Note: Descriptions are shown in the official language in which they were submitted.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
1
DEVICES AND METHODS FOR SEQUENCING
CROSS-REFERENCE TO RELATED APPLICATIONS
This application claims the benefit under 35 U.S.C. 119(e) of the filing
date of USSN
63/014,071, filed on April 22, 2020; USSN 63/014,087, filed on April 22, 2020;
USSN
63/014,093, filed on April 22, 2020; USSN 63/014,106, filed on April 22, 2020;
USSN
63/041,206, filed on June 19, 2020; USSN 63/139.339, filed on January 20,
2021; USSN
63/139,343, filed on January 20, 2021; USSN 63/139,346, filed on January 20,
2021; and USSN
63/139,348, filed on January 20, 2021; the entire contents of each application
are incorporated
herein by reference.
BACKGROUND OF INVENTION
Proteomics, genomics, and transcriptomics have emerged as important and
necessary
in the study of biological systems. These analysis of an individual organism
or sample type can
provide insights into cellular processes and response patterns, which lead to
improved diagnostic
and therapeutic strategies. The complexity surrounding nucleic acid and
protein compositions
and modification present challenges in determining large-scale sequencing
information for a
biological sample.
SUMMARY OF INVENTION
Aspects of the instant disclosure provide methods, compositions, devices,
and/or
cartridges for use in a process to prepare a sample for analysis and/or
analyze (e.g., analyze by
sequencing) one or more target molecules in a sample. In some embodiments, a
target molecule
is a nucleic acid (e.g., DNA or RNA, including without limitation, cDNA,
genomic DNA,
mRNA, and derivatives and fragments thereof). In some embodiments, a target
molecule is a
protein.
Some aspects of the disclosure provide devices for preparing a biological
sample for
sequencing. In some embodiments, the device comprises an automated module
configured to
receive two or more cartridges selected from the group consisting of (i) a
lysis cartridge; (ii) an
enrichment cartridge; (iii) a fragmentation cartridge; and (iv) a
functionalization cartridge. In
some embodiments, the device comprises an automated module comprising one or
more
microfluidic channels and configured to intake a biological sample comprising
one or more
target molecules. In some embodiments, the device comprises an automated
module configured
to receive (i) a lysis cartridge; and (ii) an enrichment cartridge. In some
embodiments, the
device comprises an automated module configured to receive (i) a lysis
cartridge; and (iii) a
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
2
fragmentation cartridge. In some embodiments, the device comprises an
automated module
configured to receive (i) a lysis cartridge; and (iv) a functionalization
cartridge. In some
embodiments, the device comprises an automated module configured to receive
(ii) an
enrichment cartridge; and (iii) a fragmentation cartridge. In some
embodiments, the device
comprises an automated module configured to receive (i) an enrichment
cartridge; and (iv) a
functionalization cartridge. In some embodiments, the device comprises an
automated module
configured to receive (i) a fragmentation cartridge; and (iv) a
functionalization cartridge. In some
embodiments, the device comprises an automated module configured to receive
(i) a
fragmentation cartridge; (ii) an enrichment cartridge; and (iii) a
fragmentation cartridge. In some
embodiments, the device comprises an automated module configured to receive
(i) a
fragmentation cartridge; (ii) an enrichment cartridge; and (iv) a
functionalization cartridge. In
some embodiments, the device comprises an automated module configured to
receive (ii) an
enrichment cartridge; (iii) a fragmentation cartridge; and (iv) a
functionalization cartridge. In
some embodiments, the device comprises an automated module configured to
receive (i) a
fragmentation cartridge; (ii) an enrichment cartridge; (iii) a fragmentation
cartridge; and (iv) a
functionalization cartridge. In some embodiments, the device produces nucleic
acids with an
average read-length that is longer than an average read-length produced using
control methods.
Further aspects of the disclosure provide devices for preparing one or more
target molecules,
configured to perform two or more of the following steps selected from (i),
(ii), (iii), and (iv),
wherein (i), (ii), (iii), and (iv) are defined as follows: (i) lyse a
biological sample comprising one
or more target molecules; (ii) enrich at least one of the one or more target
molecules and/or at
least one non-target molecule; (iii) fragment the one or more target
molecules; and (iv)
functionalize a terminal moiety of the one or more target molecules.
In some embodiments, one or more of the method steps selected from (i), (ii),
(iii), and
(iv) are performed in a cartridge. In some embodiments, the one or more steps
are performed in
the same cartridge. In some embodiments, the cartridge is a single-use
cartridge or a multi-use
cartridge. In some embodiments, the cartridge comprises one or more
microfluidic channels
configured to contain and/or transport a fluid used in any one of the
automated steps. In some
embodiments, the cartridge comprises one or more microfluidic channels
configured to contain
and/or transport the one or more target molecules between any one of the
automated steps. In
some embodiments, the cartridge comprises resin for purification of the one or
more target
molecules between any one of the automated steps. In some embodiments, the
resin is Sephadex
resin, optionally G-10 Sephadex resin. In some embodiments, the cartridge
comprises any size
exclusion medium.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
3
Still further aspects of the disclosure provide methods for preparing one or
more target
molecules. In some embodiments, methods for preparing one or more target
molecules comprise
two or more of the following steps selected from (i), (ii), (iii), and (iv),
wherein (i), (ii), (iii), and
(iv) are defined as follows: (i) lyse a biological sample comprising one or
more target molecules;
(ii) enrich at least one of the one or more target molecules and/or at least
non-target molecule;
(iii) fragment the one or more target molecules; and (iv) functionalize a
terminal moiety of the
one or more fragmented target molecules; wherein at least one of steps (i),
(ii), (iii), or (iv) is
performed in an automated sample preparation device. In some embodiments, two
steps are
performed in an automated sample preparation device. In some embodiments,
three steps are
.. performed in an automated sample preparation device. In some embodiments,
four steps are
performed in an automated sample preparation device. In some embodiments, step
(i) is
performed using a lysis cartridge. In some embodiments, step (ii) is performed
using an
enrichment cartridge. In some embodiments, step (iii) is performed using a
fragmentation
cartridge. In some embodiments, step (iv) is performed using a
functionalization cartridge.
Yet further aspects of the disclosure provide cartridges for preparing one or
more target
molecules. In some embodiments, a cartridge is configured to perform two or
more of the
following steps selected from (i), (ii), (iii), and (iv), wherein (ii), (iii),
and (iv) are defined as
follows: (i) lyse a biological sample comprising one or more target molecules;
(ii) enrich at least
one of the one or more target molecules and/or at least one non-target
molecule; (iii) fragment
the one or more target molecules; and (iv) functionalize a terminal moiety of
the one or more
target molecules. In some embodiments, the cartridge is a single-use cartridge
or a multi-use
cartridge. In some embodiments, the cartridge comprises one or more
microfluidic channels
configured to contain and/or transport a fluid used in any one of the
automated steps. In some
embodiments, the cartridge comprises one or more microfluidic channels
configured to contain
and/or transport the one or more target molecules between any one of the
automated steps. In
some embodiments, the cartridge comprises resin for purification of the one or
more target
molecules between any one of the automated steps. In some embodiments, the
resin is Sephadex
resin, optionally G-10 Sephadex resin.
In some embodiments, the biological sample is a single cell, mammalian cell
tissue,
.. animal sample, fungal sample, or plant sample. In some embodiments, the
biological sample is a
blood sample, saliva sample, sputum sample, fecal sample, urine sample, buccal
swab sample,
amniotic sample, seminal sample, synovial sample, spinal sample, or pleural
fluid sample. In
some embodiments, the one or more target molecules are nucleic acids. In some
embodiments,
the one or more target molecules are proteins.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
4
In some embodiments, a device further comprises a peristaltic pump configured
to
transport one or more fluids into, within, or out of any one of cartridges
received by the device.
In some embodiments, a device further comprises a peristaltic pump configured
to transport one
or more fluids within, or through any of the microfluidic channels of
cartridges received by the
device. In some embodiments, a device is configured to transport fluids with a
fluid flow
resolution of less than or equal to 1000 microliters, less than or equal to
100 microliters, less than
or equal to 50 microliters, or less than or equal to 10 microliters. In some
embodiments, the
device is configured to receive two or more cartridges at the same time. In
some embodiments,
the device is configured to establish fluidic communication between two or
more cartridges
received by the device at the same time. In some embodiments, the device is
configured to
receive two or more cartridges sequentially.
In some embodiments, the device further comprises a sequencing module. In some
embodiments, the device is configured to deliver the one or more target
molecules to the
sequencing module. In some embodiments, the sequencing module performs nucleic
acid
sequencing. In some embodiments, the nucleic acid sequencing comprises single-
molecule real-
time sequencing, sequencing by synthesis, sequencing by ligation, nanopore
sequencing, and/or
Sanger sequencing. In some embodiments, the sequencing module performs protein
sequencing.
In some embodiments, the protein sequencing comprises Edman degradation or
mass
spectroscopy. In some embodiments, the sequencing module performs single-
molecule protein
sequencing.
In some embodiments, a lysis cartridge comprises one or more microfluidic
channels and
configured to intake a biological sample comprising one or more target
molecules and produce a
lysed sample. In some embodiments, an enrichment cartridge comprises one or
more
microfluidic channels and is configured to enrich at least one of the one or
more target molecules
to produce an enriched sample. In some embodiments, a fragmentation cartridge
comprises one
or more microfluidic channels and is configured to digest or fragment at least
one of the one or
more target molecules to produce a fragmented sample. In some embodiments, a
functionalization cartridge comprises one or more microfluidic channels and is
configured to
functionalize a terminal moiety of at least one of the one or more target
molecules to form a
functionalized sample.
In some embodiments, any one cartridge is positioned to receive a sample or
target
molecule(s) from any other cartridge. In some embodiments, any one cartridge
is connected by
one or more microfluidic channels to any other cartridge.
In some embodiments, a lysis cartridge comprises reagents that lyse the sample
but does
not degrade or fragment the one or more target molecules. In some embodiments,
the lysis
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
cartridge comprises reagents that promote the one or more target molecules to
be at least
partially isolated or purified from non-target molecules of the sample. In
some embodiments, the
reagents comprise detergents, acids, and/or bases. In some embodiments, the
reagents comprise
a lysis buffer. In some embodiments, the lysis buffer is selected from the
group consisting of:
5 RIPA buffer, GC1 (Guanidine-HC1) buffer, and GlyNP40 buffer. In some
embodiments, the one
or more microfluidic channels in the lysis cartridge promote shearing of cells
and/or tissues (e.g.,
shear flow of cells and/or tissues). In some embodiments, the lysis cartridge
comprises a needle
passage that promotes mechanical shearing of cells and/or tissues. In some
embodiments, the
needle passage has an internal diameter of 0.1 to 1 mm. In some embodiments,
the one or more
microfluidic channels in the lysis cartridge comprise a post array. In some
embodiments, the
lysis cartridge is configured to be heated at an elevated temperature (e.g.,
20-60 C, 20-30 C,
25-40 C, 30-50 C, 35-50 C, or 50-75 C). In some embodiments, the device is
configured to
heat the lysis cartridge at an elevated temperature (e.g., 20-60 C, 20-30 C,
25-40 C, 30-50 C,
35-50 C, or 50-75 C). In some embodiments, the device is configured to
subject the lysis
cartridge to microwaves or sonication.
In some embodiments, the enrichment cartridge comprises one or more affinity
matrices.
In some embodiments, the one or more affinity matrices are in microfluidic
channels of the
enrichment cartridge. In some embodiments, the one or more target molecules
are nucleic acids,
the immobilized capture probe is an oligonucleotide capture probe, and the
oligonucleotide
capture probe comprises a sequence that is at least partially complementary to
at least one of the
one or more target molecules. In some embodiments, the oligonucleotide capture
probe
comprises a sequence that is at least 80%, 90% 95%, or 100% complementary to
the target
molecule. In some embodiments, the one or more target molecules are proteins,
and the
immobilized capture probe is a protein capture probe that binds to at least
one of the one or more
target molecules. In some embodiments, the protein capture probe is an aptamer
or an antibody.
In some embodiments, the protein capture probe binds to the target protein
with a binding
affinity of 10-9 to 10-8 M, 10-8 to 10-7 M, 10-7 to 10-6 M, 10-6 to 10-5 M, 10-
5 to 10-4 M, 10-
4 to 10-3 M, or 10-3 to 10-2 M. In some embodiments, the one or more target
molecules are
nucleic acids, the immobilized capture probe is an oligonucleotide capture
probe, and the
oligonucleotide capture probe comprises a sequence that is at least partially
complementary to at
least one non-target molecule. In some embodiments, the oligonucleotide
capture probe
comprises a sequence that is at least 80%, 90% 95%, or 100% complementary to
the non-target
molecule. In some embodiments, the oligonucleotide capture probe is not
complementary to the
one or more target molecules. In some embodiments, the one or more target
molecules are
proteins, and the immobilized capture probe is a protein capture probe that
binds to at least one
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
6
non-target molecule. In some embodiments, the protein capture probe binds to
the non-target
protein with a binding affinity of 10-9 to 10-8 M, 10-8 to 10-7 M, 10-7 to 10-
6 M, 10-6 to 10-5
M, 10-5 to 10-4 M, 10-4 to 10-3 M, or 10-3 to 10-2 M. In some embodiments, the
protein
capture probe does not bind to the one or more target molecules. In some
embodiments, the
enrichment cartridge is configured to deplete the sample of non-target
molecules.
In some embodiments, the fragmentation cartridge comprises non-enzymatic
reagents
that digest or fragment the sample and/or the one or more target molecules. In
some
embodiments, the non-enzymatic reagents that digest or fragment the sample
and/or the one or
more target molecules comprise detergents, acids, and/or bases. In some
embodiments, the non-
enzymatic reagents that digest or fragment the sample and/or the one or more
target molecules
comprise cyanogen bromide, hydroxylamine, iodosobenzoic acid, dimethyl
sulfoxide,
hydrochloric acid, BNPS-skatole [2-(2-nitrophenylsulfeny1)-3-methylindole],
and/or 2-nitro-5-
thiocyanobenzoic acid. In some embodiments, the fragmentation cartridge
comprises one or
more enzymatic reagents that digest or fragment at least one of the one or
more target molecules.
In some embodiments, the one or more enzymatic reagents comprise one or more
proteases. In
some embodiments, the one or more proteases are selected from the group
consisting of: trypsin,
chymotrypsin, LysC, LysN, AspN, GluC and ArgC. In some embodiments, the one or
more
enzymatic reagents comprise one or more endonucleases or exonucleases. In some
embodiments,
the fragmentation cartridge can be heated at an elevated temperature (e.g., 20-
60 C, 20-30 C,
25-40 C, 30-50 C, 35-50 C, or 50-75 C). In some embodiments, a device is
configured to
heat the fragmentation cartridge at an elevated temperature (e.g., 20-60 C,
20-30 C, 25-40 C,
30-50 C, 35-50 C, or 50-75 C). In some embodiments, a device is configured
to subject the
fragmentation cartridge to microwaves or sonication.
In some embodiments, the functionalization cartridge comprises a first chamber
comprising reagents that covalently modify a moiety MO of the one or more
target molecules, or
of one or more fragments thereof, to a modified moiety Ml. In some
embodiments, the reagents
are non-enzymatic. In some embodiments, the covalent modification is
regiospecific. In some
embodiments, the portion of the one or more target molecules, or of the one or
more fragments
thereof, is a C-terminal carboxylate group or a C-terminal amino group. In
some embodiments,
the reagents comprise buffers, salts, organic compounds, acids, and/or bases.
In some
embodiments, the portion of the one or more target molecules, or of the one or
more fragments
thereof, is a C-terminal amino group, and the covalent modification is diazo
transfer. In some
embodiments, moiety MO is ¨NH2 and moiety M1 is ¨N3. In some embodiments, the
reagents
comprise imidazole-l-sulfonyl azide and a copper salt (e.g., copper sulfate),
and a buffer having
a pH of about 9-11 (e.g. a potassium carbonate buffer having a pH of about 9-
11). In some
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
7
embodiments, the reagents comprise any azide transfer agent. In some
embodiments, the
reagents comprise trifluoromethanesulfonyl azide. In some embodiments, the
azide transfer agent
comprises benzenesulfonyl-azide. In some embodiments, the first chamber is
connected via one
or more microfluidic channels, and/or optionally a purification chamber, to a
second chamber. In
some embodiments, the second chamber comprises reagents that covalently modify
moiety M1
to produce a functionalized peptide. In some embodiments, the covalent
modification is an
electrocyclic click reaction. In some embodiments, the reagents comprise a
DBCO-labeled
DNA-streptavidin conjugate and a buffer, optionally wherein the DBCO-labeled
DNA-
streptavidin conjugate is immobilized to the surface of the second chamber. In
some
embodiments, the functionalized peptide is functionalized with a DBCO-labeled
DNA-
streptavidin conjugate.
In some embodiments, a purification chamber is positioned between the first
chamber
and the second chamber, comprising a resin that promotes purification or
enrichment of the
modified target molecules, or fragments thereof. In some embodiments, the
resin is Sephadex
resin, optionally G-10 Sephadex resin. In some embodiments, the
functionalization cartridge can
be heated at an elevated temperature (e.g., 20-60 C , 20-30 C, 25-40 C, 30-
50 C, 35-50 C, or
50-75 C). In some embodiments, a device is configured to heat the
functionalization cartridge at
an elevated temperature (e.g., 20-60 C, 20-30 C, 25-40 C, 30-50 C, 35-50
C, or 50-75 C).
In some embodiments, the functionalization cartridge can be subjected to
microwaves or
sonication.
In some embodiments, purifying comprises passing the functionalized sample
through a
size exclusion medium. In some embodiments, the size exclusion medium may be a
column.
The column may be a desalting column. In some embodiments, the column is a
Zeba column
(e.g. a Zeba 7 kDa or a Zeba 40 kDa column). In some embodiments, the size
exclusion medium
is part of a fluidic device. In some embodiments, the size exclusion medium is
part of a system,
but is not part of a fluidic device of that system.
In some embodiments, purifying a protein comprises purification via
immunoprecipitation. In some embodiments, immunoprecipitation comprises
precipitating a
target protein out of sample (e.g., a sample before or after
functionalization) using an antibody
that specifically binds to the target protein.
In some embodiments, the one or more microfluidic channels are configured to
contain
and/or transport fluid(s) and/or reagent(s).
In some embodiments, any one of the cartridges comprises a base layer having a
surface
comprising channels. In some embodiments, the channels include the one or more
microfluidic
channels. In some embodiments, at least a portion of at least some of the
channels have a
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
8
substantially triangularly-shaped cross-section having a single vertex at a
base of the channel and
having two other vertices at the surface of the base layer. In some
embodiments, at least a
portion of at least some of the channels of any one of the cartridges have a
surface layer,
comprising an elastomer, configured to substantially seal off a surface
opening of the channel. In
some embodiments, the elastomer comprises silicone. In some embodiments, at
least one portion
of at least some of the channels have walls and a base comprising a
substantially rigid material
compatible with biological material. In some embodiments, any one of the
cartridges comprise
one or more fluid reservoirs. In some embodiments, at least some of the
channels connect to a
reservoir in a temperature zone. In some embodiments, at least some of the
channels connect to
an electrophoresis gel.
BRIEF DESCRIPTION OF DRAWINGS
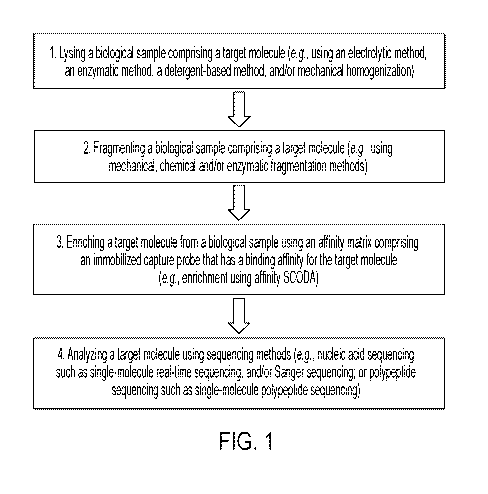
FIG. 1 shows an example method for preparing a target molecule from a
biological
sample (e.g., using an automated sample preparation device or cartridge of the
disclosure).
FIG. 2 shows an example workflow for sample preparation of a target protein
(e.g., using
an automated sample preparation device or cartridge of the disclosure).
FIG. 3 shows an example workflow for sample lysis (e.g., using an automated
device or
cartridge of the disclosure).
FIG. 4 shows an example workflow for sample enrichment of a target molecule
(e.g.,
using an automated device or cartridge of the disclosure).
FIG. 5 shows an example workflow for digestion of a target molecule (e.g.,
using an
automated device or cartridge of the disclosure).
FIGs. 6-7 shows example workflows for C-terminal functionalization of a target
protein
(e.g., using an automated device or cartridge of the disclosure).
FIG. 8 shows a schematic diagram of a cross-section view of a cartridge 100
along the
width of channels 102, in accordance with some embodiments.
FIGs. 9A-9B show a top view schematic diagram (FIG. 9A) and an image of
exemplary
cartridges of the disclosure.
FIGs. 10A-10B show sequencing data output from DNA libraries generated with
automated end-to-end (DNA extraction-to-finished library) sample preparation
using a sample
preparation device of the disclosure compared to libraries generated from
manually extracted and
purified DNA.
FIGs. 11A-11D show sequencing data output from a DNA library generated with
automated end-to-end (DNA extraction-to-finished library) sample preparation
using a sample
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
9
preparation device of the disclosure compared to DNA libraries derived from
samples that were
size selected using commercial and manual methods.
FIG. 12 shows an example of a C-terminal carboxylate coupling procedure.
FIG. 13 shows an example of a C-terminal carboxylate coupling procedure.
FIGs. 14A-14D show examples of C-terminal coupling procedures. FIG. 14A shows
representative functionalization of aspartic acid and glutamic acid terminated
peptides. FIG. 14B
shows representative functionalization of lysine and arginine terminated
peptides. FIG. 14C
shows an exemplary protection of sulfide moieties prior to functionalization
of a lysine
terminated peptide (Reaction 1), and an example of competitive intramolecular
cyclization,
which can be overcome using high concentrations of nucleophile and coupling
reagent (Reaction
2). FIG. 14D shows model functionalization of a lysine terminated peptide
(Reaction 3), and
model functionalization of an arginine terminated peptide having internal
glutamic acid and
aspartic acid residues (Reaction 4).
FIG. 15 shows a model C-terminal lysine coupling procedure.
FIGs. 16A-16C show data related to a model C-terminal lysine coupling
procedure. FIG.
16A and FIG. 16B show binding events to the N-terminus of QP126. The red arrow
denotes
when enzyme (peptidase) is added, after which a change in pulsing behavior is
observed due to
binding of the Clps to a different amino acid. FIG. 16C shows full length CRP
sequence with
bold fragments that were tagged).
FIG. 17 shows an example of a C-terminal lysine coupling procedure using the 4-
nitrovinyl sulfonamide reagent.
FIGs. 18A-18B show schemes related to an exemplary C-terminal lysine coupling
procedure using diazo transfer chemistry. FIG. 18A shows site-selective diazo
transfer. FIG. 18B
shows site-selective diazo transfer using a dipeptide followed by hydrolysis.
FIG. 19 shows an example of a lysine coupling procedure using diazo transfer.
FIG. 20 show representative schemes of solid-phase and solution-phase peptide
activation methods.
FIG. 21 shows an example of a functionalization process using an immobilized
carbodiimide reagent.
FIG. 22 shows an example of peptide surface immobilization.
FIGs. 23A-23B show representative examples of peptide sequencing. FIG. 23A
shows a
representative example of peptide sequencing by iterative cycles of terminal
amino acid
recognition and cleavage. FIG. 23B shows a representative example of dynamic
peptide
sequencing using a labeled amino acid recognition molecule and an exopeptidase
in a single
reaction mixture.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
FIGs. 24A-24F show schematic diagrams of exemplary sample preparation devices
of
the disclosure.
FIGs. 25-26 shows example workflows for C-terminal functionalization of a
target
protein (e.g., using an automated device or cartridge of the disclosure).
5 FIGs. 27A-27D show the results of sequencing peptide samples prepared in
an
exemplary fluidic device, according to certain embodiments.
DETAILED DESCRIPTION OF INVENTION
Sample Preparation Process
10 In some aspects, the disclosure provides processes for preparing a
sample, e.g., for
detection and/or analysis. In some embodiments, a process described herein may
be used to
identify properties or characteristics of a sample, including the identity or
sequence (e.g.,
nucleotide sequence or amino acid sequence) of one or more target molecules in
the sample. In
some embodiments, a process may include one or more sample transformation
steps, such as
sample lysis, sample purification, sample fragmentation, purification of a
fragmented sample,
library preparation (e.g., nucleic acid library preparation), purification of
a library preparation,
sample enrichment (e.g., using affinity SCODA), and/or detection/analysis of a
target molecule.
In some embodiments, a sample may be a purified sample, a cell lysate, a
single-cell, a
population of cells, or a tissue. In some embodiments, a sample is any
biological sample. In
some embodiments, a sample (e.g., a biological sample) is a blood, saliva,
sputum, feces, urine
or buccal swab sample. In some embodiments, a biological sample is from a
human, a non-
human primate, a rodent, a dog, a cat, a horse, or any other mammal. In some
embodiments, a
biological sample is from a bacterial cell culture (e.g., an E. coli bacterial
cell culture). A
bacterial cell culture may comprise gram positive bacterial cells and/or gram-
negative bacterial
cells. In some embodiments, a sample is a purified sample of nucleic acids or
proteins that have
been previously extracted via user-developed methods from metagenomic samples
or
environmental samples. A blood sample may be a freshly drawn blood sample from
a subject
(e.g., a human subject) or a dried blood sample (e.g., preserved on solid
media (e.g. Guthrie
cards)). A blood sample may comprise whole blood, serum, plasma, red blood
cells, and/or white
blood cells.
In some embodiments, a sample (e.g., a sample comprising cells or tissue), may
be
prepared, e.g., lysed (e.g., disrupted, degraded and/or otherwise digested) in
a process in
accordance with the instant disclosure. In some embodiments, a sample to be
prepared, e.g.,
lysed, comprises cultured cells, tissue samples from biopsies (e.g., tumor
biopsies from a cancer
patient, e.g., a human cancer patient), or any other clinical sample. In some
embodiments, a
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
11
sample comprising cells or tissue is lysed using any one of known physical or
chemical
methodologies to release a target molecule (e.g., a target nucleic acid or a
target protein) from
said cells or tissues. In some embodiments, a sample may be lysed using an
electrolytic method,
an enzymatic method, a detergent-based method, and/or mechanical
homogenization. In some
embodiments, a sample (e.g., complex tissues, gram positive or gram-negative
bacteria) may
require multiple lysis methods performed in series. In some embodiments, if a
sample does not
comprise cells or tissue (e.g., a sample comprising purified nucleic acids), a
lysis step may be
omitted. In some embodiments, lysis of a sample is performed to isolate target
nucleic acid(s).
In some embodiments, lysis of a sample is performed to isolate target
protein(s). In some
embodiments, a lysis method further includes use of a mill to grind a sample,
sonication, surface
acoustic waves (SAW), freeze-thaw cycles, heating, addition of detergents,
addition of protein
degradants (e.g., enzymes such as hydrolases or proteases), and/or addition of
cell wall digesting
enzymes (e.g., lysozyme or zymolase). Exemplary detergents (e.g., non-ionic
detergents) for
lysis include polyoxyethylene fatty alcohol ethers, polyoxyethylene
alkylphenyl ethers,
polyoxyethylene-polyoxypropylene block copolymers, polysorbates and
alkylphenol ethoxylates,
preferably nonylphenol ethoxylates, alkylglucosides and/or polyoxyethylene
alkyl phenyl ethers.
In some embodiments, lysis methods involve heating a sample for at least 1-30
min, 1-25 min, 5-
min, 5-20 min, 10-30 min, 5-10 min, 10-20 min, or at least 5 min at a desired
temperature
(e.g., at least 60 C, at least 70 C, at least 80 C, at least 90 C, or at
least 95 C).
20 In some embodiments, a sample is prepared, e.g., lysed, in the presence
of a buffer
system. This buffer system may be used to make a slurry of the sample, to
suspend the sample,
and/or to stabilize the sample during any known lysis methodology, including
those methods
described herein. In some embodiments, a sample is prepared, e.g., lysed, in
the presence of
RIPA buffer, GCI buffer that comprises Guanidine-HC1 buffer, Gly-NP40 buffer,
a TRIS buffer,
25 a HEPES buffer, or any other known buffering solution.
Many of the lysis methods described herein allow for the sample to be lysed by
mechanically
homogenizing the sample such that the cell walls of the sample break down. For
example,
methods that cause lysis by mechanical homogenization include, but are not
limited to bead-
beating, heating (e.g., to high temperatures sufficient to disrupt cell walls,
e.g., greater than 50
C, 60 C, 70 C, 80 C, 90 C, or 95 C), syringe/needle/microchannel passage
(to cause
shearing), sonication, or maceration with a grinder. In some embodiments, any
lysis
methodology may be combined with any other lysis methodology. For example, any
lysis
methodology may be combined with heating and/or sonication and/or
syringe/needle/microchannel passage to quicken the rate of lysis.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
12
In some embodiments, sample preparation comprises cell disruption (i.e.,
subsequent removal of
unwanted cell and tissue elements following lysis). In some embodiments, cell
disruption
involves protein and/or nucleic acid precipitation. In some embodiments,
following
precipitation, the lysed and disrupted sample is subjected to centrifugation.
In some
embodiments, following centrifugation, the supernatant is discarded.
Precipitation can be
accomplished through multiple processes, including but not limited to those
methods described
in Winter, D. and H. Steen (2011). "Optimization of cell lysis and protein
digestion protocols for
the analysis of HeLa S3 cells by LC-MS/MS." PROTEOMICS 11(24): 4726-4730. In
some
embodiments, proteins or peptides are immunoprecipitated. In some embodiments,
centrifugation of precipitated proteins and/or nucleic acids is followed by
discarding of the
supernatant and subsequent washing of the pellet fraction (e.g., washing using
chloroform/methanol or trichloroacetic acid).
In some embodiments, a sample is prepared using lysis in the presence of a
lysis buffer
(e.g., GCI buffer (6M Guanidine HC1, 0.1 M TEAB, 1% Triton X-100, a standard
buffer, and
1mM EDTA/EGTA)) and disrupted by needle shearing (e.g., by passage of the
sample through a
26.5 gauge needle, e.g., at 4 C). In some embodiments, a lysed and disrupted
sample is further
subjected to precipitation of proteins and/or nucleic acids (e.g., using
trichloroacetic acid at 4 C
with vortexing) and optionally followed by centrifugation. In some
embodiments, a sample is
prepared as described in FIG. 3.
In some embodiments, a sample (e.g., a sample comprising a target nucleic acid
or a
target protein) may be purified, e.g., following lysis, in a process in
accordance with the instant
disclosure. In some embodiments, a sample may be purified using chromatography
(e.g., affinity
chromatography that selectively binds the sample) or electrophoresis. In some
embodiments, a
sample may be purified in the presence of precipitating agents. In some
embodiments, after a
purification step or method, a sample may be washed and/or released from a
purification matrix
(e.g., affinity chromatography matrix) using an elution buffer. In some
embodiments, a
purification step or method may comprise the use of a reversibly switchable
polymer, such as an
electroactive polymer. In some embodiments, a sample may be purified by
electrophoretic
passage of a sample through a porous matrix (e.g., cellulose acetate, agarose,
acrylamide).
In some embodiments, a sample (e.g., a sample comprising a target nucleic acid
or a
target protein) may be fragmented (i.e., digested) in a process in accordance
with the instant
disclosure. In some embodiments, a nucleic acid sample may be fragmented to
produce small
(<1 kilobase) fragments for sequence specific identification to large (up to
10+ kilobases)
fragments for long read sequencing applications. Fragmentation of nucleic
acids or proteins
may, in some embodiments, be accomplished using mechanical (e.g., fluidic
shearing), chemical
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
13
(e.g., iron (Fe+) cleavage) and/or enzymatic (e.g., restriction enzymes,
tagmentation using
transposases) methods. In some embodiments, a protein sample may be fragmented
to produce
peptide fragments of any length. Fragmentation of proteins may, in some
embodiments, be
accomplished using chemical and/or enzymatic (e.g., proteolytic enzymes such
as trypsin)
methods. In some embodiments, mean fragment length may be controlled by
reaction time,
temperature, and concentration of sample and/or enzymes (e.g., restriction
enzymes,
transposases). In some embodiments, a nucleic acid may be fragmented by
tagmentation such
that the nucleic acid is simultaneously fragmented and labeled with a
fluorescent molecule (e.g.,
a fluorophore). In some embodiments, a fragmented sample may be subjected to a
round of
purification (e.g., chromatography or electrophoresis) to remove small and/or
undesired
fragments as well as residual payload, chemicals and/or enzymes (e.g.,
transposases) used during
the fragmentation step. For example, a fragmented sample (e.g., sample
comprising nucleic
acids) may be purified from an enzyme (e.g., a transposase), wherein the
purification comprises
denaturing the enzyme (e.g., by a combination of heat, chemical (e.g. SDS),
and enzymatic (e.g.
proteinase K) processes).
In some embodiments, the target molecule(s) is fragmented/digested prior to
enrichment.
In some embodiments, the target molecule is fragmented/digested after
enrichment. In some
embodiments, the target molecule(s) is fragmented/digested without any
enrichment of the target
molecule(s).
Fragmentation/digestion can be conducted using any known method, but typically
will
involve a non-enzymatic or enzymatic method. Non-enzymatic methods typically
have an
advantage as it relates to speed, simplicity, robustness, and ease of
automation. These
approaches include, but are not limited to, acid hydrolysis and/or cleavage
using a chemical
entity such as cyanogen bromide, hydroxylamine, iodosobenzoic acid, dimethyl
sulfoxide¨hydrochloric acid, BNPS-skatole [2-(2-nitrophenylsulfenyl) -3-
methylindole], or 2-
nitro-5-thiocyanobenzoic acid. Non-enzymatic, electro-physical digestion
methods have been
employed as well, including electrochemical oxidation and/or digestion in
conjunction with
microwaves. Enzymatic methods typically utilize proteases to fragment protein
into component
peptides. These enzymes include trypsin (which is typically favored for the
size of the peptides
.. generated and the generation of a basic residue at the carboxyl terminus of
the peptide),
chymotrypsin, LysC, LysN, AspN, GluC and/or ArgC.
Enzymatic fragmentation/digestion methods may be optimized for ease of use,
speed,
automation and/or effectiveness. In some embodiments, enzymatic methods
include enzyme
immobilization on solid substrates. In some embodiments, enzymatic methods are
performed in
flow (e.g., in a microfluidic channel).
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
14
Fragmentation/digestion methods may be performed using an automated device or
module. Alternatively, or in addition, fragmentation/digestion methods may be
performed
manually. An enzymatic digestion may utilize any number or combination of
enzymes and may
further comprise any of the known non-enzymatic methods.
In some embodiments, a fragmentation/digestion process is as described in FIG.
5. In
some embodiments, a sample comprising target protein(s) is first denatured and
reduced (e.g.,
using acetonitrile and TCEP). In some embodiments, target protein(s) to be
fragmented are
subjected to capping of an amino acid side chain (e.g., a cysteine block)
(e.g., using an amino
acid side chain capping agent). In some embodiments, target protein(s) are
fragmented using a
mixture of trypsin and LysC (e.g., for 120 minutes). Enzymatic reactions may
be quenched (e.g.,
using sodium carbonate buffer).
Any suitable reducing agent may be used to reduce a target protein within a
sample. In
some embodiments, the reducing agent is suitable for reducing a disulfide-
bond. In some
embodiments, the reducing agent may reversibly reduce a disulfide bond.
Suitable reversable
reducing agents may comprise compounds such as dithiothreitol (DTT), P-
mercaptoethanol
(BME), and/or Glutathione (GSH). In some embodiments, the reducing agent may
irreversibly
reduce a disulfide bond. Suitable irreversible reducing agents may comprise
compounds such as
tris(2-carboxyethyl)phosphine (TCEP). In some specific embodiments, the
reducing agent
comprises tris(2-carboxyethyl)phosphine (TCEP).
Any suitable amino acid side chain capping agent may be used to cap amino acid
side
chains of a protein within a peptide sample. In some embodiments, the amino
acid side chain
capping agent prevents the formation of disulfide bonds. In some embodiments,
the amino acid
side chain capping agent prevents the amino acid side chain from undergoing
further reactivity
such as nucleophile/electrophile or redox reactivity. In some embodiments, the
amino acid side
.. chain capping agent is a cysteine capping agent. In some embodiments, the
amino acid side
chain capping agent is a sulfhydryl-reactive alkylating reagent (e.g. a
cysteine alkylation agent).
For instance, in some embodiments, the amino acid side chain capping agent
comprises a
haloacetamide (e.g. chloroacetamide, iodoacetamide) or a
haloacetate/haloacetic acid (e.g.,
chloroacetate/chloroacetic acid, iodoacetate/iodoacetic acid). In some
embodiments, the amino
acid side chain capping agent is an aromatic benzyl halide. Other examples of
suitable cysteine
alkylating agents include 4-vinylpyridine, acrylamide, and
methanethiosulfonate, In some
embodiments, the amino acid side chain capping agent comprises iodoacetamide.
In some embodiments, a sample comprising a target nucleic acid may be used to
generate
a nucleic acid library for subsequent analysis (e.g., genomic sequencing) in a
process in
accordance with the instant disclosure. A nucleic acid library may be a linear
library or a
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
circular library. In some embodiments, nucleic acids of a circular library may
comprise elements
that allow for downstream linearization (e.g., endonuclease restriction sites,
incorporation of
uracil). In some embodiments, a nucleic acid library may be purified (e.g.,
using
chromatography, e.g., affinity chromatography), or electrophoresis.
5 In some embodiments, a library of nucleic acids (e.g., linear nucleic
acids) is prepared
using end-repair, a process wherein a combination of enzymes (e.g., Taq DNA
Ligase,
Endonuclease IV, Bst DNA Polymerase, Fpg, Uracil-DNA Glycosylase, T4
Endonuclease V
and/or Endonuclease VIII) extend the 3' end of the nucleic acids, generating a
complement to
the 5' payload, and repairing any abasic sites or nicks in the nucleic acids.
In some embodiments,
10 a library of linear nucleic acids is prepared using a self-priming
hairpin adaptor, a process which
may obviate the need to anneal a unique sequencing primer to an individual
nucleic acid
fragment primer prior to formation of a polymerase complex. Following end-
repair, a library of
nucleic acids (e.g., linear nucleic acids) may be purified using solid-phase
adsorption with
subsequent elution into a fresh buffer, using passage of the nucleic acids
through a size-selective
15 matrix (e.g., agarose gel). The size-selective matrix may be used to
remove nucleic acid
fragments that are smaller than the size of the target nucleic acids.
In some embodiments, a sample (e.g., a sample comprising a target nucleic acid
or a
target protein) may be enriched for a target molecule in a process in
accordance with the instant
disclosure. Enrichment is typically used when the complexity of the un-
enriched sample exceeds
the capacity of the sequencing platform, or when the target molecule is
present in the sample at a
low abundance (e.g., such that it cannot be easily detected by the sequencing
platform).
Enrichment involves the use of a mechanism that selectively amplifies the
target molecule. This
enrichment may involve the use of antibodies, aptamers, size-based selection,
or electrostatic
charge-based selection in order to selectively amplify the target molecule(s)
(e.g., target
protein(s) or target nucleic acid(s)).
Enrichment may typically be used when the intent of the sample preparation is
to
sequence specific target molecules. Enrichment may be used to perform or
conduct a proteomic,
genomic, or metagenomic analysis or survey, when the target molecules are
related or
homologous to one another.
In some embodiments, a sample is enriched for a target molecule using an
electrophoretic
method. In some embodiments, a sample is enriched for a target molecule using
affinity
SCODA. In some embodiments, a sample is enriched for a target molecule using
field inversion
gel electrophoresis (FIGE). In some embodiments, a sample is enriched for a
target molecule
using pulsed field gel electrophoresis (PFGE). In some embodiments, the matrix
used during
enrichment (e.g., a porous media, electrophoretic polymer gel) comprises
immobilized affinity
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
16
agents (also known as 'immobilized capture probes') that bind to target
molecule present in the
sample. In some embodiments, a matrix used during enrichment comprises 1, 2,
3, 4, 5, or more
unique immobilized capture probes, each of which binds to a unique target
molecule and/or bind
to the same target molecule with different binding affinities.
In some embodiments, an immobilized capture probe is an oligonucleotide
capture probe
that hybridizes to a target nucleic acid. In some embodiments, an
oligonucleotide capture probe
is at least 50%, 60%, 70%, 80%, 90% 95%, or 100% complementary to a target
nucleic acid. In
some embodiments, a single oligonucleotide capture probe may be used to enrich
a plurality of
related target nucleic acids (e.g., 2, 3, 4, 5, 6, 7, 8, 9, 10, 20, 30, 40,
50, or more related target
nucleic acids) that share at least 50%, 60%, 70%, 80%, 90% 95%, or 99%
sequence identity.
Enrichment of a plurality of related target nucleic acids may allow for the
generation of a
metagenomic library. In some embodiments, an oligonucleotide capture probe may
enable
differential enrichment of related target nucleic acids. In some embodiments,
an oligonucleotide
capture probe may enable enrichment of a target nucleic acid relative to a
nucleic acid of
identical sequence that differs in its modification state (e.g., single
nucleotide polymorphism,
methylation state, acetylation state). In some embodiments, an oligonucleotide
capture probe is
used to enrich human genomic DNA for a specific gene of interest (e.g., HLA).
A specific gene
of interest may be a gene that is relevant to a specific disease state or
disorder. In some
embodiments, an oligonucleotide capture probe is used to enrich nucleic
acid(s) of a
metagenomic sample.
In some embodiments, for the purposes of enriching nucleic acid target
molecules with a
length of 0.5-2 kilobases, oligonucleotide capture probes may be covalently
immobilized in an
acrylamide matrix using a 5' Acrydite moiety. In some embodiments, for the
purposes of
enriching larger nucleic acid target molecules (e.g., with a length of >2
kilobases),
oligonucleotide capture probes may be immobilized in an agarose matrix. In
some
embodiments, oligonucleotide capture probes may be immobilized in an agarose
matrix using
thiol-epoxide chemistries (e.g., by covalently attached thiol-modified
oligonucleotides to
crosslinked agarose beads). Oligonucleotide capture probes linked to agarose
beads can be
combined and solidified within standard agarose matrices (e.g., at the same
agarose percentage).
In some embodiments, enrichment of nucleic acids using methods described
herein (e.g.,
enrichment using SCODA) produces nucleic acid target molecules that comprise a
length of
about 0.5 kilobases (kb), about 1 kb, about 1.5 kb, about 2 kb, about 3 kb,
about 4 kb, about 5 kb,
about 6 kb, about 7 kb, about 8 kb, about 9 kb, about 10 kb, about 12 kb,
about 15 kb, about 20
kb, or more. In some embodiments, enrichment of nucleic acids using methods
described herein
(e.g., enrichment using SCODA) produces nucleic acid target molecules that
comprise a length
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
17
of about 0.5-2 kb, 0.5-5 kb, 1-2 kb, 1-3 kb, 1-4 kb, 1-5 kb, 1-10 kb, 2-10 kb,
2-5 kb, 5-10 kb, 5-
15 kb, 5-20 kb, 5-25 kb, 10-15 kb, 10-20 kb, or 10-25 kb.
In some embodiments, an immobilized capture probe is a protein capture probe
(e.g., an
aptamer or an antibody) that binds to a target protein or peptide fragment. In
some
.. embodiments, a protein capture probe binds to a target protein or peptide
fragment with a
binding affinity of 10-9 to 10-8 M, 10-8 to 10-7 M, 10-7 to 10-6 M, 10-6 to 10-
5 M, 10-5 to 10-4 M, 10-
4 to 10-3 M, or 10-3 to 10-2 M. In some embodiments, the binding affinity is
in the picomolar to
nanomolar range (e.g., between about 10-12 and about 10-9 M). In some
embodiments, the
binding affinity is in the nanomolar to micromolar range (e.g., between about
10-9 and about 10-6
M). In some embodiments, the binding affinity is in the micromolar to
millimolar range (e.g.,
between about 10-6 and about 10-3 M). In some embodiments, the binding
affinity is in the
picomolar to micromolar range (e.g., between about 10-12 and about 10-6 M). In
some
embodiments, the binding affinity is in the nanomolar to millimolar range
(e.g., between about
10-9 and about 10-3 M). In some embodiments, a single protein capture probe
may be used to
enrich a plurality of related target proteins that share at least 50%, 60%,
70%, 80%, 90% 95%, or
99% sequence identity. In some embodiments, a single protein capture probe may
be used to
enrich a plurality of related target proteins (e.g., 2, 3, 4, 5, 6, 7, 8, 9,
10, 20, 30, 40, 50, or more
related target proteins) that share at least 50%, 60%, 70%, 80%, 90% 95%, or
99% sequence
homology. Enrichment of a plurality of related target proteins may allow for
the generation of a
metaproteomics library. In some embodiments, a protein capture probe may
enable differential
enrichment of related target proteins.
In some embodiments, multiple capture probes (e.g., populations of multiple
capture
probe types, e.g., that bind to deterministic target molecules of infectious
agents such as
adenovirus, staphylococcus, pneumonia, or tuberculosis) may be immobilized in
an enrichment
matrix. Application of a sample to an enrichment matrix with multiple
deterministic capture
probes may result in diagnosis of a disease or condition (e.g., presence of an
infectious agent).
In some embodiments, a target molecule or related target molecules may be
released from the
enrichment matrix after removal of non-target molecules, in a process in
accordance with the
instant disclosure. In some embodiments, a target molecule may be released
from the
enrichment matrix by increasing the temperature of the enrichment matrix.
Adjusting the
temperature of the matrix further influences migration rate as increased
temperatures provide a
higher capture probe stringency, requiring greater binding affinities between
the target molecule
and the capture probe. In some embodiments, when enriching related target
molecules, the
matrix temperature may be gradually increased in a step-wise manner in order
to release and
isolate target molecules in steps of ever-increasing homology. In some
embodiments,
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
18
temperature is increased by about 5%, 10%, 15%, 20%, 25%, 30%, 40%, 50%, or
more in each
step or over a period of time (e.g., 1-10 min, 1-5 min, or 4-8 min). In some
embodiments,
temperature is increased by 5%-10%, 5-15%, 5%-20%, 5%-25%, 5%-30%, 5%-40%, 5%-
50%,
10%-25%, 20%-30%, 30%-40%, 35%-50%, or 40%-70% in each step or over a period
of time
.. (e.g., 1-10 min, 1-5 min, or 4-8 min). In some embodiments, temperature is
increased by about 1
C, 2 C, 3 C, 4 C, 5 C, 6 C, 7 C, 8 C, 9 C, or 10 C in each step or
over a period of time
(e.g., 1-10 min, 1-5 min, or 4-8 min). In some embodiments, temperature is
increased by 1-10
C, 1-5 C, 2-5 C, 2-10 C, 3-8 C, 4-9 C, or 5-10 C in each step or over a
period of time
(e.g., 1-10 min, 1-5 min, or 4-8 min). This may allow for the sequencing of
target proteins or
target nucleic acids that are increasingly distant in their relation to an
initial reference target
molecule, enabling discovery of novel proteins (e.g., enzymes) or functions
(e.g., enzymatic
function or gene function). In some embodiments, when using multiple capture
probes (e.g.,
multiple deterministic capture probes), the matrix temperature may be
increased in a step-wise or
gradient fashion, permitting temperature-dependent release of different target
molecules and
resulting in generation of a series of barcoded release bands that represent
the presence or
absence of control and target molecules.
Enrichment of a sample (e.g., a sample comprising a target nucleic acid or a
target
protein) allows for a reduction in the total volume of the sample. For
example, in some
embodiments, the total volume of a sample is reduced after enrichment by at
least 10%, at least
20%, at least 30%, at least 40%, at least 50%, at least 60%, at least 70%, at
least 80%, at least
90%, at least 100%, or at least 120%. In some embodiments, the total volume of
a sample is
reduced after enrichment from 1-20 mL initial volume to 100-1000 i.t.L final
volume, from 1-5
mL initial volume to 100-1000 i.t.L final volume, from 100-1000 i.t.L initial
volume to 25-100 i.t.L
final volume, from 100-500 i.t.L initial volume to 10-100 i.t.L final volume,
or from 50-200 i.t.L
initial volume to 1-25 i.t.L final volume. For example, in some embodiments,
the final volume of
a sample after enrichment is 10-100 t.L, 10-50 t.L, 10-25 t.L, 20-100 t.L, 20-
50 t.L, 25-100 t.L,
25-250 t.L, 25-1000 t.L, 100-1000 t.L, 100-500 t.L, 100-250 t.L, 200-1000 t.L,
200-500 t.L,
200-750 t.L, 500-1000 t.L, 500-1500 t.L, 500-750 t.L, 1-5 mL, 1-10 mL, 1-2 mL,
1-3 mL, or 1-4
mL.
In addition to amplification of the target molecule, or as an alternative to
amplification of
the target molecule, a sample may be enriched (e.g., for a low abundance
target molecule) by
depletion of unwanted non-target molecules (e.g., high-abundance proteins
(e.g. albumin)).
Depletion of unwanted non-target molecules may be performed using similar
capture strategies
as discussed above. When using a depletion strategy, the capture probes will
bind to unwanted,
non-target molecules and allow for target molecules to remain in solution.
This strategy equally
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
19
enables enrichment of the target molecule (i.e.., increased relative
concentrations of the target
molecule(s)).
For example, an immobilized capture probe that is used for depletion may be an
oligonucleotide capture probe that hybridizes to an unwanted non-target
nucleic acid. In some
embodiments, an oligonucleotide capture probe that is used for depletion is at
least 50%, 60%,
70%, 80%, 90% 95%, or 100% complementary to an unwanted non-target nucleic
acid. In some
embodiments, a single oligonucleotide capture probe that is used for depletion
may be used to
deplete a plurality of related target nucleic acids (e.g., 2, 3, 4, 5, 6, 7,
8, 9, 10, 20, 30, 40, 50, or
more related target nucleic acids) that share at least 50%, 60%, 70%, 80%, 90%
95%, or 99%
sequence identity.
In some embodiments, an immobilized capture probe that is used for depletion
is a
protein capture probe (e.g., an aptamer or an antibody) that binds to an
unwanted non-target
protein or peptide fragment. In some embodiments, a protein capture probe that
is used for
depletion binds to an unwanted non-target protein or peptide fragment with a
binding affinity of
10-9 to 10-8 M, 10-8 to 10-7 M, 10-7 to 10-6 M, 10-6 to 10-5 M, 10-5 to 104 M,
104 to 10-3 M, or 10-3
to 10-2 M. In some embodiments, the binding affinity is in the nanomolar to
millimolar range
(e.g., between about 10-9 and about 10-3 M). In some embodiments, a single
protein capture
probe that is used for depletion may be used to deplete a plurality of related
target proteins that
share at least 50%, 60%, 70%, 80%, 90% 95%, or 99% sequence identity. In some
embodiments, a single protein capture probe that is used for depletion may be
used to deplete a
plurality of related target proteins (e.g., 2, 3, 4, 5, 6, 7, 8, 9, 10, 20,
30, 40, 50, or more related
target proteins) that share at least 50%, 60%, 70%, 80%, 90% 95%, or 99%
sequence homology.
In some embodiments, enrichment comprises amplification of target molecule(s)
and depletion
(e.g., of high abundance proteins). In some embodiments, depletion steps are
performed before
amplification and enrichment of target molecule(s). In some embodiments, in
order to avoid
possible contamination of the target molecule(s) by the capture elements of
the enrichment
process (e.g., antibodies or aptamers), the capture elements are depleted from
an enriched sample
(i.e., after enrichment by either amplification of target molecules and/or
depletion of unwanted
non-target molecules from the original sample).
In some embodiments, a sample is first subjected to a depletion step (e.g., to
remove
unwanted non-target proteins). In some embodiments, a sample is enriched using
amplification
or immobilized target capture (e.g., using antibodies to selectively enrich
for a target protein)
following a first depletion step. Following amplification or immobilized
target capture, the
sample may then be subjected to a second depletion step (e.g., to remove
excess antibody or
capture probe). In some embodiments, a sample is enriched, for example, as
described in FIG. 4.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
In some embodiments, any number of enrichment steps (e.g., amplification of
target
molecule(s) and/or depletion(s)) can be performed by the automated device or
module (e.g., on a
chip or cartridge). In some embodiments, the enrichment steps are amenable to
automation on
the cartridge using capture elements (e.g., antibodies) immobilized on solid
phase structures.
5 In some embodiments, any immobilized capture element or probe described
herein may be on
any solid support structure or surface. The solid support structure or surface
may be magnetic
and/or may be a frit, a filter, a chip, or a cartridge surface. In some
embodiments, the capture
elements or probes for enrichment may be interchanged (e.g., using flow on a
chip).
In some embodiments, any number of the enrichment steps are performed
manually. If
10 performed manually, any enriched target molecule may be subsequently
placed into an
automated sample preparation device described herein.
In some embodiments, a target molecule or target molecules may be detected
after
enrichment and subsequent release to enable analysis of said target
molecule(s) and its upstream
sample, in a process in accordance with the instant disclosure. In some
embodiments, a target
15 nucleic acid may be detected using gene sequencing, absorbance,
fluorescence, electrical
conductivity, capacitance, surface plasmon resonance, hybrid capture,
antibodies, direct labeling
of the nucleic acid (e.g., end-labeling, labeled tagmentation payloads), non-
specific labeling with
intercalating dyes (e.g., ethidium bromide, SYBR dyes), or any other known
methodology for
nucleic acid detection. In some embodiments, a target protein or peptide
fragment may be
20 detected using absorbance, fluorescence, mass spectroscopy, amino acid
sequencing, or any
other known methodology for protein or peptide detection.
Sample Preparation Devices and Modules
Devices or modules including apparatuses, cartridges (e.g., comprising
channels (e.g.,
microfluidic channels)), and/or pumps (e.g., peristaltic pumps) for use in a
process of preparing a
sample for analysis are generally provided. Devices can be used in accordance
with the instant
disclosure to promote capture, concentration, manipulation, and/or detection
of a target molecule
from a biological sample. In some embodiments, devices and related methods are
provided for
automated processing of a sample to produce material for next generation
sequencing and/or
other downstream analytical techniques. Devices and related methods may be
used for
performing chemical and/or biological reactions, including reactions for
nucleic acid and/or
protein processing in accordance with sample preparation or sample analysis
processes described
elsewhere herein.
A sample preparation device or module may, in some embodiments, perform any
number
of the following sample preparation steps:
CA 03177368 2022-09-27
WO 2021/216763 PCT/US2021/028471
21
(1) Cell or tissue preparation (e.g., lysis); and/or
(2) Enrichment of at least one target molecule (e.g., at least one target
nucleic acid
and/or at least one target protein); and/or
(3) Digestion or fragmentation of the at least one target molecule (e.g.,
at least one
target nucleic acid and/or at least one target protein); and/or
(4) Terminal functionalization of the at least one target molecule (e.g., C-
terminal
functionalization of a target protein).
In some embodiments, a sample preparation device or module performs sample
preparation steps as shown in FIG. 1. In some embodiments, a sample
preparation device or
module performs sample preparation steps as shown in FIG. 2.
In some embodiments, a sample preparation device or module performs all of
steps (1)-
(4). In some embodiments, a sample preparation device or module performs step
(1) and
optionally performs steps (2)-(4). In some embodiments, a sample preparation
device or module
performs step (1) and optionally performs steps (2)-(3). In some embodiments,
a sample
preparation device or module performs step (1) and optionally performs step
(2). In some
embodiments, a sample preparation device or module performs step (1) and
optionally performs
steps (3)-(4). In some embodiments, a sample preparation device or module
performs step (1)
and optionally performs step (3). In some embodiments, a sample preparation
device or module
performs step (1) and optionally performs step (4). In some embodiments, a
sample preparation
device or module does not perform step (1) and only performs steps (2)-(4). In
some
embodiments, a sample preparation device or module does not perform step (1)
and only
performs steps (3)-(4). In some embodiments, a sample preparation device or
module does not
perform step (1) and only performs steps (2) and (4). In some embodiments, a
sample
preparation device or module does not perform step (1) and only performs one
of steps (2), (3),
or (4). The order of steps can be altered as necessary for an experiment. For
example, step (3) ¨
digestion or fragmentation ¨ can precede step (2) ¨ enrichment. In some
embodiments, the at
least one target molecule can be purified after step (1), and/or step (2),
and/or step (3), and/or
step 4. In some embodiments, any one of the steps is interspersed with manual
steps. This
flexibility enables the user to address multiple sample types and sequencing
platforms.
In some embodiments, a sample preparation device or module is positioned to
deliver or transfer
to a sequencing module or device a target molecule or a plurality of target
molecules (e.g., target
nucleic acids or target proteins). In some embodiments, a sample preparation
device or module
is connected directly to (e.g., physically attached to) or indirectly to a
sequencing device or
module.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
22
In some embodiments, a sample preparation device or module is used to prepare
a sample
for diagnostic purposes. In some embodiments, a sample preparation device that
is used to
prepare a sample for diagnostic purposes is positioned to deliver or transfer
to a diagnostic
module or diagnostic device a target molecule or a plurality of molecules
(e.g., target nucleic
.. acids or target proteins). In some embodiments, a sample preparation device
or module is
connected directly to (e.g., physically attached to) or indirectly to a
diagnostic device.
In some embodiments, a device comprises a cartridge housing that is configured
to receive one
or more cartridges (e.g., configured to receive one cartridge at a time). FIG.
24A shows a
schematic diagram of sample preparation device 300, in accordance with some
embodiments. A
device (e.g., a sample preparation device comprising a cartridge housing) may
be configured to
receive one or more cartridges (or two or more, or three or more, and so on)
either sequentially
or simultaneously. Sample preparation device 300, for example, can be
configured to receive
one or more of lysis cartridge 301, enrichment cartridge 302, fragmentation
cartridge 303, and/or
functionalization cartridge 304 simultaneously or sequentially. It should be
understood that the
.. device need not be configured to receive each of the four cartridges shown
in FIG. 4A in all
embodiments. For example, in some embodiments sample preparation device 300 is
configured
to receive only lysis cartridge 301 and enrichment cartridge 302, with
fragmentation and
functionalization performed manually rather than in an automated fashion.
The sample preparation device may further comprise a pump configured to
transport
components (e.g., reagents, samples) in the received cartridges (e.g., within
a channels/reservoirs
of a cartridge or into and/or out of a cartridge). For example, referring to
FIG. 24B, sample
preparation device 300 may comprise pump 305 configured to transport
components in one or
more of lysis cartridge 301, enrichment cartridge 302, fragmentation cartridge
303, and/or
functionalization cartridge 304. In some embodiments, a pump comprises an
apparatus and a
received cartridge, and an interaction between the apparatus of the pump and
cartridge causes
fluid flow. For example, pump 305 may be a peristaltic pump, and apparatus 306
may
operatively couple to a cartridge (e.g., cartridge 301) to cause fluid motion
in the cartridge (e.g.,
when apparatus 306 comprises a roller and cartridge 301 comprises a flexible
surface deformable
by the roller). Further description of exemplary peristaltic pump methods and
devices are
described in more detail below.
As mentioned elsewhere, a prepared sample from the sample preparation device
may be
transported (directly or indirectly) to a downstream detection module (e.g., a
sequencing module,
a diagnostic module). For example, FIG. 24C shows an embodiment in which
conduit 308
connects sample preparation device 300 and detection module 307 (e.g., a
sequencing module).
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
23
Sample preparation device 300 and detection module 307 may be directly
connected (e.g.,
physically attached) or may be connected indirectly (e.g., via one or more
intervening modules).
While in some embodiments various steps of the processes are performed in
separate
cartridges (e.g., a lysis step in a lysis cartridge, an enrichment step in an
enrichment cartridge, a
.. fragmentation step in a fragmentation cartridge, a functionalization step
in a functionalization
cartridge), in other embodiments two or more (or all) such steps may be
performed in a single
cartridge. For example, a cartridge may comprise different regions for
different steps of an
overall process (each region comprising various reservoirs, channels, and/or
microchannels for
performing a respective step). FIG. 24D depicts a schematic illustration of
one such
embodiment, where cartridge 401 comprises lysis region 402, enrichment region
403,
fragmentation region 404, and functionalization region 405. It should be
understood that while
cartridge 401 shows regions for four such steps, the depiction is purely
illustrative, and more or
fewer regions for more or fewer steps may be present on a given cartridge
(e.g., a cartridge may
comprise only a lysis region and an enrichment region, or various other
combinations). Sample
preparation device 400 may be configured to receive cartridge 401, as shown in
FIG. 24D
according to certain embodiments. As in the embodiments described in FIGS. 24B-
24C, sample
preparation device 400 may comprise pump 406 comprising apparatus 407 to
operatively couple
to cartridge 407 (e.g., to transport components such as fluids), as shown in
FIG. 24E. Further, as
shown in FIG. 24F, conduit 408 can connect sample preparation device 400 to
downstream
detection module 409 (e.g., a sequencing module, a diagnostic module), in
accordance with
certain embodiments. Such a connection may allow transportation of a prepared
sample from
sample preparation device 400 to detection module 409 directly or indirectly,
according to
certain embodiments.
In some embodiments, a cartridge comprises one or more reservoirs or reaction
vessels
configured to receive a fluid and/or contain one or more reagents used in a
sample preparation
process. In some embodiments, a cartridge comprises one or more channels
(e.g., microfluidic
channels) configured to contain and/or transport a fluid (e.g., a fluid
comprising one or more
reagents) used in a sample preparation process. Reagents include buffers,
enzymatic reagents,
polymer matrices, capture reagents, size-specific selection reagents, sequence-
specific selection
reagents, and/or purification reagents. Additional reagents for use in a
sample preparation
process are described elsewhere herein.
In some embodiments, a cartridge includes one or more stored reagents (e.g.,
of a liquid
or lyophilized form suitable for reconstitution to a liquid form). The stored
reagents of a
cartridge include reagents suitable for carrying out a desired process and/or
reagents suitable for
processing a desired sample type. In some embodiments, a cartridge is a single-
use cartridge
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
24
(e.g., a disposable cartridge) or a multiple-use cartridge (e.g., a reusable
cartridge). In some
embodiments, a cartridge is configured to receive a user-supplied sample. The
user-supplied
sample may be added to the cartridge before or after the cartridge is received
by the device, e.g.,
manually by the user or in an automated process. In some embodiments, a
cartridge is a sample
preparation cartridge. In some embodiments, a sample preparation cartridge is
capable of
isolating or purifying a target molecule (e.g., a target nucleic acid or
target protein) from a
sample (e.g., a biological sample).
FIG. 9A shows a top view schematic diagram of one embodiment of cartridge 200,
in
accordance with certain embodiments. Cartridge 200 may be configured to
perform one or more
of a variety of processes described in this disclosure, such a lysis,
enrichment, depletion,
fragmentation, and/or terminal functionalization of target molecules from
fluid samples (e.g.,
biological samples). Configuration of a cartridge for any of these processes
may be determined,
for example, by the presence of reagents selected for the process in the
cartridge (e.g., in a
reservoir, reaction vessel or channel of the cartridge). For example,
cartridge 200 in FIG. 9A can
comprise first reagent reservoir 201 comprising or capable of comprising
reagents for a first step
of a process (e.g., purification/size selection reagents), second reagent
reservoirs 202 comprising
or capable of comprising reagents for a second step of a process (e.g., target
molecule extraction
reagents), and third reagent reservoirs 203 comprising or capable of
comprising reagents for a
third step of a process (e.g., library preparation reagents). Some such
reagents may be stored in
.. reservoirs or channels of the cartridge (e.g., a packaged consumable
cartridge), or reagents may
be introduced into reservoirs or channels of the cartridge prior or during any
of the processes
described. A sample (e.g., biological sample) may be introduced into the
sample via, for
example, a sample inlet or port. For example, FIG. 8 shows sample input 206,
through which a
biological sample may be introduced to a network of channels 205 (e.g., in the
form of
.. microchannels) of cartridge 200. Reagents from any of the reservoirs (e.g.,
first reagent reservoir
201, etc.) may be made to flow through channels 205 to a desired region of
cartridge 200 to
perform a desire step of a process (e.g., lysis, enrichment, fragmentation,
functionalization). For
example, reagents for purification/size selection may be made to flow from
first reagent reservoir
201 to fourth reservoir 204, and the sample may be made to flow from sample
input 206 to
fourth reservoir 204, and upon interaction (e.g., via mixing), a purification
process of the sample
may proceed in fourth reservoir 204 (e.g., via purification/size selection).
Samples and reagents
may be made to flow (e.g., through channels) in the cartridge via any of a
variety of techniques.
One such technique is causing flow via peristaltic pumping. Further
description of exemplary
peristaltic pumping techniques is described below. Other regions of cartridge
may be configured
for other steps of a process, such as fifth reservoir 205, which may be
configured to perform, for
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
example, library recovery, according to some embodiments. FIG. 9B shows an
image of an
exemplary cartridge that may be configured to perform one or more processes
described herein.
It should be understood that cartridge configurations other than that shown in
FIG. 9B are
possible, and FIG. 9B is shown for illustrative purposes.
5 In some embodiments, a cartridge comprises an affinity matrix for
enrichment as
described herein. In some embodiments, a cartridge comprises an affinity
matrix for enrichment
using affinity SCODA, FIGE, or PFGE. In some embodiments, a cartridge
comprises an affinity
matrix comprising an immobilized affinity agent that has a binding affinity
for a target nucleic
acid or target protein.
10 In some embodiments, a sample preparation device of the disclosure
produces (e.g.,
enriches or purifies) target nucleic acids with an average read-length for
downstream sequencing
applications that is longer than an average read-length produced using control
methods (e.g.,
Sage BluePippin methods, manual methods (e.g., manual bead-based size
selection methods)).
In some embodiments, a sample preparation device produces target nucleic acids
with an average
15 read-length for sequencing that comprises at least 700, 800, 900, 1000,
1100, 1200, 1300, 1400,
1500, 1600, 1700, 1800, 1900, 2000, 2100, 2200, 2300, 2400, 2500, 2600, 2700,
2800, 2900, or
3000 nucleotides in length. In some embodiments, a sample preparation device
produces target
nucleic acids with an average read-length for sequencing that comprises 700-
3000, 1000-3000,
1000-2500, 1000-2400, 1000-2300, 1000-2200, 1000-2100, 1000-2000, 1000-1900,
1000-1800,
20 1000-1700, 1000-1600, 1000-1500, 1000-1400, 1000-1300, 1000-1200, 1500-
3000, 1500-2500,
1500-2000, or 2000-3000 nucleotides in length.
Devices in accordance with the instant disclosure generally contain mechanical
and
electronic and/or optical components which can be used to operate a cartridge
as described
herein. In some embodiments, the device components operate to achieve and
maintain specific
25 temperatures on a cartridge or on specific regions of the cartridge. In
some embodiments, the
device components operate to apply specific voltages for specific time
durations to electrodes of
a cartridge. In some embodiments, the device components operate to move
liquids to, from, or
between reservoirs and/or reaction vessels of a cartridge. In some
embodiments, the device
components operate to move liquids through channel(s) of a cartridge, e.g.,
to, from, or between
reservoirs and/or reaction vessels of a cartridge. In some embodiments, the
device components
move liquids via a peristaltic pumping mechanism (e.g., apparatus) that
interacts with an
elastomeric, reagent-specific reservoir or reaction vessel of a cartridge. In
some embodiments,
the device components move liquids via a peristaltic pumping mechanism (e.g.,
apparatus) that is
configured to interact with an elastomeric component (e.g., surface layer
comprising an
elastomer) associated with a channel of a cartridge to pump fluid through the
channel. Device
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
26
components can include computer resources, for example, to drive a user
interface where sample
information can be entered, specific processes can be selected, and run
results can be reported.
In some embodiments, a cartridge is capable of handling small-volume fluids
(e.g., 1-10
i.tt, 2-10 i.tt, 4-10 i.tt, 5-10 i.tt, 1-8 i.tt, or 1-6 i.it fluid). In some
embodiments, the sequencing
cartridge is physically embedded or associated with a sample preparation
device or module (e.g.,
to allow for a prepared sample to be delivered to a reaction mixture for
sequencing. In some
embodiments, a sequencing cartridge that is physically embedded or associated
with a sample
preparation device or module comprises microfluidic channels that have fluid
interfaces in the
form of face sealing gaskets or conical press fits (e.g., Luer fittings). In
some embodiments,
fluid interfaces can then be broken after delivery of the prepared sample in
order to physically
separate the sequencing cartridge from the sample preparation device or
module.
The following non-limiting example is meant to illustrate aspects of the
devices,
methods, and compositions described herein. The use of a sample preparation
device or module
in accordance with the instant disclosure may proceed with one or more of the
following
described steps. A user may open the lid of the device and insert a cartridge
that supports the
desired process. The user may then add a sample, which may be combined with a
specific lysis
solution, to a sample port on the cartridge. The user may then close the
device lid, enter any
sample specific information via a touch screen interface on the device, select
any process
specific parameters (e.g., range of desired size selection, desired degree of
homology for target
molecule capture, etc.), and initiate the sample preparation process run.
Following the run, the
user may receive relevant run data (e.g., confirmation of successful
completion of the run, run
specific metrics, etc.), as well as process specific information (e.g., amount
of sample generated,
presence or absence of specific target sequence, etc.). Data generated by the
run may be
subjected to subsequent bioinformatics analysis, which can be either local or
cloud based.
Depending on the process, a finished sample may be extracted from the
cartridge for subsequent
use (e.g., genomic sequencing, qPCR quantification, cloning, etc.). The device
may then be
opened, and the cartridge may then be removed.
In some embodiments, the sample preparation module comprises a pump. In some
embodiments, the pump is peristaltic pump. Some such pumps comprise one or
more of the
inventive components for fluid handling described herein. For example, the
pump may comprise
an apparatus and/or a cartridge. In some embodiments, the apparatus of the
pump comprises a
roller, a crank, and a rocker. In some such embodiments, the crank and the
rocker are configured
as a crank-and-rocker mechanism that is connected to the roller. The coupling
of a crank-and-
rocker mechanism with the roller of an apparatus can, in some cases, allow for
certain of the
advantages describe herein to be achieved (e.g., facile disengagement of the
apparatus from the
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
27
cartridge, well-metered stroke volumes). In certain embodiments, the cartridge
of the pump
comprises channels (e.g., microfluidic channels). In some embodiments, at
least a portion of the
channels of the cartridge have certain cross-sectional shapes and/or surface
layers that may
contribute to any of a number of advantages described herein.
One non-limiting aspect of some cartridges that may, in some cases, provide
certain
benefits is the inclusion of channels having certain cross-sectional shapes in
the cartridges. For
example, in some embodiments, the cartridge comprises v-shaped channels. One
potentially
convenient but non-limiting way to form such v-shaped channels is by molding
or machining v-
shaped grooves into the cartridge. The recognized advantages of including a v-
shaped channel
(also referred to herein as a v-groove or a channel having a substantially
triangularly-shaped
cross-section) in certain embodiments in which a roller of the apparatus
engages with the
cartridge to cause fluid flow through the channels. For example, in some
instances, a v-shaped
channel is dimensionally insensitive to the roller. In other words, in some
instances, there is no
single dimension to which the roller (e.g., a wedge shaped roller) of the
apparatus must adhere in
order to suitably engage with the v-shaped channel. In contrast, certain
conventional cross
sectional shapes of the channels, such as semi-circular, may require that the
roller have a certain
dimension (e.g., radius) in order to suitably engage with the channel (e.g.,
to create a fluidic seal
to cause a pressure differential in a peristaltic pumping process). In some
embodiments, the
inclusion of channels that are dimensionally insensitive to rollers can result
in simpler and less
expensive fabrication of hardware components and increased
configurability/flexibility.
In certain aspects, the cartridges comprise a surface layer (e.g., a flat
surface layer). One
exemplary aspect relates to potentially advantageous embodiments involving
layering a
membrane (also referred to herein as a surface layer) comprising (e.g.,
consisting essentially of)
an elastomer (e.g., silicone) above the v-groove, to produce, in effect, half
of a flexible tube.
Figure 24 depicts an exemplary cartridge 100 according to certain such
embodiments and is
described in more detail below. Then, in some embodiments, by deforming the
surface layer
comprising an elastomer into the channel to form a pinch and by then
translating the pinch,
negative pressure can be generated on the trailing edge of the pinch which
creates suction and
positive pressure can be generated on the leading edge of the pinch, pumping
fluid in the
direction of the leading edge of the pinch. In certain embodiments, this
pumping by interfacing a
cartridge (comprising channels having a surface layer) with an apparatus
comprising a roller,
which apparatus is configured to carry out a motion of the roller that
includes engaging the roller
with a portion of the surface layer to pinch the portion of the surface layer
with the walls and/or
base of the associated channel, translating the roller along the walls and/or
base of the associated
channel in a rolling motion to translate the pinch of the surface layer
against the walls and/or
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
28
base, and/or disengaging the roller with a second portion of the surface
layer. In certain
embodiments, a crank-and-rocker mechanism is incorporated into the apparatus
to carry out this
motion of the roller.
A conventional peristaltic pump generally involves tubing having been inserted
into an
apparatus comprising rollers on a rotating carriage, such that the tubing is
always engaged with
the remainder of the apparatus as the pump functions. By contrast, in certain
embodiments,
channels in cartridges herein are linear or comprise at least one linear
portion, such that the roller
engages with a horizontal surface. In certain embodiments, the roller is
connected to a small
roller arm that is spring-loaded so that the roller can track the horizontal
surface while
continuously pinching a portion of the surface layer. Spring loading the
apparatus (e.g., a roller
arm of the apparatus) can in some cases help regulate the force applied by the
apparatus (e.g.,
roller) to the surface layer and a channel of a cartridge.
In certain embodiments, each rotation of the crank in a crank-and-rocker
mechanism
connected to the roller provides a discrete pumping volume. In certain
embodiments, it is
straightforward to park the apparatus in a disengaged position, where the
roller is disengaged
from any cartridge. In certain embodiments, forward and backward pumping
motions are fairly
symmetrical as provided by apparatuses described herein, such that a similar
amount of force
(torque) (e.g., within 10%) is required for forward and backward pumping
motions.
In certain embodiments, it may be advantageous to, for a particular size of
apparatus,
have a relatively high crank radius (e.g., greater than or equal to 2 mm,
optionally including
associated linkages). Consequently, it may, in certain embodiments, also be
advantageous to
have a relatively high stroke length (e.g., greater than or equal to 10 mm) to
engage with an
associated cartridge. Having relatively high crank radius and stroke length,
in certain
embodiments, ensures no mechanical interference between the apparatus and the
cartridge when
moving components of the apparatus relative to the cartridge.
In certain embodiments, having v-shaped grooves advantageously allows for
utilization
with rollers of a variety of sizes having a wedge-shaped edge. By contrast,
for example, having a
rectangular channel rather than a v-groove results in the width of the roller
associated with the
rectangular channel needing to be more controlled and precise in relation to
the width of the
rectangular channel, and results in the forces being applied to the
rectangular channel needing to
be more precise. Similarly, the channel(s) having a semicircular cross-section
may also require
more controlled and precise dimension for the width of the associated roller.
In certain embodiments, an apparatus described herein may comprise a multi-
axis system
(e.g., robot) configured so as to move at least a portion of the apparatus in
a plurality of
dimensions (e.g., two dimensions, three dimensions). For example, the multi-
axis system may
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
29
be configured so as to move at least a portion of the apparatus to any pumping
lane location
among associated cartridge(s). For example, in certain embodiments, a carriage
herein may be
functionally connected to a multi-axis system. In certain embodiments, a
roller may be indirectly
functionally connected to a multi-axis system. In certain embodiments, an
apparatus portion,
comprising a crank-and-rocker mechanism connected to a roller, may be
functionally connected
to a multi-axis system. In certain embodiments, each pumping lane may be
addressed by
location and accessed by an apparatus described herein using a multi-axis
system.
Nucleic Acid Sequencing Process
Some aspects of the instant disclosure further involve sequencing nucleic
acids (e.g.,
deoxyribonucleic acids or ribonucleic acid). In some aspects, compositions,
devices, systems,
and techniques described herein can be used to identify a series of
nucleotides incorporated into
a nucleic acid (e.g., by detecting a time-course of incorporation of a series
of labeled
nucleotides). In some embodiments, compositions, devices, systems, and
techniques described
herein can be used to identify a series of nucleotides that are incorporated
into a template-
dependent nucleic acid sequencing reaction product synthesized by a
polymerizing enzyme (e.g.,
RNA polymerase).
Accordingly, also provided herein are methods of determining the sequence of a
target
nucleic acid. In some embodiments, the target nucleic acid is enriched (e.g.,
enriched using
electrophoretic methods, e.g., affinity SCODA) prior to determining the
sequence of the target
nucleic acid. In some embodiments, provided herein are methods of determining
the sequences
of a plurality of target nucleic acids (e.g., at least 2, 3, 4, 5, 10, 15, 20,
30, 50, or more) present in
a sample (e.g., a purified sample, a cell lysate, a single-cell, a population
of cells, or a tissue). In
some embodiments, a sample is prepared as described herein (e.g., lysed,
purified, fragmented,
and/or enriched for a target nucleic acid) prior to determining the sequence
of a target nucleic
acid or a plurality of target nucleic acids present in a sample. In some
embodiments, a target
nucleic acid is an enriched target nucleic acid (e.g., enriched using
electrophoretic methods, e.g.,
affinity SCODA).
In some embodiments, methods of sequencing comprise steps of: (i) exposing a
complex
in a target volume to one or more labeled nucleotides, the complex comprising
a target nucleic
acid or a plurality of nucleic acids present in a sample, at least one primer,
and a polymerizing
enzyme; (ii) directing one or more excitation energies, or a series of pulses
of one or more
excitation energies, towards a vicinity of the target volume; (iii) detecting
a plurality of emitted
photons from the one or more labeled nucleotides during sequential
incorporation into a nucleic
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
acid comprising one of the at least one primers; and (iv) identifying the
sequence of incorporated
nucleotides by determining one or more characteristics of the emitted photons.
In another aspect, the instant disclosure provides methods of sequencing
target nucleic
acids or a plurality of target nucleic acids present in a sample by sequencing
a plurality of
5 nucleic acid fragments, wherein the target nucleic acid(s) comprises the
fragments. In certain
embodiments, the method comprises combining a plurality of fragment sequences
to provide a
sequence or partial sequence for the parent nucleic acid (e.g., parent target
nucleic acid). In
some embodiments, the step of combining is performed by computer hardware and
software. The
methods described herein may allow for a set of related nucleic acids (e.g.,
two or more nucleic
10 acids present in a sample), such as an entire chromosome or genome to be
sequenced.
In some embodiments, a primer is a sequencing primer. In some embodiments, a
sequencing
primer can be annealed to a nucleic acid (e.g., a target nucleic acid) that
may or may not be
immobilized to a solid support. A solid support can comprise, for example, a
sample well (e.g., a
nanoaperture, a reaction chamber) on a chip or cartridge used for nucleic acid
sequencing. In
15 some embodiments, a sequencing primer may be immobilized to a solid
support and
hybridization of the nucleic acid (e.g., the target nucleic acid) further
immobilizes the nucleic
acid molecule to the solid support. In some embodiments, a polymerase (e.g.,
RNA Polymerase)
is immobilized to a solid support and soluble sequencing primer and nucleic
acid are contacted to
the polymerase. In some embodiments a complex comprising a polymerase, a
nucleic acid (e.g.,
20 a target nucleic acid) and a primer is formed in solution and the
complex is immobilized to a
solid support (e.g., via immobilization of the polymerase, primer, and/or
target nucleic acid). In
some embodiments, none of the components are immobilized to a solid support.
For example, in
some embodiments, a complex comprising a polymerase, a target nucleic acid,
and a sequencing
primer is formed in situ and the complex is not immobilized to a solid
support.
25 In some embodiments, sequencing by synthesis methods can include the
presence of a population
of target nucleic acid molecules (e.g., copies of a target nucleic acid)
and/or a step of
amplification (e.g., polymerase chain reaction (PCR)) of a target nucleic acid
to achieve a
population of target nucleic acids. However, in some embodiments, sequencing
by synthesis is
used to determine the sequence of a single nucleic acid molecule in any one
reaction that is being
30 evaluated and nucleic acid amplification may not be required to prepare
the target nucleic acid.
In some embodiments, a plurality of single molecule sequencing reactions are
performed in
parallel (e.g., on a single chip or cartridge) according to aspects of the
instant disclosure. For
example, in some embodiments, a plurality of single molecule sequencing
reactions are each
performed in separate sample wells (e.g., nanoapertures, reaction chambers) on
a single chip or
cartridge.
CA 03177368 2022-09-27
WO 2021/216763 PCT/US2021/028471
31
In some embodiments, sequencing of a target nucleic acid molecule comprises
identifying at least two (e.g., at least 3, at least 4, at least 5, at least
6, at least 7, at least 8, at least
9, at least 10, at least 11, at least 12, at least 13, at least 14, at least
15, at least 16, at least 17, at
least 18, at least 19, at least 20, at least 25, at least 30, at least 35, at
least 40, at least 45, at least
50, at least 60, at least 70, at least 80, at least 90, at least 100, or more)
nucleotides of the target
nucleic acid. In some embodiments, the at least two nucleotides are contiguous
nucleotides. In
some embodiments, the at least two amino acids are non-contiguous nucleotides.
In some embodiments, sequencing of a target nucleic acid comprises
identification of less than
100% (e.g., less than 99%, less than 95%, less than 90%, less than 85%, less
than 80%, less than
75%, less than 70%, less than 65%, less than 60%, less than 55%, less than
50%, less than 45%,
less than 40%, less than 35%, less than 30%, less than 25%, less than 20%,
less than 15%, less
than 10%, less than 5%, less than 1% or less) of all nucleotides in the target
nucleic acid. For
example, in some embodiments, sequencing of a target nucleic acid comprises
identification of
less than 100% of one type of nucleotide in the target nucleic acid. In some
embodiments,
sequencing of a target nucleic acid comprises identification of less than 100%
of each type of
nucleotide in the target nucleic acid.
Terminal functionalization
A target molecule may be functionalized at a terminal end or position. For
example, a
target protein may be functionalized at its N-terminal end or its C-terminal
end. A target nucleic
acid may be functionalized at its 5' end or its 3' end. The nucleobase (e.g.,
guanidine) or the
sugar moiety (e.g., ribose or deoxyribose) may be functionalized.
C-Terminal Carboxylate Functionalization
In one aspect, the present disclosure provides a method of selective C-
terminal
functionalization of a peptide, comprising:
a. reacting a plurality of peptides of Formula (I):
P-R(CO2H)n
or salts thereof;
with a compound of Formula (II):
HX-L1-R1
(II)
to obtain a plurality of compounds of Formula (III):
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
32
P-R CO-X-Li-Ril
'n
(III)
or salts thereof; and
b. reacting the plurality of compounds of Formula (III), or salts
thereof, with a compound
of Formula (IV):
R2-L2-Z
(IV)
to obtain a plurality of compounds of Formula (V):
P-R-ECO-x-Li-Y-L2-zIn
(V)
or salts thereof; wherein m, n, P, R(CO2H)., HX, X, Li, L2, Ri, R2, Y and Z
are defined as
follows.
m is an integer of 1-25, inclusive. In certain embodiments, m is 1-10,
inclusive. In certain
embodiments, m is 5-10, inclusive. In certain embodiments, m is 1-5,
inclusive. In certain
embodiments, m is 1, 2, 3, 4, 5, 6,7 8 9, 10, 11, 12, 13, 14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24,
or 25.
n is 1 or 2. In certain embodiments, n is 1. In certain embodiments, n is 2.
Each P independently is a peptide. In certain embodiments, P has 2-100 amino
acid
residues. In certain embodiments, P has 2-30 amino acid residues.
Each R(CO2H). independently is an amino acid residue having n carboxylate
moieties. n
is 1 or 2. In certain embodiments, n is 1. When n is 1, R(CO2H). is lysine or
arginine. In a
particular embodiment, R(CO2H). is lysine. In another particular embodiment,
R(CO2H). is
arginine. In certain embodiments, n is 2. When n is 2, R(CO2H). is glutamic
acid or aspartic acid.
In a particular embodiment, R(CO2H). is glutamic acid. In another particular
embodiment,
.. R(CO2H)n is aspartic acid.
HX is nucleophilic moiety that is capable of being acylated, wherein H is a
proton. X is
one or more heteroatoms. In certain embodiments, X is 0, S, or NH, or NO.
Li is a linker. In certain embodiments, Li is a substituted or unsubstituted
aliphatic chain,
wherein one or more carbon atoms are optionally, independently replaced by a
heteroatom, an
aryl, heteroaryl, cycloalkyl, or heterocyclyl moiety. In certain embodiments,
Li is polyethylene
glycol (PEG). In other embodiments, Li is a peptide, or an oligonucleotide. In
certain
embodiments, Li is less than 5 nm. In certain embodiments Li is less than 1
nm.
L2 is a linker, or is absent. In certain embodiments, L2 is absent. In certain
embodiments,
L2 is a substituted or unsubstituted aliphatic chain, wherein one or more
carbon atoms are
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
33
optionally, independently replaced by a heteroatom, an aryl, heteroaryl,
cycloalkyl, or
heterocyclyl moiety. In certain embodiments, L2 is polyethylene glycol (PEG).
In other
embodiments, L2 is a peptide, or an oligonucleotide. In certain embodiments L2
is between 5-20
nm, inclusive.
Ri is a moiety comprising a click chemistry handle. In certain embodiments, Ri
is a
moiety comprising an azide, tetrazine, nitrile oxide, alkyne or strained
alkene. In certain
embodiments, the alkyne is a primary alkyne. In certain embodiments, the
alkyne is a cyclic
(e.g., mono- or polycyclic) alkyne (e.g., diarylcyclooctyne, or
bicycle[6.1.0]nonyne). In certain
embodiments, the strained alkene is trans-cyclooctene. In certain embodiments,
Ri is a moiety
comprising an azide. In certain embodiments, the tetrazine comprises the
structure:
N,N .
R2 is a moiety comprising a click chemistry handle that is complementary to
Ri. The
click chemistry handle of R2 is capable of undergoing a click reaction (i.e.,
an electrocyclic
reaction to form a 5-membered heterocyclic ring) with Ri. For example, when Ri
comprises an
azide, nitrile oxide, or a tetrazine, then R2 may comprise an alkyne or a
strained alkene.
Conversely, when Ri comprises an alkyne or a strained alkene, then R2 may
comprise an azide,
nitrile oxide, or tetrazine. In certain embodiments, R2 is a moiety comprising
an azide, tetrazine,
nitrile oxide, alkyne or strained alkene. In certain embodiments, the alkyne
is a primary alkyne.
In certain embodiments, the alkyne is a cyclic (e.g., mono- or polycyclic)
alkyne (e.g.,
diarylcyclooctyne, or bicycle[6.1.0]nonyne). In certain particular
embodiments, R2 comprises
BCN. In other particular embodiments, R2 comprises DBCO. In certain
embodiments, the
strained alkene is trans-cyclooctene. In certain embodiments, the tetrazine
comprises the
structure:
N,N, .
Y is a moiety resulting from the click reaction of Ri and R2. Y is a 5-
membered
heterocyclic ring resulting from an electrocyclic reaction (e.g., 3+2
cycloaddition, or 4+2
cycloaddition) between the reactive click chemistry handles of Ri and R2. In
certain
embodiments, Y is a diradical comprising a 1,2,3-triazolyl, 4,5-dihydro-1,2,3-
triazolyl,
isoxazolyl, 4,5-dihydroisoxazolyl, or 1,4-dihydropyridazyl moiety.
Z is a water-soluble moiety. In certain embodiments, Z imparts water-
solubility to the
compound to which it is attached. In certain embodiments, Z comprises
polyethylene glycol
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
34
(PEG). In certain embodiments, Z comprises single-stranded DNA. In certain
particular
embodiments, Z comprises Q24. In certain embodiments, Z comprises double-
stranded DNA. In
certain embodiments (e.g., compounds of Formula (V)), Z further comprises
biotin (e.g.,
bisbiotin). When Z comprises biotin (e.g., bisbiotin), Z may further comprise
streptavidin. In
certain embodiments, Z comprises double-stranded DNA. In some embodiments, the
moieties of
Z are capable of intermolecularly binding another molecule or surface, e.g.,
to anchor a
compound comprising Z to the molecule or surface.
In certain embodiments, the compound of Formula (II) is of Formula (Ha):
\
H2INJO-40-1,
/ 1=13
m .
(Ha)
In certain embodiments, Formula (III) is of Formula (Ma):
1 110
P¨R
-ikfq-C)(0N3)
H i n
m .
(Ma)
In certain embodiments, n is 1. In certain embodiments, n is 2. In certain
embodiments, m
is 1. In certain embodiments, m is 5.
In certain embodiments, Formula (IV) comprises TCO, and single-stranded DNA.
In
certain embodiments, Formula (IV) further comprises biotin (e.g., bisbiotin).
In certain
embodiments, Formula (IV) is Q24-BisBt-BCN. In certain embodiments, Formula
(IV) is Q24-
BisBt-DBCO. In certain embodiments, Formula (IV) is Q24- BisBt-TCO. Generally,
Formula
(IV) may comprise a branching moiety (e.g., a 1, 3, 5-tricarboxylate moiety),
wherein two
branches are direct or indirect attachments to biotin moieties, and the third
branch is an
attachment to the water soluble moiety (e.g., a polynucleotide such as Q24).
As shown in FIG.
18B and FIG. 20, in certain embodiments Formula (IV) comprises a triazole
moiety derived from
the click-coupling of fragments comprising (i) a bisbiotin-azide
functionalized linker and (ii) an
alkyne (e.g., BCN)-functionalized polynucleotide (e.g. Q24). The click-coupled
product may be
derivatived to introduce a further click handle R2, such as BCN or DBCO.
In certain embodiments, Formula (V) is of Formula (Va):
0
p¨R ,0--li(,õ,Y¨L2¨Z )
, N
(Va)
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
wherein m, n is 1 or 2; and L2, Y, and Z are as defined above. In certain
particular embodiments,
n is 1. In certain particular embodiments, n is 2. In certain particular
embodiments, m is 1. In
certain particular embodiments, m is 5. In certain particular embodiments, L2
is absent. In certain
embodiments, Y comprises a moiety selected from 1,2,3-triazolyl, 4,5-dihydro-
1,2,3-triazolyl,
5 isoxazolyl, 4,5-dihydroisoxazolyl, and 1,4-dihydropyridazyl. In certain
embodiments, Z
comprises single-stranded DNA. In certain embodiments, Z comprises double-
stranded DNA. In
certain embodiments, Z comprises biotin (e.g., bisbiotin). In certain
embodiments, Z further
comprises streptavidin.
In certain embodiments, the reaction of step (a) is performed in the presence
of a
10 carbodiimide reagent. In certain embodiments, the carbodiimide reagent
is water soluble. In a
particular embodiment, the carbodiimide reagent is 1-Ethy1-3-(3-
dimethylaminopropyl)carbodiimide (EDC). In certain embodiments, the reaction
of step (a) is
performed at a pH in the range of 3-5. In certain embodiments (e.g., when to
total peptide
concentration below 1 mM), the concentration of EDC is about 10 mM and the
concentration of
15 the compound of Formula (II) is about 20 mM. In certain embodiments
(e.g., in connection with
Trypsin/LysC digestion, as described below) the concentration of the compound
of Formula (II)
is about may be about 50 mM and the concentration of EDC may be about 25 mM to
suppress C-
terminal intramolecular cyclization.
In certain embodiments of step (a), the plurality of compounds of Formula
(III) is
20 enriched prior to step (b), for example, by passing the compounds
through a G10 sephadex
column and/or passing the compounds through a C18 resin column. The use of C18
resin-based
enrichment is particularly useful when the compound of Formula (II) is greater
than about 200
g/mol. When G-10 sephadex is used in the enrichment, the elution buffer may be
0.5x PBS (pH
7.0). When C18 resin is used in the enrichment, the elution buffer may be 0.1%
formic acid with
25 80% acetonitrile in water. The C18 eluent may be dried and the residue
re-suspended in 0.5x
PBS prior to step (b).
In certain embodiments, the reaction of step (a) is performed in the presence
of an
immobilized carbodiimide reagent. For example, the carbodiimide reagent may be
covalently
attached to a moiety that is stationary and/or insoluble in the reaction
solvent, thereby facilitating
30 separation of excess reagent and/or reaction by-products and/or
unreacted peptides. See, for
example, Fig. 20. In certain embodiments, the immobilized carbodiimide reagent
comprises a
carbodiimide moiety that is covalently attached to a resin, such as
polystyrene (PS). In certain
embodiments, the PS-immobilized carbodiimide reagent is of the formula:
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
36
0
N N
tel / \
In certain embodiments, when the reaction of step (a) is performed in the
presence of an
immobilized carbodiimide reagent, for example, a PS-immobilized reagent as
described herein,
the reaction is performed at a pH in the range of 4 to 5 and/or at ambient
temperature and or for
.. about 20 minutes.
In certain embodiments, performing the reaction of step (a) in the presence of
an
immobilized carbodiimide reagent, for example, a PS-immobilized reagent as
described herein,
facilitates removal of all unreacted (i.e., non-acylated) peptides because the
unreacted peptides
remain covalently bound to the immobilized carbodiimide reagent.
An exemplary process using an immobilized carbodiimide reagent is shown in
Fig. 21.
An exemplary flowchart for an automation compatible process is shown in Fig.
7.
In certain embodiments of step (b), the click reaction between the plurality
of compounds of
Formula (III) and the compound of Formula (IV) is uncatalyzed. In certain
embodiments, the
click reaction is catalyzed, for example, using a copper salt (e.g., a Cu +
salt, or a Cu2+ salt that is
reduced in situ to a Cu + salt). Suitable Cu2+ salts include CuSO4. In certain
embodiments, the
reaction of step (b) comprises heating the reaction mixture.
In certain embodiments, the compound of Formula (IV) is added to the plurality
of
compounds of Formula (III). In certain embodiments, the total concentration of
the compound of
Formula (IV) and the plurality of compounds of Formula (III) is maintained in
the range
between 1011M to 1 mM.
In certain embodiments of step (b), when Z comprises single-stranded DNA, the
method
further comprises hybridizing a complementary DNA strand to the single-
stranded DNA to
obtain a compound wherein Z comprises double-stranded DNA. In certain
embodiments, the
single-stranded DNA is Q24 and the complementary DNA strand is Cy3B.
In certain embodiments of step (b), when Z comprises biotin (e.g., bisbiotin),
the method
further comprises contacting the biotin (e.g., bisbiotin) with streptavidin to
obtain a compound
wherein Z comprises biotin (e.g., bisbiotin) and streptavidin.
In certain embodiments, the plurality of peptides of Formula (I), or salts
thereof, is
obtained by subjecting a protein to enzymatic digestion to obtain a digestive
mixture comprising
.. the plurality of peptides of Formula (I), or salts thereof. In certain
embodiments, the enzymatic
digestion comprises cleaving the C-terminal bonds of aspartic acid and/or
glutamic acid residues
of the protein. In certain specific embodiments, the enzymatic digestion is
Glu-C digestion.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
37
In certain embodiments, the total concentration of the plurality of peptides
of Formula
(I), or salts thereof, after digestion of 20 pg protein is below 100 [tM.
In certain embodiments, the enzymatic digestion is performed in phosphate
buffer (pH
7.8) or ammonium bicarbonate buffer (pH 4.0).
In certain embodiments, the enzymatic digestion comprises cleaving the C-
terminal
bonds of lysine and/or arginine residues of the protein. In certain specific
embodiments, the
enzymatic digestion is Trypsin+Lys-C digestion.
In certain embodiments, the carboxylic acid moieties of the protein, if
present, are
protected prior to the enzymatic digestion. For example, the carboxylic acid
moieties of the
protein, if present, may be esterified prior to enzymatic digestion. In
certain specific
embodiments, the esterified carboxylic acids are methyl esters.
In certain embodiments, the sulfide moieties of the protein are protected
prior to
enzymatic digestion. In certain specific embodiments, the sulfide moieties are
protected by
exposing the protein to tris(carboxyethyl)phosphine (TCEP) and iodoacetamide
(ICM), or
maleimide.
In certain embodiments, the method further comprises the step of enriching the
digestive
mixture prior to step (a).
C-Terminal Amine Functionalization
In another aspect, the present disclosure provides a method of selective C-
terminal amine
functionalization of a peptide, comprising:
a. reacting a plurality of peptides of Formula (VI):
0
P,N)NH2
CO2H
(VI)
or salts thereof, with a compound of Formula (VII):
0
,L3¨R3
0 r4 =
(VII)
to obtain a plurality of compounds of Formula (VIII):
0 74
P,N)No,N
CO2H 00
, or salts thereof; and
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
38
(VIII)
b. reacting the plurality of compounds of Formula (VIII), or
salts thereof, with a
compound of Formula (IX):
R5-1_4¨Z1 ;
(IX)
to afford a plurality of compounds of Formula (X):
R4
0 I
P.N. L....,-...,0_,N,,.
H
CO2H 00
(X)
or salts thereof; wherein P, L3, L4, R3, R4, Yl, and Zi are as defined below.
Each P independently is a peptide. In certain embodiments, P has 2-100 amino
acid
residues. In certain embodiments, P has 2-30 amino acid residues.
L3 is a linker. In certain embodiments, L3 is a substituted or unsubstituted
aliphatic chain,
wherein one or more carbon atoms are optionally, independently replaced by a
heteroatom, an
aryl, heteroaryl, cycloalkyl, or heterocyclyl moiety. In certain embodiments,
L3 is polyethylene
glycol (PEG). In other embodiments, L3 is a peptide, or an oligonucleotide.
L4 is a linker, or is absent. In certain embodiments, L4 is absent. In certain
embodiments,
L4 is a substituted or unsubstituted aliphatic chain, wherein one or more
carbon atoms are
optionally, independently replaced by a heteroatom, an aryl, heteroaryl,
cycloalkyl, or
heterocyclyl moiety. In certain embodiments, L4 is polyethylene glycol (PEG).
In other
embodiments, L4 is a peptide, or an oligonucleotide.
R3 is a moiety comprising a click chemistry handle. In certain embodiments, R3
is a
moiety comprising an azide, tetrazine, nitrile oxide, alkyne or strained
alkene. In certain
embodiments, the alkyne is a primary alkyne. In certain embodiments, the
alkyne is a cyclic
(e.g., mono- or polycyclic) alkyne (e.g., diarylcyclooctyne, or
bicycle[6.1.0]nonyne). In certain
embodiments, the strained alkene is trans-cyclooctene. In certain embodiments,
Ri is a moiety
comprising an azide. In certain embodiments, the tetrazine comprises the
structure:
iN1,,N
N,N, .
R4 is substituted or unsubstituted aryl or substituted or unsubstituted
heteroaryl. In certain
embodiments, R4 is substituted or unsubstituted phenyl. In certain particular
embodiments, R4 is
.. phenyl. In certain particular embodiments, R4 is 4-nitrophenyl.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
39
Rs is a moiety comprising a click chemistry handle that is complementary to
R3. The
click chemistry handle of Rs is capable of undergoing a click reaction (i.e.,
an electrocyclic
reaction to form a 5-membered heterocyclic ring) with R3. For example, when R3
comprises an
azide, nitrile oxide, or a tetrazine, then Rs may comprise an alkyne or a
strained alkene.
Conversely, when R3 comprises an alkyne or a strained alkene, then Rs may
comprise an azide,
nitrile oxide, or tetrazine. In certain embodiments, Rs is a moiety comprising
an azide, tetrazine,
nitrile oxide, alkyne or strained alkene. In certain embodiments, the alkyne
is a primary alkyne.
In certain embodiments, the alkyne is a cyclic (e.g., mono- or polycyclic)
alkyne (e.g.,
diarylcyclooctyne, or bicycle[6.1.0]nonyne). In certain particular
embodiments, Rs comprises
BCN. In other particular embodiments, Rs comprises DBCO. In certain
embodiments, the
strained alkene is trans-cyclooctene. In certain embodiments, the tetrazine
comprises the
structure:
fN,
` N
N,N,/f.
Yi is a moiety resulting from the click reaction of R3 and Rs. Yi is a 5-
membered
heterocyclic ring resulting from an electrocyclic reaction (e.g., 3+2
cycloaddition, or 4+2
cycloaddition) between the reactive click chemistry handles of R3 and Rs. In
certain
embodiments, Yi is a diradical comprising a 1,2,3-triazolyl, 4,5-dihydro-1,2,3-
triazolyl,
isoxazolyl, 4,5-dihydroisoxazolyl, or 1,4-dihydropyridazyl moiety.
Zi is a water-soluble moiety. In certain embodiments, Zi imparts water-
solubility to the
.. compound to which it is attached. In certain embodiments, Zi comprises
polyethylene glycol
(PEG). In certain embodiments, Zi comprises single-stranded DNA. In certain
particular
embodiments, Z1 comprises Q24. In certain embodiments, Z1 comprises single-
stranded DNA.
In certain embodiments (e.g., compounds of Formula (V)), Zi further comprises
biotin (e.g.,
bisbiotin). When Zi comprises biotin (e.g., bisbiotin), Zi may further
comprise streptavidin. In
certain embodiments, Zi comprises double-stranded DNA. In some embodiments,
the moieties of
Zi are capable of intermolecularly binding another molecule or surface, e.g.,
to anchor a
compound comprising Zi to the molecule or surface.
In certain embodiments, the compound of Formula (VII) is selected from:
9 / % 9 /
`-S-N 0
ii o 8.
N N NO2 N N
and \ .
CA 03177368 2022-09-27
WO 2021/216763 PCT/US2021/028471
In certain embodiments, Formula (VIII) is of Formula (VIIIa) or Formula
(VIIIb):
0
0
N..............--, ,...N.,,,,,,,,õ,,,..N
N /S,
H
CO2H 0"O 0 NõN
I
(Villa), or
NO2
0
0
P N ,N N
N /S,
H
CO2H 00 0 ,
I N' N
N'N (VIIIb).
In certain embodiments, Formula (IX) comprises TCO, single-stranded DNA, and
biotin
5 (e.g., bisbiotin). In certain embodiments, Formula (IX) is Q24-BisBt-BCN.
In certain
embodiments, Formula (IX) is Q24- BisBt-DBCO. In certain embodiments, Formula
(IX) is
Q24- BisBt-TCO. Generally, Formula (IX) may comprise a branching moiety (e.g.,
a 1, 3, 5-
tricarboxylate moiety), wherein two branches are direct or indirect
attachments to biotin
moieties, and the third branch is an attachment to the water soluble moiety
(e.g., a polynucleotide
10 such as Q24). In certain embodiments Formula (IX) comprises a triazole
moiety derived from
the click-coupling of fragments comprising (i) a bisbiotin-azide
functionalized linker and (ii) an
alkyne (e.g., BCN)-functionalized polynucleotide (e.g. Q24). The click-coupled
product may be
derivatived to introduce a further click handle R5, such as BCN or DBCO.
In certain embodiments, the reaction of step (a) is performed in the presence
of a buffer
15 having a concentration in the range of about 20 mM-500 mM and a pH in
the range of about 9-
11, and acetonitrile in the range of about 20-70% of total volume. In certain
embodiments, the
reaction of step (a) is performed in pH 9.5 buffer/acetonitrile (1:3 v/v) at
approximately 37 C. In
certain embodiments, the reaction of step (a) is performed using a
concentration of the
compound of Formula (VII) of about 500 .IM-50 mM.
20 In certain embodiments, the plurality of compounds of Formula (VIII) is
enriched prior
to step (b). In certain embodiments, the enrichment comprises ethyl
acetate/hexane extraction.
Suitable ranges for ethyl acetate/hexane include, but are not limited to, 20
to 100 volume % ethyl
acetate in hexanes. In certain embodiments, the volume of organic solvent used
in the extraction
is about 10x the volume of aqueous layer. Other water immiscible organic
solvents can be used
25 in the extraction, e.g., diethyl ether, dichloromethane, chloroform,
benzene, toluene, and n-1-
butanol.
CA 03177368 2022-09-27
WO 2021/216763 PCT/US2021/028471
41
In certain embodiments, the reaction of step (b) comprises reacting the
compounds of
Formula (VIII) with about one equivalent of the compound of Formula (IX). In
certain
embodiments, the reaction of step (b) comprises heating the reaction mixture.
In certain embodiments of step (b), when Zi comprises single-stranded DNA, the
method
.. further comprises hybridizing a complementary DNA strand to the single-
stranded DNA to
obtain a compound wherein Zi comprises double-stranded DNA. In certain
embodiments, the
single-stranded DNA is Q24 and the complementary DNA strand is Cy3B.
In certain embodiments of step (b), when Zi comprises biotin (e.g.,
bisbiotin), the method
further comprises contacting the biotin (e.g., bisbiotin) with streptavidin to
obtain a compound
wherein Zi comprises biotin (e.g., bisbiotin) and streptavidin.
In certain embodiments, the plurality of peptides of Formula (VI), or salts
thereof, is
obtained by subjecting a protein to enzymatic digestion to obtain a digestive
mixture comprising
the plurality of peptides of Formula (VI), or salts thereof. The enzymatic
digestion comprises
cleaving the C-terminal bonds of lysine and/or arginine residues of the
protein. In certain
embodiments, the enzymatic digestion is performed using Trypsin, Lys-C, or a
combination
thereof. In certain embodiments, the enzymatic digestion comprises reacting
the protein with
Trypsin and Lys-C in Tris-HC1 buffer (pH 8.5). In certain embodiments, the
total concentration
of the plurality of peptides of Formula (VI), or salts thereof, after
digestion of 20 pg protein is
below 100 [tM.
In certain embodiments, the sulfide moieties of the protein are protected
prior to
enzymatic digestion. In certain specific embodiments, the sulfide moieties are
protected by
exposing the protein to tris(carboxyethyl)phosphine (TCEP) and iodoacetamide
(ICM), or
maleimide.
In certain embodiments, the method further comprises the step of enriching the
digestive
.. mixture prior to step (a). In certain embodiments, the digestive mixture is
used in the method of
selective C-terminal amine functionalization of a peptide without enrichment
or purification.
Selective Amine Functionalization via Diazo Transfer
Prior to sequencing, digested peptides must be functionalized with a moiety
that is
capable of immobilizing the peptides on the sequencing substrate. Accordingly,
the present
disclosure provides a method of selective N-functionalization of a peptide,
comprising reacting a
plurality of peptides of Formula (XI):
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
42
0
H2N-1:1N)NH2
CO2H
(XI)
or salts thereof, wherein each P independently is a peptide having an N-
terminal amine, with a
compound of Formula (XII):
0
N%\ ii
I .N-S-N3
0
(XII)
under conditions comprising Cu2 , or a precursor thereof, and a buffer having
a pH of about 10-
11; to obtain a plurality of c-azido compounds of the Formula (XIII):
0
N3
H2N-N).
CO2H
(XIII)
or salts thereof.
Each P independently is a peptide having an N-terminal amine. In certain
embodiments,
P has 2-100 amino acid residues. In certain embodiments, P has 2-30 amino acid
residues. In
some embodiments, the concentration of a peptide in the reaction is any
conceivable
concentration necessary.
In certain embodiments, the Cu2+ salt is CuC12, CuBr2, Cu(OH)2, or CuSO4. In a
particular embodiment, the Cu2+ salt is CuSO4. In certain embodiments, the
molar amount of the
Cu2+ salt is about 2.5 times the molar amount of the compound of Formula (XI).
In certain
particular embodiments, the concentration of the Cu2+ salt is about 250 tM. In
some
embodiments, the concentration of the Cu2+ salt is between 1-5 mM or 100-1000
In certain embodiments, the conditions further comprise reaction at about 20-
30 C, e.g.,
20-25 C, 22-27 C, 25-30 C, 20 C, 21 C, 22 C, 23 C, 24 C, 25 C, 26 C,
27 C, 28 C, 29
C, or 30 C.
In certain embodiments, the conditions further comprise reaction for about 30-
60
minutes, e.g., 30-35 minutes, 35-40 minutes, 40-45 minutes, 45-50 minutes, 50-
55 minutes, or
55-60 minutes.
In certain embodiments, the buffer has a pH of about 10.5. In certain
embodiments, the
buffer comprises bicarbonate, e.g., sodium bicarbonate. In certain
embodiments, the buffer
comprises carbonate, e.g., potassium carbonate. In certain embodiments, the
buffer comprises
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
43
phosphate, e.g., potassium phosphate. In some embodiments, the buffer does not
comprise an
amino group. In some embodiments, the buffer is a Good's buffer (e.g., HEPES,
TRIS). In
certain embodiments, the buffer has a concentration in the range of 10 mM to
1M, e.g., 10-
100mM, 50-500 mM, 50-100 mM, or 100 mM.
In certain embodiments, the concentration of the compound of Formula (XI) is
about 100
i.t.M. In some embodiments, the concentration of the compound of Formula (XI)
is about 50 t.M.
In some embodiments, the concentration of the compound of Formula (XI) is
between 1 nM and
1 mM.
In certain embodiments, the amount of the compound of Formula (XII) used in
the
reaction is 10-30 molar equivalents, e.g., about 20 molar equivalents,
relative to the amount of
the compound of Formula (XI) used in the reaction. In certain embodiments, the
concentration of
the compound of Formula (XII) is about 1-3 mM, e.g., about 2 mM.
In certain embodiments, the N-terminal: selectivity of the diazo transfer
reaction is at
least about 90%.
In some embodiments, the method further comprises enriching the plurality of
compounds of Formula (XIII), or salts thereof. In certain embodiments, excess
compound of
Formula (XII) is removed from the reaction mixture using a purification
cartridge, e.g., a G-10
sephadex column. In certain embodiments, removal of excess Formula (XIII)
using a G-10
sephadex column comprises a buffer exchange to 25 mM HEPES, 25 mM KOAc, pH
7.8.
In some embodiments, the plurality of peptides of Formula (XI), or salts
thereof, is
obtained by subjecting a protein to enzymatic digestion, as described herein,
to obtain a digestive
mixture comprising the plurality of peptides of Formula (XI), or salts
thereof. The enzymatic
digestion comprises cleaving the C-terminal bonds of aspartic acid and/or
glutamic acid residues
of the protein.
In some embodiments, the enzymatic digestion is Trypsin+Lys-C digestion. In
some
embodiments, the Trypsin+Lys-C digestion comprises reacting the protein with
Trypsin and Lys-
C at room temperature in pH 9.5 buffer.
In some embodiments, the method further comprises reacting the plurality of
compounds
of Formula (XIII) or salts thereof with a DBCO-labeled DNA-streptavidin
conjugate, such that
the azide moiety of the compounds of Formula (XIII), or salts thereof,
undergoes an
electrocyclic reaction with the alkyne moiety of DBCO (diarylcyclooctyne) to
form a plurality of
peptide-DNA-streptavidin conjugates.
In some embodiments, the DBCO-labeled DNA-streptavidin is of Formula (MV):
R6¨L5¨Z2
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
44
(XIV)
wherein R6 is DBCO; L5 is a linker or is absent; and Z2 is a dsDNA-
streptavidin
conjugate;
and the plurality of peptide-DNA-streptavidin conjugates are of Formula (XV),
or salts
thereof:
0
H2N-R,N -11Y2-1-5-Z2
H
CO2H
(XV)
wherein Y2 is a moiety resulting from a click reaction with the azide moiety
of Formula
(XIIIb) and R6.
R6 is a moiety comprising a click chemistry handle that is complementary to
the azide
moiety of Formula (XIIIb). The click chemistry handle of R6 is capable of
undergoing a click
reaction (i.e., an electrocyclic reaction to form a 5-membered heterocyclic
ring) with the azide
moiety of Formula (XIIIb). In certain embodiments, R6 comprises an alkyne or a
strained
alkene. In certain embodiments, the alkyne is a primary alkyne. In certain
embodiments, the
alkyne is a cyclic (e.g., mono- or polycyclic) alkyne (e.g.,
diarylcyclooctyne, or
bicycle[6.1.0]nonyne). In certain particular embodiments, R6 comprises BCN. In
other particular
embodiments, R6 comprises DBCO. In certain embodiments, the strained alkene is
trans-
cyclooctene.
In certain embodiments, L5 is absent. In certain embodiments, L5 is a
substituted or
.. unsubstituted aliphatic chain, wherein one or more carbon atoms are
optionally replaced by a
heteroatom, an aryl, heteroaryl, cycloalkyl, or heterocyclyl moiety. In
certain embodiments, L5 is
polyethylene glycol (PEG). In other embodiments, L5 is a peptide, or an
oligonucleotide.
In certain embodiments, Z2 is prepared from a bis-biotin tag which
specifically binds to
streptavidin in the cis form, leaving the other cis-binding sites free for
surface immobilization.
In certain embodiments, Z2 comprises PEG. In certain embodiments, Z2 further
comprises
biotin (e.g., bisbiotin). In certain embodiments, when Z2 comprises single-
stranded DNA, the
method further comprises hybridizing a complementary DNA strand to the single-
stranded DNA
to obtain a compound wherein Z2 comprises double-stranded DNA. In certain
embodiments, the
single-stranded DNA is Q24 and the complementary DNA strand is Cy3B.
In certain embodiments, Formula (XIV) is Q24-BisBt-BCN. In certain
embodiments,
Formula (XIV) is Q24- BisBt-DBCO. In certain embodiments, Formula (XIV) is Q24-
BisBt-
TCO. Generally, Formula (XIV) may comprise a branching moiety (e.g., a 1, 3, 5-
tricarboxylate
moiety), wherein two branches are direct or indirect attachments to biotin
moieties, and the third
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
branch is an attachment to the water soluble moiety (e.g., a polynucleotide
such as Q24). In
certain embodiments Formula (XIV) comprises a triazole moiety derived from the
click-coupling
of fragments comprising (i) a bisbiotin-azide functionalized linker and (ii)
an alkyne (e.g., BCN)-
functionalized polynucleotide (e.g. Q24). The click-coupled product may be
derivatived to
5 introduce a further click handle R6, such as BCN or DBCO.
In certain embodiments, when Z2 comprises biotin (e.g., bisbiotin), the method
further
comprises contacting the biotin (e.g., bisbiotin) with streptavidin to obtain
a compound wherein
Z2 comprises biotin (e.g., bisbiotin) and streptavidin.
In a particular embodiment, the method of selective N-functionalization of a
peptide is
10 carried out according to one or more steps as shown in Figure 6.
Click Chemistry
In certain embodiments, the reaction used to conjugate the host to the tag is
a "click
chemistry" reaction (e.g., the Huisgen alkyne-azide cycloaddition). It is to
be understood that any
15 "click chemistry" reaction known in the art can be used to this end.
Click chemistry is a chemical
approach introduced by Sharpless in 2001 and describes chemistry tailored to
generate
substances quickly and reliably by joining small units together. See, e.g.,
Kolb, Finn and
Sharpless, Angewandte Chemie International Edition (2001) 40: 2004-2021;
Evans, Australian
Journal of Chemistry (2007) 60: 384-395). Exemplary coupling reactions (some
of which may
20 be classified as "click chemistry") include, but are not limited to,
formation of esters, thioesters,
amides (e.g., such as peptide coupling) from activated acids or acyl halides;
nucleophilic
displacement reactions (e.g., such as nucleophilic displacement of a halide or
ring opening of
strained ring systems); azide¨alkyne Huisgen cycloaddition; thiol¨yne
addition; imine formation;
Michael additions (e.g., maleimide addition); and Diels¨Alder reactions (e.g.,
tetrazine [4 + 2]
25 cycloaddition).
The term "click chemistry" refers to a chemical synthesis technique introduced
by K.
Barry Sharpless of The Scripps Research Institute, describing chemistry
tailored to generate
covalent bonds quickly and reliably by joining small units comprising reactive
groups together.
See, e.g., Kolb, Finn and Sharpless Angewandte Chemie International Edition
(2001) 40: 2004-
30 2021; Evans, Australian Journal of Chemistry (2007) 60: 384-395).
Exemplary reactions
include, but are not limited to, azide¨alkyne Huisgen cycloaddition; and
Diels¨Alder reactions
(e.g., tetrazine [4 + 2] cycloaddition). In some embodiments, click chemistry
reactions are
modular, wide in scope, give high chemical yields, generate inoffensive
byproducts, are
stereospecific, exhibit a large thermodynamic driving force > 84 kJ/mol to
favor a reaction with a
35 .. single reaction product, and/or can be carried out under physiological
conditions. In some
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
46
embodiments, a click chemistry reaction exhibits high atom economy, can be
carried out under
simple reaction conditions, use readily available starting materials and
reagents, uses no toxic
solvents or use a solvent that is benign or easily removed (preferably water),
and/or provides
simple product isolation by non-chromatographic methods (crystallization or
distillation).
The term "click chemistry handle," as used herein, refers to a reactant, or a
reactive
group, that can partake in a click chemistry reaction. For example, a strained
alkyne, e.g., a
cyclooctyne, is a click chemistry handle, since it can partake in a strain-
promoted cycloaddition
(see, e.g., Table 1). In general, click chemistry reactions require at least
two molecules
comprising click chemistry handles that can react with each other. Such click
chemistry handle
pairs that are reactive with each other are sometimes referred to herein as
partner click chemistry
handles. For example, an azide is a partner click chemistry handle to a
cyclooctyne or any other
alkyne. Exemplary click chemistry handles suitable for use according to some
aspects of this
invention are described herein, for example, in Tables 1 and 2. Other suitable
click chemistry
handles are known to those of skill in the art.
R1.----= + NER---=i9,1¨P. __ it -
)--/ 1,3-diWar cydostictioa
wmin3i Ntoino 44114e Ri
N R,
NI N
¨
¨
+ N 4P¨W¨R-.? 0
azide _____________________ -AP (1-1\--Ri
1\¨$ Strafts-promoted cyclixiddlion
s farketlalkyne
+ r R2
.cgenco*, __________________ mu R,
Dieh.i.-Aider reodion
Rafi=+ IL X 1 Thid-erte Feadan
N3 N.3
VW
AOnti.,
Table 1: Exemplary click chemistry handles and reactions.
In some embodiments, click chemistry handles are used that can react to form
covalent
bonds in the presence of a metal catalyst, e.g., copper (II). In some
embodiments, click chemistry
handles are used that can react to form covalent bonds in the absence of a
metal catalyst. Such
click chemistry handles are well known to those of skill in the art and
include the click chemistry
handles described in Becer, Hoogenboom, and Schubert, Click Chemistry beyond
Metal-
Catalyzed Cycloaddition, Angewandte Chemie International Edition (2009) 48:
4900 ¨ 4908.
CA 03177368 2022-09-27
WO 2021/216763 - 47 -
PCT/US2021/028471
Reagent A Reagent B Mechanism Notes on reactioe
0 azide alkyne Cu=catalyzed [3+21 2 h at 60'C in H20
azide-alkyne cycloaddition
(CuAAC)
I azide cydooctyrse strain-promoted [34-21azide,alkyne cycloaddition
1 h at RI
(SPAAC)
2 azide activated Hoisgen cycioaddition 4 h at 50 C
alkyne
3 azide eiectron-deficient al- p+zi cycloaddittion 12 h at RI in Hp
kYrie
4 azide aryne [3+21 cycloaddition 4 h at RT n THF with
crown ether or
24h at RI in CH,CN
tetrazine alkene Dies¨Aider retro-1.4+4
cycioaddition 40 min at 25'C (100% yd)
Nz is the only by-product
6 tettazole alkene 1,3-dipolar
cycloa.ddition few min UV irradiation and then overnight
(photodick) at 4"C
7 dithioester ciiene iletero-Diels¨Aider cycioaddition 10 min at RT
8 anthracene maleimide [4+21 Diels---Alder reaction 2 days at reflux
in toluene
9 thiol alkene radical addition 30 min UV
(quantitative conc.) or
(thio dick) 24 h UV irradiation
(;I, 96%)
thiol enone Mt-.haei addition 24 h at RT in CH,CN
11 thioi maleimide Michael addition 1 h at 40 C in THF
16h at RI in dioxane
12 thic, poerpfluoro nucleophilic substitution overnight at RT in
DMF or
60 min at 400C in DMF
13 amine pares-fluor nudeophilic substitution 20 min MW at 95'C in
NMP as soivent
RI::: room; temperature, DM F N,N.dinlethyliotmamide, NMP
,,,N,methylpyrolidorie, THF:::tetrahydrofUran, CH,CN acetonitrile.
Table 2: Exemplary click chemistry handles and reactions.
From Becer, Hoogenboom, and Schubert, Click Chemistry Beyond Metal-Catalyzed
Cycloaddition, Angewandte Chemie International Edition (2009) 48: 4900 ¨ 4908.
5
Additional click chemistry handles suitable for use in methods of conjugation
described
herein are well known to those of skill in the art, and such click chemistry
handles include, but
are not limited to, the click chemistry reaction partners, groups, and handles
described in
PCT/US2012/044584 and references therein, which references are incorporated
herein by
10 reference for click chemistry handles and methodology.
Compounds
In certain aspects, the present disclosure provides compounds of Formulae
(II), (Ha),
(III), (Ma), (IV), (V), (Va), (VII), (VIII), (Villa), (VIIIb), (XIV), (X),
(XI), (XII), (XIIIa),
(XIIIb), (XV), and salts thereof, as described herein in various embodiments.
In certain embodiments, the compounds are water soluble.
In certain embodiments, the compounds are useful for applications relating to
the analysis
of proteins and peptides, such as peptide sequencing. For example, in certain
embodiments,
compounds of Formulae (V), (X), (XV), and salts thereof, may be covalently or
non-covalently
attached to a surface.
8856783.1
SUBSTITUTE SHEET (RULE 26)
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
48
Definitions
In the following description, certain specific details are set forth in order
to provide a
thorough understanding of various embodiments of the invention. However, one
skilled in the art
will understand that the invention may be practiced without these details.
Unless the context requires otherwise, throughout the present specification
and claims, the word
"comprise" and variations thereof, such as, "comprises" and "comprising" are
to be construed in
an open, inclusive sense (i.e., as "including, but not limited to").
Unless defined otherwise, all technical and scientific terms used herein have
the same
meaning as is commonly understood by one of skill in the art to which this
invention belongs. As
used in the specification and claims, the singular form "a", "an", and "the"
include plural
references unless the context clearly dictates otherwise.
The term "aliphatic" refers to alkyl, alkenyl, alkynyl, and carbocyclic
groups. Likewise,
the term "heteroaliphatic" refers to heteroalkyl, heteroalkenyl,
heteroalkynyl, and heterocyclic
groups.
The term "alkyl" refers to a radical of a straight¨chain or branched saturated
hydrocarbon
group having from 1 to 20 carbon atoms ("Ci_20 alkyl") In some embodiments, an
alkyl group
has 1 to 10 carbon atoms ("Ci_io alkyl"). In some embodiments, an alkyl group
has 1 to 9 carbon
atoms ("Ci_9 alkyl"). In some embodiments, an alkyl group has 1 to 8 carbon
atoms ("Ci-8
alkyl"). In some embodiments, an alkyl group has 1 to 7 carbon atoms ("Ci_7
alkyl"). In some
embodiments, an alkyl group has 1 to 6 carbon atoms ("Ci_6 alkyl"). In some
embodiments, an
alkyl group has 1 to 5 carbon atoms ("Ci_s alkyl"). In some embodiments, an
alkyl group has 1 to
4 carbon atoms ("Ci_4 alkyl"). In some embodiments, an alkyl group has 1 to 3
carbon atoms
("Ci_3 alkyl"). In some embodiments, an alkyl group has 1 to 2 carbon atoms
("Ci_2 alkyl"). In
some embodiments, an alkyl group has 1 carbon atom ("Ci alkyl"). In some
embodiments, an
alkyl group has 2 to 6 carbon atoms ("C2_6 alkyl"). Examples of C1-6 alkyl
groups include methyl
(CO, ethyl (C2), propyl (C3) (e.g., n¨propyl, isopropyl), butyl (C4) (e.g.,
n¨butyl, tert¨butyl, sec¨
butyl, iso¨butyl), pentyl (Cs) (e.g., n¨pentyl, 3¨pentanyl, amyl, neopentyl,
3¨methyl-2¨butanyl,
tertiary amyl), and hexyl (C6) (e.g., n¨hexyl). Additional examples of alkyl
groups include n¨
heptyl (C7), n¨octyl (C8), and the like. Unless otherwise specified, each
instance of an alkyl
group is independently unsubstituted (an "unsubstituted alkyl") or substituted
(a "substituted
alkyl") with one or more substituents (e.g., halogen, such as F). In certain
embodiments, the
alkyl group is an unsubstituted Ci_io alkyl (such as unsubstituted Ci_6 alkyl,
e.g., ¨CH3 (Me),
unsubstituted ethyl (Et), unsubstituted propyl (Pr, e.g., unsubstituted n-
propyl (n-Pr),
unsubstituted isopropyl (i-Pr)), unsubstituted butyl (Bu, e.g., unsubstituted
n-butyl (n-Bu),
unsubstituted tert-butyl (tert-Bu or t-Bu), unsubstituted sec-butyl (sec-Bu or
s-Bu), unsubstituted
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
49
isobutyl (i-Bu)). In certain embodiments, the alkyl group is a substituted
Ci_io alkyl (such as
substituted C1_6 alkyl, e.g., ¨CH2F, ¨CHF2, ¨CF3 or benzyl (Bn)). An alkyl
group may be
branched or unbranched.
The term "alkenyl" refers to a radical of a straight-chain or branched
hydrocarbon group
having from 1 to 20 carbon atoms and one or more carbon-carbon double bonds
(e.g., 1, 2, 3, or
4 double bonds). In some embodiments, an alkenyl group has 1 to 20 carbon
atoms ("Ci_20
alkenyl"). In some embodiments, an alkenyl group has 1 to 12 carbon atoms
("C1_12 alkenyl"). In
some embodiments, an alkenyl group has 1 to 11 carbon atoms ("C1_11 alkenyl").
In some
embodiments, an alkenyl group has 1 to 10 carbon atoms ("Ci_io alkenyl"). In
some
embodiments, an alkenyl group has 1 to 9 carbon atoms ("Ci-9 alkenyl"). In
some embodiments,
an alkenyl group has 1 to 8 carbon atoms ("C1-8 alkenyl"). In some
embodiments, an alkenyl
group has 1 to 7 carbon atoms ("Ci_7 alkenyl"). In some embodiments, an
alkenyl group has 1 to
6 carbon atoms ("C1_6 alkenyl"). In some embodiments, an alkenyl group has 1
to 5 carbon
atoms ("C1_5 alkenyl"). In some embodiments, an alkenyl group has 1 to 4
carbon atoms ("Ci_4
alkenyl"). In some embodiments, an alkenyl group has 1 to 3 carbon atoms
("C1_3 alkenyl"). In
some embodiments, an alkenyl group has 1 to 2 carbon atoms ("C1_2 alkenyl").
In some
embodiments, an alkenyl group has 1 carbon atom ("Ci alkenyl"). The one or
more carbon-
carbon double bonds can be internal (such as in 2-butenyl) or terminal (such
as in 1-buteny1).
Examples of C1-4 alkenyl groups include methylidenyl (CO, ethenyl (C2), 1-
propenyl (C3), 2-
propenyl (C3), 1-butenyl (C4), 2-butenyl (C4), butadienyl (C4), and the like.
Examples of C1-6
alkenyl groups include the aforementioned C2-4 alkenyl groups as well as
pentenyl (C5),
pentadienyl (C5), hexenyl (C6), and the like. Additional examples of alkenyl
include heptenyl
(C7), octenyl (C8), octatrienyl (C8), and the like. Unless otherwise
specified, each instance of an
alkenyl group is independently unsubstituted (an "unsubstituted alkenyl") or
substituted (a
"substituted alkenyl") with one or more substituents. In certain embodiments,
the alkenyl group
is an unsubstituted C1-20 alkenyl. In certain embodiments, the alkenyl group
is a substituted C1-20
alkenyl. In an alkenyl group, a C=C double bond for which the stereochemistry
is not specified
µ2Pr.jj
(e.g., ¨CH=CHCH3 or ) may be in the (E)- or (Z)-configuration.
The term "heteroalkenyl" refers to an alkenyl group, which further includes at
least one
heteroatom (e.g., 1, 2, 3, or 4 heteroatoms) selected from oxygen, nitrogen,
or sulfur within (e.g.,
inserted between adjacent carbon atoms of) and/or placed at one or more
terminal position(s) of
the parent chain. In certain embodiments, a heteroalkenyl group refers to a
group having from 1
to 20 carbon atoms, at least one double bond, and 1 or more heteroatoms within
the parent chain
("heteroCi_2o alkenyl"). In certain embodiments, a heteroalkenyl group refers
to a group having
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
from 1 to 12 carbon atoms, at least one double bond, and 1 or more heteroatoms
within the
parent chain ("heteroCi_12 alkenyl"). In certain embodiments, a heteroalkenyl
group refers to a
group having from 1 to 11 carbon atoms, at least one double bond, and 1 or
more heteroatoms
within the parent chain ("heteroCi_ii alkenyl"). In certain embodiments, a
heteroalkenyl group
5 refers to a group having from 1 to 10 carbon atoms, at least one double
bond, and 1 or more
heteroatoms within the parent chain ("heteroCi_io alkenyl"). In some
embodiments, a
heteroalkenyl group has 1 to 9 carbon atoms at least one double bond, and 1 or
more heteroatoms
within the parent chain ("heteroCi_9 alkenyl"). In some embodiments, a
heteroalkenyl group has
1 to 8 carbon atoms, at least one double bond, and 1 or more heteroatoms
within the parent chain
10 ("heteroCi_8 alkenyl"). In some embodiments, a heteroalkenyl group has 1
to 7 carbon atoms, at
least one double bond, and 1 or more heteroatoms within the parent chain
("heteroCi_7 alkenyl").
In some embodiments, a heteroalkenyl group has lto 6 carbon atoms, at least
one double bond,
and 1 or more heteroatoms within the parent chain ("heteroCi_6 alkenyl"). In
some embodiments,
a heteroalkenyl group has 1 to 5 carbon atoms, at least one double bond, and 1
or 2 heteroatoms
15 within the parent chain ("heteroCi_s alkenyl"). In some embodiments, a
heteroalkenyl group has
1 to 4 carbon atoms, at least one double bond, and 1 or 2 heteroatoms within
the parent chain
("heteroCi_4 alkenyl"). In some embodiments, a heteroalkenyl group has 1 to 3
carbon atoms, at
least one double bond, and 1 heteroatom within the parent chain ("heteroCi_3
alkenyl"). In some
embodiments, a heteroalkenyl group has 1 to 2 carbon atoms, at least one
double bond, and 1
20 .. heteroatom within the parent chain ("heteroCi_2 alkenyl"). In some
embodiments, a
heteroalkenyl group has 1 to 6 carbon atoms, at least one double bond, and 1
or 2 heteroatoms
within the parent chain ("heteroCi_6 alkenyl"). Unless otherwise specified,
each instance of a
heteroalkenyl group is independently unsubstituted (an "unsubstituted
heteroalkenyl") or
substituted (a "substituted heteroalkenyl") with one or more substituents. In
certain
25 embodiments, the heteroalkenyl group is an unsubstituted heteroC 1_20
alkenyl. In certain
embodiments, the heteroalkenyl group is a substituted heteroC 1-20 alkenyl.
The term "alkynyl" refers to a radical of a straight-chain or branched
hydrocarbon group
having from 1 to 20 carbon atoms and one or more carbon-carbon triple bonds
(e.g., 1, 2, 3, or 4
triple bonds) ("C1_20 alkynyl"). In some embodiments, an alkynyl group has 1
to 10 carbon atoms
30 .. ("Ci_io alkynyl"). In some embodiments, an alkynyl group has 1 to 9
carbon atoms ("C1-9
alkynyl"). In some embodiments, an alkynyl group has 1 to 8 carbon atoms
("C1_8 alkynyl"). In
some embodiments, an alkynyl group has 1 to 7 carbon atoms ("Ci_7 alkynyl").
In some
embodiments, an alkynyl group has 1 to 6 carbon atoms ("C1-6 alkynyl"). In
some embodiments,
an alkynyl group has 1 to 5 carbon atoms ("C1-5 alkynyl"). In some
embodiments, an alkynyl
35 group has 1 to 4 carbon atoms ("C1_4 alkynyl"). In some embodiments, an
alkynyl group has 1 to
CA 03177368 2022-09-27
WO 2021/216763 PCT/US2021/028471
51
3 carbon atoms ("Ci_3 alkynyl"). In some embodiments, an alkynyl group has 1
to 2 carbon
atoms ("Ci_2 alkynyl"). In some embodiments, an alkynyl group has 1 carbon
atom ("Ci
alkynyl"). The one or more carbon-carbon triple bonds can be internal (such as
in 2-butynyl) or
terminal (such as in 1-butyny1). Examples of C1-4 alkynyl groups include,
without limitation,
methylidynyl (CO, ethynyl (C2), 1-propynyl (C3), 2-propynyl (C3), 1-butynyl
(C4), 2-butynyl
(C4), and the like. Examples of C1-6 alkenyl groups include the aforementioned
C2-4 alkynyl
groups as well as pentynyl (Cs), hexynyl (C6), and the like. Additional
examples of alkynyl
include heptynyl (C7), octynyl (C8), and the like. Unless otherwise specified,
each instance of an
alkynyl group is independently unsubstituted (an "unsubstituted alkynyl") or
substituted (a
"substituted alkynyl") with one or more substituents. In certain embodiments,
the alkynyl group
is an unsubstituted C1-20 alkynyl. In certain embodiments, the alkynyl group
is a substituted C1-20
alkynyl.
The term "heteroalkynyl" refers to an alkynyl group, which further includes at
least one
heteroatom (e.g., 1, 2, 3, or 4 heteroatoms) selected from oxygen, nitrogen,
or sulfur within (e.g.,
inserted between adjacent carbon atoms of) and/or placed at one or more
terminal position(s) of
the parent chain. In certain embodiments, a heteroalkynyl group refers to a
group having from 1
to 20 carbon atoms, at least one triple bond, and 1 or more heteroatoms within
the parent chain
("heteroCi_20 alkynyl"). In certain embodiments, a heteroalkynyl group refers
to a group having
from 1 to 10 carbon atoms, at least one triple bond, and 1 or more heteroatoms
within the parent
chain ("heteroCi_io alkynyl"). In some embodiments, a heteroalkynyl group has
1 to 9 carbon
atoms, at least one triple bond, and 1 or more heteroatoms within the parent
chain ("heteroC 1-9
alkynyl"). In some embodiments, a heteroalkynyl group has 1 to 8 carbon atoms,
at least one
triple bond, and 1 or more heteroatoms within the parent chain ("heteroC 1-8
alkynyl"). In some
embodiments, a heteroalkynyl group has 1 to 7 carbon atoms, at least one
triple bond, and 1 or
more heteroatoms within the parent chain ("heteroC 1-7 alkynyl"). In some
embodiments, a
heteroalkynyl group has 1 to 6 carbon atoms, at least one triple bond, and 1
or more heteroatoms
within the parent chain ("heteroCi_6 alkynyl"). In some embodiments, a
heteroalkynyl group has
1 to 5 carbon atoms, at least one triple bond, and 1 or 2 heteroatoms within
the parent chain
("heteroCi_s alkynyl"). In some embodiments, a heteroalkynyl group has 1 to 4
carbon atoms, at
least one triple bond, and 1 or 2 heteroatoms within the parent chain
("heteroC 1-4 alkynyl"). In
some embodiments, a heteroalkynyl group has 1 to 3 carbon atoms, at least one
triple bond, and
1 heteroatom within the parent chain ("heteroCi_3 alkynyl"). In some
embodiments, a
heteroalkynyl group has 1 to 2 carbon atoms, at least one triple bond, and 1
heteroatom within
the parent chain ("heteroCi_2 alkynyl"). In some embodiments, a heteroalkynyl
group has 1 to 6
carbon atoms, at least one triple bond, and 1 or 2 heteroatoms within the
parent chain ("heteroCi_
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
52
6 alkynyl"). Unless otherwise specified, each instance of a heteroalkynyl
group is independently
unsubstituted (an "unsubstituted heteroalkynyl") or substituted (a
"substituted heteroalkynyl")
with one or more substituents. In certain embodiments, the heteroalkynyl group
is an
unsubstituted heteroCi_20 alkynyl. In certain embodiments, the heteroalkynyl
group is a
substituted heteroC1-20 alkynyl.
"Aralkyl" is a subset of "alkyl" and refers to an alkyl group substituted by
an aryl group,
wherein the point of attachment is on the alkyl moiety
The term "cycloalkyl" refers to cyclic alkyl radical having from 3 to 10 ring
carbon
atoms ("C3_io cycloalkyl"). In some embodiments, a cycloalkyl group has 3 to 8
ring carbon
atoms ("C3_8 cycloalkyl"). In some embodiments, a cycloalkyl group has 3 to 6
ring carbon
atoms ("C3_6 cycloalkyl"). In some embodiments, a cycloalkyl group has 5 to 6
ring carbon
atoms ("C5-6 cycloalkyl"). In some embodiments, a cycloalkyl group has 5 to 10
ring carbon
atoms ("Cs_io cycloalkyl"). Examples of C5-6 cycloalkyl groups include
cyclopentyl (Cs) and
cyclohexyl (Cs). Examples of C3-6 cycloalkyl groups include the aforementioned
C5-6 cycloalkyl
groups as well as cyclopropyl (C3) and cyclobutyl (C4). Examples of C3-8
cycloalkyl groups
include the aforementioned C3_6 cycloalkyl groups as well as cycloheptyl (C7)
and cyclooctyl
(C8). Unless otherwise specified, each instance of a cycloalkyl group is
independently
unsubstituted (an "unsubstituted cycloalkyl") or substituted (a "substituted
cycloalkyl") with one
or more substituents. In certain embodiments, the cycloalkyl group is
unsubstituted C3-10
cycloalkyl. In certain embodiments, the cycloalkyl group is substituted C3-10
cycloalkyl.
The term "heteroalkyl," as used herein, refers to an alkyl group, as defined
herein, in
which one or more of the constituent carbon atoms have been replaced by a
heteroatom or
RN
optionally substituted heteroatom, e.g., nitrogen (e.g.,
\( 1 ), oxygen (e.g., 0
Y), or sulfur
0 0, 0
(e.g., ( Y ( Y \S*
\S \'
, or ). Heteroalkyl groups may be optionally
substituted with one,
two, three, or, in the case of alkyl groups of two carbons or more, four,
five, or six substituents
independently selected from any of the substituents described herein.
Heteroalkyl group
substituents include: (1) carbonyl; (2) halo; (3) C6-C10 aryl; and (4) C3-C10
carbocyclyl. A
heteroalkylene is a divalent heteroalkyl group.
The term "alkoxy," as used herein, refers to _OR, where Ra is, e.g., alkyl,
alkenyl,
alkynyl, aryl, alkylaryl, carbocyclyl, heterocyclyl, or heteroaryl. Examples
of alkoxy groups
include methoxy, ethoxy, isopropoxy, tert-butoxy, phenoxy, and benzyloxy.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
53
The term "aryl" refers to a radical of a monocyclic or polycyclic (e.g.,
bicyclic or
tricyclic) 4n+2 aromatic ring system (e.g., having 6, 10, or 14 it electrons
shared in a cyclic
array) having 6-14 ring carbon atoms and zero heteroatoms provided in the
aromatic ring system
("C6-14 aryl"). In some embodiments, an aryl group has 6 ring carbon atoms
("C6 aryl"; e.g.,
phenyl). In some embodiments, an aryl group has 10 ring carbon atoms ("Cio
aryl"; e.g.,
naphthyl such as 1¨naphthyl and 2¨naphthyl). In some embodiments, an aryl
group has 14 ring
carbon atoms ("C14 aryl"; e.g., anthracyl). "Aryl" also includes ring systems
wherein the aryl
ring, as defined above, is fused with one or more carbocyclyl or heterocyclyl
groups wherein the
radical or point of attachment is on the aryl ring, and in such instances, the
number of carbon
atoms continue to designate the number of carbon atoms in the aryl ring
system. Unless
otherwise specified, each instance of an aryl group is independently
unsubstituted (an
"unsubstituted aryl") or substituted (a "substituted aryl") with one or more
substituents (e.g., -F, -
OH or -0(Ci_6 alkyl) . In certain embodiments, the aryl group is an
unsubstituted C6-14 aryl. In
certain embodiments, the aryl group is a substituted C6_14 aryl.
The term "aryloxy" refers to an -0-aryl substituent.
The term "heteroaryl" refers to a radical of a 5-14 membered monocyclic or
polycyclic
(e.g., bicyclic, tricyclic) 4n+2 aromatic ring system (e.g., having 6, 10, or
14 it electrons shared
in a cyclic array) having ring carbon atoms and 1-4 ring heteroatoms provided
in the aromatic
ring system, wherein each heteroatom is independently selected from nitrogen,
oxygen, and
sulfur ("5-14 membered heteroaryl"). In heteroaryl groups that contain one or
more nitrogen
atoms, the point of attachment can be a carbon or nitrogen atom, as valency
permits. Heteroaryl
polycyclic ring systems can include one or more heteroatoms in one or both
rings. "Heteroaryl"
includes ring systems wherein the heteroaryl ring, as defined above, is fused
with one or more
carbocyclyl or heterocyclyl groups wherein the point of attachment is on the
heteroaryl ring, and
in such instances, the number of ring members continue to designate the number
of ring
members in the heteroaryl ring system. "Heteroaryl" also includes ring systems
wherein the
heteroaryl ring, as defined above, is fused with one or more aryl groups
wherein the point of
attachment is either on the aryl or heteroaryl ring, and in such instances,
the number of ring
members designates the number of ring members in the fused polycyclic
(aryl/heteroaryl) ring
system. Polycyclic heteroaryl groups wherein one ring does not contain a
heteroatom (e.g.,
indolyl, quinolinyl, carbazolyl, and the like) the point of attachment can be
on either ring, e.g.,
either the ring bearing a heteroatom (e.g., 2-indoly1) or the ring that does
not contain a
heteroatom (e.g., 5-indoly1). In certain embodiments, the heteroaryl is
substituted or
unsubstituted, 5- or 6-membered, monocyclic heteroaryl, wherein 1, 2, 3, or 4
atoms in the
heteroaryl ring system are independently oxygen, nitrogen, or sulfur. In
certain embodiments, the
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
54
heteroaryl is substituted or unsubstituted, 9- or 10-membered, bicyclic
heteroaryl, wherein 1,2,
3, or 4 atoms in the heteroaryl ring system are independently oxygen,
nitrogen, or sulfur.
In some embodiments, a heteroaryl group is a 5-10 membered aromatic ring
system having ring
carbon atoms and 1-4 ring heteroatoms provided in the aromatic ring system,
wherein each
heteroatom is independently selected from nitrogen, oxygen, and sulfur ("5-10
membered
heteroaryl"). In some embodiments, a heteroaryl group is a 5-8 membered
aromatic ring system
having ring carbon atoms and 1-4 ring heteroatoms provided in the aromatic
ring system,
wherein each heteroatom is independently selected from nitrogen, oxygen, and
sulfur ("5-8
membered heteroaryl"). In some embodiments, a heteroaryl group is a 5-6
membered aromatic
ring system having ring carbon atoms and 1-4 ring heteroatoms provided in the
aromatic ring
system, wherein each heteroatom is independently selected from nitrogen,
oxygen, and sulfur
("5-6 membered heteroaryl"). In some embodiments, the 5-6 membered heteroaryl
has 1-3 ring
heteroatoms selected from nitrogen, oxygen, and sulfur. In some embodiments,
the 5-6
membered heteroaryl has 1-2 ring heteroatoms selected from nitrogen, oxygen,
and sulfur. In
some embodiments, the 5-6 membered heteroaryl has 1 ring heteroatom selected
from nitrogen,
oxygen, and sulfur. Unless otherwise specified, each instance of a heteroaryl
group is
independently unsubstituted (an "unsubstituted heteroaryl") or substituted (a
"substituted
heteroaryl") with one or more substituents. In certain embodiments, the
heteroaryl group is an
unsubstituted 5-14 membered heteroaryl. In certain embodiments, the heteroaryl
group is a
substituted 5-14 membered heteroaryl.
The term "heterocyclyl" or "heterocyclic" refers to a radical of a 3- to 14-
membered non-
aromatic ring system having ring carbon atoms and 1 to 4 ring heteroatoms,
wherein each
heteroatom is independently selected from nitrogen, oxygen, and sulfur ("3-14
membered
heterocyclyl"). In heterocyclyl groups that contain one or more nitrogen
atoms, the point of
attachment can be a carbon or nitrogen atom, as valency permits. A
heterocyclyl group can either
be monocyclic ("monocyclic heterocyclyl") or polycyclic (e.g., a fused,
bridged or spiro ring
system such as a bicyclic system ("bicyclic heterocyclyl") or tricyclic system
("tricyclic
heterocyclyl")), and can be saturated or can contain one or more carbon-carbon
double or triple
bonds. Heterocyclyl polycyclic ring systems can include one or more
heteroatoms in one or both
rings. "Heterocycly1" also includes ring systems wherein the heterocyclyl
ring, as defined above,
is fused with one or more carbocyclyl groups wherein the point of attachment
is either on the
carbocyclyl or heterocyclyl ring, or ring systems wherein the heterocyclyl
ring, as defined above,
is fused with one or more aryl or heteroaryl groups, wherein the point of
attachment is on the
heterocyclyl ring, and in such instances, the number of ring members continue
to designate the
number of ring members in the heterocyclyl ring system. Unless otherwise
specified, each
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
instance of heterocyclyl is independently unsubstituted (an "unsubstituted
heterocyclyl") or
substituted (a "substituted heterocyclyl") with one or more substituents. In
certain embodiments,
the heterocyclyl group is an unsubstituted 3-14 membered heterocyclyl. In
certain embodiments,
the heterocyclyl group is a substituted 3-14 membered heterocyclyl. In certain
embodiments, the
5 heterocyclyl is substituted or unsubstituted, 3- to 7-membered,
monocyclic heterocyclyl, wherein
1,2, or 3 atoms in the heterocyclic ring system are independently oxygen,
nitrogen, or sulfur, as
valency permits.
In some embodiments, a heterocyclyl group is a 5-10 membered non-aromatic ring
system having ring carbon atoms and 1-4 ring heteroatoms, wherein each
heteroatom is
10 independently selected from nitrogen, oxygen, and sulfur ("5-10 membered
heterocyclyl"). In
some embodiments, a heterocyclyl group is a 5-8 membered non-aromatic ring
system having
ring carbon atoms and 1-4 ring heteroatoms, wherein each heteroatom is
independently selected
from nitrogen, oxygen, and sulfur ("5-8 membered heterocyclyl"). In some
embodiments, a
heterocyclyl group is a 5-6 membered non-aromatic ring system having ring
carbon atoms and
15 1-4 ring heteroatoms, wherein each heteroatom is independently selected
from nitrogen, oxygen,
and sulfur ("5-6 membered heterocyclyl"). In some embodiments, the 5-6
membered
heterocyclyl has 1-3 ring heteroatoms selected from nitrogen, oxygen, and
sulfur. In some
embodiments, the 5-6 membered heterocyclyl has 1-2 ring heteroatoms selected
from nitrogen,
oxygen, and sulfur. In some embodiments, the 5-6 membered heterocyclyl has 1
ring heteroatom
20 selected from nitrogen, oxygen, and sulfur.
The term "carbonyl" refers a group wherein the carbon directly attached to the
parent
molecule is sp2 hybridized, and is substituted with an oxygen, nitrogen or
sulfur atom, e.g., a
group selected from ketones (e.g., ¨C(=0)Raa), carboxylic acids (e.g., ¨CO2H),
aldehydes (¨
CHO), esters (e.g., ¨CO2Raa, ¨C(=0)SRaa, ¨C(=S)SRaa), amides (e.g.,
¨C(=0)N(Rbb)2, ¨
25 C(=o)NRbbso2Raa, C(=S)N(Rbb)2), and imines (e.g., ¨C(=NRbb)Raa,
(=NRbb)0Raa),
(=NRbb)N(R) bbµ 2.µ
) wherein Raa and Rbb are as defined herein.
The term "amino," as used herein, represents ¨N(RN)2, wherein each RN is,
µ
independently, H, OH, NO2, N(RNo )2, S020RN0, s02RN0, soRNo, an N-protecting
group, alkyl,
alkoxy, aryl, cycloalkyl, acyl (e.g., acetyl, trifluoroacetyl, or others
described herein), wherein
30 each of these recited RN groups can be optionally substituted; or two RN
combine to form an
alkylene or heteroalkylene, and wherein each RN is, independently, H, alkyl,
or aryl. The amino
groups of the disclosure can be an unsubstituted amino (i.e., ¨NH2) or a
substituted amino (i.e., ¨
N(RN)2).
The term "substituted" as used herein means at least one hydrogen atom is
replaced by a
35 bond to a non-hydrogen atoms such as, but not limited to: a halogen atom
such as F, Cl, Br, and
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
56
I; an oxygen atom in groups such as hydroxyl groups, alkoxy groups, and ester
groups; a sulfur
atom in groups such as thiol groups, thioalkyl groups, sulfone groups,
sulfonyl groups, and
sulfoxide groups; a nitrogen atom in groups such as amines, amides,
alkylamines, dialkylamines,
arylamines, alkylarylamines, diarylamines, N-oxides, imides, and enamines; a
silicon atom in
groups such as trialkylsilyl groups, dialkylarylsilyl groups, alkyldiarylsilyl
groups, and
triarylsilyl groups; and other heteroatoms in various other groups.
"Substituted" also means one
or more hydrogen atoms are replaced by a higher-order bond (e.g., a double- or
triple-bond) to a
heteroatom such as oxygen in oxo, carbonyl, carboxyl, and ester groups; and
nitrogen in groups
such as imines, oximes, hydrazones, and nitriles. For example, in some
embodiments
"substituted" means one or more hydrogen atoms are replaced with NRgRh,
NRgC(=0)Rh,
NRgC(=0)NRgRh, NRgC(=0)ORh, NRgS02Rh, OC(=0)NRgRh, ORg, SRg, SORg, SO2Rg,
OSO2Rg, SO2ORg, =NSO2Rg, and SO2NRgRh. "Substituted also means one or more
hydrogen
atoms are replaced with C(=0)Rg, C(=0)0Rg, C(=0)NRgRh, CH2S02Rg, CH2S02NRgRh.
In the foregoing, Rg and Rh are the same or different and independently
hydrogen, alkyl, alkoxy,
.. alkylaminyl, thioalkyl, aryl, aralkyl, cycloalkyl, cycloalkylalkyl,
haloalkyl, heterocyclyl, N-
heterocyclyl, heterocyclylalkyl, heteroaryl, N-heteroaryl and/or
heteroarylalkyl. "Substituted"
further means one or more hydrogen atoms are replaced by a bond to an aminyl,
cyano,
hydroxyl, imino, nitro, oxo, thioxo, halo, alkyl, alkoxy, alkylaminyl,
thioalkyl, aryl, aralkyl,
cycloalkyl, cycloalkylalkyl, haloalkyl, heterocyclyl, N-heterocyclyl,
heterocyclylalkyl,
heteroaryl, N-heteroaryl and/or heteroarylalkyl group. In addition, each of
the foregoing
substituents may also be optionally substituted with one or more of the above
substituents.
The terms "salt thereof' or "salts thereof' as used herein refer to salts
which are well
known in the art. For example, Berge et al., describe pharmaceutically
acceptable salts in detail
in J. Pharmaceutical Sciences, 1977, 66, 1-19, incorporated herein by
reference. Additional
information on suitable salts can be found in Remington's Pharmaceutical
Sciences, 17th ed.,
Mack Publishing Company, Easton, Pa., 1985, which is incorporated herein by
reference.
Salts of the compounds of this invention include those derived from suitable
inorganic
and organic acids and bases. Examples of acid addition salts are salts of an
amino group formed
with inorganic acids such as hydrochloric acid, hydrobromic acid, phosphoric
acid, sulfuric acid
and perchloric acid or with organic acids such as acetic acid, oxalic acid,
maleic acid, tartaric
acid, citric acid, succinic acid or malonic acid or by using other methods
used in the art such as
ion exchange. Other pharmaceutically acceptable salts include adipate,
alginate, ascorbate,
aspartate, benzenesulfonate, benzoate, bisulfate, borate, butyrate,
camphorate, camphorsulfonate,
citrate, cyclopentanepropionate, digluconate, dodecylsulfate, ethanesulfonate,
formate, fumarate,
glucoheptonate, glycerophosphate, gluconate, hemisulfate, heptanoate,
hexanoate, hydroiodide,
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
57
2¨hydroxy¨ethanesulfonate, lactobionate, lactate, laurate, lauryl sulfate,
malate, maleate,
malonate, methanesulfonate, 2¨naphthalenesulfonate, nicotinate, nitrate,
oleate, oxalate,
palmitate, pamoate, pectinate, persulfate, 3¨phenylpropionate, phosphate,
picrate, pivalate,
propionate, stearate, succinate, sulfate, tartrate, thiocyanate,
p¨toluenesulfonate, undecanoate,
.. valerate salts, and the like. Salts derived from appropriate bases include
alkali metal, alkaline
earth metal, ammonium and N (C1_4alky1)4 salts. Representative alkali or
alkaline earth metal
salts include sodium, lithium, potassium, calcium, magnesium, and the like.
Further
pharmaceutically acceptable salts include, when appropriate, nontoxic
ammonium, quaternary
ammonium, and amine cations formed using counter ions such as halide,
hydroxide, carboxylate,
sulfate, phosphate, nitrate, lower alkyl sulfonate and aryl sulfonate.
A "protein," "peptide," or "polypeptide" comprises a polymer of amino acid
residues
linked together by peptide bonds. The terms refer to proteins, polypeptides,
and peptides of any
size, structure, or function. Typically, a protein or peptide will be at least
three amino acids in
length. In some embodiments, a peptide is between about 3 and about 100 amino
acids in length
(e.g., between about 5 and about 25, between about 10 and about 80, between
about 15 and about
70, or between about 20 and about 40, amino acids in length). In some
embodiments, a peptide is
between about 6 and about 40 amino acids in length (e.g., between about 6 and
about 30,
between about 10 and about 30, between about 15 and about 40, or between about
20 and about
30, amino acids in length). In some embodiments, a plurality of peptides can
refer to a plurality
of peptide molecules, where each peptide molecule of the plurality comprises
an amino acid
sequence that is different from any other peptide molecule of the plurality.
In some
embodiments, a plurality of peptides can include at least 1 peptide and up to
1,000 peptides (e.g.,
at least 1 peptide and up to 10, 50, 100, 250, or 500 peptides). In some
embodiments, a plurality
of peptides comprises 1-5, 5-10, 1-15, 15-20, 10-100, 50-250, 100-500, 500-
1,000, or more,
different peptides. A protein may refer to an individual protein or a
collection of proteins.
Inventive proteins preferably contain only natural amino acids, although non-
natural amino acids
(i.e., compounds that do not occur in nature but that can be incorporated into
a polypeptide
chain) and/or amino acid analogs as are known in the art may alternatively be
employed. Also,
one or more of the amino acids in a protein may be modified, for example, by
the addition of a
chemical entity such as a carbohydrate group, a hydroxyl group, a phosphate
group, a farnesyl
group, an isofarnesyl group, a fatty acid group, a linker for conjugation or
functionalization, or
other modification. A protein may also be a single molecule or may be a multi-
molecular
complex. A protein or peptide may be a fragment of a naturally occurring
protein or peptide. A
protein may be naturally occurring, recombinant, synthetic, or any combination
of these.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
58
With respect to the use of substantially any plural and/or singular terms
herein, those having skill
in the art can translate from the plural to the singular and/or from the
singular to plural as is
appropriate to the context and/or application. The various singular/plural
permutations can be
expressly set forth herein for sake of clarity.
It will be understood by those within the art that, in general, terms used
herein, and
especially in the appended claims (for example, bodies of the appended claims)
are generally
intended as "open" terms (for example, the term "including" should be
interpreted as "including
but not limited to," the term "having" should be interpreted as "having at
least," the term
"includes" should be interpreted as "includes but is not limited to," etc.).
It will be further
understood by those within the art that if a specific number of an introduced
claim recitation is
intended, such an intent will be explicitly recited in the claim, and in the
absence of such
recitation no such intent is present. For example, as an aid to understanding,
the following
appended claims can contain usage of the introductory phrases "at least one"
and "one or more"
to introduce claim recitations. However, the use of such phrases should not be
construed to imply
that the introduction of a claim recitation by the indefinite articles "a" or
"an" limits any
particular claim containing such introduced claim recitation to embodiments
containing only one
such recitation, even when the same claim includes the introductory phrases
"one or more" or "at
least one" and indefinite articles such as "a" or "an" (for example, "a"
and/or "an" should be
interpreted to mean "at least one" or "one or more"); the same holds true for
the use of definite
articles used to introduce claim recitations. In addition, even if a specific
number of an
introduced claim recitation is explicitly recited, those skilled in the art
will recognize that such
recitation should be interpreted to mean at least the recited number (for
example, the bare
recitation of "two recitations," without other modifiers, means at least two
recitations, or two or
more recitations). Furthermore, in those instances where a convention
analogous to "at least one
.. of A, B, and C, etc." is used, in general such a construction is intended
in the sense one having
skill in the art would understand the convention (for example, " a system
having at least one of
A, B, and C" would include but not be limited to systems that have A alone, B
alone, C alone, A
and B together, A and C together, B and C together, and/or A, B, and C
together, etc.). In those
instances where a convention analogous to "at least one of A, B, or C, etc."
is used, in general
such a construction is intended in the sense one having skill in the art would
understand the
convention (for example," a system having at least one of A, B, or C" would
include but not be
limited to systems that have A alone, B alone, C alone, A and B together, A
and C together, B
and C together, and/or A, B, and C together, etc.). It will be further
understood by those within
the art that virtually any disjunctive word and/or phrase presenting two or
more alternative terms,
whether in the description, claims, or drawings, should be understood to
contemplate the
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
59
possibilities of including one of the terms, either of the terms, or both
terms. For example, the
phrase "A or B" will be understood to include the possibilities of "A" or "B"
or "A and B."
In addition, where features or aspects of the disclosure are described in
terms of Markush groups,
those skilled in the art will recognize that the disclosure is also thereby
described in terms of any
individual member or subgroup of members of the Markush group.
As will be understood by one skilled in the art, for any and all purposes,
such as in terms
of providing a written description, all ranges disclosed herein also encompass
any and all
possible sub-ranges and combinations of sub-ranges thereof. Any listed range
can be easily
recognized as sufficiently describing and enabling the same range being broken
down into at
least equal halves, thirds, quarters, fifths, tenths, etc. As a non-limiting
example, each range
discussed herein can be readily broken down into a lower third, middle third
and upper third, etc.
As will also be understood by one skilled in the art all language such as "up
to," "at least,"
"greater than," "less than," and the like include the number recited and refer
to ranges which can
be subsequently broken down into sub-ranges as discussed above. Finally, as
will be understood
by one skilled in the art, a range includes each individual member. Thus, for
example, a group
having 1-3 articles refers to groups having 1, 2, or 3 articles. Similarly, a
group having 1-5
articles refers to groups having 1, 2, 3, 4, or 5 articles, and so forth.
Those skilled in the art will appreciate that certain compounds described
herein can exist
in one or more different isomeric (e.g., stereoisomers, geometric isomers,
tautomers) and/or
isotopic (e.g., in which one or more atoms has been substituted with a
different isotope of the
atom, such as hydrogen substituted for deuterium) forms. Unless otherwise
indicated or clear
from context, a depicted structure can be understood to represent any such
isomeric or isotopic
form, individually or in combination.
Peptide Surface Immobilization
In certain single molecule analytical methods, a molecule to be analyzed is
immobilized
onto surfaces such that the molecule may be monitored without interference
from other reaction
components in solution. In some embodiments, surface immobilization of the
molecule allows
the molecule to be confined to a desired region of a surface for real-time
monitoring of a reaction
involving the molecule.
Accordingly, in some aspects, the application provides methods of immobilizing
a
peptide to a surface by attaching any one of the compounds described herein to
a surface of a
solid support. In some embodiments, the methods comprise contacting a compound
of Formula
(V), (X), (XV), or a salt thereof, to a surface of a solid support. In some
embodiments, the
surface is functionalized with a complementary functional moiety configured
for attachment
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
(e.g., covalent or non-covalent attachment) to a functionalized terminal end
of a peptide. In some
embodiments, the solid support comprises a plurality of sample wells formed at
the surface of the
solid support. In some embodiments, the methods comprise immobilizing a single
peptide to a
surface of each of a plurality of sample wells. In some embodiments, confining
a single peptide
5 per sample well is advantageous for single molecule detection methods,
e.g., single molecule
peptide sequencing.
As used herein, in some embodiments, a surface refers to a surface of a
substrate or solid
support. In some embodiments, a solid support refers to a material, layer, or
other structure
having a surface, such as a receiving surface, that is capable of supporting a
deposited material,
10 such as a functionalized peptide described herein. In some embodiments,
a receiving surface of a
substrate may optionally have one or more features, including nanoscale or
microscale recessed
features such as an array of sample wells. In some embodiments, an array is a
planar
arrangement of elements such as sensors or sample wells. An array may be one
or two
dimensional. A one dimensional array is an array having one column or row of
elements in the
15 first dimension and a plurality of columns or rows in the second
dimension. The number of
columns or rows in the first and second dimensions may or may not be the same.
In some
embodiments, the array may include, for example, 102, 103, 104, 105, 106, or
107 sample wells.
An example scheme of peptide surface immobilization is depicted in FIG. 9. As
shown,
panels (I)-(II) depict a process of immobilizing a peptide 900 that comprises
a functionalized
20 terminal end 902. In panel (I), a solid support comprising a sample well
is shown. In some
embodiments, the sample well is formed by a bottom surface comprising a non-
metallic layer
910 and side wall surfaces comprising a metallic layer 912. In some
embodiments, non-metallic
layer 910 comprises a transparent layer (e.g., glass, silica). In some
embodiments, metallic layer
912 comprises a metal oxide surface (e.g., titanium dioxide). In some
embodiments, metallic
25 layer 912 comprises a passivation coating 914 (e.g., a phosphorus-
containing layer, such as an
organophosphonate layer). As shown, the bottom surface comprising non-metallic
layer 910
comprises a complementary functional moiety 904. Methods of selective surface
modification
and functionalization are described in further detail in U.S. Patent
Publication No. 2018/0326412
and U.S. Provisional Application No. 62/914,356, the contents of each of which
are hereby
30 incorporated by reference.
In some embodiments, peptide 900 comprising functionalized terminal end 902 is
contacted with complementary functional moiety 904 of the solid support to
form a covalent or
non-covalent linkage group. In some embodiments, functionalized terminal end
902 and
complementary functional moiety 904 comprise partner click chemistry handles,
e.g., which
35 form a covalent linkage group between peptide 900 and the solid support.
Suitable click
CA 03177368 2022-09-27
WO 2021/216763 PCT/US2021/028471
61
chemistry handles are described elsewhere herein. In some embodiments,
functionalized terminal
end 902 and complementary functional moiety 904 comprise non-covalent binding
partners, e.g.,
which form a non-covalent linkage group between peptide 900 and the solid
support. Examples
of non-covalent binding partners include complementary oligonucleotide strands
(e.g.,
complementary nucleic acid strands, including DNA, RNA, and variants thereof),
protein-protein
binding partners (e.g., barnase and barstar), and protein-ligand binding
partners (e.g., biotin and
streptavidin).
In panel (II), peptide 900 is shown immobilized to the bottom surface through
a linkage
group formed by contacting functionalized terminal end 902 and complementary
functional
moiety 904. In this example, peptide 900 is attached through a non-covalent
linkage group,
which is depicted in the zoomed region of panel (III). As shown, in some
embodiments, the non-
covalent linkage group comprises an avidin protein 920. Avidin proteins are
biotin-binding
proteins, generally having a biotin binding site at each of four subunits of
the avidin protein.
Avidin proteins include, for example, avidin, streptavidin, traptavidin,
tamavidin, bradavidin,
xenavidin, and homologs and variants thereof. In some embodiments, avidin
protein 920 is
streptavidin. The multivalency of avidin protein 920 can allow for various
linkage
configurations, as each of the four binding sites are independently capable of
binding a biotin
molecule (shown as white circles).
As shown in panel (III), in some embodiments, the non-covalent linkage is
formed by
avidin protein 920 bound to a first bis-biotin moiety 922 and a second bis-
biotin moiety 924. In
some embodiments, functionalized terminal end 902 comprises first bis-biotin
moiety 922, and
complementary functional moiety 904 comprises second bis-biotin moiety 924. In
some
embodiments, functionalized terminal end 902 comprises avidin protein 920
prior to being
contacted with complementary functional moiety 904. In some embodiments,
complementary
functional moiety 904 comprises avidin protein 920 prior to being contacted
with functionalized
terminal end 902.
In some embodiments, functionalized terminal end 902 comprises first bis-
biotin moiety
922 and a water-soluble moiety, where the water-soluble moiety forms a linkage
between first
bis-biotin moiety 922 and an amino acid (e.g., a terminal amino acid) of
peptide 900. Water-
soluble moieties are described in detail elsewhere herein.
Protein Sequencing Process
Aspects of the instant disclosure also involve methods of protein sequencing
and
identification, methods of protein sequencing and identification, methods of
amino acid
.. identification, and compositions, systems, and devices for performing such
methods. Such
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
62
protein sequencing and identification is performed, in some embodiments, with
the same
instrument that performs sample preparation and/or genome sequencing,
described in more detail
herein. In some aspects, methods of determining the sequence of a target
protein are described.
In some embodiments, the target protein is enriched (e.g., enriched using
electrophoretic
methods, e.g., affinity SCODA) prior to determining the sequence of the target
protein. In some
aspects, methods of determining the sequences of a plurality of proteins
(e.g., at least 2, 3, 4, 5,
10, 15, 20, 30, 50, or more) present in a sample (e.g., a purified sample, a
cell lysate, a single-
cell, a population of cells, or a tissue) are described. In some embodiments,
a sample is prepared
as described herein (e.g., lysed, purified, fragmented, and/or enriched for a
target protein) prior
.. to determining the sequence of a target protein or a plurality of proteins
present in a sample. In
some embodiments, a target protein is an enriched target protein (e.g.,
enriched using
electrophoretic methods, e.g., affinity SCODA)
In some embodiments, the instant disclosure provides methods of sequencing
and/or
identifying an individual protein in a sample comprising a plurality of
proteins by identifying one
or more types of amino acids of a protein from the mixture. In some
embodiments, one or more
amino acids (e.g., terminal amino acids) of the protein are labeled (e.g.,
directly or indirectly, for
example using a binding agent) and the relative positions of the labeled amino
acids in the
protein are determined. In some embodiments, the relative positions of amino
acids in a protein
are determined using a series of amino acid labeling and cleavage steps. In
some embodiments,
the relative position of labeled amino acids in a protein can be determined
without removing
amino acids from the protein but by translocating a labeled protein through a
pore (e.g., a protein
channel) and detecting a signal (e.g., a Forster resonance energy transfer
(FRET) signal) from the
labeled amino acid(s) during translocation through the pore in order to
determine the relative
position of the labeled amino acids in the protein molecule.
In some embodiments, the identity of a terminal amino acid (e.g., an N-
terminal or a C-
terminal amino acid) is determined prior to the terminal amino acid being
removed and the
identity of the next amino acid at the terminal end being assessed; this
process may be repeated
until a plurality of successive amino acids in the protein are assessed. In
some embodiments,
assessing the identity of an amino acid comprises determining the type of
amino acid that is
present. In some embodiments, determining the type of amino acid comprises
determining the
actual amino acid identity (e.g., determining which of the naturally-occurring
20 amino acids an
amino acid is, e.g., using a binding agent that is specific for an individual
terminal amino acid).
However, in some embodiments, assessing the identity of a terminal amino acid
type can
comprise determining a subset of potential amino acids that can be present at
the terminus of the
protein. In some embodiments, this can be accomplished by determining that an
amino acid is
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
63
not one or more specific amino acids (i.e., and therefore could be any of the
other amino acids).
In some embodiments, this can be accomplished by determining which of a
specified subset of
amino acids (e.g., based on size, charge, hydrophobicity, binding properties)
could be at the
terminus of the protein (e.g., using a binding agent that binds to a specified
subset of two or more
terminal amino acids).
In some embodiments, a protein can be digested into a plurality of smaller
proteins and
sequence information can be obtained from one or more of these smaller
proteins (e.g., using a
method that involves sequentially assessing a terminal amino acid of a protein
and removing that
amino acid to expose the next amino acid at the terminus).
In some embodiments, a protein is sequenced from its amino (N) terminus. In
some
embodiments, a protein is sequenced from its carboxy (C) terminus. In some
embodiments, a
first terminus (e.g., N or C terminus) of a protein is immobilized and the
other terminus (e.g., the
C or N terminus) is sequenced as described herein.
As used herein, sequencing a protein refers to determining sequence
information for a
protein. In some embodiments, this can involve determining the identity of
each sequential
amino acid for a portion (or all) of the protein. In some embodiments, this
can involve
determining the identity of a fragment (e.g., a fragment of a target protein
or a fragment of a
sample comprising a plurality of proteins). In some embodiments, this can
involve assessing the
identity of a subset of amino acids within the protein (e.g., and determining
the relative position
of one or more amino acid types without determining the identity of each amino
acid in the
protein). In some embodiments amino acid content information can be obtained
from a protein
without directly determining the relative position of different types of amino
acids in the protein.
The amino acid content alone may be used to infer the identity of the protein
that is present (e.g.,
by comparing the amino acid content to a database of protein information and
determining which
protein(s) have the same amino acid content).
In some embodiments, sequence information for a plurality of protein fragments
obtained
from a target protein or sample comprising a plurality of proteins (e.g., via
enzymatic and/or
chemical cleavage) can be analyzed to reconstruct or infer the sequence of the
target protein or
plurality of proteins present in the sample. Accordingly, in some embodiments,
the one or more
types of amino acids are identified by detecting luminescence of one or more
labeled affinity
reagents that selectively bind the one or more types of amino acids. In some
embodiments, the
one or more types of amino acids are identified by detecting luminescence of a
labeled protein.
In some embodiments, the instant disclosure provides compositions, devices,
and
methods for sequencing a protein by identifying a series of amino acids that
are present at a
terminus of a protein over time (e.g., by iterative detection and cleavage of
amino acids at the
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
64
terminus). In yet other embodiments, the instant disclosure provides
compositions, devices, and
methods for sequencing a protein by identifying labeled amino content of the
protein and
comparing to a reference sequence database.
In some embodiments, the instant disclosure provides compositions, devices,
and
methods for sequencing a protein by sequencing a plurality of fragments of the
protein. In some
embodiments, sequencing a protein comprises combining sequence information for
a plurality of
protein fragments to identify and/or determine a sequence for the protein. In
some embodiments,
combining sequence information may be performed by computer hardware and
software. The
methods described herein may allow for a set of related proteins, such as an
entire proteome of
an organism, to be sequenced. In some embodiments, a plurality of single
molecule sequencing
reactions are performed in parallel (e.g., on a single chip or cartridge)
according to aspects of the
instant disclosure. For example, in some embodiments, a plurality of single
molecule sequencing
reactions are each performed in separate sample wells on a single chip or
cartridge.
In some embodiments, methods provided herein may be used for the sequencing
and
identification of an individual protein in a sample comprising a plurality of
proteins. In some
embodiments, the instant disclosure provides methods of uniquely identifying
an individual
protein in a sample comprising a plurality of proteins. In some embodiments,
an individual
protein is detected in a mixed sample by determining a partial amino acid
sequence of the
protein. In some embodiments, the partial amino acid sequence of the protein
is within a
contiguous stretch of approximately 5-50, 10-50, 25-50, 25-100, or 50-100
amino acids.
Without wishing to be bound by any particular theory, it is expected that most
human proteins
can be identified using incomplete sequence information with reference to
proteomic databases.
For example, simple modeling of the human proteome has shown that
approximately 98% of
proteins can be uniquely identified by detecting just four types of amino
acids within a stretch of
6 to 40 amino acids (see, e.g., Swaminathan, et al. PLoS Comput Biol. 2015,
11(2):e1004080;
and Yao, et al. Phys. Biol. 2015, 12(5):055003). Therefore, a sample
comprising a plurality of
proteins can be fragmented (e.g., chemically degraded, enzymatically degraded)
into short
protein fragments of approximately 6 to 40 amino acids, and sequencing of this
protein-based
library would reveal the identity and abundance of each of the proteins
present in the original
sample. Compositions and methods for selective amino acid labeling and
identifying proteins by
determining partial sequence information are described in in detail in U.S.
Pat. Application No.
15/510,962, filed September 15, 2015, entitled "SINGLE MOLECULE PEPTIDE
SEQUENCING," which is incorporated herein by reference in its entirety.
Sequencing in accordance with the instant disclosure, in some aspects, may
involve
immobilizing a protein (e.g., a target protein) on a surface of a substrate
(e.g., of a solid support,
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
for example a chip or cartridge, for example in an sequencing device or module
as described
herein). In some embodiments, a protein may be immobilized on a surface of a
sample well
(e.g., on a bottom surface of a sample well) on a substrate. In some
embodiments, the N-
terminal amino acid of the protein is immobilized (e.g., attached to the
surface). In some
5 .. embodiments, the C-terminal amino acid of the protein is immobilized
(e.g., attached to the
surface). In some embodiments, one or more non-terminal amino acids are
immobilized (e.g.,
attached to the surface). The immobilized amino acid(s) can be attached using
any suitable
covalent or non-covalent linkage, for example as described in this disclosure.
In some
embodiments, a plurality of proteins are attached to a plurality of sample
wells (e.g., with one
10 protein attached to a surface, for example a bottom surface, of each
sample well), for example in
an array of sample wells on a substrate.
In some embodiments, the identity of a terminal amino acid (e.g., an N-
terminal or a C-
terminal amino acid) is determined, then the terminal amino acid is removed,
and the identity of
the next amino acid at the terminal end is determined. This process may be
repeated until a
15 plurality of successive amino acids in the protein are determined. In
some embodiments,
determining the identity of an amino acid comprises determining the type of
amino acid that is
present. In some embodiments, determining the type of amino acid comprises
determining the
actual amino acid identity, for example by determining which of the naturally-
occurring 20
amino acids is the terminal amino acid is (e.g., using a binding agent that is
specific for an
20 individual terminal amino acid). In some embodiments, the type of amino
acid is selected from
alanine, arginine, asparagine, aspartic acid, cysteine, glutamine, glutamic
acid, glycine, histidine,
isoleucine, leucine, lysine, methionine, phenylalanine, proline,
selenocysteine, serine, threonine,
tryptophan, tyrosine, and valine. In some embodiments, determining the
identity of a terminal
amino acid type can comprise determining a subset of potential amino acids
that can be present
25 at the terminus of the protein. In some embodiments, this can be
accomplished by determining
that an amino acid is not one or more specific amino acids (and therefore
could be any of the
other amino acids). In some embodiments, this can be accomplished by
determining which of a
specified subset of amino acids (e.g., based on size, charge, hydrophobicity,
post-translational
modification, binding properties) could be at the terminus of the protein
(e.g., using a binding
30 agent that binds to a specified subset of two or more terminal amino
acids).
In some embodiments, assessing the identity of a terminal amino acid type
comprises
determining that an amino acid comprises a post-translational modification.
Non-limiting
examples of post-translational modifications include acetylation, ADP-
ribosylation, caspase
cleavage, citrullination, formylation, N-linked glycosylation, 0-linked
glycosylation,
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
66
hydroxylation, methylation, myristoylation, neddylation, nitration, oxidation,
palmitoylation,
phosphorylation, prenylation, S-nitrosylation, sulfation, sumoylation, and
ubiquitination.
In some embodiments, a protein or protein can be digested into a plurality of
smaller
proteins and sequence information can be obtained from one or more of these
smaller proteins
(e.g., using a method that involves sequentially assessing a terminal amino
acid of a protein and
removing that amino acid to expose the next amino acid at the terminus).
In some embodiments, sequencing of a protein molecule comprises identifying at
least
two (e.g., at least 3, at least 4, at least 5, at least 6, at least 7, at
least 8, at least 9, at least 10, at
least 11, at least 12, at least 13, at least 14, at least 15, at least 16, at
least 17, at least 18, at least
19, at least 20, at least 25, at least 30, at least 35, at least 40, at least
45, at least 50, at least 60, at
least 70, at least 80, at least 90, at least 100, or more) amino acids in the
protein molecule. In
some embodiments, the at least two amino acids are contiguous amino acids. In
some
embodiments, the at least two amino acids are non-contiguous amino acids.
In some embodiments, sequencing of a protein molecule comprises identification
of less
than 100% (e.g., less than 99%, less than 95%, less than 90%, less than 85%,
less than 80%, less
than 75%, less than 70%, less than 65%, less than 60%, less than 55%, less
than 50%, less than
45%, less than 40%, less than 35%, less than 30%, less than 25%, less than
20%, less than 15%,
less than 10%, less than 5%, less than 1% or less) of all amino acids in the
protein molecule. For
example, in some embodiments, sequencing of a protein molecule comprises
identification of
less than 100% of one type of amino acid in the protein molecule (e.g.,
identification of a portion
of all amino acids of one type in the protein molecule). In some embodiments,
sequencing of a
protein molecule comprises identification of less than 100% of each type of
amino acid in the
protein molecule.
In some embodiments, sequencing of a protein molecule comprises identification
of at
least 1, at least 5, at least 10, at least 15, at least 20, at least 25, at
least 30, at least 35, at least 40,
at least 45, at least 50, at least 55, at least 60, at least 65, at least 70,
at least 75, at least 80, at
least 85, at least 90, at least 95, at least 100 or more types of amino acids
in the protein.
A non-limiting example of protein sequencing by iterative terminal amino acid
detection
and cleavage is depicted in FIG. 14A. In some embodiments, protein sequencing
comprises
providing a protein 1000 that is immobilized to a surface 1004 of a solid
support (e.g., attached
to a bottom or sidewall surface of a sample well) through a linkage group
1002. In some
embodiments, linkage group 1002 is formed by a covalent or non-covalent
linkage between a
functionalized terminal end of protein 1000 and a complementary functional
moiety of surface
1004. For example, in some embodiments, linkage group 1002 is formed by a non-
covalent
linkage between a biotin moiety of protein 1000 (e.g., functionalized in
accordance with the
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
67
disclosure) and an avidin protein of surface 1004. In some embodiments,
linkage group 1002
comprises a nucleic acid.
In some embodiments, protein 1000 is immobilized to surface 1004 through a
functionalization moiety at one terminal end such that the other terminal end
is free for detecting
and cleaving of a terminal amino acid in a sequencing reaction. Accordingly,
in some
embodiments, the reagents used in certain protein sequencing reactions
preferentially interact
with terminal amino acids at the non-immobilized (e.g., free) terminus of
protein 1000. In this
way, protein 1000 remains immobilized over repeated cycles of detecting and
cleaving. To this
end, in some embodiments, linker 1002 may be designed according to a desired
set of conditions
used for detecting and cleaving, e.g., to limit detachment of protein 1000
from surface 1004.
Suitable linker compositions and techniques for functionalizing proteins
(e.g., which may be
used for immobilizing a protein to a surface) are described in detail
elsewhere herein.
In some embodiments, as shown in FIG. 14A, protein sequencing can proceed by
(1)
contacting protein 1000 with one or more amino acid recognition molecules that
associate with
one or more types of terminal amino acids. As shown, in some embodiments, a
labeled amino
acid recognition molecule 1006 interacts with protein 1000 by associating with
the terminal
amino acid.
In some embodiments, the method further comprises identifying the amino acid
(terminal
amino acid) of protein 1000 by detecting labeled amino acid recognition
molecule 1006. In some
embodiments, detecting comprises detecting a luminescence from labeled amino
acid recognition
molecule 1006. In some embodiments, the luminescence is uniquely associated
with labeled
amino acid recognition molecule 1006, and the luminescence is thereby
associated with the type
of amino acid to which labeled amino acid recognition molecule 1006
selectively binds. As such,
in some embodiments, the type of amino acid is identified by determining one
or more
luminescence properties of labeled amino acid recognition molecule 1006.
In some embodiments, protein sequencing proceeds by (2) removing the terminal
amino
acid by contacting protein 1000 with an exopeptidase 1008 that binds and
cleaves the terminal
amino acid of protein 1000. Upon removal of the terminal amino acid by
exopeptidase 1008,
protein sequencing proceeds by (3) subjecting protein 1000 (having n-1 amino
acids) to
additional cycles of terminal amino acid recognition and cleavage. In some
embodiments, steps
(1) through (3) occur in the same reaction mixture, e.g., as in a dynamic
peptide sequencing
reaction. In some embodiments, steps (1) through (3) may be carried out using
other methods
known in the art, such as peptide sequencing by Edman degradation.
Edman degradation involves repeated cycles of modifying and cleaving the
terminal
amino acid of a protein, wherein each successively cleaved amino acid is
identified to determine
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
68
an amino acid sequence of the protein. Referring to FIG. 14A, peptide
sequencing by
conventional Edman degradation can be carried out by (1) contacting protein
1000 with one or
more amino acid recognition molecules that selectively bind one or more types
of terminal amino
acids. In some embodiments, step (1) further comprises removing any of the one
or more labeled
.. amino acid recognition molecules that do not selectively bind protein 1000.
In some
embodiments, step (2) comprises modifying the terminal amino acid (e.g., the
free terminal
amino acid) of protein 1000 by contacting the terminal amino acid with an
isothiocyanate (e.g.,
PITC) to form an isothiocyanate-modified terminal amino acid. In some
embodiments, an
isothiocyanate-modified terminal amino acid is more susceptible to removal by
a cleaving
.. reagent (e.g., a chemical or enzymatic cleaving reagent) than an unmodified
terminal amino acid.
In some embodiments, Edman degradation proceeds by (2) removing the terminal
amino
acid by contacting protein 1000 with an exopeptidase 1008 that specifically
binds and cleaves the
isothiocyanate-modified terminal amino acid. In some embodiments, exopeptidase
1008
comprises a modified cysteine protease. In some embodiments, exopeptidase 1008
comprises a
.. modified cysteine protease, such as a cysteine protease from Trypanosoma
cruzi (see, e.g.,
Borgo, et al. (2015) Protein Science 24:571-579). In yet other embodiments,
step (2) comprises
removing the terminal amino acid by subjecting protein 1000 to chemical (e.g.,
acidic, basic)
conditions sufficient to cleave the isothiocyanate-modified terminal amino
acid. In some
embodiments, Edman degradation proceeds by (3) washing protein 1000 following
terminal
.. amino acid cleavage. In some embodiments, washing comprises removing
exopeptidase 1008. In
some embodiments, washing comprises restoring protein 1000 to neutral pH
conditions (e.g.,
following chemical cleavage by acidic or basic conditions). In some
embodiments, sequencing
by Edman degradation comprises repeating steps (1) through (3) for a plurality
of cycles.
In some embodiments, peptide sequencing can be carried out in a dynamic
peptide
sequencing reaction. In some embodiments, referring again to FIG. 10A, the
reagents required to
perform step (1) and step (2) are combined within a single reaction mixture.
For example, in
some embodiments, steps (1) and (2) can occur without exchanging one reaction
mixture for
another and without a washing step as in conventional Edman degradation. Thus,
in this
embodiments, a single reaction mixture comprises labeled amino acid
recognition molecule 1006
.. and exopeptidase 1008. In some embodiments, exopeptidase 1008 is present in
the mixture at a
concentration that is less than that of labeled amino acid recognition
molecule 1006. In some
embodiments, exopeptidase 1008 binds protein 1000 with a binding affinity that
is less than that
of labeled amino acid recognition molecule 1006.
In some embodiments, dynamic protein sequencing is carried out in real-time by
evaluating
binding interactions of terminal amino acids with labeled amino acid
recognition molecules and
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
69
a cleaving reagent (e.g., an exopeptidase). FIG. 14B shows an example of a
method of
sequencing in which discrete binding events give rise to signal pulses of a
signal output. The
inset panel (left) of FIG. 14B illustrates a general scheme of real-time
sequencing by this
approach. As shown, a labeled amino acid recognition molecule associates with
(e.g., binds to)
.. and dissociates from a terminal amino acid (shown here as phenylalanine),
which gives rise to a
series of pulses in signal output which may be used to identify the terminal
amino acid. In some
embodiments, the series of pulses provide a pulsing pattern (e.g., a
characteristic pattern) which
may be diagnostic of the identity of the corresponding terminal amino acid.
As further shown in the inset panel (left) of FIG. 14B, in some embodiments, a
sequencing reaction mixture further comprises an exopeptidase. In some
embodiments, the
exopeptidase is present in the mixture at a concentration that is less than
that of the labeled
amino acid recognition molecule. In some embodiments, the exopeptidase
displays broad
specificity such that it cleaves most or all types of terminal amino acids.
Accordingly, a dynamic
sequencing approach can involve monitoring recognition molecule binding at a
terminus of a
protein over the course of a degradation reaction catalyzed by exopeptidase
cleavage activity.
FIG. 14B further shows the progress of signal output intensity over time
(right panels). In
some embodiments, terminal amino acid cleavage by exopeptidase(s) occurs with
lower
frequency than the binding pulses of a labeled amino acid recognition
molecule. In this way,
amino acids of a protein may be counted and/or identified in a real-time
sequencing process. In
some embodiments, one type of amino acid recognition molecule can associate
with more than
one type of amino acid, where different characteristic patterns correspond to
the association of
one type of labeled amino acid recognition molecule with different types of
terminal amino
acids. For example, in some embodiments, different characteristic patterns (as
illustrated by each
of phenylalanine (F, Phe), tryptophan (W, Trp), and tyrosine (Y, Tyr))
correspond to the
association of one type of labeled amino acid recognition molecule (e.g., ClpS
protein) with
different types of terminal amino acids over the course of degradation. In
some embodiments, a
plurality of labeled amino acid recognition molecules may be used, each
capable of associating
with different subsets of amino acids.
In some embodiments, dynamic peptide sequencing is performed by observing
different
association events, e.g., association events between an amino acid recognition
molecule and an
amino acid at a terminal end of a peptide, wherein each association event
produces a change in
magnitude of a signal, e.g., a luminescence signal, that persists for a
duration of time. In some
embodiments, observing different association events, e.g., association events
between an amino
acid recognition molecule and an amino acid at a terminal end of a peptide,
can be performed
.. during a peptide degradation process. In some embodiments, a transition
from one characteristic
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
signal pattern to another is indicative of amino acid cleavage (e.g., amino
acid cleavage resulting
from peptide degradation). In some embodiments, amino acid cleavage refers to
the removal of
at least one amino acid from a terminus of a protein (e.g., the removal of at
least one terminal
amino acid from the protein). In some embodiments, amino acid cleavage is
determined by
5 inference based on a time duration between characteristic signal
patterns. In some embodiments,
amino acid cleavage is determined by detecting a change in signal produced by
association of a
labeled cleaving reagent with an amino acid at the terminus of the protein. As
amino acids are
sequentially cleaved from the terminus of the protein during degradation, a
series of changes in
magnitude, or a series of signal pulses, is detected.
10 In some embodiments, signal pulse information may be used to identify an
amino acid
based on a characteristic pattern in a series of signal pulses. In some
embodiments, a
characteristic pattern comprises a plurality of signal pulses, each signal
pulse comprising a pulse
duration. In some embodiments, the plurality of signal pulses may be
characterized by a
summary statistic (e.g., mean, median, time decay constant) of the
distribution of pulse durations
15 in a characteristic pattern. In some embodiments, the mean pulse
duration of a characteristic
pattern is between about 1 millisecond and about 10 seconds (e.g., between
about 1 ms and about
1 s, between about 1 ms and about 100 ms, between about 1 ms and about 10 ms,
between about
10 ms and about 10 s, between about 100 ms and about 10 s, between about 1 s
and about 10 s,
between about 10 ms and about 100 ms, or between about 100 ms and about 500
ms). In some
20 embodiments, different characteristic patterns corresponding to
different types of amino acids in
a single protein may be distinguished from one another based on a
statistically significant
difference in the summary statistic. For example, in some embodiments, one
characteristic
pattern may be distinguishable from another characteristic pattern based on a
difference in mean
pulse duration of at least 10 milliseconds (e.g., between about 10 ms and
about 10 s, between
25 about 10 ms and about 1 s, between about 10 ms and about 100 ms, between
about 100 ms and
about 10 s, between about 1 s and about 10 s, or between about 100 ms and
about 1 s). It should
be appreciated that, in some embodiments, smaller differences in mean pulse
duration between
different characteristic patterns may require a greater number of pulse
durations within each
characteristic pattern to distinguish one from another with statistical
confidence.
Sequencing Device or Module
Sequencing of nucleic acids or proteins in accordance with the instant
disclosure, in some
aspects, may be performed using a system that permits single molecule
analysis. The system
may include a sequencing device or module and an instrument configured to
interface with the
sequencing device or module. The sequencing device or module may include an
array of pixels,
CA 03177368 2022-09-27
WO 2021/216763 PCT/US2021/028471
71
where individual pixels include a sample well and at least one photodetector.
The sample wells
of the sequencing device or module may be formed on or through a surface of
the sequencing
device or module and be configured to receive a sample placed on the surface
of the sequencing
device or module. In some embodiments, the sample wells are a component of a
cartridge (e.g.,
a disposable or single-use cartridge) that can be inserted into the device.
Collectively, the
sample wells may be considered as an array of sample wells. The plurality of
sample wells may
have a suitable size and shape such that at least a portion of the sample
wells receive a single
target molecule or sample comprising a plurality of molecules (e.g., a target
nucleic acid or a
target protein). In some embodiments, the number of molecules within a sample
well may be
distributed among the sample wells of the sequencing device or module such
that some sample
wells contain one molecule (e.g., a target nucleic acid or a target protein)
while others contain
zero, two, or a plurality of molecules.
In some embodiments, a sequencing device or module is positioned to receive a
target
molecule or sample comprising a plurality of molecules (e.g., a target nucleic
acid or a target
protein) from a sample preparation device or module. In some embodiments, a
sequencing
device or module is connected directly (e.g., physically attached to) or
indirectly to a sample
preparation device or module.
Excitation light is provided to the sequencing device or module from one or
more light
sources external to the sequencing device or module. Optical components of the
sequencing
device or module may receive the excitation light from the light source and
direct the light
towards the array of sample wells of the sequencing device or module and
illuminate an
illumination region within the sample well. In some embodiments, a sample well
may have a
configuration that allows for the target molecule or sample comprising a
plurality of molecules to
be retained in proximity to a surface of the sample well, which may ease
delivery of excitation
light to the sample well and detection of emission light from the target
molecule or sample
comprising a plurality of molecules. A target molecule or sample comprising a
plurality of
molecules positioned within the illumination region may emit emission light in
response to being
illuminated by the excitation light. For example, a nucleic acid or protein
(or pluralities thereof)
may be labeled with a fluorescent marker, which emits light in response to
achieving an excited
state through the illumination of excitation light. Emission light emitted by
a target molecule or
sample comprising a plurality of molecules may then be detected by one or more
photodetectors
within a pixel corresponding to the sample well with the target molecule or
sample comprising a
plurality of molecules being analyzed. When performed across the array of
sample wells, which
may range in number between approximately 10,000 pixels to 1,000,000 pixels
according to
some embodiments, multiple sample wells can be analyzed in parallel.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
72
The sequencing device or module may include an optical system for receiving
excitation
light and directing the excitation light among the sample well array. The
optical system may
include one or more grating couplers configured to couple excitation light to
the sequencing
device or module and direct the excitation light to other optical components.
The optical system
may include optical components that direct the excitation light from a grating
coupler towards
the sample well array. Such optical components may include optical splitters,
optical combiners,
and waveguides. In some embodiments, one or more optical splitters may couple
excitation light
from a grating coupler and deliver excitation light to at least one of the
waveguides. According
to some embodiments, the optical splitter may have a configuration that allows
for delivery of
excitation light to be substantially uniform across all the waveguides such
that each of the
waveguides receives a substantially similar amount of excitation light. Such
embodiments may
improve performance of the sequencing device or module by improving the
uniformity of
excitation light received by sample wells of the sequencing device or module.
Examples of
suitable components, e.g., for coupling excitation light to a sample well
and/or directing
emission light to a photodetector, to include in a sequencing device or module
are described in
U.S. Pat. Application No. 14/821,688, filed August 7, 2015, titled "INTEGRATED
DEVICE
FOR PROBING, DETECTING AND ANALYZING MOLECULES," and U.S. Pat. Application
No. 14/543,865, filed November 17, 2014, titled "INTEGRATED DEVICE WITH
EXTERNAL
LIGHT SOURCE FOR PROBING, DETECTING, AND ANALYZING MOLECULES," both of
.. which are incorporated herein by reference in their entirety. Examples of
suitable grating
couplers and waveguides that may be implemented in the sequencing device or
module are
described in U.S. Pat. Application No. 15/844,403, filed December 15, 2017,
titled "OPTICAL
COUPLER AND WAVEGUIDE SYSTEM," which is incorporated herein by reference in
its
entirety.
Additional photonic structures may be positioned between the sample wells and
the
photodetectors and configured to reduce or prevent excitation light from
reaching the
photodetectors, which may otherwise contribute to signal noise in detecting
emission light. In
some embodiments, metal layers which may act as a circuitry for the sequencing
device or
module, may also act as a spatial filter. Examples of suitable photonic
structures may include
spectral filters, a polarization filters, and spatial filters and are
described in U.S. Pat. Application
No. 16/042,968, filed July 23, 2018, titled "OPTICAL REJECTION PHOTONIC
STRUCTURES," which is incorporated herein by reference in its entirety.
Components located off of the sequencing device or module may be used to
position and
align an excitation source to the sequencing device or module. Such components
may include
optical components including lenses, mirrors, prisms, windows, apertures,
attenuators, and/or
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
73
optical fibers. Additional mechanical components may be included in the
instrument to allow for
control of one or more alignment components. Such mechanical components may
include
actuators, stepper motors, and/or knobs. Examples of suitable excitation
sources and alignment
mechanisms are described in U.S. Pat. Application No. 15/161,088, filed May
20, 2016, titled
"PULSED LASER AND SYSTEM," which is incorporated herein by reference in its
entirety.
Another example of a beam-steering module is described in U.S. Pat.
Application No.
15/842,720, filed December, 14, 2017, titled "COMPACT BEAM SHAPING AND
STEERING
ASSEMBLY," which is incorporated herein by reference in its entirety.
Additional examples of
suitable excitation sources are described in U.S. Pat. Application No.
14/821,688, filed August 7,
2015, titled "INTEGRATED DEVICE FOR PROBING, DETECTING AND ANALYZING
MOLECULES," which is incorporated herein by reference in its entirety.
The photodetector(s) positioned with individual pixels of the sequencing
device or
module may be configured and positioned to detect emission light from the
pixel's
corresponding sample well. Examples of suitable photodetectors are described
in U.S. Pat.
Application No. 14/821,656, filed August 7, 2015, titled "INTEGRATED DEVICE
FOR
TEMPORAL BINNING OF RECEIVED PHOTONS," which is incorporated herein by
reference in its entirety. In some embodiments, a sample well and its
respective photodetector(s)
may be aligned along a common axis. In this manner, the photodetector(s) may
overlap with the
sample well within the pixel.
Characteristics of the detected emission light may provide an indication for
identifying
the marker associated with the emission light. Such characteristics may
include any suitable type
of characteristic, including an arrival time of photons detected by a
photodetector, an amount of
photons accumulated over time by a photodetector, and/or a distribution of
photons across two or
more photodetectors. In some embodiments, a photodetector may have a
configuration that
allows for the detection of one or more timing characteristics associated with
a sample's
emission light (e.g., luminescence lifetime). The photodetector may detect a
distribution of
photon arrival times after a pulse of excitation light propagates through the
sequencing device or
module, and the distribution of arrival times may provide an indication of a
timing characteristic
of the sample's emission light (e.g., a proxy for luminescence lifetime). In
some embodiments,
.. the one or more photodetectors provide an indication of the probability of
emission light emitted
by the marker (e.g., luminescence intensity). In some embodiments, a plurality
of photodetectors
may be sized and arranged to capture a spatial distribution of the emission
light. Output signals
from the one or more photodetectors may then be used to distinguish a marker
from among a
plurality of markers, where the plurality of markers may be used to identify a
sample within the
sample. In some embodiments, a sample may be excited by multiple excitation
energies, and
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
74
emission light and/or timing characteristics of the emission light emitted by
the sample in
response to the multiple excitation energies may distinguish a marker from a
plurality of
markers.
In operation, parallel analyses of samples within the sample wells are carried
out by
exciting some or all of the samples within the wells using excitation light
and detecting signals
from sample emission with the photodetectors. Emission light from a sample may
be detected by
a corresponding photodetector and converted to at least one electrical signal.
The electrical
signals may be transmitted along conducting lines in the circuitry of the
sequencing device or
module, which may be connected to an instrument interfaced with the sequencing
device or
module. The electrical signals may be subsequently processed and/or analyzed.
Processing
and/or analyzing of electrical signals may occur on a suitable computing
device either located on
or off the instrument.
The instrument may include a user interface for controlling operation of the
instrument
and/or the sequencing device or module. The user interface may be configured
to allow a user to
input information into the instrument, such as commands and/or settings used
to control the
functioning of the instrument. In some embodiments, the user interface may
include buttons,
switches, dials, and/or a microphone for voice commands. The user interface
may allow a user
to receive feedback on the performance of the instrument and/or sequencing
device or module,
such as proper alignment and/or information obtained by readout signals from
the photodetectors
on the sequencing device or module. In some embodiments, the user interface
may provide
feedback using a speaker to provide audible feedback. In some embodiments, the
user interface
may include indicator lights and/or a display screen for providing visual
feedback to a user.
In some embodiments, the instrument or device described herein may include a
computer
interface configured to connect with a computing device. The computer
interface may be a USB
interface, a FireWire interface, or any other suitable computer interface. A
computing device
may be any general purpose computer, such as a laptop or desktop computer. In
some
embodiments, a computing device may be a server (e.g., cloud-based server)
accessible over a
wireless network via a suitable computer interface. The computer interface may
facilitate
communication of information between the instrument and the computing device.
Input
information for controlling and/or configuring the instrument may be provided
to the computing
device and transmitted to the instrument via the computer interface. Output
information
generated by the instrument may be received by the computing device via the
computer
interface. Output information may include feedback about performance of the
instrument,
performance of the sequencing device or module, and/or data generated from the
readout signals
.. of the photodetector.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
In some embodiments, the instrument may include a processing device configured
to
analyze data received from one or more photodetectors of the sequencing device
or module
and/or transmit control signals to the excitation source(s). In some
embodiments, the processing
device may comprise a general purpose processor, and/or a specially-adapted
processor (e.g., a
5 central processing unit (CPU) such as one or more microprocessor or
microcontroller cores, a
field-programmable gate array (FPGA), an application-specific integrated
circuit (ASIC), a
custom integrated circuit, a digital signal processor (DSP), or a combination
thereof). In some
embodiments, the processing of data from one or more photodetectors may be
performed by both
a processing device of the instrument and an external computing device. In
other embodiments,
10 an external computing device may be omitted and processing of data from
one or more
photodetectors may be performed solely by a processing device of the
sequencing device or
module.
According to some embodiments, the instrument that is configured to analyze
target
molecules or samples comprising a plurality of molecules based on luminescence
emission
15 characteristics may detect differences in luminescence lifetimes and/or
intensities between
different luminescent molecules, and/or differences between lifetimes and/or
intensities of the
same luminescent molecules in different environments. The inventors have
recognized and
appreciated that differences in luminescence emission lifetimes can be used to
discern between
the presence or absence of different luminescent molecules and/or to discern
between different
20 environments or conditions to which a luminescent molecule is subjected.
In some cases,
discerning luminescent molecules based on lifetime (rather than emission
wavelength, for
example) can simplify aspects of the system. As an example, wavelength-
discriminating optics
(such as wavelength filters, dedicated detectors for each wavelength,
dedicated pulsed optical
sources at different wavelengths, and/or diffractive optics) may be reduced in
number or
25 eliminated when discerning luminescent molecules based on lifetime. In
some cases, a single
pulsed optical source operating at a single characteristic wavelength may be
used to excite
different luminescent molecules that emit within a same wavelength region of
the optical
spectrum but have measurably different lifetimes. An analytic system that uses
a single pulsed
optical source, rather than multiple sources operating at different
wavelengths, to excite and
30 discern different luminescent molecules emitting in a same wavelength
region may be less
complex to operate and maintain, may be more compact, and may be manufactured
at lower cost.
Although analytic systems based on luminescence lifetime analysis may have
certain
benefits, the amount of information obtained by an analytic system and/or
detection accuracy
may be increased by allowing for additional detection techniques. For example,
some
35 embodiments of the systems may additionally be configured to discern one
or more properties of
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
76
a sample based on luminescence wavelength and/or luminescence intensity. In
some
implementations, luminescence intensity may be used additionally or
alternatively to distinguish
between different luminescent labels. For example, some luminescent labels may
emit at
significantly different intensities or have a significant difference in their
probabilities of
excitation (e.g., at least a difference of about 35%) even though their decay
rates may be similar.
By referencing binned signals to measured excitation light, it may be possible
to distinguish
different luminescent labels based on intensity levels.
According to some embodiments, different luminescence lifetimes may be
distinguished
with a photodetector that is configured to time-bin luminescence emission
events following
excitation of a luminescent label. The time binning may occur during a single
charge-
accumulation cycle for the photodetector. A charge-accumulation cycle is an
interval between
read-out events during which photo-generated carriers are accumulated in bins
of the time-
binning photodetector. Examples of a time-binning photodetector are described
in U.S. Pat.
Application No. 14/821,656, filed August 7, 2015, titled "INTEGRATED DEVICE
FOR
TEMPORAL BINNING OF RECEIVED PHOTONS," which is incorporated herein by
reference in its entirety. In some embodiments, a time-binning photodetector
may generate
charge carriers in a photon absorption/carrier generation region and directly
transfer charge
carriers to a charge carrier storage bin in a charge carrier storage region.
In such embodiments,
the time-binning photodetector may not include a carrier travel/capture
region. Such a time-
binning photodetector may be referred to as a "direct binning pixel." Examples
of time-binning
photodetectors, including direct binning pixels, are described in U.S. Pat.
Application No.
15/852,571, filed December, 22, 2017, titled "INTEGRATED PHOTODETECTOR WITH
DIRECT BINNING PIXEL," which is incorporated herein by reference in its
entirety.
In some embodiments, different numbers of fluorophores of the same type may be
linked
to different components of a target molecule (e.g., a target nucleic acid or a
target protein) or a
plurality of molecules present in a sample (e.g., a plurality of nucleic acids
or a plurality of
proteins), so that each individual molecule may be identified based on
luminescence intensity.
For example, two fluorophores may be linked to a first labeled molecule and
four or more
fluorophores may be linked to a second labeled molecule. Because of the
different numbers of
fluorophores, there may be different excitation and fluorophore emission
probabilities associated
with the different molecule. For example, there may be more emission events
for the second
labeled molecule during a signal accumulation interval, so that the apparent
intensity of the bins
is significantly higher than for the first labeled molecule.
The inventors have recognized and appreciated that distinguishing nucleic
acids or
proteins based on fluorophore decay rates and/or fluorophore intensities may
enable a
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
77
simplification of the optical excitation and detection systems. For example,
optical excitation
may be performed with a single-wavelength source (e.g., a source producing one
characteristic
wavelength rather than multiple sources or a source operating at multiple
different characteristic
wavelengths). Additionally, wavelength discriminating optics and filters may
not be needed in
the detection system. Also, a single photodetector may be used for each sample
well to detect
emission from different fluorophores. The phrase "characteristic wavelength"
or "wavelength"
is used to refer to a central or predominant wavelength within a limited
bandwidth of radiation.
For example, a limited bandwidth of radiation may include a central or peak
wavelength within a
20 nm bandwidth output by a pulsed optical source. In some cases,
"characteristic wavelength"
or "wavelength" may be used to refer to a peak wavelength within a total
bandwidth of radiation
output by a source.
Combined Sample Preparation and Sequencing Device
In some embodiments, a device herein comprising a sample preparation module
further
comprises a sequencing module. In some embodiments, a device that comprises a
sample
preparation module and a sequencing module involves a sequencing chip or
cartridge that is
embedded into a sample preparation cartridge, such that the two cartridges
comprise a single,
inseparable consumable. In some embodiments, the sequencing chip or cartridge
requires
consumable support electronics (e.g., a PCB substrate with wirebonds,
electrical contacts). The
consumable support electronics may be in direct physical contact with the
sequencing chip or
cartridge. In some embodiments, the sequencing chip or cartridge requires an
interface for a
peristaltic pump, temperature control and/or electropheresis contacts. These
interfaces may
allow for precise geometric registration for the many electrical contacts and
laser alignment. In
some embodiments, different sections of a chip or cartridge may comprise
different
temperatures, physical forces, electrical interfaces of varying voltage and
current, vibration,
and/or competing alignment requirements. In some embodiments, disparate
instrument sub-
systems associated with either the sample preparation or sequencing module
must be in close
proximity in order to share resources. In some embodiments, a device that
comprises a sample
preparation module and a sequencing module is hands-free (i.e., can be used
without the use of
hands).
In some embodiments, a device that comprises a sample preparation module and a
sequencing module produces (e.g., enriches or purifies) target nucleic acids
with an average
read-length for downstream sequencing applications that is longer than an
average read-length
produced using control methods (e.g., Sage BluePippin methods, manual methods
(e.g., manual
bead-based size selection methods)). In some embodiments, a sample preparation
device
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
78
produces target nucleic acids with an average read-length for sequencing that
comprises at least
700, 800, 900, 1000, 1100, 1200, 1300, 1400, 1500, 1600, 1700, 1800, 1900,
2000, 2100, 2200,
2300, 2400, 2500, 2600, 2700, 2800, 2900, or 3000 nucleotides in length. In
some embodiments,
a sample preparation device produces target nucleic acids with an average read-
length for
sequencing that comprises 700-3000, 1000-3000, 1000-2500, 1000-2400, 1000-
2300, 1000-
2200, 1000-2100, 1000-2000, 1000-1900, 1000-1800, 1000-1700, 1000-1600, 1000-
1500, 1000-
1400, 1000-1300, 1000-1200, 1500-3000, 1500-2500, 1500-2000, or 2000-3000
nucleotides in
length.
In some embodiments, a device that comprises a sample preparation module and a
sequencing module allows for shortened times between initiation of sample
preparation and
detection of a target molecule contained within the sample than control or
traditional methods
(e.g., Sage BluePippin methods followed by sequencing). In some embodiments, a
device that
comprises a sample preparation module and a sequencing module is capable of
detecting a target
molecule using sequencing in less time (e.g., 2-fold, 3-fold, 4-fold, 5-fold,
or 10-fold less time)
than control or traditional methods (e.g., Sage BluePippin methods followed by
sequencing).
In some embodiments, a device that comprises a sample preparation module and a
sequencing module is capable of detecting a target molecule with lower inputs
of sample than
control or traditional methods (e.g., Sage BluePippin methods followed by
sequencing). In some
embodiments, a device of the disclosure requires as little as 0.1 i.tg, 0.2
i.tg, 0.3 i.tg, 0.4 i.tg, 0.5
j..tg, 0.61.1g, 0.7 j..tg, 0.8 j..tg, 0.9 j..tg, or li.tg of sample (e.g.,
biological sample). In some
embodiments, a device of the disclosure requires as little as 10 i.t.L, 20
i.t.L, 30 i.t.L, 40 i.t.L, 50 i.t.L,
60 i.t.L, 70 i.t.L, 80 i.t.L, 90 i.t.L, 100 i.t.L, 110 i.t.L, 130 i.t.L, 150
i.t.L, 175 i.t.L, 200 i.t.L, 225 i.t.L, or 250
0_, of sample (e.g., biological sample such as blood).
Devices or Modules
In some embodiments, devices or modules (e.g., sample preparation devices;
sequencing
devices; combined sample preparation and sequencing devices) are configured to
transport small
volume(s) of fluid precisely with a well-defined fluid flow resolution, and
with a well-defined
flow rate in some cases. In some embodiments, devices or modules are
configured to transport
fluid at a flow rate of greater than or equal to 0.1 L/s, greater than or
equal to 0.5 i.tIls, greater
than or equal to 1 i.tIls, greater than or equal to 2 L/s, greater than or
equal to 5 i.tIls, or higher.
In some embodiments, devices or modules herein are configured to transport
fluid at a flow rate
of less than or equal to 100 i.tIls, less than or equal to 75 i.tIls, less
than or equal to 50 L/s, less
than or equal to 30 i.tIls, less than or equal to 20 i.tIls, less than or
equal to 15 L/s, or less.
Combinations of these ranges are possible. For example, in some embodiments,
devices or
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
79
modules herein are configured to transport fluid at a flow rate of greater
than or equal to 0.1 Lis
and less than or equal to 100 tt/s, or greater than or equal to 5 i.tt/s and
less than or equal to 15
L/s. For example, in certain embodiments, systems, devices, and modules herein
have a fluid
flow resolution on the order of tens of microliters or hundreds of
microliters. Further description
of fluid flow resolution is described elsewhere herein. In certain
embodiments, systems, devices,
and modules are configured to transport small volumes of fluid through at
least a portion of a
cartridge.
Some aspects relate to configurations of pumps and apparatuses that include a
roller (e.g.,
in combination with a crank-and-rocker mechanism). Other aspects relate to
cartridges
comprising channels (e.g., microchannels) having cross-sectional shapes (e.g.,
substantially
triangular shapes), valving, deep sections, and/or surface layers (e.g., flat
elastomer membranes).
Certain aspects relate to a decoupling of certain components of the
peristaltic pump (e.g., the
roller) from other components of the pump (e.g., pumping lanes). In some
cases, certain
elements of apparatuses (e.g., edges of the roller) are configured to interact
with elements of the
cartridge (e.g., surface layers and certain shapes of the channels) in such a
way (e.g., via
engagement and disengagement) that any of a variety of advantages are
achieved. In some non-
limiting embodiments, certain inventive features and configurations of the
apparatuses,
cartridges, and pumps described herein contribute to improved automation of
the fluid pumping
process (e.g., due to the use of a translatable roller and a separate
cartridge containing multiple
different fluidic channels that can be indexed by the roller). In some cases,
features described
herein contribute to an ability to handle a relatively high number of
different fluids (e.g., for
multiplexing with multiple samples) with a relatively high number of
configurations using a
relatively small number of hardware components (e.g., due to the use of
separate cartridges with
multiple different channels, each of which may be accessible to the roller).
As one example, in
some cases, the features described herein allow for more than one apparatus to
be paired with a
cartridge to pump more than one lane simultaneously or use two pumps in one
lane for other
functionality. In some cases, the features contribute to a reduction in
required fluid volume
and/or less stringent tolerances in roller/channel interactions (e.g., due to
inventive cross-
sectional shapes of the channels and/or the edge of the roller, and/or due to
the use of inventive
valving and/or deep sections of channels). In some cases, features described
herein result in a
reduction in required washing of hardware components (e.g., due to a
decoupling of an apparatus
and a cartridge of the peristaltic pump). In some embodiments, aspects of the
apparatuses,
cartridges, and pumps described herein are useful for preparing samples. For
example, some
such aspects may be incorporated into a sample preparation module upstream of
a detection
module (e.g., for analysis/sequencing/identification of biologically-derived
samples).
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
In another aspect, peristaltic pumps are provided. In some embodiments, a
peristaltic
pump comprises a roller and a cartridge, wherein the cartridge comprises a
base layer having a
surface comprising channels, wherein at least a portion of at least some of
the channels (1) have
a substantially triangularly-shaped cross-section having a single vertex at a
base of the channel
5 and having two other vertices at the surface of the base layer, and (2)
have a surface layer,
comprising an elastomer, configured to substantially seal off a surface
opening of the channel.
Embodiments of peristaltic pumps are further described elsewhere herein.
In some embodiments, a system (e.g., pump, device) described herein undergoes
a pump
cycle. In some embodiments, a pump cycle corresponds to one rotation of a
crank of the system.
10 In some embodiments, each pump cycle may transport greater than or equal
to 1 tL, greater than
or equal to 2 tL, greater than or equal to 4 tL, less than or equal to 10 tL,
less than or equal to 8
and/or less than or equal to 6 itt of fluid. Combinations of the above-
referenced ranges are
also possible (e.g., between or equal to 1 itt and 10 lL). Other ranges of
volumes of fluid are
also possible.
15 In some embodiments, a system described herein has a particular stroke
length. In certain
embodiments, given that each pump cycle may transport on the order of between
or equal to 1
itt and 10 itt of fluid, and/or given that channel dimensions may preferably
be on the order of 1
mm wide and on the order of 1 mm deep (e.g., depending on what can be machined
or molded to
decrease channel volume and maintain reasonable tolerances), a stroke length
may be greater
20 than or equal to 10 mm, greater than or equal to 12 mm, greater than or
equal to 14 mm, less than
or equal to 20 mm, less than or equal to 18 mm, and/or less than or equal to
16 mm.
Combinations of the above-referenced ranges are also possible (e.g., between
or equal to 10 mm
and 20 mm). Other ranges are also possible. As used herein, "stroke length"
refers to a distance
a roller travels while engaged with a substrate. In certain embodiments, the
substrate comprises
25 a cartridge.
In another aspect, cartridges are provided. In some embodiments, a cartridge
comprises a
base layer having a surface comprising channels, and at least a portion of at
least some of the
channels (1) have a substantially triangularly-shaped cross-section having a
single vertex at a
base of the channel and having two other vertices at the surface of the base
layer, and (2) have a
30 surface layer, comprising an elastomer, configured to substantially seal
off a surface opening of
the channel. Embodiments of cartridges are further described elsewhere herein.
In some embodiments, a cartridge comprises a base layer. In some embodiments,
a base layer
has a surface comprising one or more channels. For example, Figure 8 is a
schematic diagram of
a cross-section view of a cartridge 100 along the width of channels 102, in
accordance with some
35 embodiments. The depicted cartridge 100 includes a base layer 104 having
a surface 111
CA 03177368 2022-09-27
WO 2021/216763 PCT/US2021/028471
81
comprising channels 102. In certain embodiments, at least some of the channels
are
microchannels. For example, in some embodiments, at least some of channels 102
are
microchannels. In certain embodiments, all of the channels microchannels. For
example,
referring again to Figure 8, in certain embodiments, all of channels 102 are
microchannels.
As used herein, the term "channel" will be known to those of ordinary skill in
the art and may
refer to a structure configured to contain and/or transport a fluid. A channel
generally comprises:
walls; a base (e.g., a base connected to the walls and/or formed from the
walls); and a surface
opening that may be open, covered, and/or sealed off at one or more portions
of the channel.
As used herein, the term "microchannel" refers to a channel that comprises at
least one
dimension less than or equal to 1000 microns in size. For example, a
microchannel may
comprise at least one dimension (e.g., a width, a height) less than or equal
to 1000 microns (e.g.,
less than or equal to 100 microns, less than or equal to 10 microns, less than
or equal to 5
microns) in size. In some embodiments, a microchannel comprises at least one
dimension
greater than or equal to 1 micron (e.g., greater than or equal to 2 microns,
greater than or equal to
10 microns). Combinations of the above-referenced ranges are also possible
(e.g., greater than or
equal to 1 micron and less than or equal to 1000 microns, greater than or
equal to 10 micron and
less than or equal to 100 microns). Other ranges are also possible. In some
embodiments, a
microchannel has a hydraulic diameter of less than or equal to 1000 microns.
As used herein, the
term "hydraulic diameter" (DH) will be known to those of ordinary skill in the
art and may be
determined as: DH = 4A/P, wherein A is a cross-sectional area of the flow of
fluid through the
channel and P is a wetted perimeter of the cross-section (a perimeter of the
cross-section of the
channel contacted by the fluid).
In some embodiments, at least a portion of at least some channel(s) have a
substantially
triangularly-shaped cross-section. In some embodiments, at least a portion of
at least some
channel(s) have a substantially triangularly-shaped cross-section having a
single vertex at a base
of the channel and having two other vertices at the surface of the base layer.
Referring again to
Figure 24, in some embodiments, at least a portion of at least some of
channels 102 have a
substantially triangularly-shaped cross-section having a single vertex at a
base of the channel and
having two other vertices at the surface of the base layer.
As used herein, the term "triangular" is used to refer to a shape in which a
triangle can be
inscribed or circumscribed to approximate or equal the actual shape, and is
not constrained
purely to a triangle. For example, a triangular cross-section may comprise a
non-zero curvature
at one or more portions.
A triangular cross-section may comprise a wedge shape. As used herein, the
term
"wedge shape" will be known by those of ordinary skill in the art and refers
to a shape having a
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
82
thick end and tapering to a thin end. In some embodiments, a wedge shape has
an axis of
symmetry from the thick end to the thin end. For example, a wedge shape may
have a thick end
(e.g., surface opening of a channel) and taper to a thin end (e.g., base of a
channel), and may
have an axis of symmetry from the thick end to the thin end.
Additionally, in certain embodiments, substantially triangular cross-sections
(i.e., "v-
groove(s)") may have a variety of aspect ratios. As used herein, the term
"aspect ratio" for a v-
groove refers to a height-to-width ratio. For example, in some embodiments, v-
groove(s) may
have an aspect ratio of less than or equal to 2, less than or equal to 1, or
less than or equal to 0.5,
and/or greater than or equal to 0.1, greater than or equal to 0.2, or greater
than or equal to 0.3.
Combinations of the above-referenced ranges are also possible (e.g., between
or equal to 0.1 and
2, between or equal to 0.2 and 1). Other ranges are also possible.
In some embodiments, at least a portion of at least some channel(s) have a
cross-section
comprising a substantially triangular portion and a second portion opening
into the substantially
triangular portion and extending below the substantially triangular portion
relative to the surface
of the channel. In some embodiments, the second portion has a diameter (e.g.,
an average
diameter) significantly smaller than an average diameter of the substantially
triangular portion.
Referring again to Figure 24, in some embodiments, at least a portion of at
least some of
channels 102 have a cross-section comprising a substantially triangular
portion 101 and a second
portion 103 opening into substantially triangular portion 101 and extending
below substantially
triangular portion 101 relative to surface 105 of the channel, wherein second
portion 103 has a
diameter 107 significantly smaller than an average diameter 109 of
substantially triangular
portion 101. In some such cases, the second portion of a channel having a
significantly smaller
diameter than that of the average diameter of the substantially triangular
portion of the channel
can result in the substantially triangular portion being accessible to the
roller of the apparatus and
deformed portions of the surface layer, but the second portion being
inaccessible to the roller and
deformed portions of the surface layer. For example, referring again to Figure
24, substantially
triangular portion 101 of channel 102 is accessible to a roller (not pictured)
and deformed
portions of surface layer 106, while second portion 103 is inaccessible to the
roller and deformed
portions of surface layer 106, in accordance with certain embodiments. In some
such cases, a
seal with the surface layer 106 cannot be achieved in portions of the channel
102 having a
second portion 103, because fluid can still move freely in second portion 103,
even when surface
layer 106 is deformed by a roller such that it fills substantially triangular
portion 101 but not
second portion 103. In some embodiments, a portion along a length of a channel
may have both
a substantially triangular portion and a second portion ("deep section"),
while a different portion
along the length of the channel has only the substantially triangular portion.
In some such
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
83
embodiments, when the apparatus (e.g., roller) engages with the portion having
both a
substantially triangular portion and a second portion (deep section), pump
action is not started,
because a seal with the surface layer is not achieved. However, as the
apparatus engages along
the length direction of the channel, when the apparatus deforms the surface
layer at the portion of
the channel having only a substantially triangular section, pump action begins
because the lack
of second portion (deep section) at that portion allows for a seal (and
consequently a pressure
differential) to be created. Therefore, in some cases, the presence and
absence of deep sections
along the length of the channels of the cartridge can allow for control of
which portions of the
channel are capable of undergoing pump action upon engagement with the
apparatus.
The inclusion of such "deep sections" as second portions of at least some of
the channels
of the cartridge may contribute to any of a variety of potential benefits. For
example, such deep
sections (e.g., second portion 103) may, in some cases, contribute to a
reduction in pump volume
in peristaltic pumping processes. In some such cases, pump volume can be
reduced by a factor
of two or more for higher volume resolution. In some cases, such deep sections
may also
provide for a well-defined starting point for the pump volume that is not
determined by where
the roller lands on the channel. For example, the interface between a portion
of a channel having
both a substantially triangular portion and a second portion (deep section)
and a portion of a
channel having only a substantially triangular portion can, in some cases, be
used as a well-
defined starting point for the pump volume, because only fluid occupying the
volume of the
latter channel portion can be pumped. In some cases, where the rollers lands
on the channel may
have some error associated depending on any of a variety of factors, such as
cartridge
registration. The inclusion of deep sections may, in some cases, reduce or
eliminate variations in
pump volume associated with such error.
As used herein, an average diameter of a substantially triangular portion of a
channel may
be measured as an average over the z-axis from the vertex of the substantially
triangular portion
to the surface of the channel.
SCODA
SCODA can involve providing a time-varying driving field component that
applies forces
to particles in some medium in combination with a time-varying mobility-
altering field
component that affects the mobility of the particles in the medium. The
mobility-altering field
component is correlated with the driving field component so as to provide a
time-averaged net
motion of the particles. SCODA may be applied to cause selected particles to
move toward a
focus area.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
84
In one embodiment of SCODA based purification, described herein as
electrophoretic
SCODA, time varying electric fields both provide a periodic driving force and
alter the drag (or
equivalently the mobility) of molecules that have a mobility in the medium
that depends on
electric field strength, e.g. nucleic acid molecules. For example, DNA
molecules have a mobility
that depends on the magnitude of an applied electric field while migrating
through a sieving
matrix such as agarose or polyacrylamide. By applying an appropriate periodic
electric field
pattern to a separation matrix (e.g. an agarose or polyacrylamide gel) a
convergent velocity field
can be generated for all molecules in the gel whose mobility depends on
electric field. The field
dependent mobility is a result of the interaction between a repeating DNA
molecule and the
sieving matrix, and is a general feature of charged molecules with high
conformational entropy
and high charge to mass ratios moving through sieving matrices. Since nucleic
acids tend to be
the only molecules present in most biological samples that have both a high
conformational
entropy and a high charge to mass ratio, electrophoretic SCODA based
purification has been
shown to be highly selective for nucleic acids.
The ability to detect specific biomolecules in a sample has wide application
in the field of
diagnosing and treating disease. Research continues to reveal a number of
biomarkers that are
associated with various disorders. Exemplary biomarkers include genetic
mutations, the presence
or absence of a specific protein, the elevated or reduced expression of a
specific protein, elevated
or reduced levels of a specific RNA, the presence of modified biomolecules,
and the like.
Biomarkers and methods for detecting biomarkers are potentially useful in the
diagnosis,
prognosis, and monitoring the treatment of various disorders, including
cancer, disease,
infection, organ failure and the like.
The differential modification of biomolecules in vivo is an important feature
of many
biological processes, including development and disease progression. One
example of
differential modification is DNA methylation. DNA methylation involves the
addition of a
methyl group to a nucleic acid. For example a methyl group may be added at the
5' position on
the pyrimidine ring in cytosine. Methylation of cytosine in CpG islands is
commonly used in
eukaryotes for long term regulation of gene expression. Aberrant methylation
patterns have been
implicated in many human diseases including cancer. DNA can also be methylated
at the 6
nitrogen of the adenine purine ring.
Chemical modification of molecules, for example by methylation, acetylation or
other
chemical alteration, may alter the binding affinity of a target molecule and
an agent that binds
the target molecule. For example, methylation of cytosine residues increases
the binding energy
of hybridization relative to unmethylated duplexes. The effect is small.
Previous studies report an
increase in duplex melting temperature of around 0.7 C. per methylation site
in a 16 nucleotide
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
sequence when comparing duplexes with both strands unmethylated to duplexes
with both
strands methylated.
Affinity SCODA
SCODAphoresis is a method for injecting biomolecules into a gel, and
preferentially
5 concentrating nucleic acids or other biomolecules of interest in the
center of the gel. SCODA
may be applied, for example, to DNA, RNA and other molecules. Following
concentration, the
purified molecules may be removed for further analysis. In one specific
embodiment of
SCODAphoresis¨affinity SCODA¨binding sites which are specific to the
biomolecules of
interest may be immobilized in the gel. In doing so one may be able generate a
non-linear motive
10 response to an electric field for biomolecules that bind to the specific
binding sites. One specific
application of affinity SCODA is sequence-specific SCODA. Here
oligonucleotides may be
immobilized in the gel allowing for the concentration of only DNA molecules
which are
complementary to the bound oligonucleotides. All other DNA molecules which are
not
complementary may focus weakly or not at all and can therefore be washed off
the gel by the
15 application of a small DC bias.
SCODA based transport is a general technique for moving particles through a
medium by
first applying a time-varying forcing (i.e. driving) field to induce periodic
motion of the particles
and superimposing on this forcing field a time-varying perturbing field that
periodically alters
the drag (or equivalently the mobility) of the particles (i.e. a mobility-
altering field). Application
20 of the mobility-altering field is coordinated with application of the
forcing field such that the
particles will move further during one part of the forcing cycle than in other
parts of the forcing
cycle.
By varying the drag (i.e. mobility) of the particle at the same frequency as
the external
applied force, a net drift can be induced with zero time-averaged forcing. An
appropriate choice
25 of driving force and drag coefficients that vary in time and space can
generate a convergent
velocity field in one or two dimensions. A time varying drag coefficient and
driving force can be
utilized in a real system to specifically concentrate (i.e. preferentially
focus) only certain
molecules, even where the differences between the target molecule and one or
more non-target
molecules are very small, e.g. molecules that are differentially modified at
one or more locations,
30 or nucleic acids differing in sequence at one or more bases.
An affinity matrix can be generated by immobilizing an agent with a binding
affinity to
the target molecule (i.e. a probe) in a medium. Using such a matrix, operating
conditions can be
selected where the target molecules transiently bind to the affinity matrix
with the effect of
reducing the overall mobility of the target molecule as it migrates through
the affinity matrix.
35 The strength of these transient interactions is varied over time, which
has the effect of altering
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
86
the mobility of the target molecule of interest. SCODA drift can therefore be
generated. This
technique is called affinity SCODA, and is generally applicable to any target
molecule that has
an affinity to a matrix.
Affinity SCODA can selectively enrich for nucleic acids based on sequence
content, with
single nucleotide resolution. In addition, affinity S CODA can lead to
different values of k for
molecules with identical DNA sequences but subtly different chemical
modifications such as
methylation. Affinity SCODA can therefore be used to enrich for (i.e.
preferentially focus)
molecules that differ subtly in binding energy to a given probe, and
specifically can be used to
enrich for methylated, unmethylated, hypermethylated, or hypomethylated
sequences.
Exemplary media that can be used to carry out affinity SCODA include any
medium
through which the molecules of interest can move, and in which an affinity
agent can be
immobilized to provide an affinity matrix. In some embodiments, polymeric gels
including
polyacrylamide gels, agarose gels, and the like are used. In some embodiments,
microfabricated/microfluidic matrices are used.
Exemplary operating conditions that can be varied to provide a mobility
altering field
include temperature, pH, salinity, concentration of denaturants, concentration
of catalysts,
application of an electric field to physically pull duplexes apart, or the
like.
Exemplary affinity agents that can be immobilized on the matrix to provide an
affinity
matrix include nucleic acids having a sequence complementary to a nucleic acid
sequence of
interest, proteins having different binding affinities for differentially
modified molecules,
antibodies specific for modified or unmodified molecules, nucleic acid
aptamers specific for
modified or unmodified molecules, other molecules or chemical agents that
preferentially bind to
modified or unmodified molecules, or the like.
The affinity agent may be immobilized within the medium in any suitable
manner. For
example where the affinity agent is an oligonucleotide, the oligonucleotide
may be covalently
bound to the medium, acrydite modified oligonucleotides may be incorporated
directly into a
polyacrylamide gel, the oligonucleotide may be covalently bound to a bead or
other construct
that is physically entrained within the medium, or the like.
Where the affinity agent is a protein or antibody, in some embodiments the
protein may
be physically entrained within the medium (e.g. the protein may be cast
directly into an agarose
or polyacrylamide gel), covalently coupled to the medium (e.g. through use of
cyanogen bromide
to couple the protein to an agarose gel), covalently coupled to a bead that is
entrained within the
medium, bound to a second affinity agent that is directly coupled to the
medium or to beads
entrained within the medium (e.g. a hexahistidine tag bound to NTA-agarose),
or the like.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
87
Where the affinity agent is a protein, the conditions under which the affinity
matrix is
prepared and the conditions under which the sample is loaded should be
controlled so as not to
denature the protein (e.g. the temperature should be maintained below a level
that would be
likely to denature the protein, and the concentration of any denaturing agents
in the sample or in
the buffer used to prepare the medium or conduct SCODA focusing should be
maintained below
a level that would be likely to denature the protein).
Where the affinity agent is a small molecule that interacts with the molecule
of interest,
the affinity agent may be covalently coupled to the medium in any suitable
manner.
One embodiment of affinity SCODA is sequence-specific SCODA. In sequence
specific
SCODA, the target molecule is or comprises a nucleic acid molecule having a
specific sequence,
and the affinity matrix contains immobilized oligonucleotide probes that are
complementary to
the target nucleic acid molecule. In some embodiments, sequence specific SCODA
is used both
to separate a specific nucleic acid sequence from a sample, and to separate
and/or detect whether
that specific nucleic acid sequence is differentially modified within the
sample. In some such
embodiments, affinity SCODA is conducted under conditions such that both the
nucleic acid
sequence and the differentially modified nucleic acid sequence are
concentrated by the
application of SCODA fields. Contaminating molecules, including nucleic acids
having
undesired sequences, can be washed out of the affinity matrix during SCODA
focusing. A
washing bias can then be applied in conjunction with SCODA focusing fields to
separate the
differentially modified nucleic acid molecules as described below by
preferentially focusing the
molecule with a higher binding energy to the immobilized oligonucleotide
probe.
EXAMPLES
Embodiments of the invention are further described with reference to the
following
examples, which are intended to be illustrative and not restrictive in nature.
Example 1 ¨ Use of a Sample Preparation Device
An automated sample preparation device of the disclosure was used to prepare a
sample
of DNA extracted from human blood.
The sample preparation device comprised a fluidics module (comprising a
peristaltic
pumping system), a temperature control module (to provide temperature and
mechanical
precision), a touch screen interface on the device that allowed the user to
select any process-
specific parameters (e.g., range of desired size of the nucleic acids, desired
degree of homology
for target molecule capture, etc.), and a lid that the user was able open in
order to insert a sample
preparation cartridge of the disclosure. The device was powered with a 1000-
volt electrode
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
88
supply. The sample preparation cartridge comprised thirteen discrete
microfluidics channels (or
pumping lanes) and was fabricated such that it could perform end-to-end sample
preparation.
The microfluidic channels were designed to manipulate reagents and the
cartridge enabled, in
automated succession: (1) Pipet introduction of combined sample lysis using
lysis+ Lysis buffer
and subsequent extraction of target DNA; (2) DNA purification; (3) DNA
tagmentation using
transposase Tn5 succeeded by DNA repair; (4) selection of DNA fragments of
particular size
range using nucleic acid capture probes and SCODA; and (5) DNA clean-up.
100 i.it of whole human blood was mixed with lysis buffer and Proteinase K was
incubated at
55 C for 10 minutes then mixed with isopropanol; lysate mixture was
subsequently added to a
sample port in the sample preparation cartridge, the loaded cartridge was
inserted into the sample
preparation device, and DNA was extracted. The automated device, as described
above, yielded
1.2 i.t.g extracted DNA; 1 i.t.g of that extracted DNA was further processed
using the successive
steps described above to generate 530 ng of a DNA library at a concentration
of 6.5 nM. This
purified DNA library produced by the sample preparation device was then
subjected to
sequencing using a glass sequencing chip.
As a control experiment, 100 i.it of whole human blood (from the same sample
as above)
was manually processed to generate DNA library for sequencing using
traditional DNA
extraction and purification techniques.
The inventors found that sequencing data acquired using DNA library prepared
using the
automated sample preparation device was similar in quality (e.g., as assessed
by average read
length) relative to the sequencing data acquired using DNA manually prepared
using traditional
DNA extraction and purification techniques. As shown in Table 3, the automated
device
generated more total reads (72 total reads using automated process compared to
27 total reads
using manual process) and greater read lengths (1989.0 760.1 base pair read
lengths using
automated process compared to 1132.1 324.5 base pair read lengths using
manual process)
than the manual process, with no significant difference observed between the
processes in terms
of accuracy and GC content of the resulting reads.
Table 3. Sequencing results from DNA libraries generated from whole human
blood
Standard Standard
Standard
Average Average Average
Deviation Deviation
Deviation
Total Read Read GC
Read Read
GC
Reads Length Accuracy content
Length Accuracy
content
(bp) (%) (%)
Manual
27 1132.1 324.5 60.7% 4.1% 35.2% 4.5%
process
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
89
Automated
process
using
Sample
72 1989.0 760.1 59.9% 4.3% 37.0% 4.7%
Preparation
device of
this
disclosure
Example 2 ¨ Use of a Sample Preparation Device to enrich DNA for sequencing
An automated sample preparation device of the disclosure was used to prepare a
sample
of DNA extracted from cultured E. coli cells.
The sample preparation device comprised a fluidics module (comprising a
peristaltic
pumping system), a temperature control module (to provide temperature and
mechanical
precision), a touch screen interface on the device that allowed the user to
select any process-
specific parameters (e.g., range of desired size of the nucleic acids, desired
degree of homology
for target molecule capture, etc.), and a lid that the user was able open in
order to insert a sample
preparation cartridge of the disclosure. The device was powered with a 1000-
volt electrode
supply. The sample preparation cartridge comprised thirteen discrete
microfluidics channels (or
pumping lanes) and was fabricated such that it could perform end-to-end sample
preparation.
The microfluidic channels were designed to manipulate reagents and the
cartridge enabled, in
automated succession: (1) Pipet introduction of combined sample + Lysis buffer
and subsequent
extraction of target DNA; (2) DNA purification; (3) DNA tagmentation using
transposase Tn5
succeeded by DNA repair; (4) selection of DNA fragments of particular size
range using
SCODA; and (5) DNA clean-up.
A sample of seven-hundred million E.coli cells from an overnight culture mixed
with
lysis buffer and Proteinase K was incubated at 55 C for 10 minutes then mixed
with isopropanol;
lysate mixture was added to a sample port in the sample preparation cartridge,
the loaded
cartridge was inserted into the sample preparation device, and DNA was
extracted. Automated
processing continued to render the DNA into DNA library ready for sequencing
with a brief
pause for the user to add DNA Repair Enzyme and DNA Repair Buffer Mix to the
cartridge just
prior to the DNA Repair step. The automated device transported the DNA Repair
Enzyme and
DNA Repair Buffer Mix to the reaction location in the cartridge. The automated
device, as
described above, yielded 0.96 iig extracted DNA; subsequent automated steps
generated 279 ng
of a DNA library at a concentration of 2.89 nM.
As a control experiment, a sample of seven-hundred million E.coli cells (from
the same
sample as above) was manually processed to generate DNA using traditional DNA
extraction
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
and purification techniques. This manually prepared DNA was subjected to the
same automated
library preparation process on the automated device generating 199 ng of a DNA
library at a
concentration of 2.65 nM.
The purified DNA libraries produced by the sample preparation device were
concentrated
5 using Aline beads and then subjected to sequencing on a Pacific
Biosciences RSII DNA
Sequencer.
The inventors found that sequencing data acquired using DNA purified and
prepared into
library format using the automated sample preparation device generated
sequencing reads that
were slightly shorter in length, but similar in quality (as assessed by Rsq
score) relative to the
10 sequencing data acquired using DNA manually prepared with traditional
DNA extraction and
purification techniques followed by automated DNA library preparation (Figure
25).
As shown in Table 4, the fully automated library generated reads with
identical read quality (Rsq
0.82) to those generated with manual DNA extraction, with roughly equivalent
read lengths (851
base average reads lengths versus 922 for manual).
Table 4. Sequencing results from DNA libraries generated from E. coli cells
extracted and
purified via an Automated Sample Preparation Device versus manually extracted
and
purified DNA run on the same automated device.
Median
Librar
Seq Treatment Reads read
RS q
name y length
From lysate, E.coli library (Sample
C1856 E2E 5756 851 0.82
Prep device of this disclosure)
From purified DNA, E.coli library
C890 MEAL (Sample Prep device of this 7674 922
0.82
disclosure)
Example 3 ¨ Use of a Sample Preparation Device to enrich DNA for sequencing
An automated sample preparation device of the disclosure was used to select
DNA
fragments of a particular size range using SCODA for a DNA library manually
prepared from E.
coli cultured cells.
Four micrograms of manually purified E.coli DNA was subjected to Tn5a
tagmentation
and then split into four separate samples consisting of 1 i.t.g each.
Selection of DNA fragments of
a particular size was conducted separately by four different methods (1) Sage
BluePippin with
program to collect fragments from 3 kb to 10kb in size, (2) Sage BluePippin
with program to
collect fragments greater in size than 4 kb to 10 kb, (3) manual Aline bead
size selection with
0.45x bead addition, or (4) SCODA technology as in the automated sample
preparation device
(described in Example 8.0).
CA 03177368 2022-09-27
WO 2021/216763 PCT/US2021/028471
91
After size selection, each sample was separately prepared into DNA library and
sequenced on a Pacific Biosciences RSII DNA Sequencer.
The inventors found that sequencing data acquired using DNA library size
selection using the
automated sample preparation device was superior to or equivalent to replicate
DNA libraries
selected for size by the standard manual bead-based process or the automated
Sage BluePippin
size selection method (Figure 26).
As shown in Table 5 (below), the automated device generated read lengths
longer than
the manual size selection process and equivalent to the BluePippin methods
with no significant
difference observed among the processes in terms of accuracy and GC content of
the resulting
reads.
Table 5. Sequencing metrics from DNA libraries generated automated size
selection
compared to those derived from samples size selected by commercial and manual
methods
Median
Read
read
Size selection length
Sage BluePippin, selecting for 3-10kb
675 2389
range
Sage BluePippin, selecting >4-10kb high
2253 2409
pass
Manual bead-based size selection (Aline) 2296 1478
Automated size selection (Sample Prep
18707 2358
device of this disclosure)
Example 4 ¨ Preparation of a biological sample for sequencing
Sample lysis
Cultured cells or tissue samples comprising one or more target molecules
(e.g., proteins)
are lysed using any method known to a skilled person. The biological samples
are suspended in
lysis buffer (e.g., RIPA buffer, GC1 (Guanidine-HC1) buffer, GlyNP40 buffer)
and mechanically
homogenized to break down cell walls (e.g., in a lysis cartridge). Once the
cells are disrupted,
the target molecules are then precipitated and the supernatant discarded.
Precipitation can be
accomplished using centrifugation including washing steps (e.g., addition of
either a mix of
chloroform/methanol or trichloroacetic acid). See Figure 3.
Enrichment
The lysed sample is then optionally enriched (e.g., using affinity matrices)
to capture the
target molecules and discard the remaining non-target molecules (e.g., in an
enrichment
cartridge). Enrichment may include depletion strategies utilized to reduce
sample complexity by
sequestering the non-target molecules (e.g., using affinity matrices). See
Figure 4.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
92
Fragmentation
The lysed sample (if not enriched) or the enriched sample may then be
fragmented (e.g.,
digested) (e.g., in a fragmentation cartridge). This step in the sample
process converts target
molecules into smaller fragments or subunits. This step can be conducted using
non-enzymatic
and/or enzymatic processes. Non-enzymatic methods include (but are not limited
to) acid
hydrolysis, cleavage via cyanogen bromide, hydroxylamine, and 2-nitro-5-
thiocyanobenzoic
acid, and electrochemical oxidation. Enzymatic methods include (but are not
limited to) the use
of nucleases or proteases. See Figure 6.
Functionalization
Prior to sequencing, the fragmented sample may be functionalized at one of its
terminal
moieties (e.g., N-terminus or C-terminus of a protein fragment) (e.g., in a
functionalization
cartridge). For example, digested peptides may be labeled with some moiety
capable of
immobilizing the peptides on the sequencing substrate. Functionalization can
be accomplished
through a variety of chemical or enzymatic methods. See Figures 6 and 7.
Example 5 ¨ Preparation of a protein sample
This example describes the preparation of a protein sample using a device of
the
disclosure, wherein the incubation, functionalization, quenching,
immobilization complex
forming, and purifying steps were performed on a single cartridge. Proteins
were prepared by
pulldown from spiked plasma, wherein the enriched protein was purified using
either an antibody
or a DNA aptamer on a solid support. Proteins were then equilibrated with the
desired buffer,
either by gel filtration or by pH adjustment. Then, an enriched protein sample
(50-200 i.t.M in
100 i.tt) comprising an equal mixture of 2, 3, or 4 proteins was prepared in
100mM HEPES or
sodium phosphate (pH 6-9) with 10-20% acetonitrile was mixed with a solution
of tris(2-
carboxyethyl)phosphine hydrochloride (TCEP-HC1, 200 mM in water, 1 tt), to act
as a reducing
agent, freshly dissolved iodoacetamide solution (9 mg in 97.3 i.tt water for
500 mM, 2 tt), to
act as an amino acid side-chain capping agent, and Trypsin (1 iig/i.tt, 0.5-1
tt), to act as a
protein digestion agent. Next, the peptide sample was incubated at 37 C for 6
to 10 hours in the
digestion portion, wherein the protein was denatured and digested. This
resulted in the formation
of a digested peptide sample.
Next, the digested peptide sample was automatedly transported through a series
of
reservoirs, where it mixed with a functionalization agent, a first (catalytic)
reagent, and a second
(pH-adjusting) reagent. Initially, the digested peptide sample was automatedly
added to
potassium carbonate (1 M, 5 tt), to adjust the pH to a value of 10-11.
Following this, the
digested peptide sample was automatedly exposed to imidazole-l-sulfonyl azide
solution ("ISA"
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
93
200 mM in 200 mM KOH, 1.2 lL), an azide transfer agent. Next, the digested
peptide sample
was automatedly mixed with copper sulfate (a catalytic reagent) solution.
Finally, the digested
peptide sample was automatedly transferred to a functionalization portion of
the modular
cartridge where was incubated for one hour at room temperature. This resulted
in the formation
unquenched mixture comprising one or more derivatized peptides.
Following functionalization of the peptides in the functionalization region,
50 i.it of the
unquenched sample was automatedly transported to a portion of the of the
modular cartridge
where it was mixed with a plurality of polystyrene beads (a solid substrate),
and quenched using
actively mixed quench steps, with each quench step followed by a stationary
mixing step, for
10 a total of 23 minutes. Finally, the resulting quenched mixture was
passed through an on-
cartridge column to filter it from the plurality of polystyrene beads.
Next, the pH of the quenched peptide sample was adjusted to between 7 and 8
through
the addition of 6 i.it of 1 M acetic acid. Following this, the quenched
mixture was automatedly
mixed with DBCO-Q24-SV (50 t.M, 6 lL), an immobilization complex, before being
incubated
at 37 C on the device for 4 hours. Following this, the peptide sample was
automatedly
transported to a column of the modular cartridge, consisting of Zeba de-
salting column resin with
a cut off of 40 kDa that was equilibrated first with 10 mM TRIS, 10 mM
potassium acetate
buffer (pH 7.5). Finally, the purified peptide sample that resulted from this
workflow was frozen
and stored at a temperature below -20 C.
At a later time, purified peptide samples were sequenced, and observed
peptides were
identified based on their correspondence to protein sequences. FIGS. 27A-27D
present the
results in the form of bar charts. FIG. 27A corresponds to a mixture of two
proteins ¨ GIP and
ADM. FIG. 27B corresponds to a mixture of three proteins ¨ GLP1, Insulin, and
ADM. FIG.
27C corresponds to a mixture of four proteins ¨ GLP1, ADM, Insulin, and GIP.
FIG. 27D
corresponds to a mixture of four peptides ¨ GLP1, ADM, Insulin, and GIP. A few
off-target
assignments 801 are indicated, but in general the peptides sequenced were
correctly assigned to
the proteins prepared in the peptide sample. Moreover, the generated libraries
in this example
had similar or more total reads than replicate manually prepared libraries of
the same protein
mixes. This example demonstrates that a purified peptide sample can be
prepared in an
automated way on a modular cartridge of the type disclosed here.
Example 6 ¨ Use of a device of the disclosure
This example describes an exemplary device, wherein the incubation,
functionalization,
quenching, immobilization complex forming, and purifying steps may be
performed using a
device of the disclosure comprising multiple modular cartridges. Although the
modular
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
94
cartridges of this embodiment are not connected, peptide samples were prepared
by following the
protocol of Example 5. The protein sample was loaded and then incubated (e.g.
at 37 C for 5
hours), wherein the protein was denatured and digested. The cartridges further
comprised pump
lanes to facilitate pumping of the fluids within the cartridge, as well as a
reagent/sample mixture
source.
After incubation, the peptide sample became a digested peptide sample. The
digested
peptide sample was then automatedly transferred to a second cartridge, where
it was automatedly
transported through a series of reservoirs, where it mixed with a
functionalization agent, a first
(catalytic) reagent, and a second (pH-adjusting) reagent. The digested peptide
sample was
transported to the second cartridge through a sample input. The digested
peptide sample was
automatedly transported mixed with the functionalization agent, a first
(catalytic) reagent, and a
second (pH-adjusting) reagent , in sequence. Finally, the digested peptide
sample was incubated
for the period of time (e.g. one hour at room temperature). This resulted in
the formation of an
unquenched mixture. The second cartridge further comprised pump lanes.
A portion of the unquenched sample was automatedly transported to a third
cartridge
comprising a sample input, a filter for beads, a small volume acidic reagent
reservoir, and mixing
channels. Here, the unquenched mixture was quenched at room temperature.
Finally, the
resulting quenched mixture was passed through an on-cartridge column to remove
the plurality
of polystyrene beads, and the pH was adjusted to between 7 and 8 by the
addition of acetic acid
from an acidic reagent reservoir.
Following this, the quenched mixture was mixed with the DBCO-Q24-SV
immobilization complex in the mixture source of the first modular cartridge,
before it was
incubated at 37 C.
Finally, the peptide sample was automatedly transported to a fourth cartridge,
which
controlled the flow of the quenched peptide sample through a commercial Zeba
de-salting
column resin. Additional equilibration buffer was dispensed through the column
to ensure that
the peptides were transmitted through the column. The purified peptide sample
was collected
from a specific fraction of the fluid passing through the column, while the
remaining fluid was
transmitted to a waste reservoir. This example demonstrates that in some
embodiments, purified
peptide samples can be produced automatedly using devices comprising multiple
cartridges.
Additional Embodiments
Additional embodiments of the present disclosure are encompassed by the
following
numbered paragraphs:
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
1. A device for enriching a target molecule from a biological sample, the
device comprising
an automated sample preparation module comprising a cartridge housing that is
configured to
receive a removable cartridge.
2. The device of paragraph 1, wherein the removable cartridge is a single-
use cartridge or a
5 multi-use cartridge.
3. The device of paragraph 1 or 2, wherein the removable cartridge is
configured to receive
the biological sample.
4. The device of paragraph 3, wherein the removable cartridge further
comprises the
biological sample.
10 5. The device of any one of paragraphs 1-4, wherein the cartridge
comprises one or more
microfluidic channels configured to contain and/or transport a fluid used in a
sample preparation
process.
6. The device of any one of paragraphs 1-5, wherein the cartridge
comprises one or more
affinity matrices, wherein each affinity matrix comprises an immobilized
capture probe that has a
15 binding affinity for the target molecule.
7. The device of any one of paragraphs 1-6, wherein the
biological sample is a
blood, saliva, sputum, feces, urine or buccal swab sample.
8. The device of any one of paragraphs 1-7, wherein the target molecule
is a target nucleic
acid.
20 9. The device of paragraph 8, wherein the target nucleic acid is an
RNA or DNA molecule.
10. The device of any one of paragraphs 3-9, wherein the immobilized
capture probe is an
oligonucleotide capture probe, and wherein the oligonucleotide capture probe
comprises a
sequence that is at least partially complementary to the target nucleic acid.
11. The device of paragraph 10, wherein the oligonucleotide capture probe
comprises a
25 sequence that is at least 80%, 90% 95%, or 100% complementary to the
target nucleic acid.
12. The device of any one of paragraphs 8-11, wherein the device produces
target nucleic
acid with an average read-length for downstream sequencing applications that
is longer than an
average read-length produced using control methods.
13. The device of any one of paragraphs 1-7, wherein the target molecule is
a target protein.
30 14. The device of any one of paragraphs 1-7 or 13, wherein the
immobilized capture probe is
a protein capture probe that binds to the target protein.
15. The device of paragraph 13, wherein the protein capture probe is an
aptamer or an
antibody.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
96
16. The device of paragraph 14 or 15, wherein the protein capture probe
binds to the target
protein with a binding affinity of 10-9 to 10-8 M, 10-8 to 10-7 M, 10-7 to 10-
6 M, 10-6 to 10-5 M, 10-
to 104 M, 104 to 10-3 M, or 10-3 to 10-2 M.
17. The device of any one of paragraphs 1-16, wherein the device further
comprises a
5 sequencing module.
18. The device of paragraph 17, wherein the automated sample preparation
module is directly
or indirectly connected to the sequencing module.
19. The device of paragraph 17 or 18, wherein the device is configured to
deliver the target
molecule from the automated sample preparation module to the sequencing
module.
20. The device of any one of paragraphs 17-19, wherein the sequencing
module performs
nucleic acid sequencing.
21. The device of paragraph 20, wherein the nucleic acid sequencing
comprises single-
molecule real-time sequencing, sequencing by synthesis, sequencing by
ligation, nanopore
sequencing, and/or Sanger sequencing.
22. The device of any one of paragraphs 17-19, wherein the sequencing
module performs
protein sequencing.
23. The device of paragraph 22, wherein the protein sequencing comprises
Edman
degradation or mass spectroscopy.
24. The device of any one of paragraphs 17-19, wherein the sequencing
module performs
single-molecule protein sequencing.
25. A method for purifying a target molecule from a biological sample, the
method
comprising:
(i) lysing the biological sample;
(ii) fragmenting the lysed sample of (i); and
(iii) enriching the sample using an affinity matrix comprising an
immobilized capture
probe that has a binding affinity for the target molecule,
thereby purifying the target molecule.
26. The method of paragraph 25, wherein the target molecule is a molecule
is a target nucleic
acid.
27. The method of paragraph 26, wherein the target nucleic acid is an RNA
or DNA
molecule.
28. The method of any one of paragraphs 25-27, wherein the immobilized
capture probe is an
oligonucleotide capture probe, and wherein the oligonucleotide capture probe
comprises a
sequence that is at least partially complementary to the target nucleic acid.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
97
29. The method of paragraph 28 wherein the oligonucleotide capture probe
comprises a
sequence that is at least 80%, 90% 95%, or 100% complementary to the target
nucleic acid.
30. The method of paragraph 28, wherein the target molecule is a target
protein.
31. The method of paragraph 25 or 30, wherein the immobilized capture probe
is a protein
capture probe that binds to the target protein.
32. The method of paragraph 31, wherein the protein capture probe is an
aptamer or an
antibody.
33. The method of paragraph 31 or 32, wherein the protein capture probe
binds to the target
protein with a binding affinity of 10-9 to 10-8 M, 10-8 to 10-7 M, 10-7 to 10-
6 M, 10-6 to 10-5 M, 10-
5 to 104 M, 10-4 to 10-3 M, or 10-3 to 10-2 M.
34. The method of any one of paragraphs 25-33, wherein step (i) comprises
an electrolytic
method, an enzymatic method, a detergent-based method, and/or mechanical
homogenization.
35. The method of any one of paragraphs 25-34, wherein step (i) comprises
multiple lysis
methods performed in series.
36. The method of any one of paragraphs 25-35, wherein the sample is
purified following
lysis and prior to step (ii) or (iii).
37. The method of any one of paragraphs 25-36, wherein step (ii) comprises
mechanical,
chemical and/or enzymatic fragmentation methods.
38. The method of any one of paragraphs 25-37, wherein the sample is
purified following
.. fragmentation and prior to step (iii).
39. The method of any one of paragraphs 25-38, wherein step (iii) comprises
enrichment
using an electrophoretic method.
40. The method of paragraph 39, wherein the electrophoretic method is
affinity SCODA,
FIGE, or PFGE.
41. The method of any one of paragraphs 25-40, further comprising:
(iv) detecting the target molecule.
42. The method of paragraph 41, wherein step (iv) comprises detection using
absorbance,
fluorescence, mass spectroscopy, and/or sequencing methods.
43. The method of any one of paragraphs 25-42, wherein the biological
sample is a blood,
saliva, sputum, feces, urine or buccal sample.
44. The method of any one of paragraphs 25-43, wherein the biological
sample is from a
human, a non-human primate, a rodent, a dog, a cat, or a horse.
45. The method of any one of paragraphs 25-44, wherein the biological
sample comprises a
bacterial cell or a population of bacterial cells.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
98
46. A device for enriching a target molecule from a biological sample,
the device comprising
an automated sample preparation module, wherein the automated sample
preparation module
performs the following steps:
(i) receives a biological sample
(ii) lyses the biological sample;
(iii) fragments the sample of (ii); and
(iv) enriches the sample using an affinity matrix comprising an immobilized
capture
probe that has a binding affinity for the target molecule.
47. The device of paragraph 46, wherein the target molecule is a
molecule is a target nucleic
acid.
48. The device of paragraph 46, wherein the target nucleic acid is an
RNA or DNA molecule.
49. The device of any one of paragraphs 46-48, wherein the immobilized
capture probe is an
oligonucleotide capture probe, and wherein the oligonucleotide capture probe
comprises a
sequence that is at least partially complementary to the target nucleic acid.
50. The device of paragraph 49, wherein the oligonucleotide capture probe
comprises a
sequence that is at least 80%, 90% 95%, or 100% complementary to the target
nucleic acid.
51. The device of paragraph 46, wherein the target molecule is a target
protein.
52. The device of paragraph 46 or 51, wherein the immobilized capture probe
is a protein
capture probe that binds to the target protein.
53. The device of paragraph 52, wherein the protein capture probe is an
aptamer or an
antibody.
54. The device of paragraph 52 or 53, wherein the protein capture probe
binds to the target
protein with a binding affinity of 10-9 to 10-8 M, 10-8 to 10-7 M, 10-7 to 10-
6 M, 10-6 to 10-5 M, 10-
5 to 104 M, 104 to 10-3 M, or 10-3 to 10-2 M.
55. The device of any one of paragraphs 46-54, wherein the device further
comprises a
sequencing module.
56. The device of paragraph 55, wherein the automated sample preparation
module is directly
connected or indirectly connected to the sequencing module.
57. The device of paragraph 55 or 56, wherein the device is configured to
deliver the target
molecule from the automated sample preparation module to the sequencing
module.
58. The device of any one of paragraphs 55-57, wherein the sequencing
module performs
nucleic acid sequencing.
59. The device of paragraph 58, wherein the nucleic acid sequencing
comprises single-
molecule real-time sequencing, sequencing by synthesis, sequencing by
ligation, nanopore
sequencing, and/or Sanger sequencing.
CA 03177368 2022-09-27
WO 2021/216763
PCT/US2021/028471
99
60. The device of any one of paragraphs 55-57, wherein the sequencing
module performs
protein sequencing.
61. The device of paragraph 60, wherein the protein sequencing comprises
Edman
degradation or mass spectroscopy.
62. The device of any one of paragraphs 55-57, wherein the sequencing
module performs
single-molecule protein sequencing.
63. The device of any one of paragraphs 55-59, wherein the device
produces target nucleic
acids with an average sequencing read-length that is longer than an average
sequencing read-
length produced using control methods.
Further Aspects of the Invention
Aspects of the exemplary embodiments and examples described above may be
combined in
various combinations and subcombinations to yield further embodiments of the
invention. To the
extent that aspects of the exemplary embodiments and examples described above
are not
mutually exclusive, it is intended that all such combinations and
subcombinations are within the
scope of the present invention. It will be apparent to those of skill in the
art that embodiments of
the present invention include a number of aspects. Accordingly, the scope of
the claims should
not be limited by the preferred embodiments set forth in the description and
examples, but should
be given the broadest interpretation consistent with the description as a
whole.