Note: Descriptions are shown in the official language in which they were submitted.
WO 2022/006564
PCT/US2021/040538
POLYFUNCTIONAL ORTHOGONAL PROTEIN CHIMERAS
CROSS-REFERENCE TO RELATED APPLICATIONS
The present application claims priority to U.S. Provisional Patent Application
Nos.
63/047,938 filed July 3, 2020, 63/050,346 filed July 10, 2020, 63/075,388
filed September 8,
2020, 63/145,083 filed February 3,2021, and 63/189,412 filed May 17, 2021, all
of which are
incorporated herein by reference in its entirety.
SEQUENCE LISTING
The instant application contains a sequence listing which has been submitted
electronically in ASCII format and is hereby incorporated by reference in its
entirety.
BACKGROUND
Presented on most acute myeloid leukemia (AML) leukemic cells, CD33 is an
appealing target for immunotherapy, and represents a target population for
which existing
therapies are underperforming. Gerntuzurnab ozogarn ic in, a drug fusing a
monoclonal antibody
to CD33 with a DNA-scission cytotoxic calicheamicin. was available for
patients after first
AML relapse between 2000 to 2010 before being pulled for increasing patient
death rate
without discernible benefit (Lowenburg et al. 2010). In 2017, it was
reintroduced to market tor
primary CD33+ AML with modestly improved median survival in combination with
cytosine
arabinoside and daunorubicin, DNA intercalators that prevent DNA synthesis. As
a
monotherapy, it does not improve outcomes (Renneville et al. 2014). Bi-
specific T-cell
engagers (BiTE) which are currently in Phase I clinical trials, target CD33+
AML to CD3+ T-
cells (Krupka et al. 2014). BiTEs, including those approved for B-cell
malignancies such as
blinatumomab which targets CD19+ B-cells for destruction via CD3+ T-cells,
have such low
bioavailability that they are injected continuously for weeks at a time,
repeating for several
months, prompting the need for alternative approaches to immune therapy
(Nanbakhsh et al.
2014).
Many malignancies, including AML, are linked to decreased activity of the
natural
killer (NK) cytotoxic response. Expression of NK-activating ligands such as
ULBP1 are
positively correlated with survival but known to have depressed expression in
AML even in
conditions that would normally upregulate it (Elias et al. 2014). CD48, a
ligand of the NK
receptor 2B4, is downregulated on the surface of AML cells expressing fusion
proteins such as
AML1-ETO (Mastaglio et al. 2018). Conversely, NK-inhibiting ligands such as PD-
L2 are
1
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
upregulated (Dulphy et al. 2016). AML has in some cases been shown to
dysregulate
maturation of NK cells, as well as "distracting" NK cells with soluble or
cxosomc-bound
ligands of receptors such as NKG2D (Mundy-Bosse et al. 2014). Suppression of
NK
maturation, noted by low numbers of CD11b+CD27+ NK cells, is more potent with
increasing
AML burden (Stringaris et al. 2014). The activating-inhibiting balance of NK
cells where
cytotoxic function depends on expression of and engagement of inhibitory and
activating
receptors on NK cells¨is frequently disturbed by AML, reducing surface
expression of
activating receptors and increasing expression of inhibiting receptors such as
NKG2A
(Orleans-Lindsay et al. 2001). In the case of exosomes or soluble ligands, NK
cells struggle to
find appropriate targets, revealing a therapeutic opportunity if a method can
be developed that
utilizes the NK cytotoxic response without relying on natural ligands on the
surface of leukemic
cells.
T-cells and NK cells alike have reduced function in AML contexts. Both primary
blasts
and AML cell lines like HL60 have demonstrated capacity to suppress the growth
of T- and
NK cells without affecting cytolytic activity or causing the death of these
cells (LeDieu et al.
2009). A more comprehensive screen of patients suggested that T-cells in
circulation greatly
increase without an increase in activity, and others note increased proportion
of T-regulatory
cells in particular (Shenghui et al. 2011; Schnoffeil et al. 2015). Shifts
toward memory T-cells,
correlated with elevated PD-1 expression, have been demonstrated that once
again do not
suggest impairment in activity or in "exhaustion" of cytotoxic T-cells
(Chamuleau et al. 2008).
Breakdown of tryptophan and arginine via indoleamine 2,3-dioxygenase and
arginase II,
upregulated in some AMLs also contribute to poor proliferation and activation
of T-cells, and
may serve as a useful serum indicator of immune avoidance by myeloid
malignancies
(Thorsson et al. 2018). The inflammatory landscape is also implicated in AML's
escape from
T-cell activity, but the predictive and therapeutic potential of specific
interactions has not been
harnessed (Benci et al. 2016; Chen et al. 2019). Ultimately, the methods by
which myeloid
blasts avoid immune clearance
___________________________________________________ modulating immune sub-
populations, inhibiting cytotoxicity,
and masking themselves¨are widely varied and difficult to predict.
Currently, new approaches are needed for diseases such as acute myeloid
leukemia
(AML) in which the outcomes, especially in older patients who are unable to
receive
intensive chemotherapy, the current standard of care, remains very poor, with
a median survival
of only 5 to 10 months (Dohner et al. 2015).
2
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
SUMMARY
The present disclosure provides for engineered proteins or polyfunctional
orthogonal
protein chimeras.
Polyspecific or polyfunctional proteins are biomolecules that can
simultaneously
engage two or more different types of agents (proteins, DNA, RNA or cells)
based on the
specificity of interaction and affinity to each of these agents.
For example, bispecific or bifunctional antibodies genetically engineered from
two
different monoclonal antibodies, one with specificity to an immune cell (e.g.,
T or NK) and
other with specificity towards a cancer cell, are being used to enhance tumor
killing.
Traditionally, these polyspecific proteins are made as a fusion protein but
such fusion proteins
may be rendered nonfunctional due to steric inhibition of engaging sites.
Recent advances in
synthetic biology enabled creation of synthetic polypeptides that allow for
creation of
orthogonal protein heterodimers formed via non-covalent interaction based on
hydrogen
bonding, similar to that observed between two anti-parallel strands of DNA.
Shown herein are improved polyspecific or polyfunctional or hetero or
heterodimer or
heterotrimer proteins which use a non-naturally occurring polypeptide domain
comprising 1-5
alpha helices connected by amino acid linkers and an IgG2 hinge domain either
alone or in
conjunction with an IgG2 Fe domain. These polyfunctional proteins show
improved properties
over the previously described polyfunctional proteins. For one, the use of the
IgG sequences
allows for the formation of covalent disulfide bonds making the proteins more
stable. These
polyfunctional proteins have antibody-like properties and engage the body's
complement
system via the Fe domain. Furthermore, these polyfunctional proteins, like
antibodies, are not
easily destroyed in the body. Additionally, because these proteins arc
antibody-like, there are
less immunogenic and have better pharmacokinetics than the standard,
previously disclosed
bispecifi c proteins.
While there are engineered proteins exemplified herein, the innovative use of
the non-
naturally occurring polypeptide domain comprising 1-5 alpha helices connected
by amino acid
linkers and an IgG2 hinge domain either alone or in conjunction with an IgG2
Fe domain can
be expanded to construct engineered proteins with other specificities that
also have superior
binding and cytotoxic properties. These engineered proteins can comprise an
antigen-binding
fragment or other moiety which binds any lineage-specific cell surface antigen
or other antigens
expressed and/or over-expressed by cancer and tumor cells, and any polypeptide
which binds
to a molecule expressed on immune cells.
3
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
Exemplified herein is a heterodimer bispecific protein using a NKG2D binding
domain
of a protein ULBP1 to create a heterodimer with antigen recognition domain of
monoclonal
antibody that bind a CD33 protein (present on myeloid cells), to engage a NK
cell with a
myeloid cell. See Figures 1, 2 and 10.
Also, exemplified herein is a heterodimer bispecific protein using an antigen
recognition domain of monoclonal antibodies that binds CD3 protein (present on
T cells) and
a CD33 protein (present on myeloid cells) to engage a T-cell with a myeloid
cell. See Figures
1,2 and 10.
Also exemplified herein is a heterodimer bispecific protein using an antigen
recognition
domain of monoclonal antibodies that binds CD16 protein (present on NK cells)
to create a
heterodimer with antigen recognition domain of monoclonal antibody that bind a
CD33 protein
(present on myeloid cells), to engage a NK cell with a myeloid cell. See
Figure 23.
While the engineered proteins exemplified herein utilize an antigen-binding
fragment
or other moiety which hinds a lineage-specific cell surface antigen (e.g.,
CD33), the disclosure
includes engineered proteins comprising an antigen-binding fragment or other
moiety which
recognizes other antigens expressed and/or over-expressed by cancer and tumor
cells. The
disclosure also includes engineered proteins comprising polypeptides which
bind to other
molecules expressed on immune cells.
These exemplified heterodimer proteins comprise an IgG2 hinge domain. See
Figures
lA and 23A.
Also shown herein are the exemplified heterodimer proteins comprising an IgG2
hinge
domain and an IgG2 Fc domain (CH2 and CH3 domains). See Figure 1B and 23B.
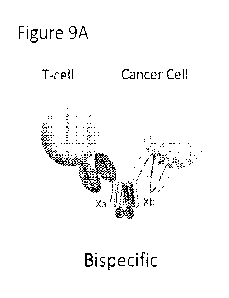
Polyfunctional proteins can also be constructcd comprising more than two
polypeptides. These proteins can be formed by combining the monomer a of each
heterodimer
X, Y or Z with a linker and monomer h fused to a target binding domain. While
this schematic
shows a trispecific protein, tetraspecific proteins as well as engineered
proteins comprising
more than four can be constructed as shown in the schematic and methods
herein. See Figure
9.
The utility of these synthetic molecules includes therapeutic targeting, gene
editing,
diagnostics, pathway manipulation by activating and or deactivating two or
more signals
simultaneously.
The present disclosure provides for an engineered heterodimer protein. In some
embodiments, a first engineered heterodimer protein comprises: a first
polypeptide comprising
an antigen-binding fragment that binds a lineage-specific cell-surface
antigen, a non-naturally
4
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
occurring polypeptide domain comprising 1-5 alpha helices connected by amino
acid linkers
and a first covalent dimerization domain; and a second polypeptide comprising
a polypeptide
that binds a molecule expressed on natural killer (NK) cells, a non- naturally
occurring
polypeptide domain comprising 1-5 alpha helices connected by amino acid
linkers, and a
second covalent dimerization domain; and wherein the first and second
polypeptides are
covalently bonded through the covalent dimerization domain.
In some embodiments, a second engineered heterodimer protein comprises: a
first
polypeptide comprising an antigen-binding fragment that binds a lineage-
specific cell-surface
antigen, a non-naturally occurring polypeptide domain comprising 1-5 alpha
helices connected
by amino acid linkers and a first covalent dimerization domain; and a second
polypeptide
comprising a polypeptide that binds a molecule expressed on T cells, a non-
naturally occurring
polypeptide domain comprising 7-5 alpha helices connected by amino acid
linkers, and a
second covalent dimerization domain; and wherein the first and second
polypeptides are
coval en tl y bonded through the covalent dimerization domain.
In some embodiments, the non-naturally occurring polypeptide domain comprising
1-
5 alpha helices connected by amino acid linkers in the first polypeptide and
the second
polypeptide are chosen from the group consisting of 6DMPb and 6DMPa.
In some embodiments, the first covalent dimerization domain and/or the second
covalent dimerization domain comprise an IgG2 hinge domain. In some
embodiments, the first
covalent dimerization domain and/or the second covalent dimerization domain
further
comprise an IgG2 Fe domain.
In some embodiments, the lineage-specific cell-surface antigen may be CD33,
CD19,
or any of the lineage-specific cell-surface antigens described herein.
In some embodiments, the molecule expressed on the NK cells may be NKG2D, and
the polypeptide that hinds a molecule expressed on NK cells is ULBP1 , ULBP2,
ULBP3,
ULBP4, ULBP5, ULBP6, MICA, MICB, or mutants or fragments thereof. In some
embodiments, the polypeptide that binds a molecule expressed on NK cells is an
ectodomain
of ULBP1, ULBP2, ULBP3, ULBP4, ULBP5, ULBP6, MICA, or MICB.
In some embodiments, the molecule expressed on NK cells is CD16, and the
polypeptide that binds a molecule expressed on NK cells is a monoclonal
antibody of CD16.
In some embodiments, the molecule expressed on T cells is CD3, and the
polypeptide
that binds a molecule expressed on T cells is a monoclonal antibody of CD3.
In some embodiments, the antigen-binding fragment is a single-chain antibody
fragment (scFv).
5
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
The present disclosure also provides for an engineered heterotrimer protein
comprising
three polypeptides, or an engineered protein comprising more than three
polypeptides,
comprising four polypeptides, comprising five polypeptides, comprising six
polypeptides, or
comprising more than six polypeptides, wherein the engineered hetero proteins
comprise an
additional polypeptide with binding domains to each of the other polypeptides
in the engineered
protein.
In some embodiments, the engineered heterotrimer protein comprises: a first
polypeptide comprising a polypeptide that binds a molecule expressed on T
cells, a non-
naturally occurring polypeptide domain comprising 1-5 alpha helices connected
by amino acid
linkers (al), and a first covalent dimerization domain; a second polypeptide
comprising an
antigen-binding fragment that binds a lineage-specific cell-surface antigen, a
non-naturally
occurring polypeptide domain comprising 1-5 alpha helices connected by amino
acid linkers
(b 1), and a second covalent dimerization domain; a third polypeptide
comprising a polypeptide
that hinds a molecule expressed on natural killer (NK) cells, a non- naturally
occurring
polypeptide domain comprising 1-5 alpha helices connected by amino acid
linkers (c 1), and a
third covalent dimerization domain; and a fourth polypeptide comprising three
non-naturally
occurring polypeptide domains comprising 1-5 alpha helices connected by amino
acid linkers,
wherein each domain is the binding domain of al, bl and cl (a2, b2 and c2),
and a fourth, fifth
and sixth covalent dimerization domain; and wherein the first and second and
third and fourth
polypeptides are covalently bonded through the covalent dimerization domain.
In some embodiments, the non-naturally occurring polypeptide domain comprising
1-
5 alpha helices connected by amino acid linkers in the first polypeptide, the
second polypeptide,
the third polypeptidc and the fourth polypeptide arc chosen from the group
consisting of
6DMPb and 6DMPa.
In some embodiments, the first covalent dimerization domain, the second
covalent
dimerization domain, the third covalent dimerization domain, the fourth
covalent dimerization
domain, the fifth covalent dimerization domain, and/or the sixth covalent
dimerization domain
comprise an IgG2 hinge domain. In some embodiments, one or more covalent
dimerization
domains further comprise an IgG2 Fc domain.
In some embodiments, the lineage-specific cell-surface antigen may be CD33,
CD19,
or any of the lineage-specific cell-surface antigens described herein.
In some embodiments, the molecule expressed on the NK cells may be NKG2D, and
the polypeptide that binds a molecule expressed on NK cells is ULBP1, ULBP2,
ULBP3,
ULBP4, ULBP5, ULBP6, MICA, MICB, or mutants or fragments thereof. In some
6
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
embodiments, the polypeptide that binds a molecule expressed on NK cells is an
ectodomain
of ULBP1, ULBP2, ULBP3, ULBP4, ULBP5, ULBP6, MICA, or MICB.
In some embodiments, the molecule expressed on the NK cells may be CD16 and
the
polypeptide that binds a molecule expressed on T cells is a monoclonal
antibody of CD16.
In some embodiments, the molecule expressed on T cells is CD3, and the
polypeptide
that binds a molecule expressed on T cells is a monoclonal antibody of CD3.
In some embodiments, the third polypeptide is a chemokine or cytokine protein,
which
increases the immune response. The chemokine or cytokine protein includes but
is not limited
to CXCLs including CXCL14, GCSF, and interleukins, including IL2 and IL16.
In some embodiments, the antigen-binding fragment is a single-chain antibody
fragment (scFv).
In certain embodiments, the first polypeptide in the first and second and
third
engineered heterodimer proteins comprises an amino acid at least 80% or at
least 90% identical
to SEQ ID NO: 1 (Figure 3). In some embodiments, the first polypeptide in the
engineered
heterodimer proteins comprises an amino acid at least 80% or at least 90%
identical to SEQ ID
NO: 4 (Figure 6).
In certain embodiments, the second polypeptide in the first engineered
heterodimer
comprises an amino acid at least 80% or at least 90% identical to SEQ Ill NO:
2 (Figure 4). In
some embodiments, the second polypeptide in the first engineered heterodimer
protein
comprises an amino acid at least 80% or at least 90% identical to SEQ ID NO: 5
(Figure 7).
In certain embodiments, the second polypeptide in the second engineered
heterodimer
protein comprises an amino acid at least 80% or at least 90% identical to SEQ
ID NO: 3 (Figure
5). In some embodiments, the second polypeptide in the second engineered
heterodimer protein
comprises an amino acid at least 80% or at least 90% identical to SEQ ID NO: 6
(Figure 8).
In certain embodiments, the second polypeptide in the third engineered
heterodimer
protein comprises an amino acid at least 80% or at least 90% identical to SEQ
ID NO: 26
(Figure 23A). In some embodiments, the second polypeptide in the second
engineered
heterodimer protein comprises an amino acid at least 80% or at least 90%
identical to SEQ ID
NO: 27 (Figure 23B).
The present disclosure provides for a composition comprising any of the
engineered
proteins, or a nucleic acid molecule encoding any of the engineered proteins.
The present disclosure also provides for a nucleic acid molecule encoding a
first
engineered heterodimer protein. The engineered dimer protein may comprise: (i)
a first
polypeptide comprising an antigen-binding fragment that binds a lineage-
specific cell-surface
7
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
antigen, a non-naturally occurring polypeptide domain comprising 1-5 alpha
helices connected
by amino acid linkers and a first covalent dimerization domain; and (ii) a
second polypeptide
comprising a polypeptide that binds a molecule expressed on natural killer
(NK) cells, a non-
naturally occurring polypeptide domain comprising 1-5 alpha helices connected
by amino acid
linkers, and a second covalent dimerization domain as described herein.
The present disclosure also provides for a nucleic acid molecule encoding a
second
engineered heterodimer protein. The engineered dimer protein may comprise: (i)
a first
polypeptide comprising an antigen-binding fragment that binds a lineage-
specific cell-surface
antigen, a non-naturally occurring polypeptide domain comprising 1-5 alpha
helices connected
by amino acid linkers and a first covalent dimerization domain; and (ii) a
second polypeptide
comprises a polypeptide that binds a molecule expressed on T cells, a non-
naturally occurring
polypeptide domain comprising 7-5 alpha helices connected by amino acid
linkers, and a
second covalent dimerization domain as described herein.
The present disclosure also provides for a nucleic acid molecule encoding a
third
engineered heterodimer protein. The engineered dimer protein may comprise: (i)
a first
polypeptide comprising an antigen-binding fragment that binds a lineage-
specific cell-surface
antigen, a non-naturally occurring polypeptide domain comprising 1-5 alpha
helices connected
by amino acid linkers and a first covalent dimerization domain; and (ii) a
second polypeptide
comprising a polypeptide that binds a molecule expressed on natural killer
(NK) cells, a non-
naturally occurring polypeptide domain comprising 1-5 alpha helices connected
by amino acid
linkers, and a second covalent dimerization domain as described herein.
The present disclosure also provides for a nucleic acid molecule encoding an
engineered trimer protein. The engineered heterotrimer protein may comprise:
(i) a first
polypeptide comprises a polypeptide that binds a molecule expressed on T
cells, a non-
naturally occurring polypeptide domain comprising 1-5 alpha helices connected
by amino acid
linkers, and a second covalent dimerization domain an antigen-binding fragment
that binds a
lineage-specific cell-surface antigen; and (ii) a second polypeptide
comprising an antigen-
binding fragment that binds a lineage-specific cell-surface antigen, a non-
naturally occurring
polypeptide domain comprising 1-5 alpha helices connected by amino acid
linkers and a first
covalent dimerization domain; and (iii) a third polypeptide comprising a
polypeptide that binds
a molecule expressed on natural killer (NK) cells, a non- naturally occurring
polypeptide
domain comprising 1-5 alpha helices connected by amino acid linkers, and a
second covalent
dimerization domain; and (iv) a fourth polypeptide comprising three non-
naturally occurring
polypeptide domains comprising 1-5 alpha helices connected by amino acid
linkers, wherein
8
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
each domain is the binding domain of al, b 1 and el (a2, b2 and c2), and a
fourth, fifth and
sixth covalent dimerization domain, as described herein.
The present disclosure provides for a vector comprising any of the present
nucleic acid
molecules, or a composition comprising any of the present nucleic acid
molecules.
The present disclosure provides for a cell comprising the present vectors or
nucleic acid
molecules.
The present disclosure provides for a composition comprising at least one
vector
encoding: (i) a first polypeptide comprising an antigen-binding fragment that
binds a lineage-
specific cell-surface antigen, a non-naturally occurring polypeptide domain
comprising 1-5
alpha helices connected by amino acid linkers and a first covalent
dimerization domain; and
(ii) a second polypeptide comprising a polypeptide that binds a molecule
expressed on natural
killer (NK) cells, a non- naturally occun-ing polypeptide domain comprising 1-
5 alpha helices
connected by amino acid linkers, and a second covalent dimerization domain as
described
herein.
The present disclosure provides for a composition comprising at least one
vector
encoding: (i) a first polypeptide comprising an antigen-binding fragment that
binds a lineage-
specific cell-surface antigen, a non-naturally occurring polypcptide domain
comprising 1-5
alpha helices connected by amino acid linkers and a first covalent
dimerization domain; and
(ii) a second polypeptide comprises a polypeptide that binds a molecule
expressed on T cells,
a non- naturally occurring polypeptide domain comprising 1-5 alpha helices
connected by
amino acid linkers, and a second covalent dimerization domain as described
herein.
The present disclosure provides for a composition comprising at least one
vector
encoding: (i) a first polypeptide comprises a polypeptide that binds a
molecule expressed on T
cells, a non- naturally occurring polypeptide domain comprising 1-5 alpha
helices connected
by amino acid linkers, and a second covalent dimerization domain an antigen-
binding fragment
that binds a lineage-specific cell-surface antigen; and (ii) a second
polypeptide comprising an
antigen-binding fragment that binds a lineage-specific cell-surface antigen, a
non-naturally
occurring polypeptide domain comprising 1-5 alpha helices connected by amino
acid linkers
and a first covalent dimerization domain and (iii) a third polypeptide
comprising a polypeptide
that binds a molecule expressed on natural killer (NK) cells, a non- naturally
occurring
polypeptide domain comprising 1-5 alpha helices connected by amino acid
linkers, and a
second covalent dimerization domain; and (iv) a fourth polypeptide comprising
three non-
naturally occurring polypeptide domains comprising 1-5 alpha helices connected
by amino acid
9
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
linkers, wherein each domain is the binding domain of al, bl and cl (a2, b2
and c2), and a
fourth, fifth and sixth covalent dimerization domain, as described herein.
The present disclosure provides for a composition comprising any of the
engineered
proteins, any of the nucleic acid molecules, any of the present vectors,
and/or any of the present
cells.
Also encompassed by the present disclosure is a kit comprising any of the
engineered
proteins, any of the nucleic acid molecules, any of the present vectors, any
of the present cells
and/or any of the present compositions.
The present disclosure provides for a method of treating cancer in a subject,
comprising
administering to the subject an effective amount of any of the engineered
proteins, any of the
nucleic acid molecules, any of the present vectors, any of the present cells
and/or any of the
present compositions disclosed or described herein.
The present disclosure provides for a method of treating a hematopoietic
malignancy
in a subject, comprising administering to the subject an effective amount of
any of the
engineered proteins, any of the nucleic acid molecules, any of the present
vectors, any of the
present cells and/or any of the present compositions disclosed or described
herein.
The hematopoictic malignancy may be a myeloid malignancy.
The hematopoietic malignancy may be Hodgkin' s lymphoma, non-Hodgkin's
lymphoma, leukemia, or multiple myeloma.
The hematopoietic malignancy may be acute myeloid leukemia, chronic
myelogenous
leukemia, acute lymphoblastic leukemia, or chronic lymphoblastic leukemia.
BRIEF DESCRIPTION OF THE FIGURES
For the purpose of illustrating the invention, there are depicted in drawings
certain
embodiments of the invention. However, the invention is not limited to the
precise
arrangements and instrumentalities of the embodiments depicted in the
drawings.
Figure 1 are schematics of the various binding proteins. Figure lA is a
schematic of the
anti-CD33, anti-CD3, and NKG2D binding proteins with the IgG2 hinge domain
only used in
the engineered proteins. Figure 1B is a schematic of the anti-CD33, anti-CD3,
and NKG2D
binding proteins with both IgG2 hinge and IgG2 Fc domains used in the
engineered proteins.
Figure 2 is a schematic of the bifunctional protein dimers engaging immune
cells with
cancer cells.
Figure 3 is the amino acid sequence of the anti-CD33 construct with the IgG2
hinge
domain only (SEQ ID NO: 1).
1 0
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
Figure 4 is the amino acid sequence of the ULBP1 construct with the IgG2 hinge
domain only (SEQ ID NO: 2).
Figure 5 is the amino acid sequence of the anti-CD3 construct with the IgG2
hinge
domain only (SEQ ID NO: 3).
Figure 6 is the amino acid sequence of the anti-CD33 construct with both IgG2
hinge
and IgG2 Fc domains (SEQ ID NO: 4).
Figure 7 is the amino acid sequence of the ULBP1 construct with both IgG2
hinge and
IgG2 Fe domains (SEQ ID NO: 5).
Figure 8 is the amino acid sequence of the anti-CD3 construct with both IgG2
hinge
and IgG2 Fe domains (SEQ ID NO: 6).
Figure 9 is a schematic of an engineered heterotrimer protein.
Figure 10 are schematics of the engineered heterodimer proteins with the IgG2-
hinge
and the 6DMPa/b heterodimerizing structure. Figure 10A is an anti-CD33/anti-
CD3
heterodimer protein. Figure 10B is an anti -CD33/ULPB 1 heterodimer protein.
Figure 11 are plasmid maps of the expression vectors used in the studies.
Figure 11A
is a map of the aCD33-6DMPa-Hinge expression vector. Figure 11B is a map of
the 1.JLBP1-
6DMPb-Hinge expression vector. Figure 11C is a map of the aCD3-6DMPb
expression vector.
Figure 12 are immunoblots showing the expression and purification of the anti-
CD33/ULBP engineered protein in 293T cells transfected with anti-CD33-ULBP1
chimeras 1
and 2 plasmids. Figure 12A shows the expression in cells. Figure 12B shows the
expression
and purification in supernatant.
Figure 13 is an immunoblot showing the expression and purification of hinge-
based
constructs in CHO cells. The proteins arc secreted into supernatant and can be
6xHis-purified
using cobalt-resin and elution with imidazole.
Figure 14 shows FACS plots of HL-60 cells incubated with either anti-CD33
construct
alone or with one of the engineered heterodimer proteins. Figure 14A shows co-
incubation
with an anti-FLAG antibody. Figure 14B shows co-incubation with an anti-MYC
antibody.
Figure 15 is a table summarizing binding experiments of the engineered
heterodimer
proteins to HL-60 cells.
Figure 16 is a table summarizing binding experiments of the engineered
heterodimer
proteins to Jurkat cells.
Figure 17 is a table summarizing binding experiments of the engineered
heterodimer
proteins to PMBCs.
11
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
Figure 18 shows the result of a cytotoxicity assay using M0LM14 cells and the
anti-
CD33/anti-CD3 engineered heterodimer protein. Figure 18A is a representative
dot plot
demonstrating flow cytometry gating scheme. Single dTomato+ (MOLM14) cells are
gated,
and % specific cells killed is assessed as total % of dTomato+ cells that are
also DAPI+. Figure
18B is a graph of the results of a cytotoxicity in MOLM14 cells using the anti-
C1133/anti-CD3
engineered heterodimer protein.
Figure 19 shows the result of a cytotoxicity assay using HL60 cells and the
anti-
CD33/anti-CD3 engineered heterodimer protein. Figure 19A is a representative
dot plot
demonstrating flow cytometry gating scheme. Single CellTrace Violet+ (HL-60)
cells are
gated, and % specific cells killed is assessed as total % of CellTrace Violet+
cells that are also
DAPI+. Figure 19B is a graph showing the results of a cytotoxicity assay in HL-
60 cells using
the anti-CD33/anti-CD3 engineered heterodimer protein.
Figure 20 is a graph showing further results of a cytotoxicity assay in HL-60
cells using
the anti-CD33/anti-CD3 engineered heterodimer protein as compared to monomers.
Figure 21 is a graph showing results of a dose dependent cytotoxicity assay
using
protein titration (i.e., increase in the anti-CD33/anti-CD3 engineered
heterodimer protein) in
HL-60 cells.
Figure 22 is a graph showing results of a dose dependent cytotoxicity assay
using
effector titration (i.e., increase in the effector/target ratio) in HL-60
cells.
Figure 23 are the amino acid sequences of the anti-CD16 construct. Figure 23A
is the
amino acid sequence of the anti-CD16 construct with IgG2 hinge domain only
(SEQ ID NO:
26). Figure 23B is the amino acid sequence of the anti-CD16 construct with
both IgG2 hinge
and IgG2 Fe domains (SEQ ID NO: 27).
Figure 24 shows the results of the in vivo anti-tumor activity of anti-CD33-
anti-CD3
engineered heterodimer protein. Figure 24A is a schematic of the experiment.
Figure 24B are
images of bioluminescence imaging (BLI) used to monitor the growth of FFluc-
dtomato
transduced MOLM14. Figure 24C is a graph of the quantification of BLI in mice
treated with
MOLM14 alone, unloaded T cells or anti-CD33-anti-CD3 engineered heterodimer
protein
loaded T cells. Figure 24D is a Kaplan-Meier survival plot. Mice treated with
anti-CD33-anti-
CD3 engineered heterodimer protein loaded T cells have better survival than
the 2 control
groups (no treatment or unloaded T cells). Log-rank test *p <0.05.
12
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
DETAILED DESCRIPTION
Definitions
The term -engineered protein" or -engineered heterodimer protein" or
"engineered
heterotrimer protein" or "engineered hetero protein" or "polyfunctional
protein" or
"polyfunctional orthogonal protein chimera" or "protein chimera" or the like
as used herein
refers to a hybrid polypeptide which comprises protein domains from at least
two different
proteins. One domain may be located at the amino-terminal (N-terminal) portion
of the fusion
protein or at the carboxy-terminal (C-terminal) portion of the fusion protein.
Any of the
proteins provided herein may be produced by any method known in the art. For
example, the
proteins provided herein may be produced via recombinant protein expression
and purification,
which is especially suited for fusion proteins comprising a peptide linker.
Methods for
recombinant protein expression and purification are well known, and include
those described
by Green and Sambrook, Molecular Cloning: A Laboratory Manual (4th ed., Cold
Spring
Harbor Laboratory Press, Cold Spring Harbor, N.Y. (2012)), the entire contents
of which are
incorporated herein by reference.
The terms "protein," "peptide," and "polypeptide" are used interchangeably
herein, and
refer to a polymer of amino acid residues linked together by peptide (amide)
bonds. The terms
refer to a protein, peptide, or polypeptide of any size, structure, Or
function. Typically, a protein,
peptide, or polypeptide will be at least three amino acids long. A protein,
peptide, or
polypeptide may refer to an individual protein or a collection of proteins.
One or more of the
amino acids in a protein, peptide, or polypeptide may be modified, for
example, by the addition
of a chemical entity such as a carbohydrate group, a hydroxyl group, a
phosphate group, a
farnesyl group, an isofarnesyl group, a fatty acid group, a linker for
conjugation,
functionalization, or other modification, etc. A protein, peptide, or
polypeptide may also be a
single molecule or may be a multi-molecular complex. A protein, peptide, or
polypeptide may
be just a fragment of a naturally occurring protein or peptide. A protein,
peptide, or polypeptide
may be naturally occurring, recombinant, or synthetic, or any combination
thereof.
The terms "subject," "individual," and "patient" are used interchangeably, and
refer to
a vertebrate, preferably a mammal such as a human. Mammals include, but are
not limited to,
human primates, non-human primates or murine, bovine, equine, canine or feline
species. In
the context of the present disclosure, the term "subject" also encompasses
tissues and cells that
can be cultured in vitro or ex vivo or manipulated in vivo. The term "subject"
can be used
interchangeably with the term "organism".
13
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
The terms "polynucleotide", "nucleotide", "nucleotide sequence", "nucleic
acid" and
"oligonucleotide" are used interchangeably. They refer to a polymeric form of
nucleotides of
any length, either deoxyribonucleotides or ribonucleotides, or analogs
thereof. Examples of
polynucleotides include, but are not limited to, coding or non-coding regions
of a gene or gene
fragment, exons, introns, messenger RNA (mRNA), transfer RNA, ribosomal RNA,
short
interfering RNA (siRNA), short-hairpin RNA (shRNA), micro-RNA (miRNA),
ribozymes,
cDNA, recombinant polynucleotides, branched polynucleotides, plasmids,
vectors, isolated
DNA of any sequence, isolated RNA of any sequence, nucleic acid probes, and
primers. One
or more nucleotides within a polynucleotide can further be modified. The
sequence of
nucleotides may be interrupted by non-nucleotide components. A polynucleotide
may also be
modified after polymerization, such as by conjugation with a labeling agent.
The term "hybridization" refers to a reaction in which one or more
polynucleotides
react to form a complex that is stabilized via hydrogen bonding between the
bases of the
nucleotide residues. The hydrogen bonding may occur by Watson Crick base
pairing,
Hoogstein binding, or in any other sequence specific manner. The complex may
comprise two
strands forming a duplex structure, three or more strands forming a multi-
stranded complex, a
single self-hybridizing strand, or any combination of these. A hybridization
reaction may
constitute a step in a more extensive process, such as the initiation of PCR,
or the cleavage of
a polynucleotide by an enzyme. A sequence capable of hybridizing with a given
sequence is
referred to as the "complement" of the given sequence.
The terms "vector", "cloning vector" and "expression vector" mean the vehicle
by
which a DNA or RNA sequence (e.g., a foreign gene) can be introduced into a
host cell, so as
to transform the host and promote expression (e.g., transcription and
translation) of the
introduced sequence. Vectors include, but are not limited to, plasmids,
phages, and viruses.
Vectors typically comprise the DNA of a transmissible agent, into which
foreign DNA is
inserted. A common way to insert one segment of DNA into another segment of
DNA involves
the use of enzymes called _restriction enzymes that cleave DNA at specific
sites (specific groups
of nucleotides) called restriction sites. A "cassette" refers to a DNA coding
sequence or
segment of DNA which codes for an expression product that can be inserted into
a vector at
defined restriction sites. The cassette restriction sites are designed to
ensure insertion of the
cassette in the proper reading frame. Generally, foreign DNA is inserted at
one or more
restriction sites of the vector DNA, and then is carried by the vector into a
host cell along with
the transmissible vector DNA. A segment or sequence of DNA having inserted or
added DNA,
such as an expression vector, can also be called a "DNA construct" or "gene
construct." A
14
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
common type of vector is a "plasmid", which generally is a self-contained
molecule of double-
stranded DNA, usually of bacterial origin, that can readily accept additional
(foreign) DNA
and which can readily introduced into a suitable host cell. A plasmid vector
often contains
coding DNA and promoter DNA and has one or more restriction sites suitable for
inserting
foreign DNA. Coding DNA is a DNA sequence that encodes a particular amino acid
sequence
for a particular protein or enzyme. Promoter DNA is a DNA sequence which
initiates,
regulates, or otherwise mediates or controls the expression of the coding DNA.
Promoter DNA
and coding DNA may be from the same gene or from different genes and may be
from the
same or different organisms. A large number of vectors, including plasmid and
fungal vectors,
have been described for replication and/or expression in a variety of
eukaryotic and prokaryotic
hosts. Non-limiting examples include pKK plasmids (Clonetech), pUC plasmids,
pET
plasmids (Novagen, Inc., Madison, WI), pRSET or pREP plasmids (Invitrogen, San
Diego,
CA), or pMAL plasmids (New England Biolabs, Beverly, MA), and many appropriate
host
cells, using methods disclosed or cited herein or otherwise known to those
skilled in the
relevant art. Recombinant cloning vectors will often include one or more
replication systems
for cloning or expression, one or more markers for selection in the host,
e.g., antibiotic
resistance, and one or more expression cassettes.
The term "recombinant expression vector" means a genetically modified
oligonucleotide or polynucleotide construct that permits the expression of an
mRNA, protein,
polypeptide, or peptide by a host cell, when the construct comprises a
nucleotide sequence
encoding the mRNA, protein, polypeptide, or peptide, and the vector is
contacted with the cell
under conditions sufficient to have the mRNA, protein, polypeptide, or peptide
expressed
within the cell. The vectors of the present disclosure are not naturally
occurring as a whole.
Parts of the vectors can be naturally occurring. The non-naturally occurring
recombinant
expression vectors of the present disclosure can comprise any type of
nucleotides, including,
but not limited to DNA and RNA, which can be single-stranded or double-
stranded,
synthesized or obtained in part from natural sources, and which can contain
natural, non-natural
or altered nucleotides.
"Transfection," "transformation," or "transduction," as used herein, refer to
the
introduction of one or more exogenous polynucleotides into a host cell by
using physical or
chemical methods.
"Antibody," "fragment of an antibody," "antibody fragment," "functional
fragment of
an antibody," or "antigen-binding portion" are used interchangeably to mean
one or more
fragments or portions of an antibody that retain the ability to specifically
bind to a specific
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
antigen (Holliger et al., Nat. Biotech. (2005) 23(9): 1126). The present
antibodies may be
antibodies and/or fragments thereof. Antibody fragments include Fab, F(ab')2,
scFv, disulfide
linked Fv, Fc, or variants and/or mixtures. The antibodies may be chimeric,
humanized, single
chain, or hi-specific. All antibody isotypes are encompassed by the present
disclosure,
including, IgA, 1gD, IgE, IgG, and IgM. Suitable IgG subtypes include 1gGl,
IgG2, IgG3 and
IgG4. An antibody light or heavy chain variable region consists of a framework
region
interrupted by three hypervariable regions, referred to as complementarity
determining regions
(CDRs). The CDRs of the present antibodies or antigen-binding portions can be
from a non-
human or a human source. The framework of the present antibodies or antigen-
binding
portions can be human, humanized, non-human (e.g., a murine framework modified
to decrease
antigenicity in humans), or a synthetic framework (e.g., a consensus
sequence).
The present antibodies or antigen-binding portions can specifically bind with
a
dissociation constant (Ku) of less than about 10-7 M, less than about 10-8M,
less than about 10-
9 M, less than about 10-1 M, less than about 10-11 M, or less than about 10-
12 M. Affinities of
the antibodies according to the present disclosure can be readily determined
using conventional
techniques (see, e.g., Scatchard et al., Ann. N.Y. Acad. Sci. (1949) 51:660;
and U.S. Patent Nos.
5,283,173, 5,468,614, or the equivalent).
The antigen recognition moiety of the engineered protein encoded by the
nucleic acid
sequence can contain any lineage antigen-specific, antigen-binding antibody
fragment. The
antibody fragment can comprise one or more CDRs, the variable region (or
portions thereof),
the constant region (or portions thereof), or combinations of any of the
foregoing.
The term ''host cell" means any cell of any organism that is selected,
modified,
transformed, grown, used or manipulated in any way, for the production of a
substance by the
cell, for example, the expression by the cell of a gene, a DNA or RNA
sequence, a protein or
an enzyme. Host cells can further he used for screening or other assays, as
described herein.
The term "cell lineage" refers to cells with a common ancestry and developing
from the
same type of identifiable cell into specific identifiable/functioning cells.
The cell lineages used
herein include, but are not limited to, respiratory, prostatic, pancreatic,
mammary, renal,
intestinal, neural, skeletal, vascular, hepatic, hematopoietic, muscle or
cardiac cell lineages.
The term "inhibition" when used in reference to gene expression Or function of
a lineage
specific antigen refers to a decrease in the level of gene expression or
function of the lineage
specific antigen, where the inhibition is a result of interference with gene
expression or
function. The inhibition may be complete, in which case there is no detectable
expression or
16
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
function, or it may be partial. Partial inhibition can range from near
complete inhibition to a
near absence of inhibition.
The terms -treat", -treatment", and the like refer to a means to slow down,
relieve,
ameliorate or alleviate at least one of the symptoms of the disease, or
reverse the disease after
its onset.
"Treating" or "treatment" of a state, disorder or condition includes:
(1) preventing or delaying the appearance of clinical symptoms of the state,
disorder,
or condition developing in a person who may be afflicted with or predisposed
to the
state, disorder or condition but does not yet experience or display clinical
symptoms of
the state, disorder or condition; or
(2) inhibiting the state, disorder or condition, i.e., arresting, reducing or
delaying the
development of the disease or a relapse thereof (in case of maintenance
treatment) or at
least one clinical symptom, sign, or test, thereof; or
(3) relieving the disease, i.e., causing regression of the state, disorder or
condition or at
least one of its clinical or sub-clinical symptoms or signs.
The benefit to a subject to be treated is either statistically significant or
at least
perceptible to the patient or to the physician.
The terms -prevent", "prevention", and the like refer to acting prior to overt
disease
onset, to prevent the disease from developing or minimize the extent of the
disease or slow its
course of development.
An "immune response" refers to the development in the host of a cellular
and/or
antibody-mediated immune response to a composition or vaccine of interest.
Such a response
usually consists of the subject producing antibodies, B cells, helper T cells,
suppressor T cells,
regulatory T cells, and/or cytotoxic T cells directed specifically to an
antigen or antigens
included in the composition or vaccine of interest.
A "therapeutically effective amount" or "effective amount" means the amount of
a
compound or agent that, when administered to an animal for treating a state,
disorder or
condition, is sufficient to affect such treatment. The "therapeutically
effective amount" will
vary depending on the compound, the disease and its severity and the age,
weight, physical
condition and responsiveness of the animal to be treated.
The compositions disclosed herein may include a -therapeutically effective
amount" or
a "prophylactically effective amount" of a compound described herein. A
"therapeutically
effective amount" refers to an amount effective, at dosages and for periods of
time necessary,
to achieve the desired therapeutic result. A therapeutically effective amount
of an antibody or
17
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
antibody portion may vary according to factors such as the disease state, age,
sex, and weight
of the individual, and the ability of the antibody or antibody portion to
elicit a desired response
in the individual. A therapeutically effective amount is also one in which any
toxic or
detrimental effects of the compound are outweighed by the therapeutically
beneficial effects.
A "prophylactically effective amount" refers to an amount effective, at
dosages and for periods
of time necessary, to achieve the desired prophylactic result. Typically,
since a prophylactic
dose is used in subjects prior to or at an earlier stage of disease, the
prophylactically effective
amount will be less than the therapeutically effective amount.
While it is possible to use a composition provided by the present disclosure
for therapy
as is, it may be preferable to administer it in a pharmaceutical formulation,
e.g., in admixture
with a suitable pharmaceutical excipient, diluent or carrier selected with
regard to the intended
route of administration and standard pharmaceutical practice. Accordingly, in
one aspect, the
present disclosure provides a pharmaceutical composition or formulation
comprising at least
one active composition, or a pharmaceutically acceptable derivative thereof,
in association with
a pharmaceutically acceptable excipient, diluent and/or carrier. The
excipient, diluent and/or
carrier must be "acceptable" in the sense of being compatible with the other
ingredients of the
formulation and not deleterious to the recipient thereof.
The compositions of the disclosure can be formulated for administration in any
convenient way for use in human or veterinary medicine. The invention
therefore includes
within its scope pharmaceutical compositions comprising a product of the
present invention
that is adapted for use in human or veterinary medicine.
The term "pharmaceutical composition," as used herein, refers to a composition
that
can be administrated to a subject in the context of treatment and/or
prevention of a disease or
disorder. In some embodiments, a pharmaceutical composition comprises an
active ingredient,
e.g., the present fusion pol ypepti de, nucleic acid molecule, vector, agent,
etc., and optionally a
pharmaceutically acceptable excipient, diluent and/or carrier.
Acceptable excipients, diluents, and carriers for therapeutic use are well
known in the
pharmaceutical art, and are described, for example, in Remington: The Science
and Practice of
Pharmacy. Lippincott Williams & Wilkins (A. R. Gennaro edit. 2005). The choice
of
pharmaceutical excipient, diluent, and carrier can be selected with regard to
the intended route
of administration and standard pharmaceutical practice.
As used herein, the phrase "pharmaceutically acceptable" refers to molecular
entities
and compositions that are "generally regarded as safe", e.g., that are
physiologically tolerable
and do not typically produce an allergic or similar untoward reaction, such as
gastric upset,
18
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
dizziness and the like, when administered to a human. Preferably, as used
herein, the term
"pharmaceutically acceptable" means approved by a regulatory agency of the
Federal or a state
government or listed in the U.S. Pharmacopoeia or other generally recognized
pharmacopeias
for use in animals, and more particularly in humans.
The dosage of the therapeutic formulation will vary widely, depending upon the
nature
of the disease, the patient's medical history, the frequency of
administration, the manner of
administration, the clearance of the agent from the host, and the like. The
initial dose may be
larger, followed by smaller maintenance doses. The dose may be administered as
infrequently
as weekly or biweekly, or fractionated into smaller doses and administered
daily, semi-weekly,
etc., to maintain an effective dosage level. In some cases, oral
administration will require a
higher dose than if administered intravenously. In some cases, topical
administration will
include application several times a day, as needed, for a number of days or
weeks in order to
provide an effective topical dose.
The term "carrier" refers to a diluent, adjuvant, excipient, or vehicle with
which the
compound is administered. Such pharmaceutical carriers can be sterile liquids,
such as water
and oils, including those of petroleum, animal, vegetable or synthetic origin,
such as peanut
oil, soybean oil, mineral oil, olive oil, sesame oil and the like. Water or
aqueous solution saline
solutions and aqueous dextrose and glycerol solutions are preferably employed
as carriers,
particularly for injectable solutions. Alternatively, the carrier can be a
solid dosage form
carrier, including but not limited to one or more of a binder (for compressed
pills), a glidant,
an encapsulating agent, a tlavorant, and a colorant. Suitable pharmaceutical
carriers are
described in "Remington's Pharmaceutical Sciences" by E. W. Martin.
The term "agent" as used herein means a substance that produces or is capable
of
producing an effect and would include, but is not limited to, chemicals,
pharmaceuticals,
biologics, small organic molecules, antibodies, nucleic acids, peptides, and
proteins.
The term "about" or "approximately" means within an acceptable error range for
the
particular value as determined by one of ordinary skill in the art, which will
depend in part on
how the value is measured or determined, i.e., the limitations of the
measurement system, i.e.,
the degree of precision required for a particular purpose, such as a
pharmaceutical formulation.
For example, "about" can mean within 1 or more than 1 standard deviations, per
the practice
in the art. Alternatively, "about" can mean a range of up to 20%, preferably
up to 10%, more
preferably up to 5%, and more preferably still up to 1% of a given value.
Alternatively,
particularly with respect to biological systems or processes, the term can
mean within an order
of magnitude, preferably within 5-fold, and more preferably within 2-fold, of
a value. Where
19
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
particular values are described in the application and claims, unless
otherwise stated, the term
"about" meaning within an acceptable error range for the particular value
should be assumed.
General techniques
The practice of the present disclosure will employ, unless otherwise
indicated,
conventional techniques of molecular biology (including recombinant
techniques),
microbiology, cell biology, biochemistry, and immunology, which are within the
skill of the
art. Such techniques are explained fully in the literature, such as Molecular
Cloning: A
Laboratory Manual, second edition (Sambrook, et al., 1989) Cold Spring Harbor
Press;
Oligonucleoticie Synthesis (M. J. Gait, ed. 1984); Methods in Molecular
Biology, Humana
Press; Cell Biology: A Laboratory Notebook (J. E. Cellis, ed., 1989) Academic
Press; Animal
Cell Culture (R. I. Freshney, ed. 1987); Introduction to Cell and Tissue
Culture (J. P. Mather
and P. E. Roberts, 1998) Plenum Press; Cell and Tissue Culture: Laboratory
Procedures (A.
Doyle, J. B. Griffiths, and D. G. Newell, eds. 1993-8) J. Wiley and Sons;
Methods in
Enzymology (Academic Press, Inc.); Handbook of Experimental Immunology (D. M.
Weir and
C. C. Blackwell, eds.): Gene Transfer Vectors for Mammalian Cells (J. M.
Miller and M. P.
Cabs, eds., 1987); Current Protocols in Molecular Biology (F. M. Ausubel, et
al. eds. 1987);
PCR: The Polymerase Chain Reaction, (Mullis, et al., eds. 1994); Current
Protocols in
Immunology (J. E. Coligan et al., eds., 1991); Short Protocols in Molecular
Biology (Wiley
and Sons, 1999); Immunobiology (C. A. Janeway and P. Travers, 1997);
Antibodies (P. Finch,
1997); Antibodies: a practice approach (D. Catty., ed., IRL Press, 1988-1989);
Monoclonal
antibodies: a practical approach (P. Shepherd and C. Dean, eds., Oxford
University Press,
2000); Using antibodies: a laboratory manual (E. Harlow and D. Lane (Cold
Spring Harbor
Laboratory Press, 1999): The Antibodies (M. Zanetti and J. D. Capra, cds.
Harwood Academic
Publishers, 1995); DNA Cloning: A practical Approach, Volumes I and II (D.N.
Glover ed.
1985); Nucleic Acid Hybridization (B.D. Hames & S.J. Higgins eds.(1985);
Transcription and
Translation (B.D. Hames & S.J. Higgins, eds. (1984 ; Animal Cell Culture (R.I.
Freshney, ed.
(1986 ; Immobilized Cells and Enzymes (1RL Press, (1986).
The present disclosure provides for agents comprising an antigen-binding
fragment that
binds a lineage-specific cell-surface antigen (e.g., CD33) which can cause
cell death of the
cells expressing the lineage-specific cell-surface antigen. Immunotherapies
involving the
combination of an antigen-binding fragment that binds a lineage-specific cell-
surface antigen
(e.g., CD33), and a polypeptide that binds a molecule expressed on an immune
cell such as a
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
natural killer (NK) cell and/or a T cell, would provide an efficacious method
of treatment for
hematopoietic malignancies.
The present disclosure provides for a first and a third engineered heterodimer
protein,
comprising (or consisting essentially of, or consisting of): (i) an antigen-
binding fragment that
binds a lineage-specific cell-surface antigen (e.g., CD33); and (ii) a
polypeptide that binds a
molecule expressed on immune cells. In some embodiments, the immune cells are
natural killer
(NK) cells. In some embodiments, the molecule is ULBP. In some embodiments,
the molecule
is CD16.
The present disclosure provides for a second engineered heterodimer protein,
comprising (or consisting essentially of, or consisting of): (i) an antigen-
binding fragment that
binds a lineage-specific cell-surface antigen (e.g., CD33); and (ii) a
polypeptide that binds a
molecule expressed on T cells. In some embodiments, the molecule is CD3.
The present disclosure further provides for an engineered heterotrimer
protein,
comprising (or consisting essentially of, or consisting of): (i) an antigen-
binding fragment that
binds a lineage-specific cell-surface antigen (e.g., CD33); and (ii) a
polypeptide that binds to
immune cells (e.g., NK cells); and (iii) a polypeptide that binds a molecule
expressed on T cells
(e.g., CD3).
See Figures 2, 9, and 10.
The present compositions and methods may help activate NKG2D-bearing immune
effector cells, such as natural killer (NK) cells and/or CD8+ T cells. The
present compositions
and methods may enhance or prompt a cellular immune response against diseased
cells (such
as tumor cells) that may induce cytotoxicity (e.g., culminate in the death of
the diseased cells
such as tumor cells). The present compositions and methods may enhance a
subject's immune
response, including, but not limited to one or more of the following:
upregulation of natural
killer (NK) cell; upregulation of T cell (e.g., gamma delta T cell, alpha beta
T cell) function;
upregulation of natural killer T (NKT) cell function; and upregulation of B
cell function. In
some embodiments, upregulation of one or more of NK cell, T cell, natural
killer T (NKT) cell,
and B cell function includes enhancement and/or endowment of activity capable
of inhibiting
or decreasing cancer progression.
In some embodiments, inhibiting cancer progression may be accomplished by
cytolysis
of tumor cells, e.g., by direct induction of tumor cell apoptosis, induction
of tumor cell cytolysis
through stimulation of intrinsic host antitumor responses, induction of tumor
cell apoptosis
through stimulation of intrinsic host antitumor responses, inhibition of tumor
cell metastasis,
inhibition of tumor cell proliferation, and induction of senescence in the
tumor cell.
21
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
The present disclosure also provides one or more nucleic acid (polynucleotide)
molecules encoding the present engineered proteins, agents or compositions.
Other aspects of the present disclosure provide vectors comprising any of the
nucleic
acid (or polynucleotide) molecules provided herein. Also, within the scope of
the present
disclosure are polynucleotides encoded by the nucleic acids described herein
and cells
expressing such polynucleotides.
In some embodiments, the cells can be obtained from a patient having a
hematopoietic
malignancy. In some embodiments, the cell is a hematopoietic cell, such as a
hematopoietic
stem cell (e.g., CD34 ). In some embodiments, cells are provided, e.g., for
recombinant
expression and purification of the engineered proteins provided herein. The
cells include any
cell suitable for recombinant protein expression, for example, cells
comprising a genetic
construct or vector expressing or capable of expressing an engineered proteins
(e.g., cells that
have been transformed or transfected with one or more vectors described
herein, or cells having
genomic modifications, for example, those that express a protein provided
herein). Methods
for transforming cells, genetically modifying cells, and expressing genes and
proteins in such
cells are well known in the art, and include those provided by, for example,
Green and
Sambrook, Molecular Cloning: A Laboratory Manual (4th ed., Cold Spring Harbor
Laboratory
Press, Cold Spring Harbor, N.Y. (2012)) and Friedman and Rossi, Gene Transfer:
Delivery and
Expression of DNA and RNA, A Laboratory Manual (1st ed., Cold Spring Harbor
Laboratory
Press, Cold Spring Harbor, N.Y. (2006)).
Further, the present disclosure provides pharmaceutical compositions
comprising the
present agents, polypeptides, nucleic acid (or polynucleotide) molecules,
vectors, cells, and/or
compositions.
The present disclosure also provides for a method of treating a hematopoietic
malignancy. The method may comprise administering to a subject in need thereof
an effective
amount of any of the disclosed engineered proteins or a polynucleotide
encoding the engineered
proteins. The method may comprise administering to a subject in need thereof
an effective
amount of the present agents or composition, or a polynucleotide encoding the
agents (e.g., a
combination of polypeptides) or compositions.
Another aspect of the present disclosure provides a method for treating a
hematopoietic
malignancy (or a hematological neoplasm), the method comprising administering
to a subject
in need thereof an effective amount of any of the disclosed engineered
proteins or a
polynucleotide encoding the engineered proteins. The method may comprise
administering to
22
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
a subject in need thereof an effective amount of the present agents or
composition, or a
polynucleotide encoding the agents (e.g., a combination of polypeptides) or
compositions.
The present disclosure also relates to methods of using the engineered
proteins to treat
hematopoietic malignancies such as myeloid malignancies.
Also, within the scope of the present disclosure are kits comprising the
present agents,
polypeptides, nucleic acid (or polynucleotide) molecules, vectors, cells,
and/or compositions.
Engineered Hetero Proteins
The current disclosure provides for engineered hetero proteins.
In one embodiment, the present disclosure provides for a first and third
engineered
heterodimer protein, comprising (or consisting essentially of, or consisting
of): (i) an antigen-
binding fragment that binds a lineage-specific cell-surface antigen (e.g.,
CD33); and (ii) a
polypeptide that binds a molecule expressed on immune cells. In some
embodiments, the
immune cells are natural killer (NK) cells. In some embodiments, the molecule
is ULB P. in
some embodiments, the molecule is CD16.
In a further embodiment, the present disclosure provides for a second
engineered
heterodimer protein, comprising (or consisting essentially of, or consisting
of): (i) an antigen-
binding fragment that binds a lineage-specific cell-surface antigen (e.g.,
CD33); and (ii) a
polypeptide that binds a molecule expressed on T cells. In some embodiments,
the molecule is
CD3.
In yet a further embodiment, the present disclosure provides for an engineered
heterotrimer protein, comprising (or consisting essentially of, or consisting
of): (i) an antigen-
binding fragment that binds a lineage-specific cell-surface antigen (e.g..
CD33); and (ii) a
polypeptide that binds to immune cells (e.g., NK cells); and (iii) a
polypeptide that binds a
molecule expressed on T cells (e.g., CD3).
In some embodiments, the third polypeptide is a chemokine or cytokine protein,
which
increases the immune response. The chemokine or cytokine protein includes but
is not limited
to CXCLs including CXCL14, GCSF, and interleukins, including IL2 and IL16.
In some embodiments, the antigen-binding fragment binds a lineage-specific
cell-
surface antigen that is a type 2 lineage-specific cell-surface antigen (e.g.,
CD33). In some
embodiments, the antigen-binding fragment binds a lineage-specific cell-
surface antigen that
is a type 1 lineage-specific cell-surface antigen (e.g., CD19). In some
embodiments, the
antigen-binding fragment binds an antigen expressed or over-expressed by
cancer and/or tumor
cells.
23
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
The polypeptide that binds a molecule expressed on natural killer (NK) cells
may be a
fragment of fragment of ULBP1, ULBP2, ULBP3, ULBP4, ULBP5, ULBP6, MICA, MICB,
Raetla, Raetlb, Raetic, Raetld, Raetle, H60b , H60c, or HCMV UL18, homologs
thereof,
mutants thereof, or fragments thereof. The polypeptide that binds a molecule
expressed on
natural killer (NK) cells may be an ectodomain of ULBP1. ULBP2, ULBP3, ULBP4,
ULBP5,
ULBP6, MICA, MICB, Raetla, Raetlb, Raetic, Raetld, Raetle, H60b, H60c, HCMV
UL18,
homologs thereof, mutants thereof, or fragments thereof.
In certain embodiments, the fragment of ULBP1, ULBP2, ULBP3, ULBP4, ULBP5,
ULBP6, MICA, MICB, Raetla, Raetlb, Raetic, Raetld, Raetle, H60b, H60c, or HCMV
UL18
comprises an ectodomain of ULBP1, ULBP2, ULBP3, ULBP4, ULBP5, ULBP6, MICA,
MICB, Raetla, Raetlb, Raetic, Raetld, Raetle, H60b, H60c, or HCMV UL18. In
certain
embodiments, the fragment of ULBP1, ULBP2, ULBP3, ULBP4, ULBP5, ULBP6, MICA,
MICB, Raetla, Raetlb, Raetic, Raetld, Raetle, H60b, H60c, or HCMV UL18
comprises an
ectodomain of ULBP1, ULBP2, ULBP3, ULBP4, ULBP5, ULBP6, MICA, MICB, Raetla,
Raetlb, Raetic, Raetld, Raetle, H60b, H60c, HCMV UL18, homologs thereof,
mutants
thereof, or fragments thereof.
In some embodiments, the polypeptidc binds a molecule expressed on NK cells is
an
antibody of a cell surface marker of NK cells. In some embodiments, the cell
surface marker
is CD16. In some embodiments, the antibody is a monoclonal antibody.
In certain embodiments, the polypeptide that binds a molecule expressed on T
cells is
an antibody of a cell surface marker of T cells. In some embodiments, the cell
surface marker
is CD3. In sonic embodiments, the antibody is a monoclonal antibody.
In certain embodiments, the first engineered heterodimer protein comprises:
(i) a first
polypeptide comprising an antigen-binding fragment that binds a lineage-
specific cell-surface
antigen, a non -naturally occun-ing polypeptide domain comprising 1-5 alpha
helices connected
by amino acid linkers and a first covalent dimerization domain; and (ii) a
second polypeptide
comprising a polypeptide that binds a molecule expressed on natural killer
(NK) cells, a non-
naturally occurring polypeptide domain comprising 1-5 alpha helices connected
by amino acid
linkers, and a second covalent dimerization domain as described herein. See
Figures 1 and
10B.
In certain embodiments, antigen-binding fragments include, but are not limited
to, Fab,
F(ab')2, Fab', F(ab)', Fv, a disulfide linked Fv, single chain Fv (scFv),
bivalent scFv (bi-scFv),
trivalent scFv (tri-scFv), Fd, dAb fragment, an isolated CDR, diabodies,
triabodies, tetrabodies,
24
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
linear antibodies, single-chain antibody molecules. In some embodiments, scFv
comprises a
heavy chain variable region (Vii), and a light chain variable region (VL).
In some embodiments, the non-naturally occurring polypeptide domain comprising
1-
alpha helices is 6DMPa (Chen et al. 2019). In some embodiments, the non-
naturally
5 occurring polypeptide domain comprising 1-5 alpha helices is 6DMPb (Chen
et al. 2019).
In some embodiments, the covalent dimerization domains are IgG2 hinge domains.
In
some embodiments, the covalent dimerization domains are IgG2 domains and IgG2
Fc
domains. In some embodiments, the Fe domains are CH2 and CH3 domains.
The first engineered heterodimer protein can be designed to place the
functional
moieties (an antigen-binding fragment that binds a lineage-specific cell-
surface antigen, and a
polypeptide that binds a molecule expressed on NK cells) in any order. In
certain embodiments,
the antigen-binding fragment that binds a lineage-specific cell-surface
antigen is located at the
N-terminus or C-terminus of the fusion polypeptide. In certain embodiments,
the polypeptide
that binds a molecule expressed on natural killer (NK) cells is located at the
C-terminus or N-
terminus of the fusion polypeptide.
In some embodiments, the first engineered heterodimer protein comprises, from
N-
terminus to C-terminus, a scFv that binds to the lineage-specific cell-surface
antigen (e.g.,
CD33 or CD19), and an ectodomain of ULBP1 (or an ectodomain of ULBP2, ULBP3,
ULBP4,
ULBP5, ULBP6, MICA, or MICB). In some embodiments, the fusion polypeptide
comprises,
from N terminus to C terminus, an ectodomain of ULBP1 (or an ectodomain of
ULBP2,
ULBP3, ULBP4, ULBP5, ULBP6, MICA, or MICB), and a scFv that binds to the
lineage-
specific cell-surface antigen (e.g., CD33 or CD19).
The first engineered heterodimer protein may further comprise a signal
sequence,
and/or one or more linkers. In the fusion polypeptide, these functional
moieties may be
coval en tl y li gated continuously or non-continuously (e.g., they may be
separated by 1 i nkers).
The linker may have up to 50, up to 40, up to 30, up to 20, up to 18, up to
15, up to 12, up to
11, or up to 10, amino acid residues in length. In certain embodiments, the
linker has about 1,
2, 3, 4, 5, 6, 7, 8, 9, 10-20, 8-10, 8-12, 8-15, 8-20, or 8-30 amino acid
residues in length. In
certain embodiments, the linker has about 7-10, 7-12, 7-15, 7-20, or 7-30
amino acid residues
in length.
One type of derivatized protein is produced by crosslinking two or more
polypeptides
(of the same type or of different types). Suitable crosslinkers include those
that are
heterobifunctional, having two distinct reactive groups separated by an
appropriate spacer (e.g.,
m-maleimidobenzoyl-N-hydroxysuccinimide ester) or homobifunctional (e.g.,
disuccinimidyl
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
suberate). Useful detectable agents with which a protein can be derivatized
(or labeled) include
fluorescent agents, various enzymes, prosthetic groups, luminescent materials,
bioluminescent
materials, and radioactive materials. Non-limiting, exemplary fluorescent
detectable agents
include fluorescein, fluorescein isothiocyanate, rhodamine, and phycoerythrin.
A polypeptide
can also be derivatized with detectable enzymes, such as alkaline phosphatase,
horseradish
peroxidase, beta-galactosidase, acetylcholinesterase, glucose oxidase and the
like. A
polypeptide can also be derivatized with a prosthetic group (e.g.,
streptavidin/biotin and
avidin/biotin).
The first engineered heterodimer protein can be derivatized or linked to
another
functional molecule. For example, first engineered heterodimer protein can be
functionally
linked (by chemical coupling, genetic fusion, noncovalent interaction) to one
or more other
molecular entities, such as an antibody or antibody fragment, a detectable
agent, an
immunosuppressant, a cytotoxic agent, a pharmaceutical agent, a protein or
peptide that can
mediate association with another molecule (such as a streptavidin core region
or a polyhistidine
tag), amino acid linkers, signal sequences, immunogenic carriers, or ligands
useful in protein
purification, such as glutathione-S-transferase, histidine tag, and
staphylococcal protein A.
Cytotoxic agents may include radioactive isotopes, chemotherapeutic agents,
and toxins such
as enzymatically active toxins of bacterial, fungal, plant, or animal origin,
and fragments
thereof.
The first engineered heterodimer protein may further comprise a fragment
(e.g., a tag)
useful for polypeptide production and/or detection, including, but not limited
to, poly-histidine
(e.g., six histidine residues), a maltose binding protein, GST, green
fluorescent protein (GFP),
hemagglutinin, or alkaline phosphatasc, secretion signal peptides (e.g.,
preprotyrypsin signal
sequence), Myc, and/or FLAG.
In one embodiment, the first engineered heterodimer protein comprises (or
consists
essentially of, or consists of) an amino acid sequence at least or about 50%,
at least about 55%,
at least or about 60%, at least or about 70%, at least or about 75%, at least
or about 80%, at
least or about 81%, at least or about 82%, at least or about 83%, at least or
about 84%, at least
or about 85%, at least or about 86%, at least or about 87%, at least or about
88%, at least or
about 89%, at least or about 90%, at least or about 91%, at least or about
92%, at least or about
93%, at least or about 94%, at least or about 95%, at least or about 96%, at
least or about 97%,
at least or about 98%, at least or about 99%, or about 100%, identical to SEQ
ID NO: 1 (Figure
3) and SEQ ID NO: 2 (Figure 4) or SEQ ID NO: 4 (Figure 6) and SEQ ID NO: 5
(Figure 7).
26
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
In one embodiment, the first engineered heterodimer protein comprises a signal
sequence comprising (or consisting essentially of, or consisting of) an amino
acid sequence at
least or about 50%, at least about 55%, at least or about 60%, at least or
about 70%, at least or
about 75%, at least or about 80%, at least or about 81%, at least or about
82%, at least or about
83%, at least or about 84%, at least or about 85%, at least or about 86%, at
least or about 87%,
at least or about 88%, at least or about 89%, at least or about 90%, at least
or about 91%, at
least or about 92%, at least or about 93%, at least or about 94%, at least or
about 95%, at least
or about 966/9, at least or about 97%, at least or about 98%, at least or
about 99%, or about
100%, identical to an IL2 secretory signal sequence: MYRMQLLSCIALSLALVTNS (SEQ
ID NO:7) (Figures 3, 4, 6 and 8).
In one embodiment, the first engineered heterodimer protein comprises one or
more
tags comprising (or consisting essentially of, or consisting of) an amino acid
sequence at least
or about 50%, at least about 55%, at least or about 60%, at least or about
70%, at least or about
75%, at least or about 80%, at least or about 81%, at least or about 82%, at
least or about 83%,
at least or about 84%, at least or about 85%, at least or about 86%, at least
or about 87%, at
least or about 88%, at least or about 89%, at least or about 90%, at least or
about 91%, at least
or about 92%, at least or about 93%, at least or about 94%, at least or about
95%, at least or
about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about 100%,
identical to Myc: EQKLISEEDL (SEQ ID NO: 8) (Figures 3 and 6).
In one embodiment, the first engineered heterodimer protein comprises one or
more
tags comprising (or consisting essentially of, or consisting of) an amino acid
sequence at least
or about 50%, at least about 55%, at least or about 60%, at least or about
70%, at least or about
75%, at least or about 80%, at least or about 81%, at least or about 82%, at
least or about 83%,
at least or about 84%, at least or about 85%, at least or about 86%, at least
or about 87%, at
least or about 88%, at least or about 89%, at least or about 90%, at least or
about 91%, at least
or about 92%, at least or about 93%, at least or about 94%, at least or about
95%, at least or
about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about 100%,
identical to FLAG: DYKDDDDK (SEQ ID NO: 14) (Figures 4 and 7)
In one embodiment, the first engineered heterodimer protein comprises an anti-
CD33
scFv comprising (or consisting essentially of, or consisting of) an amino acid
sequence at least
or about 50%, at least about 55%, at least or about 60%, at least or about
70%, at least or about
75%, at least or about 80%, at least or about 81%, at least or about 82%, at
least or about 83%,
at least or about 84%, at least or about 85%, at least or about 86%, at least
or about 87%, at
least or about 88%, at least or about 89%, at least or about 90%, at least or
about 91%, at least
27
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
or about 92%, at least or about 93%, at least or about 94%, at least or about
95%, at least or
about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about 100%,
identical to the anti-CD33 scEv disclosed in U.S. Patent Publication No.
20130078241.
In one embodiment, the first engineered heterodimer protein comprises an
antigen-
binding fragment that binds CD33, where the antigen-binding fragment comprises
a light chain
variable region (VI) comprising (or consisting essentially of, or consisting
of) an amino acid
sequence at least or about 50%, at least about 55%, at least or about 60%, at
least or about 70%,
at least or about 75%, at least or about 80%, at least or about 81%, at least
or about 82%, at
least or about 83%, at least or about 84%, at least or about 85%, at least or
about 86%, at least
or about 87%, at least or about 88%, at least or about 89%, at least or about
90%, at least or
about 91%, at least or about 92%, at least or about 93%, at least or about
94%, at least or about
95%, at least or about 96%, at least or about 97%, at least or about 98%, at
least or about 99%,
or about 100%, identical to SEQ ID NO: 9 (Figures 3 and 6).
Anti-CD33 Light Chain variable region (VI) (SEQ ID NO: 9):
EIVLTQS PGSLAVSPGERVTMSC KS S QSVFFS SS QKNYLAWYQQIPGQSPRLLIYWAS
TRESGVPDRFTGSGSGTDFTLTISSVQPEDLAIYYCHQYLSSRTFGQGTKLEIKR
In one embodiment, the first engineered heterodimer protein comprises an
antigen-
binding fragment that binds CD33, where the antigen-binding fragment comprises
a heavy
chain variable region (VH) comprising (or consisting essentially of, or
consisting of) an amino
acid sequence at least or about 50%, at least about 55%, at least or about
60%, at least or about
70%, at least or about 75%, at least or about 80%, at least or about 81%, at
least or about 82%,
at least or about 83%, at least or about 84%, at least or about 85%, at least
or about 86%, at
least or about 87%, at least or about 88%, at least or about 89%, at least or
about 90%, at least
or about 91%, at least or about 92%, at least or about 93%, at least or about
94%, at least or
about 95%, at least or about 96%, at least or about 97%, at least or about
98%, at least or about
99%, or about 100%, identical to SEQ ID NO: 10 (Figures 3 and 6):
Anti-CD33 Heavy Chain variable region (VH) (SEQ ID NO: 10):
QVQLQQPGAEVVKPGASVKMSCKASGYTFTSYYIHWIKQTPGQGLEWVGVIYPGND
DISYNQKFQGKATLTADKSSTTAYMQLSSLTSEDSAVYYCAREVRLRYFDVWGQGT
TVTVSSSSSA
28
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
In certain embodiments, the polypeptide that binds a molecule expressed on
natural
killer (NK) cells comprises (or consists essentially of, or consists of) an
amino acid sequence
at least or about 50%, at least about 55%, at least or about 60%, at least or
about 70%, at least
or about 75%, at least or about 80%, at least or about 81%, at least or about
82%, at least or
about 83%, at least or about 84%, at least or about 85%, at least or about
86%, at least or about
87%, at least or about 88%, at least or about 89%, at least or about 90%, at
least or about 91%,
at least or about 92%, at least or about 93%, at least or about 94%, at least
or about 95%, at
least or about 96%, at least or about 97%, at least or about 98%, at least or
about 99%, or about
100%, identical to the amino acid sequence of the full-length, or a fragment,
of wildtype
ULBP1, ULBP2, ULBP3, ULBP4, ULBP5, ULBP6, MICA, MICB, or HCMV UL18
(including human ULBP1, ULBP2, ULBP3, ULBP4, ULBP5, ULBP6, MICA, MICB, or
HCMV UL18), or of the full-length, or a fragment, of the human homolog of
Raetl a, Raetl h,
Raet lc, Raetld, Raetle, H60b, H60c.
In one embodiment, human ULBP1 has a UniProt accession number Q9BZM6. In one
embodiment, an ectodomain human ULBP1 comprises (or consists essentially of,
or consists
of) amino acid residues 27 to 216 of Q9BZM6-1.
In one embodiment, the first engineered heterodimer protein comprises a ULBP1
ectodomain comprising (or consisting essentially of, or consisting of) an
amino acid sequence
at least or about 50%, at least about 55%, at least or about 60%, at least or
about 70%, at least
or about 75%, at least or about 80%, at least or about 81%, at least or about
82%, at least or
about 83%, at least or about 84%, at least or about 85%, at least or about
86%, at least or about
87%, at least or about 88%, at least or about 89%, at least or about 90%, at
least or about 91%,
at least or about 92%, at least or about 93%, at least or about 94%, at least
or about 95%, at
least or about 96%, at least or about 97%, at least or about 98%, at least or
about 99%, or about
100%, identical to SEQ ID NO: 15 (Figures 4 and 7):
ULBP1 ectodomain (27 to 216 aa of Q9BZM6-1) (SEQ ID NO: 15):
WVDTHCLCYDFIITPKSRPEPQWCEVQGLVDERPFLHYDCVNHKAKAFASLGKKVN
VTKTWEE QTETLRDVVDFLKGQLLDIQVENLIPIEPLTLQARMS CEHEAHGHGRGSW
QFLFNGQKFLLFDSNNRKWTALHPGAKKMTEKWEKNRDVTMFFQKISLGDCKMW
LEEFLMYWEQMLDPTKPPSLAPG
In one embodiment, the first engineered heterodimer protein comprises one or
more
non-naturally occurring polypeptide domain comprising 1-5 alpha helices
comprising (or
29
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
consisting essentially of, or consisting of) an amino acid sequence at least
or about 50%, at
least about 55%, at least or about 60%, at least or about 70%, at least or
about 75%, at least or
about 80%, at least or about 81%, at least or about 82%, at least or about
83%, at least or about
84%, at least or about 85%, at least or about 86%, at least or about 87%, at
least or about 88%,
at least or about 89%, at least or about 90%, at least or about 91%, at least
or about 92%, at
least or about 93%, at least or about 94%, at least or about 95%, at least or
about 96%, at least
or about 97%, at least or about 98%, at least or about 99%, or about 100%,
identical to SEQ
ID NO: 12 (Figures 3 and 6).
6DMPa (SEQ ID NO: 12):
GTKEDILERQRKIIERAQEIHRRQQEILEELERIIRKPGSSEEAMKRMLKLLEESLRLLK
ELLELSEE S A QLLYEQR
In one embodiment, the first engineered heterodimer protein comprises one or
more
non-naturally occurring polypeptide domain comprising 1-5 alpha helices
comprising (or
consisting essentially of, or consisting of) an amino acid sequence at least
or about 50%, at
least about 55%, at least or about 60%, at least or about 70%, at least or
about 75%, at least or
about 80%, at least or about 81%, at least or about 82%, at least or about
83%, at least or about
84%, at least or about 85%, at least or about 86%, at least or about 87%, at
least or about 88%,
at least or about 89%, at least or about 90%, at least or about 91%, at least
or about 92%, at
least or about 93%, at least or about 94%, at least or about 95%, at least or
about 96%, at least
or about 97%, at least or about 98%, at least or about 99%, or about 100%,
identical to SEQ
ID NO: 16 (Figures 4 and 7).
6DMPb (SEQ TD NO: 16):
TEKRLLEEAERAHREQKEIIKKAQELHRRLEEIVRQSGSSEEAKKEAKKILEEIRELSK
RSLELLREILYLSQEQKGSLVPR
In one embodiment, the first engineered heterodimer protein comprises one or
more
covalent dimerization IgG2 hinge domains comprising (or consisting essentially
of or
consisting of) an amino acid sequence at least or about 50%, at least about
55%, at least or
about 60%, at least or about 70%, at least or about 75%, at least or about
80%, at least or about
81%, at least or about 82%, at least or about 83%, at least or about 84%, at
least or about 85%,
at least or about 86%, at least or about 87%, at least or about 88%, at least
or about 89%, at
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
least or about 90%, at least or about 91%, at least or about 92%, at least or
about 93%, at least
or about 94%, at least or about 95%, at least or about 96%, at least or about
97%, at least or
about 98%, at least or about 99%, or about 100%, identical to IgG2 hinge:
ERKCCVECPPCP
(SEQ ID NO: 13) (Figures 3, 4, 6, and 7).
In one embodiment, the first engineered heterodimer protein comprises one or
more
covalent dimerization IgG2 Fe domains comprising (or consisting essentially
of, or consisting
of) an amino acid sequence at least or about 50%, at least about 55%, at least
or about 60%, at
least or about 70%, at least or about 75%, at least or about 80%, at least or
about 81%, at least
or about 82%, at least or about 83%, at least or about 84%, at least or about
85%, at least or
about 86%, at least or about 87%, at least or about 88%, at least or about
89%, at least or about
90%, at least or about 91%, at least or about 92%, at least or about 93%, at
least or about 94%,
at least or about 95%, at least or about 96%, at least or about 97%, at least
or about 98%, at
least or about 99%, or about 100%, identical to SEQ ID NO: 19 (Figures 6 and
7).
IgG2 Fe Domain (SEQ ID NO: 19):
APPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNA
KTKPREEQFNS TFRVVS VLTVVHQDWLNGKEYKCKVSNKGLPAPIEKTIS KTKGQPR
EPQ V YTLPPSREEMTKN QV SLTCLV KGFYPSDIS V EWESNGQPENN Y KTTPPMLDSD
GSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
In one embodiment, the first engineered heterodimer protein comprises one or
more
linkers comprising (or consisting essentially of, or consisting of) an amino
acid sequence at
least or about 50%, at least about 55%, at least or about 60%, at least or
about 70%, at least or
about 75%, at least or about 80%, at least or about 81%, at least or about
82%, at least or about
83%, at least or about 84%, at least or about 85%, at least or about 86%, at
least or about 87%,
at least or about 88%, at least or about 89%, at least or about 90%, at least
or about 91%, at
least or about 92%, at least or about 93%, at least or about 94%, at least or
about 95%, at least
or about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about
100%, identical to SEQ ID NO: 11: GGGGSGGGGSGGGGS (Figures 3, 4, 6, and 7).
In further embodiments, the first engineered heterodimer protein comprises a
His6 tag
(HHHHHH; SEQ ID NO: 20).
In certain embodiments, the second engineered heterodimer protein comprises:
(i) a
first polypeptide comprising an antigen-binding fragment that binds a lineage-
specific cell-
surface antigen, a non-naturally occurring polypeptide domain comprising 1-5
alpha helices
31
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
connected by amino acid linkers and a first covalent dimerization domain; and
(ii) a second
polypeptide comprising a polypeptide that binds a molecule expressed on T
cells, a non-
naturally occurring polypeptide domain comprising 1-5 alpha helices connected
by amino acid
linkers, and a second covalent dimerization domain as described herein.
In certain embodiments, antigen-binding fragments include, but are not limited
to, Fab,
F(ab')2, Fab', F(ab)', Fv, a disulfide linked Fv, single chain Fv (scFv),
bivalent scFv (bi-scFv),
trivalent scFv (tri-scFv), Fd, dAb fragment, an isolated CDR, diabodies,
triabodies, tetrabodies,
linear antibodies, single-chain antibody molecules. In some embodiments, scFv
comprises a
heavy chain variable region (VH), and a light chain variable region (VL).
In some embodiments, the non-naturally occurring polypeptide domain comprising
1-
5 alpha helices is 6DMPa (Chen et al. 2019). In some embodiments, the non-
naturally
occurring polypeptide domain comprising 1-5 alpha helices is 6DMPh (Chen et
al. 2019).
In some embodiments, the covalent dimerization domains are IgG2 hinge domains.
In
some embodiments, the covalent dimerization domains are IgG2 domains and IgG2
Fe
domains. In some embodiments, the Fe domains are CH2 and CH3 domains.
The second engineered heterodimer protein can be designed to place the
functional
moieties (an antigen-binding fragment that binds a lineage-specific cell-
surface antigen, and a
polypeptide that binds a molecule expressed on T cells) in any order. In
certain embodiments,
the antigen-binding fragment that binds a lineage-specific cell-surface
antigen is located at the
N-terminus or C-terminus of the fusion polypeptide. In certain embodiments,
the polypeptide
that binds a molecule expressed on T cells is located at the C-terminus or N-
terminus of the
fusion polypeptide.
In some embodiments, the second engineered heterodimer protein comprises, from
N-
terminus to C-terminus, a scFv that binds to the lineage-specific cell-surface
antigen (e.g.,
CD33 or CD19), and a polypeptide that binds a molecule expressed on T cells
(e.g., an anti-
CD3 monoclonal antibody). In some embodiments, the fusion polypeptide
comprises, from N
terminus to C terminus, a polypeptide that binds a molecule expressed on T
cells (e.g., an anti-
CD3 mono clonal antibody). and a scFv that binds to the lineage-specific cell-
surface antigen
(e.g., CD33 or CD19).
The second engineered heterodimer protein may further comprise a signal
sequence,
and/or one or more linkers. In the second engineered heterodimer, these
functional moieties
may be covalently ligated continuously or non-continuously (e.g., they may be
separated by
linkers). The linker may have up to 50, up to 40, up to 30, up to 20, up to
18, up to 15, up to
12, up to 11, or up to 10, amino acid residues in length. In certain
embodiments, the linker has
32
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10-20, 8-10, 8-12, 8-15, 8-20, or 8-30 amino
acid residues in
length. In certain embodiments, the linker has about 7-10, 7-12, 7-15, 7-20,
or 7-30 amino acid
residues in length.
One type of derivatized protein is produced by crosslinking two or more
polypeptides
(of the same type or of different types). Suitable crosslinkers include those
that are
heterobifunctional, having two distinct reactive groups separated by an
appropriate spacer (e.g.,
m-maleimidobenzoyl-N-hydroxysuccinimide ester) or homobifunctional (e.g.,
disuccinimidyl
suberate). Useful detectable agents with which a protein can be derivatized
(or labeled) include
fluorescent agents, various enzymes, prosthetic groups, luminescent materials,
bioluminescent
materials, and radioactive materials. Non-limiting, exemplary fluorescent
detectable agents
include fluorescein, fluorescein isothiocyanate, rhodamine, and,
phycoerythrin. A polypeptide
can also be derivatized with detectable enzymes, such as alkaline phosphatase,
horseradish
peroxidase, beta-galactosidase, acetylcholinesterase, glucose oxidase and the
like. A
polypeptide can also be derivatized with a prosthetic group (e.g.,
streptavidin/biotin and
avidin/biotin).
The second engineered heterodimer protein can be derivatized or linked to
another
functional molecule. For example, second engineered heterodimer protein can be
functionally
linked (by chemical coupling, genetic fusion, noncovalent interaction, etc.)
to one or more other
molecular entities, such as an antibody or antibody fragment, a detectable
agent, an
immunosuppressant, a cytotoxic agent, a pharmaceutical agent, a protein or
peptide that can
mediate association with another molecule (such as a streptavidin core region
or a polyhistidine
tag), amino acid linkers, signal sequences, immunogenic carriers, or ligands
useful in protein
purification, such as glutathione-S-transferase, histidine tag, and
staphylococcal protein A.
Cytotoxic agents may include radioactive isotopes, chemotherapeutic agents,
and toxins such
as enzymatically active toxins of bacterial, fungal, plant, or animal origin,
and fragments
thereof.
The second engineered heterodimer protein may further comprise a fragment
(e.g., a
tag) useful for polypeptide production and/or detection, including, but not
limited to, poly-
histidine (e.g., six histidine residues), a maltose binding protein, GST,
green fluorescent protein
(GFP), hemagglutinin, or alkaline phosphatase, secretion signal peptides
(e.g., preprotyrypsin
signal sequence). Myc, and/or FLAG.
In one embodiment, the second engineered heterodimer protein comprises (or
consists
essentially of, or consists of) an amino acid sequence at least or about 50%,
at least about 55%,
at least or about 60%, at least or about 70%, at least or about 75%, at least
or about 80%, at
33
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
least or about 81%, at least or about 82%, at least or about 83%, at least or
about 84%, at least
or about 85%, at least or about 86%, at least or about 87%, at least or about
88%, at least or
about 89%, at least or about 90%, at least or about 91%, at least or about
92%, at least or about
93%, at least or about 94%, at least or about 95%, at least or about 96%, at
least or about 97%,
at least or about 98%, at least or about 99%, or about 100%, identical to SEQ
Ill NO: 1 (Figure
3) and SEQ ID NO: 3 (Figure 5) or SEQ ID NO: 4 (Figure 6) and SEQ ID NO: 6
(Figure 8).
In one embodiment, the second engineered heterodimer protein comprises a
signal
sequence comprising (or consisting essentially of, or consisting of) an amino
acid sequence at
least or about 50%, at least about 55%, at least or about 60%, at least or
about 70%, at least or
about 75%, at least or about 80%, at least or about 81%, at least or about
82%, at least or about
83%, at least or about 84%, at least or about 85%, at least or about 86%, at
least or about 87%,
at least or about 88%, at least or about 89%, at least or about 90%, at least
or about 91%, at
least or about 92%, at least or about 93%, at least or about 94%, at least or
about 95%, at least
or about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about
100%, identical to an IL2 secretory signal sequence: MYRMQLLSCIALSLALVTNS (SEQ
ID NO:7) (Figures 3, 5, 6 and 8).
In one embodiment, the second engineered heterodimer protein comprises one or
more
tags comprising (or consisting essentially of, or consisting of) an amino acid
sequence at least
or about 50%, at least about 55%, at least or about 60%, at least or about
70%, at least or about
75%, at least or about 80%, at least or about 81%, at least or about 82%, at
least or about 83%,
at least or about 84%, at least or about 85%, at least or about 86%, at least
or about 87%, at
least or about 88%, at least or about 89%, at least or about 90%, at least or
about 91%, at least
or about 92%, at least or about 93%, at least or about 94%, at least or about
95%, at least or
about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about 100%,
identical to Myc: EQKLISEEDL (SEQ ID NO: 8) (Figures 3 and 6).
In one embodiment, the second engineered heterodimer protein comprises one or
more
tags comprising (or consisting essentially of, or consisting of) an amino acid
sequence at least
or about 50%, at least about 55%, at least or about 60%, at least or about
70%, at least or about
75%, at least or about 80%, at least or about 81%, at least or about 82%, at
least or about 83%,
at least or about 84%, at least or about 85%, at least or about 86%, at least
or about 87%, at
least or about 88%, at least or about 89%, at least or about 90%, at least or
about 91%, at least
or about 92%, at least or about 93%, at least or about 94%, at least or about
95%, at least or
about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about 100%,
identical to FLAG: DYKDDDDK (SEQ ID NO: 14) (Figures 5 and 8)
34
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
In one embodiment, the second engineered heterodimer protein comprises an anti-
CD33 scFv comprising (or consisting essentially of, or consisting of) an amino
acid sequence
at least or about 50%, at least about 55%, at least or about 60%, at least or
about 70%, at least
or about 75%, at least or about 80%, at least or about 81%, at least or about
82%, at least or
about 83%, at least or about 84%, at least or about 85%, at least or about
86%, at least or about
87%, at least or about 88%, at least or about 89%, at least or about 90%, at
least or about 91%,
at least or about 92%, at least or about 93%, at least or about 94%, at least
or about 95%, at
least or about 96%, at least or about 97%, at least or about 98%, at least or
about 99%, or about
100%, identical to the anti-CD33 scFv disclosed in U.S. Patent Publication No.
20130078241.
In one embodiment, the second engineered heterodimer protein comprises an
antigen-
binding fragment that binds CD33, where the antigen-binding fragment comprises
a light chain
variable region (VI) comprising (or consisting essentially of, or consisting
of) an amino acid
sequence at least or about 50%, at least about 55%, at least or about 60%, at
least or about 70%,
at least or about 75%, at least or about 80%, at least or about 81%, at least
or about 82%, at
least or about 83%, at least or about 84%, at least or about 85%, at least or
about 86%, at least
or about 87%, at least or about 88%, at least or about 89%, at least or about
90%, at least or
about 91%, at least or about 92%, at least or about 93%, at least or about
94%, at least or about
95%, at least or about 96%, at least or about 97%, at least or about 98%, at
least or about 99%,
or about 100%, identical to SEQ ID NO: 9 (Figures 3 and 6).
Anti-CD33 Light Chain variable region (VL) (SEQ ID NO: 9):
EIVLTQSPGSLAVSPGERVTMSCKSSQSVFFSSSQKNYLAWYQQIPGQSPRLLIYWAS
TRESGVPDRFTGSGSGTDFTLTISSVQPEDLAIYYCHQYLSSRTFGQGTKLEIKR
In one embodiment, the second engineered heterodimer protein comprises an
antigen-
binding fragment that binds CD33, where the antigen-binding fragment comprises
a heavy
chain variable region (VH) comprising (or consisting essentially of, at
consisting of) an amino
acid sequence at least or about 50%, at least about 55%, at least or about
60%, at least or about
70%, at least or about 75%, at least or about 80%, at least or about 81%, at
least or about 82%,
at least or about 83%, at least or about 84%, at least or about 85%, at least
or about 86%, at
least or about 87%, at least or about 88%. at least or about 89%, at least or
about 90%, at least
or about 91%, at least or about 92%, at least or about 93%, at least or about
94%, at least or
about 95%, at least or about 96%, at least or about 97%, at least or about
98%, at least or about
99%, or about 100%, identical to SEQ ID NO: 10 (Figures 3 and 6):
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
Anti-CD33 Heavy Chain variable region (VH) (SEQ ID NO: 10):
QVQLQQPGAEVVKPGASVKMSCKASGYTFTSYYTHWIKQTPGQGLEWVGVIYPGND
DISYNQKFQGKATLTADKSSTTAYMQLSSLTSEDSAVYYCAREVRLRYFDVWGQGT
TVTVSSSSSA
In certain embodiments, the polypeptide that binds a molecule expressed on T
cells
comprises (or consists essentially of, or consists of) an amino acid sequence
at least or about
50%, at least about 55%, at least or about 60%, at least or about 70%, at
least or about 75%, at
least or about 80%, at least or about 81%, at least or about 82%, at least or
about 83%, at least
or about 84%, at least or about 85%, at least or about 86%, at least or about
87%, at least or
about 88%, at least or about 89%, at least or about 90%, at least or about
91%, at least or about
92%, at least or about 93%, at least or about 94%, at least or about 95%, at
least or about 96%,
at least or about 97%, at least or about 98%, at least or about 99%, or about
100%, identical to
the amino acid sequence of SEQ ID NOs: 17 and 18.
In one embodiment, the second engineered heterodimer protein comprises an
antigen-
binding fragment that binds CD3, where the antigen-binding fragment comprises
a heavy chain
variable region (VH) comprising (or consisting essentially of, or consisting
of) an amino acid
sequence at least or about 50%, at least about 55%, at least or about 60%, at
least or about 70%,
at least or about 75%, at least or about 80%, at least or about 81%, at least
or about 82%, at
least or about 83%, at least or about 84%, at least or about 85%, at least or
about 86%, at least
or about 87%, at least or about 88%, at least or about 89%, at least or about
90%, at least or
about 91%, at least or about 92%, at least or about 93%, at least or about
94%, at least or about
95%, at least or about 96%, at least or about 97%, at least or about 98%, at
least or about 99%,
or about 100%, identical to SEQ ID NO: 17 (Figures 5 and 8):
Anti-CD3 Heavy Chain variable region (VH) (SEQ ID NO: 17):
DIKLQQSGAELARPGASVKMSCKTSGYTFTRYTMHWVKQRPGQGLGLEWIGYINPS
RGYTNYNQKFKDKATLTTD KSSSTAYMQLSSLTSEDSAVYYCARYYDDHYCLDYW
GQGTTLTVSS
In one embodiment, the second engineered heterodimer protein comprises an
antigen-
binding fragment that binds CD3, where the antigen-binding fragment comprises
a light chain
variable region (VL) comprising (or consisting essentially of, or consisting
of) an amino acid
36
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
sequence at least or about 50%, at least about 55%, at least or about 60%, at
least or about 70%,
at least or about 75%, at least or about 80%, at least or about 81%, at least
or about 82%, at
least or about 83%, at least or about 84%, at least or about 85%, at least or
about 86%, at least
or about 87%, at least or about 88%, at least or about 89%, at least or about
90%, at least or
about 91%, at least or about 92%, at least or about 93%, at least or about
94%, at least or about
95%, at least or about 96%, at least or about 97%, at least or about 98%, at
least or about 99%,
or about 100%, identical to SEQ ID NO: 18 (Figures 5 and 8):
Anti-CD3 Light Chain variable region (VII) (SEQ ID NO: 18):
DIQLTQSPAIMSASPGGKVTMTCRASSSVSYMNWYQQKSGTSPKRWIYDTSKVASG
VPYRFTSYSLTISSMEAEDAATYYCQQWSSNPLTFGAGTKLELK
In one embodiment, the second engineered heterodimer protein comprises one or
more
non-naturally occurring pol ypepti de domain comprising 1-5 alpha helices
comprising (or
consisting essentially of, or consisting of) an amino acid sequence at least
or about 50%, at
least about 55%, at least or about 60%, at least or about 70%, at least or
about 75%, at least or
about 80%, at least or about 81%, at least or about 82%, at least or about
83%, at least or about
84%, at least or about 85%, at least or about 86%, at least or about 87%, at
least or about 88%,
at least or about 89%, at least or about 90%, at least or about 91%, at least
or about 92%, at
least or about 93%, at least or about 94%, at least or about 95%, at least or
about 96%, at least
or about 97%, at least or about 98%, at least or about 99%, or about 100%,
identical to SEQ
ID NO: 12 (Figures 3 and 6).
6DMPa (SEQ ID NO: 12):
GTKEDILERQRKIIER A QEIHRR QQEILEELERIIRKPGSSEEAMKRMLKLLEESLRLLK
ELLELSEESAQLLYEQR
In one embodiment, the second engineered heterodimer protein comprises one or
more
non-naturally occurring polypeptide domain comprising 1-5 alpha helices
comprising (or
consisting essentially of, or consisting of) an amino acid sequence at least
or about 50%, at
least about 55%, at least or about 60%, at least or about 70%, at least or
about 75%, at least or
about 80%, at least or about 81%, at least or about 82%, at least or about
83%, at least or about
84%, at least or about 85%, at least or about 86%, at least or about 87%, at
least or about 88%,
at least or about 89%, at least or about 90%, at least or about 91%, at least
or about 92%, at
37
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
least or about 93%, at least or about 94%, at least or about 95%, at least or
about 96%, at least
or about 97%, at least or about 98%, at least or about 99%, or about 100%,
identical to SEQ
ID NO: 16 (Figures 5 and 8).
6DMPb (SEQ Ill NO: 16):
TEKRLLEEAERAHREQKEIIKKAQELHRRLEEIVRQSGSSEEAKKEAKKILEEIRELSK
RS LELLREILYLS QEQ KGSLVPR
In one embodiment, the second engineered heterodimer protein comprises one or
more
covalent dimerization IgG2 hinge domains comprising (or consisting essentially
of, or
consisting of) an amino acid sequence at least or about 50%, at least about
55%, at least or
about 60%, at least or about 70%, at least or about 75%, at least or about
80%, at least or about
81%, at least or about 82%, at least or about 83%, at least or about 84%, at
least or about 85%,
at least or about 86%, at least or about 87%, at least or about 88%, at least
or about 89%, at
least or about 90%, at least or about 91%, at least or about 92%, at least or
about 93%, at least
or about 94%, at least or about 95%, at least or about 96%, at least or about
97%, at least or
about 98%, at least or about 99%, or about 100%, identical to IgG2 hinge:
ERKCCVECPPCP
(SEQ Ill NO: 13) (Figures 3, 5, 6 and 8).
In one embodiment, the second engineered heterodimer protein comprises one or
more
covalent dimerization IgG2 Fc domains comprising (or consisting essentially
of, or consisting
of) an amino acid sequence at least or about 50%, at least about 55%, at least
or about 60%, at
least or about 70%, at least or about 75%, at least or about 80%, at least or
about 81%, at least
or about 82%, at least or about 83%, at least or about 84%, at least or about
85%, at least or
about 86%, at least or about 87%, at least or about 88%, at least or about
89%, at least or about
90%, at least or about 91%, at least or about 92%, at least or about 93%, at
least or about 94%,
at least or about 95%, at least or about 96%, at least or about 97%, at least
or about 98%, at
least or about 99%, or about 100%, identical to SEQ ID NO: 19. (Figures 6 and
8).
IgG2 Fe Domain (SEQ ID NO: 19):
APPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNA
KTKPREEQFNS TFRVVS VLTVVHQDWLNGKEYKCKVSNKGLPAPIEKTIS KTKGQPR
EPQVYTLPPSREEMTKNQVSLTC LVKGFYPSDIS VEWESNGQPENNYKTTPPMLDSD
GSFFLYSKLTVDKSRWQQGNVFSCSVMHEALIINHYTQKSLSLSPGK
38
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
In one embodiment, the second engineered heterodimer protein comprises one or
more
linkers comprising (or consisting essentially of, or consisting of) an amino
acid sequence at
least or about 50%, at least about 55%, at least or about 60%, at least or
about 70%, at least or
about 75%, at least or about 80%, at least or about 81%, at least or about
82%, at least or about
83%, at least or about 84%, at least or about 85%, at least or about 86%, at
least or about 87%,
at least or about 88%, at least or about 89%, at least or about 90%, at least
or about 91%, at
least or about 92%, at least or about 93%, at least or about 94%, at least or
about 95%, at least
or about 966/9, at least or about 97%, at least or about 98%, at least or
about 99%, or about
100%, identical to SEQ ID NO: 11: GGGGSGGGGSGGGGS (Figures 3-8) or to SEQ ID
NO: 21: GGSGGSGGSGGSGG (Figures 5 and 8, CD3 VH-VL linker) or to SEQ ID NO:
22:
SGSGSG (Figures 5 and 8, linker within in CD3-VL)
In further embodiments, the second engineered heterodimer protein comprises a
His6
tag (HHHHHH; SEQ ID NO: 20).
In certain embodiments, the disclosure provides for a third engineered
heterodimer
protein which comprises: (i) a first polypeptide comprising an antigen-binding
fragment that
binds a lineage-specific cell-surface antigen, a non-naturally occurring
polypeptide domain
comprising 1-5 alpha helices connected by amino acid linkers and a first
covalent dimerization
domain; and (ii) a second polypeptide comprising a polypeptide that binds a
molecule
expressed on natural killer (NK) cells, a non- naturally occurring polypeptide
domain
comprising 1-5 alpha helices connected by amino acid linkers, and a second
covalent
dimerization domain as described herein. See Figure 23.
In certain embodiments, antigen-binding fragments include, but are not limited
to, Fab,
F(ab')2, Fab', F(ab)', Fv, a disulfide linked Fv, single chain Fv (scFv),
bivalent scFv (bi-scFv),
trivalent scFv (tri-scFv), Fd, dAb fragment, an isolated CDR, diabodies,
triabodies, tetrabodies,
linear antibodies, single-chain antibody molecules. In some embodiments, scFv
comprises a
heavy chain variable region (Vn), and a light chain variable region (VL).
In some embodiments, the non-naturally occurring polypeptide domain comprising
1-
5 alpha helices is 6DMPa (Chen et al. 2019). In some embodiments, the non-
naturally
occurring polypeptide domain comprising 1-5 alpha helices is 6DMPb (Chen et
al. 2019).
In some embodiments, the covalent dimerization domains are IgG2 hinge domains.
In
some embodiments, the covalent dimerization domains are IgG2 domains and IgG2
Fc
domains. In some embodiments, the Fe domains are CH2 and CH3 domains.
The third engineered heterodimer protein can be designed to place the
functional
moieties (an antigen-binding fragment that binds a lineage-specific cell-
surface antigen, and a
39
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
polypeptide that binds a molecule expressed on NK cells) in any order. In
certain embodiments,
the antigen-binding fragment that binds a lineage-specific cell-surface
antigen is located at the
N-terminus or C-terminus of the fusion polypeptide. In certain embodiments,
the polypeptide
that binds a molecule expressed on natural killer (NK) cells is located at the
C-terminus or N-
S terminus of the fusion polypeptide.
In some embodiments, the third engineered heterodimer protein comprises, from
N-
terminus to C-terminus, a scFv that binds to the lineage-specific cell-surface
antigen (e.g.,
CD33 or CD19), and a polypeptide that binds a molecule expressed on NK cells
(e.g., an anti-
CD16 monoclonal antibody). In some embodiments, the fusion polypeptide
comprises, from
N terminus to C terminus, a polypeptide that binds a molecule expressed on NK
cells (e.g., an
anti-CD16 monoclonal antibody) and a scFv that binds to the lineage-specific
cell-surface
antigen (e.g., CD33 or CD] 9).
The third engineered heterodimer protein may further comprise a signal
sequence,
and/or one or more linkers. In the fusion polypeptide, these functional
moieties may be
covalently ligated continuously or non-continuously (e.g., they may be
separated by linkers).
The linker may have up to 50, up to 40, up to 30, up to 20, up to 18, up to
15, up to 12, up to
11, or up to 10, amino acid residues in length. In certain embodiments, the
linker has about 1,
2, 3, 4, 5, 6, 7, 8, 9, 10-20, 8-10, 8-12, 8-15, 8-20, or 8-30 amino acid
residues in length. In
certain embodiments, the linker has about 7-10, 7-12, 7-15, 7-20, or 7-30
amino acid residues
in length.
One type of derivatized protein is produced by crosslinking two or more
polypeptides
(of the same type or of different types). Suitable crosslinkers include those
that are
heterobifunctional, having two distinct reactive groups separated by an
appropriate spacer (e.g.,
m-maleimidobenzoyl-N-hydroxysuccinimide ester) or homobifunctional (e.g.,
disuccinimidyl
suberate). Useful detectable agents with which a protein can be derivatized
(or labeled) include
fluorescent agents, various enzymes, prosthetic groups, luminescent materials,
bioluminescent
materials, and radioactive materials. Non-limiting, exemplary fluorescent
detectable agents
include fluorescein, fluorescein isothiocyanate, rhodamine, and phycoerythrin.
A polypeptide
can also be derivatized with detectable enzymes, such as alkaline phosphatase,
horseradish
peroxidase, beta-galactosidase, acetylcholinesterase, glucose oxidase and the
like. A
polypeptide can also be derivatized with a prosthetic group (e.g.,
streptavidin/biotin and
avidin/biotin).
The third engineered heterodimer protein can be derivatized or linked to
another
functional molecule. For example, the third engineered heterodimer protein can
be functionally
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
linked (by chemical coupling, genetic fusion, noncovalent interaction) to one
or more other
molecular entities, such as an antibody or antibody fragment, a detectable
agent, an
immunosuppressant, a cytotoxic agent, a pharmaceutical agent, a protein or
peptide that can
mediate association with another molecule (such as a streptavidin core region
or a polyhistidine
tag), amino acid linkers, signal sequences, immunogenic carriers, or ligands
useful in protein
purification, such as glutathione-S-transferase, histidine tag, and
staphylococcal protein A.
Cytotoxic agents may include radioactive isotopes, chemotherapeutic agents,
and toxins such
as enzymatically active toxins of bacterial, fungal, plant, or animal origin,
and fragments
thereof.
The third engineered heterodimer protein may further comprise a fragment
(e.g., a tag)
useful for polypeptide production and/or detection, including, but not limited
to, poly-histidine
(e.g., six histidine residues), a maltose binding protein, GST, green
fluorescent protein (GFP),
hemagglutinin, or alkaline phosphatase, secretion signal peptides
preprotyrypsin signal
sequence), Myc, and/or FLAG.
In one embodiment, the third engineered heterodimer protein comprises (or
consists
essentially of, or consists of) an amino acid sequence at least or about
500/c, at least about 55%,
at least or about 60%, at least or about 70%, at least or about 75%, at least
or about 80%, at
least or about 81%, at least or about 82%, at least or about 83%, at least or
about 84%, at least
or about 85%, at least or about 86%, at least or about 87%, at least or about
88%, at least or
about 89%, at least or about 90%, at least or about 91%, at least or about
92%, at least or about
93%, at least or about 94%, at least or about 95%, at least or about 96%, at
least or about 97%,
at least or about 98%, at least or about 99%, or about 100%, identical to SEQ
ID NO: 1 (Figure
3) and SEQ ID NO: 26 (Figure 23A) or SEQ ID NO: 4 (Figure 6) and SEQ ID NO: 27
(Figure
23B).
Tn one embodiment, the third engineered heterodimer protein comprises a signal
sequence comprising (or consisting essentially of, or consisting of) an amino
acid sequence at
least or about 50%, at least about 55%, at least or about 60%, at least or
about 70%, at least or
about 75%, at least or about 80%, at least or about 81%, at least or about
82%, at least or about
83%, at least or about 84%, at least or about 85%, at least or about 86%, at
least or about 87%,
at least or about 88%, at least or about 89%, at least or about 90%, at least
or about 91%, at
least or about 92%, at least or about 93%, at least or about 94%, at least or
about 95%, at least
or about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about
100%, identical to an IL2 secretory signal sequence: MYRMQLLSCIALSLALVTNS (SEQ
ID NO:7) (Figures 3, 4, and 23).
41
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
In one embodiment, the third engineered heterodimer protein comprises one or
more
tags comprising (or consisting essentially of, or consisting of) an amino acid
sequence at least
or about 50%, at least about 55%, at least or about 60%, at least or about
70%, at least or about
75%, at least or about 80%, at least or about 81%, at least or about 82%, at
least or about 83%,
at least or about 84%, at least or about 85%, at least or about 86%, at least
or about 87%, at
least or about 88%, at least or about 89%, at least or about 90%, at least or
about 91%, at least
or about 92%, at least or about 93%, at least or about 94%, at least or about
95%, at least or
about 96%, at least or about 97%, at least or about 98%, at least or about
990/c, or about 100%,
identical to Myc: EQKLISEEDL (SEQ ID NO: 8) (Figures 3 and 6).
In one embodiment, the third engineered heterodimer protein comprises one or
more
tags comprising (or consisting essentially of, or consisting of) an amino acid
sequence at least
or about 50%, at least about 55%, at least or about 60%, at least or about
70%, at least or about
75%, at least or about 80%, at least or about 81%, at least or about 82%, at
least or about 83%,
at least or about 84%, at least or about 85%, at least or about 86%, at least
or about 87%, at
least or about 88%, at least or about 89%, at least or about 90%, at least or
about 91%, at least
or about 92%, at least or about 93%, at least or about 94%, at least or about
95%, at least or
about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about 100%,
identical to FLAG: DYKDDDDK (SEQ Ill NO: 14) (Figure 23)
In one embodiment, the third engineered heterodimer protein comprises an anti-
CD33
scFv comprising (or consisting essentially of, or consisting of) an amino acid
sequence at least
or about 50%, at least about 55%, at least or about 60%, at least or about
70%, at least or about
75%, at least or about 80%, at least or about 81%, at least or about 82%, at
least or about 83%,
at least or about 84%, at least or about 85%, at least or about 86%, at least
or about 87%, at
least or about 88%, at least or about 89%, at least or about 90%, at least or
about 91%, at least
or about 92%, at least or about 93%, at least or about 94%, at least or about
95%, at least or
about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about 100%,
identical to the anti-CD33 scFv disclosed in U.S. Patent Publication No.
20130078241.
In one embodiment, the third engineered heterodimer protein comprises an
antigen-
binding fragment that binds CD33, where the antigen-binding fragment comprises
a light chain
variable region (VL) comprising (or consisting essentially of, or consisting
of) an amino acid
sequence at least or about 50%, at least about 55%, at least or about 60%, at
least or about 70%,
at least or about 75%, at least or about 80%, at least or about 81%, at least
or about 82%, at
least or about 83%, at least or about 84%, at least or about 85%, at least or
about 86%, at least
or about 87%, at least or about 88%, at least or about 89%, at least or about
90%, at least or
42
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
about 91%, at least or about 92%, at least or about 93%, at least or about
94%, at least or about
95%, at least or about 96%, at least or about 97%, at least or about 98%, at
least or about 99%,
or about 100%, identical to SEQ ID NO: 9 (Figures 3 and 6).
Anti-CD33 Light Chain variable region (VL) (SEQ Ill NO: 9):
EIVLIQSPGSLAVSPGERVTMSCKSSQSVFFSSSQKNYLAWYQQIPGQSPRLLIYWAS
TRESGVPDRFTGSGSGTDFTLTISSVQPEDLAIYYCHQYLSSRTFGQGTKLEIKR
In one embodiment, the third engineered heterodimer protein comprises an
antigen-
binding fragment that binds CD33, where the antigen-binding fragment comprises
a heavy
chain variable region (Vri) comprising (or consisting essentially of, or
consisting of) an amino
acid sequence at least or about 50%, at least about 55%, at least or about
60%, at least or about
70%, at least or about 75%, at least or about 80%, at least or about 81%, at
least or about 82%,
at least or about 83%, at least or about 84%, at least or about 85%, at least
or about 86%, at
least or about 87%, at least or about 88%, at least or about 89%, at least or
about 90%, at least
or about 91%, at least or about 92%, at least or about 93%, at least or about
94%, at least or
about 95%, at least or about 96%, at least or about 97%, at least or about
98%, at least or about
99%, or about 100%, identical to SEQ Ill NO: 10 (Figures 3 and 6):
Anti-CD33 Heavy Chain variable region (VI)) (SEQ ID NO: 10):
QVQLQQPGAEVVKPGASVKMSCKASGYTFTSYYTHWIKQTPGQGLEWVGVIYPGND
DISYNQKFQGKATLTADKSSTTAYMQLSSLTSEDSAVYYCAREVRLRYFDVWGQGT
TVTVSSSSSA
In certain embodiments, the polypeptide that hinds a molecule expressed on NK
cells
comprises (or consists essentially of, or consists of) an amino acid sequence
at least or about
50%, at least about 55%, at least or about 60%, at least or about 70%, at
least or about 75%, at
least or about 80%, at least or about 81%, at least or about 82%, at least or
about 83%, at least
or about 84%, at least or about 85%, at least or about 86%, at least or about
87%, at least or
about 88%, at least or about 89%, at least or about 90%, at least or about
91%, at least or about
92%, at least or about 93%, at least or about 94%, at least or about 95%, at
least or about 96%,
at least or about 97%, at least or about 98%, at least or about 99%, or about
100%, identical to
the amino acid sequence of SEQ ID NOs: 28 and 29.
43
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
In one embodiment, the second engineered heterodimer protein comprises an
antigen-
binding fragment that binds CD16, where the antigen-binding fragment comprises
a heavy
chain variable region (VII) comprising (or consisting essentially of, or
consisting of) an amino
acid sequence at least or about 50%, at least about 55%, at least or about
60%, at least or about
70%, at least or about 75%, at least or about 80%, at least or about 81%, at
least or about 82%,
at least or about 83%, at least or about 84%, at least or about 85%, at least
or about 86%, at
least or about 87%, at least or about 88%, at least or about 89%, at least or
about 90%, at least
or about 91%, at least or about 92%, at least or about 93%, at least or about
94%, at least or
about 95%, at least or about 96%, at least or about 97%, at least or about
98%, at least or about
99%, or about 100%, identical to SEQ ID NO: 28 (Figure 23):
Anti-CD16 Heavy Chain variable region (VI)) (SEQ ID NO: 28):
EVQLVESGGGVVRPGGSLRLSCAASGFTFDDYGMSWVRQAPGKGLEWVSGINWNG
GSTGYADSVKGRFTISRDNAKNSLYLQMNSLR AEDT AVYYCARGRSLLFDYWGQG
TLVTVSR
In one embodiment, the third engineered heterodimer protein comprises an
antigen-
binding fragment that binds CD16, where the antigen-binding fragment comprises
a light chain
variable region (VL) comprising (or consisting essentially of, or consisting
of) an amino acid
sequence at least or about 50%, at least about 55%, at least or about 60%, at
least or about 70%,
at least or about 75%, at least or about 80%, at least or about 81%, at least
or about 82%, at
least or about 83%, at least or about 84%, at least or about 85%, at least or
about 86%, at least
or about 87%, at least or about 88%, at least or about 89%, at least or about
90%, at least or
about 91%, at least or about 92%, at least or about 93%, at least or about
94%, at least or about
95%, at least or about 96%, at least or about 97%, at least or about 98%, at
least or about 99%,
or about 100%, identical to SEQ ID NO: 29 (Figure 23).
Anti-CD16 Light Chain variable region (Vii) (SEQ ID NO: 29):
SSELTQDPAVSVALGQTVRITCQGDSLRSYYASWYQQKPGQAPVLVIYGKNNRPSGI
PDRFSGSSSGNTASLTITGAQAEDEADYYCNSRDSSGNHVVFGGGTKLTVG
In one embodiment, the third engineered heterodimer protein comprises one or
more
non-naturally occurring polypeptide domain comprising 1-5 alpha helices
comprising (or
consisting essentially of, or consisting of) an amino acid sequence at least
or about 50%, at
44
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
least about 55%, at least or about 60%, at least or about 70%, at least or
about 75%, at least or
about 80%, at least or about 81%, at least or about 82%, at least or about
83%, at least or about
84%, at least or about 85%, at least or about 86%, at least or about 87%, at
least or about 88%,
at least or about 89%, at least or about 90%, at least or about 91%, at least
or about 92%, at
least or about 93%, at least or about 94%, at least or about 95%, at least or
about 96%, at least
or about 97%, at least or about 98%, at least or about 99%, or about 100%,
identical to SEQ
ID NO: 12 (Figures 3 and 6).
6DMPa (SEQ ID NO: 12):
GTKEDILERQRKIIERAQEIHRRQQEILEELERIIRKPGSSEEAMKRMLKLLEESLRLLK
ELLELSEESAQLLYEQR
In one embodiment, the third engineered heterodimer protein comprises one or
more
non-naturally occurring pol ypepti de domain comprising 1-5 alpha helices
comprising (or
consisting essentially of, or consisting of) an amino acid sequence at least
or about 50%, at
least about 55%, at least or about 60%, at least or about 70%, at least or
about 75%, at least or
about 80%, at least or about 81%, at least or about 82%, at least or about
83%, at least or about
84%, at least or about 85%, at least or about 86%, at least or about 87%, at
least or about 88%,
at least or about 89%, at least or about 90%, at least or about 91%, at least
or about 92%, at
least or about 93%, at least or about 94%, at least or about 95%, at least or
about 96%, at least
or about 97%, at least or about 98%, at least or about 99%, or about 100%,
identical to SEQ
ID NO: 16 (Figure 23).
6DMPb (SEQ ID NO: 16):
TEKRLLEEAER AHREQKEIIKK A QELHRRLEEIVR QSGS SEEA KKEA KKTLEETRELSK
RSLELLREILYLSQEQKGSLVPR
In one embodiment, the third engineered heterodimer protein comprises one or
more
covalent dimerization IgG2 hinge domains comprising (or consisting essentially
of. or
consisting of) an amino acid sequence at least or about 50%, at least about
55%, at least or
about 60%, at least or about 70%, at least or about 75%, at least or about
80%, at least or about
81%, at least or about 82%, at least or about 83%, at least or about 84%, at
least or about 85%,
at least or about 86%, at least or about 87%, at least or about 88%, at least
or about 89%, at
least or about 90%, at least or about 91%, at least or about 92%, at least or
about 93%, at least
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
or about 94%, at least or about 95%, at least or about 96%, at least or about
97%, at least or
about 98%, at least or about 99%, or about 100%, identical to IgG2 hinge:
ERKCCVECPPCP
(SEQ ID NO: 13) (Figures 3 and 23).
In one embodiment, the third engineered heterodimer protein comprises one or
more
covalent dimerization IgG2 Fc domains comprising (or consisting essentially
of, or consisting
of) an amino acid sequence at least or about 50%, at least about 55%, at least
or about 60%, at
least or about 70%, at least or about 75%, at least or about 80%, at least or
about 81%, at least
or about 82%, at least or about 83%, at least or about 84%, at least or about
85%, at least or
about 86%, at least or about 87%, at least or about 88%, at least or about
89%, at least or about
90%, at least or about 91%, at least or about 92%, at least or about 93%, at
least or about 94%,
at least or about 95%, at least or about 96%, at least or about 97%, at least
or about 98%, at
least or about 99%, or about 100%, identical to SEQ ID NO: 19 (Figures 6 and
23).
IgG2 Fc Domain (SEQ ID NO: 19):
APPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNA
KTKPREEQFNS TFRVVS VLTVVHQDWLNGKEYKCKVSNKGLPAPIEKTIS KTKGQPR
EPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIS VEWE SNGQPENNYKTTPPMLD SD
GSEELYSKLTVDKSRWQQGN VFSCSVMHEALHNHYTQKSLSLSPGK
In one embodiment, the third engineered heterodimer protein comprises one or
more
linkers comprising (or consisting essentially of, or consisting of) an amino
acid sequence at
least or about 50%, at least about 55%, at least or about 60%, at least or
about 70%, at least or
about 75%, at least or about 80%, at least or about 81%, at least or about
82%, at least or about
83%, at least or about 84%, at least or about 85%, at least or about 86%, at
least or about 87%,
at least or about 88%, at least or about 89%, at least or about 90%, at least
or about 91%, at
least or about 92%, at least or about 93%, at least or about 94%, at least or
about 95%, at least
or about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about
100%, identical to SEQ ID NO: 11: GGGGSGGGGSGGGGS (Figures 3, 6, and 23) or to
SEQ
ID NO: 30: GGGGSGGGGSGGGGSGGGGS (Figure 23, CD16 VH-VL linker).
In further embodiments, the third engineered heterodimer protein comprises a
His6 tag
(HHHHHH; SEQ ID NO: 20).
In certain embodiments, the engineered heterotrimer protein comprises: (i) a
first
polypeptide comprising a polypeptide that binds a molecule expressed on T
cells, a non-
naturally occurring polypeptide domain comprising 1-5 alpha helices connected
by amino acid
46
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
linkers (al), and a first covalent dimerization domain; and (ii) a second
polypeptide comprising
an antigen-binding fragment that binds a lineage-specific cell-surface
antigen, a non-naturally
occurring polypeptide domain comprising 1-5 alpha helices connected by amino
acid linkers
(b1) and a second covalent dimerization domain; and (iii) a third polypeptide
comprising a
polypeptide that binds a molecule expressed on natural killer (NK) cells, a
non-naturally
occurring polypeptide domain comprising 1-5 alpha helices connected by amino
acid linkers
(cl), and a third second covalent dimerization domain and (iv) a fourth
polypeptide comprising
three non- naturally occurring polypeptide domains comprising 1-5 alpha
helices connected by
amino acid linkers, wherein each domain is the binding domain of al, bl and cl
(a2, b2 and
c2), and a fourth, fifth and sixth covalent dimerization domain as described
herein.
In certain embodiments, antigen-binding fragments include, but are not limited
to, Fab,
F(ab')2, Fab', F(ab)', Fv, a disulfide linked Fv, single chain Fv (scFv),
bivalent scFv (hi-scFv),
trivalent scFv (tri-scFv), Fd, dAb fragment, an isolated CDR, diabodies,
triabodies, tetrabodies,
linear antibodies, single-chain antibody molecules. In some embodiments, scFv
comprises a
heavy chain variable region (VH), and a light chain variable region (VL).
In some embodiments, the non-naturally occurring polypeptide domain comprising
1-
5 alpha helices is 6DMPa (Chen et al. 2019). In sonic embodiments, the non-
naturally
occurring polypeptide domain comprising 1-5 alpha helices is 6DMPb (Chen et
al. 2019).
In some embodiments, the covalent dimerization domains are IgG2 hinge domains.
In
some embodiments, the covalent dimerization domains are IgG2 domains and IgG2
Fc
domains. In some embodiments, the Fc domains are CH2 and CH3 domains.
The engineered heterotrimer protein can be designed to place the functional
moieties
(an antigen-binding fragment that binds a lineage-specific cell-surface
antigen, and a
polypeptide that binds a molecule expressed on NK cells, and a polypeptide
that binds a
molecule expressed on T cells) in any order. In certain embodiments, the
polypeptide that binds
a molecule expressed on T cells antigen-binding fragment is located at the N-
terminus or C-
terminus of the fusion polypeptide. In certain embodiments, the polypeptide
that binds a
molecule expressed on natural killer (NK) cells is located at the C-terminus
or N-terminus of
the fusion polypeptide.
In some embodiments, the engineered heterotrimer protein comprises, from N-
terminus
to C-terminus, a polypeptide that binds a molecule expressed on T cells, a
scFv that binds to
the lineage-specific cell-surface antigen (e.g., CD33 or CD19) and an
ectodomain of ULBP2,
ULBP3, ULBP4, ULBP5, ULBP6, MICA, or MICB) or a polypeptide that binds CD16.
In
some embodiments, the fusion polypeptide comprises, from N terminus to C
terminus, an
47
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
ectodomain of ULBP1 (or an ectodomain of ULBP2, ULBP3, ULBP4, ULBP5, ULBP6,
MICA, or MICB), a scFv that binds to the lineage-specific cell-surface antigen
(e.g., CD33 or
CD19) and a polypeptide that binds a molecule expressed on T cells.
The engineered heterotrimer protein may further comprise a signal sequence,
and/or
one or more linkers. In the fusion polypeptide, these functional moieties may
be covalently
ligated continuously or non-continuously (e.g., they may be separated by
linkers (e.g., linker
amino acid residues)). The linker may have up to 50, up to 40, up to 30, up to
20, up to 18, up
to 15, up to 12, up to 11, or up to 10, amino acid residues in length. In
certain embodiments,
the linker has about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10-20, 8-10, 8-12, 8-15, 8-20,
or 8-30 amino acid
residues in length. In certain embodiments, the linker has about 7-10, 7-12, 7-
15, 7-20, or 7-
30 amino acid residues in length.
One type of derivatized protein is produced by crosslinking two or more
polypeptides
(of the same type or of different types). Suitable crosslinkers include those
that are
h eterobi fun cti on al , having two distinct reactive groups separated by an
appropriate spacer (e.g.,
m-maleimidobenzoyl-N-hydroxysuccinimide ester) or homobifunctional (e.g.,
disuccinimidyl
suberate). Useful detectable agents with which a protein can be derivatized
(or labeled) include
fluorescent agents, various enzymes, prosthetic groups, luminescent materials,
bioluminescent
materials, and radioactive materials. Non-limiting, exemplary fluorescent
detectable agents
include fluorescein, fluorescein isothiocyanate, rhodamine, and,
phycoerythrin. A polypeptide
can also be derivatized with detectable enzymes, such as alkaline phosphatase,
horseradish
peroxidase, beta-galactosidase, acetylcholinesterase, glucose oxidase and the
like. A
polypeptide can also be derivatized with a prosthetic group (e.g.,
streptavidin/biotin and
avidin/biotin).
The engineered heterotrimer protein can be derivatized or linked to another
functional
molecule. For example, heterotrimer protein can be functionally linked (by
chemical coupling,
genetic fusion, noncovalent interaction, etc.) to one or more other molecular
entities, such as
an antibody or antibody fragment, a detectable agent, an inamunosuppressant, a
cytotoxic agent,
a pharmaceutical agent, a protein or peptide that can mediate association with
another molecule
(such as a streptavidin core region or a polyhistidine tag), amino acid
linkers, signal sequences,
immunogenic carriers, or ligands useful in protein purification, such as
glutathione-S-
transferase. histidine tag, and staphylococcal protein A. Cytotoxic agents may
include
radioactive isotopes, chemotherapeutic agents, and toxins such as
enzymatically active toxins
of bacterial, fungal, plant, or animal origin, and fragments thereof.
48
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
The engineered heterotrimer protein may further comprise a fragment (e.g., a
tag) useful
for polypeptide production and/or detection, including, but not limited to,
poly-histidine (e.g.,
six histidine residues), a maltose binding protein, GST, green fluorescent
protein (GFP),
hemagglutinin, or alkaline phosphatase, secretion signal peptides (e.g.,
preprotyrypsin signal
sequence), Myc, and/or FLAG.
In one embodiment, the engineered heterotrimer protein comprises one or more
signal
sequences comprising (or consisting essentially of, or consisting of) an amino
acid sequence at
least or about 50%, at least about 55%, at least or about 60%, at least or
about 70%, at least or
about 75%, at least or about 80%, at least or about 81%, at least or about
82%, at least or about
83%, at least or about 84%, at least or about 85%, at least or about 86%, at
least or about 87%,
at least or about 88%, at least or about 89%, at least or about 90%, at least
or about 91%, at
least or about 92%, at least or about 93%, at least or about 94%, at least or
about 95%, at least
or about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about
100%, identical to an 1L2 secretory signal sequence: MYRMQLLSCIALSLALVTNS (SEQ
ID NO:7) (Figures 3-8).
In one embodiment, the engineered heterotrimer protein comprises one or more
tags
comprising (or consisting essentially of, or consisting of) an amino acid
sequence at least or
about 50%, at least about 55%, at least or about 60%, at least or about 70%,
at least or about
75%, at least or about 80%, at least or about 81%, at least or about 82%, at
least or about 83%,
at least or about 84%, at least or about 85%, at least or about 86%, at least
or about 87%, at
least or about 88%, at least or about 89%, at least or about 90%, at least or
about 91%, at least
or about 92%, at least or about 93%, at least or about 94%, at least or about
95%, at least or
about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about 100%,
identical to Myc: EQKLISEEDL (SEQ ID NO: 8) (Figures 3 and 6).
In one embodiment, the engineered heterotrimer protein comprises one or more
tags
comprising (or consisting essentially of, or consisting of) an amino acid
sequence at least or
about 50%. at least about 55%, at least or about 60%, at least or about 70%,
at least or about
75%, at least or about 80%, at least or about 81%, at least or about 82%, at
least or about 83%,
at least or about 84%, at least or about 85%, at least or about 86%, at least
or about 87%, at
least or about 88%, at least or about 89%, at least or about 90%, at least or
about 91%, at least
or about 92%, at least or about 93%, at least or about 94%, at least or about
95%, at least or
about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about 100%,
identical to FLAG: DYKDDDDK (SEQ ID NO: 14) (Figures 4, 5, 7 and 8)
49
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
In one embodiment, the engineered heterotrimer protein comprises a ULBP1
ectodomain comprising (or consisting essentially of, or consisting of) an
amino acid sequence
at least or about 50%, at least about 55%, at least or about 60%, at least or
about 70%, at least
or about 75%, at least or about 80%, at least or about 81%, at least or about
82%, at least or
about 83%, at least or about 84%, at least or about 85%, at least or about
86%, at least or about
87%, at least or about 88%, at least or about 89%, at least or about 90%, at
least or about 91%,
at least or about 92%, at least or about 93%, at least or about 94%, at least
or about 95%, at
least or about 96%, at least or about 97%, at least or about 98%, at least or
about 99%, or about
100%, identical to SEQ ID NO: 15 (Figures 4 and 7).
In one embodiment, the engineered heterotrimer protein comprises an anti-CD33
scFv
comprising (or consisting essentially of, or consisting of) an amino acid
sequence at least or
about 50%, at least about 55%, at least or about 60%, at least or about 70%,
at least or about
75%, at least or about 80%, at least or about 81%, at least or about 82%, at
least or about 83%,
at least or about 84%, at least or about 85%, at least or about 86%, at least
or about 87%, at
least or about 88%, at least or about 89%, at least or about 90%, at least or
about 91%, at least
or about 92%, at least or about 93%, at least or about 94%, at least or about
95%, at least or
about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about 100%,
identical to the anti-CD33 scFv disclosed in U.S. Patent Publication No.
20130078241.
In certain embodiments, the polypeptide that binds a molecule expressed on NK
cells
comprises (or consists essentially of, or consists of) an amino acid sequence
at least or about
50%, at least about 55%, at least or about 60%, at least or about 70%, at
least or about 75%, at
least or about 80%, at least or about 81%, at least or about 82%, at least or
about 83%, at least
or about 84%, at least or about 85%, at least or about 86%, at least or about
87%, at least or
about 88%, at least or about 89%, at least or about 90%, at least or about
91%, at least or about
92%, at least or about 93%, at least or about 94%, at least or about 95%, at
least or about 96%,
at least or about 97%, at least or about 98%, at least or about 99%, or about
100%, identical to
the amino acid sequence of SEQ ID NOs: 28 and 29 (Figure 23).
In one embodiment, the engineered heterotrimer protein comprises an antigen-
binding
fragment that binds CD33, where the antigen-binding fragment comprises a light
chain variable
region (VL) comprising (or consisting essentially of, or consisting of) an
amino acid sequence
at least or about 50%, at least about 55%, at least or about 60%, at least or
about 70%, at least
or about 75%, at least or about 80%, at least or about 81%, at least or about
82%, at least or
about 83%, at least or about 84%, at least or about 85%, at least or about
86%, at least or about
87%, at least or about 88%, at least or about 89%, at least or about 90%, at
least or about 91%,
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
at least or about 92%, at least or about 93%, at least or about 94%, at least
or about 95%, at
least or about 96%, at least or about 97%, at least or about 98%, at least or
about 99%, or about
100%, identical to SEQ ID NO: 9 (Figures 3 and 6).
In one embodiment, the engineered heterotrimer protein comprises an antigen-
binding
fragment that binds CD33, where the antigen-binding fragment comprises a heavy
chain
variable region (VH) comprising (or consisting essentially of, or consisting
of) an amino acid
sequence at least or about 50%, at least about 55%, at least or about 60%, at
least or about 70%,
at least or about 75%, at least or about 80%, at least or about 81%, at least
or about 820/c, at
least or about 83%, at least or about 84%, at least or about 85%, at least or
about 86%, at least
or about 87%, at least or about 88%, at least or about 89%, at least or about
90%, at least or
about 91%, at least or about 92%, at least or about 93%, at least or about
94%, at least or about
95%, at least or about 96%, at least or about 97%, at least or about 98%, at
least or about 99%,
or about 100%, identical to SEQ ID NO: 10 (Figures 3 and 6):
In certain embodiments, the polypeptide that binds a molecule expressed on T
cells
comprises (or consists essentially of, or consists of) an amino acid sequence
at least or about
50%, at least about 55%, at least or about 60%, at least or about 70%, at
least or about 75%, at
least or about 80%, at least or about 81%, at least or about 82%, at least or
about 83%, at least
or about 84%, at least or about 85%, at least or about 86%, at least or about
87%, at least or
about 88%, at least or about 89%, at least or about 90%, at least or about
91%, at least or about
92%, at least or about 93%, at least or about 94%, at least or about 95%, at
least or about 96%,
at least or about 97%, at least or about 98%, at least or about 99%, or about
100%, identical to
the amino acid sequence of SEQ ID NOs: 17 and 18 (Figures 5 and 8).
In one embodiment, the engineered heterotrimer protein comprises one or more
non-
naturally occurring polypeptide domain comprising 1-5 alpha helices comprising
(or consisting
essentially of, or consisting of) an amino acid sequence at least or about
50%, at least about
55%, at least or about 60%, at least or about 70%, at least or about 75%, at
least or about 80%,
at least or about 81%, at least or about 82%, at least or about 83%, at least
at about 84%, at
least or about 85%, at least or about 86%, at least or about 87%, at least or
about 88%, at least
or about 89%, at least or about 90%, at least or about 91%, at least or about
92%, at least or
about 93%, at least or about 94%, at least or about 95%, at least or about
96%, at least or about
97%, at least or about 98%, at least or about 99%, or about 100%, identical to
SEQ ID NO: 12
(Figures 3 and 6).
In one embodiment, the engineered heterotrimer protein comprises one or more
non-
naturally occurring polypeptide domain comprising 1-5 alpha helices comprising
(or consisting
51
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
essentially of, or consisting of) an amino acid sequence at least or about
50%, at least about
55%, at least or about 60%, at least or about 70%, at least or about 75%, at
least or about 80%,
at least or about 81%, at least or about 82%, at least or about 83%, at least
or about 84%, at
least or about 85%, at least or about 86%, at least or about 87%, at least or
about 88%, at least
or about 89%, at least or about 90%, at least or about 91%, at least or about
92%, at least or
about 93%, at least or about 94%, at least or about 95%, at least or about
96%, at least or about
97%, at least or about 98%, at least or about 99%, or about 100%, identical to
SEQ ID NO: 16
(Figures 5 and 8).
In one embodiment, the engineered heterotrimer protein comprises one or more
covalent dimerization IgG2 hinge domains comprising (or consisting essentially
of, or
consisting of) an amino acid sequence at least or about 50%, at least about
55%, at least or
about 60%, at least or about 70%, at least or about 75%, at least or about
80%, at least or about
81%, at least or about 82%, at least or about 83%, at least or about 84%, at
least or about 85%,
at least or about 86%, at least or about 87%, at least or about 88%, at least
or about 89%, at
least or about 90%, at least or about 91%, at least or about 92%, at least or
about 93%, at least
or about 94%, at least or about 95%, at least or about 96%, at least or about
97%, at least or
about 98%, at least or about 99%, or about 100%, identical to IgG2 hinge:
ERKCCVECPPCP
(SEQ Ill NO: 13) (Figures 3-8).
In one embodiment, the engineered heterotrimer protein comprises one or more
covalent dimerization IgG2 Fc domains comprising (or consisting essentially
of, or consisting
of) an amino acid sequence at least or about 50%, at least about 55%, at least
or about 60%, at
least or about 70%, at least or about 75%, at least or about 80%, at least or
about 81%, at least
or about 82%, at least or about 83%, at least or about 84%, at least or about
85%, at least or
about 86%, at least or about 87%, at least or about 88%, at least or about
89%, at least or about
90%, at least or about 91%, at least or about 92%, at least or about 93%, at
least or about 94%,
at least or about 95%, at least or about 96%, at least or about 97%, at least
or about 98%, at
least or about 99%, or about 100%, identical to SEQ ID NO: 19. (Figures 6-8).
In one embodiment, the engineered heterotrimer protein comprises one or more
linkers
comprising (or consisting essentially of, or consisting of) an amino acid
sequence at least or
about 50%. at least about 55%, at least or about 60%, at least or about 70%,
at least or about
75%, at least or about 80%, at least or about 81%, at least or about 82%, at
least or about 83%,
at least or about 84%, at least or about 85%, at least or about 86%, at least
or about 87%, at
least or about 88%, at least or about 89%, at least or about 90%, at least or
about 91%, at least
or about 92%, at least or about 93%, at least or about 94%, at least or about
95%, at least or
52
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
about 96%, at least or about 97%, at least or about 98%, at least or about
99%, or about 100%,
identical to SEQ ID NO: 11 GGGGSGGGGSGGGGS (Figures 3, 5, 6, and 8) or to SEQ
ID
NO: 21 GGGGGSGGSGGSGG or to SEQ ID NO: 22 SGSGSG (Figures 5 and 8)
In further embodiments, the engineered heterotrimer protein comprises a His6
tag
(HHHHHH; SEQ Ill NO: 20).
In some embodiments, the third polypeptide is a chemokine or cytokine protein,
which
increases the immune response. The chemokine or cytokine protein includes but
is not limited
to CXCLs including CXCL14, GCSF, and interleukins, including IL2 and ILI 6.
Shown herein is that the disclosed engineered proteins bind to and are
cytotoxic against
cells expressing CD33, such as HL60 and MOLM cells. Also shown is that when
CD33
expression is increased in a cell, the binding efficiency of the engineered
protein increases.
Additionally, the cytotoxicity of the engineered protein increases in cells
such as MOLM,
where CD33 expression is increased.
In certain embodiments, the antigen-binding fragment that hinds a lineage-
specific cell-
surface antigen (e.g., CD33) is a derivative, or a modified form, or a
variant, of a fragment of
the wildtype antigen-binding fragment.
In certain embodiments, the polypeptidc that binds a molecule expressed on
natural
killer (N K) cells is a derivative, or a modified form, or a variant, of a
fragment of the wildtype
polypeptide.
In certain embodiments, the polypeptide that binds a molecule expressed on T
cells is
a derivative, or a modified form, or a variant, of a fragment of the wildtype
polypeptide.
As used herein, the term variant also denotes any peptide, pseudopeptide
(peptide
incorporating non-biochemical elements) or protein differing from the wildtype
protein or
peptide, obtained by one or more genetic and/or chemical modifications.
Genetic and/or
chemical modification may he understood to mean any mutation, substitution,
deletion,
addition and/or modification of one or more residues of the protein or peptide
considered.
Chemical modification may refer to any modification of the peptide or protein
generated by
chemical reaction or by chemical grafting of biological or non-biological
molecule(s) onto any
number of residues of the protein.
The present polypeptides or peptides may include variants, analogs, orthologs,
homologs and derivatives of amino acids or peptides. The present polypeptides
or peptides
may contain one or more analogs of amino acids (including, for example, non-
naturally
occurring amino acids, amino acids which only occur naturally in an unrelated
biological
system, modified amino acids etc.), peptides with substituted linkages, as
well as other
53
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
modifications known in the art. The present polypeptides or peptides may
comprise a
peptidomimetic, such as a peptoid. The present polypeptides or peptides may
contain one or
more amino acid residues modified by, e.g., glycosylation, acylation (e.g.,
acetylation,
formylation, myristoylation, palmitoylation, lipoylation), alkylation (e.g.,
methylation),
isoprenylation or prenylation (e.g., farnesylation, geranylgeranylation),
sulfation, amidation,
hydroxylation, succinylation, etc. The present polypeptides and agents may be
glycosylated,
sulfonated and/or phosphorylated and/or grafted to complex sugars or to a
lipophilic compound
such as, for example, a polycarbon chain or a cholesterol derivative.
Lineage-Specific Cell-Surface Antigens
Aspects of the disclosure provide agents targeting a lineage-specific cell-
surface
antigen, for example on a target cancer cell. Such an agent may comprise an
antigen-binding
fragment that binds and targets the lineage-specific cell-surface antigen. In
some instances, the
antigen-binding fragment can be a single chain antibody (scFv) specifically
binding to the
lineage-specific antigen. As used herein, the terms "lineage-specific cell-
surface antigen"
and "cell-surface lineage-specific antigen" may be used interchangeably and
refer to any
antigen that is sufficiently present on the surface of a cell and is
associated with one or more
populations of cell lineage(s). For example, the antigen may be present on one
or more
populations of cell lineage(s) and absent (or at reduced levels) on the cell-
surface of other cell
populations.
In general, lineage-specific cell-surface antigens can be classified based on
a number
of factors such as whether the antigen and/or the populations of cells that
present the antigen
are required for survival and/or development of the host organism. A summary
of exemplary
types of lineage-specific antigens is provide in Table 1 below.
Table 1. Classification of Lineage Specific Antigens
Type of Lineage Characteristics of the Lineage Specific Antigen
Specific Antigen
Type 0 a) antigen is required for survival of an
organism
and
b) cell type carrying type 0 antigen is
required for survival of an
organism and is not unique to a tumor, or tumor-associated virus
54
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
Type 1 a) antigen is not required for survival of an
organism
and
b) cell type carrying type 1 antigen is not
required for survival of
an organism
Type 2 a) antigen is not required for survival of an
organism
and
b) cell type carrying type 2 antigen is
required for the survival of
an organism
Type 3 a) antigen is not required for the survival of
an organism
and
b) cell type carrying antigen is not required for survival of an
organism
c) The antigen is unique to a tumor, or a tumor associated virus.
An example is the LMP-2 antigen in EBV infected cells, including
EBV infected tumor cells (Nasopharyngeal carcinoma and Burldtts
Lymphoma)
Lineage specific antigens of type 1 class may be expressed in a wide variety
of different
tissues, including, ovaries, testes, prostate, breast, endometrium, and
pancreas. In some
embodiments, the agent targets a cell-surface lineage-specific antigen that is
a type 1 antigen.
In some embodiments, the agent targets a cell-surface lineage-specific antigen
that is a
type 2 antigen. For example, CD33 is a type 2 antigen expressed in both normal
myeloid cells
as well as in Acute Myeloid Leukemia (AML) cells (Dohner et al.2015).
A wide variety of antigens may be targeted by the methods and compositions of
the
present disclosure. Monoclonal antibodies to these antigens may be purchased
commercially
or generated using standard techniques, including immunization of an animal
with the antigen
of interest followed by conventional monoclonal antibody methodologies, e.g.,
the standard
somatic cell hybridization technique of Kohler and Milstein, Nature (1975)
256: 495, as
discussed above. The antibodies or nucleic acids encoding for the antibodies
may be sequenced
using any standard DNA or protein sequencing techniques.
In some embodiments, the cell-surface lineage-specific antigen that is
targeted using
the methods and compositions described herein is a cell-surface lineage-
specific antigen of
leukocytes or a subpopulation of leukocytes. In some embodiments, the cell-
surface lineage-
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
specific antigen is an antigen that is associated with myeloid cells. In some
embodiments, the
cell-surface lineage-specific antigen is a cluster of differentiation antigens
(CDs). Examples of
CD antigens include, without limitation, CD1a, CD1b, CD1c, CD1d, CD1e, CD2,
CD3, CD3d,
CD3e, CD3g, CD4, CD5, CD6, CD7, CD8a, CD8b, CD9, CD10, CD11a, CD11b, CD1 lc,
CD11d, Cllw12, CD13, CD14, CD15, CD16, CD16b, CD17, CD18, CD19, C1120, CD21,
CD22, CD23, CD24, CD25, CD26, CD27, CD28, CD29, CD30, CD31, CD32a, CD32b,
CD32e, CD33, CD34, CD35, CD36, CD37, CD38, CD39, CD40, CD41, CD42a, CD42b,
CD42c, CD42d, CD43, CD44, CD45, CD45RA, CD45RB, CD45RC, CD45RO, CD46, CD47,
CD48, CD49a, CD49b, CD49c, CD49d, CD49e, CD49f, CD50, CD51, CD52, CD53, CD54,
CD55, CD56, CD57, CD58, CD59, CD60a, CD61, CD62E, CD62L, CD62P, CD63, CD64a,
CD65, CD65s, CD66a, CD66b, CD66c, CD66F, CD68, CD69, CD70, CD71, CD72, CD73,
CD74, CD75, CD75S, CD77, CD79a, CD79b, CD80, CD81, CD82, CD83, CD84, CD85A,
CD85C, CD85D, CD85E, CD85F, CD85G, CD85H, CD85I, CD85J, CD85K, CD86, CD87,
CD88, CD89, CD90, CD91 , CD92, CD93, CD94, CD95. CD96, CD97, CD98, CD99,
CD99R,
CD100, CD101, CD102, CD103, CD104, CD105, CD106, CD107a, CD107b, CD108, CD109,
CD110, CD111, CD112, CD113, CD114, CD115, CD116, CD117, CD118, CD119, CD120a,
CD120b, CD121a, CD121b, CD121a, CD121b, CD122, CD123, CD124, CD125, CD126,
CD127, CD129, CD130, CD131, CD132, CD133, CD134, CD135, CD136, CD137, CD138,
CD139, CD140a, CD140b, CD141, CD142, CD143, CD144, CDw145, CD146, CD147,
CD148, CD150, CD152, CD152, CD153, CD154, CD155, CD156a, CD156b, CD156c,
CD157, CD158b1, CD158b2, CD158d, CD158e1/e2, CD158f, CD158g, CD158h, CD158i,
CD158j, CD158k, CD159a, CD159c, CD160, CD161, CD163, CD164, CD165, CD166,
CD167a, CD168, CD169, CD170, CD171, CD172a, CD172b, CD172g, CD173, CD174,
CD175, CD175s, CD176, CD177, CD178, CD179a, CD179b, CD180, CD181, CD182,
CD183, CD184, CD185, CD186, CD191, CD192, CD193, CD194, CD195, CD196, CD197,
CDw198, CDw199, CD200, CD201, CD202b, CD203c, CD204, CD205, CD206, CD207,
CD208, CD209, CD210a, CDw210b, CD212, CD213a1, CD213a2, CD215, CD217, CD218a,
CD218b, CD220, CD221, CD222, CD223, CD224, CD225, CD226, CD227, CD228, CD229,
CD230, CD231, CD232, CD233, CD234, CD235a, CD235b, CD236, CD236R, CD238,
CD239, CD240, CD241, CD242, CD243, CD244, CD245, CD246, CD247, CD248, CD249,
CD252, CD253, CD254, CD256, CD257, CD258, CD261, CD262, CD263, CD264, CD265,
CD266, CD267, CD268, CD269, CD270, CD271, CD272, CD273, CD274, CD275, CD276,
CD277, CD278, CD279, CD280, CD281, CD282, CD283, CD284, CD286, CD288, CD289,
CD290, CD292, CDw293, CD294, CD295, CD296, CD297, CD298, CD299, CD300a,
56
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
CD300c, CD300e, CD301, CD302, CD303, CD304, CD305, 306, CD307a, CD307b,
CD307c,
D307d, CD307e, CD309, CD312, CD314, CD315, CD316, CD317, CD318, CD319, CD320,
CD321, CD322, CD324, CD325, CD326, CD327, CD328, CD329, CD331, CD332, CD333,
CD334, CD335, CD336, CD337, CD338, CD339, CD340, CD344, CD349, CD350, CD351,
CD352, CD353, CD354, CD355, CD357, CD358, CD359, CD360, CD361, CD362 and
CD363.
In some embodiments, the cell-surface lineage-specific antigen is CD19, CD20,
CD11,
CD123, CD56, CD34, CD14, CD33, CD66b, CD41, CD61, CD62, CD235a, CD146, CD326,
LMP2, CD22, CD52, CD10, CD3/TCR, CD79/BCR, and CD26. In some embodiments, the
cell-surface lineage-specific antigen is CD33 or CD19.
Alternatively or in addition, the cell-surface lineage-specific antigen may be
a cancer
antigen, for example a cell-surface lineage-specific antigen that is
differentially present on
cancer cells. In some embodiments, the cancer antigen is an antigen that is
specific to a tissue
or cell lineage. Examples of cell -surface lineage-specific antigen that are
associated with a
specific type of cancer include, without limitation. CD20, CD22 (Non-Hodgkin's
lymphoma,
B-cell lymphoma, chronic lymphocytic leukemia (CLL)), CD52 (B-cell CLL), CD33
(Acute
myelogenous leukemia (AML)). CD10 (gp100) (Common (pre-B) acute lymphocytic
leukemia
and malignant melanoma), CD3/T-cell receptor (TCR) (T-cell lymphoma and
leukemia),
CD79/B-cell receptor (BCR) (B-cell lymphoma and leukemia), CD26 (epithelial
and lymphoid
malignancies), human leukocyte antigen (HLA)-DR, HLA-DP, and HLA-DQ (lymphoid
malignancies), CD307e and BCMA (myeloma), RCAS1 (gynecological carcinomas,
biliary
adenocarcinomas and ductal adenocarcinomas of the pancreas), C1audin3, TMPRSS3
and Her2
(ovarian cancer), Hcr2 (breast cancer) as well as prostate specific membrane
antigen. In some
embodiments, the cell-surface antigen CD33 and is associated with AML cells.
In certain embodiments, the antigen-binding fragment that binds a lineage-
specific cell-
surface antigen (e.g., CD33) has up to or about 500, up to or about 490, up to
or about 480, up
to or about 470, up to or about 460, up to or about 450, up to or about 440,
up to at about 430,
up to or about 420, up to or about 410, up to or about 400, up to or about
390, up to or about
380, up to or about 370, up to or about 360, up to or about 350, up to or
about 340, up to or
about 330, up to or about 320, up to or about 310, up to or about 200, up to
or about 190, up to
or about 180, up to or about 170, up to or about 160, up to or about 150, up
to or about 140, up
to or about 130, up to or about 120, up to or about 110, up to or about 100,
up to or about 90,
up to or about 80, up to or about 70, up to or about 60, up to or about 50, up
to or about 40, up
to or about 30, up to or about 20, up to or about 15, or up to or about 10,
amino acid residues
57
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
in length. In certain embodiments, the antigen-binding fragment that binds a
lineage-specific
cell-surface antigen (e.g., CD33) has about 100-200, 80-210, 80-250, 150-250,
100-30, 50-200,
150-250, 150-300, 300-400, 200-400, 400-500, or 150-190 amino acid residues in
length.
NK cell surface receptor-bindin2 peptides
In certain embodiments, the present fusion polypeptide or composition
comprises a
polypeptide that binds to a molecule expressed on natural killer (NK) cells,
such as a C-type
lectin-like receptor (e.g., NKG2D).
A C-type lectin-like NK cell receptor may be a receptor expressed on the
surface of
natural killer cells. Exemplary NK cell receptors of this type include, but
are not limited to,
NKG2D (GENBANK accession number BC039836), Dectin-1 (GENBANK accession number
AJ312373 or AJ312372), Mast cell function-associated antigen (GENBANK
accession number
AF097358), HNKR-PlA (GENBANK accession number U11276), LLT1 (GENBANK
accession number AF133299), CD69 (GENBANK accession number NM_001781), CD69
homolog, CD72 (GENBANK accession number NM_001782), CD94 (GENBANK accession
number NM_002262 or NM_007334), KLRF1 (GENBANK accession number NM_016523),
Oxidised LDL receptor (GENBANK accession number NM 002543), CLEC-1, CLEC-2
(GENBANK accession number NM 016509), NKG2C (GENBANK accession number
AJ001684), NKG2A (GENBANK accession number AF461812), NKG2E (GENBANK
accession number AF461157), WUGSC:H_DJ0701016.2. or Myeloid DAP12-associating
lectin (MDL-1; GENBANK accession number AJ271684). In particular embodiments,
the NK
cell receptor is human NKG2D or human NKG2C.
As used herein, the terms "Natural Killer Group 2D", "NKG2D" and "NKG2D
receptor", also known as KLRK1, refer to an activating cell surface molecule
that is found on
numerous types of immune cells, particularly NK cells, CD8+ T cells, some CD4+
T cells, and
gamma delta T cells. The terms NKG2D and NKG2D receptor include variants,
isoforms, and
homologs of human NKG2D receptor (see, e.g., the isoforms described in
Diefenbach et al.,
Nat Immunol. (2002) 3(12):1142-9). NKG2D is a type II transmembrane protein
with an
extracellular C-type (i.e., Ca2'-binding) lectin-like domain but lacking the
Ca2 binding site. It
can form heterodimers with adapter proteins such as DAP10 Or DAP12, and
recognizes protein
ligands that include, but are not limited to, MICA, MICB, ULBP1, ULBP2, ULBP3,
ULBP4,
ULBP5, and ULBP6.
58
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
In certain embodiments, the NKG2D-binding peptide is an agonist of NKG2D. In
certain embodiments, the NKG2D-binding peptide is an antagonist of NKG2D. In
certain
embodiments, the NKG2D-binding peptide is neither an antagonist nor an agonist
of NKG2D.
The polypeptide that binds a molecule expressed on NK cells may be a ligand
for a NK
cell surface receptor, such as a ligand for the N KG2D cell surface receptor.
Non-limiting
examples of the ligands for NKG2D (or the NKG2D ligands) include, an MHC class
I chain-
related (MIC) antigen such as MICA and MICB. a UL16 binding protein (ULBP)
such as
ULBP1, ULBP2, ULBP3, ULBP4, ULBP5, ULBP6, and the like (Bahram Adv. Immunol.
(2000) 76:1-60; Cerwenka and Lanier Immunol. Rev. (2001) 181:158-169; Cosman,
et al.
Immunity (2001) 14:123-133; Kubin, et al. Eur. J. Immunol. (2001) 31:1428-
1437). Murine
NKG2D ligands include, for example, the retinoic acid early inducible-1 gene
products (RAE-
1) and minor hi stocompatibility antigen peptide H60. NK cells can be
regulated by interaction
of immunomodulating polypeptide ligands with receptors on the NK cell surface.
For example,
ligands for the NKG2D receptor that can regulate NK cell activity, include
chemokines such
as muCCL21, and stress-inducible polypeptide ligands such as MHC class I chain-
related
antigens and ULL16 binding proteins. Murine H60 minor histocompatibility
antigen peptide is
reported to bind to the NKG2D receptor, as well. See, e.g., Robertson et al.
Cell Immunol.
(2000) 199(1):8-14; Choi et al. Immunity (2002) 17(5):593-603, and Farag et
al., Blood. (2002)
100(6):1935-1947. As used herein, the term "NKG2D ligand" refers to a binding
partner that
binds specifically to an NKG2D receptor. Exemplary ligands include MICA, MICB,
ULBP1,
ULBP2, ULBP3, ULBP4, ULBPS, ULBP6, and functional fragments thereof, such as
soluble
forms of MIC and ULBP proteins.
Table 2 lists exemplary NKG2D binding proteins and domains.
Table 2. NKG2D binding proteins and domains
Protein Gene names Uniport NKG2D
ID binding
domain*
UL16-binding protein ULBP1/RAET1I Q9BZM6 27-216
1
UL16-binding protein ULBP2/RAET1H Q9BZM5 26-217
2
59
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
UL16-binding protein ULBP3/RAET1N Q9BZM4 30-217
3
UL16-binding protein ULBP4/RAET1E Q8TD07 31-225
4
UL-16 binding protein ULBP5/RAET1G Q6H3X3 26-223
UL16-binding protein ULBP6/RAET1L Q5VY80 26-218
6
MHC class I MICA Q29983 24-307
polypeptide-related
sequence A
MHC class I MICB Q29980 23-309
polypeptide-related
sequence B
Retinoic acid early-
inducible protein 1-
alpha Raet 1 a 008602 29-229
Retinoic acid early-
inducible protein 1-
beta Raetlb 008603 29-229
Retinoic acid early-
inducible protein 1-
gamma Ractic 008604 29-227
Retinoic acid early-
inducible protein 1-
delta Raetld Q9JI58 29-225
Retinoic acid early-
inducible protein 1-
epsilon Raetle Q9CZQ6 29-225
Histocompatibility
antigen 60h H60b B1B212 25-251
Histocompatibility
antigen 60c H60c B1B213 18-177
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
* The start and end amino acids numbers are based on the sequence of protein
identified by
the uniport ID.
The MIC and ULBP proteins act as ligands that bind to C-type lectin-like
activating
receptor Natural Killer Group 2D (NKG2D) on immune effector cells, including
NK cells,
NKT cells, alpha beta CD8+ T cells, and gamma delta CD8+ T cells.
As used herein, the term "ULBP protein" refers to members of the MHC class I-
related
molecules having a characteristic organization for the unprocessed protein
that includes a N-
terminal signal sequence, centrally located alpha-1 and alpha-2 domains, and a
C-terminal cell
membrane association domain, which can be a glycosylphosphatidylinositol (GPI)
anchoring
domain or a transmembrane domain. Some species of ULBP protein have a
cytoplasmic
domain. Generally, ULBP proteins have weak amino acid sequence identity to MI
C A/MICB
proteins. ULBP family members are ligands for the effector cell receptor
NKG2D, and are
known to activate NK cells. As used herein. "ULBP protein" includes active
variants, isoforms,
and species homologs of human ULBP protein, and includes fragments having
NKG2D
receptor binding activity.
As used herein, the term "ULBP1", also described as "retinoie acid early
transcript 1
protein" or "RAET1", refers to a member of the MHC class I family, including
variants,
isoforms, and species homologs of human ULBP1. The protein functions as a
ligand for
receptor NKG2D. ULBP1 protein activates multiple signaling pathways in primary
NK cells.
The C terminal membrane association domain in ULBP1 comprises a GPI domain.
ULBP1 is
weakly homologous with MICA and MICB and has about 55% to 60% amino acid
sequence
identity to ULBP2 and ULBP3. Exemplary sequence of human ULBP1 is available as
NCBI
accession no. NP_079494.1. DNA and protein sequences for human ULBP1 have been
reported by Cosman et al., Immunity (2001) 14(2):123-133, DNA Accession No.
AF304377 in
the EMBL database of the European Bioinformatics Institute, Wellcome Trust
Genome
Campus, Hinxton, Cambridge CB10 1SD, UK.
As used herein, the term "ULBP2", also described as "retinoic acid early
transcript 1H
protein" or "RAET1H", refers to a member of the MHC class I family, including
variants,
isoforms, and species homologs of human ULBP2. The protein functions acts as a
ligand for
receptor NKG2D. ULBP2 activates multiple signaling pathways in primary NK
cells. The C
terminal membrane association domain in ULBP2 comprises a GPI domain. ULBP2 is
weakly
homologous with MICA and MICB and has about 55% and 60% amino acid sequence
identity
to ULBP1 and ULBP3. Exemplary sequence of human ULBP2 is available as NCBI
accession
61
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
no. NP_079493.1. DNA and protein sequences for human ULBP2 have been reported
by
Cosman et al., Immunity (2001) 14(2):123-133, DNA Accession No. AF304378 in
the EMBL
database of the European Bioinformatics Institute, Wellcome Trust Genome
Campus, Hinxton,
Cambridge CB10 1SD, UK.
As used herein, the term "ULBP3", also described as "retinoic acid early
transcript 1N
protein" or "RAET1N", refers to a member of the MHC class I family, including
variants,
isoforms, and species homologs of human ULBP3. The protein functions as a
ligand for
receptor NKG2D. The C terminal membrane association domain in ULBP2 comprises
a GPI
anchoring domain. ULBP3 activates multiple signaling pathways in primary NK
cells. ULBP3
is weakly homologous with MICA and MICB. Exemplary sequence of human ULBP3 is
available as NCBI accession no. NP_078794.1. DNA and protein sequences for
ULBP3 have
been reported by Cosman et al., Immunity (2001) 14(2):123-133, DNA Accession
No.
AF304379 in the EMBL database of the European Bioinformatics Institute,
Wellcome Trust
Genome Campus, Hinxton, Cambridge CB10 1SD, UK.
As used herein, the term "ULBP4", also described as "retinoic acid early
transcript lE
protein" or "RAET1E'', refers to a member of the MHC class I family, including
variants,
isoforms, and species homologs of human ULBP4. The protein functions as a
ligand for
receptor NKG2D. The C terminal region of ULBP4 comprises a transmembrane
domain and a
cytoplasmic domain, (see, e.g., U.S. patent publication US20090274699). ULBP4
is involved
in activating NK cells through its binding to receptor NKG2D and induces NK-
mediated lysis
(see, e.g., Kong et al., Blood (2009) 114(2):310-17). ULBP4 has higher
sequence identity to
ULBP3 than ULBP1 and ULBP2. Exemplary amino acid sequences of human ULBP4 are
available as NCBI accession nos. NP_001230254.1; NP 001230256.1; NP
001230257.1; and
NP 631904.1. As used herein, the term "ULBP5", also described as ''retinoic
acid early
transcript 1G protein" or "RAET1G", refers to a member of the MHC class I
family, including
variants, isoforms, and species homologs of human ULBP5. The C-terminal region
of the
protein has a transmembrane domain and a cytoplasmic domain. ULBP5 is involved
in
activating NK cells and NK cell-mediated cytotoxicity through its binding to
receptor NKG2D.
Exemplary sequence of human ULBP5 is available as NCBI accession no.
NP_001001788.2.
As used herein, the term "ULBP6", also described as "retinoic acid early
transcript 1L
protein" or "RAET1L'', refers to a member of the MHC class I family, including
variants,
isoforms, and species homologs of human ULBP6. ULBP6 contains a GPI anchoring
domain,
similar to ULBP1, ULBP2, and ULBP3. ULBP6 is involved in activating NK cells
and NK
62
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
cell mediated cytotoxicity through its binding to receptor NKG2D. Exemplary
sequence of
human ULBP6 is available as NCBI accession no. NP_570970.2.
As with MICA and MICB, a known function of ULBP proteins is binding to NKG2D
receptor and activating NK cell activity.
MICA is MHC class 1 chain-related gene A protein (MICA), including variants,
isoforms, and homologs of human MICA, and includes fragments of MICA having
functional
MICA activity. MICA protein comprises three extracellular Ig-like domains,
i.e., alpha-1,
alpha-2 and alpha-3, a transmembrane domain, and an intracellular domain. The
protein is
expressed at low levels in cells of the gastric epithelium, endothelial cells
and fibroblasts and
in the cytoplasm of keratinocytes and monocytes. An exemplary sequence of MICA
is available
as NCBI Accession Nos. NP_000238.1. Other exemplary MICA sequences can be
found in
U.S. patent publication 20110311561.
MICB is MHC class I chain-related gene B protein (MICB), including variants,
isoforms, and homologs of human MICB, and includes fragments of MICE having
functional
MICB activity. MICB has about 84% sequence identity to MICA. MICB protein
comprises
three extracellular Ig-like domains, i.e., alpha-1, alpha-2 and alpha-3, a
transmembrane
domain, and an intracellular domain. An exemplary sequence of MICB is
available as
UniProtKB accession number Q29980.1. Other exemplary MICB sequences can be
found in
U.S. patent publication 20110311561.
The NKG2D ligands (ligands for the NKG2D receptor) may also include an anti-
NKG2D antibody or its fragment (e.g., an antigen-binding portion or fragment
thereof),
including, but not limited to, all or part of antibody that specifically
recognizes or binds to
NKG2D. Such antibodies can be monoclonal or polyclonal antibodies. Antibodies
can also be
variant antibodies, such as chimeric antibodies, humanized antibodies, single
chain antibodies,
and hybrid antibodies comprising immunoglobulin chains capable of binding
NKG2D. in
particular embodiments, the antibody comprises a single chain variable
fragment. In particular
embodiments, the antibody is 16F16, 16F31, MS, or 21F2, as set forth in U.S.
Pat. No.
7,879,985, which is hereby incorporated by reference. The antibody fragment
can be any
suitable fragment as discussed herein.
In certain embodiments, the hetero protein or composition comprises a
polypeptide that
binds to a molecule expressed on natural killer (NK) cells that is CD16.
Such a polypeptide may comprise an antigen-binding fragment that binds and
targets
the molecule expressed on NK cells. In some instances, the antigen-binding
fragment can be
a single chain antibody (scFv) specifically binding to the molecule expressed
on NK cells
63
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
A wide variety of antigens may be targeted by the methods and compositions of
the
present disclosure. Monoclonal antibodies to these antigens may be purchased
commercially
or generated using standard techniques, including immunization of an animal
with the antigen
of interest followed by conventional monoclonal antibody methodologies, e.g.,
the standard
somatic cell hybridization technique of Kohler and Milstein, Nature (1975)
256: 495, as
discussed above. The antibodies or nucleic acids encoding for the antibodies
may be sequenced
using any standard DNA or protein sequencing techniques.
In certain embodiments, the polypeptide that binds a molecule expressed on
natural
killer (NK) cells has up to or about 500, up to or about 490, up to or about
480, up to or about
470, up to or about 460, up to or about 450, up to or about 440, up to or
about 430, up to or
about 420, up to or about 410, up to or about 400, up to or about 390, up to
or about 380, up to
or about 370, up to or about 360, up to or about 350, up to or about 340, up
to or about 330, up
to or about 320, up to or about 310, up to or about 200, up to or about 190,
up to or about 180,
up to or about 170, up to or about 160, up to or about 150, up to or about
140, up to or about
130, up to or about 120, up to or about 110, up to or about 100, up to or
about 90, up to or about
80, up to or about 70, up to or about 60, up to or about 50, up to or about
40, up to or about 30,
up to or about 20, up to or about 15, or up to or about 10, amino acid
residues in length. In
certain embodiments, the polypeptide that binds a molecule expressed on
natural killer (NK)
cells has about 100-200, 80-210, 80-250, 150-250, 100-30, 50-200, 150-250, 150-
300, or 150-
190 amino acid residues in length.
Molecules Expressed on T cells
Aspects of the disclosure provide agents targeting a molecule expressed on T
cells.
Such an agent may comprise an antigen-binding fragment that binds and targets
the molecule
expressed on T cells. In some instances, the antigen-binding fragment can be a
single chain
antibody (scFv) specifically binding to the molecule expressed on T cells
A wide variety of antigens may be targeted by the methods and compositions of
the
present disclosure. Monoclonal antibodies to these antigens may be purchased
commercially
or generated using standard techniques, including immunization of an animal
with the antigen
of interest followed by conventional monoclonal antibody methodologies, e.g.,
the standard
somatic cell hybridization technique of Kohler and Milstein, Nature (1975)
256: 495, as
discussed above. The antibodies or nucleic acids encoding for the antibodies
may be sequenced
using any standard DNA or protein sequencing techniques.
In certain embodiments, the antigen-binding fragment that binds a molecule
expressed
on T cells lineage-specific cell-surface antigen (e.g., CD3) has up to or
about 500, up to or
64
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
about 490, up to or about 480, up to or about 470, up to or about 460, up to
or about 450, up to
or about 440, up to or about 430, up to or about 420, up to or about 410, up
to or about 400, up
to or about 390, up to or about 380, up to or about 370, up to or about 360,
up to or about 350,
up to or about 340, up to or about 330, up to or about 320, up to or about
310, up to or about
200, up to or about 190, up to or about 180, up to or about 170, up to or
about 160, up to or
about 150, up to or about 140, up to or about 130, up to or about 120, up to
or about 110, up to
or about 100, up to or about 90, up to or about 80, up to or about 70, up to
or about 60, up to
or about 50, up to or about 40, up to or about 30, up to or about 20, up to or
about 15, or up to
or about 10, amino acid residues in length. In certain embodiments, the
antigen-binding
fragment that binds a molecule expressed on T cells (e.g., CD3) has about 100-
200, 80-210,
80-250, 150-250, 100-30, 50-200, 150-250, 150-300, 300-400, 200-400, 400-500,
or 150-190
amino acid residues in length.
Non-Naturally Occurring Polypeptide Domain
Aspects of the disclosure use a 6DMP heterodimer approach which is based on
four
helices
_________________________________________________________________________ in
some heterodimer designs, each protein monomer has two helices, in others, 3-
to-
1-which create four binding networks along the helix that ultimately favor
only a heterodimer
forming this network. The 6DMP heterodimerization network generates three
hydrogen-bond
networks and one hydrophobic core. The four helices are separated into two
protein sequences,
with each contributing two helices. The highly-specific four-helix structure
forms only
heterodimers between its partner proteins. It is known to be useful in
facilitating intranuclear
processes but has not been tested as a facilitator of cell-to-cell
interactions.
In some embodiments, the non-naturally occurring polypcptide domain comprising
1-
5 alpha helices is 6DMPa (SEQ ID NO: 12). In some embodiments, the non-
naturally
occurring polypeptide domain comprising 1-5 alpha helices is 6DMPh (SEQ ID NO:
16).
See Chen et al. 2019, incorporated in its entirety by reference herein.
Antigen-Binding Fragment
The antigen-binding fragment may be an antibody fragment. The antibody or
antibody
fragment may be any of the immunoglobulin classes (e.g., IgA, IgD, IgE, IgG,
and IgM) and
subclasses, so long as they are capable of binding. In certain embodiments,
the antibody
fragment has an antigen-binding portion. In certain embodiments, antibody
fragments include,
but are not limited to, Fab, F(ab')2, Fab', F(ab)', Fv, a disulfide linked Fv,
single chain Fv
(scFv), bivalent scFv (bi-scFv), trivalent scFv (tri-scFv), Fd, dAb fragment
(e.g., Ward et al.,
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
Nature, (1989) 341:544-546), an isolated CDR, diabodies, affibodies,
triabodies, tetrabodies,
linear antibodies, single-chain antibody molecules. Single chain antibodies
produced by
joining antibody fragments using recombinant methods, or a synthetic linker,
are also
encompassed by the present disclosure. Bird et al. Science, (1988), 242:423-
426. Huston et
al., Proc. Natl. Acad. Sci. USA. (1988), 85:5879-5883. Antibody fragments
comprise only a
portion of an intact antibody, generally including an antigen binding site of
the intact antibody
and thus retaining the ability to bind antigen. Examples of antibody fragments
encompassed by
the present invention include: the Fab fragment, having a light chain variable
domain (VL),
light chain constant domain (CL), heavy chain variable domain (VH), and heavy
chain constant
domain (CH); the Fab' fragment, which is a Fab fragment having one or more
cysteine residues
at the C-terminus of the Cu domain; the Fd fragment having VH and Cu domains;
the Fd'
fragment having VH and CH domains and one or more cysteine residues at the C-
terminus of
the CH domain; the Fv fragment having the VL and VH domains of a single arm of
an antibody;
the dAb fragment (Ward et al., Nature (1989) 341:544-546) which consists of a
VH domain;
isolated CDR regions; F(ab')2 fragments, a bivalent fragment including two
Fab' fragments
linked by a disulphide bridge at the hinge region; single chain antibody
molecules (Bird et al.,
Science (1988) 242:423-426; and Huston et al., PNAS (1988) 85:5879-5883;
diabodies with
two antigen binding sites, comprising a VH domain connected to a VL domain in
the same
polypeptide chain (see, e.g., WO 93/11161 to Whitlow et al. and Hollinger et
al., PNAS (1993)
90:6444-6448; affibodies which are triple helix high affinity peptides (see,
e.g., Nygren, "FEBS
Journal (2008) 275:2668-2676,), and linear antibodies comprising a pair of
tandem Fd
segments (VH-CH1-VH-CH1) which, together with complementary light chain
polypeptides,
form a pair of antigen binding regions (Zapata et al., Protein Eng. (1995)
8(10):1057-1062;
U.S. Patent No. 5,641,870; U.S. Patent No. 8,580,755).
Any antibody or an antigen-binding fragment thereof can he used for
constructing the
agent that targets a lineage-specific cell-surface antigen as described
herein. Such an antibody
or antigen-binding fragment can be prepared by a conventional method, for
example, the
hybridoma technology or recombinant technology.
For example, antibodies specific to a lineage-specific antigen of interest can
be made
by the conventional hybridoma technology. The lineage-specific antigen, which
may be
coupled to a carrier protein such as KLH, can be used to immunize a host
animal for generating
antibodies binding to that complex. The route and schedule of immunization of
the host animal
are generally in keeping with established and conventional techniques for
antibody stimulation
and production, as further described herein. General techniques for production
of mouse,
66
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
humanized, and human antibodies are known in the art and are described herein.
It is
contemplated that any mammalian subject including humans or antibody producing
cells
therefrom can be manipulated to serve as the basis for production of
mammalian, including
human hybridoma cell lines. Typically, the host animal is inoculated
intraperitoneally,
intramuscularly, orally, subcutaneously, intraplantar, and/or intradermally
with an amount of
immunogen, including as described herein.
Hybridomas can be prepared from the lymphocytes and immortalized myeloma cells
using the general somatic cell hybridization technique of Kohler, B. and
Milstein, C. (1975)
Nature 256:495-497 or as modified by Buck, et al., In Vitro, (1982)18:377-381.
Available
myeloma lines, including but not limited to X63-Ag8.653 and those from the
Salk Institute,
Cell Distribution Center, San Diego, Calif., USA, may be used in the
hybridization. Generally,
the technique involves fusing myeloma cells and lymphoid cells using a fusogen
such as
polyethylene glycol, or by electrical means well known to those skilled in the
art. After the
fusion, the cells are separated from the fusion medium and grown in a
selective growth
medium, such as hypoxanthine-aminopterin-thymidine (HAT) medium, to eliminate
unhybridized parent cells. Any of the media described herein, supplemented
with or without
scrum, can be used for culturing hybridomas that secrete monoclonal
antibodies. As another
alternative to the cell fusion technique, EB V immortalized B cells may be
used to produce the
TCR-like monoclonal antibodies described herein. The hybridomas are expanded
and
subcloned, if desired, and supernatants are assayed for anti-immunogen
activity by
conventional immunoassay procedures (e.g., radioimmunoassay, enzyme
immunoassay, or
fluorescence immunoassay).
Hybridomas that may be used as source of antibodies encompass all derivatives,
progeny cells of the parent hybridomas that produce monoclonal antibodies
capable of binding
to a lineage-specific antigen. Hyhridomas that produce such antibodies may be
grown in vitro
or in vivo using known procedures. The monoclonal antibodies may be isolated
from the
culture media or body fluids, by conventional immunoglobulin purification
procedures such as
ammonium sulfate precipitation, gel electrophoresis, dialysis, chromatography,
and
ultrafiltration, if desired. Undesired activity if present, can be removed,
for example, by
running the preparation over adsorbents made of the immunogen attached to a
solid phase and
eluting or releasing the desired antibodies off the immunogen. Immunization of
a host animal
with a target antigen or a fragment containing the target amino acid sequence
conjugated to a
protein that is immunogenic in the species to be immunized, e.g., keyhole
limpet hemocyanin,
serum albumin, bovine thyroglobulin, or soybean trypsin inhibitor using a
bifunctional or
67
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
derivatizing agent, for example maleimidobenzoyl sulfosuccinimide ester
(conjugation through
cysteine residues), N-hydroxysuccinimide (through lysine residues),
glutaraldehyde, succinic
anhydride, SOC1, or R1N=C=NR, where R and R1 are different alkyl groups, can
yield a
population of antibodies (e.g., monoclonal antibodies).
If desired, an antibody of interest (e.g., produced by a hybridoma) may be
sequenced
and the polynucleotide sequence may then be cloned into a vector for
expression or
propagation. The sequence encoding the antibody of interest may be maintained
in vector in a
host cell and the host cell can then be expanded and frozen for future use. In
an alternative,
the polynucleotide sequence may be used for genetic manipulation to "humanize"
the antibody
or to improve the affinity (affinity maturation), or other characteristics of
the antibody. For
example, the constant region may be engineered to more resemble human constant
regions to
avoid immune response if the antibody is used in clinical trials and
treatments in humans. It
may be desirable to genetically manipulate the antibody sequence to obtain
greater affinity to
the lineage-specific antigen. It will be apparent to one of skill in the art
that one or more
polynucleotide changes can be made to the antibody and still maintain its
binding specificity
to the target antigen.
In other embodiments, fully human antibodies can be obtained by using
commercially
available mice that have been engineered to express specific human
immunoglobulin proteins.
Transgenic animals that are designed to produce a more desirable (e.g., fully
human antibodies)
or more robust immune response may also be used for generation of humanized or
human
antibodies. Examples of such technology are XenomouseRTM from Amgen, Inc.
(Fremont,
Calif.) and HuMAb-MouseRTm and TC MouseTm from Medarex, Inc. (Princeton,
N.J.). In
another alternative, antibodies may be made recombinantly by phage display or
yeast
technology. See, for example, U.S. Pat. Nos. 5,565,332; 5,580,717; 5,733,743;
and 6,265,150;
and Winter et al., A nnu. Rev. Itninunol. (1994) 12:433-455. Alternatively,
the ph age display
technology (McCafferty et al., Nature (1990) 348:552-553) can be used to
produce human
antibodies and antibody fragments in vitro, from immunoglobulin variable (V)
domain gene
repertoires from unimmunized donors.
Antigen-binding fragments of an intact antibody (full-length antibody) can be
prepared
via routine methods. For example, F(ab')2 fragments can be produced by pepsin
digestion of
an antibody molecule, and Fab fragments that can be generated by reducing the
disulfide
bridges of F(ab')2 fragments.
Genetically engineered antibodies, such as humanized antibodies, chimeric
antibodies,
single-chain antibodies, and bi-specific antibodies, can be produced via,
e.g., conventional
68
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
recombinant technology. In one example, DNA encoding a monoclonal antibody
specific to
a target antigen can be readily isolated and sequenced using conventional
procedures (e.g., by
using oligonucleotide probes that are capable of binding specifically to genes
encoding the
heavy and light chains of the monoclonal antibodies). The hybridoma cells
serve as a preferred
source of such DNA. Once isolated, the DNA may be placed into one or more
expression
vectors, which are then transfected into host cells such as E. call cells,
simian COS cells,
Chinese hamster ovary (CHO) cells, or myeloma cells that do not otherwise
produce
immunoglobulin protein, to obtain the synthesis of monoclonal antibodies in
the recombinant
host cells. See, e.g., PCT Publication No. WO 87/04462. The DNA can then be
modified, for
example, by substituting the coding sequence for human heavy and light chain
constant
domains in place of the homologous murine sequences, Morrison et al., Proc.
Nat. Acad. Sci.
(1984) 81:6851, or by covalently joining to the immunoglobulin coding sequence
all or part of
the coding sequence for a non-immunoglobulin polypeptide. In that manner,
genetically
engineered antibodies, such as "chimeric" or "hybrid" antibodies; can be
prepared that have
the binding specificity of a target antigen.
Techniques developed for the production of "chimeric antibodies" are well
known in
the art. See, e.g., Morrison et al. Proc. Natl. Acad. Sci. USA (1984) 81:6851;
Neuberger et al.
Nature (1984) 312:604; and Takeda et al. Nature (1984) 314:452.
Methods for constructing humanized antibodies are also well known in the art.
See,
e.g., Queen et al., Proc. Natl. Acad. Sci. USA. (1989) 86:10029-10033. In one
example,
variable regions VI) and VL of a parent non-human antibody are subjected to
three-dimensional
molecular modeling analysis following methods known in the art. Next,
framework amino
acid residues predicted to be important for the formation of the correct CDR
structures are
identified using the same molecular modeling analysis. In parallel, human Vu
and VI_ chains
having amino acid sequences that are homologous to those of the parent non-
human antibody
are identified from any antibody gene database using the parent WI and VL
sequences as search
queries. Human VH and VL acceptor genes are then selected.
The CDR regions within the selected human acceptor genes can be replaced with
the
CDR regions from the parent non-human antibody or functional variants thereof.
When
necessary, residues within the framework regions of the parent chain that are
predicted to be
important in interacting with the CDR regions (see above description) can be
used to substitute
for the corresponding residues in the human acceptor genes.
A single-chain antibody can be prepared via recombinant technology by linking
a
nucleotide sequence coding for a heavy chain variable region and a nucleotide
sequence coding
69
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
for a light chain variable region. Preferably, a flexible linker is
incorporated between the two
variable regions. Alternatively, techniques described for the production of
single chain
antibodies (U.S. Patent Nos. 4,946,778 and 4,704,692) can be adapted to
produce a phage or
yeast scFv library and scFv clones specific to a lineage-specific antigen can
be identified from
the library following routine procedures. Positive clones can be subjected to
further screening
to identify those that bind lineage-specific antigen.
The "percent identity" of two amino acid sequences is determined using the
algorithm
of Karlin and Altschul Proc. Natl. Acad. Sci. USA (1990) 87:2264-68, modified
as in Karlin
and Altschul Proc. Natl. Acad. Sci. USA (1993) 90:5873-77. Such an algorithm
is incorporated
into the NBLAST and XBLAST programs (version 2.0) of Altschul, et al. J. Mol.
Biol. (1990)
215:403-10. BLAST protein searches can be performed with the XBLAST program,
score=50,
wordlength=3 to obtain amino acid sequences homologous to the protein
molecules of the
present disclosure. Where gaps exist between two sequences, Gapped BLAST can
be utilized
as described in Altschul et al., Nucleic Acids Res. (1997) 25(17):3389-3402.
When utilizing
BLAST and Gapped BLAST programs, the default parameters of the respective
programs (e.g.,
XBLAST and NBLAST) can be used.
Nucleic Acids and Vectors
The present disclosure provides for a nucleic acid/polynucleotide encoding any
of the
disclosed polypeptides, engineered proteins or agents. For example,
polynucleotides encoding
any of the proteins described herein are provided, e.g., for recombinant
expression and
purification. In some embodiments, an isolated polynucleotide comprises one or
more
sequences encoding the fusion proteins or agents. The nucleic acid may be
deoxyribonucleic
acid (DNA), ribonucleic acid (RNA) or a DNA/RNA hybrid. The nucleic acid may
be linear
or circular (such as a plasmid). The nucleic acid may be single-stranded,
double-stranded,
branched or modified by the ligation of non-nucleic acid molecules. The
nucleic acids include
nucleic acids produced by recombinant technology.
In certain embodiments, the nucleic acid encoding the disclosed engineered
proteins is
codon optimized. Methods for codon optimization are known in the art.
In certain embodiments, the nucleic acid encoding the aCD33-6DMPa-Hinge
polypeptide has a nucleic acid sequence that is at least 70% identical to SEQ
ID NO: 23 (e.g.,
a nucleic acid sequence that is 70%, 71%, 72%, 73%, 74%, 75%, 76%, 77%, 78%,
79%, 80%,
81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%,
96%,
97%, 98%, 99%, or 100% identical to the nucleic acid sequence in SEQ ID NO:
23).
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
In certain embodiments, the nucleic acid encoding the polypeptide ULBP1-6DMPb-
Hinge has a nucleic acid sequence that is at least 70% identical to SEQ ID NO:
24 (e.g., a
nucleic acid sequence that is 70%, 71%, 72%, 73%, 74%, 75%, 76%, 77%, 78%,
79%, 80%,
81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%,
96%,
97%, 98%, 99%, or 100% identical to the nucleic acid sequence in SEQ Ill NO:
24).
In certain embodiments, the nucleic acid encoding the aCD3-6DMPb polypeptide
has
a nucleic acid sequence that is at least 70% identical to SEQ ID NO: 25 (e.g.,
a nucleic acid
sequence that is 70%, 71%, 72%, 73%, 74%, 75%, 76%, 77%, 78%, 79%, 80%, 81%,
82%,
83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%,
98%,
99%, or 100% identical to the nucleic acid sequence in SEQ ID NO: 25).
In certain embodiments, the nucleic acid is a plasmid DNA including a coding
sequence
for the disclosed polypeptide or agents, together with flanking regulatory
sequences effective
to cause the expression of the fusion polypeptide or agents in cells. Examples
of flanking
regulatory sequences are a promoter sequence sufficient to initiate
transcription and a
terminator sequence sufficient to terminate the gene product, by termination
of transcription or
translation. Suitable transcriptional or translational enhancers can be
included in the vector to
further assist the expression of the fusion polypeptide or agents.
In some embodiments, vectors encoding any of the engineered proteins described
herein are provided, e.g., for recombinant expression and purification. In
some embodiments,
the vector comprises or is engineered to include an isolated polynucleotide,
e.g., those
described herein. Typically, the vector comprises a sequence encoding the
engineered
polypeptide or protein or agents operably linked to a promoter, such that the
engineered protein
(or agents) is (are) expressed in a host cell.
The nucleic acid may be contained within an expression vector. Thus, for
example, a
nucleic acid sequence may he included in any one of a variety of expression
vectors for
expressing one or more polypeptides, and more than one nucleic acid may be
included in one
expression vector. Alternatively, parts of one gene or nucleic acid may be
included in separate
vectors. In some embodiments, vectors include, but are not limited to,
chromosomal,
nonchromosomal and synthetic DNA sequences (e.g., derivatives of SV40,
bacterial plasmids,
phage DNA; baculovirus, yeast plasmids, vectors derived from combinations of
plasmids and
phage DNA, and derivatives of viral DNA).
Vectors of the present disclosure can drive the expression of one or more
sequences in
mammalian cells using a mammalian expression vector. Examples of mammalian
expression
vectors include pCDM8 (Seed, Nature (1987) 329:840) and pMT2PC (Kaufman, et
al., EMBO
71
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
T. (1987) 6:187). When used in mammalian cells, the expression vector's
control functions are
typically provided by one or more regulatory elements. For example, commonly
used
promoters are derived from polyoma, adenovirus 2, cytomegalovirus, simian
virus 40, and
others disclosed herein and known in the art. For other suitable expression
systems for both
prokaryotic and eukaryotic cells see, e.g., Chapters 16 and 17 of Sambrook, et
at.,
MOLECULAR CLONING: A LABORATORY MANUAL. 2nd eds., Cold Spring Harbor
Laboratory, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.,
1989.
The vectors of the present disclosure may direct expression of the nucleic
acid
preferentially in a particular cell type (e.g., tissue-specific regulatory
elements are used to
express the nucleic acid). Such regulatory elements include promoters that may
be tissue-
specific or cell type-specific. The term "tissue-specific" as it applies to a
promoter refers to a
promoter that is capable of directing selective expression of a nucleotide
sequence of interest
to a specific type of tissue in the relative absence of expression of the same
nucleotide sequence
of interest in a different type of tissue. The term "cell type-specific" as
applied to a promoter
refers to a promoter that is capable of directing selective expression of a
nucleotide sequence
of interest in a specific type of cell in the relative absence of expression
of the same nucleotide
sequence of interest in a different type of cell within the same tissue. The
term "cell type-
specific" when applied to a promoter also means a promoter capable of
promoting selective
expression of a nucleotide sequence of interest in a region within a single
tissue. Cell type
specificity of a promoter may be assessed using methods well known in the art,
e.g.,
immunohistochemical staining.
Conventional viral and non-viral based gene transfer methods can be used to
introduce
nucleic acids in mammalian cells or target tissues. Such methods can be used
to administer
nucleic acids encoding the present agents/polypeptides to cells in culture, or
in a subject. Non-
viral vector delivery systems include DNA plasmids, RNA (e.g., a transcript of
a vector
described herein), naked nucleic acid, and nucleic acid complexed with a
delivery vehicle.
Viral vector delivery systems include DNA and RNA viruses, which have either
episomal or
integrated genomes after delivery to the cell.
Viral vectors can be administered directly to patients (in vivo) or they can
be used to
manipulate cells in vitro or ex vivo, where the modified cells may be
administered to patients.
In one embodiment, the present disclosure utilizes viral based systems
including, but not
limited to retroviral, lentivirus, adenoviral, adeno-associated and herpes
simplex virus vectors
for gene transfer. Furthermore, the present disclosure provides vectors
capable of integration
in the host genome, such as retrovirus or lentivirus.
72
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
The vectors of the present disclosure may be delivered to the eukaryotic cell
in a subject.
Any of the chimeric proteins described herein can be prepared by routine
methods,
such as recombinant technology. Methods for preparing the chimeric proteins
herein
involve generation of a nucleic acid that encodes a polypeptide comprising
each of the
fragments/domains/moieties of the chimeric proteins, including the antigen-
binding
fragment and the polypeptide that binds a molecule expressed on natural killer
(NK) cells.
In some embodiments, a nucleic acid encoding each of the components of
chimeric protein
are joined together using recombinant technology.
Sequences of each of the components of the engineered proteins may be obtained
via routine technology, e.g., PCR amplification from any one of a variety of
sources known
in the art. In some embodiments, sequences of one or more of the components of
the
chimeric proteins are obtained from a human cell. Alternatively, the sequences
of one or
more components of the chimeric proteins can be synthesized. Sequences of each
of the
components (e.g., fragments/domains/moieties) can he joined directly or
indirectly (e.g.,
using a nucleic acid sequence encoding a peptide linker) to form a nucleic
acid sequence
encoding the chimeric protein, using methods such as PCR amplification or
ligation.
Alternatively, the nucleic acid encoding the chimeric protein may be
synthesized. In sonic
embodiments, the nucleic acid is DNA. In other embodiments, the nucleic acid
is RNA.
Mutation of one or more residues within one or more of the components of the
chimeric protein (e.g., the antigen-binding fragment, etc.), prior to or after
joining the
sequences of each of the components. In some embodiments, one or more
mutations in a
component of the engineered protein may be made to modulate (increase or
decrease) the
affinity of the component for a target (e.g., the antigen-binding fragment for
the target
antigen) and/or modulate the activity of the component.
Any of the engineered proteins described herein can he introduced into a
suitable cell
for expression via conventional technology.
To express the engineered proteins, expression vectors for stable or transient
expression
of the engineered proteins may be constructed via conventional methods as
described herein.
For example, nucleic acids encoding the chimeric proteins may be cloned into a
suitable
expression vector, such as a viral vector in operable linkage to a suitable
promoter. The nucleic
acids and the vector may be contacted, under suitable conditions, with a
restriction enzyme to
create complementary ends on each molecule that can pair with each other and
be joined with
a ligase. Alternatively, synthetic nucleic acid linkers can be ligated to the
termini of the nucleic
acid encoding the engineered proteins. The synthetic linkers may contain
nucleic acid
73
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
sequences that correspond to a particular restriction site in the vector. The
selection of
expression vectors/plasmids/viral vectors would depend on the type of host
cells for expression
of the chimeric proteins, but should be suitable for integration and
replication in eukaryotic
cells.
A variety of promoters can be used for expression of the engineered proteins
described
herein, including, without limitation, cytomegalovirus (CMV) intermediate
early promoter, a
viral LTR such as the Rous sarcoma virus LTR, HIV-LTR, HTLV-1 LTR, Maloney
murine
leukemia virus (MMLV) LTR, myeoloproliferative sarcoma virus (MPSV) LTR,
spleen focus-
forming virus (SFFV) LTR, the simian virus 40 (SV40) early promoter, herpes
simplex tk virus
promoter, elongation factor 1-alpha (EF1-a) promoter with or without the EF1-a
intron.
Additional promoters for expression of the chimeric proteins include any
constitutively active
promoter. Alternatively, any regulatahle promoter may be used, such that its
expression can
be modulated.
Additionally, the vector may contain, for example, some or all of the
following: a
selectable marker gene, such as the neomycin gene for selection of stable or
transient
transfectants in host cells; enhancer/promoter sequences from the immediate
early gene of
human CMV for high levels of transcription; transcription termination and RNA
processing
signals from S V40 for naRNA stability; 5'-and 3' -untranslated regions for
inRNA stability and
translation efficiency from highly-expressed genes like a-globin or f3-globin;
SV40 polyoma
origins of replication and ColE1 for proper episomal replication; internal
ribosome binding
sites (IRESes), versatile multiple cloning sites; T7 and SP6 RNA promoters for
in vitro
transcription of sense and antisense RNA; a "suicide switch" or "suicide gene"
which when
triggered causes cells carrying the vector to die (e.g., HSV thymidine kinase,
an inducible
caspase such as iCasp9), and reporter gene for assessing expression of the
chimeric protein.
See section VI below. Suitable vectors and methods for producing vectors
containing
transgenes are well known and available in the art. Examples of the
preparation of vectors for
expression of chimeric proteins can be found, for example, in US2014/0106449.
In some embodiments, the engineered protein or the nucleic acid encoding said
engineered protein is a DNA molecule. In some embodiments, the nucleic acid
encoding said
engineered protein is a DNA vector. In some embodiments, the nucleic acid
encoding the
engineered protein is an RNA molecule.
Any of the vectors comprising a nucleic acid sequence that encodes an
engineered
protein described herein is also within the scope of the present disclosure.
Such a vector may
be delivered into host cells by a suitable method. Methods of delivering
vectors to cells are
74
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
well known in the art and may include DNA, RNA, or transposon electroporation,
transfection
reagents such as liposomes or nanoparticles to delivery DNA, RNA, or
transposons; delivery
of DNA, RNA, or transposons or protein by mechanical deformation (see, e.g.,
Sharei et al.
Proc. Natl. Acad. Sci. USA (2013) 110(6):2082-2087); or viral transduction. In
some
embodiments, the vectors for expression of the chimeric proteins are delivered
to host cells by
viral transduction. Exemplary viral methods for delivery include, but are not
limited to,
recombinant retroviruses (see. e.g., PCT Publication Nos. WO 90/07936; WO
94/03622; WO
93/25698; WO 93/25234; WO 93/11230; WO 93/10218; WO 91/02805; U.S. Pat. Nos.
5,219,740 and 4,777,127; GB Patent No. 2,200,651; and EP Patent No. 0 345
242), alphavirus-
based vectors, and adeno-associated virus (AAV) vectors (see, e.g., PCT
Publication Nos. WO
94/12649, WO 93/03769; WO 93/19191; WO 94/28938; WO 95/11984 and WO 95/00655).
In some embodiments, the vectors for expression are retroviruses. In some
embodiments, the
vectors for expression are lentiviruses. In some embodiments, the vectors for
expression are
adeno-associated viruses.
In examples in which the vectors encoding engineered proteins are introduced
to the
host cells using a viral vector, viral particles that are capable of infecting
the cells and carry the
vector may be produced by any method known in the art and can be found, for
example in PCT
Application No. WO 1991/002805A2, WO 1998/009271 Al, and U.S. Patent
6,194,191. The
viral particles are harvested from the cell culture supernatant and may be
isolated and/or
purified prior to contacting the viral particles with the cells.
Therapeutic methods
Any of the disclosed engineered proteins may be administered to a subject to
treat a
condition such as hematopoietic malignancy. Additionally, any of the nucleic
acids/polynucleotides/vectors encoding the disclosed engineered proteins may
he administered
to a subject to treat a condition such as hematopoietic malignancy.
Additionally, any
compositions or pharmaceutical compositions comprising any of the disclosed
engineered
proteins or nucleic acids/polynucleotides/vectors encoding the disclosed
engineered proteins
may be administered to a subject to treat a condition such as hematopoietic
malignancy. As
used herein, "subject," "individual," and "patient" are used interchangeably,
and refer to a
vertebrate, preferably a mammal such as a human. Mammals include, but are not
limited to,
human primates, non-human primates or murine, bovine, equine, canine or feline
species. In
some embodiments, the subject is a human patient having a hematopoietic
malignancy.
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
In some embodiments, the present vectors, engineered proteins or agents may be
mixed
with a pharmaceutically acceptable carrier to form a pharmaceutical
composition, which is also
within the scope of the present disclosure.
The present disclosure provides for a vaccine suitable for eliciting an immune
response
against cancer cells. Method of inhibiting tumor growth by administering the
vaccine of the
invention to a mammal is also described.
The present composition may be delivered to, or administered to be in contact
with, any
suitable types of cells. The cell may a eukaryotic cell. The cell may a
mammalian cell, such as
a human cell or a non-human mammalian cell (e.g., a non-human primate cell).
These include
a number of cell lines that can be obtained from American Tissue Culture
Collection. In certain
embodiments, the cell is a tumor cell.
In certain embodiments, the cell is present in a subject (e.g., a mammal). The
mammal
can be a human or a non-human primate. Non-human primates include, but are not
limited to,
chimpanzees, cyn omol ogous monkeys, spider monkeys, and macaques, e.g.,
Rhesus.
In certain embodiments, the cell may be removed and maintained in tissue
culture in a
primary, secondary, immortalized or transformed state. In certain embodiments,
the cells are
cultured cells or cells freshly obtained from a source (e.g., a tissue, an
organ, a subject, etc.).
The mammalian cell can be primary or secondary which means that it has been
maintained in
culture for a relatively short time after being obtained from an animal
tissue.
To perform the methods described herein, an effective amount of the present
composition may be administered to a subject in need of the treatment. As used
herein the term
"effective amount" may be used interchangeably with the term "therapeutically
effective
amount" and refers to that quantity of a vector, an engineered protein, an
agent, or
pharmaceutical composition that is sufficient to result in a desired activity
upon administration
to a subject in need thereof. Within the context of the present disclosure,
the term "effective
amount" refers to that quantity of a vector, an engineered protein, an agent,
or pharmaceutical
composition that is sufficient to delay the manifestation, arrest the
progression, relieve or
alleviate at least one symptom of a disorder treated by the methods of the
present disclosure.
Effective amounts vary, as recognized by those skilled in the art, depending
on the
particular condition being treated, the severity of the condition, the
individual patient
parameters including age, physical condition, size, gender and weight, the
duration of the
treatment, the nature of concurrent therapy (if any), the specific route of
administration and
like factors within the knowledge and expertise of the health practitioner. In
some
embodiments, the effective amount alleviates, relieves, ameliorates, improves,
reduces the
76
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
symptoms, or delays the progression of any disease or disorder in the subject.
In some
embodiments, the subject is a human. In some embodiments, the subject is a
human patient
having a hematopoietic malignancy.
In some embodiments, the present composition is administered to a subject in
an
amount effective in to reduce the number of target cells (e.g., cancer cells)
by least 20%, e.g.,
50%, 80%, 100%, 2-fold, 5-fold, 10-fold, 20-fold, 50-fold, 100-fold or more.
In one embodiment, the present composition is administered to a subject (e.g.,
human
patient) as an initial dose. One or more subsequent administrations of the
present composition
may be provided to the patient at intervals of 15 days, 14, 13, 12, 11, 10, 9,
8,7, 6, 5,4, 3, or 2
days after the previous administration. More than one dose of the present
composition can be
administered to the subject per week, e.g., 2, 3, 4, or more administrations
of the agent. The
subject may receive more than one doses of the present composition per week,
followed by a
week of no administration of the agent, and finally followed by one or more
additional doses
of the present composition (e.g., more than one administration of the present
composition per
week). The present composition may be administered every other day for 3
administrations
per week for two, three, four, five, six, seven, eight or more weeks.
In the context of the present disclosure insofar as it relates to any of the
disease
conditions recited herein, the terms "treat," "treatment," and the like mean
to relieve or alleviate
at least one symptom associated with such condition, or to slow or reverse the
progression of
such condition. Within the meaning of the present disclosure, the term "treat"
also denotes to
arrest, delay the onset (i.e., the period prior to clinical manifestation of a
disease) and/or reduce
the risk of developing or worsening a disease. For example, in connection with
cancer the term
"treat" may mean eliminate or reduce a patient's tumor burden, or prevent,
delay or inhibit
metastasis.
In some embodiments, the present engineered proteins fusion recognizes (hinds)
a
target cell expressing the cell-surface lineage-specific antigen for targeting
killing.
The efficacy of the present therapeutic methods may be assessed by any method
known
in the art and would be evident to a skilled medical professional. For
example, the efficacy of
the therapy may be assessed by survival of the subject or cancer burden in the
subject or tissue
or sample thereof. In some embodiments, the efficacy of the therapy is
assessed by quantifying
the number of cells belonging to a particular population or lineage of cells.
In some
embodiments, the efficacy of the therapy is assessed by quantifying the number
of cells
presenting the cell-surface lineage-specific antigen.
77
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
The present composition may be administered to a subject in combination with a
second
therapy. The present composition may be administered prior to administration
of the second
therapy. In some embodiments, the present composition is administered at least
about 1 day,
2 days, 3 days, 4 days, 5 days, 6 days, 1 week, 2 weeks, 3 weeks, 4 weeks, 5
weeks, 6 weeks,
7 weeks, 8 weeks, 9 weeks, 10 weeks, 11 weeks, 12 weeks, 3 months, 4 months, 5
months, 6
months or more prior to administration of the second therapy.
In some embodiments, the second therapy is administered prior to the
administration of
the present composition. In some embodiments, the second therapy is
administered at least
about 1 day, 2 days, 3 days, 4 days, 5 days, 6 days, 1 week, 2 weeks, 3 weeks,
4 weeks, 5
weeks, 6 weeks, 7 weeks, 8 weeks, 9 weeks, 10 weeks, 11 weeks, 12 weeks, 3
months, 4
months, 5 months, 6 months or more prior to administration of the present
composition.
In some embodiments, the present composition and the second therapy are
administered
at substantially the same time. In some embodiments, the present composition
is administered,
and the patient is assessed for a period of time, after which the second
therapy is administered.
In some embodiments, the second therapy is administered, and the patient is
assessed for a
period of time, after which the present composition is administered.
Also within the scope of the present disclosure are multiple administrations
(e.g., doses)
of the present composition. In some embodiments, the present composition is
administered to
the subject once. In some embodiments, the present composition is administered
to the subject
more than once (e.g., at least 2, 3, 4, 5, or more times). In some
embodiments, the present
composition is administered to the subject at a regular interval, e.g., every
six months.
In some embodiments, the subject is a human subject having a hematopoietic
malignancy or hematological neoplasm. As used herein a hematopoietic
malignancy refers to
a malignant abnormality involving hematopoietic cells (e.g., blood cells,
including progenitor
and stem cells). Examples of hematopoietic malignancies include, without
limitation,
Hodgkin's lymphoma, non-Hodgkin's lymphoma, leukemia, or multiple myeloma.
Leukemias
include acute myeloid leukemia, acute lymphoid leukemia, chronic myelogenous
leukemia,
acute lymphoblastic leukemia or chronic lymphoblastic leukemia, and chronic
lymphoid
leukemia.
Hematological malignancies or neoplasms include but not limited to, myeloid
malignancies, lymphatic malignancies, malignant histiocytosis and mast cell
leukemia.
The hematopoietic malignancy may be a myeloid malignancy wherein the myeloid
malignancies include but not limited to myeloproliferative disorders (MPD),
myelodysplastic
78
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
syndrome (MDS), myelodysplastic/myeloproliferative disorders (MD/MPD) and
acute
myeloid leukemia (AML).
In certain embodiments, myeloid malignancies refer to a condition associated
with a
defect in the proliferation of a hematopoietic cell. In certain embodiments,
myeloid
malignancies refer to clonal hematological diseases affecting the myeloid
blood lineages,
including chronic and acute conditions. Myeloid malignancies include
myeloproliferative
neoplasms, myelodysplastic syndromes and acute myeloid leukemias. A
myeloproliferative
neoplasm may be primary myelofibrosis (PMF), or essential thrombocythemia
(ET). A
myelodysplastic syndrome may be refractory anemia with ringed sideroblasts and
thrombocythemia (RARS-T). Myeloid malignancies include, but are not limited
to,
myeloproliferative disorders (MPD), myelodysplastic
syndrome (MDS),
myelodyspl astic/myeloprol ferati ve disorders (MD/M PD), and acute myeloi d
leukemi a
(AML).
Lymphatic malignancies include, but are not limited to, T/NK cell tumor, B
cell tumor
and Hodgkin's disease.
In some embodiments, the leukemia is acute myeloid leukemia (AML). AML is
characterized as a heterogeneous, clonal, neoplastic disease that originates
from transformed
cells that have progressively acquired critical genetic changes that disrupt
key differentiation
and growth-regulatory pathways. (Dohner et al. 2015). CD33 glycoprotein is
expressed on the
majority of myeloid leukemia cells as well as on normal myeloid and monocytic
precursors
and has been considered to be an attractive target for AML therapy (Laszlo et
al., Blood Rev.
(2014) 28(4):143-53). While clinical trials using anti-CD33 monoclonal
antibody based
therapy have shown improved survival in a subset of AML patients when combined
with
standard chemotherapy, these effects were also accompanied by safety and
efficacy concerns.
Alternatively or in addition, the methods described herein may he used to
treat non-
hematopoietic cancers, including without limitation: lung cancer; ear, nose
and throat cancer;
colon cancer; melanoma; pancreatic cancer; mammary cancer; prostate cancer;
breast cancer;
ovarian cancer; basal cell carcinoma; biliary tract cancer; bladder cancer;
bone cancer; breast
cancer; cervical cancer; choriocarcinoma; colon and rectum cancer; connective
tissue cancer;
cancer of the digestive system; endometrial cancer; esophageal cancer; eye
cancer; cancer of
the head and neck; gastric cancer; intra-epithelial neoplasm; kidney cancer;
larynx cancer; liver
cancer; fibroma, neuroblastoma; oral cavity cancer (e.g., lip, tongue, mouth,
and pharynx);
ovarian cancer; pancreatic cancer; prostate cancer; retinoblastoma;
rhabdomyosarcoma; rectal
cancer; renal cancer; cancer of the respiratory system; sarcoma; skin cancer;
stomach cancer;
79
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
testicular cancer; thyroid cancer; uterine cancer; cancer of the urinary
system, as well as other
carcinomas and sarcomas.
Carcinomas are cancers of epithelial origin. Carcinomas intended for treatment
with the
methods of the present disclosure include, but are not limited to, acinar
carcinoma, acinous
carcinoma, alveolar adenocarcinoma (also called adenocystic carcinoma,
adenomyoepithelioina, cribriform carcinoma and cylindroma), carcinoma
adenomatosum,
adenocarcinoma, carcinoma of adrenal cortex, alveolar carcinoma, alveolar cell
carcinoma
(also called bronchiolar carcinoma, alveolar cell tumor and pulmonary
adenomatosis), basal
cell carcinoma, carcinoma basocellulare (also called basaloma, or basiloma,
and hair matrix
carcinoma), basaloid carcinoma, basosquamous cell carcinoma, breast carcinoma,
bronchioalveolar carcinoma, bronchiolar carcinoma, bronchogenic carcinoma,
cerebriform
carcinoma, cholangiocellular carcinoma (also called cholangioma and
cholangiocarcinoma),
chorionic carcinoma, colloid carcinoma, comedo carcinoma, corpus carcinoma,
cribriform
carcinoma, carcinoma en cuirasse, carcinoma cutaneum, cylindrical carcinoma,
cylindrical cell
carcinoma, duct carcinoma, carcinoma durum, embryonal carcinoma, encephaloid
carcinoma,
epibulbar carcinoma, epidermoid carcinoma, carcinoma epitheliale adenoides,
carcinoma
exulcere, carcinoma fibrosum, gclatiniform carcinoma, gelatinous carcinoma,
giant cell
carcinoma, gigantocellulare, glandular carcinoma, granulosa cell carcinoma,
hair-matrix
carcinoma, hematoid carcinoma, hepatocellular carcinoma (also called hepatoma,
malignant
hepatoma and hepatocarcinoma), Huirthle cell carcinoma, hyaline carcinoma,
hypernephroid
carcinoma, infantile embryonal carcinoma, carcinoma in situ, intraepidermal
carcinoma,
intraepithelial carcinoma, Krompecher's carcinoma, Kulchitzky-cell carcinoma,
lenticular
carcinoma, carcinoma lenticulare, lipomatous carcinoma, lymphoepithelial
carcinoma,
carcinoma mastitoides, carcinoma medullare, medullary carcinoma, carcinoma
melanodes,
melanotic carcinoma, mucinous carcinoma, carcinoma muciparum, carcinoma
mucocellulare,
mucoepidermoid carcinoma, carcinoma mucosum, mucous carcinoma, carcinoma
rnyxomatodes, nasopharyngeal carcinoma, carcinoma nigruni, oat cell carcinoma,
carcinoma
ossificans, osteoid carcinoma, ovarian carcinoma, papillary carcinoma,
periportal carcinoma,
preinvasive carcinoma, prostate carcinoma, renal cell carcinoma of kidney
(also called
adenocarcinoma of kidney and hypemephoroid carcinoma), reserve cell carcinoma.
carcinoma
sarcomatodes, scheinderian carcinoma, scirrhous carcinoma, carcinoma scroti,
signet-ring cell
carcinoma, carcinoma simplex, small-cell carcinoma, solanoid carcinoma,
spheroidal cell
carcinoma, spindle cell carcinoma, carcinoma spongiosum, squamous carcinoma,
squamous
cell carcinoma, string carcinoma, carcinoma telangiectaticum, carcinoma
telangiectodes,
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
transitional cell carcinoma, carcinoma tuberosum, tuberous carcinoma,
verrucous carcinoma,
carcinoma vilosum.
Sarcomas are mesenchymal neoplasms that arise in bone and soft tissues.
Different
types of sarcomas are recognized and these include: liposarcomas (including
myxoid
liposarcomas and pleiomorphic liposarcomas), leiomyosarcomas,
rhabdomyosarcomas,
malignant peripheral nerve sheath tumors (also called malignant schwannomas,
neurofibrosarcomas, or neurogenic sarcomas), Ewing's tumors (including Ewing's
sarcoma of
bone, extraskeletal (i.e., non-bone) Ewing's sarcoma, and primitive
neuroectodermal tumor
[PNET]), synovial sarcoma, angiosarcomas, hemangiosarcomas,
lymphangiosarcomas,
Kaposi's sarcoma, hemangioendothelioma, fibrosarcoma, desmoid tumor (also
called
aggressive fibromatosis), dermatofibrosarcoma protuberans (DFSP), malignant
fibrous
h isti oc ytom a (MFH) , hem angioperi cytom a, malignant m esenchymom a,
alveolar soft-part
sarcoma, epithelioid sarcoma, clear cell sarcoma, desmoplastic small cell
tumor,
gastrointestinal stromal tumor (GIST) (also known as GI stromal sarcoma),
osteosarcoma (also
known as osteogenic sarcoma)-skeletal and extraskeletal, and chondrosarcoma.
In some embodiments, the cancer to be treated can be a refractory cancer. A
"refractory
cancer," as used herein, is a cancer that is resistant to the standard of care
prescribed. These
cancers may appear initially responsive to a treatment (and then recur), or
they may be
completely non-responsive to the treatment. The ordinary standard of care will
vary depending
upon the cancer type, and the degree of progression in the subject. It may be
a chemotherapy,
or surgery, or radiation, or a combination thereof. Those of ordinary skill in
the art are aware
of such standards of care. Subjects being treated according to the present
disclosure for a
refractory cancer therefore may have already been exposed to another treatment
for their
cancer. Alternatively, if the cancer is likely to be refractory (e.g., given
an analysis of the
cancer cells or history of the subject), then the subject may not have already
been exposed to
another treatment. Examples of refractory cancers include, but are not limited
to, leukemia,
melanomas, renal cell carcinomas, colon cancer, liver (hepatic) cancers,
pancreatic cancer,
Non-Hodgkin's lymphoma and lung cancer.
Any of the present vectors, engineered proteins or agents described herein may
be
administered in a pharmaceutically acceptable carrier or excipient as a
pharmaceutical
composition.
The phrase "pharmaceutically acceptable," as used in connection with
compositions
and/or cells of the present disclosure, refers to molecular entities and other
ingredients of such
compositions that are physiologically tolerable and do not typically produce
untoward reactions
81
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
when administered to a mammal (e.g., a human). Preferably, as used herein, the
term
"pharmaceutically acceptable" means approved by a regulatory agency of the
Federal or a state
government or listed in the U.S. Pharmacopeia or other generally recognized
pharmacopeia for
use in mammals, and more particularly in humans. "Acceptable" means that the
carrier is
compatible with the active ingredient of the composition (e.g., the nucleic
acids, vectors, cells,
or therapeutic antibodies) and does not negatively affect the subject to which
the
composition(s) are administered. Any of the pharmaceutical compositions and/or
cells to be
used in the present methods can comprise pharmaceutically acceptable carriers,
excipients, or
stabilizers in the form of lyophilized formations or aqueous solutions.
Pharmaceutically acceptable carriers, including buffers, are well known in the
art, and
may comprise phosphate, citrate, and other organic acids; antioxidants
including ascorbic acid
and methi nine ; preservatives; low molecular weight polypepti des ;
proteins, such as serum
albumin, gelatin, or immunoglobulins; amino acids; hydrophobic polymers;
monosaccharides;
disaccharides; and other carbohydrates; metal complexes; and/or non-ionic
surfactants. See,
e.g. Remington: The Science and Practice of Pharmacy 20th Ed. (2000)
Lippincott Williams
and Wilkins, Ed. K. E. Hoover.
Kits
Also within the scope of the present disclosure are kits for use of the
present engineered
proteins, agents, vectors, and/or compositions. Such kits may include one or
more containers
comprising present engineered proteins, agents, vectors, and/or compositions.
Some aspects of this disclosure provide kits comprising the present engineered
proteins
or agents. In some embodiments, the kit comprises a polynucleotide encoding
the present
engineered proteins. In some embodiments, the kit comprises a vector for
recombinant protein
expression, wherein the vector comprises a polynucleotide encoding the present
engineered
proteins. In some embodiments, the kit comprises a cell that comprises a
genetic construct for
expressing the present engineered proteins. In some embodiments, the kit
comprises an
excipient and instructions for using the kit. In some embodiments, the
excipient is a
pharmaceutically acceptable excipient.
In some embodiments, the kit can comprise instructions for use in any of the
methods
described herein. The included instructions can comprise a description of
administration of the
pharmaceutical compositions to a subject to achieve the intended activity in a
subject. The kit
may further comprise a description of selecting a subject suitable for
treatment based on
identifying whether the subject is in need of the treatment. In some
embodiments, the
82
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
instructions comprise a description of administering the pharmaceutical
composition to a
subject who is in need of the treatment.
The instructions relating to the use of the pharmaceutical composition
described herein
generally include information as to dosage, dosing schedule, and route of
administration for
the intended treatment. The containers may be unit doses, bulk packages (e.g.,
multi-dose
packages) or sub-unit doses. Instructions supplied in the kits of the
disclosure are typically
written instructions on a label or package insert. The label or package insert
indicates that the
pharmaceutical compositions are used for treating, delaying the onset, and/or
alleviating a
disease or disorder in a subject.
The kits provided herein are in suitable packaging. Suitable packaging
includes, but is
not limited to, vials, bottles, jars, flexible packaging, and the like. Also
contemplated are
packages for use in combination with a specific device, such as an inhaler,
nasal administration
device, or an infusion device. A kit may have a sterile access port (for
example, the container
may be an intravenous solution hag or a vial having a stopper pierceable by a
hypodermic
injection needle). The container may also have a sterile access port.
In some embodiment, the disclosure provides articles of manufacture comprising
contents of the kits described above.
In some embodiments, the individual components of the formulation can be
provided
in one container. Alternatively, it can be desirable to provide the components
of the formulation
separately in two or more containers. The different components can be
combined, e.g.,
according to instructions provided with the kit. The components can be
combined according to
a method described herein, e.g., to prepare and administer a pharmaceutical
composition.
The present engineered proteins, agents, vectors, or compositions can be
provided in
any form, e.g., liquid, dried or lyophilized form.
EXAMPLES
The present invention may be better understood by reference to the following
non-
limiting examples, which are presented in order to more fully illustrate the
preferred
embodiments of the invention. They should in no way be construed to limit the
broad scope
of the invention.
Example 1- Construction of the Engineered Heterodimer Proteins
Initially, pcDNA3.1 expression vectors were used for the aCD33-ULBP1
engineered
heterodimer proteins. All constructs utilized an N-terminal IL-2 secretory
signal to trigger
83
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
export of the completed protein to cell supernatant, and C-terminal tnyc- and
6xHis-tags to
facilitate purification and labeling. In two constructs, ULBP1 immediately
follows the IL-2
signal, followed by a linker and then the VH and VL regions of aCD33. In
another construct,
aCD33 precedes ULBP1, in order to determine whether one orientation is
favorable.
Additionally, constructs have been developed for the aCD33-scFV or 1.1LBP I
alone.
Expression via Mirus TransIT-293 transfection reagent was confirmed using
Western blot probing for Myc following purification of 6xHis-tagged proteins
in
supernatant of transfected HEK-293T cells, as well as Myc probing of cellular
protein. See
Example 2 and Figure 12.
The 6DMPa-monomer was fused to anti-CD33, GFP, and the associated tags (6xHis
and myc) listed above. The 6DMPb monomer was fused to UPBP1, RFP, and 6xHis
and
FLAG tags to facilitate purification independently of the 6DMPa-myc (and to
allow for
separate co-immunoprecipitation experiments).
peDNA3.1 plasmids were also created that encode for the engineered protein
that
attaches anti-CD3 to 6DMPb, with tags to match those of the ULBP1-6DMPb
construct
(Figure 10). In this approach, CD3+ T-cells replaced NK cells as the relevant
effector cell,
and thus 6DMPb was attached in order to bind the anti-CD3 facilitator to the
anti-CD33
targeting moiety. The anti-CD3 sequence was taken from the Amgen CD3-CD19 BiTE
blinotumomab, and fused to 6DMPb via the same linkers used previously (Kufer
et al.
2017). This was co-expressed with aCD33-6DMPa and co-immunoprecipitated to
verify
heterodimerization. The construct was cloned into bacterial or lentiviral
vectors to generate
high-output protein expression methods as noted above.
The latest modification to these designs included the hinge region of IgG2.
With
four cysteine residues creating four disulfide bridges in the span of 12 total
residues, hinge
regions added to the C-terminus of 6DMP-based chimeras were expected to
enhance the
binding of heterodimers and prevent dissociation (Wypych et al. 2008). An
identical IgG2-
hinge was thus added to each construct, following the 6DMPa/b and preceding
purification
tags (Figures 1 and 10). Constructs have been designed that add this hinge
directly or add
this hinge after a short linker.
All constructs were expressed from pcDNA3.4-TOPO vectors with ampicillin
resistance, codon-optimized for expression in Cricetulus griseus (Chinese
hamster). See
Figure 11.
To express the polypeptides/proteins, the amino acid sequences were back
translated
to obtain the DNA sequences which were then codon optimized (SEQ ID NOs: 23-
25).
84
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
Proteins were expressed according to a modified version of the manufacturer
protocol using Lipofectamine 3000.
= aCD33-6DMPa-Hinge was co-expressed in the same dish with aCD3-6DMPb-
Hinge or ULBP1-6DMPb-Hinge
= Proteins were produced CHO-Kl cells in 15-cm tissue-culture treated dishes
10-12 x 106 cells were cultured the day before transfection, 37 C 5% CO2
Reagent volumes per plate were as follows:
135uL Lipofectamine 3000
40ug of each plasmid (aCD33-6DMPa-Hinge and aCD3-6DMPb-
Hinge)
16Oug P3000 reagent (2uL/ug DNA)
Supernatant collected 3-4 days post-transfection
= Cell culture supernatant was purified for His-tag containing proteins
(aCD33-
6DMPa-Hinge and aCD3-6DMPb-Hinge) using TALON Metal Affinity Reason
(TaKaRa Bio)
400uL of suspended resin (200uL of pelleted resin) used per 20mL
supernatant
Typically, 5 plates were used at once, resulting in two 50-mL
batches, each receiving 500uL of washed and equilibrated resin
Manufacturer protocol was followed, using the following buffers in place
of HisTALON buffers:
Wash buffer: TBST-Tris 20mM, NaCl 150mM, 0.1% Tween-20;
Equilibration buffer: Tris 20m1M, NaCl 150mM, 5mM imidazolc,
1mM PMSF;
Elution buffer: NaC1 150mM, 1mM KH2PO4, 3nrIM Na2HPO4-
7H20, 150mM imidazole
Supernatant and resin rotated 30minat 4degC before wash and elution
= 500uL of dimer-bound beads were eluted 3x with Elution Buffer to a final
volume ¨7.5mL
7.5mL of protein-containing elution buffer was diluted to 20rnL final
volume with PBS
20mL of protein-containing buffer was concentrated using Pierce Protein
Concentrator PES (30k MVVCO, 5-20mL size), following manufacturer
protocol
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
Following concentration, samples once again diluted with PBS to
10-20mL, and concentrated once more
30k MWCO cutoff is large enough to keep most of the dimer in
upper chamber, but allows most of smaller monomers to pass to
lower chamber to be discarded
= Concentrated protein was quantified using Bio-Rad Protein Assay
Samples diluted to 0.5mg/mL with PBS, and frozen at -20C until use
Using the above protocol, a third engineered heterodimer protein is made using
the
constructs shown in Figures 3 and 23A, and Figures 5 and 23B.
Example 2 ¨ Expression of the Engineered Heterodimer Proteins
The expression of anti-CD33-ULBP1 engineered heterodimer protein in 293T cells
was
tested using the following protocol. 293T cells mock transfected (No plasmid)
or transfected
with anti-CD33-ULBP1 chimeras 1 and 2 plasmids (as described in Example 1),
and the cell
pellets were lysed in 2X SDS gel loading buffer. The lysate were loaded on a
SDS-PACE gel
and protein separated was transferred to a nylon membrane. The membrane was
probed with
anti-MYC antibody to detect CD33-ULBP1 and anti-Beta Actin to detect the actin
protein
(loading control). The membrane was also probed with a fluoroclu-ome
conjugated secondary
antibody (red to detect anti-MYC; green to detect anti-beta actin) to detect
the primary
antibody.
As shown in Figure 12A, the lanes loaded with lysate from the transfected
cells had
detectable myc protein indicative of the heterodimer protein.
Expression of the anti-CD33-ULBP1 engineered heterodimer protein in the
supernatant
of the 293T cells was also tested used the following protocol. Supernatant of
2931 cells mock
transfected (No plasmid) or transfected with anti-CD33-ULBP1 chimeras 1 and 2
plasmids
were subjected to affinity purification using Talon beads. The input, flow-
through, and the
purified protein (Elute) was then separated on SDS-PAGE gel and transferred to
a nylon
membrane. The membrane was probed with ant-MYC antibody to detect. The
membrane was
also probed with a flurochrome conjugated secondary antibody (red to detect
anti-MYC to
detect the primary antibody. Again as shown in Figure 12B, the lanes loaded
with supernatant
from the transfected cells had detectable myc protein indicative of the
heterodimer protein.
Cell culture supernatants or eluate from TALON resin from the CHO-Kl cells
transfected with chimeras as listed and described in Example 1 were added to
2X SDS loading
buffer, heated, and run on an SDS Page gel. Proteins were transferred to a
PVDF membrane
86
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
and probed with anti-myc antibody to detect anti-CD33-6DMPa-Hinge-MycHis, and
anti-
FLAG antibody to detect ULBP1 -6DMPb-Hinge -FLAG His or anti-CD3-6DMPb-Hinge-
FLAGHis. Fluorochrome-conjugated secondary antibodies were used to probe for
the antiMyc
and antiFLAG antibodies (green and red respectively).
As shown in Figure 13, the cells had detectable myc or flag protein indicative
of the
heterodimer proteins.
Example 3- Binding Experiments
The binding of the engineered heterodimer proteins was tested using the
following
protocol:
Resuspended 50k cells in 50uL FACS Buffer
Add 50uL of engineered heterodimer protein, incubated 30 minutes
Wash
Resuspend in anti-MYC-FITC or anti-FLAG-PE, incubated 30 minutes
Results showed that in HL60 cells incubated with the purified engineered
heterodimer proteins and then with anti-FLAG FITC antibodies there was binding
of both
constructs (Figure 14A). When HL60 cells were incubated with the constructs
and then
anti-MYC FITC antibodies, over 99% of binding was seen for each construct as
well as the
CD33 construct alone, showing that the CD33 construct bound to the cell
surface CD33 well
and was not disrupted by the CD3 or ULBP conjugate (Figure 14B).
Similar results are summarized in Figures 15-17 for binding of the engineered
proteins to HL60 cells, Jurkat cells and PMBCs.
Example 4- Cytotoxic Activity of the Engineered Heterodimer Proteins
NK and CD3 in vitro cytotoxic activity of the engineered heterodimer proteins
was
evaluated using the following general protocol:
1. From frozen human PBMC, CD3+ cytotoxic T-cells were selected using
REAlease CD3 MicroBead Kit (Miltenyi). Dynabeads Human T-activator CD3/CD28
are
used immediately after to activate T-cells for one week.
2. One week after selection and activation, removed Dynabeads from T-cells.
3. Counted MOLM14-dTomato-luciferase cells (AML cell line) and T-cells
(see above). Co-cultured in RPMI (+20% fetal bovine serum, 1% penicillin-
streptomycin)
at a ratio of 1:5 (10,000 MOLM14 cells with 50,000 T-cells).
87
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
4. Added freshly-thawed chimeric proteins to each well (e.g., various
concentrations of anti-CD33-anti-CD3 engineered heterodimer)
5. Cultured overnight at 37 C, 5% CO2.
6. Spun plates (500rpm, 5min), remove supernatant.
7. Washed with 200uL FACS buffer (PBS with: 1% FES, 1mM EDTA)
8. Spun plates (500rpm, 5min), remove supernatant
9. Resuspended in 100uL FACS buffer, add DAPI to final concentration of
0 .lug/mL
10. Proceeded to FACS analysis; gate for single cells, and identify target
cells
as PE+DAPI+ (dTomato in MOLM14 AML cells in PE channel, dead cells in DAPI
channel) See Figure 18A.
11. Compared % of DAPI+ cells among PE+ cells in the T-cell+dimer group,
considering wells with MOLM14 and T-cells (without dimer) as baseline. % cell
death
was normalized to control group with T-cells and M0LM14 cells co-incubated
without
dimer treatment according to the formula below:
[ (death from T-cells and dimer) ¨ (death from T-cells alone) I / [ 100 ¨
(death
from T-cells alone)] *100
In one experiment, MOLM14-dTomato cells were incubated with varying amount
of anti-CD33-anti-CD3 heterodimer (10 ng, 100 ng, 1 lig and 10 g) with 5:1
ratio of T-
cells (effector) as well as control (no heterodimer, no T cells) and
incubation with 10 lag of
heterodimer only and 5:1 T cells only for 24 hours.
As shown in Figure 18B, cytotoxicity was seen for all of the doses of
heterodimer
with the T cells (effector cells) as compared to the controls, dimer alone or
T cells alone and
anti-CD33-anti-CD3 heterodimer enhanced the killing of CD33-expressing target
cells
(MOLM14) by effector T-cells in a dose dependent manner.
In a further experiment using the protocol above. activated T-cells (CD3+)
selected
from PBMCs were co-incubated with CellTrace Violet CD33 expressing HL-60
target cells
for 16 hours, in the presence of anti-CD33-anti-CD3 heterodimer at three
concentrations.
Following incubation, cells were stained with 7-AAD viability dye and analyzed
using flow
cytometry. See Figure 19A. Percent (%) cell death was normalized to control
group with
T-cells and HL-60 cells co-incubated without dimer treatment according to the
formula
below:
[ (death from T-cells and dimer) ¨ (death from T-cells alone) / [ 100 ¨ (death
from
T-cells alone)] *100
88
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
As shown in Figure 19B, anti-CD33-anti-CD3 heterodimer enhances killing of
CD33-expressing target cells (HL60) by effector T-cells in a dose dependent
manner.
An additional cytotoxicity assay was performed as follows: AML cells (HL-60)
were
co-incubated with either monomer of CD3 or CD33 or with anti-CD33-anti-CD33
heterodimer with and without T-cells for 24 hours and viability was measured
using
live/dead staining using 7AAD and flow cytometry as described. Results in
Figure 20 show
that the heterodimers but not the monomers enhanced effector cells (T-cells)
cytotoxic
activity on AML cells.
A further dose dependent cytotoxicity assay was performed as follows: AML
cells
(HL-60) were co-incubated with anti-CD3-anti-CD33 heterodimer at the indicated
amounts
(3, 30, 300 or 30000 rig of heterodimer) with or without T-cells for 24 hours
and viability
was measured using live/dead staining using 7AAD and flow cytometry as
described.
Dose dependent enhancement of effector cell cytotoxic activity of the
heterodimer
on AML cells was seen. There was no appreciable increase in cytotoxicity with
the anti-
CD33-anti-CD3 heterodimer alone (no T-cells) between 3 and 3000 ng, suggesting
the anti-
CD33-anti-CD3 heterodimer does not show cytotoxic by itself and the cytotoxic
effects are
mediated via enhancement of T-cell function. See Figure 21.
Another dose dependent cytotoxicity assay was performed as follows: AML cells
(HL-60) were incubated with 300 ng of anti-CD33-anti-CD3 heterodimer with
varying
ratios of T-cells (effector) (1:1, 2:1, 5:1, and 10:1 effector to target) for
24 hours and viability
was measured using live/dead staining using 7AAD and flow cytometry as
described.
A dose dependent increase in cytotoxicity was seen up to the 5:1 ratio of
effector to
target using 300 ng of heterodimer. There was no appreciable increase in
cytotoxicity
between the ratios of 5:1 and 10:1 effector to target, likely because a
maximal cytotoxicity
was reached. See Figure 22
All the above data suggest that heterodimer was functionally active and
enhanced
the T-cell cytotoxic function towards the AML cells. The data also shows that
the
cytotoxicity of the heterodimer is increase in cells which exhibit a higher
expression of
CD33, such as MOLM14 cells. See Figure 18B versus Figure 19B.
Example 5- In Vivo Anti-Tumor Activity of the Engineered Heterodimer Proteins
The in vivo activity of the anti-CD33-anti-CD3 engineered heterodimer protein
was
tested in mice using the following protocol. All mice received 2x105 MOLM14-
dTomato-
89
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
luciferase cells (AML cell line), then at day 2 and following the below
schedule, one group
received no treatment, one group received unloaded T cells ("unloaded T
cells") and one
group T cells loaded with the anti-CD33-anti-CD3 engineered heterodimer
protein (Bite)
("loaded T cells").
Day 2: 10 million T cells with or without 100ugr anti-CD33-anti-CD3 engineered
heterodimer protein;
Days 6 and 11: 20 million T cells with or without 200ugr anti-CD33-anti-CD3
engineered heterodimer protein
Days 15 and 20: 40 million T cells with or without 400ugr anti-CD33-anti-CD3
engineered heterodimer protein.
Bioluminescence imaging (3LI) to monitor the growth of FFluc-dtomato
transduced
MOLM14 on days 7, 17 and 24. See Figure 24A.
Mice were sacked for analysis when one or more leukemia-related symptoms were
observed such as hunch-backed, significant weight loss, ruffled coat, and limb
paralysis.
As shown in Figures 24B and C, mice treated with loaded T cells had lower
tumor
burden at days 7 and 17.
Additionally, mice treated with the loaded T cells had better survival than
the 2
control groups (no treatment or unloaded T cells). The Kaplan-Meier survival
plot was made
with starting date as the date of MOLM14 injection and end date as date of
death/sacking
for each mouse. See Figure 24D.
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
REFERENCES
Benci, et aL "Tumor Interferon Signaling Regulates a Multigenic Resistance
Program to
Immune Checkpoint Blockade." Cell. 167(6):1540-54. 2016.
Chamuleau, et al. "1-ligh INDO (indoleamine 2,3-dioxygenase) mRNA level in
blasts of
acute myeloid leukemic patients predicts poor clinical outcome."
Haematologica.
93(12):1894-8. 2008.
Chen, et al. "Programmable design of orthogonal protein heterodimers." Nature
565: 106-111.
2019.
Dohner et al., NEJM (2015) 373:1136.
Dulphy, et al. A. "Underground adaptation to a hostile environment: acute
myeloid leukemia
vs. natural killer cells." Front Immunol. 7(94):1-15. 2016.
Elias, et al. "Immune evasion by oncogenic proteins of acute myeloid
leukemia." Blood
123(10):1535-43. 2014.
Krupka, et al. "CD33 target validation and sustained depletion of AML blasts
in long-term
cultures by the hi-specific T-cell-engaging antibody AMG 330." Blood. 123:356-
365. 2014.
Kufer, et al. "Pharmaceutical compositions comprising bispccific anti-CD3,
anti-CD19
antibody constructs for the treatment of B-cell related disorders." Patent
CA2522586. 2017.
Le Dieu, et al. "Peripheral blood T cells in acute myeloid leukemia (AML)
patients at diagnosis
have abnormal phenotype and genotype and form defective immune synapses with
AML
blasts." Blood 114(18):3909-16. 2009.
Lowenberg, et al. Dutch-Belgian Hemato-Oncology Cooperative Group (HOVON).,
German
Austrian AML Study Group (AMLSG)., Swiss Group for Clinical Cancer Research
Collaborative Group (SAKK). "Gemtuzumab ozogamicin as postremission treatment
in AML
at 60 years of age or more: results of a multicenter phase 3 study." Blood.
115(13):2586-91.
2010.
91
CA 03184819 2023- 1- 3
WO 2022/006564
PCT/US2021/040538
Mastaglio, et al. "Natural killer receptor ligand expression on acute myeloid
leukemia impacts
survival and relapse after chemotherapy". Blood Adv. 2(4):335-346. 2018.
Mundy-Bosse, et al. "Acute Myeloid Leukemia Alters Natural Killer Cell
Maturation and
Functional Activation." Blood. 124(21):754. 2014.
Nanbakhsh, et al. "c-Myc regulates expression of NKG2D ligands ULBP1/2/3 in
AML and
modulates their susceptibility to NK-mediated lysis". Immunobio. 123(23):3585-
3596. 2014.
Orleans-Lindsay, et al. "Acute myeloid leukaemia cells secrete a soluble
factor that inhibits T
and NK cell proliferation but not cytolytic function ¨ implications for the
adoptive
immunotherapy of leukaemia." Clin Exp Immunol. 126(3):403-11. 2001.
Renneville, et al. "Clinical impact of gene mutations and lesions detected by
SNP-array
karyotyping in acute myeloid leukemia patients in the context of gemtuzumab
ozogamicin
treatment: results of the ALFA-0701 trial." Oncotarget. 5(4):916-932. 2014.
Schnorfeil, et al. "T cells are functionally not impaired in AML: increased PD-
1 expression is
only seen at time of relapse and correlates with a shift towards the memory T
cell
compartment." J Hematol Oncol. 8:93. 2015.
Shenghui, et al. "Elevated frequencies of CD4(+) CD25(+) CD12710 regulatory T
cells is
associated to poor prognosis in patients with acute myeloid leukemia." Int J
Cancer.
129(6) :1373-81. 2011.
Stringaris, et al. "Leukemia-induced phenotypic and functional defects in
natural killer cells
predict failure to achieve remission in acute myeloid leukemia."
Haematologica. 99(5):836-
47. 2014.
Thor sson, et al. ''The Immune Landscape of Cancer." Immunity. 48(4):812-30.
2018.
Wypych., et al. "Human IgG2 Antibodies Display Disulfide-mediated Structural
Isoforms." J
Biol Chem. 283:16194-205. 2008.
92
CA 03184819 2023- 1- 3