Note: Descriptions are shown in the official language in which they were submitted.
WO 2021/232028
PCT/US2021/034037
TITLE: FUNCTIONAL LIGANDS TO SARS-COV-2 SPIKE PROTEIN
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application claims the benefit and priority of the
following: U.S. provisional patent application
Ser. No. 63/066,417, filed August 17, 2020, entitled "FUNCTIONAL LIGANDS TO
SARS-COV-2 SPIKE
PROTEIN"; and U.S. provisional patent application Ser. No. 63/023,816, filed
May 12, 2020, entitled
"FUNCTIONAL LIGANDS TO CORONAVIRUS". The contents of the foregoing
applications are hereby
incorporated by reference in their entireties.
FIELD OF THE INVENTION
[0002] This invention relates to functional ligands to target
molecules, particularly to functional nucleic
acids and modifications thereof, more particularly to functional ligands with
binding affinity to targets such as
viruses and/or antigens of such viruses, such as the spike protein of Severe
acute respiratory syndrome
coronavirus 2 (SARS-CoV-2).
SEQUENCE LISTING
I_0003] Deoxyribonucleic acid (DNA) sequences, which are disclosed in
the ASCII text file entitled
"PCOV2PCOl_ST25.txt", created on May 25, 2021 and of 54.6 KB in size, which is
incorporated by reference
in its entirety, herein are intended to include other aptamers incorporating
modifications, truncations (e.g. trivial
truncations, such as 1-5 nucleotides removed at an end, which consist
essentially of the same sequence and
retains binding to the target molecule), incorporations into larger molecules
or complexes (e.g. the aptamer
sequence within a longer nucleic acid strand, for example and without
limitation, of about 60-100 nucleotides
or less in length), and/or other aptamers having substantial structural or
sequence homology, for example,
greater than 75-90% sequence homology or identity within a similar length of
nucleic acid (e.g. similar to within
5-10 nucleotides in length with significant sequence homology or identity
within that length, such as greater
than 75-90%), as well as RNA and/or other non-DNA/RNA aptamers, and/or reverse-
complementary sequences
thereof. The disclosed aptamers may also bind to homologous proteins or
molecules from organisms other than
the organisms listed herein, to recombinant or non-recombinant versions of the
proteins or molecules, to
modified versions of the proteins or molecules, to proteins or molecules from
sources other than the source
listed herein. The aptamers are artificial, non-naturally occurring sequences
designed and/or selected for specific
andfor high affinity binding to a target, such as, without limitation, SARS-
CoV-2, such as by binding to the
SARS-CoV-2 Spike Protein (e.g. as present in a naturally occurring form, as
expressed on a pseudovinis and/or
a recombinant version of the Spike Protein (e.g. the Si subunit + S2 subunit
ECD-His Recombinant Protein,
available from Sino Biological, Catalog No. 40589-VO8B1)). Non-naturally
occurring sequences of aptamers,
may also not be present in naturally occurring systems or situations, such as
by, for example, not -being already
present or having a pre-existing function in a naturally occurring setting.
The indication of the species and
source of the target proteins or molecules is given for reference only and is
not intended to be limiting.
1
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/US2021/034037
BACKGROUND OF THE INVENTION
[0004] Aptamers, which are nucleic acid ligands capable of binding
to molecular targets, have recently
attracted increased attention for their potential application in many areas of
biology and biotechnology. They
may be used as sensors, therapeutic tools, to regulate cellular processes, as
well as to guide drugs to their specific
cellular target(s). Contrary to the actual genetic material, their specificity
and characteristics are not directly
determined by their primary sequence, but instead by their secondary and/or
tertiary structure. Aptamers have
been recently investigated as immobilized capture elements in a microarray
format. Others have recently
selected aptamers against whole cells and complex biological mixtures.
Aptamers are typically characterized
by binding to their target molecules via non-Watson-Crick (i.e. non-
hybridization) mechanisms, such as by
intermolecular forces resulting from the secondary or tertiary structure of
the aptamer. This is especially true of
non-nucleic acid target molecules where Watson-Crick mechanisms typically do
not apply.
[0005] Aptamers are commonly identified by an in vitro method of
selection sometimes referred to as
Systematic Evolution of Ligands by EXponential enrichment or "SELEX". SELEX
typically begins with a very
large pool of randomized polynucleotides which is generally narrowed to one
aptamer ligand per molecular
target. Once multiple rounds (typically 10-15) of SELEX are completed, the
nucleic acid sequences are
identified by conventional cloning and sequencing. Aptamers have most famously
been developed as ligands to
important proteins, rivaling antibodies in both affinity and specificity, and
the first aptamer-based therapeutics
are now emerging. More recently, however, aptamers have been also developed to
bind small organic molecules
and cellular toxins, viruses, and even targets as small as heavy metal ions.
After identification of an aptamer
sequence from sequencing after SELEX, the aptamer is typically manufactured
afterwards in manners utilized
with any other oligonucleotide, such as by standard synthesis methods, such as
standard commercial nucleic
acid synthesis (e.g. oligonucleotide synthesis by phosphoramidite method,
etc.), or the sequence may be utilized
with biological synthesis methods, such polymerase chain reaction (PCR) or the
like.
[0006] Coronavirus disease 2019 (COVID-19) is an infectious disease
caused by a coronavirus called
severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The disease was
first identified in December
2019 in Wuhan, China, and has since spread globally, resulting in a
coronavirus pandemic. Common symptoms
include fever, cough, and shortness of breath. Other symptoms may include
fatigue, muscle pain, diarrhea, sore
throat, loss of smell, and abdominal pain. The time from exposure to onset of
symptoms is typically around five
days but may range from two to fourteen days. While the majority of cases
result in mild symptoms, some
patients progress to viral pneumonia and multi-organ failure. The virus is
primarily spread between people
during close contact, often via small droplets produced by coughing, sneezing,
or talking. While these droplets
are produced when breathing out, they usually fall to the ground or onto
surfaces rather than being infectious
over long distances. People may also become infected by touching a
contaminated surface and then touching
their eyes, nose, or mouth. The virus can survive on surfaces up to 72 hours.
It is most contagious during the
first three days after the onset of symptoms, although spread may be possible
before symptoms appear and in
later stages of the disease. The role of these asymptomatic carriers in
transmission is not yet fully known;
however, preliminary evidence suggests that they may contribute to the spread
of the disease.
[0007] The standard method of diagnosis is by real-time reverse
transcription polymerase chain reaction
(rRT-PCR) from a nasopharyngeal swab. Chest CT imaging may also be helpful for
diagnosis in individuals
2
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/US2021/034037
where there is a high suspicion of infection based on symptoms and risk
factors; however, it is not recommended
for routine screening. Current testing for the presence of the virus are
experiencing issues, such as due to scale
of sample collection, personnel required to perform the task, centralized lab
processing with reporting of results
subjected to extended delays, etc. Additionally, the availability of tests and
lab capacity are often limiting
availability to only those experiencing symptoms, far after they have likely
been contagious. Current more rapid
testing methods include detection of antibodies which the body produces in
response to viral infections (e.g.
IgG, IgM), but antibodies may only be typically present in detectable levels
days or weeks after infection and
may thus result in false negative results of a person being infected and
having limited usefulness in early stage
detection.
SUMMARY OF THE INVENTION
[0008] The present invention relates functional ligands to target
molecules, particularly to functional
nucleic acids and modifications thereof, and to methods for simultaneously
generating, for example, numerous
different functional biomolecules, particularly to methods for generating
numerous different functional nucleic
acids against multiple target molecules simultaneously. The present invention
further relates to functional
ligands which bind with affinity to targets, more particularly to functional
ligands with binding affinity to viruses
and/or antigens of such viruses, such as the spike protein (S protein) of
Severe acute respiratory syndrome
coronavirus 2 (SARS-CoV-2).
[0009] In general, a method for generating functional biomolecules
includes obtaining a library, such as a
diverse or randomized library, for example, of biomolecules. Biomolecules may
generally include nucleic acids,
particularly single-stranded nucleic acids, peptides, other biopolymers and/or
combinations or modifications
thereof A library of biomolecules may include nucleic acid sequences, such as
ribonucleic acid (RNA),
deoxyribonucleic acid (DNA), artificially modified nucleic acids, and/or
combinations thereof The method for
generating functional biomolecules further includes contacting the library of
biomolecules with more than one
target, such as, for example, a molecular target, material and/or substance.
In general, the members of the library
that do not bind with some affinity to the more than one target may be washed
or otherwise partitioned from the
remainder of the library, which may have a given level of binding affinity to
the more than one target. The
process may be repeated to partition the strongest binding members of the
library. Amplification of the
biomolecules may also be utilized to increase the numbers of the binding
members of the library for subsequent
repetitions and for isolation and/or purification of any final products of the
process. Embodiments of the SELEX
method may generally be utilized to achieve the generation of functional
biomolecules of a given binding
affinity, such biomolecules generally referred to as aptamers or ligands.
[0010] in one exemplary aspect of the invention, generation of
functional biomolecules may be performed
against more than one or multiple targets simultaneously within a single
system, such as the generation of
functional nucleic acid ligands within a single reaction volume. In general,
more than one or a plurality of targets
may be disposed within in a single reaction volume, and a library of
biomolecules, such as a nucleic acid library,
may he applied to the reaction volume. The members of the library that do not
bind to any of the plurality of
targets under given conditions may then be partitioned, such as by washing.
One or more rounds of binding and
partitioning of the members of the library may be performed, such as, for
example, to obtain a remainder of
3
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
members of the library with a given affinity for their targets. The remaining
members that bind to the plurality
of targets of the library may then be marked and/or tagged, such as to
identify the particular target or targets to
which the member(s) of the library binds. The binding members of the library
may then be isolated and, by
virtue of the marking or tagging, be matched to a particular target or
targets. This is desirable as high capacity,
multiplexed identification proccdurcs may save time, expense, and physical
space for the process over single
target identification processes. The present method may also be desirable as
it may be utilized to identify and/or
eliminate biomolecules that bind or have a tendency to bind to multiple
targets.
[0011] In an exemplary embodiment, a plurality of target molecules
are affixed to a substrate within a
single reaction volume, such as, for example, by attaching the targets to a
substrate of an array. It may generally
be appreciated that a single reaction volume may refer to or include multiple
reaction sub-volumes, such as, for
example, discrete or semi-discrete fluid droplets. In general, the targets may
be disposed with multiple copies
of each target in clusters or "spots" such that a given array may have an
ordered deposition of targets on the
substrate, with each target identifiable by the location of a particular spot
on the substrate. A library of nucleic
acids may then be contacted or applied to the array and the non-binding
members of the library may be
partitioned or washed off the array. The binding and washing steps may be
repeated and may also utilize an
amplification step to generate additional copies of any remaining binding
members of the library. The array
may then be marked or tagged with a plurality of identifiers, such as, for
example, a plurality of oligonucleotides
which may universally bind through Watson-Crick interactions to the members of
the library of nucleic acids.
The marking or tagging may be, for example, accomplished by manually applying
identifiers, such as by
pipetting or the like, utilizing microcontact pins, applying membranes/films
with identifiers, printing, for
example, inkjet printing, and/or other similar tagging methods, of identifier
containing solutions to the array.
The identifiers may further include a unique or semi-unique sequence which may
be utilized to correspond to
the spots and thus the targets of the array. For example, a unique or semi-
unique identifier sequence may be
utilized that identifies each spatial location on an array, such as each
particular target spot. The identifier may
then be associated with and/or attached to the nucleic acid members bound to a
particular spot. Thus, the nucleic
acids, for example, bound to a particular target spot may be identified by the
sequence of the associated
identifier. In some embodiments, the identifiers may further be primers and
may be utilized with a nucleic acid
amplification reaction on the array to generate additional copies of the bound
nucleic acids. The unique or semi-
unique identifier sequence may also be incorporated into the members of the
library amplified. This may be
desirable for associating a given member with a target or targets while
preserving the particular sequence of the
member as the locational identifying sequence is appended to the sequence of
the library member. This may be
particularly desirable for resolving multiple binders to a single target or
members of the library that bind to
multiple targets.
[0012] In general, the starting library of biomolecules, such as
nucleic acids, may be the product of at least
one round of a previous SELEX protocol. For example, at least one round of
SELEX may be performed with a
library of biomolecules against multiple targets, such as, for example, in a
solution. The targets in the solution
may be substantially identical to the targets disposed on an array. This may
be desirable as multiple rounds of
selection may be performed with a library prior to applying the remaining
members to an array for
marking/tagging. Complex target arrays may generally be more expensive and/or
difficult to make or utilize
4
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
than solutions of target molecules, so performing only the final binding and
marking,/-tagging procedure on the
array may be desirable.
[0013] In other embodiments, identifiers may be predisposed on the
array substrate in substantial proximity
to the spots such that they may bind to, for example, nucleic acids bound to
the target spots. The identifiers may,
for example, be covalently attached to the substrate. In some embodiments, the
attachments may be controllably
breakable or cleavable such that the identifiers may be released from the
substrate such that they may, for
example, more easily bind to the bound nucleic acids on the spots.
[0014] In further embodiments, identifiers may be synthesized in
situ on the array, such as by light directed
in situ nucleic acid synthesis. Appropriately sequenced identifiers may then
be synthesized in proximity to
particular spots such that the newly synthesized identifiers may bind to the
nucleic acids bound to the target
spot.
[0015] In still other embodiments, identifiers may be disposed
and/or synthesized on a separate substrate,
such as a membrane, in a spatial disposition that substantially matches the
spatial disposition of spots on the
array, i.e. the identifiers may be arranged such that they may be readily
superimposed onto the target spots on
the array. The identifier substrate may then be contacted with the array with
locational matching of the spots
with identifiers. The identifiers may then bind to the nucleic acids bound to
the target spots. Any appropriate
method of facilitating binding may be utilized, such as, for example, actions
to drive migration of the identifiers
to the array, such as capillary action, electrophoresis, pressure,
gravitational settling, and/or any other
appropriate method or combination thereof. The separate substrate may also be
soluble, erodible, substantially
permeable to the identifiers, and/or otherwise adapted for facilitating
migration of the identifiers to the array.
[0016] In yet still other embodiments, the array substrate may be
physically divided and/or partitioned for
separate collection of the, for example, nucleic acids bound to the spots. The
spots may, for example, also be
controllably removable from the substrate such that they may be individually
recovered and sorted.
[0017] In still yet other embodiments, identifiers may be disposed
and/or synthesized on a separate
substrate, such as a membrane, in a spatial disposition that substantially
matches the spatial disposition of spots
on the array, i.e. the identifiers may be arranged such that may be readily
superimposed onto the target spots on
the array. The separate substrate may be kept separately while the array
substrate maybe physically divided
and/or partitioned for separate collection of the nucleic acids bound to the
spots. In this manner, the location of
the different nucleic acids maybe maintained even when the array substrate is
no longer intact, if the locations
are of value. The identifiers may also be selectively applied to particular
locations on the array and/or applied
in a particular order or in groups.
[0018] In some embodiments, identifiers may only be applied to spots
with bound nucleic acids. The spots
with bound nucleic acids may be detected, for example, by detecting the
presence of nucleic acids, such as by
applying nucleic acid binding dyes, such as SYBR dyes, ethidium bromide and/or
other appropriate dyes. The
members of the nucleic acid library may also include detectable portions, such
as, for example, fluorescent
moieties, radioactive tags and/or other appropriate detectable portions.
[0019] In some embodiments, the identifiers may be applied to the
bound nucleic acids together with other
materials, such as for example, components of a nucleic acid amplification
reaction, a nucleic acid ligation
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
reaction, photo-linking reagents, and/or any other appropriate material, such
as those materials that may
facilitate attachment or association of the identifiers to the bound nucleic
acids.
[0020] In yet another embodiment, identifiers may be ligated to the,
for example, bound nucleic acids. For
example, a nucleic acid ligase may be utilized to covalently link an
identifier sequence to the bound nucleic
acid. Further nucleic acid fragments may be utilized to facilitate ligase
action, such as appropriate
complementary fragments that may aid the formation of a substantially double-
stranded nucleic acid complex
compatible with a ligase. For another example, photo-ligation may be used to
attach the identifiers to the, for
example, bound nucleic acids. Photo-ligation may be especially useful when
certain substrates are used. For
example, macro-porous substrates.
[0021] In general, methods may be applied that may facilitate
binding or other interactions between the
identifiers and the, for example, nucleic acids bound to the spots. For
example, the temperature may be increased
to dissociate the nucleic acids from the spots. The temperature may
subsequently be lowered such that, for
example, base pairing may occur between the nucleic acids and the identifiers.
Further in general, it may be
desirable to apply the identifiers in a manner that physically separates
and/or isolates the individual target spots
such that cross-marking due to identifier diffusion/migration may be
minimized. For example, the identifiers
may be applied in individual fluid droplets such that there is no continuous
fluid contact between individual
identifier containing fluids. For further example, the substrate of the array
may be absorbent and/or porous such
that the identifiers may be absorbed into the substrate material. The
substrate material may also block lateral
diffusion while allowing vertical diffusion, such that identifiers may be
applied and absorbed into the substrate
while minimizing diffusion across the plane of the substrate, such as to other
target spots.
[0022] In a further embodiment, a method for generating functional
biomolcculcs includes obtaining a
library of peptide sequences and contacting the library with a plurality of
targets. In some exemplary
embodiments, the peptide sequence may be tagged, linked, marked and/or
otherwise associated with a nucleic
acid sequence. The nucleic acid sequence may be, for example, representative
of the sequence of the peptide.
For example, the nucleic acid may substantially encode the peptide sequence.
Also for example, the nucleic acid
may be a unique or semi-unique identifier sequence. The nucleic acid sequence
may then be utilized to bind
another identifier, as described above, such that a peptide bound to a target
may be tagged or marked as to which
target it bound.
[0023] in an exemplary embodiment, a bacteriophage (phase) may be
generated that includes a peptide
sequence of interest in its protein coat. The phage may further include a
nucleic acid sequence that may be
representative of the peptide sequence within the nucleic acid of the phage.
The phage may then be contacted
with a plurality of targets, as above. This may generally be referred to as
phage display. Non-binding phages
may be washed and/or partitioned, while binding phages may be tagged or marked
with identifiers, as above.
As phase nucleic acids are generally contained within the protein coat of the
phage, the nucleic acid may
generally be exposed for binding to the identifier. For example, the phage may
be heated such that the protein
coat denatures and/or disassembles such that the nucleic acid is exposed. The
identifier may also be introduced
into the phage, such as by electroporation, electrophoresis, and/or any other
appropriate method.
[0024] Other methods of peptide selection may include, but are not
limited to, mRNA display, ribosome
display, and/or any other appropriate peptide display method or combination
thereof.
6
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
[0025] In another aspect of the invention, methods for handling and
sorting the resultant sequences of a
multiplexed binding process are provided. In some embodiments, the sequences
may be sorted by identifier
sequences to establish which target or targets the sequence bound. The
sequences may further be compared,
aligned and/or otherwise processed to identify features, characteristics
and/or other useful properties,
relationships to each other, and/or target properties.
[0026] In a further aspect of the invention, methods for monitoring
and/or controlling the diversity of the
library of biomolecules may be utilized. For example, too few rounds of
selection may result in a biomolecule
pool with too many weak binding members while too many rounds of selection may
result in only a few binding
members, such as members corresponding to only a few targets rather than
members corresponding to all of the
targets present. In one embodiment, Cot analysis may be employed to measure
and/or monitor the diversity of
the library of biomolecules through multiple rounds of selection. Cot, or
Concentration x time, analysis
measures the annealing time of particular oligonucleotides while in solution
with other nucleic acids, such as
the members of the library of biomolecules. In general, the annealing time
will be faster the lower the diversity
of the library.
[0027] The present invention together with the above and other
advantages may best be understood from
the following detailed description of the embodiments of the invention
illustrated in the drawings.
BRIEF DESCRIPTION OF THE FIGURES
[0028] FIG. 1 illustrates an embodiment of a multiple target array;
[0029] FIG. 2 illustrates the application of a library of
biomolecules to a target array;
[0030] FIG. 2a illustrates the binding of members of a library of
biomolecules to a target spot;
[0031] FIGs. 3 and 3a illustrate embodiments of biomolecule library
members;
[0032] FIG. 3b illustrates an embodiment of an identifier;
[0033] FIG. 4 illustrates the tagging of a library member bound to
target with an identifier;
[0034] FIG. 4a illustrates a tagged library member product;
[0035] FIG. 5 illustrates a target spot with nearby identifiers on a
substrate;
[0036] FIG. 5a illustrates the application of an identifier sheet to
a target array;
[0037] FIGs 6, 6a, 6b and 6c illustrate embodiments of identifiers
and ligation of identifiers to a library
member;
[0038] FIGs. 7 and 7a illustrate phage display for a target;
[0039] FIG. 7b illustrates an mRNA display fusion product;
[0040] FIG. 7c illustrates a ribosome display fusion product:
[0041] FIG. 8 illustrates an example of a histology section target;
and
[0042] FIG. 9 illustrates an example of binding of an aptabeacon to
a taiga molecule.
DETAILED DESCRIPTION OF THE INVENTION
[0043] The detailed description set forth below is intended as a
description of the presently exemplified
methods, devices, and compositions provided in accordance with aspects of the
present invention and is not
7
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
intended to represent the only forms in which the present invention may be
practiced or utilized. It is to be
understood, however, that the same or equivalent functions and components may
be accomplished by different
embodiments that are also intended to be encompassed within the spirit and
scope of the invention.
[0044]
Unless defined otherwise, all technical and scientific terms used herein
have the same meaning as
commonly understood to one of ordinary skill in the art to which this
invention belongs. Although any methods,
devices and materials similar or equivalent to those described herein can be
used in the practice or testing of the
invention, the exemplified methods, devices and materials are now described.
[0045]
The present invention relates functional ligands to target molecules,
particularly to functional
nucleic acids and modifications thereof, and to methods for simultaneously
generating, for example, numerous
different functional biomolecules, particularly to methods for generating
numerous different functional nucleic
acids against multiple target molecules simultaneously. The present invention
further relates to functional
ligands which bind with affinity to targets, more particularly to functional
ligands with binding affinity to viruses
and/or antigens of such viruses, such as the spike protein (S protein) of
Severe acute respiratory syndrome
coronavirus 2 (SARS-CoV-2). Functional ligands, particularly functional
nucleic acids, of the present invention
are generally artificial, non-naturally occurring sequences designed and/or
selected for specific and/or high
affinity binding to a target, such as SARS-CoV-2 or antigens thereof, such as
the S protein (S) or subunits
thereof (e.g. subunit 1 and/or subunit 2). Non-naturally occurring sequences
of functional nucleic acids, such as
aptamers, may also be useful by interacting with a target molecule in a manner
not present in naturally occurring
systems or situations, such as by, for example, not being already present or
having a pre-existing function in a
naturally occurring setting.
[0046]
In general, a method for generating functional biomolecules includes
obtaining a library, such as a
diverse or randomized library, of biomolecules. Biomolecules may generally
include nucleic acids, particularly
single-stranded nucleic acids, peptides, other biopolymers and/or combinations
or modifications thereof. A
library of biomolecules may include nucleic acid sequences, such as
ribonucleic acid (RNA), deoxyribonucleic
acid (DNA), artificially modified nucleic acids, and/or combinations thereof
In general, modified nucleic acid
bases may be utilized and may include, but are not limited to, 2'-Deoxy-P-
nucleoside-5'-Triphosphate, 2'-
Deoxyinosine-5'-Triphosphate, 2'-Deoxypseudouridine-5'-Triphosphate, 2'-
Deoxyuridine-5'-Triphosphate, 2'-
Deoxyzebularine-5'-Triphosphate, 2-Amino-2'-deoxyadenosine-5'-Triphosphate, 2-
Amino-6-chloropurine-2'-
deoxyriboside-5'-Triphosphate, 2-Aminopurine-2'-deoxyribose-5'-Triphosphate, 2-
Thio-2'-deoxycytidine-5'-
Triphosphate, 2-Thiothymidine-5'-Triphosphate, 2' -Deoxy-L-adenosine-5' -
Triphosphate, 2' -Deoxy-L-
cytidine-5' -Triphosphate, 2' -Deoxy-L-guanosine-5' -Triphosphate, 2' -Deoxy-L-
thymidine-5' -Triphosphate, 4-
Th othym idi ne-5'-Triphosphate, 5-Am noally1-2'-deoxycyti di ne-5'-
Triphosphate, 5-AminoallyI-2'-
deoxyuri di n e -5'-Tri ph osphate ,
5 -B rom o-2'-deoxycyti di n e-5'-Tri ph osphate , 5-B rom o -2'-de
oxyuri din e -5'-
Tripho sphate, S -Fluoro-2'-de oxyuridine -5'-Triphosphate, 5-Trifluoromethy1-
2-deoxyuridine-5' -Triphosphate,
2'-Fluoro-2'-deoxyuridine-5'-Triphosphate, 2'-Fluoro-2'-deoxycytidine-5'-
Triphosphate, and/or any other
appropriate modified nucleic acid base. It may generally be understood that
the nucleoside triphosphates (NTPs)
listed above may generally refer to any appropriate phosphate of the modified
base, such as additionally, for
example, monophosphates (NMPs) or diphosphates (NDPs) of the base. The method
for generating functional
biomolecules further includes contacting the library of biomolecules with at
least one target, such as, for
8
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
example, a molecular target, material and/or substance. In general, the
members of the library that do not bind
with some affinity to the target may be washed or otherwise partitioned from
the remainder of the library, which
may have a given level of binding affinity to the target. The process may be
repeated to partition the strongest
binding members of the library. Amplification of the biomolecules may also be
utilized to increase the numbers
of the binding members of the library for subsequent repetitions and for
isolation and/or purification of any final
products ofthe process. Embodiments of the SELEX method may generally be
utilized to achieve the generation
of functional biomolecules of a given binding affinity. The basic SELEX
protocol and aptamers are described
in U.S. Patent No. 5,270,163, entitled "Methods for identifying nucleic acid
ligands," the entire contents of
which are hereby incorporated by reference.
[0047] in one exemplary aspect of the invention, generation of
functional biomolecules may be performed
against multiple targets simultaneously within a single system, such as the
generation of functional nucleic acid
ligands within a single reaction volume. In general, a plurality of targets
may be disposed within in a single
reaction volume and a library of biomolecules, such as a nucleic acid library,
may be applied to the reaction
volume. The targets may be, for example, proteins, cells, small molecules,
biomolecules, and/or combinations
or portions thereof. The members of the library that do not bind to any of the
plurality of targets under given
conditions may then be partitioned, such as by washing. The remaining members
of the library may then be
marked and/or tagged, such as to identify the particular target or targets to
which the member of the library
binds. The binding members of the library may then be isolated and, by virtue
of the marking or tagging, be
matched to a particular target or targets. This may be desirable as high
capacity, multiplexed identification
procedures may save time, expense, and physical space for the process over
single target identification
processes. The present method may also be desirable as it may be utilized to
identify and/or eliminate molecules
that bind to multiple targets.
[0048] Functional ligands to SARS-CoV-2 and/or antigens thereof
(e.g. S protein, Si subunit and/or S2
subunit), without limitation and without being bound to any particular theory,
may be utilized for detection,
quantification, and/or other diagnostic applications, such as for detection of
the virus or its antigens, in body
fluids or tissues or for purification, separation or other forms of analysis
or processing, and/or for therapeutic
purposes (e.g. treatment by administering the functional ligand). For example,
detection and/or quantitation of
the virus and/or its antigens may be utilized in bodily fluids for
ascertaining exposure, infection and/or severity
of infection by a person. Forms of detection may utilize the functional
ligands, such as in backscattering
interferometry (BST), microscale thermophoresis (MST), biolayer interferometry
(BI.T), electrochemical
sensors, gold nanoparticle assays, enzyme linked aptamer sorbent assays
(ELASA), pull down assays
(immunoprecipitation), microplate/well assays, cell sorting, lateral flow
assays and/or any other appropriate
form of detection. For example, the conformational change of a functional
ligand upon binding to its target
molecule may be detected, such as, for example and without being bound to any
particular theory, due to a
change in its hydration shell or spatial volume. Functional ligands may also
be utilized in therapeutic
applications, for example and without being bound to any particular theory, to
bind to the virus and/or its
antigens, such as in a manner that disrupts the virus and/or its ability to
infect host cells. For example, binding
to the S protein of the virus may be useful in preventing the interaction of
the S protein with the host cell, such
9
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
as through binding to the receptor angiotensin converting enzyme 2 (ACE2) of
human cells, which may be
useful in impairing the virus's entry and subsequent infection of the host
cell.
[0049] In an exemplary embodiment, a plurality of target molecules
are affixed to a substrate within a
single reaction volume, such as, for example, by attaching the targets to a
substrate of an array. As illustrated in
FIG. 1, the targets may be disposed with multiple copies of each target, such
as target molecules, in clusters or
µ`spots" 110 on the substrate 102 of an array 100 such that a given array 100
may have an ordered deposition of
targets on the substrate 102, with each target identifiable by the location of
a particular spot 110 on the substrate
102. Each spot 110 may be a unique target or there may be multiple spots 110
of at least one target on a given
army 100. In general, high content target arrays, such as high content protein
or antibody arrays or other forms
of arrays recognized as high content or high density for interrogating a high
number of different targets at once
may be utilized. For example, human protein microarrays, such as ProtoArray0
from ThermoFisher containing
over 9,000 unique proteins in spotted format, may be utilized. A library 200
of, for example, nucleic acids 202
may then be applied A to array 100, as illustrated in FIG. 2. Particular
members 204 of the library 200 may then
bind to target spots 110, such as illustrated in FIG. 2a. The non-binding
members 206 of the library 200 may be
partitioned or washed off the array 100. The binding and washing steps may be
repeated and may also utilize
an amplification step to generate additional copies of any remaining binding
members 204 of the library 200.
The array 100 may then be marked or tagged with a plurality of identifiers,
such as, for example, a plurality of
oligonucleotides which may universally bind through Watson-Crick interactions
to the members of the library
of, for example, nucleic acids. In one embodiment, each member 202 of the
library 200 may include a potential
binding sequence 202a and at least one conserved region 202b which may bind an
identifier oligonucleotide,
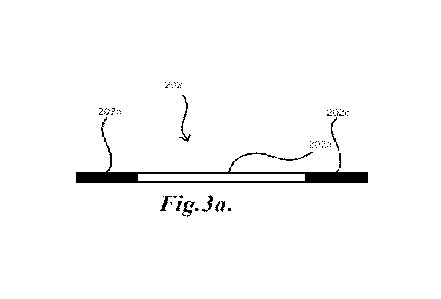
such as illustrated in FIG. 3. A further conserved region 202c may also be
included to facilitate priming for
amplification or extension reactions, such as Polymerase Chain Reaction (PCR),
as illustrated in FIG. 3a. In
general the conserved regions 202b, 202c may flank the potential binding
sequence 202a, such as to facilitate
priming for amplification. An identifier 302 may then include a unique or semi-
unique sequence 302a, such as
illustrated in FIG. 3b, which may be utilized to correspond to the spots 110
and thus the targets of the array 100
by location of the spot 110 on the substrate 102. The identifiers 302 may
further include conserved region 302b
which may bind to the conserved region 202b of the library members 202 by
Watson-Crick base pairing. The
identifiers 302 may also include a further conserved region 302c which may
facilitate priming for amplification.
The identifiers 302 may be, for example, applied to the spots 110 by printing,
for example, inkjet printing, using
micro-contact pins, and/or otherwise applying solutions containing identifiers
302 to the substrate 102 of the
array 100, such as, for example, by pipetting or the like, onto the spots 110.
A library member 202 bound to a
target spot 110 may then be tagged with an identifier 302 via base pairing B
at regions 202b, 302b, as illustrated
in FIG. 4. Thus, the nucleic acids 202 bound to a particular target spot 110
may be identified by the sequence
302a of the identifier 302. In an exemplary embodiment, nucleic acid
amplification or extension, such as PCR,
may be utilized to generate copies of the members 202 bound to the spots 110,
incorporating the identifier
sequence 302a (or more its complementary sequence) into the product 203, as
illustrated in FIG. 4a. This may
be desirable for associating a given member 202 with a target or targets 110
while preserving the particular
sequence of the member 202. This may be particularly desirable for resolving
multiple binders to a single target
or members of the library that bind to multiple targets. Subsequent
amplifications may utilize primers for the
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
sequences 202c, 302c such that only the products 203 containing both the
sequences 202a, 302a are amplified.
It may be understood that references to nucleic acid sequences, such as above,
may generally refer to either a
particular sequence or the corresponding complementary nucleic acid sequence.
In general, it may be desirable
for single droplets and/or otherwise separated volumes of solutions containing
identifiers 302 for each spot 110
on the array 100 such that the possibility of mistagging may be reduced.
[0050] In one aspect, the identifiers may be printed on all the
targets. In another aspect, the identifiers may
be printed only on targets with bound biomoleculcs.
[0051] In another embodiment, a histology section, such as the
section 110- on substrate 102- of histology
slide 100" in FIG. 8, may be utilized as a target set. The section 110" may
be, for example, a tissue section, a
cell mass, and/or any other appropriate biological sample which may generally
have structurally significant
features. As with the array 100, a library of biomolecules, such as nucleic
acids, may be applied which may
bind to specific locations on the section 110", the locations on which may,
for example, represent separate
targets to generate affinity binding nucleic acids. Identifiers may then be
disposed on the slide 100" as described
above, or as in the embodiments below, such that identifiers may be utilized
to correspond to specific features
of the section 110".
[0052] In other embodiments, identifiers may be predisposed on the
array substrate in substantial proximity
to the spots, such as illustrated with identifiers 302 disposed on substrate
102 in proximity to spot 110 in FIG.
5, such that they may bind to nucleic acids bound to the target spots. The
identifiers may, for example, be
covalently attached to the substrate. In some embodiments, the attachments may
be controllably breakable or
cleavable such that the identifiers may be released from the substrate such
that they may, for example, more
easily bind to the bound nucleic acids on the spots.
[0053] In further embodiments, identifiers may be synthesized in
situ on the array, such as by light directed
in situ nucleic acid synthesis. Appropriately sequenced identifiers may then
be synthesized in proximity to
particular spots such that the newly synthesized identifiers may bind to the
nucleic acids bound to the spot.
[0054] In still other embodiments, identifiers may be disposed
and/or synthesized on a separate substrate,
such as a membrane, in a spatial disposition that matches the spatial
disposition of spots on the array. FIG. 5a
illustrates an example of an identifier sheet 100' with membrane 102' which
may include identifier spots 110'
which may substantially correspond to target spots 110 of the array 100. The
identifier sheet 100' may then be
contacted C with the array 100 with locational matching of the target spots
110 with identifier spots 110'. The
identifiers may then bind to the nucleic acids bound to the target spots. Any
appropriate method of facilitating
binding may be utilized, such as, for example, actions to drive migration of
the identifiers to the array, such as
capillary action, electrophoresis, pressure, gravitational settling, and/or
any other appropriate method or
combination thereof.
[0055] In some embodiments, the membrane may be soluble and/or
substantially erodible. For example,
the membrane may include a film forming and/or soluble material. Identifiers
and/or other materials, such as
components of a nucleic acid amplification or ligation reaction, may be
included such that a film is formed
containing the desired materials. The membrane may then be applied to the
substrate and a suitable solvent,
such as water or ethanol, may be utilized to dissolve and/or erode the film,
which may then release the included
materials, such as the identifiers, to the substrate. Suitable materials for
the film may include hydrophilic
11
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
materials including polysaccharides such as carrageenan, chondroitin sulfate,
glucosamine, pullulan, soluble
cellulose derivatives such as hydroxypropyl cellulose and hydroxymethyl
cellulose, polyacrylic acid, polyvinyl
alcohol, polyethylene glycol (PEG), polyethylene oxide (PEO), ethylene oxide-
propylene oxide co-polymer,
polyvinylpyrrolidone (PVP), polycaprolactone, polyorthoesters,
polyphosphazene, polyvinyl acetate, and
polyisobutylcnc.
[0056] The membrane may further be adapted to have a desirable rate
of erosion and/or dissolution. The
rate may be modified by the inclusion of hydrophobic and/or less soluble
additives. Suitable materials may
include, but are not limited to, those from the family of quaternary ammonium
acrylate/methacrylate co-
polymers, (Eudragit RS), cellulose and its lower solubility derivatives, such
as butyl cellulose, hydroxybutyl
cellulose and ethylhydroxyethyl cellulose, high molecular weight PEG or PEO or
a combination thereof.
[0057] In yet still other embodiments, the array substrate may be
physically divided and/or partitioned for
separate collection of the nucleic acids bound to the spots. The spots may,
for example, also be controllably
removable from the substrate such that they may be individually recovered and
sorted. The array itself may also
be perforated and/or otherwise easily and/or conveniently partitionable.
[0058] In another embodiment, identifiers may be ligated to the
bound nucleic acids. For example, a nucleic
acid ligase may be utilized to covalently link an identifier sequence to the
bound nucleic acid. In general, nucleic
acid ligases are enzymes that covalently join two nucleic acids by catalyzing
the formation of phosphodiester
bonds at the ends of the phosphate backbone of the nucleic acids. Examples of
appropriate nucleic acid ligases
may include, but are not limited to, E. coli DNA ligase, T4 DNA ligase, T4 RNA
ligase, strand break DNA
repair enzymes, and/or any other appropriate ligase, modified enzyme, and/or a
combination thereof In general
the ligase utilized may be selected based on the form of ligation performed,
such as ligation of blunt ends,
compatible overhang ("sticky") ends, single stranded DNA, singe stranded RNA
and/or any other form of
ligation. Further in general, the steps in ligating two nucleic acids together
is a one step process that may be
carried out at or near room temperature. Further nucleic acid fragments may be
utilized to facilitate ligase action,
such as appropriate complementary fragments that may aid the formation of a
substantially double-stranded
nucleic acid complex compatible with a ligase. In general, double stranded
ligation may be employed and may
utilize substantially compatible overhang fragments to facilitate ligation, or
also blunt end ligation may be
utilized, such as with either the nucleic acid end or the identifier having a
phosphorylated end while the other is
unphosphorylated for ligation. Single stranded ligation may also be employed.
[0059] Photo ligation may also be employed. Photo ligation may, for
example, include covalently linking
adjacent nucleic acids by application of electromagnetic energy, such as
ultraviolet or visible light. Coupling
agents may also be utilized to facilitate the formation of covalent linkages.
[0060] In some embodiments, dyes may be included into the
identifiers. In one aspect, the identifiers may
be doped with dyes. In another aspect, the identifier solutions may be mixed
with dyes. According to one
embodiment, the dyes may be photosensitive and may be fluorescent. According
to another embodiment, the
dyes maybe photosensitive and may be phosphorescent.
[0061] The substrates used may be glass, ceramic or polymeric, as
long as their surfaces promote adhesion
between the substrates and the targets. Polymers may include synthetic
polymers as well as purified biological
polymers. The substrate may also be any film, which may be non-porous or
macroporous.
12
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
[0062] The substrate may be generally planar and may be of any
appropriate geometry such as, for
example, rectangular, square, circular, elliptical, triangular, other
polygonal shape, irregular and/or any other
appropriate geometry. The substrate may also be of other forms, such as
cylindrical, spherical, irregular and/or
any other appropriate form.
[0063] Appropriate ceramics may include, for example,
hydroxyapatite, alumina, graphite and pyrolytic
carbon.
[0064] Appropriate synthetic materials may include polymers such as
polyamides (e.g., nylon), polyesters,
polystyrenes, polyacrylates, vinyl polymers (e.g., polyethylene,
polytetrafluoroethylene, polypropylene and
polyvinyl chloride), polyearbonates, polyurethanes, poly dimethyl siloxanes,
cellulose acetates, polymethyl
methacrylates, ethylene vinyl acetates, polysulfones, nitrocelluloses and
similar copolymers. These synthetic
polymers may be woven or knitted into a mesh to form a matrix or similar
structure. Alternatively, the synthetic
polymer materials can be molded or cast into appropriate forms.
[0065] Biological polymers may be naturally occurring or produced in
vitro by fermentation and the like
or by recombinant genetic engineering. Recombinant DNA technology can be used
to engineer virtually any
polypeptide sequence and then amplify and express the protein in either
bacterial or mammalian cells. Purified
biological polymers can be appropriately formed into a substrate by techniques
such as weaving, kn itti
casting, molding, extrusion, cellular alignment and magnetic alignment.
Suitable biological polymers include,
without limitation, collagen, clastin, silk, keratin, gelatin, polyamino
acids, polysaccharides (e.g., cellulose and
starch) and copolymers thereof.
[0066] Any suitable substrate may be susceptible to adhesion,
attachment or adsorption by targets. The
susceptibility may be inherent or modified. In one example, the surfaces of
substrates may be susceptible to
adhesion, attachment or adsorption to proteins. In another example, the
surfaces of substrates may be susceptible
to adhesion, attachment or adsorption to proteins and not to nucleic acids.
[0067] In one exemplary embodiment, a glass substrate may have a
layer or coating of a material that
promotes adhesion with targets, such as proteins, materials that maybe
charged, such as those that are positively
charged, for binding target materials. Examples of charged materials include
cellulosic materials, for example,
nitrocellulose, methylcelluose, ethylcellulose, carboxymethyl cellulose,
hydroxyethyl cellulose,
methylhydroxypropyl cellulose; epoxies, PVDF (polyvinylidene fluoride);
partially or fully hydrolyzed
poly(vinyl alcohol); polv(vinylpyrrolidone); poly(ethyloxazoline);
poly(ethylene oxide)-co-poly(propylene
oxide) block copolymers; polyamines; polyacrylamide;
hydroxypropylmethacrylate; polysucrose; hyaluronic
acid; alginate; chitosan; dextran; gelatin and mixtures and copolymers
thereof.
[0068] In another exemplary embodiment, if the substrate is not
susceptible for attachment by charged
materials, or may be susceptible only for attachment by wrongly charged
materials, some areas of the substrate
may have adhesives, binding agents, or similar attached, adsorbed or coated
thereon. Examples of adhesives
may include any suitable adhesives that bind the charged materials.
[0069] The targets may be present on the substrate discretely or in
clusters. The distance between the
discrete targets may be close or may be far apart and may usually be of
different targets. Clusters may be used
for multiple spots of a single target.
13
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
[0070] In one embodiment, the substrate may be macroporous.
Macroporous substrates may be desirable,
for example, if the different targets are very close together. When the
targets are close by, there may not be
sufficient distance between different targets to distinguish which target a
biomolecule may be binding to.
Closely packed targets may increase the efficiency of the generating of
biomolecules. A macroporous substrate
may be suited for balancing between efficiency and separation. For a
macroporous substrate, the walls of the
pores may be sufficient to separate even closely packed targets if the pores
are large enough to enable the
binding process to occur within the pores.
[0071] Also, for macroporous substrates, the pores may have an
average diameter greater than the average
size of the target material such that the target material may enter or partly
enter the pores to anchor. Hydrogels
may also be useful for binding or anchoring targets to the pores. Hydrogels
may also fill the pores under fluid
conditions and present a smooth surface for fluid flow while at the same time
may keep the fluid from flowing
through the pores.
[0072] The plurality of targets may be arranged in any appropriate
manner such as, for example, in circular
or elliptical spots, square or rectangular spots, stripes, concentric rings
and/or any other appropriate arrangement
on the subject.
[0073] According to one exemplaty embodiment, the substrate may be
at ambient temperature throughout.
[0074] According to another exemplary embodiment, the substrate may
include a temperature affecting
system that generally produces at least one desired temperature on the surface
of the substrate and the adjacent
fluid. The desired temperature may facilitate the biomolecule generating
process.
[0075] According to a further exemplary embodiment, the substrate
may include a temperature affecting
system for producing a range of desired temperatures on the surface of the
substrate and the adjacent fluid. This
may be particularly useful when employing a set of targets having a
significant range of, for example Tms, or
melting temperatures. In one embodiment, the system may include a plurality of
temperature affecting devices
that are in thermal communication with the substrate. The plurality of devices
may generally be disposed such
that they may each produce a desired temperature in a given locality on the
surface of the substrate. The set of
targets may also be distributed on the surface of the substrate such that the
temperature at the location of a target
is substantially at the Tm of the target.
[0076] Temperature affecting devices may be any appropriate device
that may substantially produce a
desired temperature on a substrate and may include, but are not limited to,
thermoelectric devices such as Peltier
junction devices, semiconductor heating devices, resistive heating devices,
inductive healing devices,
heating/cooling pumps, electromagnetic radiation sources and/or any other
appropriate devices. Temperature
may also be affected by other systems, such as, for example, fluid flows
including, but not limited to, water
flows, air flows, and/or any other appropriate fluid flows.
[0077] In an exemplary embodiment, a plurality of Peltier junction
devices may be utilized to generate
desired temperatures at localities on the surface of the substrate. Peltier
junction devices are particularly useful
since they are able to both heat and cool using electrical current. This
enables Peltier junction devices to generate
temperatures above and below the ambient temperature of a system. They may
also be useful in maintaining
given temperature conditions at a steady state by adding and removing heat as
necessary from the system.
14
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
[0078] In general, the placement of the temperature affecting
devices may determine the temperature
profile on the surface of the substrate and the adjacent fluid in the chamber.
The temperature affecting devices
may thus be disposed at appropriate positions such that given temperatures may
be produced and maintained at
known positions on the substrate.
[0079] The substrate may in general have a given thermal
conductivity such that the application of at least
one temperature affecting device may substantially generate a temperature
gradient profile on the surface of the
substrate. In general, the temperature on the surface of the substrate may
change as a function of the distance
from the position of the at least one temperature affecting device. Substrate
materials with a relatively low
thermal conductivity may generally produce highly localized temperature
variations around a temperature
affecting device. Substrate materials with a relatively high thermal
conductivity may generally produce more
gradual variations in temperature over a given distance from a temperature
affecting device. It may be
understood that at steady state, the effect of the thermal conductivity of the
substrate may not contribute to the
temperature profile of the system.
[0080] In some embodiments, at least one temperature affecting
device may be utilized to produce a
particular temperature gradient profile on the surface of the substrate. In
general, a temperature gradient may be
generated by utilizing at least one temperature affecting device producing a
temperature different from the
ambient temperature of the system. Multiple temperature affecting devices with
at least two producing different
temperatures may be utilized to generate a temperature gradient without
reliance on the ambient temperature of
the system.
[0081] The positions and temperatures of multiple temperature
affecting devices may be utilized to
calculate a resulting temperature gradient profile on the surface of a
substrate using standard heat transfer
equations. An algorithm may then be utilized to calculate the optimal
positions and/or temperatures for a
plurality of temperature affecting devices to produce a desired temperature
gradient profile on the surface of a
substrate. The algorithm may be, for example, applied using a computational
assisting system, such as a
computer and or other calculatory device. This may be performed to tailor a
temperature gradient profile to a
particular substrate with a known disposition of targets of known and/or
calculated Tm. Similarly, a set of targets
of known and/or calculated Tm may be arranged on a substrate based on a
temperature gradient profile. This
may be desirable as placement of a target at a given location on a substrate
may be accomplished more easily
than tailoring a temperature profile to pre-existing locations of targets on a
substrate. In general, a target may
be disposed on the substrate at a temperature address within the temperature
profile gradient. The temperature
address may, for example, be substantially at the Tm of the target during
operation of the molecular
hybridization system, and/or any other appropriate temperature.
[0082] In another aspect, the molecular hybridization system
includes an adjustable system for generating
a temperature profile. The adjustable system generally includes a plurality of
temperature affecting devices,
each affecting the temperature at a particular location of a substrate.
[0083] Details of the temperature affecting systems may be found in,
for example, U.S. utility patent
application Ser. No. 12/249,525, filed on October 10, 2008, entitled "METHODS
AND DEVICES FOR
MOLECULAR ASSOCIATION AND IMAGING", the contents of all of which are hereby
incorporated by
reference.
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
[0084] FIG. 6 illustrates an example of an identifier sequence 302
and a complement identifier sequence
402. The complement sequence 402 may include a complement identifier region
402a which may be
substantially complementary to identifier region 302a such that they may base
pair bind. The complement
sequence 402 may further include a primer region 402c which may also be
complementary to primer region
302c of the identifier 302. Further, the complement sequence 402 may include a
compatible end 402b which
may be compatible with ligation to the end of another nucleic acid. As shown
in FIG. 6a, a nucleic acid library
member 202 may be bound to a spot 110. An identifier 302 and a complement
sequence 402 may then be applied
to the member 202 such that the identifier 302 binds to the member 202 at
region 202b, 302b. The complement
sequence 402 may bind to the identifier 302 at regions 302a/402a, 302c/402c.
The compatible end 402b may
then be 1 igated to the end D of the member 202 by an appropriate ligase
and/or other appropriate method. A
product 203', as illustrated in FIG. 6b, may then be generated including the
primer region 202c, binding
sequence 202a, region 202b, complement identifier region 402a, and complement
primer region 402c. The
product 203' may then be amplified, such as with the product 203 discussed
above in FIG. 4a. The product 203'
may also be generated by single-stranded ligation of the member 202 and the
complement sequence 402, where
in general the either the member 202 or the complement sequence 402 may have a
phosphorylated end while
the other may be unphosphorylated for end to end ligation.
[0085] In another example, as illustrated in FIG. 6c, a further
complementary fragment 502 may be
included that may base pair bind to a complementary region 202d of the nucleic
acid library member 202. This
may be desirable as some nucleic acid ligascs may generally join double
stranded nucleic acids. The addition
of the complementary fragment 502 may generally generate a substantially
double stranded nucleic acid, such
as illustrated spanning from region 302c to the end of complementary fragment
502. There may further be a
double stranded "break" at points D and E. In general, the sizing of the
fragments may be tailored to generate a
suitably long stretch of double stranded nucleic acid for ligase action. In
general, the complementary region
202d may be the same for all members 202 of the library 200 such that the same
complementary fragment 502
may be utilized, such as, for example, convenience, cost and/or ease of use.
[0086] In general, methods may be applied that may facilitate
binding or other interactions between the
identifiers and the nucleic acids bound to the spots. For example, the
temperature may be increased to dissociate
the nucleic acids from the spots. The temperature may subsequently be lowered
such that, for example, base
pairing may occur between the nucleic acids and the identifiers. Temperature
changes may also, for example,
denature the target such that the nucleic acids may no longer bind and/or bind
with lower affinity to the targets.
This may be desirable in that it may aid in binding of the nucleic acids to
the identifiers.
[0087] In a further aspect of the invention, methods for monitoring
and/or controlling the diversity of the
library of biomolecules may be utilized. For example, too few rounds of
selection may result in a biomolecule
pool with too many weak binding members while too many rounds of selection may
result in only a few binding
members, such as members corresponding to only a few targets rather than
members corresponding to all of the
targets present. In one embodiment, Cot analysis may be employed to measure
and/or monitor the diversity of
the library of biomolecules through multiple rounds of selection. Cot, or
Concentration x time, analysis
measures the annealing time of particular oligonucleotides while in solution
with other nucleic acids, such as
16
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
the members of the library of biomolecules. In general, the annealing time
will be faster the lower the diversity
of the library.
[0088] In one embodiment, a Cot-standard curve for measuring the
sequence diversity of the aptamer
library at any point during the multiplex SELEX process may be utilized. For
example, a group of DNA
oligonucleotides with a 5'- and 3'- constant region of.-20 bases identical to
the initial SELEX library may be
utilized. The oligos may then be converted to dsDNA by standard methods.
Briefly, after annealing a primer to
the oligos, Exo-minus Klenow Taq polymerase (Epicentre, Madison, WI) may be
used in conjunction with
dNTPs to fill in the ssDNA to create a dsDNA or mixture thereof. Using a
standard quantitative PCR thermal
cycler, a temperature profile for melting and controlled annealing of each DNA
mixture may be programmed.
Standard SYBR Green I specific for double-stranded DNA (dsDNA) may be utilized
to report the amount of
re-annealed dsDNA. At one extreme, the annealing time for a single sequence
will be measured. At the other
extreme, the annealing time for the initial SELEX pool, such as containing
approximately 1 nmol of sequence
diversity, may be measured. Annealing times of intermediate diversity may also
be measured to establish a very
specific Cot-standard-curve for the SELEX library. Using this standard curve,
at any point during SELEX, the
sequence diversity of the evolving library of aptamers may be determined by
comparison to the curve.
[0089] In a further embodiment, a method for generating functional
biomolecules includes obtaining a
library of peptide sequences and contacting the library with a plurality of
targets. In some embodiments, the
peptide sequence may be tagged, linked, marked and/or otherwise associated
with a nucleic acid sequence. The
nucleic acid sequence may be, for example, representative of the sequence of
the peptide. For example, the
nucleic acid may substantially encode the peptide sequence. Also for example,
the nucleic acid may be a unique
or semi-unique identifier sequence. The nucleic acid sequence may then be
utilized to bind another identifier,
as described above, such that a peptide bound to a target may be tagged or
marked as to which target it bound.
[0090] In an exemplary embodiment, a bacteriophage (phage) may be
generated that includes a peptide
sequence of interest in its protein coat. The phage may further include a
nucleic acid sequence that may be
representative of the peptide sequence within the nucleic acid of the phage.
The phage may then be contacted
with a plurality of targets, as above. This may generally be referred to as
phage display. Phages employed may
include, but are not limited to, M13 phage, fd filamentous phage, T4 phage, T7
phage, phage, and/or any other
appropriate phage. Non-binding phages may be washed and/or partitioned, while
binding phages may be tagged
or marked with identifiers, as above. As phage nucleic acids are generally
contained within the protein coat of
the phage, the nucleic acid may generally be exposed for binding to the
identifier. For example, the phage may
be heated such that the protein coat denatures and/or disassembles such that
the nucleic acid is exposed. The
identifier may also be introduced into the phage, such as by electroporation,
electrophoresis, and/or any other
appropriate method.
[0091] in FIG. 7, an example of a phage 600 may include a nucleic
acid 610 which may generally encode,
among other things, and be encapsulated by a protein coat 602, which may
contain a binding region for a target
110. The nucleic acid 610 may further include a region 612 which may identify
the phage and/or encode the
binding region for a target. A bound phage 600, as illustrated in FIGs. 7 and
7a, may then be heated, disrupted
and/or otherwise treated such that an identifier 302 may contact F the region
612. For example, the protein coat
602 may be broken and/or otherwise disrupted for entry of the identifier 302.
In general, an amplification
17
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
reaction and/or other method, such as those discussed above, may be utilized
to tag, mark and/or otherwise
introduce identifier information to the sequence of region 612. Further in
general, thc identifier 302 and region
612 may incorporate any, all or a combination of the elements discussed above
in regards to nucleic acid library
members, identifiers and/or other nucleic acid fragments. As also discussed
above, the phage 600 may also be
physically removed and/or partitioned in a manner that may preserve thc
idcntity of the target 110 thc phagc
600 was associated.
[0092] In other embodiments, other methods of incorporating and/or
linking nucleic acids to peptides may
be utilized, such as, for example, mRNA display, ribosome display, and/or any
other appropriate method. In
general, in mRNA display, as illustrated in FIG. 7b, a fusion product 600' of
a messenger RNA (mRNA) 610'
may be linked to a peptide 602' that the mRNA 610' encodes, such as with a
puromycin-ended mRNA 612'
which may generally cause fusion of the mRNA 610' to the nascent peptide 602'
in a ribosome, which may
then be contacted with targets such as described above with phage display.
Also in general, in ribosome display,
as illustrated in FIG. 7c, a fusion product 600- of a modified mRNA 610" may
be utilized that codes for a
peptide 602", but lacks a stop codon and may also incorporate a spacer
sequence 612" which may occupy the
channel of the ribosome 620" during translation and allow the peptide 602"
assembled at the ribosome 620" to
fold, which may result in the peptide 602" attached to the ribosome 620- and
also attached to the mRNA 610-.
This product 600" may then be contacted with targets such as described above
with phage display. Other
methods may include, but are not limited to, yeast display, bacterial display,
and/or any other appropriate
method.
[0093] In another aspect of the invention, methods for handling and
sorting the resultant sequences of a
multiplexed binding process are provided. In some embodiments, the sequences
may be sorted by identifier
sequences to establish which target or targets the sequence bound. The
sequences may further be compared,
aligned and/or otherwise processed to identify features, characteristics
and/or other useful properties,
relationships to each other, and/or target properties. For example, it may be
expected that multiple aptamer
sequences bound to a single target may potentially share sequence motifs
and/or other common features which
may be at least partially elucidated by sequence sorting and/or comparison.
Specific binding affinities of
resultant sequences may also be determined through affinity assays. In some
embodiments, surface plasmon
resonance may be utilized to determine binding of an aptamer to a target. For
example, sensors which monitor
the refractive index of a surface bound to a target may be utilized, where the
refractive index may change as a
result of binding of an aptamer to the target. In general, aside from standard
sequencing methods, parallel
sequencing methods, such as, for example, massively parallel sequencing such
as 454 Clonal Sequencing
(Roche, Branford, CT), massively parallel clonal array sequencing, Solexa
Sequencing (Illumina, San Diego,
CA), and/or any other appropriate sequencing method may be employed.
[0094] Aptamers may also be utilized to create molecular beacons
which may fluoresce and/or otherwise
produce a detectable signal when the aptamer binds to its target. Aptamers
typically undergo a conformational
change when binding a target and this conformational change may be utilized to
modulate the activity of other
molecules or components of a molecule, such as modulating the distance between
a fluorophore (fluor) and a
quencher. In general, an aptamer beacon or aptabeacon may include an aptamer
with a fluor and a quencher
attached to the 5' and 3' ends, respectively, or vice versa. The aptamer in
its unbound state may generally be
18
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/US2021/034037
designed to keep the fluor and quencher in proximity such that quenching of
the fluor occurs and thus little or
no fluorescent signal produced. Linkers and/or stem structures may also be
utilized with the base aptamer to
produce this quenching effect in the unbound state. Such linkers and/or stem
structures to produce "beacon"
structures in nucleic acids are generally well known and are standard
laboratory techniques. When the aptamer
binds to its target, its conformational change upon binding may then generally
cause spacing of the fluor and
quencher such that the fluor may undergo fluorescence without quenching by the
quencher, and such
fluorescence may then be detected as a signal to indicate binding of the
target to the aptamer. FIG. 9 illustrates
the conformational changes of an unbound aptabeacon structure 700 when binding
to a target molecule 110 at
the aptamer portion 702, showing the fluor 704 and quencher 706 in an initial
proximity resulting in quenching
A and a fluorescence emitting B conformation after binding the target molecule
110 to yield bound conformation
700'.
[0095] Aptamers may also be selected and/or designed to exhibit
large, detectable and/or specific switching
conformational changes when binding to a target molecule. In general, aptamers
frequently exhibit induced-fit
folding behavior, where the aptamer may be largely unstructured in solution
and may undergo significant
compaction and/or structural stabilization upon binding its target molecule.
In some embodiments, aptarners
may be selected and/or designed such that a portion of the aptamer may
hybridize to another nucleic acid, such
as a primer, anchoring oligo and/or other nucleic acid with a complementary
sequence, and may dehybridize
from such nucleic acid when the aptamer binds to its target molecule, such as
to release the aptamer from such
hybridized nucleic acid.
EXAMPLE OF MULTIPLEXED SELEX PROTOCOL
[0096] As a demonstration of parallel, de novo selection of aptamers
against multiple targets, a
combinatorial DNA library containing a core randomized sequence of 40 nts
flanked by two 20 nt conserved
primer binding sites is used as the starting point for an aptamer pool. The
primer sequences are designed and
optimized using Vector NTI's (Invitrogen) oligo analysis module. Typically,
such a library is expected to
contain approximately 10' unique sequences. The primer binding sites are used
to amplify the core sequences
during the SELEX process. The single stranded DNA pool dissolved in binding
buffer is denatured by heating
at 95 C for 5 mm, cooled on ice for 10 mm and exposed to multiple protein
targets fixed onto a nitrocellulose
coated glass slide (e.g., Whatman).
EXAMPLE OF DNA LIBRARY SELEX
[0097] An example DNA library consists of a random sequence of 40
nucleotides flanked by conserved
primers. In the first round of SELEX, 500 pmol of the ssDNA pool is incubated
with each slide in binding buffer
(PBS with 0.1 mg/ml yeast tRNA and 1 mg/ml BSA) for 30 minutes at 37 C. The
slide is then washed in 1 ml
of binding buffer for one minute. To elute specifically bound aptamers the
slide is heated to 95 C in binding
buffer. The eluted ssDNA is subsequently be precipitated using a high salt
solution and ethanol. After
precipitation, the aptamer pellet is resuspended in water and amplified by PCR
with a 3'- biotin-labeled primer
and a 5'-fluorescein (FITC)-labeled primer (20 cycles of 30 sec at 95 C, 30
sec at 52 C, and 30 sec at 72 C,
followed by a 10 min extension at 72 C). The selected FITC-labeled sense ssDNA
is separated from the
19
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/US2021/034037
biotinylated antisense ssDNA by streptavidin-coated Sepharose beads (Promega,
Madison, WI) for use in the
next round. Alternatively, "asymmetric PCR" may be utilized for generating a
large excess of an intended
strand of a PCR product in SELEX procedures. Also alternatively, the undesired
strand may be digested by k-
exonuclease, such as, for example, when a phosphorylated PCR primer is
employed.
[0098]
The labeling of individual aptamers with fluoresce in isoth ocyan ate
(FTTC) facilitates the
monitoring of the SELEX procedure. FITC is also compatible with scanning in
the green (cy3) channel of
standard microarray scanners. The sense primer used to amplify the ssDNA
aptamers after each round of
selection is fluorescently labeled, resulting in fluorescently labeled
aptamers. The protein spotted
nitrocellulose-coated slides are scanned in a microarray scanner.
Alternatively, proteins may be spotted on
epoxy-coated glass slides. While epoxy slides may have less protein binding
capacity than 3-D nitrocellulose
pads, it has been observed that there may be less non-specific binding of
nucleic acid aptamer pools to the
background of the slide (blocked or not). Blocking may be employed to reduce
background fluorescence.
[0099]
In each round of the SELEX process, the slide is incubated for 30 mm at
37 C to allow binding of
the aptamers to their targets. The slides are then washed in binding buffer
before the specifically bound DNAs
are eluted by heating the slide at 95 C in 7M urea. Nucleic acids from the
eluate are phenol-chloroform purified
and precipitated, and the concentrated single stranded DNA molecules will be
amplified by PCR. In order to
increase stringency throughout the SELEX process, the washes are gradually
increased in volume (from
approximately 1-10 m1). After a given point in the selection, such as, for
example, after the final round of
selection, the aptamers may be tagged, marked and/or partitioned.
EXAMPLE OF IN SITU HYBRIDIZATION OF IDENTIFIERS
[00100] An example of in situ hybridization of identifiers to aptamers was
performed with short, ssDNA
sequence tags to the 3' end of aptamers bound to their protein target. These
synthetic ssDNA tag
oligonucleotides consists of three regions, as illustrated in FIG. 3b with
identifier 302: (i) the C2 region, region
302c of the identifier 302, at the 3' end of the oligonucleotide consists of a
17-20 nucleotide sequence
complementary to a corresponding region on all of the used aptamers, (ii) the
Cl region, region 302b at the 5'
end of the oligonucleotide 302 contains a 17-20 nt primer binding site, used
during the amplification of the
tag:aptamer hybrid, prior to sequencing and (iii) a variable region 302a in
the center of the tag oligonucleotide
(V) that serves a as a unique identifier for each locus on the glass slide
surface. A variable sequence of 8
nucleotides will allow 48 (65,536) unique sequences to be generated,
sufficient for many complex protein arrays
(8000 samples) on the market.
[00101]
As outlined above, after the final round of the SELEX procedure
(typically, round 10) the specific
aptamers are bound to their protein targets, fixed to a glass slide. While the
40 nt core sequence of each aptamer
are unique, its terminal sequences have not been subject to any kind of
selection during the procedure. After
each round of binding to their protein targets, the aptamers were amplified
using conserved primers, requiring
the maintenance of corresponding regions at their distal ends (P1, P2). The 3'
-region of each aptamer, for
instance, can thus serve as a binding site (via standard hybridization) for
the C2 region of the proposed tag
oligonucleotide. Given the unique variable sequence (V) of each tag
oligonucleotide, each aptamer will now be
tagged with a sequence that can be traced back to the location of the aptamer
on the glass slide, and thus the
protein spotted at that location.
CA 03178640 2022- 11- 11
WO 2021/232028
PCT/ITS2021/034037
EXAMPLE OF SELEX AGAINST TARGETS
[00102] A SELEX procedure as described above was performed utilizing target
molecules, including SARS-
CoV-2 S Protein (e.g. as expressed on an HIV-derived pseudovirus, available
from RetroVirox. Inc. and/or a
recombinant version of the S Protein (e.g. the Si subunit + S2 subunit ECD-His
Recombinant Protein, available
from Sino Biological, Catalog No. 40589-V08B1)), to produce candidate
aptamers, and to yield aptamer
sequences given in the sequence listing above. The sequences yielded are
artificial, non-naturally occurring
sequences designed and/or selected for artificially for specific and/or high
affinity binding to the target molecule
and/or similar/related molecules, where the sequences have no known natural
function.
[00103] It will be appreciated by those of ordinary skill in the art
that the present invention can be embodied
in other specific forms without departing from the spirit or essential
character hereof The present description
is therefore considered in all respects to be illustrative and not
restrictive. The scope of the present invention is
indicated by the appended claims, and all changes that come within the meaning
and range of equivalents thereof
are intended to be embraced therein.
21
CA 03178640 2022- 11- 11