Note: Descriptions are shown in the official language in which they were submitted.
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
INIR6 TRANSGENIC MAIZE
REFERENCE TO SEQUENCE LISTING SUBMITTED
ELECTRONICALLY
100011 The sequence listing contained in the file named "10086W01 ST25.txt",
which was
created on July 28, 2021 and electronically filed via EFS-Web on July 28,
2021, is
incorporated herein by reference in its entirety.
BACKGROUND
[0002] Transgenes which are placed into different positions in the plant
genome through non-
site specific integration can exhibit different levels of expression (Weising
et al., 1988, Ann.
Rev. Genet. 22:421-477). Such transgene insertion sites can also contain
various undesirable
rearrangements of the foreign DNA elements that include deletions and/or
duplications.
Furthermore, many transgene insertion sites can also comprise selectable or
scoreable marker
genes which in some instances are no longer required once a transgenic plant
event
containing the linked transgenes which confer desirable traits are selected.
[0003] Commercial transgenic plants typically comprise one or more independent
insertions
of transgenes at specific locations in the host plant genome that have been
selected for
features that include expression of the transgene(s) of interest and the
transgene-conferred
trait(s), absence or minimization of rearrangements, and normal Mendelian
transmission of
the trait(s) to progeny. An examples of a selected transgenic corn event which
confers
lepidopteran and coleopteran insect pest tolerance is the DP-4114 transgenic
maize event
disclosed in U.S. Patent No. 8,575,434. DP-4114 transgenic maize plants
express a CrylF
protein which can confer resistance to European corn borer (ECB, Ostrinia
nubilalis)
infestations as well as cry34Ab1 and cry35Ab1 proteins which can confer
resistance to corn
rootworm (CRW; Diabrotica sp. Including Diabrotica virgifera virgifera)
infestations. DP-
4114 transgenic maize plants also express a phosphinotricin acetyl transferase
(PAT) protein
which confers tolerance to the herbicide glufosinate.
[0004] Methods for removing selectable marker genes and/or duplicated
transgenes in
transgene insertion sites in plant genomes involving use of site-specific
recombinase systems
- 1 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
(e.g., cre-lox) as well as for insertion of new genes into transgene insertion
sites have been
disclosed (Srivastava and Ow; Methods Mol Biol, 2015,1287:95-103; Dale and Ow,
1991,
Proc. Natl Acad. Sci. USA 88, 10558-10562; Srivastava and Thomson, Plant
Biotechnol
J, 2016;14(2):471-82). Such methods typically require incorporation of the
recombination site
sequences recognized by the recombinase at particular locations within the
transgene.
SUMMARY
[0005] Transgenic maize plant cells comprising an INIR6 transgenic locus
comprising an
originator guide RNA recognition site (OgRRS) in a first DNA junction
polynucleotide of a
DP-4114 transgenic locus and a cognate guide RNA recognition site (CgRRS) in a
second
DNA junction polynucleotide of the DP-4114 transgenic locus are provided.
Transgenic
maize plant cells comprising an INIR6 transgenic locus comprising an insertion
and/or
substitution in a DNA junction polynucleotide of a DP-4114 transgenic locus of
DNA
comprising a cognate guide RNA recognition site (CgRRS) are provided. In
certain
embodiments, the DP-4114 transgenic locus is set forth in SEQ ID NO:1, is
present in seed
deposited at the ATCC under accession No. PTA-11506 is present in progeny
thereof, is
present in allelic variants thereof, or is present in other variants thereof.
INIR6 transgenic
maize plant cells, transgenic maize plant seeds, and transgenic maize plants
all comprising a
transgenic locus set forth in SEQ ID NO: 20 are provided. Transgenic maize
plant parts
including seeds and transgenic maize plants comprising the maize plant cells
are also
provided.
[0006] Methods for obtaining a bulked population of inbred seed comprising
selfing the
aforementioned transgenic maize plants and harvesting seed comprising the
INIR6 transgenic
locus from the selfed maize plant are also provided.
[0007] Methods of obtaining hybrid maize seed comprising crossing the
aforementioned
transgenic maize plants to a second maize plant which is genetically distinct
from the first
maize plant and harvesting seed comprising the INIR6 transgenic locus from the
cross are
provided. Methods for obtaining a bulked population of seed comprising selfing
a transgenic
maize plant of comprising SEQ ID NO: 20 and harvesting transgenic seed
comprising the
transgenic locus set forth in SEQ ID NO: 20 are provided.
- 2 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
[0008] A DNA molecule comprising SEQ ID NO: 2, 3, 8, 9, 10, 19, 20, or an
allelic variant
thereof is provided. Processed transgenic maize plant products and biological
samples
comprising the DNA molecules are provided. Nucleic acid molecules adapted for
detection of
genomic DNA comprising the DNA molecules, wherein said nucleic acid molecule
optionally comprises a detectable label are provided. Methods of detecting a
maize plant cell
comprising the INIR6 transgenic locus of any one of claims 1 to 3, comprising
the step of
detecting a DNA molecule comprising SEQ ID NO: 2, 3, 8, 9, 10, 19, or 20 are
provided.
[0009] Methods of excising the INIR6 transgenic locus from the genome of the
aforementioned maize plant cells comprising the steps of: (a) contacting the
edited transgenic
plant genome of the plant cell with: (i) an RNA dependent DNA endonuclease
(RdDe); and
(ii) a guide RNA (gRNA) capable of hybridizing to the guide RNA hybridization
site of the
OgRRS and the CgRRS; wherein the RdDe recognizes a OgRRS/gRNA and a CgRRS/gRNA
hybridization complex; and, (b) selecting a transgenic plant cell, transgenic
plant part, or
transgenic plant wherein the INIR6 transgenic locus flanked by the OgRRS and
the CgRRS
has been excised.
BRIEF DESCRIPTION OF THE DRAWINGS/FIGURES
100101 The patent or application file contains at least one drawing executed
in color. Copies
of this patent or patent application publication with color drawing(s) will be
provided by the
Office upon request and payment of the necessary fee.
[0011] Figure 1 shows a diagram of transgene expression cassettes and
selectable markers in
the DP-4114 transgenic locus. Nucleotides 1-2422 of SEQ ID NO: 1 are 5'
flanking plant
genomic DNA sequence, nucleotides 2423 to 14347 of SEQ ID NO: 1 are transgenic
insert
DNA sequence, and nucleotides 14348 to 16752 of SEQ ID NO: 1 are 3' plant
genome
flanking DNA sequence of the DP-4114 transgenic locus.
[0012] Figure 2 shows a schematic diagram which compares current breeding
strategies for
introgression of transgenic events (i.e., transgenic loci) to alternative
breeding strategies for
introgression of transgenic events where the transgenic events (i.e.,
transgenic loci) can be
removed following introgression to provide different combinations of
transgenic traits. In
Figure 2, "GE" refers to genome editing (e.g., including introduction of
targeted genetic
changes with genome editing molecules and "Event Removal" refers to excision
of a
transgenic locus (i.e., an "Event") with genome editing molecules.
- 3 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
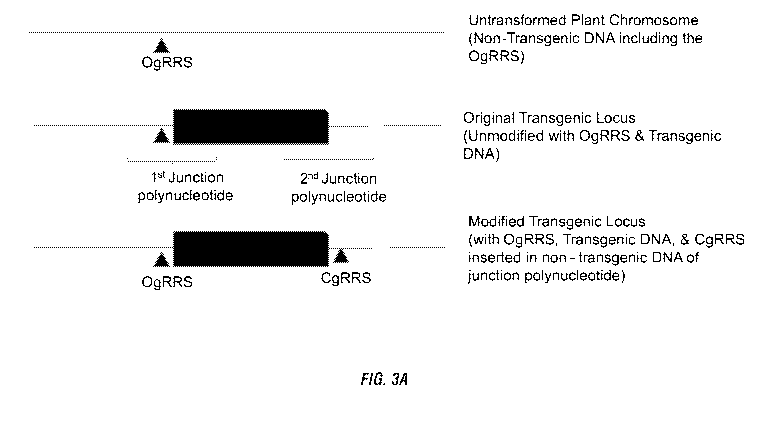
[0013] Figure 3A, B, C. Figure 3A shows a schematic diagram of a non-limiting
example of:
(i) an untransformed plant chromosome containing non-transgenic DNA which
includes the
originator guide RNA recognition site (OgRRS) (top); (ii) the original
transgenic locus with
the OgRRS in the non-transgenic DNA of the 1st junction polynucleotide
(middle); and (iii)
the modified transgenic locus with a cognate guide RNA inserted into the non-
transgenic
DNA of the 2nd junction polynucleotide (bottom). Figure 3B shows a schematic
diagram of a
non-limiting example of a process where a modified transgenic locus with a
cognate guide
RNA inserted into the non-transgenic DNA of the 2' junction polynucleotide
(top) is
subjected to cleavage at the OgRRS and CgRRS with one guide RNA (gRNA) that
hybridizes
to gRNA hybridization site in both the OgRRS and the CgRRS and an RNA
dependent DNA
endonuclease (RdDe) that recognizes and cleaves the gRNA/OgRRS and the
gRNA/CgRRS
complex followed by non-homologous end joining processes to provide a plant
chromosome
where the transgenic locus is excised. Figure 3C shows a schematic diagram of
a non-limiting
example of a process where a modified transgenic locus with a cognate guide
RNA inserted
into the non-transgenic DNA of the 2nd junction polynucleotide (top) is
subjected to cleavage
at the OgRRS and CgRRS with one guide RNA (gRNA) that hybridizes to the gRNA
hybridization site in both the OgRRS and the CgRRS and an RNA dependent DNA
endonuclease (RdDe) that recognizes and cleaves the gRNA/OgRRS and the
gRNA/CgRRS
complex in the presence of a donor DNA template. In Figure 3C, cleavage of the
modified
transgenic locus in the presence of the donor DNA template which has homology
to non-
transgenic DNA but lacks the OgRRS in the 1st and 2nd junction polynucleotides
followed by
homology-directed repair processes to provide a plant chromosome where the
transgenic
locus is excised and non-transgenic DNA present in the untransformed plant
chromosome is
at least partially restored.
DETAILED DESCRIPTION
[0014] Unless otherwise stated, nucleic acid sequences in the text of this
specification are
given, when read from left to right, in the 5' to 3' direction. Nucleic acid
sequences may be
provided as DNA or as RNA, as specified; disclosure of one necessarily defines
the other, as
well as necessarily defines the exact complements, as is known to one of
ordinary skill in the
art.
- 4 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
[0015] Where a term is provided in the singular, the inventors also
contemplate embodiments
described by the plural of that term.
[0016] The term "about" as used herein means a value or range of values which
would be
understood as an equivalent of a stated value and can be greater or lesser
than the value or
range of values stated by 10 percent. Each value or range of values preceded
by the term
"about" is also intended to encompass the embodiment of the stated absolute
value or range
of values.
[0017] The phrase "allelic variant" as used herein refers to a polynucleotide
or polypeptide
sequence variant that occurs in a different strain, variety, or isolate of a
given organism.
[0018] The term "and/or" where used herein is to be taken as specific
disclosure of each of
the two specified features or components with or without the other. Thus, the
term and/or" as
used in a phrase such as "A and/or B" herein is intended to include "A and B,"
"A or B," "A"
(alone), and "B" (alone). Likewise, the term "and/or" as used in a phrase such
as "A, B,
and/or C" is intended to encompass each of the following embodiments: A, B,
and C; A, B, or
C; A or C; A or B; B or C; A and C; A and B; B and C; A (alone); B (alone);
and C (alone).
[0019] As used herein, the phrase "approved transgenic locus" is a genetically
modified plant
event which has been authorized, approved, and/or de-regulated for any one of
field testing,
cultivation, human consumption, animal consumption, and/or import by a
governmental
body. Illustrative and non-limiting examples of governmental bodies which
provide such
approvals include the Ministry of Agriculture of Argentina, Food Standards
Australia New
Zealand, National Biosafety Technical Committee (CTNBio) of Brazil, Canadian
Food
Inspection Agency, China Ministry of Agriculture Biosafety Network, European
Food Safety
Authority, US Department of Agriculture, US Department of Environmental
Protection, and
US Food and Drug Administration.
[0020] The term "backcross", as used herein, refers to crossing an F 1 plant
or plants with one
of the original parents. A backcross is used to maintain or establish the
identity of one parent
(species) and to incorporate a particular trait from a second parent
(species). The term
"backcross generation", as used herein, refers to the offspring of a
backcross.
[0021] As used herein, the phrase "biological sample" refers to either intact
or non-intact
(e.g., milled seed or plant tissue, chopped plant tissue, lyophilized tissue)
plant tissue. It may
also be an extract comprising intact or non-intact seed or plant tissue. The
biological sample
can comprise flour, meal, syrup, oil, starch, and cereals manufactured in
whole or in part to
- 5 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
contain crop plant by-products. In certain embodiments, the biological sample
is "non-
regenerable" (i.e., incapable of being regenerated into a plant or plant
part). In certain
embodiments, the biological sample refers to a homogenate, an extract, or any
fraction
thereof containing genomic DNA of the organism from which the biological
sample was
obtained, wherein the biological sample does not comprise living cells.
[0022] As used herein, the terms "correspond," "corresponding," and the like,
when used in
the context of an nucleotide position, mutation, and/or substitution in any
given
polynucleotide (e.g., an allelic variant of SEQ ID NO: 1) with respect to the
reference
polynucleotide sequence (e.g., SEQ ID NO: 1) all refer to the position of the
polynucleotide
residue in the given sequence that has identity to the residue in the
reference nucleotide
sequence when the given polynucleotide is aligned to the reference
polynucleotide sequence
using a pairwise alignment algorithm (e.g., CLUSTAL 0 1.2.4 with default
parameters).
[0023] As used herein, the terms "Cpfl" and "Cas12a" are used interchangeably
to refer to
the same RNA dependent DNA endonuclease (RdDe). A Cas12a protein provided
herein
includes the protein of SEQ ID NO: 21.
[0024] The term "crossing" as used herein refers to the fertilization of
female plants (or
gametes) by male plants (or gametes). The term "gamete" refers to the haploid
reproductive
cell (egg or pollen) produced in plants by meiosis from a gametophyte and
involved in sexual
reproduction, during which two gametes of opposite sex fuse to form a diploid
zygote. The
term generally includes reference to a pollen (including the sperm cell) and
an ovule
(including the ovum). When referring to crossing in the context of achieving
the introgression
of a genomic region or segment, the skilled person will understand that in
order to achieve
the introgression of only a part of a chromosome of one plant into the
chromosome of another
plant, random portions of the genomes of both parental lines recombine during
the cross due
to the occurrence of crossing-over events in the production of the gametes in
the parent lines.
Therefore, the genomes of both parents must be combined in a single cell by a
cross, where
after the production of gametes from the cell and their fusion in
fertilization will result in an
introgression event.
[0025] As used herein, the phrases "DNA junction polynucleotide" and "junction
polynucleotide" refers to a polynucleotide of about 18 to about 500 base pairs
in length
comprised of both endogenous chromosomal DNA of the plant genome and
heterologous
transgenic DNA which is inserted in the plant genome. A junction
polynucleotide can thus
- 6 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
comprise about 8, 10, 20, 50, 100, 200, 250, 500, or 1000 base pairs of
endogenous
chromosomal DNA of the plant genome and about 8, 10, 20, 50, 100, 200, 250,
500, or 1000
base pairs of heterologous transgenic DNA which span the one end of the
transgene insertion
site in the plant chromosomal DNA. Transgene insertion sites in chromosomes
will typically
contain both a 5' junction polynucleotide and a 3' junction polynucleotide. In
embodiments
set forth herein in SEQ ID NO: 1, the 5' junction polynucleotide is located at
the 5' end of the
sequence and the 3' junction polynucleotide is located at the 3' end of the
sequence. In a non-
limiting and illustrative example, a 5' junction polynucleotide of a
transgenic locus is
telomere proximal in a chromosome arm and the 3' junction polynucleotide of
the transgenic
locus is centromere proximal in the same chromosome arm. In another non-
limiting and
illustrative example, a 5' junction polynucleotide of a transgenic locus is
centromere
proximal in a chromosome arm and the 3' junction polynucleotide of the
transgenic locus is
telomere proximal in the same chromosome arm. The junction polynucleotide
which is
telomere proximal and the junction polynucleotide which is centromere proximal
can be
determined by comparing non-transgenic genomic sequence of a sequenced non-
transgenic
plant genome to the non-transgenic DNA in the junction polynucleotides.
[0026] The term "donor," as used herein in the context of a plant, refers to
the plant or plant
line from which the trait, transgenic event, or genomic segment originates,
wherein the donor
can have the trait, introgression, or genomic segment in either a heterozygous
or homozygous
state.
[0027] As used herein, the term "DP-4114" is used to refer to any of a
transgenic maize
locus, transgenic maize plants and parts thereof including seed set forth in
US Patent No.
8,575,434, which is incorporated herein by reference in its entirety.
Representative DP-4114
transgenic maize seed have been deposited with American Type Culture
Collection (ATCC,
Manassas, Va. 20110-2209 USA) under Accession No. PTA-11506. DP-4114
transgenic loci
include loci having the sequence of SEQ ID NO:1, the sequence of the DP-4114
locus in the
deposited seed of Accession No. PTA-11506 and any progeny thereof, as well as
allelic
variants and other variants of SEQ ID NO:1
[0028] As used herein, the terms "excise" and "delete," when used in the
context of a DNA
molecule, are used interchangeably to refer to the removal of a given DNA
segment or
element (e.g., transgene element or transgenic locus or portion thereof) of
the DNA molecule.
- 7 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
[0029] As used herein, the phrase "elite crop plant" refers to a plant which
has undergone
breeding to provide one or more trait improvements. Elite crop plant lines
include plants
which are an essentially homozygous, e.g., inbred or doubled haploid. Elite
crop plants can
include inbred lines used as is or used as pollen donors or pollen recipients
in hybrid seed
production (e.g., used to produce Fl plants). Elite crop plants can include
inbred lines which
are selfed to produce non-hybrid cultivars or varieties or to produce (e.g.,
bulk up) pollen
donor or recipient lines for hybrid seed production. Elite crop plants can
include hybrid Fl
progeny of a cross between two distinct elite inbred or doubled haploid plant
lines.
[0030] As used herein, an "event," "a transgenic event," "a transgenic locus"
and related
phrases refer to an insertion of one or more transgenes at a unique site in
the genome of a
plant as well as to DNA fragments, plant cells, plants, and plant parts (e.g.,
seeds) comprising
genomic DNA containing the transgene insertion. Such events typically comprise
both a 5'
and a 3' DNA junction polynucleotide and confer one or more useful traits
including
herbicide tolerance, insect resistance, male sterility, and the like.
[0031] As used herein, the phrases "endogenous sequence," "endogenous gene,"
"endogenous DNA," "endogenous polynucleotide," and the like refer to the
native form of a
polynucleotide, gene or polypeptide in its natural location in the organism or
in the genome
of an organism.
[0032] The terms "exogenous" and "heterologous" as are used synonymously
herein to refer
to any polynucleotide (e.g., DNA molecule) that has been inserted into a new
location in the
genome of a plant. Non-limiting examples of an exogenous or heterologous DNA
molecule
include a synthetic DNA molecule, a non-naturally occurring DNA molecule, a
DNA
molecule found in another species, a DNA molecule found in a different
location in the same
species, and/or a DNA molecule found in the same strain or isolate of a
species, where the
DNA molecule has been inserted into a new location in the genome of a plant.
[0033] As used herein, the term "Fl" refers to any offspring of a cross
between two
genetically unlike individuals.
[0034] The term "gene," as used herein, refers to a hereditary unit consisting
of a sequence of
DNA that occupies a specific location on a chromosome and that contains the
genetic
instruction for a particular characteristics or trait in an organism. The term
"gene" thus
includes a nucleic acid (for example, DNA or RNA) sequence that comprises
coding
sequences necessary for the production of an RNA, or a polypeptide or its
precursor. A
- 8 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
functional polypeptide can be encoded by a full length coding sequence or by
any portion of
the coding sequence as long as the desired activity or functional properties
(e.g., enzymatic
activity, pesticidal activity, ligand binding, and/or signal transduction) of
the RNA or
polypeptide are retained.
[0035] The term "identifying," as used herein with respect to a plant, refers
to a process of
establishing the identity or distinguishing character of a plant, including
exhibiting a certain
trait, containing one or more transgenes, and/or containing one or more
molecular markers.
[0036] As used herein, the term "INIR6" is used to refer either individually
collectively to
items that include any or all of the DP-4114 transgenic maize loci which have
been modified
as disclosed herein, modified DP-4114 transgenic maize plants and parts
thereof including
seed, and DNA obtained therefrom.
[0037] The term "isolated" as used herein means having been removed from its
natural
environment.
[0038] As used herein, the terms "include," "includes," and "including" are to
be construed
as at least having the features to which they refer while not excluding any
additional
unspecified features.
[0039] As used herein, the phrase "introduced transgene" is a transgene not
present in the
original transgenic locus in the genome of an initial transgenic event or in
the genome of a
progeny line obtained from the initial transgenic event. Examples of
introduced transgenes
include exogenous transgenes which are inserted in a resident original
transgenic locus.
[0040] As used herein, the terms "introgression", "introgressed" and
"introgressing" refer to
both a natural and artificial process, and the resulting plants, whereby
traits, genes or DNA
sequences of one species, variety or cultivar are moved into the genome of
another species,
variety or cultivar, by crossing those species. The process may optionally be
completed by
backcrossing to the recurrent parent. Examples of introgression include entry
or introduction
of a gene, a transgene, a regulatory element, a marker, a trait, a trait
locus, or a chromosomal
segment from the genome of one plant into the genome of another plant.
[0041] The phrase "marker-assisted selection", as used herein, refers to the
diagnostic
process of identifying, optionally followed by selecting a plant from a group
of plants using
the presence of a molecular marker as the diagnostic characteristic or
selection criterion. The
process usually involves detecting the presence of a certain nucleic acid
sequence or
polymorphism in the genome of a plant.
- 9 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
[0042] The phrase "molecular marker", as used herein, refers to an indicator
that is used in
methods for visualizing differences in characteristics of nucleic acid
sequences. Examples of
such indicators are restriction fragment length polymorphism (RFLP) markers,
amplified
fragment length polymorphism (AFLP) markers, single nucleotide polymorphisms
(SNPs),
microsatellite markers (e.g. SSRs), sequence-characterized amplified region
(SCAR)
markers, Next Generation Sequencing (NGS) of a molecular marker, cleaved
amplified
polymorphic sequence (CAPS) markers or isozyme markers or combinations of the
markers
described herein which defines a specific genetic and chromosomal location.
[0043] As used herein the terms "native" or "natural" define a condition found
in nature. A
"native DNA sequence" is a DNA sequence present in nature that was produced by
natural
means or traditional breeding techniques but not generated by genetic
engineering (e.g., using
molecular biology/transformation techniques).
[0044] The term "offspring", as used herein, refers to any progeny generation
resulting from
crossing, selfing, or other propagation technique.
[0045] The phrase "operably linked" refers to a juxtaposition wherein the
components so
described are in a relationship permitting them to function in their intended
manner. For
instance, a promoter is operably linked to a coding sequence if the promoter
affects its
transcription or expression. When the phrase "operably linked" is used in the
context of a
PAM site and a guide RNA hybridization site, it refers to a PAM site which
permits cleavage
of at least one strand of DNA in a polynucleotide with an RNA dependent DNA
endonuclease or RNA dependent DNA nickase which recognize the PAM site when a
guide
RNA complementary to guide RNA hybridization site sequences adjacent to the
PAM site is
present. A OgRRS and its CgRRS are operably linked to junction polynucleotides
when they
can be recognized by a gRNA and an RdDe to provide for excision of the
transgenic locus or
portion thereof flanked by the junction polynucleotides.
[0046] As used herein, the term "plant" includes a whole plant and any
descendant, cell,
tissue, or part of a plant. The term "plant parts" include any part(s) of a
plant, including, for
example and without limitation: seed (including mature seed and immature
seed); a plant
cutting; a plant cell; a plant cell culture; or a plant organ (e.g., pollen,
embryos, flowers,
fruits, shoots, leaves, roots, stems, and explants). A plant tissue or plant
organ may be a seed,
protoplast, callus, or any other group of plant cells that is organized into a
structural or
functional unit. A plant cell or tissue culture may be capable of regenerating
a plant having
- 10 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
the physiological and morphological characteristics of the plant from which
the cell or tissue
was obtained, and of regenerating a plant having substantially the same
genotype as the plant.
Regenerable cells in a plant cell or tissue culture may be embryos,
protoplasts, meristematic
cells, callus, pollen, leaves, anthers, roots, root tips, silk, flowers,
kernels, ears, cobs, husks,
or stalks. In contrast, some plant cells are not capable of being regenerated
to produce plants
and are referred to herein as "non-regenerable" plant cells.
[0047] The term "purified," as used herein defines an isolation of a molecule
or compound in
a form that is substantially free of contaminants normally associated with the
molecule or
compound in a native or natural environment and means having been increased in
purity as a
result of being separated from other components of the original composition.
The term
"purified nucleic acid" is used herein to describe a nucleic acid sequence
which has been
separated from other compounds including, but not limited to polypeptides,
lipids and
carbohydrates.
[0048] The term "recipient", as used herein, refers to the plant or plant line
receiving the trait,
transgenic event or genomic segment from a donor, and which recipient may or
may not have
the have trait, transgenic event or genomic segment itself either in a
heterozygous or
homozygous state.
[0049] As used herein the term "recurrent parent" or "recurrent plant"
describes an elite line
that is the recipient plant line in a cross and which will be used as the
parent line for
successive backcrosses to produce the final desired line.
[0050] As used herein the term "recurrent parent percentage" relates to the
percentage that a
backcross progeny plant is identical to the recurrent parent plant used in the
backcross. The
percent identity to the recurrent parent can be determined experimentally by
measuring
genetic markers such as SNPs and/or RFLPs or can be calculated theoretically
based on a
mathematical formula.
[0051] The terms "selfed," "selfing," and "self," as used herein, refer to any
process used to
obtain progeny from the same plant or plant line as well as to plants
resulting from the
process. As used herein, the terms thus include any fertilization process
wherein both the
ovule and pollen are from the same plant or plant line and plants resulting
therefrom.
Typically, the terms refer to self-pollination processes and progeny plants
resulting from self-
pollination.
-11-
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
[0052] The term "selecting", as used herein, refers to a process of picking
out a certain
individual plant from a group of individuals, usually based on a certain
identity, trait,
characteristic, and/or molecular marker of that individual.
[0053] As used herein, the phrase "originator guide RNA recognition site" or
the acronym
"OgRRS" refers to an endogenous DNA polynucleotide comprising a protospacer
adjacent
motif (PAM) site operably linked to a guide RNA hybridization site. In certain
embodiments,
an OgRRS can be located in an untransformed plant chromosome or in non-
transgenic DNA
of a DNA junction polynucleotide of both an original transgenic locus and a
modified
transgenic locus. In certain embodiments, an OgRRS can be located in
transgenic DNA of a
DNA junction polynucleotide of both an original transgenic locus and a
modified transgenic
locus. In certain embodiments, an OgRRS can be located in both transgenic DNA
and non-
transgenic DNA of a DNA junction polynucleotide of both an original transgenic
locus and a
modified transgenic locus (i.e., can span transgenic and non-transgenic DNA in
a DNA
junction polynucleotide).
[0054] As used herein the phrase "cognate guide RNA recognition site" or the
acronym
"CgRRS" refer to a DNA polynucleotide comprising a PAM site operably linked to
a guide
RNA hybridization site, where the CgRRS is absent from transgenic plant
genomes
comprising a first original transgenic locus that is unmodified and where the
CgRRS and its
corresponding OgRRS can hybridize to a single gRNA. A CgRRS can be located in
transgenic DNA of a DNA junction polynucleotide of a modified transgenic
locus, in
transgenic DNA of a DNA junction polynucleotide of a modified transgenic
locus, or in both
transgenic and non-transgenic DNA of a modified transgenic locus (i.e., can
span transgenic
and non-transgenic DNA in a DNA junction polynucleotide).
[0055] As used herein, the phrase "a transgenic locus excision site" refers to
the DNA which
remains in the genome of a plant or in a DNA molecule (e.g., an isolated or
purified DNA
molecule) wherein a segment comprising, consisting essentially of, or
consisting of a
transgenic locus has been deleted. In a non-limiting and illustrative example,
a transgenic
locus excision site can thus comprise a contiguous segment of DNA comprising
at least 10
base pairs of DNA that is telomere proximal to the deleted transgenic locus or
to the deleted
segment of the transgenic locus and at least 10 base pairs of DNA that is
centromere proximal
to the deleted transgenic locus or to the deleted segment of the transgenic
locus.
- 12 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
[0056] As used herein, the phrase "transgene element" refers to a segment of
DNA
comprising, consisting essentially of, or consisting of a promoter, a 5' UTR,
an intron, a
coding region, a 3'UTR, or a polyadenylation signal. Polyadenylation signals
include
transgene elements referred to as "terminators" (e.g., NOS, pinII, rbcs,
Hsp17, TubA).
[0057] To the extent to which any of the preceding definitions is inconsistent
with definitions
provided in any patent or non-patent reference incorporated herein by
reference, any patent or
non-patent reference cited herein, or in any patent or non-patent reference
found elsewhere, it
is understood that the preceding definition will be used herein.
[0058] Genome editing molecules can permit introduction of targeted genetic
change
conferring desirable traits in a variety of crop plants (Zhang et al. Genome
Biol. 2018; 19:
210; Schindele et al. FEBS Lett. 2018;592(12):1954). Desirable traits
introduced into crop
plants such as maize and soybean include herbicide tolerance, improved food
and/or feed
characteristics, male-sterility, and drought stress tolerance. Nonetheless,
full realization of the
potential of genome editing methods for crop improvement will entail efficient
incorporation
of the targeted genetic changes in germplasm of different elite crop plants
adapted for distinct
growing conditions. Such elite crop plants will also desirably comprise useful
transgenic loci
which confer various traits including herbicide tolerance, pest resistance
(e.g.; insect,
nematode, fungal disease, and bacterial disease resistance), conditional male
sterility systems
for hybrid seed production, abiotic stress tolerance (e.g., drought
tolerance), improved food
and/or feed quality, and improved industrial use (e.g., biofuel). Provided
herein are methods
whereby targeted genetic changes are efficiently combined with desired subsets
of transgenic
loci in elite progeny plant lines (e.g., elite inbreds used for hybrid seed
production or for
inbred varietal production). Also provided are plant genomes containing
modified transgenic
loci which can be selectively excised with a single gRNA molecule. Such
modified
transgenic loci comprise an originator guide RNA recognition site (OgRRS)
which is
identified in non-transgenic DNA of a first junction polynucleotide of the
transgenic locus
and cognate guide RNA recognition site (CgRRS) which is introduced (e.g., by
genome
editing methods) into a second junction polynucleotide of the transgenic locus
and which can
hybridize to the same gRNA as the OgRRS, thereby permitting excision of the
modified
transgenic locus with a single guide RNA. An originator guide RNA recognition
site
(OgRRS) comprises endogenous DNA found in untransformed plants and in
endogenous
non-transgenic DNA of junction polynucleotides of transgenic plants containing
a modified
- 13 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
or unmodified transgenic locus. The OgRRS located in non-transgenic DNA of a
first DNA
junction polynucleotide is used to design a related cognate guide RNA
recognition site
(CgRRS) which is introduced (e.g., by genome editing methods) into the second
junction
polynucleotide of the transgenic locus. A CgRRS is thus present in junction
polynucleotides
of modified transgenic loci provided herein and is absent from endogenous DNA
found in
untransformed plants and absent from endogenous non-transgenic DNA found in
junction
sequences of transgenic plants containing an unmodified transgenic locus. Also
provided are
unique transgenic locus excision sites created by excision of such modified
transgenic loci,
DNA molecules comprising the modified transgenic loci, unique transgenic locus
excision
sites and/or plants comprising the same, biological samples containing the
DNA, nucleic acid
markers adapted for detecting the DNA molecules, and related methods of
identifying the
elite crop plants comprising unique transgenic locus excision sites.
[0059] Also provided herein are methods whereby targeted genetic changes are
efficiently
combined with desired subsets of transgenic loci in elite progeny plant lines
(e.g., elite
inbreds used for hybrid seed production or for inbred varietal production).
Examples of such
methods include those illustrated in Figure 2. In certain embodiments, INIR6
transgenic loci
provided here are characterized by polynucleotide sequences that can
facilitate as necessary
the removal of the INIR6 transgenic loci from the genome. Useful applications
of such INIR6
transgenic loci and related methods of making include targeted excision of a
INIR6
transgenic locus or portion thereof in certain breeding lines to facilitate
recovery of
germplasm with subsets of transgenic traits tailored for specific geographic
locations and/or
grower preferences. Other useful applications of such INIR6 transgenic loci
and related
methods of making include removal of transgenic traits from certain breeding
lines when it is
desirable to replace the trait in the breeding line without disrupting other
transgenic loci
and/or non-transgenic loci. In certain embodiments, maize genomes containing
INIR6
transgenic loci or portions thereof which can be selectively excised with one
or more gRNA
molecules and RdDe (RNA dependent DNA endonucleases) which form gRNA/target
DNA
complexes. Such selectively excisable INIR6 transgenic loci can comprise an
originator guide
RNA recognition site (OgRRS) which is identified in non-transgenic DNA,
transgenic DNA,
or a combination thereof in of a first junction polynucleotide of the
transgenic locus and
cognate guide RNA recognition site (CgRRS) which is introduced (e.g., by
genome editing
methods) into a second junction polynucleotide of the transgenic locus and
which can
- 14 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
hybridize to the same gRNA as the OgRRS, thereby permitting excision of the
modified
transgenic locus or portions thereof with a single guide RNA (e.g., as shown
in Figures 3A
and B). In certain embodiments, an originator guide RNA recognition site
(OgRRS)
comprises endogenous DNA found in untransformed plants and in endogenous non-
transgenic DNA of junction polynucleotides of transgenic plants containing a
modified or
unmodified transgenic locus. In certain embodiments, an originator guide RNA
recognition
site (OgRRS) comprises exogenous transgenic DNA of junction polynucleotides of
transgenic plants containing a modified or unmodified transgenic locus. The
OgRRS located
in non-transgenic DNA transgenic DNA, or a combination thereof in of a first
DNA junction
polynucleotide is used to design a related cognate guide RNA recognition site
(CgRRS)
which is introduced (e.g., by genome editing methods) into the second junction
polynucleotide of the transgenic locus. A CgRRS is thus present in junction
polynucleotides
of modified transgenic loci provided herein and is absent from endogenous DNA
found in
untransformed plants and absent from junction sequences of transgenic plants
containing an
unmodified transgenic locus. A CgRRS is also absent from a combination of non-
transgenic
and transgenic DNA found in junction sequences of transgenic plants containing
an
unmodified transgenic locus. Examples of OgRRS polynucleotide sequences in or
near a 5'
junction polynucleotide in an DP-4114 transgenic locus include SEQ ID NO: 7.
OgRRS
polynucleotide sequences located in a first junction polynucleotide can be
introduced into the
second junction polynucleotide using donor DNA templates as illustrated in
Figure 3C and as
elsewhere described herein. A donor DNA template for introducing the SEQ ID
NO: 7
OgRRS into the 3' junction polynucleotide of an DP-4114 locus includes the
donor DNA
template formed by annealing SEQ ID NO: 11 and 12 or by annealing SEQ ID NO:
11 and
13. Double stranded breaks in a 3' junction polynucleotide of SEQ ID NO: 1 can
be
introduced with gRNAs encoded by SEQ ID NO: 4, 5, and/or 6 and a Cas12a
nuclease.
Integration of the SEQ ID NO: 11/12 or 11/13 donor DNA template into the 3'
junction
polynucleotide of an DP-4114 locus at the double stranded breaks introduced by
the gRNAs
encoded by SEQ ID NO: 4, 5, and/or 6 and a Cas12a nuclease can provide an
INIR6 locus
comprising the CgRRS sequence set forth in SEQ ID NO: 8, 9, or 10. Double
stranded
breaks in a 3' junction polynucleotide of SEQ ID NO: 1 can be introduced with
gRNAs
encoded by SEQ ID NO: 4, 5, and/or 6. Another donor DNA template adapted for
insertion
of the OgRRS of SEQ ID NO: 7 in a 3' junction polynucleotide of a DP-4114
transgenic
- 15 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
locus can comprise SEQ ID NO: 14. Double stranded breaks in a 3' junction
polynucleotide
of SEQ ID NO: 1 can be introduced with gRNAs encoded by SEQ ID NO: 5 and a
Cas12a
nuclease. A donor DNA template of SEQ ID NO: 14 or the equivalent thereof
having longer
or shorter homology arms can be used to obtain the CgRRS insertion in the 3'
junction
polynucleotide that is set forth in SEQ ID NO: 19. An INIR6 transgenic locus
containing this
CgRRS insertion is set forth in SEQ ID NO: 20.
[0060] Also provided herein are allelic variants of any of the INIR6
transgenic loci or DNA
molecules provided herein. In certain embodiments, such allelic variants of
INIR6 transgenic loci
include sequences having at least 85%, 90%, 95%, 98%, or 99% sequence identity
across the
entire length or at least 20, 40, 100, 500, 1,000, 2,000, 4,000, 6,000, 8,000,
9,000, 10,000,
12,000, 14,000, 16,000, or 16,752 nucleotides of SEQ ID NO: 2, 3, or 20. In
certain
embodiments, such allelic variants of INIR6 DNA molecules include sequences
having at least
85%, 90%, 95%, 98%, or 99% sequence identity across the entire length of SEQ
ID NO: 2, 3, 8,
9, 10, 19, or 20.
[0061] Also provided are unique transgenic locus excision sites created by
excision of INIR6
transgenic loci or selectively excisable INIR6 transgenic loci, DNA molecules
comprising the
INIR6 transgenic loci or unique fragments thereof (i.e., fragments of an INIR6
locus which
are not found in an DP-4114 transgenic locus). INIR6 plants comprising the
same, biological
samples containing the DNA, nucleic acid markers adapted for detecting the DNA
molecules,
and related methods of identifying maizew plants comprising unique INIR6
transgenic locus
excision sites and unique fragments of a INIR6 transgenic locus. DNA molecules
comprising
unique fragments of an INIR6 transgenic locus are diagnostic for the presence
of an INIR6
transgenic locus or fragments thereof in a maize plant, maize cell, maize
seed, products
obtained therefrom (e.g., seed meal or stover), and biological samples. DNA
molecules
comprising unique fragments of an INIR6 transgenic locus include DNA molecules
comprising
[0062] Methods provided herein can be used to excise any transgenic locus
where the first
and second junction sequences comprising the endogenous non-transgenic genomic
DNA and
the heterologous transgenic DNA which are joined at the site of transgene
insertion in the
plant genome are known or have been determined. In certain embodiments
provided herein,
transgenic loci can be removed from crop plant lines to obtain crop plant
lines with tailored
combinations of transgenic loci and optionally targeted genetic changes. Such
first and
- 16 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
second junction sequences are readily identified in new transgenic events by
inverse PCR
techniques using primers which are complementary the inserted transgenic
sequences. In
certain embodiments, the first and second junction sequences of transgenic
loci are published.
An example of a transgenic locus which can be improved and used in the methods
provided
herein is the maize DP-4114 transgenic locus. The maize DP-4114 transgenic
locus and its
transgenic junction sequences are also depicted in Figure 1. Maize plants
comprising the DP-
4114 transgenic locus and seed thereof have been cultivated, been placed in
commerce, and
have been described in a variety of publications by various governmental
bodies. Databases
which have compiled descriptions of the DP-4114 transgenic locus include the
International
Service for the Acquisition of Agri-biotech Applications (ISAAA) database
(available on the
world wide web internet site "isaaa.org/gmapprovaldatabase/event"), the GenBit
LLC
database (available on the world wide web internet
site
"genbitgroup.com/en/gmo/gmodatabase"), and the Biosafety Clearing-House (BCH)
database
(available on the http internet site "bch.cbd.int/database/organisms").
[0063] Sequences of the junction polynucleotides as well as the transgenic
insert(s) of the
DP-4114 transgenic locus which can be improved by the methods provided herein
are set
forth or otherwise provided in SEQ ID NO: 1, US 8,575,434, the sequence of the
DP-4114
locus in the deposited seed of ATCC accession No. PTA-11506, and elsewhere in
this
disclosure. In certain embodiments provided herein, the DP-4114 transgenic
locus set forth in
SEQ ID NO: 1 or present in the deposited seed of ATCC accession No. PTA-11506
is
referred to as an "original DP-4114 transgenic locus." Allelic or other
variants of the
sequence set forth SEQ ID NO: 1, the patent references set forth therein and
incorporated
herein by reference in their entireties, and elsewhere in this disclosure
which may be present
in certain variant DP-4114 transgenic plant loci (e.g., progeny of deposited
seed of accession
No. PTA-11506 which contain allelic variants of SEQ ID NO:1 or progeny
originating from
transgenic plant cells comprising the original MIR162 transgenic set forth in
US 8,575,434)
can also be improved by identifying sequences in the variants that correspond
to the SEQ ID
NO: 1 by performing a pairwise alignment (e.g., using CLUSTAL 0 1.2.4 with
default
parameters) and making corresponding changes in the allelic or other variant
sequences.
Such allelic or other variant sequences include sequences having at least 85%,
90%, 95%,
98%, or 99% sequence identity across the entire length or at least 20, 40,
100, 500, 1,000,
2,000, 4,000, 8,000, 10,000, 12,000, 14,350, 15,000, or 16,652 nucleotides of
SEQ ID NO: 1.
- 17 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
Also provided are plants, plant parts including seeds, genomic DNA, and/or DNA
obtained
from INIR6 plants which comprise one or more modifications (e.g., via
insertion of a CgRRS
in a junction polynucleotide sequence) which provide for selective excision of
the INIR6
transgenic locus or a portion thereof Also provided herein are methods of
detecting plants,
genomic DNA, and/or DNA obtained from plants comprising a INIR6 transgenic
locus which
contains one or more of a CgRRS, deletions of selectable marker genes,
deletions of non-
essential DNA, and/or a transgenic locus excision site. A first junction
polynucleotide of a
DP-4114 transgenic locus can comprise either one of the junction
polynucleotides found at
the 5' end or the 3' end of any one of the sequences set forth in SEQ ID NO:
1, allelic
variants thereof, or other variants thereof. In certain embodiments, a 5'
junction
polynucleotide of a DP-4114 transgenic locus can comprise DNA spanning the
plant genomic
DNA/ transgene insert junction at nucleotides 2422 and 2423 of SEQ ID NO: 1.
In certain
embodiments, a 3' junction polynucleotide of a DP-4114 transgenic locus can
comprise DNA
spanning the plant genomic DNA/ transgene insert junction at nucleotides 14347
and 14348
of SEQ ID NO: 1. An OgRRS can be found within non-transgenic DNA, transgenic
DNA, or
a combination thereof in either one of the junction polynucleotides of any one
of SEQ ID
NO: 1, allelic variants thereof, or other variants thereof A second junction
polynucleotide of
a transgenic locus can comprise either one of the junction polynucleotides
found at the 5' or
3' end of any one of the sequences set forth in SEQ ID NO: 1, allelic variants
thereof, or
other variants thereof. A CgRRS can be introduced within transgenic, non-
transgenic DNA,
or a combination thereof of either one of the junction polynucleotides of any
one of SEQ ID
NO: 1, allelic variants thereof, or other variants thereof to obtain an INIR6
transgenic locus.
In certain embodiments, the OgRRS is found in non-transgenic DNA or transgenic
DNA of
the 5' junction polynucleotide of a transgenic locus of any one of SEQ ID NO:
1, allelic
variants thereof, or other variants thereof and the corresponding CgRRS is
introduced into the
transgenic DNA, non-transgenic DNA, or a combination thereof in the 3'
junction
polynucleotide of the DP-4114 transgenic locus of SEQ ID NO: 1, allelic
variants thereof, or
other variants thereof to obtain an INIR6 transgenic locus. In other
embodiments, the OgRRS
is found in non-transgenic DNA or transgenic DNA of the 3' junction
polynucleotide of the
DP-4114 transgenic locus of any one of SEQ ID NO: 1, allelic variants thereof,
or other
variants thereof and the corresponding CgRRS is introduced into the transgenic
DNA,non-
transgenic DNA, or a combination thereof in the 5' junction polynucleotide of
the transgenic
- 18 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
locus of SEQ ID NO: 1, allelic variants thereof, or other variants thereof to
obtain an INIR6
transgenic locus.
[0064] In certain embodiments, the CgRRS is comprised in whole or in part of
an exogenous
DNA molecule that is introduced into a DNA junction polynucleotide by genome
editing. In
certain embodiments, the guide RNA hybridization site of the CgRRS is operably
linked to a
pre-existing PAM site in the transgenic DNA or non-transgenic DNA of the
transgenic plant
genome. In other embodiments, the guide RNA hybridization site of the CgRRS is
operably
linked to a new PAM site that is introduced in the DNA junction polynucleotide
by genome
editing. A CgRRS can be located in non-transgenic plant genomic DNA of a DNA
junction
polynucleotide of an INIR6 transgenic locus, in transgenic DNA of a DNA
junction
polynucleotide of an INIR6 transgenic locus or can span the junction of the
transgenic and
non-transgenic DNA of a DNA junction polynucleotide of an INIR6 transgenic
locus. An
OgRRS can likewise be located in non-transgenic plant genomic DNA of a DNA
junction
polynucleotide of an INIR6 transgenic locus, in transgenic DNA of a DNA
junction
polynucleotide of an INIR6 transgenic locus, or can span the junction of the
transgenic and
non-transgenic DNA of a DNA junction polynucleotide of an INIR6 transgenic
locus
[0065] Methods provided herein can be used in a variety of breeding schemes to
obtain elite
crop plants comprising subsets of desired modified transgenic loci comprising
an OgRRS and
a CgRRS operably linked to junction polynucleotide sequences and transgenic
loci excision
sites where undesired transgenic loci or portions thereof have been removed
(e.g., by use of
the OgRRS and a CgRRS). Such methods are useful at least insofar as they allow
for
production of distinct useful donor plant lines each having unique sets of
modified transgenic
loci and, in some instances, targeted genetic changes that are tailored for
distinct geographies
and/or product offerings. In an illustrative and non-limiting example, a
different product lines
comprising transgenic loci conferring only two of three types of herbicide
tolerance (e.g.,
glyphosate, glufosinate, and dicamba) can be obtained from a single donor line
comprising
three distinct transgenic loci conferring resistance to all three herbicides.
In certain aspects,
plants comprising the subsets of undesired transgenic loci and transgenic loci
excision sites
can further comprise targeted genetic changes. Such elite crop plants can be
inbred plant lines
or can be hybrid plant lines. In certain embodiments, at least two transgenic
loci (e.g.,
transgenic loci including an INIR6 and another modified transgenic locus
wherein an
OgRRS and a CgRRS site is operably linked to a first and a second junction
sequence and
- 19 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
optionally a selectable marker gene and/or non-essential DNA are deleted) are
introgressed
into a desired donor line comprising elite crop plant germplasm and then
subjected to genome
editing molecules to recover plants comprising one of the two introgressed
transgenic loci as
well as a transgenic loci excision site introduced by excision of the other
transgenic locus or
portion thereof by the genome editing molecules. In certain embodiments, the
genome editing
molecules can be used to remove a transgenic locus and introduce targeted
genetic changes in
the crop plant genome. Introgression can be achieved by backcrossing plants
comprising the
transgenic loci to a recurrent parent comprising the desired elite germplasm
and selecting
progeny with the transgenic loci and recurrent parent germplasm. Such
backcrosses can be
repeated and/or supplemented by molecular assisted breeding techniques using
SNP or other
nucleic acid markers to select for recurrent parent germplasm until a desired
recurrent parent
percentage is obtained (e.g., at least about 95%, 96%, 97%, 98%, or 99%
recurrent parent
percentage). A non-limiting, illustrative depiction of a scheme for obtaining
plants with both
subsets of transgenic loci and the targeted genetic changes is shown in the
Figure 2 (bottom
"Alternative" panel), where two or more of the transgenic loci ("Event" in
Figure 2) are
provided in Line A and then moved into elite crop plant germplasm by
introgression. In the
non-limiting Figure 2 illustration, introgression can be achieved by crossing
a "Line A"
comprising two or more of the modified transgenic loci to the elite germplasm
and then
backcrossing progeny of the cross comprising the transgenic loci to the elite
germplasm as
the recurrent parent) to obtain a "Universal Donor" (e.g., Line A+ in Figure
2) comprising
two or more of the modified transgenic loci. This elite germplasm containing
the modified
transgenic loci (e.g., "Universal Donor" of Figure 2) can then be subjected to
genome editing
molecules which can excise at least one of the transgenic loci ("Event
Removal" in Figure 2)
and introduce other targeted genetic changes ("GE" in Figure 2) in the genomes
of the elite
crop plants containing one of the transgenic loci and a transgenic locus
excision site
corresponding to the removal site of one of the transgenic loci. Such
selective excision of
transgenic loci or portion thereof can be effected by contacting the genome of
the plant
comprising two transgenic loci with gene editing molecules (e.g., RdDe and
gRNAs,
TALENS, and/or ZFN) which recognize one transgenic loci but not another
transgenic loci.
Genome editing molecules that provide for selective excision of a first
modified transgenic
locus comprising an OgRRS and a CgRRS include a gRNA that hybridizes to the
OgRRS and
CgRRS of the first modified transgenic locus and an RdDe that recognizes the
gRNA/OgRRS
- 20 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
and gRNA/CgRRS complexes. Distinct plant lines with different subsets of
transgenic loci
and desired targeted genetic changes are thus recovered (e.g., "Line B-1,"
"Line B-2," and
"Line B-3" in Figure 2). In certain embodiments, it is also desirable to bulk
up populations of
inbred elite crop plants or their seed comprising the subset of transgenic
loci and a transgenic
locus excision site by selfing. In certain embodiments, inbred progeny of the
selfed maize
plants comprising the INIR6 transgenic loci can be used as a pollen donor or
recipient for
hybrid seed production. Such hybrid seed and the progeny grown therefrom can
comprise a
subset of desired transgenic loci and a transgenic loci excision site.
[0066] Hybrid plant lines comprising elite crop plant germplasm, at least one
transgenic
locus and at least one transgenic locus excision site, and in certain aspects,
additional targeted
genetic changes are also provided herein. Methods for production of such
hybrid seed can
comprise crossing elite crop plant lines where at least one of the pollen
donor or recipient
comprises at least the transgenic locus and a transgenic locus excision site
and/or additional
targeted genetic changes. In certain embodiments, the pollen donor and
recipient will
comprise germplasm of distinct heterotic groups and provide hybrid seed and
plants
exhibiting heterosis. In certain embodiments, the pollen donor and recipient
can each
comprise a distinct transgenic locus which confers either a distinct trait
(e.g., herbicide
tolerance or insect resistance), a different type of trait (e.g., tolerance to
distinct herbicides or
to distinct insects such as coleopteran or lepidopteran insects), or a
different mode-of-action
for the same trait (e.g., resistance to coleopteran insects by two distinct
modes-of-action or
resistance to lepidopteran insects by two distinct modes-of-action). In
certain embodiments,
the pollen recipient will be rendered male sterile or conditionally male
sterile. Methods for
inducing male sterility or conditional male sterility include emasculation
(e.g., detasseling),
cytoplasmic male sterility, chemical hybridizing agents or systems, a
transgenes or transgene
systems, and/or mutation(s) in one or more endogenous plant genes.
Descriptions of various
male sterility systems that can be adapted for use with the elite crop plants
provided herein
are described in Wan et al. Molecular Plant; 12, 3, (2019):321-342 as well as
in US
8,618,358; US 20130031674; and US 2003188347.
[0067] In certain embodiments, it will be desirable to use genome editing
molecules to make
modified transgenic loci by introducing a CgRRS into the transgenic loci, to
excise modified
transgenic loci comprising an OgRRS and a CgRRS, and/or to make targeted
genetic changes
in elite crop plant or other germplasm. Techniques for effecting genome
editing in crop plants
- 21 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
(e.g., maize,) include use of morphogenic factors such as Wuschel (WUS), Ovule
Development Protein (ODP), and/or Babyboom (BBM) which can improve the
efficiency of
recovering plants with desired genome edits. In some aspects, the morphogenic
factor
comprises WUS1, WUS2, WUS3, WOX2A, WOX4, WOX5, WOX9, BBM2, BMN2,
BMN3, and/or ODP2. In certain embodiments, compositions and methods for using
WUS,
BBM, and/or ODP, as well as other techniques which can be adapted for
effecting genome
edits in elite crop plant and other germplasm, are set forth in US
20030082813, US
20080134353, US 20090328252, US 20100100981, US 20110165679, US 20140157453,
US
20140173775, and US 20170240911, which are each incorporated by reference in
their
entireties. In certain embodiments, the genome edits can be effected in
regenerable plant parts
(e.g., plant embryos) of elite crop plants by transient provision of gene
editing molecules or
polynucleotides encoding the same and do not necessarily require incorporating
a selectable
marker gene into the plant genome (e.g., US 20160208271 and US 20180273960,
both
incorporated herein by reference in their entireties; Svitashev et al. Nat
Commun. 2016;
7:13274).
[0068] In certain embodiments, edited transgenic plant genomes, transgenic
plant cells, parts,
or plants containing those genomes, and DNA molecules obtained therefrom, can
comprise a
desired subset of transgenic loci and/or comprise at least one transgenic
locus excision site. In
certain embodiments, a segment comprising an INIR6 transgenic locus comprising
an
OgRRS in non-transgenic DNA of a 1st junction polynucleotide sequence and a
CgRRS in a
2nd junction polynucleotide sequence is deleted with a gRNA and RdDe that
recognize the
OgRRS and the CgRRS to produce an INIR6 transgenic locus excision site. In
certain
embodiments, the transgenic locus excision site can comprise a contiguous
segment of DNA
comprising at least 10 base pairs of DNA that is telomere proximal to the
deleted segment of
the transgenic locus and at least 10 base pairs of DNA that is centromere
proximal to the
deleted segment of the transgenic locus wherein the transgenic DNA (i.e., the
heterologous
DNA) that has been inserted into the crop plant genome has been deleted. In
certain
embodiments where a segment comprising a transgenic locus has been deleted,
the transgenic
locus excision site can comprise a contiguous segment of DNA comprising at
least 10 base
pairs DNA that is telomere proximal to the deleted segment of the transgenic
locus and at
least 10 base pairs of DNA that is centromere proximal DNA to the deleted
segment of the
transgenic locus wherein the heterologous transgenic DNA and at least 1, 2, 5,
10, 20, 50, or
- 22 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
more base pairs of endogenous DNA located in a 5' junction sequence and/or in
a 3' junction
sequence of the original transgenic locus that has been deleted. In such
embodiments where
DNA comprising the transgenic locus is deleted, a transgenic locus excision
site can comprise
at least 10 base pairs of DNA that is telomere proximal to the deleted segment
of the
transgenic locus and at least 10 base pairs of DNA that is centromere proximal
to the deleted
segment of the transgenic locus wherein all of the transgenic DNA is absent
and either all or
less than all of the endogenous DNA flanking the transgenic DNA sequences are
present. In
certain embodiments where a segment consisting essentially of an original
transgenic locus
has been deleted, the transgenic locus excision site can be a contiguous
segment of at least 10
base pairs of DNA that is telomere proximal to the deleted segment of the
transgenic locus
and at least 10 base pairs of DNA that is centromere proximal to the deleted
segment of the
transgenic locus wherein less than all of the heterologous transgenic DNA that
has been
inserted into the crop plant genome is excised. In certain aforementioned
embodiments where
a segment consisting essentially of an original transgenic locus has been
deleted, the
transgenic locus excision site can thus contain at least 1 base pair of DNA or
1 to about 2 or
5, 8, 10, 20, or 50 base pairs of DNA comprising the telomere proximal and/or
centromere
proximal heterologous transgenic DNA that has been inserted into the crop
plant genome. In
certain embodiments where a segment consisting of an original transgenic locus
has been
deleted, the transgenic locus excision site can contain a contiguous segment
of DNA
comprising at least 10 base pairs of DNA that is telomere proximal to the
deleted segment of
the transgenic locus and at least 10 base pairs of DNA that is centromere
proximal to the
deleted segment of the transgenic locus wherein the heterologous transgenic
DNA that has
been inserted into the crop plant genome is deleted. In certain embodiments
where DNA
consisting of the transgenic locus is deleted, a transgenic locus excision
site can comprise at
least 10 base pairs of DNA that is telomere proximal to the deleted segment of
the transgenic
locus and at least 10 base pairs of DNA that is centromere proximal to the
deleted segment of
the transgenic locus wherein all of the heterologous transgenic DNA that has
been inserted
into the crop plant genome is deleted and all of the endogenous DNA flanking
the
heterologous sequences of the transgenic locus is present. In any of the
aforementioned
embodiments or in other embodiments, the continuous segment of DNA comprising
the
transgenic locus excision site can further comprise an insertion of 1 to about
2, 5, 10, 20, or
more nucleotides between the DNA that is telomere proximal to the deleted
segment of the
- 23 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
transgenic locus and the DNA that is centromere proximal to the deleted
segment of the
transgenic locus. Such insertions can result either from endogenous DNA repair
and/or
recombination activities at the double stranded breaks introduced at the
excision site and/or
from deliberate insertion of an oligonucleotide. Plants, edited plant genomes,
biological
samples, and DNA molecules (e.g., including isolated or purified DNA
molecules)
comprising the INIR6 transgenic loci excision sites are provided herein.
[0069] In other embodiments, a segment comprising a INIR6 transgenic locus
(e.g., a
transgenic locus comprising an OgRRS in non-transgenic DNA of a 1" junction
sequence and
a CgRRS in a 2nd junction sequence) can be deleted with a gRNA and RdDe that
recognize
the OgRRS and the CgRRS and replaced with DNA comprising the endogenous non-
transgenic plant genomic DNA present in the genome prior to transgene
insertion. A non-
limiting example of such replacements can be visualized in Figure 3C, where
the donor DNA
template can comprise the endogenous non-transgenic plant genomic DNA present
in the
genome prior to transgene insertion along with sufficient homology to non-
transgenic DNA
on each side of the excision site to permit homology-directed repair. In
certain embodiments,
the endogenous non-transgenic plant genomic DNA present in the genome prior to
transgene
insertion can be at least partially restored. In certain embodiments, the
endogenous non-
transgenic plant genomic DNA present in the genome prior to transgene
insertion can be
essentially restored such that no more than about 5, 10, or 20 to about 50,
80, or 100
nucleotides are changed relative to the endogenous DNA at the essentially
restored excision
site.
[0070] In certain embodiments, edited transgenic plant genomes and transgenic
plant cells,
plant parts, or plants containing those edited genomes, comprising a
modification of an
original transgenic locus, where the modification comprises an OgRRS and a
CgRRS which
are operably linked to a 1" and a 2' junction sequence, respectively or
irrespectively, and
optionally further comprise a deletion of a segment of the original transgenic
locus. In certain
embodiments, the modification comprises two or more separate deletions and/or
there is a
modification in two or more original transgenic plant loci. In certain
embodiments, the
deleted segment comprises, consists essentially of, or consists of a segment
of non-essential
DNA in the transgenic locus. Illustrative examples of non-essential DNA
include but are not
limited to synthetic cloning site sequences, duplications of transgene
sequences; fragments of
transgene sequences, and Agrobacterium right and/or left border sequences. In
certain
- 24 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
embodiments, the non-essential DNA is a duplication and/or fragment of a
promoter
sequence and/or is not the promoter sequence operably linked in the cassette
to drive
expression of a transgene. In certain embodiments, excision of the non-
essential DNA
improves a characteristic, functionality, and/or expression of a transgene of
the transgenic
locus or otherwise confers a recognized improvement in a transgenic plant
comprising the
edited transgenic plant genome. In certain embodiments, the non-essential DNA
does not
comprise DNA encoding a selectable marker gene. In certain embodiments of an
edited
transgenic plant genome, the modification comprises a deletion of the non-
essential DNA and
a deletion of a selectable marker gene. The modification producing the edited
transgenic plant
genome could occur by excising both the non-essential DNA and the selectable
marker gene
at the same time, e.g., in the same modification step, or the modification
could occur step-
wise. For example, an edited transgenic plant genome in which a selectable
marker gene has
previously been removed from the transgenic locus can comprise an original
transgenic locus
from which a non-essential DNA is further excised and vice versa. In certain
embodiments,
the modification comprising deletion of the non-essential DNA and deletion of
the selectable
marker gene comprises excising a single segment of the original transgenic
locus that
comprises both the non-essential DNA and the selectable marker gene. Such
modification
would result in one excision site in the edited transgenic genome
corresponding to the
deletion of both the non-essential DNA and the selectable marker gene. In
certain
embodiments, the modification comprising deletion of the non-essential DNA and
deletion of
the selectable marker gene comprises excising two or more segments of the
original
transgenic locus to achieve deletion of both the non-essential DNA and the
selectable marker
gene. Such modification would result in at least two excision sites in the
edited transgenic
genome corresponding to the deletion of both the non-essential DNA and the
selectable
marker gene. In certain embodiments of an edited transgenic plant genome,
prior to excision,
the segment to be deleted is flanked by operably linked protospacer adjacent
motif (PAM)
sites in the original or unmodified transgenic locus and/or the segment to be
deleted
encompasses an operably linked PAM site in the original or unmodified
transgenic locus. In
certain embodiments, following excision of the segment, the resulting edited
transgenic plant
genome comprises PAM sites flanking the deletion site in the modified
transgenic locus. In
certain embodiments of an edited transgenic plant genome, the modification
comprises a
modification of a DP-4114 transgenic locus.
- 25 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
[0071] In certain embodiments, improvements in a transgenic plant locus are
obtained by
introducing a new cognate guide RNA recognition site (CgRRS) which is operably
linked to
a DNA junction polynucleotide of the transgenic locus in the transgenic plant
genome. Such
CgRRS sites can be recognized by RdDe and a single suitable guide RNA directed
to the
CgRRS and the originator gRNA Recognition Site (OgRRS) to provide for cleavage
within
the junction polynucleotides which flank an INIR6 transgenic locus. In certain
embodiments,
the CgRRS/gRNA and OgRRS/gRNA hybridization complexes are recognized by the
same
class of RdDe (e.g., Class 2 type II or Class 2 type V) or by the same RdDe
(e.g., both the
CgRRS/gRNA and OgRRS/gRNA hybridization complexes recognized by the same Cas9
or
Cas 12 RdDe). Such CgRRS and OgRRS can be recognized by RdDe and suitable
guide
RNAs containing crRNA sufficiently complementary to the guide RNA
hybridization site
DNA sequences adjacent to the PAM site of the CgRRS and the OgRRS to provide
for
cleavage within or near the two junction polynucleotides. Suitable guide RNAs
can be in the
form of a single gRNA comprising a crRNA or in the form of a crRNA/tracrRNA
complex.
In the case of the OgRRS site, the PAM and guide RNA hybridization site are
endogenous
DNA polynucleotide molecules found in the plant genome. In certain embodiments
where the
CgRRS is introduced into the plant genome by genome editing, gRNA
hybridization site
polynucleotides introduced at the CgRRS are at least 17 or 18 nucleotides in
length and are
complementary to the crRNA of a guide RNA. In certain embodiments, the gRNA
hybridization site sequence of the OgRRS and/or the CgRRS is about 17 or 18 to
about 24
nucleotides in length. The gRNA hybridization site sequence of the OgRRS and
the gRNA
hybridization site of the CgRRS can be of different lengths or comprise
different sequences
so long as there is sufficient complementarity to permit hybridization by a
single gRNA and
recognition by a RdDe that recognizes and cleaves DNA at the gRNA/OgRRS and
gRNA/CgRRS complex. In certain embodiments, the guide RNA hybridization site
of the
CgRRS comprise about a 17 or 18 to about 24 nucleotide sequence which is
identical to the
guide RNA hybridization site of the OgRRS. In other embodiments, the guide RNA
hybridization site of the CgRRS comprise about a 17 or 18 to about 24
nucleotide sequence
which has one, two, three, four, or five nucleotide insertions, deletions or
substitutions when
compared to the guide RNA hybridization site of the OgRRS. Certain CgRRS
comprising a
gRNA hybridization site containing has one, two, three, four, or five
nucleotide insertions,
deletions or substitutions when compared to the guide RNA hybridization site
of the OgRRS
- 26 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
can undergo hybridization with a gRNA which is complementary to the OgRRS gRNA
hybridization site and be cleaved by certain RdDe. Examples of mismatches
between gRNAs
and guide RNA hybridization sites which allow for RdDe recognition and
cleavage include
mismatches resulting from both nucleotide insertions and deletions in the DNA
which is
hybridized to the gRNA (e.g., Lin et al., doi: 10.1093/nar/gku402). In certain
embodiments,
an operably linked PAM site is co-introduced with the gRNA hybridization site
polynucleotide at the CgRRS. In certain embodiments, the gRNA hybridization
site
polynucleotides are introduced at a position adjacent to a resident endogenous
PAM sequence
in the junction polynucleotide sequence to form a CgRRS where the gRNA
hybridization site
polynucleotides are operably linked to the endogenous PAM site. In certain
embodiments,
non-limiting features of the OgRRS, CgRRS, and/or the gRNA hybridization site
polynucleotides thereof include: (i) absence of significant homology or
sequence identity
(e.g., less than 50% sequence identity across the entire length of the OgRRS,
CgRRS, and/or
the gRNA hybridization site sequence) to any other endogenous or transgenic
sequences
present in the transgenic plant genome or in other transgenic genomes of the
maize plant
being transformed and edited; (ii) absence of significant homology or sequence
identity (e.g.,
less than 50% sequence identity across the entire length of the sequence) of a
sequence of a
first OgRRS and a first CgRRS to a second OgRRS and a second CgRRS which are
operably
linked to junction polynucleotides of a distinct transgenic locus; (iii) the
presence of some
sequence identity (e.g., about 25%, 40%, or 50% to about 60%, 70%, or 80%)
between the
OgRRS sequence and endogenous sequences present at the site where the CgRRS
sequence is
introduced; and/or (iv) optimization of the gRNA hybridization site
polynucleotides for
recognition by the RdDe and guide RNA when used in conjunction with a
particular PAM
sequence. In certain embodiments, the first and second OgRRS as well as the
first and second
CgRRS are recognized by the same class of RdDe (e.g., Class 2 type II or Class
2 type V) or
by the same RdDe (e.g., Cas9 or Cas 12 RdDe). In certain embodiments, the
first OgRRS
site in a first junction polynucleotide and the CgRRS introduced in the second
junction
polynucleotide to permit excision of a first transgenic locus by a first
single guide RNA and a
single RdDe. Such nucleotide insertions or genome edits used to introduce
CgRRS in a
transgenic plant genome can be effected in the plant genome by using gene
editing molecules
(e.g., RdDe and guide RNAs, RNA dependent nickases and guide RNAs, Zinc Finger
nucleases or nickases, or TALE nucleases or nickases) which introduce blunt
double stranded
- 27 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
breaks or staggered double stranded breaks in the DNA junction
polynucleotides. In the case
of DNA insertions, the genome editing molecules can also in certain
embodiments further
comprise a donor DNA template or other DNA template which comprises the
heterologous
nucleotides for insertion to form the CgRRS. Guide RNAs can be directed to the
junction
polynucleotides by using a pre-existing PAM site located within or adjacent to
a junction
polynucleotide of the transgenic locus. Non-limiting examples of such pre-
existing PAM sites
present in junction polynucleotides, which can be used either in conjunction
with an inserted
heterologous sequence to form a CgRRS or which can be used to create a double
stranded
break to insert or create a CgRRS, include PAM sites recognized by a Cas12a
enzyme. Non-
limiting examples where a CgRRS are created in a DNA sequence are illustrated
in Example
2.
[0072] Transgenic loci comprising OgRRS and CgRRS in a first and a second
junction
polynucleotides can be excised from the genomes of transgenic plants by
contacting the
transgenic loci with RdDe or RNA directed nickases, and a suitable guide RNA
directed to
the OgRRS and CgRRS. A non-limiting example where a modified transgenic locus
is
excised from a plant genome by use of a gRNA and an RdDe that recognizes an
OgRRS/gRNA and a CgRRS/gRNA complex and introduces dsDNA breaks in both
junction
polynucleotides nd repaired by NHEJ is depicted in Figure 3B. In the depicted
example set
forth in Figure 3B, the OgRRS site and the CgRRS site are absent from the
plant
chromosome comprising the transgene excision site that results from the
process. In other
embodiments provided herein where a modified transgenic locus is excised from
a plant
genome by use of a gRNA and an RdDe that recognizes an OgRRS/gRNA and a
CgRRS/gRNA complex and repaired by NHEJ or microhomology-mediated end joining
(MMEJ), the OgRRS and/or other non-transgenic sequences that were originally
present prior
to transgene insertion are partially or essentially restored.
[0073] In certain embodiments, edited transgenic plant genomes provided herein
can lack one
or more selectable and/or scoreable markers found in an original event
(transgenic locus).
Original DP-4114 transgenic loci (events), including those set forth in SEQ ID
NO: 1), US
8,575,434, the sequence of the DP-4114 locus in the deposited seed of
accession No. PTA-
11506 and progeny thereof, contain a selectable marker gene encoding a
phosphinotricin
acetyl transferase (PAT) protein which confers tolerance to the herbicide
glufosinate. In
certain embodiments provided herein, the DNA element comprising, consisting
essentially of,
- 28 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
or consisting of the PAT selectable marker gene of an DP-4114 transgenic locus
is absent
from an INIR6 transgenic locus. The PAT selectable marker cassette can be
excised from an
original DP-4114 transgenic locus by contacting the transgenic locus with one
or more gene
editing molecules which introduce double stranded breaks in the transgenic
locus at the 5'
and 3' end of the expression cassette comprising the PAT selectable marker
transgene (e.g.,
an RdDe and guide RNAs directed to PAM sites located at the 5' and 3' end of
the expression
cassette comprising the PAT selectable marker transgene) and selecting for
plant cells, plant
parts, or plants wherein the selectable marker has been excised. In certain
embodiments, the
selectable or scoreable marker transgene can be inactivated. Inactivation can
be achieved by
modifications including insertion, deletion, and/or substitution of one or
more nucleotides in
a promoter element, 5' or 3' untranslated region (UTRs), intron, coding
region, and/or 3'
terminator and/or polyadenylation site of the selectable marker transgene.
Such
modifications can inactivate the selectable marker transgene by eliminating or
reducing
promoter activity, introducing a missense mutation, and/or introducing a pre-
mature stop
codon. In certain embodiments, the selectable PAT marker transgene can be
replaced by an
introduced transgene. In certain embodiments, an original transgenic locus
that was
contacted with gene editing molecules which introduce double stranded breaks
in the
transgenic locus at the 5' and 3' end of the expression cassette comprising
the PAT selectable
marker transgene can also be contacted with a suitable donor DNA template
comprising an
expression cassette flanked by DNA homologous to remaining DNA in the
transgenic locus
located 5' and 3' to the selectable marker excision site. In certain
embodiments, a coding
region of the PAT selectable marker transgene can be replaced with another
coding region
such that the replacement coding region is operably linked to the promoter and
3' terminator
or polyadenylation site of the PAT selectable marker transgene.
[0074] In certain embodiments, edited transgenic plant genomes provided herein
can
comprise additional new introduced transgenes (e.g., expression cassettes)
inserted into the
transgenic locus of a given event. Introduced transgenes inserted at the
transgenic locus of an
event subsequent to the event's original isolation can be obtained by inducing
a double
stranded break at a site within an original transgenic locus (e.g., with
genome editing
molecules including an RdDe and suitable guide RNA(s); a suitable engineered
zinc-finger
nuclease; a TALEN protein and the like) and providing an exogenous transgene
in a donor
DNA template which can be integrated at the site of the double stranded break
(e.g. by
- 29 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
homology-directed repair (HDR) or by non-homologous end-joining (NHEJ)). In
certain
embodiments, an OgRRS and a CgRRS located in a 1st junction polynucleotide and
a 2nd
junction polynucleotide, respectively, can be used to delete the transgenic
locus and replace it
with one or more new expression cassettes. In certain embodiments, such
deletions and
replacements are effected by introducing dsDNA breaks in both junction
polynucleotides and
providing the new expression cassettes on a donor DNA template (e.g., in
Figure 3C, the
donor DNA template can comprise an expression cassette flanked by DNA
homologous to
non-transgenic DNA located telomere proximal and centromere proximal to the
excision
site). Suitable expression cassettes for insertion include DNA molecules
comprising
promoters which are operably linked to DNA encoding proteins and/or RNA
molecules
which confer useful traits which are in turn operably linked to
polyadenylation sites or
terminator elements. In certain embodiments, such expression cassettes can
also comprise 5'
UTRs, 3' UTRs, and/or introns. Useful traits include biotic stress tolerance
(e.g., insect
resistance, nematode resistance, or disease resistance), abiotic stress
tolerance (e.g., heat,
cold, drought, and/or salt tolerance), herbicide tolerance, and quality traits
(e.g., improved
fatty acid compositions, protein content, starch content, and the like).
Suitable expression
cassettes for insertion include expression cassettes which confer insect
resistance, herbicide
tolerance, biofuel use, or male sterility traits contained in any of the
transgenic events set
forth in US Patent Application Public. Nos. 20090038026, 20130031674,
20150361446,
20170088904, 20150267221, 201662346688, and 20200190533 as well as in US
Patent Nos.
6342660, 7323556, 8575434, 6040497, 8759618, 7157281, 6852915, 7705216,
10316330,
8618358, 8450561, 8212113, 9428765, 7897748, 8273959, 8093453,8901378,
9994863,
7928296, and 8466346, each of which are incorporated herein by reference in
their entireties.
[0075] In certain embodiments, INIR6 plants provided herein, including plants
with one or
more transgenic loci, modified transgenic loci, and/or comprising transgenic
loci excision
sites can further comprise one or more targeted genetic changes introduced by
one or more of
gene editing molecules or systems. Also provided are methods where the
targeted genetic
changes are introduced and one or more transgenic loci are removed from plants
either in
series or in parallel (e.g., as set forth in the non-limiting illustration in
Figure 2, bottom
"Alternative" panel, where "GE" can represent targeted genetic changes induced
by gene
editing molecules and "Event Removal" represents excision of one or more
transgenic loci
with gene editing molecules). Such targeted genetic changes include those
conferring traits
- 30 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
such as improved yield, improved food and/or feed characteristics (e.g.,
improved oil, starch,
protein, or amino acid quality or quantity), improved nitrogen use efficiency,
improved
biofuel use characteristics (e.g., improved ethanol production), male
sterility/conditional male
sterility systems (e.g., by targeting endogenous MS26, MS45 and MSCA1 genes),
herbicide
tolerance (e.g., by targeting endogenous ALS, EPSPS, HPPD, or other herbicide
target
genes), delayed flowering, non-flowering, increased biotic stress resistance
(e.g., resistance to
insect, nematode, bacterial, or fungal damage), increased abiotic stress
resistance (e.g.,
resistance to drought, cold, heat, metal, or salt ), enhanced lodging
resistance, enhanced
growth rate, enhanced biomass, enhanced tillering, enhanced branching, delayed
flowering
time, delayed senescence, increased flower number, improved architecture for
high density
planting, improved photosynthesis, increased root mass, increased cell number,
improved
seedling vigor, improved seedling size, increased rate of cell division,
improved metabolic
efficiency, and increased meristem size in comparison to a control plant
lacking the targeted
genetic change. Types of targeted genetic changes that can be introduced
include insertions,
deletions, and substitutions of one or more nucleotides in the crop plant
genome. Sites in
endogenous plant genes for the targeted genetic changes include promoter,
coding, and non-
coding regions (e.g., 5' UTRs, introns, splice donor and acceptor sites and 3'
UTRs). In
certain embodiments, the targeted genetic change comprises an insertion of a
regulatory or
other DNA sequence in an endogenous plant gene. Non-limiting examples of
regulatory
sequences which can be inserted into endogenous plant genes with gene editing
molecules to
effect targeted genetic changes which confer useful phenotypes include those
set forth in US
Patent Application Publication 20190352655, which is incorporated herein by
reference in its
entirety, such as: (a) auxin response element (AuxRE) sequence; (b) at least
one D1-4
sequence (Ulmasov et al. (1997) Plant Cell, 9:1963-1971), (c) at least one DRS
sequence
(Ulmasov et al. (1997) Plant Cell, 9:1963-1971); (d) at least one m5-DRS
sequence (Ulmasov
et al. (1997) Plant Cell, 9:1963-1971); (e) at least one P3 sequence; (I) a
small RNA
recognition site sequence bound by a corresponding small RNA (e.g., an siRNA,
a
microRNA (miRNA), a trans-acting siRNA as described in U.S. Patent No.
8,030,473, or a
phased sRNA as described in U.S. Patent No. 8,404,928; both of these cited
patents are
incorporated by reference herein); (g) a microRNA (miRNA) recognition site
sequence; (h)
the sequence recognizable by a specific binding agent includes a microRNA
(miRNA)
recognition sequence for an engineered miRNA wherein the specific binding
agent is the
-31-
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
corresponding engineered mature miRNA; (i) a transposon recognition sequence;
(j) a
sequence recognized by an ethylene-responsive element binding-factor-
associated
amphiphilic repression (EAR) motif; (k) a splice site sequence (e.g., a donor
site, a branching
site, or an acceptor site; see, for example, the splice sites and splicing
signals set forth in the
internet site lemur[dot]amu[dot]edu[dot]pl/share/ERISdb/home.html); (1) a
recombinase
recognition site sequence that is recognized by a site-specific recombinase;
(m) a sequence
encoding an RNA or amino acid aptamer or an RNA riboswitch, the specific
binding agent is
the corresponding ligand, and the change in expression is upregulation or
downregulation; (n)
a hormone responsive element recognized by a nuclear receptor or a hormone-
binding
domain thereof; (o) a transcription factor binding sequence; and (p) a
polycomb response
element (see Xiao et al. (2017) Nature Genetics, 49:1546-1552, doi:
10.1038/ng.3937). Non
limiting examples of target maize genes that can be subjected to targeted gene
edits to confer
useful traits include: (a) ZmIPK1 (herbicide tolerant and phytate reduced
maize; Shukla et al.,
Nature. 2009;459:437-41); (b) ZmGL2 (reduced epicuticular wax in leaves; Char
et al. Plant
Biotechnol J. 2015;13:1002); (c) ZmMTL (induction of haploid plants; Kelliher
et al. Nature.
2017;542:105); (d) Wxl (high amylopectin content; US 20190032070; incorporated
herein
by reference in its entirety); (e) TMS5 (thermosensitive male sterile; Li et
al. J Genet
Genomics. 2017;44:465-8); (f) ALS (herbicide tolerance; Svitashev et al.;
Plant Physiol.
2015;169:931-45); and (g) ARGOS8 (drought stress tolerance; Shi et al., Plant
Biotechnol J.
2017;15:207-16). Non-limiting examples of target genes in crop plants
including maize
which can be subjected to targeted genetic changes which confer useful
phenotypes include
those set forth in US Patent Application Nos. 20190352655, 20200199609,
20200157554,
and 20200231982, which are each incorporated herein in their entireties; and
Zhang et al.
(Genome Biol. 2018; 19: 210).
[0076] Gene editing molecules of use in methods provided herein include
molecules capable
of introducing a double-strand break ("DSB") or single-strand break ("SSB") in
double-
stranded DNA, such as in genomic DNA or in a target gene located within the
genomic DNA
as well as accompanying guide RNA or donor DNA template polynucleotides.
Examples of
such gene editing molecules include: (a) a nuclease comprising an RNA-guided
nuclease, an
RNA-guided DNA endonuclease or RNA directed DNA endonuclease (RdDe), a class 1
CRISPR type nuclease system, a type II Cas nuclease, a Cas9, a nCas9 nickase,
a type V Cas
nuclease, a Cas12a nuclease, a nCas12a nickase, a Cas12d (CasY), a Cas12e
(CasX), a
- 32 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
Cas12b (C2c1), a Cas12c (C2c3), a Cas12i, a Cas12j, a Cas14, an engineered
nuclease, a
codon-optimized nuclease, a zinc-finger nuclease (ZFN) or nickase, a
transcription activator-
like effector nuclease (TAL-effector nuclease or TALEN) or nickase (TALE-
nickase), an
Argonaute, and a meganuclease or engineered meganuclease; (b) a polynucleotide
encoding
one or more nucleases capable of effectuating site-specific alteration
(including introduction
of a DSB or SSB) of a target nucleotide sequence; (c) a guide RNA (gRNA) for
an RNA-
guided nuclease, or a DNA encoding a gRNA for an RNA-guided nuclease; (d)
donor DNA
template polynucleotides; and (e) other DNA templates (dsDNA, ssDNA, or
combinations
thereof) suitable for insertion at a break in genomic DNA (e.g., by non-
homologous end
joining (NHEJ) or microhomology-mediated end joining (MMEJ).
[0077] CRISPR-type genome editing can be adapted for use in the plant cells
and methods
provided herein in several ways. CRISPR elements, e.g., gene editing molecules
comprising
CRISPR endonucleases and CRISPR guide RNAs including single guide RNAs or
guide
RNAs in combination with tracrRNAs or scoutRNA, or polynucleotides encoding
the same,
are useful in effectuating genome editing without remnants of the CRISPR
elements or
selective genetic markers occurring in progeny. In certain embodiments, the
CRISPR
elements are provided directly to the eukaryotic cell (e.g., plant cells),
systems, methods, and
compositions as isolated molecules, as isolated or semi-purified products of a
cell free
synthetic process (e.g., in vitro translation), or as isolated or semi-
purified products of in a
cell-based synthetic process (e.g., such as in a bacterial or other cell
lysate). In certain
embodiments, genome-inserted CRISPR elements are useful in plant lines adapted
for use in
the methods provide herein. In certain embodiments, plants or plant cells used
in the
systems, methods, and compositions provided herein can comprise a transgene
that expresses
a CRISPR endonuclease (e.g., a Cas9, a Cpfl-type or other CRISPR
endonuclease). In
certain embodiments, one or more CRISPR endonucleases with unique PAM
recognition
sites can be used. Guide RNAs (sgRNAs or crRNAs and a tracrRNA) to form an RNA-
guided endonuclease/guide RNA complex which can specifically bind sequences in
the
gDNA target site that are adjacent to a protospacer adjacent motif (PAM)
sequence. The type
of RNA-guided endonuclease typically informs the location of suitable PAM
sites and design
of crRNAs or sgRNAs. G-rich PAM sites, e.g., 5'-NGG are typically targeted for
design of
crRNAs or sgRNAs used with Cas9 proteins. Examples of PAM sequences include 5'-
NGG
(Streptococcus pyogenes), 5'-NNAGAA (Streptococcus thermophilus CRISPR1), 5'-
- 33 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
NGGNG (Streptococcus thermophilus CRISPR3), 5' -NNGRRT or 5' -NNGRR
(Staphylococcus aureus Cas9, SaCas9), and 5'-NNNGATT (Neisseria meningitidis).
T-rich
PAM sites (e.g., 5'-TTN or 5'-TTTV, where "V" is A, C, or G) are typically
targeted for
design of crRNAs or sgRNAs used with Cas12a proteins. In some instances,
Cas12a can also
recognize a 5'-CTA PAM motif Other examples of potential Cas12a PAM sequences
include TTN, CTN, TCN, CCN, TTTN, TCTN, TTCN, CTTN, ATTN, TCCN, TTGN,
GTTN, CCCN, CCTN, TTAN, TCGN, CTCN, ACTN, GCTN, TCAN, GCCN, and CCGN
(wherein N is defined as any nucleotide).
Cpfl (i.e., Cas12a) endonuclease and
corresponding guide RNAs and PAM sites are disclosed in US Patent Application
Publication
2016/0208243 Al, which is incorporated herein by reference for its disclosure
of DNA
encoding Cpfl endonucleases and guide RNAs and PAM sites. Introduction of one
or more
of a wide variety of CRISPR guide RNAs that interact with CRISPR endonucleases
integrated into a plant genome or otherwise provided to a plant is useful for
genetic editing
for providing desired phenotypes or traits, for trait screening, or for gene
editing mediated
trait introgression (e.g., for introducing a trait into a new genotype without
backcrossing to a
recurrent parent or with limited backcrossing to a recurrent parent). Multiple
endonucleases
can be provided in expression cassettes with the appropriate promoters to
allow multiple
genome site editing.
[0078] CRISPR technology for editing the genes of eukaryotes is disclosed in
US Patent
Application Publications 2016/0138008A1 and U52015/0344912A1, and in US
Patents
8,697,359, 8,771,945, 8,945,839, 8,999,641, 8,993,233, 8,895,308, 8,865,406,
8,889,418,
8,871,445, 8,889,356, 8,932,814, 8,795,965, and 8,906,616.
Cpfl endonuclease and
corresponding guide RNAs and PAM sites are disclosed in US Patent Application
Publication
2016/0208243 Al. Other CRISPR nucleases useful for editing genomes include
Cas12b and
Cas12c (see Shmakov et al. (2015) Mol. Cell, 60:385 ¨ 397; Harrington et al.
(2020)
Molecular Cell doi:10.1016/j.molce1.2020.06.022) and CasX and CasY (see
Burstein et al.
(2016) Nature, doi:10.1038/nature21059; Harrington et al. (2020) Molecular
Cell
doi:10.1016/j.molce1.2020.06.022), or Cas12j (Pausch et al, (2020) Science
10.1126/science.abb1400). Plant RNA promoters for expressing CRISPR guide RNA
and
plant codon-optimized CRISPR Cas9 endonuclease are disclosed in International
Patent
Application PCT/U52015/018104 (published as WO 2015/131101 and claiming
priority to
US Provisional Patent Application 61/945,700). Methods of using CRISPR
technology for
- 34 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
genome editing in plants are disclosed in US Patent Application Publications
US
2015/0082478A1 and US 2015/0059010A1 and in International Patent Application
PCT/U52015/038767 Al (published as WO 2016/007347 and claiming priority to US
Provisional Patent Application 62/023,246). All of the patent publications
referenced in this
paragraph are incorporated herein by reference in their entirety. In certain
embodiments, an
RNA-guided endonuclease that leaves a blunt end following cleavage of the
target site is
used. Blunt-end cutting RNA-guided endonucleases include Cas9, Cas12c, and Cas
12h
(Yan et al., 2019). In certain embodiments, an RNA-guided endonuclease that
leaves a
staggered single stranded DNA overhanging end following cleavage of the target
site
following cleavage of the target site is used.
Staggered-end cutting RNA-guided
endonucleases include Cas12a, Cas12b, and Cas12e.
[0079] The methods can also use sequence-specific endonucleases or sequence-
specific
endonucleases and guide RNAs that cleave a single DNA strand in a dsDNA target
site.
Such cleavage of a single DNA strand in a dsDNA target site is also referred
to herein and
elsewhere as "nicking" and can be effected by various "nickases" or systems
that provide for
nicking. Nickases that can be used include nCas9 (Cas9 comprising a DlOA amino
acid
substitution), nCas12a (e.g., Cas12a comprising an R1226A amino acid
substitution; Yamano
et al., 2016), Cas12i (Yan et al. 2019), a zinc finger nickase e.g., as
disclosed in Kim et al.,
2012), a TALE nickase (e.g., as disclosed in Wu et al., 2014), or a
combination thereof In
certain embodiments, systems that provide for nicking can comprise a Cas
nuclease (e.g.,
Cas9 and/or Cas12a) and guide RNA molecules that have at least one base
mismatch to DNA
sequences in the target editing site (Fu et al., 2019). In certain
embodiments, genome
modifications can be introduced into the target editing site by creating
single stranded breaks
(i.e., "nicks") in genomic locations separated by no more than about 10, 20,
30, 40, 50, 60,
80, 100, 150, or 200 base pairs of DNA. In
certain illustrative and non-limiting
embodiments, two nickases (i.e., a CAS nuclease which introduces a single
stranded DNA
break including nCas9, nCas12a, Cas12i, zinc finger nickases, TALE nickases,
combinations
thereof, and the like) or nickase systems can directed to make cuts to nearby
sites separated
by no more than about 10, 20, 30, 40, 50, 60, 80 or 100 base pairs of DNA. In
instances
where an RNA guided nickase and an RNA guide are used, the RNA guides are
adjacent to
PAM sequences that are sufficiently close (i.e., separated by no more than
about 10, 20, 30,
40, 50, 60, 80, 100, 150, or 200 base pairs of DNA). For the purposes of gene
editing,
- 35 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
CRISPR arrays can be designed to contain one or multiple guide RNA sequences
corresponding to a desired target DNA sequence; see, for example, Cong et at.
(2013)
Science, 339:819-823; Ran et at. (2013) Nature Protocols, 8:2281 ¨ 2308. At
least 16 or 17
nucleotides of gRNA sequence are required by Cas9 for DNA cleavage to occur;
for Cpfl at
least 16 nucleotides of gRNA sequence are needed to achieve detectable DNA
cleavage and
at least 18 nucleotides of gRNA sequence were reported necessary for efficient
DNA
cleavage in vitro; see Zetsche et at. (2015) Cell, 163:759 ¨ 771. In practice,
guide RNA
sequences are generally designed to have a length of 17 ¨ 24 nucleotides
(frequently 19, 20,
or 21 nucleotides) and exact complementarity (i.e., perfect base-pairing) to
the targeted gene
or nucleic acid sequence; guide RNAs having less than 100% complementarity to
the target
sequence can be used (e.g., a gRNA with a length of 20 nucleotides and 1 ¨4
mismatches to
the target sequence) but can increase the potential for off-target effects.
The design of
effective guide RNAs for use in plant genome editing is disclosed in US Patent
Application
Publication 2015/0082478 Al, the entire specification of which is incorporated
herein by
reference. More recently, efficient gene editing has been achieved using a
chimeric "single
guide RNA" ("sgRNA"), an engineered (synthetic) single RNA molecule that
mimics a
naturally occurring crRNA-tracrRNA complex and contains both a tracrRNA (for
binding the
nuclease) and at least one crRNA (to guide the nuclease to the sequence
targeted for editing);
see, for example, Cong et at. (2013) Science, 339:819 ¨ 823; Xing et at.
(2014) BMC Plant
Biol., 14:327 ¨ 340. Chemically modified sgRNAs have been demonstrated to be
effective in
genome editing; see, for example, Hendel et at. (2015) Nature Biotechnol., 985
¨ 991. The
design of effective gRNAs for use in plant genome editing is disclosed in US
Patent
Application Publication 2015/0082478 Al, the entire specification of which is
incorporated
herein by reference.
[0080] Genomic DNA may also be modified via base editing. Both adenine base
editors
(ABE) which convert A/T base pairs to G/C base pairs in genomic DNA as well as
cytosine
base pair editors (CBE) which effect C to T substitutions can be used in
certain embodiments
of the methods provided herein. In certain embodiments, useful ABE and CBE can
comprise
genome site specific DNA binding elements (e.g., RNA-dependent DNA binding
proteins
including catalytically inactive Cas9 and Cas12 proteins or Cas9 and Cas12
nickases)
operably linked to adenine or cytidine deaminases and used with guide RNAs
which position
the protein near the nucleotide targeted for substitution. Suitable ABE and
CBE disclosed in
- 36 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
the literature (Kim, Nat Plants, 2018 Mar;4(3):148-151) can be adapted for use
in the
methods set forth herein. In certain embodiments, a CBE can comprise a fusion
between a
catalytically inactive Cas9 (dCas9) RNA dependent DNA binding protein fused to
a cytidine
deaminase which converts cytosine (C) to uridine (U) and selected guide RNAs,
thereby
effecting a C to T substitution; see Komor et at. (2016) Nature, 533:420 ¨
424. In other
embodiments, C to T substitutions are effected with Cas9 nickase [Cas9n(D10A)]
fused to an
improved cytidine deaminase and optionally a bacteriophage Mu dsDNA (double-
stranded
DNA) end-binding protein Gam; see Komor et at., Sci Adv. 2017 Aug;
3(8):eaa04774. In
other embodiments, adenine base editors (ABEs) comprising an adenine deaminase
fused to
catalytically inactive Cas9 (dCas9) or a Cas9 DlOA nickase can be used to
convert A/T base
pairs to G/C base pairs in genomic DNA (Gaudelli et al., (2017)Nature
551(7681):464-471.
[0081] In certain embodiments, zinc finger nucleases or zinc finger nickases
can also be used
in the methods provided herein. Zinc-finger nucleases are site-specific
endonucleases
comprising two protein domains: a DNA-binding domain, comprising a plurality
of
individual zinc finger repeats that each recognize between 9 and 18 base
pairs, and a DNA-
cleavage domain that comprises a nuclease domain (typically Fokl). The
cleavage domain
dimerizes in order to cleave DNA; therefore, a pair of ZFNs are required to
target non-
palindromic target polynucleotides. In certain embodiments, zinc finger
nuclease and zinc
finger nickase design methods which have been described (Urnov et at. (2010)
Nature Rev.
Genet., 11:636 ¨ 646; Mohanta et al. (2017) Genes vol. 8,12: 399; Ramirez et
al. Nucleic
Acids Res. (2012); 40(12): 5560-5568; Liu et al. (2013) Nature Communications,
4: 2565)
can be adapted for use in the methods set forth herein. The zinc finger
binding domains of the
zinc finger nuclease or nickase provide specificity and can be engineered to
specifically
recognize any desired target DNA sequence. The zinc finger DNA binding domains
are
derived from the DNA-binding domain of a large class of eukaryotic
transcription factors
called zinc finger proteins (ZFPs). The DNA-binding domain of ZFPs typically
contains a
tandem array of at least three zinc "fingers" each recognizing a specific
triplet of DNA. A
number of strategies can be used to design the binding specificity of the zinc
finger binding
domain. One approach, termed "modular assembly", relies on the functional
autonomy of
individual zinc fingers with DNA. In this approach, a given sequence is
targeted by
identifying zinc fingers for each component triplet in the sequence and
linking them into a
multifinger peptide. Several alternative strategies for designing zinc finger
DNA binding
- 37 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
domains have also been developed. These methods are designed to accommodate
the ability
of zinc fingers to contact neighboring fingers as well as nucleotide bases
outside their target
triplet. Typically, the engineered zinc finger DNA binding domain has a novel
binding
specificity, compared to a naturally-occurring zinc finger protein.
Engineering methods
include, for example, rational design and various types of selection. Rational
design
includes, for example, the use of databases of triplet (or quadruplet)
nucleotide sequences and
individual zinc finger amino acid sequences, in which each triplet or
quadruplet nucleotide
sequence is associated with one or more amino acid sequences of zinc fingers
which bind the
particular triplet or quadruplet sequence. See, e.g., US Patents 6,453,242 and
6,534,261, both
incorporated herein by reference in their entirety. Exemplary selection
methods (e.g., phage
display and yeast two-hybrid systems) can be adapted for use in the methods
described
herein. In addition, enhancement of binding specificity for zinc finger
binding domains has
been described in US Patent 6,794,136, incorporated herein by reference in its
entirety. In
addition, individual zinc finger domains may be linked together using any
suitable linker
sequences. Examples of linker sequences are publicly known, e.g., see US
Patents 6,479,626;
6,903,185; and 7,153,949, incorporated herein by reference in their entirety.
The nucleic acid
cleavage domain is non-specific and is typically a restriction endonuclease,
such as Fokl.
This endonuclease must dimerize to cleave DNA. Thus, cleavage by Fokl as part
of a ZFN
requires two adjacent and independent binding events, which must occur in both
the correct
orientation and with appropriate spacing to permit dimer formation. The
requirement for two
DNA binding events enables more specific targeting of long and potentially
unique
recognition sites. Fokl variants with enhanced activities have been described
and can be
adapted for use in the methods described herein; see, e.g., Guo et at. (2010)
1 Mol. Biol.,
400:96 - 107.
[0082] Transcription activator like effectors (TALEs) are proteins secreted by
certain
Xanthomonas species to modulate gene expression in host plants and to
facilitate the
colonization by and survival of the bacterium. TALEs act as transcription
factors and
modulate expression of resistance genes in the plants. Recent studies of TALEs
have
revealed the code linking the repetitive region of TALEs with their target DNA-
binding sites.
TALEs comprise a highly conserved and repetitive region consisting of tandem
repeats of
mostly 33 or 34 amino acid segments. The repeat monomers differ from each
other mainly at
amino acid positions 12 and 13. A strong correlation between unique pairs of
amino acids at
- 38 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
positions 12 and 13 and the corresponding nucleotide in the TALE-binding site
has been
found. The simple relationship between amino acid sequence and DNA recognition
of the
TALE binding domain allows for the design of DNA binding domains of any
desired
specificity. TALEs can be linked to a non-specific DNA cleavage domain to
prepare genome
editing proteins, referred to as TAL-effector nucleases or TALENs. As in the
case of ZFNs, a
restriction endonuclease, such as Fokl, can be conveniently used. Methods for
use of
TALENs in plants have been described and can be adapted for use in the methods
described
herein, see Mahfouz et al. (2011) Proc. Natl. Acad. Sci. USA, 108:2623 ¨ 2628;
Mahfouz
(2011) GM Crops, 2:99 ¨ 103; and Mohanta et al. (2017) Genes vol. 8,12: 399).
TALE
nickases have also been described and can be adapted for use in methods
described herein
(Wu et al.; Biochem Biophys Res Commun. (2014);446(1):261-6; Luo et al;
Scientific
Reports 6, Article number: 20657 (2016)).
[0083] Embodiments of the donor DNA template molecule having a sequence that
is
integrated at the site of at least one double-strand break (DSB) in a genome
include double-
stranded DNA, a single-stranded DNA, a single-stranded DNA/RNA hybrid, and a
double-
stranded DNA/RNA hybrid. In embodiments, a donor DNA template molecule that is
a
double-stranded (e.g., a dsDNA or dsDNA/RNA hybrid) molecule is provided
directly to the
plant protoplast or plant cell in the form of a double-stranded DNA or a
double-stranded
DNA/RNA hybrid, or as two single-stranded DNA (ssDNA) molecules that are
capable of
hybridizing to form dsDNA, or as a single-stranded DNA molecule and a single-
stranded
RNA (ssRNA) molecule that are capable of hybridizing to form a double-stranded
DNA/RNA hybrid; that is to say, the double-stranded polynucleotide molecule is
not
provided indirectly, for example, by expression in the cell of a dsDNA encoded
by a plasmid
or other vector. In various non-limiting embodiments of the method, the donor
DNA template
molecule that is integrated (or that has a sequence that is integrated) at the
site of at least one
double-strand break (DSB) in a genome is double-stranded and blunt-ended; in
other
embodiments the donor DNA template molecule is double-stranded and has an
overhang or
"sticky end" consisting of unpaired nucleotides (e.g., 1, 2, 3, 4, 5, or 6
unpaired nucleotides)
at one terminus or both termini. In an embodiment, the DSB in the genome has
no unpaired
nucleotides at the cleavage site, and the donor DNA template molecule that is
integrated (or
that has a sequence that is integrated) at the site of the DSB is a blunt-
ended double-stranded
DNA or blunt-ended double-stranded DNA/RNA hybrid molecule, or alternatively
is a
- 39 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
single-stranded DNA or a single-stranded DNA/RNA hybrid molecule. In another
embodiment, the DSB in the genome has one or more unpaired nucleotides at one
or both
sides of the cleavage site, and the donor DNA template molecule that is
integrated (or that
has a sequence that is integrated) at the site of the DSB is a double-stranded
DNA or double-
stranded DNA/RNA hybrid molecule with an overhang or "sticky end" consisting
of unpaired
nucleotides at one or both termini, or alternatively is a single-stranded DNA
or a single-
stranded DNA/RNA hybrid molecule; in embodiments, the donor DNA template
molecule
DSB is a double-stranded DNA or double-stranded DNA/RNA hybrid molecule that
includes
an overhang at one or at both termini, wherein the overhang consists of the
same number of
unpaired nucleotides as the number of unpaired nucleotides created at the site
of a DSB by a
nuclease that cuts in an off-set fashion (e.g., where a Cas12 nuclease effects
an off-set DSB
with 5-nucleotide overhangs in the genomic sequence, the donor DNA template
molecule
that is to be integrated (or that has a sequence that is to be integrated) at
the site of the DSB is
double-stranded and has 5 unpaired nucleotides at one or both termini). In
certain
embodiments, one or both termini of the donor DNA template molecule contain no
regions of
sequence homology (identity or complementarity) to genomic regions flanking
the DSB; that
is to say, one or both termini of the donor DNA template molecule contain no
regions of
sequence that is sufficiently complementary to permit hybridization to genomic
regions
immediately adjacent to the location of the DSB. In embodiments, the donor DNA
template
molecule contains no homology to the locus of the DSB, that is to say, the
donor DNA
template molecule contains no nucleotide sequence that is sufficiently
complementary to
permit hybridization to genomic regions immediately adjacent to the location
of the DSB. In
embodiments, the donor DNA template molecule is at least partially double-
stranded and
includes 2-20 base-pairs, e. g., 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19, or 20
base-pairs; in embodiments, the donor DNA template molecule is double-stranded
and blunt-
ended and consists of 2-20 base-pairs, e.g., 2, 3,4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17,
18, 19, or 20 base-pairs; in other embodiments, the donor DNA template
molecule is double-
stranded and includes 2-20 base-pairs, e.g., 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17,
18, 19, or 20 base-pairs and in addition has at least one overhang or "sticky
end" consisting of
at least one additional, unpaired nucleotide at one or at both termini. In an
embodiment, the
donor DNA template molecule that is integrated (or that has a sequence that is
integrated) at
the site of at least one double-strand break (DSB) in a genome is a blunt-
ended double-
- 40 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
stranded DNA or a blunt-ended double-stranded DNA/RNA hybrid molecule of about
18 to
about 300 base-pairs, or about 20 to about 200 base-pairs, or about 30 to
about 100 base-
pairs, and having at least one phosphorothioate bond between adjacent
nucleotides at a 5' end,
3' end, or both 5' and 3' ends. In embodiments, the donor DNA template
molecule includes
single strands of at least 11, at least 18, at least 20, at least 30, at least
40, at least 60, at least
80, at least 100, at least 120, at least 140, at least 160, at least 180, at
least 200, at least 240, at
about 280, or at least 320 nucleotides. In embodiments, the donor DNA template
molecule
has a length of at least 2, at least 3, at least 4, at least 5, at least 6, at
least 7, at least 8, at least
9, at least 10, or at least 11 base-pairs if double-stranded (or nucleotides
if single-stranded),
or between about 2 to about 320 base-pairs if double-stranded (or nucleotides
if single-
stranded), or between about 2 to about 500 base-pairs if double-stranded (or
nucleotides if
single-stranded), or between about 5 to about 500 base-pairs if double-
stranded (or
nucleotides if single-stranded), or between about 5 to about 300 base-pairs if
double-stranded
(or nucleotides if single-stranded), or between about 11 to about 300 base-
pairs if double-
stranded (or nucleotides if single-stranded), or about 18 to about 300 base-
pairs if double-
stranded (or nucleotides if single-stranded), or between about 30 to about 100
base-pairs if
double-stranded (or nucleotides if single-stranded). In embodiments, the donor
DNA template
molecule includes chemically modified nucleotides (see, e.g., the various
modifications of
internucleotide linkages, bases, and sugars described in Verma and Eckstein
(1998) Annu.
Rev. Biochem., 67:99-134); in embodiments, the naturally occurring
phosphodiester
backbone of the donor DNA template molecule is partially or completely
modified with
phosphorothioate, phosphorodithioate, or methylphosphonate internucleotide
linkage
modifications, or the donor DNA template molecule includes modified nucleoside
bases or
modified sugars, or the donor DNA template molecule is labelled with a
fluorescent moiety
(e.g., fluorescein or rhodamine or a fluorescent nucleoside analogue) or other
detectable label
(e.g., biotin or an isotope). In another embodiment, the donor DNA template
molecule
contains secondary structure that provides stability or acts as an aptamer.
Other related
embodiments include double-stranded DNA/RNA hybrid molecules, single-stranded
DNA/RNA hybrid donor molecules, and single-stranded donor DNA template
molecules
(including single-stranded, chemically modified donor DNA template molecules),
which in
analogous procedures are integrated (or have a sequence that is integrated) at
the site of a
double-strand break. Donor DNA templates provided herein include those
comprising
-41 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
CgRRS sequences flanked by DNA with homology to a donor polynucleotide and
include the
donor DNA template set forth in SEQ ID NO: 14 and equivalents thereof with
longer or
shorter homology arms. In certain embodiments, a donor DNA template can
comprise an
adapter molecule (e.g., a donor DNA template formed by annealing SEQ ID NO: 11
and 12
or by annealing SEQ ID NO: 11 and 13) with cohesive ends which can anneal to
an
overhanging cleavage site (e.g., introduced by a Cas12a nuclease and suitable
gRNAs). In
certain embodiments, integration of the donor DNA templates can be facilitated
by use of a
bacteriophage lambda exonuclease, a bacteriophage lambda beta SSAP protein,
and an E. coli
SSB essentially as set forth in US Patent Application Publication 20200407754,
which is
incorporated herein by reference in its entirety.
[0084] Donor DNA template molecules used in the methods provided herein
include DNA
molecules comprising, from 5' to 3', a first homology arm, a replacement DNA,
and a second
homology arm, wherein the homology arms containing sequences that are
partially or
completely homologous to genomic DNA (gDNA) sequences flanking a target site-
specific
endonuclease cleavage site in the gDNA. In certain embodiments, the
replacement DNA can
comprise an insertion, deletion, or substitution of 1 or more DNA base pairs
relative to the
target gDNA. In an embodiment, the donor DNA template molecule is double-
stranded and
perfectly base-paired through all or most of its length, with the possible
exception of any
unpaired nucleotides at either terminus or both termini. In another
embodiment, the donor
DNA template molecule is double-stranded and includes one or more non-terminal
mismatches or non-terminal unpaired nucleotides within the otherwise double-
stranded
duplex. In an embodiment, the donor DNA template molecule that is integrated
at the site of
at least one double-strand break (DSB) includes between 2-20 nucleotides in
one (if single-
stranded) or in both strands (if double-stranded), e. g., 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19, or 20 nucleotides on one or on both strands, each of which
can be base-
paired to a nucleotide on the opposite strand (in the case of a perfectly base-
paired double-
stranded polynucleotide molecule). Such donor DNA templates can be integrated
in genomic
DNA containing blunt and/or staggered double stranded DNA breaks by homology-
directed
repair (HDR). In certain embodiments, a donor DNA template homology arm can be
about
20, 50, 100, 200, 400, or 600 to about 800, or 1000 base pairs in length. In
certain
embodiments, a donor DNA template molecule can be delivered to a plant cell)
in a circular
(e.g., a plasmid or a viral vector including a geminivirus vector) or a linear
DNA molecule. In
- 42 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
certain embodiments, a circular or linear DNA molecule that is used can
comprise a modified
donor DNA template molecule comprising, from 5' to 3', a first copy of the
target sequence-
specific endonuclease cleavage site sequence, the first homology arm, the
replacement DNA,
the second homology arm, and a second copy of the target sequence-specific
endonuclease
cleavage site sequence. Without seeking to be limited by theory, such modified
donor DNA
template molecules can be cleaved by the same sequence-specific endonuclease
that is used
to cleave the target site gDNA of the eukaryotic cell to release a donor DNA
template
molecule that can participate in HDR-mediated genome modification of the
target editing site
in the plant cell genome. In certain embodiments, the donor DNA template can
comprise a
linear DNA molecule comprising, from 5' to 3', a cleaved target sequence-
specific
endonuclease cleavage site sequence, the first homology arm, the replacement
DNA, the
second homology arm, and a cleaved target sequence-specific endonuclease
cleavage site
sequence. In certain embodiments, the cleaved target sequence-specific
endonuclease
sequence can comprise a blunt DNA end or a blunt DNA end that can optionally
comprise a
5' phosphate group. In certain embodiments, the cleaved target sequence-
specific
endonuclease sequence comprises a DNA end having a single-stranded 5' or 3'
DNA
overhang. Such cleaved target sequence-specific endonuclease cleavage site
sequences can
be produced by either cleaving an intact target sequence-specific endonuclease
cleavage site
sequence or by synthesizing a copy of the cleaved target sequence-specific
endonuclease
cleavage site sequence. Donor DNA templates can be synthesized either
chemically or
enzymatically (e.g., in a polymerase chain reaction (PCR)). Donor DNA
templates provided
herein include those comprising CgRRS sequences flanked by DNA with homology
to a
donor polynucleotide.
[0085] Various treatments are useful in delivery of gene editing molecules
and/or other
molecules to a DP-4114 or INIR6 plant cell. In certain embodiments, one or
more treatments
is employed to deliver the gene editing or other molecules (e.g., comprising a
polynucleotide,
polypeptide or combination thereof) into a eukaryotic or plant cell, e.g.,
through barriers such
as a cell wall, a plasma membrane, a nuclear envelope, and/or other lipid
bilayer. In certain
embodiments, a polynucleotide-, polypeptide-, or RNP-containing composition
comprising
the molecules are delivered directly, for example by direct contact of the
composition with a
plant cell. Aforementioned compositions can be provided in the form of a
liquid, a solution,
a suspension, an emulsion, a reverse emulsion, a colloid, a dispersion, a gel,
liposomes,
- 43 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
micelles, an injectable material, an aerosol, a solid, a powder, a
particulate, a nanoparticle, or
a combination thereof can be applied directly to a plant, plant part, plant
cell, or plant explant
(e.g., through abrasion or puncture or otherwise disruption of the cell wall
or cell membrane,
by spraying or dipping or soaking or otherwise directly contacting, by
microinjection). For
example, a plant cell or plant protoplast is soaked in a liquid genome editing
molecule-
containing composition, whereby the agent is delivered to the plant cell. In
certain
embodiments, the agent-containing composition is delivered using negative or
positive
pressure, for example, using vacuum infiltration or application of
hydrodynamic or fluid
pressure. In certain embodiments, the agent-containing composition is
introduced into a plant
cell or plant protoplast, e.g., by microinjection or by disruption or
deformation of the cell
wall or cell membrane, for example by physical treatments such as by
application of negative
or positive pressure, shear forces, or treatment with a chemical or physical
delivery agent
such as surfactants, liposomes, or nanoparticles; see, e.g., delivery of
materials to cells
employing microfluidic flow through a cell-deforming constriction as described
in US
Published Patent Application 2014/0287509, incorporated by reference in its
entirety herein.
Other techniques useful for delivering the agent-containing composition to a
eukaryotic cell,
plant cell or plant protoplast include: ultrasound or sonication; vibration,
friction, shear stress,
vortexing, cavitation; centrifugation or application of mechanical force;
mechanical cell wall
or cell membrane deformation or breakage; enzymatic cell wall or cell membrane
breakage or
permeabilization; abrasion or mechanical scarification (e.g., abrasion with
carborundum or
other particulate abrasive or scarification with a file or sandpaper) or
chemical scarification
(e.g., treatment with an acid or caustic agent); and electroporation. In
certain embodiments,
the agent-containing composition is provided by bacterially mediated (e.g.,
Agrobacterium
sp., Rhizobium sp., Sinorhizobium sp., Mesorhizobium sp., Bradyrhizobium sp.,
Azobacter
sp., Phyllobacterium sp.) transfection of the plant cell or plant protoplast
with a
polynucleotide encoding the genome editing molecules (e.g., RNA dependent DNA
endonuclease, RNA dependent DNA binding protein, RNA dependent nickase, ABE,
or
CBE, and/or guide RNA); see, e.g., Broothaerts et at. (2005) Nature, 433:629 ¨
633). Any of
these techniques or a combination thereof are alternatively employed on the
plant explant,
plant part or tissue or intact plant (or seed) from which a plant cell is
optionally subsequently
obtained or isolated; in certain embodiments, the agent-containing composition
is delivered in
a separate step after the plant cell has been isolated.
- 44 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
[0086] In some embodiments, one or more polynucleotides or vectors driving
expression of
one or more genome editing molecules or trait-conferring genes (e.g.,
herbicide tolerance,
insect resistance, and/or male sterility) are introduced into a DP-4114 or
INIR6 plant cell. In
certain embodiments, a polynucleotide vector comprises a regulatory element
such as a
promoter operably linked to one or more polynucleotides encoding genome
editing molecules
and/or trait-conferring genes. In such embodiments, expression of these
polynucleotides can
be controlled by selection of the appropriate promoter, particularly promoters
functional in a
eukaryotic cell (e.g., plant cell); useful promoters include constitutive,
conditional, inducible,
and temporally or spatially specific promoters (e.g., a tissue specific
promoter, a
developmentally regulated promoter, or a cell cycle regulated promoter).
Developmentally
regulated promoters that can be used in plant cells include Phospholipid
Transfer Protein
(PLTP), fructose-1,6-bisphosphatase protein, NAD(P)-binding Rossmann-Fold
protein,
adipocyte plasma membrane-associated protein-like protein, Rieske [2Fe-2S]
iron-sulfur
domain protein, chlororespiratory reduction 6 protein, D-glycerate 3-kinase,
chloroplastic-
like protein, chlorophyll a-b binding protein 7, chloroplastic-like protein,
ultraviolet-B-
repressible protein, Soul heme-binding family protein, Photosystem I reaction
center subunit
psi-N protein, and short-chain dehydrogenase/reductase protein that are
disclosed in US
Patent Application Publication No. 20170121722, which is incorporated herein
by reference
in its entirety and specifically with respect to such disclosure. In certain
embodiments, the
promoter is operably linked to nucleotide sequences encoding multiple guide
RNAs, wherein
the sequences encoding guide RNAs are separated by a cleavage site such as a
nucleotide
sequence encoding a microRNA recognition/cleavage site or a self-cleaving
ribozyme (see,
e.g., Ferre-D'Amare and Scott (2014) Cold Spring Harbor Perspectives Biol.,
2:a003574). In
certain embodiments, the promoter is an RNA polymerase III promoter operably
linked to a
nucleotide sequence encoding one or more guide RNAs. In certain embodiments,
the RNA
polymerase III promoter is a plant U6 spliceosomal RNA promoter, which can be
native to
the genome of the plant cell or from a different species, e.g., a U6 promoter
from maize,
tomato, or soybean such as those disclosed U.S. Patent Application Publication
2017/0166912, or a homologue thereof; in an example, such a promoter is
operably linked to
DNA sequence encoding a first RNA molecule including a Cas12a gRNA followed by
an
operably linked and suitable 3' element such as a U6 poly-T terminator. In
another
embodiment, the RNA polymerase III promoter is a plant U3, 75L (signal
recognition
- 45 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
particle RNA), U2, or U5 promoter, or chimerics thereof, e.g., as described in
U.S. Patent
Application Publication 20170166912. In certain embodiments, the promoter
operably linked
to one or more polynucleotides is a constitutive promoter that drives gene
expression in
eukaryotic cells (e.g., plant cells). In certain embodiments, the promoter
drives gene
expression in the nucleus or in an organelle such as a chloroplast or
mitochondrion.
Examples of constitutive promoters for use in plants include a CaMV 35S
promoter as
disclosed in US Patents 5,858,742 and 5,322,938, a rice actin promoter as
disclosed in US
Patent 5,641,876, a maize chloroplast aldolase promoter as disclosed in US
Patent 7,151,204,
and the nopaline synthase (NOS) and octopine synthase (OCS) promoters from
Agrobacterium tumefaciens. In certain embodiments, the promoter operably
linked to one or
more polynucleotides encoding elements of a genome-editing system is a
promoter from
figwort mosaic virus (FMV), a RUBISCO promoter, or a pyruvate phosphate
dikinase
(PPDK) promoter, which is active in photosynthetic tissues. Other contemplated
promoters
include cell-specific or tissue-specific or developmentally regulated
promoters, for example,
a promoter that limits the expression of the nucleic acid targeting system to
germline or
reproductive cells (e.g., promoters of genes encoding DNA ligases,
recombinases, replicases,
or other genes specifically expressed in germline or reproductive cells). In
certain
embodiments, the genome alteration is limited only to those cells from which
DNA is
inherited in subsequent generations, which is advantageous where it is
desirable that
expression of the genome-editing system be limited in order to avoid
genotoxicity or other
unwanted effects. All of the patent publications referenced in this paragraph
are incorporated
herein by reference in their entirety.
[0087] Expression vectors or polynucleotides provided herein may contain a DNA
segment
near the 3' end of an expression cassette that acts as a signal to terminate
transcription and
directs polyadenylation of the resultant mRNA and may also support promoter
activity. Such
a 3' element is commonly referred to as a "3'-untranslated region" or "3'-UTR"
or a
c`polyadenylation signal." In some cases, plant gene-based 3' elements (or
terminators)
consist of both the 3'-UTR and downstream non-transcribed sequence (Nuccio et
al., 2015).
Useful 3' elements include: Agrobacterium tumefaciens nos 3', tml 3', tmr 3',
tms 3', ocs 3',
and tr7 3' elements disclosed in US Patent No. 6,090,627, incorporated herein
by reference,
and 3' elements from plant genes such as the heat shock protein 17, ubiquitin,
and fructose-
1,6-biphosphatase genes from wheat (Triticum aestivum), and the glutelin,
lactate
- 46 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
dehydrogenase, and beta-tubulin genes from rice (Oryza sativa), disclosed in
US Patent
Application Publication 2002/0192813 Al. All of the patent publications
referenced in this
paragraph are incorporated herein by reference in their entireties.
[0088] In certain embodiments, the DP-4114 or INIR6 plant cells used herein
can comprise
haploid, diploid, or polyploid plant cells or plant protoplasts, for example,
those obtained
from a haploid, diploid, or polyploid plant, plant part or tissue, or callus.
In certain
embodiments, plant cells in culture (or the regenerated plant, progeny seed,
and progeny
plant) are haploid or can be induced to become haploid; techniques for making
and using
haploid plants and plant cells are known in the art, see, e.g., methods for
generating haploids
in Arabidopsis thaliana by crossing of a wild-type strain to a haploid-
inducing strain that
expresses altered forms of the centromere-specific histone CENH3, as described
by
Maruthachalam and Chan in "How to make haploid Arabidopsis thaliana", protocol
available
at www [dot] op enwetware [dot] org/images/d/d3/Hapl oi d Arab i dop si
s_protoc ol [dot] p df; (Ravi
et at. (2014) Nature Communications, 5:5334, doi: 10.1038/nc0mm56334).
Haploids can
also be obtained in a wide variety of monocot plants (e.g., maize, wheat,
rice, sorghum,
barley) by crossing a plant comprising a mutated CENH3 gene with a wildtype
diploid plant
to generate haploid progeny as disclosed in US Patent No. 9,215,849, which is
incorporated
herein by reference in its entirety. Haploid-inducing maize lines that can be
used to obtain
haploid maize plants and/or cells include Stock 6, MI-II (Moldovian Haploid
Inducer),
indeterminate gametophyte (ig) mutation, KEMS, RWK, ZEM, ZMS, KMS, and well as
transgenic haploid inducer lines disclosed in US Patent No. 9,677,082, which
is incorporated
herein by reference in its entirety. Examples of haploid cells include but are
not limited to
plant cells obtained from haploid plants and plant cells obtained from
reproductive tissues,
e.g., from flowers, developing flowers or flower buds, ovaries, ovules,
megaspores, anthers,
pollen, megagametophyte, and microspores. In certain embodiments where the
plant cell or
plant protoplast is haploid, the genetic complement can be doubled by
chromosome doubling
(e.g., by spontaneous chromosomal doubling by meiotic non-reduction, or by
using a
chromosome doubling agent such as colchicine, oryzalin, trifluralin,
pronamide, nitrous oxide
gas, anti-microtubule herbicides, anti-microtubule agents, and mitotic
inhibitors) in the plant
cell or plant protoplast to produce a doubled haploid plant cell or plant
protoplast wherein the
complement of genes or alleles is homozygous; yet other embodiments include
regeneration
of a doubled haploid plant from the doubled haploid plant cell or plant
protoplast. Another
- 47 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
embodiment is related to a hybrid plant having at least one parent plant that
is a doubled
haploid plant provided by this approach. Production of doubled haploid plants
provides
homozygosity in one generation, instead of requiring several generations of
self-crossing to
obtain homozygous plants. The use of doubled haploids is advantageous in any
situation
where there is a desire to establish genetic purity (i.e., homozygosity) in
the least possible
time. Doubled haploid production can be particularly advantageous in slow-
growing plants or
for producing hybrid plants that are offspring of at least one doubled-haploid
plant.
[0089] In certain embodiments, the DP-4114 or INIR6 plant cells used in the
methods
provided herein can include non-dividing cells. Such non-dividing cells can
include plant cell
protoplasts, plant cells subjected to one or more of a genetic and/or
pharmaceutically-induced
cell-cycle blockage, and the like.
[0090] In certain embodiments, the DP-4114 or INIR6 plant cells in used in the
methods
provided herein can include dividing cells. Dividing cells can include those
cells found in
various plant tissues including leaves, meristems, and embryos. These tissues
include
dividing cells from young maize leaf, meristems and scutellar tissue from
about 8 or 10 to
about 12 or 14 days after pollination (DAP) embryos. The isolation of maize
embryos has
been described in several publications (Brettschneider, Becker, and Lorz 1997;
Leduc et al.
1996; Frame et al. 2011; K. Wang and Frame 2009). In certain embodiments,
basal leaf
tissues (e.g., leaf tissues located about 0 to 3 cm from the ligule of a maize
plant; Kirienko,
Luo, and Sylvester 2012) are targeted for HDR-mediated gene editing. Methods
for obtaining
regenerable plant structures and regenerating plants from the NHEJ-, MMEJ-, or
HDR-
mediated gene editing of plant cells provided herein can be adapted from
methods disclosed
in US Patent Application Publication No. 20170121722, which is incorporated
herein by
reference in its entirety and specifically with respect to such disclosure. In
certain
embodiments, single plant cells subjected to the HDR-mediated gene editing
will give rise to
single regenerable plant structures. In certain embodiments, the single
regenerable plant cell
structure can form from a single cell on, or within, an explant that has been
subjected to the
NHEJ-, MMEJ-, or HDR-mediated gene editing.
[0091] In some embodiments, methods provided herein can include the additional
step of
growing or regenerating an INIR6 plant from a INIR6 plant cell that had been
subjected to
the gene editing or from a regenerable plant structure obtained from that
INIR6 plant cell. In
certain embodiments, the plant can further comprise an inserted transgene, a
target gene edit,
- 48 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
or genome edit as provided by the methods and compositions disclosed herein.
In certain
embodiments, callus is produced from the plant cell, and plantlets and plants
produced from
such callus. In other embodiments, whole seedlings or plants are grown
directly from the
plant cell without a callus stage. Thus, additional related aspects are
directed to whole
seedlings and plants grown or regenerated from the plant cell or plant
protoplast having a
target gene edit or genome edit, as well as the seeds of such plants. In
certain embodiments
wherein the plant cell or plant protoplast is subjected to genetic
modification (for example,
genome editing by means of, e.g., an RdDe), the grown or regenerated plant
exhibits a
phenotype associated with the genetic modification. In certain embodiments,
the grown or
regenerated plant includes in its genome two or more genetic or epigenetic
modifications that
in combination provide at least one phenotype of interest. In certain
embodiments, a
heterogeneous population of plant cells having a target gene edit or genome
edit, at least
some of which include at least one genetic or epigenetic modification, is
provided by the
method; related aspects include a plant having a phenotype of interest
associated with the
genetic or epigenetic modification, provided by either regeneration of a plant
having the
phenotype of interest from a plant cell or plant protoplast selected from the
heterogeneous
population of plant cells having a target gene or genome edit, or by selection
of a plant
having the phenotype of interest from a heterogeneous population of plants
grown or
regenerated from the population of plant cells having a targeted genetic edit
or genome edit.
Examples of phenotypes of interest include herbicide resistance, improved
tolerance of
abiotic stress (e.g., tolerance of temperature extremes, drought, or salt) or
biotic stress (e.g.,
resistance to nematode, bacterial, or fungal pathogens), improved utilization
of nutrients or
water, modified lipid, carbohydrate, or protein composition, improved flavor
or appearance,
improved storage characteristics (e.g., resistance to bruising, browning, or
softening),
increased yield, altered morphology (e.g., floral architecture or color, plant
height, branching,
root structure). In an embodiment, a heterogeneous population of plant cells
having a target
gene edit or genome edit (or seedlings or plants grown or regenerated
therefrom) is exposed
to conditions permitting expression of the phenotype of interest; e.g.,
selection for herbicide
resistance can include exposing the population of plant cells having a target
gene edit or
genome edit (or seedlings or plants grown or regenerated therefrom) to an
amount of
herbicide or other substance that inhibits growth or is toxic, allowing
identification and
selection of those resistant plant cells (or seedlings or plants) that survive
treatment. Methods
- 49 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
for obtaining regenerable plant structures and regenerating plants from plant
cells or
regenerable plant structures can be adapted from published procedures (Roest
and Gilissen,
Acta Bot. Neerl., 1989, 38(1), 1-23; Bhaskaran and Smith, Crop Sci. 30(6):1328-
1337;
Ikeuchi et al., Development, 2016, 143: 1442-1451). Methods for obtaining
regenerable
plant structures and regenerating plants from plant cells or regenerable plant
structures can
also be adapted from US Patent Application Publication No. 20170121722, which
is
incorporated herein by reference in its entirety and specifically with respect
to such
disclosure. Also provided are heterogeneous or homogeneous populations of such
plants or
parts thereof (e.g., seeds), succeeding generations or seeds of such plants
grown or
regenerated from the plant cells or plant protoplasts, having a target gene
edit or genome edit.
Additional related aspects include a hybrid plant provided by crossing a first
plant grown or
regenerated from a plant cell or plant protoplast having a target gene edit or
genome edit and
having at least one genetic or epigenetic modification, with a second plant,
wherein the
hybrid plant contains the genetic or epigenetic modification; also
contemplated is seed
produced by the hybrid plant. Also envisioned as related aspects are progeny
seed and
progeny plants, including hybrid seed and hybrid plants, having the
regenerated plant as a
parent or ancestor. The plant cells and derivative plants and seeds disclosed
herein can be
used for various purposes useful to the consumer or grower. In other
embodiments,
processed products are made from the INIR6 plant or its seeds, including: (a)
maize seed
meal (defatted or non-defatted); (b) extracted proteins, oils, sugars, and
starches; (c)
fermentation products; (d) animal feed or human food products (e.g., feed and
food
comprising maize seed meal (defatted or non-defatted) and other ingredients
(e.g., other
cereal grains, other seed meal, other protein meal, other oil, other starch,
other sugar, a
binder, a preservative, a humectant, a vitamin, and/or mineral; (e) a
pharmaceutical; (f) raw
or processed biomass (e.g., cellulosic and/or lignocellulosic material); and
(g) various
industrial products.
EMBODIMENTS
[0092] Various embodiments of the plants, genomes, methods, biological
samples, and other
compositions described herein are set forth in the following sets of numbered
embodiments.
[0093] la. A transgenic maize plant cell comprising an INIR6 transgenic locus
comprising an
originator guide RNA recognition site (OgRRS) in a first DNA junction
polynucleotide of a
- 50 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
DP-4114 transgenic locus and a cognate guide RNA recognition site (CgRRS) in a
second
DNA junction polynucleotide of the DP-4114 transgenic locus.
[0094] lb. A transgenic maize plant cell comprising an INIR6 transgenic locus
comprising an
insertion and/or substitution of DNA in a DNA junction polynucleotide of a DP-
4114
transgenic locus with DNA comprising a cognate guide RNA recognition site
(CgRRS).
[0095] 2. The transgenic maize plant cell of embodiment la or lb, wherein said
CgRRS
comprises the DNA molecule set forth in SEQ ID NO: 8, 9, 10, or 19; and/or
wherein said
DP-4114 transgenic locus is set forth in SEQ ID NO:1, is present in seed
deposited at the
ATCC under accession No. PTA-11506, is present in progeny thereof, is present
in allelic
variants thereof, or is present in other variants thereof
[0096] 3. The transgenic maize plant cell of embodiments la, lb, or 2, wherein
said INIR6
transgenic locus comprises the DNA molecule set forth in SEQ ID NO: 2, 3, 20,
or an allelic
variant thereof.
[0097] 4. A transgenic maize plant part comprising the maize plant cell of any
one of
embodiments la, lb, 2, or 3, wherein said maize plant part is optionally a
seed.
[0098] 5. A transgenic maize plant comprising the maize plant cell of any one
of
embodiments la, lb, 2, or 3.
[0099] 6. A method for obtaining a bulked population of inbred seed comprising
selfing the
transgenic maize plant of embodiment 5 and harvesting seed comprising the
INIR6 transgenic
locus from the selfed maize plant.
[00100] 7. A method of obtaining hybrid maize seed comprising crossing the
transgenic maize plant of embodiment 5 to a second maize plant which is
genetically distinct
from the first maize plant and harvesting seed comprising the INIR6 transgenic
locus from
the cross.
[00101] 8. A DNA molecule comprising SEQ ID NO: 2, 3, 8, 9, 10, 19, 20, or
an
allelic variant thereof.
[00102] 9. A processed transgenic maize plant product comprising the DNA
molecule
of embodiment 8.
[00103] 10. A biological sample containing the DNA molecule of embodiment
8.
[00104] 11. A nucleic acid molecule adapted for detection of genomic DNA
comprising the DNA molecule of embodiment 8, wherein said nucleic acid
molecule
optionally comprises a detectable label.
-51 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
[00105] 12. A method of detecting a maize plant cell comprising the INIR6
transgenic
locus of any one of embodiments la, lb, 2, or 3, comprising the step of
detecting DNA
molecule comprising SEQ ID NO: 2, 3, 8, 9, 10, 19, 20, or an allelic variant
thereof
[00106] 13. A method of excising the INIR6 transgenic locus from the
genome of the
maize plant cell of any one of embodiments la, lb, 2, or 3, comprising the
steps of:
(a) contacting the edited transgenic plant genome of the plant cell of
embodiment 5
with: (i) an RNA dependent DNA endonuclease (RdDe); and (ii) a guide RNA
(gRNA)
capable of hybridizing to the guide RNA hybridization site of the OgRRS and
the CgRRS;
wherein the RdDe recognizes a OgRRS/gRNA and a CgRRS/gRNA hybridization
complex;
and,
(b) selecting a transgenic plant cell, transgenic plant part, or transgenic
plant wherein
the INIR6 transgenic locus flanked by the OgRRS and the CgRRS has been
excised.
Examples
[00107] Example 1. Introduction of a CgRRS in a 3' junction
polynucleotides of a
DP-4114 Transgenic Locus
[00108] Transgenic plant genomes containing one or more of the following
transgenic
loci (events) are contacted with:
(i) an ABE or CBE and guide RNAs which recognize the indicated target DNA
sites
(protospacer (guide RNA coding) plus PAM site) in the 5' or3' junction
polynucleotides of
the event to introduce a CgRRS in the junction polynucleotide;
(ii) an RdDe and guide RNAs which recognize the indicated target DNA site
(guide RNA
coding plus PAM site) in the 5' or 3' junction polynucleotides of the event as
well as a donor
DNA template spanning the double stranded DNA break site in the junction
polynucleotide to
introduce a CgRRS in a junction polynucleotide.
Plant cells, callus, parts, or whole plants comprising the introduced CgRRS in
the transgenic
plant genome are selected.
[00109] Table 1. Examples of OgRRS and CgRRS in DP-4114
CORN
EVEN OgRRS CgRRS
NAME
DP- tttgtagcacttgcacgtagtta cgcttttgtagcacttgcacgtagttacccggata
- 52 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
4114 cccg (SEQ ID NO: 7; (SEQ ID NO: 8; inserted into 3'
located in 5' junction junction polynucleotide)
polynucleotide of SEQ
ID NO: 1)
aacgtgcaagcgcttttgtagcacttgcacgtagtt
acccggatataagaacttcgatccgaaa (SEQ
ID NO: 9; inserted into 3' junction
polynucleotide)
aacgtgcaagcgcttttgtagcacttgcacgtagtt
acccggccagatataagaacttcgatccgaaa
(SEQ ID NO: 10; inserted into 3'
junction polynucleotide)
[00110] Example 2. Insertion of a CgRRS element in the 3'-junction of the
DP-4114
event.
[00111] Two plant gene expression vectors are prepared. Plant expression
cassettes for
expressing a bacteriophage lambda exonuclease, a bacteriophage lambda beta
SSAP protein,
and an E. coli SSB are constructed essentially as set forth in US Patent
Application
Publication 20200407754, which is incorporated herein by reference in its
entirety. A DNA
sequence encoding a tobacco c2 nuclear localization signal (NLS) is fused in-
frame to the
DNA sequences encoding the exonuclease, the bacteriophage lambda beta SSAP
protein, and
the E. coli SSB to provide a DNA sequence encoding the c2 NLS-Exo, c2 NLS
lambda beta
SSAP, and c2 NLS-SSB fusion proteins that are set forth in SEQ ID NO: 135, SEQ
ID NO:
134, and SEQ ID NO: 133 of US Patent Application Publication 20200407754,
respectively,
and incorporated herein by reference in its entirety. DNA sequences encoding
the c2 NLS-
Exo, c2 NLS lambda beta SSAP, and c2NLS-SSB fusion proteins are operably
linked to a
OsUBIL ZmUBIL OsACT promoter and a OsUbi 1, ZmUBIL OsACT polyadenylation site
respectively, to provide the exonuclease, SSAP, and SSB plant expression
cassettes.
[00112] A DNA donor template sequence (SEQ ID NO: 14) that targets the 3'-T-
DNA
junction polynucleotide of the DP-4114 event (SEQ ID NO:1; Figure 1) for HDR-
mediated
insertion of a 27 base pair OgRRS sequence (SEQ ID NO: 7) that is identical to
a Cas12a
recognition site at the 5'-junction polynucleotide of the DP-4114 T-DNA insert
is
constructed. The DNA donor sequence includes a replacement template with
desired insertion
region (27 base pairs long) flanked on both sides by homology arms about 500-
635 bp in
length. The homology arms match (i.e., are homologous to) gDNA (genomic DNA)
regions
flanking the target gDNA insertion site (SEQ ID NO: 15). The replacement
template region
- 53 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
comprising the donor DNA is flanked at each end by DNA sequences identical to
the DP-
4114 3'-T-DNA junction polynucleotide sequence recognized by a Cas12a RNA-
guided
nuclease and a gRNA (e.g., encoded by SEQ ID NO: 5).
[00113] A plant expression cassette that provides for expression of the
RNA-guided
sequence-specific Cas12a endonuclease is constructed. A plant expression
cassette that
provides for expression of a guide RNA (e.g., encoded by SEQ ID NO: 5)
complementary to
sequences adjacent to the insertion site is constructed. An Agrobacterium
superbinary
plasmid transformation vector containing a cassette that provides for the
expression of the
phosphinothricin N-acetyltransferasesynthase (PAT) protein is constructed.
Once the
cassettes, donor sequence and Agrobacterium superbinary plasmid transformation
vector are
constructed, they are combined to generate two maize transformation plasmids.
[00114] A maize transformation plasmid is constructed with the PAT
cassette, the
RNA-guided sequence-specific endonuclease cassette, the guide RNA cassette,
and the DP-
4114 3'-T DNA junction sequence DNA donor sequence into the Agrobacterium
superbinary
plasmid transformation vector (the control vector).
[00115] A maize transformation plasmid is constructed with the PAT
cassette, the
RNA-guided sequence-specific endonuclease cassette, the guide RNA cassette,
the SSB
cassette, the lambda beta SSAP cassette, the Exo cassette, and the DP-4114 3'-
T DNA
junction sequence donor DNA template sequence (SEQ ID NO: 14) into the
Agrobacterium
superbinary plasmid transformation vector (the lambda red vector).
[00116] All constructs are transformed into Agrobacterium strain LBA4404.
[00117] Maize transformations are performed based on published methods
(Ishida et.
al, Nature Protocols 2007; 2, 1614-1621). Briefly, immature embryos from
inbred line
GIBE0104, approximately 1.8-2.2 mm in size, are isolated from surface
sterilized ears 10-14
days after pollination. Embryos are placed in an Agrobacterium suspension made
with
infection medium at a concentration of OD 600=1Ø Acetosyringone (200 [tM) is
added to
the infection medium at the time of use. Embryos and Agrobacterium are placed
on a rocker
shaker at slow speed for 15 minutes. Embryos are then poured onto the surface
of a plate of
co-culture medium. Excess liquid media is removed by tilting the plate and
drawing off all
liquid with a pipette. Embryos are flipped as necessary to maintain a scutelum
up orientation.
Co-culture plates are placed in a box with a lid and cultured in the dark at
22 C for 3 days.
Embryos are then transferred to resting medium, maintaining the scutellum up
orientation.
- 54 -
CA 03188323 2022-12-23
WO 2022/026574 PCT/US2021/043496
Embryos remain on resting medium for 7 days at 27-28 C. Embryos that produced
callus are
transferred to Selection 1 medium with 7.5 mg/L phosphinothricin (PPT) and
cultured for an
additional 7 days. Callused embryos are placed on Selection 2 medium with 10
mg/L PPT
and cultured for 14 days at 27-28 C. Growing calli resistant to the selection
agent are
transferred to Pre-Regeneration media with 10 mg/L PPT to initiate shoot
development. Calli
remained on Pre-Regeneration media for 7 days. Calli beginning to initiate
shoots are
transferred to Regeneration medium with 7.5 mg/L PPT in Phytatrays and
cultured in light at
27-28 C. Shoots that reached the top of the Phytatray with intact roots are
isolated into Shoot
Elongation medium prior to transplant into soil and gradual acclimatization to
greenhouse
conditions.
[00118] When a sufficient amount of viable tissue is obtained, it can be
screened for
insertion at the DP-4114 junction sequence, using a PCR-based approach. The
PCR primer
on the 5'-end is 5'-tacgctgggccctggaaggctagga-3' (SEQ ID NO: 17). The PCR
primer on the
3'-end is 5'-gatggacgagacgaggcggtggaga-3' (SEQ ID NO: 18). The above primers
that flank
donor DNA homology arms are used to amplify the DP-4114 3'-junction
polynucleotide
sequence. The correct donor sequence insertion will produce a 1563 bp product.
A unique
DNA fragment comprising the CgRRS in the DP-4114 3' junction polynucleotide is
set forth
in SEQ ID NO: 19. Amplicons can be sequenced directly using an amplicon
sequencing
approach or ligated to a convenient plasmid vector for Sanger sequencing.
Those plants in
which the DP-4114 junction sequence now contains the intended Cas12a
recognition
sequence are selected and grown to maturity. The T-DNA encoding the Cas12a
reagents can
be segregated away from the modified junction sequence in a subsequent
generation. The
resultant INIR6 transgenic locus (SEQ ID NO: 20) comprising the CgRRS and
OgRRS (e.g.,
which each comprise SEQ ID NO: 7) can be excised using Cas12a and a suitable
gRNA
which hybridizes to DNA comprising SEQ ID NO: 7 at both the OgRRS and the
CgRRS.
[00119] The breadth and scope of the present disclosure should not be
limited by any
of the above-described embodiments.
- 55 -