Note: Descriptions are shown in the official language in which they were submitted.
WO 2022/006246
PCT/US2021/039849
METHODS AND SYSTEMS FOR EFFICIENT SAMPLE POOLING FOR
DIAGNOSTIC TESTING
CROSS-REFERENCE
100011 This application claims the benefit of U.S. Provisional
Patent Application No.
63/047,630, filed July 2, 2020, which is incorporated by reference herein in
its entirety.
SUMMARY
100021 Methods and systems are provided for performing or directing
the pooling of a
plurality of bodily samples. Bodily samples, health data, contact tracing
data, location data,
and/or movement data may be collected from a plurality of subjects (e.g.,
patients), and
trained computer algorithms may be used to efficiently perform or direct the
pooling of the
bodily samples into a plurality of sample pools for diagnostic testing. Such
efficient sample
pooling can be used to perform frequent and widespread diagnostic testing of
an infectious
disease across a population, which may be essential for containment and
mitigation,
especially in cases of pandemic outbreaks (e.g., COVID-19).
100031 In an aspect, the present disclosure provides a method
comprising: (a) obtaining a
plurality of: health data, contact tracing data, location data, movement data,
or any
combination thereof associated with a plurality of subjects; and (b)
processing the plurality
of: health data, contact tracing data, location data, movement data, or any
combination
thereof with a trained computer algorithm to at least some individual subjects
of the plurality
of subjects to a pool from among a plurality of pools, wherein a number of
pools of the
plurality of pools is less than a number of subjects of the plurality of
subjects.
100041 In some embodiments, the method further comprises outputting
an electronic
recommendation to create, for each of at least two given pools of the
plurality of pools, a
pooled sample by combining bodily samples or portions thereof obtained from
subjects in the
at least two given pools. In some embodiments, the method further comprises
creating, for
each of at least two given pools of the plurality of pools, a pooled sample by
combining
bodily samples or portions thereof obtained from subjects in the at least two
given pools. In
some embodiments, the method further comprises obtaining the bodily samples or
portions
thereof from the plurality of subjects.
100051 In some embodiments, the bodily samples are individually selected from
the group
consisting of: nasopharyngeal swab, oropharyngeal swab, blood, serum, plasma,
vitreous,
sputum, urine, stool, tears, perspiration, saliva, semen, mucosal excretions,
mucus, spinal
-1 -
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
fluid, cerebrospinal fluid (CSF), pleural fluid, peritoneal fluid, amniotic
fluid, lymph fluid,
eye swab, cheek swab, vaginal swab, cervical swab, rectal swab, cells, and
tissue.
100061 In some embodiments, the method further comprises isolating nucleic
acids from the
bodily samples, and creating, for a given pool of the plurality of pools, the
pooled sample by
combining at least some of nucleic acids isolated from bodily samples obtained
from the
subjects in the given pool. In some embodiments, the method further comprises
enriching the
nucleic acids for a plurality of genomic regions. In some embodiments, the
method further
comprises amplifying at least some of the nucleic acids. In some embodiments,
the
amplification comprises selective amplification. In some embodiments, the
amplification
comprises universal amplification. In some embodiments, enriching at least
some of the
nucleic acids for the plurality of genomic regions comprises contacting the
nucleic acids with
a plurality of probes, each of the plurality of probes having sequence
complementarity with at
least a portion of a genomic region of the plurality of genomic regions In
some
embodiments, the plurality of genomic regions comprises genomic regions
associated with a
disease or disorder. In some embodiments, the disease or disorder comprises
coronavirus
disease 2019 (COVID-19), human immunodeficiency virus (HIV), or malaria. In
some
embodiments, the disease or disorder comprises COVID-19.
100071 In some embodiments, the method further comprises performing a
plurality of
diagnostic tests on the plurality of pooled samples to obtain a plurality of
diagnostic results
associated with the plurality of pooled samples. In some embodiments, the
plurality of
diagnostic tests are configured to detect a presence or absence of a disease
or disorder based
on analyzing at least the plurality of pooled samples. In some embodiments,
the disease or
disorder comprises coronavirus disease 2019 (COVID-19), human immunodeficiency
virus
(HIV), or malaria. In some embodiments, the disease or disorder comprises
COV1D-19.
100081 In some embodiments, the method further comprises, for a given pool
among the
plurality of pools, detecting the absence of the disease or disorder in each
of the individual
subjects of the given pool when the absence of the disease or disorder is
detected based on
analyzing the pooled sample corresponding to the given pool. In some
embodiments, the
method further comprises, for a given pool among the plurality of pools,
testing each of the
individual subjects of the given pool for the disease or disorder when the
presence of the
disease or disorder is detected based on analyzing the pooled sample
corresponding to the
given pool. In some embodiments, the method further comprises, for a given
pool among the
plurality of pools, testing each of a plurality of sub-pools of the given pool
for the disease or
-2-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
disorder when the presence of the disease or disorder is detected based on
analyzing the
pooled sample corresponding to the given pool.
100091 In some embodiments, the method further comprises detecting the
presence or
absence of the disease or disorder in the plurality of subjects with a
clinical sensitivity of at
least about 50%. In some embodiments, the method further comprises detecting
the presence
or absence of the disease or disorder in the plurality of subjects with a
clinical sensitivity of at
least about 70%. In some embodiments, the method further comprises detecting
the presence
or absence of the disease or disorder in the plurality of subjects with a
clinical sensitivity of at
least about 90%. In some embodiments, the method further comprises detecting
the presence
or absence of the disease or disorder in the plurality of subjects with a
clinical sensitivity of at
least about 50%, at least about 55%, at least about 60%, at least about 65%,
at least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least
about 91%, at least about 92%, at least about 93%, at least about 94%, at
least about 95%, at
least about 96%, at least about 97%, at least about 98%, at least about 99%,
or 100%.
100101 In some embodiments, the method further comprises detecting the
presence or
absence of the disease or disorder in the plurality of subjects with a
clinical specificity of at
least about 50%. In some embodiments, the method further comprises detecting
the presence
or absence of the disease or disorder in the plurality of subjects with a
clinical specificity of
at least about 70%. In some embodiments, the method further comprises
detecting the
presence or absence of the disease or disorder in the plurality of subjects
with a clinical
specificity of at least about 90%. In some embodiments, the method further
comprises
detecting the presence or absence of the disease or disorder in the plurality
of subjects with a
clinical specificity of at least about 50%, at least about 55%, at least about
60%, at least about
65%, at least about 70%, at least about 75%, at least about 80%, at least
about 85%, at least
about 90%, at least about 91%, at least about 92%, at least about 93%, at
least about 94%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
at least about
99%, or 100%.
100111 In some embodiments, the method further comprises detecting the
presence or
absence of the disease or disorder in the plurality of subjects with a
positive predictive value
(PPV) of at least about 50%. In some embodiments, the method further comprises
detecting
the presence or absence of the disease or disorder in the plurality of
subjects with a positive
predictive value (PPV) of at least about 70%. In some embodiments, the method
further
comprises detecting the presence or absence of the disease or disorder in the
plurality of
subjects with a positive predictive value (PPV) of at least about 90%. In some
embodiments,
-3 -
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
the method further comprises detecting the presence or absence of the disease
or disorder in
the plurality of subjects with a positive predictive value (PPV) of at least
about 50%, at least
about 55%, at least about 60%, at least about 65%, at least about 70%, at
least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about
92%, at least about 93%, at least about 94%, at least about 95%, at least
about 96%, at least
about 97%, at least about 98%, at least about 99%, or 100%.
100121 In some embodiments, the method further comprises detecting the
presence or
absence of the disease or disorder in the plurality of subjects with a
negative predictive value
(NPV) of at least about 50%. In some embodiments, the method further comprises
detecting
the presence or absence of the disease or disorder in the plurality of
subjects with a negative
predictive value (NPV) of at least about 70%. In some embodiments, the method
further
comprises detecting the presence or absence of the disease or disorder in the
plurality of
subjects with a negative predictive value (NPV) of at least about 90% In some
embodiments,
the method further comprises detecting the presence or absence of the disease
or disorder in
the plurality of subjects with a negative predictive value (NPV) of at least
about 50%, at least
about 55%, at least about 60%, at least about 65%, at least about 70%, at
least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about
92%, at least about 93%, at least about 94%, at least about 95%, at least
about 96%, at least
about 97%, at least about 98%, at least about 99%, or 100%.
100131 In some embodiments, the method further comprises detecting the
presence or
absence of the disease or disorder in the plurality of subjects with an area
under the curve
(AUC) of at least about 0.60. In some embodiments, the method further
comprises detecting
the presence or absence of the disease or disorder in the plurality of
subjects with an area
under the curve (AUC) of at least about 0.70. In some embodiments, the method
further
comprises detecting the presence or absence of the disease or disorder in the
plurality of
subjects with an area under the curve (AUC) of at least about 0.80. In some
embodiments, the
method further comprises detecting the presence or absence of the disease or
disorder in the
plurality of subjects with an area under the curve (AUC) of at least about
0.90. In some
embodiments, the method further comprises detecting the presence or absence of
the disease
or disorder in the plurality of subjects with an area under the curve (AUC) of
at least about
0.50, at least about 0.55, at least about 0.60, at least about 0.65, at least
about 0.70, at least
about 0.75, at least about 0.80, at least about 0.85, at least about 0.90, at
least about 0.91, at
least about 0.92, at least about 0.93, at least about 0.94, at least about
0.95, at least about
0.96, at least about 0.97, at least about 0.98, or at least about 0.99.
-4-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
100141 In some embodiments, the plurality of health data, contact tracing
data, location data,
movement data, or any combination thereof associated with the plurality of
subjects
comprises de-identified data. In some embodiments, the plurality of health
data comprises a
diagnosis of a disease or disorder, a prognosis of a disease or disorder, a
risk of having a
disease or disorder, a treatment history of a disease or disorder, a history
of previous
treatment for a disease or disorder, a history of prescribed medications, a
history of
prescribed medical devices, age, height, weight, sex, smoking status, one or
more symptoms,
and one or more vital signs. In some embodiments, the one or more vital signs
comprise one
or more of: heart rate, heart rate variability, blood pressure, respiratory
rate, blood oxygen
concentration (Sp02), carbon dioxide concentration in respiratory gases, a
hormone level,
sweat analysis, blood glucose, body temperature, impedance, conductivity,
capacitance,
resistivity, electromyography, galvanic skin response, neurological signals,
el ectroencephal ography, electrocardiography, immunology markers, and other
physiological
measurements.
100151 In some embodiments, the trained computer algorithm comprises a trained
machine
learning classifier. In some embodiments, the trained machine learning
classifier comprises
an algorithm selected from the group consisting of: a support vector machine,
a neural
network, a random forest, a linear regression, a logistic regression, a
Bayesian classifier, a
boosted classifier, a gradient boosting algorithm, an adaptive boosting
(AdaBoost) algorithm,
and an extreme gradient boosting (XGBoost) algorithm.
100161 In some embodiments, the method further comprises processing health
data, contact
tracing data, location data, or movement data of the individual subject with
the trained
computer algorithm to determine an expected prevalence of a disease or
disorder, and
assigning the individual subject of the plurality of subjects to the pool from
among the
plurality of pools based at least in part on the determined expected
prevalence of the disease
or disorder. In some embodiments, the method further comprises assigning the
individual
subject of the plurality of subjects to the pool from among the plurality of
pools when the
determined expected prevalence of the disease or disorder is less than a pre-
determined
prevalence threshold. In some embodiments, the pre-determined prevalence
threshold is
about 5%, about 10%, about 15%, about 20%, about 25%, about 30%, about 35%,
about
40%, about 45%, or about 50%. In some embodiments, the method further
comprises
determining a maximum pool size based on the determined expected prevalence of
the
disease or disorder, and assigning the individual subject of the plurality of
subjects to the pool
from among the plurality of pools based on the maximum pool size. In some
embodiments,
-5-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
the method further comprises determining an expected level of severity of
symptoms of the
plurality of subjects. In some embodiments, the expected level of severity of
symptoms
comprises a moderate level of symptoms, an intermediate level of symptoms, or
a severe
level of symptoms.
[0017] In some embodiments, the number of pools of the plurality of pools is
reduced
relative to the number of subjects of the plurality of subjects by at least
50%. In some
embodiments, the number of pools of the plurality of pools is reduced relative
to the number
of subjects of the plurality of subjects by at least 100%. In some
embodiments, the number of
pools of the plurality of pools is reduced relative to the number of subjects
of the plurality of
subjects by at least 200%. In some embodiments, the number of pools of the
plurality of
pools is reduced relative to the number of subjects of the plurality of
subjects by at least
300%. In some embodiments, the number of pools of the plurality of pools is
reduced relative
to the number of subjects of the plurality of subjects by at least about 10%,
at least about
20%, at least about 30%, at least about 40%, at least about 50%, at least
about 60%, at least
about 70, at least about 80%, at least about 90%, at least about 100%, at
least about 110%, at
least about 120%, at least about 130%, at least about 140%, at least about
150%, at least
about 160%, at least about 170%, at least about 180%, at least about 190%, at
least about
200%, at least about 210%, at least about 220%, at least about 230%, at least
about 240%, at
least about 250%, at least about 260%, at least about 270%, at least about
280%, at least
about 290%, at least about 300%, at least about 310%, at least about 320%, at
least about
330%, at least about 340%, at least about 350%, at least about 360%, at least
about 370%, at
least about 380%, at least about 390%, at least about 400%, or more than about
400%.
[0018] In some embodiments, the method further comprises, based on the
detected presence
or absence of the disease or disorder in the plurality of subjects,
administering a
therapeutically effective dose of a treatment to treat the disease or disorder
detected in at least
a subset of the plurality of subjects.
[0019] In another aspect, the present disclosure provides a computer system,
comprising: a
database that is configured to store a plurality of: health data, contact
tracing data, location
data, movement data, or any combination thereof associated with a plurality of
subjects; and
one or more computer processors operatively coupled to the database, wherein
the one or
more computer processors are individually or collectively programmed to:
process the
plurality of: health data, contact tracing data, location data, movement data,
or any
combination thereof with a trained computer algorithm to assign at least some
individual
subjects of the plurality of subjects to a pool from among a plurality of
pools, wherein a
-6-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
number of pools of the plurality of pools is less than a number of subjects of
the plurality of
subjects.
[0020] Another aspect of the present disclosure provides a non-
transitory computer-
readable medium comprising machine-executable code that, upon execution by one
or more
computer processors, implements any of the methods above or elsewhere herein.
[0021] Another aspect of the present disclosure provides a system
comprising one or
more computer processors and computer memory coupled thereto. The computer
memory
comprises machine-executable code that, upon execution by the one or more
computer
processors, implements any of the methods above or elsewhere herein.
[0022] Additional aspects and advantages of the present disclosure
will become readily
apparent to those skilled in this art from the following detailed description,
wherein only
illustrative embodiments of the present disclosure are shown and described. As
will be
realized, the present disclosure is capable of other and different
embodiments, and its several
details are capable of modifications in various obvious respects, all without
departing from
the disclosure. Accordingly, the drawings and description are to be regarded
as illustrative in
nature, and not as restrictive.
INCORPORATION BY REFERENCE
[0023] All publications, patents, and patent applications mentioned
in this specification
are herein incorporated by reference to the same extent as if each individual
publication,
patent, or patent application was specifically and individually indicated to
be incorporated by
reference. To the extent publications and patents or patent applications
incorporated by
reference contradict the disclosure contained in the specification, the
specification is intended
to supersede and/or take precedence over any such contradictory material.
BRIEF DESCRIPTION OF THE DRAWINGS
[0024] The novel features of the invention are set forth with
particularity in the appended
claims. A better understanding of the features and advantages of the present
invention will be
obtained by reference to the following detailed description that sets forth
illustrative
embodiments, in which the principles of the invention are utilized, and the
accompanying
drawings (also "Figure" and "FIG." herein), of which.
[0025] FIG. lA shows an example method for performing sample
pooling of a plurality
of clinical samples for diagnostic testing.
-7-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
100261 FIG. 1B shows a general workflow for performing sample
pooling of a plurality
of clinical samples for diagnostic testing, including data input, consuming
and consolidating
data, performing a method for sample pooling, recommending pooling protocols,
and
performing diagnostic testing on the sample pools.
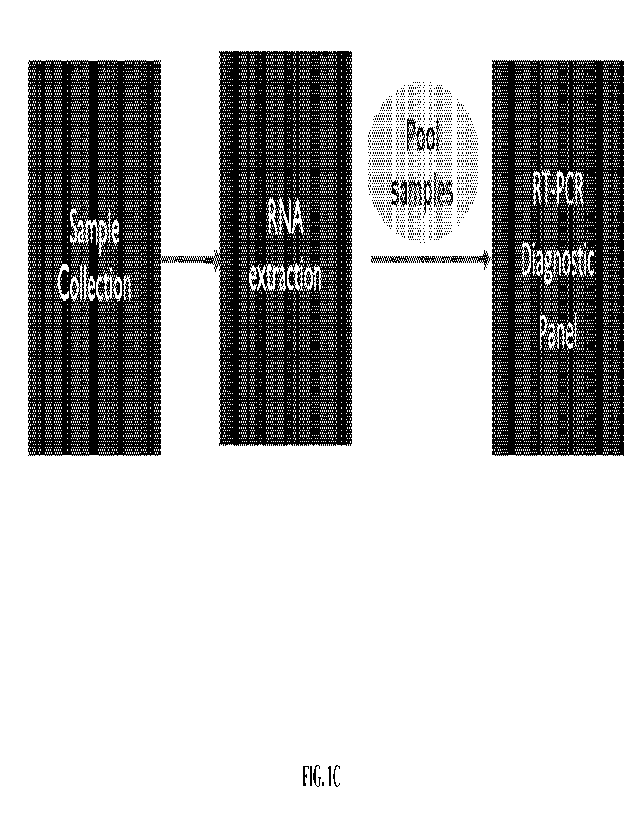
100271 FIG. IC shows a workflow for performing sample handling and
testing.
100281 FIG. ID shows an example of a system procedure diagram of
methods and
systems of the present disclosure.
100291 FIG. IE shows an example of a user activity diagram of
methods and systems of
the present disclosure.
100301 FIG. 2A shows an example of sample pooling of a plurality of
clinical samples
for diagnostic testing If a sample pool comprising a plurality of individual
clinical samples
receives a negative clinical test outcome, then all the individual clinical
samples in the
sample pool must also have a negative clinical test outcome Conversely, if the
sample pool
comprising the plurality of individual clinical samples receives a positive
clinical test
outcome, then either the individual clinical samples can be individually
tested, or the sample
pool can be further sub-divided into smaller subsets and the sample pooling
process can be
repeated.
100311 FIG. 2B shows an example of how methods and systems of the
present disclosure
advantageously use sample pooling. For example, currently about 2 out of 10
test results
return positive; patterns can be learned via machine learning about the 8 out
of 10 negative
patients. Using methods and systems of the present disclosure, predictive
analytics and
machine learning may be applied to identify patients with a lower prevalence
rate. A plurality
of predicted negative samples may be pooled together to create sample pool,
and a single
diagnostic test may be performed on the sample pool.
100321 FIG. 3 shows a computer system that is programmed or
otherwise configured to
implement methods provided herein
100331 FIG. 4A shows the number of people who were able to be
tested with 1,000
diagnostic tests, using Boston vs. Cambridge cohort information (left 3
columns), clinical
symptom information (middle 3 columns), and contact tracing information (right
3 columns)
Within each set of 3 columns, the number of people who were able to be tested
with 1,000
diagnostic tests is indicated using no sample pooling (left), simple sample
pooling (middle),
and intelligent sample pooling based on methods and systems of the present
disclosure
(right).
-8-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
100341 FIG. 4B shows that using intelligent sample pooling based on
methods and
systems of the present disclosure, the rules for diagnostic testing can be
redefined.
100351 FIG. 5A shows the percentage of test savings that is
achieved using simple
sample pooling (gray), using intelligent sample pooling based on methods and
systems of the
present disclosure (purple), and an upper bound achieved using intelligent
sample pooling
(dashed).
100361 FIG. 5B shows the percentage increase in diagnostic testing
capacity that is
achieved using simple sample pooling (gray), using intelligent sample pooling
based on
methods and systems of the present disclosure (purple), and an upper bound
achieved using
intelligent sample pooling (dashed).
100371 FIG. 6 shows a relative number of diagnostic tests that is
required versus the
sample pool size (ranging from 1 sample to 20 samples per pool), at a
prevalence rate (PR) of
1% (light blue), 5% (orange), 10% (gray), 20% (yellow), and 30% (dark blue)
100381 FIG. 7 shows that using methods and systems of the present
disclosure, an
optimal pool size of 4 samples per pool was selected, and a 40% reduction in
diagnostic test
kit utilization was achieved, along with a clinical sensitivity of 80% and a
clinical specificity
of 96%.
DETAILED DESCRIPTION
100391 The term "nucleic acid," or "polynucleotide," as used
herein, generally refers to a
molecule comprising one or more nucleic acid subunits, or nucleotides. A
nucleic acid may
include one or more nucleotides selected from adenosine (A), cytosine (C),
guanine (G),
thymine (T) and uracil (U), or variants thereof A nucleotide generally
includes a nucleoside
and at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more phosphate (P03) groups. A
nucleotide can
include a nucleobase, a five-carbon sugar (either ribose or deoxyribose), and
one or more
phosphate groups, individually or in combination.
100401 Ribonucleotides are nucleotides in which the sugar is
ribose.
Deoxyribonucleotides are nucleotides in which the sugar is deoxyribose A
nucleotide can be
a nucleoside monophosphate or a nucleoside polyphosphate A nucleotide can be a
deoxyribonucleoside polyphosphate, such as, e.g., a deoxyribonucleoside
triphosphate
(dNTP), which can be selected from deoxyadenosine triphosphate (dATP),
deoxycytosine
triphosphate (dCTP), deoxyguanosine triphosphate (dGTP), uridine triphosphate
(dUTP) and
deoxythymidine triphosphate (dTTP) dNTPs, that include detectable tags, such
as
luminescent tags or markers (e.g., fluorophores). A nucleotide can include any
subunit that
-9-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
can be incorporated into a growing nucleic acid strand. Such subunit can be an
A, C, G, T, or
U, or any other subunit that is specific to one or more complementary A, C, G,
T or U, or
complementary to a purine (i.e., A or G, or variant thereof) or a pyrimidine
(i.e., C, T or U, or
variant thereof). In some examples, a nucleic acid is deoxyribonucleic acid
(DNA),
ribonucleic acid (RNA), or derivatives or variants thereof. A nucleic acid may
be single-
stranded or double stranded. A nucleic acid molecule may be linear, curved, or
circular or any
combination thereof
100411 The terms "nucleic acid molecule," "nucleic acid sequence,"
"nucleic acid
fragment," "oligonucleotide" and "polynucleotide," as used herein, generally
refer to a
polynucleotide that may have various lengths, such as either
deoxyribonucleotides or
ribonucleotides (RNA), or analogs thereof. A nucleic acid molecule can have a
length of at
least about 5 bases, 10 bases, 20 bases, 30 bases, 40 bases, 50 bases, 60
bases, 70 bases, 80
bases, 90, 100 bases, 110 bases, 120 bases, 130 bases, 140 bases, 150 bases,
160 bases, 170
bases, 180 bases, 190 bases, 200 bases, 300 bases, 400 bases, 500 bases, 1
kilobase (kb), 2
kb, 3, kb, 4 kb, 5 kb, 10 kb, or 50 kb or it may have any number of bases
between any two of
the aforementioned values. An oligonucleotide is typically composed of a
specific sequence
of four nucleotide bases: adenine (A); cytosine (C); guanine (G); and thymine
(T) (uracil (U)
for thymine (T) when the polynucleotide is RNA). Thus, the terms "nucleic acid
molecule,"
"nucleic acid sequence," "nucleic acid fragment," "oligonucleotide" and
"polynucleotide" are
at least in part intended to be the alphabetical representation of a
polynucleotide molecule.
Alternatively, the terms may be applied to the polynucleotide molecule itself.
This
alphabetical representation can be input into databases in a computer having a
central
processing unit and/or used for bioinformatics applications such as functional
genomics and
homology searching. Oligonucleotides may include one or more nonstandard
nucleotide(s),
nucleotide analog(s) and/or modified nucleotides.
100421 The term "sample," as used herein, generally refers to a
biological sample.
Examples of biological samples include nucleic acid molecules, amino acids,
polypeptides,
proteins, carbohydrates, fats, or viruses. In an example, a biological sample
is a nucleic acid
sample including one or more nucleic acid molecules. The nucleic acid
molecules may be
cell-free or cell-free nucleic acid molecules, such as cell-free DNA (cfDNA)
or cell-free
RNA (cfRNA). The nucleic acid molecules may be derived from a variety of
sources
including human, mammal, non-human mammal, ape, monkey, chimpanzee, reptilian,
amphibian, or avian, sources. Further, samples may be extracted from a variety
of animal
fluids containing cell-free sequences, including but not limited to bodily
fluid samples such
-10-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
as blood, serum, plasma, vitreous, sputum, urine, tears, perspiration, saliva,
semen, mucosal
excretions, mucus, spinal fluid, cerebrospinal fluid (CSF), pleural fluid,
peritoneal fluid,
amniotic fluid, lymph fluid, and the like. Cell free polynucleotides (e.g.,
cfDNA) may be
fetal in origin (via fluid taken from a pregnant subject), or may be derived
from tissue of the
subject itself.
100431 The term "subject," as used herein, generally refers to an
individual having a
biological sample that is undergoing processing or analysis. A subject can be
an animal or
plant. The subject can be a mammal, such as a human, dog, cat, horse, pig or
rodent. The
subject can be a patient, e.g., have or be suspected of having a disease, such
as one or more
cancers (e.g., brain cancer, breast cancer, cervical cancer, colorectal
cancer, endometrial
cancer, esophageal cancer, gastric cancer, hepatobiliary tract cancer,
leukemia, liver cancer,
lung cancer, lymphoma, ovarian cancer, pancreatic cancer, skin cancer, urinary
tract cancer),
one or more infectious diseases, one or more genetic disorder, or one or more
tumors, or any
combination thereof. For subjects having or suspected of having one or more
tumors, the
tumors may be of one or more types.
100441 The term "whole blood,- as used herein, generally refers to
a blood sample that
has not been separated into sub-components (e.g., by centrifugation). The
whole blood of a
blood sample may contain cfDNA and/or germline DNA. Whole blood DNA (which may
contain cfDNA and/or germline DNA) may be extracted from a blood sample. Whole
blood
DNA sequencing reads (which may contain cfDNA sequencing reads and/or germline
DNA
sequencing reads) may be extracted from whole blood DNA.
100451 Diagnostic testing of subjects (e.g., patients) for a
disease or disorder may be
limited or scarce. In some cases, diagnostic testing may be saved for
symptomatic subjects or
high-risk subjects; this can be less than ideal because asymptomatic subjects
continue to be
contagious and spread infectious diseases such as viruses (e.g., COVID-19 or
HIV), bacteria,
or parasites (e.g., malaria). Therefore, frequent and widespread diagnostic
testing of an
infectious disease across a population may be essential for containment and
mitigation,
especially in cases of pandemic outbreaks.
100461 Methods and systems are provided for pooling a plurality of
bodily samples.
Bodily samples, health data, contact tracing data, location data, and/or
movement data may
be collected from a plurality of subjects (e.g., patients), and trained
computer algorithms may
be used to efficiently pool the bodily samples into a plurality of sample
pools for diagnostic
testing. Such efficient sample pooling can be used to perform frequent and
widespread
diagnostic testing of an infectious disease across a population, which may be
essential for
-11-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
containment and mitigation, especially in cases of pandemic outbreaks (e.g.,
COVID-19,
HIV, or malaria).
[0047] In an aspect, the present disclosure provides a method
comprising: (a) obtaining a
plurality of: health data, contact tracing data, location data, movement data,
or any
combination thereof associated with a plurality of subjects; and (b)
processing the plurality
of: health data, contact tracing data, location data, movement data, or any
combination
thereof with a trained computer algorithm to at least some individual subjects
of the plurality
of subjects to a pool from among a plurality of pools, wherein a number of
pools of the
plurality of pools is less than a number of subjects of the plurality of
subjects.
[0048] In some embodiments, the method further comprises outputting
an electronic
recommendation to create, for each of at least two given pools of the
plurality of pools, a
pooled sample by combining bodily samples or portions thereof obtained from
subjects in the
at least two given pools In some embodiments, the method further comprises
creating, for
each of at least two given pools of the plurality of pools, a pooled sample by
combining
bodily samples or portions thereof obtained from subjects in the at least two
given pools. In
some embodiments, the method further comprises obtaining the bodily samples or
portions
thereof from the plurality of subjects.
100491 In some embodiments, the bodily samples are individually selected from
the group
consisting of: nasopharyngeal swab, oropharyngeal swab, blood, serum, plasma,
vitreous,
sputum, urine, stool, tears, perspiration, saliva, semen, mucosal excretions,
mucus, spinal
fluid, cerebrospinal fluid (CSF), pleural fluid, peritoneal fluid, amniotic
fluid, lymph fluid,
eye swab, cheek swab, vaginal swab, cervical swab, rectal swab, cells, and
tissue.
[0050] In some embodiments, the method further comprises isolating nucleic
acids from the
bodily samples, and creating, for a given pool of the plurality of pools, the
pooled sample by
combining at least some of nucleic acids isolated from bodily samples obtained
from the
subjects in the given pool. In some embodiments, the method further comprises
enriching the
nucleic acids for a plurality of genomic regions. In some embodiments, the
method further
comprises amplifying at least some of the nucleic acids. In some embodiments,
the
amplification comprises selective amplification. In some embodiments, the
amplification
comprises universal amplification. In some embodiments, enriching the nucleic
acids for the
plurality of genomic regions comprises contacting the nucleic acids with a
plurality of probes,
each of the plurality of probes having sequence complementarity with at least
a portion of a
genomic region of the plurality of genomic regions. In some embodiments, the
plurality of
genomic regions comprises genomic regions associated with a disease or
disorder. In some
-12-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
embodiments, the disease or disorder comprises coronavirus disease 2019 (COVID-
19),
human immunodeficiency virus (HIV), or malaria. In some embodiments, the
disease or
disorder comprises COVID-19.
100511 In some embodiments, the method further comprises performing a
plurality of
diagnostic tests on the plurality of pooled samples to obtain a plurality of
diagnostic results
associated with the plurality of pooled samples. In some embodiments, the
plurality of
diagnostic tests are configured to detect a presence or absence of a disease
or disorder based
on analyzing at least the plurality of pooled samples. In some embodiments,
the disease or
disorder comprises coronavirus disease 2019 (COVID-19), human immunodeficiency
virus
(HIV), or malaria. In some embodiments, the disease or disorder comprises
COVID-19.
100521 In some embodiments, the method further comprises, for a given pool
among the
plurality of pools, detecting the absence of the disease or disorder in each
of the individual
subjects of the given pool when the absence of the disease or disorder is
detected based on
analyzing the pooled sample corresponding to the given pool. In some
embodiments, the
method further comprises, for a given pool among the plurality of pools,
testing each of the
individual subjects of the given pool for the disease or disorder when the
presence of the
disease or disorder is detected based on analyzing the pooled sample
corresponding to the
given pool. In some embodiments, the method further comprises, for a given
pool among the
plurality of pools, testing each of a plurality of sub-pools of the given pool
for the disease or
disorder when the presence of the disease or disorder is detected based on
analyzing the
pooled sample corresponding to the given pool.
100531 In some embodiments, the method further comprises detecting the
presence or
absence of the disease or disorder in the plurality of subjects with a
clinical sensitivity of at
least about 50%. In some embodiments, the method further comprises detecting
the presence
or absence of the disease or disorder in the plurality of subjects with a
clinical sensitivity of at
least about 70% In some embodiments, the method further comprises detecting
the presence
or absence of the disease or disorder in the plurality of subjects with a
clinical sensitivity of at
least about 90%. In some embodiments, the method further comprises detecting
the presence
or absence of the disease or disorder in the plurality of subjects with a
clinical sensitivity of at
least about 50%, at least about 55%, at least about 60%, at least about 65%,
at least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least
about 91%, at least about 92%, at least about 93%, at least about 94%, at
least about 95%, at
least about 96%, at least about 97%, at least about 98%, at least about 99%,
or 100%.
-13-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
100541 In some embodiments, the method further comprises detecting the
presence or
absence of the disease or disorder in the plurality of subjects with a
clinical specificity of at
least about 50%. In some embodiments, the method further comprises detecting
the presence
or absence of the disease or disorder in the plurality of subjects with a
clinical specificity of
at least about 70%. In some embodiments, the method further comprises
detecting the
presence or absence of the disease or disorder in the plurality of subjects
with a clinical
specificity of at least about 90%. In some embodiments, the method further
comprises
detecting the presence or absence of the disease or disorder in the plurality
of subjects with a
clinical specificity of at least about 50%, at least about 55%, at least about
60%, at least about
65%, at least about 70%, at least about 75%, at least about 80%, at least
about 85%, at least
about 90%, at least about 91%, at least about 92%, at least about 93%, at
least about 94%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
at least about
99%, or 100%
100551 In some embodiments, the method further comprises detecting the
presence or
absence of the disease or disorder in the plurality of subjects with a
positive predictive value
(PPV) of at least about 50%. In some embodiments, the method further comprises
detecting
the presence or absence of the disease or disorder in the plurality of
subjects with a positive
predictive value (PPV) of at least about 70%. In some embodiments, the method
further
comprises detecting the presence or absence of the disease or disorder in the
plurality of
subjects with a positive predictive value (PPV) of at least about 90%. In some
embodiments,
the method further comprises detecting the presence or absence of the disease
or disorder in
the plurality of subjects with a positive predictive value (PPV) of at least
about 50%, at least
about 55%, at least about 60%, at least about 65%, at least about 70%, at
least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about
92%, at least about 93%, at least about 94%, at least about 95%, at least
about 96%, at least
about 97%, at least about 98%, at least about 99%, or 100%
100561 In some embodiments, the method further comprises detecting the
presence or
absence of the disease or disorder in the plurality of subjects with a
negative predictive value
(NPV) of at least about 50% In some embodiments, the method further comprises
detecting
the presence or absence of the disease or disorder in the plurality of
subjects with a negative
predictive value (NPV) of at least about 70%. In some embodiments, the method
further
comprises detecting the presence or absence of the disease or disorder in the
plurality of
subjects with a negative predictive value (NPV) of at least about 90%. In some
embodiments,
the method further comprises detecting the presence or absence of the disease
or disorder in
-14-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
the plurality of subjects with a negative predictive value (NPV) of at least
about 50%, at least
about 55%, at least about 60%, at least about 65%, at least about 70%, at
least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about
92%, at least about 93%, at least about 94%, at least about 95%, at least
about 96%, at least
about 97%, at least about 98%, at least about 99%, or 100%.
100571 In some embodiments, the method further comprises detecting the
presence or
absence of the disease or disorder in the plurality of subjects with an area
under the curve
(AUC) of at least about 0.60. In some embodiments, the method further
comprises detecting
the presence or absence of the disease or disorder in the plurality of
subjects with an area
under the curve (AUC) of at least about 0.70. In some embodiments, the method
further
comprises detecting the presence or absence of the disease or disorder in the
plurality of
subjects with an area under the curve (AUC) of at least about 0.80. In some
embodiments, the
method further comprises detecting the presence or absence of the disease or
disorder in the
plurality of subjects with an area under the curve (AUC) of at least about
0.90. In some
embodiments, the method further comprises detecting the presence or absence of
the disease
or disorder in the plurality of subjects with an area under the curve (AUC) of
at least about
0.50, at least about 0.55, at least about 0.60, at least about 0.65, at least
about 0.70, at least
about 0.75, at least about 0.80, at least about 0.85, at least about 0.90, at
least about 0.91, at
least about 0.92, at least about 0.93, at least about 0.94, at least about
0.95, at least about
0.96, at least about 0.97, at least about 0.98, or at least about 0.99.
100581 In some embodiments, the plurality of health data comprises a diagnosis
of a disease
or disorder, a prognosis of a disease or disorder, a risk of having a disease
or disorder, a
treatment history of a disease or disorder, a history of previous treatment
for a disease or
disorder, a history of prescribed medications, a history of prescribed medical
devices, age,
height, weight, sex, smoking status, one or more symptoms, and one or more
vital signs. In
some embodiments, the one or more vital signs comprise one or more of: heart
rate, heart rate
variability, blood pressure, respiratory rate, blood oxygen concentration
(Sp02), carbon
dioxide concentration in respiratory gases, a hormone level, sweat analysis,
blood glucose,
body temperature, impedance, conductivity, capacitance, resistivity,
electromyography,
galvanic skin response, neurological signals, electroencephalography,
electrocardiography,
immunology markers, and other physiological measurements.
100591 In some embodiments, the trained computer algorithm comprises a trained
machine
learning classifier. In some embodiments, the trained machine learning
classifier comprises
an algorithm selected from the group consisting of: a support vector machine,
a neural
-15-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
network, a random forest, a linear regression, a logistic regression, a
Bayesian classifier, a
boosted classifier, a gradient boosting algorithm, an adaptive boosting
(AdaBoost) algorithm,
and an extreme gradient boosting (XGBoost) algorithm.
100601 In some embodiments, the method further comprises processing health
data, contact
tracing data, location data, or movement data of the individual subject with
the trained
computer algorithm to determine an expected prevalence of a disease or
disorder, and
assigning the individual subject of the plurality of subjects to the pool from
among the
plurality of pools based at least in part on the determined expected
prevalence of the disease
or disorder. In some embodiments, the method further comprises assigning the
individual
subject of the plurality of subjects to the pool from among the plurality of
pools when the
determined expected prevalence of the disease or disorder is less than a pre-
determined
prevalence threshold. In some embodiments, the pre-determined prevalence
threshold is
about 5%, about 10%, about 15%, about 20%, about 25%, about 30%, about 35%,
about
40%, about 45%, or about 50%. In some embodiments, the method further
comprises
determining a maximum pool size based on the determined expected prevalence of
the
disease or disorder, and assigning the individual subject of the plurality of
subjects to the pool
from among the plurality of pools based on the maximum pool size.
100611 In some embodiments, the number of pools of the plurality of pools is
reduced
relative to the number of subjects of the plurality of subjects by at least
50%. In some
embodiments, the number of pools of the plurality of pools is reduced relative
to the number
of subjects of the plurality of subjects by at least 100%. In some
embodiments, the number of
pools of the plurality of pools is reduced relative to the number of subjects
of the plurality of
subjects by at least 200%. In some embodiments, the number of pools of the
plurality of
pools is reduced relative to the number of subjects of the plurality of
subjects by at least
300%. In some embodiments, the number of pools of the plurality of pools is
reduced relative
to the number of subjects of the plurality of subjects by at least about 10%,
at least about
20%, at least about 30%, at least about 40%, at least about 50%, at least
about 60%, at least
about 70, at least about 80%, at least about 90%, at least about 100%, at
least about 110%, at
least about 120%, at least about 130%, at least about 140%, at least about
150%, at least
about 160%, at least about 170%, at least about 180%, at least about 190%, at
least about
200%, at least about 210%, at least about 220%, at least about 230%, at least
about 240%, at
least about 250%, at least about 260%, at least about 270%, at least about
280%, at least
about 290%, at least about 300%, at least about 310%, at least about 320%, at
least about
-16-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
330%, at least about 340%, at least about 350%, at least about 360%, at least
about 370%, at
least about 380%, at least about 390%, at least about 400%, or more than about
400%.
100621 In some embodiments, the method further comprises, based on the
detected presence
or absence of the disease or disorder in the plurality of subjects,
administering a
therapeutically effective dose of a treatment to treat the disease or disorder
detected in at least
a subset of the plurality of subjects.
100631 FIG. 1A shows an example method 100 for performing sample
pooling of a
plurality of clinical samples for diagnostic testing. The method 100 may
comprise obtaining a
plurality of bodily samples from a plurality of subjects (as in operation
102). In some
embodiments, the bodily samples comprises a nasopharyngeal swab, oropharyngeal
swab,
blood, serum, plasma, vitreous, sputum, urine, stool, tears, perspiration,
saliva, semen,
mucosal excretions, mucus, spinal fluid, cerebrospinal fluid (CSF), pleural
fluid, peritoneal
fluid, amniotic fluid, lymph fluid, eye swab, cheek swab, vaginal swab,
cervical swab, rectal
swab, cells, or tissue. Next, the method 100 may comprise obtaining a
plurality of: health
data, contact tracing data, location data, movement data, or any combination
thereof
associated with the plurality of subjects (as in operation 104). In some
instances, any of the
data herein (e.g., health data, contact tracing data, location data, or
movement data associated
with the plurality of subjects) comprises de-identified data. In some
embodiments, the
plurality of health data comprises a diagnosis of a disease or disorder, a
prognosis of a
disease or disorder, a risk of having a disease or disorder, a treatment
history of a disease or
disorder, a history of previous treatment for a disease or disorder, a history
of prescribed
medications, a history of prescribed medical devices, age, height, weight,
sex, smoking
status, one or more symptoms, and one or more vital signs. In some
embodiments, the one or
more vital signs comprise one or more of: heart rate, heart rate variability,
blood pressure,
respiratory rate, blood oxygen concentration (Sp02), carbon dioxide
concentration in
respiratory gases, a hormone level, sweat analysis, blood glucose, body
temperature,
impedance, conductivity, capacitance, resistivity, electromyography, galvanic
skin response,
neurological signals, electroencephalography, electrocardiography, immunology
markers, and
other physiological measurements. In some embodiments, the plurality of
contact tracing
data, location data, or movement data comprises data associated with subjects'
environment,
location, movement, or daily schedules. For example, for a delivery warehouse
that is
performing diagnostic testing on its employees, intelligent pooling may be
performed based
on the employees' delivery routes, shift schedules, contact tracing
information, etc. In some
embodiments, the method 100 is applied to genetic sequencing of samples, such
that pooling
-17-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
of samples is performed where the mutation rate is low. Next, the method 100
may comprise
processing the plurality of: health data, contact tracing data, location data,
movement data, or
any combination thereof with a trained computer algorithm to at least some
individual
subjects of the plurality of subjects to a pool from among a plurality of
pools (as in operation
106). In some embodiments, a number of pools of the plurality of pools is less
than a number
of subjects of the plurality of subjects. In some embodiments, the trained
computer algorithm
comprises a trained machine learning classifier. In some embodiments, the
trained machine
learning classifier comprises an algorithm selected from the group consisting
of: a support
vector machine, a neural network, a random forest, a linear regression, a
logistic regression, a
Bayesian classifier, a boosted classifier, a gradient boosting algorithm, an
adaptive boosting
(AdaBoost) algorithm, and an extreme gradient boosting (XGBoost) algorithm.
100641 FIG. 1B shows a general workflow for performing sample
pooling of a plurality
of clinical samples for diagnostic testing, including data input, consuming
and consolidating
data, performing a method for sample pooling, recommending pooling protocols,
and
performing diagnostic testing on the sample pools. In some embodiments,
performing the
diagnostic testing on the sample pools comprises using an integrated robot
and/or robotics
application programming interface (API) to perform liquid handling of a
plurality of bodily
samples and/or sample pools. In some embodiments, performing the diagnostic
testing on the
sample pools comprises detecting a plurality of diseases or disorders (e.g.,
common cold,
influenza, COVID-19 infection, and/or COVID-19 immunity) using a same sample
pool. In
some embodiments, performing the diagnostic testing on the sample pools
comprises tagging
a plurality of bodily samples and/or sample pools (e.g., using sample tags or
sample
barcodes) to increase a multiplexity of the diagnostic testing (e.g., via DNA
sequencing,
RNA sequencing, or reverse-transcription polymerase chain reaction (RT-PCR)).
In some
embodiments, performing the diagnostic testing on the sample pools comprises
performing
genetic sequencing on a plurality of bodily samples and/or sample pools (e.g.,
in cases where
a mutation rate is low).
100651 FIG. 1C shows a workflow for performing sample handling and
testing. The
workflow for performing sample handling and testing may comprise sample
collection of a
plurality of bodily samples from a plurality of subjects. Logistical synergies
(or
disadvantages) may be associated with the handling of the bodily samples.
Seamless
information flow may be required for the software (e.g., barcodes may be used
to identify
subject samples through a connected EHR system, if the bodily samples at the
collection sites
lack the required data). Next, the workflow for performing sample handling and
testing may
-18-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
comprise extracting nucleic acids (e.g., DNA or RNA) from the bodily fluid
samples. Next,
the workflow for performing sample handling and testing may comprise pooling
the samples,
where the pools are selected using methods and systems of the present
disclosure. Such pools
are selected using machine learning to inform decisions on which samples to
pool together
and/or the sample size to pool together. Next, the workflow for performing
sample handling
and testing may comprise performing a diagnostic test or panel on the pooled
samples (e.g.,
an RT-PCR diagnostic panel).
100661 FIG. 113 shows an example of a system procedure diagram of
methods and
systems of the present disclosure.
100671 FIG. lE shows an example of a user activity diagram of
methods and systems of
the present disclosure.
100681 FIG. 2A shows an example of sample pooling of a plurality of
clinical samples
for diagnostic testing If a sample pool comprising a plurality of individual
clinical samples
receives a negative clinical test outcome, then all the individual clinical
samples in the
sample pool must also have a negative clinical test outcome. Conversely, if
the sample pool
comprising the plurality of individual clinical samples receives a positive
clinical test
outcome, then either the individual clinical samples can be individually
tested, or the sample
pool can be further sub-divided into smaller subsets and the sample pooling
process can be
repeated.
100691 FIG. 2B shows an example of how methods and systems of the
present disclosure
advantageously use sample pooling. For example, currently about 2 out of 10
test results
return positive; patterns can be learned via machine learning about the 8 out
of 10 negative
patients. Using methods and systems of the present disclosure, predictive
analytics and
machine learning may be applied to identify patients with a lower prevalence
rate. A plurality
of predicted negative samples may be pooled together to create sample pool,
and a single
diagnostic test may be performed on the sample pool. As a result, the same
number of
subjects may be tested using a smaller number of diagnostic tests; the saved
diagnostic tests
may be used to test a broader population of subjects. Generally, when a
disease or disorder
has a high enough prevalence, the expected probability of encountering at
least one positive
sample among a sample pool may increase to a high enough value such that the
savings from
sample pooling may be spoiled. Therefore, intelligent pooling strategies of
the methods and
systems of the present disclosure enables the efficient use of pool testing.
100701 In some embodiments, nucleic acids may be extracted from
bodily samples and
analyzed for diagnostic testing. For example, sequencing reads may be
generated from the
-19-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
nucleic acids using any suitable sequencing method. The sequencing method can
be a first-
generation sequencing method, such as Maxam-Gilbert or Sanger sequencing, or a
high-
throughput sequencing (e.g., next-generation sequencing or NGS) method. A high-
throughput
sequencing method may sequence simultaneously (or substantially
simultaneously) at least
10,000, 100,000, 1 million, 10 million, 100 million, 1 billion, or more
polynucleotide
molecules. Sequencing methods may include, but are not limited to:
pyrosequencing,
sequencing-by-synthesis, single-molecule sequencing, nanopore sequencing,
semiconductor
sequencing, sequencing-by-ligation, sequencing-by-hybridization, Digital Gene
Expression
(Helicos), massively parallel sequencing, e.g., Helicos, Clonal Single
Molecule Array
(Solexa/Illumina), sequencing using PacBio, SOLiD, Ion Torrent, or Nanopore
platforms.
100711 In some embodiments, the sequencing comprises whole genome
sequencing
(WGS). The sequencing may be performed at a depth sufficient to assess tumor
progression
or tumor non-progression in a subject with a desired performance (e g ,
accuracy, sensitivity,
specificity, positive predictive value (PPV), negative predictive value (NPV),
or the area
under curve (AUC) of a receiver operator characteristic (ROC)).
100721 In some embodiments, the sequencing reads may be aligned or
mapped to a
reference genome. The reference genome may comprise at least a portion of a
genome (e.g.,
the human genome). The reference genome may comprise an entire genome (e.g.,
the entire
human genome). The reference genome may comprise an entire genome with certain
base
conversions applied (e.g., the entire human genome with cytosines converted to
thymines), as
may be used for methylation data alignment. The reference genome may comprise
a database
comprising a plurality of genomic regions that correspond to coding and/or non-
coding
genomic regions of a genome. The database may comprise a plurality of genomic
regions that
correspond to disease-associated coding and/or non-coding genomic regions of a
genome,
such as single nucleotide variants (SNVs), copy number variants (CNVs),
insertions or
deletions (indels), and fusion genes. The alignment may be performed using a
Burrows-
Wheeler algorithm (BWA), a sambamba algorithm, a samtools algorithm, or any
other
suitable alignment algorithm.
100731 In some embodiments, a quantitative measure of the
sequencing reads may be
generated for each of a plurality of genomic regions. Quantitative measures of
the sequencing
reads may be generated, such as counts of sequencing reads that are aligned
with a given
genomic region. Sequencing reads having a portion or all of the sequencing
read aligning
with a given genomic region may be counted toward the quantitative measure for
that
genomic region.
-20-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
100741 In some embodiments, genomic regions may comprise disease
markers. Patterns
of specific and non-specific genomic regions may be indicative of disease
progression or
disease non-progression status. Changes over time in these patterns of genomic
regions may
be indicative of changes in disease progression or disease non-progression
status.
100751 In some embodiments, nucleic acids may be assayed by
performing binding
measurements of the nucleic acids at each of a plurality of genomic regions.
In some
embodiments, performing the binding measurements comprises assaying the
nucleic acids
using probes that are selective for at least a portion of a plurality of
genomic regions in the
plurality of nucleic acids. In some embodiments, the probes are nucleic acid
molecules
having sequence complementarity with nucleic acid sequences of the plurality
of genomic
regions. In some embodiments, the nucleic acid molecules are primers or
enrichment
sequences. In some embodiments, the assaying comprises use of array
hybridization or
polymerase chain reaction (PCR), or nucleic acid sequencing
100761 In some embodiments, the nucleic acids are enriched for at
least a portion of the
plurality of genomic regions. In some embodiments, the enrichment comprises
amplifying at
least some of the nucleic acids. For example, the nucleic acids may be
amplified by selective
amplification (e.g., by using a set of primers or probes comprising nucleic
acid molecules
having sequence complementarity with nucleic acid sequences of the plurality
of genomic
regions). Alternatively or in combination, the nucleic acids may be amplified
by universal
amplification (e.g., by using universal primers). In some embodiments, the
enrichment
comprises selectively isolating at least a portion of the plurality of nucleic
acids.
100771 In some embodiments, the sequencing reads may be normalized
or corrected. For
example, the sequencing reads may be de-deduplicated, normalized, and/or
corrected to
account for known biases in sequencing and library preparation and/or known
biases in
sequencing and library preparation. In some embodiments, a subset of the
quantitative
measures (e.g., statistical measures) may be filtered out, e.g., based on
whether the changes in
such quantitative measures (e.g., across different time points) are
significantly different from
those observed in unaffected subjects (e.g., a background profile of nucleic
acids).
100781 The plurality of genomic regions may comprise at least about
10 distinct genomic
regions, at least about 50 distinct genomic regions, at least about 100
distinct genomic
regions, at least about 500 distinct genomic regions, at least about 1
thousand distinct
genomic regions, at least about 5 thousand distinct genomic regions, at least
about 10
thousand distinct genomic regions, at least about 50 thousand distinct genomic
regions, at
least about 100 thousand distinct genomic regions, at least about 500 thousand
distinct
-21-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
genomic regions, at least about 1 million distinct genomic regions, at least
about 2 million
distinct genomic regions, at least about 3 million distinct genomic regions,
at least about 4
million distinct genomic regions, at least about 5 million distinct genomic
regions, at least
about 10 million distinct genomic regions, at least about 15 million distinct
genomic regions,
at least about 20 million distinct genomic regions, at least about 25 million
distinct genomic
regions, at least about 30 million distinct genomic regions, or more than 30
million distinct
genomic regions.
Computer systems
100791 The present disclosure provides computer systems that are
programmed to
implement methods of the disclosure. FIG. 3 shows a computer system 301 that
is
programmed or otherwise configured to, for example, direct the obtaining of
bodily samples
from subjects, obtain health data, contact tracing data, location data, or
movement data
associated with subjects, process health data, contact tracing data, location
data, or movement
data with a trained computer algorithm to assign individual subjects to a
pool, output an
electronic recommendation to create a pooled sample by combining bodily
samples or
portions thereof obtained from subjects in a pool, direct the creating of a
pooled sample by
combining bodily samples or portions thereof obtained from subjects in a pool,
direct the
isolating of nucleic acids from bodily samples, direct the enriching of
nucleic acids, direct the
amplifying of nucleic acids, direct the performing of diagnostic tests on
pooled samples to
obtain diagnostic results associated with the pooled samples, and detect the
absence of the
disease or disorder in individual subjects of a pool when an absence of a
disease or disorder is
detected based on analyzing a pooled sample corresponding to the pool. The
computer system
301 can regulate various aspects of analysis, calculation, and generation of
the present
disclosure, such as, for example, directing the obtaining of bodily samples
from subjects,
obtaining health data, contact tracing data, location data, or movement data
associated with
subjects, processing health data, contact tracing data, location data, or
movement data with a
trained computer algorithm to assign individual subjects to a pool, outputting
an electronic
recommendation to create a pooled sample by combining bodily samples or
portions thereof
obtained from subjects in a pool, directing the creating of a pooled sample by
combining
bodily samples or portions thereof obtained from subjects in a pool, directing
the isolating of
nucleic acids from bodily samples, directing the enriching of nucleic acids,
directing the
amplifying of nucleic acids, directing the performing of diagnostic tests on
pooled samples to
obtain diagnostic results associated with the pooled samples, and detecting
the absence of the
disease or disorder in individual subjects of a pool when an absence of a
disease or disorder is
-22-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
detected based on analyzing a pooled sample corresponding to the pool. The
computer system
301 can be an electronic device of a user or a computer system that is
remotely located with
respect to the electronic device. The electronic device can be a mobile
electronic device.
100801 The computer system 301 includes a central processing unit
(CPU, also
"processor" and "computer processor" herein) 305, which can be a single core
or multi core
processor, or a plurality of processors for parallel processing. The computer
system 301 also
includes memory or memory location 310 (e.g., random-access memory, read-only
memory,
flash memory), electronic storage unit 315 (e.g., hard disk), communication
interface 320
(e.g., network adapter) for communicating with one or more other systems, and
peripheral
devices 325, such as cache, other memory, data storage and/or electronic
display adapters.
The memory 310, storage unit 315, interface 320 and peripheral devices 325 are
in
communication with the CPU 305 through a communication bus (solid lines), such
as a
motherboard The storage unit 315 can be a data storage unit (or data
repository) for storing
data. The computer system 301 can be operatively coupled to a computer network
("network") 330 with the aid of the communication interface 320. The network
330 can be
the Internet, an internet and/or extranet, or an intranet and/or extranet that
is in
communication with the Internet. The network 330 in some cases is a
telecommunication
and/or data network. The network 330 can include one or more computer servers,
which can
enable distributed computing, such as cloud computing. For example, one or
more computer
servers may enable cloud computing over the network 330 ("the cloud") to
perform various
aspects of analysis, calculation, and generation of the present disclosure,
such as, for
example, directing the obtaining of bodily samples from subjects, obtaining
health data,
contact tracing data, location data, or movement data associated with
subjects, processing
health data, contact tracing data, location data, or movement data with a
trained computer
algorithm to assign individual subjects to a pool, outputting an electronic
recommendation to
create a pooled sample by combining bodily samples or portions thereof
obtained from
subjects in a pool, directing the creating of a pooled sample by combining
bodily samples or
portions thereof obtained from subjects in a pool, directing the isolating of
nucleic acids from
bodily samples, directing the enriching of nucleic acids, directing the
amplifying of nucleic
acids, directing the performing of diagnostic tests on pooled samples to
obtain diagnostic
results associated with the pooled samples, and detecting the absence of the
disease or
disorder in individual subjects of a pool when an absence of a disease or
disorder is detected
based on analyzing a pooled sample corresponding to the pool. Such cloud
computing may be
provided by cloud computing platforms such as, for example, Amazon Web
Services (AWS),
-23 -
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
Microsoft Azure, Google Cloud Platform, and IBM cloud. The network 330, in
some cases
with the aid of the computer system 301, can implement a peer-to-peer network,
which may
enable devices coupled to the computer system 301 to behave as a client or a
server.
100811 The CPU 305 can execute a sequence of machine-readable
instructions, which can
be embodied in a program or software. The instructions may be stored in a
memory location,
such as the memory 310. The instructions can be directed to the CPU 305, which
can
subsequently program or otherwise configure the CPU 305 to implement methods
of the
present disclosure. Examples of operations performed by the CPU 305 can
include fetch,
decode, execute, and writeback.
100821 The CPU 305 can be part of a circuit, such as an integrated
circuit. One or more
other components of the system 301 can be included in the circuit. In some
cases, the circuit
is an application specific integrated circuit (ASIC).
100831 The storage unit 315 can store files, such as drivers,
libraries and saved programs
The storage unit 315 can store user data, e.g., user preferences and user
programs. The
computer system 301 in some cases can include one or more additional data
storage units that
are external to the computer system 301, such as located on a remote server
that is in
communication with the computer system 301 through an intranet or the
Internet.
100841 The computer system 301 can communicate with one or more
remote computer
systems through the network 330. For instance, the computer system 301 can
communicate
with a remote computer system of a user (e.g., a physician, a nurse, a
caretaker, a patient, or a
subject). Examples of remote computer systems include personal computers
(e.g., portable
PC), slate or tablet PC's (e.g., Apple iPad, Samsung Galaxy Tab),
telephones, Smart
phones (e.g., Apple iPhone, Android-enabled device, Blackberry ), or personal
digital
assistants. The user can access the computer system 301 via the network 330.
100851 Methods as described herein can be implemented by way of
machine (e.g.,
computer processor) executable code stored on an electronic storage location
of the computer
system 301, such as, for example, on the memory 310 or electronic storage unit
315. The
machine-executable or machine-readable code can be provided in the form of
software.
During use, the code can be executed by the processor 305. In some cases, the
code can be
retrieved from the storage unit 315 and stored on the memory 310 for ready
access by the
processor 305. In some situations, the electronic storage unit 315 can be
precluded, and
machine-executable instructions are stored on memory 310.
100861 The code can be pre-compiled and configured for use with a
machine having a
processor adapted to execute the code, or can be compiled during runtime. The
code can be
-24-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
supplied in a programming language that can be selected to enable the code to
execute in a
pre-compiled or as-compiled fashion.
100871 Aspects of the systems and methods provided herein, such as
the computer system
301, can be embodied in programming. Various aspects of the technology may be
thought of
as "products" or "articles of manufacture" typically in the form of machine
(or processor)
executable code and/or associated data that is carried on or embodied in a
type of machine-
readable medium. Machine-executable code can be stored on an electronic
storage unit, such
as memory (e.g., read-only memory, random-access memory, flash memory) or a
hard disk.
"Storage" type media can include any or all of the tangible memory of the
computers,
processors or the like, or associated modules thereof, such as various
semiconductor
memories, tape drives, disk drives and the like, which may provide non-
transitory storage at
any time for the software programming. All or portions of the software may at
times be
communicated through the Internet or various other telecommunication networks
Such
communications, for example, may enable loading of the software from one
computer or
processor into another, for example, from a management server or host computer
into the
computer platform of an application server. Thus, another type of media that
may bear the
software elements includes optical, electrical and electromagnetic waves, such
as used across
physical interfaces between local devices, through wired and optical landline
networks and
over various air-links. The physical elements that carry such waves, such as
wired or
wireless links, optical links or the like, also may be considered as media
bearing the software.
As used herein, unless restricted to non-transitory, tangible "storage" media,
terms such as
computer or machine "readable medium" refer to any medium that participates in
providing
instructions to a processor for execution.
100881 Hence, a machine-readable medium, such as computer-
executable code, may take
many forms, including but not limited to, a tangible storage medium, a carrier
wave medium
or physical transmission medium. Non-volatile storage media include, for
example, optical
or magnetic disks, such as any of the storage devices in any computer(s) or
the like, such as
may be used to implement the databases, etc. shown in the drawings. Volatile
storage media
include dynamic memory, such as main memory of such a computer platform.
Tangible
transmission media include coaxial cables; copper wire and fiber optics,
including the wires
that comprise a bus within a computer system. Carrier-wave transmission media
may take
the form of electric or electromagnetic signals, or acoustic or light waves
such as those
generated during radio frequency (RF) and infrared (IR) data communications.
Common
forms of computer-readable media therefore include for example: a floppy disk,
a flexible
-25-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
disk, hard disk, magnetic tape, any other magnetic medium, a CD-ROM, DVD or
DVD-
ROM, any other optical medium, punch cards paper tape, any other physical
storage medium
with patterns of holes, a RAM, a ROM, a PROM and EPROM, a FLASH-EPROM, any
other
memory chip or cartridge, a carrier wave transporting data or instructions,
cables or links
transporting such a carrier wave, or any other medium from which a computer
may read
programming code and/or data. Many of these forms of computer-readable media
may be
involved in carrying one or more sequences of one or more instructions to a
processor for
execution.
100891 The computer system 301 can include or be in communication
with an electronic
display 335 that comprises a user interface (UI) 340 for providing, for
example, health data,
contact tracing data, location data, or movement data, recommendations to
create pooled
samples, and diagnostic results of pooled samples Examples of UIs include,
without
limitation, a graphical user interface (GUI) and web-based user interface
100901 Methods and systems of the present disclosure can be
implemented by way of one
or more algorithms. An algorithm can be implemented by way of software upon
execution
by the central processing unit 305. The algorithm can, for example, direct the
obtaining of
bodily samples from subjects, obtain health data, contact tracing data,
location data, or
movement data associated with subjects, process health data, contact tracing
data, location
data, or movement data with a trained computer algorithm to assign individual
subjects to a
pool, output an electronic recommendation to create a pooled sample by
combining bodily
samples or portions thereof obtained from subjects in a pool, direct the
creating of a pooled
sample by combining bodily samples or portions thereof obtained from subjects
in a pool,
direct the isolating of nucleic acids from bodily samples, direct the
enriching of nucleic acids,
direct the amplifying of nucleic acids, direct the performing of diagnostic
tests on pooled
samples to obtain diagnostic results associated with the pooled samples, and
detect the
absence of the disease or disorder in individual subjects of a pool when an
absence of a
disease or disorder is detected based on analyzing a pooled sample
corresponding to the pool.
EXAMPLES
Example 1: Efficient sample pooling for increasing diagnostic testing capacity
of
COVID-19
100911 Using methods and systems of the present disclosure, efficient sample
pooling was
performed for increasing diagnostic testing capacity of coronavirus disease
2019 (COVID-
19), which is caused by the severe acute respiratory syndrome coronavirus 2
(SARS-CoV-2)
-26-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
virus. The sample pooling was performed using one of three different types of
information to
perform the classification of a plurality of individual samples each into a
respective sample
pool from among a plurality of sample pools, including Boston vs. Cambridge
cohort
information, clinical symptom information, and contact tracing information.
FIG. 4A shows
the number of people who were able to be tested with 1,000 diagnostic tests,
using Boston vs.
Cambridge cohort information (left 3 columns), clinical symptom information
(middle 3
columns), and contact tracing information (right 3 columns). Within each set
of 3 columns,
the number of people who were able to be tested with 1,000 diagnostic tests is
indicated using
no sample pooling (left), simple sample pooling (middle), and intelligent
sample pooling
based on methods and systems of the present disclosure (right).
100921 As shown in FIG. 4A, simple sample pooling outperformed no sample
pooling for all
three different types of information used; further, intelligent sample pooling
based on
methods and systems of the present disclosure outperformed simple sample
pooling for all
three different types of information used (and significantly so for the cases
of clinical
symptom information and contact tracing information. Therefore, using
intelligent sample
pooling based on methods and systems of the present disclosure, diagnostic
testing capacity
for COVID-19 can be increased by about 3X, with at least a 40% cost savings at
96%
specificity.
100931 FIG. 4B shows that using intelligent sample pooling based on methods
and systems
of the present disclosure, the rules for diagnostic testing can be redefined.
Based on the
current state (left side), symptoms alone are the prevalence assessment. Some
subjects who
meet a set of certain criteria receive testing, while the others are denied
testing (e.g., because
of rationing of diagnostic testing due to a shortage of testing capacity).
Using intelligent
sample pooling based on methods and systems of the present disclosure (right
side), the ratio
of diagnostic tests available to individuals tested for COV1D-19 is no longer
1:1. This ratio
can be effectively increased, for example, to about 1.1, about 1.2, about 1.3,
about 1.4, about
1.5, about 1.6, about 1.7, about 1.8, about 1.9, about 2.0, about 2.1, about
2.2, about 2.3,
about 2.4, about 2.5, about 2.6, about 2.7, about 2.8, about 2.9, about 3.0,
or more than about
3Ø
Example 2: Efficient sample pooling using PrecisionFDA data
100941 Using methods and systems of the present disclosure, efficient sample
pooling was
performed using PrecisionFDA data for increasing diagnostic testing capacity
of COV1D-19.
-27-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
The PrecisionFDA data was analyzed as follows. First, the data set was
obtained. Next, the
COVID-19 related observations were filtered.
100951 Next, patient details regarding those observations were obtained (e.g.,
clinical health
data, contact tracing data, location data, movement data, and demographic
data, including
age). Next, diagnostic test outcomes were obtained (represented in binary
format, where
values of 1 indicate positive test results and values of 0 indicates negative
test results). Next,
gender data was obtained (represented in binary format, where values of 0
indicate male and
values of 1 indicate female). Next, race and ethnicity data was obtained
(represented in one
hot encoding via 6 binary values corresponding to each race/ethnicity
category). Next, the
data were filtered to keep certain columns corresponding to pertinent data
features, including
marital status, race, ethnicity, gender, healthcare expenses, age, counts by
county, positive
cases by county, counts by city, positive cases by city, prevalence by county,
prevalence by
city. As an example of an input data structure, the data for each subject or
patient may
comprise one or more of: a diagnostic test result, an age, sex/gender, zip
code (or another de-
identified geolocator), reason for diagnostic testing, vital signs (if
available and accessible),
and symptoms (if available). For example, symptoms may include fever, cough,
sore throat,
tiredness, body temperature, respiratory rate, loss of taste, loss of smell,
vomiting, shortness
of breath, chills, hypoxemia, etc. Next, prevalence data was obtained from the
Starschema
COVID-19 epidemiological data set.
100961 Next, the data was split into a training dataset and a testing dataset.
Next, a supervised
machine learning classifier (e.g., Random Forest classifier, extreme gradient
boosting
(XGBoost), and gradient boosting) was created and trained using the training
dataset. Next,
predicted outcomes for the testing dataset were obtained. Next, a
classification report and
confusion matrix were generated to evaluate the classifier performance. Next,
the total
numbers of positive cases and negative cases in the test dataset were
obtained. Next, a set of
feature importance values (e.g., weights) were calculated.
100971 Table 1 shows relative performance metrics obtained using three
different supervised
machine learning classifiers (Random Forest classifier, extreme gradient
boosting
(XGBoost), and gradient boosting). The Random Forest classifier had the
following top 5
weighted features: fever, cough, body temperature, respiratory rate, and loss
of taste. The
XGBoost classifier had the following top 5 weighted features: fever, cough,
hypoxemia, body
temperature, and loss of taste. The gradient boosting classifier had the
following top 5
weighted features: fever, cough, body temperature, respiratory rate, and loss
of taste.
-28-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
Model Name Precision
Recall fl Score
0
Random Forest 0.988292 G.980624 0.984221
XGBoost 0.988992 0.985825 0.987366
2 Gradient Boosting 0.986808 0.980375 0.983421
[0098] Table 1: Performance comparison across machine learning classifier
models
[0099] FIG. 5A shows the percentage of test savings that is achieved using
simple sample
pooling (gray), using intelligent sample pooling based on methods and systems
of the present
disclosure (purple), and an upper bound achieved using intelligent sample
pooling (dashed).
As shown in FIG. SA, the percentage of test savings achieved using simple
pooling
diminishes rapidly as the prevalence rate of the disease increases, and simple
pooling is not
viable above a prevalence rate of about 25%, further, a greater percentage of
test savings is
achieved using intelligent sample pooling based on methods and systems of the
present
disclosure, as compared to using simple sample pooling, across all prevalence
rates of
disease.
101001 FIG. 5B shows the percentage increase in diagnostic testing capacity
that is achieved
using simple sample pooling (gray), using intelligent sample pooling based on
methods and
systems of the present disclosure (purple), and an upper bound achieved using
intelligent
sample pooling (dashed). As shown in FIG. 5B, the percentage increase in
diagnostic testing
capacity achieved using simple pooling diminishes rapidly as the prevalence
rate of the
disease increases, and simple pooling is not viable above a prevalence rate of
about 25%;
further, a greater percentage increase in diagnostic testing capacity is
achieved using
intelligent sample pooling based on methods and systems of the present
disclosure, as
compared to using simple sample pooling, across all prevalence rates of
disease. Therefore,
using intelligent sample pooling based on methods and systems of the present
disclosure, a
consistent increase in testing capacity is achieved in all prevalence
environments, but also a
pool prevalence below 30% is strategically and actively maintained.
-29-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
Example 3: Number of diagnostic tests needed with intelligent sample pool
testing
101011 Using methods and systems of the present disclosure, efficient sample
pooling is
performed using for increasing diagnostic testing capacity of COVID-19. FIG. 6
shows a
relative number of diagnostic tests that is required versus the sample pool
size (ranging from
1 sample to 20 samples per pool), at a prevalence rate (PR) of 1% (light
blue), 5% (orange),
10% (gray), 20% (yellow), and 30% (dark blue). The relative number of tests is
calculated by
mimicking the utilization of resources. The relative number of tests is
minimized based on
two approaches. First, the best sample pool size or sample pooling strategy is
selected. By
referring to the resulting graph, the sample pool size that provides the
lowest number of
diagnostic tests is determined. Second, the prevalence rate (PR) is reduced.
Lower prevalence
rates within sample pools yield a lower number of diagnostic tests required,
thereby
improving upon the first strategy of selecting the best sample pool size or
sample pooling
strategy Using methods and systems of the present disclosure, the prevalence
rate of sample
pools is reduced through intelligent segmentation using machine learning
methodologies.
Example 4: Increase in diagnostic testing capacity at high clinical
sensitivity
101021 A population of 40,000 clinical test samples (e.g., which corresponds
to Quest
Diagnostic's daily testing capacity) was simulated, assuming a disease
prevalence rate of
10%, a clinical sensitivity of 90%, and a clinical specificity of 90%. Using
methods and
systems of the present disclosure, an optimal pool size of 4 samples per pool
was selected,
and a 40% reduction in diagnostic test kit utilization was achieved, along
with a clinical
sensitivity of 80% and a clinical specificity of 96% (as shown in FIG. 7).
Therefore, these
results represent an increase in Quest Diagnostic's testing capacity by 16,000
diagnostic tests,
without changing their initial kit availability.
101031 While preferred embodiments of the present invention have been shown
and
described herein, it will be obvious to those skilled in the art that such
embodiments are
provided by way of example only. It is not intended that the invention be
limited by the
specific examples provided within the specification. While the invention has
been described
with reference to the aforementioned specification, the descriptions and
illustrations of the
embodiments herein are not meant to be construed in a limiting sense. Numerous
variations,
changes, and substitutions will now occur to those skilled in the art without
departing from
the invention. Furthermore, it shall be understood that all aspects of the
invention are not
limited to the specific depictions, configurations or relative proportions set
forth herein which
depend upon a variety of conditions and variables. It should be understood
that various
-30-
CA 03187387 2023- 1- 26
WO 2022/006246
PCT/US2021/039849
alternatives to the embodiments of the invention described herein may be
employed in
practicing the invention. It is therefore contemplated that the invention
shall also cover any
such alternatives, modifications, variations or equivalents. It is intended
that the following
claims define the scope of the invention and that methods and structures
within the scope of
these claims and their equivalents be covered thereby.
-31-
CA 03187387 2023- 1- 26