Note: Descriptions are shown in the official language in which they were submitted.
WO 2021/225952
PCT/US2021/030452
LARGE SCALE PRODUCTION OF OLIVETOL, OLIVETOLIC ACID AND OTHER ALKYL
RESORCINOLS
BY FERMENTATION
PRIORITY CLAIM
This application claims priority to US provisional application nos. US
63/122õ369 filed on
December 7, 2020; US 63/089,736 filed on October 9, 2020; 63/079,390 filed on
September 16,
2020; US 63/070,513 filed on August 26, 2020; and US 63/022,038 filed on May
8, 2020, each of
which is incorporated herein in its entirety by reference.
STATEMENT ABOUT FEDERAL FUNDING
Not applicable.
FIELD
Provided herein are processes, preferably scalable, commercially relevant
processes, of
producing olivetol and olivetolic acid, or an analog thereof, or a salt of
each thereof by
fermentation employing a recombinant heterologous host microorganism.
BACKGROUND
Olivetol and olivetolic acid are key gateway molecules for preparing
cannabinoids. And
yet, there is very little if any report of a scalable, commercially viable,
production of olivetol and
olivetolic acid by fermentation.
Further, divarin, which is 1,3-dihydroxy-5-propylbenzene and divarinic acid,
which is 2,4-
dihydroxy-6-propylbenzoic acid, are key gateway molecules for preparing
certain minor
cannabinoids. Minor cannabinoids are naturally obtained in quantities smaller
than major
cannabinoids such as tetrahydrocannabinol (THC). And yet, there is very little
if any report of
producing divarin and divarinic acid by fermentation.
SUMMARY
In one aspect provided herein are processes of producing a compound of formula
(IA)
and/or (IB)
1
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
OH OH
CO2H
HO R HO 40i
IA IB
or a salt thereof wherein R is optionally substituted CI-Cs alkyl, optionally
substituted C2-C6
alkenyl, or optionally substituted C2-Cs alkynyl. In certain embodiments,
other R groups such as
optionally substituted cycloalkyl, preferably optionally substituted C3-Cs
cycloalkyl; optionally
substituted heterocyclyl; optionally substituted aryl, preferably optionally
substituted phenyl;
and optionally substituted heteroaryl are contemplated as employed according
to the present
invention. In one embodiment, the compound produced is of formula IA. In
another
embodiment, the compound produced is of formula IB.
Certain of these processes utilize a recombinant, heterologous, host cells or
host
microorganism. Certain of the host microorganisms comprise a recombinant
olivetol synthase
(OLS or OS), which is a tetraketide synthase (TKS). Certain of the host
microorganisms comprise
a recombinant Cannabis sativa olivetol synthase (which is a tetraketide
synthase, csOLS).
Certain of the host microorganisms comprise a recombinant olivetolic acid
cyclase (OAC).
Certain of the heterologous microorganisms comprise a recombinant Cannabis
sativa olivetolic
acid cyclase (csOAC). Certain of the host microorganisms comprise an acyl
activating enzyme
(AAE). Certain of the heterologous microorganisms comprise a recombinant
Cannabis sativa
acyl activating enzyme (csAAE), such as, without limitation, csAAE1.
In another embodiment, the recombinant host microorganism comprises 4-20, or 6-
16
copies of csOLS. In another embodiment, the recombinant host microorganism
comprises 4-20,
or 6-16 copies of csOAC. In another embodiment, the recombinant host
microorganism
comprises 4-20, or 6-16 copies of csAAE1.
In certain of these processes, glucose is fermented. In certain of these
processes,
galactose is fermented. The process further comprises a carboxylic acid of
formula R-CO2H or a
salt thereof. In certain of the processes, the microorganism is a yeast. In
certain of the
2
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
processes, the microorganism is Saccharomyces cereyisiae. In certain of the
processes, the
microorganism is a bacteria. In certain of the processes, the microorganism is
Escherichia co/i.
In some embodiments, the process further comprises contacting: an aqueous
phase
comprising glucose and RCO2H or a salt thereof and an organic phase immiscible
with the
aqueous phase.
In one embodiment, R is CI-Cs alkyl. In another embodiment, R is Ci-C4 alkyl.
In another
embodiment, R is C6-C8 alkyl. In another embodiment, R is substituted CI-Cs
alkyl.
In another embodiment, R is C2-C8 alkenyl. In another embodiment, R is
substituted C2-
C8 alkenyl.
In another embodiment, R is C2-C8 alkynyl. In another embodiment, R is
substituted C2-
C8 alkynyl.
In another embodiment, the compounds of formula IA and IB are provided in a
combined amount of at least about 2 g/liter over about 4 to about 7 days. In
another
embodiment, the compounds of formula IA and IB are provided in a combined
amount of at
least about 3 g/liter over about 4 to about 7 days. In another embodiment, the
compounds of
formula IA and IB are provided in a combined amount of at least about4 g/liter
over about 4 to
about 7 days. In another embodiment, the compounds of formula IA and IB are
provided in a
combined amount of at least about 5 g/liter over about 4 to about 7 days. In
another
embodiment, the compounds of formula IA and IB are provided in a combined
amount of at
least about 10 g/liter over about 4 to about 7 days.
This invention arises in part from the surprising discovery that recombinant
host
microorganisms produce commercially relevant amounts of olivetol and
olivetolic acid by
fermentation. In some aspects, provided herein are processes of producing
olivetol, olivetolic
acid, or a salt thereof. Certain of these processes are commercially viable
for producing
olivetol, olivetolic acid or a salt thereof, which are key gateway compounds
for preparing a
variety of cannabinoids. Certain of these processes utilize a recombinant,
heterologous, host
microorganism. Certain of the host microorganisms include a recombinant
Cannabis satiya
olivetol synthase (which is a tetraketide synthase, csOLS). Certain of the
heterologous
microorganisms include a recombinant Cannabis satiya olivetolic acid cyclase
(csOAC). Certain
3
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
of the heterologous microorganisms include a recombinant Cannabis sativa acyl
activating
enzyme (csAAE), such as, without limitation, csAAE1. In another embodiment,
the recombinant
host microorganism comprises 4-20, or 6-16 copies of csOLS. In another
embodiment, the
recombinant host microorganism comprises 4-20, or 6-16 copies of csOAC. In
another
embodiment, the recombinant host microorganism comprises 4-20, or 6-16 copies
of csAAE1.
In certain of these processes, glucose is fermented. In certain of these
processes, galactose is
fermented. In certain of these processes, the fermentation further comprises
hexanoic acid or a
salt thereof. Certain of these processes provide olivetol and olivetolic acid
in a combined
amount of at least 3 g/liter. In certain of the processes, the microorganism
is Saccharomyces
cereyisiae.
This invention arises in another part from the surprising discovery that
recombinant
host microorganisms produce divarin and divarinic acid by fermentation. In
some aspects,
provided herein are processes of producing divarin and/or divarinic acid or a
salt thereof.
Certain of these processes are commercially viable for producing divarin and
divarinic acid,
which are key gateway compounds for preparing a variety of minor cannabinoids.
Certain of
these processes utilize a recombinant, heterologous, host microorganism.
Certain of the host
microorganisms include a recombinant Cannabis sativa olivetol synthase (which
is a tetraketide
synthase, csOLS). Certain of the heterologous microorganisms include a
recombinant Cannabis
sativa olivetolic acid cyclase (csOAC). Certain of the heterologous
microorganisms include a
recombinant Cannabis sativa acyl activating enzyme (csAAE), such as, without
limitation,
csAAE1. In another embodiment, the recombinant host microorganism comprises 4-
20, or 6-16
copies of csOLS. In another embodiment, the recombinant host microorganism
comprises 4-20,
or 6-16 copies of csOAC. In another embodiment, the recombinant host
microorganism
comprises 4-20, or 6-16 copies of csAAE1. In certain of these processes,
glucose is fermented. In
certain of these processes, the fermentation further comprises butyric acid or
a salt thereof.
Certain of these processes provide divarin and/or divarinic acid or a salt
thereof in a combined
amount of at least about 0.25 ¨ about 8 g/liter, about 1 ¨ about 7 g/liter,
about 0.25 ¨ about 2
g/liter, about 0.25 ¨ about 2 g/liter, about 0.5 ¨ about 1 g/liter, or about 2
¨ about 4 g/liter.
Certain of these processes provide divarin and/or divarinic acid or a salt
thereof in a combined
4
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
amount of at least about 2 ¨ about 5, preferably about 3 ¨ about 4 g/liter. In
one embodiment,
the combined amount of divarin and/or divarinic acid is provided over 2-7 or 4-
7 days, such as
2,3,4, 5, 6, or 7 days. In certain of the processes, the microorganism is
Saccharomyces
cerevisiae.
In one embodiment, the fermentation is performed as a batch/fed batch
fermentation
with a fixed batch duration. In one embodiment, the fermentation is performed
as a "semi-
continuous" fermentation operating mode. In one embodiment, the fermentation
is performed
as a continuous fermentation operating mode. The continuous mode may be a fill-
and-draw, or
a true continuous operation.
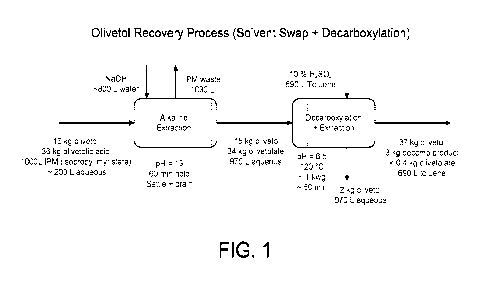
An illustrative and non-limiting process of isolating olivetol or another
compound of
formula IA or IB is schematically illustrated in Figure 1.
In one embodiment, a mixture of compounds of formula IA and IB provided by
fermentation is extracted from a fermentation media by alkaline extraction. In
some
embodiments, the alkaline extraction is an aqueous alkaline extraction. In
some embodiments,
the alkaline extraction is performed at a pH of about 12 - about 14. In some
embodiments, the
alkaline extraction is performed at a pH of about 13. In some embodiments, the
alkaline
extraction is performed under milder alkaline conditions. In some embodiments,
the alkaline
extraction is performed at a pH of about 7 - about 12. In some embodiments,
the alkaline
extraction is performed at a pH of about 7 - about 10. Without being bound by
theory, under
the milder alkaline extraction, a compound of formula IB is preferentially
extracted. The
compound of formula IA can thereafter be extracted under stronger alkaline
conditions, e.g., as
described herein.
In some embodiments, the extracted mixture of compounds of formula IA and IB
are
decarboxylated to provide a compound of formula IA. In some embodiments, the
decarboxylation is performed by heating. In some embodiments, the heating is
performed at
about 100nC ¨ about 140nC, or preferably at about 110 C¨ about 130 C. In some
embodiments,
the heating is performed at about 120 C. Post decarboxylation, the compound of
formula IA
provided, comprises by weight about 2% or less, or preferably about 1% or less
of a compound
of formula IB or a salt thereof. In some embodiments, the extracted mixture of
compounds of
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
formula IA and IB are acidified before decarboxylation. In some embodiments,
the
decarboxylation is performed at a pH of about 5 - about 8. In some
embodiments, the
decarboxylation is performed at a pH of about 6.5.
In one embodiment, the compound of formula IA provided by decarboxylation is
extracted into an organic solvent (e.g., a water immiscible organic solvent)
to provide a solution
of the compound of formula IA in the organic solvent. In some embodiments, the
organic
solvent is a solvent capable of dissolving a compound of formula IA; formula
IA comprises an
aromatic ring and polar hydroxy groups. In one embodiment, the organic solvent
comprises an
aromatic hydrocarbon solvent. In one embodiment, the organic solvent comprises
toluene. In
one embodiment, the organic solvent is toluene. In some embodiments, the
organic solvent
comprises aliphatic or alicyclic hydrocarbon solvents.
In some embodiments, the compound of formula IA, present as a solution in the
organic
solvent, is reacted with a terpene alcohol, a terpenal (i.e., a terpene
aldehyde), and the likes. In
some embodiments, the solution of the compound of formula IA in the organic
solvent is
employed for reacting the compound of formula IA with a terpene alcohol. In
some
embodiments, the solution of the compound of formula IA in the organic solvent
is employed
for reacting the compound of formula IA with a terpenal. In one embodiment,
the terpene
alcohol is geraniol. In one embodiment, the terpene alcohol is farnesol. In
one embodiment,
the terpene alcohol is menthadienol (trans 2,8-menthadienol or PMD). In one
embodiment, the
terpene alcohol is
HO
or a diastereomer thereof, or an ester of each thereof. In one embodiment, the
the hydroxy
form (unesterified) is employed. In one embodiment, the terpene alcohol is:
6
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
// \_/
'2'om
(1R,4R)-4-lsopropeny1-1-methyl-2-cyclohexen-1-ol
In one embodiment, the terpenal is citral. In some embodiments, the reaction
with a terpenal
further comprises a primary amine. In one embodiment, the primary amine is
tertiary butyl
amine.
In some embodiments, the reaction of a compound of formula IA with a terpene
alcohol, a terpenal, or the likes provides a cannabinoid. In one embodiment,
the cannabinoid is
cannabigerol (CBG). In another embodiment, the cannabinoid is cannabichromene
(CBC). In
another embodiment, the cannabinoid is cannabidiol (CBD). In another
embodiment, the
cannabinoid is tetrahydrocannabinol (THC). In another embodiment, the
cannabinoid is
cannabinol (CBN). In another embodiment, the cannabinoid is the varin analog
(CBGV, CBCV,
CBDV, THCV, CBNV) of CBG, CBC, CBD, THC, CBN. A varin analog is a compound
where the n-
pentyl chain of a cannabinoid, e.g., and without limitation, CBG, CBC, CBD, or
THC is replaced by
an n-propyl chain. The cannabinoids obtained are purified by a variety of
purification methods.
In one embodiment, the purification method comprises chromatography. In one
embodiment
the purification method comprises distillation. In one embodiment, the
chromatography
comprises a reverse phase chromatography.
In one embodiment, R is n-pentyl. In another embodiment, R is n-propyl. In
another
embodiment, R is n-heptyl.
A non-limiting example of reacting (prenylating) olivetol with the terpene
alcohol,
geraniol, is schematically illustrated in Figure 2.
A non-limiting example of reacting (prenylating) olivetol with the terpenal,
citral, is
schematically illustrated in Figure 3.
The initial engineering of Saccharomyces cerevisiae was done by introducing a
gene
fragment containing csOLS, csOAC, and csAAE1 under the control of galactose
regulatable
7
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
elements called promoters. The csOLS and csOAC were physically linked to each
other on the
gene with a genetic element called T2A in all examples.
To select for Saccharomyces cerevisiae cells that efficiently incorporated the
foreign
DNA, but removed Saccharomyces cerevisiae that had no foreign genes, a
standard protocol
method was utilized that allows growth on nutrient preferred media. One way to
construct
gene fragments that are functional in an organism is to generate individual
gene fragments by a
polymerase chain reaction (PCR). This creates individual gene fragments from
simple smaller
DNA sequences and a well defined DNA fragment called a 'template' that
contains pieces of
your final gene fragment. The smaller pieces of DNA are called 'primers'.
These primers flank
the DNA you want to generate from various templates to generate the final
product you desire
by PCR. These final products are called 'amplicons'. One process to 'stitch'
together various
amplicons is called Gibson Assembly. Many gene fragments disclosed in this
method were first
generated by Gibson Assembly of several amplicons generated by PCR. These
assembled gene
fragments were then allowed to be uptaken iteratively into a wild type
Saccharomyces
cerevisiae cell called JK9-3d. In some embodiments, CEN.PK is useful as a wild
type
Saccharomyces cerevisiae.
The process by which Saccharomyces cerevisiae uptakes foreign DNA and stably
utilize
the foreign DNA is called recombination. The final Saccharomyces cerevisiae
strains that took
up the foreign DNA and utilized the DNA are called recombinants. Saccharomyces
cerevisiae
that did not undergo recombination are the wild type. The process of selecting
recombinants in
a preferred media is termed prototrophy rescue. To separate recombinants from
wild type
prototrophy rescue was utilized.
Examples herein below provide a method to create recombinants that produce
various
levels of 0/0A by varying how many of those gene fragments are uptaken by JK9-
3d. In one
example, recombinants that produce 0/0A under the control of galactose are
disclosed.
Another example discloses, how the number of exogenous fragments taken up from
Saccharomyces cerevisiae correlates with 0/0A concentrations in the media. The
number of
genetic fragments recombinants contain can be determined by sequencing the DNA
of the
recombinant and by using the PCR method to quantify the number of amplicons
generated.
8
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Amp!icons are quantified by quantitative real time polymerase chain reaction
(qPCR). qPCR and
direct sequencing, and how those quantitative values of the genetic elements
relate to the
quantitative levels of 0 and 0A are exemplified and provided herein.
DESCRIPTION OF THE FIGURES
Figure 1 schematically illustrates recovery of olivetol and other compounds of
formula
IA in accordance with the present invention.
Figure 2 schematically illustrates the semisynthesis of cannabinoids (CBG) by
prenylation
of fermented olivetol.
Figure 3 schematically illustrates the semisynthesis of cannabinoids (CBC) by
prenylation
of fermented olivetol.
Figure 4A graphically illustrates the time course of total product titer
(olivetol and
olivetolic acid) in g/L.
Figure 4B graphically illustrates the time course of titer/time (g/L/day).
Figure 5A graphically illustrates the time course of total product titer
(divarin and
divarinic acid) in g/L.
Figure 5B graphically illustrates the time course of titer/time (g/L/day).
DETAILED DESCRIPTION
While the present invention is described herein with reference to aspects and
specific
embodiments thereof, those skilled in the art will recognize that various
changes may be made
and equivalents may be substituted without departing from the invention. The
present
invention is not limited to particular nucleic acids, expression vectors,
enzymes, host
microorganisms, or processes, as such may vary. The terminology used herein is
for purposes of
describing particular aspects and embodiments only, and is not to be construed
as limiting. In
addition, many modifications may be made to adapt a particular situation,
material,
composition of matter, process, process step or steps, in accordance with the
invention. All
such modifications are within the scope of the claims appended hereto. Headers
are used solely
9
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
for readers' convenience, and disclosure found under any header is understood
in the context
of and applicable to the entire disclosure.
Definitions
In this specification and in the claims that follow, reference will be made to
a number of
terms that shall be defined to have the following meanings.
As used in the specification and the appended claims, the singular forms "a",
"an", and
"the" include plural referents unless the context clearly dictates otherwise.
Thus, for example,
reference to an "expression vector" includes a single expression vector as
well as a plurality of
expression vectors, either the same (e.g., the same operon) or different;
reference to "cell"
includes a single cell as well as a plurality of cells; and the like.
As used herein, the term "comprising" is intended to mean that the compounds,
compositions and processes include the recited elements, but not exclude
others.
"Consisting essentially of" when used to define compounds, compositions and
processes,
shall mean excluding other elements of any essential significance to the
combination.
Thus, a composition consisting essentially of the elements as defined herein
would not
exclude trace contaminants, e.g., from the isolation and purification method.
"Consisting
of" shall mean excluding more than trace elements of other ingredients.
Embodiments
defined by each of these transition terms are within the scope of this
technology.
All numerical designations, e.g., pH, temperature, time, concentration, and
molecular weight, including ranges, are approximations which are varied (+) or
(-) by
increments of 1, 5, or 10%, e.g., by using the prefix, "about." It is to be
understood,
although not always explicitly stated that all numerical designations are
preceded by the
term "about." It also is to be understood, although not always explicitly
stated, that the
reagents described herein are merely exemplary and that equivalents of such
are known
in the art.
"Alkyl" refers to monovalent saturated aliphatic hydrocarbyl groups having
from 1 to 10
carbon atoms and preferably 1 to 6 carbon atoms. Higher carbon atom containing
alkyl groups
are also contemplated in certain embodiments, as the context will indicate.
This term includes,
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
by way of example, linear and branched hydrocarbyl groups such as methyl (CH3-
), ethyl
(CH3CH2), -n- propyl- (CH3CH2CH2-), isopropyl ((CH3)2CH), -n-butyl-
(CH3CH2CH2CH2-), isobutyl
((CH3)2CHCH2-), sec-butyl ((CH3)(CH3CH2)CH), -t-butyl- ((CH3)3C), -n- pentyl-
(CH3CH2CH2CH2CH2-
), and neopentyl ((CH3)3CCH2-).
"Alkenyl" refers to monovalent straight or branched hydrocarbyl groups having
from 2
to 10 carbon atoms and preferably 2 to 6 carbon atoms or preferably 2 to 4
carbon atoms and
having at least 1 and preferably from 1 to 2 sites of vinyl (>C=C<)
unsaturation. Higher carbon
atom containing alkenyl groups are also contemplated in certain embodiments,
as the context
will indicate. Such groups are exemplified, for example, by vinyl, ally!, and
but-3-en-1yI.
Included within this term are the cis and trans isomers or mixtures of these
isomers.
"Alkynyl" refers to straight or branched monovalent hydrocarbyl groups having
from
2 to 10 carbon atoms and preferably 2 to 6 carbon atoms or preferably 2 to 3
carbon atoms and
having at least 1 and preferably from 1 to 2 sites of acetylenic (-C=C-)
unsaturation. Higher
carbon atom containing alkynyl groups are also contemplated in certain
embodiments, as the
context will indicate. Examples of such alkynyl groups include acetylenyl (-
CCH), and
propargyl (-CH2C=CH).
"Substituted alkyl" refers to an alkyl group having from 1 to 5, preferably 1
to 3, or more
preferably 1 to 2 substituents selected from the group consisting of alkoxy,
substituted alkoxy,
acyl, acylamino, acyloxy, amino, substituted amino, aminocarbonyl,
aminothiocarbonyl,
aminocarbonylamino, aminothiocarbonylamino, aminocarbonyloxy, aminosulfonyl,
aminosulfonyloxy, aminosulfonylamino, amidino, aryl, substituted aryl,
aryloxy, substituted
aryloxy, arylthio, substituted arylthio, carboxyl, carboxyl ester, (carboxyl
ester)amino, (carboxyl
ester)oxy, cyano, cycloalkyl, substituted cycloalkyl, cycloalkyloxy,
substituted cycloalkyloxy,
cycloalkylthio, substituted cycloalkylthio, cycloalkenyl, substituted
cycloalkenyl,
cycloalkenyloxy, substituted cycloalkenyloxy, cycloalkenylthio, substituted
cycloalkenylthio,
guanidino, substituted guanidino, halo, hydroxy, heteroaryl, substituted
heteroaryl,
heteroaryloxy, substituted heteroaryloxy, heteroarylthio, substituted
heteroarylthio,
heterocyclic, substituted heterocyclic, heterocyclyloxy, substituted
heterocyclyloxy,
heterocyclylthio, substituted heterocyclylthio, nitro, SO3H, substituted
sulfonyl, substituted
11
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
sulfonyloxy, thioacyl, thiol, alkylthio, and substituted alkylthio, wherein
said substituents are as
defined herein.
"Heteroalkyl" refers to an alkyl group one or more carbons is replaced with -0-
, -S-, SO2,
a phosphorous (P) containing moiety, or -NRQ- moieties where IRQ is H or C1-C6
alkyl. Substituted
heteroalkyl refers to a heteroalkyl group having from Ito 5, preferably Ito 3,
or more
preferably 1 to 2 substituents selected from the group consisting of alkoxy,
substituted alkoxy,
acyl, acylamino, acyloxy, amino, substituted amino, aminocarbonyl,
aminothiocarbonyl,
aminocarbonylamino, aminothiocarbonylamino, aminocarbonyloxy, aminosulfonyl,
aminosulfonyloxy, aminosulfonylamino, amidino, aryl, substituted aryl,
aryloxy, substituted
aryloxy, arylthio, substituted arylthio, carboxyl, carboxyl ester, (carboxyl
ester)amino, (carboxyl
ester)oxy, cyano, cycloalkyl, substituted cycloalkyl, cycloalkyloxy,
substituted cycloalkyloxy,
cycloalkylthio, substituted cycloalkylthio, cycloalkenyl, substituted
cycloalkenyl,
cycloalkenyloxy, substituted cycloalkenyloxy, cycloalkenylthio, substituted
cycloalkenylthio,
guanidino, substituted guanidino, halo, hydroxy, heteroaryl, substituted
heteroaryl,
heteroaryloxy, substituted heteroaryloxy, heteroarylthio, substituted
heteroarylthio,
heterocyclic, substituted heterocyclic, heterocyclyloxy, substituted
heterocyclyloxy,
heterocyclylthio, substituted heterocyclylthio, nitro, SO3H, substituted
sulfonyl, substituted
sulfonyloxy, thioacyl, thiol, alkylthio, and substituted alkylthio, wherein
said substituents are as
defined herein.
"Substituted alkenyl" refers to alkenyl groups having from 1 to 3
substituents, and
preferably 1 to 2 substituents, selected from the group consisting of alkoxy,
substituted alkoxy,
acyl, acylamino, acyloxy, amino, substituted amino, aminocarbonyl,
aminothiocarbonyl,
aminocarbonylamino, aminothiocarbonylamino, aminocarbonyloxy, aminosulfonyl,
aminosulfonyloxy, aminosulfonylamino, amidino, aryl, substituted aryl,
aryloxy, substituted
aryloxy, arylthio, substituted arylthio, carboxyl, carboxyl ester, (carboxyl
ester)amino, (carboxyl
ester)oxy, cyano, cycloalkyl, substituted cycloalkyl, cycloalkyloxy,
substituted cycloalkyloxy,
cycloalkylthio, substituted cycloalkylthio, cycloalkenyl, substituted
cycloalkenyl,
cycloalkenyloxy, substituted cycloalkenyloxy, cycloalkenylthio, substituted
cycloalkenylthio,
guanidino, substituted guanidino, halo, hydroxyl, heteroaryl, substituted
heteroaryl,
12
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
heteroaryloxy, substituted heteroaryloxy, heteroarylthio, substituted
heteroarylthio,
heterocyclic, substituted heterocyclic, heterocyclyloxy, substituted
heterocyclyloxy,
heterocyclylthio, substituted heterocyclylthio, nitro, SO3H, substituted
sulfonyl, substituted
sulfonyloxy, thioacyl, thiol, alkylthio, and substituted alkylthio, wherein
said substituents are as
defined herein and with the proviso that any hydroxyl or thiol substitution is
not attached to a
vinyl (unsaturated) carbon atom.
"Heteroalkenyl" refers to an alkenyl group where one or more carbons is
replaced with
one or more -0-, -S-, SO2, P containing moiety, or -NRQ- moieties where R0 is
H or Ci-C6 alkyl.
Substituted heteroalkenyl refers to a heteroalkenyl group having from 1 to 5,
preferably 1 to 3,
or more preferably Ito 2 substituents selected from the group consisting of
alkoxy, substituted
alkoxy, acyl, acylamino, acyloxy, amino, substituted amino, aminocarbonyl,
aminothiocarbonyl,
aminocarbonylamino, aminothiocarbonylamino, aminocarbonyloxy, aminosulfonyl,
aminosulfonyloxy, aminosulfonylamino, amidino, aryl, substituted aryl,
aryloxy, substituted
aryloxy, arylthio, substituted arylthio, carboxyl, carboxyl ester, (carboxyl
ester)amino, (carboxyl
ester)oxy, cyano, cycloalkyl, substituted cycloalkyl, cycloalkyloxy,
substituted cycloalkyloxy,
cycloalkylthio, substituted cycloalkylthio, cycloalkenyl, substituted
cycloalkenyl,
cycloalkenyloxy, substituted cycloalkenyloxy, cycloalkenylthio, substituted
cycloalkenylthio,
guanidino, substituted guanidino, halo, hydroxy, heteroaryl, substituted
heteroaryl,
heteroaryloxy, substituted heteroaryloxy, heteroarylthio, substituted
heteroarylthio,
heterocyclic, substituted heterocyclic, heterocyclyloxy, substituted
heterocyclyloxy,
heterocyclylthio, substituted heterocyclylthio, nitro, SO3H, substituted
sulfonyl, substituted
sulfonyloxy, thioacyl, thiol, alkylthio, and substituted alkylthio, wherein
said substituents are as
defined herein.
"Substituted alkynyl" refers to alkynyl groups having from Ito 3 substituents,
and
preferably Ito 2 substituents, selected from the group consisting of alkoxy,
substituted alkoxy,
acyl, acylamino, acyloxy, amino, substituted amino, aminocarbonyl,
aminothiocarbonyl,
aminocarbonylamino, aminothiocarbonylamino, aminocarbonyloxy, aminosulfonyl,
aminosulfonyloxy, aminosulfonylamino, amidino, aryl, substituted aryl,
aryloxy, substituted
aryloxy, arylthio, substituted arylthio, carboxyl, carboxyl ester, (carboxyl
ester)amino, (carboxyl
13
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
ester)oxy, cyano, cycloalkyl, substituted cycloalkyl, cycloalkyloxy,
substituted cycloalkyloxy,
cycloalkylthio, substituted cycloalkylthio, cycloalkenyl, substituted
cycloalkenyl,
cycloalkenyloxy, substituted cycloalkenyloxy, cycloalkenylthio, substituted
cycloalkenylthio,
guanidino, substituted guanidino, halo, hydroxy, heteroaryl, substituted
heteroaryl,
heteroaryloxy, substituted heteroaryloxy, heteroarylthio, substituted
heteroarylthio,
heterocyclic, substituted heterocyclic, heterocyclyloxy, substituted
heterocyclyloxy,
heterocyclylthio, substituted heterocyclylthio, nitro, SO3H, substituted
sulfonyl, substituted
sulfonyloxy, thioacyl, thiol, alkylthio, and substituted alkylthio, wherein
said substituents are as
defined herein and with the proviso that any hydroxyl or thiol substitution is
not attached to an
acetylenic carbon atom.
"Heteroalkynyl" refers to an alkynyl group one or more carbons is replaced
with -0-, -S-
SO2, P containing moiety, or -NRQ- moieties where IRQ is H or C1-C6 alkyl.
Substituted
heteroalkynyl refers to a heteroalkynyl group having from 1 to 5, preferably 1
to 3, or more
preferably 1 to 2 substituents selected from the group consisting of alkoxy,
substituted alkoxy,
acyl, acylamino, acyloxy, amino, substituted amino, aminocarbonyl,
aminothiocarbonyl,
anninocarbonylannino, anninothiocarbonylannino, anninocarbonyloxy,
anninosulfonyl,
aminosulfonyloxy, aminosulfonylamino, amidino, aryl, substituted aryl,
aryloxy, substituted
aryloxy, arylthio, substituted arylthio, carboxyl, carboxyl ester, (carboxyl
ester)amino, (carboxyl
ester)oxy, cyano, cycloalkyl, substituted cycloalkyl, cycloalkyloxy,
substituted cycloalkyloxy,
cycloalkylthio, substituted cycloalkylthio, cycloalkenyl, substituted
cycloalkenyl,
cycloalkenyloxy, substituted cycloalkenyloxy, cycloalkenylthio, substituted
cycloalkenylthio,
guanidino, substituted guanidino, halo, hydroxy, heteroaryl, substituted
heteroaryl,
heteroaryloxy, substituted heteroaryloxy, heteroarylthio, substituted
heteroarylthio,
heterocyclic, substituted heterocyclic, heterocyclyloxy, substituted
heterocyclyloxy,
heterocyclylthio, substituted heterocyclylthio, nitro, SO3H, substituted
sulfonyl, substituted
sulfonyloxy, thioacyl, thiol, alkylthio, and substituted alkylthio, wherein
said substituents are as
defined herein.
"Alkylene" refers to divalent saturated aliphatic hydrocarbyl groups having
from 1 to 10
carbon atoms, preferably having from 1 to 6 and more preferably 1 to 3 carbon
atoms that are
14
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
either straight-chained- or branched. Higher carbon atom containing alkenyl
groups are also
contemplated in certain embodiments, as the context will indicate. This term
is exemplified by
groups such as methylene (-CH2-), ethylene (-CH2CH2-), n-propylene (-CH2CH2CH2-
), iso-
propylene (-CH2CH(CH3)- or -CH(CH3)CH2-), butylene (-CH2CH2CH2CH2-),
isobutylene (-
CH2CH(CH3)CH2-), sec-butylene (-CH2CH2(CH3)CH), and the like. Similarly,
"alkenylene" and
"alkynylene" refer to an alkylene moiety containing respective 1 or 2 carbon-
carbon double
bonds or a carbon-carbon triple bond.
"Substituted alkylene" refers to an alkylene group having from 1 to 3
hydrogens
replaced with substituents selected from the group consisting of alkyl,
substituted alkyl, alkoxy,
substituted alkoxy, acyl, acylamino, acyloxy, amino, substituted amino,
aminoacyl, aryl,
substituted aryl, aryloxy, substituted aryloxy, cyano, halogen, hydroxyl,
nitro, carboxyl, carboxyl
ester, cycloalkyl, substituted cycloalkyl, heteroaryl, substituted heteroaryl,
heterocyclic,
substituted heterocyclic, and oxo wherein said substituents are defined
herein. In some
embodiments, the alkylene has 1 to 2 of the aforementioned groups, or having
from 1-3 carbon
atoms replaced with ¨0-, -S-, SO2, P containing moiety or ¨NV- moieties where
Ir is H or Ci-C6
alkyl. It is to be noted that when the alkylene is substituted by an oxo
group, 2 hydrogens
attached to the same carbon of the alkylene group are replaced by "=0."
"Substituted
alkenylene" and "substituted alkynylene" refer to alkenylene and alkynylene
moieties
substituted with substituents as described for substituted alkylene.
"Alkynylene " refers to straight or branched divalent hydrocarbyl groups
having from 2
to 10 carbon atoms and preferably 2 to 6 carbon atoms or preferably 2 to 3
carbon atoms and
having at least 1 and preferably from 1 to 2 sites of acetylenic (-C=C-)
unsaturation. Higher
carbon atom containing alkynylene groups are also contemplated in certain
embodiments, as
the context will indicate. Examples of such alkynylene groups include -CC- and
-CH2CC-.
"Substituted alkynylene" refers to alkynylene groups having from 1 to 3
substituents,
and preferably 1 to 2 substituents, selected from the group consisting of
alkoxy, substituted
alkoxy, acyl, acylamino, acyloxy, amino, substituted amino, aminocarbonyl,
aminothiocarbonyl,
aminocarbonylamino, aminothiocarbonylamino, aminocarbonyloxy, aminosulfonyl,
aminosulfonyloxy, aminosulfonylamino, amidino, aryl, substituted aryl,
aryloxy, substituted
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
aryloxy, arylthio, substituted arylthio, carboxyl, carboxyl ester, (carboxyl
ester)amino, (carboxyl
ester)oxy, cyano, cycloalkyl, substituted cycloalkyl, cycloalkyloxy,
substituted cycloalkyloxy,
cycloalkylthio, substituted cycloalkylthio, cycloalkenyl, substituted
cycloalkenyl,
cycloalkenyloxy, substituted cycloalkenyloxy, cycloalkenylthio, substituted
cycloalkenylthio,
guanidino, substituted guanidino, halo, hydroxy, heteroaryl, substituted
heteroaryl,
heteroaryloxy, substituted heteroaryloxy, heteroarylthio, substituted
heteroarylthio,
heterocyclic, substituted heterocyclic, heterocyclyloxy, substituted
heterocyclyloxy,
heterocyclylthio, substituted heterocyclylthio, nitro, SO3H, substituted
sulfonyl, substituted
sulfonyloxy, thioacyl, thiol, alkylthio, and substituted alkylthio, wherein
said substituents are as
defined herein and with the proviso that any hydroxyl or thiol substitution is
not attached to an
acetylenic carbon atom.
"Heteroalkylene" refers to an alkylene group wherein one or more carbons is
replaced
with -0-, -S-, SO2, a P containing moiety, or -NRQ- moieties where RQ is H or
C1-C6 alkyl.
"Substituted heteroalkylene" refers to heteroalkynylene groups having from Ito
3
substituents, and preferably Ito 2 substituents, selected from the
substituents disclosed for
substituted alkylene.
"Heteroalkenylene" refers to an alkenylene group wherein one or more carbons
is
replaced with -0-, -S-, SO2, a P containing moiety, or -NRQ- moieties where RQ
is H or Ci-C6 alkyl.
"Substituted heteroalkenylene" refers to heteroalkynylene groups having from
Ito 3
substituents, and preferably 1 to 2 substituents, selected from the
substituents disclosed for
substituted alkenylene.
"Heteroalkynylene" refers to an alkynylene group wherein one or more carbons
is
replaced with -0-, -S-, SO2, a P containing moiety, or -NR - moieties where IR
is H or Ci-C6 alkyl.
"Substituted heteroalkynylene" refers to heteroalkynylene groups having from 1
to 3
substituents, and preferably Ito 2 substituents, selected from the
substituents disclosed for
substituted alkynylene.
"Alkoxy" refers to the group ¨0-alkyl wherein alkyl is defined herein. Alkoxy
includes, by
way of example, methoxy-, ethoxy, n-propoxy, isopropoxy, n-butoxy, t- butoxy, -
sec-butoxy,
and- n-pentoxy.
16
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
"Substituted alkoxy" refers to the group -0-(substituted alkyl) wherein
substituted alkyl
is defined herein.
"Acyl" refers to the groups H-C(0), -alkyl-C-(0)-, substituted alkyl-C(0)-,
alkenyl-C(0)-,
substituted alkenyl-C(0)-, alkynyl-C(0)-, substituted alkynyl-C(0)-,
cycloalkyl-C(0)-, substituted
cycloalkyl-C(0)-, cycloalkenyl-C(0)-, substituted cycloalkenyl-C(0)-, aryl-
C(0)-, substituted aryl-
C(0)-, heteroaryl-C(0)-, substituted heteroaryl-C(0)-, heterocyclic-C(0), and
substituted-
heterocyclic-C-(0)-, wherein alkyl, substituted alkyl, alkenyl, substituted
alkenyl, alkynyl,
substituted alkynyl, cycloalkyl, substituted cycloalkyl, cycloalkenyl,
substituted cycloalkenyl,
aryl, substituted aryl, heteroaryl, substituted heteroaryl, heterocyclic, and
substituted
heterocyclic are as defined herein. Acyl includes the "acetyl" group CH3C(0)-.
"Acylamino" refers to the groups -NR40C(0)alkyl, -NR40C(0)substituted alkyl, -
NR4 C(0)cycloalkyl, -NR4 C(0)substituted cycloalkyl, -NR4 C(0)cycloalkenyl, -
NR4 C(0)substituted cycloalkenyl, -NR40C(0)alkenyl, -NR4 C(0)substituted
alkenyl, -
NR40C(0)alkynyl, -NR40C(0)substituted alkynyl, -NR40C(0)aryl, -
NR40C(0)substituted aryl, -
NR4 C(0)heteroaryl, -NR4 C(0)substituted heteroaryl, -NR4 C(0)heterocyclic,
and -
NR40C(0)substituted heterocyclic wherein R4 is hydrogen or alkyl and wherein
alkyl,
substituted alkyl, alkenyl, substituted alkenyl, alkynyl, substituted alkynyl,
cycloalkyl,
substituted cycloalkyl, cycloalkenyl, substituted cycloalkenyl, aryl,
substituted aryl, heteroaryl,
substituted heteroaryl, heterocyclic, and substituted heterocyclic are as
defined herein.
"Acyloxy" refers to the groups alkyl-C-(0)0, substituted-alkyl-C-(0)0-,
alkenyl-C(0)O-,
substituted alkenyl-C(0)O-, alkynyl-C(0)O-, substituted alkynyl-C(0)O-, aryl-
C(0)0, substituted-
aryl-C-(0)0-, cycloalkyl-C(0)O-, substituted cycloalkyl-C(0)O-, cycloalkenyl-
C(0)O-, substituted
cycloalkenyl-C(0)O-, heteroaryl-C(0)O-, substituted heteroaryl-C(0)0, -
heterocyclic-C-(0)0, and
substituted-heterocyclic-C-(0)0- wherein alkyl, substituted alkyl, alkenyl,
substituted alkenyl,
alkynyl, substituted alkynyl, cycloalkyl, substituted cycloalkyl,
cycloalkenyl, substituted
cycloalkenyl, aryl, substituted aryl, heteroaryl, substituted heteroaryl,
heterocyclic, and
substituted heterocyclic are as defined herein.
"Amino" refers to the group -NH2.
17
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
"Substituted amino" refers to the group -NR41R42 where R41 and R4 are
independently
selected from the group consisting of hydrogen, alkyl, substituted alkyl,
alkenyl, substituted
alkenyl, alkynyl, substituted alkynyl, aryl, substituted aryl, cycloalkyl,
substituted cycloalkyl,
cycloalkenyl, substituted cycloalkenyl, heteroaryl, substituted heteroaryl,
heterocyclic,
substituted heterocyclic, -502-alkyl, -502-substituted alkyl, -502-alkenyl, -
502-substituted
alkenyl, -502-cycloalkyl, -502-substituted cylcoalkyl, -502-cycloalkenyl, -502-
substituted
cylcoalkenyl, -502-aryl, -502-substituted aryl, -502-heteroaryl, -502-
substituted heteroaryl, -502-
heterocyclic, and -502-substituted heterocyclic and wherein R41 and R42 are
optionally joined,
together with the nitrogen bound thereto to form a heterocyclic or substituted
heterocyclic
group, provided that R41 and R42 are both not hydrogen, and wherein alkyl,
substituted alkyl,
alkenyl, substituted alkenyl, alkynyl, substituted alkynyl, cycloalkyl,
substituted cycloalkyl,
cycloalkenyl, substituted cycloalkenyl, aryl, substituted aryl, heteroaryl,
substituted heteroaryl,
heterocyclic, and substituted heterocyclic are as defined herein. When R4' is
hydrogen and R42
is alkyl, the substituted amino group is sometimes referred to herein as
alkylamino. When R41
and R42 are alkyl, the substituted amino group is sometimes referred to herein
as dialkylamino.
When referring to a nnonosubstituted amino, it is meant that either R41 or R42
is hydrogen but
not both. When referring to a disubstituted amino, it is meant that neither
R41 nor R42 are
hydrogen.
"Aminocarbonyl" refers to the group -C(0)NR50R51 where R5 and R51 are
independently
selected from the group consisting of hydrogen, alkyl, substituted alkyl,
alkenyl, substituted
alkenyl, alkynyl, substituted alkynyl, aryl, substituted aryl, cycloalkyl,
substituted cycloalkyl,
cycloalkenyl, substituted cycloalkenyl, heteroaryl, substituted heteroaryl,
heterocyclic, and
substituted heterocyclic and where R5 and R51 are optionally joined together
with the nitrogen
bound thereto to form a heterocyclic or substituted heterocyclic group, and
wherein alkyl,
substituted alkyl, alkenyl, substituted alkenyl, alkynyl, substituted alkynyl,
cycloalkyl,
substituted cycloalkyl, cycloalkenyl, substituted cycloalkenyl, aryl,
substituted aryl, heteroaryl,
substituted heteroaryl, heterocyclic, and substituted heterocyclic are as
defined herein.
"Aminothiocarbonyl" refers to the group -C(S)NR50R51 where R5 and R51 are
independently selected from the group consisting of hydrogen, alkyl,
substituted alkyl, alkenyl,
18
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
substituted alkenyl, alkynyl, substituted alkynyl, aryl, substituted aryl,
cycloalkyl, substituted
cycloalkyl, cycloalkenyl, substituted cycloalkenyl, heteroaryl, substituted
heteroaryl,
heterocyclic, and substituted heterocyclic and where R5 and R51 are
optionally joined together
with the nitrogen bound thereto to form a heterocyclic or substituted
heterocyclic group, and
wherein alkyl, substituted alkyl, alkenyl, substituted alkenyl, alkynyl,
substituted alkynyl,
cycloalkyl, substituted cycloalkyl, cycloalkenyl, substituted cycloalkenyl,
aryl, substituted aryl,
heteroaryl, substituted heteroaryl, heterocyclic, and substituted heterocyclic
are as defined
herein.
"Aminocarbonylamino" refers to the group -NR40C(0)NR50R51 where R4 is
hydrogen or
alkyl and R5 and R51 are independently selected from the group consisting of
hydrogen, alkyl,
substituted alkyl, alkenyl, substituted alkenyl, alkynyl, substituted alkynyl,
aryl, substituted aryl,
cycloalkyl, substituted cycloalkyl, cycloalkenyl, substituted cycloalkenyl,
heteroaryl, substituted
heteroaryl, heterocyclic, and substituted heterocyclic, and where R5 and R51
are optionally
joined together with the nitrogen bound thereto to form a heterocyclic or
substituted
heterocyclic group, and wherein alkyl, substituted alkyl, alkenyl, substituted
alkenyl, alkynyl,
substituted alkynyl, cycloalkyl, substituted cycloalkyl, cycloalkenyl,
substituted cycloalkenyl,
aryl, substituted aryl, heteroaryl, substituted heteroaryl, heterocyclic, and
substituted
heterocyclic are as defined herein.
"Aminothiocarbonylamino" refers to the group -NR40C(S)NR50R51 where R4 is
hydrogen
or alkyl and R5 and R51 are independently selected from the group consisting
of hydrogen,
alkyl, substituted alkyl, alkenyl, substituted alkenyl, alkynyl, substituted
alkynyl, aryl,
substituted aryl, cycloalkyl, substituted cycloalkyl, cycloalkenyl,
substituted cycloalkenyl,
heteroaryl, substituted heteroaryl, heterocyclic, and substituted heterocyclic
and where R5 and
R51 are optionally joined together with the nitrogen bound thereto to form a
heterocyclic or
substituted heterocyclic group, and wherein alkyl, substituted alkyl, alkenyl,
substituted alkenyl,
alkynyl, substituted alkynyl, cycloalkyl, substituted cycloalkyl,
cycloalkenyl, substituted
cycloalkenyl, aryl, substituted aryl, heteroaryl, substituted heteroaryl,
heterocyclic, and
substituted heterocyclic are as defined herein.
19
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
"Aminocarbonyloxy" refers to the group -0-C(0)NR50R51 where Rs and Rs' are
independently selected from the group consisting of hydrogen, alkyl,
substituted alkyl, alkenyl,
substituted alkenyl, alkynyl, substituted alkynyl, aryl, substituted aryl,
cycloalkyl, substituted
cycloalkyl, cycloalkenyl, substituted cycloalkenyl, heteroaryl, substituted
heteroaryl,
heterocyclic, and substituted heterocyclic and where R50 and R51 are
optionally joined
together with the nitrogen bound thereto to form a heterocyclic or substituted
heterocyclic
group, and wherein alkyl, substituted alkyl, alkenyl, substituted alkenyl,
alkynyl, substituted
alkynyl, cycloalkyl, substituted cycloalkyl, cycloalkenyl, substituted
cycloalkenyl, aryl,
substituted aryl, heteroaryl, substituted heteroaryl, heterocyclic, and
substituted heterocyclic
are as defined herein.
"Aminosulfonyl" refers to the group -S02NR50R51 where Rs and Rs' are
independently
selected from the group consisting of hydrogen, alkyl, substituted alkyl,
alkenyl, substituted
alkenyl, alkynyl, substituted alkynyl, aryl, substituted aryl, cycloalkyl,
substituted cycloalkyl,
cycloalkenyl, substituted cycloalkenyl, heteroaryl, substituted heteroaryl,
heterocyclic, and
substituted heterocyclic and where Rs and R51 are optionally joined together
with the nitrogen
bound thereto to form a heterocyclic or substituted heterocyclic group, and
wherein alkyl,
substituted alkyl, alkenyl, substituted alkenyl, alkynyl, substituted alkynyl,
cycloalkyl,
substituted cycloalkyl, cycloalkenyl, substituted cycloalkenyl, aryl,
substituted aryl, heteroaryl,
substituted heteroaryl, heterocyclic, and substituted heterocyclic are as
defined herein.
"Aminosulfonyloxy" refers to the group -0-S02NFOR51 where Rs and Rs' are
independently selected from the group consisting of hydrogen, alkyl,
substituted alkyl, alkenyl,
substituted alkenyl, alkynyl, substituted alkynyl, aryl, substituted aryl,
cycloalkyl, substituted
cycloalkyl, cycloalkenyl, substituted cycloalkenyl, heteroaryl, substituted
heteroaryl,
heterocyclic, and substituted heterocyclic and where Rs and Rs' are
optionally joined together
with the nitrogen bound thereto to form a heterocyclic or substituted
heterocyclic group, and
wherein alkyl, substituted alkyl, alkenyl, substituted alkenyl, alkynyl,
substituted alkynyl,
cycloalkyl, substituted cycloalkyl, cycloalkenyl, substituted cycloalkenyl,
aryl, substituted aryl,
heteroaryl, substituted heteroaryl, heterocyclic, and substituted heterocyclic
are as defined
herein.
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
"Aminosulfonylamino" refers to the group -NR40S02NR50R51 where R4 is hydrogen
or
alkyl and R5 and R51 are independently selected from the group consisting of
hydrogen, alkyl,
substituted alkyl, alkenyl, substituted alkenyl, alkynyl, substituted alkynyl,
aryl, substituted aryl,
cycloalkyl, substituted cycloalkyl, cycloalkenyl, substituted cycloalkenyl,
heteroaryl, substituted
heteroaryl, heterocyclic, and substituted heterocyclic and where R5 and R"
are optionally
joined together with the nitrogen bound thereto to form a heterocyclic or
substituted
heterocyclic group, and wherein alkyl, substituted alkyl, alkenyl, substituted
alkenyl, alkynyl,
substituted alkynyl, cycloalkyl, substituted cycloalkyl, cycloalkenyl,
substituted cycloalkenyl,
aryl, substituted aryl, heteroaryl, substituted heteroaryl, heterocyclic, and
substituted
heterocyclic are as defined herein.
"Amidino" refers to the group -C(=NR52)NR50R51 where R50, R51, and R52 are
independently selected from the group consisting of hydrogen, alkyl,
substituted alkyl, alkenyl,
substituted alkenyl, alkynyl, substituted alkynyl, aryl, substituted aryl,
cycloalkyl, substituted
cycloalkyl, cycloalkenyl, substituted cycloalkenyl, heteroaryl, substituted
heteroaryl,
heterocyclic, and substituted heterocyclic and where R50 and R51 are
optionally joined
together with the nitrogen bound thereto to form a heterocyclic or substituted
heterocyclic
group, and wherein alkyl, substituted alkyl, alkenyl, substituted alkenyl,
alkynyl, substituted
alkynyl, cycloalkyl, substituted cycloalkyl, cycloalkenyl, substituted
cycloalkenyl, aryl,
substituted aryl, heteroaryl, substituted heteroaryl, heterocyclic, and
substituted heterocyclic
are as defined herein.
"Aryl" or "Ar" refers to an aromatic carbocyclic group of from 6 to 14 carbon
atoms
having a single ring (e.g., phenyl) or multiple condensed rings (e.g.,
naphthyl or anthryl) which
condensed rings may or may not be aromatic (e.g., 2-benzoxazolinone, 2H-1,4-
benzoxazin-
3(4H)-one-7-yl, and the like) provided that the point of attachment is at an
aromatic carbon
atom. Certain, preferred aryl groups include phenyl and naphthyl.
"Substituted aryl" refers to aryl groups which are substituted with 1 to 5,
preferably 1 to
3, or more preferably 1 to 2 substituents selected from the group consisting
of alkyl,
substituted alkyl, alkenyl, substituted alkenyl, alkynyl, substituted alkynyl,
alkoxy, substituted
alkoxy, acyl, acylamino, acyloxy, amino, substituted amino, aminocarbonyl,
aminothiocarbonyl,
21
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
aminocarbonylamino, aminothiocarbonylamino, aminocarbonyloxy, aminosulfonyl,
aminosulfonyloxy, aminosulfonylamino, amidino, aryl, substituted aryl,
aryloxy, substituted
aryloxy, arylthio, substituted arylthio, carboxyl, carboxyl ester, (carboxyl
ester)amino, (carboxyl
ester)oxy, cyano, cycloalkyl, substituted cycloalkyl, cycloalkyloxy,
substituted cycloalkyloxy,
cycloalkylthio, substituted cycloalkylthio, cycloalkenyl, substituted
cycloalkenyl,
cycloalkenyloxy, substituted cycloalkenyloxy, cycloalkenylthio, substituted
cycloalkenylthio,
guanidino, substituted guanidino, halo, hydroxy, heteroaryl, substituted
heteroaryl,
heteroaryloxy, substituted heteroaryloxy, heteroarylthio, substituted
heteroarylthio,
heterocyclic, substituted heterocyclic, heterocyclyloxy, substituted
heterocyclyloxy,
heterocyclylthio, substituted heterocyclylthio, nitro, SO3H, substituted
sulfonyl, substituted
sulfonyloxy, thioacyl, thiol, alkylthio, and substituted alkylthio, wherein
said substituents are as
defined herein.
"Arylene" refers to a divalent aromatic carbocyclic group of from 6 to 14
carbon atoms
having a single ring or multiple condensed rings. "Substituted arylene" refers
to an arylene
having from 1 to 5, preferably 1 to 3, or more preferably 1 to 2 substituents
as defined for aryl
groups.
"Heteroarylene" refers to a divalent aromatic group of from 1 to 10 carbon
atoms and 1
to 4 heteroatoms selected from the group consisting of oxygen, nitrogen and
sulfur within the
ring. "Substituted heteroarylene" refers to heteroarylene groups that are
substituted with from
1 to 5, preferably 1 to 3, or more preferably 1 to 2 substituents selected
from the group
consisting of the same group of substituents defined for substituted aryl.
Unless otherwise
noted, the context will clearly indicate, whether an aryl or heteroaryl moiety
is monovalent or
divalent.
"Aryloxy" refers to the group¨O-aryl-, where aryl is as defined herein, that
includes, by
way of example, phenoxy and naphthoxy.
"Substituted aryloxy" refers to the group -0-(substituted aryl) where
substituted aryl is
as defined herein.
"Arylthio" refers to the group¨S-aryl-, where aryl is as defined herein.
22
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
"Substituted arylthio" refers to the group -S-(substituted aryl), where
substituted aryl is
as defined herein.
"Carbonyl" refers to the divalent group -C(0)- which is equivalent to -C(=0)-.
"Carboxyl" or"carboxy" refers to -COOH or salts thereof.
"Carboxyl ester" or "carboxy ester" refers to the group -C(0)(0)-alkyl, -
C(0)(0)-
substituted alkyl, -C(0)0-alkenyl, -C(0)(0)-substituted alkenyl, -C(0)(0)-
alkynyl, - C(0)(0)-
substituted alkynyl, -C(0)(0)-aryl, -C(0)(0)-substituted-aryl, -C(0)(0)-
cycloalkyl, -C(0)(0)-
substituted cycloalkyl, -C(0)(0)-cycloalkenyl, -C(0)(0)-substituted
cycloalkenyl, -C(0)(0)-
heteroaryl, -C(0)(0)-substituted heteroaryl, -C(0)(0)-heterocyclic, and -
C(0)(0)- substituted
heterocyclic wherein alkyl, substituted alkyl, alkenyl, substituted alkenyl,
alkynyl, substituted
alkynyl, cycloalkyl, substituted cycloalkyl, cycloalkenyl, substituted
cycloalkenyl, aryl,
substituted aryl, heteroaryl, substituted heteroaryl, heterocyclic, and
substituted heterocyclic
are as defined herein.
"(Carboxyl ester)amino" refers to the group¨NR40C(0)(0)-alkyl, -NR4 C(0)(0)-
substituted alkyl, -NR4 C(0)0-alkenyl, -NR40C(0)(0)-substituted alkenyl, -
NR40C(0)(0)- alkynyl, -
NR4 C(0)(0)-substituted alkynyl, -NR4 C(0)(0)-aryl, -NR4 C(0)(0)- substituted-
aryl, -
NR4 C(0)(0)-cycloalkyl, -NR40C(0)(0)-substituted cycloalkyl, -NR40C(0)(0)-
cycloalkenyl, -
NR4 C(0)(0)-substituted cycloalkenyl, -NR4 C(0)(0)- heteroaryl, -NR4 C(0)(0)-
substituted
heteroaryl, -NR40C(0)(0)-heterocyclic, and -NR40C(0)(0)-substituted
heterocyclic wherein R4 is
alkyl or hydrogen, and wherein alkyl, substituted alkyl, alkenyl, substituted
alkenyl, alkynyl,
substituted alkynyl, cycloalkyl, substituted cycloalkyl, cycloalkenyl,
substituted cycloalkenyl,
aryl, substituted aryl, heteroaryl, substituted heteroaryl, heterocyclic, and
substituted
heterocyclic are as defined herein.
"(Carboxyl ester)oxy" refers to the group -0-C(0)0-alkyl, -0-C(0)0-substituted
alkyl, -0-
C(0)0-alkenyl, -0-C(0)0-substituted alkenyl, -0-C(0)0-alkynyl, -0-C(0)(0)-
substituted alkynyl,
-0-C(0)0-aryl, -0-C(0)0-substituted-aryl, -0-C(0)0-cycloalkyl, - 0-C(0)0-
substituted cycloalkyl,
-0-C(0)0-cycloalkenyl, -0-C(0)0-substituted cycloalkenyl, -0-C(0)0-heteroaryl,
-0C(0)-0-
substituted heteroaryl, -0C(0)-0- heterocyclic-, and -0C(0)-0-substituted
heterocyclic wherein
alkyl, substituted alkyl, alkenyl, substituted alkenyl, alkynyl, substituted
alkynyl, cycloalkyl,
23
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
substituted cycloalkyl, cycloalkenyl, substituted cycloalkenyl, aryl,
substituted aryl, heteroaryl,
substituted heteroaryl, heterocyclic, and substituted heterocyclic are as
defined herein.
"Cyano" refers to the group -CN.
"Cycloalkyl" refers to cyclic alkyl groups of from 3 to 10 carbon atoms having
single or
multiple cyclic rings including fused, bridged, and spiro ring systems, and
further includes
cycloalkenyl. The fused ring can be an aryl ring provided that the non aryl
part is joined to the
rest of the molecule. Examples of suitable cycloalkyl groups include, for
instance, adamantyl,
cyclopropyl, cyclobutyl, cyclopentyl, and cyclooctyl.
"Cycloalkenyl" refers to nonaromatic- cyclic alkyl groups of from 3 to 10
carbon atoms
having single or multiple cyclic rings and having at least one >C=C< ring
unsaturation and
preferably from 1 to 2 sites of >C=C< ring unsaturation.
"Substituted cycloalkyl" and "substituted cycloalkenyl" refers to a cycloalkyl
or
cycloalkenyl group having from 1 to 5 or preferably 1 to 3 substituents
selected from the group
consisting of oxo, thioxo, alkyl, substituted alkyl, alkenyl, substituted
alkenyl, alkynyl,
substituted alkynyl, alkoxy, substituted alkoxy, acyl, acylamino, acyloxy,
amino, substituted
amino, anninocarbonyl, anninothiocarbonyl, anninocarbonylannino,
anninothiocarbonylannino,
aminocarbonyloxy, aminosulfonyl, aminosulfonyloxy, aminosulfonylamino,
amidino, aryl,
substituted aryl, aryloxy, substituted aryloxy, arylthio, substituted
arylthio, carboxyl, carboxyl
ester, (carboxyl ester)amino, (carboxyl ester)oxy, cyano, cycloalkyl,
substituted cycloalkyl,
cycloalkyloxy, substituted cycloalkyloxy, cycloalkylthio, substituted
cycloalkylthio, cycloalkenyl,
substituted cycloalkenyl, cycloalkenyloxy, substituted cycloalkenyloxy,
cycloalkenylthio,
substituted cycloalkenylthio, guanidino, substituted guanidino, halo, hydroxy,
heteroaryl,
substituted heteroaryl, heteroaryloxy, substituted heteroaryloxy,
heteroarylthio, substituted
heteroarylthio, heterocyclic, substituted heterocyclic, heterocyclyloxy,
substituted
heterocyclyloxy, heterocyclylthio, substituted heterocyclylthio, nitro, SO3H,
substituted
sulfonyl, substituted sulfonyloxy, thioacyl, thiol, alkylthio, and substituted
alkylthio, wherein
said substituents are as defined herein.
"Cycloalkyloxy" refers to¨O-cycloalkyl-.
"Substituted cycloalkyloxy refers to -0-(substituted cycloalkyl).
24
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
"Cycloalkylthio" refers to ¨S-cycloalkyl-.
"Substituted cycloalkylthio" refers to -S-(substituted cycloalkyl).
"Cycloalkenyloxy" refers to¨O-cycloalkenyl-.
"Substituted cycloalkenyloxy" refers to -0-(substituted cycloalkenyl).
"Cycloalkenylthio" refers to¨S-cycloalkenyl-.
"Substituted cycloalkenylthio" refers to -S-(substituted cycloalkenyl).
"Guanidino" refers to the group -NHC(=NH)NH2.
Substituted guanidino" refers to -NR53C(=NR53)N(R53)2 where each R53 is
independently
selected from the group consisting of hydrogen, alkyl, substituted alkyl,
aryl, substituted aryl,
heteroaryl, substituted heteroaryl, cycloalkyl, substituted cycloalkyl,
heterocyclic, and
substituted heterocyclic and two R53 groups attached to a common guanidino
nitrogen atom
are optionally joined together with the nitrogen bound thereto to form a
heterocyclic or
substituted heterocyclic group, provided that at least one R53 is not
hydrogen, and wherein said
substituents are as defined herein.
"Halo" or "halogen" refers to fluoro, chloro, bromo and iodo.
"Hydroxy" or "hydroxyl" refers to the group -OH.
"Heteroaryl" refers to an aromatic group of from 1 to 10 carbon atoms and 1 to
4
heteroatoms selected from the group consisting of oxygen, nitrogen and sulfur
within the ring.
Such heteroaryl groups can have a single ring (e.g., pyridinyl or furyl) or
multiple condensed
rings (e.g., indolizinyl or benzothienyl) wherein the condensed rings may or
may not be
aromatic and/or contain a heteroatom provided that the point of attachment is
through an
atom of the aromatic heteroaryl group. In one embodiment, the nitrogen and/or
the sulfur ring
atom(s) of the heteroaryl group are optionally oxidized to provide for the N-
oxide (N-0),
sulfinyl, or sulfonyl moieties. Certain non-limiting examples include
pyridinyl, pyrrolyl, indolyl,
thiophenyl, oxazolyl, thizolyl, and- furanyl.
"Substituted heteroaryl" refers to heteroaryl groups that are substituted with
from Ito
5, preferably Ito 3, or more preferably 1 to 2 substituents selected from the
group consisting
of the same group of substituents defined for substituted aryl.
"Heteroaryloxy" refers to -0-heteroaryl.
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
"Substituted heteroaryloxy" refers to the group -0-(substituted heteroaryl).
"Heteroarylthio" refers to the group¨S-heteroaryl-.
"Substituted heteroarylthio" refers to the group -S-(substituted heteroaryl).
"Heterocycle" or "heterocyclic" or "heterocycloalkyl" or "heterocycly1" refers
to a
saturated or partially saturated, but not aromatic, group having from 1 to 10
ring carbon atoms
and from 1 to 4 ring heteroatoms selected from the group consisting of
nitrogen, sulfur, or
oxygen. Heterocycle encompasses single ring or multiple condensed rings,
including fused
bridged and Spiro ring systems. In fused ring systems, one or more the rings
can be cycloalkyl,
aryl, or heteroaryl provided that the point of attachment is through a
nonaromatic ring. In one
embodiment, the nitrogen and/or sulfur atom(s) of the heterocyclic group are
optionally
oxidized to provide for the¨N-oxide, sulfinyl-, or sulfonyl moieties.
"Substituted heterocyclic" or "substituted heterocycloalkyl" or "substituted
heterocycly1" refers to heterocyclyl groups that are substituted with from 1
to 5 or preferably 1
to 3 of the same substituents as defined for substituted cycloalkyl.
"Heterocyclyloxy" refers to the group -0-heterocycyl.
"Substituted heterocyclyloxy" refers to the group -0-(substituted
heterocycyl).
"Heterocyclylthio" refers to the group -S-heterocycyl.
"Substituted heterocyclylthio" refers to the group -S-(substituted
heterocycyl).
Examples of heterocycle and heteroaryls include, but are not limited to,
azetidine,
pyrrole, furan, thiophene, imidazole, pyrazole, pyridine, pyrazine,
pyrimidine, pyridazine,
indolizine, isoindole, indole, dihydroindole, indazole, purine, quinolizine,
isoquinoline,
quinoline, phthalazine, naphthylpyridine, quinoxaline, quinazoline, cinnoline,
pteridine,
carbazole, carboline, phenanthridine, acridine, phenanthroline, isothiazole,
phenazine,
isoxazole, phenoxazine, phenothiazine, imidazolidine, imidazoline, piperidine,
piperazine,
indoline, phthalimide, 1,2,3,4-tetrahydroisoquinoline, 4,5,6,7-tetrahydrobenzo-
[b]thiophene,
thiazole, thiazolidine, thiophene, benzo[b]thiophene, morpholinyl,
thiomorpholinyl (also
referred to as thiamorpholinyl), 1,1-dioxothiomorpholinyl, piperidinyl,
pyrrolidine, and
tetrahydrofuranyl.
"Nitro" refers to the group -NO2.
26
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
"Oxo" refers to the atom (=0).
Phenylene refers to a divalent aryl ring, where the ring contains 6 carbon
atoms.
Substituted phenylene refers to phenylenes which are substituted with 1 to 4,
preferably 1 to 3, or more preferably 1 to 2 substituents selected from the
group consisting of
alkyl, substituted alkyl, alkenyl, substituted alkenyl, alkynyl, substituted
alkynyl, alkoxy,
substituted alkoxy, acyl, acylamino, acyloxy, amino, substituted amino,
aminocarbonyl,
aminothiocarbonyl, aminocarbonylamino, aminothiocarbonylamino,
aminocarbonyloxy,
aminosulfonyl, aminosulfonyloxy, aminosulfonylamino, amidino, aryl,
substituted aryl, aryloxy,
substituted aryloxy, arylthio, substituted arylthio, carboxyl, carboxyl ester,
(carboxyl
ester)amino, (carboxyl ester)oxy, cyano, cycloalkyl, substituted cycloalkyl,
cycloalkyloxy,
substituted cycloalkyloxy, cycloalkylthio, substituted cycloalkylthio,
cycloalkenyl, substituted
cycloalkenyl, cycloalkenyloxy, substituted cycloalkenyloxy, cycloalkenylthio,
substituted
cycloalkenylthio, guanidino, substituted guanidino, halo, hydroxy, heteroaryl,
substituted
heteroaryl, heteroaryloxy, substituted heteroaryloxy, heteroarylthio,
substituted
heteroarylthio, heterocyclic, substituted heterocyclic, heterocyclyloxy,
substituted
heterocyclyloxy, heterocyclylthio, substituted heterocyclylthio, nitro, 503H,
substituted
sulfonyl, substituted sulfonyloxy, thioacyl, thiol, alkylthio, and substituted
alkylthio, wherein
said substituents are as defined herein.
"Spirocycloalkyl" and "spiro ring systems" refers to divalent cyclic groups
from 3 to 10
carbon atoms having a cycloalkyl or heterocycloalkyl ring with a Spiro union
(the union formed
by a single atom which is the only common member of the rings).
"Sulfonyl" refers to the divalent group -S(0)2-.
"Substituted sulfonyl" refers to the group -502-alkyl-, -SO2- substituted-
alkyl, -502-
alkenyl, -502-substituted alkenyl, -502-cycloalkyl, -502-substituted
cylcoalkyl, -502-cycloalkenyl,
-502-substituted cylcoalkenyl, -502-aryl, -502-substituted aryl, -502-
heteroaryl, -502-substituted
heteroaryl, -502-heterocyclic, -502-substituted-heterocyclic, wherein alkyl,
substituted alkyl,
alkenyl, substituted alkenyl, alkynyl, substituted alkynyl, cycloalkyl,
substituted cycloalkyl,
cycloalkenyl, substituted cycloalkenyl, aryl, substituted aryl, heteroaryl,
substituted heteroaryl,
27
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
heterocyclic, and substituted heterocyclic are as defined herein. Substituted
sulfonyl includes
groups such as methyl-S02-, phenyl-S02-, and 4-methylphenyl-S02-.
"Substituted sulfonyloxy" refers to the group -0S02-alkyl, -0S02-substituted-
alkyl, -
0S02-alkenyl, -0502-substituted alkenyl, -0S02-cycloalkyl, -0S02-substituted
cylcoalkyl, -0S02-
cycloalkenyl, -0S02-substituted cylcoalkeny1,-0S02-aryl, -0S02-substituted
aryl, -0S02-
heteroaryl, -0S02-substituted heteroaryl, -0S02-heterocyclic, -0S02-
substituted heterocyclic,
wherein alkyl, substituted alkyl, alkenyl, substituted alkenyl, alkynyl,
substituted alkynyl,
cycloalkyl, substituted cycloalkyl, cycloalkenyl, substituted cycloalkenyl,
aryl, substituted aryl,
heteroaryl, substituted heteroaryl, heterocyclic, and substituted heterocyclic
are as defined
herein.
"Thioacyl" refers to the groups H-C(S)-, alkyl-C(S)-, substituted alkyl-C(S), -
alkenyl-C-(S),
substituted¨alkenyl-C-(S)-, alkynyl-C(S)-, substituted alkynyl-C(S)-,
cycloalkyl-C(S)-, substituted
cycloalkyl-C(S)-, cycloalkenyl-C(S)-, substituted cycloalkenyl-C(S)-, aryl-
C(S)-, substituted aryl-
C(S)-, heteroaryl-C(S)-, substituted heteroaryl-C(S)-, heterocyclic-C(S), and
substituted¨
heterocyclic-C-(S)-, wherein alkyl, substituted alkyl, alkenyl, substituted
alkenyl, alkynyl,
substituted alkynyl, cycloalkyl, substituted cycloalkyl, cycloalkenyl,
substituted cycloalkenyl,
aryl, substituted aryl, heteroaryl, substituted heteroaryl, heterocyclic, and
substitutedheterocyclic are as defined herein.
"Thiol" refers to the group -SH.
"Thiocarbonyl" refers to the divalent group -C(S)- which is equivalent to -
C(=S)-.
"Thioxo" refers to the atom (=S).
"Alkylthio" refers to the group¨S-alkyl- wherein alkyl is as defined herein.
"Substituted alkylthio" refers to the group -S-(substituted alkyl) wherein
substituted
alkyl is as defined herein.
"Optionally substituted" refers to a group selected from that group and a
substituted
form of that group. Substituents are such as those defined hereinabove. E.g.,
and without
limitation, substituents can be selected from monovalent and divalent groups,
such as, Ci-Cio or
C1-C6 alkyl, substituted CI-CI or Ci-C6 alkyl, C2-C6 alkenyl, C2-C6 alkynyl,
C6-Clo aryl, C3-C8
cycloalkyl, C2-Cio heterocyclyl,
heteroaryl, substituted C2-C6 alkenyl, substituted C2-C6
28
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
alkynyl, substituted C6-C10 aryl, substituted C3-Cs cycloalkyl, substituted C2-
C10 heterocyclyl,
substituted Ci-Clo heteroaryl, halo, nitro, cyano, oxo (=0), -CO2H or a Ci-C6
alkyl ester thereof.
Unless indicated otherwise, the nomenclature of substituents that are not
explicitly
defined herein are arrived at by naming the terminal portion of the
functionality followed by
the adjacent functionality toward the point of attachment. For example, the
substituent
"alkoxycarbonylalkyl" refers to the group (alkoxy)-C(0)-(alkyl)
It is understood that in all substituted groups defined above, polymers
arrived at by
defining substituents with further substituents to themselves (e.g.,
substituted aryl having a
substituted aryl group as a substituent which is itself substituted with a
substituted aryl group,
etc.) are not intended for inclusion herein. In such cases, the maximum number
of such
substituents is three. That is to say that each of the above definitions is
constrained by a
limitation that, for example, substituted aryl groups are limited
to¨substituted aryl-
(substituted aryl)-substituted aryl.
It is understood that the above definitions are not intended to include
impermissible
substitution patterns (e.g., methyl substituted with 5 fluoro groups). Such
impermissible
substitution patterns are well known to the skilled artisan.
A "salt" is derived from a variety of organic and inorganic counter ions well
known in the
art and include, when the compound has an acidic functionality, by way of
example only,
sodium, potassium, calcium, magnesium, ammonium, and tetraalkylammonium; and
when the
molecule has a basic functionality, salts of organic or inorganic acids, such
as hydrochloride,
hydrobromide, tartrate, mesylate, acetate, maleate, and oxalate. Salts include
acid addition
salts formed with inorganic acids or organic acids. Inorganic acids suitable
for forming acid
addition salts include, by way of example and not limitation, hydrohalide
acids (e.g.,
hydrochloric acid, hydrobromic acid, hydroiodic acid, etc.), sulfuric acid,
nitric acid, phosphoric
acid, and the like.
Organic acids suitable for forming acid addition salts include, by way of
example and not
limitation, acetic acid, trifluoroacetic acid, propionic acid, hexanoic acid,
cyclopentanepropionic
acid, glycolic acid, oxalic acid, pyruvic acid, lactic acid, malonic acid,
succinic acid, malic acid,
maleic acid, fumaric acid, tartaric acid, citric acid, palmitic acid, benzoic
acid, 3-(4-
29
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
hydroxybenzoyl) benzoic acid, cinnamic acid, mandelic acid, alkylsulfonic
acids (e.g.,
methanesulfonic acid, ethanesulfonic acid, 1,2- ethane-disulfonic acid, 2-
hydroxyethanesulfonic
acid, etc.), arylsulfonic acids (e.g., benzenesulfonic acid, 4-
chlorobenzenesulfonic acid, 2-
naphthalenesulfonic acid, 4- toluenesulfonic acid, camphorsulfonic acid,
etc.), glutamic acid,
hydroxynaphthoic- acid, salicylic acid, stearic acid, muconic acid, and the
like.
Salts also include salts formed when an acidic proton present in the parent
compound is
either replaced by a metal ion (e.g., an alkali metal ion, an alkaline earth
metal ion, or an
aluminum ion) or by an ammonium ion (e.g., an ammonium ion derived from an
organic base,
such as, ethanolamine, diethanolamine, triethanolamine, morpholine,
piperidine,
dimethylamine, diethylamine, triethylamine, and ammonia).
Amino acids in a protein coding sequence are identified herein by the
following
abbreviations and symbols. Specific amino acids are identified by a three-
letter abbreviation, as
follows: Ala is alanine, Arg is arginine, Asn is asparagine, Asp is aspartic
acid, Cys is cysteine, Gln
is glutamine, Glu is glutamic acid, Gly is glycine, His is histidine, Leu is
leucine, Ile is isoleucine,
Lys is lysine, Met is methionine, Phe is phenylalanine, Pro is proline, Ser is
serine, Thr is
threonine, Trp is tryptophan, Tyr is tyrosine, and Val is valine, or by a one-
letter abbreviation, as
follows: A is alanine, R is arginine, N is asparagine, D is aspartic acid, C
is cysteine, Q is
glutamine, E is glutamic acid, G is glycine, H is histidine, L is leucine, I
is isoleucine, K is lysine, 0
is pyrrolysine, M is methionine, F is phenylalanine, P is proline, S is
serine, T is threonine, W is
tryptophan, Y is tyrosine, and V is valine. A dash (-) in a consensus sequence
indicates that there
is no amino acid at the specified position. A plus (+) in a consensus sequence
indicates any
amino acid may be present at the specified position. Thus, a plus in a
consensus sequence
herein indicates a position at which the amino acid is generally non-
conserved; a homologous
enzyme sequence, when aligned with the consensus sequence, can have any amino
acid at the
indicated "+" position. Specific amino acids in a protein coding sequence are
identified by their
respective one-letter abbreviation followed by the amino acid position in the
protein coding
sequence where 1 corresponds to the amino acid (typically methionine) at the N-
terminus of
the protein. For example, G204 in C. sativa wild type OLS refers to the
glycine at position 204
from the OLS N-terminal methionine (i.e., M1). Amino acid substitutions (i.e.,
point mutations)
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
are indicated by identifying the mutated (i.e., progeny) amino acid after the
one-letter code and
number in the parental protein coding sequence; for example, G204A in C.
sativa OLS refers to
substitution of glycine by alanine at position 204 in the OLS protein coding
sequence. The
mutation may also be identified in parentheticals, for example OLS (G204A).
Multiple point
mutations in the protein coding sequence are separated by a backslash (/); for
example, OLS
G204A/Q205N indicates that mutations G204A and Q205N are both present in the
OLS protein
coding sequence. The number of mutations introduced into some examples has
been
annotated by a dash followed by the number of mutations, preceding the
parenthetical
identification of the mutation (e.g., B1Q2B6-1 (G204A)). The Uniprot IDs with
and without the
dash and number are used interchangeably herein (i.e., B1Q2B6-1 (G204A) =
B1Q2B6 (G204A)).
As used herein, the term "express", when used in connection with a nucleic
acid
encoding an enzyme or an enzyme itself in a cell, means that the enzyme, which
may be an
endogenous or exogenous (heterologous) enzyme, is produced in the cell. The
term
"overexpress", in these contexts, means that the enzyme is produced at a
higher level, i.e.,
enzyme levels are increased, as compared to the wild type, in the case of an
endogenous
enzyme. Those skilled in the art appreciate that overexpression of an enzyme
can be achieved
by increasing the strength or changing the type of the promoter used to drive
expression of a
coding sequence, increasing the strength of the ribosome binding site or Kozak
sequence,
increasing the stability of the mRNA transcript, altering the codon usage,
increasing the stability
of the enzyme, and the like.
The term "expression vector" or "vector" refer to a nucleic acid and/or a
composition
comprising a nucleic acid that can be introduced into a host cell, e.g., by
transduction,
transformation, or infection, such that the cell then produces ("expresses")
nucleic acids and/or
proteins other than those native to the cell, or in a manner not native to the
cell, that are
contained in or encoded by the nucleic acid so introduced. Thus, an
"expression vector"
contains nucleic acids (ordinarily DNA) to be expressed by the host cell.
Optionally, the
expression vector can be contained in materials to aid in achieving entry of
the nucleic acid into
the host cell, such as the materials associated with a virus, liposome,
protein coating, or the
like. Expression vectors suitable for use in various aspects and embodiments
include those into
31
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
which a nucleic acid sequence can be, or has been, inserted, along with any
preferred or
required operational elements. Thus, an expression vector can be transferred
into a host cell
and, typically, replicated therein (although, one can also employ, in some
embodiments, non-
replicable vectors that provide for "transient" expression). In some
embodiments, an
expression vector that integrates into chromosomal, mitochondria!, or plastid
DNA is employed.
In other embodiments, an expression vector that replicates extrachromosomally
is employed.
Typical expression vectors include plasmids, and expression vectors typically
contain the
operational elements required for transcription of a nucleic acid in the
vector. Such plasmids, as
well as other expression vectors, are described herein or are well known to
those of ordinary
skill in the art.
The terms "ferment", "fermentative", and "fermentation" are used herein to
describe
culturing host cells and microbes under conditions to produce useful
chemicals, including but
not limited to conditions under which microbial growth, be it aerobic or
anaerobic, occurs.
The term "heterologous" as used herein refers to a material that is non-native
to a cell.
For example, a nucleic acid is heterologous to a cell, and so is a
"heterologous nucleic acid" with
respect to that cell, if at least one of the following is true: (a) the
nucleic acid is not naturally
found in that cell (that is, it is an "exogenous" nucleic acid); (b) the
nucleic acid is naturally
found in a given host cell (that is, "endogenous to"), but the nucleic acid or
the RNA or protein
resulting from transcription and translation of this nucleic acid is produced
or present in the
host cell in an unnatural (e.g., greater or lesser than naturally present)
amount; (c) the nucleic
acid comprises a nucleotide sequence that encodes a protein endogenous to a
host cell but
differs in sequence from the endogenous nucleotide sequence that encodes that
same protein
(having the same or substantially the same amino acid sequence), typically
resulting in the
protein being produced in a greater amount in the cell, or in the case of an
enzyme, producing a
mutant version possessing altered (e.g., higher or lower or different)
activity; and/or (d) the
nucleic acid comprises two or more nucleotide sequences that are not found in
the same
relationship to each other in the cell. As another example, a protein is
heterologous to a host
cell if it is produced by translation of RNA or the corresponding RNA is
produced by
transcription of a heterologous nucleic acid; a protein is also heterologous
to a host cell if it is a
32
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
mutated version of an endogenous protein, and the mutation was introduced by
genetic
engineering.
The terms "host cell" and "host microorganism" are used interchangeably herein
to
refer to a living cell that can perform one or more steps of the cannabinoid
pathway, e.g. and
without limitation, converting malonyl-CoA and hexanoyl-CoA (or another acyl-
CoA) to olivetol
and olivetolic acid. A host cell can be (or is) transformed via insertion of
an expression vector. A
host microorganism or cell as described herein may be a prokaryotic cell
(e.g., a microorganism
of the kingdom Eubacteria) or a eukaryotic cell. As will be appreciated by one
of skill in the art,
a prokaryotic cell lacks a membrane-bound nucleus, while a eukaryotic cell has
a membrane-
bound nucleus. In certain instances, a host cell is part of a multi-cellular
organism.
Polyketide synthases (PKSs) are a family of multi-domain enzymes or enzyme
complexes
that produce polyketides, a large class of secondary metabolites, in bacteria,
fungi, plants, and
a few animal lineages. The terms "polyketide synthase", "PKS", "olivetol
synthase" ("OLS" or
"OS"), "tetraketide synthase", TKS, and olivetolic synthase as described
herein or elsewhere
typically refers to any enzyme capable of converting three molecules of
malonyl-CoA and one
molecule of hexanoyl-CoA or another acyl-CoA to olivetol or an olivetol
analog. A wild type
example of an OLS is the native C. sativa OLS enzyme (UniProt ID: B1Q2B6; SEQ
ID NO: 1).
Sequence ID 1: OS
MN HLRAEGPASVLAIGTAN PENILIQDEFPDYYFRVTKSEHMTQLKEKFRKICDKSMIRKRNCFLN EEHLKQN
PRLVEHEMCITLDARQDMINVEVPKLGKDACAKAIKEWGQPKSKITHLIFTSASTTDMPGADYHCAKLIGLSP
WKRVMMYQLGCYGGGTVLRIAKDIAENNI<GARVLAVCCDIMACLFRGPSDSDLELLVGQA1FGDGAAAVI
VGAEPDESVGERPIFELVSTGQTILPNSEGTIGGHIREAGLIFDLHKDVPMLISNNIEKCLIEAFTPIGISDWNSIF
WITHPGGKAILDKVEEKLDLKKEKR/DSRHVLSEHGNMSSSTVLFVMDELRKRSLEEGKS
_______________________ I I GDGFEWGVLF
GFGPGLTVERVVVRSVPIKY
Olivetolic acid cyclase ("OAC", EC: 4.4.1.26) is a polyketide cyclase derived
from C. sativa
which functions in concert with an OLS enzyme or a tetraketide synthase
("TKS") to form OLA.
See, e.g.:
Sequence D 2.4: OAC
33
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
MAVKH Li VLKFKDE1TEAQKEEFFKTYVN LVN 1 I PAM KDVYWG
KDVIQKKEEGYTHIVEVTFESVETIODYI1H P
AHVGFGDVYRSFWEKLLIFDYTPRK
The terms "cannabinoid pathway", "cannabinoid production", "cannabinoid
compound
production", "cannabinoid synthesis", "THC synthesis", and the like, refer
generally to a
biosynthetic pathway that facilitates the synthesis and production of
olivetol, olivetolic acid,
and olivetolic acid-derived compounds. This biosynthetic pathway utilizes a
variety of enzymes,
catalysts, and intermediate compounds. For example, cannabigerolic acid
synthase (EC:
2.5.1.102) is used to convert OA to cannabigerolic acid, which is a key
intermediate acted upon
by a variety of enzymes during THC synthesis. Cannabidiolic acid synthase (EC:
1.21.3.7) is used
to convert cannabigerolic acid into cannabidiolic acid. Tetrahydrocannabinolic
acid synthase
(EC: 1.21.3.8) is used to convert cannabigerolic acid into A9-
tetrahydrocannabinolic acid. A
cannabichronnenic acid synthase is used to convert cannabigerolic acid into
cannabichronnenic
acid (CAS# 20408-52-0). These three olivetolic acid-derived compounds (i.e.,
cannabidiolic acid,
A9-tetrahydrocannabinolic acid, and cannabichromenic acid) are themselves
converted to even
more diverse cannabinoids via a combination of oxidation, decarboxylation, and
isomerization
reactions, which can be catalyzed using either biological or synthetic
catalysts, or can also occur
spontaneously following heating and/or application of UV light. For example,
cannabidiol
results from cannabidiolic acid decarboxylation, 1x9-tetrahydrocannabinol
results from A9-
tetrahydrocannabinolic acid decarboxylation, and subsequent isomerization of
A9-
tetrahydrocannabinol results in A6-tetrahydrocannabinol.
As used herein, "recombinant" refers to the alteration of genetic material by
human
intervention. Typically, recombinant refers to the manipulation of DNA or RNA
in a cell or virus
or expression vector by molecular biology (recombinant DNA technology)
methods, including
cloning and recombination. Recombinant can also refer to manipulation of DNA
or RNA in a cell
or virus by random or directed mutagenesis. A "recombinant" cell or nucleic
acid can typically
be described with reference to how it differs from a naturally occurring
counterpart (the "wild
type"). In addition, any reference to a cell or nucleic acid that has been
"engineered" or
"modified" and variations of those terms, is intended to refer to a
recombinant cell or nucleic
acid.
34
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
The terms "transduce", "transform", "transfect", and variations thereof as
used herein
refers to the introduction of one or more nucleic acids into a cell. For
practical purposes, the
nucleic acid must be stably maintained or replicated by the cell for a
sufficient period of time to
enable the function(s) or product(s) it encodes to be expressed for the cell
to be referred to as
"transduced", "transformed", or "transfected". As will be appreciated by those
of skill in the
art, stable maintenance or replication of a nucleic acid may take place either
by incorporation
of the sequence of nucleic acids into the cellular chromosomal DNA, e.g., the
genome, as occurs
by chromosomal integration, or by replication extrachromosomally, as occurs
with a freely-
replicating plasmid. A virus can be stably maintained or replicated when it is
"infective": when it
transduces a host microorganism, replicates, and (without the benefit of any
complementary
virus or vector) spreads progeny expression vectors, e.g., viruses, of the
same type as the
original transducing expression vector to other microorganisms, wherein the
progeny
expression vectors possess the same ability to reproduce.
Descriptive Embodiments
In one aspect, provided herein is a process comprising:
contacting an aqueous phase comprising glucose and hexanoic acid or a salt
thereof and
an organic phase immiscible with the aqueous phase
with a heterologous microorganism comprising a Cannabis sativa olivetol
synthase
(which is a tetraketide synthase, csOLS), Cannabis sativa olivetolic acid
cyclase (csOAC), and a
Cannabis sativa acyl activating enzyme (csAAE)
to provide olivetol and olivetolic acid or a salt thereof,
wherein the olivetol and olivetolic acid is provided in a combined amount of
at least
about 3 g/liter over about 4 to about 7 days.
In accordance with certain embodiments of this process, other microorganisms,
such as
those utilized herein are useful.
In one aspect, provided herein is a process comprising:
contacting an aqueous phase comprising glucose and butyric acid or a salt
thereof and
an organic phase immiscible with the aqueous phase
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
with a heterologous microorganism comprising a Cannabis sativa olivetol
synthase
(which is a tetraketide synthase, csOLS), Cannabis sativa olivetolic acid
cyclase (csOAC), and a
Cannabis sativa acyl activating enzyme (csAAE)
to provide divarin and/or divarinic acid or a salt thereof.
Other microorganisms, such as those utilize herein, utilized herein are useful
in
accordance with certain embodiments of this process.
In one embodiment, the divarin and/or divarinic acid is provided in a combined
amount
of at least about 0.25 ¨ about 2 g/liter, or about 0.5 ¨ about 1g/liter over
about 4 to about 7
days.
In one embodiment, the fermenting is performed in the absence of galactose. In
another embodiment, the aqueous phase comprises galactose.
In one embodiment, the fermenting is performed in the absence of galactose. In
another embodiment, the aqueous phase comprises galactose.
Organic Phase Immiscible With Aqueous Phase
In one embodiment, the organic phase immiscible with aqueous phase, or simply
the
organic phase, comprises an alkane, an alcohol with carbon number greater than
4, an ester
(such as isopropyl myristate), a triglyceride (including commercially
available vegetable oils
such as sunflower oil, soybean oil, or olive oil), a diester (such as dialkyl
malonate), a ketone, or
a glyme. Other organic solvents immiscible with water or the aqueous phase
employed can be
utilized. In another embodiment, the organic phase comprises isopropyl
myristate. Suitable
solvents include without limitation, other esters, aromatic solvents, and the
likes. In one
embodiment, the organic phase comprises an aromatic solvent. Non limiting
examples of
aromatic hydrocarbon solvents include benzene, toluene, other alkylated
benzenes, anisole and
the likes, and mixtures thereof. In one embodiment, the organic phase
comprises toluene.
In-situ liquid-liquid extraction (biphasic fermentation) is a strategy that
can be employed
in accordance with the present invention for physical separation of product
from
microorganisms via partitioning into the water immiscible organic liquid phase
from an aqueous
culture phase. The organic liquid phase or organic phase is present as either
an overlay if its
density is less than that of the aqueous phase, or an underlay if its density
is greater than that
36
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
of the aqueous phase. Certain properties of the overlay or underlay are
considered for
production of olivetolic acid/olivetol and other resorcinols such as of
formulas IA and IB: (1)
non-toxic or low toxicity for growth of the host strain, and/or (2) a
favorable partition
coefficient of the product in the organic phase vs. the aqueous phase, and/or
(3) preferably a
lower partition coefficient for fed hexanoic acid (for olivetolic
acid/olivetol) or other fatty acid
such as RCO2H (for other resorcinols) in the organic phase vs. the aqueous
phase. Additional
properties of the organic phase enhance its suitability for downstream
conversion, e.g. and
without limitation, to cannabigerol and other cannabinoid compounds, including
suitability as a
solvent or co-solvent during downstream prenylation or other reactions, and
boiling point if
downstream separation by distillation is employed.
The performance of various classes of organic phase compounds are provided
herein.
Among the diesters tested, certain may be toxic to growth under the test
conditions. Certain
diethyl esters were toxic under the test conditions with the exception of
modest growth by
most strains in the presence of diethyl sebacate and diethyl diethylmalonate
(with glucose only,
with galactose strains appeared to exhibit substantial lag). For malonate
diesters, under the test
conditions, di-tert-butyl nnalonate supported growth of all strains with
glucose addition, again
appearing toxic or to induce substantial lag with galactose addition.
Increasing the dialkyl ester chain length from diethyl to diisopropyl to
dibutyl in a dialkyl
adipate series reduced toxicity. Growth was observed with diisopropyl adipate
and no apparent
toxicity observed in dibutyl adipate. Dibutyl sebacate was also completely non-
inhibitory to
growth and accordingly, non-toxic. In certain embodiments, the minimum non-
toxic internal
alkyl chain length of diethyl diesters is sebacate. In certain embodiments,
shorter internal alkyl
chain length down to adipate is possible with diisopropyl diesters.
For monoester compounds, under the test conditions, octyl acetate was toxic
and for
the hexanoate series, growth was only observed starting with hexyl hexanoate,
which was
moderately non-toxic. Isopropyl octanoate was moderately inhibitory but
allowed for some
growth. For the decanoate series, methyl decanoate was moderately inhibitory
to growth but
still allowed for growth. Texanol, a monoester alcohol (2,2,4-trimethy1-1,3-
pentanediol
monoisobutyrate), was inhibitory to growth under the conditions tested.
37
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
However, ethyl decanoate and higher alkyl chains were increasingly non-toxic.
Both
ethyl and butyl laurate were non-toxic, as well as methyl and ethyl myristate.
In certain
embodiments, growth-suitable monoester overlays for resorcinol or cannabinoid
production
include hexyl hexanoate or any higher chain length alkyl hexanoate ester, C3
chain-length or
higher (e.g., and without limitation C6-Cs or higher) alkyl octanoate esters,
and methyl (C1) or
higher (e.g., and without limitation C6-C8 or higher) alkyl decanoates,
laurates, or myristates.
In various embodiments, esters and diesters are employed as the organic phase
in
accordance with the present invention.
Fatty alcohols are mostly solids above C10 saturated chain length. Decanol, a
liquid, was
toxic to growth under test conditions. However, leyl alcohol supported robust
growth. In
certain embodiments, longer chain length (C12 or higher) unsaturated fatty
alcohols can be
suitable overlays supporting S. cerevisiae or another fermenting organism's
growth. In various
embodiments, fatty alcohols, preferably C12 or higher alcohols, are employed
as the organic
phase in accordance with the present invention.
In certain embodiments, alkanes and paraffins support robust growth. Lack of
toxicity
was observed for dodecane, tetradecane, hexadecane, light and heavy paraffin
oils, and isopar
M. In certain embodiments, C12 and higher paraffins are suitable overlays
supporting S.
cerevisiae or another fermenting organism's growth. In various embodiments,
fatty alcohols,
preferably C12 or higher alcohols, are employed as the organic phase in
accordance with the
present invention.
Certain triacylglycerols were tested, including tricaprylin, coconut oil, and
canola oil
(vegetable oils having different average chain length compositions of fatty
acid chains, with
coconut oil being predominantly C12-C14 saturated fatty acids, and canola
being predominantly
C16-Cis and a mixture of saturated and unsaturated fatty acids). Tricaprylin,
a synthetic oil
containing three C8 fatty acid chains, was fairly toxic, however allowed some
growth of strains.
In certain embodiments, coconut and canola oil were non-toxic to growth.
Mixtures of isopropyl myristate (IPM) and isopar M with different diesters ¨
dibasic
esters (DBE), diethyl sebacate, and di-tert-butyl malonate were explored to
investigate if lower
percentage mixtures of these compounds in non-toxic IPM or isopar M would
mitigate their
38
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
toxicity toward growth of a microorganism such as S. cerevisiae, as they may
also
advantageously alter partitioning properties of olivetolic acid, olivetol, and
other analogues into
the overlay and could offer advantages with alternative downstream separations
processes. For
example, and without limitation, a DBE, which may be toxic by itself as an
underlay, was much
less toxic at concentrations of between 1 and 2.5% (v/v) in IPM and especially
isopar M. Di-tert-
butyl malonate also exhibited much lower toxicity at 1-10% (v/v), and
particularly 1-2.5% (v/v),
in IPM and isopar M. In certain embodiments, mixtures of longer chain
monoesters or paraffins
with moderately to very toxic diesters are useful according to the present
invention.
In another embodiment, the aqueous phase further comprises histidine. In
another
embodiment, the pH of the aqueous phase is at a pH of about 4 to about 8.
In another embodiment, the olivetol and olivetolic acid is provided in a
combined
amount of at least about 4 g/liter over about 4 to about 7 days. In another
embodiment, the
olivetol and olivetolic acid is provided in a combined amount of at least
about 4.5 g/liter over
about 4 to about 7 days. In another embodiment, the olivetol and olivetolic
acid is provided in
a combined amount of at least about 5 g/liter over about 4 to about 7 days. In
another
embodiment, the olivetol and olivetolic acid is provided in a combined amount
of at least about
7 g/liter over about 4 to about 7 days. In another embodiment, the olivetol
and olivetolic acid
is provided in a combined amount of at least about 9 g/liter over about 4 to
about 7 days. In
another embodiment, the combined amount of olivetol and olivetolic acid
provided herein, is
provided over 4 days. In another embodiment, the combined amount of olivetol
and olivetolic
acid provided herein, is provided over 5 days. In another embodiment, the
combined amount
of olivetol and olivetolic acid provided herein, is provided over 6 days. In
another embodiment,
the combined amount of olivetol and olivetolic acid provided herein, is
provided over 7 days.
In one embodiment, the fermentation is performed in a semi-continuous mode
("fill-
and-draw"). In another embodiment, the fermentation is performed in a
continuous mode. In
one embodiment, the overall combined productivity of olivetol and olivetolic
acid is greater
than 0.3 g per L of total volume (including aqueous and immiscible liquid
phases) per day of
operation. In another embodiment, the fermentation is performed in a total
volume of 15 liters
or a larger volume such as 1,000 liters, 10,000 liters, 20,000 or 50,000
liters, or an even larger
39
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
volume. The combined yield of 0 and OA obtained in such large scale
fermentation performed
according to the present invention over 2-7 or 4-7 days, such as over 2, 3, 4,
5, 6, or 7 days is
unexpectedly high. In some embodiment, the combined amount of 0/0A obtained,
even in
large scale fermentations, e.g., and without limitation in 10,000 liters
fermentations, is about 7
¨ about 10 g/liter. In some embodiment, the combined amount of 0/0A obtained,
even in large
scale fermentations, e.g., and without limitation in 20,000 liters
fermentations, is about 7 ¨
about 10 g/liter.
In one embodiment, the functional OLS has a Sequence ID 1. In another
embodiment,
the functional OLS has an at least 95% sequence identity with Sequence ID 1.
In another
embodiment, the functional olivetolic acid cyclase has at least 50%, at least
75%, or at least
95% sequence identity to SEQ ID 1.
In one embodiment, the functional OAC has a Sequence ID 2A. In another
embodiment,
the functional OAC has an at least 95% sequence identity with Sequence ID 2A.
In another
embodiment, the functional olivetolic acid cyclase has at least 50%, at least
75%, or at least
95% sequence identity to SEQ ID NO: 2A. In another embodiment, the functional
olivetolic acid
cyclase is of SEQ ID NO: 2B. In another embodiment, the functional olivetolic
acid cyclase has at
least 50%, at least 75%, or at least 95% sequence identity to SEQ ID NO: 2B.
SEQ ID NO: 2B
MAVKHLIVLKFKDEITEAQKEEFFKTYVNLVN II PAM KDVYWG KDVTQKN KEEGYTH
IVEVTFESVETIQDYI I
HPAHVGFGDVYRSFWEKLLIFDYTPRK.
In one embodiment, the functional AAE has a Sequence ID 3A. In another
embodiment,
the functional AAE has an at least 95% sequence identity with Sequence ID 3A.
Sequence ID 3A: Cannabis sativa acyl activating enzyme (CsAAE1)
MGKNYKSLDSWASDFIALGITSEVAETLAGRLAEIVCNYGAATPOTWINIANHILSPDLPFSLFICIMLFYGGYK
DEGPAPPAWIPDPFKVKSTNI_GALLEKRGKEELGVKYKDPISSFSHFOFFSVRNPEVYWRIVLIVIDFMKISFSK
DPECIL.FIRDDINNPGGSEWLPGGYLNSAKNCL.NVNSNKKLNDTMIVWRDEGNDDL_PLNKL.TI_DO,LRKRVW
LVGYALEEMGLEKGCAIAIDMPMHVDAVVIYLAIVLAGYVVVSIADSFSAPEISTRLRLSKAKAIFTQDHIIRGK
KRIPLYSRVVEAKSPMAIVIPCSGSNIGAELRDGDISWDYFLERAKEFKNCEFTAREQPVDAYTNILFSSG H
GE
PKAIPWTQATPLKAAADGWS1--ILDIRKGDVIVWPTNLGWMMGPWLWASLINGASIALYNGSPLVSGFAKE
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
VQDAKVTMLGVVPSIVRSWKSTNCVSGYDWSTIRCFSSSGEASNVDEYLWLMGRANYKPVIEMCGGTEIG
GAFSAGSFLOAQSLSSFSSQCMGCTLYILDKNGYPMPKNKPGIGELALGPVMFGASKTLLNGNHHDVYFKG
MPUNGEVLRRFIGDIFELTSNGYYHAFIGRADDTMNIGGIKISSIEIERVCNEVDDRVFE
_______________________ I I AIGVPPLGGGPE
QLVIFFVLKDSND _______ I I
IDLNQLRLSFNLGLQKKLNPLFKVTRVVPLSSLPRTATNKIMRRVLRQQFSHFE
In another embodiment, the functional AAE polypeptide comprises an amino acid
sequence SEQ ID NO: 3B. In another embodiment, the functional AAE polypeptide
comprises an
amino acid sequence that has at least 50%, at least 75%, or at least 95%
sequence identity to SEQ
ID NO: 3B. In another embodiment, the functional AAE polypeptide comprises an
amino acid
sequence SEQ ID NO: 3C. In another embodiment, the functional AAE polypeptide
comprises an
amino acid sequence that has at least 50%, at least 75%, or at least 95%
sequence identity to SEQ
ID NO: 3C. In another embodiment, the functional AAE polypeptide comprises an
amino acid
sequence that is SEQ ID NO: 3D. In another embodiment, the functional AAE
polypeptide has at
least 50%, at least 75%, or at least 95% sequence identity to SEQ ID NO: 3D.
SEQ ID NO 3B: Cannabis sativa acyl activating enzyme (CsAAE3)
MEKSGYGRDGIYRSLRPPLHLPNNNNLSMVSFLFRNSSSYPQKPALIDS
ETNQILSFSHFKSTVIKVSHGFLNLGIKKNDWLIYAPNSIHFPVCFLGIIA
SGAIATTSNPLYTVSELSKQVKDSNPKLIITVPQLLEKVKGFNLPTILIGP
DSEQESSSDKVMTFNDLVNLGGSSGSEFPIVDDFKQSDTAALLYSSGTT
GMSKGWLTHKN FIASSLMVTM EQDLVGEMDNVFLCFLPM FHVFG LAI
ITYAQLQRGNTVISARFDLEKMLKDVEKYVTHLWWPPVILALSKNSM
VKFN LSSIKYIGSGAAPLGKDLMEECSKWPYGIVAQGYGMTETCGIVS
MEDI RGGKRNSGSAGM LASGVEAQIVSVDTLKPLPPNQLGEIWVKGPN
MMQGYFNN PQATKLTIDKKGWVHTGDLGYFDEDGHLYWDRIKELIK
YKGFQVAPAELEGLLVSHPEILDAWIPFPDAEAGEVPVAYWRSPNSSL TEN DVKKFIAGQV
ASFKRLRKVTFINSVPKSASGKILRRELIQKVRSN M
SEQ ID NO 3C: Truncated Cannabis sativa acyl activating enzyme
MEKSGYGRDGIYRSLRPPLHLPNNNNLSMVSFLFRNSSSYPQKPALIDS
ETNQILSFSHIKSTVIKVSHGFLNLGIKKNDWLIYAPNSIHIPVCILGIIA
SGAIATTSNPLYTVSELSKQVKDSNPKLIITVPQLLEKVKGFNLPTILIGP
41
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
DSEQESSSDKVMTFNDLVNLGGSSGSEFPIVDDFKQSDTAALLYSSGTT
GMSKGWLTHKNFIASSLMVTMEQDLVGEMDNVFLCFLPMFHVFGLAI
ITYAQLQRGNTVISARFDLEKMLKDVEKYVTHLWWPPVILALSKNSM
VKFNLSSIKYIGSGAAPLGKDLMEECSKWPYGIVAQGYGMTETCGIVS
MEDIRGGKRNSGSAGMLASGVEAQIVSVDTLKPLPPNQLGEIWVKGPN
MMQGYFNNPQATKLTIDKKGWVHTGDLGYFDEDGHLYWDRIKELIK
YKGFQVAPAELEGLLVSHPEILDAWIPFPDAFAGEVPVAYWRSPNSSL
TEN DVKKFIAGQVASFKRLRKVTFINSVPKSASGKIL.
SEQ ID NO 3D: Escherichia coli hexanoyl-CoA synthetase amino acid sequence
MHPTGPHLGPDVLFRESNMKVTLTFNEQRRAAYRQQGLWGDASLADYWQQTARAMPDKIAVVDNHGA
SYTYSALDHAASCLANWMLAKGIESGDRIAFQLPGWCEFTVIYLACLKIGAVSVPLLPSWREAELVWVLNKC
QAKMFFAPTLFKQTRPVDLILPLQNQLPQLQQIVGVDKLAPATSSLSLSQIIADNTSLTTAITTHGDELAAVLFT
SGTEGLPKGVMLTHNNILASERAYCARLNLTWQDVFMMPAPLGHATGFLHGVTAPFLIGARSVLLDIFTPDA
CLALLEQQRCTCMLGATPFVYDLLNVLEKQPADLSALRFFLCGGTTIPKKVARECQQRGIKLLSVYGSTESSPH
AVVNLDDPLSRFMHTDGYAAAGVEIKVVDDARKTLPPGCEGEEASRGPNVFMGYFDEPELTARALDEEGW
YYSGDLCRMDEAGYIKITGRKKDIIVRGGENISSREVEDILLQHPKIHDACVVAMSDERLGERSCAYVVLKAPH
HSLSLEEVVAFFSRKRVAKYKYPEHIVVIEKLPRTTSGKIQKFLLRKDIMRRLTQDVCEEIE
In one embodiment, the sequence identity is at least 50%. In another
embodiment, the
sequence identity is at least 75%. In another embodiment, the sequence
identity is at least 95%.
In another embodiment, the sequence identity is at least 99% with a protein
sequence
utilized herein. In another embodiment, the sequence identity is at least 99%
with a nucleic
acid sequence utilized herein.
In another embodiment, the heterologous microorganism is antibiotic-marker
free. In
another embodiment, the heterologous microorganism is an FAA2 (peroxisomal
medium chain
fatty acyl-CoA synthetase) knock out or has a lowered FAA2 activity. In
another embodiment,
the heterologous microorganism is a PXA1 (part of the heterodimeric
peroxisomal fatty acid
and/or acyl-CoA ABC transport complex with PXA2) knockout or has a lowered
PXA2 activity. In
another embodiment, the heterologous microorganism is a PEX11 (peroxisomal
protein
required for medium-chain fatty acid oxidation) knockout or has a lowered
PEX11 activity. In
42
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
another embodiment, the heterologous microorganism is an ANT1 (peroxisomal
adenine
nucleotide transporter, which exchanges AMP generated in peroxisomes by acyl-
CoA
synthetases for ATP, that is consumed in that reaction, from the cytosol)
knockout or has a
lowered ANT1 activity.
In another embodiment, the microorganism is Saccharomyces cerevisiae. In
another
embodiment, the Saccharomyces cerevisiae comprises galactose regulatable
promoters for the
heterologous genes (csOLS, csOAC, csAAE, and the likes). In another
embodiment, the
Saccharomyces cerevisiae does not include galactose regulatable promoters for
the
heterologous genes. In another embodiment, the Saccharomyces cerevisiae is
haploid. In
another embodiment, the Saccharomyces cerevisiae is diploid.
Initial construction of an olivetol/olivetolic acid (0/0A) producing line was
done by
introducing 3 genes (cannabis olivetol synthase csOLS, cannabis olivetolic
acid cyclase csOAC,
and a cannabis acyl-activating enzyme csAAE1) from the Cannabis sativa plant
under the
control of genetic elements that are regulated by a galactose carbon source.
It is well known
that galactose regulates gene expression in Saccharomyces cerevisiae.
Introduction of foreign
genes into Saccharomyces cerevisiae can be toxic to Saccharomyces cerevisiae
for a variety of
reasons. One way to regulate toxicity of foreign genes is to produce their
expression under the
control of galactose. Glucose is used under normal Saccharomyces cerevisiae
growth
conditions. However, during the course of growth or at the beginning of
growth, glucose can
then be exchanged with galactose to tightly control the expression of foreign
genes.
The initial engineering of Saccharomyces cerevisiae was done by introducing a
gene
fragment containing csOLS, csOAC, and csAAE1 under the control of galactose
regulatable
elements called promoters. The csOLS and csOAC were physically linked to each
other on the
gene with a genetic element called T2A in all examples. In order to select for
Saccharomyces
cerevisiae cells that efficiently incorporated the foreign DNA, but removed
Saccharomyces
cerevisiae that had no foreign genes, a method that allows growth on nutrient
preferred media
was utilized. The process by which Saccharomyces cerevisiae uptake foreign DNA
and stably
utilize the foreign DNA is called recombination. The final Saccharomyces
cerevisiae strains that
took up the foreign DNA and utilized the DNA are called recombinants.
Saccharomyces
43
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
cereyisioe that did not undergo recombination is called wild type. The process
of selecting in
preferred media is defined as prototrophy rescue. In order to separate
recombinants from wild
type we utilized prototrophy rescue. In some cases, recombinants we added
foreign genes that
contained genetic elements that controlled the resistance to antibiotics.
Antibiotics such as
G418 or hygromycin will normally kill Soccharomyces cerevisioe. In some
instances, foreign
genes were introduced into 0/0A producing lines in order to rescue survival of
G418 or
hygromycin for the purpose of removing genes native to the Saccharomyces
cerevisiae and
allowing them to survive in antibiotics while decreasing the need for
galactose utilization.
Expression Vectors
In various aspects, provided herein is a recombinant host cell modified by
genetic
engineering as disclosed herein. In one embodiment, a recombinant polyketide
synthase, such
as an OLS enzyme, is introduced. In another embodiment, an aromatic
prenyltransferase is
introduced. In another embodiment, the modification increases the production
of malonyl-CoA,
hexanoyl-CoA or a R-CoA. In some embodiments, the host cell is engineered via
recombinant
DNA technology to express heterologous nucleic acids that encode a cannabinoid
pathway
enzyme such as an OLS enzyme, which is either a mutated version of a naturally
occurring
enzyme, or a non-naturally occurring enzyme as provided herein.
In one preferred embodiment, the invention includes methods of generating a
polynucleotide that expresses one or more of the SEQ IDs related to a mutant
or modified OLS
provided or utilized herein. In certain preferred embodiments, the proteins of
the invention are
expressed using any of a number of systems, such as in whole plants, as well
as plant cell
and/or yeast suspension cultures. E.g., the polynucleotide that encodes the
OLS is placed under
the control of a promoter that is functional in the desired host cell. An
extremely wide variety
of promoters may be available and can be used in the expression vectors of the
invention,
depending on the particular application. Ordinarily, the promoter selected
depends on the cell
in which the promoter is to be active. Other expression control sequences such
as ribosome
binding sites, transcription termination sites and the like are also
optionally included.
Nucleic acid constructs provided and utilized herein include expression
vectors that
comprise nucleic acids encoding one or more polyketide synthase enzymes. The
nucleic acids
44
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
encoding the enzymes are operably linked to promoters and optionally other
control sequences
such that the subject enzymes are expressed in a host cell containing the
expression vector
when cultured under suitable conditions. The promoters and control sequences
employed
depend on the host cell selected for the production of olivetol, olivetolic
acid (OLA or OA), OLA-
derived compound, or another cannabinoid or cannabinoid derivative. Thus, the
invention
provides not only expression vectors but also nucleic acid constructs useful
in the construction
of expression vectors. Methods for designing and making nucleic acid
constructs and expression
vectors generally are well known to those skilled in the art and so are only
briefly reviewed
herein.
Nucleic acids encoding the polyketide synthase enzymes can be prepared by any
suitable method known to those of ordinary skill in the art, including, for
example, direct
chemical synthesis and cloning. Further, nucleic acid sequences for use in the
invention can be
obtained from commercial vendors that provide de nova synthesis of the nucleic
acids.
A nucleic acid encoding the desired enzyme can be incorporated into an
expression
vector by known methods that include, for example, the use of restriction
enzymes to cleave
specific sites in an expression vector, e.g., plasmid, thereby producing an
expression vector of
the invention. Some restriction enzymes produce single stranded ends that may
be annealed to
a nucleic acid sequence having, or synthesized to have, a terminus with a
sequence
complementary to the ends of the cleaved expression vector. The ends are then
covalently
linked using an appropriate enzyme, e.g., DNA ligase. DNA linkers may be used
to facilitate
linking of nucleic acids sequences into an expression vector.
A set of individual nucleic acid sequences can also be combined by utilizing
polymerase
chain reaction (PCR)-based methods known to those of skill in the art. For
example, each of the
desired nucleic acid sequences can be initially generated in a separate PCR.
Thereafter, specific
primers are designed such that the ends of the PCR products contain
complementary
sequences. When the PCR products are mixed, denatured, and reannealed, the
strands having
the matching sequences at their 3' ends overlap and can act as primers for
each other.
Extension of this overlap by DNA polymerase produces a molecule in which the
original
sequences are "spliced" together. In this way, a series of individual nucleic
acid sequences may
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
be joined and subsequently transduced into a host cell simultaneously. Thus,
expression of each
of the plurality of nucleic acid sequences is affected.
A typical expression vector contains the desired nucleic acid sequence
preceded and
optionally followed by one or more control sequences or regulatory regions,
including a
promoter and, when the gene product is a protein, ribosome binding site, e.g.,
a nucleotide
sequence that is generally 3-9 nucleotides in length and generally located 3-
11 nucleotides
upstream of the initiation codon that precede the coding sequence, which is
followed by a
transcription terminator in the case of E. coli or other prokaryotic hosts.
See Shine et al., Nature
254:34 (1975) and Steitz, in Biological Regulation and Development: Gene
Expression (ed. R. F.
Goldberger), vol. 1, p. 349 (1979) Plenum Publishing, N.Y. In the case of
eukaryotic hosts like
yeast, a typical expression vector contains the desired nucleic acid coding
sequence preceded
by one or more regulatory regions, along with a Kozak sequence to initiate
translation and
followed by a terminator. See Kozak, Nature 308:241-246 (1984).
Regulatory regions or control sequences include, for example, those regions
that
contain a promoter and an operator. A promoter is operably linked to the
desired nucleic acid
coding sequence, thereby initiating transcription of the nucleic acid sequence
via an RNA
polymerase. An operator is a sequence of nucleic acids adjacent to the
promoter, which
contains a protein-binding domain where a transcription factor can bind.
Transcription factors
activate or repress transcription initiation from a promoter. In this way,
control of transcription
is accomplished, based upon the particular regulatory regions used and the
presence or
absence of the corresponding transcription factor. Non-limiting examples for
prokaryotic
expression include lactose promoters (Lad l repressor protein changes
conformation when
contacted with lactose, thereby preventing the Lad l repressor protein from
binding to the
operator) and tryptophan promoters (when complexed with tryptophan, TrpR
repressor protein
has a conformation that binds the operator; in the absence of tryptophan, the
TrpR repressor
protein has a conformation that does not bind to the operator). Non-limiting
examples of
promoters to use for eukaryotic expression include pTDH3, pTEF1, pTEF2, pRNR2,
pRPL18B,
pREV1, pGAL1, pGAL10, pGAPDH, pCUP1, pMET3, pPGK1, pPYK1, pHXT7, pPDC1, pFBA1,
46
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
pTDH2, pPGI1, pPDC1, pTPI1, pEN02, pADH1, and pADH2. As will be appreciated by
those of
ordinary skill in the art, a variety of expression vectors and components
thereof are useful.
Although any suitable expression vector are useful to incorporate the desired
sequences, readily available expression vectors include, without limitation:
plasmids, such as
pESC, pTEF, p414CYC1, p414GALS, pSC101, pBR322, pBBR1MCS-3, pUR, pEX, pMR100,
pCR4,
pBAD24, pUC19, pRS series; and bacteriophages, such as M13 phage and X phage.
Of course,
such expression vectors may only be suitable for particular host cells or for
expression of
particular polyketide synthases. One of ordinary skill in the art, however,
can readily determine
through routine experimentation whether any particular expression vector is
suited for any
given host cell or protein. For example, the expression vector can be
introduced into the host
cell, which is then monitored for viability and expression of the sequences
contained in the
vector. In addition, relevant texts and literature describe expression vectors
and their suitability
to any particular host cell. In addition to the use of expression vectors,
strains are built where
expression cassettes are directly integrated into the host genome.
The expression vectors are introduced or transferred, e.g., by transduction,
transfection,
or transformation, into the host cell. Such methods for introducing expression
vectors into host
cells are well known to those of ordinary skill in the art. For example, one
method for
transforming P. kudriayzevii with an expression vector involves a calcium
chloride treatment
wherein the expression vector is introduced via a calcium precipitate.
For identifying whether a nucleic acid has been successfully introduced or
into a host
cell, a variety of methods are available. For example, potentially transformed
host cells in a
culture are separated, using a suitable dilution, into individual cells and
thereafter individually
grown and tested for expression of a desired gene product of a gene contained
in the
introduced nucleic acid. For example, an often-used practice involves the
selection of cells
based upon antibiotic resistance that has been conferred by antibiotic
resistance-conferring
genes in the expression vector, such as the beta lactamase (amp),
aminoglycoside
phosphotransferase (neo), and hygromycin phosphotransferase (hyg, hph, hpt)
genes.
In one embodiment, a host cell of the disclosure is transformed with at least
one
expression vector. When only a single expression vector is used, the vector
will typically contain
47
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
a polyketide synthase gene. Once the host cell has been transformed with the
expression
vector, the host cell is cultured in a suitable medium containing a carbon
source, such as a
sugar (e.g., glucose). As the host cell is cultured, expression of the
polyketide synthase
enzyme(s) occurs. Once expressed, these OLS(s) and other enzymes provided and
utilized
herein convert three molecules of malonyl-CoA and one molecule of hexanoyl-CoA
or R-CoA,
wherein R is defined as herein, to olivetol or a compound of formula (I).
If a host cell of the invention is to include more than one heterologous gene,
the
multiple genes can be expressed from one or more vectors. For example, a
single expression
vector can comprise one, two, or more genes encoding one, two, or more mutant
OLS
enzyme(s), other enzymes of the cannabinoid pathway, e.g., improved malonyl-
CoA production,
hexanoyl-CoA, or R-CoA production, etc. The heterologous genes can be
contained in a vector
replicated episomally or in a vector integrated into the host cell genome, and
where more than
one vector is employed, then all vectors may replicate episomally
(extrachromasomally), or all
vectors may integrate, or some may integrate and some may replicate
episomally. While a
"gene" is generally composed of a single promoter and a single coding
sequence, in certain host
cells, two or more coding sequences are controlled by one promoter in an
operon. In some
embodiments, a two or three operon system is used.
In some embodiments, the coding sequences employed have been modified,
relative to
some reference sequence, to reflect the codon preference of a selected host
cell. Codon usage
tables for numerous organisms are readily available and can be used to guide
sequence design.
The use of prevalent codons of a given host organism generally improves
translation of the
target sequence in the host cell. As one non-limiting example, in some
embodiments the
subject nucleic acid sequences will be modified for yeast codon preference
(see, for example,
Bennetzen etal., J. Biol. Chem. 257: 3026-3031 (1982)). In some embodiments,
the nucleotide
sequences will be modified for P. kudriavzevii codon preference (see, for
example, Nakamura et
al., Nucleic Acids Res. 28:292 (2000)). In other embodiments, the nucleotide
sequences are
modified to include codons optimized for S. cerevisiae codon preference.
Nucleic acids can be prepared by a variety of routine recombinant techniques.
Briefly,
the subject nucleic acids can be prepared from genomic DNA fragments, cDNAs,
and RNAs, all
48
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
of which can be extracted directly from a cell or recombinantly produced by
various
amplification processes including but not limited to PCR and rt-PCR. Subject
nucleic acids can
also be prepared by a direct chemical synthesis.
The nucleic acid transcription levels in a host microorganism can be increased
(or
decreased) using numerous techniques. For example, the copy number of the
nucleic acid can
be increased through use of higher copy number expression vectors comprising
the nucleic acid
sequence, or through integration of multiple copies of the desired nucleic
acid into the host
microorganism's genome. Non-limiting examples of integrating a desired nucleic
acid sequence
onto the host chromosome include recA-mediated recombination, lambda phage
recombinase-
mediated recombination and transposon insertion. Nucleic acid transcript
levels can be
increased by changing the order of the coding regions on a polycistronic mRNA
or breaking up a
polycistronic operon into multiple poly- or mono-cistronic operons each with
its own promoter.
RNA levels can be increased (or decreased) by increasing (or decreasing) the
strength of the
promoter to which the protein-coding region is operably linked.
The translation level of a desired polypeptide sequence in a host
microorganism can
also be increased in a number of ways. Non-limiting examples include
increasing the mRNA
stability, modifying the ribosome binding site (or Kozak) sequence, modifying
the distance or
sequence between the ribosome binding site (or Kozak sequence) and the start
codon of the
nucleic acid sequence coding for the desired polypeptide, modifying the
intercistronic region
located 5' to the start codon of the nucleic acid sequence coding for the
desired polypeptide,
stabilizing the 3'-end of the mRNA transcript, modifying the codon usage of
the polypeptide,
altering expression of low-use/rare codon tRNAs used in the biosynthesis of
the polypeptide.
Determination of preferred codons and low-use/rare codon tRNAs can be based on
a sequence
analysis of genes derived from the host microorganism.
The polypeptide half-life, or stability, can be increased through mutation of
the nucleic
acid sequence coding for the desired polypeptide, resulting in modification of
the desired
polypeptide sequence relative to the control polypeptide sequence. When the
modified
polypeptide is an enzyme, the activity of the enzyme in a host is altered due
to increased
solubility in the host cell, improved function at the desired pH, removal of a
domain inhibiting
49
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
enzyme activity, improved kinetic parameters (lower Km or higher kcat values)
for the desired
substrate, removal of allosteric regulation by an intracellular metabolite,
and the like.
Altered/modified enzymes can also be isolated through random mutagenesis of an
enzyme,
such that the altered/modified enzyme can be expressed from an episomal vector
or from a
recombinant gene integrated into the genome of a host microorganism.
Host Cells
Provided herein are host cells, preferably recombinant host cells, more
preferably
heterologous recombinant host cells for performing one or more steps of the
cannabinoid
pathway. In some embodiments, the recombinant host cell is a eukaryote. In
various
embodiments, the eukaryote is a yeast strain selected from the non-limiting
list of example
genera: Can dida, Cryptococcus, Hansenula, lssatchenkia, Kluyveromyces,
Komagataella,
Lipomyces, Pichia, Rhodosporidium, Rhodotorula, Saccharomyces, or Yarrowia.
Those skilled in
the art will recognize that these genera broadly encompass yeast, including
those distinguished
as oleaginous yeast. In some embodiments, the host cell is Saccharomyces
cerevisiae. In other
embodiments, the host cell is Pichia kudriavzevii. In other embodiments of the
invention, the
eukaryotic host cell is a fungus or algae. In yet other embodiments, the
recombinant host cell is
a prokaryote selected from the non-limited example genera: Bacillus,
Clostridium,
Corynebacterium, Escherichia, Pseudomonas, Rhodobacter, and Streptomyces. In
various
embodiments, the host cell is P. kudriavzevii.
In one embodiment, the host cell is part of a multicellular organism. In one
embodiment, the multicellular organism is a plant. In one embodiment, the
plant is a cannabis
plant. In one embodiment, the plant is a tobacco plant.
As utilized herein, a number of genetic modifications are further useful for
increasing
microbial biosynthesis of malonyl-CoA. For example, in some embodiments a host
cell provided
or utilized herein is further engineered to include a genetic modification
useful for converting
pyruvate to malonyl-CoA, wherein the genetic modification produces and/or
provides a
pyruvate decarboxylase, an acetaldehyde dehydrogenase, an acetyl-CoA
synthetase, an acetyl-
CoA carboxylase, and a carbonic anhydrase.
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
In some embodiments, an engineered host cell provided or utilized herein is a
Saccharomyces cerevisiae host cell. In some embodiments, an engineered host
cell comprises
heterologous enzymes that are overexpressed to increase malonyl-CoA
production, thereby
facilitating production of olivetol, OLA, OLA-derived compound, or another
cannabinoid or
cannabinoid derivative. In some embodiments, the engineered host cell
comprises
heterologous enzymes selected from the group consisting of an acetyl Co-A
carboxylase, such
as P. kudriavzevii acetyl-CoA carboxylase, S. cerevisiae aldehyde
dehydrogenase, Yarrowia
lipolytica acetyl-CoA synthetase, and S. cerevisiae pyruvate decarboxylase.
In some embodiments, the host cell is a Saccharomyces cerevisiae host cell. In
some
embodiments, a yeast host cell expressing an OLS is used to produce olivetol,
OLA, OLA-derived
compound, or another cannabinoid or cannabinoid derivative. In some
embodiments, an
oleaginous yeast host cell expressing an OLS is used to produce olivetol, OLA,
OLA-derived
compound, or another cannabinoid or cannabinoid derivative.
Also provided herein is a mutated OLS comprising a mutated active site,
vectors for
expressing the mutant, and host cells that express the mutant. In another
embodiment, the
host cell further produces olivetol, OLA, OLA-derived compound, or another
cannabinoid or
cannabinoid derivative. Introduction of mutations in the region comprising
D198 to G209 of
OLA increases the turnover rate (i.e., !Qat values) of the mutated OLS. One or
more point
mutations at amino acid positions D198 to G209 can be introduced alone or in
any desired
combination. In these embodiments, the recombinant host cell can be, without
limitation, a P.
kudriavzevii or yeast, including but not limited to S. cerevisiae or other
yeast, host cell.
In some aspects, provided herein are recombinant host cells, preferably host
cells
suitable for producing olivetol (including OLA and/or OLA-derived compounds)
and other
cannabinoids and cannabinoid derivatives in accordance with the methods
provided herein, the
host cells comprising one or more heterologous OLS enzymes, preferably OLS
enzymes having
an increased !cat value as compared to wild type or homologous OLS enzymes,
wherein the
recombinant host cells provide increase olivetol titer, yield, and/or
productivity relative to a
host cell not comprising a heterologous OLS enzyme. In some aspects, provided
herein are
recombinant host cells suitable for producing olivetol in accordance with the
methods of the
51
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
invention comprising increased malonyl-CoA biosynthesis. In some aspects,
provided herein are
recombinant host cells suitable for producing olivetol in accordance with the
methods of the
invention comprising increased hexanoyl-CoA synthetase biosynthesis. In some
aspects,
provided herein are recombinant host cells suitable for producing olivetol in
accordance with
the methods of the invention comprising increased pyruvate dehydrogenase
biosynthesis. In
some aspects, provided herein are recombinant host cells suitable for
producing olivetol in
accordance with the methods of the invention comprising increased acetaldehyde
dehydrogenase biosynthesis. In some aspects, provided herein are recombinant
host cells
suitable for producing olivetol in accordance with the methods of the
invention comprising
increased acetyl-CoA synthetase biosynthesis. In some aspects, provided herein
are
recombinant host cells suitable for producing olivetol in accordance with the
methods of the
invention comprising increased acetyl-CoA carboxylase biosynthesis. In some
aspects, provided
herein are recombinant host cells suitable for producing olivetol in
accordance with the
methods of the invention comprising increased carbonic anhydrase biosynthesis.
In accordance with the invention, increased olivetol titer, yield, and/or
productivity can
be achieved through increased OLS enzymatic activity, which may require
increased malonyl-
CoA biosynthesis, and the invention provides host cells, vectors, enzymes, and
methods relating
thereto. Malonyl-CoA is produced in host cells through the activity of an
acetyl-CoA carboxylase
(EC 6.4.1.2) catalyzing the formation of malonyl-CoA from acetyl-CoA and
carbon dioxide. The
invention provides recombinant host cells for producing olivetol that express
a heterologous
acetyl-CoA carboxylase (ACC). In some embodiments, the host cell is a S.
cerevisiae cell
comprising a heterologous S. cerevisiae acetyl-CoA carboxylase ACC1 or an
enzyme homologous
thereto. In some embodiments, the host cell modified for heterologous
expression of an ACC
such as S. cerevisiae ACC1 is further modified to eliminate ACC1 post-
translational regulation by
genetic modification of S. cerevisiae SN F1 protein kinase or an enzyme
homologous thereto.
The disclosure also provides a recombinant host cell suitable for producing
olivetol in
accordance with the invention that is an E. coil cell that comprises a
heterologous nucleic acid
coding for expression of E. coli acetyl-CoA carboxylase complex proteins AccA,
AccB, AccC and
AccD or one or more enzymes homologous thereto.
52
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Thus, in one aspect of the invention, the recombinant host cell comprises a
heterologous nucleic acid encoding a mutant OLS enzyme or another mutant
cannabinoid
pathway enzyme, that results in increased production of olivetol, OLA, OLA-
derived compound,
or another cannabinoid or cannabinoid derivative relative to host cells not
comprising the
mutant OLS enzyme and/or an OLS enzyme.
Thus, in accordance with the invention an OLS enzyme other than, or in
addition to, OLS
derived from C. sativa can be used for biological synthesis of olivetol, OLA,
OLA-derived
compound, or another cannabinoid or cannabinoid derivative in a recombinant
host. In some
embodiments, the recombinant host is P. kudriavzevii. In some embodiments, the
recombinant
host is S. cerevisiae. In other embodiments, the recombinant host is E. co/i.
In other
embodiments, the recombinant host is a yeast other than P. kudriavzevii. In
various
embodiments, the host is modified to express a mutated OLS enzyme and/or an
OLS enzyme
provided or utilized herein. In various embodiments, the host is further
modified to express one
or more heterologous enzymes that are overexpressed to increase malonyl-CoA
production. In
various embodiments, the host is further modified to express or overexpress a
functional
hexanoyl-CoA synthetase.
Moreover, additional enzymes and catalysts other than those specifically
disclosed
herein can be utilized in mutated or heterologously expressed form. It will be
well understood
to those skilled in the art in view of this disclosure how other appropriate
enzymes can be
identified, modified, and expressed to achieve the desired olivetol, OLA, OLA-
derived
compound, or another cannabinoid or cannabinoid derivative production, as
disclosed herein.
In one aspect, provided herein are recombinant host cells suitable for
biological
production of cannabinoids and derivatives, such as without limitation
olivetol, OLA, OLA-
derived compound, or another cannabinoid or cannabinoid derivative. Any
suitable host cell is
useful in practice of the methods provided herein. In some embodiments, the
host cell is a
recombinant host microorganism in which nucleic acid molecules have been
inserted, deleted
or modified (i.e., mutated; e.g., by insertion, deletion, substitution, and/or
inversion of
nucleotides), either to produce olivetol, or to increase yield, titer, and/or
productivity of
olivetol relative to a "wild type", "control cell", "parental cell", or
"reference cell". A "control
53
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
cell" can be used for comparative purposes, and is typically a wild type or
recombinant parental
cell that does not contain one or more of the modification(s) made to the host
cell of interest.
In some embodiments, the invention provides a recombinant host cell that has
been
modified to produce one or more enzymes that facilitate malonyl-CoA
production. In some
embodiments, the invention provides a recombinant host cell that has been
modified to
produce one or more enzymes of the cannabinoid pathway. In some embodiments,
the
invention provides a recombinant host cell that has been modified to produce
an OLS, such as,
without limitation, an engineered or modified OLS, for example, olivetol
synthase, having
improved kcat values. In some embodiments, the invention provides a
recombinant host cell
that has been modified to produce an OLS, such as, without limitation, an
engineered or
modified OLS having improved solubility in the host. In some embodiments, the
invention
provides a recombinant host cell that has been modified to produce an OLS,
such as, without
limitation, an engineered or modified OLS or having improved stability in the
host. Thus, various
embodiments of the invention provide recombinant host cells capable of
producing increased
amounts of olivetol, OLA, OLA-derived compound, or another cannabinoid or
cannabinoid
derivative (i.e., product) per unit time. Accordingly, various embodiments of
the invention
provide recombinant host cells capable of achieving higher titers of product
over shorter
fermentation run times.
With respect to production titer levels, the recombinant host cells provided
or utilized
herein produce titer levels that exceed production titer levels of control
cells. In some
embodiments, the recombinant host cells provided or utilized herein produce
titer levels that
are suitable for commercial production, for example approximately 1-20 g/L,
such as 2-10 g/L or
3-8 g/L, or greater. The recombinant host cells described herein promote high
titer levels of
product(s) in at least two ways. First, the recombinant host cells produce
mutated OLS enzymes
having improved synthetase kinetics (i.e., an increase in kcat), which allows
for faster product
production, thereby increasing the rate and ease at which a desired titer
level can be achieved.
Secondly, the materials and methods provided or utilized herein provide and
facilitate in situ
extraction of the product into an organic phase, as described below. By adding
the organic
phase directly to the broth during the fermentation, the product can be
quickly and
54
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
continuously separated from the fermentation process, thereby decreasing
undesirable effects
of the product on the fermentation process, such as toxicity and product
inhibition feedback on
the pathway enzymes, thereby further increasing the titer levels of the
product(s). Additionally,
various genetic modifications provided or utilized herein are useful for
increasing the provision
of malonyl-CoA, which is a substrate for OLS.
In one embodiment, provided herein are recombinant yeast cells suitable for
the
production of cannabinoids and derivatives such as, without limitation,
olivetol, at levels
sufficient for subsequent purification and use as described herein. Yeast host
cells are excellent
host cells for construction of recombinant metabolic pathways comprising
heterologous
enzymes catalyzing production of small molecule products. There are
established molecular
biology techniques and nucleic acids encoding genetic elements necessary for
construction of
yeast expression vectors, including, but not limited to, promoters, origins of
replication,
antibiotic resistance markers, auxotrophic markers, terminators, and the like.
Second,
techniques for integration of nucleic acids into the yeast chromosome are well
established.
Yeast also offers a number of advantages as an industrial fermentation host.
Yeast can tolerate
high concentrations of organic acids and maintain cell viability at low pH and
can grow under
both aerobic and anaerobic culture conditions, and there are established
fermentation broths
and fermentation protocols. The ability of a strain to propagate and/or
produce desired
product under low pH provides a number of advantages. First, this
characteristic provides
tolerance to the environment created by the production of malonic acid.
Second, from a
process standpoint, the ability to maintain a low pH environment limits the
number of
organisms that are able to contaminate and spoil a batch.
In some embodiments of the invention, the recombinant host cell comprising a
heterologous nucleic acid provided or utilized herein is a eukaryote. In
various embodiments,
the eukaryote is a yeast selected from the non-limiting list of genera;
Saccharomyces, Candida,
Cryptococcus, Hansenula, lssatchenki, Kluyveromyces, Komagataella, Lipomyces,
Pichia,
Rhodosporidium, Rhodotorula, or Yarrowia species. In various embodiments, the
yeast is of a
species selected from the group consisting of Candida albicans, Candida
ethanolica, Candida
krusei, Candida methanosorbosa, Candida son orensis, Candida tropicalis,
Cryptococcus
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
curvatus, Hansenula polymorpha, lssatchenkia orientalis, Kluyveromyces lactis,
Kluyveromyces
marxianus, Kluyveromyces thermotolerans, Komagataella pastoris, Lipomyces
starkeyi, Pichia
an gusto, Pichia deserticola, Pichia galeiformis, Pichia kodamae, Pichia
kudriavzevii, Pichia
membranaefaciens, Pichia methanolica, Pichia pastoris, Pichia salictaria,
Pichia stipitis, Pichia
thermotolerans, Pichia trehalophila, Rhodosporidium toruloides, Rhodotorula
glutinis,
Rhodotorula graminis, Saccharomyces bayan us, Saccharomyces boulardi,
Saccharomyces
cerevisiae, Saccharomyces kluyveri, and Yarrowia lipolytica. One skilled in
the art will recognize
that this list encompasses yeast in the broadest sense, including both
oleaginous and non-
oleaginous strains.
Other recombinant host cells provided or utilized herein include without
limitation,
eukaryotic, prokaryotic, and archaea cells. Illustrative examples of
eukaryotic cells include, but
are not limited to: Aspergillus niger, Aspergillus oryzae, Crypthecodinium
cohnii,
Cunninghamella japonica, Entomophthora coronata, Mortierella alpina, Mucor
circinelloides,
Neurospora crassa, Pythium ultimum, Schizochytrium limacinum, Thraustochytrium
aureum,
Trichoderma reesei and Xanthophyllomyces dendrorhous. In general, if a
eukaryotic cell is used,
a non-pathogenic strain is employed. Illustrative examples of non-pathogenic
strains include
but are not limited to: Pichia pastoris and Saccharomyces cerevisiae. In
addition, certain strains,
including Saccharomyces cerevisiae, have been designated by the Food and Drug
Administration as Generally Regarded As Safe (or GRAS) and so can be
conveniently employed
in various embodiments of the methods of the invention.
Illustrative and non-limiting examples of recombinant prokaryotic host cells
provided or
utilized herein include, Bacillus subtilis, Brevibacterium ammonia genes,
Clostridium beigerinckii,
Corynebacterium glutamicum, Escherichia coli, Enterobacter sakazakii,
Lactobacillus
acidophilus, Lactococcus lactis, Mesorhizobium loti, Pseudomonas aeruginosa,
Pseudomonas
putida, Rhodobacter capsulatus, Rhodobacter sphaeroides, Salmonella enterica,
Salmonella
typhi, Salmonella typhimurium, Shigella flexneri, Staphylococcus aureus,
Streptomyces
ambofaciens, Streptomyces aureofaciens, Streptomyces aureus, Streptomyces fun
gicidicus,
Streptomyces griseochromogenes, Streptomyces griseus, Streptomyces lividans,
Streptomyces
olivogriseus, Streptomyces rameus, Streptomyces tanashiensis, and Streptomyces
vinaceus.
56
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Certain of these cells, including Bacillus subtilis, Corynebacterium
glutamicum, and Lactobacillus
acidophilus, have been designated by the Food and Drug Administration as
Generally Regarded
As Safe (or GRAS) and so are employed in various embodiments of the methods of
the
invention. While desirable from public safety and regulatory standpoints, GRAS
status does not
impact the ability of a host strain to be used in the practice of this
invention; hence, non-GRAS
and even pathogenic organisms are included in the list of illustrative host
strains suitable for
use in the practice of this invention.
Escherichia coli and Corynebacterium glutamicum are suitable prokaryotic host
cells for
metabolic pathway construction. Wild type E. coli can catabolize both pentose
and hexose
sugars as carbon sources. Provided herein are variety of E. coli host cells
suitable for the
production of malonate as described herein. In various embodiments, the
recombinant host cell
comprising a heterologous nucleic acid provided or utilized herein is an E.
coli cell. In various
embodiments of the methods of the invention, the recombinant host cell
comprising a
heterologous nucleic acid provided or utilized herein is a C. glutamicum cell.
Fermentation
In one embodiment, the fermentation is performed at a pH of about 5-6,
preferably at
about 5.5. In one embodiment, the fermentation is performed at a temperature
of about 30 C.
In one embodiment, the organic solvent immiscible with aqueous phase employed
in the
fermentation is loaded at about 26% of total fermentation tank volume. In one
embodiment,
the organic solvent immiscible with aqueous phase employed in the fermentation
is loaded at
about 40% of initial fermentation tank volume. In one embodiment, Isopropyl
myristate is the
aqueous phase immiscible organic solvent. In one embodiment, the aqueous phase
immiscible
organic solvent is added about 12 to about 36 hours post inoculation.
In one embodiment, the fermentation is performed wherein the compound of
formula
RCO2H or a salt thereof is present in an amount of 0.1-0.3 moles/500 g of
glucose in feed. In
one embodiment, the fermentation is performed wherein the compound of formula
RCO2H or a
salt thereof is present in an amount of 0.14-0.25 moles/500 g of glucose in
feed. In one
embodiment, the fermentation is performed wherein the compound of formula
RCO2H or a salt
thereof is present in an amount of 0.14-0.21 moles/500 g of glucose in feed.
In one
57
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
embodiment, the fermentation is performed wherein the sodium hexanoate/glucose
in feed
ratio was in the range of about 20 to about 28 g sodium hexanoate/ 500 g
glucose. In one
embodiment, the fermentation is performed wherein the sodium hexanoate/glucose
in feed
ratio was in the range of about 23 to about 28 g sodium hexanoate/ 500 g
glucose.
In one embodiment, the oxygen transmission rate (OTR) is about 60 ¨ about 80
mmoles/L/hr. In one embodiment, an oxygen uptake rate (OUR) of about 100¨
about 110
mmoles/L/hr is achieved. In one embodiment, the pulse parameter was about 1.7
g glucose/L
initial tank volume/pulse with a feed rate of about 10 g/L of initial tank
volume/hr. In one
embodiment, the batch glucose concentration employed in the fermentation was
about 10 ¨
about 20 g/L.
Synthesis, Utilization, and Purification of Cannabinoids and Derivatives
In some aspects, provided herein are methods of producing a cannabinoid, a
cannabinoid derivative, a cannabinoid precursor, or a cannabinoid precursor
derivative. In
some embodiments, the methods may involve culturing a genetically modified
host cell of the
present disclosure in a suitable medium and recovering the produced
cannabinoid, the
cannabinoid precursor, the cannabinoid precursor derivative, or the
cannabinoid derivative.
The methods may also involve cell-free production of cannabinoids, cannabinoid
precursors,
cannabinoid precursor derivatives, or cannabinoid derivatives using one or
more polypeptides
disclosed herein expressed or overexpressed by a genetically modified host
cell of the
disclosure.
In some embodiments, provided herein are methods of producing a cannabinoid or
a
cannabinoid derivative. The methods may involve culturing a genetically
modified host cell of
the present disclosure in a suitable medium and recovering the produced
cannabinoid or
cannabinoid derivative. The methods may also involve cell-free production of
cannabinoids or
cannabinoid derivatives using one or more polypeptides disclosed herein
expressed or
overexpressed by a genetically modified host cell of the disclosure.
Cannabinoids, cannabinoid derivatives, cannabinoid precursors, or cannabinoid
precursor derivatives that can be produced according to the present disclosure
may include,
58
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
but are not limited to, cannabichromene (CBC) type (e.g., cannabichromenic
acid), cannabigerol
(CBG) type (e.g., cannabigerolic acid), cannabidiol (CBD) type (e.g.,
cannabidiolic acid), A9-trans-
tetrahydrocannabinol (A9 -THC) type (e.g., A9-tetrahydrocannabinolic acid), A8-
trans-
tetrahydrocannabinol (A8 -THC) type, cannabicyclol (CBL) type, cannabielsoin
(CBE) type,
cannabinol (CBN) type, cannabinodiol (CBND) type, cannabitriol (CBT) type,
olivetolic acid, GPP,
derivatives of any of the foregoing, and others as listed in Elsohly M.A. and
Slade D., Life
Sci.2005 Dec 22;78N:539-48. [pub 2005 Sep 30.
Cannabinoids or cannabinoid derivatives that can be produced with the methods
or
genetically modified host cells of the present disclosure may also include,
but are not limited to,
cannabigerolic acid (CBGA), cannabigerolic acid monomethylether, (CBGAM),
cannabigerol
(CBG), cannabigerol monomethylether (CBGM), cannabigerovarinic acid (CBGVA),
cannabigerovarin (CBGV), cannabichromenic acid (CBCA), cannabichromene (CBC),
cannabichromevarinic acid (CBCVA), cannabichromevarin (CBCV), cannabidiolic
acid (CBDA),
cannabidiol (CBD), cannabidiol monomethylether (CBDM), cannabidiol-C4 (CBD-
C4),
cannabidivarinic acid (CBDVA), cannabidivarin (CBDV), cannabidiorcol (CBD-C1),
tetrahydrocannabinolic acid A (THCA-A), A9¨tetrahydrocannabinolic acid B (THCA-
B), A9¨
tetrahydrocannabinol (THC), A9¨tetrahydrocannabinolic acid-C4(THCA-C4), A9¨
tetrahydrocannabinol-C4 (THC-C4), ¨tetrahydrocannabivarinic acid (THCVA),
A9¨
tetrahydrocannabivarin (THCV), A9¨ tetrahydrocannabiorcolic acid (THCA-C1),
A9¨
tetrahydrocannabiorcol (THC-C1), A7¨cis-iso-tetrahydrocannabivarin, A8¨
tetrahydrocannabinolic acid (A8¨THCA), A8¨tetrahydrocannabinol (A8¨THC),
cannabicyclolic acid
(CBLA), cannabicyclol (CBL), cannabicyclovarin (CBLV), cannabielsoic acid A
(CBEA-A),
cannabielsoic acid B (CBEA- B), cannabielsoin (CBE), canna bielsoinic acid,
cannabicitranic acid,
cannabinolic acid (CBNA), cannabinol (CBN), cannabinol methylether (CBNM),
cannabinol-C4,
(CBN-C4), cannabivarin (CBV), cannabinol-C2(CNB-C2), cannabiorcol (CBN-C1),
cannabinodiol
(CBND), cannabinodivarin (CBVD), cannabitriol (CBT), 10-ethyoxy-9-hydroxy-
delta-6a-
tetrahydrocannabinol, 8,9-dihydroxyl-delta-6a-tetrahydrocannabinol,
cannabitriolvarin
(CBTVE), dehydrocannabifuran (DCBF), cannabifuran (CBF), cannabichromanon
(CBCN),
cannabicitran (CBT), 10-oxo-delta-6a-tetrahydrocannabinol (OTHC), delta-9-cis-
59
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
tetrahydrocannabinol (cis-THC), 3,4,5,6-tetrahydro-7-hydroxy-alpha-alpha-2-
trimethy1-9-n-
propy1-2,6-methano-2H-1-benzoxocin-5-methanol (OH-iso-HHCV), cannabiripsol
(CBR),
trihydroxy-delta-9-tetrahydrocannabinol (tri0H-THC), and derivatives of any of
the foregoing.
Additional cannabinoid derivatives that can be produced with the methods or
genetically modified host cells of the present disclosure may also include,
but are not limited to,
2-gerany1-5-pentyl-resorcylic acid, 2-gerany1-5-(4-pentyny1)-resorcylic acid,
2-gerany1-5- (trans-
2-penteny1)-resorcylic acid, 2-gerany1-5-(4-methylhexyl)-resorcylic acid, 2-
gerany1-5- (5-hexynyl)
resorcylic acid, 2-gerany1-5-(trans-2-hexeny1)-resorcylic acid, 2-gerany1-5-(5-
hexenyI)-resorcylic
acid, 2-gerany1-5-heptyl-resorcylic acid, 2-gerany1-5-(6-heptynoic)-
resorcylic acid, 2-gerany1-5-
octyl-resorcylic acid, 2-gerany1-5-(trans-2-octeny1)-resorcylic acid, 2-
gerany1-5-nonyl-resorcylic
acid, 2-gerany1-5-(trans-2-nonenyl) resorcylic acid, 2- gerany1-5-decyl-
resorcylic acid, 2-gerany1-
5-(4-phenylbuty1)-resorcylic acid, 2-gerany1-5-(5- phenylpentyI)-resorcylic
acid, 2-gerany1-5-(6-
phenylhexyl)-resorcylic acid, 2-gerany1-5-(7- phenylheptyI)-resorcylic acid,
(6aR,10aR)-1-
hydroxy-6,6,9-trimethy1-3-propy1-6a,7,8,10a- tetrahydro-6H-dibenzo[b,d]pyran-2-
carboxylic
acid, (6aR,10aR)-1-hydroxy-6,6,9-trimethyl- 3-(4-methylhexyl)-6a,7,8,10a-
tetrahydro-6H-
dibenzo[b,d]pyran-2-carboxylic acid, (6aR,10aR)-1-hydroxy-6,6,9-trinnethy1-3-
(5-hexeny1)-
6a,7,8,10a-tetrahydro-6H- dibenzo[b,d]pyran-2-carboxylic acid, (6aR,10aR)-1-
hydroxy-6,6,9-
trimethy1-3-(5-hexeny1)- 6a,7,8,10a-tetrahydro-6H-dibenzo[b,d]pyran-2-
carboxylic acid,
(6aR,10aR)-1-hydroxy- 6,6,9-trimethy1-3-(6-heptyny1)-6a,7,8,10a-tetrahydro-6H-
dibenzo[b,d]pyran-2-carboxylic acid, 3-[(2E)-3,7-dimethylocta-2,6-dien-1-y1]-6-
(hexan-2-y1)-2,4-
dihydroxybenzoic acid, 3- [(2E)-3,7-dimethylocta-2,6-dien-1-yI]-2,4-dihydroxy-
6-(2-
methylpentypbenzoic acid, 3- [(2E)-3,7-dimethylocta-2,6-dien-1-yI]-2,4-
dihydroxy-6-(3-
methylpentypbenzoic acid, 3- [(2E)-3,7-dimethylocta-2,6-dien-1-yI]-2,4-
dihydroxy-6-(4-
methylpentypbenzoic acid, 3- [(2E)-3,7-dimethylocta-2,6-dien-1-yI]-2,4-
dihydroxy-6-[(1E)-pent-
1-en-1-yl]benzoic acid, 3- [(2E)-3,7-dimethylocta-2,6-dien-1-y1]-2,4-dihydroxy-
6-[(2E)-pent-2-en-
1-yl]benzoic acid, 3- [(2E)-3,7-dimethylocta-2,6-dien-1-yI]-2,4-dihydroxy-6-
[(2E)-pent-3-en-1-
yl]benzoic acid, 3- [(2E)-3,7-dimethylocta-2,6-dien-1-y1]-2,4-dihydroxy-6-
(pent-4-en-1-yObenzoic
acid, 3- [(2E)-3,7-dimethylocta-2,6-dien-1-yI]-2,4-dihydroxy-6-propylbenzoic
acid, 3-[(2E)-3,7-
dimethylocta-2,6-dien-1-y1]-2,4-dihydroxy-6-butylbenzoic acid, 3-[(2E)-3,7-
dimethylocta- 2,6-
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
dien-1-yI]-2,4-dihydroxy-6-hexylbenzoic acid, 3-[(2E)-3,7-dimethylocta-2,6-
dien-1-0]- 2,4-
dihydroxy-6-heptylbenzoic acid, 3-[(2E)-3,7-dimethylocta-2,6-dien-1-yI]-2,4-
dihydroxy- 6-
octylbenzoic acid, 3-[(2E)-3,7-dimethylocta-2,6-dien-1-yI]-2,4-dihydroxy-6-
nonanylbenzoic acid,
3-[(2E)-3,7-dimethylocta-2,6-dien-1-yI]-2,4-dihydroxy-6- decanylbenzoic acid,
3-[(2E)-3,7-
dimethylocta-2,6-dien-1-y1]-2,4-dihydroxy-6- undecanylbenzoic acid, 6-(4-
chlorobuty1)-3-[(2E)-
3,7-dimethylocta-2,6-dien-1-y1]-2,4- dihydroxybenzoic acid, 3-[(2E)-3,7-
dimethylocta-2,6-dien-
1-yI]-2,4-dihydroxy-6-[4- (methylsulfanyl)butyl]benzoic acid, and others as
listed in Bow, E. W.
and Rimoldi, J. M., "The Structure¨Function Relationships of Classical
Cannabinoids: CB1/CB2
Modulation," Perspectives in Medicinal Chemistry 2016:8, 17-39 doi:
10.4137/PMC.S32171,
incorporated herein by reference. Methods of determining the activity and
properties of
cannabinoids and cannabinoid derivatives are well known (see, e.g., Bow and
Rimoldi, supra),
and can be adapted in view of the present disclosure by the skilled artisan.
Certain non-limiting processes for producing certain compounds of formulas IA
and IB
are exemplified. A skilled artisan will be able to prepare other compounds of
formulas IA and IB
based on the disclosure provided herein.
EXAMPLES
These examples illustrate but do not limit the disclosed invention. Methods
and strains
useful in accordance with this invention can be adapted by a skilled artisan
from US 10,392,635
(incorporated herein by reference).
Strain Construction Examples
Certain strains may be renumbered over time for convenience, as will be
apparent to
the skilled artisan.
Example 1A: Construction of LSC3-16 and LSC3-2
LSC3-2 was iteratively constructed by transforming chemically competent JK9-3d
(LSC3-
1) with pAG304Ga11100SOACCSAAE1, pAG305Ga11100SOACCSAAE1 and
pAG306Ga11100SOACCSAAE1 and controlling their genomic copy number at specific
genomic
loci. pAG304Ga11100SOACCSAAE1 and pAG306Ga11100SOACCSAAE1 were constructed by
first
61
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
amplifying the yeast shuttle vectors designated as pAG304 and pAG306 with the
primers
pAG304_fwd and pAG304_rev to amplify the Saccharomyces cerevisiae prototrophy
genetic
elements in addition, E. coli origins of replication and an ampicillin
resistant expression
cassette. The dual promoter system pGal1 and pGa110 was amplified from genomic
DNA of JK9-
3d with primers Ga11_10_fwd and Ga11_10_rev. The csAAE1 and 0S-T2A-OAC
fragments were
amplified from sequences that were stored in pUC19 subcloning vectors.
Amplified DNA
fragments were mixed at equimolar concentrations with their respective shuttle
vector
sequences (pAG304 or pAG306) and preassembled by Gibson Assembly using the
NEBuilder HiFi
DNA Assembly Mix (NEB E5520S). The final sequences pAG304Ga11100SOACCSAAE1 and
pAG306Ga11100SOACCSAAE1. To generate the DNA fragment pAG305Ga11100SOACCSAAE1,
the template Ga1110CBGA was amplified with Ga11_10_fwd and Ga11_10_rev to
generate the
yeast shuttle vector containing leucine prototrophy, dual expression promoter
pGal1 and
pGa110, and the 0S-T2A-OAC fragment. The csAAE1 fragment was amplified from a
pUC19
subcloning containing the csAAE1 gene fragment using primers CB CSAAE1 fwd and
CB_CSAAE1_rev. The amplified sequences were mixed at equimolar concentrations
and
assembled by Gibson Assembly using the NEBuilder HiFi DNA Assembly Mix.
First a parental strain to LSC3-2, designated as LSC3-16, was generated. LSC3-
16 was
generated by transforming 2 microgram (ug) of AfIll (NEB R05205) linearized
pAG305Ga11100SOACCSAAE1 into chemically competent JK9-3d mating type alpha
cells that
are auxotrophic to leucine, histidine, tryptophan, and uracil. Selection for
pAG305Ga11100SOACCSAAE1 integration was done with leucine prototrophy rescue
on yeast
nitrogen base agar plates with dropout amino acid mixes deficient in leucine
supplemented
with 100 mg/L glucose. Genetic copy number of pAG305Ga11100SOACCSAAE1
integrated at
chromosome III was initially quantitated by qPCR through isolation of sister
clones from the
transformation. The highest copy integrant was taken and designated as LSC3-
16.
Chemically competent LSC3-16 were co-transformed with both
pAG304Ga11100SOACCSAAE1 and pAG306Ga11100SOACCSAAE1 and selected on yeast
nitrogen base agar plates with dropout amino acid mixes deficient in leucine,
uracil and
tryptophan supplemented with 100 mg/L glucose. Genetic copy number for
62
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
pAG304Ga11100SOACCSAAE1, pAG305Ga11100SOACCSAAE1 and pAG306Ga11100SOACCSAAE1
integrated at chromosomes 4, 3 and 5 were quantitated by qPCR through
isolation of genomic
DNA of sister clones on selection plates. A correlation of both total
polyketide and OA:0 molar
ratio was observed. Whole genomic sequencing was done on LSC3-16 and LSC3-2
which had
achieved titers of 150 mg/L and 350 mg/L in shake flask experiments,
respectively. Genetic copy
numbers were determined to be 6 (LSC3-16) and 16 (LSC3-2).
Table of Primers Used
pAG304_fwd AAG AAA GTG ACG ATA CCG TCG
ACC TCG AG
pAG304_rev TTT AAT HG CTA CTA GAG CTC CAA
TTC GCC
CsAAEl_fwd AGC TCT AGT AGC AAA HA AAG CCT
TCG AG
CsAAE1_rev AAT TTT TGA AGG ATC CAC GAT
TAA AAG AAT
GGG TAA AAA CTA TAA GTC C
Ga11_10_fwd TTT TAC CCA TTC ITT TAA TCG
TGG ATC CTT CAA
AAA TTC HA CH UT TTT TGG
Ga11_10_rev GGT GGC GGC GGG GTT TTT TCT
CCT TGA CGT
TAA AG
OSOAC _fwd GAG AAA AAA CCC CGC CGC CAC
CAT GAA CCA
TTT GAG AGC C
OSOAC_rev GAC GGT ATC GTC ACT TTC TTG
GGG TGT AAT C
ga110_rev TCA TGT AAT TAG HA TGT CAC GCT
TAC AU C
ga110_fwd TCT ITT AAT CGT GGA TCC TIC
AAA AAT TCT TAC
TTT UT TTT GG
CB_CSAAE1 _fwd GGA GGG CGT GAA TGT AAG CGT
GAC ATA ACT
AAT TAC ATG ATC All CGA AAT GAC TGA ATT G
CB_CSAAEl_rev TTC UT GCG TCC ATC CAA AAA AAA
AGT AAG
AAT TTT TGA AGG ATC CAC GAT TAA AAG AAT
GGG TAA AAA CTA TAA GTC C
Table of Sequences
pAG30
tcgcgcgtttcggtgatgacggtgaaaacctctgacacatgcagctcccggagacggtcacagcttgtctgtaagcgga
tgccggg
4G3Ill
agcagacaagcccgtcagggcgcgtcagcgggtgttggcgggtgtcggggctggcttaactatgcggcatcagagcaga
ttgtact
00S0A
gagagtgcaccaaacgacattactatatatataatataggaagcatttaatagacagcatcgtaatatatgtgtacttt
gcagttat
CCSAAE
gacgccagatggcagtagtggaagatattctttattgaaaaatagcttgtcaccttacgtacaatcttgatccggagct
tttcttthtt
1
gccgattaagaattaattcggtcgaaaaaagaaaaggagagggccaagagggagggcattggtgactattgagcacgtg
agtat
acgtgattaagcacacaaaggcagcttggagtatgtctgttattaatttcacaggtagttctggtccattggtgaaagt
ttgcggctt
gcagagcacagaggccgcagaatgtgctctagattccgatgctgacttgctgggtattatatgtgtgcccaatagaaag
agaacaa
ttgacccggttattgcaaggaaaatttcaagtcttgtaaaagcatataaaaatagttcaggcactccgaaatacttggt
tggcgtgtt
tcgtaatcaacctaaggaggatgttttggctctggtcaatgattacggcattgatatcgtccaactgcatggagatgag
tcgtggca
63
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
agaataccaagagttcctcggtttgccagttattaaaagactcgtatttccaaaagactgcaacatactactcagtgca
gcttcaca
gaaacctcattcgtttattcccttgtttgattcagaagcaggtgggacaggtgaacttttggattggaactcgatttct
gactgggttg
gaaggcaagagagccccgaaagcttacattttatgttagctggtggactgacgccagaaaatgttggtgatgcgcttag
attaaat
ggcgttattggtgttgatgtaagcggaggtgtggagacaaatggtgtaaaagactctaacaaaatagcaaatttcgtca
aaaatgc
taagaaataggttattactgagtagtatttatttaagtattgtttgtgcacttgcctgcggtgtgaaataccgcacaga
tgcgtaagg
agaaaataccgcatcaggaaattgtaaacgttaatattttgttaaaattcgcgttaaatttttgttaaatcagctcatt
ttttaaccaa
taggccgaaatcggcaaaatcccttataaatcaaaagaatagaccgagatagggttgagtgttgttccagtttggaaca
agagtcc
actattaaagaacgtggactccaacgtcaaagggcgaaaaaccgtctatcagggcgatggcccactacgtgaaccatca
ccctaa
tcaagttttttggggtcgaggtgccgtaaagcactaaatcggaaccctaaagggagcccccgatttagagcttgacggg
gaaagcc
ggcgaacgtggcgagaaaggaagggaagaaagcgaaaggagcgggcgctagggcgctggcaagtgtagcggtcacgctg
cgc
gtaaccaccacacccgccgcgcttaatgcgccgctacagggcgcgtcgcgccattcgccattcaggctgcgcaactgtt
gggaagg
gcgatcggtgcgggcctcttcgctattacgccagctggcgaaggggggatgtgctgcaaggcgattaagttgggtaacg
ccagggt
tttcccagtcacgacgttgtaaaacgacggccagtgaattgtaatacgactcactatagggcgaattggagctcgcaaa
ttaaagc
cttcgagcgtcccaaaaccttctcaagcaaggttttcagtataatgttacatgcgtacacgcgtctgtacagaaaaaaa
agaaaaa
tttgaaatataaataacgttcttaatactaacataactataaaaaaataaatagggacctagacttcaggttgtctaac
tccttcctt
ttcggttagagcggatgtggggggagggcgtgaatgtaagcgtgacataactaattacatgatcattcgaaatgactga
attgttgt
ctcaaaactcttctcatgatcttgtttgttgcagttctaggtaaggatgacaatgggacaactctagtaactttgaata
atgggttcaa
tttcttttgcaaacccaagttaaaggataatctcaattggttcaaatcaatggttgtgtcgtttgaatccttcaatacg
aaaaatatga
ccaattgttctggaccaccacccaaaggtggaacaccaatagcagtggtttcaaaaactctgtcatctacttcattaca
gactctttc
gatttcgatagaactaattttgataccaccgatgttcatagtgtcatcggctctaccgtgtgcatggtagtaaccgtta
gaggtcaatt
cgaaaatgtcaccatgtcttctcaatacttcaccattcaaggttggcatacccttgaaatagacatcgtgatgattacc
gtttaacaa
tgtttttgaggcaccaaacataacaggacctaatgccaattcaccgatacctggcttatttttaggcattgggtaaccg
ttcttatcta
atatgtacaaggtgcaacccatacattgggatgaaaaagaacttaaagattgagcttgcaaaaatgaaccagcagaaaa
agcac
caccgatttctgtaccaccacacatttctataactggcttgtagttagctctacccattaaccacaaatattcgtctac
attagaggct
tcaccggatgaagaaaagcatcttatggtggaccaatcgtaacctgaaacacaatttgtggatttccatgatcttacaa
tagatggt
acgacacccaacattgtgacctttgcatcttgaacaaatttagcgaaaccagagactaaaggactaccgttgtacaagg
caataga
tgcaccatttaacaaactagcataaaccaaccaaggacccatcatccaacccaaattagttggccatactataacgtca
ccttttct
aatatccaaatgagaccaaccatcagcagcagccttcaatggggtggcttgtgtccaaggaattgcttttggttcacct
gtagtacc
actggagaataagatgttagtataagcatcaacaggttgttctctggcagtaaactcgcagtttttaaactccttggct
ctttctaaa
aagtaatcccaagatatgtcaccatctctcaattctgcaccaatgttagaaccactacaagggataactattgccattg
gggattta
gcttcaactactcttgaatacaatggtattctctttttacctctgatgatgtgatcttgtgtgaaaattgccttagctt
tggataatctca
atctagttgagatttcaggggcggaaaatgaatctgctatagagacaactacgtaaccagccaatactatggccaaata
tataaca
acagcatcaacatgcattggcatatcgatggctattgcacaacctttttctaaacccatttcttccaatgcataaccaa
ccaaccaaa
ctctctUctcaattgatctaatgtcaacttattcaaaggcaagtcatcgttaccctcgtctctccaaacgatcatagta
tcgttcaatt
tcttattggagtttacgttcaagcaatttttagctgagttcaagtaaccaccaggtaaccattcagaaccacctgggtt
gttgatgtca
tctcttctcaagatacattctgggtccttagagaaactaattttcatttcatccatcaatactgttctccaatagactt
cagggtttcta
acagaaaattcttggaagtgagaaaaagaagaaattggatctttgtactttacacccaaaaattctttacctctctttt
ccaacaaa
gcacccaaattagttgacttgactttttcagggtctggaatccaagcaggtggggctggaccgaaatccttgtagcaac
cataaaac
aacatttggtgtaaggagaaaggcaaatctggtgacaagatatggttagcgatgttgatccaagtttgaggggttgcag
caccata
attacaaacgatttctgccaatctaccatgtaatgtttctgctacttctgaggtgatacccaatgcgatgaaatctgag
gcaacgact
gaatccaaggacttatagtttttacccattcttttaatcgtggatccttcaaaaattcttactttttttttggatggac
gcaaagaagttt
aataatcatattacatggcattaccaccatatacatatccatatacatatccatatctaatcttacttatatgttgtgg
aaatgtaaag
agccccattatcttagcctaaaaaaaccttctctttggaactttcagtaatacgcttaactgctcattgctatattgaa
gtacggatta
gaagccgccgagcgggtgacagccctccgaaggaagactctcctccgtgcgtcctcgtcttcaccggtcgcgttcctga
aacgcag
atgtgcctcgcgccgcactgctccgaacaataaagattctacaatactagcttttatggttatgaagaggaaaaattgg
cagtaacc
tggccccacaaaccttcaaatgaacgaatcaaattaacaaccataggatgataatgcgattagttttttagccttattt
ctggggtaa
ttaatcagcgaagcgatgatttttgatctattaacagatatataaatgcaaaaactgcataaccactttaactaatact
ttcaacatt
ttcggtttgtattacttcttattcaaatgtaataaaagtatcaacaaaaaattgttaatatacctctatactttaacgt
caaggagaaa
64
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
aaaccccggatccgtaatacgactcactataggatgaaccatttgagagccgaaggtcctgcctccgtattagccatag
gtacagc
caacccagaaaacatattgatccaagatgaatttcctgattattacttcagagttaccaagagtgaacacatgactcaa
ttgaagg
aaaagtttagaaaaatatgtgataagtctatgatcagaaagagaaactgcttcttgaacgaagaacatttgaagcaaaa
tccaag
attggtagaacacgaaatgcaaacattggatgccagacaagacatgttagttgtcgaagttcctaaattgggtaaagat
gcttgtg
caaaagccattaaggaatggggtcaaccaaagtcaaagatcactcatttgatttttacaagtgcatctactacagatat
gcctggtg
cagactaccactgtgccaaattgttaggtttgtcaccatccgttaagagagtcatgatgtatcaattaggttgctacgg
tggtggtac
tgttttgagaatcgctaaggatattgcagaaaacaacaagggtgccagagtattagctgtttgttgcgacattatggct
tgcttgttt
agaggtccaagtgattctgacttggaattgttagttggtcaagctatcttcggtgacggtgctgctgctgttattgttg
gtgcagaacc
tgacgaatctgttggtgaaagaccaatatttgaattagtcagtacaggtcaaaccatcttgcctaattctgaaggtaca
attggtggt
catataagagaagcaggtttgatcttcgatttgcacaaagacgttccaatgttaatctctaacaacatagaaaagtgtt
tgatagaa
gcattcactcctataggtatctcagattggaactctattttctggataacacatccaggtggtaaagccattttggata
aggttgaag
aaaaattggatttgaagaaagaaaagtttgtagatagtagacatgttttatctgaacacggtaacatgtcttcatccac
tgtcttgtt
cgtaatggatgaattgagaaagagatcattagaagagggtaaatctactactggtgacggttttgaatggggtgtctta
tttggtttc
ggtcctggtttgaccgtcgaaagagtagttgtcagatcagtaccaattaaatatgaaggtagaggttccttgttaactt
gtggtgacg
ttgaagaaaacccaggtcctatggccgtcaagcatttgatagtattgaagtttaaagatgaaatcacagaagctcaaaa
ggaaga
atttttcaagacctacgttaatttggtcaacattatacctgctatgaaagatgtatactggggtaaagacgttacacaa
aagaaaga
agaaggttatacacacattgtcgaagtaaccttcgaatcagttgaaactatccaagattacatcattcatccagctcac
gttgettt
ggtgacgtttacagatccttctgggaaaaattgttgatcttcgattacaccccaagaaagtgatgatgggctgcaggaa
ttcgatat
caagcttatcgataccgtcgacctcgagtcatgtaattagttatgtcacgcttacattcacgccctccccccacatccg
ctctaaccg
aaaaggaaggagttagacaacctgaagtctaggtccctatttatttttttatagttatgttagtattaagaacgttatt
tatatttcaa
atttttcttttttttctgtacagacgcgtgtacgcatgtaacattatactgaaaaccttgcttgagaaggttttgggac
gctcgaaggct
ttaatttgcggccggtacccagcttttgttccctttagtgagggttaattccgagcttggcgtaatcatggtcatagct
gtttcctgtgt
gaaattgttatccgctcacaattccacacaacataggagccggaagcataaagtgtaaagcctggggtgcctaatgagt
gaggta
actcacattaattgcgttgcgctcactgcccgctttccagtcgggaaacctgtcgtgccagctgcattaatgaatcggc
caacgcgc
ggggagaggcggtttgcgtattgggcgctcttccgcttcctcgctcactgactcgctgcgctcggtcgttcggctgcgg
cgagcggta
tcagctcactcaaaggcggtaatacggttatccacagaatcaggggataacgcaggaaagaacatgtgagcaaaaggcc
agcaa
aaggccaggaaccgtaaaaaggccgcgttgctggcgtttttccataggctcggcccccctgacgagcatcacaaaaatc
gacgctc
aagtcagaggtggcgaaacccgacaggactataaagataccaggcgttcccccctggaagctccctcgtgcgctctcct
gttccga
ccctgccgcttaccggatacctgtccgcctttctcccttcgggaagcgtggcgctttctcaatgctcacgctgtaggta
tctcagttcg
gtgtaggtcgttcgctccaagctgggctgtgtgcacgaaccccccgttcagcccgaccgctgcgccttatccggtaact
atcgtcttg
agtccaacccggtaagacacgacttatcgccactggcagcagccactggtaacaggattagcagagcgaggtatgtagg
cggtgc
tacagagttcttgaagtggtggcctaactacggctacactagaaggacagtatttggtatctgcgctctgctgaagcca
gttaccttc
ggaaaaagagttggtagctcttgatccggcaaacaaaccaccgctggtagcggtggtttttttgtttgcaagcagcaga
ttacgcgc
agaaaaaaaggatctcaagaagatcctttgatcUttctacggggtctgacgctcagtggaacgaaaactcacgttaagg
gattttg
gtcatgagattatcaaaaaggatcttcacctagatccttttaaattaaaaatgaagttttaaatcaatctaaagtatat
atgagtaaa
cttggtctgacagttaccaatgcttaatcagtgaggcacctatctcagcgatctgtctatttcgttcatccatagttgc
ctgactgccc
gtcgtgtagataactacgatacgggagggcttaccatctggccccagtgctgcaatgataccgcgagacccacgctcac
cggctcc
agatttatcagcaataaaccagccagccggaagggccgagcgcagaagtggtcctgcaactttatccgcctccatccag
tctatta
attgttgccgggaagctagagtaagtagttcgccagttaatagtttgcgcaacgttgttgccattgctacaggcatcgt
ggtgtcacg
ctcgtcgtttggtatggcttcattcagctccggttcccaacgatcaaggcgagttacatgatcccccatgttgtgaaaa
aaagcggtt
agctccttcggtcctccgatcgttgtcagaagtaagttggccgcagtgttatcactcatggttatggcagcactgcata
attctcttac
tgtcatgccatccgtaagatgcttttctgtgactggtgagtactcaaccaagtcattctgagaatagtgtatgcggcga
ccgagttgc
tcttgcccggcgtcaatacgggataataccgcgccacatagcagaactttaaaagtgctcatcattggaaaacgttctt
cggggcg
aaaactctcaaggatcttaccgctgttgagatccagttcgatgtaacccactcgtgcacccaactgatcttcagcatca
ttactttca
ccagcgtttctgggtgagcaaaaacaggaaggcaaaatgccgcaaaaaagggaataagggcgacacggaaatgttgaat
actc
atactcttcctttttcaatattattgaagcatttatcagggttattgtctcatgagcggatacatatttgaatgtattt
agaaaaataaa
caaataggggttccgcgcacatttccccgaaaagtgccacctgacgtctaagaaaccattattatcatgacattaacct
ataaaaat
aggcgtatcacgaggccctttcgtc
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
pAG30
attcagctccggttcccaacgatcaaggcgagttacatgatcccccatgttgtgaaaaaaagcggttagctccttcggt
cctccgat
5Ga111
cgttgtcagaagtaagttggccgcagtgttatcactcatggttatggcagcactgcataattctcttactgtcatgcca
tccgtaagat
00S0A
gcttttctgtgactggtgagtactcaaccaagtcattctgagaatagtgtatgcggcgaccgagttgctcttgcccggc
gtcaatacg
CCSAAE
ggataataccgcgccacatagcagaactttaaaagtgctcatcattggaaaacgttcttcggggcgaaaactctcaagg
atcttac
1
cgctgttgagatccagttcgatgtaacccactcgtgcacccaactgatcttcagcatcttttactttcaccagcgtttc
tgggtgagca
aaaacaggaaggcaaaatgccgcaaaaaagggaataagggcgacacggaaatgttgaatactcatactcttcctttttc
aatatt
attgaagcatttatcagggttattgtctcatgagcggatacatatttgaatgtatttagaaaaataaacaaataggggt
tccgcgca
catttccccgaaaagtgccacctgacgtctaagaaaccattattatcatgacattaacctataaaaataggcgtatcac
gaggccct
ttcgtctcgcgcgtttcggtgatgacggtgaaaacctctgacacatgcagctcccggagacggtcacagcttgtctgta
agcggatg
ccgggagcagacaagcccgtcagggcgcgtcagcgggtgttggcgggtgtcggggctggcttaactatgcggcatcaga
gcagat
tgtactgagagtgcaccatatcgactacgtcgtaaggccgtttctgacagagtaaaattcttgagggaactttcaccat
tatgggaa
atggttcaagaaggtattgacttaaactccatcaaatggtcaggtcattgagtgttttttatttgttgtattttttttt
ttttagagaaaa
tcctccaatatcaaattaggaatcgtagtttcatgattttctgttacacctaactttttgtgtggtgccctcctccttg
tcaatattaatg
ttaaagtgcaattctttttccttatcacgttgagccattagtatcaatttgcttacctgtattcctttactatcctcct
ttttctccttcttga
taaatgtatgtagattgcgtatatagtttcgtctaccctatgaacatattccattttgtaatttcgtgtcgtttctatt
atgaatttcattt
ataaagtttatgtacaaatatcataaaaaaagagaatcUtttaagcaaggattttcttaacacttcggcgacagcatca
ccgactt
cggtggtactgttggaaccacctaaatcaccagttctgatacctgcatccaaaacctttttaactgcatcttcaatggc
cttaccttct
tcaggcaagttcaatgacaatttcaacatcattgcagcagacaagatagtggcgatagggtcaaccttattctttggca
aatctgga
gcagaaccgtggcatggttcgtacaaaccaaatgcggtgttcttgtctggcaaagaggccaaggacgcagatggcaaca
aaccca
aggaacctgggataacggaggcttcatcggagatgatatcaccaaacatgttgctggtgattataataccatttaggtg
ggttgggt
tcttaactaggatcatggcggcagaatcaatcaattgatgttgaaccttcaatgtagggaattcgttcttgatggtttc
ctccacagtt
tttctccataatcttgaagaggccaaaacattagctttatccaaggaccaaataggcaatggtggctcatgttgtaggg
ccatgaaa
gcggccattcttgtgattctttgcacttctggaacggtgtattgttcactatcccaagcgacaccatcaccatcgtctt
cctttctcttac
caaagtaaatacctcccactaattctctgacaacaacgaagtcagtacctttagcaaattgtggcttgattggagataa
gtctaaaa
gagagtcggatgcaaagttacatggtcttaagttggcgtacaattgaagttctttacggatttttagtaaaccttgttc
aggtctaac
actaccggtaccccatttaggaccacccacagcacctaacaaaacggcatcaaccttcttggaggcttccagcgcctca
tctggaa
gtgggacacctgtagcatcgatagcagcaccaccaattaaatgattttcgaaatcgaacttgacattggaacgaacatc
agaaata
gctttaagaaccttaatggcttcggctgtgatttcttgaccaacgtggtcacctggcaaaacgacgatcttcttagggg
cagacata
ggggcagacattagaatggtatatccttgaaatatatatatatattgctgaaatgtaaaaggtaagaaaagttagaaag
taagac
gattgctaaccacctattggaaaaaacaataggtccttaaataatattgtcaacttcaagtattgtgatgcaagcattt
agtcatga
acgcttctctattctatatgaaaagccggttccggcctctcacctttcctttttctcccaatttttcagttgaaaaagg
tatatgcgtca
ggcgacctctgaaattaacaaaaaatttccagtcatcgaatttgattctgtgcgatagcgcccctgtgtgttctcgtta
tgttgagga
aaaaaataatggttgctaagagattcgaactcttgcatcttacgatacctgagtattcccacagttaactgcggtcaag
atatttctt
gaatcaggcgccttagaccgctcggccaaacaaccaattacttgttgagaaatagagtataattatcctataaatataa
cgtUttg
aacacacatgaacaaggaagtacaggacaattgattttgaagagaatgtggattttgatgtaattgttgggattccatt
tttaataa
ggcaataatattaggtatgtggatatactagaagttctcctcgagggtcgatatgcggtgtgaaataccgcacagatgc
gtaagga
gaaaataccgcatcaggaaattgtaaacgttaatattttgttaaaattcgcgttaaatttttgttaaatcagctcattt
tttaaccaat
aggccgaaatcggcaaaatcccttataaatcaaaagaatagaccgagatagggttgagtgttgttccagtttggaacaa
gagtcc
actattaaagaacgtggactccaacgtcaaagggcgaaaaaccgtctatcagggcgatggcccactacgtgaaccatca
ccctaa
tcaagttUttggggtcgaggtgccgtaaagcactaaatcggaaccctaaagggagcccccgatttagagcttgacgggg
aaagcc
ggcgaacgtggcgagaaaggaagggaagaaagcgaaaggagcgggcgctagggcgctggcaagtgtagcggtcacgctg
cgc
gtaaccaccacacccgccgcgcttaatgcgccgctacagggcgcgtcgcgccattcgccattcaggctgcgcaactgtt
gggaagg
gcgatcggtgcgggcctcttcgctattacgccagctggcgaaggggggatgtgctgcaaggcgattaagttgggtaacg
ccagggt
Mcccagtcacgacgttgtaaaacgacggccagtgaattgtaatacgactcactatagggcgaattggagctctagtcgc
aaatta
aagccttcgagcgtcccaaaaccttctcaagcaagglittcagtataatgttacatgcgtacacgcgtctgtacagaaa
aaaaaga
aaaatttgaaatataaataacgttcttaatactaacataactataaaaaaataaatagggacctagacttcaggttgtc
taactcct
tccttttcggttagagcggatgtggggggagggcgtgaatgtaagcgtgacataactaattacatgatcattcgaaatg
actgaatt
gttgtctcaaaactcttctcatgatcttgtttgttgcagttctaggtaaggatgacaatgggacaactctagtaacttt
gaataatggg
66
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
ttcaatttctMgcaaacccaagttaaaggataatctcaattggttcaaatcaatggttgtgtcgtttgaatccttcaat
acgaaaaa
tatgaccaattgttctggaccaccacccaaaggtggaacaccaatagcagtggatcaaaaactctgtcatctacttcat
tacagac
tctttcgatttcgatagaactaattttgataccaccgatgttcatagtgtcatcggctctaccgtgtgcatggtagtaa
ccgttagagg
tcaattcgaaaatgtcaccatgtcttctcaatacttcaccattcaaggttggcatacccttgaaatagacatcgtgatg
attaccgttt
aacaatgtttttgaggcaccaaacataacaggacctaatgccaattcaccgatacctggcttatttttaggcattgggt
aaccgttct
tatctaatatgtacaaggtgcaacccatacattgggatgaaaaagaacttaaagattgagcttgcaaaaatgaaccagc
agaaaa
agcaccaccgatttctgtaccaccacacatttctataactggcttgtagttagctctacccattaaccacaaatattcg
tctacattag
aggcttcaccggatgaagaaaagcatcttatggtggaccaatcgtaacctgaaacacaatttgtggatttccatgatct
tacaatag
atggtacgacacccaacattgtgacctttgcatcttgaacaaatttagcgaaaccagagactaaaggactaccgttgta
caaggca
atagatgcaccatttaacaaactagcataaaccaaccaaggacccatcatccaacccaaattagttggccatactataa
cgtcacc
ttttctaatatccaaatgagaccaaccatcagcagcagccttcaatggggtggcttgtgtccaaggaattgcttttggt
tcacctgta
gtaccactggagaataagatgttagtataagcatcaacaggttgttctctggcagtaaactcgcagtttttaaactcct
tggctctttc
taaaaagtaatcccaagatatgtcaccatctctcaattctgcaccaatgttagaaccactacaagggataactattgcc
attgggga
tttagcttcaactactcttgaatacaatggtattctctttttacctctgatgatgtgatcttgtgtgaaaattgcctta
gctttggataat
ctcaatctagttgagatttcaggggcggaaaatgaatctgctatagagacaactacgtaaccagccaatactatggcca
aatatat
aacaacagcatcaacatgcattggcatatcgatggctattgcacaacctUttctaaacccatttcttccaatgcataac
caaccaac
caaactctctttctcaattgatctaatgtcaacttattcaaaggcaagtcatcgttaccctcgtctctccaaacgatca
tagtatcgttc
aatttcttattggagtttacgttcaagcaatttttagctgagttcaagtaaccaccaggtaaccattcagaaccacctg
ggttgttgat
gtcatctcttctcaagatacattctgggtccttagagaaactaattttcatttcatccatcaatactgttctccaatag
acttcagggtt
tctaacagaaaattcttggaagtgagaaaaagaagaaattggatctttgtactttacacccaaaaattctttacctctc
ttttccaac
aaagcacccaaattagttgacttgacttlitcagggtctggaatccaagcaggtggggctggaccgaaatccttgtagc
aaccata
aaacaacatttggtgtaaggagaaaggcaaatctggtgacaagatatggttagcgatgttgatccaagtttgaggggtt
gcagcac
cataattacaaacgatttctgccaatctaccatgtaatgtttctgctacttctgaggtgatacccaatgcgatgaaatc
tgaggcaac
gactgaatccaaggacttatagtttttacccattcttttaatcgtggatccttcaaaaattcttactttttttttggat
ggacgcaaaga
agtttaataatcatattacatggcattaccaccatatacatatccatatacatatccatatctaatcttacttatatgt
tgtggaaatgt
aaagagccccattatcttagcctaaaaaaaccttctctttggaactttcagtaatacgcttaactgctcattgctatat
tgaagtacg
gattagaagccgccgagcgggtgacagccctccgaaggaagactctcctccgtgcgtcctcgtcttcaccggtcgcgtt
cctgaaac
gcagatgtgcctcgcgccgcactgctccgaacaataaagattctacaatactagcttttatggttatgaagaggaaaaa
ttggcagt
aacctggccccacaaaccttcaaatgaacgaatcaaattaacaaccataggatgataatgcgattagttttttagcctt
atttctgg
ggtaattaatcagcgaagcgatgatttttgatctattaacagatatataaatgcaaaaactgcataaccactttaacta
atactttca
acattttcggtttgtattacttcttattcaaatgtaataaaagtatcaacaaaaaattgttaatatacctctatacttt
aacgtcaagg
agaaaaaaccccggatccgtaatacgactcactataggatgaaccatttgagagccgaaggtcctgcctccgtattagc
cataggt
acagccaacccagaaaacatattgatccaagatgaatttcctgattattacttcagagttaccaagagtgaacacatga
ctcaattg
aaggaaaagtttagaaaaatatgtgataagtctatgatcagaaagagaaactgcttcttgaacgaagaacatttgaagc
aaaatc
caagattggtagaacacgaaatgcaaacattggatgccagacaagacatgttagttgtcgaagttcctaaattgggtaa
agatgct
tgtgcaaaagccattaaggaatggggtcaaccaaagtcaaagatcactcatttgatttttacaagtgcatctactacag
atatgcct
ggtgcagactaccactgtgccaaattgttaggtttgtcaccatccgttaagagagtcatgatgtatcaattaggttgct
acggtggtg
gtactgttttgagaatcgctaaggatattgcagaaaacaacaagggtgccagagtattagctgtttgttgcgacattat
ggcttgctt
gtttagaggtccaagtgattctgacttggaattgttagttggtcaagctatcttcggtgacggtgctgctgctgttatt
gttggtgcag
aacctgacgaatctgttggtgaaagaccaatatttgaattagtcagtacaggtcaaaccatcttgcctaattctgaagg
tacaattg
gtggtcatataagagaagcaggtttgatcttcgatttgcacaaagacgttccaatgttaatctctaacaacatagaaaa
gtgtttga
tagaagcattcactcctataggtatctcagattggaactctattttctggataacacatccaggtggtaaagccatttt
ggataaggt
tgaagaaaaattggatttgaagaaagaaaagtttgtagatagtagacatgttttatctgaacacggtaacatgtcttca
tccactgt
cttgttcgtaatggatgaattgagaaagagatcattagaagagggtaaatctactactggtgacggttttgaatggggt
gtcttattt
ggtttcggtcctggtttgaccgtcgaaagagtagttgtcagatcagtaccaattaaatatgaaggtagaggttccttgt
taacttgtg
gtgacgttgaagaaaacccaggtcctatggccgtcaagcatttgatagtattgaagtttaaagatgaaatcacagaagc
tcaaaa
ggaagaatttttcaagacctacgttaatttggtcaacattatacctgctatgaaagatgtatactggggtaaagacgtt
acacaaaa
gaaagaagaaggttatacacacattgtcgaagtaaccttcgaatcagttgaaactatccaagattacatcattcatcca
gctcacgt
67
CA 03177968 2022- 11-4
17-TT -ZZOZ 996LL10 VD
89
aeme222e4eeeweeeeee4epee4eDeepewe4p440Deewee4e4eee2444eeeee2eeeeeeee2eDe424D
42DODepeT2o2i.epe442TeeTeT2eDiTT422eeD2eeopTpoeeee3o342o2e2DTTDD2eeeTTeeea2DTDO
e224Tee
8o8SSelelpeoloe8pelee1011eeS18eop8&e8peeemB110DeSpeolSenolmOSSepoOpee1505118eel
le
SASeeDSTDSTSTeSS9SSSeeSABTDSeDDSDeTTepS3443433SSSDSTSS3TeBASSeeSSSTTSpeeDSDSTDS
2
eolleop2olleop2o2D12D2D022eDep2o32o2leelp2o2Do2oppeoeopeopeel2o2D2p2DeD102DOe12
12ee
320p2o022e4D2D2OODOeneee2DOeee0eanee2Oeee2e0D0242Dee0onoo2eee2022DeOTTD2e2eTT
leSopoopSenSeeeloopeeSSDleeelpeoSeeelSpoS1SSeSoMSSmmSeepleeloopeoleopeeS1Speloe
DDD224e0D2S2eD4e4D4Sppeeeee8D222eee342Deepppe2242DeeSeee44e4DeDD42e2eepee224442
eDD44
11010e511020e1e0e0opeOelee0eeeepleeelelpopleeeepOODleee2DDOOeleepoeellmleoloOeo
lee
el104111leeell2o2olleeee1124111eleell2Jeee1214eee02eDle32ooeleeee2e22eeVo2le2eo
eo2ooele
ee212122321333elle112eDlelelleemeepp32e2elleeeDeDpeeelDele121eAleeel2emelle121D
eee
eeei.oeeeeo2e3a22324e2ee2e2T4TeTeD2ee2224322eD2eeee2eoeTT2Dee24222e2e422eep2Te2
02ee
000ece301.11epe02e0ce0011011ellellepeOple00eDepp102101e0le0010opecOele15epeeD10
0011e
32De2e222emale2e1.1422212422Dopepalelle2112211e2oe1122eale2e2ee2212221e3e2e2eee
op
244e1.44D22D4e4424444e2eeepe2D2e2ee2D241eDe21.424De4222eepemee2e224DememD4D222e
eD24
eo421.Tee2eo2e4424e2TiTToD22e2epoee22eemeeVee2ee2e322o22eD2ee244422D2e44244e422
e0002
SSI2S121SSDepeDSleeSpelleoeSeDS2SleeSeoSeleeSeoele1S1SSSDSloloel2eAlleeeolSepel
eelSS1
4e3e2432444eeee2e3e2ee2341343e444144eme42ee332334e44e322eee432332ee442e3e3H2e20
4e33444
11e2pe21plele22121eDepeeeeepem2meeeepo3102elleo2ee0112m2e22pellee22eeopeopel2o
4101e0011eolp0401.01peemeemOeeee0ocoOleoleleeplelogemo0p0110pDVemleDpepOp010
DeeneelelepeloSeeeSolSleoleeeleSeeSpeeeSSeDSlopeeeeepeeSepeoSloeepopeellollelSe
opoS
peee24epeeegee244242e424e4eD2De4e4e4e42244e2e44DameD2e2Seenee2Deaee22eamee2DD
3vvsjj
PleeMplle2111411leee21113111000111e011111111311e1111111111eDleolleeDlleeD111100
DeDDeD210e0e2 VOS00
pe1.21.1e8eDSe8eole3S8oSlepee4p2SpSSSSolS12SSDSSI.121SSSDBeolSoSoSSSED1Sopo8eep
e8eD8e iiieD9
2223321e2232ee1213121132e3e31223e2e22333132e321eDeDa1Jpopeee2122Dale21223111232
D231 0EDed
DTTD02124224442DVDTD2DepT2
1SS1SpleoneoeloSlleDDSpS11SpeeDSDSmSeleenSepoSollSelSeelSeSeloSeeSSSDDS11Slleel
lelo
42e334e3343)2334e4443ee324334221.2ee2e3232e233222ee22332e332e33eeewe32e34e444e2
e334322
Do2D132Deopoe22232Doele21223213212epooD2213123Delp222222232122Depeele224212312D
oo2p
e2TDD2442eTeDDTeD442341.4epTOTD4e2DOmpTeppeD20e2i2epTeeTTD2TeeppeTT2eDe2TD42244
DeeeVe
SlelelelSeeeloleepleeelmSeeSleeeeelleeemmleSelDoeolmeSSeeepeolelleSeSleoMmle
222ee442Dep4Jeeee2Dee2242eD4D2De24340222De4D4444D4e24443D4e2ee2ee3434e22eeeeeee
2eD2D2D
eTTe2eD2eD2eeD244421.44444422422o2e422p2DoeopeeepeeeD22DoTe2pop2e422442e0eeeee2
234Too
e442e3D2ee2p2p4D2D24D4e422444e42eDe22ee2e4Depep22Depeepp2242242ee244D442e2eDep2
42
2322e124e422e232e2e32e44e22p3ep42243p332e32e32243e33234e443e23e3e2pe422333pp334
2e2443
42DTepee422DDTeipp2D2p2DDe2DD32eD442Dpnppee2DeD242424D222p2eeDDTD2D442D422e4242
2DTT
5eome105e151D23eolo5leeplomo5D501535ee525omoommo5DoMoDelenopem5Do2loope5Do
41243343432aS1234333432ee3S433333341239Se3De4eSeee4e4DeSSe3eS33DeeeSDSS49Sege34
See3433
De2oTeepeepeoTeD2e2De2TopooDoMp22eTeop4444423224D244232Do2OpeepeT2Doep22eponeep
eo
2eDDOeeeeD2eWTepee2eee22e32DeeTe2222epTee2eDe3DTeiT22DeTeeT2OD22eeeppepTD2epTeT
O
goSeSoSSoSloSSonSD1SSoloSoSloSoloeSlopoloSolDomBopmloSoSSS1lelSoSmSSDSSeSeSSSSA
D
2Dee33223Tee24ee44e32432e33242J4243Deee202J42e3344432333243e3432324423244eepeDe
Dpee422
e212e2leepo2122221n2epe121222eleD2e222332222elepeeD2Deoppeepeop2Dole142112e2212
121
DD4442p2e4eD422TeDwei2D224p2e2DATeeTT222e242eTTiDDD442444p2eDDDeT22DD22D21.44ee
TTTD22e
e8olo8oe88S1111S8ee8e8uo8moeeee8loeleueoemSleoSpelS1SoSpeSepelSloummollmeeeoll
4e4e444e442Dee2ee44e42e4424e442e4e4444444e444e4D3D422e4D42ee2ppeepe2e442e22ee22
eeee2Dpee
lop2DowDeapooDoppApeollepelp2DeD121211221122121.2o1222Dpo2OD12Do2122olelp2eeple
le2D
44eeneD24D2221e24e242eee2eeDDDDepe44e2D4p4e244244eeeee2224D4m4e2eDe4442De242244
44224
ZSt00/IZOZSI1IIci ZS6SZZ/IZOZ OAA
WO 2021/225952
PCT/US2021/030452
cttcaggttgtctaactccttccttttcggttagagcggatgtggggggagggcgtgaatgtaagcgtgacataactaa
ttacatgat
cattcgaaatgactgaattgttgtctcaaaactcactcatgatcttgtttgttgcagttctaggtaaggatgacaatgg
gacaactct
agtaactttgaataatgggttcaatttcttttgcaaacccaagttaaaggataatctcaattggttcaaatcaatggtt
gtgtcgtttg
aatccttcaatacgaaaaatatgaccaattgttctggaccaccacccaaaggtggaacaccaatagcagtggtttcaaa
aactctg
tcatctacttcattacagactctttcgatttcgatagaactaattttgataccaccgatgttcatagtgtcatcggctc
taccgtgtgca
tggtagtaaccgttagaggtcaattcgaaaatgtcaccatgtcttctcaatacttcaccattcaaggttggcataccct
tgaaataga
catcgtgatgattaccgtttaacaatgtttttgaggcaccaaacataacaggacctaatgccaattcaccgatacctgg
cttattttta
ggcattgggtaaccgttcttatctaatatgtacaaggtgcaacccatacattgggatgaaaaagaacttaaagattgag
cttgcaa
aaatgaaccagcagaaaaagcaccaccgatttctgtaccaccacacatttctataactggcttgtagttagctctaccc
attaacca
caaatattcgtctacattagaggcttcaccggatgaagaaaagcatcttatggtggaccaatcgtaacctgaaacacaa
tttgtgg
atttccatgatcttacaatagatggtacgacacccaacattgtgacctttgcatcttgaacaaatttagcgaaaccaga
gactaaag
gactaccgttgtacaaggcaatagatgcaccatttaacaaactagcataaaccaaccaaggacccatcatccaacccaa
attagtt
ggccatactataacgtcaccttttctaatatccaaatgagaccaaccatcagcagcagccttcaatggggtggcttgtg
tccaagga
attgcttttggttcacctgtagtaccactggagaataagatgttagtataagcatcaacaggttgttctctggcagtaa
actcgcagt
ttttaaactccttggctctttctaaaaagtaatcccaagatatgtcaccatctctcaattctgcaccaatgttagaacc
actacaagg
gataactattgccattggggatttagcttcaactactcttgaatacaatggtattctctUttacctctgatgatgtgat
cttgtgtgaa
aattgccttagctttggataatctcaatctagttgagatttcaggggcggaaaatgaatctgctatagagacaactacg
taaccagc
caatactatggccaaatatataacaacagcatcaacatgcattggcatatcgatggctattgcacaacctttttctaaa
cccatttct
tccaatgcataaccaaccaaccaaactctctttctcaattgatctaatgtcaacttattcaaaggcaagtcatcgttac
cctcgtctct
ccaaacgatcatagtatcgttcaatttcttattggagtttacgttcaagcaatttttagctgagttcaagtaaccacca
ggtaaccatt
cagaaccacctgggttgttgatgtcatctcttctcaagatacattctgggtccttagagaaactaattttcatttcatc
catcaatactg
ttctccaatagacttcagggtttctaacagaaaattcttggaagtgagaaaaagaagaaattggatctttgtactttac
acccaaaa
attctttacctctcttttccaacaaagcacccaaattagttgacttgactttttcagggtctggaatccaagcaggtgg
ggctggaccg
aaatccttgtagcaaccataaaacaacatttggtgtaaggagaaaggcaaatctggtgacaagatatggttagcgatgt
tgatcca
agtttgaggggttgcagcaccataattacaaacgatttctgccaatctaccatgtaatgtttctgctacttctgaggtg
atacccaat
gcgatgaaatctgaggcaacgactgaatccaaggacttatagtattacccattcttttaatcgtggatccttcaaaaat
tcttacat
thtttggatggacgcaaagaagtttaataatcatattacatggcattaccaccatatacatatccatatacatatccat
atctaatct
tacttatatgttgtggaaatgtaaagagccccattatcttagcctaaaaaaaccttctctttggaactttcagtaatac
gcttaactgc
tcattgctatattgaagtacggattagaagccgccgagcgggtgacagccctccgaaggaagactctcctccgtgcgtc
ctcgtctt
caccggtcgcgttcctgaaacgcagatgtgcctcgcgccgcactgctccgaacaataaagattctacaatactagcttt
tatggttat
gaagaggaaaaattggcagtaacctggccccacaaaccttcaaatgaacgaatcaaattaacaaccataggatgataat
gcgatt
agttttttagccttatttctggggtaattaatcagcgaagcgatgatttttgatctattaacagatatataaatgcaaa
aactgcataa
ccactttaactaatactttcaacattttcggtttgtattacttcttattcaaatgtaataaaagtatcaacaaaaaatt
gttaatatacc
tctatactttaacgtcaaggagaaaaaaccccggatccgtaatacgactcactataggatgaaccatttgagagccgaa
ggtcctg
cctccgtattagccataggtacagccaacccagaaaacatattgatccaagatgaatttcctgattattacttcagagt
taccaaga
gtgaacacatgactcaattgaaggaaaagtttagaaaaatatgtgataagtctatgatcagaaagagaaactgcttctt
gaacga
agaacatttgaagcaaaatccaagattggtagaacacgaaatgcaaacattggatgccagacaagacatgttagttgtc
gaagtt
cctaaattgggtaaagatgcttgtgcaaaagccattaaggaatggggtcaaccaaagtcaaagatcactcatttgattt
ttacaagt
gcatctactacagatatgcctggtgcagactaccactgtgccaaattgttaggtttgtcaccatccgttaagagagtca
tgatgtatc
aattaggttgctacggtggtggtactgttttgagaatcgctaaggatattgcagaaaacaacaagggtgccagagtatt
agctgttt
gttgcgacattatggcttgcttgtttagaggtccaagtgattctgacttggaattgttagttggtcaagctatcttcgg
tgacggtgctg
ctgctgttattgttggtgcagaacctgacgaatctgttggtgaaagaccaatatttgaattagtcagtacaggtcaaac
catcttgcc
taattctgaaggtacaattggtggtcatataagagaagcaggtttgatcttcgatttgcacaaagacgttccaatgtta
atctctaac
aacatagaaaagtgtttgatagaagcattcactcctataggtatctcagattggaactctattttctggataacacatc
caggtggta
aagccattttggataaggttgaagaaaaattggatttgaagaaagaaaagtttgtagatagtagacatgttttatctga
acacggt
aacatgtcttcatccactgtcttgttcgtaatggatgaattgagaaagagatcattagaagagggtaaatctactactg
gtgacggtt
ttgaatggggtgtcttatttggtttcggtcctggtttgaccgtcgaaagagtagttgtcagatcagtaccaattaaata
tgaaggtag
aggttccttgttaacttgtggtgacgttgaagaaaacccaggtcctatggccgtcaagcatttgatagtattgaagttt
aaagatgaa
69
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
atcacagaagctcaaaaggaagaatttttcaagacctacgttaatttggtcaacattatacctgctatgaaagatgtat
actggggt
aaagacgttacacaaaagaaagaagaaggttatacacacattgtcgaagtaaccttcgaatcagttgaaactatccaag
attaca
tcattcatccagctcacgttggttttggtgacgtttacagatccttctgggaaaaattgttgatcttcgattacacccc
aagaaagtga
tgatgggctgcaggaattcgatatcaagcttatcgataccgtcgacctcgagtcatgtaattagttatgtcacgcttac
attcacgcc
ctccccccacatccgctctaaccgaaaaggaaggagttagacaacctgaagtctaggtccctatttatttttttatagt
tatgttagta
ttaagaacgttatttatatttcaaatttacttttttttctgtacagacgcgtgtacgcatgtaacattatactgaaaac
cttgcttgaga
aggttttgggacgctcgaaggctttaatttgccggccggtacccagcttttgttccctttagtgagggttaattccgag
cttggcgtaa
tcatggtcatagctgtttcctgtgtgaaattgttatccgctcacaattccacacaacataggagccggaagcataaagt
gtaaagcc
tggggtgcctaatgagtgaggtaactcacattaattgcgttgcgctcactgcccgctttccagtcgggaaacctgtcgt
gccagctgc
attaatgaatcggccaacgcgcggggagaggcggtttgcgtattgggcgctcttccgcttcctcgctcactgactcgct
gcgctcggt
cgttcggctgcggcgagcggtatcagctcactcaaaggcggtaatacggttatccacagaatcaggggataacgcagga
aagaa
catgtgagcaaaaggccagcaaaaggccaggaaccgtaaaaaggccgcgttgctggcgtttttccataggctcggcccc
cctgac
gagcatcacaaaaatcgacgctcaagtcagaggtggcgaaacccgacaggactataaagataccaggcgttcccccctg
gaagc
tccctcgtgcgctctcctgttccgaccctgccgcttaccggatacctgtccgcctttctcccttcgggaagcgtggcgc
tttctcaatgc
tcacgctgtaggtatctcagttcggtgtaggtcgttcgctccaagctgggctgtgtgcacgaaccccccgttcagcccg
accgctgcg
ccttatccggtaactatcgtcttgagtccaacccggtaagacacgacttatcgccactggcagcagccactggtaacag
gattagca
gagcgaggtatgtaggcggtgctacagagttcttgaagtggtggcctaactacggctacactagaaggacagtatttgg
tatctgc
gctctgctgaagccagttaccttcggaaaaagagttggtagctcttgatccggcaaacaaaccaccgctggtagcggtg
gttttttt
gtttgcaagcagcagattacgcgcagaaaaaaaggatctcaaga
agatcctttgatcttttctacggggtctgacgctcagtggaa
cgaaaactcacgttaagggattttggtcatgagattatcaaaaaggatcttcacctagatccttttaaattaaaaatga
agattaaa
tca atcta a agtatatatgagta a acttggtctga cagtta ccaatgctta atcagtgaggc a
cctatctcagcgatctgtctatttcg
ttcatccatagttgcctgactgcccgtcgtgtagataactacgatacgggagggcttaccatctggccccagtgctgca
atgata cc
gcgagacccacgctcaccggctccagatttatcagcaataaaccagccagccggaagggccgagcgcagaagtggtcct
gcaac
tttatccgcctccatccagtctattaattgttgccgggaagctagagtaagtagttcgccagttaatagtttgcgcaac
gttgttgcca
ttgctacaggcatcgtggtgtcacgctcgtcgtttggtatggcttcattcagctccggttcccaacgatcaaggcgagt
tacatgatcc
cccatgttgtgaaaaaaagcggttagctccttcggtcctccgatcgttgtcagaagtaagaggccgcagtgttatcact
catggtta
tggcagcactgcataattctcttactgtcatgccatccgtaagatgcttttctgtgactggtgagta ctca a cc
a agtcattctgaga a
tagtgtatgcggcgaccgagttgctcttgcccggcgtcaatacgggataataccgcgccacatagcagaactttaaaag
tgctcatc
attggaaaacgttcttcggggcgaaaactctcaaggatcttaccgctgttgagatccagttcgatgtaacccactcgtg
cacccaac
tgatcttcagcatcUttactttcaccagcgtttctgggtgagcaaaaacaggaaggcaaaatgccgcaaaaaagggaat
aagggc
gacacggaaatgttgaatactcatactcttcctttttcaatattattgaagcatttatcagggttattgtctcatgagc
ggatacatatt
tga atgtatttaga a a a ata a a ca a ataggggttccgcgca catttccccgaa a
agtgccacctga cgtcta agaa a ccattatta
tcatgacattaacctataaaaataggcgtatcacgaggccctttcgtc
Ga1110 TAAACTCCATCAAATGGTCAGGTCATTGAGTGTTTTTTATTTGTTGTATTTTTTTTTTTTTAGAGAAAA
cbga
TCCTCCAATATCAAATTAGGAATCGTAGTTTCATGATTTTCTGTTACACCTAACTTTTTGTGTGGTGCC
CTCCTCCTTGTCAATATTAATGTTAAAGTGCAA1TC11111CCITATCACGTTGAGCCATTAGTATCAA
TTTGCTTACCTGTATTCCTTTACTATCCTCCTTTTTCTCCTTCTTGATAAATGTATGTAGATTGCGTATA
TAGTTTCGTCTACCCTATGAACATATTCCATTTTGTAATTTCGTGICGTTTCTATTATGAATTTCATTTA
TAAAGTTTATGTACAAATATCATAAAAAAAGAGAATCTTTTTAAGCAAGGATTTTCTTAACTTCTTCG
GCGACAGCATCACCGACTTCGGTGGTACTGTTGGAACCACCTAAATCACCAGTTCTGATACCTGCAT
CCAAAACCTTTTTAACTGCATCTTCAATGGCCTTACCTTCTTCAGGCAAGTTCAATGACAATTTCAAC
ATCATTGCAGCAGACAAGATAGTGGCGATAGGGTCAACCTTATTCTTTGGCAAATCTGGAGCAGAA
CCGTGGCATGGTTCGTACAAACCAAATGCGGTGTTCTTGTCTGGCAAAGAGGCCAAGGACGCAGAT
GGCAACAAACCCAAGGAACCTGGGATAACGGAGGCTTCATCGGAGATGATATCACCAAACATGTT
GCTGGTGATTATAATACCATTTAGGTGGGTTGGGTTCTTAACTAGGATCATGGCGGCAGAATCAAT
CAATTGATGTTGAACCTTCAATGTAGGGAATTCGTTCTTGATGGTTTCCTCCACAG11111CTCCATA
ATCTTGAAGAGGCCAAAACATTAGCTTTATCCAAGGACCAAATAGGCAATGGTGGCTCATGTTGTA
GGGCCATGAAAGCGGCCATTCTTGTGATTCTTTGCACTTCTGGAACGGTGTATTGTTCACTATCCCA
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
AGCGACACCATCACCATCGTCTTCCTTTCTCTTACCAAAGTAAATACCTCCCACTAATTCTCTGACAAC
AACGAAGTCAGTACCTTTAGCAAATTGTGGCTTGATTGGAGATAAGTCTAAAAGAGAGTCGGATGC
AAAGTTACATGGTCTTAAGTTGGCGTACAATTGAAGTTCTTTACGGATTTTTAGTAAACCTTGTTCAG
GTCTAACACTACCGGTACCCCATTTAGGACCACCCACAGCACCTAACAAAACGGCATCAACCTTCTT
GGAGGCTTCCAGCGCCTCATCTGGAAGTGGGACACCTGTAGCATCGATAGCAGCACCACCAATTAA
ATGATTTTCGAAATCGAACTTGACATTGGAACGAACATCAGAAATAGCTTTAAGAACCTTAATGGCT
TCGGCTGTGATTTCTTGACCAACGTGGTCACCTGGCAAAACGACGATCTTCTTAGGGGCAGACATA
GGGGCAGACATTAGAATGGTATATCCTTGAAATATATATATATATTGCTGAAATGTAAAAGGTAAG
AAAAGTTAGAAAGTAAGACGATTGCTAACCACCTATTGGAAAAAACAATAGGTCCTTAAATAATATT
GTCAACTTCAAGTATTGTGATGCAAGCATTTAGTCATGAACGCTTCTCTATTCTATATGAAAAGCCG
GTTCCGGCCTCTCACCTTTCCTTTTTCTCCCAATTTTTCAGTTGAAAAAGGTATATGCGTCAGGCGAC
CTCTGAAATTAACAAAAAATTTCCAGTCATCGAATTTGATTCTGTGCGATAGCGCCCCTGTGTGTTCT
CGTTATGTTGAGGAAAAAAATAATGGTTGCTAAGAGATTCGAACTCTTGCATCTTACGATACCTGAG
TATTCCCACAGTTAACTGCGGTCAAGATATTTCTTGAATCAGGCGCCTTAGACCGCTCGGCCAAACA
ACCAATTACTTGTTGAGAAATAGAGTATAATTATCCTATAAATATAACGTTTTTGAACACACATGAAC
AAGGAAGTACAGGACAATTGATTTTGAAGAGAATGTGGATTTTGATGTAATTGTTGGGATTCCATTT
TTAATAAGGCAATAATATTAGGTATGTGGATATACTAGAAGTTCTCCTCGAGGGTCGATATGCGGT
GTGAAATACCGCACAGATGCGTAAGGAGAAAATACCGCATCAGGAAATTGTAAACGTTAATATTTT
GTTAAAATTCGCGTTAAATTTTTGTTAAATCAGCTCATTTTTTAACCAATAGGCCGAAATCGGCAAAA
TCCCTTATAAATCAAAAGAATAGACCGAGATAGGGTTGAGTGTTGTTCCAGTTTGGAACAAGAGTC
CACTATTAAAGAACGTGGACTCCAACGTCAAAGGGCGAAAAACCGTCTATCAGGGCGATGGCCCAC
TACGTGAACCATCACCCTAATCAAGTTTTTTGGGGTCGAGGTGCCGTAAAGCACTAAATCGGAACCC
TAAAGGGAGCCCCCGATTTAGAGCTTGACGGGGAAAGCCGGCGAACGTGGCGAGAAAGGAAGGG
AAGAAAGCGAAAGGAGCGGGCGCTAGGGCGCTGGCAAGTGTAGCGGTCACGCTGCGCGTAACCA
CCACACCCGCCGCGCTTAATGCGCCGCTACAGGGCGCGTCGCGCCATTCGCCATTCAGGCTGCGCA
ACTGTTGGGAAGGGCGATCGGTGCGGGCCTCTTCGCTATTACGCCAGCTGGCGAAGGGGGGATGT
GCTGCAAGGCGATTAAGTTGGGTAACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCC
AGTGAATTGTAATACGACTCACTATAGGGCGAATTGGAGCTCTAGTCGCAAATTAAAGCCTTCGAG
CGTCCCAAAACCTTCTCAAGCAAGGTTTTCAGTATAATGTTACATGCGTACACGCGTCTGTACAGAA
AAAAAAGAAAAATTTGAAATATAAATAACGTTCTTAATACTAACATAACTATAAAAAAATAAATAGG
GACCTAGACTTCAGGTTGTCTAACTCCTTCCTTTTCGGTTAGAGCGGATGTGGGGGGAGGGCGTGA
ATGTAAGCGTGACATAACTAATTACATGACTCGAGGTCGACGGTATCGTTAAATAAAAACGTATACC
AAATATTCAGCGTAGTACAATTICCACATAAACTCGTAGAATCTTCTACCTGCTICAGGGICATAATT
TGTCAAAGCGAAATCTCTAGTTTGCAAGATCAACCAGAAAGCCAAGATGGCATGTGACAACAACAT
AACGTTAGAATTAAAGGCTTGTGGCCAAATGATACCTGCCAAAATGGCTGCGACGTAACTTAACAA
AACGATACCGGAGCAGAACAAAGTCAAATTTCTTGAACCGTACTTAGAAGCCAAGGTACTAATACC
GAACTTTGTGTCACCTTCAACGTCAGAGGCATCCTTGATCAAGGCTAATGCAGAACCCATACTTTTC
ATGAATGCCAACAAAAATGTGAATGAAGGTCTCAATTCGAATGGCAAACCTAAAGCAGCTCTTGAA
GCGTAGTAGAAGGTGAAGTTTGTGATGATATGAGCTAAGAAATTCAACAAAAAGGCAGTACTAGG
GTTTTGTTTCCATCTAAAAGGTGGTACGGAATAGACAATACCACCGAAGATACCGAAACAGTAACC
GAAGATGTACAATGGACCACCCTTCATTTTAATTGTGATGATCAAACCGAACAAGGCTACTATGATA
GACATGATCCATGCAGTATTGACGGATATTTCACCTGAAGCCAAAGGCAAATCTGGTTTGTTAATTC
TGTCGATGTGCAAATCGTATATTTGATTAATTGTAGTGGTGAATGAAGCGATGCACAAGATGGCAA
CTAAAAAGAAAAATGCCTTGAACATCAAGGACCATGAAATTAAGTTAGTGTTATGCAACAATTCTTT
ACCGAATAAACCGCATGCACAAGAAGTAAAAGCGATTATGGTGTATGGTCTTTGCAACTTCCAACAT
GCTTTACCGAAGTTCAAAATTTTTGTGGCAACAGAGTGATTATCACTTTCAGGTGGTTCAGTTTGATT
TGTAGTTGCAGCTCTGATAGAGTTCTTAGCTATAGACAAACTTTCGGAGCACTTATTTTGTAAGTGG
AAGGACTTGGTTGAACAATGTTTTGATGGAAAGTIGTTGTAAGAGTACTTAATAGGTGTCTTTGGAT
71
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
GTCTGTAACACAACAATGATGTTTTTGGATTGTTGTTGTGAGGATTCAATAAGGTATGATAGTTAGT
TTGGAAGGAGAAAGTACAGACGGATGATAAACCCATTTTCAAAAATTCTTACTTTTTTTTTGGATGG
ACGCAAAGAAGTTTAATAATCATATTACATGGCATTACCACCATATACATATCCATATACATATCCAT
ATCTAATCTTACTTATATGTTGTGGAAATGTAAAGAGCCCCATTATCTTAGCCTAAAAAAACCTTCTC
TTTGGAACTTTCAGTAATACGCTTAACTGCTCATTGCTATATTGAAGTACGGATTAGAAGCCGCCGA
GCGGGTGACAGCCCTCCGAAGGAAGACTCTCCTCCGTGCGTCCTCGTCTTCACCGGTCGCGTTCCTG
AAACGCAGATGTGCCTCGCGCCGCACTGCTCCGAACAATAAAGATTCTACAATACTAGCTTTTATGG
TTATGAAGAGGAAAAATTGGCAGTAACCTGGCCCCACAAACCTTCAAATGAACGAATCAAATTAAC
AACCATAGGATGATAATGCGATTAGTTTTTTAGCCTTATTTCTGGGGTAATTAATCAGCGAAGCGAT
GATTTTTGATCTATTAACAGATATATAAATGCAAAAACTGCATAACCACTTTAACTAATACTTTCAAC
ATTTTCGGTTTGTATTACTTCTTATTCAAATGTAATAAAAGTATCAACAAAAAATTGTTAATATACCTC
TATACTITAACGTCAAGGAGAAAAAACCCCGGATCCGTAATACGACTCACTATAGGATGAACCATTT
GAGAGCCGAAGGTCCTGCCTCCGTATTAGCCATAGGTACAGCCAACCCAGAAAACATATTGATCCA
AGATGAATTTCCTGATTATTACTTCAGAGTTACCAAGAGTGAACACATGACTCAATTGAAGGAAAAG
TTTAGAAAAATATGTGATAAGTCTATGATCAGAAAGAGAAACTGCTTCTTGAACGAAGAACATTTG
AAGCAAAATCCAAGATTGGTAGAACACGAAATGCAAACATTGGATGCCAGACAAGACATGTTAGTT
GTCGAAGTTCCTAAATTGGGTAAAGATGCTTGTGCAAAAGCCATTAAGGAATGGGGTCAACCAAAG
TCAAAGATCACTCATTTGATTITTACAAGTGCATCTACTACAGATATGCCTGGTGCAGACTACCACTG
TGCCAAATTGTTAGGTTTGTCACCATCCGTTAAGAGAGTCATGATGTATCAATTAGGTTGCTACGGT
GGTGGTACTGTTTTGAGAATCGCTAAGGATATTGCAGAAAACAACAAGGGTGCCAGAGTATTAGCT
GTTTGTTGCGACATTATGGCTTGCTTGTTTAGAGGTCCAAGTGATTCTGACTTGGAATTGTTAGTTG
GTCAAGCTATCTTCGGTGACGGTGCTGCTGCTGTTATTGTTGGTGCAGAACCTGACGAATCTGTTGG
TGAAAGACCAATATTTGAATTAGTCAGTACAGGTCAAACCATCTTGCCTAATTCTGAAGGTACAATT
GGTGGTCATATAAGAGAAGCAGGTTTGATCTTCGATTTGCACAAAGACGTTCCAATGTTAATCTCTA
ACAACATAGAAAAGTGTTTGATAGAAGCATTCACTCCTATAGGTATCTCAGATTGGAACTCTATITT
CTGGATAACACATCCAGGTGGTAAAGCCATTTTGGATAAGGTTGAAGAAAAATTGGATTTGAAGAA
AGAAAAGTTTGTAGATAGTAGACATGTTTTATCTGAACACGGTAACATGTCTTCATCCACTGTCTTGT
TCGTAATGGATGAATTGAGAAAGAGATCATTAGAAGAGGGTAAATCTACTACTGGTGACGGTTTTG
AATGGGGTGTCTTATTTGGITTCGGTCCTGGTTTGACCGTCGAAAGAGTAGTTGTCAGATCAGTACC
AATTAAATATGAAGGTAGAGGTTCCTTGTTAACTTGTGGTGACGTTGAAGAAAACCCAGGTCCTAT
GGCCGTCAAGCATTTGATAGTATTGAAGTTTAAAGATGAAATCACAGAAGCTCAAAAGGAAGAATT
TTTCAAGACCTACGTTAATTTGGTCAACATTATACCTGCTATGAAAGATGTATACTGGGGTAAAGAC
GTTACACAAAAGAAAGAAGAAGGITATACACACATTGICGAAGTAACCITCGAATCAGTTGAAACT
ATCCAAGATTACATCATTCATCCAGCTCACGTTGGTTTTGGTGACGTTTACAGATCCTTCTGGGAAAA
ATTGTTGATCTTCGATTACACCCCAAGAAAGTGATGATGGGCTGCAGGAATTCGATATCAAGCTTAT
CGATACCGTCGACCTCGAGTCATGTAATTAGTTATGTCACGCTTACATTCACGCCCTCCCCCCACATC
CGCTCTAACCGAAAAGGAAGGAGTTAGACAACCTGAAGTCTAGGTCCCTATTTATTTTTTTATAGTT
ATGTTAGTATTAAGAACGTTATTTATATTTCAAATTTTTCTTTTTTTTCTGTACAGACGCGTGTACGCA
TGTAACATTATACTGAAAACCTTGCTTGAGAAGGTTTTGGGACGCTCGAAGGCTTTAATTTGCGGCC
GGTACCCAGCTITTGITCCCITTAGTGAGGGITAATTCCGAGCTIGGCGTAATCATGGICATAGCTG
TTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAACATAGGAGCCGGAAGCATAAAGTGTA
AAGCCTGGGGTGCCTAATGAGTGAGGTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCA
GTCGGGAAACCTGTCGTGCCAGCTGCATTAATGAATCGGCCAACGCGCGGGGAGAGGCGGTTTGC
GTATTGGGCGCTCTTCCGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCTGCGGCGAGC
GGTATCAGCTCACTCAAAGGCGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAGA
ACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTC
CATAGGCTCGGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCG
ACAGGACTATAAAGATACCAGGCGTTCCCCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCC
72
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
TGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCAATGCTCACGC
TGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTG GGCTGTGTGCACGAACCCCCCGTTC
AGCCCGACCGCTGCGCCTTATCCGGTAACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATC
GCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGT
TCTTGAAGIGGIGGCCTAACTACGGCTACACTAGAAGGACAGTATTIGGTATCTGCGCTCTGCTGAA
GCCAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGG
TGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATC
TTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGAGATTAT
CAAAAAGGATCTTCACCTAGATCCTTTTAAATTAAAAATGAAGTTTTAAATCAATCTAAAGTATATAT
GAGTAAACTTGGTCTGACAGTTACCAATGCTTAATCAGTGAGGCACCTATCTCAGCGATCTGTCTAT
TTCGTTCATCCATAGTTGCCTGACTGCCCGTCGTGTAGATAACTACGATACGGGAGGGCTTACCATC
TGGCCCCAGTGCTGCAATGATACCGCGAGACCCACG CTCACCGGCTCCAGATTTATCAGCAATAAAC
CAGCCAGCCGGAAGGGCCGAGCGCAGAAGTGGTCCTGCAACTTTATCCGCCTCCATCCAGTCTATT
AATTGTTGCCGGGAAGCTAGAGTAAGTAGTTCGCCAGTTAATAGTTTGCGCAACGTTGTTGCCATTG
CTACAGG CATCGTGGTGTCACGCTCGTCGTTTGGTATGGCTTCATTCAG CTCCGGTTCCCAACGATC
AAGGCGAGTTACATGATCCCCCATGTTGTGAAAAAAAGCGGTTAGCTCCTTCGGTCCTCCGATCGTT
GTCAGAAGTAAGTTGGCCGCAGTGTTATCACTCATGGTTATGGCAGCACTGCATAATTCTCTTACTG
TCATGCCATCCGTAAGATGCTTTTCTGTGACTGGTGAGTACTCAACCAAGTCATTCTGAGAATAGTG
TATGCGGCGACCGAGTTGCTCTTGCCCGGCGTCAATACGGGATAATACCGCGCCACATAGCAGAAC
TTTAAAAGTGCTCATCATTGGAAAACGTTCTTCGGGGCGAAAACTCTCAAGGATCTTACCGCTGTTG
AGATCCAGTTCGATGTAACCCACTCGTGCACCCAACTGATCTTCAGCATCTTTTACTTTCACCAGCGT
TTCTGGGTGAGCAAAAACAGGAAGGCAAAATGCCGCAAAAAAGGGAATAAGGGCGACACGGAAA
TGTTGAATACTCATACTCTTCCTTTTTCAATATTATTGAAGCATTTATCAG GGTTATTGTCTCATGAGC
GGATACATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAA
GTGCCACCTGACGTCTAAGAAACCATTATTATCATGACATTAACCTATAAAAATAGGCGTATCACGA
GGCCCTTTCGTCTCGCGCGTTTCGGTGATGACGGTGAAAACCTCTGACACATGCAGCTCCCGGAGA
CGGTCACAGCTTGTCTGTAAGCGGATGCCGGGAGCAGACAAGCCCGTCAGGGCGCGTCAGCGGGT
GTTGGCGGGTGTCGGGGCTGGCTTAACTATGCGGCATCAGAGCAGATTGTACTGAGAGTGCACCA
TATCGACTACGTCGTAAGGCCGTTTCTGACAGAGTAAAATTCTTGAGGGAACTTTCACCATTATGGG
AAATGGTTCAAGAAGGTATTGACT
pAG30 tcgcgcgtttcggtgatgacggtgaaaa cctctgacacatgcagctcccggaga
cggtcacagcttgtctgtaagcggatgccggg
4
agcagacaagcccgtcagggcgcgtcagcgggtgttggcgggtgtcggggctggcttaactatgcggcatcagagcaga
ttgtact
gagagtgcaccaaacgacattactatatatataatataggaagcatttaatagacagcatcgtaatatatgtgtacttt
gcagttat
gacgccagatggcagtagtggaagatattctttattgaaaaatagcttgtcaccttacgtacaatcttgatccggagct
tttcttttttt
gccgattaagaattaattcggtcgaaaaaagaaaaggagagggccaagagggagggcattggtgactattgagcacgtg
agtat
acgtgattaagcacacaaaggcagcttggagtatgtctgttattaatttcacaggtagttctggtccattggtgaaagt
ttgcggctt
gcagagcacagaggccgcagaatgtgctctagattccgatgctgacttgctgggtattatatgtgtgcccaatagaa
agagaaca a
ttgacccggttattgcaaggaaaatttcaagtcttgtaaaagcatataaaaatagttcaggcactccgaaatacttggt
tggcgtgtt
tcgtaatcaacctaaggaggatgattggctctggtcaatgattacggcattgatatcgtccaactgcatggagatgagt
cgtggca
agaataccaagagttcctcggtttgccagttattaaaagactcgtatttccaaaagactgcaacatactactcagtgca
gcttcaca
gaaacctcattcgtttattcccttgtttgattcagaagcaggtgggacaggtgaacttttggattggaactcgatttct
gactgggttg
gaaggcaagagagccccgaaagcttacattttatgttagctggtggactgacgccagaaaatgttggtgatgcgcttag
attaaat
ggcgttattggtgttgatgtaagcggaggtgtggagacaaatggtgtaaaagactctaacaaaatagcaaatttcgtca
aaaatgc
taagaaataggttattactgagtagtatttatttaagtattgtttgtgcacttgcctgcggtgtgaaataccgcacaga
tgcgtaagg
agaaaataccgcatcaggaaattgtaaacgttaatattagttaaaattcgcgttaaatttttgttaaatcagctcattt
ataaccaa
taggccgaaatcggcaaaatcccttataaatcaaaagaatagaccgagatagggttgagtgttgttccagtttggaaca
agagtcc
actattaaagaacgtggactccaacgtcaaagggcgaaa
aaccgtctatcagggcgatggcccactacgtgaaccatcaccctaa
tcaagttttttggggtcgaggtgccgtaaagcactaaatcggaaccctaaagggagcccccgatttagagcttgacggg
gaaagcc
73
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
ggcgaacgtggcgagaaaggaagggaagaaagcgaaaggagcgggcgctagggcgctggcaagtgtagcggtcacgctg
cgc
gtaaccaccacacccgccgcgcttaatgcgccgctacagggcgcgtcgcgccattcgccattcaggctgcgcaactgtt
gggaagg
gcgatcggtgcgggcctcttcgctattacgccagctggcgaaggggggatgtgctgcaaggcgattaagttgggtaacg
ccagggt
Mcccagtcacgacgttgtaaaacgacggccagtgaattgtaatacgactcactatagggcgaattggagctctagtacg
gattag
aagccgccgagcgggcgacagccctccgacggaagactctcctccgtgcgtcctcgtcttcaccggtcgcgttcctgaa
acgcaga
tgtgcctcgcgccgcactgctccgaacaataaagattctacaatactagcttttatggttatgaagaggaaaaattggc
agtaacct
ggccccacaaaccttcaaattaacgaatcaaattaacaaccataggatgataatgcgattagttttttagccttatttc
tggggtaat
taatcagcgaagcgatgatttttgatctattaacagatatataaatggaaaagctgcataaccactttaactaatactt
tcaacattt
tcagtttgtattacttcttattcaaatgtcataaaagtatcaacaaaaaattgttaatatacctctatactttaacgtc
aaggagaaa
aaaccccggattctagaactagtggatcccccatcacaagtttgtacaaaaaagctgaacgagaaacgtaaaatgatat
aaatat
caatatattaaattagattttgcataaaaaacagactacataatactgtaaaacacaacatatccagtcactatggcgg
ccgcatta
ggcaccccaggctttacactttatgcttccggctcgtataatgtgtggattttgagttaggatccgtcgagattttcag
gagctaagg
aagctaaaatggagaaaaaaatcactggatataccaccgttgatatatcccaatggcatcgtaaagaacattttgaggc
atttcag
tcagttgctcaatgtacctataaccagaccgttcagctggatattacggcctttttaaagaccgtaaagaaaaataagc
acaagttt
tatccggcctttattcacattcttgcccgcctgatgaatgctcatccggaattccgtatggcaatgaaagacggtgagc
tggtgatat
gggatagtgttcacccttgttacaccgttttccatgagcaaactgaaacgttttcatcgctctggagtgaataccacga
cgatttccg
gcagtttctacacatatattcgcaagatgtggcgtgttacggtgaaaacctggcctatttccctaaagggtttattgag
aatatgtttt
tcgtctcagccaatccctgggtgagtttcaccagttttgatttaaacgtggccaatatggacaacttcttcgcccccgt
tttcaccatg
ggcaaatattatacgcaaggcgacaaggtgctgatgccgctggcgattcaggttcatcatgccgtctgtgatggcttcc
atgtcggc
agaatgcttaatgaattacaacagtactgcgatgagtggcagggcggggcgtaaacgccgcgtggatccggcttactaa
aagcca
gataacagtatgcgtatttgcgcgctgatttttgcggtataagaatatatactgatatgtatacccgaagtatgtcaaa
aagaggtat
gctatgaagcagcgtattacagtgacagttgacagcgacagctatcagttgctcaaggcatatatgatgtcaatatctc
cggtctgg
taagcacaaccatgcagaatgaagcccgtcgtctgcgtgccgaacgctggaaagcggaaaatcaggaagggatggctga
ggtcg
cccggtttattgaaatgaacggctcttttgctgacgagaacaggggctggtgaaatgcagtttaaggtttacacctata
aaagaga
gagccgttatcgtctgtttgtggatgtacagagtgatattattgacacgcccgggcgacggatggtgatccccctggcc
agtgcacg
tctgctgtcagataaagtctcccgtgaactttacccggtggtgcatatcggggatgaaagctggcgcatgatgaccacc
gatatggc
cagtgtgccggtctccgttatcggggaagaagtggctgatctcagccaccgcgaaaatgacatcaaaaacgccattaac
ctgatgt
tctggggaatataaatgtcaggctcccttatacacagccagtctgcaggtcgaccatagtgactggatatgttgtgttt
tacagtatt
atgtagtctgttttttatgcaaaatctaatttaatatattgatatttatatcattttacgtttctcgttcagctttctt
gtacaaagtggtg
atgggctgcaggaattcgatatcaagcttatcgataccgtcgacctcgagtcatgtaattagttatgtcacgcttacat
tcacgccct
ccccccacatccgctctaaccgaaaaggaaggagttagacaacctgaagtctaggtccctatttattthttatagttat
gttagtatt
aagaacgttatttatatttcaaatttttcttttttttctgtacagacgcgtgtacgcatgtaacattatactgaaaacc
ttgcttgagaa
ggttttgggacgctcgaaggctttaatttgcggccggtacccagcttttgttccctttagtgagggttaattccgagct
tggcgtaatc
atggtcatagctgtttcctgtgtgaaattgttatccgctcacaattccacacaacataggagccggaagcataaagtgt
aaagcctg
gggtgcctaatgagtgaggtaactcacattaattgcgttgcgctcactgcccgctttccagtcgggaaacctgtcgtgc
cagctgcat
taatgaatcggccaacgcgcggggagaggcggtttgcgtattgggcgctcttccgcttcctcgctcactgactcgctgc
gctcggtc
gttcggctgcggcgagcggtatcagctcactcaaaggcggtaatacggttatccacagaatcaggggataacgcaggaa
agaac
atgtgagcaaaaggccagcaaaaggccaggaaccgtaaaaaggccgcgttgctggcgtttttccataggctcggccccc
ctgacg
agcatcacaaaaatcgacgctcaagtcagaggtggcgaaacccgacaggactataaagataccaggcgttcccccctgg
aagct
ccctcgtgcgctctcctgttccgaccctgccgcttaccggatacctgtccgcctttctcccttcgggaagcgtggcgct
ttctcaatgct
cacgctgtaggtatctcagttcggtgtaggtcgttcgctccaagctgggctgtgtgcacgaaccccccgttcagcccga
ccgctgcg
ccttatccggtaactatcgtcttgagtccaacccggtaagacacgacttatcgccactggcagcagccactggtaacag
gattagca
gagcgaggtatgtaggcggtgctacagagttcttgaagtggtggcctaactacggctacactagaaggacagtatttgg
tatctgc
gctctgctgaagccagttaccttcggaaaaagagttggtagctcttgatccggcaaacaaaccaccgctggtagcggtg
gttttttt
gtttgcaagcagcagattacgcgcagaaaaaaaggatctcaaga
agatcctttgatcttttctacggggtctgacgctcagtggaa
cgaaaactcacgttaagggattttggtcatgagattatcaaaaaggatcttcacctagatccttttaaattaaaaatga
agttttaaa
tcaatctaa agtatatatgagtaaacttggtctgacagtta
ccaatgcttaatcagtgaggcacctatctcagcgatctgtctatttcg
ttcatccatagttgcctgactgcccgtcgtgtagataactacgatacgggagggcttaccatctggccccagtgctgca
atgata cc
74
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
gcgagacccacgctcaccggctccagatttatcagcaataaaccagccagccggaagggccgagcgcagaagtggtcct
gcaac
tttatccgcctccatccagtctattaattgttgccgggaagctagagtaagtagttcgccagttaatagtttgcgcaac
gttgttgcca
ttgctacaggcatcgtggtgtcacgctcgtcgtttggtatggcttcattcagctccggttcccaacgatcaaggcgagt
tacatgatcc
cccatgttgtgaaaaaaagcggttagctccttcggtcctccgatcgttgtcagaagtaagttggccgcagtgttatcac
tcatggtta
tggcagcactgcataattctcttactgtcatgccatccgtaagatgcttttctgtgactggtgagtactcaaccaagtc
attctgagaa
tagtgtatgcggcgaccgagttgctcttgcccggcgtcaatacgggataataccgcgccacatagcagaactttaaaag
tgctcatc
attggaaaacgttcttcggggcgaaaactctcaaggatcttaccgctgttgagatccagttcgatgtaacccactcgtg
cacccaac
tgatcttcagcatcttttactttcaccagcgtttctgggtgagcaaaaacaggaaggcaaaatgccgcaaaaaagggaa
taagggc
gacacggaaatgttgaatactcatactcttcctttttcaatattattgaagcatttatcagggttattgtctcatgagc
ggatacatatt
tgaatgtatttagaaaaataaacaaataggggttccgcgcacatttccccgaaaagtgccacctgacgtctaagaaacc
attatta
tcatgacattaacctataaaaataggcgtatcacgaggccctttcgtc
pAG30
tcgcgcgtttcggtgatgacggtgaaaacctctgacacatgcagctcccggagacggtcacagcttgtctgtaagcgga
tgccggg
6
agcagacaagcccgtcagggcgcgtcagcgggtgttggcgggtgtcggggctggcttaactatgcggcatcagagcaga
ttgtact
gagagtgcaccacgcattcaattcaattcatcattttttttttattcttttttttgatttcggtttctttgaaattttt
ttgattcggtaatct
ccgaacagaaggaagaacgaaggaaggagcacagacttagattggtatatatacgcatatgtagtgttgaagaaacatg
aaatt
gcccagtattcttaacccaactgcacagaacaaaaacctgcaggaaacgaagataaatcatgtcgaaagctacatataa
ggaac
gtgctgctactcatcctagtcctgttgctgccaagctatttaatatcatgcacgaaaagcaaacaaacttgtgtgcttc
attggatgtt
cgtaccaccaaggaattactggagttagttgaagcattaggtcccaaaatttgtttactaaaaacacatgtggatatct
tgactgatt
tttccatggagggcacagttaagccgctaaaggcattatccgccaagtacaatatttactcttcgaagacagaaaattt
gctgacat
tggtaatacagtcaaattgcagtactctgcgggtgtatacagaatagcagaatgggcagacattacgaatgcacacggt
gtggtgg
gcccaggtattgttagcggtttgaagcaggcggcagaagaagtaacaaaggaacctagaggccttttgatgttagcaga
attgtca
tgcaagggctccctatctactggagaatatactaagggtactgttgacattgcgaagagcgacaaagattttgttatcg
gctttattg
ctcaaagagacatgggtggaagagatgaaggttacgattggttgattatgacacccggtgtgggtttagatgacaaggg
agacgc
attgggtcaacagtatagaaccgtggatgatgtggtctctacaggatctgacattattattgttggaagaggactattt
gcaaaggg
aagggatgctaaggtagagggtgaacgttacagaaaagcaggctgggaagcatatttgagaagatgcggccagcaaaac
taaa
aaactgtattataagtaaatgcatgtatactaaactcacaaattagagcttcaatttaattatatcagttattaccctg
cggtgtgaa
ataccgcacagatgcgtaaggagaaaataccgcatcaggaaattgtaaacgttaatattttgttaaaattcgcgttaaa
tttttgtta
aatcagctcattttttaaccaataggccgaaatcggcaaaatcccttataaatcaaaagaatagaccgagatagggttg
agtgttg
ttccagtaggaacaagagtccactattaaagaacgtggactccaacgtcaaagggcgaaaaaccgtctatcagggcgat
ggccc
actacgtgaaccatcaccctaatcaagttttttggggtcgaggtgccgtaaagcactaaatcggaaccctaaagggagc
ccccgat
ttagagcttgacggggaaagccggcgaacgtggcgagaaaggaagggaagaaagcgaaaggagcgggcgctagggcgct
ggc
aagtgtagcggtcacgctgcgcgtaaccaccacacccgccgcgcttaatgcgccgctacagggcgcgtcgcgccattcg
ccattca
ggctgcgcaactgttgggaagggcgatcggtgcgggcctcttcgctattacgccagctggcgaaggggggatgtgctgc
aaggcg
attaagttgggtaacgccagggttttcccagtcacgacgttgtaaaacgacggccagtgaattgtaatacgactcacta
tagggcg
aattggagctctagtacggattagaagccgccgagcgggcgacagccctccgacggaagactctcctccgtgcgtcctc
gtcttca
ccggtcgcgttcctgaaacgcagatgtgcctcgcgccgcactgctccgaacaataaagattctacaatactagctttta
tggttatga
agaggaaaaattggcagtaacctggccccacaaaccttcaaattaacgaatcaaattaacaaccataggatgataatgc
gattag
Mtttagccttatttctggggtaattaatcagcgaagcgatgatttttgatctattaacagatatataaatggaaaagct
gcataacc
actttaactaatactacaacatatcagatgtattacttcttattcaaatgtcataaaagtatcaacaaaaaattgttaa
tatacctc
tatactttaacgtcaaggagaaaaaaccccggattctagaactagtggatcccccatcacaagtttgtacaaaaaagct
gaacga
gaaacgtaaaatgatataaatatcaatatattaaattagattttgcataaaaaacagactacataatactgtaaaacac
aacatat
ccagtcactatggcggccgcattaggcaccccaggctttacactttatgcttccggctcgtataatgtgtggattttga
gttaggatcc
gtcgagattttcaggagctaaggaagctaaaatggagaaaaaaatcactggatataccaccgttgatatatcccaatgg
catcgta
aagaacattttgaggcatttcagtcagttgctcaatgtacctataaccagaccgttcagctggatattacggccttttt
aaagaccgt
aaagaaaaataagcacaagttttatccggcctttattcacattcttgcccgcctgatgaatgctcatccggaattccgt
atggcaatg
aaagacggtgagctggtgatatgggatagtgttcacccttgttacaccgttttccatgagcaaactgaaacgttttcat
cgctctgga
gtgaataccacgacgatttccggcagtttctacacatatattcgcaagatgtggcgtgttacggtgaaaacctggccta
tttccctaa
agggtttattgagaatatgtttttcgtctcagccaatccctgggtgagtttcaccagttttgatttaaacgtggccaat
atggacaact
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
tcttcgcccccgttttcaccatgggcaaatattatacgcaaggcgacaaggtgctgatgccgctggcgattcaggttca
tcatgccgt
ctgtgatggcttccatgtcggcagaatgcttaatgaattacaacagtactgcgatgagtggcagggcggggcgtaaacg
ccgcgtg
gatccggcttactaaaagccagataacagtatgcgtatttgcgcgctgatttttgcggtataagaatatatactgatat
gtatacccg
aagtatgtcaaaaagaggtatgctatgaagcagcgtattacagtgacagttgacagcgacagctatcagttgctcaagg
catatat
gatgtcaatatctccggtctggtaagcacaaccatgcagaatgaagcccgtcgtctgcgtgccgaacgctggaaagcgg
aaaatc
aggaagggatggctgaggtcgcccggtttattgaaatgaacggctcttttgctgacgagaacaggggctggtgaaatgc
agtttaa
ggtttacacctataaaagagagagccgttatcgtctgtttgtggatgtacagagtgatattattgacacgcccgggcga
cggatggt
gatccccctggccagtgcacgtctgctgtcagataaagtctcccgtgaactttacccggtggtgcatatcggggatgaa
agctggcg
catgatgaccaccgatatggccagtgtgccggtctccgttatcggggaagaagtggctgatctcagccaccgcgaaaat
gacatca
aaaacgccattaacctgatgttctggggaatataaatgtcaggctcccttatacacagccagtctgcaggtcgaccata
gtgactgg
atatgttgtgttttacagtattatgtagtctgttttttatgcaaaatctaatttaatatattgatatttatatcatttt
acgtttctcgttca
gctttcttgtacaaagtggtgatgggctgcaggaattcgatatcaagcttatcgataccgtcgacctcgagtcatgtaa
ttagttatg
tcacgcttacattcacgccctccccccacatccgctctaaccgaaaaggaaggagttagacaacctgaagtctaggtcc
ctatttat
ttttttatagttatgttagtattaagaacgttatttatatttcaaatttttcttttttttctgtacagacgcgtgtacg
catgtaacattata
ctgaaaaccttgcttgagaaggttttgggacgctcgaaggctttaatttgcggccggtacccagcttttgttcccttta
gtgagggtta
attccgagcttggcgtaatcatggtcatagctgtacctgtgtgaaattgttatccgctcacaattccacacaacatagg
agccggaa
gcataaagtgtaaagcctggggtgcctaatgagtgaggtaactcacattaattgcgttgcgctcactgcccgctttcca
gtcgggaa
acctgtcgtgccagctgcattaatgaatcggccaacgcgcggggagaggcggtttgcgtattgggcgctcttccgcttc
ctcgctca
ctgactcgctgcgctcggtcgttcggctgcggcgagcggtatcagctcactcaaaggcggtaatacggttatccacaga
atcaggg
gataacgcaggaaagaacatgtgagcaaaaggccagcaaaaggccaggaaccgtaaaaaggccgcgttgctggcgtttt
tccat
aggctcggcccccctgacgagcatcacaaaaatcgacgctcaagtcagaggtggcgaaacccgacaggactataaagat
accag
gcgttcccccctggaagctccctcgtgcgctctcctgttccgaccctgccgcttaccggatacctgtccgcctttctcc
cttcgggaag
cgtggcgctttctcaatgctcacgctgtaggtatctcagttcggtgtaggtcgttcgctccaagctgggctgtgtgcac
gaacccccc
gttcagcccgaccgctgcgccttatccggtaactatcgtcttgagtccaacccggtaagacacgacttatcgccactgg
cagcagcc
actggtaacaggattagcagagcgaggtatgtaggcggtgctacagagttcttgaagtggtggcctaactacggctaca
ctagaa
ggacagtatttggtatctgcgctctgctgaagccagttaccttcggaaaaagagttggtagctcttgatccggcaaaca
aaccaccg
ctggtagcggtggtttttttgtttgcaagcagcagattacgcgcagaaaaaaaggatctcaagaagatcctttgatctt
ttctacggg
gtctgacgctcagtggaacgaaaactcacgttaagggattttggtcatgagattatcaaaaaggatcttcacctagatc
cttttaaa
ttaaaaatgaagttttaaatcaatctaaagtatatatgagtaaacttggtctgacagttaccaatgcttaatcagtgag
gcacctatc
tcagcgatctgtctatttcgttcatccatagttgcctgactgcccgtcgtgtagataactacgatacgggagggcttac
catctggccc
cagtgctgcaatgataccgcgagacccacgctcaccggctccagatttatcagcaataaaccagccagccggaagggcc
gagcgc
agaagtggtcctgcaactttatccgcctccatccagtctattaattgttgccgggaagctagagtaagtagttcgccag
ttaatagtt
tgcgcaacgttgttgccattgctacaggcatcgtggtgtcacgctcgtcgtaggtatggcttcattcagctccggttcc
caacgatca
aggcgagttacatgatcccccatgttgtgaaaaaaagcggttagctccttcggtcctccgatcgttgtcagaagtaagt
tggccgca
gtgttatcactcatggttatggcagcactgcataattctcttactgtcatgccatccgtaagatgcttttctgtgactg
gtgagtactca
accaagtcattctgagaatagtgtatgcggcgaccgagttgctcttgcccggcgtcaatacgggataataccgcgccac
atagcag
aactttaaaagtgctcatcattggaaaacgttcttcggggcgaaaactctcaaggatcttaccgctgttgagatccagt
tcgatgta
acccactcgtgcacccaactgatcttcagcatcttttactttcaccagcgtttctgggtgagcaaaaacaggaaggcaa
aatgccgc
aaaaaagggaataagggcgacacggaaatgttgaatactcatactcttcctttttcaatattattgaagcatttatcag
ggttattgt
ctcatgagcggatacatatttgaatgtatttagaaaaataaacaaataggggaccgcgcacatttccccgaaaagtgcc
acctga
cgtctaagaaaccattattatcatgacattaacctataaaaataggcgtatcacgaggccctttcgtc
Gal110
gcaaattaaagccttcgagcgtcccaaaaccttctcaagcaaggttttcagtataatgttacatgcgtacacgcgtctg
tacagaaa
OSOAC
aaaaagaaaaatttgaaatataaataacgttcttaatactaacataactataaaaaaataaatagggacctagacttca
ggttgtc
CSAAE1
taactccttcctMcggttagagcggatgtggggggagggcgtgaatgtaagcgtgacataactaattacatgatcattc
gaaatg
actgaattgttgtctcaaaactcttctcatgatcttgtttgttgcagttctaggtaaggatgacaatgggacaactcta
gtaactttga
ataatgggttcaatttcttttgcaaacccaagttaaaggataatctcaattggttcaaatcaatggttgtgtcgtttga
atccttcaat
acgaaaaatatgaccaattgttctggaccaccacccaaaggtggaacaccaatagcagtggtttcaaaaactctgtcat
ctacttc
attacagactctttcgatttcgatagaactaattttgataccaccgatgttcatagtgtcatcggctctaccgtgtgca
tggtagtaac
76
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
cgttagaggtcaattcgaaaatgtcaccatgtcttctcaatacttcaccattcaaggttggcatacccttgaaatagac
atcgtgatg
attaccgtttaacaatgtttttgaggcaccaaacataacaggacctaatgccaattcaccgatacctggcttatattag
gcattggg
taaccgttcttatctaatatgtacaaggtgcaacccatacattgggatgaaaaagaacttaaagattgagcttgcaaaa
atgaacc
agcagaaaaagcaccaccgatttctgtaccaccacacatttctataactggcttgtagttagctctacccattaaccac
aaatattcg
tctacattagaggcttcaccggatgaagaaaagcatcttatggtggaccaatcgtaacctgaaacacaatttgtggatt
tccatgat
cttacaatagatggtacgacacccaacattgtgaccMgcatcttgaacaaatttagcgaaaccagagactaaaggacta
ccgtt
gtacaaggcaatagatgcaccatttaacaaactagcataaaccaaccaaggacccatcatccaacccaaattagttggc
catact
ataacgtcaccttttctaatatccaaatgagaccaaccatcagcagcagccttcaatggggtggcttgtgtccaaggaa
ttgcttttg
gttcacctgtagtaccactggagaataagatgttagtataagcatcaacaggttgttctctggcagtaaactcgcagtt
tttaaactc
cttggctctttctaaaaagtaatcccaagatatgtcaccatctctcaattctgcaccaatgttagaaccactacaaggg
ataactatt
gccattggggatttagcttcaactactcttgaatacaatggtattctctttttacctctgatgatgtgatcttgtgtga
aaattgccttag
ctttggataatctcaatctagttgagatttcaggggcggaaaatgaatctgctatagagacaactacgtaaccagccaa
tactatgg
ccaaatatataacaacagcatcaacatgcattggcatatcgatggctattgcacaacctttttctaaacccatttcttc
caatgcata
accaaccaaccaaactctctttctcaattgatctaatgtcaacttattcaaaggcaagtcatcgttaccctcgtctctc
caaacgatc
atagtatcgttcaatttcttattggagtttacgttcaagcaatttttagctgagttcaagtaaccaccaggtaaccatt
cagaaccacc
tgggttgttgatgtcatctcttctcaagatacattctgggtccttagagaaactaattttcatttcatccatcaatact
gttctccaata
gacttcagggtttctaacagaaaattcttggaagtgagaaaaagaagaaattggatctttgtactttacacccaaaaat
tctttacct
ctcttttccaacaaagcacccaaattagttgacttgactttttcagggtctggaatccaagcaggtggggctggaccga
aatccttgt
agcaaccataaaacaacatttggtgtaaggagaaaggcaaatctggtgacaagatatggttagcgatgttgatccaaga
tgagg
ggttgcagcaccataattacaaacgatttctgccaatctaccatgtaatgtttctgctacttctgaggtgatacccaat
gcgatgaaa
tctgaggcaacgactgaatccaaggacttatagtttttacccattcttttaatcgtggatccttcaaaaattcttactt
atttttggatg
gacgcaaagaagtttaataatcatattacatggcattaccaccatatacatatccatatacatatccatatctaatctt
acttatatgt
tgtggaaatgtaaagagccccattatcttagcctaaaaaaaccttctctttggaactttcagtaatacgcttaactgct
cattgctata
ttgaagtacggattagaagccgccgagcgggtgacagccctccgaaggaagactctcctccgtgcgtcctcgtcttcac
cggtcgc
gttcctgaaacgcagatgtgcctcgcgccgcactgctccgaacaataaagattctacaatactagcttttatggttatg
aagaggaa
aaattggcagtaacctggccccacaaaccttcaaatgaacgaatcaaattaacaaccataggatgataatgcgattagt
tttttag
ccttatttctggggtaattaatcagcgaagcgatgatttttgatctattaacagatatataaatgcaaaaactgcataa
ccactttaa
ctaatactttcaacattttcggtttgtattacttcttattcaaatgtaataaaagtatcaacaaaaaattgttaatata
cctctatacttt
aacgtcaaggagaaaaaaccccggatccgtaatacgactcactataggatgaaccatttgagagccgaaggtcctgcct
ccgtatt
agccataggtacagccaacccagaaaacatattgatccaagatgaatttcctgattattacttcagagttaccaagagt
gaacaca
tgactcaattgaaggaaaagtttagaaaaatatgtgataagtctatgatcagaaagagaaactgcttcttgaacgaaga
acatttg
aagcaaaatccaagattggtagaacacgaaatgcaaacattggatgccagacaagacatgttagttgtcgaagttccta
aattgg
gtaaagatgcttgtgcaaaagccattaaggaatggggtcaaccaaagtcaaagatcactcatttgatttttacaagtgc
atctacta
cagatatgcctggtgcagactaccactgtgccaaattgttaggtttgtcaccatccgttaagagagtcatgatgtatca
attaggttg
ctacggtggtggtactgttttgagaatcgctaaggatattgcagaaaacaacaagggtgccagagtattagctgtttgt
tgcgacat
tatggcttgcttgtttagaggtccaagtgattctgacttggaattgttagttggtcaagctatcttcggtgacggtgct
gctgctgttat
tgttggtgcagaacctgacgaatctgttggtgaaagaccaatatttgaattagtcagtacaggtcaaaccatcttgcct
aattctgaa
ggtacaattggtggtcatataagagaagcaggtttgatcttcgatttgcacaaagacgttccaatgttaatctctaaca
acatagaa
aagtgtttgatagaagcattcactcctataggtatctcagattggaactctattttctggataacacatccaggtggta
aagccatttt
ggataaggttgaagaaaaattggatttgaagaaagaaaagtttgtagatagtagacatgUttatctgaacacggtaaca
tgtctt
catccactgtcttgttcgtaatggatgaattgagaaagagatcattagaagagggtaaatctactactggtgacggttt
tgaatggg
gtgtcttatttggtttcggtcctggtttgaccgtcgaaagagtagttgtcagatcagtaccaattaaatatgaaggtag
aggttccttg
ttaacttgtggtgacgttgaagaaaacccaggtcctatggccgtcaagcatttgatagtattgaagtttaaagatgaaa
tcacagaa
gctcaaaaggaagaatttttcaagacctacgttaatttggtcaacattatacctgctatgaaagatgtatactggggta
aagacgtt
acacaaaagaaagaagaaggttatacacacattgtcgaagtaaccttcgaatcagttgaaactatccaagattacatca
ttcatcc
agctcacgttggttttggtgacgtttacagatccttctgggaaaaattgttgatcttcgattacaccccaagaaagtga
tgatgggct
gcaggaattcgatatcaagcttatcgataccgtcgacctcgagtcatgtaattagttatgtcacgcttacattcacgcc
ctcccccca
catccgctctaaccgaaaaggaaggagttagacaacctgaagtctaggtccctatttattUtttatagttatgttagta
ttaagaac
77
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
gttatttatatttcaaatttttcttttttttctgtacagacgcgtgtacgcatgtaacattatactgaaaaccttgctt
gagaaggttttg
ggacgctcgaaggctttaatttgc
Example 1B: LSC3-4 strain
Table of primers used in examples 1B through 1F
Primer Sequence (5'->31
name
Y0316 caagtgagaaatcaccatgagtg
Y0949 CGCGTATTTCGTCTCGCTCA
Y011307 CTCGATAGTTGGTTTCCCGTTCTTTCCACTCCCGTCATAGCTTCAAAATGTTTCTACTCC
Y011308 GACTCATAAAGTGGGAGTACAGGAAATACCATAAACTTAGATTAGATTGCTATGCTTTC
Y011334 CTCATTAGAAAGAAAGCATAGCAATCTAATCTAAGTTTATGGTATTTCCTGTACTCCCAC
Y011335 ACGTTCGCTGCACTGGGGGCCAAGCACAGGGCAAGATGCTTTCAGTATCCTTCAGGGAGC
Y011337 TACACGAGAGTTGAGTATAGTGGAGACGACATACTACCATAGCCAGCTTGCCTTGTCCCC
Y011338 TGTCTTTTGATTTATCTGCACCGCCAAAAACTTGTCAGCGTATCGACACTGGATGGCGGC
Y011431 GAAGTCTTTTGGATTGGTCTGCTC
Y011432 CAACCAAAGGCTGAAGAAGAAAAAC
Y011436 CTTTCAGTAATACGCTTAACTGCTCATTGCTATATTGAAGTagcttgccttgtccccgcc
Y011437 ATGAGAAGTTGTTCTGAACAAAGTAAAAAAAAGAAGTATACtcgacactggatggcggcg
Y011438 CGATAGTTGGTTTCCCGTTCTTTCCACTCCCGTCtaccgttcgtataatgtatgctatac
Y011439 GCTGCACTGGGGGCCAAGCACAGGGCAAGATGCTTtaccgttcgtatagcatacattata
Y011478
CAGGCACCTGGGAGGAAACATTCCGTTTCGAGTCGTACTCCACGGTATCTtgatgataccgttcgtataatgtatgc
tatacg
Y011479 GCTTTCACTAATTGATCCTCATATATTATAGAAtaccgttcgtatagcatacattatacg
Y011488 GGGTCCCGGGAGGAGAAAAAACGAGGGCTGGGAtaccgttcgtataatgtatgctatacg
Y011489 ACCACACACGAAAACGAAAACATTTGATCAGATtaccgttcgtatagcatacattatacg
Y011498 AGATATAAAAGGGAAGTGACTCCAACAACTGAAtaccgttcgtataatgtatgctatacg
Y011499 TTACTITAAAGATAGTTAGTTAGTTATTAATGGtaccgttcgtatagcatacattatacg
Y011680 CGGCATCTATGATACTTAGAGGGCAATTGCATTtaccgttcgtataatgtatgctatacg
Y011681 GCAATGTGCTTATTTCAGTAATAGTAAGGATTCtaccgttcgtatagcatacattatacg
Y011687
CcaaataaaattcaaacaaaaacCAAAACTAACtaccgttcgtataatgtatgctatacg
Y011688
AAGGATAGGGCGGAGaagtaagaaaagtttAGCtaccgttcgtatagcatacattatacg
Y011709 CGATACCACGGCAGGAAGACAACAGTGGTGTGAGCATTGCGATACGATGGGTCATAATACAGCAGAAT
GCCagcttgccttgtccccgcc
Y011710
CTTACGTCGTTCGAAGTGATGACAATAAGGATATTCATTTATTAATCGCTATTTGATACCCACTCTTGCTAt
cgacactggatggcggcg
Y011791
TAAAAAAACCTTCTCTTTGGAACTTTCAGTAATACGCTTAACTGCTCATTGCTATATTGAAGTtaccgttcgtat
aatgtatgctatacg
Y011792 CTAAAGTTATGAGTAGAAAAAAATGAGAAGTTGTTCTGAACAAAGTAAAAAAAAGAAG
TATACtaccgttcgtatagcatacattatacg
Y011795
ATTAAGAAATTATTCTTGACGCAATATTCAGCTATATGTTGATCGGGCTTAACCGCATAAGTTTtaccgttcgt
ataatgtatgctatac
Y011796
TTGTTTCATTTTCTTTTCGTTCTCTGCCCTTTTCTAGTTTGAGAGGGCATTCCCATGTCGATATtaccgttcgtat
agcatacattatac
78
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Y011970
GTCTTTGGCCTATCTTGTTTTGICCTCGGTAGATCAGGTCAGTACAAACGCAACACGAAAGAACtaccgttc
gtataatgtatgctatac
Y011971
GGCACTTTGTTTATTATTTAAAATACACCCATACATACGGACGCCAGATGCTAGAAGCAACTGTGtaccgtt
cgtatagcatacattata
Table of strain names and parents/differential genotypes used in examples 1B
through 1F
Strain Parent/genotype
name
LSC3-1 JK9-3d MATa wild-type (leu2 h1s4 trpl ura3 defective)
LSC3-2 LSC3-1 with 15-16 copies of pathway gene cassette (h1s4
defective)
LSC3-4 LSC3-2 Aga180::PkHIS4
LSC3-13 LSC3-4 Amig1::HygR
LSC3-18 LSC3-4 Aga11::HygR
LSC3-46 LSC3-2 Aga180::loxHIS4
LSC3-48, LSC3-2 Afaa2::loxHIS4
LSC3-49
LSC3-52 LSC3-2 his4::HygR
LSC3-63 LSC3-2 Apxa1:doxHIS4
LSC3-64, LSC3-13 Agall::loxKanMX
LSC3-65
LSC3-74, LSC3-52 1pex11::loxHIS4
LSC3-75
LSC3-76, LSC3-52 Aant1::loxHIS4
LSC3-77
LSC3-89, LSC3-13 Agpd1::loxKanMX
LSC3-90
LSC3-91, LSC3-18 Agpd1::loxKanMX
LSC3-92
LSC3-103 LSC3-2 Aga180::1ox72
LSC3-133, LSC3-103 Amig1::loxPkHIS4
LSC3-134
LSC3-133A, LSC3-4 mig1::lox72
LSC3-134A
LSC3-4 was generated from LSC3-2 by integrating a cassette containing PkHIS4
(HIS4
from Pichia kudriavzevii) preceded by the TEF1 promoter from Saccharomyces
cerevisiae
(pScTEF1) into the GAL80 locus using PCR fragments amplified from Pichia
kudriavzevii genomic
DNA using primers Y011334 and Y011335, and from S. cerevisiae genomic DNA
using primers
Y011307 and Y011308. The two PCR fragments were transformed into chemically
competent
LSC3-2, and colonies were selected on defined media containing Complete
Supplement Mixture
79
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
(CSM; Formedium, Hunstanton, UK) without histidine (CSM-His). Integration of
the cassette at
the GAL80 locus was confirmed by colony PCR.
Example IC: LSC3-13 strain
LSC3-13 was generated from LSC3-4 by integrating a cassette (HygR) containing
the hph
gene encoding hygromycin-B 4-0-kinase from Escherichia coli (with a GGT codon
inserted
immediately after the start codon) flanked by the 379 bp TEF1 promoter and 240
bp TEF1
terminator from Ashbya gossypn, into the MIG1 locus. A PCR fragment containing
the HygR
cassette was amplified from an in-house plasmid (pLYG-001) using primers
Y011337 and
Y011338. The PCR fragment was transformed into chemically competent LSC3-4,
and colonies
were selected on YPD medium containing 300 i.t.g/mL hygromycin B. Integration
of the cassette
at the MIG1 locus was confirmed by colony PCR.
Example ID: LSC3-18 strain
LSC3-18 was generated from LSC3-4 by integrating the HygR cassette into the
GAL1
locus. A PCR fragment containing the HygR cassette was amplified from an in-
house plasmid
using primers Y011436 and Y011437. The PCR fragment was transformed into
chemically
competent LSC3-4, and colonies were selected on YPD medium containing 300
pg/mL
hygromycin. Integration of the cassette at the GAL1 locus was confirmed by
colony PCR.
Example IE: LSC3-13 gall^ strain (LSC3-64, LSC3-65)
LSC3-64 and LSC3-65 were generated by transforming two PCR fragments into
chemically competent LSC3-13 that together compose a split KanMX marker (with
a TEF1
promoter and terminator from Ashbya gossypii flanking a kanR gene encoding an
aminoglycoside phosphotransferase conferring G418 resistance) flanked by 1ox66
and lox71
recombination sites, replacing the GAL1 gene region. This cassette is
hereafter referred to as a
loxKanMX cassette. The first fragment was amplified from an internal plasmid
containing the
assembled KanMX cassette flanked by lox sites (pLOA-058) using primers Y011791
and Y0316,
binding internally in the KanMX cassette. The second PCR fragment, was
generated by PCR from
the same internal plasmid template using primers Y0949, binding internally in
the KanMX
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
cassette, and Y011792. Colonies were selected on YPD agar plates containing
2001..tg/mL G418,
and two isolates (LSC3-64 and LSC3-65) containing the full length integration
at the desired
locus were confirmed by colony PCR.
Example 1F: Additional strain construction examples
An antibiotic marker-free version of LSC3-13 (LSC3-133 and LSC3-134) was
generated by
first integrating a cassette containing the HIS4 gene from S. cerevisiae
CEN.PK2-1C MATa,
together with its native upstream promoter and downstream terminator regions,
and flanked
by 1ox66 and lox71 recombination sites, into the GAL80 locus. This "loxHIS4"
cassette was
amplified in two fragments from an in-house constructed vector (pLOA-027)
using primers
Y011438 and Y011431, and Y011432 and Y011439. The PCR fragments were
transformed into
chemically competent LSC3-2, and colonies were selected on CSM-His agar
plates. Integration
of the cassette at the GAL80 locus was confirmed by colony PCR, and the strain
was designated
LSC3-46. Subsequently, the integrated functional HIS4 marker was looped out by
transforming
an in-house vector (pLYG-005) expressing Cre recombinase and harboring the
CEN/ARS origin of
replication. Transformants were selected on YPD plates containing 200 [tg/mL
G418 and up to
50 colonies were restruck on both G418 and YPD plates to screen for colonies
that were
spontaneously cured of pLYG-005 (grow on YPD but not on YPD plus G418). Cured
isolates were
then confirmed for loss of HIS4 by colony PCR and checking for lack of growth
on CSM-His
plates. One confirmed isolate was designated LSC3-103. Following this, a
cassette consisting of
the PkHIS4 marker with promoter and terminator as previously described,
flanked with 1ox66
and lox71 recombination sites (hereafter referred to as a loxPkHIS4 cassette),
was amplified
from an in-house vector (pLOA-093) in two PCR fragments and integrated into
the MIG1 locus
of LSC3-103. The first fragment was amplified using primers Y012096 and
Y012098, and the
second fragment was amplified using primers Y012018 and Y012097. Both
fragments were
transformed into the chemically competent loopout strain and colonies were
selected on CSM-
His agar plates. Integration of the cassette into the MIG1 locus was confirmed
by colony PCR.
An alternative marker-free version of LSC3-13 (LSC3-133A and LSC3-134A) was
generated by integrating a cassette containing the HygR cassette previously
described, flanked
81
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
by the 1ox66 and lox71 recombination sites (hereafter referred to as a loxHygR
cassette). Two
PCR fragments were amplified from an in-house vector containing the loxHygR
cassette (pLOA-
094) using primers Y012096 and Y0343, and Y0189 and Y012097. The two PCR
fragments were
transformed into chemically competent LSC3-4 and colonies selected on YPD
medium
containing 300 pg/mL hygromycin B. Integration of the cassette at the MIG1
locus was
confirmed by colony PCR. Subsequently, the HygR cassette was looped out by
transforming the
resulting strain above with pLYG-005, expressing Cre recombinase and harboring
the CEN/ARS
origin of replication. Transformants were selected on YPD plates containing
200 g/mL G418
and up to 50 colonies were restruck on both YPD plus G418 and YPD plates to
screen for
colonies that were spontaneously cured of pLYG-005. Cured isolates were
confirmed for loss of
HygR by colony PCR and checking for lack of growth on YPD plus 300 p,g/mL
hygromycin-B
plates.
LSC3-89 and LSC3-90 were generated by transforming two PCR fragments into
chemically competent LSC3-13 that together comprise a split loxl<anMX cassette
as described
above into the GPD1 locus. The first fragment was amplified as described above
using primers
Y011970 and Y0316, and the second PCR fragment was amplified as described
above using
primers Y0949 and Y011971. Similarly, LSC3-91 and LSC3-92 were generated in an
identical
way except into chemically competent LSC3-18. Colonies were selected on YPD
medium
containing 200 pg/mL G418 and integration in the GPD1 locus was confirmed by
colony PCR.
To prevent degradation of hexanoic acid and hexanoyl-CoA through native
peroxisomal
13-oxidation pathways, genes were individually disrupted and tested in the
LSC3-2 background.
These included FAA2 (peroxisomal medium chain fatty acyl-CoA synthetase), PXA1
(part of the
heterodimeric peroxisomal fatty acid and/or acyl-CoA ABC transport complex
with PXA2),
PEX11 (peroxisomal protein required for medium-chain fatty acid oxidation),
and ANT1
(peroxisomal adenine nucleotide transporter, which exchanges AMP generated in
peroxisomes
by acyl-CoA synthetases for ATP, that is consumed in that reaction, from the
cytosol). LSC3-48
and LSC3-49 (FAA2 knockouts) were generated by integrating a loxHIS4 cassette
as 2 PCR
fragments in the 3' portion (starting at nucleotide position 412) of the FAA2
locus. The
immediate 5' portion of the gene containing the first 411 nucleotides of FAA2
and its upstream
82
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
region were preserved due to overlap with the BUD25 locus transcribed from the
complement
strand. The two PCR fragments were amplified from pLOA-027 using primers
Y011478 and
Y011431, and Y011432 and Y011479, transformed into chemically competent LSC3-
2, and
colonies were selected CSM-His agar plates. LSC3-63 (PXA1 knockout) was
generated by
integrating a loxHIS4 cassette as 2 PCR fragments in the PXA1 locus. The two
PCR fragments
were amplified from pLOA-027 using primers Y011795 and Y011431, and Y011432
and
Y011796, transformed into chemically competent LSC3-2, and colonies were
selected on CSM-
His agar plates. To generate the PEX11 and ANT1 knockouts, the native non-
functional HIS4
locus was first knocked out in LSC3-2 by integrating a HygR cassette,
generating strain LSC3-52.
A PCR fragment was amplified from pLYG-001 using primers Y011709 and Y011710,
transformed into chemically competent LSC3-2, and colonies were selected on
YPD plus 300
1..tg/mL hygromycin B, with integration at the HIS4 locus confirmed by colony
PCR. This strain
exhibited enhanced efficiency of desired integrations using the loxHIS4
cassette, due to
reduced homology with the native HIS4 locus. LSC3-74 and LSC3-75 (PEX11
knockouts) were
subsequently generated by integrating a loxHIS4 cassette as 2 PCR fragments
into the PEX11
locus of LSC3-52. The two PCR fragments were amplified from pLOA-027 using
primers
Y011498 and Y011431, and Y011432 and Y011499, transformed into chemically
competent
LSC3-52, and colonies were selected on CSM-His agar plates. LSC3-76 and LSC3-
77 (ANT1
knockouts) were generated by integrating a loxHIS4 cassette as 2 PCR fragments
into the ANT1
locus of LSC3-52. The two PCR fragments were amplified from pLOA-027 using
primers
Y011680 and Y011431, and Y011432 and Y011681, transformed into chemically
competent
LSC3-52, and colonies were selected on CSM-His agar plates. Integrations of
cassettes into the
desired loci were all confirmed by colony PCR for all strains.
To reduce proteolysis of the heterologously expressed pathway proteins, common
proteases were additionally deleted in the LSC3-2 background. LSC3-47
(harboring a knockout
of PRB1, encoding vacuolar proteinase B) was generated by integrating a
loxHIS4 cassette in the
PRB1 locus using 2 PCR fragments. The two PCR fragments were amplified from
pLOA-027 using
primers Y011488 and Y011431, and Y011432 and Y011489, transformed into
chemically
competent LSC3-2, and colonies were selected on CSM-His agar plates. LSC3-87
and LSC3-88
83
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
(harboring knockouts of PEP4, encoding vacuolar aspartyl protease/proteinase
A) were
generated by integrating a loxHIS4 cassette at the PEP4 locus in LSC3-52 using
two PCR
fragments. The two PCR fragments were amplified from pLOA-027 using primers
Y011687 and
Y011431, and Y011432 and Y011688, transformed into chemically competent LSC3-
52, and
colonies were selected on CSM-His agar plates. Integrations of cassettes into
both desired loci
were confirmed by colony PCR.
Additional knockouts can subsequently be combined from any combination of
integrated 1ox66/Iox71 flanked cassettes by transforming into strains where
the previous
marker was looped out by transforming pLYG-005, isolating colonies
spontaneously cured for
pLYG-005 with confirmed loopout by colony PCR and phenotypic checks, and
integrating the
next lox site flanked marker into a new locus. For example, a strain can
harbor knockouts in
modifications that allow production from glucose (GAL80 knockout in
combination with either
MIG1 or GAL1 knockouts), knockouts in genes involved in hexanoic acid or
hexanoyl-CoA
degradation (e.g. FAA2 and ANT1 knockouts), and/or knockouts in one or
multiple proteases
involved in degradation of expressed heterologous pathway proteins (e.g. PRB1
and PEP4
knockouts).
Example 1G: Small-scale strain screening examples
Strains were tested either in shake flasks or an adapted protocol scaling down
to 96 well
plates. For shake flask testing, precultures were grown overnight
(approximately 16-24 hours)
in 15 or 30 mL of VP + 2% (w/v) glucose in 250 mL baffled shake flasks at 30 C
with 200 rpm
shaking and 80% humidity. Main cultures were inoculated using between 1 to 3
mL of
preculture in 250 mL baffled shake flasks containing 30 mL of YP + 0.02-0.04%
(w/v) hexanoic
acid + 2% (w/v) galactose or 2% (w/v) glucose + 5 mL of isopropyl myristate
(IPM). In some
experiments, the percentage of galactose or glucose was altered, the percent
hexanoic acid
added was modified, or the overlay was intentionally not added or was replaced
with
alternative overlay candidates, such as diethyl sebacate, di-tert-butyl
malonate, or methyl
soyate. Sampling time was between 24 to 50 hours as indicated.
84
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
The shake flask experiments were scaled down to 96 well deepwell plate format.
Precultures from colonies of each strain were grown in 300 p.L VP + 2% (w/v)
glucose. Main
cultures containing 300 pl VP + 2% (w/v) galactose (or glucose or combinations
of galactose and
glucose) + 0.04% (w/v) hexanoic acid + 20% (v/v) IPM (60 p.L) or alternative
overlay candidates,
were grown at 30 C with 950 rpm shaking and 80% humidity in an Infors
Multitron plate
shaker, and the IPM or diethyl sebacate overlay was sampled at different
elapsed times
between 18 and 48 hours post-inoculation, following acidification of the media
with 10 p.L of 5
M phosphoric acid. Overlay from the cultures was diluted 2:1 with methanol
prior to HPLC
analysis.
Additional defined media optimization and production experiments were
conducted
with YNB or Delft medium base. YNB medium was initially optimized and
consisted of 100 mL/L
of a 10X YNB stock solution (containing 68 g/L yeast nitrogen base without
amino acids from
Sigma-Aldrich, product number Y0626), optionally 1 mL/L of 10% BactoTM
casamino acids (BD
Biosciences), 300 mL/L of 1 M MES buffer (pH 6.5), optionally 3.6 mL/L of a
trace element
solution (containing 130 g/L citric acid monohydrate, 0.574 g/L copper (II)
sulfate pentahydrate,
8.07 g/L iron (III) chloride hexahydrate, 0.5 g/L boric acid, 0.333 g/L
manganese (II) chloride, 0.2
g/L sodium molybdate, and 4.67 g/L zinc sulfate heptahydrate), and optionally
1 mL/L of a
vitamin solution (containing 0.008 g/L biotin, 1.6 g/L calcium pantothenate,
0.008 g/L folic acid,
8 g/L myo-inositol, 1.6 g/L nicotinic acid, 0.8 g/L p-aminobenzoic acid, 1.6
g/L pyridoxal
hydrochloride, 0.8 g/L riboflavin, 1.6 g/L thiamine hydrochloride, adjusted to
pH 10.5 with
sodium hydroxide.
Delft CSM medium, consisted of (per liter solution) 7.5 g ammonium sulfate,
14.4 g
potassium phosphate monobasic, 0.5 g magnesium sulfate heptahydrate (with
these first three
components prepared as an 0.9X solution and adjusted to pH 6.5 with sodium
hydroxide prior
to autoclaving), 3.6 mL of a trace metal solution (consisting of 130 g/L
citric acid monohydrate,
0.574 g/L copper (II) sulfate pentahydrate, 8.07 g/L iron (III) chloride
hexahydrate, 0.5 g/L boric
acid, 0.333 g/L manganese (II) chloride, 0.2 g/L sodium molybdate, and 4.67
g/L zinc sulfate
heptahydrate), 1.0 mL of a vitamin solution (0.008 g/L biotin, 1.6 g/L calcium
pantothenate,
0.008 g/L folic acid, 8 g/L myo-inositol, 1.6 g/L nicotinic acid, 0.8 g/L p-
aminobenzoic acid, 1.6
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
g/L pyridoxal hydrochloride, 0.8 g/L riboflavin, 1.6 g/L thiamine
hydrochloride, adjusted to pH
10.5 with sodium hydroxide), 0.79 g of Complete Supplement Mixture (Formedium,
Norfolk,
UK), and 2% (w/v) of either galactose or glucose where specified. The final
media was filter-
sterilized, and hexanoic acid was added to 0.04% (w/v) for production.
Titers for both screening methods are presented as mg/L values on the basis of
the
entire volume of broth and overlay (total volume of 35 mL for shake flasks and
0.36 mL for 96
well deep well plates). In some plots, "olivetol equivalents" are depicted,
which is the titer of
olivetol, plus the titer of olivetolic acid multiplied by the molecular weight
of olivetol divided by
the molecular weight of olivetolic acid. In some plots, "olivetolic acid
equivalents" are depicted,
which is the titer of olivetolic acid, plus the titer of olivetol multiplied
by molecular weight of
olivetolic acid divided by the molecular weight of olivetol.
50 hour shake flask production of olivetolic acid and olivetol from LSC3-2
("3X"), LSC3-4 ("3X
ga180^::HIS4"), and LSC3-13 ("3X ga180^::HIS4 mig1A") from YP + 2% (w/v)
galactose + 0.02%
(w/v) hexanoic acid and YP + 2% (w/v) glucose + 0.02% (w/v) hexanoic acid for
LSC3-2 were
determined.
Example 1J
50 hour 96 well deepwell plate production of olivetolic acid equivalents
(total
olivetolates) and corresponding measured optical density (600 nm) values from
the aqueous
phase of the culture for LSC3-2 (pink) and LSC3-4 (blue) cultivated in YP + 2%
0r4% (w/v)
galactose + varying concentrations of hexanoic acid were determined. A
tradeoff between
hexanoic acid toxicity and conversion efficiency of hexanoic acid to end
product could be
observed, with an optimum between 0.08 to 0.1% (w/v) for 50 hour sampling.
Lower
concentrations of hexanoic acid can be employed to minimize cellular toxicity
with earlier
sampling points.
Example 1K
50 hour 96 well deepwell plate production of olivetolic acid equivalents
(total
olivetolates) and corresponding measured optical density (600 nm) values from
the aqueous
phase of the culture for, from left to right in each column, LSC3-13 (pink),
LSC3-18 (green),
LSC3-2 (blue), and LSC3-4 (purple) cultivated in YNB base medium plus 2% (w/v)
galactose
("gal") or 2% (w/v) glucose ("glu"), with or without casamino acids ("a.a") or
casamino acids
86
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
plus trace element and vitamin solutions ("a.a v.t") + 0.04% (w/v) hexanoic
acid. Optimal
production levels were observed in YNB medium supplemented with casamino acids
and
vitamin plus trace element solutions.
Example 1L
Further defined media optimization with alternative defined amino acid
compositions
and vitamin/trace solutions for LSC3-4 and LSC3-18, with total olivetolate
equivalents after 24
hours and optical density (600 nm), were determined. Glucose was added to 2%
(w/v),
galactose to 0.05% (w/v), and hexanoic acid to 0.04% (w/v). For these fully
amino acid
prototrophic strains, optimal growth and production were observed in Delft
medium base
containing CSM supplement and the trace and vitamin solution (T05 and V01) for
which the
composition is described above.
Example 1M
18 hour and 48 hour 96 well deepwell plate titers of olivetolic acid and
olivetol (and
byproducts PDAL and HTAL) for strains LSC3-2, LSC3-48 and LSC3-49 (FAA2
knockouts in LSC3-
2), LSC3-63 (PXA1 knockout in LSC3-2), LSC3-74 and LSC3-75 (PEX11 knockouts in
LSC3-2), LSC3-
76 and LSC3-77 (ANTI knockouts in LSC3-2), and LSC3-47 (PRB1 knockout in LSC3-
2) in YP
medium + 2% (w/v) galactose + 0.04% (w/v) hexanoic acid + 20% (v/v) IPM. The
sampling time
at 18 hours is indicative of productivity/rate of product formation due to
hexanoic acid not yet
being depleted. The sampling time at 48 hours represents a total conversion of
hexanoic acid
after hexanoic acid is fully utilized. For LSC3-48, LSC3-74, LSC3-76, and LSC3-
77 in particular,
both improved 18 and 48 hour titers were observed, indicating more efficient
incorporation of
hexanoic acid into olivetolic acid and olivetol. LSC3-48 and LSC3-76/77 had
the highest overall
conversions of hexanoic acid to olivetolic acid and olivetol, therefore these
FAA2 and ANT1
were selected for further combinatorial knockouts and introduction into
galactose independent
strains. LSC3-47 had a higher 48 hour titer of olivetolic acid, indicating a
potential role in
proteolysis of CsOAC.
87
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Example 1N
24 hour and 48 hour 96 well deepwell plate titers of olivetolic acid and
olivetol (and
byproducts PDAL and HTAL) for strains LSC3-2, LSC3-50 and LSC3-51 (LSC3-2
his4::loxHIS4 as
HI54 prototrophic controls), LSC3-48 and LSC3-49 (FAA2 knockouts in LSC3-2),
LSC3-77 (ANT1
knockout in LSC3-2), LSC3-47 (PRB1 knockout in LSC3-2), and LSC3-87 and LSC3-
88 (PEP4
knockouts in LSC3-2) in VP medium + 2% (w/v) galactose + 0.04% (w/v) hexanoic
acid + 20%
(v/v) IPM. The sampling time at 24 hours is indicative of productivity/rate of
product formation
due to hexanoic acid not yet being fully depleted. Higher 24 hour
productivities were again
observed for LSC3-48 and LSC3-77, indicating more efficient incorporation of
hexanoic acid into
olivetolic acid and olivetol. LSC3-87 and LSC3-88 also had higher 24 hour
titers, than LSC3-2 or
LSC3-50/51, indicating a higher pathway flux to olivetolic acid and olivetol
from the PEP4
knockout. PEP4 and PRB1 are additionally selected for further combinatorial
knockouts and
introduction into galactose-independent strain.
Example 10
24 and 48 hour total olivetolate titers for strains LSC3-2, LSC3-4, LSC3-46,
LSC3-18, and
LSC3-64 and LSC3-65 (GAL80, MIG1, GAL1 triple knockout strains) tested in VP
medium + 2%
(w/v) glucose + different indicated galactose concentrations (0, 0.05, 0.25,
or 1.0% (w/v)). LSC3-
64 and LSC3-65 combine the features of LSC3-13 and LSC3-18, with greatly
enhanced
productivities up to at least 24 hours compared to LSC3-13 in VP + 2% glucose
that are more
similar to productivities from LSC3-18, as well as reducing the galactose-
dependent inhibition of
production of LSC3-13 after 48 hours.
Example 1P
(Left) 24 and 48 hour total olivetolate titers for strains LSC3-13, LSC3-18,
LSC3-89 and
LSC3-90 (GPD1 knockouts in LSC3-13), and LSC3-91 and LSC3-92 (GPD1 knockouts
in LSC3-18) in
VP + 2% (w/v) glucose + 0.04% (w/v) hexanoic acid + 20% (v/v) IPM (left side),
or the same but
with an additional 0.05% (w/v) galactose (right side). In the presence of
0.05% galactose, LSC3-
89 and LSC3-90 have slightly increased final titers compared to LSC3-13.
(Right) glycerol titers
after 48 hours indicate greatly reduced glycerol formation in all GPD1
knockout strains.
88
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Example 1Q
48 hour 96 well deepwell plate production of LSC3-2 in YP + 2% (w/v) galactose
+ 0.04%
(w/v) hexanoic acid + 20% (v/v) of different overlays (IPM, di-tert-butyl
malonate, diethyl
sebacate, and methyl soyate). Enhanced production was observed with the
diethyl sebacate
overlay compared to IPM.
Sequences for Examples 1B through 1F
pLYG-001
tcgcgcgtttcggtgatgacggtgaaaacctctgacacatgcagctcccggagacggtcacagcttgtctgtaagcgga
tgccgggagca
gacaagcccgtcagggcgcgtcagcgggtgttggcgggtgtcggggctggcttaactatgcggcatcagagcagattgt
actgagagtg
caccacgcttttcaattcaattcatcattttttttttattcttttttttgatttcggtttctttgaaatttttttgatt
cggtaatctccgaacagaa
ggaagaacgaaggaaggagcacagacttagattggtatatatacgcatatgtagtgttgaagaaacatgaaattgccca
gtattcttaa
cccaactgcacagaacaaaaacctgcaggaaacgaagataaatcatgtcgaaagctacatataaggaacgtgctgctac
tcatcctagt
cctgttgctgcca agctattta atatcatgca ego a a agca a a ca a
acttgtgtgcttcattggatgttcgta cca ccaagga atta ctgg
agttagttgaagcattaggtcccaaaatttgtttactaaaaacacatgtggatatcttgactgatttttccatggaggg
cacagttaagccg
ctaaaggcattatccgccaagtacaattttttactcttcgaagacagaaaatttgctgacattggtaatacagtcaaat
tgcagtactctgc
gggtgtatacagaatagcagaatgggcagacattacgaatgcacacggtgtggtgggcccaggtattgttagcggtttg
aagcaggcgg
cagaagaagtaacaaaggaacctagaggccttttgatgttagcagaattgtcatgcaagggctccctatctactggaga
atatactaag
ggta ctgttga cattgcga agagcga ca aagattttgttatcggctttattgctca a agaga
catgggtgga agagatga aggtta cgat
tggttgattatgacacccggtgtgggtttagatgacaagggagacgcattgggtcaacagtatagaaccgtggatgatg
tggtctctaca
ggatctgacattattattgttggaagaggactatttgcaaagggaagggatgctaaggtagagggtgaacgttacagaa
aagcaggctg
ggaagcatatttgagaagatgcggccagcaaaactaaaaaactgtattataagtaaatgcatgtatactaaactcacaa
attagagctt
caatttaattatatcagttattaccctgcggtgtgaaataccgcacagatgcgtaaggagaaaataccgcatcaggaaa
ttgtaaacgtt
aatattttgttaaaattcgcgttaaatttttgttaaatcagctcatatttaaccaataggccgaaatcggcaaaatccc
ttataaatcaaaa
gaatagaccgagatagggttgagtgttgttccagtttggaacaagagtccactattaaagaacgtggactccaacgtca
aagggcgaaa
a a ccgtctatcagggcgatggccca cta cgtga a ccatca cccta atca
agttttttggggtcgaggtgccgta a agca cta a atcgga a
ccctaaagggagcccccgatttagagcttgacggggaaagccggcgaacgtggcgagaaaggaagggaagaaagcgaaa
ggagcgg
gcgctagggcgctggcaagtgtagcggtcacgctgcgcgtaaccaccacacccgccgcgcttaatgcgccgctacaggg
cgcgtcgcgc
cattcgccattcaggctgcgca a ctgttggga agggcgatcggtgcgggcctatcgctatta
cgccagctggcga aggggggatgtgct
gca aggcgatta agttgggta a cgccagggttttcccagtca cga cgttgtaa a acgacggccagtga
attgtaata cga ctca ctatag
ggcgaattggagctccaccgcggtggcggccgcataggccactagtggatctgatatcatcgatgaattcgagctcgtM
cgacactgg
atggcggcgttagtatcga atcga cagcagtatagcga ccagcattca cata cgattga
cgcatgatattactttctgcgca ctta a cttc
gcatctgggcagatgatgtcgaggcgaaaaaaaatataaatcacgctaacatttgattaaaatagaacaactacaatat
aaaaaaact
atacaaatgacaagttcttgaaaacaagaatctttttattgtcagtactgattattcctttgccctcggacgagtgctg
gggcgtcggtttcc
actatcggcgagtacttctacacagccatcggtccagacggccgcgcttctgcgggcgatttgtgtacgcccgacagtc
ccggctccggat
cgga cgattgcgtcgcatcga ccctgcgccca agctgcatcatcga a attgccgtca a cca
agctctgatagagttggtca aga cca atg
cggagcatatacgcccggagccgcggcgatcctgcaagctccggatgcctccgctcgaagtagcgcgtctgctgctcca
tacaagccaa
cca cggcctccaga aga agatgttggcga cctcgtattggga atccccga a catcgcctcgctccagtca
atga ccgctgttatgcggcc
attgtccgtcaggacattgttggagccgaaatccgcgtgcacgaggtgccggacttcggggcagtcctcggcccaaagc
atcagctcatc
gagagcctgcgcga cgga cgca ctga cggtgtcgtccatca cagtttgccagtgata ca
catggggatcagca atcgcgca tatga a at
89
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
cacgccatgtagtgtattgaccgattccttgcggtccgaatgggccgaacccgctcgtctggctaagatcggccgcagc
gatcgcatccat
ggcctccgcgaccggctgcagaacagcgggcagttcggtttcaggcaggtcttgcaacgtgacaccctgtgcacggcgg
gagatgcaat
aggtcaggctctcgctgaattccccaatgtcaagcacttccggaatcgggagcgcggccgatgcaaagtgccgataaac
ataacgatctt
tgtagaaaccatcggcgcagctatttacccgcaggacatatccacgccctcctacatcgaagctgaaagcacgagattc
ttcgccctccg
agagctgcatcaggtcggagacgctgtcgaacttttcgatcagaaacttctcgacagacgtcgcggtgagttcaggctt
tttacccatggt
tgtttatgttcggatgtgatgtgagaactgtatcctagcaagattttaaaaggaagtatatgaaagaagaacctcagtg
gcaaatcctaac
cttttatatttctctacaggggcgcggcgtggggacaattcaacgcgtctgtgaggggagcgtttccctgctcgcaggt
ctgcagcgagga
gccgtaatttttgcttcgcgccgtgcggccatcaaaatgtatggatgcaaatgattatacatggggatgtatgggctaa
atgtacgggcga
cagtcacatcatgcccctgagctgcgcacgtcaagactgtcaaggagggtattctgggcctccatgtcgctggccgggt
gacccggcggg
gacaaggcaagctaaacagatctggcgcgccttaattaacccggggatccgtcgacctgcagcgtacgaagcttcagct
ggcggccgct
ctagccagcttttgttccattagtgagggttaattccgagcttggcgtaatcatggtcatagctgtttcctgtgtgaaa
ttgttatccgctca
caattccacacaacataggagccggaagcataaagtgtaaagcctggggtgcctaatgagtgaggtaactcacattaat
tgcgttgcgct
cactgcccgctttccagtcgggaaacctgtcgtgccagctgcattaatgaatcggccaacgcgcggggagaggcggttt
gcgtattgggc
gctcttccgcttcctcgctcactgactcgctgcgctcggtcgttcggctgcggcgagcggtatcagctcactcaaaggc
ggtaatacggtta
tccacagaatcaggggataacgcaggaaagaacatgtgagcaaaaggccagcaaaaggccaggaaccgtaaaaaggccg
cgttgct
ggcgtttttccataggctcggcccccctgacgagcatcacaaaaatcgacgctcaagtcagaggtggcgaaacccgaca
ggactataaa
gataccaggcgttcccccctggaagctccctcgtgcgctctcctgttccgaccctgccgcttaccggatacctgtccgc
ctttctcccttcgg
gaagcgtggcgctttctcaatgctcacgctgtaggtatctcagttcggtgtaggtcgttcgctccaagctgggctgtgt
gcacgaacccccc
gttcagcccgaccgctgcgccttatccggtaactatcgtcttgagtccaacccggtaagacacgacttatcgccactgg
cagcagccactg
gtaacaggattagcagagcgaggtatgtaggcggtgctacagagttcttgaagtggtggcctaactacggctacactag
aaggacagta
tttggtatctgcgctctgctgaagccagttaccttcggaaaaagagttggtagctcttgatccggcaaacaaaccaccg
ctggtagcggtg
gtttttttgtttgcaagcagcagattacgcgcagaaaaaaaggatctcaagaagatcctttgatcttttctacggggtc
tgacgctcagtgg
aacgaaaactcacgttaagggattttggtcatgagattatcaaaaaggatcttcacctagatccttttaaattaaaaat
gaagttttaaat
caatctaaagtatatatgagtaaacttggtctgacagttaccaatgcttaatcagtgaggcacctatctcagcgatctg
tctatttcgttcat
ccatagttgcctgactgcccgtcgtgtagataactacgatacgggagggcttaccatctggccccagtgctgcaatgat
accgcgagacc
cacgctcaccggctccagatttatcagca ata a a ccagccagccgga agggccgagcgcaga
agtggtcctgca a ctttatccgcctcc
atccagtctattaattgttgccgggaagctagagtaagtagttcgccagttaatagtttgcgcaacgttgttgccattg
ctacaggcatcgt
ggtgtcacgctcgtcgtttggtatggcttcattcagctccggttcccaacgatcaaggcgagttacatgatcccccatg
ttgtgaaaaaaag
cggttagctccttcggtcctccgatcgttgtcagaagtaagttggccgcagtgttatcactcatggttatggcagcact
gcataattctctta
ctgtcatgccatccgtaagatgcttttctgtgactggtgagtactcaaccaagtcattctgagaatagtgtatgcggcg
accgagttgctctt
gcccggcgtcaatacgggataataccgcgccacatagcagaactttaaaagtgctcatcattggaaaacgttcttcggg
gcgaaaactct
caaggatcttaccgctgttgagatccagttcgatgtaacccactcgtgcacccaactgatcttcagcatcttttacttt
caccagcgtttctg
ggtgagcaaaaacaggaaggcaaaatgccgcaaaaaagggaataagggcgacacggaaatgttgaatactcatactctt
cctttttca
atattattgaagcatttatcagggttattgtctcatgagcggatacatatttgaatgtatttagaaaaataaacaaata
ggggttccgcgca
cataccccgaaaagtgccacctgggtccattcatcacgtgctataaaaataattataatttaaatttataatataaata
tataaattaaa
aatagaaagtaaaaaaagaaattaaagaaaaaatagtttttgttttccgaagatgtaaaagactctagggggatcgcca
acaaatacta
ccttttatcttgctcttcctgctctcaggtattaatgccgaattgtttcatcttgtctgtgtagaagaccacacacgaa
aatcctgtgattttac
attttacttatcgttaatcgaatgtatatctatttaatctgcttttcttgtctaataaatatatatgtaaagtacgctt
tttgttgaaattttttaa
acctttgtttatttttttttcttcattccgtaactcttctaccttctttatttactttctaaaatccaaatacaaaaca
taaaaataaataaacac
agagtaaattcccaaattattccatcattaaaagatacgaggcgcgtgtaagttacaggcaagcgatccgtcctaagaa
accattattat
catgacattaacctataaaaataggcgtatcacgaggccctttcgtc
pLYG-005
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
agatcctttgatcttttctacggggtctgacgctcagtggaacgaaaactcacgttaagggattttggtcatgagatta
tcaaaaaggatct
tcacctagatccttttaaattaaaaatgaagttttaaatcaatctaaagtatatatgagtaaacttggtctgacagtta
ccaatgcttaatc
agtgaggcacctatctcagcgatctgtctatttcgttcatccatagttgcctgactgcccgtcgtgtagataactacga
tacgggagggctt
accatctggccccagtgctgcaatgataccgcgagacccacgctcaccggctccagatttatcagcaataaaccagcca
gccggaaggg
ccgagcgcagaagtggtcctgcaactttatccgcctccatccagtctattaattgttgccgggaagctagagtaagtag
ttcgccagttaat
agtttgcgcaacgttgttgccattgctacaggcatcgtggtgtcacgctcgtcgtttggtatggcttcattcagctccg
gttcccaacgatca
aggcgagttacatgatcccccatgttgtgaaaaaaagcggttagctccttcggtcctccgatcgttgtcagaagtaagt
tggccgcagtgt
tatcactcatggttatggcagcactgcataattctcttactgtcatgccatccgtaagatgcttttctgtgactggtga
gtactcaaccaagt
cattctgagaatagtgtatgcggcgaccgagttgctcttgcccggcgtcaatacgggataataccgcgccacatagcag
aactttaaaag
tgctcatcattggaaaacgacttcggggcgaaaactctcaaggatcttaccgctgttgagatccagttcgatgtaaccc
actcgtgcaccc
aactgatcttcagcatcttttactttcaccagcgtttctgggtgagcaaaaacaggaaggcaaaatgccgcaaaaaagg
gaataagggc
gacacggaaatgttgaatactcatactcttcctttttcaatattattgaagcatttatcagggttattgtctcatgagc
ggatacatatttga
atgtatttagaaaaataaacaaataggggttccgcgcacataccccgaaaagtgccacctgggtccattcatcacgtgc
tataaaaata
attataatttaaattttttaatataaatatataaattaaaaatagaaagtaaaaaaagaaattaaagaaaaaatagttt
ttgttttccgaa
gatgtaaaagactctagggggatcgccaacaaatactaccttttatcttgctcttcctgctctcaggtattaatgccga
attgtttcatcttgt
ctgtgtagaagaccacacacgaaaatcctgtgattttacattnacttatcgttaatcgaatgtatatctatttaatctg
cttttcttgtctaat
aaatatatatgtaaagtacgctttttgttgaaattttttaaacctttgtttatttttttttcttcattccgtaactctt
ctaccttctttatttacttt
ctaaaatccaaatacaaaacataaaaataaataaacacagagtaaattcccaaattattccatcattaaaagatacgag
gcgcgtgtaa
gttacaggcaagcgatcatccgtcctaagaaaccattattatcatgacattaacctataaaaataggcgtatcacgagg
ccctttcgtctc
gcgcgtttcggtgatgacggtgaaaacctctgacacatgcagctcccggagacggtcacagcttgtctgtaagcggatg
ccgggagcag
acaagcccgtcagggcgcgtcagcgggtgttggcgggtgtcggggctggcttaactatgcggcatcagagcagattgta
ctgagagtgc
accacgcttttcaattcaattcatcattttttttttattcttttttttgatttcggtttctttgaaatttttttgattc
ggtaatctccgaacagaag
gaagaacgaaggaaggagcacagacttagattggtatatatacgcatatgtagtgttgaagaaacatgaaattgcccag
tattcttaac
ccaactgcacagaacaaaaacctgcaggaaacgaagataaatcgaaacatcatgaaaactgtttcaccctctgtgaagc
ataaacact
agaaagccaatgaagagctctacaagcctcttatgggttcaatgggtctgcaatgaccgcatacgggcttggacaatta
ccttctattgaa
tttctgagaagagatacatctcaccagcaatgtaagcagacaatcccaattctgtaaacaacctctttgtccataattc
cccatcagaaga
gtgaaaaatgccctcaaaatgcatgcgccacacccatctttcaactgcactgcgccacctctgagggtcttttcagggg
tcgactaccccg
gacacctcgcagaggagcgaggtcacgtacttttaaaatggcagagacgcgcagtttcttgaagaaaggataaaaatga
aatggtgcg
gaaatgcgaaaatgatgaaaaattttcttggtggcgaggaaattgagtgcaataattggcacgaggttgttgccacccg
agtgtgagtat
atatcctagtttctgcacttttcttcttcttttctttaccttncttttcaacttttttttactttttccttcaacagac
aaatctaacttatatatca
caATGGGTAAGGAAAAGACTCACGTTTCGAGGCCGCGATTAAATTCCAACATGGATGCTGATTTATATG
GGTATAAATGGGCTCGCGATAATGTCGGGCAATCAGGTGCGACAATCTATCGATTGTATGGGAAGCCC
GATGCGCCAGAGTTGTTTCTGAAACATGGCAAAGGTAGCGTTGCCAATGATGITACAGATGAGATGGIC
AGACTAAACTGGCTGACGGAATTTATGCCTCTTCCGACCATCAAGCATTTTATCCGTACTCCTGATGATGC
ATGGTTACTCACCACTGCGATCCCCGGCAAAACAGCATTCCAGGTATTAGAAGAATATCCTGATTCAGGT
GAAAATATTGTTGATGCGCTGGCAGTGTTCCTGCGCCGGTTGCATTCGATTCCTGTTTGTAATTGTCCTTT
TAACAGCGATCGCGTATTTCGTCTCGCTCAGGCGCAATCACGAATGAATAACGGTTTGGTTGATGCGAG
TGATTTTGATGACGAGCGTAATGGCTGGCCTGTTGAACAAGTCTGGAAAGAAATGCATAAGCTTTTGCC
ATTCTCACCGGATTCAGTCGTCACTCATGGTGATTTCTCACTTGATAACCTTATTTTTGACGAGGGGAAAT
TAATAGGTTGTATTGATGTTGGACGAGTCGGAATCGCAGACCGATACCAGGATCTTGCCATCCTATGGA
ACTGCCTCGGTGAGTTTTCTCCTTCATTACAGAAACGGCTTTTTCAAAAATATGGTATTGATAATCCTGAT
ATGAATAAATTGCAGTTTCATTTGATGCTCGATGAGTTITTCTAAgtgaatttactttaaatcttgcatttaaataaat
t
ttctttttatagctttatgacttagtttcaatttatatactattttaatgacattttcgattcattgattgaaagcttt
gtgttttttcttgatgcgc
tattgcattgttcttgtctttttcgccacatgtaatatctgtagtagatacctgatacattgtggataaaactgtatta
taagtaaatgcatgt
91
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
atactaaactcacaaattagagcttcaatttaattatatcagttattaccctgcggtgtgaaataccgcacagatgcgt
aaggagaaaata
ccgcatcaggaaattgtaaacgttaatattttgttaaa attcgcgttaaatttttgttaaatcagctcatttttta
accaataggccgaaatc
ggcaaaatcccttataaatcaaaagaatagaccgagatagggttgagtgttgttccagtttggaacaagagtccactat
taaagaacgtg
gactccaacgtcaaagggcgaaaaaccgtctatcagggcgatggcccactacgtgaaccatcacccta
atcaagttttttggggtcgagg
tgccgtaaagcactaaatcggaaccctaaagggagcccccgatttagagcttgacggggaaagccggcgaacgtggcga
gaaaggaa
gggaagaaagcgaaaggagcgggcgctagggcgctggcaagtgtagcggtcacgctgcgcgtaacca
ccacacccgccgcgcttaat
gcgccgctacagggcgcgtcgcgccattcgccattcaggctgcgcaactgttgggaagggcgatcggtgcgggcctctt
cgctattacgc
cagctggcgaaggggggatgtgctgcaaggcgattaagttgggtaacgccagggttttcccagtcacgacgttgtaaaa
cgacggccag
tgaattgtaatacgactcactatagggcgaattggagctccaccgcggtggcggccgcataggccactagtggatctga
tatcatcgatg
aattcgagctcgtttgggcccgctacttagcttctatagttagttaatgcactcacgatattcaaaattgacacccttc
aactactccctact
attgtctactactgtctactactcctctttactatagctgctcccaataggctccaccaataggctctgccaatacatt
ttgcgccgccacctt
tcaggttgtgtcactcctgaaggaccatattgggtaatcgtgcaatttctggaagagagtccgcgagaagtgaggcccc
cactgtaaatc
ctcgagggggcatggagtatggggcatggaggatggaggatggggggggggcgaaaaataggtagcaaaaggacccgct
atcacccc
acccggagaactcgttgccgggaagtcatatttcgacactccggggagtctataaaaggcgggttttgtcttttgccag
ttgatgttgctga
aaggacttgtttgccgtttcttccgatttaacagtatagaaatca
accactgttaattatacacgttatactaacacaacaaaaacaaaaa
caacgacaacaacaacaacaATGTCCAATTTACTGACCGTACACCAAAATTTGCCTGCATTACCGGTCGATGCA
ACGAGTGATGAGGTTCGCAAGAACCTGATGGACATGTTCAGGGATCGCCAGGCGTTTTCTGAGCATACC
TGGAAAATGCTTCTGTCCGTTTGCCGGTCGTGGGCGGCATGGIGCAAGTTGAATAACCGGAAATGGTTT
CCCGCAGAACCTGAAGATGTTCGCGATTATCTTCTATATCTTCAGGCGCGCGGTCTGGCAGTAAAAACTA
TCCAG CAACATTTG GG CCAG CTAAACATG CTTCATCGTCGGTCCG GG CTGCCACGACCAAGTGACAGCA
ATGCTGTTTCACTGGTTATGCGGCGGATCCGAAAAGAAAACGTTGATGCCGGTGAACGTGCAAAACAG
G CTCTAG CGTTCGAACG CACTGATTTCGACCAGGTTCGTTCACTCATG GAAAATAGCGATCG CTGCCAG
GATATACGTAATCTG G CATTTCTGG GGATTGCTTATAACACCCTGTTACGTATAGCCGAAATTG CCAG GA
TCAGGGTTAAAGATATCTCACGTACTGACGGTGGGAGAATGTTAATCCATATTGGCAGAACGAAAACGC
TGGTTAGCACCGCAGGTGTAGAGAAGGCACTTAGCCTGGGGGTAACTAAACTGGTCGAGCGATGGATT
TCCGTCTCTG GTGTAGCTGATGATCCGAATAACTACCTGTTTTG CCGG GTCAGAAAAAATGGTGTTGCCG
CG CCATCTG CCACCAG CCAGCTATCAACTCGCGCCCTG GAAG GGATTTTTGAAGCAACTCATCGATTGAT
TTACG GCGCTAAG GATGACTCTG GTCAGAGATACCTG GCCTG GTCTG GACACAGTGCCCGTGTCG GAG C
CGCGCGAGATATGGCCCGCGCTGGAGTTTCAATACCGGAGATCATGCAAGCTGGTGGCTGGACCAATG
TAAATATTGTCATGAACTATATCCGTACCCTG GATAGTGAAACAGGGGCAATGGTGCGCCTGCTGGAAG
ATGGCGATTAGtcatgta attagttatgtca cgctta
cattcacgccctccccccacatccgctctaaccgaaaaggaaggagttag
acaacctgaagtctaggtccctatttatttttttatagttatgttagtattaagaacgttatttatatttcaaattttt
cttttttttctgtacaga
cgcgtgtacgcatgtaacattatactgaaaaccttgcttgagaaggttttgggacgctcgaaggctttaatttgcggcc
ggcgcgccttaat
taacccggggatccgtcgacctgcagcgtacgaagcttcagctggcggccgctctagccagcttttgttccctttagtg
agggttaattccg
agcaggcgtaatcatggtcatagctgatcctgtgtgaaattgttatccgctcacaattccacacaacataggagccgga
agcataaagt
gtaaagcctggggtgcctaatgagtgaggtaactcacattaattgcgttgcgctcactgcccgctttccagtcgggaaa
cctgtcgtgcca
gctgcattaatgaatcggccaacgcgcggggagaggcggtttgcgtattgggcgctcttccgcttcctcgctcactgac
tcgctgcgctcg
gtcgttcggctgcggcgagcggtatcagctcactcaaaggcggta
atacggttatccacagaatcaggggataacgcaggaaagaacat
gtgagcaaaaggccagcaaaaggccaggaaccgtaaaaaggccgcgttgctggcgtttttccataggctcggcccccct
gacgagcatc
acaaaaatcgacgctcaagtcagaggtggcgaaacccgacaggactataaagataccaggcgttcccccctggaagctc
cctcgtgcg
ctctcctgttccgaccctgccgcttaccggatacctgtccgcctttctcccttcgggaagcgtggcgctttctcaatgc
tcacgctgtaggtat
ctcagttcggtgtaggtcgttcgctccaagctgggctgtgtgcacgaaccccccgttcagcccgaccgctgcgccttat
ccggtaactatcg
tcttgagtccaacccggtaagacacgacttatcgccactggcagcagccactggtaacaggattagcagagcgaggtat
gtaggcggtg
ctacagagttcttgaagtggtggcctaactacggctacactagaaggacagtatttggtatctgcgctctgctgaagcc
agttaccttcgg
92
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
aaaaagagttggtagctcttgatccggcaaacaaaccaccgctggtagcggtggttntttgtttgcaagcagcagatta
cgcgcagaaa
aaaaggatctcaaga
pLOA-027
gacgaaagggcctcgtgatacgcctatttttataggttaatgtcatgataataatggtttcttagacgtcaggtggcac
ttttcggggaaat
gtgcgcggaacccctatttgtttatttttctaaatacattcaaatatgtatccgctcatgagacaataaccctgataaa
tgcttcaataatat
tgaaaaaggaagagtatgagtattcaacatttccgtgtcgcccttattcccttttttgcggcattttgccttcctgttt
ttgctcacccagaaa
cgctggtgaaagtaaaagatgctgaagatcagttgggtgcacgagtgggttacatcgaactggatctcaacagcggtaa
gatccttgag
agttttcgccccgaagaacgttttccaatgatgagcacttttaaagttctgctatgtggcgcggtattatcccgtattg
acgccgggcaaga
gcaactcggtcgccgcatacactattctcagaatgacttggttgagtactcaccagtcacagaaaagcatcttacggat
ggcatgacagt
aagagaattatgcagtgctgccataaccatgagtgataacactgcggccaacttacttctgacaacgatcggaggaccg
aaggagctaa
ccgcttttttgcacaacatgggggatcatgtaactcgccttgatcgttgggaaccggagctgaatgaagccataccaaa
cgacgagcgtg
acaccacgatgcctgtagcaatggcaacaacgagcgcaaactattaactggcgaactacttactctagcttcccggcaa
caattaatag
actggatggaggcggataaagttgcaggaccacttctgcgctcggcccttccggctggctggtttattgctgataaatc
tggagccggtga
gcgtgggtcCcgcggtatcattgcagcactggggccagatggtaagccctcccgtatcgtagttatctacacgacgggg
agtcaggcaac
tatggatgaacgaaatagacagatcgctgagataggtgcctcactgattaagcattggtaactgtcagaccaagtttac
tcatatatactt
tagattgatttaaaacttcatttttaatttaaaaggatctaggtgaagatcctttttgataatctcatgaccaaaatcc
cttaacgtgagtttt
cgttccactgagcgtcagaccccgtagaaaagatcaaaggatcttcttgagatcctttttttctgcgcgtaatctgctg
cttgcaaacaaaa
aaaccaccgctaccagcggtggiftgtttgccggatcaagagctaccaactctattccgaaggtaactggcttcagcag
agcgcagatac
caaatactgttcttctagtgtagccgtagttaggccaccacttcaagaactctgtagcaccgcctacatacctcgctct
gctaatcctgttac
cagtggctgctgccagtggcgataagtcgtgtcttaccgggttggactcaagacgatagttaccggataaggcgcagcg
gtcgggctgaa
cggggggttcgtgcacacagcccagcttggagcgaacgacctacaccgaactgagatacctacagcgtgagctatgaga
aagcgccac
gcttcccgaagggagaaaggcggacaggtatccggtaagcggcagggtcggaacaggagagcgcacgagggagcttcca
gggggaa
acgcctggtatctttatagtcctgtcgggtttcgccacctctgacttgagcgtcgatttttgtgatgctcgtcaggggg
gcggagcctatgga
aaaacgccagcaacgcggccttatacggttcctggccttttgctggccttttgctcacatgttctttcctgcgttatcc
cctgattctgtggat
aaccgtattaccgcctttgagtgagctgataccgctcgccgcagccgaacgaccgagcgcagcgagtcagtgagcgagg
aagcggaag
agcgcccaatacgcaaaccgcctctccccgcgcgttggccgattcattaatgcaggtttaaacAGGTGGTAATAATCGC
GCGAT
TCAATTGCATTCATTAAAGACAGATAATTCGCAAGACCTTCTCCCTCCAGATCAACTTGTATCAATGATTC
ACTTGTTCATCAACGATGAAAGGTTTACCTCCGGTATAACGAGITTTGACATTGATTTTTCTAGAATGAAA
ATGCCATAGAAATTTCTAAATTTAGACTGAATCCCTACGTCACTGGTTTAAAAATTGAGTGGTGCTTACTA
ATTATTACATTCGGAAACGTCTCATCAAGTGTTTCCGAAAAAATGAGGGTTTTTCTAAAGCTTCTTTCTTT
CACGGATATCACCGGGITTAAGATGTATTITTITTITCCACAGAAATTAAAGTTCCAGCGTTTACCAAAGT
AGATCGTTCAATAATATGGATGGTGTTATAAGAAGACGACCACTATCCCCCATGAATTCTCACATGATAC
TTTCTTTTACTTTATTTACAGAGGCAGTAACATCCAAGAAGAAtaccgttcgtataatgtatgctatacgaagttataa
c
cggcgttgccagcgataaacggCCCATCACAATCCTGACAACCAGCAGTTCTTCTAGGCAGTCGAACTGACTCTA
ATAGTGACTCCGGTAAATTAGTTAATTAATTGCTAAACCCATGCACAGTGACTCACGTTTTTITATCAGTC
ATTCGATATAGAAGGTAAGAAAAGGATATGACTATGAACAGTAGTATACTGTGTATATAATAGATATGG
AACGTTATATTCACCTCCGATGTGTGTTGTACATACATAAAAATATCATAGCACAACTGCGCTGTGTAAT
AGTAATACAATAGTTTACAAAATTTTTTTTCTGAATAATGGTTTTGCCGATTCTACCGTTAATTGATGATCT
GGCCTCATGGAATAGTAAGAAGGAATACGTTTCACTTGTTGGTCAGGTACTTTTGGATGGCTCGAGCCT
GAGTAATGAAGAGATTCTCCAGTTCTCCAAAGAGGAAGAAGTTCCATTGGTGGCTTTGTCCTTGCCAAG
TGGTAAATTCAGCGATGATGAAATCATTGCCTTCTTGAACAACGGAGTTTCTTCTCTGTTCATTGCTAGCC
AAGATGCTAAAACAGCCGAACACTTGGTTGAACAATTGAATGTACCAAAGGAGCGTGTTGTTGTGGAA
GAGAACGGTGTTTTCTCCAATCAATTCATGGTAAAACAAAAATTCTCGCAAGATAAAATTGTGTCCATAA
93
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
AGAAATTAAGCAAGGATATGTTGACCAAAGAAGTGCTTG GTGAAGTACGTACAGACCGTCCTGACG GTT
TATATACCACCCTAGTTGTCGACCAATATG AGCGTTGTCTAG GGTTGGTGTATTCTTCGAAG AAATCTAT
AG CAAAG GCCATCG ATTTG GGTCGTG G CGTTTATTATTCTCGTTCTAG GAATGAAATCTG G ATCAAGGG
TG AAACTTCTG GCAATG GCCAAAAG CTTTTACAAATCTCTACTG ACTGTGATTCGG ATG CCTTAAAGTTT
ATCGTTGAACAAGAAAACGTTGGATTTTGCCACTTGGAGACCATGTCTTGCTTTGGTGAATTCAAGCATG
GTTTGGTG GG GCTAG AATCTTTACTAAAACAAAGGCTACAGG ACG CTCCAGAGG AATCTTATACTAG AA
GACTATTCAACGACTCTGCATTGTTAGATG CCAAGATCAAGGAAGAAGCTGAAGAACTGACTGAGG CAA
AG GGTAAGAAGGAGCTTTCTTG GGAGG CTGCCGATTTGTTCTACTTTGCACTGG CCAAATTAGTG GCCA
ACGATGTTTCATTGAAG GACGTCGAGAATAATCTGAATATGAAGCATCTGAAGGTTACAAGACG GAAAG
GTGATGCTAAGCCAAAGTTTGTTGGACAACCAAAGGCTGAAGAAGAAAAACTGACCGGTCCAATTCACT
TGGACGTGGTGAAGGCTTCCGACAAAGTTGGTGTGCAGAAGGCTTTGAGCAGACCAATCCAAAAGACT
TCTG AAATTATG CATTTAGTCAATCCGATCATCG AAAATGTTAGAG ACAAAG GTAACTCTG CCCTTTTG G
AGTACACAGAAAAGTTTGATG GTGTAAAATTATCCAATCCTGTTCTTAATGCTCCATTCCCAG AAGAATA
CTTTGAAGGTTTAACCGAGGAAATGAAGGAAGCTTTGGACCITTCAATTGAAAACGTCCGCAAATTCCAT
GCTGCTCAATTGCCAACAGAGACTCTTGAAGTTGAAACCCAACCTGGTGTCTTGTGTTCCAGATTCCCTC
GTCCTATTGAAAAAGTTGGTTTGTATATCCCTGGTGGCACTGCCATTTTACCAAGTACTGCATTAATGCTT
GGTGTTCCAGCACAAGTTGCCCAATGTAAGGAGATTGIGTTTGCATCTCCACCAAGAAAATCTGATGGT
AAAGTTTCACCCGAAGTTGTTTATGTCG CAGAAAAAGTTG GCGCTTCCAAGATTGTTCTAG CTGGTG GTG
CCCAAGCCGTTGCTGCTATGGCTTACGGGACAGAAACTATTCCTAAAGTGGATAAGATCTTGGGTCCAG
GTAATCAATTTGTGACTGCCG CCAAAATGTATGTTCAAAATG ACACTCAAG CTCTATGTTCCATTG ATATG
CCAGCTGGCCCAAGTGAAGTTTTGGTTATTGCCGATGAAGATGCCGATGTGGATTTTGTTGCAAGTGAT
TTGCTATCG CAAGCTGAACACG GTATTGACTCCCAAGTTATCCTTGTTG GTGTTAACTTGAG CGAAAAGA
AAATTCAAGAGATTCAAGATGCTGTCCACAATCAAGCTTTACAACTGCCACGTGTGGATATTGTTCGTAA
ATGTATTGCTCACAGTACGATCGTTCTTTGTGACGGTTACGAAGAAGCCCTTGAAATGTCCAACCAATAT
G CACCAG AACATTTGATTCTACAAATCG CCAATG CTAACG ATTATGTTAAATTGGTTG ACAATGCAGG GT
CCGTATTTGTG GGTG CTTACACTCCAG AATCGTG CG GTGACTATTCAAGTG GTACTAACCATACATTACC
AACCTATGGTTACGCTAGGCAGTACAGTGGTGCCAACACTGCAACCTTCCAAAAGTTTATCACTGCCCAA
AACATTACCCCTGAAGGTTTAGAAAACATCG GTAGAGCTGTTATGTG CGTTG CCAAG AAGGAGGGTCTA
GACGGTCACAGAAACGCTGTGAAAATCAGAATGAGTAAGCTTGGGTTGATCCCAAAGGATTTCCAGTA
G ATTATTTCTAACTTGG AAACCG AACACTAACG AAAATAATATGTATATATACATATATATATCAAACAA
AATACAGTCTTG AATGAATAG AGATACACTATGTAATGAATGGTAACGTAAAAATTGTAATTTTGG ATTA
AAAGAGAGGTAGcgcctggcagcagggcgata a cctcata a cttcgtata atgtatgctata cga
acggtaTTIGGIGTIGTT
TTCTATTGCATACGAATTAG AATG CCCAG ACTTGTTTATATACTACGCTG AATGTTTGTACATTTATACTTA
AAACAAAATGCTAGTCAGCCATATTAAACAGAGCCGTTTAGCAACATTTCAATAGCACCTTCCACAGATC
CACCG CTACGTYTCAATG CG GCAATGTTACG GTCGAAGTCAAAGAAG CCCATATCGTTTAATTGACGTAA
TTGTGTTG CATAGACTTCTTCTGGAGG CCTTGTATCGGAAGCAGAAG GTGCAGTTGAACCAGTACCAGT
ACTGGCACCACCAAAGAGATTCATTAAATTAGGATTTGCCAAAATG GGATTACCTCCTAAACCTGCTCCA
GGAACACCTGCTCCAAATAGTGACGCAAATGGATTGCTTGGTACAGAGGAACCTGAAGAGTTTGAAGTT
GAATTACGTGGCGAATCCGTGTTTGCAGTGTTAGAGTCAGATGGATTTCCGGGTGATGGGAAGTgttta a
a cctggcgta atagcga agaggcccgca ccgatcgcccttccca a cagttgcgcagcctga a tggcga
atggcgcctgatgcggtatttt
ctcctta cgcatctgtgcggtatttca ca ccgcatatggtgca ctctcagtaca atctgctctga
tgccgcatagtta agccagccccga ca
cccgcca a ca cccgctga cgcgccctga cgggcttgtctgctcccggcatccgctta caga ca
agctgtga ccgtctccgggagctgcat
gtgtcagaggttttca ccgtcatca ccga a a cgcgcga
94
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
p LOA-058
atgaccatgattacgccactagtccgaggcctcgagatccgatatcgccgtggcggccgccagctgaagcttaattatc
ctgggcacgag
tgaaacaaagctaaaacctttatttagcatggccattgaatgtaacaattatatatatcgcaagcacaaaaaatcaagg
agagagaact
accactttgttcatgtgtacaatgttcattatctccataagcaaaaaaaaaaaaatagaaaacatatgctataaggttg
atattctcacga
gtaagcggcacttgctacttattgacattgcagatttttggctacagaaatagtatattagagattataattgctaatc
aaatcaaaatata
aaattagtaaaccaaaccatttatacccttccttagtagttatggattgttttttaatgatatttctgcaaaccaaaga
aagattgttatcca
gatagaatttagttttgatattcatttttttgttgaagattgaacgccatatctgggcctcataattcaaaagacggtg
ccattatcggtagc
gtttcgcattgtactggatttcagaaatttcacagttgatgaatcgaaaagaatggtctcattgcaacacgtaaggtta
agatgtcccttttt
accattataggcaataaatgaatcataaaacgaccgtatactggtgaaatagtagggagaacgagtacctgtagtaaaa
agtataaatc
atagttaatcgggcaatgtccctcgatcaaggagtattgtgtcatgttcgagacaaacgccaacatttttgtttctttt
ggacaaatgttgtt
tgcatttatgatccgttatattttgatctaatgtagagttgcacgtagttcttactggcaaagaaatcgatgcatacca
aaaaagaataaa
ggtgatatttgatctttaccgtttagttccaacgtaaaattgtgcctttggacttaaaatggcgtcgtacgctgcaggt
cgacggatccccg
ggttaattaaggcgcgccagatctgatagcttgcctcgtccccgccgggtcactaccgttcgtataatgtatgctatac
gaagttatgaca
tggaggcccagaataccctccttgacagtcttgacgtgcgcagctcaggggcatgatgtgactgtcgcccgtacattta
gcccatacatcc
ccatgtataatcatttgcatccatacattttgatggccgcacggcgcgaagcaaaaattacggctcctcgctgcagacc
tgcgagcaggg
aaacgctcccctcacagacgcgttgaattgtccccacgccgcgcccctgtagagaaatataaaaggttaggatttgcca
ctgaggttcttc
tttcatatacttccttttaaaatcttgctaggatacagttctcacatcacatccgaacataaacaaccatgggtaagga
aaagactcacgtt
tcgaggccgcgattaaattccaacatggatgctgatttatatgggtataaatgggctcgcgataatgtcgggcaatcag
gtgcgacaatct
atcgattgtatgggaagcccgatgcgccagagttgtttctgaaacatggcaaaggtagcgttgccaatgatgttacaga
tgagatggtca
gactaaactggctgacggaatttatgcctcttccgaccatcaagcattttatccgtactcctgatgatgcatggttact
caccactgcgatc
cccggcaaaacagcattccaggtattagaagaatatcctgattcaggtgaaaatattgttgatgcgctggcagtgttcc
tgcgccggttgc
attcgattcctgtttgtaattgtccttttaacagcgatcgcgtatttcgtctcgctcaggcgcaatcacgaatgaataa
cggtttggttgatgc
gagtgattttgatgacgagcgtaatggctggcctgttgaacaagtctggaaagaaatgcataagcttttgccattctca
ccggattcagtc
gtcactcatggtgatttctcacttgataaccttatttttgacgaggggaaattaataggttgtattgatgttggacgag
tcggaatcgcaga
ccgataccaggatcttgccatcctatggaactgcctcggtgagttttctccttcattacagaaacggctttttcaaaaa
tatggtattgataa
tcctgatatgaataaattgcagtttcatttgatgctcgatgagtttttctaatcagtactgacaataaaaagattcttg
ttttcaagaacttgt
catttgtatagtttttttatattgtagttgttctattttaatcaaatgttagcgtgatttatattttttttcgcctcga
catcatctgcccagatgc
gaagttaagtgcgcagaaagtaatatcatgcgtcaatcgtatgtgaatgctggtcgctatactgataacttcgtataat
gtatgctatacg
aacggtaaattcctgggggaacaacttcacagaatgttttgtcatattgtcgaagtggtcacaaaacaagagaagttcc
gccaattataa
aaagggaacccgtatatttcagcttcacggatgatttccagggtgagagtactgtatatgggcttacgatagaaggcca
taaaaatttctt
gcttggcaacaaaatagaagtgaaatcatgtcgaggctgctgtgtgggagaacagcataaaatatcacaaaaaaagaat
ctaaaacac
tgtgttgcttgtcccagaaagggaatcaagtatttttataaagattggagtggtaaaaatcgagtatgtgctagatgct
atggaagataca
aattcagcggtcatcactgtataaattgcaagtatgtaccagaagcacgtgaagtgaaaaaggcaaaagacaaaggcga
aaaattggg
cattacgcccgaaggtttgccagttaaaggaccagagtgtataaaatgtggcggaatcttacagtggcctatgcggccg
ctctagaacta
gtggatcgatccccaattcgccctatagtgagtcgtattacaattcactggccgtcgattacaacgtcgtgactgggaa
aaccctggcgtt
acccaacttaatcgccttgcagcacatccccctttcgccagctggcgtaatagcgaagaggcccgcaccgatcgccctt
cccaacagttgc
gcagcctgaatggcgaatggcgcctgatgcggtattttctccttacgcatctgtgcggtatttcacaccgcatacgtca
aagcaaccatag
tacgcgccctgtagcggcgcattaagcgcggcgggtgtggtggttacgcgcagcgtgaccgctacacttgccagcgccc
tagcgcccgct
cctttcgctttcttcccttcctttctcgccacgttcgccggctttccccgtcaagctctaaatcggggtgggccatcgc
cctgatagacggtttt
tcgccctttgacgttggagtccacgttctttaatagtggactcttgttccaaactggaacaacactcaaccctatctcg
ggctattctatgat
ttataagggattttgccgatttcggcctattggttaaaaaatgagctgatttaacaaaaatttaacgcgaattttaaca
aaatattaacgtt
tacaattttatggtgcactctcagtacaatctgctctgatgccgcatagttaagccagccccgacacccgccaacaccc
gctgacgcgccc
tgacgggcttgtctgctcccggcatccgcttacagacaagctgtgaccgtctccgggagctgcatgtgtcagaggtttt
caccgtcatcacc
gaaacgcgcgagacgaaagggcctcgtgatacgcctatttttataggttaatgtcatgataataatggtttcttagacg
tcaggtggcact
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Mcggggaaatgtgcgcggaacccctatttgtttatttttctaaatacattcaaatatgtatccgctcatgagacaataa
ccctgataaatg
cttcaataatattgaaaaaggaagagtatgagtattcaacatttccgtgtcgcccttattcccttttttgcggcatttt
gccttcctgtttttgc
tcacccagaaacgctggtgaaagtaaaagatgctgaagatcagttgggtgcacgagtgggttacatcgaactggatctc
aacagcggta
agatccttgagagttttcgccccgaagaacgttttccaatgatgagcacttttaaagttctgctatgtggcgcggtatt
atcccgtattgacg
ccgggcaagagcaactcggtcgccgcatacactattctcagaatgacttggttgagtactcaccagtcacagaaaagca
tcttacggatg
gcatgacagtaagagaattatgcagtgctgccataaccatgagtgataacactgcggccaacttacttctgacaacgat
cggaggaccg
aaggagctaaccgcttttttgcacaacatgggggatcatgtaactcgccttgatcgttgggaaccggagctgaatgaag
ccataccaaac
gacgagcgtgacaccacgatgcctgtagcaatggcaacaacgttgcgcaaactattaactggcgaactacttactctag
cttcccggcaa
caattaatagactggatggaggcggataaagttgcaggaccacttctgcgctcggcccttccggctggctggtttattg
ctgataaatctg
gagccggtgagcgtgggtctcgcggtatcattgcagcactggggccagatggtaagccctcccgtatcgtagttatcta
cacgacgggga
gtcaggcaactatggatgaacgaaatagacagatcgctgagataggtgcctcactgattaagcattggtaactgtcaga
ccaagtttact
catatatactttagattgatttaaaacttcatttttaatttaaaaggatctaggtgaagatcctttttgataatctcat
gaccaaaatccctta
acgtgagattcgttccactgagcgtcagaccccgtagaaaagatcaaaggatcttcttgagatccattatctgcgcgta
atctgctgctt
gcaaacaaaaaaaccaccgcta ccagcggtggtttgtttgccggatcaagagctaccaactctttttccga
aggtaactggcttcagcag
agcgcagataccaaatactgtccttctagtgtagccgtagttaggccaccacttcaagaactctgtagcaccgcctaca
tacctcgctctg
ctaatcctgttaccagtggctgctgccagtggcgataagtcgtgtcttaccgggttggactcaagacgatagttaccgg
ataaggcgcagc
ggtcgggctgaacggggggttcgtgcacacagcccagcttggagcgaacgacctacaccgaactgagatacctacagcg
tgagcattg
agaaagcgccacgcttcccgaagggagaaaggcggacaggtatccggtaagcggcagggtcggaacaggagagcgcacg
agggagc
ttccagggggaaacgcctggtatctttatagtcctgtcgggtttcgccacctctgacttgagcgtcgatttttgtgatg
ctcgtcaggggggc
ggagcctatggaaaaacgccagcaacgcggcctttttacggttcctggccttttgctggccttttgctcacatgttctt
tcctgcgttatcccc
tgattctgtggataaccgtattaccgcctttgagtgagctgataccgctcgccgcagccgaacgaccgagcgcagcgag
tcagtgagcga
ggaagcggaagagcgcccaatacgcaaaccgcctctccccgcgcgttggccgattcattaatgcagctggcacgacagg
tttcccgact
ggaaagcgggcagtgagcgcaacgcaattaatgtgagttagctcactcattaggcaccccaggctttacactttatgct
tccgcggctcgt
atgttgtgtggaattgtgagcggataacaatttcacacaggaaacagct
p LOA-093
gacgaaagggcctcgtgatacgcctatttttataggttaatgtcatgataataatggtttcttagacgtcaggtggcac
ttttcggggaaat
gtgcgcggaacccctatttgtttatttttctaaatacattcaaatatgtatccgctcatgagacaataaccctgataaa
tgcttcaataatat
tgaaaaaggaagagtatgagtattcaacatttccgtgtcgcccttattcccttttttgcggcattttgccttcctgttt
ttgctcacccagaaa
cgctggtgaaagtaaaagatgctgaagatcagttgggtgcacgagtgggttacatcgaactggatctcaacagcggtaa
gatccttgag
agttttcgccccgaagaacgttttccaatgatgagcacttttaaagttctgctatgtggcgcggtattatcccgtattg
acgccgggcaaga
gcaactcggtcgccgcatacactattctcagaatgacttggttgagtactcaccagtcacagaaaagcatcttacggat
ggcatgacagt
aagagaattatgcagtgctgccataaccatgagtgataacactgcggccaacttacttctgacaacgatcggaggaccg
aaggagctaa
ccgcttttttgcacaacatgggggatcatgtaactcgccttgatcgttgggaaccggagctgaatgaagccataccaaa
cgacgagcgtg
acaccacgatgcctgtagcaatggcaacaacgagcgcaaactattaactggcgaactacttactctagcttcccggcaa
caattaatag
actggatggaggcggataaagttgcaggaccacttctgcgctcggcccttccggctggctggtttattgctgataaatc
tggagccggtga
gcgtgggtcCcgcggtatcattgcagcactggggccagatggtaagccctcccgtatcgtagttatctacacgacgggg
agtcaggcaac
tatggatgaacgaaatagacagatcgctgagataggtgcctcactgattaagcattggtaactgtcagaccaagtttac
tcatatatactt
tagattgatttaaaacttcatttttaatttaaaaggatctaggtgaagatcctttttgataatctcatgaccaaaatcc
cttaacgtgagtttt
cgttccactgagcgtcagaccccgtagaaaagatcaaaggatcttcttgagatcctttttttctgcgcgtaatctgctg
cttgcaaacaaaa
aaaccaccgctaccagcggtggtttgtttgccggatcaagagctaccaactctttttccgaaggtaactggcttcagca
gagcgcagatac
caaatactgttcttctagtgtagccgtagttaggccaccacttcaagaactctgtagcaccgcctacatacctcgctct
gctaatcctgttac
cagtggctgctgccagtggcgataagtcgtgtcttaccgggttggactcaagacgatagttaccggataaggcgcagcg
gtcgggctgaa
cggggggttcgtgcacacagcccagcttggagcgaacgacctacaccgaactgagatacctacagcgtgagctatgaga
aagcgccac
96
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
gcttcccga agggaga a aggcgga caggtatccggta agcggcagggtcgga acaggagagcgca
cgagggagcttccaggggga a
a cgcctggta tctttatagtcctgtcgggtttcgcca cctctga cttga
gcgtcgatttttgtgatgctcgtcaggggggcggagcctatgga
aaaacgccagcaacgcggcctttttacggttcctggccttttgctggccttttgctcacatgttattcctgcgttatcc
cctgattctgtggat
a a ccgtatta ccgcctttgagtgagctgata ccgctcgccgcagccga a
cgaccgagcgcagcgagtcagtgagcgagga agcgga ag
agcgccca ata cgca a a ccgcctctccccgcgcgttggccgattcatta atgcaggttta a a
cAGGTGGTAATAATCGCG CGAT
TCAATTGCATTCATTAAAGACAGATAATTCGCAAGACCTTCTCCCTCCAGATCAACTTGTATCAATGATTC
ACTTGTTCATCAACGATGAAAGGTTTACCTCCGGTATAACGAGITTTGACATTGATTTTTCTAGAATGAAA
ATGCCATAGAAATTTCTAAATTTAGACTGAATCCCTACGTCACTGGTTTAAAAATTGAGTGGTGCTTACTA
ATTATTACATTCGGAAACGTCTCATCAAGTGTTTCCGAAAAAATGAGGGTTTTTCTAAAGCTTCTTTCTTT
CACGGATATCACCGGGITTAAGATGTATTITTITTITCCACAGAAATTAAAGTTCCAGCGTTTACCAAAGT
AGATCGTTCAATAATATGGATGGTGTTATAAGAAGACGACCACTATCCCCCATGAATTCTCACATGATAC
TTTCTTTTACTTTATTTACAGAGGCAGTAACATCCAAGAAGAAta ccgttcgtata atgtatgctata cga
agttata a c
cggcgttgccagcgata aa cggatagcttca a a atgtttcta ctccttnttactcttccagattnctcgga
ctccgcgcatcgccgta cca
cttcaaaacacccaagcacagcatactaaatttcccctctttcttcctctagggtgtcgttaattacccgtactaaagg
tttggaaaagaaa
a a agaga ccgcctcgtttctttttcttcgtcga a a a aggca ata aa a
atttttatcacgtttctttttcttgaa a attttttttttgatttttttct
ctttcgatga cctcccattgatattta agtta ata a a cggtcttca atttctca agtttca
gtttcatttttcttgttctatta ca a cttttttta c
ttcttgctcattaga a aga a agcatagca atcta atcta
agtttATGGTATTTCCTGTACTCCCACTTTATGAGTCCACAG
GCAAACCTCTGTTGTCTGTTGTTGGCCAAGCTCTTTACAGATTTAATGGATCTAATACTGATGCAATAGTT
CAAATTTCCAAGTACACTCCAAACTTGAATGTGITTGTCGAATTGGCGGTCAATGAGATATCTGATGCTA
TTGTTGAACAATTACTCTCATTATACAACAATGGAGTTTGTTCGGTTTTAGCAACTCCCGAACAAGGAAG
TGTTATTCTTGAAAAAATCCCAAATGCAAGAATTACATATAAGGCATCCGAGAACAAACAATATCAGTCT
ATTGCCTACGTTTTGGGATCATCATTACCGCAGACCATTGATGAAAAGATTACTGCTTTTGTTTATGTTGA
AGACACGTTATCCCTTGAGGAATTACAAAAGCTCGTTAAGTCAGGTTATATTCCGATTGTCAAATCAGAC
TTATTGACAAATGAGTACGAAGATGTCAAGGGICAATATCCATTAGTTGATTTTATTATCCCTAAAATTGT
CACCGATCGTGCAGATGGACTATACACTACTCTGGTTGTGGATTCATCAAATCAATCTTTGGGTCTCGTG
TATTCATCTGTAACTTCTATTTCCGAATCAATTAGAACCGGTACAGGTGTTTATCAATCCAGGAAACATGG
TTTATGGTATAAGGGCAAAACCTCTGGTGCAACCCAAAAATTAATTTCTTTTGACCTAGACTGCGATTCC
GACTGTTTGAAGGCAATAGTCGAGCAAACTGGATCTGGATTTTGCCACCTGAGTACTAACTCATGTTTTG
GCAATTTTACAGGCTTGAAAGCCCTGGAAGCGACTCTATTTCAACGTAAAACAGATGCACCAGAAGGIT
CGTACACTAAACGTCTTTTTGATGATGAATCGTTGTTGAACGCTAAGATCAAAGAGGAAGCAGAAGAGT
TAGCAGATGCCAAAACCAAGGAAGAAATTGCTTGGGAGGCTGCTGATCTGTTTTATITTGCATTGGCTA
GATGTGCGAAATATAATGTTACCITAGCTGATATTGAAAAGAACTTGGACATGAAAGCATTGAAAG TIT
CAAGAAGAAAGGGCGATGCGAAACCTAAGTTTATTGAAAAGAAGCAATCAACAGAGAAGTCGGAAATT
AATGAAAGACATATTGGCCCAGATGACAAGATTTATTTGAATAGAATCAATGCTGCAACTGCATCAAAG
GAAGAAGTTGAAGCGTGCTTGGAAAGACCAATCCAGAAATCGGCAGATATTATGTCATTGGTTACTCCG
ATTGTTGAGAATGTCAAGGCGAACGGAGATAAGGCTCTATTGGAGTTAACTGCTAAGTTTGACAGGGTT
CAGTTAGATTCACCCGTTTTATTTGCTCCTTACAAGCCTGATATGATGCAAATCTCAGAGAAGCTAAAAA
AGGCGATCGATGTATCATTTGAAAATATCAGGATTITCCACGAAGCTCAAAATCAAAAGGATATTCTAAC
GGIGGAAACATCGCCAGGAGTTTACTGTTCTAGATTTGCTAGGCCTATCGAGAAGGTTGGTTGTTATATT
CCAGGIGGAACTGCTGTTTTGCCATCAACATCGTTGATGTTATCTGTTCCAGCATTAGTTGCTGGTTGCA
AGGAGATTATCTTTGCTTCTCCACCTG GTAAGGATGGTAAACTAACTCCAGAGGTTGTTTATGTAGCACA
CAAGGTTGGCGCCAAGTGTATTGTTATGGCAGGTGGAGCACAGGCTGTAGCAGCTATGGCTTATGGTAC
CGAGAGTGTTCCAAAATGTGATAAAATTATGGGTCCGGGTAATCAATTTGTCACTGCTGCTAAGATGTTA
GTCTCTAATGATTCCAATGCATTATGTGCCATTGATATGCCAGCAGGTCCATCTGAAGTATTAGTCATTGC
97
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
TGATAAGCATGCCGATCCTGATTTTGTTGCCAGCGATTTACTCTCACAAGCTGAACATGGTATCGATTCCC
AGGTCATTCTACTGGCTGTTGACATGACTGACGCAGAAGTTGATGCCATTGACGAAGCTGTCCATAGAC
AAGCTTTAGCGCTACCGAGAGTCGACATTGTTAGAAAATGTATTGCACATTCCACTACAATTGTAGTCAA
AACGCTGGATGAGGCATTTGAAATGTCCAACAAATATGCTCCAGAGCATTTGATTTTGCAGATTGAGAA
CGCAGAAGAATGGGTTCCTAAGGTTGACAATGCAGGTTCTGTCTTTGTTGGCGCATTATCGCCAGAATCT
TGTGGTGATTATTCCTCCGGTACTAACCATACATTACCTACGTATGGTTATGCTAGGATGTACAGCGGAG
TGAACACAGCAACCTTCCAAAAGTTCATCACCICTCAGGTTGTCACAAGGGAAGGITTGAAGAACATCG
GTCCTGCAGTTATGGATTTGGCTGAGGTTGAAGGTCTTGATGGCCACCGTAACGCCGTTAGGGTGAGAA
TGGATAAACTTGGTATGCTCCCTGAAGGATACTGAcgcctggcagcagggcgataacctcataacttcgtataatgtat
g
ctatacgaacggtaTTTGGTGTTGTTTTCTATTGCATACGAATTAGAATGCCCAGACTTGITTATATACTACGC
TGAATGTTTGTACATTTATACTTAAAACAAAATGCTAGTCAGCCATATTAAACAGAGCCGTTTAGCAACA
TTTCAATAGCACCTTCCACAGATCCACCGCTACGTYTCAATGCGGCAATGTTACGGTCGAAGTCAAAGAA
GCCCATATCGTTTAATTGACGTAATTGTGTTGCATAGACTTCTTCTGGAGGCCTTGTATCGGAAGCAGAA
GGTGCAGTTGAACCAGTACCAGTACTGGCACCACCAAAGAGATTCATTAAATTAGGATTTGCCAAAATG
GGATTACCTCCTAAACCTGCTCCAGGAACACCTGCTCCAAATAGTGACGCAAATGGATTGCTTGGTACAG
AGGAACCTGAAGAGTTTGAAGTTGAATTACGTGGCGAATCCGTGTTTG CAGTGTTAGAGTCAGATG GAT
TTCCGGGTGATGGGAAGTgttta a a cctggcgta atagcga agaggcccgca ccgatcgcccttccca a
cagttgcgca gcctg
a atggcga atggcgcctgatgcggtattttctcctta cgcatctgtgcggtatttcaca ccgcatatggtgca
ctctcagta ca atctgctct
gatgccgcatagttaagccagccccgacacccgccaacacccgctgacgcgccctgacgggcttgtctgctcccggcat
ccgcttacaga
caagctgtgaccgtctccgggagctgcatgtgtcagaggttttcaccgtcatcaccgaaacgcgcga
pLOA-094
gacgaaagggcctcgtgatacgcctatttttataggttaatgtcatgataataatggtttcttagacgtcaggtggcac
ttttcggggaaat
gtgcgcggaacccctatttgtttattntctaaatacattcaaatatgtatccgctcatgagacaataaccctgataaat
gcttcaataatat
tgaaaaaggaagagtatgagtattcaacataccgtgtcgcccttattcccttnttgcggcattttgccttcctgttttt
gctcacccagaaa
cgctggtgaaagtaaaagatgctgaagatcagttgggtgcacgagtgggttacatcgaactggatctcaacagcggtaa
gatccttgag
agttttcgccccgaagaacgttttccaatgatgagcacttttaaagactgctatgtggcgcggtattatcccgtattga
cgccgggcaaga
gcaactcggtcgccgcatacactattctcagaatgacttggttgagtactcaccagtcacagaaaagcatcttacggat
ggcatgacagt
aagagaattatgcagtgctgccataaccatgagtgataacactgcggccaacttacttctgacaacgatcggaggaccg
aaggagctaa
ccgcttttttgcacaacatgggggatcatgtaactcgccttgatcgttgggaaccggagctgaatgaagccataccaaa
cgacgagcgtg
acaccacgatgcctgtagcaatggcaacaacgttgcgcaaactattaactggcgaactacttactctagcttcccggca
acaattaatag
actggatggaggcggataaagttgcaggaccacttctgcgctcggccatccggctggctggtttattgctgataaatct
ggagccggtga
gcgtgggtcCcgcggtatcattgcagcactggggccagatggtaagccctcccgtatcgtagttatctacacgacgggg
agtcaggcaac
tatggatgaacgaaatagacagatcgctgagataggtgcctcactgattaagcattggtaactgtcagaccaagtttac
tcatatatactt
tagattgatttaaaacttcatttttaatttaaaaggatctaggtgaagatccatttgataatctcatgaccaaaatccc
ttaacgtgagatt
cgttccactgagcgtcagaccccgtagaaaagatcaaaggatcttcttgagatcctttttttctgcgcgtaatctgctg
cttgcaaacaaaa
aaaccaccgctaccagcggtggtttgtttgccggatcaagagctaccaactattttccgaaggtaactggcttcagcag
agcgcagatac
caaatactgttcnctagtgtagccgtagttaggccaccacttcaagaactctgtagcaccgcctacatacctcgctctg
ctaatcctgttac
cagtggctgctgccagtggcgataagtcgtgtcttaccgggttggactcaagacgatagttaccggataaggcgcagcg
gtcgggctgaa
cggggggttcgtgcacacagcccagcttggagcgaacgacctacaccgaactgagatacctacagcgtgagctatgaga
aagcgccac
gcttcccgaagggagaaaggcggacaggtatccggtaagcggcagggtcggaacaggagagcgcacgagggagcttcca
gggggaa
acgcctggtatctttatagtcctgtcgggtttcgccacctctgacttgagcgtcgatttttgtgatgctcgtcaggggg
gcggagcctatgga
aaaacgccagcaacgcggcctttttacggttcctggccttttgctggccttttgctcacatgttctttcctgcgttatc
ccctgattctgtggat
aaccgtattaccgcctttgagtgagctgataccgctcgccgcagccgaacgaccgagcgcagcgagtcagtgagcgagg
aagcggaag
98
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
agcgcccaatacgcaaaccgcctctccccgcgcgttggccgattcattaatgcaggtttaaacAGGTGGTAATAATCGC
GCGAT
TCAATTGCATTCATTAAAGACAGATAATTCGCAAGACCTTCTCCCTCCAGATCAACTTGTATCAATGATTC
ACTTGTTCATCAACGATGAAAGGTTTACCTCCGGTATAACGAGITTTGACATTGATTTTTCTAGAATGAAA
ATGCCATAGAAATTTCTAAATTTAGACTGAATCCCTACGTCACTGGTTTAAAAATTGAGTGGTGCTTACTA
ATTATTACATTCGGAAACGTCTCATCAAGTGTTTCCGAAAAAATGAGGGTTTTTCTAAAGCTTCTTTCTTT
CACGGATATCACCGGGTTTAAGATGTATTTTTTTTTTCCACAGAAATTAAAGTTCCAGCGTTTACCAAAGT
AGATCGTTCAATAATATGGATGGTGTTATAAGAAGACGACCACTATCCCCCATGAATTCTCACATGATAC
TTTCTTTTACTTTATTTACAGAGGCAGTAACATCCAAGAAGAAtaccgttcgtataatgtatgctatacgaagttataa
c
cggcgttgccagcgataaacggagcttgccttgtccccgccgggtcacccggccagcgacatggaggcccagaataccc
tccttgacagt
cttgacgtgcgcagctcaggggcatgatgtgactgtcgcccgtacatttagcccatacatccccatgtataatcatttg
catccatacatttt
gatggccgcacggcgcgaagca aaaattacggctcctcgctgcagacctgcgagcagggaa
acgctcccctcacaga cgcgttga attg
tccccacgccgcgcccctgtagagaaatataaaaggttaggatttgccactttttaaaatcttgctaggatacagttct
cacatcacatccg
aacataaacaaccatgggtaaaaagcctgaactcaccgcgacgtctgtcgagaagatctgatcgaaaagttcgacagcg
tctccgacc
tgatgcagctctcggagggcgaagaatctcgtgctttcagcttcgatgtaggagggcgtggatatgtcctgcgggtaaa
tagctgcgccg
atggtttctacaaagatcgttatgtttatcggcactttgcatcggccgcgctcccgattccggaagtgcttgacattgg
ggaattcagcgag
agcctgacctattgcatctcccgccgtgcacagggtgtcacgttgcaagacctgcctgaaaccgaactgcccgctgttc
tgcagccggtcg
cggaggccatggatgcgatcgctgcggccgatcttagccagacgagcgggttcggcccattcggaccgcaaggaatcgg
tcaatacact
acatggcgtgatttcatatgcgcgattgctgatccccatgtgtatcactggcaaactgtgatggacgacaccgtcagtg
cgtccgtcgcgc
aggctctcgatgagctgatgctttgggccgagga
ctgccccgaagtccggcacctcgtgcacgcggatttcggctccaa caatgtcctgac
ggacaatggccgcataacagcggtcattgactggagcgaggcgatgttcggggattcccaatacgaggtcgccaacatc
ttcttctggag
gccgtggttggcttgtatggagcagcagacgcgctacttcgagcggaggcatccggagcttgcaggatcgccgcggctc
cgggcgtatat
gctccgcattggtcttgaccaactctatcagagcttggttgacggcaatttcgatgatgcagcttgggcgcagggtcga
tgcgacgcaatc
gtccgatccggagccgggactgtcgggcgtacaca
aatcgcccgcagaagcgcggccgtctggaccgatggctgtgtagaagtactcgc
cgatagtggaaaccgacgccccagcactcgtccgagggcaaaggaataatcagtactgacaataaaaagattcttgttt
tcaagaactt
gtcatttgtatagtattttatattgtagttgactattttaatcaaatgttagcgtgatttatattattttcgcctcgac
atcatctgcccagat
gcgaagttaagtgcgcagaaagtaatatcatgcgtcaatcgtatgtgaatgctggtcgctatactgctgtcgattcgat
actaacgccgcc
atccagtgtcgacgcctggcagcagggcgataacctcataacttcgtataatgtatgctatacgaacggtaTTTGGTGT
TGTTTTCT
ATTGCATACGAATTAGAATGCCCAGACTTGTTTATATACTACGCTGAATGTTTGTACATTTATACTTAAAA
CAAAATGCTAGTCAGCCATATTAAACAGAGCCGTTTAGCAACATTTCAATAGCACCTTCCACAGATCCAC
CGCTACGTYTCAATGCGGCAATGTTACGGTCGAAGTCAAAGAAGCCCATATCGTTTAATTGACGTAATTG
TGTTGCATAGACTTCTTCTGGAGGCCTTGTATCGGAAGCAGAAGGTGCAGTTGAACCAGTACCAGTACT
GGCACCACCAAAGAGATTCATTAAATTAGGATTTGCCAAAATGGGATTACCTCCTAAACCTGCTCCAGGA
ACACCTGCTCCAAATAGTGACGCAAATGGATTGCTTGGTACAGAGGAACCTGAAGAGTTTGAAGTTGAA
TTACGTGGCGAATCCGTGTTTGCAGTGTTAGAGTCAGATGGATTTCCGGGTGATGGGAAGTgtttaaacctg
gcgtaatagcgaagaggcccgcaccgatcgccatcccaacagagcgcagcctgaatggcgaatggcgcctgatgcggta
ttttctcctt
acgcatctgtgcggtatttcacaccgcatatggtgcactctcagtacaatctgctctgatgccgcatagttaagccagc
cccgacacccgc
caacacccgctgacgcgccctgacgggcttgtctgctcccggcatccgcttacagacaagctgtgaccgtctccgggag
ctgcatgtgtca
gaggttttcaccgtcatcaccgaaacgcgcga
Fermentation Examples
99
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Example 2A
This example demonstrates producing a surprisingly high and commercially
relevant
yield of a combined amount of olivetol and olivetolic acid of about 4.5
g/liter over 4-5 days.
= Strain: LSC3-4
= Genotype: ga180^::pScTEF1>PkHIS4<tScGAL80
= Parent strain: LSC3-2
= Genotype of parent strain:
o pGa110-CsAAE1-tCyc1-pGa11-0ST2A0AC-tCyc1::1eu2-3,
pGa110-CsAAE1-tCyc1-pGa11-0ST2A0AC-tCyc1::ura3-52,
pGa110-CsAAE1-tCyc1-pGa11-0ST2A0AC-tCyc1::trp1,
pGa110-HMGIQR-tADH1-pGa11-1D11-tCyc1-KanMX::YORWA22
Fermentation Process Summary:
Seed Train:
A shake flask containing 50 mL YPD with 20 g/L glucose was inoculated with
freshly streaked
LSC3-4. Strain grew at 30 C for 24 hours to an 0D600 of 8. 40 mL of this
culture was used to
inoculate the fermentation tank.
Media:
Seed Media: YP with 20 g/L glucose
Batch media:
= lx YP + 55g/L glucose + 500mg/L histidine+12 mg/L rnyo-inosito1+12 mg/L
thiamin
hydrochloride+12 mg/L pyridoxal hydrochloride+12 mg/L nicotinic acid+12 mg/L
calcium
pantothenate+0.6 mg/L biotin+12 mg/L p-aminobenzoic acid+0.15 g/L EDTA+7.8
mg/L
CuSO4-5H20+0.0512 g/L FeSO4-7H20+0.0032 g/L MnC12+4.77 mg/L Na2Mo04+0.102 g/L
ZnSO4-7H20+0.0086 g/L CoC12-6H20+0.0384 g/L CaCl2-2H20+5.5 g/L KH2PO4+2.9 g/L
MgSO4-7H20+45.1 g/L (NH4)2504
Growth media:
= 600 g/L glucose+500 mg/L histidine
Production Media:
100
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
= 650 g/L glucose+10 g/L hexanoic acid
Base (for pH Control)
= 5M NH4OH
Galactose Addition
= 4 g galactose was added to tank at 24, 48 and 120 hours, respectively.
Overlay
= 100 mL isopropyl myristate (25% V/VO) was added to tank at 24 hours.
Additionally, 10
mL (2.5% V/Vo) isopropyl myristate was added to tank at 48 and 120 hours,
respectively.
Antifoam
= Struktol SB2121 (0.1 mL/L) at the beginning of the run
= Struktol SB509 (0.5 mL/day)
Fermentation Run Condition:
Pulse feeding was used for both growth phase and production phase during the
run.
Fermentation batch was inoculated with 40 mL of inoculum. Feeding was
triggered at the end
of batch phase when batch glucose was completely exhausted and p02 was
increased by 20%
(or more). Feed media (growth or production media) was delivered in pulses.
Each pulse
delivered 2 g glucose /starting batch volume with maximum feed rate not
exceeding 20 g
glucose/L/hr. pH was maintained at 6 throughout the run. Temperature of
fermenter was
maintained at 30 C. Air flow rate was maintained at 1.25 L/min. Agitation was
800 rpm.
Summary of Metrices:
= Maximum Olivetolic acid titer: 3.48 g/L
= Maximum Oliveto!: 0.9 g/L
= Total product titer: 4.36 g/L
101
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
= Titer/time at 119 hours: 0.86 g/L/day
= Cumulative yield of olivetolic acid /HA consumed: 0.49 mol/rriol
= Cumulative yield of olivetol/HA consumed: 0.15 mol/mol
Example 2B
In this example, a different strain was used compared to that used in example
2A, and no
galactose was added in this run. Surprisingly, despite the absence of
galactose, the combined
amount of olivetol and olivetolic acid obtained was about 3.5 g/liter over a
period of 4-5 days.
= Strain: LSC3-13
= Genotype: mig1^::HygR
= Parent strain: LSC3-5 (sister clone of LSC3-4)
= Genotype of parent strain: See example 2A
Fermentation Process Summary:
Seed Train:
A shake flask containing 50 mL YPD with 20 g/L glucose was inoculated with
freshly streaked
LSC3-13. Strain grew at 30 C for 24 hours to an 0D600 of 8. 40 mL of this
culture was used to
inoculate the fermentation tank.
Media:
Seed Media: YP with 20 g/L glucose
Batch media:
= lx YP + 55g/L glucose + 500mg/L histidine+12 mg/L myo-inosito1+12 mg/L
thiamin
hydrochloride+12 mg/L pyridoxal hydrochloride+12 mg/L nicotinic acid+12 mg/L
calcium
pantothenate+0.6 mg/L biotin+12 mg/L p-aminobenzoic acid+0.15 g/L EDTA+7.8
mg/L
CuSO4-5H20+0.0512 g/L FeSO4-7H20+0.0032 g/L MnC12+4.77 mg/L Na2Mo04+0.102 g/L
ZnSO4-7H20+0.0086 g/L C0C12-6H20+0.0384 g/L CaCl2-2H20+5.5 g/L KH2PO4+2.9 g/L
MgSO4-7H20+45.1 g/L (NH4)2504
Growth media:
= 600 g/L glucose+500 mg/L histidine
Production Media:
102
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
= 650 g/L glucose+10 g/L hexanoic acid
Base (for pH Control)
= 5M NH4OH
Overlay:
= 100 mL isopropyl myristate (25% V/VO) was added to tank at 24 hours.
Additionally, 10
mL (2.5% V/Vo) isopropyl myristate was added to tank at 48 and 120 hours,
respectively.
Antifoam
= Struktol SB2121 (0.1 mL/L) at the beginning of the run
= Struktol SB509 (0.5 mL/day)
Fermentation Run Condition:
Pulse feeding was used for both growth phase and production phase during the
run.
Fermentation batch was inoculated with 40 mL of inoculum. Feeding was
triggered at the end
of batch phase when batch glucose was completely exhausted and p02 was
increased by 20%
(or more). Feed media (growth or production media) was delivered in pulses.
Each pulse
delivered 2 g glucose /starting batch volume with maximum feed rate not
exceeding 20 g
glucose/L/hr. pH was maintained at 6 throughout the run. Temperature of
fermenter was
maintained at 30 C. Air flow rate was maintained at 1.25 L/min. Agitation was
800 rpm.
Summary of Metrices:
= Maximum Olivetolic acid titer: 2.92 g/L
= Maximum Oliveto!: 0.65 g/L
= Total product titer: 3.53 g/L
= Titer/time at 119 hours: 0.71 g/L/day
= Cumulative yield of olivetolic acid /HA consumed: 0.49 mol/mol
103
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
= Cumulative yield of olivetol/HA consumed: 0.10 mol/mol
Example 2C
This example demonstrates a high yielding fermentation of 0 and OA.
= Strain: LSC3-13
= Genotype: mig1^::HygR
= Parent strain: LSC3-5 (sister clone of LSC3-4)
= Genotype of parent strain: See example 2A
Fermentation Process Summary:
Seed Train:
A shake flask containing 50 mL YPD with 20 g/L glucose was inoculated with
freshly streaked
LSC3-13. Strain grew at 30 C for 24 hours to an 0D600 of 4. 40 mL of this
culture was used to
inoculate the fermentation tank.
Media:
Seed Media: YP with 20 g/L glucose
Batch media:
= lx YP + 55g/L glucose + 500mg/L histidine+12 mg/L myo-inosito1+12 mg/L
thiamin
hydrochloride+12 mg/L pyridoxal hydrochloride+12 mg/L nicotinic acid+12 mg/L
calcium
pantothenate+0.6 mg/L biotin+12 mg/L p-aminobenzoic acid+0.15 g/L EDTA+7.8
mg/L
CuSO4-5H20+0.0512 g/L FeSO4-7H20+0.0032 g/L MnC12+4.77 mg/L Na2Mo04+0.102
g/L ZnSO4-7H20+0.0086 g/L CoC12-6H20+0.0384 g/L CaCl2-2H20+5.5 g/L KH2PO4+2.9
g/L MgSO4-7H20+45.1 g/L (NH4)2504
Production Media:
= 650 g/L glucose +438 mg/L citric acid monohydrate+2 mg/L H3B03+1.3 mg/L
CuSO4-
5H20+22.4 mg/L FeC13-6H20+1.33 mg/L MnC12+0.8 mg/L Na2Mo04+10.8 mg/L ZnSO4-
7H20+12 mg/L nnyo-inosito1+12 mg/L thiamin hydrochloride+12 mg/L pyridoxal
hydrochloride+12 mg/L nicotinic acid+12 mg/L calcium pantothenate+12 mg/L
biotin+12
mg/L p-aminobenzoic acid+12 mg/L folic acid+12mg/L riboflavin+2.5 g/L KH2PO4+1
g/L
MgSO4-7H20+20 g/L (NH4)2SO4+17.8 g/L sodium hexanoate
= Base (for pH Control): 5M NH4OH
Overlay:
104
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
= 100 mL isopropyl myristate (25% V/VO) was added to tank at 24 hours.
Additionally, 10
mL (2.5% V/Vo) isopropyl myristate was added to tank at 96, 120 and 144 hours,
respectively.
Antifoam
= Struktol SB2121 (0.1 mL/L) at the beginning of the run
= Struktol SB509 (0.5 mL/day)
Fermentation run condition:
We used pulse feeding for both growth phase and production phase during the
run.
Fermentation batch was inoculated with 40 mL of inoculum. Feeding was
triggered at the end
of batch phase when batch glucose was completely exhausted and p02 was
increased by 20%
(or more). Feed media (growth or production media) was delivered in pulses.
Each pulse
delivered 2 g glucose /starting batch volume with maximum feed rate not
exceeding 40 g
glucose/L/hr. pH was maintained at 6 throughout the run. Temperature of
fermenter was
maintained at 30C. Air flow rate was maintained at 1.25 L/min. Agitation was
800 rpm. This
fermentation works at pH range of 5-6. It is contemplated that in some
embodiments, the
fermentation is carried out at about pH 5Ø It is contemplated that in some
embodiments, a
pulse rate of about 1.7 g/L/pulse, with maximum feed rate of about 10 g/L/hr
is employed. It is
contemplated that in some embodiments, a pulse rate of about 1.7 g/L/pulse is
employed. It is
contemplated that in some embodiments a maximum feed rate of about 10 g/L/hr
is employed.
Summary of metrics:
= Maximum Olivetolic acid titer: 6.07 g/L
= Maximum Oliveto!: 2.03 g/L
= Total product titer: 8 g/L (see Figure 4A)
= Titer/time at 119 hours: 1.5 g/L/day (see Figure 4B)
= Cumulative yield of olivetolic acid /HA consumed: 0.93 g/g
= Cumulative yield of olivetol/HA consumed: 0.27 g/g
105
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Example 2D
= Strain: LSC3-134
Genotype: ga180^::(1oxHIS4)/ his4^/ mig1^::(1oxPkHIS4)
= Parent strain: LSC300002
= Genotype of parent strain:
le L1i 2"::ScLEU2<pScLELI2AScaC1>CsAEE1<pScGAL10,/pScGAL1>CsTKS-T2A-CsOAC<LSca
ClipAG305-backbonei1eu2(defective)ura3A::pScURA3>ScURA3/tScCYC1>CsAEE1<pScG
AL10/pScGAL1>CsTKS-T2A-CsOAC<tScCYClipAG306-backbonelura3(defective)trplA::p
ScTRP1>ScTRPVtScCYC1_>CsAEE1<pScGAL10/pScGAL1>CsTKS-T2A-CsOAC<tScCYCljpAG
304-backboneArpl(defective)yorWdelta22A:tScADH1>FIMGK2R<pScGAL10/pScGAL1>1
Dil<tScCYCIIKanNIX
Fermentation Process Summary:
Seed Train:
A shake flask containing 50 mL YPD (10 g/L yeast extract, 20 g/L peptones and
20 g/L glucose)
was inoculated with freshly streaked LSC3-134. Strain grew at 30 C for 24
hours to an 0D600 of
4. 17 mL of this culture was used to inoculate the fermentation tank (3.5% of
initial tank
volume).
Media:
Seed Media: YPD (10 g/L yeast extract+ 20 g/L peptones+ 20 g/L glucose)
Batch media:
= 10 g/L yeast extract+ 20 g/L peptones+ 20 g/L glucose + 500mg/L
histidine+12 mg/L
myo-inosito1+12 mg/L thiamin hydrochloride+12 mg/L pyridoxal hydrochloride+12
mg/L
nicotinic acid+12 mg/L calcium pantothenate+0.6 mg/L biotin+12 mg/L p-
aminobenzoic
acid+0.15 g/L EDTA+7.8 mg/L CuSO4-5H20+0.0512 g/L FeSO4-7H20+0.0032 g/L
106
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
MnC12+4.77 mg/L Na2Mo04+0.102 g/L ZnSO4-7H20+0.0086 g/L CoC12-6H20+0.0384
g/L CaCl2-2H20+5.5 g/L KH2PO4+2.9 g/L MgSO4-7H20+45.1 g/L (NH4)2504
Production Media:
= 650 g/L glucose +438 mg/L citric acid nnonohydrate+2 mg/L H3B03+1.3 mg/L
CuSO4-
5H20+22.4 mg/L FeC13-6H20+1.33 mg/L MnC12+0.8 mg/L Na2Mo04+10.8 mg/L ZnSO4-
7H20+12 mg/L myo-inosito1+12 mg/L thiamin hydrochloride+12 mg/L pyridoxal
hydrochloride+12 mg/L nicotinic acid+12 mg/L calcium pantothenate+12 mg/L
biotin+12
mg/L p-aminobenzoic acid+12 mg/L folic acid+12mg/L riboflavin+2.5 g/L KH2PO4+1
g/L
MgSO4-7H20+20 g/L (NH4)2504+36 g/L sodium hexanoate
= Base (for pH Control): 5M NH4OH
Overlay:
= 182 mL isopropyl myristate (IPM, 40% V/VO) was added to tank at 24 hours.
Stir rate
was reduced to 500 rpm before addition of IPM at 24 hours and was increased to
800
rpm at around 48 hours. This step was performed to eliminate the risk of
foaming after
IPM addition. 30% to 50% positive p02 was maintained between 24 and 48 hours
runs
time. 1.6% )V/V) antifoam was added to IPM before addition to tank.
= Antifoam: Struktol SB2121 (0.1 mL/L) at the beginning of the run
Fermentation run condition:
We used pulse feeding for both growth phase and production phase during the
run.
Fermentation batch was inoculated with 17 mL of inoculum. Feeding was
triggered at the end
of batch phase when batch glucose was completely exhausted and p02 was
increased by 10%
(or more). Feed media (production media) was delivered in pulses. Each pulse
delivered 1.7 g
glucose /starting batch volume with maximum feed rate not exceeding 10 g
glucose/L/hr. pH
was maintained at 5.5 throughout the run. Temperature of fermenter was
maintained at 30C.
Air flow rate was maintained at 1.25 L/min. Agitation was 800 rpm. Median
oxygen uptake rate
107
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
(OUR) was 60-80 mmoles/L/hr. A maximum OUR of 100-110 moles/L/hr was achieved
during
the process.
Summary of metrics:
= Maximum Olivetolic acid titer: 6.95 0.22
= Maximum Olivetol: 2.68 0.24
= Titer/time at 96 hours: 2.2 g/L/day (0A+0)
= Cumulative yield of olivetolic acid /Hexanoic Acid consumed:1.4 g/g
= Cumulative yield of olivetol/Hexanoic Acid consumed:0.56 g/g
Effect of pH on process metrics
We found that optimum pH for our process is 5.5 (+/-) 0.3.
Effect of temperature on process metrics
We found that optimum temperature for our process is 30(+/-) 2
Optimum time to add IPM: We found that the optimum time to add IPM is between
12 and 36
hours post inoculation.
Effect of sodium hexanoate/glucose ratio in feed: In a series of experiments,
we tested
sensitivity of metrics (titer and productivity) to the ratio of sodium
hexanoate to glucose in
feed. We found that the maximum olivetol equivalent titer was achieved when
sodium
hexanoate/glucose in feed ratio was in the range of 20 to 28 g sodium
hexanoate/ 500 g
glucose. Maximum productivity was achieved at a range of 23 to 28 g sodium
hexanoate/500 g
glucose in feed.
Effect of Oxygen transfer rate on metrics: We found that the optimum median
OTR for our
process is 60-80 mmoles/L/hr and a maximum OUR of 100-110 mmoles/L/hr is
achieved in our
process.
Pulse metric parameters: We found that the optimum pulse parameters for our
process was
1.7 g glucose/L initial tank volume/pulse with a maximum feed rate of 10 g/L
of initial tank
volume/hr.
108
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Effect of overlay: Isopropyl myristate is used as an overlay in our process.
The optimum IPM
loading for our process at pH 5.5 is 26% of total tank volume or 40% of
initial tank volume.
There was a clear negative effect when no IPM was used.
Note: Percentages of IPM reported here in this figure are based on total tank
volume.
Effect of batch glucose concentration: We found that the optimum batch glucose
concentration for our process was 10-20 g/L.
Seed train condition: We found that the optimum seed train condition was to
inoculate an
initial flask containing YPD (10 g/L yeast extract, 20 g/L peptones and 20 g/L
glucose) with 1 mL
seed vial. All subsequent seed tanks would be inoculated with 2% inoculum and
will run as
batch tanks with pH control (pH set at 5.5). The production tank will be
inoculated with 3.5%
inoculum from the last seed train stage.
= Batch media composition for seed tanks: lx VP + 55g/L glucose + 500mg/L
histidine+12
mg/L myo-inosito1+12 mg/L thiamin hydrochloride+12 mg/L pyridoxal
hydrochloride+12
mg/L nicotinic acid+12 mg/L calcium pantothenate+0.6 mg/L biotin+12 mg/L p-
aminobenzoic acid+0.15 g/L EDTA+7.8 mg/L CuSO4-5H20+0.0512 g/L FeSO4-
7H20+0.0032 g/L MnC12+4.77 mg/L Na2Mo04+0.102 g/L ZnSO4-7H20+0.0086 g/L
CoC12-6H20+0.0384 g/L CaCl2-2H20+5.5 g/L KI-12PO4+2.9 g/L MgSO4-7H20+45.1 g/L
(N H4)2504
= Batch media composition for production tanks: lx VP + 17-20g/L glucose +
500mg/L
histidine+12 mg/L myo-inosito1+12 mg/L thiamin hydrochloride+12 mg/L pyridoxal
hydrochloride+12 mg/L nicotinic acid+12 mg/L calcium pantothenate+0.6 mg/L
biotin+12 mg/L p-aminobenzoic acid+0.15 g/L EDTA+7.8 mg/L CuSO4-5H20+0.0512
g/L
FeSO4-7H20+0.0032 g/L MnC12+4.77 mg/L Na2Mo04+0.102 g/L ZnSO4-7H20+0.0086
g/L CoC12-6H20+0.0384 g/L CaCl2-2H20+5.5 g/L KH2PO4+2.9 g/L MgSO4-7H20+45.1
g/L
(N H4)2504
= Production Media (for production tank): 650 g/L glucose +438 mg/L citric
acid
monohydrate+2 mg/L H3B03+1.3 mg/L CuSO4-5H20+22.4 mg/L FeC13-6H20+1.33 mg/L
MnC12+0.8 mg/L Na2Mo04+10.8 mg/L ZnSO4-7H20+12 mg/L myo-inosito1+12 mg/L
thiamin hydrochloride+12 mg/L pyridoxal hydrochloride+12 mg/L nicotinic
acid+12 mg/L
109
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
calcium pantothenate+12 mg/L biotin+12 mg/L p-aminobenzoic acid+12 mg/L folic
acid+12mg/L riboflavin+2.5 g/L KH2PO4+1 g/L MgSO4-7H20+20 g/L (NH4)2504+36 g/L
sodium hexanoate
= Base (for pH Control): 5-10 M NH4OH
Example 3: Divarinic acid/Divarin Production in LSC3-2 and derived strains
Precultures of LSC3-2 were grown in 50 mL tubes containing 10 mL of YP glucose
overnight. These were used to inoculate 50 mL tubes containing 10 mL of YP +
2% (w/v)
galactose + 2 mM (176 mg/L) butyric acid (BA) + 20% (v/v) isopropyl myristate
(IPM) and were
grown for 48 hours with 180 rpm shaking at 30 C. D/DA present in IPM layer
only, is tabulated
below based on standard curves run for DA and D, and titers are based on the
whole volume of
broth plus overlay (12 mL total).
: ]!:
ffeplicate-,-Divarinic-and (mg/L) = =Divann (rng/L)'-DivariTT equivalents
(mg/4
1 43.5 16.1 49.9
2 36.8 16.1 44.5
Additionally, the same experiment was conducted with 50 mL of YP + 2% (w/v)
galactose
+ 2 mM BA + 20% (v/v) IPM in 250 mL shake flasks. D/DA was quantified in both
the IPM and
aqueous layers in two biological replicates, and summed between the phases:
Divarinic acid (mg/L) : Divarrn (mg/L) Mann equivalents (rng/L)Replicate
1 12.9 7.6 17.6
2 16.1 8.2 20.8
110
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
In another experiment, the same growth conditions were employed with 50 mL of
YP +
2% (w/v) galactose + 2 mM BA with no overlay in 250 mL shake flasks. D/DA was
quantified in
the aqueous culture broth:
:::Divarinic acid (mg/L) Diva rin (rng/L): Divarin equivalents (mg/4
miummonumamionmiongimonimi=mium:
1 30.0 9.4 32.6
1
In a final shake flask/tube experiment, the same growth conditions were
employed with
mL of VP + 2% (w/v) galactose + 4 mM BA + 20% (v/v) IPM in 50 mL Falcon tubes.
D/DA was
quantified in both the IPM and aqueous layers, and summed between phases:
bivarinic acid (mg/L) tivarin (mg/L) bivarin equivalents (mgja:i
Replicate
1 I 30.0 7.2 30.5
The shake flask/tube experiments were scaled down to 96 well deepwell plate
format.
Precultures from colonies of each strain were grown in 300 p.L VP + 2% (w/v)
glucose. Main
cultures containing 300 p.L VP + 2% (w/v) galactose + 0.02-0.08% (w/v) butyric
acid + 20% (v/v)
IPM (60 p.L) or 20% (v/v) diethyl sebacate (60 L) for strain LSC3-2, or the
same but with 2%
(w/v) glucose instead of galactose for strains LSC3-4, LSC3-13, and LSC3-18,
were grown for 48
hours and the IPM or diethyl sebacate overlay was sampled at 24 and 48 hours
following
acidification of the media with 10 [IL of 5 M phosphoric acid. Three
replicates of each
strain/media condition were tested and averages and standard deviations for
detected divarinic
acid, divarin, and total divarin equivalents are reported in the table below
for 24 hour and 48
hour sampling:
24 hour sampling:
111
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
diva rinic acid total
divarin
strain medium overlay divarin (mg/L) (mg/L) eq
(mg/L)
LSC3-2 VP gal + 0.02% BA IPM 0.000 0.000 0.000
LSC3-2 VP gal + 0.04% BA IPM 0.000 0.000 0.000
LSC3-2 VP gal + 0.08% BA IPM 0.000 0.000 0.000
LSC3-4 VP glu 0.02 BA IPM 0.000 0.000 0.000
LSC3-4 VP glu 0.04 BA IPM 0.000 0.000 0.000
LSC3-4 VP glu 0.08 BA IPM 0.000 0.000 0.000
LSC3-13 VP glu 0.02 BA IPM 0.000 0.000 0.000
LSC3-13 VP glu 0.04 BA IPM 0.000 0.000 0.000
LSC3-13 VP glu 0.08 BA IPM 0.000 0.000 0.000
LSC3-18 VP glu 0.02 BA IPM 0.000 0.000 0.000
LSC3-18 VP glu 0.04 BA IPM 0.000 0.000 0.000
LSC3-18 VP glu 0.08 BA IPM 0.000 0.000 0.000
LSC3-48 VP gal 0.02 BA IPM 0.000 0.000 0.000
LSC3-48 VP gal 0.04 BA IPM 0.000 0.000 0.000
LSC3-48 VP gal 0.08 BA IPM 0.000 0.000 0.000
LSC3-77 VP gal 0.02 BA IPM 0.000 0.000 0.000
LSC3-77 VP gal 0.04 BA IPM 0.000 0.000 0.000
LSC3-77 VP gal 0.08 BA IPM 0.000 0.000 0.000
LSC3-2 VP gal 0.02 BA DESeb 0.000 0.000 0.000
LSC3-2 VP gal 0.04 BA DESeb 0.000 2.973 2.306
LSC3-2 VP gal 0.08 BA DESeb 0.000 0.715 0.555
48 hour sampling:
diva rinic acid total
divarin
strain medium overlay divarin (mg/L) (mg/L) eq
(mg/L)
LSC3-2 VP gal 0.04 BA IPM 0.000 1.213 0.941
LSC3-2 VP gal 0.08 BA IPM 1.438 2.211 3.153
LSC3-4 VP glu 0.02 BA IPM 0.000 0.000 0.000
LSC3-4 VP glu 0.04 BA IPM 0.383 1.529 1.569
LSC3-4 VP glu 0.08 BA IPM 6.330 13.526 16.822
LSC3-13 VP glu 0.02 BA IPM 3.119 10.223 11.049
LSC3-13 VP glu 0.04 BA IPM 4.437 13.383 14.817
LSC3-13 VP glu 0.08 BA IPM 3.686 11.012 12.228
LSC3-18 VP glu 0.02 BA IPM 0.000 0.000 0.000
LSC3-18 VP glu 0.04 BA IPM 1.307 6.087 6.029
LSC3-18 VP glu 0.08 BA IPM 8.293 17.935 22.205
LSC3-48 VP gal 0.02 BA IPM 0.000 0.000 0.000
LSC3-48 VP gal 0.04 BA IPM 0.000 0.000 0.000
LSC3-48 VP gal 0.08 BA IPM 0.000 2.300 1.784
112
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
LSC3-77 VP gal 0.02 BA IPM 0.000 1.631 1.265
LSC3-77 VP gal 0.04 BA IPM 0.528 0.846 1.184
LSC3-77 VP gal 0.08 BA IPM 3.720 5.787 8.209
LSC3-2 VP gal 0.02 BA DESeb 2.759 6.103 7.492
LSC3-2 VP gal 0.04 BA DESeb 2.872 5.688 7.284
LSC3-2 VP gal 0.08 BA DESeb 2.176 3.291 4.729
Example 3A: Divarinic acid/divarin production in LSC3-2 and derived strains in
defined media
with varying pH
One step in optimizing divarinic acid/divarin production was to identify the
optimum pH
for production and to investigate titers in alternative media. pH was adjusted
using a defined
medium with buffers added that were pre-adjusted to different pH values. A
defined medium,
Delft CSM medium, consisted of (per liter solution) 7.5 g ammonium sulfate,
14.4 g potassium
phosphate monobasic, 0.5 g magnesium sulfate heptahydrate (with these first
three
components prepared as an 0.9X solution and adjusted to pH 6.5 with sodium
hydroxide prior
to autoclaving), 3.6 mL of a trace metal solution (consisting of 130 g/L
citric acid monohydrate,
0.574 g/L copper (II) sulfate pentahydrate, 8.07 g/L iron (III) chloride
hexahydrate, 0.5 g/L boric
acid, 0.333 g/L manganese (II) chloride, 0.2 g/L sodium molybdate, and 4.67
g/L zinc sulfate
heptahydrate), 1.0 mL of a vitamin solution (0.008 g/L biotin, 1.6 g/L calcium
pantothenate,
0.008 g/L folic acid, 8 g/L myo-inositol, 1.6 g/L nicotinic acid, 0.8 g/L p-
aminobenzoic acid, 1.6
g/L pyridoxal hydrochloride, 0.8 g/L riboflavin, 1.6 g/L thiamine
hydrochloride, adjusted to pH
10.5 with sodium hydroxide), 0.79 g of Complete Supplement Mixture (Formedium,
Norfolk,
UK), and 2% (w/v) of either galactose or glucose where specified. The final
media was filter-
sterilized, and butyric acid was added to 0.04% (w/v) for production. Media
with different pHs
were prepared according to the same recipe, only with 3.75 g/L ammonium
sulfate, 7.2 g/L
potassium phosphate monobasic, and 0.25 g magnesium sulfate heptahydrate,
adjusted to pH
6.5 and autoclaved (the pH the next day was measured to be 6.63), with other
components the
same as above, plus 300 nnM of 2-(N-nnorpholino)ethanesulfonic acid (MES) from
a 1 M stock
adjusted to pH 5.0, 5.75, 6.0, 6.25, or 6.5. The final pH of each media
formulation with different
pH MES buffers added are shown in the table below:
113
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
MES buffer added (medium name) pH of final medium
MES pH 5.0 (Delft CSM pH 5.0) 5.69
MES pH 5.75 (Delft CSM pH 5.75) 6.08
MES pH 6.0 (Delft CSM pH 6.0) 6.20
MES pH 6.25 (Delft CSM pH 6.25) 6.42
MES pH 6.5 (Delft CSM pH 6.5) 6.60
LSC3-2, LSC3-4, LSC3-13, and LSC3-18 were then tested in 96 well deepwell
plates as described
in the last experiment in Example 2, with precultures containing 2% (w/v)
glucose as carbon
source, and production cultures containing 2% (w/v) galactose for LSC3-2, 2%
(w/v) glucose for
LSC3-4, LSC3-13, and LSC3-18, 20% (v/v) IPM overlay, plus 0.04% (w/v) butyric
acid as substrate.
After 48 hours, production cultures were acidified with 10 p.L of 5 M
phosphoric acid, and the
IPM layer was sampled. Divarinic acid and divarin were quantified by HPLC and
detected whole
broth + overlay titers, averaged across 3 biological replicates. Up to 59.6
mg/L divarinic acid
plus 19.0 mg/L divarin were produced by strain LSC3-13 in full-strength Delft
CSM medium plus
2% (w/v) glucose. In reduced salt strength buffered media, addition of MES pH
5.0 (leading to a
final medium pH of 5.69) appeared optimal for most strains, with less of a pH
dependence in
production observed for LSC3-13. When normalized to optical density (600 nm),
a measure of
cell density, it is clear that the medium with MES pH 5.0 also resulted in the
highest yield of
divarinic acid/divarin (expressed as "divarin equivalents", which are equal to
the titer of divarin
plus the titer of divarinic acid multiplied by the ratio of the molecular
weight of divarin to
divarinic acid) per unit of biomass. LSC3-13 exhibited the highest yield per
unit of biomass of
the 4 tested strains.
Example 3B
This example provides a process of producing divarin and/or divarinic acid .
= Strain: LSC3-4
= Genotype: ga180^::pScTEF1>PkHIS4<tScGAL80
= Parent strain: LSC3-2
= Genotype of parent strain:
114
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
o pGa110-CsAAE1-tCyc1-pGa11-0ST2A0AC-tCyc1::1eu2-3,
pGa110-CsAAE1-tCyc1-pGa11-0ST2A0ACACyc1::ura3-52,
pGa110-CsAAE1-tCyc1-pGa11-0ST2A0AC-tCyc1::trp1,
pGa110-HMGK2R-tADH1-pGa11-1D11-tCyc1-KanMX::YORWA22
Fermentation Process Summary:
Seed Train:
A shake flask containing 50 mL YPD with 20 g/L glucose is inoculated with
freshly streaked LSC3-
4. Strain grows at 30 C for 24 hours to an 0D600 of 8. 40 mL of this culture
is used to inoculate
the fermentation tank.
Media:
Seed Media: YP with 20 g/L glucose
Batch media:
= lx YP + 55g/L glucose + 500mg/L histidine+12 mg/L myo-inosito1+12 mg/L
thiamin
hydrochloride+12 mg/L pyridoxal hydrochloride+12 mg/L nicotinic acid+12 mg/L
calcium
pantothenate+0.6 mg/L biotin+12 mg/L p-aminobenzoic acid+0.15 g/L EDTA+7.8
mg/L
CuSO4-5H20+0.0512 g/L FeSO4-7H20+0.0032 g/L MnC12+4.77 mg/L Na2Mo04+0.102 g/L
ZnSO4-7H20+0.0086 g/L CoC12-6H20+0.0384 g/L CaCl2-2H20+5.5 g/L KH2PO4+2.9 g/L
MgSO4-7H20+45.1 g/L (NH4)2504
Growth media:
= 600 g/L glucose+500 mg/L histidine
Production Media:
= 650 g/L glucose+10 g/L hexanoic acid
Base (for pH Control)
= 5M NH4OH
Galactose Addition
= 4 g galactose was added to tank at 24, 48 and 120 hours, respectively.
115
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Overlay
= 100 mL isopropyl myristate (25% V/VO) is added to tank at 24 hours.
Additionally, 10 mL
(2.5% V/Vo) isopropyl myristate is added to tank at 48 and 120 hours,
respectively.
Antifoam
= Struktol SB2121 (0.1 mL/L) at the beginning of the run
= Struktol SB509 (0.5 mL/day)
Fermentation Run Condition:
Pulse feeding is used for both growth phase and production phase during the
run.
Fermentation batch is inoculated with 40 mL of inoculum. Feeding is triggered
at the end of
batch phase when batch glucose is completely exhausted and p02 is increased by
20% (or
more). Feed media (growth or production media) is delivered in pulses. Each
pulse delivered 2 g
glucose /starting batch volume with maximum feed rate not exceeding 20 g
glucose/L/hr. pH is
maintained at 6 throughout the run. Temperature of fernnenter is maintained at
30C. Air flow
rate is maintained at 1.25 L/min. Agitation is 800 rpm.
Example 3C
In this example, a different strain is used compared to that used in example
3A, and no
galactose is added in this run.
= Strain: LSC3-13
= Genotype: mig1^::HygR
= Parent strain: LSC3-5 (sister clone of LSC3-4)
= Genotype of parent strain: See example 3A
Fermentation Process Summary:
116
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Seed Train:
A shake flask containing 50 mL YPD with 20 g/L glucose is inoculated with
freshly streaked LSC3-
13. Strain grew at 30 C for 24 hours to an 0D600 of 8. 40 mL of this culture
is used to inoculate
the fermentation tank.
Media:
Seed Media: YP with 20 g/L glucose
Batch media:
= lx YP + 55g/L glucose + 500mg/L histidine+12 mg/L myo-inosito1+12 mg/L
thiamin
hydrochloride+12 mg/L pyridoxal hydrochloride+12 mg/L nicotinic acid+12 mg/L
calcium
pantothenate+0.6 mg/L biotin+12 mg/L p-aminobenzoic acid+0.15 g/L EDTA+7.8
mg/L
CuSO4-5H20+0.0512 g/L FeSO4-7H20+0.0032 g/L MnC12+4.77 mg/L Na2Mo04+0.102 g/L
ZnSO4-7H20+0.0086 g/L CoC12-6H20+0.0384 g/L CaCl2-2H20+5.5 g/L KH2PO4+2.9 g/L
MgSO4-7H20+45.1 g/L (NH4)2SO4
Growth media:
= 600 g/L glucose+500 mg/L histidine
Production Media:
= 650 g/L glucose+10 g/L hexanoic acid
Base (for pH Control)
= 5M NH4OH
Overlay:
= 100 mL isopropyl myristate (25% V/VO) is added to tank at 24 hours.
Additionally, 10 mL
(2.5% V/Vo) isopropyl nnyristate is added to tank at 48 and 120 hours,
respectively.
Antifoam
= Struktol SB2121 (0.1 mL/L) at the beginning of the run
= Struktol SB509 (0.5 mL/day)
117
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
Fermentation Run Condition:
Pulse feeding is used for both growth phase and production phase during the
run.
Fermentation batch is inoculated with 40 mL of inoculum. Feeding is triggered
at the end of
batch phase when batch glucose is completely exhausted and p02 is increased by
20% (or
more). Feed media (growth or production media) is delivered in pulses. Each
pulse delivered 2 g
glucose /starting batch volume with maximum feed rate not exceeding 20 g
glucose/L/hr. pH is
maintained at 6 throughout the run. Temperature of fermenter is maintained at
30 C. Air flow
rate is maintained at 1.25 L/min. Agitation is 800 rpm.
In a fermentation experiment run substantially as Example 3B, and employing
strain
LSC3-134A or LSC3-134 as disclosed here, surprisingly, a titer of 2 g/L of
divarin equivalent was
obtained.
Example 4A: Growth of S. cerevisiae strains in overlay/underlay candidates
In-situ liquid-liquid extraction (biphasic fermentation) is a strategy that
can be employed
in accordance with the present invention for physical separation of product
from cells via
partitioning into a second liquid phase from an aqueous culture phase. The
second or organic
liquid phase is present as either an overlay if its density is less than that
of the aqueous phase,
or underlay if its density is greater than that of the aqueous phase. Certain
properties of the
overlay or underlay are considered for production of olivetolic acid/olivetol
and other
resorcinols such as formulas IA and IB: (1) non-toxic or low toxicity for
growth of the host strain,
(2) a favorable partition coefficient of the product in the organic phase vs.
the aqueous phase,
and (3) preferably a lower partition coefficient for fed hexanoic acid (for
olivetolic acid/olivetol)
or other fatty acid such as RCO2H (for other resorcinols) in the organic phase
vs. the aqueous
phase. Additional properties of the organic phase enhance its suitability for
downstream
conversion, e.g. and without limitation, to cannabigerol and other cannabinoid
compounds,
including suitability as a solvent or co-solvent during downstream prenylation
or other
reactions, and boiling point if downstream separation by distillation is
employed.
To test non-toxic organic overlays/underlays, different non-production
background and
production strains of S. cerevisiae (Table 1) were inoculated from colonies on
YPO agar plates
118
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
and grown for approximately 24 hours in 96 well deepwell plates containing 300
L YP + 2%
(w/v) glucose at 30 C with 950 rpm shaking (3 mm throw) and maintained at 85%
humidity.
These cultures were then used to inoculate 96 well deepwell plates containing
3004 YP + 2%
(w/v) galactose (YP gal) or glucose (YP glu), with or without addition of
0.04% (w/v) hexanoic
acid (HA) plus 20% (v/v) (604) of different overlays/underlays or no organic
phase, under the
same conditions described for precultures. At three different times (specified
in plots),
biological triplicate cultures were sampled and optical density at 600 nm
(0D600) was
measured as a proxy for biomass growth on a SpectraMax plate reader, with
dilution in water
to allow measurements to be within the linear range of instrumental readings.
Averaged
OD600s across at least three biological replicates of each strain/overlay
condition at each
sampling time. Multiple sets of measurements were performed on large groups of
overlay and
underlay candidates in different batches.
Table 1: Strain IDs and description/genotype features. Double colons (::)
indicate replacement of the
indicated locus to the left of the colons with the integration cassette to the
right of the colons. PkHIS4 is
the HIS4 gene from Pichia kudriavzevii under control of a TEF1 promoter from
S. cerevisiae. HygR is a
hygromycin resistance cassette. Defective genes that generate auxotrophies are
indicated in
parentheses and strains that have none listed are fully prototrophic.
Strain ID Description/genotype
LSC3-1 JK9-3d "wild-type" (HIS4- LEU2- TRP1- URA3-)
LSC3-2 ¨15 copies of OA production pathway under pGAL1-10
bidirectional promoter
in LSC3-1 (HIS4-)
LSC3-4 LSC3-2 GAL80::PkHIS4
LSC3-13 LSC3-4 MIG1::HygR
LSC3-18 LSC3-4 GAL1::HygR
LSC7-1 CEN.PK2-1C MAToc (HIS3- LEU2-TRP1- URA3-)
The performance of various classes of organic phase compounds are provided
herein.
Among the diesters tested, certain were toxic to growth under the test
conditions. Diethyl
esters were toxic under the test conditions with the exception of modest
growth by most
strains in the presence of diethyl sebacate and diethyl diethylmalonate (with
glucose only, with
galactose strains appeared to exhibit substantial lag). For malonate diesters,
under the test
119
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
conditions, di-tert-butyl malonate supported growth of all strains with
glucose addition, again
appearing toxic or to induce substantial lag with galactose addition.
Increasing the dialkyl ester chain length from diethyl to diisopropyl to
dibutyl in a dialkyl
adipate series reduced toxicity. Some growth was observed with diisopropyl
adipate and no
apparent toxicity in dibutyl adipate. Dibutyl sebacate was also completely non-
inhibitory to
growth and accordingly, non-toxic. In certain embodiments, the minimum non-
toxic internal
alkyl chain length of diethyl diesters is sebacate. In certain
embodiments,shorter internal alkyl
chain length down to adipate is possible with diisopropyl diesters.
For monoester compounds, under the test conditions, octyl acetate was toxic
and for
the hexanoate series, growth was only observed starting with hexyl hexanoate,
which was
moderately non-toxic. Isopropyl octanoate was moderately inhibitory but
allowed for some
growth. For the decanoate series, methyl decanoate was moderately inhibitory
to growth but
still allowed for growth. Texanol, a monoester alcohol (2,2,4-trimethy1-1,3-
pentanediol
monoisobutyrate), was inhibitory to growth under the conditions tested.
However, ethyl decanoate and higher alkyl chains were increasingly non-toxic.
Both
ethyl and butyl laurate were non-toxic, as well as methyl and ethyl
nnyristate. In certain
embodiments, growth-suitable monoester overlays for resorcinol or cannabinoid
production
include hexyl hexanoate or any higher chain length alkyl hexanoate ester, C3
chain-length or
higher alkyl octanoate esters, and methyl (C1) or higher alkyl decanoates,
laurates, or
myristates.
In various embodiments, esters and diesters are employed as the organic phase
in
accordance with the present invention.
Fatty alcohols are mostly solids above Clo saturated chain length. Decanol is
a liquid
however it was toxic to growth. However, oleyl alcohol supports robust growth.
In certain
embodiments, longer chain length (C12 or higher) unsaturated fatty alcohols
can be suitable
overlays supporting S. cerevisiae or another fermenting organism's growth. In
various
embodiments, fatty alcohols, preferably C12 or higher alcohols, are employed
as the organic
phase in accordance with the present invention.
120
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
In certain embodiments, alkanes and paraffins support robust growth. Lack of
toxicity
was observed for dodecane, tetradecane, hexadecane, light and heavy paraffin
oils, and isopar
M. In certain embodiments, all C12 and higher paraffins are suitable overlays
supporting S.
cerevisiae or another fermenting organism's growth. In various embodiments,
fatty alcohols,
preferably C12 or higher alcohols, are employed as the organic phase in
accordance with the
present invention.
Certain triacylglycerols were tested, including tricaprylin, coconut oil, and
canola oil
(vegetable oils having different average chain length compositions of fatty
acid chains, with
coconut oil being predominantly C12-C14 saturated fatty acids, and canola
being predominantly
C16-Cis and a mixture of saturated and unsaturated fatty acids). Tricaprylin,
a synthetic oil
containing three C8 fatty acid chains, was fairly toxic, however allowed some
growth of all
strains in VP + 2% glucose. In certain embodiments, coconut and canola oil
were non-toxic to
growth.
Mixtures of IPM and isopar M with different diesters ¨ dibasic esters (DBE),
diethyl
sebacate, and di-tert-butyl malonate were explored to investigate if lower
percentage mixtures
of these compounds in non-toxic IPM or isopar M would mitigate their toxicity
toward growth
of S. cerevisiae, as they may also advantageously alter partitioning
properties of olivetolic acid,
olivetol, and other analogues into the overlay and could offer advantages with
alternative
downstream separations processes. DBE, which is highly toxic by itself as an
underlay, was
much less toxic at concentrations of between 1 and 2.5% (v/v) in IPM and
especially isopar M.
Di-tert-butyl malonate also exhibited much lower toxicity at 1-10% (v/v), and
particularly 1-
2.5% (v/v), in IPM and isopar M. In certain embodiments, mixtures of longer
chain monoesters
or paraffins with moderately to very toxic diesters are useful according to
the present
invention.
Example 4B: Olivetolic acid and olivetol production with organic solvent
overlays
Precultures of LSC3-2 and LSC3-18 were inoculated from YPD streak plates and
grown in
30 mL of YP + 2% (w/v) glucose in baffled 250 mL shake flasks for
approximately 18 hours
overnight at 30 C with 200 rpm shaking. Baffled 250 mL shake flasks containing
30 mL of VP +
121
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
2% (w/v) galactose, 5 mL of IPM or diethyl sebacate, and 0.04% (w/v) hexanoic
acid, were
inoculated with 1 mL of preculture and grown for at 30 C with 200 rpm shaking.
After 24 and 48
hours, samples of cell culture broth plus overlay were sampled into
microcentrifuge tubes and
stored at -20 C at least overnight. Sample tubes were thawed and aqueous
sample and overlay
sample were pipetted into plates for HPLC analysis. Overlay samples were
diluted 1:1 v/v with
methanol in the HPLC plate prior to analysis.
Total olivetol equivalents are defined as the concentration of olivetol in
mg/L, plus the
concentration of olivetolic acid in mg/L multiplied by the ratio of the
molecular weight of
olivetol to olivetolic acid. Lower total olivetol equivalents were observed
with diethyl sebacate
overlayer as compared to IPM. With IPM overlay, OA partitioned between the IPM
and aqueous
phases, with a substantial amount of OA remaining in the aqueous phase in
these culturing
conditions. By contrast, OA entirely partitioned into diethyl sebacate with
none present in the
aqueous phase. The reduction in total production levels with a diethyl
sebacate overlay may
occur due to a reduction in 0D600 due to moderately growth inhibitory
properties of diethyl
sebacate.
In another experiment, LSC3-2 precultures were inoculated from YPD streak
plates and
grown in 300 i.tLYP + 2% (w/v) glucose in round-bottom square well 96 well
deepwell plates for
approximately 18 hours overnight at 30 C with 950 rpm shaking in an Infors
plate shaker. 96
well deepwell plate wells containing 3004 of YP + 2% (w/v) galactose, 604 of
IPM, diethyl
sebacate, di-tert-butyl malonate, or methyl soyate, and 0.04% (w/v) hexanoic
acid, were
inoculated with 10 tL of preculture and grown for 30 C with 950 rpm shaking.
After 48 hours,
cultures were acidified with 104 of 5 M phosphoric acid to enhance
partitioning of olivetolic
acid into the organic phase (as the free acid), and overlays were sampled on a
Bravo automated
liquid handling platform (Agilent) by first adding 120 L of IPM, mixing on a
shaking platform
for several minutes, centrifuging the plate at 3000 rpm for 5 minutes to
separate phases, and
pipetting 100 L of overlay from each well into an HPLC plate. Overlay samples
were diluted 1:1
v/v with methanol in the HPLC plate, sealed and analyzed by HPLC.
Under these conditions, higher production levels were observed with a diethyl
sebacate
overlayer as compared with IPM. No product was observed in the overlayer
(aqueous samples
122
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
were not measured) with a di-tert-butyl malonate overlayer. Substantial
production was
observed in methyl soyate, however at slightly lower levels than with IPM.
In another experiment, several monoester overlay candidates and one diester
candidate
were compared to IPM. LSC3-13 precultures were inoculated from YPD streak
plates and grown
in 300 Delft medium + 0/9 g/L complete supplement mixture (CSM)
(ForMedium, Norfolk,
UK) + 2% (w/v) glucose in round-bottom square well 96 well deepwell plates for
approximately
18 hours overnight at 30 C with 950 rpm shaking in an lnfors plate shaker. 96
well deepwell
plate wells containing 300 L of Delft medium + CSM + 2% (w/v) glucose, 604 of
IPM, diethyl
sebacate, di-tert-butyl malonate, or methyl soyate, and 0.04% (w/v) hexanoic
acid, were
inoculated with 10 L of preculture and grown for 30 C with 950 rpm shaking.
Delft medium
contains (per liter solution) 7.5 g ammonium sulfate, 14.4 g potassium
phosphate monobasic
(added from a 1 M stock solution adjusted to pH 6.5 with sodium hydroxide),
0.5 g magnesium
sulfate heptahydrate, 3.6 mL of a trace metal solution (consisting of 130 g/L
citric acid
monohydrate, 0.574 g/L copper (II) sulfate pentahydrate, 8.07 g/L iron (III)
chloride
hexahydrate, 0.5 g/L boric acid, 0.333 g/L manganese (II) chloride, 0.2 g/L
sodium molybdate,
and 4.67 g/L zinc sulfate heptahydrate), and 1.0 mL of a vitamin solution
(0.008 g/L biotin, 1.6
g/L calcium pantothenate, 0.008 g/L folic acid, 8 g/L myo-inositol, 1.6 g/L
nicotinic acid, 0.8 g/L
p-aminobenzoic acid, 1.6 g/L pyridoxal hydrochloride, 0.8 g/L riboflavin, 1.6
g/L thiamine
hydrochloride, adjusted to pH 10.5 with sodium hydroxide). After 48 hours, the
aqueous layer
and overlay were sampled on a Bravo automated liquid handling platform
(Agilent) by first
removing 200 i.tL of aqueous sample into a 96 well filter plate, adding 180
jiL of IPM to each
well, mixing on a shaking platform for several minutes, centrifuging the plate
at 3000 rpm for 5
minutes to separate phases, and pipetting 1004 of overlay from each well into
an HPLC plate.
Overlay samples were diluted 1:1 v/v with methanol in the HPLC plate, sealed
and analyzed by
HPLC. Aqueous samples were centrifuged in the 96 well filter plate at 3000 rpm
for 5 minutes
into an HPLC plate, sealed, and analyzed by HPLC.
Titers were calculated in each phase on the basis of the volume of the full
broth plus
overlay, thus concentrations reported correspond to actual concentrations in
the full liquid
volume of each production well. Ethyl myristate exhibited approximately equal
aqueous phase
123
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
concentrations of olivetolic acid and olivetol product as IPM, but with
slightly higher overlay
concentrations. Other monoesters also supported robust production slightly
lower than that of
IPM, including methyl decanoate and hexyl hexanoate. The results demonstrate
that
monoester overlays that are not inhibitory to growth support robust production
of olivetolic
acid and olivetol.
Example 4C: Divarinic acid and divarin production with organic solvent
overlays
Several monoester and one diester overlay candidate were compared to IPM for
production of divarinic acid and divarin. LSC3-13 precultures were inoculated
from YPD streak
plates and grown in 300 jiL Delft medium + 0.79 g/L complete supplement
mixture (CSM)
(ForMedium, Norfolk, UK) + 2% (w/v) glucose in round-bottom square well 96
well deepwell
plates for approximately 18 hours overnight at 30 C with 950 rpm shaking in an
lnfors plate
shaker. 96 well deepwell plate wells containing 3004 of Delft medium + CSM +
2% (w/v)
glucose, 60 jiL of different overlay solvents, and 0.08% (w/v) butyric acid,
were inoculated with
104 of preculture and grown for 30 C with 950 rpm shaking. After 48 hours, the
aqueous layer
and overlay were sampled on a Bravo automated liquid handling platform
(Agilent) and samples
from the aqueous and organic overlay phases were subjected to HPLC analysis to
measure
divarinic acid and divarin production.
Multiple overlays support production of divarinic acid and divarin. Under the
test
conditions, divarinic acid and divarin partition less effectively into
monoester overlays than
olivetolic acid and olivetol. IPM allowed for higher production levels than
other tested
monesters without branched chain substituents.
Example 5: Fermentation of glucose to produce Divarin and Divarinic Acid
= Strain: LSC3-134
= Genotype: ga180^::(1oxHIS4)/his4A/mig1^::(1oxPkHIS4)
= Parent strain: LSC300002
= Genotype of parent strain:
leu2^::ScLEU2<pScLEU2/tScCYC1>CsAEE1<pScGAL10/pScGAL1>CsTKS-T2A-CsOAC<tScCY
C1/pAG305-backbone/1eu2(defective)_ura3 A : :
pScURA3>ScURA3/tScCYC1>CsAEE1<pScG
124
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
AL10/pScGAL1>CsTKS-T2A-CsOAC<tScCYC1/pAG306-backbone/ura3(defective) trp1^::p
ScTRP1>ScTRP1/tScCYC1>CsAEE1<pScGAL10/pScGAL1>CsTKS-T2A-CsOAC<tScCYC1/pAG
304-backbone/trp1(defective)_yorWdelta22^:tScADH1>HMGK2R<pScGAL10/pScGAL1>1
D11<tScCYC1/KanMX
Fermentation Process Summary:
Seed Train:
A shake flask containing 50 mL YPD with 20 g/L glucose was inoculated with a
seed vial
containing LSC3-134. Strain grew at 30 C for 24 hours to an 0D600 of 3-4. 15
mL of this culture
was used to inoculate the fermentation tank.
Media:
Seed Media: YP with 20 g/L glucose
Batch media:
= 10 g/L yeast extract+20g/L peptones + 20g/L glucose + 500mg/L
histidine+12 mg/L myo-
inosito1+12 mg/L thiamin hydrochloride+12 mg/L pyridoxal hydrochloride+12 mg/L
nicotinic acid+12 mg/L calcium pantothenate+0.6 mg/L biotin+12 mg/L p-
aminobenzoic
acid+0.15 g/L EDTA+7.8 mg/L CuSO4-5H20+0.0512 g/L FeSO4-7H20+0.0032 g/L
MnC12+4.77 mg/L Na2Mo04+0.102 g/L ZnSO4-7H20+0.0086 g/L CoC12-6H20+0.0384
g/L CaCl2-2H20+5.5 g/L KH2PO4+2.9 g/L MgSO4-7H20+45.1 g/L (NH4)2504
Production Media:
= 650 g/L glucose +438 mg/L citric acid monohydrate+2 mg/L H3B03+1.3 mg/L
CuSO4-
5H20+22.4 mg/L FeC13-6H20+1.33 mg/L MnC12+0.8 mg/L Na2Mo04+10.8 mg/L ZnSO4-
7H20+12 mg/L myo-inosito1+12 mg/L thiamin hydrochloride+12 mg/L pyridoxal
hydrochloride+12 mg/L nicotinic acid+12 mg/L calcium pantothenate+12 mg/L
biotin+12
mg/L p-aminobenzoic acid-F12 mg/L folic acid+12mg/L riboflavin-'-2.5 g/L
KH2PO4+1 g/L
MgSO4-7H20+20 g/L (NH4)2504+20 g/L sodium butyrate
Base (for pH Control)
125
CA 03177968 2022- 11-4
WO 2021/225952
PCT/US2021/030452
= 5M NH4OH
Overlay
= 170 mL isopropyl myristate (40% V/VO) was added to tank at 24 hours.
= Antifoam Struktol SB2121 (0.1 mL/L) at the beginning of the run
Fermentation run condition:
We used pulse feeding during the run. Fermentation batch was inoculated with
15 mL of
inoculum. Feeding was triggered at the end of batch phase when batch glucose
was completely
exhausted and p02 was increased by 10% (or more). Feed media (growth or
production media)
was delivered in pulses. Each pulse delivered 1.7 g glucose /starting batch
volume with
maximum feed rate not exceeding 10 g glucose/L/hr. pH was maintained at 5.5
throughout the
run. Temperature of fermenter was maintained at 30 C. Air flow rate was
maintained at 1.25
L/min. Agitation was 800 rpm.
Summary of metrics:
= Total product titer: 5 g/L (D+DA; See Figure 5A)
= Titer/time: 0.74 g Divarin equivalent/L/day (see Figure 5B)
= Maximum Divarinic acid titer: 2.5 g/L
= Maximum Divarin: 2.5 g/L
Also evaluated was the effect of sodium butyrate concentration in feed in a
set of three runs
(10, 14.3 and 20 g/L sodium butyrate in feed, all other nutrients remained the
same as stated
above) and we observed that titer and productivity increased as we increased
sodium butyrate
concentration in feed. The highest titer (and productivity) was observed in
the run with 20g/L
sodium butyrate in feed. 10g/L sodium butyrate, while producing an appreciable
amount of the
products, demonstrated the lowest titer.
126
CA 03177968 2022- 11-4