Note: Descriptions are shown in the official language in which they were submitted.
1
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
Description
Title of Invention: ANTI-MERS-COV ANTIBODY AND USE
THEREOF
Technical Field
[1] Provided is an anti-MERS-CoV antibody or an antigen-binding fragment
thereof, and
medical uses thereof.
[2]
Background Art
[31 Middle East Respiratory Syndrome Coronavirus (MERS-CoV) was first
identified in
Saudi Arabia in 2012 from a patient who suffered acute pneumonia and
subsequent
renal failure. Since then, the World Health Organization has reported 2,254
laboratory-
confirmed cases of MERS-CoV infections in 27 different countries around the
world,
and South Korea has recorded the highest number of cases outside of the Middle
East.
Despite resilient efforts throughout the scientific and medical communities,
no vaccine
or antiviral agent for MERS-CoV is currently available.
[4] MERS-CoV is a large (30 kb), enveloped, single-stranded, positive-
sense RNA virus.
The viral genome encodes four major structural proteins: spike (S), envelope
(E),
membrane (M), and nucleocapsid (N) proteins. The S glycoprotein is a major
envelope
protein and interacts with the cellular receptor dipeptidyl peptidase 4 (DPP4)
for entry
into the host cell. This protein consists of the 51 and S2 subunits. The
receptor-binding
domain (RBD) within the 51 subunit mediates receptor binding, whereas the S2
subunit facilitates membrane fusion. DPP4 is expressed on a variety of human
cells,
including fibroblasts, intestinal epithelial cells, and hepatocytes, as well
as in the lung
parenchyma and interstitium. MERS-CoV is detected in respiratory secretions
and the
lower respiratory tract of the infected patients. In the most severe cases of
MERS-CoV
infection, aggravating respiratory failure ultimately results in mechanical
ventilation.
These observations suggest that the MERS-CoV virus primarily infects the human
res-
piratory tract and replicates within the human airway epithelium.
[51 Antibodies play a crucial role in the prevention and treatment of
viral infection.
Polysera taken from recovered patients and vaccinated donors have been used as
pro-
phylactic agents for hepatitis B, rabies, and other viral diseases.
Palivizumab (Synagis,
Medimmune, Gaithersburg, MD, USA) was approved for the prophylaxis of RSV
(Respiratory Syndrome Virus) in 1998, and ibalizumab-uiyk (Trogarzo, TailMed
Biologics, Taiwan) became clinically available in 2018 for the treatment of
human im-
munodeficiency virus type 1 (HIV-1) infection in treatment-experienced adults
with
multi-drug-resistant HIV-1 and failure to respond to the current
antiretroviral regimen.
2
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[6] In response to the ongoing epidemic, several groups have developed
anti-
MERS-CoV neutralizing monoclonal or polyclonal antibodies that target RBD.
These
antibodies were generated from B cells derived from convalescent patients,
nonimmune human antibody phage-display libraries, fully humanized mice, tran-
schromosomic bovines, or hybridomas from mice that were immunized with MERS-
CoV S. These antibodies potently inhibit RBD binding to the DPP4 receptor. Fur-
thermore, therapeutic effects of RBD-specific neutralizing antibodies were
evaluated in
several animal models, including Ad5/hDPP4-trasduced mice, humanized DPP4
mice,
and hDPP4-transgenic mice as well as hDPP4 knockin mice, rabbits, and rhesus
monkeys.
171 All MERS-CoV neutralizing antibodies were developed for intravenous
(i. v.)
delivery; however, recent reports indicate that the amount of antibody
delivered to lung
tissue is often quite limited following systemic delivery (Hart TK, Cook RM,
Zia-
Amirhosseini P, Minthorn E, Sellers TS, Maleeff BE, et al. Preclinical
efficacy and
safety of mepolizumab (SB-240563), a humanized monoclonal antibody to IL-5, in
cynomolgus monkeys. J Allergy Clin Immunol. 2001;108(2):250-257; Koleba T,
Ensom MH. Pharmacokinetics of intravenous immunoglobulin: a systematic review.
Pharmacotherapy. 2006;26(6):813-827). In cynomolgus monkeys, bronchoalveolar
lavage fluid contained dose-proportional concentrations of systemically
administrated
antibody, and these concentrations were approximately 500-fold less than those
in
plasma. Therefore, delivery of therapeutic antibody to lung tissues via
inhalation has
garnered considerable interest. Following delivery via the airway, cetuximab,
an anti-
epidermal growth factor receptor (EGFR) antibody, accumulated in normal and
cancerous tissues in the lung at a concentration that was twice that achieved
after i. v.
delivery. In addition, recent studies showed that Fc fusion proteins and
nanobodies are
also efficiently delivered via the pulmonary route (Bitonti AJ, Dumont JA.
Pulmonary
administration of therapeutic proteins using an immunoglobulin transport
pathway.
Adv Drug Deliv Rev. 2006;58(9-10):1106-1118). Therefore, MERS-CoV neutralizing
antibody may also accumulate at higher concentrations following delivery via a
pulmonary route, guaranteeing higher efficacy. In order for this pulmonary
delivery to
be successful, the antibody must be sufficiently stable to resist denaturation
during the
process of nebulization.
[81
Disclosure of Invention
Technical Problem
[91 Provided an antibody against a coronavirus, MERS-CoV, or an antigen-
binding
fragment thereof, having improved stability and efficacy, and uses thereof.
3
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[10] An embodiment provides an anti-MERS-CoV antibody or an antigen-binding
fragment thereof comprising:
[11] a VL-CDR1(complementarity determining region 1) comprising an amino
acid
sequence of SEQ ID NO: 1, 2, 3, 4, 5, 6, or 7;
[12] a VL-CDR2 comprising an amino acid sequence of SEQ ID NO: 8, 9, 10,
11, 12, 13,
14, or 71;
[13] a VL-CDR3 comprising an amino acid sequence of SEQ ID NO: 15, 16, 17,
18, 19,
20, 21, 22, 23, or 72;
[14] a VH-CDR1 comprising an amino acid sequence of SEQ ID NO: 24, 25, 26,
27, 28,
29, 30, or 73;
[15] a VH-CDR2 comprising an amino acid sequence of SEQ ID NO: 31, 32, 33,
34, 35,
36, 37, or 74; and
[16] a VH-CDR3 comprising an amino acid sequence of SEQ ID NO: 38, 39, 40,
41, 42,
43, or 75.
[17] Another embodiment provides an anti-MERS-CoV antibody or an antigen-
binding
fragment thereof comprising:
[18] a light chain variable region comprising a VL-CDR1 comprising an amino
acid
sequence of SEQ ID NO: 1, 2, 3, 4, 5, 6, or 7, a VL-CDR2 comprising an amino
acid
sequence of SEQ ID NO: 8,9, 10, 11, 12, 13, 14, or 71, and a VL-CDR3
comprising
an amino acid sequence of SEQ ID NO: 15, 16, 17, 18, 19, 20, 21, 22, 23, or
72; and
[19] a heavy chain variable region comprising a VH-CDR1 comprising an amino
acid
sequence of SEQ ID NO: 24, 25, 26, 27, 28, 29, 30, or 73, a VH-CDR2 comprising
an
amino acid sequence of SEQ ID NO: 31, 32, 33, 34, 35, 36, 37, or 74, and a VH-
CDR3
comprising an amino acid sequence of SEQ ID NO: 38, 39, 40, 41, 42, 43, or 75.
[20] Another embodiment provides an anti-MERS-CoV antibody or an antigen-
binding
fragment thereof comprising:
[21] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and
[22] a heavy chain variable region comprising or consisting essentially of
an amino acid
sequence of SEQ ID NO: 55, 56, 57, 58, 59, 60, 61, 62, 63, or 77.
[23] Another embodiment provides a pharmaceutical composition comprising
the anti-
MERS-CoV antibody or an antigen-binding fragment thereof. The pharmaceutical
composition may further comprise a pharmaceutically acceptable carrier. The
pharma-
ceutical composition may be used for preventing and/or treating MERS-CoV
infection
and/or a disease associated with MERS-CoV infection.
[24] Another embodiment provides a method of treating and/or preventing
MERS-CoV
infection and/or a disease associated with MERS-CoV infection, comprising
admin-
istering a pharmaceutically effective amount of the anti-MERS-CoV antibody or
an
4
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
antigen-binding fragment thereof or the pharmaceutical composition to a
subject in
need of treating and/or preventing the MERS-CoV infection and/or the disease.
The
method may further comprise a step of identifying the subject in need of
treating and/
or preventing MERS-CoV infection and/or a disease associated with MERS-CoV
infection, prior to the administering step.
[25] Another embodiment provides a use of the anti-MERS-CoV antibody or an
antigen-
binding fragment thereof or the pharmaceutical composition in treating and/or
preventing MERS-CoV infection and/or a disease associated with MERS-CoV
infection. Another embodiment provides a use of the anti-MERS-CoV antibody or
an
antigen-binding fragment thereof in preparing a pharmaceutical composition for
treating and/or preventing MERS-CoV infection and/or a disease associated with
MERS-CoV infection.
[26] An embodiment provides a polynucleotide encoding the anti-MERS-CoV
antibody
or an antigen-binding fragment thereof. In particular, an embodiment provides
a first
polynucleotide encoding a combination of VL-CDR1, VL-CDR2, and VL-CDR3, a
heavy chain variable region, or a heavy chain of the anti-MERS-CoV antibody or
an
antigen-binding fragment thereof. Another embodiment provides a second polynu-
cleotide encoding a combination of VH-CDR1, VH-CDR2, and VH-CDR3, a light
chain variable region, or a light chain of the anti-MERS-CoV antibody or an
antigen-
binding fragment thereof.
[27] An embodiment provides a recombinant vector comprising the first
polynucleotide,
the second polynucleotide, or a combination thereof. The recombinant vector
may be
used as an expression vector of the polynucleotide. Another embodiment
provides a re-
combinant cell transfected with the recombinant vector.
[28] Another embodiment provides a method of preparing the anti-MERS-CoV
antibody
or an antigen-binding fragment thereof, comprising expressing the first
polynucleotide
and the second polynucleotide in a cell. The step of expressing the
polynucleotides
may be conducted by culturing the cell comprising the polynucleotides (for
example,
each of the polynucleotides is carried by each recombinant vector, or both of
the
polynucleotides are carried by one recombinant vector) under a condition
allowing the
expression of the polynucleotides. The method may further comprise isolating
and/or
purifying the anti-MERS-CoV antibody or an antigen-binding fragment thereof
from
the cell culture, after the step of expressing or culturing.
[29]
Solution to Problem
[30] In this disclosure, an Ab-phage library was constructed using as a
source peripheral
blood mononuclear cell (PMBC) isolated from biological specimen of recovered
5
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
patients from MERS-CoV; and from convalescent MERS-CoV infected patients,
several potent human neutralizing antibodies that specifically bind to RBD in
the Si
domain of S glycoprotein of MERS-CoV were successfully identified and char-
acterized. More specifically, a phage-display library was constructed from two
con-
valescent MERS-CoV-infected patients and nine MERS-CoV RBD-specific neu-
tralizing monoclonal antibodies (mAbs) were successfully isolated therefrom.
After ad-
ministration (e.g. nebulization), these antibodies showed significant
aggregation and
reduced reactivity to recombinant S glycoprotein. In this disclosure, the
number of hy-
drophobic residues was reduced and solubilizing mutations were introduced
within the
complementarity-determining regions (CDRs), thereby generating an antibody
that is
resistant to aggregation during administration (e.g. nebulization) and retains
its MERS-
CoV neutralizing activity.
[31] One embodiment provides a MERS-CoV neutralizing antibody that bind to
RBD in
the Si domain of S glycoprotein of MERS-CoV or an antigen-binding fragment
thereof. In an embodiment, the antibody or antigen-binding fragment thereof
may be
for delivery via nebulization.
[32] Another embodiment provides medical uses of the anti- MERS-CoV
antibody or an
antigen-binding fragment thereof for treating and/or preventing MERS-CoV
infection
and/or a disease associated with MERS-CoV infection.
[33]
[34] Hereinafter, more detailed descriptions are provided.
[35]
[36] Definitions
[37] As used herein, 'consisting of a sequence,' consisting essentially of
a sequence,' or
'comprising a sequence' may refer to any case comprising the sequence, but it
may not
be intended to exclude a case comprising further sequence other than the
sequence.
[38] As used herein, the term 'a protein or polypeptide comprising or
consisting of an
amino acid sequence identified by SEQ ID NO' and 'a gene or polynucleotide
comprising or consisting of a nucleic acid sequence identified by SEQ ID NO'
may
refer to a protein (or polypeptide) or gene (or polynucleotide), which
consists es-
sentially of the amino acid sequence or nucleic acid sequence, or which has at
least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%,
at least 97%, at least 98%, or at least 99% sequence identity with the amino
acid
sequence or nucleic acid sequence with maintaining its inherent activity
and/or
function.
[39] As used herein, the term "antibody" may encompass various broad
classes of
polypeptides that can be distinguished biochemically. Those skilled in the art
will ap-
preciate that heavy chains are classified as gamma, mu, alpha, delta, or
epsilon (7, [I, a,
6
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
6, E) with some subclasses among them (e.g., 71-74), and light chains are
classified as
either kappa or lambda (K, X). It is the nature of this chain that determines
the "class"
of the antibody as IgG, IgM, IgA IgG, or IgE, respectively. The immunoglobulin
subclasses (isotypes), e.g., IgGl, IgG2, IgG3, IgG4, IgG5, etc., are well
characterized
and are known to confer functional specialization.
[40] An intact antibody includes two full-length light chains and two full-
length heavy
chains, in which each light chain is linked to a heavy chain by disulfide
bonds. The
antibody has a heavy chain constant region and a light chain constant region.
The
heavy chain constant region is of a gamma (7), mu (pc), alpha (a), delta (6),
or epsilon
(E) type, which may be further categorized as gamma 1 (71), gamma 2(72), gamma
3(73), gamma 4(74), alpha 1(a1), or alpha 2(a2). The light chain constant
region is of
either a kappa (cc) or lambda (X) type.
[41] The term "heavy chain" refers to a full-length heavy chain or a
fragment thereof,
including a variable region VH that includes amino acid sequences sufficient
to provide
specificity to antigens, and three constant regions, CHI, CH2, and CH3, and a
hinge. The
term "light chain" refers to a full-length light chain or a fragment thereof,
including a
variable region VL that includes amino acid sequences sufficient to provide
specificity
to antigens, and a constant region CL.
[42] The term "complementarity determining region (CDR)" refers to an amino
acid
sequence found in a hyper variable region of a heavy chain or a light chain of
im-
munoglobulin. The light and heavy chains may respectively include three CDRs
(light
chain: VL-CDR1, VL-CDR2, and VL-CDR3; heavy chain: HL-CDR1, HL-CDR2, and
HL-CDR3). The CDR may provide residues that play an important role in the
binding
of antibodies to an antigens or epitope. The terms "specifically binding" or
"specifically recognized" is well known to one of ordinary skill in the art,
and indicates
that an antibody and an antigen specifically interact with each other to lead
to an im-
munological activity.
[43] In this disclosure, the term "epitope" may refer to a site of an
antigen where an
antibody or an antigen-binding fragment binds, recognizes, targets, and/or
interacts.
[44] In this disclosure, the antibody may include, but not be limited to,
polyclonal or
monoclonal; and/or human, humanized, animal (e.g., mouse, rabbit, etc.)
derived
antibody, or chimeric antibodies (e.g., mouse-human chimeric antibody, rabbit-
human
chimeric antibody, etc.).
[45] An animal-derived antibody which is produced by immunizing an animal
with a
desired antigen may generally trigger an immune rejection response when
administered
to humans for treatment purpose, and a chimeric antibody has been developed to
suppress such immune rejection response. A chimeric antibody is formed by
replacing
the constant region of an animal-derived antibody, which is a cause of anti-
isotype
7
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
response, with the constant region of a human antibody using genetic
engineering
methods. The chimeric antibody has considerably improved anti-isotype response
in
comparison with animal-derived antibodies, but animal-derived amino acids are
still
present in its variable regions and thus it still contains potential side
effects resulting
from an anti-idiotypic response. It is a humanized antibody that has been
developed to
improve such side effects. This is manufactured by grafting CDR
(complementarity de-
termining regions) which, of the variable regions of a chimeric antibody, has
an
important role in antigen binding into a human antibody framework.
[46] As used herein, the term "antigen binding fragment" refers to a
fragment derived
from a full immunoglobulin structure including a portion capable of binding to
an
antigen such as CDRs. For example, the antigen binding fragment may be scFv,
(scFv)
2, Fab, Fab', or F(abt)2, but not be limited thereto. In the present
disclosure, the antigen
binding fragment may be a fragment derived from an antibody, including at
least one
complementarity determining region, for example, selected from the group
consisting
of scFv, (scFv)2, scFv-Fc, Fab, Fab' and F(ab')2.
[47] Of the antigen binding fragments, Fab is a structure having variable
regions of a light
chain and a heavy chain, a constant region of the light chain, and the first
constant
region (CHI) of the heavy chain, and it has one antigen binding site.
[48] Fab' is different from Fab in that it has a hinge region including one
or more cysteine
residues at the C-terminal of heavy chain CHI domain. An F(ab')2antibody is
formed
through disulfide bond of the cysteine residues at the hinge region of Fab'.
[49] Fv is a minimal antibody piece having only a heavy chain variable
region and light
chain variable region, and a recombinant technique for producing the Fv
fragment is
well known in the pertinent art. Two-chain Fv may have a structure in which
the heavy
chain variable region is linked to the light chain variable region by a non-
covalent
bond, and single-chain Fv (scFv) may generally have a dimer structure in which
the
variable region of a heavy chain and the variable region of a light chain are
covalently
linked via a peptide linker or they are directly (i.e., without a peptide
linker) linked to
each other. The peptide linker may comprise about 1 to about 100 amino acids,
about 2
to about 50 amino acids, or about 5 to about 25 amino acids, and any kinds of
amino
acids may be included therein without any restrictions.
[50] The antigen binding fragments may be obtained using proteases (for
example, a
whole antibody is digested with papain to obtain Fab fragments, and is
digested with
pepsin to obtain F(abt)2 fragments), and may be prepared by a genetic
recombinant
technique.
[51] Immunoglobulin (e.g., a human immunoglobulin) or antibody molecules of
the
disclosure can be of any type (e.g., IgG, IgE, IgM, IgD, IgA, IgY, etc.),
class (e.g.,
IgGl, IgG2, IgG3, IgG4, IgG5, IgAl, IgA2, etc.), or subclass of immunoglobulin
8
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
molecule.
[52] In the antibody or antibody fragment, portions(e.g., frameworks,
constant regions,
etc.) except the CDRs or variable regions may be derived from an
immunoglobulin
(e.g., a human immunoglobulin), and particularly, they may be derived from
IgG, IgA,
IgD, IgE, IgM, or IgY, for example, IgGl, IgG2, IgG 3, or IgG4.
[53] In this disclosure, the term "antibody" may include an antigen binding
fragment
thereof comprising any combination of 6 CDRs or VH (heavy chain variable
region)
and VL (light chain variable region) as well as a complete (full length)
antibody (e.g.,
IgG), unless otherwise indicated or clearly contradicted by context.
[54] The antibody or antigen binding fragment may be chemically or
recombinantly syn-
thesized (not naturally occurring).
[55] The term "subject" may refer to any subject, particularly a mammalian
subject, for
whom diagnosis, prophylaxis, and/or therapy is desired. Mammalian subjects may
include humans, domestic animals, farm animals, and zoo, sport, or pet animals
such as
dogs, cats, guinea pigs, rabbits, rats, mice, horses, cattle, cows, and so on.
[56]
[57] RBD and/or S protein of MERS-CoV as an epitope
[58] One embodiment provides an anti-MERS-CoV antibody that binds to
(and/or
recognizes and/or targets) a receptor-binding domain (RBD) in the Si domain of
S
protein (spike protein) of MERS-CoV or an antigen-binding fragment thereof.
[59] An epitope for an anti-MERS-CoV antibody or an antigen-binding
fragment thereof
provided herein may be positioned in RBD of S protein of MERS-CoV. The epitope
may be positioned at least partially overlapping or adjacent to a receptor
binding site of
S protein of MERS-CoV, thereby allowing the antibody or an antigen-binding
fragment thereof to inhibit the binding of S protein of MERS-CoV to a cell
(hereinafter, "host cell") of a subject infected by MERS-CoV.
[60] The RBD or S protein of MERS-CoV which is targeted by an anti-MERS-CoV
antibody or an antigen-binding fragment thereof provided herein may be a wild-
type
and/or variants thereof.
[61] For example, the S protein of MERS-CoV may be represented by GenBank
Accession No. AF588936.1 (SEQ ID NO: 70), wherein the region from E367 to Y606
amino acid residues corresponds to the RBD. For example, the variant of S
protein of
MERS-CoV may have a polymorphism (mutation) from a wild type. In a specific em-
bodiment, the variant of S protein may have mutation (e.g., amino acid
substitution) at
position 510, 529, or both thereof, in SEQ ID NO: 70. For example, the variant
of S
protein may have one or both of mutation D5 10G (wherein the amino acid (D)
corre-
sponding to position 510 is substituted with G) and I529T(wherein the amino
acid (I)
corresponding to position 529 is substituted with T) in SEQ ID NO: 70.
CA 03152245 12022-02-23
WO 2021/060837 PCT/KR2020/012887
[62] The receptor binding site of the S protein may be a binding site with
a receptor on a
host cell. In an embodiment, the receptor on a host cell may be dipeptidyl
peptidase IV
(DPP4). For example, when the subject infected by MERS-CoV is a human being
(i.e.,
the host cell is a human cell), and the receptor on a host cell is human DPP4
(e.g.,
NCBI Accession No. NP 001926.2 etc.), the receptor binding site of the S
protein with
DPP4 may comprise at least one selected from amino acid residues corresponding
to
positions 452, 454, 460, 461, 462, 463, 466, 499, 501, 502, 504, 505, 506,
510, 511,
512, 513, 515, 535, 536, 537, 538, 539, 540, 541, 542, 553, 555, 557, 559, and
562 of
SEQ ID NO: 70 (https://www.nature.com/articlesincomms9223?origin=ppub).
[63] The epitope may be at least partially overlapping the receptor binding
site of S
protein (SEQ ID NO: 70) as described above. For example, an anti-MERS-CoV
antibody or an antigen-binding fragment thereof provided herein may bind to an
epitope comprising at least one (e.g., at least two, at least three, at least
four, at least
five, at least six, at least seven, at least eight, at least nine, at least
ten, at least eleven,
at least twelve, at least thirteen, at least fourteen, at least fifteen, at
least sixteen, at
least seventeen, at least eighteen, at least nineteen, or at least twenty,
etc.) selected
from amino acid residues corresponding to positions 498 to 554 of the S
protein (SEQ
ID NO: 70) or variants thereof.
[64] In a specific embodiment, the epitope for an anti-MERS-CoV antibody or
an antigen-
binding fragment thereof provided herein may be:
[65] (a) an epitope comprising at least one (e.g., 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19, 20, 21, 22 or 23) selected from amino acid residues
corresponding
to positions 498 to 520 (e.g., SYINKCSRLLSDDRTEVPQLVNA (SEQ ID NO: 67;
wild-type) or SYINKCSRLLSDGRTEVPQLVNA (SEQ ID NO: 68; variant (including
mutation D5 10G)) of the S protein (SEQ ID NO: 70) or variants thereof;
[66] (b) an epitope comprising at least one (e.g., 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14,
or 15) selected from amino acid residues corresponding to positions 540 to 554
(YYRKQLSPLEGGGWL (SEQ ID NO: 69)) of the S protein (SEQ ID NO: 70) or
variants thereof; or
[67] (c) both of (a) and (b).
[68] The epitope and receptor binding site positioned in S protein (SEQ ID
NO: 70) are il-
lustrated below.
[69] 1 MIHSVFLLMF LLTPTESYVD VGPDSVKSAC IEVDIQQTFF DKTWPRPIDV
SKADGIIYPQ
[70] 61 GRTYSNITIT YQGLFPYQGD HGDMYVYSAG HATGITPQKL
FVANYSQDVK QFANGFVVRI
[71] 121 GAAANSTGTV IISPSTSATI RKIYPAFMLG SSVGNFSDGK
MGRFFNHTLV LLPDGCGTLL
10
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[72] 181 RAFYCILEPR SGNHCPAGNS YTSFATYHTP ATDCSDGNYN
RNASLNSFKE YFNLRNCTFM
[73] 241 YTYNITEDEI LEWFGITQTA QGVHLFSSRY VDLYGGNMFQ
FATLPVYDTI KYYSIIPHSI
[74] 301 RSIQSDRKAW AAFYVYKLQP LTFLLDFSVD GYIRRAIDCG
FNDLSQLHCS YESFDVESGV
[75] 361 YSVSSFEAKP SGSVVEQAEG VECDFSPLLS GTPPQVYNFK
RLVFTNCNYN LTKLLSLFSV
[76] 421 NDFTCSQISP AAIASNCYSS LILDYFSYPL SMKSDLSVSS AGPISQFNYK
QSFSNPTCLI
[77] 481 LATVPHNLTT ITKPLKYSYI NKCSRLLSDD RTEVPQLVNA
NQYSPCVSIV PSTVWEDGDY
[78] 541 YRKQLSPLEG GGWLVASGST VAMTEQLQMG FGITVQYGTD
TNSVCPKLEF ANDTKIASQL
[79] 601 GNCVEYSLYG VSGRGVFQNC TAVGVRQQRF VYDAYQNLVG
YYSDDGNYYC LRACVSVPVS
[80] 661 VIYDKETKTH ATLFGSVACE HISSTMSQYS RSTRSMLKRR
DSTYGPLQTP VGCVLGLVNS
[81] 721 SLFVEDCKLP LGQSLCALPD TPSTLTPRSV RSVPGEMRLA
SIAFNHPIQV DQLNSSYFKL
[82] 781 SIPTNFSFGV TQEYIQTTIQ KVTVDCKQYV CNGFQKCEQL
LREYGQFCSK INQALHGANL
[83] 841 RQDDSVRNLF ASVKSSQSSP IIPGFGGDFN LTLLEPVSIS TGSRSARSAI
EDLLFDKVTI
[84] 901 ADPGYMQGYD DCMQQGPASA RDLICAQYVA GYKVLPPLMD VN-
MEAAYTSS LLGSIAGVGW
[85] 961 TAGLSSFAAI PFAQSIFYRL NGVGITQQVL SENQKLIANK FN-
QALGAMQT GFTTTNEAFR
[86] 1021 KVQDAVNNNA QALSKLASEL SNTFGAISAS IGDIIQRLDV
LEQDAQIDRL INGRLTTLNA
[87] 1081 FVAQQLVRSE SAALSAQLAK DKVNECVKAQ SKRSGFCGQG
THIVSFVVNA PNGLYFMHVG
[88] 1141 YYPSNHIEVV SAYGLCDAAN PTNCIAPVNG YFIKTNNTRI
VDEWSYTGSS FYAPEPITSL
[89] 1201 NTKYVAPQVT YQNISTNLPP PLLGNSTGID FQDELDEFFK
NVSTSIPNFG SLTQINTTLL
[90] 1261 DLTYEMLSLQ QVVKALNESY IDLKELGNYT YYNKWPWYIW
LGFIAGLVAL ALCVFFILCC
11
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[91] 1321 TGCGTNCMGK LKCNRCCDRY EEYDLEPHKV HVH
[92] (the amino sequence of GenBank Accession No. AFS88936.1 (SEQ ID NO:
70),
wherein the possible region where the epitope may position is indicated in
bold and the
receptor binding site is underlined)
[93]
[94] Anti-MERS-CoV antibody or antigen-binding fragment thereof
[95] Provided an antibody against a coronavirus, MERS-CoV, or an antigen-
binding
fragment thereof, having improved stability and efficacy, and uses thereof.
The
antibody may be a neutralizing antibody against MERS-CoV. The antibody or an
antigen-binding fragment thereof may bind to an epitope positioned at least
partially
overlapping or adjacent to a receptor binding site of S protein of MERS-CoV,
to
competing with RBD and/or S protein) of MERS-CoV for binding a receptor on a
host
cell, thereby inhibiting the binding of S protein of MERS-CoV to the host
cell. The S
protein of MERS-CoV may be a wild type and/or a variant (e.g., including
D510G,
I529T, or both thereof).
[96] An embodiment provides an anti-MERS-CoV antibody or an antigen-binding
fragment thereof, which binds to an epitope positioned in a RBD in the 51
domain of S
protein of MERS-CoV or an antigen-binding fragment thereof.
[97] In a specific embodiment, an anti-MERS-CoV antibody or an antigen-
binding
fragment thereof provided herein may bind to
[98] (a) an epitope comprising at least one (e.g., 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19, 20, 21, 22 or 23) selected from amino acid residues
corresponding
to positions 498 to 520 (e.g., SYINKCSRLLSDDRTEVPQLVNA (SEQ ID NO: 67;
wild-type) or SYINKCSRLLSDGRTEVPQLVNA (SEQ ID NO: 68; variant (including
D5 10G)) of the S protein (SEQ ID NO: 70) or variants thereof;
[99] (b) an epitope comprising at least one (e.g., 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14,
or 15) selected from amino acid residues corresponding to positions 540 to 554
(YYRKQLSPLEGGGWL (SEQ ID NO: 69)) of the S protein (SEQ ID NO: 70) or
variants thereof; or
[100] (c) both of (a) and (b).
[101]
[102] Another embodiment provides an anti-MERS-CoV antibody or an antigen-
binding
fragment thereof comprising:
[103] a VL-CDR1 comprising an amino acid sequence of SEQ ID NO: 1, 2, 3, 4,
5, 6, or 7;
[104] a VL-CDR2 comprising an amino acid sequence of SEQ ID NO: 8, 9, 10,
11, 12, 13,
14 or 71;
[105] a VL-CDR3 comprising an amino acid sequence of SEQ ID NO: 15, 16, 17,
18, 19,
20, 21, 22, 23, or 72;
12
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[106] a VH-CDR1 comprising an amino acid sequence of SEQ ID NO: 24, 25, 26,
27, 28,
29, 30, or 73;
[107] a VH-CDR2 comprising an amino acid sequence of SEQ ID NO: 31, 32, 33,
34, 35,
36, 37, or 74; and
[108] a VH-CDR3 comprising an amino acid sequence of SEQ ID NO: 38, 39, 40,
41, 42,
43, or 75.
[109] Amino acid sequences of CDRs of the anti-MERS-CoV antibody or an
antigen-
binding fragment thereof are illustrated in Table 1, wherein the CDRs are
determined
according to IMGT numbering:
13
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[110] [Table 11
CDR Amino Acid Sequence SEQ
ID NO
VL-CDR1 QSLLHSNGYNY 1
QSLVHSNGYNY 2
SSNIGSNY 3
QSVLYSSNNKNY 4
SSNIGAGYD 5
SSSVGNNY 6
KLGEKY 7
VL CDR2 LGS 8
EGS 9
SNN 10
WAS 11
GNS 12
SNS 13
QDS 14
QDT 71
VL CDR3 MQALQTPLT 15
MQAVQTPLT 16
ATWDDNLSGPV 17
QQYYGSPYT 18
QQYYSTPPT 19
AAWDDSLSGPV 20
AAWDDSLSGVV 21
AAWDDSLNGPV 22
QAWDSRRAV 23
QAWDNNFYV 72
14
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
VH CDR1 GGTFSSYA 24
DGKEKREA 25
EGNESKEA 26
GGSISSSSYY 27
GGTFSSYG 28
GGTFSSFT 29
GFTFSSYS 30
GFSIDDYA 73
VH CDR2 IIPFFGTA 31
IIPFFDKA 32
IFYIGNT 33
IIPILGIA 34
IIPIFGIA 35
IYYTGNT 36
ISTTGSYI 37
ISWDSGSI 74
VH CDR3 ARDGRKDYYGSGSYLHYYGMDV 38
ARQEGSSIIRFDP 39
ASLFDSSGYYPYYFDY 40
ATHFGASGYDPYYFDY 41
ARQVADLGYFDY 42
AKGTAFDGGLAFDI 43
AREKQLVPYYYYGMDV 75
[111] Another embodiment provides an anti-MERS-CoV antibody or an antigen-
binding
fragment thereof comprising:a light chain variable region comprising a VL-CDR1
comprising an amino acid sequence of SEQ ID NO: 1, 2, 3, 4, 5, 6, or 7, a VL-
CDR2
comprising an amino acid sequence of SEQ ID NO: 8,9, 10, 11, 12, 13, 14, or
71, and
a VL-CDR3 comprising an amino acid sequence of SEQ ID NO: 15, 16, 17, 18, 19,
20,
21, 22, 23, or 72; and
[112] a heavy chain variable region comprising a VH-CDR1 comprising an
amino acid
sequence of SEQ ID NO: 24, 25, 26, 27, 28, 29, 30, or 73, a VH-CDR2 comprising
an
amino acid sequence of SEQ ID NO: 31, 32, 33, 34, 35, 36, 37, or 74, and a VH-
CDR3
15
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
comprising an amino acid sequence of SEQ ID NO: 38, 39, 40, 41, 42, 43, or 75.
[113] Another embodiment provides an anti-MERS-CoV antibody or an antigen-
binding
fragment thereof comprising:
[114] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and
[115] a heavy chain variable region comprising or consisting essentially of
an amino acid
sequence of SEQ ID NO: 55, 56, 57, 58, 59, 60, 61, 62, 63, or 77.
[116] The amino acid sequences of the variable regions of the anti-MERS-CoV
antibody or
an antigen-binding fragment are illustrated in Table 2:
16
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[117] [Table 2]
SEQ ID Light chain variable region targeting MERS-CoV (e.g., RBD in Si
NO domain of S glycoprotein of MERS-CoV)
44 ELVMTQSPLSLPVTPGEPASISCRSSQSLLHSNGYNYLDWYLQRPG
QSPQLLIYLGSNRASGVPDRFS GS GS GTDFTLKIGRVEAEDVGIYYC
MQALQTPLTFGGGTKVEIK
45 ELVMTQSPLSLPVTPGEPASISCRSSQSLVHSNGYNYLDWYLQRPG
QSPQLLIYLGSNRASGVPDRFS GS GS GTDFTLKIGRVEAEDVGIYYC
MQAVQTPLTFGGGTKVEIK
46 ELVMTQSPLSLPVTPGEPASISCRSSQSLLHSNGYNYLDWYLQRPG
QSPQLLIYEGSNRASGVPDRFS GS GS GTDFTLKIGRVEAEDVGIYYC
MQALQTPLTFGGGTKVEIK
47 ELELT QPPS VS GTPGQRVTISC S GS SSNIGSNYVYWYQQLPGTAPKL
LIYSNNQRPS GVPDRFS GS KS GT S ASLAIS GLRSEDEADYYCA TWD
DNLSGPVFGGGTKVTVLG
48 ELVMTQSPDSLAVSLGERATINCKSSQSVLYSSNNKNYLAWYQQK
PGQPPKLLIYWASTRES GVPDRFS GS GSGTDFTLTIS SVQTEDVAVY
YCQQYYGSPYTFGQGTKLEIK
49 ELQMTQSPDSLAVSLGERATINCKSSQSVLYSSNNKNYLAWYQQK
PGQPPKLLIS WA STRES GVPERFS GS GS GTDFTLTIS GLQAEDVAVY
YCQQYYSTPPTFGQGTKVDIK
50 ELELT QPPS VS GAPGQRVTIS CT GS SSNIGAGYDVHWYQQLPGTAP
KLLIYGNSNRPSGVPDRFS GS KS GTSASLAIS GLQSEDEGDYYCAA
WDDSLSGPVFGGGTELTVLG
51 ELVVT QPPS AS GTPGQRVAIS CS GS SSNIGSNYVYWYQQLPGTAPK
LLIYSNNHRPS GVPDRFS GS KS GTSASLAIS GLRSEDEAVYYCAAW
DDSLSGVVFGGGTELTVLG
52 ELMLT QPHS AS GTPGQRVAIS CS GRSSSVGNNYVYWYQQLPGAAP
KLLIYSNSQRPSGVPDRFS GS KS GTSASLVIS GLRSEDEADYYCAA
WDDSLSGPVFGGGTQLTVLG
53 ELELT QPPS VS GTPGQRVTISC S GS SSNIGSNYVYWYQQLPGTAPKL
LIYSNNQRPS GVPDRFS GS KS GT S ASLAINGLQSED EADYYCAAWD
DSLNGPVFGGGTKLTVLG
54 QAAELVLTQSPSVSVSPGQTATITCSGDKLGEKYASWYQQRPGQS
PVLVIYQDSRRASGIPERFS GSNS GNTATLTIS GT QAMDEADYYCQ.
17
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
AWDSRRAVFGGGTELTVLG
76 ELVLTQPPSVSVSPGQAASITCSADKLGEKYVFWYQQKPGQSPVL
AIYODTKRPS GIPERFS GS NS GNTATLTIS GTQPMDEADYYC QAWD
NNFYVFGTGTKLTVLG
SEQ ID Heavy chain variable region targeting MERS-CoV (e.g., RBD in Si
NO domain of S glycoprotein of MERS-CoV)
55 EVQLVQSGAEVKKPGSSVKVSCKASGGTFSSYAISWVRQAPGQGL
EWMGGIIPFFGTA NYAQKFQGRVTITADES TS TAYMELS S LRS EDT
AVYYCARDGRKDYYGSGSYLHYYGMDVWGQGTTVTVSS
56 EVQLVQSGAEVKKPGSSVKVSCKASDGKEKREAISWVRQAPGQG
LEWM GGIIPFFDKANYAQKFQGRVTITADES TS TAYMELS SLRS ED
TAVYYCARDGRKDYYGSGSYLHYYGMDVWGQGTTVTVSS
57 EVQLVQSGAEVKKPGSSVKVSCKASEGNESKEAISWVRQAPGQG
LEWM GGIIPFFDKANYAQKFQGRVTITADES TS TAYMELS SLRS ED
TAVYYCARDGRKDYYGSGSYLHYYGMDVWGQGTTVTVSS
58 QVQLQES GPGLVKPSETLSLTCTVS GGSI SS S SYYWGWIRQPPGKG
LEWIGS IFYIGNTYYNPS LKS RVTIS VDT S KNQFS LRLS S VTAADTA
VYYCARQEGSSIIRFDPWGQGTLVTVSS
59 QVQLVQSGAEVKKPGSSVKVSCKASGGTFSSYGISWVRQAPGQG
LEWM GRIIPILGIANYAQKFQGRVTITAD KS TS TAYMELS SLRS ED
TAVYYCASLFDSSGYYPYYFDYWGQGTLVTVSS
60 EVQLVQSGAEVKKPGSSVKVSCKASGGTFSSFTISWVRQAPGQGL
EWMGRIIPIFGIA NYAQKFQ GRVTITAD KST GTAYMELS SLRS EDT
AVYYCATHFGASGYDPYYFDYWGQGTLVTVSS
61 QVQLQES GPGLVKPSETLSLTCTVS GGSI SS S SYYWGWIRQPPGKG
LEWIGS IFYIGNTYYNPS LKS RVTIS VDT S KNQFS LKLS S VTAADTA
VYYCARQEGSSIIRFDPWGQGTLVTVSS
62 QVQLQES GPGLVKPSETLSLTCTVS GGSI SS S SYYWGWIRQPPGKG
LEWIGS IYYTGNTYYNPS LKS RLTIS VDT S KNQFS LKLS S VTAADT
AVYYCARQVADLGYFDYWGQGTLVTVSS
63 EVQLLES GGGLVKPGGSLRLSCAAS GFTFSSYSMNWVRQAPGKGL
EWVS SI ST TGSYIFYAD S VKGRFTIS RDNAKNS LYLQMNTLRPEDT
ALYYCAKGTAFDGGLAFDIWGQGTIVTVSS
77 EVQLVQS GGGLVRPGRSLRLSCVAPGFSIDDYAMHWVRQTPGKG
LEWVS GISWDSGSIAYADSVKGRFTISRDNAKNSLYLQMNSLRAE
18
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
DTAVYYCAREKQLVPYYYYGMDVWGQGTTVTVSS
[118] (CDR1, CDR2, and CDR3, which are determined by IMGT numbering, are un-
derlined in order)In a specific embodiment, the anti-MERS-CoV antibody or an
antigen-binding fragment thereof may comprise:
[119] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and a heavy chain variable
region
comprising or consisting essentially of an amino acid sequence of SEQ ID NO:
55;
[120] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and a heavy chain variable
region
comprising or consisting essentially of an amino acid sequence of SEQ ID NO:
56;
[121] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and a heavy chain variable
region
comprising or consisting essentially of an amino acid sequence of SEQ ID NO:
57;
[122] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and a heavy chain variable
region
comprising or consisting essentially of an amino acid sequence of SEQ ID NO:
58;
[123] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and a heavy chain variable
region
comprising or consisting essentially of an amino acid sequence of SEQ ID NO:
59;
[124] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and a heavy chain variable
region
comprising or consisting essentially of an amino acid sequence of SEQ ID NO:
60;
[125] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and a heavy chain variable
region
comprising or consisting essentially of an amino acid sequence of SEQ ID NO:
61;
[126] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and a heavy chain variable
region
comprising or consisting essentially of an amino acid sequence of SEQ ID NO:
62;
[127] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and a heavy chain variable
region
comprising or consisting essentially of an amino acid sequence of SEQ ID NO:
63; or
[128] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and a heavy chain variable
region
comprising or consisting essentially of an amino acid sequence of SEQ ID NO:
77.
[129]
[130] For example, the anti-MERS-CoV antibody or an antigen-binding
fragment thereof
may comprise:
19
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[131] (1) a VL-CDR1 of SEQ ID NO: 1, a VL-CDR2 of SEQ ID NO: 8, a VL-CDR3
of
SEQ ID NO: 15, a VH-CDR1 of SEQ ID NO: 24, a VH-CDR2 of SEQ ID NO: 31, and
a VH-CDR3 of SEQ ID NO: 38;
[132] (2) a VL-CDR1 of SEQ ID NO: 1, a VL-CDR2 of SEQ ID NO: 8, a VL-CDR3
of
SEQ ID NO: 15, a VH-CDR1 of SEQ ID NO: 25, a VH-CDR2 of SEQ ID NO: 32, and
a VH-CDR3 of SEQ ID NO: 38;
[133] (3) a VL-CDR1 of SEQ ID NO: 2, a VL-CDR2 of SEQ ID NO: 8, a VL-CDR3
of
SEQ ID NO: 16, a VH-CDR1 of SEQ ID NO: 25, a VH-CDR2 of SEQ ID NO: 32, and
a VH-CDR3 of SEQ ID NO: 38;
[134] (4) a VL-CDR1 of SEQ ID NO: 1, a VL-CDR2 of SEQ ID NO: 8, a VL-CDR3
of
SEQ ID NO: 15, a VH-CDR1 of SEQ ID NO: 26, a VH-CDR2 of SEQ ID NO: 32, and
a VH-CDR3 of SEQ ID NO: 38;
[135] (5) a VL-CDR1 of SEQ ID NO: 1, a VL-CDR2 of SEQ ID NO: 9, a VL-CDR3
of
SEQ ID NO: 15, a VH-CDR1 of SEQ ID NO: 26, a VH-CDR2 of SEQ ID NO: 32, and
a VH-CDR3 of SEQ ID NO: 38;
[136] (6) a VL-CDR1 of SEQ ID NO: 3, a VL-CDR2 of SEQ ID NO: 10, a VL-CDR3
of
SEQ ID NO: 17, a VH-CDR1 of SEQ ID NO: 27, a VH-CDR2 of SEQ ID NO: 33, and
a VH-CDR3 of SEQ ID NO: 39;
[137] (7) a VL-CDR1 of SEQ ID NO: 4, a VL-CDR2 of SEQ ID NO: 11, a VL-CDR3
of
SEQ ID NO: 18, a VH-CDR1 of SEQ ID NO: 28, a VH-CDR2 of SEQ ID NO: 34, and
a VH-CDR3 of SEQ ID NO: 40;
[138] (8) a VL-CDR1 of SEQ ID NO: 4, a VL-CDR2 of SEQ ID NO: 11, a VL-CDR3
of
SEQ ID NO: 19, a VH-CDR1 of SEQ ID NO: 29, a VH-CDR2 of SEQ ID NO: 35, and
a VH-CDR3 of SEQ ID NO: 41;
[139] (9) a VL-CDR1 of SEQ ID NO: 5, a VL-CDR2 of SEQ ID NO: 12, a VL-CDR3
of
SEQ ID NO: 20, a VH-CDR1 of SEQ ID NO: 27, a VH-CDR2 of SEQ ID NO: 33, and
a VH-CDR3 of SEQ ID NO: 39;
[140] (10) a VL-CDR1 of SEQ ID NO: 3, a VL-CDR2 of SEQ ID NO: 10, a VL-CDR3
of
SEQ ID NO: 21, a VH-CDR1 of SEQ ID NO: 27, a VH-CDR2 of SEQ ID NO: 36, and
a VH-CDR3 of SEQ ID NO: 42;
[141] (11) a VL-CDR1 of SEQ ID NO: 6, a VL-CDR2 of SEQ ID NO: 13, a VL-CDR3
of
SEQ ID NO: 20, a VH-CDR1 of SEQ ID NO: 27, a VH-CDR2 of SEQ ID NO: 36, and
a VH-CDR3 of SEQ ID NO: 42;
[142] (12) a VL-CDR1 of SEQ ID NO: 3, a VL-CDR2 of SEQ ID NO: 10, a VL-CDR3
of
SEQ ID NO: 22, a VH-CDR1 of SEQ ID NO: 27, a VH-CDR2 of SEQ ID NO: 36, and
a VH-CDR3 of SEQ ID NO: 42;
[143] (13) a VL-CDR1 of SEQ ID NO: 7, a VL-CDR2 of SEQ ID NO: 14, a VL-CDR3
of
SEQ ID NO: 23, a VH-CDR1 of SEQ ID NO: 30, a VH-CDR2 of SEQ ID NO: 37, and
20
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
a VH-CDR3 of SEQ ID NO: 43; or
[144] (14) a VL-CDR1 of SEQ ID NO: 7, a VL-CDR2 of SEQ ID NO: 71, a VL-CDR3
of
SEQ ID NO: 72, a VH-CDR1 of SEQ ID NO: 73, a VH-CDR2 of SEQ ID NO: 74, and
a VH-CDR3 of SEQ ID NO: 75.
[145] For example, the anti-MERS-CoV antibody or an antigen-binding
fragment thereof
may comprise:
[146] (1) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
44 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 55;
[147] (2) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
44 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 56;
[148] (3) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
45 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 56;
[149] (4) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
44 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 57;
[150] (5) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
46 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 55;
[151] (6) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
47 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 58;
[152] (7) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
48 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 59;
[153] (8) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
49 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 60;
[154] (9) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
50 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 61;
[155] (10) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
51 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 62;
[156] (11) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
52 and a heavy chain variable region comprising or consisting essentially of
an amino
21
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
acid sequence of SEQ ID NO: 62;
[157] (12) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
53 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 62;
[158] (13) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
54 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 63; or
[159] (14) a light chain variable region comprising or consisting
essentially of SEQ ID NO:
76 and a heavy chain variable region comprising or consisting essentially of
an amino
acid sequence of SEQ ID NO: 77.
[160]
[161] The antibody provided herein may include an antigen binding fragment
thereof
comprising any combination of VH (heavy chain variable region) and VL (light
chain
variable region), such as a scFv or a scFv linked with Fc and/or hinge region,
as well
as a complete (full length) antibody (e.g., IgGl, IgG2, IgG3, IgG4, etc.).
[162] In an embodiment, the anti-MERS-CoV antibody or antigen-binding
fragment
thereof may be in a full-length form of IgGl, IgG2, IgG3, or IgG4, comprising:
[163] a light chain comprising a light chain variable region comprising or
consisting es-
sentially of SEQ ID NO: 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and
[164] a heavy chain comprising a heavy chain variable region comprising or
consisting es-
sentially of an amino acid sequence of SEQ ID NO: 55, 56, 57, 58, 59, 60, 61,
62, 63,
or 77.
[165] In another embodiment, anti-MERS-CoV antibody or antigen-binding
fragment
thereof may be in a scFv (single chain variable fragment) form, comprising:
[166] a light chain variable region comprising a VL-CDR1 comprising an
amino acid
sequence of SEQ ID NO: 1, 2, 3, 4, 5, 6, or 7, a VL-CDR2 comprising an amino
acid
sequence of SEQ ID NO: 8,9, 10, 11, 12, 13, 14, or 71, and a VL-CDR3
comprising
an amino acid sequence of SEQ ID NO: 15, 16, 17, 18, 19, 20, 21, 22, 23, or
72; and
[167] a heavy chain variable region comprising a VH-CDR1 comprising an
amino acid
sequence of SEQ ID NO: 24, 25, 26, 27, 28, 29, 30, or 73, a VH-CDR2 comprising
an
amino acid sequence of SEQ ID NO: 31, 32, 33, 34, 35, 36, 37, or 74, and a VH-
CDR3
comprising an amino acid sequence of SEQ ID NO: 38, 39, 40, 41, 42, 43, or 75.
[168] Another embodiment provides an anti-MERS-CoV antibody or an antigen-
binding
fragment thereof in a scFv form, comprising:
[169] a light chain variable region comprising or consisting essentially of
SEQ ID NO: 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, or 76; and
[170] a heavy chain variable region comprising or consisting essentially of
an amino acid
sequence of SEQ ID NO: 55, 56, 57, 58, 59, 60, 61, 62, 63, or 77.
22
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[171] More specific description of the light chain variable and the heavy
chain variable
region is the same as above.
[172] In the scFv, the light chain variable region and the heavy chain
variable region may
be linked in any order. For example, the scFv may comprise the light chain
variable
region and the heavy chain variable region in a direction of N- to C-terminus
(i.e., the
C-terminus of the light chain variable region and the N-terminus of the heavy
chain
variable region are linked; N-VL-VH-C). Alternatively, the scFv may comprise
the
heavy chain variable region and the light chain variable region in a direction
of N- to
C-terminus (i.e., the C-terminus of the heavy chain variable region and the N-
terminus
of the light chain variable region are linked; N-VH-VL-C).
[173] In the scFv, the light chain variable region and the heavy chain
variable region may
be linked via a peptide linker (e.g., N-VL-linker-VH-C or N-VH-linker-VL-C) or
directly (without a peptide linker). The peptide linker may be an oligopeptide
including
1 to 100 amino acids or 5 to 25 amino acids, each of which may be any kind of
amino
acids without any restrictions. Any conventional peptide linker may be used
with or
without an appropriate modification to comply with specific purposes. In a
specific
embodiment, the peptide linker may comprise, for example, at least one Gly, at
least
one Ser, and/or at least one Arg residues, in any order. The amino acid
sequences
suitable for the peptide linker may be known in the relevant art. The length
of the
peptide linker can be properly determined within such a limit that the
functions of the
polypeptide and/or scFv will not be affected. For instance, the peptide linker
may be
formed by including a total of about 1 to about 100 amino acids, about 2 to
about 50
amino acids, about 5 to about 30 amino acids, or about 5 to about 25 (e.g., 1,
2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, or 25)
amino acids,
each of which is independently selected from the group consisting of Gly, Ser,
Arg,
and the like, in any order. In one embodiment, the peptide linker may be
represented as
"GGSSRSSSSGGGGSGGGG" (SEQ IF NO: 65), or (GmS1)õ (m, 1, and n are the
number of "G", "S", and "(GmS1)", respectively, and independently selected
from
integers of about 1 to about 10, particularly, 1, 2, 3, 4, 5, 6, 7, 8, 9, or
10). For
example, the peptide linker can be amino acids of SEQ ID NO: 65, (GGGGS)2,
(GGGGS)3, (GGGGS)4, or (GS)9, but not be limited thereto.
[174] In an embodiment, the anti-MERS-CoV antibody or antigen-binding
fragment
thereof may be in a scFv-Fc form, wherein a scFv and Fc region are linked to
each
other via a peptide linker or directly (without a peptide linker). In the scFv-
Fc, the scFv
and the peptide linker are described as above; the Fc region may be derived
from IgG
(e.g., IgGl, IgG2, IgG3, or IgG4), for example, represented by SEQ ID NO: 66;
and
the Fc region may be linked to the N-terminus or C-terminus of the scFv in
direction of
N- to C-terminus or C- to N-terminus (i.e., the N-terminus of the scFv and the
N-
23
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
terminus of the Fc region are linked, the N-terminus of the scFv and the C-
terminus of
the Fc region are linked, the C-terminus of the scFv and the N-terminus of the
Fc
region are linked, or the C-terminus of the scFv and the C-terminus of the Fc
region
are linked).
[175] In an embodiment, the scFv and/or the scFv-Fc antibody may further
comprise a
hinge region. In this case, the scFv and/or scFv-Fc are described as above;
the hinge
region may be derived from IgG (e.g., IgGl, IgG2, IgG3, or IgG4), for example,
rep-
resented by SEQ ID NO: 65; and the hinge region may be linked to the N-
terminus or
C-terminus (e.g., N-terminus) of the Fc region in the scFv and/or the scFv-Fc.
[176] As used herein, the immunoglobulin such as IgG (e.g., IgGl, IgG2,
IgG3, or IgG4)
may be a human or murine immunoglobulin, for example, a human immunoglobulin
(hIgG; e.g., hIgGl, hIgG2, hIgG3, or hIgG4).
[177]
[178] The binding affinity of an antibody or its antigen-binding fragment
provided in this
disclosure to RBD and/or S protein of MERS-CoV can be assessed using one or
more
techniques well established in the art. For example, the binding affinity may
be
assessed by a binding assay such as ELISA assays using a recombinant RBD
and/or S
protein of MERS-CoV, but not be limited thereto. For example, the binding
affinity of
an antibody or its antigen-binding fragment provided in this disclosure to RBD
and/or
S protein of MERS-CoV may be KD of 1 x 106 M or less, 1 x 10 7 M or less, or 1
x 10
M or less, for example, 1.01 x 10 9 M or less, but not be limited thereto.
[179] Without limitation, the anti-MERS-CoV antibody or fragment thereof is
a chimeric
antibody, a humanized antibody, or a fully human antibody. In one aspect,
antibody or
fragment thereof is not naturally occurring, or chemically or recombinantly
syn-
thesized.
[180] Given that each of antibodies can bind to MERS-CoV, for example,
receptor-binding
domain (RBD) of MERS-CoV, the CDR sequences, or VL (heavy chain variable
region) and VL (light chain variable region) sequences as disclosed herein can
be
"mixed and matched" to create other Anti-MERS-CoV binding molecules.
[181]
[182] Medical use
[183] Provided is a medical use of an anti-MERS-CoV antibody or an antigen-
binding
fragment thereof provided herein for treating and/or preventing MERS-CoV
infection
and/or a disease associated with MERS-CoV infection. Such medical use of the
antibody or antigen-binding fragment may be due to the capability to compete
with
MERS-CoV (RBD and or S protein) for binding to a receptor on a host cell,
thereby in-
hibiting the binding of MERS-CoV to a receptor on a host cell and inhibiting
infection
of MERS-CoV.
24
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[184] More specifically, an embodiment provides a pharmaceutical
composition
comprising the anti-MERS-CoV antibody or an antigen-binding fragment thereof
as an
active ingredient. The pharmaceutical composition may further comprise a
pharma-
ceutically acceptable carrier. The pharmaceutical composition may be used for
preventing and/or treating MERS-CoV infection and/or a disease associated with
MERS-CoV infection.
[185] Another embodiment provides a method of treating and/or preventing
MERS-CoV
infection and/or a disease associated with MERS-CoV infection, comprising
admin-
istering a pharmaceutically effective amount of the anti-MERS-CoV antibody or
an
antigen-binding fragment thereof or the pharmaceutical composition to a
subject in
need of treating and/or preventing the MERS-CoV infection and/or the disease.
The
method may further comprise a step of identifying the subject in need of
treating and/
or preventing MERS-CoV infection and/or a disease associated with MERS-CoV
infection, prior to the administering step.
[186] Another embodiment provides a use of the anti-MERS-CoV antibody or an
antigen-
binding fragment thereof or the pharmaceutical composition in treating and/or
preventing MERS-CoV infection and/or a disease associated with MERS-CoV
infection. Another embodiment provides a use of the anti-MERS-CoV antibody or
an
antigen-binding fragment thereof in preparing a pharmaceutical composition for
treating and/or preventing MERS-CoV infection and/or a disease associated with
MERS-CoV infection.
[187] The disease associated with MERS-CoV infection may be selected from
any res-
piratory syndrome caused by MERS-CoV infection (e.g., pneumonia, acute upper
res-
piratory infection, etc.) and/or complications thereof (e.g., respiratory
failure, septic
shock, etc.) and/or symptoms (e.g., cough, fever, shortness of breath,
headache, chill,
sore throat, rhinorrhea, muscular pain, inappetence, nausea, stomachache,
emesis,
diarrhea, etc.) associated with the respiratory syndrome.
[188] The pharmaceutical composition may further comprise a pharmaceutical
acceptable
carrier, solubilizer, diluent, emulsifier, preservative, excipient, and/or
adjuvant, in
addition to the active ingredient. For example, the pharmaceutically
acceptable carrier
may be one or more selected from the group consisting of lactose, dextrose,
sucrose,
sorbitol, mannitol, starch, gum acacia, calcium phosphate, alginates, gelatin,
calcium
silicate, micro-crystalline cellulose, polyvinylpyrrolidone, cellulose, water,
syrup,
methyl cellulose, methylhydroxy benzoate, propylhydroxy benzoate, talc,
magnesium
stearate, and mineral oil, but are not limited thereto. In certain
embodiments, the phar-
maceutical composition may further comprise formulation materials for
modifying,
maintaining or preserving, for example, the pH, osmolality, viscosity,
clarity, color,
isotonicity, odor, sterility, stability, rate of dissolution or release,
adsorption or pen-
25
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
etration of the composition. In such embodiments, suitable formulation
materials may
be at least one selected from the group consisting of, but are not limited to,
amino acids
(such as glycine, glutamine, asparagine, arginine or lysine); antimicrobials;
an-
tioxidants (such as ascorbic acid, sodium sulfite or sodium hydrogen-sulfite);
buffers
(such as borate, bicarbonate, Tris-HC1, citrates, phosphates or other organic
acids);
bulking agents (such as mannitol or glycine); chelating agents (such as
ethylenediamine tetraacetic acid (EDTA)); complexing agents (such as caffeine,
polyvinylpyrrolidone, beta-cyclodextrin or hydroxypropyl-beta-cyclodextrin);
fillers;
monosaccharides; disaccharides; and other carbohydrates (such as glucose,
sucrose,
mannose or dextrins); proteins (such as serum albumin, gelatin or
immunoglobulins);
coloring, flavoring and diluting agents; emulsifying agents; hydrophilic
polymers (such
as polyvinylpyrrolidone); low molecular weight polypeptides; salt-forming
counterions
(such as sodium); preservatives (such as benzalkonium chloride, benzoic acid,
salicylic
acid, thimerosal, phenethyl alcohol, methylparaben, propylparaben,
chlorhexidine,
sorbic acid or hydrogen peroxide); solvents (such as glycerin, propylene
glycol or
polyethylene glycol); sugar alcohols (such as mannitol or sorbitol);
suspending agents;
surfactants or wetting agents (such as pluronics, PEG, sorbitan esters,
polysorbates
such as polysorbate 20, polysorbate, triton, tromethamine, lecithin,
cholesterol,
tyloxapal); stability enhancing agents (such as sucrose or sorbitol); tonicity
enhancing
agents (such as alkali metal halides, preferably sodium or potassium chloride,
mannitol
sorbitol); delivery vehicles; diluents; and excipients and/or pharmaceutical
adjuvants.
[189] The antibody or antigen-binding fragment, or the pharmaceutical
composition may
be administered to a subject orally or parenterally. The parenteral
administration may
be intranasal administration, intrapulmonary administration, intravenous
injection, sub-
cutaneous injection, muscular injection, intraperitoneal injection,
endothelial admin-
istration, or local administration. Since oral administration leads to
digestion of
proteins or peptides, an active ingredient in the compositions for oral
administration
must be coated or formulated to prevent digestion in stomach. In addition, the
com-
positions may be administered using an optional device that enables the active
in-
gredient to be delivered to target cells or organ.
[190] For example, the antibody or antigen-binding fragment may be
administered by in-
tranasal or intrapulmonary route, such as nasal spray, inhalation, or
nebulization.
[191] As used herein, the term "the pharmaceutically effective amount" may
refer to an
amount at which the active ingredient (the antibody or antigen-binding
fragment) can
exert pharmaceutically meaningful effects in preventing or treating MERS-CoV
infection and/or a disease associated with MERS-CoV infection. The
pharmaceutically
effective amount of the active ingredient, or a suitable dosage of the
pharmaceutical
composition indicated by an amount of the active ingredient, may be prescribed
in a
26
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
variety of ways, depending on various factors, such as age, body weight,
gender,
pathologic conditions, diets, excretion speed, and/or reaction sensitivity of
a patient,
formulation types, administration time, administration route, administration
manner,
and the like. For example, the pharmaceutically effective amount of the active
in-
gredient, or a suitable dosage of the pharmaceutical composition, may be in
the range
from about 0.001 to about 1000 mg(amount of the antibody or antigen-binding
fragment)/kg(body weight), about 0.01 to about 100 mg/kg, or 0.1 to 50 mg/kg
per day
for an adult.
[192] The subject to which the antibody or antigen-binding fragment or the
pharmaceutical
composition is administered may be one selected from mammals, for example,
humans, monkeys, rats, mice, dogs, cats, guinea pigs, rabbits, rats, mice,
horses, cattle,
cows, and so on, or a cell or tissue obtained therefrom, but are not limited
thereto. The
subject may be one having a risk of MERS-CoV infection or suffering from MERS-
CoV infection or a disease associated with MERS-CoV infection.
[193] The pharmaceutical composition may be formulated with a
pharmaceutically ac-
ceptable carrier and/or excipient into a unit or a multiple dosage form by a
method
easily carried out by a skilled person in the pertinent art. The dosage form
may be a
solution in oil or an aqueous medium, a suspension, syrup, an emulsifying
solution, an
extract, powder, granules, a tablet, or a capsule, and may further include a
dispersing
or a stabilizing agent. For example, the pharmaceutical composition may be
formulated
for intranasal or intrapulmonary delivery of the antibody or antigen-binding
fragment,
such as nasal spray, inhalation, or nebulization formulation.
[194]
[195] Polynucleotide, recombinant vector, and preparation of antibody
[196] An embodiment provides a polynucleotide encoding the anti-MERS-CoV
antibody
or an antigen-binding fragment thereof. In particular, an embodiment provides
a first
polynucleotide encoding a combination of VL-CDR1, VL-CDR2, and VL-CDR3, a
heavy chain variable region, or a heavy chain of the anti-MERS-CoV antibody or
an
antigen-binding fragment thereof. Another embodiment provides a second polynu-
cleotide encoding a combination of VH-CDR1, VH-CDR2, and VH-CDR3, a light
chain variable region, or a light chain of the anti-MERS-CoV antibody or an
antigen-
binding fragment thereof.
[197] An embodiment provides a recombinant vector comprising the first
polynucleotide,
the second polynucleotide, or a combination thereof. The recombinant vector
may be
used as an expression vector of the polynucleotide. Another embodiment
provides a re-
combinant cell transfected with the recombinant vector.
[198] Another embodiment provides a method of preparing the anti-MERS-CoV
antibody
or an antigen-binding fragment thereof, comprising expressing the first
polynucleotide
27
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
and the second polynucleotide in a cell. The step of expressing the
polynucleotides
may be conducted by culturing the cell comprising the polynucleotides (for
example,
each of the polynucleotides is carried by each recombinant vector, or both of
the
polynucleotides are carried by one recombinant vector) under a condition
allowing the
expression of the polynucleotides. The method may further comprise isolating
and/or
purifying the anti-MERS-CoV antibody or an antigen-binding fragment thereof
from
the cell culture, after the step of expressing or culturing.
[199] The term "vector" refers to a means for expressing a target gene in a
host cell, as ex-
emplified by a plasmid vector, a cosmid vector, and a viral vector such as a
bacte-
riophage vector, an adenovirus vector, a retrovirus vector, and an adeno-
associated
virus vector. The recombinant vector may be constructed from plasmids
frequently
used in the art (for example, pComb3XSS vector, pSC101, pGV1106, pACYC177,
ColEL pKT230, pME290, pBR322, pUC8/9, pUC6, pBD9, pHC79, p1161, pLAFR1,
pHV14, pGEX series, pET series, and pUC19), phages (for example, Xgt4XB, X-
Charon, XAzl, and M13) or by manipulating viruses (for example, SV40, etc.).
[200] In the recombinant vector, the polynucleotide may be operatively
linked to a
promoter. The term "operatively linked" is intended to pertain to a functional
linkage
between a nucleotide sequence of interest and an expression regulatory
sequence (for
example, a promoter sequence). When being "operatively linked", the regulatory
element can control the transcription and/or translation of the nucleotide of
interest.
[201] The recombinant vector may be constructed typically as a cloning
vector or an ex-
pression vector. For recombinant expression vectors, a vector generally
available in the
relevant art for expressing a foreign protein in plant, animal, or microbial
cells may be
employed. Various methods well known in the art may be used for the
construction of
recombinant vectors.
[202] For use in hosts, such as prokaryotic or eukaryotic cells, the
recombinant vector may
be constructed accordingly. For example, when a vector is constructed as an
expression
vector for use in a prokaryotic host, the vector typically includes a strong
promoter for
transcription (e.g., a pLia promoter, a CMV promoter, a trp promoter, a lac
promoter,
a tac promoter, a T7 promoter, etc.), a ribosomal binding site for initiating
translation,
and transcriptional/translational termination sequences. On the other hand, an
ex-
pression vector for use in a eukaryotic host includes an origin of replication
operable in
a eukaryotic cell, such as an fl origin of replication, an SV40 origin of
replication, a
pMB1 origin of replication, an adeno origin of replication, an AAV origin of
replication, and a BBV origin of replication, but is not limited thereto. In
addition, the
expression vector typically includes a promoter derived from genomes of
mammalian
cells (for example, metallothionein promoter) or from mammalian viruses (for
example, adenovirus late promoter, vaccinia virus 7.5K promoter, SV40
promoter, cy-
28
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
tomegalovirus promoter, and tk promoter of HSV), and a polyadenylation
sequence as
a transcription termination sequence.
[203] The recombinant cell may be prepared by introducing the recombinant
vector into a
suitable host cell. As long as it allows the sequential cloning and expression
of the re-
combinant vector in a stable manner, any host cell known in the art may be
employed
in the present disclosure. Examples of the prokaryotic host cell available for
the
present disclosure may be selected from E. coli, Bacillus spp. such as
Bacillussubtilis
and Bacillus thuringiensis, and enterobacteriaceae strains such as Salmonella
ty-
phimurium, Serratia marcescens and various Pseudomonas species. Eukaryotic
host
cells that may be used for transformation may selected from, but are not
limited to,
Saccharomyce cerevisiae, insect cells, and animal cells, such as Sp2/0, CHO
(Chinese
hamster ovary) Kl, CHO DG44, PER.C6, W138, BHK, COS-7, 293, HepG2, Huh7,
3T3, RIN, and MDCK.
[204] The polynucleotide or a recombinant vector carrying the same may be
introduced
(transfected) into a host cell using a method well known in the relevant art.
For
example, this transfection may be carried out using a CaCl2 or electroporation
method
when the host cell is prokaryotic. For eukaryotic host cells, the genetic
introduction
may be achieved using, but not limited to, microinjection, calcium phosphate
pre-
cipitation, electroporation, liposome-mediated transfection, or particle
bombardment.
[205] To select a transformed host cell, advantage may be taken of a
phenotype associated
with a selection marker according to methods well known in the art. For
example,
when the selection marker is a gene conferring resistance to a certain
antibiotic, the
host cells may be grown in the presence of the antibiotic in a medium to
select a
transformant of interest.
[206] Another embodiment provides a method for production of the antibody
or antigen-
binding fragment, the method comprising a step of expressing the
polynucleotide or
the recombinant vector in a host cell. In one embodiment, the production
method may
comprise culturing a recombinant cell harboring the polynucleotide or the
recombinant
vector thereat, and optionally isolating and/or purifying the antibody from
the culture
medium.
[207]
Advantageous Effects of Invention
[208] Middle East respiratory syndrome coronavirus (MERS-CoV) induces
severe ag-
gravating respiratory failure in infected patients, frequently resulting in
mechanical
ventilation. As limited therapeutic antibody is accumulated in lung tissue
following
systemic administration, inhalation is newly recognized as an alternative,
possibly
better, route of therapeutic antibody for pulmonary diseases. The nebulization
process,
29
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
however, generates diverse physiological stresses, and thus, the therapeutic
antibody
must be resistant to these stresses, remain stable, and form minimal
aggregates. The
disclosure provides antibodies against MERS-CoV with significantly greater
stability,
reactivity and neutralizing activity following nebulization.
[209]
Brief Description of Drawings
[210] Figures la and lb are graphs showing reactivity of scFv clones before
and after neb-
ulization.
[211] Figure 2 (A-D) shows results of flow cytometry analysis of the
inhibition of re-
combinant S glycoprotein binding to hDPP4-expressing cells.
[212] Figure 3 (A-B) schematically shows sequential randomization of CDR
residues of the
C-8 clone.
[213] Figure 4 (A-D) shows graphs showing reactivity of C-8, C-8-2, and C-8-
2-4B clones
in scFv-Fc fusion protein form or IgG1 (full-length) form before and after
nebu-
lization.
[214] Figure 5 (A-C) shows graphs showing Reactivity of anti-MERS-CoV IgG1
an-
tibodies before and after nebulization.
[215] Figure 6 shows graphs showing DLS analysis results.
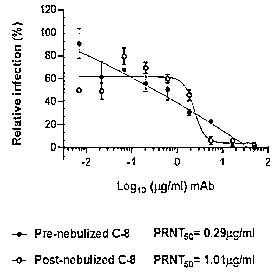
[216] Figures 7a-7c are graphs showing relative infection (%), indicating
neutralization of
MERS-CoV by pre- and post-nebulized IgGl.
[217] Figure 8 shows graphs showing reactivity of anti-MERS-CoV IgG1
antibodies
against recombinant MERS-CoV S RBD mutants.
[218] Figures 9a and 9b are graphs showing results of flow cytometry
analysis of the in-
hibition of recombinant mutant MERS-CoV RBD protein binding to
hDPP4-expressing cells.
[219] Figures 10a-10c show HDX profiles of free- and C-8 IgGi-bound MERS-
CoV RBD,
wherein 10a shows three-dimensional structure of MERS-CoV RBD, 10b shows
graphs corresponding to (A) region (5er498-Ala520) indicated in 10a (upper
graph:
deuterium uptake of 5498-L506; lower graph: deuterium uptake of RBD L507-
A520),
and 10c shows a graph corresponding to (B) region (Tyr540-Leu554) indicated in
10a,
wherein "Single" indicates MERS-CoV RBD only (C-8 IgG1 antibody does not bind
to MERS-CoV RBD), and "complex" indicates a complex of MERS-CoV RBD and C-
8 IgG1 antibody.
[220] Figure 11 illustrates mapping of the C-8 epitope and DPP4 binding
site on MERS-
CoV RBD sequence.
[221]
Mode for the Invention
30
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[222] Hereafter, the present invention will be described in detail by
examples. The
following examples are intended merely to illustrate the invention and are not
construed to restrict the invention.
[223]
[224] Example 1. Construction of a human scFv phage-display library and
three ran-
domization libraries
[225] PBMCs were isolated from two MERS-CoV-infected convalescent patients
using a
Ficoll-Paque density gradient medium (GE Healthcare, Pittsburgh, PA, USA) as
described previously (Kanof ME, Smith PD, Zola H. Isolation of whole
mononuclear
cells from peripheral blood and cord blood. Curr Protoc Immunol. 2001;Chapter
7:Unit
7 1). The PBMCs were subjected to total RNA isolation using the TRI Reagent
(Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. The
RNA
was used to synthesize cDNA using Superscript III First-Strand Synthesis
system
(Invitrogen) with oligo(dT) primers according to the manufacturer's
instructions.
Using the cDNA as a template, the genes encoding the variable regions of heavy
and
light chains (VH and Vic/V),) were amplified and used for the construction of
a human
scFv phage-display libraries as described previously (Phage Display: A
Laboratory
Manual. Carlos F. Barbas III, Dennis R. Burton, Jamie K. Scott, Gregg J.
Silverman.
2001;76(4):487-488; Andris-Widhopf J, Steinberger P, Fuller R, Rader C, Barbas
CF,
3rd. Generation of human scFv antibody libraries: PCR amplification and
assembly of
light- and heavy-chain coding sequences. Cold Spring Harb Protoc.
2011;2011(9)).
[226] For the construction of the first randomization library, a set of
degenerate Ultramer
DNA oligonucleotides (Integrated DNA Technologies, Coralville, IA, USA)
encoding
residues from H1 to H65 of clone C-8 (VH,H) was chemically synthesized to
contain
either a codon encoding the wild-type amino acid or a GAK degenerate codon at
the
H29, H32, H51, H52, H53, and H54 residues (Table 3). Then, the gene fragment
(VHc)
encoding residues from H58 to H113 of clone C-8 was amplified by PCR in a T100
Thermal Cycler (Bio-Rad, Carlsbad, CA, USA). The PCR conditions were as
follows:
preliminary denaturation at 94 C for 5 min, followed by 25 cycles of 15 s at
94 C, 15 s
at 56 C and 90 s at 72 C. A final extension was then conducted for 10 min at
72 C.
[227] After electrophoresis on a 1% agarose gel, the PCR products were
purified using
QIAquick gel extraction kit (Qiagen Inc., Valencia, CA, USA) according to the
manu-
facturer's instructions. The purified VHNT1 and Vfic gene fragments were mixed
at a con-
centration of 100 ng and subjected to linker PCR in a T100 Thermal Cycler to
yield the
VH1 fragment. The PCR conditions were as follows: preliminary denaturation at
94 C
for 5 min, followed by 25 cycles of 15 s at 94 C, 15 s at 56 C and 120 s at 72
C. The
reaction was ended with an extension step for 10 min at 72 C. The gene
fragment
encoding VL (VIA) of clone C-8 was amplified by PCR with the same PCR
conditions
31
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
described above for amplification of VHc. Then, the VH1 and VII fragments were
subjected to electrophoresis on a 1% agarose gel, and excised bands were
purified
using the QIAquick gel extraction kit. The purified VH1 and VII fragments were
used
for the synthesis of the scFv gene (scFv) using PCR as described previously
(Phage
Display: A Laboratory Manual. Carlos F. Barbas III, Dennis R. Burton, Jamie K.
Scott,
Gregg J. Silverman. 2001;76(4):487-488). The amplified scFv i fragment was
purified
and cloned into the phagemid vector as described (Phage Display: A Laboratory
Manual. Carlos F. Barbas III, Dennis R. Burton, Jamie K. Scott, Gregg J.
Silverman.
2001;76(4):487-488; Andris-Widhopf J, Steinberger P, Fuller R, Rader C, Barbas
CF,
3rd. Generation of human scFv antibody libraries: PCR amplification and
assembly of
light- and heavy-chain coding sequences. Cold Spring Harb Protoc.
2011;2011(9)).
[228] For the construction of the second randomization library, a set of
degenerate
Ultramer DNA oligonucleotides encoding residues from H1 to H65 of clone C-8-2
(V
HN2) was chemically synthesized to contain either a codon encoding the wild-
type
amino acid or a GAK degenerate codon at the H26 to H33 (HCDR1) and H51 to H57
(HCDR2) residues (Table 3), excluding the previously randomized residues. The
VHN2
and VHc gene fragments were mixed at equal ratios at 100 ng and subjected to
linker
PCR in a T100 Thermal Cycler to yield the VH2 gene fragment as described
above. The
VH2 gene fragment was purified as described above and subjected to linker PCR
with V
Li fragments to yield the scFv2 gene fragment, which was cloned into the
phagemid
vector as described above.
[229] For the construction of the third randomization library, two sets of
degenerate
Ultramer DNA oligonucleotides with a length of 200 nucleotides were chemically
syn-
thesized. One set encoded from Li to L61 residues of clone C-8 (VLN), while
the other
one encoded from L56 to L107 of clone C-8 (Vir). These degenerate
oligonucleotides
contained either a codon encoding the wild-type amino acid or a GAK degenerate
codon at L27B, L27C, L30, L32, L50, L89, L92, and L96 residues (Table 3). The
VLN
and Vir gene fragments (100 ng each) were subjected to a linker PCR in a T100
Thermal Cycler to produce the VL2 gene fragment using the same PCR conditions
as
described above for the amplification of the VH1 gene fragment. The gene
fragment
encoding VH of C-8-2-4B (VH3) was amplified by PCR using the same PCR
conditions
used for the amplification of the Vfic gene fragment as described above. After
pu-
rification, VL2 and VH3 gene fragments were used to produce the scFv3 gene
fragment,
which was cloned into the phagemid vector as described above.
32
CA 03152245 2022-02-23
WO 2021/060837 PCTXR2020/012887
[230] [Table 31
Degenerate ccidoiic in7ed in the randomized libraries
The fir.: H:
HCDR1 HCDR2
- H26 H27 H28 H29 H30 H31 H32 H33 H51 H52 H52A H53 H54 H55 H56 H57
flu -r
GGTFSSYA 1 PF F GT A
acid
De-Jen
KW
erate KAK RVVK RVVK KWK KWK
cidon
--11-10
acids
encode D,E, bill I 7'1 N F.L Y FLY
he .
F DE
,LV ,V.b \e õ ,b,
! ly"
DE DE
de-Jene
ra7:e.
codon
The second library
HCDR1 HCDR2
KaI.r.a H52 H26 H27 H28 H29 H30 H31
H32 H33 H51 H52 H53 H54 H55 H56 H57
nu i-ieisr A
GGTESSEA I I P F F GT A
Dn
acid
era:e GRK GRF lb RRK RRK GRK i:YAK
aciciD
P R,
G.D G.D, T KH A.D G. .D
K,A .
!he E E E
D.E
degene E E
ra,.e
en
The third library
LCDR1 LCDR2 LCDR3
Kai:a4, L2 L2 L2 L2 L2 L2 L2 L2 L3 L3 L31 L8 L9 L9 L9 L9 L9 L9 L9 L9
50 L51 L52
nu rie'sr 7 Th7B70707E 8 9 0 1 2 9 0 1 2 3 4 5 6 7
A
LL HS NGYN Y L GS MDALIDTPLI
acid
erx.e. K K
K K K
cocion K
Eno
acids LF L,F d LF M.1 L.F L,F
.
encoe ,HõH, , .H,
Y, Y, H
d 00. D N. 0. 0,
D
V, V , , V, V,
E ,
ID, 0, E ,D
D. 0, 0,
EE
codon
[231]
[232] Example 2. Biopanning
[233] The human scFv phage-display libraries were subjected to four rounds
of biopanning
against recombinant MERS-CoV S RBD protein (Sino Biological Inc., Beijing,
China)
33
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
as described previously (Lee Y, Kim H, Chung J. An antibody reactive to the
Gly63-Lys68 epitope of NT-proBNP exhibits 0-glycosylation-independent binding.
Exp Mol Med. 2014;46:e114). Briefly, the scFv phage-display libraries (-10"
phage)
were added to 3[1g of the recombinant MERS-CoV S RBD protein conjugated to 5.0
x
106 magnetic beads (Dynabeads M-270 epoxy, Invitrogen) and incubated with
rotation
for 2 h at 37 C. The beads were washed once with 500 [IL of 0.05% (v/v) Tween-
20
(Sigma-Aldrich, St. Louis, MO, USA) in PBS (PBST) during the first round of
biopanning. The number of washes was increased to three for the other rounds.
Phages
bound to beads were eluted, neutralized, allowed to infect E. coli ER2738 (New
England Biolabs, Ipswich, MA, USA), and rescued as described previously (Lee
Y,
Kim H, Chung J. An antibody reactive to the Gly63-Lys68 epitope of NT-proBNP
exhibits 0-glycosylation-independent binding. Exp Mol Med. 2014;46:e114).
[234] The first randomized scFv library was subjected to two rounds of
biopanning against
recombinant MERS-CoV S RBD protein. The scFv phage-display library (-10"
phage) was added to 1.5[1g of the recombinant MERS-CoV S RBD protein
conjugated
to 2.5 x 106 magnetic beads and incubated with rotation for 2 h at 37 C. The
beads
were washed once with 500 [AL of 0.5% PBST and three times with 500 [IL of
0.5%
PBST during the first and second rounds of biopanning, respectively. After
each round
of washing, bound phages were eluted and rescued as described above.
[235] For first round of biopanning for the second and third randomized
scFv libraries, the
scFv phage-display libraries (-10" phage) were added to 1.5[1g of the
recombinant
MERS-CoV S RBD protein conjugated to 2.5 x 106 magnetic beads and incubated
with
rotation for 2 h at 37 C. After washing three times with 500 [IL of 0.5% PBST,
bound
phages were eluted and rescued as described above.
[236] Before the second round of biopanning of the second and third
randomized scFv
libraries, 10 [tg of recombinant MERS-CoV S RBD protein was conjugated to 200
[tg
of non-magnetic beads (Nacalai, San Diego, CA, USA) following the
manufacturer's
instructions. Then, the scFv phage-display libraries (-10" phage) were added
to 1.5[1g
of recombinant MERS-CoV S RBD protein conjugated to 2.5 x 106 magnetic beads
and incubated on a rotator for 2 h at 37 C. After washing three times with 500
[IL of
0.5% PBST, magnetic beads were resuspended in 100 [AL of PBS and transferred
to a
microtube (microTUBE AFA Fiber Pre-Slit Snap-Cap, 520045, Covaris, Woburn, MA,
USA) along with the recombinant MERS-COV S RBD protein-conjugated non-
magnetic beads resuspended in 30 [IL of PBS at a concentration of 0.33 [tg/mL.
Then,
these bead mixtures were subjected to an ultrasound washing step using an
ultra-
sonicator (M220, Covaris) with the following conditions: duty factor (DF) 20%,
peak
incident power (PIP) 12.5 W, cycles/burst 50, 20 min, and 24 C. After
ultrasonication,
magnetic beads were transferred to 1.5-mL microcentrifuge tube and washed
three
34
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
times with 0.5% PBST. Then, the bound phages were eluted and rescued as
described
above.
[237]
[238] Example 3. High-throughput retrieval of scFv clones and phage ELISA
[239] After the fourth round of biopanning of human scFv phage-display
libraries, the
plasmid DNA was obtained from overnight cultures of E. coli cells and
subjected to
high-throughput retrieval of scFv clones by TrueRepertoire analysis as
described
previously (Celemics, Seoul, Republic of Korea) (Noh J, Kim 0, Jung Y, Han H,
Kim
JE, Kim S, et al. High-throughput retrieval of physical DNA for NGS-
identifiable
clones in phage display library. MAbs. 2019;11(3):532-545).
[240] To select reactive clones to recombinant MERS-CoV S RBD protein, the
scFv genes
obtained from TrueRepertoire were cloned into the pComb3XSS vector (Phage
Display: A Laboratory Manual. Carlos F. Barbas III, Dennis R. Burton, Jamie K.
Scott,
Gregg J. Silverman. 2001;76(4):487-488) and used to transform E. coli ER2738
cells.
After overnight culture, the phages were rescued from individual colonies
using the
M13K07 helper phage and subjected to phage ELISA as described previously
(Phage
Display: A Laboratory Manual. Carlos F. Barbas III, Dennis R. Burton, Jamie K.
Scott,
Gregg J. Silverman. 2001;76(4):487-488). Microtiter plates (Costar, Cambridge,
MA,
USA) were coated with 100 ng of recombinant MERS-CoV S RBD protein in coating
buffer (0.1 M sodium bicarbonate, pH 8.6) at 4 C overnight. The wells were
blocked
with 3% (w/v) bovine serum albumin (BSA; Thermo Scientific, Waltham, MA, USA)
dissolved in PBS for 1 h at 37 C, and culture supernatant containing scFv-
displayed
phages that were rescued from individual colonies were added into each well.
After in-
cubation for 2 h at 37 C, the microtiter plates were washed three times with
0.05%
PBST. Then, horseradish peroxidase (HRP)-conjugated anti-M13 monoclonal
antibody
(GE Healthcare) in 3% BSA/PBS was added into wells, and the plate was
incubated
for 1 h at 37 C. After washing three times with PBST,
2,2'-azino-bis-3-ethylbenzothiazoline-6-sulfonic acid solution (Thermo
Scientific) was
used as the substrate for HRP. Absorbance was measured at 405 nm with a
Multiskan
Ascent microplate reader (Labsystems, Helsinki, Finland).
[241] To select reactive clones from the randomized libraries, phage ELISA
was performed
as described previously (Phage Display: A Laboratory Manual. Carlos F. Barbas
III,
Dennis R. Burton, Jamie K. Scott, Gregg J. Silverman. 2001;76(4):487-488)
using re-
combinant MERS-CoV S RBD protein-coated microtiter plates. The nucleotide
sequences of positive scFv clones were determined by Sanger sequencing
(Cosmogenetech, Seoul, Republic of Korea).
[242]
[243] Example 4. Expression of scFv-hFc and IgG1
35
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[244] The genes encoding the selected scFv clones were cloned into a
modified
mammalian expression vector containing the hIgGi Fc regions (hFc) at the C-
terminus
as described previously (Lee S, Yoon IH, Yoon A, Cook-Mills JM, Park CG, Chung
J.
An antibody to the sixth Ig-like domain of VCAM-1 inhibits leukocyte
transendothelial migration without affecting adhesion. J. Immunol.
2012;189(9):4592-4601). The expression vectors were transfected into HEK293F
cells
(Invitrogen), and the fusion proteins were purified by Protein A affinity chro-
matography as described previously (Lee S, Yoon IH, Yoon A, Cook-Mills JM,
Park
CG, Chung J. An antibody to the sixth Ig-like domain of VCAM-1 inhibits
leukocyte
transendothelial migration without affecting adhesion. J. Immunol.
2012;189(9):4592-4601).
[245] For the expression of IgGI (with kappa light chain), genes encoding
VH and V, were
amplified from the phage clones, cloned into a mammalian expression vector,
and
transfected into HEK293F cells. Then, IgGI was purified by Protein A affinity
chro-
matography as described previously (Jin J, Park G, Park JB, Kim S, Kim H,
Chung J.
An anti-EGFR x cotinine bispecific antibody complexed with cotinine-conjugated
duo-
carmycin inhibits growth of EGFR-positive cancer cells with KRAS mutations.
Exp
Mol Med. 2018;50(5):67). Then the eluate containing IgGI was subjected to gel
filtration chromatography. A total of 4 mg of IgGI was injected at a flow rate
of 1 mL/
min and purified by gel filtration using a XK16/100 column packed with
Superdex 200
pg at pH 7.4 (AKTA pure, GE Healthcare). The chromatogram was recorded at a UV
absorbance of 280nm. The fractions containing IgGI were pooled by collection
criteria
and concentrated.
[246]
[247] Example 5. Experimental methods
[248] 5.1. ELISA
[249] Microtiter plates (Costar) were coated with 100 ng of recombinant S
glycoprotein
(GenBank Accession No. AF588936.1; wild type) in coating buffer at 4 C
overnight.
The wells were blocked with 3% BSA/PBS for 1 h at 37 C. Both nebulized and non-
nebulized scFv-hFc or IgGI were serially diluted (5-fold, 12 dilutions
starting from 500
nM for scFv-hFc fusion protein or 1,000 nM for IgGI) in blocking buffer and
added
into individual wells. After incubation for 1 h at 37 C, the microtiter plates
were
washed three times with 0.05% PBST. Then, HRP-conjugated rabbit anti-human IgG
antibody (Invitrogen) in blocking buffer (1:5,000) was added into wells, and
the plate
was incubated for 1 h at 37 C. After washing three times with PBST,
2,2'-azino-bis-3-ethylbenzothiazoline-6-sulfonic acid solution (Thermo
Scientific) was
used as the substrate. Absorbance was measured at 405 nm using a microplate
spec-
trophotometer (Multiskan GO; Thermo Scientific).
36
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[250]
[251] 5.2. Nebulization
[252] A nebulizer (Aerogen Pro, Aerogen, Galway, Ireland) was used for all
experiments
following the manufacturer's instructions. The nebulizer containing 1 mL of
scFv-hFc
fusion proteins or IgGI antibodies was placed on top of a 50-mL conical tube
(SPL Life
Sciences, Pocheon, Republic of Korea) and nebulized at a concentration of
either 0.1,
0.3, or 1 mg/mL in phosphate-buffered saline (PBS).
[253]
[254] 5.3. Microneutralization assay
[255] The virus (MERS-CoV/KOR/KNIH/002 05 2015, accession number
KT029139.1)
was obtained from the Korea National Institute of Health (kindly provided by
Dr. Sung
Soon Kim) and propagated in Vero cells (ATCC CCL-81) in Dulbecco's Modified
Eagle's Medium (DMEM, Welgene, Gyeongsan, Republic of Korea) in the presence
of
2% fetal bovine serum (Gibco). The cells were grown in T-75 flasks, inoculated
with
MERS-CoV, and incubated at 37 C in a 5% CO2 environment. Then 3 days after in-
oculation, the viruses were harvested and stored at -80 C. The virus titer was
de-
termined via a TCID50 assay (Reed LJ, Muench H. Lancaster Press, In-
corporated:1938).
[256] A neutralization assay was performed as previously described (Jiang
L, Wang N, Zuo
T, Shi X, Poon KM, Wu Y, et al. Potent neutralization of MERS-CoV by human neu-
tralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl
Med.
2014;6(234):234ra259). Briefly, Vero cells were seeded in 96-well plates (1 x
104
cells/well) in Opti-PRO SFM (Thermo Scientific) supplemented with 4 mM L-
glutamine and lx Antibiotics-Antimycotic (Thermo Scientific) and grown for 24
h at
37 C in a 5% CO2 environment. Two-fold serially diluted scFv-hFc fusion
proteins
were mixed with 100 TCID50 of MERS-CoV, and the mixture was incubated for 30
min at 37 C. Then, the mixture was added to the Vero cells in tetrad and
incubated for
4 days at 37 C in a 5% CO2 environment. The cytopathic effect (CPE) in each
well
was visualized following crystal violet staining 4 days post-infection. The
IC50 values
were calculated using the dose-response inhibition equation of GraphPad Prism
6
(GraphPad Software, La Jolla, CA, USA).
[257]
[258] 5.4. Flow cytometry
[259] The scFv-hFc fusion proteins (2,000, 1000, 250, or 200 nM) were
incubated either
with 200 nM of the recombinant S glycoprotein fused with a polyhistidine tag
at the C-
terminus (Sino Biological Inc.) or without S protein in 50 [IL of 1% (w/v) BSA
in PBS
containing 0.02% (w/v) sodium azide (FACS buffer) at 37 C for 1 h. The m336
scFv-
hFc and irrelevant scFv-hFc fusion proteins were used as positive and negative
37
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
controls, respectively. Huh-7 cells (hDPP4+)(KCLB (Korean Cell Line Bank)
Accession number KCLB #60104) were added into v-bottom 96-well plates
(Corning,
Corning, NY, USA) at a density of 3 x 105 cells per well, and then, the
mixture was
added to the wells. After incubation at 37 C for 1 h, cells were washed three
times with
FACS buffer and incubated with FITC-labeled rabbit anti-HIS Ab (Abcam,
Cambridge, UK) at 37 C for 1 h. Then, the cells were washed three times with
FACS
buffer, resuspended in 200 [AL of PBS, and subjected to analysis by flow
cytometry
using a FACS Canto II instrument (BD Bioscience, San Jose, CA, USA). For each
sample, 10,000 cells were assessed, and the data were analyzed using the
FlowJo
software (TreeStar, Ashland, OR, USA).
[260]
[261] 5.5. SE-HPLC (size exclusion HPLC)
[262] Non-nebulized and nebulized samples were analyzed using Waters e2695
HPLC
system (Waters Corporation, Milford, MA, USA) equipped with a BioSuite high-
resolution size-exclusion chromatography column (250 A 7.5 mm x 300 mm). Each
sample (10 [tg) was injected at a flow rate of 1 mL/min. The mobile phase was
PBS
(pH 7.4), and UV detection was performed at 280 nm/220 nm. The sample tray and
column holder were maintained at 4 and 30 C, respectively, throughout data ac-
quisition. The molecular weights corresponding to the antibody peaks were
calculated
using the Empower software (Waters Corporation, USA).
[263]
[264] 5.6. DLS (Dynamic light scattering) assay
[265] DLS experiments were performed using a Zetasizer Nano S (Malvern
Panalytical
Ltd, Malvern, UK) and a 633-nm/4-mW laser at a 173 detection angle as
described
previously (Van Heeke G, Allosery K, De Brabandere V, De Smedt T, Detalle L,
de
Fougerolles A. Nanobodies(R) as inhaled biotherapeutics for lung diseases.
Pharmacol
Ther. 2017;169:47-56). Non-nebulized and nebulized samples were analyzed by
performing three acquisitions per sample. PBS (pH 7.4) was used as the
reference
solvent. The results were evaluated with the Zetasizer software 7.02.
[266]
[267] 5.7. PRNT (plaque reduction neutralization test) assay
[268] Vero cells were seeded in 12-well plates (3.5 x 105 cells/well) in
Opti-PRO SFM
supplemented with 4 mM L-glutamine and lx Antibiotics-Antimycotic (Thermo
Scientific) and grown for 24 h at 37 C in a 5% CO2 environment. IgGI
antibodies were
serially diluted three-fold in Dulbecco's PBS (Welgene) and mixed with an
equal
volume of culture media containing MERS-CoV/KOR/KNIH/002 05 2015 (100 pfu).
After incubation for 1 h at 37 C in a 5% CO2 environment, the virus-antibody
mixture
was added to the cells and maintained for 1 h at room temperature. The mixture
was
38
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
then removed, and the cells were overlaid with 1% agarose in DMEM. After in-
cubation for 2 days at 37 C in a 5% CO2 environment, the cells were washed
with PBS
and fixed for 24 h with 4% paraformaldehyde. The agarose overlay was removed,
and
the cell monolayer was gently washed with water to remove residual agarose.
The cells
were stained with 0.5% crystal violet solution, and the plaques were counted
manually.
The number of plaques was plotted as a function of IgGI antibodies, and the
con-
centration of IgGI at which the number of plaques was reduced by 50% compared
to
that in the absence of IgGI (PRNT50) was calculated using GraphPad Prism 6.
[269]
[270] Example 6. Generation of antibodies reactive to recombinant MERS-CoV
RBD
protein from patients
[271] Human single-chain variable fragment (scFv) phage-display libraries
were generated
using peripheral blood mononuclear cells (PBMCs) isolated from two MERS-
CoV-infected convalescent patients. One patient (P014) was considered to be
the super
spreader, and the other patient (P002) was the wife of the index patient. The
complexity of the libraries exceeded 3.6 x 109 and 1.9 x 109 colony-forming
units for
patients P002 and P014, respectively. After the third and fourth rounds of
biopanning
against recombinant MERS-CoV S RBD protein, the scFv clones were retrieved in
a
high-throughput manner as described previously (Noh J, Kim 0, Jung Y, Han H,
Kim
JE, Kim S, et al. High-throughput retrieval of physical DNA for NGS-
identifiable
clones in phage display library. MAbs. 2019;11(3):532-545). Briefly, 1,800
micro-
colonies formed on the TR chip, and of these, 542 clones with unique VH and
Vic/V),
were identified. In these clones, 44 unique hCDR3 sequences were identified.
We
selected 44 clones encoding unique hCDR3 sequences and rescued phages for
phage
enzyme-linked immunosorbent assay (ELISA) analysis. A total of 36 unique scFv
clones were highly reactive to recombinant MERS-CoV S RBD protein (data not
shown). These clones were prepared as scFv fused with human Fc (scFv-hFc)
using a
eukaryotic expression vector and HEK293F cells. A human anti-MERS-CoV neu-
tralizing mAb reported previously, m336
(https://www.nature.com/articlesincomms9223?origin=ppub), was also prepared in
this
same form for use as a positive control.
[272] Amino acid sequences of prepared antibodies (scFv or scFv-Fc) are
illustrated in
Tables 4 to 17:
39
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[273] [Table 41
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
C-8 VL-CDR1 QSLLHSNGYNY 1
(N-VL-V VL CDR2 LGS 8
H-C)
VL CDR3 MQALQTPLT 15
VH CDR1 GGTFSSYA 24
VH CDR2 IIPFFGTA 31
VH CDR3 ARDGRKDYYGSGSYLHYYGMDV 38
VL
ELVMTQSPLSLPVTPGEPASISCRSSQSLLHS 44
NGYNYLDWYLQRPGQSPQLLIYLGSNRASG
VPDRFSGSGSGTDFTLKIGRVEAEDVGIYYC
MQALQTPLTFGGGTKVEIK
VH
EVQLVQSGAEVKKPGSSVKVSCKASGGTFS 55
SYAISWVRQAPGQGLEWMGGIIPFFGTANY
AQKFQGRVTITADESTSTAYMELSSLRSEDT
AVYYCARDGRKDYYGSGSYLHYYGMDV
WGQGTTVTVSS
40
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[274] [Table 51
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
C-8-2-4B VL-CDR1 QSLLHSNGYNY 1
(N-VL-li VL CDR2 LGS 8
nker-VH
VL CDR3 MQALQTPLT 15
-hinge-F
c-C)( see VH¨CDR1 DGKEKREA 25
Example VH CDR2 IIPFFDKA 32
8) VH CDR3 ARDGRKDYYGSGSYLHYYGMDV 38
Linker of GGSSRSSSSGGGGSGGGG 64
VL-VH
hinge EPKSSDKTHTSPPCP 65
Fc
APELLGGPSVFLFPPKPKDTLMISRTPEVTCV 66
VVDVSHEDPEVKFNWYVDGVEVHNAKTKP
REEQYNSTYRVVSVLTVLHQDWLNGKEYK
CKVSNKALPAPIEKTISKAKGQPREPQVYTLP
PSRDELTKNQVSLTCLVKGFYPSDIAVEWES
NGQPENNYKTTPPVLDSDGSFFLYSKLTVDK
SRWQQGNVFSCSVMHEALHNHYTQKSLSLS
PGK
VH
EVQLVQSGAEVKKPGSSVKVSCKASDGKEK 56
REAISWVRQAPGQGLEWMGGIIPFFDKANY
AQKFQGRVTITADESTSTAYMELSSLRSEDT
AVYYCARDGRKDYYGSGSYLHYYGMDV
WGQGTTVTVSS
VL
ELVMTQSPLSLPVTPGEPASISCRSSQSLLHS 44
NGYNYLDWYLQRPGQSPQLLIYLGSNRASG
VPDRFSGSGSGTDFTLKIGRVEAEDVGIYYC
MQALQTPLTFGGGTKVEIK
41
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[275] [Table 61
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
C-8-2-4B VL-CDR1 QSLVHSNGYNY 2
-10D VL CDR2 LGS 8
(N-VL-li
VL CDR3 MQAVQTPLT 16
nker-VH-
hinge-Fc- VH¨CDR1 DGKEKREA 25
C) (see VH CDR2 IIPFFDKA 32
Example VH CDR3 DGRKDYYGSGSYLHYYGMDV 38
8)
Linker of VL- GGSSRSSSSGGGGSGGGG 64
VH
hinge EPKSSDKTHTSPPCP 65
Fc
APELLGGPSVFLFPPKPKDTLMISRTPEVTCV 66
VVDVSHEDPEVKFNWYVDGVEVHNAKTKP
REEQYNSTYRVVSVLTVLHQDWLNGKEYK
CKVSNKALPAPIEKTISKAKGQPREPQVYTL
PPSRDELTKNQVSLTCLVKGFYPSDIAVEWE
SNGQPENNYKTTPPVLDSDGSFFLYSKLTVD
KSRWQQGNVFSCSVMHEALHNHYTQKSLS
LSPGK
VH
EVQLVQSGAEVKKPGSSVKVSCKASDGKE 56
KREAISWVRQAPGQGLEWMGGIIPFFDKA
NYAQKFQGRVTITADESTSTAYMELSSLRSE
DTAVYYCARDGRKDYYGSGSYLHYYGMD
VWGQGTTVTVSS
VL
ELVMTQSPLSLPVTPGEPASISCRSSQSLVHS 45
NGYNYLDWYLQRPGQSPQLLIYLGSNRASG
VPDRFSGSGSGTDFTLKIGRVEAEDVGIYYC
MQAVQTPLTFGGGTKVEIK
42
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[276] [Table 71
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
C-8-2-2E VL-CDR1 QSLLHSNGYNY 1
(N-VL-V VL CDR2 LGS 8
H-C)
VL CDR3 MQALQTPLT 15
VH CDR1 EGNESKEA 26
VH CDR2 IIPFFDKA 32
VH CDR3 ARDGRKDYYGSGSYLHYYGMDV 38
VL
ELVMTQSPLSLPVTPGEPASISCRSSQSLLHS 44
NGYNYLDWYLQRPGQSPQLLIYLGSNRASG
VPDRFSGSGSGTDFTLKIGRVEAEDVGIYYC
MQALQTPLTFGGGTKVEIK
VH
EVQLVQSGAEVKKPGSSVKVSCKASEGNES 57
KEAISWVRQAPGQGLEWMGGIIPFFDKAN
YAQKFQGRVTITADESTSTAYMELSSLRSED
TAVYYCARDGRKDYYGSGSYLHYYGMDV
WGQGTTVTVSS
43
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[277] [Table 81
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
C-8-2-2E VL-CDR1 QSLLHSNGYNY 1
-5G VL CDR2 EGS 9
(N-VL-V
VL CDR3 MQALQTPLT 15
H-C)
VH CDR1 EGNESKEA 26
VH CDR2 IIPFFDKA 32
VH CDR3 ARDGRKDYYGSGSYLHYYGMDV 38
VL
ELVMTQSPLSLPVTPGEPASISCRSSQSLLHS 46
NGYNYLDWYLQRPGQSPQLLIYEGSNRASG
VPDRFSGSGSGTDFTLKIGRVEAEDVGIYYC
MQALQTPLTFGGGTKVEIK
VH
EVQLVQSGAEVKKPGSSVKVSCKASEGNES 57
KEAISWVRQAPGQGLEWMGGIIPFFDKAN
YAQKFQGRVTITADESTSTAYMELSSLRSED
TAVYYCARDGRKDYYGSGSYLHYYGMDV
WGQGTTVTVSS
44
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[278] [Table 9]
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
VL-CDR1 SSNIGSNY 3
(N-VL-V VL CDR2 SNN 10
H-C)
VL CDR3 ATWDDNLSGPV 17
VH CDR1 GGSISSSSYY 27
VH CDR2 IFYIGNT 33
VH CDR3 ARQEGSSIIRFDP 39
VL
ELELTQPPSVSGTPGQRVTISCSGSSSNIGSN 47
YVYWYQQLPGTAPKLLIYSNNQRPSGVPDR
FSGSKSGTSASLAISGLRSEDEADYYCATWD
DNLSGPVFGGGTKVTVLG
VH
QVQLQESGPGLVKPSETLSLTCTVSGGSISSS 58
SYYWGWIRQPPGKGLEWIGSIFYIGNTYYN
PSLKSRVTISVDTSKNQFSLRLSSVTAADTA
VYYCARQEGSSIIRFDPWGQGTLVTVSS
45
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[279] [Table 101
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
15 VL-CDR1 QSVLYSSNNKNY 4
(N-VL-V VL CDR2 WAS 11
H-C)
VL CDR3 QQYYGSPYT 18
VH CDR1 GGTFSSYG 28
VH CDR2 IIPILGIA 34
VH CDR3 ASLFDSSGYYPYYFDY 40
VL
ELVMTQSPDSLAVSLGERATINCKSSQSVLY 48
SSNNKNYLAWYQQKPGQPPKLLIYWASTRE
SGVPDRFSGSGSGTDFTLTISSVQTEDVAVY
YCQQYYGSPYTFGQGTKLEIK
VH
QVQLVQSGAEVKKPGSSVKVSCKASGGTFS 59
SYGISWVRQAPGQGLEWMGRIIPILGIANY
AQKFQGRVTITADKSTSTAYMELSSLRSEDT
AVYYCASLFDSSGYYPYYFDYWGQGTLVT
VSS
46
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[280] [Table 111
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
20 VL-CDR1 QSVLYSSNNKNY 4
(N-VL-V VL CDR2 WAS 11
H-C)
VL CDR3 QQYYSTPPT 19
VH CDR1 GGTFSSFT 29
VH CDR2 IIPIFGIA 35
VH CDR3 ATHFGASGYDPYYFDY 41
VL
ELQMTQSPDSLAVSLGERATINCKSSQSVLY 49
SSNNKNYLAWYQQKPGQPPKLLISWASTRE
SGVPERFSGSGSGTDFTLTISGLQAEDVAVY
YCQQYYSTPPTFGQGTKVDIK
VH
EVQLVQSGAEVKKPGSSVKVSCKASGGTFS 60
SFTISWVRQAPGQGLEWMGRIIPIFGIANYA
QKFQGRVTITADKSTGTAYMELSSLRSEDTA
VYYCATHFGASGYDPYYFDYWGQGTLVT
VSS
47
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[281] [Table 121
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
42 VL-CDR1 SSNIGAGYD 5
(N-VL-V VL CDR2 GNS 12
H-C)
VL CDR3 AAWDDSLSGPV 20
VH CDR1 GGSISSSSYY 27
VH CDR2 IFYIGNT 33
VH CDR3 ARQEGSSIIRFDP 39
VL
ELELTQPPSVSGAPGQRVTISCTGSSSNIGAG 50
YDVHWYQQLPGTAPKLLIYGNSNRPSGVPD
RFSGSKSGTSASLAISGLQSEDEGDYYCAA
WDDSLSGPVFGGGTELTVLG
VH
QVQLQESGPGLVKPSETLSLTCTVSGGSISSS 61
SYYWGWIRQPPGKGLEWIGSIFYIGNTYYN
PSLKSRVTISVDTSKNQFSLKLSSVTAADTA
VYYCARQEGSSIIRFDPWGQGTLVTVSS
48
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[282] [Table 131
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
46 VL-CDR1 SSNIGSNY 3
(N-VL-V VL CDR2 SNN 10
H-C)
VL CDR3 AAWDDSLSGVV 21
VH CDR1 GGSISSSSYY 27
VH CDR2 IYYTGNT 36
VH CDR3 ARQVADLGYFDY 42
VL
ELVVTQPPSASGTPGQRVAISCSGSSSNIGSN 51
YVYWYQQLPGTAPKLLIYSNNHRPSGVPDR
FSGSKSGTSASLAISGLRSEDEAVYYCAAW
DDSLSGVVFGGGTELTVLG
VH
QVQLQESGPGLVKPSETLSLTCTVSGGSISSS 62
SYYWGWIRQPPGKGLEWIGSIYYTGNTYYN
PSLKSRLTISVDTSKNQFSLKLSSVTAADTA
VYYCARQVADLGYFDYWGQGTLVTVSS
49
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[283] [Table 141
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
47 VL-CDR1 SSSVGNNY 6
(N-VL-V VL CDR2 SNS 13
H-C)
VL CDR3 AAWDDSLSGPV 20
VH CDR1 GGSISSSSYY 27
VH CDR2 IYYTGNT 36
VH CDR3 ARQVADLGYFDY 42
VL
ELMLTQPHSASGTPGQRVAISCSGRSSSVGN 52
NYVYWYQQLPGAAPKLLIYSNSQRPSGVPD
RFSGSKSGTSASLVISGLRSEDEADYYCAAW
DDSLSGPVFGGGTQLTVLG
VH
QVQLQESGPGLVKPSETLSLTCTVSGGSISSS 62
SYYWGWIRQPPGKGLEWIGSIYYTGNTYYN
PSLKSRLTISVDTSKNQFSLKLSSVTAADTA
VYYCARQVADLGYFDYWGQGTLVTVSS
50
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[284] [Table 151
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
48 VL-CDR1 SSNIGSNY 3
(N-VL-V VL CDR2 SNN 10
H-C)
VL CDR3 AAWDDSLNGPV 22
VH CDR1 GGSISSSSYY 27
VH CDR2 IYYTGNT 36
VH CDR3 ARQVADLGYFDY 42
VL
ELELTQPPSVSGTPGQRVTISCSGSSSNIGSN 53
YVYWYQQLPGTAPKLLIYSNNQRPSGVPDR
FSGSKSGTSASLAINGLQSEDEADYYCAAW
DDSLNGPVFGGGTKLTVLG
VH
QVQLQESGPGLVKPSETLSLTCTVSGGSISSS 62
SYYWGWIRQPPGKGLEWIGSIYYTGNTYYN
PSLKSRLTISVDTSKNQFSLKLSSVTAADTA
VYYCARQVADLGYFDYWGQGTLVTVSS
51
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[285] [Table 161
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
34 VL-CDR1 KLGEKY 7
(N-VL-V VL CDR2 QDS 14
H-C)
VL CDR3 QAWDSRRAV 23
VH CDR1 GFTFSSYS 30
VH CDR2 ISTTGSYI 37
VH CDR3 AKGTAFDGGLAFDI 43
VL
QAAELVLTQSPSVSVSPGQTATITCSGDKLG 54
EKYASWYQQRPGQSPVLVIYQDSRRASGIP
ERFS GSNS GNTATLTIS GT QAMDEADYYCQ
AWDSRRAVFGGGTELTVLG
VH
EVQLLESGGGLVKPGGSLRLSCAASGFTFSS 63
YSMNWVRQAPGKGLEWVSSISTTGSYIFYA
DSVKGRFTISRDNAKNSLYLQMNTLRPEDT
ALYYCAKGTAFDGGLAFDIWGQGTIVTVS
S
52
CA 03152245 2022-02-23
WO 2021/060837
PCT/KR2020/012887
[286] [Table 171
Clone/ Part Amino Acid Sequence (N¨>C) SEQ ID
name NO
119 VL-CDR1 KLGEKY 7
(N-VL-V VL CDR2 QDT 71
H-C)
VL CDR3 QAWDNNFYV 72
VH CDR1 GFSIDDYA 73
VH CDR2 ISWDSGSI 74
VH CDR3 AREKQLVPYYYYGMDV 75
VL
ELVLTQPPSVSVSPGQAASITCSADKLGEKY 76
VFWYQQKPGQSPVLAIYQDTKRPSGIPERFS
GSNSGNTATLTISGTQPMDEADYYCQAWD
NNFYVFGTGTKLTVLG
VH
EVQLVQSGGGLVRPGRSLRLSCVAPGFSIDD 77
YAMHWVRQTPGKGLEWVSGISWDSGSIAY
ADS VKGRFTISRDNAKNSLYLQMNSLRAED
TAVYYCAREKQLVPYYYYGMDVWGQGTT
VTVSS
[287]
[288] Example 7. Selection of MERS-CoV neutralizing antibodies
[289] We performed a microneutralization assay (Example 5.3) to test the
neutralizing
activity of the 36 identified scFv clones against MERS-CoV
(MERS-CoV/KOR/KNIH/002 05 2015). The obtained results are shown in Table 18
(CPE inhibition by scFv clones)
53
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[290] [Table 181
Clone IC50 (n/mL)
C-8 6.45
10 9.61
15 2.78
20 3.22
34 4.43
119 9.61
42 4.67
46 4.67
47 3.03
48 2.40
m336 3.71
[291] as shown in Table 18, among the clones, scFV clones
10, 15, 20, C-8, 34, 42, 46, 47,
and 48 potently inhibited MERS-CoV replication, with half-maximal inhibitory
con-
centration (IC50) values ranging from 2.40 to 9.61 [tg/mL.
[292] Next, the stability of these clones during nebulization was tested.
The inventors
nebulized the fusion proteins, each of which comprises the scFv clone and hFc
(SEQ
ID NO: 66) (N-VL-(linker)-VH-hinge-Fc-C; hereinafter, "scFv-hFc"), at a con-
centration of 100 [tg/mL (each scFv-hFc) in PBS using a vibrating mesh
nebulizer and
then collected the aerosol. All the collected samples showed clearly visible
ag-
gregation. After centrifugation to remove the aggregated material, the
supernatant
(post-nebulized scFv-hFc fusion proteins) and pre-nebulized scFv-hFc fusion
proteins
were subjected to ELISA using recombinant S glycoprotein-coated microtiter
plates
(Example 5.1). The amount of bound scFv-hFc fusion protein was determined
using
HRP-conjugated anti-human IgG antibody and ABTS(2,2'-Azinobis
[3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt) by measuring
absorbance
at 405 nm, the results obtained from post- and pre-nebulized scFv-hFc fusion
proteins
were compared.
[293] The obtained results are shown in Figures la and lb. Figures la and
lb shows re-
activity of scFv clones before and after nebulization. As shown in Figures la
and lb,
all nine scFv-hFc fusion proteins showed significantly reduced reactivity
against re-
combinant S glycoprotein after nebulization.
[294]
54
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[295] The clones C-8 and 48 were selected, as these antibodies exhibited
the lowest IC50
values among the antibodies derived from patients P002 and P014, respectively.
Before performing further studies, the mechanism underlying inhibition of
viral
infection on cells was examined. The antibodies were mixed and incubated with
re-
combinant S glycoprotein fused with a polyhistidine tag at the C-terminus.
After in-
cubation with hDPP4-expres sing Huh-7 cells (hDPP4+), the relative amount of
bound
recombinant S glycoprotein was measured by flow cytometry analysis (Example
5.4)
using FITC-conjugated anti-HIS antibody. Per each sample, 10,000 cells were
monitored, and the data were analyzed using FlowJo software.
[296] The obtained results are shown in Figure 2 (C-8 (A), 48 (B), m336
(C), or negative
control (D) scFv-hFc). As shown in Figure 2, both C-8 and 48 scFv-hFc nearly
completely blocked binding of recombinant S glycoprotein to cells at equimolar
con-
centration of 100 nM, indicating that the antibodies block the initial
interaction of the
virus with cells.
[297]
[298] Example 8. Modification of CDR residues to enhance antibody stability
[299] To enhance the stability of the C-8 and 48 clones, the inventors
sought to introduce
mutations in CDRs, except for heavy chain CDR3 (HCDR3), for replacement of hy-
drophobic residues with hydrophilic residues. We defined CDRs according to the
Inter-
national Immunogenetics Information System (IMGT) and targeted Phe, Ile, Leu,
Val,
Met, Trp, and Tyr which were defined as hydrophobic amino acids. For the C-8
clone,
the F29, Y32, 151, 152, F53, and F54 hydrophobic residues in HCDR1 and HCDR2
were selected for randomization (Figure 3A). These six residues were designed
to
encode the wild-type amino acid, Asp, Glu, or redundant amino acids depending
on the
degenerate codon in the first scFv phage-display library. The inventors
preferred
negatively charged amino acids to positively charged amino acids as lowering
the iso-
electric point of an antibody may reduce the non-specific in vivo clearance.
The
randomized scFv phage-display library had a complexity of 2.6 x 109 colony-
forming
units, which exceeded the theoretical complexity of 1.3 x 105 on the
nucleotide level.
After two rounds of biopanning on recombinant MERS-CoV S RBD protein, the
inventors randomly rescued phage clones and performed phage ELISA. Eleven scFv
clones showed reactivity to recombinant MERS-CoV S RBD protein similar to or
higher than that of the original C-8 clone. The C-8-2 clone harbored F29E and
Y32E
replacements, while the other 10 clones had only one residue replaced with
either Asp,
Glu, or redundant amino acids, depending on the degenerate codon.
[300] Figure 3 shows sequential randomization of CDR residues of the C-8
clone. As
shown in Figure 3 (A), in the first randomized library, six hydrophobic amino
acid
residues (asterisks) in HCDR1 and HCDR2 were targeted. The second library was
55
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
prepared in the C-8-2 clone by randomizing nine amino acid residues
(asterisks) that
were not randomized in the first randomized library. As shown in Figure 3 (B),
eight
amino acid residues (asterisks) in LCDRs of the C-8-2-4B clone selected from
the
second library were randomized in the third randomized library.
[301]
[302] Biophysical characterization of C-8, C-8-2, and C-8-2-4B clones were
conducted.
Following nebulization at a concentration of 100 [tg/mL or 300 [tg/mL for scFv-
hFc or
IgG1 full length antibody (with kappa light chain) (Example 4), respectively,
aerosol
was collected and subjected to ELISA (Figure 4A-C) and flow cytometry (Figure
4D).
C-8-2 scFv-hFc (Figure 4A), C-8-2-4B scFv-hFc (Figure 4B), and C-8-2-4B IgG1
(Figure 4C) were serially diluted and incubated with recombinant S
glycoprotein-
coated microtiter plates. In Figure 4D, C-8 IgG1 and C-8-2-4B IgG1 were
incubated
with recombinant S glycoprotein fused with a polyhistidine tag at the C-
terminus, and
the complex was allowed to react with hDPP4-expressing cells. The amount of
bound
recombinant S glycoprotein was measured using FITC-conjugated anti-HIS
antibody
(absorbance at 405 nm). Data are representative of 10,000 cells for each
sample.
[303]
[304] More detailed description on the above biophysical characterization
is provided as
follows.
[305]
[306] Biophysical characterization of scFv-hFc fusion proteins
[307] To test the stability of the C-8-2 clone during nebulization, a scFv-
hFc fusion protein
was prepared and subjected to ELISA following nebulization as above. As shown
in
Figure 4A, the reactivity of C-8-2 scFv-hFc to recombinant S glycoprotein was
much
less affected by nebulization than that of C-8 scFv-hFc; however, the
reactivity of the
C-8-2 clone was somewhat reduced compared with that of the C-8 clone.
[308] To achieve further stabilization and affinity maturation, a second
scFv phage-display
library was generated using the same strategy to randomize nine residues in
HCDR1
and HCDR2 of the C-8-2 clone to introduce more negatively charged residues
(Figure
3A). The proline at H52A was excluded from the randomization, as proline
frequently
forms a unique structure essential for antibody reactivity. The second
randomized scFv
phage-display library had a complexity of 1.0 x 109 colony-forming units,
which
exceeded the theoretical complexity of 4.2 x 106 on the nucleotide level.
After the
second round of biopanning on recombinant MERS-CoV S RBD protein, we selected
12 clones that displayed greater reactivity to recombinant MERS-CoV S RBD
protein
than the C-8-2 clone in phage ELISA analysis. Clone C-8-2-4B contained
replacement
at six residues (G26D, T28K, S30K, 531R, G55D, and T56K; Figure 3A) and showed
the highest intrinsic solubility score among the 12 tested clones.
Interestingly, only two
56
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
residues were replaced with Asp, and four residues were replaced with
positively
charged amino acids, as allowed by the degenerate codons (Figure 3A). Then, C-
8-2-4B scFv-hFc fusion protein was prepared using a eukaryotic expression
system.
After nebulization, the reactivity of C-8-2-4B scFv-hFc to recombinant S
glycoprotein
was less affected than either C-8 or C-8-2 scFv-hFc (Figure 4A, B). In
addition, the re-
activity of C-8-2-4B scFv-hFc was enhanced compared to that of C-8-2 scFv-hFc
and
comparable to that of C-8 scFv-hFc.
[309]
[310] Biophysical characterization of IgG1 antibodies
[311] ELISA
[312] Next, C-8 and C-8-2-4B IgGI were prepared using a eukaryotic
expression system
and compared the reactivity of these immunoglobulins to recombinant S
glycoprotein
before and after nebulization. As expected, the reactivity of C-8-2-4B IgGI
was better
retained following nebulization than that of C-8 IgGI (Figure 4C). It was
tested
whether C-8-2-4B IgGI effectively blocked the interaction between recombinant
S gly-
coprotein and hDPP4-expressing Huh-7 cells after nebulization. In flow
cytometry
analysis, we found that C-8-2-4B IgGI almost completely blocked the binding of
re-
combinant S glycoprotein to hDPP4-expressing cells following nebulization,
while C-8
IgGI failed to block this interaction after nebulization (Figure 4D).
[313] As C-8-2-4B IgGI showed a somewhat reduced reactivity after
nebulization, we
sought to confer additional stability by randomizing eight hydrophobic
residues in
LCDRs using the same randomization scheme. The inventors achieved 2.0 x 109
colony-forming units in the third randomized scFv phage-display library,
exceeding
the theoretical complexity of 2.1 x 106 (Figure 3B). After two rounds of
biopanning on
recombinant MERS-CoV S RBD protein, clones in a phage ELISA with reactivity
similar to or greater than that of C-8-2-4B were selected. Sanger sequencing
revealed
that a single clone was repetitively selected. The selected clone, C-8-2-4B-
10D,
harbored replacements at L27C and L92V with valine (Figure 3B). C-8-2-4B-10D
IgG
1 was prepared using a eukaryotic expression system and analyzed the
characteristics
using ELISA, size-exclusion high-performance liquid chromatography (SE-HPLC),
dynamic light scattering (DLS), and plaque reduction neutralization tests
(PRNT50).
[314] Following nebulization at a concentration of 1 mg/mL of each of C-8
IgGI, C-
8-2-4B-10D IgGI and m336, aerosol was collected and subjected to ELISA. Re-
combinant S glycoprotein-coated microtiter plates were incubated with pre-
nebulized
and post-nebulized C-8 IgGi (Figure 5A), C-8-2-4B-10D IgGi (Figure 5B), and
m336
(Figure 5C). HRP-conjugated anti-human IgG antibody was used as the probe, and
ABTS was used as the substrate. All experiments were performed in duplicate,
and the
data indicate mean SD.
57
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[315] The obtained results of ELISA as above are shown in Figure 5. As
shown in Figure
5, a noticeable decline in reactivity to recombinant S glycoprotein by C-8
IgGI and
m336 IgGI after nebulization was observed; yet, the change in reactivity of C-
8-2-4B-10D IgGI after nebulization was negligible.
[316]
[317] SE-HPLC & DLS analysis (Stability)
[318] To examine the stability of test antibodies, SE-HPLC and DLS analysis
for C-8 IgGI,
C-8-2-4B-10D IgGi, and m336 IgGi were conducted referring to Examples 5.5 and
5.6, respectively.
[319] More specifically, to evaluate the size distribution profile of pre-
nebulized C-8 IgGI
(Figure 6A), pre-nebulized C-8-2-4B-10D IgGi (Figure 6B), pre-nebulized m336
IgGi
(Figure 6C), post-nebulized C-8 IgGI (Figure 6D), post-nebulized C-8-2-4B-10D
IgGI
(Figure 6E), and post-nebulized m336 IgGI (Figure 6F) antibodies, DLS was
performed using 633-nm/4-mW laser at a 173 detection angle. PBS was used as
the
reference solvent, and the results were evaluated with Zetasizer software
7.02. All ex-
periments were performed in triplicate, and representative results are shown
for each
antibody.
[320] The obtained results are shown in Table 19 and Figure 6.
[321] [Table 191
SE-HPLC (% monomer / % DLS (% monomer SD / % aggregates
aggregates) SD)
Antibody Pre-nebulizat Post-nebulizat Pre-nebulization Post-nebulization
ion ion
C-8 100.0 / 0 97.9 / 2.1 100.0 0 /0 78.4 3.5 / 21.6
3.5
C-8-2-4B- 100.0 / 0 100.0 / 0 99.2 0.7 / 0.8 98.6 0.4 / 1.4
0.4
10D 0.7
m336 100.0 / 0 99.4 / 0.6 96.6 0.6 / 3.4 77.5 2.3 /
22.5 2.3
0.6
[322] As shown in Table 19, in SE-HPLC analysis, high-molecular weight
aggregates were
detected in post-nebulization samples of C-8 and m336 IgGi; however, no
aggregate
was found in post-nebulized samples of C-8-2-4B-10D IgGi as shown om Table 17
above. As shown in Table 18 and Figure 6,in accordance with these SE-HPLC
data,
DLS analysis showed that the nebulization process converted 21.6% and 22.5% of
C-8
and m336 IgGi, respectively, into high-molecular-weight aggregates, while nebu-
lization resulted in <1% aggregates for C-8-2-4B-10D IgGi.
[323]
58
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
[324] Example 9. Neutralizing potency after nebulization
[325] The neutralizing activities of pre- and post-nebulized C-8 IgGI and C-
8-2-4B-10D
IgGI were evaluated in PRNT50 using the live MERS-CoV referring to Example
5.7.
Antibodies were mixed with live MERS-CoV, and then the antibody-virus mixture
was
allowed to infect Vero cells.
[326] More specifically, culture media containing 100 PFU live MERS-CoV
(MERS-CoV/KOR/KNIH/002 05 2015) was mixed with equal volume of serially
diluted C-8 IgGI (Figure 7a), C-8-2-4B-10D IgGI (Figure 7b), and palivizumab
(Figure
7c). After incubation for 1 h, the mixture was added to Vero cells (ATCC CCL-
81).
After 2 days, the plaques were counted. The inhibition of virus infection was
plotted as
a function of IgGI antibody concentration, and PRNT50 values were calculated
by
GraphPad Prism 6. All experiments were performed in quadruplicate, and the
data
indicate mean SD.
[327] The obtained results (Relative infection (%)) are shown in Figures 7a-
7c. As shown
in Figures 7a-7c, C-8 IgGI and C-8-2-4B-10D IgGI exhibited effective
inhibitory
activity against MERS-CoV, with IC50 values of 0.29 and 0.28 [tg/mL,
respectively.
After nebulization, C-8-2-4B-10D showed an IC50 value similar to that of pre-
nebulized IgGI, but the IC50 value of C-8 IgGI was dramatically increased
following
nebulization.
[328]
[329] Example 10. Test of binding ability of CDR-modified neutralizing
antibody to
MERS-CoV mutants
[330] In this example, it was tested whether the CDR-modified neutralizing
antibodies of
Example 8 can retain their ability to bind to MERS-CoV mutants. As several
poly-
morphisms within S protein (D5 10G and I529T) of MERS-CoV were identified
during
the MERS outbreak in South Korea, the inventors examined binding activity of C-
8
and C-8-2-4B-10D antibodies against recombinant wild-type or mutant MERS-CoV
RBD (including mutation of D5 10G, I529T, or D5 10G & I529T) protein using
ELISA
analysis referring to Example 5.1.
[331] More specifically, recombinant wild-type or mutant MERS-CoV RBD
protein-coated
microtiter plates were incubated with varying concentration of C-8 IgGl, C-
8-2-4B-10D IgGl, or irrelevant IgGl(negative control). HRP-conjugated anti-
human
IgG antibody was used as the probe, and ABTS was used as the substrate. All ex-
periments were performed in duplicate, and the data indicate mean SD.
[332] The obtained results are shown in Figure 8. As shown in Figure 8,
antibody clone C-
8 as well as CDR-modified C-8-2-4B-10D successfully bound to recombinant
mutant
RBD proteins (D5 10G, I529T, or D5 10G & I529T) in a dose-dependent manner
without any noticeable decline in reactivity compared to that of recombinant
wild-type
59
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
RBD protein.
[333] Furthermore, it was tested whether the antibody can inhibit the
interaction between
RBD mutants and DPP4.
[334] Flow cytometry analysis of the inhibition of recombinant mutant MERS-
CoV RBD
protein binding to hDPP4-expressing cells were performed referring to Example
5.4.
Antibodies C-8 IgGI, C-8-2-4B-10D IgGI, m336 IgGI, or negative control IgGI
(each 1
or 4 [LM) were mixed and incubated with recombinant wild type and mutant (D5
10G,
I529T, or D510G/I529T) MERS-CoV RBD protein (each 1 [LM) fused with a poly-
histidine tag at the C-terminus. After incubation with Huh-7 (hDPP4+) cells,
the
relative amount of bound recombinant mutant MERS-CoV RBD protein was measured
using FITC-conjugated anti-HIS antibody. Per each sample, 10,000 cells were
monitored, and the data were analyzed using FlowJo software.
[335] The obtained results are shown in Figures 9a and 9b. As shown in
Figures 9a and 9b,
both C-8 IgGI and C-8-2-4B-10D IgGI nearly completely blocked binding of re-
combinant mutant MERS-CoV RBD protein (D5 10G and I529T) to cells at equimolar
concentration of 1000 nM (Figure 2), indicating that the antibodies block the
initial in-
teraction of the virus with cells regardless of mutations (D5 10G or I529T) in
the RBD.
In case of double mutant carrying both D5 10G and I529T, this mutant did not
bind to
hDPP4-expressing Huh-7 cells which prevent us from conducting further studies.
[336]
[337] Example 11. Conformational epitope mapping
[338] Conformational epitope mapping was performed by hydrogen/deuterium
exchange
mass spectrometry. To elucidate the site where C-8 binds, hydrogen/deuterium
exchange mass spectrometry (HDX-MS) was performed and the kinetics of
hydrogen/
deuterium exchange at protein backbone amides was evaluated. More
specifically, the
proteins were deglycosylated by PNGase-F and diluted to 40 pmole in a buffer
composed of 10mM potassium phosphate (pH 7.0). Hydrogen/deuterium exchange was
performed by mixing 2.5[cL of proteins with 37.5 [LL of D 20 buffer (10 mM
potassium phosphate pD 7.0) and incubating for 0, 0.33 min (20 sec), 10 min,
60 min
and 240 min on ice. The incubation of samples was ended by adding 40 [LL of
ice-cold
quench buffer (1 M TCEP, 2 M Urea, pH 2.66). The quenched samples were quickly
thawed and digested by treating pepsin enzyme for 5 min on ice. Rapidly,
pepsin
treated samples were injected into nanoACQUITY with HDX Technology (Waters,
Milford, MA, USA) and online-digested by passing through an Enzymate pepsin
column (2.1 mm x 30 mm, 300A, 5 [cm, Waters). Peptide fragments were
subsequently
trapped and desalted in VanGuard BEH C18 trap column (2.1 mm x 5 mm, 1.7 [cm,
Waters) at a flow rate of 100 [cUmin 5% ACN for 3 min and then separated by
C18
analytical column (1.0 x 100 mm, 1.7 [cm, Waters, Milford, MA, USA) at a flow
rate
60
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
of 40 [tt/min with gradient for 13 min (started with 5% B for 1 min and
increased to
40% B for 7 min). The mobile phase A was 0.1% formic acid in H 2 0, and B was
ACN containing 0.1 % formic acid. To minimize the back-exchange of deuterium
to
hydrogen, the sample, solvents, trap and analytical columns were all
maintained at pH
of 2.66 and 0 C during analysis. Mass spectrometric analyses was performed
with a
SYNAPT G2-Si (Waters, Milford, MA, USA) equipped with Ion Mobility Separation
(IMS, Waters, Milford, MA, USA) and standard ESI source. The mass spectra were
acquired in the range of m/z 50-2000 in the positive ion mode for 10.5 min.
[339] The obtained HDX profiles of free- and C-8 IgGl-bound MERS-CoV RBD
were
shown in Figures 10a-10c, wherein Figure 10b shows a graph corresponding to
(A) of
Figure 10a and Figure 10c shows graphs corresponding to (B) of Figure 10a. The
deuterium uptake graphs (Figures 10b and 10c) for peptides show the relative
deuterium incorporation as a function of exposure time. The peptides that had
reduction in deuterium incorporation in RBD and C-8 IgG1 complex are
highlighted in
(A) (5er498-Ala520) and (B) (Tyr540-Leu554) and on the X-ray crystal structure
of
MERS-CoV RBD (PDB: 4KQZ) (Figure 10a).
[340] Then, mapping of the C-8 epitope and DPP4 binding site on MERS-CoV
RBD
sequence was performed. By monitoring the exchange of backbone amide hydrogen
with deuterium within MERS-CoV RBD, the inventors observed reduction in
deuterium incorporation in the RBD-C-8 IgGI complex in residues between
5er498-Ala520 (A in Figure 10a) and Tyr540-Leu554 ((B) in Figure 10a),
indicating
that these residues are involved in C-8 binding. The RBD residues interacting
with C-8
and DPP4 are marked in Figure 11 with orange and blue dots, respectively. In
contrast,
other regions shared almost identical deuterium levels in free and bound RBD,
suggesting that these regions do not engage in protein-protein interactions.
Then, we
compared the C-8 epitope with the DPP4 binding site that was reported as shown
in
Figure 11. Remarkably, the C-8 epitope overlapped extensively with the DPP4
binding
site indicating that the C-8 block the initial interaction of the virus with
natural
receptor DPP4 (Figure 11).
[341]
[342] All references, including publications, patent applications, and
patents, cited herein
are hereby incorporated by reference to the same extent as if each reference
were indi-
vidually and specifically indicated to be incorporated by reference and were
set forth in
its entirety herein.
[343] The use of the terms "a" and "an" and "the" and "at least one" and
"one or more" and
similar referents in the context of describing the invention (especially in
the context of
the following claims) are to be construed to cover both the singular and the
plural,
unless otherwise indicated herein or clearly contradicted by context. The use
of the
61
CA 03152245 2022-02-23
WO 2021/060837 PCT/KR2020/012887
term "at least one" (or "one or more") followed by a list of one or more items
(for
example, "at least one of A and B") is to be construed to mean one item
selected from
the listed items (A or B) or any combination of two or more of the listed
items (A and
B), unless otherwise indicated herein or clearly contradicted by context. The
terms
"comprising," "having," "including," and "containing" are to be construed as
open-
ended terms (i.e., meaning "comprising, but not limited to,") unless otherwise
noted.
Recitation of ranges of values herein are merely intended to serve as a
shorthand
method of referring individually to each separate value falling within the
range, unless
otherwise indicated herein, and each separate value is incorporated into the
speci-
fication as if it were individually recited herein. All methods described
herein can be
performed in any suitable order unless otherwise indicated herein or otherwise
clearly
contradicted by context. The use of any and all examples, or exemplary
language (e.g.,
"such as") provided herein, is intended merely to better illuminate the
invention and
does not pose a limitation on the scope of the invention unless otherwise
claimed. No
language in the specification should be construed as indicating any non-
claimed
element as essential to the practice of the invention.
[344] Preferred embodiments of this invention are described herein,
including the best
mode known to the inventors for carrying out the invention. Variations of
those
preferred embodiments may become apparent to those of ordinary skill in the
art upon
reading the foregoing description. The inventors expect skilled artisans to
employ such
variations as appropriate, and the inventors intend for the invention to be
practiced
otherwise than as specifically described herein. Accordingly, this invention
includes all
modifications and equivalents of the subject matter recited in the claims
appended
hereto as permitted by applicable law. Moreover, any combination of the above-
described elements in all possible variations thereof is encompassed by the
invention
unless otherwise indicated herein or otherwise clearly contradicted by
context.
[345] The present disclosure is not to be limited in scope by the specific
embodiments
described which are intended as single illustrations of individual aspects of
the
disclosure, and any compositions or methods which are functionally equivalent
are
within the scope of this disclosure. It will be apparent to those skilled in
the art that
various modifications and variations can be made in the methods and
compositions of
the present disclosure without departing from the spirit or scope of the
disclosure.
Thus, it is intended that the present disclosure cover the modifications and
variations of
this disclosure provided they come within the scope of the appended claims and
their
equivalents.
[346] All publications and patent applications mentioned in this
specification are herein in-
corporated by reference to the same extent as if each individual publication
or patent
application was specifically and individually indicated to be incorporated by
reference.