Note: Descriptions are shown in the official language in which they were submitted.
WO 2020/252208
PCT/U52020/037312
MACROPHAGE SPECIFIC ENGAGER COMPOSITIONS AND METHODS OF USE
THEREOF
CROSS REFERNCE
[0001] This application claims the benefit of U.S. Provisional Application No,
62/860,055, filed on June
11, 2019, and U.S. Provisional Application No. 62/908,978, filed on October 1,
2019, each of which is
incorporated herein by reference in its entirety.
BACKGROUND
100021 Cellular immunotherapy is a promising new technology
for fighting difficult to treat diseases,
such as cancer, persistent infections and diseases that are refractory to
other forms of treatment.
Macrophages represent the dominant cell type present in a tumor or an
infection site and possess several
strategic advantages such that they can be potentially utilized to treat the
disease most effectively. As
natural sentinels of the immune system, these cells can sense and eliminate
aberrant and non-healthy cell
types, including cancer cells. However, potential use of macrophages for
irrununotherapy has not been fully
explored. Newer avenues are sought for using these cell types towards
development of improved
therapeutics, including but not limited to T cell malignancies.
SUMMARY
[0003] The present disclosure relates to new compositions and
methods that initiate a target cell
destruction pathway through phagocytosis. This application is based on an
unexpected finding that when a
phagocytic receptor is triggered with at least a second concurrent or
subsequent activation signal in
addition to binding to its classical ligand, the second or additional
signal(s) can lead to efficient
destruction of a target cell by phagocytosis. Presented herein are chimeric
receptors, and chimeric
receptor-binding extracellular elements designed for enhancing phagocytosis of
a cell, such as a myeloid
cell, or a monocyte or macrophage. Careful design and/or manipulation of the
at least second concurrent
or subsequent signal is useful for successful activation of a chimeric
phagocytic receptor such as those
described herein, such that the target cell is effectively destroyed
thereafter. For example, the first signal
(signal 1) can be mediated via phagocytosis/tethering receptors and the second
signal (signal 2) can by
mediated danger signals such as pathogen-associated molecular patterns
(DAMPs), or cytokines that
trigger nuclear factor-ic.B (NF-x11)-mediated upregulation of inflammatory
genes. As described in the
following section, triggering phagocytosis alone may be insufficient to
activate monocytes or
macrophages in the context of harnessing the phagocytic ability of monocytes
or macrophages to kill
cancer cells, and to drive an effective anti-tumor response.
[0004] One of the specific advantages of the inventions
described here is that the compositions for
effective cellular inununotherapy disclosed herein are cost-effective and
efficient.
-1-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[0005] In some aspects, provided herein are new chimeric cell
surface binding elements or
"engagers" that bind to an extracellular portion of a chimeric phagocytic
receptor, and bind additionally to
at least a cell surface component on a target cell such as a cancer cell.
[0006] In one embodiment, the new chimeric engagers can bind
to an extracellular portion of a
chimeric phagocytic receptor, and additionally bind to one or more cell
surface components, at least one
of which is on a target cancer cell. Accordingly, an engager may be a bi-
specific monocyte or macrophage
engager (BiME) and have two binding portions, wherein one binding portion
binds to an extracellular
portion of a chimeric phagocytic receptor, and the other binds to the cell
surface component on a target
cell. Likewise, an engager may be a trispecific monocyte or macrophage engager
(TriME) and have three
binding portions, wherein one binding portion binds to an extracellular
portion of a chimeric phagocytic
receptor, another binding portion binds to the cell surface component on a
target cell and the third binding
portion binds to the cell surface component on the phagocytic cell.
[0007] In one aspect, the engager is a synthetic protein or a
peptide, a conjugated protein or
conjugated peptide. Provided herein is a composition comprising: a first
therapeutic agent, wherein the
therapeutic agent comprises: (a) a first binding domain, wherein the first
binding domain is a first
antibody or functional fragment thereof that specifically interacts with an
antigen of a target cell, and (b)
a second binding domain, wherein the second binding domain is a second
antibody or functional fragment
thereof that specifically interacts with a myeloid cell; wherein, (i) the
first therapeutic agent is coupled to
a first component, wherein the first component is an additional therapeutic
agent or a third binding
domain, or (ii) the composition comprises an additional therapeutic agent.
[0008] In one aspect, provided herein is a composition
comprising: a therapeutic agent, wherein the
therapeutic agent is an engager that comprises: (a) a first binding domain
that specifically interacts with
an antigen of a target cell, (b) a second binding domain that specifically
interacts with a myeloid cell, and
(c) a third binding domain that specifically interacts with the myeloid cell.
[0009] In one aspect, provided herein is a composition
comprising: a therapeutic agent, wherein the
therapeutic agent is an engager that comprises: (a) a first binding domain
that specifically interacts with an
antigen of a target cell, (b) a second binding domain, wherein the second
binding domain: (i) specifically
interacts with a myeloid cell (e.g., a monocyte or macrophage, or a dendritic
cell) and promotes
phagocytosis activity of the myeloid cell, or, (ii) specifically interacts
with a myeloid cell and promotes
inflammatory signaling of the myeloid cell, or (iii) specifically interacts
with a myeloid cell or an
adhesion molecule and promotes adhesion of the myeloid cell to the target
cell, and (c) a third binding
domain, wherein the third binding domain (i) specifically interacts with the
myeloid cell and promotes
phagocytic activity of the myeloid cell, or, (ii) specifically interacts with
the myeloid cell and promotes
inflammatory signaling of the myeloid cell, or, (iii) specifically interacts
with the myeloid cell and
promotes adhesion of the myeloid cell to the target cell, or, (iv)
specifically interacts with the myeloid cell
-2-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
and inhibits anti-phagocytic activity of the myeloid cell mediated by the
target cell, or (v) specifically
interacts with the myeloid cell and inhibits anti-inflammatory activity of the
myeloid cell mediated by the
target cell.
[0010] In some embodiments, the myeloid cell is a monocyte or
macrophage cell.
[0011] In some embodiments, the target cell is a cancer cell.
[0012] In some embodiments, the second binding domain that
specifically interacts with a myeloid
cell interacts with a phagocytic or tethering receptor of the myeloid cell or
monocyte or macrophage cell.
[0013] In some embodiments, the third binding domain that
specifically interacts with a myeloid cell
interacts with an extracellular region of a first phagocytic or tethering
receptor of the myeloid cell or
monocyte or macrophage cell.
[0014] The composition of any one of the preceding claims
wherein any one of binding domains of
the therapeutic agent comprises the binding domain of an antibody, a
functional fragment of an antibody,
a variable domain thereof, a Vii domain, a VI, domain, a VNAR domain, a Vial
domain, a single chain
variable fragment (scFv), an Fab, a single-domain antibody (sdAb), a nanobody,
a bispecific antibody, a
diabody, or a functional fragment or a combination thereof.
[0015] In some embodiments, the therapeutic agent is a
recombinant protein or more than one
recombinant proteins.
[0016] In some embodiments, the therapeutic agent comprises
recombinant proteins comprising one
or more fusion proteins.
[0017] In some embodiments, the therapeutic agent is a
recombinant protein comprising an antibody,
a functional fragment of an antibody, a variable domain thereof, a VH domain,
a Vi. domain, a VNAR
domain, a Vim domain, a single chain variable fragment (scFv), an Fab, a
single-domain antibody (sdAb),
a nanobody, a bispecific antibody, a diabody, or a functional fragment or a
combination thereof In some
embodiments, the therapeutic agent is a recombinant protein or more than one
recombinant proteins, each
comprising multiple binding fragments, each binding fragment constituting a
functional fragment of an
antibody, a variable domain thereof, a VH domain, a VI, domain, a VNAR domain,
a VHH domain, a single
chain variable fragment (scFv), an Fab, a single-domain antibody (sdAb), a
nanobody, a bispecific
antibody, a diabody, or a functional fragment or a combination thereof
[0018] In some embodiments, the therapeutic agent is a
recombinant protein (the engager) comprising
multiple binding domains, each having individual binding specificities, that
are each linked together by
linkers (e.g., peptide linkers) that exhibit complementary binding with each
other. For example, one
binding domain of the recombinant protein is fused with the first of a pair of
linker peptides, and the other
binding domain is fused with the second of the pair of linker peptides,
wherein, the pair of linker peptides
exhibit complementary binding with each other, wherein the pair of linker
peptides comprise: (a) leucine
zipper domains that exhibit complementary binding with each other, for
example, leucine zippers in
-3-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
naturally occurring protein-protein interactions, such as the zipper sequences
within the binding regions of
c-Fos and c-Jun proteins, (b) synthetic peptides designed to specifically bind
to each other via designed
affinities, such as synthetic clasps.
[0019] In some embodiments, the therapeutic agent is a
recombinant protein comprising multiple
binding fragments configured to facilitate accelerated association with each
other by means of leucine
zipper peptide pairs comprised in the recombinant proteins.
[0020] In some embodiments, the therapeutic agent is a
recombinant protein comprising multiple
binding fragments configured to facilitate accelerated association with each
other by means of c-Fos/c-Jun
binding domains in the peptide pairs comprised within the recombinant
proteins.
[0021] In some embodiments, the therapeutic agent is a
recombinant protein comprising multiple
binding fragments configured to facilitate accelerated association with each
other by means of synthetic
clasps.
[0022] In some embodiments, the antigen on the target cell to
which the first binding domain binds, is
a cancer antigen or a pathogenic antigen on the target cell or an autoimmune
antigen.
[0023] In some embodiments, the antigen on the target cell to
which the first binding domain binds, is
a viral antigen
[0024] In some embodiments, the antigen on the target cell to
which the first binding domain binds is
a T-lymphocyte antigen.
[0025] In some embodiments, the antigen on the target cell to
which the first binding domain binds is
an extracellular antigen.
[0026] In some embodiments, the antigen on the target cell to
which the first binding domain binds is
an intracellular antigen.
[0027] In some embodiments, the antigen on the target cell to
which the first binding domain binds is
selected from the group consisting of Thymidine Kinase (TK1), Hypoxanthine-
Guanine
Phosphoribosyltransferase (HPRT), Receptor Tyrosine Kinase-Like Orphan
Receptor 1 (ROR1), Mucin-
1, Mucin-16 (MUC16), MUC1, Epidermal Growth Factor Receptor An (EGFRvIC),
Mesothelin, Human
Epidermal Growth Factor Receptor 2 (HER2), Mesothelin, EBNA-1, LEMD1,
Phosphatidyl Serine,
Carcinoembryonic Antigen (CEA), B-Cell Maturation Antigen (BCMA), Glypican 3
(GPC3), Follicular
Stimulating Hormone receptor, Fibroblast Activation Protein (FAP),
Erythropoietin-Producing
Hepatocellular Carcinoma A2 (EphA2), EphB2, a Natural Killer Group 2D (NKG2D)
ligand,
Disialoganglioside 2 (GD2), CD2, CD3, CD4, CD5, CD7, CD8, CD19, CD20, CD22,
CD24, CD30,
CD33, CD38, CD44v6, CD45, CD56CD79b, CD97, CD117, CD123, CD133, CD138, CD171,
CD179a,
CD213A2, CD248, CD276, PSCA, CS-1, CLECL1, GD3, PSMA, FLT3, TAG72, EPCAM,M-1,
an
integrin receptor, PRSS21, VEGFR2, PDGFR-beta, SSEA-4, EGFR, NCAM, prostase,
PAP, ELF2M,
GM3, TEM7R, CLDN6, TSHR, GPRC5D, ALE, IGLL1 and combinations thereof
-4-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[0028] In some embodiments, the antigen on the target cell to
which the first binding domain binds is
selected from the group consisting of CD2, CD3, CD4, CD5, CD7, CCR4, CD8,
CD30, CD45, CD56.
[0029] In some embodiments, the antigen on the target cell to
which the first binding domain binds is
an ovarian cancer antigen or a T lymphoma antigen.
[0030] In some embodiments, the antigen on the target cell to
which the first binding domain binds is
an integrin receptor.
[0031] In some embodiments, the second binding domain or the
third binding domain binds to an
integrin receptor.
[0032] In some embodiments, the second binding domain or the
third binding domain binds to an
integrin receptor selected from the group consisting of al, a2, allb, a3, a4,
a5, a6, a7, a8, a9, al , all,
aD, aE, aL, aM, aV, aX, 13 1, (32, (3 3,13 4,13 5,13 6,13 7, and 138.
[0033] In some embodiments, the therapeutic agent binds to a
phagocytic or tethering receptor that
comprises a phagocytosis activation domain.
[0034] In some embodiments, the therapeutic agent binds to a
receptor or a protein selected from the
group consisting the receptors listed in Table 2A and Table 2B, or a fragment
thereof
[0035] In some embodiments, the therapeutic agent binds to a
phagocytic receptor selected from the
group consisting of lectin, dectin 1, CD206, scavenger receptor Al (SRA1),
MARCO, CD36, CD163,
MSR1, SCARA3, COLEC12, SCARA5, SCARB1, SCARB2, CD68, OLR1, SCARF], SCARF2,
CXCL16, STAB1, STAB2, SRCRB4D, SSC5D, CD205, CD207, CD209, RAGE, CD14, CD64,
F4/80,
CCR2, CX3CR1, CSF1R, Tie2, HuCRIg(L), CD64, CD32a, CD16a, CD89, Fc-alpha
receptor I, CR1,
CD35, CR3, CR4, Tim-1, Tim-4 and CD169.
[0036] In some embodiments, the therapeutic agent binds to a
receptor comprising an intracellular
signaling domain that comprises a pro-inflammatory signaling domain.
[0037] In some embodiments, the first therapeutic agent
comprises a polypeptide that is less than
1000 amino acids or 1000 nm in length or 1000 nm.
[0038] In some embodiments, the first therapeutic agent
comprises a polypeptide that is less than 500
amino acids or 500 nm in length.
[0039] In some embodiments, the first therapeutic agent
comprises a polypeptide that is 200-1000
amino acids or 200-1000 nm in length.
100401 In some embodiments, engagement of the binding domains
of the first therapeutic agent
contacts the cancer cell to the myeloid cell.
[0041] In some embodiments, the second binding domain
specifically interacts with a myeloid cell
and promotes phagocytosis activity of the myeloid cell.
-5-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[0042] In some embodiments, the second binding domain
specifically interacts with a myeloid cell
and promotes inflammatory signaling of the myeloid cell.
[0043] In some embodiments, the second binding domain
specifically interacts with a myeloid cell or
an adhesion molecule and promotes adhesion of the myeloid cell to the target
cell.
[0044] In some embodiments, the second binding domain
specifically interacts with a myeloid cell
and inhibits anti-phagocytic activity of the myeloid cell mediated by the
target cell.
[0045] In some embodiments, the second binding domain
specifically interacts with a myeloid cell
and inhibits anti-inflammatory activity of the myeloid cell mediated by the
target cell.
[0046] In some embodiments, the second and/or the third
binding domain promotes phagocytic
activity of the myeloid cell.
[0047] In some embodiments, the second and/or the third
binding domain promotes inflammatory
signaling of the myeloid cell.
[0048] In some embodiments, the second and/or the third
binding domain specifically interacts with a
myeloid cell or an adhesion molecule and promotes adhesion of the myeloid cell
to the target cell.
[0049] In some embodiments, the second and/or the third
binding domain inhibits anti-phagocytic
activity of the myeloid cell mediated by the target cell.
[0050] In some embodiments, the second and/or the third
binding domain inhibits anti-inflammatory
activity of the myeloid cell mediated by the target cell.
[0051] In some embodiments, the therapeutic agent comprises a
therapeutic polypeptide.
[0052] In some embodiments, the therapeutic agent comprises a
recombinant nucleic acid encoding
the therapeutic polypeptide.
[0053] In some embodiments, the third binding domain or the
additional therapeutic agent comprises
a CD47 antagonist, a CD47 blocker, an antibody, a chimeric CD47 receptor, a
sialidase, a cytokine, a
proinflammatory gene, a procaspase, or an anti-cancer agent.
[0054] In some embodiments, the first binding domain, the
second binding domain and the third
binding domain bind to distinct non-identical target antigens.
[0055] In some embodiments, the first binding domain, the
second binding domain or the third
binding domain is a ligand binding domain.
[0056] In some embodiments, the first, the second or the
third binding domains are operably linked
by one or more linkers.
[0057] In some embodiments, the linker is a polypeptide. In
some embodiments, the linker is a
functional peptide. In some embodiments, the linker is a ligand for a
receptor. In some embodiments, the
linker is a ligand for a monocyte or macrophage receptor. In some embodiments,
the linker activates the
receptor. In some embodiments, the linker inhibits the receptor. In some
embodiments, the linker is a
-6-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
ligand for a M2 monocyte or macrophage. In some embodiments, the linker is a
ligand for a TLR
receptor. In some embodiments, the linker activates the TLR receptor.
[0058] In some embodiments, the first, the second and /or the
third binding domains are associated
with a mask that binds to the binding domain.
[0059] In some embodiments, the mask is an inhibitor that
inhibits the interaction of binding domain
to its target when the mask remains associated with the respective binding
domain.
[0060] In some embodiments, the mask is associated with the
binding domain via a peptide linker.
[0061] In some embodiments, the peptide linker comprises a
cleavable moiety.
[0062] In some embodiments, the cleavable moiety is cleaved
by a protein or an enzyme selectively
abundant in the site of the cancer or tumor.
[0063] In some embodiments, the third binding domain that
specifically interacts with an
extracellular region of a second receptor of the monocyte or macrophage
activates the monocyte or
macrophage.
[0064] In some embodiments, upon binding of the therapeutic
agent to the myeloid cell, the killing or
phagocytosis activity of the myeloid cell is increased by at least 10%, or
20%, or 30%, or 40%, or 50%,
or 60%, or 70% or 90% or 100% compared to a myeloid cell not bound by the
therapeutic agent, as
measured by a particle uptake assay.
[0065] In some embodiments, engagement of the binding domains
of first therapeutic agent triggers
phagocytosis of the cancer cell by the myeloid cell.
[0066] In some embodiments, engagement of the second
therapeutic agent potentiates or increases the
phagocytic killing of the cancer cell by the myeloid cell.
[0067] In some embodiments, the second or third binding
domain binds to an extracellular of IgA,
IgD, IgE, IgG, IgM, FcTRI, FcyR-11A, FcyR1113, FcyRIIC, FcyRIRA, FcyRIHIB,
FcRn, TREV121, FcRL5.
[0068] In some embodiments, the second or the third binding
domain comprises an M2 domain.
[0069] In some embodiments, the second or the third binding
domain comprises a LIGHT domain or
an HI/EM binding domain.
[0070] In some embodiments, the second or the third binding
domain comprises a HVEM binding
domain.
[0071] In some embodiments, the second or the third binding
domain comprises a GITR binding
domain.
[0072] In some embodiments, the first binding domain
comprises a sequence having an amino acid
sequence with at least 80%, 85%, 90%, 95% or 100% sequence identity to a
sequence selected from the
group consisting of SEQ ID NOs: 27, 28, 111, 112, 113, 115, 143 and 144.
-7-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[0073] In some embodiments, the second binding domain
comprises a sequence having an amino acid
sequence with at least 80%, 85%, 90%, 95% or 100% sequence identity to a
sequence selected from the
group 141 and 142.
[0074] In some embodiments, the first component comprises an
amino acid sequence
GGQEINSSYGG (SEQ ID NO: 105) or QEINSSY (SEQ ID NO: 129).
[0075] In some embodiments, the first component comprises an
amino acid sequence
GGAPPHALSGG (SEQ ID NO: 109) or APPHALS (SEQ ID NO: 137).
[0076] In some embodiments, the linker comprises an amino
acid sequence GGQEINSSYGG (SEQ
ID NO: 105), or QEINSSY (SEQ ID NO: 129) or GGAPPHALSGG (SEQ ID NO: 109) or
APPHALS
(SEQ ID NO: 137).
[0077] In some embodiments, provided herein is a bispecific
or trispecific engager, comprising a
sequence having an amino acid sequence with at least 80%, 85%, 90%, 95% or
1000/c sequence identity to
SEQ ID NO: 151.
[0078] In some embodiments, provided herein is a hi specific
or trispecific engager, comprising a
sequence having an amino acid sequence with at least 80%, 85%, 90%, 95% or
100% sequence identity to
SEQ ID NO: 152.
[0079] Provided herein is a pharmaceutical composition
comprising: a first therapeutic agent, wherein
the therapeutic agent comprises one or more polypeptides or recombinant
nucleic acids encoding the one
or more polypeptides, wherein the one or more polypeptides comprise: a first
binding domain, wherein the
first binding domain is a first antibody or functional fragment thereof that
specifically interacts with an
antigen of a target cell, and a second binding domain, wherein the second
binding domain is a second
antibody or functional fragment thereof that specifically interacts with a
myeloid cell; wherein, (i) the first
therapeutic agent is coupled to a first component, wherein the first component
is an additional therapeutic
agent or a third binding domain, or (ii) the composition comprises an
additional therapeutic agent; and an
acceptable pharmaceutical salt or excipient.
[0080] In some embodiments, the first therapeutic agent of
the pharmaceutical composition comprises
a single polypeptide. In some embodiments, the first therapeutic agent of the
pharmaceutical composition
comprises multiple polypeptides. In some embodiments, the first therapeutic
agent of the pharmaceutical
composition is a recombinant nucleic acid encoding the one or more
polypeptides. In some embodiments,
the pharmaceutical composition further comprises a second therapeutic agent.
[0081] Provided herein is a method of treatment, comprising:
administering to the subject in need
thereof, a pharmaceutical composition, comprising: a first therapeutic agent,
wherein the therapeutic
agent comprises one or more polypeptides or recombinant nucleic acids encoding
the one or more
polypeptides, wherein the one or more polypeptides comprise: a first binding
domain, wherein the first
binding domain is a first antibody or functional fragment thereof that
specifically interacts with an antigen
-8-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
of a target cell, and a second binding domain, wherein the second binding
domain is a second antibody or
functional fragment thereof that specifically interacts with a myeloid cell;
wherein, (i) the first therapeutic
agent is coupled to a first component, wherein the first component is an
additional therapeutic agent or a
third binding domain, or (ii) the composition comprises an additional
therapeutic agent; and an acceptable
pharmaceutical salt or excipient_
[0082] In some embodiments, the method of treatment further
comprises administering a second
therapeutic agent. In some embodiments, the method of treatment further
comprises administering the
pharmaceutical composition comprises administering the pharmaceutical
composition intravenously.
[0083] In some embodiments, the method of treatment further
comprises the administering the
pharmaceutical composition comprises administering the pharmaceutical
composition subcutaneously. In
some embodiments, the method of treatment further comprises administering the
pharmaceutical
composition comprises injecting the pharmaceutical composition.
In some embodiments, the target cell is a cancer cell.
[0084] In some embodiments, the target cell is a cancer cell
that is a lymphocyte,
[0085] In some embodiments, the target cell is a cancer cell
that is an ovarian cancer cell.
[0086] In some embodiments, the target cell is a cancer cell
that is an ovarian pancreatic cell.
[0087] In some embodiments, the target cell is a cancer cell
that is a glioblastoma cell.
[0088] In some embodiments, the recombinant nucleic acid is
DNA.
[0089] In some embodiments, the recombinant nucleic acid is
RNA.
[0090] In some embodiments, the recombinant nucleic acid is
mRNA.
[0091] In some embodiments, the recombinant nucleic acid is a
circRNA.
[0092] In some embodiments, the recombinant nucleic acid is a
tRNA.
[0093] In some embodiments, the recombinant nucleic acid is a
microRNA.
[0094] Provided herein is a vector, comprising the
composition described above.
[0095] In some embodiments, vector is viral vector. In some
embodiments, the viral vector is retroviral
vector or a lentiviral vector. In some embodiments, the vector further
comprises a promoter operably linked
to at least one nucleic acid sequence encoding one or more polypeptides. In
some embodiments, the vector
is polycistronic. In some embodiments, each of the at least one nucleic acid
sequence is operably linked to
a separate promoter. In some embodiments, the vector further comprises one or
more internal ribosome
entry sites (IRESs). In some embodiments, the vector further comprises a
5'LTTR and/or a 3'1UTR flanking
the at least one nucleic acid sequence encoding one or more polypeptides. In
some embodiments, the vector
further comprises one or more regulatory regions_
[0096] Provided herein is a polypeptide encoded by the
recombinant nucleic acid of the composition
described above.
-9-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
100971 Additional aspects and advantages of the present
disclosure will become readily apparent to
those skilled in this art from the following detailed description, wherein
only illustrative embodiments of
the present disclosure are shown and described. As will be realized, the
present disclosure is capable of
other and different embodiments, and its several details are capable of
modifications in various obvious
respects, all without departing from the disclosure. Accordingly, the drawings
and description are to be
regarded as illustrative in nature, and not as restrictive.
INCORPORATION BY REFERENCE
100981 All publications, patents, and patent applications
mentioned in this specification are herein
incorporated by reference to the same extent as if each individual
publication, patent, or patent application
was specifically and individually indicated to be incorporated by reference.
To the extent publications and
patents or patent applications incorporated by reference contradict the
disclosure contained in the
specification, the specification is intended to supersede and/or take
precedence over any such contradictory
material.
BRIEF DESCRIPTION OF THE DRAWINGS
100991 The novel features of the invention are set forth with
particularity in the appended claims. A
better understanding of the features and advantages of the present invention
will be obtained by reference
to the following detailed description that sets forth illustrative
embodiments, in which the principles of the
invention are utilized, and the accompanying drawings (also "FIG." herein), of
which:
1001001 FIG. 1A is a graphical representation of the exemplary extracellular
stimulants, receptors and
pathways generating a dual signal for a myeloid cell: a signal 1 and a signal
2.
[00101] FIG. 1B is a graphical representation of a simplified engager
construct with a binder A, a liner
L and a second binder B. In this simplified diagram, binder A binds to a cell
surface biomolecule on a
target cell; binder B binds to a cell surface biomolecule on a myeloid cell.
[00102] FIG. 2A is a graphical representation of bispecific scFv engager with
protease cleavable
masked site, that are the antigen binding domains.
[00103] FIG. 213 is a graphical representation of bispecific \Tun engager with
protease cleavable masked
antigen binding domains.
1001041 FIG. 3A is a graphical representation of an exemplary bispecific scFy
engager with protease
cleavable masked site, and a peptide linker connecting the two scFy engagers
that are the antigen binding
domains; in this case, the peptide linker is an additional therapeutic agent
or a third binding domain,
specifically a TLR4 ligand peptide.
1001051 FIG. 3B is a graphical representation of an exemplary bispecific VHF;
enuager with protease
cleavable masked antigen binding domains, and a peptide linker connecting the
two protease cleavable
-10-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
masked antigen binding domains; in this case, the peptide linker is an
additional therapeutic agent or a third
binding domain, specifically a TLR4 ligand peptide.
[00106] FIG. 3C is a graphical representation of an exemplary bispecific scFv
engager with protease
cleavable masked site, and a peptide linker connecting the two scFv engagers
that are the antigen binding
domains; in this case, the peptide linker is an additional therapeutic agent
or a third binding domain,
specifically a M2 targeting peptide.
[00107] FIG. 3D is a graphical representation of an exemplary bispecific Vim
engager with protease
cleavable masked antigen binding domains, and a peptide linker connecting the
two protease cleavable
masked antigen binding domains; in this case, the peptide linker is an
additional therapeutic agent or a third
binding domain, specifically a M2 targeting peptide.
[001081 FIG. 3E depicts data indicating cytokine production by monocytes
cultured overnight in the
presence of each TLR peptide indicated.
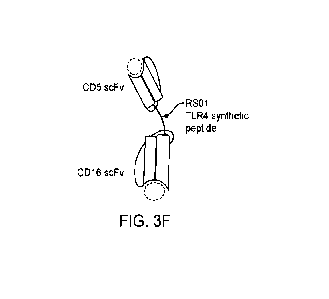
[00109] FIG. 3F shows a graphical illustration of the protein structure of a
bispecific binder construct
CD5-RS01-CD16, having two scFv binders specific for CD5 and CD16 respectively,
and a TLR4 synthetic
peptide linker (RS01).
[00110] FIG. 3G shows expression data of the CD5-RS01-CD16 demonstrated in
FIG. 3F. Lanes M1
and M2, Commercially available protein molecular weight marker from TaKaRa,
Cat No. 3452 and
GenScript, Cat No. M00521 respectively. Lanes 1 and 2 are SDS PAGE results or
western blot results as
indicated, under reducing and non-reducing conditions respectively. Lane P,
positive control (Multiple lag,
Gene Script, Cat No, M0101). Primary Antibody used for Western Blot: Mouse
anti-His mAb (GenScripts,
Cat. No. A00186).
[00111] FIG. 3H shows a graphical illustration of a protein structure of
bispecific binder construct
CD5-RS09-CD16, having two scFv binders specific for CD5 and CD16 respectively,
and a TLR4 synthetic
peptide linker (RS09).
[00112] FIG. 31 depicts expression data of the CD5-RS09-CD16 demonstrated in
FIG. 311. Lane
annotation and indices are as indicated in description for FIG. 3G.
[00113] FIG. 4A is a graphical representation of an exemplary trispecific scFv
engager.
[00114] FIG. 4B is a graphical representation of an exemplary trispecific
V}I}I engager.
[00115] FIG. 4C is a graphical representation of an exemplary mode of action
of a trispecific engager.
[00116] FIG. 5Ai depicts a graphical representation of the structural
configuration of a recombinant
bispecific scFv engager, where each of the binding domains is masked by an
agent (a mask), that prevents
interaction of the binding domain with its cognate substrate. The mask is
attached with the terminal section
of each light chain by a cleavable linker, in the example a metalloprotease
(MMP2) substrate. Arrows point
to the structural components of the recombinant bispecific scFv engager, which
are as follows: 1, mask; 2,
MMP2 substrate linker; 3, ABDI (antigen binding domain 1)-light chain; 3',
ABD2 (antigen binding
-11-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
domain 2)-light chain; 4, a linker connecting the binding domain light chain
and the binding domain heavy
chain; 5, ABD1 (antigen binding domain 1) heavy chain; 5', ABD2 (antigen
binding domain 2) heavy
chain..
[00117] FIG. 5Aii depicts a graphical representation of the structural
configuration of a recombinant
bispecific diabody engager, where each of the binding domains is masked by an
agent (a mask), that
prevents interaction of the binding domain with its cognate substrate. The
mask is attached with the
terminal section of each light chain by a cleavable linker, in the example a
metalloprotease (MMP2)
substrate. Arrows point to the structural components of the recombinant
bispecific diabody engager, which
are as follows: 1, mask; 2, MMP2 substrate linker; 3, ABD1 (antigen binding
domain 1)-light chain; 3',
ABD2 (antigen binding domain 2)-light chain; 4, linker connecting the ABD1
light chain and the ABD1
heavy chain; 4', linker connecting the ABD2 light chain and the ABD2 heavy
chain; 5, ABD2 heavy chain;
5', ABD1 heavy chain.
[00118] FIG. 5B depicts a graphical representation of the linear construct for
a single chain of the
bispecific scFv. The parts are corresponding to FIG. 5Ai or FIG. 5Aii are
depicted within the linearized
diagram from N-terminal to C-terminal.
[00119] FIG. 6 depicts exemplary modular constructs comprising two or three
binding domains to
utilize as bispecific and trispecific engagers.
1001201 FIG. 7A upper panel is a graphical representation of MD2 mediated
dimerization of TLR4
receptor, which leads to TLR activation. FIG. 7A lower panel is an exemplary
design of a monocyte or
macrophage specific engager, where one binding domain can bind to a tumor cell
associated molecule
(tumor antigen), another binding domain can bind to a monocyte or macrophage
receptor, in this case an
FcR, The third domain is an MD2 domain, which can bind to and dimerize TLR4
receptors, to activate
them.
[00121] FIG. 7B is a graphical representation that shows the mode of action of
the monocyte or
macrophage specific engager of FIG. 7A.
[00122] FIG. 8A is an exemplary design of a monocyte or macrophage specific
engager, where one
binding domain can bind to a tumor cell associated molecule (tumor antigen), a
second binding domain
can bind to a monocyte or macrophage receptor, in this case an FcR. The third
domain is an LIGHT domain,
which can engage with monocyte or macrophage HVEM and activate an inflammatory
signal in the
monocyte or macrophage.
[00123] FIG. 8B is a graphical representation that shows the mode of action of
the monocyte or
macrophage specific engager of FIG. SA.
1001241 FIG. 9A is an exemplary design of a monocyte or macrophage specific
engager, where one
binding domain can bind to a tumor cell associated molecule (tumor antigen),
and a second binding domain
can bind to a monocyte or macrophage receptor, in this case an FcR. The third
domain is a GIRT ligand
-12-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
(GIRTL) domain or alternatively an antigen binding domain of anti-GITR
antibody that can activate
monocyte or macrophage receptor GITR, and can induce an inflammatory signal in
the monocyte or
macrophage.
[00125] FIG. 9B is a graphical representation that shows the mode of action of
the monocyte or
macrophage specific engager of FIG. 9A.
1001261 FIG. 10A shows exemplary heterodimeric antibody-based engager molecule
designs,
comprising peptides with leucine zipper domains. Li, L2 indicate ligands.
[00127] FIG. 10B shows exemplary heteromultimeric antibody-based engager
molecule designs,
comprising peptides with leucine zipper domains. Li-IA indicate ligands.
[00128] FIG. 10C shows exemplary heterodimeric antibody-based engager molecule
designs,
comprising peptides having synthetic anchoring design. Li, L2 indicate
ligands, and 'In' and 'n' indicate
synthetic binding designs.
DETAILED DESCRIPTION
[00129] All terms are intended to be understood as they would be understood by
a person skilled in the
art. Unless defined otherwise, all technical and scientific terms used herein
have the same meaning as
commonly understood by one of ordinary skill in the art to which the
disclosure pertains.
[00130] The section headings used herein are for organizational purposes only
and are not to be
construed as limiting the subject matter described.
[00131] Although various features of the present disclosure can be described
in the context of a single
embodiment, the features can also be provided separately or in any suitable
combination. Conversely,
although the present disclosure can be described herein in the context of
separate embodiments for clarity,
the disclosure can also be implemented in a single embodiment.
[00132] As used in this specification and claim(s), the words "comprising"
(and any form of comprising,
such as "comprise" and "comprises"), "having" (and any form of having, such as
"have" and "has"),
"including" (and any form of including, such as "includes" and "include") or
"containing" (and any form
of containing, such as "contains" and "contain") are inclusive or open-ended
and do not exclude additional,
unrecited elements or method steps. It is contemplated that any embodiment
discussed in this specification
can be implemented with respect to any method or composition of the
disclosure, and vice versa
Furthermore, compositions of the disclosure can be used to achieve methods of
the disclosure.
[00133] The term "about" or "approximately" as used herein when referring to a
measurable value such
as a parameter, an amount, a temporal duration, and the like, is meant to
encompass variations of +/-20%
or less, +/-10% or less, +1-5% or less, or +/-1% or less of and from the
specified value, insofar such
variations are appropriate to perform in the present disclosure. It is to be
understood that the value to which
the modifier "about" or "approximately" refers is itself also specifically
disclosed.
-13-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00134] An "agent" can include any type of molecule and includes, but is not
limited to, an antibody, a
peptide, a protein, a polynucleofide (e.g., an oligonucleotide, RNA, or DNA),
a small molecule, derivatives
thereof and analogs thereof
[00135] A "biologic sample" is any tissue, cell, fluid, or other material
derived from an organism. As
used herein, the term "sample" includes a biologic sample such as any tissue,
cell, fluid, or other material
derived from an organism.
[00136] "Specifically binds" refers to a condition in which a compound (e.g.,
peptide) recognizes and
binds to a molecule (e.g., peptide or polypeptide), but does not substantially
recognize and bind other
molecules in a sample, for example, a biological sample, that is, the compound
exhibits a selective binding
to a molecule. A "binder" as described herein includes, but is not limited to,
a protein, a polypeptide or
fragments thereof, that exhibits specific binding to a cognate molecule. A
binder may refer to an antigen
binding domain, such as the first binding domain of a bispecific or
trispecific engager, or the second antigen
binding domain of a bispecific or trispecific engager, and so on. In some
cases, a binder may be any
biomolecule or fragment thereof, such as a peptide or conjugated peptide or a
ligand that can specifically
bind to a receptor on a cell and therefore exhibits specific binding of one
portion of an exemplary engager.
[00137] The term "immune response" includes T cell mediated and/or B cell
mediated immune
responses that are influenced by modulation of T cell costimulation. Exemplary
immune responses include
T cell responses, e.g., cytokine production, and cellular cytotoxicity. In
addition, the term immune response
includes immune responses that are indirectly affected by T cell activation,
e.g., antibody production
(htunoral responses) and activation of cytokine responsive cells, e.g.,
monocytes or macrophages.
[00138] A "functional derivative" of a native sequence polypeptide is a
compound having a qualitative
biological property in common with a native sequence polypeptide. "Functional
derivatives" include, but
are not limited to, fragments of a native sequence and derivatives of a native
sequence polypeptide and its
fragments, provided that they have a biological activity in common with a
corresponding native sequence
polypeptide. The term "derivative" encompasses both amino acid sequence
variants of polypeptide and
covalent modifications thereof
[00139] The terms "phagocytic cells" and "phagocytes" are used interchangeably
herein to refer to a
cell that is capable of phagocytosis. There are three main categories of
phagocytes: macrophages,
mononuclear cells (histiocytes and monocytes); polymorphonuclear leukocytes
(neutrophils) and dendritic
cells.
[00140] The term "biological sample" encompasses a variety of sample types
obtained from an
organism and can be used in a diagnostic or monitoring assay. The term
encompasses blood and other
liquid samples of biological origin, solid tissue samples, such as a biopsy
specimen or tissue cultures or
cells derived therefrom and the progeny thereof. The term encompasses samples
that have been
manipulated in any way after their procurement, such as by treatment with
reagents, solubilization, or
-14-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
enrichment for certain components. The term encompasses a clinical sample, and
also includes cells in cell
culture, cell supernatants, cell lysates, serum, plasma, biological fluids,
and tissue samples.
[00141] As used herein, the term "antigen-presenting cell" or "antigen-
presenting cells" or its
abbreviation "APC" or "APCs" refers to a cell or cells capable of endocytosis
adsorption, processing and
presenting of an antigen. The term includes professional antigen presenting
cells for example; B
lymphocytes, monocytes, dendritic cells (DCs) and Langerhans cells, as well as
other antigen presenting
cells such as keratinocytes, endothelial cells, glial cells, fibroblasts and
oligodendrocytes. The term
"antigen presenting" means the display of antigen as peptide fragments bound
to MHC molecules, on the
cell surface. Many different kinds of cells may function as APCs including,
for example, monocytes or
macrophages, B cells, follicular dendritic cells and dendritic cells. APCs can
also cross-present peptide
antigens by processing exogenous antigens and presenting the processed
antigens on class I MEW
molecules. Antigens that give rise to proteins that are recognized in
association with class I MHC molecules
are generally proteins that are produced within the cells, and these antigens
are processed and associate
with class I WIC molecules.
[00142] An "epitope" refers to a portion of an antigen or other macromolecule
capable of forming a
binding interaction with the variable region binding pocket of an antibody or
TCR. The term includes any
protein determinant capable of specific binding to an antibody, antibody
peptide, and/or antibody-like
molecule (including but not limited to a T cell receptor) as defined herein.
Epitopic determinants typically
consist of chemically active surface groups of molecules such as amino acids
or sugar side chains and
generally have specific three dimensional structural characteristics as well
as specific charge
characteristics.
[00143] In some embodiments, the phagocytic receptor fusion protein (PFP)
comprises an extracellular
antigen binding domain specific to an antigen of a target cell, fused to the
phagocytic receptor. A target
cell is, for example, a cancer cell. In some embodiments, the phagocytic cell,
after engulfment of the cancer
cell may present the cancer antigen on its cell surface to activate a T cell.
[00144] As used herein the term "antigen" is any organic or inorganic molecule
capable of stimulating
an immune response. The term "antigen" as used herein extends to any molecule
such as, but not limited,
to a peptide, polypeptide, protein, nucleic acid molecule, carbohydrate
molecule, organic or inorganic
molecule capable of stimulating an immune response.
[00145] In some embodiments, the phagocytic receptor fusion protein may
comprise an extracellular
domain, which comprises an antibody domain or a antigen binding portion
thereof that can bind to a cancer
antigen or a cell surface molecule on a cancer cell. The term "antibody" or
"antibody moiety" is includes,
but is not limited to any polypeptide chain-containing molecular structure
that recognizes an epitope.
Antibodies utilized in the present invention may be polyclonal antibodies,
although monoclonal antibodies
are preferred because they may be reproduced by cell culture or recombinantly,
and can be modified to
-15-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
reduce their antigenicity. The term includes IgG (including IgGl, igG2, IgG3,
and IgG4), IgA (including
IgAl and IgA2), IgD, IgE, IgM, and IgY, and is meant to include whole
antibodies, including single-chain
whole antibodies, and antigen-binding (Fab) fragments thereof Antigen-binding
antibody fragments
include, but are not limited to, Fab, Fab' and F(ab')2, Fd (consisting of Vii
and CH1), single-chain variable
fragment (scFv), single-chain antibodies, disulfide-linked variable fragment
(dsFv) and fragments
comprising either a Vi. or VII domain. The antibodies can be from any animal
origin. Antigen-binding
antibody fragments, including single-chain antibodies, can comprise the
variable region(s) alone or in
combination with the entire or partial of the following: hinge region, CH1,
CH2, and CH3 domains. Also
included are any combinations of variable region(s) and hinge region, CHI,
CH2, and CH3 domains.
Antibodies can be monoclonal, polyclonal, chimeric, humanized, and human
monoclonal and polyclonal
antibodies which, e.g., specifically bind an HLA-associated polypeptide or an
HLA-peptide complex. A
person of skill in the art will recognize that a variety of immunoaffinity
techniques are suitable to enrich
soluble proteins, such as soluble HLA-peptide complexes or membrane bound HLA-
associated
polypeptides, e.g., which have been proteolytically cleaved from the membrane.
These include techniques
in which (1) one or more antibodies capable of specifically binding to the
soluble protein are immobilized
to a fixed or mobile substrate (e.g., plastic wells or resin, latex or
paramagnetic beads), and (2) a solution
containing the soluble protein from a biological sample is passed over the
antibody coated substrate,
allowing the soluble protein to bind to the antibodies. The substrate with the
antibody and bound soluble
protein is separated from the solution, and optionally the antibody and
soluble protein are disassociated,
for example by varying the pH and/or the ionic strength and/or ionic
composition of the solution bathing
the antibodies. Alternatively, immunoprecipitation techniques in which the
antibody and soluble protein
are combined and allowed to form macromolecular aggregates can be used. The
macromolecular
aggregates can be separated from the solution by size exclusion techniques or
by centrifugation.
[00146] The adaptive immune system reacts to molecular structures, referred to
as antigens, of the
intruding organism. Unlike the innate immune system, the adaptive immune
system is highly specific to a
pathogen. Adaptive immunity can also provide long-lasting protection; for
example, someone who recovers
from measles is now protected against measles for their lifetime. There are
two types of adaptive immune
reactions, which include the humoral immune reaction and the cell-mediated
immune reaction. In the
htunoral immune reaction, antibodies secreted by B cells into bodily fluids
bind to pathogen-derived
antigens, leading to the elimination of the pathogen through a variety of
mechanisms, e.g. complement-
mediated lysis. In the cell-mediated immune reaction, T cells capable of
destroying other cells are activated.
For example, if proteins associated with a disease are present in a cell, they
are fragmented proteolytically
to peptides within the cell. Specific cell proteins then attach themselves to
the antigen or peptide formed in
this manner and transport them to the surface of the cell, where they are
presented to the molecular defense
-16-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
mechanisms, in T cells, of the body. Cytotoxic T cells recognize these
antigens and kill the cells that harbor
the antigens.
1001471 The term "major histocompatibility complex (MHC', "MHC molecules", or
"MHC proteins"
refers to proteins capable of binding antigenic peptides resulting from the
proteolytic cleavage of protein
antigens inside phagocytes or antigen presenting cells and for the purpose of
presentation to and activation
of T lymphocytes. Such antigenic peptides represent T cell epitopes. The human
MHC is also called the
HLA complex. Thus, the term "human leukocyte antigen (HLA) system", "111,A
molecules" or "HLA
proteins" refers to a gene complex encoding the MHC proteins in humans. The
term MHC is referred as
the "11-2" complex in murine species. Those of ordinary skill in the art will
recognize that the terms" major
histocompatibility complex (MHC)", "MHC molecules", "MHC proteins" and "human
leukocyte antigen
(HLA) system", "HLA molecules", "LILA proteins" are used interchangeably
herein.
[00148] HLA proteins are tylically classified into two types, referred to as
HLA class I and HLA class
II. The structures of the proteins of the two HLA classes are very similar;
however, they can have different
functions. Class I IlLA proteins are present on the surface of almost all
cells of the body, including most
tumor cells. Class I 111,A proteins are loaded with antigens that usually
originate from endogenous proteins
or from pathogens present inside cells, and are then presented to naive or
cytotoxic T-lymphocytes (CTLs).
HLA class II proteins are present on antigen presenting cells (APCs),
including but not limited to dendritic
cells, B cells, and monocytes or macrophages. They mainly present peptides,
which are processed from
external antigen sources, e.g. outside of the cells, to helper T cells. Most
of the peptides bound by the HLA
class I proteins originate from cytoplasmic proteins produced in the healthy
host cells of an organism itself,
and do not normally stimulate an immune reaction.
[00149] In HLA class II system, phagocytes such as monocytes or macrophagesand
immature dendritic
cells take up entities by phagocytosis into phagosomes ¨ though B cells
exhibit the more general
endocytosis into endosomes which fuse with lysosomes whose acidic enzymes
cleave the uptaken protein
into many different peptides. Autophagy is a source of HLA class 11 peptides.
Via physicochemical
dynamics in molecular interaction with the HLA class 11 variants borne by the
host, encoded in the hoses
genome, a particular peptide exhibits immunodominance and loads onto HLA class
II molecules. These
are trafficked to and externalized on the cell surface. The most studied
subclass II HLA genes are: HLA-
DPA1, HILA-DPB1, HLA-DQA1, HLA-DQB1, HLA-DRA, and HLA-DRE1.
[00150] Presentation of peptides by HLA class 11 molecules to CD4+ helper T
cells is required for
immune responses to foreign antigens. Once activated, CD4+ T cells promote B
cell differentiation and
antibody production, as well as CD8+ T cell (CTL) responses. CD4+ T cells also
secrete cytokines and
chemokines that activate and induce differentiation of other immune cells. HLA
clogs 11 molecules are
heterodimers of a- and 3-chains that interact to form a peptide-binding groove
that is more open than class
I peptide-binding grooves. Peptides bound to VILA class 11 molecules are
believed to have a 9-amino acid
-17-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
binding core with flanking residues on either N- or C-terminal side that
overhang from the groove. These
peptides are usually 12-16 amino acids in length and often contain 3-4 anchor
residues at positions Pl, P4,
P6/7 and P9 of the binding register (Rossjohn et at., 2015).
[00151] HLA alleles are expressed in codominant fashion, meaning that the
alleles (variants) inherited
from both parents are expressed equally. For example, each person carries 2
alleles of each of the 3 class I
genes, (HLA-A, HLA-B and HLA-C) and so can express six different types of
class II HLA. In the class
II HLA locus, each person inherits a pair of HLA-DP genes (DPA1 and DPB1,
which encode a and (3
chains), HLA-DQ (DQA1 and DQB1, for a and p chains), one gene HLA-DRa (DRA1),
and one or more
genes 1-11A-DR13 (DRB1 and DRB3, -401 -5). EILA-DRB1, for example, has more
than nearly 400 known
alleles. That means that one heterozygous individual can inherit six or eight
functioning class 11 HLA
alleles: three or more from each parent. Thus, the HLA genes are highly
polymorphic; many different
alleles exist in the different individuals inside a population. Genes encoding
HLA proteins have many
possible variations, allowing each person's immune system to react to a wide
range of foreign invaders.
Some HLA genes have hundreds of identified versions (alleles), each of which
is given a particular number.
In some embodiments, the class I HLA alleles are HLA-A02:01, HLA-B*14:02, MLA-
A*23:01, HLA-
E*01:01 (non-classical). In some embodiments, class 1.1 HLA alleles are HILA-
DRB*01:01, HLA-
DRB*01: 02, HLA-DRB*11 :01, HLA-DRB*15: 01, and 1{LA-DRB*07: 0 L
[00152] In some embodiments, the phagocytic cell is administered to a patient
or a subject. A cell
administered to a human subject must be immunocompatible to the subject,
having a matching HLA
subtype that is naturally expressed in the subject. Subject specific HLA
alleles or HLA genotype of a
subject can be determined by any method known in the art. In exemplary
embodiments, the methods include
determining polymorphic gene types that can comprise generating an alignment
of reads extracted from a
sequencing data set to a gene reference set comprising allele variants of the
polymorphic gene, determining
a first posterior probability or a posterior probability derived score for
each allele variant in the alignment,
identifying the allele variant with a maximum first posterior probability or
posterior probability derived
score as a first allele variant, identifying one or more overlapping reads
that aligned with the first allele
variant and one or more other allele variants, determining a second posterior
probability or posterior
probability derived score for the one or more other allele variants using a
weighting factor, identifying a
second allele variant by selecting the allele variant with a maximum second
posterior probability or
posterior probability derived score, the first and second allele variant
defining the gene type for the
polymorphic gene, and providing an output of the first and second allele
variant.The expression "amino
acid" as used herein is intended to include both natural and synthetic amino
acids, and both D and L amino
acids. A synthetic amino acid also encompasses chemically modified amino
acids, including, but not
limited to salts, and amino acid derivatives such as amides. Amino acids
present within the polypeptides
of the present invention can be modified by methylation, amidation,
acetylation or substitution with other
-18-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
chemical groups which can change the circulating half-life without adversely
affecting their biological
activity.
[001531 The terms "peptide", "polypeptide" and "protein" are used herein
interchangeably to describe
a series of at least two amino acids covalently linked by peptide bonds or
modified peptide bonds such as
isosteres. No limitation is placed on the maximum number of amino acids which
may comprise a peptide
or protein. The terms "oligomer" and "oligopeptide" are also intended to mean
a peptide as described
herein. Furthermore, the term polypeptide extends to fragments, analogues and
derivatives of a peptide,
wherein said fragment, analogue or derivative retains the same biological
functional activity as the peptide
from which the fragment, derivative or analogue is derived.
1001541 A polypeptide as used herein can be a "protein", including but not
limited to a glycoprotein, a
lipoprotein, a cellular protein or a membrane protein. A polypeptide may
comprise one or more subunits
of a protein. A polypeptide may be encoded by a recombinant nucleic acid. In
some embodiments,
polypeptide may comprise more than one peptides in a single amino acid chain,
which may be separated
by a spacer, a linker or peptide cleavage sequence. A polypeptide may be a
fused polypeptide. A
polypeptide or a protein may comprise one or more domains. A domain is a
structural portion of a protein
with a defined function, a polypeptide or a protein may comprise one or more
modules. A module is domain
or a portion of the domain or portion of a protein with a specific function. A
module may be a structural
module of a protein, designated by its structural embodiments. A moiety is a
portion of polypeptide, a
protein or a nucleic acid, having a specific structure or perform a specific
function. For example, a signaling
moiety is a specific unit within the larger structure of the polypeptide or
protein or a recombinant nucleic
acid, which (or the protein portion encoded by it in case of a nucleic acid)
engages in a signal transduction
process, for example a phosphorylation. A module, a domain and a moiety, as
used herein, can be used
interchangeably, unless a specific structural or functional orientation is
otherwise defined in the text. A
motif is a structural entity in a biomolecule. A signaling motif in a protein
or polypeptide, for example,
refers to a stretch of amino acids on the protein or polypeptide which contain
an amino acid which may be
phosphorylated, dephosphorylated or can serve as a binding site of another
signaling molecule. Similarly,
in case of nucleic acids, for example, TNF mRNA has a conserved motif,
1UUAUUTJAUU, in the 3'UTR
to which mRNA destabilizing enzymes such as zinc-finger binding protein 36
family members bind.
[001551 The term "pro-antibody" as used herein may refer to an antibody, an
scFv, a Vim, single domain
antibody, or a protein or polypeptide that comprises an inactive antigen
binding domain; wherein the
antigen binding capability is designed to be blocked or inactive e.g. by
binding a cleavable antigen domain
binding polypeptide, until an active step is performed to convert the pro-
antibody to its active form. In
some embodiments, the active step involves a protease cleavage of the entity
that block the antigen binding
domain.
-19-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00156] As used herein, the term "recombinant nucleic acid molecule" refers to
a recombinant DNA
molecule or a recombinant RNA molecule. A recombinant nucleic acid molecule is
any nucleic acid
molecule containing joined nucleic acid molecules from different original
sources and not naturally
attached together. A recombinant nucleic acid may be synthesized in the
laboratory. A recombinant nucleic
acid can be prepared by using recombinant DNA technology by using enzymatic
modification of DNA,
such as enzymatic restriction digestion, ligation, and DNA cloning. A
recombinant nucleic acid as used
herein can be DNA, or RNA. A recombinant DNA may be transcribed in vitro, to
generate a messenger
RNA (mRNA), the recombinant mRNA may be isolated, purified and used to
transfect a cell. A
recombinant nucleic acid may encode a protein or a polypeptide. A recombinant
nucleic acid, under suitable
conditions, can be incorporated into a living cell, and can be expressed
inside the living cell As used herein,
"expression" of a nucleic acid usually refers to transcription and/or
translation of the nucleic acid. The
product of a nucleic acid expression is usually a protein but can also be an
mRNA. Detection of an mRNA
encoded by a recombinant nucleic acid in a cell that has incorporated the
recombinant nucleic acid, is
considered positive proof that the nucleic acid is "expressed" in the cell.
[00157] The process of inserting or incorporating a nucleic acid into a cell
can be via transformation,
transfection or transduction. Transformation is the process of uptake of
foreign nucleic acid by a bacterial
cell. This process is adapted for propagation of plasmid DNA, protein
production, and other applications.
Transformation introduces recombinant plasmid DNA into competent bacterial
cells that take up
extracellular DNA from the environment. Some bacterial species are naturally
competent under certain
environmental conditions, but competence is artificially induced in a
laboratory setting. Transfection is the
forced introduction of small molecules such as DNA, RNA, or antibodies into
eukaryotic cells. Just to
make life confusing, `transfection' also refers to the introduction of
bacteriophage into bacterial cells.
'Transduction' is mostly used to describe the introduction of recombinant
viral vector particles into target
cells, while 'infection' refers to natural infections of humans or animals
with wild-type viruses.
1001581 As used herein, the term "vector" means any genetic construct, such as
a plasmid, phage,
transposon, cosmid, chromosome, virus, virion, etc., which is capable
transferring nucleic acids between
cells. Vectors may be capable of one or more of replication, expression,
recombination, insertion or
integration, but need not possess each of these capabilities. A plasmid is a
species of the genus encompassed
by the term "vector." A vector typically refers to a nucleic acid sequence
containing an origin of replication
and other entities necessary for replication and/or maintenance in a host
cell. Vectors capable of directing
the expression of genes and/or nucleic acid sequence to which they are
operatively linked are referred to
herein as "expression vectors", In general, expression vectors of utility are
often in the form of "plasmids"
which refer to circular double stranded DNA molecules which, in their vector
form are not bound to the
chromosome, and typically comprise entities for stable or transient expression
or the encoded DNA. Other
expression vectors that can be used in the methods as disclosed herein
include, but are not limited to
-20-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
plasmids, episomes, bacterial artificial chromosomes, yeast artificial
chromosomes, bacteriophages or viral
vectors, and such vectors can integrate into the host's genome or replicate
autonomously in the cell. A
vector can be a DNA or RNA vector. Other forms of expression vectors known by
those skilled in the art
which serve the equivalent functions can also be used, for example, self-
replicating extrachromosomal
vectors or vectors capable of integrating into a host genome. Exemplary
vectors are those capable of
autonomous replication and/or expression of nucleic acids to which they are
linked.
[00159] The terms "spacer" or "linker" as used in reference to a fusion
protein refers to a peptide that
joins the proteins comprising a fusion protein. In some embodiments, the
constituent amino acids of a
spacer can be selected to influence some property of the molecule such as the
folding, net charge, or
hydrophobicity of the molecule. Suitable linkers for use in an embodiment of
the present disclosure are
well known to those of skill in the art and include, but are not limited to,
straight or branched-chain carbon
linkers, heterocyclic carbon linkers, or peptide linkers. The linker is used
to separate two antigenic peptides
by a distance sufficient to ensure that, in some embodiments, each antigenic
peptide properly folds.
Exemplary peptide linker sequences adopt a flexible extended conformation and
do not exhibit a propensity
for developing an ordered secondary structure. Typical amino acids in flexible
protein regions include ay,
Asn and Ser. Virtually any permutation of amino acid sequences containing Gly,
Asir and Ser would be
expected to satisfy the above criteria for a linker sequence. Other near
neutral amino acids, such as Thr and
Ala, also can be used in the linker sequence.
[00160] In some embodiments, the peptide linkers have more than one functional
properties, such as
the ones described herein. For example, the peptide linker links two or more
functional domains, such as
binding domains. Additionally, the peptide linker may be a specific signal
inducer when the linker contacts
an extracellular portion of a cell, such as a receptor or a ligand binding
protein.
[00161] The term "imrnunopurification (IP)" (or immunoaffinity purification or
irrununoprecipitation)
is a process well known in the art and is widely used for the isolation of a
desired antigen from a sample.
In general, the process involves contacting a sample containing a desired
antigen with an affinity matrix
comprising an antibody to the antigen covalently attached to a solid phase.
The antigen in the sample
becomes bound to the affinity matrix through an immunochemical bond. The
affinity matrix is then washed
to remove any unbound species. The antigen is removed from the affinity matrix
by altering the chemical
composition of a solution in contact with the affinity matrix. The
immunopurification can be conducted on
a column containing the affinity matrix, in which case the solution is an
eluent. Alternatively, the
immunopurification can be in a batch process, in which case the affinity
matrix is maintained as a
suspension in the solution. An important step in the process is the removal of
antigen from the matrix_ This
is commonly achieved by increasing the ionic strength of the solution in
contact with the affinity matrix,
for example, by the addition of an inorganic salt. An alteration of pH can
also be effective to dissociate the
immunochemical bond between antigen and the affinity matrix.
-21-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00162] As used herein, the terms "determining", "assessing", "assaying",
"measuring", "detecting"
and their grammatical equivalents refer to both quantitative and qualitative
determinations, and as such,
the term "determining" is used interchangeably herein with "assaying,"
"measuring," and the like. Where
a quantitative determination is intended, the phrase "determining an amount"
of an analyte and the like is
used. Where a qualitative and/or quantitative determination is intended, the
phrase "determining a level"
of an analyte or "detecting" an analyte is used.
[001631 A "fragment" is a portion of a protein or nucleic acid that is
substantially identical to a reference
protein or nucleic acid. In some embodiments, the portion retains at least
50%, 75%, or 80%, or 90%, 95%,
or even 99% of the biological activity of the reference protein or nucleic
acid described herein.
[001641 The terms "isolated," "purified", "biologically pure" and their
grammatical equivalents refer to
material that is free to varying degrees from components which normally
accompany it as found in its
native state. "Isolate" denotes a degree of separation from original source or
surroundings. "Purify" denotes
a degree of separation that is higher than isolation. A "purified" or
"biologically pure" protein is sufficiently
free of other materials such that any impurities do not materially affect the
biological properties of the
protein or cause other adverse consequences. That is, a nucleic acid or
peptide of the present disclosure is
purified if it is substantially free of cellular material, viral material, or
culture medium when produced by
recombinant DNA techniques, or chemical precursors or other chemicals when
chemically synthesized.
Purity and homogeneity are typically determined using analytical chemistry
techniques, for example,
polyacrylamide gel electrophoresis or high performance liquid chromatography.
The term "purified" can
denote that a nucleic acid or protein gives rise to essentially one band in an
electrophoretic gel. For a protein
that can be subjected to modifications, for example, phosphorylation or
glycosylation, different
modifications can give rise to different isolated proteins, which can be
separately purified.
[00165] The terms "neoplasia" and "cancer" refers to any disease that is
caused by or results in
inappropriately high levels of cell division, inappropriately low levels of
apoptosis, or both. Glioblastoma
is one non-limiting example of a neoplasia or cancer. The terms "cancer" or
"tumor" or "hyperproliferative
disorder" refer to the presence of cells possessing characteristics typical of
cancer-causing cells, such as
uncontrolled proliferation, immortality, metastatic potential, rapid growth
and proliferation rate, and
certain characteristic morphological features. Cancer cells are often in the
form of a tumor, but such cells
can exist alone within an animal, or can be a non-tumorigenic cancer cell,
such as a leukemia cell.
[00166] The term "vaccine" is to be understood as meaning a composition for
generating immunity for
the prophylaxis and/or treatment of diseases (e.g., neoplasia/tumor/infectious
agents/autoiinmune
diseases). Accordingly, vaccines as used herein are medicaments which comprise
recombinant nucleic
acids, or cells comprising and expressing a recombinant nucleic acid and are
intended to be used in humans
or animals for generating specific defense and protective substance by
vaccination. A "vaccine
-22-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
composition" can include a pharmaceutically acceptable excipient, carrier or
diluent. Aspects of the present
disclosure relate to use of the technology in preparing a phagocytic cell-
based vaccine.
[001671 The term "pharmaceutically acceptable" refers to approved or
approvable by a regulatory
agency of the Federal or a state government or listed in the U.S. Pharmacopeia
or other generally
recognized pharmacopeia for use in animals, including humans. A
"pharmaceutically acceptable excipient,
carrier or diluent" refers to an excipient, carrier or diluent that can be
administered to a subject, together
with an agent, and which does not destroy the pharmacological activity thereof
and is nontoxic when
administered in doses sufficient to deliver a therapeutic amount of the agent.
A "pharmaceutically
acceptable salt" of pooled disease specific antigens as recited herein can be
an acid or base salt that is
generally considered in the art to be suitable for use in contact with the
tissues of human beings or animals
without excessive toxicity, irritation, allergic response, or other problem or
complication. Such salts include
mineral and organic acid salts of basic residues such as amines, as well as
alkali or organic salts of acidic
residues such as carboxylic acids. Specific pharmaceutical salts include, but
are not limited to, salts of acids
such as hydrochloric, phosphoric, hydrobromic, malic, glycolic, fumaric,
sulfuric, sulfamic, sulfanilic,
formic, toluene sulfonic, methane sulfonic, benzene sulfonic, ethane
disulfonic, 2-hydroxyethylsulfonic,
nitric, benzoic, 2-acetoxybenzoic, citric, tartaric, lactic, stearic,
salicylic, glutamic, ascorbic, pamoic,
succinic, furnaric, maleic, propionic, hydroxymaleic, hydroiodic,
phenylacetic, alkanoic such as acetic,
HOOC-(CH2)n-COOH where n is 0-4, and the like. Similarly, pharmaceutically
acceptable cations include,
but are not limited to sodium, potassium, calcium, aluminum, lithium and
anrunonium. Those of ordinary
skill in the art will recognize from this disclosure and the knowledge in the
art that further pharmaceutically
acceptable salts for the pooled disease specific antigens provided herein,
including those listed by
Remington's Pharmaceutical Sciences, 17th ed., Mack Publishing Company,
Easton, PA, p. 1418 (1985).
[001681 Nucleic acid molecules useful in the methods of the disclosure include
any nucleic acid
molecule that encodes a polypeptide of the disclosure or a fragment thereof
Such nucleic acid molecules
need not be 100% identical with an endogenous nucleic acid sequence, but will
typically exhibit substantial
identity. Polynucleotides having substantial identity to an endogenous
sequence are typically capable of
hybridizing with at least one strand of a double-stranded nucleic acid
molecule. "Hybridize" refers to when
nucleic acid molecules pair to form a double-stranded molecule between
complementary polynucleotide
sequences, or portions thereof, under various conditions of stringency. For
example, stringent salt
concentration can ordinarily be less than about 750 mM NaCl and 75 mM
trisodium citrate, less than about
500 mM NaC1 and 50 mM trisodium citrate, or less than about 250 mM NaC1 and 25
mM trisodium citrate.
Low stringency hybridization can be obtained in the absence of organic
solvent, e.g., formamide, while
high stringency hybridization can be obtained in the presence of at least
about 35% formamide, or at least
about 50% formamide. Stringent temperature conditions can ordinarily include
temperatures of at least
about 30 C, at least about 37 C, or at least about 42 C. Varying additional
parameters, such as
-23-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
hybridization time, the concentration of detergent, e.g., sodium dodecyl
sulfate (SDS), and the inclusion or
exclusion of carrier DNA, are well known to those skilled in the art. Various
levels of stringency are
accomplished by combining these various conditions as needed. In an exemplary
embodiment,
hybridization can occur at 30 C in 750 mM NaC1, 75 na.M trisodium citrate,
and 1% SDS. In another
exemplary embodiment, hybridization can occur at 37 C in 500 mM NaC1, 50 mM
trisodium citrate, 1%
SDS, 35% formamide, and 100
denatured salmon sperm
DNA (ssDNA). In another exemplary
embodiment, hybridization can occur at 42 C in 250 mM NaCl, 25 mM trisodium
citrate, 1% SDS, 50%
fonrnamide, and 200 pg/m1 ssDNA. Useful variations on these conditions will be
readily apparent to those
skilled in the art. For most applications, washing steps that follow
hybridization can also vary in stringency.
Wash stringency conditions can be defined by salt concentration and by
temperature. As above, wash
stringency can be increased by decreasing salt concentration or by increasing
temperature. For example,
stringent salt concentration for the wash steps can be less than about 30 mM
Na.C1 and 3 mM trisodium
citrate, or less than about 15 m114 NaC1 and 1.5 mM trisodium citrate.
Stringent temperature conditions for
the wash steps can include a temperature of at least about 25 C, of at least
about 42 C, or at least about
68 C. In exemplary embodiments, wash steps can occur at 25 C in 30 mlµel
NaCl, 3 in.M trisodium citrate,
and 0.1% SDS. In other exemplary embodiments, wash steps can occur at 42 C in
15 mM NaCl, 1.5 mM
trisodium citrate, and 0.1% SDS. In another exemplary embodiment, wash steps
can occur at 68 C in 15
mM NaC1, 1.5 mM trisodium citrate, and 0.1% SDS. Additional variations on
these conditions will be
readily apparent to those skilled in the art. Hybridization techniques are
well known to those skilled in the
art and are described, for example, in Benton and Davis (Science 196:180,
1977); Grunstein and Hogness
(Proc. Natl. Acad. Sci., USA 72:3961, 1975); Ausubel et al. (Current Protocols
in Molecular Biology,
Wiley Interscience, New York, 2001); Berger and Kimmel (Guide to Molecular
Cloning Techniques, 1987,
Academic Press, New York); and Sambrook et al., Molecular Cloning: A
Laboratory Manual, Cold Spring
Harbor Laboratory Press, New York.
[00169]
"Substantially identical"
refers to a polypeptide or nucleic acid molecule exhibiting at least
50% identity to a reference amino acid sequence (for example, any one of the
amino acid sequences
described herein) or nucleic acid sequence (for example, any one of the
nucleic acid sequences described
herein). Such a sequence can be at least 60%, 8004 or 85%, 90%, 95%, 96%, 97%,
98%, or even 99% or
more identical at the amino acid level or nucleic acid to the sequence used
for comparison. Sequence
identity is typically measured using sequence analysis software (for example,
Sequence Analysis Software
Package of the Genetics Computer Group, University of Wisconsin Biotechnology
Center, 1710 University
Avenue, Madison, Wis. 53705, BLAST, BESTF1T, GAP, or P1LEUP/PRETTYBOX
programs). Such
software matches identical or similar sequences by assigning degrees of
homology to various substitutions,
deletions, and/or other modifications. Conservative substitutions typically
include substitutions within the
following groups: glycine, alanine; valine, isoleucine, leucine; aspartic
acid, glutatnic acid, asparagine,
-24-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
glutamine; serine, threonine; lysine, arginine; and phenylalanine, tyrosine.
In an exemplary approach to
determining the degree of identity, a BLAST program can be used, with a
probability score between e-3
and e-m indicating a closely related sequence. A "reference" is a standard of
comparison.
[00170] The term "subject" or "patient" refers to an animal which is the
object of treatment, observation,
or experiment. By way of example only, a subject includes, but is not limited
to, a mammal, including, but
not limited to, a human or a non-human mammal, such as a non-human primate,
murine, bovine, equine,
canine, ovine, or feline.
1001711 The terms "treat," "treated," "treating," "treatment," and the like
are meant to refer to reducing,
preventing, or ameliorating a disorder and/or symptoms associated therewith
(e.g., a neoplasia or tumor or
infectious agent or an autoinunune disease). "Treating" can refer to
administration of the therapy to a
subject after the onset, or suspected onset, of a disease (e.g., cancer or
infection by an infectious agent or
an autoimmune disease). "Treating" includes the concepts of "alleviating",
which refers to lessening the
frequency of occurrence or recurrence, or the severity, of any symptoms or
other ill effects related to the
disease and/or the side effects associated with therapy. The term "treating"
also encompasses the concept
of "managing" which refers to reducing the severity of a disease or disorder
in a patient, e.g., extending the
life or prolonging the survivability of a patient with the disease, or
delaying its recurrence, e.g., lengthening
the period of remission in a patient who had suffered from the disease. It is
appreciated that, although not
precluded, treating a disorder or condition does not require that the
disorder, condition, or symptoms
associated therewith be completely eliminated.
[00172] The term "prevent", "preventing", "prevention" and their grammatical
equivalents as used
herein, means avoiding or delaying the onset of symptoms associated with a
disease or condition in a
subject that has not developed such symptoms at the time the administering of
an agent or compound
commences.
[00173] The term "therapeutic effect" refers to some extent of relief of one
or more of the symptoms of
a disorder (e.g., a neoplasia, tumor, or infection by an infectious agent or
an autoimmune disease) or its
associated pathology. "Therapeutically effective amount" as used herein refers
to an amount of an agent
which is effective, upon single or multiple dose administration to the cell or
subject, in prolonging the
survivability of the patient with such a disorder, reducing one or more signs
or symptoms of the disorder,
preventing or delaying, and the like beyond that expected in the absence of
such treatment. "Therapeutically
effective amount" is intended to qualify the amount required to achieve a
therapeutic effect. A physician
or veterinarian having ordinary skill in the art can readily determine and
prescribe the "therapeutically
effective amount" (e.g., ED50) of the pharmaceutical composition required.
[00174] As used herein, the term "affinity molecule" refers to a molecule or a
ligand that binds with
chemical specificity to an affinity acceptor peptide. Chemical specificity is
the ability of a protein's binding
site to bind specific ligands. The fewer ligands a protein can bind, the
greater its specificity. Specificity
-25-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
describes the strength of binding between a given protein and ligand. This
relationship can be described by
a first scFv specific to a cell surface component on a dissociation constant
O(D), which characterizes the
balance between bound and unbound states for the protein-ligand system.
[00175] Reference in the specification to "some embodiments," "an embodiment,"
"one embodiment"
or "other embodiments" means that a feature, structure, or characteristic
described in connection with the
embodiments is included in at least some embodiments, but not necessarily all
embodiments, of the present
disclosure.
1001761 The term "myeloid cells" refers to normal or neoplastic cells found in
the blood, bone marrow,
other hematopoietic or other non-hematopoietic compartments of the body. In
particular, the term "myeloid
cells" is used herein to mean the cell lineage originating from the bone
marrow that includes
polymotphonuclear neutrophils, eosinophils, basophils, and mast cells, as well
as the
monocyte/macrophage lineage and different dendritic cell lineages. Myeloid
cells are not capable of
differentiating into lymphoid cells (e.g., NKr, B- and T-lymphocytes). The
term refers to cells of the
myeloid lineages in all stages of their differentiation and therefore includes
hematopoietic blast cells, i.e.,
hematopoietic cells that are committed to the myeloid cell lineage, but that
are in early stages of
differentiation. When stimulated with appropriate growth factors,
hematopoietic blast cells divide to
produce a large number of cells that are more differentiated than the blast
stage of differentiation. Examples
are inter alia myeloblasts. Although monocytes or macrophages are exemplified
throughout the
specification, the compositions and methods described here are applicable to
cells of a myeloid cell lineage,
such as a dendritic cell. Minor optimizations and changes are envisioned on a
cell to cell basis as is known
to one of skill in the art, and is contemplated within the scope of the
invention.
[00177] Cells that are more differentiated than blasts but not yet fully
differentiated are appended with
the prefix "pro" and are also intended to fall under the definition of
"myeloid cells." Examples are
promyelocytes.
[00178] The term "myeloid cells" also includes myeloid
progenitor cells, i.e., cell lineages, e.g., in the
bone marrow, that are capable of differentiating in cells such as
myelomonocytic progenitor cells,
proerythroblasts or immature megakaryoblasts. Myeloid progenitor cells are not
capable of giving rise to
lymphoid cells.
[00179] The term "myeloid cells" does not include lympho-hematopoietic stem
cells. Lympho-
hematopoietic stem cells are defined as those cells that are capable of both
self-renewal and differentiation
into the two principle precursor components, the myeloid and lymphoid lines.
Such stem cells are said to
be totipotent. Stem cells that are less general but that can still
differentiate into several lines are called
pluripotent.
-26-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00180] The term "monocyte or macrophage specific engagers" applies to not
only monocyte or
macrophage cells, but to all myeloid cells, and therefore a monocyte or
macrophage specific engager is
similar to a "myeloid cell specific engager."
[00181] Phagocytes are the natural sentinels of the immune system and form the
first line of defense in
the body. They engulf a pathogen, a pathogen infected cell a foreign body or a
cancerous cell and remove
it from the body. Most potential pathogens are rapidly neutralized by this
system before they can cause, for
example, a noticeable infection. This can involve receptor-mediated uptake
through the clathrin-coated pit
system, pinocytosis, particularly macropinocytosis as a consequence of
membrane ruffling and
phagocytosis. The phagocytes therefore can be activated by a variety of non-
self (and self) elements and
exhibit a level of plasticity in recognition of their "targets". Most
phagocytes express scavenger receptors
on their surface which are pattern recognition molecules and can bind to a
wide range of foreign particles
as well as dead cell, debris and unwanted particles within the body.
[00182] Myeloid cells, such as, monocytes and macrophages are also one of the
most abundant cell
types in the site of an infection, inflammation or in a tumor. Therefore,
monocytes or macrophages can be
attractive cell therapy vehicles. Provided herein are mechanisms to modify a
monocyte or macrophage or
a phagocytic cell to enhance phagocytic killing of a diseased cell, such as a
tumor or an infected cell.
[00183] Although a monocyte/macrophage is described in detail in the
disclosure the composition and
methods can be applicable for use in any phagocytic cell type, or applicable
towards myeloid cell types
including and not limited to neutrophil and dendritic cells with minor
optimizations if applicable, as is
known to one of skill in the art. Likewise, although cancer is described in
detail as the indication for a
myeloid cell therapy in the disclosure, the composition and methods can be
made applicable to infections
and autoimmune conditions, with minor modifications as deemed necessary by a
person of skill in the art.
[00184] Phagocytosis, defined as the cellular uptake of particulates (>0.5 Um)
within a plasma-
membrane envelope, is closely relate to and partly overlaps the endocytosis of
soluble ligands by fluid-
phase macropinocytic and receptor pathways. Variants associated with the
uptake of apoptotic cells, also
known as efferocytosis, and that of necrotic cells arising from infection and
inflammation (necroptosis and
pyroptosis). The uptake of exogenous particles (heterophagy) has features in
common with autophagy, an
endogenous process of sequestration and lysosomal disposal of damaged
intracellular organelles. Uptake
mechanisms vary depending on the particle size, multiplicity of receptor-
ligand interactions, and
involvement of the cytoskeleton. Once internalized, the phagosome vacuole can
fuse selectively with
primary lysosomes, or the product of the endoplasmic reticulum (ER) and Golgi
complex, to form a
secondary phagolysosome. This pathway is dynamic in that it undergoes fusion
and fission with endocytic
and secretory vesicles, macrophages, DCs, osteoclasts, and eosinophils. Anti-
microbe phagocytosis clears
and degrades disease-causing microbes, induces pro-inflammatory signaling
through cytokine and
chemokine secretion, and recruits immune cells to mount an effective
inflammatory response. This type of
-27-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
phagocytosis is often referred to as "inflammatory phagocytosis" (or
"immunogenic phagocytosis").
However, in some instances, such as with certain persistent infections, anti-
inflammatory responses may
follow microbial uptake. Anti-microbe phagocytosis is commonly performed by
professional phagocytes
of the myeloid lineage, such as immature dendritic cells (DCs) and monocytes
or macrophages and by
tissue-resident immune cells. Phagocytosis of damaged, self-derived apoptotic
cells or cell debris (e.g.,
efferocytosis), in contrast, is typically a non-inflammatory (also referred to
as a "nonimmunogenic")
process. Billions of damaged, dying, and unwanted cells undergo apoptosis each
day. Unwanted cells
include, for example, excess cells generated during development, senescent
cells, infected cells
(intracellular bacteria or viruses), transformed or malignant cells, and cells
irreversibly damaged by
cytotoxic agents.
[00185] The bone marrow is the source of circulating neutrophils and monocytes
that will replace
selected tissue-resident monocytes or macrophages and amplify tissue myeloid
populations during
inflammation and infection. After phagocytosis, newly recruited monocytes and
tissue macrophages
secrete their products by generating them from pre-existing phospholipids and
arachidonates in the plasma
membrane and by releasing radicals generated by activation of a respiratory
burst or induction of inducible
nitric oxide synthesis; apart from being achieved by synthesis of the low-
molecular-weight products
(arachidonate metabolites, superoxide anions, and nitric oxide) generated as
above, secretion induced by
phagocytosis in monocytes or macrophages is mainly achieved by new synthesis
of RNA and changes in
pH, resulting in progressive acidification. Highly phagocytic macrophages
appear to be MARCO+
SignR1+ and are found in the outer marginal zone rapidly clear capsulated
bacteria. Similar CD169+ F4/80-
macrophages line the subcapsular sinus in lymph nodes and have been implicated
in virus infection. It was
noted that endothelial macrophages, including Kupffer cells in the liver,
clear microbial and antigenic
ligands from blood and lymph nodes to provide a sinusoidal immune function
comparable to but distinct
from mucosal immunity. Not all tissue macrophages are constitutively
phagocytic, even though they still
express typical macrophage markers. In the marginal zone of the rodent spleen,
metallophilic macrophages,
which lack F4/80, strongly express CD169, sialic acid-binding immunoglobulin
(Ig)-like lectin 1
(SIGLEC1 [sialoadhesin]}, but are poorly phagocytic. Non-professional
phagocytes include epithelial cells,
and fibroblasts. Fibroblasts are "working-class phagocytes" clear apoptotic
debris by using integrins other
than CD11b-CD18 through adhesion molecules ICAM and vitronectin receptors.
Astrocytes have also been
reported to engulf, even if not efficiently degrade, apoptotic corpses. Plasma-
membrane receptors relevant
to phagocytosis can be opsonic, FeRs (activating or inhibitory) for mainly the
conserved domain of IgG
antibodies, and complement receptors, such as CR3 for iC3b deposited by
classical (IgNI or IgG) or
alternative lectin pathways of complement activation. CR3 can also mediate
recognition in the absence of
opsonins, perhaps by depositing macrophage-derived complement. Anti-microbe
phagocytosis is
-28-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
commonly performed by professional phagocytes of the myeloid lineage, such as
immature dendritic cells
(DCs) and macrophages and by tissue-resident immune cells.
[00186] In cancer, monocytes, attracted by numerous factors including CCL2,
ATP, etc., migrate into
the tumor microenvironment. However, a majority of these monocytes can then
differentiate into tumor
associated monocytes or macrophages and or myeloid suppressor cells. In order
to generate monocytes or
macrophages and myeloid cells that are potent in killing tumor cells, as
opposed to being myeloid
suppressor cells and tumor associated monocytes or macrophages, the present
composition provide means
for enhancing phagocytosis of the tumor cells by resident monocytes or
macrophages, and also mount a
successful and stable immune response.
[00187] Investigations on monocyte or macrophage function in a tumor
environment indicated that at
least two signals are required for the activation of monocytes or macrophages.
The first signal (signal 1) is
mediated via phagocytosis/tethering receptors and the second signal (signal 2)
by danger signals such as
pathogen-associated molecular patterns (DAMPs), or cytokines that trigger
nuclear factor-KB (NF-KB)-
mediated upregulation of inflanunatory genes (FIG. 1A). Triggering
phagocytosis alone may be
insufficient to activate monocytes or macrophages and in the context of
harnessing macrophages to kill
cancer, as it is insufficient to drive an anti-tumor response with a
phagocytosis triggering signal alone
generated by binding to a cancer cell.
[00188] Whereas a cancer cell or a tumor cell is repeatedly referred here as
the target cell, the concepts
described here are suitable for any type of a target cell, such as an infected
cell, or a specific disease cell
type that needs to be eliminated by phagocytosis, as long as the binding
domain for a cell surface
component of a cancer cell is suitably replaced by a binding domain for a cell
surface component of the
specific for the target cell.
[00189] The present disclosure is based on a number of endeavors that address
effective ways to trigger
a myeloid cell to mount a strong response to a target cell, for example, a
cancer cell or tumor cell, such that
the myeloid cell destroys the target upon contact, as well as trigger an
immune response that activates other
immune cells, for example, T lymphocytes, B lymphocytes and NK cells. One
aspect of the endeavor is to
generate therapeutically effective myeloid cells in the patient, in situ. In
another aspect, therapeutic myeloid
cells are generated ex vivo, and introduced into a patient in need thereof.
[00190] In one aspect, the disclosure provides one or more synthetic or
recombinant biomolecules, such
as proteins or polypeptides, that are capable of binding to and activating a
myeloid cell to trigger phagocytic
killing and immune response against a target cell, such as a tumor cell. In
some embodiments, the synthetic
or recombinant biomolecule can bind (a) on one hand, a cell surface molecule
(i.e. and antigen) on a
myeloid cell, and on the other hand (b) a cell surface molecule (i.e. and
antigen) on a target cell, thereby
effectively, at least, bringing the two cells (an effector and a target), in
close proximity, such that other
cellular receptors and membrane components on either cell can interact and the
effector myeloid cell can
-29-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
thereby trigger engulfment of the target cell. A simplified graphical
representation is depicted in FIG. 1B.
Structurally, such a molecule would have two arms, one specific for each cell
surface molecule, for
example, a first binding domain (e.g, A), and a second binding domain (e.g.,
B) connected by a linker (e.g.
L) (FIG. 1B). Such synthetic or recombinant biomolecules can be called
bispecific engagers, or, bispecific
myeloid cell engagers, or BiMEs. In one or more embodiments, the bispecific
engagers comprise two
antigen binding domains ("binders"). One of the two binders is designed to
bind to an antigen on the surface
of an effector myeloid cell; the other is designed to bind to an antigen on a
target cell. In some embodiments
the antigen binding domains are antibodies or fragments thereof In some
embodiments, a binder may be a
ligand, binding to a receptor on a cell surface, such as a receptor on a
myeloid cell or on a target cell.
[00191] In one aspect, the present disclosure provides a therapeutic
composition comprising one or
more synthetic or recombinant biomolecules, such as proteins or polypeptides,
that are capable of binding
to and activating a myeloid cell to trigger phagocytic killing and immune
response ,iginst a target cell,
such as a cancer cell, and the synthetic or recombinant biomolecule comprises
more than two binders.
Accordingly, in some embodiments, provided herein is a first therapeutic
agent, wherein the therapeutic
agent comprises: a first binding domain (or, a first binder), wherein the
first binding domain may be a first
antibody Of functional fragment thereof that specifically interacts with an
antigen or a surface molecule on
a target cell, and a second binding domain (or, a second binder), wherein the
second binding domain may
a second antibody or functional fragment thereof that specifically interacts
with a myeloid cell. In one or
more embodiments, the first therapeutic agent is coupled to a first component
such as a linker or another
bioactive peptide that may offer a third binding domain; or an activator
molecule, or an additional
therapeutic agent. In some embodiments, the composition comprises an
additional therapeutic agent.
[00192] In one aspect, the disclosure provides one or more synthetic or
recombinant biomolecules, such
as proteins or polypeptides, that are capable of binding to and activating a
myeloid cell to trigger phagocytic
killing and immune response against a target cell, such as a cancer cell, and
the synthetic or recombinant
biomolecule comprises more than two binders. In one embodiment, the
recombinant biomolecule
comprises three binders each of exhibit specific binding to a surface
molecule, and therefore the
recombinant biomolecule can exhibit binding to three elements on two or more
cells. In one embodiment,
the recombinant biomolecule having three binders is capable of binding to more
than one antigens on a
myeloid cell or on a target cell. A recombinant biomolecule as described here,
having three binders is
termed a trispecific myeloid cell engager (TriME). In some embodiments, a
TriME may bind to, or engage
three different cells, for example, a myeloid cell, a target cell such as a
cancer cell, and a third cell, or a
target. In some embodiments, the BiME or TriME may engage more than one
antigens or surface molecules
on either a myeloid cell or on a cancer cell that either activates the myeloid
cell or inhibits a function of a
cancer cell, such as engaging to and inducing tolerance or inununosuppression
on a myeloid cell. In some
embodiments, a bispecific, trispecific or a multispecific engager may comprise
a second trigger, i.e., a
-30-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
second signal that not only induces phagocytosis of the target cell by the
myeloid cell, but also initiates an
immune response or inflammatory response that activates other inurtune cells
for a prolonged response and
generation of immunological memory. In some embodiments, a bispecific,
trispecific or a multispecific
engager is a chimeric molecule. Accordingly, provided herein is a composition
comprising a therapeutic
agent, wherein the therapeutic agent comprises: (a) a first binding domain
that specifically interacts with
an antigen of a target cell, (14 a second binding domain that specifically
interacts with a myeloid cell, and
(c) a third binding domain that specifically interacts with the myeloid cell.
In some embodiments, the
composition comprises an additional therapeutic agent. In some embodiments, a
binder may also be an
activator of a receptor, such as a scavenger receptor, or a TLR receptor. In
some embodiments, a binder
may be an inhibitor or blocker of a virulent agent on a pathogenic target, or
an immune suppressor molecule
on a pathogen cell or a tumor cell. In some embodiments, a binding domain or a
binder or any part of an
engager that may perform a function as described above may be a therapeutic
element of the binder. In
some embodiments, the engager may comprise one or more therapeutic agents. In
some embodiments, the
therapeutic composition may comprise one or more engagers, and one or more
therapeutic agents, such as
a separate recombinant protein, or nucleic acid encoding the same, a
pharmaceutical product or a small
molecule.
[00193] In some aspects as described herein, a second therapeutic agent may be
required_ A second
therapeutic agent may be a second recombinant protein. In some embodiments,
the second therapeutic
agent can suppress a tumor-mediated iinmunosuppressor. In some embodiments,
the second therapeutic
agent is necessary for evading a myeloid cell suppressor function, or a
tolerogenic response on myeloid
cells. In some embodiments, the second therapeutic is necessary to evade the
anti-phagocytic, "don't-eat-
me" signals by a tumor cell towards a phagocyte. For example, the second
therapeutic may comprise a
CD47 antagonist, a CD47 blocker, an antibody, a chimeric CD47 receptor, a
sialidase, a cytokine, a
proinflammatory gene, a procaspase, or an anti-cancer agent. In some
embodiments, the second therapeutic
agent can provide the second signal for the phagocytic cell mediated immune
response.
[00194] Using the methods and compositions described herein, a myeloid cell
can be directed to activate
the immune response cycle irrespective of the effects in a tumor
microenvironment. A phagocytic cell can
be directed to phagocytose and kill a target cell, and activate the immune
response sequelae that generates
successful and sustained adaptive immune response and immunological memory
against the target.
[00195] In the following section, compositions comprising therapeutic agents
are described.
FIRST THERAPEUTIC AGENT
1001961 In one aspect, a first therapeutic agent is described, wherein the
first therapeutic agent
comprises: (i) a first antigen binding domain that specifically interacts with
an antigen of a target cell, and
(ii) a second antigen binding domain that specifically interacts with an
extracellular region of a receptor of
a myeloid cell, such as a monocyte or a macrophage cell. The first therapeutic
agent is a recombinant
-31-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
chimeric protein, which can bind to at least a target cell, such as a tumor
cell, and a monocyte or
macrophage cell and attach the two cell types to facilitate phagocytosis of
the cancer cell by the monocyte
or macrophage. In some embodiments, the first therapeutic agent is a chimeric
hi- or trispecific engager.
In some embodiments, the first therapeutic agent is coupled to a first
component, wherein the first
component is an additional therapeutic agent or a third binding domain. An
additional therapeutic agent
may be a peptide, a protein, a conjugated protein, an antibody, a functional
derivative of an antibody such
as an scFv, a ligand, a receptor or a functional fragment thereof for a
ligand, or a small molecule. In several
examples in the preceding sentence, the first therapeutic agent is coupled to
a first component, wherein the
first component is a binding element that can associate with a cell surface
component of the target cell or
the monocyte or macrophage cell.
[00197] In some embodiments, the first therapeutic agent comprises an
additional therapeutic agent. An
additional therapeutic agent as described herein can be a small molecule. In
some embodiments, the
additional therapeutic agent is a peptide binding domain. In some embodiments,
the additional therapeutic
agent is a cell surface binding domain. In some embodiments, the additional
therapeutic agent is a target
cell binding domain. In some embodiments, the additional therapeutic agent may
be an antibody, a
functional derivative of an antibody, such as an scFv. In some embodiments,
the additional therapeutic
agent is a ligand, a peptide. In some embodiments, the additional therapeutic
agent is a protein, a conjugated
protein, a receptor or a functional fragment thereof for a ligand. In some
embodiments, the additional
therapeutic agent is an inhibitor of the myeloid cell, e.g., the monocyte or
macrophage cell mediated by the
target cell.
1001981 In some embodiments, the therapeutic agent is a recombinant protein.
The therapeutic agent as
described herein is a recombinant protein that not only binds a tumor or a
cancer cell and a monocyte or
macrophage thereby providing a first signal (signal 1) for triggering
phagocytosis of the tumor cell by the
monocyte or macrophage, but also provides a second signal (signal 2) to
enhance phagocytic killing by the
monocyte or macrophage.
[00199] In one embodiment the first therapeutic agent is an extracellular
protein.
[00200] In some embodiments, the first therapeutic agent is a secreted
protein.
[00201] In some embodiments, the first therapeutic agent is encoded by a
recombinant nucleic acid
encoding one or more nucleic acid sequences encoding a first antigen binding
domain that specifically
interacts with an antigen of a target cell, and (ii) a second antigen binding
domain that specifically interacts
with an extracellular region of a receptor of a monocyte or macrophage cell.
[00202] In some embodiments, the first therapeutic agent is encoded by a
vector expressing a
recombinant nucleic acid encoding one or more polypeptides comprising a first
antigen binding domain
that specifically interacts with an antigen of a target cell, and (ii) a
second antigen binding domain that
specifically interacts with an extracellular region of a receptor of a myeloid
cell.
-32-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00203] In some embodiments, a binder is selected on the basis of its binding
specificity to a its target
or cognate element. A binding domain may be derived from a protein that is an
antibody or a functional
fragment thereof, that binds to the target antigen or the cognate molecule.
The binding domain is one that
has high specificity, high binding affinity or both, towards its target.
[00204] In some embodiments, the binding affinity to its target or cognate
molecule is 10-8 M or less,
le NI or less 1 0-1 M or less or 14Y11 M or less, 1 0-12M or less. In some
embodiments, the binding domain
may further be modified to increase its binding specificity or binding
affinity or both. One of skill in the
art can use existing technology to enhance the binding properties of a binder
region, and such modifications
are contemplated within the scope of this disclosure.
Bi- and Trisnecific Monocvte or Macrooliate Enza2ers
A. Binding Target Cell and Effector Cell
[00205] Provided herein are recombinant bi- and trispecific engagers designed
to anchor a target cell
with an effector cell, such that the effector cell attack the target cell, and
kill the specific target cell. In
some embodiments, the effector cell is a myeloid cell. In some embodiments,
the myeloid cell is a monocyte
or macrophage cell. In some embodiments, the target cell is a cancer cell.
[00206] While cancer is one exemplary embodiment described in exclusive detail
in the instant
disclosure, the methods and technologies described herein are contemplated to
be useful in targeting an
infected or otherwise diseased cell inside the body.
[00207] In some embodiments, the present disclosure provides compositions and
methods for cancer
immunotherapy. The methods provided herein help design tools that can induce
resident human monocytes
or macrophages to become efficient killer cells that target cancer cells and
eliminate them by efficient
phagocytosis. In some embodiments, the monocytes or macrophages provide
sustained immunological
response against the cancer cell. Various embodiments are described herein.
[00208] Provided herein are specific constructs and designs are disclosed for
such chimeric proteins,
termed chimeric "engagers".
[00209] In some embodiments, the chimeric engagers comprise two or more fused
antibodies, each
having a specific binding region on the target cell, such as cancer cell or on
the monocyte or macrophage.
In certain embodiments, the two or more fused antibodies or the inununofusion
comprises a target binding
domain operably linked by a hinge-CH2-CH3 domain or a hinge-CH3 domain of an
inununoglobulin
constant region to an effector binding domain that specifically binds a cell
surface component of the
monocyte or macrophage.
[00210] In one aspect the chimeric protein is a bispecific monocyte or
macrophage engager.
[00211] In some embodiments, a bispecific engager comprises a first
therapeutic agent, wherein the
first therapeutic agent comprises: (i) a first antigen binding domain that
specifically interacts with an
antigen of a target cell, and (ii) a second antigen binding domain that
specifically interacts with an
-33-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
extracellular region of a receptor of a monocyte or macrophage cell. In one
embodiment, the therapeutic
agent is a bispecific engager. In one embodiment, the bispecific monocyte or
macrophage engager
comprises two antibody single chain variable regions (scFv) only (no Fc amino
acid segments were
included) with a flexible linker, one scFv binds a cell surface component of a
target cell and the other binds
a receptor on monocyte or macrophage cell surface. In full unmodified forms of
IgG, the variable light
chain domain (VL ) and the variable heavy chain domain (VH) are separate
polypeptide chains, i.e., are
located in the light chain and heavy chain, respectively. Interaction of the
antibody light chain and an
antibody heavy chain, in particular the interaction of the VL and VH domains,
one of the epitope binding
site of the antibody is formed. In contrast, in the scFv construct, but VL and
Vu domains of antibodies are
included in a single polypeptide chain. The two domains are separated by
flexible linkers long enough to
allow self-assembly of the VL and VH domains into functional epitope binding
site.
[00212] In some embodiments, a bispecific monocyte or macrophage engager
comprises: (a) a single
chain variable fragment (scFv) that binds to a cell surface component of a
target cell, e.g., a cancer antigen,
(b) a single chain variable fragment (scFv) that binds to a cell surface
component of an effector cell, e.g.
the monocyte or macrophage, (c) a short linker operably linking (a) and (b).
In some embodiments, the
scFvs are fused at their C-termini. Each scFv comprises a light chain variable
domain, and a heavy chain
variable domain, operably linked by a peptide linker. In certain embodiments,
the scFvs are humanized.
Humanized scFvs comprise "complementarity determining regions" (CDR) that are
present on a
framework of an inrununoglobulin of a different species as compared to that of
the parent immunoglobulin
from which the CDR was derived. For example, a murine CDR may be grafted into
the framework region
of a human antibody to prepare the "humanized antibody." The design of an
exemplary bispecific engager
comprising two scFvs can be represented by the simplified formula
NH2-[Target cell binding scFv]-COOH ¨ [Linker] ¨COOH- [Effector cell binding
scFv]-N}[2 [I]
[00213] In some embodiments, the bispecific engager is a diabody. The
bispecific diabody is
constructed with a VL and a Vii domain on a single polypeptide chain have
binding specificities to different
(non-identical) epitopes. Additionally, the linker connecting VL and VH is
shorter than 12 amino acid in
length that is insufficient for reassembly into a functional epitope.
Generally, one polypeptide chain
construct comprises VL having binding specificity to a first antigen and VII
having binding specificity to a
second antigen, and another polypeptide chain construct comprises VL having
binding specificity to the
second antigen and VII having binding specificity to the first antigen; the
two polypeptide chains are
allowed to self-assemble into a hi-specific diabody. In some embodiments, a
cysteine residue may be
introduced at the C terminus of the construct that can allow disulfide bond
formation between two chains
without interfering with the binding properties of the
-34-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
1002141 In some embodiments, the bispecific engager is a tandem-di-scFv.
[00215] In some embodiments, recombinant nucleic acid constructs can be
prepared encoding the
bispecific scFv engager. The recombinant nucleic acid constructs for
expressing a bispecific scFv engager
comprises one or more polypeptides encoding (a) a nucleic acid sequence
encoding a variable domain of
the target cell binding scFv light chain, a linker, a variable domain of the
target cell binding scFv heavy
chain; (b) a nucleic acid sequence encoding a linker; (c) a nucleic acid
sequence encoding a variable domain
of the effector (monocyte or macrophage) cell binding scFv light chain, a
linker, a variable domain of the
effector (monocyte or macrophage) cell binding scFv heavy chain. In some
embodiments, the nucleic acid
constructs for expressing a bispecific scFv engager comprises an N-terminal
signal peptide sequence for
secretion of the bispecific scFv engager.
1002161 In some embodiments, a bispecific engager comprises two single domain
antibodies (Vim)
operably linked with a flexible linker, one Viai binds a cell surface
component of a target cell, and the other
V1111 binds a receptor on a monocyte or macrophage cell surface. In some
embodiments, a chimeric
bispecific monocyte or macrophage engager comprises: (a) a Vim domain that
binds to a cell surface
component of a target cell, e.g., a cancer antigen, (b) a Vim domain that
binds to a cell surface component
of an effector cell, e.g. the monocyte or macrophage, (c) a short linker
operably linking (a) and (b). In some
embodiments the engager comprising two single domain antibodies is a nanobody.
The design of an
exemplary bispecific engiager comprising two Vim domains can be represented by
the simplified formula:
NH24Target cell binding single domainkCOOH - [Linker] -0001-1- [Effector cell
binding single
domain] NH2
[III
[00217] In some embodiments, the short linker operably linking (a) and (b) may
further have additional
functions. In some embodiments, the peptides can bind to a specific cell
surface receptor, such as, for
example, a TLR receptor, and can activate a receptor mediated cell signaling
pathway in the monocyte or
macrophage cell. In some embodiments, the linker is designed such as to be
able to bind and activate at
least an inflammatory pathway in the monocyte or macrophage cell, or
potentiate monocyte or macrophage
mediated phagocytosis and killing of a target cell. In some embodiments, the
linker peptide may have a
function of blocking or inhibiting a target cell mediated downregulation of a
monocyte or macrophage cell
function.
[00218] In some embodiments, nucleic acid constructs for a bispecific VH:F{
engager can be generated,
which comprises: (a) a nucleic acid sequence encoding a (a) a Vim domain that
binds to a cell surface
component of a target cell, e.g., a cancer antigen, (b) a Vim domain that
binds to a cell surface component
of an effector cell, e.g. the monocyte or macrophage, (c) a short linker
operably linking (a) and (b). In some
-35-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
embodiments, the nucleic acid constructs for expressing a bispecific scFv
engager comprises an N-terminal
signal peptide sequence for secretion of the bispecific scFv engager.
[00219] As is known to one of skill in the art, the nucleic acid sequences
encoding the polypeptides
comprising the Vim or scFv binding domains can be inserted in a suitable
expression vector under one or
more promoters, e.g. CMV at the 5'end, and a polyadenylation signal at the 3'-
end of the sequences
encoding the polypeptides.
[00220] In some embodiments, the constructs may comprise internal ribosomal
entry site (IRES), e.g.,
a nucleic acid sequences encoding one or more polypeptides may be preceded by
an IRES.
[00221] In some embodiments, the nucleic acid sequences encoding one of the
polypeptides may be
placed under a separate promoter control than the remaining of the expressed
sequences.
[00222] In some embodiments, a bispecific engager may further comprise an
antibody or a fragment
thereof that binds to a cell surface component of a target cell, and an
antibody or a fragment thereof that
binds to a cell surface component of an effector cell.
[00223] Provided herein are further variations of an engager, a trispecific
engager. A trispecific engager
comprises a first therapeutic agent, wherein the first therapeutic agent
comprises: a first antigen binding
domain that specifically interacts with an antigen of a target cell; a second
antigen binding domain that
specifically interacts with an extracellular region of a first receptor of a
monocyte or macrophage cell; and
a third antigen binding domain that specifically interacts with an
extracellular region of a second receptor
of the monocyte or macrophage cell.
[00224] In some embodiments, the trispecific engager is a fused construct of
three scFvs, comprising a
first scFv specific to a cell surface component on a target cancer cell, a
second scFv specific to a cell surface
component on the monocyte or macrophage, for example, the chimeric phagocytic
receptor, and a third
scFv specific to another cell surface component on the monocyte or macrophage.
In some embodiments,
the trispecific engager is designed such that the cell surface component on
the monocyte or macrophage to
which the third scFv can bind, provides an additional activation signal for
the monocyte or macrophage to
trigger phagocytosis and killing of the target cell. In some embodiments the
third scFv binds to another
phagocytic receptor on the monocyte or macrophage. In some embodiments the
third scFv binds to a danger
associated monocyte or macrophage signaling pathway (DAMP). In some
embodiments, the third scFv
binds to a TLR receptor. In some embodiments, the third scFv binds to a
cytokine receptor which activates
the receptor and triggers monocyte or macrophage intracellular signaling In
some embodiments, the third
scFv binds to a monocyte or macrophage receptor known to generate a
phagocytosis inhibitory signal and
that binding of the third scFv to the receptor blocks the receptor, enabling
enhanced phagocytosis. In some
embodiments, the third scFv binds to a receptor that engages with one or more
transmembrane domains
and enhances phagocytic signaling. Various designs of trispecific engagers
have been contemplated herein,
-36-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
of which an exemplary trispecific engager comprising two scFvs can be
represented by the simplified
formulae:
(i) NH2-[Target cell binding scFv]-COOH- [Linker] -COOH- [Effector cell
binding first scFv]-NH2
[Linker] -COOH- [Effector cell binding second scFv1-
NH2;
[III]
OR
(ii) NH2-[Target cell binding scFv]-COOH- [Linker] -COOH- [Effector cell
binding first scFv]-NH2-
[Linker] - COOH- [Effector cell binding second sell-N112.
[IV]
[00225] In some embodiments, each of the three binding domains of the
trispecific engager comprises
the antigen binding domain of an antibody, a functional fragment of an
antibody, a variable domain thereof,
a Vii domain, a VI, domain, a VNAR domain, a VHF{ domain, a single chain
variable fragment (scFv), an
Fab, a single-domain antibody (sdAb), a nanobody, a bispecific antibody, a
diabody, or a functional
fragment or a combination thereof
[00226] In some embodiments, the binding domains of the trispecific engager
are operably linked by
one or more peptide linkers. In some embodiments, the one or more peptide
linkers may be functional
peptides that can bind to a specific cell surface receptor, such as, for
example, a TLR receptor, and can
activate a receptor mediated cell signaling pathway in the monocyte or
macrophage cell. In some
embodiments, the linker is designed such as to be able to bind and activate at
least an inflammatory pathway
in the monocyte or macrophage cell, or potentiate monocyte or macrophage
mediated phagocytosis and
killing of a target cell. In some embodiments, the linker peptide may have a
function of blocking or
inhibiting a target cell mediated downregulation of a monocyte or macrophage
cell function.
[00227] In some embodiments, a nucleic acid constructs encoding a trispecific
engager comprises one
or more nucleic acid encoding (a) a polypeptide comprising an scFv domain that
binds to a cell surface
component of a target cell, e.g., a cancer antigen, (b) a polypeptide
comprising an scFv domain that binds
to a first cell surface component of an effector cell, e.g. the monocyte or
macrophage, (c) a polypeptide
comprising an scFv domain that binds to a second cell surface component of the
monocyte or macrophage,
for example, the chimeric construct constituting the second therapeutic agent;
or a native monocyte or
macrophage cell surface receptor, wherein each of the polypeptides are
operably linked to one another. In
some embodiments, a nucleic acid constructs encoding a trispecific engager
comprises one or more nucleic
acid encoding (a) a polypeptide comprising a VIM domain that binds to a cell
surface component of a target
-37-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
cell, e.g., a cancer antigen, (b) a polypeptide comprising a VIM domain that
binds to a first cell surface
component of an effector cell, e.g. the monocyte or macrophage, (c) a
polypeptide comprising a Vim
domain that binds to a second cell surface component of the monocyte or
macrophage. In some
embodiments, the nucleic acid constructs for expressing a bispecific scFv
engager comprises an N-terminal
signal peptide sequence for secretion of the bispecific scEv engager. As
contemplated herein, a skilled
artisan can exchange the scEv or Vial binding sequences with a nucleic acid
sequence of a short peptide
encoding any suitable target region binding element. In some embodiments, the
polypeptide constructs are
encoded in a monocistronic construct. In some embodiments, the polypeptide
constructs are encoded in a
polycistronic construct. In some embodiments, one or more nucleic acid
sequences encoding short linker
polypeptides are inserted in between sequences encoding two polypeptides. In
some embodiments, the
expression of the nucleic acid sequence encoding each polypeptide is driven by
a separate promoter. In
some embodiments, the nucleic acid sequence encoding each polypeptide is
driven by a single promoter.
In some embodiments one or more 1RES sequences are introduced into the
construct
[002281 In some embodiments, one or more polypeptides may be expressed
separately within a cell,
and which may assemble post-translationally.
[002291 In some embodiments, polypeptides may be designed to assemble on
special peptide scaffolds
upon secretion outside the cell.
[002301 In some embodiments, the hi- or trispecific engagers bind to an
antigen on a cancer cell,
selected from the group consisting of Thymidine Kinase (TK1), Hypoxanthine-
Guanine
Phosphoribosyltransferase (HPRT), Receptor Tyrosine Kinase-Like Orphan
Receptor 1 (ROR1), Mucin-
1, Mucin-16 (MUC16), MUC1, Epidermal Growth Factor Receptor vlII (EGFRv[II),
Mesothelin, Human
Epidermal Growth Factor Receptor 2 (ITER2), Mesothelin, EBNA-1, LEMD1,
Phosphatidyl Serine,
Carcinoembryonic Antigen (CEA), B-Cell Maturation Antigen (BCMA), Cdypican 3
(GPC3), Follicular
Stimulating Hormone receptor, Fibroblast Activation Protein (FAT'),
Erythropoietin-Producing
Hepatocellular Carcinoma A2 (EphA2), EphB2, a Natural Killer Group 2D (NKG2D)
ligand,
Disialoganglioside 2 (GD2), CD2, CD3, CD4, CD5, CD7, CD8, CD19, CD20, CD22,
CD24, CD30, CD33,
CD38, CD44v6, CD45, CD56CD79b, CD97, CD1I7, CD123, CD133, CD138, CD171,
CD179a,
CD213A2, CD248, CD276, PSCA, CS-1, CLECL1, GD3, PSMA, FLT3, TAG72, EPCAM, IL-
1, an
integrin receptor, PRSS21, VEGFR.2, PDGFR-beta, SSEA-4, EGER, NCAM, prostase,
PAP, ELF2M,
GM3, TEM7R, CLDN6, TSHR,, GPRC5D, ALK, IGLL1 and combinations thereof In some
embodiments,
for example, the cancer antigen for a target cancer cell can be one or more of
the mutated/cancer antigens:
MUC16, CCAT2, CTAG1A, CTAG1B, MAGE Al, MAGrEA2, MAGEA3, MAGE A4, MAGEA6,
FRAME, PCA3, MACE Cl, MAGEC2, MAGED2, AFP, MAGEA8, MAGE9, MAGEA1 1, MAGEA12,
11,13RA2, PLAC1, SDCCAG8, LSP1, CT45A1, CT45A2, CT45A3, CT45A5, CT45A6,
CT45A8,
-38-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
CT45A10, CT47A1, CT47A2, CT47A3, CT47A4, CT47A5, CT47A6, CT47A8, CT47A9,
CT47A10,
CT47A1 1, CT47Al2, CT47B1, SAGE1, and CT55.
[00231] In some embodiments, the antigen on a cancer cell is selected from the
group consisting of
CD2, CD3, CD4, CD5, CD7, CCR4, CDS, CD3O, CD45, CD56µ
[00232] In some embodiments, the antigen is an ovarian cancer antigen or a T
lymphoma antigen.
1002331 In some embodiments, for example, the cancer antigen for a target
cancer cell can be one or
more of the mutated/cancer antigens: 1DH1, ATRX, PRL3, or ETBR, where the
cancer is a glioblastoma.
[00234] In some embodiments, for example, the cancer antigen for a target
cancer cell can be one or
more of the mutated/cancer antigens: CA125, beta-hCG, urinary gonadotropin
fragment, AFP, CEA, SCC,
inhibin or extradiol, where the cancer is ovarian cancer.
[00235] In some embodiments the cancer antigen for a target cancer cell may be
CD5.
[00236] In some embodiments the cancer antigen for a target cancer cell may be
HER2.
[00237] In some embodiments the cancer antigen for a target cancer cell may be
EGFR Variant DI.
[00238] In some embodiments the cancer antigen for a target cancer cell may be
CD19.
[00239] In some embodiments, the antigen is an integrin receptor.
1002401 In some embodiments, the antigen is an integrin receptor selected from
the group consisting of
a1, a2, a4, a5, a6, a7, a8, a9, al 0, al 1, aD, aE, aL,
aM, aV, cr.X, 13 1, 13 2, 13 3, 13 4, [3 5, 13 6, 13 7,
and 138. In some embodiments, the bi- or trispecific engager binds to an
extracelfular domain of a monocyte
or macrophage receptor from a member of the integrin 132 subfamily a432 (CD1
lb/CD18, Mac-1, CR3,
Mo-1), cul32 (CD1 la/CD18, LEA-1), ax132 (CD11c/CD1 8), and 0E032
(CD11d/CD18).
[00241] Provided herein are exemplary target cell binders (e.g., engagers)
that can specifically bind to
a cell surface molecule (such as a cell surface antigen) on a cancer cell. In
some embodiments, the binder
is an antibody specific to the antigen, or a fragment thereof In some
embodiments, the binder comprises a
scFv, or a fragment thereof, that specifically binds to an antigen on a tumor
cell. In some embodiments, the
antigen on a tumor cell is CD5. The binder comprises a heavy chain (HC)
sequence and a light chain (LC)
sequence. An scFv specific for CD5 (anti-CD5 scFv) comprises an amino acid
sequence corresponding to
a variable heavy chain (VH) domain and an amino acid sequence corresponding to
a (Vi,). In some
embodiments, a first binding domain, which is a CD5 binder can be an scFv
having comprising a sequence
of SEQ ID NO: 27, and a sequence of SEQ ID NO: 28, joined by a linker peptide.
Provided herein in Table
1A are exemplary anti-CD5 HC and LC variable domains.
-39-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
Table IA. Exemplary CD5 binder domains
Domain Sequence
Anti-CD5
EIQLVQSGGGLVICPGGSVRISCAASGYTITNYGMNWVRQAPGK
heavy chain GLEWMGWINTHTGEPTYADSFKGRFTFSLDDSKNTAYLQINSLR
variable AEDTAVYFCTRRGYDWYFDVWGQGTTVTV
domain (SEQ ID NO: 27).
Anti-CD5
DIQMTQSPSSLSASVGDRVTITCRASQD1NSYLSWFQQKPGKAPK
light chain
TLIYRANRI,ESGVPSRFSGSGSGTDYTLTISSLQYEDEGIYYCQQY
variable DESPWTEGGGTKLEIK
domain (SEQ ID NO: 28).
[00242] In one embodiment, the target cell binder is a single domain antibody
that binds CD5. In some
embodiments the target cell binder is a CD5-binding Vim. In some embodiments,
the target cell binder or
a first binding domain can be a CD5 binding Vim comprising a sequence of SEQ
ID NO: 27.
[00243] In some embodiments, an exemplary target cell binder (e.g., an
engager) is a HER2 engager,
that can specifically bind to cell surface antigen HER2 on a HER2 positive
cancer cell. In some
embodiments, the binder is an antibody specific to the antigen, or a fragment
thereof In some embodiments,
the binder comprises a scFv, or a fragment thereof, that specifically binds to
HER2. The binder comprises
a heavy chain (HC) sequence and a light chain (LC) sequence. An scFv specific
for HER2 (anti-HER2
scFv) comprises an amino acid sequence corresponding to a variable heavy chain
(VII) domain and an
amino acid sequence corresponding to a (VL ). In some embodiments, a first
binding domain may be an
scFv having a HER2 binder comprising a sequence of SEQ ID NO: 29, and a
sequence of SEQ ID NO: 30,
joined by a linker peptide. In some embodiments, the target cell binder or a
first binding domain can be a
HER2 binding Vifi4 comprising a sequence of SEQ ID NO: 29.
11002441 Provided herein in Table 1B are exemplary anti-HER2 HC
and LC variable domains.
Table IB. Exemplary HER2 binder domains
Domain Sequence
Anti-HER2 DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKA
heavy chain PKLLIYSASFLYSGVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQ
variable QHYTTPPTFGQGTKVE1KRTGSTSGSGKPGSGEGSEVQLVE
domain (SEQ ID NO: 29).
-40-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
Anti-HER2 LVQPGGSLRLSCAASGFNIKDTYIHAVVRQAPGKGLEWVARIYPT
light chain
NGYTR.YADSVKGRETISADTSICNTAYLQMNSLRAEDTAVYYCS
variable RWGGDGFYAIvIDVWGQGTLVTV
domain (SEQ ID NO: 30).
[00245] In some embodiments, the tumor associated macrophages may be
characterized largely as
having an M2 phenotype. Human M2 macrophages can be identified as nearing the
cell surface markers
CD14+CD163+CD206+ CD80- phenotype. Hence, a bi-or trispecific engager that
specifically binds to the
myeloid cell, e.g., a monocyte or macrophage associated with a tumor can
comprise one or more binding
domains that can bind to one or more of: CD14, CD163, and CD206 cell surface
molecules.
[00246] Typically, the M2-like tumor associated macrophage (TAM) population
lacks expression of
reactive nitrogen intermediates, less efficiently presents antigen, displays
little tumoricidal activity, and
produces angiogenic factors, metalloproteases, and cathepsins. Matrix
metalloproteinases, e.g., MMP2 is
readily expressed in TAMs. Classical activation of macrophages up-regulate MMP-
1, -3, -7, -10, -12, -14
and -25 and decrease TIMP-3 (tissue inhibitors of metalloproteinase-3) levels.
Bacterial
lipopolysaccharide, IL-1 and TNFa are found to be more effective than IFN-
gamma except for the effects
on MMP-25, and TIMP-3. By contrast, alternative activation decrease MMP-2, -8
and -19 but increase
MMP -11, -12, -25 and TIMP-3 steady-state mRNA levels. Up-regulation of MMPs
during classical
activation depends on mitogen activated protein kinases, phosphoinositide-3-
kinase and inhibitor of KB
kinase-2. Therefore, depending on the target monocyte or macrophage
population, an engager may be
designed such that a metalloproteinase can be a binding moiety for the
monocyte or macrophage engager.
MMP2 being one of the readily expressed TAM markers, a tumor specific myeloid
cell engager comprises
a MMP2 binding domain.
[00247] Hypoxia, or cytokines produced secondary to hypoxia, attract
macrophages which
subsequently up-regulate hypoxia inducible factor 2-alpha (HIF-2a).
Accordingly, a binding domain on a
bi- or trispecific engager that specifically binds to a tumor associated
macrophage can bind to HIF-2a
which is upregulated in these cells.
[00248] Monocyte/Macrophage cell-surface markers include LPS co-receptor
(CD14), HLA-DR (MUG
class II), CD312, CD115, the Fey-receptor FcyRBI (CD16). Subset-specific
markers include CD163 and
CD204, both scavenger receptors expressed by M2 macrophages, CD301, a
galactose-type C-type lectin
expressed by M2 macrophages.
[00249] In some embodiments, the bi- or trispecific engager binds to an
extracellular domain of a
phagocytic receptor, selected from the extracellular domains of any one of the
proteins in Table 2A.
-41-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
Table 2A. Exemplary receptors on phagocytes
Gene names, aliases
NCBI Acc #
MSR1 , SR-AIõ CD204, SCARA1, SR-Al
NM_138715
Alternatively spliced form of SR-AI SR-All SR-A1.1
NM 002445
MARCO, SCARA2, SR-A6
NM 006770
SCARA3, MSRL1, SR-A3
NM 016240
C0LEC12, SCARA4, SRCLI, SRCLII, CL-P1, SR-A4
NM _130386
SCARA5, TESR, NET33 SR-A5
NM_173833
CD36 SCARB3, FAT, GPIV, PAM SR-B2
NM 001001548
SCARB1 SR-BI, CD36L1 SR-B1
NM_005505
CD68 gp110, SCARD1, LAMP4 SR-D1
NM 001251
OLR1 LOX-1, SCARE1, CLEC8A SR-E1
NM _002543
Alternatively spliced form of SRE-1 LOX1N SR-E1.1
NM_001172632
CLEC7A, Dectin-1, SCARE2, CD369, SR-E2
NM 197947
CD206/MRCL Mannose receptor 1 SR-E3
NM_002438
ASGPR ASGR1, CLEC4H1, HL-1 SR-E4
NM 001197216
SCARF1, SREC-I, SR-El
NM 003693
MEGF10, EMARDD, SR-F2
NM 032446
CXCL16, SR-PSOX SR-G1
NM 001100812
STAB!, FEEL-1, SR-H1
NIVI_015136
STAB2, FEEL-2, SR-H2
NM_017564
CD163 M130, CD163A, sit-n
NM 004244
CD163L1 CD163B, M160 SR-I2
NM 001297650
SCART1 CD163c-a SR-I3
NR 002934.3
RAGE (membrane form) AGER SR-J1
NM 001136
RAGE (soluble form) AGER SR-J1.1
AB061668
CD44 Pgp-1 SR-K1
NM 000610
LRP1 A2MR, APOER, CD91 SR-L1
NM 002332
LRP2 Megalin, gp330 SR-L2
NM 004525
SRCRB4D
NM 080744
SSC5D
NM_001144950
CD14
NM 000591
Ly75/CD205
NM_002349
-42-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
CD207/Langerin
NM 015717
CD209/DC-SIGN CLEC4L
NW021155
[00250] In some embodiments, the bi- or trispecific engager binds to an
extracellular domain of a PEP,
selected from the extracellular domains of any one of the proteins in Table
2B.
[00251] Table 2B provides exemplary surface markers and phenotypic
characteristics of monocytes,
macrophages and DCs.
Table 2B.
Molecularly defined
Other characteristics
Monocytes CD14*WD16-
CD16+ monocytes (undefined as to
CD14-HFCD16+
whether they CD14-H-CD16+ or
DC-like phenotype -
CD16+CD14dim) possess superior
High
phagocytosis compared to blood
DR, CD80+
monocytes and can efficiently activate
Macrophage-like
CD4+ T cells
phenotype - CD163+,
CD68+
CD16+CD14dim
CD14 "DC"-Postulated
to
be monocyte derived
Macrophages in Pan CD68
Liver Macrophages appear to
the liver
be predominantly tolerogenic
in nature, with a regulatory and
scavenging role
Dendritic cells BDCA1 (CD1c+) DC
Tolerogenic in nature; Lower
BDCA2 (CD303+) DC expression of costimulation markers
BDCA3 (CD141hi) DC compared to
spleen; Produce 1L-10 on LPS
stimulation; Stimulate T-cells that are
IL-10 producing and hypo-responsive
on re-stimulation ; Produce higher
numbers of FoxP3+ Treg cells on naive
T cell stimulation; Weak MLR
response compared
-43-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
to blood.
[00252] In some embodiments, the bi- or trispecific engager binds to an
extracellular domain of a
myeloid cell receptor, e.g., a monocyte receptor, a macrophage receptor, for
examples, a receptor selected
from the extracellular domain comprises an Ig binding domain.
[00253] In some embodiments, the bi- or trispecific engager binds to an
extracellular domain of a
macrophage receptor e.g., an IgA, IgD, IgE, IgG, IgM, FcyRI, FcyRIIA, FcyRBE,
FcyRIIC, FcyRIIIA
(CD16), FcyRIBE, FcRn, FcRL5 binding domain. A CD16 receptor referred to
herein can be a CDI6A
receptor or a CD16B receptor.
[00254] In some embodiments, the bi- or trispecific engager binds to an
extracellular domain of a an
FcR extracellular domain.
[00255] In some embodiments, the bi- or trispecific engager binds to an
extracellular domain of a
macrophage receptor selected from the extracellular domains of an FcR-alpha,
an FcR-beta, an FcR-
Epsilon or an FcR-gamma.
[00256] In some embodiments, the bi- or trispecific engager binds to an
extracellular domain of an
FcccR (FCAR).
[00257] In some embodiments, the bi- or trispecific engager binds to an
extracellular domain of an FcR-
beta.
[00258] In some embodiments, the bi- or trispecific engager binds to an
extracellular domain of an FceR
(FCER1A).
[00259] In some embodiments, bi- or trispecific engager binds to the
extracellular domain comprises an
FcyR (FDGR1A, FCGR2A, FCGR2B, FCGR2C, FCGR3A, FCGR3B) receptor.
[00260] In some embodiments, the bi- or trispecific engager binds to an
extracellular domain of a
monocyte or macrophage phagocytic receptor selected from selected from lectin,
dectin 1, mannose
receptor (CD206), scavenger receptor Al (SRA1), MARCO, CD36, CD163, MSR1,
SCAR.A3, COLEC12,
SCARA5, SCARB1, SC ARB2, CD68, OLR1, SCARF1, SCARF2, CXCL16, STAB1, STAB2,
SRCRB4D, SSC5D, CD205, CD207, CD209, RAGE, CD14, CD64, F4/80, CCR2, CX3CR1,
CSF1R,
Tie2, HuCRIg(L), and CD169 receptor.
1002611 In some embodiments, the bi- or trispecific engager binds to the
extracellular domain of a
TREM protein. In some embodiments, the extracellular domain of a TREM protein
is a TREM 1 protein
extracellular domain. In some embodiments, the extracellular domain of a TREM
protein is a TREM 2
protein extracellular domain. In some embodiments, the extracellular domain of
a TREM protein is a
TREM 3 protein extracellular domain.
-44-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00262] In some embodiments, the bi- or trispecific engager binds to an
extracellular domain of a
monocyte or macrophage receptor selected from a group consisting of lectin,
dectin 1, mannose receptor
(CD206), scavenger receptor Al (SRA1), MARCO, CD36, CD163, MSR1, SC ARA3,
COLEC12,
SCARA5, SCAR131, SC ARB2, CD68, OLR1, SCARF1, SCARF2, CXCL16, STAB1, STAB2,
SRCRB4D, SSC5D, CD205, CD207, CD209, RAGE, CD14, CD64, CCR2, CX3CR1, CSF1R,
Tie2,
HuCRIg(L), and CD169 receptor.
[002631 It may be understood that in some embodiments, a binder or any part of
an engager can be a
molecule other than an antibody or a fragment thereof For example, a binder
that binds to a surface
molecule of a cell, such as a target cell or an effector myeloid cell, and may
be a ligand for a receptor,
where the cell surface molecule is a receptor specific for the ligand. In some
embodiments a ligand may be
a chimeric protein or a fusion protein, or a naturally occurring ligand of the
receptor. In some embodiments
one binder in an engager recombinant protein may be a ligand, and another may
be an antibody or a
fragment thereof
[00264] In some embodiments, an engager molecule may comprise one or more
linkers or spacers.
Linkers or spacers may be made up of 2-50 amino acids. In some embodiments,
the linker is a 3-30 amino
acid spacer. In some embodiments, the linker is a 4-20 or 5-10 amino acid long
peptide. In some
embodiments, the spacer may be made of nonreactive amino acid moieties, for
example, a series of glycine
or serine or alanine residues. An exemplary linker may comprise an amino acid
sequence GSGS, or SGGG,
or SGGGGSG. An exemplary linker may comprise an amino acid sequence
SSGGGGSGGGGSGGGGS.
A linker may link the Vii and VL domains of an scFv. A linker may link the
binder domains of a bi- or
trispecific engager. The linkers generally serve as structural elements that
connect the effective binder sites.
In some embodiments, the linker may be flexible. In some embodiments, the
linker may be rigid. The
length of the linker is adjusted as per the need of the design and the length
that is optimal or necessary to
space the binders at the opposite ends. In some embodiments, the linker may
comprise a peptide that has a
unique function, other than linking two domains. Exemplary peptides discussed
below may be part of the
design of a bi- or trispecific or multispecific engager, and may or may not be
a part of a linker.
[00265] In some embodiments, the bi- or trispecific engager comprises a
peptide that specifically targets
the tumor associated macrophages, such as, for instance, the M2 macrophages.
In some embodiments, the
M2-specific peptide is M2-pep, having an amino acid sequence, YEQDPWGVKWWY.
(SEQ NO:
116).
[00266] In some embodiments, an M2-specific peptide may comprise a sequence
HLSWLPDVVYAW,
(hereafter, HLS pep) (SEQ ID NO: 117).
[00267] A peptide such as the M2- pep or the HLS pep described above can form
a part of the bi-specific
engager or a trispecific engager, such as a linker between two binding domains
or a part of a linker. In
some embodiments, the first and the second binding domains of an engager are
coupled to an M2- pep or
-45-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
an HLS pep, whereas the M2-pep or the HLS pep further target the engager to
the tumor associated
macrophages, and help tether the engager to the tumor associated macrophages.
[00268] In some embodiments, an exemplary engager may comprise a CD5 binding
binding domain
and a CD16 binding domain, connected by a linker. In some embodiments an
exemplary engager comprises
TLR activation peptide, such as a TLR4 peptide.
B. Engagers with Masked Antigen Binding Domains
[002691 Provided herein are compositions for a therapeutic agent that
comprising bispecific or
trispecific engagers comprising one or more pro-antibodies. In some
embodiments, a pro-antibody is an
inactive form of an antibody, or fragments or variants thereof, whose antigen
binding domain is blocked
or "masked" from interacting with the antigen. In some embodiments, the pro-
antibody comprises a
substrate peptide or a conjugate that remains associated with the antigen
binding domain by a proteace
cleavable linker peptide and "mask" the antigen binding domain from binding to
its cognate antigen. Under
suitable condition, the substrate peptide is cleaved to release the mask, and
promote antigen binding at the
antigen binding domain.
11002701 In some embodiments, the bispecific or trispecific antibody comprises
one or more scfv that is
a pro-antibody, that is, the antigen binding domain of the scFv is masked with
a cleavable blocker. In some
embodiments the blocker comprises a substrate peptide and a protease cleavable
linker. In some
embodiments, the bispecific or trispecific antibody comprises a Vint pro-
antibody, wherein, the Vi. domain
or the VH domain or both of the VHH antibody is masked with a cleavable
blocker. In some embodiments,
the bispecific or trispecific antibody comprises a nanobody where one or more
of antigen binding domains
are masked by association with a substrate peptide or conjugate linked to the
antibody by a cleavable linker.
[00271] In some embodiments, the cleavable linker is designed such that it is
cleaved when the
therapeutic agent reaches the target site of its action. For example, the pro-
antibody for a cancer therapeutic
agent can be designed to contain a protease cleavable linker where the
protease that cleaves the protease
cleavable linker is abundant in the tumor microenvironment and relatively
absent or negligible in the non-
tumor tissue, and the therapeutic agent is activated by the protease when the
therapeutic agent reaches the
tumor microenvironment when administered systemically. In some embodiments,
therapeutic agents are
developed comprising protease-activated pro-antibodies to direct antibody
action solely to disease sites.
[00272] In some embodiments, the cleavable linker is a matrix metalloprotease-
2 (MMP2) cleavable
peptide having the amino acid sequence GPLGVR (SEQ ID NO: 118).
[00273] In some embodiments, the cleavable linker is a M2-specific peptide,
having the amino acid
sequence YEQDPWGYKWWY (SEQ ID NO: 116), or the amino acid sequence
HLSWLPDVVYAW
(SEQ ID NO: 117).
[00274] In some embodiments, the cleavable linker comprises a hypoxia
inducible protein mediated
cleavage site.
-46-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00275] In some embodiments, the cleavable linker is a non-naturally occurring
synthetic peptide, and
comprises a protease cleavable site. In some embodiments, the cleavage site
can be cleaved by a protease
that is administered exogenously. In some embodiments, the cleavage site can
be cleaved by a protease that
is associated with a cancer targeted drug.
[00276] In some embodiments, the cleavable linker is a mutated peptide, where
the mutated peptide
contains a protease cleavable site, not occurring in the corresponding non-
mutated peptide.
[00277] In some embodiments, the purpose of the instant program disclosed
herein is generating
therapeutic products for use in immunotherapy.
C. Engagers with domains that promotes enhanced phagocytic activity and immune
response of the
myeloid cell
[00278] The tumor microenvironment (TIME) constitutes an immunosuppressive
environment.
Influence of IL-10, glucocorticoid hormones, apoptotic cells, and immune
complexes can interfere with
innate immune cell function. Immune cells, including phagocytic cells settle
into a tolerogenic phenotype.
In macrophages, this phenotype, commonly known as the M2 phenotype is distinct
from the M1 phenotype,
where the macrophages are potent and capable of killing pathogens. Macrophages
exposed to LPS or IFN-
gamma, for example, can polarize towards an MI phenotype, whereas macrophages
exposed to IL-4 or 1L-
13 will polarize towards an M2 phenotype. LPS or TEN-gamma can interact with
Toll-like receptor 4
(TLR4) on the surface of macrophages inducing the Trif and MyD88 pathways,
inducing the activation of
transcription factors IRF3, AP-1, and NFKB and thus activating TNF-D genes,
interferon genes, CXCL10,
14052, IL-12, etc., which are necessary in a pro-inflammatory MI macrophage
response. Similarly, IL-4
and IL-13 bind to IL-4R, activation the Jak/Stat6 pathway, which regulates the
expression of CCL17,
ARG1, IRF4, IL-10, SOCS3, etc., which are genes associated with an anti-
inflammatory response (M2
response). Expression of CD14, CD80, D206 and low expression of CD163 are
indicators of macrophage
polarization towards the M1 phenotype.
[00279] In some embodiments, the engagers comprise a binding domain that can
bind to the
extracellular domain of a receptor, such as a phagocytic receptor. Engagement
with the monocyte or
macrophage phagocytic receptor, for example, at a specific site may activate
the receptor by enhancing the
intracellular signaling mediated by the intracellular domain of the receptor.
In some embodiments, the
binding domain of the engager comprises a ligand for the phagocytic receptor.
In some embodiments the
binding domain binds to the ligand which then binds to the phagocytic
receptor.
[00280] Some phagocytic receptors are more potent in activating phagocytosis
than the others, and can
induce rapid phagocytosis of the target cell. It is necessary to identify the
potent phagocytic receptors. Most
macrophage scavenger have broad binding specificity that may be used to
discriminate between self and
non-self in the nonspecific antibody-independent recognition of foreign
substances. The type I and H class
A scavenger receptors (SR-All and SR-All) are trimeric membrane glycoproteins
with a small N112-
-47-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
terminal intracellular domain, and an extra/cellular portion containing a
short spacer domain, an a-helical
coiled-coil domain, and a triple-helical collagenous domain. The type I
receptor additionally contains a
cysteine-rich COOH-terminal (SRCR) domain. These receptors are present in
macrophages in diverse
tissues throughout the body and exhibit an unusually broad ligand binding
specificity. They bind a wide
variety of polyanions, including chemically modified proteins, such as
modified LDL, and they have been
implicated in cholesterol deposition during atherogenesis. They may also play
a role in cell adhesion
processes in macrophage-associated host defense and inflammatory conditions.
[00281] Table 2A and Table 2B exemplify a non-extensive list of receptors or
surface antigens
associated with different myeloid cells, wherein the cells have a range of
characteristics ranging from
highly phagocytic to tolerogenic. Even within macrophages, some receptors are
associated with the actively
phagocytic MI phenotype, while others are associated with the anti-
inflammatory M2 phenotype which
has dampened phagocytic response. Activation of the MI-associated receptors by
engaging with an MI
receptor can generate a characteristic shift in the macrophage type, from M2
towards M1 phenotype.
[00282] Macrophage receptors that activate phagocytosis comprise an
intracellular phagocytosis
signaling domain that comprises a domain having one or more Immunoreceptor
Tyrosine-based Activation
Motif (ITAM) motifs. ITAMs are conserved sequences present in the cytoplasmic
tails of several receptors
of the immune system, such as T cell receptors, immunoglobulins (Ig) and FcRs.
They have a conserved
amino acid sequence motif consisting of paired YXXL/I motifs (Y= Tyrosine,
1_,= Lysine and I= Isoleucine)
separated by a defined interval (YXXL/1-X64-YXXL/1). In addition, most ITAMs
contain a negatively
charged amino acid (DIE) in the +2 position relative to the first ITAM
tyrosine. Phosphorylation of residues
within the ITAM recruits several signaling molecules that activate
phagocytosis. ITAM motifs are also
present in the intracellular adapter protein, DNAX Activating Protein of 12
kDa (DAP12).
[00283] In some embodiments, the phagocytic signaling domain in the
intracellular region can comprise
a PI3kinase (PI3K) recruitment domain (also called PI3K binding domain). CD19,
CD28, CSFR or PDGFR
receptors comprise PI3 kinase recruitment to the binding domain. In some
embodiments, the bi- or
trispecific engager binds to a receptor such as any one or more of CD19, CD28,
CSFR or PDGFR receptors.
Engaging with such receptors lead to Akt mediated signaling cascade and
activation of phagocytosis. The
PI3K-Akt signaling pathway is important in phagocytosis, regulation of the
inflammatory response, and
other activities, including vesicle trafficking and cytoskeletal
reorganization. The PI3kinase recruitment
domain is an intracellular domain in a plasma membrane protein, which has
tyrosine residues that can be
phosphorylated, and which can in turn be recognized by the Src homology domain
(S112) domain of
PI3Kp85. The SH2 domain of p85 recognizes the phosphorylated tyrosines on the
cytosolic domain of the
receptor. This causes an allosteric activation of p110 and the production of
phosphatidylinosito1-3,4,5-
trisphosphate (PIP3) that is recognized by the enzymes Akt and the
constitutively active 3'-
phosphoinositide-dependent kinase 1 (PDK1) through their plekstrin homology
domains. The interaction
-48-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
of Akt with P1P3 causes a change in the Akt conformation and phosphorylation
of the residues Thr308 and
Ser473 by PDK1 and rictor-mTOR complex, respectively. Phosphorylation of these
two residues causes
the activation of Akt which in turn phosphorylates, among other substrates,
the enzyme glycogen synthase
kinase-3 (GSK-3). GSK-3 has two isoforins, GSK-3a and GSK-3I3 both of which
are constitutively active.
The isoforms are structurally related but functionally nonredundant.
Inactivation of GSK-3 is observed
when the residues Ser21 in GSK-3a or Ser9 in GSK-3I3, located in their
regulatory N-terminal domains,
are phosphorylated by Akt and other kinases. Inhibition of GSK-3 by
phosphorylation is important for the
modulation of the inflammation and in phagocytosis processes.
[00284] In some embodiments, a bi- or trispecific engager comprises a binding
domain that binds to a
receptor or part thereof that can activate pro-phagocytic signaling by
engaging DAP12 activation.
[00285] In some embodiments, a bi- or trispecific engager comprises a binding
domain that binds to a
receptor or part thereof that promotes clustering of a group of receptors on a
monocyte or macrophage or
phagocytic cell, and potentiates phagocytosis. In some embodiments, clustering
of receptors activate
intracellular signaling pathways.
1002861 In some embodiments, the bi- or trispecific engager comprises a
binding domain for Fc nRi
(CD89). Fc n R1 receptor engagement or cross-linking activates antigen
mediated cytotoxicity, and activate
phagocytosis on monocytes or macrophages. Fe URI is expressed constitutively
in macrophages as well as
some other cells such as neutrophils and eosinophils. It is especially
advantageous in a trispecific engager
when the engager comprises a binding domain that binds to an antigen on a
target cell, such as a tumor cell,
a binding domain specific for a monocyte or macrophage receptor such as CD206,
and a binding domain
for CD89. Given the length and flexibility of the design of the engager
molecule, the CD206 and Fc 111R1
(CD89) binding domains could engage and provide multiple activation signals by
cross-linking with more
than one phagocytosis specific receptors, and crosslinking with the target
cell. Fc R1 activation redirects
monocytes or macrophages from M2 phenotype to killer M1 phenotype and can
therefore having an Fc DR1
binding domain in a bi- or trispecific engager can be a powerful tool in
repurposing tumor associated
macrophages for tumor cytotoxicity.
[00287] In some embodiments, the bi- and trispecific engager comprises a
binding domain that binds to
a monocyte or macrophage scavenger receptor. There are currently eight clacses
of scavenger receptors
(classes A¨I-I). In some cases, multiple names have been assigned to the same
receptor (e.g., MSR1, SR-
AI, CD204, and SCARA1). In addition, there are proteins exhibiting scavenger
receptor activity that have
been named based on other criteria and have not been included in a general
scavenger receptor
nomenclature. Some examples include RAGE (SR-E1), LRP1, LRP2, ASGP, CD163, SR-
PSOX, and
CXCL16. In some embodiments, the bi- or trispecific engager comprises a
binding domain that binds to a
scavenger receptor, selected from lectin, dectin 1, mannose receptor (CD206),
scavenger receptor Al
(SRA1), MARCO, CD36, CD163, MSR1, SCARA3, C0LEC12, SCARA5, SCARB 1, SCARB2,
CD68,
-49-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
OLR1, SCARF1, SCARF2, CXCL16, STAB1, STAB2, SRCRB4D, SSC5D, CD205, CD207,
CD209,
RAGE, CD14, CD64, CCR2, CX3CR1, CSF1R, Tie2, HuCR1g(L), and CD169 receptor.
The binding
domains bind to their respective ligands with a dissociation constant (KO of
104 to 10-12M or less, or, 101
7 to 10-12 M or less or, 104 to 10-12M (i.e. with an association constant (KA)
of 105 to 1012M or more, or,
107 to 1012 M Of more Of 108 to 1012M).
[00288] In some embodiments, an exemplary binding domain of an engager that
binds to the
scavenger receptor SRA1 comprises a variable region having an amino acid
sequence or a portion
thereof, or a sequence having at least 95% sequence identity to a sequence:
EVQLVESGGGLVQAGGSLRLSCTASGRAVSTYAMGWERQAPGKEREFVAAMISSLSSKSYADT
VKGRFTISRDYAKNTVYXQMINSLKPEDTADYYCAADLLPYSSSRSLPMGYDYWGQGTQVTVSS
(SEQ ID NO: I).
[00289] Exemplary binding domains of an engager that binds to the scavenger
receptor SRA1 can
comprise a binding domain having an amino acid sequence of any one of SEQ ID
NOs 2-7, or a portion
thereof, or a sequence having at least 95% sequence identity to any one of the
sequences:
1002901 EVQLVESGGGLVQAGGSLRLSCTASGRAVSTYAMGWFRQAPGKEREFVAAMISSLS
SKSYADSVKGRFTISRDYAKNTVYLQMINSLICPEDTADYYCAADLLPYSSTRSLPMGYDYWGQG
TQVTVSS (SEQ ID NO: 2).
1002911 EVQLVESGGGLVQAGGSLRLSCAASGSFSLYDMGWESQAPGKEREEVAAINWSGGS
TAYADSVKGRITISRDSAKNTVYLQMNSLICPEDTAVYYCAAKPAKYHEGSGYRDFAEYPYWG
QGTQVTVSS (SEQ ID NO: 3).
[00292] EVQLVESGGGLVQAGGSLRLSCAASGRTFSRYAMAWFRHAPGKDREFIVAAVSQSGL
LTFYADSVKGRFTISRDNAKNTVYLQMNSLKPEDTAVYDCAAXSRFPLVVPVAYENWGQGTQ
VTVSS (SEQ ID NO: 4); wherein X can be any naturally occurring amino acid.
[00293] EVQLVESGGGLVQAGGSLRLSCAASGRTFSRYAMAWFRHAPGKDREFVAAVSQSGL
LTFYADSVKGRFTISRDNAKNTVYLQMNSLKPEDTAVYDCAADSRFPLVVPVAYENWGQGTQ
VTVSS (SEQ ID NO: 5).
[00294] EVQLVESGGGLVQVGGSLRLSCAASGISIRTHANIGWYRQAPGKQRELVATITSVTSG
GSLNYADSVKGRFTISRDNAKNTVYLQMNSLICPEDTAVYYCKLLGFDYRGQGTQVTVSS (SEQ
ID NO: 6).
[00295] EVQLVESGGGLVQPGGSLRLSCAASGSIGRFVAMGWYRQAPGKQRELVATITSITSG
GRTNYADSVKGRFTISRDNAKNTVYLQMNSLICPEDTAVYYCNVVPYVNDYWGQGTQVTVSS
(SEQ ID NO: 7).
[00296] In some embodiments, an exemplary binding domain of an engager that
binds to the
scavenger receptor RAGE can comprise a binding domain having an amino acid
sequence of any one of
-50-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
SEQ ID NOs 8-15, or a portion thereof, or a sequence having at least 95%
sequence identity to any one
of the sequences.
EVQLVESGGGLVQAGDSLRLSCIASGRTFTMGWFRQAPGKEREFVAAISWSGGRTYYADSVKG
RFTISRENAKNTVYLQIVINSLIKPEDTAVYCCATENLASSGSAYSDDRYNACGQGTQWVSS (SEQ
ID NO: 8).
[00297] EVQLVESGGEVVQPGGSLRLSCAASGF
_______________________________________________________________________________
_______________ I liDDRAIGWFRQAPGICEREGVACSANNDN
RAFYEDSVICGRFAVSRDNAKNTVYLQMNSLKPEDTAVYYCATRCAAGRVNLYYGMDYWGKG
TLVTVSS (SEQ ID NO: 9).
[00298] EVQLVESGGGLVQPGGSLRLSCAASGFTLGNYAIGWFRQAPGKEREGVSCVDRDGG
STYYLDSVTGRFTTSRDDAENTVYLQMNSLIPDDTAVYYCATRLYGCSGYGRDYADWGQGTQ
VTVSS (SEQ ID NO: 10).
[00299] EVQLVESGGGLVQAGGSLRLSCAVSGRTFSTDAFGWFRQAPG10EREFVSAM1flWNGS
SSYYADLVICGRFTISRDNAKNTVYLLMNSLICPEDTAVYYCTAGKRYGYYDYWGQGTQVIVSS
(SEQ ID NO: 11).
[00300] EVQLVESGGGLVQAGGSLRLSCAASGRTFSNYSMGWFRQAPGKEREFVATISWSGA
LTHYTDSVICGRFTISRDNAKNTVYLQMNSLICPEDTAVYYCAASDSDYGNICYDYWGQGTQVTV
SS (SEQ ID NO: 12).
[00301] EVQLVESGGGLVQAGGSLRLSCAASGRTVSDMTMGWFRQAPGKERVFVAAISNSGL
STYYQDSVKGRFTISRDTANNTVALQMNSLICPEDTAVYFCAARSGWSGQYDYWGQGTQVTVS
S (SEQ ID NO: 13).
[00302] EVQLVESGGGLVQAGGSLRLSCAASGRIFNNYAMGWFRQAPGKEREFVAGISWSGD
STLYADSVKGRETTSRDNAKNTVYLQMNSLKPEDTANYYCAEKQGADWAPYDYWGQGTQVT
VSS (SEQ ID NO: 14).
[00303] EVQLVESGGGLVQAGGSLRLSCVASELTFSLYRMGWFRQAPGKEREFVSAMSTSGA
GTYYADSVKGRFTISRDNPICNTVYL,QMNSLICPEDTAVYYCVAGVRFGVYDYWGQGTQVTV SS
(SEQ ID NO: 15).
[00304] In some embodiments, an exemplary binding domain of an engager that
binds to the
scavenger receptor Lox-1 can comprise a binding domain having an amino acid
sequence of any one of
SEQ ID NOs 16-26, or a portion thereof, or a sequence having at least 95%
sequence identity to any one
of the sequences.
[00305] EVQLVES6GGLVQPGGSLRLSCAASGFTLDDYAIGWFRQAPGKEREGVSCISRTDGS
TDVADSVKGRITISRDNAKNTVYLQMNSLKPEDTAVYYCAAGRTYYSGSYYFGLGSDEYDYW
GQGTQVTVSS (SEQ 11) NO: 16).
1003061 Sequences of additional exemplary binding domains of an engager that
binds to the scavenger
receptor Lox-1 comprises a variable region having an amino acid sequence are
given below:
-51-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
1003071 EVQLVESGGGLVQPGGSLRLSCAASGSIFTINAMAWYRQAPGKQRELVAHLTNSGRT
GYADSVKGRFTISSDNAKNTVYLQMNSLKPEDTAVYYCNRLGUIWSWGQGTQVTVSS (SEQ
ID NO: 17).
[00308] EVQLVESGGGLVQAGGSLRLSCAASIGTFSAYHMGWFRQAPGKERELVAAISWSVSS
TYYADSVKGRFTISRDNAKRTVSLQMDSLICPEDTAVYYCAARSGERYDYYKAQYEYWGQGTQ
VTVSS (SEQ ID NO: 18).
[00309] EVQLVESGGGLVQPGGSLRLSCAAYGSFFSIGTMGWYRQPPGNQRELVAVTYGLGS
TNYAESVKGRFTISRDNAKNTVSLQMNSLKPEDTAVYYCYAEIDTDPRSGEWDYWGQGTQVTV
SS (SEQ ID NO: 19).
[00310] EVQLVESGGGLVQPGGSLRLSCLPSTSTSSLRTVGWYRQGPGKQRDLVAIMSAGTTR
YADSVKGRITISLDDAKNTVYLQMNSLICPEDTAVYICNGRPVFSNVDWGQGTQVTVSS (S EQ
ID NO: 20).
[00311] EVQLVESGGGLVQAGGSLRLSCAASGFTFDDYAIGWFRQAP'GKEREGVSCVSRDGG
STYYLDSVKGRFTISSDNAKNTVYLQMNSLKPEDAAVYYCAASRYDCSKYLIDYNYRGQGTQV
TVSS (SEQ ID NO: 21).
[00312] EVQLVICSGGGLVQAGGSLRLSCAASGRRFSTSGMGWFRQAPGREREFVXGIXWNSR
XTYYAESVKGRFTISRDNSKNTVYLQMNSLKPEDTAVYYCATNYYGSXWSVNSDDYDYWXQG
XQVTVSS (SEQ ID NO: 22).
[00313] EVQLVESGGGLVQAGGSLRLSCAASGRTFSNYAMGWFRQAPGKEREFVAAITWSGS
STYYADSVKGRFTISRDNAKNTVYLQMNSLICPEDTAVYYCAAAQRGRYYYLDRNVEYDYWG
QGTQVTVSS (SEQ ID NO: 23).
[00314] EVQLVESGGGLVQPGGSLRLSCAASGFTLDDYGIGWFRQAPGKEREGVSCISSSDGST
DYADSVKGRFTISRDNAKNTVYLQIV1NNLKPEDTAVYYCAAGRTYYSGSYYFGLGSDEYDYWG
QGTQVTVSS (SEQ ID NO: 24).
[00315] EVQLVESGGNLVQAGGSLRLSCAASGFTFDDYVIGWFR.QAPGKEREGVSCISSVEGS
TYYADSVKGRITISGDNAKNTVYLQMNSLKPEDTAVYYCAAGTWLDCSGYGSYDIVIDYWGKG
TLVTVSS (SEQ ID NO: 25).
[00316] EVQLVESGGGLVQAGGSLRLSCAASGFTFDDYVIGWFRQAPGKEREGVSCISSSEGST
YYAESVKGRFTISSDNAKNTVYLQMNSLKPEDTAVYYCAASTWLDFVHGNEYDYRGQGTQVT
VSS (SEQ ID NO: 26).
[00317] In some embodiments, an exemplary SRA-1 binding CDR1 sequence can be
any one of the
amino acid sequences: TYAMG (SEQ ID NO: 31), YDMG (SEQ ID NO: 32), RYAMA (SEQ
ID NO:
33), THAMG (SEQ ID NO: 34), FVAMG (SEQ ID NO: 35).
[00318] In some embodiments, an exemplary SRA-1 binding CDR2 sequence can be
any one of the
amino acid sequence;
-52-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
AMISSLSSKSYADTVKG (SEQ ID NO: 36).
AMISSLSSKSYADSVKG (SEQ ID NO: 37).
A1NWSGGSTAYADSVKG (SEQ ID NO: 38).
AVSQSGLLTFYADSVKG (SEQ ID NO: 39).
AVSQSGLLTFYADSVKG (SEQ ID NO: 40).
TITSVTSGGSLNYADSVKG (SEQ ID NO: 41).
TITSITSGGRTNYADSVKG (SEQ ID NO: 42).
[00319] In some embodiments, an exemplary SRA-1 binding CDR3 sequence
sequences can be any
one of the amino acid sequences:
DLLPYSSSRSLPMGYD (SEQ ID NO: 43).
DLLPYSSTRSLPMGYDY (SEQ ID NO: 44).
KPAKYHFGSGYRDFAE (SEQ ID NO: 45).
XSRFPLVVPVAYEN (SEQ ID NO: 46).
DSRFPLVVPVAYEN (SEQ ID NO: 47).
LGFDY (SEQ ID NO: 48).
VPYVNDY (SEQ ID NO: 49).
wherein, X is a naturally occurring amino acid.
[00320] In some embodiments, an exemplary RAGE binding CDR1 sequence can be
any one of the
amino acid sequences: DRA1G (SEQ ID NO: 50), NYA1G (SEQ ID NO: 51), TDAFG (SEQ
ID NO:
52), NYSMG (SEQ ID NO: 53), DMTMG (SEQ ID NO: 54), NYAMG (SEQ ID NO: 55), or
LYRMG
(SEQ ID NO: 56).
[00321] In some embodiments, an exemplary RAGE binding CDR2 sequence can be
any one of the
amino acid sequences:
AISWSGGRTYYADSVKG (SEQ ID NO: 57).
CSANNDNRAFYEDSVKG (SEQ ID NO: 58).
CVDRDGGSTYYLDSVTG (SEQ ID NO: 59).
AMRWNGSSSYYADLVKG (SEQ ID NO: 60).
TTSWSGALTHYTDSVKG (SEQ ID NO: 61).
AISNSGLSTYYQDSVKG (SEQ ID NO: 62).
GISWSGDSTLYADSVKG (SEQ ID NO: 63).
AMSTSGAGTYYADSVKG (SEQ ID NO: 64).
CISRTDGSTDYADSVKG (SEQ ID NO: 65).
[00322] In some embodiments, an exemplary RAGE binding CDR3 sequence can be
any one of the
amino acid sequences:
ENLASSGSAYSDDRYN (SEQ ID NO: 66)
-53-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
RCAAGRVNLYYGMDY (SEQ ID NO: 67),
RLYGCSGYGRDYAD (SEQ ID NO: 68).
GKRYGYYDY (SEQ ID NO: 69).
SDSDYGNKYDY (SEQ ID NO: 70).
RSGWSGQYDY (SEQ ID NO: 71).
KQGADWAPYDY (SEQ ID NO: 72).
GVRFGVYDY (SEQ ID NO: 73).
[00323] In some embodiments, Lox-1 binding CDR1 sequences can be any one of
the amino acid
sequences: DYAIG (SEQ ID NO: 74), 1NAMA (SEQ ID NO: 75), AYHMG (SEQ ID NO.:
76), IGTMG
(SEQ ID NO: 77), LRTVG (SEQ ID NO: 78), DYAIG (SEQ ID NO: 79), TSGMG (SEQ ID
NO: 80),
NYAMG (SEQ ID NO: 81), DYGIG (SEQ ID NO: 82), DYVIG (SEQ ID NO: 83).
[00324] In some embodiments, Lox-1 binding CDR2 sequences can be any one of
the amino acids
sequences:
HLTNSGRTGYADSVKG (SEQ ID NO: 84)
AISWSVSSTYYADSVKG (SEQ ID NO: 85)
VTYGLGSTNYAESVKG (SEQ ID NO: 86)
IMSAGTTRYADSVKG (SEQ ID NO: 87)
CVSRDGGSTYYLDSVKG (SEQ ID NO: 88)
GIXWNSRXTYYAESVKG (SEQ ID NO: 89)
AITWSGSSTYYADSVKG (SEQ ID NO: 90)
CISSSDGSTDYADSVKG (SEQ ID NO: 91)
CISSVEGSTYYADSVKG (SEQ ID NO: 92)
CISSSEGSTYYAESVKG (SEQ ID NO: 93)
wherein, X is a naturally occurring amino acid.
1003251 In some embodiments, Lox-lbinding CDR3 sequences can be any one of the
amino acid
sequences:
GRTYYSGSYYFGLGSD (SEQ ID NO: 94)
LGLHWS (SEQ ID NO: 95)
RSGERYDYYKAQYEY (SEQ ID NO: 96)
EIDTDPRSGEWDY (SEQ ID NO: 97)
RPVFSNVDY (SEQ ID NO: 98)
SRYDCSKYLIDYNY (SEQ ID NO: 99)
NYYGSXWSVNSDDYDY (SEQ ID NO: 100)
AQRGRYYYLDRNVEYD (SEQ ID NO: 101)
GRTYYSGSYYFGLGSDEYDY (SEQ ID NO: 102)
-54-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
GTWLDCSGYGSYDMDY (SEQ ID NO: 103)
STWLDFVHGNEYDY (SEQ ID NO: 104)
[00326] In some embodiments, the bi- or trispecific engager comprises a
binding domain that binds to
a protein that can generate phagocytosis activation signals or pro-
inflammatory signals, for example via
activation of any one of: MRC1, Itg115, MERTK, ELMO, BAIL Tyro3, Axl, Traf6,
Syk, MyD88, Zap70,
PI3K, FcyR1, FcyR2A, FcyR2B2, FcyR2C, FcyR3A, FcER1, FcccRI, BAFF-R, DAP12,
NFAM1, and
CD79b.
[00327] In some embodiments, the bi- or trispecific engager comprises a
binding domain that binds to
the extracellular domain of a TREM protein. TREM 1, 2, 3. TREMs share common
structural properties,
including the presence of a single extracellular immunoglobulin-like domain of
the V-type, a trans-
membrane domain and a short cytoplasmic tail. In particular, the TREM trans-
membrane domain (TM)
possesses negatively charged residues that interact with the positively
charged residues of the DNAX
Activating Protein of 12 lcDa (DAP12), a trans-membrane adaptor containing an
immunoreceptor tyrosine-
based activation motif (ITAM).
D. Engagers with domains that promote inflammatory activity of the myeloid
cell
[00328] Activation of monocytes or macrophages can lead to increase in
inflammatory activity.
Activated M1 monocytes or macrophages are characterized by 1FN-gamma
production, as well as
production of pro-inflammatory cytolcines,
1L-6, CSF, GMCSF, and TNF
to name a few. In some
embodiments, a monocyte or macrophage M1 phenotype is associated with potent
pro-inflammatory
response associated with IL-1 signaling cascade and inflammasome activation.
[00329] In some embodiments, a bi- or trispecific engager may comprise a
domain that generates a
signal is necessary to trigger inflammasomes and pro-inflammatory signals.
Toll-like receptors, TLRs are
known to induce inflammasome activation. TLRs elicit conserved inflammatory
pathways culminating in
the activation of NF-KB and activating protein-1 (AP-1). TLR ligands include
high-mobility group B1
(HMGB1), heat shock proteins (FISP60, HSP70), endotoxins, and extracellular
matrix components. TLR2
and TLR4, for example comprise extracellular domains which are activated by
ligand binding, and which
is turn activates a pro-inflammatory cascade associated with inflammasome
activation. Intracellular
signaling pathway is mediated by signaling proteins e.g., Nod-like receptors
(NLRs) that recruit
proinflammatory caspases and induce their cleavage and activation. This leads
to direct activation of ROS,
and often results in a violent cell death known as pyroptosis. There are four
inflammasome complexes,
NLRP1m, NLRP3, IPAF and AIEV12.
[00330] In some embodiments, a bi- or trispecific engager may comprise a
binding domain that
generates a signal is necessary to trigger inflarnmasomes and pro-inflammatory
signal binds to TLRs, such
as TLR4. TLR4 is expressed in monocytes or macrophages and is induced by LPS
and other ligands. In
some embodiments, a bi- or trispecific engager may bind to a TLR ligand which
then binds to the TLR.
-55-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
E Engagers with domains that promote cell adhesion and inflammatory activity
of the myeloid cell
[00331] Cell-cell and cell-substratum adhesion is mediated by the binding of
integrin extracellular
domains to diverse protein ligands; however, cellular control of these
adhesive interactions and their
translation into dynamic cellular responses, such as cell spreading or
migration, requires the integrin
cytoplasmic tails. These short tails bind to intracellular ligands that
connect the receptors to signaling
pathways and cytoskeletal networks. Integrins are heterodimeric adhesion
receptors formed by the non-
covalent association of a and p subunits. Each subunit is a type I
transmembrane glycoprotein that has
relatively large extracellular domains and, with the exception of the 134
subunit, a short cytoplasmic tail.
Individual integrin family members have the ability to recognize multiple
ligands. Integrins can bind to a
large number of extracellular matrix proteins (bone matrix proteins,
collagens, fibronectins, fibrinogen,
laminins, thrombospondins, vitronectin, and von Willebrand factor), reflecting
the primary function of
integrins in cell adhesion to extracellular matrices. Many "counter-receptors"
are ligands, reflecting the
role of integrins in mediating cell-cell interactions. Integrins undergo
conformational changes to increase
ligand affinity.
[00332] The Integrin 132 subfamily consists of four different integrin
receptors, amI32 (CD1 lb/CD1 8,
Mac-1, CR3, Mo-1), a1,132 (CD1 1 a/CD 18, LFA- 1), ax132 (CD 1 1 c/CD1 8), and
an32 (CD 1 1 cVCD1 8). These
leukocyte integrins are involved in virtually every aspect of leukocyte
function, including the immune
response, adhesion to and transmigration through the endothelium, phagocytosis
of pathogens, and
leukocyte activation.
[00333] The a subunits of all 132 integrins contain an inserted region of ¨200
amino acids, termed the I
or A domain. Highly conserved I domains are found in several other integrin a
subunits and other proteins,
such as certain coagulation and complement proteins. I domains mediate protein-
protein interactions, and
in integrins, they are integrally involved in the binding of protein ligands.
Although the I domains dominate
the ligand binding functions of their integrins, other regions of the a
subunits do influence ligand
recognition. As examples, in am132 a mAb (0K1v11) recognizing an epitope
outside the I domain but in the
cmt subunit inhibits ligand binding; and the EF-hand regions in aLl32 and
a.2131, integrins with I domains in
their a subunits, contribute to ligand recognition. The am subunit, and
perhaps other a subunits, contains a
lectin-like domain, which is involved in engagement of non-protein ligands,
and occupancy may modulate
the function of the I domain.
[00334] As integrins lack enzymatic activity, signaling is instead induced by
the assembly of signaling
complexes on the cytoplasmic face of the plasma membrane. Formation of these
complexes is achieved in
two ways; first, by receptor clustering, which increases the avidity of
molecular interactions thereby
increasing the on-rate of binding of effector molecules, and second, by
induction of conformational changes
in receptors that creates or exposes effector binding sites. Within the ECM,
integrins have the ability to
bind fibronectin, laminins, collagens, tenascin, vitronectin and
thrombospondin. Clusters of integrin/ECM
-56-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
interactions form focal adhesions, concentrating cytoskeletal components and
signaling molecules within
the cell. The cytoplasmic tail of integrins serve as a binding site for a-
actinin and talin which then recruit
vinculin, a protein involved in anchoring F-actin to the membrane. Talin is
activated by kinases such as
protein kinase C (PKCILI).
[00335] Integrins are activated by selectins. Leucocytes express L-selectin,
activated platelets express
P-selectin, and activated endothelial cells express E- and P-selectin. P-
selectin-mediated adhesion enables
chemokine- or platelet-activating factor-triggered activation of 112
integrins, which stabilizes adhesion. It
also facilitates release of chemokines from adherent leucocytes. The
cytoplasmic domain of P-selectin
glycoprotein ligand 1 formed a constitutive complex with Nef-associated factor
1. After binding of P-
selectin, Src kinases phosphorylated Nef-associated factor 1, which recruit
the phosphoinositide-3-0H
kinase p85-pl 1 05 heterodimer and result in activation of leukocyte
integrins. E-selectin ligands transduce
signals that also affect 132 integrin function. Selectins trigger activation
of Src family kinases. SFKs
activated by selectin engagement phosphorylate the immunoreceptor tyrosine-
based activation motifs
(ITAMs) in the cytoplasmic domains of DAP12 and FcRy. In some respects, CD44
is sufficient to transduce
signals from E-selectin. CD44 triggers the inside-out signaling of integrins.
A final common step in integrin
activation is binding of talin to the cytoplasmic tail of the (3 subunit
Kindlins, another group of cytoplasmic
adaptors, bind to a different region of integrin tails. Kindlins increase the
clustering of talin-activated
integrins. Kindlins are responsive to selectin signaling, however, kindlins
are found mostly in
hematopoietic cells, such as neutrophils. Selectin signaling as well as
signaling upon integrin activation by
chemokines components have shared components, including SFKs, Syk, and SLP-76.
1003361 In some embodiments, the engagers comprise a binding domain that can
bind to the
extracellular domain of an adhesion molecule such as an integrin or a
selectin, for example, a P-selectin,
L-selectin or E-selectin.
F. Engagers with binding domains that inhibits anti-phagocytic and anti-
inflammatory activity of
the myeloid cell
[00337] In one aspect, a hi- or a trispecific engager may comprise an
additional functional domain that
inhibits CD47 mediated downregulation of monocyte or macrophage phagocytosis.
Tumor cells typically
express the "don't eat me" signal CD47 that binds to a monocyte or macrophage
receptor S1RP-a, and
inhibits phagocytosis. Inhibition of CD47 therefore counteracts the tumor cell
mediated anti-phagocytosis
activity. One arm of a bi-or trispecific engager may comprise a CD47 blocker.
The CD47 blocker associated
with the engager may be the extracellular CD47-binding domain of SIRP-a,
acting as a decoy receptor or
neutralizing receptor.
[00338] In one aspect, disclosed herein are compositions that can inhibit
phagocytosis regulatory signal
transduction by members of the Siglec family of membrane proteins that are
expressed on immune cells.
Various members of the family transduce checkpoint signal upon contact with
sialylated glycans on
-57-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
membrane proteins. In some members, the intracellular domains of the Siglec
proteins comprise multiple
immunoreceptor tyrosine-based inhibitory motifs (ITIMs). ITIMs share a
consensus amino acid sequence
in their cytoplasmic tail, namely (IN/L/S)-X-Y-X-X-(LN), where X denotes any
amino acid,
Isoleucine, V=valine, L=Lysine, S=Serine, Y=Tyrosine. Phosphorylation of the
Tyrosine residues at the
motif recruit either of two SH2 domain¨containing negative regulators: the
inositol phosphatase
SHIP (Src homology 2¨containing inositol polyphosphate 5-phosphatase) or the
tyrosine phosphatase
SHP-1 (Src homology 2¨containing protein tyrosine phosphatase-1). A leucine in
the (Y+2) position favors
binding to SHIP, whereas an isoleucine in the (Y-2) position favors SHP-1
binding. ITIMs can also bind
to another tyrosine phosphatase, SHP-2, but evidence for SHP-2 playing a
functional role in ITIM-mediated
inhibition is less clear than for the other mediators. Therefore, activation
of the Siglec membrane proteins
at the extracellular ligand binding domain by binding with a sialic acid
residue, (e.g. in sialylated membrane
glycan proteins), the ITIMs receive the intracellular signals, which are
phosphorylated, and initiate the SLIP
mediated signaling for immunomodulation, including reduction in phagocytic
potential.
1003391 Siglec family receptors comprise the membrane proteins, siglec 1
(CD169), siglec 2 (CD22),
siglec 3 (CD33), siglec 4 (MAG), siglec 5, siglec 6, siglec 7, siglec 8,
siglec 9, siglec 10, siglec 11, siglec
12, siglec 13, siglec 14, siglec 15, siglec 16.
[00340] In some embodiments the composition described herein may comprise a
binding domain for a
Siglec receptor (SgR) such that the SgR receptor is blocked, and SgR induced
immunoregulatoiy
intracellular signaling is inhibited.
Specific multimerization domains for engagers
[00341] Also envisioned in the molecular design of bi- and trispecific
engagers are additional structures
and helpers that assist in the engager's capability to modularly and
concomitantly engage with multiple
targets. These designs include additional anchoring or clasping elements for
two or more binding domains
separated by linkers, such as in a hi- or trispecific antibody, a tribody or a
triple body formats. These higher
order multi-specific binding domains often require inclusion of the
multimerization domains to improve
stability and flexibility in binding the multiple domains on the same and
different cells. The additional
anchoring or clasping elements occur in cognate pairs, such that one of the
cognate pair of the anchoring
or clasping modality is attached to one of the binding domains of an engager
and the other of the pair is
attached to the other of the cognate pair.
[00342] In some embodiments, the engager is a recombinant protein comprising
multiple binding
domains as described throughout the specification, each having individual
binding specificities, that are
each linked together by linkers having cognate peptide anchoring or clasping
elements that exhibit
complementary binding with each other. For example, one binding domain of the
recombinant protein is
fused with the first of a pair of cognate peptides, and the other binding
domain is fused with the second of
the pair of peptides, wherein, the pair of peptides exhibit complementary
binding with each other, wherein
-58-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
the pair of cognate peptides comprise: (a) leucine zipper domains that exhibit
complementary binding
with each other, for example, leucine zippers in naturally occurring protein-
protein interactions, such as
the zipper sequences within the binding regions of c-Fos and c-Jun proteins,
(b) synthetic peptides
designed to specifically bind to each other via synthetic clasps.
[00343] In some embodiments, the therapeutic agent is a recombinant protein
comprising multiple
binding fragments configured to facilitate accelerated association with each
other by means of leucine
zipper peptide pairs comprised in the recombinant proteins. Leucine zipper
sequences often comprise a
heptad leucine repeat and constitute adhesive peptide pairs when two peptides
possess the leucine zipper
structures. Among the naturally occurring leucine zippers, the c-Fos and c-
June pairs are most widely
known. They exhibit a strong binding affinity with KD: 5.4 x 104 M. They form
parallel coils. In some
embodiments, the leucine zipper coil is the coil of the c-Fos: c-Jun pair. In
some embodiments, exemplary
cognate pair anchoring or clasping elements include the ACID-pl(LZA) and BASE-
p1 (LZB) pair; which
are prevented from homodimerizing because of the electrostatic repulsion
between the charges among the
amino acid side chains. Prevention of homodimerization can be beneficial in a
number of embodiments.
[00344] Exemplary c-Fos leucine zipper domain comprises an amino acid sequence
as follows:
IARLEEKVICTLKAQNSELASTANMLREQVAQLKQKVMNH (SEQ ID NO: 119).
[00345] Exemplary c-Jun leucine zipper domain comprises an amino acid sequence
as follows:
TDTLQAETDQLEDEKSALQTEIANLLICEICEKLEFTLAAH (SEQ ID NO: 120).
[00346] Exemplary LZA leucine zipper domain comprises an amino acid sequence
as follows:
AQLEICELQALEICENAQLEWELQALEKELAQK (SEQ ID NO: 121).
[00347] Exemplary LZB leucine zipper domain comprises an amino acid sequence
as follows:
AQLKKKLQALKKKNAQLKWKLQALKKKLAQK (SEQ ID NO: 122).
[00348] In some embodiments, the therapeutic agent is a recombinant protein
comprising multiple
binding fragments configured to facilitate accelerated association with each
other by means of c-Fosic-Jun
binding domains in the peptide pairs comprised within the recombinant
proteins.
[00349] In some embodiments, the anchoring or clasping elements exhibit
specific heterodimerizing
capabilities and do not exhibit homodimerization.
[00350] In some embodiments, the therapeutic agent is a recombinant protein
comprising multiple
binding fragments configured to facilitate accelerated association with each
other by means of synthetic
clasps. In some embodiments, the synthetic anchoring or clasping elements are
designed to heterodimerize
and prevent homodimerization.
[00351] In some embodiments the synthetic clasps of the linkers are non-
peptide crosslinkers.
[00352] In some embodiments the complementary binding of cognate peptides with
each other can be
via chemical binding, such as crosslinking. Chemical crosslinkers can be
useful for activating the
crosslinking in vitro. There are home- and heterobifunctional protein
crosslinkers that can be commercially
-59-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
available. Examples include BS2G crosslinker (BS2G; Bis[Sulfosuccinimidyl]
glutarate) is an amine-
reactive, water soluble, homobifunctional protein crosslinker (both binding
units at the opposite ends of a
spacer arm have the identical reactive groups), or its membrane permeable
version, DSG (Disuccinimidyl
glutarate;Di(N-succinimidyl) glutarate); BS3 crosslinker
(Bis[sulfosuccinimidyl] suberate; Sulfo-DSS;
BSSS) or DST crosslinker (Disuccinimidyl tartrate), are among other
homobifuncfional crosslinkers for
peptides; whereas BMPS (N-(13-Maleimidopropyloxy) succinirnide ester; NIBS
crosslinker (m-
Maleirnidobenzoy1-14-hydroxysuccinimide ester); PDPH crosslinker (3[2-
Pyridyldithioipmpionyl
hydrazide) provide examples of some heterobifunctional crosslinkers.
[00353] In some embodiments, the variable light chain (VI) subunit and the
variable heavy chain (VH)
regions arranged in tandem within a multi-specific engager may be linked via
two linkers having the
cognate peptide anchoring or clasping elements. The length of the linkers can
limit or facilitate specific VL
-VII associations. For example, limiting the linker peptide length to less
than 10 amino acids restricts the
association between two adjacent VL and V11 domains.
[00354] In some embodiments, the anchoring or clasping elements exhibit an
affinity having a KD: less
than 5 x 10-6M, or less than 104M, less than 5 x 10-7M, or less than 4 x 10-
7M, or less than 3 x 10-7M, or
less than 2 x 10-7M; or less than 10-7M, or less than 9 x 10-8M, or less than
8 x 104M, or or less than 7 x
104M, or less than 6 x 104M, or less than 5 x 104M, or less than 4 x 104M, or
less than 3 x 104M, or less
than 2 x 10-8M, or less than 104M, or less than 104M, or less than 10-1 M, or
higher affinity.
[00355] Additionally, inclusion of the additional anchoring or hetero-
multimerization domains in these
higher order multi-specific engagers (e.g., engagers with multiple binding
domains) that are formed by the
assembly of heterodimeric or heteromultimeric units assist in the production,
folding, stability and tissue
availability of the multi-specific engagers.
Co-expression of an Inflammatoty Gene
[00356] In one aspect, the recombinant nucleic acid comprises a coding
sequence for a pro-
inflammatory gene, which is expressed in an engineered cell. In some
embodiments, the pro-inflammatory
gene is a cytokine. Examples include but not limited to TNF-oc, IL-la, IL-1f3,
IL-6, CSF, GMCSF, or 11,-
12 or interferons. In some embodiments, the recombinant nucleic acid encoding
a coding sequence of a
proinflammatory gene is a therapeutic agent, such as an additional therapeutic
agent to accompany at least
the first therapeutic agent.
Peptide Linker
[00357] In some embodiments, the extracellular antigen binding domains, scFvs
or binding domains
are linked with each other by a linker. In some embodiments, where there are
more than one scFv at the
extracellular antigen binding domain the more than scFvs are linked with each
other by linkers.
[00358] In some embodiments the linkers are flexible, In some embodiments the
linkers comprise a
hinge region. Linkers are usually short peptide sequences. In some embodiments
the linkers are stretches
-60-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
of Glycine and one or more Serine residues. Other amino acids preferred for a
peptide linker include but
are not limited to threonine (Thr), serine (Ser), proline (Pro), glycine (0y),
aspartic acid (Asp), lysine
(Lys), glutamine (Gin), asparagine (Mn), and alanine (Ala) arginine (Arg),
phenyialanine (Phe), giutamic
acid (Glu). Of these Pro, Thr, and Gin are frequently used amino acids for
natural linkers. Pro is a unique
amino acid with a cyclic side chain which causes a very restricted
conformation. Pro-rich sequences are
used as interdomain linkers, including the linker between the lipoyl and E3
binding domain in pyruvate
dehydrogenase (GA2PA3PAKQEA3PAPA2ICAEAPA3PA21CA). For the purpose of the
disclosure, the
empirical linkers may be flexible linkers, rigid linkers, and cleavable
linkers. Sequences such as (G4S)x
(where x is multiple copies of the moiety, designated as 1, 2, 3, 4, and so
on) comprise a flexible linker
sequence. Other flexible sequences used herein include several repeats of
glycine, e.g., (G1y)6 or (Gly)8µ
On the other hand, a rigid linker may be used, for example, a linker (EAAAK)x,
where x is an integer, 1,
2, 3, 4 etc. gives rise to a rigid linker.
[00359] The length of a linker peptide can be crucial in the design of a multi-
specific engager. For
example, limiting the linker peptide length to less than 10 amino acids
restricts the association between
two adjacent domains. In some embodiments, the linker may comprise a anchoring
or clasping function
and may comprise a crosslinking moiety. The cross linking moiety may be a
peptide or a chemical cross
linking moiety, several of which are described in the previous section.
[00360] Specific peptides with specific functions have been discussed
elsewhere in the document. In
some embodiments, a peptide linker may further function as an activator or a
signal in a myeloid cell. For
example, a TLR4 activation peptide may be incorporated within the linker
between two binding domains
of an engager, and the TLR4 activation peptide binds to and activates a TLR4
signal in a monocyte or
macrophage.
[00361] In some embodiments, a peptide linker may further function as a
conditionally cleavable linker.
By conditionally cleavable it is understood that the peptide is cleaved when
the agent that cleaves it is
available. For example, an MMP2 cleavable peptide is described herein, which
is readily cleaved only
when the peptide is in a region rich in MMP2. An exemplary MMP2 cleavable
peptide is GPLGVR.
[00362] In some embodiments, a peptide linker may further function as a
targeting peptide. For
example, an M2 peptide is described, which is can bind to a M2 monocyte or
macrophage, which is the
predominant tumor associated monocyte or macrophage phenotype. An exemplary
peptide is
YEQDPWGVICWWY.
[00363] Any one or more peptide linkers may comprise specialized functions,
such as they can dimerize,
trimerize or multimerize. In some embodiments, one or more linkers may
comprise leucine zipper
sequences.
[00364] In some embodiments, the peptide linker is 5, 6, 7, 8,
9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19
or 20 amino acids in length. In some embodiments, a peptide linker or the two
linker peptides with an
-61-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
anchor or a clasp together span a length of 50 amino acids or less, 45 amino
acids or less, 40 amino acids
or less, 35 amino acids or less, 30 amino acids or less, 25 amino acids or
less, 20 amino acids or less, 15
amino acids or less, 10 amino acids or less, or 5 amino acids or less. In some
embodiments, a peptide linker
or the two linker peptides with an anchor or a clasp together span a length of
25 amino acids or less, 24
amino acids or less, 23 amino acids or less, 22 amino acids or less, 21 amino
acids or less, 20 amino acids
or less, 19 amino acids or less, 19 amino acids or less, 18 amino acids or
less, 17 amino acids or less, 16
amino acids or less, 15 amino acids or less, 14 amino acids or less, 13 amino
acids or less, 12 amino acids
or less, 11 amino acids or less, 10 amino acids or less, 9 amino acids or
less, 8 amino acids or less, 7 amino
acids or less, 6 amino acids or less, or 5 amino acids or less.
Methods for Preparing Monocvte or macrophage Specific Engagers
[00365] The engagers described herein are produced as recombinant proteins.
Generally, a polynucleotide
sequence is constructed that encodes the recombinant protein is prepared and
inserted into an expression
vector, such as a plasmid, in proper orientation and correct reading frame for
expression, if necessary, the
DNA may be linked to the appropriate transcriptional and translational
regulatory control nucleotide
sequences recognized by the desired host (e.g., bacteria), although such
controls are generally available in
the expression vector. The vector is then introduced into the host bacteria
for cloning using standard
techniques (see, e.g., Sambrook et al. (1989) Molecular Cloning, A Laboratory
Manual, Cold Spring
Harbor Laboratory, Cold Spring Harbor, N.Y.).
[00366] The recombinant polynucleotide is synthesized by ligating DNA
encoding, for example, a first
binding domain, a linker, and a second binding domain in the same open reading
frame using the molecular
cloning techniques well known to one of skill in the art. In some embodiments,
one or more polynucleotide
sequences are arranged under the same promoter and regulatory elements for
generation of a single
polypeptide. In some embodiments, a short spacer may be inserted between two
adjacent polynucleotides
encoding two polypeptides wherein the spacer may encode a post translational
cleavage site. The two
polypeptides can be separated after translation by induction of the cleavage
at the specific cleavage site. In
some embodiments, the construct may be monocistronic or polycistronic. In some
embodiments, more than
one polypeptides are generated which then reassemble after translation. For
example, light chain and heavy
chain domains of an antibody or parts thereof can be generated by translation
from two independent
polynucleotide sequences, which are allowed to freely assemble with each other
post-translationally.
Alternatively, multiple polypeptide chains containing LC and HC variable
domains that bind with each
other are transcribed and translated from a single polynucleotide, which is
cleaved after translation into
respective peptide chains which can then reassemble. The polypeptide having a
leader sequence is a
preprotein and can have the leader sequence cleaved by the host cell to form
the mature form of the
polypeptide.
-62-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00367] In some embodiments, the polynucleotide construct encodes an N-
terminal signal sequence
upstream of the polypeptide for secretion of the polypeptides. In some
embodiments, the N terminal signal
sequence comprises a secretion sequence. The resulting translated protein
product having the N-terminal
signal sequence for secretion would be secreted by the cell.
[00368] In some embodiments the plasmid vector is introduced or incorporated
in the cell by known
methods of transfection, such as using lipofectamine, or calcium phosphate, or
via physical means such as
electroporation or nucleofection. In some embodiments the viral vector is
introduced or incorporated in the
cell by infection, a process commonly known as viral transduction.
[00369] In some embodiments, recombinant nucleic acid is integrated or
incorporated in an expression
vector. A vector comprises one or more promoters, and other regulatory
components, including enhancer
binding sequence, initiation and terminal codons, a 51.1TR, a 31TIR comprising
a transcript stabilization
element, optional conserved regulatory protein binding sequences and others.
[00370] In some embodiments the vectors of use in the application are
specifically enhanced for
expression. Other exemplary vectors of use throughout the process include
phages, cosmids, or artificial
chromosomes.
[00371] It is understood that any one of the first binder domains (domain
binding to a target cell such
as a cancer cell or a diseased cell or a pathogen) can be designed in
combination with a second binder
domain that binds to a myeloid cell or a third binding domain described
anywhere in the specification.
[00372] Viral Vectors: In some embodiments, the vector for expression of the
recombinant protein is of
a viral origin, namely a lentiviral vector or an adenoviral vector. In some
embodiments, the nucleic acid
encoding the recombinant nucleic acid is encoded by a lentiviral vector. In
some embodiments the lentiviral
vector is prepared in-house and manufactured in large scale for the purpose.
In some embodiments,
commercially available lentiviral vectors are utilized, as is known to one of
skill in the art.
[00373] In some embodiments the viral vector is an Adeno-Associated Virus
(AAV) vector.
[00374] Lipid nanoparticle mediated delivery: Lipid nanoparticles (LNP) may
comprise a polar and or
a nonpolar lipid. In some embodiments cholesterol is present in the LNPs for
efficient delivery. LNPs are
100-300 nm in diameter provide efficient means of mRNA delivery to various
cell types, including
monocytes or macrophages. In some embodiments, LNP may be used to introduce
the recombinant nucleic
acids into a cell in in vitro cell culture. In some embodiments, the LNP
encapsulates the nucleic acid
wherein the nucleic acid is a naked DNA molecule. In some embodiments, the LNP
encapsulates the
nucleic acid wherein the nucleic acid is an mRNA molecule. In some
embodiments, the LNP encapsulates
the nucleic acid wherein the nucleic acid is inserted in a vector, such as a
plasmid vector. In some
embodiments, the LNP encapsulates the nucleic acid wherein the nucleic acid is
a circRNA molecule.
[00375] In some embodiments, the LNP is used to deliver the nucleic acid into
a subject. LNP can be
used to deliver nucleic acid systemically in a subject. It can be delivered by
injection. In some
-63-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
embodiments, the LNP comprising the nucleic acid is injected by intravenous
route. In some embodiments
the LNP is injected subcutaneously.
[00376] Microbubble mediated delivery: In some embodiments, microbubbles can
be used for delivery
of a composition comprising e.g., a nucleic acid in a subject. Perfluorocarbon-
filled microbubbles are stable
for circulating in the vasculature as blood pool agents, they act as carriers
of these agents until the site of
interest is reached. Ultrasound applied over the skin surface can then be used
to burst the microbubbles at
this site, causing localized release of the drug. Various other forms of
microbubbles include Sonazoid
Optison, gas-filled albumin microbubble, and PESDA. Optimization of the
composition of the microbubble
with respect to the composition of the therapeutic agent that is delivered,
along with the site of delivery
intended is necessary.
[00377] In some embodiments, the recombinant proteins, for example the
engagers, or the inflammatory
proteins that are co-expressed, or any associated protein designed to be
expressed in a myeloid cell may be
encoded by a recombinant nucleic acid, wherein the recombinant nucleic acid is
an RNA. In some
embodiments, the recombinant nucleic acid is an mRNA. In some embodiments, the
mRNA comprises one
or more modifications for enhanced expression and stability. In some
embodiments, the mRNA may be
circularized. In some embodiments, the modifications may include but are not
limited to: replacement of a
nucleobase with a base analog, or a modified nucleotide; inserting one or more
motifs within the mRNA,
and introducing modifications in the 5'- and 3' UTRs. In some embodiments, the
recombinant nucleic acid
may be administered directly in a subject in need thereof
Pharmaceutical Composition
[00378] Provided herein is a pharmaceutical composition, comprising at least a
first therapeutic agent
which comprises monocyte or macrophage specific engagers. The monocyte or
macrophage specific
engagers in the composition may be in the form of peptides or polypeptides or
a complex of multiple
peptides. The monocyte or macrophage specific engagers may be provided in a
composition as purified
recombinant proteins. The monocyte or macrophage specific engagers may be
provided in a composition
as conjugated recombinant proteins, VHH complexes, scEv complexes or
nanobodies. The monocyte or
macrophage specific engagers may be in the form of a polynucleotide encoding
the recombinant monocyte
or macrophage specific engager& In some embodiments, polynucleofide encoding
the monocyte or
macrophage specific engagers may comprise DNA, mRNA or circRNA or a liposomal
composition of any
one of these. The liposome is a LNP.
[00379] Pharmaceutical compositions can include, in addition to active
ingredient, a pharmaceutically
acceptable excipient, carrier, buffer, stabilizer or other materials well
known to those skilled in the art.
Such materials should be non-toxic and should not interfere with the efficacy
of the active ingredient. The
precise nature of the carrier or other material will depend on the route of
administration.
-64-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00380] Acceptable carriers, excipients, or stabilizers are those that are non-
toxic to recipients at the
dosages and concentrations employed, and include buffers such as phosphate,
citrate, and other organic
acids; antioxidants including ascorbic acid and methionine; preservatives
(such as
octadecyldimethylbenzyl ammonium chloride; hexamethonium chloride;
benzalkonium chloride,
benzethonium chloride; phenol, butyl or benzyl alcohol; alkyl parabens such as
methyl or propyl paraben;
catechol; resorcinol; cyclohexanol; 3-pentanol; and m-cresol); low molecular
weight (less than about 10
residues) polypeptides; proteins, such as serum albumin, gelatin, or
immunoglobulins; hydrophilic
polymers such as polyvinylpyrrolidone; amino acids such as glycine, glutamine,
asparagine, histidine,
arginine, or lysine; monosaccharides, disaccharides, and other carbohydrates
including glucose, mannose,
or dextrins; chelating agents such as EDTA; sugars such as sucrose, mannitol,
trehalose or sorbitol; salt-
forming counter-ions such as sodium; metal complexes (e.g., Zn-protein
complexes); and/or non-ionic
surfactants such as TWEEN*, PLURONICS* or polyethylene glycol (PEG).
[00381] Acceptable carriers are physiologically acceptable to the administered
patient and retain the
therapeutic properties of the compounds with/in which it is administered.
Acceptable carriers and their
formulations are generally described in, for example, Remington'
pharmaceutical Sciences ( 1 8th ed_ A.
Gennaro, Mack Publishing Co., Easton, PA 1990). One example of carrier is
physiological saline. A
pharmaceutically acceptable carrier is a pharmaceutically acceptable material,
composition or vehicle, such
as a liquid or solid filler, diluent, excipient, solvent or encapsulating
material, involved in carrying or
transporting the subject compounds from the administration site of one organ,
or portion of the body, to
another organ, or portion of the body, or in an in vitro assay system.
Acceptable carriers are compatible
with the other ingredients of the formulation and not injurious to a subject
to whom it is administered. Nor
should an acceptable carrier alter the specific activity of the neoantigens.
[00382] In one aspect, provided herein are pharmaceutically acceptable or
physiologically acceptable
compositions including solvents (aqueous or non-aqueous), solutions,
emulsions, dispersion media,
coatings, isotonic and absorption promoting or delaying agents, compatible
with pharmaceutical
administration. Pharmaceutical compositions or pharmaceutical formulations
therefore refer to a
composition suitable for pharmaceutical use in a subject. Compositions can be
formulated to be compatible
with a particular route of administration (i.e., systemic or local). Thus,
compositions include carriers,
diluents, or excipients suitable for administration by various routes.
[00383] In some embodiments, a composition can further comprise an acceptable
additive in order to
improve the stability of immune cells in the composition. Acceptable additives
may not alter the specific
activity of the immune cells. Examples of acceptable additives include, but
are not limited to, a sugar such
as mannitol, sorbitol, glucose, xylitol, trehalose, sorbose, sucrose,
galactose, dextran, dextrose, fructose,
lactose and mixtures thereof. Acceptable additives can be combined with
acceptable carriers and/or
excipients such as dextrose. Alternatively, examples of acceptable additives
include, but are not limited to,
-65-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
a surfactant such as polysorbate 20 or polysorbate 80 to increase stability of
the peptide and decrease gelling
of the solution. The surfactant can be added to the composition in an amount
of 0.01% to 5% of the solution.
Addition of such acceptable additives increases the stability and half-life of
the composition in storage.
[00384] The pharmaceutical composition can be administered, for example, by
injection. Compositions
for injection include aqueous solutions (where water soluble) or dispersions
and sterile powders for the
extemporaneous preparation of sterile injectable solutions or dispersion. For
intravenous administration,
suitable carriers include physiological saline, bacteriostatic water, or
phosphate buffered saline (PBS). The
carrier can be a solvent or dispersion medium containing, for example, water,
ethanol, polyol (for example,
glycerol, propylene glycol, and liquid polyethylene glycol, and the like), and
suitable mixtures thereof
Fluidity can be maintained, for example, by the use of a coating such as
lecithin, by the maintenance of the
required particle size in the case of dispersion and by the use of
surfactants. Antibacterial and antifungal
agents include, for example, parabens, chlorobutanol, phenol, ascorbic acid
and thimerosal. Isotonic agents,
for example, sugars, polyalcohols such as mannitol, sorbitol, and sodium
chloride can be included in the
composition. The resulting solutions can be packaged for use as is, or
lyophilized; the lyophilized
preparation can later be combined with a sterile solution prior to
administration. For intravenous, injection,
or injection at the site of affliction, the active ingredient will be in the
form of a parenterally acceptable
aqueous solution which is pyrogen-free and has suitable pH, isotonicity and
stability. Those of relevant
skill in the art are well able to prepare suitable solutions using, for
example, isotonic vehicles such as
Sodium Chloride Injection, Ringer's Injection, Lactated Ringer's Injection.
Preservatives, stabilizers,
buffers, antioxidants and/or other additives can be included, as needed.
Sterile injectable solutions can be
prepared by incorporating an active ingredient in the required amount in an
appropriate solvent with one
or a combination of ingredients enumerated above, as required, followed by
filtered sterilization. Generally,
dispersions are prepared by incorporating the active ingredient into a sterile
vehicle which contains a basic
dispersion medium and the required other ingredients from those enumerated
above. In the case of sterile
powders for the preparation of sterile injectable solutions, the preferred
methods of preparation can be
vacuum drying and freeze drying which yields a powder of the active ingredient
plus any additional desired
ingredient from a previously sterile-filtered solution thereof.
[00385] Compositions can be conventionally administered intravenously, such as
by injection of a unit
dose, for example. For injection, an active ingredient can be in the form of a
parenterally acceptable
aqueous solution which is substantially pyrogen-free and has suitable pH.,
isotonicity and stability. One can
prepare suitable solutions using, for example, isotonic vehicles such as
Sodium Chloride Injection, Ringer's
Injection, Lactated Ringer's Injection. Preservatives, stabilizers, buffers,
antioxidants and/or other
additives can be included, as required. Additionally, compositions can be
administered via aerosolization.
[00386] When the compositions are considered for use in medicaments or any of
the methods provided
herein, it is contemplated that the composition can be substantially free of
pyrogens such that the
-66-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
composition will not cause an inflammatory reaction or an unsafe allergic
reaction when administered to a
human patient. Testing compositions for pyrogens and preparing compositions
substantially free of
pyrogens are well understood to one or ordinary skill of the art and can be
accomplished using
commercially available kits.
1003871 Acceptable carriers can contain a compound that stabilizes, increases
or delays absorption, or
increases or delays clearance. Such compounds include, for example,
carbohydrates, such as glucose,
sucrose, or dextrans; low molecular weight proteins; compositions that reduce
the clearance or hydrolysis
of peptides; or excipients or other stabilizers and/or buffers. Agents that
delay absorption include, for
example, aluminum monostearate and gelatin. Detergents can also be used to
stabilize or to increase or
decrease the absorption of the pharmaceutical composition, including liposomal
carriers. To protect from
digestion the compound can be complexed with a composition to render it
resistant to acidic and enzymatic
hydrolysis, or the compound can be complexed in an appropriately resistant
carrier such as a liposome.
Means of protecting compounds from digestion are known in the art (e.g., Fix
(1996) Pharm Res. 13:1760
1764; Sarnanen (1996) .1. Pharm. Phartrtacol. 48:119 135; and U.S. Pat. No.
5,391,377).
1003881 The compositions can be administered in a manner compatible with the
dosage formulation,
and in a therapeutically effective amount. The quantity to be administered
depends on the subject to be
treated, capacity of the subject's immune system to utilize the active
ingredient, and degree of binding
capacity desired. Precise amounts of active ingredient required to be
administered depend on the judgment
of the practitioner and are peculiar to each individual. Suitable regimes for
initial administration and
booster shots are also variable, but are typified by an initial administration
followed by repeated doses at
one or more hour intervals by a subsequent injection or other administration.
Alternatively, continuous
intravenous infusions sufficient to maintain concentrations in the blood are
contemplated.
Treatment Methods
1003891 The instant disclosure comprises methods of treatment for diseases
such as cancer, and
infection, where enhanced phagocytosis by myeloid cells can be beneficial to
remove diseased cells, or
infected cells.
[00390] Cancers include, but are not limited to T cell lymphoma, cutaneous
lymphoma, B cell cancer
(e.g., multiple myeloma, Waldenstrom's macroglobulinemia), the heavy chain
diseases (such as, for
example, alpha chain disease, gamma chain disease, and mu chain disease),
benign monoclonal
gammopathy, and immunocytic amyloidosis, melanomas, breast cancer, lung
cancer, bronchus cancer,
colorectal cancer, prostate cancer (e.g., metastatic, hormone refractory
prostate cancer), pancreatic cancer,
stomach cancer, ovarian cancer, urinary bladder cancer, brain or central
nervous system cancer, peripheral
nervous system cancer, esophageal cancer, cervical cancer, uterine or
endometrial cancer, cancer of the
oral cavity or pharynx, liver cancer, kidney cancer, testicular cancer,
biliary tract cancer, small bowel or
appendix cancer, salivary gland cancer, thyroid gland cancer, adrenal gland
cancer, osteosarcoma,
-67-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
chondrosarcoma, cancer of hematological tissues, and the like. Other non-
limiting examples of types of
cancers applicable to the methods encompassed by the present disclosure
include human sarcomas and
carcinomas, e.g., fibrosarcoma, myxosarcoma, liposarcoma, chondrosarcoma,
osteogenic sarcoma,
chordoma, angiosarcoma, endotheliosarcoma, lymphangiosarcoma,
lymphangioendotheliosarcoma,
synovioma, mesothelioma, Ewing's tumor, leiomyosarcoma, rhabdomyosarcoma,
colon carcinoma,
colorectal cancer, pancreatic cancer, breast cancer, ovarian cancer, squamous
cell carcinoma, basal cell
carcinoma, adenocarcinoma, sweat gland carcinoma, sebaceous gland carcinoma,
papillary carcinoma,
papillary adenocarcinomas, cystadenocarcinoma, medullary carcinoma,
bronchogenic carcinoma, renal
cell carcinoma, hepatoma, bile duct carcinoma, liver cancer, choriocarcinoma,
seminoma, embryonal
carcinoma, Wilms' tumor, cervical cancer, bone cancer, brain tumor, testicular
cancer, lung carcinoma,
small cell lung carcinoma, bladder carcinoma, epithelial carcinoma, glioma,
astrocytoma,
medulloblastoma, craniopharyngioma, ependymoma, pinealoma, hemangioblastoma,
acoustic neuroma,
oligodendrogliorna, meningioma, melanoma, neuroblastoma, retinoblastoma;
leukemias, e.g., acute
lymphocytic leukemia and acute myelocytic leukemia (myeloblastic,
promyelocytic, myelomonocytic,
monocytic and erythroleukemia); chronic leukemia (chronic myelocyfic
(granulocytic) leukemia and
chronic lymphocytic leukemia); and polycythemia vera, lymphoma (Hodgkin's
disease and non-Hodgkin's
disease), multiple myeloma, Waldenstrom's macroglobulinemia, and heavy chain
disease. In some
embodiments, the cancer is an epithelial cancer such as, but not limited to,
bladder cancer, breast cancer,
cervical cancer, colon cancer, gynecologic cancers, renal cancer, laryngeal
cancer, lung cancer, oral cancer,
head and neck cancer, ovarian cancer, pancreatic cancer, prostate cancer, or
skin cancer. In other
embodiments, the cancer is breast cancer, prostate cancer, lung cancer, or
colon cancer. In still other
embodiments, the epithelial cancer is non-small-cell lung cancer, nonpapillary
renal cell carcinoma,
cervical carcinoma, ovarian carcinoma (e.g., serous ovarian carcinoma), or
breast carcinoma The epithelial
cancers can be characterized in various other ways including, but not limited
to, serous, endometrioid,
mucinous, clear cell, or undifferentiated. In some embodiments, the present
disclosure is used in the
treatment, diagnosis, and/or prognosis of lymphoma or its subtypes, including,
but not limited to, mantle
cell lymphoma. Lymphoproliferative disorders are also considered to be
proliferative diseases.
[003911 In some embodiments, a composition comprising at least a first
therapeutic agent, comprising
a monocyte or macrophage specific engager is administered per administration
dose. In some embodiments,
a composition the first therapeutic agent is administered in combination with
a second or a third therapy.
[003921 In some embodiments, the composition comprising at least a first
therapeutic agent comprising
a monocyte or macrophage specific engager is administered once. In some
embodiments, the composition
comprising at least a first therapeutic agent is administered more than once.
In some embodiments, the
composition comprising at least a first therapeutic agent comprising a
monocyte or macrophage specific
engager is administered repeatedly, multiple times over a span of the therapy.
-68-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00393] In some embodiments, the composition comprising at least a first
therapeutic agent comprising
a monocyte or macrophage specific engager is administered twice, thrice, four
times, five times, six times,
seven times, eight times, nine times, or ten times or more to a subject over a
span of time comprising a few
months, a year or more.
[00394] In some embodiments, the composition comprising at least a first
therapeutic agent comprising
a monocyte or macrophage specific engager is administered once weekly. In some
embodiments, the
composition comprising at least a first therapeutic agent comprising a
monocyte or macrophage specific
engager is administered twice weekly.
[00395] In some embodiments, the composition comprising at least a first
therapeutic agent comprising
a monocyte or macrophage specific engager is administered once every two
weeks.
[00396] In some embodiments, the composition comprising at least a first
therapeutic agent comprising
a monocyte or macrophage specific engager is administered once every three
weeks.
[00397] In some embodiments, the composition comprising at least a first
therapeutic agent comprising
a monocyte or macrophage specific engager is administered once monthly.
[00398] In some embodiments, the composition comprising at least a first
therapeutic agent comprising
a monocyte or macrophage specific engager is administered once in every 2
months, once in every 3
months, once in every 4 months, once in every 5 months or once in every 6
months.
[00399] In some embodiments, the composition comprising at least a first
therapeutic agent comprising
a monocyte or macrophage specific engager is administered by injection.
[00400] In some embodiments, the composition comprising at least a first
therapeutic agent comprising
a monocyte or macrophage specific engager is administered by infusion.
[00401] In some embodiments, the composition comprising at least a first
therapeutic agent comprising
a monocyte or macrophage specific engager is administered by intravenous
infusion.
[00402] In some embodiments, the composition comprising at least a first
therapeutic agent comprising
a monocyte or macrophage specific engager is administered by subcutaneous
infusion.
[00403] In some embodiments, treatment with monocyte or macrophage specific
engagers increase the
phagocytic ability and monocyte or macrophage mediated target cell killing,
compared to a case where no
monocyte or macrophage specific engagers were used. Monocytes or macrophages
retrieved from the
tumor site after treatment with monocyte or macrophage specific engagers may
demonstrate a greater than
10%, or greater than 20%, or greater than 30%, or greater than 40%, or greater
than 50%, or greater than
60%, or greater than 70%, or greater than 80%, or greater than 90%, or greater
than 100%, or greater than
150%, or greater than 200%, or greater than 250%, or greater than 300%, or
greater than 350%, or greater
than 400%, or greater than 450%, or greater than 500%, or greater than 600%,
or greater than 700%, or
greater than 800%, or greater than 900%, or greater than 1000% increase in
phagocytosis.
-69-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00404] In some embodiments, treatment with monocyte or macrophage specific
engagers increases
ROS production in associated monocytes or macrophages that may be retrieved
from the tumor site,
compared to a case with no monocyte or macrophage specific engager treatment.
Monocytes or
macrophages retrieved from the tumor site after treatment with monocyte or
macrophage specific
engagers may demonstrate a greater than 2-fold, or greater than 3-fold, or
greater than 4-fold, or greater
than 5-fold, or greater than 6-fold, or greater than 7-fold, or greater than 8-
fold, or greater than 9-fold, or
greater than 10-fold, or greater than 20-fold, or greater than 30-fold, or
greater than 40-fold, or greater
than 50-fold, or greater than 60-fold, or greater than 70-fold, or greater
than 80-fold, or greater than 90-
fold, or greater than 100-fold, or greater than 200-fold, or greater than 300-
fold, or greater than 400-fold,
or greater than 500-fold, or greater than 700-fold, or greater than 800-fold,
or greater than 900-fold, or
greater than 1000-fold increase in ROS compared to a case with no monocyte or
macrophage specific
engager treatment. In some embodiments, treatment with monocyte or macrophage
specific engagers
increases iNOS production in associated monocytes or macrophages that may be
retrieved from the tumor
site, compared to a case with no monocyte or macrophage specific engager
treatment. In some
embodiments, treatment with monocyte or macrophage specific engagers increases
respiratory burst in
associated monocytes or macrophages that may be retrieved from the tumor site,
compared to a case with
no monocyte or macrophage specific engager treatment.
[00405] In some embodiments, treatment with monocyte or macrophage specific
engagers may
increase the expression of CD80 in the associated monocytes or macrophages. In
some embodiments,
treatment with monocyte or macrophage specific engagers may increase the
expression of CD86 in the
associated monocytes or macrophages compared to a no treatment. In some
embodiments, treatment with
monocyte or macrophage specific engagers may increase the expression of
TRAlL/TNF Family death
receptors in associated monocytes or macrophages compared to a case with no
monocyte or macrophage
specific engager treatment. In some embodiments, treatment with monocyte or
macrophage specific
engagers may increase the expression of LIGHT in associated monocytes or
macrophages compared to a
case with no monocyte or macrophage specific engager treatment. In some
embodiments, treatment with
monocyte or macrophage specific engagers may increase the expression of HVEM
in associated
monocytes or macrophages compared to a case with no monocyte or macrophage
specific engager
treatment. In some embodiments, treatment with monocyte or macrophage specific
engagers may increase
the expression of CD40 in associated monocytes or macrophages compared to a
case with no monocyte or
macrophage specific engager treatment. In some embodiments, treatment with
monocyte or macrophage
specific engagers may increase the expression of 'FHA in associated monocytes
or macrophages
compared to a case with no monocyte or macrophage specific engager treatment.
In some embodiments,
treatment with monocyte or macrophage specific engagers may increase the
expression of OX4OL in
associated monocytes or macrophages compared to a case with no monocytes or
macrophages specific
-70-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
engager treatment. In some embodiments, treatment with monocyte or macrophage
specific engagers may
increase the expression of GITR in associated monocytes or macrophages
compared to a case with no
monocytes or macrophages specific engager treatment. In some embodiments,
treatment with monocyte
or macrophage specific engagers may increase the expression of SLAM in
associated monocytes or
macrophages compared to a case with no monocyte or macrophage specific engager
treatment. In some
embodiments, treatment with monocyte or macrophage specific engagers may
increase the expression of
CD58 in associated monocytes or macrophages compared to a case with no
monocyte or macrophage
specific engager treatment. In some embodiments, treatment with monocyte or
macrophage specific
engagers may increase the expression of CD155 in associated monocytes or
macrophages compared to a
case with no monocyte or macrophage specific engager treatment. In some
embodiments, treatment with
monocyte or macrophage specific engagers may increase the expression of CD112
in associated
monocytes or macrophages compared to a case with no monocyte or macrophage
specific engager
treatment. In some embodiments, treatment with monocyte or macrophage specific
engagers may increase
the expression of B7-DC in a tumor associated myeloid cell compared to a case
with no monocyte or
macrophage specific engager treatment.
EXAMPLES
Example 1. Materials and Methods:
[00406] Dulbecco modified Eagle medium, trypsin-EDTA, wortmannin (W), LY294002
(LY),
Bradford reagent, and lysostaphin are purchased from Sigma-Aldrich, Inc. (St.
Louis, MO). Reduced serum
Opti-MEM I medium are purchased from Gibco-BRL (Gaithersburg, MD). SH-5 was
acquired from Enzo
Life Sciences (Plymouth, PA), and OSU-03012 (OSU) was purchased from Cedarlane
Labs (Burlington,
NC). FuGENE transfection reagent and the 50x EDTA-free protease inhibitor
cocktail are purchased from
Roche Applied Science (Manheim, Germany). Cells are grown in 24-well plates to
60 to 70% confluence,
and the culture medium was changed to DMEM 10% FCS. Then, in order to have a
similar protein
expression 5 ng of pCMV5-Akt-CA or 200 ng of pCMV5-Akt-DN in 1.2 pi of FuGENE
transfection
reagent (ratio, 4:1 [FuGENE-plasmid]) are added to BEC in reduced serum Opti-
MEM I medium according
to the manufacturer's instructions.
1004071 Cloning and characterization of the BiME, TriME and multi-specific
engagers is performed in
a bacterial expression system, such as in an E coil system for test and
screening purposes. Briefly, following
screening of the binding domains to incorporate in an engager design,
polynucleotide sequences encoding
specific variable light chain and/or variable heavy chain domains are
individually amplified by PCR from
respective antibody-expressing clones. In case the binding domains comprise
entire Fab regions, the
respective regions are amplified by PCR from the respective antibody-
expressing clones_ The linkers are
either enzymatically ligated typically to the C-terminal end of the encoded
Fab or the variable domain.
-71-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
Alternatively, polynucleotides encoding the binding domains and the linker
sequences are incorporated
into plasmid by sequential cloning. In yet another alternative method, the
specific sequences encoding the
binding domains (Fab or variable regions) are ligated to each other by
overlapping PCR, and larger inserts
comprising Fab-linker-Fab designs are cloned into the expression vector to
express chimeric proteins
comprising the engagers.
[00408] Expressed proteins are purified and concentrated by commonly known
techniques and the
products are tested in experimental animals for tumor targeting and toxicity.
[004091 In some examples, a lentiviral construct comprising the chimeric
proteins are used to transduce
the chimeric construct in a monocyte or macrophage.
Example 2. Construction of a bispecific engager (BiME) platform
[004101 In this example, a monocyte or macrophage and tumor targets with
protease masking site is
designed. The bispecific engager comprises a monocyte or macrophage binding
domain, which is a
scavenger receptor (SRA1) binding domain. The target cell binding domain is a
tumor recognition domain
(e.g., TROP2). An scFv construct polypeptide design is designated in FIG. 24 A
VITH construct
polypeptide design is designated in FIG. 218. In another exemplary design, the
antigen binding domains
are occupied with protease cleavable masking elements. Additionally, the two
binding domains are linked
by a TLR4 synthetic peptide. An scFv construct polypeptide design is
designated in FIG. 34. A VHF{
construct polypeptide design is designated in FIG. 3B. The synthetic peptide
is bound to two scFvs (FIG.
3A) or across two single domains (FIG 3B), The specific TLR4 synthetic peptide
linker activates TLR4
receptor on monocytes or macrophages and provides the second activation signal-
Signal 2 that potentiates
a monocyte or macrophage phagocytic and pro-inflammatory activity. In another
exemplary design, the
two binding domains of a monocyte or macrophage specific engager comprises a
M2 targeting peptide.
The M2 targeting peptide has an amino acid sequence of YEQDPWGVKWWY (SEQ ID
NO: 116) (M2-
pep) or HLSWLPDVVYAW (BLS pep) (SEQ ID NO: 117), which specifically target and
bind to an M2
monocyte or macrophage which is the predominant phenotype of tumor associated
monocytes or
macrophages. Thus, in addition to the binding and activation by the specific
binding domain, in this design
a bi-specific engager can further be employed to target the engager to the
specific cell, in this case an M2
monocyte or macrophage cell (FIG. 3C and FIG. 3D).
Example 3. Construction and expression of bispecific engagers.
[00411] This example describes construction, expression and testing of BiMEs
having activator peptide
sequences within the linkers. First, a peptides sequences that were derived
from different TLR activators
were tested for immune activation on monocytes in culture. Exemplary TLR
peptide sequences that were
tested are listed below:
[00412] Table 3, TLR activator peptide sequences used as part of a linker
sequences to generate
bispecific engager constructs exemplified in FIG. 3A and FIG 3B are shown in
Table 3.
-72-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
Table 3.
Sequence Amino Acid Sequence
Name
RS01 GGQEINSSYGG (SEQ ID NO: 105) or QE1NSSY (SEQ ID
NO: 129)
RS02 GGSHPRLSAGG (SEQ ID NO: 123) or SHPRLSA (SEQ ID
NO: 130)
RS03 GGSMPNPMVGG (SEQ ID NO: 106) or SMPNPMV (SEQ ID
NO: 131)
RS04 GGGLQQVLLGG (SEQ ID NO: 107) or GLQQVLL (SEQ ID
NO: 132)
RS05 GGIIELSVLLGG (SEQ ID NO: 124) or HELSVLL (SEQ ID
NO: 133)
RS06 GGYAPQRLPGG (SEQ ID NO: 108) or YAPQRLP (SEQ ID
NO: 134)
RS07 GGTPRTLPTGG (SEQ ID NO: 125) or TPRTLPT (SEQ ID
NO: 135)
RS08 GGAPVHSSIGG (SEQ ID NO: 126) or APVHSSI (SEQ ID
NO: 136)
RS09 GGAPPHALSGG (SEQ ID NO: 109) or APPHALS (SEQ ID
NO: 137)
RS10 GGTFSNRFIGG (SEQ ID NO: 127) or TFSNRFI (SEQ ID
NO: 138)
RS11 GGVVPTPPYGG (SEQ ID NO: 110) or VVPTPPY (SEQ ID
NO: 139)
RS12 GGELAPDSPGG (SEQ ID NO: 128) or ELAPDSP (SEQ ID
NO: 140)
1004131 For testing immune response of each of the peptides, 2 x 10A6
monocytes were incubated
overnight with 1 microgram/ml of a peptide from Table 3, and IL6 and TNF-alpha
release was measured
using fluorimetric detection using Luminex 200. As shown in FIG. 3E, RS01 and
RS09 peptides induced
higher IL6 release. These two peptide sequences were next selected from the
pool above and utilized to
generate bispecific engagers. Similarly, several more are being tested.
[00414] The bispecific or trispecific engagers can be constructed by molecular
cloning. Upon generation
of successful clones, each clone can be sequenced and the sequence validated.
In some embodiments, a
bispecific or trispecific engager comprises (i) an anti-CD5 scFv capable of
binding to a CD5-1- tumor cell,
and (ii) an anti-CD16 scFv capable of binding to a CD16 surface molecule on a
monocyte, or
macrophage cell.
[00415] An exemplary anti-CD5 binder comprises a heavy chain comprising the
sequence:
MWLQSLLLLGTVACSISEIQLVQSGGGLVKPGGSVRISCAASGYTFTNYGMNWVRQAPGICGLE
WMGWINTHTGEPTYADSFKGRFTFSLDDSKNTAYLQINSLRAEDTAVYFCTRRGYDWYFDVW
GQGTTVTV (SEQ ID NO: 111) or
MWLQSLLLLGTVACSISEIQLVQSGGGLVICPGGSVRISCAASGYTFTNYGMNWYRQAPGKGLE
WMGWINTHTGEPTYADSFKGRFTFSLDDSKNTAYLQINSLRAEDTAVYFCTRRGYDWYFDVW
GQGTTVTVSS (SEQ ID NO: 144).
-73-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00416] Another exemplary anti-CD5 binder comprises a heavy chain comprising
the sequence:
EIQLVQSGGGLVKPGGSVRISCAASGYTFTNYGMNWVRQAPGKGLEWMGWIINTHTGEIYTYAD
SFKGRFTFSLDDS1CNTAYLQINSLRAEDTAVYFCTRRGYDWYFDVWGQGTTVTV
(SEQ ID NO: 112) or
EIQLVQSGGGLVKPGGSVRISCAASGYTFTNYGMNWVRQAPGKGLEWMGWINTHTG
EPTYADSFKGRFTFSLDDSICNTAYLQINSLRAEDTAVYFCTRRGYDWYFDVWGQGTTVTVSS
(SEQ ID NO: 143).
1004171 An exemplary anti-CD5 binder may comprise a light chain comprising the
amino acid
sequence:
DIQMTQSPSSLSASVGDRVTITCRASQDINSYLSWFQQ1CPGICAPICTLIYRANRLESGVPSRFSGSG
SGTDYTLTISSLQICEDFGIYYCQQYDESPWITGGGTKLEIK (SEQ ID NO:113).
[00418] An exemplary a bispecific or trispecific engager, such as bispecific
or trispecific engager
containing an anti-CD5 scFv may comprise a short peptide linker connecting an
exemplary heavy chain
and an exemplary light chain, having a sequence: SSGGGGSGGGGSGGC_TGS (SEQ ID
NO: 114) or
SGGGGS (SEQ ID NO: 145) or GGGGS (SEQ ID NO: 146) or GGGG (SEQ ID NO: 147).
[00419] An exemplary anti-CDS scFv comprises an amino acid sequence:
MWLQSLLLLGTVACSISEIQLVQSGGGLVKPGGSVRISCAASGYTFTNYGMNWVRQAPGKGLE
WMGW1NTHTGEPTYADSFICGRFTFSLDDSICNTAYLQINSLRAEDTAVYFCTRRGYDWYFDVW
GQGTTVTVSSGGGGSGGGGSGGGGSDIQMTQSPSSLSASVGDRVTITCRASQDINSYLSWFQQK
PG1CAPKTLIYRANRLESGVPSRFSGSGSGTDYTLTISSLQYEDFGIYYCQQYDESPWTFGGGTKLE
1K (SEQ ID NO: 115).
1004201 An exemplary anti-CD16 scFv can comprise a heavy chain variable
sequence comprising
the amino acid sequence:
QVQLVQSGAEVKICPGESLKVSCKASGYTFTSYYMEWVRQAPGQGLEWMGDNPSGGSTSY
AQKFQGRVTMTRDTSTSTVYWIELSSLRSEDTAVYYCARGSAYYYDFADYWGQGTLVTVSS
(SEQ ID NO: 141).
[00421] An exemplary anti-CD16 scFv can comprise a light chain variable
sequence comprising the
amino acid sequence:
SYVLTQPSSVSVAPGQTATISCCIGUNIGSKNVHWYQQRPGQSPVLVIYQDNICRPSG1PERFSGSN
SGNTATLTISGTQAMDEADYYCQVWDNYSVLFGGGTKLTVL (SEQ ID NO: 142).
1004221 The two scFvs can be linked by a synthetic peptide linker that
comprises one of the TLR
activating peptide sequences, such as those described in Table 3. The
constructs are thereafter named
as Binder 1-linker-Binder 2, such as, CD5-RS01-CD16, having an RS01 TLR
activating peptide
sequence in the linker; or, CD5-RS09-CD16, having an RS01 TLR activating
peptide sequence in the
-74-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
linker, as shown in FIG. 3F, or FIG. 313 respectively. Sequence verified
clones are then expressed in
a suitable cell and the protein is detected by gel migration using molecular
markers and western blot,
using a suitable positive control.
1004231 An exemplary bispecific or trispecific engager can comprise the
sequence:
METDTLLLWVLLLWVPGSTG (SEQ 1.1) NO: 148) or any other useful leader sequence.
1004241 An exemplary bispecific or trispecific engager can comprise the
sequence: IIHRE-MH (SEQ
ID NO: 149) or any other useful affinity tag.
[00425] An exemplary bispecific or trispecific engager can comprise the
sequence: ENLYFQG
(SEQ ID NO: 150) or any other useful protease cleavage sequence.
[00426] An exemplary bispecific or trispecific engager can comprise a first
scFv comprising a
variable heavy chain linked to a variable light chain via a first linker,
which can be linked to a second
scFv via a second linker, wherein the second scFv comprises a variable heavy
chain linked to a
variable light chain via a third linker. In some embodiments the second linker
comprises a TLR
activating peptide sequence, such as those described in Table 3. In some
embodiments, the first
linker has a length of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, or 20 or more
amino acids. In some embodiments, the second linker has a length of 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 or 30
or more amino acids. In
some embodiments, the third linker has a length of 1, 2, 3, 4, 5, 6, 7, 8,9,
10, 11, 12, 13, 14, 15, 16,
17, 18, 19, or 20 or more amino acids.
[00427] An exemplary bispecific or trispecific engager can comprise the
sequence:
METDTLLLWVLLLWVPGSTGIMH-LHENLYFQGEIQLVQSGGGLVKPGGSVRISCAASGYTFT
NYGMNWVRQAPGICGLEWMGWINTHTGEPTYADSFICGRFTFSLDDSICNTAYLQINSLRAEDTA
VYFCTRIZGYDWYFDVWGQGTTVTVSSsggggsSYVLTQPSSVSVAPGQTATISCGGHNIGSKNVH
WYQQRPGQSPVLVIYQDNKRPSGIPERFSGSNSGNTATLTISGTQAMDEADYYCQVWDNYSVLF
GGGTICLTVLggggQEINSSYggggsQVQLVQSGAEVKICPGESLKVSCICASGYTFTSYYMIHWVRQA
PGQGLEWMOINPSGGSTSYAQKFQGRVFMTRDTSTSTVYMELSSLRSEDTAVYYCARGSAYY
YDFADYWGQGTLVTVSSsggggsDIQMTQSPSSLSASVGDRVTITCRASQD1NSYLSWFQQKPGICA
PKTLIYRANRLESGVPSRFSGSGSGTDYTLTISSLQYEDFGIYYCQQYDESPWTFGGGTKLEIK.
(SEQ ID NO: 151).
[00428] An exemplary bispecific or trispecific engager can comprise the
sequence:
METDTLLLWVLLLWVPGSTGHREHILHENLYFQGEIQLVQSGGGLVKPGGSVRISCAASGY
TFTNYGMNWVRQAPGKGLEWMGWINTHTGEPTYADSFKGRFTFSLDDSKNTAYLQINSLR
AEDTAVYFCTRRGYDWYFDVWGQGTTVTVSSsggggsSYVLTQPSSVSVAPGQTATISCGGH
NIGSKNVHWYQQRPGQSPVLVIYQDNICRPSGIPERFSGSNSGNTATLTISGTQAMDEADYYC
-75-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
QVWDNYSVLFGGGTKLTVLggggAPPHALSggggsQVQLVQSGAEVICKPGESLKVSCKASGY
TFTSYYMIIWVRQAPGQGLEWMGIINP SGGST SYAQICFQGRVTMTRDT STSTVYMELS SLRS
EDTAVYYCARGSAYYYDFADYWGQGTLVTVSS sggggsDIQMTQSP SSLSASVGDRVTITCR
ASQDINSYLSWFQQKPGKAPKTLINRANRLESGVPSRFSGSGSGTDYTLTISSLQYEDFGIYY
CQQYDESPWTFGGGTKLEIK (SEQ ID NO: 152).
[004291 Expression of the CD5-RS01-CD16 BiME is shown by SDS PAGE FIG. 3G
(left) and by
western blot in FIG. 3G (right) under reducing and non-reding gel
electrophoresis as indicated.
Expression of the CD5-RS09-CD16 BiME is shown by SDS PAGE FIG. 31 (left) and
by western blot in
FIG. 31 (right) under reducing and non-reding gel electrophoresis as
indicated. These data demonstrate
successful generation and expression of the TLR- activating sequence
containing bispecific engagers.
The engagers described above are tested in in vitro. A microparticle based
phagocytosis assay was used
to examine changes in phagocytosis. Briefly, streptavidin coupled fluorescent
polystyrene microparticles
(6 gm diameter) are conjugated with biotinylated recombinantly expressed and
purified cancer ligand, in
this case CD5. Myeloid cells cultured in presence of the beads and an engager
protein, are incubated with
the ligand coated microparticles for 1-4 h and the amount of phagocytosis was
analyzed and quantified
using flow cytometry.
Example 4. Trispecifc engager (TriME) design
[004301 In this example a design of a trispecific antigen-binding protein is
set forth as follows. The
trispecific antigen-binding protein comprises (a) a first domain (A) which
specifically binds to human
Scavenger/Phagocytic receptor; (b) a second domain (B) which is a danger
signal receptor; and (c) a third
domain (C) which specifically binds to a target antigen, wherein the domains
are linked in the order H2N-
(A)-(B)-(C)-COOH, H2N-(A)-(C)-(B)-COOH, H2N-(B)-(A)-(C)-COOH, H2N-(B)-(C)-(A)-
COOH, H2N-
(C)-(B)-(A)-COOH, or H2N-(C)-(A)-(B)-COOH by linkers Li and L2 (FIG. 4A). The
antigen binding
domains are occupied with protease cleavable masking elements, which are
activated by availability and
contact with the protease. An exemplary nanobody design is shown in FIG. 4B.
FIG. 4C provides a
graphical view of a trispecific engager and an exemplary nature of function on
a target tumor cell and a
monocyte or macrophage. The trispecific engager structurally comprises a tumor
recognition or tumor
binding domain, a monocyte or macrophage receptor 1 binding domain, and a
monocyte or macrophage
receptor 2 binding domain (shown in inset on the top right comer of Fig. 4C).
As an expected functional
mode, the tumor binding domain binds to the target surface molecule (tumor
antigen) of a tumor cell, while
the two monocyte or macrophage receptors, B and A are bound respectively by
the monocyte or
macrophage receptor 1 binding domain, and the monocyte or macrophage receptor
2 binding domain of
the trispecific engager. As shown in the figure, engagement of a receptor A
and B, by the trispecific engager
also operably linked with the tumor antigen by the tumor recognition domain,
provides a Signal 1 and a
Signal 2 to the monocyte or macrophage. The dual signal (Signal 1 + Signal 2)
activates the monocyte or
-76-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
macrophage thereby enhancing phagocytosis and activating an inflammatory
cascade in this exemplary
figure, which lead to phagocytic killing of the target cell.
Example 5. Antigen binding domain masking design
[00431] In this example, a generalized exemplary design for an engager having
masked antigen binding
domains is described in further detail. FIG. 5Ai is a diagrammatic
representation of a bispecific engager
with two scFV binders, scFv1, and scFv2. SdAb or diabody engagers can also be
likewise constructed with
necessary structural modifications, an exemplary diabody construct with two
binders is represented in FIG.
5Aii. The antigen binding domains are masked by a peptide mask (1) that
remains bound to the antigen
binding portions of the diabody ABD1 and ABD2, linked at the N terminal
portion of the light chain
variable domain of ABD1 (3) of the first chain, or the light chain variable
region of ABD2 of the second
chain by a peptide linker (2). The peptide linker joining the mask with the
light chain variable domains is
a substrate for matrix metalloproteinase 2 (MMP2) substrate, having an amino
acid sequence GPLGVR.
The design allows passing of the masked diabody engager to pass through the
circulation without binding
to any substrate until MMP2 is available to cleave the linking peptide. It is
understood that the cancer
microenvirortment is rich in MIMP2. Therefore, the diabody engager is
activated in a cancer
microenvironment to bind its target cancer cell and the monocyte or macrophage
with ABD1 and ABD2
respectively in a tumor environment. FIG. 5B exemplifies the nucleic acid
construct of a single chain of a
diabody. The nucleic acid construct comprises from 5'-3' end a nucleic acid
sequence encoding the mask
peptide, a MMP2 linker, a sequence encoding ABD1 light chain (ABD1-LC), which
is linked to a nucleic
acid sequence encoding a peptide linker that joins with the ABD1-LC and ABD2-
14C; followed by the
nucleic acid sequence encoding the ABD2 HC
Example 6. Modular antigen binding engager designs
[00432] In this example, several modular designs of binding domains
represented by light chain heavy
chain domains arranged on an antibody-like polypeptide structure as shown in
FIG. 6. In one design, a
common light chain is used to pair with two non-identical heavy chains in an
IgG like structure, thereby
rendering a bispecific binding domain that could be used in a bispecific or a
trispecific engager design. In
another model depicted herein, a chimeric bi- or trispecific engager uses a
combination of an scFv joined
to one arm of an usual antibody light and heavy chain combination. In one
design, two scFvs replace the
heavy chain-light chain paired regions, while the scFvs are connected by the
constant regions of the heavy
chains. In other designs as depicted herein, one or more scFvs may be
conjugated to the Fc region. In yet
other designs, one or more scFvs may be conjugated to the constant regions as
side chains of an IgG like
polypeptide.
Example 7. Use of monocyte or macrophage specific activators in an engager for
activating inflammatory
signal in monocyte or macrophage an potentiating phagocytosis
-77-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00433] MD2 can bind to and activate TLR4 in response to LPS, as shown in the
diagrammatic
representation in FIG. 7A, upper panel. In an exemplary design, MD2 is
constructed into a monocyte or
macrophage specific engager, where the MD2, in addition to the tumor specific
binding domain and the
monocyte or macrophage specific binding domain associates with TLR4 receptors,
and help in the
dimerization of TLR4 receptors on the monocyte or macrophage thereby sending a
monocyte or
macrophage activating signal (Signal 2) that further potentiates the
inflammatory activation and phagocytic
killing of a target cancer cell by the monocyte or macrophage (FIG. 7B).
1004341 In another example, Herpes Virus Entry Mediator (HVEM) and its
association with tumor
necrosis factor (TNF)-related 2 (LIGHT) is exploited in designing an exemplary
monocyte or macrophage
specific engager that potentiates monocyte or macrophage effector functions.
HVEM is a member of the
TNFR superfamily and has two more ligands: HSV surface envelope gD and LTD. It
is expressed on T
cells, B cells, NK cells, monocytes, neutrophils, and DC. The LIGHT-HVEM
interaction increased levels
of phagocytosis, interleukin (lIL)-8, TNF-D, nitric oxide (NO), and reactive
oxygen species (ROS) in
monocytes and neutrophils. In an exemplary design, a monocyte or macrophage
specific engager comprises
a LIGHT domain that can bind to HVEM. In a variation of the design, the LIGHT
domain that binds to
HVEM may be replaced by an agonist antibody of antigen binding domain that
binds to HVEM, as shown
in FIG. 8A. The corresponding mode of function of the engager is depicted
graphically in FIG. 8B, where
binding of the LIGHT domain with monocyte or macrophage associated HVEM
activates an inflammatory
signal (Signal 2) in the monocyte or macrophage, that potentiates its effector
functions as a phagocytic cell.
[00435] In another example, GIRT associated activation signal is exploited in
an exemplary monocyte
or macrophage specific engager design that is shown in FIG. 9A. GIRT is
expressed on monocytes or
macrophages, and when bound by its ligand, ORTL, it generates an inflammatory
signal in the monocyte
or macrophage, as depicted graphically in FIG. 9B.
[00436] Example 8. Use of linkers with sequences to facilitate accelerated
association with in an
engager
[00437] In this example, monocyte or macrophage specific engagers are designed
to have linkers
between multi-specific binding domains that have complementarity to each
other. FIG. 10A-FIG. 10C
demonstrates exemplary designs which include leucine zipper domains, (FIG. 10A
and 10B) or rationally
designed synthetic sequences comprising a complementary binding region (FIG.
10C). Exemplary linker
sequences disclosed in the specification are used. In specific constructs
linker domains are utilized that
dimerize of trimerize, bringing useful domains in closer proximity. Shown in
these figures are exemplary
use of leucine zipper domains and coupling protein domains in binding
heteromeric binder domains closer
together.
Example 9. Screen for selecting a myeloid cell binder domains
-78-
CA 03140875 2021-12-7
WO 2020/252208
PCT/US2020/037312
[00438] In this example, a screen is undertaken to select the cell surface
molecules on a myeloid cell,
or functional fragments thereof, that can be useful to design binding domains
for engagers described herein.
A binding domain can be a phagocytosis receptor engager or activator. As is
now understood, not all
phagocytic cell surface receptors on a phagocytic cell have equal ability to
be induced or activated to
generate proinflarnmatory signaling or in any way potentiate monocyte or
macrophage effector functions.
Hence, to harness monocytes or macrophages and other myeloid cells to kill
cancer, a series of signal 1
and signal 2 targets are generated on myeloid cells. These targets were
identified through the screening of
materials associated with inflammation as well as immune tolerance.
[00439] This is done using a unique tool that uses proprietary arrays of
expression vectors ¨ encoding over
5,500 full-length human plasma membrane and tethered secreted proteins ¨
spotted onto slides. Human cells are
grown over the top become reverse-transfected resulting in cell surface
expression of each respective protein at
distinct slide locations. The test formulation is then applied and specific
binding analyzed and confirmed using an
appropriate detection system. These hits were then interrogated and examined
as potential targets for monocyte or
macrophage binding and modulation.
[0440] Specific useful binding agents, or domains identified
from the screens are then reverse
transcribed, and cloned into lentiviral expression vectors to generate the
second binding domain or an
engager BiME or TriME constructs. A recombinant nucleic acid encoding a BiMEs
or TriMEs can
generated using one or more domains from highly phagocytic receptor binding
domains generated from
the screen.
-79-
CA 03140875 2021-12-7