Note: Descriptions are shown in the official language in which they were submitted.
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
Multiplexed Assay Using Differential Fragment Size to Identify Cancer Specific
Cell-Free DNA
[0001] CROSS REFERENCE TO RELATED APPLICATIONS
[0002] This application claims priority to U.S. Provisional Application No.
62/950,830, filed
December 19, 2019, which is incorporated herein by reference in its entirety.
[0003] SEQUENCE LISTING
[0004] The instant application contains a Sequence Listing which has been
submitted
electronically in ASCII format and is hereby incorporated by reference in its
entirety. Said
ASCII copy, created on December 14, 2020, is named 116352 PD012W0 SL.txt and
is
21,067 bytes in size.
[0005] BACKGROUND
[0006] Cancer is one of the leading causes of death in developed, and
increasingly also
developing, nations. According to the World Health Organization, in 2012, over
14 million
new cases were reported and over 8 million people died worldwide (Atlanta:
American
Cancer Society, Cancer Facts & Figures, 2014). Colorectal cancer (CRC) is the
third most
commonly diagnosed cancer and third-leading cause of cancer deaths in the
United States. In
2014, nearly 140,000 diagnoses and 50,000 deaths are expected in the U.S.
(Atlanta:
American Cancer Society, Colorectal Cancer: Facts & Figures 2014-2016). CRC is
often
curable if detected early, and outcomes can be improved with post-treatment
monitoring and
surveillance for recurrence.
[0007] Effective cancer management depends on early diagnosis, accurate tumor
staging, and
consistent monitoring. While many methods have been developed for the
detection and
quantification of nucleic acids, e.g., NanoDrop Microvolume Spectrophotomer
and
Fluorometer, many current diagnostic procedures are invasive, expensive and
unpleasant. In
multiple recent published studies, circulating cell-free DNA (cfDNA)
concentration and
integrity (fragmentation pattern) has shown promise as a highly sensitive and
specific,
minimally invasive blood biomarker for multiple cancer types (see, e.g., Hao,
T B, et al.,
British Journal of Cancer 2014: 1-2, doi 10.1038/bjc.2014.470; Gonzalez-Masia,
et al., Onco.
1
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
Targets Ther. 6:819-832 (2013); Yu, J, et al., Lab Med. 45(1): 6-12 (2014)). A
number of
these studies have indicated the utility of a highly sensitive assay to
measure cfDNA integrity
(fragmentation pattern) and concentration based on quantitation of an ALU
element, the most
common type of retrotransposable elements (RE) in the human genome (Table 1).
RE-based
methods for quantitating DNA are attractive due to their superior sensitivity
(multi-copy
representation in the genome) and robustness. The sequence of ALU Yb8 and
other ALU Yb
subfamilies are known in the art, see e.g., Ahmed et al., Mob DNA 2013, 4: 25
incorporated
herein by reference in its entirety.
[0008] The most commonly employed cfDNA integrity/concentration assessment
method,
the ALU 247/115 bp index, targets sequences of a single ALU element, and thus
the two
fragments analyzed are not independent. This precludes use of these targets in
a single
multiplexed assay for maximum accuracy, efficiency and practical clinical use.
This prior art
method poses several particular problems. First, evaluating the first sequence
and the second
sequence in conventional single-plex polymerase chain reactions (PCR) wherein
a single
target is amplified in a single reaction well rather than multiplexing the two
sequences into a
single reaction mixture introduces well-to-well variability into the results.
Every PCR
reaction is somewhat different from every other PCR reaction, and experimental
variation in
set-up steps, such as variation in pipetting volumes, introduces error and can
impact the
results. Secondly, it has been shown (see U.S. Patent Application Publication
No. US
2016/0186239 Al, published June 30, 2016, incorporated herein in its entirety)
that the
primers used in prior art studies to amplify these specific 247 bp/115 bp
sequences show poor
primer specificity, with false signals being generated from non-template
controls. Thirdly,
single-plex amplification prohibits the incorporation of an internal PCR
control. The use of
an internal PCR control is useful for confirming the success of the reaction
and for providing
confidence that other experimental factors such as the presence of PCR
inhibitors in the
sample have not interfered with sample integrity. Additionally, single-plex
amplification of
each target is cumbersome, more labor-intensive and less cost effective than
is running a
multiplexed amplification.
[0009] One of the cancer types studied using cell free DNA integrity is
colorectal cancer
(CRC). The current gold-standard for CRC diagnosis and staging is colonoscopy
and
subsequent histological examination. While specific and accurate, colonoscopy
is invasive,
2
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
expensive, and poses some risks; all of which decrease patient compliance to
screening
recommendations and discourage routine monitoring. In CRC and a few other
cancer types,
tissue biopsy is supplemented with detection of cancer protein biomarkers in
blood serum,
e.g. carcinoembryonic antigen (CEA). Such assays have the significant
advantage of being
minimally invasive and also do not require immediate localization of the
tumor. Nevertheless,
these assays suffer from limited sensitivity. CEA, one component of the
current standard of
care for CRC post-treatment monitoring, has relatively low sensitivity and
specificity for
early (stages I and II) and late (stages III and IV) disease (early: 36%
sensitivity and 87%
specificity; late: 74% sensitivity and 83% specificity) (Fakih, M. G.;
Padmanabhan, A.,
Oncology 20(6): 579-587 (2006)). Given this performance, CEA is not
recommended for
CRC diagnosis according to the National Comprehensive Cancer Network
guidelines for
CRC (Ms-PSEE, Hunt, S., NCCN, Clinical Practice Guidelines in Oncology (NCCN
Guidelines ) Colon Cancer, 2013).
[0010] Furthermore, imaging tests such as computerized tomography (CT) scans,
bone scan,
magnetic resonance imaging (MRI), positron emission tomography (PET) scan,
ultrasound,
and x-ray, among other radiological imaging, may be used to monitor disease
progression and
therapeutic effectiveness. Such tests have the downside of exposing patients
to large amounts
of radiation over the course of their treatment and while they are in
remission, are costly, and
may be burdensome.
[0011] cfDNA: A Brief Overview of Biology and Physiology
[0012] Characterization of cell-free DNA (cfDNA), DNA found in circulation in
human
blood plasma and serum, has emerged as an exciting prospect for a new
generation of blood-
based tools for cancer detection, monitoring and surveillance. It is also an
exciting prospect
for monitoring minimum residual disease, therapeutic effectiveness, and
disease progression.
Nucleic acid circulation in human blood plasma was first reported in 1948
(Mandel P; Metais
P., C.R. Acad. Sci. Paris 142: 241-243 (1948)). Leon, et al., (1977) were the
first to report
that mean cfDNA levels were significantly higher in the serum of patients with
malignant
cancers versus healthy patients (Leon, SA; Shapiro, B; Sklaroff, D M; Yarns, M
J, Cancer
Research 1977: 646-650). In the past two decades, many details of cfDNA
biology, and the
3
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
relationship between cfDNA and disease, have been elucidated. A brief primer
of these
studies is provided below.
[0013] Circulating cfDNA is derived from both the nuclear and mitochondrial
genomes of
normal and tumor cells (Mandel and Matais 1948, referenced supra; Zhong, S;
Ng, M CY;
Lo, Y MD; Chan, JC N; Johnson, P J; Kong H., J. Clin. Pathol. 53: 466-469
(2000)). Both
coding and noncoding portions of the genome are represented among circulating
cfDNA
(Bettegowda, C, et al., Sci. Transl. Med. 6(224): 224ra24 (2014), doi:
10.1126/
scitranslmed.3007094.Detection). Although several mechanisms are believed to
contribute to
the circulating cfDNA pool, including spontaneous release of free, exosome
encapsulated,
and microvesicle-encapsulated DNA into the bloodstream, cell death is the
major generator
of circulating cfDNAs (Jahr, S; Hentze, H; Englisch, S; Hardt, D; Fackelmayer,
F 0; Hesch,
R, Cancer Research 61:1659-1665(2001)). Cell turnover in normal cells is
ordinarily due to
apoptosis, which results in stereotyped sized fragments of DNA: a monomeric
form
composed of ¨ 180 bp fragments of DNA and associated nucleosomes, and reduced
amounts
of oligomeric forms. Id. Alternatively, tumor cells turn over using a
diversity of cell death
pathways, not only apoptosis, but also necrosis, autophagy, and mitotic
catastrophe (Jin, Z;
El-Deiry, W S, Cancer Biology & Therapy 4(2): 139-163 (2005), available at
http ://fly-
bay.net/ journals/cbt/jin4-2.pdf (accessed 15 Dec. 2014)). Non-apoptotic
pathways non-
specifically and incompletely degrade DNA, generating substantially longer DNA
fragments,
up to 21 kilo bases in the case of necrosis (Jahr, S., cited supra).
Differences in the rate of
cell death and type of cell death pathway utilized between normal and cancer
cells lead to
distinct characteristics of cfDNA pools that distinguish patients with and
without cancer.
cfDNAs have variable half-life within the body, ranging from minutes to hours
(Lo Y MD;
Zhang J; Leung TN; Lau T K; Chang AM Z; Hj elm NM, Am. J. Hum. Genet. 64: 218-
224(1999); Emlen W; Mannik M., Clin. Exp. Immunol. 56(1): 185-192 (1984);
Corcoran
and Chabner, N Engl J Med 2018;379:1754-65). Short half-life implies that
circulating
cfDNA levels provide a dynamic measure of the physiological and pathological
state of an
individual. Finally, there is evidence that a small fraction of circulating
cfDNA from blood is
able to pass the kidney barrier and enter urine. These cfDNAs are called
'trans-renal' cfDNAs
(Su Y-H, et al., J. Molecular Diagnostics, 6(2): 101-107 (2004); Botezatu I,
et al., Clin.
Chem. 46(8): 1078-1084 (2000)). The specific physiology of transrenal cfDNAs
awaits
detailed exploration.
4
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0014] Circulating cfDNAs from patients with and without cancer differ in a
number of
ways. Tumor genomes harbor specific genetic and epigenetic alterations that
distinguish them
from normal genomes, and these differences are reflected in cfDNAs.
Nonspecific
characteristics of cfDNA, such as concentration and integrity, differ between
cancer patients
and control subjects due to the specific mechanisms of cfDNA release into the
blood by
normal versus tumor cells. cfDNA concentration and integrity have often been
found to be
elevated in patients with cancer due to high rate of tumor cell death
(reviewed in
Schwarzenbach H; Hoon D S B; Pantel K., Nature Reviews Cancer 11: 426-437
(2011 ), doi:
10. 1038/nrc3066; Gonzalez-Masia, J A; Garcia-Olmo, D; Garcia- Olmo, D C,
Onco.
Targets. Ther. 6: 819-832 (2013)). However, absolute cfDNA concentration
significantly
varies among currently employed assays, significantly hampering the ability to
compare
results across studies. There is currently no standardized, validated,
commercially available
cfDNA concentration and integrity assay. There are no reports in the prior art
of using a
multiplexed quantitative polymerase chain reaction (qPCR) system of the kind
described
herein for accurate simultaneous measurement of concentration and integrity of
cfDNA.
[0015] Efforts in cell-free DNA (cfDNA) testing using blood samples focus
almost
exclusively on mutations which are not tumor-type agnostic and lack the
analytic sensitivity
required for therapy monitoring. cfDNA was detected in blood as early as the
1940s by
Madel and Metais (McLarty, J.L., Yeh, C.-H. Circulating Cell-Free DNA: The
Blood Biopsy
in Cancer Management. 2015 (cited 2020 Nov. 6); Available from: https:
//circulogene.
com/wp-content/uploads /2015/10/MOJCSR-02-00021.pdf.) and since this time,
many
studies have shown that the presence of elevated levels of cfDNA in the blood
of patients
with several cancer types including colorectal cancer (CRC) have poor
prognosis (Bortner, et
al., Trends Cell Biol, 1995. 5(1): p. 21-6.; Liu, X., et al., Enrichment of
short mutant cell-free
DNA fragments enhanced detection of pancreatic cancer. EBioMedicine, 2019. 41:
p. 345-
356). The elevation in cfDNA levels originates through the release of DNA from
cellular
necrosis and apoptosis of tumor cells. In healthy individuals, the main source
of cfDNA in
circulating blood is through necrosis. Necrosis generates a spectrum of DNA
fragments of
different sizes, due to random digestion by DNases (up to several kbp).
Apoptosis generates
small and uniform DNA fragments (less than <180bp) (Lapin, M., et al., J
Transl Med, 2018.
16(1): p. 300). A recent study of fragment length of cfDNA shows mutant KRAS
alleles tend
to be significantly shorter when compared with DNA fragments bearing wild-type
allele by
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
densitometry in pancreatic cancer (Dasari, A., et al., Nat Rev Clin Oncol,
2020). Older
publications often reached unreliable conclusions about correlation of cfDNA
fragment size
concentration and cancer progression due to inadequate cell free DNA
purification protocols
for small DNA fragments, as well as sample collection, plasma separation and
storage
protocols causing contamination of large DNA fragments from whole cell
degradation.
However, with the availability of new sample collection tubes with
preservatives to prevent
cellular contamination, such as Streck tubes, as well as availability of
improved cfDNA
extraction methods, several recent published studies have clearly established
that cfDNA
from cancer cells are smaller in fragment size as compared to cfDNA produced
by
noncancerous cells (Lin, S.Y., et al., JCO Precis Oncol, 2018. 2).
[0016] In clinical oncology, cfDNA has been suggested as a new surrogate
marker for
therapy response, disease progression and/or detecting early relapse. More
recent studies
have begun to explore the potential use of circulating tumor DNA (ctDNA) and
oncogene
biomarkers for prognostic uses (Elazezy, M. and S.A. Joosse, Comput Struct
Biotechnol J,
2018. 16: p. 370-378.); Tie, J., et al., Ann Oncol, 2015. 26(8): p. 1715-22).
Although ctDNA
markers can provide great information in cancer biology, challenges and
limitations have
arisen when working with it as ctDNA can be as little as 0.01% of the entire
cfDNA in
plasma (Tie, J., et al., Ann Oncol, 2015. 26(8): p. 1715-22). Due to
heterogeneity
intratumorally as well as between tumors and metastatic lesions make it
difficult to detect the
cancer progression within individual patients. Additionally, the use of
oncogene biomarkers
may only represent a subpopulation of patients expressing these genes
resulting in the missed
opportunity to monitor an entire population of patients undergoing treatment.
For ctDNA
analysis, the use of sophisticated instrumentation by highly trained
personnel, high blood
volume requirements and cost are prohibitive factors especially in low
resource areas and for
economically disadvantaged patients. On the contrary, cfDNA is circulating in
every
individual's blood while elevated in cancer patients. A recent study comparing
RECIST
results to Carcinoembryonic Antigen (CEA), cfDNA or ctDNA levels demonstrated
cfDNA
had the highest correlation compared to RECIST for tumor burden and tumor
volume of the
main lesion (Henley, et al., Invasive Cancer Incidence, 2004-2013, and Deaths,
2006-2015,
in Nonmetropolitan and Metropolitan Counties ¨ United States. 2017 (cited 2020
Nov. 6);
Available from: https ://www. cdc.gov/ mmwr/volumes /66/ss/ss6614a1 .htm ).
This study
6
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
highlights a great potential use of cfDNA for cancer monitoring, which can
track the change
in tumor burden.
[0017] Overview of Retrotransposable Elements (REs)
[0018] Retrotransposable Elements (REs) are mobile element insertion
polymorphisms that
are essentially homoplasy-free characters, identical by descent and easy to
genotype
(reviewed in Batzer M A; Deininger, P L, Nat. Rev. Genet. 3(5): 370-9 (2002),
doi:10.1038/nrg798). ALUs are REs that are approximately 300 bp insertions and
are
distributed throughout the human genome in large copy number. In addition to
the major
retrotransposon families, REs include smaller families of transposons such as
SVA or long
interspersed element ("LINE"). SVA elements, named after its main components,
short
interspersed element ("SINE"), variable number tandem repeat ("VNTR") and Alu
element
("ALU"), contain the hallmarks of retrotransposons, in that they are flanked
by target site
duplications ("TSDs"), terminate in a poly(A) tail and they are occasionally
truncated and
inverted during their integration into the genome (Ono, M; Kawakami, M;
Takezawa, T,
Nucleic Acids Res. 15(21): 8725-8737 (1987); Wang, H, et al., J. Mol. Biol.
354( 4): 994-
1007 (2005), doi: 10.1016/j jmb.2005.09.085). Long-interspersed Elements
(LINE) are
similar to ALU and SVA in that they also contain the hallmarks of
retrotransposons and are
high copy number, but differ in size, being up to several kilo bases in length
(Deininger, P L;
Batzer, M A, Genome Res. 12(10): 1455-65 (2002), doi:10.1101/ gr.282402).
[0019] RE-Based DNA Quantitation
[0020] RE-based quantitation methods are advantageous when compared to
current,
commercially available systems due to the presence of a large number of fixed
insertions.
With a high copy number of subfamily-specific RE repeats within the human
genome, these
human-specific DNA assays have a very sensitive dynamic range of 1 pg to 100
ng (Nicklas,
J A; Buel, E., J. Forensic Sci. 48(5): 1-9 (2003)). For example, the ALUYb
lineage contains
approximately 1800 copies per genome and SVA contains approximately 1700 full
length
element copies per genome (Wang, H., referenced supra; Carter, A B, et al.,
Hum. Genomics
1(3): 167-178 (2004)). This large copy number minimizes the effect of
variation between
individuals, resulting in highly reproducible quantitation values.
7
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0021] U.S. Patent Publication 2014/0051075 Al, to Sudhir K. Sinha, is
entitled
"Development of a Highly Sensitive Quantification System for Assessing DNA
Degradation
and Quality in Forensic Samples" and describes the detection of DNA quality
with a
multiplex reaction using ALU and SVA for human DNA quantification. Though very
useful
for forensic purposes, the described method does not detail specific
application to cell free
DNA from plasma and/or serum. The amplicon sizes needed for a cfDNA assay are
different
from those needed for forensic applications, and other details of the two
methods such as
amplification conditions and primer/probe concentrations differ as well.
[0022] SUMMARY OF THE INVENTION
[0023] A majority of healthy (non-cancer) human cfDNA fragment sizes are
around 140-
180bp long. Cell free DNA released from cancer cells (often called circulating
tumor DNA or
ctDNA) are shorter than the cfDNA released from normal cells. In contrast to
cfDNA in
samples from cancer patients, the majority of cfDNA fragments from non-cancer
humans are
generated from apoptotic cells, generating around 180 bp-long (or 140-180bp)
fragments
equivalent to the length of DNA that wraps around 1 nucleosome, and sometimes
accompanied by DNA fragments with sizes in multiples of 180 bp. The qPCR
method of
measurement of any retrotransposable element (RE) target sequence quantitates
cfDNA
fragments equal to or longer than the size of the RE target sequence. For
example, an qPCR
measurement of Yb8 ALU target sequence of 80bp quantitates cfDNA fragments of
>80bp in
length, including both short and long cfDNA fragments that comprise the 80 bp
RE target
sequence. On the other hand, qPCR measurement of a 265bp SVA target sequence
quantitates cfDNA fragments of >265bp, those comprising the 265 bp RE target
sequence,
which does not include cfDNA fragments of less than 265bp in length
[0024] Herein we discuss the development and evaluation of a retrotransposable
interspersed
element (RE) based multiplexed qPCR assays to robustly quantitate and
distinguish cfDNA
integrity and concentration in test and control subjects' bodily fluids, e.g.,
plasma and serum.
The assays provide accurate, minimally invasive, rapid, high-throughput, and
cost-effective
methods with the potential to complement for characterizing minimum residual
disease,
therapeutic effectiveness, and disease, e.g., cancer, progression in humans,
thereby improving
patient outcomes.
8
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0025] The methods described herein for assessing the cfDNA and ctDNA
integrity and
concentration and thereby assessing the presence of cancer cells do not depend
on a clonal
mutation being present in the cancer cells. As such, the methods are
"agnostic" in that the
methods can be applied to samples from patients having many different types of
cancers.
Moreover, the sensitivity of the methods described herein is far greater than
other cfDNA and
ctDNA assays as the levels of cfDNA and ctDNA above a normal threshold are
detected in
virtually all cancer patients tested. In addition, the methods described
herein have low Cost
of Goods Sold, are based on commonly used qPCR lab methodology and have a fast
Turnaround Time (TAT), e.g., the DNA integrity and concentration can be
assayed and a
conclusion as to the presence of cancer cells or if a cancer therapy is
ineffective and whether
a patient has progressive disease can be completed quickly, e.g., in less than
24 hours, less
than 18 hours, less than 12 hours, or less than about 4 hours.
[0026] An embodiment of the invention is a method whereby RE targets are
simultaneously
assayed in a single, highly sensitive qPCR reaction, wherein a single RE
target is amplified in
a single qPCR reaction vessel, e.g., a well, (singleplex qPCR) or wherein
multiple RE targets
are amplified in a single qPCR reaction vessel, e.g., a well (multiplex qPCR)
, optionally
including an internal positive control to monitor the presence of PCR
inhibitors potentially
present in the sample of blood serum, plasma, urine, or other biological
fluid. This method
enables development of an accurate, rapid, affordable, minimally invasive,
high throughput,
cost effective clinical test to complement or replace existing procedures and
improve
characterizing minimum residual disease, therapeutic effectiveness, and
disease progression
in humans.
[0027] Accordingly, one embodiment of the invention is a qPCR method that
accurately
quantitates cfDNA in a patient's biological fluids including, e.g., blood
plasma or serum as an
indication of cancer cells present in the patient or as an indication of the
ineffectiveness of a
neoadjuvant or a cancer therapy or as an indication the patient has
progressive disease. The
method may be singleplex wherein a single RE target is amplified in a single
qPCR reaction
well or the method may be multiplex wherein multiple RE targets are amplified
in a single
qPCR reaction well.
9
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0028] Another embodiment of the invention is a qPCR method that accurately
provides a
determination of the extent of fragmentation or integrity of cfDNA in
biological fluids
including, e.g., blood plasma or serum, as an indication in the level of
"minimum residual
disease" ("MRD"). The method may be singleplex wherein a single RE target is
amplified in
a single qPCR reaction well or the method may be multiplex wherein multiple RE
targets are
amplified in a single qPCR reaction well.
[0029] Another embodiment of the invention is a three RE target (a first
"short" RE target, a
second "short" RE target, and one "long" RE target) multiplex RE-qPCR assay to
accurately
and robustly obtain cfDNA concentration, a determination of fragmentation and
integrity, and
DNA integrity index ("DII" or "DI") of biological samples from normal controls
and patients
having cancer, e.g. colorectal cancer (CRC), by direct qPCR from plasma or
serum samples
with or without DNA purification. The assay may also include one internal
positive control
synthetic target. The short RE targets are preferably about 60bp to about
135bp in length,
about 70 bp to about 130bp, or about 60 to about 120 bp in length with the
proviso that the
short RE targets differ sufficiently, e.g., in length and sequence, so that
their amplification
products generated in the qPCR assay can be distinguished from each other,
e.g., the short RE
targets may differ at least by about 10bp, at least by about 15 bp, or at
least by about 20 bp in
length. The third long RE target is preferably about 200 bp to 300 bp in
length, or 207 bp to
270 bp, e.g., about 260 bp to 267 bp. DII indicates a level of cfDNA
fragmentation and is a
ratio of long target quantities to a short target quantity. DII as used herein
is a ratio of the
long RE target, e.g., 265 base-pairs to the short RE targets, e.g., 80 base-
pairs (265 bp/80 bp).
When DII (265 bp/80 bp) is lower than 0.4, it indicates the major source of
cfDNA is from
apoptotic cells. When DII (265 bp/80 bp) is above 0.4, cfDNA are also
generated through
necrosis. Information on DNA integrity and DII, is found in Sinha et al.,
Surgery, DOT: https
:// doi. org/10.1016/j.surg.2019.06.004 (2019) and Madhavan, Dharanij a, et
al.,
Epidemiology, DOT 10.1007/s10549-014-2946-2 (2013) incorporated herein in
their entirety
by reference.
[0030] An embodiment of the invention is a multiplexed method to quantitate
the integrity of
circulating cell free human DNA in a test subject, comprising providing a
sample of serum,
plasma, urine, or other biological fluid from the test subject, the sample
comprising cell free
human DNA, and the cell free human DNA comprising a first and second short RE
target
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
each having a length of between about 60 and 135bp, about 70 to about 130 bp
or about 60
and 120 base pairs, then using a multiplex quantitative polymerase chain
reaction (qPCR)
method to quantitate the short RE targets, obtaining for the quantitated RE
targets a threshold
cycle number, comparing each threshold cycle number with a standard curve to
determine a
quantity of the RE targets that was present in the sample, and determining the
quantity of
each of the RE targets is higher in the test subject's sample as compared to a
control sample,
e.g. a sample from a healthy subject, concluding the test subject should
receive a treatment
and administering the treatment to the test subject.. For example, the cfDNA
concentration
measured for a first short RE target of 80 bp (Yb-8-80bp), and a second RE
target of 120 bp
(Yb-8-120bp), and a third RE target of 265 bp (SVA 265), in plasma samples
from 40 healthy
controls and 39 cancer patients is set forth in Table 1. The RE targets were
amplified using
the primer pairs for Yb-8-80bp, Yb-8-120bp and SVA 265 set forth in Tables 2A
and 2B.
The data in Table 1 demonstrate that while the absolute levels of the
retrotransposable
element targets are all different in each sample the amount of cfDNA in cancer
patients is
greater than that in control subjects. The concentration of the shortest 80 bp
target is
consistently higher than the longer the 120 bp target and the 265 bp target
indicating that the
cfDNA is highly degraded (apoptotic cell death).. The method may further
comprise the step
of concluding the subject is in need of a cancer therapy or has progressive
disease based on
the difference in the amount of the short targets being above the threshold
amount in a control
sample, and optionally also based on the DII of the sample, and then
administering the
treatment to the subject.
[0031] Another embodiment of the invention is a multiplexed method to
quantitate the
integrity of circulating cell free human DNA, comprising providing a sample of
serum,
plasma, urine, or other biological fluid, preferably a plasma sample, the
sample comprising
cell free human DNA, the cell free human DNA comprising two retrotransposable
element
(RE) targets, a short RE target sequence between 60bp and 135 base pairs or
between,
60bp and 120 base pairs, or about 70bp to about 130bp, and a long RE target
sequence
between 200bp-300bp, about 207bp to about 270bp, or about 260 bp to about 265
base pairs,
the retrotransposable element genomic targets are preferably independent of
each other, using
a multiplex quantitative polymerase chain reaction (qPCR) method to separately
and
simultaneously quantitate the short and long RE targets, obtaining for each
quantitated RE
target a threshold cycle number, comparing each threshold cycle number with a
standard
11
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
curve to determine for each quantitated RE target a quantity of the RE targets
that were
present in the sample, and (i) calculating a ratio of the quantity of the long
RE target to the
quantity of the short RE target, and concluding based on the long RE
target/short RE target
ratio the subject should receive a treatment and administering the treatment
to the subject.
[0032] Another embodiment of the invention is a multiplexed method to identify
a subject
who has cancer or MRD comprising,
providing a sample of serum, plasma, urine, or other biological fluid, from
the subject,
the sample comprising cell free human DNA, the cell free human DNA comprising
(a) a first short RE target and a second short RE target, the short target
being between
about 60 and about 135 base pairs, between about 70bp and about 130bp, or
between
about 60 bp and about 120 bp in length with the proviso the first and short RE
targets
differ in size and (b) a long RE target being between about 200bp and about
300 bp in
length,
using a quantitative polymerase chain reaction (qPCR) method to quantitate the
first
and second short targets in the sample and obtaining for each of the
quantitated targets
a threshold cycle number,
comparing each threshold cycle numbers with a standard curve to determine the
amounts of the RE targets that were present in the sample,
determining the difference in the amounts of the two short RE targets,
identifying the subject as having cancer or MRD by determining the difference
between the quantity of the two short RE targets is increased as compared to a
control.
The method may further comprise determining the DII for the DNA in the sample.
The method may further comprise administering an appropriate treatment to the
subject identified as having cancer or MRD.
[0033] Another embodiment of the invention is a multiplexed method to identify
a
neoadjuvant or cancer therapy as ineffective or identify a subject who has a
progressive
cancer, is in remission, or has MRD comprising,
12
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
providing a first sample of serum, plasma, urine, or other biological fluid,
taken from
a subject before administering a cycle of cancer therapy and a second sample
of
serum, plasma, urine, or other biological fluid from the subject after the
cycle of
cancer therapy, the first and second samples comprising cell free human DNA,
the
cell free human DNA comprising (a) two short RE target between about 60 and
about
135 base pairs, between about 70bp and about 130bp, or between about 60 bp and
about 120 bp, and (b) a long RE target between about 200 bp to about 300 bp,
between about 207 to about 265 bp, between about 260 to about 265bp in length
using a quantitative polymerase chain reaction (qPCR) method to quantitate the
two
short RE targets and the long RE target in the first and second samples and
obtaining
for the quantitated short and long RE targets a threshold cycle number,
comparing each threshold cycle number with a standard curve to determine the
amounts of the short and long RE targets that were present in the first and
second
samples,
determining the difference between the quantity of the short targets is
increased in the
second sample
identifying the subject having an increased the short RE targets in the second
sample
as having received an ineffective neoadjuvant or cancer therapy or as having a
progressive cancer or MRD. In this method the samples may be obtained from the
subject one week apart, at least 2 weeks apart, at least 3 weeks apart or at
least 4
weeks apart, e.g., 12 to 21 days apart, provided the second sample is obtained
from
the subject before the next cycle of therapy. The steps of this method may be
repeated throughout multiple cycles of therapy, wherein a sample is obtained
before
and after each cycle of therapy and subjected to this method. The method may
further comprise calculating a DII of the DNA in the samples. The method may
further comprise administering an appropriate treatment to the subject
identified as
having the progressive cancer or MRD.
[0034] In the multiplexed method to identify a cancer therapy as ineffective
or identify a
subject who has a progressive cancer, the difference between the quantity of
the short RE
13
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
targets in each of the first and second samples may be determined by
determining the value of
the amount of the shorter of the short targets minus the amount of the other
short RE target in
the first sample (said value = Frag 1) and determining the value of the amount
of the shorter
of the short targets minus the amount of the other short RE target in the
second sample (said
value = Frag 2) and determining the value of Fragl minus Frag2 (said value =
FragDiff)
wherein a FragDiff of greater than a threshold value identifies the cancer
therapy as
ineffective or the subject as having progressive cancer or MRD.
[0035] In certain embodiments of the multiplexed method of the present
invention, the
retrotransposable element genomic targets may be an interspersed ALU, SVA or
LINE1
element. In certain embodiments of the multiplexed method of the present
invention, the
retrotransposable element genomic targets may be each independently an
interspersed ALU,
SVA, or LINE element. In certain embodiments, these retrotransposable element
genomic
targets may each have a copy number in excess of 1000 copies per genome.
[0036] Some embodiments of the multiplexed method of the present invention
further
comprise a step of adding a synthetic DNA sequence to the sample as an
internal positive
control (IPC) prior to the using step/ qPCR quantitation step, quantitating
the internal positive
control in the using step, and utilizing the quantitative internal positive
control result in the
comparing step to improve the accuracy and reliability of the comparing step
to determine the
amounts of the RE targets.
[0037] In embodiments of the multiplexed method of the present invention, the
use of an
internal positive control enables a determination of the concentration of cell
free DNA in the
sample.
[0038] In some embodiments of the multiplexed method of the present invention,
the sample
of serum, plasma, urine, or other biological fluid may be placed in a single
tube, and the
qPCR reactions for quantitation of the nucleic acid fragments may be carried
out in that same
single tube. Alternatively, each nucleic acid fragment may be separately and
simultaneously
amplified in separate tubes.
[0039] In some embodiments of the multiplexed method of the present invention,
the ratio of
the quantity of the longer RE target to the quantity of a shorter RE target
may serve as the DII
14
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
of circulating cell free DNA for diagnostic and therapeutic applications.
These diagnostic
applications may include one or more of the characterizing minimum residual
disease,
therapeutic effectiveness, and disease progression in human patients, and
treating such
patients.
[0040] In certain embodiments, the multiplexed method of the present invention
may include
a step of deactivating or eliminating proteins that bind to the short nucleic
acid fragment or
the long nucleic acid fragment. This may be done by mixing the sample with a
buffer
including a surfactant and chelating agent, enzymatically digesting the
protein, then using
heat to deactivate and inactivate the digested protein, followed by
centrifugation.
Alternatively, dilution of the sample using 40 parts sterile water to one part
sample by
volume may have the effect of deactivating or eliminating these proteins.
[0041] In some embodiments, the multiplexed methods of the present invention
may include
a step of separating amplification products obtained from the qPCR reaction
using
electrophoresis. In some embodiments of this invention, the amplification
products of the
qPCR method used in the methods of this invention may be detected and/or
quantified using
electrical biosensors (see Liu, et al., Single-Nucleotide Polymorphism
Genotyping Using a
Novel Multiplexed Electrochemical Biosensor with Nonfouling Surface. Biosens.
Bioelectron. 2013, 42, 516-521).
[0042] In some embodiments, the multiplexed methods of the present invention
may include
a step of determining an optimum temperature for the qPCR reaction.
[0043] The multiplexed methods of the present invention may include a sample
that comes
from an individual who is suffering from cancer, is in remission from cancer,
or who is at risk
for developing cancer, who has received a treatment for cancer, e.g., a
targeted therapy,
chemotherapy, immunotherapy, targeted-immunotherapy, surgery to remove a
tumor, or a
radiotherapy. Targeted therapy is a type of cancer treatment that uses drugs
or other
substances to precisely identify and attack certain types of cancer cells. A
targeted therapy
can be used by itself or in combination with other treatments, such as
traditional or standard
chemotherapy, surgery, or radiation therapy.
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0044] In certain embodiments, the present invention may take the form of a
multiplexed
system for evaluating the effectiveness or ineffectiveness of a cancer therapy
or for
characterizing cancer in humans, the system including a sample of serum,
plasma, urine, or
other biological fluid, preferably a plasma sample, the sample comprising cell
free DNA. The
cell free DNA comprises one short retrotransposable element targets, or two
short
retrotransposable element targets, and optionally a long retrotranspoable
element target. The
short retrotransposable element targets may have a length in the range of
about 60 base pairs
to about 135 base pairs, about 70 to about 130bp, or 60 base pairs to about
120 base pairs,
and two short retrotransposable element targets, may each with a length of 60
base pairs to
135 base pairs, or about 70 bp to about 130 bp, or about 60 base pairs to
about 120 base pairs,
preferably the two short RE targets differ in size and sequence sufficiently
to distinguish their
amplification products generated in the qPCR assay, e.g., the short RE targets
differ in size
by at least about 10 bp, at least about 15 bp, or at least about 20. The third
RE target being a
fragment of another multi-copy retrotransposon with a length of about 200 bp
to about 300bp,
e.g., 207 bp to 265 base pairs. In an embodiment, the retrotransposable
element targets are
independent of one another. The system may further comprise an internal
positive control
(IPC) comprising synthetic DNA, a TaqMan probe corresponding to each RE
target and
IPC, each probe comprising a detectable label that is distinct from the labels
incorporated into
the other probes, a forward primer and a reverse primer pair for amplifying
each RE target
and IPC, a DNA standard for generating standard curves for each RE target and
IPC, a qPCR
system for simultaneously amplifying each RE target and IPC and for producing
a threshold
cycle number for each RE target and IPC, and a qPCR data analysis system for
producing
DNA quantitation values for each RE target by interpolation using threshold
cycle numbers
and linear standard curves and for using the DNA quantitation values to
produce an
indication of the integrity of the cell free DNA and for characterizing cancer
in a human.
[0045] In one embodiment, one or more of the retrotransposable element targets
used in the
methods and systems of the invention described herein, are an ALU (e.g. ALU-
Yb8) target or
an SVA. The ALU target may be, for example, a 60 bp target, a 65 bp target, a
71bp target,
an 80 bp target, a 97 bp target, a 105 bp target. The targets may be amplified
with forward
and reverse primers. In embodiments comprising multiple RE targets from the
same RE, e.g.,
ALU Yb-8õ PCR blockers/PNA clamping may be included to limit extension from
the
primers beyond the position of the blockers, thus limiting the extension from
a primer pair
16
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
used to amplify one RE target into the other RE target and thereby enhancing
the specificity
by limiting the production of extraneous or overlapping products. Preferably
the PCR
blockers are peptide nucleic acid (PNA) oligos and bind to the
retrotransposable element
between the targets to be amplified. See e.g., Figure 2 depicting the position
of the forward
and reverse primers used to amplify an 80 base pair target and 97 bp target on
an ALU, e.g.,
ALU-Yb8, and the position of the 80 bp blocker that limits extension from the
80 bp forward
primer, and a 97 bp blocker that limits extension from the 97 bp blocker. In
this embodiment,
during qPCR the 80 base pair and 97 base pair specific forward primer and
reverse primer
hybridize to their respective sites on the ALU, but extension from the primers
is limited by
the presence of the PCR blocker at their respective sites.
[0046] BRIEF DESCRIPTION OF THE FIGURES
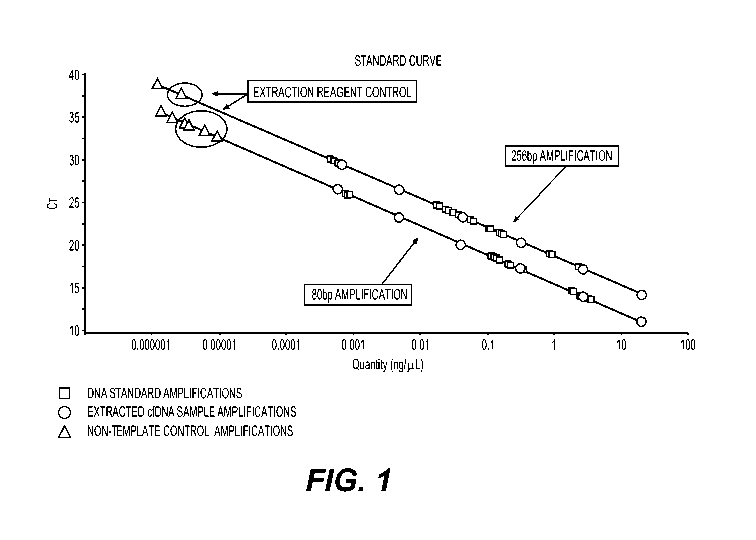
[0047] Figure 1. Titration illustration using a standard curve of known
quantities of cfDNA
with concentration measured in ng/pL using the test described in US Patent
Application
Publication No. US 2016/0186239 Al, incorporated by reference herein in its
entirety. The x-
axis depicts the concentration of DNA fragments longer than 80 bp and longer
than 265 bp
measured in ng/pL. The y-axis depicts the number of the PCR amplification
cycle.
[0048] Figure 2. Diagram showing two PCR target regions, 80 bp and 97 bp, on
the Alu-Yb8
sequence using two peptide nucleic acid (PNA) oligos to block PCR extension
beyond the
target regions.
[0049] Figures 3A and 3B present the Log-odds (y-axis) vs Fragl (x-axis) (FIG.
3A) and Log-
odds (y-axis) vs FragDff (ng/m1)(x-axis) (FIG. 3B).
[0050] Figure 4 is an illustration of the specificity of a method described
herein used in
identifying samples from patients with progressive disease.
[0051] DETAILED DESCRIPTION OF THE INVENTION
[0052] There is a clear need in cancer management, and colorectal cancer (CRC)
treatment
specifically, for a standardized and validated blood test to sensitively and
robustly quantitate
cfDNA integrity and concentration. The present application addresses this need
by creating a
multiplex qPCR assay for quantitating cfDNA integrity and concentration based
on
17
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
retrotransposable element targets to identify, characterize and/or
appropriately treat the
patient having cancer, e.g., progressive disease, or MRD. The assays are also
useful in
indentifying a cancer therapy's effectiveness or ineffectiveness.
[0053] The most commonly employed method conducted by others in the field of
cfDNA
integrity and concentration assessment for cancer detection and monitoring is
qPCR using the
ALU 247/115 index. The methods described herein for assessing integrity and
concentration
of cfDNA and ctDNA quantitates "short" retrotransposable element targets
having lengths
between 60bp and 135bp, 70 bp to 130bp, or between 60bp and 120 bp to reliably
indicate
therapy effectiveness or ineffectiveness. The ranges between 60bp and 135bp,
between 70 to
about 130bp, e.g., 71bp to 132bp, or between 60bp and 120 bp ranges of ALU,
SVA and
LINE1 retrotransposable elements targets are also useful for discriminating
between normal
(non-cancer) human and humans with cancer, particularly progressive disease,
or the
presence of MRD. Preferably the retrotransposable elements are ALU, e.g., Yb-8
ALU, SVA,
or LINE1.
[0054] MRD refers to the small number of malignant cancer cells that remain in
the body
during or after treatment (see NCI Dictionary of Cancer Terms,
https://www.cancer.
gov/publications/dictionaries/ cancer-terms/def/797386). Even when a patient
is in remission
from cancer and the solid tumor has shrunk beyond detection, the patient may
still have
MRD. The MRD assessment is used to determine if additional treatment is
necessary, if a
treatment already administered has been effective in reducing tumor load, or
to select and
administer a particular treatment of the subject. MRD assessment is mainly
used in blood
cancers (leukemia, lymphoma and myeloma), but is being studied in other solid
cancers.
MRD assessment has been used in guiding the treatment of cancer patients in
cases of, e.g.,
resected hepatoma, resection of mastectomy, esophageal cancer, rectal cancer,
anal cancer,
head and neck cancer, colon cancer, lung cancer, breast cancer, neu metastatic
breast cancer.
[0055] Cancer patients in remission must undergo quarterly imaging (e.g. MRI,
x-ray, CT
scan, or other radiology studies) to determine whether the cancer has
returned. However,
some patients in remission may not have a solid tumor that is detectable by
imaging studies,
but may still have MRD. The methods described herein for quantitating the
integrity and
concentration of cfDNA by using short retrotrasposable elements target(s)
having a length
18
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
between 60bp and 135bp, 70bp to 130bp, or 60 to 120bp, may be used to
characterize
cancer or MRD. The change in the amount of the quantitated short RE target
sequence
between 60bp and 135bp, 70 bp to 130bp or 60 bp to 120 bp over time may be
used alone or
in conjunction with standard assays to reliably identify subjects who have MRD
or cancer
progression or evaluate the ineffectiveness of a cancer therapy. Based upon a
determination
that the subject has MRD or progressive cancer, or the ineffectiveness of a
therapy, additional
rounds of therapy or another therapy may be administered to the subject. We
demonstrate
herein that cfDNA comprising elevated or increasing amounts of short ALU Yb8
targets of
60 base pair to 135 base pair, about 70bp to about 130bp, or 60bp to about
120bp sequence as
compared to the amount of long RE targets, e.g., SVA or LINE targets, between
200 bp and
about 300 bp, or between 207 bp and about 270 bp, between 260bp and 265bp,
e.g., 265bp or
267 to be highly effective in discriminating between normal humans (non-
cancer) and
humans with cancer (see e.g., Figure 3 and Figure 4). And because the methods
herein do not
rely on detecting CEA, the methods are "agnostic" and can be applied to
samples from
patients having or suspected of having any type of cancer, e.g., colorectal
cancer (CRC),
hepatoma, esophageal cancer, rectal cancer, anal cancer, head and neck cancer,
colon cancer,
lung cancer, e.g., non-small cell lung cancer (NSCLC), small cell lung cancer
(SCLC) breast
cancer, and blood cancers, e.g., leukemia.
[0056] The methods described herein for assessing cfDNA integrity and
concentration, using
a sample from a subject, e.g., a plasma or serum sample or another bodily
fluid sample, and
RE targets, are useful in detecting, measuring, or monitoring cancer and are
an additional
parameter for use in the assessment of tumor load, cancer progression, therapy
ineffectiveness and or MRD such that an appropriate treatment is administered
to the subject.
The methods described herein allow for detection of cancer cells in patients
who have a
nearly undetectable level as determined by standard clinical tests, such as
imaging assays,
e.g., CT scans or Xrays, or detection of cancer cells in a blood or tissue
sample. The patients
may be a patient suspected of having or treated for hepatoma, esophageal
cancer, rectal
cancer, anal cancer, head and neck cancer, colon cancer, colorectal cancer
(CRC), lung
cancer, e.g., non-small cell lung cancer (NSCLC), small cell lung cancer
(SCLC) breast
cancer, and blood cancers, e.g., leukemia. Thus, the methods described herein
are an
improvement over existing methods because they reduce patients' exposure to
radiation from
imaging studies.
19
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0057] Patients diagnosed with cancer, including patients receiving a cancer
therapy, may be
categorized based on their disease progression, e.g., following a cycle of
chemotherapy or
immunotherapy or other therapeutic regime. A "complete response" ("CR")
patient is one
where there is no evidence of the disease due to a disappearance of all target
lesions as
determined by standard methods, e.g., such as CT scans or detection of cancer
cells in a blood
or tissue sample. A "stable disease" ("SD") patient is one where there is
neither sufficient
shrinkage of cancer lesion size to qualify for partial response ("PR") nor
sufficient increase in
lesion size to qualify for "progressive disease" ("PD") using as a reference
the smallest sum
of diameter of target lesions. A PR patient is one who demonstrates at least a
30% decrease in
the sum of the diameters of target lesions vs. the baseline sum of the
diameters of the target
lesions. Additionally, the sum of the diameters of the target lesions must
demonstrate an
absolute increase of at least 5 mm or one or more new lesions have been
detected to be
considered PR. A PD patient is one where there is at least a 20% increase in
the sum of the
diameters of the target lesions vs. the smallest sum of target lesions, which
may be the
baseline sum.
[0058] The present invention is non-invasive and may also be used for
screening high risk
patients for onset of cancer, e.g., hepatoma, esophageal cancer, rectal
cancer, anal cancer,
head and neck cancer, colon cancer, colorectal cancer, lung cancer, breast
cancer, neu
metastatic breast cancer and blood cancers, e.g., leukemia. Patients may be
considered "high
risk" for a variety of reasons including past family history of cancer,
environmental exposure,
and lifestyle. However, it is not feasible, highly wasteful, and harmful for
patients to be
exposed to radiological scans to screen them for cancer.
[0059] The present invention may be used to distinguish between therapy
ineffectiveness or
futility and therapies that are partially ineffective. Current methods make it
burdensome,
costly, and inefficient to determine whether a therapy is ineffective in a
patient or the patient
experience a partial response to a therapy. The present invention allows
clinical providers to
detect noninvasively and quickly whether the therapy is entirely ineffective
or partially
ineffective. This allows providers to make quicker and better informed
clinical decisions
about patient therapy and administer an appropriate therapy.
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0060] One drawback of many currently available methods is the inability to
identify cell
necrosis. One method for identifying cell necrosis is the DII. DII is a ratio
of long fragments
quantities to short fragment quantities. DII indicates a level of cfDNA
fragmentation. When
the DII using the ratio of 265bp to 80bp targets is calculated and determined
to be lower than
0.4, it indicates the major source of cfDNA is from apoptotic cells. When the
DII using the
ratio of 265bp to 80bp targets is calculated and determined to be above 0.4,
cfDNA are also
generated through necrosis. This DII may be used in the methods of this
invention to assess
cell necrosis.
[0061] Described herein are methods and systems for quantitating the integrity
of circulating
cell free human DNA and implementing a treatment of a patient. An embodiment
of this
invention is a method for quantitating the integrity of circulating cell free
human DNA and
implementing a treatment of a patient comprising:
(a) providing a sample of a bodily fluid comprising cell free human DNA, the
cell free
human DNA comprising an RE target of between 60 base pairs to 135 base pairs,
about 70 to about 130bp, or 60 to about 120 bp in length;
(b) using a quantitative polymerase chain reaction (qPCR) method to quantitate
the RE
target;
(c) obtaining for the quantitated RE target a threshold cycle number;
(d) comparing the threshold cycle number with a standard curve to determine a
quantity
of the RE target that was present in the sample; and
(e) determining the quantitated RE target amount in the patient sample is
higher than
present in a control subject, and concluding the patient is in need of a
treatment, and
implementing the treatment of the patient. The method may be singleplex
wherein a
single RE target is amplified in a single qPCR reaction well or the method may
be
multiplex wherein multiple RE targets are amplified in a single qPCR reaction
well.
[0062] The bodily fluid samples used in the methods of this invention should
be treated so as
to remove cells. Suitable bodily fluids include, e.g., serum, plasma, urine,
saliva, tears or
21
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
other biological fluid. Preferably the sample used in the methods and system
of this invention
is a plasma sample.
[0063] In the methods of this invention a single short retrotransposable
element target of
between 60 to 135bp, about 70 to about 130 bp or 60 bp to about 120 bp, may be
subjected to
quantitative polymerase chain reaction (qPCR) method to quantitate the single
target.
Alternatively, a multiple retrotransposable element targets, e.g., two or more
short RE targets,
and/or a long RE target of between 200 bp and 300bp or 207 bp to about 300 bp,
and 265-
267 bp, may be subjected to the quantitative polymerase chain reaction (qPCR)
method to
quantitate the targets.
[0064] The methods of this invention may further comprise a step of adding a
synthetic DNA
sequence to the sample as an internal positive control (IPC) and quantitating
the
retrotransposable element targets and the IPC, and utilizing the quantitative
IPC result in the
step of comparing the qPCR threshold cycle numbers to a standard curve to
improve the
accuracy and reliability of the comparing step. The IPC also enables a
determination of a
concentration of cell free DNA in the sample when quantitating the RE targets
by qPCR in a
single tube.
[0065] The methods of this invention may further comprise a step of adding a
hybridization
probe that hybridizes to the RE targets to detect the targets. The probe may
be added to the
sample before the target(s) are subject to q-PCR or thereafter. The probe may
include an
observable label. Any observable label routinely used in the art for labeling
nucleic acid
probes could be used to label the probe, e.g., a fluorescent label. Suitable
fluorescent probes
include, e.g., FAM, Cy5, Hex, or Cy3). The observable label may be detected
using a
microfluidic device.
[0066] The retrotransposable elements of the methods of this invention include
e.g., an ALU,
particularly ALU Yb8, an SVA, or a LINE element. The retrotransposable element
may have
a copy number in excess of 1000 copies per genome.
[0067] In the methods of this invention the short retrotransposable element
targets may have
a length from about 60 base pairs to about 135 base pairs, about 60 base pairs
to about 120
base pairs, about 60 base pairs to about 120 base pairs, and about 70 bp to
about 130 bp,. For
22
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
example, the retrotransposable element target may have a length of e.g. 60bp,
65bp, 71 bp, 80
bp, 97 bp, 105 bp, or 120 bp. In the methods of this invention the long
retrotransposable
element target may have a length from about 200bp to about 300 bp, or about
207 pb to about
270 bp, e.g. 265 bp -267bp. The RE targets may be amplified with the forward
and reverse
primer pairs set forth in Table 2A, 2B and/or 2C:
Table 2A: ALU-Yb8 targets' primer and probe sequences
Name Size Primer Type Primer & Probe Sequence SEQ ID NO
Forward GGAAGCGGAGCTTGCAGTGA 1
Yb8-80bp 80bp Reverse AGACGGAGTCTCGCTCTGTCGC 2
Probe AGATTGCGCCACTGCAGTCCGCAGT 3
Forward CTTGCAGTGAGCCGAGATT 4
Yb8-71bp 71bp Reverse GAGACGGAGTCTCGCTCTGTC 5
Probe ACTGCAGTCCGCAGTCCGGCCT 6
Forward GTGGCTCACGCCTGTAAT 7
Yb8-97bp 97bp Reverse GGGTTTCACCTTGTTAGCCA 8
Probe TGGATCATGAGGTCAGGAGAT 9
Forward AGGCAGGAGAATGGCGTGAACC 10
Yb8-105bp 105bp Reverse AGACGGAGTCTCGCTCTGTCGC 11
Probe AGATTGCGCCACTGCAGTCCGCAGT 12
Forward AGACCATCCTGGCTAACAA 13
Yb8-119bp 119 Reverse GCCATTCTCCTGCCTCA 14
Probe
Forward TGGATCATGAGGTCAGGAGAT 15
Yb8-120bp 120bp Reverse CCGAGTAGCTGGGACTACA 16
Probe ACCATCCTGGCTAACAAGGTGAAACC 17
Forward ATCCTGGCTAACAAGGTCAAA 18
Yb8-123bp 123bp Reverse CGGGTTCACGCCATTCT 19
Probe
23
CA 03164737 2022-06-14
WO 2021/127462
PCT/US2020/066048
Table 2B: SVA targets' primer and probe sequences
Name Size Primer Type Primer & Probe Sequence SEQ ID NO
Forward AATGGCGGCTTTGTGGAATA 20
SVA-100bp 100bp Reverse GTCTCCCATGTCTACTTCTTTCTAC 21
Probe AGAAATCGGATGGTTGCCGTGTCT 22
SVA-101bp 101bp Forward AACCCTGTGCTCTCTGAAAC 23
Reverse ACGCTGCCTTCAAGCAT 24
Probe
SVA-103bp 103bp Forward GCCCAACAGCTCATTGAGAA 25
Reverse ACGGCAACCATCCGATTT 26
Probe
Forward TGTCCACTCAGGGTTAAATGG 27
SVA-104bp 104bp Reverse GATTAGGGATTGGTGATAACTCTTA 28
Probe AAGGGCGGTGCAAGATGTGCTTTGTT 29
Forward TGTGTCCACTCAGGGTTAAAT 30
SVA-106bp 106bp Reverse GATTAGGGATTGGTGATGACTCT 31
Probe AAGGGCGGTGCAAGATGTGCTTTGTT 32
Forward TGTGCCCAACAGCTCATT 33
SVA-106bp-v2 106bp Reverse ACGGCAACCATCCGATTT 34
Probe
Forward CTGTGTCCACTCAGGGTTAAATG 35
SVA-116bp 116bp Reverse ATTACTTGAGATTAGGGATTGGTGATG 36
Probe AAGGGCGGTGCAAGATGTGCTTTGTT 37
Forward CCCAACAGCTCATTGAGAACG 38
SVA-116bp-v2 116bp Reverse CTTTCTACACAGACACGGCAA 39
Probe
Forward CTCTCTGAAACATGTGCTGTGT 40
SVA-118bp 118bp Reverse GGGATTGGTGATGACTCTTAACG 41
Probe AAGGGCGGTGCAAGATGTGCTTTGTT 42
Forward CTGTGTCCACTCAGGGTTAAAT 43
SVA-118bp-v2 118bp
Reverse TGATTACTTGAGATTAGGGATTGGT 44
24
CA 03164737 2022-06-14
WO 2021/127462
PCT/US2020/066048
Table 2B: SVA targets' primer and probe sequences
Name Size Primer Type Primer & Probe Sequence SEQ ID NO
Probe AAGGGCGGTGCAAGATGTGCTTTGTT 45
SVA-126bp 126bp Forward CTGTGTCCACTCAGGGTTAAAT 46
Reverse TGTGTCCCTGATTACTTGAGATTAG 47
Probe
SVA-126bp-V2 126bp Forward CCTGTTGATCTGTGACCTTACC 48
Reverse ACGCTGCCTTCAAGCAT 49
Probe AAGGGCGGTGCAAGATGTGCTTTGTT 50
Forward GTTGCCGTGTCTGTGTAGAA 51
SVA-128bp 128bp Reverse TTTCAGAGAGCACAGGGTTG 52
Probe AAGGGCGGTGCAAGATGTGCTTTGTT 53
Forward AACCCTGTGCTCTCTGAAAC 54
SVA-132bp 132bp Reverse GATTAGGGATTGGTGATAACTCTTA 55
Probe AAGGGCGGTGCAAGATGTGCTTTGTT 56
Forward CTGTGTCCACTCAGGGTTAAAT 57
SVA-207bp 207bp Reverse GAGGGAAGGTCAGCAGATAAAC 58
Probe AAGGGCGGTGCAAGATGTGCTTTGTT 59
Forward CCTGTGCTCTCTGAAACATGTGCT 60
SVA-257bp 257bp Reverse GATTTGGCAGGGTCATGGGACAAT 61
Probe AAGGGCGGTGCAAGATGTGCTTTGTT 62
Forward ATGTGCTGTGTCCACTCAGGGTTA 63
SVA-265bp 265bp Reverse ATTCTTGGGTGTTTCTCACAGAGG 64
Probe AAGGGCGGTGCAAGATGTGCTTTGTT 65
Forward TGGGATCCTGTTGATCTGTGACCT 66
SVA-290bp 290bp Reverse GATTTGGCAGGGTCATGGGACAAT 67
Probe
Forward GTTGCCGTGTCTGTGTAGAA. 68
SVA-355bp 355bp Reverse ATGGGACAATAGTGGAGGGA 69
Probe
Forward CCGTGTCTGTGTAGAAAGAAGTAG 70
SVA-367bp 367bp
Reverse GGGATTTGGCAGGGTCAT 71
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
Table 2B: SVA targets' primer and probe sequences
Name Size Primer Type Primer & Probe Sequence SEQ ID NO
Probe
Forward GGCGGCTTTGTGGAATAGA 72
SVA-399bp 399bp Reverse GAGGGAAGGTCAGCAGATAAAC 73
Probe ATCAGGGACACAAACACTGCGGAA 74
Forward TGGAATAGAAAGGCAGGAAAGG 75
SVA-411bp 411bp Reverse GCAGGGTCATGGGACAATAG 76
Probe
Table 2C: Linel targets' primer and probe sequences
Name Size Primer Type Primer & Probe Sequence SEQ ID NO
Forward CACAATAGCAAAGACTTGGAACC 77
Line1-252bp 252bp Reverse CCCTTCCTGTGTCCATGTG 78
Probe CCTTTGTAGGGACATGGATGAAAGTGGA 79
Forward GACTTGGAACCAACCCAAATG 80
Line1-257bp 257bp Reverse CCCAGAGTGTGACGTTCC 81
Probe AGTGAGAACACATGGACACAGGAAGG 82
Line1-262bp 262bp Forward GTGGCACATATACACCATGGAA 83
Reverse CGTTAGGTATATCTCCCAATGCTATC 84
Probe TGAGAACACATGGACACAGGAAGGG 85
Line1-266bp 266bp Forward ACTTGGAACCAACCCAAATG 86
Reverse CACAACAGTCCCCAGAGTG 87
Probe TGAGAACACATGGACACAGGAAGGG 88
Forward CATGGAATACTATGCAGCCATAAA 89
Line1-267bp 267bp Reverse CCCACTAACTCGTCATCTAGC 90
Probe TGAGAACACATGGACACAGGAAGGG 91
[0068] The samples used in the methods of this invention may be from a patient
has been
diagnosed as having a has stage I, stage II, stage III or stage IV cancer, is
suffering from
cancer, is in remission from cancer, is at risk for developing cancer, has had
surgery to
26
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
remove a tumor, has undergone a neoadjuvant therapy, a targeted therapy, a
chemotherapy,
immunotherapy and/or radiotherapy to treat a cancer.
[0069] The methods of this invention are also useful in further evaluating the
patient having a
minimum residual disease diagnosis to implement a disease treatment. For
example, in an
embodiment of this invention a determination is made that the quantity of the
short RE
targets as compared to the long Re targets is higher in the sample from the
patient than that of
a control sample, e.g., a sample from a healthy subject, and in view of that
determination an
appropriate treatment of the patient is instituted, e.g., a targeted therapy,
cancer
chemotherapy, immunotherapy, or radiotherapy is administered. Such treatment
might
include e.g., antineoplastic agents, alkylating agents, topoisomerase
inhibitors, mitotic
inhibitors, methotrexate, vinca alkaloids, antimetabolites, antifolates,
pyrimidine antagonists,
purine analogs, purine antagonists, proteasome inhibitors, tyrosine kinase
inhibitors, nitrogen
mustards, or another cancer therapy. Alternatively, a determination of a
threshold cycle
number of the quantitated nucleic acid fragment, is made and based on that
number the
clinical provider administers the treatment to the patient.
[0070] Multiplex methods
[0071] An embodiment of this invention is a method to quantitate the integrity
of circulating
cell free human DNA and optionally to implement a treatment of a subject,
comprising:
providing a sample from a subject, preferably a sample that has been treated
to remove cells,
the sample comprising cell free human DNA comprising a first RE target being
97 base pairs
and the second RE target having a length between 260 and 265 base pairs, e.g.,
263 bp; using
a quantitative polymerase chain reaction (qPCR) method to quantitate the first
and second RE
targets; obtaining for the quantitated RE targets a threshold cycle number;
comparing the
threshold cycle number with a standard curve to determine a quantity of each
of the RE
targets that was present in the sample; calculating a ratio of the quantity of
the 97 RE target to
the quantity of the between 260 and 265 base pair nucleic acid fragment; and
using the
quantitated nucleic acid fragment to quantitate the integrity of the
circulating cell free human
DNA and optionally to implement treatment of a patient. The subject's sample
may be
serum, plasma, urine, or other biological fluid from a human, preferably the
sample is a
plasma sample. The targets may be amplified in singleplex qPCR wherein a
single target is
27
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
amplified in a single reaction well or the targets may be amplified in a
multiplex qPCR
wherein all the targets are amplified in a single reaction well.
[0072] Also an embodiment of this invention is a method to quantitate the
integrity of
circulating cell free human DNA and optionally to implement a treatment of a
subject,
comprising: providing a sample from a subject, preferably a sample that has
been treated to
remove cellsõ the sample comprising cell free human DNA comprising a first
short RE
nucleic acid target having a length between 60 and 135 base pairs, 70bp and
about 130bp,
e.g., 71 and 132 base pairs, or between 60 and 120bp (the first RE target),
and the second RE
nucleic acid target having a length between 200 to 300 base pairs, between
about 207 and 270
bp, or between 260 and 265 base pairs; using a quantitative polymerase chain
reaction
(qPCR) method to quantitate the first and second RE targets; obtaining for the
quantitated RE
nucleic acid targets a threshold cycle number; comparing the threshold cycle
number with a
standard curve to determine a quantity of each of the RE nucleic acid targets
that was present
in the sample; calculating a ratio of the quantity of the short RE target to
the quantity of
second RE target; and using the quantitated nucleic acid targets to quantitate
the integrity of
the circulating cell free human DNA and to implement treatment of a patient.
The subject's
sample may be serum, plasma, urine, or other biological fluid from a human,
preferably the
sample is a plasma sample. The first and second RE target may be a target of
the same
retrotransposable element or may be different retrotransposable elements. If
they are from
the same retrotransposable element then PCR blockers may be included to limit
extension
from the primers beyond the position of the blockers, thus limiting the
extension from a
primer pair used to amplify one RE target into the other RE target and thereby
enhancing the
specificity by limiting the production of extraneous or overlapping products.
In an
embodiment the first and second RE targets are targets of an ALU, an SVA or a
LINE1
target. In an embodiment the first and second RE targets are targets of an ALU
or SVA
target or a LINE1 target. Preferably the short RE target is an ALU or an SVA
target, e.g., a
Yb8 ALU target, and the long RE element is an SVA or LINE1 target. In an
embodiment
the prime pairs used in the qPCR to quantitate the RE targets are selected
from the primer
pairs of Table 2A and 2B and 2C . The targets may be amplified in singleplex
qPCR wherein
a single target is amplified in a single reaction well or the targets may be
amplified in a
multiplex qPCR wherein all the targets are amplified in a single reaction
well.
28
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0073] An embodiment of this invention is a method to quantitate the integrity
of circulating
cell free human DNA and optionally to implement a treatment of a subject,
comprising:
providing a sample from a subject, preferably a sample that has been treated
to
remove cells, the sample comprising cell free human DNA comprising a two short
RE
targets, i.e., first RE nucleic acid target and a second RE nucleic acid
target having a
length of having a length between 60 and 135 base pairs, e.g., 71 and 132 base
pairs
or between 60 and 120bp, and optionally a third long RE target having a length
of
between 200bp and 300 bp, between about 207bp to about 270 base pairs, between
260 and 265 base pairs, e.g., 263 bp;
using a quantitative polymerase chain reaction (qPCR) method to quantitate the
three
targets;
obtaining for the targets a threshold cycle number; comparing the threshold
cycle
number with a standard curve to determine a quantity of each of the targets
that was
present in the sample;
calculating the difference between the quantity of the first target and the
second target
and optionally calculating a ratio of the quantity of the first or second
target to the
quantity of the third target; and
using the quantitated targets to implement treatment of a patient. The
subject's
sample may be serum, plasma, urine, or other biological fluid from a human,
preferably the sample is a plasma sample.
The RE targets may be amplified in singleplex qPCR wherein a single target is
amplified in a
single reaction well or the targets may be amplified in a multiplex qPCR
wherein all the
targets are amplified in a single reaction well.
[0074] The methods of this invention are contemplated to be useful in
identifying a subject
having progressive disease or MRD. Accordingly, an embodiment of this
invention is a
method for identifying a subject having progressive cancer or MRD, said method
comprising:
29
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
(a) providing a first and second sample of serum, plasma, urine, or other
biological fluid from a subject wherein the first and second samples are
obtained at least one week apart, at least 2 weeks apart, at least 3 weeks
apart
or at least 4 weeks apart, e.g., 12 to 21 days apart,
the samples comprising cell free human DNA (cfDNA), the cfDNA
comprising (i) a first and second short retrotransposable interspersed element
(RE) target sequence having a length of between about 60 base pairs to about
135 base pairs and (ii) a long RE target having a length of between 200 base
pairs and about 300 base pairs, wherein the first short target is shorter than
the
second short RE target;
(b) quantitating each of the short and long RE targets in the first and second
samples using a quantitative polymerase chain reaction (qPCR) method;
(c) obtaining for each of the quantitated RE targets in the first and second
samples
a threshold cycle number;
(d) comparing the threshold cycle number of each quantitated RE target with a
standard curve to determine an amount of each of the quantitated RE targets
that were present in the samples;
(e) determining the amount of first short RE target less the amount of the
second
RE target in the first sample (Fragl) and the amount of first short RE target
less the amount of the second RE target in the second sample (Frag2) wherein
in increase in Frag2 as compared to Fragl over a threshold level identifies
the
subject as having progressive disease or MRD.
The targets may be amplified in singleplex PCR wherein a single target is
amplified in a
single reaction well or the targets may be amplified in a multiplex PCR
wherein all the targets
are amplified in a single reaction well.
[0075] A subject identified as having progressive disease or MRD may be
administered a
cancer therapy or MRD therapy. The method may further comprise the step of
determining
the DNA integrity index (DII) of the cfDNA in the sample
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0076] In an embodiment of this method, an increase in Frag2 as compared to
Fragl may be
determined by subtracting Fragl from Frag2 to generate a value, FragDiff, that
is compared
to a threshold value and based on that comparison it is concluded that the
ctDNA has
increased and identifies the subject as having progressive disease or MRD and
an appropriate
therapy may be administered.
[0077] Neoadjuvant therapies, which include, e.g., chemotherapy, hormone
therapy,
immunotherapy, radiation therapy, and targeted therapy are delivered to a
subject before the
main treatment is administered to help reduce the size of a tumor or kill
cancer cells that have
spread. Neoadjuvant therapies are recommended when a patient with early-stage
cancer,
stage I, stage II or stage III, undergoes surgery or radiation therapy. The
methods of this
invention may be applied to a sample of subject having a stage I, stage II,
stage III or stage
IV cancer wherein the samples are obtained from the subject before and after
the neoadjuvant
therapy to quantitate the integrity of circulating cell free human DNA and to
implement a
treatment of a subject. The methods of this invention may also be applied to
samples from a
subject who has had a therapy for hepatoma, esophageal cancer, rectal cancer,
anal cancer,
head and neck cancer, colon cancer, colorectal cancer, lung cancer, breast
cancer, neu
metastatic breast cancer or a blood cancer, e.g., leukemia, and the first
sample was taken from
the subject before administering the a first cycle of therapy and the second
sample was taken
from the subject after administering the first therapy cycle, but before the
administration of
another cycle of therapy, and as such the first and second samples may be
obtained from the
subject at least 1 week apart, at least 2 weeks apart, at least 3 weeks apart,
at least 4 weeks
apart, e.g. 12 to 21 days apart. The therapy may be a targeted therapy, a
chemotherapy,
immunotherapy or radiotherapy. The therapy may be treatment with an
antineoplastic agents,
alkylating agents, topoisomerase inhibitors, mitotic inhibitors, methotrexate,
vinca alkaloids,
antimetabolites, antifolates, pyrimidine antagonists, purine analogs, purine
antagonists,
proteasome inhibitors, tyrosine kinase inhibitors, nitrogen mustards,
immunotherapy, or
another cancer therapy.
[0078] In the methods described herein for identifying the patient as having
progressive
disease or MRD, The short and long retrotransposable elements may have a copy
number in
excess of 1000 copies per genome, e.g., the short retrotransposable
interspersed element may
be an ALU or an SVA and the long RE may be an ALU, SVA or LINE.
31
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0079] The short RE targets may be from about 60 base pairs to about 135 base
pairs, or from
about 60 base pairs to about 120 base pairs, or from about 70 base pairs to
about 130 base
pairs, or from about 80 base pairs to about 100 base pairs. The long RE target
may be about
200 bp to about 300 bp or about 207 bp to about 270 bp, or about 260 bp to
about 265 bp in
length.
[0080] The forward and reverse primer pairs used to amplify the short and long
target
sequences in the qPCR may be selected from the following forward and reverse
primer pairs
of Tables 2A, 2B, or 2C.
[0081] The samples used in the methods described herein may be a sample of
serum, plasma,
urine, or other biological fluid, preferably the sample is a plasma sample.
[0082] The method may further comprise a step of adding a synthetic DNA
sequence as an
internal positive control (IPC) to the samples prior to quantitating each of
the short and long
RE targets in the first and second samples by qPCR, and then quantitating the
IPC and
utilizing the quantitative IPC result in the step of comparing the threshold
cycle number of
each quantitated RE target with a standard curve to improve the accuracy and
reliability of
the comparing step. For example, the use of the IPC enables a determination of
a
concentration of cell free DNA in the sample.
[0083] It is specifically contemplated that the quantitation of the short and
long
retrotransposable interspersed elements of each sample by qPCR may be carried
out in a
single tube or well.
[0084] The amplified RE targets may be detected with one or more hybridization
probes that
hybridize specifically to the RE targets sequences. The probes may comprise an
observable
label, e.g., a fluorescent label, e.g., FAM, Cy5, Hex, or Cy3. The observable
label could be
detected using a microfluidic device. In some embodiments of this invention,
the
amplification products of the qPCR method used in the methods of this
invention may be
detected and/or quantified using electrical biosensors (see Liu et al. Single-
Nucleotide
Polymorphism Genotyping Using a Novel Multiplexed Electrochemical Biosensor
with
Nonfouling Surface. Biosens. Bioelectron. 2013, 42, 516-521).
32
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0085] Also an embodiment of this invention is a system for characterizing
cancer or MRD
in a patient, the system comprising:
(a) a sample of serum, plasma, urine, or other biological fluid from a
patient, the
sample comprising cell free DNA, the cell free DNA comprising a first and
second short retrotransposable element targets, the short targets having a
length of from about 60 bp to about 135 bp, the sample further comprising an
added third target, the third target being an internal positive control (IPC)
comprising synthetic DNA;
(b) a TaqMan probe corresponding to each of the two short targets, and the
third
target, each probe comprising a detectable label that is distinct from the
labels
incorporated into the other probes;
(c) a forward primer and a reverse primer pair for amplifying each of the
short
targets, and the third target;
(d) a DNA standard for generating standard curves for the two short targets;
(e) a qPCR system for simultaneously amplifying the short targets, and the
third
target and for producing a threshold cycle number for each the short targets
and the third target; and
(f) a qPCR data analysis system for producing DNA quantitation values for each
retrotransposable element target by interpolation using threshold cycle
numbers and standard curves and for using the DNA quantitation values to
produce an indication of the amount of the two short retrotransposable element
target, and determining a difference in the amount of the first short RE
target
less the amount of the second RE target compared to a threshold value and
characterizing the cancer or MRD.
The system may be a singleplex system wherein a single target is amplified in
a single
reaction well or the system may be a multiplex system where multiple target
are amplified in
a single reaction well.
33
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0086] In the system of this invention for characterizing cancer or MRD in a
patient the
patient may be a patient who is suffering from a cancer, e.g., is diagnosed as
having a stage 1,
stage II or stage III cancer, is in remission from cancer, is at high risk for
developing cancer,
has been categorized by another method as having a complete response ("CR"), a
stable
disease ("SD"), a partial response ("PR"), or progressive disease ("PD"), or
has had a
neoadjuvant therapy, has had surgery to remove a tumor, or has undergone
chemotherapy,
immunotherapy or radiotherapy to treat the cancer or MRD. The cfDNA in the
multiplex
system may further comprise cfDNA comprising a long retrotransposable element
target
having a length of between 200 bp and 300 bp, or 207 bp to 270 bp, e.g., 260-
267 bp, a
TaqMan probe corresponding to the long RE target, and forward and reverse
primers for
amplifying the long RE target. In the system, the forward primer and reverse
primer pair for
amplifying the RE targets are selected from Table 2A, 2B or 2C.
[0087] The method of this invention are contemplated for allow for the
quantitated RE target
amounts to be correlated to one cancer cell in 500,000 total cells or greater,
one cancer cell in
1,000,000 total cells or greater, one cancer cell in 1,500,000 cells or
greater.
[0088] EXAMPLES
[0089] Example 1 - Protocol for Serum and Plasma Separation
[0090] Serum and plasma separation were performed according to the standard
protocol and
within four hours of collection, and stored at -80 C until they were
processed. Care was taken
to avoid freeze-thaw cycles. For serum specimens, whole blood is collected in
the
commercially available red-topped test tube Vacutainer (Becton Dickinson). For
plasma
specimens, whole blood is collected in the commercially available
anticoagulant-treated tubes
e.g., EDTA-treated or citrate-treated.
[0091] Example 2 - Protocol for Direct DNA Quantitation
[0092] Two separate protocols have previously been described for direct DNA
quantification
from either human serum (Umetani, N., et al., Increased integrity of free
circulating DNA in
sera of patients with colorectal or periampullary cancer: Direct quantitative
PCR for ALU
repeats, Clin. Chem. 52(6): 1062-1069 (2006),
doi:10.1373/clinchem.2006.068577,
incorporated herein in its entirety) or plasma (Breitbach, S, et al., Direct
quantification of
34
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
cell-free, circulating DNA from unpurified plasma, PLOS One 9(3): e87838
(2014),
doi:10.1371/journal.pone.0087838, incorporated herein in its entirety). We
used both of these
methods on serum and plasma and compared the amplification efficiency from
both methods.
The first method includes deactivation or elimination of proteins that bind to
template DNA
or DNA polymerase and might invalidate qPCR results. Briefly, a volume of 20
tL of each
serum or plasma sample was mixed with 20 tL of a preparation buffer that
contains 25 mL/L
Tween 20, 50 mM Tris, and 1 mM EDTA. This mixture was then digested with 16
[tg of
proteinase K solution (Qiagen) at 50 C. for 20 min, followed by 5 min of heat
deactivation
and inactivation at 95 C. After subsequent centrifugation at 10,000 g for 5
min, 0.2 tL of the
supernatant (containing 0.1-4, equivalent volume of serum or plasma) was used
as a
template for each direct RE-qPCR reaction. The second method bypasses the
protein removal
step and only requires 1:40 dilution of the serum/plasma sample with sterile
H20.
[0093] Example 3 - Procedure for DNA Purification
[0094] For comparison to and validation of direct quantification of cfDNA, RE-
qPCR has
been performed on isolated, purified cfDNA. cfDNA was purified by magnetic
bead
extraction or by using the silica based membrane QIAamp DNA Investigator Kit
(Qiagen).
[0095] Example 4 - Design of Primers and TaqMang Probes for ALU Yb8 and SVA
targets
[0096] In general primers and labeled probes used in the qPCR reactions may be
obtained
from Eurofins MWG/Operon, Integrated DNA Technologies, or a variety of other
vendors.
[0097] Short ALU primer sets were designed to produce amplicon lengths of 80
bp, 97 bp,
105bp, 120 bp, and 123 bp among others, were developed for use in the assays
of the present
invention. The primer sequences are shown in Table 2A. Primer pairs to produce
amplicon
lengths from SVA of 100bp, 104bp, 106 bp, 116bp, 118bp, 126bp, 132bp, 207 bp,
257 bp,
265 bp, 290 bp, or to produce amplicon lengths of 252bp, 257bp, 262bp, and
267bp from
LINE1, among others are set forth in Table 2B and 2C. The primer pairs were
developed
using Primer 3 software and an SVA or LINE1 (genebank ID: AH005269 (PUBMED
10655552) retrotransposon sequence. Because the SVA and LINE1 sequences are
truncated
in many individuals and also have sequence similarities with ALU sequences in
certain
regions, the target SVA sequences were selected from the SVA-R region, and the
target
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
LINE1 sequence was selected from the LINE1 ORF2 region, which have no or
minimal
sequence similarity as compared with the ALU sequence. The primer sequences
and probes
that hybridize to the amplified targets are shown in Table 2A, 2B and 2C.
[0098] Additional primer design based on ALU Yb8, SVA and LINE1 may be done
using
Primer software (Koressaar, T; Remm, M, Bioinformatics 23(10): 1289-91 (2007),
doi:10.1093/bioinformatics /btm091; Untergasser A, et al., Nucleic Acids Res.
40(15): el 15
(2012), doi:10.1093/nar/gks596).
[0099] Example 5 - Procedure for qPCR
[0100] The qPCR assays were run on an Applied Biosystems 7500 Real Time PCR
instrument and/or the Biorad CFX, but useful instrument platforms are not
limited thereto.
The qPCR assays of the present invention may be adapted to work on most Real-
Time PCR
instruments. To assess the concentration and integrity index of serum and
plasma circulating
cfDNA, both short and long fragments may be amplified and quantified. The
short fragment
primer sets may amplify the short (apoptotic) DNA fragments, whereas the long
fragment
primer sets may amplify the long (non-apoptotic) DNA fragments. The RE-qPCR
multiplex
reaction may contain three targets in a Taqman based assay: a short RE target,
a long RE
target, and a synthetic IPC sequence. The hybridization probes detecting each
target may be
labeled with different fluorophores (e.g. FAM, Cy5, Hex, or Cy3) to enable
simultaneous
detection. The following PCR conditions may be used, but they can be modified
as necessary:
min 95 C denaturation cycle, followed by 32 cycles of 2-step qPCR (15 s at 95
C and 2
min at 61 C combined annealing/extension time) at maximum ramp speed.
Additional PCR
parameters (i.e. cycle number, denaturation and annealing/extension times and
temperatures)
are investigated to obtain a robust, sensitive qPCR multiplex.
[0101] Short Yb8 and long SVA primer pairs selected from those shown in Table
2A and 2B
were combined into eight different multiplex sets (Yb8-80 & SVA-207, Yb8-80 &
SVA-257,
Yb8-80 & SVA-265, Yb8-80 & SVA-290, Yb8-120 & SVA-207, Yb8-120 & SVA-257,
Yb8-120 & SVA-265, and Yb8-120 & SVA-290). The optimal temperature for each
multiplex was determined by a temperature gradient ranging from 64.0 C to
55.0 C. The
concentration of primers and additives including DMSO and additional MgCl2
were
optimized for each multiplex set.
36
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
[0102] The reaction mixture of each multiplex Yb8-SVA-qPCR included a
template, forward
primer and reverse primer pairs, fluorescent probe, Brilliant Multiplex QPCR
Master Mix
(Agilent) and the additives bovine serum albumin (BSA), dimethyl sulfoxide
(DMSO), and
magnesium chloride (MgCl2). Real-time PCR amplification was performed with pre-
cycling
heat activation of DNA polymerase at 95 C for 10 min followed by 32 cycles of
denaturation
at 95 C for 15 sec and extension at 61-62.5 C (adapted to the multiplex set)
in a CFX96
Touch Real-Time PCR Detection System (Bio-Rad Laboratories). The
quantification of DNA
in each sample was determined by use of a calibration curve with serial
dilutions (20ng/u1 to
0.6pg/u1).
[0103] Example 6 - Procedure for qPCR ¨ 97 bp fragment of ALU Yb8
[0104] The qPCR assays may be run on an Applied Biosystems 7500 Real Time PCR
instrument and/or the Biorad CFX, but useful instrument platforms are not
limited thereto.
The qPCR assays of the present invention may be adapted to work on most Real-
Time PCR
instruments. To assess the concentration of a 97bp target of ALU Y8b in plasma
circulating
cfDNA in control healthy subjects compared to cancer patients, plasma samples
of control
(healthy subjects) and test (samples from patients with metastatic colorectal
cancer (mCRC))
containing cfDNA was combined with the 97 bp forward primer
(GTGGCTCACGCCTGTAAT)(SEQ ID NO: 7) , 97 bp reverse primer
(GGGTTTCACCTTGTTAGCCA) (SEQ ID NO: 8), a fluorescent probe comprising
TGGATCATGAGGTCAGGAGAT (SEQ ID NO: 9), Brilliant Multiplex QPCR Master Mix
(Agilent) and the additives bovine serum albumin (BSA), dimethyl sulfoxide
(DMSO), and
magnesium chloride (MgCl2). Real-time PCR amplification was performed with pre-
cycling
heat activation of DNA polymerase at 95 C for 10 min followed by 40 cycles of
denaturation
at 90-95 C for 10-15 sec and extension at 61-64 C (depending on the
multiplex set) in an
ABI 7500 Instrument (ThermoFisher Scientific). The quantification of DNA in
each sample
was determined by use of a calibration curve with serial dilutions (20 ng/ul
to 0.6 pg/ul)
[0105] Example 7 - Procedure for qPCR 97 bp ALU Yb8 and 80 bp ALU Yb8
[0106] To assess the concentration of plasma circulating cfDNA, both 80bp and
97bp
fragments of ALU Yb8 are amplified and quantified. The RE-qPCR multiplex
reaction
contains three targets in a TaqMang based assay: 80 bp ALU Yb8 forward and
reverse
37
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
primers (GGAAGCGGAGCTTGCAGTGA (SEQ ID NO:1) and
AGACGGAGTCTCGCTCTGT CGC (SEQ ID NO: 2)) , the 97 bp ALU Yb8 RE forward
and reverse primers GTGGCTCACGCCTGTAAT (SEQ ID NO: 7) and
GGGTTTCACCTTGTTAGCCA(SEQ ID NO:8)), an 80R- blocker, peptide nucleic acid
(PNA) oligo which binds to the 80 bp ALU Yb8 fragment, a 97R-PNA blocker a PNA
which
binds to the 97 bp ALU Yb8 fragment and probes that hybridize to the 80 bp and
the 97 bp
sequences (e.g., Fluorophore- TGAGGTCAGGAGATCGAGACCATCC-Quencher)(SEQ ID
NO: 92), and in some instances a synthetic IPC sequence. PNA oligo mimics DNA.
In PNA,
the negatively-charged sugar phosphate backbone of DNA is replaced with an
uncharged
pseudo-peptide backbone. The two strands of a PNA/DNA hybrid therefore lack
the
electrostatic repulsion as observed for DNA/DNA duplexes, giving rise to
thermal stability.
Hybridization probes are also included in some instances for detecting each
target and the
probes are labeled with different fluorophores (e.g. FAM, Cy5, Hex, or Cy3) to
enable
simultaneous detection. The reaction mixture includes the forward primers,
reverse primers,
the blockers, the fluorescent probe, Brilliant Multiplex QPCR Master Mix
(Agilent) and the
additives bovine serum albumin (BSA), dimethyl sulfoxide (DMSO), and magnesium
chloride (MgCl2). Real-time PCR amplification is performed with pre-cycling
heat activation
of DNA polymerase at 95 C for 10 min followed by 32 cycles of denaturation at
95 C for 15
sec and extension at 61-62.5 C (depending on the multiplex set) in a CFX96
Touch Real-
Time PCR Detection System (Bio-Rad Laboratories). The quantification of DNA in
each
sample is determined by use of a calibration curve with serial dilutions
(20ng/u1 to 0.6pg/u1).
[0107] Figure 2 shows the two PCR target regions of 80bp and 97bp on the Yb8
sequence. In
order to amplify the 97bp and 80bp separately, we use two peptide nucleic acid
(PNA) oligos
to block PCR extension beyond the target regions. PNA oligos are used as
sequence specific
PCR blockers because PNA probes have strong binding affinity and specificity
to its target
DNA and are not recognized by DNA polymerase as primer. In the diagram 97F-
Blocker
binds the complement sequence of Alu-Yb8 between 97bp and 80bp target regions
and
prevents DNA elongation from the 97bp forward primer beyond the region where
the 97bp
reverse primer binds. In a similar way, 80R-Blocker binds Alu-Yb8 sequence
between 97bp
and 80bp target regions and prevents DNA elongation from the 80bp reverse
primer beyond
the region where the 80bp forward primer binds. By incorporating these two PCR
blockers, it
is possible to prevent PCR amplification of nearly entire Alu-Yb8 sequence
that can occur
38
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
with the 97F/80R primer pair. We are then able to compare the amounts of the
80 bp and 97
bp fragments in the plasma of control samples from healthy subjects and the
plasma from
cancer patients.
[0108] Example 8 - Procedure for qPCR 80 bp ALU Yb8 and 120 bp ALU Yb8 and 265
SVA
[0109] Plasma from 40 control subjects, healthy subjects without cancer and 39
subjects
having cancer were subjected to qPCR assays to assess the level of ctDNA. The
qPCR
assays were run on an Applied Biosystems 7500 Real Time PCR instrument and/or
the
Biorad CFX, but useful instrument platforms are not limited thereto. The qPCR
assays of the
present invention may be adapted to work on most Real-Time PCR instruments. To
assess the
concentration and integrity index of serum circulating cfDNA in the samples
from control
and cancer subject, a first ALU Yb8 target of 80 bp, and a second ALU Yb8
target of 120 bp
and an SVA target of 265 bp were amplified and quantified in a RE-qPCR
multiplex reaction.
The RE-qPCR multiplex reaction contained three targets in a Taqman based
assay: the first
ALU Yb8 target of 80 bp , the second ALU Yb8 target of 120 bp, a third SVA
target of 265
bp, and a synthetic internal positive control (IPC) sequence. The
hybridization probes
detecting each amplified target were labeled with different fluorophores (FAM,
Cy5, or Hex)
to enable simultaneous detection. The following PCR conditions are used: 10
min 95 C
denaturation cycle, followed by 40 cycles of 2-step qPCR (15 s at 96 C and 2
min at 64 C
combined annealing/extension time) at maximum ramp speed.
[0110] The reaction mixture of each multiplex Yb8-qPCR included the forward
primers and
reverse primers for the first Yb8-80 target and for the second ALU Yb8-120
target, and the
long SVA 265 target (see Table 2A and 3B for primer pair sequences). the
fluorescent probes
for detecting the amplified fragments, Brilliant Multiplex QPCR Master Mix
(Agilent) and
the additives bovine serum albumin (BSA), dimethyl sulfoxide (DMSO), and
magnesium
chloride (MgCl2). Real-time PCR amplification was performed with pre-cycling
heat
activation of DNA polymerase at 95 C for 10 min followed by 40 cycles of
denaturation at
95 C for 15 sec and extension at 61-62.5 C (depending on the multiplex set)
in a CFX96
Touch Real-Time PCR Detection System (Bio-Rad Laboratories). The
quantification of the
targets in each sample was determined by use of a calibration curve with
serial dilutions
39
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
(20ng/u1 to 0.6pg/u1). Table 3 sets forth the quantitated amounts of the 80bp
target, the 120
bp target and the 265 bp target in each sample and the difference in the
quantitated amount of
80 bp Yb8 target and the quantitated amount of 120 bp Yb8 target.
Table 3
Healthy Control (ng/mL) Cancer patient (ng/mL)
80bp- 80bp-
120bp 120bp
Median 1.85 Median 25.60
Average 2.22 Average 217.52
SD 1.87 SD 793.35
Target Size (bp) Target Size (bp)
Sample 120b 265b 80bp- Sample
80bp-
ID# 80bp p P 120bp ED# 80bp 120bp 265bp 120bp
1287 4.68 2.98 1.06 1.69 1683 16.13 10.07 3.19
6.06
1125.5 114.4
1289 4.77 3.24 1.01 1.53 1685 2 468.29 7
657.23
1291 5.51 3.64 0.79 1.88 1689 24.00 14.81 3.54
9.19
1293 9.93 6.13 2.04 3.80 1691 108.65 50.63
8.78 58.02
20.8
1295 2 9.56 2.25 11.26 1693 30.47 17.16 3.94
13.31
11.7
1299 9 6.52 1.99 5.26 1695 109.66 61.52 12.89
48.14
1301 7.40 5.32 2.02 2.08 1697 142.73 71.97
16.07 70.76
12.1
1303 9 5.93 2.37 6.26 1699 98.21 46.29 10.31
51.92
1305 5.07 3.73 1.26 1.34 1701 458.87 163.28
36.89 295.60
1307 3.71 2.17 0.96 1.53 1703 19.19 8.19 2.30
11.00
1311 8.89 4.87 1.40 4.02 1705 36.65 17.82 6.06
18.84
1313 5.47 3.03 0.88 2.44 1707 25.60 12.62 2.13
12.98
1315 4.60 2.59 1.04 2.01 1709 14.10 8.33 2.83
5.77
1316 3.07 1.72 0.59 1.35 1711 66.40 30.19 8.93
36.21
1317 4.17 2.19 0.73 1.98 1713 116.21 67.75
16.99 48.47
1318 5.92 3.09 1.18 2.84 1715 86.71 39.03 3.47
47.68
1319 4.15 2.98 1.38 1.17 1717 154.01 56.55
16.67 97.46
1320 6.82 4.77 1.73 2.05 1719 69.53 26.53 6.86
43.00
1321 3.09 1.93 0.65 1.16 1721 22.51 11.52 3.22
10.98
7395.8 2497.7 372.1
1322 2.25 1.61 0.64 0.65 1723 7 1 1
4898.16
1323 3.97 2.09 0.81 1.88 1725 182.55 95.65
32.66 86.91
1324 5.49 3.63 1.55 1.86 1727 10.26 6.80 2.77
3.46
CA 03164737 2022-06-14
WO 2021/127462
PCT/US2020/066048
Table 3
Healthy Control (ng/mL) Cancer patient (ng/mL)
80bp- 80bp-
120bp 120bp
Median 1.85 Median 25.60
Average 2.22 Average 217.52
SD 1.87 SD 793.35
Target Size (bp) Target Size (bp)
Sample 120b 265b 80bp- Sample
80bp-
ID# 80bp p P 120bp ED# 80bp 120bp 265bp 120bp
1325 4.74 3.13 0.90 1.61 1729 281.95 104.02
31.17 177.93
1329 2.27 1.69 0.63 0.58 1731 401.77 168.31
39.10 233.46
1331 3.29 1.99 0.34 1.31 1733 17.73 8.26 2.78
9.47
1333 8.64 6.80 1.94 1.84 1735 21.73 11.47 2.26
10.26
1421 6.73 4.30 1.72 2.43 1737 67.34 29.16 8.38
38.18
10.3
1431 9 8.24 2.77 2.15 1739 55.40 29.80 8.20
25.60
1435 5.00 3.93 1.24 1.07 1741 381.36 178.68
46.72 202.68
1439 7.39 4.24 1.13 3.15 1743 38.51 15.86 3.56
22.66
1643.1 129.6
1441 2.56 1.62 0.54 0.94 1745 9 612.70 __ 8 1030.49
1443 6.03 4.12 1.30 1.91 1747 18.35 8.52 2.51
9.83
1447 2.51 1.97 0.67 0.54 1749 23.30 14.88 5.17
8.43
1449 4.52 2.80 1.15 1.72 1751 53.04 46.31 31.81
6.74
10.1
1451 7 8.20 2.65 1.98 1753 31.03 17.16 4.19
13.87
1453 4.73 3.49 1.09 1.24 1755 22.51 13.15 4.35
9.37
1455 7.34 5.81 1.63 1.54 1757 8.00 5.37 1.85
2.64
1457 7.43 6.18 1.23 1.25 1759 46.28 36.00 8.47
10.28
1459 7.61 5.60 1.90 2.00 1761 282.85 142.45
25.42 140.40
1765 4.77 3.36 1.16 1.41
[0111] Example 9 - Procedure for qPCR Data Analysis and Quality Control
[0112] Data analysis was performed utilizing the respective AB 7500,
QuantStudio-5 or
BioRad CFX instrument software. Melt curve analysis was generated using
Qiagen's
QuantiTect 1 SYBR1 Green PCR Kit (Cat#204141) and operated using the Applied
Biosystems 7500 Real Time PCR instrument. For each experiment, a freshly
prepared 3-fold
serial dilution of high molecular weight standard DNA (ranging from 10 ng/IIL
to 0.004
41
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
ng/IlL) was run in duplicate on each plate to generate standard curves for the
long and short
targets. The standard curves are plotted CT vs. Delta R. (the fluorescence
emission intensity
of the reporter dye divided by the fluorescence emission intensity of the
passive reference
dye). Resultant DNA quantitation values are interpolated from the resulting
linear standard
curves. At least one negative No Template Control (NTC) was run on each plate.
[0113] In experiments where a ratio between DNA concentration of a 265bp SVA
long target
and 80bp ALU short target were calculated, the DNA concentration of the long
target divided
by DNA concentration of the short target provides an indication as to the
degree of DNA
integrity for the quantified sample. DNA integrity index is calculated as the
ratio of
concentrations ([concentration of long RE marker]/[concentration of short RE
marker]).
Quality metrics, including PCR efficiencies (i.e. slope) of both short and
long targets, Y-
intercept values, and verification of no true amplification in negative
controls was assessed.
[0114] Example 10 - Identifying Patients with Increasing ctDNA
[0115] Blinded samples of plasma from 66 cancer patients were subjected to a
qPCR to
assess the level of ctDNA using the methods described herein and identify
patients as having
progressive disease. The plasma samples were from patients who had been
previously
diagnosed as having either colorectal cancer, non-small cell lung cancer,
small cell lung
cancer or breast cancer and had received either chemotherapy, targeted
therapy,
immunotherapy, or a combination of therapies.
[0116] A first plasma sample was obtained from the patients before receiving a
cycle of
therapy and a second plasma sample was obtained 12 days to 21 days after the
cycle of
therapy and before receiving another cycle of therapy. The qPCR assays were
run on an
Applied Biosystems 7500 Real Time PCR instrument and/or the Biorad CFX, but
useful
instrument platforms are not limited thereto and the qPCR assays of the
present invention
may be adapted to work on most Real-Time PCR instruments.
[0117] To assess the concentration and integrity index of the cfDNA in the
samples from
control and cancer patients, a first ALU Yb8 target of 80 bp, and a second ALU
Yb8 target of
105 bp and an SVA target of 265 bp were amplified and quantified in a RE-qPCR
multiplex
reaction. The sequence of the primer pairs used to amplify yb-8-80, yb-8 105
and SVA 265
42
CA 03164737 2022-06-14
WO 2021/127462 PCT/US2020/066048
are set forth in Table 1. The RE-qPCR multiplex reaction was a Taqmang based
assay
comprising Brilliant Multiplex QPCR Master Mix (Agilent), bovine serum albumin
(BSA),
dimethyl sulfoxide (DMSO), and magnesium chloride (MgCl2), and comprised the
plasma
sample, a primer pair for amplifying the first ALU yb-8 target of 80 bp, a
primer pair for
amplifying the second ALU Yb8 target of 105 bp, and a primer pair for
amplifying a third
SVA target of 265 bp, and a synthetic internal positive control (IPC). The
amplification
products were detected with hybridization probes for the Yb-8 80bp target, the
Yb8-105bp
target and the SVA 265 bp target, each labeled with a different fluorophore to
enable
simultaneous detection of the different amplified targets.
[0118] Real-time PCR amplification was performed with pre-cycling heat
activation of DNA
polymerase in a QuantStudio-5 (Thermofisher Scientifics). The quantification
of the RE
targets in each sample was determined by use of a calibration curve with
serial dilutions
(20ng/u1 to 0.6pg/u1) and the difference between the amount of the first Yb8
80bp target and
the amount of the second Yb8 105bp target in the first sample (Fragl) and the
amount of the
first Yb8 80bp target and the amount of the second Yb8 105bp target in the
second samples
of plasma (Frag2) were calculated. The difference between Frag2 and Frag I
(Frag Diff) was
also calculated. An increase in the amount of the 80bp yb-8 target as compared
to the 105bp
yb-8 target in the second sample as compared to the first sample indicated an
increase in the
ctDNA.
[0119] Figure 3 depicts the FragDiff of the 66 patients, who had been
classified as having
progressive disease (triangles), or having non-progressive disease (circles).
Figure 3
demonstrates that the method disclosed herein rapidly assesses cfDNA
integrity. Figure 3
also demonstrates that based upon the FragDiff being above a threshold level,
the method
rapidly and reliably identifies a patient as having progressive disease see
also Figure 4. Thus,
the method described herein can also be used as a factor for rapidly
concluding the patient
has progressive disease or the cancer treatment was not effective. This is in
contrast to other
standard assays, e.g., CT scans, Xrays, and CEA measurements, that require
weeks, if not
months, before it is determined the patient has progressive disease and a
therapy can be
identified as ineffective.
43
CA 03164737 2022-06-14
WO 2021/127462
PCT/US2020/066048
[0120] While the invention has been described and illustrated with reference
to certain
particular embodiments thereof, those skilled in the art will appreciate that
various
adaptations, changes, modifications, substitutions, deletions, or additions of
procedures and
protocols may be made without departing from the spirit and scope of the
invention. It is
intended, therefore, that the invention be defined by the scope of the claims
that follow and
that such claims be interpreted as broadly as is reasonable. All references
cited herein are
incorporated by reference.
44