Note: Descriptions are shown in the official language in which they were submitted.
WO 2021/163706
PCT/US2021/018263
PANOMIC GENOMIC PREVALENCE SCORE
CLAIM OF PRIORITY
This application claims the benefit of U.S. Provisional Patent Application
Serial Nos.
62/977,015, filed on February 14, 2020; 63/014,515, filed on April 23, 2020;
63/052,363, filed on July
5 15, 2020; and 63/145,305, filed on February 3, 2021; the entire contents
of which applications are
hereby incorporated by reference in their entirety.
This application is related to International Patent Publication
WO/2020/146554, entitled
Genomic Profiling Similarity and based on International Patent Application
PCT/US2020/012815
filed on January 8, 2020, the entire contents of which application is hereby
incorporated by reference
10 in its entirety
TECHNICAL FIELD
The present disclosure relates to the fields of data structures, data
processing, and machine
learning, and their use in precision medicine, e.g., tumor characterization
including without lirnitation
the usc of molecular profiling to predict an attribute of a biological sample
such as the primary origin,
15 organ type, histology and/or cancer type.
BACKGROUND
Carcinoma of Unknown Primary (CUP) represents a clinically challenging
heterogeneous
group of metastatic malignancies in which a primary tumor remains elusive
despite extensive clinical
and pathologic evaluation. Approximately 2-4% of cancer diagnoses worldwide
comprise CUP. See,
20 e.g., Varadhachary. New Strategies for Carcinoma of Unknown Primary: the
role of tissue of origin
molecular profiling. Clin Cancer Res. 2013 Aug 1;19(15) A027-33. In addition,
some level of
diagnostic uncertainty with respect to an exact tumor type classification is a
frequent occurrence
across oncologic subspecialties. Efforts to secure a definitive diagnosis can
prolong the diagnostic
process and delay treatment initiation. Furthermore, CI IP is associated with
poor outcome which
25 might be explained by use of suboptirnal therapeutic intervention.
Inununohistochemical (MC)
testing is the gold standard method to diagnose the site of tumor origin,
especially in cases of poorly
differentiated or undifferentiated tumors. Assessing the accuracy in
challenging cases and performing
a meta-analysis of these studies reported that MC analysis had an accuracy of
66% in the
characterization of metastatic tumors. See, e.g., Brown RW, et al.
Immunohistochemical identification
30 of tumor markers in metastatic adenocarcinoma: a diagnostic adjunct in
the determination of primary
site. Am J Clin Pathol 1997, 107:12e19; Dennis JL, et al. Markers of
adenocarcinoma characteristic of
the site of origin: development of a diagnostic algorithm. Clin Cancer Res
2005, 11:3766e3772;
Gamble AR, et al. Use of tumour marker imrnunoreactivity to identify primary
site of metastatic
cancer. BMJ 1993, 306:295e298; Park SY, et al. Panels of immunohistochemical
markers help
1
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
determine primary sites of metastatic adenocarcinoma. Arch Pathol Lab Med
2007, 131:1561e1567;
DeYoung BR, Wick MR. Irnmunohistologic evaluation of metastatic carcinomas of
unknown origin:
an algorithmic approach. Semin Diagn Pathol 2000, 17:184e193; Anderson CIG,
Weiss T.M.
Determining tissue of origin for metastatic cancers: meta-analysis and
literature review of
5 irnmunohistochemistry performance. App! Irnmunohistochem Mol Morphol
2010, 18:3e8. Since
therapeutic regimes can be dependent upon diagnosis, this represents an
important unmet clinical
need.
To address these challenges, assays aiming at tissue-of-origin (TOO)
identification based on
assessment of differential gene expression have been developed and tested
clinically. However,
10 integration of such assays into clinical practice is hampered by
relatively poor performance
characteristics (from 83% to 89%) and limited sample availability. See, e.g.,
Pillai R, et al. Validation
and reproducibility of a microarray-based gene expression test for tumor
identification in formalin-
fixed, paraffm-embedded specimens. J Mol Diagn 2011, 13:48e56; Rosenwald S,
etal. Validation of a
microRNA-based qRT-PCR test for accurate identification of tumor tissue
origin. Mod Pathol 2010,
15 23:814e823; Kerr SE, et al. Multisite validation study to determine
performance characteristics of a
92-gene molecular cancer classifier. Clin Cancer Res 2012, 18:3952e3960; Kucab
JE, etal. A
Compendium of Mutational Signatures of Environmental Agents. Cell. 2019 May
2;177(4):821-
836.e16. For example, a recent commercial RNA-based assay has a sensitivity of
83% in a test set of
187 tumors and confirmed results on only 78% of a separate 300 sample
validation set. See
20 Hainsworth JD, et al, Molecular gene expression profiling to predict the
tissue of origin and direct
site-specific therapy in patients with carcinoma of unknown primary site: a
prospective trial of the
Sarah Cannon research institute. J Chin Oncol. 2013 Jan 10;31(2):217-23. This
may, at least in part, be
a consequence of limitations of typical RNA-based assays in regards to normal
cell contamination,
RNA stability, and dynamics of RNA expression. Thus, there is a need for more
robust approaches to
25 TOO identification to aid cancer patients, particularly but not limited
to CUP.
Machine learning models can be configured to analyze labeled training data and
then draw
inferences from the training data. Once the machine learning model has been
trained, sets of data that
are not labeled may be provided to the machine learning model as an input. The
machine learning
model may process the input data, e.g., molecular profiling data, and make
predictions about the input
30 based on inferences learned during training. The present disclosure
further provides a voting
methodology to combine multiple classifier models to achieve more accurate
classification than that
achieved by use a single model.
Comprehensive molecular profiling provides a wealth of data concerning the
molecular status
of patient samples. We have performed such profiling on well over 100,000
tumor patients from
35 practically all cancer lineages. Patient and molecular data can be
processed using machine learning
algorithms to identity additional biomarker signatures that can be used to
characterize various
phenotypes of interest. Here, this "next generation profiling" (NGP) approach
has been applied to
2
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
build models to predict an attribute of a biological sample, including without
limitation such as the
primary origin, organ type, histology and/or cancer type.
SUMIVIARY
Comprehensive molecular profiling provides a wealth of data concerning the
molecular status
5 of patient samples. Such data can be compared to patient response to
treatments to identify biomarker
signatures that predict response or non-response to such treatments. Herein we
provide systems and
methods to predict attributes of a patient sample, including without
limitation a tissue-of-origin
(TOO).
In an aspect, the disclosure provides a data processing apparatus for
generating input data
10 structure for use in training a machine learning model to predict at
least one attribute of a biological
sample, wherein the at least one attribute is selected from the group
comprising a primary tumor
origin, cancer/disease type, organ group, histology, and any combination
thereof, the data processing
apparatus including one or more processors and one or more storage devices
storing instructions that
when executed by the one or more processors cause the one or more processors
to perform operations,
15 the operations comprising: obtaining, by the data processing apparatus
one or more biomarker data
structures and one or more sample data structures; extracting, by the data
processing apparatus, first
data representing one or more biomarkers associated with the sample from the
one or more biomarker
data structures, second data representing the sample data from the one or more
sample data structures,
and third data representing a predicted at least one attribute; generating, by
the data processing
20 apparatus, a data structure, for input to a machine learning model,
based on the first data representing
the one or more biomarkers and the second data representing the predicted at
least one attribute and
sample; providing, by the data processing apparatus, the generated data
structure as an input to the
machine learning model; obtaining, by the data processing apparatus, an output
generated by the
machine learning model based on the machine learning model's processing of the
generated data
25 structure; determining, by the data processing apparatus, a difference
between the third data
representing a predicted predicted at least one attribute for the sample and
the output generated by the
machine learning model; and adjusting, by the data processing apparatus, one
or more parameters of
the machine learning model based on the difference between the third data
representing a predicted
predicted at least one attribute for the sample and the output generated by
the machine learning model.
30 In some embodiments, the set of one or more biomarkers include one or
more biomarkers listed in any
one of Tables 121-129, Tables 117-120, ENTSM1, any table selected from Tables
2-116, and any
combination thereof, optionally wherein the set of one or more biomarkers
comprises one or more
biomarkers listed in any one of Table 117, Table 118, Table 119, Table 120,
INSM1, or any
combination thereof. In some embodiments, the set of one or more biomarkers
include each of the
35 biomarkers. In some embodiments, the set of one or more biomarkers
includes at least one of these
biomarkers, optionally wherein the set of one or more biomarkers comprises
each of the biomarkers in
3
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Table 118, Table 119, Table 120, and INSM1, and wherein optionally the set of
one or more
biomarkers further comprises the markers in any table selected from Tables 2-
116.
In an aspect, the disclosure provides a data processing apparatus for
generating input data
structure for use in training a machine learning model to predict at least one
attribute of a biological
5 sample, wherein the at least one attribute is selected from the group
comprising a primary tumor
origin, cancer/disease type, organ group, histology, and any combination
thereof, the data processing
apparatus including one or more processors and one or more storage devices
storing instructions that
when executed by the one or more processors cause the one or more processors
to perform operations,
the operations comprising: obtaining, by the data processing apparatus, a
first data structure that
10 structures data representing a set of one or more biomarkers associated
with a biological sample from
a first distributed data source, wherein the first data structure includes a
key value that identifies the
sample; storing, by the data processing apparatus, the first data structure in
one or more memory
devices; obtaining, by the data processing apparatus, a second data structure
that structures data
representing data for the at least one attribute for the sample having the one
or more biomarkers from
15 a second distributed data source, wherein the data for the at least one
attribute includes data
identifying a sample, at least one attribute, and an indication of the
predicted at least one attribute,
wherein second data structure also includes a key value that identifies the
sample; storing, by the data
processing apparatus, the second data structure in the one or more memory
devices; generating, by the
data processing apparatus and using the first data structure and the second
data structure stored in the
20 memory devices, a labeled training data structure that includes (i) data
representing the set of one or
more biomarkers and the sample, and (ii) a label that provides an indication
of a predicted at least one
attribute, wherein generating, by the data processing apparatus and using the
first data structure and
the second data structure includes correlating, by the data processing
apparatus, the first data structure
that structures the data representing the set of one or more biomarkers
associated with the sample with
25 the second data structure representing predicted at least one attribute
data for the sample having the
one or more biomarkers based on the key value that identifies the subject: and
training, by the data
processing apparatus, a machine learning model using the generated label
training data structure,
wherein training the machine learning model using the generated labeled
training data structure
includes providing, by the data processing apparatus and to the machine
learning model, the generated
30 label training data structure as an input to the machine learning model.
In some embodiments, the
operations further comprise: obtaining, by the data processing apparatus and
from the machine
learning model, an output generated by the machine learning model based on the
machine learning
model's processing of the generated labeled training data structure; and
determining, by the data
processing apparatus, a difference between the output generated by the machine
learning model and
35 the label that provides an indication of the predicted at least one
attribute. In some embodiments, the
operations further comprise: adjusting, by the data processing apparatus, one
or more parameters of
the machine learning model based on the determined difference between the
output generated by the
4
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
machine learning model and the label that provides an indication of the
predicted at least one attribute.
In some embodiments, the set of one or more biomarkers include one or more
biomarkers listed in any
one of Tables 121-129, Tables 117-120, INSM1, any table selected from Tables 2-
116, and any
combination thereof, optionally wherein the set of one or more biomarkers
comprises one or more
5 biomarkers listed in any one of Table 117, Table 118, Table 119, Table
120, INSM1, or any
combination thereof. In some embodiments, the set of one or more hiomarkers
include each of the
biomarkers. In some embodiments, the set of one or more biomarkers includes at
least one of these
biomarkers, optionally wherein the set of one or more biomarkers comprises
each of the biomarkers in
Table 118, Table 119, Table 120, and INTSM1, and wherein optionally the set of
one or more
10 biomarkers further comprises the markers in any table selected from
Tables 2-116.
The disclosure also provides a method comprising steps that correspond to each
of the
operations described above. The disclosure also provides a system comprising
one or more computers
and one or more storage media storing instructions that, when executed by the
one or more computers,
cause the one or more computers to perform each of the operations described
above. The disclosure
15 also provides a non-transitory computer-readable medium storing software
comprising instructions
executable by one or more computers which, upon such execution, cause the one
or more computers
to perform the operations described above.
In an aspect, the disclosure provides a method for determining at least one
attribute of a
biological sample, wherein the at least one attribute is selected from the
group comprising a primary
20 tumor origin, cancer/disease type, organ group, histology, and any
combination thereof, the method
comprising: for each particular machine learning model of a plurality of
machine learning models that
have each been trained to perform an prediction operation between received
input data representing a
sample and the at least one attribute: providing, to the particular machine
learning model, input data
representing a sample of a subject, wherein the sample was obtained from
tissue or an organ of the
25 subject; and obtaining output data, generated by the particular machine
learning model based on the
particular machine learning model's processing the provided input data, that
represents a probability
or likelihood that the sample represented by the provided input data
corresponds to the at least one
attribute; providing, to a voting unit, the output data obtained for each of
the plurality of machine
learning models, wherein the provided output data includes data representing
initial sample attributes
30 determined by each of the plurality of machine learning models; and
determining, by the voting unit
and based on the provided output data, the predicted at least one attribute.
In some embodiments, the
predicted at least one attribute is determined by applying a majority rule to
the provided output data,
by using the provided output data as input into a dynamic voting model, or a
combination thereof In
some embodiments, the determining, by the voting unit and based on the
provided output data, the
35 predicted at least one attribute comprises: determining, by the voting
unit, a number of occurrences of
each initial attribute class of the multiple candidate attribute classes; and
selecting, by the voting unit,
the initial attribute class of the multiple candidate attribute classes having
the highest number of
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
occurrences. In some embodiments, each machine learning model of the plurality
of machine learning
models comprises a random forest classification algorithm, boosted tree,
support vector machine,
logistic regression, k-nearest neighbor model, artificial neural network,
naive flayes model, quadratic
discriminant analysis, Gaussian processes model, or any combination thereof In
some embodiments,
5 each machine learning model of the plurality of machine learning models
comprises a random forest
classification algorithm. In some embodiments, each machine learning model of
the plurality of
machine learning models comprises a boosted tree classification algorithm. In
some embodiments,
the plurality of machine learning models includes multiple representations of
a same type of
classification algorithm. In some embodiments, the input data represents a
description of (i) sample
10 attributes and (ii) origins. In some embodiments, the multiple candidate
attribute classes include at
least one class for prostate, bladder, endocervix, peritoneum, stomach,
esophagus, ovary, parietal
lobe, cervix, endometriurn, liver, sigmoid colon, upper-outer quadrant of
breast, uterus, pancreas,
head of pancreas, rectum, colon, breast, intrahepatic bile duct, cecum,
gastroesophageal junction,
frontal lobe, kidney, tail of pancreas, ascending colon, descending colon,
gallbladder, appendix,
15 rectosigmoid colon, fallopian tube, brain, lung, temporal lobe, lower
third of esophagus, upper-inner
quadrant of breast, transverse colon, and skin. In some embodiments, the
multiple candidate attribute
classes include at least at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, or all
21 of breast adenocarcinoma, central nervous system cancer, cervical
adenocarcinoma,
cholangiocarcinoma, colon adenocarcinoma, gastrocsophageal adenocarcinoma,
gastrointestinal
20 stromal tumor (GIST), hepatocellular carcinoma, lung adenocarcinoma,
melanoma, meningioma,
ovarian granulosa cell tumor, ovarian & fallopian tube adenocarcinoma,
pancreas adenocarcinoma,
prostate adenocarcinoma, renal cell carcinoma, squamous cell carcinoma,
thyroid cancer, urothelial
carcinoma, uterine endometrial adenocarcinoma, and uterine sarcoma. In some
embodiments, the
sample attributes includes one or more biomarkers for the sample, wherein
optionally the one or more
25 biomarkers comprises one or more biomarkers listed in any one of Tables
121-129, Tables 117-120,
INISM1, any table selected from Tables 2-116, and any combination thereof,
optionally wherein the
set of one or more biomarkers comprises one or more biomarkers listed in any
one of Table 117,
Table 118, Table 119, Table 120, INSM1, or any combination thereof In some
embodiments, the set
of one or more biomarkers include each of the biomarkers. In some embodiments,
the set of one or
30 more biomarkers includes at least one of these biomarkers, optionally
wherein the set of one or more
biomarkers comprises each of the biomarkers in Table 118, Table 119, Table
120, and INSM1, and
wherein optionally the set of one or more biomarkers further comprises the
markers in any table
selected from Tables 2-116. In some embodiments, the input data further
includes data representing a
description of the sample and/or subject. The disclosure also provides a
system comprising one or
35 more computers and one or more storage media storing instructions that,
when executed by the one or
more computers, cause the one or more computers to perform each of the
operations described above.
The disclosure also provides a non-transitory computer-readable medium storing
software comprising
6
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
instructions executable by one or more computers which, upon such execution,
cause the one or more
computers to perform the operations described above.
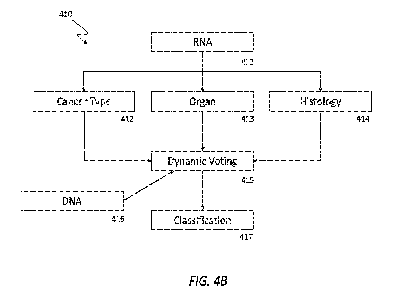
1. In an aspect, the disclosure provides a method for
classifying a biological sample, the
method comprising: obtaining, by one or more computers, first data
representing one or more initial
5 classifications for the biological sample that were previously determined
based on RNA sequences of
the biological sample; obtaining, by one or more computers, second data
representing another initial
classification for the biological sample that were previously determined based
on DNA sequences of
the biological sample; providing, by one or more computers, at least a portion
of the first data and the
second data as an input to a dynamic voting engine that has been trained to
predict a target biological
10 sample classification based on processing of multiple initial biological
sample classifications;
processing, by one or more computers, the provided input data through the
dynamic voting engine;
obtaining, by one or more computers, output data generated by the dynamic
voting engine based on
the dynamic voting engine's processing of the provided input data; and
determining, by one or more
computers, a target biological sample classification for the biological sample
based on the obtained
15 output data. In some embodiments, the obtaining, by one or more
computers, first data representing
one or more initial classifications for the biological sample that were
previously determined based on
RNA sequences of the biological sample comprises: obtaining data representing
a cancer type
classification for the biological sample based the RNA sequences of the
biological sample; obtaining
data representing an organ from which the biological sample originated based
on the RNA sequences
20 of the biological sample; and obtaining data representing a histology
for the biological sample based
on the RNA sequences of the biological sample, and wherein providing at least
a portion of the first
data and the second data as an input to the dynamic voting engine comprises:
providing the obtained
data representing the cancer type classification, the obtained data
representing the organ from which
the biological sample originated, the obtained data representing the
histology, and the second data as
25 an input to the dynamic voting engine. In some embodiments, the dynamic
voting engine comprises
one or more machine learning model. In some embodiments, training the dynamic
voting engine
comprises: obtaining a labeled training data item that includes (I) one or
more initial classifications
that include data indicating a cancer classification type, data indicating an
initial organ of origin, data
indicating a histology, or data indicating output of a DNA analysis engine and
(H) a target biological
30 sample classification, generating training input data for input to the
dynamic voting engine based on
the obtained training data item, processing the generated training input data
through the dynamic
voting engine, obtaining output data generated by the dynamic voting engine
based on the dynamic
voting engine's processing of the generated training input data, and adjusting
one or more parameters
of the dynamic voting engine based on the level of similarity between the
output data and the label of
35 the obtained training data item.
7
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
In some embodiments, previously determining an initial classification for the
biological
sample based on DNA sequences of the biological sample comprises: receiving,
by one or more
computers, a biological signature representing the biological sample that was
obtained from a
cancerous neoplasm in a first portion of a body, wherein the model includes a
cancerous biological
5 signature for each of multiple different types of cancerous biological
samples, wherein each of the
cancerous biological signatures include at least a first cancerous biological
signature representing a
molecular profile of a cancerous biological sample from the first portion of
one or more other bodies
and a second cancerous biological signature representing a molecular profile
of a cancerous biological
sample from a second portion of one or more other bodies; performing, by one
or more computers and
10 using a pairwise-analysis model, pairwise analysis of the biological
signature using the first cancerous
biological signature and the second cancerous biological signature;
generating, by one or more
computers and based on the performed pairwise analysis, a likelihood that the
cancerous neoplasm in
the first portion of the body was caused by cancer in a second portion of the
body; and storing, by one
or more computers, the generated likelihood in a memory device. The disclosure
also provides a
15 system comprising one or more computers and one or more storage media
storing instructions that,
when executed by the one or more computers, cause the one or more computers to
perform each of the
operations described above. The disclosure also provides a non-transitory
computer-readable medium
storing software comprising instructions executable by one or more computers
which, upon such
execution, cause the one or more computers to perform the operations described
above.
20 In an aspect, the disclosure provides a method comprising: (a) obtaining
a biological sample
from a subject having a cancer; (b) performing at least one assay on the
sample to assess one or more
biomarkers, thereby obtaining a biosignature for the sample; (c) providing the
biosignature into a
model that has been trained to predict at least one attribute of the cancer,
wherein the model
comprises at least one pre-determined biosignature indicative of at least one
attribute, and wherein the
25 at least one attribute of the cancer is selected from the group
comprising primary tumor origin,
cancer/disease type, organ group, histology, and any combination thereof; (d)
processing, by one or
more computers, the provided biosignature through the model; and (e)
outputting from the model a
prediction of the at least one attribute of the cancer.
In the methods provided herein, the biological sample may comprise forrnalin-
fixed paraffm-
30 embedded (1414PE) tissue, fixed tissue, a core needle biopsy, a fine
needle aspirate, unstained slides,
fresh frozen (FF) tissue, formalin samples, tissue comprised in a solution
that preserves nucleic acid
or protein molecules, a fresh sample, a malignant fluid, a bodily fluid, a
tumor sample, a tissue
sample, or any combination thereof In some embodiments, the biological sample
comprises cells
from a solid tumor, a bodily fluid, or a combination thereof. In some
embodiments, the bodily fluid
35 comprises a malignant fluid, a pleural fluid, a peritoneal fluid, or any
combination thereof. In some
embodiments, the bodily fluid comprises peripheral blood, sera, plasma,
ascites, urine, cerebrospinal
fluid (CSF), sputum, saliva, bone marrow, synovial fluid, aqueous humor,
amniotic fluid, cerumen,
8
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
breast milk, broncheoalveolar lavage fluid, semen, prostatic fluid, Cowper's
fluid, pre-ejaculatory
fluid, female ejaculate, sweat, fecal matter, tears, cyst fluid, pleural
fluid, peritoneal fluid, pericardial
fluid, lymph, chyme, chyle, bile, interstitial fluid, menses, pus, sebum,
vomit, vaginal secretions,
mueosal secretion, stool water, pancreatic juice, lavage fluids from sinus
cavities, bronehopulmonary
5 aspirates, blastocyst cavity fluid, or umbilical cord blood.
In the methods provided herein, performing the at least one assay in step (b)
may comprise
determining a presence, level, or state of a protein or nucleic acid for each
of the one or more
biomarkers, wherein optionally the nucleic acid comprises deoxyribonucleic
acid (DNA), ribonucleic
acid (RNA), or a combination thereof. In some embodiments, the presence, level
or state of at least
10 one of the proteins is determined using a technique selected from
itnmunohistochemistry (11-1C), flow
cytometry, an i llllll unoassay, an antibody or functional fragment thereof,
an aptamer, mass
spectrometry, or any combination thereof, wherein optionally the presence,
level or state of all of the
proteins is determined using the technique; and/or the presence, level or
state of at least one of the
nucleic acids is determined using a technique selected from polymerase chain
reaction (PCR), in situ
15 hybridization, amplification, hybridization, microarray, nucleic acid
sequencing, dye termination
sequencing, pyrosequencing, next generation sequencing (NGS; high-throughput
sequencing), whole
exome sequencing, whole genome sequencing, whole transcriptome sequencing, or
any combination
thereof, wherein optionally the presence, level or state of all of the nucleic
acids is determined using
the technique. In some embodiments, the state of the nucleic acid comprises a
sequence, mutation,
20 polymorphism, deletion, insertion, substitution, translocation, fusion,
break, duplication,
amplification, repeat, copy number, copy number variation (CNV; copy number
alteration; CNA), or
any combination thereof. In some embodiments, the state of the nucleic acid
consists of or comprises
a copy number. In some embodiments, the at least one assay comprises next-
generation sequencing,
wherein optionally the next-generation sequencing is used to assess: i) at
least one of the genes,
25 genomic information / signatures, and fusion transcripts in any of
Tables 121-130, or any
combination thereof ii) at least one of the genes and/or transcripts in any
table selected from Tables
117-120, IN SM1, and any combination thereof; in) the whole exome or
substantially the whole
exome; iv) the whole transcriptome or substantially the whole transcriptome;
v) at least one gene in
any table selected from Tables 2-116, and any combination thereof; or vi) any
combination thereof.
30 In the methods provided herein, predicting the at least one attribute of
the cancer may
comprise determining a probability that the attribute is each member of a
plurality of such attributes
and selecting the attribute with the highest probability.
In some embodiments of the methods provided herein, the primary tumor origin
or plurality
of primary tumor origins consists of, comprises, or comprises at least 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11,
35 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, or
all 38 of prostate, bladder, endocervix, peritoneum, stomach, esophagus,
ovary, parietal lobe, cervix,
endometrium, liver, sigrnoid colon, upper-outer quadrant of breast, uterus,
pancreas, head of pancreas,
9
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
rectum, colon, breast, intrahepatic bile duct, cecum, gastroesophageal
junction, frontal lobe, kidney,
tail of pancreas, ascending colon, descending colon, gallbladder, appendix,
rectosigmoid colon,
fallopian tube, brain, lung, temporal lobe, lower third of esophagus, upper-
inner quadrant of breast,
transverse colon, and skin. In some embodiments, the primary tumor origin or
plurality of primary
5 tumor origins consists of, comprises, or comprises at least 1, 2, 3, 4,
5, 6, 7, 8,9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19,20, or all 21 of breast adenocarcinoma, central nervous
system cancer, cervical
adenocarcinoma, cholangiocarcinoma, colon adenocarcinoma, gastroesophageal
adenocarcinoma,
gastrointestinal stromal tumor (GIST), hepatocellular carcinoma, lung
adenocarcinoma, melanoma,
meningioma, ovarian granulosa cell tumor, ovarian & fallopian tube
adenocarcinoma, pancreas
10 adenocarcinoma, prostate adenocarcinoma, renal cell carcinoma, squamous
cell carcinoma, thyroid
cancer, urothelial carcinoma, uterine endometrial adenocarcinoma, and uterine
sarcoma. In some
embodiments, the cancer/disease type consists of, comprises, or comprises at
least 1, 2, 3,4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, or
all 28 of adrenal cortical
carcinoma; bile duct, cholangiocarcinoma; breast carcinoma; central nervous
system (CNS); cervix
15 carcinoma; colon carcinoma; endometrium carcinoma; gastrointestinal
stromal tumor (GIST);
gastroesophageal carcinoma; kidney renal cell carcinoma; liver hepatocellular
carcinoma; lung
carcinoma; melanoma; meningioma; Merkel; neuroendocrine; ovary granulosa cell
tumor; ovary,
fallopian, peritoneum; pancreas carcinoma; pleural mesothelioma; prostate
adenocarcinoma;
retroperitoneum; salivary and parotid; small intestine adenocarcinoma;
squamous cell carcinoma;
20 thyroid carcinoma; urothelial carcinoma; uterus. In some embodiments,
the organ group consists of,
comprises, or comprises at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, or all 17 of adrenal
gland; bladder; brain; breast; colon; eye; female genital tract and peritoneum
(FGTP);
gastroesophageal; head, face or neck, NOS; kidney; liver, gallbladder, ducts;
lung; pancreas; prostate;
skin; small intestine; thyroid. In some embodiments, the histology consists
of, comprises, or
25 comprises at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, or all 29 of adenocarcinoma, adenoid cystic carcinoma,
adenosquamous carcinoma,
adrenal cortical carcinoma, astrocytoma, carcinoma, carcinosarcoma,
cholangiocarcinoma, clear cell
carcinoma, ductal carcinoma in situ (DCIS), glioblastoma (GBM), GIST, glioma,
granulosa cell
tumor, infiltrating lobular carcinoma, leiomyosarcoma, liposarcoma, melanoma,
meningioma, Merkel
30 cell carcinoma, mesothelioma, neuroendocrine, non-small cell carcinoma,
oligodendroglioma,
sarcoma, sarcomatoid carcinoma, serous, small cell carcinoma, squamous.
In some embodiments of the methods provided herein, the at least one pre-
determined
biosignature indicative of the at least one attribute of the cancer, wherein
optionally the at least one
attribute is a cancer/disease type, comprises selections of biomarkers
according to Table 118, wherein
35 optionally: i. a pre-determined biosignature indicative of adrenal
cortical carcinoma consists of,
comprises, or comprises at least, 1,2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, or 20
features selected from [MIA, MD31, SYP, CDII1, NKX3-1, CALB2, KRT19, MUC1,
5100A5,
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CD34, TNIPRSS2, KRT8, NCANI2, ARG1, TG, NCANI1, SERPINA1, PSAP, TPM3, and
ACVRL1;
ii. a pre-determined biosignature indicative of bile duct, cholangiocarcinoma
consists of, comprises,
or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15,
16,17, 18, 19, or 20 features
selected from HNF1B, VTL1, SERPINA1, ESR1, AN01, SOX2, MUC4, S100A2, KRT5,
KRT7,
5 CNN1, AR, EN02, S100A9, NKX2-2, SATB2, PSAP, S100A6, CALB2, and TMPRSS2;
iii. a pre-
determined biosignature indicative of breast carcinoma consists of, comprises,
or comprises at least, 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features
selected from GATA3,
ANKRD30A, KRT15, KRT7, S100A2, PAX8, MUC4, KRT18, HNF1B, S100A1, PIP, SOX2,
MDM2, MUCSAC, PMEL, TFF1, KRT16, KRT6B, S100A6, and SERPINB5; iv. a pre-
determined
10 biosignature indicative of central nervous system (CNS) consists of,
comprises, or comprises at least,
1, 2, 3, 4, 5, 6, 7, 3, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20
features selected from S100B,
KRT18, KRT8, SOX2, AN01, NCANIL PDPN, NKX2-2, KRT19, S100A14, S100A11, S100A1,
MSH2, CEACANIL GPC3, ERBB2, TG, KRT7, CGB3, and S100A2; v. a pre-determined
biosignature indicative of cervix carcinoma consists oT comprises, or
comprises at least, 1, 2, 3, 4, 5,
15 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features
selected from ESR1, CDKN2A,
CCND1, LIN28A, PGR, SMARCB1, CEACANI4, S100B, FUT4, PSAP, MUC2, MDM2, NCANI1,
SATB2, TNFRSF8, CD79A, S100A13, VFIL, CD3G, and TPSAB1; vi. a pre-determined
biosignature
indicative of colon carcinoma consists of, comprises, or comprises at least,
1, 2, 3, 4, 5, 6, 7, 8,9, 10,
11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected from CDX2, KRT7,
MUC2, KRT20, MUC1,
20 SATB2, vllA, CEACAM5, CDH17, S100A6, CEACANI20, KRT6B, TFF3, FUT4, BCL2,
KRT6A,
KRT18, CEACAM18, TFF1, and MLH1; vii. a pre-determined biosignature indicative
of
endometrium carcinoma consists of, comprises, or comprises at least, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected from PAX8, PGR, ESR1,
VIIL, CALD1,
LIN28B, NAPSA, KRT5, S100A6, DES, FLI1, DSC3, SlOOP, CEACAM16, PDPN, ARG1,
TLE1,
25 WT1, BCL6, and MLH1; viii. a pre-determined biosignature indicative of
gastrointestinal stromal
tumor (GIST) consists of, comprises, or comprises at least, 1, 2, 3, 4, 5, 6,
7, 8,9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19, or 20 features selected from ANOL SDC1, KR:119, MUC1,
KR.18, ACVRL1,
KIT, CDH1, S100A2, KRT7, ERBB2, SI00A16, EN02, S100A9, TPSAB1, KRT17, PAX8,
PGR,
ESR1, and VTIL; ix. a pre-determined biosignature indicative of
gastroesophageal carcinoma consists
30 of, comprises, or comprises at least, 1,2, 3,4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, or 20
features selected from FUT4, CDX2, SERPENB5, MUC5AC, AR, TFF1, NCANI2, TFF3,
ISL1,
AN01, VILl, PAX8, SOX2, CEACA_M6, 5100A13, EN02, NAPSA, TPSAB1, S100B, and
CD34; x.
a pre-determined biosignature indicative of kidney renal cell carcinoma
consists oT comprises, or
comprises at least, 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, or 20 features selected
35 from PAX8, CDH1, CDKN2A, SlOOP, S100A14, HAVCR1, KL, KRT7, MUC1,
POU5F1,
WEL, PAX2, AMACR, BCL6, S100A13, CA9, MDM2, SALL4, and SYP; xi. a pre-
determined
biosignature indicative of liver hepatocellular carcinoma consists of,
comprises, or comprises at least,
11
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20
features selected from SEMINAL
CEACAM16, KRT19, AFP, MUC4, CEACAM5, MSII2, BCL6, DSC3, ICRT15, S100A6,
CEACAM20, GPC3, MUC I , CD34, VITT , ERBB2, POU5F1, KRT18, and KRT16; xii. a
pre-
determined biosignature indicative of lung carcinoma consists of, comprises,
or comprises at least, 1,
5 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20
features selected from NAPSA, SOX2,
CEACAM7, KRT7, S100A10, CEACAM6, S100A1, PAX8, AR, VT-H., S100A13, CD991,2,
KRT5,
MUC1, CEACAME SFTPA1, TMPRSS2, TFF1, KRT15, and MUC4; xiii. a pre-determined
biosignature indicative of melanoma consists of, comprises, or comprises at
least, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected from S100B,
KRT8, PMEL, KRT19,
10 MUC1, MLANA, S100A14, S100A13, MITE, S100A1, VIM, CDKN2A, ACVRL1, MS4A1,
POIJ5F1, TPM1, IJPK3A, SlOOP, CiATA3, and CEACAM1; xiv. a pre-determined
biosignature
indicative of meningioma consists of, comprises, or comprises at least, 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected from SDC1, KRT8, AN01,
VIM, S100A14,
S100A2, CEACAM1, MSH2, PGR, KRT10, TP63, CD5, ENTIA, CDH1, CCND1, MDM2, KRT16,
15 SPN, SMARCB1, and S100A9; xv a pre-determined biosignature indicative of
Merkel cell carcinoma
consists of, comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8,9, 10,
11, 12, 13, 14, 15, 16, 17, 18,
19, or 20 features selected from ISL1, ERBB2, S100Al2, S100A14, MYOG, SDC1,
KRT7,
SlOOPBP, MME, TMPRSS2, CEACAM5, CPS1, CR1, MUC4, CEACAM4, CA9, EN02, FLI1,
LI1,128B, and MLANA; xvi. a pre-determined biosignature indicative of
neuroendocrine consists of,
20 comprises, or comprises at least, 1,2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, or 20
features selected from NCAME ISL1, EN02, POU5F1, TFF3, SYP, TPM4, S100A1,
S1OOZ, MUC4,
MPO, DSC3, CEACAM4, S100A7, ERBB2, CDX2, S100A11, KRT10, CEACAM5, and
CEACAM3; xvii. a pre-determined biosignature indicative of ovary granulosa
cell tumor consists of,
comprises, or comprises at least, at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19,
25 or 20 features selected from FOXL2, SDC1, MSH6, MUC1, KRT8, PGR, MME,
SERPINA1, FLI1,
S100B, CEACAM21, AMACR, KRT1, SETPA1, TPM1, CALCA, S100A11, NCAME ISL1, and
EN 02; xviii. a pre-determined biosignature indicative of ovary, fallopian,
peritoneum consists of,
comprises, or comprises at least, 1,2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, or 20
features selected from WT1, PAX8,1NHA, TFE3, S100A13, FOXL2, TLE1, MSLN,
POU5F1,
30 CEACAM3, ALPP, S100A10, FUT4, NKX3-1, CEACAM5, SOX2, ESR1, EN02,
ACVICL1, and
SYP; xix. a pre-determined biosignature indicative of pancreas carcinoma
consists of, comprises, or
comprises at least, 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, or 20 features selected
trom PDX1, GNIA3, AN01, SERPINA1, ISLE MUC5AC, FUT4, SMAD4, CD5, CALB2,
S100A4,
SMN1, ESR1, IIN1711.1, AMACR, MSII2, PDPN, MSLN, T17171, and KRT6C; xx. a pre-
determined
35 biosignature indicative of pleural mesothelioma consists of, comprises,
or comprises at least, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features
selected from UPK3B, CALB2,
WT1, SMARCB1, PDPN, INIIA, CEACAME MSLN, ICRT5, CA9, S100A13, SF1, CDII1,
12
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CDKN2A, FLI1, SYP, CEACAM3, CPS1, SATB2, and BCL6; xxi. a pre-determined
biosignature
indicative of prostate adenocarcinoma consists of, comprises, or comprises at
least, 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected from
KRT7, KT,K3, NKX3-1,
AMACR, SIO0A5, MUC1, MUC2, UPK3A, ICL, CPS1, MSLN, PMEL, CNN1, SERPINA1, KRT2,
5 CGB3, TMPRSS2, CEACAM6, SDC1, and AR; xxii. a pre-determined biosignature
indicative of
retroperitoneum consists of, comprises, or comprises at least, 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19, or 20 features selected from KRT19, KRT18, KRT8, TPM1,
S100A14, CD34,
TPM4, CDH1, CNN1, SDC1, AR, MDM2, KIT, TLE1, CPS1, CDK4, UPK3A, TMPRSS2, TPM3,
and CEACAM1; xxiii a pre-determined biosignature indicative of salivary and
parotid consists of,
10 comprises, or comprises at least, 1,2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, or 20
features selected from EN02, PIP, TPM1, KRT14, S100A1, ERFIT12, TFF1, AT.PP,
DSC3, CTNNB1,
CALB2, SALL4, AN01, CEACAN116, BINF1B, KIT, ARG1, CLACAN118, TMPRSS2, and
HAVCR1; xxiv a pre-determined biosignature indicative of small intestine
adenocarcinoma consists
of, comprises, or comprises at least, 1,2, 3,4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, or 20
15 features selected from PDX1, DES, MUC2, CDH17, CEACAM5, SERPINA1, KRT20,
HNF 1B,
ESR1, ARG1, CD5, TLE1, PMEL, SOX2, SETPA1, MME, CD99L2, MPO, SlOOP, and CA9;
xxv. a
pre-detennined biosignature indicative of squamous cell carcinoma consists of,
comprises, or
comprises at least, 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, or 20 features selected
from TP63, SOX2, KRT6A, KRT17, S100A1, CD3G, SETPA1, AR, KRT5, SDC1, KRT20,
DSC3,
20 CNN1, MSH2, ESR1, S100A2, SERPINB5, PDPN, S100A14, and TPM3; xxvi. a pre-
determined
biosignature indicative of thyroid carcinoma consists of, comprises, or
comprises at least, 1, 2, 3,4, 5,
6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected
from TG, PAX8, CPS1,
S100A2, TPSAB1, CALB2, IINT1B, [NITA, ARG1, CNN1, CDK4, VIM, CEACAM5, TLE1,
TFF3,
KRT8, SlOOP, FOXL2, MUC1, and GATA3; xxvii. a pre-determined biosignature
indicative of
25 urothelial carcinoma consists of, comprises, or comprises at least, 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, or 20 features selected from GATA3, UPK2, KRT20,
MUC1, S100A2,
CPS', "IP63, CALB2, MITE, SlOOP, SERPINA1, DES, CTNN131, MSLN, SALTA, VHL,
KR:17,
CD2, PAX8, and UPK3A; and/or xxviii. a pre-determined biosignature indicative
of uterus consists
of, comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, or 20
30 features selected from KRT19, KRT18, NCAM1, DES, FOXL2, CD79A, S100A14,
ESR1, MSLN,
MITE, UPK3B, TPM1, EN02, SlOOP, MLH1, KRT8, CDH1, TPM4, SATB2, and MDM2.
In some embodiments of the methods provided herein, the at least one pre-
determined
biosignature indicative of the at least one attribute of the cancer, wherein
optionally the at least one
attribute is an organ type, comprises selections of biomarkers according to
Table 119; wherein
35 optionally: i. a pre-determined biosignature indicative of adrenal gland
consists of, comprises, or
comprises at least, 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, or 20 features selected
from INHA, CDII1, SYP, MB31, CALB2, KRT8, PSAP, KRT19, NCAM2, NKX3-1, ARG1,
13
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
SERPINA1, CD34, TPM3, S100A7, ACVRL1, PMEL, CR1, ERG, and PECAM1; ii. a pre-
dektinined biosignature indicative of bladder consists of, comprises, or
comprises at least, 1,2, 3,4,
5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected
from GATA3, KRT20,
UPK2, CPS1, SALL4, SERPI:C.1AI, DES, CALB2, MUC1, 5100A2, MSLN, MITE, PAX8,
S100A10,
5 CNN1, UPK3A, CD3G, NAPSA, CD2, and MME; iii. a pre-determined
biosignature indicative of
brain consists of, comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, or 20 features selected from KRT8, AN01, S100B, S100A14, SOX2,
PDPN, CEACAML
S100A2, NCAM1, MSH2, KRT18, NKX2-2, WT1, S100A1, GPC3, TLE1, CD5, SlOOZ,
S100A16,
and PGR; iv. a pre-determined biosignature indicative of breast consists of,
comprises, or comprises at
10 least, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18,
19, or 20 features selected from
GATA3, ANKRD30A, KRT15, KRT7, S100A2, S100A1, MIJC4, LINF1B, KRTI 8, SOX2,
PIP,
PAX8, MDM2, KRT16, MUC5AC, S100A6, TP63, TFF1, KRT5, and SERPINAl; v. a pre-
determined biosignature indicative of colon consists of, comprises, or
comprises at least, 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected
from CDX2, KRT7, MUC2,
15 KRT20, MUC1, CEACAM5, CDH17, TFF3, KRT18, KRT6B, VILl, SATB2, S100A6,
SOX2,
S100A14, HAVCR1, FUT4, ERG, EINF1B, and PTPRC; vi. a pre-determined
biosignature indicative
of eye consists of, comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, or 20 features selected from PMEL, MLANA, MITE, BCL2, S100A13,
S100A2,
S100A10, S100A1, MD31, SOX2, EN02, S100A16, VIM, VUL, PDPN, WT1, S100B, KRT7,
20 KRT10, and PSAP; vii. a pre-determined biosignature indicative of female
genital tract and
peritoneum (FGTP) consists of, comprises, or comprises at least, 1, 2, 3, 4,
5,6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, or 20 features selected from PAX8, ESR1, WT1, PGR,
CDKN2A, FOXL2,
KRT5, TPM4, SMARCB1, DES, TMPRSS2, CDK4, GATA3, AR, S100A13, MSII2, AN01,
CALB2, MS4A1, and CCND1; viii. a pre-determined biosignature indicative of
gastroesophageal
25 consists of, comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14, 15, 16, 17, 18,
19, or 20 features selected from CDX2, AN01, FUT4, SERPINB5, SPN, NCAM2, vrLi
, CD34,
EN02, 1EE3, AR, S100A13, 1PM1, CEACAM6, SOX2, PAX8, MUC5AC, CURE S100A11, and
ISL1; ix. a pre-determined biosignature indicative of head, face or neck, NOS
consists of, comprises,
or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, or 20 features
30 selected from KR15, DSC3, 1P63, 1-INE113, MUC5AC, PAX5, KR115, PGR,
S100A6, TMPRSS2,
MME, S100B, EN02, CEACAM8, SALL4, ANOI, GATA3, LENI28B, CD99L2, and UPK3A; x.
a
pre-determined biosignature indicative of kidney consists of, comprises, or
comprises at least, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features
selected from PAX8, CDHE
IINF113, S100A14, ILAVCR1, CDKN2A, SlOOP, KL, KRT7, 5100A13, VILL, PAX2,
POU51,1,
35 MUC1, AMACR, EN02, MDM2, WT1, SYP, and AR; xi. a pre-determined
biosignature indicative
of liver, gallbladder, ducts consists of, comprises, or comprises at least, 1,
2, 3, 4, 5, 6, 7, 8,9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected from SERPINAL VELE
IlINFiB, AN01, ESR1,
14
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
SOX2, MUC4, S100A2, EN02, CNN1, POU5F1, ICRT5, S100A9, UPK3B, PSAP, KRT7, KL,
TMPRSS2, SATB2, and S100A14; xii. a pre-determined biosignature indicative of
lung consists of,
comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, or 20
features selected from NAPSA, SOX2, SFTPA1, VFIIL, S100A1, S100A10, AR,
TMPRSS2, CD99L2,
5 CEACAM7, CEACAM6, KRT6A, KRT7, NCAM2, TP63, CEACAM1, MUC4, KRT20, CNN1,
and
IST,1; xiii. a pre-determined biosignature indicative of pancreas consists of,
comprises, or comprises
at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19,
or 20 features selected from
PDX1, AN01, SERPINTA1, GATA3, ISL1, MUC5AC, SMAD4, FUT4, CD5, SNIN1, NIOC2-2,
TFF1, AMACR, SOX2, HINF1B, S1OOZ, MSLN, DES, 5100A4, and CALB2; xiv. a pre-
determined
10 biosignature indicative of prostate consists of, comprises, or comprises
at least, 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 13, 19, or 20 features selected from KT,K3,
KRT7, NICX3-1, AMACR,
CPS1, S100A5, UPK3A, ICE, MUC1, CGB3, MUC2, TMPRSS2, MSLN, PMEL, S100A10,
SERPINA1, KRT20, SFTPA1, BCL6, and TFF1; xv. a pre-determined biosignature
indicative of skin
consists of, comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17, 18,
15 19, or 20 features selected from S100B, KRT8, PMEL, KRT7, KRT19, GATA3,
MDM2, AMACR,
TPM1, TLE1, CEACAM19, CEACAM16, MLANA, TMPRSS2, AR, TFF3, BCL6, CR1, NCAM1,
and MS4A1; xvi. a pre-determined biosignature indicative of small intestine
consists of, comprises, or
comprises at least, 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, or 20 features selected
from MUC2, CDH17, FLI1, KRT20, CDX2, CD5, KRT7, MPO, CNN1, DSC3, DES, AN01,
20 S100A1, CALD1, TFF1, SPN, MITF, TMPRSS2, CALB2, and CEACAM16; and/or
xvii. a pre-
determined biosignature indicative of thyroid consists of, comprises, or
comprises at least, 1,2, 3,4,
5, 6,7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected
from PAX8, TG, CPS1,
SERP1NB5, INTIA, AR.G1, CNN1, CEACANI5, TPSAB1, CALB2, I1NF1B, VIM, CDK4,
SlOOP,
S100A2, L1N28B, TFF3, CGA, TLE1, and TPM3.
25 In some embodiments of the methods provided herein, the at least one pre-
determined
biosignature indicative of the at least one attribute of the cancer, wherein
optionally the at least one
attribute is a histology, comprises selections of biomarkers according to
Table 120; wherein
optionally: i. a pre-determined biosignature indicative of adenocarcinoma
consists of, comprises, or
comprises at least, 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, or 20 features selected
30 from TMPRSS2, fiNF113, KR15, MUC1, CFACAM5, MUC5AC, CDH17, 1P63, ALPP,
GA1A3,
CEACAM1, TFF3, S100A1, KRT8, PDX1, KRT17, CDH1, KLK3, CPS1, and S100A2; ii. a
pre-
determined biosignature indicative of adenoid cystic carcinoma consists of,
comprises, or comprises
at least, 1,2, 3, 4, 5, 6,7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or
20 features selected from
KRT14, KIT, TPM3, CGA, SMAD4, CTNNI31, DSC3, S100A6, TP63, TPM1, CALD1, MiIBi,
CD2,
35 CDH1, AN01, EN02, CD3G, TPM2, CEACAM1, and BCL2; iii. a pre-determined
biosignature
indicative of adenosquamous carcinoma consists of, comprises, or comprises at
least, 1, 2, 3, 4, 5, 6,
7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected from
TP63, SFTPA1, OSCAR,
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
KRT19, KRT15, NAPSA, GPC3, MS4A1, S100Al2, ERG, CEACAM6, VHL, SOX2, SERPEcAl,
KRT6A, CDKN2A, CD3C, PIP, NCAM2, and CEACAIV17; iv. a pre-determined
biosignature
indicative of adrenal cortical carcinoma consists of, comprises, or comprises
at least, 1, 2, 3,4, 5, 6, 7,
8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected from
MB31, INHA, CDH1, SYP,
5 CALB2, NKX3-1, KRT19, ERBB2, MUC1, ARG1, VIM, CD34, CALD1, S100A9, MSLN,
S100A10, CD5, PMETõ SDC1, and TP63; v. a pre-determined biosignature
indicative of astrocytoma
consists of, comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17, 18,
19, or 20 features selected from S100B, SOX2, NCAME MUC1, S100A4, KRT17, KRT8,
S100A1,
TPM4, CNN1, TPM2, OSCAR, AR, SDC1, SALL4, SMN1, SETPA1, KIT, CA9, and S100A9;
vi. a
10 pre-determined biosignature indicative of carcinoma consists of,
comprises, or comprises at least, 1, 2,
3, 4, 5, 6, 7, 9, 10, 11, 12, 13, 14,15, 16,17, 19, or 20 features selected
from GATA3, MITE,
MUC5AC, PDPN, VILE CEACAM5, CDH1, CDH17,1L12B, SlOOP, ICRT20, KRT7, SPN,
TMPRSS2, EN02, NKX2-2, PMEL, IMP3, BCL6, and S100A8; vii. a pre-determined
biosignature
indicative of carcinosarcoina consists of, comprises, or comprises at least,
1, 2, 3, 4, 5, 6,7, 8,9, 10,
15 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected from KRT6B,
GPC3, MSLN, MUC I,
S100A6, S100A2, MME, CDKN2A, CDH1, FOXL2, KRT7, CALB2, SETPA1, ERG, PGR,
KRT17,
NAPSA, CALD1, LIT428B, and KIT; viii. a pre-determined biosignature indicative
of
cholangiocarcinoma consists of, comprises, or comprises at least, 1, 2, 3, 4,
5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, or 20 features selected from SERPINA1, FINF1B,
VIL1, TFF1, EN02,
20 NKX2-2, FUT4, MUC4, MLH1, TMPRSS2, WT1, KL, KRT7, ESR1, MDM2, SETPA1,
SMN1,
KRT18, UPK3B, and COQ2; ix. a pre-determined biosignature indicative of clear
cell carcinoma
consists of, comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17, 18,
19, or 20 features selected from P0U5F1, ITAVCRE CEACAM6, ITNE1B, PAX8, NAPSA,
CD34,
MYOG, FOXL2, MITE, SlOOP, 5100A9, 5100A14, SlOOZ, WT1, CDH1, TTF1, SYP, MLH1,
and
25 KRT16; x. a pre-determined biosignature indicative of ductal carcinoma
in situ (DCIS) consists of,
comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, or 20
features selected from GA1'A3, FINF113, DES, MME, ANKRD30A, SA1132, 50X2,
NCAL\42, PAX8,
CEACAM4, PIP, MUC4, NKX3-1, SERPINA1, KRT20, KIT, NCAME KRT14, S100A2, and
CDKN2A; xi. a pre-determined biosignature indicative of glioblastoma (GBM)
consists of,
30 comprises, or comprises at least, 1,2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, or 20
features selected from S100B, KRT18, PDPN, NKX2-2, SOX2, NCAME KRT8, ERBB2,
KRT15,
KRT19, GATA3, CDKN2A, BCL6, S100A14, KRT10, UPK3A, SF1, CA9, CCND1, and KRT5;
xii.
a pre-determined biosignature indicative of GIST consists of, comprises, or
comprises at least, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features
selected from AN01, SDC1,
35 MUC1, KRT19, KRT8, ACVRL1, KIT, ERBB2, CDH1, CEACAM19, FUT4, TFF3,
S100A16,
S100A13, ISL1, S100A9, TPSAB1, KRT18, IMP3, and KRT3; xiii. a pre-determined
biosignature
indicative of glioma consists of, comprises, or comprises at least, 1, 2, 3,4,
5, 6, 7, 8,9, 10, 11, 12,
16
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
13, 14, 15, 16, 17, 18, 19, or 20 features selected from KRT8, S100B, SYP,
NCAM2, CD3G, SDC1,
SOX2, CEACAM1, POU5I71, MITIL SATB2, MDM2, NCAM1, KRT7, CCB3, CPS1, PDPN,
CAT,CA, ERBB2, and TNERSF8; xiv. a pre-determined biosignature indicative of
granulosa cell
tumor consists of, comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14, 15, 16,
5 17, 18, 19, or 20 features selected from FOXL2, SDC1, MSII6, KRT18, KRT8,
MIME, FLI1,
S100A9, CAT,CA, S100B, CCND1, CEACAM21, TIE], SERPINA1, S100A1 1, SETPA1, SYP,
NCAM2, CD3G, and SOX2; xv. a pre-determined biosignature indicative of
infiltrating lobular
carcinoma consists of, comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19, or 20 features selected from CDH1, GATA3, S100A1, TFF3, CA9,
M1JC1,NKX3-1,
10 ANKRD30A, SOX2, S100A5, MUC4, KRT7, OSCAR, MME, SERP1NA1, CDK4, AR,
CEACAM3,
FICT,6, and KRT5; xvi, a pre-determined biosignature indicative of
leioniyosarcoma consists of,
comprises, or comprises at least, 1,2, 3,4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19, or 20
features selected from KRT19, KRT8, KRT18, CNN1, TPM4, FOXL2, TPM2, TPM1,
CD79A,
CALB2, SATB2, S100A5, DES, S100A14, KRT2, ERBB2, PDPN, EN02, CD2, and CALD1;
xvii. a
15 pre-determined biosignature indicative of liposarcoma consists of,
comprises, or comprises at least, 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features
selected from KRT18,
MDM2, CDK4, CDH1, KRT19, KRT7, PDPN, CD34, TPM4, CR1, ACVRL1, MME, KRT8,
AIVIACR, CEACAM5, S100B, OSCAR, LIN28A, 5100Al2, and SDC1; xviii. a pre-
determined
biosignature indicative of melanoma consists of, comprises, or comprises at
least, 1, 2, 3, 4, 5, 6, 7, 8,
20 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected from
S100B, PMEL, KRT19, KRT8,
MUC1, 5100A14, MLANA, 5100A13, TPM1, MITE, VIM, CEACAM19, POU5F1, SATB2, CPS1,
CDKN2A, KRT10, AR, ACVRL1, and E1N28A; xix. a pre-determined biosignature
indicative of
meningioma consists of, comprises, or comprises at least, 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19, or 20 features selected from SDC1, KRT8, 5100A14, ANOI,
CEACAM1, VIM,
25 KRT10, PGR, MSH2, CD5, S100A2, CDH1, TP63, SMARCB1, KRT16, S100A10,
S100A4, DSC3,
CCND1, and GATA3: xx. a pre-determined biosignature indicative of Merkel cell
carcinoma consists
of, comprises, or comprises at least, 1,2, 3,4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, or 20
features selected from ISL1, ERBB2, MME, MYOG, CPS1, KRT7, SALL4, S100Al2,
S100A14,
SlOOPBP, CR1, SMAD4, CEACAM5, MUC4, CA9, KRT10, SYP, CCND1, MSLN, and MLANA;
30 xxi. a pre-determined biosignature indicative of mesothelioma consists
of, comprises, or comprises at
least, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or
20 features selected from
UPK3B, CALB2, PDPN, SMARCB1, MSLN, KRT5, CEACAM3, WT1, INHA, CEACAM1, CA9,
TEFL SA1I32, CDH1, MUC2, CDKN2A, CEACALV118, MSH2, DSC3, and PTPRC; xxn. a pre-
determined biosignature indicative of neuroendocrine consists of, comprises,
or comprises at least, 1,
35 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20
features selected from ISL1, NCAM1,
S100A11, EN02, S100A1, SYP, MUC1, TFF3, SlOOZ, PAX8, ERBB2, ESR1, S100A10,
CLACAM5, SDC1, MUC4, MPO, S100A4, S100A7, and TP63; xxiii. a pre-determined
biosignature
17
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
indicative of non-small cell carcinoma consists of, comprises, or comprises at
least, 1, 2, 3, 4, 5, 6,7,
8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected from
ESR1, TMPRSS2, AR,
S100A1, SFTPA1, MSLN, SOX2, EN02, TP63, SMAD4, PTPRC, IST,1, CEACAM7,
CEACAM20,
SlOOZ, INHA, NCAME MUC2, TFF3, and PAX8; xxiv. a pre-determined biosignature
indicative of
5 oligodendroglioma consists of, comprises, or comprises at least, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, or 20 features selected from NCAM1, KRT18, CD2,
S100A11, SYP, CD1-11,
S100A4, S100A14, CEACAM1, SlOOPBP, SDC1, SALL4, UPK2, COQ2, TPM2, CD99L2,
TTF1,
CD79A, MBA, and VIM; xxv. a pre-determined biosignature indicative of sarcoma
consists of,
comprises, or comprises at least, 1,2, 3,4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19, or 20
10 features selected from NCAME KRT19, S100A14, NKX2-2, KRT2, KRT7, SATB2,
MYOG,
CAT,D1, CEACAM19, CA9, KRT15, CDKN2A, SlOOP, WTI, TMPRSS2, S100A7, SERPINF15,
DSC3, and EN02; xxvi. a pre-determined biosignature indicative of sarcomatoid
carcinoma consists
of, comprises, or comprises at least, 1,2, 3,4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, or 20
features selected from MME, VIM, S100A14, CD99L2, S100A11, NKX3-1, SATB2,
CPS1, MSLN,
15 SFTPA1, POU5F1, CDH1, OSCAR, S100A5, IMP3, CEACA_Ml, PMS2, NCA_M2,
ICRT15, and
Si 00Al2; xxvii. a pre-determined biosignature indicative of serous consists
of, comprises, or
comprises at least, 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, or 20 features selected
from WT1, PAX8, KRT7, CDKN2A, MSLN, ACVRL1, SATB2, CDK4, DSC3, AR, S100A16,
AN01, S100A5, SDC1, IMP3, SERPINA1, KRT4, ESR1, FOXL2, and KRT15; xxviii. a
pre-
20 determined biosignature indicative of small cell carcinoma consists of,
comprises, or comprises at
least, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or
20 features selected from
NCAM1, ISL1, PAX5, KIT, MUC4, S100A10, MUC1, CTNN131, MITE, NKX2-2, S100A11,
SMN1,
MSLN, S100A6, BCL2, SYP, KL, CGB3, TPSAB1, TEF3; and/or xxix. a pre-determined
biosignature indicative of squamous consists of, comprises, or comprises at
least, 1,2, 3,4, 5,6, 7, 8,
25 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 features selected from
TP63, KRT5, KRT17, SOX2,
AR, CD3G, KRT6A, S100A1, DSC3, SERPINB5, FINF1B, SDC1, S100A6, TPSAB1, KRT20,
HAVCR1,11141, MSH2, PMS2, and CNN 1. The system and methods provided herein
envision any
combination of predetermined biosignatures above. See, e.g., FIGs. 4A-C and
related text.
If making selections of biomarkers from within the pre-determined
biosignatures provided
30 herein, one may choose biomarkers that provide the most informative
predictions. For example, one
may choose the top 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, or 20 features, e.g., 3
or 5 or 10 or 20 features, or at least 3 or 5 or 10 or 20 features, with the
highest Importance value for
each pre-determined biosignature listed in Tables 118-120.
In some embodiments of the methods provided herein, performing the at least
one assay to
35 assess the one or more biomarkers in step (b), including without
limitation those described above with
respect to Tables 118-120, comprises assessing the markers in the at least one
pre-determined
biosignature using DNA analysis and/or expression analysis, wherein: i. the
DNA analysis consists of
18
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
or comprises determining a sequence, mutation, polymorphism, deletion,
insertion, substitution,
translocation, fusion, break, duplication, amplification, repeat, copy number,
copy number variation
(CNV; copy number alteration; CNA), or any combination thereof; ii the DNA
analysis is performed
using polymerase chain reaction (PCR), in situ hybridization, amplification,
hybridization,
5 microarray, nucleic acid sequencing, dye termination sequencing,
pyrosequencing, next generation
sequencing (NCIS; high-throughput sequencing), whole exome sequencing, or any
combination
thereof; and/or iii. the expression analysis consists of or comprises analysis
of RNA, where
optionally: i. the RNA analysis consists of or comprises determining a
sequence, mutation,
polymorphism, deletion, insertion, substitution, translocation, fusion, break,
duplication,
10 amplification, repeat, copy number, amount, level, expression level,
presence, or any combination
thereof; and/or ii. the RNA analysis is performed using polymerase chain
reaction (PCR), in situ
hybridization, amplification, hybridization, microarray, nucleic acid
sequencing, dye termination
sequencing, pyrosequencing, next generation sequencing (NGS; high-throughput
sequencing), whole
transcriptome sequencing, or any combination thereof; iv. the expression
analysis consists of or
15 comprises analysis of protein, where optionally: i. the protein analysis
consists of or comprises
determining a sequence, mutation, polymorphism, deletion, insertion,
substitution, fusion,
amplification, amount, level, expression level, presence, or any combination
thereof; and/or ii. the
protein analysis is performed using imrnunohistochemistry (1E1C), flow
cytometry, an immunoassay,
an antibody or functional fragment thereof, an aptamer, mass spectrometry, or
any combination
20 thereof, and/or v. any combination thereof. In some embodiments,
performing the assay to assess the
one or more biomarkers in step (b) comprises assessing the markers in the at
least one pre-determined
biosignature using: a combination of the DNA analysis and the RNA analysis; a
combination of the
DNA analysis and the protein analysis; a combination of the RNA analysis and
the protein analysis;
or a combination of the DNA analysis, the RNA analysis, and the protein
analysis. In some
25 embodiments, performing the assay to assess the one or more biomarkers
in step (b) comprises RNA
analysis of messenger RNA transcripts.
In some embodiments of the methods provided herein, the at least one pre-
determined
biosignature indicative of the at least one attribute of the cancer,
optionally a cancer type or primary
tumor origin, comprises selections of biomarkers according to at least one of
FIGs. 6I-AC; wherein
30 optionally: i. a pre-determined biosignature indicative of breast
adenocarcinoma comprises DNA
analysis of at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 features selected from
GATA3, CDH1, PAX8,
KRAS, ELK4, CCND1, MECOM, PBX1, CREBBP, and/or expression analysis of at
least, 1, 2, 3,4,
5, 6,7, 8,9, or 10 features selected from GA1'A3, NY-BR-1, KR115, CK7, S100A2,
RCCMa,
MUC4, CK18, IINFlII and S1 00A1; ii. a pre-determined biosignature indicative
of central nervous
35 system cancer comprises DNA analysis of at least, 1, 2, 3, 4, 5, 6,7, 8,
9, or 10 features selected from
ID111, SOX2, OLIG2, MYC, CREB3L2, SPECC1, EGFR, FGFR2, SETBP1, and ZNF217,
and/or
expression analysis of at least, 1, 2, 3,4, 5, 6, 7, 8, 9, or 10 features
selected from S100B, CK18,
19
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CK8, SOX2, DOG1, CD56, PDPN, NKX2-2, CK19, and S100A14; iii. a pre-deterrnined
biosignature
indicative of cervical adenocarcinoma comprises DNA analysis of at least, 1,
2, 3,4, 5, 6, 7, 8,9, or
features selected from TP53, MF,COM, RPN1, U2AF1, GNAS, RAC], KRAS, FT,11,
EXT1, and
CDK6, and/or expression analysis of at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10
features selected from ER,
5 p16, CYCLIND1, LIN28A, PR, SMARCB1, CEACAM4, S100B, CD15, and PSAP; iv. a
pre-
determined biosignature indicative of cholangiocarcinoma comprises DNA
analysis of at least, 1, 2, 3,
4, 5, 6, 7, 8,9, or 10 features selected from TP53, ART:DIA, MAF, KRAS,
CACNA1D, SPEN,
SETBP1, CDK12, LI-IFPL6, and MDS2, and/or expression analysis of at least, 1,
2, 3, 4, 5, 6, 7, 8,9,
or 10 features selected from FINF1B, VILLIN, ANTITRYPSIN, ER, DOG1, SOX2,
MUC4, S100A2,
10 KRT5, and CK7; v. a pre-determined biosignature indicative of colon
adenocarcinoma comprises
DNA analysis of at least, 1, 2, 3, 4, 5, 6, 7, A, 9, or 10 features selected
from APC, CDX2, KRAS,
SETBP1, FLT3, LITTPL6, CDKN2A, FLT1, ASXL1, and CDKN2B, and/or expression
analysis of at
least, 1,2, 3, 4, 5, 6,7, 8, 9, or 10 features selected from CDX2, CK7, MUC2,
CK20, MUC1, SATB2,
vmurg, CEACAM5, CDK17, and S100A6; vi. a pre-determined biosignature
indicative of
15 gastroesophageal adenocarcinoma comprises DNA analysis of at least, 1,
2, 3, 4, 5, 6,7, 8,9, or 10
features selected from CDX2, ERG, TP53, KRAS, U2AF1, ZNF217, CREB3L2, 1RF4,
TCF7L2, and
LFIFPL6, and/or expression analysis of at least, 1, 2, 3,4, 5, 6, 7, 8, 9, or
10 features selected from
CD15, CDX2, MASPIN, MUC5AC, AR, TFF1, NCAM2, TFF3, ISL1, and DOG1; vii. a pre-
determined biosignature indicative of gastrointestinal stromal tumor (GIST)
comprises DNA analysis
20 of at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 features selected from c-
KIT (KIT), TP53, MAX, PDGFRA,
TSF1R, MSI2, SPEN, JAKE SETBP1, and CDH11, and/or expression analysis of at
least, 1, 2, 3, 4, 5,
6, 7, 8, 9, or 10 features selected from DOG1, CD138, CK19, MUC1, CK8, ACVRL1,
KIT, E-
CADITERIN, S100A2, and CK7; viii. a pre-determined biosignature indicative of
hepatocellular
carcinoma comprises DNA analysis of at least, 1, 2, 3, 4, 5, 6,7, 8, 9, or 10
features selected from
25 FILE, CACNA1D, HMGN2P46, KRAS, FANCF, PRCC, ERG, FLT1, FGER1, and ACSL6,
and/or
expression analysis of at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 features
selected from ANTITRYPS1N,
CEACAM16, CK19, AFP, MUC4, CEACAM5, MSH2, BCL6, DSC3, and KRT15; ix. a pre-
determined biosignature indicative of lung adenocarcinoma comprises DNA
analysis of at least, 1, 2,
3, 4, 5, 6, 7, 8,9, or 10 features selected from NKX-2, KRAS, TP53, TPM4,
CDX2, TERT, FOXA1,
30 SETBP1, CDKN2A, and LHEPL6, and/or expression analysis of at least, 1,
2, 3, 4, 5, 6, 7, 8, 9, or 10
features selected from Napsin A, SOX2, CEACAM7, CK7, S100A10, CEACAM6, S1
00A1, RCCMa,
AR and VHIL; x. a pre-determined biosignature indicative of melanoma comprises
DNA analysis of
at least, 1,2, 3, 4, 5, 6,7, 8, 9, or 10 features selected from 112E4, SOX10,
TP53, BRAE, FGER2,
TRIM27, EP300, CDKN2A, LRP113, and NRAS, and/or expression analysis of at
least, 1, 2, 3, 4, 5, 6,
35 7, 8,9, or 10 features selected from S100B, CK8, HMB-45, CD19, MUC1,
MLANA, S100A14,
S100A13, MITE, and S100A1; xi. a pre-determined biosignature indicative of
meningioma comprises
DNA analysis of at least, 1,2, 3,4, 5, 6, 7, 8, 9, or 10 features selected
from CHEK2, TP53, MYCL,
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
THRAP3, MPL, EBF1, EWSR1, PMS2, FLI1, and NTRK2, and/or expression analysis of
at least, 1,
2, 3, 4, 5, 6, 7, 8,9, or 10 features selected from CD138, CK8, DOG1, VIM,
S100A14, S100A2,
CEACAM1, MSH2, PR, and KRT10; xii a pre-determined biosignature indicative of
ovarian
granulosa cell tumor comprises DNA analysis of at least, 1,2, 3, 4, 5, 6, 7,
8, 9, or 10 features selected
5 from FOXL2, TP53, EWSR1, C13113, SPECC1, BCL3, MYII9, TSITR, GID4, and
SOX2, and/or
expression analysis of at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 features
selected from FOXT,2, CD138,
MSH6, MUC1, CK8, PR, MME, ANTITRYPSIN, FLI1, and S100B; xiii. a pre-determined
biosignature indicative of ovarian & fallopian tube adenocarcinoma comprises
DNA analysis of at
least, 1,2, 3,4, 5, 6,7, 8, 9, or 10 features selected from TP53, MECOM, KRAS,
TPM4, RAC1,
10 ASXL1, EP300, CDX2, RPN1, and WTI, and/or expression analysis of at
least, 1, 2, 3, 4, 5, 6, 7, 8,9,
or 10 features selected from WTI, RCCMa, TNHIBIN-alpha, TFE3, S100A13, FOT,X2,
TT,E1,
MSLN, POU5F1, and CEACAN13; xiv. a pre-determined biosignature indicative of
pancreas
adenocarcinoma comprises DNA analysis of at least, 1,2, 3, 4, 5, 6, 7, 8,9, or
10 features selected
from KRAS, CDKN2A, CDKN2B, FANCF, IRF4, TP53, ASXL1, SETBP1, APC, and FOX01,
15 and/or expression analysis of at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10
features selected from PDX1,
GATA3, DOG1, ANTITRYPSIN, ISL1, MUC5AC, CD15, SMAD4, CD5, and CALB2; xv. a pre-
determined biosignature indicative of prostate adenocarcinoma comprises DNA
analysis of at least, 1,
2, 3, 4, 5, 6, 7, 8,9, or 10 features selected from FOXAL PTEN, KLK2, FOX01,
GATA2, FANCA,
LIAFPL6, KRAS, ETV6, and ERCC3, and/or expression analysis of at least, 1, 2,
3, 4, 5, 6, 7, 8, 9, or
20 10 features selected from CK7, PSA, NKX3-1, AMACR, S100A5, MUC1, MUC2,
UPK3A, KL and
FIEPPAR-1; xvi a pre-determined biosignature indicative of renal cell
carcinoma comprises DNA
analysis of at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 features selected from
VHF, TP53, EBF1, MAF,
RAF1, CTNNA1, XPC, MUC1, KRAS, and BTG1, and/or expression analysis of at
least, 1, 2, 3, 4, 5,
6, 7, 8, 9, or 10 features selected from RCCMa, E-CADHERTN, p16, SlOOP,
S100A14, HAVCR1,
25 FTNF1B, KL, CK7, and MUC1; xvii. a pre-determined biosignature
indicative of squamous cell
carcinoma comprises DNA analysis of at least, 1, 2, 3, 4, 5, 6,7, 8, 9, or 10
features selected from
TP53, SOX2, KLHL6, CDKN2A, LPP, CACNA1D, TEKC, KRAS, RPN1, and CDX2, and/or
expression analysis of at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 features
selected from P63, SOX2, CK6,
KRT17, S100A1, CD3G, SETPA1, AR, KRT5, and CD138; xviii. a pre-determined
biosignature
30 indicative of thyroid cancer comprises DNA analysis of at least, 1, 2,
3, 4, 5, 6, 7, 8, 9, or 10 features
selected from BRAF, NKX2-1, TP53, MYC, KDSR, TRRAP, CDX2, KRAS, FHIT, and
SETBP1,
and/or expression analysis of at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10
features selected from
THYROGE013ULAN, RCCMa, HEPPAR-1, S100A2, 1'PSAB1, CAL132, 11NE113,
ARG1, and CNN1; xix. a pre-determined biosignature indicative of urothelial
carcinoma comprises
35 DNA analysis of at least, 1,2, 3,4, 5, 6, 7, 8, 9, or 10 features
selected from GATA3, ASXL1,
CDKN2B, TP53, CTNNAL CDKN2A, KRAS, IL7R, CREBBP, and VFIL, and/or expression
analysis of at least, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 features selected from
GATA3, UPII, CK20, MUC1,
21
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Si 00A2, HEPPAR-1, P63, CALB2, MITE, and SlOOP; xx. a pre-determined
biosignature indicative
of uterine endometrial adenocarcinoma comprises DNA analysis of at least, 1,
2, 3, 4, 5, 6, 7, 8, 9, or
features selected from PTEN, PAX8, PIK3CA, CCNF,1, TP53, MECOM, ESRI, CDX2,
CDKN2A, and KRAS, and/or expression analysis of at least, 1, 2, 3, 4, 5, 6, 7,
8,9, or 10 features
5 selected from RCCMa, PR, ER, VI1L, CALD1, LIN28B, Napsin A, KRT5, S100A6,
and DES; and/or
xxi. a pre-determined biosignature indicative of uterine sarcoma comprises DNA
analysis of at least,
1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 features selected from RBI, SPECC1, FANCC,
TP53, CACNA1D,
JAKE ETV1, PRRX1, PTCH1, and HOXD13, and/or expression analysis of at least,
1, 2, 3, 4, 5, 6, 7,
8, 9, or 10 features selected from CK19, CK18, CD56, DES, FOXL2, CD79A,
S100A14, ER, MSLN,
10 and MITE In some embodiments, the DNA analysis consists of or comprises
determining a sequence,
mutation, polymorphism, deletion, insertion, substitution, translocation,
fusion, break, duplication,
amplification, repeat, copy number, copy number variation (CNV; copy number
alteration; CNA), or
any combination thereof In some embodiments, the DNA analysis is performed
using polymerase
chain reaction (PCR), in situ hybridization, amplification, hybridization,
microarray, nucleic acid
15 sequencing, dye termination sequencing, pyrosequencing, next generation
sequencing (NGS; high-
throughput sequencing), whole exome sequencing, or any combination thereof. In
some embodiments,
the expression analysis consists of or comprises analysis of RNA. In some
embodiments, the RNA
analysis consists of or comprises determining a sequence, mutation,
polymorphism, deletion,
insertion, substitution, translocation, fusion, break, duplication,
amplification, repeat, copy number,
20 amount, level, expression level, presence, or any combination thereof.
In some embodiments, the
RNA analysis is performed using polymerase chain reaction (PCR), in situ
hybridization,
amplification, hybridization, microarray, nucleic acid sequencing, dye
termination sequencing,
pyrosequencing, next generation sequencing (NOS; high-throughput sequencing),
whole
transcriptome sequencing, or any combination thereof. In some embodiments, the
expression analysis
25 consists of or comprises analysis of protein. In some embodiments, the
protein analysis consists of or
comprises determining a sequence, mutation, polymorphism, deletion, insertion,
substitution, fusion,
amplification, amount, level, expression level, presence, or any combination
thereof in some
embodiments, the protein analysis is performed using immunohistochemistry (II-
IC), flow eytometry,
an immunoassay, an antibody or functional fragment thereof, an aptamer, mass
spectrometry, or any
30 combination thereof Any useful combination of such analyses is
contemplated by the invention.
In the methods provided herein, the at least one pre-determined biosignature
may comprise or
may further comprise, as the case may be, selections of biomarkers according
to any one of Tables 2-
116 assessed using DNA analysis. In some embodiments, the DNA analysis
consists of or comprises
determining a sequence, mutation, polymorphism, deletion, insertion,
substitution, translocation,
35 fusion, break, duplication, amplification, repeat, copy number, copy
number variation (CNV; copy
number alteration; CNA) or any combination thereof In some embodiments, the
DNA analysis is
performed using polymerase chain reaction (PCR), in situ hybridization,
amplification, hybridization,
22
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
microarray, nucleic acid sequencing, dye termination sequencing,
pyrosequencing, next generation
sequencing (NGS; high-throughput sequencing), whole exome sequencing, or any
combination
thereof In some embodiments, the at least one pre-determined biosignature
comprising selections of
biomarkers according to any one of Tables 2-116 comprises:
5i. a pre-determined biosignature indicative of
adrenal cortical carcinoma origin
consisting of, comprising, or comprising at least 1,2, 3, 4, 5, 6, 7, 8,9, 10,
11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44,
45, 46,47, 48,49, or at least 50 features selected from Table 2; ii. a pre-
determined biosignature
indicative of anus squamous carcinoma origin consisting of, comprising, or
comprising at least 1, 2, 3,
10 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31, 32,
33, 34, 35, 36, 37, 33, 39, 40, 41, 42, 43, 44, 45, 46, 47, 43, 49, or at
least 50 features selected from
Table 3; iii. a pre-determined biosignature indicative of appendix
adenocarcinoma origin consisting
of, comprising, or comprising at least 1, 2, 3, 4, 5, 6,7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19,20,
21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47,
15 48, 49, or at least 50 features selected from Table 4; iv a pre-
determined biosignature indicative of
appendix mucinous adenocarcinoma NOS origin consisting of, comprising, or
comprising at least 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at
least 50 features selected
from Table 5; v. a pre-determined biosignature indicative of bile duct NOS
cholangiocarcinoma
20 origin consisting of, comprising, or comprising at least 1, 2, 3, 4,
5,6, 7, 8,9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19, 20, 21, 22, 23, 24,25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42,
43, 44,45, 46, 47, 48, 49, or at least 50 features selected from Table 6; vi.
a pre-determined
biosignature indicative of brain astrocytoma NOS origin consisting of,
comprising, or comprising at
least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24, 25, 26, 27, 28, 29,
25 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47,
48, 49, or at least 50 features
selected from Table 7; vii. a pre-determined biosignature indicative of brain
astrocytoma anaplastic
origin consisting of, comprising, or comprising at least 1, 2, 3,4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19, 20, 21, 22, 23, 24,25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42,
43, 44, 45, 46, 47, 48, 49, or at least 50 features selected from Table 8;
viii. a pre-determined
30 biosignature indicative of breast adenocarcinoma NOS origin consisting
of, comprising, or comprising
at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47,
48, 49, or at least 50 features
selected from Table 9; ix. a pre-determined biosignature indicative of breast
carcinoma NOS
consisting of, comprising, or comprising at least 1,2, 3, 4, 5, 6, 7, 8,9, 10,
11, 12, 13, 14, 15, 16, 17,
35 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44,
45, 46, 47, 48, 49, or at least 50 features selected from Table 10; x. a pre-
determined biosignature
indicative of breast infiltrating duct adenocarcinoma origin consisting of,
comprising, or comprising
23
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47,
48, 49, or at least 50 features
selected from Table 11; xi a pre-determined biosignature indicative of breast
infiltrating lobular
adenocareinoma NOS origin consisting of, comprising, or comprising at least 1,
2, 3, 4, 5, 6, 7, 8,9,
5 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50 features
selected from Table 12; xii. a
pre-detennined biosignature indicative of breast metaplastie carcinoma NOS
origin consisting of,
comprising, or comprising at least 1,2, 3,4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19,20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
10 49, or at least 50 features selected from Table 13; xiii. a pre-
determined biosignature indicative of
cervix adenocarcinoma NOS origin consisting of, comprising, or comprising at
least 1, 2, 3, 4, 5, 6, 7,
8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table 14;
xiv. a pre-determined biosignature indicative of cervix carcinoma NOS origin
consisting of,
15 comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 15; xv. a pre-determined
biosignature indicative of
cervix squamous carcinoma NOS origin consisting of, comprising, or comprising
at least 1, 2, 3,4, 5,
6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33,
20 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at
least 50 features selected from Table
16; xvi a pre-determined biosignature indicative of colon adenocarcinoma NOS
origin consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 17; xvii. a pre-determined
biosignature indicative of
25 colon carcinoma NOS origin consisting of, comprising, or comprising at
luist 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44, 45,46, 47,48, 49, or at least 50 features
selected from Table 18; xviii.
a pre-determined biosignature indicative of colon mucinous adenocarcinoma
origin consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
30 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 19; xix. a pre-determined
biosignature indicative of
conjunctiva malignant melanoma NOS origin consisting of, comprising, or
comprising at least 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,
25, 26, 27, 28, 29, 30, 31, 32,
33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at
least 50 features selected from
35 Table 20; xx. a pre-determined biosignature indicative of duodenum and
ampulla adenocarcinoma
NOS origin consisting of, comprising, or comprising at least 1,2, 3, 4, 5, 6,
7, 8, 9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41,
24
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
42, 43,44, 45, 46, 47, 48, 49, or at least 50 features selected from Table 21;
xxi. a pre-determined
biosignature indicative of endometrial endometrioid adenocarcinoma origin
consisting of, comprising,
or comprising at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14,15, 16,
17, 18, 19, 20, 21, 22, 23, 24,
25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47, 48, 49, or at
5 least 50 features selected from Table 22; xxii. a pre-determined
biosignature indicative of endometrial
adenocarcinoma NOS origin consisting of, comprising, or comprising at least 1,
2, 3,4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44, 45,46, 47,48, 49, or at least 50 features
selected from Table 23; xxiii. a
pre-detennined biosignature indicative of endometrial carcinosarcoma origin
consisting of,
10 comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 23, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 43,
49, or at least 50 features selected from Table 24; xxiv. a pre-determined
biosignature indicative of
endometrial serous carcinoma origin consisting of, comprising, or comprising
at least 1, 2, 3,4, 5, 6,
7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26,
27, 28, 29, 30, 31, 32, 33, 34,
15 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least
50 features selected from Table 25;
xxv. a pre-determined biosignature indicative of endometrium carcinoma NOS
origin consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 26; xxvi. a pre-determined
biosignature indicative of
20 endometrium carcinoma undifferentiated origin consisting of, comprising,
or comprising at least 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at
least 50 features selected
from Table 27; xxvii. a pre-determined biosignature indicative of endometrium
clear cell carcinoma
origin consisting of, comprising, or comprising at least 1, 2, 3, 4, 5,6, 7,
8,9, 10, 11, 12, 13, 14, 15,
25 16, 17, 18, 19, 20, 21, 22, 23, 24,25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42,
43, 44,45, 46, 47, 48, 49, or at least 50 features selected from Table 28;
xxviii. a pre-determined
biosignature indicative of esophagus adenocarcinoma NOS origin consisting of,
comprising, or
comprising at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44,
45, 46, 47, 48, 49, or at least
30 50 features selected from Table 29; xxix. a pre-determined biosignature
indicative of esophagus
carcinoma NOS origin consisting of, comprising, or comprising at least 1,2, 3,
4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30,
31, 32, 33, 34, 35, 36, 37, 38,
39, 40,41, 42, 43, 44, 45, 46, 47,48, 49, or at least 50 features selected
from Table 30; xxx. a pre-
determined biosignature indicative of esophagus squamous carcinoma origin
consisting of,
35 comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 31; xxxi. a pre-determined
biosignature indicative of
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
extrahepatic cholangio common bile gallbladder adenocarcinoma NOS origin
consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 32; xxxii. a pre-determined
biosignature indicative of
5 fallopian tube adenocarcinoma NOS origin consisting of, comprising, or
comprising at least 1, 2, 3, 4,
5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table
33; xxxiii. a pre-determined biosignature indicative of fallopian tube
carcinoma NOS origin consisting
of, comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20,
10 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38,
39, 40, 41, 42, 43, 44, 45, 46, 47,
43, 49, or at least 50 features selected from Table 34; xxxiv a pre-
determined hiosignature indicative
of fallopian tube carcinosarcoma NOS origin consisting of, comprising, or
comprising at least 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,
25, 26, 27, 28, 29, 30, 31, 32,
33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at
least 50 features selected from
15 Table 35; xxxv a pre-determined biosignature indicative of fallopian
tube serous carcinoma origin
consisting of, comprising, or comprising at least 1,2, 3, 4, 5, 6, 7, 8,9, 10,
11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44,
45, 46,47, 48,49, or at least 50 features selected from Table 36; xxxvi. a pre-
determined biosignature
indicative of gastric adenocarcinoma origin consisting of, comprising, or
comprising at least 1, 2, 3, 4,
20 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table
37; xxxvii. a pre-determined biosignature indicative of gastroesophageal
junction adenocarcinoma
NOS origin consisting of, comprising, or comprising at least 1,2, 3, 4, 5, 6,
7, 8, 9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41,
25 42, 43,44, 45, 46, 47, 48, 49, or at least 50 features selected from
Table 38; xxxviii. a pre-determined
biosignature indicative of glioblastoma origin consisting of, comprising, or
comprising at least 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,
25, 26, 27, 28, 29, 30, 31, 32,
33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at
least 50 features selected from
Table 39; xxxix. a pre-determined biosignature indicative of glioma NOS origin
consisting of,
30 comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 40; xi a pre-determined
biosignature indicative of
ghosarcoma origin consisting of, comprising, or comprising at least 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35, 36, 37,38, 39,
35 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50 features selected
from Table 41; xli. a pre-
determined biosignature indicative of head, face or neck NOS squamous
carcinoma origin consisting
of, comprising, or comprising at least 1, 2, 3, 4, 5, 6,7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19,20,
26
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47,
48, 49, or at least 50 features selected from Table 42; xlii. a pre-determined
biosignature indicative of
intrahepatic bile duct cholangiocarcinoma origin consisting of, comprising, or
comprising at least 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28, 29, 30, 31,
5 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49,
or at least 50 features selected
from Table 43; xliii a pre-determined biosignature indicative of kidney
carcinoma NOS origin
consisting of, comprising, or comprising at least 1, 2, 3,4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44,
45, 46,47, 48,49, or at least 50 features selected from Table 44; xliv. a pre-
determined biosignature
10 indicative of kidney clear cell carcinoma origin consisting of,
comprising, or comprising at least 1, 2,
3, 4,5, 6, 7, 5, 9, 10, 11, 12, 13, 14, 15, 16, 17, IS, 19, 20, 21, 22, 23,
24, 25, 26, 27, 25, 29, 30, 31,
32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at
least 50 features selected
from Table 45; xlv a pre-determined biosignature indicative of kidney
papillary renal cell carcinoma
origin consisting of comprising, or comprising at least 1, 2, 3, 4, 5,6, 7,
8,9, 10, 11, 12, 13, 14, 15,
15 16, 17, 18, 19, 20, 21, 22, 23, 24,25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42,
43, 44,45, 46, 47, 48, 49, or at least 50 features selected from Table 46;
xlvi. a pre-determined
biosignature indicative of kidney renal cell carcinoma NOS origin consisting
of, comprising, or
comprising at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44,
45, 46, 47, 48, 49, or at least
20 50 features selected from Table 47; xlvii. a pre-determined biosignature
indicative of larynx NOS
squamous carcinoma origin consisting of, comprising, or comprising at least 1,
2, 3,4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50 features
selected from Table 48; xlviii.
a pre-determined biosignature indicative of left colon adenocarcinoma NOS
origin consisting of,
25 comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 49; xlix. a pre-determined
biosignature indicative of
left colon mucinous adenocarcinoma origin consisting of, comprising, or
comprising at least 1, 2, 3, 4,
5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33,
30 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at
least 50 features selected from Table
50; 1. a pre-determined biosignature indicative of liver hepatocellular
carcinoma NOS origin
consisting of, comprising, or comprising at least 1,2, 3, 4, 5, 6, 7, 8,9, 10,
11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44,
45, 46,47, 48,49, or at least 50 features selected from Table 51; li. a pre-
determined biosignature
35 indicative of lung adenocarcinoma NOS origin consisting of, comprising,
or comprising at least 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at
least 50 features selected
27
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
from Table 52; lii. a pre-determined biosignature indicative of lung
adenosquamous carcinoma origin
consisting of, comprising, or comprising at least 1, 2, 3,4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39,40, 41, 42, 43, 44,
45, 46,47, 48,49, or at least 50 features selected from Table 53; liii. a pre-
determined biosignature
5 indicative of lung carcinoma NOS origin consisting of, comprising, or
comprising at least 1,2, 3,4, 5,
6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table
54; liv. a pre-determined biosignature indicative of lung mucinous carcinoma
origin consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
10 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 55; Iv a pre-determined
biosignature indicative of lung
neuroendocrine carcinoma NOS origin consisting of, comprising, or comprising
at least 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table
15 56; lvi a pre-determined biosignature indicative of lung non-small cell
carcinoma origin consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 57; lvii. a pre-determined
biosignature indicative of
lung sarcomatoid carcinoma origin consisting of, comprising, or comprising at
least 1, 2, 3, 4, 5, 6, 7,
20 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table 58;
lviii. a pre-determined biosignature indicative of lung small cell carcinoma
NOS origin consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30,31, 32, 33, 34,35, 36, 37, 38, 39, 40, 41,
42, 43, 44, 45, 46, 47, 48,
25 49, or at least 50 features selected from Table 59; lix. a pre-
delt:rmined biosignature indicative of
lung squamous carcinoma origin consisting of, comprising, or comprising at
least 1, 2, 3,4. 5, 6, 7, 8,
9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table 60; lx.
a pre-determined biosignature indicative of meninges meningioma NOS origin
consisting of,
30 comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 61; bi a pre-determined
biosignature indicative of
nasopharynx NOS squamous carcinoma origin consisting of, comprising, or
comprising at least 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28, 29, 30, 31,
35 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49,
or at least 50 features selected
from Table 62; lxii. a pre-determined biosignature indicative of
oligodendroglioma NOS origin
consisting of, comprising, or comprising at least 1, 2, 3,4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17,
28
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44,
45, 46,47, 48,49, or at least 50 features selected from Table 63; lxiii. a pre-
determined biosignature
indicative of oligodendroglioma aplastic origin consisting of, comprising, or
comprising at least 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28, 29, 30, 31,
5 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49,
or at least 50 features selected
from Table 64; lxiv a pre-determined biosignature indicative of ovary
adenocarcinoma NOS origin
consisting of, comprising, or comprising at least 1, 2, 3,4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44,
45, 46,47, 48,49, or at least 50 features selected from Table 65; lxv. a pre-
determined biosignature
10 indicative of ovary carcinoma NOS origin consisting of, comprising, or
comprising at least 1, 2, 3,4,
5, 6, 7, A, 9, 10, 11, 12, 13, 14,15, 16,17, 18, 19, 20, 21,22, 23, 24, 25,
26, 27, 28, 29, 30, 31,32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table
66; lxvi a pre-determined biosignature indicative of ovary carcinosarcoma
origin consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
15 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 67; lxvii. a pre-deterrnined
biosignature indicative of
ovary clear cell carcinoma NOS origin consisting of, comprising, or comprising
at least 1, 2, 3, 4, 5, 6,
7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26,
27, 28, 29, 30, 31, 32, 33, 34,
35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table 68;
20 lxviii. a pre-determined biosignature indicative of ovary endometrioid
adenocarcinoma origin
consisting of, comprising, or comprising at least 1,2, 3,4, 5, 6, 7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44,
45, 46,47, 48,49, or at least 50 features selected from Table 69; lxix. a pre-
determined biosignature
indicative of ovary granulosa cell tumor NOS origin consisting of, comprising,
or comprising at least
25 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24, 25, 26, 27, 28, 29, 30,
31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or
at least 50 features selected
from Table 70; but. a pre-determined biosignature indicative of ovary high-
grade serous carcinoma
origin consisting of, comprising, or comprising at least 1, 2, 3, 4, 5, 6,
7,8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19, 20, 21, 22, 23, 24,25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42,
30 43, 44,45, 46, 47, 48, 49, or at least 50 features selected from Table
71; lxxi. a pre-determined
biosignature indicative of ovary low-grade serous carcinoma origin consisting
of, comprising, or
comprising at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44,
45, 46, 47, 48, 49, or at least
50 features selected from Table 72; lx.xii. a pre-determined biosignature
indicative of ovary mucinous
35 adenocarcinoma origin consisting of, comprising, or comprising at least
1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30,
31, 32, 33, 34, 35, 36, 37, 38,
39, 40,41, 42, 43, 44, 45, 46, 47,48, 49, or at least 50 features selected
from Table 73; 1xxiii. a pre-
29
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
determined biosignature indicative of ovary serous carcinoma origin consisting
of, comprising, or
comprising at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44,
45, 46, 47, 48, 49, or at least
50 features selected from Table 74; lxxiv. a pre-determined biosignature
indicative of pancreas
5 adenocarcinoma NOS origin consisting of, comprising, or comprising at
least 1, 2, 3, 4, 5, 6, 7, 8,9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34,35, 36,
37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50 features
selected from Table 75; lxxv. a
pre-determined biosignature indicative of pancreas carcinoma NOS origin
consisting of, comprising,
or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24,
10 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42,
43, 44, 45, 46, 47, 48, 49, or at
least 50 features selected from Table 76; lxxvi a pre-determined biosignature
indicative of pancreas
mucinous adenocarcinoma origin consisting of, comprising, or comprising at
least 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table 77;
15 lxxvii a pre-determined biosignature indicative of pancreas
neuroendocrine carcinoma NOS origin
consisting of, comprising, or comprising at least 1, 2, 3,4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44,
45, 46,47, 48,49, or at least 50 features selected from Table 78; lxxviii. a
pre-determined
biosignature indicative of parotid gland carcinoma NOS origin consisting of,
comprising, or
20 comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44,
45, 46, 47, 48, 49, or at least
50 features selected from Table 79; kxix. a pre-determined biosignature
indicative of peritoneum
adenocarcinoma NOS origin consisting of, comprising, or comprising at least 1,
2, 3, 4, 5, 6, 7, 8,9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35, 36,
25 37, 38, 39, 40, 41, 42, 43, 44, 45,46, 47,48, 49, or at least 50
features selected from Table 89; lxxx.
pre-detennined biosignature indicative of peritoneum carcinoma NOS origin
consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 81; lxxxi. a pre-determined
biosignature indicative of
30 peritoneum serous carcinoma origin consisting of, comprising, or
comprising at least 1, 2, 3,4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table 82;
lxxxn. a pre-determined biosignature indicative of pleural mesothekoma NOS
origin consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
35 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 83; lxxxiii. a pre-determined
biosignature indicative of
prostate adenocarcinoma NOS origin consisting of, comprising, or comprising at
least 1, 2, 3, 4, 5, 6,
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26,
27, 28, 29, 30, 31, 32, 33, 34,
35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table 84;
lxxxiv a pre-determined biosignature indicative of rectosigmoid adenocarcinoma
NOS origin
consisting of, comprising, or comprising at least 1, 2, 3,4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17,
5 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44,
45, 46,47, 48,49, or at least 50 features selected from Table 85; lxxxv a pre-
determined biosignature
indicative of rectum adenocarcinoma NOS origin consisting of, comprising, or
comprising at least 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at
least 50 features selected
10 from Table 86; lxxxvi. a pre-determined biosignature indicative of
rectum mucinous adenocarcinoma
origin consisting of, comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34,
35, 36, 37, 38, 39, 40, 41, 42,
43, 44,45, 46, 47, 48, 49, or at least 50 features selected from Table 87;
lxxxvii a pre-determined
biosignature indicative of retroperitoneuna dedifferentiated liposarcoma
origin consisting of,
15 comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 88; lxxxviii. a pre-determined
biosignature indicative
of retroperitoneum leiomyosarcoma NOS origin consisting of, comprising, or
comprising at least 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28, 29, 30, 31,
20 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49,
or at least 50 features selected
from Table 89; lxxxix. a pre-determined biosignature indicative of right colon
adenocarcinoma NOS
origin consisting of, comprising, or comprising at least 1, 2, 3,4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19, 20, 21, 22, 23, 24,25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42,
43, 44,45, 46, 47, 48, 49, or at least 50 features selected from Table 90; xc.
a pre-determined
25 biosignature indicative of right colon mucinous adenocarcinoma origin
consisting of, comprising, or
comprising at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44,
45, 46, 47, 48, 49, or at least
50 features selected from Table 91; xci. a pre-determined biosignature
indicative of salivary gland
adenoidcystic carcinoma origin consisting of, comprising, or comprising at
least 1, 2, 3, 4, 5, 6, 7, 8,
30 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26,
27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table 92;
xcii a pre-determined biosignature indicative of skin Merkel cell carcinoma
origin consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
35 49, or at least 50 features selected from Table 93; xciii. a pre-
determined biosignature indicative of
skin nodular melanoma origin consisting of, comprising, or comprising at least
1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35, 36,
31
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
37, 38, 39, 40, 41, 42, 43, 44, 45,46, 47,48, 49, or at least 50 features
selected from Table 94; xciv. a
pre-determined biosignature indicative of skin squamous carcinoma origin
consisting of, comprising,
or comprising at least I, 2, 3, 4, 5, 6, 7, A, 9, 10, I I, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24,
25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47, 48, 49, or at
5 least 50 features selected from Table 95; xcv. a pre-determined
biosignature indicative of skin
melanoma origin consisting of, comprising, or comprising at least 1, 2, 3,4,
5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50 features selected from
Table 96; xcvi. a pre-
determined biosignature indicative of small intestine gastrointestinal stromal
tumor (GIST) NOS
10 origin consisting of, comprising, or comprising at least 1, 2, 3, 4,
5,6, 7, 8,9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34,
35, 36, 37, 31, 39, 40, 41, 42,
43, 44,45, 46, 47, 48, 49, or at least 50 features selected from Table 97;
xcvii. a pre-determined
biosignature indicative of small intestine adenocarcinoma origin consisting
of, comprising, or
comprising at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25,
15 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47, 48, 49, or at least
50 features selected from Table 98; xcviii. a pre-determined biosignature
indicative of stomach
gastrointestinal stromal tumor (GIST) NOS origin consisting of, comprising, or
comprising at least 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at
least 50 features selected
20 from Table 99; xcix. a pre-determined biosignature indicative of stomach
signet ring cell
adenocarcinoma origin consisting of, comprising, or comprising at least 1, 2,
3, 4, 5, 6, 7, 8,9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30,
31, 32, 33, 34, 35, 36, 37, 38,
39, 40,41, 42, 43, 44, 45, 46, 47,48, 49, or at least 50 features selected
from Table 100; c. a pre-
determined biosignature indicative of thyroid carcinoma NOS origin consisting
of, comprising, or
25 comprising at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44,
45, 46, 47, 48, 49, or at least
50 features selected from Table 101; ci. a pre-determined biosignature
indicative of thyroid
carcinoma anaplastic NOS origin consisting of, comprising, or comprising at
least 1, 2, 3,4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35,
30 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table 102;
cii. a pre-determined biosignature indicative of papillary carcinoma of
thyroid origin consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 103; ciii. a pre-determined
biosignature indicative of
35 tonsil oropharynx tongue squamous carcinoma origin consisting of,
comprising, or comprising at least
1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30,
31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or
at least 50 features selected
32
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
from Table 104; civ. a pre-determined biosignature indicative of transverse
colon adenocarcinoma
NOS origin consisting of, comprising, or comprising at least 1, 2, 3, 4, 5, 6,
7, 8,9, 10, 11, 12, 13, 14,
15, 16,17, 18, 19,20, 21,22, 23,24, 25,26, 27,28, 29, 30, 31, 32, 33, 34,35,
36,37, 38, 39, 40, 41,
42, 43,44, 45, 46, 47, 48, 49, or at least 50 features selected from Table
105; cv. a pre-detemiined
5 biosignature indicative of urothelial bladder adenocarcinoma NOS origin
consisting of, comprising, or
comprising at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44,
45, 46, 47, 48, 49, or at least
50 features selected from Table 106; cvi. a pre-determined biosignature
indicative of urothelial
bladder carcinoma NOS origin consisting of, comprising, or comprising at least
1,2, 3,4, 5, 6, 7, 8,9,
10 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 33, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50 features
selected from Table 107; cvii.
a pre-determined biosignature indicative of urothelial bladder squamous
carcinoma origin consisting
of, comprising, or comprising at least 1, 2, 3, 4, 5, 6,7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19,20,
21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47,
15 48, 49, or at least 50 features selected from Table 108; eviii a pre-
determined biosignature indicative
of urothelial carcinoma NOS origin consisting of, comprising, or comprising at
least 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table 109;
eix. a pre-determined biosignature indicative of uterine endometrial stromal
sarcoma NOS origin
20 consisting of, comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44,
45, 46,47, 48,49, or at least 50 features selected from Table 110; cx. a pre-
determined biosignature
indicative of uterus leiomyosarcoma NOS origin consisting of, comprising, or
comprising at least 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28, 29, 30, 31,
25 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49,
or at least 50 features selected
from Table 111; cxi. a pre-determined biosignature indicative of uterus
sarcoma NOS origin
consisting of, comprising, or comprising at least 1, 2, 3,4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44,
45, 46,47, 48,49, or at least 50 features selected from Table 112; cxii. a pre-
determined biosignature
30 indicative of uveal melanoma origin consisting of, comprising, or
comprising at least 1, 2, 3, 4, 5, 6,7,
8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table 113;
emir. a pre-determined biosignature indicative of vaginal squamous carcinoma
origin consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
35 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 114; cxiv. a pre-determined
biosignature indicative of
vulvar squamous carcinoma origin consisting of, comprising, or comprising at
least 1, 2, 3, 4, 5, 6, 7,
33
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or at least 50
features selected from Table 115;
and/or cxy a pre-determined biosignature indicative of skin trunk melanoma
origin consisting of,
comprising, or comprising at least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21,
5 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48,
49, or at least 50 features selected from Table 116. In some embodiments, the
selections of
biomarkers according to any one of Tables 2-116 comprises the top 1%, 2%, 3%,
4%, 5%, 6%, 7%,
8%, 9%, 10%, 11%, 12%, 13%, 14%, 15%, 16%, 17%, 18%, 19%, 20%, 21%, 22%, 23%,
24%, 25%,
26%, 27%, 28%, 29%, 30%, 31%, 32%, 33%, 34%, 35%, 36%, 37%, 38%, 39%, 40%,
41%, 42%,
10 43%, 44%, 45%, 46%, 47%, 48%, 49%, 50%, 55%, 60%, 65%, 70%, 75%, 80%,
85%, 90%, 95%, or
100% of the feature biomarkers with the highest Importance value in the
corresponding table/s. In
some embodiments, the selections of biomarkers according to any one of Tables
2-116 comprises the
top 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49
or 50 feature biomarkers
15 with the highest Importance value in the corresponding table/s. In some
embodiments, the selections
of biomarkers according to any one of Tables 2-116 comprises at least 1%, 2%,
3%, 4%, 5%, 6%,
7%, 8%, 9%, 10%, 11%, 12%, 13%, 14%, 15%, 16%, 17%, 18%, 19%, 20%, 21%, 22%,
23%, 24%,
25%, 26%, 27%, 28%, 29%, 30%, 31%, 32%, 33%, 34%, 35%, 36%, 37%, 38%, 39%,
40%, 41%,
42%, 43%, 44%, 45%, 46%, 47%, 48%, 40%, 50%, 55%, 60%, 65%, 70%, 75%, 80%,
85%, 90%,
20 95%, or 100% of the top 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28, 29, 30,31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47, 48, 49, or
50 feature biomarkers with the highest Importance value in the corresponding
table/s. In some
embodiments, the selections of biomarkers according to any one of Tables 2-116
comprises at least
50%, 60%, 70%, 75%, 80%, 85%, 90%, 95%, or 100% of the top 5, 10, 15, 20, 25,
30, 35, 40, 45, 50,
25 60, 65, 70, 75, 80, 85, 90, 95, or 100 feature biomarkers with the
highest Importance value in the
corresponding table.
If making selections of biomarkers from within the pre-determined
biosignatures provided
herein, one may choose biomarkers that provide the most informative
predictions. For example, one
may choose the top 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25,
30 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47, 48, 49, or 50
features, e.g., 3 or 5 or 10 or 20 or 25 features, or at least 3 or 5 or 10 or
20 or 25 features, with the
highest Importance value for each pre-determined biosignature listed in Tables
2-116.
In some embodiments of the methods provided herein, step (b) comprises
detennining a gene
copy number for at least one member of the biosignature, and step (d)
comprises processing the gene
35 copy number. In some embodiments, step (b) comprises determining a
sequence for at least one
member of the biosignature, and step (d) comprises processing the sequence. In
some embodiments,
step (b) comprises determining a sequence for a plurality of members of the
biosignature, and step (d)
34
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
comprises comparing the sequence to a reference sequence (e.g., wild type) to
identify microsatellite
repeats, and identifying members of the biosignature that have microsatellite
instability (NISI. In some
embodiments, step (b) comprises determining a sequence for a plurality of
members of the
biosignature, and step (d) comprises comparing the sequence to a reference
sequence (e.g., wild type)
5 to identify a tumor mutational burden (TMB. In some embodiments, step (b)
comprises determining
an mRNA transcript level for at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47,
48, 49, or at least 50 genes in any one of Tables 117-120, and/or INSM1, and
step (d) comprises
processing the transcript levels. In some embodiments, a gene copy number, CNV
or CNA of a gene
10 in the biosignature is determined by measuring the copy number of at
least one proximate region to
the gene, wherein optionally the proximate region comprises at least one
location in the same sub-
band, band, or arm of the chromosome wherein the gene is located.
In some embodiments of the methods provided herein, the one or more biomarkers
in the
biosignature are assessed as described in their corresponding table, including
without limitation
15 Tables 2-116 or Tables 117-120.
In some embodiments of the methods provided herein, the model comprises a
plurality of
intermediate models, wherein the plurality of intermediate models comprises at
least one pairwise
comparison module and/or at least one multi-class classification model. In
some embodiments, the
model calculates a statistical measure that the biosignature corresponds to at
least one of the at least
20 one pre-determined biosignatures. In some embodiments, the processing in
step (d) comprises a
pairwise comparison between candidate pre-determined biosignatures, and a
probability is calculated
that the biosignature corresponds to either one of the pairs of the at least
one pre-determined
biosignatures; and/or using at least one multi-class classification model to
assess the biosignature. In
some embodiments, the pairwise comparison between the two candidate primary
tumor origins and/or
25 the multi-class classification model is determined using a machine
learning classification algorithm,
wherein optionally the machine learning classification algorithm comprises a
boosted tree. In some
embodiments, the pairwise comparison between the two candidate primary tumor
origins is applied to
at least one pre-determined biosignature supplied herein, e.g., with respect
to Tables 2-116; and/or the
multi-class classification model is applied to at least one pre-determined
biosignature supplied herein,
30 e.g., with respect to Tables 118-120.
In some embodiments, the methods supplied herein further comprise determining
intermediate model predictions, wherein the intermediate model predictions
comprise: a cancer type
determined by the joint pairwise comparisons between at least one pair of pre-
determined
biosignatures supplied herein, e.g., with respect to Tables 2-116; a
cancer/disease type determined by
35 an intermediate multi-class model applied to at least one pre-determined
biosignature supplied herein,
e.g., with respect to Table 118, wherein optionally the intermediate multi-
class model is applied to at
least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24, 25, 26, 27, or 28
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
of the pre-determined biosignatures in Table 118; an organ group type
determined by an intermediate
multi-class model applied to at least one pre-determined biosignature supplied
herein, e.g., with
respect to Table 119, wherein optionally the intermediate multi-class model is
applied to at least 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, or 27 of the pre-
5 determined biosignatures in Table 119; and/or a histology determined by
an intermediate multi-class
model applied to at least one pre-determined biosignature supplied herein,
e.g., with respect to Table
120, wherein optionally the intermediate multi-class model is applied to at
least 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, or
29 of the pre-determined
biosignatures in Table 120. In some embodiments, the processing in step (d)
comprises inputting the
10 outputs of each of the utilized intermediate multi-class models into a
fmal predictor model that
provides the prediction in step (e), wherein optionally the final predictor
model comprises a machine
learning algorithm, wherein optionally the machine learning algorithm
comprises a boosted tree.
As described herein, the predicted at least one attribute of the cancer
provided by the systems
and methods herein can be provided at a desired level of granularity. In some
embodiments, the
15 predicted at least one attribute of the cancer comprises at least one of
adrenal cortical carcinoma; anus
squamous carcinoma; appendix adenocarcinoma, NOS; appendix mucinous
adenocarcinoma; bile
duct, NOS, eholangiocareinoma; brain astrocytoma, anaplastie; brain
astroeytoma, NOS; breast
adenocarcinoma, NOS; breast carcinoma, NOS; breast infiltrating duct
adenocarcinoma; breast
infiltrating lobular carcinoma, NOS; breast metaplastic carcinoma, NOS; cervix
adenocareinoma,
20 NOS; cervix carcinoma, NOS; cervix squamous carcinoma; colon
adenocarcinoma, NOS; colon
carcinoma, NOS; colon mucinous adenocarcinoma; conjunctiva malignant melanoma,
NOS;
duodenum and ampulla adenocarcinoma, NOS; endometrial adenocarcinoma, NOS;
endometrial
carcinosarcoma; endometrial endometrioid adenocarcinoma; endometrial serous
carcinoma;
endometrium carcinoma, NOS; endometrium carcinoma, undifferentiated;
endometrium clear cell
25 carcinoma; esophagus adenocarcinoma, NOS; esophagus carcinoma, NOS;
esophagus squamous
carcinoma; extrahepatic cholangio, common bile, gallbladder adenocarcinoma,
NOS; fallopian tube
adenocarcinoma, NOS; fallopian tube carcinoma, NOS; fallopian tube
carcinosarcoma, NOS;
fallopian tube serous carcinoma; gastric adenocarcinoma; gastroesophageal
junction adenocarcinoma,
NOS; glioblastoma; glioma, NOS; gliosarcoma; head, face or neck, NOS squamous
carcinoma;
30 intrahepatic bile duct cholangiocarcinoma; kidney carcinoma, NOS; kidney
clear cell carcinoma;
kidney papillary renal cell carcinoma; kidney renal cell carcinoma, NOS;
larynx, NOS squamous
carcinoma; left colon adenocarcinoma, NOS; left colon mucinous adenocarcinoma;
liver
hepatocellular carcinoma, NOS; lung adenocarcmoma, NOS; lung adenosquamous
carcinoma; lung
carcinoma, NOS; lung mucinous adenocarcinoma; lung neuroendocrine carcinoma,
NOS; lung non-
35 small cell carcinoma; lung sarcomatoid carcinoma; lung small cell
carcinoma, NOS; lung squamous
carcinoma; meninges meningioma, NOS; nasopharynx, NOS squamous carcinoma;
oligodendroglioma, anaplastic; oligodendroglioma, NOS; ovary adenocarcinoma,
NOS; ovary
36
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
carcinoma, NOS; ovary carcinosarcoma; ovary clear cell carcinoma; ovary
endometrioid
adenocarcinoma; ovary granulosa cell tumor, NOS; ovary high-grade serous
carcinoma; ovary low-
grade serous carcinoma; ovary mucinous adenocarcinoma; ovary serous carcinoma;
pancreas
adenocarcinoma, NOS; pancreas carcinoma, NOS; pancreas mucinous
adenocarcinoma; pancreas
5 neuroendocrine carcinoma, NOS; parotid gland carcinoma, NOS; peritoneum
adenocarcinoma, NOS;
peritoneum carcinoma, NOS; peritoneum serous carcinoma; pleural mesothelioma,
NOS; prostate
adenocarcinoma, NOS; rectosigmoid adenocarcinoma, NOS; rectum adenocarcinoma,
NOS; rectum
mucinous adenocarcinoma; retroperitoneum dedifferentiated liposarcoma;
retroperitoneum
leiomyosarcoma, NOS; right colon adenocarcinoma, NOS; right colon mucinous
adenocarcinoma;
10 salivary gland adenoid cystic carcinoma; skin melanoma; skin melanoma;
skin merkel cell carcinoma;
skin nodular melanoma; skin squamous carcinoma; skin trunk melanoma; small
intestine
adenocarcinoma; small intestine gastrointestinal stromal tumor, NOS; stomach
gastrointestinal
stromal tumor, NOS; stomach signet ring cell adenocarcinoma; thyroid
carcinoma, anaplastic, NOS;
thyroid carcinoma, NOS; thyroid papillary carcinoma of thyroid; tonsil,
oropharynic, tongue squamous
15 carcinoma; transverse colon adenocarcinoma, NOS; urothelial bladder
adenocarcinoma, NOS;
urothelial bladder carcinoma, NOS; urothelial bladder squamous carcinoma;
urothelial carcinoma,
NOS; uterine endometrial stromal sarcoma, NOS; uterus leiomyosarcoma, NOS;
uterus sarcoma,
NOS; uveal melanoma; vaginal squamous carcinoma; vulvar squamous carcinoma;
and any
combination thereof. In some embodiments, the predicted at least one attribute
of the cancer
20 comprises at least one of breast adenocarcinoma, central nervous system
cancer, cervical
adenocarcinoma, cholangiocarcinoma, colon adenocarcinoma, gastroesophageal
adenocarcinoma,
gastrointestinal stromal tumor (GIST), hepatocellular carcinoma, lung
adenocarcinoma, melanoma,
meningiorna, ovarian granulosit cell tumor, ovarian & fallopian tube
adenocarcinoma, pancreas
adenocarcinoma, prostate adenocarcinoma, renal cell carcinoma, squamous cell
carcinoma, thyroid
25 cancer, urothclial carcinoma, uterine endometrial adenocarcinoma, and
uterine sarcoma. In some
embodiments, the predicted at least one attribute of the cancer comprises at
least one of bladder; skin;
lung; head, face or neck (NOS); esophagus; female genital tract (FM); brain;
colon; prostate; liver,
gall bladder, ducts; breast; eye; stomach; kidney; and pancreas. In some
embodiments, the sample
comprises a cancer of unlmown primary (CUP).
30 in an aspect, provided herein is a method of predicting at least one
attribute of a cancer, the
method comprising: (a) obtaining a biological sample from a subject having a
cancer, wherein the
biological sample can be a biological sample such as described above; (b)
performing at least one
assay to assess one or more lmomarkers m the biological sample to obtain a
biosignature tor the
sample, wherein the at least one assay can be as described above; (c)
providing the biosignature into a
35 model that has been trained to predict at least one attribute of the
cancer, wherein the model
comprises at least one intermediate model, wherein the at least one
intermediate model comprises: (1)
an first intermediate model trained to process DNA data using the
predetermined biosignatures
37
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
supplied herein with respect to Tables 2-116; (2) a second intermediate model
trained to process RNA
data using the predetermined biosignatures supplied herein with respect to
Table 118; (3) a third
intermediate model trained to process RNA data using the predetermined
biosignatures supplied
herein with respect to Table 119; and/or (4) a fourth intermediate model
trained to process RNA data
5 using the predetermined biosignatures supplied herein with respect to
Table 120; (d) processing, by
one or more computers, the provided biosignamre through each of the plurality
of intermediate
models in part (c), providing the output of each of the plurality of
intermediate models into a fmal
predictor model, and processing by one or more computers, the output of each
of the plurality of
intermediate models through the fmal predictor model; and (e) outputting from
the final predictor
10 model a prediction of the at least one attribute of the cancer. In some
embodiments, the predicted at
least one attribute of the cancer is a tissue-of-origin selected from the
group consisting of breast
adenocarcinoma, central nervous system cancer, cervical adenocarcinoma,
cholangiocarcinoma, colon
adenocarcinoma, gastroesophageal adenocarcinoma, gastrointestinal stromal
tumor (GIST),
hepatocellular carcinoma, lung adenocarcinoma, melanoma, meningioma, ovarian
granulosa cell
15 tumor, ovarian & fallopian tube adenocarcinoma, pancreas adenocarcinoma,
prostate adenocarcinoma,
renal cell carcinoma, squamous cell carcinoma, thyroid cancer, urothelial
carcinoma, uterine
endometrial adenocarcinoma, uterine sarcoma, and any combination thereof. In
some embodiments,
step (b) comprises performing DNA analysis by sequencing genomic DNA from the
biological
sample, wherein the DNA analysis is performed for the genes in Tables 2-116.
In some embodiments,
20 step (b) comprises performing RNA analysis by sequencing messenger RNA
transcripts from the
biological sample, wherein the RNA analysis is performed for the genes in
Table 117 or Tables 118-
120. In some embodiments, the at least one of the at least one intermediate
model and fmal predictor
model comprises a machine learning module, wherein optionally the machine
learning module
comprises one or more of a random forest, support vector machine, logistic
regression, K-nearest
25 neighbor, artificial neural network, naive Bayes, quadratic discriminant
analysis, and Gaussian
processes models, wherein optionally the machine learning module comprises an
XGBoost decision-
tree-based ensemble machine learning algorithm.
The prediction of the at least one attribute of the cancer made using the
systems and methods
provided herein may be used in various settings. See, e.g., Example 3 herein.
In some embodiments,
30 the prediction is used to confirm a diagnosis. In some embodiments, the
prediction is used to change a
diagnosis. In some embodiments, the prediction is used to perform a quality
check. In some
embodiments, the prediction is used to indicate additional molecular testing
to be performed.
In some embodiments of the methods of the invention, the predicted at least
one attribute
comprises an ordered list, wherein optionally the list is ordered using a
statistical measure. For
35 example, the list may be ordered by confidence in the prediction. In
some embodiments, the methods
provided herein further comprise determining whether the prediction of the at
least one attribute meets
38
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
a threshold level, wherein optionally the threshold level is related to a
probability of the prediction
and/or a confidence in the prediction.
In some embodiments, the methods provided herein further comprise generating a
molecular
profile that identifies the presence, level, or state of the biomarkers in the
biosignature, e.g., whether
5 each biomarker has a copy number alteration and/or mutation; and/or a TMB
level, MSI, LOH, or
TVIMR status; and/or expression level, wherein the expression level comprises
that of at least one
transcript and/or protein level. See, e.g., Example 1 for more details.
In some embodiments, the methods provided herein further comprise selecting at
least one
treatment for the patient based at least in part upon the classified at least
one attribute of the cancer,
10 wherein optionally the treatment comprises administration of
immunotherapy, chemotherapy, or a
combination thereof
In an aspect, provided herein is a method comprising preparing a report,
wherein the report
comprises a summary or overview of the molecular profile generated herein,
e.g., as described above,
wherein the report identifies the classified at least one attribute of the
cancer, wherein optionally the
15 report further identifies the at least one treatment selected according
to the methods provided herein,
e.g., as described above. In some embodiments, the report is computer
generated, is a printed report
and/or a computer file, and/or is accessible via a web portal.
Further provided herein is a system comprising one or more computers and one
or more
storage media storing instructions that, when executed by the one or more
computers, cause the one or
20 more computers to perform operations described with reference to the
methods described above.
Relatedly, also provided herein is a non-transitory computer-readable medium
storing software
comprising instructions executable by one or more computers which, upon such
execution, cause the
one or more computers to perform operations with reference to the methods
described above.
In an aspect, provided herein is a system for identifying a lineage for a
cancer, the system
25 comprising: (a) at least one host server; (b) at least one user
interface for accessing the at least one
host server to access and input data; (c) at least one processor for
processing the inputted data: (d) at
least one memory coupled to the processor for storing the processed data and
instructions for carrying
out operations with reference to the methods described above; and (e) at least
one display for
displaying the classified primary origin of the cancer. In some embodiments,
the system further
30 comprise at least one memory coupled to the processor for storing the
processed data and instructions
for selecting treatment and/or generating molecular profiling reports as
described herein. In some
embodiments, the at least one display comprises a report comprising the
classified at least one
attribute of the cancer.
In an aspect, provided herein is a system for identifying at least one
attribute of a sample
35 obtained from a body, wherein the at least one attribute is selected
from the group consisting of a
primary tumor origin, cancer/disease type, organ group, histology, and any
combination thereof the
system comprising: one or more processors and one or more memory units storing
instructions that,
39
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
when executed by the one or more processors, cause the one or more processors
to perform
operations, the operations comprising: obtaining, by the system, a sample
biological signature
representing the sample that was obtained from the body, wherein the sample
comprises cancer cells;
providing, by the system, the sample biological signature as an input to a
model, wherein: the model
5 is configured to perform analysis between the sample biological signature
and each of multiple
different biological signatures, wherein each of the multiple different
biological signatures
corresponds to a different attribute; and/or the model is a multi-class model
wherein the classes
comprise different attributes; and receiving, by the system, an output
generated by the model that
represents data indicating a likely attribute of the sample obtained from the
body based on the
10 pairwise analysis. In another aspect, provided herein is a system for
identifying at least one attribute
of a sample obtained from a body, wherein the at least one attribute is
selected from the group
consisting of a primary tumor origin, cancer/disease type, organ group,
histology, and any
combination thereof, the system comprising: one or more processors and one or
more memory units
storing instructions that, when executed by the one or more processors, cause
the one or more
15 processors to perform operations, the operations comprising: obtaining,
by the system, a sample
biological signature representing the sample that was obtained from the body;
providing, by the
system, the sample biological signature as an input to a model, wherein: the
model is configured to
perform analysis between the sample biological signature and each of multiple
different biological
signatures, wherein each of the multiple different biological signatures
corresponds to a different
20 attribute; and/or the model is a multi-class model wherein the classes
comprise different attributes;
and receiving, by the system, an output generated by the model that represents
data indicating a
probability that an attribute identified by the particular biological
signature identifies a likely attribute
of the sample. In still another aspect, provided herein is a system for
identifying at least one attribute
of a sample obtained from a body, wherein the at least one attribute is
selected from the group
25 consisting of a primary tumor origin, cancer/disease type, organ group,
histology, and any
combination thereof, the system comprising: one or more processors and one or
more memory units
storing instructions that, when executed by the one or more processors, cause
the one or more
processors to perform operations, the operations comprising: obtaining, by the
system, a sample
biological signature representing a biological sample that was obtained from
the cancer sample in a
30 first portion of the body, wherein the sample biological signature
includes data describing a plurality
of features of the biological sample, wherein the plurality of features
include data describing the first
portion of the body; providing, by the system, the sample biological signature
as an input to a model,
wherein: the model is configured to perform analysis between the sample
biological signature and
each of multiple different biological signatures, wherein each of the multiple
different biological
35 signatures corresponds to a different attribute; and/or the model is a
multi-class model wherein the
classes comprise different attributes; and receiving, by the system, an output
generated by the model
that represents data indicating a likely attribute of the sample obtained from
the body. In some
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
embodiments, the sample obtained from the body is a biological sample as
described above. In some
embodiment, the at least one attribute is a primary tumor origin,
cancer/disease type, organ group,
and/or histology as described above. In some embodiments, the sample
biological signature includes
data representing features obtained based on performance of an assay to assess
one or more
5 biomarkers in the cancer sample, wherein optionally the assay is
according to at least one assay
described above. In some embodiments, the operations further comprise:
determining, based on the
output generated by the model, a proposed cancer treatment. In some
embodiments, each of the
multiple different biological signatures comprise pre-identified biosignatures
as described above, e.g.,
with respect to Tables 2-116 or Tabled 118-120. In some embodiments, the
operations further
10 comprise: receiving, by the system, an output generated by the model
that represents a likelihood that
the sample obtained from the body in a first portion of the body originated
from a cancer in a second
portion of the body. In some embodiments, further comprising determining, by
the system and based
on the received output, whether the received output generated by the model
satisfies one or more
predetermined thresholds; and based on the detemnning, by the system, that the
received output
15 satisfies the one or more predetermined thresholds, determining, by the
system, that the cancerous
neoplasm in the first portion of the body originated from a cancer in a second
portion of the body or
that the cancerous neoplasm in the first portion of the body did not originate
from a cancer in a second
portion of the body. In some embodiments, the received output generated by the
model includes a
matrix data structure, wherein the matrix data structure includes a cell for
each feature of the plurality
20 of features evaluated by the pairwise model, wherein each of the cells
includes data describing a
probability that the corresponding feature indicates that the cancerous
neoplasm in the first portion of
the body was caused by cancer in the second portion of the first body.
In an aspect, provided herein is a system for identifying at least one
attribute of a cancer,
wherein the at least one attribute is selected from the group consisting of a
primary tumor origin,
25 cancer/disease type, organ group, histology, and any combination
thereof, the system comprising: one
or more processors and one or more memory units storing instructions that,
when executed by the one
or more processors, cause the one or more processors to perform operations,
the operations
comprising: receiving, by the system storing a model that is configured to
perform analysis of a
biological signature, a sample biological signature representing a biological
sample that was obtained
30 from a cancerous neoplasm in a first portion of a body, wherein the
model includes a cancerous
biological signature for each of multiple different types of cancerous
biological samples, wherein the
cancerous biological signatures include at least a first cancerous biological
signature representing a
molecular profile of a cancerous biological sample from the first portion of
one or more other bodies;
performing, by the system and using the model, analysis of the sample
biological signature using the
35 cancerous biological signatures; generating, by the system and based on
the performed analysis, a
likelihood that the cancerous neoplasm in the first portion of the body was
caused by cancer in a
41
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
second portion of the body; providing, by the system, the generated likelihood
to another device for
display on the other device.
In an aspect, provided herein is a system for training an analysis model for
identifying at least
one attribute of a cancer sample obtained from a body, wherein the at least
one attribute is selected
5 from the group consisting of a primary tumor origin, cancer/disease type,
organ group, histology, and
any combination thereof, the system comprising: one or more processors and one
or more memory
units storing instructions that, when executed by the one or more processors,
cause the one or more
processors to perform operations, the operations comprising: generating, by
the system, an analysis
model, wherein generating the analysis model includes generating a plurality
of model signatures,
10 wherein each model signature is configured to differentiate between at
least one attribute within each
of the at least one attribute; obtaining, by the system, a set of training
data items, wherein each
training data item represents DNA or RNA sequencing results and includes data
indicating (i) whether
or not a variant was detected in the sequencing results and (ii) a number of
copies of a gene or
transcript in the sequencing results; and training, by the system, an analysis
model using the obtained
15 set of training data items_ In some embodiments, the plurality of model
signatures are generated using
random forest models, wherein optionally the random forest models comprise
gradient boosted
forests.
Unless otherwise defined, all technical and scientific terms used herein have
the same
meaning as commonly understood by one of ordinary skill in the art to which
this invention belongs.
20 Methods and materials are described herein for use in the present
invention; other, suitable methods
and materials known in the art can also be used. The materials, methods, and
examples are illustrative
only and not intended to be limiting. All publications, patent applications,
patents, sequences,
database entries, and other references mentioned herein are incorporated by
reference in their entirety.
In case of conflict, the present specification, including definitions, will
control.
25 Other features and advantages of the invention will be apparent from the
following detailed
description and figures, and from the claims.
DESCRIPTION OF DRAWINGS
FIG. 1A is a block diagram of an example of a prior art system for training a
machine
learning model.
30 FIG. 1B is a block diagram of a system that generates training data
structures for training a
machine learning model to predict a sample origin.
FIG. IC is a block diagram of a system for using a trained machine learning
model to predict
a sample origin of sample data from a subject.
FIG. 1D is a flowchart of a process for generating training data structures
for training a
35 machine learning model to predict sample origin.
42
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
FIG. 1E is a flowchart of a process for using a trained machine learning model
to predict
sample origin of sample data from a subject.
FIG. IF is an example of a system for performing pairwise to predict a sample
origin.
FIG. 1G is a block diagram of a system for predicting a sample origin using a
voting unit to
5 interpret output generated by multiple machine learning models that are
each trained to perform
pairwise analysis.
FIG. 1H is a block diagram of system components that can be used to implement
systems of
FIGs. 1B, 1C, 1G, 1F, and 1G.
FIG. 11 illustrates a block diagram of an exemplary embodiment of a system for
determining
10 individualized medical intervention for cancer that utilizes molecular
profiling of a patient's
biological specimen.
FIGs. 2A-C are flowcharts of exemplary embodiments of (FIG. 2A) a method for
determining individualized medical intervention for cancer that utilizes
molecular profiling of a
patient's biological specimen, (FIG. 2B) a method for identifying signatures
or molecular profiles that
15 can be used to predict benefit from therapy, and (FIG. 2C) an alternate
version of (FIG. 2B).
FIGs. 3A-B use of biosignatures to predict a primary tumor lineage from a
cancer sample.
FIGs. 4A-B show schemes for classifying a tissue sample using RNA transcript
analysis
(FIG. 4A) or combined RNA and DNA analysis (FIG. 4B). FIG. 4C is flowchart of
an example of a
process 400C for training a dynamic voting engine.
20 FIGs. 5A-E illustrate performance of the MDC/GPS to classify cancers
using analysis of
genomic DNA.
FIGs. 6A-AL show further development of GPS using combined RNA and DNA
analysis.
FIGs. 7A-Q show an exemplary molecular profiling report that incorporates the
Genomic
Prevalence Score (GPS; also Genomic Profiling Similarity) information
according to the systems and
25 methods provided herein.
FIGs. 8A-M show another exemplary molecular profiling report that incorporates
the
Genomic Prevalence Score information according to the systems and methods
provided herein.
DETAILED DESCRIPTION
30 Described herein are methods and systems for characterizing various
phenotypes of biological
systems, organisms, cells, samples, or the like, by using molecular profiling,
including systems,
methods, apparatuses, and computer programs for training a machine learning
model and then using
the trained machine learning model to characterize such phenotypes. The term
"phenotype" as used
herein can mean any trait or characteristic that can be identified in part or
in whole by using the
35 systems and/or methods provided herein_ In some implementations, the
systems can include one or
43
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
more computer programs on one or more computers in one or more locations,
e.g., configured for use
in a method described herein.
Phenotypes to be characterized can be any phenotype of interest, including
without limitation
a tissue of origin, anatomical origin, histology, organ, medical condition,
ailment, disease, disorder, or
5 useful combinations thereof. A phenotype can be any observable
characteristic or trait of, such as a
disease or condition, a stage of a disease or condition, susceptibility to a
disease or condition,
prognosis of a disease stage or condition, a physiological state, or response
/ potential response (or
lack thereof) to interventions such as therapeutics. A phenotype can result
from a subject's genetic
makeup as well as the influence of enviromnental factors and the interactions
between the two, as well
10 as from epigenetic modifications to nucleic acid sequences.
In various embodiments, a phenotype in a subject is characterized by obtaining
a biological
sample from a subject and analyzing the sample using the systems and/or
methods provided herein.
For example, characterizing a phenotype for a subject or individual can
include detecting a disease or
condition (including pre-symptomatic early stage detection), determining a
prognosis, diagnosis, or
15 theranosis of a disease or condition, or determining the stage or
progression of a disease or condition.
Characterizing a phenotype can include identifying appropriate treatments or
treatment efficacy for
specific diseases, conditions, disease stages and condition stages,
predictions and likelihood analysis
of disease progression, particularly disease recurrence, metastatic spread or
disease relapse. A
phenotype can also be a clinically distinct type or subtype of a condition or
disease, such as a cancer
20 or tumor. Phenotype determination can also be a determination of a
physiological condition, or an
assessment of organ distress or organ rejection, such as post-transplantation
The compositions and
methods described herein allow assessment of a subject on an individual basis,
which can provide
benefits of more efficient and economical decisions in treatment.
Theranostics includes diagnostic testing that provides the ability to affect
therapy or treatment
25 of a medical condition such as a disease or disease state. Thcminosties
testing provides a thcranosis in
a similar manner that diagnostics or prognostic testing provides a diagnosis
or prognosis, respectively.
As used herein, theranostics encompasses any desired form of therapy related
testing, including
predictive medicine, personalized medicine, precision medicine, integrated
medicine,
pharrnacodiagnostics and Dx/Rx partnering. Therapy related tests can be used
to predict and assess
30 drug response in individual subjects, thereby providing personalized
medical recommendations.
Predicting a likelihood of response can be determining whether a subject is a
likely responder or a
likely non-responder to a candidate therapeutic agent, e.g., before the
subject has been exposed or
otherwise treated with the treatment. Assessing a therapeutic response can be
mondormg a response to
a treatment, e.g., monitoring the subject's improvement or lack thereof over a
time course after
35 initiating the treatment. Therapy related tests are useful to select a
subject for treatment who is
particularly likely to benefit or lack benefit from the treatment or to
provide an early and objective
indication of treatment efficacy in an individual subject. Characterization
using the systems and
44
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
methods provided herein may indicate that treatment should be altered to
select a more promising
treatment, thereby avoiding the expense of delaying beneficial treatment and
avoiding the financial
and morbidity costs of less efficacious or ineffective treatment(s).
In various embodiments, a theranosis comprises predicting a treatment efficacy
or lack
5 thereof, classifying a patient as a responder or non-responder to
treatment. A predicted "responder"
can refer to a patient likely to receive a benefit from a treatment whereas a
predicted "non-responder"
can be a patient unlikely to receive a benefit from the treatment. Unless
specified otherwise, a benefit
can be any clinical benefit of interest, including without limitation cure in
whole or in part, remission,
or any improvement, reduction or decline in progression of the condition or
symptoms. The theranosis
10 can be directed to any appropriate treatment, e.g., the treatment may
comprise at least one of
chemotherapy, immunotherapy, targeted cancer therapy, a monoclonal antibody,
small molecule, or
any useful combinations thereof.
The phenotype can comprise detecting the presence of or likelihood of
developing a tumor,
neoplasm, or cancer, or characterizing the tumor, neoplasm, or cancer (e.g.,
stage, grade,
15 aggressiveness, likelihood of metastatis or recurrence, etc). In some
embodiments, the cancer
comprises an acute myeloid leukemia (AML), breast carcinoma,
cholangiocarcinoma, colorectal
adenocarcinoma, extrahepatic bile duct adenocarcinoma, female genital tract
malignancy, gastric
adenocarcinoma, gastroesophageal adenocarcinoma, gastrointestinal stromal
tumors (GIST),
glioblastoma, head and neck squamous carcinoma, leukemia, liver hepatocellular
carcinoma, low
20 grade glioma, lung bronchioloalveolar carcinoma (BAC), lung non-small
cell lung cancer (NSCLC),
lung small cell cancer (SCLC), lymphoma, male genital tract malignancy,
malignant solitary fibrous
tumor of the pleura (MSFT), melanoma, multiple myeloma, neuroendocrine tumor,
nodal diffuse large
B-cell lymphoma, non epithelial ovarian cancer (non-LOC), ovarian surface
epithelial carcinoma,
pancreatic adenocarcinoma, pituitary carcinomas, oligodendroglioma, prostatic
adenocarcinoma,
25 retroperitoneal or peritoneal carcinoma, retropL-ritoneal or peritoneal
sarcoma, small intestinal
malignancy, soft tissue tumor, thymic carcinoma, thyroid carcinoma, or uveal
melanoma. The systems
and methods herein can be used to characterize these and other cancers. Thus,
characterizing a
phenotype can be providing a diagnosis, prognosis or theranosis of one of the
cancers disclosed
herein.
30 In various embodiments, the phenotype comprises a tissue or anatomical
origin. For example,
the tissue can be muscle, epithelial, connective tissue, nervous tissue, or
any combination thereof. For
example, the anatomical origin can be the stomach, liver, small intestine,
large intestine, rectum, anus,
lungs, nose, bronchi, kidneys, urinary bladder, urethra, pituitary gland,
pineal gland, adrenal gland,
thyroid, pancreas, parathyroid, prostate, heart, blood vessels, lymph node,
bone marrow, thymus,
35 spleen, skin, tongue, nose, eyes, ears, teeth, uterus, vagina, testis,
penis, ovaries, breast, mammary
glands, brain, spinal cord, nerve, bone, ligament, tendon, or any combination
thereof Additional non-
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
limiting examples of phenotypes of interest include clinical characteristics,
such as a stage or grade of
a tumor, or the tumor's origin, e.g., the tissue origin.
In various embodiments, phenotypes are determined by analyzing a biological
sample
obtained from a subject. A subject (individual, patient, or the like) can
include, but is not limited to,
5 mammals such as bovine, avian, canine, equine, feline, ovine, porcine, or
primate animals (including
humans and non-human primates). In preferred embodiments, the subject is a
human subject. A
subject can also include a mammal of importance due to being endangered, such
as a Siberian tiger; or
economic importance, such as an animal raised on a farm for consumption by
humans, or an animal of
social importance to humans, such as an animal kept as a pet or in a zoo.
Examples of such animals
10 include, but are not limited to, carnivores such as cats and dogs; swine
including pigs, hogs and wild
boars; ruminants or ungulates such as cattle, oxen, sheep, giraffes, deer,
goats, bison, camels or
horses. Also included are birds that are endangered or kept in zoos, as well
as fowl and more
particularly domesticated fowl, e.g., poultry, such as turkeys and chickens,
ducks, geese, guinea fowl.
Also included are domesticated swine and horses (including race horses). In
addition, any animal
15 species connected to commercial activities are also included such as
those animals connected to
agriculture and aquaculture and other activities in which disease monitoring,
diagnosis, and therapy
selection are routine practice in husbandry for economic productivity and/or
safety of the food chain.
The subject can have a pre-existing disease or condition, including without
limitation cancer.
Alternatively, the subject may not have any known pre-existing condition. The
subject may also be
20 non-responsive to an existing or past treatment, such as a treatment for
cancer.
Data Analysis and Machine Learning
Aspects of the present disclosure are directed towards a system that generates
a set of one or
more training data structures that can be used to train a machine learning
model to provide various
classifications, such as characterizing a phenotype of a biological sample. As
described above,
25 characterizing a phenotype can include providing a diagnosis, prognosis,
theranosis or other relevant
classification. For example, the classification may include a disease state, a
predicted efficacy of a
treatment for a disease or disorder of a subject, or the anatomical origin of
a sample having a
particular set of biomarkers. Once trained, the trained machine learning model
can then be used to
process input data provided by the system and make predictions based on the
processed input data.
30 The input data may include a set of features related to a subject such
as data representing one or more
subject biomarkers and data representing a phenotype of interest, e.g., a
disease and/or anatomical
origin In some embodiments, the input data may further include features
representing an anatomical
origin and the system may make a prediction describing whether the sample is
from that anatomical
origin. The prediction may include data that is output by the machine learning
model based on the
35 machine learning model's processing of a specific set of features
provided as an input to the machine
learning model. The data may include without limitation data representing one
or more subject
46
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
biomarkers, data representing a disease or anatomical origin, and data
representing a proposed
treatment type as desired.
As used herein, "biomarkers" or "sets of biomarkers" are used to train and
test machine
learning models and classify naïve samples. Such references include particular
biomarkers such as
5 particular nucleic acids or proteins, and optionally also include a state
of such nucleic acids or
proteins. Examples of the state of a biomarker include various aspects that
can he queried such as
presence, level (quantity, concentration, etc), sequence, location, activity,
structure, modifications,
covalent or non-covalent binding partners, and the like. As a non-limiting
examples, a set of
biomarkers may include a gene or gene product (i.e., mRNA or protein) having a
specified sequence
10 (e.g., KRAS mutant), and/or a gene or gene product and a level thereof
(e.g., amplified ERBB2 gene
or oyerexpressed HER2 protein). Useful biomarkers and aspects thereof are
further described below.
Innovative aspects of the present disclosure include the extraction of
specific data from
incoming data streams for use in generating training data structures. An
important aspect may be the
selection of a specific set of one or more biomarkers for inclusion in the
training data structure. This is
15 because the presence, absence or other state of particular biomarkers
may be indicative of the desired
classification. For example, certain biomarkers may be selected to determine a
desired phenotype,
such as whether a treatment for a disease or disorder is of likely benefit, or
a tumor origin. By way of
example, in the present disclosure, the Applicant puts forth specific sets of
biomarkers that, when used
to train a machine learning model, result in a trained model that can more
accurately predict a tumor
20 origin than using a different set of biomarkers. See, e.g., Examples 1-
3, Tables 121-130.
The system is configured to obtain output data generated by the trained
machine learning
model based on the machine learning model's processing of the input data. In
various embodiments,
the input data comprises biological data representing one or more biomarkers,
data representing a
disease or disorder, data representing a sample, data representing sample
origins, or any combination
25 thereof. The system may then predict an anatomical origin of a
biological sample haying a particular
set of biomarkers. In some implementations, the disease or disorder may
include a type of cancer and
the anatomical origins can include various tissues and organs. In this
setting, output of the trained
machine learning model that is generated based on trained machine learning
model processing of the
input data that includes the set of biomarkers, the disease or disorder and
various anatomical origins
30 includes data representing the predicted anatomical origin of the
biological sample.
In some implementations, the output data generated by the trained machine
learning model
includes a probability of the desired classification. By way of illustration,
such probability may be a
probability that the biological sample is derived from tissue from a
particular organ. In other
implementations, the output data may include any output data generated by the
trained machine
35 learning model based on the trained machine learning model's processing
of the input data. In some
embodiments, the input data comprises set of biomarkers, data representing the
disease or disorder,
data representing a sample, the data representing the sample origin, or any
combination thereof.
47
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
In some implementations, the training data structures generated by the present
disclosure may
include a plurality of training data structures that each include fields
representing feature vector
corresponding to a particular training sample. The feature vector includes a
set of features derived
from, and representative of, a training sample. The training sample may
include, for example, one or
5 more biomarkers of a biological sample, a disease or disorder associated
with the biological sample,
and an anatomical origin from the biological sample. The training data
structures are flexible because
each respective training data structure may be assigned a weight representing
each respective feature
of the feature vector. Thus, each training data structure of the plurality of
training data structures can
be particularly configured to cause certain inferences to be made by a machine
learning model during
10 training.
Consider a non-limiting example wherein the model is trained to make a
prediction of likely
anatomical origin of a biological sample, e.g., a tumor sample. As a result,
the novel training data
structures that are generated in accordance with this specification are
designed to improve the
performance of a machine learning model because they can be used to train a
machine learning model
15 to predict an anatomical origin of a biological sample having a
particular set of biomarkers. By way of
example, a machine learning model that could not perform predictions regarding
the anatomical origin
of a biological sample having a particular set of biomarkers prior to being
trained using the training
data structures, system, and operations described by this disclosure can learn
to make predictions
regarding the anatomical origin of a biological sample having a particular set
of biomarkers by being
20 trained using the training data structures, systems and operations
described by the present disclosure.
Accordingly, this process takes an otherwise general purpose machine learning
model and changes the
general purpose machine leaning model into a specific computer for perform a
specific task of
performing predicting the anatomical origin of a biological sample having
particular set of
biomarkers.
25 FIG. lA is a block diagram of an example of a prior art system 100 for
training a machine
learning model 110. In some implementations, the machine learning model may
be, for example, a
support vector machine. Alternatively, the machine learning model may include
a neural network
model, a linear regression model, a random forest model, a logistic regression
model, a naive Bayes
model, a quadratic discriminant analysis model, a K-nearest neighbor model, a
support vector
30 machine, or the like. 'Me machine learning model training system 100 may
be implemented as
computer programs on one or more computers in one or more locations, in which
the systems,
components, and techniques described below can be implemented. The machine
learning model
training system 100 trams the machine learning model 110 using training data
items from a database
(or data set) 120 of training data items. The training data items may include
a plurality of feature
35 vectors. Each training vector may include a plurality of values that
each correspond to a particular
feature of a training sample that the training vector represents. The training
features may be referred
48
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
to as independent variables. In addition, the system 100 maintains a
respective weight for each feature
that is included in the feature vectors.
The machine learning model 110 is configured to receive an input training data
item 122 and
to process the input training data item 122 to generate an output 118. The
input training data item may
5 include a plurality of features (or independent variables "X") and a
training label (or dependent
variable "Y"). The machine learning model may be trained using the training
items, and once trained,
is capable of predicting X =f(Y).
To enable machine learning model 110 to generate accurate outputs for received
data items,
the machine learning model training system 100 may train the machine learning
model 110 to adjust
10 the values of the parameters of the machine learning model 110, e.g., to
determine trained values of
the parameters from initial values. These parameters derived from the trai i
g steps may include
weights that can be used during the prediction stage using the fully trained
machine learning model
110.
In training, the machine learning model 110, the machine learning model
training system 100
15 uses training data items stored in the database (data set) 120 of
labeled training data items. The
database 120 stores a set of multiple training data items, with each training
data item in the set of
multiple training items being associated with a respective label. Generally,
the label for the training
data item identifies a correct classification (or prediction) for the training
data item, i.e., the
classification that should be identified as the classification of the training
data item by the output
20 values generated by the machine learning model 110. With reference to
FIG. IA, a training data item
122 may be associated with a training label 122a.
The machine learning model training system 100 trains the machine learning
model 110 to
optimize an objective function. Optimizing an objective function may include,
for example,
minimizing a loss function 130. Generally, the loss function 130 is a function
that depends on the (i)
25 output 118 generated by the machine learning model 110 by processing a
given training data item 122
and (ii) the label 122a for the training data item 122, i.e., the target
output that the machine learning
model 110 should have generated by processing the training data item 122.
Conventional machine learning model training system 100 can train the machine
learning
model 110 to minimize the (cumulative) loss function 130 by performing
multiple iterations of
30 conventional machine learning model training techniques on training data
items from the database
120, e.g., hinge loss, stochastic gradient methods, stochastic gradient
descent with backpropagation,
or the like, to iteratively adjust the values of the parameters of the machine
learning model 110. A
fully trained machine learning model 110 may then be deployed as a predicting
model that can be
used to make predictions based on input data that is not labeled.
35 FIG. 1B is a block diagram of a system that generates training data
structures for training a
machine learning model to predict a sample origin.
49
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
The system 200 includes two or more distributed computers 210, 310, a network
230, and an
application server 240. The application server 240 includes an extraction unit
242, a memory unit 244,
a vector generation unit 250, and a machine learning model 270. The machine
learning model 270
may include one or more of a neural network model, a linear regression model,
a random forest
5 model, a logistic regression model, a naive Bayes model, a quadratic
discriminant analysis, model, a
K-nearest neighbor model, a support vector machine, or the like. Each
distributed computer 210, 310
may include a smartphone, a tablet computer, laptop computer, or a desktop
computer, or the like.
Alternatively, the distributed computers 210, 310 may include server computers
that receive data input
by one or more terminals 205, 305, respectively. The terminal computers 205,
305 may include any
10 user device including a smartphone, a tablet computer, a laptop
computer, a desktop computer or the
like. The network 230 may include one or more networks 230 such as a T,AN, a
WAN, a wired
Ethernet network, a wireless network, a cellular network, the Internet, or any
combination thereof.
The application server 240 is configured to obtain, or otherwise receive, data
records 220,
222, 224, 320 provided by one or more distributed computers such as the first
distributed computer
15 210 and the second distributed computer 310 using the network 230. In
some implementations, each
respective distributed computer 210, 310 may provide different types of data
records 220, 222, 224,
320. For example, the first distributed computer 210 may provide biomarker
data records 220, 222,
224 representing biomarkers for a biological sample from a subject and the
second distributed
computer 310 may provide sample data 320 representing anatomical origin or
other sample data for a
20 subject obtained from the sample database 312. However, the present
disclosure need not be limited to
two computers 210, 310 providing data records 220, 222, 224, 230. Though such
implementations
can provide technical advantages such as load balancing, bandwidth
optimization, or both, it is also
contemplated that the data records 220, 222, 224, 230 can each be provided by
the same computer.
The biomarker data records 220, 222, 224 may include any type of biomarker
data that
25 describes biometric attributes of a biological sample. By way of
example, the example of FIG. 1B
shows the biomarker data records as including data records representing DNA
biomarkers 220,
protein biomarkers 222, and RNA data biomarkers 224. These biomarker data
records may each
include data structures having fields that structure information 220a, 222a,
224a describing
biomarkers of a subject such as a subject's DNA biomarkers 220a, protein
biomarkers 222a, or RNA
30 biomarkers 224a. However, the present disclosure need not be so limited
and any useful biomarkers
can be assessed. In some embodiments, the biomarker data records 220, 222, 224
include next
generation sequencing data from DNA and/or RNA, including without limitation
single variants,
insertions and deletions, substitution, translocabon, fusion, break,
duplication, amplification, loss,
copy number, repeat, total mutational burden, microsatellite instability, or
the like. Alternatively, or in
35 addition, the biomarker data records 220, 222, 224 may also include in
situ hybridization data. Such in
situ hybridization data may include DNA copy numbers, translocations, or the
like. Alternatively, or in
addition, the biomarker data records 220, 222, 224 may include RNA data such
as gene expression or
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
gene fusion, including without limitation data derived from whole
transcriptome sequencing.
Alternatively, or in addition, the biomarker data records 220, 222, 224 may
include protein expression
data such as obtained using immunohistochemistry (NC). Alternatively, or in
addition, the biomarker
data records 220, 222, 224 may include ADAPT data such as complexes.
5 In some implementations, the biomarker data records 220, 222, 224
include one or more
biomarkers and attributes listed in any one of Tables 2-116, Tables 117-120,
ISSIM1, Tables 121-
130. However, the present disclosure need not be so limited, and other types
of biomarkers may be
used as desired. For example, the biomarker data may be obtained by whole
exome sequencing, whole
transcriptome sequencing, whole genome sequencing, or a combination thereof.
10 The sample data records 320 may describe various aspects of a biological
sample, e.g., a
tissue and/or organ from which the sample is derived. For example, the sample
data records 320
obtained from the sample database 312 may include one or more data structures
having fields that
structure data attributes of a biological sample such as a disease or disorder
320a-1 ("ailment"), a
tissue or organ 320a-2 where the sample was obtained, a sample type 320a-3, a
verified sample origin
15 label 320a-4, or any combination thereof The sample record 320 can
include up to n data records
describing a sample, where n is any positive integer greater than 0. For
example, though the example
of FIG. 1B trains the machine learning model using patient sample data
describing disease / disorder,
tissue / organ where sample was obtained, and sample type, the present
disclosure is not so limited.
For example, in some implementations, the machine learning model 370 can be
trained to predict the
20 origin of sample using patient sample information that includes the
tissue or organ 320a-2 where the
sample was obtained and sample type 320a-3 without including the ailment or
disorder 320a-1.
Alternatively, or in addition, the sample data records 320 may also include
fields that structure
data attributes describing details of the biological sample, including
attributes of a subject from which
the sample is derived. An example of a disease or disorder may include, for
example, a type of cancer.
25 A tissue or organ may include, for example, a type of tissue (e.g.,
muscle tissue, epithelial tissue,
connective tissue, nervous tissue, etc.) or organ (e.g., colon, lung, brain,
etc.). A sample type may
include data representing the type of sample, such as tumor sample, bodily
fluid, fresh or frozen,
biopsy, FFPE, or the like. In some implementations, attributes of a subject
from which the sample is
derived include clinical attributes such as pathology details of the sample,
subject age and/or sex,
30 prior subject treatments, or the like. If the sample is a metastatic
sample of unknown primary origin
(i.e., a cancer of unknown primary (CUPS)), the attributes may include the
location from which the
sample was taken. As a non-limiting example, a metastatic lesion of unknown
primary origin may be
found in the liver or brain. Accordingly, though the example of FIG. 1B shows
that sample data may
include a disease or disorder, a tissue or organ, and a sample type, the
sample data may include other
35 types of information, as described herein. Moreover, there is no
requirements that the sample data be
limited to human "patients." Instead, the sample data records 220, 222, 224
and biometric data records
320 may be associated with any desired subject including any non-human
organism.
51
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
In some implementations, each of the data records 220, 222, 224, 320 may
include keyed data
that enables the data records from each respective distributed computer to be
correlated by application
server 240. The keyed data may include, for example, data representing a
subject identifier The
subject identifier may include any form of data that identifies a subject and
that can associate
5 biomarker for the subject with sample data for the subject.
The first distributed computer 210 may provide 208 the biomarker data records
220, 222, 224
to the application server 240. The second distributed computer 310 may provide
210 the sample data
records 320 to the application server 240. The application server 240 can
provide the biomarker data
records 220 and the sample data records 220, 222, 224 to the extraction unit
242.
10 The extraction unit 242 can process the received biomarker data 220,
222, 224 and sample
data records 320 in order to extract data 220a-1, 222a-1, 224a-1, 320a-1, 320a-
2, 320a-3 that can he
used to train the machine learning model. For example, the extraction unit 242
can obtain data
structured by fields of the data structures of the biometric data records 220,
222, 224, obtain data
structured by fields of the data structures of the outcome data records 320,
or a combination thereof
15 The extraction unit 242 may perform one or more information extraction
algorithms such as keyed
data extraction, pattern matching, natural language processing, or the like to
identify and obtain data
220a-1, 222a-1, 224a-1, 320a-1, 320a-2, 320a-3 from the biometric data records
220, 222, 224 and
sample data records 320, respectively. The extraction unit 242 may provide the
extracted data to the
memory unit 244. The extracted data unit may be stored in the memory unit 244
such as flash memory
20 (as opposed to a hard disk) to improve data access times and reduce
latency in accessing the extracted
data to improve system performance. In some implementations, the extracted
data may be stored in
the memory unit 244 as an in-memory data grid.
In more detail, the extraction unit 242 may be configured to filter a portion
of the biomarker
data records 220, 222, 224 and the sample data records 320 such as 220a-1,
222a-1, 224a-1, 320a-1,
25 320a-2, 320a-3 that will be used to generate an input data structure 260
for processing by the machine
learning model 270 from the portion of the sample data records 320a-4 that
will be used as a label for
the generated input data structure 260. Such filtering includes the extraction
unit 242 separating the
biomarker data and a first portion of the sample data that includes a disease
or disorder 320a-1, tissue
/ organ 320a-1 where sample was obtained (e.g., biopsied), sample type 320a-3
details, or any
30 combination thereof, from the verified origin of the sample 320a-4. 'The
verified sample origin of the
sample may be a different tissue / organ or the same tissue / organ than the
sample was obtained from.
An example of who the tissue / organ that the sample was obtained from can be
different than the
verified origin can include instances where the disease or disorder has spread
from a first tissue /
organ to a second tissue / organ from which the sample was then obtained. The
application server 240
35 can then use the biomarker data 220a-1, 222a-1, 224a-1, and the first
portion of the sample data that
includes the disease or disorder 320a-1, tissue or organ 320a-2, sample type
details (not shown in
FIG. 1B), or a combination thereof, to generate the input data structure 260.
In addition, the
52
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
application server 240 can use the second portion of the sample data
describing the verified origin of
the sample 320a-4 as the label for the generated data structure.
The application server 240 may process the extracted data stored in the memory
unit 244
correlate the biomarker data 220a-1, 222a-1, 224a-1 extracted from biomarker
data records 220, 222,
5 224 with the first portion of the sample data 320a-1, 320a-2, 320a-3. The
purpose of this correlation is
Lo cluster biomarker data with sample data so that the sample data for the
biological sample is
clustered with the biomarker data for the same biological sample. In some
implementations, the
correlation of the biomarker data and the first portion of the sample data may
be based on keyed data
associated with each of the biomarker data records 220, 222, 224 and the
sample data records 320. For
10 example, the keyed data may include a sample identifier or a subject
identifier, e.g., a subject from
which the sample is derived.
The application server 240 provides the extracted biomarker data 220a-1, 222a-
1, 224a-I and
the extracted first portion of the sample data 320a-1, 320a-2, 320a-3 as an
input to a vector generation
unit 250. The vector generation unit 250 is used to generate a data structure
based on the extracted
15 biomarker data 220a-1, 222a-1, 224a-1 and the extracted first portion of
the sample data 320a-1,
320a-2, 320a-3. The generated data structure is a feature vector 260 that
includes a plurality of values
that numerical represents the extracted biomarker data 220a-1, 222a-1, 224a-1
and the extracted first
portion of the sample data 320a-1, 320a-2, 320a-3. The feature vector 260 may
include a field for each
type of biomarker and each type of sample data. For example, the feature
vector 260 may include one
20 or more fields corresponding to (i) one or more types of next generation
sequencing data such as
single variants, insertions and deletions, substitution, translocation,
fusion, break, duplication,
amplification, loss, copy number, repeat, total mutational burden,
microsatellite instability, (ii) one or
more types of in situ hybridization data such as DNA copy number, gene copies,
gene translocations,
(iii) one or more types of RNA data such as gene expression or gene fusion,
(iv) one or more types of
25 protein data siu:h as presence, level or cellular location obtained
using irnmunohistochemistry, (v) one
or more types of ADAPT data such as complexes, and (vi) one or more types of
sample data such as
disease or disorder, sample type, each sample details, or the like.
The vector generation unit 250 is configured to assign a weight to each field
of the feature
vector 260 that indicates an extent to which the extracted biomarker data 220a-
1, 222a-1, 224a-1 and
30 the extracted first portion of the sample data 320a-1, 320a-2, 320a-3
includes the data represented by
each field. In one implementation, for example, the vector generation unit 250
may assign a '1' to
each field of the feature vector that corresponds to a feature found in the
extracted biomarker data
220a-1, 222a-1, 224a-1 and the extracted tirst portion of the sample data 320a-
1, 320a-2, 320a-3. In
such implementations, the vector generation unit 250 may, for example, also
assign a '0' to each field
35 of the feature vector that corresponds to a feature not found in the
extracted biomarker data 220a-1,
222a-1, 224a-1 and the extracted first portion of the sample data 320a-1, 320a-
2, 320a-3. The output
53
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
of the vector generation unit 250 may include a data structures such as a
feature vector 260 that can be
used to train the machine learning model 270.
The application server 240 can label the training feature vector 260.
Specifically, the
application server can use the extracted second portion of the sample data
320a-4 to label the
5 generated feature vector 260 with a verified sample origin 320a-4. The
label of the training feature
vector 260 generated based on the verified sample origin 320a-4 can be used to
predict the tissue or
organ that was the origin for a biological sample represented by the sample
record 320 and having
disease or disorder 320a-1 defined by the specific set of biomarkers 220a-1,
222a-1, 224a-1, each of
which is described by described in the training data structure 260.
10 The application server 240 can train the machine learning model 270 by
providing the feature
vector 260 as an input to the machine learning model 270. The machine learning
model 270 may
process the generated feature vector 260 and generate an output 272. The
application server 240 can
use a loss function 280 to determine the amount of error between the output
272 of the machine
learning model 280 and the value specified by the training label, which is
generated based on the
15 second portion of the extracted sample data describing the verified
sample origin 320a-4. The output
282 of the loss function 280 can be used to adjust the parameters of the
machine learning model 282.
In some implementations, adjusting the parameters of the machine learning
model 270 may
include manually tuning of the machine learning model parameters model
parameters. Alternatively,
in some implementations, the parameters of the machine learning model 270 may
be automatically
20 tuned by one or more algorithms of executed by the application server
242.
The application server 240 may perform multiple iterations of the process
described above
with reference to FIG. 1B for each sample data record 320 stored in the sample
database that
correspond to a set of biomarker data for a biological sample. This may
include hundreds of iterations,
thousands of iterations, tens of thousands of iterations, hundreds of
thousands of iterations, millions of
25 iterations, or more, until each of the sample data records 320 stored in
the sample database 312 and
having a corresponding set of biomarker data for a biological sample are
exhausted, until the machine
learning model 270 is trained to within a particular margin of error, or a
combination thereof. A
machine learning model 270 is trained within a particular margin of error
when, for example, the
machine learning model 270 is able to predict, based upon a set of unlabeled
biomarker data, disease
30 or disorder data, and sample type data, an origin of an sample having
the biomarker data. The origin
may include, for example, a probability, a general indication of the
confidence in the origin
classification, or the like.
FIG. 1C is a block diagram of a system for using a trained machine learning
model 370 to
predict a sample origin of sample data from a subject.
35 The machine learning model 370 includes a machine learning model that
has been trained
using the process described with reference to the system of FIG. 1B above. For
example, FIG. 1B is
an example of a machine learning model 370 that has been trained to predict
sample origin using
54
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
patient sample data that comprises data representing a tissue / organ 422a
where the sample was
obtained and a sample type 420a. In the example of FIG. 111, a disease,
disorder, or ailment was not
used to train the model ¨ though there may be implementations of the present
disclosure where the
machine learning model 370 can be trained using an ailment or disorder in
addition to a tissue / organ
5 422a where the sample was obtained and a sample type 420a. The trained
machine learning model
370 is capable of predicting, based on an input feature vector representative
of a set of one or more
biomarkers, a disease or disorder, and other relevant sample data such as
sample type, a origin of a
biological sample having the biomarkers. In some implementations, the "origin"
may include an
anatomical system, location, organ, tissue type, and the like.
10 The application server 240 hosting the machine learning model 370 is
configured to receive
unlabeled biomarker data records 320, 322, 324. The biomarker data records
320, 322, 324 include
one or more data structures that have fields structuring data that represents
one or more particular
biomarkers such as DNA biomarkers 320a, protein biomarkers 322a, RNA
biomarkers 324a, or any
combination thereof As discussed above, the received biomarker data records
may include various
15 types of biomarkers not explicitly depicted by FIG. 1C such as (i) next
generation sequencing data
from DNA and/or RNA, including without limitation single variants, insertions
and deletions,
substitution, translocation, fusion, break, duplication, amplification, loss,
copy number, repeat, total
mutational burden, microsatellite instability, or the like, (ii) one or more
types of in situ hybridization
data such as DNA copies, gene copies, gene translocations, (iii) one or more
types of RNA data such
20 as gene expression or gene fusion, (iv) one or more types of protein
data such as presence, level or
location obtained using immunohistochemistry, or (v) one or more types of
ADAPT data such as
complexes. In some implementations, the biomarker data records 320, 322, 324
include one or more
biomarkers and attributes listed in any one of Tables 2-116, Tables 117-120,
ISNIVI1, and/or Tables
121-130. However, the present disclosure need not be so limited, and other
biomarkers may be used
25 as desired. For example, the biomarker data may be obtained by whole
exome sequencing, whole
transcriptome sequencing, or a combination thereof
The application server 240 hosting the machine learning model 370 is also
configured to
receive sample data 420 representing a proposed origin data 422a for a
biological sample described by
the sample data 420a of the biological sample having biomarkers represented by
the received
30 biomarker data records 320, 322, 324. The proposed origin data 422a for
the biological sample 420a
are also unlabeled and merely a suggestion for the origin of a biological
sample having biomarkers
representing by biomarker data records 320, 322, 324. However, as discussed
elsewhere herein, due to
the potential tor disease (e.g., cancer) to spread from, e.g., organ to organ,
the tissue / organ 422a
where a sample was obtained may not be the actual sample origin.
35 In some implementations, the sample data 420 is received or provided 305
by a terminal 405
over the network 230 and the biomarker data is obtained from a second
distributed computer 310. The
biomarker data may be derived from laboratory machinery used to perform
various assays. See, e.g.,
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Example 1 herein. The sample data 420 can include data representing a tissue /
organ 422a where the
sample was obtained and a sample type 420a. The tissue / organ 422a from where
the sample was
obtained may be referred to as the proposed origin of the sample. In other
implementations, the
sample data 420a, the proposed origin 422a, and the biomarker data 320, 322,
324 may each be
5 received from the terminal 405. For example, the terminal 405 may be user
device of a doctor, an
employee or agent of the doctor working at the doctor's office, or other human
entity that inputs data
representing a sample, data representing a proposed origin, and a data
representing patient attributes
for a the biological sample. In some implementations, the sample data 420 may
include data structures
structuring fields of data representing a proposed origin described by a
tissue or organ name. In other
10 implementations, the sample data 420 may include data structures
structuring fields of data
representing more complex sample data such as sample type, age and/or sex of
the patient from which
the sample is derived, or the like.
The application server 240 receives the biomarker data records 320, 322, 324,
the sample data
420, and the proposed origin data 422. The application server 240 provides the
biomarker data records
15 320, 322, 324, the sample data 420, and the origin data 422 to an
extraction unit 242 that is configured
to extract (i) particular biomarker data such as DNA biomarker data 320a-1,
protein expression data
322a-1, 324a-1, (ii) sample data 420a-1, and (iii) proposed origin data 422a-1
from the fields of the
biomarker data records 320, 322, 324 and the sample data records 420, 422. In
some implementations,
the extracted data is stored in the memory unit 244 as a buffer, cache or the
like, and then provided as
20 an input to the vector generation unit 250 when the vector generation
unit 250 has bandwidth to
receive an input for processing. In other implementations, the extracted data
is provided directly to a
vector generation unit 250 for processing. For example, in some
implementations, multiple vector
generation units 250 may be employed to enable parallel processing of inputs
to reduce latency.
The vector generation unit 250 can generate a data structure such as a feature
vector 360 that
25 includes a plurality of fields and includes one or more fields for each
type of biomarker data and one
or more fields for each type of origin data. For example, each field of the
feature vector 360 may
correspond to (i) each type of extracted biomarker data that can be extracted
from the biomarker data
records 320, 322, 324 such as each type of next generation sequencing data,
each type of in situ
hybridization data, each type of RNA or DNA data, each type of protein (e.g.,
imrnunohistochemistry)
30 data, and each type of ADAPT data and (ii) each type of sample data that
can be extracted from the
sample data records 420, 422 such as each type of disease or disorder, each
type of sample, and each
type of origin details.
The vector generation unit 250 is configured to assign a weight to each field
of the feature
vector 360 that indicates an extent to which the extracted biomarker data 320a-
1, 322a-1, 324a-1, the
35 extracted sample 420a-1, and the extracted origin 422a-1 includes the
data represented by each field.
In one implementation, for example, the vector generation unit 250 may assign
a '1' to each field of
the feature vector 360 that corresponds to a feature found in the extracted
biomarker data 320a-1,
56
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
322a-1, 324a-1, the extracted sample 420a-1, and the extracted origin 422a-1.
In such
implementations, the vector generation unit 250 may, for example, also assign
a '0' to each field of
the feature vector that corresponds to a feature not found in the extracted
biomarker data 320a-1,
322a-1, 324a-1, the extracted sample 420a-1, and the extracted origin 422a-1.
The output of the vector
5 generation unit 250 may include a data structure such as a feature vector
360 that can be provided as
an input to the trained machine learning model 370.
The trained machine learning model 370 process the generated feature vector
360 based on
the adjusted parameters that were determining during the training stage and
described with reference
to FIG. IB. The output 272 of the trained machine learning model provides an
indication of the origin
10 422a-1 of the sample 420a-1 for the biological sample having biomarkers
320a-1, 322a-1, 324a-1. In
some implementations, the output 272 may include a probability that is
indicative of the origin 422a-1
of the sample 420a-1 for the biological sample having biomarkers 320a-1, 322a-
1, 324a-1. In such
implementations, the output 272 may be provided 311 to the terminal 405 using
the network 230. The
terminal 405 may then generate output on a user interface 420 that indicates a
predicted origin for the
15 biological sample having the biomarkers represented by the feature
vector 360.
In other implementations, the output 272 may be provided to a prediction unit
380 that is
configured to decipher the meaning of the output 272. For example, the
prediction unit 380 can be
configured to map the output 272 to one or more categories of effectiveness.
Then, the output of the
prediction unit 328 can be used as part of message 390 that is provided 311 to
the terminal 305 using
20 the network 230 for review by laboratory staff, a healthcare provider, a
subject, a guardian of the
subject, a nurse, a doctor, or the like.
FIG. 113 is a flowchart of a process 400 for generating training data
structures for training a
machine learning model to predict sample origin. In one aspect, the process
400 may include
obtaining, from a first distributed data source, a first data structure that
includes fields structuring data
25 representing a set of one or more biomarkers associated with a
biological sample (410), storing the
first data structure in one or more memory devices (420), obtaining from a
second distributed data
source, a second data structure that includes fields structuring data
representing the biological sample
and origin data for the biological sample having the one or more biomarkers
(430), storing the second
data structure in the one or more memory devices (440), generating a labeled
training data structure
30 that structures data representing (i) the one or more biomarkers, (ii) a
biological sample, (iii) an
origin, and (iv) a predicted origin for the biological sample based on the
first data structure and the
second data structure (450), and training a machine learning model using the
generated labeled
training data (460).
FIG. lE is a flowchart of a process 500 for using a trained machine learning
model to predict
35 sample origin of sample data from a subject. In one aspect, the process
500 may include obtaining a
data structure representing a set of one or more biomarkers associated with a
biological sample (510),
obtaining data representing sample data for the biological sample (520),
obtaining data representing a
57
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
origin type for the biological sample (530), generating a data structure for
input to a machine learning
model that structures data representing (i) the one or more biomarkers, (ii)
the biological sample, and
(iii) the origin type (540), providing the generated data structure as an
input to the machine learning
model that has been trained to predict sample origins using labeled training
data structures structuring
5 data representing one or more obtained biomarkers, one or more sample
types, and one or more
origins (550), and obtaining an output generated by the machine learning model
based on the machine
learning model processing of the provided data structure (560), and
determining a predicted origin for
the biological sample having the one or more biomarkers based on the obtained
output generated by
the machine learning model (570).
10 Provided herein are methods of employing multiple machine learning
models to improve
classification performance. Conventionally, a single model is chosen to
perform a desired
prediction/classification. For example, one may compare different model
parameters or types of
models, e.g., random forests, support vector machines, logistic regression, k-
nearest neighbors,
artificial neural network, naive Bayes, quadratic discriminant analysis, or
Gaussian processes models,
15 during the training stage in order to identify the model haying the
optimal desired performance.
Applicant realized that selection of a single model may not provide optimal
performance in all
settings. Instead, multiple models can be trained to perform the
prediction/classification and the joint
predictions can be used to make the classification. In this scenario, each
model is allowed to "vote"
and the classification receiving the majority of the votes is deemed the
winner.
20 This voting scheme disclosed herein can be applied to any machine
learning classification,
including both model building (e.g., using training data) and application to
classify naive samples.
Such settings include without limitation data in the fields of biology,
finance, communications, media
and entertainment. In some preferred embodiments, the data is highly
dimensional "big data." In some
embodiments, the data comprises biological data, including without limitation
biological data
25 obtained via molecular profiling such as described herein. See, e.g.,
Example 1. The molecular
profiling data can include without limitation highly dimensional next-
generation sequencing data, e.g.,
for particular biomarker panels (see, e.g., Example 1) or whole exome and/or
whole transcriptome
data. The classification can be any useful classification, e.g., to
characterize a phenotype. For
example, the classification may provide a diagnosis (e.g., disease or
healthy), prognosis (e.g., predict
30 a better or worse outcome), theranosis (e.g., predict or monitor
therapeutic efficacy or lack thereof), or
other phenotypic characterization (e.g., origin of a CUPs tumor sample).
FIG. 1F is an example of a system for performing pairwise analysis to predict
a sample
origin. A disease type can include, tor example, an origin of a subject sample
processed by the
system. An origin of a subject sample can include, for example location of a
subject's body where a
35 disease, such as cancer, originated. With reference to a practical
example, a biopsy of a subject tumor
may be obtained from a subject's liver. Then, input data can be generated
based on the biopsied
tumor and provided as an input to the pairwise analysis model 340. The model
can compare the
58
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
generated input data to a corresponding biological signature of each known
type of disease (e.g.,
different cancer types). Based on the output generated by the pairwise
analysis model 340, the
computer 310 can determine whether biopsied tumor represented by the input
data originated in the
liver or in some other portion of the subject's body such as the pancreas. One
or more treatments can
5 then be determined based on the origin of the disease as opposed to the
treatments being based on the
biopsied tumor, alone.
In more detail, the system 300 can include one or more processors and one or
more memory
units 320 storing instructions that, when executed by the one or more
processors, cause the one or
more processors to perform operations. In some implementations, the one or
more processors and the
10 one or memories 320 may be implemented in a computer such as a computer
310.
The system 300 can obtain first biological signature data 322, 324 as an
input. The first
biological signature 322, 324 data can include one or more biomarkers 322,
sample data 324, or both.
Sample data 324 can include data representing the sample that was obtained
from the body, e.g., a
tissue sample, tumor sample, malignant fluid, or other sample such as
described herein. In some
15 implementations, the biological signature 322, 324 represents features
of a disease, e.g., a cancer. In
some implementations, the features may represent molecular data obtained using
next generation
sequencing (NGS). In some implementations, the features may be present in the
DNA of a disease
sample, including without limitation mutations, polymorphisms, deletions,
insertions, substitutions,
translocations, fusions, breaks, duplications, loss, amplification, repeats,
or gene copy numbers. In
20 some implementations, the features may be present in the RNA of a
disease.
The system can generate input data for input to a machine learning model 340
that has been
trained to perform pairwise analysis. The machine learning model can include a
neural network
model, a linear regression model, a random forest model, a logistic regression
model, a naive Bayes
model, a quadratic discrirninant analysis model, a K-nearest neighbor model, a
support vector
25 machine, or the like. The machine learning model 340 can be implemented
as one or more computer
programs on one or more computers in one or more locations.
In some implementations, the generated input data may include data
representing the
biological signature 322, 324. In other implementations, the generated data
that represents the
biological signature can include a vector 332 generated using a vector
generation unit 330. For
30 example, the vector generation unit 330 can obtain biological signature
data 322, 324 from the
memory unit 320 and generate an input vector 333, based on the biological
signature data 322, 324
that represents the biological signature data 322, 324 in a vector space. The
generated vector 332
can be provided, as an input, to the pairwise analysis model 340.
The pairwise analysis model 340 can be configured to perform pairwise analysis
of the input
35 vector 352 representing the biological signature 322, 324 with each
biological signature 341-1, 341-2,
341-n, where n is any positive, non-zero integer. Each of the multiple
different biological signatures
correspond to a different type of disease, e.g., a different type of cancer.
In some implementations,
59
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
the model 340 can be a single model that is trained to determine a source of a
sample based on in
input sample by determining a level of similarity of features of an input
sample to each of a plurality
of biological signature classifications represented by biological signatures
341-1, 341-2, 341-n. In
other implementations, the model 340 can include multiple different models
that each perfomi a
5 pairwise comparison between an input vector 332 and one biological
signature such as 341-1. In such
instances, output data generated by each of the models can he evaluated by a
voting unit to determine
a source of a sample represented by the processed input vector 332.
The pairwise analysis model 340 can generate an output 342 that can be
obtained by the
system such as computer 310. The output 342 can indicate a likely disease type
of the sample based
10 on the pairwise analysis. In some implementations, the output 342 can
include a matrix such as the
matrix described in FIG. 511. The system can determine, based on the generated
matrix and using the
prediction unit 350, data 360 indicating a likely disease type.
Example 2 herein provides an implementation of such a system In the Example,
the models
are trained to distinguish 115 disease types, where each disease type
comprises a primary tumor origin
15 and histology. In some embodiments, the data 360 provides a list of
disease types ranked by
probability. If desired, the data 360 can be presented as an aggregate of
various disease types. In the
Example, such aggregation of Organ Groups is presented, wherein each Organ
Group comprises
appropriate disease types. As an example, the Organ Group "colon" comprises
the disease types
"colon adenocarcinoma, NOS; colon carcinoma, NOS; colon mucinous
adenocarcinoma" and the like.
20 FIG. 1G is a block diagram of a system for predicting a sample origin
using a voting unit to
interpret output generated by multiple machine learning models that are each
trained to perform
pairwise analysis. The system 600 is similar to the system 300 of FIG. IF.
However, instead of a
single machine learning model 340 trained to perform pairwise analysis, the
system 600 includes
multiple machine learning models 340-0, 340-1 ... 340-x, where x is any non-
zero integer greater than
25 1, that have been trained to perform pairwise analysis. The system 600
also include a voting unit 480.
As a non-limiting example, system 600 can be used for predicting origin and
related attributes of a
biological sample having a particular set of biomarkers. See, e.g., Examples 2-
3.
Each machine learning model 370-0, 370-1, 370-x can include a machine learning
model that
has been trained to classify a particular type of input data 320-0, 320-1 ...
320-x, wherein xis any
30 non-zero integer greater than 1 and equal to the number x of machine
learning models. In some
implementations, each machine learning models 340-0, 340-1, 340-x (labeled PW
Compare Models in
FIG. 1G) can be trained, or otherwise configured, to perform a particular
pairwise comparison
between (1) an input vector including data representing the sample data and
(n) another vector
representing a particular biological signature including data representing a
known disease type,
35 portion of a subject body, or a both. Accordingly, in such
implementations, the classification
operation can include classifying (i) an input data vector including data
representing sample data (e.g.,
sample origin, sample type, or the like) and (ii) one or more biomarkers
associated with the sample as
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
being sufficiently similar to a biological signature associated with the
particular machine learning
model or not sufficiently similar to the biological signature associated with
the particular machine
learning model. In some implementations, an input vector may be sufficiently
similar to a biological
signature if a similarity between the input vector and biological signature
satisfies a predetermined
5 threshold.
In some implementations, each of the machine learning models 340-0, 340-1, 340-
x can be of
the same type. For example, each of the machine learning models 340-0, 340-1,
340-x can be a
random forest classification algorithm, e.g., trained using differing
parameters. In other
implementations, the machine learning models 340-0, 340-1, 340-x can be of
different types. For
10 example, there can be one or more random forest classifiers, one or more
neural networks, one or
more K-nearest neighbor classifiers, other types of machine learning models,
or any combination
thereof
Input data such as 420 representing sample data and one or more biomarkers
associated with
the sample can be obtained by the application server 240. The sample data can
include a sample type,
15 sample origin, or the like, as described herein. In some
implementations, the input data 420 is
obtained across the network 230 from one or more distributed computers 310,
405. By way of
example, one or more of the input data items 420 can be generated by
correlating data from multiple
different data sources 210, 405. In such an implementation, (i) first data
describing biomarkers for a
biological sample can be obtained from the first distributed computer 310 and
(ii) second data
20 describing a biological sample and related data can be obtained from the
second computer 405. The
application server 240 can correlate the first data and the second data to
generate an input data
structure such as input data structure 420. This process is described in more
detail in FIG. IC. The
input data 420 can be provided to the vector generation unit 250. The vector
generation unit 250 can
generate input vectors 360-0, 360-1, 360-x that that each represent the input
data 420. While some
25 implementations may generate vectors 360-0, 360-1, 360-x serially, the
present disclosure need not be
so limited.
In some implementations, each input data structure 320-0, 320-1, 320-x can
include data
representing biomarkers of a biological sample, data describing a biological
sample and related data
(e.g., a sample type, disease or disorder associated with the sample, and/or
patient characteristics from
30 which the sample is derived), or any combination thereof. 'The data
representing the biomarkers of a
biological sample can include data describing a specific subset or panel of
genes or gene products.
Alternatively, in some implementations, the data representing biomarkers of
the biological sample can
include data representing complete set of known genes or gene products, e.g.,
via whole exome
sequencing and/or whole transcriptome sequencing. The complete set of known
genes can include all
35 of the genes of the subject from which the biological sample is derived.
In some implementations,
each of the machine learning models 340-0, 340-1, 340-x are the same type
machine learning model
such as a random forest model trained to classify the input data vectors as
corresponding to a sample
61
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
origin (e.g., tissue or organ) associated by the vector processed by the
machine learning model. In
such implementations, though each of the machine learning models 340-0, 340-1,
340-x is the same
type of machine learning model, each of the machine learning models 340-0, 340-
1, 340-x may be
trained in different ways. The machine learning models 340-0, 340-1, 340-x can
generate output data
5 372-0, 372-1, 372-x, respectively, representing whether a biological
sample associated with input
vectors 360-0, 360-1, 360-x is likely to he derived from an anatomical origin
associated with the input
vectors 360-0, 360-1, 360-x. In this example, the input data sets, and their
corresponding input
vectors, are the same - e.g., each set of input data has the same biomarkers,
same sample type, same
origin, or any combination thereof. Nonetheless, given the different training
methods used to train
10 each respective machine learning model 340-0, 340-1, 340-x may generate
different outputs 372-0,
372-1, 372-x, respectively, based on each machine learning model 370-0, 370-1,
370-x processing the
input vector 360-0, 361-1, 361-x, as shown in FIG. 1G.
Alternatively, each of the machine learning models 340-0, 340-1, 340-x can be
a different
type of machine learning model that has been trained, or otherwise configured,
to classify input data
15 as most likely origin of a biological sample. For example, the first
machine learning model 340-1 can
include a neural network, the machine learning model 340-1 can include a
random forest classification
algorithm, and the machine learning model 340-x can include a K-nearest
neighbor algorithm. In this
example, each of these different types of machine learning models 340-0, 340-
1, 340-x can be trained,
or otherwise configured, to receive and process an input vector and determine
whether the input
20 vector is associated with to a sample origin also associated with the
input vector. In this example, the
input data sets, and their corresponding input vectors, can be the same -
e.g., each set of input data
has the same biomarkers, same sample type, same origin, or any combination
thereof Accordingly,
the machine learning model 340-0 can be a neural network trained to process
input vector 360-0 and
generate output data 372-0 indicating whether the biological associated with
the input vector 360-0 is
25 likely to be from an origin also associated with input vector 360-0. In
addition, the machine learning
model 340-1 can be a random forest classification algorithm trained to process
input vector 360-1,
which for purposes of this example is the same as input vector 360-0, and
generate output data 372-1
indicating whether the biological sample associated with the input vector 360-
1 is likely to be from an
origin also associated with the input vector 360-1. This method of input
vector analysis can continue
30 for each of the x inputs, x input vectors, and x machine learning
models. Continuing with this
example with reference to FIG. 1G the machine learning model 340-x can be a K-
nearest neighbor
algorithm trained to process input vector 360-x, which for purposes of this
example is the same as
input vector 360-0 and 360-1, and generate output data 372-x indicating
whether the subject
associated with the input vector 360-x is likely to be responsive or non-
responsive to the treatment
35 also associated with the input vector 360-x.
Alternatively, each of the machine learning models 340-0, 340-1, 340-x can be
the same type
of machine learning models or different type of machine learning models that
are each configured to
62
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
receive different inputs. For example, the input to the first machine learning
model 340-0 can include
a vector 360-0 that includes data representing a first subset or first panel
of biomarkers from a
biological sample and then predict, based on the machine learning models 340-0
processing of vector
360-0 whether the sample is more or less likely to be from a number of
origins. In addition, in this
5 example, an input to the second machine learning model 340-1 can include
a vector 360-1 that
includes data representing a second subset or second panel ofbiomarkers from
the biological sample
that is different than the first subset or first panel of biomarkers. Then,
the second machine learning
model can generate second output data 372-1 that is indicative of whether the
sample associated with
the input vector 360-1 is likely to be responsive or likely to be of an origin
associated with the input
10 vector 360-2. This method of input vector analysis can continue for each
of the x inputs, x input
vectors, and x machine learning models. The input to the xth machine learning
model 340-x can
include a vector 360-x that includes data representing an xth subset or xth
panel of biomarkers of a
subject that is different than (i) at least one, (i) two or more, or (iii)
each of the other x-1 input data
vectors 340-0 to 340-x-1. In some implementations, at least one of the x input
data vectors can
15 include data representing a complete set of biomarkers from the sample,
e.g., next generation
sequencing data. Then, the xth machine learning model 340-x can generate
second output data 372-x,
the second output data 372-x being indicative of whether the sample associated
with the input vector
360-x is likely of an origin associated with the input vector 360-x.
Multiple implementations of system 400 described above are not intended to be
limiting, and
20 instead, are merely examples of configurations of the multiple machine
learning models 340-0, 340-1,
340-x, and their respective inputs, that can be employed using the present
disclosure. With reference
to these examples, the subject can be any human, non-human animal, plant, or
other subject such as
described herein. As described above, the input feature vectors can be
generated, based on the input
data, and represent the input data. Accordingly, each input vector can
represent data that includes one
25 or more biomarkers, a disease or disorder, a sample type, an origin,
patient data, an origin of a sample
having the biomarkers.
In the implementation of FM. 1G, the output data 372-0, 372-1, 372-x can be
analyzed using
a voting unit 480. For example, the output data 372-0, 372-1, 372-x can be
input into the vote unit
480. In some implementations, the output data 372-0, 372-1, 372-x can be data
indicating whether the
30 biological sample associated with the input vector processed by the
machine learning model is likely
to be from a certain origin associated with the vector processed by the
machine learning model. Data
indicating whether the sample associated with the input vector, and generated
by each machine
learning model, can include a "0" or a "1." A "0," produced by a machine
learning model 340-0
based on the machine learning model's 340-0 processing of an input vector 360-
0, can indicate that
35 the sample associated with the input vector 360-0 is not likely to be
from an origin associated with
input vector 360-0. Similarity, as "1," produced by a machine learning model
360-0 based on the
machine learning model's 370-0 processing of an input vector 360-0, can
indicate that the sample
63
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
associated with the input vector 360-0 is likely to be of an origin associated
with the input vector 360-
0. Though the example uses "0" as not likely and "1" as likely, the present
disclosure is not so
limited. Instead, any value can be generated as output data to represent the
output classes. For
example, in some implementations "1" can be used to represent the "not likely"
class and "0" to
5 represent the "likely" class. In yet other implementations, the output
data 372-0, 372-1, 372-x can
include probabilities that indicate a likelihood that the sample associated
with an input vector
processed by a machine learning model is associated with a given origin (e.g.,
a given organ). In such
implementations, for example, the generated probability can be applied to a
threshold, and if the
threshold is satisfied, then the subject associated with an input vector
processed by the machine
10 learning model can be determined to be likely to be of that origin.
In some implementations, the machine learning models output an indication
whether the
sample is more likely to be from one origin versus another, instead of or in
addition to indicating that
the sample is more of less likely to be from a certain origin. For example,
the machine learning model
may indicate that the sample is more or less likely to be of prostatic origin
(i.e., from the prostate), or
15 the machine learning module may indicate whether the sample is most
likely derived from the prostate
or from the colon. Any such origins can be so compared.
The voting unit 480 can evaluate the received output data 370-0, 372-1, 372-x
and determine
whether the sample associated with the processed input vectors 360-0, 360-1,
360-x is likely to be of
an origin associated with the processed input vectors 360-0, 360-1, 360-x. The
voting unit 480 can
20 then determine, based on the set of received output data 370-0, 372-1,
372-x, whether the sample
associated with input vectors 360-0, 360-1, 360-x is likely to be from an
origin associated with the
input vectors 360-0, 360-2, 360-x. In some implementations, the voting unit
480 can apply a
"majority rule." Applying a majority rule, the voting unit 480 can tally the
outputs 372-0, 372-1, and
372-x indicating that the sample is from a given origin and outputs 372-0, 372-
1, 372-x indicating that
25 the sample is not from that origin (or is from a different origin as
described above). Then, the class -
e.g., from origin A or not from origin A. or from origin A and not from origin
B. etc - having the
majority predictions or votes is selected as the appropriate classification
for the subject associated
with the input vector 360-0, 360-1, 360-x. For example, the majority may
determine that the sample is
from origin A or is not from origin A, or alternately the majority may
determine that the sample is
30 from origin A or is from origin B.
In some implementations, the voting unit 480 can complete a more nuanced
analysis. For
example, in some implementations, the voting unit 480 can store a confidence
score for each machine
learning model 340-0, 340-1, 340-x. This confidence score, for each machine
learning model 340-0,
340-1, 340-x, can be initially set to a default value such as 0, 1, or the
like. Then, with each round of
35 processing of input vectors, the voting unit 480, or other module of the
application server 240, can
adjust the confidence score for the machine learning model 340-0, 340-1, 340-x
based on whether the
machine learning model accurately predicted the sample classification selected
by the voting unit 480
64
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
during a previous iteration. Accordingly, the stored confidence score, for
each machine learning
model, can provide an indication of the historical accuracy for each machine
learning model.
In the more nuanced approached, the voting unit 480 can adjust output data 372-
0, 372-0,
372-x produced by each machine learning model 340-0, 340-1, 340-x,
respectively, based on the
5 confidence score calculated for the machine learning model. Accordingly,
a confidence score
indicating that a machine learning mode is historically accurate can he used
to boost a value of output
data generated by the machine learning model. Similarly, a confidence score
indicating that a
machine learning model is historically inaccurate can be used to reduce a
value of output data
generated by the machine learning model. Such boosting or reducing of the
value of output data
10 generated by a machine learning model can be achieved, for example, by
using the confidence score
as a multiplier of less than one for reduction and more than 1 for boosting.
Other operations can also
be used to adjust the value of output data such as subtracting a confidence
score from the value of the
output data to reduce the value of the output data or adding the confidence
score to the value of the
output data to boost the value of the output data. Use of confidence scores to
boost or reduce the
15 value of output data generated by the machine learning models is
particularly useful when the
machine learning models are configured to output probabilities that will be
applied to one or more
thresholds to determine whether a sample is or is not from an origin, or is
from one of two possible
origins. This is because using the confidence score to adjust the output of a
machine learning model
can be used to move a generated output value above or below a class threshold,
thereby altering a
20 prediction by a machine learning model based on its historical accuracy.
Use of the voting unit 480 to evaluate outputs of multiple machine learning
models can lead
to greater accuracy in prediction of the origin of a sample for a particular
set of subject biomarkers, as
the consensus amongst multiple machine learning models can be evaluated
instead of the output of
only a single machine learning model.
25 FIG. 1H is a block diagram of system components that can be used to
implement systems of
FIGs. 1B, 1C, 1G, 1F, and 1G.
Computing device 600 is intended to represent various forms of digital
computers, such as
laptops, desktops, workstations, personal digital assistants, servers, blade
servers, mainframes, and
other appropriate computers. Computing device 650 is intended to represent
various forms of mobile
30 devices, such as personal digital assistants, cellular telephones,
smartphones, and other similar
computing devices. Additionally, computing device 600 or 650 can include
Universal Serial Bus
(USB) flash drives. The USB flash drives can store operating systems and other
applications. The
US13 flash drives can include mput/output components, such as a wireless
transmitter or US13
connector that can be inserted into a USB port of another computing device.
The components shown
35 here, their connections and relationships, and their functions, are
meant to be exemplary only, and are
not meant to limit implementations of the inventions described and/or claimed
in this document.
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Computing device 600 includes a processor 602, memory 604, a storage device
608, a high-
speed interface 608 connecting to memory 604 and high-speed expansion ports
610, and a low speed
interface 612 connecting to low speed bus 614 and storage device 608. Each of
the components 602,
604, 608, 608, 610, and 612, are interconnected using various busses, and can
be mounted on a
5 common motherboard or in other manners as appropriate. The processor 602
can process instructions
for execution within the computing device 600, including instructions stored
in the memory 604 or on
the storage device 608 to display graphical information for a GUI on an
external input/output device,
such as display 616 coupled to high speed interface 608. In other
implementations, multiple
processors and/or multiple buses can be used, as appropriate, along with
multiple memories and types
10 of memory. Also, multiple computing devices 600 can be connected, with
each device providing
portions of the necessary operations, e.g., as a server bank, a group of blade
servers, or a multi-
processor system.
The memory 604 stores information within the computing device 600. In one
implementation,
the memory 604 is a volatile memory unit or units. In another implementation,
the memory 604 is a
15 non-volatile memory unit or units. The memory 604 can also be another
form of computer-readable
medium, such as a magnetic or optical disk.
The storage device 608 is capable of providing mass storage for the computing
device 600. In
one implementation, the storage device 608 can be or contain a computer-
readable medium, such as a
floppy disk device, a hard disk device, an optical disk device, or a tape
device, a flash memory or
20 other similar solid state memory device, or an array of devices,
including devices in a storage area
network or other configurations. A computer program product can be tangibly
embodied in an
information carrier. The computer program product can also contain
instructions that, when executed,
perform one or more methods, such as those described above. The infonnation
carrier is a computer-
or machine-readable medium, such as the memory 604, the storage device 608, or
memory on
25 processor 602.
The high speed controller 608 manages bandwidth-intensive operations for the
computing
device 600, while the low speed controller 612 manages lower bandwidth
intensive operations. Such
allocation of functions is exemplary only. In one implementation, the high-
speed controller 608 is
coupled to memory 604, display 616, e.g., through a graphics processor or
accelerator, and to high-
30 speed expansion ports 610, which can accept various expansion cards (not
shown). In the
implementation, low-speed controller 612 is coupled to storage device 608 and
low-speed expansion
port 614. The low-speed expansion port, which can include various
communication ports, e.g., USB,
Bluetooth, Ethernet, wireless Ethernet can be coupled to one or more
input/output devices, such as a
keyboard, a pointing device, microphone/speaker pair, a scanner, or a
networking device such as a
35 switch or router, e.g., through a network adapter. The computing device
600 can be implemented in a
number of different forms, as shown in the figure. For example, it can be
implemented as a standard
server 620, or multiple times in a group of such servers. It can also be
implemented as part of a rack
66
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
server system 624. In addition, it can be implemented in a personal computer
such as a laptop
computer 622. Alternatively, components from computing device 600 can be
combined with other
components in a mobile device (not shown), such as device 650. Each of such
devices can contain one
or more of computing device 600, 650, and an entire system can be made up of
multiple computing
5 devices 600, 650 communicating with each other.
The computing device 600 can he implemented in a number of different forms, as
shown in
the figure. For example, it can be implemented as a standard server 620, or
multiple times in a group
of such servers. It can also be implemented as part of a rack server system
624. In addition, it can be
implemented in a personal computer such as a laptop computer 622.
Alternatively, components from
10 computing device 600 can be combined with other components in a mobile
device (not shown), such
as device 650. Each of such devices can contain one or more of computing
device 600, 650, and an
entire system can be made up of multiple computing devices 600, 650
communicating with each
other.
Computing device 650 includes a processor 652, memory 664, and an input/output
device
15 such as a display 654, a communication interface 666, and a transceiver
668, among other
components. The device 650 can also be provided with a storage device, such as
a micro-drive or
other device, to provide additional storage. Each of the components 650, 652,
664, 654, 666, and 668,
are interconnected using various buses, and several of the components can be
mounted on a common
motherboard or in other manners as appropriate.
20 The processor 652 can execute instructions within the computing device
650, including
instructions stored in the memory 664. The processor can be implemented as a
chipset of chips that
include separate and multiple analog and digital processors. Additionally, the
processor can be
implemented using any of a number of architectures. For example, the processor
610 can be a CISC
(Complex Instruction Set Computers) processor, a RISC (Reduced Instruction Set
Computer)
25 processor, or a MISC (Minimal Instruction Set Computer) processor. The
processor can provide, for
example, for coordination of the other components of the device 650, such as
control of user
interfaces, applications run by device 650, and wireless communication by
device 650.
Processor 652 can communicate with a user through control interface 658 and
display
interface 656 coupled to a display 654. The display 654 can be, for example, a
TFT (Thin-Film-
30 Transistor Liquid Crystal Display) display or an OLED (Organic Light
Emitting Diode) display, or
other appropriate display technology. The display interface 656 can comprise
appropriate circuitry for
driving the display 654 to present graphical and other information to a user.
The control interface 658
can receive commands from a user and convert them tor submission to the
processor 652. In addition,
an external interface 662 can be provide in communication with processor 652,
so as to enable near
35 area communication of device 650 with other devices. External interface
662 can provide, for
example, for wired communication in some implementations, or for wireless
communication in other
implementations, and multiple interfaces can also be used.
67
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
The memory 664 stores information within the computing device 650. The memory
664 can
be implemented as one or more of a computer-readable medium or media, a
volatile memory unit or
units, or a non-volatile memory unit or units. Expansion memory 674 can also
be provided and
connected to device 650 through expansion interface 672, which can include,
for example, a SIMM
5 (Single In Line Memory Module) card interface. Such expansion memory 674
can provide extra
storage space for device 650, or can also store applications or other
information for device 650.
Specifically, expansion memory 674 can include instructions to carry out or
supplement the processes
described above, and can include secure information also. Thus, for example,
expansion memory 674
can be provide as a security module for device 650, and can be programmed with
instructions that
10 permit secure use of device 650. In addition, secure applications can be
provided via the SlMM cards,
along with additional information, such as placing identifying information on
the SIMM card in a
non-hackable manner.
The memory can include, for example, flash memory and/or NVRAM memory, as
discussed
below. In one implementation, a computer program product is tangibly embodied
in an information
15 carrier The computer program product contains instructions that, when
executed, perform one or
more methods, such as those described above. The information carrier is a
computer- or machine-
readable medium, such as the memory 664, expansion memory 674, or memory on
processor 652 that
can be received, for example, over transceiver 668 or external interface 662.
Device 650 can communicate wirelessly through communication interface 666,
which can
20 include digital signal processing circuitry where necessary.
Communication interface 666 can provide
for communications under various modes or protocols, such as GSM voice calls,
SMS, EMS, or MMS
messaging, CDMA, TDMA, PDC, WCDMA, CDMA2000, or GPRS, among others. Such
communication can occur, for example, through radio-frequency transceiver 668.
In addition, short-
range communication can occur, such as using a Bluetooth, Wi-Fi, or other such
transceiver (not
25 shown). In addition, GPS (Global Positioning System) receiver module 670
can provide additional
navigation- and location-related wireless data to device 650, which can be
used as appropriate by
applications running on device 650.
Device 650 can also communicate audibly using audio codec 660, which can
receive spoken
information from a user and convert it to usable digital information. Audio
codec 660 can likewise
30 generate audible sound for a user, such as through a speaker, e.g., in a
handset of device 650. Such
sound can include sound from voice telephone calls, can include recorded
sound, e.g., voice
messages, music files, etc and can also include sound generated by
applications operating on device
650.
The computing device 650 can be implemented in a number of different forms, as
shown in
35 the figure. For example, it can be implemented as a cellular telephone
680. It can also be implemented
as part of a smartphone 682, personal digital assistant, or other similar
mobile device.
68
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Various implementations of the systems and methods described here can be
realized in digital
electronic circuitry, integrated circuitry, specially designed ASICs
(application specific integrated
circuits), computer hardware, firmware, software, and/or combinations of such
implementations.
These various implementations can include implementation in one or more
computer programs that
5 are executable and/or interpretable on a programmable system including at
least one programmable
processor, which can be special or general purpose, coupled to receive data
and instructions from, and
to transmit data and instructions to, a storage system, at least one input
device, and at least one output
device.
These computer programs (also known as programs, software, software
applications or code)
10 include machine instructions for a programmable processor, and can be
implemented in a high-level
procedural and/or object-oriented progra llllll ing language, and/or in
assembly/machine language. As
used herein, the terms "machine-readable medium" or "computer-readable medium"
refers to any
computer program product, apparatus and/or device, e.g., magnetic discs,
optical disks, memory,
Programmable Logic Devices (PLDs), used to provide machine instructions and/or
data to a
15 programmable processor, including a machine-readable medium that
receives machine instructions as
a machine-readable signal. The term "machine-readable signal" refers to any
signal used to provide
machine instructions and/or data to a programmable processor.
To provide for interaction with a user, the systems and techniques described
here can be
implemented on a computer having a display device, e.g., a CRT (cathode ray
tube) or LCD (liquid
20 crystal display) monitor for displaying information to the user and a
keyboard and a pointing device,
e.g., a mouse or a trackball by which the user can provide input to the
computer_ Other kinds of
devices can be used to provide for interaction with a user as well; for
example, feedback provided to
the user can be any form of sensory feedback, e.g., visual feedback, auditory
feedback, or tactile
feedback; and input from the user can be received in any form, including
acoustic, speech, or tactile
25 input.
The systems and techniques described here can be implemented in a computing
system that
includes a back end component, e.g., as a data server, or that includes a
middlevvare component, e.g.,
an application server, or that includes a front end component, e.g., a client
computer having a
graphical user interface or a Web browser through which a user can interact
with an implementation
30 of the systems and techniques described here, or any combination of such
back end, middleware, or
front end components. The components of the system can be interconnected by
any form or medium
of digital data communication, e.g., a communication network Examples of
communication networks
include a local area network ("LAN"), a wide area network ("WAN"), and the
Internet.
The computing system can include clients and servers. A client and server are
generally
35 remote from each other and typically interact through a communication
network. The relationship of
client and server arises by virtue of computer programs running on the
respective computers and
having a client-server relationship to each other.
69
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Computer Systems
The practice of the present methods may also employ computer related software
and systems.
Computer software products as described herein typically include computer
readable medium having
computer-executable instructions for performing the logic steps of the method
as described herein.
5 Suitable computer readable medium include floppy disk, CD-ROM/DVD/DVD-
ROM, hard-disk
drive, flash memory, ROM/RAM, magnetic tapes and etc. The computer executable
instructions may
be written in a suitable computer language or combination of several
languages. Basic computational
biology methods are described in, for example Setubal and Meidanis et al.,
Introduction to
Computational Biology Methods (PWS Publishing Company, Boston, 1997);
Salzberg, Searles, Kasif,
10 (Ed.), Computational Methods in Molecular Biology, (Elsevier, Amsterdam,
1998); Rashidi and
Buehler, Bioinformatics Basics: Application in Biological Science and Medicine
(CRC Press, London,
2000) and Ouelette and Bzevanis Bioinforniatics: A Practical Guide for
Analysis of Gene and Proteins
(Wiley & Sons, Inc., 2<sup>nd</sup> ed., 2001). See U.S. Pat. No. 6,420,108.
The present methods may also make use of various computer program products and
software
15 for a variety of purposes, such as probe design, management of data,
analysis, and instrument
operation. See, U.S. Pat. Nos. 5,593,839, 5,795,716, 5,733,729, 5,974,164,
6,066,454,6,090,555,
6,185,561, 6,188,783, 6,223,127, 6,229,911 and 6,308,170.
Additionally, the present methods relates to embodiments that include methods
for providing
genetic information over networks such as the Internet as shown in U.S. Ser.
Nos. 10/197,621,
20 10/063,559 (U.S. Publication Number 20020183936), 10/065,856,
10/065,868, 10/328,818,
10/328,872, 10/423,403, and 60/482,389. For example, one or more molecular
profiling techniques
can be performed in one location, e.g., a city, state, country or continent,
and the results can be
transmitted to a different city, state, country or continent. Treatment
selection can then be made in
whole or in part in the second location. The methods as described herein
comprise transmittal of
25 information between different locations.
Conventional data networking, application development and other functional
aspects of the
systems (and components of the individual operating components of the systems)
may not be
described in detail herein but are part as described herein. Furthermore, the
connecting lines shown in
the various figures contained herein are intended to represent illustrative
functional relationships
30 and/or physical couplings between the various elements. It should be
noted that many alternative or
additional functional relationships or physical connections may be present in
a practical system.
The various system components discussed herein may include one or more of the
following: a
host server or other computing systems including a processor tor processing
digital data; a memory
coupled to the processor for storing digital data; an input digitizer coupled
to the processor for
35 inputting digital data; an application program stored in the memory and
accessible by the processor
for directing processing of digital data by the processor; a display device
coupled to the processor and
memory for displaying information derived from digital data processed by the
processor; and a
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
plurality of databases. Various databases used herein may include: patient
data such as family history,
demography and environmental data, biological sample data, prior treatment and
protocol data, patient
clinical data, molecular profiling data of biological samples, data on
therapeutic drug agents and/or
investigative drugs, a gene library, a disease library, a drug library,
patient tracking data, file
5 management data, financial management data, billing data and/or like data
useful in the operation of
the system. As those skilled in the art will appreciate, user computer may
include an operating system
(e.g., Windows NT, 95/98/2000, 0S2, UNIX, Linux, Solaris, MacOS, etc.) as well
as various
conventional support software and drivers typically associated with computers.
The computer may
include any suitable personal computer, network computer, workstation,
minicomputer, mainframe or
10 the like. User computer can be in a home or medical/business environment
with access to a network.
In an illustrative embodiment, access is through a network or the Internet
through a commercially-
available web-browser software package.
As used herein, the term "network" shall include any electronic communications
means which
incorporates both hardware and software components of such. Communication
among the parties may
15 be accomplished through any suitable communication channels, such as,
for example, a telephone
network, an extranet, an intranet, Internet, point of interaction device,
personal digital assistant (e.g.,
Palm Pilot , Blackberry ), cellular phone, kiosk, etc.), online
communications, satellite
communications, off-line communications, wireless communications, transponder
communications,
local area network (LAN), wide area network (WAN), networked or linked
devices, keyboard, mouse
20 and/or any suitable communication or data input modality. Moreover,
although the system is
frequently described herein as being implemented with TCP/IP communications
protocols, the system
may also be implemented using 1PX, Appletalk, 113-6, NetBIOS, OSI or any
number of existing or
future protocols. If the network is in the nature of a public network, such as
the Internet, it may be
advantageous to presume the network to be insecure and open to eavesdroppers.
Specific information
25 related to the protocols, standards, and application software used in
connection with the Internet is
generally known to those skilled in the art and, as such, need not be detailed
herein. See, for example,
Dilip Naik, Internet Standards and Protocols (1998); Java 2 Complete, various
authors, (Sybex 1999);
Deborah Ray and Eric Ray, Mastering HTML 4.0 (1997); and Loshin, TCP/IP
Clearly Explained
(1997) and David Gourley and Brian Totty, HTTP, The Defmitive Guide (2002),
the contents of which
30 are hereby incorporated by reference.
The various system components may be independently, separately or collectively
suitably
coupled to the network via data links which includes, for example, a
connection to an Internet Service
Provider (1SP) over the local loop as is typically used m connection with
standard modem
communication, cable modem, Dish networks, ISDN, Digital Subscriber Line
(DSL), or various
35 wireless communication methods, see, e.g., Gilbert Held, Understanding
Data Communications
(1996), which is hereby incorporated by reference. It is noted that the
network may be implemented as
other types of networks, such as an interactive television (ITV) network.
Moreover, the system
71
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
contemplates the use, sale or distribution of any goods, services or
information over any network
having similar functionality described herein.
As used herein, "transmit" may include sending electronic data from one system
component
to another over a network connection. Additionally, as used herein, "data" may
include encompassing
5 information such as commands, queries, files, data for storage, and the
like in digital or any other
form.
The system contemplates uses in association with web services, utility
computing, pervasive
and individualized computing, security and identity solutions, autonomic
computing, commodity
computing, mobility and wireless solutions, open source, biometrics, grid
computing and/or mesh
10 computing.
Any databases discussed herein may include relational, hierarchical,
graphical, or object-
oriented structure and/or any other database configurations. Common database
products that may be
used to implement the databases include DB2 by IBM (White Plains, NY), various
database products
available from Oracle Corporation (Redwood Shores, CA), Microsoft Access or
Microsoft SQL
15 Server by Microsoft Corporation (Redmond, Washington), or any other
suitable database product
Moreover, the databases may be organized in any suitable manner, for example,
as data tables or
lookup tables. Lath record may be a single file, a series of files, a linked
series of data fields or any
other data structure. Association of certain data may be accomplished through
any desired data
association technique such as those known or practiced in the art. For
example, the association may be
20 accomplished either manually or automatically. Automatic association
techniques may include, for
example, a database search, a database merge, GREP, AGREP, SQL, using a key
field in the tables to
speed searches, sequential searches through all the tables and files, sorting
records in the file
according to a known order to simplify lookup, and/or the like. The
association step may be
accomplished by a database merge function, for example, using a "key field" in
pre-selected databases
25 or data sectors.
More particularly, a "key field" partitions the database according to the high-
level class of
objects defmed by the key field. For example, certain types of data may be
designated as a key field in
a plurality of related data tables and the data tables may then be linked on
the basis of the type of data
in the key field. The data corresponding to the key field in each of the
linked data tables is preferably
30 the same or of the same type. However, data tables having similar,
though not identical, data in the
key fields may also be linked by using AGREP, for example. In accordance with
one embodiment, any
suitable data storage technique may be used to store data without a standard
format. Data sets may be
stored using any suitable technique, including, for example, storing
individual tiles using an ISO/1EC
7816-4 file structure; implementing a domain whereby a dedicated file is
selected that exposes one or
35 more elementary files containing one or more data sets; using data sets
stored in individual files using
a hierarchical filing system; data sets stored as records in a single file
(including compression, SQL
accessible, hashed vione or more keys, numeric, alphabetical by first tuple,
etc.); Binary Large Object
72
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
(BLOB); stored as ungrouped data elements encoded using ISO/WC 7816-6 data
elements; stored as
ungrouped data elements encoded using IsurEc Abstract Syntax Notation (ASN.1)
as in IsurEc
8824 and 8825; and/or other proprietary techniques that may include fractal
compression methods,
image compression methods, etc.
5 In one illustrative embodiment, the ability to store a wide variety of
information in different
formats is facilitated by storing the information as a BLOB. Thus, any binary
information can be
stored in a storage space associated with a data set. The BLOB method may
store data sets as
ungrouped data elements formatted as a block of binary via a fixed memory
offset using either fixed
storage allocation, circular queue techniques, or best practices with respect
to memory management
10 (e.g., paged memory, least recently used, etc.). By using BLOB methods,
the ability to store various
data sets that have different formats facilitates the storage of data by
multiple and unrelated owners of
the data sets. For example, a first data set which may be stored may be
provided by a first party, a
second data set which may be stored may be provided by an unrelated second
party, and yet a third
data set which may be stored, may be provided by a third party unrelated to
the first and second party.
15 Each of these three illustrative data sets may contain different
information that is stored using
different data storage formats and/or techniques. Further, each data set may
contain subsets of data
that also may be distinct from other subsets.
As stated above, in various embodiments, the data can be stored without regard
to a common
format However, in one illustrative embodiment, the data set (e.g., BLOB) may
be annotated in a
20 standard manner when provided for manipulating the data. The annotation
may comprise a short
header, trailer, or other appropriate indicator related to each data set that
is configured to convey
information useful in managing the various data sets. For example, the
annotation may be called a
"condition header", "header", "trailer", or "status", herein, and may comprise
an indication of the
status of the data set or may include an identifier correlated to a specific
issuer or owner of the data.
25 Subsequent bytes of data may be used to indicate for example, the
identity of the issuer or owner of
the data, user, transaction/membership account identifier or the like. Each of
these condition
annotations are further discussed herein.
The data set annotation may also be used for other types of status information
as well as
various other purposes. For example, the data set annotation may include
security information
30 establishing access levels. The access levels may, for example, be
configured to permit only certain
individuals, levels of employees, companies, or other entities to access data
sets, or to permit access to
specific data sets based on the transaction, issuer or owner of data, user or
the like. Furthermore, the
security information may restrict/permit only certain actions such as
accessing, modifying, and/or
deleting data sets. In one example, the data set annotation indicates that
only the data set owner or the
35 user are permitted to delete a data set, various identified users may be
permitted to access the data set
for reading, and others are altogether excluded from accessing the data set.
However, other access
restriction parameters may also be used allowing various entities to access a
data set with various
73
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
permission levels as appropriate. The data, including the header or trailer
may be received by a
standalone interaction device configured to add, delete, modify, or augment
the data in accordance
with the header or trailer
One skilled in the art will also appreciate that, for security reasons, any
databases, systems,
5 devices, servers or other components of the system may consist of any
combination thereof at a single
location or at multiple locations, wherein each database or system includes
any of various suitable
security features, such as firewalls, access codes, encryption, decryption,
compression,
decompression, and/or the like.
The computing unit of the web client may be further equipped with an Internet
browser
10 connected to the Internet or an intranet using standard dial-up, cable,
DSL or any other Internet
protocol known in the art. Transactions originating at a web client may pass
through a firewall in
order to prevent unauthorized access from users of other networks. Further,
additional fuewalls may
be deployed between the varying components of CMS to further enhance security
Firewall may include any hardware and/or software suitably configured to
protect CMS
15 components and/or enterprise computing resources from users of other
networks. Further, a firewall
may be configured to limit or restrict access to various systems and
components behind the firewall
for web clients connecting through a web server. Firewall may reside in
varying configurations
including Stateful Inspection, Proxy based and Packet Filtering among others.
Firewall may be
integrated within an web server or any other CMS components or may further
reside as a separate
20 entity.
The computers discussed herein may provide a suitable website or other
Internet-based
graphical user interface which is accessible by users. In one embodiment, the
Microsoft Internet
Information Server (HS), Microsoft Transaction Server (MTS), and Microsoft SQL
Server, are used in
conjunction with the Microsoft operating system, Microsoft NT web server
software, a Microsoft SQL
25 Server database system, and a Microsoft Commerce Server. Additionally,
components such as Access
or Microsoft SQL Server, Oracle, Sybase, Informix MySQL, Interbase, etc., may
be used to provide
an Active Data Object (ADO) compliant database management system.
Any of the communications, inputs, storage, databases or displays discussed
herein may be
facilitated through a website having web pages. The term "web page" as it is
used herein is not meant
30 to limit the type of documents and applications that might be used to
interact with the user. For
example, a typical website might include, in addition to standard HTML
documents, various forms,
Java applets, JavaScript, active server pages (ASP), common gateway interface
scripts (COI),
extensible markup language (XML), dynamic HTML, cascading style sheets (CSS),
helper
applications, plug-ins, and the like. A server may include a web service that
receives a request from a
35 web server, the request including a URL
(http://yahoo.comistockquotes/ge) and an IF address
(123.56.789.234). The web server retrieves the appropriate web pages and sends
the data or
applications for the web pages to the IP address. Web services are
applications that are capable of
74
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
interacting with other applications over a communications means, such as the
internet. Web services
are typically based on standards or protocols such as XML, XSLT, SOAP, WSDL
and UDDI. Web
services methods are well known in the art, and are covered in many standard
texts. See, e.g., Alex
Nghiem, IT Web Services: A Roadmap for the Enterprise (2003), hereby
incorporated by reference.
5 The web-based clinical database for the system and method of the present
methods preferably
has the ability to upload and store clinical data files in native formats and
is searchable on any clinical
parameter. The database is also scalable and may use an EAV data model
(metadata) to enter clinical
annotations from any study for easy integration with other studies. In
addition, the web-based clinical
database is flexible and may be XNIL and XSLT enabled to be able to add user
customized questions
10 dynamically. Further, the database includes exportability to CDISC ODM.
Practitioners will also appreciate that there are a number of methods for
displaying data
within a browser-based document. Data may be represented as standard text or
within a fixed list,
scrollable list, drop-down list, editable text field, fixed text field, pop-up
window, and the like.
Likewise, there are a number of methods available for modifying data in a web
page such as, for
15 example, free text entry using a keyboard, selection of menu items,
cheek boxes, option boxes, and
the like.
The system and method may be described herein in terms of functional block
components,
screen shots, optional selections and various processing steps. It should be
appreciated that such
functional blocks may be realized by any number of hardware and/or software
components configured
20 to perform the specified functions. For example, the system may employ
various integrated circuit
components, e.g., memory elements, processing elements, logic elements, look-
up tables, and the like,
which may carry out a variety of functions under the control of one or more
microprocessors or other
control devices. Similarly, the software elements of the system may be
implemented with any
programming or scripting language such as C, C++, Macromedia Cold Fusion,
Microsoft Active
25 Server Pages, Java, COBOL, assembler, PERL, Visual Basic, SQL Stored
Procedures, extensible
markup language (XNIL), with the various algorithms being implemented with any
combination of
data structures, objects, processes, routines or other programming elements.
Further, it should be
noted that the system may employ any number of conventional techniques for
data transmission,
signaling, data processing, network control, and the like. Still further, the
system could be used to
30 detect or prevent security issues with a client-side scripting language,
such as JavaScript, VBScript or
the like. For a basic introduction of cryptography and network security, see
any of the following
references: (1) "Applied Cryptography: Protocols, Algorithms, And Source Code
In C," by Bruce
Schneier, published by John Wiley & Sons (second edition, 1995); (2) "Java
Cryptography" by
Jonathan Knudson, published by O'Reilly & Associates (1998); (3) "Cryptography
& Network
35 Security: Principles & Practice" by William Stallings, published by
Prentice Hall; all of which are
hereby incorporated by reference.
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
As used herein, the term "end user", "consumer", "customer", "client",
"treating physician",
"hospital", or "business" may be used interchangeably with each other, and
each shall mean any
person, entity, machine, hardware, software or business. Each participant is
equipped with a
computing device in order to interact with the system and facilitate online
data access and data input.
5 The customer has a computing unit in the form of a personal computer,
although other types of
computing units may be used including laptops, notebooks, hand held computers,
set-top boxes,
cellular telephones, touch-tone telephones and the like. The owner/operator of
the system and method
of the present methods has a computing unit implemented in the form of a
computer-server, although
other implementations are contemplated by the system including a computing
center shown as a main
10 frame computer, a mini-computer, a PC server, a network of computers
located in the same of
different geographic locations, or the like. Moreover, the system contemplates
the use, sale or
distribution of any goods, services or information over any network having
similar functionality
described herein.
In one illustrative embodiment, each client customer may be issued an
"account" or "account
15 number". As used herein, the account or account number may include any
device, code, number,
letter, symbol, digital certificate, smart chip, digital signal, analog
signal, biometric or other
identifierindicia suitably configured to allow the consumer to access,
interact with or communicate
with the system (e.g., one or more of an authorizatiortfaccess code, personal
identification number
(PIN), Internet code, other identification code, and/or the like). The account
number may optionally
20 be located on or associated with a charge card, credit card, debit card,
prepaid card, embossed card,
smart card, magnetic stripe card, bar code card, transponder, radio frequency
card or an associated
account. The system may include or interface with any of the foregoing cards
or devices, or a fob
having a transponder and REID reader in RE communication with the fob.
Although the system may
include a fob embodiment, the methods is not to be so limited. Indeed, system
may include any device
25 having a transponder which is configured to communicate with REID reader
via RF communication.
Typical devices may include, for example, a key ring, tag, card, cell phone,
wristwatch or any such
form capable of being presented for interrogation. Moreover, the system,
computing unit or device
discussed herein may include a "pervasive computing device," which may include
a traditionally non-
computerized device that is embedded with a computing unit. The account number
may be distributed
30 and stored in any form of plastic, electronic, magnetic, radio
frequency, wireless, audio and/or optical
device capable of transmitting or downloading data from itself to a second
device.
As will be appreciated by one of ordinary skill in the art, the system may be
embodied as a
customization of an existing system, an add-on product, upgraded software, a
standalone system, a
distributed system, a method, a data processing system, a device for data
processing, and/or a
35 computer program product. Accordingly, the system may take the form of
an entirely software
embodiment, an entirely hardware embodiment, or an embodiment combining
aspects of both
software and hardware. Furthermore, the system may take the form of a computer
program product on
76
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
a computer-readable storage medium haying computer-readable program code means
embodied in the
storage medium. Any suitable computer-readable storage medium may be used,
including hard disks,
CD-ROM, optical storage devices, magnetic storage devices, and/or the like.
The system and method is described herein with reference to screen shots,
block diagrams and
5 flowchart illustrations of methods, apparatus (e.g., systems), and
computer program products
according to various embodiments. It will be understood that each Functional
block of the block
diagrams and the flowchart illustrations, and combinations of functional
blocks in the block diagrams
and flowchart illustrations, respectively, can be implemented by computer
program instructions.
These computer program instructions may be loaded onto a general purpose
computer, special
10 purpose computer, or other programmable data processing apparatus to
produce a machine, such that
the instructions that execute on the computer or other programmable data
processing apparatus create
means for implementing the functions specified in the flowchart block or
blocks. These computer
program instructions may also be stored in a computer-readable memory that can
direct a computer or
other programmable data processing apparatus to function in a particular
manner, such that the
15 instructions stored in the computer-readable memory produce an article
of manufacture including
instruction means which implement the function specified in the flowchart
block or blocks. The
computer program instructions may also be loaded onto a computer or other
programmable data
processing apparatus to cause a series of operational steps to be performed on
the computer or other
programmable apparatus to produce a computer-implemented process such that the
instructions which
20 execute on the computer or other programmable apparatus provide steps
for implementing the
functions specified in the flowchart block or blocks.
Accordingly, functional blocks of the block diagrams and flowchart
illustrations support
combinations of means for performing the specified functions, combinations of
steps for performing
the specified functions, and program instruction means for performing the
specified functions. It will
25 also be understood that each functional block of the block diagrams and
flowchart illustrations, and
combinations of functional blocks in the block diagrams and flowchart
illustrations, can be
implemented by either special purpose hardware-based computer systems which
perform the specified
functions or steps, or suitable combinations of special purpose hardware and
computer instructions.
Further, illustrations of the process flows and the descriptions thereof may
make reference to user
30 windows, web pages, websites, web forms, prompts, etc. Practitioners
will appreciate that the
illustrated steps described herein may comprise in any number of
configurations including the use of
windows, web pages, web forms, popup windows, prompts and the like. It should
be further
appreciated that the multiple steps as illustrated and described may be
combined into single web pages
and/or windows but have been expanded for the sake of simplicity. In other
cases, steps illustrated and
35 described as single process steps may be separated into multiple web
pages and/or windows but have
been combined for simplicity.
77
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Molecular Profiling
The molecular profiling approach provides a method for selecting a candidate
treatment for an
individual that could favorably change the clinical course for the individual
with a condition or
disease, such as cancer. The molecular profiling approach provides clinical
benefit for individuals,
5 such as identifying therapeutic regimens that provide a longer
progression free survival (PI'S), longer
disease free survival (DFS), longer overall survival (OS) or extended
lifespan. Methods and systems
as described herein are directed to molecular profiling of cancer on an
individual basis that can
identify optimal therapeutic regimens. Molecular profiling provides a
personalized approach to
selecting candidate treatments that are likely to benefit a cancer. The
molecular profiling methods
10 described herein can be used to guide treatment in any desired setting,
including without limitation the
front-line / standard of care setting, or for patients with poor prognosis,
such as those with metastatic
disease or those whose cancer has progressed on standard front line therapies,
or whose cancer has
progressed on previous chemotherapeutic or hormonal regimens.
The systems and methods of the invention may be used to classify patients as
more or less
15 likely to benefit or respond to various treatments. Unless otherwise
noted, the terms "response" or
"non-response," as used herein, refer to any appropriate indication that a
treatment provides a benefit
to a patient (a "responder" or "benefiter") or has a lack of benefit to the
patient (a "non-responder" or
"non-benefiter"). Such an indication may be determined using accepted clinical
response criteria such
as the standard Response Evaluation Criteria in Solid Tumors (RECIST)
criteria, or any other useful
20 patient response criteria such as progression free survival (PFS), time
to progression (TTP), disease
free survival (DFS), time-to-next treatment (TNT, TTNT), time-to-treatment
failure (TTF, TTTF),
tumor shrinkage or disappearance, or the like. RECIST is a set of rules
published by an international
consortium that define when tumors improve ("respond"), stay the same
("stabilize"), or worsen
("progress") during treatment of a cancer patient. As used herein and unless
otherwise noted, a patient
25 "benefit" from a treatment may refer to any appropriate measure of
improvement, including without
limitation a RECIST response or longer PFS/TTP/DFS/TNT/TTNT, whereas "lack of
benefit" from a
treatment may refer to any appropriate measure of worsening disease during
treatment. Generally
disease stabilization is considered a benefit, although in certain
circumstances, if so noted herein,
stabilization may be considered a lack of benefit. A predicted or indicated
benefit may be described as
30 "indeterminate" if there is not an acceptable level of prediction of
benefit or lack of benefit. In some
cases, benefit is considered indeterminate if it cannot be calculated, e.g.,
due to lack of necessary data.
Personalized medicine based on pharrnacogenetic insights, such as those
provided by
molecular profiling as described herein, is increasingly taken for granted by
some practitioners and
the lay press, but forms the basis of hope for improved cancer therapy.
However, molecular profiling
35 as taught herein represents a fundamental departure from the traditional
approach to oncologic therapy
where for the most part, patients are grouped together and treated with
approaches that are based on
fmdings from light microscopy and disease stage. Traditionally, differential
response to a particular
78
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
therapeutic strategy has only been determined after the treatment was given,
i.e., a posteriori. The
"standard" approach to disease treatment relies on what is generally true
about a given cancer
diagnosis and treatment response has been vetted by randomized phase III
clinical trials and forms the
"standard of care" in medical practice. The results of these trials have been
codified in consensus
5 statements by guidelines organizations such as the National Comprehensive
Cancer Network and The
American Society of Clinical Oncology. The NCCN Compendium irm contains
authoritative,
scientifically derived information designed to support decision-making about
the appropriate use of
drugs and biologics in patients with cancer. The NCCN Compendiunirm is
recognized by the Centers
for Medicare and Medicaid Services (CMS) and United Healthcare as an
authoritative reference for
10 oncology coverage policy. On-compendium treatments are those recommended
by such guides. The
biostatistical methods used to validate the results of clinical trials rely on
minimizing differences
between patients, and are based on declaring the likelihood of error that one
approach is better than
another for a patient group defmed only by light microscopy and stage, not by
individual differences
in tumors. The molecular profiling methods described herein exploit such
individual differences. The
15 methods can provide candidate treatments that can be then selected by a
physician for treating a
patient.
Molecular profiling can be used to provide a comprehensive view of the
biological state of a
sample. In an embodiment, molecular profiling is used for whole tumor
profiling. Accordingly, a
number of molecular approaches are used to assess the state of a tumor. The
whole tumor profiling
20 can be used for selecting a candidate treatment for a tumor. Molecular
profiling can be used to select
candidate therapeutics on any sample for any stage of a disease. In
embodiment, the methods as
described herein are used to profile a newly diagnosed cancer. The candidate
treatments indicated by
the molecular profiling can be used to select a therapy for treating the newly
diagnosed cancer. In
other embodiments, the methods as described herein are used to profile a
cancer that has already been
25 treated, e.g., with one or more standard-of-care therapy. In
embodiments, the cancer is refractory to
the prior treatment/s. For example, the cancer may be refractory to the
standard of care treatments for
the cancer. The cancer can be a metastatic cancer or other recurrent cancer.
The treatments can be on-
compendium or off-compendium treatments.
Molecular profiling can be performed by any known means for detecting a
molecule in a
30 biological sample. Molecular profiling comprises methods that include
but are not limited to, nucleic
acid sequencing, such as a DNA sequencing or RNA sequencing;
irmnunohistochemistry (ffIC); in
situ hybridization (ISH); fluorescent in situ hybridization (FISH);
chromogenic in situ hybridization
(CISH); PCR amplification (e.g., qPCR or RI-PCR); various types of microarray
(mRNA expression
arrays, low density arrays, protein arrays, etc); various types of sequencing
(Sanger, pyrosequencing,
35 etc); comparative genomic hybridization (CGH); high throughput or next
generation sequencing
(NGS); Northern blot; Southern blot; immunoassay; and any other appropriate
technique to assay the
presence or quantity of a biological molecule of interest. In various
embodiments, any one or more of
79
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
these methods can be used concurrently or subsequent to each other for
assessing target genes
disclosed herein.
Molecular profiling of individual samples is used to select one or more
candidate treatments
for a disorder in a subject, e.g., by identifying targets for drugs that may
be effective for a given
5 cancer. For example, the candidate treatment can be a treatment known to
have an effect on cells that
di fferentially express genes as identified by molecular profiling techniques,
an experimental drug, a
government or regulatory approved drug or any combination of such drugs, which
may have been
studied and approved for a particular indication that is the same as or
different from the indication of
the subject from whom a biological sample is obtain and molecularly profiled.
10 When multiple biomarker targets are revealed by assessing target genes
by molecular
profiling, one or more decision rules can be put in place to prioritize the
selection of certain
therapeutic agent for treatment of an individual on a personalized basis.
Rules as described herein aide
prioritizing treatment, e.g., direct results of molecular profiling,
anticipated efficacy of therapeutic
agent, prior history with the same or other treatments, expected side effects,
availability of therapeutic
15 agent, cost of therapeutic agent, drug-drug interactions, and other
factors considered by a treating
physician. Based on the recommended and prioritized therapeutic agent targets,
a physician can
decide on the course of treatment for a particular individual. Accordingly,
molecular profiling
methods and systems as described herein can select candidate treatments based
on individual
characteristics of diseased cells, e.g., tumor cells, and other personalized
factors in a subject in need of
20 treatment, as opposed to relying on a traditional one-size fits all
approach that is conventionally used
to treat individuals suffering from a disease, especially cancer. In some
cases, the recommended
treatments are those not typically used to treat the disease or disorder
inflicting the subject. In some
cases, the recommended treatments are used after standard-of-care therapies
are no longer providing
adequate efficacy.
25 The treating physician can use the results of the molecular profiling
methods to optimize a
treatment regimen for a patient. The candidate treatment identified by the
methods as described herein
can be used to treat a patient; however, such treatment is not required of the
methods. Indeed, the
analysis of molecular profiling results and identification of candidate
treatments based on those results
can be automated and does not require physician involvement.
30 Biological Entities
Nucleic acids include deoxyribonucleotides or ribonucleotides and polymers
thereof in either
single- or double-stranded form, or complements thereof Nucleic acids can
contain known nucleotide
analogs or modified backbone residues or linkages, which are synthetic,
naturally occurring, and non-
naturally occurring, which have similar binding properties as the reference
nucleic acid, and which are
35 metabolized in a manner similar to the reference nucleotides. Examples
of such analogs include,
without limitation, phosphorothioates, phosphoramidates, methyl phosphonates,
chiral-methyl
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
phosphonates, 2-0-methyl ribonucleotides, peptide-nucleic acids (PNAs).
Nucleic acid sequence can
encompass conservatively modified variants thereof (e.g., degenerate codon
substitutions) and
complementary sequences, as well as the sequence explicitly indicated.
Specifically, degenerate codon
substitutions may be achieved by generating sequences in which the third
position of one or more
5 selected (or all) codons is substituted with mixed-base and/or
deoxyinosine residues (Batzer et al.,
Nucleic Acid Res. 19:5081 (1991); Ohtsuka et al., J. Biol. Chem. 260:2605-2608
(1985); Rossolini et
al., Mol. Cell Probes 8:91-98 (1994)). The tenn nucleic acid can be used
interchangeably with gene,
cDNA, mRNA, oligonucleotide, and polynucleotide.
A particular nucleic acid sequence may implicitly encompass the particular
sequence and
10 "splice variants" and nucleic acid sequences encoding truncated forms.
Similarly, a particular protein
encoded by a nucleic acid can encompass any protein encoded by a splice
variant or truncated form of
that nucleic acid. "Splice variants," as the name suggests, are products of
alternative splicing of a
gene. After transcription, an initial nucleic acid transcript may be spliced
such that different (alternate)
nucleic acid splice products encode different polypeptides. Mechanisms for the
production of splice
15 variants vary, but include alternate splicing of exons. Alternate
polypeptides derived from the same
nucleic acid by read-through transcription are also encompassed by this
definition. Any products of a
splicing reaction, including recombinant forms of the splice products, are
included in this definition.
Nucleic acids can be truncated at the 5' end or at the 3' end. Polypeptides
can be truncated at the N-
terminal end or the C-terminal end. Truncated versions of nucleic acid or
polypeptidc sequences can
20 be naturally occurring or created using recombinant techniques.
The terms "genetic variant" and "nucleotide variant" are used herein
interchangeably to refer
to changes or alterations to the reference human gene or cDNA sequence at a
particular locus,
including, but not limited to, nucleotide base deletions, insertions,
inversions, and substitutions in the
coding and non-coding regions. Deletions may be of a single nucleotide base, a
portion or a region of
25 the nucleotide sequence of the gene, or of the entire gene sequence.
Insertions may be of one or more
nucleotide bases. The genetic variant or nucleotide variant may occur in
transcriptional regulatory
regions, untranslated regions of mKNA, exons, introns, exonAntron junctions,
etc. The genetic variant
or nucleotide variant can potentially result in stop codons, frame shifts,
deletions of amino acids,
altered gene transcript splice forms or altered amino acid sequence.
30 An allele or gene allele comprises generally a naturally occurring gene
having a reference
sequence or a gene containing a specific nucleotide variant.
A haplotype refers to a combination of genetic (nucleotide) variants in a
region of an mR_NA
or a genonuc DNA on a chromosome found in an individual. Thus, a haplotype
includes a number of
genetically linked polymorphic variants which are typically inherited together
as a unit.
35 As used herein, the term "amino acid variant" is used to refer to an
amino acid change to a
reference human protein sequence resulting from genetic variants or nucleotide
variants to the
reference human gene encoding the reference protein. The term "amino acid
variant" is intended to
81
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
encompass not only single amino acid substitutions, but also amino acid
deletions, insertions, and
other significant changes of amino acid sequence in the reference protein.
The term "genotype" as used herein means the nucleotide characters at a
particular nucleotide
variant marker (or locus) in either one allele or both alleles of a gene (or a
particular chromosome
5 region). With respect to a particular nucleotide position of a gene of
interest, the nucleotide(s) at that
locus or equivalent thereof in one or both alleles form the genotype of the
gene at that locus. A
genotype can be homozygous or heterozygous. Accordingly, "genotyping" means
determining the
genotype, that is, the nucleotide(s) at a particular gene locus. Genotyping
can also be done by
determining the amino acid variant at a particular position of a protein which
can be used to deduce
10 the corresponding nucleotide variant(s).
The term "locus" refers to a specific position or site in a gene sequence or
protein. Thus, there
may be one or more contiguous nucleotides in a particular gene locus, or one
or more amino acids at a
particular locus in a polypeptide. Moreover, a locus may refer to a particular
position in a gene where
one or more nucleotides have been deleted, inserted, or inverted.
15 Unless specified otherwise or understood by one of skill in art, the
terms "polypeptide,"
"protein," and "peptide" are used interchangeably herein to refer to an amino
acid chain in which the
amino acid residues are linked by covalent peptide bonds. The amino acid chain
can be of any length
of at least two amino acids, including full-length proteins. Unless otherwise
specified, polypeptide,
protein, and peptide also encompass various modified forms thereof, including
but not limited to
20 glycosylated forms, phosphorylated forms, etc. A polypeptide, protein or
peptide can also be referred
to as a gene product
Lists of gene and gene products that can be assayed by molecular profiling
techniques are
presented herein. Lists of genes may be presented in the context of molecular
profiling techniques that
detect a gene product (e.g., an mR_NA or protein). One of skill will
understand that this implies
25 detection of the gene product of the listed genes. Similarly, lists of
gene products may be presented in
the context of molecular profiling techniques that detect a gene sequence or
copy number. One of skill
will understand that this implies detection of the gene corresponding to the
gene products, including
as an example DNA encoding the gene products. As will be appreciated by those
skilled in the art, a
"biomarker" or "marker" comprises a gene and/or gene product depending on the
context.
30 The terms "label" and "detectable label" can refer to any composition
detectable by
spectroscopic, photochemical, biochemical, immunochemical, electrical,
optical, chemical or similar
methods. Such labels include biotin for staining with labeled streptavidin
conjugate, magnetic beads
(e.g., DYNABEADSTm), fluorescent dyes (e.g., fluorescein, 'Texas red,
rhodamme, green fluorescent
protein, and the like), radiolabels (e.g., 311, 1251, 35s, 14C, or 32F.),
enzymes k 'e.g.,
horse radish
35 peroxidase, alkaline phosphatase and others commonly used in an ELISA),
and calorimetric labels
such as colloidal gold or colored glass or plastic (e.g., polystyrene,
polypropylene, latex, etc) beads.
Patents teaching the use of such labels include U.S. Pat. Nos. 3,817,837;
3,850,752; 3,939,350;
82
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
3,996,345; 4,277,437; 4,275,149; and 4,366,241. Means of detecting such labels
are well known to
those of skill in the art. Thus, for example, radiolabels may be detected
using photographic film or
scintillation counters, fluorescent markers may be detected ming a
photodetector to detect emitted
light. Enzymatic labels are typically detected by providing the enzyme with a
substrate and detecting
5 the reaction product produced by the action of the enzyme on the
substrate, and calorimetric labels are
detected by simply visualizing the colored label. Labels can include, e.g.,
ligands that bind to labeled
antibodies, fluorophores, chemiluminescent agents, enzymes, and antibodies
which can serve as
specific binding pair members for a labeled ligand. An introduction to labels,
labeling procedures and
detection of labels is found in Polak and Van Noorden Introduction to
Immunocytochemistry, 2nd ed.,
10 Springer Verlag, NY (1997); and in Haugland Handbook of Fluorescent
Probes and Research
Chemicals, a combined handbook and catalogue Published by Molecular Probes,
Inc. (1996).
Detectable labels include, but are not limited to, nucleotides (labeled or
unlabelled),
compomers, sugars, peptides, proteins, antibodies, chemical compounds,
conducting polymers,
binding moieties such as biotin, mass tags, calorimetric agents, light
emitting agents,
15 chemiluminescent agents, light scattering agents, fluorescent tags,
radioactive tags, charge tags
(electrical or magnetic charge), volatile tags and hydrophobic tags,
biomolecules (e.g., members of a
binding pair antibody/antigen, antibody/antibody, antibody/antibody fragment,
antibody/antibody
receptor, antibody/protein A or protein G, hapten/anti-hapten, biotin/avidin,
biotin/streptavidin, folic
acidIfolate binding protein, vitamin B12/intrinsic factor, chemical reactive
group/complementary
20 chemical reactive group (e.g., sulfhydryl/maleimide,
sulfhydryl/haloacetyl derivative,
amine/isotriocyanate, amine/succinimidyl ester, and arnine/sulfonyl halides)
and the like.
The terms "primer", "probe," and "oligonucleotide" are used herein
interchangeably to refer
to a relatively short nucleic acid fragment or sequence. They can comprise
DNA, RNA, or a hybrid
thereof, or chemically modified analog or derivatives thereof. Typically, they
are single-stranded.
25 However, they can also be double-stranded having two complementing
strands which can be
separated by denaturation. Normally, primers, probes and oligonucleotides have
a length of from
about 8 nucleotides to about 200 nucleotides, preferably from about 12
nucleotides to about 100
nucleotides, and more preferably about 18 to about 50 nucleotides. They can be
labeled with
detectable markers or modified using conventional manners for various
molecular biological
30 applications.
The term "isolated" when used in reference to nucleic acids (e.g., genomic
DNAs, cDNAs,
mRNAs, or fragments thereof) is intended to mean that a nucleic acid molecule
is present in a form
that is substantially separated from other naturally occurring nucleic acids
that are normally associated
with the molecule. Because a naturally existing chromosome (or a viral
equivalent thereof) includes a
35 long nucleic acid sequence, an isolated nucleic acid can be a nucleic
acid molecule having only a
portion of the nucleic acid sequence in the chromosome but not one or more
other portions present on
the same chromosome. More specifically, an isolated nucleic acid can include
naturally occurring
83
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
nucleic acid sequences that flank the nucleic acid in the naturally existing
chromosome (or a viral
equivalent thereof). An isolated nucleic acid can be substantially separated
from other naturally
occurring nucleic acids that are on a different chromosome of the same
organism. An isolated nucleic
acid can also be a composition in which the specified nucleic acid molecule is
significantly enriched
5 so as to constitute at least 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%,
95%, or at least 99% of
the total nucleic acids in the composition.
An isolated nucleic acid can be a hybrid nucleic acid having the specified
nucleic acid
molecule covalently linked to one or more nucleic acid molecules that are not
the nucleic acids
naturally flanking the specified nucleic acid. For example, an isolated
nucleic acid can be in a vector.
10 In addition, the specified nucleic acid may have a nucleotide sequence
that is identical to a naturally
occurring nucleic acid or a modified form or mutein thereof having one or more
mutations such as
nucleotide substitution, deletion/insertion, inversion, and the like.
An isolated nucleic acid can be prepared from a recombinant host cell (in
which the nucleic
acids have been recombinantly amplified and/or expressed), or can be a
chemically synthesized
15 nucleic acid having a naturally occurring nucleotide sequence or an
artificially modified form thereof
The term "high stringency hybridization conditions," when used in connection
with nucleic
acid hybridization, includes hybridization conducted overnight at 42 C in a
solution containing 50%
formamide, 5 xSSC (750 mIVINaCI, 75 mI\4 sodium citrate), 50 m1\4 sodium
phosphate, pH 7.6,
xDenhardt's solution, 10% dextran sulfate, and 20 microgram/nil denatured and
sheared salmon
20 sperm DNA, with hybridization filters washed in 0.1xSSC at about 65 'C.
The tenn "moderate
stringent hybridization conditions," when used in connection with nucleic acid
hybridization, includes
hybridization conducted overnight at 37 C in a solution containing 50%
forniamide, 5x SSC (750
mA4 NaCl, 75 mM sodium citrate), 50 mA4 sodium phosphate, pII 7.6, 5
xDenhardt's solution, 10%
dextran sulfate, and 20 microgram/mi. denatured and sheared salmon sperm DNA,
with hybridization
25 filters washed in 1 xSSC at about 50 'C. It is noted that many other
hybridization methods, solutions
and temperatures can be used to achieve comparable stringent hybridization
conditions as will be
apparent to skilled artisans.
For the purpose of comparing two different nucleic acid or polypeptide
sequences, one
sequence (test sequence) may be described to be a specific percentage
identical to another sequence
30 (comparison sequence). The percentage identity can be determined by the
algorithm of Karlin and
Altschul, Proc. Natl. Acad. Sci. USA, 90:5873-5877 (1993), which is
incorporated into various
BLAST programs. The percentage identity can be determined by the "BLAST 2
Sequences" tool,
which is available at the National Center for Biotechnology Information
(NCI31) website. See
Tatusova and Madden, FEMS Microbiol. Lett., 174(2):247-250 (1999). For
pairwise DNA-DNA
35 comparison, the BLASTN program is used with default parameters (e.g.,
Match: 1; Mismatch: -2;
Open gap: 5 penalties; extension gap: 2 penalties; gap x dropolif 50; expect:
10; and word size: 11,
with filter). For pairwise protein-protein sequence comparison, the BLASTP
program can be
84
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
employed using default parameters (e.g., Matrix: BLOSUM62; gap open: 11; gap
extension: 1;
x dropoff: 15; expect: 10.0; and wordsize: 3, with filter). Percent identity
of two sequences is
calculated by aligning a test sequence with a comparison sequence using BLAST,
determining the
number of amino acids or nucleotides in the aligned test sequence that are
identical to amino acids or
5 nucleotides in the same position of the comparison sequence, and dividing
the number of identical
ammo acids or nucleotides by the number of amino acids or nucleotides in the
comparison sequence.
When BLAST is used to compare two sequences, it aligns the sequences and
yields the percent
identity over defined, aligned regions. If the two sequences are aligned
across their entire length, the
percent identity yielded by the BLAST is the percent identity of the two
sequences. If BLAST does
10 not align the two sequences over their entire length, then the number of
identical amino acids or
nucleotides in the unaligned regions of the test sequence and comparison
sequence is considered to be
zero and the percent identity is calculated by adding the number of identical
amino acids or
nucleotides in the aligned regions and dividing that number by the length of
the comparison sequence.
Various versions of the BLAST programs can be used to compare sequences, e.g.,
BLAST 2.1.2 or
15 BLAST-I- 2.2.22.
A subject or individual can be any animal which may benefit from the methods
described
herein, including, e.g., humans and non-human mammals, such as primates,
rodents, horses, dogs and
cats. Subjects include without limitation a eukaryotic organisms, most
preferably a mammal such as a
primate, e.g., chimpanzee or human, cow; dog; cat; a rodent, e.g., guinea pig,
nit, mouse; rabbit; or a
20 bird; reptile; or fish. Subjects specifically intended for treatment
using the methods described herein
include humans. A subject may also be referred to herein as an individual or a
patient. In the present
methods the subject has colorectal cancer, e.g., has been diagnosed with
colorectal cancer. Methods
for identifying subjects with colorectal cancer are known in the art, e.g.,
using a biopsy. See, e.g.,
Fleming et al., J Gastrointest Oncol. 2012 Sep; 3(3): 153-173; Chang et al.,
Dis Colon Rectum. 2012;
25 55(8):831-43.
Treatment of a disease or individual according to the methods described herein
is an approach
for obtaining beneficial or desired medical results, including clinical
results, but not necessarily a
cure. For purposes of the methods described herein, beneficial or desired
clinical results include, but
are not limited to, alleviation or amelioration of one or more symptoms,
diminishment of extent of
30 disease, stabilized (i.e., not worsening) state of disease, preventing
spread of disease, delay or slowing
of disease progression, amelioration or palliation of the disease state, and
remission (whether partial
or total), whether detectable or undetectable. Treatment also includes
prolonging survival as compared
to expected survival if not receiving treatment or if receivmg a different
treatment. A treatment can
include administration of various small molecule drugs or biologics such as
irnmunotherapies, e.g.,
35 checkpoint inhibitor therapies. A biomarker refers generally to a
molecule, including without
limitation a gene or product thereof, nucleic acids (e.g., DNA, RNA),
proteiMpeptide/polypeptide,
carbohydrate structure, lipid, glycolipid, characteristics of which can be
detected in a tissue or cell to
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
provide information that is predictive, diagnostic, prognostic and/or
theranostic for sensitivity or
resistance to candidate treatment.
Biological Samples
A sample as used herein includes any relevant biological sample that can be
used for
5 molecular profiling, e.g., sections of tissues such as biopsy or tissue
removed during surgical or other
procedures, bodily fluids, autopsy samples, and frozen sections taken for
histological purposes. Such
samples include blood and blood fractions or products (e.g., serum, buffy
coat, plasma, platelets, red
blood cells, and the like), sputum, malignant effusion, cheek cells tissue,
cultured cells (e.g., primary
cultures, explants, and transformed cells), stool, urine, other biological or
bodily fluids (e.g., prostatic
10 fluid, gastric fluid, intestinal fluid, renal fluid, lung fluid,
cerebrospinal fluid, and the like), etc. The
sample can comprise biological material that is a fresh frozen & formalin
fixed paraffm embedded
(FFPE) block, formalin-fixed paraffm embedded, or is within an RNA
preservative + formalin
fixative. More than one sample of more than one type can be used for each
patient. In a preferred
embodiment, the sample comprises a fixed tumor sample.
15 The sample used in the systems and methods of the invention can be a
formalin fixed paraffm
embedded (FFPE) sample. The FFPE sample can be one or more of fixed tissue,
unstained slides,
bone marrow core or clot, core needle biopsy, malignant fluids and fme needle
aspirate (FNA). In an
embodiment, the fixed tissue comprises a tumor containing formalin fixed
paraffin embedded (FFPE)
block from a surgery or biopsy. In another embodiment, the unstained slides
comprise unstained,
20 charged, unbaked slides from a paraffm block. In another embodiment,
bone marrow core or clot
comprises a decalcified core. A formalin fixed core and/or clot can be
paraffin-embedded. In still
another embodiment, the core needle biopsy comprises 1, 2, 3, 4, 5, 6, 7, 8,
9, 10 or more, e.g., 3-4,
paraffm embedded biopsy samples. An 18 gauge needle biopsy can be used. The
malignant fluid can
comprise a sufficient volume of fresh pleural/medic fluid to produce a
5x5x2mrn cell pellet. The fluid
25 can be formalin fixed in a paraffm block. In an embodiment, the core
needle biopsy comprises 1, 2, 3,
4, 5, 6, 7, 8, 9, 1001 more, e.g., 4-6, paraffin embedded aspirates.
A sample may be processed according to techniques understood by those in the
art. A sample
can be without limitation fresh, frozen or fixed cells or tissue. In some
embodiments, a sample
comprises formalin-fixed paraffm-embedded (FFPE) tissue, fresh tissue or fresh
frozen (FF) tissue. A
30 sample can comprise cultured cells, including primary or immortalized
cell lines derived from a
subject sample. A sample can also refer to an extract from a sample from a
subject. For example, a
sample can comprise DNA, RNA or protein extracted from a tissue or a bodily
fluid_ Many techniques
and commercial kits are available for such purposes. The fresh sample from the
individual can be
treated with an agent to preserve RNA prior to further processing, e.g., cell
lysis and extraction.
35 Samples can include frozen samples collected for other purposes. Samples
can be associated with
relevant information such as age, gender, and clinical symptoms present in the
subject; source of the
86
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
sample; and methods of collection and storage of the sample. A sample is
typically obtained from a
subject.
A biopsy comprises the process of removing a tissue sample for diagnostic or
prognostic
evaluation, and to the tissue specimen itself Any biopsy technique known in
the art can be applied to
5 the molecular profiling methods of the present disclosure. The biopsy
technique applied can depend
on the tissue type to be evaluated (e.g., colon, prostate, kidney, bladder,
lymph node, liver, bone
marrow, blood cell, lung, breast, etc.), the size and type of the tumor (e.g.,
solid or suspended, blood
or ascites), among other factors. Representative biopsy techniques include,
but are not limited to,
excisional biopsy, incisional biopsy, needle biopsy, surgical biopsy, and bone
marrow biopsy. An
10 "excisional biopsy" refers to the removal of an entire tumor mass with a
small margin of normal tissue
surrounding it An "incisional biopsy" refers to the removal of a wedge of
tissue that includes a cross-
sectional diameter of the tumor. Molecular profiling can use a "core-needle
biopsy" of the tumor
mass, or a "fme-needle aspiration biopsy" which generally obtains a suspension
of cells from within
the tumor mass. Biopsy techniques are discussed, for example, in Harrison's
Principles of Internal
15 Medicine, Kasper, et al., eds., 16th ed., 2005, Chapter 70, and
throughout Part V.
Unless otherwise noted, a "sample" as referred to herein for molecular
profiling of a patient
may comprise more than one physical specimen. As one non-limiting example, a
"sample" may
comprise multiple sections from a tumor, e.g., multiple sections of an FFPE
block or multiple core-
needle biopsy sections. As another non-limiting example, a "sample" may
comprise multiple biopsy
20 specimens, e.g., one or more surgical biopsy specimen, one or more core-
needle biopsy specimen, one
or more fme-needle aspiration biopsy specimen, or any useful combination
thereof As still another
non-limiting example, a molecular profile may be generated for a subject using
a "sample"
comprising a solid tumor specimen and a bodily fluid specimen. In some
embodiments, a sample is a
unitary sample, i.e., a single physical specimen.
25 Standard molecular biology techniques known in the art and not
specifically described are
generally followed as in Sambrook et al., Molecular Cloning: A Laboratory
Manual, Cold Spring
Harbor Laboratory Press, New York (1989), and as in Ausubel et al., Current
Protocols in Molecular
Biology, John Wiley and Sons, Baltimore, Md. (1989) and as in Perbal, A
Practical Guide to
Molecular Cloning, John Wiley & Sons, New York (1988), and as in Watson et
al., Recombinant
30 DIVA, Scientific American Books, New York and in Birren et al (eds)
Genome Analysis: A Laboratory
Manual Series, Vols. 1-4 Cold Spring Harbor Laboratory Press, New York (1998)
and methodology as
set forth in U.S. Pat Nos. 4,666,828; 4,683,202; 4,801,531; 5,192,659 and
5,272,057 and incorporated
herein by reference. Polymerase chain reaction (PCR) can be earned out
generally as m PCR
Protocols: A Guide to Methods and Applications, Academic Press, San Diego,
Calif. (1990).
35 Vesicles
The sample can comprise vesicles. Methods as described herein can include
assessing one or
more vesicles, including assessing vesicle populations. A vesicle, as used
herein, is a membrane
87
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
vesicle that is shed from cells. Vesicles or membrane vesicles include without
limitation: circulating
microvesicles (cMVs), microvesicle, exosome, nanovesicle, dexosome, bleb,
blebby, prostasome,
microparticle, intralumenal vesicle, membrane fragment, intralumenal endosomal
vesicle, endosomal-
like vesicle, exocytosis vehicle, endosome vesicle, endosomal vesicle,
apoptotic body, multivesicular
body, secretory vesicle, phospholipid vesicle, liposomal vesicle, argosome,
texasome, secresome,
tolerosome, melanosome, oncosome, or exocytosed vehicle. Furthermore, although
vesicles may be
produced by different cellular processes, the methods as described herein are
not limited to or reliant
on any one mechanism, insofar as such vesicles are present in a biological
sample and are capable of
being characterized by the methods disclosed herein. Unless otherwise
specified, methods that make
use of a species of vesicle can be applied to other types of vesicles.
Vesicles comprise spherical
structures with a lipid bilayer similar to cell membranes which surrounds an
inner compartment which
can contain soluble components, sometimes referred to as the payload. In some
embodiments, the
methods as described herein make use of exosomes, which are small secreted
vesicles of about 40
100 rim in diameter. For a review of membrane vesicles, including types and
characterizations, see
Thely et al., Nat Rev Immunol. 2009 Aug;9(8):581-93. Some properties of
different types of vesicles
include those in Table 1:
Table 1: Vesicle Properties
Feature Exosomes Micro- Ectosomes Mem- Exosome-
Apoptotic
vesicles brane like
vesicles
particles vesicles
Size 50-100 rim 100-1,000 50-200 nm 50-80
nrn 20-50 nrn 50-500 nm
rilT1
Density in 1.13-1.19g/ml 1.04-1.07 1.1 g/ml
1.16-1.28
sucrose g/ml g/m1
EM Cup shape Irregular Bilamellar Round
Irregular Hetero-
appearance shape, round shape geneous
electron structures
dense
Sedimen- 100,000 g 10,000 g 160,000- 100,000-
175,000 g 1,200 g,
tation 200,000 g 200,000 g 10,000
g,
100,000 g
Lipid com- Enriched in Expose PPS
Enriched in No lipid
position cholesterol, cholesterol rafts
sphingomyelin and
and ceramide; diacylglycero
contains lipid 1; expose PPS
rafts; expose
PPS
Major Tetraspanins Integrins, CR1 and
CD133; no TNFRI Histones
protein (e.g., CD63, selectins and
proteolytic CD63
markers CD9), Alix, CD40 ligand enzymes; no
TSG101 CD63
Infra-cellular Internal Plasma Plasma Plasma
origin compartments membrane membrane membrane
(endosomes)
Abbreviations: phosphatidylserine (PPS); electron microscopy (EM)
88
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Vesicles include shed membrane bound particles, or "microparticles," that are
derived from
either the plasma membrane or an internal membrane. Vesicles can be released
into the extracellular
environment from cells. Cells releasing vesicles include without limitation
cells that originate from, or
are derived from, the ectoderm, endoderm, or mesoderm The cells may have
undergone genetic,
5 environmental, and/or any other variations or alterations. For example,
the cell can be tumor cells. A
vesicle can reflect any changes in the source cell, and thereby reflect
changes in the originating cells,
e.g., cells having various genetic mutations. In one mechanism, a vesicle is
generated intracellularly
when a segment of the cell membrane spontaneously invaginates and is
ultimately exocytosed (see for
example, Keller et al., Immunol. Lett. 107 (2): 102-8 (2006)). Vesicles also
include cell-derived
10 structures bounded by a lipid bilayer membrane arising from both
herniated evagination (blebbing)
separation and sealing of portions of the plasma membrane or from the export
of any intracellular
membrane-bounded vesicular structure containing various membrane-associated
proteins of tumor
origin, including surface-bound molecules derived from the host circulation
that bind selectively to
the tumor-derived proteins together with molecules contained in the vesicle
lumen, including but not
15 limited to tumor-derived microRNAs or intracellular proteins_ Blebs and
blebbing are further
described in Charras et al., Nature Reviews Molecular and Cell Biology, Vol.
9, No. 11, p. 730-736
(2008). A vesicle shed into circulation or bodily fluids from tumor cells may
be referred to as a
"circulating tumor-derived vesicle." When such vesicle is an exosome, it may
be referred to as a
circulating-tumor derived exosome (CTE). In some instances, a vesicle can be
derived from a specific
20 cell of origin. CTE, as with a cell-of-origin specific vesicle,
typically have one or more unique
biomarkers that permit isolation of the CTE or cell-of-origin specific
vesicle, e.g., from a bodily fluid
and sometimes in a specific manner. For example, a cell or tissue specific
markers are used to identify
the cell of origin. Examples of such cell or tissue specific markers are
disclosed herein and can further
be accessed in the Tissue-specific Gene Expression and Regulation (TiGER)
Database, available at
25 bioinfo.wilmerjhu.edu/tiger/; Liu et al. (2008) TiGER: a database for
tissue-specific gene expression
and regulation. BMC Bioinforrnatics. 9:271: TissueDistributionDBs, available
at genome.dlth-
heidelberg.de/menultissuedb/index.html.
A vesicle can have a diameter of greater than about 10 nm, 20 nm, or 30 nm. A
vesicle can
have a diameter of greater than 40 nm, 50 nm, 100 nm, 200 nin, 500 nm, 1000 nm
or greater than
30 10,000 nm. A vesicle can have a diameter of about 30-1000 nm, about 30-
800 nm, about 30-200 nm,
or about 30-100 rim. In some embodiments, the vesicle has a diameter of less
than 10,000 nm, 1000
nm, 800 rim, 500 nm, 200 nm, 100 nm, 50 nrn, 40 nm, 30 nm, 20 nm or less than
10 run. As used
herein the term "about" in reference to a numerical value means that
variations of 10% above or
below the numerical value are within the range ascribed to the specified
value. Typical sizes for
35 various types of vesicles are shown in Table 1. Vesicles can be assessed
to measure the diameter of a
single vesicle or any number of vesicles. For example, the range of diameters
of a vesicle population
or an average diameter of a vesicle population can be determined. Vesicle
diameter can be assessed
89
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
using methods known in the art, e.g., imaging technologies such as electron
microscopy. In an
embodiment, a diameter of one or more vesicles is dekauained using optical
particle detection. See,
e.g., US. Patent 7,751,053, entitled "Optical Detection and Analysis of
Particles" and issued July 6,
2010; and U.S. Patent 7,399,600, entitled "Optical Detection and Analysis of
Particles" and issued
5 July 15, 2010.
In some embodiments, vesicles are directly assayed from a biological sample
without prior
isolation, purification, or concentration from the biological sample. For
example, the amount of
vesicles in the sample can by itself provide a biosignature that provides a
diagnostic, prognostic or
theranostic determination. Alternatively, the vesicle in the sample may be
isolated, captured, purified,
10 or concentrated from a sample prior to analysis. As noted, isolation,
capture or purification as used
herein comprises partial isolation, partial capture or partial purification
apart from other components
in the sample. Vesicle isolation can be performed using various techniques as
described herein or
known in the art, including without limitation size exclusion chromatography,
density gradient
centrifugation, differential centrifugation, nanomembrane ultrafiltration,
imnaunoabsorbent capture,
15 affinity purification, affinity capture, immunoassay,
immunoprecipitation, microfluidic separation,
flow cytometry or combinations thereof.
Vesicles can be assessed to provide a phenotypic characterization by comparing
vesicle
characteristics to a reference. In some embodiments, surface antigens on a
vesicle are assessed. A
vesicle or vesicle population carrying a specific marker can be referred to as
a positive (biomarker+)
20 vesicle or vesicle population. For example, a DLL4+ population refers to
a vesicle population
associated with DLL4. Conversely, a DLL4- population would not be associated
with DLL4. The
surface antigens can provide an indication of the anatomical origin and/or
cellular of the vesicles and
other phenotypic information, e.g., tumor status. For example, vesicles found
in a patient sample can
be assessed for surface antigens indicative of colorectal origin and the
presence of cancer, thereby
25 identifying vesicles associated with colorectal cancer cells. The
surface antigens may comprise any
informative biological entity that can be detected on the vesicle membrane
surface, including without
limitation surface proteins, lipids, carbohydrates, and other membrane
components. For example,
positive detection of colon derived vesicles expressing tumor antigens can
indicate that the patient has
colorectal cancer. As such, methods as described herein can be used to
characterize any disease or
30 condition associated with an anatomical or cellular origin, by
assessing, for example, disease-specific
and cell-specific biomarkers of one or more vesicles obtained from a subject.
In embodiments, one or more vesicle payloads are assessed to provide a
phenotypic
characterization. The payload with a vesicle comprises any informative
biological entity that can be
detected as encapsulated within the vesicle, including without limitation
proteins and nucleic acids,
35 e.g., genomic or cDNA, mRNA, or functional fragments thereof, as well as
microRNAs (miRs). In
addition, methods as described herein are directed to detecting vesicle
surface antigens (in addition or
exclusive to vesicle payload) to provide a phenotypic characterization. For
example, vesicles can be
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
characterized by using binding agents (e.g., antibodies or aptamers) that are
specific to vesicle surface
antigens, and the bound vesicles can be further assessed to identify one or
more payload components
disclosed therein_ As described herein, the levels of vesicles with surface
antigens of interest or with
payload of interest can be compared to a reference to characterize a
phenotype. For example,
5 overexpression in a sample of cancer-related surface antigens or vesicle
payload, e.g., a tumor
associated mRNA or microRNA, as compared to a reference, can indicate the
presence of cancer in
the sample. The biomarkers assessed can be present or absent, increased or
reduced based on the
selection of the desired target sample and comparison of the target sample to
the desired reference
sample. Non-limiting examples of target samples include: disease; treated/not-
treated; different time
10 points, such as a in a longitudinal study; and non-limiting examples of
reference sample: non-disease;
normal; different time points; and sensitive or resistant to candidate
treatment(s).
In an embodiment, molecular profiling as described herein comprises analysis
of
microvesicles, such as circulating microvesicles.
MicroRNA
15 Various biomarker molecules can be assessed in biological samples or
vesicles obtained from
such biological samples. MicroRNAs comprise one class biomarkers assessed via
methods as
described herein. MicroRNAs, also referred to herein as miRNAs or miRs, are
short RNA strands
approximately 21-23 nucleotides in length. MiRNAs are encoded by genes that
are transcribed from
DNA but arc not translated into protein and thus comprise non-coding RNA. The
milts are processed
20 from primary transcripts known as pri-miRNA to short stem-loop
structures called pre-miRNA and
fmally to the resulting single strand miRNA. The pre-miRNA typically forms a
structure that folds
back on itself in self-complementary regions. These structures are then
processed by the nuclease
Dicer in animals or DCL1 in plants. Mature miRNA molecules are partially
complementary to one or
more messenger RNA (mRNA) molecules and can function to regulate translation
of proteins.
25 Identified sequences of miRNA can be accessed at publicly available
databases, such as
www.microRNA.org, www.mirbase.org, or www.mirz.unibas.ch/cgi/miRNA.cgi.
miRNAs are generally assigned a number according to the naming convention "
mir-
[number]." The number of a miRNA is assigned according to its order of
discovery relative to
previously identified miRNA species. For example, if the last published miRNA
was mir-121, the next
30 discovered miRNA will be named mir-122, etc. When a miRNA is discovered
that is homologous to a
known miRNA from a different organism, the name can be given an optional
organism identifier, of
the form [organism identifier]- mir-[number]. Identifiers include hsa for Homo
sapiens and mrnu for
Mus Musculus. For example, a human homolog to nur-121 might be referred to as
hsa-nur-121
whereas the mouse homolog can be referred to as mmu-mir-121.
35 Mature microRNA is commonly designated with the prefix "miR" whereas the
gene or
precursor miRNA is designated with the prefix "mir." For example, mir-121 is a
precursor for miR-
121. When differing miRNA genes or precursors are processed into identical
mature miRNAs, the
91
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
genes/precursors can be delineated by a numbered suffix. For example, mir-121-
1 and mir-121-2 can
refer to distinct genes or precursors that are processed into miR-121.
Lettered suffixes are used to
indicate closely related mature sequences. For example, mur-121a and mir-12111
can be processed to
closely related miRNAs miR-121a and miR-121b, respectively. In the context of
the present
5 disclosure, any microRNA (miRNA or miR) designated herein with the prefix
mir-* or miR-* is
understood to encompass both the precursor and/or mature species, unless
otherwise explicitly stated
otherwise.
Sometimes it is observed that two mature naiRNA sequences originate from the
same
precursor. When one of the sequences is more abundant that the other, a "*"
suffix can be used to
10 designate the less common variant. For example, miR-121 would be the
predominant product whereas
miR-121* is the less co llllll on variant found on the opposite arm of the
precursor. If the predominant
variant is not identified, the miRs can be distinguished by the suffix "5p"
for the variant from the 5'
arm of the precursor and the suffix "3p" for the variant from the 3' arm For
example, miR-121-5p
originates from the 5' arm of the precursor whereas miR-121-3p originates from
the 3' ann. Less
15 commonly, the 5p and 3p variants are referred to as the sense ("s") and
anti-sense ("as") forms,
respectively. For example, miR-121-5p may be referred to as miR-121-s whereas
miR-121-3p may be
referred to as miR-121-as.
The above naming conventions have evolved over time and are general guidelines
rather than
absolute rules. For example, the let- and lin- families of miRNAs continue to
be referred to by these
20 monikers. The mir/miR convention for precursor/mature forms is also a
guideline and context should
be taken into account to determine which form is referred to. Further details
of miR naming can be
found at www.mirbase.org or Ambros et al., A uniform system for microRNA
annotation, RNA 9:277-
279 (2003).
Plant miRNAs follow a different naming convention as described in Meyers et
al., Plant Cell.
25 2008 20(12):3186-3190.
A number of miRNAs are involved in gene regulation, and miRNAs are part of a
growing
class of non-coding RNAs that is now recognized as a major tier of gene
control. in some cases,
miRNAs can interrupt translation by binding to regulatory sites embedded in
the 3'-UTRs of their
target mRNAs, leading to the repression of translation. Target recognition
involves complementary
30 base pairing of the target site with the miRNA's seed region (positions
2-8 at the miRNA's 5' end),
although the exact extent of seed complementarity is not precisely determined
and can be modified by
3' pairing. In other cases, miRNAs function like small interfering RNAs
(siR_NA) and bind to
perfectly complementary mR.NA sequences to destroy the target transcript.
Characterization of a number of miRNAs indicates that they influence a variety
of processes,
35 including early development, cell proliferation and cell death,
apoptosis and fat metabolism. For
example, some miRNAs, such as lm-4, let-7, mir-14, mir-23, and bantam, have
been shown to play
92
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
critical roles in cell differentiation and tissue development. Others are
believed to have similarly
important roles because of their differential spatial and temporal expression
patterns.
The miRNA database available at miRBase (wwvv.mirbase.org) comprises a
searchable
database of published miRNA sequences and annotation. Further information
about miRBase can be
5 found in the following articles, each of which is incorporated by
reference in its entirety herein:
Ciriffiths-Jones et al., miRBase: tools for microRNA genomics. NAR 2008
36(Database Issue):D154-
D158; Griffiths-Jones et al., miRBase: microRNA sequences, targets and gene
nomenclature. NAR
2006 34(Database Issue):D140-D144; and Griffiths-Jones, S. The microRNA
Registry. NAR 2004
32(Database Issue):D109-D111. Representative miRNAs contained in Release 16 of
miRBase, made
10 available September 2010.
As described herein, microRNAs are known to be involved in cancer and other
diseases and
can be assessed in order to characterize a phenotype in a sample. See, e.g.,
Ferracin et al.,
Micromarkers: miRNAs in cancer diagnosis and prognosis, Exp Rev Mol Diag, Apr
2010, Vol. 10,
No. 3, Pages 297-308; Fabbri, miRNAs as molecular biomarkers of cancer, Exp
Rev Mol Diag, May
15 2010, Vol. 10, No. 4, Pages 435-444.
In an embodiment, molecular profiling as described herein comprises analysis
of microRNA.
Techniques to isolate and characterize vesicles and miRs are known to those of
skill in the art.
In addition to the methodology presented herein, additional methods can be
found in U.S. Patent Nos.
7,888,035, entitled "METHODS FOR ASSESSING RNA PATTERNS" and issued February
15, 2011;
20 and 7,897,356, entitled "METHODS AND SYSTEMS OF USING EXOSOMES FOR
DETERMINING PHENOTYPES" and issued March 1, 2011; and International Patent
Publication
Nos. WO/2011/066589, entitled "METHODS AND SYSTEMS FOR ISOLATING, STORING, AND
ANALYZING VESICLES" and filed November 30, 2010; WO/2011/088226, entitled
"DETECTION
OF GASTROINTESTINAL DISORDERS" and filed January 13, 2011; WO/2011/109440,
entitled
25 "BIOMARKERS FOR TFIERANOSTICS" and filed March 1, 2011; and
WO/2011/127219, entitled
"CIRCULATING BIOMARKERS FOR DISEASE" and filed April 6, 2011, each of which
applications are incorporated by reference herein in their entirety.
Circulating Biomarkers
Circulating biomarkers include biomarkers that are detectable in body fluids,
such as blood,
30 plasma, serum. Examples of circulating cancer biomarkers include cardiac
troponin '1 (cTif1), prostate
specific antigen (PSA) for prostate cancer and CA125 for ovarian cancer.
Circulating biomarkers
according to the present disclosure include any appropriate biomarker that can
be detected in bodily
thud, including without limitation protein, nucleic acids, e.g., DNA, mR.NA
and microRNA,
carbohydrates and metabolites. Circulating biomarkers can include biomarkers
that are not associated
35 with cells, such as biomarkers that are membrane associated, embedded in
membrane fragments, part
of a biological complex, or free in solution. In one embodiment, circulating
biomarkers are
biomarkers that are associated with one or more vesicles present in the
biological fluid of a subject.
93
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Circulating biomarkers have been identified for use in characterization of
various phenotypes,
such as detection of a cancer. See, e.g., Ahmed N, etal., Proteomic-based
identification of
haptoglobin-1 precursor as a novel circulating biomarker of ovarian cancer Br.
J. Cancer 2004;
Mathelin _et al., Circulating proteinic biomarkers and breast cancer, Gynecol
Obstet Feral. 2006 Jul-
5 Aug;34(7-8):638-46. Epub 2006 Jul 28; Ye et al., Recent technical
strategies to identify diagnostic
biomarkers for ovarian cancer_ Expert Rev Proteomics. 2007 Feb;4(1):121-31;
Carney, Circulating
oncoproteins HER2/neu, EGFR and CAIX (MN) as novel cancer biomarkers. Expert
Rev Mol Diagn.
2007 May;7(3):309-19; Gagnon, Discovery and application of protein biomarkers
for ovarian cancer,
Curr Opin Obstet Gynecol. 2008 Feb;20(1):9-13; Pasterkamp et al., Immune
regulatory cells:
10 circulating biomarker factories in cardiovascular disease. Chin Sci
(Lond). 2008 Aug;115(4):129-31;
Fabbri, miRNAs as molecular biomarkers of cancer, Exp Rev Mol Diag, May 2010,
Vol. 10, No. 4,
Pages 435-444; PCT Patent Publication WO/2007/088537; U.S. Patents 7,745,150
and 7,655,479;
U.S. Patent Publications 20110008808, 20100330683, 20100248290, 20100222230,
20100203566,
20100173788, 20090291932, 20090239246, 20090226937, 20090111121, 20090004687,
15 20080261258, 20080213907, 20060003465, 20050124071, and 20040096915,
each of which
publication is incorporated herein by reference in its entirety. In an
embodiment, molecular profiling
as described herein comprises analysis of circulating biomarkers.
Gene Expression Profiling
The methods and systems as described herein comprise expression profiling,
which includes
20 assessing differential expression of one or more target genes disclosed
herein. Differential expression
can include overexpression and/or underexpression of a biological product,
e.g., a gene, mR_NA or
protein, compared to a control (or a reference). The control can include
similar cells to the sample but
without the disease (e.g., expression profiles obtained from samples from
healthy individuals). A
control can be a previously determined level that is indicative of a drug
target efficacy associated with
25 the particular disease and the particular drug target. The control can
be derived from the same patient,
e.g., a normal adjacent portion of the same organ as the diseased cells, the
control can be derived from
healthy tissues from other patients, or previously determined thresholds that
are indicative of a disease
responding or not-responding to a particular drug target. The control can also
be a control found in the
same sample, e.g. a housekeeping gene or a product thereof (e.g., mRNA or
protein). For example, a
30 control nucleic acid can be one which is known not to differ depending
on the cancerous or non-
cancerous state of the cell. The expression level of a control nucleic acid
can be used to normalize
signal levels in the test and reference populations. Illustrative control
genes include, but are not
limited to, e.g., 13-actin, glyceraldehyde 3-phosphate dehydrogenase and
ribosomal protein Pl.
Multiple controls or types of controls can be used. The source of differential
expression can vary. For
35 example, a gene copy number may be increased in a cell, thereby
resulting in increased expression of
the gene. Alternately, transcription of the gene may be modified, e.g., by
chromatin remodeling,
differential methylation, differential expression or activity of transcription
factors, etc. Translation
94
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
may also be modified, e.g., by differential expression of factors that degrade
mRNA, translate mRNA,
or silence translation, e.g., microRNAs or siRNAs. In some embodiments,
differential expression
comprises differential activity For example, a protein may carry a mutation
that increases the activity
of the protein, such as constitutive activation, thereby contributing to a
diseased state. Molecular
5 profiling that reveals changes in activity can be used to guide treatment
selection.
Methods of gene expression profiling include methods based on hybridization
analysis of
polynucleotides, and methods based on sequencing of polynucleotides. Commonly
used methods
known in the art for the quantification of mRNA expression in a sample include
northern blotting and
in situ hybridization (Parker & Barnes (1999) Methods in Molecular Biology
106:247-283); RNAse
10 protection assays (Hod (1992) Biotechniques 13:852-854); and reverse
transcription polymerase chain
reaction (RT-PCR) (Weis et al. (1992) Trends in Genetics A:263-264).
Alternatively, antibodies may be
employed that can recognize specific duplexes, including DNA duplexes, RNA
duplexes, and DNA-
RNA hybrid duplexes or DNA-protein duplexes. Representative methods for
sequencing-based gene
expression analysis include Serial Analysis of Gene Expression (SAGE), gene
expression analysis by
15 massively parallel signature sequencing (MPSS) and/or next generation
sequencing.
RT-PCR
Reverse transcription polymerase chain reaction (RT-PCR) is a variant of
polymerase chain
reaction (PCR). According to this technique, a RNA strand is reverse
transcribed into its DNA
complement (i.e., complementary DNA, or cDNA) using the enzyme reverse
transeriptase, and the
20 resulting cDNA is amplified using PCR. Real-time polymerase chain
reaction is another PCR variant,
which is also referred to as quantitative PCR, Q-PCR, qRT-PCR, or sometimes as
RT-PCR. Either the
reverse transcription PCR method or the real-time PCR method can be used for
molecular profiling
according to the present disclosure, and RT-PCR can refer to either unless
otherwise specified or as
understood by one of skill in the art.
25 RT-PCR can be used to determine RNA levels, e.g., mRNA or miRNA levels,
of the
biomarkers as described herein. RT-PCR can be used to compare such RNA levels
of the biomarkers
as described herein in different sample populations, in normal and tumor
tissues, with or without drug
treatment, to characterize patterns of gene expression, to discriminate
between closely related RNAs,
and to analyze RNA structure.
30 'The first step is the isolation of RNA, e.g., mRNA, from a sample. The
starting material can
be total RNA isolated from human tumors or tumor cell lines, and corresponding
normal tissues or
cell lines, respectively. Thus RNA can be isolated from a sample, e.g., tumor
cells or tumor cell lines,
and compared with pooled DNA from healthy donors. If the source of mRNA is a
primary tumor,
mRNA can be extracted, for example, from frozen or archived paraffin-embedded
and fixed (e.g.
35 formalin-fixed) tissue samples.
General methods for mRNA extraction are well known in the art and are
disclosed in standard
textbooks of molecular biology, including Ausubel et al. (1997) Current
Protocols of Molecular
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Biology, John Wiley and Sons. Methods for RNA extraction from paraffin
embedded tissues are
disclosed, for example, in Rupp & Locker (1987) Lab Invest. 56:A67, and De
Andres et al.,
flioTechniques 18:42044 (1995). In particular, RNA isolation can be performed
using purification kit,
buffer set and protease from commercial manufacturers, such as Qiagen,
according to the
5 manufacturer's instructions (QIAGEN Inc., Valencia, CA). For example,
total RNA from cells in
culture can be isolated using Qiagen RNeasy mini-columns. Numerous RNA
isolation kits are
commercially available and can be used in the methods as described herein.
In the alternative, the first step is the isolation of miRNA from a target
sample. The starting
material is typically total RNA isolated from human tumors or tumor cell
lines, and corresponding
10 normal tissues or cell lines, respectively. Thus RNA can be isolated
from a variety of primary tumors
or tumor cell hues, with pooled DNA from healthy donors. If the source of
miRNA is a primary
tumor, miRNA can be extracted, for example, from frozen or archived paraffm-
embedded and fixed
(e.g. formalin-fixed) tissue samples.
General methods for miRNA extraction are well known in the art and are
disclosed in
15 standard textbooks of molecular biology, including Ausubel et al. (1997)
Current Protocols of
Molecular Biology, John Wiley and Sons. Methods for RNA extraction from
paraffm embedded
tissues are disclosed, for example, in Rupp & Locker (1987) Lab Invest.
56:A67, and De Andres et al.,
BioTechniques 18:42044 (1995). In particular, RNA isolation can be performed
using purification kit,
buffer set and protease from commercial manufacturers, such as Qiagen,
according to the
20 manufacturer's instructions. For example, total RNA from cells in
culture can be isolated using
Qiagen RNeasy mini-columns. Numerous miRNA isolation kits are commercially
available and can
be used in the methods as described herein.
Whether the RNA comprises mRNA, miRNA or other types of RNA, gene expression
profiling by RT-PCR can include reverse transcription of the RNA template into
cDNA, followed by
25 amplification in a PCR reaction. Commonly used reverse transeriptases
include, but are not limited to,
avilo myeloblastosis virus reverse transcriptase (AMV-RT) and Moloney murine
leukemia virus
reverse transcriptase (MMLV-12:1). 'The reverse transcription step is
typically primed using specific
primers, random hexamers, or oligo-dT primers, depending on the circumstances
and the goal of
expression profiling. For example, extracted RNA can be reverse-transcribed
using a GeneAmp RNA
30 PCR kit (Perkin Elmer, Calif , USA), following the manufacturer's
instructions. The derived cllNA
can then be used as a template in the subsequent PCR reaction.
Although the PCR step can use a variety of thermostable DNA-dependent DNA
polymerases,
it typically employs the Taq DNA polymerase, which has a 5' -3' nuclease
activity but lacks a 3 '-5 '
proofreading endonuclease activity. TaqMan PCR typically uses the 5'-nuclease
activity of Taq or Tth
35 polymerase to hydrolyze a hybridization probe bound to its target
amplicon, but any enzyme with
equivalent 5' nuclease activity can be used. Two oligonucleotide primers are
used to generate an
amplicon typical of a PCR reaction. A third oligonucleotide, or probe, is
designed to detect nucleotide
96
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
sequence located between the two PCR primers. The probe is non-extendible by
Taq DNA polymerase
enzyme, and is labeled with a reporter fluorescent dye and a quencher
fluorescent dye. Any laser-
induced emission from the reporter dye is quenched by the quenching dye when
the two dyes are
located close together as they are on the probe. During the amplification
reaction, the Taq DNA
5 polymerase enzyme cleaves the probe in a template-dependent manner. The
resultant probe fragments
disassociate in solution, and signal from the released reporter dye is free
from the quenching effect of
the second fluorophore. One molecule of reporter dye is liberated for each new
molecule synthesized,
and detection of the unquenched reporter dye provides the basis for
quantitative interpretation of the
data.
10 TaqManTm RT-PCR can be performed using commercially available equipment,
such as, for
example, AIM PRISM 7700TM Sequence Detection SystemTM (Perkin-Elmer-Applied
Biosystems,
Foster City, Calif., USA), or EightCycler (Roche Molecular Biochemicals,
Mannheim, Germany). In
one specific embodiment, the 5' nuclease procedure is run on a real-time
quantitative PCR device
such as the ABI PRISM 7700 Sequence Detection System. The system consists of a
thermocycler,
15 laser, charge-coupled device (CCD), camera and computer. The system
amplifies samples in a 96-well
format on a therrnocycler. During amplification, laser-induced fluorescent
signal is collected in real-
time through fiber optic cables for all 96 wells, and detected at the CCD. The
system includes
software for running the instrument and for analyzing the data.
TaqMan data are initially expressed as Ct, or the threshold cycle. As
discussed above,
20 fluorescence values are recorded during every cycle and represent the
amount of product amplified to
that point in the amplification reaction. The point when the fluorescent
signal is first recorded as
statistically significant is the threshold cycle (Ct).
To minimize errors and the effect of sample-to-sample variation, RT-PCR is
usually
performed using an internal standard. The ideal internal standard is expressed
at a constant level
25 among different tissues, and is unaffected by the experimental
treatment. RNAs most frequently used
to normalize patterns of gene expression are mRNAs for the housekeeping genes
glyceraldehyde-3-
phosphate-dehydrogenase (CIAPDH) and I3-actin.
Real time quantitative PCR (also quantitative real time polymerase chain
reaction, QRT-PCR
or Q-PCR) is a more recent variation of the RT-PCR technique. Q-PCR can
measure PCR product
30 accumulation through a dual-labeled fluorigenic probe (i.e., TaqMan
probe). Real time PCR is
compatible both with quantitative competitive PCR, where internal competitor
for each target
sequence is used for normalization, and with quantitative comparative PCR
using a normalization
gene contained within the sample, or a housekeeping gene for RI-PCR. See, e.g.
Held et al. (1996)
Genome Research 6:986-994.
35 Protein-based detection techniques are also useful for molecular
profiling, especially when
the nucleotide variant causes amino acid substitutions or deletions or
insertions or frame shift that
affect the protein primary, secondary or tertiary structure. To detect the
amino acid variations, protein
97
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
sequencing techniques may be used. For example, a protein or fragment thereof
corresponding to a
gene can be synthesized by recombinant expression using a DNA fragment
isolated from an
individual to be tested_ Preferably, a cDNA fragment of no more than 100 to
150 base pairs
encompassing the polymorphic locus to be determined is used. The amino acid
sequence of the
5 peptide can then be deterrnined by conventional protein sequencing
methods. Alternatively, the
HPT,C-microscopy tandem mass spectrometry technique can be used for
determining the amino acid
sequence variations. In this technique, proteolytic digestion is performed on
a protein, and the
resulting peptide mixture is separated by reversed-phase chromatographic
separation. Tandem mass
spectrometry is then performed and the data collected is analyzed. See Gatlin
et al., Anal. Chem.,
10 72:757-763 (2000).
Microarray
The biomarkers as described herein can also be identified, confirmed, and/or
measured using
the microarray technique. Thus, the expression profile biomarkers can be
measured in cancer samples
using microarray technology. In this method, polynucleotide sequences of
interest are plated, or
15 arrayed, on a microchip substrate. The arrayed sequences are then
hybridized with specific DNA
probes from cells or tissues of interest. The source of mRNA can be total RNA
isolated from a sample,
e.g., human tumors or tumor cell lines and corresponding normal tissues or
cell lines. Thus RNA can
be isolated from a variety of primary tumors or tumor cell lines. If the
source of mRNA is a primary
tumor, mRNA can be extracted, for example, from frozen or archived paraffm-
embedded and fixed
20 (e.g. fonnalin-fixed) tissue samples, which are routinely prepared and
preserved in everyday clinical
practice.
The expression profile of biomarkers can be measured in either fresh or
parafFm-embedded
tumor tissue, or body fluids using microarray technology. In this method,
polynucleotide sequences of
interest are plated, or arrayed, on a microchip substrate. The arrayed
sequences are then hybridized
25 with specific DNA probes from cells or tissues of interest. As with the
RT-PCR method, the source of
miRNA typically is total RNA isolated from human tumors or tumor cell lines,
including body fluids,
such as serum, urine, tears, and exosomes and corresponding normal tissues or
cell lines. Thus RNA
can be isolated from a variety of sources. If the source of miRNA is a primary
tumor, miRNA can be
extracted, for example, from frozen tissue samples, which are routinely
prepared and preserved in
30 everyday clinical practice.
Also known as biochip, DNA chip, or gene array, cDNA microarray technology
allows for
identification of gene expression levels in a biologic sample cDNAs or
oligonucleotides, each
representing a given gene, are immobilized on a substrate, e.g., a small chip,
bead or nylon membrane,
tagged, and serve as probes that will indicate whether they are expressed in
biologic samples of
35 interest. The simultaneous expression of thousands of genes can be
monitored simultaneously.
In a specific embodiment of the microarray technique, PCR amplified inserts of
cDNA clones
are applied to a substrate in a dense array. In one aspect, at least 100, 200,
300, 400, 500, 600, 700,
98
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
800, 900, 1,000, 1,500, 2,000, 3000, 4000, 5000, 6000, 7000, 8000, 9000,
10,000, 15,000, 20,000,
25,000, 30,000, 35,000, 40,000, 45,000 or at least 50,000 nucleotide sequences
are applied to the
substrate. Each sequence can correspond to a different gene, or multiple
sequences can be arrayed per
gene. The microarrayed genes, immobilized on the microchip, are suitable for
hybridization under
5 stringent conditions. Fluorescently labeled cDNA probes may be generated
through incorporation of
fluorescent nucleotides by reverse transcription of RNA extracted from tissues
of interest. -Labeled
cDNA probes applied to the chip hybridize with specificity to each spot of DNA
on the array. After
stringent washing to remove non-specifically bound probes, die chip is scanned
by confocal laser
microscopy or by another detection method, such as a CCD camera. Quantitation
of hybridization of
10 each arrayed element allows for assessment of corresponding mRNA
abundance. With dual color
fluorescence, separately labeled cDNA probes generated from two sources of RNA
are hybridized
pairvvise to the array. The relative abundance of the transcripts from the two
sources corresponding to
each specified gene is thus determined simultaneously The miniaturized scale
of the hybridization
affords a convenient and rapid evaluation of the expression pattern for large
numbers of genes. Such
15 methods have been shown to have the sensitivity required to detect rare
transcripts, which are
expressed at a few copies per cell, and to reproducibly detect at least
approximately two-fold
differences in the expression levels (Schena et al. (1996) Proc. Natl. Acad.
Sci. USA 93(2):106-149).
Microarray analysis can be performed by commercially available equipment
following manufacturer's
protocols, including without limitation the Affymetrix GeneChip technology
(Affymetrix, Santa
20 Clara, CA), Agilent (Agilent Technologies, Inc., Santa Clara, CA), or
Illumina (Illianina, Inc., San
Diego, CA) microarray technology
The development of microarray methods for large-scale analysis of gene
expression makes it
possible to search systematically for molecular markers of cancer
classification and outcome
prediction in a variety of tumor types.
25 In some embodiments, the Agilent Whole Human Genome Microarray Kit
(Agilent
Technologies, Inc., Santa Clara, CA). The system can analyze more than 41,000
unique human genes
and transcripts represented, all with public domain annotations. The system is
used according to the
manufacturer's instructions.
In some embodiments, the Illumina Whole Genome DASL assay (Illumina Inc., San
Diego,
30 CA) is used. The system offers a method to simultaneously profile over
24,000 transcripts from
minimal RNA input, from both fresh frozen (FF) and formalin-fixed paraffin
embedded (FFPE) tissue
sources, in a high throughput fashion.
Microarray expression analysis comprises identifying whether a gene or gene
product is up-
regulated or down-regulated relative to a reference. The identification can be
performed using a
35 statistical test to determine statistical significance of any
differential expression observed. In some
embodiments, statistical significance is determined using a parametric
statistical test. The parametric
statistical test can comprise, for example, a fractional factorial design,
analysis of variance (ANOVA),
99
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
a t-test, least squares, a Pearson correlation, simple linear regression,
nonlinear regression, multiple
linear regression, or multiple nonlinear regression. Alternatively, the
parametric statistical test can
comprise a one-way analysis of variance, two-way analysis of variance, or
repeated measures analysis
of variance. In other embodiments, statistical significance is determined
using a nonparametric
5 statistical test. Examples include, but are not limited to, a Wilcoxon
signed-rank test, a Mann-Whitney
test, a Kruskal-Wallis test, a Friedman test, a Spearman ranked order
correlation coefficient, a Kendall
Tau analysis, and a nonparametric regression test. In some embodiments,
statistical significance is
determined at a p-value of less than about 0.05, 0.01, 0.005, 0.001, 0.0005,
or 0.0001. Although the
microarray systems used in the methods as described herein may assay thousands
of transcripts, data
10 analysis need only be performed on the transcripts of interest, thereby
reducing the problem of
multiple comparisons inherent in performing multiple statistical tests. The p-
values can also be
corrected for multiple comparisons, e.g., using a Bonferroni correction, a
modification thereof, or
other technique known to those in the art, e.g., the Hochberg correction,
Holni-Bonferroni correction,
iclak correction, or Dunnett's correction. The degree of differential
expression can also be taken into
15 account. For example, a gene can be considered as differentially
expressed when the fold-change in
expression compared to control level is at least 1.2, 1.3, 1.4, 1.5, 1.6, 1.7,
1.8, 1.9, 2.0, 2.2, 2.5, 2.7,
3.0, 4, 5, 6, 7, 8, 9 or 10-fold different in the sample versus the control.
The differential expression
takes into account both overexpression and underexpression. A gene or gene
product can be
considered up or down-regulated if the differential expression meets a
statistical threshold, a fold-
20 change threshold, or both. For example, the criteria for identifying
differential expression can
comprise both a p-value of 0.001 and fold change of at least I.5-fold (up or
down). One of skill will
understand that such statistical and threshold measures can be adapted to
determine differential
expression by any molecular profiling technique disclosed herein.
Various methods as described herein make use of many types of microarrays that
detect the
25 presence and potentially the amount of biological entities in a sample.
Arrays typically contain
addressable moieties that can detect the presence of the entity in the sample,
e.g., via a binding event.
Microarrays include without limitation DNA microarrays, such as cllNA
microarrays, oligonucleotide
microarrays and SNP microarrays, microRNA arrays, protein microarrays,
antibody microarrays,
tissue microarrays, cellular microarrays (also called transfection
microarrays), chemical compound
30 microarrays, and carbohydrate arrays (glycoarrays). DNA arrays typically
comprise addressable
nucleotide sequences that can bind to sequences present in a sample. MicroRNA
arrays, e.g., the
MNIChips array from the University of Louisville or commercial systems from
Agilent, can be used to
detect microRNAs. Protein microarrays can be used to identify protem¨protem
interactions, including
without limitation identifying substrates of protein kinases, transcription
factor protein-activation, or
35 to identify the targets of biologically active small molecules. Protein
arrays may comprise an array of
different protein molecules, commonly antibodies, or nucleotide sequences that
bind to proteins of
interest. Antibody microarrays comprise antibodies spotted onto the protein
chip that are used as
100
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
capture molecules to detect proteins or other biological materials from a
sample, e.g., from cell or
tissue lysate solutions. For example, antibody arrays can be used to detect
biomarkers from bodily
fluids, e.g., serum or urine, for diagnostic applications. Tissue microarrays
comprise separate tissue
cores assembled in array fashion to allow multiplex histological analysis.
Cellular microarrays, also
5 called transfection microarrays, comprise various capture agents, such as
antibodies, proteins, or
lipids, which can interact with cells to facilitate their capture cm
addressable locations. Chemical
compound microarrays comprise arrays of chemical compounds and can be used to
detect protein or
other biological materials that bind the compounds. Carbohydrate arrays
(glycoarrays) comprise
arrays of carbohydrates and can detect, e.g., protein that bind sugar
moieties. One of skill will
10 appreciate that similar technologies or improvements can be used
according to the methods as
described herein.
Certain embodiments of the current methods comprise a multi-well reaction
vessel, including
without limitation, a multi-well plate or a multi-chambered microfluidic
device, in which a
multiplicity of amplification reactions and, in some embodiments, detection
are performed, typically
15 in parallel. In certain embodiments, one or more multiplex reactions for
generating amplicons are
performed in the same reaction vessel, including without limitation, a multi-
well plate, such as a 96-
well, a 384-well, a 1536-well plate, and so forth; or a microfluidie device,
for example but not limited
to, a TaqManTm Low Density Array (Applied Biosystems, Foster City, CA). In
some embodiments, a
massively parallel amplifying step comprises a multi-well reaction vessel,
including a plate
20 comprising multiple reaction wells, for example but not limited to, a 24-
well plate, a 96-well plate, a
384-well plate, or a 1536-well plate; or a multi-chamber microfluidics device,
for example but not
limited to a low density array wherein each chamber or well comprises an
appropriate primer(s),
primer set(s), and/or reporter probe(s), as appropriate. Typically such
amplification steps occur in a
series of parallel single-plex, two-plex, three-plex, four-plex, five-plex, or
six-plex reactions, although
25 higher levels of parallel multiplexing are also within the intended
scope of the current teachings.
These methods can comprise PCR methodology, such as RT-PCR, in each of the
wells or chambers to
amplify and/or detect nucleic acid molecules of interest.
Low density arrays can include arrays that detect lOs or 100s of molecules as
opposed to
1000s of molecules. These arrays can be more sensitive than high density
arrays. In embodiments, a
30 low density array such as a TaqMari'm Low Density Array is used to
detect one or more gene or gene
product in any of Tables 5-12 of W02018175501. For example, the low density
array can be used to
detect at least 1, 2, 3,4, 5, 6, 7, 8, 9, 10, 15, 20, 25, 30, 40, 50, 60, 70,
80,90 or 100 genes or gene
products selected from any of Tables 5-12 of W02018175501.
In some embodiments, the disclosed methods comprise a microfluidics device,
"lab on a
35 chip," or micrototal analytical system (pTAS). In some embodiments,
sample preparation is
performed using a microfluidics device. In some embodiments, an amplification
reaction is performed
using a microfluidics device. In some embodiments, a sequencing or PCR
reaction is performed using
101
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
a microfluidic device. In some embodiments, the nucleotide sequence of at
least a part of an amplified
product is obtained using a microfluidics device. In some embodiments,
detecting comprises a
microfluidic device, including without limitation, a low density array, such
as a TaqManTm T,ow
Density Array. Descriptions of exemplary microfluidic devices can be found in,
among other places,
5 Published PCT Application Nos. WO/0185341 and WO 04/011666; Kartalov and
Quake, Nucl. Acids
Res. 32:2873-79, 2004; and Fiorini and Chiu, Rio Techniques 38:429-46, 2005.
Any appropriate microfluidic device can be used in the methods as described
herein.
Examples of microfluidic devices that may be used, or adapted for use with
molecular profiling,
include but are not limited to those described in U.S. Pat. Nos. 7,591,936,
7,581,429, 7,579,136,
10 7,575,722, 7,568,399, 7,552,741, 7,544,506, 7,541,578, 7,518,726,
7,488,596, 7,485,214, 7,467,928,
7,452,713, 7,452,509, 7,449,096, 7,431,887, 7,422,725, 7,422,669, 7,419,822,
7,419,639, 7,413,709,
7,411,184, 7,402,229, 7,390,463, 7,381,471, 7,357,864, 7,351,592, 7,351,380,
7,338,637, 7,329,391,
7,323,140, 7,261,824, 7,258,837, 7,253,003, 7,238,324, 7,238,255, 7,233,865,
7,229,538, 7,201,881,
7,195,986, 7,189,581, 7,189,580, 7,189,368, 7,141,978, 7,138,062, 7,135,147,
7,125,711, 7,118,910,
15 7,118,661, 7,640,947, 7,666,361, 7,704,735; U.S. Patent Application
Publication 20060035243; and
International Patent Publication WO 2010/072410; each of which patents or
applications are
incorporated herein by reference in their entirety. Another example for use
with methods disclosed
herein is described in Chen et al., "Mierofluidie isolation and transeriptome
analysis of serum
vesicles," Lab on a chip, Dec. 8, 2009 DOI: 10.1039/b916199f.
20 Gene Expression Analysis by Massively Parallel Signature Sequencing
(MPSS)
This method, described by Brenner et al. (2000) Nature Biotechnology 18:630-
634, is a
sequencing approach that combines non-gel-based signature sequencing with in
vitro cloning of
millions of templates on separate microbeads. First, a microbead library of
DNA templates is
constructed by in vitro cloning. This is followed by the assembly of a planar
array of the template-
25 containing microbeads in a flow cell at a high density. The free ends of
the cloned templates on each
microbead are analyzed simultaneously, using a fluorescence-based signature
sequencing method that
does not require DNA fragment separation. This method has been shown to
simultaneously and
accurately provide, in a single operation, hundreds of thousands of gene
signature sequences from a
cDNA library.
30 MPSS data has many uses. The expression levels of nearly all transcripts
can be quantitatively
determined; the abundance of signatures is representative of the expression
level of the gene in the
analyzed tissue Quantitative methods for the analysis of tag frequencies and
detection of differences
among libraries have been published and incorporated into public databases for
SAGETM data and are
applicable to MPSS data. The availability of complete genome sequences permits
the direct
35 comparison of signatures to genomic sequences and further extends the
utility of MPSS data. Because
the targets for MPSS analysis are not pre-selected (like on a microarray),
MPSS data can characterize
102
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
the full complexity of transcriptomes. This is analogous to sequencing
millions of ESTs at once, and
genomic sequence data can be used so that the source of the MPSS signature can
be readily identified
by computational means.
Serial Analysis of Gene Expression (SAGE)
5 Serial analysis of gene expression (SAGE) is a method that allows the
simultaneous and
quantitative analysis of a large number of gene transcripts, without the need
of providing an individual
hybridization probe for each transcript. First, a short sequence tag (e.g.,
about 10-14 bp) is generated
that contains sufficient information to uniquely identify a transcript,
provided that the tag is obtained
from a unique position within each transcript. Then, many transcripts are
linked together to form long
10 serial molecules, that can be sequenced, revealing the identity of the
multiple tags simultaneously. The
expression pattern of any population of transcripts can be quantitatively
evaluated by determining the
abundance of individual tags, and identifying the gene corresponding to each
tag. See, e.g. Velculescu
et al. (1995) Science 270:484-487; and Veleuleseu et al. (1997) Cell 88:243-
51.
DNA Copy Number Profiling
15 Any method capable of determining a DNA copy number profile of a
particular sample can be
used for molecular profiling according to the methods described herein as long
as the resolution is
sufficient to identify a copy number variation in the biomarkers as described
herein. The skilled
artisan is aware of and capable of using a number of different platforms for
assessing whole genome
copy number changes at a resolution sufficient to identify the copy number of
the one or more
20 biomarkers of the methods described herein. Some of the platforms and
techniques are described in
the embodiments below. In some embodiments as described herein, next
generation sequencing or
ISH techniques as described herein or known in the art are used for
determining copy number / gene
amplification.
In some embodiments, the copy number profile analysis involves amplification
of whole
25 genome DNA by a whole genome amplification method. The whole genome
amplification method can
use a strand displacing polymerase and random primers.
In some aspects of these embodiments, the copy number profile analysis
involves
hybridization of whole genome amplified DNA with a high density array. In a
more specific aspect,
the high density array has 5,000 or more different probes. In another specific
aspect, the high density
30 array has 5,000, 10,000, 20,000, 50,000, 100,000, 200,000, 300,000,
400,000, 500,000, 600,000,
700,000, 800,000, 900,000, or 1,000,000 or more different probes. In another
specific aspect, each of
the different probes on the array is an oligonucleotide having from about 15
to 200 bases in length. In
another specific aspect, each of the different probes on the array is an
oligonucleotide having from
about 15 to 200, 15 to 150, 15 to 100, 15 to 75, 15 to 60, or 20 to 55 bases
in length.
103
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
In some embodiments, a microarray is employed to aid in determining the copy
number
profile for a sample, e.g., cells from a tumor. Microarrays typically comprise
a plurality of oligomers
(e.g., DNA or RNA polynucleotides or oligonucleotides, or other polymers),
synthesized or deposited
on a substrate (e.g., glass support) in an array pattern. The support-bound
oligomers are "probes",
5 which function to hybridize or bind with a sample material (e.g., nucleic
acids prepared or obtained
from the tumor samples), in hybridization experiments. The reverse situation
can also he applied: the
sample can be bound to the microanuy substrate and the oligomer probes are in
solution for the
hybridization. In use, the array surface is contacted with one or more targets
under conditions that
promote specific, high-affinity binding of the target to one or more of the
probes. In some
10 configurations, the sample nucleic acid is labeled with a detectable
label, such as a fluorescent tag, so
that the hybridized sample and probes are detectable with scanning equipment.
DNA array technology
offers the potential of using a multitude (e.g., hundreds of thousands) of
different oligonucleotides to
analyze DNA copy number profiles. In some embodiments, the substrates used for
arrays are surface-
derivatized glass or silica, or polymer membrane surfaces (see e.g., in Z.
Guo, et al., Nucleic Acids
15 Res, 22, 5456-65 (1994); U. Maskos, E. M Southern, Nucleic Acids Res,
20, 1679-84 (1992), and R
M. Southern, et al., Nucleic Acids Res, 22, 1368-73 (1994), each incorporated
by reference herein).
Modification of surfaces of array substrates can be accomplished by many
techniques. For example,
siliceous or metal oxide surfaces can be derivatized with bifunctional
silanes, i.e., silanes having a
first functional group enabling covalent binding to the surface (e.g., Si-
halogen or Si-alkoxy group, as
20 in --SiC13 or --Si(OCH3) 3, respectively) and a second functional group
that can impart the desired
chemical and/or physical modifications to the surface to covalently or non-
covalently attach ligands
and/or the polymers or monomers for the biological probe array. Silylated
derivatizations and other
surface derivatizations that are known in the art (see for example U.S. Pat.
No. 5,624,711 to Sundberg,
U.S. Pat. No. 5,266,222 to Willis, and U.S. Pat. No. 5,137,765 to Farnsworth,
each incorporated by
25 reference herein). Other processes for preparing arrays are described in
U.S. Pat. No. 6,649,348, to
Bass et. al., assigned to Agilent Corp., which disclose DNA arrays created by
in situ synthesis
methods.
Polymer array synthesis is also described extensively in the literature
including in the
following: WO 00/58516, U.S. Pat. Nos. 5,143,854, 5,242,974, 5,252,743,
5,324,633, 5,384,261,
30 5,405,783, 5,424,186, 5,451,683, 5,482,867, 5,491,074, 5,527,681,
5,550,215, 5,571,639, 5,578,832,
5,593,839, 5,599,695, 5,624,711, 5,631,734, 5,795,716, 5,831,070, 5,837,832,
5,856,101, 5,858,659,
5,936,324, 5,968,740, 5,974,164,5,981,185, 5,981,956, 6,025,601, 6,033,860,
6,040,193, 6,090,555,
6,136,269, 6,269,846 and 6,428,752, 5,412,087, 6,147,205, 6,262,216,
6,310,189, 5,889,165, and
5,959,098 in PCT Applications Nos. PCT/US99/00730 (International Publication
No. WO 99/36760)
35 and PCT/US01/04285 (International Publication No. WO 01/58593), which
are all incorporated
herein by reference in their entirety for all purposes.
104
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Nucleic acid arrays that are useful in the present disclosure include, but are
not limited to,
those that are commercially available from Affymetrix (Santa Clara, Calif.)
under the brand name
GeneChipTM Example arrays are shown on the website at affymetrix.com. Another
microarray
supplier is lumina, Inc., of San Diego, Calif with example arrays shown on
their website at
5 illumina.com.
In some embodiments, the inventive methods provide for sample preparation.
Depending on
the microarray and experiment to be performed, sample nucleic acid can be
prepared in a number of
ways by methods known to the skilled artisan. In some aspects as described
herein, prior to or
concurrent with genotyping (analysis of copy number profiles), the sample may
be amplified any
10 number of mechanisms. The most common amplification procedure used
involves PCR. See, for
example, PCR Technology: Principles and Applications for DNA Amplification
(Ed. H. A. Erlich,
Freeman Press, NY, N.Y, 1992); PCR Protocols: A Guide to Methods and
Applications (Eds. Innis, et
aL, Academic Press, San Diego, Calif, 1990); Mattila et aL, Nucleic Acids Res.
19,4967 (1991);
Eckert et al., PCR Methods and Applications 1, 17 (1991); PCR (Eds. McPherson
et al., 1RL Press,
15 Oxford); and U.S. Pat. Nos. 4,683,202,4,683,195, 4,800,159 4,965,188,
and 5,333,675, and each of
which is incorporated herein by reference in their entireties for all
purposes. In some embodiments,
the sample may be amplified on the array (e.g., U.S. Pat. No. 6,300,070 which
is incorporated herein
by reference).
Other suitable amplification methods include the ligasc chain reaction (LCR)
(for example,
20 Wu and Wallace, Genomics 4, 560 (1989), Landegren et al., Science 241,
1077 (1988) and Barringer
et al. Gene 89:117 (1990)), transcription amplification (Kwoh et al., Proc.
Natl. Acad. Sci. USA 86,
1173 (1989) and W088/10315), self-sustained sequence replication (Guatelli et
al., Proc. Nat. Acad.
Sci. USA, 87, 1874 (1990) and W090/06995), selective amplification of target
polynucleotide
sequences (U.S. Pat. No. 6,410,276), consensus sequence primed polymerase
chain reaction (CP-
25 PCR) (U.S. Pat. No. 4,437,975), arbitrarily primed polymerase chain
reaction (AP-PCR) (U.S. Pat.
Nos. 5,413,909, 5,861,245) and nucleic acid based sequence amplification
(NABSA). (See, U.S. Pat.
Nos. 5,409,818, 5,554,517, and 6,063,603, each of which is incorporated herein
by reference). Other
amplification methods that may be used are described in, U.S. Pat. Nos.
5,242,794, 5,494,810,
4,988,617 and in U.S. Ser. No. 09/854,317, each of which is incorporated
herein by reference.
30 Additional methods of sample preparation and techniques for reducing the
complexity of a
nucleic sample are described in Dong et al., Genome Research 11, 1418 (2001),
in U.S. Pat. Nos.
6,361,947, 6,391,592 and U.S. Ser. Nos. 09/916,135, 09/920,491 (U.S. Patent
Application Publication
20030096235), 09/910,292 (U.S. Patent Application Publication 20030082543),
and 10/013,598.
Methods for conducting polynucleotide hybridization assays are well developed
in the art.
35 Hybridization assay procedures and conditions used in the methods as
described herein will vary
depending on the application and are selected in accordance with the general
binding methods known
including those referred to in: Maniatis et al. Molecular Cloning: A
Laboratory Manual (2<sup>nd</sup> Ed.
105
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Cold Spring Harbor, N.Y., 1989); Berger and Kimmel Methods in Enzymology, Vol.
152, Guide to
Molecular Cloning Techniques (Academic Press, Inc., San Diego, Calif , 1987);
Young and Davism,
P.N.A.S, 80: 1194 (1983). Methods and apparatus for carrying out repeated and
controlled
hybridization reactions have been described in U.S. Pat. Nos. 5,871,928,
5,874,219, 6,045,996 and
5 6,386,749, 6,391,623 each of which are incorporated herein by reference.
The methods as described herein may also involve signal detection of
hybridization between
ligands in after (and/or during) hybridization. See U.S. Pat. Nos. 5,143,854,
5,578,832; 5,631,734;
5,834,758; 5,936,324; 5,981,956; 6,025,601; 6,141,096; 6,185,030; 6,201,639;
6,218,803; and
6,225,625, in U.S. Ser. No. 10/389,194 and in PCT Application PCT/1JS99/06097
(published as
10 W099/47964), each of which also is hereby incorporated by reference in
its entirety for all purposes.
Methods and appararns for signal detection and processing of intensity data
are disclosed in,
for example, U.S. Pat. Nos. 5,143,854, 5,547,839, 5,578,832, 5,631,734,
5,800,992, 5,834,758;
5,856,092, 5,902,723, 5,936,324, 5,981,956, 6,025,601, 6,090,555, 6,141,096,
6,185,030, 6,201,639;
6,218,803; and 6,225,625, in U.S. Ser. Nos. 10/389,194, 60/493,495 and in PCT
Application
15 PCT/US99/06097 (published as W099/47964), each of which also is hereby
incorporated by
reference in its entirety for all purposes.
Immuno-based Assays
Protein-based detection molecular profiling techniques include irnmunoaffinity
assays based
on antibodies selectively immunoreactive with mutant gene encoded protein
according to the present
20 methods. These techniques include without limitation
imrnunoprecipitation, Western blot analysis,
molecular binding assays, enzyme-linked immunosorbent assay (FLISA), enzyme-
linked
irnmunofiltration assay (EL1FA), fluorescence activated cell sorting (FACS)
and the like. For
example, an optional method of detecting the expression of a biomarker in a
sample comprises
contacting the sample with an antibody against the biomarker, or an
imrnunoreactive fragment of the
25 antibody thereof, or a recombinant protein containing an antigen binding
region of an antibody against
the biomarker; and then detecting the binding of the biomarker in the sample.
Methods for producing
such antibodies are known in the art. Antibodies can be used to
imrnunoprecipitate specific proteins
from solution samples or to immunoblot proteins separated by, e.g.,
polyacrylamide gels.
Immunocytochemical methods can also be used in detecting specific protein
polymorphisms in tissues
30 or cells. Other well-known antibody-based techniques can also be used
including, e.g., ELISA,
radioinununoassay (RIA), inununoradiometric assays (IRMA) and inununoenzymatic
assays (IEMA),
including sandwich assays using monoclonal or polyclonal antibodies See, e g
IT S Pat Nos
4,376,110 and 4,486,530, both of which are incorporated herein by reference.
In alternative methods, the sample may be contacted with an antibody specific
for a
35 biomarker under conditions sufficient for an antibody-biomarker complex
to form, and then detecting
said complex. The presence of the biomarker may be detected in a number of
ways, such as by
106
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Western blotting and ELISA procedures for assaying a wide variety of tissues
and samples, including
plasma or serum. A wide range of immunoassay techniques using such an assay
format are available,
see, e.g., U.S. Pat Nos. 4,016,043,4,424,279 and 4,018,653. These include both
single-site and two-
site or "sandwich" assays of the non-competitive types, as well as in the
traditional competitive
5 binding assays. These assays also include direct binding of a labelled
antibody to a target biomarker.
A number of variations of the sandwich assay technique exist, and all are
intended Lobe
encompassed by the present methods. Briefly, in a typical forward assay, an
unlabelled antibody is
immobilized on a solid substrate, and the sample to be tested brought into
contact with the bound
molecule. After a suitable period of incubation, for a period of time
sufficient to allow formation of an
10 antibody-antigen complex, a second antibody specific to the antigen,
labelled with a reporter molecule
capable of producing a detectable signal is then added and incubated, allowing
time sufficient for the
formation of another complex of antibody-antigen-labelled antibody. Any
unreacted material is
washed away, and the presence of the antigen is determined by observation of a
signal produced by
the reporter molecule. The results may either be qualitative, by simple
observation of the visible
15 signal, or may be quantitated by comparing with a control sample
containing known amounts of
biomarker.
Variations on the forward assay include a simultaneous assay, in which both
sample and
labelled antibody are added simultaneously to the bound antibody. These
techniques are well known
to those skilled in the art, including any minor variations as will be readily
apparent. In a typical
20 forward sandwich assay, a first antibody having specificity for the
biomarker is either covalently or
passively bound to a solid surface. The solid surface is typically glass or a
polymer, the most
commonly used polymers being cellulose, polyacrylamide, nylon, polystyrene,
polyvinyl chloride or
polypropylene. The solid supports may be in the form of tubes, beads, discs of
microplates, or any
other surface suitable for conducting an immunoassay. The binding processes
are well-known in the
25 art and generally consist of cross-linking covalently binding or
physically adsorbing, the polymer-
antibody complex is washed in preparation for the test sample. An aliquot of
the sample to be tested is
then added to the solid phase complex and incubated for a period of time
sufficient (e.g. 2-40 minutes
or overnight if more convenient) and under suitable conditions (e.g. from room
temperature to 40 C
such as between 25 C and 32 C inclusive) to allow binding of any subunit
present in the antibody.
30 Following the incubation period, the antibody subunit solid phase is
washed and dried and incubated
with a second antibody specific for a portion of the biomarker. The second
antibody is linked to a
reporter molecule which is used to indicate the binding of the second antibody
to the molecular
marker.
An alternative method involves immobilizing the target biomarkers in the
sample and then
35 exposing the immobilized target to specific antibody which may or may
not be labelled with a reporter
molecule. Depending on the amount of target and the strength of the reporter
molecule signal, a bound
target may be detectable by direct labelling with the antibody. Alternatively,
a second labelled
107
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
antibody, specific to the first antibody is exposed to the target-first
antibody complex to form a target-
first antibody-second antibody tertiary complex. The complex is detected by
the signal emitted by the
reporter molecule. By "reporter molecule", as used in the present
specification, is meant a molecule
which, by its chemical nature, provides an analytically identifiable signal
which allows the detection
5 of antigen-bound antibody. The most commonly used reporter molecules in
this type of assay are
either enzymes, fluorophores or radionuclide containing molecules (i.e.
radioisotopes) and
chemiluminescent molecules.
In the case of an enzyme immunoassay, an enzyme is conjugated to the second
antibody,
generally by means of glutaraldehyde or periodate. As will be readily
recognized, however, a wide
10 variety of different conjugation techniques exist, which are readily
available to the skilled artisan.
Commonly used enzymes include horseradish peroxidase, glucose oxidase, ii-
galactosidase and
alkaline phosphatase, amongst others. The substrates to be used with the
specific enzymes are
generally chosen for the production, upon hydrolysis by the corresponding
enzyme, of a detectable
color change. Examples of suitable enzymes include alkaline phosphatase and
peroxidase. It is also
15 possible to employ fluorogenic substrates, which yield a fluorescent
product rather than the
chromogenic substrates noted above. In all cases, the enzyme-labelled antibody
is added to the first
antibody-molecular marker complex, allowed to bind, and then the excess
reagent is washed away. A
solution containing the appropriate substrate is then added to the complex of
antibody-antigen-
antibody. The substrate will react with the enzyme linked to the second
antibody, giving a qualitative
20 visual signal, which may be further quantitated, usually
spectrophotometrically, to give an indication
of the amount of biomarker which was present in the sample. Alternately,
fluorescent compounds,
such as fluorescein and rhodamine, may be chemically coupled to antibodies
without altering their
binding capacity. When activated by illumination with light of a particular
wavelength, the
fluorochrome-labelled antibody adsorbs the light energy, inducing a state to
excitability in the
25 molecule, followed by emission of the light at a characteristic color
visually detectable with a light
microscope. As in the EIA, the fluorescent labelled antibody is allowed to
bind to the first antibody-
molecular marker complex. After washing off the unbound reagent, the remaining
tertiary complex is
then exposed to the light of the appropriate wavelength, the fluorescence
observed indicates the
presence of the molecular marker of interest. Irmnunofluorescence and EIA
techniques are both very
30 well established in the art. However, other reporter molecules, such as
radioisotope, chemiluminescent
or bioluminescent molecules, may also be employed.
Immunohistochemistry (IHC)
IFIC is a process of localizing antigens (e.g., proteins) in cells of a tissue
binding antibodies
specifically to antigens in the tissues. The antigen-binding antibody can be
conjugated or fused to a
35 tag that allows its detection, e.g., via visualization. In some
embodiments, the tag is an enzyme that
can catalyze a color-producing reaction, such as alkaline phosphatase or
horseradish peroxidase. The
108
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
enzyme can be fused to the antibody or non-covalently bound, e.g., using a
biotin-avadin system.
Alternatively, the antibody can be tagged with a fluorophore, such as
fluorescein, rhodamine, DyLight
Fluor or Alexa Fluor. The antigen-binding antibody can be directly tagged or
it can itself be
recognized by a detection antibody that carries the tag. Using ILIC, one or
more proteins may be
5 detected. The expression of a gene product can be related to its staining
intensity compared to control
levels. In some embodiments, the gene product is considered di fTerentially
expressed if its staining
varies at least 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9, 2.0, 2.2, 2.5, 2.7,
3.0, 4, 5, 6, 7, 8, 9 or 10-fold in the
sample versus the control.
ITIC comprises the application of antigen-antibody interactions to
histochemical techniques.
10 In an illustrative example, a tissue section is mounted on a slide and
is incubated with antibodies
(polyclonal or monoclonal) specific to the antigen (primary reaction). The
antigen-antibody signal is
then amplified using a second antibody conjugated to a complex of peroxidase
antiperoxidase (PAP),
avidin-biotin-peroxidase (ABC) or avidin-biotin alkaline phosphatase. In the
presence of substrate and
chromogen, the enzyme forms a colored deposit at the sites of antibody-antigen
binding.
15 Immunofluorescence is an alternate approach to visualize antigens. In
this technique, the primary
antigen-antibody signal is amplified using a second antibody conjugated to a
fluorochrome. On UV
light absorption, the fluorochrome emits its own light at a longer wavelength
(fluorescence), thus
allowing localization of antibody-antigen complexes.
Epigenetic Status
20 Molecular profiling methods according to the present disclosure also
comprise measuring
epigenetic change, i.e., modification in a gene caused by an epigenetic
mechanism, such as a change
in methylation status or histone acetylation. Frequently, the epigenetic
change will result in an
alteration in the levels of expression of the gene which may be detected (at
the RNA or protein level
as appropriate) as an indication of the epigenetic change. Often the
epigenetic change results in
25 silencing or down regulation of the gene, referred to as "epigenetic
silencing." The most frequently
investigated epigenetic change in the methods as described herein involves
determining the DNA
methylation status of a gene, where an increased level of methylation is
typically associated with the
relevant cancer (since it may cause down regulation of gene expression).
Aberrant methylation, which
may be referred to as hypermethylation, of the gene or genes can be detected.
Typically, the
30 methylation status is determined in suitable CpG islands which are often
found in the promoter region
of the gene(s). The term "methylation," "methylation state" or "methylation
status" may refers to the
presence or absence of 5-methylcytosine at one or a plurality of CpG
dinucleotides within a DNA
sequence. CpG dinucleotides are typically concentrated in the promoter regions
and exons of human
genes.
35 Diminished gene expression can be assessed in terms of DNA methylation
status or in terms
of expression levels as determined by the methylation status of the gene. One
method to detect
109
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
epigenetic silencing is to determine that a gene which is expressed in normal
cells is less expressed or
not expressed in tumor cells. Accordingly, the present disclosure provides for
a method of molecular
profiling comprising detecting epigenetic silencing.
Various assay procedures to directly detect methylation are known in the art,
and can be used
5 in conjunction with the present methods. These assays rely onto two
distinct approaches: bisulphite
conversion based approaches and non-bisulphite based approaches. Non-
bisulphite based methods for
analysis of DNA methylation rely on the inability of methylation-sensitive
enzymes to cleave
methylation cytosines in their restriction. The bisulphite conversion relies
on treatment of DNA
samples with sodium bisulphite which converts unmethylated cytosine to uracil,
while methylated
10 cytosines are maintained (Furuichi Y, Wataya Y, Hayatsu H, Ukita T.
Biochem Biophys Res Comrnun.
1970 Dec 9;41(5):1185-91). This conversion results in a change in the sequence
of the original DNA.
Methods to detect such changes include MS AP-PCR (Methylation-Sensitive
Arbitrarily-Primed
Polymerase Chain Reaction), a technology that allows for a global scan of the
genome using CG-rich
primers to focus on the regions most likely to contain CpG dinucleotides, and
described by Gonzalgo
15 et al., Cancer Research 57:594-599, 1997; MethyLightTM, which refers to
the art-recognized
fluorescence-based real-time PCR technique described by Eads et al., Cancer
Res. 59:2302-2306,
1999; the HeavyMethylTmassay, in the embodiment thereof implemented herein, is
an assay, wherein
methylation specific blocking probes (also referred to herein as blockers)
covering CpG positions
between, or covered by the amplification primers enable methylation-specific
selective amplification
20 of a nucleic acid sample; HeavyMethylTmMethyLightTm is a variation of
the MethyLightTM assay
wherein the MethyLighlrm assay is combined with methylation specific blocking
probes covering
CpG positions between the amplification primers; Ms-SNuPE (Methylation-
sensitive Single
Nucleotide Primer Extension) is an assay described by Gonzalgo & Jones,
Nucleic Acids Res.
25:2529-2531, 1997; MSP (Methylation-specific PCR) is a methylation assay
described by Herman et
25 al. Proc. Natl. Acad. Sci. USA 93:9821-9826, 1996, and by U.S. Pat. No.
5,786,146; COBRA
(Combined Bisulfite Restriction Analysis) is a methylation assay described by
Xiong & Laird,
Nucleic Acids Res. 25:2532-2534, 1997; MCA (Methylated CpG Island
Amplification) is a
methylation assay described by Toyota et al., Cancer Res. 59:2307-12, 1999,
and in WO 00/26401A1.
Other techniques for DNA methylation analysis include sequencing, methylation-
specific
30 PCR (MS-PCR), melting curve methylation-specific PCR (McMS-PCR), MLPA
with or without
bisulfite treatment, QAMA, MSRE-PCR, MethyLight, ConLight-MSP, bisulfite
conversion-specific
methylation-specific PCR (BS-MSP), COBRA (which relies upon use of restriction
enzymes to reveal
methylation dependent sequence differences m PCR products of sodium bisulfite-
treated DNA),
methylation-sensitive single-nucleotide primer extension conformation (MS-
SNuPE), methylation-
35 sensitive single-strand conformation analysis (MS-SSCA), Melting curve
combined bisulfite
restriction analysis (MeCOBRA), PyroMethA, HeavyMethyl, MALDI-TOF, MassARRAY,
Quantitative analysis of methylated alleles (QAMA), enzymatic regional
methylation assay (ERMA),
110
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
QBSUPT, MethylQuant, Quantitative PCR sequencing and oligonucleotide-based
microarray systems,
Pyrosequencing, Meth-DOP-PCR. A review of some useful techniques is provided
in Nucleic acids
research, 1998, Vol. 26, No. 10, 2255-2264; Nature Reviews, 2003, Vol.3, 253-
266; Oral Oncology,
2006, Vol. 42, 5-13, which references are incorporated herein in their
entirety. Any of these techniques
5 may be used in accordance with the present methods, as appropriate. Other
techniques are described
in U.S. Patent Publications 20100144836; and 20100184027, which applications
are incorporated
herein by reference in their entirety.
Through the activity of various acetylases and deacetylylases the DNA binding
function of
histone proteins is tightly regulated. Furthermore, histone acetylation and
histone deactelyation have
10 been linked with malignant progression. See Nature, 429: 457-63, 2004.
Methods to analyze histone
acetylation are described in U.S. Patent Publications 20100144543 and
20100151468, which
applications are incorporated herein by reference in their entirety.
Sequence Analysis
Molecular profiling according to the present disclosure comprises methods for
genotyping
15 one or more biomarkers by determining whether an individual has one or
more nucleotide variants (or
amino acid variants) in one or more of the genes or gene products. Genotyping
one or more genes
according to the methods as described herein in some embodiments, can provide
more evidence for
selecting a treatment.
The biomarkers as described herein can be analyzed by any method useful for
determining
20 alterations in nucleic acids or the proteins they encode. According to
one embodiment, the ordinary
skilled artisan can analyze the one or more genes for mutations including
deletion mutants, insertion
mutants, frame shift mutants, nonsense mutants, missense mutant, and splice
mutants.
Nucleic acid used for analysis of the one or more genes can be isolated from
cells in the
sample according to standard methodologies (Sambrook et al., 1989). The
nucleic acid, for example,
25 may be genonaic DNA or fractionated or whole cell RNA, or miRNA acquired
from exosomes or cell
surfaces. Where RNA is used, it may be desired to convert the RNA to a
complementary DNA. In one
embodiment, the RNA is whole cell RNA; in another, it is poly-A RNA; in
another, it is exosomal
RNA. Normally, the nucleic acid is amplified. Depending on the format of the
assay for analyzing the
one or more genes, the specific nucleic acid of interest is identified in the
sample directly using
30 amplification or with a second, known nucleic acid following
amplification. Next, the identified
product is detected. In certain applications, the detection may be performed
by visual means (e.g.,
ethidium bromide staining of a gel). Alternatively, the detection may involve
indirect identification of
the product via chemiluminescence, radioactive scintigraphy of radiolabel or
fluorescent label or even
via a system using electrical or thermal impulse signals (Affymax Technology;
Bellus, 1994).
35 Various types of defects are known to occur in the biomarkers as
described herein. Alterations
include without limitation deletions, insertions, point mutations, and
duplications. Point mutations can
111
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
be silent or can result in stop codons, flume shift mutations or amino acid
substitutions. Mutations in
and outside the coding region of the one or more genes may occur and can be
analyzed according to
the methods as described herein. The target site of a nucleic acid of interest
can include the region
wherein the sequence varies. Examples include, but are not limited to,
polymorphisms which exist in
5 different forms such as single nucleotide variations, nucleotide repeats,
multibase deletion (more than
one nucleotide deleted from the consensus sequence), multibase insertion (more
than one nucleotide
inserted from the consensus sequence), microsatellite repeats (small numbers
of nucleotide repeats
with a typical 5-1000 repeat units), di-nucleotide repeats, tri-nucleotide
repeats, sequence
rearrangements (including translocation and duplication), chimeric sequence
(two sequences from
10 different gene origins are fused together), and the like. Among sequence
polymorphisms, the most
frequent polymorphisms in the human genome are single-base variations, also
called single-nucleotide
polymorphisms (SNPs). SNPs are abundant, stable and widely distributed across
the genome.
Molecular profiling includes methods for haplotyping one or more genes. The
haplotype is a
set of genetic determinants located on a single chromosome and it typically
contains a particular
15 combination of alleles (all the alternative sequences of a gene) in a
region of a chromosome. In other
words, the haplotype is phased sequence information on individual chromosomes.
Very often, phased
SNPs on a chromosome define a haplotype. A combination of haplotypes on
chromosomes can
determine a genetic profile of a cell. It is the haplotype that determines a
linkage between a specific
genetic marker and a disease mutation. Haplotyping can be done by any methods
known in the art.
20 Common methods of scoring SNPs include hybridization microarray or
direct gel sequencing,
reviewed in Landgren et al., Genome Research, 8:769-776, 1998. For example,
only one copy of one
or more genes can be isolated from an individual and the nucleotide at each of
the variant positions is
determined. Alternatively, an allele specific PCR or a similar method can be
used to amplify only one
copy of the one or more genes in an individual, and SNPs at the variant
positions of the present
25 disclosure are determined. The Clark method known in the art can also be
employed for haplotyping.
A high throughput molecular haplotyping method is also disclosed in Tost et
al., Nucleic Acids Res.,
30(19):e96 (2002), which is incorporated herein by reference.
Thus, additional variant(s) that are in linkage disequilibrium with the
variants and/or
haplotypes of the present disclosure can be identified by a haplotyping method
known in the art, as
30 will be apparent to a skilled artisan in the field of genetics and
haplotyping. The additional variants
that are in linkage disequilibrium with a variant or haplotype of the present
disclosure can also be
useful in the various applications as described below.
For purposes of genotypmg and haplotyping, both genomic DNA and mR.NA/cDNA can
be
used, and both are herein referred to generically as "gene."
35 Numerous techniques for detecting nucleotide variants are known in the
art and can all be
used for the method of this disclosure. The techniques can be protein-based or
nucleic acid-based. In
either case, the techniques used must be sufficiently sensitive so as to
accurately detect the small
112
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
nucleotide or amino acid variations. Very often, a probe is used which is
labeled with a detectable
marker. Unless otherwise specified in a particular technique described below,
any suitable marker
known in the art can be used, including but not limited to, radioactive
isotopes, fluorescent
compounds, biotin which is detectable using streptavidin, enzymes (e.g.,
alkaline phosphatase),
5 substrates of an enzyme, ligands and antibodies, etc. See Jablonski et
al., Nucleic Acids Res.,
14:6115-6128 (1986); Nguyen et al., Biotechniques, 13:116-123 (1992); Rigby et
al., J. Mol. Biol.,
113:237-251 (1977).
In a nucleic acid-based detection method, target DNA sample, i.e., a sample
containing
genomic DNA, cDNA, mR_NA and/or miRNA, corresponding to the one or more genes
must be
10 obtained from the individual to be tested. Any tissue or cell sample
containing the genomic DNA,
miRNA, m-RNA, and/or cDNA (or a portion thereof) corresponding to the one or
more genes can he
used. For this purpose, a tissue sample containing cell nucleus and thus
genomic DNA can be
obtained from the individual. Blood samples can also be useful except that
only white blood cells and
other lymphocytes have cell nucleus, while red blood cells are without a
nucleus and contain only
15 mR_NA or miRNA. Nevertheless, miRNA and mR_NA are also useful as either
can be analyzed for the
presence of nucleotide variants in its sequence or serve as template for cDNA
synthesis. The tissue or
cell samples can be analyzed directly without much processing. Alternatively,
nucleic acids including
the target sequence can be extracted, purified, and/or amplified before they
are subject to the various
detecting procedures discussed below. Other than tissue or cell samples, eDNAs
or genomic DNAs
20 from a cDNA or genomic DNA library constructed using a tissue or cell
sample obtained from the
individual to be tested are also useful.
To determine the presence or absence of a particular nucleotide variant,
sequencing of the
target genomic DNA or cDNA, particularly the region encompassing the
nucleotide variant locus to
be detected. Various sequencing techniques are generally known and widely used
in the art including
25 the Stinger method and Gilbert chemical method. The pyrosequencing
method monitors DNA
synthesis in real time using a luminometric detection system. Pyrosequencing
has been shown to be
effective in analyzing genetic polymorphisms such as single-nucleotide
polymorphisms and can also
be used in the present methods. See Nordstrom et al., Biotechnol. Appl.
Biochem., 31(2):107-112
(2000); Ahmadian et al., Anal. Biochem., 280:103-110 (2000).
30 Nucleic acid variants can be detected by a suitable detection process.
Non limiting examples
of methods of detection, quantification, sequencing and the like are; mass
detection of mass modified
amplicons (e.g., matrix-assisted laser desorption ionization (MALD1) mass
spectrometry and
electrospray (ES) mass spectrometry), a pruner extension method (e.g.,
iPLEXIm; Sequenom, Inc.),
microsequencing methods (e.g., a modification of primer extension
methodology), ligase sequence
35 determination methods (e.g., U.S. Pat. Nos. 5,679,524 and 5,952,174, and
WO 01/27326), mismatch
sequence determination methods (e.g., U.S. Pat. Nos. 5,851,770; 5,958,692;
6,110,684; and
6,183,958), direct DNA sequencing, fragment analysis (EA), restriction
fragment length
113
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
polymorphism (RFLP analysis), allele specific oligonucleotide (ASO) analysis,
methylation-specific
PCR (MSPCR), pyrosequencing analysis, acycloprime analysis, Reverse dot blot,
GeneChip
microarrays, Dynamic allele-specific hybridization (DASH), Peptide nucleic
acid (PNA) and locked
nucleic acids (LNA) probes, TaqMan, Molecular Beacons, Intercalating dye, FRET
primers,
5 AlphaScreen, SNPstream, genetic bit analysis (GBA), Multiplex
minisequencing, SNaPshot, GOOD
assay, Microarray miniseq, arrayed primer extension (APEX), Microarray primer
extension (e.g.,
microarray sequence determination methods), Tag arrays, Coded microspheres,
Template-directed
incorporation (TDI), fluorescence polarization, Colorimetric oligonucleotide
ligation assay (OLA),
Sequence-coded OLA, Microarray ligation, Ligase chain reaction, Padlock
probes, Invader assay,
10 hybridization methods (e.g., hybridization using at least one probe,
hybridization using at least one
fluorescently labeled probe, and the like), conventional dot blot analyses,
single strand conformational
polymorphism analysis (SSCP, e.g., U.S. Pat. Nos. 5,891,625 and 6,013,499;
Orita et al., Proc. Natl.
Acad. Sci. U.S.A. 86: 27776-2770 (1989)), denaturing gradient gel
electrophoresis (DGGE),
heteroduplex analysis, mismatch cleavage detection, and techniques described
in Sheffield et al., Proc.
15 Natl. Acad. Sci. USA 49: 699-706 (1991), White et al., Genomics 12: 301-
306 (1992), Grompe et al.,
Proc. Natl. Acad. Sci. USA 86: 5855-5892 (1989), and Grompe, Nature Genetics
5: 111-117 (1993),
cloning and sequencing, electrophoresis, the use of hybridization probes and
quantitative real time
polymerase chain reaction (QRT-PCR), digital PCR, nanopore sequencing, chips
and combinations
thereof The detection and quantification of alleles or paralogs can be carried
out using the "closed-
20 tube" methods described in U.S. patent application Ser. No. 11/950,395,
filed on Dec. 4,2007. In
some embodiments the amount of a nucleic acid species is determined by mass
spectrometry, primer
extension, sequencing (e.g., any suitable method, for example nanopore or
pyrosequencing),
Quantitative PCR (Q-PCR or QRT-PCR), digital PCR, combinations thereof, and
the like.
The term "sequence analysis" as used herein refers to determining a nucleotide
sequence, e.g.,
25 that of an amplification product. The entire sequence or a partial
sequence of a polynuckolide, e.g.,
DNA or mRNA, can be determined, and the determined nucleotide sequence can be
referred to as a
"read" or "sequence read." For example, linear amplification products may be
analyzed directly
without further amplification in some embodiments (e.g., by using single-
molecule sequencing
methodology). In certain embodiments, linear amplification products may be
subject to further
30 amplification and then analyzed (e.g., using sequencing by ligation or
pyrosequencing methodology).
Reads may be subject to different types of sequence analysis. Any suitable
sequencing method can be
used to detect, and determine the amount of, nucleotide sequence species,
amplified nucleic acid
species, or detectable products generated from the foregoing. Examples of
certain sequencing
methods are described hereafter.
35 A sequence analysis apparatus or sequence analysis component(s) includes
an apparatus, and
one or more components used in conjunction with such apparatus, that can be
used by a person of
ordinary skill to determine a nucleotide sequence resulting from processes
described herein (e.g.,
114
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
linear and/or exponential amplification products). Examples of sequencing
platforms include, without
limitation, the 454 platform (Roche) (Margulies, M. et al. 2005 Nature 437,
376-380), Illumina
Genomic Analyzer (or Solexa platform) or SOLID System (Applied Biosystems; see
PCT patent
application publications WO 06/084132 entitled "Reagents, Methods, and
Libraries For Bead-Based
5 Sequencing" and W007/121,489 entitled "Reagents, Methods, and Libraries
for Gel-Free Bead-Based
Sequencing"), the Helicos True Single Molecule DNA sequencing technology
(Harris TD et al. 2008
Science, 320, 106-109), the single molecule, real-time (SMRTTm) technology of
Pacific Biosciences,
and nanopore sequencing (Soni G V and Meller A. 2007 Clin Chem 53: 1996-2001),
Ion
semiconductor sequencing (Ion Torrent Systems, Inc, San Francisco, CA), or DNA
nanoball
10 sequencing (Complete Genomics, Mountain View, CA), VisiGen
Biotechnologies approach
(Invitrogen) and polony sequencing. Such platforms allow sequencing of many
nucleic acid molecules
isolated from a specimen at high orders of multiplexing in a parallel manner
(Dear Brief Funct
Genomic Proteomic 2003; 1: 397-416; Haimovich, Methods, challenges, and
promise of next-
generation sequencing in cancer biology. Yale J Biol Med. 2011 Dec;84(4):439-
46). These non-
15 Sanger-based sequencing technologies are sometimes referred to as
NextCren sequencing, NGS, next-
generation sequencing, next generation sequencing, and variations thereof.
Typically they allow much
higher throughput than the traditional Sanger approach. See Schuster, Next-
generation sequencing
transforms today's biology, Nature Methods 5:16-18 (2008); Metzker, Sequencing
technologies - the
next generation. Nat Rev Genet. 2010 Jan;11(1):31-46; Levy and Myers,
Advancements in Next-
20 Generation Sequencing. Annu Rev Genomics Hum Genet. 2016 Aug 31;17:95-
115. These platforms
can allow sequencing of clonally expanded or non-amplified single molecules of
nucleic acid
fragments. Certain platforms involve, for example, sequencing by ligation of
dye-modified probes
(including cyclic ligation and cleavage), pyrosequencing, and single-molecule
sequencing. Nucleotide
sequence species, amplification nucleic acid species and detectable products
generated there from can
25 be analyzed by such sequence analysis platforms. Next-generation
sequencing can be used in the
methods as described herein, e.g., to determine mutations, copy number, or
expression levels, as
appropriate. The methods can be used to perform whole genome sequencing or
sequencing of specific
sequences of interest, such as a gene of interest or a fragment thereof.
Sequencing by ligation is a nucleic acid sequencing method that relies on the
sensitivity of
30 DNA ligase to base-pairing mismatch. DNA ligase joins together ends of
DNA that are correctly base
paired. Combining the ability of DNA ligase to join together only correctly
base paired DNA ends,
with mixed pools of fluorescently labeled oligonucleotides or primers, enables
sequence
determination by fluorescence detection. Longer sequence reads may be obtained
by including
primers containing cleavable linkages that can be cleaved after label
identification. Cleavage at the
35 linker removes the label and regenerates the 5' phosphate on the end of
the ligated primer, preparing
the primer for another round of ligation. In some embodiments primers may be
labeled with more than
one fluorescent label, e.g., at least 1, 2, 3, 4, or 5 fluorescent labels.
115
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Sequencing by ligation generally involves the following steps. Clonal bead
populations can be
prepared in emulsion microreactors containing target nucleic acid template
sequences, amplification
reaction components, beads and primers. After amplification, templates are
denatured and bead
enrichment is performed to separate beads with extended templates from
undesired beads (e.g., beads
5 with no extended templates). The template on the selected beads undergoes
a 3' modification to allow
covalent bonding to the slide, and modified heads can he deposited onto a
glass slide_ Deposition
chambers offer the ability to segment a slide into one, four or eight chambers
during the bead loading
process. For sequence analysis, primers hybridize to the adapter sequence. A
set of four color dye-
labeled probes competes for ligation to the sequencing primer. Specificity of
probe ligation is
10 achieved by interrogating every 4th and 5th base during the ligation
series. Five to seven rounds of
ligation, detection and cleavage record the color at every 5th position with
the number of rounds
determined by the type of library used. Following each round of ligation, a
new complimentary primer
offset by one base in the 5' direction is laid down for another series of
ligations. Primer reset and
ligation rounds (5-7 ligation cycles per round) are repeated sequentially five
times to generate 25-35
15 base pairs of sequence for a single tag. With mate-paired sequencing,
this process is repeated for a
second tag.
Pyrosequencing is a nucleic acid sequencing method based on sequencing by
synthesis, which
relies on detection of a pyrophosphate released on nucleotide incorporation.
Generally, sequencing by
synthesis involves synthesizing, one nucleotide at a time, a DNA strand
complimentary to the strand
20 whose sequence is being sought. Target nucleic acids may be immobilized
to a solid support,
hybridized with a sequencing primer, incubated with DNA polymerase, ATP
sulfurylase, luciferase,
apyrase, adenosine 5' phosphosulfate and luciferin. Nucleotide solutions are
sequentially added and
removed. Correct incorporation of a nucleotide releases a pyrophosphate, which
interacts with ATP
sulfurylase and produces ATP in the presence of adenosine 5' phosphosulfate,
fueling the luciferin
25 reaction, which produces a chemilumincseent signal allowing sequence
determination. The amount of
light generated is proportional to the number of bases added. Accordingly, the
sequence downstream
of the sequencing primer can be determined. An illustrative system for
pyrosequencing involves the
following steps: ligating an adaptor nucleic acid to a nucleic acid under
investigation and hybridizing
the resulting nucleic acid to a bead; amplifying a nucleotide sequence in an
emulsion; sorting beads
30 using a picoliter multiwell solid support; and sequencing amplified
nucleotide sequences by
pyrosequencing methodology (e.g., Nakano et al., "Single-molecule PCR using
water-in-oil
emulsion;" Journal of Biotechnology 102: 117-124 (2003)).
Certain single-molecule sequencing embodiments are based on the principal of
sequencing by
synthesis, and use single-pair Fluorescence Resonance Energy Transfer (single
pair FRET) as a
35 mechanism by which photons are emitted as a result of successful
nucleotide incorporation. The
emitted photons often are detected using intensified or high sensitivity
cooled charge-couple-devices
in conjunction with total internal reflection microscopy (T1RM). Photons are
only emitted when the
116
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
introduced reaction solution contains the correct nucleotide for incorporation
into the growing nucleic
acid chain that is synthesized as a result of the sequencing process. In FRET
based single-molecule
sequencing, energy is transferred between two fluorescent dyes, sometimes
polymethine cyanine dyes
Cy3 and Cy5, through long-range dipole interactions. The donor is excited at
its specific excitation
5 wavelength and the excited state energy is transferred, non-radiatively
to the acceptor dye, which in
turn becomes excited. The acceptor dye eventually returns to the ground state
by radiative emission of
a photon. The two dyes used in the energy transfer process represent the
"single pair" in single pair
FRET. Cy3 often is used as the donor fluorophore and often is incorporated as
the first labeled
nucleotide. Cy5 often is used as the acceptor fluorophore and is used as the
nucleotide label for
10 successive nucleotide additions after incorporation of a first Cy3
labeled nucleotide. The fluorophores
generally are within 10 nanometers of each for energy transfer to occur
successfully
An example of a system that can be used based on single-molecule sequencing
generally
involves hybridizing a primer to a target nucleic acid sequence to generate a
complex; associating the
complex with a solid phase; iteratively extending the primer by a nucleotide
tagged with a fluorescent
15 molecule; and capturing an image of fluorescence resonance energy
transfer signals after each
iteration (e.g., U.S. Pat. No. 7,169,314; Braslaysky et al., PNAS 100(7): 3960-
3964 (2003)). Such a
system can be used to directly sequence amplification products (linearly or
exponentially amplified
products) generated by processes described herein. In some embodiments the
amplification products
can be hybridized to a primer that contains sequences complementary to
immobilized capture
20 sequences present on a solid support, a bead or glass slide for example.
Hybridization of the primer-
amplification product complexes with the immobilized capture sequences,
immobilizes amplification
products to solid supports for single pair FRET based sequencing by synthesis.
The primer often is
fluorescent, so that an initial reference image of the surface of the slide
with immobilized nucleic
acids can be generated. The initial reference image is useful for determining
locations at which true
25 nucleotide incorporation is occurring. Fluorescence signals detected in
array locations not initially
identified in the "primer only" reference image are discarded as non-specific
fluorescence. Following
immobilization of the primer-amplification product complexes, the bound
nucleic acids often are
sequenced in parallel by the iterative steps of, a) polymerase extension in
the presence of one
fluorescently labeled nucleotide, b) detection of fluorescence using
appropriate microscopy, TIRNI for
30 example, c) removal of fluorescent nucleotide, and d) return to step a
with a different fluorescently
labeled nucleotide.
In some embodiments, nucleotide sequencing may be by solid phase single
nucleotide
sequencing methods and processes. Solid phase single nucleotide sequencing
methods involve
contacting target nucleic acid and solid support under conditions in which a
single molecule of sample
35 nucleic acid hybridizes to a single molecule of a solid support. Such
conditions can include providing
the solid support molecules and a single molecule of target nucleic acid in a
"microreactor." Such
conditions also can include providing a mixture in which the target nucleic
acid molecule can
117
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
hybridize to solid phase nucleic acid on the solid support. Single nucleotide
sequencing methods
useful in the embodiments described herein are described in U.S. Provisional
Patent Application Ser.
No. 61/021,871 filed Jan. 17,200S
In certain embodiments, nanopore sequencing detection methods include (a)
contacting a
5 target nucleic acid for sequencing ("base nucleic acid," e.g., linked
probe molecule) with sequence-
specific detectors, under conditions in which the detectors specifically
hybridize to substantially
complementary subsequences of the base nucleic acid; (b) detecting signals
from the detectors and (c)
determining the sequence of the base nucleic acid according to the signals
detected. In certain
embodiments, the detectors hybridized to the base nucleic acid are
disassociated from the base nucleic
10 acid (e.g., sequentially dissociated) when the detectors interfere with
a nanopore structure as the base
nucleic acid passes through a pore, and the detectors disassociated from the
base sequence are
detected. In some embodiments, a detector disassociated from a base nucleic
acid emits a detectable
signal, and the detector hybridized to the base nucleic acid emits a different
detectable signal or no
detectable signal. In certain embodiments, nucleotides in a nucleic acid
(e.g., linked probe molecule)
15 are substituted with specific nucleotide sequences corresponding to
specific nucleotides ("nucleotide
representatives"), thereby giving rise to an expanded nucleic acid (e.g., U.S.
Pat. No. 6,723,513), and
the detectors hybridize to the nucleotide representatives in the expanded
nucleic acid, which serves as
a base nucleic acid. In such embodiments, nucleotide representatives may be
arranged in a binary or
higher order arrangement (e.g., Soni and Moller, Clinical Chemistry 53(11):
1996-2001 (2007)). In
20 some embodiments, a nucleic acid is not expanded, does not give rise to
an expanded nucleic acid,
and directly serves a base nucleic acid (e.g., a linked probe molecule serves
as a non-expanded base
nucleic acid), and detectors are directly contacted with the base nucleic
acid. For example, a first
detector may hybridize to a first subsequence and a second detector may
hybridize to a second
subsequence, where the first detector and second detector each have detectable
labels that can be
25 distinguished from one another, and where the signals from the first
detector and second detector can
be distinguished from one another when the detectors are disassociated from
the base nucleic acid. In
certain embodiments, detectors include a region that hybridizes to the base
nucleic acid (e.g., two
regions), which can be about 3 to about 100 nucleotides in length (e.g., about
4, 5, 6, 7, 8,9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 25, 30, 35, 40,50, 55, 60, 65, 70, 75, 80,
85, 90, or 95 nucleotides in
30 length). A detector also may include one or more regions of nucleotides
that do not hybridize to the
base nucleic acid. In some embodiments, a detector is a molecular beacon. A
detector often comprises
one or more detectable labels independently selected from those described
herein. Each detectable
label can be detected by any convenient detection process capable of detecting
a signal generated by
each label (e.g., magnetic, electric, chemical, optical and the like). For
example, a CD camera can be
35 used to detect signals from one or more distinguishable quantum dots
linked to a detector.
In certain sequence analysis embodiments, reads may be used to construct a
larger nucleotide
sequence, which can be facilitated by identifying overlapping sequences in
different reads and by
118
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
using identification sequences in the reads. Such sequence analysis methods
and software for
constructing larger sequences from reads are known to the person of ordinary
skill (e.g., Venter at al.,
Science 291: 1304-1351 (2001)). Specific reads, partial nucleotide sequence
constructs, and full
nucleotide sequence constructs may be compared between nucleotide sequences
within a sample
5 nucleic acid (i.e., internal comparison) or may be compared with a
reference sequence (i.e., reference
comparison) in certain sequence analysis embodiments. Internal comparisons can
be performed in
situations where a sample nucleic acid is prepared from multiple samples or
from a single sample
source that contains sequence variations. Reference comparisons sometimes are
performed when a
reference nucleotide sequence is known and an objective is to determine
whether a sample nucleic
10 acid contains a nucleotide sequence that is substantially similar or the
same, or different, than a
reference nucleotide sequence. Sequence analysis can be facilitated by the use
of sequence analysis
apparatus and components described above.
Primer extension polymorphism detection methods, also referred to herein as
"microsequencing" methods, typically are carried out by hybridizing a
complementary
15 oligonucleotide to a nucleic acid carrying the polymorphic site. In
these methods, the oligonucleotide
typically hybridizes adjacent to the polymorphic site. The term "adjacent" as
used in reference to
"microsequencing" methods, refers to the 3' end of the extension
oligonucleotide being sometimes 1
nucleotide from the 5' end of the polymorphic site, often 2 or 3, and at times
4, 5, 6, 7, 8, 9, or 10
nucleotides from the 5' end of the polymorphic site, in the nucleic acid when
the extension
20 oligonucleotide is hybridized to the nucleic acid. The extension
oligonucleotide then is extended by
one or more nucleotides, often 1, 2, or 3 nucleotides, and the number and/or
type of nucleotides that
are added to the extension oligonucleotide determine which polymorphic variant
or variants are
present. Oligonueleotide extension methods are disclosed, for example, in U.S.
Pat. Nos. 4,656,127;
4,851,331; 5,679,524; 5,834,189; 5,876,934; 5,908,755; 5,912,118; 5,976,802;
5,981,186; 6,004,744;
25 6,013,431; 6,017,702; 6,046,005; 6,087,095; 6,210,891; and WO 01/20039.
The extension products
can be detected in any manner, such as by fluorescence methods (see, e.g.,
Chen & Kwok, Nucleic
Acids Research 25: 347-353 (1997) and Chen et al., Proc. Natl. Acad. Sci. USA
94/20: 10756-10761
(1997)) or by mass spectrometric methods (e.g., MALDI-TOF mass spectrometry)
and other methods
described herein. Oligonucleotide extension methods using mass spectrometry
are described, for
30 example, in U.S. Pat. Nos. 5,547,835; 5,605,798; 5,691,141; 5,849,542;
5,869,242; 5,928,906;
6,043,031; 6,194,144; and 6,258,538.
Microsequencing detection methods often incorporate an amplification process
that proceeds
the extension step. 'Me amplification process typically amplifies a region
from a nucleic acid sample
that comprises the polymorphic site. Amplification can be carried out using
methods described above,
35 or for example using a pair of oligonucleotide primers in a polymerase
chain reaction (PCR), in which
one oligonucleotide primer typically is complementary to a region 3' of the
polymorphism and the
other typically is complementary to a region 5' of the polymorphism. A PCR
primer pair may be used
119
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
in methods disclosed in U.S. Pat. Nos. 4,683,195; 4,683,202, 4,965,188;
5,656,493; 5,998,143;
6,140,054; WO 01/27327; and WO 01/27329 for example. PCR primer pairs may also
be used in any
commercially available machines that perform PCR, such as any of the GeneAmpTM
Systems
available from Applied Biosystems.
5 Other appropriate sequencing methods include multiplex polony sequencing
(as described in
Shendure et al., Accurate Multiplex Polony Sequencing of an Evolved Bacterial
Genome,
Sciencexpress, Aug. 4, 2005, pg 1 available at www.sciencexpress.org/4 Aug.
2005/Page1/10.1126/science.1117389, incorporated herein by reference), which
employs immobilized
microbeads, and sequencing in microfabricated picoliter reactors (as described
in Margulies et al.,
10 Genome Sequencing in Microfabricated High-Density Picolitre Reactors,
Nature, August 2005,
available at vvww.nature.com/nature (published online 31 Jul. 2005,
doi:10.1038/nature03959,
incorporated herein by reference).
Whole genome sequencing may also be used for discriminating alleles of RNA
transcripts, in
some embodiments. Examples of whole genome sequencing methods include, but are
not limited to,
15 nanopore-based sequencing methods, sequencing by synthesis and
sequencing by ligation, as
described above.
Nucleic acid variants can also be detected using standard electrophoretic
techniques.
Although the detection step can sometimes be preceded by an amplification
step, amplification is not
required in the embodiments described herein. Examples of methods for
detection and quantification
20 of a nucleic acid using electrophoretic techniques can be found in the
art. A non-limiting example
comprises running a sample (e.g., mixed nucleic acid sample isolated from
maternal serum, or
amplification nucleic acid species, for example) in an agarose or
polyacrylamide gel. The gel may be
labeled (e.g., stained) with ethidium bromide (see, Sambrook and Russell,
Molecular Cloning: A
Laboratory Manual 3d ed., 2001). The presence of a band of the same size as
the standard control is
25 an indication of the presence of a target nucleic acid sequence, the
amount of which may then be
compared to the control based on the intensity of the band, thus detecting and
quantifying the target
sequence of interest. In some embodiments, restriction enzymes capable of
distinguishing between
maternal and paternal alleles may be used to detect and quantify target
nucleic acid species. In certain
embodiments, oligonucleotide probes specific to a sequence of interest are
used to detect the presence
30 of the target sequence of interest. The oligonucleotides can also be
used to indicate the amount of the
target nucleic acid molecules in comparison to the standard control, based on
the intensity of signal
imparted by the probe.
Sequence-specific probe hybridization can be used to detect a particular
nucleic acid m a
mixture or mixed population comprising other species of nucleic acids. Under
sufficiently stringent
35 hybridization conditions, the probes hybridize specifically only to
substantially complementary
sequences. The stringency of the hybridization conditions can be relaxed to
tolerate varying amounts
of sequence mismatch. A number of hybridization formats are known in the art,
which include but are
120
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
not limited to, solution phase, solid phase, or mixed phase hybridization
assays. The following articles
provide an overview of the various hybridization assay formats: Singer et al.,
Biotechniques 4:230,
1986; Haase et al., Methods in Virology, pp. 189-226, 1984; Wilkinson, In situ
Hybridization,
Wilkinson ed., IRL Press, Oxford University Press, Oxford; and Haines and
Higgins eds., Nucleic
5 Acid hybridization: A Practical Approach, IRL Press, 1987.
Hybridization complexes can be detected by techniques known in the art.
Nucleic acid probes
capable of specifically hybridizing to a target nucleic acid (e.g., mRNA or
DNA) can be labeled by
any suitable method, and the labeled probe used to detect the presence of
hybridized nucleic acids.
One commonly used method of detection is autoradiography, using probes labeled
with 41-1, 1251, 35S,
10 '4C, 3214, 33P, or the like. The choice of radioactive isotope depends
on research preferences due to ease
of synthesis, stability, and half-lives of the selected isotopes. Other labels
include compounds (e.g.,
biotin and digoxigenin), which bind to antiligands or antibodies labeled with
fluorophores,
chemiluminescent agents, and enzymes. In some embodiments, probes can be
conjugated directly
with labels such as fluorophores, chemilunninescent agents or enzyimes. The
choice of label depends
15 on sensitivity required, ease of conjugation with the probe, stability
requirements, and available
instrumentation.
In embodiments, fragment analysis (referred to herein as "FA") methods are
used for
molecular profiling. Fragment analysis (FA) includes techniques such as
restriction fragment length
polymorphism (RFLP) and/or (amplified fragment length polymorphism). If a
nucleotide variant in
20 the target DNA corresponding to the one or more genes results in the
elimination or creation of a
restriction enzyme recognition site, then digestion of the target DNA with
that particular restriction
enzyme will generate an altered restriction fragment length pattern. Thus, a
detected RFLP or AFLP
will indicate the presence of a particular nucleotide variant.
Terminal restriction fragment length polymorphism (TRFLP) works by PCR
amplification of
25 DNA using primer pairs that have been labeled with fluorescent tags. The
PCR products are digested
using RFLP enzymes and the resulting patterns are visualized using a DNA
sequencer. The results are
analyzed either by counting and comparing bands or peaks in the TRFLP profile,
or by comparing
bands from one or more TRFLP runs in a database.
The sequence changes directly involved with an RFLP can also be analyzed more
quickly by
30 PCR. Amplification can be directed across the altered restriction site,
and the products digested with
the restriction enzyme. This method has been called Cleaved Amplified
Polymorphic Sequence
(CAPS). Alternatively, the amplified segment can be analyzed by Allele
specific oligonucleotide
(ASO) probes, a process that is sometimes assessed using a Dot blot.
A variation on AFLP is cDNA-AFLP, which can be used to quantify differences in
gene
35 expression levels.
Another useful approach is the single-stranded conformation polymorphism assay
(SSCA),
which is based on the altered mobility of a single-stranded target DNA
spanning the nucleotide variant
121
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
of interest. A single nucleotide change in the target sequence can result in
different intramolecular
base pairing pattern, and thus different secondary structure of the single-
stranded DNA, which can be
detected in a non-denaturing gel. See Orita et al., Proc. Natl. Acad. Sci.
USA, 86:2776-2770 (1989).
Denaturing gel-based techniques such as clamped denaturing gel electrophoresis
(CDGE) and
5 denaturing gradient gel electrophoresis (DGGE) detect differences in
migration rates of mutant
sequences as compared to wild-type sequences in denaturing gel. See Miller et
al., Biotechniques,
5:1016-24 (1999); Sheffield et al., Am. J. Hum, Genet., 49:699-706 (1991);
Wartell et al., Nucleic
Acids Res., 18:2699-2705 (1990); and Sheffield et al., Proc. Natl. Acad. Sci.
USA, 86:232-236
(1989). In addition, the double-strand conformation analysis (DSCA) can also
be useful in the present
10 methods. See Arguello et al., Nat. Genet., 18:192-194 (1998).
The presence or absence of a nucleotide variant at a particular locus in the
one or more genes
of an individual can also be detected using the amplification refractory
mutation system (ARMS)
technique. See e.g., European Patent No. 0,332,435; Newton et al., Nucleic
Acids Res., 17:2503-2515
(1989); Fox et al., Br. J. Cancer, 77:1267-1274 (1998); Robertson et al., Fur.
Respir. J., 12:477-482
15 (1998). In the ARMS method, a primer is synthesized matching the
nucleotide sequence immediately
5' upstream from the locus being tested except that the 3'-end nucleotide
which corresponds to the
nucleotide at the locus is a predetermined nucleotide. For example, the 3 '-
end nucleotide can be the
same as that in the mutated locus. The primer can be of any suitable length so
long as it hybridizes to
the target DNA under stringent conditions only when its 3'-end nucleotide
matches the nucleotide at
20 the locus being tested. Preferably the primer has at least 12
nucleotides, more preferably from about
18 to 50 nucleotides. If the individual tested has a mutation at the locus and
the nucleotide therein
matches the 3'-end nucleotide of the primer, then the primer can be further
extended upon hybridizing
to the target DNA template, and the primer can initiate a PCR amplification
reaction in conjunction
with another suitable PCR primer. In contrast, if the nucleotide at the locus
is of wild type, then
25 primer extension cannot be achieved. Various forms of ARMS techniques
developed in the past few
years can be used. See e.g., Gibson et al., Clin. Chem. 43:1336-1341(1997).
Similar to the ARMS technique is the mini sequencing or single nucleotide
primer extension
method, which is based on the incorporation of a single nucleotide. An
oligonucleotide primer
matching the nucleotide sequence immediately 5' to the locus being tested is
hybridized to the target
30 DNA, mRNA or miRNA in the presence of labeled dideoxyribonucleotides. A
labeled nucleotide is
incorporated or linked to the primer only when the dideoxyribonucleotides
matches the nucleotide at
the variant locus being detected. Thus, the identity of the nucleotide at the
variant locus can be
revealed based on the detection label attached to the incorporated
dideoxyribonucleotides. See
Syvanen et al., Genomics, 8:684-692 (1990); Shumaker et al., Hum. Mutat.,
7:346-354 (1996); Chen
35 et al., Genome Res., 10:549-547 (2000).
Another set of techniques useful in the present methods is the so-called
"oligonucleotide
ligation assay" (OLA) in which differentiation between a wild-type locus and a
mutation is based on
122
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
the ability of two oligonucleotides to anneal adjacent to each other on the
target DNA molecule
allowing the two oligonucleotides joined together by a DNA ligase. See
Landergren et al., Science,
241:1077-1080 (1988); Chen et al, Cienome Res., 8:549-556 (1998); iannone et
al., Cytometry,
39:131-140 (2000). Thus, for example, to detect a single-nucleotide mutation
at a particular locus in
5 the one or more genes, two oligonucleotides can be synthesized, one
having the sequence just 5'
upstream from the locus with its 3' end nucleotide being identical to the
nucleotide in the variant
locus of the particular gene, the other having a nucleotide sequence matching
the sequence
immediately 3' downstream from the locus in the gene. The oligonucleotides can
be labeled for the
purpose of detection. Upon hybridizing to the target gene under a stringent
condition, the two
10 oligonucleotides are subject to ligation in the presence of a suitable
ligase. The ligation of the two
oligonucleotides would indicate that the target DNA has a nucleotide variant
at the locus being
detected.
Detection of small genetic variations can also be accomplished by a variety of
hybridization-
based approaches. Allele-specific oligonucleotides are most useful. See Conner
et al., Proc. Natl.
15 Acad. Sci. USA, 80:278-282 (1983); Saiki et al, Proc. Natl. Acad. Sci.
USA, 86:6230-6234 (1989).
Oligonucleotide probes (allele-specific) hybridizing specifically to a gene
allele having a particular
gene variant at a particular locus but not to other alleles can be designed by
methods known in the art.
The probes can have a length of, e.g., from 10 to about 50 nucleotide bases.
The target DNA and the
oligonucleotide probe can be contacted with each other under conditions
sufficiently stringent such
20 that the nucleotide variant can be distinguished from the wild-type gene
based on the presence or
absence of hybridization. The probe can be labeled to provide detection
signals. Alternatively, the
allele-specific oligonucleotide probe can be used as a PCR amplification
primer in an "allele-specific
PCR" and the presence or absence of a PCR product of the expected length would
indicate the
presence or absence of a particular nucleotide variant.
25 Other useful hybridization-based techniques allow two single-stranded
nucleic acids annealed
together even in the presence of mismatch due to nucleotide substitution,
insertion or deletion. The
mismatch can then be detected using various techniques. For example, the
annealed duplexes can be
subject to electrophoresis. The mismatched duplexes can be detected based on
their electrophoretic
mobility that is different from the perfectly matched duplexes. See Cariello,
Human Genetics, 42:726
30 (1988). Alternatively, in an RNase protection assay, a RNA probe can be
prepared spanning the
nucleotide variant site to be detected and having a detection marker. See
Giunta et al., Diagn. Mol.
Path., 5:265-270 (1996); Finkelstein et al., Genomics, 7:167-172 (1990);
Kinszler et al., Science
251:1366-1370 (1991). The RNA probe can be hybridized to the target DNA or
mRNA forming a
heteroduplex that is then subject to the ribonuclease RNase A digestion. RNase
A digests the RNA
35 probe in the heteroduplex only at the site of mismatch. The digestion
can be determined on a
denaturing electrophoresis gel based on size variations. In addition,
mismatches can also be detected
123
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
by chemical cleavage methods known in the art. See e.g., Roberts etal.,
Nucleic Acids Res., 25:3377-
3378 (1997).
In the mutS assay, a probe can be prepared matching the gene sequence
surrounding the locus
at which the presence or absence of a mutation is to be detected, except that
a predetermined
5 nucleotide is used at the variant locus. Upon annealing the probe to the
target DNA to form a duplex,
the E. coli mutS protein is contacted with the duplex. Since the mutS protein
hinds only to
heteroduplex sequences containing a nucleotide mismatch, the binding of the
mutS protein will be
indicative of the presence of a mutation. See Modrith etal., Ann. Rev. Genet.,
25:229-253 (1991).
A great variety of improvements and variations have been developed in the art
on the basis of
10 the above-described basic techniques which can be useful in detecting
mutations or nucleotide
variants in the present methods. For example, the "sunrise probes" or
"molecular beacons" use the
fluorescence resonance energy transfer (FRET) property and give rise to high
sensitivity. See Wolf et
al., Proc. Nat Acad. Sei. USA, 85:8790-8794 (1988). Typically, a probe
spanning the nucleotide locus
to be detected are designed into a hairpin-shaped structure and labeled with a
quenching fluorophore
15 at one end and a reporter fluorophore at the other end. In its natural
state, the fluorescence from the
reporter fluorophore is quenched by the quenching fluorophore due to the
proximity of one
fluorophore to the other. Upon hybridization of the probe to the target DNA,
the 5' end is separated
apart from the 3'-end and thus fluorescence signal is regenerated. See
Nazarenko et al., Nucleic Acids
Res., 25:2516-2521(1997); Ryehlik et al., Nucleic Acids Res., 17:8543-
8551(1989); Sharkey et al.,
20 Bio/Technology 12:506-509 (1994); Tyagi etal., Nat. Biotechnol., 14:303-
308 (1996); Tyagi et al.,
Nat Biotechnol., 16:49-53 (1998). The homo-tag assisted non-dimer system
(HANDS) can be used in
combination with the molecular beacon methods to suppress primer-dimer
accumulation. See Brownie
etal., Nucleic Acids Res., 25:3235-3241(1997).
Dye-labeled oligonucleotide ligation assay is a FRET-based method, which
combines the
25 OLA assay and PCR. See Chen et at, Genome Res. 8:549-556 (1998). TaqMan
is another FRET-
based method for detecting nucleotide variants. A TaqMan probe can be
oligonucleotides designed to
have the nucleotide sequence of the gene spanning the variant locus of
interest and to differentially
hybridize with different alleles. The two ends of the probe are labeled with a
quenching fluorophore
and a reporter fluorophore, respectively. The TaqMan probe is incorporated
into a PCR reaction for
30 the amplification of a target gene region containing the locus of
interest using Taq polymerase. As Taq
polymerase exhibits 5'-3' exonuclease activity but has no 3'-5' exonuclease
activity, if the TaqMan
probe is annealed to the target DNA template, the 5'-end of the TaqMan probe
will be degraded by
Taq polymerase during the PCR reaction thus separating the reporting
fluorophore from the quenching
fluorophore and releasing fluorescence signals. See Ifolland et al., Proc.
Natl. Acad. Sci. USA,
35 88:7276-7280 (1991); Kalinina et al., Nucleic Acids Res., 25:1999-2004
(1997); Whitcombe et al.,
Clin. Chem., 44:918-923 (1998).
124
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
In addition, the detection in the present methods can also employ a
chemiluminescence-based
technique. For example, an oligonucleotide probe can be designed to hybridize
to either the wild-type
or a variant gene locus but not both. The probe is labeled with a highly
chemiluminescent acridinium
ester. Hydrolysis of the acridinium ester destroys chemiluminescence. The
hybridization of the probe
5 to the target DNA prevents the hydrolysis of the acridinium ester.
Therefore, the presence or absence
of a particular mutation in the target DNA is determined by measuring
chemiluminescence changes.
See Nelson et al., Nucleic Acids Res., 24:4998-5003 (1996).
The detection of genetic variation in the gene in accordance with the present
methods can also
be based on the "base excision sequence scanning" (BESS) technique. The BESS
method is a PCR-
10 based mutation scanning method. BESS T-Scan and BESS G-Tracker are
generated which are
analogous to T and G ladders of dideoxy sequencing. Mutations are detected by
comparing the
sequence of normal and mutant DNA. See, e.g., Hawkins et al., Electrophoresis,
20:1171-1176 (1999).
Mass spectrometry can be used for molecular profiling according to the present
methods. See
Graber et al., Cun-. Opin. Biotechnol., 9:14-18 (1998). For example, in the
primer oligo base extension
15 (PROBETm) method, a target nucleic acid is immobilized to a solid-phase
support. A primer is
annealed to the target immediately 5' upstream from the locus to be analyzed.
Primer extension is
carried out in the presence of a selected mixture of deoxyribonueleolides and
dideoxyribonueleolides.
The resulting mixture of newly extended primers is then analyzed by MALDI-TOF.
See e.g.,
Monfortc ct al., Nat. Mcd., 3:360-362 (1997).
20 In addition, the microchip or microarray technologies are also
applicable to the detection
method of the present methods. Essentially, in microchips, a large number of
different oligonucleotide
probes are immobilized in an array on a substrate or carrier, e.g., a silicon
chip or glass slide. Target
nucleic acid sequences to be analyzed can be contacted with the immobilized
oligonucleotide probes
on the microchip. See Lipshutz et al., Biotechniques, 19:442-447 (1995); Chee
et al., Science,
25 274:610-614 (1996); Kozal et al., Nat. Med. 2:753-759 (1996); Hacia et
al., Nat. Genet., 14:441-447
(1996); Saiki et al., Proc. Natl. Acad. Sci. USA, 86:6230-6234 (1989);
Gingeras et al., Genome Res.,
8:435-448 (1998). Alternatively, the multiple target nucleic acid sequences to
be studied are fixed onto
a substrate and an array of probes is contacted with the immobilized target
sequences. See Drrnanac et
al., Nat. Biotechnol., 16:54-58 (1998). Numerous microchip technologies have
been developed
30 incorporating one or more of the above described techniques for
detecting mutations. The microchip
technologies combined with computerized analysis tools allow fast screening in
a large scale. The
adaptation of the microchip technologies to the present methods will be
apparent to a person of skill in
the art apprised of the present disclosure. See, e.g., U.S. Pat. No. 5,925,525
to Fodor et al; Wilgenbus
et al., J. Mol. Med., 77:761-786 (1999); Graber et al., Curr. Opin.
Biotechnol., 9:14-18 (1998); Hacia
35 et al., Nat. Genet., 14:441-447 (1996); Shoemaker et al., Nat. Genet.,
14:450-456 (1996); DeRisi et
al., Nat. Genet., 14:457-460(1996); Chee et al., Nat. Genet., 14:610-614
(1996); Lockhart et al., Nat.
Genet., 14:675-680 (1996); Drobyshev et al., Gene, 188:45-52 (1997).
125
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
As is apparent from the above survey of the suitable detection techniques, it
may or may not
be necessary to amplify the target DNA, i.e., the gene, cDNA, mRNA, miRNA, or
a portion thereof to
increase the number of target DNA molecule, depending on the detection
techniques used. For
example, most PCR-based techniques combine the amplification of a portion of
the target and the
5 detection of the mutations. PCR amplification is well known in the art
and is disclosed in U.S. Pat.
Nos. 4,683,195 and 4,800,1.59, both which are incorporated herein by
reference. For non-PCR-based
detection techniques, if necessary, the amplification can be achieved by,
e.g., in vivo plasmid
multiplication, or by purifying the target DNA from a large amount of tissue
or cell samples. See
generally, Sambrook et al., Molecular Cloning: A Laboratory Manual, 2'd ed.,
Cold Spring Harbor
10 Laboratory, Cold Spring Harbor, N.Y., 1989. However, even with scarce
samples, many sensitive
techniques have been developed in which small genetic variations such as
single-nucleotide
substitutions can be detected without having to amplify the target DNA in the
sample. For example,
techniques have been developed that amplify the signal as opposed to the
target DNA by, e.g_,
employing branched DNA or dendrimers that can hybridize to the target DNA. The
branched or
15 dendrimer DNAs provide multiple hybridization sites for hybridization
probes to attach thereto thus
amplifying the detection signals. See Detmer et al., J. Clin. Microbiol.,
34:901-907 (1996); Collins et
al., Nucleic Acids Res., 25:2979-2984 (1997); Horn et al., Nucleic Acids Res.,
25:4835-4841 (1997);
Horn et al., Nucleic Acids Res., 25:4842-4849 (1997); Nilsen et al., J. Theor.
Biol., 187:273-284
(1997).
20 The lnvaderTM assay is another technique for detecting single nucleotide
variations that can be
used for molecular profiling according to the methods. The Invader'. assay
uses a novel linear signal
amplification technology that improves upon the long turnaround times required
of the typical PCR
DNA sequenced-based analysis. See Cooksey et al., Antimicrobial Agents and
Chemotherapy
44:1296-1301 (2000). This assay is based on cleavage of a unique secondary
structure formed
25 between two overlapping oligonueleolides that hybridize to the target
sequence of interest to form a
"flap." Each "flap" then generates thousands of signals per hour. Thus, the
results of this technique
can be easily read, and the methods do not require exponential amplification
of the DNA target. The
lnvaderTM system uses two short DNA probes, which are hybridized to a DNA
target. The structure
formed by the hybridization event is recognized by a special cleavase enzyme
that cuts one of the
30 probes to release a short DNA "flap." Each released "flap" then binds to
a fluorescently-labeled probe
to form another cleavage structure. When the cleavase enzyme cuts the labeled
probe, the probe emits
a detectable fluorescence signal. See e.g. Lyamichev et al., Nat. Biotechnol.,
17:292-296 (1999).
'The rolling circle method is another method that avoids exponential
amplification. Lizard' et
al., Nature Genetics, 19:225-232 (1998) (which is incorporated herein by
reference). For example,
35 Sniper"., a commercial embodiment of this method, is a sensitive, high-
throughput SNP scoring
system designed for the accurate fluorescent detection of specific variants.
For each nucleotide
variant, two linear, allele-specific probes are designed. The two allele-
specific probes are identical
126
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
with the exception of the 3'-base, which is varied to complement the variant
site. In the first stage of
the assay, target DNA is denatured and then hybridized with a pair of single,
allele-specific, open-
circle oligonucleotide probes. When the 3'-base exactly complements the target
DNA, ligation of the
probe will preferentially occur. Subsequent detection of the circularized
oligonucleotide probes is by
5 rolling circle amplification, whereupon the amplified probe products are
detected by fluorescence. See
Clark arid Pickering, Life Science News 6, 2000, Amersham Pharmacia Biotech
(2000).
A number of other techniques that avoid amplification all together include,
e.g., surface-
enhanced resonance Raman scattering (SERRS), fluorescence correlation
spectroscopy, and single-
molecule electrophoresis. In SERRS, a chromophore-nucleic acid conjugate is
absorbed onto colloidal
10 silver and is irradiated with laser light at a resonant frequency of the
chromophore. See Graham et al.,
Anal. Chem., 69:4703-4707 (1997). The fluorescence correlation spectroscopy is
based on the spatio-
temporal correlations among fluctuating light signals and trapping single
molecules in an electric
field. See Eigen et aL, Proc. Natl. Acad. Sci. USA, 91:5740-5747 (1994). In
single-molecule
electrophoresis, the electrophoretic velocity of a fluorescently tagged
nucleic acid is determined by
15 measuring the time required for the molecule to travel a predetermined
distance between two laser
beams. See Castro et al., Anal. Chem., 67:3181-3186 (1995).
In addition, the allele-specific oligonucleotides (ASO) can also be used in in
situ
hybridization using tissues or cells as samples. The oligonucleotide probes
which can hybridize
differentially with the wild-type gene sequence or the gene sequence harboring
a mutation may be
20 labeled with radioactive isotopes, fluorescence, or other detectable
markers. In situ hybridization
techniques are well known in the art and their adaptation to the present
methods for detecting the
presence or absence of a nucleotide variant in the one or more gene of a
particular individual should
be apparent to a skilled artisan apprised of this disclosure.
Accordingly, the presence or absence of one or more genes nucleotide variant
or amino acid
25 variant in an individual can be determined using any of the detection
methods described above.
Typically, once the presence or absence of one or more gene nucleotide
variants or amino acid
variants is determined, physicians or genetic counselors or patients or other
researchers may be
informed of the result. Specifically the result can be cast in a transmittable
form that can be
communicated or transmitted to other researchers or physicians or genetic
counselors or patients.
30 Such a form can vary and can be tangible or intangible. 't he result
with regard to the presence or
absence of a nucleotide variant of the present methods in the individual
tested can be embodied in
descriptive statements, diagrams, photographs, charts, images or any other
visual forms. For example,
images of gel electrophoresis of PCR products can be used m explaining the
results. Diagrams
showing where a variant occurs in an individual's gene are also useful in
indicating the testing results.
35 The statements and visual forms can be recorded on a tangible media such
as papers, computer
readable media such as floppy disks, compact disks, etc., or on an intangible
media, e.g., an electronic
media in the form of email or website on intern& or intranet. In addition, the
result with regard to the
127
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
presence or absence of a nucleotide variant or amino acid variant in the
individual tested can also be
recorded in a sound form and transmitted through any suitable media, e.g.,
analog or digital cable
lines, fiber optic cables, etc., via telephone, facsimile, wireless mobile
phone, internet phone and the
like.
5 Thus, the information and data on a test result can be produced anywhere
in the world and
transmitted to a different location. For example, when a genotyping assay is
conducted offshore, the
information and data on a test result may be generated and cast in a
transmittable form as described
above. The test result in a transmittable form thus can be imported into the
U.S. Accordingly, the
present methods also encompasses a method for producing a transmittable form
of information on the
10 genotype of the two or more suspected cancer samples from an individual.
The method comprises the
steps of (1) determining the genotype of the DNA from the samples according to
methods of the
present methods; and (2) embodying the result of the determining step in a
transmittable form. The
transmittable form is the product of the production method.
In Situ Hybridization
15 In situ hybridization assays are well known and are generally described
in Angerer et al.,
Methods Enzymol. 152:649-660 (1987). In an in situ hybridization assay, cells,
e.g., from a biopsy,
are fixed to a solid support, typically a glass slide. If DNA is to be probed,
the cells are denatured with
heat or alkali. The cells are then contacted with a hybridization solution at
a moderate temperature to
permit annealing of specific probes that are labeled. The probes are
preferably labeled, e.g., with
20 radioisotopes or fluorescent reporters, or enzymatically. FISH
(fluorescence in situ hybridization) uses
fluorescent probes that bind to only those parts of a sequence with which they
show a high degree of
sequence similarity. CISH (chromogenic in situ hybridization) uses
conventional peroxidase or
alkaline phosphatase reactions visualized under a standard bright-field
microscope.
In situ hybridization can be used to detect specific gene sequences in tissue
sections or cell
25 preparations by hybridizing the complementary strand of a nucleotide
probe to the sequence of
interest Fluorescent in situ hybridization (FISH) uses a fluorescent probe to
increase the sensitivity of
in situ hybridization.
FISH is a cytogenetic technique used to detect and localize specific
polynucleotide sequences
in cells. For example, FISH can be used to detect DNA sequences on
chromosomes. FISH can also be
30 used to detect and localize specific RNAs, e.g., mRNAs, within tissue
samples. In FISH uses
fluorescent probes that bind to specific nucleotide sequences to which they
show a high degree of
sequence similarity_ Fluorescence microscopy can be used to find out whether
and where the
fluorescent probes are bound. In addition to detecting specific nucleotide
sequences, e.g.,
translocations, fusion, breaks, duplications and other chromosomal
abnormalities, FISH can help
35 define the spatial-temporal patterns of specific gene copy number and/or
gene expression within cells
and tissues.
128
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Various types of FISH probes can be used to detect chromosome translocations.
Dual color,
single fusion probes can be useful in detecting cells possessing a specific
chromosomal translocation.
The DNA probe hybridization targets are located on one side of each of the two
genetic breakpoints.
"Extra signal" probes can reduce the frequency of normal cells exhibiting an
abnormal FISH pattern
5 due to the random co-localization of probe signals in a normal nucleus.
One large probe spans one
breakpoint, while the other probe flanks the breakpoint on the other gene.
Dual color, break apart
probes are useful in cases where there may be multiple translocation partners
associated with a known
genetic breakpoint. This labeling scheme features two differently colored
probes that hybridize to
targets on opposite sides of a breakpoint in one gene. Dual color, dual fusion
probes can reduce the
10 number of normal nuclei exhibiting abnormal signal patterns. The probe
offers advantages in
detecting low levels of nuclei possessing a simple balanced translocation.
Large probes span two
breakpoints on different chromosomes. Such probes are available as Vysis
probes from Abbott
Laboratories, Abbott Park, IL.
CISH, or chromogenic in situ hybridization, is a process in which a labeled
complementary
15 DNA or RNA strand is used to localize a specific DNA or RNA sequence in
a tissue specimen. CISH
methodology can be used to evaluate gene amplification, gene deletion,
chromosome translocation,
and chromosome number. CISH can use conventional enzymatic detection
methodology, e.g.,
horseradish peroxidase or alkaline phosphatase reactions, visualized under a
standard bright-field
microscope. In a common embodiment, a probe that recognizes the sequence of
interest is contacted
20 with a sample. An antibody or other binding agent that recognizes the
probe, e.g., via a label carried
by the probe, can be used to target an enzymatic detection system to the site
of the probe. In some
systems, the antibody can recognize the label of a FISH probe, thereby
allowing a sample to be
analyzed using both FISH and CISII detection. CISII can be used to evaluate
nucleic acids in multiple
settings, e.g., forrnalin-fixed, paraffin-embedded (FFPE) tissue, blood or
bone marrow smear,
25 metaphase chromosome spread, and/or fixed cells. In an embodiment, CISH
is performed following
the methodology in the SPOT-Light HER2 CISH Kit available from Life
Technologies (Carlsbad,
CA) or similar CISH products available from Life Technologies. 'The SPoT-Light
HER2 CISH Kit
itself is FDA approved for in vitro diagnostics and can be used for molecular
profiling of HER2.
CISH can be used in similar applications as FISH. Thus, one of skill will
appreciate that reference to
30 molecular profiling using FISH herein can be performed using CISH,
unless otherwise specified.
Silver-enhanced in situ hybridization (SISH) is similar to CISH, but with SISH
the signal
appears as a black coloration due to silver precipitation instead of the
chromogen precipitates of
CISH.
Modifications of the in situ hybridization techniques can be used for
molecular profiling
35 according to the methods. Such modifications comprise simultaneous
detection of multiple targets,
e.g., Dual ISH, Dual color CISH, bright field double in situ hybridization
(BDISH). See e.g., the FDA
129
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
approved INFORM HER2 Dual ISH DNA Probe Cocktail kit from Ventana Medical
Systems, Inc.
(Tucson, AZ); DuoCISHrm, a dual color CISII kit developed by Dako Denmark A/S
(Denmark).
Comparative Ctenomic Hybridization (CGH) comprises a molecular cytogenetic
method of
screening tumor samples for genetic changes showing characteristic patterns
for copy number changes
5 at chromosomal and subchromosomal levels. Alterations in patterns can be
classified as DNA gains
and losses. CGH employs the kinetics of in situ hybridization to compare the
copy numbers of
different DNA or RNA sequences from a sample, or the copy numbers of different
DNA or RNA
sequences in one sample to the copy numbers of the substantially identical
sequences in another
sample. In many useful applications of CGH, the DNA or RNA is isolated from a
subject cell or cell
10 population. The comparisons can be qualitative or quantitative.
Procedures are described that permit
determination of the absolute copy numbers of DNA sequences throughout the
genome of a cell or
cell population if the absolute copy number is known or determined for one or
several sequences. The
different sequences are discriminated from each other by the different
locations of their binding sites
when hybridized to a reference genome, usually metaphase chromosomes but in
certain cases
15 interphase nuclei. The copy number information originates from
comparisons of the intensities of the
hybridization signals among the different locations on the reference genome.
The methods, techniques
and applications of CGH are known, such as described in U.S. Pat. No.
6,335,167, and in U.S. App.
Ser. No. 60/804,818, the relevant parts of which are herein incorporated by
reference.
In an embodiment, CGH used to compare nucleic acids between diseased and
healthy tissues.
20 The method comprises isolating DNA from disease tissues (e.g., tumors)
and reference tissues (e.g.,
healthy tissue) and labeling each with a different "color" or fluor. The two
samples are mixed and
hybridized to normal metaphase chromosomes. In the case of array or matrix
CGH, the hybridization
mixing is done on a slide with thousands of DNA probes. A variety of detection
system can be used
that basically determine the color ratio along the chromosomes to determine
DNA regions that might
25 be gained or lost in the diseased samples as compared to the reference.
Molecular Profiling Methods
FIG. 11 illustrates a block diagram of an illustrative embodiment of a system
10 for
determining individualized medical intervention for a particular disease state
that uses molecular
profiling of a patient's biological specimen. System 10 includes a user
interface 12, a host server 14
30 including a processor 16 for processing data, a memory 18 coupled to the
processor, an application
program 20 stored in the memory 18 and accessible by the processor 16 for
directing processing of the
data by the processor 16, a plurality of internal databases 22 and external
databases 24, and an
interface with a wired or wireless communications network 26 (such as the
Internet, for example).
System 10 may also include an input digitizer 28 coupled to the processor 16
for inputting digital data
35 from data that is received from user interface 12.
130
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
User interface 12 includes an input device 30 and a display 32 for inputting
data into system
and for displaying information derived from the data processed by processor
16. User interface 12
may also include a printer 34 for printing the information derived from the
data processed by the
processor 16 such as patient reports that may include test results for targets
and proposed drug
5 therapies based on the test results.
Internal databases 22 may include, but are not limited to, patient biological
sample/specimen
information and tracking, clinical data, patient data, patient tracking, file
management, study
protocols, patient test results from molecular profiling, and billing
information and tracking. External
databases 24 nay include, but are not limited to, drug libraries, gene
libraries, disease libraries, and
10 public and private databases such as UniGene, OMIM, GO, TIGR, GenBank,
KEGG and Biocarta.
Various methods may be used in accordance with system 10. FIGs. 2A-C shows a
flowchart
of an illustrative embodiment of a method for determining individualized
medical intervention for a
particular disease state that uses molecular profiling of a patient's
biological specimen that is non
disease specific. In order to determine a medical intervention for a
particular disease state using
15 molecular profiling that is independent of disease lineage diagnosis
(i.e., not single disease restricted),
at least one molecular test is performed on the biological sample of a
diseased patient. Biological
samples are obtained from diseased patients by taking a biopsy of a tumor,
conducting minimally
invasive surgery if no recent tumor is available, obtaining a sample of the
patient's blood, or a sample
of any other biological fluid including, but not limited to, cell extracts,
nuclear extracts, cell lysates or
20 biological products or substances of biological origin such as
excretions, blood, sera, plasma, urine,
sputum, tears, feces, saliva, membrane extracts, and the like.
A target can be any molecular finding that may be obtained from molecular
testing. For
example, a target may include one or more genes or proteins. For example, the
presence of a copy
number variation of a gene can be determined. As shown in FIG. 2, tests for
fmding such targets can
25 include, but are not limited to, NGS, ILIC, fluorescent in-situ
hybridization (FISH), in-situ
hybridization (ISH), and other molecular tests known to those skilled in the
art.
Furthermore, the methods disclosed herein include profiling more than one
target. As a non-
limiting example, the copy number, or presence of a copy number variation
(CNV), of a plurality of
genes can be identified. Furthermore, identification of a plurality of targets
in a sample can be by one
30 method or by various means. For example, the presence of a CN V of a
first gene can be determined
by one method, e.g., NGS, and the presence of a CNV of a second gene
determined by a different
method, e.g., fragment analysis. Alternatively, the same method can be used to
detect the presence of a
CNV in both the tirst and second gene, e.g., using NGS.
The test results can be compiled to determine the individual characteristics
of the cancer.
35 After determining the characteristics of the cancer, a therapeutic
regimen may be identified, e.g.,
comprising treatments of likely benefit as well as treatments of unlikely
benefit.
131
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Finally, a patient profile report may be provided which includes the patient's
test results for
various targets and any proposed therapies based on those results.
The systems as described herein can be used to automate the steps of
identifying a molecular
profile to assess a cancer. In an aspect, the present methods can be used for
generating a report
5 comprising a molecular profile. The methods can comprise: performing
molecular profiling on a
sample from a subject to assess characteristics of a plurality of cancer
biomarkers, and compiling a
report comprising the assessed characteristics into a list, thereby generating
a report that identifies a
molecular profile for the sample. The report can further comprise a list
describing the potential benefit
of the plurality of treatment options based on the assessed characteristics,
thereby identifying
10 candidate treatment options for the subject. The report can also suggest
treatments of potential
unlikely benefit, or indeterminate benefit, based on the assessed
characteristics.
Molecular Profiling for "treatment Selection
The methods as described herein provide a candidate treatment selection for a
subject in need
thereof Molecular profiling can be used to identify one or more candidate
therapeutic agents for an
15 individual suffering from a condition in which one or more of the
biomarkers disclosed herein are
targets for treatment. For example, the method can identify one or more
chemotherapy treatments for
a cancer. In an aspect, the methods provides a method comprising: performing
at least one molecular
profiling technique on at least one biomarker. Any relevant biomarker can be
assessed using one or
more of the molecular profiling techniques described herein or known in the
art. The marker need
20 only have some direct or indirect association with a treatment to be
useful. Any relevant molecular
profiling technique can be performed, such as those disclosed here. These can
include without
limitation, protein and nucleic acid analysis techniques. Protein analysis
techniques include, by way
of non-limiting examples, immunoassays, imrnunohistochemistry, and mass
spectrometry. Nucleic
acid analysis techniques include, by way of non-limiting examples,
amplification, polymerase chain
25 amplification, hybridization, microarrays, in situ hybridization,
sequencing, dye-terminator
sequencing, next generation sequencing, pyrosequencing, and restriction
fragment analysis.
Molecular profiling may comprise the profiling of at least one gene (or gene
product) for each
assay technique that is performed. Different numbers of genes can be assayed
with different
techniques. Any marker disclosed herein that is associated directly or
indirectly with a target
30 therapeutic can be assessed. For example, any "druggable target"
comprising a target that can be
modulated with a therapeutic agent such as a small molecule or binding agent
such as an antibody, is a
candidate for inclusion in the molecular profiling methods as described
herein_ The target can also be
indirectly drug associated, such as a component of a biological pathway that
is affected by the
associated drug. The molecular profiling can be based on either the gene,
e.g., DNA sequence, and/or
35 gene product, e.g., mRNA or protein. Such nucleic acid and/or
polypeptide can be profiled as
applicable as to presence or absence, level or amount, activity, mutation,
sequence, haplotype,
132
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
rearrangement, copy number, or other measurable characteristic. In some
embodiments, a single gene
and/or one or more corresponding gene products is assayed by more than one
molecular profiling
technique. A gene or gene product (also referred to herein as "marker" or
"biomarker"), e.g., an
mRNA or protein, is assessed using applicable techniques (e.g., to assess DNA,
RNA, protein),
5 including without limitation ISII, gene expression, MC, sequencing or
immunoassay. Therefore, any
of the markers disclosed herein can be assayed by a single molecular profiling
technique or by
multiple methods disclosed herein (e.g., a single marker is profiled by one or
more of ILIC, ISH,
sequencing, microarray, etc.). In some embodiments, at least about 1, 2, 3, 4,
5, 6,7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 35,
40,45, 50, 55, 60, 65, 70, 75,
10 80, 85, 90, 95 or at least about 100 genes or gene products are profiled
by at least one technique, a
plurality of techniques, or using any desired combination of TSB, IBC, gene
expression, gene copy,
and sequencing. In some embodiments, at least about 100, 200, 300, 400, 500,
600, 700, 800, 900,
1000, 2000, 3000, 4000, 5000, 6000, 7000, 8000, 9000, 10,000, 11,000, 12,000,
13,000, 14,000,
15,000, 16,000, 17,000, 18,000, 19,000, 20,000, 21,000, 22,000, 23,000,
24,000,25,000, 26,000,
15 27,000, 28,000, 29,000, 30,000, 31,000, 32,000, 33,000, 34,000, 35,000,
36,000,37,000, 38,000,
39,000, 40,000, 41,000, 42,000, 43,000, 44,000,45,000, 46,000, 47,000, 48,000,
49,000, or at least
50,000 genes or gene products are profiled using various techniques. The
number of markers assayed
can depend on the technique used. For example, microarray and massively
parallel sequencing lend
themselves to high throughput analysis. Because molecular profiling queries
molecular characteristics
20 of the tumor itself, this approach provides information on therapies
that might not otherwise be
considered based on the lineage of the tumor.
In some embodiments, a sample from a subject in need thereof is profiled using
methods
which include but are not limited to BIC analysis, gene expression analysis,
LSE analysis, and/or
sequencing analysis (such as by PCR, RT-PCR, pyrosequencing, NGS) for one or
more of the
25 following: ABCC1, ABCG2, ACE2, ADA, ADH1C, ADH4, AGT, AR, AREG, ASNS,
BCL2, BCRP,
BDCA1, beta III tubulin, B1RC5, B-RAF, BRCA1, BRCA2, CA2, caveolin, CD20,
CD25, CD33,
CD52, CDA, CDKN2A, CDKN1A, CDKN113, CDK2, CDW52, CES2, CK 14, CK 17, CK 5/6, c-
KIT, c-Met, c-Myc, COX-2, Cyclin DI, DCK, DHFR, DNMT1, DNMT3A, DNMT3B, E-
Cadherin,
ECGF1, EGFR, EML4-ALK fusion, EPHA2, Epiregulin, ER, ERBR2, ERCC1, ERCC3,
EREG,
30 ESR1, FLI'l , folate receptor, FOLR1, FOLR2, FSHB, FSHPRH1, FSHR, FYN,
GARY, GNAll,
GNAQ, GNRI-11, GNRIIR1, GSTP1, HCK, HDAC1, hENT-1, Her2/Neu, HGF, HIF1A, IRGE
HSP90, HSP9OAA1, HSPCA, IGF-1R, IGFRBP, IGERBP3, IGERBP4, IGFRBP5,1L13RA1,
IL2RA,
KDR, Ki67, KIT, K-RAS, LCK, LIB, Lymphotoxm Beta Receptor, LYN, MET, WWI,
MLH1,
MMR, MRP1, MS4A1, MSII2, MSII5, Myc, NEK131, NEKB2, NEK131A, NRAS, ODC1, OGFR,
35 p16, p21, p27, p53, p95, PARP-1, PDGFC, PDGFR, PDGFRA, PDGFRB, PGP, PGR,
PI3K, POLA,
POLA1, PPARG, PPARGC1, PR, PTEN, PTGS2, PTPN12, RAF1, RARA, ROS1, RRM1, RRM2,
RRM2B, RXRB, RXRG, SIK2, SPARC, SRC, SSTR1, SSTR2, SSTR3, SSTR4, SSTR5,
Survivin,
133
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
TK1, TLE3, TNF, TOP1, TOP2A, TOP2B, TS, TUBB3, TXN, TXNRD I, TYMS, VDR, VEGF,
VEGFA, VEGFC, VIIIL, YES1, ZAP70, a biomarker listed in any one of Tables 2-
116, Tables 117-
120, ISNIVII , Tables 121-130, and any useful combination thereof
As understood by those of skill in the art, genes and proteins have developed
a number of
5 alternative names in the scientific literature. Listing of gene aliases
and descriptions used herein can
be found using a variety of online databases, including GeneCards
(www.genecards.org), HIJGO
Gene Nomenclature (wvvw.genenames.org), Entrez Gene
(wrvw.ncbisihn.nih.goy/entrez/query.fcgi?db=gene), UniProtKB/Swiss-Prot (vvww-
.uniprot.org),
UniProtK13/TrEMBL (www.uniprotorg), OMI1V1
10 (www.ncbi.n1m.nih.goy/entrez/query.fcgi?db=0MEM), GeneLoc
(genecards.weizrnann.ac.d/geneloc/),
and Ensembl (wrywensembl.org). For example, gene symbols and names used herein
can correspond
to those approved by HUGO, and protein names can be those recommended by
UniProtKB/Swiss-
Prot. In the specification, where a protein name indicates a precursor, the
mature protein is also
implied. Throughout the application, gene and protein symbols may be used
interchangeably and the
15 meaning can be derived from context, e.g., 1SH or NGS can be used to
analyze nucleic acids whereas
[RC is used to analyze protein.
The choice of genes and gene products to be assessed to provide molecular
profiles as
described herein can be updated over time as new treatments and new drug
targets are identified. For
example, once the expression or mutation of a biomarker is correlated with a
treatment option, it can
20 be assessed by molecular profiling. One of skill will appreciate that
such molecular profiling is not
limited to those techniques disclosed herein but comprises any methodology
conventional for
assessing nucleic acid or protein levels, sequence information, or both. The
methods as described
herein can also take advantage of any improvements to current methods or new
molecular profiling
techniques developed in the future. In some embodiments, a gene or gene
product is assessed by a
25 single molecular profiling technique. In other embodiments, a gene
and/or gene product is assessed by
multiple molecular profiling techniques. In a non-limiting example, a gene
sequence can be assayed
by one or more of NGS, 1SH and pyrosequencing analysis, the mRNA gene product
can be assayed by
one or more of NGS, RT-PCR and microarray, and the protein gene product can be
assayed by one or
more of RIC and immunoassay. One of skill will appreciate that any combination
of biomarkers and
30 molecular profiling techniques that will benefit disease treatment are
contemplated by the present
methods.
Genes and gene products that are known to play a role in cancer and can be
assayed by any of
the molecular profiling techniques as described harem include without
limitation those listed m any of
International Patent Publications WO/2007/137187 (Intl Appl. No.
PCT/US2007/069286), published
35 November 29, 2007; WO/2010/045318 (Intl Appl. No. PCT/US2009/060630),
published April 22,
2010; WO/2010/093465 (Int'lAppl. No. PCT/US2010/000407), published August 19,
2010;
WO/2012/170715 (InelAppl. No. PCT/US2012/041393), published December 13, 2012;
134
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
WO/2014/089241 (Int'lAppl. No. PCT/US2013/073184), published June 12, 2014;
WO/2011/056688
(Int'lAppl. No. PCT/US2010/054366), published May 12, 2011; WO/2012/092336
(Tnt'lAppl. No.
PCTAIS2011/067527), published July 5, 2012; WO/2015/116868 (Tnt'lAppl. No.
PCT/US2015/013618), published August 6, 2015; WO/2017/053915 (hit'! App!. No.
5 PCT/US2016/053614), published March 30, 2017; WO/2016/141169 (Int'lAppl.
No.
PCTAI52016/020657), published September 9, 2016; and W02018175501 (Intl Appl.
No.
PCT/US2018/023438), published September 27, 2018; each of which publications
is incorporated by
reference herein in its entirety.
Mutation profiling can be determined by sequencing, including Sanger
sequencing, array
10 sequencing, pyrosequencing, high-throughput or next generation (NGS,
NextGen) sequencing, etc.
Sequence analysis may reveal that genes harbor activating mutations so that
drugs that inhibit activity
are indicated for treatment. Alternately, sequence analysis may reveal that
genes harbor mutations that
inhibit or eliminate activity, thereby indicating treatment for compensating
therapies. In some
embodiments, sequence analysis comprises that of exon 9 and 11 of c-KIT.
Sequencing may also be
15 performed on EGFR-kinase domain exons 18, 19,20, and 2L Mutations,
amplifications or
misregulations of EGER or its family members are implicated in about 30% of
all epithelial cancers.
Sequencing can also be performed on PI3K, encoded by the PIK3CA gene. This
gene is a found
mutated in many cancers. Sequencing analysis can also comprise assessing
mutations in one or more
ABCC1, ABCG2, ADA, AR, ASNS, BCL2, BIRC5, BRCA1, BRCA2, CD33, CD52, CDA, CES2,
20 DCK, DILFR, DNA4T1, DNA4T3A, DNMT3B, ECGF1, EGFR, EPHA2, ERBB2, ERCC1,
ERCC3,
ESRI, FLTI, FOLR2, FYN, GART, GNRHI, GSTP I, HCK, HDAC I, H1F IA, HSP9OAA1,
IGEBP3,
IGEBP4, IGEBP5, EL2RA, KDR, KIT, LCK, LYN, MET, MGMT, MLH1, MS4A1, MSH2,
NEKB1,
NEK132, NEK131A, NRAS, OGFR, PARP1, PDGFC, PDGFRA, PDGER13, PGP, PGR, POLA1,
PTEN, PTGS2, PTPN12, RAF1, RARA, RRIVI1, RRIVI2, RRM2B, RXRB, RXRG, SIK2,
SPARC,
25 SRC, SSTR1, SSTR2, SSTR3, SSTR4, SSTR5, TK1, TNF, TOP1, TOP2A, TOP2B,
TXNRD1,
TYMS, VDR, VEGFA, VIAL, YES1, and ZAP70. One or more of the following genes
can also be
assessed by sequence analysis: ALK, EML4, hENT- I, IGE-1R, HSP9OAA1, MMR, p16,
p21, p27,
PARP-1, PI3K and TLE3. The genes and/or gene products used for mutation or
sequence analysis can
be at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25, 30, 40, 50, 60, 70, 80,
90, 100, 200, 300, 400, 500 or
30 all of the genes and/or gene products listed many of Tables 4-12 of
W02018175501, e.g., many of
Tables 5-10 of W02018175501, or in any of Tables 7-10 of W02018175501.
In embodiments, the methods as described herein are used detect gene fusions,
such as those
listed in any of International Patent Publications WO/2007/137187 (Int'lAppl.
No.
PCT/1352007/069286), published November 29, 2007; WO/2010/045318 (Int'lAppl.
No.
35 PCT/US2009/060630), published April 22, 2010; WO/2010/093465 (Intl Appl.
No.
PCT/US2010/000407), published August 19, 2010; WO/2012/170715 (hit'! App!. No.
PCT/US2012/041393), published December 13, 2012; WO/2014/089241 ant'lAppl. No.
135
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
PCT/US2013/073184), published June 12, 2014; WO/2011/056688 (Inel App!. No.
PCT/US2010/054366), published May 12, 2011; WO/2012/092336 (Inel App!. No.
PCT/I1S2011/067527), published July 5, 2012; W0/2015/116868 (Tnt'lAppl. No.
PCT/US2015/013618), published August 6, 2015; WO/2017/053915 (InelAppl. No.
5 PCT/US2016/053614), published March 30, 2017; WO/2016/141169 (Int'lAppl.
No.
PCT/IT52016/020657), published September 9,2016; and W0/2018/175501 (In t'l
Appl. No.
PCT/US2018/023438), published September 27, 2018; each of which publications
is incorporated by
reference herein in its entirety. A fusion gene is a hybrid gene created by
the juxtaposition of two
previously separate genes. This can occur by chromosomal translocation or
inversion, deletion or via
10 trans-splicing. The resulting fusion gene can cause abnormal temporal
and spatial expression of genes,
leading to abnormal expression of cell growth factors, angiogenesis factors,
tumor promoters or other
factors contributing to the neoplastic transformation of the cell and the
creation of a tumor. For
example, such fusion genes can be oncogenic due to the juxtaposition of: 1) a
strong promoter region
of one gene next to the coding region of a cell growth factor, tumor promoter
or other gene promoting
15 oncogenesis leading to elevated gene expression, or 2) due to the fusion
of coding regions of two
different genes, giving rise to a chimeric gene and thus a chimeric protein
with abnormal activity.
Fusion genes are characteristic of many cancers. Once a therapeutic
intervention is associated with a
fusion, the presence of that fusion in any type of cancer identifies the
therapeutic intervention as a
candidate therapy for treating the cancer.
20 The presence of fusion genes can be used to guide therapeutic selection.
For example, the
BCR-ABL gene fusion is a characteristic molecular aberration in ¨90% of
chronic myelogenous
leukemia (CML) and in a subset of acute leukemias (Kurzrock et al., Annals of
Internal Medicine
2003; 138:819-830). The BCR-ABL results from a translocation between
chromosomes 9 and 22,
commonly referred to as the Philadelphia chromosome or Philadelphia
translocation. The
25 translocation brings together the 5' region of the BCR gene and the 3'
region of ABL1, generating a
chimeric BCR-ABL1 gene, which encodes a protein with constitutively active
tyrosine kinase activity
(Mittleman et al., Nature Reviews Cancer 2007; 7:233-245). The aberrant
tyrosine kinase activity
leads to de-regulated cell signaling, cell growth and cell survival, apoptosis
resistance and growth
factor independence, all of which contribute to the pathophysiology of
leukemia (Kurzrock et al.,
30 Annals of Internal Medicine 2003; 138:819-830). Patients with the
Philadelphia chromosome are
treated with imatinib and other targeted therapies. Imatinib binds to the site
of the constitutive tyrosine
kinase activity of the fusion protein and prevents its activity. Imatinib
treatment has led to molecular
responses (disappearance of BCR-ABL+ blood cells) and improved progression-
free survival intICR-
ABL-E CML patients (Kantarjian et al., Clinical Cancer Research 2007; 13:1089-
1097).
35 Another fusion gene, IGH-MYC, is a defining feature of ¨80% of Burkitt's
lymphoma (Ferry
et al. Oncologist 2006; 11:375-83). The causal event for this is a
translocation between chromosomes
8 and 14, bringing the c-Myc oncogene adjacent to the strong promoter of the
immunoglobulin heavy
136
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
chain gene, causing c-myc overexpression (Mittleman et al., Nature Reviews
Cancer 2007; 7:233-
245). The c-myc rearrangement is a pivotal event in lymphomagenesis as it
results in a perpetually
proliferative state. It has wide ranging effects on progression through the
cell cycle, cellular
differentiation, apoptosis, and cell adhesion (Ferry et al. Oncologist 2006;
11:375-83).
5 A number of recurrent fusion genes have been catalogued in the Mittleman
database
(cgap.nci.nih.gov/ChromosomesiMitelman). The gene fusions can be used to
characterize neoplasms
and cancers and guide therapy using the subject methods described herein. For
example, TMPRSS2-
ERG, 114PRSS2-ETV and SLC45A3-ELK4 fusions can be detected to characterize
prostate cancer;
and ETV6-NTRK3 and ODZ4-NRG1 can be used to characterize breast cancer. The
EML4-ALK,
10 RLF-MYCL1, TGF-ALK, or CD74-ROS1 fusions can be used to characterize a
lung cancer. The
ACST3-ETV1, C150RF21-ETV1, FT135294-ETV1, 1-IF,RV-ETV1, TMPRSS2-ERG, TMPRSS2-
ETV1/4/5, TMPRSS2-ETV4/5, SLC5A3-ERG, SLC5A3-ETV1, SLC5A3-ETV5 or KLK2-ETV4
fusions can be used to characterize a prostate cancer. The GOPC-ROS1 fusion
can be used to
characterize a brain cancer. The CHCHD7-PLAG1, CTNNB1-PLAG1, FHIT-TIMGA2,
TEMGA2-
15 NFEB, LIFR-PLAGL or TCEAL-PLAGI fusions can be used to characterize a
head and neck cancer_
The ALPHA-TFEB, NONO-TFE3, PRCC-TFE3, SFPQ-TFE3, CLTC-TFE3, or MALAT1-TFEB
fusions can be used to characterize a renal cell carcinoma (RCC). The AKAP9-
BRAF, CCDC6-RET,
ERC1-RETM, GOLGA5-RET, HOOK3-RET, URH4-RET, KTN1-RET, NCOA4-RET, PCM1-RET,
PRKARA1A-RET, RFG-RET, REG9-RET, Ria-RET, TGF-NTRK1, TPM3-NTRK1, TPM3-TPR,
20 TPR-MET, TPR-NTRK1, TRIM24-RET, TRIM27-RET or TREM33-RET fusions can be
used to
characterize a thyroid cancer and/or papillary thyroid carcinoma; and the PAX8-
PPARy fusion can be
analyzed to characterize a follicular thyroid cancer. Fusions that are
associated with hematological
malignancies include without limitation TTL-ETV6, CDK6-MLL, CDK6-TLX3, ETV6-
FLT3, ETV6-
RUNX1, ETV6-TTL, MLL-AFF1, MiLL-AFF3, MLL-AFF4, MLL-GAS7, TCBA1-ETV6, TCF3-
25 PBX1 or TCF3-TEPT, which are characteristic of acute lymphocytic
leukemia (ALL); BCL11B-
TLX3, EL2-TNFRFS17, NUP214-ABL1, NUP98-CCDC28A, TALL-STET, or ETV6-ABL2, which
are characteristic of 'T-cell acute lymphocytic leukemia (T-ALL); Al'IC-ALK,
KLAA1618-ALK,
MSN-ALK, MYH9-ALK, NPM1-ALK, TGF-ALK or TPM3-ALK, which are characteristic of
anaplastic large cell lymphoma (ALCL); BCR-ABL1, BCR-JAK2, ETV6-EVI1, ETV6-
MIN1 or
30 E1V6-1C13A1, characteristic of chronic myelogenous leukemia (CML);
C131413-MYH11, CH1C2-
ETV6, ETV6-ABL1, ETV6-ABL2, ETV6-ARNT, ETV6-CDX2, ETV6-13LXB9, ETV6-PER1,
MEF2D-DAZAP1, AML-AFF1, MLL-ARHGAP26, MLL-ARHGEF12, MLL-CASC5, MILL-
Cl3L,MLL-CRE1313P, MLL-DAB21P, MLL-ELL, MLL-EP300, MLL-EPS15, MLL-INBP1, MLL-
FOX03A, MLL-GMPS, MLL-GPIIN, MLL-MLLT1, MLL-MLLT11, MLL-MLLT3, MLL-MLLT6,
35 MLL-MYOLF, MLL-PICALM, MLL-SEPT2, MLL-SEPT6, MLL-SORBS2, MYST3-SORBS2,
MYST-CREBBP, NPM1-MLF1, NUP98-HOXA13, PRDM16-EVI1, RABEP1-PDGFR13, RUNX1-
EVI1, RUNX1-MD S1 , RUNX1-RPL22, RUNX1-RUNX1T1, RUNX1-SII3D19, RUNX1-USP42,
137
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
RUNX1-YTHDF2, RUNX1-ZNF687, or TAF15-ZNF-384, which are characteristic of
acute myeloid
leukemia (AML); CCND1-FSTL3, which is characteristic of chronic lymphocytic
leukemia (CLL);
LICT3-MYC, MYCJFITG1, LICT,7A-MYC, BRWD3-ARTIGAP20 or LITG1-MYC, which are
characteristic of B-cell chronic lymphocytic leukemia (B-CLL); CITTA-BCL6,
CLTC-ALK, EL21R-
BCL6, P1M1-BCL6, TFCR-BCL6, IKZF1-BCL6 or SEC31A-ALK, which are characteristic
of
diffuse large B-cell lymphomas (DTBCL); FLIP1-PDGFRA, FIT3-ETV6, KTAA1509-
PDGFRA,
PDE4DEP-PDGFRB, NThi-PDGFRB, TP53BP1-PDGFRB, or TPM3-PDGFRB, which are
characteristic of hyper eosinophilia / chronic eosinophilia; and IGH-MYC or
LCP1-BCL6, which are
characteristic of Burkitt's lymphoma. One of skill will understand that
additional fusions, including
those yet to be identified to date, can be used to guide treatment once their
presence is associated with
a therapeutic intervention.
The fusion genes and gene products can be detected using one or more
techniques described
herein. In some embodiments, the sequence of the gene or corresponding mRNA is
determined, e.g.,
using Sanger sequencing, NGS, pyrosequencing, DNA microarrays, etc.
Chromosomal abnormalities
can be assessed using ISH, NGS or PCR techniques, among others. For example, a
break apart probe
can be used for ISH detection of ALK fusions such as EML4-ALK, KIF5B-ALK
and/or TFG-ALK. As
an alternate, PCR can be used to amplify the fusion product, wherein
amplification or lack thereof
indicates the presence or absence of the fusion, respectively. mRNA can be
sequenced, e.g., using
NGS to detect such fusions. See, e.g., Table 9 or Table 12 of W02018175501 or
Tables 126-127
herein. In some embodiments, the fusion protein fusion is detected.
Appropriate methods for protein
analysis include without limitation mass spectroscopy, electrophoresis (e.g.,
2D gel electrophoresis or
SDS-PAGE) or antibody related techniques, including immunoassay, protein array
or
iminunohistochemistry. The techniques can be combined. As a non-limiting
example, indication of an
ALK fusion by NGS can be confirmed by ISH or ALK expression using 111C, or
vice versa.
Molecular Profiling Targets for Treatment Selection
The systems and methods described herein allow identification of one or more
therapeutic
regimes with projected therapeutic efficacy, based on the molecular profiling.
Illustrative schemes for
using molecular profiling to identify a treatment regime are provided
throughout. Additional schemes
are described in International Patent Publications WO/2007/137187 (Intl Appl.
No.
PCT/US2007/069286), published November 29, 2007; WO/2010/045318 (InElAppl. No.
PCT/US2009/060630), published April 22, 2010; WO/2010/093465 (Intl Appl. No.
PCT/T TS2010/000407), published August 19, 2010; WO/2012/170711 (Tnt'l Appl No
PCT/US2012/041393), published December 13, 2012; WO/2014/089241 (InelAppl. No.
PCT/US2013/073184), published June 12, 2014; WO/2011/056688 (Mel Appl. No.
PCT/US2010/054366), published May 12, 2011; WO/2012/092336 (Inel Appl. No.
PCT/US2011/067527), published July 5, 2012; WO/2015/116868 (InflAppl. No.
138
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
PCT/US2015/013618), published August 6, 2015; WO/2017/053915 (InClAppl. No.
PCT/US2016/053614), published March 30, 2017; WO/2016/141169 (Int'lAppl. No.
PCT/1152016/020657), published September 9, 2016; and W02018175501 (Intl Appl.
No.
PCT/US2018/023438), published September 27, 2018; each of which publications
is incorporated by
5 reference herein in its entirety.
The methods described herein comprise use or molecular profiling results to
suggest
associations with treatment benefit. In some embodiments, rules are used to
provide the suggested
chemotherapy treatments based on the molecular profiling test results. Rules
can be constructed in a
format such as "if biomarker positive then treatment option one, else
treatment option two," or
10 variations thereof. Treatment options comprise treatment with a single
therapy (e.g., 5-FU) or
treatment with a combination regimen (e.g., FOT,FOX or FOT,FIRI regimens for
colorectal cancer). In
some embodiments, more complex rules are constructed that involve the
interaction of two or more
biomarkers. Finally, a report can be generated that describes the association
of the predicted benefit of
a treatment and the biomarker and optionally a sununary statement of the best
evidence supporting the
15 treatments selected. Ultimately, the treating physician will decide on
the best course of treatment. The
report may also list treatments with predicted lack of benefit. See, e.g.,
Examples 4-5.
The selection of a candidate treatment for an individual can be based on
molecular profiling
results from any one or more of the methods described.
In some embodiments, molecular profiling assays arc performed to determine
whether is copy
20 number or copy number variation (CNV; also copy number alteration, CNA)
of one or more genes is
present in a sample as compared to a control, e.g., diploid level. The CNV of
the gene or genes can be
used to select a regimen that is predicted to be of benefit or lack of benefit
for treating the patient. The
methods can also include detection of mutations, indels, fusions, and the like
in other genes and/or
gene products, e.g., as described in Example 1 herein, and International
Patent Publications
25 WO/2007/137187 (Intl App!. No. PCT/US2007/069286), published November
29, 2007;
WO/2010/045318 (Inel App!. No. PCT/US2009/060630), published April 22, 2010;
WO/2010/093465 (Intl App!. No. PCT/US2010/000407), published August 19, 2010;
WO/2012/170715 (InelAppl. No. PCT/US2012/041393), published December 13, 2012;
WO/2014/089241 (InelAppl. No. PCT/US2013/073184), published June 12, 2014;
WO/2011/056688
30 (Int'lAppl. No. PC1/1JS2010/054366), published May 12, 2011;
WO/2012/092336 (1nt'lAppl. No.
PCT/US2011/067527), published July 5, 2012; WO/2015/116868 (Int'lAppl. No.
PCT/U52015/013618), published August 6, 2015; WO/2017/053915 (InelAppl. No.
PC1/1JS2016/053614), published March 30, 2017; WO/2016/141169 (InelAppl. No.
PCT/US2016/020657), published September 9, 2016; and W02018175501 (Inel App!.
No.
35 PCT/US2018/023438), published September 27, 2018; each of which
publications is incorporated by
reference herein in its entirety.
139
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
The methods described herein are intended to prolong survival of a subject
with cancer by
providing personalized treatment. In some embodiments, the subject has been
previously treated with
one or more therapeutic agents to treat the cancer. The cancer may be
refractory to one of these
agents, e.g., by acquiring drug resistance mutations. In some embodiments,
there is no known
5 standard of care agent for the cancer or the cancer may be resistant to
all known standard of care
agent. Such standard of care agents may include "on label" agents, or those
with an indication in a
drug label. In some embodiments, the cancer is metastatic. In some
embodiments, the subject has not
previously been treated with one or more therapeutic agents identified by the
method. Using
molecular profiling, candidate treatments can be selected regardless of the
stage, progression,
10 anatomical location, or anatomical origin of the cancer cells.
The present disclosure provides methods and systems for analyzing diseased
tissue using
molecular profiling as previously described above. Because the methods rely on
analysis of the
characteristics of the tumor under analysis, the methods can be applied in for
any tumor or any stage
of disease, such an advanced stage of disease or a metastatic tumor of unknown
origin. As described
15 herein, a tumor or cancer sample is analyzed for one or more biomarkers
in order to predict or identify
a candidate therapeutic treatment.
The present methods can be used for selecting a treatment of primary or
metastatic cancer.
The biomarker patterns and/or biomarker signature sets can comprise
pluralities of
biomarkers. In yet other embodiments, the biomarker patterns or signature sets
can comprise at least
20 6, 7, 8, 9, or 10 biomarkers. In some embodiments, the biomarker
signature sets or biomarker patterns
can comprise at least 15, 20, 30,40, 50, or 60 biomarkers. In some
embodiments, the biomarker
signature sets or biomarker patterns can comprise at least 70, 80, 90, 100, or
200, biomarkers. In some
embodiments, the biomarker signature sets or biomarker patterns can comprise
at least 100, 200, 300,
400, 500, 600, 700, or at least 800 biomarkers. In some embodiments, the
biomarker signature sets or
25 biomarker patterns can comprise at least 1000, 2000, 3000, 4000, 5000,
6000, 7000, 8000, 9000,
10,000, 20,000, or at least 30,000 biomarkers. For example, the biomarkers may
comprise whole
exome sequencing and/or whole transeriptome sequencing and thus comprise all
genes and gene
products. Analysis of the one or more biomarkers can be by one or more
methods, e.g., as described
herein. See, e.g., Example 1.
30 As described herein, the molecular profiling of one or more targets can
be used to determine
or identify a therapeutic for an individual. For example, the presence, level
or state of one or more
biomarkers can be used to determine or identify a therapeutic for an
individual. The one or more
biomarkers, such as those disclosed herein, can be used to form a biomarker
pattern or biomarker
signature set, which is used to identify a therapeutic for an individual. In
some embodiments, the
35 therapeutic identified is one that the individual has not previously
been treated with. For example, a
reference biomarker pattern has been established for a particular therapeutic,
such that individuals
with the reference biomarker pattern will be responsive to that therapeutic.
An individual with a
140
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
biomarker pattern that differs from the reference, for example the expression
of a gene in the
biomarker pattern is changed or different from that of the reference, would
not be administered that
therapeutic. In another example, an individual exhibiting a biomarker pattern
that is the same or
substantially the same as the reference is advised to be treated with that
therapeutic. In some
5 embodiments, the individual has not previously been treated with that
therapeutic and thus a new
therapeutic has been identified for the individual. The biomarker pattern may
be based on a single
biomarker (e.g., expression of HER2 suggests treatment with anti-HER2 therapy)
or multiple
biomarkers.
The genes used for molecular profiling, e.g., by IFIC, ISH, sequencing (e.g.,
NGS), and/or
10 PCR (e.g., qPCR), can be selected from those listed in Example 1 herein,
or as described in
W02015175501, e.g., in Tables 5-10 therein. Assessing one or more biomarkers
disclosed herein can
be used for characterizing a cancer.
A cancer in a subject can be characterized by obtaining a biological sample
from a subject and
analyzing one or more biomarkers from the sample. For example, characterizing
a cancer for a subject
15 or individual can include identifying appropriate treatments or
treatment efficacy for specific diseases,
conditions, disease stages and condition stages, predictions and likelihood
analysis of disease
progression, particularly disease recurrence, metastatic spread or disease
relapse. The products and
processes described herein allow assessment of a subject on an individual
basis, which can provide
benefits of more efficient and economical decisions in treatment.
20 In an aspect, characterizing a cancer includes predicting whether a
subject is likely to benefit
from a treatment for the cancer. Biomarkers can be analyzed in the subject and
compared to biomarker
profiles of previous subjects that were known to benefit or not from a
treatment. If the biomarker
profile in a subject more closely aligns with that of previous subjects that
were known to benefit from
the treatment, the subject can be characterized, or predicted, as one who
benefits from the treatment.
25 Similarly, if the biomarker profile in the subject more closely aligns
with that of previous subjects that
did not benefit from the treatment, the subject can be characterized, or
predicted as one who does not
benefit from the treatment. 'Me sample used for characterizing a cancer can be
any useful sample,
including without limitation those disclosed herein.
The methods can further include administering the selected treatment to the
subject.
30 'Me treatment can be any beneficial treatment, e.g., small molecule
drugs or biologics.
Various inununotherapies, e.g., checkpoint inhibitor therapies such as
ipilitnumab, nivolumab,
pembrolizumab, atezolizurnab, avelumab, and durvalumab, are FDA approved and
others are in
clinical trials or developmental stages.
35 Genomic Prevalence Score (GPS)
The present disclosure provides systems, methods, and computer programs for
determining
attributes (phenotypes) of a biological sample, including without limitation a
tissue of origin (TOO).
141
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
The present disclosure can determine such attribute for a biological sample in
a number of different
ways. For example, in some implementations, a first type of analysis can be
performed on a biological
sample to generate attributes of the DNA of the biological sample and then a
trained model can be
used to predict an attribute of the biological sample based on the assessment
of the sample's DNA. In
5 some embodiments, the model comprises a dynamic voting engine such as
provided herein. By way of
another example, a second type of analysis can he performed on a biological
sample to generate
attributes of the RNA of the biological sample and then a trained model can be
used to predict the
attributes for the biological sample based on the assessment of the sample's
RNA. In some
embodiments, the model may also comprise a dynamic voting engine such as
provided herein. In
10 other implementations, the first type of analysis and the second type of
analysis can be performed in
order to generate first biological data based on the biological sample's DNA
and second biological
data based on the biological sample's RNA and then use the trained model to
predict an attribute for
the biological sample based on the first biological data and the second
biological data. In some
embodiments, the model may also comprise a dynamic voting engine such as
provided herein. In
15 some implementations, the biological sample may be a cancer sample,
e.g., tumor sample or bodily
fluid comprising shed tumor cells or nucleic acids, and the attributed tissue
of origin may be the origin
where the tumor originated.
There are many technical advantages that are achieved through use of the
systems, methods,
and computer programs of the present disclosure. By way of example, the
present disclosure provides
20 a machine learning model in the form of a dynamic voting engine that can
more accurately classify
data a biological sample relative to conventional analyses. In some
implementations, such accuracy
increases can be achieved by training the machine learning model to
dynamically vote a plurality of
initial input tissue classifications and then select a target or fmal tissue
classification indicative of an
attribute (phenotype) tissue of origin for the biological sample such as the
tissue of origin. The
25 training processes employed to achieve such increases in accuracy are
described in more detail herein.
The first step in treating cancer is diagnosis. Diagnosis may include physical
exam (e.g., to
detect an enlarged origin or suspicious skin lesion or discoloration),
laboratory testing (e.g., urine or
blood tests), medical imaging (e.g., computerized tomography (CT), bone scans,
magnetic resonance
imaging (MRI), positron emission tomography (PET), ultrasound and/or X-ray),
and biopsy, which
30 may be the preferred means to provide a definitive diagnosis. However, 3-
9% of cases are
misdiagnosed. See, e.g., Peck, M. et al, Review of diagnostic error in
anatomical pathology and the
role and value of second opinions in error prevention. J Clin Pathol, 2018,
71: p. 995-1000, which
reference is incorporated herein m its entirety. In addition, 5-10% of a
Cancer of Occult/Unknown
Primary (CUP). See vvww.mdanderson.org/cancer-types/cancer-of-unknown-
prirnary.htmL
35 www.cancer.govitypes/unknown-primary/hp/unknown-prirnary-treatment-pdq#
1. Thus there is a
need for improved methods of determining and/or verifying the tissue of origin
(TOO) of a substantial
142
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
number of cancers. Automated verification of TOO may also identify laboratory
errors in rare cases
(e.g., switched samples).
The diagnosis of a malignancy is typically informed by clinical presentation
and tumor tissue
features including cell morphology, irnmunohistochemistry, cytogenetics, and
molecular markers.
5 Lack of reliable classification of a tumor poses a significant treatment
dilemma for the oncologist
leading to inappropriate and/or delayed treatment. Gene expression profiling
has been used to try to
identify the tumor type for CUP patients, but suffers from a number of
inherent limitations.
Specifically, tumor percentage, variation in expression, and the dynamic
nature of RNA all contribute
to suboptimal performance. For example, one commercial RNA-based assay has
sensitivity of 83% in
10 a test set of 187 tumors and confirmed results on only 78% of a separate
300 sample validation set.
See Erlander MG, et al. Performance and clinical evaluation of the 92-gene
real-time PCR assay for
tumor classification. J Mol Diagn. 2011 Sep;13(5):493-503; which reference is
incorporated herein by
reference in its entirety. Moreover, the diagnosis for any cancer may be
mistaken in some cases.
Herein we provide systems and methods to predict attributes (phenotypes) of a
biological
15 sample, including primary location, histology, disease/cancer, and/or
organ group. The granularity of
the attribute can be chosen at a desired level such as described herein. We
used molecular profiling
(see, e.g., Example 1; FIGs. 2B-C) and machine learning to construct models
and biosignatures for
predicting such attributes. As a non-limiting example, such information can be
used to identify the
primary tumor site of a metastatic cancer of unknown primary (CUPS). In some
embodiments, the
20 predictions can be used to assist in planning treatment of cancer
patients. In some embodiments, such
information is used to verify the original diagnosis of a cancer at the same
time molecular profiling is
used to identify treatment options. If the information differs from the
original diagnosis, additional
inquiry may be performed (e.g., pathologist review) to verify the diagnosis
and thus benefit patient
treatment.
25 A general approach is as follows. First, we obtain a sample comprising
cells from a cancer in
a subject, e.g., a tumor sample or bodily fluid sample such as described
herein. In some embodiments,
the sample comprises metastatic cells. We perform molecular profiling assays
on the sample to assess
one or more biomarkers and thereby obtain a molecular profile, or
biosignature, for the sample. See,
e.g., Example 1. The sample biosignature can be input into a statistical model
such as described
30 herein. In some embodiments, this comprises comparing the sample
biosignature to a number of
biosignatures indicative of a plurality of attributes of interest. As a non-
limiting example, one may
compare the sample biosignature to each of a plurality of pre-determined
biosignatures indicative of
various attributes, e.g., various primary tumor origins. A probability or
similar metric can be
calculated that the sample biosignature corresponds to each of the pre-
determined biosignatures. In
35 some embodiments, the sample biosignature is used as an input into one
or more machine learning
models that are trained to take part in the overall prediction of the
attribute's of interest. Such models
may calculate the probability or similarity metric described above. In some
embodiments, one may
143
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
assign the attribute with the highest confidence, e.g., the highest
probability. A threshold may be set
such that the strength of assignment is determined.
The statistical models, e.g., machine learning models, are trained to the
different attributes of
interest. Herein, we demonstrate our approach using next-generation sequencing
results for thousands
5 of patient tumor samples. See, e.g., Examples 2-3. As a non-limiting
example, consider that such data
is used to identify a pre-determined biosignature for each of a plurality of
tumor lineages,such as
prostate, bladder, endocervix, peritoneum, stomach, esophagus, ovary, parietal
lobe, cervix,
endometrium, liver, sigmoid colon, upper-outer quadrant of breast, uterus,
pancreas, head of pancreas,
rectum, colon, breast, intrahepatic bile duct, cecum, gastroesophageal
junction, frontal lobe, kidney,
10 tail of pancreas, ascending colon, descending colon, gallbladder,
appendix, rectosigmoid colon,
fallopian tube, brain, lung, temporal lobe, lower third of esophagus, upper-
inner quadrant of breast,
transverse colon, and skin. The biosignatures and models for each of the
lineage predictors can
comprise any number of features, here biomarkers, to achieve the desired level
of performance. As
will be understood by those of skill in the art, multiple features may provide
a more robust prediction,
15 but too many may lead to overfitting. Such parameters can be optimized
in the training and testing
phases of model development. As an non-limiting example, a biosignature for
prostate may comprise
DNA copy number for one or more of the genes FOXAL PTEN, KLK2, GATA2, LCP1,
ETV6,
ERCC3, FANCA, MLLT3, MLH1, NCOA4, NCOA2, CCDC6, PTCH1, FOX01, and IRF4.
FIGs. 3A and 3B provide examples of the classification of individual tumor
samples of
20 known origin as test cases. FIG. 3A shows the prediction of a prostate
cancer sample, correctly
classified as of prostatic origin with high confidence as indicated by the
tight shaded area. FIG. 3B
shows the prediction of a tumor with a primary site as unknown but lineage as
pancreatic. The
predictor correctly identified the tumor as a pancreatic tumor although the
site within the pancreas
was indeterminate as indicated by the shaded region covering "Pancreas," "Head
of pancreas," and
25 "Tail of pancreas."
Provided herein is a method comprising obtaining a biological sample
comprising cells from a
cancer in a subject; performing an assay to assess one or more biomarkers in
the sample to obtain a
biosignature (also referred to as a molecular profile) for the sample; using
the biosignature for the
sample as an input into at least one statistical model, wherein the one or
more statistical model may
30 comprise at least one pre-determined biosignature; and (d) classifying
or predicting an attribute of the
sample based on the comparison, wherein the attribute comprises a primary
origin, an organ type, a
histology, and disease/cancer type, or any useful combination thereof
Similarly, provided herein is a
method comprising: (a) obtaining a biological sample comprising cells from a
subject; (b) performing
an assay to assess one or more biomarkers in the sample to obtain a
biosignature for the sample; (c)
35 generating an input data based on the obtained sample and the one or
more biomarkers; (d) providing
the input data to a machine learning model that has been trained to predict an
attribute of the sample
using the input data, wherein the attribute is selected from the group
consisting of a primary tumor
144
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
origin, cancer/disease type, organ group, histology, and any combination
thereof (e) obtaining output
data generated by the machine learning model based on the machine learning
models processing of
the input data; and (f) classifying the attribute of the sample based on the
output data.
In some embodiments, the model is configured to perform pairwise analysis
between the
5 sample's biosignature and each of multiple different pre-determined (or
trained) biosignatures,
wherein each of the multiple different pre-determined biosignatures
corresponds to a different
attribute. See Examples 2-3, wherein performing pairwise analysis includes the
machine learning
model determining a level of similarity between the input data and
biosignature for one or more of a
plurality of disease types.
10 The desired attributes to be predicted may be determined at varying
levels of specificity. For
example, a tumor origin may he determined as a primary tumor location and a
histology, which may
be combined. For example, primary origin of a sample determined to be prostate
and histology
determined to be adenocarcinoma may combined as prostate adenocareinoma. The
models employed
herein can be trained to such different specificities as desired. For example,
a predictor model may be
15 trained to recognize samples of prostatic origin, or may be trained to
recognize prostate
adenocarcinoma. In some embodiments, multiple models are trained at different
attributes, e.g., organ
or histology, and the results are combined to predict the desired level of
attribute. As desired, the
predictor models may be trained at a highly granular level, and the output can
be identified in a less
granular category of interest. See, e.g., more granular disease types and less
granular organ groups in
20 Tables 2-116 below. In some embodiments, the predictor models are
trained at such less granular
level. In some embodiments, the predictor models are trained to different
attributes (e.g., organ versus
histology) which are then combined to provide the final predicted attribute.
In some embodiments, the systems and methods incorporate analysis of genomic
DNA.
Genomic abnormalities are a hallmark of cancer tissue. For example, 1p19q is
indicative of certain
25 cancers such as oligodendriogliomas. A single chromosome loss of 17 is
the most frequent early
occurrence in ovarian cancer, and 3p deletion in clear cell kidney and trisomy
7 and 17 in papillary
renal cancer are established predictors. Chromosome 6 loss, 8 gain is a marker
of eye cancers. Her2
amplification is observed in breast cancer. We hypothesized that the phenomena
of genomic
abnormalities such as gene copy number and mutational signatures may be
predictive of many, if not
30 all, types of cancers. DNA has certain advantages as an analyte
biomarker as it can be robust to tumor
percentage, metastasis, and sequencing depth, and can be analyzed efficiently
using next-generation
sequencing approaches. See, e.g., Example 1. In an aspect, we used the systems
and methods
provided herein to determine features of genomic DNA that are part of pre-
deterrnmed biosignatures
for 115 different granular disease/cancer types, including adrenal cortical
carcinoma; anus squamous
35 carcinoma; appendix adenocarcinoma, NOS; appendix mucinous
adenocarcinoma; bile duct, NOS,
cholangiocarcinoma; brain astrocytoma, anaplastic; brain astrocytoma, NOS;
breast adenocarcinoma,
NOS; breast carcinoma, NOS; breast infiltrating duct adenocarcinoma; breast
infiltrating lobular
145
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
carcinoma, NOS; breast metaplastic carcinoma, NOS; cervix adenocarcinoma, NOS;
cervix
carcinoma, NOS; cervix squamous carcinoma; colon adenocarcinoma, NOS; colon
carcinoma, NOS;
colon mucinous adenocarcinoma; conjunctiva malignant melanoma, NOS; duodenum
and ampulla
adenocarcinoma, NOS; endometrial adenocarcinoma, NOS; endometrial
carcinosarcoma; endometrial
5 endometrioid adenocarcinoma; endometrial serous carcinoma; endometrium
carcinoma, NOS;
endometrium carcinoma, undifferentiated; endometrium clear cell carcinoma;
esophagus
adenocarcinoma, NOS; esophagus carcinoma, NOS; esophagus squamous carcinoma;
extrahepatic
cholangio, common bile, gallbladder adenocarcinoma, NOS; fallopian tube
adenocarcinoma, NOS;
fallopian tube carcinoma, NOS; fallopian tube carcinosarcoma, NOS; fallopian
tube serous
10 carcinoma; gastric adenocarcinoma; gastroesophageal junction
adenocarcinoma, NOS; glioblastoma;
glionia, NOS; gliosarcoma; head, face or neck, NOS squamons carcinoma;
intrahepatic bile duct
cholangiocarcinoma; kidney carcinoma, NOS; kidney clear cell carcinoma; kidney
papillary renal cell
carcinoma; kidney renal cell carcinoma, NOS; larynx, NOS squamous carcinoma;
left colon
adenocarcinoma, NOS; left colon mucinous adenocarcinoma; liver hepatocellular
carcinoma, NOS;
15 lung adenocarcinoma, NOS; lung adenosquamous carcinoma; lung carcinoma,
NOS; lung mucinous
adenocarcinoma; lung neuroendocrine carcinoma, NOS; lung non-small cell
carcinoma; lung
sarcomatoid carcinoma; lung small cell carcinoma, NOS; lung squamous
carcinoma; meninges
meningioma, NOS; nasopharynx, NOS squamous carcinoma; oligodendroglioma,
anaplastic;
oligodendroglioma, NOS; ovary adenocarcinoma, NOS; ovary carcinoma, NOS; ovary
20 carcinosarcoma; ovary clear cell carcinoma; ovary endometrioid
adenocarcinoma; ovary granulosa
cell tumor, NOS; ovary high-grade serous carcinoma; ovary low-grade serous
carcinoma; ovary
mucinous adenocarcinoma; ovary serous carcinoma; pancreas adenocarcinoma, NOS;
pancreas
carcinoma, NOS; pancreas mucinous adenocarcinoma; pancreas neuroendocrine
carcinoma, NOS;
parotid gland carcinoma, NOS; peritoneum adenocarcinoma, NOS; peritoneum
carcinoma, NOS;
25 peritoneum serous carcinoma; pleural mcsotheliormt, NOS; prostate
adenocarcinoma, NOS;
rectosigmoid adenocarcinoma, NOS; rectum adenocarcinoma, NOS; rectum mucinous
adenocarcinoma; retroperitoneum dedifferentiated liposarcoma; retroperitoneum
leiomyosarcoma,
NOS; right colon adenocarcinoma, NOS; right colon mucinous adenocarcinoma;
salivary gland
adenoid cystic carcinoma; skin melanoma; skin melanoma; skin merkel cell
carcinoma; skin nodular
30 melanoma; skin squamous carcinoma; skin trunk melanoma; small intestine
adenocarcinoma; small
intestine gastrointestinal stromal tumor, NOS; stomach gastrointestinal
stromal tumor, NOS; stomach
signet ring cell adenocarcinoma; thyroid carcinoma, anaplastic, NOS; thyroid
carcinoma, NOS;
thyroid papillary carcinoma of thyroid; tonsil, oropharynx, tongue squamous
carcinoma; transverse
colon adenocarcinoma, NOS; urothelial bladder adenocarcinoma, NOS; urothelial
bladder carcinoma,
35 NOS; urothelial bladder squamous carcinoma; urothelial carcinoma, NOS;
uterine endometrial
stromal sarcoma, NOS; uterus leiomyosarcoma, NOS; uterus sarcoma, NOS; uveal
melanoma;
vaginal squamous carcinoma; vulvar squamous carcinoma; and any combination
thereof. Note that
146
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
NOS, or "Not Otherwise Specified," is a subcategory in systems of
disease/disorder classification
such as ICD-9, ICD-10, or DSM-IV, and is generally but not exclusively used
where a more specific
diagnosis was not made. The models for these disease types were trained using
NGS data for a
specified gene panel (see Example 1, Tables 123-125) obtained for tens of
thousands of patient
5 samples. Training of the models is further described in Examples 2-3.
Tables 2-116 list selections of features that contribute to the 115 disease
type predictions,
where each row in the table represents a feature ranked by Importance. In the
tables, the column
"GENE" is the identifier for the feature, which is a typically a gene 1D;
column "TECH" is the
technology used to assess the biomarker, where "CNA" refers to copy number
alteration as assessed
10 by NGS, "NGS" is
mutational analysis using next-generation sequencing, and "META" is a patient
characteristic such as age at time of specimen collection ("Age") or gender
("Gender"); and col
"IMP" is a normalized Importance score for the feature. A row in the tables
where the GENE column
is MSI and the TECH column is NGS refers to the feature microsatellite
instability (MSI) as assessed
by next-generation sequencing. The table headers indicate the more granular
disease type (see above)
15 and less granular organ group in the format "disease type - organ
group". There are such 15 such
organ groups indicated that each contain disease types originating in
different organs or organ
systems: bladder; skin; lung; head, face or neck (NOS); esophagus; female
genital tract and
peritoneum (FGTP); brain; colon; prostate; liver, gall bladder, ducts; breast;
eye; stomach; kidney;
and pancreas. A biological specimen can be grouped into one of the less
granular 15 organ groups
20 according to its more
granular predicted disease type. As noted, the rows in the tables are sorted
by
importance. The higher the importance score the more important or relevant the
feature is in making
the disease type prediction. As indicated in the tables, in most cases we
observed that gene copy
numbers were driving the predictions.
Table 2: Adrenal Cortical Carcinoma - Adrenal Gland
GENE TECH IMP CCNF,1 CNA 0.492 T,PP CNA 0.357
HIMGA2 CNA 1.000 c-KIT NGS 0.486 ABL1 NGS 0.355
FOXL2 NGS 0.900 CDH11 CNA 0.480 LGR5 CNA 0.338
CTCE CNA 0.886 TSC1 CNA 0.450 BTG1 CNA 0.338
WW1 CNA 0.768 NR4A3 CNA 0.448 11'M3 CNA 0.335
DDIT3 CNA 0.698 CTNNA1 CNA 0.441 EP300 CNA 0.307
PTPN11 CNA 0.689 FGFR2 CNA 0.439 SRSF2 CNA 0.306
EWSR1 CNA 0.664 ATF1 CNA 0.438 1CRAS NGS 0.298
PPP2R1A CNA 0.640 ATP1A1 CNA 0.428 RBM15 CNA 0.290
EBF1 CNA 0.637 FOX01 CNA 0.401 ABL2 CNA 0.288
CDH1 CNA 0.633 ACSL6 CNA 0.394 VIM NGS 0.284
CDK4 CNA 0.607 BRCA2 CNA 0.374 MYCL CNA 0.279
Age META 0.599 CHEK2 CNA 0.374
ITK CNA 0.278
NUP93 CNA 0.507 SOX2 CNA 0.373 ZNF331 CNA 0.273
CRKL CNA 0.499 ENBP1 CNA 0.361 TEPT CNA 0.268
147
CA 03167694 2022- 8- 11
WO 2021/163706 PCT/US2021/018263
ARNT CNA 0.267 BCT ,9 CNA 0.265 El K4 CNA
0.263
ALDH2 CNA 0.265 MECOM CNA 0.264 RBI CNA 0.261
Table 3: Anus Squamous carcinoma - Colon
GENE TECH IMP CDKN2I3 CNA 0.782 SRGA
P3 CNA 0.652
LPP CNA
1.000 Gender META 0.781 NTRK2 CNA 0.646
FOXL2 NGS 0.956 ARID lA CNA 0.771
FINIGN2P46 CNA 0.641
CDKN2A CNA 0.894 13CL6 CNA 0.759 A14143
CNA 0.636
SOX2 CNA 0.872 SDHD CNA 0.746 10141K CNA 0.631
CACNA1D CNA 0.852 PAX3 CNA 0.745 MDS2
CNA 0.630
CNBP CNA 0.852 XPC CNA 0.710 BARDI CNA 0.624
KLIIL6 CNA 0.843 KDSR CNA 0.707 EXT1 CNA 0.618
TFRC CNA 0.842 TGEBR2 CNA 0.705 MECOM CNA 0.617
SPEN CNA 0.805 WIVTR1 CNA 0.701 TRIM27 CNA 0.615
TP53 NGS 0.804 FLII CNA 0.697 KMT2A CNA 0.614
Age META
0.803 PCSK7 CNA 0.693 GNAS CNA 0.597
VIIL CNA 0.797 BCL2 CNA 0.683 ATIC CNA 0.594
PPARG CNA 0.794 PAFAH1B2 CNA 0.674
MAX CNA 0.569
RPN1 CNA 0.794 CBI CNA 0.667 FHIT CNA 0.563
Z11T1316 CNA 0.786 CRE1131,2 CNA 0.664 SDHB CNA
0.552
FANCC CNA 0.785 CCNEI CNA 0.654 PRDMI CNA 0.550
Table 4: Appendix Adenocarcinoma NOS - Colon
GENE TECH IMP C1C14 CNA 0.678 FAN CC CNA
0.570
KRAS NGS 1.000 SOX2 CNA 0.671 CHEK2
CNA 0.566
FOXL2 NGS 0.948 HEYI CNA 0.664 CCNEI
CNA 0.564
CDX2 CNA 0.916 NE113 CNA 0.658 HOXA9 CNA
0.563
LFIEPL6 CNA 0.901 ESRI CNA 0.656 CBFB
CNA 0.557
Age META 0.873 NUP214 CNA 0.645 BTGI CNA
0.556
FLTI CNA 0.807 LCPI CNA 0.639 CACNAID CNA
0.555
CDKN2A CNA 0.781 SMAD4 CNA 0.635 F0X03 CNA
0.554
SRSF2 CNA 0.772 FGFI4 CNA 0.617 PSIF'l
CNA 0.554
BCL2 CNA 0.768 IGF IR CNA 0.615 RB1
CNA 0.554
Gender META 0.744 TSCI CNA 0.606 ERCC5
CNA 0.544
SETBPI CNA 0.728 MAP2K1 CNA 0.604 PTCHI CNA
0.542
FLT3 CNA 0.728 WAVTR1 CNA 0.599 CDKN IB CNA 0.538
CR1CL CNA 0.722 FCRL4 CNA 0.597 BAPI
CNA 0.533
CDKN2B CNA 0.698 CNBP CNA 0.590 SS18
CNA 0.533
KDSR CNA 0.688 CDH1 I CNA 0.588 APC
NUS 0.533
PDCD1LG2 CNA 0.687 MLLT3 CNA 0.575 ARNT CNA
0.533
Table 5; Appendix Mueinous adenocarcinoma - Colon
GENE TECH IMP KRAS NGS 1.000 GNAS
NGS 0.828
148
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
FOXL2 NGS 0.804 T,HEPT,6 CNA 0.472 ESR1 CNA
0.383
Age META 0.682 NR4A3 CNA 0.471 EBF 1 CNA
0.382
APC NGS 0.657 GNA13 CNA 0.464 CDH1 CNA
0.382
CDX2 CNA 0.657 c-KIT NGS 0.455 NF2 CNA
0.374
EPHA3 CNA 0.629 NSDI CNA 0.449 SETBPI CNA 0.372
PDCD1LG2 CNA 0.605 HERPUD1 CNA 0.442 W1F1 CNA 0.371
CDKN2A CNA 0.603 Gender META 0.439 HOXD13 CNA
0.370
CDKN2B CNA 0.598 WWTRI CNA 0.433 HOXAll CNA 0.366
CDIIII CNA 0.597 RPNI CNA 0.427 AFF4 CNA 0.365
1-1MGN2P46 CNA 0.514 TTL CNA 0.412 TSC1 CNA 0.358
CACNA1D CNA 0.506 FLT1 CNA 0407 KLHL6 CNA 0.356
ERCC5 CNA 0.500 AFF3 CNA 0.396 VIAL CNA 0.352
TAL2 CNA 0.493 CD274 CNA 0.392 PBXI CNA
0.350
MSI2 CNA 0.488 CREB3L2 CNA 0.391 KD SR CNA
0.348
FANCG CNA 0.481 NUP214 CNA 0.389 SPECC1 CNA 0.345
FNBPI CNA 0.472 EXTI CNA 0.385 SRSF2 CNA
0.342
Table 6: Bile duct NOS, cholangiocarcinoma - Liver, GaMadder, Ducts
GENE TECH IMP SRGAP3 CNA 0.704 1ITG1 CNA 0.618
SPEN CNA 1.000 CDKN2T1 CNA 0.698 KD SR CNA
0.611
FOXL2 NGS 0.944 MDS2 CNA 0.695 MAF CNA 0.606
C15orf65 CNA 0.923 PBXI CNA 0.681 MAML2 CNA 0.595
ARTD1A CNA 0.906 ERE1 CNA 0.680 TS1-112 CNA
0.585
CAMTA1 CNA 0.884 ERG CNA 0.674 CDKN2A CNA 0.575
VANCE CNA 0.803 VI1L NGS 0.669 ARI IGAP26 NGS
0.570
Gender META 0.802 TP53 NGS 0.651 FLT3 CNA
0.562
Age META 0.794 MTOR CNA 0.650 NTRK2 CNA
0.559
CDK12 CNA 0.769 FANCC CNA 0.648 LHFPL6 CNA 0.546
CHIC2 CNA 0.761 MCL1 CNA 0.646 CDH1 NGS 0.545
FHIT CNA 0.759 VEIL CNA 0.643 ELF CNA 0.544
SDHB CNA 0.753 LPP CNA 0.638 BCL6 CNA 0.544
PTPRC NGS 0.742 FOXA1 CNA 0.634 MYD88 CNA 0.542
NOTCH2 CNA 0.734 SUZ12 CNA 0.630 FSTL3 CNA
0.535
XPC CNA 0.714 PRDM1 CNA 0.629 PPARG CNA
0.532
APC NGS 0.706 WISP3 CNA 0.624 PDCD1LG2 CNA
0.532
Table 7: Brain Astrocytoma NOS - Brain
GENE TECH IMP SOX2 CNA 0.722 ZNF217 CNA 0.587
IDH1 NGS 1.000 SPECC1 CNA 0.705 HIST1H3B CNA 0.575
Age META 0.867 CREB3L2 CNA 0.651 PDGERA CNA 0.556
E0XL2 NGS 0.856 NDRG1 CNA 0.647 HMGA2 CNA 0.552
EGFR CNA 0.769 CDK6 CNA 0.625 MSI2 CNA 0.548
FGFR2 CNA 0.755 ATRX NGS 0.604 AKAP9 CNA 0.534
MYC CNA 0.722 KA16B CNA 0.598 OLIG2 CNA 0.533
149
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Gender META 0.528 GID4 CNA 0.438 NCOA2 CNA 0.390
TP53 NGS 0 514 SRSF2 CNA 0 435 STKII CNA 0
387
DDX6 CNA 0.508 CBL CNA 0.424 PIK3CG CNA 0.387
TRRAP CNA 0.501 N UP93 CNA 0.424 LPP CNA 0.387
TET 1 CNA 0.493 CHIC2 CNA 0.414 MECOM CNA 0.383
MCT A CNA 0 480 SR GAP3 CNA 0 414 CDX2 CNA 0381
ZBTB16 CNA 0.472 ECT2L CNA 0.413 SPEN CNA 0.378
BTGI CNA 0.458 KRAS NGS 0.410 TCLIA CNA 0.376
NEKB2 CNA 0.451 CCDC6 CNA 0.409 RABEPI CNA 0.375
CDKN2B CNA 0.447 ACSL6 CNA 0.405 PMS2 CNA 0.370
Table 8: Brain Astrocytoma anaplastic - Brain
GENE TECH IMP MSI NGS 0.519 KRAS NGS 0.405
Age META 1.000 NTRK2 CNA 0.499 MLLT11 CNA 0.403
IDH1 NGS 0.864 SDHD CNA 0.481 FGFR2 CNA
0.401
FOXL2 NGS 0.847 TETI CNA 0.470 EGFR CNA 0.394
HIVIGA2 CNA 0.709 OLIG2 CNA 0.451 RUNX 1 TI CNA
0.394
SOX2 CNA 0.709 CLPI CNA 0.445 NFICBLA CNA 0.391
MYC CNA 0.695 VIAL NGS 0.432 e-KIT NGS
0.382
SPECCI CNA 0.675 CTCF CNA 0.432 FAM46C CNA 0.380
CREB3L2 CNA 0.672 VTII A CNA 0.427 BCL9 CNA
0.377
MSI2 CNA 0.617 PMS2 CNA 0.423 FG1410 CNA
0.376
ZNF217 CNA 0.593 CDK6 CNA 0.422 CDKN2B CNA 0.374
EXTI CNA 0.582 CBFB CNA 0.420 MLHI CNA 0.374
1PM3 CNA 0.572 NUP93 CNA 0.419 CCDC6 CNA 0.373
SETBPI CNA 0.548 ELK4 CNA 0.416 PDE4DIP CNA 0.372
CACNA1D CNA 0.536 ENBP1 CNA 0.409 H3F3A CNA 0.370
NR4A3 CNA 0.524 TP53 NGS 0.409 MECOM CNA 0.368
Gender META 0.523 PBXI CNA 0.406 N1JP214 CNA
0.366
Table 9: Breast Adenocarcinoma NOS - Breast
GENE TECH IMP GNAQ NGS 0.588 SDI ID CNA
0.535
GATA3 CNA 1.000 EWSR1 CNA 0.579 FHIT CNA 0.533
Gender META 0.906 BCL9 CNA 0.571 CACNAID CNA
0.528
Age META 0.811 MYC CNA 0.569 MECOM CNA 0.526
ELK4 CNA 0.773 HIST1H4I NGS 0.556 YWHAE CNA 0.522
FUS CNA 0.739 CDHI NGS 0.556 AKT3 CNA 0.522
CCND I CNA 0.698 LITFPL6 CNA 0.555 CDK_N2A CNA 0.521
KRAS NGS 0.682 VHL NGS 0.551 SDHC CNA 0.518
FOXE2 NGS 0.646 PRCC CNA 0.550 RPL22 CNA
0.513
P13X1 CNA 0.631 CRE131313 CNA 0.545 FOX01 CNA
0.512
MCEI CNA 0.625 PDGFRA NGS 0.539 TRIM 27 CNA 0.511
APC NGS 0.602 FLII CNA 0.536 TNERSF17 CNA
0.511
PAX8 CNA 0.592 CDX2 CNA 0.535 S1A13 CNA
0.506
150
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
RMI2 CNA 0.506 TPM3 CNA 0.498 FH CNA 0.494
PAFAHIB2 CNA 0 504 MUC1 CNA 0 498 ITIVIGA2 CNA 0 493
ZNE217 CNA 0.499 EXT I CNA 0.498 RUNXIT1 CNA 0.492
CDKN2I3 CNA 0.498 CCND2 CNA 0.496 POU2AF 1 CNA 0.490
Table 10: Breast Carcinoma NOS - Breast
GENE TECH IMP BCL9 CNA 0.734 SPECCI CNA 0.671
GATA3 CNA 1.000 TNERSF17 CNA 0.734 H3F3A CNA 0.670
Age META 0.974 CREBBP CNA 0.725 SDHC CNA 0.665
ELK4 CNA 0.922 CACNA1D CNA 0.723 SETBP1 CNA 0.659
Gender META 0.908 EXT I CNA 0.721 YWTIAE CNA
0.658
FOXL2 NGS 0.898 MECOM CNA 0.700 TGFBR2 CNA 0.656
MCLI CNA 0.886 PAX8 CNA 0.699 CDKN2A CNA 0.656
MYC CNA 0.865 FUS CNA 0.698 PDE4DIP CNA 0.651
CCND1 CNA 0.845 FLI1 CNA 0.694 FHIT CNA 0.650
RMI2 CNA 0.807 HMGA2 CNA 0.689 GA S7 CNA 0.648
LITFPL6 CNA 0.790 ARID 1 A CNA 0.689 ARNT CNA 0.647
PBXI CNA 0.789 TP53 NGS 0.685 CDKN2B CNA 0.642
USP6 CNA 0.776 PRCC CNA 0.684 CDH1 CNA 0.639
FOXAI CNA 0.760 STAT3 CNA 0.681 MAML2 CNA 0.634
MUC1 CNA 0.757 FOX01 CNA 0.677 GID4 CNA 0.632
MLL111 CNA 0.752 CDH11 CNA 0.672 1PM3 CNA 0.630
COX6C CNA 0.738 ZNF217 CNA 0.672 RPN1 CNA 0.626
Table 11: Breast Inffitrating Duct Adenocarcinoma - Breast
GENE TECH IMP 1NERSF17 CNA 0.617 MS12 CNA 0.563
CiATA3 CNA 1.000 I JSP6 CNA 0.604 CilD4 CNA
0.562
Age META 0.841 RAD21 CNA 0.604 ZNF217 CNA
0.561
FOXL2 NGS 0.833 STAT5B CNA 0.603 MAML2 CNA 0.556
MYC CNA 0.797 ELI CNA 0.595 TPM3 CNA 0.554
EXTI CNA 0.796 SNX29 CNA 0.592 BRCAI CNA
0.554
Gender META 0.786 HI CNA 0.590 PAFAI Il B2 CNA
0.553
PBX1 CNA 0.778 P1K3CA NGS 0.584 IKBKE CNA 0.553
MCL 1 CNA 0.727 SLC34A2 CNA 0.580 MUC1 CNA 0.552
ELK4 CNA 0.692 CACNA ID CNA 0.578 RMI2 CNA
0.547
COX6C CNA 0.683 PAX8 CNA 0.578 FOX01 CNA 0.547
CDH1 NGS 0.671 CREBBP CNA 0.576 CDKN2B CNA 0.547
CCND1 CNA 0.667 CDKN2A CNA 0.574 ITMGA2 CNA 0.546
FUS CNA 0.665 PCMI CNA 0.571 MDM4 CNA 0.546
RUNXI11 CNA 0.647 SPECCI CNA 0.571 ESRI NGS 0.545
13CL9 CNA 0.640 U2AE1 CNA 0.568 HOXD13 CNA 0.544
ITIFPL6 CNA 0.624 TP53 NGS 0.564 FANCC CNA 0.538
151
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Table 12: Breast Infiltrating Lobular Carcinoma NOS - Breast
GENE TECH IMP FANCA CNA 0.377 NUP93 CNA
0.282
CDHI NGS 1.000 YWHAE CNA 0.361 ARNT CNA
0.282
CDHI CNA 0.684 Age META 0.344 VI-IL NGS
0.281
CTCF CNA 0.649 BCL2 CNA 0.343 ABL2 CNA
0.280
CDT Il 1 CNA 0.640 TP53 NGS 0.342 TRIM 33 NGS 0.273
ELK4 CNA 0.600 MECOM CNA 0.339 PAX8 CNA
0.271
FOXL2 NGS 0.590 FH CNA 0.332 KDM5C NGS 0.270
CAMTA1 CNA 0.563 USP6 CNA 0.331 PAFAHI B2 CNA
0.270
Gender META 0.535 PCSK7 CNA 0.330 HOXD11 CNA
0.269
IKBKE CNA 0.478 AKT3 CNA 0.328 APC NGS
0.269
FLU_ CNA 0.477 KCNT5 CNA 0.323 ALTRKB CNA 0.269
CBFB CNA 0.474 CDKN2B CNA 0.314 TFRC CNA
0.267
PBXI CNA 0.450 CBL CNA 0.302 KRAS NGS
0.266
CDC73 CNA 0.438 E1V5 CNA 0.302 CDKN 2A CNA 0.265
GATA3 CNA 0.394 1vlDM4 CNA 0.295 KT ,HT
,6 CNA 0.262
BCL9 CNA 0.387 FUS CNA 0.292 CTNNAI CNA 0.261
CREB13P CNA 0.385 CDX2 CNA 0.285 DDR2 CNA
0.261
Table 13: Breast Metaplastic Carcinoma NOS - Breast
GENE TECH BIP EWSRI CNA 0.733 ARHGAP26 CNA
0.595
Gender META 1.000 ERCC3 CNA 0.728 TP53
NGS 0.592
MAF CNA 0.966 TP.11\427 CNA 0.723 PLAGI
CNA 0.592
FOXL2 NGS 0.919 PRKDC CNA 0.718 ATFI
CNA 0.562
NUTM2B CNA 0.916 MYC CNA 0.714 CDK4 CNA
0.561
EP300 CNA 0.906 COX6C CNA 0.714 WISP3
CNA 0.560
CDKN2A CNA 0.880 HEY]. CNA 0.701 CDH11 CNA
0.558
Age META 0.873 PDCD1LG2 CNA 0.697 FANCC
CNA 0.557
ERBB3 CNA 0.855 14 G1410 CNA 0.695 RN1443
CNA 0.555
DDIT3 CNA 0.849 ITK CNA 0.688 CITEK2 CNA
0.555
PIK3 CA NGS 0.816 NR4A3 CNA 0.687 EIMGN2P46 CNA
0.551
MSI2 CNA 0.815 N172 CNA 0.684 ERG CNA
0.546
PRRX1 CNA 0.791 PIK3R1 NGS 0.661 CHCI-TD7 CNA
0.543
NTRK2 CNA 0.755 SMARCB1 CNA 0.632 PMS2 CNA
0.538
CDKN2B CNA 0.748 EXT1 CNA 0.629 TAL2 CNA
0.537
I IMGA2 CNA 0.744 CCNEI CNA 0.629 SDI ID CNA
0.531
STAT5B CNA 0.735 CLTCLI CNA 0.626 NFIEI CNA
0.531
Table 14: Cervix Adenocarc-inoma NOS - FGTP
GENE TECH IMP Gender META 0.704 SDC4 CNA
0.626
Age META 1.000 GNAS CNA 0.695 CDK6 CNA 0.601
FOXL2 NGS 0.815 FLII CNA 0.692 LPP CNA
0.599
TP53 NGS 0.718 KRAS NGS 0.641 MECOM CNA 0.596
152
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
LHEPL6 CNA 0.593 MYC CNA 0.511 CDHI CNA 0.463
KLITL6 CNA 0 570 HEY]. CNA 0 504 TRRAP CNA 0 452
KDSR CNA 0.566 MLFI CNA 0.498 CBL CNA 0.451
CRE133L2 CNA 0.548 PDGERA CNA 0.496 Ul3R5 CNA 0.451
RACI CNA 0.548 PAM CNA 0.493 PIK3 CA NGS 0.446
PBX1 CNA 0538 CTNNA 1 CNA 0488 F,WSR 1 CNA 0 444
ETV5 CNA 0.534 CDKN2A CNA 0.483 IKZE1 CNA 0.441
MLLTII CNA 0.531 TFRC CNA 0.481 ARID I A CNA 0.430
BCL6 CNA 0.526 W WTR1 CNA 0.477 ASXLI CNA 0.427
MUC1 CNA 0.526 SETBPI CNA 0.471 CCNEI CNA 0.427
PLAGI CNA 0.522 SDHAF2 CNA 0.471 KIAA1549 CNA 0.425
TPM3 CNA 0.521 EXTI CNA 0.470 PRRXI CNA 0.425
ZNF217 CNA 0.517 APC NGS 0.466 FGFR2 CNA 0.425
Table 15: Cervix Carcinoma NOS - FGTP
GENE TECII MU' WWTRI CNA 0.714 NDRG1 CNA 0.568
MECOM CNA 1.000 CCNE1 CNA 0.692 YWHAE CNA 0.567
FOXL2 NGS 0.973 SRSF2 CNA 0.683 ZNF217 CNA
0.558
Gender META 0.973 PDGFRA CNA 0.673 FOXL2 CNA
0.555
Age META 0.972 SEPT5 CNA 0.671 EGFR CNA
0.549
RPN1 CNA 0.950 BTG1 CNA 0.668 ACSL3 NGS
0.546
312A141 CNA 0.900 CDK12 CNA 0.654 ERCC3 CNA
0.541
SOX2 CNA 0.856 CDKN2B CNA 0.647 IKZE1 CNA 0.539
BCL6 CNA 0.832 RAD50 CNA 0.624 SDHC CNA
0.536
EXTI CNA 0.819 RNE213 NOS 0.615 SDC4 CNA 0.535
IRVIGN21'46 CNA 0.802 TP53 NGS 0.600 CREB3L2 CNA 0.525
ATIC CNA 0.761 DAXX CNA 0.598 TFRC CNA 0.522
RACI CNA 0.750 MLF 1 CNA 0.596 CACNA ID CNA
0.519
KLI11.6 CNA 0.748 BCL2 CNA 0.585 CCND2 CNA
0.517
ECT2L CNA 0.747 ETV5 CNA 0.585 MUCI CNA 0.510
LPP CNA 0.741 ARERPI CNA 0.579 BCL9 CNA 0.508
USP6 CNA 0.740 GMPS CNA 0.569 MYCL CNA 0.505
Table 16: Cervix Squamous Carcinoma - FGTP
GENE TECH IMP WWTRI CNA 0.739 MAX CNA 0.553
Age META 1.000 ARID 1A CNA 0.736 PAX3 CNA
0.548
TP53 NGS 0.863 Gender META 0.724 CACNAID
CNA 0.539
CNBP CNA 0.851 SOX2 CNA 0.722 FOXP1 CNA
0.527
TFRC CNA 0.838 CREB3L2 CNA 0.699 ERBB3 CNA 0.526
FOXL2 NGS 0.828 CDKN 2B CNA 0.663 PMS2 CNA 0.513
RPN 1 CNA 0.794 CDKN 2A CNA 0.614 MDS2 CNA 0.507
LPP CNA 0.758 SPEN CNA 0.600 ATIC CNA 0.502
BCL6 CNA 0.751 MECOM CNA 0.595 RUNXI CNA 0.500
KEHL6 CNA 0.740 ETV5 CNA 0.578 SYK CNA 0.498
153
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
SETBP1 CNA 0.495 SLC34A2 CNA 0.464 KIAA1549 CNA 0.447
IGF IR CNA 0 494 SFPQ CNA 0 463 GSK3B CNA 0 444
ERBB4 CNA 0.478 EPITLI1 CNA 0.454 NSD2
CNA 0.441
KDSR CNA 0.473 NIKIIIA CNA 0.453 SPLCCI CNA 0.437
ZNF384 CNA 0.470 TRIM27 CNA 0.450 EXT1 CNA 0.430
BCT ,2 CNA 0 467 MITE CNA 0 410 T ILFPT ,6 CNA 0
426
FGF10 CNA 0.464 ERG CNA 0.449 BCLI IA CNA 0.421
Table 17: Colon Adenocarcinoma NOS - Colon
GENE TECH IMP GNAS CNA 0.620 FGFR2 CNA 0.512
CDX2 CNA 1.000 Gender META 0.615 WWTR1 CNA 0.512
APC NGS 0.912 ERG CNA 0.600 RACI CNA 0.511
FOXL2 NGS 0.801 CDKN2B CNA 0.592 TP53 NGS 0.511
KRAS NGS 0.781 ERCC5 CNA 0.587 MYC CNA 0.509
SETBP1 CNA 0.764 NSD2 CNA 0.580 JAK1 CNA 0.508
ASXLI CNA 0.715 IRS2 CNA 0.577 SPEN CNA 0.508
LHFPL6 CNA 0.713 SMAD4 CNA 0.574 SPECCI CNA 0.505
FLT3 CNA 0.707 TOPI CNA 0.574 TP53 CNA 0.505
BCL2 CNA 0.704 EPHA5 CNA 0.564 MSI2 CNA 0.499
FOX01 CNA 0.703 HOXA9 CNA 0.552 EWSRI CNA 0497
SDC4 CNA 0.693 CDHI CNA 0.551 CCNEI CNA 0.496
KDSR CNA 0.691 CDKN 2A CNA 0.548 ARIDIA CNA 0.494
ZNF217 CNA 0.686 CBFB CNA 0.537 CDK6 CNA 0.491
Age META 0.660 ZNF521 CNA 0.536 MAML2 CNA 0.490
FLT1 CNA 0.639 CDK8 CNA 0.533 RB1 CNA 0.489
EBF I CNA 0.627 USP6 CNA 0.529 U2AF I CNA 0.485
Table 18: Colon Carcinoma NOS - Colon
GENE TECH IMP ZNF217 CNA 0.507 PML CNA 0.430
APC NGS 1.000 SETBP1 CNA 0.496 BCL2L11 CNA 0.428
SDC4 CNA 0.773 FOXL2 NGS 0.487 CDK12 CNA
0.427
VITT, NGS 0.715 ARID 1A NGS 0.482 CYP2D6 CNA 0.424
CDH1 CNA 0.683 FANCF CNA 0.480 TTL CNA
0.423
GNAS CNA 0.676 CTCF CNA 0.478 KDM5C NGS 0.422
IDI-11 NGS 0.676 TOPI CNA 0.475 BCL6 CNA 0.421
HMGN2P46 CNA 0.647 KRAS NGS 0.472 CASP 8 CNA
0.416
Gender META 0.634 TP53 NGS 0.465 ACKR3 NGS
0.415
CDX2 CNA 0.616 U2AF1 CNA 0.463 KLAA1549 CNA
0.414
c-KIT NGS 0.601 MYC CNA 0.451 RPL22 CNA 0.408
Age META 0.574 CDKN 2C CNA 0.438 FLT3 CNA
0.408
LHEPL6 CNA 0.554 AURKA CNA 0.437 1PM3 CNA 0.407
CDHI NGS 0.553 HOXA9 CNA 0.435 STAT3 CNA 0.404
ASXLI CNA 0.522 KII-11,6 CNA 0.434 FOX01
CNA 0.393
SMAD4 CNA 0.520 BCL9 CNA 0.431 FKIBP I CNA
0.392
154
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
PTEN NGS 0.390 PTCH1 CNA 0.383 MECOM CNA 0.381
Table 19: Colon Mucinous Adenocarcinoma - Colon
GENE TECH IMP TFRC CNA 0.533 STAT3 CNA 0.474
KRAS NGS 1.000 SRSF2 CNA 0.527 EPHA5 CNA
0.454
APC NGS 0.778 ALDH2 CNA 0.513 SLC34A2 CNA
0.450
RPNI CNA 0.745 SDHAF2 CNA 0.511 HEYI CNA 0.449
FOXL2 NGS 0.727 PTEN CNA 0.504 MSI2 CNA 0.449
Age META 0.686 TSCI CNA 0.501 CAMTAI CNA 0.448
CDX2 CNA 0.668 SMAD4 CNA 0.500 FGF 14 CNA 0.442
NI JP214 CNA 0.638 WWTR I CNA 0.492 MAX CNA 0.441
CDKN2B CNA 0.632 IDH1 NGS 0.492 TPM4 CNA 0.441
LUFPL6 CNA 0.620 KDSR CNA 0.491 BCL2 CNA 0.426
SETBPI CNA 0.619 VIIL NGS 0.485 LPP CNA 0.423
Gender META 0.608 NFIB CNA 0.485 KLF4 CNA
0.420
TP53 NGS 0.571 MAF CNA 0.481 BTGI CNA 0.420
FGFR2 CNA 0.568 BCL6 CNA 0.481 CDH 1 1 CNA
0.417
RETNXIT1 CNA 0.558 FLT3 CNA 0.479 FANCG CNA 0.409
PTEN NGS 0.554 PDCD1LG2 CNA 0.478 H3F3B CNA
0.405
CDKN2A CNA 0.553 GID4 CNA 0.475 PRKDC CNA 0.402
Table 20: Conjunctiva Malignant melanoma NOS - Skin
GENE TECH IMP Age META 0.465 GNAQ NGS 0.301
IRF4 CNA 1.000 VIAL NGS 0.465 CCND3 CNA 0.300
ACSL6 NGS 0.847 POU2AF1 CNA 0.463 LPP CNA 0.283
FEU CNA 0.837 DAXX CNA 0.454 KRAS NGS 0.282
WWTR1 CNA 0.810 NRA S NOS 0.436 PDGFRA CNA 0.279
TRIIV127 CNA 0.763 PMS2 CNA 0.421 SOX2 CNA 0.277
RPN1 CNA 0.762 KLUL6 CNA 0.411 EPUB1 CNA 0.275
CDHI NGS 0.738 ZBTB16 CNA 0.378 AFF3 CNA 0.275
FOXL2 NGS 0.738 APC NGS 0.370 ESRI CNA 0.274
TP53 NGS 0.602 EBF1 CNA 0.367 CTNNB1 NGS 0.273
KCNJ5 CNA 0.593 PRKAR1A CNA 0.351 KIT CNA 0.257
SOX10 CNA 0.575 ETVI CNA 0.339 CLPI CNA 0.251
DEK CNA 0.557 SRSF3 CNA 0.338 GATA2 CNA 0.246
MLF1 CNA 0.519 TRIM26 CNA 0.328 SDUD CNA 0.245
EP300 CNA 0.491 WT1 CNA 0.328 CBL CNA 0.244
CNBP CNA 0.484 BCL6 CNA 0.321 WIFI CNA 0.233
Gender META 0.482 BRAF NGS 0.306 KD SR CNA 0.230
Table 21: Duodenum and Ampulla Adenocarcinoma NOS - Colon
GENE TECH IMP FOXL2 NGS 0.926 CDX2 CNA 0.870
KRAS NGS 1.000 SETTIPI CNA 0.902 Age META 0.842
155
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
FLT3 CNA 0.837 CBFB CNA 0.657 BCL6 CNA 0.564
KDSR CNA 0 829 PMS2 CNA 0 648 EXTI CNA 0 559
JAZEI CNA 0.807 U2AE1 CNA 0.631 PRRXI CNA 0.557
ELT1 CNA 0.804 CACNA ID CNA 0.623 PTPN11 CNA 0.557
USP6 CNA 0.769 CDK8 CNA 0.620 CALR CNA 0.556
APC NGS 0768 CR TC3 CNA 0 620 VT-H. NGS 0 112
CDKN2A CNA 0.741 LCPI CNA 0.604 CTCF CNA 0.551
LI-IFPL6 CNA 0.741 RB 1 CNA 0.604 CRKL CNA 0.548
13CL2 CNA 0.725 CDHI CNA 0.603 GNAS CNA 0.547
SPECCI CNA 0.704 ERCC5 CNA 0.602 CHEK2 CNA 0.545
Gender META 0.695 TP53 NGS 0.600 HOXA9 CNA 0.543
G1D4 CNA 0.691 SDHEI CNA 0.598 SDC4 CNA
0.543
TCF7L2 CNA 0.685 ETV6 CNA 0.584 ARIDIA CNA 0.542
CDKN2B CNA 0.681 CDH1 NGS 0.568 FH1T CNA 0.537
FOX01 CNA 0.665 EGE6 CNA 0.565 NE2 CNA 0.537
Table 22: Endometrial Endometroid Adenocardnoma - FGTP
GENE TECH IMP IKZE1 CNA 0.520 PAX8 CNA 0.488
PTEN NGS 1.000 MUC1 CNA 0.516 HNIGN2P46 NGS
0.485
ESRI CNA 0.807 CDKN2A CNA 0.513 CCDC6 CNA 0.481
Gender META 0.759 FGFR2 CNA 0.513 FGER1
CNA 0.479
CDHI NGS 0.696 N1JP214 CNA 0.513 CDKN 213
CNA 0.472
Age META 0.683 RACI CNA 0.512 FHIT CNA
0.472
FOXL2 NGS 0.641 HOXA13 CNA 0.511 SOX2 CNA 0.462
PIK3CA NGS 0.600 TP53 NGS 0.509 MYC CNA 0.457
APC NGS 0.589 PBX 1 CNA 0.503 SETBPI CNA 0.456
ARID1A NGS 0.586 GNAS CNA 0.503 EWSRI CNA 0.454
GATA2 CNA 0.575 MLLT11 CNA 0.502 LHFPL6 CNA 0.452
CDX2 CNA 0.562 CRKL CNA 0.495 PIK3R1 NGS
0.451
CBFB CNA 0.558 MECOM CNA 0.493 PRRXI CNA 0.444
CTNNBI NGS 0.551 AFF3 CNA 0.493 CDII11 CNA
0.444
ZNF217 CNA 0.529 HIVIGN2P46 CNA 0.491 STAT3 CNA
0.439
ENBP1 CNA 0.528 ELK4 CNA 0.491 MDM4 CNA 0.434
FANCF CNA 0.526 U2AFI CNA 0.488 BCL9 CNA
0.434
Table 23: Endometrial Adenocarcinoma NOS - FGTP
GENE TECH IMP KAT6B CNA 0.707 ELK4 CNA 0.619
Age META 1.000 CDHI NGS 0.700 MUCI CNA
0.602
PTEN NGS 0.967 MLLT11 CNA 0.684 CDHI CNA 0.597
Gender META 0.852 ESRI CNA 0.664 TP53 NGS
0.594
MECOM CNA 0.801 CDH11 CNA 0.648 NR4A3 CNA
0.593
APC NGS 0.779 CDX2 CNA 0.647 BCL9 CNA 0.589
PAX8 CNA 0.742 FGFR2 CNA 0.646 LEIFPL6 CNA
0.587
PIK3CA NGS 0.737 HIVIGN2P46 CNA 0.627 .. CDKN 213 CNA
0.583
156
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CDKN2A CNA 0.580 GNAQ NGS 0.549 KLUL6 CNA 0.523
ARID1A NGS 0 580 MAP2K1 CNA 0 548 CCDC6 CNA 0 523
KRAS NGS 0.575 ETV5 CNA 0.547 MAP CNA 0.521
CCNE 1 CNA 0.571 CBFB CNA 0.546 SEIB?' CNA
0.520
NUTM1 CNA 0.566 IKZE1 CNA 0.536 EXT1 CNA
0.519
GATA3 CNA 0 163 AR TD1 A CNA 0 133 CDK6 CNA 0
517
FOXL2 NGS 0.562 EBFI CNA 0.530 HOOK3 CNA
0.517
CTCF CNA 0.561 RACI CNA 0.527 ERBB3 CNA
0.514
PRAM. CNA 0.556 NUP214 CNA 0.526 VI-IL CNA 0.505
Table 24: Endometrial Carcinosarcoma - FGTP
GENE TECII PVIP FGER1 CNA 0.687 IKZE1 CNA
0.609
CCNEI CNA 1.000 XPA CNA 0.682 NCOA2 CNA 0.607
FOXL2 NGS 0.961 MAP CNA 0.672 FSTL3 CNA 0.606
Age META 0.906 BCL9 CNA 0.672 NTRK2 CNA
0.603
Gender META 0.819 PRRX1 CNA 0.654 HOXD13 CNA
0.596
MAP2K2 CNA 0.814 ENBP1 CNA 0.654 FANCF CNA
0.595
ASXL1 CNA 0.799 SYK CNA 0.647 TAL2 CNA 0.589
HMGN2P46 CNA 0.792 CBFB CNA 0.646 MECOM CNA 0.588
MLLT11 CNA 0.785 PIK3CA NGS 0.641 DDR2 CNA 0.588
KLF4 CNA 0.777 ALK CNA 0.633 PRKDC CNA 0.581
PTEN NGS 0.742 "TP53 NGS 0.631 FAN CC CNA
0.571
AFF3 CNA 0.734 TR1M27 CNA 0.626 CDKN2B CNA 0.570
WDCP CNA 0.723 ETV6 CNA 0.623 EWSR1 CNA
0.569
NR4A3 CNA 0.721 RAC 1 CNA 0.622 BTGI CNA
0.566
RPNI CNA 0.707 CDKN2A CNA 0.621 GATA2 CNA 0.563
WISP3 CNA 0.705 EP300 CNA 0.616 GNAQ CNA
0.561
CDHI CNA 0.694 ETVI CNA 0.611 FOXAI CNA
0.554
Table 25: Endometrial Serous Carcinoma - FGTP
GENE TECH IMP STAT3 CNA 0.702 KAT6B CNA 0.633
CCNE1 CNA 1.000 CBFB CNA 0.696 ESR1 CNA 0.633
Age META 0.984 RAC1 CNA 0.695 KLF4 CNA
0.632
MECOM CNA 0.959 CDKN2A CNA 0.685 CREBBP CNA 0.632
TP53 NGS 0.955 CREB3L2 CNA 0.683 FGER2 CNA
0.628
FOXL2 NGS 0.910 CDK6 CNA 0.674 P1K3CA NGS 0.628
PAX8 CNA 0.908 FSTL3 CNA 0.666 MAP2K1 CNA 0.627
NUTM1 CNA 0.865 BCL6 CNA 0.665 IK2F1 CNA
0.614
Gender META 0.854 MAP2K2 CNA 0.663 NR4A3 CNA
0.611
KLHL6 CNA 0.826 _LANCE CNA 0.661 LPP CNA
0.611
CDH1 CNA 0.776 C15 orf65 CNA 0.653 CDH11 CNA
0.607
111VIGN2P46 CNA 0.765 GATA2 CNA 0.648 ETV1 CNA 0.604
MAP CNA 0.716 SS18 CNA 0.634 TAL2 CNA 0.600
E1V5 CNA 0.705 AF 143 CNA 0.634 S'1K11 CNA
0.590
157
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
TPM4 CNA 0.590 INHBA CNA 0.582 LIIFPL6 CNA
0.578
NT_TP214 CNA 0 585 CTCF CNA 0 581 ALK CNA 0 578
MLLT11 CNA 0.584 GID4 CNA 0.581 CALR CNA 0.573
Table 26: Endometrium Carcinoma NOS - FGTP
GENE TECH IMP KLF4 CNA 0.601 CBFB CNA 0.526
PTEN NGS 1.000 RAC1 CNA 0.592 CDK6 CNA 0.524
FOXL2 NGS 0.896 CDHI CNA 0.590 ARID1A NGS 0.524
Age META 0.804 IIKZE 1 CNA 0.578 BCL9
CNA 0.523
JAZE1 CNA 0.797 SDHC CNA 0.573 NUP214 CNA 0.517
Gender META 0.766 CDKN2A CNA 0.570 FANCL CNA
0.510
C 15erf65 CNA 0.725 ELK4 CNA 0.564 NTRK2 CNA 0.508
PIK3 CA NGS 0.724 PIK3R1 NGS 0.560 EP300 CNA
0.504
LUTPL6 CNA 0.710 MAP2K1 CNA 0.559 VITT CNA 0.500
FGFR2 CNA 0.665 PPARG CNA 0.557 GID4 CNA
0.499
TET 1 CNA 0.654 FLT3 CNA 0.553 ETVI CNA 0.499
TP53 NGS 0.651 PAX8 CNA 0.552 GNAS CNA 0.499
MLLTII CNA 0.650 BMPRI A CNA 0.545 EWSRI CNA 0.498
FNBP1 CNA 0.647 FLI1 CNA 0.542 NR4A3 CNA
0.497
GNAQ CNA 0.635 CCNEI CNA 0.534 CTNNA1 CNA 0.495
EGFR CNA 0.633 HMGN2P46 CNA 0.534 TAF15 CNA
0.494
FAN CC CNA 0.604 PMS2 CNA 0.532 MLCOM CNA 0.491
Table 27: Endometrium Carcinoma Undifferentiated - FGTP
GENE TECH IMP PRRX1 CNA 0.718 GATA2 CNA 0.547
PIK3 CA NGS 1.000 IKZE1 CNA 0.717 PCM1 NGS 0.533
MAF CNA 0.994 ST ,C45A3 CNA 0.713 WTSP3 CNA
0.523
Gender META 0.991 RIVII2 CNA 0.705 CCNB11P1
CNA 0.520
FOXL2 NGS 0.976 TP53 NGS 0.688 CCDC6 CNA
0.518
ELK4 CNA 0.971 CDK6 CNA 0.670 PDE4DTP CNA 0.504
GID4 CNA 0.952 GNA13 CNA 0.663 ARHGAP26 CNA
0.499
ARID1A NGS 0.932 AURKB CNA 0.619 PMS2 CNA 0.493
PTEN NGS 0.881 KDM5C NGS 0.605 FGER1 CNA 0.486
H3F3A CNA 0.873 NTRK1 CNA 0.603 GNAQ CNA
0.484
PRCC CNA 0.804 MLLT10 CNA 0.589 ETV6 CNA 0.477
IIMGN2P46 CNA 0.775 RPL22 NGS 0.587 SOX2 CNA 0.472
HSP9OAA1 CNA 0.765 TGFBR2 CNA 0.587 CDK8 CNA 0.470
HIST1H3B CNA 0.753 SDC4 CNA 0.579 HEY1 CNA 0.468
SMARCA4 NGS 0.750 MYC CNA 0.574 SPEN CNA 0.468
PRKDC CNA 0.737 HIS11H41 CNA 0.571 EXT1 CNA
0.466
Age META 0.727 TETI. CNA 0.560 EP300 CNA
0.465
Table 28: Endometrium Clear Cell Carcinoma - FGTP
158
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
GENE TECH IMP CLTCL1 CNA 0.637 CRKL CNA 0.511
PAX8 CNA 1 000 CALR CNA 0 628 GNAS CNA 0 501
FOXL2 NGS 0.950 CNTRL CNA 0.626 FGFR2 CNA 0.499
CDK12 CNA 0.941 S1A13 CNA 0.625 FUS CNA
0.498
Gender META 0.871 FANCC CNA 0.617 RAC1 CNA 0.496
Age META 0853 CCNE1 CNA 0 600 7NF217 CNA 0 495
KLF4 CNA 0.823 NR4A3 CNA 0.600 N1RG1 CNA 0.490
FNBP1 CNA 0.780 TPM4 CNA 0.597 KRAS NGS 0.489
N142 CNA 0.754 OMD CNA 0.596 SE113P1 CNA 0.488
WWTR1 CNA 0.735 ERBB2 CNA 0.589 PMS2 CNA 0.488
MECOM CNA 0.728 MKL1 CNA 0.577 FANCF CNA 0.486
CHEK2 CNA 0.716 EP300 CNA 0.557 PIK3 CA NGS
0.476
YWHAE CNA 0.680 TSC1 CNA 0.555 CDKN2A CNA 0.474
KAT6A CNA 0.679 XPA CNA 0.534 CREB3T,2 CNA 0.472
SUFU CNA 0.675 PCSK7 CNA 0.532 TRIP11 CNA
0.461
AFF3 CNA 0.655 PAFAH1B2 CNA 0.521 GNA13 CNA
0.460
EWSR1 CNA 0.646 BCL6 CNA 0.518 RNF213 NGS 0.459
Table 29: Esophagus Adenocarcinoma NOS - Esophagus
GENE TECH IMP ERBB2 CNA 0.757 SMAD4 CNA 0.631
Gender META 1.000 BCL2 CNA 0.757 SMAD2 CNA 0.630
SE113P1 CNA 0.943 PITIT CNA 0.743 CACNA1D CNA 0.629
APC NGS 0.932 KIAA1549 CNA 0.726 HSP90AB1 CNA
0.629
ZNF217 CNA 0.931 CDKN2A CNA 0.694 WWTR1 CNA 0.620
ERG CNA 0.922 CDKN 213 CNA 0.693 14G1412.2
CNA 0.612
TP53 NGS 0.908 RUNX1 CNA 0.693 ASXL1 CNA 0.605
Age META 0.904 GNAS CNA 0.672 RAC1 CNA
0.602
CDX2 CNA 0.856 TRR_AP CNA 0.671 MLLT11 CNA 0.601
SDC4 CNA 0.849 AFFI CNA 0.671 ELIF 1 CNA
0.600
CDK12 CNA 0.827 FLT3 CNA 0.670 KRAS NGS 0.600
IRF4 CNA 0.818 ERBB3 CNA 0.655 TCF7L2 CNA 0.595
CREB3L2 CNA 0.803 CREBBP CNA 0.652 MALT1 CNA 0.593
U2AF1 CNA 0.802 JAZF 1 CNA 0.651 CTCF CNA
0.593
KDSR CNA 0.801 CTNNA1 CNA 0.650 PRRX1 CNA 0.591
KRAS CNA 0.796 FOX01 CNA 0.633 ARID1A CNA 0.583
MYC CNA 0.758 T .HEPT .6 CNA 0.633 KMT2C CNA
0.573
Table 30: Esophagus Carcinoma NOS - Esophagus
GENE TECH IMP PRRX1 CNA 0.740 FGER1OP CNA 0.658
ERG CNA 1.000 XPC CNA 0.740 1A11-g 19 CNA
0.642
FOXE2 NGS 0.946 RUNX1 CNA 0.707 MEF1 CNA 0.629
Gender META 0.878 TP53 NGS 0.697 APC NGS
0.624
PDGFRA CNA 0.873 TCF7L2 CNA 0.674 WIT CNA 0.602
Age META 0.753 YWHAE CNA 0.665 IDH1 NITS 0.585
159
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
VEIL NGS 0.572 CACNA ID CNA 0.479 NOTCH2 CNA 0.449
FHIT CNA 0 569 CBFB CNA 0 475 CDK_N2B CNA 0 447
KIT CNA 0.544 CREB3L2 CNA 0.473 CCNDI CNA 0.446
"[FRC CNA 0.532 NU1M213 CNA 0.470 CDK4 CNA 0.446
KRAS NGS 0.519 SETBPI CNA 0.467 RHOH CNA 0.442
WWTR 1 CNA 0 507 F ANCC CNA 0 466 D A XX CNA 0 440
RPNI CNA 0.494 AURKB CNA 0.462 FLTI CNA 0.435
LFIEPL6 CNA 0.486 USP6 CNA 0.460 FGFR2 CNA 0.434
FUE3 CNA 0.485 U2AE1 CNA 0.456 SRGAP3 CNA 0.431
JAKI CNA 0.484 SOX2 CNA 0.455 TGEBR2 CNA 0.431
PHOX2B CNA 0.482 FOXPI CNA 0.453 MLLT11 CNA 0.428
Table 31: Esophagus Squamous Carcinoma - Esophagus
GENE TECII IMP FGF19 CNA 0.655 EP300 CNA
0.510
KLIAL6 CNA 1.000 CDKN2A CNA 0.647 BCL6 CNA 0.499
TFRC CNA 0.969 PPARG CNA 0.637 CDICN2B CNA 0.498
SOX2 CNA 0.923 SRGAP3 CNA 0.637 XPC CNA 0.495
FOXL2 NGS 0.913 YWHAE CNA 0.610 EBF 1 CNA 0.472
EPH_A3 CNA 0.898 CTNNAI CNA 0.609 IDHI NGS 0.471
FHIT CNA 0.879 FGF4 CNA 0.609 KRAS NGS 0.470
FGF3 CNA 0.869 EWSRI CNA 0.591 WWTRI CNA 0.464
CCND1 CNA 0.811 MAML2 CNA 0.588 N UP214 CNA 0.462
TGFBR2 CNA 0.804 Age META 0.571 EZR CNA 0.440
LPP CNA 0.799 ERG CNA 0.560 FOXPI CNA 0.436
MIFF CNA 0.783 RACI CNA 0.556 VI-IL CNA
0.434
Gender META 0.750 VI-IL NGS 0.535 MYC
CNA 0.432
TP53 NGS 0.708 RPNI CNA 0.531 RABEP1 CNA 0.431
CACNAID CNA 0.706 APC NGS 0.527 RAFI CNA 0.430
LI IFPE6 CNA 0.700 FANCC CNA 0.524 GID4 CNA 0.428
ETV5 CNA 0.666 TP53 CNA 0.511 BCL2 NGS 0.423
Table 32: Extrahepatic Cholangio Common Bile Gallbladder Adenocarcinoma NOS -
Liver, Gallbladder, Ducts
GENE TECH IMP KDSR CNA 0.760 JAZF 1 CNA
0.686
Age META 1.000 CDKN2B CNA 0.751 ZNF217 CNA 0.685
Gender ME l'A 0.953 CACNA ID CNA 0.744 CD274 CNA
0.683
CDK12 CNA 0.868 LUFPL6 CNA 0.733 HEYI CNA 0.651
PDCD1LG2 CNA 0.847 ERG CNA 0.729 WWTRI CNA 0.649
APC NGS 0.842 TP53 NGS 0.724 CALR CNA 0.647
USP6 CNA 0.841 PTPN11 CNA 0.719 CCNEI CNA 0.644
YWHAE CNA 0.780 VEIL NGS 0.713 KRAS NGS 0.640
SETBPI CNA 0.776 CDKN2A CNA 0.710 TPM4 CNA 0.639
STAT3 CNA 0.772 FOXL2 NGS 0.686 TAF15 CNA
0.631
160
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
PRRX1 CNA 0.628 WISP3 CNA 0.617 PAX3 CNA
0.583
SPEN CNA 0 627 CBFB CNA 0 614 RABEP I CNA 0 583
LPP CNA 0.626 MDM2 CNA 0.614 EXTI CNA 0.583
MAME2 CNA 0.626 I ISP9OAA1 CNA 0.606 1131'313
CNA 0.582
FANCC CNA 0.624 RAC1 CNA 0.593 ARID1A CNA 0.580
NFTB CNA 0 620 BCT ,6 CNA 0 192 SIT7,12 CNA
0 580
KLEIL6 CNA 0.619 BCL2 CNA 0.584 ETV5 CNA 0.578
Table 33: Fallopian tube Adenocarcinoma NOS - FGTP
GENE TECH IMP WDCP CNA 0.568 CACNAID CNA 0.444
EWSR1 CNA 1.000 TP53 NGS 0.551 KMT2D CNA 0.444
CDK12 CNA 0.973 PSEPI CNA 0.545 ELF CNA 0.437
FOXL2 NGS 0.942 CDHI NGS 0.522 NF2 CNA 0.428
STAT3 CNA 0.915 KLETL6 CNA 0.506 GNAS CNA 0.428
ETV6 CNA 0.910 MKL1 CNA 0.502 CDH1 CNA 0.423
ICAT6B CNA 0.851 AFF3 CNA 0.496 e-KIT NGS 0.421
ABLI NGS 0.815 CDH11 CNA 0.496 STAT5B CNA 0.411
SMARCEI CNA 0.788 NUTMI CNA 0.495 SSI8 CNA 0.411
Gender META 0.778 CBFB CNA 0.493 ASXL1 CNA 0.410
RPN1 CNA 0.724 EP300 CNA 0.491 BMPR1A CNA 0.409
TFRC CNA 0.692 SDHC CNA 0.478 ZNF521 CNA 0.405
CCNE1 CNA 0.670 CDKN 113 CNA 0.478 LI SP6 CNA
0.401
LPP CNA 0.663 PMS2 CNA 0.475 ETV5 CNA 0.398
WWTR1 CNA 0.655 MYCN CNA 0.466 MYD88 CNA 0.397
Age META 0.629 MSH2 CNA 0.465 MAE CNA
0.396
MAP2K1 CNA 0.616 EPHBI CNA 0.463 DAXX CNA 0.394
Table 34: Fallopian tube Carcinoma NOS - FGTP
GENE TECH IMP PICALM CNA 0.556 EMSY CNA 0.466
RPNI CNA 1.000 WWTRI CNA 0.554 GMPS CNA 0.463
MUC1 CNA 0.926 LYLI CNA 0.547 BCL2 CNA 0.456
FOXL2 NGS 0.926 EP300 CNA 0.546 SPECC1 CNA 0.448
ETV5 CNA 0.919 ELK4 CNA 0.545 SLC45A3 CNA 0.448
Gender META 0.871 CARS CNA 0.540 T SCI CNA
0.447
STAT3 CNA 0.772 PDCD1LG2 CNA 0.539 TNFAIP3 CNA
0.446
TP53 NGS 0.718 FOXL2 CNA 0.522 STAT5B CNA 0.445
SMARCE1 CNA 0.708 ABL1 NGS 0.518 CDK12 CNA 0.444
NF1 CNA 0.672 NUMAI CNA 0.515 NUP214 CNA 0.440
CDHI NGS 0.668 MECOM CNA 0.514 c -KIT NGS 0.436
Age ME TA 0.658 N1RK3 CNA 0.499 N UP93
CNA 0.436
SOX2 CNA 0.625 KLEIL6 CNA 0.494 C15orf65 CNA
0.429
BCL6 CNA 0.608 RACI CNA 0.491 LPP CNA 0.426
NUP98 CNA 0.608 NDRG1 CNA 0.478 PSEP1 CNA
0.422
MAP2K1 CNA 0.593 RECQL4 CNA 0.467 VEIL CNA 0.418
161
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
MSI2 CNA 0.414 APC NGS 0.412 FGF 10 CNA
0.411
Table 35: Fallopian tube Carcinosarcoma NOS - FGTP
GENE TECH IMP WIF 1 CNA 0.481 CDK12 CNA
0.346
ASXL1 CNA 1.000 BRD4 CNA 0.466 STK11 CNA
0.345
ABL2 NGS 0.855 ERCI CNA 0.458 CNBP CNA 0.340
WDCP CNA 0.795 ATIC CNA 0.443 WISP3 CNA
0.338
MECOM CNA 0.768 HMGN2P46 CNA 0.432 FSTL3 CNA
0.333
BCL1 lA CNA 0.724 CDH1 NGS 0.428 GATA3 CNA 0.317
FOXL2 NGS 0.703 BRCAI CNA 0.397 MLLT11 CNA 0.315
KT ,F4 CNA 0.661 ARNT CNA 0.396 GNA 13 CNA
0.312
AFF3 CNA 0.643 KRAS NGS 0.375 PMS2 CNA 0.308
DDR2 CNA 0.598 MAP2K1 CNA 0.374 MLLT3 CNA 0.302
BCL9 CNA 0.592 CTLA4 CNA 0.367 KD SR CNA
0.301
NUTMI CNA 0.544 VHE NGS 0.367 FGF23 CNA 0.299
Gender META 0.531 HMGA2 CNA 0.365 KAT6A CNA 0.293
GNAS CNA 0.516 PAX3 CNA 0.364 BCL2 CNA 0.286
CDKN2A CNA 0.493 CASP8 CNA 0.354 ASPSCRI NGS 0.277
TP53 NGS 0.493 RET CNA 0.352 NOTCH2 CNA 0.276
APC NGS 0.488 CCND2 CNA 0.349 CALR CNA
0.274
Table 36: Fallopian tube Serous Carcinoma - FGTP
GENE TECH IMP CDH11 CNA 0.660 GNAS CNA 0.552
MECOM CNA 1.000 WWTRI CNA 0.643 SMARCE 1 CNA 0.550
TP53 NGS 0.955 RACI CNA 0.630 MLLT11 CNA 0.549
FOXL2 NGS 0.912 RPN 1 CNA 0.629 S1A1513 CNA
0.545
TPM4 CNA 0.847 ASXT,1 CNA 0.625 WTI CNA 0.543
Gender META 0.815 CDK12 CNA 0.613 FGFR2 CNA 0.538
CCNEI CNA 0.812 NUP214 CNA 0.604 HEY]. CNA 0.531
CBFB CNA 0.795 TSC1 CNA 0.600 KRAS NGS 0.531
EP300 CNA 0.753 SUZ12 CNA 0.596 CDX2 CNA
0.528
Age META 0.753 ETV5 CNA 0.590 CACNAID CNA
0.528
MAF CNA 0.750 ZNF217 CNA 0.580 NFI CNA 0.526
CTCF CNA 0.738 BCL9 CNA 0.578 GID4 CNA 0.519
STAT3 CNA 0.735 FSTL3 CNA 0.576 BRD4 CNA
0.516
BCL6 CNA 0.700 TET2 CNA 0.573 CRKL CNA 0.516
KLI-11,6 CNA 0.696 GNAll CNA 0.572 KLF4 CNA 0.507
TAF 15 CNA 0.675 PMS2 CNA 0.562 SRSF2 CNA
0.505
CDH1 CNA 0.671 EWSR1 CNA 0.560 AFF3 CNA 0.502
Table 37: Gastric Adenocarcinoma - Stomach
GENE TECH IMP ERG CNA 0.989 U2AF 1 CNA 0.956
Age META 1.000 FOXT ,2 NGS 0.962 CDX2
CNA 0.881
162
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CDKN2B CNA 0.866 TP53 NGS 0.738 KD SR CNA 0.703
ZNF217 CNA 0 850 IKZE1 CNA 0 737 CDK6 CNA 0 701
EXTI CNA 0.840 TCF7L2 CNA 0.736 USP6 CNA 0.697
CACNA ID CNA 0.825 EWSRI CNA 0.725 RAC 1 CNA
0.690
LL1FPL6 CNA 0.820 CBFB CNA 0.725 FGFR2 CNA 0.685
Gender MF,T A 0 815 WWTR 1 CNA 0 723 F ANCC CNA
0 679
CDHI NGS 0.807 MYC CNA 0.721 CDH11 CNA 0.678
SPECCI CNA 0.799 KLEIL6 CNA 0.719 XPC CNA
0.677
FOX01 CNA 0.795 EL13 CNA 0.717 CREB3L2 CNA 0.676
CDKN2A CNA 0.779 HLVIGN2P46 CNA 0.716 BCL2 CNA
0.673
KRAS NGS 0.751 RUNX1 CNA 0.715 FANCF CNA
0.672
FHIT CNA 0.749 PMS2 CNA 0.713 SBDS CNA 0.670
SETBPI CNA 0.745 MLLT11 CNA 0.709 CDK12 CNA 0.670
PRRX1 CNA 0.742 .IA7E1 CNA 0.704 PPARG CNA
0.669
SDC4 CNA 0.739 EBFI CNA 0.703 TGIBR2 CNA 0.665
Table 38: Gastroesophageal junction Adenocarcinoma NOS - Esophagus
GENE TECH IMP KDSR CNA 0.720 LHFPL6 CNA 0.634
ERG CNA 1.000 EWSR1 CNA 0.712 CHEK2 CNA 0.621
FOXL2 NGS 0.979 RACI CNA 0.709 PCMI CNA 0.619
U2AFI CNA 0.966 SETBPI CNA 0.702 RPNI CNA 0.618
Gender META 0.902 "TP53 NGS 0.692 HOXAll CNA
0.614
CDK12 CNA 0.896 ARID 1A CNA 0.682 TCF7L2 CNA 0.612
Age META 0.858 JAZE1 CNA 0.679 SRGAP3 CNA
0.595
ZNF217 CNA 0.830 FfifT CNA 0.676 KEHE6 CNA 0.593
CREB3L2 CNA 0.828 CTNNAI CNA 0.675 FGER2 CNA 0.592
ERBB2 CNA 0.793 CD1CN2A CNA 0.670 HOXD13 CNA 0.584
SDC4 CNA 0.778 GNAS CNA 0.662 HOXA13 CNA 0.583
CDX2 CNA 0.776 KRAS NGS 0.661 CRTC3 CNA 0.580
RUNXI CNA 0.764 IRF4 CNA 0.660 TOPI CNA 0.576
ASXLI CNA 0.742 MYC CNA 0.654 WRN CNA 0.575
EBFI CNA 0.735 ACSL6 CNA 0.638 CCNEI CNA 0.574
CACNA1D CNA 0.734 FNBP1 CNA 0.636 CDKN2B CNA 0.571
KIAA1549 CNA 0.730 CBFB CNA 0.636 CDHII CNA 0.566
Table 39: Glioblastoma - Brain
GENE TECH IMP Age META 0.870 EXTI CNA 0.756
FGER2 CNA 1.000 CDK_N2A CNA 0.820 TR_RAP CNA 0.755
EGFR CNA 0.993 PDGFRA CNA 0.809 CDKN2B CNA 0.749
FOXE2 NGS 0.953 TETI. CNA 0.801 KA16B CNA 0.741
1CF712 CNA 0.912 MYC CNA 0.791 CDK6 CNA 0.738
OLIG2 CNA 0.910 CREB3L2 CNA 0.787 SPECCI CNA 0.734
VTIIA CNA 0.896 CCDC6 CNA 0.779 JAZF 1 CNA 0.719
S13DS CNA 0.889 SOX2 CNA 0.773 NFICB2 CNA 0.713
163
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
NDRG1 CNA 0.711 Gender META 0.618 NUP214 CNA 0.570
GATA3 CNA 0 684 SPEN CNA 0 614 IDH1 NGS 0 568
TPM3 CNA 0.683 ETV1 CNA 0.605 MET CNA 0.568
N T5 C2 CNA 0.668 MCL1 CNA 0.598 'EP53 NGS 0.564
1-1MGA2 CNA 0.660 NCOA2 CNA 0.594 HEP1 CNA 0.558
KIT CNA 0658 FGF14 CNA 0588 PTEN CNA 0 550
ZNF217 CNA 0.658 SUFU CNA 0.585 PTEN NGS 0.542
FOX01 CNA 0.657 KMT2C CNA 0.582 LCP1 CNA 0.528
KIAA1549 CNA 0.633 PIK3CG CNA 0.576 LEIFPE6 CNA 0.522
Table 40: Glioma NOS - Brain
GENE TECII IMP OLIG2 CNA 0.549 KDR CNA 0.448
Age META 1.000 KIAA1549 CNA 0.537 MCL1 CNA
0.432
IDH1 NGS 0.871 CDX2 CNA 0.536 FAM46C CNA 0.425
FOXL2 NGS 0.738 VTI1A CNA 0.533 NR4A3 CNA 0.421
Gender META 0.709 ICRAS NGS 0.532 RPL22 CNA 0.420
CREB3L2 CNA 0.685 CDKN2B CNA 0.531 CDK6 CNA 0.406
SETBP1 CNA 0.657 CDKN2A CNA 0.521 MYCL CNA 0406
SOX2 CNA 0.656 PEK3R1 CNA 0.515 PDE4DEF' CNA 0.405
PDGFRA CNA 0.645 EGFR CNA 0.513 KAT613 CNA 0402
e-KIT NGS 0.640 APC NGS 0.493 1RF4 CNA 0.397
PDGFRA NGS 0.612 1CF7L2 CNA 0.482 NFKB2 CNA 0.391
TPM3 CNA 0.605 TP53 NGS 0.480 H3F3A CNA 0.387
VEIL NGS 0.594 NDRG1 CNA 0.471 LIMGA2 CNA 0.387
SPECC1 CNA 0.588 TERT CNA 0.464 KIT CNA 0.374
CDH1 NGS 0.571 MSI2 CNA 0.459 EIF4A2 CNA 0.374
STK11 CNA 0.567 SBDS CNA 0.458 EZI-12 CNA
0.372
MYC CNA 0.556 PMS2 CNA 0.449 NT5C2 CNA 0.361
Table 41: Gliosarcoma - Brain
GENE TECH IMP ETV1 CNA 0.549 NTRK2 CNA 0.448
IKZE1 CNA 1.000 KAT6D CNA 0.540 ELK4 CNA 0.425
PTEN NGS 0.916 FGFR2 CNA 0.531 FHIT CNA 0.423
FOXL2 NGS 0.899 CDK12 CNA 0.510 ABIl CNA 0.421
CDH1 NGS 0.817 SS18 CNA 0.504 SOX10 CNA 0.416
CREB3L2 CNA 0.774 EGFR CNA 0.503 Gender META 0.416
TRRAP CNA 0.732 GATA3 CNA 0.492 ERG CNA 0.415
NF1 NGS 0.713 EBF1 CNA 0.489 e-KIT NGS
0.409
CCDC6 CNA 0.703 MYC CNA 0.482 TCF7L2 CNA 0.405
JAZE1 CNA 0.619 PDGERA CNA 0.480 MSH2 NGS 0.404
TETI CNA 0.604 VI-IL NGS 0.477 VTI1A CNA 0.402
Age META 0.582 RAC1 CNA 0.474 KIAA1549 CNA
0.401
CDK6 CNA 0.575 KRAS NGS 0.466 NR4A3 CNA 0.397
MLLT10 CNA 0.550 KIF513 CNA 0.461 COX6C CNA 0.396
164
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CBFB CNA 0.390 STAT3 CNA 0.376
RBM15 CNA 0.368
FOXP1 CNA 0 380 APC NGS 0 371 IRF4
CNA 0 368
CDX2 CNA 0.378 ATP1A1 CNA 0.371
SOX2 CNA 0.360
Table 42: Head, face or neck NOS Squamous carcinoma - Head, face or neck, NOS
GENE TECH IMP TFRC CNA 0.666 TP53
NGS 0.501
Gender META 1.000 MLF 1 CNA 0.655 CRKL
CNA 0.498
ETV5 CNA 0.977 FNBP1 CNA 0.648
SETBP1 CNA 0.494
KLBL6 CNA 0.947 ARID 1A CNA 0.609 MAF CNA
0.493
NOTCH1 NGS 0.930 CDH1 CNA 0.609 FAS CNA
0.491
FOXL2 NGS 0.922 NOTCH2 NGS 0.589 NTRK2 CNA 0.485
MN1 CNA 0.898 PAFAH1B2 CNA 0.584
CREB3L2 CNA 0.484
EWSR1 CNA 0.891 SET CNA 0.563 FOXP1
CNA 0.483
LPP CNA 0.846 NDRG1 CNA 0.563
JUN CNA 0.482
NF2 CNA 0.824 CDKN2A CNA 0.560
PAX3 CNA 0.473
BCL6 CNA 0.786 GMPS CNA 0.557 FLT1
CNA 0.466
WWTR1 CNA 0.728 FGF3 CNA 0.552 GID4 CNA
0.464
Age META 0.712 CDKN2A NGS 0.535
DDX6 CNA 0.458
SOX2 CNA 0.704 TBL DCR1 CNA 0.534
FLI1 CNA 0.451
MAML2 CNA 0.697 SPEN CNA 0.523 FGF 19
CNA 0.451
ATIC CNA 0.689 KRAS NGS 0.516 TSC1
CNA 0.447
MECOM CNA 0.684 130-9 CNA 0.503 Z13T1316 CNA
0.442
Table 43: Intrahepatic bile duct Cholangiocarcinoma - Liver, Gallbladder,
Ducts
GENE TECH IMP CDKN2A CNA 0.808 FANCF CNA 0.705
MDS2 CNA 1.000 SPEN CNA 0.799 W1SP3
CNA 0.698
Age META 0.992 I J2 AF1 CNA 0.799
TGEBR2 CNA 0.696
ARID1A CNA 0.983 PBRIVI1 CNA 0.794 FOXP1 CNA
0.696
CACNA1D CNA 0.975 NOTCH2 CNA 0.760 NR4A3 CNA 0.694
FHIT CNA 0.957 ELK4 CNA 0.755 EXT1
CNA 0.692
APC NGS 0.952 ERG CNA 0.747 CBFB
CNA 0.691
MAF CNA 0.948 MSI2 CNA 0.742 ECT2L
CNA 0.686
CAMTA1 CNA 0.921 SDUB CNA 0.740 MYB
CNA 0.686
TP53 NGS 0.898 TAF15 CNA 0.733
FOXL2 NGS 0.686
MTOR CNA 0.857 CDK12 CNA 0.733
ZNF331 CNA 0.683
VI-IL NGS 0.851 FANCC CNA 0.730
ETV5 CNA 0.683
ESR1 CNA 0.851 RPL22 CNA 0.725
NTRK2 CNA 0.683
STAT3 CNA 0.834 LIFFPL6 CNA 0.725
SRGAP3 CNA 0.681
CDKN2B CNA 0.834 PTCH1 CNA 0.722 ZNF217 CNA 0.676
EZR CNA 0.832 SETBP I CNA 0.714
MYC CNA 0.673
TSHR CNA 0.829 13CL3 CNA 0.713 LPP
CNA 0.673
Gender META 0.821 KRAS NGS 0.712 EL2
CNA 0.673
165
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Table 44: Kidney Carcinoma NOS - Kidney
GENE TECH IMP CDH11 CNA 0.593 ITK CNA 0.505
EBFI CNA 1.000 CDKN 113 CNA 0.580 HOXD13 CNA 0.502
BTG 1 CNA 0.971 MAML2 CNA 0.564 SPEN CNA 0.501
FOXL2 NGS 0.931 CBFB CNA 0.560 RMI2 CNA 0497
HITT CNA 0.817 EGF23 CNA 0.558 CD74 CNA 0.494
VT-IL NGS 0.810 Age META 0.558 HOXA13 CNA 0.494
TP53 NGS 0.797 CNBP CNA 0.555 MYC CNA 0489
XPC CNA 0.772 FGF14 CNA 0.553 CREBBP CNA 0477
MAF CNA 0.765 FGER1OP CNA 0.544 c-KIT NGS 0.475
GID4 CNA 0.712 FAM46C CNA 0.540 ARIDIA CNA 0.467
MYCN CNA 0.671 WWTRI CNA 0.533 EXTI CNA 0.457
SDHAF2 CNA 0.639 MTOR CNA 0.528 KRAS NGS 0.452
Gender META 0.633 USP6 CNA 0.520 ACSL6 CNA 0.452
FAN CC CNA 0.626 TERC CNA 0.520 CKKL CNA 0.451
CTNNA1 CNA 0.624 SPECC1 CNA 0.518 RAF I CNA 0.446
FANCA CNA 0.622 PAX3 CNA 0.516 BCL9 CNA 0.439
SDUB CNA 0.608 I IMGA2 CNA 0.513 GNA13 CNA 0.437
Table 45: Kidney Clear Cell Carcinoma - Kidney
GENE TECH IMP MT ,T.T I 1 CNA 0.403
CDH I 1 CNA 0.264
VIAL NGS 1.000 PRCC CNA 0.382 ABL2
CNA 0.264
FOXL2 NGS 0.743 Age META 0.366
HIVIGN2P46 CNA 0.261
TP53 NGS 0.618 MAF CNA 0.357 CBLB
CNA 0.260
EBF1 CNA 0.577 KRAS NGS 0.349 TSHR
CNA 0.259
VI-IL CNA 0.569 APC NGS 0.338 YWHAE
CNA 0.254
XPC CNA 0.535 USP6 CNA 0.325 SETD2
NGS 0.254
MYD88 CNA 0.517 CDICN2A CNA 0.319 PPARG CNA 0.252
Gender META 0.495 PTPN 11 CNA 0.312 ZNE217
CNA 0.247
c-KIT NGS 0.490 MCL1 CNA 0.298 TR1M33
NGS 0.247
11K CNA 0.481 1L21R CNA 0.296 SE1BP1
CNA 0.245
SRGAP3 CNA 0.446 RPNI CNA 0.291 CACNAID CNA 0.244
MDM4 CNA 0.431 ICDSR CNA 0.289 BTGI
CNA 0.242
RAF1 CNA 0.430 PAX3 CNA 0.275 CYP2D6
CNA 0.240
ARNT CNA 0.428 MUC1 CNA 0.273 NUTM2B
CNA 0.239
CTNNA1 CNA 0.411 STAT5B NGS 0.265 FANCD2 CNA 0.238
TGEBR2 CNA 0.405 MAX CNA 0.265 BCL2 CNA 0.238
Table 46: Kidney Papillary Renal Cell Carcinoma - Kidney
GENE TECH IMP c-KIT NGS 0.899 SRSF2 CNA 0.763
MSI2 CNA 1.000 TP53 NGS 0.890 TDI-11
NGS 0.739
Gender META 0.945 CREB3L2 CNA 0.873 GNA13 CNA 0.717
FOXL2 NGS 0.914 ULF CNA 0.825 AURKB CNA 0.661
166
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
VEIL NGS 0.652 IRF4 CNA 0.478 CBFB CNA 0.397
CDX2 CNA 0 619 STAT3 CNA 0 477 CRKL CNA 0 392
APC NGS 0.592 BRAT' CNA 0.476 COX6C CNA 0.391
CNA 0.591 EXTI CNA 0.452 DDX5 CNA 0.387
SNX29 CNA 0.584 NUP93 CNA 0.451 BCL7A CNA 0.387
KRAS NGS 0 168 SOX10 CNA 0 440 SR SF3 CNA 0 381
H3F3B CNA 0.561 TAF15 CNA 0.428 ERCC4 CNA 0.380
TPM3 CNA 0.559 RECQL4 CNA 0.425 MAP2K4 CNA 0.367
PERI. CNA 0.525 Age META 0.419 SMARCEI CNA 0.366
KIAA1549 CNA 0.513 PRCC CNA 0.419 MLLT11 CNA 0.366
YWHAE CNA 0.505 RNF 213 CNA 0.411 PRKAR1A CNA 0.366
NKX2-I CNA 0.491 SPEN CNA 0.411 BRIP1 CNA 0.365
CLTC CNA 0.488 RMI2 CNA 0.402 ASXLI CNA 0.365
Table 47: Kidney Renal Cell Carcinoma NOS - Kidney
GENE TECII MU' ITK CNA 0.683 TSCI CNA 0.566
VIIL NGS 1.000 FLII CNA 0.666 NUP214 CNA 0.563
RAFI CNA 0.977 CDHII CNA 0.660 K1AA1549 CNA
0.560
EBF1 CNA 0.971 CACNA1D CNA 0.654 HSP9OAA1 CNA
0.559
MAF CNA 0.968 FANCC CNA 0.648 TPM3 CNA 0.556
CTNNA1 CNA 0.939 ACSL6 CNA 0.647 ABL2 CNA 0.554
FOXE2 NGS 0.916 TRIM27 CNA 0.637 APC NGS 0.548
TP53 NGS 0.898 FANCF CNA 0.630 SPEN CNA
0.544
e-KIT NGS 0.870 FNBPI CNA 0.623 ETV5 CNA
0.540
SRGAP3 CNA 0.852 C131413 CNA 0.605 BTG1 CNA
0.535
MUC1 CNA 0.831 PDGFRA NGS 0.598 ZNF217 CNA
0.532
XPC CNA 0.826 CDX2 CNA 0.598 CD74 CNA 0.518
Gender META 0.807 MLLT11 CNA 0.594 SNX29 CNA
0.513
N1JP93 CNA 0.760 KRAS NGS 0.577 PPAR_G CNA
0.510
VUL CNA 0.740 CREB3L2 CNA 0.574 RANBP17 CNA 0.508
MTOR CNA 0.710 FANCD2 CNA 0.573 ARIIGAP26 CNA
0.507
Age META 0.709 FHIT CNA 0.573 ARFRPI NGS 0.505
Table 48: Larynx NOS Squamous carcinoma - Head, Face or Neck, NOS
GENE TECH IMP EGFR CNA 0.727 P1K3 CA CNA 0.592
TGFBR2 CNA 1.000 USP6 CNA 0.723 LPP CNA 0.589
Gender META 0.979 WWTR1 CNA 0.698 N THE CNA 0.561
FOXL2 NGS 0.949 VEIL NGS 0.697 CREB3L2 CNA 0.557
ETV5 CNA 0.896 RAF 1 CNA 0.683 Age META
0.557
KLHL6 CNA 0.803 SOX2 CNA 0.682 CACNA1D CNA 0.551
13CL6 CNA 0.787 14 0 XPI CNA 0.673 TP53
NGS 0.534
HMGN2P46 CNA 0.755 SETD2 CNA 0.660 GNAS CNA 0.533
YWHAE CNA 0.749 NF2 CNA 0.644 FHIT CNA 0.528
TERC CNA 0.745 MYD88 CNA 0.601 KRAS NGS
0.525
167
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
MECOM CNA 0.511 TOP1 CNA 0.438 CAMTA1 CNA 0.394
GID4 CNA 0 511 EWSR1 CNA 0 433 FANCC
CNA 0 390
TBL1X121 CNA 0.474 ZNE217 CNA 0.419 CI IEK2 CNA
0.389
ELT3 CNA 0.473 EXTI CNA 0.415 CDKN2A
NUS 0.385
SPECC1 CNA 0.470 XPC CNA 0.412 CDH1 CNA 0.384
CDKN2A CNA 0 466 CTNNT11 CNA 0 402 RI TNX1 CNA 0
371
RABEP1 CNA 0.445 PPAR.G CNA 0.396 SETBP1 CNA 0.369
Table 49: Left Colon Adenocarcinoma NOS - Colon
GENE TECH IMP CDH1 CNA 0.595 TP53 NGS 0.485
CDX2 CNA 1.000 ZNF217 CNA 0.585 COX6C CNA 0482
APC NGS 0.989 ZMYM2 CNA 0.585 CDKN2A CNA 0.479
FLT1 CNA 0.824 CDKN2B CNA 0.575 LCP1 CNA 0.478
FOXL2 NGS 0.821 RB1 CNA 0.566 ETV5 CNA 0.475
FLT3 CNA 0.793 GNAS CNA 0.557 PDE4DEP CNA 0.467
SETBP1 CNA 0.773 HOXA9 CNA 0.548 PMS2 CNA 0.465
BCL2 CNA 0.738 SMAD4 CNA 0.547 U2AF 1 CNA 0.463
KRAS NGS 0.733 SOX2 CNA 0.543 AURKA CNA 0460
Age META 0.708 WWTR1 CNA 0.536 RAC1 CNA 0.453
LITFPL6 CNA 0.696 JAZT 1 CNA 0.530 EBF 1 CNA 0452
ZNF521 CNA 0.664 Gender META 0.518 BCL6 CNA 0.447
ASXL1 CNA 0.649 ERCC5 CNA 0.505 SPECC1 CNA 0.444
SDC4 CNA 0.649 HOXA 1 1 CNA 0.498 EP300 CNA 0.443
KDSR CNA 0.644 MSI2 CNA 0.497 SS18 CNA 0.439
CDK8 CNA 0.644 FOX01 CNA 0.492 PTC1-11 CNA 0.434
TOP1 CNA 0.621 WRN CNA 0.487 H0XA13 CNA 0.433
Table 50: Left Colon Mucinous Adenocarcinoma - Colon
GENE TECH IMP TOP1 CNA 0.586 MYD88 CNA 0.421
APC NGS 1.000 WWTR1 CNA 0.582
MUC1 CNA 0.414
FOXL2 NGS 0.909 SDHAF2 CNA 0.563 CACNA1D CNA 0.412
CDX2 CNA 0.902 CDKN2A CNA 0.527
WISP3 CNA 0.403
KRAS NGS 0.845 HOXA9 CNA 0.525
AFF3 CNA 0.396
LITFPL6 CNA 0.814 SETBP1 CNA 0.522 MLLT11 CNA 0.395
CDK8 CNA 0.688 SOX2 CNA 0.519 RNF213
CNA 0.391
Age META 0.661 ABL1 CNA 0.510 SDHB
CNA 0.384
Gender META 0.658 CAMTA1 CNA 0.497 ASXL1 CNA 0.384
FLT1 CNA 0.657 CDK_N2B CNA 0.494
TP53 NGS 0.382
FLT3 CNA 0.638 SYK CNA 0.484 ZNF217
CNA 0.379
ETV5 CNA 0.609 PTCH1 CNA 0.472 FGF 14
CNA 0.378
FAN CC CNA 0.605 VI-IL NGS 0.455 NE2 CNA 0.377
SMAD4 NGS 0.594 MLLT3 CNA 0.446 CDK12 CNA 0.376
SET CNA 0.592 BCL2 CNA 0.439 CCNE1
CNA 0.370
N TRK2 CNA 0.586 MAX CNA 0.430 1RS2 CNA 0.368
168
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
RPN1 CNA 0.366 ERG CNA 0.365 GATA3 CNA 0.359
Table 51: Liver Hepatocellular Carcinoma NOS - Liver, Gallbladder, Ducts
GENE TECH IMP COX6C CNA 0.742 ETV6 CNA 0.651
PRCC CNA 1.000 NSDI CNA 0.741 FLTI CNA 0.637
FILE CNA 0.992 HMGN2P46 CNA 0.732 KRAS NGS
0.636
FOXL2 NGS 0.981 YWHAE CNA 0.727 ABL2 CNA 0.636
SDHC CNA 0.955 TRH\ 426 CNA 0.713 HIST1H4I CNA
0.636
Gender META 0.901 SPEN CNA 0.707 HEYI CNA
0.636
BCL9 CNA 0.894 CACNA1D CNA 0.706 BTG1 CNA 0.633
ET K4 CNA 0.863 TPM3 CNA 0.704 AFF 1 CNA
0.633
ERG CNA 0.852 H3F3A CNA 0.698 ZNE703 CNA
0.631
MLLT11 CNA 0.834 ACSL6 CNA 0.691 TP53 NGS 0.630
FGFRI CNA 0.814 NCOA2 CNA 0.678 APC NGS
0.627
WRN CNA 0.813 TRIM27 CNA 0.675 CDH11 CNA 0.617
Age META 0.802 USP6 CNA 0.674 CDICN2A CNA
0.613
CAMTAI CNA 0.771 LI-IFPL6 CNA 0.669 MCLI CNA 0.612
FANCF CNA 0.763 MTOR CNA 0.669 KLHL6 CNA
0.610
PCM1 CNA 0.762 EXTI CNA 0.667 IRF4 CNA 0.601
NSD3 CNA 0.746 MECOM CNA 0.651 ADGRA2 CNA 0.600
Table 52: Lung Adenocarcinoma NOS - Lung
GENE TECH IMP HMGN2P46 CNA 0.578 CDK12 CNA
0.543
NKX2-1 CNA 1.000 FANCC CNA 0.577 FLI1 CNA 0.542
Age META 0.890 PPARG CNA 0.575 YWHAE CNA 0.540
1PM4 CNA 0.707 CDKN 213 CNA 0.574 RAC 1 CNA
0.540
TERT CNA 0.685 SDHC CNA 0.572 XPC CNA 0.535
KRAS NGS 0.671 TL7R CNA 0.571 APC NGS 0.529
CALR CNA 0.667 EGE10 CNA 0.571 TP53 NGS
0.525
MUC1 CNA 0.660 CACNA1D CNA 0.571 WWTR1 CNA 0.522
Gender META 0.656 KDSR CNA 0.562 FHIT CNA
0.522
VIII, NGS 0.655 TPM3 CNA 0.559 JAZE1 CNA
0.520
NEKBIA CNA 0.625 ASXL1 CNA 0.557 IKZE1 CNA
0.519
USP6 CNA 0.624 BCL2 CNA 0.555 NUTM2B CNA 0.516
FOXAI CNA 0.608 SLC34A2 CNA 0.554 CCNEI CNA
0.515
CDKN2A CNA 0.607 EWSR1 CNA 0.550 CDKN1B CNA 0.515
LI-IFPL6 CNA 0.606 WISP3 CNA 0.547 ELK4 CNA 0.514
ESR1 CNA 0.588 PTCH1 CNA 0.547 LIFR CNA
0.514
FGFR2 CNA 0.585 MLLT11 CNA 0.547 SYK CNA 0.513
PMS2 CNA 0.579 MCLI CNA 0.546 LRP113 NGS
0.512
13CL9 CNA 0.579 SRGAY3 CNA 0.543 5
SETBPI CNA 0.578 CDX2 CNA 0.543
169
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Table 53: Lung Adenosquamous Carcinoma - Lung
GENE TECH IMP FNBPI CNA 0.614 GNAS CNA 0.511
Age META 1.000 Eliff CNA 0.599 KIT
CNA 0.509
F0XL2 NGS 0.928 NKX2-I CNA 0.583 PPARG CNA 0.509
TERT CNA 0.848 MYD88 CNA 0.573 SOX2 CNA 0.503
CDKN2A CNA 0.795 ERBB3 CNA 0.557 CDX2 CNA 0.498
LRP IB NGS 0.788 RHOH CNA 0.556 C15orf65 CNA 0.496
RUNX1 CNA 0.756 PTPN11 CNA 0.549 GNA13 CNA 0.496
FLU. CNA 0.756 TP53 NGS 0.549 EPHA3 CNA 0.483
CALR CNA 0.746 LIFFPL6 CNA 0.546 APC NGS 0.472
ELK4 CNA 0.709 CDK4 CNA 0.541 MLHI CNA 0.470
CACNAID CNA 0.707 NTRK2 CNA 0.541 RAFI CNA 0.470
CDKN2B CNA 0.699 FOXA1 CNA 0.537 RPNI CNA 0.468
EL7R CNA 0.695 SDI ID CNA 0.536 MLLT11
CNA 0.465
MAML2 CNA 0.666 MAX CNA 0.533 VI-[L NGS 0.462
FANCC CNA 0.645 CBFB CNA 0.528 FTMGA2 CNA 0.457
HIST1H3B CNA 0.634 USP6 CNA 0.520 MECOM CNA 0.457
Gender META 0.631 KRAS NGS 0.512 FLTI CNA
0.456
Table 54: Lung Carcinoma NOS - Lung
GENE TECH IMP XPC CNA 0.647 IT ,7R CNA 0.603
Age META 1.000 SRGAP3 CNA 0.642 HNIGN2P46 CNA
0.597
CDX2 CNA 0.870 FHIT CNA 0.641 CDK4 CNA 0.594
FOXA1 CNA 0.798 FOXL2 NGS 0.640 SETBP1 CNA 0.594
VIAL NGS 0.777 TERT CNA 0.628 FLT1 CNA 0.592
KRAS NGS 0.756 ARID 1A CNA 0.627 RBM15 CNA
0.591
NKX2-1 CNA 0.742 LRPIB NGS 0.625 USP6 CNA 0.590
APC NGS 0.741 BRD4 CNA 0.620 TRIIVI27 CNA
0.583
11'53 NUS 0.731 MSI2 CNA 0.620 CDK12 CNA
0.581
CALR CNA 0.728 FGF10 CNA 0.616 TGFBR2 CNA 0.580
1PM4 CNA 0.726 CDKN 213 CNA 0.614 RAC 1 CNA
0.577
CTNNA1 CNA 0.720 LI LFPL6 CNA 0.613 PPAR.G CNA
0.574
CACNAID CNA 0.719 RPNI CNA 0.613 FANCC CNA 0.573
Gender META 0.687 PBXI CNA 0.608 CDKN1B CNA 0.569
FGFR2 CNA 0.672 PCM1 CNA 0.607 MYC CNA 0.566
ATP 1A1 CNA 0.672 WWTRI CNA 0.606 STAT3 CNA 0.566
CDKN2A CNA 0.660 FLT3 CNA 0.605 MLLT11 CNA 0.564
Table 55: Lung Mucinous Adenocarcinoma - Lung
GENE TECH IMP CDKN2B CNA 0.687 ASXLI CNA 0.624
KRAS NGS 1.000 TP53 NGS 0.636 Gender META
0.614
Age META 0.880 CDKN2A CNA 0.634 IGFIR CNA
0.596
FOXL2 NGS 0.818 TPM4 CNA 0.626 C15orf65 CNA
0.593
170
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
BCL6 CNA 0.587 CTNNAI CNA 0.483 CCNEI CNA 0.448
CRICL CNA 0 586 FLII CNA 0 481 ACSL6 CNA 0
446
11MGN2P46 CNA 0.550 AR1IGAP26 CNA 0.477 BTG1 CNA
0.443
El311 CNA 0.534 CR1C3 CNA 0.474 CD111 CNA
0.437
ETV5 CNA 0.526 EI1F4A2 CNA 0.472 EPHBI
CNA 0.436
RPN1 CNA 0 519 CBFFI CNA 0 469 STK]] NGS
0428
LPP CNA 0.518 NUTM2B CNA 0.468 TPM3 CNA 0.427
EXTI CNA 0.512 ZNF521 CNA 0.467 GID4 CNA
0.419
SE113P1 CNA 0.512 CDK6 CNA 0.457 N UTM 1 CNA
0.417
LHFPL6 CNA 0.511 FANCC CNA 0.456 TR1M33 NGS 0.416
MAP2K1 CNA 0.509 FOXA1 CNA 0.456 EP300 CNA
0.416
ELK4 CNA 0.501 MLF 1 CNA 0.450 FLT3 CNA
0.413
SDHC CNA 0.484 APC NGS 0.450 MUCI CNA 0.408
Table 56: Lung Neuroendocrine Carcinoma NOS - Lung
GENE TECII MU' RPL22 CNA 0.681 MSI2 CNA
0.580
NKX2-1 CNA 1.000 FANCC CNA 0.680 FOX01 CNA
0.578
FOXL2 NGS 0.955 MYD88 CNA 0.677 FLTI CNA
0.574
CAMTAI CNA 0.870 PRF1 CNA 0.653 CDKN2C CNA 0.562
VIIL CNA 0.813 FANCD2 CNA 0.650 ZNF217 CNA
0.553
PBRM1 CNA 0.801 RB1 NGS 0.645 MYC CNA 0.528
1GE1312.2 CNA 0.798 13Hil CNA 0.640 13CL2 CNA
0.515
KDSR CNA 0.752 HMGN2P46 CNA 0.634 CACNAID CNA
0.487
SFPQ CNA 0.751 TCF7L2 CNA 0.631 FLU. CNA 0.481
EANCG CNA 0.746 LHEPL6 CNA 0.626 RA141 CNA 0.481
FOXA1 CNA 0.739 WWTRI CNA 0.623 CDKN IB CNA 0.477
SUFU CNA 0.731 FHIT CNA 0.622 CD1CN2A CNA 0.463
SETBP1 CNA 0.730 Age META 0.616 CDK4 CNA 0.462
PRRXI CNA 0.702 MYCL CNA 0.612 DDX5 CNA 0.461
XPC CNA 0.701 HIST1H3B CNA 0.603 BCL9 CNA
0.460
BAPI CNA 0.691 PPARG CNA 0.599 FLT3 CNA
0.451
FGFR2 CNA 0.682 Gender META 0.598 CDX2 CNA
0.451
Table 57: Lung Non-small Cell Carcinoma - Lung
GENE TECH IMP FLT1 CNA 0.722 CDKN2A CNA 0.650
Age META 1.000 Gender META 0.706 FGFR2
CNA 0.647
NKX2-1 CNA 0.831 LTIFPL6 CNA 0.697 BCL9 CNA 0.643
TP53 NGS 0.827 HMGN2P46 CNA 0.692 KRAS NGS
0.625
CDX2 CNA 0.800 FLT3 CNA 0.682 CALR CNA 0.624
TERT CNA 0.786 EW SRI CNA 0.677 PTCHI CNA
0.621
1PM4 CNA 0.783 FANCC CNA 0.667 CDKN 213 CNA
0.620
VI-EL NGS 0.764 FOXA1 CNA 0.662 GNA13 CNA
0.611
CTNNA1 CNA 0.741 FGF10 CNA 0.661 LRP1B NGS
0.603
APC NGS 0.735 CACNA ID CNA 0.660 IKZ141 CNA
0.603
171
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
ARID1A CNA 0.602 LIFR CNA 0.576 PPARG CNA 0.564
MSI2 CNA 0 601 EBFI CNA 0 575 WWTRI CNA 0 556
SRSF2 CNA 0.599 IL7R CNA 0.573 KLIrL6 CNA
0.556
SET13P I CNA 0.593 MCL I CNA 0.572 HEY 1 CNA
0.550
RACI CNA 0.591 SPECCI CNA 0.569 MUCI CNA 0.547
MITT CNA 0 190 VTTI A CNA 0 167 SR GAP3 CNA 0
546
TGFBR2 CNA 0.590 BRD4 CNA 0.566 HVIGA2 CNA 0.546
ZNF2 1 7 CNA 0.579 CCNEI CNA 0.565 BTGI CNA
0.545
Mil CNA 0.577 PAX8 CNA 0.565
XPC CNA 0.576 IRF4 CNA 0.565
Table 58: Lung Sarcomatoid Carcinoma - Lung
GENE TECH IMP BTGI CNA 0.618 FCRL4 CNA 0.509
Age META 1.000 FANCC CNA 0.617 JAK2 CNA 0.502
YWHAE CNA 0.964 PRCC CNA 0.614 MAME2 CNA 0.494
FOXL2 NGS 0.930 LRPIB NGS 0.602 WRN NGS 0.486
RACI CNA 0.915 PBXI CNA 0.600 FANCF CNA
0.481
KRAS NGS 0.857 c-KIT NGS 0.588 KDM5C NGS 0.472
RHOH CNA 0.855 SPECCI CNA 0.587 SRSF2 CNA 0.466
CNBP CNA 0.788 FOXPI CNA 0.586 CCNEI CNA
0.461
CD274 CNA 0.775 ELK4 CNA 0.584 GNAS NGS 0.455
RPN 1 CNA 0.769 ERAS CNA 0.573 H3F3A CNA
0.455
CTNNA1 CNA 0.737 MECOM CNA 0.570 LHFPL6 CNA 0.451
POTI NGS 0.731 CREB3L2 CNA 0.563 IRF4 CNA 0.449
PDCD1LG2 CNA 0.707 C13L CNA 0.556 EH CNA 0.446
TP53 NGS 0.689 FHIT CNA 0.544 GMPS CNA 0.443
GSK3B CNA 0.662 VTII A CNA 0.541 FLII CNA
0.441
CRKL CNA 0.655 WWTR1 CNA 0.533 TRRAP CNA 0.440
Gender META 0.624 CTCF CNA 0.518 APC NGS
0.440
Table 59: Lung Small Cell Carcinoma NOS - Lung
GENE TECH IMP SRGAP3 CNA 0.701 BAPI CNA 0.618
RBI NGS 1.000 ARID 1A CNA 0.699 KD SR CNA
0.616
NKX2-1 CNA 0.924 SS18 CNA 0.699 BCL9 CNA 0.612
FOXL2 NGS 0.918 RBI CNA 0.693 MYCL CNA 0.605
SETBPI CNA 0.892 CBFB CNA 0.691 SOX2 CNA 0.595
VI-IL CNA 0.832 PBRM1 CNA 0.688 HA 4GN2P46 CNA
0.588
MSI2 CNA 0.829 CDK_N2C CNA 0.685 HIST1H3B CNA
0.576
TGFER2 CNA 0.807 FOXAI CNA 0.672 LHFPL6 CNA 0.567
MITE CNA 0.797 CDKN 2B CNA 0.665 KLHL6 CNA
0.560
XP C CNA 0.793 13CL2 CNA 0.656 PPARG CNA
0.550
FOXPI CNA 0.778 Age META 0.652 FH1T CNA 0.548
CACNAID CNA 0.743 FLT3 CNA 0.640 FOX01 CNA 0.535
SMAD4 CNA 0.729 PBX 1 CNA 0.625 DEK CNA 0.532
172
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
TTL CNA 0.527 JAK1 CNA 0.509 SFPQ CNA 0.498
Gender META 0 518 FGER2 CNA 0 509 CDH11 CNA 0
498
FLT1 CNA 0.515 MYD88 CNA 0.507 DAXX CNA 0.497
IIIS111141 CNA 0.514 JUN CNA 0.505 FAN CD2 CNA 0.496
Table 60: Lung Squantous Carcinoma - Lung
GENE TECH IMP FGF10 CNA 0.717 SRGAP3 CNA 0.652
Age META 1.000 BTG1 CNA 0.716 GNAS CNA
0.649
SOX2 CNA 0.971 TERT CNA 0.708 MAF CNA 0.645
FOXL2 NGS 0.917 WWTR1 CNA 0.700 CALR CNA 0.645
CACNA 1D CNA 0.899 EWSR1 CNA 0.700 BCI6 CNA 0.644
KITIL6 CNA 0.895 ETV5 CNA 0.698 EBF 1 CNA
0.644
CTNNA1 CNA 0.865 MECOM CNA 0.692 EL7R CNA 0.637
XPC CNA 0.826 TGFBR2 CNA 0.691 FGER2 CNA 0.632
CDKN2A CNA 0.791 Gender META 0.685 U2AF1 CNA
0.629
LPP CNA 0.789 PPARG CNA 0.678 BCL11A CNA 0.629
TP53 NGS 0.786 FLT1 CNA 0.677 HMGN2P46 CNA
0.627
TFRC CNA 0.783 CDX2 CNA 0.674 ERG CNA 0.625
CRKL CNA 0.750 FOXPI CNA 0.669 HMGA2 CNA 0.624
FRIT CNA 0.748 SPECC1 CNA 0.669 EP300 CNA 0.622
CDKN2B CNA 0.740 RAC1 CNA 0.664 NF2 CNA 0.621
RPN 1 CNA 0.739 LTIFPL6 CNA 0.657 ACSL6 CNA
0.617
FLT3 CNA 0.728 RAF 1 CNA 0.655 ELK4 CNA
0.617
Table 61: Meninges Meningioma NOS - Brain
GENE TECH IMP N1RK2 CNA 0.609 MAP2K4 CNA 0.478
CHEK2 CNA 1.000 HOXA9 CNA 0.601 MYC CNA 0.477
MYCL CNA 0.986 CDKN2C CNA 0.601 ELK4 CNA 0.473
THRAP3 CNA 0.959 RPL22 CNA 0.599 CTNNA 1 CNA 0.471
FOXE2 NGS 0.948 USP6 CNA 0.584 FANCF CNA 0.466
EWSR1 CNA 0.905 ZNF217 CNA 0.566 SDBB CNA 0.465
E13171 CNA 0.863 LI LEPL6 CNA 0.553 c-KIT NGS 0.458
TP53 NGS 0.857 EP300 CNA 0.550 SPECC 1 CNA 0.457
MPL CNA 0.823 Gender META 0.538 PDGFRB CNA 0.455
PMS2 CNA 0.734 NTRK3 CNA 0.538 GAS7 CNA 0.435
NF2 CNA 0.678 HOXA13 CNA 0.537 ZBTB16 CNA 0.435
SPEN CNA 0.661 RAC1 CNA 0.518 U2AF 1 CNA 0.433
Age META 0.640 ERG CNA 0.517 RABEP1 CNA 0.427
STIL CNA 0.639 LCK CNA 0.505 FHIT CNA 0.425
HLF CNA 0.636 EC12L CNA 0.493 CSF3R CNA 0.413
CDH11 CNA 0.628 MTOR CNA 0.484 Y WHAE CNA 0.408
FLI1 CNA 0.610 SETBP1 CNA 0.483 IGF1R CNA 0.406
173
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Table 62: Nasopharynx NOS Squamous Carcinoma - Head, Face or Neck, NOS
GENE TECH IMP PTPN11 CNA 0.673 W1F1 CNA 0.537
CTCE CNA 1.000 E1V6 CNA 0.641 TSC1 CNA 0.534
FOXL2 NGS 0.955 C15 orf65 CNA 0.632 USP6 CNA
0.523
TP53 NGS 0.870 JAZF 1 CNA 0.621 REL CNA 0.509
SOX2 CNA 0.842 BCL6 CNA 0.612 CDK4 CNA 0.506
GNAS CNA 0.838 TFRC CNA 0.612 NUTM1 CNA 0.500
CDH1 CNA 0.834 KDSR CNA 0.598 CYP2D6 CNA 0496
RPN1 CNA 0.833 MAML2 CNA 0.586 CDX2 CNA 0481
Gender META 0.828 MLLT11 CNA 0.584 LHFPL6 CNA 0.478
KMT2A CNA 0.770 CBL CNA 0.580 SDHB CNA 0.477
ASXL1 CNA 0.739 BUB1B CNA 0.563 KRAS NGS 0.460
MAP3K1 NGS 0.713 ABL2 NGS 0.553 RB1 NGS 0.453
TGEBR2 CNA 0.703 EPIT131 CNA 0.550 PMS2 CNA 0.447
SDHD CNA 0.690 APC NUS 0.547 WRN CNA 0.441
Age META 0.690 VHT, NGS 0.541 EGER CNA 0.441
CDICN2B CNA 0.685 BTG1 CNA 0.540 CCDC6 CNA 0.432
CBEB CNA 0.680 PCM1 CNA 0.538 MECOM CNA 0.428
Table 63: Oligodendroglioma NOS - Brain
GENE TECH IMP .TUN CNA 0.485 SPECC1 CNA 0.351
TDH1 NGS 1.000 CD79A CNA 0.463 ATP 1A1 CNA
0.343
Age META 0.871 MYCL CNA 0.452 e-KIT NGS
0.339
FOXL2 NGS 0.846 NUP93 CNA 0.450 VIAL NGS 0.339
MPL CNA 0.689 PDE4DIP CNA 0.432 HIST1H4I CNA
0.321
BCL3 CNA 0.651 RAD51 CNA 0.432 PAFAH1B2 CNA
0.320
FAM46C CNA 0.640 CTCF CNA 0.399 MSI NGS 0.320
ACSL6 CNA 0.624 TP53 NGS 0.396 EXT1 CNA 0.316
RHOH CNA 0.591 PALB2 CNA 0.372 AXL CNA
0.312
MLLT11 CNA 0.574 ERCC1 CNA 0.359 APC NGS 0.309
JAK1 CNA 0.564 PPP2R1A CNA 0.358 NEKBIA CNA 0.309
ZNE331 CNA 0.560 CSE3R CNA 0.358 CACNA1D CNA 0.306
OLIG2 CNA 0.560 ZNE217 CNA 0.356 RPL22 CNA 0.305
ATP 1A1 NGS 0.529 CBL CNA 0.354 ELK4 CNA 0.304
MCL1 CNA 0.498 MYC CNA 0.352 MSI2 CNA 0.301
Gender META 0.486 ELT1 CNA 0.352 CCNE1 CNA
0.299
KLK2 CNA 0.486 SETBP1 CNA 0.351 ARID1A CNA 0.298
Table 64: Oligodendroglioma Anaplastic - Brain
GENE TECH IMP FOXL2 NGS 0.916 RPL22 CNA 0.694
IDH1 NGS 1.000 ZNF703 CNA 0.844 THRAP3 CNA 0.647
CCNE1 CNA 0.933 JUN CNA 0.763 BCL3 CNA 0.619
Age META 0.917 SFPQ CNA 0.752 ZNF331 CNA 0.610
174
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
SDHB CNA 0.610 MUTYH CNA 0.373 SYK CNA 0.334
MPL CNA 0 582 CDKN2C CNA 0 373 C1TEK2 CNA 0 332
MCLI CNA 0.564 AFF3 CNA 0.369 EWSRI CNA 0.325
ERCCI CNA 0.555 MYCL CNA 0.366 PTEN NOS 0.323
CDHI NGS 0.482 NR4A3 CNA 0.359 U2AF 1 CNA 0.321
ERG CNA 0 464 ET K4 CNA 0 318 SETBP1 CNA 0 319
TNERSF14 CNA 0.436 ACSL6 CNA 0.358 MDM4 NGS 0.318
NF2 CNA 0.414 MUCI CNA 0.354 SPECCI CNA 0.316
e-KIT NGS 0.410 APC NGS 0.349 AT131A1 CNA 0.316
GR1N2A CNA 0.409 CSF3R CNA 0.348 CBLC CNA 0.312
RPL5 CNA 0.406 MLLT11 CNA 0.347 AR1D1A CNA 0.307
USP6 CNA 0.391 TETI NGS 0.345 SOXIO CNA 0.304
ZNF217 CNA 0.378 KRAS NGS 0.341 TP53 NGS 0.302
Table 65: Ovary Adenocarcinoma NOS - FGTP
GENE TECII MU' CDH11 CNA 0.660 CNBP CNA
0.607
Age META 1.000 MLLT11 CNA 0.659 NUP214 CNA 0.605
Gender META 0.986 SUZI2 CNA 0.657 SOX2 CNA
0.604
MECOM CNA 0.875 CDKN2B CNA 0.652 GATA3 CNA 0.604
KLITL6 CNA 0.834 CDKN2A CNA 0.649 BCL2 CNA 0.603
APC NGS 0.827 HMGN2P46 CNA 0.649 ETV5 CNA
0.601
MYC CNA 0.784 1PM4 CNA 0.644 GNAS CNA 0.600
BCL6 CNA 0.761 RPNI CNA 0.644 PAX8 CNA 0.596
TP53 NGS 0.760 CDKN2C CNA 0.644 CDHI NGS 0.595
ERAS NGS 0.752 WT1 CNA 0.642 C15orf65 CNA 0.595
SPECCI CNA 0.748 SETBP1 CNA 0.640 ZNF331 CNA 0.594
VIAL NGS 0.740 BCL9 CNA 0.640 CDKN IB CNA 0.594
WWTR1 CNA 0.728 FANCC CNA 0.637 EWSR1 CNA 0.593
ZN17217 CNA 0.720 EP300 CNA 0.633 NDRG1 CNA
0.591
CBFB CNA 0.703 NTRK2 CNA 0.633 KD SR CNA
0.584
MUC1 CNA 0.700 LI IFPL6 CNA 0.630 EBFI CNA
0.583
CDH1 CNA 0.691 CACNA1D CNA 0.625 PMS2 CNA 0.582
c-KIT NGS 0.680 ARID 1A CNA 0.625 MSI2 CNA 0.581
CCNEI CNA 0.678 CDX2 CNA 0.624 ASXLI CNA
0.579
KAT6B CNA 0.671 CTCF CNA 0.624
GID4 CNA 0.665 RAC I CNA 0.611
Table 66: Ovary Carcinoma NOS - FGTP
GENE TECH IMP KIBL6 CNA 0.824 SUZ12 CNA
0.768
Age META 1.000 '11353 NGS 0.815 JAZE1
CNA 0.766
Gender META 0.996 CDH11 CNA 0.797 N141 CNA
0.756
MECOM CNA 0.973 RACI CNA 0.794 ETV5 CNA 0.754
FOXL2 NGS 0.875 CDHI CNA 0.788 CBFB CNA 0.753
HNIGN2P46 CNA 0.826 RPN I CNA 0.769 ERAS NGS 0.753
175
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
ZNF217 CNA 0.748 PMS2 CNA 0.689 MUCI CNA 0.646
ETVI CNA 0 747 TET2 CNA 0 681 EWSRI CNA 0
645
LI IFPL6 CNA 0.732 C15 orf65 CNA 0.673 CDKN2B CNA 0.645
MYC CNA 0.731 FANCC CNA 0.669 FOXAI CNA 0.644
MAF CNA 0.731 CDKN2A CNA 0.668 PDE4DEF' CNA 0.640
AR TD1 A CNA 0 716 CCNE1 CNA 0 664 APC NGS 0 639
TAF 15 CNA 0.715 NUP98 CNA 0.656 MCLI CNA
0.638
WWTRI CNA 0.715 HOXD13 CNA 0.651 CDK12 CNA 0.630
EP300 CNA 0.700 CACNA ID CNA 0.650 CDX2 CNA
0.628
CARS CNA 0.694 N1IJP214 CNA 0.650 PRCC CNA
0.627
FGFR2 CNA 0.693 FANCF CNA 0.648
SPECCI CNA 0.690 CTCF CNA 0.647
Table 67: Ovary Carcinosarcoma - FGTP
GENE TECH IMP MYCN CNA 0.666 BCL2 NGS 0.571
ASXLI CNA 1.000 AFFI CNA 0.662 PIK3 CA NGS 0.570
STK11 CNA 0.951 TRH\ 427 CNA 0.649 STAT3 CNA
0.568
FOXL2 NGS 0.945 ALK CNA 0.644 CRKL CNA 0.566
MECOM CNA 0.925 RACI CNA 0.642 HVIGN2P46 CNA
0.561
ZN1F384 CNA 0.917 BCLIIA CNA 0.640 FGER1 CNA 0.553
Gender META 0.895 CBFB CNA 0.640 ERBB2 CNA
0.552
"TP53 NGS 0.822 PRRXI CNA 0.633 14 GI423
CNA 0.550
ETV5 CNA 0.815 LfIFPL6 CNA 0.630 ELK4 CNA 0.538
GNAS CNA 0.795 CCND2 CNA 0.630 MAX CNA
0.533
Age META 0.783 HMGA2 CNA 0.622 CCNE 1 CNA
0.533
WDCP CNA 0.778 IVIAF CNA 0.619 FANCF CNA
0.532
EP300 CNA 0.762 CDHI CNA 0.606 PMS2 CNA 0.529
FGF6 CNA 0.715 TCF3 CNA 0.602 VEGFA CNA
0.527
FSTL3 CNA 0.708 ETV6 CNA 0.600 KLIIL6 CNA
0.524
EWSRI CNA 0.691 NUTMI CNA 0.592 AURKA CNA 0.522
PBXI CNA 0.672 DDR2 CNA 0.584 NCOAI CNA
0.516
Table 68: Ovary Clear Cell Carcinoma - FGTP
GENE TECH IMP MECOM CNA 0.639 CDKN2B CNA 0.589
ZNF217 CNA 1.000 NF2 CNA 0.635 PAX8 CNA 0.588
Age META 0.965 KAT6A CNA 0.625 FANCC CNA 0.587
FOXL2 NGS 0.935 TRIM 27 CNA 0.623 PLAGI CNA
0.586
ARIDIA NGS 0.920 ERBB3 CNA 0.611 MED12 NGS
0.582
TP53 NGS 0.887 EXTI CNA 0.610 TSCI CNA 0.581
PIK3 CA NGS 0.853 ERCC5 CNA 0.608 CDKN 2A CNA 0.574
S1A13 CNA 0.826 NCOA2 CNA 0.597 CCNE 1 CNA
0.570
Gender META 0.810 FHIT CNA 0.594 ACKR3 CNA
0.567
ULF CNA 0.755 STAT5B CNA 0.593 NR4A3 CNA 0.563
EP300 CNA 0.743 CDK12 CNA 0.592 BCL2 CNA
0.560
176
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
WWTR1 CNA 0.558 FLI1 CNA 0.514 EWSR1 CNA
0.503
IRS2 CNA 0 553 NUTM1 CNA 0 510
SUFU CNA 0 502
RACI CNA 0.537 BRCAI CNA 0.509 PBXI
CNA 0.500
PDCD LEG2 CNA 0.531 13TG1 CNA 0.508 I IMGN 2P46 CNA
0.494
HSP90AB1 CNA 0.531 MSI2 CNA 0.508 CDH11 CNA
0.490
CBT , CNA 0 123 NI1P214 CNA 0 103
APC NGS 0 489
Table 69: Ovary Endometrioid Adenocarcinoma - FGTP
GENE TECH IMP CDKN2A CNA 0.604
CRKL CNA 0.526
Age META 1.000 MDM4 CNA 0.596 FLI1
CNA 0.526
FOXL2 NGS 0.951 AI,K CNA 0.594 NUP98
CNA 0.526
CTNNBI NGS 0.936 VTII A CNA 0.582 CBL
CNA 0.524
ARIDIA NGS 0.879 ZNF331 CNA 0.581 BCL6
CNA 0.524
CHIC2 CNA 0.848 CCDC6 CNA 0.578 PTEN
NGS 0.522
FGFR2 CNA 0.834 LIIFPL6 CNA 0.575
MYCL CNA 0.517
Gender META 0.809 BCL9 CNA 0.562 RACI
CNA 0.517
FANCF CNA 0.791 HMGN2P46 CNA 0.560
ARIDIA CNA 0.516
MUC1 CNA 0.774 CTNNA1 CNA 0.555
BCL1 IA CNA 0.515
ELK4 CNA 0.675 CDK12 CNA 0.547 TETI
CNA 0.509
TP53 NGS 0.667 CACNA ID CNA 0.541
FRIT CNA 0.506
PBXI CNA 0.662 ZNF384 CNA 0.540
CDKN IB CNA 0.501
C131413 CNA 0.656 HOXA13 CNA 0.535
S1A13 CNA 0.499
AFF3 CNA 0.655 PPAR.G CNA 0.534 CDKN2B
CNA 0.494
MAF CNA 0.655 WWTRI CNA 0.532
SETBPI CNA 0.489
H314313 CNA 0.605 RIK3CA NGS 0.528
U2AE1 CNA 0.488
Table 70: Ovary Granulosa Cell Tumor - FGTP
GENE TECH IMP CYP2D6 CNA 0.319 NR4A3 CNA 0.248
FOXL2 NGS 1.000 CHIEK2 CNA 0.317 CACNAID CNA 0.244
EWSR1 CNA 0.475 RIVII2 CNA 0.317 MN1
CNA 0.242
Gender META 0.455 GID4 CNA 0.312 BCR
CNA 0.241
NT2 CNA 0.454 SOX2 CNA 0.306 ALDII2
CNA 0.237
MYH9 CNA 0.450 CRKL CNA 0.301 CEBPA
CNA 0.231
TP53 NGS 0.425 HIVIGA2 CNA 0.290
IDH1 NGS 0.229
Age META 0.422 PATZI CNA 0.281 TSCI
CNA 0.225
CBFB CNA 0.408 SOX10 CNA 0.276
PTCH1 CNA 0.225
MKL1 CNA 0.388 ZNF217 CNA 0.276
APC NGS 0.222
BCL3 CNA 0.377 EP300 CNA 0.274 KRAS
NGS 0.220
TSTIR CNA 0.368 PTPN11 CNA 0.270
BLM NGS 0.215
SPECCI CNA 0.355 ATE' CNA 0.267 ERG NUS
0.215
Mil CNA 0.346 PCMI CNA 0.266 HEE
NGS 0.215
SMARCBI CNA 0.346 IGF IR CNA 0.266 NUP214 CNA 0.212
FANCC CNA 0.331 CCND2 CNA 0.261 PTEN NGS
0.211
SOCS1 CNA 0.324 FETI CNA 0.254 HOXA13
CNA 0.205
177
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Table 71: Ovary High-grade Serous Carcinoma - FGTP
GENE TECH IMP ETVI CNA 0.615 ABLI NGS 0.472
MECOM CNA 1.000 ALDH2 NGS 0.607 AK13 NGS 0.463
MLL111 NGS 0.987 AURKB NGS 0.606 Gender META 0.459
KLI31.6 CNA 0.984 ACSL3 NGS 0.589 HOXA9 CNA 0.448
ETV5 CNA 0.942 CBEB NGS 0.589 RPNI CNA 0.445
HIST1H4I NGS 0.927 H3F3B NGS 0.584 CBFB CNA 0.434
BTGI NGS 0.881 WWT121 CNA 0.577 ATP lA 1 NGS 0.433
EZR CNA 0.791 ALK NGS 0.554 RAP1GDS1 CNA 0.430
C15orf65 NGS 0.779 BRCAI NGS 0.554 MAF CNA 0.429
BCL2L11 NGS 0.776 AKTI NGS 0.547 ASXLI CNA 0.407
LIIVIGN2P46 NGS 0.769 BCL6 CNA 0.536 GSK3B CNA 0.402
AKT2 NGS 0.728 ACSL6 NGS 0.522 LEVI CNA
0.390
ARERPI NGS 0.671 DDIT3 NGS 0.520 WRN CNA 0.384
13AP1 NGS 0.658 ARHGAP26 NGS 0.502 140X01 CNA
0.376
BCI2 NGS 0.637 ABT,2 NGS 0.500 SUZ12 CNA
0.372
ZNE384 CNA 0.635 NEI CNA 0.486 GNAll NGS 0.366
TAE15 CNA 0.615 TERC CNA 0.472 PIK3CA CNA 0.366
Table 72: Ovary Low-grade Serous Carcinoma - FGTP
GENE TECH IMP GNA I 1 NGS 0.544 SDHC CNA
0.358
RPL22 CNA 1.000 H3F3A CNA 0.484 ERAS NGS
0.358
L1MGN2P46 NGS 0.898 G1D4 CNA 0.477 LIIVIGN2P46 CNA
0.352
CDKN2A CNA 0.780 ARERPI NGS 0.466 AURKB NGS 0.350
CDKN2B CNA 0.752 TNERSE14 CNA 0.464 COX6C CNA
0.343
WRN CNA 0.712 DDIT3 NGS 0.456 ABL1 NGS
0.330
HOOK3 CNA 0.667 BCL2 NGS 0.451 ACKR3 NGS 0.329
PCMI CNA 0.631 PSIPI CNA 0.431 SBDS CNA
0.325
BCL2L1 I NUS 0.613 ALDH2 NGS 0.424 TCL1A CNA
0.321
H3F3B NGS 0.604 MCL1 CNA 0.423 CACNA ID CNA
0.321
13TG1 NGS 0.598 AK12 NGS 0.404 MLLT3 CNA
0.318
HIST 1II4I NGS 0.584 C15 orf65 NGS 0.403 USP6 CNA
0.318
PLAGI CNA 0.578 MLLT11 CNA 0.400 SDHB CNA 0.312
NUTM2B CNA 0.562 PRKDC CNA 0.395 ABL2 NGS 0.312
SOX2 CNA 0.558 MAP2K1 CNA 0.389 AC SL6 NGS
0.310
WISP3 CNA 0.547 CDK4 NGS 0.387 AKTI NGS 0.303
R1JNXIT1 CNA 0.545 NRAS NGS 0.362 RBM15 CNA 0.299
Table 73: Ovary Mucinous Adenocarcinoma - FGTP
GENE TECH IMP Gender META 0.784 CDKN2B CNA
0.579
KRAS NGS 1.000 CDKN2A CNA 0.628 YWHAE CNA 0.569
Age META 0.941 HMGN2P46 CNA 0.620 TPM4 CNA
0.566
FOXL2 NGS 0.896 FUS CNA 0.618 BCL6 CNA 0.565
178
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
LHFPL6 CNA 0.558 C15 orf65 CNA 0.464 STAT3 CNA
0.424
SRGAP3 CNA 0 538 AS X_Ll CNA 0 456 ENT1BA CNA
0 418
ZNI1217 CNA 0.534 APC NGS 0.447 EB111 CNA 0.418
e-K!! NGS 0.524 N UTMI CNA 0.447 RACI CNA
0.416
HEY1 CNA 0.523 BCL2 CNA 0.443 U2AF 1 CNA
0.415
FNBP1 CNA 0 511 KT HT A CNA 0 440 WT1
CNA 0 411
CDKN2C CNA 0.506 MSI NGS 0.438 CDX2 CNA 0.410
CTNNA1 CNA 0.502 NTRK2 CNA 0.436 CRKL CNA 0.409
CACNAID CNA 0.495 RMI2 CNA 0.434 ERB134 CNA
0.406
SETBPI CNA 0.481 BRCA2 CNA 0.434 SDC4 CNA 0.404
SOX2 CNA 0.474 PDCD1LG2 CNA 0.432 SPECCI CNA 0.401
KDM5 C NGS 0.471 FHIT CNA 0.432 CDHI CNA 0.394
MYC CNA 0.470 PPARG CNA 0.425 TP53 NGS
0.389
Table 74: Ovary Serous Carcinoma - FGTP
GENE TECII IA/P FANCF CNA 0.689 MLLT11 CNA
0.639
WTI CNA 1.000 PAX8 CNA 0.686 HIMGN2P46 CNA
0.634
Gender META 0.988 CDHI CNA 0.685 NDRGI CNA
0.634
Age META 0.933 P1K3CA NGS 0.672 MYC CNA 0.633
EP300 CNA 0.821 CDKN1B CNA 0.671 CTCF CNA 0.632
MECOM CNA 0.819 ARID 1A CNA 0.669 e-KIT NGS 0.629
APC NGS 0.791 RAC 1 CNA 0.660 HOOK3 CNA
0.626
RPN1 CNA 0.778 TAF15 CNA 0.657 CDKN2A CNA 0.625
CBFB CNA 0.773 CDH11 CNA 0.653 SUZ12 CNA
0.616
1PM4 CNA 0.754 JAZ141 CNA 0.650 ZNE384 CNA
0.616
TP53 NGS 0.748 ETVI CNA 0.649 CDKN2B CNA 0.614
KRAS NGS 0.735 FOXL2 NGS 0.646 SMARCE1 CNA
0.608
MUC1 CNA 0.729 CRKL CNA 0.645 BCL9 CNA 0.606
KLIIL6 CNA 0.718 ETV6 CNA 0.644 STAT3 CNA
0.602
PMS2 CNA 0.712 CDX2 CNA 0.643 ZNF331 CNA
0.601
MAI' CNA 0.709 CDK12 CNA 0.640 ETV5 CNA
0.596
BCL6 CNA 0.698 CCNEI CNA 0.639 EWSRI CNA
0.593
Table 75: Pancreas Adenocarcinoma NOS - Pancreas
GENE TECH IMP USP6 CNA 0.588 FOX01 CNA 0.546
KRAS NGS 1.000 1RF4 CNA 0.584 BCL2 CNA 0.541
APC NGS 0.731 TP53 NGS 0.584 SPEN CNA 0.537
Age META 0.706 SPECC1 CNA 0.582 LITFPL6 CNA 0.536
SETBPI CNA 0.676 CACNA ID CNA 0.577 HIMGN2P46 CNA
0.536
CDK.N2A CNA 0.649 C11113 CNA 0.567 Y WHAE CNA 0.524
FAN CF CNA 0.633 MDS2 CNA 0.561 ARIDIA CNA 0.513
CDKN2B CNA 0.621 Gender META 0.561 CDX2 CNA
0.511
ERG CNA 0.610 SMAD4 CNA 0.559 RABEPI CNA 0.509
KDSR CNA 0.594 SMAD2 CNA 0.556 PDCD1LG2 CNA 0.508
179
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CRTC3 CNA 0.507 SDHB CNA 0.493 HOXA9
CNA 0.477
MAE CNA 0 504 RACI CNA 0 493 EXTI
CNA 0 476
WWTRI CNA 0.502 FLII CNA 0.490 ELK4 CNA 0.475
VI IL NGS 0.502 CDI111 CNA 0.482 CR1(1,
CNA 0.469
CDHI CNA 0.500 EWSRI CNA 0.481 RPNI
CNA 0.468
TGFBR 2 CNA 0 497 MST2 CNA 0 479 A SXT .1 CNA
0468
EP300 CNA 0.493 FHIT CNA 0.478 PMS2
CNA 0.468
Table 76: Pancreas Carcinoma NOS - Pancreas
GENE TECH IMP FCRL4 CNA 0.483 PBXI CNA 0.443
KRA S NGS 1.000 RPN I CNA 0482 BTG1
CNA 0.440
FOXL2 NGS 0.850 ACSL6 CNA 0.481 ERG CNA 0.440
CDKN2A CNA 0.748 IRF4 CNA 0.475 EBF 1 CNA 0.436
FHIT CNA 0.724 TNERSF17 CNA 0.472
TFRC CNA 0.435
CDKN2B CNA 0.617 ASXLI CNA 0.471 CDH11 CNA 0.432
SETBPI CNA 0.595 CBFB CNA 0.466 JAZF 1 CNA
0.431
Gender META 0.591 KLUL6 CNA 0.465 ZNF217 CNA 0.425
TP53 NGS 0.585 CTNNAI CNA 0461
CTCF CNA 0.424
YWHAE CNA 0.576 FAM46C CNA 0.456 MYC CNA 0.424
Age META 0.576 EP300 CNA 0454 GNAS
CNA 0.423
PDE4D1T' CNA 0.553 BCLIIA CNA 0.454 ESRI CNA 0.421
RPL22 CNA 0.547 ZN14521 CNA 0.452
N142 CNA 0.418
RMI2 CNA 0.530 USP6 CNA 0.452 CDHI
CNA 0.416
CAMTA1 CNA 0.528 IL6ST CNA 0.450 HEY1 CNA 0.409
FSTL3 CNA 0.507 FANCE CNA 0.447
CACNAID CNA 0.407
CREB3L2 CNA 0.499 MAML2 CNA 0.444 SOX2 CNA 0.404
Table 77: Pancreas Mucinous Adenocarcinoma - Pancreas
GENE TECH IMP STAT3 NGS 0.372 FAM46C
CNA 0.277
KRAS NGS 1.000 ZNE331 CNA 0.369 C15orf65
CNA 0.273
APC NGS 0.568 CDKN2A CNA 0.369
AFF4 NGS 0.268
FOXL2 NGS 0.516 TP53 NGS 0.367 SDI LB
CNA 0.264
ASXLI CNA 0.489 RMI2 CNA 0.356 MSI2
CNA 0.264
JUN CNA 0.487 ERCC3 NGS 0.340 TAL2
CNA 0.257
Gender META 0.455 VHL NGS 0.332 RUNXI
CNA 0.247
GNAS NGS 0.442 CDHI NGS 0.332 SOCSI
CNA 0.242
FOX01 CNA 0.436 NTRK2 CNA 0.327 COX6C
CNA 0.235
NUTM I CNA 0.429 CDK_N2B CNA 0.327 SMAD4 CNA 0.235
STK11 NGS 0.425 RACI CNA 0.314 CREB3L2
CNA 0.234
ACKR3 NGS 0.406 HIVIGN2P46 CNA 0.311
RPNI CNA 0.232
CACNA ID CNA 0.386 LLK4 CNA 0.306 KD SR CNA 0.229
MUC1 CNA 0.382 Age META 0.305 EBF 1
CNA 0.228
SETBPI CNA 0.379 FANCE CNA 0.302 FANCC CNA
0.226
ARID I A CNA 0.373 JAKI CNA 0.281 FCRIA CNA 0.224
180
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
USP6 CNA 0.224 EZR CNA 0.222 CCDC6
CNA 0.222
Table 78: Pancreas Neuroendocrine Carcinoma - Pancreas
GENE TECH IMP ZNF217 CNA 0.722 MYC CNA 0.592
JAZE1 CNA 1.000 BTGI CNA 0.718 DICERI CNA 0.589
GATA3 CNA 0.992 FCRL4 CNA 0.695 MN CNA 0.576
FOXL2 NGS 0.973 EBFI CNA 0.678 CD79A NGS 0.567
WWTRI CNA 0.962 NOTCH2 CNA 0.677 SPECCI CNA 0.565
Age META 0.904 STAT5B CNA 0.672 ITK CNA 0.541
MECOM CNA 0.874 INTIBA CNA 0.665 ETV1 CNA 0.530
FOXA 1 CNA 0.856 TCT ,1A CNA 0.657 KD SR CNA 0.525
EPHA3 CNA 0.825 KLI-11,6 CNA 0.646 PMS2 CNA 0.522
MELT3 CNA 0.774 SMAD4 CNA 0.635 CTCF CNA 0.509
BCL6 CNA 0.770 MLF 1 CNA 0.632 FGFR2 CNA 0.508
LHEPL6 CNA 0.769 TP53 NGS 0.631 FLT1 CNA 0.508
PTPRC CNA 0.764 SETBP1 CNA 0.630 DDIT3 CNA 0.507
CDK4 CNA 0.761 SOX2 CNA 0.610 NR4A3 CNA 0.507
PTPNII CNA 0.754 TCEAI CNA 0.609 I1L7R CNA 0.507
LPP CNA 0.749 GMPS CNA 0.600 RUNX1 CNA 0.505
TFRC CNA 0.730 Gender META 0.596 H3F3A CNA 0.505
Table 79: Parotid Gland Carcinoma NOS - Head, Face or Neck, NOS
GENE TECH IMP Age META 0.690 CREBBP CNA
0.530
ERBB2 CNA 1.000 PTEN NGS 0.686 FUS
CNA 0.526
FOXL2 NGS 0.974 CDICN2A CNA 0.676 MDM2 CNA 0.509
CACNA I D CNA 0.864 VEGEA CNA 0.673 GNA13 CNA 0.507
CRTC3 CNA 0.829 T,HEPT,6 CNA 0.671 GNAS CNA
0.505
RMI2 CNA 0.801 IGF 1R CNA 0.658 NTRK3 CNA
0.504
TRRAP CNA 0.793 TFRC CNA 0.638 TP53
NGS 0.504
RUNX1 CNA 0.782 SMAD2 CNA 0.632 CYLD CNA
0.496
LRPIB NOS 0.764 HOXD13 CNA 0.621 ASXLI CNA 0.494
RPL22 CNA 0.754 CDII11 CNA 0.614 GRIN2A CNA 0.494
Gender META 0.749 CDH1 NGS 0.609 CDK6
CNA 0.480
SBDS CNA 0.719 HEY1 CNA 0.591 ELK4 CNA
0.479
NDRGI NGS 0.715 ACKR3 CNA 0.580 VTII A CNA
0.474
CBFB CNA 0.701 SOX2 CNA 0.565 PRDM1 CNA
0.473
GATA3 CNA 0.696 c-KIT NGS 0.560 ZRSR2 NGS
0.460
NSD3 CNA 0.695 HMGA2 CNA 0.535 BCL I lA CNA 0.456
APC NGS 0.693 TL7R NGS 0.535 JAZF 1
CNA 0.456
Table 80: Peritoneum Adenocarcinoma NOS - FGTP
GENE TECH IMP Gender META 0.948 EWSR1 CNA 0.869
Age META 1.000 FOXT ,2 NGS 0.921 ETV5 CNA 0.830
181
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
EPHA3 CNA 0.828 SETBPI CNA 0.648 TRIM27 CNA 0.564
GMT'S CNA 0 826 TNF384 CNA 0 635 MAME2 CNA 0 556
SYK CNA 0.821 SOX2 CNA 0.632 MELT 1 1 CNA 0.555
CCNEI CNA 0.799 LI IFPL6 CNA 0.628 1PM4 CNA 0.551
TP53 NGS 0.768 JAZE1 CNA 0.626 TAF15 CNA 0.550
F ANCC CNA 0 767 RAC] CNA 0618 CCNT)1 CNA 0 548
CDHI CNA 0.742 NUP214 CNA 0.615 NSDI CNA 0.548
MECOM CNA 0.741 PRCC CNA 0.615 RNF213 NGS 0.545
LPP CNA 0.734 CALR CNA 0.612 13CL9 CNA 0.540
FGER2 CNA 0.734 CHEK2 CNA 0.602 MYC CNA 0.537
FNBPI CNA 0.679 KIITL6 CNA 0.586 WWTRI CNA 0.535
TFRC CNA 0.677 PTCHI CNA 0.582 MEDI2 NGS 0.535
MAF CNA 0.676 WTI CNA 0.582 CAMTAI CNA 0.531
NTRK2 CNA 0.675 ERCC4 CNA 0.577 BCI A CNA 0.531
RPNI CNA 0.653 CDKN2A CNA 0.571 FIIIT CNA 0.526
Table 81: Peritoneum Carcinoma NOS - FGTP
GENE TECH IMP WR_N CNA 0.631 APC
NGS 0.537
Age META 1.000 CDK6 CNA 0.628
STAT5B CNA 0.534
FOXL2 NGS 0.940 CDH11 CNA 0.624
ETVI CNA 0.530
Gender META 0.875 VHE CNA 0.604 KRAS
NGS 0.522
1E53 NGS 0.777 LPP CNA 0.597 1PM4
CNA 0.522
KAT6B CNA 0.772 SRGAP3 CNA 0.592
CHEK2 CNA 0.521
WWTRI CNA 0.757 GMPS CNA 0.589 BCL6 CNA
0.521
CDK12 CNA 0.732 MLL'1'3 CNA 0.579
HIVIGN2P46 CNA 0.519
RPNI CNA 0.687 CDHI CNA 0.571
PAFAHI B2 CNA 0.505
MLFI CNA 0.681 NUTM2B CNA 0.570
CRTC3 CNA 0.505
TFRC CNA 0.679 EP300 CNA 0.558
LITFPL6 CNA 0.500
RAC1 CNA 0.679 INT IBA CNA 0.557
SOX2 CNA 0.497
XPC CNA 0.675 MECOM CNA 0.550
FGER2 CNA 0.496
NTRK2 CNA 0.669 CTCF CNA 0.549
MAME2 CNA 0.494
NF1 CNA 0.662 SUZ12 CNA 0.548
PAX5 CNA 0.493
EWSR1 CNA 0.660 HOXA9 CNA 0.545 KD
SR CNA 0.483
EXT I CNA 0.647 ETV5 CNA 0.545
NDRGI CNA 0.479
Table 82: Peritoneum Serous Carcinoma - FGTP
GENE TECH IMP TP53 NGS 0.933 PMS2 CNA 0.853
TPM4 CNA 1.000 TAF15 CNA 0.902
WWTR1 CNA 0.845
BCL6 CNA 0.984 RACI CNA 0.877 ETVI
CNA 0.838
FOXE2 NGS 0.978 CDK12 CNA 0.875 CDHI CNA 0.822
SUZ12 CNA 0.978 EP300 CNA 0.866
LPP CNA 0.807
Gender META 0.973 CDKN2B CNA 0.865 ASXLI CNA 0.794
Age META 0.955 MECOM CNA 0.865
CDH11 CNA 0.793
CTCF CNA 0.940 RPN 1 CNA 0.863 KLHL6
CNA 0.793
182
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
FANCA CNA 0.786 EWSR1 CNA 0.726 EBF 1 CNA 0.681
CBFB CNA 0 786 TAL2 CNA 0 716 TFRC
CNA 0 677
FANCF CNA 0.784 CDKN2A CNA 0.713 SMARCEI CNA 0.676
ETV5 CNA 0.778 GMPS CNA 0.711 CCNE
1 CNA 0.671
NUP93 CNA 0.766 NFI CNA 0.710 WTI CNA
0.668
EGER 2 CNA 0 760 NI TP214 CNA 0 706 7NE217 CNA 0 666
JAZF I CNA 0.753 CRKL CNA 0.702 MLFI
CNA 0.665
FHIT CNA 0.740 SPECCI CNA 0.700
ETV6 CNA 0.664
CYP2D6 CNA 0.738 KLE4 CNA 0.700 BCL9 CNA
0.664
Table 83: Pleural Mesothelioma NOS - Lung
GENE TECII PVIP ASXL1 CNA 0.684 PBRMI
CNA 0.488
Age META 1.000 FOXPI CNA 0.658 CDX2
CNA 0.487
FOXL2 NGS 0.954 RACI CNA 0.630 CALR
CNA 0.484
EWSRI CNA 0.938 FSTL3 CNA 0.619 BAPI
CNA 0.484
CD1CN2B CNA 0.909 ARID 1A CNA 0.602 ITK CNA 0.484
TP53 NGS 0.849 NUTM2B CNA 0.550
CDHI CNA 0.483
EPHA3 CNA 0.848 LYLI CNA 0.543 CDHII
CNA 0.482
CDKN2A CNA 0.834 EGFR CNA 0.528 KRAS NGS 0.479
Gender META 0.834 CDKN2C CNA 0.526 c-
KIT NGS 0.477
WTI CNA 0.825 HMGN2P46 CNA 0.520
NEM CNA 0.473
MAE CNA 0.822 W1SP3 CNA 0.516 MAP2K1
CNA 0.471
EBFI CNA 0.778 KDR CNA 0.513
C15orf65 CNA 0.468
NF2 CNA 0.754 NTRK3 CNA 0.504 VIIL
NGS 0.465
PRDMI CNA 0.714 RUNX111 CNA 0.502
FGE 10 CNA 0.461
MSI2 CNA 0.712 FGER2 CNA 0.500 ULF
CNA 0.460
ACSL6 CNA 0.707 TPM4 CNA 0.497 ERG
CNA 0.454
EP300 CNA 0.698 FAM46C CNA 0.491
CREB3L2 CNA 0.452
Table 84: Prostate Adenocarcinoma NOS - Prostate
GENE TECH IMP GNAll NGS 0.562 ELK4 CNA 0.430
Gender META 1.000 NCOA2 CNA 0.537 SDC4 CNA 0.430
FOXAI CNA 0.875 LCPI CNA 0.531 MAF CNA
0.411
PTEN CNA 0.825 PTCH1 CNA 0.530 FGF
14 CNA 0.404
KRAS NGS 0.783 c-KIT NGS 0.510 RB1
CNA 0.403
Age META 0.697 TP53 NGS 0.500
CACNAID CNA 0.401
KLK2 CNA 0.693 CDKN1B CNA 0.491
CDKN2B CNA 0.394
FOX01 CNA 0.675 HOXAll CNA 0.466 HEY1 CNA 0.388
FANCA CNA 0.664 FGFR2 CNA 0.457 TP53 CNA 0.384
GA1A2 CNA 0.663 ID H1 NGS 0.456 COX6C CNA
0.381
APC NGS 0.623 1RF4 CNA 0.454 CDX2
CNA 0.377
LIIFPL6 CNA 0.608 PCMI CNA 0.452 SOX 1 0
CNA 0.376
ETV6 CNA 0.580 CDKN2A CNA 0.442
BRAE NGS 0.374
ERCC3 CNA 0.579 VI-It NGS 0.431 SRGAP3 CNA
0.373
183
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
FGFRI CNA 0.371 CREBBP CNA 0.366 MLHI CNA 0.364
CDHI 1 CNA 0 370 TGFBR2 CNA 0 366 PRDMI CNA 0 363
SPECC1 CNA 0.368 CLIFB CNA 0.365 I IOXA13 CNA
0.355
Table 85: Rectosigmoid Adenocarcinoma NOS - Colon
GENE TECH IMP Age META 0.561 SS18 CNA 0.449
APC NGS 1.000 R_AC1 CNA 0.550 CAMTA1 CNA 0.440
CDX2 CNA 0.877 TOP1 CNA 0.540 BRAF NGS 0.437
FOXL2 NGS 0.771 CDKN2A CNA 0.532 NSD3 CNA 0.437
FLT3 CNA 0.769 FOX01 CNA 0.523 MTOR CNA 0.432
BGL2 CNA 0.750 KRAS NGS 0.521 CTCF CNA 0.420
FLT1 CNA 0.705 ZMYM2 CNA 0.518 SOX2 CNA 0.419
SETBP1 CNA 0.704 SDC4 CNA 0.515 Wit NGS 0.418
ZNF521 CNA 0.657 ZNF217 CNA 0.510 PRRX1 CNA 0.412
CDK8 CNA 0.645 CDKN2B CNA 0.500 GNAS CNA 0.405
ICDSR CNA 0.638 BRCA2 CNA 0.492 PIK3 CA NGS 0.404
LHFPL6 CNA 0.628 HOXAll CNA 0.491 FANCF CNA 0.398
ASXLI CNA 0.603 Gender META 0.488 MECOM CNA 0.397
SMAD4 CNA 0.584 PMS2 CNA 0.477 LCPI CNA 0.397
RB1 CNA 0.578 FCRL4 CNA 0.475 HOXA13 CNA 0.396
MALT1 CNA 0.568 WWTR1 CNA 0.471 CARS CNA 0.396
HOXA9 CNA 0.563 130-2 NGS 0.454 ERCC5 CNA 0.393
Table 86: Rectum Adenocarcinoma NOS - Colon
GENE TECH IMP TP53 NGS 0.521 PTCHI CNA 0.438
APC NGS 1.000 SPECC1 CNA 0.519 E0X01 CNA 0.435
CDX2 CNA 0.904 SMAD4 CNA 0.514 SS18 CNA 0.427
SETBPI CNA 0.745 AMER1 NGS 0.503 WWTRI CNA 0.424
KRAS NGS 0.738 FOXL2 NGS 0.503 CCNEI CNA
0.424
ASXLI CNA 0.701 ERCC5 CNA 0.499 USP6 CNA
0.423
FLT3 CNA 0.698 GNAS CNA 0.498 JAZF 1 CNA
0.422
Age META 0.669 CDKN2B CNA 0.493 CAMTAI CNA 0.421
SDC4 CNA 0.663 RBI CNA 0.481 CDKN2A CNA 0.417
KDSR CNA 0.649 HOXA9 CNA 0.458 EXTI CNA 0.417
FLT1 CNA 0.649 VHL NGS 0.456 ERG CNA 0.416
ZNF217 CNA 0.631 HOXAll CNA 0.455 CDHI CNA 0.415
CDK8 CNA 0.614 TOPI CNA 0.449 ENBP1 CNA
0.413
BCL2 CNA 0.601 MALT1 CNA 0.443 BRCA2 CNA
0.413
LHFPL6 CNA 0.583 EBF1 CNA 0.442 NSD2 CNA 0.412
Gender META 0.545 RAC 1 CNA 0.441
HIVIGN2P46 CNA 0.406
ZNE521 CNA 0.536 13CL9 CNA 0.441 ABL1 CNA
0.403
Table 87: Rectum Mucinous Adenocarcinoma - Colon
184
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
GENE TECH IMP SDC4 CNA 0.498 PDGFRA CNA
0.395
KRAS NGS 1 000 RPL22 CNA 0 471 EPHA3 CNA 0
394
APC NGS 0.917 SOX2 CNA 0.469 VTII A
CNA 0.394
EOXL2 NUS 0.887 PPARG CNA 0.466 RI\ 412 CNA
0.394
CDKN2A CNA 0.665 CTCF CNA 0.456 NDRGI CNA
0.394
CDKN2B CNA 0 643 T ,HEPT ,6 CNA 0 416 I TSP6
CNA 0 393
NUP214 CNA 0.641 ARFRPI CNA 0.449 WWTRI CNA 0.389
GPTIN CNA 0.625 TAL2 CNA 0.441 EXTI
CNA 0.384
TSCI CNA 0.605 SETBPI CNA 0.441 PMS2 CNA
0.380
KLF4 CNA 0.554 SYK CNA 0.440 RAFI
CNA 0.369
CDHI NGS 0.550 CACNA ID CNA 0.415 TGFBR2 CNA 0.363
PRKDC CNA 0.542 LIER CNA 0.413 SMAD4 NGS
0.360
Gender META 0.538 NTRK2 CNA 0.411 ARIDIA CNA 0.359
A SPSCR1 NGS 0.521 TP53 NGS 0.403 JAK2
CNA 0.355
Age META 0.519 1RS2 CNA 0.403 CCND2 CNA
0.352
CDX2 CNA 0.512 KDSR CNA 0.400 HOXD13 CNA
0.352
BCL2 CNA 0.503 FHIT CNA 0.397 TRIN127 CNA
0.350
'Fable 88: Iletroperitoneum DeditTerentiated Liposarcoma - ['GIP
GENE TECH IMP USP6 CNA 0.120 KAT6B CNA
0.079
CDK4 CNA 1.000 MUC1 CNA 0.116 ZNF521 CNA 0.079
MDM2 CNA 0.760 S1A15B NGS 0.114 1E2 CNA
0.079
RET CNA 0.379 BCL9 CNA 0.112 KDM5C NGS
0.079
SBDS CNA 0.334 PAX3 CNA 0.112 TRS2
CNA 0.078
ASXLI CNA 0.245 1P53 NGS 0.107 13CE6
CNA 0.077
VTIIA CNA 0.216 FGF4 CNA 0.106 ELK4
CNA 0.076
KMT2D CNA 0.212 SOX2 CNA 0.091 MNX1 CNA 0.070
GRIN2A CNA 0.178 RABEPI CNA 0.090 WRN CNA
0.068
IIMGA2 CNA 0.173 PTEN CNA 0.090 CDK6 CNA 0.068
PTCHI CNA 0.156 FUBPI NGS 0.089 AFDN CNA 0.068
CYP2D6 CNA 0.156 RAD51 CNA 0.089 POU2AF 1 CNA 0.068
BMPRIA CNA 0.145 MLLT11 CNA 0.089 ESRI NGS
0.067
CDX2 CNA 0.137 ACKR3 NGS 0.089 ELN CNA
0.067
GID4 CNA 0.134 ZNF 217 CNA 0.089 NTRK2 CNA 0.067
ETVI CNA 0.134 NF2 CNA 0.087 NUMAI CNA
0.067
GATA2 CNA 0.128 Age META 0.082 SRC
CNA 0.067
Table 89: Retroperitoneum Leiomyosarcoma NOS - FGTP
GENE TECH IMP SPECCI CNA 0.817 BCL11A CNA 0.662
GID4 CNA 1.000 TET1 CNA 0.786 JUN
CNA 0.659
FOXE2 NGS 0.916 TCE7L2 CNA 0.763 RET CNA
0.620
NFKB2 CNA 0.905 PDGFRA CNA 0.727 MAP2K4 CNA 0.614
SUFU CNA 0.874 MSH2 CNA 0.696 CHIC2
CNA 0.586
1GEBR2 CNA 0.870 EGER2 CNA 0.670 ALK
CNA 0.585
185
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
NT5 C2 CNA 0.578 CRKL CNA 0.466 BAP1 CNA 0.365
ATIC CNA 0 572 vfm NGS 0 458 NCOA4 CNA 0 356
E13171 CNA 0.535 LI LFPL6 CNA 0.457 CDII1 NOS
0.354
PRF1 CNA 0.521 WDCP CNA 0.438 "[P53 NOS
0.351
KAT6B CNA 0.506 LCP1 CNA 0.422 EML4 CNA 0.345
TP13 CNA 0 502 CC-DC6 CNA 0 416 KTA A1149 CNA 0
337
FHIT CNA 0.500 m2 CNA 0.414 KRAS NGS 0.336
EP300 CNA 0.491 FUBPI CNA 0.406 RB1 CNA
0.335
Gender META 0.480 N1RK3 CNA 0.384 GNAll CNA 0.328
JAKI CNA 0.478 CRTC3 CNA 0.382 FLCN CNA 0.326
MLHI CNA 0.471 CDX2 CNA 0.368 CACNAID CNA 0.323
Table 90: Right Colon Adenocarcinoma NOS - Colon
GENE TECII IMP EBFI CNA 0.626 ERCC5 CNA 0.513
CDX2 CNA 1.000 MYC CNA 0.619 SDC4 CNA 0.512
APC NGS 0.952 HOXAll CNA 0.584 BRCA2 CNA 0.509
FLT3 CNA 0.842 ASXLI CNA 0.583 USP6 CNA 0.506
FOXL2 NGS 0.827 U2AFI CNA 0.577 RB1 CNA 0.503
KRAS NGS 0.823 Gender META 0.574 CTCF
CNA 0.503
FLTI CNA 0.798 CDKN2A CNA 0.570 PDGFRA CNA 0.503
BRAF NGS 0.784 CDK8 CNA 0.565 RACI CNA 0.502
RNI443 NGS 0.770 W WTR1 CNA 0.563 FOX01 CNA 0.498
LLIFPL6 CNA 0.759 SPECC1 CNA 0.560 TR1M27 CNA 0.495
SETBPI CNA 0.748 CDHI CNA 0.551 ZNF217 CNA 0.495
HOXA9 CNA 0.705 ZNI4521 CNA 0.551 CACNA I D CNA
0.490
Age META 0.703 ETV5 CNA 0.548 ERG CNA
0.488
GID4 CNA 0.659 LCP1 CNA 0.533 FGF 14 CNA
0.482
SOX2 CNA 0.634 ZIVIYM2 CNA 0.526 PMS2 CNA 0.481
CDKN2B CNA 0.631 KDSR CNA 0.526 SLC34A2 CNA 0.479
BCL2 CNA 0.629 SMAD4 CNA 0.522 LIFR CNA 0.477
Table 91: Right Colon Mucinous Adenocarcinoma - Colon
GENE TECH IMP WWTR1 CNA 0.634 CBFB CNA 0.520
KRAS NGS 1.000 H[VIGN2P46 CNA 0.610 PDGFRA CNA
0.513
CDX2 CNA 0.891 Gender META 0.606 GNA13
CNA 0.506
FOXL2 NGS 0.876 PRRX1 CNA 0.591 TCF7L2 CNA 0.499
APC NGS 0.864 RPL22 NGS 0.591 FOXL2 CNA
0.494
Age META 0.864 MYC CNA 0.575 FLT1 CNA 0.492
RNF43 NGS 0.793 BRAF NGS 0.568 SETBPI CNA 0.487
LILFPL6 CNA 0.730 HOXA9 CNA 0.564 KLF4 CNA 0.484
CDK6 CNA 0.685 ASXL1 CNA 0.553 E1V5 CNA
0.481
RPNI CNA 0.678 FLT3 CNA 0.543 SOX2 CNA 0.481
PTCHI CNA 0.670 CDKN2B CNA 0.543 ELK4 CNA 0.479
CDKN2A CNA 0.668 GP FIN CNA 0.537 E13141 CNA
0.479
186
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
SPEN CNA 0.478 BRAF CNA 0.467 MAML2 CNA
0.448
HOXA13 CNA 0 477 MSI2 CNA 0 466 PDCD1LG2 CNA
0 447
RPL22 CNA 0.472 EZII2 CNA 0.457 RUNX1T1 CNA
0.446
KIAA1549 CNA 0.469 12M12 CNA 0.453 "[CEA'
CNA 0.445
KMT2C CNA 0.468 CDHI CNA 0.453 GATA2
CNA 0.443
Table 92: Salivary Gland Adenoid Cystic Carcinoma - Head, Face or Neck, NOS
GENE TECH IMP MDS2 CNA 0.553 TRRAP CNA
0.451
SOX10 CNA 1.000 ERBB3 CNA 0.548 TGFBR2 CNA
0.446
TP53 NGS 0.825 BTGI CNA 0.532 PDGFRA NGS
0.441
FICI2 CNA 0.791 RUNX1 CNA 0.531 WDCP CNA
0435
Age META 0.771 PMS2 CNA 0.531 TLX1
CNA 0.427
ATF 1 CNA 0.742 CEBPA CNA 0.527 CDH11
CNA 0.421
FOXL2 NGS 0.736 HOXC11 CNA 0.519 ABL1 NGS
0.412
IDH1 NGS 0.684 DDIT3 CNA 0.515 ENBP1
CNA 0.412
c-KIT NGS 0.677 PTEN NGS 0.512 NCOA1 NGS
0.412
APC NGS 0.669 ASXL1 CNA 0.510 MAF
CNA 0.409
CDK4 CNA 0.653 MYH9 CNA 0.502 BCL6
CNA 0405
FANCF CNA 0.624 RPNI CNA 0.501 BCLI IA CNA
0.405
FANCC CNA 0.605 PDCD1LG2 CNA 0498 SDC4 CNA
0404
Gender META 0.603 1RF4 CNA 0.474 FGER2
CNA 0.404
KRAS NGS 0.591 LHERL6 CNA 0.471 SET13P1 CNA 0.403
VIAL NGS 0.579 PAX3 CNA 0.452 HEYI
CNA 0.403
KMT2D CNA 0.554 CDH1 NGS 0.452 IKZE1
CNA 0.400
Table 93: Skin Merkel Cell Carcinoma - Skin
GENE TECH IMP SMAD2 CNA 0.495 SDC4 CNA
0.355
Age META 1.000 KRAS NGS 0.493 HOOK3 CNA
0.353
RBI NGS 0.980 FOX01 CNA 0.468 SDHB CNA
0.352
AKT1 NGS 0.902 MAX CNA 0.462 VITT
CNA 0.346
SFPQ CNA 0.881 MDS2 CNA 0.452 PBX1
CNA 0.344
FOXL2 NGS 0.874 ECT2L CNA 0.452 GOPC NOS
0.344
WWTRI CNA 0.843 PRKDC CNA 0.439 MYCL CNA 0.335
TGFBR2 CNA 0.799 CBFB CNA 0.438 L CP1
CNA 0.332
Gender META 0.795 STAT5B CNA 0.423 RB1 CNA
0.327
JAKI CNA 0.719 HMGA2 CNA 0.419 PTCHI CNA 0.323
WISP3 CNA 0.716 MYC CNA 0.413 ELL
NGS 0.318
SETBP1 CNA 0.694 RAC1 CNA 0.401 SRSF3
CNA 0.317
CHIC2 CNA 0.632 MSI2 CNA 0.399 TP53
NGS 0.315
AEDN CNA 0.615 ZNE217 CNA 0.388 LMO1 CNA 0.311
VIAL NGS 0.592 HIE CNA 0.379 ERB133 CNA
0.308
CDKN2C CNA 0.518 CALR CNA 0.362 .. ARID1A CNA
0.307
HSP90AB1 CNA 0.507 CAMTA1 CNA 0.361 SPEN CNA
0.304
187
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Table 94: Skin Nodular Melanoma - Skin
GENE TECH IMP PDCD1LG2 CNA 0.614 ESRI CNA 0.459
CDKN2A CNA 1.000 CDKN 213 CNA 0.609 HIS11H4I CNA
0.457
EZR CNA 0.956 NEM CNA 0.603 ABL1 CNA 0.456
FOXL2 NGS 0.946 ZNF217 CNA 0.598 TNFAIP3 CNA 0.449
DAXX CNA 0.833 SDI IA172 CNA 0.574 Age META
0.447
BRAF NGS 0.792 SOX10 CNA 0.573 N1JP214 CNA
0.421
ABL1 NGS 0.752 POT! CNA 0.544 MTOR CNA 0421
CREB3L2 CNA 0.729 Gender META 0.513 GMPS CNA
0.418
TP53 NGS 0.725 SOX2 CNA 0.497 CACNAID CNA 0.403
KIAA1549 CNA 0.722 MLLT10 CNA 0.489 BTGI CNA 0.402
CD274 CNA 0.710 BRAF CNA 0.488 SMAD2 CNA 0.400
NRAS NGS 0.697 1RF4 CNA 0.482 KRAS NGS 0.397
CDII1 NGS 0.679 FOXL2 CNA 0.478 MLLT11 CNA 0.395
e-KIT NUS 0.655 FANCCi CNA 0.478 CARS CNA 0.391
FOX03 CNA 0.634 ENTIP I CNA 0.472 TCF7I 2
CNA 0.389
EBFI CNA 0.624 FGER2 CNA 0.468 PRDMI CNA 0.386
TRIM 27 CNA 0.624 CCDC6 CNA 0.466 I ISP9OAA1 CNA
0.384
Table 95: Skin Squamous Carcinoma - Skin
GENE TECH IMP AR1D1A CNA 0.576 NR4A3 CNA 0.499
Age META 1.000 CHEK2 CNA 0.574 JAZE1 CNA
0.495
NOTCH1 NGS 0.943 TAL2 CNA 0.554 RABEPI CNA 0.491
LRPIB NGS 0.884 FHIT CNA 0.547 GNAS CNA 0.490
FOXL2 NGS 0.873 CAMTA1 CNA 0.536 NOTCH2 NGS 0.487
Gender META 0.765 SPECC1 CNA 0.536 FANCC CNA
0.486
CACNAID CNA 0.744 FOXPI CNA 0.532 CDH11 CNA
0.485
EWSRI CNA 0.726 PPAR_G CNA 0.530 SPEN CNA
0.484
ARFRP1 NOS 0.698 ASXL1 NOS 0.528 OPEIN CNA
0.483
DDIT3 CNA 0.687 ABL1 CNA 0.518 ATR NGS 0.483
"IP53 NGS 0.672 SD!-II) CNA 0.514 TGE13R2
CNA 0.481
ENBP1 CNA 0.668 VIET NGS 0.511 SETD2 CNA
0.474
CDK4 CNA 0.647 CCNEI CNA 0.511 1-1MON2P46 CNA
0.471
KMT2D NGS 0.646 HOXD13 CNA 0.508 GREQA NGS 0.467
MEW_ CNA 0.636 RAF 1 CNA 0.507 ZNF217 CNA
0.459
NTRK2 CNA 0.627 KRAS NGS 0.505 XPC CNA 0.457
KLILL6 CNA 0.626 NUP214 CNA 0.500 SDIB3 CNA 0.455
Table 96: Skin Melanoma - Skin
GENE TECH IMP FOXL2 NGS 0.799 LRP IB NGS 0.738
11RF4 CNA 1.000 EP300 CNA 0.785 CCDC6 CNA
0.731
SOX10 CNA 0.977 BRAF NGS 0.772 MITT CNA 0.675
FGFR2 CNA 0.807 TP53 NGS 0.744 CREB3L2 CNA 0.645
188
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Age META 0.636 GRIN2A NGS 0.542 SOX2 CNA 0.482
TRIM27 CNA 0 632 NF1 NGS 0 536 LHFPL6 CNA 0 478
Gender META 0.624 CCND2 CNA 0.534 CI TEK2 CNA
0.478
PDCD1LG2 CNA 0.620 PRDM1 CNA 0.531 MLL13 CNA 0.477
CDKN2A CNA 0.615 KRAS NGS 0.528 VTII A CNA
0.472
NR AS NGS 0 609 EZR CNA 0 121 CTNNA 1 CNA 0 471
TCF7L2 CNA 0.597 MECOM CNA 0.502 KIAA1549 CNA 0.471
MTOR CNA 0.594 PAX3 CNA 0.497 ARID I A CNA
0.466
N142 CNA 0.590 NFIB CNA 0.497 CDX2 CNA 0.459
CDKN2B CNA 0.575 CNBP CNA 0.494 DEK CNA 0.458
ESRI CNA 0.562 CAMTAI CNA 0.486 CD274 CNA 0.453
GATA3 CNA 0.560 TNFAIP3 CNA 0.485 CRKL CNA 0.453
FOXAI CNA 0.547 KIF5B CNA 0.483 BTGI CNA
0.453
Table 97: Small Intestine Gastrointestinal Stromal Tumor NOS - Small Intestine
GENE TECII Mil' MYCL CNA 0.538 SETBPI CNA 0.382
c-KIT NGS 1.000 ATPIAI CNA 0.532 C15orf65 CNA 0.372
ABLI NGS 0.908 TNFAIP3 CNA 0.521 ARID I A CNA
0.370
JAKI CNA 0.861 SFPQ CNA 0.480 CDKN2B CNA 0.361
SPEN CNA 0.836 APC NGS 0.471 MPL CNA 0.338
FOXL2 NGS 0.766 ERG CNA 0.450 CACNA ID CNA 0.320
EPS15 CNA 0.732 NOTCH2 CNA 0.441 EGER CNA 0.319
STIL CNA 0.727 RBI NGS 0.426 JUN CNA 0.318
UNIGN2P46 CNA 0.721 CAMTAI CNA 0.421 TSUR CNA 0.305
Age META 0.713 RPL22 CNA 0.413 SUE U CNA
0.303
TP53 NGS 0.641 PIK3CG CNA 0.410 AMERI NGS 0.297
BLM CNA 0.615 PTCHI CNA 0.403 MTOR CNA
0.297
TITRAP3 CNA 0.602 KNLI CNA 0.398 FGER2 CNA 0.293
CDI 111 CNA 0.602 ABL2 CNA 0.390 NUP93 CNA
0.290
MSI2 CNA 0.578 BTGI CNA 0.389 BCL9 CNA 0.286
CRTC3 CNA 0.550 ACSL6 CNA 0.386 VIII, NGS
0.284
MYCL NGS 0.543 ELK4 CNA 0.386 U2AF 1 CNA
0.281
Table 98: Small Intestine Adenoeareinoma - Small Intestine
GENE TECH IMP LTIFPL6 CNA 0.620 Gender META 0.535
KRAS NGS 1.000 LPP CNA 0.619 RPNI CNA 0.510
CDX2 CNA 0.866 POU2AF1 CNA 0.613 EBF 1 CNA 0.499
FOXL2 NGS 0.862 Age META 0.602 ERCC5 CNA 0.497
SETBPI CNA 0.853 CDK8 CNA 0.590 KD SR CNA 0.493
ELT3 CNA 0.837 BCL2 CNA 0.573 SDHC CNA 0.488
AURKB CNA 0.762 R131 CNA 0.559 HOXA 1 1 CNA 0.479
FLTI CNA 0.733 TP53 NGS 0.552 SD1-41) CNA 0.477
LCPI CNA 0.691 MYC CNA 0.552 AFF3 CNA 0.474
SPECCI CNA 0.621 APC NGS 0.551 GID4 CNA 0.473
189
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
ASXL1 CNA 0.469 MLF 1 CNA 0.441 FHIT CNA 0.422
GMPS CNA 0 468 FGF14 CNA 0_437 ATP 1A1 CNA 0 422
CDII1 CNA 0.465 ABL2 CNA 0.435 JAZ171 CNA 0.418
ZNF217 CNA 0.457 CTCF CNA 0.433 CDKN2A CNA 0.417
FOX01 CNA 0.456 ARNT CNA 0.428 EWSR1 CNA 0.410
CCNF.1 CNA 04S C 1 5 orf6I CNA 0_427 CHTC2 CNA 0408
EXTI CNA 0.448 CDKN2B CNA 0.427 MLLT11 CNA 0.407
Table 99: Stomach Gastrointestinal Stromal Tumor NOS - Stomach
GENE TECH IMP CCN1311131 CNA 0.440 VI-[L NGS
0.292
c-KIT NGS 1.000 ROS I CNA 0.439 KTN1 CNA
0.292
PDGFRA NGS 0.838 BCLI1B CNA 0.438 USP6 CNA 0.274
MAX CNA 0.815 CDHI NGS 0.438 ADGRA2 CNA
0.272
FOXL2 NGS 0.802 HSP9OAA1 CNA 0.419 GPILN CNA
0.271
TSFIR CNA 0.684 BCL2 CNA 0.405 TPM3 CNA
0.266
BCL2L2 CNA 0.628 CHEK2 CNA 0.391 LPP CNA 0.262
TP53 NGS 0.610 ECT2L CNA 0.371 APC NGS
0.261
FOXAI CNA 0.601 NFKBIA CNA 0.348 BCL6 CNA 0.258
MST2 CNA 0.591 RADS 1B CNA 0.329 PMS2 NGS 0.255
N IN CNA 0.578 KRAS NGS 0.301 AKTI CNA
0.255
NKX2-1 CNA 0.568 JUN CNA 0.300 CTCF CNA
0.254
PDGERA CNA 0.536 PER1 CNA 0.299 GOLGAS CNA
0.247
SETBP1 CNA 0.460 PTEN NGS 0.298 FGFR4 CNA
0.246
CDHI 1 CNA 0.451 MPL CNA 0.297 MUCI CNA
0.244
Age META 0.449 PDGE13 CNA 0.295 TCL1A CNA 0.240
Gender META 0.440 FGFR1 CNA 0.293 PDE4DIP CNA
0.240
Table 100: Stomach Signet Ring Cell Adenocarcinoma - Stomach
GENE TECH IMP TGFBR2 CNA 0.616 USP6 CNA 0.546
Age META 1.000 BCL2 CNA 0.598 FGFR2 CNA
0.543
CDX2 CNA 0.936 PRCC CNA 0.595 FANCF CNA
0.531
FOXL2 NGS 0.911 NSD2 CNA 0.583 SETBP1 CNA
0.531
CDH1 NGS 0.898 FN13P1 CNA 0.579 HOXD11 CNA
0.516
LLIFPL6 CNA 0.858 RPN I CNA 0.578 CDKN2A CNA
0.514
AFF3 CNA 0.815 MLLT11 CNA 0.577 WWTRI CNA 0.513
BCL3 CNA 0.790 CDK4 CNA 0.562 MYC CNA
0.509
ERG CNA 0.783 CTNNA1 CNA 0.561 CCNE1 CNA 0.499
HOXD13 CNA 0.755 c-KIT NGS 0.554 CALR CNA
0.485
Gendet META 0.709 HIVIGN2P46 CNA 0.552 HIMGA2 CNA
0.483
FAN CC CNA 0.686 1C.F7L2 CNA 0.550 LPP CNA
0.473
EXT1 CNA 0.674 HIST1H41 CNA 0.549 'IP53 NGS
0.466
PBXI CNA 0.664 H3F3B CNA 0.549 CITEK2 CNA
0.464
RUNXI CNA 0.663 U2AF1 CNA 0.546 NUTM2B CNA
0.462
CDKN213 CNA 0.622 ERAS NGS 0.546 CDH11 CNA
0.461
190
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
BTG1 CNA 0.459 GID4 CNA 0.457 WRN CNA 0.457
Table 101: Thyroid Carcinoma NOS - Thyroid
GENE TECH IMP HOXA13 CNA 0.612 FH1T CNA 0.533
NKX2-1 CNA 1.000 DDX6 CNA 0.600 TMPRS S2 CNA 0.531
Age META 0.988 NDRG1 CNA 0.577 FANCF CNA
0.530
FOXL2 NGS 0.980 CRKL CNA 0.574 MUC1 CNA 0.524
HOXA9 CNA 0.756 BCL2 CNA 0.570 HOXAll CNA 0.520
SBDS CNA 0.750 CDH11 CNA 0.566 CARS CNA
0.518
TP53 NGS 0.740 EBF1 CNA 0.559 DAXX CNA 0.514
SOX10 CNA 0.728 KNI,1 CNA 0.558 MYC CNA
0.510
NF2 CNA 0.726 RADS 1 CNA 0.554 HIST1H3B CNA
0.506
ERG CNA 0.719 HMGN2P46 CNA 0.553 DDIT3 CNA
0.497
111\4GA2 CNA 0.686 CD274 CNA 0.553 LCPI CNA 0.493
EWSR1 CNA 0.683 STAT5B CNA 0.541 ERC1 CNA 0.492
GNAS CNA 0.671 TSHR CNA 0.541 SETBPI CNA 0.489
MLLT11 CNA 0.662 CRTC3 CNA 0.534 TRIM 33 NGS 0.488
KDSR CNA 0.646 FANCA CNA 0.533 TTL CNA 0.481
Gender META 0.636 AKAP9 NGS 0.533 PAK3 NGS
0.479
LI1FPL6 CNA 0.628 BRCAI CNA 0.533 PAX8 CNA 0.478
Table 102: Thyroid Carcinoma Anaplastic NOS - Thyroid
GENE TECH IMP ERBB3 CNA 0.603 FLT1 CNA 0.474
TRRAP CNA 1.000 KIAA1549 CNA 0.594 BCL9 CNA 0.469
BRAF NGS 0.847 FUS CNA 0.578 CBFB CNA 0.463
CDR]. NGS 0.842 SPEN CNA 0.559 13CL1 IA NGS 0.459
W1SP3 CNA 0.832 PDGFRA CNA 0.548 CDKN2A CNA 0.453
Age META 0.782 NRAS NGS 0.547 MN1 CNA 0.451
Gender META 0.744 KDSR CNA 0.534 AFF3 CNA 0.448
MYC CNA 0.706 LIFFPL6 CNA 0.533 BAP1 CNA 0.434
VHL NGS 0.705 FGF14 CNA 0.520 CDKN2B CNA 0.433
CDX2 CNA 0.680 IGF 1R CNA 0.517 I IOXA9 CNA 0.432
PDE4DTP CNA 0.670 EBF1 CNA 0.515 RB1 NGS 0.431
SBDS CNA 0.666 HOOK3 CNA 0.510 PTCH1 CNA 0.424
KRAS NGS 0.637 NCKEPSD CNA 0.494 TP53 NGS 0.421
113H1 NGS 0.636 ARID 1A CNA 0.490 PBRM1 CNA 0.417
FHIT CNA 0.636 PBX1 CNA 0.482 CHIC2 CNA 0.412
PTEN NGS 0.629 SPECC1 CNA 0.479 ABL2 NGS 0.412
ELK4 CNA 0.619 CLPI CNA 0.475 HOXA13 CNA 0.409
Table 103: Thyroid Papillary Carcinoma of Thyroid - Thyroid
GENE TECH IMP FOXL2 NGS 0.922 MYC CNA 0.752
BRAF NGS 1.000 NKX2-1 CNA 0.798 RAT ,GDS NGS 0.728
191
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
TP53 NGS 0.727 CTNNA1 CNA 0.477 FNBP1 CNA 0.405
SETBP1 CNA 0 642 H3F3B CNA 0 465 TPR CNA 0 404
EXT1 CNA 0.608 AFF1 CNA 0.465 TCEA1 CNA 0.404
KDSR CNA 0.604 APC CNA 0.460 MAF CNA 0.399
KLLIL6 CNA 0.560 ITK CNA 0.452 WWTR1 CNA 0.395
EBF1 CNA 0 160 ABT ,1 CNA 0 441 I TSP6 CNA
0 395
YWHAE CNA 0.555 Gender META 0.440 PRKDC CNA 0.385
FHIT CNA 0.529 NR4A3 CNA 0.431 TAL2 CNA 0.383
Age META 0.515 NDRG1 CNA 0.431 SET CNA 0.379
U2AF1 CNA 0.512 IGF 1R CNA 0.429 MCL1 CNA 0.372
SLC34A2 CNA 0.498 FBXW7 CNA 0.422 CRKL CNA 0.371
SRSF2 CNA 0.498 RUNX1T1 CNA 0.422 ZNF521 CNA 0.370
AKT3 CNA 0.492 FANCF CNA 0.421 ETV5 CNA 0.367
COX6C CNA 0.490 PDE4D IP CNA 0.414 CDX2 CNA 0.365
TERC CNA 0.485 IKZE1 CNA 0.411 ERG CNA 0.361
Table 104: Tonsil Oropharynx Tongue Squamous Carcinoma - Head, Face or Neck,
NOS
GENE TECH IMP FHIT CNA 0.773 TPM3 CNA 0.675
SOX2 CNA 1.000 PRCC CNA 0.768 NF2 CNA 0.667
LPP CNA 0.999 CHEK2 CNA 0.758 FGF 10 CNA
0.661
KLBL6 CNA 0.995 FLI1 CNA 0.757 MITE CNA 0.661
FOXL2 NGS 0.977 CKKL CNA 0.757 VI-[L CNA
0.660
Gender META 0.897 TP53 NGS 0.740 BCL9 CNA
0.660
CACNA1D CNA 0.888 PPARG CNA 0.736 CREB3L2 CNA 0.659
SDHD CNA 0.860 CM, CNA 0.729 EWSR1 CNA 0.658
ZBTB16 CNA 0.859 FANCG CNA 0.727 HSP9OAA1 CNA 0.658
BCL6 CNA 0.851 NTRK2 CNA 0.716 FANCC CNA 0.658
RPN1 CNA 0.846 PBRIVI1 CNA 0.715 NDRG1
CNA 0.644
TGEBR2 CNA 0.845 POU2A111 CNA 0.705 CDKN2A CNA 0.641
Age META 0.810 PRKDC CNA 0.705 ETV5 CNA 0.639
SYK CNA 0.807 KIAA1549 CNA 0.699 RAF1 CNA
0.633
TFRC CNA 0.793 EGFR CNA 0.692 EPUB1 CNA
0.628
PCSK7 CNA 0.789 WWTR1 CNA 0.691 PAFAH1B2 CNA 0.628
KMT2A CNA 0.780 TRIM27 CNA 0.680 ASXL1 CNA 0.618
Table 105: Transverse Colon Adenocarcinoma NOS - Colon
GENE TECH IMP BCL2 CNA 0.763 SOX2 CNA 0.574
APC NGS 1.000 Age META 0.732 ERCC5 CNA
0.568
CDX2 CNA 0.969 KRAS NGS 0.701 ZNF217 CNA
0.563
EL13 CNA 0.902 BRAE NGS 0.637 TRRAP NGS
0.554
FOXL2 NGS 0.880 KDSR CNA 0.637 EPHA5 CNA
0.552
SETBP1 CNA 0.842 ASXL1 CNA 0.620 MCL1 CNA 0.550
LHFPL6 CNA 0.778 HOXA9 CNA 0.595 SFPQ CNA 0.548
FLT1 CNA 0.769 AURKA CNA 0.584 LCP1 CNA 0.547
192
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
KLHL6 CNA 0.538 FOX01 CNA 0.487 CBFB CNA
0.450
EBFI CNA 0 528 CDKN2B CNA 0 479 GNAI3 CNA 0 447
WWTRI CNA 0.521 SMAD4 CNA 0.477 SDC4 CNA 0.443
ZNE521 NGS 0.516 COX6C CNA 0.469 CACNAID CNA
0.442
CCNE1 CNA 0.511 SPEN CNA 0.465 RB1 CNA 0.442
GNA S CNA 0 101 PRR X1 CNA 0 464 TOP 1 CNA 0
437
Gender META 0.501 U2AF1 CNA 0.464 JAZF 1 CNA
0.436
CDHI CNA 0.493 CDKN2A CNA 0.455 RUNXI CNA 0.436
ZMYM2 CNA 0.492 1P53 NGS 0.453 HIVIGN2P46 CNA
0.422
Table 106: Urothelial Bladder Adenocarcinoma NOS - Bladder
GENE TECII IMP IKZE1 CNA 0.546 RACI CNA
0.453
CTNNA1 CNA 1.000 Gender META 0.544 CEBPA CNA 0.451
FOXL2 NGS 0.945 EGE10 CNA 0.533 PCSK7 CNA
0.448
ZNF217 CNA 0.770 SDC4 CNA 0.533 CBFB CNA 0.447
ENBP1 CNA 0.693 HOXA13 CNA 0.518 SET CNA 0.445
EWSRI CNA 0.687 WWTRI CNA 0.517 STAT3 CNA 0.441
11,7R CNA 0.686 ARID2 NGS 0.513 RICTOR CNA 0.439
TP53 NGS 0.643 APC NGS 0.508 STAT5B CNA 0.433
ACSL6 CNA 0.642 MTOR CNA 0.497 MYC CNA 0.432
CTCF CNA 0.639 ACSL3 CNA 0.497 SDBB CNA
0.425
13CL3 CNA 0.637 CRE133L2 CNA 0.496 HOXAll CNA 0.425
LIFR CNA 0.636 EPHA3 CNA 0.475 SETBP1 CNA 0.422
CHEK2 CNA 0.628 EP300 CNA 0.468 FILE CNA
0.418
Age META 0.606 DDX6 CNA 0.461 PAFAH1132 CNA
0.410
CDHI NGS 0.577 CDK4 CNA 0.457 FANCD2 NGS 0.410
VIAL NGS 0.577 BCL2L11 CNA 0.455 CDK6 CNA 0.404
CD79A NGS 0.562 CDX2 CNA 0.455 GNAS CNA 0.391
Table 107: Urothelial Bladder Carcinoma NOS - Bladder
GENE TECH IMP MYCL CNA 0.709 USP6 CNA 0.574
Age META 1.000 FGFR2 CNA 0.694 OMB CNA
0.559
VT-IL CNA 0.971 KDM6A NGS 0.658 MDS2 CNA 0.558
CREBBP CNA 0.939 TP53 NGS 0.656 HEYI CNA 0.556
FOXE2 NGS 0.912 CTNNAI CNA 0.648 EWSRI CNA 0.554
Gender META 0.836 KRAS NGS 0.623 ZNF331 CNA
0.551
CDKN2B CNA 0.835 XPC CNA 0.612 CARS CNA 0.550
FANCC CNA 0.806 LIMPL6 CNA 0.612 FBXW7 CNA 0.545
GATA3 CNA 0.797 CCNEI CNA 0.608 TMPRS S2 CNA
0.544
GNA13 CNA 0.755 U2AF1 CNA 0.602 ARIDIA CNA 0.539
IL7R CNA 0.748 PPARG CNA 0.602 PAX3 CNA
0.533
RAFI CNA 0.736 ERG CNA 0.596 MECOM CNA 0.526
WISP3 CNA 0.728 ACKR3 CNA 0.580 CACNAID CNA
0.524
ASXL1 CNA 0.722 CDKN 2A CNA 0.579 W WTR1 CNA 0.523
193
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CTCF CNA 0.520 CDH1 CNA 0.515
LIIVIGN2P46 CNA 0.501
CDH11 CNA 0 518 ABL2 NGS 0 510
FANCD2 CNA 0 501
RPNI CNA 0.518 ETV5 CNA 0.505
VIII, NGS 0.500
Table 108: Urothelial Bladder Squamous Carcinoma- Bladder
GENE TECH IMP FHIT CNA 0.522 EPHB1 CNA
0.448
Age META 1.000 KRAS NGS 0.519
COX6C CNA 0.445
FOXL2 NGS 0.934 TP53 NGS 0.512 ARIDIA CNA
0.445
EL7R CNA 0.857 SOX2 CNA 0.510
CTLA4 CNA 0.443
CDH1 NGS 0.808 MLLT11 CNA 0.506
CACNA1D CNA 0.439
ABT,2 NGS 0.808 FANCF CNA 0.503
BAP1 CNA 0.433
TFRC CNA 0.785 CDKN2A CNA 0.501
EXTI CNA 0.432
KITIL6 CNA 0.733 EPS15 CNA 0.497 NUP98 CNA
0.431
LPP CNA 0.696 RPN1 CNA 0.484 NPMI
CNA 0.429
WWTR1 CNA 0.696 CDH1 CNA 0.478 GID4 CNA
0.429
EBFI CNA 0.689 CDK4 CNA 0.474 LIFR
CNA 0.425
CDKN2C CNA 0.665 INHBA CNA 0.474 FANCC CNA 0.425
c-KIT NGS 0.656 MLF 1 CNA 0.467
NOTCH I NGS 0.422
AFF1 CNA 0.591 JAK2 CNA 0.467
GRIN2A CNA 0.420
ETV5 CNA 0.574 PRKDC CNA 0.463
MAML2 CNA 0.416
Gender META 0.566 JAZF 1 CNA 0.458
STAT3 CNA 0.412
CN13P CNA 0.559 KMT2A CNA 0.452
'[ER! CNA 0.410
Table 109: Urothelial Carcinoma NOS - Bladder
GENE TECH IMP KMT2D NGS 0.510 ELK4 CNA 0.455
GA1A3 CNA 1.000 FGFR2 CNA 0.501 13ARD1 CNA 0.454
Age META 0.820 EWSR1 CNA 0.492
T,HFPI6 CNA 0.453
ASXL1 CNA 0.698 VIAL CNA 0.491 KLIAL6 CNA 0.452
CDKN2A CNA 0.637 NR4A3 CNA 0.482 APC NGS 0.449
Gender META 0.637 FGFR3 NGS 0.481 CCNE1 CNA 0.445
CDKN2B CNA 0.634 c-KIT NGS 0.479 EL7R CNA 0.441
ATIC CNA 0.577 PAX3 CNA 0.479 DDB2 CNA 0.440
EBF1 CNA 0.575 CTNNA1 CNA 0.477
PTCH1 CNA 0.440
NSD1 CNA 0.567 ZNF217 CNA 0.475 ARID1A CNA 0.438
PPAR.G CNA 0.550 XPC CNA 0.473 PBXI CNA 0.432
ZNF331 CNA 0.545 FGF10 CNA 0.473 FLT1 CNA 0.432
ACSL6 CNA 0.535 MYC CNA 0.465 MLLT11 CNA 0.431
TP53 NGS 0.532 MYCL CNA 0.463
BCL6 CNA 0.431
RAFI CNA 0.517 KDM6A NGS 0.461 CASP 8 CNA 0.426
KRAS NGS 0.517 LXT2 CNA 0.459 l'IK CNA 0.424
CARS CNA 0.511 CTLA4 CNA 0.457 FAN CF CNA 0.422
Table 110: Uterine Endometrial Stromal Sarcoma NOS - FGTP
194
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
GENE TECH IMP CDH1 NGS 0.539 KRAS NGS 0.360
ETV I CNA 1000 AFF I CNA 0 520 FAM46C CNA
0 359
FOXL2 NGS 0.967 ERG CNA 0.512 FCRL4 CNA
0.357
IINRNPA2131 CNA 0.957 DDR2 CNA 0.507 1103(1)13 CNA
0.341
PMS2 CNA 0.809 TERT CNA 0.498 FH CNA
0.337
TGEFIR 2 CNA 0 734 NR4 A3 CNA 0 497 CDX2
CNA 0328
Gender META 0.726 SDC4 CNA 0.483 CACNA I D CNA
0.327
TP53 NGS 0.690 VBE NGS 0.447 CNBP CNA 0.326
Age META 0.688 RPN 1 CNA 0.440 13CL6
CNA 0.325
SPECCI CNA 0.684 FANCE CNA 0.430 NDRGI CNA
0.321
FANCC CNA 0.683 PCMI NGS 0.415 XPC CNA
0.310
ENTIBA CNA 0.601 TOPI CNA 0.414 PTEN NGS
0.310
CDHI CNA 0.570 ZNF 217 CNA 0.409 CDK12
CNA 0.308
RAC I CNA 0.570 PPARG CNA 0.396 WRN CNA
0.306
PTCII1 CNA 0.569 PDCD1LG2 CNA 0.396 SRGAP3 CNA
0.302
PDE4D1T' CNA 0.565 RUNX1 CNA 0.368 JAKI CNA
0.289
MAP2K4 CNA 0.541 RAPIGDS1 CNA 0.367 ESRI CNA
0.289
'I'able 111: Uterine Leiomyosarcoma NOS - FGTP
GENE TECH IMP PTCHI CNA 0.686 LRIG3 CNA 0.547
RBI CNA 1.000 PAX3 CNA 0.676 PDGFRA CNA 0.540
FOXE2 NGS 0.966 E13141 CNA 0.665 P13X1 CNA
0.538
SPECC1 CNA 0.943 SYK CNA 0.659 NTRK3 CNA 0.531
Age META 0.868 WDCP CNA 0.619 IGFIR CNA 0.530
JAK1 CNA 0.830 C131413 CNA 0.612 MAP2K4
CNA 0.522
PDCDI CNA 0.825 ESRI CNA 0.605 KDR CNA 0.518
PRRX1 CNA 0.795 KLBL6 CNA 0.604 DNIMT3A CNA 0.494
Gender META 0.790 NTRK2 CNA 0.587 CDKN2B CNA 0.491
ACKR3 CNA 0.771 MYCN CNA 0.578 11)II1 CNA 0.482
ATIC CNA 0.767 JUN CNA 0.574 BIM:1'MA CNA 0.478
LCP1 CNA 0.762 CTCF CNA 0.573 NUTM2B CNA 0.477
HERPUD1 CNA 0.740 CRTC3 CNA 0.566 KD SR CNA 0.475
FANCC CNA 0.739 SOX2 CNA 0.560 KIT CNA 0.474
0ID4 CNA 0.728 RPNI CNA 0.559 AFF3 CNA 0.470
NUP93 CNA 0.716 FOX01 CNA 0.556 TP53 NGS 0.467
CDHI CNA 0.692 ITIFPI6 CNA 0.548 TPM4 CNA 0.462
Table 112: Uterine Sarcoma NOS - FGTP
GENE TECH IMP TTL CNA 0.778 PRDMI CNA 0.718
HOXD13 CNA 1.000 Age META 0.773 PML CNA 0.697
FOXE2 NGS 0.972 HMGA2 CNA 0.751 RI31 CNA 0.678
CACNAID CNA 0.887 MITE CNA 0.739 CDKN2B CNA 0.677
Gender META 0.870 PRRXI CNA 0.736 DDR2 CNA 0.676
MAX CNA 0.799 N142 CNA 0.728 HOXAll CNA 0.665
195
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
HOXA9 CNA 0.645 USP6 CNA 0.545 ADGRA2 CNA 0.473
KIT CNA 0 643 PDE4DEP CNA 0 538 SEPT5 CNA 0
463
CDKN2A CNA 0.630 IDII2 CNA 0.537 FGER2 CNA
0.454
PDGERA CNA 0.614 1P53 NGS 0.534 PSIP 1 CNA
0.441
ALK NGS 0.610 MYC CNA 0.531 FGER1 CNA 0.439
FNBP1 CNA 0 600 PEACH CNA 0 119 FH1T CNA
0438
CDHI CNA 0.597 ERCC3 CNA 0.497 ZNF217 CNA 0.433
WRN CNA 0.593 HOXD11 CNA 0.495 RALGDS CNA 0.431
SNX29 CNA 0.574 EANCA CNA 0.487 AEE3 CNA 0.428
GID4 CNA 0.572 FCRL4 CNA 0.485 SFPQ CNA
0.421
BCL 1 lA CNA 0.559 JAZE1 CNA 0.484 MAP2K4 CNA 0.417
Table 113: Uveal Melanoma - Eye
GENE TECII IMP LPP CNA 0.565 ETV5 CNA 0.419
IRF4 CNA 1.000 MLF 1 CNA 0.525 UBR5 CNA
0.415
HEY1 CNA 0.873 ICLI1L6 CNA 0.523 FOXL2 CNA 0.406
FOXL2 NGS 0.858 NCOA2 CNA 0.522 HSP90AB1 CNA 0.401
EXTI CNA 0.826 c-KIT NGS 0.519 HIST1H4I CNA
0.401
PAX3 CNA 0.785 TFRC CNA 0.511 SETBPI CNA 0.389
TRIM27 CNA 0.780 WWTRI CNA 0.509 KRAS NGS 0.383
TP53 NGS 0.730 COX6C CNA 0.507 NR4A3 CNA 0.378
GNAll NGS 0.710 H1ST1H3B CNA 0.503 DEK CNA 0.372
GNAQ NGS 0.707 BAP1 NGS 0.491 TCEAI CNA 0.362
RUNX 1 T1 CNA 0.679 SF3B1 NGS 0.466 MUCI CNA 0.354
SOX 1 0 CNA 0.668 GA1A2 CNA 0.465 USP6 CNA 0.351
MYC CNA 0.658 EWSRI CNA 0.457 YWHAE CNA 0.348
BCL6 CNA 0.650 GMPS CNA 0.456 SOX2 CNA 0.345
RPNI CNA 0.616 BCL2 CNA 0.453 IDHI NGS 0.341
ABL2 NGS 0.598 CNBP CNA 0.452 VIII. NOS
0.340
SRGAP3 CNA 0.570 DAXX CNA 0.427 CDX2 CNA 0.333
Table 114: Vaginal Squamous Carcinoma - FGTP
GENE TECH IMP FNBPI CNA 0.792 AIHD2 NGS
0.623
CNBP CNA 1.000 CD274 CNA 0.778 WT1 CNA
0.605
RPNI CNA 0.985 CBFB CNA 0.774 ABIl CNA 0.602
FOXL2 NGS 0.980 PPARG CNA 0.755 KMT2C NGS 0.586
KMT2D NGS 0.961 MLLT3 CNA 0.750 TFRC CNA 0.578
VILL NGS 0.927 WWTR1 CNA 0.749 RAF1 CNA 0.560
SPEN CNA 0.917 FANCC CNA 0.682 SOX2 CNA 0.552
Gender ME EA 0.909 PDCD1EG2 CNA 0.661 ETV5 CNA
0.548
EMT CNA 0.894 PAX3 CNA 0.651 CDKN 2C CNA 0.546
CDHI NGS 0.874 KLE1L6 CNA 0.640 BARDI
CNA 0.545
TP53 NGS 0.872 SDHC CNA 0.629 Age META 0.531
JUN CNA 0.807 HOXD13 CNA 0.626 MAE CNA 0.523
196
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
MECOM CNA 0.514 LPP CNA 0.474 MLLT10 CNA 0.454
SDBB CNA 0 511 ESR1 CNA 0 472 KD SR CNA 0
450
MDS2 CNA 0.498 CDIIII CNA 0.467 CDKN2B CNA
0.447
ASXL 1 CNA 0.492 GSK3I3 CNA 0.466 TRRAP CNA
0.447
EP300 CNA 0.481 CLPI CNA 0.464 HOXD11 CNA 0.446
Table 115: Vulvar Squamous Carcinoma - EGTP
GENE TECH IMP KITIL6 CNA 0.674 U2AF 1 CNA 0.596
CNBP CNA 1.000 SPECCI CNA 0.666 PRDMI CNA 0.592
CACNAID CNA 0.975 EXTI CNA 0.665 SET CNA 0.591
FOXL2 NGS 0.973 CDKN2B CNA 0.653 NTRK2 CNA 0.590
Gender META 0.967 CAMTAI CNA 0.651 GNAS CNA 0.583
SDBB CNA 0.928 CHEK2 CNA 0.642 ENBP1 CNA 0.579
SYK CNA 0.924 RPL22 CNA 0.641 PDCD1LG2 CNA
0.579
Age META 0.832 RPNI CNA 0.641 PBXI CNA
0.579
TAL2 CNA 0.817 NR4A3 CNA 0.634 TR1M27 CNA 0.578
TGFBR2 CNA 0.807 CREB3L2 CNA 0.629 CD274 CNA 0.576
MTOR CNA 0.807 TP53 NGS 0.629 TFRC CNA 0.567
HOOK3 CNA 0.802 NUP93 CNA 0.624 sTEL CNA 0.566
SETD2 CNA 0.773 ARID 1A CNA 0.623 PAX3 CNA 0.559
PRKDC CNA 0.729 CBFB CNA 0.623 ETV5 CNA 0.556
1313KM1 CNA 0.709 FANCC CNA 0.614 EWSR1 CNA 0.555
MDS2 CNA 0.704 BCL9 CNA 0.614 BCL 1 IA CNA
0.555
KAT6A CNA 0.699 FGF4 CNA 0.604 XPC CNA 0.554
Table 116: Skin Trunk Melanoma - Skin
GENE TECH IMP Gender META 0.558 FHIT CNA 0.453
IRF4 CNA 1.000 SDHAF2 CNA 0.547 BCL6 CNA 0.444
FOXL2 NGS 0.900 HIST1H4I CNA 0.540 MKLI CNA 0.442
BRAF NGS 0.853 ELK4 CNA 0.519 DAXX CNA 0.428
SOX10 CNA 0.842 NRAS NGS 0.518 KRAS NGS 0.419
TP53 NGS 0.777 CCDC6 CNA 0.518 Age META 0.414
TCF7L2 CNA 0.757 FLII CNA 0.517 PTCHI CNA 0.409
FGFR2 CNA 0.734 SOX2 CNA 0.516 e-KIT NGS
0.401
CDKN2A CNA 0.734 TET 1 CNA 0.511 NF2 CNA 0.399
EP300 CNA 0.686 TRIM26 CNA 0.509 BRAF CNA 0.394
CDKN2B CNA 0.669 CREB3L2 CNA 0.506 POTI CNA 0.392
DEK CNA 0.660 NOTCH2 CNA 0.505 MYCN CNA 0.388
SYK CNA 0.644 KIAA1549 CNA 0.504 CACNAID CNA
0.383
1RIM27 CNA 0.607 USP6 CNA 0.500 APC NUS 0.378
LILFPL6 CNA 0.580 FOXY' CNA 0.482 LRP113 NGS
0.376
CRTC3 CNA 0.575 ESRI CNA 0.466 TET 1 NGS 0.372
FANCC CNA 0.572 SDHD CNA 0.458 BCL2 CNA 0.363
197
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
In many cases, the features in the biosignatures in Tables 2-116 comprise gene
copy number
(CNA or CNV). Cells are typically diploid with two copies of each gene.
Ifowever, cancer may lead
to various genomic alterations which can alter copy number. In some instances,
copies of genes are
amplified (gained), whereas in other instances copies of genes are lost.
Genomic alterations can affect
5 different regions of a chromosome. For example, gain or loss may occur
within a gene, at the gene
level, or within groups of neighboring genes. Gain or loss may also be
observed at the level of
cytogenetic bands or even larger portions of chromosomal arms. Thus, analysis
of such proximate
regions to a gene may provide similar or even identical information to the
gene itself. Accordingly,
the methods provided herein are not limited to determining copy number of the
specified genes, but
10 also expressly contemplate the analysis of proximate regions to the
genes, wherein such proximate
regions provide similar or the same level of information. Copy analysis of
genes, SNPs or other
features within the band may be used within the scope of the systems and
methods described herein.
As described in the Examples herein, the methods for classifying the
attributes of the cancer
may calculate a probability that the biosignature corresponds to the at least
one pre-determined
15 biosignature. In some embodiments, the method comprises a pairvvise
comparison between two
candidate attributes, and a probability is calculated that the sample
biosignature corresponds to either
one of the at least one pre-determined biosignatures. In some embodiments, the
pairwise comparison
between the two candidate attributes is determined using a machine learning
classification algorithm,
wherein optionally the machine learning classification algorithm comprises a
voting module. In some
20 embodiments, the voting module is as provided herein, e.g., as described
above. In some
embodiments, a plurality of probabilities are calculated for a plurality of
pre-determined
biosignatures. In some embodiments, the probabilities are ranked. In some
embodiments, the
probabilities are compared to a threshold, wherein optionally the comparison
to the threshold is used
to determine whether the classification of the desired attribute of the cancer
is likely, unlikely, or
25 indeterminate. Systems and methods for implementing the classifications
are provided herein. For
example, see FIGs. 1A-I and related text.
In some embodiments, the levels of specificity for the attributes of the
patient sample are
determined at the level of an organ group. In one non-limiting example, the
organ group that is
predicted may be selected from bladder; skin; lung; head, face or neck (NOS);
esophagus; female
30 genital tract (FGT); brain; colon; prostate; liver, gall bladder, ducts;
breast; eye; stomach; kidney; and
pancreas. As desired, the systems and methods provided herein may employ
biosignatures determined
at the level of a primary tumor location and a histology, see, e.g., Tables 2-
116, and the organ group
is then determined based on the most probable primary tumor location +
histology. As a non-limiting
example, Tables 2-116 herein provide biosignatures for primary tumor location
+ histology, and the
35 table headers report both the primary tumor location + histology and
corresponding organ group.
The disclosure contemplates that selections may be made from the biosignatures
provided
herein, e.g., in Tables 2-116 for primary tumor location + histology. Use of
the features in the tables
198
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
may provide optimal origin prediction, although selection may be made so long
as the selections
retain the ability to meet desired performance criteria, such as but not
limited to accuracy of at least
50%, 60%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or
at least 99%.
In some embodiments, the biosignature comprises the top 1%, 2%, 3%, 4%, 5%,
6%, 7%, 8%, 9%,
5 10%, 11%, 12%, 13%, 14%, 15%, 16%, 17%, 18%, 19%, 20%, 21%, 22%, 23%,
24%, 25%, 26%,
27%, 28%, 29%, 30%, 31%, 32%, 33%, 34%, 35%, 36%, 37%, 38%, 39%, 40%, 41%,
42%, 43%,
44%, 45%, 46%, 47%, 48%, 49%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%,
95%, or 100%
of the feature biomarkers with the highest Importance value in the
corresponding table (i.e., Tables 2-
116). In some embodiments, the biosignature comprises the top 1, 2, 3, 4, 5,
6,7, 8,9, 10, 11, 12, 13,
10 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48, 49 or 50 feature biomarkers with the highest
Importance value in the
corresponding table (i.e., 'rabies 2-116). In some embodiments, the
biosignature comprises at least
1%, 2%, 3%, 4%, 5%, 6%, 7%, 8%, 9%, 10%, 11%, 12%, 13%, 14%, 15%, 16%, 17%,
18%, 19%,
20%, 21%, 22%, 23%, 24%, 25%, 26%, 27%, 28%, 29%, 30%, 31%, 32%, 33%, 34%,
35%, 36%,
15 37%, 38%, 39%, 40%, 41%, 42%, 43%, 44%, 45%, 46%, 47%, 48%, 40%, 50%,
55%, 60%, 65%,
70%, 75%, 80%, 85%, 90%, 95%, or 100% of the top 1, 2, 3, 4, 5, 6,7, 8,9, 10,
11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40, 41, 42, 43,
44, 45,46, 47, 48, 49, or 50 feature biomarkers with the highest Importance
value in the
corresponding table (i.e., Tables 2-116). In some embodiments, the
biosignaturc comprises at least
20 50%, 60%, 70%, 75%, 80%, 85%, 90%, 95%, or 100% of the top 5, 10, 15,
20, 25, 30, 35, 40, 45, 50,
60, 65, 70, 75, 80, 85, 90, 95, or 100 feature biomarkers with the highest
Importance value in the
corresponding table. As a non-limiting example, the biosignature may comprise
at least 1, 2, 3, 4, or 5
of the top 10, 20 or 50 features. Provided herein is any selection of
biomarkers that can be used to
obtain a desired performance for predicting the attribute of interest, be it a
primary location, organ
25 group, histology, or disease/cancer type.
Systems for implementing the methods are also provided herein. See, e.g.,
FIGs. 1F-1G and
related disclosure.
In some embodiments, the systems and methods of the invention implement
systems and
methods for predicting sample attributes as detailed in International Patent
Publication
30 W0/2020/146554, entitled Genomic Profiling Similarity and based on
International Patent
Application PCT/US2020/012815 filed on January 8,2020, the entire contents of
which application is
hereby incorporated by reference in its entirety.
Expression-based Predictor of Disease Type
35 The section above provides a machine learning based classifier to
predict attributes of a
cancer sample based on molecular analysis of the sample, such attributes
comprising a primary tumor
origin, cancer/disease type, organ group, histology, and any combination
thereof. The methods and
199
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
systems provided accordingly can be applied with various biological analytes
as desired, e.g., nucleic
acids, e.g., DNA and RNA, and protein. The section above and WO/2020/146554
demonstrated such
analysis using genomic DNA. There have been attempts to use mRNA expression
profiling to build
classifiers or predictors of such attributes. mRNA is an attractive analyte
because it can be assessed
5 using well established techniques, e.g.. PCR or microarray. mRNA
sequences and expression can also
be assessed in a high throughput manner using next generation sequencing,
including without
limitation whole transcriptome sequencing. However, RNA also has drawbacks.
Consider analysis of
a tumor sample using ILIC for protein expression. A stained RIC slide will
show areas of normal
versus tumor tissue, and also other features such as nuclear or membrane
staining of the protein. Thus
10 a pathologist can focus on areas of interest for analysis of the protein
expression levels and patterns.
However, RNA would comprise a mix of RNA from different cells and cell types
within the sample,
without cellular location, and wherein background amounts of various RNA
transcripts may vary
greatly between cells. In particular, RNA classifiers may struggle with low
neoplastic percentage in
metastatic sites which is where TOO identification is often most needed.
Accordingly, an RNA
15 expression based assay may be confounded by the particular sample and
cells from which the RNA is
extracted. See, e.g., Hayashi et al., Randomized Phase II Trial Comparing Site-
Specific Treatment
Based on Gene Expression Profiling with Carboplatin and Paclitaxel for
Patients with Cancer of
Unknown Primary Site, J Clin Oncol 37:57-579 (finding no significant
improvement in one-year
survival based on site-specific treatment as determined by gene expression
profiling). Thus, there is a
20 need to improve analysis of RNA based characterization of cancer
samples.
Herein, we provide systems and methods to predict sample origin of a tumor
sample based on
RNA expression analysis with much higher accuracy than previously achieved.
The general scheme
400 for performing the prediction is shown in FIG. 4A. RNA expression data 401
is collected for the
desired transcripts. Any useful method of acquiring such data can be employed.
For example, we used
25 whole transcriptome sequencing analysis (WTS; RNA-seq) using the
Illumina NGS platform, which
methodology queries over 22,000 transcripts in a single assay. The raw
expression data is processed
via any desired methodology for processing. See, e.g., Li et al., Comparing
the Normalization
Methods for the Differential Analysis of Illumina High-Throughput RNA-Seq
Data, BMC
Bioinforrnatics. 2015 Oct 28;16:347. doi: 10.1186/s12859-015-0778-7; Abbas-
Aghababazadeh and
30 Fridley, Comparison of normalization approaches for gene expression
studies completed with high-
throughput sequencing, PLoS One. 2018; 13(10): e0206312. In some embodiments,
the RNA
expression data 402 is normalized using Trimmed Mean of M-values (TA4M). See
Robinson and
Oshlack, A Scaling Normalization Method for Differential Expression Analysis
of RNA-seq Data,
Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. Epub 2010 Mar 2.
35 Continuing with FIG. 4A, normalized expression data for the target
transcripts can be used to
train machine learning models for various attributes of interest, including
without limitation a primary
tumor origin, cancer/disease type 403, organ group 404, and/or histology 405.
In some embodiments,
200
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
the primary tumor origin or plurality of primary tumor origins consists of,
comprises, or comprises at
least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24, 25, 26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, or all 38 of prostate, bladder, endocervix,
peritoneum, stomach,
esophagus, ovary, parietal lobe, cervix, endometrium, liver, sigmoid colon,
upper-outer quadrant of
5 breast, uterus, pancreas, head of pancreas, rectum, colon, breast,
intrahepatic bile duct, cecum,
gastroesophageal junction, frontal lobe, kidney, tail of pancreas, ascending
colon, descending colon,
gallbladder, appendix, rectosigrnoid colon, fallopian tube, brain, lung,
temporal lobe, lower third of
esophagus, upper-inner quadrant of breast, transverse colon, and skin. In some
embodiments, the
primary tumor origin or plurality of primary tumor origins consists of,
comprises, or comprises at
10 least 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18,
19,20, or all 21 of breast
adenocarcinoma, central nervous system cancer, cervical adenocarcinoma,
cholangiocarcinoma, colon
adenocarcinoma, gastroesophageal adenocarcinoma, gastrointestinal stromal
tumor (GIST),
hepatoeellular carcinoma, lung adenocarcinoma, melanoma, meningioma, ovarian
granulosa cell
tumor, ovarian & fallopian tube adenocarcinoma, pancreas adenocarcinoma,
prostate adenocarcinoma,
15 renal cell carcinoma, squamous cell carcinoma, thyroid cancer,
urothelial carcinoma, uterine
endometrial adenocarcinoma, and uterine sarcoma. In some embodiments, the
cancer/disease type 403
consists of, comprises, or comprises at least 1, 2, 3, 4, 5, 6,7, 8,9, 10, 11,
12, 13, 14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24, 25, 26, 27, or all 28 of adrenal cortical carcinoma;
bile duct,
cholangiocarcinoma; breast carcinoma; central nervous system (CNS); cervix
carcinoma; colon
20 carcinoma; endometrium carcinoma; gastrointestinal stromal tumor (GIST);
gastroesophageal
carcinoma; kidney renal cell carcinoma; liver hepatocellular carcinoma; lung
carcinoma; melanoma;
mcningioma; Merkel; neuroendocrine; ovary granulosa cell tumor; ovary,
fallopian, peritoneum;
pancreas carcinoma; pleural mesothelioma; prostate adenocarcinoma;
retroperitoneum; salivary and
parotid; small intestine adenocarcinoma; squamous cell carcinoma; thyroid
carcinoma; urothelial
25 carcinoma; uterus. In some embodiments, the organ group 404 consists of,
comprises, or comprises at
least 1, 2, 3,4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, or all 17 of
adrenal gland; bladder; brain; breast;
colon; eye; female genital tract and peritoneum (FOOT gastroesophageal; head,
face or neck, NOS;
kidney; liver, gallbladder, ducts; lung; pancreas; prostate; skin; small
intestine; thyroid. In some
embodiments, the histology 405 consists of, comprises, or comprises at least
1, 2, 3,4, 5, 6, 7, 8, 9,
30 10, 11, 12, 13, 14, 15, 16, 17, IS, 19, 20, 21, 22, 23, 24, 25, 26, 27,
2g, or all 29 of adenocarcinoma,
adenoid cystic carcinoma, adenosquamous carcinoma, adrenal cortical carcinoma,
astrocytoma,
carcinoma, carcinosarcoma, cholangiocarcinoma, clear cell carcinoma, ductal
carcinoma in situ
(DCIS), glioblastoma (GBM), GIST, glioma, granulosa cell tumor, infiltrating
lobular carcinoma,
leiomyosarcoma, liposarcoma, melanoma, meningioma, Merkel cell carcinoma,
mesothelioma,
201
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
neuroendocrine, non-small cell carcinoma, oligodendroglioma, sarcoma,
sarcomatoid carcinoma,
serous, small cell carcinoma, squamous.
Various classification methodology can be applied to the chosen attributes as
desired,
including without limitation a neural network model, a linear regression
model, a random forest
5 model, a logistic regression model, a naive Bayes model, a quadratic
discriminant analysis model, a
K-nearest neighbor model, a support vector machine, or various forms of or
combinations thereof. In
some embodiments, the machine learning approach comprises an XGBoost multi-
class classification.
XGBoost is a decision-tree-based ensemble machine learning algorithm that uses
a gradient boosting
framework. Combinations of classification methods can be employed.
Calculations can be performed
10 using various statistical analysis platforms, including without
limitation R.
FIG. 4A illustrates a scenario wherein three different classifications 403-405
performed on
the same transcript expression data. The classifications from each of these
three models can be
combined using another model, such as those described above. In some
embodiments, the
combination is also made using an XGBoost model. This mechanism of combining
intermediate
15 classifications of the chose attributes, such as the illustrated 403-
405, is an implementation of the
voting scheme described herein (see, e.g., FIG. 1F and related text) and
provides for dynamic voting
406. As a non-limiting example, consider that one of the intermediate models
403-405 is very
accurate at making a given classification. In such case, that single model's
classification may carry
more weight than the two other intermediate models when making the fmal
classification 407. In such
20 case, that model's classification may dominate the other intermediate
models when making the final
classification 407. The various intermediate models can be assigned different
weights when
performing the dynamic voting 406. Any such combination of one or more of the
intermediate models
can outweigh others. Thus the dynamic voting 406 can provide classification
407 based on trained and
optimized contributions from each of the intermediate models.
25 In some embodiments, analysis of different types of analytes are
combined in order to classify
the input sample and estimate the desired one or more attributes. In this
regard. FIG. 4B presents an
exemplary variation 410 of scheme 400 that is shown in FIG. 4A. In this
variation, both RNA
transcript levels 411 and DNA 416 are used to classify the input sample. As
noted herein, DNA and
RNA have various strengths and weaknesses for predicting attributes of a
biological sample. For
30 example, DNA is relatively more stable and more uniform amongst
different types of cells, whereas
RNA is more dynamic and may be more indicative of differences within
individual cells. Without
being bound by theory, we hypothesized that a combination of genomic DNA
analysis with RNA
transcriptome analysis may provide optimal results. We term this combined
classifier a "panomic"
predictor. As desired, analysis from additional analytes such as other types
of RNA and/or protein
35 could also be input into the system in a similar manner. In the
embodiment illustrated in FIG. 4B, the
three intermediate RNA transcript models 412 414 are identical to FIG. 4A 403
405 as described
above, respectively. In addition, the figure shows DNA 416 input into the
system. In some
202
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
embodiments, the DNA is processed using the 115 disease types as described
above. See, e.g., Tables
2-116 and related discussion; see also Examples 2-3. In this case, the dynamic
voting 415 is applied
to the four intermediate models comprising RNA 412-414 and DNA 416. Models
assessing attributes
based on alternate analytes may also be input into the dynamic voting module
415 in a similar
5 manner. As described above, the dynamic voting mechanism is a variation
of the voting scheme
described herein (see, e.g., FIG. IF and related text) and provides for
essentially dynamic voting
between the inputs into the dynamic voting module 415 in order to provide the
prediction/classification 417. As a non-limiting example, consider that one of
the intermediate models
412-414 or 416 are very accurate at making a given classification. In such
case, that model's
10 classification may outweigh the other intermediate models when making
the fmal classification 417.
Similarly, two of the intermediate models may outperform the two other
intermediate models for a
given classification and may thus dominate in that setting, or three of the
intermediate models may
combine to provide a better classification with lesser input from the
remaining model. Thus the
dynamic voting 415 can provide classification 417 based on trained and
optimized contributions from
15 each of the intermediate models.
FIG. 4C illustrates a flowchart of an example of a process 400C for training a
dynamic
voting engine. Process 400C may be performed by a system such as the system
400 of FIG. 4A or
410 of FIG. 4B.
The dynamic voting engine such as the dynamic voting engine of FIG. 4A, 406,
FIG. 4B,
20 415 or FIG. 1G, 400 can be trained in a number of different ways. In one
implementation, the
dynamic voting engine can be trained to predict a target classification for a
biological sample based
on processing, by the dynamic voting engine, data corresponding to one or more
initial classifications
that were previously determined for a biological sample. In some
implementations, the biological
sample can include a cancer sample and the target classification can include
an attribute for the
25 cancer, including without limitation a TOO. In some implementations, the
one or more previously
determined classifications can be based on processing of DNA sequences of the
biological sample,
RNA sequences of the biological sample, or both.
The system can begin performance of the process 400C by using one or more
computers to
obtain 410C, from a database of labeled training data items, a labeled
training data item Each labeled
30 training data item can include one or more initial classifications and a
target classification. The one or
more initial classifications can be based on or derived from actual data
generated by one or more
initial classification engines such as cancer type classification engine
(e.g., FIG. 4A, 403 or FIG. 4B,
412), an initial organ of origin engine (e.g., FIG. 4A, 404 or FIG. 4B, 413),
a histology engine (e.g.,
FIG. 4A, 405 or FIG. 4B, 414), or a DNA analysis engine (e.g., FIG. 4B, 416),
based on processing,
35 by one or more of the respective initial classification engines, data
derived from the biological sample.
The data derived from the biological sample can include DNA sequences of the
sample, RNA
sequences of the sample, or both. In other implementations, the one or more
initial classifications can
203
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
be based on or derived from simulated data that is generated to represent
initial classifications that
ought to be generated by such initial classification models when such initial
classification models
process data such as DNA sequences, RNA sequences, or both, derived from the
biological sample.
The system can continue performance of the process 400C by using one or more
computers to
5 generate 420C training input data for input to the dynamic voting engine.
In some implementations,
the training input data can include, for example, a numerical representation
of the one or more initial
classifications. For example, data that represents each of the initial
classifications can be encoded into
one or more fields of a data structure that is formatted for input to the
dynamic voting engine.
The system can continue performance of the process 400C by using one or more
computers to
10 process 430C the generated training input data through the dynamic
voting engine. in some
implementations, the dynamic voting engine can include one or more machine
learning models, e.g.,
one or more of a random forests, support vector machines, logistic
regressions, K-nearest neighbors,
artificial neural networks, naïve Bayes, quadratic discriminant analysis,
Gaussian processes models,
decision trees, or any combination thereof. In such implementations,
processing the generated training
15 input data through the dynamic voting engine can include processing the
generated training input data
through each layer of the one or more machine learning models. In some
implementations, the
dynamic voting engine includes an XGBoost decision-tree-based ensemble machine
learning
algorithm.
The system can continue performance of the process 400C by using one or more
computers to
20 obtain 440C the output data generated by the dynamic voting engine based
on the dynamic voting
engine's processing of the training input data generated at stage 420C. The
system can then use one or
more computers to determine a level of similarity between the output data
generated by the dynamic
voting engine that is obtained at stage 440C and the label for the training
data item obtained at stage
410C. In some implementations, the level of similarity between the label of
the training data item
25 obtained at stage 410C and the output data that is obtained at stage
440C can include the difference
between the label and the output data.
The system can continue performance of the process 400C by using one or more
computers to
adjust 460C one or more parameters of the dynamic voting engine based on the
level of similarity
between the output data and the label of the training data item obtained at
stage 410C. The system can
30 then continue to iteratively perform the process 400C until the output
data generated by the system
and obtained at stage 440C begins to match the label for the training data
item obtained at stage 410C
within a threshold amount of error. In some implementations, the threshold of
error can be zero error.
In other implementations, the threshold can include less than 1% error, less
than 2 % error, less than
5% error, less than 10% error, or the like. Once the system begins to detect
that the dynamic voting
35 engine is predicting output data that matches the label for the training
input data processed by the
204
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
dynamic voting engine within a threshold amount of error, then the dynamic
voting engine may be
considered to be fully trained.
The systems 400, 410 and variations thereof can be trained to desired panels
of RNA
transcripts in order to classify the at least one attribute of the cancer of
interest. In some embodiments,
5 the systems are trained using NGS based whole transcriptome sequencing
data, e.g., mRNA from
22,000 genes. To avoid overfitting or similar error, analysis of such panels
may require training data
on tens of thousands of tumor samples. To further avoid issues faced relying
on RNA transcript
analysis, such as overfitting of data based on the high number of total mRNAs,
we may train the
systems using more limited sets of transcripts. Traditionally, proteins that
have been used in 11-IC
10 based tumor classification. See, e.g., Lin and Liu, Immunohistochemistry
in Undifferentiated
Neoplasm/Tumor of Uncertain Origin, Arch Pathol Lab Med. 2014;138:1583-1610,
which reference
is incorporated herein by reference in its entirety. In some embodiments, the
panel of mRNA
transcripts used to implement the system comprise the mRNA encoding such
proteins, and may
further include various isoforms or related family members thereof. The
correlation between RNA
15 transcript expression and protein expression levels is noisy and tissue
dependent, and thus one would
not be able to predict a priori whether such an approach would yield
acceptable results. See, e.g.,
Edfors et al, Gene-specific correlation of RNA and protein levels in human
cells and tissues, Mol Syst
Biol. (2016) 12: 883; Franks A, et al (2017) Post-transcriptional regulation
across human tissues.
PLoS Comput Biol 13(5): el005535. However, we hypothesized that the analysis
of multiple genes
20 would improve noise levels to achieve acceptable accuracy and
unexpectedly found our approach to
perform with high levels of accuracy.
Based on the above rational for identifying a subset of potentially useful RNA
transcripts, we
constructed a list of candidate biomarkers shown in Table 117. The table
provides the official gene
symbol and full name as reported by the National Center for Biotechnology
Information (NOM) Gene
25 database with reference to the HUGO Gene Nomenclature Committee (HGNC)
database. See
www.nebi.nlm.nih.gov/gene (NCBI Gene); www.genenames.org (HGNC). The NCBI's
Gene ID is
also provided. The "Aliases" column provides a non-exhaustive list of
alternate descriptions for the
genes such as alternate gene names, e.g., that may also be used herein.
Comprehensive listings of
alternate symbols are provided by the NCBI and HGNC databases, among others
available and known
30 to those of skill in the art (e.g., Ensembl, Crenecards, etc).
Table 117¨ RNA Transcripts used to Characterize Tumor Sample
Gene Symbol Full Name Aliases NCBI Gene
ID
ACVRL1 activin A receptor like type 1 94
AFP alpha fetoprotein 174
ALPP alkaline phosphatase, placental 250
AMACR alpha-methylacyl-CoA racemase 23600
ANKRD30A ankyrin repeat domain 30A NY-BR-1 91074
205
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
ANO1 anoctamin 1 DOG1 55107
AR androgen receptor 367
ARG1 arginase 1 383
13CL2 13CL2 apoptosis regulator 596
BCL6 BCL6 transcription repressor 604
CA9 carbonic anhydrase 9 768
CALB2 calbindin 2 794
CALCA calcitonin related polypeptide alpha 796
CALD1 caldesmon 1 800
CCNT)1 cyclin D1 CYCT ,IND1 595
CD 1A CD1a molecule 909
CD2 CD2 molecule 914
CD34 CD34 molecule 947
CD3G CD3g molecule 917
CD5 CD5 molecule 921
CD79A CD79a molecule 973
CD99L2 CD99 molecule like 2 83692
CDH1 cadherin 1 E-cadherin 999
CDH17 cadhcrin 17 1015
CDK4 cyclin dependent kinase 4 1019
CDKN2A cyclin dependent kinase inhibitor 2A p16
1029
CDX2 caudal type homeobox 2 1806
CEACAM1 CEA cell adhesion molecule 1 634
CEACAM16 CEA cell adhesion molecule 16, tectorial 388551
membrane component
CEACAM18 CEA cell adhesion molecule 18 729767
CEACAM19 CEA cell adhesion molecule 19 56971
CEACALV120 CEA cell adhesion molecule 20 125931
CEACAM21 CEA cell adhesion molecule 21 90273
CEACAM3 CEA cell adhesion molecule 3 1084
CEACAM4 CEA cell adhesion molecule 4 1089
CEACAIVI5 CEA cell adhesion molecule 5 1048
CEACAM6 CEA cell adhesion molecule 6 4680
CEACAM7 CEA cell adhesion molecule 7 1087
CEACAM8 CEA cell adhesion molecule 8 1088
CGA glycoprotein hormones, alpha polypeptide 1081
CGB3 chorionic gonadotropin subunit beta 3 1082
CNN1 calponin 1 1264
COQ2 coenzyme Q2, polyprenyltransferase 27235
CPS] carbamoyl -phosphate synthase 1 HepPar-1
1373
antibody target
CR1 complement C3b/C4b receptor 1 (Knops 1378
blood group)
CR2 complement C3d receptor 2 1380
CTNNB1 catenin beta 1 1499
206
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
DES desmin 1674
DSC3 desmocollin 3 1825
EN02 enolase 2 2026
E1213132 erb-b2 receptor tyrosine kinase 2 11E122,
2064
HER2/neu
ERG ETS transcription factor ERG 2078
ESR1 estrogen receptor 1 ER 2099
FLI1 Fli-1 proto-oncogene, ETS transcription 2313
factor
FOXL2 forkhead box L2 668
FUT4 fucosyltransferase 4 CD15 2526
GATA3 GATA binding protein 3 2625
GPC3 glypican 3 2719
HAVCR1 hepatitis A virus cellular receptor 1 26762
ETNTF1B EINF1 homeobox B 6928
EL12B interleukin 12B 3593
IMP3 IMP U3 small nucleolar ribonucleoprotein
55272
3
INHA inhibin subunit alpha Inhibin-alpha 3623
ISL1 ISL LIM homcobox 1 3670
KIT KIT proto-oncogene, receptor tyrosine 3815
kinasc
KL klotho 9365
KLK3 kallikrein related peptidase 3 PSA
354
KRT1 keratin 1 3848
KRT10 keratin 10 3858
KR114 keratin 14 3861
KRT15 keratin 15 3866
KI2116 keratin 16 3868
KRT17 keratin 17 CK17 3872
KRT18 keratin 18 CK18 3875
KRT19 keratin 19 CK19 3880
KRT2 keratin 2 3849
KRT20 keratin 20 CK20 54474
KRT3 keratin 3 3850
KRT4 keratin 4 3851
KRT5 keratin 5 3852
KRT6A keratin 6A CK6A 3853
KRT6B keratin 6B CK6B 3854
KRT6C keratin 6C CK6C 28688
KRT7 keratin 7 CK7 3855
KRT8 keratin 8 CK8 3856
LIN28A lin-28 homolog A 79727
LINT28B lin-28 homolog B 389421
MAGEA2 MAGE family member A2 4101
MDM2 MDM2 proto-oncogene 4193
207
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
MD31 mindbomb E3 ubiquitin protein ligase 1 57534
Milt. melanocyte inducing transcription factor 4286
MLANA melan-A 2315
ML111 mutL homolog 1 4292
MME membrane metalloendopeptidase 4311
MIPO myeloperoxidase 4353
MS4A1 membrane spanning 4-domains Al 931
MSH2 mutS homolog 2 4436
MSH6 mutS homolog 6 2956
MSTN mesothelin 10232
MEI IFR methylenetetrahydrofolate reductase 4524
MUC1 mucin 1, cell surface associated 4582
MUC2 mucin 2, oligomeric mucus/gel-forming 4583
MUC4 mucin 4, cell surface associated 4585
MUC5AC mucin 5AC, oligomcric mucus/gel-forming 4586
MY0D1 myogenic differentiation 1 4654
MYOG myogenin 4656
NANOG Nanog homeobox 79923
NAPSA napsin A aspartic pcptidasc Napsin A
9476
NCAM1 neural cell adhesion molecule 1 CD56
4684
NCAM2 neural cell adhesion molecule 2 4685
NKX2-2 NK2 homeobox 2 4821
NKX3-1 NK3 homcobox 1 4824
OSCAR osteoclast associated 1g-like receptor 126014
PAX2 paired box 2 5076
PAX5 paired box 5 5079
PAX8 paired box 8 7849
PDPN podoplanin 10630
PDX1 pancreatic and duodenal homeobox 1 3651
PECAM1 platelet and endothelial cell adhesion 5175
molecule 1
PGR progesterone receptor PR 5241
PIP prolactin induced protein 5304
PMEL premelanosome protein (gp100) GP100,
6490
PMEL17,
S1LV, filV113-45
target
PMS2 PMS1 homolog 2, mismatch repair system 5395
component
POU5F 1 POU class 5 homeobox 1 5460
PSAP prosaposin 5660
PTPRC protein tyrosine phosphatase receptor type
5788
S100A1 S100 calcium binding protein Al 6271
S100A10 S100 calcium binding protein A10 6281
S100All S100 calcium binding protein All 6282
208
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
S100Al2 S100 calcium binding protein Al2 6283
S100A13 S100 calcium binding protein A13 6284
S100A14 S100 calcium binding protein A14 57402
S100A16 S100 calcium binding protein A16 140576
S100A2 S100 calcium binding protein A2 6273
S100A4 S100 calcium binding protein A4 6275
S100A5 S100 calcium binding protein A5 6276
S100A6 S100 calcium binding protein A6 6277
S100A7 S100 calcium binding protein A7 6278
S100A7A 5100 calcium binding protein A7A 338324
S100A7L2 5100 calcium binding protein A7 like 2 645922
S100A8 S100 calcium binding protein A8 6279
S100A9 S100 calcium binding protein A9 6280
S 100B S100 calcium binding protein B 6285
S 100P S100 calcium binding protein P 6286
Si 00PBP SlOOP binding protein 64766
SlOOZ S100 calcium binding protein Z 170591
SALL4 spalt like transcription factor 4 57167
SATB2 SATB homcobox 2 23314
SDC1 syndecan 1 CD138 6382
SERPINA1 serpin family A member 1 al -antitrypsin,
5265
antitrypsin
SERP1NB5 serpin family B member 5 P15, maspin 5268
SF1 splicing factor 1 7536
SF TPA1 surfactant protein Al 653509
SMAD4 SMAD family member 4 4089
SMARCB1 SWI/SNF related, matrix associated, actin
6598
dependent regulator of chromatin,
subfamily b, member 1
SMN1 survival of motor neuron 1, telomeric 6606
SOX2 SRY-box transcription factor 2 6657
SPN sialophorin 6693
SYP synaptophysin 6855
1EE3 transcription factor binding to IGIAM 7030
enhancer 3
TFF1 trefoil factor 1 7031
TFF3 trefoil factor 3 7033
TO thyroglobulin 7038
TLE1 TT,E family member 1, transcriptional 7088
corepressor
TME'RSS2 transmembrane scrine protease 2 7113
1NERS148 TN14 receptor superfamily member 8 943
TP63 tumor protein p63 P63 8626
TPM1 tropomyosin 1 7168
TPM2 tropomyosin 2 7169
TPM3 tropomyosin 3 7170
209
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
TPM4 tropomyosin 4 7171
TPSAB1 tryptase alphaibeta 1 7177
TTF1 transcription termination factor 1 7270
1JPK2 uroplakin 2 UPll 7379
UPK3A uroplakin 3A 7380
LTPK3B uroplakin 3B 105375355
VIEL von Hippel-Lindau tumor suppressor 7428
VIL1 villin 1 Villin 7429
VIM vimentin 7431
WTI WTI transcription factor 7490
In some embodiments, data for the chosen features, here transcript expression
levels, is used
to train the prediction models for the attributes of interest, e.g., as in
FIG. 4B 412-414 or FIG. 4A
403-405. Although we rationalized selection of the group of transcripts in
Table 117 by tissue
5 classification based on LEIC protein expression, we did not replicate
classification schemes based on
the protein ¨ tissue correlations. Rather, expression data for the RNA
transcripts in Table 117 were
used to build machine learning models to predict tissue characteristics. The
machine learning
algorithms selected the appropriate transcript features during the training
phase. The transcript INSM1
(Full name: INSM transcriptional repressor 1; NCBI Gene 11): 3642) was also
used as a verification
10 for neuroendocrine tumors but was not included when training the machine
learning framework. See,
e.g., Mukhopadhyay, M et al., Insulinoma-associated protein 1 HNSM is a
sensitive and highly
specific marker of neuroendocrine differentiation in primary lung neoplasms:
an
immunohistochemical study of 345 cases, including 292 whole-tissue sections,
Modern Pathology
(2019) 32:100-109.
15 The models were trained as described herein. See, e.g., FIGs. 4A-B and
related discussion;
Examples 2-3. The training was performed using all transcript features in
Table 117. Features of
most importance for each prediction of the attributes cancer type, organ
group, and histology are listed
in Tables 118-120, respectively. In some embodiments, the prediction models
for individual attributes
use features found to contribute most to the predictions. In Tables 118-120,
the "importance" values
20 represent the relative contribution of each corresponding transcript to
the noted classification model.
Higher values indicate greater importance. Abbreviations in Table 118 include
ACC (adrenal cortical
carcinoma), BDC (bile duct, cholangiocarcinoma), BC (breast cancer), Cery
(cervix carcinoma),
Colon (colon carcinoma), EC (endometrium carcinoma), GC (gastroesophageal
carcinoma), KRCC
(kidney renal cell carcinoma), LIIC (liver hepatocellular carcinoma), Lung
(lung carcinoma), Mel
25 (melanoma), Men (meningioma), Merk (Merkel), Neu (neuroendocrine), OGCT
(ovary granulosa cell
tumor), OFP (ovary, fallopian, peritoneum), Pane (pancreas carcinoma), PM
(pleural mesothelioma),
PA (prostate adenocarcinoma), Ret (retroperitoneum), SP (salivary and
parotid), SIA (small intestine
adenocarcinoma), SCC (squamous cell carcinoma), TC (thyroid carcinoma), IJC
(urothelial
210
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
carcinoma), Ute (uterus). Abbreviations in Table 119 include AG (adrenal
gland), Bla (bladder), Br
(breast), Gast (Gastroesophageal), IIIN (head, face or neck, NOS), Kid
(kidney), LCD (liver,
gallbladder, ducts), Pane (pancreas), Pros (prostate), SI (small intestine),
Thy (thyroid). Table 119
omits leading zeros before the decimal for brevity. Abbreviations in Table 120
include Adeno
(adenocarcinoma), ACyC (Adenoid cystic carcinoma), AC (adenosquamous
carcinoma), ACC
(adrenal cortical carcinoma), Astro (astrocytoma), Care (carcinoma), CS
(careinosarcoma), Chol
(cholangiocarcinoma), CCC (clear cell carcinoma), DCIS (ductal carcinoma in
situ), GBM
(glioblastoma), GIST (gastrointestinal stromal tumor), Gli (glioma), GCT
(granulosa cell tumor), TLC
(infiltrating lobular carcinoma), Lei (leiomyosarcoma), Lipo (liposarcoma),
Mel (melanoma), Men
(meningioma), Merk (Merkel cell carcinoma), Meso (mesothelioma), Neuro
(neuroendocrine), NSCC
(non-small cell carcinoma), Oligo (oligodendroglioma), Sarc (sarcoma), SerC
(sarcomatoid
carcinoma), SCC (small cell carcinoma), Sq (squamous).
Table 118- Importance of RNA Transcripts used to Classify Cancer/Disease Type
Transcript ACC BDC BC CNS Carr Colon EC GIST GC KRCC LIIC Lung Mel Men
ACVRL1 0.0004 0.1199 0.0248 0.0000 0.0040 0.0230 0.2195
0.0976 0.0108 0.0470 0.0000 00301 0.1601 0.0000
AFP 0.0000 0 0571 00321 0.0019 0.0517 0.1342 0.1118
0.0000 00883 0.0000 0.3803 00209 0.0000 0.0000
AT,PP 0_0000 0 0609 0 1331 0 0000 0_0820 0_1160 0_1729 0
0000 0 0256 0_0107 0 0000 0_0050 0 0000 0_0000
AMACR 0.0000 0.0712 0.1790 0.0000 0.0459 0.0142 0.0219
0.0000 0.0882 0.2849 0.0154 0.0116 1/0005 0.0000
ANKRD30A 0.0000 0 0758 0.7886 0.0000 0.1003 0.0019 0.0370 0.0000 00189 0.0000
00019 00762 0.0000 0.0000
ANOI 0.0000 0.3746 0.0930 0.5582 0.0019 0.0349 0.2271
0.4210 0.3991 0.0424 0.0000 0.1994 0.0000 0.3991
ARG1 0.0282 0.0159 0.1184 0.0000 0.0283 0.1287 0.2650
0.0000 0.0299 0.0073 0.0668 0.1887 0.0371 0.0000
AR 0.0000 0.2429 0.1239 0.0020 0.0000 0.0612 0.1165
0.0000 0.4879 0.0346 0.0000 03547 0.0242 0.0099
BCL2 0.0000 00847 00213 0.0169 0.0092 02816 0.1625
0.0000 0.1195 0.0038 00000 0.0585 0.0000 0.0000
TICIA 0_0000 0.1002 0.0250 0.0000 0_0231 0_0347 0_2506
0.0000 0.1025 0.2594 0.2069 0_0962 5.0625 0_0211
CA9 0.0000 0.1177 0.1194 0.0102 0.1060 0.0113 0.0136
0.0000 0.0518 0.1982 0.0000 0.0247 0.0073 0.0000
CALB2 0.0706 0.1980 0.1016 0.0000 0.0087 00390 0.0345
0.0000 00509 0.0000 00000 00571 00071 0.0000
CALCA 0.0000 0.0940 0.0409 0.0000 0.0054 0.0173 0.0291
0.0000 0.0737 0.1475 0.0000 01323 0.0000 0.0000
CALD1 0.0000 0.1236 0.0360 0.0251 0.0086 0.0145 0.4457
0.0000 0.0079 0.0959 0.0005 00906 0.0008 0.0068
CCNDI 0_0000 0 0379 0 1132 0 0089 0_3474 0_0401 0_1933 0
0000 0 0121 0_0296 0 0166 0_0612 0 0949 0_0549
CDIA 0.0000 00580 0.1178 0.0000 0.0814 0.0362 0.0680
0.0000 0.2925 0.0000 00054 00327 0.0000 0.0000
CD2 0.0000 0.0484 0.0221 0.0393 0.0715 0.0662 0.0299
0.0000 0.0187 0.0000 0.0000 0.0615 0.0434 0.0194
CD34 0.0306 0.0250 0.0079 0.0000 0.0026 0.1113 0.1006
0.0000 0.2945 0.1061 0.1227 0.0378 0.0000 0.0000
CD3G 0.0000 0.0054 0.0465 0.0391 0.2238 00182 0.0326
0.0000 0.0453 0.0021 0.0246 00313 0.0247 0.0000
CD5 0.0000 0.1825 0.1934 0.0000 0.0554 01106 0.0434
0.0000 0.0416 0.0000 0.0071 00879 0.0004 0.0777
CD79A 0.0000 0.0582 0.1118 0.0000 0.2401 0.0662 0.0711
0.0000 0.0238 0.0046 0.0000 00242 0.0113 0.0000
CD99L2 0.0000 0.0427 0.1201 0.0579 0.0221 0.0134 0.0553
0.0000 0.0594 0.0000 0.0022 0.2901 0.0064 0.0000
CDH17 0.0000 0.0835 00034 0.0000 0.0018 04591 0.0785
0.0000 00357 0.0070 0.0055 01139 0.0000 0.0000
CDH1 0.0771 0.0161 0.1336 0.0544 0.0152 0.0166 0.0474
0.0320 0.2661 0.6591 0.0000 00191 0.0000 0.0563
CDK I 0_0000 0 1843 0 0275 0 0000 0_1197 0_0310 0_0171 0
0000 0 0430 0_0037 0 0000 0_1193 0 0000 0_0000
CDKN2A 0.0000 0.09!2 0.1531 0.0093 1/3759 012/0 0.1142
00000 0.0196 0.5109 0.0000 0.1210 1/161/6 1/0086
211
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CDX2 00000 0 0206 0 1544 0 0000 00300 16334 00274 0 0000
0 7635 00000 0 0000 0_0740 0 0000 00000
CEACAM16 0.0000 0.0676 0.1928 0.0000 0.0755 0.0727 0.2698 0.0000 0.0194 0.0000
0.5075 0.1828 0.0000 0.0000
CEACAM18 0.0000 0.0365 0.1524 0.0000 0.0000 02429 0.0217 0.0000 0.0788 0.0000
0.0000 0.0262 0.0000 0.0000
CEACAM19 0.0000 0.0464 0.0252 0.0038 0.1472 0.0772 0.1867 0.0000 0.1050 0.0656
0.0109 0.0851 0.0677 0.0000
CEACAM1 0.0000 0.0654 0.0122 0.1894 0.0085 0.0939 0.1046
0.0000 0.0521 0.0363 0.0389 0.2672 0.1125 0.2127
CEACAM20 0.0000 0.0059 0.0003 0.0000 0.0142 03682 0.0789 0.0000 0.0508 0.0000
0.1473 0.0159 0.0020 0.0000
CEACAM21 0.0000 0.0538 0.0382 0.0000 0.1321 0.0130 0.0591 0.0000 0.0035 0.0000
0.0000 0.0286 0.0000 0.0000
CEACAM3 00000 0 0270 0 0197 0.0000 00000 0_0169 00403 0
0000 0.0302 00000 0.0010 0_0340 0 0066 00000
CEACAM4 0.0000 0.0434 0.2064 0.0000 0.2952 0.0293 0.0162
0.0000 0.0622 0.0033 0.0000 0.0449 0.0149 0.0000
CEACAM5 0.0000 0.0342 0.0884 0.0016 0.0573 04906 0.0259
0.0000 0.0291 0.0783 0.2582 00113 0.0000 0.0061
CEACAM6 0.0000 0.0119 0.0048 0.0000 0.0065 0.0995 0.1930
0.0000 0.3695 0.0202 0.0160 04092 0.0020 0.0000
CEACAM7 0.0000 0.1211 0.1673 0.0000 0.1162 0.0211 0.0715
0.0000 0.0231 0.0023 0.0000 0.5022 0.0000 0.0000
CEACAM8 0.0000 0.0331 0.0057 0.0000 0.0361 0.0392 0.0932
0.0000 0.0093 0.0311 0.0078 00264 0.0046 0.0000
CGA 0.0000 00561 0.0075 0.0000 0.0083 0.0392 0.1350
0.0000 00293 0.0000 00000 00149 00000 0.0039
CGB3 00000 0.1212 0.0666 0.0987 00144 0_0253 00389
0.0000 0.1087 00064 0.0000 0_0295 0.0063 00000
CNN1 0.0000 0.2455 0.1790 0.0000 0.0246 0.1649 0.1165
0.0000 0.0061 0.0043 0.0000 0.1622 0.0000 0.0000
COQ2 0.0000 0.1545 00434 0.0000 0.0460 0.0509 0.0186
0.0000 00911 0.0454 00000 00338 0.0000 0.0000
CPS1 0.0000 00376 0.0288 0.0000 0.0337 02157 0.0971
0.0000 00678 0.1034 00030 0.1469 0.0815 0.0000
CR1 0.0000 00067 00219 0.0000 0.0680 0.1208 0.0306
0.0000 00547 0.0000 00000 00552 0.0160 0.0017
CR2 0.0000 00702 00070 0.0000 0.0613 0.1518 0.1308
0.0000 00320 0.0000 00010 00254 00081 0.0000
CTNNB1 0.0000 0.0503 0.0/77 0.0027 0.1224 0.0602 0.0430
0.0000 0.1372 0.0000 0.0000 0.1204 0.0081 0.0000
DES 043000 0.1269 0.2030 0.0019 00049 0_0554 03589
0.0000 0.2451 00278 0.0047 0_0532 0.0000 00000
DSC3 0.0000 0.0947 0.0479 0.0240 0.2025 0.1638 0.2982
0.0000 0.0491 0.0146 0.1840 00709 0.0055 0.0174
EN02 0.0000 0.2213 0.1018 0.0484 0.0245 0.1621 0.0513
0.0025 0.3330 0.1448 0.0021 00740 0.0155 0.0000
ERBB2 0.0000 0.0523 0.0108 0.1156 0.0067 0.0140 0.1281
0.0145 0.0472 0.0674 0.1205 0.1194 0.0050 0.0021
ERG 0.0000 0.0378 0.0427 0.0071 0.1084 0.1028 0.0444
0.0000 0.0110 0.0037 0.0097 0.0424 0.0000 0.0000
ESR1 0.0000 0.4155 0.0774 0.0000 0.6968 0.1522 0.5633
0.0000 0.0694 0.0454 0.0191 0.1661 0.0141 0.0000
FLI1 0.0003 00191 00309 0.0037 0.0111 0.0253 0.3088
0.0000 0.0185 0.0108 00000 0.1259 00007 0.0000
FOXL2 0.0000 00337 00212 0.0000 0.1575 0.1196 0.0875
0.0000 0.1158 0.0000 0.0380 0.0138 0.0000 0.0000
FIFT4 00000 0.0441 0.0859 0.0000 02820 0_3326 00713
0.0000 0.7653 0_1120 0.0447 0_0897 0.0148 00000
GATA3 0.0000 0.1473 1.9751 0.0409 0.0403 0.1323 0.1365
0.0000 0.0156 0.0369 0.0086 0.1119 0.1175 0.0234
GPC3 0.0000 0.0757 0.0184 0.1721 0.0000 0.1183 0.1398
0.0000 0.0291 0.0271 0.1407 0.1804 0.0000 0.0003
HAVCR1 0.0000 0.0760 0.0267 0.0000 0.0102 00567 0.0489
0.0000 0.0167 0.4287 0.0121 0.1936 9.0000 0.0000
1INF1B 0.0000 0.9014 0.4113 0.0000 0.0330 02249 0.0448
0.0000 0.0365 0.3831 0.0073 00741 0.0000 0.0000
11,1211 00000 0.0407 0.0351 0.0000 00778 0_0270 00236
0.0000 0.0367 00026 0.0800 0_1086 0.0000 00000
IMP3 0.0000 0.0395 00232 0.0000 0.0363 02060 0.0144
0.0000 00197 0.0000 00006 0.1069 0.0000 0.0000
INKS. 01270 0.1763 0.0491 0.0337 00644 0_1489 01608
0.0000 0.1896 00112 0.0000 0_0843 0.0610 00769
ISL1 0.0000 0.0894 0.1559 0.0043 0.1671 0.0771 0.0211
0.0000 0.4124 0.0081 0.0187 0.1219 0.0000 0.0000
KIT 0.0000 0.0272 0.1239 0.0000 0.0029 0.0612 0.0580
0.0677 0.1704 0.0761 0.0026 0.1541 0.0000 0.0000
KLK3 0.0000 00507 0.0645 0.0000 0.0174 0.1677 0.0545
0.0000 00066 0.0558 00000 0.0553 0.0000 0.0000
KL 0.0000 0.1828 0.1707 0.0000 0.0316 0.0214 0.0754
0.0000 0.0900 0.3624 0.0000 0.0176 0.0024 0.0000
KRT10 0.0000 0.0200 0.0073 0.0000 0.0214 0.1886 0.0352
0.0000 0.0303 0.0000 0.0076 0.2021 0.0267 0.1797
212
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
KRT14 0_0000 0 1351 0 1226 0 0047 0_0079 0_0936 0_1089 0
0000 0 1042 0_0000 0 0000 0_0556 80000 0_0000
KRT15 0.0000 0 0453 0.6266 0.0156 0.0438 0.0457 0.0559
0.0000 0.1042 0.0032 0.1799 0.2116 0.0000 0.0000
Is1RT16 0.0000 0.0358 0.2420 0.0008 0.0467 0.0180 0.0128
0.0000 00260 0.0000 00792 0.0515 0.0000 0.0452
KRT17 0.0000 0.1331 00193 0.0061 0.1592 0.0570 0.0143
0.0008 00463 0.0581 00004 0.1115 0.0349 0.0000
KRT18 0.0000 0.0201 0.4157 1.0434 0.0172 02612 0.0282
0.0000 0.0531 0.0007 0.0831 00396 0.0586 0.0000
KRT19 0.0670 0.0128 0.0/89 0.3758 0.0000 0.0356 0.0527
0.3005 0.0545 0.0108 0.4374 00656 0.5359 0.0000
KATI 0.0000 00148 00119 0.0008 0.0177 0.0026 0.0414
0.0000 00274 0.0043 00037 00204 0.0000 0.0000
KRT20 0_0000 0 0344 0 0077 0.0000 0_0826 0_7625 0_0481
0.0000 0.0898 0_0000 0.0031 0_1707 8.0000 0_0000
KRT2 0.0000 0.0212 0.0551 0.0000 0.0544 0.0247 0.0444
0.0000 0.1291 0.0657 0.0000 00423 0.0000 0.0000
KRT3 0.0000 00490 0.0538 0.0000 0.0224 00041 0.0061
0.0000 00014 0.0000 00000 00127 0.0807 0.0000
KRT4 0.0000 0.1454 0.0520 0.0000 0.0932 0.1828 0.0783
0.0000 0.0421 0.0000 0.0024 0.0245 0.0000 0.0000
KRT5 0.0000 0.2816 0.1591 0.0042 0.0038 0.0270 0.3821
0.0000 0.0270 0.0033 0.0000 0.2748 0.0000 0.0000
KRT6A 0.0000 0.0124 0.0774 0.0010 0.0022 02649 0.0206
0.0000 0.0639 0.0000 0.0446 0.1030 0.0006 0.0000
KRT6B 0.0000 0.0895 0.2370 0.0000 0.0026 03555 0.0083
0.0000 00319 0.0084 00000 00573 00007 0.0000
KRT6C 0_0000 0.0171 0.0874 0.0000 0_0809 0_0272 0_0616
0.0000 0.0422 0_0000 0.0000 0_0705 8.0007 0_0000
KRT7 0.0000 0.2611 0.5100 0.1042 0.0374 14166 0.0785
0.0164 0.0742 0.3134 0.0000 04525 0.0000 0.0051
KRT8 0.0295 0.1635 0.0546 1.0032 0.0436 0.0185 0.0389
0.2585 0.0500 0.0092 0.0000 0.1172 0.8518 0.4163
L1N28A 0.0000 0.0122 0.0287 0.0000 0.3409 0.0741 0.0268
0.0000 0.0244 0.0000 0.0150 00186 0.0975 0.0000
L1N28B 0.0000 0.0373 0.0/32 0.0021 0.0000 0.0228 0.4217
0.0000 0.0021 0.0000 0.0000 00462 0.0000 0.0000
MAGEA2 0.0000 0.1055 0.0066 0.0000 0.0013 0.0025 0.0102
0.0000 0.0554 0.0000 0.0000 00529 0.0123 0.0126
MDM2 0.0000 0.1220 0.2848 0.0019 0.2589 0.0265 0.1140
0.0000 0.0116 0.1901 0.0000 00210 0.0000 0.0471
84031 0_1185 0 0235 0 1144 0 0000 0_0718 0_0828 0_0719 0
0000 0 0092 013410 0 0000 0_0132 80080 0_0000
MITE 0.0000 0.0981 0.0159 0.0053 0.1067 0.0571 0.2480
0.0000 0.0311 0.0005 0.0040 0.1927 0.2270 0.0108
MLANA 0.0000 0.0948 0.0481 0.0132 0.1234 0.0678 0.0679
0.0000 0.0640 0.0174 0.0000 0.1531 0.4586 0.0000
MLH1 0.0000 00557 00199 0.0000 0.0783 02382 0.2500
0.0000 00131 0.0100 00000 00699 0.0000 0.0000
MME 0.0000 0.0823 0.0803 0.0000 0.1093 0.1141 0.0662
0.0000 0.0227 0.0685 0.0000 0.0496 0.0000 0.0000
MPO 0.0000 0.0714 0.0100 0.0000 0.0560 0.0020 0.0441
0.0000 0.0248 0.0075 0.0000 00580 0.0000 0.0165
MS/A1 0.0000 0.1279 0.0/70 0.0000 0.0626 0.0565 0.0126
0.0000 0.0050 0.0113 0.0033 0.1088 0.1585 0.0000
MS112 0.0000 00366 0.0268 0.2361 0.0199 0.0610 0.0421
0.0000 00532 0.0544 0.2183 00431 0.0000 0.2008
MSH6 0_0000 0 0193 0 0137 00039 0_0148 0_0060 0_0889 0
0000 0 0919 0_0000 0 0033 0_0740 80065 0_0000
MSL_N 0.0000 0.0536 0.0586 0.0000 0.0148 0.1393 0.1502
0.0000 0.0249 0.1571 0.0576 0.1468 0.0000 0.0094
MTHFR 0.0000 00140 0.2133 0.0000 0.0400 0.0393 0.0463
0.0000 0.1256 0.0406 00027 00453 0.0095 0.0000
MUC1 0.0535 0.0929 0.0032 0.0061 0.0649 05842 0.0903
0.2777 0.1772 0.2964 0.1388 0.2699 05180 0.0000
MUC2 0.0000 0.0219 0.0125 0.0000 0.2677 1.1616 0.0161
0.0000 0.0173 0.0018 0.0000 00526 0.0000 0.0000
MUC4 0_0000 0.3099 0.4270 0.0035 0_1352 0_1016 0_1268
0.0000 0.2198 0_0443 0.3336 0_2033 8.0000 0_0147
MTJC5AC 0.0000 0.1903 0.2662 0.0000 0.1500 0.0143 0.1385
0.0000 0.5114 0.0777 00118 0.1097 0.0000 0.0000
MYOD1 0_0000 0 0345 00864 0 0000 0_0359 0_0120 0_1814 0
0000 0 0446 0_0000 0 0276 0_0376 80035 0_0000
MYOG 0.0000 0.0217 0.0755 0.0059 0.0020 0.0333 0.0947
0.0000 0.1759 0.0000 0.0011 00228 0.0997 0.0000
NANOG 0.0000 0.0207 0.0311 0.0079 0.0975 00155 0.1539
0.0000 0.1042 0.0055 0.0000 00586 0.0000 0.0000
NAPSA 0.0000 0.0940 0.0983 0.0102 0.0449 0.0454 0.3890
0.0000 0.3190 0.0000 0.0000 10851 0.0042 0.0022
NCAM1 0.0161 0.0385 0.0786 0.5217 0.2480 0.0031 0.0604
0.0000 0.0083 0.0022 0.0000 00437 0.0660 0.0000
NCAM2 0.0294 0.1541 0.0382 0.0000 0.0480 02094 0.0676
0.0000 0.4229 0.0000 0.0000 0.1625 0.0466 0.0000
213
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
NKX2-2 0_0000 0 2202 0 0439 0 4077 0_0319 0_0222 0_1920 0
0000 0 0088 0_0000 0 0000 0_0601 0 0310 0_0000
NKX3-1 0.0715 0.1334 0.0299 0.0000 0.0489 02269 0.0418
0.0000 0.1014 0.0067 0.0048 0.1436 0.0000 0.0000
OSCAR 0.0000 0 0762 00949 0.0396 0.0145 0.1087 0.0906
0.0000 0 0190 0.0000 00000 0.0515 0.0000 0.0000
PAX2 0.0000 0.0091 0.0384 0.0000 0.0227 0.0384 0.1052
0.0000 0.0748 0.2851 0.0000 0.1045 9.0000 0.0000
PAX5 0.0000 0.0863 0.0813 0.0000 0.0260 0.0289 0.2066
0.0000 0.0915 0.0000 0.0000 00110 0.0256 0.0023
PAX8 0.0000 0.1905 0.4312 0.0000 0.1539 0.1731 1.6954
0.0000 0.3831 0.7741 0.0000 0.3878 9.0006 0.0082
PDPN 0.0000 0.0141 0.1592 0.4476 0.0048 0.0262 0.2675
0.0000 0.1346 0.0000 0.0000 00637 0.1012 0.0017
PDX1 0_0000 0 0993 00502 0 0000 0_0847 0_0691 0_0120 0
0000 0 1910 0_0000 00202 0_1244 90000 0_0000
PECAM I 0.0000 0.1201 0.1237 00000 0.0051 0.0367 0.0310
0.0000 0.1697 0.0504 00000 00164 00011 0.0000
PGR 0.0000 0.0619 0.1286 0.0000 0.3198 0.1078 0.5994
0.0000 0.0301 0.0000 0.0032 00448 9.0020 0.1911
PIP 0.0000 0.0909 0.3383 0.0000 0.0293 0.0208 0.1348
0.0000 0.0375 0.0072 0.0026 00842 9.0000 0.0000
PMEL 0.0000 0.0805 0.2466 0.0000 0.2023 0.0290 0.0776
0.0000 0.2113 0.0038 0.0297 00551 0.6758 0.0000
PMS2 0.0000 0.0404 0.0188 0.0000 0.0266 0.0101 0.0546
0.0000 0.1613 0.0000 0.0155 00196 9.0020 0.0000
POU5FI 0.0000 0.1802 00734 0.0000 0.0068 0.0667 0.0884
0.0000 00566 0.2956 0.1149 01029 0.1426 0.0000
PSAP 0_0153 0.2165 0.0939 0.0000 0_2756 0_0281 0_0901
0.0000 0.0982 0_0120 0.0000 0_0394 0.0000 0_0000
PTPRC 0.0000 0.0430 0.0243 0.0185 0.0000 0.0497 0.1087
0.0000 0.0321 0.0060 0.0000 00206 0.0055 0.0000
S100A10 0.0000 0.0535 0.1032 0.0048 0.1155 0.0099 0.0497
0.0000 0.0309 0.0598 0.0000 0.4226 0.0000 0.0067
S100A1 1 0.0000 0.0266 0.0222 0.2679 0.0665 0.0535 0.1391
0.0000 0.2227 0.0069 0.0095 00586 9.0137 0.0000
5100Al2 0.0000 00118 0.1145 0.0000 0.1333 0.1050 0.0291
0.0000 0.1106 0.0000 00010 00800 0.0000 0.0000
S100A13 0.0000 0.0531 0.1346 0.0000 0.2296 0.0142 0.0090
0.0000 0.3664 0.2409 0.0097 03093 0.2785 0.0000
S100A14 0.0000 0.1249 0.2299 0.2962 0.0198 02156 0.0664
0.0000 0.0307 0.1307 0.0000 00213 0.3043 0.2359
S100A16 0_0000 0.0250 0.0146 0.0024 0_0054 0_0070 0_2035
0.0046 0.0300 0_0000 0.0000 0_0073 0.0000 0_0000
S100A1 0.0000 0.0617 0.3432 0.2453 0.1060 0.0155 0.0530
0.0000 0.0570 0.0082 0.0002 03935 0.2097 0.0000
S100A2 0.0000 0.2901 0.4465 0.0903 0.1006 0.1114 0.1342
0.0180 0.1053 0.0000 0.0680 00470 0.0117 0.2339
S100A4 0.0000 0.0947 0.0464 0.0483 0.0028 0.0979 0.0217
0.0000 0.0110 0.0032 0.0000 00296 9.0153 0.0183
S100A5 0.0464 0.0693 0.0477 0.0241 0.0479 0.0165 0.1167
0.0000 0.1373 0.0225 0.0000 0.0717 0.0227 0.0018
S100A6 0.0000 0.2004 0.2369 0.0000 0.1529 04517 0.3725
0.0000 0.0480 0.0000 0.1595 0.1261 0.0000 0.0153
S100A7A 0.0000 0.1159 0.0065 0.0000 0.0334 0.0696 0.0677
0.0000 00632 0.0000 00061 0.0250 0.0000 0.0000
S100A7L2 0.0000 0.0094 0.1057 0.0000 0.0290 0.0075 0.0166
0.0000 0.0077 0.0000 0.0000 0.0041 0.0000 0.0000
S100A7 0_0000 0.0148 0.0100 0.0000 0_0419 0_0515 0_1609
0.0000 0.2783 0_0000 0.0000 0_1521 0.0007 0_0000
SIO0A8 0.0000 0.0450 0.0116 0.0000 0.0080 0.0427 0.0198
0.0000 0.0256 0.0018 0.0029 00366 0.0000 0.0175
S100A9 0.0000 0.2209 0.0939 0.0000 0.0765 0.0773 0.2121
0.0020 0.2167 0.0000 0.0000 00603 0.0010 0.0322
S10013 0.0000 0.0517 0.0971 1.0716 0.2872 0.0174 0.0168
0.0000 0.3090 0.0480 0.0154 0.0283 1.2799 0.0000
SlOOPBP 0.0000 0.1183 00459 0.0002 0.0442 0.0178 0.0391
0.0000 0.0150 0.0044 00000 01418 00161 0.0000
SlOOP 0_0000 0.0464 0.1035 0.0000 0_0458 0_0154 0_2953
0.0000 0.0415 0.4360 0.0020 0_0287 0.1176 0_0031
SlOOZ 0.0000 00392 0.0013 0.0061 0.0019 0.0148 0.0261
0.0000 00333 0.0678 00000 01288 0.0000 0.0000
SALL4 0_0000 0.1235 0.1416 0.0314 0_1017 0_0255 0_1639
0.0000 0.1336 0_1856 0.0029 0_0184 0.0000 0_0155
SATB2 0.0000 0.2178 0.0032 0.0000 0.2461 0.5521 0.0431
0.0000 0.1301 0.0017 0.0588 00746 0.1050 0.0000
SDC1 0.0000 0.0448 0.0625 0.0024 0.0561 0.0818 0.0334
0.4088 0.0614 0.0000 0.0000 0.1180 0.0000 0.6138
SERPINA1 0.0158 0.5546 0.1814 0.0000 0.0515 0.0237 0.0520
0.0000 0.0987 0.0859 0.7962 00604 0.0000 0.0000
SERPINB5 0.0000 0.0840 0.2329 0.0000 0.0082 0.1128 0.0562
0.0000 0.5175 0.0280 0.0141 01436 0.0000 0.0018
SF! 0.0000 0.0445 0.0725 0.0000 0.0242 0.0260 0.0164
0.0000 0.0592 0.1009 0.0067 0.1398 0.0000 0.0015
214
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
SFTPA1 00000 01372 0 0461 0 0000 0_0110 0.0188 0_0331 0
0000 0 0953 0_0151 0 0000 0_2640 00020 0_0000
SMAD4 0.0000 0.0423 0.0369 0.0000 0.0093 0.0888 0.0668
0.0000 0.0800 0.0033 0.0081 00067 0.0000 0.0000
SMARCB1 0.0000 0.0753 0.0065 0.0325 0.3181 0.0016 0.2247
0.0000 0.0813 0.0096 0.0063 0.1316 0.0000 0.0333
SMNI 0.0000 0.1124 0.0081 0.0027 0.0768 0.0181 0.1144
0.0000 0.0492 0.0082 0.0000 00576 0.0000 0.0000
SOX2 0.0003 0.3363 0.3114 0.7907 0.0563 0.1969 0.0355
0.0000 0.3802 0.0220 0.0161 05792 0.0062 0.0000
SPN 0.0000 0.0141 0.0546 0.0000 0.0030 0.0777 0.0667
0.0000 0.2709 0.0000 0.0006 00173 0.0000 0.0398
SYP 0.1109 0.0444 0.0986 0.0000 0.0074 0.0356 0.0852
0.0000 0.1467 0.1603 0.0000 00204 0.0046 0.0000
TFE3 0_0000 01307 0 1111 0 0000 0.0183 0_0067 0_0179 0
0000 0 0119 0_0340 0 0000 0_0313 0 0034 0_0000
TFF1 0.0000 0.1821 0.2434 0.0000 0.0033 02416 0.0509
0.0000 0.4452 0.0000 0.0229 0.2230 0.0000 0.0000
TFF3 0.0000 00476 0.1606 0.0000 0.0381 03417 0.1866
0.0000 04172 0.0689 00000 00481 0.0021 0.0000
TO 0.0279 0.1321 0.0160 0.1140 0.0092 0.0808 0.0674
0.0000 0.0637 0.0481 0.0000 0.1287 0.0000 0.0008
TLEI 0.0000 0.1445 0.0225 0.0018 0.0051 0.0395 0.2590
0.0000 0.0294 0.0695 0.0000 0.1319 0.0032 0.0000
TMPRSS2 0.0297 0.1909 0.0829 0.0430 0.0078 0.1968 0.0803
0.0000 0.2937 0.0505 0.0000 0.2302 0.0000 0.0000
TNFRSF8 0.0004 0.0265 0.1215 0.0000 0.2457 0.0337 0.0043
0.0000 0.0157 0.0005 0.0054 0.1232 0.0020 0.0000
TP63 0_0000 0 0365 0 1117 00007 0_1018 0_0123 0_0739 0
0000 0 0123 0_0054 0 0000 0_0642 0 1038 0_1028
TPMI 0.0000 0.1078 0.0858 0.0045 0.0382 0.0673 0.0464
0.0000 0.2065 0.0011 0.0000 0.1372 0.1401 0.0021
TPM2 0.0000 0.0575 0.0205 0.0050 0.1451 0.0259 0.0845
0.0000 0.1216 0.0090 0.0149 00342 0.0000 0.0000
TPM3 0.0120 0.0484 0.0228 0.0048 0.0748 0.0085 0.0712
0.0000 0.0092 0.0519 0.0000 0.1855 0.0091 0.0082
TPM4 0.0000 0.0822 0.0866 0.0000 0.0337 0.0916 0.0518
0.0000 0.0468 0.0411 0.0549 0.1722 0.0000 0.0000
TPSABI 0.0000 0.1863 0.0758 0.0028 0.2121 0.1570 0.0613
0.0018 0.3180 0.1164 0.0000 00876 0.0000 0.0000
TTFI 0.0000 0.0503 0.0094 0.0812 0.1321 0.0279 0.1320
0.0000 0.1,192 0.0803 0.0215 00727 0.0215 0.0000
UPIC2 0_0000 0.0412 0.0201 0.0222 0_1078 0_1170 0_0764
0.0000 0.1224 0_0000 0.0000 0_0776 0.0000 0_0000
UPK3A 0.0000 0.0213 0.1437 0.0017 0.0078 0.0162 0.2065
0.0000 0.0446 0.0000 0.0698 00076 0.1314 0.0000
UPK3B 0.0000 0.1889 0.2206 0.0169 0.1160 0.0398 0.0594
0.0000 00467 0.0148 00042 0.1143 00036 0.0000
VEL 0.0003 0.0806 0.0534 0.0000 0.2247 0.0285 0.4873
0.0000 0.0736 0.2955 0.0000 0.3369 0.0000 0.0067
VIL1 0.0000 0.5994 0.0240 0.0000 0.0848 05227 0.0238
0.0000 0.3881 0.0064 0.1221 0.0326 0.0682 0.0000
VIM 0.0000 0.0188 0.0328 0.0000 0.0033 0.0468 0.0369
0.0000 0.0438 0.0765 0.0000 00137 0.1803 0.2430
WTI 0.0000 0.0811 0.0/66 0.0160 0.0391 0.0392 0.2561
0.0000 0.0696 0.0411 0.0000 0.17,18 0.0000 0.0216
Table 118 continued
Transcript Merk Neu OGCT OFF Panc PM PA Ret SP SIA SCC TC UC Ute
ACVRL1 0.0000 0.0000 0.0000 0.2065 0.0367 0.0000 0.0000
0.0022 0.0000 0.0096 0.0034 00000 0.0587 0.0100
APP 0.0000 0.0047 0.0000 0.0347 0.0163 0.0000 0.0000
0.0346 0.0000 0.0633 0.0672 00000 0.0249 0.0000
ALPP 0.0000 0.0000 0.0000 0.2427 0.0571 0.0000 0.0214
0.0000 0.2317 0.1172 0.0751 0.0000 0.0233 0.0000
AMACR 0.0000 0.0028 0.0033 0.1114 0.2357 0.0008 0.5918
0.0000 0.0000 0.0164 0.0335 00044 0.0899 0.0025
ANKRD30A 0.0000 0.0061 0.0000 0.0726 0.1040 0.0000 0.0000 0.0000 00064 0.0118
0.0134 00000 00109 0.0019
ANO1 0.0000 0.0183 0.0000 0.1417 0.7039 0.0000 0.0177
0.0074 0.1828 0.0138 0.1547 0.0052 0.1598 0.0055
ARGI 0_0000 0_1080 0.0000 0.1220 0.2156 0_0000 0_0000
0_0497 0.1190 0_2540 0_0613 0_2657 0.0133 0_0300
AR 0.0000 0.0181 0.0000 0.1520 00692 0.0000 0.1169
0.1206 00000 0.1860 0.4215 00031 00096 0.0465
BCL2 0_0000 0_0000 00500 0 0560 0 0404 0_0000 0_0140
0_0014 0 0321 0_0398 0_0403 0_0014 80029 0_0091
BCL6 0.0000 0.0100 0.0000 0.0155 0.0300 0.0027 0.0718
0.0330 0.0000 0.0157 0.0300 00032 0.0671 0.0623
CAS 0.0013 0.0612 0.0000 0.1736 0.0732 0.0321 0.0211
0.0000 0.0098 0.1940 0.0569 00237 0.0861 0.0000
215
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CAL132 00000 00035 0 0000 00618 0 3098 0_5246 00076 00156
0 1907 01585 00587 02775 0 3746 00372
CALCA 0.0000 0.0206 0.0018 0.1032 0.0794 0.0000 0.0050
0.0015 0.0028 0.0181 0.1741 00000 0.0055 0.0000
CALD1 0.0000 0.0438 0.0000 0.0481 0.0228 0.0000 0.0002
0.0166 0.0000 0.0237 0.0778 00000 0.0352 0.0325
CCNDI 0.0000 0.0316 0.0000 0.1941 0.0634 0.0000 0.0000
0.0017 0.0056 0.0445 0.0409 00799 0.0752 0.0000
CDIA 0.0000 0.0006 0.0000 0.0712 0.1698 0.0000 0.0036
0.0000 00000 0.0480 0.1672 00047 00610 0.0116
CD2 0.0000 0.0198 0.0000 0.0205 0.0681 0.0000 0.0032
0.0000 0.0040 0.0202 0.0112 00000 0.2658 0.0909
CD34 0.0000 0.0069 0.0000 0.0231 0.1297 0.0000 0.1084
0.2570 0.0005 0.0463 0.1436 00016 0.0352 0.0000
CD3G 00000 00333 0 0000 0 0154 0 0372 0_0000 00625 00000
0 0000 00306 04505 00077 0 2254 00069
CD5 0.0000 0.0224 0.0000 0.0271 0.3262 0.0000 0.0217
0.0035 0.0000 0.2452 0.0437 00189 0.1800 0.0177
CD79A 0.0000 0.0002 0.0000 0.0564 0.0607 00000 0.0000
0.0203 0.0088 0.0188 0.0938 00136 0.0361 0.4022
CD99L2 0.0000 0.0313 0.0000 0.1654 0.0522 0.0000 0.0119
0.0000 0.0000 0.2136 0.0335 00302 0.1242 0.0008
CDII17 0.0000 0.0270 0.0000 0.0926 0.1250 0.0000 0.0146
0.0076 0.0081 0.3786 0.0426 00000 0.0237 0.0687
CDH1 0.0000 0.0070 0.0000 0.0031 0.0312 0.0113 0.0772
0.1926 0.0074 0.0000 0.0790 0.1070 0.0024 0.1516
CDK4 0.0000 0.0000 0.0000 0.0402 0.0479 0.0000 0.0135
0.0780 0.0060 0.0515 0.1250 0.2140 0.1472 0.0444
CDKN2A 00000 00678 0 0000 0 0425 0 1363 00105 00475 00113
0 0061 01300 00548 00138 0 1118 00069
CDX2 0.0000 0.1367 0.0000 0.0507 0.1207 0.0000 0.0325
0.0176 0.0000 0.0253 0.0662 00000 0.0222 0.0000
CEACAMI 6 0.0000 0.0000 0.0000 0.0865 0.0625 0.0000 0.0025 0.0000 0.1820
0.0526 0.0256 00237 0.1766 0.0104
CEACAM18 0.0000 0.0270 0.0000 0.0307 0.1543 0.0000 0.0923 0.0095 0.1035 0.1317
0.0344 00488 0.0016 0.0045
CEACAM19 0.0000 0.0018 0.0000 0.1167 0.0660 0.0000 0.0045 0.0212 0.0000 0.0280
0.0753 0.0176 0.0388 0.0097
CEACAMI 0.0000 0.0000 0.0000 0.0246 0.0927 0.1300 0.1096
0.0563 0.0014 0.1391 0.1982 00111 0.0651 0.0554
CEACAM20 0.0000 0.0000 0.0000 0.0136 0.0637 0.0000 0.0028 0.0000 0.0000 0.0223
0.0393 00000 0.0000 0.0000
CEACAM21 00000 00000 0 0035 0 1164 0 0118 00000 0_1023 00000 0 0056 00265
00104 00000 0 0456 00000
CEACAM3 0.0000 0.1156 0.0000 0.2474 0.1011 0.0057 0.0373
0.0000 0.0020 0.0944 0.0497 0.0715 0.0567 0.0265
CEACAM4 0.0013 0.1420 0.0000 0.0370 0.0907 0.0000 0.0047
0.0000 0.0000 0.1055 0.0318 00463 0.1265 0.0000
CEACAM5 0.0473 0.1210 0.0000 0.2252 0.0651 0.0000 0.0792
0.0043 0.0000 0.3319 0.0687 0.2028 0.0849 0.0000
CEACAM6 0.0000 0.0044 0.0000 0.1199 0.1324 0.0000 0.1188
0.0062 0.0000 0.0081 0.1136 0.0340 0.1440 0.0000
CEACAM7 0.0000 0.0007 0.0000 0.0685 0.1338 0.0000 0.0011
0.0000 0.0000 0.0537 0.0276 00000 0.0443 0.0000
CEACAM8 0.0000 0.0085 0.0000 0.0469 0.0591 0.0000 0.0076
0.0000 0.0007 0.0485 0.1073 00000 0.0411 0.0019
CGA 0.0000 0.0132 0.0000 0.0208 0.1910 0.0000 0.0094
0.0076 0.0000 0.0873 0.0434 00477 0.0426 0.0000
CG133 00000 00000 0 0000 00668 0 0102 0_0000 0_1259 00071
0 0000 0_1308 02238 00000 0 0368 00503
CNN1 0.0000 0.0065 0.0000 0.0826 0.0256 0.0000 0.1392
0.1850 0.0135 0.1274 0.2971 0.2199 0.1757 0.0918
COQ2 0.0000 0.0049 00000 0.0162 0.1601 0.0000 0.0000
0.0000 00000 0.0096 0.0972 00000 0.0268 0.0062
CPS1 0.0306 0.0010 0.0000 0.1042 0.2197 0.0030 0.1975
0.0849 0.0308 0.1777 0.0843 04173 0.4016 0.0000
CRI 0.0175 0.0010 0.0000 0.2003 0.0521 0.0000 0.0238
0.0206 0.0150 0.1249 0.1301 00029 0.0314 0.0092
CR2 00000 00000 0 0000 0 1221 01608 0_0000 00502 00000
0 0052 01074 00474 00000 0 0217 00000
CTNNB1 0.0000 0.0038 0.0000 0.0528 0.0185 0.0000 0.0000
0.0000 0.1967 0.0000 0.1189 00000 0.3425 0.0000
DES 00000 00555 0 0000 0 0907 0 2096 00000 00000 00014
0 0022 04895 01498 00000 0 3442 0_5577
DSC3 0.0000 0.1499 0.0000 0.1993 0.0164 0.0000 0.0430
0.0024 0.2247 0.1327 0.3182 00958 0.0009 0.0011
EN02 0.0012 0.4094 0.0000 0.2069 0.0417 00000 0.0527
0.0019 0.6462 0.0198 0.0625 00171 0.0286 0.2003
ERBB2 0.2359 0.1385 0.0000 0.1432 0.1510 0.0000 0.0049
0.0000 0.2965 0.1034 0.0228 0.0380 0.0421 0.0895
ERG 0.0000 0.0572 0.0000 0.0488 0.0708 0.0000 0.0275
0.0107 0.0000 0.1162 0.0789 00044 0.0956 0.0495
ESR1 0.0000 0.0700 0.0000 0.2085 0.2562 0.0000 0.0145
0.0053 0.0000 0.2587 0.2922 0.0007 0.1219 0.3616
216
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
FIJI 0_0007 00119 0 0062 0 0702 0 0237 0_0091 0_0071
0_0048 0 0056 0_0931 0_0471 0_0126 0 0186 0_0910
FOXL2 0.0000 0.0000 0.6541 0.3268 0.0217 0.0000 0.0038
0.0068 0.0000 0.0073 0.1735 0.1298 0.0158 0.4519
EUT4 0.0000 0.0355 0.0000 0.2257 0.4461 0.0000 0.0217
0.0000 0.0000 0.0113 0.1870 00056 0.0874 0.0034
GAT_A3 0.0000 0.0087 0.0000 0.0255 0.7533 0.0000 0.0126
0.0035 0.0000 0.1591 0.0991 0.1194 1.3531 0.0416
GPC3 0.0000 0.0483 0.0000 0.1366 00427 0.0000 0.0030
0.0061 00000 0.1143 0.0288 00000 0.1322 0.0038
HAVCR1 0.0000 0.0244 0.0000 0.0296 00290 0.0008 0.0000
0.0000 00997 0.1009 0.1116 00356 0.0612 0.0017
TINF1B 0.0000 0.0097 0.0000 0.0412 0.2391 0.0000 0.0117
0.0000 0.1674 0.2912 0.1936 0.2745 0.1571 0.0000
TI12B 0_0000 0_0270 0.0000 01U2 0 0112 0_0000 0_0545
0_0016 0.0086 0_0484 0_0191 0_0000 0.0067 0_0000
IM1P3 0.0000 0.0000 0.0000 01021 00161 0.0000 0.0068
0.0000 00000 0.0256 0.1442 0.0083 0.0145 0.0110
1NHA 0.0000 0.1020 0.0000 0.5386 0.0755 0.1400 0.0474
0.0000 0.0687 0.0125 0.0112 0.2668 0.0717 0.0000
ISL1 0.2415 0.5980 0.0000 0.1816 0.6570 0.0000 0.0000
0.0000 0.0000 0.0468 0.0848 00062 0.1594 0.0000
KIT 0.0000 0.0140 0.0000 0.0467 0.0867 0.0000 0.0043
0.1085 0.1652 0.0227 0.0778 00000 0.0080 0.0058
KLK3 0.0000 0.0140 0.0000 0.0130 0.0244 0.0000 1.2859
0.0000 00000 0.0032 0.0845 00000 0.0148 0.0000
ICL 0.0000 0.0000 0.0000 0.1202 0.0208 0.0000 0.2215
0.0345 0.0000 0.0091 0.0269 00349 0.1833 0.0000
ICRT10 0_0000 0_1224 0.0000 0.0549 0.1298 0_0000 0_0055
0_0177 0.0000 0_0952 0_0443 0_0044 0.0308 0_0076
KRT14 0.0000 0.0120 0.0000 0.0077 0.0418 0.0003 0.0028
0.0000 0.3191 0.0859 0.0383 00053 0.1801 0.0000
KRT15 0.0000 0.0241 0.0000 0.1212 0.0182 0.0000 0.0443
0.0081 0.0000 0.0737 0.1695 00000 0.0225 0.0000
KRT16 0.0000 0.0000 0.0000 0.0369 00679 0.0000 0.0000
0.0026 00163 0.0053 0.0550 00488 0.0050 0.0000
KRT17 0.0000 0.0183 0.0000 0.1493 0.0220 0.0000 0.0508
0.0000 0.0000 0.0417 0.5310 00329 0.1235 0.0010
KRT18 0.0000 0.0000 0.0000 0.1602 00248 0.0000 0.0772
0.6936 00110 0.1117 0.0600 00000 00102 0.7609
ICRT19 0.0000 0.0000 0.0000 0.0251 0.1952 0.0013 0.0515
0.7039 00276 0.0514 0.0339 0.0085 0.2366 1.0412
ICRT1 0_0000 0_0018 0 0031 0 0649 0 0446 0_0000 0_0021
0_0000 0 0167 0_0090 0_0199 0_0004 0 0298 0_0933
KRT20 0.0000 0.0000 0.0000 0.0395 00796 0.0000 0.0521
0.0000 00000 0.2969 0.3367 00000 0.5293 0.0015
KRT2 0.0000 0.0000 0.0000 0.0261 0.0074 0.0000 0.1371
0.0000 0.0000 0.0201 0.0433 00512 0.0236 0.0444
KRT3 0.0000 0.0000 0.0000 0.0489 0.1180 0.0006 0.0037
0.0000 00000 0.0072 0.0322 00000 0.0393 0.0129
KRT4 0.0000 0.0000 0.0000 0.0691 00339 0.0000 0.0000
0.0053 00107 0.0972 0.1146 0.0000 0.1128 0.0086
KRT5 0.0000 0.0000 0.0000 0.0525 0.0342 0.0464 0.0544
0.0000 0.0019 0.0574 0.4137 00000 0.0165 0.0000
ICRT6A 0.0000 0.0000 0.0000 0.0507 00534 0.0000 0.0755
0.0000 00000 0.0051 0.5694 00000 00213 0.0000
ICRT6B 0.0000 0.0011 0.0000 0.0278 0.2216 0.0000 0.0048
0.0042 0.0000 0.0341 0.1458 0.0000 0.0290 0.0903
KRT6C 0_0000 0_0000 0 0000 0 0387 0 2225 0_0000 0_0020
0_0000 0 0000 0_0400 0_1469 0_0000 0 0071 0_0000
KRT7 0.0660 0.0102 0.0000 0.0490 0.1859 0.0005 1.3765
0.0022 0.0544 0.0283 0.0844 00521 0.2697 0.0066
ICRT8 0.0000 0.0000 0.1357 0.0468 0.1697 0.0000 0.0534
0.6236 00000 0.0915 0.0253 0.1412 0.0053 0.1662
L1N28A 0.0000 0.0780 0.0000 0.1663 0.0102 0.0000 0.0186
0.0000 0.0255 0.0894 0.0626 0.0028 0.0074 0.0043
LIN28B 0.0007 0.0527 0.0000 0.0413 0.0414 0.0000 0.0025
0.0000 0.0000 0.0229 0.0846 0.1007 0.0607 0.0000
MAGEA2 0_0000 0_0000 0.0000 0.0006 0.0882 0_0000 0_0000
0_0000 0.0009 0_0000 0_0079 0_0000 0.0031 0_0000
MDM2 0.0000 0.1009 0.0000 0.0494 0.1451 0.0000 0.0000
0.1194 0.0224 0.1082 0.0439 00000 0.0195 0.1168
MI131 0_0000 0_0000 0 0000 0 0799 0 0341 0_0000 0_0075
0_0000 0 0000 0_0306 0_0208 0_0000 0 0021 0_0052
MIFF 0.0000 0.0000 0.0000 0.1419 0.0700 0.0000 0.0864
0.0017 0.0000 0.0541 0.0143 00720 0.3510 0.2870
MLANA 0.0006 0.0000 0.0000 0.0667 0.0316 0.0000 0.0027
0.0000 0.0444 0.0496 0.0525 00053 0.1215 0.0470
MLH1 0.0000 0.0626 0.0000 0.0548 0.1467 0.0000 0.0000
0.0000 0.0000 0.0187 0.0212 00773 0.0245 0.1779
MME 0.0532 0.0052 0.0112 0.0410 0.0900 0.0000 0.0346
0.0004 0.0000 0.2221 0.0427 00781 0.1436 0.0163
MPG 0.0000 0.1720 0.0000 0.0319 0.0217 0.0000 0.0005
0.0000 0.0000 0.2111 0.0431 0.1047 0.0350 0.0061
217
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
MS4A1 0_0000 0_0173 0 0000 0 0720 0 0001 0_0000 0_0000
0_0113 0 0000 00174 0_0021 00029 0 0050 0_0000
MSH2 0.0000 0.0039 0.0000 0.0545 0.2342 0.0027 0.0000
0.0060 0.0035 0.0118 0.2956 0.0045 0.0144 0.0591
MSII6 0.0000 0.0347 0.1914 0.0060 0.0730 0.0000 0.0000
0.0000 0.0125 0.0258 0.1152 0.0385 0.0057 0.0000
MSLN 0.0000 0.0000 0.0000 0.2905 0.2293 0.0843 0.1757
0.0000 0.0000 0.0904 0.0835 00353 0.3326 0.3346
MTHER 0.0000 0.0399 0.0000 0.0657 0.0602 0.0000 0.0020
0.0015 0.0000 0.0247 0.0902 00093 0.0718 0.0006
MTJC1 0.0000 0.1051 0.1647 0.1800 0.0815 0.0000 0.2526
0.0000 0.0253 0.0179 0.0801 0.1233 0.5292 0.0276
MUC2 0.0000 0.0000 0.0000 0.0507 0.0817 0.0000 0.2307
0.0000 0.0000 0.4382 0.0224 00056 0.0018 0.0049
MUC4 0_0066 0_1878 0.0000 0.0428 0 1120 0_0000 0_0217
0_0000 0.0000 0_1516 0_0536 0_1056 0.0034 0_0801
MUC5AC 0.0000 0.0000 0.0000 0 1069 0.5233 0.0000 0.1067
0.0000 00000 0.0320 0.0637 0.0000 0.1855 0.0000
MY0D1 0.0000 0.0004 0.0000 0.1284 00361 0.0000 0.0000
0.0000 00000 0.0328 0.0178 00000 0.0752 0.0049
MYOG 0.0767 0.0000 0.0000 0.0218 0.0141 0.0000 0.0021
0.0000 0.0043 0.0015 0.0644 00000 0.0291 0.0873
NANOG 0.0000 0.0064 0.0000 0.0363 0.0361 0.0000 0.0000
0.0000 0.0000 0.0123 0.0411 00073 0.0478 0.0308
NAPSA 0.0000 0.0406 0.0000 0.0559 0.2030 0.0000 0.0200
0.0007 0.0022 0.1853 0.1043 00003 0.2322 0.0000
NCAM1 0.0000 0.6042 0.0000 0.1455 0.0044 0.0000 0.0000
0.0000 0.0000 0.1297 0.0456 00132 0.0253 0.6726
NCAM2 0_0000 0_0000 0.0000 0.1088 0.1730 0_0006 0_0543
0_0000 0.0000 0_1071 0_0958 0_0103 0.0727 0_0321
NKX2-2 0.0000 0.0469 0.0000 0.1041 0.1918 0.0000 0.0406
0.0000 0.0579 0.0976 0.0559 00000 0.0855 0.0838
NKX3-1 0.0000 0.0162 0.0000 0.2255 0.0636 0.0000 1.2703
0.0000 0.0000 0.0145 0.0570 00286 0.0659 0.0010
OSCAR 0.0000 0.0008 0.0000 0.0600 0.2009 0.0000 0.0099
0.0026 0.0000 0.0245 0.1075 0.1099 0.0620 0.0284
PAX2 0.0000 0.0103 0.0000 0.0552 0.0219 0.0000 0.0000
0.0000 0.0000 0.0737 0.0483 00000 0.0477 0.0000
PAX5 0.0000 0.0000 0.0000 0.0671 00196 0.0000 0.0542
0.0000 00040 0.0528 0.0503 00162 0.1061 0.0000
PAYS 0.0000 0.1138 0.0000 0.8760 00330 0.0000 0.0026
0.0000 00892 0.0869 0.1754 0.6914 0.2608 0.0000
PDPN 0_0000 0_0000 0.0000 0.1066 0.2313 0_1504 0_0037
0_0078 0.0000 0_1543 0_2600 0_0025 0.0932 0_0256
PDX1 0.0000 0.0127 0.0000 0.1495 0.8076 0.0000 0.0202
0.0000 0.0000 0.7265 0.0707 00316 0.0336 0.0032
PECAM1 0.0000 0.0141 0.0000 0.0918 0.0178 0.0000 0.0730
0.0072 0.0000 0.0082 0.0297 00000 0.0080 0.0256
PGR 0.0000 0.0154 0.1352 0.1223 0.0433 0.0000 0.0214
0.0096 0.0000 0.0230 0.0572 00000 0.0142 0.0000
P11' 0.0000 0.0091 0.0000 0.0373 00157 0.0000 0.0799
0.0098 0.5509 0.0078 0.0342 0.0141 0.1562 0.0000
PMEL 0.0000 0.0000 0.0000 0.1900 0.0832 0.0000 0.1445
0.0000 0.0000 0.2305 0.0862 00058 0.0520 0.0740
PMS2 0.0000 0.0471 0.0000 0.0221 0.1820 0.0000 0.0438
0.0000 0.0000 0.0560 0.1036 00000 0.0549 0.0000
POU5F1 0.0004 0.3770 0.0000 0.2549 0.1719 0.0000 0.0000
0.0028 0.0000 0.0305 0.0599 0.0425 0.0268 0.0211
PSAP 0_0000 0_0000 0 0000 0 0594 0 0153 0_0000 0_0000
0_0000 0 0061 0_0384 0_1554 0_0155 0 0005 0_0000
PTPRC 0.0000 0.0129 0.0000 0.1692 0.0172 0.0024 0.0061
0.0000 0.0000 0.1415 0.0390 00028 0.0000 0.1112
S100A10 0.0000 0.0263 0.0000 0.2405 0.0918 0.0000 0.1119
0.0054 0.0000 0.0692 0.0531 00230 0.2036 0.0346
S100A1 I 0.0000 0.1247 0.0011 0.0184 0.1784 0.0007 0.0295
0.0000 0.0000 0.0037 0.0163 0.0006 0.0173 0.0112
S100Al2 0.0846 0.0066 0.0000 0.0844 0.0266 0.0000 0.0781
0.0000 0.0000 0.0582 0.0304 00000 0.0088 0.1121
S100A13 0_0000 0_0067 0.0000 0.3704 0.0017 0_0239 0_0601
0_0000 0.0000 0_0328 0_0461 0_0050 0.0091 0_0000
S100A14 0.0787 0.0124 0.0000 0.0590 0.1071 0.0000 0.0434
0.2697 0.0000 0.1100 0.2446 00683 0.1086 0.3884
S100A16 0_0000 0_0243 0.0000 0.0818 0.0216 0_0000 0_0600
0_0000 0.0047 0_0123 0_0207 0_0019 0.1370 0_0289
S100A1 0.0000 0.2747 0.0000 0.1272 0.0683 0.0000 0.0000
0.0000 0.3037 0.1091 0.4703 00000 0.0297 0.0107
S100A2 0.0000 0.0000 0.0000 0.0214 0.1344 0.0000 0.0271
0.0000 0.0027 0.1516 0.2694 0.2900 0.4107 0.0000
5100A4 0.0000 0.0068 0.0000 0.0840 0.2693 0.0000 0.0328
0.0000 0.0137 0.0158 0.0583 00000 0.1036 0.0168
S100A5 0.0000 0.0020 0.0000 0.0335 0.0678 0.0000 0.3275
0.0000 0.0000 0.0634 0.0096 00041 0.1003 0.0000
S100A6 0.0000 0.0127 0.0000 0.0136 0.0168 0.0000 0.0967
0.0000 0.0073 0.0402 0.2069 0.0200 0.0475 0.0000
218
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
S100A7A 0_0000 00000 0 0000 0 0492 0 1427 0_0004 0_0171
0_0000 0 0109 00029 0_0318 0_0021 1)0063 0_0115
SIO0A7L2 0.0000 0.0066 0.0000 0.0042 0.0012 0.0000 0.0000
0.0000 0.0000 0.0390 0.0553 00314 0.0173 0.0000
S100A7 0.0000 0.1408 0.0000 0.0500 0.0629 0.0000 0.0042
0.0000 0.0037 0.0085 0.0360 00000 0.0029 0.0000
S100A8 0.0000 0.0000 0.0000 0.0504 0.0777 0.0000 0.0043
0.0450 0.0082 0.1005 0.0850 00000 0.0119 0.0000
S100A9 0.0000 0.0436 0.0000 0.0086 00392 0.0000 0.0000
0.0082 00009 0.0330 0.0185 00047 00027 0.0000
S10013 0.0000 0.0000 0.0036 0.0204 0.0343 0.0000 0.0042
0.0272 0.0518 0.0473 0.0446 00082 0.0706 0.0833
SlOOPBP 0.0650 0.0176 0.0000 0.0800 0.0832 0.0000 0.0057
0.0142 0.0032 0.0051 0.0238 00204 0.0673 0.0144
SlOOP 0_0000 0_0000 0 0000 0 0740 0 2088 0_0000 0_0047
0_0218 0 0051 0_1975 00230 0_1375 0 3496 0_1993
S1OOZ 0.0000 0.1949 0.0000 00160 0.2012 0.0000 0.0125
0.0026 00000 0.0496 0.0178 0.0066 0.0035 0.0000
SALL4 0.0000 0.0000 0.0000 0.0322 0.2072 0.0000 0.0208
0.0000 0.1862 0.0444 0.0452 0.0292 0.3200 0.0245
SATB2 0.0000 0.0050 0.0000 0.0988 0.1879 0.0029 0.0332
0.0113 0.0128 0.0693 0.1365 00066 0.1447 0.1369
SDC1 0.0681 0.0167 0.2236 0.1215 0.0221 0.0000 0.1176
0.1562 0.0113 0.0265 0.3517 00279 0.0329 0.0632
SERPINA1 0.0000 0.0069 0.0076 0.1785 0.6933 0.0000 0.1383
0.0000 0.0000 0.3080 0.0627 00051 0.3476 0.0082
SERPIN115 0.0000 0.0607 0.0000 0.0683 0.1196 0.0000 0.0042
0.0012 0.0000 0.0982 0.2638 01166 0.0712 0.0000
SF! 0_0000 00000 0 0000 0 1115 0 1241 0_0163 00434
0_0000 0 0000 0_0401 00082 0_0047 0 0028 00000
SFTPA1 0.0000 0.0321 0.0028 0.1190 0.1051 0.0000 0.0945
0.0000 0.0000 0.2277 0.4403 0.0505 0.0514 0.0000
SMAD4 0.0000 0.0168 0.0000 0.0566 0.4264 0.0000 0.0020
0.0523 0.0181 0.0162 0.0363 00000 0.0314 0.0045
SMARCBI 0.0000 0.0000 0.0000 0.1221 0.2192 0.1813 0.0000
0.0000 0.0000 0.0136 0.0824 00183 0.0000 0.0000
SMN1 0.0000 0.0090 0.0000 0.0235 0.2683 0.0000 0.0000
0.0000 0.0000 0.1115 0.0403 0.0125 0.0218 0.0472
SOX2 0.0000 0.0342 0.0000 0.2216 0.2178 0.0000 0.0115
0.0031 0.0419 0.2305 0.6443 00000 0.1667 0.0869
SPN 0.0000 0.0223 0.0000 0.1472 0.1709 0.0000 0.0000
0.0000 0.01,16 0.1605 0.0583 00211 0.0367 0.0265
SYP 00000 0_3155 0 0000 0 2023 0 0230 0_0087 0_0283
00007 0 0000 0_1538 00614 0_0493 1)0275 0_0117
TFE3 0.0000 0.0000 0.0000 0.3920 0.0098 0.0000 0.0210
0.0060 0.0000 0.0933 0.0856 00000 0.0137 0.0012
TFFI 0.0000 0.0045 0.0000 0.0313 0.2263 0.0000 0.0840
0.0061 0.2886 0.1426 0.0275 00008 0.1139 0.0141
TFF3 0.0000 0.3324 0.0000 0.1789 0.1254 0.0000 0.0000
0.0000 0.0110 0.1575 0.0444 0.1715 0.0229 0.0162
TO 0.0000 0.0457 0.0000 0.1462 0.0907 0.0000 0.0763
0.0000 0.0000 0.0046 0.0501 0.8319 0.0058 0.0026
TLEI 0.0000 0.0000 0.0000 0.3220 0.0808 0.0000 0.0184
0.0851 0.0000 0.2334 0.1047 0.1768 0.0664 0.0000
TMPRSS2 0.0475 0.0061 0.0000 0.1440 0.1280 0.0000 0.1206
0.0720 0.1013 0.0610 0.1099 00003 0.0443 0.0089
TNFRSF8 0.0000 0.0492 0.0000 0.0109 0.0088 0.0004 0.0728
0.0093 0.0000 0.0617 0.0232 0.0000 0.0062 0.0015
TP63 0_0000 00335 0 0000 0 0277 0 1223 0_0000 0_0000
0_0000 0 0061 00907 2_3082 0_0000 0 3923 0_0014
TPM I 0.0000 0.0000 0.0020 0.0425 0.2042 0.0000 0.0132
0.3712 0.5131 0.0215 0.1198 00391 0.0075 0.2254
TPM2 0.0000 0.0247 0.0000 0.0497 00282 0.0000 0.0093
0.0050 00111 0.0265 0.0889 00038 0.0689 0.0100
'11'M3 0.0006 0.0528 0.0000 0.0773 0.0662 0.0000 0.0794
0.0713 0.0129 0.0567 0.2273 0.0725 0.0227 0.0079
TPM4 0.0000 0.2880 0.0000 0.1518 0.0796 0.0000 0.0521
0.2444 0.0015 0.1282 0.0779 00004 0.0386 0.1426
TPSAR1 00000 0_0428 0 0000 0 1971 0 1180 0_0012 0_0668
00114 0 0000 01520 0_1283 0_2829 1)0985 0_0155
TTF1 0.0000 0.0000 0.0000 0.0127 00491 0.0000 0.0088
0.0000 00000 0.0786 0.2237 00000 0.0194 0.0000
LIPIC2 00000 00000 0 0000 0 0039 0 0129 0_0000 0_0058
0_0000 0 0000 00826 00436 0_0000 0 5618 0_0000
UPK3A 0.0000 0.0727 0.0000 0.0806 0.0537 0.0000 0.2229
0.0736 0.0000 0.0270 0.0645 00960 0.2551 0.0062
UPK3B 0.0000 0.0000 0.0000 0.0668 0.0437 03605 0.0272
0.0017 0.0135 0.0289 0.0574 0.0268 0.0952 0.2858
VEL 0.0000 0.0393 0.0000 0.1045 0.0238 0.0000 0.0052
0.0000 0.0075 0.0042 0.0913 00059 0.2840 0.0023
V1LI 0.0000 0.1146 0.0000 0.1179 0.0235 0.0000 0.0000
0.0000 00000 0.0289 0.0364 00000 0.2484 0.1114
VIM 0.0000 0.0000 0.0000 0.0857 0.0377 0.0000 0.0413
0.0000 0.0012 0.0425 0.0817 0.2083 0.2505 0.0040
219
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
WT1 0 0000 0 0173 00000 2 0090 0 0094 03347 00022 00110
0 0000 0 0346 0 0731 0 0072 01307 0 0315
Table 119- Importance of RNA Transcripts used to Classify Organ Type
Transcript AG Ma Brain Br Colon Eye FGTP Gast HFN Kid LGC Lung Pant Pros Skin
SI Thy
ACVRL1 0003 0671 0000 0475 0222 0000 0056 0236 0064 0680
0876 0352 .0320 0005 0272 0094 .0000
AFP 0000 0096 0000 0369 1508 0000 0130 1900 0214 0000
0740 0188 0423 0019 0028 0427 0012
ALPP 0000 0096 0000 0724 1021 0000 1964 0383 0181 0172
0522 0222 .1045 0269 0104 0000 .0000
AMACR 0000 0913 0000 1646 0941 0005 0430 1599 0887 2368
1110 0666 2646 5598 3141 0064 0000
ANKRD30A 0000 0124 0000 8385 0095 0000 0209 0134 0004 0000 1418 0822 .1093
0000 0045 0000 .0000
ANO1 0000 1123 1.0334 1658 0384 0000 2532 6185 2232 0825
4571 1535 .7984 0207 0738 2189 .0014
ARGI 0313 0395 0000 0809 1492 0000 1317 0390 0177 0488
0170 0735 .1897 0000 0252 0469 3135
AR 0000 0745 0679 1416 0317 0000 2628 3634 0504 1697
1404 4098 .1246 0766 2539 0690 .0000
BCL2 0000 0627 0850 0299 0123 3040 2323 1117 0239 0200
1067 0598 .0308 0589 0184 0060 .0040
BCL6 0000 0723 0279 0000 0422 0002 1007 0607 0158 1668
1525 1039 .0186 1279 2406 1593 .0000
CA9 0000 1180 0000 1187 1010 0007 0292 1173 0200 1638
1019 0117 .0125 0181 0406 0452 .0608
CALB2 0882 3649 0000 0711 0760 0000 2521 0375 0236 0000
1588 0353 .2212 0156 0274 1687 .2420
CALCA 0000 0092 0000 0622 0957 0000 0353 0744 0032 0953
0859 0437 .0637 0021 0768 0072 .0000
CALDI 0000 0055 0391 0768 0371 0000 1536 0040 0025 0110
1722 1287 .0349 0000 0732 2104 .0003
CCNDI 0000 0979 0147 1192 0074 0056 2440 1178 0452 0208
0268 0110 .0890 0000 0288 0589 .0851
CD 1 A 0000 0757 0000 0888 0243 0000 0162 2311 0789 0000
0915 0221 .1749 0205 0518 0338 .0103
CD2 0000 2638 0096 0297 1065 0000 0481 0622 0384 0000
0510 0071 0942 0167 0935 0242 0153
CD34 0282 0182 0016 0150 1194 0000 0274 3914 0189 1022
0415 0971 .0999 1035 1163 0000 .0000
CD3G 0000 2669 0157 0464 0414 0000 1717 0928 0025 0000
0031 0387 .0419 0224 0874 0018 .0000
CD5 0000 2324 1592 1878 0535 0000 0275 0993 0954 0000
1891 0497 3574 0052 0345 3299 .0062
CD79A 0000 0133 0000 0729 0477 0020 0423 1161 0386 0000
1012 0752 .0642 0025 1694 0592 .0098
CD99L2 0000 0754 0123 1116 0727 0000 1779 0798 1949 0000
0917 3663 .0641 0045 0071 0049 .0007
CDH17 0000 0423 0033 0032 3831 0000 0184 0422 0172 0000
0189 0817 .0842 0108 0334 4462 .0000
CD1-11 1257 0168 0399 1486 0120 0000 1459 3014 0925 7014
0143 0326 .0373 0667 0966 0000 .0322
CDK4 0000 1171 0018 0056 0590 0000 2757 0669 0363 0000
1529 0802 .0494 0161 0046 0000 .2172
CDKN2A 0000 1014 0453 2024 1300 0000 4237 0981 0318 4499
1653 1417 .1154 0370 0037 0634 .0172
CDX2 0000 0502 0047 1807 1.3118 0000 1523 7682 0101 0000
0409 0862 .1480 0085 0040 3510 .0000
CEACAM 1 6 0000 1401 0000 1643 0981 0000 0547 0539 0290 0096 1304 1034 0742
0072 2789 1652 0050
CEACAM18 0000 0097 0003 0977 1766 0000 0426 0255 0055 0000 0392 0807 .1546
0422 0000 1313 .0488
CEACAM19 0000 0328 0000 0222 0298 0000 0437 2109 0297 0378 0833 1299 .0743
0132 2811 0099 .0167
CEACAM 1 0000 1303 5129 0081 1826 0000 0548 0400 1096 0096 0813 2729 .0858
0877 1139 0000 .0159
CF ACAM20 0000 0022 0000 0018 1326 0000 0038 0505 1120 0046 0392 0026 0285
0000 0114 0000 0000
CEACAM21 0000 0152 0000 0329 0114 0000 1227 0088 0744 0000 1198 0040 .0026
0839 0093 0167 .0000
CEACAM3 0000 0312 0059 0372 0454 0000 0089 1434 0223 0000 0909 0587 .1765 0244
0084 0121 .0584
CEACAM4 0000 0812 0675 1648 0174 0000 0276 0942 0046 0000 0487 0132 .1209 0000
0834 1479 .0189
CEACAM5 0000 9332 0000 0755 4657 0000 1099 0082 1680 0825 1855 0166 .0626 0518
0388 0260 .2552
CEACAM6 0000 1477 0000 0124 0330 0000 1584 3346 0446 0170 0117 3440 .1333 0965
0000 0246 .0039
CEACAM7 0000 0128 0000 2111 1943 0000 1543 0694 0782 0037 1400 3624 .1242 0151
0259 1387 .0000
CEACAM8 0000 0666 0000 0080 1539 0000 1574 0168 2591 0040 0254 1268 1016 0000
0000 0095 0000
CGA 0000 0482 0000 0109 0306 0000 0434 0112 0056 0000
0458 0190 .1832 0000 0177 0942 .1288
220
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CGB3 0000 A477 0085 0198 A598 A000 A676 1499 A030 A000
1153 0650 .0147 2017 A542 0268 _0000
G74N1 .0000 2837 .0179 1656 A832 A000 A795 A394 1034 A000
2537 2339 .0232 .0806 1730 .2583 .2661
COW .0000 A445 .0060 A623 A028 A002 A235 .1307 A422 A538
A192 0157 .1701 .0072 A956 .0000 .0000
CPS1 .0000 A645 .0000 A101 A177 A000 1630 A638 A412 1171
A499 0792 .2032 .3389 A451 .0038 .3436
CR1 .0002 A075 .0317 A205 .1081 A000 .1264 A577 A068
A362 A119 0909 .0211 .0000 .1970 .1178 .0025
CR2 .0000 A099 .0000 A120 A336 A003 A377 A600 A356 A002
A466 0196 .1997 .0860 A047 .0106 .0000
CTNN131 .0000 A319 .0000 A328 A840 A043 A529 .1220 A080 A000
A696 0631 .0404 .0000 A105 .1604 .0098
DES 0000 4203 0279 2248 A060 0000 3107 2486 0051 0097
A672 1804 1281 0000 1019 2349 0030
DSC3 .0000 A068 .0118 A430 A329 A000 A392 A577 .7147 A027
A996 0414 .0225 .0057 A000 .2462 .0833
EN02 .0000 A167 .0391 A912 A702 A379 A214 .3843 2596 2268
2694 1003 .0542 .0415 A051 .0332 A0127
ERBB2 .0000 A365 .0215 A124 .1209 A000 .1466 A053 A397
A138 A167 2024 .1639 .0000 A154 .0398 .0229
ERG 0002 A992 0152 0179 2343 0055 0912 0249 0127 0120
0242 0392 _0743 0370 A403 0363 _0000
ESR1 .0000 1535 .0652 1127 A408 A000 1.0530 A577 1233
A391 A028 1011 .1813 .0210 .1503 .0167 .0000
EMI .0000 A665 .0074 A187 A942 A000 A424 A080 1055 A145
A456 1075 43187 .0317 A157 .4217 .0358
FONT/ 0000 0094 0131 0225 A601 0000 4227 .1.110 0621 0000
0669 0549 43137 0024 0297 0452 1166
FUT4 .0000 1533 .0749 A810 2366 A000 A897 J438 A129 A963
A524 1631 .3926 .0295 A072 .1623 .0615
CA.T.A3 .0000 1.3362 .0360 2.0010 A265 A000 2732 A478 2203
A386 .1597 1885 .6680 .0035 .3548 .0047 .0887
CdN23 .0000 A924 .1749 A215 A034 A000 1597 A236 A336 A773
.1257 0690 .0641 .0000 A846 .0601 .0000
EUVA/CI21 0000 0285 0000 0259 2369 0017 0156 0702 1647 4680
0909 0878 0346 0000 0055 0016 0163
1460113 .0000 1637 .0266 A322 2227 A008 1474 A309 3677 A912
.7119 0808 .2556 .0061 .0959 .0171 .2405
1L12B .0000 A205 .0000 A478 A434 A000 1123 A416 1894 A024
A282 1107 .0043 .0498 A148 .0370 .0000
EVIP3 .0000 A818 .0000 A050 A307 A000 A080 A336 .0100 A000
A504 0384 .0222 .0000 A195 .0000 .0000
TNIIA 1494 0375 1251 0987 0321 0000 0473 1673 0870 0000
1546 0468 0057 0294 0311 0017 3150
ISIA .0000 2428 .0260 1131 M911 A000 A789 2998 A819 A000
A930 2304 45155 .0020 A238 .0300 .0000
KIT .0000 A213 .0000 1038 A682 A000 1478 1008 A510 A256
A399 1076 .1514 .0166 A142 .0077 .0000
KJAC3 .0000 A610 .0000 A352 A028 A000 A257 A090 A512 A152
A014 0322 .0469 1.2958 A281 .0051 .0000
RI. 0000 1684 0000 1550 A225 A000 A553 A273 1720 3120
2054 0375 _0267 2279 A025 0000 _0359
KAT1 .0000 A291 .1109 A050 A625 A080 A437 A150 A548 A000
A103 2288 .1276 .0175 A061 .0757 .0042
KRT14 .0000 2083 .0115 A979 A050 A000 1055 A955 1525 A024
A009 0884 .0272 .0000 1471 .0062 .0000
KAZT15 .0000 A687 .1006 .5284 A836 A000 2371 A422 2901 A096
A613 1612 .0350 .0282 1112 .0227 .0000
KR 116 0000 A089 0331 2914 (1147 A000 1705 M346 (1179 M007
A354 0804 _0616 0000 M611 0371 (1580
KRT17 .0000 A528 .0170 A347 A050 A000 A713 A267 A407 A431
A401 0749 .0457 .0283 A842 .0167 .0000
KRT18 .0000 A043 .2272 A277 3549 A000 1155 A070 A830 A004
A609 0817 .0206 .0776 1036 .0018 .0000
KRT19 0524 2239 0315 0629 A533 0000 0312 0394 0225 0184
0307 1090 A840 0517 3821 0000 0044
KRT1 .0000 A547 .0000 A268 A407 A000 A190 A299 A197 A000
A246 0396 .0360 .0133 .1066 .0117 .0000
KRT20 .0000 .5602 .0000 A009 .6969 A000 M228 A630 M523
M001 A346 2407 .0662 .1508 M657 .3990 .0004
KRT2 .0000 A174 .0000 A222 A340 A005 A429 A963 A930 A452
A181 0410 43107 .0947 A243 .0202 .0438
KIZT3 0000 A459 0000 A410 A097 A000 A436 A106 A721 A096
A929 0205 1160 0022 A018 0000 _0000
KRT4 .0000 A579 .0000 A604 A359 A000 A581 A740 1764 A000
.1881 0467 .0230 .0158 A114 .0309 .0000
KRT5 .0000 A561 .0448 2414 M894 A000 3243 M082 .7575 M018
2450 0642 .0502 .0617 M730 .0137 .0000
ICR:f6A .0000 A183 .0018 A846 A164 A000 A237 A195 A203 A000
A114 3301 .0551 .0683 A067 .0202 .0042
K.12.1613 .0000 A209 .0000 2187 3467 A000 M287 M547 M743 M033
A520 0848 .2088 .0106 M086 .1043 .0000
ItiRDSC .0000 A067 .0000 A556 A036 A000 A762 1064 A047 A000
A110 0227 .1520 .0476 A049 .0000 .0000
KRT7 .0000 2521 .0628 .5254 1.2701 A080 M557 M694 M345
2875 2164 3106 .1843 1.2860 A042 .3030 .0339
221
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
KIZT8 0570 0070 10342 0194 0289 0005 0726 0753 1716 0324
1153 0006 _1772 1102 6755 1144 _0822
LIN28A .0000 M072 .0000 M096 M637 M000 M120 M076 M156 M000
M260 0175 .0343 .0261 A665 .0280 .0000
L1N28B .0000 A592 .0000 M351 M450 M000 .1485 M676 .2085
M000 M138 0315 .0429 .0041 M147 .0000 .1655
MAGEA2 .0000 M013 .0000 M117 M020 M000 M060 M392 .0000 M000
M856 0709 .0683 .0000 M000 .0000 .0000
MD1612 .0000 M140 .0020 2969 M579 M000 2265 M276 .1408
.1983 .1261 0509 .1656 .0000 .3251 .0574 .0000
MB31 .0962 M048 .0331 M884 A189 .0544 M323 M366 A373 M253
M806 0671 .0396 .0052 M199 .0036 .0000
MITF .0000 .3069 .0213 M226 M196 3109 M792 M714 M180 M000
M450 1549 .0408 .1111 A420 .1808 .0054
MAANA 0000 0648 0041 0475 0192 3318 0533 0368 0555 0234
0977 1835 _0200 0072 2699 0143 0161
MLH1 .0000 M189 .0069 M156 A564 M003 M830 M191 A273 M162
M594 2300 .1279 .0034 M534 .0000 .0822
MME .0000 2636 .0013 M735 A515 .0000 M462 M055 2608 A049
M880 0335 .0956 .0654 M839 .1181 .1127
MPO .0000 M352 .0000 M071 M438 M000 M034 M363 M201 M108
M795 0499 .0263 .0000 M029 .2622 .0509
MS4A1 0000 0071 0102 0584 1182 0003 2448 0091 0186 0113
1348 1566 _0104 0027 1812 0078 _0001
MSH2 .0000 M083 .3471 M284 M135 M000 2538 M432 M156 M318
M345 0813 .1875 .0000 M084 .0423 .0000
.0000 M000 .0098 M012 M104 M000 M526 M790 A828 M000 M206 1600 .0389 .0056 M105
.0000 0148
MSLN 0000 3432 0000 0438 1143 0000 1068 0310 0971 1380
0957 0482 _2315 1680 0169 0940 _0803
MTHFR .0000 M064 .0053 2116 M403 .0000 M226 A700 M053 M275
M372 1302 .0500 .0170 M283 .0324 .0186
MUC1 .0000 .3594 .0728 M028 .5746 .0000 2050 .1341 M888
2678 M567 1148 .0732 .2098 M722 .0115 .0312
MUC2 .0000 M392 .0000 M017 .8717 M000 M130 M027 M146 M000
M172 0546 .0829 .1871 M133 .5774 .0340
MUC4 0000 0522 0179 4349 0926 0006 0528 2242 1497 0215
3392 2554 _1277 0737 1638 0050 _0487
MUC5AC .0000 2247 .0024 2808 M850 M000 M566 3093 2958 M637
.1325 1807 .4736 .0776 M581 .0596 .0000
MYOD1 .0000 A281 .0218 M555 M196 .0000 M231 M213 M067 M000
M058 0145 .0439 .0000 M102 .0300 .0000
MYOG .0000 M302 .0000 M768 M186 .0000 M094 2205 A699 M290
M118 0649 0165 .0028 M306 .0000 .0014
NANOG 0000 0777 0173 0107 0337 0000 0263 0704 0080 0000
0574 0119 0507 0000 0797 0000 0000
NAPSA .0001 .2645 .0063 A281 M415 M000 A032 A494 M847 M063
M746 9241 .1344 .0284 M339 .0111 43169
NCAM1 .0000 M409 .3968 M429 M122 M055 M204 M202 M186 M072
M580 0368 .0088 .0000 .1824 .0036 .0494
NCAM2 .0437 M730 .0000 M737 A190 .0000 M972 A127 A296 M000
A791 3102 .1403 .0558 M556 .1095 .0143
NKX2-2 0000 1005 2205 0522 0990 0000 1576 0511 0114 0000
1899 0210 _2672 0444 1354 0048 _0000
NKX3-1 .0425 M429 .0000 M292 A744 M000 M960 .1352 M110 M000
A139 1494 .0219 1.1378 M109 .0042 .0231
OSCAR .0000 M124 .0034 M532 A362 M000 M294 M562 M392 M016
M739 0732 .1713 .0084 M677 .0391 .1180
PAX2 .0000 M122 .0000 M370 M207 .0000 A434 M926 M067 2834
M730 1325 .0367 .0000 M162 .0033 .0000
PAX5 0000 0924 0000 1044 0086 0006 1276 0185 2914 0000
0005 0118 .0179 0557 0000 0511 _0056
PAX8 .0000 .3050 .0132 3208 M373 M000 1.2795 .3209 A479
.8966 .1523 2109 .0231 .0065 M731 .1650 .8590
praw .0000 M124 .6385 A994 A385 M210 A941 2792 M548 M096
M053 0253 .1933 .0000 M576 .0015 .0019
PIDC1 0000 0366 0060 0316 0984 0000 0538 1423 0072 0078
0506 2131 _8132 0085 0013 1270 _0295
PECAM1 .0002 M141 .0000 .1046 M353 M000 M067 .1972 M374
M463 M920 0147 .0234 .0973 M252 .0923 .0000
PGR .0000 M186 .1330 A311 A656 M000 3083 M444 2894 M000
M100 0978 .0183 .0296 M437 .0100 .0000
PIP .0000 A526 .0000 3285 M380 .0057 M558 A931 A178 M073
M483 0620 .0254 .1123 M396 .0000 0155
PATEL 0003 0356 0129 1972 1023 10156 0518 1773 0228 0000
1240 0124 _1000 1675 5473 1542 _0027
PMS2 .0000 M287 .0000 M191 M260 M037 A119 A046 M365 M000
M377 0748 .1378 .0177 M600 .0027 .0000
PCgJ5F1 .0000 M362 .0000 M681 M283 M000 A162 M538 M786 2831
2509 1150 .2034 .0103 M055 .0119 .0679
PSAP .0563 M265 .0000 M065 M869 .0063 M702 A636 M091 M077
2201 0257 .0072 .0003 M305 .0359 .0162
P1PRC .0000 M058 .0000 M337 2122 M000 M800 M318 M066 M000
M523 0629 .0387 .0336 M000 .0720 .0021
S100A10 .0000 2972 .0019 A128 M151 A215 A124 M085 M391 M138
M175 4153 .0864 .1658 .1544 .0469 .0782
S100A1l .0000 M113 .0106 M099 M300 M000 M426 3009 A101 M000
M155 0579 .1451 .0015 A747 .0000 .0174
222
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
S100Al2 0000 _0297 0036 A926 1323 A000 A492 A293 A774 A000
A337 0770 _0091 0803 A804 0070 _0000
S100A13 .0000 .0057 .0066 1174 A270 1525 2538 3404 A622 2862
A851 2209 .0091 .0197 .1541 .0093 .0106
S100A14 .0000 A720 .8152 A965 2377 A000 A929 A084 .1456 A861
A913 0189 .1482 .0681 A377 .0124 .0618
S100A16 .0000 1208 .1491 A259 A510 A310 1116 A267 A073 A000
A420 0424 .0161 .0580 A579 .0000 .0007
S100A1 .0000 A444 .1976 A451 A344 A673 A775 .1901 .1661
A164 A598 4323 .0931 .0000 .1450 .2117 A1128
S100A2 .0001 .3483 .4600 A888 A843 A423 A662 A832 A175 A000
3213 0589 .1294 .0129 A093 .0260 .1894
S100A4 .0000 A493 .1041 A242 A409 A000 A464 A080 A180 A236
A917 0350 .2247 .0253 A231 .0080 .0163
S100A5 0000 D429 0000 0424 0227 0000 0761 0986 1627 0165
0511 1205 _1296 3310 0247 0553 _0053
S100A6 .0000 1034 .0067 2751 2919 A000 A925 A465 .2660 A000
A196 0394 0183 .0907 A238 .0206 .0421
S100A7A .0000 A312 .0029 A106 A538 A000 A444 A724 A214 A000
A421 0288 .1400 .0000 A000 .0000 0191
S100A7L2 .0000 A166 .0022 .1401 A685 A000 A074 A299 A164 A000
A000 0042 .0000 .0086 A000 .0000 .0433
S100A7 0005 A076 0165 _0118 _0166 A000 1777 2378 _0951
_0012 A149 0637 _0359 0132 A032 0000 _0141
S100A8 .0000 A114 .1244 A143 A796 A000 1051 A029 1445 A000
A538 0194 .0946 .0195 A000 .0236 .0000
S100A9 .0000 A745 .0184 A696 A332 A000 1800 2175 A316 A000
2408 0603 .0295 .0136 A018 .0265 .0026
S10013 0000 1028 9620 1504 0476 0147 0782 2350 2606 0381
0658 0815 _0460 0101 _8089 0116 _0270
SlOOPBP .0000 A981 .0301 A615 A249 A000 A751 A220 A301 A281
A467 0860 .1319 .0000 A862 .0132 .0158
SlOOP .0000 2341 .0121 1709 A183 A000 1015 A753 A791 A178
A718 0110 .0724 .0207 A289 .0078 .2033
SlOOZ .0000 A187 .1509 A003 A101 A022 A343 A934 A089 A189
A111 1308 .2410 .0419 .1333 .0241 0153
SALL4 0000 A484 0000 1879 0377 0000 2077 0702 2586 1135
0942 0459 _1665 0567 0235 0040 1158
SA1B2 .0000 2100 .0196 M157 3127 A036 M687 A100 M978 M070
A929 0649 .2148 .0420 M683 .0284 .0033
SE.C1 .0000 A480 .0442 A335 A946 A000 A525 1007 A971 A000
A066 0872 0177 .0760 A779 .1141 0150
SERPINA1 .0297 A227 .0000 2262 A950 A000 2388 A393 A243 A568 .7522 0195 .7488
.1644 A341 .0653 .0039
SERPTNTIS 0000 0369 0109 1940 1776 0000 0596 4347 0117 0599
0663 0783 0690 0000 0019 0145 3405
SF1 .0000 A049 .0000 A792 A235 A000 A335 A198 A655 1336
A670 0822 .1559 .0473 1015 .1107 .0000
SFTPA1 .0000 1543 .0051 A297 A753 A000 1514 .1391 A353 A000
A969 5577 .0979 .1310 A365 .0295 .0244
SMAD4 .0000 A259 .0000 A259 A948 A000 A713 A336 A542 A000
A119 0468 .4014 .0205 A936 .0000 .0138
SMARCR1 0000 A041 0037 A317 A247 A003 _3124 A567 A059 A000
A740 0380 _1731 0000 A035 0000 _0161
SN11,11 .0000 A294 .0000 A241 A636 A015 A893 A755 A065 M067
A227 0686 .2914 .0048 A977 .0000 .0104
607(2 .0000 2171 .6623 3559 2748 A379 1072 .3247 A164 A373
3972 6865 .2639 .0029 A966 .0875 .0000
SPN .0000 A442 .0704 A443 A209 A000 A745 A132 1534 A000
A176 0390 .1740 .0000 M020 .1942 0189
SYP 1104 A457 0037 0826 0476 0052 0610 1916 1654 1942
0233 0281 _0659 0809 M443 0725 M114
TFE3 .0000 A803 .0000 A118 A113 A000 A354 A475 A683 A202
A734 0574 0120 .0297 A134 .0206 .0000
TFF1 .0000 1299 .0032 2456 A615 A005 1175 2323 1540 A017
A709 1328 .2668 .1127 A500 .1950 .0005
TFF3 0000 0279 0000 1382 3563 0000 1708 3722 0261 0318
0719 1564 _0725 0019 2413 0547 _1485
TO .0000 A355 .0099 M492 M655 A000 M691 .1482 M778 M887
.1582 0215 .0877 .0445 M560 .0000 .8142
TLE1 .0000 A385 .1665 M147 M724 M000 A913 M174 M494 M407
A724 0918 .0440 .0458 2932 .0053 .1212
TMPRSS2 .0000 M226 .0087 A828 A775 A000 2887 .1526 2659 A407
A977 3973 .1369 .1683 2548 .1761 .0000
T/011SF8 0000 M113 0137 M889 M461 A000 M310 M119 M652 M000
M268 1567 _0085 0960 M070 0082 .0014
TP63 .0000 A924 .0006 2707 A365 A000 1571 A534 .6012 A000
A126 2757 .0482 .0188 A035 .0479 .0000
TP1,41 .0000 A159 .0000 A240 A292 A000 A741 .3391 A776 A000
A453 0435 .0910 .0000 2978 .0714 .0000
TP1k42 .0000 M435 .0017 M348 M418 .0000 M327 M658 M814 M159
M844 0291 0107 .0116 A418 .0531 .0000
APM3 .0013 A104 .0079 A530 .0137 A000 A876 .0162 A559
.0360 A586 1213 .0796 .0707 M705 .0065 .1187
TPN14 .0000 A306 .0039 A407 A157 A006 3221 A346 1068 A346
A870 2280 .0772 .0650 A380 .0007 .0055
TPSAB1 .0000 A685 .0012 A699 1828 A000 A772 .1892 A338 A225
.1826 0258 .1529 .0686 A322 .0023 .2542
223
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
TTF1 0002 _0150 0000 0049 0467 0000 0502 _1130 _1137 0795
0534 1594 0845 0078 _0320 0128 _0000
UP ICI .0000 .4937 .0294 .0494 .0552 .0000 .0300 .0671
.1641 .0000 .0426 0210 0284 .0000 .0000 .1051 0000
UPIC3A .0000 .2728 .0000 .1923 .0305 .0000 .0340 .1116
.1914 .0000 .0519 0066 0172 .2308 .0111 .0000 0358
UPIC3B .0000 .1254 .0222 .1994 .0554 .0019 .0649 .0380
.0985 .0000 .2264 0429 0867 .0255 .0417 .0053 0575
VHL .0000 .2155 .0000 .0953 .0091 .0241 .1718 .0635
.0495 .2838 .0118 4338 0433 .0115 .0085 .0013 0022
V1L1 .0000 .2557 .0000 .0205 .3151 .0000 .0469 .3934
.0105 .0000 .7444 0218 0261 .0000 .1729 .0023 0000
VIM .0000 .2238 .0137 .0638 .0562 .0287 .0547 .0598
.0266 .0709 .0205 0273 0512 .0000 .0065 .0421 .2279
WTI 0000 _0189 2166 _0572 _0610 _0166 _8319 _1361 _0467
_1979 _0161 0840 _0163 0118 _0000 0108 _0432
Table 120- RNA Transcripts used to Classify Histology
Transcript Aden ACyC AC ACC Astro Carc CS Choi CCC DCIS GBM GIST Gil GCT ILC
ACVRL1 0.0303 0.0000 0.0299 0.0000 0.0000 00827 0.0117
0.0849 00254 0.0643 0.0130 01231 0.0104 0.0000 0.1148
AFP 0.0097 0.0001 0_0192 0_0000 0_0000 0_0419 0_0264
0_0589 0_0430 0_1092 0_0732 0_0000 0.0110 0_0000 0_0242
ALPP 0.1621 0.0012 00367 0.0000 0.0000 00801 0.0955
0.0200 00438 0.1049 0.0224 00000 0.0323 0.0000 0.0068
AMACR 0.0431 00000 0.1815 0.0000 0.0391 00957 0.0739
0.0513 00544 0.2248 0.0691 00000 00197 0.0000 00738
ANKRD30A 0.0788 00000 0.0000 0.0000 0.0000 00646 0.0929 0.2001 00015 0.5130
0.0620 00000 00000 0.0000 03323
ANOI 0.0390 0.0144 0_0084 0_0000 0_0970 0_0730 0_1301
0_2250 0_0095 0_0309 0_0361 0_4708 0.0000 0_0000 0_0607
ARCH 0.0144 00000 0.0133 0.0311 0.0000 00591 0.1486
0.2801 0.1504 0.0684 0.0498 00000 00800 0.0000 0.0948
AR 0.0725 0.0000 0.0192 0.0000 0.1852 00345 0.1132
0.0710 00476 0.1823 0.1346 00000 0.0046 0.0000 0.2347
BCL2 0.0655 00067 0.0462 0.0000 0.0000 0.0823 0.0186
0.1332 0.1135 0.1671 0.0424 0.0000 00800 0.0000 00050
BCL6 0.0785 0.0000 0.0176 0.0000 0.0234 0.1209 0.0273
0.0588 00667 0.0772 0.3243 00000 0.0028 0.0000 0.2172
CAS 0.0485 0.0000 0.0204 0.0000 0.1205 00361 0.0124
0.0523 0.2053 0.0456 0.1995 00000 0.0072 0.0000 0.5629
CALB2 0.0304 0.0000 0.0394 0.0998 0.0389 00707 0.3244
0.2297 0.1158 0.2715 0.0038 00000 0.0000 0.0000 00000
CALCA 0.0611 0.0000 0_1202 0_0000 0_0000 0_0254 0_1765
0_0759 0_0249 0_0842 0_0938 0_0000 0.0896 0_0022 0_0022
CALD1 0.0704 0.0186 0.0855 0.0150 0.0247 00366 0.2868
0.0325 00644 0.0220 0.0130 00000 0.0000 0.0000 00385
CCND1 0.0283 0.0000 0.1805 0.0000 0.0151 0.0220 0.1704
0.1537 0.0896 0.0739 0.1834 0.0000 0.0086 0.0020 00000
CD1A 0.0826 00000 0.0207 0.0000 0.0021 00186 0.0642
0.1054 00014 0.0760 0.0065 00000 00800 0.0000 00629
CD2 0.0517 0.0171 0_0775 0_0000 0_0571 0_0381 0_0423
0_0094 0_0114 0_0879 0_0000 0_0000 0.0000 0_0000 0_0325
CD34 0.0620 0.0000 0.0245 0.0156 0.0000 0.0569 0.0266
0.1230 04295 0.0929 0.0294 0.0000 0.0197 0.0000 00420
CD3G 0.0755 00109 0.1986 0.0000 0.0000 0.0436 0.0356
0.0364 0.0268 0.0741 0.0156 0.0000 0.5012 0.0000 00069
CD5 0.0229 00000 0.0020 0.0006 0.0000 0.0203 0.1804
0.0810 0.0082 0.1923 0.0162 0.0000 00540 0.0000 00353
CD79A 0.0278 0.0000 0.0138 0.0000 0.0024 00307 0.0384
0.0068 00809 0.0982 0.0105 00000 00057 0.0000 0.2020
CD99L2 0.0447 00000 0.1820 0.0000 0.0008 0.1029 0.0336
0.1561 0.0940 0.0767 0.0144 0.0000 00070 0.0000 0.0408
CDH17 0.2193 0.0000 0.0227 0.0000 0.0648 0.1989 0.0473
0.0596 0.0393 0.1289 0.0817 0.0000 0.0238 0.0000 00769
CDH1 0.1336 0.0165 0.0070 0.1443 0.0031 0.2006 0.3718
0.0454 0.2874 0.2352 0.0000 0.0731 0.0700 0.0000 08042
CDK4 0.0521 00000 0.0000 0.0000 0.0070 00503 0.1631
0.2535 00440 0.0260 0.0119 00000 00064 0.0000 0.2456
CDICN2A 0.0356 00000 0.1996 0.0000 0.0064 0.0491 0.3736
0.2100 0.1382 0.3090 0.3358 0.0000 00060 0.0000 0.0259
CDX2 01164 00000 0.0048 0.0000 0.0037 0.0204 0.1191
0.0765 0.0449 0.1066 0.0049 0.0000 0 A 000 0.0000 00097
CEACAM16 0.0387 0.0002 0_0609 0_0000 0_0283 0_1009 0_0115 0_0250 0_0479 0_0903
0_0223 0_0000 0.0000 0_0000 0_0031
CEACAM18 0.0532 0.0000 0.0050 0.0000 0.0091 00418 0.0232 0.0174 00000 0.1086
0.0000 00000 0.0000 0.0000 0.1954
CEACAM19 0.0363 0.0000 0_0000 0_0000 0_0035 0_0754 0_0971 0_0277 0_0663 0_0993
0_0211 0_0068 0.0273 0_0000 0_0245
CEACAM1 0.1527 0.0074 0.0014 0.0000 0.0022 0.0574 0.0788 0.0648 0.0977 0.0860
0.0928 0.0000 0.2759 0.0000 0.1013
CEACAM20 0.0377 0.0000 0.0000 0.0000 0.0153 00530 0.0281 0.0225 0.0200 0.1251
0.0000 00000 0.0000 0.0000 00000
CEACAM21 0.1119 0.0000 0.0614 0.0000 0.0148 00496 0.0103 0.0655 00594 0.0656
0.0020 00000 0.0000 0.0017 00100
224
CA 03167694 2022- 8- 11
TT 13 -000Z V69L910 VD
SZZ
8000-0 00000 6100'0 0000-0 1100 L1500 0LI-0 50100 LVOIM 8060-0 81000 00000
0000-0 00000 01000 ZLIDI
5500-0 00000 1100.0 0000-0 0001.0 8/010 -11Z0-0 ZLOZM 517/0.0 1091-0 IZOOM
00000 90/0-0 00000 1060.0 00.19101
0000-0 0000.0 00000 0000-0 0000.0 11010.0 0E0-0 00010 OZZOM 0600-0 00000 00000
0500-0 00000 0650.0 119101
9100-0 00000 65I0.0 180-0 51,1,1/0 1151.0 080-0 801,0.0 E111.0 0101-0 00000
6950.0 6660-0 00000 0150.0 6111D1
0000-0 9L010 98100 8100-0 5-18Z1 46100 9610-0 1,96TO 96000 10-00-0 1120'0
00000 0500-0 10000 90010 81101)1
9/00-0 00000 00000 0000-0 IZIOM 0950.0 9000-0 65/0.0 0100.0 L600-0 69800 00000
0/50-0 00000 09010 L111131
00000 0000-0 9/00-0 0000-0 8810-0 00600 0100-0 96000 0000-0 14700-0 1100-0
0000-0 09000 0000-0 0900-0 91.111,1
110110 00000 81500 0000-0 I551/0 160.0 5810-0 0101.0 E1-60.0 1600-0 60000
00000 0080-0 00000 1250.0 5119101
1800-0 00000 Z00'0 0000-0 90000 09Z0'0 8800-0 90000 64400 1610-0 00000 00000
0001-0 91190 5600'0 01031)1
6000-0 00000 0810.0 0000-0 0000.0 50610 L501-0 09510 0180.0 6000-0 0900.0
00000 8010-0 00000 5/50'0 011.1131
88010 0000-0 5900-0 0000-0 5060-0 5090-0 L610-0 LLE-0 0500-0 9050-0 0000-0
0000-0 0000-0 0000-0 0.000-0 11)1
5/0-0 00000 00000 0000-0 00000 64010 L900-0 HEIM L400.0 L9110 80000 00000
Z851-0 00000 00010 0)01x
8910-0 00000 Z600'0 680-0 60000 6600'0 0060-0 GLUM 00800 1260-0 01910 00000
95E0-0 1115'0 90900 SIN
1050-0 0000-0 6000-0 5000-0 091-0 011-0 0110-0 /091-0 1001-o 6000-0 0000-0
0000-0 0000-0 000090 55/0-0 1181
0110-0 00000 00000 0000-0 18000 /0010 5050-0 6500.0 1580.0 11780-0 00000 01810
0000-0 00000 9100.0 VH01
0000-0 00000 85100 9100-0 19000 1110.0 f910-0 0510.0 8/50.0 50010 00000 00000
8000-0 11000 8000.0 ULU
0000-0 00000 ZLIOM 0000-0 ZLZOM 10000 0090-0 ZZOOM 09000 E001-0 51000 00000
1000-0 11000 LCZOM 1100-11
L L0-0 0000-0 9000-0 0000-0 0000-0 LEUM 0009-0 1008-0 85/0-0 90L0-0 0000-0
0000-0 0010-0 0000-0 69/0-0 1 LAM!
0000-0 00000 00000 0000-0 00000 0000.0 8689-0 1100.0 0000.0 0590-0 0510.0
00000 0010-0 00000 MOM 1113AVH
9L80-0 00000 4000.0 0000-0 1110 8/10.0 5191-0 8940.0 6009.0 5640-0 00000
00000 1880-0 00000 6/00.0 3.10
000L-0 00000 00000 0000-0 65LC0 LLOVI 1120-0 I09Z'0 9990 6982-0 00000 00000
9000-0 60000 0121'0 VIVO
0000 0 00000 6L000 1600 0 00000 9060 0 50900 009011 9000 0 06000 HU 11 00000
00980 9611011 0-0/1111
1001-0 6059.0 1110.0 0000-0 9900.0 96000 0090-0 0000.0 0090.0 6800-0 00000
00000 50110 00000 19L00 lIXOd
1000-0 1200.0 1000.0 0000-0 0010 16/1/0 0000-0 L040.0 0910.0 0600-0 00000
00000 8000-0 00000 65000 Ina
9100-0 00000 75000 0000-0 ZL0f0 96000 95/0-0 00900 017000 9090-0 00000 00000
1100-0 60000 0000 1110q-
8900-0 0000-0 8120-0 0000-0 00010 1001-0 90010 5/10-0 0610-0 1900-0 0000-0
0000-0 56000 0000-0 8050-0 0213
0010-0 00000 40800 51/0-0 4055.0 000.0 L040-0 91100 510.0 500-0 86100
0160.0 8500-0 00000 5001.0 0110213
51-0 00000 171700.0 0000-0 06/0.0 L0910 IIL0-0 615.0 ZZ0.0 5901-0 1,000.0
00000 05E0-0 010.0 IVLOM ZONIa
0910-0 00000 00000 0000-0 96000 L980.0 1000-0 60000 0110.0 bLZ0-0 00000 00000
0000-0 66/0.0 LL80.0 300
5610-0 0000-0 L10-0 0000-0 9000-0 1615-0 0910-0 6000-0 801-0 0100-0 0100-0
0000-0 H00-0 0000-0 0880-0 930
0000-0 00000 00000 0000-0 00000 8040.0 5610-0 ZIIIM 9/10.0 0190-0 00000 00000
1050-0 8/1.0 000.0 111010113
L100-0 00000 00000 0000-0 00000 6900.0 11020-0 0800.0 LIZOM 8090-0 00000 00000
0000-0 00000 0100.0 0113
171000 0000-0 IL WM 0000-0 1.6/0-0 170170-0 6600-0 0000-0 6000-0 L81.0-0 L91.0-
0 0000-0 0800-0 0000-0 00170-0 1913
0000-0 00000 81010 0000-0 1,500 91100 1,50I-0 0100 1,0600 /950-0 00000 00000
0901-0 00000 86110 1803
1000-0 00000 6010.0 0000-0 I550.0 96600 6610-0 10800 0910.0 Z550-0 05/0.0
00000 800-0 00000 5010.0 ZoOD
0000-0 00000 9110.0 0000-0 05800 1705Z.0 0175-1-0 51610 19800 1610-0 6000
00000 1090-0 00000 01900 I NI,L3
0000-0 0000-0 Oat -0 0000-0 1911-0 590-0 6810-0 0060-0 5900-0 6000-0 0000-0
0000-0 E110-0 0000-0 0000-0 0193
0651-0 00000 110.0 0000-0 06000 8900 4600-0 6610 LOIOM 5000-0 00000 00000
8001-0 00110 09000 VDD
9090-0 00000 1810.0 0000-0 0900.0 10010 6010-0 66000 IZOIM 9040-0 00000 00000
0100-0 00000 100.0 0I'8V3V113
10110 00000 910.0 0000-0 5060.0 6010 0100-0 0100.0 0610.0 0000-0 09000 00000
5081-0 00000 01/0.0 awavao
5E00-0 00000 00000 0000-0 19900 01010 0059-0 06000 61410 0000-0 01500 00000
0/000 90000 56900 9101V3V113
050-0 00000 00000 0000-0 OZOOM 89000 9110-0 11500 00000 0502-0 00000 00000
8180-0 00000 00900 5191V3VH3
9500-0 00000 16000 0000-0 6010 0980.0 9500-0 06010 05010 0000-0 L900.0 00000
917/0-0 10000 5850.0 HAIV3VHD
911-0 0000-0 0010-0 0000-0 1000-0 9000-0 8560-0 1910-0 0560-0 1110-0 000-0
0000-0 5601-0 0000-0 9010-0 0.111939113
9Z8IO/IZOZSI1/Icl 90LE9I/IZOZ OAA
WO 2021/163706
PCT/US2021/018263
KRT3 0 0379 0 0000 0_0000 0_0000 0_0000 0_0202 0_0249
0_0456 0_2079 0_1026 0_1005 00013 00002 0_0000 0_0085
KRT4 0.0505 00009 0.0787 0.0000 0.0000 00499 0.2731
0.0584 0.0950 0.2321 0.0085 00000 00019 0.0000 00107
KRT5 0.3419 0.0000 0.0000 0.0000 0.0000 00573 0.0889
0.2456 00739 0.1943 0.1791 00000 0.0045 0.0000 0.2134
KRT6A 0.1105 0.0000 02033 0.0000 0.0000 00205 0.0541
0.0918 00059 0.0258 0.0872 00000 0 0064 0.0000 00206
KRT6B 0.0351 0.0000 0.0612 0.0000 0.0000 00470 0.6646
0.1217 00000 0.2434 0.0028 00000 0.0078 0.0000 00410
KRT6C 0.0131 0.0000 0.0714 0.0000 0.0000 00190 0.0745
0.1042 00116 0.0550 0.0000 00000 0.0000 0.0000 00117
KRT7 0.0993 0.0000 0.0313 0.0000 0.0000 0.1598 0.3404
0.3663 00671 0.2393 0.1495 00000 0.1437 0.0000 0.3083
KRT8 0 1448 0 0000 0_0008 0_0000 03103 0_0998 00099 00352
00267 0_1120 06446 02529 1 0337 00814 00243
LIN28A 0.0374 0.0000 0.1733 0.0000 0.0041 00323 0.0179
0.0100 00049 0.0343 0.0000 00000 0.0005 0.0000 00000
LIN28B 0.0357 0.0000 0.0093 0.0000 0.0179 00839 0.2837
0.0597 00123 0.0180 0.0029 00000 0.0227 0.0000 00061
MAGEA2 0.0035 0.0000 0.0197 0.0000 0.0000 0.0204 0.0069
0.1478 00000 0.0021 0.0000 00000 0.0000 0.0000 00000
MDM2 0 0571 0 0000 0_0294 0_0000 0_0635 00405 00294 03571
0_0681 01443 0_0482 0_0000 0 1915 0_0000 00020
MR31 0.0393 00184 0.0401 0.1948 0.0000 0.0171 0.1304
0.0378 0.1385 0.1610 0.0167 00000 0.2388 0.0000 00733
mrrr 0.0699 0.0000 0.0173 0.0000 0.0013 03192 0.0583
0.2196 03497 0.1355 0.0262 00000 0.0000 0.0000 00183
MLANA 0 0447 0 0000 00127 0_0000 0_0179 00565 01727 00166
00494 aceco 00566 0_0000 00240 0_0000 00527
MLH1 0.0607 0.0000 0.0142 0.0000 0.0000 0.0451 0.1695
0.4392 0.2528 0.0188 0.0000 00000 00110 0.0000 00000
MME 0.0285 0.0000 0.0186 0.0000 0.0015 0.0381 0.3911
0.0668 0.0968 0.5786 0.0026 00000 0.0009 0.0119 0.2762
MPO 0.0443 0.0000 0.0084 0.0000 0.0043 00538 0.0064
0.1377 00221 0.0417 0.0000 00000 0.0262 0.0000 00477
MS4A1 0 0791 0 0011 02588 0_0000 0_0000 00784 01161 00195
00032 0_1795 0_0705 0_0000 0 0429 0_0000 0_0398
MSH2 0.0443 0.0000 0.0045 0.0000 0.0937 0.0650 0.0930
0.1603 0.1040 0.0834 0.0324 00000 0.0000 0.0000 0.0000
MSH6 0.0980 0.0000 0.0087 0.0000 0.0595 00347 0.0549
0.0329 0.0048 0.0808 0.0000 00000 00017 0.1466 0.0150
MSLN 0.1086 0.0000 0.0503 0.0007 0.0053 0.0995 0.4299
0.1498 00399 0.1063 0.0000 00000 00123 0.0000 0.0145
MTHFR 00001 0 0000 0 0699 0 0000 0 0054 0 1041 0 0713 0
0333 0 0400 00740 0 0065 0 0000 0 0006 0 0000 0 0979
MTJC1 0.2924 0.0000 0.0180 0.0347 0.4498 00514 0.4092
0.1764 00989 0.1107 0.1503 0.2889 0.0000 0.0000 04940
MTJC2 0.0353 00000 0.0754 0.0000 0.0000 00332 0.0638
0.1168 0.0550 0.0935 0.0030 00000 00397 0.0000 00071
MTJC4 0.0366 0.0000 0.0051 0.0000 0.0007 00656 0.0282
0.4620 0.0344 0.3633 0.0035 00000 0.0000 0.0000 0.3175
MIJC5AC 02431 0 0001 0_0000 0_0000 0_0187 02406 00232 0_1563
00342 00897 0_0062 0_0000 0 0000 0_0000 00047
MYOD1 0.0305 0.0000 0.0210 0.0000 0.0029 0.0185 0.0467
0.0214 0.0648 0.2351 0.0000 00000 0.0004 0.0000 00149
MYOG 0.0455 00000 0.0067 0.0000 0.0000 00320 0.1141
0.0112 03825 0.0447 0.0083 00000 00023 0.0000 00000
NANOG 0.0626 0.0008 0.0000 0.0000 0.0366 00890 0.0342
0.0827 00213 0.1847 0.0063 00000 0.0050 0.0000 00068
NAPSA 0 0778 00(80) 03319 0_0000 0_0264 00897 0_2899
0_1382 0_5083 0_1269 041075 00000 0 0112 041000 0_1109
NCAM1 0.0416 0.0000 0.0090 0.0000 0.8230 0.0815 0.1464
0.0515 0.0815 0.3384 0.6458 00000 0.1516 0.0000 00333
NCAM2 0.0301 0.0001 0.1840 0.0000 0.0159 0.0380 0.0101
0.0125 00482 0.4548 0.0177 00000 0.5388 0.0000 0.1293
NKX2-2 0 0956 0 0001 0_0132 0_0000 0_0423 0_1316 0_0206
0_4682 0_0287 0_0153 0_8243 0_0000 0 0000 0_0000 0_0526
NKX3-1 0.0973 0.0000 0.0531 0.0928 0.0208 0.0685 0.0220
0.0607 0.1823 0.3601 0.0108 00000 0.0204 0.0000 0.3430
OSCAR 0.0590 0.0000 0.4226 0.0000 0.2128 00372 0.1323
0.0883 00846 0.0841 0.0027 00000 0.0058 0.0000 0.3083
PAX2 0.0508 0.0000 0.0000 0.0000 0.0012 0.0661 0.0235
0.0025 00700 0.0779 0.0022 00000 0.0000 0.0000 0.1699
PAX5 0 0361 0 0011 0_0453 0_0000 0_0000 0_1033 0_1375
0_0562 0_0045 0_0351 0_0478 0_0000 0 0164 0_0000 0_0013
PAX8 0.0266 0.0000 0.1035 0.0000 0.0000 00576 0.2124
0.0975 0.5638 0.4051 0.1016 00000 00060 0.0000 00566
PDPN 0.0517 0.0002 0.1428 0.0000 0.0000 0.2347 0.0552
0.0881 00134 0.0517 0.8837 00000 0.0921 0.0000 0.0036
PDX1 01379 0.0000 0.0300 0.0000 0.0000 0.0138 0.2562
0.0455 0.1878 0.0341 0.0240 00000 0.0000 0.0000 0.0476
PECAM1 0.0456 0.0000 0.0281 0.0000 0.0000 0.1047 0.1991
0.0221 00164 0.0408 0.0442 00000 0.0010 0.0000 0.0122
PGR 0.1144 0.0000 0.0000 0.0000 0.0814 00904 0.3056
0.0105 00577 0.0548 0.0138 00000 0.0000 0.0000 00277
PIP 0.0782 0.0000 0.1859 0.0000 0.0060 00669 0.0364
0.0588 00512 0.3791 0.0476 00000 00566 0.0000 0.0037
226
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
0 0237 0 0000 0_0722 0_0004 0_0031 0_1230 0_0154 0_0270 0_0402 0_0637 0_1061
0_0000 0 0644 0_0000 0_0205
PMS2 0.0263 0.0000 0.0082 0.0000 0.0036 0.0330 0.0100
0.0652 0.1249 0.0776 0.0003 0.0000 0.0139 0.0000 0.0000
POU5F1 0.0513 0.0000 0.0469 0.0000 0.0253 0.0651 0.0310
0.2375 1.0489 0.0274 0.0899 0.0000 0.2486 0.0000 0.0000
PSAP 0.0563 0.0000 0.0986 0.0000 0.0014 0.0484 0.0258
0.0861 0.0767 0.0328 0.0000 0.0000 0.0013 0.0000 0.0006
PTPRC 0.0406 0.0000 0.0018 0.0000 0.0395 0.0291 0.0029
0.0682 0.0882 0.0180 0.0054 0.0008 0.0000 0.0000 0.0000
S100A10 0.0953 0.0007 0.0043 0.0007 0.0120 0.0737 0.0519
0.0085 0.0443 0.0282 0.0583 0.0010 0.0000 0.0000 0.0420
S100A1 1 0.0415 0.0000 0.0359 0.0000 0.0946 0.0492 0.0923
0.0226 0.0177 0.2103 0.1027 0.0000 0.01300 0.0009 0.0000
S100Al2 0 0990 0 0000 0_2534 0_0000 0_0016 0_0337 0_0676
0_1337 0_1261 0_2927 0_0027 0_0000 0 0000 0_0000 0_0052
S100A13 0.0627 0.0000 0.0092 0.0000 0.0072 0.0473 0.0561
0.0384 0.0495 0.0449 0.0176 0.0037 0.0179 0.0000 0.0598
S100A14 0.0916 0.0000 0.0077 0.0000 0.0000 0.0551 0.0570
0.0609 03262 0.0332 0.3067 0.0000 0.0543 0.0000 0.0104
S100A16 0.0103 0.0000 0.0244 0.0000 0.0124 0.0251 0.1989
0.0028 0.0133 0.0157 0.0051 0.0045 0.0269 0.0000 0.0115
S100A1 0 1471 0 0000 0_0347 0_0000 0_2960 0_1011 0_0759
0_0203 0_1372 0_0820 0_0123 0_0011 0 0506 0_0000 0_7448
S100A2 0.1293 0.0000 0.0024 0.0000 0.0101 0.0448 0.4043
0.2608 0.0354 0.3199 0.0757 0.0000 0.0402 0.0000 0.0000
S100A4 0.0814 0.0018 0.0184 0.0000 0.4240 0.0280 0.2036
0.0107 0.0383 0.0648 0.0067 0.0000 0.0003 0.0000 0.0123
S100A5 0.0919 0.0000 0_0052 0_0000 0_0000 0_1135 0_0383
0_0445 0_1217 0_0388 0_0045 0_0000 0.0000 0_0000 0_3229
S100A6 0.0433 0.0778 0.0276 0.0000 0.0078 0.0550 0.4067
0.0420 0.1706 0.0491 0.0004 0.0000 0.0000 0.0000 0.0025
S100A7A 0.0955 0.0000 0.0000 0.0000 0.0000 0.0572 0.0462
0.0593 0.0674 0.0408 0.0196 0.0000 0.0000 0.0000 0.0525
S100A7L2 0.0353 0.0000 0.0000 0.0000 0.0000 0.0207 0.0056
0.0110 0.1647 0.1410 0.0474 0.0000 0.0000 0.0000 0.0014
S100A7 00033 0 0000 0_0596 0_0000 0_0000 0_0707 0_0636
0_1336 0_0364 0_1516 0_0000 0_0000 00000 0_0000 0_0062
S100A8 0.0547 0.0000 0.0036 0.0000 0.0000 0.1201 0.0045
0.1331 0.0457 0.1995 0.0874 0.0000 0.0071 0.0000 0.0051
S100A9 0.0607 0.0000 0.0135 0.0008 0.1144 0.0552 0.1603
0.1628 03308 0.0883 0.0865 0.0023 0.0113 0.0029 0.1154
SlOOB 0.0969 0.0000 0.0000 0.0000 1.2677 0.0487 0.1932
0.2718 0.0452 0.0153 1.3235 0.0000 0.8497 0.0020 0.0131
S1 OOPTIP 0 0573 0 0000 0 0105 0 0000 0 0070 0 0075 00399
00038 0 1370 0 1767 0 0091 0 0000 0 0000 0 0000 0 0000
SlOOP 0.0563 0.0000 0.0245 0.0000 0.0000 0.1691 0.0412
0.0962 03398 0.1459 0.0278 0.0000 0.0000 0.0000 0.0614
SlOOZ 0.0297 0.0000 0.0153 0.0000 0.0000 0.0196 0.1191
0.0282 03076 0.0134 0.0298 0.0000 0.0163 0.0000 0.0546
SALL4 0.0262 0.0000 0.0478 0.0000 0.1795 0.0298 0.0753
0.0297 0.0643 0.1220 0.1034 0.0000 0.0000 0.0000 0.0172
SATB2 0.0706 0.0000 0_0162 0_0000 0_0051 0_0423 0_0309
0_1550 0_0932 0_4879 0_0171 0_0000 0.2276 0_0000 0_0178
SDC1 0.0380 0.0006 0.0485 0.0003 0.1795 0.1022 0.0254
0.1856 0.0363 0.2517 0.1621 04088 0.4023 0.3116 0.0428
SERPINA1 0.1070 0.0000 02130 0.0000 0.0000 0.1024 0.2714
0.9927 0.0186 0.3578 0.0056 0.0000 0.0000 0.0011 0.2646
SERP1NB5 0.0612 0.0000 0.0086 0.0000 0.0000 0.0605 0.0455
0.0930 0.1141 0.1290 0.0113 0.0000 0.0000 0.0000 0.1706
SF1 0(271 00(08) 0_0000 0_0000 0_0000 0_0837 0_0073
0_1912 0_0991 0_0312 0_2400 0_0000 0 0029 0_0000 0_0095
SFTPA1 0.0546 0.0000 0.6110 0.0000 0.1626 0.0961 0.3220
0.3272 0.1281 0.2402 0.1506 0.0000 0.0000 0.0008 0.1089
SMAD4 0.0481 0.1555 0.0372 0.0000 0.0013 0.0814 0.0000
0.1728 0.0350 0.1275 0.0374 0.0000 0.0000 0.0000 0.0071
SMARCB1 0.0425 0.0000 0_0000 0_0000 0_0065 0_0810 0_1929
0_0100 0_0531 0_0912 0_1776 0_0000 0.0000 0_0000 0_0120
SMiN1 0.0542 0.0003 0.0772 0.0000 0.1768 0.0509 0.0372
0.3121 0.0172 0.0351 0.0000 0.0000 0.0000 0.0000 0.0000
SOX2 0.0542 0.0001 02163 0.0000 0.8539 0.0592 0.1296
0.1575 0.0550 0.4843 0.8152 0.0000 0.3863 0.0000 0.3317
SPN 0.0240 0.0000 0.0039 0.0000 0.0026 0.1516 0.0569
0.0418 0.0289 0.1275 0.0449 0.0000 0.0405 0.0000 0.0276
SYP 0.0038 0.0000 0_1574 0_1257 0_0000 0.0658 0_0040
0_0746 0_2606 0_1050 0_0155 0_0000 0.6098 0_0000 0_0100
TFE3 0.0203 0.0000 0.0000 0.0000 0.0000 0.0098 0.0412
0.1226 0.0350 0.0896 0.0024 0.0000 0.0000 0.0000 0.0000
TFFl 0.0448 0.0000 0.0000 0.0000 0.0000 0.1024 0.0123
0.7223 0.0839 0.1383 0.0864 0.0000 0.0421 0.0000 0.0227
TFF3 0.1486 0.0001 0.0340 0.0000 0.1101 0.0959 0.0123
0.1150 0.0679 0.1779 0.0482 0.0049 0.0000 0.0000 0.6256
IU 0.0923 0.0000 0.1325 0.0000 0.0000 0.0819 0.0249
0.0615 0.0465 0.0063 0.0981 0.0000 0.0000 0.0000 0.0072
TLE1 0.0352 0.0000 0.0000 0.0000 0.0276 0.0495 0.1203
0.1772 0.0407 0.1247 0.0082 0.0000 0.0082 0.0016 0.0541
TMPRSS2 0.6698 0.0000 0.0000 0.0000 0.0628 0.1438 0.0027
0.4135 0.0487 0.0494 0.0522 0.0000 0.0000 0.0000 0.0068
227
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
TNFRSFR 0 0267 0 0000 0_0064 0_0000 0_0000 0_0290 0_0114
0_0934 0_0251 0_0364 0_0040 0_0000 0 0784 0_0000 0_0925
TP63 0.1645 0.0611 0.6474 0.0000 0.0004 0.0343 0.0290
0.0225 0.0170 0.1422 0.0203 0.0000 0.0000 0.0000 0.0000
TPM I 0.0811 0.0224 0.0156 0.0000 0.0401 0.0421 0.0915
0.1594 0.0846 0.0519 0.0831 0.0000 00137 0.0000 0.0101
TPM2 0.0292 0.0089 0.0279 0.0000 0.2139 0.0753 0.2048
0.0287 0.0740 0.0239 0.0061 0.0000 0.0000 0.0000 0.0000
TPM3 0.0646 0.3315 0.1448 0.0000 0.0037 00271 0.0915
0.0435 0.1476 0.2891 0.0445 0.0000 0.0235 0.0000 00117
TPM4 0.0898 0.0015 0.0308 0.0000 0.2819 00630 0.0354
0.0467 0.0585 0.1126 0.0038 0.0000 00072 0.0000 00104
TPSA131 0.0366 0.0000 0.0804 0.0000 0.0000 0.1052 0.2333
0.0450 0.1244 0.2030 0.0252 0.0020 0.01300 0.0000 0.1027
TTF 1 0 0242 0 0000 0_0763 0_0000 0_0080 0_0191 0_0685
0_0046 0_2690 0_1715 0_0785 0_0000 0 0133 0_0000 0_0036
UP K2 0.1191 0.0000 0.0033 0.0000 0.0588 0.0950 0.0166
0.0254 0.0105 0.1552 0.0215 0.0000 0.0000 0.0000 00628
UPK3A 0.0580 0.0000 0.0000 0.0000 0.0145 00630 0.0643
0.0643 00170 0.0860 0.2445 0.0000 00067 0.0000 00503
UPIC313 0.0462 0.0000 0.0441 0.0000 0.0000 0.0721 0.0469
0.2848 0.1285 0.2996 0.0280 0.0000 0.0380 0.0000 00516
VIII, 0 0547 0 0000 0_2177 0_0000 0_0000 0_0370 0_0286
0_1825 0_0086 0_0334 0_0041 0_0000 0 0183 0_0000 0_0035
VITA 0.0791 0.0000 0.0405 0.0000 0.0034 0.2266 0.1460
0.8138 0.1260 0.0962 0.0055 0.0000 0.0000 0.0000 0.0991
VIM 0.0264 0.0030 0.0154 0.0287 0.0069 0.0364 0.0376
0.0135 0.0362 0.1135 0.0432 0.0000 0.0094 0.0000 0.1413
WTI 0.0391 0.0000 0_1805 0_0000 0_0189 0_0552 0_1780
0_4010 0_3054 0_2016 0_0114 0_0000 0.0030 0_0000 0_0432
Table 120 continued
Transcript Lei Lipo Mel Mn. Merk Meso Neuro NSCC Ito Sarc
SerC Serous SCC Sq
ACVRL1 0.0000 0.0194 0.1326 0.0000 0.0000 0.0000 0.0000
0.0702 0.0000 0.0771 0.0000 0.4134 0.0040 0.0337
AFP 0.0000 0.0001 0.0000 0.0000 00000 0.0000 0.0005
0.0253 0.0001 0.0000 0.0038 0.0198 0.0000 0.0648
ALPP 0.0000 0.0000 0.0000 0.0000 00000 0.0000 0.0000
0.0892 0.0000 0.0037 0.0000 0.2362 0.0062 0.0440
AMACR 0.0000 0.0083 0.0000 0.0000 0.0000 0.0006 0.0021
0.0446 0.0000 0.0000 0.0182 0.0705 0.0106 0.0517
ANKRD30A 0.0000 0.0000 0.0000 0.0000 00000 0.0000 0.0413 0.2199 0.0001 0.0020
0.0061 0.0338 0.0000 0.0988
47801 0_0346 0_0000 0.0191 0.2936 0.0000 0_0000 0_0266
0_0683 0_0000 0_0035 0_0000 0_3164 0_1499 0_1244
ARGI 0.0000 0.0000 00540 0.0000 00000 0.0000 0.0820
0.1353 0.0000 0.0129 0.0371 0.2312 0.0000 0.0600
AR 0.1166 0.0000 0.1381 0.0104 00000 0.0000 0.0989
0.3680 0.0013 0.0611 0.0000 0.3377 0.0000 0.5690
BCL2 0.0000 0.0000 0.0118 0.0023 0.0000 0.0000 0.0024
0.1045 0.0098 0.0750 0.0031 0.0690 0.2242 0.0549
13CL6 0_0945 0_0000 0.0944 0.0137 0.0000 0_0000 0_0009
0_1674 0_0000 0_0081 0_0000 0_0433 0_0000 0_0006
CA9 0.0017 0.0000 0.0090 0.0000 0.0037 0.0218 0.0104
0.0924 0.0000 0.1524 0.0434 0.0773 0.1230 0.1082
CALB2 0.2303 0.0000 0.0005 0.0000 0.0000 0.5584 0.0008
0.0728 0.0000 0.0028 0.0020 0.0507 0.0324 0.0603
CALCA 0.0113 0.0000 0.0110 0.0087 0.0000 0.0000 0.0089
0.0900 0.0110 0.0156 0.0000 0.0275 0.1383 0.0353
CALD1 0.1347 0.0000 0.0000 0.0022 0.0000 0.0000 0.0000
0.0849 0.0000 0.2135 0.0026 0.0323 0.0000 0.0252
CCNDI 0.0783 0.0005 0.0871 0.0379 0.0010 0.0000 0.0163
0.0786 0.0000 0.0278 0.0061 0.0941 0.0681 0.0925
CDIA 0.0080 0.0000 0.0195 0.0000 0.0000 0.0000 0.0000
0.0402 0.0000 0.0021 0.0130 0.0628 0.0456 0.0585
CD2 0.1357 0.0000 0.0781 0.0056 0.0000 0.0000 0.0239
0.0885 0.4549 0.0000 0.0016 0.0645 0.0235 0.0578
CD34 0.0239 0.0701 0.0000 0.0000 0.0000 0.0019 0.0130
0.0189 0.0016 0.0077 0.0022 0.1071 0.1177 0.1263
CD3G 0.0000 0.0003 00512 0.0000 00000 0.0000 0.0590
0.0867 0.0000 0.0790 0.0396 0.0868 0.0454 0.5591
CD5 0.0000 0.0000 0.0103 0.1699 0.0000 0.0000 0.0341
0.0347 0.0000 0.0020 0.0335 0.0627 0.0235 0.0750
CD79A 0_2340 0_0000 0.0969 0.0000 0.0000 0_0000 0_0000
0_1930 0_0334 0_0199 0_0000 0_1609 0_0175 0_0902
CD99L2 0.0032 0.0000 0.0209 0.0084 0.0000 0.0026 0.0029
0.0775 0.0343 0.0052 0.3332 0.1470 0.0261 0.0884
C0H17 0_0000 0_0000 0.0000 0.0000 0.0000 0_0000 0_0237
0_0704 0_0000 0_0186 0_0334 0_0384 0_0621 0_1226
CDH1 0.1206 0.2631 0.0000 0.1095 0.0000 0.0099 0.0000
0.0216 0.2687 0.0658 0.1951 0.1450 0.0053 0.0934
CDK4 0.0000 0.3028 0.0000 0.0000 0.0000 0.0006 0.0000
0.1002 0.0000 0.0002 0.0169 0.3539 0.0000 0.1079
CDKN2A 0.0000 0.0000 0.1460 0.0000 0.0000 0.0074 0.0324
0.1523 0.0000 0.1410 0.0978 0.5257 0.0393 0.0527
228
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CDX2 0_0000 0_0000 0 0003 0 0000 0 0000 0_0000 0_0088
00826 00010 0_0000 00219 0_2185 00013 0_0904
CEACAM16 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0005 0.2136 0.0000 00016
0.0000 0.0791 0.0925 0.0515
CEACAM18 0.0000 0.0000 0.0000 0.0000 0.0000 0.0073 0.0112 0.0415 0.0103 00077
0.0333 0.0223 0.0057 0.0827
CEACAM19 0.0617 0.0000 0.1690 0.0000 0.0000 0.0000 0.0619 0.0226 0.0000 0.1683
0.0056 0.1586 0.1520 0.1541
CEACAM1 0.0655 0.0004 0.0912 0.2840 0.0000 0.0387 0.0000
0.1772 0.1025 00060 0.1514 0.1488 0.0070 0.0627
CEACAM20 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2582 0.0000 00044
0.0000 0.0307 0.0402 0.0383
CEACAM21 0.0026 0.0000 0.0000 0.0000 0.0000 0.0000 0.0022 0.0596 0.0000 00089
0.0005 0.1190 0.0857 0.0604
CEACAM3 00000 00000 0 0107 0 0000 0 0000 00817 0_0578 01906
00000 0_0162 00000 02166 00070 0_0680
CEACAM4 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0522
0.0429 0.0054 00000 0.0081 0.0275 0.0000 0.0212
CEACAM5 0.0000 0.0081 0.0028 0.0026 0.0147 0.0000 0.1568
0.0377 0.0000 00662 0.0711 0.1794 0.0455 0.0328
CEACAM6 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0276
0.1025 0.0000 0.0069 0.0255 0.1754 0.0067 0.0508
CEACAM7 0_0000 0_0000 00000 00000 00000 0_0000 00026 0_2715
0_0000 0_0200 0_0000 0_0211 0_0000 0_0243
CEACAM8 0.0000 0.0007 0.0091 0.0000 0.0000 0.0000 0.0246
0.0523 0.0023 0.0235 0.0000 0.0688 0.0260 0.1095
CGA 0.0000 0.0000 0.0000 0.0000 0.0900 0.0000 0.0453
0.0756 0.0000 00000 0.0000 0.1266 0.1477 0.0620
CG133 0_0000 00000 0074g 0 0000 0 0000 00000 00430 00694
00000 0_0000 0_0128 0_0323 0_1818 0_1826
C4N1 0.4602 0.0000 0.0000 0.0000 0.0000 0.0000 0.0333
0.1607 0.0000 00000 0.0035 0.0938 0.0141 0.2457
COQ2 0.0199 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
0.1271 0.0404 00000 0.0117 0.0425 0.0095 0.0577
CPS1 0.0615 0.0000 0.1500 0.0000 0.0603 0.0000 0.0096
0.0797 0.0000 0.0156 0.2381 0.2112 0.0068 0.1204
CR1 0_0067 0_0328 00000 0 0013 0 0295 00000 00087 00211
00000 0_0000 00369 0_0407 00000 0_1642
CR2 0.0900 0.0900 0.0000 0.0000 00900 0.0900 9.9999
0.0648 9.0090 00408 9.0090 0.2135 0.0954 0.0319
CTNNB1 0.0004 0.0000 0.0195 0.0000 0.0000 0.0000 0.0031
0.2061 0.0000 00000 0.0025 0.0811 0.4604 0.1853
DES 0.2105 0.0000 0.0000 0.0000 0.0900 0.0000 0.0759
0.0584 0.0000 00169 0.0077 0.1431 0.0023 0.2380
DSC3 0 001 0 0017 0071? 0 0409 0 0000 0 0060 0 0189 0
0766 0 0001 0 0986 00000 04Q6 0 0000 0 4745
EN02 0.1487 0.0014 0.0196 0.0000 0.0005 0.0000 0.3925
0.2998 0.0000 00869 0.0156 0.1923 0.0020 0.0446
ERBB2 0.1595 0.0000 00139 0.0000 0.2850 0.0000 0.2159
0.1602 0.0000 00000 0.0998 0.0337 0.0695 0.0392
ERG 0.0000 0.0000 0.0000 0.0000 0.0900 0.0000 0.0189
0.0739 0.0181 00000 0.0000 0.0666 0.0000 0.1302
F.SR1 00156 0_0027 0 0592 0 0011 0 0000 0_0000 02086
0_4605 00000 0_0164 00000 0_2626 00044 0_1409
FLI1 0.0000 0.0000 0.0007 0.0000 0.0000 0.0017 0.0043
0.1105 0.0000 00703 0.0009 0.0206 0.0145 0.0784
FOXL2 0.3188 0.0000 0.0000 0.0086 0.0900 0.0000 0.0000
0.1655 0.0048 0.0848 0.0222 0.2622 0.0000 0.1393
FUT4 0.0064 0.0000 0.0090 0.0000 0.0900 0.0000 0.0000
0.2052 0.0102 0.0115 0.0000 0.0738 0.0536 0.1795
(IA I A3 0_0000 00000 0 0000 0 0355 0 0000 0_0027 0_0000
0_2180 00000 0_0000 0_0086 0_0616 00000 0_2132
GPC3 0.0002 0.0004 0.0907 0.0000 0.0000 0.0000 0.0179
0.0852 0.0002 00000 0.0038 0.0770 0.0000 0.0689
HAVCR1 0.0000 0.0000 0.0000 0.0000 0.0900 0.0004 0.0000
0.1343 0.0000 00114 0.0008 0.0647 0.0820 0.2677
IINF1B 00000 00000 00000 0 0000 0 0000 00000 00000 00600
00007 0_0314 00169 02549 00000 0_3320
IL12B 0.0000 0.0003 0.0000 0.0000 0.0000 0.0000 0.0032
0.1805 0.0000 00000 0.1007 0.0838 0.0032 0.0147
IMP3 0.0335 0.0000 0.0000 0.0004 0.0000 0.0000 0.0000
0.0119 0.0000 00249 0.1609 0.2859 0.0025 0.2011
1NHA 0.0026 0.0000 0.1065 0.0078 0.0900 0.0449 0.0543
0.2378 0.0313 00000 0.0021 0.0268 0.0710 0.0468
0_0225 00000 0 0179 0 0000 0 2910 00000 0_6480 0_2721 00016 0_0000 00000
0_1192 0_6379 0_0354
KIT 0.0202 0.0039 0.0098 0.0025 0.0000 0.0000 0.0068
0.0719 0.0000 00059 0.0000 0.0714 0.5444 0.0694
KLK3 0.0000 0.0000 00000 0.0000 0.0000 0.0000 0.0116
0.1098 0.0000 00000 0.0000 0.1166 0.0390 0.0410
KL 0.0022 0.0009 0.0000 0.0007 0.0900 0.0000 0.0136
0.0578 0.0000 00000 0.0806 0.0659 0.1887 0.0594
KR110 0.0000 0.0000 0.1388 0.2300 0.0925 0.0000 0.0289
0.1095 0.0000 00000 0.0346 0.0197 0.0045 0.0588
KRT14 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0250
0.2027 0.0000 0.0085 0.0104 0.0400 0.0579 0.1112
KRT15 0.0000 0.0013 0.0106 0.0000 0.0900 0.0000 0.0298
0.0779 0.0186 0.1461 0.1244 0.2614 0.0476 0.0824
229
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
KRT16 00658 00000 0 0000 0 0628 0 0000 00000 00000 00400
00000 00000 00000 01296 00104 00396
KRT17 0.0025 0.0000 0.0662 0.0000 0.0000 0.0000 0.0051
0.0572 0.0021 00097 0.0000 0.1598 0.0181 0.8321
KRT18 0.7156 0.5117 0.1018 0.0000 0.0000 0.0000 0.0049
0.1243 0.7509 0.0054 0.0005 0.0210 0.0000 0.0879
KRT19 1.2857 0.2603 0.7118 0.0000 00000 0.0000 0.0560
0.0352 0.0000 0.8934 0.0009 0.0659 0.0677 0.1021
KATI 0.0000 0.0000 0.0207 0.0000 0.0000 0.0000 0.0000
0.0879 0.0000 00370 0.0000 0.2108 0.0062 0.0187
KRT20 0.0000 0.0000 0.0000 0.0020 0.0000 0.0008 0.0000
0.0449 0.0036 00000 0.0000 0.0337 0.0586 0.2718
KRT2 0.1623 0.0000 0.0000 0.0000 00000 0.0000 0.0003
0.1053 0.0000 0.2684 0.0000 0.0523 0.0000 0.1150
KRT3 00212 00000 0 0000 0 0000 0 0000 00002 00049 01919
00010 00000 00014 0_1282 00000 00591
KRT4 0.0023 0.0000 0.0072 0.0079 0.0000 0.0000 0.0106
0.1192 0.0000 00000 0.0067 0.2677 0.0000 0.0307
KRT5 0.0000 0.0000 0.0000 0.0000 0.0900 0.1402 0.0000
0.1377 0.0000 00000 0.0238 0.1224 0.1361 0.8787
KRT6A 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
0.1167 0.0000 00000 0.0004 0.0457 0.1171 0.5259
KRT613 0_0000 00000 0 0000 0 0000 0 0000 00000 00000
0_1034 00000 00000 00000 01588 00066 0.1718
KRT6C 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
0.0685 0.0000 00330 0.0000 0.1959 0.0000 0.1249
KRT7 0.0195 0.1825 0.0000 0.0083 0.0494 0.0006 0.0120
0.0605 0.0000 0.2594 0.0054 0.5886 0.0162 0.2365
1CRT8 0_7388 00129 0 6362 09124 0 0000 00000 00116 00870
00000 00137 00064 0_1210 00000 00509
LIN28A 0.0000 0.0065 0.1182 0.0000 0.0000 0.0000 0.0313
0.0317 0.0000 00203 0.0066 0.1835 0.0043 0.0266
LIN28B 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0344
0.0430 0.0000 00000 0.0000 0.0736 0.0036 0.1618
MAGEA2 0.0000 0.0000 0.0000 0.0138 0.0900 0.0000 0.0000
0.2146 0.0000 00000 0.0000 0.0097 0.0025 0.0028
M13M2 00218 03254 0 0036 0 0294 0 0000 00000 00171 0_1187
00000 00032 00700 0_1588 00072 00718
M1131 0.0000 0.0000 0.0108 0.0000 0.0900 0.0000 0.0900
0.0455 0.0000 00000 9.0285 0.0891 0.0040 0.0089
MITE 0.1166 0.0000 0.2020 0.0175 0.0000 0.0000 0.0316
0.1076 0.0000 00000 0.0378 0.0334 0.3685 0.0255
MLANA 0.0067 0.0000 0.4617 0.0000 0.0905 0.0000 0.0000
0.0703 0.0027 00006 0.0000 0.1913 0.0330 0.0778
MLH1 0 0773 0 0000 0 0000 0 0000 0 0000 0 0000 0 0149 0
0573 00779 00085 80154 0 1703 80063 00700
MME 0.0000 0.0132 0.0006 0.0038 0.0944 0.0000 0.0034
0.1307 0.0000 0.0780 0.5287 0.1239 0.1573 0.0488
MPO 0.0000 0.0000 0.0000 0.0121 0.0900 0.0000 0.1090
0.0260 0.0000 00039 0.0736 0.0854 0.0465 0.0205
MS4A1 0.0000 0.0003 0.0924 0.0000 0.0000 0.0000 0.0388
0.0339 0.0000 0.0048 0.0010 0.0097 0.0267 0.0285
MS142 00042 00007 0 0000 0 2136 0 0000 00067 00000 00991
00037 00239 00013 00607 00933 02610
MSH6 0.0165 0.0000 0.0000 0.0000 0.0000 0.0000 0.0319
0.0930 0.0048 0.0028 0.0024 0.0959 0.0120 0.1485
MSLN 0.0011 0.0003 0.0390 0.0048 0.0905 0.1462 0.0000
0.3377 0.0000 00000 0.2129 0.4918 0.2586 0.0372
MTHFR 0.0008 0.0000 00619 0.0000 0.0000 0.0000 0.0534
0.0806 0.0000 00000 0.0039 0.0644 0.0538 0.1563
M11C1 00166 00000 0 5181 0 0000 018)00 00000 0_2996
0_1200 00000 00000 00016 00753 04778 00987
MTJC2 0.0000 0.0000 0.0058 0.0000 0.0000 0.0080 0.0000
0.2272 0.0001 00081 0.0000 0.1580 0.0071 0.1316
MTJC4 0.0105 0.0000 0.0000 0.0184 0.0953 0.0000 0.1225
0.0448 0.0000 00564 0.0143 0.1906 0.5281 0.1882
MUC5AC 00000 00000 0 0000 0 0000 0 0000 00000 00085 00686
00000 00041 00000 01796 00208 00524
MYOD1 0.0000 0.0000 0.0003 0.0000 0.0000 0.0000 0.0000
0.1587 0.0000 0.0480 0.0000 0.0310 0.0159 0.0153
MYOG 0.0286 0.0000 0.0519 0.0000 0.0744 0.0000 0.0084
0.1007 0.0000 0.2284 0.0000 0.0937 0.0000 0.0954
NANOG 0.0000 0.0003 0.0000 0.0000 0.0000 0.0000 0.0052
0.1241 0.0000 0.0245 0.0302 0.1074 0.0000 0.0590
NAPSA 00000 00000 0 0036 0 0047 0 0004 00000 00748 00731
00000 00024 0_1033 01671 00175 00281
NCAM1 0.1329 0.0008 0.0514 0.0000 0.0000 0.0000 0.5313
0.2375 0.8634 10584 0.0003 0.0514 1.5638 0.0364
NCAM2 0.0000 0.0000 0.0456 0.0000 0.0000 0.0000 0.0175
0.1092 0.0062 00237 0.1308 0.0401 0.0045 0.1502
NKX2-2 0.0109 0.0037 0.0122 0.0000 0.0000 0.0000 0.0891
0.0926 0.0000 03744 0.0181 0.1279 0.3525 0.0191
NKX3-1 0.0126 0.0000 0.0000 0.0000 0.0000 0.0000 0.0107
0.0656 0.0069 00176 0.2486 0.0740 0.0146 0.0173
OSCAR 0.0000 0.0071 0.0072 0.0000 0.0000 0.0000 0.0126
0.1076 0.0000 00319 0.1949 0.0401 0.0000 0.1076
PAX2 0.0000 0.0000 0.0003 0.0003 0.0000 0.0000 0.0000
0.1114 0.0000 00037 0.0000 0.1480 0.0207 0.0752
230
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
PAX5 0_0000 0_0000 0 0000 0 0000 0 0000 0_0000 0_0000
00109 0_0048 00026 0_0000 0_0328 0_5490 0_1451
PAX8 0.0000 0.0000 0.0000 0.0000 0.0900 0.0000 0.2204
0.2207 0.0014 00833 0.0000 1.4219 0.0000 0.2317
PDPN 0.1577 0.1071 0.1112 0.0014 0.0000 0.2774 0.0000
0.0653 0.0172 00021 0.0196 0.1240 0.0099 0.1429
PDX1 0.0000 0.0000 00049 0.0000 00000 0.0019 0.0079
0.0181 0.0000 00044 0.0420 0.0515 0.0000 0.0471
PECAM I 0.0030 0.0000 0.0013 0.0000 0.0900 0.0000 0.0140
0.0596 0.0000 00000 0.0032 0.1528 0.0616 0.0700
PGR 0.0143 0.0038 0.0021 0.2152 0.0900 0.0000 0.0277
0.0757 0.0000 00000 0.0085 0.1129 0.0000 0.1692
PIP 0.0000 0.0000 0.0006 00000 00000 0.0000 0.0011
0.2079 0.0000 00069 0.0000 0.1061 0.1434 0.0904
PMEL 0_0000 0_0000 0 8212 0 0000 0 0000 0_0000 0_0000
0_0754 0_0000 0_0512 0_0081 0_1625 0_0066 0_1642
PMS2 0.0000 0.0000 0.0000 0.0000 0.0900 0.0000 0.0362
0.0717 0.0000 00000 0.1479 0.0439 0.0069 0.2477
POU5F1 0.0000 0.0000 0.1686 0.0000 0.0900 0.0000 0.0668
0.0951 0.0000 00524 0.2000 0.0356 0.0037 0.0889
PSAP 0.0007 0.0000 0.0000 0.0000 00000 0.0000 0.0000
0.0954 0.0000 00000 0.0064 0.0877 0.0087 0.1666
PTPRC 0_0312 0_0007 0.0192 0.0000 0.0000 0_0053 0_0471
0_2771 0_0000 0_0000 0_0101 0_0394 0_0298 0_0290
S100A10 0.0360 0.0054 0.0027 0.0524 0.0900 0.0000 0.1669
0.0953 0.0000 00000 0.0263 0.0565 0.5088 0.0466
S100A1 1 0.0048 0.0000 0.0021 0.0000 0.0900 0.0015 0.4565
0.0661 0.4309 00000 0.2571 0.0551 0.3458 0.0141
S100Al2 0_0000 0_0063 0.0000 0.0000 0.0470 0_0000 0_0000
0_1326 0_0007 0_0000 0_1065 0_0747 0_1577 0_0311
S100A13 0.0000 0.0000 0.3703 0.0000 0.0900 0.0000 0.0000
0.0789 0.0031 00054 0.0000 0.2269 0.0530 0.0504
S100A14 0.1648 0.0037 0.4983 0.3337 0.0468 0.0000 0.0065
0.0342 0.1434 04994 0.4276 0.2245 0.0048 0.1856
S100A16 0.0096 0.0000 0.0000 0.0000 0.0900 0.0052 0.0319
0.0602 0.0000 00000 0.0404 0.3255 0.0000 0.0306
S100A1 0_0197 0_0000 0.0740 0.0000 0.0000 0_0000 0_3546
0_3587 0_0009 0_0408 0_0114 0_0937 0_0130 0_4877
S100A2 0.0007 0.0000 0.0049 0.1196 0.0900 0.0000 0.0000
0.1330 0.0088 00000 9.0274 0.0863 0.0095 9.1590
S100A4 0.0061 0.0000 0.0194 0.0416 0.0900 0.0000 0.1067
0.1375 0.2105 00000 0.0883 0.0472 0.0224 0.0687
S100A5 0.2135 0.0000 0.0000 0.0003 0.0900 0.0000 0.0095
0.1069 0.0000 00071 0.1755 0.3122 0.0849 0.0309
S10046 0 0000 0 0000 00070 0 0176 0 0000 0 0000 00711 0
0941 0 0000 0 0000 80000 08775 0 7475 0 7907
S100A7A 0.0030 0.0000 0.0000 0.0000 0.0000 0.0019 0.0000
0.1654 0.0000 00021 0.0262 0.0538 0.0094 0.0455
S100A7L2 0.0088 0.0000 00000 0.0000 0.0900 0.0000 0.0110
0.0095 0.0000 00000 0.0000 0.0351 0.0000 0.1266
S100A7 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1054
0.0370 0.0034 0.1035 0.0451 0.0240 0.0201 0.0404
S100A8 0_0000 0_0000 0.0100 0.0227 0.0000 0_0000 0_0022
0_0855 0_0000 0_0000 0_0158 0_0895 0_0423 0_1287
S100A9 0.0212 0.0059 0.0029 0.0231 0.0000 0.0000 0.0141
0.0342 0.0000 00000 0.0260 0.1034 0.0029 0.0356
SlOOB 0.0497 0.0074 1.2133 0.0000 0.0900 0.0000 0.0134
0.1238 0.0000 00251 0.0010 0.0817 0.0020 0.0271
SlOOPBP 0.0004 0.0000 0.0041 0.0000 0.0314 0.0000 0.0264
0.0240 0.1020 00509 0.0058 0.0677 0.0165 0.0468
S100 P 0_1138 0_0000 0.0135 0 0000 0.000(1 0_0000 0_0088
0_1531 0_0000 0_1394 0_0000 0_2549 0_0792 0_0417
SlOOZ 0.0044 0.0000 0.0000 0.0000 0.0000 0.0000 0.2346
0.2556 0.0000 00293 0.0546 0.0849 0.0647 0.0274
SALL4 0.0507 0.0000 0.0072 0.0184 0.0478 0.0000 0.0000
0.0931 0.0625 00000 0.0000 0.1662 0.0420 0.0445
SATB2 0_2218 0_0002 0.1597 0.0000 0.0000 0_0119 0_0651
0_0424 0_0000 0_2507 0_2480 0_4029 0_0038 0_1155
SDC1 0.0622 0.0060 0.0000 0.5929 0.0900 0.0000 0.1322
0.1158 0.1000 00191 0.0238 0.3000 0.0297 0.3134
SERPINA1 0.0000 0.0006 0.0000 0.0002 0.0000 0.0000 0.0081
0.1930 0.0000 00000 0.0000 0.2772 0.0000 0.1166
SERPIN115 0.0000 0.0000 0.0000 0.0019 0.0900 0.0000 0.0174
0.0932 0.0000 0.1004 0.0000 0.1800 0.0829 0.3867
SF1 0_0047 0_0000 0.0062 0.0014 0.0000 0_0023 0_0000
0_1650 0_0000 0_0000 0_0125 0_1431 0_0000 0_0197
SFTPA1 0.0000 0.0000 0.0076 0.0000 0.0900 0.0000 0.0270
0.3428 0.0008 00000 0.2125 0.1150 0.0059 0.2155
SMAD4 0.0272 0.0000 0.0000 0.0000 0.0150 0.0000 0.0116
0.2866 0.0000 00000 0.0496 0.1447 0.0127 0.0617
SMARCB1 0.0000 0.0000 0.0000 0.0701 0.0000 0.2646 0.0000
0.0166 0.0000 00000 0.0000 0.0312 0.0049 0.0798
SMN 1 0.0000 0.0005 0.0000 0.0000 0.0900 0.0000 0.0250
0.0541 0.0003 00000 0.0157 0.0584 0.2638 0.0639
SOX2 0.0607 0.0042 0.0777 0.0000 0.0900 0.0000 0.0509
0.3111 0.0095 00209 0.0380 0.2204 0.0025 0.7663
SPN 0.0000 0.0006 0.0000 0.0227 0.0000 0.0000 0.0087
0.0644 0.0000 00000 0.0061 0.0449 0.0101 0.0201
231
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
SYP 0_0414 0_0013 0 0020 0 0000 0 0014 0_0000 0_3135
00395 0_3229 0_0545 00297 0_0218 0_2181 0_0676
TFE3 0.0015 0.0000 0.0049 0.0075 0.0900 0.0000 0.0065
0.0676 0.0000 00609 0.0029 0.0983 0.0146 0.1474
TTF1 0.0000 0.0000 0.0000 0.0000 00000 0.0000 0.0096
0.1063 0.0276 00209 0.0071 0.1115 0.0952 0.1028
TFF3 0.0000 0.0000 0.0006 0.0000 0.0000 0.0000 0.2867
0.2256 0.0000 00066 0.0000 0.2560 0.1633 0.0155
TO 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
0.1004 0.0071 00119 0.0023 0.2005 0.0956 0.1166
TLE1 0.0052 0.0000 0.0000 0.0030 0.0900 0.0168 0.0000
0.0810 0.0000 00000 0.0122 0.1071 0.0034 0.0873
TMPRSS2 0.0147 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000
0.4196 0.0000 0.1294 0.0000 0.0587 0.0000 0.2092
TNFRSF8 00000 00000 00045 0 0074 0 0000 0_0002 00000 0_0272
00000 0_0070 00186 0_0668 00006 0_0338
TP63 0.0087 0.0000 0.1029 0.0828 0.0900 0.0000 0.1021
0.2985 0.0000 00084 0.0688 0.0563 0.0073 2.1955
TPM1 0.2399 0.0034 0.2265 0.0024 0.0900 0.0000 0.0000
0.0414 0.0000 0.0578 0.0000 0.1404 0.0000 0.0940
TPM2 0.2544 0.0000 0.0000 0.0280 0.0000 0.0000 0.0355
0.1050 0.0386 0.0359 0.0000 0.0472 0.0000 0.0962
TPM3 0_0006 00000 0 0091 0 0103 0 0000 00000 00094
0_1137 00000 0_0083 0_0768 0_0791 00185 0_1827
TPM4 0.3360 0.0658 0.0000 0.0000 0.0900 0.0000 0.0246
0.1235 0.0004 00074 0.0028 0.1710 0.0015 0.1585
TPSA131 0.0000 0.0000 0.0039 0.0000 0.0900 0.0000 0.0054
0.0588 0.0000 0.0016 0.0000 0.0877 0.1779 0.2889
TTF1 0_0000 00000 0 0267 0 0093 0 0000 00000 00027 00819
00342 0_0000 0_0515 0_0738 00969 02675
UPIC2 0.0000 0.0000 0.0000 0.0000 0.0900 0.0000 0.0065
0.0354 0.0579 00000 0.0058 0.0145 0.0888 0.0697
UPK3A 0.0055 0.0000 0.0000 0.0000 0.0900 0.0000 0.0772
0.0381 0.0008 00000 0.0000 0.0576 0.0211 0.0987
UPK3B 0.0014 0.0018 0.0055 0.0000 0.0900 0.5617 0.0000
0.0308 0.0000 00000 0.0022 0.0295 0.0004 0.1637
VIM 0_0000 0_0008 0 0000 0 0000 0 0000 00000 0_0599
01707 00000 0_0000 00686 0_0794 00631 0_0949
VIL1 0.0921 0.0900 9.9832 0.0900 9.9990 0.0900 9.9138
0.0637 9.0090 00055 9.0115 0.1072 0.0339 0.0583
VIM 0.0000 0.0000 0.1933 0.2832 0.0900 0.0000 0.0000
0.1175 0.0301 00000 0.4466 0.0938 0.0036 0.0684
WTI 0.0063 0.0017 0.0011 0.0099 0.0900 0.0771 0.0034
0.0333 0.0000 0.1347 0.0000 2.1030 0.0205 0.0966
As noted, the transcripts provided in Tables 117-120 can be used in the
systems and
processes outlined in FIGs. 4A-B. For example, the disclosure provides a
method for classifying a
biological sample 400, 410, the method comprising: obtaining, by one or more
computers, first data
representing one or more initial classifications for the biological sample
that were previously
determined based on RNA sequences of the biological sample 401,411; obtaining,
as desired, by one
or more computers, second data representing another initial classification for
the biological sample
that were previously determined based on DNA sequences of the biological
sample 416 (see, e.g.,
Tables 2-16 and related text); providing, by one or more computers, at least a
portion of the first data
and the second data as an input to a dynamic voting engine 406, 415 that has
been trained to predict a
target biological sample classification based on processing of multiple
initial biological sample
classifications; processing, by one or more computers, the provided input data
through the dynamic
voting engine; obtaining, by one or more computers, output data generated by
the dynamic voting
engine based on the dynamic voting engine's processing of the provided input
data; and determining,
by one or more computers, a target biological sample classification for the
biological sample based on
the obtained output data 407, 417. In some embodiments, obtaining, by one or
more computers, first
data representing one or more initial classifications for the biological
sample that were previously
determined based on RNA sequences of the biological sample comprises:
obtaining data representing
a cancer type classification for the biological sample based the RNA sequences
of the biological
232
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
sample 403, 412 (see, e.g., Table 118 and related text); obtaining data
representing an organ from
which the biological sample originated based on the RNA sequences of the
biological sample 404,
413 (see, e.g., Table 119 and related text); and obtaining data representing a
histology for the
biological sample based on the RNA sequences of the biological sample 405, 414
(see, e.g., Table
5 120 and related text), and wherein providing at least a portion of the
first data and the second data as
an input to the dynamic voting engine 406, 415 comprises: providing the
obtained data representing
the cancer type 403, 412, the obtained data representing the organ from which
the biological sample
originated 404, 413, the obtained data representing the histology 405, 414,
and the second data as an
input to the dynamic voting engine 406, 415. In some embodiments, the dynamic
voting engine 406,
10 415 comprises one or more machine learning model. In some embodiments,
previously determining
an initial classification for the biological sample based on DNA sequences of
the biological sample
comprises 416: receiving, by one or more computers, a biological signature
representing the
biological sample that was obtained from a cancerous neoplasm in a first
portion of a body, wherein
the model includes a cancerous biological signature for each of multiple
different types of cancerous
15 biological samples, wherein each of the cancerous biological signatures
include at least a first
cancerous biological signature representing a molecular profile of a cancerous
biological sample from
the first portion of one or more other bodies and a second cancerous
biological signature representing
a molecular profile of a cancerous biological sample from a second portion of
one or more other
bodies; performing, by one or more computers and using a pairwisc-analysis
model, pairvvisc analysis
20 of the biological signature using the first cancerous biological
signature and the second cancerous
biological signature; generating, by one or more computers and based on the
performed pairvvise
analysis, a likelihood that the cancerous neoplasm in the first portion of the
body was caused by
cancer in a second portion of the body; and storing, by one or more computers,
the generated
likelihood in a memory device.
25 Relatedly, the disclosure also a method comprising: (a) obtaining a
biological sample from a
subject having a cancer; (b) performing at least one assay on the sample to
assess one or more
biomarkers, thereby obtaining a biosignature for the sample; (c) providing the
biosignature into a
model that has been trained to predict at least one attribute of the cancer,
wherein the model
comprises at least one pre-determined biosignature indicative of at least one
attribute, and wherein the
30 at least one attribute of the cancer is selected from the group
comprising primary tumor origin,
cancer/disease type, organ group, histology, and any combination thereof; (d)
processing, by one or
more computers, the provided biosignature through the model; and (e)
outputting from the model a
prediction of the at least one attribute of the cancer. The assays may
comprise next generation
sequencing of DNA and RNA, e.g., as described in Example 1. The assays can be
performed to
35 measure the same inputs as those used to train the models, e.g., based
on Tables 2-116 and/or Tables
118 120. Therefore the data for the sample from the subject can be processed
to determine the
attribute. For example, the models may be trained using data for DNA analysis
of groups of genes
233
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
selected from Tables 123-125 and/or Tables 128-129, or selections thereof. For
example, the models
may also be trained using data for RNA analysis of groups of genes selected
from Table 117, or
selections thereof. The biomarkers within the models thereby provide
predetermined biosignatures.
Then the assays performed on the samples for the subject can query those same
biomarkers within the
5 predetermined biosignatures. As a non-limiting example, predetermined
biosignatures trained to
predict a cancer or disease type may be according to Table 118, predetermined
biosignatures trained
to predict an organ type may be according to Table 119, and/or predetermined
biosignatures trained
to predict a histology may be according to Table 120. Following this example,
a sample from a
subject would then be assayed in order to determine a biosignature comprising
the genes in Table
10 118, Table 119, and or Table 120. Accordingly, the sample biosignature
can be processed by the
models comprising the corresponding predetermined biosignatures.
As a further illustration of the method of predicting the at least one
attribute of a cancer, the
disclosure provides a method such as outlined in FIGs. 4A-B 400, 410
comprising: (a) obtaining a
biological sample from a subject haying a cancer, wherein the biological
sample comprises a tumor
15 sample, bodily fluid, or other obtainable sample such as described
herein; (b) performing at least one
assay to assess one or more biomarkers in the biological sample to obtain a
biosignature for the
sample, e.g., performing DNA analysis by sequencing genomic DNA from the
biological sample 416,
wherein the DNA analysis can be performed for selections of the genes in
Tables 2-116; and/or
performing RNA analysis by sequencing messenger RNA transcripts from the
biological sample 410,
20 411, wherein the RNA analysis is performed for selections of the genes
in Table 117 or Tables 118-
120; (c) providing the biosignature into a model that has been trained to
predict at least one attribute
of the cancer, wherein the model comprises a plurality of intermediate models,
wherein the plurality
of intermediate models comprises: (1) an first intermediate model trained to
process DNA data using
the predetermined biosignatures according to Tables 2-116 (416); (2) a second
intermediate model
25 trained to process RNA data using predetermined biosignatures according
to Table 118 (403, 412);
(3) a third intermediate model trained to process RNA data using predetermined
biosignatures
according to Table 119 (403, 412); and (4) a fourth intermediate model trained
to process RNA data
using the predetermined biosignatures according to Table 120 (404, 413); (d)
processing, by one or
more computers, the provided biosignature through each of the plurality of
intermediate models in
30 part (c), providing the output of each of the plurality of intermediate
models into a final predictor
model, e.g. dynamic voting module 415, and processing by one or more
computers, the output of each
of the plurality of intermediate models through the final predictor model; and
(e) outputting from the
final predictor model a prediction of the at least one attribute of the cancer
417. As described herein,
the attribute is related to a tissue characteristic, such as TOO, and can be
output at a desired level of
35 granularity. In some embodiments, the predicted at least one attribute
of the cancer is a tissue-of-
origin selected from the group consisting of breast adenocarcinoma, central
nervous system cancer,
cervical adenocarcinoma, cholangiocarcinoma, colon adenocarcinoma,
gastroesophageal
234
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
adenocarcinoma, gastrointestinal stromal tumor (GIST), hepatocellular
carcinoma, lung
adenocarcinoma, melanoma, meningioma, ovarian granulosa cell tumor, ovarian &
fallopian tube
adenocarcinoma, pancreas adenocarcinoma, prostate adenocarcinoma, renal cell
carcinoma, squamous
cell carcinoma, thyroid cancer, urothelial carcinoma, uterine endometrial
adenocarcinoma, uterine
5 sarcoma, and a combination thereof. As desired, the models can be trained
to output the TOO at
different levels of granularity as described herein. See, e.g., the disease
types and organ groups
denoted in Tables 2-116 and related discussion.
The predicted at least one attribute of the cancer may be compared to a
threshold. For
example, the prediction or classification provided by the systems and methods
herein may comprise a
10 probability, likelihood, or similar statistical measure that indicates a
confidence level in the predicted
attribute. Such confidence level may be determined for each potential
attribute. See, e.g., discussion in
Example 3 and in the exemplar reports in Examples 4-5. The confidence in the
prediction may be
particularly important when assisting in treatment decision making for cancer
patients. As desired, the
disclosure contemplates additional clinical testing or review to confirm or
not the predicted attribute.
15 The disclosure further provides a system comprising one or more
computers and one or more
storage media storing instructions that, when executed by the one or more
computers, cause the one or
more computers to perform each of the operations described in the paragraphs
above. The disclosure
also provides a non-transitory computer-readable medium storing software
comprising instructions
executable by one or more computers which, upon such execution, cause the one
or more computers
20 to perform the operations described in the paragraphs above.
Advantageously, the systems and methods provided herein can be performed using
the
molecular profiling data that is used to help guide treatment selection for
cancer patients. See, e.g.,
Example 1. The predicted attributes may help provide a diagnosis of a CUP
sample, or provide a
quality check and potentially adjusted diagnosis for any profiled sample. 'The
latter may be
25 particularly desirable to verify the origin of a metastatic sample, or
other remote sample such as a
blood sample or other bodily fluid. Thus, the systems and methods provided
herein provide an
efficient means to help improve treatment of cancer patients.
Example 3 provides further details and demonstration of RNA and panomic
classifiers 400
and 410.
30 Report
In an embodiment, the methods as described herein comprise generating a
molecular profile
report_ The report can be delivered to the treating physician or other
caregiver of the subject whose
cancer has been profiled. The report can comprise multiple sections of
relevant information, including
without limitation: 1) a list of the biomarkers that were profiled (i.e.,
subject to molecular testing); 2)
35 a description of the molecular profile comprising characteristics of the
genes and/or gene products as
determined for the subject; 3) a treatment associated with the characteristics
of the genes and/or gene
235
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
products that were profiled; and 4) and an indication whether each treatment
is likely to benefit the
patient, not benefit the patient, or has indeterminate benefit. The list of
the genes in the molecular
profile can be those presented herein. See, e.g., Example 1. The description
of the biomarkers
assessed may include such information as the laboratory technique used to
assess each biomarker
5 (e.g., RT-PCR, FISH/CISH, PCR, FA/RFLP, NGS, etc) as well as the result
and criteria used to score
each technique. By way of example, the criteria for scoring a CNV may be a
presence (i.e., a copy
number that is greater or lower than the "normal" copy number present in a
subject who does not have
cancer, or statistically identified as present in the general population,
typically diploid) or absence
(i.e., a copy number that is the same as the "normal" copy number present in a
subject who does not
10 have cancer, or statistically identified as present in the general
population, typically diploid) The
treatment associated with one or more of the genes and/or gene products in the
molecular profile can
be determined using a biomarker-treatment association rule set such as in
'rabies 2-116, Tables 117-
120, ISNM1, or Tables 121-130 herein or any of International Patent
Publications WO/2007/137187
(Int'l Appl. No. PCT/US2007/069286), published November 29, 2007;
WO/2010/045318 (Int'l Appl.
15 No. PCT/US2009/060630), published April 22, 2010; WO/2010/093465 (Int'l
App!. No.
PCT/US2010/000407), published August 19, 2010; WO/2012/170715 (Int'l Appl. No.
PCT/US2012/041393), published December 13, 2012; WO/2014/089241 (Int'l Appl.
No.
PCT/US2013/073184), published June 12, 2014; WO/2011/056688 (Int'l Appl. No.
PCT/US2010/054366), published May 12, 2011; WO/2012/092336 (Int'l Appl. No.
20 PCT/US2011/067527), published July 5, 2012; WO/2015/116868 (Int'l Appl.
No.
PCT/US2015/013618), published August 6, 2015; WO/2017/053915 (Int'l Appl. No.
PCT/US2016/053614), published March 30, 2017; WO/2016/141169 (InflAppl. No.
PCT/US2016/020657), published September 9, 2016; and W02018175501 (Intl Appl.
No.
PC1/US2018/023438), published September 27, 2018; each of which publications
is incorporated by
25 reference herein in its entirety. Such biomarker-treatment associations
can be updated over time, e.g.,
as associations are refuted or as new associations are discovered. The
indication whether each
treatment is likely to benefit the patient, not benefit the patient, or has
indeterminate benefit may be
weighted. For example, a potential benefit may be a strong potential benefit
or a lesser potential
benefit. Such weighting can be based on any appropriate criteria, e.g., the
strength of the evidence of
30 the biomarker-treatment association, or the results of the profiling,
e.g., a degree of over- or
underexpression.
Various additional components can be added to the report as desired. In
preferred
embodiments, the report comprises a section detailing results of tissue
classification, e.g., as described
for determining one or more of a primary tumor local, cancer category,
cancer/disease type, organ
35 type, and/or histology. See, e.g., FIGs. 7E, 8C. Such attribute can be
provided at a desired level of
granularity, e.g., at a level that may alter treatment if the predicted
attribute differs from the original
attribution. See, e.g., FIGs. 6A11-AL and related discussion.
236
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
In some embodiments, the report comprises a list having an indication of
whether a presence,
level or state of an assessed biomarker is associated with an ongoing clinical
trial. The report may
include identifiers for any such trials, e.g., to facilitate the treating
physician's investigation of
potential enrollment of the subject in the trial. In some embodiments, the
report provides a list of
5 evidence supporting the association of the assessed biomarker with the
reported treatment. The list
can contain citations to the evidentiary literature and/or an indication of
the strength of the evidence
for the particular biomarker-treatment association. In some embodiments, the
report comprises a
description of the genes and gene products that were profiled. The description
of the genes in the
molecular profile can comprise without limitation the biological function
and/or various treatment
10 associations.
The molecular profiling report can be delivered to the caregiver for the
subject, e.g., the
oncologist or other treating physician. The caregiver can use the results of
the report to guide a
treatment regimen for the subject. For example, the caregiver may use one or
more treatments
indicated as likely benefit in the report to treat the patient. Similarly, the
caregiver may avoid treating
15 the patient with one or more treatments indicated as likely lack of
benefit in the report.
In some embodiments of the method of identifying at least one therapy of
potential benefit,
the subject has not previously been treated with the at least one therapy of
potential benefit. The
cancer may comprise a metastatic cancer, a recurrent cancer, or any
combination thereof. In some
cases, the cancer is refractory to a prior therapy, including without
limitation front-line or standard of
20 care therapy for the cancer. In some embodiments, the cancer is
refractory to all known standard of
care therapies. In other embodiments, the subject has not previously been
treated for the cancer. The
method may further comprise administering the at least one therapy of
potential benefit to the
individual. Progression free survival (PFS), disease free survival (DFS), or
lifespan can be extended
by the administration.
25 Exemplary reports are provided herein in FIGs. 7 and 8, which are
detailed in Examples 4
and 5, respectively.
The report can be computer generated, and can be a printed report, a computer
file or both.
The report can be made accessible via a secure web portal.
In an aspect, the disclosure provides use of a reagent in carrying out the
methods as described
30 herein as described above. In a related aspect, the disclosure provides
of a reagent in the manufacture
of a reagent or kit for carrying out the methods as described herein as
described herein. In still another
related aspect, the disclosure provides a kit comprising a reagent for
carrying out the methods as
described herein as described herein. The reagent can be any useful and
desired reagent. In preferred
embodiments, the reagent comprises at least one of a reagent for extracting
nucleic acid from a
35 sample, and a reagent for performing next-generation sequencing.
The disclosure also provides systems for performing molecular profiling and
generating a
report comprising results and analysis thereof. In an aspect, the disclosure
provides a system for
237
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
identifying at least one therapy associated with a cancer in an individual,
comprising: (a) at least one
host server; (b) at least one user interface for accessing the at least one
host server to access and input
data; (c) at least one processor for processing the inputted data; (d) at
least one memory coupled to the
processor for storing the processed data and instructions for: i) accessing a
molecular profile, e.g.,
5 according to Example 1; and ii) identifying, based on the status of
various biomarkers within the
molecular profile, at least one therapy with potential benefit for treatment
of the cancer; and (e) at
least one display for displaying the identified therapy with potential benefit
for treatment of the
cancer. In some embodiments, the system further comprises at least one memory
coupled to the
processor for storing the processed data and instructions for identifying,
based on the generated
10 molecular profile according to the methods above, at least one therapy
with potential benefit for
treatment of the cancer; and at least one display for display thereof The
system may further comprise
at least one database comprising references for various biomarker states, data
for drug/biomarker
associations, or both. The at least one display can be a report provided by
the present disclosure.
15 EXAMPLES
The invention is further described in the following examples, which do not
limit the scope as
described herein described in the claims.
Example 1: Molecular Profiling
Comprehensive molecular profiling provides a wealth of data concerning the
molecular status
20 of patient samples. We have performed such profiling on well over
100,000 tumor patients from
practically all cancer lineages using various profiling technologies. To date,
we have tracked the
benefit or lack of benefit from treatments in over 20,000 of these patients.
Our molecular profiling
data can thus be compared to patient benefit to treatments to identify
additional biomarker signatures
that predict the benefit to various treatments in additional cancer patients.
We have applied this "next
25 generation profiling" (NGP) approach to identify biomarker signatures
that correlate with patient
benefit (including positive, negative, or indeterminate benefit) to various
cancer therapeutics.
The general approach to NGP is as follows. Over several years we have
performed
comprehensive molecular profiling of tens of thousands of patients using
various molecular profiling
techniques. As further outlined in FIG. 2C, these techniques include without
limitation next
30 generation sequencing (NGS) of DNA to assess various attributes 2301,
gene expression and gene
fusion analysis of RNA 2302, II-IC analysis of protein expression 2303, and
ISH to assess gene copy
number and chromosomal aberrations such as translocations 2304. We currently
have matched patient
clinical outcomes data for over 20,000 patients of various cancer lineages
2305. We use cognitive
computing approaches 2306 to correlate the comprehensive molecular profiling
results against the
35 actual patient outcomes data for various treatments as desired. Clinical
outcome may be determined
238
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
using the surrogate endpoint time-on-treatment (TOT) or time-to-next-treatment
(TTNT or TNT). See,
e.g., Roever L (2016) Endpoints in Clinical Trials: Advantages and
Limitations. Evidence Based
Medicine and Practice 1: ern doi:10.4172/ebmp.1000e111. The results provide a
biosignature
comprising a panel of biomarkers 2307, wherein the biosignature is indicative
of benefit or lack of
5 benefit from the treatment under investigation. The biosignature can be
applied to molecular profiling
results for new patients in order to predict benefit from the applicable
treatment and thus guide
treatment decisions. Such personalized guidance can improve the selection of
efficacious treatments
and also avoid treatments with lesser clinical benefit, if any.
Table 121 lists numerous biomarkers we have profiled over the past several
years. As
10 relevant molecular profiling and patient outcomes are available, any or
all of these biomarkers can
serve as features to input into the cognitive computing environment to develop
a biosignature of
interest The table shows molecular profiling techniques and various biomarkers
assessed using those
techniques. The listing is non-exhaustive, and data for all of the listed
biomarkers will not be available
for every patient. It will further be appreciated that various biomarker have
been profiled using
15 multiple methods. As a non-limiting example, consider the EGFR gene
expressing the Epidermal
Growth Factor Receptor (EGFR) protein. As shown in Table 121, expression of
EGFR protein has
been detected using ITIC; EGFR gene amplification, gene rearrangements,
mutations and alterations
have been detected with ISH, Sanger sequencing, NGS, fragment analysis, and
PCR such as qPCR;
and EGFR RNA expression has been detected using PCR techniques, e.g., qPCR,
and DNA
20 microarray. As a further non-limiting example, molecular profiling
results for the presence of the
EGFR variant I11 (EGERvIII) transcript has been collected using fragment
analysis (e.g., REIT) and
sequencing (e.g., NGS).
Table 122 shows exemplary molecular profiles for various tumor lineages. Data
from these
molecular profiles may be used as the input for NGP in order to identify one
or more biosignatures of
25 interest. In the table, the cancer lineage is shown in the column "Tumor
Type." The remaining
columns show various biomarkers that can be assessed using the indicated
methodology (i.e.,
irnmunohistochemistry (111C), in situ hybridization (1SH), or other
techniques). As explained above,
the biomarkers are identified using symbols known to those of skill in the
art. Under the ITIC column,
"MMR" refers to the mismatch repair proteins MLH1, MSH2, MSH6, and PMS2, which
are each
30 individually assessed using Under the WF.S column "DNA Alterations,"
"CNA" refers to copy
number alteration, which is also referred to herein as copy number variation
(CNV). Under the WES
column "Genomic Signatures," "MST refers to microsatellite instability; "TAB"
refers to tumor
mutational burden, which may be referred to as tumor mutational load or EVIL;
"LOH" refers to loss
of heterozygosity; and "FOLFOX" refers to a predictor of FOLFOX response in
metastatic colorectal
35 adenocarcinoma as described in Intl Patent Publication W02020113237,
titled "NEXT-
GENERATION MOLECULAR PROFILING" and based on InElPatent Application No.
PCT/US2019/064078, filed December 2, 2019, which publication is hereby
incorporated by reference
239
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
in its entirety. Whole transcriptome sequencing (WTS) is used to assess all
RNA transcripts in the
specimen and can detect, inter alia, fusions and variant transcripts. Under
the column "Other,"
abbreviations include EBER for Epstein-Barr encoding region; and HPV is human
papilloma virus.
One of skill will appreciate that molecular profiling technologies may be
substituted as desired and/or
5 interchangeable. For example, other suitable protein analysis methods can
be used instead of ILIC
(e.g., alternate immunoassay formats), other suitable nucleic acid analysis
methods can be used
instead of ISH (e.g., that assess copy number and/or rearrangements,
translocations and the like), and
other suitable nucleic acid analysis methods can be used instead of fragment
analysis. Similarly, FISH
and CISH are generally interchangeable and the choice may be made based upon
probe availability
10 and the like. Tables 123-125 and 128-129 present panels of genomic
analysis and genes that have
been assessed using Next Generation Sequencing (NGS) analysis of DNA such as
genomic DNA.
Whole exome sequencing (WES) can be used to analyze the genomic DNA. One of
skill will
appreciate that other nucleic acid analysis methods can be used instead of NGS
analysis, e.g., other
sequencing (e.g., Sanger), hybridization (e.g., microarray, Nanostring) and/or
amplification (e.g., PCR
15 based) methods. The biomarkers listed in Tables 126-127 can be assessed
by RNA sequencing, such
as WTS. Using WTS, any fusions, splice variants, or the like can be detected.
Tables 126-127 list
biomarkers with commonly detected alterations in cancer.
Nucleic acid analysis may be performed to assess various aspects of a gene.
For example,
nucleic acid analysis can include, but is not limited to, mutational analysis,
fusion analysis, variant
20 analysis, splice variants, SNP analysis and gene copy
number/amplification. Such analysis can be
performed using any number of techniques described herein or known in the art,
including without
limitation sequencing (e.g., Sanger, Next Generation, pyrosequencing), PCR,
variants of PCR such as
RT-PCR, fragment analysis, and the like. NGS techniques may be used to detect
mutations, fusions,
variants and copy number of multiple genes in a single assay. Unless otherwise
stated or obvious in
25 context, a "mutation" as used herein may comprise any change in a gene
or genome as compared to
wild type, including without limitation a mutation, polymorphism, deletion,
insertion, indels (i.e.,
insertions or deletions), substitution, translocation, fusion, break,
duplication, loss, amplification,
repeat, or copy number variation. Different analyses may be available for
different genomic
alterations and/or sets of genes. For example, Table 123 lists attributes of
genomic stability that can
30 be measured with NGS, Table 124 lists various genes that may be assessed
for point mutations and
indels, Table 125 lists various genes that may be assessed for point
mutations, indels and copy
number variations, Table 126 lists various genes that may be assessed for gene
fusions via RNA
analysis, e.g., via WTS, and similarly Table 127 lists genes that can be
assessed for transcript variants
via RNA. Molecular profiling results for additional genes can be used to
identify an NGP biosignature
35 as such data is available.
240
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Table 121 - Molecular Profiling Biomarkers
Technique Biomarkers
11-IC ABLE ACPP (PAP), Actin (ACTA), ADA, APP, AKT
I, ALK, ALPP
(PLAP-1), APC, AR, ASNS, ATM, BAP1, BCL2, BCRP, BRAF,
BRCA I, BRCA2, CA19-9, CALCA, CCNDI (BCL I), CCR7, CD19,
CD276, CD3, CD33, CD52, CD80, CD86, CD8A, CDH1 (ECAD),
CDW52, CEACAM5 (CEA; CD66e), CES2, CHGA (CGA), CK 14, CK
17, CK 5/6, CK1, CK10, CK14, CK15, CK16, CK19, CK2, CK3, CK4,
CK5, CK6, CK7, CK8, COX2, CSE1R, C1L4A, C1LA4, C1NN131,
Cytokeratin, DCK, DES, DNMT I, EGFR, EGFR H-score, ERBB2
(HER2), ERBB4 (ITER4), ERCC1, ERCC3, ESR1 (ER), F8 (FACTOR8),
FBXW7, FGER1, FGFR2, FLT3, FOLR2, GART, GNAI 1, GNAQ,
GNAS, Granzyme A, Granzyme B, GSTP1, IIDAC1, HIE1A, UN-171A,
HPTõ HRAS, HSP9OAA1 (HSPCA), 11)H1, IDOL IT,2, IT,2RA (CD25),
JAK2, JAK3, KDR (VEGFR2), KI67, KIT (cKIT), KLK3 (PSA), KRAS,
KRT20 (CK20), KRT7 (CK7), KRT8 (CYK8), LAG-3, MAGE-A, MAP
KINASE PROTEIN (MAPK I /3), MDM2, MET (cMET), MGMT,
MLHI, MPL, MRP I, MS4A1 (CD20), MSH2, MSH4, MSH6, MSI,
MTAP, MUC1, MUC16, NEM:11, NEKB1A, NEM-32, NGF, NOTCII1,
NPM1, NRAS, NY-ESO-1, ODC1 (ODC), OGFR, p16, p95, PARP-1,
PBRM1, PD-1, PDGF, PDGFC, PDGFR, PDGFRA, PDGFRA
(PDGFR2), PDGFRB (PDGFRI), PD-L1, PD-L2, PGR (PR), PIK3CA,
PIP, PMEL, PMS2, POLA1 (POLA), PR, PTEN, PTGS2 (COX2),
PTPN11, RAF I, RARA (RAR), Rh, RET, RHOH, ROSI, RRMI, PAR,
RXRB, S100B, SETD2, SMAD4, SMARCB1, SMO, SPARC, SST,
SSTR I , STK I 1, SYP, TAG-72, TIM-3, TK I , TLE3, TNF, TOP1
(TOPO I), TOP2A (TOP2), TOP2B (TOPO2B), TP, TP53 (p53),
TRKA/B/C, TS, TUBB3, TXNRD1, TYMP (PDECGF), TYMS (TS),
VDR, VEGT,A (VEGE), VHL, XDH, ZAP70
ISH (CISH/FISH) 1p19q, ALK, EML4-ALK, EGFR, ERCC1, UER2, UPV (human
papilloma virus), MDM2, MET, MYC, PIK3CA, ROS1, TOP2A,
chromosome 17, chromosome 12
Pyrosequencing MGMT promoter methylation
Sanger sequencing BRAE, EGFR, GNA11, GNAQ, ILRAS, IDII2, KIT, KRAS, NRAS,
PIK3CA
NGS See genes and types of testing in Tables 122-
129, MSI, TMB, LOH
241
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
WES, WTS
Fragment Analysis ALK, EML4-ALK, EGFR Variant III, HERZ exon 20, ROS 1, MSI
PCR ALK, AREG, BRAF, BRCA1, EGFR, EML4, ERBB3,
ERCC1, EREG,
hENT-1, HSP9OAA1, IGF-1R, KRAS, MMR, p16, p21, p27, PARP-1,
PGP (MDR-1), PIK3CA, RRM1, EFF3, TOP01, TOPO2A, TS, TUBB3
Microarray ABCC I, ABCG2, ADA, AR, ASNS,13CL2,13IRC5, BRCA1,13RCA2,
CD33, CD52, CDA, CES2, DCK, DHFR, DNMT1, DNMT3A,
DNMT3B, ECGF1, EGFR, EPHA2, ERBB2, ERCC1, ERCC3, ESR1,
FLT1, FOLR2, FYN, GART, GNR141, GSTP1, HCK, UDACT, HIF1A,
HSP9OAA1 (HSPCA), fL2RA, HSP9OAA1, KDR, KIT, LCK, LYN,
MGMT, MLH1, MS4A1, MSH2, NFKB1, NFKB2, OGFR, PDGFC,
PDGFRA, PDGFRB, PGR, POLA1, PTEN, PTGS2, RAF1, RARA,
RRM1, RRM2, RRM2B, RXRB, RXRG, SPARC, SRC, SSTR1, SSTR2,
SSTR3, SSTR4, SSTR5, TK1, TNF, TOPT, TOP2A, TOP2B, TXNRD1,
TYMS, VDR, VEGFA, VHL, YES1, ZAP70
Table 122 - Molecular Profiles
Whole
Whole Exome Transcriptome
Sequencing (VVF,S) Sequencing
Tumor Type RIC (WTS) Other
DNA Genomic
RNA
alterations Signatures
Bladder MMR, PD-Li Mutation, MSI, Fusions,
Variant
Indels, TMB, Transcripts
CNA LOH
Breast AR, ER, Mutation, MSI, Fusions,
Variant IIer2, TOP2A
Her2/Neu, Indels, TMB, Transcripts
(CISH)
MMR, PD-Li, CNA LOH
PR, PTEN
Cancer of Unknown AR, ER, IIER2, Mutation, MSI, Fusions, Variant
Primary - Female MMR, PD-Li Indels, TMB,
Transcripts
CNA LOH
Cancer of Unknown AR, HER2, Mutation, MSI, Fusions,
Variant
Primary - Male MMR, PD-Li Indels, TMB,
Transcripts
CNA LOH
Cervical ER, MMR, PD- Mutation, MSI,
Fusions, Variant
LE PR Indels, TMB, Transcripts
CNA LOII
Cholangiocarcinoma/ Her2/Neu, Mutation, MST, Fusions,
Variant Her2 (CTSH)
Hepatobiliary MMR, PD-Li Indels, TMB,
Transcripts
CNA LOH
Colorectal and Small Her2/Neu, Mutation, MSI, Fusions,
Variant
Intestinal MMR, PD-II, Indels, TMB, Transcripts
PTEN CNA
242
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
LOH,
FOLFOX
Endometrial ER, MMR, PD- Mutation, MSI,
Fusions, Variant
Li, PR, PTEN Indels, TMB, Transcripts
CNA LOH
Esophageal IIer2/Neu, Mutation, MSI, Fusions,
Variant EBER (CISH)
MMR, PD-Li Indels, TMB, Transcripts
CNA LOH
Gastric/GEJ Her2/Neu, Mutation, MSI, Fusions,
Variant EBER, Her2
MMR, PD-Li Indels, TMB, Transcripts
CNA LOH
GIST MMR, PD-L1, Mutation, MSI,
Fusions, Variant
PTEN Indels, TMB, Transcripts
CNA LOII
Glioma MMR, PD-L1 Mutation, MSI,
Fusions, Variant MGMT
Indels, TMB, Transcripts
Methylation
CNA LOH
(Pyrosequencing)
Itead & Neck MMR, p16, PD- Mutation,
MSI, Fusions, Variant EBER, IEPV
Ll Indels, TMB, Transcripts
(ME), reflex to
CNA LOH confirm p 16
result
Kidney MMR, PD-Li Mutation, MSI,
Fusions, Variant
Indels, TMB, Transcripts
CNA LOH
Lymphoma / Mutation, TMB Fusions,
Variant
Leukemia Indels, Transcripts
CNA
Melanoma MMR, PD-Li Mutation, MSI,
Fusions, Variant
Indels, TMB, Transcripts
CNA LOH
Merkel Cell MMR, PD-Li Mutation, MSI, Fusions,
Variant
Indels, TMB, Transcripts
CNA LOH
Neuroendoerine MMR, PD-Li Mutation, MSI, Fusions,
Variant
Indels, TMB, Transcripts
CNA LOH
Non-Small Cell Lung ALK, MMR, Mutation, MSI, Fusions,
Variant
PD-L1, PTEN Indels, TMB, Transcripts
CNA LOH
Ovarian ER, MMR, PD- Mutation, MSI,
Fusions, Variant
El, PR Indels, TMB, Transcripts
CNA LOH
Pancreatic MMR, PD-Li Mutation, MSI, Fusions,
Variant
Indels, TMB, Transcripts
CNA LOII
Prostate AR, MMR, PD- Mutation, MSI,
Fusions, Variant
Li Indels, TMB, Transcripts
CNA LOH
Salivary Gland AR, Her2/Neu, Mutation,
MSI, Fusions, Variant
MMR, PD-Li Indels, TMB, Transcripts
CNA LOH
Sarcoma MMR, PD-Li Mutation, MSI,
Fusions, Variant
Indels, TMB, Transcripts
CNA LOH
243
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Small Cell Lung MMR, PD-Li Mutation, MSI, Fusions,
Variant
Indels, TMB, Transcripts
CNA LOH
Thyroid MMR, PD-L1 Mutation, MSI, Fusions,
Variant
Indels, TMB, Transcripts
CNA LOH
Uterine Serous ER, Her2/Neu, Mutation,
MSI, Fusions, Variant Her2 (CISH)
MMR, PD-L1, Indels, TMB, Transcripts
PR, PTEN CNA LOH
Vulvar Cancer (SCC) ER, MMR, PD- Mutation, MSI, Fusions, Variant
Li, PR, TRK Indels, TMB, Transcripts
AJB/C CNA LOH
Other Tumors MMR, PD-Li Mutation, MSI,
Fusions, Variant
Indels, TMB, Transcripts
CNA LOH
Table 123 ¨ Genomic Stability Testing (DNA)
Microsatellite Instability Tumor Mutational Burden
Loss of Heterozygosity (LOH)
(MSI) (TMB)
Table 124 ¨ Point Mutations and Indels (DNA)
ABB CDK12 FOX03 INTITIA MSN PHOX213 SSX1
ABL1 CDKN2B FOX04 IRS2 MTCP1 PIK3CG STAG2
ACKR3 CDKN2C FST13 JUN M t JC1 PLAGI
TALI.
AKT1 CEBPA GATA1 KAT6A MUTYH PMS1 TAL2
(MYST3)
ANIER1 CI ICI TD7 GATA2 KAT6B MYCL P0U5F1
TBL TXR1
(FAM12313) (MYCT ,1)
AR CNOT3 GNAll KCNJ5 NBN PPP2R1A TCEA1
ARAF COT,1 Al GPC3 KDM5 C NDRG1 PRF1 TCT,1A
ATP2B3 COX6C HEY' KDM6A NKX2-1 PRKDC TERT
ATRX CRLF2 HIST1H3B KDSR NONO RAD21 TFE3
BCL11B DDB2 HIST1H4I KLF4 NOTCH1 RECQL4 TFPT
BCL2 DDIT3 ELF KLK2 NRAS RHOH THRAP3
244
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
BCL2L2 DNIVI2 HMGN2P46 LASP I NUMA I RNF213 TLX3
BCOR DNIVIT3A UNE 1 A LMO1 NUTM2B
RPL10 TMPRS S2
BCORL I E1F4A2 HOXA II LMO2 OLIG2 SEPT5 UBR5
BRD3 ELF4 HOXA13 MAFB OMD SEPT6 VIAL
BRD4 ELN HOXA9 MAX P2RY8 SFPQ WAS
BTG1 ERCC1 HOXC11 MECOM PAFAH1B2 SLC45A3 ZBTB16
131K ET V4 HOXC13 MED12 PAK3 SMARCA4 ZRSR2
C15 orf65 FAM46C HOXD11 MKLI PATZI SOCSI
CBLC FANCF HOXD13 MLLT11 PAX8 SOX2
CD79B FEV fIRAS MNI PDE4DIP SPOP
CDH1 F OXL2 11(BKE MPL PUF6 SRC
Table 125 ¨ Point Mutations, Indels and Copy Number Variations (DNA)
ABL2 CCND2 ERBB4 GSK3B MLFI PML SRGAP3
(HER4)
ACSL3 CCND3 ERC1 H3F3A MLH1 PMS2 SRSF2
ACSL6 CCNE1 ERCC2 H3F3B MLLT1 POLE SRSF3
ADGRA2 CD274 ERCC3 HERPUD1 MLLT10 POT1 SS18
(PDLI)
AFDN CD74 ERCC4 HGF MLLT3 POU2AF1 SS18L1
AFF1 CD79A ERCC5 HIM MLLT6 PPARG STAT3
AFF3 CDC73 ERG HA 4GA1 MNX1 PRCC STAT4
AFF4 CDH11 ESR1 HIVEGA2 MREll PRDM1 S
TA T5I3
AKAP9 CDK4 ETV1 UN1NPA2B1 MSH2 PRDM16 sTEL
AKT2 CDK6 ETV5 HOOK3 MSH6 PRKAR1A STK11
AKT3 CDK8 E1V6 HSP9OAA1 MSI2 PRIO(1 SUFU
ALDH2 CDKN1B EWSR1 HSP90AB1 MTOR PSIP1 SUZ12
ALK CDKN2A EXT1 IDH1 MYB PTCH1 SYK
APC CDX2 EXT2 1DII2 MYC PTEN TAF15
ARFRP1 CHEK1 EZII2 IGF1R MYCN PTPN11 TCF12
ARHGAP26 CHEK2 Ent 1KZE1 MYD88 PTPRC TCF3
ARHGEF12 CHIC2 FANCA EL2 MYH1 I RABEPI TCF7L2
245
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
ARID 1 A CITN1 FANCC EL21R MYH9 RAC1 TETI
AR1D2 CIC FANCD2 EL6ST NACA RAD50 TET2
ARNT CIITA FANCE EL7R NCKEF'SD RADS 1 TFEB
ASPSCRI CLPI FANCG TRF4 NCOAI RADS IB TFG
ASXL I CLTC FANCL ITK NCOA2 RAF I TFRC
ATF1 CLTCL1 FAS JAK1 NCOA4 RALGDS TGFBR2
A TIC CNBP FBX011 JAK2 NF1 RANBP17 TT ,X1
ATM CNTRL FBXW7 JAK3 NF2 RAP1GDS1 INFAE133
ATPIA 1 COPBI FCRL4 JAZF 1 NFE2L2 RARA TNFRSF 14
ATR CREB1 F GF10 KDM5A NFIB RB1 INFRSF 17
AURKA CREB3L1 F GF14 KDR NFKB2 RBM15 TOPI
(VFGFR2)
AURKB CREB3L2 I, CIF 19 KEAY 1 NIK131A REL
11353
AXINI CREBBP FGF23 KIAA1549 NIN RET TPM3
AXL CRKL FGF3 KIF5B NOTCH2 RICTOR TPM4
BAPI CRTCI FGF4 KIT NPMI RMI2 TPR
BARD1 CR l'C3 I, GF 6 KLHL6 N SD1 RN F 43
.. [RAF 7
BCL10 CSFIR F GFRI KMT2A NSD2 ROSI TRIM26
(MIT)
BCL 1 lA CSF3R F GFRI OP KMT2C NSD3 RPL22
TRIM27
(MT ,T 3)
13CL2E I 1 C I Ch I, GI, R2 KM I 2D N I 5C2 RPL5
I RIM33
(MLL2)
BCT ,3 CTT ,A4 FGFR3 KNT,1 NTRK1 RPN1 ..
TRIP 1 1
BCL6 CTNNAI FGFR4 KRAS NTRK2 RPTOR TRRAP
BCL7A CTNNB1 FH KIN1 NTRK3 RUNX1 TSC1
BCL9 CYLD FHIT LCK NUP214 RUNX 1 TI TSC2
BCR CYP2D6 FIP1L1 LCP1 NUP93 SBDS TSHR
BIRC3 DAXX FLCN LGR5 NUP98 SDC4 TTL
BLM DDR2 FLII LIEFPL6 NUTMI SDITAF 2 U2AF
1
BMPRI A DDX10 FLTI LIFR PALB2 SDIIB USP6
BRAF DDX5 FLT3 LPP PAX3 SDHC VEGFA
BRCA1 DDX6 FLT4 LRIG3 PAX5 SDELD VEGFB
BRCA2 DEK FNBPI LRP IB PAX7 SEPT9 VTIIA
BRIP1 DICER1 FOXA1 LYL1 PBRM1 SET WDCP
246
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
BUB 1 B DOT 1L F OX01 MAF PBX 1 SETBP 1 W IF 1
CACNA 1D EBF 1 F OXP 1 MALT 1 PCM 1 SETD2 W
ISP3
CALR ECT2L FUBP 1 MAML2 PCSK7 SF 3B 1 WRN
CAIVITA 1 EGFR FUS MAP2K 1 PDCD 1 SH2B3 WT1
(MEK1) (PD1)
CANTI ELK4 GAS7 MAP2K2 PDCD 1LG2 SH3 GL 1
WWTRI
(MEK2) (PDL2)
CARD 1 1 ELL GATA3 MAP2K4 PDGFB SLC3 4A2 XPA
CARS EME4 GID4 MAP3K 1 PDGFRA SMAD2 XPC
(C17orf39)
CASP8 EMSY GMPS MCLI PDGFRB SMAD4 XPO1
CBFA2T3 EP300 GNA13 MDM2 PDK I SMARCB1 YWHAE
CBFB EPHA3 GNAQ MDM4 PERI SMARCEI ZMYM2
CBL EPHA5 GNAS MDS2 PICALM SMO ZNF217
C13L13 EPH131 GOLGA5 MLF2B PIK3CA SN X29
ZN14331
CCDC6 EPS 15 GOPC MENI PIK3R1 SOX10
ZNF384
CCNB1IP1 ERBB2 GPI-1N MET PIK3R2 SPECCI ZNF521
(ITER2/NEU)
CCND I ERBB3 GRIN2A MITF PIM I SPEN ZNF703
(HER3)
Table 126 ¨ Gene Fusions (RNA)
ABL FUR MAML2 N 1RK2 RELA
AKT3 F GERI MASTI NTRK3 RET
ALK FGFR2 MAST2 NUMBL ROSI
ARHGAP26 FGFR3 MET PDGFRA RSPO2
AXT. ERG MSMB PDGFRB R SPO3
BCR ESRI MUSK PiK3 CA TERT
BRAF ETV I MYB PKN1 TFE3
BRD3 ETV4 NOTCHI PPARG TFEB
BRD4 ETV5 NOTCH2 PRKCA THADA
EGFR ETV6 NRGI PRKCB TMPRS S2
EWSRI INTSR NTRKI RAFI
Table 127¨ Variant Transcripts
AR-V7 EGFR vIII MET Exon 14 Skipping
247
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Abbreviations used in this Example and throughout the specification, e.g.,
IIIC:
irnmunohistochemistry; ISH: in situ hybridization; CISH: colorimetric in situ
hybridization; FISH:
fluorescent in situ hybridization; NGS: next generation sequencing; PCR:
polymerase chain reaction;
CNA: copy number alteration; CNV: copy number variation; MSI: microsatellite
instability; TNIB:
tumor mutational burden.
With whole exome sequencing (WES) and whole transcriptome sequencing (WTS),
quantitative sequencing data is available for practically all known genes and
transcripts. For example,
WES and WTS may query 22,000 or more sequences of interest. In addition to the
genes in Tables
124-125, Tables 128-129 provide additional selections of genes of interest,
e.g. genes most
commonly associated with cancer, that may be of particular interest in
molecular profiling cancer
samples.
Table 128 ¨ Point Mutations and Indels (DNA)
ABL1 CDK12 HDAC MAX PMS1 SDHAF2
ATP CXCR4 HIST1H3B MED12 POLD1 SETD2
AKT1 DNNIT3A HIST1H3C MPL PPP2R1A SMARCA4
AMER1 EPHA2 ITIVE1A MSH3 PPP2R2A SO C S1
AR FANCB I IOXB13 MST1R PRKACA SPOP
ARAF FANCF HRAS JTYFT PRKDC SRC
ATRX FANCI KDM5C NBN RABL3 TERT
B2M FANCM KDM6A NOTCH1 RAD51B TNIEM127
BCL2 FAT1 KDR NRAS RAD51C VHL
BCOR FOXL2 LYN NTHL1 RAD51D XRCC1
BTK FYN LZTR1 PARP1 RAD54L YES1
CD79B GLI2 MAPK1 PII0X2B RII0A
CDH1 GNAll MAPK3 PIK3CB SDHA
Table 129¨ Point Mutations, Indels and Copy Number Variations (DNA)
ALK CDK4 FANCA H3F3A MTIE P1K3R1 SMAD2
APC CDK6 FANCC H3F3B MLH1 PIM1 SMAD4
ARID 1 A CDICN1B FANCD2 TDH1 MREll PMS2 SMARCB1
ARID2 CDKN2A FANCE IDH2 MSH2 POLE SMARCE1
A SXT,1 CHFK1 FANCCi TRF4 MSH6 POT1 SMO
ATM CHEK2 FANCL JAK1 MTOR PPARG SPEN
ATR CIC FAS JAK2 MYCN PRDM1 STAT3
BAP1 CREBBP FBXW7 JAK3 MYD88 PRKAR1A S'1K11
248
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
BARD1 CSF1R FGFR1 KEAP1 NF1 PTCH1 SUFU
BCL9 CTNNA1 FGFR2 KIT NF2 PTEN TNFAIP3
BLM CTNNB1 FGFR3 KMT2A NFE2L2 PTPN11 TNFRSF 14
BMPR1A CYLD FGFR4 KMT2C NFKBIA RAD50 TP53
BRAF DDR2 FH KMT2D NPM1 RAF1
TSC1
BRCA1 DICER1 FLCN KRAS NSD1 RB1 TSC2
BRCA2 EGFR FLT1 T,CK NTRK1 RET 1J2AF1
BRIP1 EP300 FLT3 MAP2K1 NTRK2 RNF43
WRN
CARD11 ERBB2 FLT4 MAP2K2 NTRK3 ROS1
WT1
CBFB ERBB3 FUBP1 MAP2K4 PALB2 RUNX1
CCND1 ERBB4 GATA3 MAP3K1 PBRM1 SDHB
CCND2 ERCC2 GNA13 MEF2B PDGFRA SDHC
CCND3 ESR1 GNAQ MEN1 PDGFRB
SDIED
CDC73 EZH2 GNAS MET PIK3 CA SF3B1
The precise molecular profiles in this Example have been and are adjusted over
time,
including without limitation reasons such as the development of new and
updated technologies,
biomarker tests and companion diagnostics, and new or updated evidence for
biomarker ¨ treatment
5 associations. Thus, for some patient molecular profiles gathered in the
past, data for various
biomarkers tested with other methods than those in Tables 122-129 is available
and can be used for
NGP.
Table 130 presents a view of associations between the biomarkers assessed and
various
therapeutic agents. Such associations can be determined by correlating the
biomarker assessment
10 results with drug associations from sources such as the NCCN, literature
reports and clinical trials.
The column headed "Agent" provides candidate agents (e.g., drugs or biologics)
or biomarker status.
In some cases, the agent comprises clinical trials that can be matched to a
biomarker status. In some
cases, multiple biomarkers are associated with an agent or group of agents.
Platform abbreviations are
as used throughout the application, e.g., 11-IC: immunohistochemistry; C1SH:
colorimetric m situ
15 hybridization; NGS: next generation sequencing; PCR: polymerase chain
reaction; CNA: copy
number alteration. Tumor Type abbreviations include: TNBC: triple negative
breast cancer; NSCLC:
non-small cell lung cancer; CRC: colorectal cancer; GET: gastroesophageal
junction, EBDA:
extrahepatic bile duct adenocarcinoma. Biomarker abbreviations include: URR:
Homologous
Recombination Repair, which includes the genes ATM, ElARD1, BRCA1, BRCA2,
BRIP1, CDK12,
20 CHEK1, CHEK2, FANCL, PALB2, RAD51B, RAD51C, RAD51D, RAD54L; MST:
mierosatellite
instability; MSS: microsatellite stable; MMR: mismatch repair; TMB: tumor
mutational burden.
249
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Agents for biomarker PD-Li identify specific antibodies used in detection
assays in the
parentheticals.
Table 130 ¨ Biomarker ¨ Treatment Associations
Biomarker Technology / Agent
Alteration
ALK IBC, RNA fusion crizotinib, ceritinib,
alectmib, brigatinib (NSCLC),
lorlatinib (NSCLC)
DNA mutation resistance to crizotinib, alectinib
AR 1:11C bicalutamide, leuprolide (salivary
gland tumors)
enzalutamide, bicalutamide (TNBC)
ATM DNA mutation carboplatin, cisplatin,
oxaliplatin
olaparib (prostate)
BRAF DNA mutation vemurafenib, dabrafenib,
cobimetinib, trametinib
vemurafenib I (cetuximab or panitumumab) I irinotecan
(CRC)
encorafenib + binimetinib (melanoma)
clabrafenib+trametinib (anaphistic thyroid and NSCLC)
atezolizumab + cobirnetinib + vemurafenib (melanoma)
cetuximab + encorafenib (CRC)
cetuximab, panituimunab with BRA_F and or MEK
inhibitors (CRC)
BRCA 1 /2 DNA mutation carboplatin, cisplatin,
oxaliplatin
niraparib (ovarian, prostate), olaparib (breast,
cholangiocarcinoma, ovarian, pancreatic, prostate),
rucaparib (ovarian, pancreatic, prostate), talazoparib
(breast), veliparib combination (pancreatic)
resistance to olaparib, niraparib, rucaparib with reversion
mutation
EGFR DNA mutation afatinib (NSCLC)
afatinib + cetuximab (T790M; NSCLC)
erlotinib, gefitinib (NSCLC and CUP)
osimertinib, dacomitinib (NSCLC)
ER II-IC endocrine therapies
everolimus, temsirolimus (breast)
palbociclib, ribociclib, abemaciclib (breast)
ERBB2 CISH, DNA trastuzurnab, lapatinib, ncratinib
(breast), pertuzumab, T-
250
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
(HER2) mutation, CNA DM1, fam-trastuzumab deruxtecan-
nxki, tucatinib
DNA mutation T-DM1 (NSCLC)
EPJPR/ERBB2 CISH sacituzumab govitecan (TNBC)
(HER2)
ESR1 DNA mutation exemestane + everolimus,
fidvestrant, palbociclib
combination therapy (breast)
resistance to aromatase inhibitors (breast)
FGER2/3 DNA mutation, erdafitinib (urothelial
bladder), pemigatinib
RNA fusion (cholangiocarcinoma)
HRR DNA mutation olaparib (prostate)
IDH1 DNA mutation temozolomide (high grade glioma)
ivosidenib (cholangiocarcinoma and EBDA)
KIT DNA mutation irnatinib
regorafenib, sunitinib (both GIST)
KRAS DNA mutation resistance to cetuximab,
panitumumab (CRC)
resistance to erlotinibigefitinib (NSCLC)
resistance to trastuzumab, lapatinib, pertuzumab (CRC)
MET RNA exon cabozantinib, crizotinib
(NSCLC)
skipping, DNA
exon skipping,
CNA
MGMT Pyrosequencing temozolomide (high grade glioma)
(Methylation)
MMR 111 C, DNA pembrolizumab
Deficiency mutation
MSI pcmbrolizumab, nivolumab (CRC, small
bowel
adenocarcinoma), nivolumab-Fipilimumab (CRC, small
bowel adenocarcinoma)
MMR HIC, DNA pcmbrolizumab + lenvatinib
(endometrial)
Proficiency mutation
MSS
NRAS DNA mutation resistance to cetuximab,
panitumumab (CRC)
resistance to trastuzumab, lapatinib, pertuzumab (CRC)
NTRK 1 /2/3 RNA fusion entrectinib,
larotrectinib
DNA mutation resistance to entrectinib,
larotrectinib
PALB2 DNA mutation olaparib (pancreatic and
prostate), veliparib combination
2S1
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
(pancreatic)
PDGFRA DNA mutation imatinib , avapritinib (GIST),
sunitinib
PD-Li 11-IC pembrolizumab (22c3 TPS in NSCLC;
22c3 CPS in
cervical, GEJ/gastric, head & neck, urothelial and non-
urothelial bladder, vulvar)
atezolizumab (SP142 IC urothelial bladder cancer and
SP142 IC & TC NSCLC)
pembrolizumab + chemotherapy (22c3 CPS in TNBC)
atezolizumab I nab-paclitaxel (SP142 IC in TNBC)
nivolumab/ipilimurnab combination (28-8 NSCLC)
avelunaab (non-urothelial bladder and Merkel cell)
PIK3CA DNA mutation alpelisib + fulvestrant
(breast)
POLE DNA mutation pembrolizumab (endometrial and
CRC)
PR RIC endocrine therapies
RE! RNA fusion cabozantinib, vandetanib,
selpercatinib, pralsetinib
(NSCLC)
DNA mutation vandetanib, cabozantinib,
selpercatinib (thyroid); resistance
to vandetanib, cabozantinib
ROS1 RIC, RNA fusion crizotinib, ceritinib,
entrectinib, lorlatinib (NSCLC)
TMB DNA mutation pembrolizumab
TOP2A CISH doxorubicin, liposomal doxorubicin,
epirubicin (all breast)
Example 2: Genomic Prevalence Score (GPS) using a DNA NGS Panel to Predict
Tumor
Types
This Example describes the development of a Genomic Prevalence Score system
(which may
also be referred to herein as GPS; Genomic Profiling Similarity; Molecular
Disease Classifier; MDC)
to predict tumor type of a biological sample using a next generation
sequencing panel to assess
genomic DNA. This Example further applies GPS to the prediction of tumor types
for an expanded
specimen cohort, with closer analysis of Carcinoma of Unknown Primary (CUP;
aka Cancer of
Unknown Primary).
Current standard histological diagnostic tests are not able to determine the
origin of metastatic
cancer in as many as 10% of patients', leading to a diagnosis of cancer of
unknown primary (CUP).
The lack of a definitive diagnosis can result in administration of suboptimal
treatment regimens and
poor outcomes. Gene expression profiling has been used to identify the tissue
of origin but suffers
from a number of inherent limitations. These limitations impair performance in
identifying tumors
252
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
with low neoplastic percentage in metastatic sites which is where
identification is often most needed2.
The GPS system provided herein was developed using data for genomic DNA
sequencing of a 592
gene panel (see description in Example 1, with panel comprises of biomarkers
in Tables 123-125)
coupled with a machine learning platform to aid in the diagnosis of cancer.
The algorithm created was
5 trained on 34,352 cases and tested on 15,473 unambiguously diagnosed
cases. The performance of the
algorithm was then assessed on 1,662 CUP cases. The GPS accurately predicted
the tumor type in the
labeled data set with sensitivity, specificity, PPV, and NPV of 90.5%, 99.2%,
90.5% and 99.2%
respectively. Performance was consistent regardless of the percentage of tumor
nuclei or whether or
not the specimen had been obtained from a site of metastasis. Pathologic re-
evaluation of selected
10 discordant cases resulted in confirmation of GPS results and clinical
utility. Moreover, all genomic
markers essential for therapy selection are assessed in this assay, maximizing
the clinical utility for
patients within a single test.
Introduction
Carcinoma of Unknown Primary (CUP) represents a clinically challenging
heterogeneous
15 group of metastatic malignancies in which a primary tumor remains
elusive despite extensive clinical
and pathologic evaluation. Approximately 24% of cancer diagnoses worldwide
comprise CUP'. In
addition, some level of diagnostic uncertainty with respect to an exact tumor
type classification is a
frequent occurrence across oncologic subspecialties. Efforts to secure a
definitive diagnosis can
prolong the diagnostic process and delay treatment initiation. Furthermore,
CUP is associated with
20 poor outcome which might be explained by use of suboptimal therapeutic
intervention.
Immunohistochemical (H IC) testing is the gold standard method to diagnose the
site of tumor origin,
especially in eases of poorly differentiated or undifferentiated tumors.
Assessing the accuracy in
challenging cases and performing a meta-analysis of these studies reported
that 1HC analysis had an
accuracy of 66% in the characterization of metastatic tumors'. Since
therapeutic regimes are highly
25 dependent upon diagnosis, this represents an important unmet clinical
need. To address these
challenges, assays aiming at tissue-of-origin (TOO) identification based on
assessment of differential
gene expression have been developed and tested clinically. However,
integration of such assays into
clinical practice is hampered by relatively poor performance characteristics
(from 83% to 89%11.14)
and limited sample availability. For example, a recent commercial RNA-based
assay has a sensitivity
30 of A3% in a test set of IA7 tumors and confirmed results on only 7A% of
a separate 300 sample
validation set". This may, at least in part, be a consequence of limitations
of typical RNA-based
assays in regards to normal cell contamination, RNA stability, and dynamics of
RNA expression.
Nevertheless, initial clinical studies demonstrate possible benefit of
matching treatments to tumor
types predicted by the assay". With increasing availability of comprehensive
molecular profiling
35 assays, in particular next-generation DNA sequencing, genomic features
have been incorporated in
CUP treatment strategies". While this approach rarely supports unambiguous
identification of the
TOO, it does reveal targetable molecular alterations in some of the patients'.
253
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
In this Example, we pursued a different strategy of TOO identification by
using a novel
machine-learning approach as provided herein to build TOO classifiers based on
data from a large
NGS genomic DNA panel that assesses hundreds of gene sequences and various
attributes thereof
(see Example 1) and has been broadly used in clinical treatment of cancer
patients. This
5 computational classification system identified TOO at an accuracy
significantly exceeding that of
previously published technologies. Moreover, the 592-gene NGS assay
simultaneously determines the
GPS and presence of underlying genetic abnormalities that guide treatment
selection (see Example
1), thus generating substantially increased clinical utility in a single test.
Methodology
10 Study Design
GPS can be used with patients previously diagnosed with cancer in various
settings, including
without limitation as a confirmatory or quality control (QC) measure for every
case wherein
molecular profiling is performed. GPS may also be particularly useful in
guiding treatment of cases
having a diagnosis of cancer of unknown primary (CUP) or any cases having an
uncertain diagnosis.
15 From a database of cases that have profiled with the 592-gene NGS assay,
we selected 55,780 cases
with a pathology report available. This study was performed with 1RB approval.
This data set was
split into three cohorts: 34,352 cases with an unambiguous diagnosis; 15,473
cases with an
unambiguous diagnosis reserved as an independent validation set; and 1,662 CUP
cases. All cases
were de-identified prior to analysis.
20 The general study design 500 is shown in FIG. 5A. Starting with the
34,352 cases with an
unambiguous diagnosis, the machine learning algorithms were trained 501 using
27,439 samples at a
training cohort and 6,913 samples were used for validation. Once models were
trained and optimized,
the algorithm was locked 502. The 15,473 cases with an unambiguous diagnosis
were used as an
independent validation set 503. 1,662 CUP cases 504 were used to assess
classification and
25 prospective validation 505 was performed with over 10,000 clinical
cases.
592 NGS Panel
Next generation sequencing (NGS) was performed on genomic DNA isolated from
forrnalin-
fixed paraffm-embedded (FFPE) tumor samples using the NextSeq platform
(Illumina, Inc., San
Diego, CA). Matched normal tissue was not sequenced. A custom-designed
SureSelect XT assay was
30 used to enrich 592 whole-gene targets (Agilent Technologies, Santa
Clara, CA). The particular targets
are listed in Tables 123-125 above. All variants were detected with > 99%
confidence based on allele
frequency and amplicon coverage, with an average sequencing depth of coverage
of > 500 and an
analytic sensitivity of 5%. Prior to molecular testing, tumor enrichment was
achieved by harvesting
targeted tissue using manual microdissection techniques. Genetic variants
identified were interpreted
35 by hoard-certified molecular geneticists and categorized as
'pathogenic,' prasumed pathogenic,'
'variant of unknown significance,' presumed benign,' or 'benign,' according to
the American
College of Medical Genetics and Genomics (ACMG) standards. When assessing
mutation frequencies
254
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
of individual genes, 'pathogenic,' and 'presumed pathogenic' were counted as
mutations while
'benign', 'presumed benign' variants and 'variants of unknown significance'
were excluded.
Tumor Mutation Load (TML) was measured (592 genes and 1.4 megabases [MB]
sequenced
per tumor) by counting all non-synonymous missense mutations found per tumor
that had not been
5 previously described as germline alterations. The threshold to define TML-
high was greater than or
equal to 17 mutations/NIB and was established by comparing TML with MSI by
fragment analysis in
CRC cases, based on reports of TML having high concordance with MSI in CRC.
Microsatellite Instability (NISI) was examined using over 7,000 target
microsatellite loci and
compared to the reference genome hg19 from the University of California, Santa
Cruz (UCSC)
10 Genome Browser database. The number of microsatellite loci that were
altered by somatic insertion or
deletion was counted for each sample. Only insertions or deletions that
increased or decreased the
number of repeats were considered. Cienomic variants in the microsatellite
loci were detected using
the same depth and frequency criteria as used for mutation detection. MSI-NGS
results were
compared with results from over 2,000 matching clinical cases analyzed with
traditional PCR-based
15 methods. The threshold to determine MSI by NGS was determined to be 46
or more loci with
insertions or deletions to generate a sensitivity of > 95% and specificity of
> 99%.
Copy number alteration (CNA, also referred to as copy number variation or CNV
herein) was
tested using the NGS panel and was determined by comparing the depth of
sequencing of genomic
loci to a diploid control as well as the known performance of these genomic
loci. Calculated gains of
20 6 copies or greater were considered amplified.
For further description of the 592 NGS panel and MSI and TML calling, see
Example 1; and
International Patent Publication WO 2018/175501 Al, published September 27,
2018 and based on
Intl Patent Application PCT/US2018/023438 filed March 20, 2018, which is
incorporated by
reference herein in its entirety.
25 Machine Learning
The GPS system was built using an artificial intelligence platform leveraging
the framework
provided herein, which uses multiple models to vote against one another to
determine a fmal result.
See, e.g., FIGs. 1F-1G and accompanying text. A set of 115 distinct tumor site
and histology classes
were used to generate subpopulations of patients, stratified by primary
location (e.g., prostate) and
30 histology (e.g., adenocarcinoma), and combined as "disease type" or
"cancer type" (e.g., prostate
adenocarcinoma). The 115 disease/cancer types included: adrenal cortical
carcinoma; anus squamous
carcinoma; appendix adenocarcinoma, NOS; appendix mucinous adenocarcinoma;
bile duct, NOS,
cholangiocarcinoma; brain astrocytoma, anaplastic; brain astrocytoma, NOS;
breast adenocarcinoma,
NOS; breast carcinoma, NOS; breast infiltrating duct adenocarcinoma; breast
infiltrating lobular
35 carcinoma, NOS; breast metaplastic carcinoma, NOS; cervix
adenocarcinoma, NOS; cervix
carcinoma, NOS; cervix squamous carcinoma; colon adenocarcinoma, NOS; colon
carcinoma, NOS;
colon mucinous adenocarcinoma; conjunctiva malignant melanoma, NOS; duodenum
and ampulla
255
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
adenocarcinoma, NOS; endometrial adenocarcinoma, NOS; endometrial
carcinosarcoma; endometrial
endometrioid adenocarcinoma; endometrial serous carcinoma; endometrium
carcinoma, NOS;
endometriurn carcinoma, undifferentiated; endometriurn clear cell carcinoma;
esophagus
adenocarcinoma, NOS; esophagus carcinoma, NOS; esophagus squamous carcinoma;
extrahepatic
5 cholangio, common bile, gallbladder adenocarcinoma, NOS; fallopian tube
adenocarcinoma, NOS;
fallopian tube carcinoma, NOS; fallopian tube carcinosarcoma, NOS; fallopian
tube serous
carcinoma; gastric adenocarcinoma; gastroesophageal junction adenocarcinoma,
NOS; glioblastoma;
glioma, NOS; gliosarcoma; head, face or neck, NOS squamous carcinoma;
intrahepatic bile duct
cholangiocarcinoma; kidney carcinoma, NOS; kidney clear cell carcinoma; kidney
papillary renal cell
10 carcinoma; kidney renal cell carcinoma, NOS; larynx, NOS squamous
carcinoma; left colon
adenocarcinoma, NOS; left colon mucinous adenocarcinoma; liver hepatocellular
carcinoma, NOS;
lung adenocarcinoma, NOS; lung adenosquamous carcinoma; lung carcinoma, NOS;
lung mucinous
adenocarcinoma; lung neuroendocrine carcinoma, NOS; lung non-small cell
carcinoma; lung
sarcomatoid carcinoma; lung small cell carcinoma, NOS; lung squamous
carcinoma; meninges
15 meningioma, NOS; nasopharynx, NOS squamous carcinoma; oligodendroglioma,
anaplastic;
oligodendroglioma, NOS; ovary adcnocarcinoma, NOS; ovary carcinoma, NOS; ovary
carcinosarcoma; ovary clear cell carcinoma; ovary endometrioid adenocarcinoma;
ovary granulosa
cell tumor, NOS; ovary high-grade serous carcinoma; ovary low-grade serous
carcinoma; ovary
mucinous adenocarcinoma; ovary serous carcinoma; pancreas adenocarcinoma, NOS;
pancreas
20 carcinoma, NOS; pancreas mucinous adenocarcinoma; pancreas
neuroendocrine carcinoma, NOS;
parotid gland carcinoma, NOS; peritoneum adenocarcinoma, NOS; peritoneum
carcinoma, NOS;
peritoneum serous carcinoma; pleural mesothelioma, NOS; prostate
adenocarcinoma, NOS;
rectosigmoid adenocarcinoma, NOS; rectum adenocarcinoma, NOS; rectum mucinous
adenocarcinoma; retroperitoneum dedifferentiated liposarcoma; retroperitoneum
leiomyosarcoma,
25 NOS; right colon adenocarcinoma, NOS; right colon mucinous
adenocarcinoma; salivary gland
adenoid cystic carcinoma; skin melanoma; skin melanoma; skin merkel cell
carcinoma; skin nodular
melanoma; skin squamous carcinoma; skin trunk melanoma; small intestine
adenocarcinoma; small
intestine gastrointestinal stromal tumor, NOS; stomach gastrointestinal
stromal tumor, NOS; stomach
signet ring cell adenocarcinoma; thyroid carcinoma, anaplastic, NOS; thyroid
carcinoma, NOS;
30 thyroid papillary carcinoma of thyroid; tonsil, oropharynx, tongue
squamous carcinoma; transverse
colon adenocarcinoma, NOS; urothelial bladder adenocarcinoma, NOS; urothelial
bladder carcinoma,
NOS; urothelial bladder squamous carcinoma; urothelial carcinoma, NOS; uterine
endometrial
stromal sarcoma, NOS; uterus leiomyosarcoma, NOS; uterus sarcoma, NOS; uveal
melanoma;
vaginal squamous carcinoma; vulvar squamous carcinoma. Note that NOS, or "Not
Otherwise
35 Specified," is a subcategory in systems of disease/disorder
classification such as TCD-9, TCD-10, or
DSM-1V, and is generally but not exclusively used where a more specific
diagnosis was not made.
256
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
For training the GPS, all 115 disease types were trained against each other in
a pairwise
comparison approach using the training set to generate 6555 model signatures,
where each signature is
built to differentiate between a pair of disease types. The signatures were
generated using Gradient
Boosted Forests and applied a voting module approach as described herein.
5 The models were validated using the test cases. Each test case was
processed individually
through all 6555 signatures, thereby providing a pairvvise analysis between
every disease type for
every case. The results are analyzed in a 115 x 115 matrix where each column
and each row is a
single disease type and the cell at the intersection is the probability that a
case is one disease type or
the other. The probabilities for each disease type are summed for each column
which results in 115
10 disease types with their probability sums. These disease types are
ranked by their probability sums.
The disease types were then used to determine a fmal probability for each case
belonging to a
superset of 15 distinct organ groups, which include the following: Colon;
Liver, Gall Bladder, Ducts;
Brain; Breast; Female Genital Tract and Peritoneum (FGTP); Esophagus; Stomach;
Head, Face or
Neck, not otherwise specified (NOS); Kidney; Lung; Pancreas; Prostate;
Skin/Melanoma; and
15 Bladder. For each case, each of these organs can be assigned a
probability which will be used to make
the primary origin prediction(s). Tables 2-116 above list selections of
features that contribute to the
disease type predictions, where each row in the table represents a feature
ranked by Importance. As
noted, the titles of Tables 2-116 indicate how the 115 disease types relate to
the 15 organ groups, as
the tables are titled in the format "disease type ¨ organ group." As an
example, the title heading of
20 Table 2 is "Adrenal Cortical Carcinoma ¨ Adrenal Gland," indicating that
the disease type is adrenal
cortical carcinoma, which is placed within the organ group is adrenal gland.
FIG. 5B shows an example 115x115 matrix generated for a test ease of prostate
origin (i.e.,
Primary Site: Prostate Gland; Histology: Adenocarcinoma). In the figure, the X
and Y legends are the
115 disease types listed above. Each row is the probability of a "negative"
call (probability < 0.5) and
25 each column is the probability of a positive call, as noted above. The
shaded squares in the matrix
represent probability scores 0.98. The arrow indicates disease type "prostate
adenocarcinoma." The
probability sum for this case for prostate was 114.3 out of a possible 115.
Further details can be found in Abraham J., et al. Genomic Profiling
Similarity, Intl Patent
Publication W02020146554, which publication is herein incorporated by
reference in its entirety.
30 Results
Retrospective Validation
Using the machine learning approach, a probability was assigned to each case
that the case
was from one of the 15 distinct organ groups. The probability may be referred
to as the GPS Score. Of
the 15,473 cases with an unambiguous diagnosis used as an independent
validation set (see FIG. 5A
35 503), 6229 cases that had a GPS Score of >0.95. Of those, 98_4% were
concordant with the case-
assigned result. The 98.1% concordance exceeded our acceptance criteria for
validating the GPS
Scores >0.95. This criteria was greater than 95% accuracy when presenting a
score >0.95. The CPS
257
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Score had extremely high performance when assigning scores of 0 to organ
groups (i.e., probability of
the tumor sample being from that organ group is determined by GPS as zero).
The percentage of the
time that a tumor type that does not match the case was given a zero GPS Score
(12270/12279) was
99.92%.
5 FIG. SC shows the Scores for the 6229 cases with GPS Scores > 0.95
plotted against the
probability of match for each sample. The resulting correlation coefficient of
0.990 indicates GPS
Score is highly correlated to accuracy.
Analytical sensitivity of the GPS Score was determined by evaluating
performance relative to
two distinct parameters: (1) tumor percentage, and (2) average read depth per
sample. To evaluate
10 tumor percentage, accuracy of the GPS relative to the ease-assigned
organ type was determined. FIG.
5D shows a correlation chart for the data grouped into ranges of 20-49%, 50-
80% and >80% tumor
content The figure indicates that the UPS Score is insensitive to tumor
percentage. FIG. SE shows a
correlation chart for the data used to evaluate read depth. The accuracy of
the GPS Score relative to
the case-assigned organ type was determined with classification of read depths
between 300-500X
15 and >500X. As with tumor percentage, the figure indicates that the GPS
Score was insensitive to read
depth. In both cases, the correlation coefficient according to Pearson's r
remained greater than 98%
for each data grouping.
We also found that the GPS Score was robust to metastasis. Table 131 shows
performance
metrics on subsets of the test data from a primary site (N - 8,437),
metastatic site (6,690), and
20 samples with low (9,492) and high tumor percentages (5,945).
Table 131 - Performance metrics of assay with noted characteristics
Sensitivity Specificity PPV NPV Accuracy Call Rate
Primary 90.9% 98.0% 91.1% 98.9% 97.6%
97.3%
Metastatic 89.0% 97.9% 89.3% 98.2% 96.9%
97.6%
20-50% 90.3% 98.2% 90.6% 98.5% 97.5%
97.1%
Tumor
>50% 90.3% 98.2% 90.6% 98.5% 97.5%
97.1%
Tumor
The performance held across multiple tumor types. Table 132 shows performance
metrics
and cohort sizes of subsets of the independent test dataset where the primary
tumor site was known.
25 FGTP represents female genital tract and peritoneum.
Table 132 - Performance metrics of assay across tumor types
Tumor Type Train Test Sensitivity Specificity PPV NPV
Accuracy Call
N N Rate
Head, Face, Neck 299 144 45.4% 100.0% 96.4% 99.6% 99.6%
82.6%
258
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Melanoma 976 402
85.0% 99.9% 94.3% 99.6% 99.5% 96.3%
FGTP 8,872 4,115 93.4% 98.3% 95.4%
97.6% 97.0% 98.8%
Prostate 785 477
96.1% 99.8% 94.7% 99.9% 99.7% 96.6%
Brain 1,554 479 93.3% 99.8% 93.5%
99.8% 99.6% 96.0%
Colon 5,805 2,532 94.5% 98.5% 92.9%
98.9% 97.9% 98.9%
Kidney 426 178
84.1% 99.9% 91.7% 99.8% 99.8% 88.2%
Bladder 447 304
60.6% 99.9% 89.4% 99.3% 99.1% 91.8%
Breast 3,324 1,386 90.9% 98.7% 87.9%
99.1% 98.0% 98.3%
Lung 7,744 3,540 96.0% 95.4% 86.3%
98.7% 95.5% 98.2%
Pancreas 1,637 708 83.7% 99.3% 84.6%
99.2% 98.5% 98.3%
Gastroesophageal 1,521 743 72.0% 99.3% 82.6% 98.6%
98.0% 93.8%
Liver, 734 364
57.7% 99.7% 82.2% 99.0% 98.8% 92.6%
Gallbladder,
Ducts
The GPS Score had extremely high performance when assigning scores of 0 to
organ groups
(i.e., probability of the tumor sample being from that organ group is
determined by GPS as less than
0.001). Of the 15,473 validation cases evaluated, 12,279 had a GPS Score of 0
for one or more organ
5 types. The percentage of the time that a tumor type that did not match
the case was given a zero GPS
Score (12270/12279) was 99.92 A,, which exceeded our acceptance criteria for
validating the CB'S
Zero% scores. The criteria was greater than 99.9% accuracy when presenting a
score of 0. Thus, the
zero score was highly accurate. There were only nine cases that had a GPS
Score of 0 for the case-
assigned organ result case.
10 Table 133 shows performance metrics of the GPS algorithm on the
independent test set of
15,473 cases as compared to other methods currently available. In the table
and those below,
"Sensitivity" is the probability of getting a positive test result for tumors
with the tumor type and
therefore relates to the potential of GPS to recognize the tumor type;
"Specificity" is the probability of
a negative result in a subject without the tumor type and therefore relates to
the GPS' ability to
15 recognize subjects without the tumor type, i.e. to exclude the tumor
type; Positive Predictive Value
("PPV") is the probability of having the tumor type of interest in a subject
with positive result for that
tumor type, and therefore PPV represents a proportion of patients with
positive test result in total of
subjects with positive result; NPV is the probability of not having the tumor
type in a subject with a
negative test result, and therefore provides a proportion of subjects without
the tumor type with a
20 negative test result in total of subjects with negative test results;
Accuracy represents the proportion of
true positives and true negatives in the text population; and Call Rate is the
proportion of samples for
which GPS is able to provide a prediction.
259
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Table 133 ¨ Performance of GPS on Validation Set
Assay Overall PPV NPV Sensitivity
Specificity / Call
Accuracy / PPA NPA Rate
MDC/GPS 98.4% 90.5% 99.2% 90.5% 99.2% 97.5% 15,473
Cancer 94.1%19 NR NR 88.5% 17 99.1% 17
89% 18 46219
Genetics 3618
"[issue of
Origin
CancerTYPE NR 83% 99% 83% 99% 78% 187
11)2
Gamble AR, NR NR NR 64% NR 100% 90
199319
Brown, RW, NR NR NR 66% NR 87% 128
199729
Dennis, JL, NR NR NR 67% NR 100% 452
200521
Park SY, NR NR NR 65% NR 78 A1 374
2007'
Prospective Validation
A target of 10,000 prospective samples were evaluated by the GPS Score
platform based on
clinical samples incoming for molecular profiling using the 592 NGS gene
panel. The GPS Score for
an organ group was >0.95 for 2857 cases. Of those, 54 cases had a GPS Score
which differed from the
organ group listed on the incoming case (i.e., as listed by the ordering
physician) and were flagged for
further pathological review. Pathologists reviewed those 54 cases, plus an
additional 12 cases with
GPS scores <0.95 and requested by the pathologist for various reasons (Score
close to 0.95,
suspicious IDC findings, etc). There was a 43.9% (29/66) response from
pathology review that the
results obtained via the GPS system were considered "reasonable." The
pathology review resulted in
changes to the tumor type from what was originally reported from the ordering
physician for 11 cases.
The results of this evaluation exceeded our acceptance criteria for validating
the capability of the GPS
Score to provide evidence to support a new diagnosis. This acceptance criteria
was whether
pathologists consider the information reasonable in greater than 25% of the
cases and the information
results in any change in diagnosis that may affect patient treatment. In these
cases, a change in tumor
origin may affect such treatment. Thus, automated flagging of discordant tumor
type by GPS may
positively influence the course of treatment of a substantial number of
patients.
Analysis of CLIP
260
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Validation of a CUP assay at the individual patient level is a fundamentally
difficult as the
"truth" may be unknown. however, population based methods can be used to gain
greater insight into
the performance of the GPS classifier and generally validate its performance.
To accomplish this, we
compared the frequency of mutations across known patient populations to the
frequency in the
predicted group. For example, the frequency of BRAF mutations in colon cancer
in the known patient
cohort is 10.3% and is 4.8% in all non-colon cancer patients. The frequency of
BRAF in the CUP
cases that the classifier called colon is 10.3% and is 4.9% in the CUP cases
the classifier called as
non-colon. In this way we can show that the population of CUP cases that are
classified as a specific
cancer type matches the population of each specific tumor type. A subset of
markers we used in this
manner are shown in Table 134, demonstrating the similarities of the GPS
predicted CUP populations
to the actual populations. The data for correlation of between the frequencies
for the predicted CUP
cases and the training set show that the predicted populations most closely
resemble the actual
population with the exception of brain cancer, which, without being bound by
theory, may be due to
small sample size, with only 17 CUP cases predicted to be brain. These data
together show that the
GPS can classify CUP at the population level into classes consistent with
other molecular
characteristics of the tumors.
Table 134- Frequencies of variants detected or observed medians among
notable biomarkers per tumor type
Of This Tumor Type Not Of This Tumor Type
Marker Tumor Type Train + Test* CUP** Train
+ Test* Cur*
BRAF Colon 10.3% 10.3% 4.8% 4.9%
BRAF Lung 6.2% 6.3% 5.6% 5.7%
BRAF Melanoma 39.1% 38.4% 4.8% 4.9%
BRCA1 Breast 7.0% 7.1% 6.4% 6.4%
BRCA1 FGTP 8.6% 8.6% 5.7% 5.8%
BRCA1 Melanoma 9.9% 10.3% 6.4% 6.4%
BRCA1 Prostate 4.1% 4.2% 6.5% 6.5%
cKIT Gastroesophageal 5.8% 5.5% 3.4%
3.4%
cKIT Lung 4.3% 4.3% 3.3% 3.3%
EGFR Brain 17.6% 17.2% 6.5% 6.5%
EGFR Lung 16.1% 15.4% 4.3% 4.4%
KRAS Colon 50.0% 49.1% 16.4% 16.6%
K_RAS Lung 26.4% 26.1% 20.8% 20.7%
K_RAS Pancreas 84.2% 83.3% 19.0% 18.8%
PIK3CA Breast 31.5% 31.1% 13.5% 13.5%
PIK3CA FGTP 21.3% 21.1% 13.1% 13.0%
261
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
PIK3CA Lung 6.3% 6.6% 17.8% 17.7%
TP53 Head and Neck 45.4% 45.4% 61.8% 61.1%
TP53 Melanoma 28.2% 29.9% 62.6% 61.9%
* Represents the observed value among the known tumor type of the combined
training and testing
datasets_
¨ Represents the observed value among CUP cases predicted to be of the tumor
type in each row.
5 Cancer of unknown primary remains a substantial problem for both
clinicians and patients,
diagnosis can be aided with the GPS algorithms provided herein. The tumor type
predictors can
render a histologic diagnosis to CUP cases that can inform treatment and
potentially improve
outcomes. Our NGS analysis of tumors (see Example 1) and GPS provided here
return both
diagnostic and therapeutic information that optimize patient treatment
strategy from a single test. This
10 method provides a
substantial improvement over the current standard of multiple tests that
require
more tissue.
References (as indicated by superscripted numbers in the text of the Example)
1. Haskell CM, et al. Metastasis of unknown origin. Curr Probl Cancer. 1988
Jan-
Feb;12(1):5-58. Review. PubMed PMID: 3067982.
15 2. Erlander MG, et al.
Performance and clinical evaluation of the 92-gene real-time PCR
assay for tumor classification. J Mol Diagn. 2011 Sep;13(5):493-503. doi:
10.1016/j.jmoldx.2011.04.004. Epub 2011 Jun 25.
3. Varadhachary. New Strategies for Carcinoma of Unknown Primary: the role of
tissue of
origin molecular profiling. Clin Cancer Res. 2013 Aug 1;19(15):4027-33. DOI:
10.1158/1078-
20 0432.CCR-12-3030
4. Brown RW, et al. Immunohistochemical identification of tumor markers in
metastatic
adenocarcinoma: a diagnostic adjunct in the determination of primary site. Am
J Clin Pathol 1997,
107:12e19
5. Dennis JL, et al. Markers of adenocarcinoma characteristic of the site of
origin:
25 development of a diagnostic algorithm. Chin Cancer Res 2005,
11:3766e3772
6. Gamble AR, et al. Use of tumour marker immunoreactivity to identity primary
site of
metastatic cancer. IIMJ 1993, 306:295e298
7. Park SY, et al. Panels of immunohistochemical markers help determine
primary sites of
metastatic adenocarcinoma. Arch Pathol Lab Med 2007, 131:1561c1567
30 8. DeYoung BR, Wick MR_ Inununohistologic evaluation of metastatic
carcinomas of
unknown origin: an algorithmic approach. Semin Diagn Pathol 2000, 17:184e193
262
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
9. Anderson GG, Weiss LM. Determining tissue of origin for metastatic cancers:
meta-
analysis and literature review of immunohistochemistry performance. Appl
Lininunohistochem Mol
Morphol 2010, 18:3e8
10. Erlander MG, etal. Performance and clinical evaluation of the 92-gene real-
time PCR
5 assay for tumor classification. J Mol Diagn 2011, 13:493e503
11. Pillai R, et al. Validation and reproducibility of a microarray-based gene
expression test
for tumor identification in forrnalin-fixed, paraffin-embedded specimens. J
Mol Diagn 2011, 13:48e56
12. Rosenwald S. et al. Validation of a microRNA-based qRT-PCR test for
accurate
identification of tumor tissue origin. Mod Pathol 2010, 23:814e823
10 13. Kerr SE, et al. Multisite validation study to determine performance
characteristics of a 92-
gene molecular cancer classifier. Clin Cancer Res 2012, 18:3952e3960
14. Kucab .1E, et al. A Compendium of Mutational Signatures of Environmental
Agents. Cell.
2019 May 2;177(4):821-836.e16. doi: 10.1016/j.ce11.2019.03.001. Epub 2019 Apr
11. PubMed PMID:
30982602; PubMed Central PMCID: PMC6506336.
15 15. Hainsworth JD, et al, Molecular gene expression profiling to predict
the tissue of origin
and direct site-specific therapy in patients with carcinoma of unknown primary
site: a prospective trial
of the Sarah Cannon research institute. J Chin Oncol. 2013 Jan 10;31(2):217-
23. doi:
10.12003C0.2012.43.3755. Epub 2012 Oct 1.
16. Ross JS, et al. Comprehensive Genomie Profiling of Carcinoma of Unlmown
Primary Site
20 New Routes to Targeted Therapies. JAMA Oncol. 2015;1(1):40-49.
doi:10.1001/jamaonco1.2014.216
Example 3: Machine learning analysis using genomic and transcriptomic profiles
to
accurately predict tumor attributes
This disclosure provides a machine learning based classifiers to predict the
origin of a tumor
25 sample, or TOO (tissue-of-origin), and related attributes based on
analysis of genomic DNA (see, e.g.,
Example 2) and based on analysis of transcriptome analysis. See, e.g., FIG.
4A, Tables 117-120, and
accompanying description. As noted herein, DNA and RNA each have advantages
and disadvantages
as biological analytes. Without being bound by theory, we hypothesized that a
combination of
genomic DNA analysis with RNA transeriptome analysis may provide optimal
results. Advanced
30 machine learning analysis may take advantage of the strengths of each
analyte while curtailing the
weaknesses. We term this combined classifier a "panomic" predictor. This
Example details this
panomic classifier, which may be referred to as "MT GPSai" in this Example.
Cancer of Unknown Primary (CUP) occurs in 3-5% of patients when standard
histological
diagnostic tests are unable to determine the origin of metastatic cancer.
Typically, a CUP diagnosis is
35 treated empirically and has poor outcome, with median overall survival
less than one year. Gene
expression profiling alone has been used to identify the tissue of origin
(TOO) but struggles with low
neoplastic percentage in metastatic sites which is where identification is
often most needed. This
263
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Example provides a "Genomic Prevalence Score," or "GPS," which uses DNA
sequencing and whole
transcriptome data coupled with machine learning to aid in the diagnosis of
cancer. The system
implementing the GPS, termed "MI GPSai," was trained on genomic data from
34,352 cases and
genomic and transcriptomic data from 23,137 cases and was validated on 19,555
cases. MI GPSai
5 predicted the tumor type in the labeled data set with an accuracy of over
94% on 93% of cases while
deliberating amongst 21 possible categories of cancer: breast adenocarcinoma,
central nervous
system cancer, cervical adenocarcinoma, cholangiocarcinoma, colon
adenocarcinoma,
gastroesophageal adenocarcinoma, gastrointestinal stromal tumor (GIST),
hepatocellular carcinoma,
lung adenocarcinoma, melanoma, meningioma, ovarian granulosa cell tumor,
ovarian & fallopian tube
10 adenocarcinoma, pancreas adenocarcinoma, prostate adenocarcinoma, renal
cell carcinoma, squamous
cell carcinoma, thyroid cancer, tifothelial carcinoma, uterine endometrial
adenocarcinoma, and uterine
sarcoma. When also considering the second highest prediction, the accuracy
increased to 97%.
Additionally, MI GPSai rendered a prediction for 71.7% of CUP cases.
Pathologist evaluation of
discrepancies between submitted diagnosis and MI GPSai predictions resulted in
change of diagnosis
15 in 41.3% of the time. MI GPSai provides clinically meaningful
information in a large proportion of
CUP cases and inclusion of MI GPSai in clinical routine could improve
diagnostic fidelity. Moreover,
all genomic markers essential for therapy selection are assessed in this
assay, maximizing the clinical
utility for patients within a single test.
Introduction
20 Carcinoma of Unknown Primary (CUP) represents a clinically challenging
heterogeneous
group of metastatic malignancies in which a primary tumor remains elusive
despite extensive clinical
and pathologic evaluation. CUPs comprise approximately 3-5% of cancer
diagnoses worldwide [1]
and efforts to secure a definitive diagnosis can prolong the diagnostic
process and delay treatment
initiation. Furthermore, CUP is associated with poor outcome which may be at
least partially
25 explained by use of suboptimal therapeutic interventions since there is
general agreement that CUP
tumors retain the biologic properties of the putative primary malignancy [1],
[2].
lmrnunohistochemical (11-IC) testing has long been the gold standard method to
diagnose the site of
tumor origin, especially in cases of poorly-differentiated or undifferentiated
tumors. Meta-analysis of
studies assessing the accuracy of 111-IC in challenging cases reported an
accuracy of 60-70% in the
30 characterization of metastatic tumors [3], [4], [5], [6]. Since
therapeutic regimens may depend upon
diagnosis, there is a need for improved diagnosis of CUP. To address these
challenges, assays aiming
at tissue-of-origin (TOO) identification based on assessment of differential
gene expression have been
developed and tested clinically. However, integration of such assays into
clinical practice is hampered
by relatively poor performance characteristics, e.g., low accuracy such as <
90% combined with high
35 call rate such as 100% or higher accuracy such as <-90% combined with
low call rate such as <90%,
and limited sample availability. See Table 135. Nevertheless, initial clinical
studies demonstrate
possible benefit of matching treatments to tumor types predicted by the assay
[8]. With increasing
264
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
availability of comprehensive molecular profiling assays, particularly next-
generation DNA
sequencing, genomic features have been incorporated in CUP treatment
strategies [9]. Although this
approach has not been a panacea for unambiguous identification of the TOO, it
has revealed
targetable molecular alterations in some patients [9].
Table 135. Landscape of tissue of origin approaches
Assay Cancer N Independent Accuracy (%)
Cases Called
Categories Test Set (%)
MI GPSai 21 13,661 94.7 93
PCAWG 2020 14 1436 88 100
[32]
MSK IMPACT 22 11,644 74.1 100
2019 [10]
Cancer Genetics 9 27 94.1 89
"[issue of Origin
2012 [111
Biotheranostics 30 187 83 100
CancerTYPE ID
2011 [7]
Park SY 2007 [5] 7 60 75 78
Dermis JL 2005 7 130 88 100
[12]
Brown RW 1997 5 128 66 86
[6]
Gamble AR 1993 14 100 70 100
[13]
As described above and further detailed in this Example, we used a machine-
learning
approach to build TOO classifiers based on data from a large next-generation
DNA sequencing panel
in conjunction with data from whole transcriptome sequencing, which are both
used broadly for
routine molecular tumor profiling. See, e.g., Example 1. This panomic
computational classification
system identified TOO at an accuracy significantly exceeding that of other
currently available
technologies. See Table 135. Moreover, this assay simultaneously determines
the presence of genetic
abnormalities that guide treatment selection, thus generating substantial
clinical utility in a single test.
Methods
Next-Generation Sequencing NGS) - DNA
265
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Genomic DNA was isolated from formalin-fixed paraffm-embedded (FFPE) tumor
samples
which were microdissected to enrich tumor purity. FFPE specimens underwent
pathology review to
measure percent tumor content and tumor size; a minimum of 20% of tumor
content in the area for
microdissection was set as a threshold to enable enrichment and extraction of
tumor-specific DNA.
5 Matched normal tissue was not routinely sequenced. A custom-designed
SureSelect XT assay was
used to enrich 592 or whole exome whole-gene targets (Agilent Technologies,
Santa Clara, CA). See
Example 1 for further details. Enriched DNA was subjected to NGS using the
NextSeq platform
(Illumina, Inc., San Diego, CA). All variants were detected with > 99%
confidence based on allele
frequency and probe panel coverage, with an average sequencing depth of
coverage of > 500 and an
10 analytic sensitivity of 5%. Genetic variants identified were interpreted
by board-certified molecular
geneticists and categorized as 'pathogenic,' presumed pathogenic,' yariant of
unknown
significance,' presumed benign,' or 'benign,' according to the American
College of Medical Genetics
and Genomics (ACMG) standards. When assessing mutation frequencies of
individual genes,
'pathogenic,' presumed pathogenic,' and 'variants of unknown significance'
were counted as
15 mutations while 'benign' and 'presumed benign' variants were excluded.
Copy number alteration
(CNA; also commonly referred to as copy number variation (CNV) herein) was
simultaneously
determined by NGS by comparing the depth of sequencing of genomic loci to a
diploid control as well
as the known performance of the genomic loci. Calculated gains of 6 copies or
greater were
considered amplified.
20 Next-Generation Sequencing (NGS) - RNA
171713E specimens were microdissected as described above prior to enrichment
and extraction
of tumor-specific RNA. Qiagcn RNA FFPE tissue extraction kit was used for
extraction (Qiagcn
LLC, Germantown, MD), and the RNA quality and quantity were determined using
the Agilent
TapeStation. Biotinylated RNA baits were hybridized to the synthesized and
purified cllNA targets
25 and the bait-target complexes were amplified in a post capture PCR
reaction. The Illumina NovaSeq
6500 was used to sequence the whole transcriptome from patients to an average
of 60 M reads. Raw
data was demultiplexed by Illumma Dragen13iolf accelerator, trimmed, counted,
PCR-duplicates
removed and aligned to human reference genome hg19 by STAR aligner [141 For
transcription
counting, transcripts per million molecules was generated using the Salmon
expression pipeline [15].
30 RNA expression
RNA expression, as defmed by transcripts per million (TPM) from the Salmon RNA
expression pipeline [15] using our whole transcriptome sequencing assay (WTS;
see Example 1), was
validated using 1HC results from over 5000 human breast adenocarcinoma cases.
Protein amounts
were measured by FDA-approved antibodies using standard quantitative 1HC
assays. 11-IC scores
35 come directly from histopathology review by board-certified pathologists
for ER/ESR1 (human
estrogen receptor), PR/PGR (human progesterone receptor), AR (human androgen
receptor), and
I1ER2/neu/ERBB2 (human Herceptin, receptor tyrosine kinase CD340). 501IIC
'positive' and 50 [(IC
266
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
'negative' cases were used to decide the TPM thresholds corresponding to IHC
positive and H-IC
negative for these 4 genes. The thresholds were evaluated on 5197 independent
cases and all four
markers had a sensitivity > 86% with specificities ranging from 85% to 99%.
Validation results are
shown in Table 136 and FIGs. 6A-D, which show ROC curves for calculating ILIC
result from WTS
5 expression for the indicated biomarkers.
Table 136- Results of independent validation of IIIC result derivation from
WTS expression data
Category N Sensitivity Specificity PPV
NPV Accuracy
ER (FIG. 6A) 5098 93.5% 90.7% 94.6% 88.8%
92.5%
PR (FIG. 6B) 5024 86.3% 85.1% 79.6% 90.3%
85.6%
HER2 (FIG. 6C) 5197 91.0% 99.7% 97.8% 98.6%
98.5%
AR (FIG. 6D) 5142 88.5% 88_5% 94.4% 77.9%
88.5%
Additionally, we compared data between our WTS expression assay to the
Illumina DASL
10 Expression Microarray and publicly available Affymetrix U133A expression
arrays from the expO
project (Gene Expression Omnibus accession GSE2109) in a cross-platform
comparison method [33].
We selected 10 eases from each dataset from a diagnosed Stage IV uterine
carcinoma and 10 cases
diagnosed with Stage IV colon adenocareinoma. We identified 14,473 genes which
are common
across these three platforms. Although these cases are from different people,
without being bound by
15 theory, we hypothesized
that the gene expression profiles from uterine tumors and colon tumors are
sufficiently different from each other and sufficiently common within a tumor
type that common
patterns of over- and under-expression would be detectable. To visualize this,
we took the 1og2 ratio
of the 14,473 genes between uterine (numerator) and colon (denominator) cancer
and plotted the
ratios. FIGs. 6E-G show the ratios plotted against each other with R2 listed
in FIGs. 6E (WTS (X
20 axis) and Illumina (Y
axis)), 9F (Illumina (X axis) and Affymetrix (Y axis)) and 9G (WTS (X axis)
and Affymetrix (Y axis)). Note that the expression data was averaged across 10
patients. The
Pearson's correlation coefficient for each is 0.68, 0.75 and 0.73
respectively.
Results
Patients
25 To identify patients
for this Example, we used a database of over 200,000 samples analyzed
from 2008 to 2020 as described in Example 1. We identified 77,044 cases that
had next-generation
DNA and RNA sequencing results with an available pathology diagnosis including
CUP. CUP cases
were defined as those assigned a primary tumor site of "Unknown primary site"
and for which the
"Cancer of Unknown Primary" lineage was selected by the submitting site. The
submitted
30 pathological diagnosis was used as the training label. Subsequent
independent validation of the
classifier was accomplished by including 13,661 cases with a known primary and
1,107 CUP cases
that were analyzed prospectively as part of routine tumor profiling. See FIG.
6H, which shows a
267
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
CONSORT diagram 600 (www.consort-statement.org/consort-statement/flow-
diagram). The DNA
and RNA components of MI GPSai were trained 603 using a combined 57,489
patients (601 + 602),
which were then locked 604 and validated 011 4,602 non-CUP 605 and 185 CUP
patients 606 to
determine optimal performance settings. Following this evaluation, MI GPSai
rendered a prediction
5 on routinely profiled cases resulting in the final prospective validation
set 608 and CUP cases 609.
Artificial intelligence training
Molecular profiles from 57,489 patients were used for initial training of the
global tumor
classification algorithm designated MI GPSai. This panomic dataset was
comprised of 34,352 cases
with genomic data (FIG. 6H 601) and 23,137 with both genomic and
transcriptomic data (FIG. 611
10 602). MI GPSai was generated using an artificial intelligence platform
that leverages the
"Deliberation Analytics" (DEAN) framework as described herein. DEAN uses
biomarker data as
feature inputs into an ensemble of over 300 well-established machine learning
algorithms, including
random forest, support vector machine, logistic regression, K-nearest
neighbor, artificial neural
network, naïve Bayes, quadratic discriminant analysis, and Gaussian processes
models. Multiple
15 feature selection methods were employed to build models along with 5-
fold cross validation during
training to assess performance. High-performing models deliberate against one
another to determine a
fmal result. For DNA, a set of 115 distinct primary tumor site and histology
classes were defmed and
used to generate subpopulations of patients. For training the GPS, all 115
disease types were trained
against each other using the training set to generate 6,555 model signatures,
where each signature is
20 built to differentiate between a pair of disease types. The signatures
were generated using Gradient
Boosted Forests. The models were validated using the test cases where each
test case was processed
individually through all 6,555 signatures, thereby providing a pairwise
analysis between every disease
type for every case. The results are analyzed in a 115 x 115 matrix where each
column and each row
is a single disease type and the cell at the intersection is the probability
that a case is one disease type
25 or the other. The probabilities for each disease type are summed for
each column which results in 115
disease types with their probability sums. These disease types are ranked by
their probability sums.
See Example 2 and Tables 2-116 and related discussion for details. For RNA,
gradient boosted
forests were trained using a selection of RNA transcripts to separately
determine a cancer type, organ
group and histology. See FIGs. 4A-B, and Tables 117-120 and related discussion
for additional
30 details.
The scheme set forth in FIG. 4B was used to obtain a final prediction. The 115
x 115 matrix
described above is used as an intermediate model to assess DNA 416 and the
gradient boosted forests
were applied to the transcripts in Table 117 to build intermediate models to
assess cancer type 412,
organ group 413 and histology 414. A gradient boosted forest was applied to
the outputs of the
35 intermediate models to dynamically combine the results 415. Using this
approach, a total of 6,559
models were generated and used to determine a final probability (termed a MI
GPS Score) for each
case belonging to each of the fmal desired cancer categories. These MI GPS
Scores were then
268
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
clustered into multidimensional signatures which were empirically evaluated in
our molecular
profiling database to determine the predicted prevalence in each cancer
category. The prevalence is
the final output of the MI GPSai machine learning platform 417. The desired
cancer categories
comprised 21 broad cancer categories selected in order to achieve the highest
predictive power for a
5 clinically relevant category that would assist with therapy selection in
challenging cases. These 21
cancer categories include breast adenocarcinoma; central nervous system
cancer; cervical
adenocarcinoma; cholangiocarcinoma; colon adenocarcinoma; gastroesophageal
adenocarcinoma;
gastrointestinal stromal tumor (GIST): hepatocellular carcinoma; lung
adenocarcinoma; melanoma;
meningioma; ovarian granulosa cell tumor; ovarian, fallopian tube
adenocarcinoma; pancreas
10 adenocarcinoma; prostate adenocarcinoma; renal cell carcinoma; squamous
cell carcinoma; thyroid
cancer; urothelial carcinoma; uterine endometrial adenocarcinoma; and uterine
sarcoma.
The top DNA and RNA features that contribute the largest amount of information
to the
predictions nrtdo for each of the 21 cancer categories are shown in FIGs. 6I-
6AC. In each figure, the
leftmost biomarkers are the top contributors based on DNA analysis whereas the
10 rightmost
15 biomarkers are the top contributors based on RNA analysis. In some
cases, e.g., GATA3 in breast
carcinoma in FIG. 61, the same gene was identified as a top contributor by
both DNA and RNA.
Without being bound by theory, much of the DNA results are copy number
alterations (see, e.g,
Tables 2-116), and copy number may have a direct impact on transcript levels.
Without being bound by theory, several observations can be made regarding the
biomarkers
20 in F1Gs. 6I-6AC. For example, various canonical driver mutations are
found among the top
contributing biomarkers. Examples include IDII1 and EGER for gliomas,
cKIT/PDGERA in
gastrointestinal stromal tumors (GIST), BRAF/NRAS in melanoma, KRAS/CDKN2A in
pancreatic
cancer, GATA3 and CDH1 in breast cancer, WIT in renal cell carcinoma, BRAF in
thyroid, PTEN in
endometrial cancer, and FOXL2 in ovarian granulosa cell tumors [16], [17],
[18], [19], pot [21].
25 Expression of genes relatively specific to tissue lineage are also among
the top contributors, e.g.,
CDX2 in gastroesophageal cancer, KIT in GIST. MITF in melanoma and NKX3-1 in
prostate cancer
[22], [23], [24], [25]. Without being bound by theory, markers in the figures
were most useful for
differentiating TOO are found in these lists, canonical cancer markers such as
BRCA1 are not in the
top 10 for the machine learning as they may be found in a number of cancer
categories. Additional
30 biomarkers that have not been explicitly associated with the particular
cancer types are also included
in the algorithm, revealing previously uncovered linkages with biomarkers and
pathways. Additional
details of the machine learning configurations and inputs are described here
[26].
Validation of algorithmic disease classification in independent cohorts
Following the lock of the algorithm (FIG. 611 604), predictions made by the MI
GPSai
35 platform were first validated in an independent set of 4,602 patients
with known cancer category
(FIG. 611 605) and 185 patients with CUP (FIG. 6H 606). MI GPSai provided a
top prediction for
each case along with a score related to the confidence in the call. When
evaluating the MI GPSai top
269
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
prediction on every case in the cohort irrespective of the score, the top
prediction was concordant with
the pathologist-assigned disease type in 90.3% of cases. An assessment of the
scores in this dataset
led us to select a threshold of 0.835 as a minimum score to report a result as
it was the intersection of
accuracy of the top prediction and the call rate (percentage of cases
resulted), resulting in 93.3%
5 accuracy on 93.3% of cases with a defined primary and 75.6% of CUP cases.
See FIG. 6AD, which
shows selection of this threshold in the independent validation set. The x-
axis represents all cases with
that MI GPSai Score and greater. In the non-CUP cases (N=4,602), the predictor
demonstrates a
93.3% sensitivity on 93.3% of cases at the selected threshold of 0.835,
annotated as the upper asterisk.
In the CUP cases (N=185), 75.6% of cases exceeded the selected threshold,
annotated as the lower
10 asterisk. At this threshold, the assay was robust within both primary
and metastatic tumors as well as
various ranges of tumor purity. See, e.g., Table 137.
Table 137. Summary of performance in the independent validation cohort at the
selected
threshold
Category n Call Rate (%) Sensitivity
(%)
Global 4602 93.3 93.3
Primary Specimen 2544 94 94.1
Metastatic Specimen 1969 92.2 92.5
Percent Tumor >= 20, 2885 92.7 93.4
<= 50
Percent Tumor > 50, 1657 94.1 93_1
<=80
Percent Tumor > 80 54 100 100
15 Prospective validation
Subsequently, the assay was used in clinical testing to prospectively evaluate
the tumor of
each patient with molecular profiling performed (FIG. 611 607). Pathologists
were notified of the MI
GPSai score and empirical prevalence tables if the assay returned a MI GPSai
Score of >= 0.835 for
any cancer category. The tumors of 13,661 non-CUP patients were evaluated by
the algorithm as a
20 prospective validation cohort. See Table 138, wherein sensitivity is
abbreviated as "Sens." Globally,
this cohort exhibited a similar call rate compared to the initial independent
validation cohort (93.0%
vs 93.3%) and exhibited a higher sensitivity (94.7% vs 93.3%). The sensitivity
of the assay remained
above 93% in both primary and metastatic tumors regardless of tumor purity
(Table 138).
Table 138. Summary of algorithm performance in the prospective validation
cohort.
Category n Above Call Sens. in Sens. in Sens. in
Sens. in Sens. in Rule
Threshold Rate Top 1 Top 2 Top 3 Top 4 Top 5 Outs /
(%) (%) (%) ("/0) ('Vo)
(%) Case
270
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Global 13,661 12,699 93 94.7 97.2 97.9
98.1 98.2 17.6
Primary 7521 7087 94.2 96.1 98.2 98.7
98.8 98.9 17.8
Specimen
5942 5426 91.3 93 96 97 97.2
97.4 17.4
Metastatic
Specimen
Percent 4 3 75 100 100 100 100
100 18.7
Tumor <
Percent 8227 7636 92.8 94.5 97 97.8
97.9 98 17.4
Tumor >=
20, <= 50
Percent 5189 4835 93.2 95 97.7 98.2 98.4
98.5 17_9
Tumor >
50, <= 80
This prospective dataset also allowed us to evaluate the diagnostic rule-out
power (i.e.,
negative predictive value) of the assay. For all patients, the empirical
prevalence tables yielded an
average of 17.6 cancer categories that had not been observed per patient
(i.e., could be ruled out) for
5 their respective MI GPSai scores. The correct cancer
category had a non-zero empirical probability in
98.9% of all cases, and the 1.1% of observations in which the true cancer
category was incorrectly
ruled out represents less than 0.1% of the total disease types ruled out.
Thus, the rule out accuracy
exceeds 99.9%.
Each of the 21 cancer categories was represented in the prospective validation
dataset both
10 with respect to true tumor type and highest prediction.
See Table 139. Sixteen of the 21 cancer
categories had an observed positive predictive value (PPV) of >= 90% and three
had a PPV of >=
99%. The minimum rule-out accuracy was 98.0%. Five cancer categories (e.g.
central nervous system
cancers, GIS'1', melanoma, mcningioma, and prostate) each exhibited > 99%
sensitivity while twelve
(e.g., breast, colon, gastroesophageal, hepatocellular, lung, two subtypes of
ovarian, pancreatic, renal,
15 squamous cell, uterine adenocarcinoma, and uterine
sarcoma) achieved > 90% sensitivity.
Table 139. Summary of algorithm performance in the prospective validation
cohort by cancer
category
Category n Call Rate (%) Sensitivity PPV
(Y0) Rule Out
(%) Accuracy (%)
Breast 1533 98 98.4 99 100
Adenocarcinoma
271
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Central Nervous 445 99.8 99.8 100 100
System Cancel
Cervical 60 51.7 38.7 66.7 98
Adenocarcinoma
Cholangiocarcinoma 363 73.8 69A 83 99.7
Colon 2119 97 98.5 98.2 100
Adenocarcinoma
Gastroesophageal 613 84.5 90.9 89.5 99.9
Adenocarcinoma
GIST 23 95.7 100 95.7 100
Hepatocellular 66 84.9 92.9 96.3 99.7
Carcinoma
Lung 2287 95 96.4 93.6 100
Adenocarcinoma
Melanoma 373 96.5 99.7 99.7 100
Meningioma 21 90.5 100 95 100
Ovarian Granulosa 25 88 95.5 95.5 100
Cell Tumor
Ovarian, Fallopian 1493 91.6 92.5 94.3 99.9
Tube
Adenocarcinoma
Pancreas 815 87.6 91.9 87.7 100
Adenocarcinoma
Prostate 556 97.1 99.1 98.7 100
Adenocarcinoma
Renal Cell 176 92.6 95.7 96.9 99.8
Carcinoma
Squamous Cell 1193 93 93.5 93.4 99.9
Carcinoma
Thyroid Cancer 74 85.1 85.7 91.5 99.2
Urothelial 354 90.7 85.4 96.1 99.9
Carcinoma
IJterine Endometrial 989 894 914 89.7 100
Adenocarcinoma
Uterine Sarcoma 83 83.1 98.6 94.4 100
272
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
FIG. AE and FIG. AF show confusion matrices with respect to prediction and
truth for the
cancer categories, respectively. FIG. AE shows a prediction matrix in the
prospective validation set.
Each row shows the percentage of the actual disease types observed when a MI
GPSai achieves a
score > 0.835. The diagonal represents the PPV for the given disease type.
Blank cells have values
5 between 0 and 1. FIG.
AE shows a confusion matrix in the prospective validation set. Each column
shows observed predictions for each disease type when a MI GPSai achieves a
score > 0.835. The
diagonal represents the sensitivity for the given disease type. Blank cells
have values between 0 and 1.
Analysis of CUP
Of the 1292 CUP cases analyzed by MI GPSai, 71.7% achieved a score exceeding
the
10 reportable threshold.
See FIG. 6AG, which shows the distribution of MI GPSai predictions in CUP
cases. The top panel in the figure shows the score distributions, where 71.7%
of cases return a
reportable result, and the bottom panel represents the predictions that were
made. Validation of a CUP
assay at the individual patient level is fundamentally uncertain as the
"truth" is unknown. As such,
comparing the populations generated by MI GPSai for each cancer category in
terms of mutation
15 frequencies against the mutation frequencies in populations of known
primaries yields insight into the
similarities of these populations. The genes with mutation frequencies with a
95% confidence interval
which does not overlap with that of any other cancer category along with their
frequencies in the
populations created by MI GPSai can be seen in Table 140. In the table, "*"
represents the observed
value among the known cancer category of the combined training and testing
datascts, and "*"
20 represents the observed
value among CUP cases predicted to be of the cancer category in each row.
Many of the pathogenic mutation frequencies were similar in the labeled and
CUP predicted
populations, but not all. In particular, VDT pathogenic mutations were not
seen in the 18 CUP cases
classified as Renal Cell Carcinoma. This could potentially be due to lower
proportions of clear cell
carcinoma in CUP [27].
25 Table 140 - Percentages of pathogenic variants detected among biomarkers
per cancer category
Of This Cancer Category Not Of This Cancer Category
Biomarker Train + Test* CUP** Train + Test
CUP**
Breast Adenocarcinoma
10_7% (9.7-
CDH1 11.1% (3.4-18.6) 0.8% (0.7-0.9) 0.8% (0.2-1.4)
11.7)
ESR1 9.2% (8.2-10.1) 0.0% (0.0-0.0)
0.2%(0.2-0.3) 0.1% (0.0-0.4)
CIATA3 9.5% (8.6-10.5) 1.8% (0.0-5.1)
0.1% (0.1-0.1) 0.0% (0.0-0.0)
MAP3K1 5.2% (4.5-5.9) 2.6% (0.0-6.8)
0.8% (0.7-0.9) 0.3% (0.0-0.7)
Cholangiocarcinoma
IDHI 8.6% (7.0-104) 19.5% (13.2-
25.7) 04% (0.3-04) 0.4% (0.0-0.9)
Colon Adenocarcinoma
273
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
AMER1 6.5% (5.9-7.1) 4.7% (1.2-9.3)
0.4%(0.3-0.4) 0.6% (0.1-1.2)
APC 763% (753- 34.1% (24.4-44.2)
2.4%(2.2-2.6) 2.5% (1.5-3.6)
77.3)
Lung Ad enocarcinoma
14.7% (13.8-
1.5% (0.4-3.2) 0.3% (0.2-0.3) 0.5%
(0.0-1.1)
EGER 15.6)
KEAP1
9.3%(8.7-10.0) 20.2%(15.8-25.1) 0.9%(0.8-1.0) 1.2%(0.3-2.2)
SMARCA4 5.8% (5.3-6.4) 19.9% (15.1-24.4)
1.3% (1.2-1.5) 2.4% (1.3-3.6)
14.4% (13.5-
26.9% (21.5-31.9) 0.9% (0.8-1.0) 1.3%
(0.5-2.2)
STK]] 15.2)
Ovarian, Fallopian Tube Adenocarcinoma
BRCA1 8-8% (7-9-9-7) 4.8% (0.0-11.6) 1.3%
(1.2-1.4) 1.4% (0.7-2.2)
81.9% (80.6- 61.9% (61.4- 51.8%
(48.2-
90.5% (81.4-97.7)
TP53 83.1) 62.5) 55.2)
Pancreas Adenocarcinoma
CDKN2A 24.2% (22.3-
18.1% (10.0-27.2) 4.8% (4.5-5.0) 7.8%
(6.1-9.8)
26.3)
KRAS 88-9% (87-5- 94.2% (88-6-98-6)
19.0% (18.6- 18.1% (15.4-
90.3) 19.4) 20.8)
18.1% (16.4-
SMAD4 25.6% (15.7-37.1) 4.0% (3.8-4.2) 3.5% (2.3-4.9)
19.8)
Renal Cell Carcinoma
17.7% (13.1-
KDM5C 0.0% (0.0-0.0) 1.2% (1.1-1.4) 1.5% (0.6-2.6)
22.4)
35.1% (31. 1-
PBRM1 21.4% (5.6-39.0) 1.3% (1.2-1.4) 3.8% (2.5-5.2)
39.3)
25.5% (21.5-
SETD2 33.1% (11.1-55.6) 1.4% (1.3-1.5) 1.7% (0.8-2.6)
29.1)
59.7% (55.4-
VI EL 0.0% (0.0-0.0) 0.0%(0.0-0.1) 0.1% (0.0-0.3)
64.1)
S uamous Cell Carcinoma
NFE2L2 7.6% (6.7-8.4) 6.9% (2.5-11.9) 0.6%
(0.5-0.7) 0.4% (0.0-0.9)
NOTCH] 7.2% (6.3-8.0) 6.8% (2.5-11.9)
0.8%(0.7-0.9) 1.3% (0.6-2.2)
Urothelial Carcinoma
CREBBP 6.9%(5.4-8.4) 12.5%(0.0-29.4)
1.5%(1.4-1.7) 2.3%(1.4-3.4)
EP300 5.8%(4.4-7.2) 6.6%(0.0-17.6)
1.2%(1.1-1.3) 1.5%(0.8-2.3)
ERBB2
7.8% (6.2-9.3) 6.4% (0.0-17.6) 1.5%
(1.3-1.6) 2.4% (1.5-3.5)
(Her2/Neu)
14.6% (12.5-
FGER3 6.5% (0.0-17.6) 0.2% (0.2-0.3) 0.6% (0.1-1.1)
16.8)
21.9% (19.5-
KDM6A 13.2% (0.0-35.3) 1.3% (1.2-1.5) 2.4% (1.4-3.4)
24.5)
26.9% (24.3-
KMT2D 14.5% (0.0-29.6) 5.3% (5.0-5.5) 6.5% (4.9-8.3)
29.8)
TS Cl 9.2%(7.6-10.9) 0.0%(0.0-0.0)
0.7%(0.6-0.8) 0.9%(0.3-1.6)
274
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Uterine Endometrial Adenocarcinoma
ARID1A
824% (80.2- 100.0% (1000- 27.8%
(26.9- 25.1% (20.1-
84.6) 100.0) 28.8) 30.2)
22.6% (19.3-
ASXL1 20.0% (5.3-36.8) 6.9% (6.4-7.4) 5.9% (2.9-9.2)
26.1)
BCOR 8.5% (7.5-9.6) 17.0% (0.0-36.8)
0.9% (0.8-1.0) 1.2% (0.6-1.9)
13.7% (12.5-
FBXW7 214% (5.3-42.1) 3.7% (3.5-3.9) 2.5% (1.5-3.6)
15.0)
FGFR2 5.9% (5.1-6.8) 11.0% (0.0-26.3)
0.4% (0.3-0.4) 1.4% (0.7-2.3)
10A% 9.3-
JAK1 ( 22.5% (5.3-42.1) 0.7%
(0.7-0.8) 0.4% (0.0-0.8)
11.5)
MSH6 5.2% (4.5-6.0) 10.8% (0.0-26.3)
1.1% (1.0-1.2) 1.5% (0.8-2.3)
20.1% (18.7-
MSI 28.2% (10.5-47.4) 2.2% (2.0-2.4) 2.6% (1.7-3.7)
21.7)
39.3% (37.5- 12.2% (11.9-
PIK3CA 41.1) 52.8% (31.6-73.7) 12.6) 6.0% (4.5-7.5)
21.7% (20.1-
PIK3R1 22.4% (5.3-42.1) 1.5%
(1.4-1.6) 0.9% (0.3-1.6)
23.2)
11.7% (10.6-
PPP2R1A 11.2% (0.0-26.3) 0.4%(0.3-0.5) 0.2% (0.0-0.6)
12.9)
Fruni 6.7% (5.5-8.1) 18.2% (5.3-36.8)
1.3% (1.1-1.5) 2.2% (1.1-3.4)
42.9% (41.0-
PTEN 49.9% (26.3-73.7) 4.5% (4.2-4.7) 3.7% (2.6-5.0)
44.8)
RNE43 7.8% (6.8-8.8) 15.7% (0.0-31.6)
1.9%(1.8-2.1) 1.1% (0.5-1.8)
Clinical utility and case examples
In a non-limiting real world example, we received an inguinal lymph node
biopsy on an 82-
year-old man which was sent for molecular profiling (see Example 1). At the
time of biopsy, the
scrum PSA was not elevated, and workup had not identified the primary tumor.
Evaluation by the
refeffing pathologist included negative I,EIC stains with CK7, CK20, PSA,
PSAP, CDX2, p40,
GA1A3, SOX10, and CD45. A eytokeratin stain was positive (AE1/3) and ease was
diagnosed as
carcinoma of unknown primary. Notably, this carcinoma was evaluated
appropriately for prostatic
lineage with PSA and PSAP BIC, and given the concurrent low serum PSA,
prostatic adenocarcinoma
was considered ruled out.
MI GPSai predicted with high probability that the sample was prostate
adenocarcinoma (MI
(iPSai score 0.9998) and review of the gene expression data showed high
expression of androgen
receptor (AR). IHC of AR protein was performed and AR was found highly
expressed, which
supported the MI GPSai call. The patient had a follow-up biopsy of the
prostate which confirmed
prostatic adenocarcinoma. After discussion with the ordering physician, the
diagnosis was changed
from CUP to metastatic prostatic adenocarcinoma. Importantly, the patient's
molecular profiling also
identified pathogenic variants in BRCA2 and PTEN, highlighting the utility of
diagnosis and
biomarker analysis from the same platform_
275
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
In addition to assigning lineage and identifying biomarker data with CUP
cases, MI GPSai
can assist with pathologic diagnosis fidelity. We prospectively monitored
discrepancies between MI
GPSai and the pathologist-assigned diagnoses in 1292 cases. In cases where the
pathologist-assigned
diagnosis was different than the top MI GPSai prediction and the MI GPSai
score for the top
5 prediction exceeded 0.999, an automated email was sent to the pathologist
in charge of the case
alerting them to this discrepancy. The pathology group was previously educated
on the design and
performance of MI GPSai and instructed to consider the discrepant cases with
their medical
judgement. The pathologists were able to review patient clinical history,
imaging results if available,
order imrnunohistochemistry, and discuss the case with the referring
oncologist and/or pathologist.
10 There were 46 cases with a MI GPSai score greater than 0.999 where
pathologists were
alerted. After review with additional inamunohistochemistry and consultation
with the referring
physician, the diagnosis was changed in 19 cases (41.3%). In 11 cases (23.9%),
where the submitted
diagnosis was not changed despite MI GPSai predictions, the predicted
diagnosis was pancreatic
adenocarcinoma, a cancer with limited specific RIC markers for confirmation.
All cases did not result
15 in a diagnosis revision for various reasons ranging from a lack of
diagnostic IFICs to verify the
prediction (such as cholangiocarcinoma vs pancreatic carcinoma) to a lack of
response from the
oncologist.
In one non-limiting real world example, the patient's treatment course was
altered based on
MI GPSai. See FIGs. 6AH-AL. We received a cervical lymph node from a 61-year-
old man for
20 molecular profiling. The referring pathologist assigned a diagnosis of
poorly-differentiated squamous
cell carcinoma (FIG. 6A.1I). The patient had systemic metastasis and had not
responded well to
squamous cell carcinoma directed therapy. The MI GPSai predicted diagnosis was
urothelial
carcinoma (MI GPSai score 0.9999). Our whole transcriptome expression data was
used to select for
lineage specific gene expression to guide imrnunohistochemical antibody
selection, the current gold-
25 standard for lineage assignment. The mean RNA expression of Uroplakin II
and GATA3 of the
urothelial carcinoma cases in our database is relatively high based on WTS
data across numerous
cancers, both relatively specific for urothelial carcinoma and not typically
expressed m squamous cell
carcinoma. See FIGs. 6A1 and 9AJ, respectively. Thus the patient sample was
probed with
antibodies to these proteins. This additional IFIC was positive for Uroplakin
II and GATA3. See
30 FIGs. 6AK and 9AIõ respectively. Importantly, the choice of the PD-T,1
clone and scoring system
was affected by the lineage of cancer being tested. In this case, the
referring pathologist and
oncologist asked to change the diagnosis to urothelial carcinoma and run the
5P142 PD-TT antibody
according to the label indications for atezolizumab. This PD-L1 score was
positive and the patient
therapy was changed. These non-limiting real world patient examples show that
MI GPSai has
35 significant clinical utility with both CUP and diagnostic fidelity.
Discussion
276
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Cancer of unknown primary remains a major clinical challenge and outcomes are
poor.
Molecular predictors of tumor origin can assist in addressing this problem by
providing critical
information in CUP cases that can inform treatment decisions and potentially
improve outcomes.
Herein we provide an artificial intelligence-derived panomic molecular
classifier that uses DNA and
5 RNA information to make tumor type predictions across a broad spectrum of
diagnostic classes with
high accuracy.
Prior molecular assays for the identification of cancers of unknown primary
have focused on
RNA profiles which have degraded performance in situations where the tumor is
from a site of
metastasis or if the tumor percentage is low [7]. Our method is robust to
these limitations. Without
10 being bound by theory, this is at least in part because we isolate
nucleic acid from microdissected
material, thus enriching for tumor cells, and because we use combined analysis
of DNA and RNA,
which further reduces susceptibility to the effects of normal cell
contamination. As demonstrated in
the case examples above, availability of mutational and gene expression
analysis data further
enhances the clinical utility of our approach from a diagnostic and
therapeutic perspective.
15 The accuracy of MI GPSai surpasses recently reported uses of DNA NGS
panels for tissue of
origin identification or guidance of utilization of targeted- and
irninunotherapies [10], [28]. Moreover,
overall accuracy of these approaches may be limited. For example, predictions
made by a Random
Forest Classifier using results from a 468-gene NGS panel as input, resulted
in an overall accuracy of
74.1% [10]. Analysis of circulating tumor DNA data from a commercial 70-gene
NGS panel revealed
20 potentially targetable mutations. However, an attempt to identify the
underlying TOO was not made
[28], possibly due to the limited number of genes analyzed. In contrast,
analysis of DNA methylation
across the genome might add additional information to above-mentioned assays,
as it has been shown
to predict a primary tumor in 87% of CUP cases [29].
In addition to its role in understanding CUP, MI GPSai provides a quality
control tool that can
25 be integrated into a pathology laboratory workflow. As part of our
prospective evaluation of MI
GPSai, pathologists were alerted to discrepancies between submitted diagnosis
and MI GPSai
prediction, resulting in change in diagnosis in 41.3% of these cases.
Considering that the rate of
inaccurate diagnosis ranges between 3% and 9% [30], inclusion of MI GPSai in
clinical routine could
improve diagnostic fidelity overall.
30 In summary, MI GPSai displayed robust performance in the diagnostic
workup of CUP cases
that was consistent across 13,661 cases including both metastatic and low
percentage tumors. At the
same time, MI GPSai can also play an important role in quality control of
anatomical pathology
laboratories. Since the MI GPSai analysis uses the results of DNA and RNA
profiles obtained as part
of routine clinical tumor profiling, both diagnostic and therapeutic
information can be returned that
35 optimize patients' treatment strategy from a single test This workflow
improves the current standard
of multiple tests that require more tissue and increased turnaround time,
which can delay treatment.
277
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Our approach aims to utilize the context-specific information gained by
lineage assignment when
considering biomarker-directed therapy.
References (bracketed numbers All correspond to those in the text of this
Example)
[1] C. Massard, etal. Carcinomas of an unknown primary origin¨diagnosis and
treatment.
5 Nat. Rev. Clin. Oncol., 8 (12) (2011), pp. 701-710
[2] G.R. Varadhachary, M.N. Raber. Cancer of unknown primary site. N. Engl. J.
Med., 371
(8) (2014), pp. 757-765
[3] B.R. DeYoung, M.R. Wick. Immunohistologic evaluation of metastatic
carcinomas of
unknown origin: an algorithmic approach. Semin. Diagn. Pathol., 17 (3) (2000),
pp. 184-193
10 [4] G.G. Anderson, L.M. Weiss. Determining tissue of origin for
metastatic cancers: meta-
analysis and literature review of inamunohistochemistry performance. Appl.
Irnmunohistochem. Mol.
Morphol., 18 (1) (2010), pp. 3-8
[5] S.Y. Park, et al. Panels of immunohistochemical markers help determine
primary sites of
metastatic adenocarcinoma. Arch. Pathol. Lab. Med., 131(10) (2007), pp. 1561-
1567
15 [6] R.W. Brown, et al. Irnmunohistochemical identification of tumor
markers in metastatic
adenocarcinoma. A diagnostic adjunct in the determination of primary site. Am.
J. Clin. Pathol., 107
(1) (1997), pp. 12-19
[7] M.G. Erlander, et al. Performance and clinical evaluation of the 92-gene
real-time PCR
assay for tumor classification. J. Mol. Diagn., 13 (5) (2011), pp. 493-503
20 [8] J.D. Hainsworth, et al. Molecular gene expression profiling to
predict the tissue of origin
and direct site-specific therapy in patients with carcinoma of unknown primary
site: a prospective trial
of the Sarah Cannon research institute. J. Clin. Oncol., 31(2) (2013), pp. 217-
223
[9] J. S. Ross, et al. Comprehensive genomic profiling of carcinoma of unknown
primary site:
new routes to targeted therapies. JAMA Oncol., 1(1) (2015), pp. 40-49
25 [10] A. Penson, et al. Development of genome-derived tumor type
prediction to inform
clinical cancer care. JAMA Oncol., 6(1) (2019), pp. 84-91
[11] GA. Stancel, etal. Identification of tissue of origin in body fluid
specimens using a gene
expression microarray assay. Cancer Cytopathol., 120 (1) (2012), pp. 62-70
[12] J.L. Dennis, et al. Markers of adenocarcinoma characteristic of the site
of origin:
30 development of a diagnostic algorithm. Clin. Cancer Res., 11(10) (2005),
pp. 3766-3772
[13] A.R. Gamble, et al. Use of tumour marker immunoreactivity to identify
primary site of
metastatic cancer. TIME 306 (6873) (1993), pp. 295-29t1
[14] A. Dobin, etal. STAR: ultrafast universal RNA-seq aligner.
Bioinfonnatics, 29 (1)
(2013), pp. 15-21
35 [15] R. Patro, et al. Salmon provides fast and bias-aware quantification
of transcript
expression. Nat. Methods, 11 (1) (2017), pp. 117-119
278
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
[16] C.W. Brennan, et al. The somatic genomic landscape of glioblastoma. Cell,
155 (2)
(2013), pp. 462-477
[17] S.P. Shah, et al. Mutation of FOXL2 in granulosa-cell tumors of the
ovary. N. Engl. J.
Med., 360 (26) (2009), pp. 2719-2729
5 [18] ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-
cancer analysis
of whole genomes. Nature, 578 (7793) (2020), pp. 82-93
[19] F. Sanchez-Vega, et al. Oncogenic signaling pathways in the cancer genome
atlas. Cell,
173 (2) (2018), pp. 321-337.e10
[20] M.C. Heinrich, et al. Kinase mutations and imatinib response in patients
with metastatic
10 gastrointestinal stromal tumor. J. Chin. Oncol., 21(23) (2003), pp. 4342-
4349
[21] Cancer Genome Atlas Network. Comprehensive molecular portraits of human
breast
tumours. Nature, 490 (7418) (2012), pp. 61-70
[22] P. Tan, et al. Genetics and molecular pathogenesis of gastric
adenocarcinoma.
Gastroenterology, 149 (5) (2015), pp. 1153-1162
15 [23] M. Miettinen, et al. Immunohistochemical spectrum of GISTs at
different sites and their
differential diagnosis with a reference to CD117 (KIT). Mod. Pathol., 13 (10)
(2000), pp. 1134-1142
[24] L.A. Garraway, et al. Integrative genomic analyses identify MITF as a
lineage survival
oncogene amplified in malignant melanoma. Nature, 436 (7047) (2005), pp. 117-
122
[25] M.C. Markowski, et al. Inflammatory cytokines induce phosphorylation and
20 ubiquitination of prostate suppressor protein NKX3.1. Cancer Res.,
68(17) (2008), pp. 6896-6901
[26] Abraham J., et al. Genomic Profiling Similarity. W02020146554.
[27] F.A. Greco, J.D. Hainsworth. Renal cell carcinoma presenting as carcinoma
of unknown
primary site: recognition of a treatable patient subset. Clin. Genitourin.
Cancer, 16 (4) (2018), pp.
e893-e898
25 [28] S. Kato, et al. Utility of genomic analysis in circulating tumor
DNA from patients with
carcinoma of unknown primary. Cancer Res., 77 (16) (2017), pp. 4238-4246
[29] S. Moran, et al. Epigenetic profiling to classify cancer of unknown
primary: a
multicentre, retrospective analysis. Lancet Oncol., 17 (10) (2016), pp. 1386-
1395
[30] M. Peck, et al. Review of diagnostic error in anatomical pathology and
the role and value
30 of second opinions in error prevention. J. Clin. Pathol., 71(11) (2018),
pp. 995-1000
[31] K. Bera, et al. Artificial intelligence in digital pathology - new tools
for diagnosis and
precision oncology. Nat. Rev. Clin. Oncol., 16(11) (2019), pp. 703-715
[32] W. Jiao, G. Atwal, P. Polak, et al. A deep learning system accurately
classifies primary
and metastatic cancers using passenger mutation patterns. Nat. Conunun., 11
(2020), p. 728
35 [33] P. Stafford, M. Brun. Three methods for optimization of cross-
laboratory and cross-
platform microarray expression data. Nucl. Acids Res., 35 (10) (2007), p. e72
279
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
[34] Haskell CM, et al. Metastasis of unknown origin. Curr Probl Cancer. 1988
Jan-
Feb;12(1):5-58. Review. PubMed PMID: 3067982.
[35] Haigis KM, etal. Tissue-specificity in cancer: The rule, not the
exception. Science. 2019
Mar 15;363(6432):1150-1151. doi: 10.1126/seienee.aaw3472. P ubMed PMID:
30872507.
Example 4: Molecular Profiling Report and Use for Patient with Metastatic
Adenocarcinoma
FIGs. 7A-P present a molecular profiling report which is de-identified but
from molecular
profiling of a real life patient according to the systems and methods provided
herein.
FIG. 7A illustrates page 1 of the report indicating the specimen as reported
in the test
requisition from the ordering physician was taken from the liver and was
presented with primary
tumor site as ascending colon. The diagnosis was metastatic adenocarcinoma. In
the "Results with
Therapy Associations" section, FIG. 7A further displays a summary of therapies
associated with
potential benefit and therapies associated with potential lack of benefit
based on the relevant
biomarkers for the therapeutic associations. Here, the report notes that
mutations were not detected in
KRAS, NRAS and BRAF, thereby indicated potential benefit of cetuxirnab or
panitumurnab.
Conversely, lack of expression of HER2 protein indicates potential lack of
benefit from anti-ITER2
therapies (lapatinib, pertuzumab, trastuzamab). The section "Cancer Type
Relevant Biomarkers"
highlights certain of the molecular profiling results for particularly
relevant biomarkers. The
"Genomic Signatures" section indicates the results of microsatellite
instability (191S1) and tumor
mutational burden (TMB). Note both characteristics were also highlighted in
the section just above.
This patient was found to be MSI stable and TA/B3 low.
FIG. 7B is page 2 of the report and lists a summary of biomarker results from
the indicated
assays. Of note, APC and 1P53 were found to have known pathogenic mutations
via sequencing of
tumor genomic DNA. The section "Other Findings" notes a number of genes with
indeterminate
sequencing results due to low coverage.
FIG. 7C is page 3 of the report and continues the list of "Other Findings"
with genes where
genomic DNA sequencing (by NGS) did not find point mutations, indels, or copy
number
amplification.
FIG. 713 is page 4 of the report and further continues the list of "Other
Findings" with genes
where RNA sequencing (by NGS) did not find alterations (e.g., no fusion genes
detected).
FIG. 7E is page 5 of the report and shows the results of the Cienomic
Profiling Similarity
(GPS) analysis as provided herein performed on the specimen. Recall the
specimen comprises a
metastatic lesion taken from the liver and was reported to be an
adenocarcinoma of the ascending
colon by the ordering physician (see FIG. 7A). As shown in the figure, the
report provides a
probability that the specimen is from each of the listed organ groups (i.e.,
Bladder; Brain; Breast;
Colon; Female Genital Tract & Peritoneum; Gastroesophageal; Head, Face or
Neck, NOS; Kidney;
280
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
Liver, Gall Bladder, Ducts; Lung; Melanoma/Skin; Pancreas; Prostate; Other).
The Similarity for each
Organ type shown is in the vertical bars. In this case, GPS assigned a score
of 97 to Organ type
"Colon," and the starred shape indicates a probability of correct match > 98%.
See "Legend" box. The
Organ group Gastroesophageal had a similarity of 1, and the circular shape
indicates that the
5 probability is inconclusive. All other organs had a similarity of less
than 1 or 0, indicating that those
Organ groups were excluded with a> 99% probability.
FIG. 7F is page 6 of the report and provides a listing of "Notes of
Significance," here an
available clinical trial based on the profiling results, and additional
specimen information.
FIG. 7G is page 7 of the report and provides a "Clinical Trial Connector,"
which identifies
10 potential clinical trials for the patient based on the molecular
profiling results. A trial connected to the
APC gene mutation (see FIG. 7B) is noted.
FIG. 7H presents a disclaimer. For example, that decisions on patient care and
treatment must
be based on the independent medical judgment of the treating physician, taking
into consideration all
available information concerning the patient's condition. This page ends the
main body of the report
15 and an Appendix follows.
FIGs. 7I-M provide more details about results obtained using Next-Generation
Sequencing
(NGS). FIG. 71 is page 1 of the appendix and provides information about the
Tumor Mutational
Burden (TMB) and Microsatellite Instability (MSI) analyses and results. The
report notes that high
mutational load is a potential indicator of immunothcrapy response (Lc et at,
PD-1 Blockade in
20 Tumors with Mismatch-Repair Deficiency, N Engl J Med 2015; 372:2509-
2520; Rizvi et al.,
Mutational landscape determines sensitivity to PD-1 blockade in non¨small cell
lung cancer. Science.
2015 Apr 3; 348(6230): 124-128; Rosenberg et al., Atczolizumab in patients
with locally advanced
and metastatic urothelial carcinoma who have progressed following treatment
with platinum-based
chemotherapy: a single arm, phase 2 trial. Lancet. 2016 May 7; 387(10031):
1909-1920; Snyder et
25 al., Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma.
N Engl J Med. 2014 Dec
4; 371(23): 2189-2199; all of which references are incorporated by reference
herein in their entirety).
FIG. 7.1 is page 2 of the appendix and lists details concerning the genes
found to harbor alterations,
namely APC and TP53. See also FIG. 7B. FIG. 7K is page 3 of the appendix and
notes genes that
were tested by NGS with either indeterminate results due to low coverage for
some or all exons, or no
30 detected mutations. FIG. 7L is page 4 of the appendix and continues the
listing of genes that were
tested by NGS with no detected mutations and adds more information about how
Next Generation
Sequencing was performed. FIG. 7M is page 5 of the appendix and provides
information about copy
number alterations (CNA; copy number variation; CNV), e.g., gene
amplification, detected by NGS
analysis and corresponding methodology. FIG. 7N is page 6 of the appendix and
provides
35 information about gene fusion and transcript variant detection by RNA
Sequencing analysis and
corresponding methodology. In this specimen, no fusions or variant transcripts
were detected. FIG.
70 is page 7 of the appendix and provides more information about the BIC
analysis performed on the
281
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
patient specimen, e.g., the staining threshold and results for each marker.
FIG. 7P and FIG. 7Q are
pages 8 and 9 of the appendix, respectively, and provide a listing of
references used to provide
evidence of the biomarker ¨ agent association rules used to construct the
therapy recommendations.
5 Example 5: Molecular Profiling Report ¨ Metastatic Ovarian Carcinoma
FIGs. 8A-P present another molecular profiling report which is de-identified
but from
molecular profiling of a real life patient according to the systems and
methods provided herein.
FIG. 8A illustrates page 1 of the report indicating the specimen as reported
in the test
requisition from the ordering physician was taken from the ascending colon and
was presented with
10 primary tumor site as ovary. The diagnosis was carcinoma, NOS. In the
"Results with Therapy
Associations" section, FIG. 8A further displays a summary of therapies
associated with potential
benefit and therapies associated with potential lack of benefit based on the
relevant biomarkers for the
therapeutic associations. Here, the report notes that the sample was
identified as PD-Li positive by
111C, thereby indicated potential benefit of pembroliziunab. Conversely, lack
of expression of HER2
15 protein indicates potential lack of benefit from anti-HER2 therapies
pertuzumab or trastuzamab. The
section "Cancer Type Relevant Biomarkers" highlights certain of the molecular
profiling results for
particularly relevant biomarkers, including results from various analytes:
genomic DNA
(microsatellite instability (MSI), mismatch repair status, tumor mutational
burden (TMB), and ATM
and BRCA1/2 status); whole transeriptome sequencing (NTRK1/2/3 fusion); and 11-
IC (ER/PR protein
20 status). The sample was found to be MSI stable, MMR proficient, TMB low,
no NTRK fusions
detected, no mutation detected in ATM or BRCA1/2, and ER/PR negative. The
section "Other
Findings" notes that a pathogenic variant was found in the TP53 gene by NGS of
genomic DNA.
FIG. 8B is page 2 of the report and lists additional summary of biomarker
results from the
indicated assays. "Genomic Signatures" provides additional insight into the
MS1 and 1M13 results.
25 "Genes Tested with Pathogenic or Likely Pathogenic Alterations" provides
further detail about the
TP53 pathogenic mutation detected via sequencing of tumor genomic DNA. The
section
"Immunohistochemistry Results" provides further detail about the protein
expression results, e.g.,
criteria used to determine the result, and details results of the MMR genes
(MLH1, MSH2, MSH6,
PMS2). "Genes Tested with Indeterminate Results by Tumor DNA Sequencing" notes
certain genes
30 of interest with indeterminate results due to low sequencing coverage of
some or all exons.
FIG. 8C is page 3 of the report and shows the results of the MI GPSai (GPS)
analysis as
provided herein performed on the specimen. See, e.g., Example 3. Recall the
specimen comprises a
metastatic lesion taken from the ascending colon and was reported to be an
ovarian carcinoma by the
ordering physician (see FIG. 8A). As shown in FIG. 8C, the report provides a
probability that the
35 specimen is from each of the listed cancer categories (i.e., breast
adenocarcinoma, central nervous
system cancer, cervical adenocarcinoma, cholangiocarcinoma, colon
adenocarcinoma,
gastroesophageal adenocarcinoma, gastrointestinal stromal tumor (GIST),
hepatocellular carcinoma,
282
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
lung adenocarcinoma, melanoma, meningioma, ovarian granulosa cell tumor,
ovarian & fallopian tube
adenocarcinoma, pancreas adenocarcinoma, prostate adenocarcinoma, renal cell
carcinoma, squamous
cell carcinoma, thyroid cancer, urothelial carcinoma, uterine endometrial
adenocarcinoma, and uterine
sarcoma). The predicted Prevalence for each cancer category is shown is in the
horizontal bars. In this
5 case, GPS assigned a prevalence of 96% to cancer category "Ovarian,
Fallopian Tube
Adenocarcinoma." The cancer category "Uterine Endometrial Adenocarcinoma" had
a prevalence of
3%, and "Cervical Adenocarcinoma" had a prevalence of <1%. All other
categories had a prevalence
of ¨0%. Thus, the UPS result was consistent with the original diagnosis.
FIG. 8D is page 4 of the report and provides a listing of "Notes of
Significance," here an
10 available clinical trial based on the profiling results, and additional
specimen information.
FIG. SE is page 5 of the report and provides a "Clinical Trial Connector,"
which identifies
potential clinical trials for the patient based on the molecular profiling
results. A trial connected to the
PD-Li II4C result (see FIG. 8A) is noted.
FIG. 8F is page 6 of the report and presents a disclaimer. For example, that
decisions on
15 patient care and treatment must be based on the independent medical
judgment of the treating
physician, taking into consideration all available information concerning the
patient's condition. This
page ends the main body of the report and an Appendix follows.
FIGs. 8G-I are pages 7-9 of the report (and 1-3 of the Appendix) and provide
more details
about results obtained using Next-Generation Sequencing (NGS) of genomic tumor
DNA. FIG. 8G is
20 page 1 of the appendix and provides information about the Tumor
Mutational Burden (T1VB3) and
Microsatellite Instability (MSD analyses and results, and provides details
concerning mutations in
genes found to harbor alterations, here TP53. FIG. 8H is page 2 of the
appendix and notes genes that
were tested by NGS with either indeterminate results due to low coverage for
some or all exons and
provides details about the NGS assay. FIG. 81 is page 3 of the appendix and
provides information
25 about copy number alterations (CNA; copy number variation; CNV), e.g.,
gene amplification,
detected by NGS analysis and corresponding methodology. FIG. 8J is page 4 of
the appendix and
provides information about gene fusion and transcript variant detection by RNA
Sequencing analysis
and corresponding methodology. In this specimen, no fusions or variant
transcripts were detected.
FIGs. 8K-L are pages 5-6 of the appendix, respectively, and provides more
information about the
30 TUC analysis performed on the patient specimen, e.g., the staining
threshold and results for each
marker. FIG. 8M is page 7 of the appendix, and provide a listing of references
used to provide
evidence of the biomarker ¨ agent association rules used to construct the
therapy recommendations.
Example 6: Selecting Treatment for a Cancer
35 An oncologist is treating a cancer patient with a metastatic tumor of
unknown primary and
desires to perform molecular profiling on the tumor sample to assist in
selecting a treatment regimen
for the patient. A biological sample is collected from a tumor located in the
retroperitoneum. The
283
CA 03167694 2022- 8- 11
WO 2021/163706
PCT/US2021/018263
oncologist's pathology report states that the specimen is adenocarcinoma, NOS
with unknown
primary origin, i.e., CUP. The oncologist requisitions a molecular profiling
panel to be performed on
the tumor sample. The sample is sent to our laboratory for molecular profiling
according to Example
1 herein.
5 We perform molecular profiling comprising NGS of genomic DNA. NGS of RNA
transcripts,
and ITIC analysis on the tumor specimen. A molecular profile is generated for
the sample. The
machine learning models described in Examples 2-3 are used to predict the
primary site of the tumor.
The classification leans strongly towards "ovarian, fallopian, retroperitoneal
adenocarcinoma."
Mutations in APC and TP53 are identified. No mutations in KRAS, BRAF, and NRAS
are found.
10 IiER2 is not overexpressed. The molecular profiling results are included
in the report such as in the
Examples above. The report suggests treatment with cetuximab or panitumumab
but not anti-FIER2
therapy. 'Me report is provided to the oncologist The oncologist uses the
information provided in the
report to assist in determining a treatment regimen for the patient.
OTIIER EMBODIMENTS
It is to be understood that while the invention has been described in
conjunction with the
detailed description thereof, the foregoing description is intended to
illustrate and not limit the scope
as described herein, which is defmed by the scope of the appended claims.
Other aspects, advantages,
20 and modifications are within the scope of the following claims.
284
CA 03167694 2022- 8- 11