab without a
wash-out
period.
[0028] In some embodiments, the third antigen binding site binds a human
HER2
polypeptide, and wherein cancer cells from the individual or patient express
HER2. In
23
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
some embodiments, the cancer is breast cancer, colorectal cancer, gastric
cancer, or non-
small cell lung cancer (NSCLC).
[0029] In
some embodiments, provided herein is a method for expanding virus-specific
memory T cells, comprising contacting a virus-specific memory T cell with a
binding
protein, wherein the binding protein comprises four polypeptide chains that
form the three
antigen binding sites, wherein a first polypeptide chain comprises a structure
represented by
the formula:
VL2-L1-VL1-L2-CL [I]
and a second polypeptide chain comprises a structure represented by the
formula:
VH1-L3-VH2-L4-CH1-hinge-CH2-CH3 [II]
and a third polypeptide chain comprises a structure represented by the
formula:
VH3-CH1-hinge-CH2-CH3 [III]
and a fourth polypeptide chain comprises a structure represented by the
formula:
VL3-CL [IV]
wherein:
VLi is a first immunoglobulin light chain variable domain;
VL2 is a second immunoglobulin light chain variable domain;
VL3 is a third immunoglobulin light chain variable domain;
VHI is a first immunoglobulin heavy chain variable domain;
VH2 is a second immunoglobulin heavy chain variable domain;
VH3 is a third immunoglobulin heavy chain variable domain;
CL is an immunoglobulin light chain constant domain;
CHi is an immunoglobulin CHi heavy chain constant domain;
CH2 is an immunoglobulin CH2 heavy chain constant domain;
CH3 is an immunoglobulin CH3 heavy chain constant domain;
hinge is an immunoglobulin hinge region connecting the CHi and CH2 domains;
and
Li, L2, L3 and L4 are amino acid linkers;
wherein the polypeptide of formula I and the polypeptide of formula II form a
cross-over
light chain-heavy chain pair; and
wherein VHI and VLi form a first antigen binding site that binds a CD28
polypeptide, wherein
VH2 and VL2 form a second antigen binding site that binds a CD3 polypeptide,
wherein the
VH2 domain comprises a CDR-Ell sequence comprising the amino acid sequence of
GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the amino acid sequence
of
IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising the amino acid
24
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
sequence of RGVYYALSPFDY (SEQ ID NO:57), and the VL2 domain comprises a CDR-L1
sequence comprising the amino acid sequence of QSLVHX1NX2X3TY, wherein Xi is E
or
Q, X2 is A or L, and X3 is Q, R, or F (SEQ ID NO:180), a CDR-L2 sequence
comprising the
amino acid sequence of KVS (SEQ ID NO:64), and a CDR-L3 sequence comprising
the
amino acid sequence of GQGTQYPFT (SEQ ID NO:65), and wherein VH3 and VL3 form
a
third antigen binding site that binds a CD38 polypeptide
[0030] In some embodiments, provided herein is a binding protein that
comprises four
polypeptide chains that form the three antigen binding sites, wherein a first
polypeptide
chain comprises a structure represented by the formula:
VL2-L i-VLi-L2-CL [I]
and a second polypeptide chain comprises a structure represented by the
formula:
VH1-L3-VH2-L4-CH1-hinge-CH2-CH3 [II]
and a third polypeptide chain comprises a structure represented by the
formula:
VH3-CH1-hinge-CH2-CH3 [III]
and a fourth polypeptide chain comprises a structure represented by the
formula:
VL3-CL [IV]
wherein:
VLi is a first immunoglobulin light chain variable domain;
VL2 is a second immunoglobulin light chain variable domain;
VL3 is a third immunoglobulin light chain variable domain;
Vni is a first immunoglobulin heavy chain variable domain;
VH2 is a second immunoglobulin heavy chain variable domain;
VH3 is a third immunoglobulin heavy chain variable domain;
CL is an immunoglobulin light chain constant domain;
Cni is an immunoglobulin Cni heavy chain constant domain;
CH2 is an immunoglobulin CH2 heavy chain constant domain;
CH3 is an immunoglobulin CH3 heavy chain constant domain;
hinge is an immunoglobulin hinge region connecting the CFn and CH2 domains;
and
Li, L2, L3 and L4 are amino acid linkers;
wherein the polypeptide of formula I and the polypeptide of formula II form a
cross-over
light chain-heavy chain pair; and
wherein VH1 and VLi form a first antigen binding site that binds a CD28
polypeptide, wherein
VH2 and VL2 form a second antigen binding site that binds a CD3 polypeptide,
wherein the
VH2 domain comprises a CDR-H1 sequence comprising the amino acid sequence of
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the amino acid sequence
of
IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising the amino acid
sequence of RGVYYALSPFDY (SEQ ID NO:57), and the VL2 domain comprises a CDR-L1
sequence comprising the amino acid sequence of QSLVHX1NX2X3TY, wherein Xi is E
or
Q, X2 is A or L, and X3 is Q, R, or F (SEQ ID NO:180), a CDR-L2 sequence
comprising the
amino acid sequence of KVS (SEQ ID NO:64), and a CDR-L3 sequence comprising
the
amino acid sequence of GQGTQYPFT (SEQ ID NO:65), and wherein VH3 and VL3 form
a
third antigen binding site that binds a CD38 polypeptide for use in expanding
virus-specific
memory T cells.
[0031] In some embodiments, the virus-specific memory T cell is contacted
with the
binding protein in vitro or ex vivo. In some embodiments, contacting the virus-
specific
memory T cell with the binding protein causes activation and/or proliferation
of virus-
specific memory T cells.
[0032] In some embodiments, provided herein is a method for expanding T
cells,
comprising contacting a T cell with a binding protein in vitro or ex vivo,
wherein the
binding protein comprises four polypeptide chains that form the three antigen
binding sites,
wherein a first polypeptide chain comprises a structure represented by the
formula:
VL2-Li-VLi-L2-CL [I]
and a second polypeptide chain comprises a structure represented by the
formula:
VH1-L3-VH2-L4-CH1-hinge-CH2-CH3 [II]
and a third polypeptide chain comprises a structure represented by the
formula:
VH3-CH1-hinge-CH2-CH3 [III]
and a fourth polypeptide chain comprises a structure represented by the
formula:
VL3-CL [IV]
wherein:
VLi is a first immunoglobulin light chain variable domain;
VL2 is a second immunoglobulin light chain variable domain;
VL3 is a third immunoglobulin light chain variable domain;
VHI is a first immunoglobulin heavy chain variable domain;
VH2 is a second immunoglobulin heavy chain variable domain;
VH3 is a third immunoglobulin heavy chain variable domain;
CL is an immunoglobulin light chain constant domain;
CHi is an immunoglobulin CHi heavy chain constant domain;
CH2 is an immunoglobulin CH2 heavy chain constant domain;
26
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
CH3 is an immunoglobulin CH3 heavy chain constant domain;
hinge is an immunoglobulin hinge region connecting the CHi and CH2 domains;
and
Li, L2, L3 and L4 are amino acid linkers;
wherein the polypeptide of formula I and the polypeptide of formula II form a
cross-over
light chain-heavy chain pair; and
wherein VH1 and VLi form a first antigen binding site that binds a CD28
polypeptide, wherein
VH2 and VL2 form a second antigen binding site that binds a CD3 polypeptide,
wherein the
VH2 domain comprises a CDR-H1 sequence comprising the amino acid sequence of
GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the amino acid sequence
of
IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising the amino acid
sequence of RGVYYALSPFDY (SEQ ID NO:57), and the VL2 domain comprises a CDR-L1
sequence comprising the amino acid sequence of QSLVHX1NX2X3TY, wherein Xi is E
or
Q, X2 is A or L, and X3 is Q, R, or F (SEQ ID NO:180), a CDR-L2 sequence
comprising the
amino acid sequence of KVS (SEQ ID NO:64), and a CDR-L3 sequence comprising
the
amino acid sequence of GQGTQYPFT (SEQ ID NO:65), and wherein VH3 and VL3 form
a
third antigen binding site that binds a CD38 polypeptide.
[0033] In some embodiments, provided herein is a binding protein that
comprises four
polypeptide chains that form the three antigen binding sites, wherein a first
polypeptide
chain comprises a structure represented by the formula:
VL2-L 1-VL1-L2-CL [I]
and a second polypeptide chain comprises a structure represented by the
formula:
VH1-L3-VH2-L4-CH1-hinge-CH2-CH3 [II]
and a third polypeptide chain comprises a structure represented by the
formula:
VH3-CH1-hinge-CH2-CH3 [III]
and a fourth polypeptide chain comprises a structure represented by the
formula:
VL3-CL [IV]
wherein:
VLi is a first immunoglobulin light chain variable domain;
VL2 is a second immunoglobulin light chain variable domain;
VL3 is a third immunoglobulin light chain variable domain;
VHI is a first immunoglobulin heavy chain variable domain;
VH2 is a second immunoglobulin heavy chain variable domain;
VH3 is a third immunoglobulin heavy chain variable domain;
CL is an immunoglobulin light chain constant domain;
27
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
CHi is an immunoglobulin CHi heavy chain constant domain;
CH2 is an immunoglobulin CH2 heavy chain constant domain;
CH3 is an immunoglobulin CH3 heavy chain constant domain;
hinge is an immunoglobulin hinge region connecting the CHi and CH2 domains;
and
Li, L2, L3 and L4 are amino acid linkers;
wherein the polypeptide of formula I and the polypeptide of formula II form a
cross-over
light chain-heavy chain pair; and
wherein VHI and VIA form a first antigen binding site that binds a CD28
polypeptide, wherein
VH2 and VL2 form a second antigen binding site that binds a CD3 polypeptide,
wherein the
VH2 domain comprises a CDR-H1 sequence comprising the amino acid sequence of
GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the amino acid sequence
of
IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising the amino acid
sequence of RGVYYALSPFDY (SEQ ID NO:57), and the VL2 domain comprises a CDR-L1
sequence comprising the amino acid sequence of QSLVHX1NX2X3TY, wherein Xi is E
or
Q, X2 is A or L, and X3 is Q, R, or F (SEQ ID NO:180), a CDR-L2 sequence
comprising the
amino acid sequence of KVS (SEQ ID NO:64), and a CDR-L3 sequence comprising
the
amino acid sequence of GQGTQYPFT (SEQ ID NO:65), and wherein VH3 and VL3 form
a
third antigen binding site that binds a CD38 polypeptide for use in a method
for expanding T
cells.
[0034] In some embodiments, the T cell is a memory T cell or an effector T
cell. In
some embodiments, the T cell expresses a chimeric antigen receptor (CAR) on
its cell
surface or comprises a polynucleotide encoding a CAR.
[0035] In some embodiments, provided herein is a method for treating
chronic viral
infection, comprising administering to an individual or patient in need
thereof an effective
amount of a binding protein, wherein the binding protein comprises four
polypeptide chains
that form the three antigen binding sites, wherein a first polypeptide chain
comprises a
structure represented by the formula:
VL2-L 1-VLi-L2-CL [I]
and a second polypeptide chain comprises a structure represented by the
formula:
VH1-L3-VH2-L4-CH 1-hi nge-CH2-CH3 [II]
and a third polypeptide chain comprises a structure represented by the
formula:
VH3-CH1-hinge-CH2-CH3 [III]
and a fourth polypeptide chain comprises a structure represented by the
formula:
VL3-CL [IV]
28
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
wherein:
VLi is a first immunoglobulin light chain variable domain;
VL2 is a second immunoglobulin light chain variable domain;
VL3 is a third immunoglobulin light chain variable domain;
VH1 is a first immunoglobulin heavy chain variable domain;
VH2 is a second immunoglobulin heavy chain variable domain;
VH3 is a third immunoglobulin heavy chain variable domain;
CL is an immunoglobulin light chain constant domain;
CHi is an immunoglobulin CHi heavy chain constant domain;
CH2 is an immunoglobulin CH2 heavy chain constant domain;
CH3 is an immunoglobulin CH3 heavy chain constant domain;
hinge is an immunoglobulin hinge region connecting the CHi and CH2 domains;
and
Li, L2, L3 and L4 are amino acid linkers;
wherein the polypeptide of formula I and the polypeptide of formula II form a
cross-over
light chain-heavy chain pair; and
wherein VH1 and VLi form a first antigen binding site that binds a CD28
polypeptide, wherein
VH2 and VL2 form a second antigen binding site that binds a CD3 polypeptide,
wherein the
VH2 domain comprises a CDR-H1 sequence comprising the amino acid sequence of
GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the amino acid sequence
of
IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising the amino acid
sequence of RGVYYALSPFDY (SEQ ID NO:57), and the VL2 domain comprises a CDR-L1
sequence comprising the amino acid sequence of QSLVHX1NX2X3TY, wherein Xi is E
or
Q, X2 is A or L, and X3 is Q, R, or F (SEQ ID NO:180), a CDR-L2 sequence
comprising the
amino acid sequence of KVS (SEQ ID NO:64), and a CDR-L3 sequence comprising
the
amino acid sequence of GQGTQYPFT (SEQ ID NO:65), and wherein VH3 and VL3 form
a
third antigen binding site that binds a CD38 polypeptide.
[0036] In some embodiments, provided herein is a binding protein that
comprises four
polypeptide chains that form the three antigen binding sites, wherein a first
polypeptide
chain comprises a structure represented by the formula:
VL2-L i-VL -L2-CL [I]
and a second polypeptide chain comprises a structure represented by the
formula:
VH1-L3-VH2-L4-CH1-hinge-CH2-CH3 [II]
and a third polypeptide chain comprises a structure represented by the
formula:
VH3-CHi-hinge-CH2-CH3 [III]
29
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
and a fourth polypeptide chain comprises a structure represented by the
formula:
VL3-CL [IV]
wherein:
VLi is a first immunoglobulin light chain variable domain;
VL2 is a second immunoglobulin light chain variable domain;
VL3 is a third immunoglobulin light chain variable domain;
VH1 is a first immunoglobulin heavy chain variable domain;
VH2 is a second immunoglobulin heavy chain variable domain;
VH3 is a third immunoglobulin heavy chain variable domain;
CL is an immunoglobulin light chain constant domain;
CHi is an immunoglobulin CHi heavy chain constant domain;
CH2 is an immunoglobulin CH2 heavy chain constant domain;
CH3 is an immunoglobulin CH3 heavy chain constant domain;
hinge is an immunoglobulin hinge region connecting the CHi and CH2 domains;
and
Li, L2, L3 and L4 are amino acid linkers;
wherein the polypeptide of formula I and the polypeptide of formula II form a
cross-over
light chain-heavy chain pair; and
wherein VH1 and VLi form a first antigen binding site that binds a CD28
polypeptide, wherein
VH2 and VL2 form a second antigen binding site that binds a CD3 polypeptide,
wherein the
VH2 domain comprises a CDR-H1 sequence comprising the amino acid sequence of
GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the amino acid sequence
of
IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising the amino acid
sequence of RGVYYALSPFDY (SEQ ID NO:57), and the VL2 domain comprises a CDR-L1
sequence comprising the amino acid sequence of QSLVHX1NX2X3TY, wherein Xi is E
or
Q, X2 is A or L, and X3 is Q, R, or F (SEQ ID NO:180), a CDR-L2 sequence
comprising the
amino acid sequence of KVS (SEQ ID NO:64), and a CDR-L3 sequence comprising
the
amino acid sequence of GQGTQYPFT (SEQ ID NO:65), and wherein VH3 and VL3 form
a
third antigen binding site that binds a CD38 polypeptide for use in a method
for treating
chronic viral infection, wherein said method comprises administering to an
individual or
patient in need thereof an effective amount of the binding protein.
[0037] In some embodiments, the individual or patient is a human. In some
embodiments, the binding protein is administered to the individual or patient
in
pharmaceutical formulation comprising the binding protein and a
pharmaceutically
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
acceptable carrier. In some embodiments, administration of the binding protein
results in
activation and/or proliferation of virus-specific memory T cells in the
individual or patient.
[0038] In some embodiments that may be combined with any other embodiments
described herein, the memory T cells are CD8+ or CD4+ memory T cells. In some
embodiments, the memory T cells are central memory T cells (Tcm) or effector
memory T
cells (TEm).
[0039] In some embodiments that may be combined with any other embodiments
described herein, the virus is a human immunodeficiency virus (HIV), influenza
virus,
cytomegalovirus (CMV), hepatitis B virus (HBV), human papillomavirus (HPV),
Epstein-
barr virus (EBV), human foamy virus (HFV), herpes simplex virus 1 (HSV-1), or
herpes
simplex virus 1 (HSV-2).
[0040] In some embodiments that may be combined with any other embodiments
described herein, the CD28 polypeptide is a human CD28 polypeptide, wherein
the CD3
polypeptide is a human CD3 polypeptide, and wherein the CD38 polypeptide is a
human
CD38 polypeptide.
[0041] In some embodiments, provided herein is a vector system comprising
one or
more vectors encoding a first, second, third, and fourth polypeptide chain of
a binding
protein of any one of the above embodiments. In some embodiments, the vector
system
comprises a first vector encoding the first polypeptide chain of the binding
protein, a
second vector encoding the second polypeptide chain of the binding protein, a
third vector
encoding the third polypeptide chain of the binding protein, and a fourth
vector encoding
the fourth polypeptide chain of the binding protein.
[0042] In some embodiments, provided herein are kits comprising one, two,
three, or
four polypeptide chains of a binding protein according to any one of the above
embodiments. In some embodiments, the kits further comprise instructions for
using the
polypeptide chain or binding protein according to any of the methods or uses
described
herein, e.g., supra.
[0043] In some embodiments, provided herein are kits comprising one, two,
three, or
four polynucleotides according to any one of the above embodiments. In some
embodiments, provided herein are kits of polynucleotides comprising one, two,
three, or
four polynucleotides of a kit of polynucleotides comprising: (a) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:189, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:190, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:191, and a fourth
polynucleotide
31
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
comprising the polynucleotide sequence of SEQ ID NO:192; (b) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:193, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:194, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:195, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:196; (c) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:197, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:198, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:199, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:200; (d) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:201, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:202, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:203, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:204; (e) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:205, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:206, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:207, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:208; (f) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:209, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:210, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:211, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:212; (g) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:213, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:214, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:215, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:216; (h) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:217, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:218, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:219, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:220; (i) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:221, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:222, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:223, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:224; (j) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:225, a second
polynucleotide
32
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
comprising the polynucleotide sequence of SEQ ID NO:226, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:227, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:228; (k) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:229, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:230, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:231, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:232; (1) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:233, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:234, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:235, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:236; (m) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:237, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:238, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:239, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:240; (n) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:241, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:242, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:243, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:244; (o) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:245, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:246, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:247, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:248; (p) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:249, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:250, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:251, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:252; (q) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:253, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:254, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:255, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:256; (r) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:257, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:258, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:259, and a fourth
polynucleotide
33
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
comprising the polynucleotide sequence of SEQ ID NO:260; (s) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:261, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:262, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:263, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:264; (t) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:265, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:266, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:267, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:268; (u) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:269, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:270, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:271, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:272; or (v) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:273, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:274, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:275, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:276.
[0044] It is to be understood that one, some, or all of the properties of
the various
embodiments described herein may be combined to form other embodiments of the
present
invention. These and other aspects of the invention will become apparent to
one of skill in
the art. These and other embodiments of the invention are further described by
the detailed
description that follows.
BRIEF DESCRIPTION OF THE DRAWINGS
[0045] FIG. lA provides a schematic representation of a trispecific binding
protein
comprising four polypeptide chains that form three antigen binding sites that
binds three
target proteins: CD28, CD3, and HER2. A first pair of polypeptides possess
dual variable
domains having a cross-over orientation (VH1-VH2 and VL2-VL1) forming two
antigen
binding sites that recognize CD3 and CD28, and a second pair of polypeptides
possess a
single variable domain (VH3 and VL3) forming a single antigen binding site
that
recognizes HER2. The trispecific binding protein shown in FIG. lA uses a
constant region
with a "knobs-into-holes" mutation, where the knob is on the second pair of
polypeptides
with a single variable domain.
34
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
[0046] FIG. 1B provides the fold change (vs. parental) in binding
affinities of anti-
CD28/CD3/HER2 trispecific antibody variants using the indicated anti-HER2,
anti-CD3,
and anti-CD28 binding domains Mutations 3233QQ to QEQ (top to bottom) refer to
mutations introduced into residues 32-35 of the VL domain of the anti-CD3
binding site
(indicated by *); the remaining mutations were introduced into the VH or VL
domain of the
trastuzumab anti-HER2 binding site (indicated by #; numbering according to
Kabat). For
the mutations in the anti-HER2 binding site, mutation 30Q was introduced into
the VL
domain, and the remaining mutations were introduced into the VH domain. The
binding
affinities were measured by ELISA, and the values provided are relative to
parental
trispecific antibody.
[0047] FIG. 1C provides binding curves for the indicated trispecific
antibodies binding
to human HER2, human CD28, and CD3, as determined by ELISA.
[0048] FIG. 1D provides a proposed mechanism of action for HER2/CD28xCD3
trispecific antibody-mediated T cell activation and HER2+ cancer cell killing.
[0049] FIG. 2A provides a schematic representation of a trispecific binding
protein
comprising four polypeptide chains that form three antigen binding sites that
binds three
target proteins. CD28, CD3, and CD38 A first pair of polypeptides possess dual
variable
domains having a cross-over orientation (VH1-VH2 and VL2-VL1) forming two
antigen
binding sites that recognize CD3 and CD28, and a second pair of polypeptides
possess a
single variable domain (VH3 and VL3) forming a single antigen binding site
that
recognizes CD38. The trispecific binding protein shown in FIG. 2A uses an IgG4
constant
region with a "knobs-into-holes" mutation, where the knob is on the second
pair of
polypeptides with a single variable domain.
[0050] FIGS. 2B-2E show the binding affinities, as measured by ELISA, of
CD38/CD28sup x CD3mid ENLQ DKTHT IgG4 FALA trispecific antibodies with the
indicated anti-CD38 binding domains for the target antigens human CD38 (FIG.
2B),
cynomolgus monkey CD38 (FIG. 2C), human CD3 (FIG. 2D), and human CD28 (FIG.
2E).
[0051] FIG. 3 shows SPR competition assays for binding to CD38 by
Daratumumab
and anti-CD38 monospecific antibodies with the indicated anti-CD38 binding
domains If
an antibody recognized an epitope on CD38 which was different from that of
Daratumumab, injection of the antibody resulted in an increased SPR signal. If
an antibody
recognized an overlapping epitope as Daratumumab, injection of the antibody
did not
increase SPR signal.
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
[0052] FIGS. 4A-4B show the in vitro cell killing activity of CD38/CD28sup
x
CD3mid ENLQ DKTHT IgG4 FALA trispecific antibodies with the indicated anti-
CD38
binding domains against human multiple myeloma NCI-H929 cells (CD38+/CD28+).
The
assays were carried out in the presence of 5 nM isotype control antibody (FIG.
4A) or
Daratumumab (FIG. 4B). In the presence of daratumumab, the trispecific
antibodies
continued to exhibit cell killing activity.
[0053] FIGS. 5A-5B show the in vitro cell killing activity of CD38/CD28sup
x
CD3mid ENLQ DKTHT IgG4 FALA trispecific antibodies with the indicated anti-
CD38
binding domains against human lymphoma OCI-Ly19 cells (CD38+/CD28-). The
assays
were carried out in the presence of 5 nM isotype control antibody (FIG. 5A) or
Daratumumab (FIG. 5B). Daratumumab caused a decrease in the cell killing
activity of
anti-CD38/CD28xCD3 trispecific antibodies.
[0054] FIGS. 6A-6J show the characterization of in vitro T cell subset
expansion in
PBMCs collected from CMV-infected Donor D in response to CD38/CD28sup x
CD3mid ENLQ DKTHT IgG4 FALA trispecific antibodies with the indicated
alternative
anti-CD38 binding domains. A trispecific antibody lacking the CD38VH1 anti-
CD38
binding domain was used as a negative control (ACD38VH1/ACD28sup x ACD3mid
IgG4
FALA). T cell populations were measured at indicated time points (D3 refers to
day 3; D7
refers to day 7). The indicated trispecific antibodies were tested at the
indicated
concentrations of 0.2 nIVI and 1 nM. Flow cytometry was used to quantify CMV-
specific
CD8+ T cells (FIGS. 6A-6B), CMV-specific Tun CD8+ cells (FIGS. 6C-6D), and CMV-
specific Tern CD8+ cells (FIGS. 6E-6F). In addition, the percentages of CMV-
specific Tun
(FIGS. 6G-61I) and Tun (FIGS. 6I-6J) CD8+ cells were quantified at the
indicated time
points. All tested trispecific antibodies promoted the proliferation of CMV-
specific memory
CD8+ T cells with different potency and kinetics in a dose-responsive manner.
[0055] FIGS. 7A-7J show the characterization of in vitro T cell subset
expansion in
PBMCs collected from CMV-infected Donor E in response to CD38/CD28sup x
CD3mid ENLQ DKTHT IgG4 FALA trispecific antibodies with the indicated anti-
CD38
binding domains. A trispecific antibody lacking the CD38VH1 anti-CD38 binding
domain
was used as a negative control (ACD38VH1/ACD28sup x ACD3mid IgG4 FALA).
Antibodies shown in legend from top to bottom are shown in the graphs from
left to right.
T cell populations were measured at indicated time points (D3 refers to day 3;
D7 refers to
day 7). The indicated trispecific antibodies were tested at the indicated
concentrations of 0.2
nM, 1 nM, and 2 nM. Flow cytometry was used to quantify CMV-specific CD8+ T
cells
36
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
(FIGS. 7A-7B), CMV-specific Tcm CD8+ cells (FIGS. 7C-7D), and CMV-specific
Tern
CD8+ cells (FIGS. 7E-7F). In addition, the percentages of CMV-specific Tun
(FIGS. 7G-
711) and Tun (FIGS. 7I-7J) CD8+ cells were quantified at the indicated time
points. All
tested trispecific antibodies promoted the proliferation of CMV-specific
memory CD8+ T
cells with different potency and kinetics in dose response manner.
[0056] FIGS. 8A-8J show the characterization of in vitro T cell subset
expansion in
PBMCs collected from EBV-infected Donor C in response to CD38/CD28sup x
CD3mid ENLQ DKTHT IgG4 FALA trispecific antibodies with the indicated
alternative
anti-CD38 binding domains. A trispecific antibody lacking the CD38VH1 anti-
CD38
binding domain was used as a negative control (ACD38VH1/ACD28sup x ACD3mid
IgG4
FALA). T cell populations were measured at indicated time points (D3 refers to
day 3; D7
refers to day 7). The indicated trispecific antibodies were tested at the
indicated
concentrations of 0.2 nIVI and 1 nM. Flow cytometry was used to quantify EBV-
specific
CD8+ T cells (FIGS. 8A-8B), CMV-specific Tcm CD8+ cells (FIGS. 8C-8D), and CMV-
specific Tern CD8+ cells (FIGS. 8E-8F). In addition, the percentages of EBV-
specific Tun
(FIGS. 8G-81I) and Tun (FIGS. 8I-8J) CD8+ cells were quantified at the
indicated time
points. All tested trispecific antibodies promoted the proliferation of CMV-
specific memory
CD8+ T cells with different potency and kinetics in dose response manner.
[0057] FIGS. 9A-12 show the characterization of in vitro T cell subset
expansion in
PBMCs collected from EBV-infected Donor D in response to CD38/CD28sup x
CD3mid ENLQ DKTHT IgG4 FALA trispecific antibodies with the indicated
alternative
anti-CD38 binding domains. A trispecific antibody lacking the CD38VH1 anti-
CD38
binding domain was used as a negative control (ACD38VH1/ACD28sup x ACD3mid
IgG4
FALA). T cell populations were measured at indicated time points (D3 refers to
day 3; D7
refers to day 7). The indicated trispecific antibodies were tested at the
indicated
concentrations of 0.2 nIVI and 1 nM. Flow cytometry was used to quantify EBV-
specific
CD8+ T cells (FIGS. 9A-9B), EBV-specific Tun CD8+ cells (FIGS. 9C-9D), and EBV-
specific Tern CD8+ cells (FIGS. 9E-9F). In addition, the percentages of EBV-
specific Tun
(FIGS. 9G-10) and Tun (FIGS. 11-12) CD8+ cells were quantified at the
indicated time
points. All tested trispecific antibodies promoted the proliferation of EBV-
specific memory
CD8+ T cells with different potency and kinetics in dose response manner.
[0058] FIGS. 13A-13D show the change over time (days) in tumor volume (FIG.
13A)
and body weight (FIG. 13B) in ZR-75-1 tumor bearing NSG mice engrafted with in
vitro
expanded human CD3+ T cells. Groups of 10 mice were either treated with
vehicle or
37
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
Her2/CD28 x CD3 trispecific antibody at the indicated dosages. Arrow heads
indicate days
of administration. Tumor volume is depicted as mean SEM, mm3. Body weight
change
is depicted as % change, mean + SEM. X-axis shows days after implantation with
ZR-75-1
cells. Tumor volume (mm3) over time for individual mice in each treatment
group are
shown in FIG. 13C. Tumor weight (mg) for each treatment group is shown in FIG.
13D.
** = p<0.001; *** = p<0.0003 (two-way ANOVA, control vs. 100 & bug/kg).
[0059] FIGS. 14A-14C show the effect of Her2/CD28 x CD3 trispecific
antibody
treatment on T cells from whole blood. FIG. 14A shows the analysis of hCD45+,
CD8+,
CD4+, and mCD45+ cells by flow cytometry. FIG. 14B shows the effect of control
or
Her2/CD28 x CD3 trispecific antibody treatment (at the indicated doses) on
hCD45+,
CD8+, CD4+, and mCD45+ cell counts. FIG. 14C shows the effect of control or
Her2/CD28 x CD3 trispecific antibody treatment (at the indicated doses) on
human cell
ratios (CD4+/CD45+ and CD8+/CD45+). For each x-axis parameter shown in FIGS.
14B
& 14C, conditions are (left to right): control, 100ug/kg trispecific antibody,
lOug/kg
trispecific antibody, lug/kg trispecific antibody, and 0.1ug/kg trispecific
antibody.
Percentages shown in FIGS. 14B & 14C are based on control sample vs. 100ug/kg.
[0060] FIGS. 15A-15C show the effect of Her2/CD28 x CD3 trispecific
antibody
treatment on tumor infiltrating lymphocytes (TILs), as examined by
immunohistochemistry
(IHC). Arrows indicate tumor infiltrating T cells identified in ZR-75-1 breast
tumors.
Upper images are at 1X magnification; lower images are at 20X magnification.
In both sets
of images, staining for human CD45, human CD4, and human CD8 are shown from
left to
right. Shown are tumors from mice treated with vehicle control (FIG. 15A),
100ug/kg
trispecific antibody (FIG. 15B), or 0.1ug/kg trispecific antibody (FIG. 15C).
[0061] FIGS. 16A-16C show quantitation of the effect of Her2/CD28 x CD3
trispecific
antibody treatment on TILs as measured by IHC. Each dot represents one tumor
from an
individual mouse; rectangles represent group means; and error bars indicate
standard
deviation. * = p<0.05 compared to vehicle control group (ANOVA). Numbers of
CD45+
(FIG. 16A), CD4+ (FIG. 16B), or CD8+ (FIG. 16C) cells are shown. In FIG. 16C,
a $
area quantitation approach was used for CD8+ cells instead of cell counting
algorithm due
to excessive non-specific signal in the CD8 IHC slide.
[0062] FIGS. 17A-17F show in vitro cell lysis of HER2+ breast cancer target
cells in
the presence of human CD8+ T cells by Her2/CD28 x CD3 trispecific antibody
with wild-
type trastuzumab antigen binding domain and an anti-CD3 antigen binding domain
without
without 32/35 QQ mutations in the VL domain ("ctl") as compared to a Her2/CD28
x CD3
38
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
trispecific antibodies having mutations in the anti-HER2 arm and the VL domain
of the
anti-CD3 arm (numbering as shown in Table 1). Cell killing activities against
cell lines
with varying expression of HER2 are depicted: HCC1954 for high HER2 expression
(FIG.
17A), BT20 for intermediate HER2 expression (FIG. 17C), and MDA-MD-231 for low
HER2 expression (FIG. 17E). Graphs depicting cell killing as a function of
antibody
concentration against target cells HCC1954 (FIG. 17B), BT20 (FIG. 17D), and
MDA-MD-
231 (FIG. 17F) are shown, comparing binding protein #2 vs. ctl or binding
protein #1 and
#5 vs. ctl.
[0063] FIGS. 18A & 18B summarize the mean EC50 (pM) of in vitro cell
killing by
experimental or control Her2/CD28 x CD3 trispecific antibodies against the
indicated
breast cancer (FIG. 18A) or gastric cancer cell lines (FIG. 18B). Amino acid
sequences of
the indicated trispecific antibodies are provided in Table 1.
DETAILED DESCRIPTION
[0064] The disclosure provides trispecific and/or trivalent binding
proteins comprising
four polypeptide chains that form three antigen binding sites that
specifically bind to one or
more target proteins, wherein a first pair of polypeptides forming the binding
protein
possess dual variable domains having a cross-over orientation.
General Definitions
[0065] As utilized in accordance with the present disclosure, the following
terms,
unless otherwise indicated, shall be understood to have the following
meanings. Unless
otherwise required by context, singular terms shall include pluralities and
plural terms shall
include the singular.
[0066] It is understood that aspects and embodiments of the disclosure
described herein
include "comprising," "consisting," and "consisting essentially of' aspects
and
embodiments.
[0067] The term "polynucleotide" as used herein refers to single-stranded
or double-
stranded nucleic acid polymers of at least 10 nucleotides in length. In
certain embodiments,
the nucleotides comprising the polynucleotide can be ribonucleotides or
deoxyribonucleotides or a modified form of either type of nucleotide. Such
modifications
include base modifications such as bromuridine, ribose modifications such as
arabinoside
and 2',3'-dideoxyribose, and internucleotide linkage modifications such as
phosphorothioate, phosphorodithioate, phosphoroselenoate,
phosphorodiselenoate,
39
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
phosphoroanilothioate, phoshoraniladate and phosphoroamidate. The term
"polynucleotide" specifically includes single-stranded and double-stranded
forms of DNA.
[0068] An "isolated polynucleotide" is a polynucleotide of genomic, cDNA,
or
synthetic origin or some combination thereof, which: (1) is not associated
with all or a
portion of a polynucleotide in which the isolated polynucleotide is found in
nature, (2) is
linked to a polynucleotide to which it is not linked in nature, or (3) does
not occur in nature
as part of a larger sequence.
[0069] An "isolated polypeptide" is one that (1) is free of at least some
other
polypeptides with which it would normally be found, (2) is essentially free of
other
polypeptides from the same source, e.g., from the same species, (3) is
expressed by a cell
from a different species, (4) has been separated from at least about 50
percent of
polynucleotides, lipids, carbohydrates, or other materials with which it is
associated in
nature, (5) is not associated (by covalent or noncovalent interaction) with
portions of a
polypeptide with which the "isolated polypeptide" is associated in nature, (6)
is operably
associated (by covalent or noncovalent interaction) with a polypeptide with
which it is not
associated in nature, or (7) does not occur in nature. Such an isolated
polypeptide can be
encoded by genomic DNA, cDNA, mRNA or other RNA, of synthetic origin, or any
combination thereof. Preferably, the isolated polypeptide is substantially
free from
polypeptides or other contaminants that are found in its natural environment
that would
interfere with its use (therapeutic, diagnostic, prophylactic, research or
otherwise).
[0070] Naturally occurring antibodies typically comprise a tetramer. Each
such
tetramer is typically composed of two identical pairs of polypeptide chains,
each pair
having one full-length "light" chain (typically having a molecular weight of
about 25 kDa)
and one full-length "heavy" chain (typically having a molecular weight of
about 50-70
kDa). The terms "heavy chain" and "light chain" as used herein refer to any
immunoglobulin polypeptide having sufficient variable domain sequence to
confer
specificity for a target antigen. The amino-terminal portion of each light and
heavy chain
typically includes a variable domain of about 100 to 110 or more amino acids
that typically
is responsible for antigen recognition. The carboxy-terminal portion of each
chain typically
defines a constant domain responsible for effector function Thus, in a
naturally occurring
antibody, a full-length heavy chain immunoglobulin polypeptide includes a
variable domain
(VH) and three constant domains (Cm, CH2, and CH3), wherein the VH domain is
at the
amino-terminus of the polypeptide and the CH3 domain is at the carboxyl-
terminus, and a
full-length light chain immunoglobulin polypeptide includes a variable domain
(VI) and a
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
constant domain (CL), wherein the VL domain is at the amino-terminus of the
polypeptide
and the CL domain is at the carboxyl-terminus.
[0071] Human light chains are typically classified as kappa and lambda
light chains,
and human heavy chains are typically classified as mu, delta, gamma, alpha, or
epsilon, and
define the antibody's isotype as IgM, IgD, IgG, IgA, and IgE, respectively.
IgG has several
subclasses, including, but not limited to, IgGl, IgG2, IgG3, and IgG4. IgM has
subclasses
including, but not limited to, IgMl and IgM2. IgA is similarly subdivided into
subclasses
including, but not limited to, IgAl and IgA2. Within full-length light and
heavy chains, the
variable and constant domains typically are joined by a "J" region of about 12
or more
amino acids, with the heavy chain also including a "D" region of about 10 more
amino
acids. See, e.g., FUNDAMENTAL IMMUNOLOGY (Paul, W., ed., Raven Press, 2nd ed.,
1989),
which is incorporated by reference in its entirety for all purposes. The
variable regions of
each light/heavy chain pair typically form an antigen binding site. The
variable domains of
naturally occurring antibodies typically exhibit the same general structure of
relatively
conserved framework regions (FR) joined by three hypervariable regions, also
called
complementarity determining regions or CDRs. The CDRs from the two chains of
each
pair typically are aligned by the framework regions, which may enable binding
to a specific
epitope. From the amino-terminus to the carboxyl-terminus, both light and
heavy chain
variable domains typically comprise the domains FR1, CDR1, FR2, CDR2, FR3,
CDR3,
and FR4.
[0072] The term "CDR set" refers to a group of three CDRs that occur in a
single
variable region capable of binding the antigen. The exact boundaries of these
CDRs have
been defined differently according to different systems. The system described
by Kabat
(Kabat et at., SEQUENCES OF PROTEINS OF IMMUNOLOGICAL INTEREST (National
Institutes of
Health, Bethesda, Md. (1987) and (1991)) not only provides an unambiguous
residue
numbering system applicable to any variable region of an antibody, but also
provides
precise residue boundaries defining the three CDRs. These CDRs may be referred
to as
Kabat CDRs. Chothia and coworkers (Chothia and Lesk, 1987, 1 Mot. Biol. 196:
901-17;
Chothia et at., 1989, Nature 342: 877-83) found that certain sub-portions
within Kabat
CDRs adopt nearly identical peptide backbone conformations, despite having
great
diversity at the level of amino acid sequence. These sub-portions were
designated as Li,
L2, and L3 or H1, H2, and H3 where the "L" and the "H" designates the light
chain and the
heavy chain regions, respectively. These regions may be referred to as Chothia
CDRs,
which have boundaries that overlap with Kabat CDRs. Other boundaries defining
CDRs
41
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
overlapping with the Kabat CDRs have been described by Padlan, 1995, FASEB J.
9: 133-
39; MacCallum, 1996, 1 Mot. Biol. 262(5): 732-45; and Lefranc, 2003, Dev.
Comp.
Immunol. 27: 55-77. Still other CDR boundary definitions may not strictly
follow one of
the herein systems, but will nonetheless overlap with the Kabat CDRs, although
they may
be shortened or lengthened in light of prediction or experimental findings
that particular
residues or groups of residues or even entire CDRs do not significantly impact
antigen
binding. The methods used herein may utilize CDRs defined according to any of
these
systems, although certain embodiments use Kabat or Chothia defined CDRs.
Identification
of predicted CDRs using the amino acid sequence is well known in the field,
such as in
Martin, A.C. "Protein sequence and structure analysis of antibody variable
domains," In
Antibody Engineering, Vol. 2. Kontermann R., Dilbel S., eds. Springer-Verlag,
Berlin, p.
33-51 (2010). The amino acid sequence of the heavy and/or light chain variable
domain
may be also inspected to identify the sequences of the CDRs by other
conventional
methods, e.g., by comparison to known amino acid sequences of other heavy and
light
chain variable regions to determine the regions of sequence hypervariability.
The
numbered sequences may be aligned by eye, or by employing an alignment program
such
as one of the CLUSTAL suite of programs, as described in Thompson, 1994,
Nucleic Acids
Res. 22: 4673-80. Molecular models are conventionally used to correctly
delineate
framework and CDR regions and thus correct the sequence-based assignments.
[0073] The term "Fc" as used herein refers to a molecule comprising the
sequence of a
non-antigen-binding fragment resulting from digestion of an antibody or
produced by other
means, whether in monomeric or multimeric form, and can contain the hinge
region. The
original immunoglobulin source of the native Fc is preferably of human origin
and can be
any of the immunoglobulins, although IgG1 and IgG2 are preferred. Fc molecules
are
made up of monomeric polypeptides that can be linked into dimeric or
multimeric forms by
covalent (i.e., disulfide bonds) and non-covalent association. The number of
intermolecular
disulfide bonds between monomeric subunits of native Fc molecules ranges from
1 to 4
depending on class (e.g., IgG, IgA, and IgE) or subclass (e.g., IgGl, IgG2,
IgG3, IgAl, and
IgGA2). One example of a Fc is a disulfide-bonded dimer resulting from papain
digestion
of an IgG. The term "native Fc" as used herein is generic to the monomeric,
dimeric, and
multimeric forms.
[0074] A F(ab) fragment typically includes one light chain and the VH and
CHi domains
of one heavy chain, wherein the VH-CHi heavy chain portion of the F(ab)
fragment cannot
form a disulfide bond with another heavy chain polypeptide. As used herein, a
F(ab)
42
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
fragment can also include one light chain containing two variable domains
separated by an
amino acid linker and one heavy chain containing two variable domains
separated by an
amino acid linker and a Cm domain.
[0075] A F(ab') fragment typically includes one light chain and a portion
of one heavy
chain that contains more of the constant region (between the CHi and CH2
domains), such
that an interchain disulfide bond can be formed between two heavy chains to
form a F(a1302
molecule.
[0076] The term "binding protein" as used herein refers to a non-naturally
occurring (or
recombinant or engineered) molecule that specifically binds to at least one
target antigen.
A trispecific binding protein of the present disclosure, unless otherwise
specified, typically
comprises four polypeptide chains that form at least three antigen binding
sites, wherein a
first polypeptide chain has a structure represented by the formula:
VL2- L2-CL [I]
and a second polypeptide chain has a structure represented by the formula:
VH1-L3-VH2-L4-CH1-hinge-CH2-CH3 [II]
and a third polypeptide chain has a structure represented by the formula:
VH3-CH1 [III]
and a fourth polypeptide chain has a structure represented by the formula:
VL3-CL [IV]
wherein:
VLi is a first immunoglobulin light chain variable domain;
VL2 is a second immunoglobulin light chain variable domain;
VL3 is a third immunoglobulin light chain variable domain;
is a first immunoglobulin heavy chain variable domain;
VH2 is a second immunoglobulin heavy chain variable domain;
VH3 is a third immunoglobulin heavy chain variable domain;
CL is an immunoglobulin light chain constant domain;
CHi is the immunoglobulin CHi heavy chain constant domain; and
hinge is an immunoglobulin hinge region connecting the CHi and CH2 domains;
Li, L2, L3 and L4 are amino acid linkers;
nd wherein the polypeptide of formula I and the polypeptide of formula II form
a cross-over
light chain-heavy chain pair.
[0077] A "recombinant" molecule is one that has been prepared, expressed,
created, or
isolated by recombinant means.
43
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
[0078] One embodiment of the disclosure provides binding proteins having
biological
and immunological specificity to between one and three target antigens.
Another
embodiment of the disclosure provides nucleic acid molecules comprising
nucleotide
sequences encoding polypeptide chains that form such binding proteins. Another
embodiment of the disclosure provides expression vectors comprising nucleic
acid
molecules comprising nucleotide sequences encoding polypeptide chains that
form such
binding proteins. Yet another embodiment of the disclosure provides host cells
that express
such binding proteins (i.e., comprising nucleic acid molecules or vectors
encoding
polypeptide chains that form such binding proteins).
[0079] The term "swapability" as used herein refers to the
interchangeability of variable
domains within the binding protein format and with retention of folding and
ultimate
binding affinity. "Full swapability" refers to the ability to swap the order
of both VH1 and
VH2 domains, and therefore the order of VIA and VL2 domains, in the
polypeptide chain of
formula I or the polypeptide chain of formula II (i.e., to reverse the order)
while
maintaining full functionality of the binding protein as evidenced by the
retention of
binding affinity. Furthermore, it should be noted that the designations VH and
VL refer only
to the domain's location on a particular protein chain in the final format.
For example, VH1
and VH2 could be derived from Vu and VL2 domains in parent antibodies and
placed into
the VH1 and VH2 positions in the binding protein. Likewise, VIA and VL2 could
be derived
from VH1 and VH2 domains in parent antibodies and placed in the VH1 and VH2
positions in
the binding protein. Thus, the VH and VL designations refer to the present
location and not
the original location in a parent antibody. VH and VL domains are therefore
"swappable."
[0080] The term "antigen" or "target antigen" or "antigen target" as used
herein refers to
a molecule or a portion of a molecule that is capable of being bound by a
binding protein,
and additionally is capable of being used in an animal to produce antibodies
capable of
binding to an epitope of that antigen. A target antigen may have one or more
epitopes.
With respect to each target antigen recognized by a binding protein, the
binding protein is
capable of competing with an intact antibody that recognizes the target
antigen.
[0081] The term "Her2" refers to human epidermal growth factor receptor 2
which is a
member of the epidermal growth factor receptor family.
[0082] "CD3" is cluster of differentiation factor 3 polypeptide and is a T-
cell surface
protein that is typically part of the T cell receptor (TCR) complex.
[0083] "CD28" is cluster of differentiation 28 polypeptide and is a T-cell
surface
protein that provides co-stimulatory signals for T-cell activation and
survival.
44
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
[0084] "CD38" is cluster of differentiation 38 polypeptide and is a
glycoprotein found
on the surface of many immune cells.
[0085] The term "T-cell engager" refers to binding proteins directed to a
host's immune
system, more specifically the T cells' cytotoxic activity as well as directed
to a tumor target
protein.
[0086] The term "monospecific binding protein" refers to a binding protein
that
specifically binds to one antigen target.
[0087] The term "monovalent binding protein" refers to a binding protein
that has one
antigen binding site.
[0088] The term "bispecific binding protein" refers to a binding protein
that specifically
binds to two different antigen targets.
[0089] The term "bivalent binding protein" refers to a binding protein that
has two
binding sites.
[0090] The term "trispecific binding protein" refers to a binding protein
that specifically
binds to three different antigen targets.
[0091] The term "trivalent binding protein" refers to a binding protein
that has three
binding sites. In particular embodiments the trivalent binding protein can
bind to one
antigen target. In other embodiments, the trivalent binding protein can bind
to two antigen
targets. In other embodiments, the trivalent binding protein can bind to three
antigen
targets.
[0092] An "isolated" binding protein is one that has been identified and
separated
and/or recovered from a component of its natural environment. Contaminant
components
of its natural environment are materials that would interfere with diagnostic
or therapeutic
uses for the binding protein, and may include enzymes, hormones, and other
proteinaceous
or non-proteinaceous solutes. In some embodiments, the binding protein will be
purified:
(1) to greater than 95% by weight of antibody as determined by the Lowry
method, and
most preferably more than 99% by weight, (2) to a degree sufficient to obtain
at least 15
residues of N-terminal or internal amino acid sequence by use of a spinning
cup sequenator,
or (3) to homogeneity by SDS-PAGE under reducing or nonreducing conditions
using
Coomassie blue or, preferably, silver stain. Isolated binding proteins include
the binding
protein in situ within recombinant cells since at least one component of the
binding
protein's natural environment will not be present.
[0093] The terms "substantially pure" or "substantially purified" as used
herein refer to
a compound or species that is the predominant species present (i.e., on a
molar basis it is
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
more abundant than any other individual species in the composition). In some
embodiments, a substantially purified fraction is a composition wherein the
species
comprises at least about 50% (on a molar basis) of all macromolecular species
present. In
other embodiments, a substantially pure composition will comprise more than
about 80%,
85%, 90%, 95%, or 99% of all macromolar species present in the composition. In
still
other embodiments, the species is purified to essential homogeneity
(contaminant species
cannot be detected in the composition by conventional detection methods)
wherein the
composition consists essentially of a single macromolecular species.
[0094] The term "epitope" includes any determinant, preferably a
polypeptide
determinant, capable of specifically binding to an immunoglobulin or T-cell
receptor. In
certain embodiments, epitope determinants include chemically active surface
groupings of
molecules such as amino acids, sugar side chains, phosphoryl groups, or
sulfonyl groups,
and, in certain embodiments, may have specific three-dimensional structural
characteristics
and/or specific charge characteristics. An epitope is a region of an antigen
that is bound by
an antibody or binding protein. In certain embodiments, a binding protein is
said to
specifically bind an antigen when it preferentially recognizes its target
antigen in a complex
mixture of proteins and/or macromolecules In some embodiments, a binding
protein is
said to specifically bind an antigen when the equilibrium dissociation
constant is < 10' M,
more preferably when the equilibrium dissociation constant is < 10-9 M, and
most
preferably when the dissociation constant is < 10-10 M.
[0095] The dissociation constant (KD) of a binding protein can be
determined, for
example, by surface plasmon resonance. Generally, surface plasmon resonance
analysis
measures real-time binding interactions between ligand (a target antigen on a
biosensor
matrix) and analyte (a binding protein in solution) by surface plasmon
resonance (SPR) using
the BIAcore system (Pharmacia Biosensor; Piscataway, NJ). Surface plasmon
analysis can
also be performed by immobilizing the analyte (binding protein on a biosensor
matrix) and
presenting the ligand (target antigen). The term "KID," as used herein refers
to the dissociation
constant of the interaction between a particular binding protein and a target
antigen.
[0096] The term "specifically binds" as used herein refers to the ability
of a binding
protein or an antigen-binding fragment thereof to bind to an antigen
containing an epitope
with an Kd of at least about lx 106M, lx 107M, lx 108M, lx 109M, lx 10' M, lx
10-11M, 1 x 10-12M, or more, and/or to bind to an epitope with an affinity
that is at least two-
fold greater than its affinity for a nonspecific antigen.
46
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
[0097] In some embodiments, an antigen binding domain and/or binding
protein of the
present disclosure "cross reacts" with human and cynomolgus monkey CD38
polypeptides,
e.g., CD38 extracellular domains, human CD38 isoform A, human CD38 isoform E,
and
cynomolgus monkey CD38. A binding protein binding to antigen 1 (Agl) is "cross-
reactive"
to antigen 2 (Ag2) when the EC50s are in a similar range for both antigens. In
the
present application, a binding protein binding to Ag 1 is cross-reactive to
Ag2 when the
ratio of affinity of Ag2 to affinity of Agl is equal or less than 20,
affinities being measured
with the same method for both antigens.
[0098] The term "linker" as used herein refers to one or more amino acid
residues inserted
between immunoglobulin domains to provide sufficient mobility for the domains
of the light
and heavy chains to fold into cross over dual variable region immunoglobulins.
A linker is
inserted at the transition between variable domains or between variable and
constant domains,
respectively, at the sequence level. The transition between domains can be
identified because
the approximate size of the immunoglobulin domains are well understood. The
precise
location of a domain transition can be determined by locating peptide
stretches that do not
form secondary structural elements such as beta-sheets or alpha-helices as
demonstrated by
experimental data or as can be assumed by techniques of modeling or secondary
structure
prediction. The linkers described herein are referred to as Li, which is
located on the light
chain between the C-terminus of the VL2 and the N-terminus of the VIA domain;
and L2,
which is located on the light chain between the C-terminus of the VLi and the
N-terminus of
the CL domain. The heavy chain linkers are known as L3, which is located
between the C-
terminus of the VHI and the N-terminus of the VH2 domain; and L4, which is
located between
the C-terminus of the VH2 and the N-terminus of the CHi domain.
[0099] The term "vector" as used herein refers to any molecule (e.g.,
nucleic acid,
plasmid, or virus) that is used to transfer coding information to a host cell.
The term "vector"
includes a nucleic acid molecule that is capable of transporting another
nucleic acid to which
it has been linked. One type of vector is a "plasmid," which refers to a
circular double-
stranded DNA molecule into which additional DNA segments may be inserted.
Another type
of vector is a viral vector, wherein additional DNA segments may be inserted
into the viral
genome. Certain vectors are capable of autonomous replication in a host cell
into which they
are introduced (e.g., bacterial vectors having a bacterial origin of
replication and episomal
mammalian vectors). Other vectors (e.g., non-episomal mammalian vectors) can
be
integrated into the genome of a host cell upon introduction into the host cell
and thereby are
47
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
replicated along with the host genome. In addition, certain vectors are
capable of directing
the expression of genes to which they are operatively linked. Such vectors are
referred to
herein as "recombinant expression vectors" (or simply, "expression vectors").
In general,
expression vectors of utility in recombinant DNA techniques are often in the
form of
plasmids. The terms "plasmid" and "vector" may be used interchangeably herein,
as a
plasmid is the most commonly used form of vector. However, the disclosure is
intended to
include other forms of expression vectors, such as viral vectors (e.g.,
replication defective
retroviruses, adenoviruses, and adeno-associated viruses), which serve
equivalent functions.
[0100] The phrase "recombinant host cell" (or "host cell") as used herein
refers to a cell
into which a recombinant expression vector has been introduced. A recombinant
host cell or
host cell is intended to refer not only to the particular subject cell, but
also to the progeny of
such a cell. Because certain modifications may occur in succeeding generations
due to either
mutation or environmental influences, such progeny may not, in fact, be
identical to the parent
cell, but such cells are still included within the scope of the term "host
cell" as used herein. A
wide variety of host cell expression systems can be used to express the
binding proteins,
including bacterial, yeast, baculoviral, and mammalian expression systems (as
well as phage
display expression systems). An example of a suitable bacterial expression
vector is pUC19.
To express a binding protein recombinantly, a host cell is transformed or
transfected with one
or more recombinant expression vectors carrying DNA fragments encoding the
polypeptide
chains of the binding protein such that the polypeptide chains are expressed
in the host cell
and, preferably, secreted into the medium in which the host cells are
cultured, from which
medium the binding protein can be recovered.
[0101] The term "transformation" as used herein refers to a change in a
cell's genetic
characteristics, and a cell has been transformed when it has been modified to
contain a new
DNA. For example, a cell is transformed where it is genetically modified from
its native state.
Following transformation, the transforming DNA may recombine with that of the
cell by
physically integrating into a chromosome of the cell, or may be maintained
transiently as an
episomal element without being replicated, or may replicate independently as a
plasmid. A
cell is considered to have been stably transformed when the DNA is replicated
with the division
of the cell. The term "transfecti on" as used herein refers to the uptake of
foreign or exogenous
DNA by a cell, and a cell has been "transfected" when the exogenous DNA has
been introduced
inside the cell membrane. A number of transfection techniques are well known
in the art. Such
techniques can be used to introduce one or more exogenous DNA molecules into
suitable host
cells.
48
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
[0102] The term "naturally occurring" as used herein and applied to an
object refers to the
fact that the object can be found in nature and has not been manipulated by
man. For example,
a polynucleotide or polypeptide that is present in an organism (including
viruses) that can be
isolated from a source in nature and that has not been intentionally modified
by man is
naturally-occurring. Similarly, "non-naturally occurring" as used herein
refers to an object that
is not found in nature or that has been structurally modified or synthesized
by man.
[0103] As used herein, the twenty conventional amino acids and their
abbreviations follow
conventional usage. Stereoisomers (e.g., D-amino acids) of the twenty
conventional amino
acids; unnatural amino acids and analogs such as a-, a-disubstituted amino
acids, N-alkyl
amino acids, lactic acid, and other unconventional amino acids may also be
suitable
components for the polypeptide chains of the binding proteins. Examples of
unconventional
amino acids include: 4-hydroxyproline, y-carboxyglutamate, c-N,N,N-
trimethyllysine, c-N-
acetyllysine, 0-phosphoserine, N-acetylserine, N-formylmethionine, 3 -
methylhistidine, 5-
hydroxylysine, u-N-methylarginine, and other similar amino acids and imino
acids (e.g., 4-
hydroxyproline). In the polypeptide notation used herein, the left-hand
direction is the amino
terminal direction and the right-hand direction is the carboxyl-terminal
direction, in accordance
with standard usage and convention.
[0104] Naturally occurring residues may be divided into classes based on
common side
chain properties:
(1) hydrophobic: Met, Ala, Val, Leu, Ile, Phe, Trp, Tyr, Pro;
(2) polar hydrophilic: Arg, Asn, Asp, Gln, Glu, His, Lys, Ser, Thr ;
(3) aliphatic: Ala, Gly, Ile, Leu, Val, Pro;
(4) aliphatic hydrophobic: Ala, Ile, Leu, Val, Pro;
(5) neutral hydrophilic: Cys, Ser, Thr, Asn, Gln;
(6) acidic: Asp, Glu;
(7) basic: His, Lys, Arg;
(8) residues that influence chain orientation: Gly, Pro;
(9) aromatic: His, Trp, Tyr, Phe; and
(10) aromatic hydrophobic: Phe, Trp, Tyr.
[0105] Conservative amino acid substitutions may involve exchange of a
member of one
of these classes with another member of the same class. Non-conservative
substitutions may
involve the exchange of a member of one of these classes for a member from
another class.
49
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
[0106] A skilled artisan will be able to determine suitable variants of the
polypeptide chains
of the binding proteins using well-known techniques. For example, one skilled
in the art may
identify suitable areas of a polypeptide chain that may be changed without
destroying activity
by targeting regions not believed to be important for activity. Alternatively,
one skilled in the
art can identify residues and portions of the molecules that are conserved
among similar
polypeptides. In addition, even areas that may be important for biological
activity or for
structure may be subject to conservative amino acid substitutions without
destroying the
biological activity or without adversely affecting the polypeptide structure.
[0107] The term "patient" as used herein includes human and animal
subjects.
[0108] The terms "treatment" or "treat" as used herein refer to both
therapeutic treatment
and prophylactic or preventative measures. Those in need of treatment include
those having
a disorder as well as those prone to have the disorder or those in which the
disorder is to be
prevented. In particular embodiments, binding proteins can be used to treat
humans with
cancer, or humans susceptible to cancer, or ameliorate cancer in a human
subject. The
binding proteins can also be used to prevent cancer in a human patient. In
particular
embodiments, the cancer is multiple myeloma, acute lymphoblastic leukemia,
chronic
lymphocytic leukemia, acute myeloid leukemia, lymphoma, breast cancer such as
Her2+
breast cancer, germinal center B-cell lympohoma or B-cell acute lymphoblastic
leukemia, In
other embodiments, the binding proteins can be used to treat humans with
inflammatory
disorders, or humans susceptible to inflammatory disorders, or ameliorate
inflammatory
disorders in a human subject.
[0109] The terms "pharmaceutical composition" or "therapeutic composition"
as used
herein refer to a compound or composition capable of inducing a desired
therapeutic effect
when properly administered to a patient.
[0110] The term "pharmaceutically acceptable carrier" or "physiologically
acceptable
carrier" as used herein refers to one or more formulation materials suitable
for accomplishing
or enhancing the delivery of a binding protein.
[0111] The terms "effective amount" and "therapeutically effective amount"
when used in
reference to a pharmaceutical composition comprising one or more binding
proteins refer to
an amount or dosage sufficient to produce a desired therapeutic result More
specifically, a
therapeutically effective amount is an amount of a binding protein sufficient
to inhibit, for
some period of time, one or more of the clinically defined pathological
processes associated
with the condition being treated. The effective amount may vary depending on
the specific
binding protein that is being used, and also depends on a variety of factors
and conditions
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
related to the patient being treated and the severity of the disorder. For
example, if the
binding protein is to be administered in vivo, factors such as the age,
weight, and health of the
patient as well as dose response curves and toxicity data obtained in
preclinical animal work
would be among those factors considered. The determination of an effective
amount or
therapeutically effective amount of a given pharmaceutical composition is well
within the
ability of those skilled in the art.
[0112] One embodiment of the disclosure provides a pharmaceutical
composition
comprising a pharmaceutically acceptable carrier and a therapeutically
effective amount of a
binding protein.
Trispecific and/or Trivalent Binding Proteins
[0113] Certain aspects of the present disclosure relate to trispecific
and/or trivalent binding
proteins comprising four polypeptide chains that form three antigen binding
sites that
specifically bind to one or more target proteins, wherein a first pair of
polypeptides forming
the binding protein possess dual variable domains having a cross-over
orientation and wherein
a second pair of polypeptides forming the binding protein possess a single
variable domain.
Any of the CDRs or variable domains of any of the antigen binding proteins
described herein
may find use in a trispecific binding protein of the present disclosure.
[0114] In some embodiments, each of the three antigen binding sites binds a
different target
(e.g., polypeptide antigen). In some embodiments, the trispecific binding
protein comprises
four polypeptide chains that form the three antigen binding sites, wherein a
first polypeptide
chain comprises a structure represented by the formula:
VL2-L1-VL1-L2-CL [I]
and a second polypeptide chain comprises a structure represented by the
formula:
VH1-L3-VH2-L4-CH1-hinge-CH2-CH3 [II]
and a third polypeptide chain comprises a structure represented by the
formula:
VH3-CH1-hinge-CH2-CH3 [III]
and a fourth polypeptide chain comprises a structure represented by the
formula:
VL3-CL [IV]
wherein:
VIA is a first immunoglobulin light chain variable domain;
VL2 is a second immunoglobulin light chain variable domain;
VL3 is a third immunoglobulin light chain variable domain;
is a first immunoglobulin heavy chain variable domain;
51
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
VH2 is a second immunoglobulin heavy chain variable domain;
VH3 is a third immunoglobulin heavy chain variable domain;
CL is an immunoglobulin light chain constant domain;
CHi is an immunoglobulin CHi heavy chain constant domain;
CH2 is an immunoglobulin CH2 heavy chain constant domain;
CH3 is an immunoglobulin CH3 heavy chain constant domain;
hinge is an immunoglobulin hinge region connecting the CHi and CH2 domains;
and
Li, L2, L3 and L4 are amino acid linkers;
wherein the polypeptide of formula I and the polypeptide of formula II form a
cross-over light
chain-heavy chain pair.
[0115] It is contemplated that any of the antigen binding sites described
herein may find
use in a trispecific binding protein of the present disclosure, e.g.,
comprising four polypeptide
chains having the structures described supra. For example, in some
embodiments, a trispecific
binding protein of the present disclosure comprises a VHi and VLi domain pair
that form a first
antigen binding site, a VH2 and VL2 domain pair that form a second antigen
binding site that
binds a CD3 polypeptide, and a VH3 and VL3 domain pair that form a third
antigen binding site.
In some embodiments, a trispecific binding protein of the present disclosure
comprises a VH1
and VLi domain pair that form a first antigen binding site that binds a CD28
polypeptide, a VH2
and VL2 domain pair that form a second antigen binding site that binds a CD3
polypeptide, and
a VH3 and VL3 domain pair that form a third antigen binding site. In some
embodiments, a
trispecific binding protein of the present disclosure comprises a VH1 and VLi
domain pair that
form a first antigen binding site, a VH2 and VL2 domain pair that form a
second antigen binding
site that binds a CD3 polypeptide, and a VH3 and VL3 domain pair that form a
third antigen
binding site that binds a tumor target protein. In some embodiments, a
trispecific binding
protein of the present disclosure comprises a VH1 and VLi domain pair that
form a first antigen
binding site that binds a CD28 polypeptide, a VH2 and VL2 domain pair that
form a second
antigen binding site that binds a CD3 polypeptide, and a VH3 and VL3 domain
pair that form a
third antigen binding site that binds a tumor target protein. In some
embodiments, a trispecific
binding protein of the present disclosure comprises a VHi and VLi domain pair
that form a first
antigen binding site that binds a CD28 polypeptide, a VH2 and VL2 domain pair
that form a
second antigen binding site that binds a CD3 polypeptide, and a VH3 and VL3
domain pair that
form a third antigen binding site that binds a CD38 polypeptide. In some
embodiments, a
trispecific binding protein of the present disclosure comprises a VH1 and VLi
domain pair that
form a first antigen binding site that binds a CD28 polypeptide, a VH2 and VL2
domain pair that
52
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
form a second antigen binding site that binds a CD3 polypeptide, and a VH3 and
VL3 domain
pair that form a third antigen binding site that binds a HER2 polypeptide.
[0116] In some embodiments, a binding protein of the present disclosure
binds one or more
tumor target proteins and one or more T cell target proteins. In some
embodiments, the binding
protein is capable of specifically binding one tumor target protein and two
different epitopes
on a single T cell target protein. In some embodiments, the binding protein is
capable of
specifically binding one tumor target protein and two different T cell target
proteins (e.g., CD28
and CD3). In some embodiments, the first and second polypeptide chains of the
binding protein
form two antigen binding sites that specifically target two T cell target
proteins, and the third
and fourth polypeptide chains of the binding protein form an antigen binding
site that
specifically binds a tumor target protein. In some embodiments, the target
protein is CD38 or
HER2. Additional tumor target proteins are provided infra. In some
embodiments, the one or
more T cell target proteins are one or more of CD3 and CD28. Exemplary and non-
limiting
polypeptides that may find use in any of the trispecific binding proteins
described herein are
provided in Table 1.
[0117] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:156 or an amino acid sequence
that is at
least 959/0 identical to the amino acid sequence of SEQ ID NO:156; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:157 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:157; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:158 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:158; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:159 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:159.
[0118] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:160 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:160; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:161 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:161; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:162 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:162; and the
fourth
53
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
polypeptide chain comprises the amino acid sequence of SEQ ID NO:163 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:163.
[0119] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:164 or an amino acid sequence
that is at
least 959/0 identical to the amino acid sequence of SEQ ID NO:164; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:165 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:165; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:166 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:166; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:167 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:167.
[0120] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:168 or an amino acid sequence
that is at
least 959/o identical to the amino acid sequence of SEQ ID NO:168; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:169 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:169; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:170 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:170; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:171 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:171.
[0121] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:172 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:172; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:173 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:173; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:174 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:174; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:175 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:175.
[0122] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
54
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
comprises the amino acid sequence of SEQ ID NO:176 or an amino acid sequence
that is at
least 959/0 identical to the amino acid sequence of SEQ ID NO:176; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:177 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:177; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:178 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:178; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:179 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:179.
[0123] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:181 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:181; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:182 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:182; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:183 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:183; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:184 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:184.
[0124] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:185 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:185; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:186 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:186; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:187 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:187; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:188 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:188.
[0125] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:100 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:100; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:101 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:101; the
third polypeptide
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
chain comprises the amino acid sequence of SEQ ID NO:102 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:102; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:103 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:103.
[0126] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:104 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:104; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:105 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:105; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:106 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:106; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:107 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:107.
[0127] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:112 or an amino acid sequence
that is at
least 959/0 identical to the amino acid sequence of SEQ ID NO:112; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:113 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:113; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:114 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:114; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:115 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:115.
[0128] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:116 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:116; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:117 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:117; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:118 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:118; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:119 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:119.
56
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
[0129] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:120 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:120; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:121 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:121; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:122 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:122; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:123 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:123.
[0130] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:124 or an amino acid sequence
that is at
least 959/0 identical to the amino acid sequence of SEQ ID NO:124; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:125 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:125; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:126 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:126; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:127 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:127.
[0131] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:128 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:128; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:129 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:129; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:130 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:130; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:131 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:131.
[0132] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:132 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:132; the second
polypeptide
57
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
chain comprises the amino acid sequence of SEQ ID NO:133 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:133; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:134 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:134; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:135 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:135.
[0133] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:136 or an amino acid sequence
that is at
least 959/0 identical to the amino acid sequence of SEQ ID NO:136; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:137 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:137; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:138 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:138; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:139 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:139.
[0134] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:140 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:140; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:141 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:141; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:142 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:142; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:143 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:143.
[0135] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:144 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:144; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:145 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:145; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:146 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:146; and the
fourth
58
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
polypeptide chain comprises the amino acid sequence of SEQ ID NO:147 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:147.
[0136] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:148 or an amino acid sequence
that is at
least 959/0 identical to the amino acid sequence of SEQ ID NO:148; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:149 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:149; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:150 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:150; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:151 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:151.
[0137] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:152 or an amino acid sequence
that is at
least 959/o identical to the amino acid sequence of SEQ ID NO:152; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:153 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:153; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:154 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:154; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:155 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:155.
[0138] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:286 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:286; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:287 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:287; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:288 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:288; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:289 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:289.
[0139] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein .the first
polypeptide chain
59
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
comprises the amino acid sequence of SEQ ID NO:290 or an amino acid sequence
that is at
least 959/0 identical to the amino acid sequence of SEQ ID NO:290; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:291 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:291; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:292 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:292; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:293 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:293.
[0140] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:294 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:294; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:295 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:295; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:296 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:296; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:297 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:297.
[0141] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites, wherein the first
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:298 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:298; the second
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:299 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:299; the
third polypeptide
chain comprises the amino acid sequence of SEQ ID NO:300 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:300; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:301 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:301.
Anti-CD38 Binding Sites
[0142] Certain aspects of the present disclosure relate to binding proteins
that comprise an
antigen binding site that binds a CD38 polypeptide. In some embodiments, the
CD38
polypeptide is a human CD38 polypeptide, also known as ADPRC1. Human CD38
polypeptides are known in the art and include, without limitation, the
polypeptide represented
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
by NCBI Accession Number NP 001766.2, or a polypeptide produced from NCBI Gene
ID
Number 952. In some embodiments, the antigen binding site binds a human CD38
polypeptide,
a non-human primate (e.g., cynomolgus monkey) CD38 polypeptide, or a human
CD38
polypeptide and a non-human primate (e.g., cynomolgus monkey) CD38
polypeptide. In some
embodiments, a binding protein comprising an antigen binding site that binds a
CD38
polypeptide is monospecific and/or monovalent, bispecific and/or bivalent,
trispecific and/or
trivalent, or multispecific and/or multivalent.
[0143] In some embodiments, any of the CDRs and/or variable domains of the
anti-CD38
binding sites described below can be used in a monospecific antibody.
[0144] In other embodiments, any of the CDRs and/or variable domains of the
anti-CD38
binding sites described below can be used in any binding site of a trispecific
binding protein
comprising four polypeptides that form three antigen binding sites, e.g., as
described supra. In
certain embodiments, a binding protein that comprises an antigen binding site
that binds a
CD38 polypeptide is a trispecific binding protein comprising four polypeptides
that form three
antigen binding sites as described supra, wherein the VH3 and VL3 domains pair
and form a
third antigen binding site that binds a CD38 polypeptide.
[0145] A variety of features of exemplary binding sites and binding
proteins are described
herein. For example, in some embodiments, an anti-CD38 binding site cross-
reacts with human
CD38 (e.g., a human CD38 isoform A and/or isoform E polypeptide) and
cynomolgus monkey
CD38. In some embodiments, a binding protein comprising an anti-CD38 binding
site induces
apoptosis of a CD38+ cell. In some embodiments, a binding protein comprising
an anti-CD38
binding site recruits a T cell to a CD38+ cell and optionally activates the T
cell (e.g., though
TCR stimulation and/or costimulation).
[0146] In some embodiments, a binding site that binds CD38 comprises: an
antibody heavy
chain variable (VH) domain comprising a CDR-H1 sequence comprising the amino
acid
sequence of GYTFTSYA (SEQ ID NO:13), a CDR-H2 sequence comprising the amino
acid
sequence of IYPGQGGT (SEQ ID NO:14), and a CDR-H3 sequence comprising the
amino
acid sequence of ARTGGLRRAYFTY (SEQ ID NO:15); and/or an antibody light chain
variable (VL) domain comprising a CDR-L1 sequence comprising the amino acid
sequence of
QSVSSYGQGF (SEQ ID NO:16), a CDR-L2 sequence comprising the amino acid
sequence
of GAS (SEQ ID NO:17), and a CDR-L3 sequence comprising the amino acid
sequence of
QQNKEDPWT (SEQ ID NO:18). In some embodiments, a binding site that binds CD38
comprises: an antibody heavy chain variable (VH) domain comprising a CDR-H1
sequence
comprising the amino acid sequence of GYTFTSYA (SEQ ID NO:13), a CDR-H2
sequence
61
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
comprising the amino acid sequence of IYPGQGGT (SEQ ID NO:14), and a CDR-H3
sequence comprising the amino acid sequence of ARTGGLRRAYFTY (SEQ ID NO:15);
and
an antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the
amino acid sequence of QSVSSYGQGF (SEQ ID NO:16), a CDR-L2 sequence comprising
the amino acid sequence of GAS (SEQ ID NO:17), and a CDR-L3 sequence
comprising the
amino acid sequence of QQNKEDPWT (SEQ ID NO:18).
[0147] In some embodiments, a binding site that binds CD38 comprises: an
antibody heavy
chain variable (VH) domain comprising a CDR-H1 sequence comprising the amino
acid
sequence of GYTLTEFS (SEQ ID NO:19), a CDR-H2 sequence comprising the amino
acid
sequence of FDPEDGET (SEQ ID NO:20), and a CDR-H3 sequence comprising the
amino
acid sequence of TTGRFFDWF (SEQ ID NO:21); and/or an antibody light chain
variable (VL)
domain comprising a CDR-L1 sequence comprising the amino acid sequence of
QSVISRF
(SEQ ID NO:22), a CDR-L2 sequence comprising the amino acid sequence of GAS
(SEQ ID
NO:23), and a CDR-L3 sequence comprising the amino acid sequence of QQDSNLPIT
(SEQ
ID NO:24). In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino acid
sequence of GYTLTEFS (SEQ ID NO:19), a CDR-H2 sequence comprising the amino
acid
sequence of FDPEDGET (SEQ ID NO:20), and a CDR-H3 sequence comprising the
amino
acid sequence of TTGRFFDWF (SEQ ID NO:21); and an antibody light chain
variable (VL)
domain comprising a CDR-L1 sequence comprising the amino acid sequence of
QSVISRF
(SEQ ID NO:22), a CDR-L2 sequence comprising the amino acid sequence of GAS
(SEQ ID
NO:23), and a CDR-L3 sequence comprising the amino acid sequence of QQDSNLPIT
(SEQ
ID NO:24).
[0148] In some embodiments, a binding site that binds CD38 comprises: an
antibody heavy
chain variable (VH) domain comprising a CDR-H1 sequence comprising the amino
acid
sequence of GYAFTTYL (SEQ ID NO:25), a CDR-H2 sequence comprising the amino
acid
sequence of INPGSGST (SEQ ID NO:26), and a CDR-H3 sequence comprising the
amino acid
sequence of ARYAYGY (SEQ ID NO:27); and/or an antibody light chain variable
(VL)
domain comprising a CDR-L1 sequence comprising the amino acid sequence of
QNVGTA
(SEQ ID NO:28), a CDR-L2 sequence comprising the amino acid sequence of SAS
(SEQ ID
NO:29), and a CDR-L3 sequence comprising the amino acid sequence of QQYSTYPFT
(SEQ
ID NO:30). In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino acid
sequence of GYAFTTYL (SEQ ID NO:25), a CDR-H2 sequence comprising the amino
acid
62
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
sequence of INPGSGST (SEQ ID NO:26), and a CDR-H3 sequence comprising the
amino acid
sequence of ARYAYGY (SEQ ID NO:27); and an antibody light chain variable (VL)
domain
comprising a CDR-L1 sequence comprising the amino acid sequence of QNVGTA (SEQ
ID
NO:28), a CDR-L2 sequence comprising the amino acid sequence of SAS (SEQ ID
NO:29),
and a CDR-L3 sequence comprising the amino acid sequence of QQYSTYPFT (SEQ ID
NO:30).
[0149] In some embodiments, a binding site that binds CD38 comprises: an
antibody heavy
chain variable (VH) domain comprising a CDR-H1 sequence comprising the amino
acid
sequence of GYSFTNYA (SEQ ID NO:31), a CDR-H2 sequence comprising the amino
acid
sequence of ISPYYGDT (SEQ ID NO:32), and a CDR-H3 sequence comprising the
amino
acid sequence of ARRFEGFYYSMDY (SEQ ID NO:33); and/or an antibody light chain
variable (VL) domain comprising a CDR-L1 sequence comprising the amino acid
sequence of
QSLVHSNGNTY (SEQ ID NO:34), a CDR-L2 sequence comprising the amino acid
sequence
of KVS (SEQ ID NO:35), and a CDR-L3 sequence comprising the amino acid
sequence of
SQSTHVPLT (SEQ ID NO:36). In some embodiments, a binding site that binds CD38
comprises: an antibody heavy chain variable (VH) domain comprising a CDR-H1
sequence
comprising the amino acid sequence of GYSFTNYA (SEQ ID NO:31), a CDR-H2
sequence
comprising the amino acid sequence of ISPYYGDT (SEQ ID NO:32), and a CDR-H3
sequence
comprising the amino acid sequence of ARRFEGFYYSMDY (SEQ ID NO:33); and an
antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the
amino acid sequence of QSLVHSNGNTY (SEQ ID NO:34), a CDR-L2 sequence
comprising
the amino acid sequence of KVS (SEQ ID NO:35), and a CDR-L3 sequence
comprising the
amino acid sequence of SQSTHVPLT (SEQ ID NO:36).
[0150] In some embodiments, a binding site that binds CD38 comprises: an
antibody heavy
chain variable (VH) domain comprising a CDR-H1 sequence comprising the amino
acid
sequence of GFTFSSYG (SEQ ID NO:37), a CDR-H2 sequence comprising the amino
acid
sequence of IWYDGSNK (SEQ ID NO:38), and a CDR-H3 sequence comprising the
amino
acid sequence of ARDPGLRYFDGGMDV (SEQ ID NO:39); and/or an antibody light
chain
variable (VL) domain comprising a CDR-L1 sequence comprising the amino acid
sequence of
QGISSY (SEQ ID NO:40), a CDR-L2 sequence comprising the amino acid sequence of
AAS
(SEQ ID NO:41), and a CDR-L3 sequence comprising the amino acid sequence of
QQLNSFPYT (SEQ ID NO:42). In some embodiments, a binding site that binds CD38
comprises: an antibody heavy chain variable (VH) domain comprising a CDR-H1
sequence
comprising the amino acid sequence of GFTFSSYG (SEQ ID NO:37), a CDR-H2
sequence
63
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
comprising the amino acid sequence of IWYDGSNK (SEQ ID NO:38), and a CDR-H3
sequence comprising the amino acid sequence of ARDPGLRYFDGGMDV (SEQ ID NO:39);
and an antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising
the amino acid sequence of QGISSY (SEQ ID NO:40), a CDR-L2 sequence comprising
the
amino acid sequence of AAS (SEQ ID NO:41), and a CDR-L3 sequence comprising
the amino
acid sequence of QQLNSFPYT (SEQ ID NO:42).
[0151] In some embodiments, a binding site that binds CD38 comprises: an
antibody heavy
chain variable (VH) domain comprising a CDR-H1 sequence comprising the amino
acid
sequence of GFTFSSYG (SEQ ID NO:43), a CDR-H2 sequence comprising the amino
acid
sequence of IWYDGSNK (SEQ ID NO:44), and a CDR-H3 sequence comprising the
amino
acid sequence of ARMFRGAFDY (SEQ ID NO:45); and/or an antibody light chain
variable
(VL) domain comprising a CDR-L1 sequence comprising the amino acid sequence of
QGIRND (SEQ ID NO:46), a CDR-L2 sequence comprising the amino acid sequence of
AAS
(SEQ ID NO:47), and a CDR-L3 sequence comprising the amino acid sequence of
LQDYIYYPT (SEQ ID NO:48). In some embodiments, a binding site that binds CD38
comprises: an antibody heavy chain variable (VH) domain comprising a CDR-H1
sequence
comprising the amino acid sequence of GFTFSSYG (SEQ ID NO:43), a CDR-H2
sequence
comprising the amino acid sequence of IWYDGSNK (SEQ ID NO:44), and a CDR-H3
sequence comprising the amino acid sequence of ARMFRGAFDY (SEQ ID NO:45); and
an
antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the
amino acid sequence of QGIRND (SEQ ID NO:46), a CDR-L2 sequence comprising the
amino
acid sequence of AAS (SEQ ID NO:47), and a CDR-L3 sequence comprising the
amino acid
sequence of LQDYIYYPT (SEQ ID NO:48).
[0152] In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLVQ S GAEVVKP GA S VKVS CKA S GYTF T SYAMHWVKEAPGQRLEWIGYIYPGQ
GG'TNYNQKF QGR A TLTADT S A STAYMEL S SLRSEDTAVYFCARTGGLRRAYFTYWG
QGTLVTVSS (SEQ ID NO:79), and/or an antibody light chain variable (VL) domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
64
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
acid sequence of
DIVLTQSPATLSLSPGERATISCRASQSVSSYGQGFMHWYQQKPGQPPRLLIYGASSR
ATGIPARF SGSGSGTDFTLTISPLEPEDFAVYYCQQNKEDPWTEGGGTKLEIK (SEQ ID
NO:80). In some embodiments, a binding site that binds CD38 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:79,
and/or
an antibody light chain variable (VL) domain comprising the amino acid
sequence of SEQ ID
NO:80. In some embodiments, a binding site that binds CD38 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:79,
and an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:80.
[0153] In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLVQSGAEVKKPGASVKVSCKVSGYTLTEF SIHWVRQAPGQGLEWMGGFDPED
GETIYAQKFQGRVIMTEDTSTDTAYMEMNSLRSEDTAIYYCTTGRFEDWFWGQGTL
VTVSS (SEQ ID NO:81), and/or an antibody light chain variable (VL) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
of
EIILTQSPAILSLSPGERATLSCRASQSVISRFLSWYQVKPGLAPRLLIYGASTRATGIPV
RFSGSGSGTDFSLTISSLQPEDCAVYYCQQDSNLPITFGQGTRLEIK (SEQ ID NO:82).
In some embodiments, a binding site that binds CD38 comprises: an antibody
heavy chain
variable (VH) domain comprising the amino acid sequence of SEQ ID NO:81,
and/or an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:82. In some embodiments, a binding site that binds CD38 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:81,
and an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:82.
[0154] In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLVQSGAEVKKPGASVKVSCKASGYAFTTYLVEWIRQRPGQGLEWMGVINPGS
GSTNYAQKFQGRVTMTVDRSSTTAYMELSRLRSDDTAVYYCARYAYGYWGQGTL
VTVSS (SEQ ID NO:83), and/or an antibody light chain variable (VL) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
of
DIQMTQSPSSLSASVGDRVTITCRASQNVGTAVAWYQQKPGKSPKQUYSASNRYTG
VPSRF SGSGSGTDFTLTISSLQPEDLATYYCQQYSTYPFTFGQGTKLEIK (SEQ ID
NO:84). In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:83,
and/or an antibody light chain variable (VL) domain comprising the amino acid
sequence of
SEQ ID NO:84. In some embodiments, a binding site that binds CD38 comprises:
an
antibody heavy chain variable (VH) domain comprising the amino acid sequence
of SEQ ID
NO:83, and an antibody light chain variable (VL) domain comprising the amino
acid
sequence of SEQ ID NO:84.
[0155] In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLVESGGGVVQPGRSLRLSCAASGFTF SSYGMYWVRQAPGKGLEWVAVIWYDG
SNKYYADSVKGRF TISRDNSKNTLYLQMNSLRAEDTAVYHCARDPGLRYFDGGMD
VWGQGTTVTVSS (SEQ ID NO:87), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIQLTQSPSFLSASVGDRVTITCRASQGISSYLAWYQQKPGKAPKLLIFAASTLHSGVP
SRFSGSGSGTEFTLTISSLQPEDFATYYCQQLNSFPYTFGQGTKLEIK (SEQ ID NO:88).
In some embodiments, a binding site that binds CD38 comprises: an antibody
heavy chain
variable (VH) domain comprising the amino acid sequence of SEQ ID NO:87,
and/or an
66
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:88. In some embodiments, a binding site that binds CD38 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:87,
and an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:88.
[0156] In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLVESGGGVVQPGRSLRLSCAASGFTF SSYGMHWVRQAPGKGLEWVAVIWYDG
SNKYYADSVKGRFTISGDNSKNTLYLQMNSLRAEDTAVYYCARMERGAFDYWGQG
TLVTVSS (SEQ ID NO:89), and/or an antibody light chain variable (VL) domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
AIQMTQSPSSLSASVGDRVTITCRASQGIRNDLGWYQQKPGKAPKLLIYAASSLQSG
VPSRF SGSGSGTDFTLTISGLQPEDSATYYCLQDYIYYPTFGQGTKVEIK (SEQ ID
NO:90). In some embodiments, a binding site that binds CD38 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:89,
and/or
an antibody light chain variable (VL) domain comprising the amino acid
sequence of SEQ ID
NO:90. In some embodiments, a binding site that binds CD38 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:89,
and an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:90.
[0157] In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLVQSGAEVKKPGASVKVSCKASGYSFTNYAVHWVRQAPGQGLEWMGVISPY
YGDTTYAQKFQGRVTMTVDKSSSTAYMELSRLRSDDTAVYYCARRFEGFYYSMDY
WGQGTLVTVSS (SEQ ID NO:85), and/or an antibody light chain variable (VL)
domain
67
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89 /0, at least 90%, at least 91%, at least 92%, at least 930 o,
at least 940 o, at least
9500, at least 96 /0, at least 970 o, at least 98%, at least 99%, or 100 A
identical to the amino
acid sequence of
DVVMTQSPLSLPVTLGQPASISCRPSQSLVHSNGNTYLNWYQQRPGQSPKWYKVS
KRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCSQSTHVPLTFGGGTKVEIK
(SEQ ID NO:86). In some embodiments, a binding site that binds CD38 comprises:
an
antibody heavy chain variable (VH) domain comprising the amino acid sequence
of SEQ ID
NO:85, and/or an antibody light chain variable (VL) domain comprising the
amino acid
sequence of SEQ ID NO:86. In some embodiments, a binding site that binds CD38
comprises: an antibody heavy chain variable (VH) domain comprising the amino
acid
sequence of SEQ ID NO:85, and an antibody light chain variable (VL) domain
comprising
the amino acid sequence of SEQ ID NO:86.
[0158] In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 869/0, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 9394., at least 94 /o, at least 95 /o, at least 96%, at least 97 /o, at
least 98 /o, at least 99 /o, or
100% identical to the amino acid sequence of
QVQLQQSGPELVRPGTSVKVSCKASGYAF TTYLVEWIKQRPGQGLEWIGVINPGSGS
TNYNEKFKGKATLTVDRSSTTAYMHLSGLTSDDSAVYFCARYAYGYWGQGTTLTV
SS (SEQ ID NO:277), and/or an antibody light chain variable (VL) domain
comprising an
amino acid sequence that is at least 85%, at least 86%, at least 87%, at least
88%, at least
89%, at least 90 /o, at least 91%, at least 92%, at least 93%, at least 94%,
at least 95%, at least
96%, at least 97 /0, at least 98%, at least 99%, or 100% identical to the
amino acid sequence
of
DIVMTQSQKFMSASVGDRVSITCKASQNVGTAVAWYQQQPGHSPKQLIYSASNRYT
GVPDRFTGSGAGTDFTLTISNIQSEDLADYFCQQYSTYPFTFGSGTKLEIK (SEQ ID
NO:278). In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:277,
and/or an antibody light chain variable (VL) domain comprising the amino acid
sequence of
SEQ ID NO:278. In some embodiments, a binding site that binds CD38 comprises:
an
antibody heavy chain variable (VH) domain comprising the amino acid sequence
of SEQ ID
NO:277, and an antibody light chain variable (VL) domain comprising the amino
acid
68
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
sequence of SEQ ID NO:278. In some embodiments, the VH and/or VL domains are
humanized.
[0159] In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLLQSGAELVRPGVSVKISCTGSGYSFTNYAVHWVKQSHVKSLEWIGVISPYYGD
TTYNQKFTGKATMTVDKSSSTAYMELARLTSEDSAIYFCARRFEGFYYSMDYWGQG
TSVTVSS (SEQ ID NO:279), and/or an antibody light chain variable (VL) domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DVVMIQTPLSLPVSLGDQASISCRPSQSLVHSNGNTYLNWYLQRPGQSPKLLIYKVSK
RFSGVPDRFSGSGSGTDFTLKISRVEAEDLGVYLCSQSTHVPLTFGSGTQLEIK (SEQ
ID NO:280). In some embodiments, a binding site that binds CD38 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:279,
and/or an antibody light chain variable (VL) domain comprising the amino acid
sequence of
SEQ ID NO:280. In some embodiments, a binding site that binds CD38 comprises:
an
antibody heavy chain variable (VH) domain comprising the amino acid sequence
of SEQ ID
NO:279, and an antibody light chain variable (VL) domain comprising the amino
acid
sequence of SEQ ID NO:280. In some embodiments, the VH and/or VL domains are
humanized.
[0160] In some embodiments of any of the above embodiments, the binding
protein is a
trispecific binding protein. In some embodiments, the trispecific binding
protein comprising
an antigen binding site that binds a CD38 polypeptide, an antigen binding site
that binds a
CD28 polypeptide, and an antigen binding site that binds a CD3 polypeptide. In
some
embodiments, the binding protein is a trispecific binding protein comprising
four
polypeptides comprising three antigen binding sites, wherein the polypeptide
of formula I and
the polypeptide of formula II form a cross-over light chain-heavy chain pair
(e.g., as
described herein). In some embodiments, the VH and VL domains of any of the
anti-CD38
antigen binding sites described above represent VH3 and VL3 and form a third
antigen binding
site that binds a CD38 polypeptide. In some embodiments, VH1 and VIA form a
first antigen
69
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
binding site that binds a CD28 polypeptide, VH2 and VL2 form a second antigen
binding site
that binds a CD3 polypeptide, and the VH and VL domains of any of the anti-
CD38 antigen
binding sites described above and/or in Table 2 represent VH3 and VL3 and form
a third
antigen binding site that binds a CD38 polypeptide.
[0161] Sequences of exemplary anti-CD38 antigen binding sites are provided
in Table 2.
In some embodiments, a binding protein comprising an anti-CD38 antigen binding
site of the
present disclosure comprises 1, 2, 3, 4, 5, or all 6 CDR sequences of an anti-
CD38 antibody
described in Table 2. In some embodiments, a binding protein comprising an
anti-CD38
antigen binding site of the present disclosure comprises a VH domain sequence
and/or VL
domain sequence of an anti-CD38 antibody described in Table 2.
Table 2. Anti-CD38 binding protein sequences.
Sequence Molecule Description SEQ ID Sequence
Type NO
CDR Anti-CD38 CDR-H1 13 GYTFTSYA
(VH1) CDR-H2 14 IYPGQGGT
CDR-H3 15 ARTGGLRRAYFTY
CDR-L1 16 QSVSSYGQGF
CDR-L2 17 GAS
CDR-L3 18 QQNKEDPWT
Anti-CD38 CDR-H1 19 GYTLTEFS
(hhy992) CDR-H2 20 FDPEDGET
CDR-H3 21 TTGRFFDWF
CDR-L1 22 QSVISRF
CDR-L2 23 GAS
CDR-L3 24 QQDSNLPIT
Anti-CD38 CDR-H1 25 GYAFTTYL
(hyb5739) CDR-H2 26 INPGSGST
CDR-H3 27 ARYAYGY
CDR-L1 28 QNVGTA
CDR-L2 29 SAS
CDR-L3 30 QQYSTYPFT
Anti-CD38 CDR-H1 31 GYSFTNYA
(hyb6284) CDR-H2 32 ISPYYGDT
CDR-H3 33 ARRFEGFYYSMDY
CDR-L1 34 QSLVHSNGNTY
CDR-L2 35 KVS
CDR-L3 36 SQSTHVPLT
Anti-CD38 CDR-H1 37 GFTFSSYG
(hhy1195) CDR-H2 38 IWYDGSNK
CDR-H3 39 ARDPGLRYFDGGMDV
CDR-L1 40 QGIS SY
CDR-L2 41 A A S
CDR-L3 42 QQLNSFPYT
Anti-CD38 CDR-H1 43 GFTFSSYG
(hhy1370) CDR-H2 44 IWYDGSNK
Date Recue/Date Received 2021-10-04
170-01.-1=ZOZ panieoe eeo/enóej ele0
IL
ATINNSNCIDSIIRIDNASCIVAANNSDCI
AMIAVAMTIMIDdVONAMHIAIDASSAI OLEIICTI
ADSVV3SIIIISIIDdOAADDDSHAIOA6 68 HA 8 ECID
NITINIDO-DILAddSNTOODAA
IVACIadOTSSIITIAHIDSDSDS,111SdAD
SIMSVVAITIMV)IDd-)166AMICIASS
IDOSVIDIIIAIICIDASVSTASdSOITOICI 88 TA
SSAIAIIDODMACRAIDD
CHAIIIDd(RIVDHAAVICHVIIISNIARYI
ATINDISNCIIISIIRIDNASCIVAANNSDCI
AMIAVAMTIMIDdVONAMAIAIDASSAI S61IiC1111
ADSVVOSIIVISIIDdOAADDDSHAIOAO L8 HA SEGO
)1I
HANIDDDILTdAHISOSDAAADACEVH
ANSIXIIACIIDSDSDS,411C1c1ADSAIDISA
NAITINdSODclITOOAANTAINDNSHAT
SoSclIDSISVdODTIAdTS'IdSOIBIAACI 98 TA
SSAINILDODMAGIAI
SAMDMIIIIIVDAAAVICICISIVRISTHIAIA
VISSSNCIAIIALLAIIDOANOVAIICIDAAd
SIADIALATIDODdVOITAANHAVANIASA 178Z9114
DSVMDSANASVDMIAHVDSOATOAO S8 HA 8D
NITINIDODAIddAISAOODAAI
VICIadOiSSIrLIACIIDSDS9S,411SdADI
AIINSVSAITO)IdS)1DdNOOAMVAVIDA
NOSVIDILLAIICIDASVSTSSdSOIBIOICI 178 TA
SSAINILD
ODAUDAVAIIVOAAAVICICISIVIIISIHIA1
AVLISSIICIAIINIANDOANOVANISDSD
dNIADIALATIDODdITOITIAOKMIAVA 6EL Sliti
DSVMDSANASVDMIAHVDSOATOAO 8 HA 8D
AAVDCIadOlSSIIISACIIDSDSDS,411AdI
DIVITISVDAITTIMVIDdNAOAMSTRIS
IASOSVIDSIIVIODdSTSTIVdSOIIIIH ZS TA
SSAIKIIDO
DAUMCIARIDIIDAAIVICHSIFISNIAIHIAI
AVICIISICHIINIANDOANOVALLADCH
daiDDIALATIDODdVOITAMHISdarlIA
DSANDSANASVDMIAHVDSOATOAO 18 HA 8D
)1IHMIIDDDILAkcICIHNNOODAAAVA
CladalcISIIIIACIIDS9SDS,411VdIDIVITS
SVDAITIliddODdNOOAMHINADODASS
ASOSVIDSLINITHDdSTSTIVdSOITAICI 08 TA
SSAIATIDODAWA
AVIIIITDDIIIVOAAAVICHSIVISSTHIAIA
VISVSICIVITIVIMOANONANILD9OD
dA1A9IAMITODdV3)1AMHINIVASIAIA IHA uTui9P
DSVMDSANASVDc1)1AAHVDSOATOAO 6L HA 8D OicpnA
IdAAIACIOT 817 1-11(13
SVV LV Z1-11(13
CINITIDO 917
ACIAVDITAIAIIIV S17
OZELZO/OZOZSIVIDd Z6COIZ/OZOZ OM
WO 2020/210392
PCT/US2020/027320
LQMNSLRAEDTAVYYCARMERGAEDY
WGQGTLVTVSS
VL 90 AIQMTQSPSSLSASVGDRVTITCRASQGI
RNDLGWYQQKPGKAPKLLIYAASSLQS
GVPSRESGSGSGTDETLTISGLQPEDSAT
YYCLQDYIYYPTFGQGTKVEIK
CD38 VH 277
QVQLQQSGPELVRPGTSVKVSCKASGY
hyb5739 AFTTYLVEWIKQRPGQGLEWIGVINPGS
GSTNYNEKFKGKATLTVDRSSTTAYMH
LSGLTSDDSAVYFCARYAYGYWGQGT
TLTVSS
VL 278 DIVMTQSQKFMSASVGDRVSITCKASQ
NVGTAVAWYQQQPGHSPKQLIYSASNR
YTGVPDRFTGSGAGTDETLTISNIQSEDL
ADYECQQYSTYPETEGSGTKLEIK
CD38 VH 279
QVQLLQSGAELVRPGVSVKISCTGSGYS
hyb6284 FTNYAVHWVKQSHVKSLEWIGVISPYY
GDTTYNQKFTGKATMTVDKSSSTAYM
ELARLTSEDSAIYECARREEGFYYSMDY
WGQGTSVTVSS
VL 280 DVVMIQTPLSLPVSLGDQASISCRPSQSL
VHSNGNTYLNWYLQRPGQSPKLLIYKV
SKRESGVPDRESGSGSGTDETLKISRVE
AEDLGVYLCSQSTHVPLTFGSGTQLEIK
[0162] Further provided herein are antibodies (e.g., monospecific antibodies)
comprising any
of the anti-CD38 CDRs and/or variable domains described supra.
[0163] In some embodiments, a binding protein of the present disclosure
comprises an antigen
binding site that binds an extracellular domain of a human CD38 polypeptide
and an
extracellular domain of a cynomolgus monkey CD38 polypeptide. Exemplary assays
for
determining whether an antigen binding site binds an antigen are described
herein and known
in the art, including (without limitation) ELISA, SPR, and flow cytometry
assays.
Anti-HER2 Binding Sites
[0164]
Certain aspects of the present disclosure relate to binding proteins that
comprise
an antigen binding site that binds a HER2 polypeptide. In some embodiments,
the HER2
polypeptide is a human HER2 polypeptide, also known as NEU, NGL, ERBB2, TKR1,
CD340, HER-2, MLN19, and HER-2/neu. Human HER2 polypeptides are known in the
art
and include, without limitation, the polypeptides represented by NCBI
Accession Numbers
XP 024306411.1 XP 024306410.1 XP 024306409.1 NP 001276867.1, NP 001276866.1,
_ _ _
NP 001276865.1, NP 001005862.1, or NP 004439.2, or a polypeptide produced from
NCBI
Gene ID Number 2064. In some embodiments, a binding protein comprising an
antigen
binding site that binds a HER2 polypeptide is monospecific and/or monovalent,
bispecific
72
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
and/or bivalent, trispecific and/or trivalent, or multispecific and/or
multivalent. In some
embodiments, a binding protein that comprises an antigen binding site that
binds a HER2
polypeptide is a trispecific binding protein comprising four polypeptides that
form three
antigen binding sites as described supra, wherein VH3 and VL3 domain pair that
form a third
antigen binding site that binds a HER2 polypeptide.
[0165] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFNIKDTY (SEQ ID NO:1) or GFNIRDTY (SEQ ID NO:2), a CDR-H2
sequence comprising the amino acid sequence of IYPTNGYT (SEQ ID NO:3),
IYPTQGYT
(SEQ ID NO:4), or IYPTNAYT (SEQ ID NO:5), and a CDR-H3 sequence comprising the
amino
acid sequence of SRWGGDGFYAMDY (SEQ ID NO:6), SRWGGEGFYAMDY (SEQ ID
NO:7), or SRWGGSGFYAMDY (SEQ ID NO:8); and/or an antibody light chain variable
(VL)
domain comprising a CDR-L1 sequence comprising the amino acid sequence of
QDVNTA
(SEQ ID NO:9) or QDVQTA (SEQ ID NO:10), a CDR-L2 sequence comprising the amino
acid
sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the amino
acid
sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, a binding site that
binds
HER2 comprises: an antibody heavy chain variable (VH) domain comprising a CDR-
H1
sequence comprising the amino acid sequence of GFNIKDTY (SEQ ID NO:1) or
GFNIRDTY
(SEQ ID NO:2), a CDR-H2 sequence comprising the amino acid sequence of
IYPTNGYT
(SEQ ID NO:3), IYPTQGYT (SEQ ID NO:4), or IYPTNAYT (SEQ ID NO:5), and a CDR-H3
sequence comprising the amino acid sequence of SRWGGDGFYAMDY (SEQ ID NO:6),
SRWGGEGFYAMDY (SEQ ID NO:7), or SRWGGSGFYAMDY (SEQ ID NO:8); and an
antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the
amino acid sequence of QDVNTA (SEQ ID NO:9) or QDVQTA (SEQ ID NO:10), a CDR-L2
sequence comprising the amino acid sequence of SAS (SEQ ID NO:11), and a CDR-
L3
sequence comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12).
[0166] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFNIKDTY (SEQ ID NO:1), a CDR-H2 sequence comprising the
amino
acid sequence of IYPTNGYT (SEQ ID NO:3), and a CDR-H3 sequence comprising the
amino acid sequence of SRWGGDGFYAMDY (SEQ ID NO:6); and/or an antibody light
chain variable (VL) domain comprising a CDR-L1 sequence comprising the amino
acid
sequence of QDVNTA (SEQ ID NO:9), a CDR-L2 sequence comprising the amino acid
sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the amino
acid
73
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, a binding site that
binds
HER2 comprises: an antibody heavy chain variable (VH) domain comprising a CDR-
H1
sequence comprising the amino acid sequence of GFNIKDTY (SEQ ID NO:1), a CDR-
H2
sequence comprising the amino acid sequence of IYPTNGYT (SEQ ID NO:3), and a
CDR-
H3 sequence comprising the amino acid sequence of SRWGGDGFYAMDY (SEQ ID NO:6);
and an antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the amino acid sequence of QDVNTA (SEQ ID NO:9), a CDR-L2 sequence
comprising the amino acid sequence of SAS (SEQ ID NO:11), and a CDR-L3
sequence
comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12).
[0167] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-H2 sequence comprising the
amino
acid sequence of IYPTQGYT (SEQ ID NO:4), and a CDR-H3 sequence comprising the
amino acid sequence of SRWGGEGFYAMDY (SEQ ID NO:7); and/or an antibody light
chain variable (VL) domain comprising a CDR-L1 sequence comprising the amino
acid
sequence of QDVNTA (SEQ ID NO:9), a CDR-L2 sequence comprising the amino acid
sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the amino
acid
sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, a binding site that
binds
HER2 comprises: an antibody heavy chain variable (VH) domain comprising a CDR-
H1
sequence comprising the amino acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-
H2
sequence comprising the amino acid sequence of IYPTQGYT (SEQ ID NO:4), and a
CDR-
H3 sequence comprising the amino acid sequence of SRWGGEGFYAMDY (SEQ ID NO:7);
and an antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the amino acid sequence of QDVNTA (SEQ ID NO:9), a CDR-L2 sequence
comprising the amino acid sequence of SAS (SEQ ID NO:11), and a CDR-L3
sequence
comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12).
[0168] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-H2 sequence comprising the
amino
acid sequence of IYPTNAYT (SEQ ID NO:5), and a CDR-H3 sequence comprising the
amino acid sequence of SRWGGSGFYAMDY (SEQ ID NO:8); and/or an antibody light
chain variable (VL) domain comprising a CDR-L1 sequence comprising the amino
acid
sequence of QDVNTA (SEQ ID NO:9), a CDR-L2 sequence comprising the amino acid
sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the amino
acid
74
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, a binding site that
binds
HER2 comprises: an antibody heavy chain variable (VH) domain comprising a CDR-
H1
sequence comprising the amino acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-
H2
sequence comprising the amino acid sequence of IYPTNAYT (SEQ ID NO:5), and a
CDR-
H3 sequence comprising the amino acid sequence of SRWGGSGFYAMDY (SEQ ID NO:8);
and an antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the amino acid sequence of QDVNTA (SEQ ID NO:9), a CDR-L2 sequence
comprising the amino acid sequence of SAS (SEQ ID NO:11), and a CDR-L3
sequence
comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12).
[0169] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-H2 sequence comprising the
amino
acid sequence of IYPTQGYT (SEQ ID NO:4), and a CDR-H3 sequence comprising the
amino acid sequence of SRWGGSGFYAMDY (SEQ ID NO:8); and/or an antibody light
chain variable (VL) domain comprising a CDR-L1 sequence comprising the amino
acid
sequence of QDVNTA (SEQ ID NO:9), a CDR-L2 sequence comprising the amino acid
sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the amino
acid
sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, a binding site that
binds
HER2 comprises: an antibody heavy chain variable (VH) domain comprising a CDR-
H1
sequence comprising the amino acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-
H2
sequence comprising the amino acid sequence of IYPTQGYT (SEQ ID NO:4), and a
CDR-
H3 sequence comprising the amino acid sequence of SRWGGSGFYAMDY (SEQ ID NO:8);
and an antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the amino acid sequence of QDVNTA (SEQ ID NO:9), a CDR-L2 sequence
comprising the amino acid sequence of SAS (SEQ ID NO:11), and a CDR-L3
sequence
comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12).
[0170] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-H2 sequence comprising the
amino
acid sequence of IYPTNAYT (SEQ ID NO:5), and a CDR-H3 sequence comprising the
amino acid sequence of SRWGGEGFYAMDY (SEQ ID NO:7); and/or an antibody light
chain variable (VL) domain comprising a CDR-L1 sequence comprising the amino
acid
sequence of QDVNTA (SEQ ID NO:9), a CDR-L2 sequence comprising the amino acid
sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the amino
acid
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, a binding site that
binds
HER2 comprises: an antibody heavy chain variable (VH) domain comprising a CDR-
H1
sequence comprising the amino acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-
H2
sequence comprising the amino acid sequence of IYPTNAYT (SEQ ID NO:5), and a
CDR-
H3 sequence comprising the amino acid sequence of SRWGGEGFYAMDY (SEQ ID NO:7);
and an antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the amino acid sequence of QDVNTA (SEQ ID NO:9), a CDR-L2 sequence
comprising the amino acid sequence of SAS (SEQ ID NO:11), and a CDR-L3
sequence
comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12).
[0171] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFNIKDTY (SEQ ID NO:1), a CDR-H2 sequence comprising the
amino
acid sequence of IYPTNGYT (SEQ ID NO:3), and a CDR-H3 sequence comprising the
amino acid sequence of SRWGGDGFYAMDY (SEQ ID NO:6); and/or an antibody light
chain variable (VL) domain comprising a CDR-L1 sequence comprising the amino
acid
sequence of QDVQTA (SEQ ID NO:10), a CDR-L2 sequence comprising the amino acid
sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the amino
acid
sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, a binding site that
binds
HER2 comprises: an antibody heavy chain variable (VH) domain comprising a CDR-
H1
sequence comprising the amino acid sequence of GFNIKDTY (SEQ ID NO:1), a CDR-
H2
sequence comprising the amino acid sequence of IYPTNGYT (SEQ ID NO:3), and a
CDR-
H3 sequence comprising the amino acid sequence of SRWGGDGFYAMDY (SEQ ID NO:6);
and an antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the amino acid sequence of QDVQTA (SEQ ID NO:10), a CDR-L2 sequence
comprising the amino acid sequence of SAS (SEQ ID NO:11), and a CDR-L3
sequence
comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12).
[0172] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-H2 sequence comprising the
amino
acid sequence of IYPTQGYT (SEQ ID NO:4), and a CDR-H3 sequence comprising the
amino acid sequence of SRWGGEGFYAMDY (SEQ ID NO:7), and a CDR-H3 sequence
comprising the amino acid sequence of SRWGGSGFYAMDY (SEQ ID NO:8); and/or an
antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the
amino acid sequence of QDVQTA (SEQ ID NO:10), a CDR-L2 sequence comprising the
76
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
amino acid sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising
the
amino acid sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, a binding
site
that binds HER2 comprises: an antibody heavy chain variable (VH) domain
comprising a
CDR-Ell sequence comprising the amino acid sequence of GFNIRDTY (SEQ m NO:2),
a
CDR-H2 sequence comprising the amino acid sequence of IYPTQGYT (SEQ ID NO:4),
and
a CDR-H3 sequence comprising the amino acid sequence of SRWGGEGFYAMDY (SEQ ID
NO:7), and a CDR-H3 sequence comprising the amino acid sequence of
SRWGGSGFYAMDY (SEQ ID NO:8); and an antibody light chain variable (VL) domain
comprising a CDR-L1 sequence comprising the amino acid sequence of QDVQTA (SEQ
ID
NO:10), a CDR-L2 sequence comprising the amino acid sequence of SAS (SEQ ID
NO:11),
and a CDR-L3 sequence comprising the amino acid sequence of QQHYTTP (SEQ ID
NO:12).
[0173] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-H2 sequence comprising the
amino
acid sequence of IYPTNAYT (SEQ ID NO:5), and a CDR-H3 sequence comprising the
amino acid sequence of SRWGGSGFYAMDY (SEQ ID NO:8); and/or an antibody light
chain variable (VL) domain comprising a CDR-L1 sequence comprising the amino
acid
sequence of QDVQTA (SEQ ID NO:10), a CDR-L2 sequence comprising the amino acid
sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the amino
acid
sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, a binding site that
binds
HER2 comprises: an antibody heavy chain variable (VH) domain comprising a CDR-
H1
sequence comprising the amino acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-
H2
sequence comprising the amino acid sequence of IYPTNAYT (SEQ ID NO:5), and a
CDR-
H3 sequence comprising the amino acid sequence of SRWGGSGFYAMDY (SEQ ID NO:8);
and an antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the amino acid sequence of QDVQTA (SEQ ID NO:10), a CDR-L2 sequence
comprising the amino acid sequence of SAS (SEQ ID NO:11), and a CDR-L3
sequence
comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12).
[0174] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-H2 sequence comprising the
amino
acid sequence of IYPTQGYT (SEQ ID NO:4), and a CDR-H3 sequence comprising the
amino acid sequence of SRWGGSGFYAMDY (SEQ ID NO:8); and/or an antibody light
77
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
chain variable (VL) domain comprising a CDR-L1 sequence comprising the amino
acid
sequence of QDVQTA (SEQ ID NO:10), a CDR-L2 sequence comprising the amino acid
sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the amino
acid
sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, a binding site that
binds
HER2 comprises: an antibody heavy chain variable (VH) domain comprising a CDR-
H1
sequence comprising the amino acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-
H2
sequence comprising the amino acid sequence of IYPTQGYT (SEQ ID NO:4), and a
CDR-
H3 sequence comprising the amino acid sequence of SRWGGSGFYAMDY (SEQ ID NO:8);
and an antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the amino acid sequence of QDVQTA (SEQ ID NO:10), a CDR-L2 sequence
comprising the amino acid sequence of SAS (SEQ ID NO:11), and a CDR-L3
sequence
comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12).
[0175] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-H2 sequence comprising the
amino
acid sequence of IYPTNAYT (SEQ ID NO:5), and a CDR-H3 sequence comprising the
amino acid sequence of SRWGGEGFYAMDY (SEQ ID NO:7); and/or an antibody light
chain variable (VL) domain comprising a CDR-L1 sequence comprising the amino
acid
sequence of QDVQTA (SEQ ID NO:10), a CDR-L2 sequence comprising the amino acid
sequence of SAS (SEQ ID NO:11), and a CDR-L3 sequence comprising the amino
acid
sequence of QQHYTTP (SEQ ID NO:12). In some embodiments, a binding site that
binds
HER2 comprises: an antibody heavy chain variable (VH) domain comprising a CDR-
H1
sequence comprising the amino acid sequence of GFNIRDTY (SEQ ID NO:2), a CDR-
H2
sequence comprising the amino acid sequence of IYPTNAYT (SEQ ID NO:5), and a
CDR-
H3 sequence comprising the amino acid sequence of SRWGGEGFYAMDY (SEQ ID NO:7);
and an antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the amino acid sequence of QDVQTA (SEQ ID NO:10), a CDR-L2 sequence
comprising the amino acid sequence of SAS (SEQ ID NO:11), and a CDR-L3
sequence
comprising the amino acid sequence of QQHYTTP (SEQ ID NO:12).
[0176] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
78
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
EVQLVESGGGLVQPGGSLRL S C AA S GFNIKD TYIHWVRQAP GKGLEWVARIYPTNG
YTRYAD S VKGRF TI S AD T SKNTAYLQMNSLRAEDTAVYYC SRWGGD GFYAMDYW
GQGTLVTVSS (SEQ ID NO:72),
EVQLVESGGGLVQPGGSLRL S C AA S GFNIRD TYIHWVRQAP GKGLEWVARIYP TQ G
YTRYAD S VKGRF TI S AD T SKNTAYLQMNSLRAEDTAVYYC SRWGGEGFYAMDYW
GQGTLVTVSS (SEQ ID NO:73),
EVQLVESGGGLVQPGGSLRL S C AA S GFNIRD TYIHWVRQAP GKGLEWVARIYP TQ G
YTRYAD S VKGRF TI S AD T SKNTAYLQMNSLRAEDTAVYYC SRWGGS GF YAMDYW
GQGTLVTVSS (SEQ ID NO:74),
EVQLVESGGGLVQPGGSLRL S C AA S GFNIRD TYIHWVRQAP GKGLEWVARIYP TNA
YTRYAD S VKGRF TI S AD T SKNTAYLQMNSLRAEDTAVYYC SRWGGS GF YAMDYW
GQGTLVTVSS (SEQ ID NO:75), or
EVQLVESGGGLVQPGGSLRL S C AA S GFNIRD TYIHWVRQAP GKGLEWVARIYP TNA
YTRYAD S VKGRF TI S AD T SKNTAYLQMNSLRAEDTAVYYC SRWGGEGFYAMDYW
GQGTLVTVSS (SEQ ID NO:76); and/or an antibody light chain variable (VL) domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIQMTQ SP S SLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSG
VP SRF SGSRSGTDFTLTIS SL QPEDF ATYYC Q QHYT TPP TF GQ GTKVEIK (SEQ ID
NO:77) or
DIQMTQ SP S SLSASVGDRVTITCRASQDVQTAVAWYQQKPGKAPKWYSASFLYSG
VP SRF SGSRSGTDFTLTIS SL QPEDF ATYYC Q QHYT TPP TF GQ GTKVEIK (SEQ ID
NO:78). In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:72,
SEQ ID
NO:73, SEQ ID NO:74, SEQ ID NO:75, or SEQ ID NO:76; and/or an antibody light
chain
variable (VL) domain comprising the amino acid sequence of SEQ ID NO:77 or SEQ
ID
NO:78. In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:72,
SEQ ID
NO:73, SEQ ID NO:74, SEQ ID NO:75, or SEQ ID NO:76; and an antibody light
chain
variable (VL) domain comprising the amino acid sequence of SEQ ID NO:77 or SEQ
ID
NO:78.
79
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
[0177] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 869/0, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 9394., at least 940 o, at least 950 o, at least 96%, at least 970 o, at
least 98%, at least 990 o, or
1000o identical to the amino acid sequence of
EVQLVESGGGLVQPGGSLRLSCAASGENIKDTYTHWVRQAPGKGLEWVARTYPTNG
YTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGDGFYAMDYW
GQGTLVTVSS (SEQ ID NO:72), and/or an antibody light chain variable (VL) domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89 /o, at least 90%, at least 91%, at least 92%, at least 930 o,
at least 940 o, at least
9500, at least 96 /o, at least 970 o, at least 98%, at least 99%, or 100%
identical to the amino
acid sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKLLIYSASFLYSG
VPSRF SGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:77). In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:72,
and/or
an antibody light chain variable (VL) domain comprising the amino acid
sequence of SEQ ID
NO:77. In some embodiments, a binding site that binds HER2 comprises:: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:72,
and an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:77.
[0178] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 869/0, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 939/0, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
1000o identical to the amino acid sequence of
EVQLVESGGGLVQPGGSLRLSCAASGENIRDTYTHWVRQAPGKGLEWVARTYPTQG
YTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGEGFYAMDYW
GQGTLVTVSS (SEQ ID NO:73), and/or an antibody light chain variable (VL) domain
comprising an amino acid sequence that is at least 85 /0, at least 86 /0, at
least 87%, at least
88%, at least 89 /0, at least 90%, at least 91%, at least 92%, at least 93%,
at least 94%, at least
950o, at least 96 /0, at least 97%, at least 98%, at least 99%, or 1000o
identical to the amino
acid sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKLLTYSASELYSG
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
VPSRF SGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:77). In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:73,
and/or
an antibody light chain variable (VL) domain comprising the amino acid
sequence of SEQ ID
NO:77. In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:73,
and an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:77.
[0179] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
EVQLVESGGGLVQPGGSLRL SCAASGENIRDTYIHWVRQAPGKGLEWVARTYPTNA
YTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGSGFYAMDYW
GQGTLVTVSS (SEQ ID NO:75), and/or an antibody light chain variable (VL) domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSG
VPSRF SGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:77). In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:75,
and/or
an antibody light chain variable (VL) domain comprising the amino acid
sequence of SEQ ID
NO:77. In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:75,
and an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:77.
[0180] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
81
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
EVQLVESGGGLVQPGGSLRLSCAASGENIRDTYIHWVRQAPGKGLEWVARIYPTQG
YTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGSGFYAMDYW
GQGTLVTVSS (SEQ ID NO:74), and/or an antibody light chain variable (VL) domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSG
VPSRF SGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:77). In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:74,
and/or
an antibody light chain variable (VL) domain comprising the amino acid
sequence of SEQ ID
NO:77. In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:74,
and an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:77.
[0181] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
EVQLVESGGGLVQPGGSLRLSCAASGENIRDTYIHWVRQAPGKGLEWVARIYPTNA
YTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGEGFYAMDYW
GQGTLVTVSS (SEQ ID NO:76), and/or an antibody light chain variable (VL) domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSG
VPSRF SGSRSGTDFTLTISSLQPEDF ATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:77). In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:76,
and/or
an antibody light chain variable (VL) domain comprising the amino acid
sequence of SEQ ID
NO:77. In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
82
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:76,
and an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:77.
[0182] In some embodiments, a binding site that binds HER2 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
EVQLVESGGGLVQPGGSLRLSCAASGFNIKDTVIHWVRQAPGKGLEWVARTYPTNG
YTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGDGFYAMDYW
GQGTLVTVSS (SEQ ID NO:72), and/or an antibody light chain variable (VL) domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIQMTQSPSSLSASVGDRVTITCRASQDVQTAVAWYQQKPGKAPKLLIYSASFLYSG
VPSRF SGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIK (SEQ ID
NO:78). In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:72,
and/or
an antibody light chain variable (VL) domain comprising the amino acid
sequence of SEQ ID
NO:78. In some embodiments, a binding site that binds HER2 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:72,
and an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:78.
[0183] In some embodiments, an anti-HER2 antigen binding site of the
present disclosure
comprises 1, 2, 3, 4, 5, or all 6 CDR sequences of anti-HER2 antibody
trastuzumab,
30R/55Q/102E, 30R/56A/1025, 30R/55Q/1025, 30R/56A/102E, or 30Q. In some
embodiments, an anti-HER2 antigen binding site of the present disclosure
comprises a VH
domain sequence and/or VL domain sequence of anti-HER2 antibody trastuzumab,
30R/55Q/102E, 30R/56A/102S, 30R/55Q/102S, 30R/56A/102E, or 30Q.
[0184] Sequences of exemplary anti-HER2 antigen binding sites are provided
in Table 3.
In some embodiments, an anti-HER2 antigen binding site of the present
disclosure comprises
1, 2, 3, 4, 5, or all 6 CDR sequences of an anti-HER2 antibody described in
Table 3. In some
embodiments, an anti-HER2 antigen binding site of the present disclosure
comprises a VH
83
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
domain sequence and/or VL domain sequence of an anti-HER2 antibody described
in Table
3.
Table 3. Anti-HER2 binding protein sequences.
Sequence Molecule Description SEQ Sequence
Type ID NO
CDR Anti-Her2 CDR-HI 1 GFNIKDTY
(trastuzumab) (original)
Heavy chain CDR-H1 2 GFNIRDTY
CDRs 3OR
CDR-H2 3 IYPTNGYT
(original)
CDR-H2 4 IYPTQGYT
55Q
CDR-H2 5 IYPTNAYT
56A
CDR-H3 6 SRWGGDGFYAMDY
(original)
CDR-H3 7 SRWGGEGFYAMDY
102E
CDR-H3 8 SRWGGSGFYAMDY
102S
Anti-Her2 CDR-L1 9 QDVNTA
(trastuzumab) (original)
Light chain CDR-L1 10 QDVQTA
CDRs 30Q
CDR-L2 11 SAS
(original)
CDR-L3 12 QQHYTTP
(original)
Variable Anti-Her2 VH wt 72 EVQLVESGGGLVQPGGSLRLSCAASGF
domain Trastuzumab NIKDTYIHWVRQAPGKGLEWVARIYP
and variant TNGYTRYADSVKGRFTISADTSKNTAY
VH LQMNSLRAEDTAVYYCSRWGGDGFY
AMDYWGQGTLVTVSS
VH 73 EVQLVESGGGLVQPGGSLRLSCAASGF
30R/55Q/102E NIRDTYIHWVRQAPGKGLEWVARIYPT
QGYTRYADSVKGRFTISADTSKNTAYL
QMNSLRAEDTAVYYCSRWGGEGFYA
MDYWGQGTLVTVSS
VH 74 EVQLVESGGGLVQPGGSLRLSCAASGF
30R/55Q/102S NIRDTYIHWVRQAPGKGLEWVARIYPT
QGYTRYADSVKGRFTISADTSKNTAYL
QMNSLRAEDTAVYYCSRWGGSGFYA
MDYWGQGTLVTVSS
VH 75 EVQLVESGGGLVQPGGSLRLSCAASGF
30R/56A/102S NIRDTYIHWVRQAPGKGLEWVARIYPT
NAYTRYADSVKGRFTISADTSKNTAYL
QMNSLRAEDTAVYYCSRWGGSGFYA
MDYWGQGTLVTVSS
84
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
VH 76
EVQLVESGGGLVQPGGSLRLSCAASGF
30R/56A/102E
NIRDTYIHWVRQAPGKGLEWVARIYPT
NAYTRYADSVKGRFTISADTSKNTAYL
QMNSLRAEDTAVYYCSRWGGEGFYA
MDYWGQGTLVTVSS
Anti-Her2 VL wt 77
DIQMTQSPSSLSASVGDRVTITCRASQ
Trastuzumab
DVNTAVAWYQQKPGKAPKLLIYSASF
and variant
LYSGVPSRFSGSRSGTDFTLTISSLQPE
VL
DFATYYCQQHYTTPPTFGQGTKVEIK
VL 30Q 78
DIQMTQSPSSLSASVGDRVTITCRASQ
DVQTAVAWYQQKPGKAPKLLIYSASF
LYSGVPSRFSGSRSGTDFTLTISSLQPE
DFATYYCQQHYTTPPTFGQGTKVEIK
Other Anti-Tumor Target Binding Sites
[0185] In some embodiments, a binding protein of the present disclosures
comprises an antigen
binding site that binds a tumor target protein. In some embodiments, the tumor
target protein
is a CD38 polypeptide (e.g., a human CD38 polypeptide). In some embodiments,
the tumor
target protein is a HER2 polypeptide (e.g., a human HER2 polypeptide). In some
embodiments, a tumor target protein of the present disclosure includes,
without limitation,
A2AR, APRIL, ATPDase, BAFF, BAFFR, BCMA, BlyS, BTK, BTLA, B7DC, B7H1, B7H4
(also known as VTCN1), B7H5, B7H6, B7H7, B7RP1, B7-4, C3, C5, CCL2 (also known
as
MCP-1), CCL3 (also known as MIP-1a), CCL4 (also known as MIP-1b), CCL5 (also
known
as RANTES), CCL7 (also known as MCP-3), CCL8 (also known as mcp-2), CCL11
(also
known as eotaxin), CCL15 (also known as MIP-1d), CCL17 (also known as TARC),
CCL19
(also known as MIP-3b), CCL20 (also known as MIP-3a), CCL21 (also known as MIP-
2),
CCL24 (also known as MPIF-2/eotaxin-2), CCL25 (also known as TECK), CCL26
(also
known as eotaxin-3), CCR3, CCR4, CD3, CD19, CD20, CD23 (also known as FCER2, a
receptor for IgE), CD24, CD27, CD28, CD38, CD39, CD40, CD70, CD80 (also known
as B7-
1), CD86 (also known as B7-2), CD122, CD137 (also known as 41BB), CD137L,
CD152 (also
known as CTLA4), CD154 (also known as CD4OL), CD160, CD272, CD273 (also known
as
PDL2), CD274 (also known as PDL1), CD275 (also known as B7H2), CD276 (also
known as
B7H3), CD278 (also known as ICOS), CD279 (also known as PD-1), CDH1 (also
known as
E-cadherin), chitinase, CLEC9, CLEC91, CRTH2, CSF-1 (also known as M-CSF), CSF-
2
(also known as GM-CSF), CSF-3 (also known as GCSF), CX3CL1 (also known as
SCYD1),
CXCL12 (also known as SDF1), CXCL13, CXCR3, DNGR-1, ectonucleoside
triphosphate
diphosphohydrolase 1, EGFR, ENTPD1, FCER1A, FCER1, FLAP, FOLH1, Gi24, GITR,
GITRL, GM-CSF, Her2, HHLA2, HMGB1, HVEM, ICOSLG, IDO, IFNa, IgE, IGF1R,
IL2Rbeta, ILl, ILIA, IL1B, IL1F10, IL2, IL4, IL4Ra, IL5, IL5R, IL6, IL7,
IL7Ra, IL8, IL9,
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
IL9R, IL10, rhIL10, IL12, IL13, IL13Ra1, IL13Ra2, IL15, IL17, IL17Rb (also
known as a
receptor for IL25), IL18, IL22, IL23, IL25, IL27, IL33, IL35, ITGB4 (also
known as b4
integrin), ITK, KIR, LAG3, LAMP1, leptin, LPFS2, MEW class II, MUC-1, NCR3LG1,
NKG2D, NTPDase-1, 0X40, OX4OL, PD-1H, platelet receptor, PROM1, S152, SISP1,
SLC,
SPG64, ST2 (also known as a receptor for IL33), STEAP2, Syk kinase, TACI, TDO,
T14,
TIGIT, TIM3, TLR, TLR2, TLR4, TLR5, TLR9, TMEF1, TNFa, TNFRSF7, Tp55, TREM1,
TSLP (also known as a co-receptor for IL7Ra), TSLPR, TWEAK, VEGF, VISTA,
Vstm3,
WUCAM, and XCR1 (also known as GPR5/CCXCR1). In some embodiments, one or more
of the above antigen targets are human antigen targets.
Anti-CD28 Binding Sites
[0186]
Certain aspects of the present disclosure relate to binding proteins that
comprise
an antigen binding site that binds a CD28 polypeptide. In some embodiments,
the CD28
polypeptide is a human CD28 polypeptide, also known as Tp44. Human CD28
polypeptides
are known in the art and include, without limitation, the polypeptides
represented by NCBI
Accession Numbers XP 011510499.1, XP 011510497.1 XP 011510496.1,
_
NP 001230007.1, NP 001230006.1, or NP 006130.1, or a polypeptide produced from
NCBI
Gene ID Number 940. In some embodiments, a binding protein comprising an
antigen
binding site that binds a CD28 polypeptide is monospecific and/or monovalent,
bispecific
and/or bivalent, trispecific and/or trivalent, or multispecific and/or
multivalent. In some
embodiments, a binding protein that comprises an antigen binding site that
binds a CD28
polypeptide is a trispecific binding protein comprising four polypeptides that
form three
antigen binding sites. In some embodiments, a binding protein that comprises
an antigen
binding site that binds a CD28 polypeptide is a trispecific binding protein
comprising four
polypeptides that form three antigen binding sites, one of which binds a CD28
polypeptide,
and one of which binds a CD3 polypeptide. In some embodiments, a binding
protein that
comprises an antigen binding site that binds a CD3 polypeptide is a
trispecific binding protein
comprising four polypeptides that form three antigen binding sites, one of
which binds a
CD28 polypeptide, one of which binds a CD3 polypeptide, and one of which binds
a CD38
polypeptide. In some embodiments, a binding protein that comprises an antigen
binding site
that binds a CD3 polypeptide is a trispecific binding protein comprising four
polypeptides
that form three antigen binding sites, one of which binds a CD28 polypeptide,
one of which
binds a CD3 polypeptide, and one of which binds a HER2 polypeptide. In some
embodiments, a binding protein that comprises an antigen binding site that
binds a CD3
86
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
polypeptide is a trispecific binding protein comprising four polypeptides that
form three
antigen binding sites, one of which binds a CD28 polypeptide, one of which
binds a CD3
polypeptide, and one of which binds a tumor target protein.
[0187] In some embodiments, a binding site that binds CD28 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GYTFTSYY (SEQ ID NO:49), a CDR-H2 sequence comprising the
amino
acid sequence of IYPGNVNT (SEQ ID NO:50), and a CDR-H3 sequence comprising the
amino acid sequence of TRSHYGLDWNFDV (SEQ ID NO:51) and/or an antibody light
chain variable (VL) domain comprising a CDR-L1 sequence comprising the amino
acid
sequence of QNIYVW (SEQ ID NO:52), a CDR-L2 sequence comprising the amino acid
sequence of KAS (SEQ ID NO:53), and a CDR-L3 sequence comprising the amino
acid
sequence of QQGQTYPY (SEQ ID NO:54). In some embodiments, a binding site that
binds
CD28 comprises: an antibody heavy chain variable (VH) domain comprising a CDR-
H1
sequence comprising the amino acid sequence of GYTFTSYY (SEQ ID NO:49), a CDR-
H2
sequence comprising the amino acid sequence of IYPGNVNT (SEQ ID NO:50), and a
CDR-
H3 sequence comprising the amino acid sequence of TRSHYGLDWNFDV (SEQ ID
NO:51); and an antibody light chain variable (VL) domain comprising a CDR-L1
sequence
comprising the amino acid sequence of QNIYVW (SEQ ID NO:52), a CDR-L2 sequence
comprising the amino acid sequence of KAS (SEQ ID NO:53), and a CDR-L3
sequence
comprising the amino acid sequence of QQGQTYPY (SEQ ID NO:54).
[0188] In some embodiments, a binding site that binds CD28 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSTYPGNV
NTNYAQKFQGRATLTVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNFDVWG
KGTTVTVSS (SEQ ID NO:91), and/or an antibody light chain variable (VL) domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIQMTQSPSSLSASVGDRVTITCQASQNIYVWLNWYQQKPGKAPKWYKASNLHTG
VPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIK (SEQ ID
87
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
NO:92). In some embodiments, a binding site that binds CD28 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:91,
and/or
an antibody light chain variable (VL) domain comprising the amino acid
sequence of SEQ ID
NO:92. In some embodiments, a binding site that binds CD28 comprises: an
antibody heavy
chain variable (VH) domain comprising the amino acid sequence of SEQ ID NO:91,
and an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:92.
[0189] In some embodiments of any of the above embodiments, the binding
protein is a
trispecific binding protein. In some embodiments, the trispecific binding
protein comprising
an antigen binding site that binds a tumor target protein (including, without
limitation, CD38
or HER2), an antigen binding site that binds a CD28 polypeptide, and an
antigen binding site
that binds a CD3 polypeptide. In some embodiments, the binding protein is a
trispecific
binding protein comprising four polypeptides comprising three antigen binding
sites, wherein
the polypeptide of formula I and the polypeptide of formula II form a cross-
over light chain-
heavy chain pair (e.g., as described herein). In some embodiments, the VH and
VL domains
of any of the anti-CD28 antigen binding sites described above represent VHI
and VIA and
form a first antigen binding site that binds a CD28 polypeptide. In some
embodiments, the
VH and VL domains of any of the anti-CD28 antigen binding sites described
above and/or in
Table 4 represent VH1 and VIA and form a first antigen binding site that binds
a CD28
polypeptide, VH2 and VL2 form a second antigen binding site that binds a CD3
polypeptide,
and VH3 and VL3 and form a third antigen binding site that binds a tumor
target protein
(including, without limitation, CD38 or HER2).
[0190] Sequences of exemplary anti-CD28 antigen binding sites are provided
in Table 4.
In some embodiments, an anti-CD28 antigen binding site of the present
disclosure comprises
1, 2, 3, 4, 5, or all 6 CDR sequences of an anti-CD28 antibody described in
Table 4. In some
embodiments, an anti-CD28 antigen binding site of the present disclosure
comprises a VH
domain sequence and/or VL domain sequence of an anti-CD28 antibody described
in Table 4.
Table 4. Anti-CD28 binding protein sequences.
Sequence Molecule Description SEQ Sequence
Type ID NO
CDR Anti-CD28 CDR-H1 49 GYTFTSYY
(sup)
CDR-H2 50 IYPGNVNT
88
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
CDR-H3 51 TRSHYGLDWNFDV
CDR-L1 52 QNIYVW
CDR-L2 53 KAS
CDR-L3 54 QQGQTYPY
Variable Anti-CD28 VH 91 QVQLVQSGAEVVKPGASVKVSCKASGYT
domain (sup) FTSYYTHWVRQAPGQGLEWIGSTYPGNVN
TNYAQKFQGRATLTVDTSISTAYMELSRL
RSDDTAVYYCTRSHYGLDWNFDVWGKG
TTVTVSS
VL 92 DIQMTQSPSSLSASVGDRVTITCQASQNIY
VWLNWYQQKPGKAPKLLIYKASNLHTGV
PSRFSGSGSGTDFTLTIS SLQPEDIATYYCQ
QGQTYPYTFGQGTKLEIK
Anti-CD3 Binding Sites
[0191]
Certain aspects of the present disclosure relate to binding proteins that
comprise
an antigen binding site that binds a CD3 polypeptide. In some embodiments, the
CD3
polypeptide is a human CD3 polypeptide, including CD3-delta (also known as
T3D, IMD19,
and CD3-DELTA), CD3-epsilon (also known as T3E, IMD18, and TCRE), and CD3-
gamma
(also known as T3G, IMD17, and CD3-GAMMA). Human CD3 polypeptides are known in
the art and include, without limitation, the polypeptides represented by NCBI
Accession
Numbers XP 006510029.1 or NP 031674.1, or a polypeptide produced from NCBI
Gene ID
Numbers 915, 916, or 917. In some embodiments, a binding protein comprising an
antigen
binding site that binds a CD3 polypeptide is monospecific and/or monovalent,
bispecific
and/or bivalent, trispecific and/or trivalent, or multispecific and/or
multivalent. In some
embodiments, a binding protein that comprises an antigen binding site that
binds a CD3
polypeptide is a trispecific binding protein comprising four polypeptides that
form three
antigen binding sites. In some embodiments, a binding protein that comprises
an antigen
binding site that binds a CD3 polypeptide is a trispecific binding protein
comprising four
polypeptides that form three antigen binding sites, one of which binds a CD28
polypeptide,
and one of which binds a CD3 polypeptide. In some embodiments, a binding
protein that
comprises an antigen binding site that binds a CD3 polypeptide is a
trispecific binding protein
comprising four polypeptides that form three antigen binding sites, one of
which binds a
CD28 polypeptide, one of which binds a CD3 polypeptide, and one of which binds
a CD38
polypeptide. In some embodiments, a binding protein that comprises an antigen
binding site
that binds a CD3 polypeptide is a trispecific binding protein comprising four
polypeptides
89
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
that form three antigen binding sites, one of which binds a CD28 polypeptide,
one of which
binds a CD3 polypeptide, and one of which binds a HER2 polypeptide. In some
embodiments, a binding protein that comprises an antigen binding site that
binds a CD3
polypeptide is a trispecific binding protein comprising four polypeptides that
form three
antigen binding sites, one of which binds a CD28 polypeptide, one of which
binds a CD3
polypeptide, and one of which binds a tumor target protein.
[0192] In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the
amino
acid sequence of IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising
the
amino acid sequence of RGVYYALSPFDY (SEQ ID NO:57); and/or an antibody light
chain
variable (VL) domain comprising a CDR-L1 sequence comprising the amino acid
sequence
of QSLVHX1NX2X3TY, wherein Xi is E or Q, X2 is A or L, and X3 is Q, R, or F
(SEQ ID
NO:180), a CDR-L2 sequence comprising the amino acid sequence of KVS (SEQ ID
NO:64), and a CDR-L3 sequence comprising the amino acid sequence of GQGTQYPFT
(SEQ ID NO:65). In some embodiments, the CDR-L1 sequence of the VL2 domain
comprises an amino acid sequence selected from the group consisting of
QSLVHQNAQTY
(SEQ ID NO:59), QSLVHENLQTY (SEQ ID NO:60), QSLVHENLFTY (SEQ ID NO:61),
and QSLVHENLRTY (SEQ ID NO:62).
[0193] In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the
amino
acid sequence of IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising
the
amino acid sequence of RGVYYALSPFDY (SEQ ID NO:57); and/or an antibody light
chain
variable (VL) domain comprising a CDR-L1 sequence comprising the amino acid
sequence
of QSLVHQNAQTY (SEQ ID NO:59), a CDR-L2 sequence comprising the amino acid
sequence of KVS (SEQ ID NO:64), and a CDR-L3 sequence comprising the amino
acid
sequence of GQGTQYPFT (SEQ ID NO:65). In some embodiments, a binding site that
binds CD3 comprises: an antibody heavy chain variable (VH) domain comprising a
CDR-H1
sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:55), a CDR-
H2
sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID NO:56), and
a
CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ ID
NO:57); and an antibody light chain variable (VL) domain comprising a CDR-L1
sequence
comprising the amino acid sequence of QSLVHQNAQTY (SEQ ID NO:59), a CDR-L2
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
sequence comprising the amino acid sequence of KVS (SEQ ID NO:64), and a CDR-
L3
sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID NO:65).
[0194] In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the
amino
acid sequence of IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising
the
amino acid sequence of RGVYYALSPFDY (SEQ ID NO:57); and/or an antibody light
chain
variable (VL) domain comprising a CDR-L1 sequence comprising the amino acid
sequence
of QSLVHENLQTY (SEQ ID NO:60), a CDR-L2 sequence comprising the amino acid
sequence of KVS (SEQ ID NO:64), and a CDR-L3 sequence comprising the amino
acid
sequence of GQGTQYPFT (SEQ ID NO:65). In some embodiments, a binding site that
binds CD3 comprises: an antibody heavy chain variable (VH) domain comprising a
CDR-H1
sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:55), a CDR-
H2
sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID NO:56), and
a
CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ ID
NO:57); and an antibody light chain variable (VL) domain comprising a CDR-L1
sequence
comprising the amino acid sequence of QSLVHENLQTY (SEQ ID NO:60), a CDR-L2
sequence comprising the amino acid sequence of KVS (SEQ ID NO:64), and a CDR-
L3
sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID NO:65).
[0195] In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the
amino
acid sequence of IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising
the
amino acid sequence of RGVYYALSPFDY (SEQ ID NO:57); and/or an antibody light
chain
variable (VL) domain comprising a CDR-L1 sequence comprising the amino acid
sequence
of QSLVHENLFTY (SEQ ID NO:61), a CDR-L2 sequence comprising the amino acid
sequence of KVS (SEQ ID NO:64), and a CDR-L3 sequence comprising the amino
acid
sequence of GQGTQYPFT (SEQ ID NO:65). In some embodiments, a binding site that
binds CD3 comprises: an antibody heavy chain variable (VH) domain comprising a
CDR-H1
sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:55), a CDR-
H2
sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID NO:56), and
a
CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ ID
NO:57); and an antibody light chain variable (VL) domain comprising a CDR-L1
sequence
comprising the amino acid sequence of QSLVHENLFTY (SEQ ID NO:61), a CDR-L2
91
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
sequence comprising the amino acid sequence of KVS (SEQ ID NO:64), and a CDR-
L3
sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID NO:65).
[0196] In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GFTFTKAW (SEQ ID NO:55), a CDR-H2 sequence comprising the
amino
acid sequence of IKDKSNSYAT (SEQ ID NO:56), and a CDR-H3 sequence comprising
the
amino acid sequence of RGVYYALSPFDY (SEQ ID NO:57); and/or an antibody light
chain
variable (VL) domain comprising a CDR-L1 sequence comprising the amino acid
sequence
of QSLVHENLRTY (SEQ ID NO:62), a CDR-L2 sequence comprising the amino acid
sequence of KVS (SEQ ID NO:64), and a CDR-L3 sequence comprising the amino
acid
sequence of GQGTQYPFT (SEQ ID NO:65). In some embodiments, a binding site that
binds CD3 comprises: an antibody heavy chain variable (VH) domain comprising a
CDR-H1
sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:55), a CDR-
H2
sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID NO:56), and
a
CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ ID
NO:57); and an antibody light chain variable (VL) domain comprising a CDR-L1
sequence
comprising the amino acid sequence of QSLVHENLRTY (SEQ ID NO:62), a CDR-L2
sequence comprising the amino acid sequence of KVS (SEQ ID NO:64), and a CDR-
L3
sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID NO:65).
[0197] In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLVESGGGVVQPGRSLRLSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKDKS
NSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDY
WGQGTLVTVSS (SEQ ID NO:93), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to an amino
acid sequence selected from the group consisting of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIK (SEQ
ID NO:95),
92
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK (SEQ
ID NO:96),
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLFTYLSWYLQKPGQSPQSLIYKVSNR
F SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK (SEQ
ID NO:97), and
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLRTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK (SEQ
ID NO:98). In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:93,
and/or an antibody light chain variable (VL) domain comprising an amino acid
sequence
selected from the group consisting of SEQ ID NO:95, SEQ ID NO:96, SEQ ID
NO:97, and
SEQ ID NO:98. In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:93,
and an antibody light chain variable (VL) domain comprising an amino acid
sequence
selected from the group consisting of SEQ ID NO:95, SEQ ID NO:96, SEQ ID
NO:97, and
SEQ ID NO:98.
[0198] In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLVESGGGVVQPGRSLRLSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKDKS
NSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDY
WGQGTLVTVSS (SEQ ID NO:93), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK (SEQ
ID NO:95). In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:93,
and/or an antibody light chain variable (VL) domain comprising the amino acid
sequence of
93
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
SEQ ID NO:95. In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:93,
and an antibody light chain variable (VL) domain comprising the amino acid
sequence of
SEQ ID NO:95.
[0199] In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMHWVRQAPGKQLEWVAQIKDKS
NSYATYYADSVKGRETISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDY
WGQGTLVTVSS (SEQ ID NO:93), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK (SEQ
ID NO:96). In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:93,
and/or an antibody light chain variable (VL) domain comprising the amino acid
sequence of
SEQ ID NO:96. In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:93,
and an antibody light chain variable (VL) domain comprising the amino acid
sequence of
SEQ ID NO:96.
[0200] In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMHWVRQAPGKQLEWVAQIKDKS
NSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDY
WGQGTLVTVSS (SEQ ID NO:93), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
94
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
88%, at least 89 /o, at least 90%, at least 91%, at least 92%, at least 930 o,
at least 940 o, at least
9500, at least 96 /o, at least 970 o, at least 98%, at least 99%, or 10000
identical to the amino
acid sequence of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLFTYLSWYLQKPGQSPQSLIYKVSNR
F SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIK (SEQ
ID NO:97). In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:93,
and/or an antibody light chain variable (VL) domain comprising the amino acid
sequence of
SEQ ID NO:97. In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:93,
and an antibody light chain variable (VL) domain comprising the amino acid
sequence of
SEQ ID NO:97.
[0201] In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 869/0, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 939/0, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%, or
100% identical to the amino acid sequence of
QVQLVESGGGVVQPGRSLRLSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKDKS
NSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDY
WGQGTLVTVSS (SEQ ID NO:93), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89 /o, at least 90%, at least 91%, at least 92%, at least 93%,
at least 94%, at least
95%, at least 96 /o, at least 97%, at least 98%, at least 99%, or 100%
identical to the amino
acid sequence of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLRTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK (SEQ
ID NO:98). In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:93,
and/or an antibody light chain variable (VL) domain comprising the amino acid
sequence of
SEQ ID NO:98. In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:93,
and an antibody light chain variable (VL) domain comprising the amino acid
sequence of
SEQ ID NO:98.
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
[0202] In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising an amino acid sequence that is at
least 85%, at
least 869/0, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%, at
least 9394., at least 940 o, at least 950 o, at least 96%, at least 970 o, at
least 98%, at least 990 o, or
1000o identical to the amino acid sequence of
QVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMETWVRQAPGKQLEWVAQIKDKS
NSYATYYASSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDYW
GQGTLVTVSS (SEQ ID NO:302), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89 /0, at least 90%, at least 91%, at least 92%, at least 930 o,
at least 940 o, at least
9500, at least 96 /0, at least 970 o, at least 98%, at least 99%, or 100%
identical to the amino
acid sequence of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK (SEQ
ID NO:95). In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:302,
and/or an antibody light chain variable (VL) domain comprising the amino acid
sequence of
SEQ ID NO:95. In some embodiments, a binding site that binds CD3 comprises: an
antibody
heavy chain variable (VH) domain comprising the amino acid sequence of SEQ ID
NO:302,
and an antibody light chain variable (VL) domain comprising the amino acid
sequence of
SEQ ID NO:95.
[0203] In some embodiments of any of the above embodiments, the binding
protein is a
trispecific binding protein. In some embodiments, the trispecific binding
protein comprising
an antigen binding site that binds a tumor target protein (including, without
limitation, CD38
or HER2), an antigen binding site that binds a CD28 polypeptide, and an
antigen binding site
that binds a CD3 polypeptide. In some embodiments, the binding protein is a
trispecific
binding protein comprising four polypeptides comprising three antigen binding
sites, wherein
the polypeptide of formula I and the polypeptide of formula II form a cross-
over light chain-
heavy chain pair (e.g., as described herein). In some embodiments, the VH and
VL domains
of any of the anti-CD3 antigen binding sites described above represent VH2 and
VL2 and form
a second antigen binding site that binds a CD3 polypeptide. In some
embodiments, VH1 and
VIA form a first antigen binding site that binds a CD28 polypeptide, the VH
and VL domains
of any of the anti-CD3 antigen binding sites described above and/or in Table 5
represent VH2
and VL2 and form a second antigen binding site that binds a CD3 polypeptide,
and VH3 and
96
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
VL3 form a third antigen binding site that binds a tumor target protein
(including, without
limitation, CD38 or HER2).
[0204]
Sequences of exemplary anti-CD3 antigen binding sites are provided in Table 5.
In some embodiments, an anti-CD3 antigen binding site of the present
disclosure comprises
1, 2, 3, 4, 5, or all 6 CDR sequences of an anti-CD3 antibody described in
Table 5. In some
embodiments, an anti-CD3 antigen binding site of the present disclosure
comprises a VH
domain sequence and/or VL domain sequence of an anti-CD3 antibody described in
Table 5.
Table 5. Anti-CD3 binding protein sequences.
Sequence Molecule Descriptio SEQ Sequence
Type n ID NO
CDR Anti-CD3 CDR-H1 55 GFTFTKAW
(mid) original
CDR-H2 56 IKDKSNSYAT
original
CDR-H3 57 RGVYYALSPFDY
original
CDR-L1 58 QSLVHNNANTY
original
CDR-L1 59 QSLVHQNAQTY
QQ
CDR-L1 60 QSLVHENLQTY
ENLQ
CDR-L1 61 QSLVHENLFTY
ENLF
CDR-L1 62 QSLVHENLRTY
ENLR
CDR-L1 63 QSLVHDNAQTY
DNAQ
CDR-L2 64 KVS
original
CDR-L3 65 GQGTQYPFT
Original
CD3mid 180 QSLVHX1NX2X3TY,
consensus wherein Xi is E or Q, X2 is A or L,
and X3 is
CDR-L1 Q,R,orF
Variable Anti-CD3 VH 93 QVQLVESGGGVVQPGRSLRLSCAASGFT
Domain (mid) FTKAWMHWVRQAPGKQLEWVAQIKDK
SNSYATYYADSVKGRFTISRDDSKNTLY
LQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVSS
VL 94 DIVMTQTPLSLSVTPGQPASISCKSSQSL
Original VHNNANTYLSWYLQKPGQSPQSLIYKVS
NRFSGVPDRFSGSGSGTDFTLKISRVEAE
DVGVYYCGQGTQYPFTFGSGTKVEIK
VL 95 DIVMTQTPLSLSVTPGQPASISCKSSQSL
32/35 QQ VHQNAQTYLSWYLQKPGQSPQSLIYKVS
97
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
NRFSGVPDRFSGSGSGTDFTLKISRVEAE
DVGVYYCGQGTQYPFTFGSGTKVEIK
VL 96
DIVMTQTPLSLSVTPGQPASISCKSSQSL
ENLQ VHENLQTYLSWYLQKPGQSPQSLIYKVS
NRFSGVPDRFSGSGSGTDFTLKISRVEAE
DVGVYYCGQGTQYPFTFGSGTKVEIK
VL 97
DIVMTQTPLSLSVTPGQPASISCKSSQSL
ENLF
VHENLFTYLSWYLQKPGQSPQSLIYKVS
NRFSGVPDRFSGSGSGTDFTLKISRVEAE
DVGVYYCGQGTQYPFTFGSGTKVEIK
VL 98
DIVMTQTPLSLSVTPGQPASISCKSSQSL
ENLR VHENLRTYLSWYLQKPGQSPQSLIYKVS
NRFSGVPDRFSGSGSGTDFTLKISRVEAE
DVGVYYCGQGTQYPFTFGSGTKVEIK
VL 99
DIVMTQTPLSLSVTPGQPASISCKSSQSL
DNAQ VHDNAQTYLSWYLQKPGQSPQSLIYKVS
NRFSGVPDRFSGSGSGTDFTLKISRVEAE
DVGVYYCGQGTQYPFTFGSGTKVEIK
VH 185S 302 QVQLVESGGGVVQPGRSLRLSCAASGFT
FTKAWMHWVRQAPGKQLEWVAQIKDK
SNSYATYYASSVKGRFTISRDDSKNTLY
LQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVSS
Linkers
[0205] In some embodiments, the linkers Li, L2, L3, and L4 range from no
amino acids
(length=0) to about 100 amino acids long, or less than 100, 50, 40, 30, 20, or
15 amino acids
or less. The linkers can also be 10, 9, 8, 7, 6, 5, 4, 3, 2, or 1 amino acids
long. Li, L2, L3, and
L4 in one binding protein may all have the same amino acid sequence or may all
have
different amino acid sequences.
[0206] Examples of suitable linkers include, for example, GGGGSGGGGS (SEQ
ID
NO:69), GGGGSGGGGSGGGGS (SEQ ID NO: 70), S, RT, TKGPS (SEQ ID NO: 68),
GQPKAAP (SEQ ID NO: 67), GGSGSSGSGG (SEQ ID NO: 71), and DKTHT (SEQ ID
NO:66), as well as those disclosed in International Publication Nos.
W02017/074878 and
W02017/180913. The examples listed above are not intended to limit the scope
of the
disclosure in any way, and linkers comprising randomly selected amino acids
selected from
the group consisting of valine, leucine, isoleucine, serine, threonine,
lysine, arginine,
histidine, aspartate, glutamate, asparagine, glutamine, glycine, and proline
have been shown
to be suitable in the binding proteins.
[0207] The identity and sequence of amino acid residues in the linker may
vary
depending on the type of secondary structural element necessary to achieve in
the linker. For
example, glycine, serine, and alanine are best for linkers having maximum
flexibility. Some
98
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
combination of glycine, proline, threonine, and serine are useful if a more
rigid and extended
linker is necessary. Any amino acid residue may be considered as a linker in
combination
with other amino acid residues to construct larger peptide linkers as
necessary depending on
the desired properties.
[0208] In some embodiments, the length of Li is at least twice the length
of L3. In some
embodiments, the length of L2 is at least twice the length of L4. In some
embodiments, the
length of Li is at least twice the length of L3, and the length of L2 is at
least twice the length
of L4. In some embodiments, Li is 3 to 12 amino acid residues in length, L2 is
3 to 14 amino
acid residues in length, L3 is 1 to 8 amino acid residues in length, and L4 is
1 to 3 amino acid
residues in length. In some embodiments, Li is 5 to 10 amino acid residues in
length, L2 is 5
to 8 amino acid residues in length, L3 is 1 to 5 amino acid residues in
length, and L4 is 1 to 2
amino acid residues in length. In some embodiments, Li is 7 amino acid
residues in length, L2
is 5 amino acid residues in length, L3 is 1 amino acid residue in length, and
L4 is 2 amino acid
residues in length.
[0209] In some embodiments, Li, L2, L3 and L4 each independently are zero
amino acids in
length or comprise a sequence selected from the group consisting of GGGGSGGGGS
(SEQ
ID NO:69), GGGGSGGGGSGGGGS (SEQ ID NO: 70), S, RT, TKGPS (SEQ ID NO: 68),
GQPKAAP (SEQ ID NO: 67), and GGSGSSGSGG (SEQ ID NO: 71). In some
embodiments, Li, L2, L3 and L4 each independently comprise a sequence selected
from the
group consisting of GGGGSGGGGS (SEQ ID NO:69), GGGGSGGGGSGGGGS (SEQ ID
NO: 70), S, RT, TKGPS (SEQ ID NO: 68), GQPKAAP (SEQ ID NO: 67), and
GGSGSSGSGG (SEQ ID NO: 71). In some embodiments, Li comprises the sequence
GQPKAAP (SEQ ID NO: 67), L2 comprises the sequence TKGPS (SEQ ID NO:68), L3
comprises the sequence S, and L4 comprises the sequence RT.
[0210] In some embodiments, at least one of Li, L2, L3 or L4 comprises the
sequence DKTHT
(SEQ ID NO:66). In some embodiments, Li, L2, L3 and L4 comprise the sequence
DKTHT
(SEQ ID NO:66).
Fe regions and constant domains
[0211] In some embodiments, a binding protein of the present disclosure
comprises a
second polypeptide chain further comprising an Fc region linked to CHi, the Fc
region
comprising an immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy
chain
constant domains. In some embodiments, a binding protein of the present
disclosure
comprises a third polypeptide chain further comprising an Fc region linked to
CHi, the Fc
99
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
region comprising an immunoglobulin hinge region and CH2 and CH3
immunoglobulin heavy
chain constant domains. In some embodiments, a binding protein of the present
disclosure
comprises a second polypeptide chain further comprising an Fc region linked to
Cul, the Fc
region comprising an immunoglobulin hinge region and CH2 and CH3
immunoglobulin heavy
chain constant domains, and a third polypeptide chain further comprising an Fc
region linked
to CH1, the Fc region comprising an immunoglobulin hinge region and CH2 and
CH3
immunoglobulin heavy chain constant domains.
[0212] In some embodiments, a binding protein of the present disclosure
comprises a
full-length antibody heavy chain or a polypeptide chain comprising an Fc
region. In some
embodiments, the Fc region is a human Fc region, e.g., a human IgGl, IgG2,
IgG3, or IgG4
Fc region. In some embodiments, the Fc region includes an antibody hinge, Cul,
CH2, CH3,
and optionally CH4 domains. In some embodiments, the Fc region is a human IgG1
Fc
region. In some embodiments, the Fc region is a human IgG4 Fc region. In some
embodiments, the Fc region includes one or more of the mutations described
infra. In some
embodiments, the Fc region is an Fc region of one of the heavy chain
polypeptides (e.g.,
polypeptide 2 or 3) of a binding protein shown in Table 4. In some
embodiments, the heavy
chain constant region is a constant region of one of the heavy chain
polypeptides (e.g.,
polypeptide 2 or 3) of a binding protein shown in Table 4. In some
embodiments, the light
chain constant region is a constant region of one of the light chain
polypeptides (e.g.,
polypeptide 1 or 4) of a binding protein shown in Table 4.
[0213] In some embodiments, a binding protein of the present disclosure
includes one or
two Fc variants. The term "Fc variant" as used herein refers to a molecule or
sequence that is
modified from a native Fc but still comprises a binding site for the salvage
receptor, FcRn
(neonatal Fc receptor). Exemplary Fc variants, and their interaction with the
salvage receptor,
are known in the art. Thus, the term "Fc variant" can comprise a molecule or
sequence that is
humanized from a non-human native Fc. Furthermore, a native Fc comprises
regions that can
be removed because they provide structural features or biological activity
that are not
required for the antibody-like binding proteins of the invention. Thus, the
term "Fc variant"
comprises a molecule or sequence that lacks one or more native Fc sites or
residues, or in
which one or more Fc sites or residues has be modified, that affect or are
involved in: (1)
disulfide bond formation, (2) incompatibility with a selected host cell, (3) N-
terminal
heterogeneity upon expression in a selected host cell, (4) glycosylation, (5)
interaction with
complement, (6) binding to an Fc receptor other than a salvage receptor, or
(7) antibody-
dependent cellular cytotoxicity (ADCC).
100
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
[0214] In some embodiments, a binding protein of the present disclosure
(e.g., a trispecific
binding protein) comprises a "knob" mutation on the second polypeptide chain
and a "hole"
mutation on the third polypeptide chain. In some embodiments, a binding
protein of the present
disclosure comprises a "knob" mutation on the third polypeptide chain and a
"hole" mutation
on the second polypeptide chain. In some embodiments, the "knob" mutation
comprises
substitution(s) at positions corresponding to positions 354 and/or 366 of
human IgG1 or IgG4
according to EU Index In some embodiments, the amino acid substitutions are
S354C,
T366W, T366Y, S354C and T366W, or S354C and T366Y. In some embodiments, the
"knob"
mutation comprises substitutions at positions corresponding to positions 354
and 366 of human
IgG1 or IgG4 according to EU Index. In some embodiments, the amino acid
substitutions are
S354C and T366W. In some embodiments, the "hole" mutation comprises
substitution(s) at
positions corresponding to positions 407 and, optionally, 349, 366, and/or 368
and of human
IgG1 or IgG4 according to EU Index. In some embodiments, the amino acid
substitutions are
Y407V or Y407T and optionally Y349C, T366S, and/or L368A. In some embodiments,
the
"hole" mutation comprises substitutions at positions corresponding to
positions 349, 366, 368,
and 407 of human IgG1 or IgG4 according to EU Index. In some embodiments, the
amino acid
substitutions are Y349C, T366S, L368A, and Y407V
[0215] In some embodiments, the second polypeptide chain further comprises
a first Fc
region linked to CH1, the first Fc region comprising an immunoglobulin hinge
region and CH2
and CH3 immunoglobulin heavy chain constant domains, wherein the first Fc
region comprises
amino acid substitution(s) at positions corresponding to positions 366 and
optionally 354 of
human IgG1 or IgG4 according to EU Index, wherein the amino acid substitutions
are T366W
or T366Y and optionally S354C; and wherein the third polypeptide chain further
comprises a
second Fc region linked to CH1, the second Fc region comprising an
immunoglobulin hinge
region and CH2 and CH3 immunoglobulin heavy chain constant domains, wherein
the second
Fc region comprises amino acid substitution(s) at positions corresponding to
positions 407 and
optionally 349, 366, and/or 368 and of human IgG1 or IgG4 according to EU
Index, wherein
the amino acid substitutions are Y407V or Y407T and optionally Y349C, T366S,
and/or
L368A.
[0216] In some embodiments, the second polypeptide chain further comprises
a first Fc
region linked to CH1, the first Fc region comprising an immunoglobulin hinge
region and CH2
and CH3 immunoglobulin heavy chain constant domains, wherein the first Fc
region comprises
amino acid substitution(s) at positions corresponding to positions 407 and
optionally 349, 366,
and/or 368 and of human IgG1 or IgG4 according to EU Index, wherein the amino
acid
101
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
substitutions are Y407V or Y407T and optionally Y349C, T366S, and/or L368A;
and wherein
the third polypeptide chain further comprises a second Fc region linked to
CH1, the second Fc
region comprising an immunoglobulin hinge region and CH2 and CH3
immunoglobulin heavy
chain constant domains, wherein the second Fc region comprises amino acid
substitution(s) at
positions corresponding to positions 366 and optionally 354 of human IgG1 or
IgG4 according
to EU Index, wherein the amino acid substitutions are T366W or T366Y and
optionally S354C.
[0217] In some embodiments, the second polypeptide chain further comprises
a first Fc
region linked to CH1, the first Fc region comprising an immunoglobulin hinge
region and CH2
and CH3 immunoglobulin heavy chain constant domains, wherein the first Fc
region comprises
amino acid substitution at position corresponding to position 366 of human
IgG1 or IgG4
according to EU Index, wherein the amino acid substitution is T366W; and
wherein the third
polypeptide chain further comprises a second Fc region linked to CH1, the
second Fc region
comprising an immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy
chain
constant domains, wherein the second Fc region comprises amino acid
substitution(s) at
positions corresponding to positions 366, 368, and/or 407 and of human IgG1 or
IgG4
according to EU Index, wherein the amino acid substitutions are T366S, L368A,
and/or
Y407V
[0218] In some embodiments, the second polypeptide chain further comprises
a first Fc
region linked to CH1, the first Fc region comprising an immunoglobulin hinge
region and CH2
and CH3 immunoglobulin heavy chain constant domains, wherein the first Fc
region comprises
amino acid substitution(s) at positions corresponding to positions 366, 368,
and/or 407 and of
human IgG1 or IgG4 according to EU Index, wherein the amino acid substitutions
are T366S,
L368A, and/or Y407V; and wherein the third polypeptide chain further comprises
a second Fc
region linked to CH1, the second Fc region comprising an immunoglobulin hinge
region and
CH2 and CH3 immunoglobulin heavy chain constant domains, wherein the second Fc
region
comprises amino acid substitution at position corresponding to position 366 of
human IgG1 or
IgG4 according to EU Index, wherein the amino acid substitution is T366W.
[0219] In some embodiments, the second polypeptide chain further comprises
a first Fc
region linked to CH1, the first Fc region comprising an immunoglobulin hinge
region and CH2
and CH3 immunoglobulin heavy chain constant domains, wherein the first Fc
region comprises
amino acid substitutions at positions corresponding to positions 354 and 366
of human IgG1
or IgG4 according to EU Index, wherein the amino acid substitutions are S354C
and T366W;
and wherein the third polypeptide chain further comprises a second Fc region
linked to CH1,
the second Fc region comprising an immunoglobulin hinge region and CH2 and CH3
102
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
immunoglobulin heavy chain constant domains, wherein the second Fc region
comprises amino
acid substitutions at positions corresponding to positions 349, 366, 368, and
407 of human
IgG1 or IgG4 according to EU Index, wherein the amino acid substitutions are
Y349C, T366S,
L368A, and Y407V In some embodiments, the second polypeptide chain further
comprises a
first Fc region linked to CH1, the first Fc region comprising an
immunoglobulin hinge region
and CH2 and CH3 immunoglobulin heavy chain constant domains, wherein the first
Fc region
comprises amino acid substitutions at positions corresponding to positions
349, 366, 368, and
407 of human IgG1 or IgG4 according to EU Index, wherein the amino acid
substitutions are
Y349C, T366S, L368A, and Y407V; and wherein the third polypeptide chain
further comprises
a second Fc region linked to CH1, the second Fc region comprising an
immunoglobulin hinge
region and CH2 and CH3 immunoglobulin heavy chain constant domains, wherein
the second
Fc region comprises amino acid substitutions at positions corresponding to
positions 354 and
366 of human IgG1 or IgG4 according to EU Index, wherein the amino acid
substitutions are
S354C and T366W. In some embodiments, the first and/or second Fc regions are
human IgG1
Fc regions. In some embodiments, the first and/or second Fc regions are human
IgG4 Fc
regions.
[0220] In some embodiments, the second polypeptide chain further comprises
a first Fc
region linked to CH1, wherein the first Fc region is a human IgG4 Fc region
comprising an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains, wherein the first Fc region comprises amino acid substitutions at
positions
corresponding to positions 228, 354, 366, and 409 of human IgG4 according to
EU Index,
wherein the amino acid substitutions are S228P, S354C, T366W, and R409K; and
wherein the
third polypeptide chain further comprises a second Fc region linked to CH1,
wherein the second
Fc region is a human IgG4 Fc region comprising an immunoglobulin hinge region
and CH2
and CH3 immunoglobulin heavy chain constant domains, wherein the second Fc
region
comprises amino acid substitutions at positions corresponding to positions
228, 349, 366, 368,
407, and 409 of human IgG4 according to EU Index, wherein the amino acid
substitutions are
S228P, Y349C, T366S, L368A, Y407V, and R409K. In some embodiments, the second
polypeptide chain further comprises a first Fc region linked to CH1, wherein
the first Fc region
is a human IgG4 Fc region comprising an immunoglobulin hinge region and CH2
and CH3
immunoglobulin heavy chain constant domains, wherein the first Fc region
comprises amino
acid substitutions at positions corresponding to positions 228, 349, 366, 368,
407, and 409 of
human IgG4 according to EU Index, wherein the amino acid substitutions are
S228P, Y349C,
T366S, L368A, Y407V, and R409K; and wherein the third polypeptide chain
further comprises
103
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
a second Fc region linked to CH1, wherein the second Fc region is a human IgG4
Fc region
comprising an immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy
chain
constant domains, wherein the second Fc region comprises amino acid
substitutions at positions
corresponding to positions 228, 354, 366, and 409 of human IgG4 according to
EU Index,
wherein the amino acid substitutions are S228P, S354C, T366W, and R409K.
[0221] In some embodiments, the second polypeptide chain further comprises
a first Fc
region linked to CH1, wherein the first Fc region is a human IgG4 Fc region
comprising an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains, wherein the first Fc region comprises amino acid substitutions at
positions
corresponding to positions 234, 235, 354, and 366 of human IgG4 according to
EU Index,
wherein the amino acid substitutions are F234A, L235A, S354C, and T366W; and
wherein the
third polypeptide chain further comprises a second Fc region linked to CH1,
wherein the second
Fc region is a human IgG4 Fc region comprising an immunoglobulin hinge region
and CH2
and CH3 immunoglobulin heavy chain constant domains, wherein the second Fc
region
comprises amino acid substitutions at positions corresponding to positions
234, 235, 349, 366,
368, and 407 of human IgG4 according to EU Index, wherein the amino acid
substitutions are
F234A, L235A, Y349C, T366S, L368A, and Y407V In some embodiments, the second
polypeptide chain further comprises a first Fc region linked to CH1, wherein
the first Fc region
is a human IgG4 Fc region comprising an immunoglobulin hinge region and CH2
and CH3
immunoglobulin heavy chain constant domains, wherein the first Fc region
comprises amino
acid substitutions at positions corresponding to positions 234, 235, 349, 366,
368, and 407 of
human IgG4 according to EU Index, wherein the amino acid substitutions are
F234A, L235A,
Y349C, T366S, L368A, and Y407V; and wherein the third polypeptide chain
further comprises
a second Fc region linked to CH1, wherein the second Fc region is a human IgG4
Fc region
comprising an immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy
chain
constant domains, wherein the second Fc region comprises amino acid
substitutions at positions
corresponding to positions 234, 235, 354, and 366 of human IgG4 according to
EU Index,
wherein the amino acid substitutions are F234A, L235A, S354C, and T366W.
[0222] In some embodiments, a binding protein of the present disclosure
comprises one
or more mutations to reduce effector function, e.g., Fc receptor-mediated
antibody-dependent
cellular phagocytosis (ADCP), complement-dependent cytotoxicity (CDC), and/or
antibody-
dependent cellular cytotoxicity (ADCC) In some embodiments, the second
polypeptide
chain further comprises a first Fc region linked to CH1, the first Fc region
comprising an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
104
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
domains; wherein the third polypeptide chain further comprises a second Fc
region linked to
Cm, the second Fc region comprising an immunoglobulin hinge region and CH2 and
CH3
immunoglobulin heavy chain constant domains; wherein the first and second Fc
regions are
human IgG1 Fc regions; and wherein the first and the second Fc regions each
comprise
amino acid substitutions at positions corresponding to positions 234 and 235
of human IgG1
according to EU Index, wherein the amino acid substitutions are L234A and
L235A. In some
embodiments, the Fc regions of the second and the third polypeptide chains are
human IgG1
Fc regions, and wherein the Fc regions each comprise amino acid substitutions
at positions
corresponding to positions 234 and 235 of human IgG1 according to EU Index,
wherein the
amino acid substitutions are L234A and L235A. In some embodiments, the second
polypeptide chain further comprises a first Fc region linked to Cm, the first
Fc region
comprising an immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy
chain
constant domains; wherein the third polypeptide chain further comprises a
second Fc region
linked to Cm, the second Fc region comprising an immunoglobulin hinge region
and CH2 and
CH3 immunoglobulin heavy chain constant domains; wherein the first and second
Fc regions
are human IgG1 Fc regions; and wherein the first and the second Fc regions
each comprise
amino acid substitutions at positions corresponding to positions 234, 235, and
329 of human
IgG1 according to EU Index, wherein the amino acid substitutions are L234A,
L235A, and
P329A. In some embodiments, the Fc regions of the second and the third
polypeptide chains
are human IgG1 Fc regions, and wherein the Fc regions each comprise amino acid
substitutions at positions corresponding to positions 234, 235, and 329 of
human IgG1
according to EU Index, wherein the amino acid substitutions are L234A, L235A,
and P329A.
In some embodiments, the Fc regions of the second and the third polypeptide
chains are
human IgG4 Fc regions, and the Fc regions each comprise amino acid
substitutions at
positions corresponding to positions 234 and 235 of human IgG4 according to EU
Index,
wherein the amino acid substitutions are F234A and L235A. In some embodiments,
the
binding protein comprises a second polypeptide chain further comprising a
first Fc region
linked to Cm, the first Fc region comprising an immunoglobulin hinge region
and CH2 and
CH3 immunoglobulin heavy chain constant domains, and a third polypeptide chain
further
comprising a second Fc region linked to Cm, the second Fc region comprising an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains; and wherein the first and the second Fc regions each comprise amino
acid
substitutions at positions corresponding to positions 234 and 235 of human
IgG4 according to
EU Index, wherein the amino acid substitutions are F234A and L235A.
105
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
[0223] In some embodiments, the second polypeptide chain further comprises
a first Fc
region linked to CH1, wherein the first Fc region is a human IgG4 Fc region
comprising an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains, wherein the first Fc region comprises amino acid substitutions at
positions
corresponding to positions 228, 234, 235, 354, 366, and 409 of human IgG4
according to EU
Index, wherein the amino acid substitutions are S228P, F234A, L235A, S354C,
T366W, and
R409K; and wherein the third polypeptide chain further comprises a second Fc
region linked
to CH1, wherein the second Fc region is a human IgG4 Fc region comprising an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains, wherein the second Fc region comprises amino acid substitutions at
positions
corresponding to positions 228, 234, 235, 349, 366, 368, 407, and 409 of human
IgG4
according to EU Index, wherein the amino acid substitutions are S228P, F234A,
L235A,
Y349C, T366S, L368A, Y407V, and R409K. In some embodiments, the second
polypeptide
chain further comprises a first Fc region linked to CH1, wherein the first Fc
region is a
human IgG4 Fc region comprising an immunoglobulin hinge region and CH2 and CH3
immunoglobulin heavy chain constant domains, wherein the first Fc region
comprises amino
acid substitutions at positions corresponding to positions 228, 234, 235, 349,
366, 368, 407,
and 409 of human IgG4 according to EU Index, wherein the amino acid
substitutions are
S228P, F234A, L235A, Y349C, T366S, L368A, Y407V, and R409K; and wherein the
third
polypeptide chain further comprises a second Fc region linked to CH1, wherein
the second Fc
region is a human IgG4 Fc region comprising an immunoglobulin hinge region and
CH2 and
CH3 immunoglobulin heavy chain constant domains, wherein the second Fc region
comprises amino acid substitutions at positions corresponding to positions
228, 234, 235,
354, 366, and 409 of human IgG4 according to EU Index, wherein the amino acid
substitutions are S228P, F234A, L235A, S354C, T366W, and R409K.
[0224] In some embodiments, the Fc region is a human IgG4 Fc region
comprising one or
more mutations that reduce or eliminate FcyI and/or Fcyll binding. In some
embodiments,
the Fc region is a human IgG4 Fc region comprising one or more mutations that
reduce or
eliminate FcyI and/or FcyII binding but do not affect FcRn binding. In some
embodiments,
the Fc region is a human IgG4 Fc region comprising amino acid substitutions at
positions
corresponding to positions 228 and/or 409 of human IgG4 according to EU Index.
In some
embodiments, the amino acid substitutions are S228P and /or R409K. In some
embodiments,
the Fc region is a human IgG4 Fc region comprising amino acid substitutions at
positions
106
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
corresponding to positions 234 and/or 235 of human IgG4 according to EU Index.
In some
embodiments, the amino acid substitutions are F234A and/or L235A. In some
embodiments,
the Fc region is a human IgG4 Fc region comprising amino acid substitutions at
positions
corresponding to positions 228, 234, 235, and/or 409 of human IgG4 according
to EU Index.
In some embodiments, the amino acid substitutions are S228P, F234A, L235A, and
/or
R409K. In some embodiments, the Fc region is a human IgG4 Fc region comprising
amino
acid substitutions at positions corresponding to positions 233-236 of human
IgG4 according
to EU Index. In some embodiments, the amino acid substitutions are E233P,
F234V, L235A,
and a deletion at 236. In some embodiments, the Fc region is a human IgG4 Fc
region
comprising amino acid mutations at substitutions corresponding to positions
228, 233-236,
and/or 409 of human IgG4 according to EU Index. In some embodiments, the amino
acid
mutations are S228P; E233P, F234V, L235A, and a deletion at 236; and /or
R409K.
[0225] In some embodiments, the Fc region comprises one or more mutations
that reduce
or eliminate Fc receptor binding and/or effector function of the Fc region
(e.g., Fc receptor-
mediated antibody-dependent cellular phagocytosis (ADCP), complement-dependent
cytotoxicity (CDC), and/or antibody-dependent cellular cytotoxicity (ADCC)).
[0226] In some embodiments, the Fc region is a human IgG1 Fc region
comprising one or
more amino acid substitutions at positions corresponding to positions 234,
235, and/or 329 of
human IgG1 according to EU Index. In some embodiments, the amino acid
substitutions are
L234A, L235A, and/or P329A. In some embodiments, the Fc region is a human IgG1
Fc
region comprising amino acid substitutions at positions corresponding to
positions 298, 299,
and/or 300 of human IgG1 according to EU Index. In some embodiments, the amino
acid
substitutions are S298N, T299A, and/or Y300S.
[0227] In some embodiments, a binding protein of the present disclosure
comprises one
or more mutations to improve stability, e.g., of the hinge region and/or dimer
interface of
IgG4 (See e.g., Spiess, C. et al. (2013)1 Biol. Chem. 288:26583-26593). In
some
embodiments, the mutation comprises substitutions at positions corresponding
to positions
228 and 409 of human IgG4 according to EU Index, wherein the amino acid
substitutions are
S228P and R409K. In some embodiments, the binding protein comprises a second
polypeptide chain further comprising a first Fc region linked to CFn, the
first Fc region
comprising an immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy
chain
constant domains, and a third polypeptide chain further comprising a second Fc
region linked
to Cm, the second Fc region comprising an immunoglobulin hinge region and CH2
and CH3
immunoglobulin heavy chain constant domains; wherein the first and second Fc
regions are
107
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
human IgG4 Fc regions; and wherein the first and the second Fc regions each
comprise
amino acid substitutions at positions corresponding to positions 228 and 409
of human IgG4
according to EU Index, wherein the amino acid substitutions are S228P and
R409K. In some
embodiments, a binding protein of the present disclosure comprises knob and
hole mutations
and one or more mutations to improve stability. In some embodiments, the first
and/or second
Fc regions are human IgG4 Fc regions.
[0228] In some embodiments, the Fc region is a human IgG1 Fc region
comprising one or
more amino acid substitutions at positions corresponding to positions 234,
235, and/or 329 of
human IgG1 according to EU Index. In some embodiments, the amino acid
substitutions are
L234A, L235A, and/or P329A. In some embodiments, the Fc region is a human IgG1
Fc
region comprising amino acid substitutions at positions corresponding to
positions 298, 299,
and/or 300 of human IgG1 according to EU Index. In some embodiments, the amino
acid
substitutions are S298N, T299A, and/or Y300S.
Nucleic acids
[0229] Other aspects of the present disclosure relate to isolated nucleic
acid molecules
comprising a nucleotide sequence encoding any of the binding proteins
described herein.
Exemplary and non-limiting nucleic acid sequences are provided in Table 5.
[0230] Other aspects of the present disclosure relate to kits of
polynucleotides, e.g., that
encode one or more polypeptides of a binding protein as described herein. In
some
embodiments, a kit of polynucleotides of the present disclosure comprises one,
two, three, or
four polynucleotides of a kit of polynucleotides comprising: (a) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:189, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:190, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:191, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:192; (b) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:193, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:194, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:195, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:196; (c) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:197, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:198, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:199, and a fourth
polynucleotide
108
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
comprising the polynucleotide sequence of SEQ ID NO:200; (d) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:201, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:202, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:203, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:204; (e) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:205, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:206, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:207, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:208; (f) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:209, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:210, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:211, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:212; (g) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:213, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:214, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:215, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:216; (h) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:217, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:218, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:219, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:220; (i) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:221, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:222, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:223, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:224; (j) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:225, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:226, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:227, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:228; (k) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:229, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:230, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:231, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:232; (1) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:233, a second
polynucleotide
109
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
comprising the polynucleotide sequence of SEQ ID NO:234, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:235, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:236; (m) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:237, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:238, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:239, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:240; (n) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:241, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:242, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:243, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:244; (o) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:245, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:246, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:247, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:248; (p) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:249, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:250, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:251, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:252; (q) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:253, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:254, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:255, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:256; (r) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:257, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:258, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:259, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:260; (s) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:261, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:262, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:263, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:264; (t) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:265, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:266, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:267, and a fourth
polynucleotide
110
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
comprising the polynucleotide sequence of SEQ ID NO:268; (u) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:269, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:270, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:271, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:272; or (v) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:273, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:274, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:275, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:276.
[0231] Other aspects of the present disclosure relate to a vector system
comprising one or
more vectors encoding a first, second, third, and fourth polypeptide chain of
any of the
binding proteins described herein. In some embodiments, the vector system
comprises a first
vector encoding the first polypeptide chain of the binding protein, a second
vector encoding
the second polypeptide chain of the binding protein, a third vector encoding
the third
polypeptide chain of the binding protein, and a fourth vector encoding the
fourth polypeptide
chain of the binding protein, e.g., as shown in the polynucleotides of Table
6. In some
embodiments, the vector system comprises a first vector encoding the first and
second
polypeptide chains of the binding protein, and a second vector encoding the
third and fourth
polypeptide chains of the binding protein. In some embodiments, the vector
system comprises
a first vector encoding the first and third polypeptide chains of the binding
protein, and a
second vector encoding the second and fourth polypeptide chains of the binding
protein. In
some embodiments, the vector system comprises a first vector encoding the
first and fourth
polypeptide chains of the binding protein, and a second vector encoding the
second and third
polypeptide chains of the binding protein. In some embodiments, the vector
system comprises
a first vector encoding the first, second, third, and fourth polypeptide
chains of the binding
protein. The one or more vectors of the vector system may be any of the
vectors described
herein. In some embodiments, the one or more vectors are expression vectors.
In some
embodiments, the first, second, third, and fourth polynucleotides are present
on one or more
expression vectors, e.g., one, two, three, or four expression vectors.
[0232] Standard recombinant DNA methodologies are used to construct the
polynucleotides that encode the polypeptides which form the binding proteins,
incorporate
these polynucleotides into recombinant expression vectors, and introduce such
vectors into
host cells. See e.g., Sambrook et at., 2001, MOLECULAR CLONING: A LABORATORY
MANUAL
(Cold Spring Harbor Laboratory Press, 3rd ed.). Enzymatic reactions and
purification
111
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
techniques may be performed according to manufacturer's specifications, as
commonly
accomplished in the art, or as described herein. Unless specific definitions
are provided, the
nomenclature utilized in connection with, and the laboratory procedures and
techniques of,
analytical chemistry, synthetic organic chemistry, and medicinal and
pharmaceutical
chemistry described herein are those well-known and commonly used in the art.
Similarly,
conventional techniques may be used for chemical syntheses, chemical analyses,
pharmaceutical preparation, formulation, delivery, and treatment of patients.
[0233] In some embodiments, the isolated nucleic acid is operably linked to a
heterologous
promoter to direct transcription of the binding protein-coding nucleic acid
sequence. A
promoter may refer to nucleic acid control sequences which direct
transcription of a nucleic
acid. A first nucleic acid sequence is operably linked to a second nucleic
acid sequence when
the first nucleic acid sequence is placed in a functional relationship with
the second nucleic
acid sequence. For instance, a promoter is operably linked to a coding
sequence of a binding
protein if the promoter affects the transcription or expression of the coding
sequence.
Examples of promoters may include, but are not limited to, promoters obtained
from the
genomes of viruses (such as polyoma virus, fowlpox virus, adenovirus (such as
Adenovirus
2), bovine papilloma virus, avian sarcoma virus, cytomegalovirus, a
retrovirus, hepatitis-B
virus, Simian Virus 40 (SV40), and the like), from heterologous eukaryotic
promoters (such
as the actin promoter, an immunoglobulin promoter, from heat-shock promoters,
and the
like), the CAG-promoter (Niwa et al., Gene 108(2):193-9, 1991), the
phosphoglycerate
kinase (PGK)-promoter, a tetracycline-inducible promoter (Masui et al.,
Nucleic Acids Res.
33:e43, 2005), the lac system, the trp system, the tac system, the trc system,
major operator
and promoter regions of phage lambda, the promoter for 3-phosphoglycerate
kinase, the
promoters of yeast acid phosphatase, and the promoter of the yeast alpha-
mating factors.
Polynucleotides encoding binding proteins of the present disclosure may be
under the control
of a constitutive promoter, an inducible promoter, or any other suitable
promoter described
herein or other suitable promoter that will be readily recognized by one
skilled in the art.
[0234] In some embodiments, the isolated nucleic acid is incorporated into a
vector. In some
embodiments, the vector is an expression vector. Expression vectors may
include one or more
regulatory sequences operatively linked to the polynucleotide to be expressed
The term
"regulatory sequence" includes promoters, enhancers and other expression
control elements
(e.g., polyadenylation signals). Examples of suitable enhancers may include,
but are not
limited to, enhancer sequences from mammalian genes (such as globin, elastase,
albumin, a-
fetoprotein, insulin and the like), and enhancer sequences from a eukaryotic
cell virus (such
112
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
as SV40 enhancer on the late side of the replication origin (bp 100-270), the
cytomegalovirus
early promoter enhancer, the polyoma enhancer on the late side of the
replication origin,
adenovirus enhancers, and the like). Examples of suitable vectors may include,
for example,
plasmids, cosmids, episomes, transposons, and viral vectors (e.g., adenoviral,
vaccinia viral,
Sindbis-viral, measles, herpes viral, lentiviral, retroviral, adeno-associated
viral vectors, etc.).
Expression vectors can be used to transfect host cells, such as, for example,
bacterial cells,
yeast cells, insect cells, and mammalian cells. Biologically functional viral
and plasmid
DNA vectors capable of expression and replication in a host are known in the
art, and can be
used to transfect any cell of interest.
Host cells
[0235] Other aspects of the present disclosure relate to a host cell (e.g., an
isolated host cell)
comprising one or more isolated polynucleotides, vectors, and/or vector
systems described
herein. In some embodiments, an isolated host cell of the present disclosure
is cultured in
vitro. In some embodiments, the host cell is a bacterial cell (e.g., an E.
coil cell). In some
embodiments, the host cell is a yeast cell (e.g., an S. cerevisiae cell). In
some embodiments,
the host cell is an insect cell. Examples of insect host cells may include,
for example,
Drosophila cells (e.g., S2 cells), Trichoplusia ni cells (e.g., High FiveTM
cells), and
Spodoptera frugtperda cells (e.g., Sf21 or Sf9 cells). In some embodiments,
the host cell is a
mammalian cell. Examples of mammalian host cells may include, for example,
human
embryonic kidney cells (e.g., 293 or 293 cells subcloned for growth in
suspension culture),
Expi293 cells, CHO cells, baby hamster kidney cells (e.g., BEM, ATCC CCL 10),
mouse
sertoli cells (e.g., TM4 cells), monkey kidney cells (e.g., CV1 ATCC CCL 70),
African green
monkey kidney cells (e.g., VERO-76, ATCC CRL-1587), human cervical carcinoma
cells
(e.g., HELA, ATCC CCL 2), canine kidney cells (e.g., MDCK, ATCC CCL 34),
buffalo rat
liver cells (e.g., BRL 3A, ATCC CRL 1442), human lung cells (e.g., W138, ATCC
CCL 75),
human liver cells (e.g., Hep G2, HB 8065), mouse mammary tumor cells (e.g.,
MMT 060562,
ATCC CCL51), TRI cells, MRC 5 cells, FS4 cells, a human hepatoma line (e.g.,
Hep G2),
and myeloma cells (e.g., NSO and Sp2/0 cells).
[0236] Other aspects of the present disclosure relate to a method of producing
any of the
binding proteins described herein. In some embodiments, the method includes a)
culturing a
host cell (e.g., any of the host cells described herein) comprising an
isolated nucleic acid,
vector, and/or vector system (e.g., any of the isolated nucleic acids,
vectors, and/or vector
113
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
systems described herein) under conditions such that the host cell expresses
the binding
protein; and b) isolating the binding protein from the host cell. Methods of
culturing host
cells under conditions to express a protein are well known to one of ordinary
skill in the art.
Methods of isolating proteins from cultured host cells are well known to one
of ordinary skill
in the art, including, for example, by affinity chromatography (e.g., two step
affinity
chromatography comprising protein A affinity chromatography followed by size
exclusion
chromatography).
Pharmaceutical Compositions
[0237] Therapeutic or pharmaceutical compositions comprising binding
proteins are
within the scope of the disclosure. Such therapeutic or pharmaceutical
compositions can
comprise a therapeutically effective amount of a binding protein, or binding
protein-drug
conjugate, in admixture with a pharmaceutically or physiologically acceptable
formulation
agent selected for suitability with the mode of administration.
[0238] Acceptable formulation materials are nontoxic to recipients at the
dosages and
concentrations employed.
[0239] The pharmaceutical composition can contain formulation materials for
modifying,
maintaining, or preserving, for example, the pH, osmolarity, viscosity,
clarity, color,
isotonicity, odor, sterility, stability, rate of dissolution or release,
adsorption, or penetration of
the composition. Suitable formulation materials include, but are not limited
to, amino acids
(such as glycine, glutamine, asparagine, arginine, or lysine), antimicrobials,
antioxidants
(such as ascorbic acid, sodium sulfite, or sodium hydrogen-sulfite), buffers
(such as borate,
bicarbonate, Tris-HCl, citrates, phosphates, or other organic acids), bulking
agents (such as
mannitol or glycine), chelating agents (such as ethylenediamine tetraacetic
acid (EDTA)),
complexing agents (such as caffeine, polyvinylpyrrolidone, beta-cyclodextrin,
or
hydroxypropyl-beta-cyclodextrin), fillers, monosaccharides, disaccharides, and
other
carbohydrates (such as glucose, mannose, or dextrins), proteins (such as serum
albumin,
gelatin, or immunoglobulins), coloring, flavoring and diluting agents,
emulsifying agents,
hydrophilic polymers (such as polyvinylpyrrolidone), low molecular weight
polypeptides,
salt-forming counterions (such as sodium), preservatives (such as benzalkonium
chloride,
benzoic acid, salicylic acid, thimerosal, phenethyl alcohol, methylparaben,
propylparaben,
chlorhexidine, sorbic acid, or hydrogen peroxide), solvents (such as glycerin,
propylene
glycol, or polyethylene glycol), sugar alcohols (such as mannitol or
sorbitol), suspending
agents, surfactants or wetting agents (such as pluronics; PEG; sorbitan
esters; polysorbates
114
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
such as polysorbate 20 or polysorbate 80; triton; tromethamine; lecithin;
cholesterol or
tyloxapal), stability enhancing agents (such as sucrose or sorbitol), tonicity
enhancing agents
(such as alkali metal halides ¨ e.g., sodium or potassium chloride ¨ or
mannitol sorbitol),
delivery vehicles, diluents, excipients and/or pharmaceutical adjuvants (see,
e.g.,
REMINGTON'S PHARMACEUTICAL SCIENCES (18th Ed., A.R. Gennaro, ed., Mack
Publishing
Company 1990), and subsequent editions of the same, incorporated herein by
reference for
any purpose).
[0240] The optimal pharmaceutical composition will be determined by a
skilled artisan
depending upon, for example, the intended route of administration, delivery
format, and
desired dosage. Such compositions can influence the physical state, stability,
rate of in vivo
release, and rate of in vivo clearance of the binding protein.
[0241] The primary vehicle or carrier in a pharmaceutical composition can
be either
aqueous or non-aqueous in nature. For example, a suitable vehicle or carrier
for injection can
be water, physiological saline solution, or artificial cerebrospinal fluid,
possibly
supplemented with other materials common in compositions for parenteral
administration.
Neutral buffered saline or saline mixed with serum albumin are further
exemplary vehicles.
Other exemplary pharmaceutical compositions comprise Tris buffer of about pH
7.0-8.5, or
acetate buffer of about pH 4.0-5.5, which can further include sorbitol or a
suitable substitute.
In one embodiment of the disclosure, binding protein compositions can be
prepared for
storage by mixing the selected composition having the desired degree of purity
with optional
formulation agents in the form of a lyophilized cake or an aqueous solution.
Further, the
binding protein can be formulated as a lyophilizate using appropriate
excipients such as
sucrose.
[0242] The pharmaceutical compositions of the disclosure can be selected
for parenteral
delivery or subcutaneous. Alternatively, the compositions can be selected for
inhalation or
for delivery through the digestive tract, such as orally. The preparation of
such
pharmaceutically acceptable compositions is within the skill of the art.
[0243] The formulation components are present in concentrations that are
acceptable to
the site of administration. For example, buffers are used to maintain the
composition at
physiological pH or at a slightly lower pH, typically within a pH range of
from about 5 to
about 8.
[0244] When parenteral administration is contemplated, the therapeutic
compositions for
use can be in the form of a pyrogen-free, parenterally acceptable, aqueous
solution
comprising the desired binding protein in a pharmaceutically acceptable
vehicle. A
115
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
particularly suitable vehicle for parenteral injection is sterile distilled
water in which a
binding protein is formulated as a sterile, isotonic solution, properly
preserved. Yet another
preparation can involve the formulation of the desired molecule with an agent,
such as
injectable microspheres, bio-erodible particles, polymeric compounds (such as
polylactic acid
or polyglycolic acid), beads, or liposomes, that provides for the controlled
or sustained
release of the product which can then be delivered via a depot injection.
Hyaluronic acid can
also be used, and this can have the effect of promoting sustained duration in
the circulation.
Other suitable means for the introduction of the desired molecule include
implantable drug
delivery devices.
[0245] In one embodiment, a pharmaceutical composition can be formulated
for
inhalation. For example, a binding protein can be formulated as a dry powder
for inhalation.
Binding protein inhalation solutions can also be formulated with a propellant
for aerosol
delivery. In yet another embodiment, solutions can be nebulized.
[0246] It is also contemplated that certain formulations can be
administered orally. In
one embodiment of the disclosure, binding proteins that are administered in
this fashion can
be formulated with or without those carriers customarily used in the
compounding of solid
dosage forms such as tablets and capsules For example, a capsule can be
designed to release
the active portion of the formulation at the point in the gastrointestinal
tract where
bioavailability is maximized and pre-systemic degradation is minimized
Additional agents
can be included to facilitate absorption of the binding protein. Diluents,
flavorings, low
melting point waxes, vegetable oils, lubricants, suspending agents, tablet
disintegrating
agents, and binders can also be employed.
[0247] Another pharmaceutical composition can involve an effective quantity
of binding
proteins in a mixture with non-toxic excipients that are suitable for the
manufacture of
tablets. By dissolving the tablets in sterile water, or another appropriate
vehicle, solutions
can be prepared in unit-dose form. Suitable excipients include, but are not
limited to, inert
diluents, such as calcium carbonate, sodium carbonate or bicarbonate, lactose,
or calcium
phosphate; or binding agents, such as starch, gelatin, or acacia; or
lubricating agents such as
magnesium stearate, stearic acid, or talc.
[0248] Additional pharmaceutical compositions of the disclosure will be
evident to those
skilled in the art, including formulations involving binding proteins in
sustained- or
controlled-delivery formulations. Techniques for formulating a variety of
other sustained- or
controlled-delivery means, such as liposome carriers, bio-erodible
microparticles or porous
beads and depot injections, are also known to those skilled in the art.
Additional examples of
116
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
sustained-release preparations include semipermeable polymer matrices in the
form of shaped
articles, e.g. films, or microcapsules. Sustained release matrices can include
polyesters,
hydrogels, polylactides, copolymers of L-glutamic acid and gamma ethyl-L-
glutamate,
poly(2-hydroxyethyl-methacrylate), ethylene vinyl acetate, or poly-D(-)-3-
hydroxybutyric
acid. Sustained-release compositions can also include liposomes, which can be
prepared by
any of several methods known in the art.
[0249] Pharmaceutical compositions to be used for in vivo administration
typically must
be sterile. This can be accomplished by filtration through sterile filtration
membranes.
Where the composition is lyophilized, sterilization using this method can be
conducted either
prior to, or following, lyophilization and reconstitution. The composition for
parenteral
administration can be stored in lyophilized form or in a solution. In
addition, parenteral
compositions generally are placed into a container having a sterile access
port, for example,
an intravenous solution bag or vial having a stopper pierceable by a
hypodermic injection
needle.
[0250] Once the pharmaceutical composition has been formulated, it can be
stored in
sterile vials as a solution, suspension, gel, emulsion, solid, or as a
dehydrated or lyophilized
powder. Such formulations can be stored either in a ready-to-use form or in a
form (e.g.,
lyophilized) requiring reconstitution prior to administration.
[0251] The disclosure also encompasses kits for producing a single-dose
administration
unit. The kits can each contain both a first container having a dried protein
and a second
container having an aqueous formulation. Also included within the scope of
this disclosure
are kits containing single and multi-chambered pre-filled syringes (e.g.,
liquid syringes and
lyosyringes).
[0252] The effective amount of a binding protein pharmaceutical composition
to be
employed therapeutically will depend, for example, upon the therapeutic
context and
objectives. One skilled in the art will appreciate that the appropriate dosage
levels for
treatment will thus vary depending, in part, upon the molecule delivered, the
indication for
which the binding protein is being used, the route of administration, and the
size (body
weight, body surface, or organ size) and condition (the age and general
health) of the patient.
Accordingly, the clinician can titer the dosage and modify the route of
administration to
obtain the optimal therapeutic effect.
[0253] Dosing frequency will depend upon the pharmacokinetic parameters of
the
binding protein in the formulation being used. Typically, a clinician will
administer the
composition until a dosage is reached that achieves the desired effect. The
composition can
117
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
therefore be administered as a single dose, as two or more doses (which may or
may not
contain the same amount of the desired molecule) over time, or as a continuous
infusion via
an implantation device or catheter. Further refinement of the appropriate
dosage is routinely
made by those of ordinary skill in the art and is within the ambit of tasks
routinely performed
by them Appropriate dosages can be ascertained through use of appropriate dose-
response
data.
[0254] The route of administration of the pharmaceutical composition is in
accord with
known methods, e.g., orally; through injection by intravenous,
intraperitoneal, intracerebral
(intraparenchymal), intracerebroventricular, intramuscular, intraocular,
intraarterial,
intraportal, or intralesional routes; by sustained release systems; or by
implantation devices.
Where desired, the compositions can be administered by bolus injection or
continuously by
infusion, or by implantation device.
[0255] The composition can also be administered locally via implantation of
a membrane,
sponge, or other appropriate material onto which the desired molecule has been
absorbed or
encapsulated. Where an implantation device is used, the device can be
implanted into any
suitable tissue or organ, and delivery of the desired molecule can be via
diffusion, timed-
rel ease bolus, or continuous administration
[0256] The pharmaceutical compositions can be used to prevent and/or treat
HIV
infection. The pharmaceutical compositions can be used as a standalone therapy
or in
combination with standard anti-retroviral therapy.
The disclosure also relates to a kit comprising a binding protein and other
reagents useful for
detecting target antigen levels in biological samples. Such reagents can
include a detectable
label, blocking serum, positive and negative control samples, and detection
reagents. In some
embodiments, the kit comprises a composition comprising any binding protein,
polynucleotide, vector, vector system, and/or host cell described herein. In
some
embodiments, the kit comprises a container and a label or package insert on or
associated
with the container. Suitable containers include, for example, bottles, vials,
syringes, IV
solution bags, etc. The containers may be formed from a variety of materials
such as glass or
plastic. The container holds a composition which is by itself or combined with
another
composition effective for treating, preventing and/or diagnosing a condition
(e.g., HIV
infection) and may have a sterile access port (for example the container may
be an
intravenous solution bag or a vial having a stopper pierceable by a hypodermic
injection
needle). In some embodiments, the label or package insert indicates that the
composition is
used for preventing, diagnosing, and/or treating the condition of choice.
Alternatively, or
118
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
additionally, the article of manufacture or kit may further comprise a second
(or third)
container comprising a pharmaceutically-acceptable buffer, such as
bacteriostatic water for
injection (BWFI), phosphate-buffered saline, Ringer's solution and dextrose
solution. It may
further include other materials desirable from a commercial and user
standpoint, including
other buffers, diluents, filters, needles, and syringes.
Methods and Uses for Binding Proteins
Virus
[0257] Certain aspects of the present disclosure relate to methods for
expanding virus-
specific memory T cells. In some embodiments, the methods comprise contacting
a virus-
specific memory T cell with a binding protein of the present disclosure, e.g.,
a trispecific
binding protein that comprises a first antigen binding site that binds a CD28
polypeptide, a
second antigen binding site that binds a CD3 polypeptide, and a third antigen
binding site that
binds a CD38 polypeptide.
[0258] In some embodiments, the virus-specific memory T cell is contacted
with the
binding protein in vitro or ex vivo.
[0259] In some embodiments, contacting the virus-specific memory T cell
with the
binding protein causes activation and/or proliferation of virus-specific
memory T cells.
[0260] Other aspects of the present disclosure relate to methods for
expanding T cells. In
some embodiments, the methods comprise contacting a T cell with a binding
protein of the
present disclosure, e.g., a trispecific binding protein that comprises a first
antigen binding site
that binds a CD28 polypeptide, a second antigen binding site that binds a CD3
polypeptide,
and a third antigen binding site that binds a CD38 polypeptide.
[0261] In some embodiments, the T cell is a memory T cell or an effector T
cell.
[0262] In some embodiments, the T cell expresses a chimeric antigen
receptor (CAR) on
its cell surface or comprises a polynucleotide encoding a CAR.
[0263] Other aspects of the present disclosure relate to methods for
treating chronic viral
infection, e.g., in an individual in need thereof In some embodiments, the
methods comprise
administering to an individual in need thereof an effective amount of a
binding protein of the
present disclosure, e.g., a trispecific binding protein that comprises a first
antigen binding site
that binds a CD28 polypeptide, a second antigen binding site that binds a CD3
polypeptide,
and a third antigen binding site that binds a CD38 polypeptide.
[0264] In some embodiments, the individual is a human.
119
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
[0265] In some embodiments, the binding protein is administered to the
individual in
pharmaceutical formulation comprising the binding protein and a
pharmaceutically
acceptable carrier.
[0266] In some embodiments, administration of the binding protein results
in activation
and/or proliferation of virus-specific memory T cells in the individual.
[0267] In any of the above methods, memory T cells can be CD8+ or CD4+
memory T
cells. In any of the above methods, memory T cells can be central memory T
cells (Tcm) or
effector memory T cells (TEm).
Cancer
[0268] Certain aspects of the present disclosure relate to methods for
preventing and/or
treating cancer in a patient. In some embodiments, the methods comprise
administering to
the patient a therapeutically effective amount of a binding protein or
pharmaceutical
composition of the present disclosure.
[0269] In some embodiments, a binding protein of the present disclosure is
administered
to a patient in need thereof for the treatment or prevention of cancer. In
some embodiments,
the present disclosure relates to a method of preventing and/or treating a
proliferative disease
or disorder (e.g., cancer). In some embodiments, the method comprises
administering to a
patient a therapeutically effective amount of at least one of the binding
proteins, or
pharmaceutical compositions related thereto, described herein. In some
embodiments, the
present disclosure relates to uses of at least one of the binding proteins, or
pharmaceutical
compositions related thereto, described herein for preventing and/or treating
a proliferative
disease or disorder (e.g., cancer) in a patient in need thereof. In some
embodiments, the
present disclosure relates to at least one of the binding proteins, or
pharmaceutical
compositions related thereto, described herein for use in the manufacture of a
medicament for
preventing and/or treating a proliferative disease or disorder (e.g., cancer)
in a patient in need
thereof. In some embodiments, the patient is a human.
[0270] In some embodiments, the at least one binding protein is
administered (or is to be
administered) in combination with one or more anti-cancer therapies (e.g., any
anti-cancer
therapy known in the art, such as a chemotherapeutic agent or therapy) In some
embodiments, the at least one binding protein is administered (or is to be
administered)
before the one or more anti-cancer therapies. In some embodiments, the at
least one binding
protein is administered (or is to be administered) concurrently with the one
or more anti-
120
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
cancer therapies. In some embodiments, the at least one binding protein is
administered (or is
to be administered) after the one or more anti-cancer therapies.
[0271] In some embodiments, the binding protein comprises one or two
antigen binding
site(s) that binds a T-cell surface protein and another antigen binding site
that binds the
extracellular domain of a human HER2 polypeptide. In some embodiments, the
binding
protein comprises an antigen binding site that binds the extracellular domain
of a human
HER2 polypeptide, an antigen binding site that binds a human CD28 polypeptide,
and an
antigen binding site that binds a human CD3 polypeptide.
[0272] In some embodiments, cancer cells from the individual express HER2.
In some
embodiments, the patient is selected for treatment on the basis that the cells
of the cancer
express a human HER2 polypeptide. Assays known in the art suitable for
detecting HER2
expression by cancer cells include, without limitation, immunohistochemical
(MC) and
fluorescence in situ hybridization (FISH) assays.
[0273] In some embodiments, the cancer (e.g., HER2-positive cancer) is
breast cancer,
colorectal cancer, gastric cancer, or non-small cell lung cancer (NSCLC).
[0274] In some embodiments, the binding protein comprises one or two
antigen binding
site(s) that binds a T-cell surface protein and another antigen binding site
that binds the
extracellular domain of a human CD38 polypeptide. In some embodiments, the
binding
protein comprises an antigen binding site that binds the extracellular domain
of a human
CD38 polypeptide, an antigen binding site that binds a human CD28 polypeptide,
and an
antigen binding site that binds a human CD3 polypeptide.
[0275] In some embodiments, cancer cells from the individual express CD38.
In some
embodiments, cells of the cancer express a human CD38 isoform A polypeptide on
their cell
surface. In some embodiments, cells of the cancer express a human CD38 isoform
E
polypeptide on their cell surface. In some embodiments, the patient is
selected for treatment
on the basis that the cells of the cancer express a human CD38 isoform E
polypeptide on their
cell surface. In some embodiments, the cancer cells express CD38 and CD28. In
some
embodiments, the cancer cells express CD38 and do not express CD28.
[0276] In some embodiments, the cancer (e.g., CD38-positive cancer) is
multiple
myeloma, acute lymphoblastic leukemia, chronic lymphocytic leukemia, acute
myeloid
leukemia, lymphoma, breast cancer such as Her2+ breast cancer, prostate
cancer, germinal
center B-cell lympohoma or B-cell acute lymphoblastic leukemia. In certain
embodiments,
the cancer is multiple myeloma. In certain embodiments, the cancer is acute
myeloid
121
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
leukemia (AML), acute lymphoblastic leukemia (ALL), chronic lymphocytic
leukemia
(CLL), or a B cell lymphoma.
[0277] In certain embodiments, the cancer is multiple myeloma. Anti-CD38
antibodies
have been tested for the treatment of multiple myeloma, such as daratumumab.
However,
while multiple myeloma is considered treatable, relapse is inevitable in
almost all patients,
leading to the development of treatment-refractory disease. In some
embodiments, the cancer
is relapsed or refractory multiple myeloma. In some embodiments, the patient
has been
treated with a prior multiple myeloma treatment. In some embodiments, a
binding protein of
the present disclosure is administered to the patient as a 1st, 2nd, or 3rd
line treatment for
multiple myeloma. Without wishing to be bound to theory, it is thought that an
anti-
CD38xanti-CD28xanti-CD3 binding protein of the present disclosure may be
useful in
treating multiple myeloma, e.g., by recruiting T cells to tumor cells via anti-
CD38 (or anti-
CD28/anti-CD38), activation of engaged T cells via anti-CD3/anti-CD28, and/or
killing of
tumor cells through perforin/granzyme-based mechanisms. CD28 has been reported
as a
novel cancer marker for multiple myeloma. See Nair, J.R. et at. (2011)J.
Immunol.
187:1243-1253.
[0278] Any of the binding proteins described herein may find use in the
methods of the
present disclosure.
[0279] In some embodiments of any of the methods of the present disclosure,
prior to
administration of the binding protein, the patient has been treated with
daratumumab. As
described herein, the present disclosure provides anti-CD38 binding proteins
and sites that do
not compete for binding CD38 with daratumumab. Without wishing to be bound to
theory, it
is thought that this is advantageous because a patient previously treated with
daratumumab
can be treated with a binding protein of the present disclosure, e.g., without
a wash-out period
prior to treatment.
[0280] The binding proteins can be employed in any known assay method, such
as
competitive binding assays, direct and indirect sandwich assays, and
immunoprecipitation
assays for the detection and quantitation of one or more target antigens. The
binding proteins
will bind the one or more target antigens with an affinity that is appropriate
for the assay
method being employed
[0281] For diagnostic applications, in certain embodiments, binding
proteins can be
labeled with a detectable moiety. The detectable moiety can be any one that is
capable of
producing, either directly or indirectly, a detectable signal. For example,
the detectable
, , 32p 35s, 1251 , 99Te, 111=n,
moiety can be a radioisotope, such as 3H, 14C or
"Ga; a fluorescent
122
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
or chemiluminescent compound, such as fluorescein isothiocyanate, rhodamine,
or luciferin;
or an enzyme, such as alkaline phosphatase, P-galactosidase, or horseradish
peroxidase.
[0282] The binding proteins are also useful for in vivo imaging. A binding
protein
labeled with a detectable moiety can be administered to an animal, preferably
into the
bloodstream, and the presence and location of the labeled antibody in the host
assayed. The
binding protein can be labeled with any moiety that is detectable in an
animal, whether by
nuclear magnetic resonance, radiology, or other detection means known in the
art.
[0283] For clinical or research applications, in certain embodiments,
binding proteins can
be conjugated to a cytotoxic agent. A variety of antibodies coupled to
cytotoxic agents (i.e.,
antibody-drug conjugates) have been used to target cytotoxic payloads to
specific tumor cells.
Cytotoxic agents and linkers that conjugate the agents to an antibody are
known in the art;
see, e.g., Parslow, A.C. et at. (2016) Biomedicines 4:14 and Kalim, M. et at.
(2017) Drug
Des. Devel. Ther. 11:2265-2276.
Binding protein therapeutic compositions and administration thereof
[0284] Therapeutic or pharmaceutical compositions comprising binding
proteins are
within the scope of the disclosure. Such therapeutic or pharmaceutical
compositions can
comprise a therapeutically effective amount of a binding protein, or binding
protein-drug
conjugate, in admixture with a pharmaceutically or physiologically acceptable
formulation
agent selected for suitability with the mode of administration. These
pharmaceutical
compositions may find use in any of the methods and uses described herein
(e.g., ex vivo, in
vitro, and/or in vivo).
[0285] Acceptable formulation materials preferably are nontoxic to
recipients at the
dosages and concentrations employed.
[0286] The pharmaceutical composition can contain formulation materials for
modifying,
maintaining, or preserving, for example, the pH, osmolarity, viscosity,
clarity, color,
isotonicity, odor, sterility, stability, rate of dissolution or release,
adsorption, or penetration of
the composition. Suitable formulation materials include, but are not limited
to, amino acids
(such as glycine, glutamine, asparagine, arginine, or lysine), antimicrobials,
antioxidants
(such as ascorbic acid, sodium sulfite, or sodium hydrogen-sulfite), buffers
(such as borate,
bicarbonate, Tris-HC1, citrates, phosphates, or other organic acids), bulking
agents (such as
mannitol or glycine), chelating agents (such as ethylenediamine tetraacetic
acid (EDTA)),
complexing agents (such as caffeine, polyvinylpyrrolidone, beta-cyclodextrin,
or
hydroxypropyl-beta-cyclodextrin), fillers, monosaccharides, disaccharides, and
other
123
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
carbohydrates (such as glucose, mannose, or dextrins), proteins (such as serum
albumin,
gelatin, or immunoglobulins), coloring, flavoring and diluting agents,
emulsifying agents,
hydrophilic polymers (such as polyvinylpyrrolidone), low molecular weight
polypeptides,
salt-forming counterions (such as sodium), preservatives (such as benzalkonium
chloride,
benzoic acid, salicylic acid, thimerosal, phenethyl alcohol, methylparaben,
propylparaben,
chlorhexidine, sorbic acid, or hydrogen peroxide), solvents (such as glycerin,
propylene
glycol, or polyethylene glycol), sugar alcohols (such as mannitol or
sorbitol), suspending
agents, surfactants or wetting agents (such as pluronics; PEG; sorbitan
esters; polysorbates
such as polysorbate 20 or polysorbate 80; triton; tromethamine; lecithin;
cholesterol or
tyloxapal), stability enhancing agents (such as sucrose or sorbitol), tonicity
enhancing agents
(such as alkali metal halides ¨ preferably sodium or potassium chloride ¨ or
mannitol
sorbitol), delivery vehicles, diluents, excipients and/or pharmaceutical
adjuvants (see, e.g.,
REMINGTON'S PHARMACEUTICAL SCIENCES (18th Ed., A.R. Gennaro, ed., Mack
Publishing
Company 1990), and subsequent editions of the same, incorporated herein by
reference for
any purpose).
[0287] The optimal pharmaceutical composition will be determined by a
skilled artisan
depending upon, for example, the intended route of administration, delivery
format, and
desired dosage. Such compositions can influence the physical state, stability,
rate of in vivo
release, and rate of in vivo clearance of the binding protein.
[0288] The primary vehicle or carrier in a pharmaceutical composition can
be either
aqueous or non-aqueous in nature. For example, a suitable vehicle or carrier
for injection can
be water, physiological saline solution, or artificial cerebrospinal fluid,
possibly
supplemented with other materials common in compositions for parenteral
administration.
Neutral buffered saline or saline mixed with serum albumin are further
exemplary vehicles.
Other exemplary pharmaceutical compositions comprise Tris buffer of about pH
7.0-8.5, or
acetate buffer of about pH 4.0-5.5, which can further include sorbitol or a
suitable substitute.
In one embodiment of the disclosure, binding protein compositions can be
prepared for
storage by mixing the selected composition having the desired degree of purity
with optional
formulation agents in the form of a lyophilized cake or an aqueous solution.
Further, the
binding protein can be formulated as a lyophilizate using appropriate
excipients such as
sucrose.
[0289] The pharmaceutical compositions of the disclosure can be selected
for parenteral
delivery or subcutaneous. Alternatively, the compositions can be selected for
inhalation or
124
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
for delivery through the digestive tract, such as orally. The preparation of
such
pharmaceutically acceptable compositions is within the skill of the art.
[0290] The formulation components are present in concentrations that are
acceptable to
the site of administration. For example, buffers are used to maintain the
composition at
physiological pH or at a slightly lower pH, typically within a pH range of
from about 5 to
about 8.
[0291] When parenteral administration is contemplated, the therapeutic
compositions for
use can be in the form of a pyrogen-free, parenterally acceptable, aqueous
solution
comprising the desired binding protein in a pharmaceutically acceptable
vehicle. A
particularly suitable vehicle for parenteral injection is sterile distilled
water in which a
binding protein is formulated as a sterile, isotonic solution, properly
preserved. Yet another
preparation can involve the formulation of the desired molecule with an agent,
such as
injectable microspheres, bio-erodible particles, polymeric compounds (such as
polylactic acid
or polyglycolic acid), beads, or liposomes, that provides for the controlled
or sustained
release of the product which can then be delivered via a depot injection.
Hyaluronic acid can
also be used, and this can have the effect of promoting sustained duration in
the circulation.
Other suitable means for the introduction of the desired molecule include
implantable drug
delivery devices.
[0292] In one embodiment, a pharmaceutical composition can be formulated
for
inhalation. For example, a binding protein can be formulated as a dry powder
for inhalation.
Binding protein inhalation solutions can also be formulated with a propellant
for aerosol
delivery. In yet another embodiment, solutions can be nebulized.
[0293] It is also contemplated that certain formulations can be
administered orally. In
one embodiment of the disclosure, binding proteins that are administered in
this fashion can
be formulated with or without those carriers customarily used in the
compounding of solid
dosage forms such as tablets and capsules. For example, a capsule can be
designed to release
the active portion of the formulation at the point in the gastrointestinal
tract when
bioavailability is maximized and pre-systemic degradation is minimized.
Additional agents
can be included to facilitate absorption of the binding protein. Diluents,
flavorings, low
melting point waxes, vegetable oils, lubricants, suspending agents, tablet
disintegrating
agents, and binders can also be employed.
[0294] Another pharmaceutical composition can involve an effective quantity
of binding
proteins in a mixture with non-toxic excipients that are suitable for the
manufacture of
tablets. By dissolving the tablets in sterile water, or another appropriate
vehicle, solutions
125
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
can be prepared in unit-dose form. Suitable excipients include, but are not
limited to, inert
diluents, such as calcium carbonate, sodium carbonate or bicarbonate, lactose,
or calcium
phosphate; or binding agents, such as starch, gelatin, or acacia; or
lubricating agents such as
magnesium stearate, stearic acid, or talc.
[0295] Additional pharmaceutical compositions of the disclosure will be
evident to those
skilled in the art, including formulations involving binding proteins in
sustained- or
controlled-delivery formulations. Techniques for formulating a variety of
other sustained- or
controlled-delivery means, such as liposome carriers, bio-erodible
microparticles or porous
beads and depot injections, are also known to those skilled in the art.
Additional examples of
sustained-release preparations include semipermeable polymer matrices in the
form of shaped
articles, e.g. films, or microcapsules. Sustained release matrices can include
polyesters,
hydrogels, polylactides, copolymers of L-glutamic acid and gamma ethyl-L-
glutamate,
poly(2-hydroxyethyl-methacrylate), ethylene vinyl acetate, or poly-D(-)-3-
hydroxybutyric
acid. Sustained-release compositions can also include liposomes, which can be
prepared by
any of several methods known in the art.
[0296] Pharmaceutical compositions to be used for in vivo administration
typically must
be sterile. This can be accomplished by filtration through sterile filtration
membranes.
Where the composition is lyophilized, sterilization using this method can be
conducted either
prior to, or following, lyophilization and reconstitution. The composition for
parenteral
administration can be stored in lyophilized form or in a solution. In
addition, parenteral
compositions generally are placed into a container having a sterile access
port, for example,
an intravenous solution bag or vial having a stopper pierceable by a
hypodermic injection
needle.
[0297] Once the pharmaceutical composition has been formulated, it can be
stored in
sterile vials as a solution, suspension, gel, emulsion, solid, or as a
dehydrated or lyophilized
powder. Such formulations can be stored either in a ready-to-use form or in a
form (e.g.,
lyophilized) requiring reconstitution prior to administration.
[0298] The disclosure also relates to a kit comprising a binding protein
and other reagents
useful for detecting target antigen levels in biological samples. Such
reagents can include a
detectable label, blocking serum, positive and negative control samples, and
detection
reagents. In some embodiments, the kit comprises a composition comprising any
binding
protein, polynucleotide, vector, vector system, and/or host cell described
herein. In some
embodiments, the kit comprises a container and a label or package insert on or
associated
with the container. Suitable containers include, for example, bottles, vials,
syringes, IV
126
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
solution bags, etc. The containers may be formed from a variety of materials
such as glass or
plastic. The container holds a composition which is by itself or combined with
another
composition effective for treating, preventing and/or diagnosing a condition
and may have a
sterile access port (for example the container may be an intravenous solution
bag or a vial
having a stopper pierceable by a hypodermic injection needle). In some
embodiments, the
label or package insert indicates that the composition is used for preventing,
diagnosing,
and/or treating the condition of choice. Alternatively, or additionally, the
article of
manufacture or kit may further comprise a second (or third) container
comprising a
pharmaceutically-acceptable buffer, such as bacteriostatic water for injection
(BWFI),
phosphate-buffered saline, Ringer's solution and dextrose solution. It may
further include
other materials desirable from a commercial and user standpoint, including
other buffers,
diluents, filters, needles, and syringes.
[0299] The disclosure also encompasses kits for producing a single-dose
administration
unit. The kits can each contain both a first container having a dried protein
and a second
container having an aqueous formulation. Also included within the scope of
this disclosure
are kits containing single and multi-chambered pre-filled syringes (e.g.,
liquid syringes and
lyosyringes).
[0300] The effective amount of a binding protein pharmaceutical composition
to be
employed therapeutically will depend, for example, upon the therapeutic
context and
objectives. One skilled in the art will appreciate that the appropriate dosage
levels for
treatment will thus vary depending, in part, upon the molecule delivered, the
indication for
which the binding protein is being used, the route of administration, and the
size (body
weight, body surface, or organ size) and condition (the age and general
health) of the patient.
Accordingly, the clinician can titer the dosage and modify the route of
administration to
obtain the optimal therapeutic effect.
[0301] Dosing frequency will depend upon the pharmacokinetic parameters of
the
binding protein in the formulation being used. Typically, a clinician will
administer the
composition until a dosage is reached that achieves the desired effect. The
composition can
therefore be administered as a single dose, as two or more doses (which may or
may not
contain the same amount of the desired molecule) over time, or as a continuous
infusion via
an implantation device or catheter. Further refinement of the appropriate
dosage is routinely
made by those of ordinary skill in the art and is within the ambit of tasks
routinely performed
by them. Appropriate dosages can be ascertained through use of appropriate
dose-response
data.
127
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
[0302] The route of administration of the pharmaceutical composition is in
accord with
known methods, e.g., orally; through injection by intravenous,
intraperitoneal, intracerebral
(intraparenchymal), intracerebroventricular, intramuscular, intraocular,
intraarterial,
intraportal, or intralesional routes; by sustained release systems; or by
implantation devices.
Where desired, the compositions can be administered by bolus injection or
continuously by
infusion, or by implantation device.
[0303] The composition can also be administered locally via implantation of
a membrane,
sponge, or other appropriate material onto which the desired molecule has been
absorbed or
encapsulated. Where an implantation device is used, the device can be
implanted into any
suitable tissue or organ, and delivery of the desired molecule can be via
diffusion, timed-
release bolus, or continuous administration.
128
Date Recue/Date Received 2021-10-04
Table 1. Trispecific binding protein polypeptide sequences
Molecule Polyp epti SEQ Sequence
0
de ID
Number NO
(acc. to
formula)
0
HER2 (WT- 1
100
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSG
trastuzumab) /
TDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNI
0
CD28supxCD3
YVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTEGQG
mid (32/35 QQ
TKLEIKDKTHTRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQWKVDNALQSGNSQESVTEQDS
(LC); DKTHT
KDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
linkers on
HC/LC) IgG4
FALA
BP # 1
2 101
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATL
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNEDVWGKGTTVTVSSDKTHTQVQLVESGGGVVQPG
RSLRLSCAASGETFTKAWMEIWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKY
GPPCPPCPAPEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLS
CAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFELVSKLTVDKSRWQEGNVESCSVMHEALHNHYT
QKSLSLSLG
3 102
EVQLVESGGGLVQPGGSLRLSCAASGENIKDTYIHWVRQAPGKGLEWVARIYPTNGYTRYADSVKGRFTIS
ADTSKNTAYLQMNSLRAEDTAVYYCSRWGGDGFYAMDYWGQGTLVTVSSASTKGPSVFPLAPCSRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEV
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQE c7,
EMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFELYSKLTVDKSRWQEGNVESCSV
tµ.)
MHEALHNHYTQKSLSLSLG
4
103
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSGVPSRFSGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTEGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQ
WKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
HE R2 1
104 DIVMTQTPLSLSVTPGQPASISCKS
SQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSG
(30R/55Q/102E
TDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKDKTHTDIQMTQ SP S SL SASVGDRVTITCQAS QNI
0
+ LC-WT-
YVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTEGQG
trastuzumab) /
TKLEIKDKTHTRTVAAPSVFIFPP
SDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNSQESVTEQD S
0
KDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
CD28supxCD3
mid (32/35 QQ
0
(LC); DKTHT
linkers on
0
HC/LC) IgG4
8 FALA
BP #
2 105
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATL
TVDTSI S TAYMEL S RLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTV S SDKTHTQVQLVESGGGVVQPG
RS LRL SCAASGF TFTKAWMEIWVRQAPGKQLEWVAQIKDKSN SYATYYAD SVKGRFTISRDD SKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVPSS SLGTKTYTCNVDHKP SNTKVDKRVE SKY
GPP CPPCPAPEAAGGP SVFLEPPKPKDTLMI SRTPEVTCVVVDV S QEDPEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLS
CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFELVSKLTVDKSRWQEGNVF SCSVMHEALHNHYT
QKSLSLSLG
3 106 EVQLVE S GGGLVQPGGSLRL S CAA SGFNIRDTYIHWVRQAPGKGLEWVARIYPTQ
GYTRYAD S VKGRFTI S
ADTSKNTAYLQMNSLRAEDTAVYYCSRWGGEGFYAMDYWGQGTLVTVS SASTKGPSVFPLAPCSRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVPSS SLGTKTYTCNVDHKPSNTK
VDKRVE S KYGPP CPPCPAPEAAGGP SVFLEPPKPKDTLMI S RTPEVTCVVVDV S
QEDPEVQFNWYVDGVEV
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQE
EMTKNQVSLWCLVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFELYSKLTVDKSRWQEGNVESC SV
010
MHEALHNHYTQ KS L S L SLG
4 107
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSGVPSRFSGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTEGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQ
WKVDNALQSGNSQESVTEQDSKDSTYSLS STLTLSKADYEKHKVYACEVTHQGLS SPVTKSFNRGEC
HER2 1
108 DIVMTQTPLSLSVTPGQPASISCKS
SQSLVHDNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSG
(30R/55Q/102E
TDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKDKTHTDIQMTQ SP S SL SASVGDRVTITCQAS QNI
+ LC-WT-
YVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTEGQG
trastuzuma b) /
TKLEIKDKTHTRTVAAPSVFIFPP
SDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNS QESVTEQDS
o
CD28õpxCD3mid
KDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
o
(DNAQ (LC);
o
0 DKTHT linkers
on HC/LC) IgG4
FALA
0
BP # 8
0
2
109
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKFQGRATL
8
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTVSSDKTHTQVQLVESGGGVVQPG
RSLRLSCAASGETFTKAWMEIWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSDKTHTASTKGP SVFPLAPCSRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVP SS SLGTKTYTCNVDHKP SNTKVDKRVES KY
GPPCPPCPAPEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREE
QFN S TYRVV S VLTVLHQDWLNGKEYKC KV SNKGLP S SIEKTISKAKGQPREPQVCTLPP
SQEEMTKNQVSLS
CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFELVSKLTVDKSRWQEGNVES CSVMHEALHNHYT
QKSLSLSLG
3 110
EVQLVESGGGLVQPGGSLRLSCAASGENIRDTYIHWVRQAPGKGLEWVARTYPTQGYTRYADSVKGRFTIS
ADTSKNTAYLQMNSLRAEDTAVYYCSRWGGEGFYAMDYWGQGTLVTVSSASTKGPSVFPLAPCSRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVP SS SLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEAAGGP SVFLEPPKPKDTLMI S RTPEVTCVVVDV S QEDPEVQFNWYVDGVEV
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQE
EMTKNQVSLWCLVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFELYSKLTVDKSRWQEGNVESC SV
MHEALHNHYTQKSLSLSLG
4 111
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSGVPSRFSGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQ
WKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
HER2 1
286
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSG
(3 OR/5 6A/ 1 02 S
TDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNI
+LC-WT-
YVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTEGQG
trastuzumab) /
TKLEIKDKTHTRTVAAPSVFIFPP
SDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNS QESVTEQD S
CD28supxCD 3
KDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
mid
(32/35QQ185E
) IgG4 FALA
0
BP # 3
2 287 QVQLVQ SGAEVVKPGA SVKV S CKA
SGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKF QGRATL
TVDTSI S TAYMEL S RLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTV S SDKTHTQVQLVESGGGVVQPG
0
RS LRL S CAA SGFTFTKAWMHWVRQAPGKQLEWVAQIKDKSN SYATYYAD SVKGRFTISRDD SKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVS SDKTHTASTKGP SVFPLAPC SRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQ S SGLYSLS SVVTVPS S SLGTKTYTCNVDHKP SNTKVDKRVE
SKY
GPP CPPCPAPEAAGGP SVFLEPPKPKDTLMI SRTPEVTCVVVDV S QEDPEVQFNWYVDGVEVHNAKTKPREE
8
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVCTLPP SQEEMTKNQVSLS
CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGS FFLV SKLTVDK SRWQEGNVF S
CSVMHEALHNHYT
QKSLSLSLG
3 288 EVQLVE S GGGLVQPGGSLRL S CAA
SGFNIRDTYIHWVRQAPGKGLEWVARIYPTNAYTRYAD SVKGRFTI S
ADTSKNTAYLQMNSLRAEDTAVYYC SRWGGSGFYAMDYWGQGTLVTVS SA STKGP SVFPLAPC SRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQ S SGLYSLS SVVTVPS S SLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEFLGGP SVFLEPPKPKDTLMISRTPEVTCVVVDVS QEDPEVQFNWYVDGVEVH
NAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS SIEKTISKAKGQPREPQVYTLPPCQEE
MTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLD S DGSFFLY SKLTVDKS RWQEGNVF S C SV
MHEALHNHYTQKSLSLSLG
4 289 DI QMTQ SP S SL SA SVGDRVTITCRA S QDVNTAVAWY Q QKPGKAPKLLIY SA
SFLY SGVP SRF SGSRSGTDFTL
TIS SLQPEDFATYYCQQHYTTPPTFGQGTKVEIKRTVAAP SVFIFPP S DEQLKSGTA
SVVCLLNNFYPREAKVQ
WKVDNALQ SGNSQESVTEQD SKD S TY SL S STLTLSKADYEKHKVYACEVTHQGLS SPVTKSFNRGEC
HER2 1
290 DIVMTQTPLSLSVTPGQPASISCKS
SQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSG
(30R/55Q/102
TDFTLKI S
RVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKGQPKAAPDIQMTQ SP S S L SA SVGDRVTITCQAS Q
E +LC-WT-
NIYVWLNWYQQKPGKAPKLLIYKASNLHTGVP
SRFSGSGSGTDFTLTIS SLQPEDIATYYCQQGQTYPYTFG
trastuzumab) /
QGTKLEIKTKGP SRTVAAPSVFIFPP
SDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNS QESVTEQ
CD28supxCD3 D SKD STY SL S STLTLSKADYEKHKVYACEVTHQGLS
SPVTKSFNRGEC
mid
(32/35QQ185E
) IgG4 FALA
BP # 4
2 291 QVQLVQ SGAEVVKPGA SVKV S CKA
SGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKF QGRATL
TVDTSI S TAYMEL S RLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTV S SDKTHTQVQLVESGGGVVQPG
RSLRLSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKDKSNSYATYYAESVKGRFTISRDDSKNTLYLQM
N SLRAED TAVYY CRGVYYAL SP FDYWGQ GTLVTV S SDKTHTA S TKGP SVFP LAP C
SRSTSESTAALGCLVK
0
DYFPEPVTVSWNSGALTSGVHTFPAVLQ SSGLYSLS SVVTVP SS SLGTKTYTCNVDHKP SNTKVDKRVES KY
GPPCPPCPAPEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLS
CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFELVSKLTVDKSRWQEGNVES CSVMHEALHNHYT
0
QKSLSLSLG
3 292
EVQLVESGGGLVQPGGSLRLSCAASGENIRDTYIHWVRQAPGKGLEWVARIYPTQGYTRYADSVKGRFTIS
ADTSKNTAYLQMNSLRAEDTAVYYCSRWGGEGFYAMDYWGQGTLVTVSSASTKGPSVFPLAPCSRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQ SSGLYSLS SVVTVP SS SLGTKTYTCNVDHKPSNTK
VDKRVE S KYGPP CP P C PAP EAAGGP SVFLF PP KPKDTLMI S RTP EVTCVVVDV S Q EDP EV
Q FNWYVDGVEV
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQE
EMTKN QV SLWCLVKGFYP SDIAVEWESNGQPENNYKTTPPVLD SDGSFELYSKLTVDKSRWQEGNVESC SV
MHEALHNHYTQKSLSLSLG
4 293 DI QMTQ SP S SL SA SVGDRVTITCRA S
QDVNTAVAWY Q QKPGKAPKLLIY SA SFLY SGVP SRF SGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTEGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQ
WKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
HER2 1 294
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSG
(30R/55Q/102
TDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKGQPKAAPDIQMTQSPSSLSASVGDRVTITCQASQ
E +LC-WT-
NIYVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTEG
trastuzumab) / QGTKLEIKTKGP SRTVAAP SVF IF PP
SDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNS QESVTEQ
CD28supxCD3
DSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
mid
(32/35QQ185S
) IgG4 FALA
BP # 5
2 295
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSTYPGNVNTNYAQKFQGRATL
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNEDVWGKGTTVTVSSSQVQLVESGGGVVQPGRSLR
LSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKDKSNSYATYYASSVKGRFTISRDDSKNTLYLQMNSLR
AEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSRTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVT
VSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCP
APEAAGGP SVF LFPP KP KD TLMI S RTP EVTCVVVDV S Q EDP EV Q FNWYVDGVEVHNAKTKP
REE Q FN S TYR it
VVSVLTVLHQDWLNGKEYKCKVSNKGLP S S IEKTI S KAKGQ PREP QV C TLPP S Q EEMTKN QV S
L S CAVKGF
YPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSL
SLG
0
3 296
EVQLVESGGGLVQPGGSLRLSCAASGENIRDTYIHWVRQAPGKGLEWVARIYPTQGYTRYADSVKGRFTIS
ADTSKNTAYLQMNSLRAEDTAVYYCSRWGGEGFYAMDYWGQGTLVTVSSASTKGPSVFPLAPCSRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEV
0
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQE
EMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFELYSKLTVDKSRWQEGNVESCSV
MHEALHNHYTQKSLSLSLG
4 297
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSGVPSRFSGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTEGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQ
WKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
HER2 1 298
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHQSAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGT
(30R/55Q/102
DFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKGQPKAAPDIQMTQSPSSLSASVGDRVTITCQASQN
E +LC-WT-
IYVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTEGQ
trastuzumab) /
GTKLEIKTKGPSRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQWKVDNALQSGNSQESVTEQD
CD28supxCD3
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
mid
(32/33/35QSQ
185S) IgG4
FALA
BP # 6
2 299
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKFQGRATL
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNEDVWGKGTTVTVSSSQVQLVESGGGVVQPGRSLR
LSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKDKSNSYATYYASSVKGRFTISRDDSKNTLYLQMNSLR
AEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSRTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVT
VSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCP
APEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYR
VVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLSCAVKGF
YPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSL
SLG
3 300
EVQLVESGGGLVQPGGSLRLSCAASGENIRDTYIHWVRQAPGKGLEWVARIYPTQGYTRYADSVKGRFTIS
ADTSKNTAYLQMNSLRAEDTAVYYCSRWGGEGFYAMDYWGQGTLVTVSSASTKGPSVFPLAPCSRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEAAGGP SVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEV
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVYTLPPCQE
0
EMTKNQVSLWCLVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SD GSFFLY SKLTVDKS RWQEGNVF S C
SV
MHEALHNHYTQKSLSLSLG
4 301
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSGVPSRFSGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTEGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQ
0
WKVDNALQSGNSQESVTEQDSKDSTYSLS STLTLSKADYEKHKVYACEVTHQGLS SPVTKSFNRGEC
HE R2 1
112 DIVMTQTPLSLSVTPGQPASISCKS
SQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSG
(30R/55Q/102E
TDFTLKIS
RVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKGQPKAAPDIQMTQ SP S S L SA SVGD RVTITCQA S Q
+ LC-WT-
NIYVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTEG
trastuzu m a b) /
QGTKLEIKTKGP SRTVAAPSVFIFPP
SDEQLKS GTA SVVCLLNNFYPREAKVQWKVDNALQ S GN S QE SVTEQ
CD28CD3mid DSKDSTYSLS STLTLSKADYEKHKVYACEVTHQGLS
SPVTKSFNRGEC
(32/35Q0 (LC);
L1 linker) IgG4
FALA
BP # 9
2 113
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKFQGRATL
TVDTSIS TAYMEL S RLRSDDTAVYY CTRSHYGLDWNFDVWGKGTTVTV S SSQVQLVESGGGVVQPGRSLR
L S CAA S GFTFTKAWMHWVRQAPGKQLEWVAQIKDKSN SYATYYAD SVKGRFTI SRDD SKNTLYLQMN
SLR
AEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSRTASTKGP SVFPLAPCSRSTSESTAALGCLVKDYFPEPVT
V SWN S GALTSGVHTFPAVLQ S SGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCP
APEAAGGP SVFLEPPKPKDTLMI SRTPEVTCVVVDV S QEDPEVQFNWYVDGVEVHNAKTKPREEQFN STYR
VV SVLTVLHQDWLNGKEYKC KV SNKGLP SSIEKTISKAKGQPREPQVCTLPP SQEEMTKNQVSLSCAVKGF
YP SD IAVEWE SNGQPENNYKTTPPVLD S DGS FFLV SKLTVDKS RWQEGNVFS C SVMHEALHNHYTQKS
L SL
SLG
3 114 EVQLVE S GGGLVQPGGSLRL S CAA
SGFNIRDTYIHWVRQAPGKGLEWVARIYPTQ GYTRYAD SVKGRFTIS
ADTSKNTAYLQMN SLRAEDTAVYYC SRWGGEGFYAMDYWGQGTLVTV S SA S TKGP SVFPLAPC S RS
TSE S
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVPSS SLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEAAGGP SVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEV
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVYTLPPCQE
EMTKNQVSLWCLVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD S DGSFFLY SKLTVDKSRWQEGNVF S C
SV
MHEALHNHYTQKSLSLSLG
O 4 115
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSGVPSRFSGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTEGQGTKVETKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQ
0
WKVDNALQSGNSQESVTEQDSKDSTYSLS STLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
O HER2-30R/
1 116
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHNNANTYLSWYLQKPGQSPQSLTYKVSNRFSGVPDRFSGSGSG
FD. 55Q/102S+LC-
TDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKGQPKAAPDIQMTQSPSSLSASVGDRVTITCQASQ
WT-
NTYVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTEG
0
trastuzumab/
QGTKLEIKTKGPSRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQ
CD28supxCD
DSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
0
3mid Li linker
IgG4 FALA
BP # 10
2 117
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSTYPGNVNTNYAQKFQGRATL
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNEDVWGKGTTVTVS SSQVQLVESGGGVVQPGRSLR
LSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLR
AEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSRTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVT
VSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCP
APEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYR
VVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLSCAVKGF
YPSDIAVEWESNGQPENNYKTTPPVLDSDGSFELVSKLTVDKSRWQEGNVESCSVMHEALHNHYTQKSLSL
SLG
3 118
EVQLVESGGGLVQPGGSLRLSCAASGENIRDTYTHWVRQAPGKGLEWVARTYPTQGYTRYADSVKGRETTS
ADTSKNTAYLQMNSLRAEDTAVYYCSRWGGSGFYAMDYWGQGTLVTVSSASTKGPSVFPLAPCSRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEFLGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVH
NAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEE
MTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFELYSKLTVDKSRWQEGNVESCSV
MHEALHNHYTQKSLSLSLG
4 119
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSGVPSRFSGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTEGQGTKVETKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQ c7,
WKVDNALQSGNSQESVTEQDSKDSTYSLS STLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
HE R2-30R/ 1
120
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHNNANTYLSWYLQKPGQSPQSLTYKVSNRFSGVPDRFSGSGSG
56A/102S+LC-
TDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKGQPKAAPDIQMTQSPSSLSASVGDRVTITCQASQ
WT-
NTYVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTEG
trastuzumab/
QGTKLEIKTKGP SRTVAAP SV FIF PP
SDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNSQESVTEQ
CD28supxCD3
DSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
(D
mid L1 linker
(D
0 IgG4 FALA
BP # 11
0
2
121
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATL
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNEDVWGKGTTVTVSSSQVQLVESGGGVVQPGRSLR
0
LSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLR
AEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSRTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVT
VSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCP
APEAAGGP SVF LFP PKP KD TLMI S RTPEVT CVVVDV S Q ED PEV Q FNWYVD
GVEVHNAKTKPREE Q FN S TYR
VV SVLTVLHQ DWLNGKEYKC KV SNKGLP S S IEKTI SKA KGQ PREP QV C TLPP S Q EEMTKN
QV S L S CAVKGF
YPSDIAVEWESNGQPENNYKTTPPVLDSDGSFELVSKLTVDKSRWQEGNVESCSVMHEALHNHYTQKSLSL
SLG
3 122
EVQLVESGGGLVQPGGSLRLSCAASGENIRDTYIHWVRQAPGKGLEWVARIYPTNAYTRYADSVKGRFTIS
ADTSKNTAYLQMNSLRAEDTAVYYCSRWGGSGFYAMDYWGQGTLVTVSSASTKGPSVFPLAPCSRSTSES
TAALGCLV KDYFPEPVTV SWN S GALT S GVHTFPAVL Q SSGLYSLS SVVTVP SS
SLGTKTYTCNVDHKPSNTK
VD KRVE S KYGPP CP P C PAP EFLGGP SVF LFPP KP KD TLMI S RTPEVTCVVVDV S Q ED
PEV Q FNWYVD GVEVH
NAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEE
MTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFELYSKLTVDKSRWQEGNVESCSV
MHEALHNHYTQKSLSLSLG
4 123 DI QMTQ SP S SL SA SVGDRVTITCRA S QDVNTAVAWY Q QKPGKAPKLLIY SA
SFLY SGVPSRF SGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTEGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQ
WKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
HE R2-30R/ 1
124
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHNNANTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSG
56A/102 E/
TDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKGQPKAAPDIQMTQSPSSLSASVGDRVTITCQASQ
CD28supxCD3
NIYVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTEG
mid L1 linker
QGTKLEIKTKGP SRTVAAPSVFIFPP
SDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNS QESVTEQ
IgG4 FALA
DSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
BP # 12
2 125
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKFQGRATL
TVDTSIS TAYMEL SRLRSDDTAVYY CTRSHYGL DWNFDVWGKGTTVTV S SS QVQLVESGGGVVQPGRSLR
L S CAA S GFTFTKAWMHWVRQAPGKQLEWVAQIKDKSN SYATYYAD SVKGRFTI SRDD SKNTLYLQMN
SLR
AEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSRTASTKGP SVFPLAPCSRSTSESTAALGCLVKDYFPEPVT
0
V SWN S GALTSGVHTFPAVLQ S SGLYSLSSVVTVP SSSLGTKTYTCNVDHKP
SNTKVDKRVESKYGPPCPPCP
APEAAGGP SVFLEPPKPKDTLMISRTPEVTCVVVDVS QEDPEVQFNWYVDGVEVHNAKTKPREEQFN STYR
2
VVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVCTLPP SQEEMTKNQVSLSCAVKGF
YP SD IAVEWE SNGQPENNYKTTPPVLD S DGS FFLV S KLTVDKS RWQEGNVFS C
SVMHEALHNHYTQKS L SL
0
SLG
3 126 EVQLVE S GGGLVQPGGSLRL S CAA
SGENIRDTYTHWVRQAPGKGLEWVARTYPTNAYTRYAD SVKGRFTI S
ADTSKNTAYLQMN SLRAEDTAVYYC SRWGGEGFYAMDYWGQGTLVTV S SA S TKGP SVFPLAP C S RS
TSE S
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQ SSGLYSLS SVVTVP SS SLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEFLGGP SVFLEPPKPKDTLMISRTPEVTCVVVDVS QEDPEVQFNWYVDGVEVH
NAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS SIEKTISKAKGQPREPQVYTLPPCQEE
MTKNQV S LWCLVKGFYP S DIAVEWE SNGQPENNYKTTPPVLD S DGSFFLY SKLTVDKS RWQEGNVF S
C SV
MHEALHNHYTQKSLSLSLG
4 127 DIQMTQ SP S SL SASVGDRVTITCRAS QDVNTAVAWYQQKPGKAPKWYSASFLY
SGVPSRF SGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIKRTVAAP SVFIFPP S DEQLKSGTA SVVCLLNNFYPREAKVQ
WKVDNALQ S GN S QE SVTEQD SKD S TY SL S STLTLSKADYEKHKVYACEVTHQGLS
SPVTKSFNRGEC
HE R2- 1
128 DIVMTQTPLSLSVTPGQPASISCKS SQ
SLVHQNAQTYLSWYLQKPGQ SPQ SLIYKVSNRF SGVPDRFSGSGSG
oc
WT+trastuzum
TDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIKGQPKAAPDIQMTQSPSSLSASVGDRVTITCQASQ
a b/
NIYVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFG
CD28supxCD3
QGTKLEIKTKGPSRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQ
mid (32/35QQ)
DSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Li linker IgG4
FALA
BP # 15
2 129 QVQLVQ
SGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSTYPGNVNTNYAQKFQGRATL
TVDTSI S TAYMEL S RLRSDDTAVYY CTRSHYGLDWNFDVWGKGTTVTV S SSQVQLVESGGGVVQPGRSLR
L S CAA S GFTFTKAWMHWVRQAPGKQLEWVAQIKDKSN SYATYYAD SVKGRFTI SRDD SKNTLYLQMN
SLR
A EDTAVYYCRGVYYA L S PFDYWGQGTLVTV S S RTA STKGP SVFPLA P C S RSTSE STA A LGC
LVKDYFPEPVT
V SWN S GALTSGVHTFPAVLQ S SGLYSLSSVVTVP SSSLGTKTYTCNVDHKP
SNTKVDKRVESKYGPPCPPCP
APEAAGGP SVFLEPPKPKDTLMISRTPEVTCVVVDVS QEDPEVQFNWYVDGVEVHNAKTKPREEQFN STYR
VVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVCTLPP SQEEMTKNQVSLSCAVKGF
YPSDIAVEWESNGQPENNYKTTPPVLD SDGSFFLVSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSL
SLG
0
3 130 EVQLVE S GGGLVQPGGSLRL S CAA
SGFNIKDTYIHWVRQ APGKGLEWVARIYPTNGYTRYAD SVKGRF TIS
ADTSKNTAYLQMN SLRAEDTAVYYC SRWGGDGFYAMDYWGQGTLVTV S SA S TKGP SVFPLAPC S RS
TSE S
FD.
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVP SS SLGTKTYTCNVDHKPSNTK
x VDKRVESKYGPPCPPCPAPEAAGGP
SVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEV
0
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVYTLPPCQE
EMTKNQVSLWCLVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFELYSKLTVDKSRWQEGNVESC SV
MHEALHNHYTQKSLSLSLG
4 131
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSGVPSRFSGSRSGTDFTL
8
TISSLQPEDFATYYCQQHYTTPPTEGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQ
WKVDNALQSGNSQESVTEQDSKDSTYSLS STLTLSKADYEKHKVYACEVTHQGLS SPVTKSFNRGEC
HE R2/ 1 132 DIVMTQTPLSLSVTPGQPASISCKS
SQSLVHNNANTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSG
CD28õpxCD3mid
TDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKDKTHTDIQMTQ SP SSLSASVGDRVTITCQASQNI
DKTHT linkers
YVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQG
on HC/LC) IgG4
TKLEIKDKTHTRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDS
FALA
KDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
BP # 25
2 133
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKFQGRATL
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTVS SDKTHTQVQLVESGGGVVQPG
RS LRLSCAASGF TFTKAWMHWVRQAPGKQLEWVAQIKDKSN SYATYYAD SVKGRFTISRDD SKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVPSS SLGTKTYTCNVDHKP SNTKVDKRVE SKY
GPP CPPCPAPEAAGGP SVFLEPPKPKDTLMISRTPEVTCVVVDV S QEDPEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVCTLPP SQEEMTKNQVSLS
CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFELVSKLTVDKSRWQEGNVF SCSVMHEALHNHYT
QKSLSLSLG
3 134 EVQLVE S GGGLVQPGGSLRL S CAA
SGFNIKDTYIHWVRQ APGKGLEWVARIYPTNGYTRYAD SVKGRF TIS
ADTSKNTAYLQMN SLRAEDTAVYYC SRWGGDGFYAMDYWGQGTLVTV S SA S TKGP SVFPLAPC S RS
TSE S
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVP SS SLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEAAGGP SVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEV
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVYTLPPCQE
EMTKNQVSLWCLVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SD GSFFLY SKLTVDKS RWQEGNVF SCSV
MHEALHNHYTQ KS L S L SLG
0
4 135 DIQMTQ SP S SL SASVGDRVTITCRAS
QDVNTAVAWYQQKPGKAPKWYSASFLY SGVP SRF SGSRSGTDFTL õ
TISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIKRTVAAP SVFIFPP S DEQLKSGTA SVVCLLNNFYPREAKVQ
WKVDNALQ S GN S QE SVTEQD SKD S TY SL S STLTLSKADYEKHKVYACEVTHQGLS
SPVTKSFNRGEC
HE R2/ 1 136 DIVMTQTPLSLSVTPGQPASISCKSSQ
SLVHENLRTYLSWYLQKPGQ SPQ SLIYKVSNRFSGVPDRF SGSGSGT
0
CD28õpxCD3mid DFTLKI S
RVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKDKTHTDI QMTQ SPS SL SA SVGDRVTITCQA S QNIY
(32/33/3435 VWLNWYQ QKPGKAPKLLIYKA SNLHTGVP SRF SGS
GSGTDFTLTI S S LQPEDIATYYCQ QGQTYPYTFGQGT
ENLR (LC); KLEIKDKTHTRTVAAP SVFIFPP SDEQLKS GTA
SVVCLLNNFYPREAKVQWKVDNALQ SGNSQESVTEQDSK
DKTHT linkers DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
SPVTKSFNRGEC
on HC/LC) IgG4
FALA
BP # 26
2 137 QVQLVQ SGAEVVKPGA SVKV S CKA
SGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKF QGRATL
TVDTSI S TAYMEL S RLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTV S SDKTHTQVQLVESGGGVVQPG
RS LRL S CAA SGFTFTKAWMHWVRQAPGKQLEWVAQIKD KSN SYATYYAD SVKGRFTI S RDD S
KNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSDKTHTASTKGP SVFPLAPCSRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQ SSGLYSLS SVVTVPSS SLGTKTYTCNVDHKP SNTKVDKRVE SKY
GPP CPPCPAPEAAGGP SVFLEPPKPKDTLMI SRTPEVTCVVVDV S QEDPEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVCTLPP SQEEMTKNQVSLS
CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGS FFLV SKLTVDK SRWQEGNVF SCSVMHEALHNHYT
QKSLSLSLG
3 138 EVQLVE S GGGLVQPGGSLRL S CAA
SGFNIKDTYIHWVRQAPGKGLEWVARIYPTNGYTRYAD SVKGRFTI S
ADTSKNTAYLQMN SLRAEDTAVYYC SRWGGDGFYAMDYWGQGTLVTV S SA S TKGP SVFPLAPC S RS
TSE S
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQ SSGLYSLS SVVTVP SS SLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEAAGGP SVFLEPPKPKDTLMI S RTPEVTCVVVDV S QEDPEVQFNWYVDGVEV
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVYTLPPCQE
EMTKNQVSLWCLVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD S DGSFFLY S KLTVDKSRWQEGNVF S C
SV
MHEALHNHYTQ KS L S L SLG
4 139 DIQMTQ SP S SL SASVGDRVTITCRAS
QDVNTAVAWYQQKPGKAPKWYSASFLY SGVP SRF SGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIKRTVAAP SVFIFPP S DEQLKSGTA SVVCLLNNFYPREAKVQ
WKVDNALQ S GN S QE SVTEQD SKD S TY SL S STLTLSKADYEKHKVYACEVTHQGLS
SPVTKSFNRGEC
HE R2/ 1
140
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGT
CD28õpxCD3mid
DFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKDKTHTDIQMTQ SPS SLSASVGDRVTITCQASQNIY
0
(32/33/3435
VWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGT
ENLQ (LC);
KLEIKDKTHTRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQWKVDNALQSGNSQESVTEQDSK
DKTHT linkers DSTYSLS STLTLSKADYEKHKVYACEVTHQGLS
SPVTKSFNRGEC
on HC/LC) IgG4
0
FALA
0
BP # 27
8 2 141
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKFQGRATL
TVDTSI S TAYMEL S RLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTV S SDKTHTQVQLVESGGGVVQPG
RS LRL SCAASGF TFTKAWMEIWVRQAPGKQLEWVAQIKDKSN SYATYYAD SVKGRFTISRDD SKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSDKTHTASTKGP SVFPLAPCSRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVP SS SLGTKTYTCNVDHKP SNTKVDKRVES KY
GPP CPPCPAPEAAGGP SVFLEPPKPKDTLMI S RTPEVTCVVVDV S QEDPEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS SIEKTISKAKGQPREPQVCTLPP SQEEMTKNQVSLS
CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFELVSKLTVDKSRWQEGNVES CSVMHEALHNHYT
QKSLSLSLG
3 142 EVQLVE S GGGLVQPGGSLRL S CAA
SGFNIKDTYIHWVRQ APGKGLEWVARIYPTNGYTRYAD S VKGRF TI S
ADTSKNTAYLQMNSLRAEDTAVYYC SRWGGDGFYAMDYWGQGTLVTVS SASTKGPSVFPLAPC SRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVP SS SLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEAAGGP SVFLEPPKPKDTLMI S RTPEVTCVVVDV S QEDPEVQFNWYVDGVEV
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVYTLPPCQE
EMTKNQVSLWCLVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFELYSKLTVDKSRWQEGNVESC SV
MHEALHNHYTQKSLSLSLG
4 143
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSGVPSRFSGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIKRTVAAP SVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQ
WKVDNALQSGNSQESVTEQDSKDSTYSLS STLTLSKADYEKHKVYACEVTHQGLS SPVTKSFNRGEC
HE R2/ 1
144 DIVMTQTPLSLSVTPGQPASISCKS
SQSLVHENLFTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGT -
CD28,,,pxCD3rnid
DFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKDKTHTDIQMTQ SPS SLSASVGDRVTITCQASQNIY
(32/33/3435
VWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGT
ENLF (LC);
KLEIKDKTHTRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQWKVDNALQSGNSQESVTEQDSK
DKTHT linkers DSTYSLS STLTLSKADYEKHKVYACEVTHQGLS
SPVTKSFNRGEC
on HC/LC) IgG4
FALA
0
0 BP # 28
2
145
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATL
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNEDVWGKGTTVTVSSDKTHTQVQLVESGGGVVQPG
0
RSLRLSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVK
0
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKY
GPPCPPCPAPEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS SIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLS
CAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFELVSKLTVDKSRWQEGNVESCSVMHEALHNHYT
QKSLSLSLG
3 146
EVQLVESGGGLVQPGGSLRLSCAASGENIKDTYIHWVRQAPGKGLEWVARIYPTNGYTRYADSVKGRFTIS
ADTSKNTAYLQMNSLRAEDTAVYYCSRWGGDGFYAMDYWGQGTLVTVSSASTKGPSVFPLAPCSRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEV
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQE
EMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFELYSKLTVDKSRWQEGNVESCSV
MHEALHNHYTQKSLSLSLG
4 147
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSGVPSRFSGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTEGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQ
WKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSENRGEC
anti- 1
148 DIVMTQTPLSLSVTPGQPASISCKS
SQSLVHNNANTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSG
Her2/CD3/3CD
TDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIKGQPKAAPDIQMTQSPSSLSASVGDRVTITCQASQ
28 IgG4 FALA
NIYVWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFG
QGTKLEIKTKGPSRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQ
BP #29
DSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
2
149
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATL
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTVS SSQVQLVESGGGVVQPGRSLR
LSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLR
AEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSRTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVT
VSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCP
APEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYR
VVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVCTLPP SQEEMTKNQVSLSCAVKGF
YPSDIAVEWESNGQPENNYKTTPPVLDSDGSFELVSKLTVDKSRWQEGNVESCSVMHEALHNHYTQKSLSL
0
SLG
3 150 EVQLVE S GGGLVQPGGSLRL S CAA
SGFNIKDTYIHWVRQ APGKGLEWVARIYPTNGYTRYAD SVKGRF TIS
ADTSKNTAYLQMNSLRAEDTAVYYC SRWGGDGFYAMDYWGQGTLVTVS SASTKGP SVFPLAPCSRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVP SS SLGTKTYTCNVDHKPSNTK
0
VDKRVESKYGPPCPPCPAPEAAGGP SVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEV
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP SSIEKTISKAKGQPREPQVYTLPPCQE
EMTKNQVSLWCLVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFELYSKLTVDKSRWQEGNVESC SV
MHEALHNHYTQKSLSLSLG
8
4 151
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSGVPSRFSGSRSGTDFTL
TISSLQPEDFATYYCQQHYTTPPTEGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNEYPREAKVQ
WKVDNALQSGNSQESVTEQDSKDSTYSLS STLTLSKADYEKHKVYACEVTHQGLS SPVTKSFNRGEC
HE R230 R/55Q/ 1 152
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLRTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGT
102E/
DFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKDKTHTDIQMTQ SPS SLSASVGDRVTITCQASQNIY
CD28õpxCD3mid
VWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGT
(32/33/3435
KLEIKDKTHTRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSK
ENLR (LC); DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
SPVTKSFNRGEC
DKTHT linkers
on H C/ LC) IgG4
FALA
BP # 31
2 153
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKFQGRATL
TVDTSIS TAYMEL S RLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTV S SDKTHTQVQLVESGGGVVQPG
RS LRLSCAASGF TFTKAWMHWVRQAPGKQLEWVAQIKDKSNSYATYYAD SVKGRFTISRDD SKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSDKTHTASTKGP SVFPLAPCSRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVPSS SLGTKTYTCNVDHKP SNTKVDKRVE SKY
'A
GPP CPPCPAPEAAGGP SVFLEPPKPKDTLMISRTPEVTCVVVDV S QED PEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS SIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLS
CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFELVSKLTVDKSRWQEGNVES CSVMHEALHNHYT
QKSLSLSLG
3 154 EVQLVE S GGGLVQPGGSLRL S CAA
SGFNIRDTYIHWVRQAPGKGLEWVARIYPTQ GYTRYAD SVKGRFTIS
ADTSKNTAYLQMNSLRAEDTAVYYCSRWGGEGFYAMDYWGQGTLVTVSSASTKGPSVFPLAPCSRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEV
0
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQE
EMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSV
MHEALHNHYTQKSLSLSLG
4 155
DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKWYSASFLYSGVPSRFSGSRSGTDFTL a,
0
TISSLQPEDFATYYCQQHYTTPPTFGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQ
WKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
CD38VH1 / 1 156
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGT
CD28sup x
DFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIY
CD3mid ENL
VWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGT
Q DKTHT
KLEIKDKTHTRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSK
IgG4 FALA
DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
BP # 1
2 157
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKFQGRATL
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTVSSDKTHTQVQLVESGGGVVQPG
RSLRLSCAASGFTFTKAWMEIWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKY
GPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLS
CAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVFSCSVMHEALHNHYT
QKSLSLSLG
3 158
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYAMHWVKEAPGQRLEWIGYIYPGQGGTNYNQKFQGRAT
LTADTSASTAYMELSSLRSEDTAVYFCARTGGLRRAYFTYWGQGTLVTVSSASTKGPSVFPLAPCSRSTSES
TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTK
VDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEV
HNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQE
EMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSV c7,
MHEALHNHYTQKSLSLSLG
4 159
DIVLTQSPATLSLSPGERATISCRASQSVSSYGQGFMHWYQQKPGQPPRLLIYGASSRATGIPARFSGSGSGTD
FTLTISPLEPEDFAVYYCQQNKEDPWTFGGGTKLEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREA
KVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
tµ.)
DdOAADDDSHAIOAOIHINGS SAIALLDNDMAGANMUIDAHSNIDAAAVIGGSNINSIMATAVISISIGAI
,e4 lIVNDOANOVANLLNANDdAISDIM3IDODdVONAA11-
1TAASIAIADSVNDSANASVDdNAA3VDSONIOAO S91
.9 # dEL
el=
D3DITNASNIAdS SIDOHIA3DVAANT-DHACIVNSIIIIS sisiusa VIVA 17DSI
ci)
=
NS GOTLASHOSNDS
OIVNGANMOANVM1dAINNIIDAAS VIDSNIOHUSddA HAS dVVADIIHINGNIHIN IHING 6
IDODILAdAIOD 6 ODAAIVIGHdOIS
GIDSDSDS ANSdADIHINS VNAIIINdVND dNO6AMKIMA 1MPU1TEUD
:" Al S VODILLANCLDAS VS'IS SdSOIINOICURINCINTRANIDSDAIAdAbIDODD
AAADMIAVRANSINIIA x driss zap
IDSDSDSANCIdADS ANNSANAFIS (MS ODdNOIAMSIAIOINHHAIS S SNDS
VdODdIASISIdIOIINAIG 1791 1 /6 EL STC118 CD
DaDNNASNIAdSSIDOHIAHDVAANHNHAGVNSIIIIS SISAIS CINS GO HIASHO SNDS OIVNGA
NMo ANVANdAANNTID AAS VIDSNIOACISddA HAS dVVAINNITRILDODILIdi CLO ODAAAVD
aldO1S
S LUIS A aLD SDSDS ANAdIDIVILL S VDATTRIdVIDdNA OAAksIAN S S 6svN3 STIV2IHDdS1
TH 91 17
DISISISNOIAHN
1-TIVAHINAS DS AA NID3 oMNSNCLAEINSKIAASDUS alAd dIINANNAdoD AMAAVICIS
dAIDNAIDAVISA
ONNIV\IHHODddlIAAOdaNdODNVNSTINHISSdIDNNSANDNAHNDNIMCIOHIAIIASAANAISNAOHMId
NINVNHAHADGAAMNAOAUCHO S ACIAAADIAadDIS ITAITICINdNddAIAAS dODVVadVdD
ddDddDANS
ANNUANINS dNHGANDIAINIDI S S S dAIAA S SIS AID S S OIAVdAIHAD SIIVDS NA1S
AlAda d AAGNAID
DIVVISHSISNSDdVId AAS dDNIS VS
SAIAIIDODANAMGAAIIDIIDAAIVICHSNISNIAIHINAVIGISICHI
INIANDOANO VAIIHDGadiJADDIAIMHIDOD dVONAA1HI S ATIIIADS AND S ANAS VD dNNAHVDS
6A16A6 z9 I
DISISISNO
IAHNHIVHHIATASDSAANDHOMNSNUAIINSAIAASDUSGIAddIINANNadODNSHAGAVHISdAADNAVD
SI S AONNITAIHHO S ddlID AO daNdODNVNS
S S d'IDNNS ANDNAHNDNIMGOHIAIIA S AANAI S MAO
HaNdNINVNHAHADGAAA1N,16AUCHOSAGAAADIAadDISITAIIICINdNddAIAASdDDVVadVdDddDddD
ANS HANNUANINS dNHGANDIAINIDIS SSdAIAAS SIS AID S S OIAVdAIHAD SIIVDS NAkS
AlAda dAAG =:r
NAIDDIVVISHSISNSDdVIdAASdDNISVIHINGSSAIAIIDODMAGAdSIVAAADIIDAAAVICHVNISN
9
0
TAT S GMT S LLANDNA S GVAAIVAS NSN (INTO
VAMHIONDdVONAMIAINAWNIAIADS VVD
(7,
DdOAADDDSHAIOAOIHINGS SAIALLDNDMAGANMUIDAHSNIDAAAVERISNIIISIHINAVISISIGAI
0
IIVNDOANOVANINANDdAISDIMHIDODdVONAMHIAASIAIADSVNDSANASVDdNAAHVDSOAIOAO 191
Z-0
.>
# dEL
0
DaD2INASNIAdS SIDOHIAHDVAANHNHACIVNSIIIIS SISAIS
VIVA 170S1
s; NS GOTLASHOSNDS OIVNGANMOANVM1dAINNTIDAAS VIDSNIOHUSddA HAS
dVVADIIHINGNIHIN IHING 6
=
IDODILAdAIODOODAAIVIGHdOIS
SIIIIAGIDSDSDSANSdADIHINSVNAFTINdVNDdNO6AMNIMA MulECID
AINOSVODILLANGDASVSIS SdSOITAIOICIIHINCINTHANIDSDAIAdAOIDODDAAADACHVHANSINIIAG
x dris zap
IDSDSDSANCIdADS ANNSANAFIS (MS ODdNOIAMSIAIOINHHAIS S SNDS
VdODdIASISIdIOIINAIG 091 1 / Z66JCIIII8
CID CS
RS LRL S CAA SGFTFTKAWMHWVRQAPGKQLEWVAQIKD KSN SYATYYAD SVKGRFTISRDD SKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVS SDKTHTASTKGP SVFPLAPC SRSTSESTAALGCLVK
0
DYFPEPVTVSWNSGALTSGVHTFPAVLQ S SGLYSLS SVVTVP S S SLGTKTYTCNVDHKP SNTKVDKRVES
KY
GPPCPPCPAPEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREE
0
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLS
CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGS FFLV S KLTVDKSRWQEGNVF S
CSVMHEALHNHYT
0
QKSLSLSLG
tµ.)
3 166 QVQLQQ SGPELVRPGTSVKVS CKA
SGYAFTTYLVEWIKQRPGQGLEWIGVINPGSGSTNYNEKFKGKATLT
VDRS STTAYMHLSGLTSDDSAVYFCARYAYGYWGQGTTLTVS SA S TKGP SVFPLAPC SRSTSESTAALGCLV
KDYFPEPVTVSWNSGALTSGVHTFPAVLQ S SGLYSLS SVVTVP S S SLGTKTYTCNVDHKP
SNTKVDKRVESK
YGPPCPPCPAPEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPRE
EQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS SIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVS
LWCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLD SDGS FFLY SKLTVDKS RWQEGNVF S CSVMHEALHN
HYTQKSLSLSLG
4 167 DIVMTQ S QKFMSASVGDRVSITCKAS QNVGTAVAWY Q Q QPGHS PKQLIY SA
SNRYTGVPDRFTGSGAGTDF
TLTISNIQ SEDLADYFCQQYSTYPFTEGSGTKLEIKRTVAAP SVFIFPP SDEQLKSGTASVVCLLNNFYPREAK
VQWKVDNALQ SGNSQESVTEQD SKD S TY SL S STLTLSKADYEKHKVYACEVTHQGLS SPVTKSFNRGEC
CD38hyb6284 / 1
168
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGT
cs,
CD28sup x
DFTLKI S
RVEAEDVGVYYCGQGTQYPFTEGSGTKVEIKDKTHTDI QMTQ SPS SL SA SVGDRVTITCQA S QNIY
CD3mid ENL
VWLNWYQ QKPGKAPKLLIYKA SNLHTGVP
SRF SGSGSGTDFTLTIS SLQPEDIATYYCQQGQTYPYTFGQGT
Q DKTHT
KLEIKDKTHTRTVAAP SVFIFPP SDEQLKS
GTA SVVCLLNNFYPREAKVQWKVDNALQ SGNSQESVTEQD SK
IgG4 FALA
DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
BP # 7
2
169 QVQLVQ SGAEVVKPGA SVKV S CKA
SGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKF QGRATL
TVDTSI S TAYMEL S RLRSDDTAVYYCTRSHYGLDWNEDVWGKGTTVTV S SDKTHTQVQLVESGGGVVQPG
RS LRL S CAA SGFTFTKAWMHWVRQAPGKQLEWVAQIKD KSN SYATYYAD SVKGRFTISRDD SKNTLYLQM
N SLRA ED TAVYY CRGVYYAL SP FDYWGQ GTLVTV S SDKTHTASTKGP SVFP LAP C
SRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQ S SGLYSLS SVVTVP S S SLGTKTYTCNVDHKP SNTKVDKRVES
KY
GPPCPPCPAPEAAGGPSVFLEPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLS
CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGS FFLV SKLTVDK SRWQEGNVF S
CSVMHEALHNHYT
QKSLSLSLG
tµ.)
3 170 QVQLLQ SGAELVRPGV SVKIS CTGSGYSFTNYAVHWVKQ
SHVKSLEWIGVISPYYGDTTYNQKFTGKATMT
VDKS S STAYMELARLTS ED SAIYFCARRFEGFYYSMDYWGQGTSVTVS SA STKGP S VFPLAPC S RS
TSE S TA
ALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVD
KRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHN
0
AKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS SIEKTISKAKGQPREPQVYTLPPCQEEM
TKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSVMH
0
EALHNHYTQKSLSLSLG
4
171
DVVMIQTPLSLPVSLGDQASISCRPSQSLVHSNGNTYLNWYLQRPGQSPKWYKVSKRFSGVPDRFSGSGSG
0
TDFTLKISRVEAEDLGVYLCSQSTHVPLTFGSGTQLEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPR
EAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGE
CD38hhy1195 / 1
172
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGT
CD28sup x
DFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIY
CD3mid ENL
VWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGT
Q DKTHT
KLEIKDKTHTRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSK
IgG4 FALA
DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
BP # 8
2 173
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKFQGRATL
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTVSSDKTHTQVQLVESGGGVVQPG
RSLRLSCAASGFTFTKAWM.HWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKY
GPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLS
CAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVFSCSVMHEALHNHYT
QKSLSLSLG
3 174
QVQLVESGGGVVQPGRSLRLSCAASGFTFSSYGMYWVRQAPGKGLEWVAVIWYDGSNKYYADSVKGRFTI
SRDNSKNTLYLQMNSLRAEDTAVYHCARDPGLRYFDGGMDVWGQGTTVTVSSASTKGPSVFPLAPCSRST
SESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPS
NTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDG
VEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPP
CQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFELYSKLTVDKSRWQEGNVES
CSVMHEALHNHYTQKSLSLSLG
tµ.)
4 175
DIQLTQSPSFLSASVGDRVTITCRASQGISSYLAWYQQKPGKAPKLLIFAASTLHSGVPSRFSGSGSGTEFTLTI
SSLQPEDFATYYCQQLNSFPYTEGQGTKLEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQW
KVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
CD38hhy1370 / 1
176
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGT
CD28sup x
DFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQN1Y
CD3mid ENL 0
VWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGT
CDC
Q DKTHT
KLEIKDKTHTRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSK
FD' ) IgG4 FALA
DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
,D0a)
BP # 9
tµ.)
2 177
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATL
0"
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTVSSDKTHTQVQLVESGGGVVQPG
RSLRLSCAASGFTFTKAWMEIWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQM
8
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSDKTHTASTKGP SVFPLAPCSRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKY
GPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS SIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLS
CAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVFSCSVMHEALHNHYT
QKSLSLSLG
3 178
QVQLVESGGGVVQPGRSLRLSCAASGFTFSSYGMHWVRQAPGKGLEWVAVIWYDGSNKYYADSVKGRFTI
SGDNSKNTLYLQMNSLRAEDTAVYYCARMFRGAFDYWGQGTLVTVS SASTKGPSVFPLAPCSRSTSESTAA
oc
LGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDK
RVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNA
KTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS SIEKTISKAKGQPREPQVYTLPPCQEEMT
KNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSVMHE
ALHNHYTQKSLSLSLG
4 179
AIQMTQSPSSLSASVGDRVTITCRASQGIRNDLGWYQQKPGKAPKLLIYAASSLQSGVPSRFSGSGSGTDFTL
TISGLQPEDSATYYCLQDYIYYPTFGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKV
QWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
CD38hu5739 / 1
181
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGT
CD28sup x
DFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQN1Y
CD3mid ENL
VWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGT
Q DKTHT
KLEIKDKTHTRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSK
cpw
IgG4 FALA
DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
2 182
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATL
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTVSSDKTHTQVQLVESGGGVVQPG
RSLRLSCAASGFTFTKAWMFIWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVS SDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQ SSGLYSLS SVVTVP SS SLGTKTYTCNVDHKP SNTKVDKRVE SKY
GPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREE
0
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLS
CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFFLVSKLTVDKSRWQEGNVFS CSVMHEALHNHYT
0
QKSLSLSLG
3 183
QVQLVQSGAEVKKPGASVKVSCKASGYAFTTYLVEWIRQRPGQGLEWMGVINPGSGSTNYAQKFQGRVT
0
MTVDRSSTTAYMELSRLRSDDTAVYYCARYAYGYWGQGTLVTVSSASTKGPSVFPLAPCSRSTSESTAALG
CLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRV
ESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTK
PREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQ
V SLWC LVKGFYP SDIAVEWESNGQPENNYKTTPPVLD SDGSFFLYSKLTVDKSRWQEGNVF SC SVMHEALH
NHYTQKSLSLSLG
4 184
DIQMTQSPSSLSASVGDRVTITCRASQNVGTAVAWYQQKPGKSPKQLIYSASNRYTGVPSRFSGSGSGTDFT
LTISSLQPEDLATYYCQQYSTYPFTFGQGTKLEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKV
QWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
CD38hu6284 / 1 185
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGT
CD28sup x
DFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIY
CD3mid ENL
VWLNWYQQKPGKAPKWYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGT
Q DKTHT
KLEIKDKTHTRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSK
IgG4 FALA
DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
2 186
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYTHWVRQAPGQGLEWIGSWPGNVNTNYAQKFQGRATL
TVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNFDVWGKGTTVTVSSDKTHTQVQLVESGGGVVQPG
RSLRLSCAASGFTFTKAWMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQM
NSLRAEDTAVYYCRGVYYALSPFDYWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVK
DYFPEPVTVSWNSGALTSGVHTFPAVLQ SSGLYSLS SVVTVPSS SLGTKTYTCNVDHKP SNTKVDKRVE SKY
GPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREE
QFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVCTLPPSQEEMTKNQVSLS
CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SDGSFFLVSKLTVDKSRWQEGNVFS CSVMHEALHNHYT
QKSLSLSLG
3 187
QVQLVQSGAEVKKPGASVKVSCKASGYSFTNYAVHWVRQAPGQGLEWMGVISPYYGDTTYAQKFQGRVT c7,
MTVDKSSSTAYMELSRLRSDDTAVYYCARRFEGFYYSMDYWGQGTLVTVSSASTKGPSVFPLAPCSRSTSE
STAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQ SSGLYSLSSVVTVP SS SLGTKTYTCNVDHKPSNT
KVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVE
tµ.)
VHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPC
tµ.)
QEEMTKNQVSLWCLVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD S DGSFFLY S KLTVD KSRWQEGNVF S
C
SVMHEALHNHYTQKSLSLSLG
0
4
188 DVVMTQ
SPLSLPVTLGQPASISCRP SQ SLVHSNGNTYLNWYQQRPGQ SPKLLIYKVSKRFSGVPDRFSGSGSG õ
TDFTLKISRVEAEDVGVYYCSQ STHVPLTFGGGTKVEIKRTVAAPSVFIFPP SDEQLKSGTASVVCLLNNFYP
REAKVQWKVDNALQ SGNS QE SVTEQD SKD STY SL S STLTLSKADYEKHKVYACEVTHQGLS
SPVTKSFNRG
EC
0
0
Table 6. Trispecific binding protein polynucleotide sequences
Molecule Polyp eptid SEQ Sequence
e Number ID
(acc. to NO
formula)
HER2 (WT- 1
189
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
trastuzumab) /
AGCCAGAGCCTGGTGCACCAGAACGCCCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
CD28supxCD3m
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
id (32/35 QQ
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
(LC); DKTHT
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
linkers on
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
HC/LC) IgG4
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
FALA
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
BP # 1 GACCCACACCCGTACGGTGGCCGCTCCCAGCGTG
TTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2 190
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC ..
o
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT o
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
0
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 191
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCAAGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
ATCTACCCCACCAACGGCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA
GGCGACGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGA
CTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTAC
ACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGC
CCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGA
TGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATT
GGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGG
GTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTG
CCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACA
010
TTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACG
GCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGT
GATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 192
GACATCCAGATGACCCAGAGCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTAGAGCC
AGCCAGGACGTGAACACCGCCGTGGCCTGGTATCAGCAGAAGCCTGGCAAGGCCCCCAAGCTGCTGATCTACAGC
GCCAGCTTCCTGTACAGCGGCGTGCCCAGCAGATTCAGCGGAAGCAGAAGCGGCACCGACTTCACCCTGACCATC
AGCTCCCTGCAGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACCCCCCCCACATTTGGCCAGG
GCACCAAGGTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGA
AGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGG
ACAACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTG
AGCAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGG
0
CCTGTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
H ER2 1 193
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
(30R/550/102E
AGCCAGAGCCTGGTGCACCAGAACGCCCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
0 +LC-WT-
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
trastuzuma b) /
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
CD28supxCD3nni
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
d (32/35 QQ (LC);
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
8 DKTHT linkers on
HC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
/LC) IgG4
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
FALA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
BP #2
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2 194
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
010
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA ei
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 195
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCCGGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
ATCTACCCCACCCAGGGCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA
0
GGCGAAGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGC
CCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGG
ACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGT
GCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTA
CACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTG
CCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTG
ATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAAT
TGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCG
GGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAA
GGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCT
GCCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGAC
ATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGAC
GGCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCG
TGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 196
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
010
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
ei
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
HE R2 1
197
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
(30R/550/102 E+
AGCCAGAGCCTGGTGCACGACAACGCCCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
0 LC-WT-
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
trastuzu ma b) /
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
CD28õpxCD3mid
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
(DNAQ (LC);
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
8 DKTHT linkers on
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
HC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
/LC) IgG4
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
FALA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
BP #8
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGC C GACTAC GAGAAGCACAAGGTGTAC GC CTGC GAAGTGAC C CACCAGGGC
CTGTCTAGC CC C
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2 198
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACAC C CTGTAC CTGCAGATGAACAGC CTGC GGGC C GAGGACAC C GC
CGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
010
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA ei
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTC CAACAAGGGCCTGC CCAGCTC CATCGAGAAAACCATCAGCAAGGC CAAGGGC CAGC C C CGC
GAGC CT d
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 199
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC 0
GGCTTCAACATCCGGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
ATCTACCCCACCCAGGGCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA
0
GGCGAAGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGC
CCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGG
ACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGT
GCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTA
CACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTG
CCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTG
ATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAAT
TGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCG
GGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAA
GGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCT
GCCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGAC
ATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGAC
GGCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCG
TGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 200
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
010
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
ei
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
HE R2 1
201
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
(30R/550/102 E
AGCCAGAGCCTGGTGCACCAGAACGCCCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
0 + LC-WT-
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
trastuzu ma b) /
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
CD28õpxCD3mid
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGA
(32/35QQ ( LC);
GCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGT
GGCTGAACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCG
8 L1 linker) IgG4
FALA
GCGTGCCCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGA
CATTGCCACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAG
ACCAAGGGCCCCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCG
BP #9
GCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACG
CCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGC
ACC CTGACACTGAGCAAGGC C GACTAC GAGAAGCACAAGGTGTAC GC CTGC GAAGTGACC CAC CAGGGC
CTGTCT
AGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
2
202
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
cs,
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTG
GTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTC
ACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAA
CAC C CTGTAC CTGCAGATGAACAGCCTGC GGGCC GAGGACAC C GCC GTGTACTACTGTC GGGGC
GTGTACTATGC C
CTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCAT
CGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTA
CTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACC
010
TGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
ei
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGA
TCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGT
ACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTG
GTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGC
CTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCC
CCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATT
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGG
CTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTG
ATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 203
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCCGGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
ATCTACCCCACCCAGGGCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA
0
GGCGAAGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGC
CCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGG
ACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGT
GCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTA
CACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTG
CCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTG
ATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAAT
TGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCG
GGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAA
GGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCT
GCCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGAC
ATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGAC
GGCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCG
TGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 204
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
010
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
ei
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
HE R2-30R/ 1
205
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
550/102S+LC-
AGCCAGAGCCTGGTGCACAACAACGCCAACACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
0 WT-
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
trastuzumab/
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGA
CD28supxCD3
0
mid Ll linker
GCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGT
IgG4 FALA
GGCTGAACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCG
GCGTGCCCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGA
BP #10
CATTGCCACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAG
ACCAAGGGCCCCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCG
GCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACG
CCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGC
ACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCT
AGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
2 206
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
oc
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTG
GTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTC
ACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAA
CACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCC
CTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCAT
CGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTA
CTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACC
010
TGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
ei
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGA
TCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGT
ACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTG
GTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGC
CTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCC
CCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATT
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGG
CTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTG
ATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 207
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCCGGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
ATCTACCCCACCCAGGGCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA
0
GGCTCCGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGCC
=))
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGA
CTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTAC
ACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGC
CCTCCTTGCCCAGCCCCTGAATTTCTGGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGAT
GATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTG
GTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGG
TGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGG
GCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGC
CCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACAT
TGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGG
CTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTG
ATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 208
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
010
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
ei
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
0
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
HE R2-30R/ 1
209
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
56A/102S+ LC-
AGCCAGAGCCTGGTGCACAACAACGCCAACACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
0 WT-
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
trastuzu ma b/
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
CD28supxCD3nni
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGA
d L1 linker IgG4
GCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGT
GGCTGAACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCG
FALA
GCGTGCCCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGA
BP #11
CATTGCCACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAG
ACCAAGGGCCCCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCG
GCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACG
CCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGC
ACC CTGACACTGAGCAAGGC C GACTAC GAGAAGCACAAGGTGTAC GC CTGC GAAGTGACC CAC CAGGGC
CTGTCT
AGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
2 210
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTG
GTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTC
ACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAA
CAC C CTGTAC CTGCAGATGAACAGCCTGC GGGCC GAGGACAC C GCC GTGTACTACTGTC GGGGC
GTGTACTATGC C
CTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCAT
CGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTA
CTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACC
TGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
ei
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGA
TCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGT
ACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTG
GTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGC
CTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCC
CCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATT
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGG
CTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTG
ATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 211
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCCGGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
ATCTACCCCACCAACGCCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA
0
GGCTCCGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGCC
=))
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGA
CTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTAC
ACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGC
CCTCCTTGCCCAGCCCCTGAATTTCTGGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGAT
GATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTG
GTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGG
TGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGG
GCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGC
CCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACAT
TGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGG
CTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTG
ATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 212
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
010
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
ei
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
HE R2-30R/ 1
213
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
56A/102 E/
AGCCAGAGCCTGGTGCACAACAACGCCAACACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
0 CD28supxCD3nni
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
d L1 linker IgG4
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
FALA
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGA
GCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGT
8 BP # 12
GGCTGAACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCG
GCGTGCCCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGA
CATTGCCACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAG
ACCAAGGGCCCCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCG
GCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACG
CCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGC
ACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCT
AGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
2
214
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
t.)
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTG
GTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTC
ACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAA
CACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCC
CTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCAT
CGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTA
CTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACC
TGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
ei
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGA
TCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGT
ACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTG
GTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGC
CTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCC
CCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATT
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGG
CTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTG
ATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 215
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCCGGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
ATCTACCCCACCAACGCCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA
0
GGCGAAGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGC
CCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGG
ACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGT
GCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTA
CACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTG
CCCTCCTTGCCCAGCCCCTGAATTTCTGGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGA
TGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATT
GGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGG
GTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTG
CCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACA
TTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACG
GCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGT
GATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 216
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
010
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
ei
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
HE R2- 1
217
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
WT+trastuzu ma b
AGCCAGAGCCTGGTGCACCAGAACGCCCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
0
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
CD28supxCD3nni
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
d (32/350Q) Li
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGA
linker IgG4 FALA
GCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGT
GGCTGAACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCG
BP # 15
GCGTGCCCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGA
CATTGCCACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAG
ACCAAGGGCCCCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCG
GCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACG
CCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGC
ACC CTGACACTGAGCAAGGC C GACTAC GAGAAGCACAAGGTGTAC GCCTGC GAAGTGACC CAC
CAGGGCCTGTCT
AGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
2 218
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTG
GTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTC
ACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAA
CAC C CTGTAC CTGCAGATGAACAGCCTGC GGGCC GAGGACAC C GCC GTGTACTACTGTC GGGGC
GTGTACTATGC C
CTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCAT
CGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTA
CTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACC
TGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
ei
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGA
TCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGT
ACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTG
GTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGC
CTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCC
CCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATT
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGG
CTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTG
ATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3
219
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCAAGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
o
ATCTACCCCACCAACGGCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA
0
GGCGACGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGA
CTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTAC
ACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGC
CCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGA
TGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATT
GGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGG
GTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTG
CCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACA
TTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACG
GCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGT
GATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 220
GACATCCAGATGACCCAGAGCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTAGAGCC
AGCCAGGACGTGAACACCGCCGTGGCCTGGTATCAGCAGAAGCCTGGCAAGGCCCCCAAGCTGCTGATCTACAGC
GCCAGCTTCCTGTACAGCGGCGTGCCCAGCAGATTCAGCGGAAGCAGAAGCGGCACCGACTTCACCCTGACCATC
AGCTCCCTGCAGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACCCCCCCCACATTTGGCCAGG
GCACCAAGGTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGA
AGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGG
ACAACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTG
AGCAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGG
CCTGTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
HE R2/ 1
221
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
CD28õ,xCD3rnid
AGCCAGAGCCTGGTGCACAACAACGCCAACACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
DKTHT linkers on
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
HC/LC) IgG4
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
FALA
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
BP #25
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2
222
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
0
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
cs,
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 223
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCAAGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
ATCTACCCCACCAACGGCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC ..
010
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA ei
GGCGACGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGA
CTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTAC
ACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGC
CCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGA
TGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATT
GGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGG
GTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTG
CCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACA o
TTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACG
GCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGT
GATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
0
4 224
GACATCCAGATGACCCAGAGCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTAGAGCC
AGCCAGGACGTGAACACCGCCGTGGCCTGGTATCAGCAGAAGCCTGGCAAGGCCCCCAAGCTGCTGATCTACAGC
GCCAGCTTCCTGTACAGCGGCGTGCCCAGCAGATTCAGCGGAAGCAGAAGCGGCACCGACTTCACCCTGACCATC
AGCTCCCTGCAGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACCCCCCCCACATTTGGCCAGG
GCACCAAGGTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGA
AGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGG
ACAACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTG
AGCAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGG
CCTGTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
HE R2/ 1
225
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
CD28õpxCD3rnid
AGCCAGAGCCTGGTGCACGAGAACCTGCGTACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
(32/33/3435
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
EN LR (LC); DKTHT
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
linkers on HC/LC)
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
IgG4 FALA
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
BP #26
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2
226
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
1-3
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
wow
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
0
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 227
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCAAGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
ATCTACCCCACCAACGGCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA
GGCGACGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGA
CTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTAC
ACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGC
CCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGA
TGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATT
GGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGG
GTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTG
CCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACA
TTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACG
GCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGT
GATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 228
GACATCCAGATGACCCAGAGCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTAGAGCC
AGCCAGGACGTGAACACCGCCGTGGCCTGGTATCAGCAGAAGCCTGGCAAGGCCCCCAAGCTGCTGATCTACAGC
GCCAGCTTCCTGTACAGCGGCGTGCCCAGCAGATTCAGCGGAAGCAGAAGCGGCACCGACTTCACCCTGACCATC
AGCTCCCTGCAGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACCCCCCCCACATTTGGCCAGG
GCACCAAGGTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGA
AGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGG
ACAACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTG
AGCAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGG
CCTGTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
0
HE R2/ 1
229
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
o
CD28õpxCD3rnid
AGCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
(32/33/3435
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
0 ENLQ (LC);
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
DKTHT linkers on
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
HC/LC) IgG4
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
FALA
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
BP #27
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2 230
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
010
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
ei
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3
231
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCAAGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
o
ATCTACCCCACCAACGGCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA
0
GGCGACGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGA
CTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTAC
ACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGC
CCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGA
TGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATT
GGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGG
GTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTG
CCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACA
TTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACG
GCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGT
GATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 232
GACATCCAGATGACCCAGAGCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTAGAGCC
AGCCAGGACGTGAACACCGCCGTGGCCTGGTATCAGCAGAAGCCTGGCAAGGCCCCCAAGCTGCTGATCTACAGC
GCCAGCTTCCTGTACAGCGGCGTGCCCAGCAGATTCAGCGGAAGCAGAAGCGGCACCGACTTCACCCTGACCATC
AGCTCCCTGCAGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACCCCCCCCACATTTGGCCAGG
GCACCAAGGTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGA
AGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGG
ACAACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTG
AGCAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGG
CCTGTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
HE R2/ 1
233
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
CD28õ,xCD3rnid
AGCCAGAGCCTGGTGCACGAGAACCTGTTCACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
(32/33/3435
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
EN LE (LC); DKTHT
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
linkers on HC/LC)
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
IgG4 FALA
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
BP #28
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC o
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2
234
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
0
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 235
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCAAGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
ATCTACCCCACCAACGGCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
010
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA ei
GGCGACGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGA
CTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTAC
ACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGC
CCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGA
TGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATT
GGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGG
GTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTG
CCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACA o
TTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACG
GCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGT
GATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
0
4 236
GACATCCAGATGACCCAGAGCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTAGAGCC
AGCCAGGACGTGAACACCGCCGTGGCCTGGTATCAGCAGAAGCCTGGCAAGGCCCCCAAGCTGCTGATCTACAGC
GCCAGCTTCCTGTACAGCGGCGTGCCCAGCAGATTCAGCGGAAGCAGAAGCGGCACCGACTTCACCCTGACCATC
AGCTCCCTGCAGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACCCCCCCCACATTTGGCCAGG
GCACCAAGGTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGA
AGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGG
ACAACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTG
AGCAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGG
CCTGTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
anti- 1
237
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
H e r2/C D3/3C D2
AGCCAGAGCCTGGTGCACAACAACGCCAACACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
8 IgG4 FALA
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
BP #29
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGA
GCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGT
GGCTGAACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCG
GCGTGCCCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGA
CATTGCCACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAG
ACCAAGGGCCCCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCG
GCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACG
CCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGC
ACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCT
AGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
2
238
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
1-3
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTG
GTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTC
AC CAAGGCCTGGATGCACTGGGTGC GC CAGGCC C CTGGAAAGCAGCTGGAATGGGTGGCC
CAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAA
CACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCC
CTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCAT
CGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTA
CTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACC
TGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGA
0
TCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGT
ACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTG
GTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGC
CTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCC
CCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATT
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGG
CTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTG
ATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 239
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCAAGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
ATCTACCCCACCAACGGCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA
GGCGACGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGA
CTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTAC
ACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGC
CCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGA
TGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATT
GGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGG
GTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTG
CCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACA
TTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACG
GCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGT
GATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 240
GACATCCAGATGACCCAGAGCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTAGAGCC
AGCCAGGACGTGAACACCGCCGTGGCCTGGTATCAGCAGAAGCCTGGCAAGGCCCCCAAGCTGCTGATCTACAGC
GCCAGCTTCCTGTACAGCGGCGTGCCCAGCAGATTCAGCGGAAGCAGAAGCGGCACCGACTTCACCCTGACCATC
AGCTCCCTGCAGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACCCCCCCCACATTTGGCCAGG
GCACCAAGGTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGA
AGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGG
ACAACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTG
AGCAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGG
CCTGTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
0
HE R230R/554/1 1
241
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
o
02E/
AGCCAGAGCCTGGTGCACGAGAACCTGCGTACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
CD28õpxCD3rnid
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
0 (32/33/3435
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
EN LR (LC); DKTHT
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
linkers on HC/LC)
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
0
I gG4 FALA
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
BP #31
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2 242
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
ei
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3
243
GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTGGTGCAGCCTGGCGGATCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCAACATCCGGGACACCTACATCCACTGGGTGCGCCAGGCCCCTGGCAAGGGACTGGAATGGGTGGCCAGA
ATCTACCCCACCCAGGGCTACACCAGATACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGCCGACACCAGC
AAGAACACCGCCTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTAGTAGATGGGGA
0
GGCGAAGGCTTCTACGCCATGGACTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGTGCGTCGACCAAGGGC
CCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGG
ACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGT
GCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTA
CACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTG
CCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTG
ATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAAT
TGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCG
GGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAA
GGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCT
GCCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGAC
ATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGAC
GGCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCG
TGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4
244
GACATCCAGATGACCCAGAGCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTAGAGCC
AGCCAGGACGTGAACACCGCCGTGGCCTGGTATCAGCAGAAGCCTGGCAAGGCCCCCAAGCTGCTGATCTACAGC
GCCAGCTTCCTGTACAGCGGCGTGCCCAGCAGATTCAGCGGAAGCAGAAGCGGCACCGACTTCACCCTGACCATC
AGCTCCCTGCAGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACCCCCCCCACATTTGGCCAGG
GCACCAAGGTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGA
AGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGG
ACAACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTG
AGCAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGG
CCTGTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
CD38VH1 / 1
245
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
CD28sup x
AGCCAGAGCCTGGTGCACGAGAACCTGTTCACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
CD3mid_ENLQ
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
DKTHT IgG4
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
FALA
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
BP # 1
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC o
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2
246
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
0
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
cs,
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 247
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAAGTCGTGAAACCTGGCGCCTCCGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACGCCATGCACTGGGTCAAAGAGGCCCCTGGCCAGAGACTGGAATGGATCGGCTAC
ATCTACCCCGGCCAGGGCGGCACCAACTACAACCAGAAGTTCCAGGGCAGAGCCACCCTGACCGCCGATACAAGC
010
GCCAGCACCGCCTACATGGAACTGAGCAGCCTGCGGAGCGAGGATACCGCCGTGTACTTCTGTGCCAGAACAGGC
ei
GGCCTGAGGCGGGCCTACTTTACCTATTGGGGCCAGGGCACCCTCGTGACCGTGTCTAGCGCTAGCACAAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGA
CTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTAC
ACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGC
CCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGA
TGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATT
GGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGG
GTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTG
CCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACA o
TTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACG
GCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGT
GATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
0
4 248
GACATCGTGCTGACACAGAGCCCTGCCACCCTGTCTCTGAGCCCTGGCGAGAGAGCCACCATCAGCTGTAGAGCC
AGCCAGAGCGTGTCCAGCTACGGCCAGGGCTTCATGCACTGGTATCAGCAGAAGCCCGGCCAGCCCCCCAGACTG
CTGATCTATGGCGCCAGCAGCAGAGCCACAGGCATCCCCGCCAGATTTTCTGGCTCTGGCAGCGGCACCGACTTCA
CCCTGACAATCAGCCCCCTGGAACCCGAGGACTTCGCCGTGTACTACTGCCAGCAGAACAAAGAGGACCCCTGGA
CCTTCGGCGGAGGCACCAAGCTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGA
CGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAGGTGCAG
TGGAAGGTGGACAATGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCAC
CTACAGCCTGAGCAGCACCCTGACCCTGTCCAAGGCCGATTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGAC
CCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGC
CD38hhy992 / 1 249
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
CD28sup x
AGCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
CD3mid ENLQ
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
DKTHT IgG4
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
FALA
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
BP #5
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2 250
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
1-3
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
wow
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
0
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 251
CAGGTTCAGCTGGTTCAGTCTGGCGCCGAAGTGAAGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGTGTCCG
GCTACACCCTGACCGAGTTCAGCATCCACTGGGTCCGACAGGCTCCAGGACAAGGCTTGGAATGGATGGGCGGCT
TCGATCCCGAGGACGGCGAAACAATCTACGCCCAGAAATTCCAGGGCCGCGTGATCATGACCGAGGACACCTCTA
CCGACACCGCCTACATGGAAATGAACAGCCTGCGGAGCGAGGATACCGCCATCTACTACTGTACCACCGGCAGAT
TCTTCGACTGGTTCTGGGGCCAGGGCACCCTGGTTACAGTCTCTTCTGCGTCGACCAAGGGCCCATCGGTGTTCCCT
CTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGC
CCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTCCAGAGCAGCGG
CCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTGTAACGTGGAC
CACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTTGCCCAGCC
CCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCC
CCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGACGGCG
TGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTGCTGA
CCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCTCCA
TCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGAAG
AGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGG
AGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGT
ACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCC
TGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
cpw
4 252
GAGATCATCCTGACACAGAGCCCCGCCATCCTGTCACTGTCTCCAGGCGAAAGAGCCACACTGAGCTGTAGAGCC
AGCCAGAGCGTGATCAGCAGATTCCTGAGCTGGTATCAAGTGAAGCCCGGACTGGCCCCTCGGCTGTTGATATATG
GCGCCTCTACACGCGCCACAGGCATCCCTGTTAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCAGCCTGACAAT
TAGCAGCCTGCAGCCTGAGGACTGCGCCGTGTACTACTGTCAGCAGGACAGCAACCTGCCTATCACCTTCGGCCAG
GGCACCAGACTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTG
AAGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTG
GACAACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCT
GAGCAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGG
GCCTGTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
0
CD38hyb5739 / 1
253
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
CD28sup x
AGCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
CD3mid_ENLQ
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
0 DKTHT IgG4
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
FALA
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
BP #6
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2 254
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
ei
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 255
CAGGTTCAGCTGCAGCAGTCTGGCCCCGAACTCGTTAGACCTGGCACCTCTGTGAAGGTGTCCTGCAAGGCCAGCG
GCTACGCCTTTACCACCTACCTGGTGGAATGGATCAAGCAGAGGCCTGGACAGGGCCTCGAGTGGATCGGAGTGA
o
TCAATCCTGGCAGCGGCAGCACCAACTACAACGAGAAGTTCAAGGGCAAAGCCACACTGACCGTGGACAGAAGC
AGCACCACAGCCTACATGCACCTGAGCGGCCTGACCTCTGATGACAGCGCCGTGTACTTCTGCGCCAGATACGCCT
0
ATGGCTATTGGGGCCAGGGCACAACCCTGACCGTTAGCTCTGCGTCGACCAAGGGCCCATCGGTGTTCCCTCTGGC
CCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTG
ACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGT
ACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTGTAACGTGGACCACA
AGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTG
AAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCCCCGA
AGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGA
AGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTGCTGACCGT
GCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGA
GAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGAAGAGAT
GACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGGAGAG
CAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTACTCC
AAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCAC
AACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 256
GACATCGTGATGACCCAGAGCCAGAAATTCATGAGCGCCAGCGTGGGCGACAGAGTGTCCATCACATGTAAAGCC
AGCCAGAACGTGGGCACAGCCGTGGCTTGGTATCAGCAGCAGCCTGGCCACTCTCCTAAGCAGCTGATCTACAGC
GCCAGCAACAGATACACCGGCGTGCCCGATAGATTCACAGGATCTGGCGCCGGAACCGACTTCACCCTGACCATC
AGCAACATCCAGAGCGAGGACCTGGCCGACTACTTCTGCCAGCAGTACAGCACATACCCCTTCACCTTTGGCAGCG
GCACCAAGCTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGA
AGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGG
ACAACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTG
AGCAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGG
CCTGTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
CD38hyb6284 / 1 257
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
CD28sup x
AGCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
CD3mid_ENLQ
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
DKTHT IgG4
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
FALA
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
BP #7
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
CDC
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
wow
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
FD'n)
2 258
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
,D0a)
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
0"
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
cc,
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 259
CAGGTTCAGCTGCTGCAGTCTGGCGCCGAACTTGTCAGACCTGGCGTGTCCGTGAAGATCAGCTGTACAGGCAGCG
GCTACAGCTTCACCAACTACGCCGTGCACTGGGTCAAGCAGAGCCACGTGAAGTCCCTGGAATGGATCGGCGTGA
TCAGCCCCTACTACGGCGACACCACCTACAACCAGAAGTTCACCGGCAAGGCCACCATGACCGTGGACAAGTCTA
GCAGCACCGCCTACATGGAACTGGCCAGACTGACCAGCGAGGACAGCGCCATCTACTTTTGCGCCAGAAGATTCG
ei
AGGGCTTCTACTACAGCATGGACTACTGGGGCCAGGGCACCAGCGTGACAGTTTCTTCTGCGTCGACCAAGGGCCC
ATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGAC
TACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGC
TCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACA
CCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCC
CTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGAT
GATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTG
GTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGG
TGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGG
GCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGC
CCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACAT o
TGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGG
CTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTG
ATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
0
4 260
GACGTGGTCATGATCCAGACACCTCTGAGCCTGCCTGTGTCTCTGGGAGATCAGGCCAGCATCAGCTGCAGACCTA
GCCAGTCTCTGGTGCACAGCAACGGCAACACCTACCTGAACTGGTATCTGCAGAGGCCCGGACAGAGCCCCAAGC
TGCTGATCTACAAGGTGTCCAAGCGGTTCAGCGGCGTGCCCGATAGATTTTCTGGCAGCGGCTCTGGCACCGACTT
CACCCTGAAGATTAGCAGAGTGGAAGCCGAGGACCTGGGCGTGTACCTGTGTTCTCAGAGCACACACGTGCCCCT
GACCTTTGGCAGCGGAACCCAGCTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGC
GACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGC
AGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCC
ACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTG
ACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
CD38hhy1195 / 1
261
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
CD28sup x
AGCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
CD3mid ENLQ
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
DKTHT IgG4
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
FALA
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
BP #8
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2
262
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
1-3
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
wow
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
0
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 263
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCAGCAGCTACGGCATGTACTGGGTGCGCCAGGCCCCTGGCAAAGGCCTGGAATGGGTGGCCGTG
ATTTGGTACGACGGCAGCAACAAGTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCTCCCGGGACAACAGC
AAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACAGCCGTGTATCACTGCGCCAGAGATCCC
GGCCTGCGGTACTTTGACGGCGGCATGGATGTGTGGGGCCAGGGCACAACCGTGACCGTGTCATCTGCGTCGACC
AAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCG
TGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCC
AGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAA
GACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCC
TCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGAC
ACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAG
TTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACC
TACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCC
AACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTAT
ACCCTGCCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCA
GCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACA
GCGACGGCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCT
GCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 264
GACATCCAGCTGACCCAGAGCCCCAGCTTTCTGAGCGCCAGCGTGGGCGACAGAGTGACCATCACCTGTAGAGCC
AGCCAGGGCATCAGCAGCTACCTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAACTGCTGATCTTTGCC
GCCAGCACACTGCACAGCGGCGTGCCCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGAGTTCACCCTGACAATCA
GCAGCCTGCAGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCTGAACAGCTTCCCCTACACCTTCGGCCAGGG
CACCAAGCTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAA
GTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGAC
AACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAG
CAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCT
GTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
0
CD38hhy1370 / 1
265
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
CD28sup x
AGCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
CD3mid_ENLQ
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
0 DKTHT IgG4
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
FALA
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
BP #9
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2 266
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
ei
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 267
CAGGTGCAGCTGGTGGAAAGCGGCGGAGGCGTGGTGCAGCCTGGCAGGTCTCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCAGCAGCTACGGAATGCACTGGGTGCGCCAGGCCCCTGGCAAAGGACTGGAATGGGTGGCCGTG
ATTTGGTACGACGGCAGCAACAAGTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCGGCGACAACAGC
AAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGCGCCAGAATGTTC
0
AGAGGCGCCTTCGACTACTGGGGCCAGGGCACACTCGTGACCGTGTCTAGTGCGTCGACCAAGGGCCCATCGGTG
TTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTC
CCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTCCAGAG
CAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTGTAA
CGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTTG
CCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGC
CGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTG
GACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTC
CGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCC
CAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTG
CCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGT
GGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATT
CTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCAC
GAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 268
GCCATCCAGATGACCCAGAGCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTAGAGCC
AGCCAGGGCATCCGGAACGACCTGGGCTGGTATCAGCAGAAGCCTGGCAAGGCCCCCAAGCTGCTGATCTACGCC
GCTAGCTCTCTGCAGTCCGGCGTGCCCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCT
CTGGCCTGCAGCCCGAGGACAGCGCCACCTACTACTGTCTGCAAGACTACATCTACTACCCCACCTTCGGCCAGGG
CACCAAGGTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAA
GTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGAC
AACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAG
CAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCT
GTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
CD38hu5739 / 1
269
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
CD28sup x
AGCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
CD3mid_ENLQ
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
DKTHT IgG4
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
FALA
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
CDC
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
FD'n)
2
270
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
,D0a)
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
0"
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
cs,
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 271
CAGGTTCAGCTGGTTCAGTCTGGCGCCGAAGTGAAGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCTCTG
GCTACGCCTTCACCACCTACCTGGTGGAATGGATCAGACAGAGGCCTGGACAGGGCCTCGAATGGATGGGCGTGA
TCAATCCTGGCAGCGGCAGCACCAATTACGCCCAGAAATTCCAGGGCAGAGTGACCATGACCGTGGACAGAAGCA
GCACCACCGCCTACATGGAACTGAGCAGACTGAGAAGCGACGACACCGCCGTGTACTACTGTGCCAGATACGCCT
ei
ACGGCTATTGGGGCCAGGGAACCCTGGTTACCGTTAGCTCTGCGTCGACCAAGGGCCCATCGGTGTTCCCTCTGGC
CCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTG
ACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGT
ACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTGTAACGTGGACCACA ..
CBtµ
AGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTG
AAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCCCCGA
AGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGA
AGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTGCTGACCGT
GCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGA
GAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGAAGAGAT
GACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGGAGAG
CAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTACTCC
AAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCAC
AACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
0
CD
4
272
GACATCCAGATGACACAGAGCCCTAGCAGCCTGTCTGCCAGCGTGGGAGACAGAGTGACCATCACCTGTAGAGCC
AGCCAGAATGTGGGAACAGCCGTGGCCTGGTATCAGCAGAAGCCTGGCAAGAGCCCCAAGCAGCTGATCTACAGC
GCCAGCAACAGATACACCGGCGTGCCCAGCAGATTTTCTGGCAGCGGCTCTGGCACCGACTTCACCCTGACCATAT
CTAGCCTGCAGCCTGAGGACCTGGCCACCTACTACTGTCAGCAGTACAGCACATACCCCTTCACCTTCGGCCAGGG
CACCAAGCTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAA
GTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGAC
AACGCCCTGCAGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAG
CAGCACCCTGACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCT
GTCTAGCCCCGTGACCAAGAGCTTCAACCGGGGCGAGTGT
CD38hu6284 / 1
273
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGC
CD28sup x
AGCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAG
CD3mid ENLQ
TCCCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACT
DKTHT IgG4
TCACCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCT
FALA
TCACCTTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCA
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGA
ACTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGC
CCAGCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGC
CACCTACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAA
GACCCACACCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACA
GCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGC
AGAGCGGCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTG
ACACTGAGCAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCC
GTGACCAAGAGCTTCAACCGGGGCGAGTGT
2
274
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGC
1-3
ATCTACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGC
ATCAGCACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCAC
TACGGCCTGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACC
CAGGTGCAGCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGC
GGCTTCACCTTCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAG
ATCAAGGACAAGAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGAC
GACAGCAAGAACACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGC
GTGTACTATGCCCTGAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACA
CCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCT
GGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTG
CACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTA
AGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAG
0
CCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCC
GAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTC
AACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGC
AAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCT
CAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGC
TTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAAC
GTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 275
CAGGTTCAGCTGGTTCAGTCTGGCGCCGAAGTGAAGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGC
GGCTACAGCTTCACCAATTACGCCGTGCACTGGGTCCGACAGGCTCCAGGACAAGGACTGGAATGGATGGGCGTG
ATCAGCCCCTACTACGGCGATACCACATACGCCCAGAAATTCCAGGGCAGAGTGACCATGACCGTGGACAAGAGC
AGCAGCACCGCCTACATGGAACTGAGCAGACTGAGAAGCGACGACACCGCCGTGTACTACTGCGCCAGAAGATTC
GAGGGCTTCTACTACAGCATGGACTACTGGGGCCAGGGCACCCTGGTTACAGTCTCTTCTGCGTCGACCAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGA
CTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTAC
ACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGC
CCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGA
TGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATT
GGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGG
GTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTG
CCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACA
TTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACG
010
GCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGT
GATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 276
GACGTGGTCATGACACAGAGCCCTCTGAGCCTGCCTGTGACACTGGGACAGCCTGCCAGCATCAGCTGTAGACCTA
GCCAGAGCCTGGTGCACAGCAACGGCAACACCTACCTGAACTGGTATCAGCAGAGGCCCGGACAGAGCCCCAAGC
TGCTGATCTACAAGGTGTCCAAGCGGTTCAGCGGCGTGCCCGATAGATTTTCTGGCAGCGGCTCTGGCACCGACTT
CACCCTGAAGATTAGCAGAGTGGAAGCCGAGGACGTGGGCGTGTACTACTGTAGCCAGTCTACCCACGTGCCACT
GACCTTTGGCGGCGGAACAAAGGTGGAAATCAAGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGC
GACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGC
WO 2020/210392
PCT/US2020/027320
L) E.
HL
L)
C7 C7
(DL)
= C7
= E.
L) L)
= c-)
= C=
= H
Q7 EY
UH
Q7
Q7 LI
L) L) Q7
Q7
Q7 ,e
E.
Q7 Q7
Q) 0 Q)
Q7 Q7
Q7
L) Q7
Q7
Q7 tj
Q7
L.) L)
Q7
_de
C)
L.) HL)
Lõ)
0.(¨)(DL)
= HL)
E" L) L)
L) L) L)
L) L)
L)
CD L)
= L)
L)
H
(=
(.7
189
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
EXAMPLES
[0304] The Examples that follow are illustrative of specific embodiments of
the
disclosure, and various uses thereof. They are set forth for explanatory
purposes only, and
should not be construed as limiting the scope of the invention in any way.
Example 1: Development of trispecific HER2/CD28xCD3 antibodies and variant
anti-
CD3 binding sites
[0305] Immuno-oncology is a promising, emerging therapeutic approach to
disease
management in cancer. The immune system is the first line of defense against
cancer
development and progression. There is now large evidence that T cells are able
to control
tumor growth and prolong the survival of cancer patients in both early and
late stages of
disease. However, T cells specific for tumors can be limited in a number of
ways preventing
them from controlling the disease.
[0306] In order to remove the limitations on T cells induced by
uncontrolled tumors,
novel antibodies were developed in the trispecific antibody format depicted in
FIG. 1A to
specifically activate the T cells to engage HER2 expressing cancer cells.
These novel
trispecific antibodies are able to bind to three targets: HER2, CD3, and CD28.
Anti-HER2
and anti-CD3 binding sites were further optimized for high affinity binding
and reduction in
potential manufacturing liabilities.
[0307] HER2 amplification and overexpression can be found in molecular
subtypes of
breast cancer, and also in gastric, ovarian, lung and prostate carcinomas.
Optimal activation
of T cells requires two factors: (1) Antigen recognition and (2) Co-
stimulation. Using the
trispecific HER2/CD28xCD3 trispecific binding proteins described herein,
Signal 1 is
provided by an agonist anti-CD3 binding site, and Signal 2 is provided by an
agonist anti-
CD28 binding site (see, e.g., FIG. 1D). It is thought that the trispecific
antibodies described
in the subsequent Examples recruit T cells to the tumor via HER2 and activate
the engaged T
cells by binding to CD3 and CD28. The resulting activation induces the killing
potential of
the immune cells against the nearby tumor cells.
Materials and Methods
Production and characterization of antibodies
[0308] Trispecific antibody variants were produced by transient
transfection of
expression plasmids into Expi293 cells. 5 days after transfection, the
supernatant from
190
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
transfected cells was collected, quantified and normalized by absorbance at
280 nm on Nano
Drop. The binding of supernatant to corresponding antigens were determined by
ELISA and
the absorbance of parental HER2 WT tri Ab was set as 1Ø The fold changes of
other
variants were calculated by dividing the corresponding absorbance to that of
parental Ab.
[0309] Trispecific antibody variants were purified using protein A affinity
purification
followed by SEC purification. The binding of purified antibodies to
corresponding antigens
were determined by ELISA. The EC50 were determined based on the binding curve
generated
by Graphpad Prism7.
Results
[0310] Trispecific Ab variants were produced with several mutations in the
binding arms
in order to mitigate potential manufacturing liabilities, e.g., deamidation
sites. A binding
ELISA assay was performed to assess binding of the indicated trispecific
antibodies to each
of the three targets: HER2, CD3, and CD28. In FIG. 1B, HER2/CD3xCD28
trispecific
antibodies with the indicated anti-HER2 or anti-CD3 variants were compared to
parental
Trispecific Ab. Introducing some sets of mutations (e.g., 32/33 QQ and
33/35QQ) into the
VL domain of the anti-CD3 binding site led to dramatically reduced binding to
CD3, whereas
32/35 QQ mutations retained near wild-type binding. MS peptide analyses showed
that
binding sites with the DNAQ mutations in CDR-L1 (SEQ ID NO:63) were still
subject to
greater than 15% deamidation, whereas ENLQ (SEQ ID NO:281), ENLF (SEQ ID
NO:282),
and ENLR (SEQ ID NO:283) led to less than 5% deamidation. Importantly, these
variants
also retained binding to CD3.
[0311] In addition, binding curves for the indicated antibodies binding to
human HER2,
human CD28, and CD3 are provided in FIG. 1C. The EC50 values of selected
trispecific
antibody variants are provided in Table E.
191
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
Table E. A binding ELISA assay was performed on purified trispecific
antibodies to
determine their binding affinities for HER2, human CD3, and human CD28.
Binding Affinity (ELISA)(nM)
EC50
Trispecific antibody HER2 Human CD3
Human
CD28
HER2 (WT-trastuzumab) / CD28supxCD3mid
(32/35 QQ (LC); DKTHT linkers on HC/LC) IgG4 162.3 566.4 1321
FALA
HER2 (30R/55Q/102E +LC-WT-trastuzumab) /
CD28supxCD3mid (32/35 QQ (LC); DKTHT linkers 93.66 364.8 871.9
on HC/LC) IgG4 FALA
HER2-30R/55Q/102E/ CD28supxCD3mid
83.89 3222 1024
(32/33/35QSQ) DKTHT linker IgG4 FALA
HER2 (30R/55Q/102E+LC-WT-trastuzumab) /
CD28supxCD3mid (DNAQ (LC); DKTHT linkers on 111.2 725.7 1053
HC/LC) IgG4 FALA
HER2 (30R/55Q/102E +LC-WT-trastuzumab) /
CD28supxCD3mid (32/35QQ (LC); Li linker) IgG4 111.5 412.5 1345
FALA
HER230R/55Q/102E/ CD28supxCD3mid (32/33/3435
ENLR (LC); DKTHT linkers on HC/LC) IgG4 123.9 81.53 878.8
FALA
HER2 (30R/56A/102S +LC-WT-trastuzumab) /
516.0 5494 3631
CD28supxCD3mid (32/35QQ185E) IgG4 FALA
HER2-30R/55Q/102E+LC-30Q/ CD28supxCD3mid
1540 10616 2036
(32/35QQ) 185S Li linker IgG4 FALA
HER2-30R/55Q/102E/ CD28supxCD3mid
467.0 19382 1814
(32/33/35QSQ) 185S Li linker IgG4 FALA
HER2-30R/55Q/102E/ CD28supxCD3mid
478.6 19756 1739
(32/33/35QSQ) 185E Li linker IgG4 FALA
192
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
HER2/ CD28supxCD3mid DKTHT linkers on
228.9 671.2 752
HC/LC) IgG4 FALA
HER2/ CD28supxCD3mid (32/33/3435 ENLF (LC);
195.9 773.3 1466
DKTHT linkers on HC/LC) IgG4 FALA
HER2/ CD28supxCD3mid (32/33/3435 ENLQ (LC);
212.1 10558 2405
DKTHT linkers on HC/LC) IgG4 FALA
HER2/ CD28supxCD3mid (32/33/3435 ENLR (LC);
166.2 381.9 1051
DKTHT linkers on HC/LC) IgG4 FALA
anti-Her2/CD3/3CD28 IgG4 FALA 176.1 516.2 870.6
[0312] Without wishing to be bound by theory, as depicted in FIG. 1D, it is
believed that
HER2/CD3/CD28 trispecific antibodies recruit T cells to cancer cells through
the anti-HER2
and anti-CD3/CD28 arms. Further, it is believed that engaged T cells are
activated by the
anti-CD28/CD3 arms. Killing of cancer cells is believed, without wishing to be
bound by
theory, to occur through T cell mediated mechanisms (e.g., Perforin,
granzyme). Without
wishing to be bound to theory, it is contemplated that similar mechanisms may
allow for
killing of other types of tumors by substituting antigen binding sites that
recognize other
tumor target proteins.
Example 2: Development of trispecific CD38/CD3xCD28 antibodies
[0313] Trispecific CD38/CD3xCD28 antibodies were developed and
characterized for
binding to CD38, CD3 and CD28 polypeptides.
Materials and Methods
Generation of CD38/CD28xCD3 trispecific antibodies
[0314] A panel of anti-CD38, anti-CD3, and anti-CD28 antibodies, as well as
human
IgG4 Fc domains were used to generate CD38/CD28xCD3 trispecific antibodies in
the
trispecific antibody format depicted in FIG. 2A.
[0315] Trispecific binding proteins were produced by transient transfection
of 4
expression plasmids into Expi293 cells using ExpiFectamineTM 293 Transfection
Kit
(Thermo Fisher Scientific) according to manufacturer's protocol. Briefly, 25%
(w/w) of each
plasmid was diluted into Opti-MEM, mixed with pre-diluted ExpiFectamine
reagent for 20-
193
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
30 minutes at room temperature (RT), and added into Expi293 cells (2.5x106
cells/nil). An
optimization of transfection to determine the best ratio of plasmids was often
used in order to
produce the trispecific binding protein with good yield and purity.
[0316] 4-5 days post transfection, the supernatant from transfected cells
was collected
and filtered through 0.45 p.m filter unit (Nalgene). The trispecific binding
protein in the
supernatant was purified using a 3-step procedure. First, protein A affinity
purification was
used, and the bound Ab was eluted using "IgG Elution Buffer"(Thermo Fisher
Scientific).
Second, product was dialyzed against PBS (pH7.4) overnight with 2 changes of
PBS buffer.
Any precipitate was cleared by filtration through 0.45 p.m filter unit
(Nalgene) before next
step. Third, size-exclusion chromatography (SEC) purification (Hiload 16/600
Superdex
200pg, or Hiload 26/600 Superdex 200pg, GE Healthcare) was used to remove
aggregates
and different species in the prep. The fractions were analyzed on reduced and
non-reduced
SDS-PAGE to identify the fractions that contained the monomeric trispecific
binding protein
before combining them. The purified antibody can be aliquoted and stored at -
80 C long
term.
ELISA binding assay
[0317] Binding affinities to each target antigen by the CD38/CD28xCD3 T
cell engagers
were measured by ELISA. Briefly, each antigen was used to coat the 96-well
Immuno Plate
(Thermo Fisher Scientific) overnight at 4 C using 200 ng/well in PBS(pH7.4) of
each
antigen. The coated plate was blocked using 5% skim milk+2% BSA in PBS for one
hour at
RT, followed by washing with PBS+0.25% Tween 20 three times (Aqua Max 400,
Molecular
Devices). Serial dilution of antibodies (trispecific and control Abs) were
prepared and added
onto the ELISA plates (100 I/well in duplicate), incubated at room
temperature (RT) for one
hour, followed by washing 5 times with PBS+0.25% Tween 20. After washing, the
HRP
conjugated secondary anti-human Fab (1:5000, Cat. No. 109-035-097, Jackson
ImmunoResearch Inc) was added to each well and incubated at RT for 30 minutes.
After
washing 5 times with PBS+0.25% Tween 20, 100 IA of TMB Microwell Peroxidase
Substrate
(KPL, Gaithersburg, MID, USA) was added to each well. The reaction was
terminated by
adding 50 IA 1M H2504, and 0D450 was measured using SpectraMax M5 (Molecular
Devices) and analyzed using SoftMax Pro6.3 software (Molecular Devices). The
final data
was transferred to GraphPad Prism software (GraphPad Software, CA, USA), and
plotted.
EC50 was calculated using the same software.
194
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
Measurement of trispecific antibody binding using SPR
[0318] Human CD38-His antigens were used (Cambridge Biologics, Cambridge,
MA) for
full kinetic analysis. Kinetic characterization of purified antibodies was
performed using
SPR technology on a BIACORE 3000 (GE Healthcare). A capture assay using human
IgG1
specific antibody capture and orientation of the investigated antibodies was
used. For capture
of Fc containing protein constructs the human antibody capture kit (GE
Healthcare) was used.
For capture of His tagged antigen, anti-His antibody capture kit (GE
Healthcare) was used.
The capture antibody was immobilized via primary amine groups (11000 RU) on a
research
grade CM5 chip (GE Life Sciences) using standard procedures. The analyzed
antibody was
captured at a flow rate of 10 lL/min with an adjusted RU value that would
result in maximal
analyte binding signal of typically 30 RU. Binding kinetics were measured
against the
trispecific antibodies. Assay buffer HBS EP (10 mM HEPES, pH 7.4, 150 mM NaCl,
3 mM
EDTA, and 0.005 % Surfactant P20) was used at a flow rate of 30 11.1/min. Chip
surfaces
were regenerated with the regeneration solution of the respective capture kit.
Kinetic
parameters were analyzed and calculated in the BIA evaluation program package
v4.1 using a
flow cell without captured antibody as reference and the 1:1 Langmuir binding
model with
mass transfer.
Daratumumab competition binding assay
[0319] For Daratumumab competition binding assay, Daratumumab was amine
coupled
to the active surface of CMS chip. Reference surface was left blank and used
to subtract any
non-specific binding of injected molecules. Recombinant CD38-His (Sino
Biological, Part#
10818-H08H) was injected over the Daratumumab surface followed by injection of
test
antibodies. If a monospecific anti-CD38 antibody recognized an epitope on CD38
which was
different from that of Daratumumab, injection of the antibody resulted in an
increased SPR
signal. If an antibody recognized an overlapping epitope as Daratumumab,
injection of the
antibody did not increase SPR signal.
Results
[0320] The binding affinities of selected CD38/CD28sup x CD3mid ENLQ DKTHT
IgG4 FALA trispecific antibodies with alternative anti-CD38 binding domains
for human
CD38 were determined by SPR. The association rate constant (Kon), dissociation
rate
constant (Koff), and the KD of the selected trispecific antibodies are
provided in Table A. The
selected trispecific antibodies showed various degrees of affinities against
human CD38
antigen.
195
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
Table A. Binding characteristics of selected CD38/CD28sup x CD3mid ENLQ DKTHT
IgG4 FALA trispecific antibodies with alternative anti-CD38 binding domains
for human
CD38 determined by SPR.
Anti-CD38 binding domain kon (M's') koff (0) KD (M)
CD38VH1 5.55E+05 1.58E-03
2.85E-09
CD38hhy992 1.35E+06 1.75E-04
1.29E-10
CD38hyb6284 7.85E+05 5.12E-04
6.52E-10
CD38hyb5739 9.80E+05 5.46E-03
5.57E-09
CD38hhy1195 1.27E+06 1.80E-02
1.42E-08
CD38hhy1370 3.76E+05 3.29E-04
8.76E-10
[0321] The
binding affinities of selected CD38/CD28sup x CD3mid ENLQ DKTHT
IgG4 FALA trispecific antibodies with alternative anti-CD38 binding domains
for human
CD3, human CD28, human CD38 and cynomolgus monkey CD38 were then determined by
ELISA as described above. As shown in FIGS. 2B-2E, the selected CD38/CD28sup x
CD3mid ENLQ DKTHT IgG4 FALA trispecific antibodies with alternative anti-CD38
binding domains showed various affinities to human (FIG. 2B) and cynomolgus
monkey
CD38 (FIG. 2C), but similar affinity to human CD3 (FIG. 2D) and CD28 (FIG.
2E). EC50
values were then calculated by GraphPad Prism 7.02 using variable slope model
with four-
parameter logistic curve. The EC50 values of the selected trispecific
antibodies for human
CD3, human CD28, human CD38 and cynomolgus monkey CD38 are provided in Table
B.
Control antibody was a human IgG4 isotype control.
Table B. EC50 values of selected CD38/CD28sup x CD3mid ENLQ DKTHT IgG4 FALA
trispecific antibodies with alternative anti-CD38 binding domains for human
CD3, human
CD28, human CD38 and cynomolgus monkey CD38.
E
Anti-CD38 C50 (pM)
binding
domain hCD38 Cyno CD38 hCD3 hCD28
CD38VH1 7244 2128 6742 725
196
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
CD38hhy992 229 912 33593 809
CD38hyb6284 253 290 8222 711
CD38hyb5739 984 1628 10791 825
CD38hhy1195 102537 37701 7631 697
CD38hhy1370 587 553 24968 1257
[0322] An SPR
competition assay was carried out to determine whether anti-CD38
antibodies hhy6284, hhy992, hhy5379, or hhy1195 (tested in monospecific
antibody format)
compete with Daratumumab for binding to CD38. Following CD38 injection over
Daratumumab (immobilized on SPR sensor chip), the test antibodies (or
Daratumumab) were
injected over the Daratumumab/CD38 complex. As shown in FIG. 3, injection of
Hyb6264,
hhy992, Hyb5379, and Hhy1195 increased SPR signal, indicating that these
antibodies
recognized the epitopes on CD38 which are different from the epitope which
Daratumumab
recognizes. As expected, injection of free Daratumumab (a competitive binding
control) did
not increase the SPR signal.
[0323]
Binding of anti-CD38 antibodies to human or cynomolgus CD38 polypeptides is
summarized in Table B2.
Table B2. Summary of anti-CD38 binding characteristics to human or cynomolgus
CD38.
ELISA ELISA FACS FACS SPR SPR
Name huCD38 cynoCD38 huCD38 cynoCD38 huCD38 cynoCD38
EC50 nM EC50 nM EC50 nM EC50 nM KD M KD M
0
AntiCD38_hyb_5739 .12 0.09 0.3 0.5
AntiCD38_hyb_6284 an 0.13 0.4 0.7
AntiCD38_hhy_992 0.09 0.08 100 288 3.65E-10 6.12E-09
AntiCD38_hhy_1195 1.4 0.86 38 15
4.00E-08 2.60E-08
[0324] Anti-CD38 antibodies were also tested for competitive binding to
daratumumab in
SPR assay. For daratumumab competition binding assay, daratumumab was amine
coupled to
the active surface of CMS chip. Reference surface was left blank and used to
subtract any
non-specific binding of injected molecules. Recombinant CD38-His (Sino
Biological, Part#
10818-H08H) was injected over the daratumumab surface followed by injection of
test
antibodies. If an antibody recognizes an epitope on CD38 which is different
from that of
daratumumab, injection of the antibody will result in an increased SPR signal.
If an antibody
recognizes an overlapping epitope as daratumumab, injection of the antibody
will not
197
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
increase SPR signal. According to the results of these assays the tested
antibodies hhy992,
hyb6284, hhy1195 and hhy1370 did not compete with daratumumab.
Example 3: Trispecific CD38/CD3xCD28 antibodies promote lysis of human
multiple
myeloma and lymphoma tumor cells.
[0325] An in vitro cell lysis assay was used to determine whether
trispecific
CD38/CD3xCD28 antibodies had anti-tumor cell activity using human multiple
myeloma and
lymphoma cells.
Materials and Methods
In vitro killing assay against tumor cells using human T cells
[0326] Target tumor cells were labeled with the membrane dye PKH-26 (Sigma)
and co-
cultured for 24 hours with human PBMC or enriched CD8 T cells as effector
cells at E:T
ratio of 10:1(E:T=3:1 using enriched CD8 T cells) in the presence of indicated
concentrations
of tri-specific or relevant control antibodies. Peripheral blood mononuclear
cells were
isolated from normal human donors by Ficoll separation, and autologous CD8+ or
pan¨T
cells were enriched using kits from Miltenyi Biotech (San Diego, CA). The
extent of cell
lysis in the target cells was determined by staining with a LIVE/DEADTM
Fixable Violet
Dead Cell Stain Kit (Life Technologies) and measured by the number of dead
cells in the
labelled target cell population by running the samples on an LSRFortessa
instrument (BD
Biosciences) followed by analysis using the Flowjo software (Treestar).
In vitro killing assay against tumor cells using human T cells in the presence
of
Daratumumab
[0327] 5 nM Daratumumab or isotype control antibodies were pre-incubated
with PKH-
26 labeled target tumor cells (105 cells/well) for 30 minutes, followed by
addition of
trispecific TCEs at indicated concentrations, and human PBMCs (E:T=10:1). 24
hours later,
the extent of cell lysis in the target cells was determined by staining with a
LIVE/DEADTM
Fixable Violet Dead Cell Stain Kit (Life Technologies) and measured by the
number of dead
cells in the labelled target cell population by running the samples on an
LSRFortessa
instrument (BD Biosciences) followed by analysis using the Flowjo software
(Treestar).
Results
[0328] The in vitro cell killing activity of CD38/CD28sup x CD3mid ENLQ
DKTHT
IgG4 FALA trispecific antibodies with alternative anti-CD38 binding domains
was
determined using a human multiple myeloma cell line NCI-H929 that expresses
both CD38
198
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
and CD28. The assay was carried out in the presence of 5nM Daratumumab or
isotype
control antibodies (present during the assay period). As shown in FIGS. 4A-4B,
all tested
trispecific antibodies led to cell lysis in a concentration-dependent manner
in the presence
and absence of Daratumumab. The EC50 values were then calculated in the
presence and
absence of Daratumumab (Table C). The cell killing activities of trispecific
antibodies
CD38/CD28sup x CD3mid ENLQ DKTHT IgG4 FALA with the CD38VH1 or
CD38hhy1370 anti-CD38 binding domains were reduced by Daratumumab, while
trispecific
antibodies with the CD38hyb5739, CD38hyb6284, or CD38hhy1195 anti-CD38 binding
domains exhibited between 3-8 fold reductions in cell killing activity in the
presence of
Daratumumab (Table C).
Table C. In vitro killing activity against human multiple myeloma cell line
NCI-H929
(CD38+/CD28+) by CD38/CD28sup x CD3mid ENLQ DKTHT IgG4 FALA trispecific
antibodies with alternative anti-CD38 binding domains in the presence of
Daratumumab.
Anti-CD38 binding domains
EC50 CD38VH CD38hhy99 CD38hyb573 CD38hyb628 CD38hhy119 CD38hhy137
(PM) 1 2 9 4 5
With
29.82 125.8 9.115 33.65 89.27 255.4
Dara
With
huma
1.063 13.43 2.736 4.37 16.97 9.599
IgG1
[0329] In addition, an in vitro cell lysis assay was used to
measure the cell killing activity
of selected CD38/CD28sup x CD3mid ENLQ DKTHT IgG4 FALA trispecific antibodies
with alternative anti-CD38 binding domains using a human lymphoma cell line
OCI-LY19
that expresses CD38 but not CD28. The assay was carried out in the presence of
5nM
Daratumumab or isotype control antibodies which were present in the assay
period. As shown
in FIGS. 5A-5B, all tested trispecific led to cell lysis in a concentration-
dependent manner in
the presence and absence of Daratumumab. The EC50 values were then calculated
in the
presence and absence of Daratumumab (Table D). The cell killing activity of
199
Date Recue/Date Received 2021-10-04
WO 2020/210392
PCT/US2020/027320
CD38/CD28sup x CD3mid ENLQ DKTHT IgG4 FALA trispecific antibodies with
CD38VH1 anti-CD38 binding domain was reduced by about 24 fold by Daratumumab,
while
trispecific antibodies with the CD38hhy992, CD38hyb5739, CD38hyb6284,
CD38hhy1195,
or CD38hhy1370 anti-CD38 binding domains also exhibited reductions in cell
killing activity
in the presence of Daratumumab (Table D).
Table D. In vitro killing activity against human lymphoma cell line OCI-LY19
(CD38+/CD28-) by selected CD38/CD28sup x CD3mid ENLQ DKTHT IgG4 FALA
trispecific antibodies with alternative anti-CD38 binding domains in the
presence of
Daratumumab.
Anti-CD38 binding domains
EC50 CD38V CD38hhy9 CD38hyb57 CD38hyb62 CD38hhyll CD38hhy13
(PM) H1 92 39 84 95 70
With
135.9 133.3 219.1 81.05 715.2 209.8
Dara
With
huma
5.662 57.32 60.97 42.07 296.4 58.54
IgG1
Example 4: CD38/CD28xCD3 trispecific antibodies promote CMV-specific immune
response
[0330] As
part of adaptive immunity, T cell immunity plays a crucial role in controlling
viral infection and cancer, possibly eliminating infected cells and malignant
cells which result
in clearance of viral infection or cure of cancer. In chronic infectious
diseases such as Herpes
viral infection (HSV, CMV, EBV, etc.), HIV, and HBV viruses establish their
persistence in
humans by various mechanisms including immune suppression, T cell exhaustion,
and
latency establishment. Nevertheless, viral infection generally induces viral
antigen specific
immunity including antigen specific CD8 T cells that can readily recognize
infected cells for
controlling or killing through cytokine release or cytotoxic T cell (CTL)
mediated killing
200
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
processes. Thus, viral antigen specific T cell activation and/or amplification
in vivo and/or ex
vivo provide therapeutic strategies against chronic viral infections.
[0331] Anti-CD38/CD28xCD3 trispecific antibodies were developed and
evaluated for
their potential in activating T cells, and promoting proliferation and/or
amplification of
antigen specific T cells. These trispecific Abs can effectively expand CD4 and
CD8 effector
and memory populations, including antigen specific CD8 T central memory and
effector
memory cells in vitro. Specifically, in vitro expansion of CMV and EBV
specific CD8 central
memory and effector memory cells were demonstrated. The anti-CD38/CD28xCD3
trispecific antibodies described herein exhibited novel properties by engaging
CD3/CD28/CD38, providing signaling pathways to stimulate and expand T cells,
which may
offer an effective strategy treating chronic infectious diseases such as HSV,
CMV, EBV,
HIV-1, and HBV infections.
[0332] In this Example, the ability of CD38/CD28xCD3 trispecific antibodies
to promote
activation and expansion of CMV-specific T cells was determined.
Materials and Methods
In vitro T cell proliferation measurement
[0333] T cells were isolated from human PBMC donors by negative selection
using a
magnetic Pan T Cell Isolation Kit (Miltenyi Biotec GmbH, Germany). Antibodies
were
coated onto 96-well cell culture plates by preparing the antibodies in sterile
PBS and
dispensing 50 [EL into each well (350 ng/well). The plates were then incubated
at 37 C for at
least 2 hours and then washed with sterile PBS. The untouched T cells were
added to the
antibody-coated plates (5 x 105 cells/mL) and incubated at 37 C for multiple
days. The cells
were passaged with new cell culture media onto fresh antibody-coated plates on
Day 4. In
certain experiments with 7 days incubation, only fresh medium was added
without changing
to fresh antibody-coated plate. The cells were collected at specific time
points and cell
numbers calculated using CountBrightTM counting beads.
In vitro T cell proliferation assay and T cell subset determination
[0334] Peripheral blood mononuclear cells were isolated from blood of
healthy human
donors collected by Research Blood Components, LLC (Boston, MA). The PBMCs
were
added to antibody-coated plates (350 ng/well) (5 x 105 cells/mL), as
previously described
above, and incubated at 37 C for 3 and 7days. The cells were collected at
specific time points
and analyzed by flow cytometry for T cell subsets: naive (CCR7+ CD45R0-), Tcm
(CCR7+
201
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
CD45R0+), Tern (CCR7- CD45R0+), Tregs (CD4+ Foxp3+ CD25hi). CMV pp65-specific
and EBV BMLF-specific CD8+ T cells were detected using fluorescent-conjugated
pentamer
restricted to the PBMC donors' HLA/viral peptide (A*02:01/NLVPMVATV, SEQ ID
NO:284), (A*02:01/GLCTLVAML, SEQ ID NO:285), respectively (ProImmune, Oxford,
UK). PBMC was obtained from HemaCare (Van Nuys, CA) for donors with known CMV
or
EBV infection. PMBC from donors negative for the restricting HLA type was used
as
negative control. Staining was done as per manufacturer's protocol.
Quantification of CMV-specific T-Cells
[0335] As indicated above Peripheral blood mononuclear cells (PBMCs) were
isolated
from blood of known CMV-infected human donors and added to plates containing
the
trispecific antibody or control antibody. The plates were incubated at 37 C.
The cells were
collected at specific time points and analyzed by flow cytometry.
Results
[0336] CD38/CD28sup x CD3mid ENLQ DKTHT IgG4 FALA trispecific antibodies
with alternative anti-CD38 binding domains AVH1CD38 (control), CD38VH1,
CD38hhy992,
CD38hyb5739, CD38hyb6284, CD38hhy1195, and CD38hhy1370 were tested as
described
above using PBMCs isolated from CMV-infected human donor D (FIGS. 6A-6J) and
CMV-
infected human donor E (FIGS. 7A-7J). All tested CD38 trispecific Abs
activated and
promoted the proliferation of CMV-specific T cells, leading to increases in
CMV-specific
CD8+ T cells (cells/well) with different potency and kinetics in a dose
response manner over
the 7 day experiment (CMV Donor D, FIGS. 6A-6B; CMV Donor E, FIGS. 7A-7B). In
addition, all tested CD38 trispecific Abs promoted the amplification
(cells/well) of CMV-
specific central memory (Tcm) (CMV Donor D, FIGS. 6C-6D; CMV Donor E, FIGS. 7C-
7D) and effector memory (Tern) CD8+ T cells (CMV Donor D, FIGS. 6E-6F; CMV
Donor E,
FIGS. 7E-7F), which were both amplified dramatically in 7 days. FIGS. 6G-6J
(CMV
Donor D) and FIGS. 7G-7J (CMV Donor E) provide time courses showing the
percent of
CMV-specific Tun and Tern cells at days 0, 3, and 7 of the 7-day experiments
described above.
[0337] Taken together, these data indicate that CD38/CD28xCD3 trispecific
antibodies
promote activation and expansion of CMV-specific T cells, such as CMV-specific
CD8+ T
cells, CMV-specific effector memory (Tern) CD8+ T cells, and CMV-specific
central memory
(Tun) CD8+ T cells.
202
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
Example 5: CD38/CD28xCD3 trispecific antibodies promote EBV-specific immune
response
[0338] Next, the ability of CD38/CD28xCD3 trispecific antibodies to promote
activation
and expansion of Epstein-Barr virus (EBV)-specific T cells was determined.
Materials and Methods
Quantification of EBV-specific T-Cells
[0339] As indicated above, peripheral blood mononuclear cells (PBMCs) were
isolated
from blood of known EBV-infected human donors and added to plates containing
the
trispecific antibody or control antibody. The plates were incubated at 37 C
for up to 11 days.
The cells were collected at specific time points and analyzed by flow
cytometry.
Results
[0340] CD38/CD28sup x CD3mid ENLQ DKTHT IgG4 FALA trispecific antibodies
with alternative anti-CD38 binding domains AVH1CD38 (control), CD38VH1,
CD38hhy992,
CD38hyb5739, CD38hyb6284, CD38hhy1195, and CD38hhy1370 were also tested as
described above using PBMCs isolated from EBV-infected donor C (FIGS. 8A-8J)
and
EBV-infected donor D (FIGS. 9A-12). All tested CD38 trispecific Abs activated
T cells and
promoted the proliferation of EBV-specific T cells, leading to increases in
EBV-specific
CD8+ T cells (cells/well) with different potency and kinetics in a dose
response manner over
the 7 day experiment (EBV Donor C, FIGS. 8A-8B; EBV Donor D, FIGS. 9A-9B). In
addition, all tested CD38 trispecific Abs promoted the amplification
(cells/well) of EBV-
specific central memory (Tcm) (EBV Donor C, FIGS. 8C-8D; EBV Donor D, FIGS. 9C-
9D)
and effector memory (Tern) CD8+ T cells (EBV Donor C, FIGS.8E-8F; EBV Donor D,
FIGS. 9E-9F), which were both amplified dramatically in 7 days. FIGS. 8G-8J
(EBV Donor
C) and FIGS. 9G-12 (EBV Donor D) provide time courses showing the percent of
EBV-
specific Tcm and Tern cells at days 0, 3, and 7 of the 7-day experiments
described above.
[0341] Taken together, these data indicate that CD38/CD28xCD3 trispecific
antibodies
promote activation and expansion of EBV-specific T cells, such as EBV-specific
CD8+ T
cells, EBV-specific effector memory (Tern) CD8+ T cells, and EBV-specific
central memory
(Tern) CD8+ T cells.
203
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
Example 6: Anti-tumor effects of Her2/CD28 x CD3 trispecific antibody in tumor-
bearing
mice
[0342] In this Example, the Her2/CD28 x CD3 trispecific antibody was tested
for anti-
tumor effects in a ZR-75-1 tumor bearing Nod scid gamma (NSG) mouse model
engrafted
with in vitro expanded T cells.
Materials and Methods
[0343] NSG mice were divided into 5 groups of 10 mice each. On Day 0, ZR-75-
1
human breast cancer cells were implanted into the mammary fat pad with 50%
matrigel into
each mouse at 5 million cells/mouse. On Days 17/18, expansion of human CD3+ T
cells was
begun. Randomization of mice occurred on Day 24 when tumors were approximately
150mm3. On Day 25, all mice were engrafted with in vitro expanded human CD3+ T
cells at
million cells in 300pL/mouse (1QW, 1 IP injection).
[0344] Starting on Day 25, one group of mice received doses of vehicle
alone (8% w/v
sucrose, 0.05% w/v polysorbate 80, 10mM histidine, pH 5.5), while the other 4
groups
received Her2/CD28 x CD3 trispecific antibody, both at 10mL/kg. Groups
receiving
trispecific antibody were dosed at 100, 10, 1, or 0.1 [tg/kg. Antibody or
vehicle was
administered 1QW intravenously in 2 doses (e.g., Days 25 and 32). Blood and
tumor tissue
was collected on Day 38 or 39.
Results
[0345] Her2/CD28 x CD3 trispecific antibody (binding protein #2 from Table
1,
corresponding to SEQ ID Nos:104-107) was compared to vehicle control for its
effects on
human breast tumor growth in the NSG mouse model engrafted with in vitro
expanded
human T cells described above. Treatment with Her2/CD28 x CD3 trispecific
antibody at the
highest dose (10Oug/kg) led to the most significant inhibition of tumor growth
and regression,
although the bug/kg dose also showed anti-tumor effects (FIGS. 13A & 13D). No
significant body weight loss was observed (FIG. 13B). Individual tumor volumes
over time
from each trispecific antibody treatment group are provided in FIG. 13C.
[0346] Next, the effect of trispecific antibody treatment on individual
immune cell
subsets was examined. Human CD45+, human CD8+, and human CD4+ cell populations
were measured by flow cytometry, as well as mouse CD45+ cells (FIGS. 14A-14C).
Highest
dose (10Oug/kg) of trispecific antibody led to depletion of human CD4+ cells,
and this effect
was dose dependent (FIGS. 14B & 14C). Counts of human CD8+ cells were largely
unaffected by trispecific antibody administration.
204
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
[0347] The effect of trispecific antibody treatment on tumor infiltrating
lymphocytes
(TILs) was also assessed by immunohistochemical (IHC) staining for human CD45,
CD4,
and CD8. Using H&E staining, tumors from the low dose groups (lug/kg or 0.
lug/kg
trispecific antibody) were generally of comparable size as the vehicle control
group. As
shown in FIGS. 15A-15C, human TILs were increased in the group receiving the
low dose of
Her2/CD28 x CD3 trispecific antibody, but human TILs were sparse in the high
dose group.
IHC images were also examined quantitatively (FIGS. 16A-16C). These results
indicated
significant reductions in CD45+ and CD8+ cells in the higher trispecific
antibody dose
groups (10Oug/kg and bug/kg).
[0348] Compared to vehicle control, tumors from the high dose trispecific
antibody
treatment groups (10Oug/kg or bug/kg) were characterized by sparse TILs.
Moderate to
large numbers of CD45+, CD4+, or CD8+ human TILs were observed in the lug/kg
and
0. lug/kg trispecific antibody treatment groups. These TILs were mostly
present at the tumor
edges but occasionally extended deeper into the tumor core.
[0349] In conclusion, these results demonstrate that treatment of ZR-75-1
breast tumor
bearing NSG mice engrafted with in vitro activated T cells using 2 intravenous
doses of
HER2-targeting, T cell-engaging trispecific antibody at 10Oug/kg or bug/kg
resulted in
significant reductions in tumor volume and, concomitantly, a significant
decrease in TILs. At
the lug/kg trispecific antibody dose, there was a marginal and inconsistent
trend for increased
TILs as compared to vehicle control.
Example 7: Effect of anti-HER2 and anti-CD3 antigen binding domain sequences
in
Her2/CD28 x CD3 trispecific antibody on cancer cell killing
[0350] This Example describes the effect of anti-Her2 and anti-CD3 variable
domain
sequences on target cell killing. In this Example, a Her2/CD28 x CD3
trispecific antibody
("control") with wild-type trastuzumab antigen binding domain and an anti-CD3
antigen
binding domain without 32/35 QQ mutations in the VL domain (see Example 1) was
compared with Her2/CD28 x CD3 trispecific antibodies #1-6 from Table 1,
corresponding to
SEQ ID Nos:100-103, 104-107, 286-289, 290-293, 294-297, and 298-301,
respectively.
Materials and Methods
[0351] CD8+ T cells were isolated from human PBMCs from healthy donor using
a
magnetic bead isolation kit (Miltenyi Biotec). The T cells were used as
effector cells against
breast cancer cell lines expressing various levels of HER2 at 3:1
(Effector:Target) ratio. The
205
Date Recue/Date Received 2021-10-04
WO 2020/210392 PCT/US2020/027320
cells were incubated with experimental or control trispecific antibody for 2
days before flow
cytometry acquisition using viability dye (Invitrogen) and PKH26 target cell
staining
(Sigma). Mean EC50 for target cell lysis was calculated from 2-3 PBMC donors
for each
trispecific Ab.
Results
[0352] All trispecific antibodies were characterized for in vitro cell
lysis of three HER2+
breast cancer target cell lines: HCC1954, BT20, and MDA-MB-231. HCC1954 breast
cancer
cells were found to express high levels of HER2, as assessed by flow cytometry
(up to
¨150,000 receptors/cell), IHC (3+), or the HercepTest HER2 expression assay
(3+) (FIG.
17A). BT20 breast cancer cells were found to express intermediate levels of
HER2, as
assessed by flow cytometry (-60,000 receptors/cell), IHC (1+), or the
HercepTest HER2
expression assay (1+) (FIG. 17C). MDA-MD-231 breast cancer cells were found to
express
low levels of HER2, as assessed by flow cytometry (-9,000 receptors/cell), IHC
(0+), or the
HercepTest HER2 expression assay (0) (FIG. 17E). Results of the cell killing
assays
targeting HCC1954, BT20, or MDA-MB-231 are shown in FIGS. 17B, 17D, and 17F,
respectively, comparing binding protein #2 vs. control or binding proteins #1
and #5 vs.
control. The results demonstrated that the Her2/CD28 x CD3 trispecific
antibodies having
30R/55Q/102E mutations in the anti-HER2 arm and 32/35 QQ mutations in the VL
domain
of the anti-CD3 arm showed improved target cell killing against all three cell
lines,
particularly at lower antibody concentrations.
[0353] Mean EC50 (pM) for in vitro cell killing was determined for all
trispecific
antibodies targeting the three breast cancer cell lines noted above (HCC1954,
BT20, and
MDA-MB-231) as well as the gastric cancer cell lines 0E19 (high HER2
expression) and
GSU (intermediate HER2 expression). Generally, the Her2/CD28 x CD3 trispecific
antibodies having mutations in the anti-HER2 arm and in the VL domain of the
anti-CD3 arm
showed a lower EC50 (and thus superior cell killing) against all three breast
cancer cell lines
(FIG. 18A) and both gastric cancer cell lines (FIG. 18B). These results
demonstrate that,
while all trispecific antibodies are able to induce cell killing of EIER2+
cells, the mutated
trispecific antibodies consistently displayed improved cell killing efficacy
against multiple
target cell types.
206
Date Recue/Date Received 2021-10-04