Note: Descriptions are shown in the official language in which they were submitted.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
PRODUCTION OF OXYGENATED DITERPENOID COMPOUNDS
REFERENCE TO SEQUENCE LISTING
The present application contains a sequence listing in computer readable form,
which
is incorporated by reference.
FIELD OF INVENTION
The invention relates to the production of oxygenated diterpenoid compounds in
recom-
binant cells, such as yeast cells. The oxygenated diterpenoid compounds are
useful as
intermediate or final compounds in the synthesis of useful bioactive compounds
for use
in e.g. pharmaceutical treatment of diseases such as cancer. The invention
further re-
lates to genes, enzymes and cells, such as yeast cells, particularly suited
for production
of such compounds.
BACKGROUND FOR THE INVENTION
Terpenes is a diverse group of compounds generated from a basic 5-carbon
structure,
isoprene (2-methyl-1,3-butadiene). Diterpenes are compounds having a 20-carbon
structure, generated by the action of an enzyme, diterpene synthase, that
converts the
compound geranyl-geranyl-diphosphate (GGPP) into a diterpene structure, which
can
by further modified forming a vast range of diterpene or diterpenoid
compounds.
Diterpenes, diterpenoids, derivatives thereof are widely used, e.g. as
pharmaceuticals,
cosmetics, nutraceuticals, flavors, fragrances and pesticides. Methods for
increasing the
production of these compounds in natural or engineered cells are abundant in
the art.
The Chinese medicinal plant, Triptetygium wilfordii, is known to produce
several sesquit-
erpenoids, diterpenoids and triterpenoids with potential pharmacological
properties, in-
cluding the diterpenoid compounds triptonide and triptolide. Triptolide, an
oxygenated
diterpenoid compound, and derivatives thereof, has been identified as
potential valuable
pharmacological compounds and is under investigation as immunosuppressant and
for
treatment of cancer. Triptolide may further be used in treatment against COVID-
19. Trip-
tonide may be useful as male contraceptive agent.
Using engineered microorganisms for producing valuable molecules from
renewable
feedstock is a desirable alternative from conventional means of production.
However,
achieving economically viable yield, titers and productivity is a major
roadblock towards
industrialisation.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
2
N L Hansen et al (2017) in The Plant Journal 2017, 89, 429-441, described a
diterpene
synthase capable of converting GGPP into the dipterpene, miltiradiene, which
is a pre-
curser for triptolide. The findings were confirmed by P Su et al (2018) in The
plant Jour-
nal 2018, 50-65: and by J Guo et al. PNAS 2013, 110, 12108-12113.
The complete pathway for converting miltiradiene into other diterpenoid
compounds,
such as triptolide, has not yet been elucidated.
Cytochrome P450 enzymes (CYPs) are involved in the biosynthesis of terpenoids,
and
for many cytochrome P450 enzymes nothing is known with respect to the
substrate
they are acting on, which compounds they are generating, or their role in the
biosynthe-
sis of specific compounds.
US 20190270971A1 discloses methods for increasing productivity of microbial
host
cells that functionally express p450 enzymes. The document describes how P450
genes can be modified in order to improve performance in microorganisms, such
as
yeast, and it mentions that co-expression with cytochrome P450 reductase can
be ben-
eficial to improve the yield. It is mentioned that triptolide may be the
subject of P450
chemistry, but the document does not provide any link between triptolide and
any spe-
cific P450 enzymes or cytochromes.
ON 108395997A describes yeast with increased GGPP production. The yeast is
trans-
formed with different diterpene synthases and P450 enzymes to synthesize
diterpenoid
compounds. The scientist team behind this patent is also behind more patents
and pa-
tent applications disclosing the synthesis of different di- and triterpenoid
compounds
using suitable terpene synthases and P450 enzymes e.g. ON 108866029
(friedelin),
CN107058419 (Kauren-type) and WO 2020029564 (Fridelin and amyrins).
ON 110747178A describes the P450 gene TwCYP728B70 as encoding a Cytochrome
P450 enzyme having a role in triptolide synthesis.
SHORT DESCRIPTION OF THE INVENTION
The inventors have solved the problem of providing an improved method of
producing
oxygenated diterpenoid compounds, such as triptophenolide, triptonide and
triptolide.
In a first aspect the invention relates to a method for producing an
oxygenated diterpe-
noid compound is disclosed, the method comprising the steps of:
a. providing a host cell capable of producing miltiradiene and/or dehydro-
abietadiene;
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
3
b. transforming the host cell with a first gene encoding an enzyme having
cytochrome P450 activity;
c. growing the transformed cell under conditions leading to expression of
the
transformed gene; whereby the oxygenated diterpenoid compound is
formed;
wherein:
the first gene encoding an enzyme having cytochrome P450 activity encodes a
pol-
ypeptide comprising an amino acid sequence having at least 80 % sequence
identity,
preferably at least 85% sequence identity, preferably at least 90% sequence
identity,
preferably at least 95% sequence identity, more preferred at least 98%
sequence identity
to SEQ ID NO: 1 (TwCYP82D274v1), SEQ ID NO: 2 (TwCYP82D274v2), SEQ ID NO:
74 (TwCYP82D274v3) or SEQ ID NO: 75 (TwCYP82D274v4), or the mature polypeptide
thereof.
In a second aspect, the invention relates to methods for producing oxygenated
diterpe-
noid compound, and comprises transforming the host cell with the first gene
encoding
an enzyme having cytochrome P450 activity and further with a second gene
encoding a
second enzyme having cytochrome P450 activity and with a third gene encoding a
third
enzyme having cytochrome P450 activity
wherein:
the second gene encoding an enzyme having cytochrome P450 activity encodes a
polypeptide comprising an amino acid sequence having at least 80 % sequence
identity,
preferably at least 85% sequence identity, preferably at least 90% sequence
identity,
preferably at least 95% sequence identity, more preferred at least 98%
sequence identity
to SEQ ID NO: 3 (TwCYP71BE85) or the mature polypeptide thereof; and
the third gene encoding an enzyme having cytochrome P450 activity encodes a
polypeptide comprising an amino acid sequence having at least 80 % sequence
identity,
preferably at least 85% sequence identity, preferably at least 90% sequence
identity,
preferably at least 95% sequence identity, more preferred at least 98%
sequence identity
to SEQ ID NO: 4 (TwCYP71BE85) or the mature polypeptide thereof.
In a third aspect, the invention relates to methods for producing oxygenated
diterpenoid
compound, wherein said method comprises that the host cell is transformed with
the first,
second and third genes encoding enzymes having cytochrome P450 activities, and
fur-
ther with a fourth gene encoding a fourth enzyme having cytochrome P450
activity;
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
4
wherein:
the fourth gene encoding a fourth enzyme having cytochrome P450 activity en-
codes a polypeptide comprising an amino acid sequence having at least 80 %
sequence
identity, preferably at least 85% sequence identity, preferably at least 90%
sequence
identity, preferably at least 95% sequence identity, more preferred at least
98% se-
quence identity to SEQ ID NO: 5 (TwCYP82D213v1) or to SEQ ID NO: 76
(TwCYP82D213v2), or the mature polypeptide thereof.
According to the invention the useful oxygenated diterpenoid compounds,
triptophe-
nolide, triptonide or triptolide, may be provided.
The invention further relates to polypeptides, polynucleotides, plasmids and
expression
constructs as well as recombinant host cells useful in the methods of the
invention.
SHORT DESCRIPTION OF THE DRAWINGS
Figure 1 shows LCMS profiles of extracts from N. benthamiana leafs expressing
mil-
tirdiene biosynthesis genes and selected T. wilfordii CYPs.
TwCYPs was co-expressed with CfDXS, CfGGPPS, CfTPS1 and CfTPS3. "3xSTD" rep-
resents a LCMS run of a sample of three mixed authentic standards: Triptolide,
trip-
tonide, and triptophenolide. Solid lines represent ion chromatograms at range
m/z 280-
380. Dashed lines (- ------ -) represent extracted ion chromatograms at m/z
313.1800 0.015 which corresponds to the parental ion of triptophenolide [M+H].
Dashed
lines (- - -) represents extracted ion chromatograms at m/z 359.1490 0.0 15
which cor-
responds to the parental ion of triptonide [M+H]. LC protocol 1 used. For more
details
see example 1.
Figure 2 shows LCMS profiles of extracts of genetically engineered
Saccharomyces
cerevisiae (S. cerevisiae) strain.
In the background strain (-), genes encoding the diterpene biosynthetic
enzymes SPGG-
PPS7, CftTPS1, CftTPS3 and TwCPR1 were integrated into genome of wild type S.
cerevisiae. In the TwCYP82D274v1 strain, the diterpene biosynthetic enzymes
were ex-
pressed with TwCYP82D274v1, resulting in the formation of compound (3)
identified as
14-0H-dehydroabietadiene marked in grey. LC method1 was used for the analysis.
For
more details see example 3.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
Figure 3 shows the 1H NMR spectrum of 14-0H-dehydroabietadiene in CDCI3 at
599.85
MHz.. For more details see example 4.
Figure 4 shows the 130 NMR spectrum of 14-0H-dehydroabietadiene in CDCI3 at
150.83
MHz. For more details see example 4.
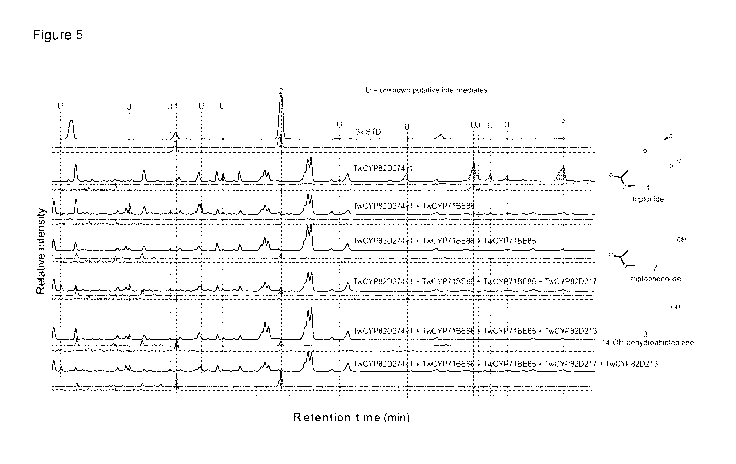
5 Figure 5 shows LCMS profiles of extracts of yeast having denoted gene
combinations
genome integrated.
All yeast strains have genome integrated spGGPPs7, CftTPS1, CftTPS3 and
TwCPR1.
"0.5ppm 3xSTD" represents a LCMS run of a sample of three mixed authentic
standards:
Triptolide, triptonide and triptophenolide. Non-dashed lines represent ion
chromato-
grams at range m/z 280-380. Dashed lines represents extracted ion
chromatograms at
m/z 359.1490 0.015 which corresponds to the parental ion of triptonide
([M+H]+). LC
protocol 2 used. For more details see example 5.
Figure 6 shows co-expression of TwCYPs and different variants of B5 proteins
isolated
from Triptetygium wilfordii
Levels of triptophenolide, triptonide and 14-0H-dehydroabietadiene quantified
from cul-
tures of engineered S. cerevisiae strains. Each column represents an
engineered yeast
strain and their output of selected compounds. The genes integrated into each
of the
individual strains are denoted in the lower panel. Quantification was based on
the areas
of the peaks representing each of the compounds of interest. Individual scales
applies
for each of the compounds. For more details see example 5.
Figure 7 shows relative quantity (bars) of key intermediates in the proposed
biosyn-
thetic pathway of triptonide, when established in vivo, via heterologous gene
expres-
sion in N. benthamiana (panel A and D) and S. cerevisiae (panel B and E,
strains listed
in table 3). Gene expression is indicated by black squares to the left, while
relative
quantity is indicated by bars (average of 3-4 biological replicates; black
diamond
squares) with white- and grey fill color distinguishing expression and no
expression of
Twb5#1, respectively. Error bars represent standard deviation. "DiTPSs"
reflects Cft-
TPS1 and CftTPS3. In quantification of peak areas, the signature mass
tolerance was
0.1m/z for GCMS (miltiradiene and 14-0H-dehydroabietadiene) and 0.005m/z for
LCMS (all other compounds). Panel C: A hypothetized biosynthetic pathway from
milti-
radiene to triptonide in vivo in N. benthamiana and S. cerevisiae that include
a Wagner-
Meerwein rearrangement reaction to account for a methyl shift of 0-19 or 0-18
to 0-3
in the abietane carbon backbone.
Figure 8 shows accumulation over 7 days of triptophenolide and triptonide
produced
with yeast strain NVJ8.15 when grown in bioreactor. Level of triptonide (solid
black line)
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
6
and triptophenolide (dotted black line) shows absolute quantity (ppm, w/v) in
samples
of the culture taken each day. Biomass was quantified by absorbance at 600nm
(grey
dotted line).
Figure 9 shows yeast strains expressing the genes needed for triptonide
biosynthesis,
but with genes variants substituting TwCYP82D274v1 or TwCYP82D213v1, pertain
the
ability to produce triptophenolide (panel A) and triptonide (panel B) and
results in simi-
lar LCMS profiles (panel C). Genes present in the engineered strains are
represented
by the black squares. Panel A and B: Bars represent the average relative
quantity (2-3
biological replicates, crosses) with error bars showing std. error. From left
to right, bars
represent yeast strains: NVJ10-1, NVJ10-3, NVJ10-6, NVJ10-8 (see table 3).
Panel C:
EICs (m/z 280-360) of LCMS analyzed yeast cultures. From top and down pairs of
chromatograms represent yeast strains NVJ 10-1, NVJ 10-3, NVJ 10-6 and NVJ 10-
8.
SHORT DESCRIPTION OF THE SEQUENCES
SEQ ID NO: 1 shows the amino acid sequence of a cytochrome P450 enzyme derived
from T. wilfordii. The enzyme is also known as TwCYP82D274v1.
SEQ ID NO: 2 shows the amino acid sequence of a cytochrome P450 enzyme derived
from T. wilfordii. The enzyme is also known as TwCYP82D274v2. SEQ ID NO: 2
differs
from SEQ ID NO: 1 in only three positions, and it is therefore assumed that
SEQ ID NO:
1 and SEQ ID NO: 2 represent different alleles of the same gene.
SEQ ID NO: 3 shows the amino acid sequence of a cytochrome P450 enzyme derived
from T. wilfordii. The enzyme is also known as TwCYP71BE85.
SEQ ID NO: 4 shows the amino acid sequence of a cytochrome P450 enzyme derived
from T. wilfordii. The enzyme is also known as TwCYP71BE86.
SEQ ID NO: 5 shows the amino acid sequence of a cytochrome P450 enzyme derived
from T. wilfordii. The enzyme is also known as TwCYP82D213v1.
SEQ ID NO: 6 shows the amino acid sequence of a cytochrome P450 enzyme derived
from T. wilfordii. The enzyme is also known as TwCYP82D217.
SEQ ID NO: 7 shows the amino acid sequence of a cytochrome P450 enzyme derived
from T. wilfordii. The enzyme is also known as TwCYP82D275.
SEQ ID NO: 8 shows the amino acid sequence of a cytochrome B5 enzyme derived
from
T. wilfordii. This enzyme is also known as TwB5#1.
SEQ ID NO: 9 shows the amino acid sequence of a cytochrome P450 reductase
enzyme
derived from T. wilfordii. This enzyme is also known as TwCPR1.
SEQ ID NO: 10-66 show PCR primers as further described in Example 2.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
7
SEQ ID NO: 67 shows the amino acid sequence of a diterpene synthase TPS1
derived
from Plectranthus barbatus. The enzyme is also known as CfTPS1.
SEQ ID NO: 68 shows the amino acid sequence of a diterpene synthase TPS3
derived
from Plectranthus barbatus. The enzyme is also known as CfTPS3.
SEQ ID NO: 69 shows the amino acid sequence of a terpene synthase TPS9,
derived
from T. wilfordii. The enzyme is also known as TwTPS9.
SEQ ID NO: 70 shows the amino acid sequence of a terpene synthase derived from
T.
wilfordii. The enzyme is also known as TwTPS27.
SEQ ID NO: 71 shows the amino acid sequence of a copalyl diphosphate synthase
CPS1
derived from Salvia miltiorrhiza. The enzyme is also known as SmCPS.
SEQ ID NO: 72 shows the amino acid sequence of a miltiradiene synthase KSL1
derived
from Salvia miltiorrhiza. The enzyme is also known as SmKSL.
SEQ ID NO: 73 shows the amino acid sequence of a geranyl geranyl diphosphate
syn-
thase derived from Synechococcus sp. The enzyme is also known as SpGGPPs7v1.
SEQ ID NO: 74 shows the amino acid sequence of a cytochrome P450 enzyme
derived
from T. wilfordii. The enzyme is also known as TwCYP82D274v3.
SEQ ID NO: 75 shows the amino acid sequence of a cytochrome P450 enzyme
derived
from T. wilfordii. The enzyme is also known as TwCYP82D274v4.
SEQ ID NO: 76 shows the amino acid sequence of a cytochrome P450 enzyme
derived
from T. wilfordii. The enzyme is also known as TwCYP82D213v2.
SEQ ID NO: 77 shows the truncated amino acid sequence of a diterpene synthase
TPS1
derived from Plectranthus barbatus. The amino acid sequence was truncated to
remove
a transit peptide. The enzyme is also known as CftTPS1.
SEQ ID NO: 78 shows the truncated amino acid sequence of a diterpene synthase
TPS3
derived from Plectranthus barbatus. The amino acid sequence was truncated to
remove
a transit peptide. The enzyme is also known as CftTPS3.
SEQ ID NO: 79 shows the amino acid sequence of a DXS enzyme derived from Plec-
tranthus barbatus. The enzyme is also known as CfDXS.
SEQ ID NO: 80 shows the amino acid sequence of a truncated HMGR enzyme derived
from S. cerevisiae. The enzyme is also known as SctHMGR.
SEQ ID NO: 81 shows the amino acid sequence of a geranyl geranyl diphosphate
syn-
thase derived from Synechococcus sp. The enzyme is also known as SpGGPPs7v2.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
8
DETAILED DESCRIPTION OF THE INVENTION
According to a first aspect of the invention, a method for producing an
oxygenated
diterpenoid compound is disclosed, the method comprising the steps of:
a. providing a host cell capable of producing miltiradiene and/or dehydro-
abietadiene;
b. transforming the host cell with a first gene encoding an enzyme having
cytochrome P450 activity;
c. growing the transformed cell under conditions leading to expression of
the
transformed gene; whereby the oxygenated diterpenoid compound is
formed;
wherein:
the first gene encoding an enzyme having cytochrome P450 activity encodes a
pol-
ypeptide comprising or consisting of SEQ ID NO:1 or an amino acid sequence
having at
least 80 % sequence identity, preferably at least 85% sequence identity,
preferably at
least 90% sequence identity, preferably at least 95% sequence identity more
preferred
at least 98% sequence identity to SEQ ID NO: 1 (TwCYP82D274V1) or the mature
pol-
ypeptide thereof.
The polypeptide having SEQ ID NO: 1 is a preferred example of a first gene
having
cytochrome P450 activity, the polypeptides having SEQ ID NO: 2, SEQ ID NO: 74
and
SEQ ID NO: 75 are other examples of such a polypeptide.
Thus, the enzyme encoded by the first gene has the ability to convert
miltiradiene and/or
dehydroabietadiene into 14-0H-dehydroabietadiene by inserting an OH group in
position
14 of the diterpene skeleton of miltiradiene.
In some embodiments, the synthesis of 14-0H-dehydroabietadiene takes place via
the
compound 14-0H-miltiradiene that subsequently is converted into 14-0H-dehydro-
abietadiene. However, the invention is not limited to any particular mechanism
for con-
verting miltiradiene into 14-0H-dehydroabietadiene.
Using the method according to the first aspect of the invention leads to the
formation of
the oxygenated diterpenoid compound, 14-0H-dehydroabietadiene,
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
9
OH
14-0H-dehydroabietadiene
that is a useful intermediate in the synthesis of oxygenated diterpenoid
compounds of
pharmaceutical use, including well known compounds such as triptophenolide,
triptonide
and triptolide.
In a second aspect of the invention, the method step b. comprises transforming
the host
cell with the first gene encoding an enzyme having cytochrome P450 activity
and further
with a second gene encoding a second enzyme having cytochrome P450 activity
and a
third gene encoding a third enzyme having cytochrome P450 activity
wherein:
the second gene encoding an enzyme having cytochrome P450 activity encodes a
polypeptide comprising or consisting of SEQ ID NO:4 or an amino acid sequence
having
at least 80 % sequence identity, preferably at least 85% sequence identity,
preferably at
least 90% sequence identity, preferably at least 95% sequence identity, more
preferred
at least 98% sequence identity to SEQ ID NO: 4 (TwCYP71BE86) or the mature
poly-
peptide thereof; and
the third gene encoding an enzyme having cytochrome P450 activity encodes a
polypeptide comprising or consisting of SEQ ID NO: 3 or an amino acid sequence
having
at least 80 % sequence identity, preferably at least 85% sequence identity,
preferably at
least 90% sequence identity, preferably at least 95% sequence identity, more
preferred
at least 98% sequence identity to SEQ ID NO: 3 (TwCYP71BE85) or the mature
poly-
peptide thereof.
In this second aspect, the host cell preferably further produces the
oxygenated diterpe-
noid compound, triptophenolide, (3bR,9b5)-6-hydroxy-9b-methyl-7-propan-2-y1-
3,3b,4,5,10,11-hexahydronaphtho[2,1-e]isobenzofuran-1-one.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
011
0 --
H
0 --
triptophenolide
Triptophenolide is a valuable compound that has been identified as an
antiandrogen. In
5 addition, it may be useful as a starting point for further modifications
leading to further
bioactive compounds.
In a preferred embodiment of the second aspect of the invention, the host cell
is further
transformed with a fifth gene encoding a polypeptide having cytochrome B5
activity and
10 comprising or consisting of SEQ ID NO:8 or an amino acid sequence having
at least 80
% sequence identity, preferably at least 85% sequence identity, preferably at
least 90%
sequence identity, preferably at least 95% sequence identity, more preferred
at least
98% sequence identity to SEQ ID NO: 8 (TwB5#1) or the mature polypeptide
thereof. It
has surprisingly been found that expressing the polypeptide having cytochrome
B5 ac-
tivity in the same cell that expresses the first, second and third genes
encoding enzymes
having cytochrome P450 activity, leads to a significantly higher production of
the oxy-
genated diterpenoid compound. The production is increased at least 50%
compared with
the production of a similar cell without the polypeptide having cytochrome B5
activity,
preferably increased at least 100%, preferably at least 200% or even more.
In a third aspect of the invention, the host cell is transformed with the
first, second and
third genes encoding enzymes having cytochrome P450 activities, and further
with a
fourth gene encoding a fourth enzyme having cytochrome P450 activity;
wherein:
the fourth gene encoding a fourth enzyme having cytochrome P450 activity en-
codes a polypeptide comprising or consisting of SEQ ID NO:5 or an amino acid
sequence
having at least 80 % sequence identity, preferably at least 85% sequence
identity, pref-
erably at least 90% sequence identity, preferably at least 95% sequence
identity, more
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
11
preferred at least 98% sequence identity to SEQ ID NO: 5 (TwCYP82D213v1) or
the
mature polypeptide thereof. The polypeptide having SEQ ID NO: 5 is a preferred
exam-
ple of a fourth gene having cytochrome P450 activity; the polypeptide having
SEQ ID
NO: 76 is another example of such a polypeptide.
In the third aspect of the invention, the transformed eukaryotic cell
preferably produces
the oxygenated diterpenoid compound, triptonide.
0
0
0
111114111. 0
0
lo 0
triptonide
The compound triptonide has been reported to have a strong inhibition activity
in cancers
(Fulu Dong et al 2019, The Prostate, Volume 19, issue 11, pages 1284-1293).
The com-
pound is also useful as male contraceptive agent. Further, the compound is
useful as a
starting point for further modifications leading to further bioactive
compounds.
In a preferred embodiment of the third aspect of the invention, the host cell
is further
transformed with a fifth gene encoding a polypeptide having cytochrome B5
activity and
comprising or consisting of SEQ ID NO:8 or an amino acid sequence having at
least 80
% sequence identity, preferably at least 85% sequence identity, preferably at
least 90%
sequence identity, preferably at least 95% sequence identity, more preferred
at least
98% sequence identity to SEQ ID NO: 8 (TwB5#1) or the mature polypeptide
thereof. It
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
12
has surprisingly been found that expressing the polypeptide having cytochrome
B5 ac-
tivity in the same cell that expresses the first, second, third and fourth
genes encoding
enzymes having cytochrome P450 activity, leads to a significantly higher
production of
the oxygenated diterpenoid compound. The production is increased at least 50%
com-
pared with the production of a similar cell without the polypeptide having
cytochrome B5
activity, preferably increased at least 100%, preferably at least 200% or even
more.
In a further preferred embodiment of the third aspect of the invention, the
host cell is
further transformed with a sixth gene encoding a fifth enzyme having
cytochrome P450
activity and/or a seventh gene encoding a sixth enzyme having cytochrome P450
activity
wherein:
the sixth gene encoding a fifth enzyme having cytochrome P450 activity encodes
a
polypeptide comprising or consisting of SEQ ID NO:6 or an amino acid sequence
having
at least 80 % sequence identity, preferably at least 85% sequence identity,
preferably at
least 90% sequence identity, preferably at least 95% sequence identity, more
preferred
at least 98% sequence identity to SEQ ID NO: 6 (TwCYP82D217) or the mature
poly-
peptide thereof; and
the seventh gene encoding a sixth enzyme having cytochrome P450 activity en-
codes a polypeptide comprising an amino acid sequence having at least 80 %
sequence
identity, preferably at least 85% sequence identity, preferably at least 90%
sequence
identity, preferably at least 95% sequence identity, more preferred at least
98% se-
quence identity to SEQ ID NO: 7 (TwCYP82D275) or the mature polypeptide
thereof.
It has surprisingly been found that expressing the sixth and/or seventh genes
encoding
enzymes having cytochrome P450 activity, leads to a higher production of the
oxygen-
ated diterpenoid compound. Preferably, the production is increased at least
10% com-
pared with the production of a similar cell without sixth and/or seventh genes
encoding
enzymes having cytochrome P450 activity, preferably increased at least 20%,
even more
preferred at least 50% or even more.
.. The first, second, third, fourth, fifth, sixth and seventh gene may be
comprised in one or
more nucleic acid molecules, such as one or more heterologous nucleic acids.
The het-
erologous nucleic acid encoding the first enzyme having cytochrome P450
activity may
herein be referred to as the "first heterologous nucleic acid". The
heterologous nucleic
acid encoding the second enzyme having cytochrome P450 activity may herein be
re-
ferred to as the "second heterologous nucleic acid". The heterologous nucleic
acid en-
coding the third enzyme having cytochrome P450 activity may herein be referred
to as
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
13
the "third heterologous nucleic acid". The heterologous nucleic acid encoding
the fourth
enzyme having cytochrome P450 activity may herein be referred to as the
"fourth het-
erologous nucleic acid". The heterologous nucleic acid encoding the fifth
enzyme hav-
ing cytochrome P450 activity may herein be referred to as the "fifth
heterologous nu-
cleic acid". The heterologous nucleic acid encoding the sixth enzyme having
cyto-
chrome P450 activity may herein be referred to as the "sixth heterologous
nucleic acid".
The heterologous nucleic acid encoding the enzyme having cytochrome B5
activity
may herein be referred to as the "seventh heterologous nucleic acid". This
does not im-
ply that the recombinant host cell must comprises seven heterologous nucleic
acids in
total; in some embodiments, the cell comprises only one or more of the first,
second,
third, fourth, fifth, sixth and seventh heterologous nucleic acids.
The oxygenated diterpenoid compounds produced according to the methods of the
in-
vention may be further modified by biological or chemical synthesis. In
connection with
this, biological synthesis is understood as a method where the host cell
comprising the
genes of the invention is further provided with one of more additional genes
encoding
further enzymes having the capability of modifying the oxygenated diterpenoid
com-
pounds produced according to the methods of the invention.
Chemical modification of the oxygenated diterpenoid compounds produced
according to
the methods of the invention may be performed directly on the culture broth
before re-
covery of the oxygenated diterpenoid compounds or it may be performed on the
recov-
ered oxygenated diterpenoid compounds.
Reduction of triptonide to triptolide can be achieved by organic synthesis. An
example of
such synthesis is the reduction by a nucleophilic attack by a hydride on C-14
ketone. For
this reaction Sodium borohydride is a suitable agent for catalyzing this
reaction at neutral
pH in the appropriate solvent e.g. water or Met0H.
In one preferred embodiment, triptonide produced according to the methods of
the in-
vention is converted into the compound triptolide, that is reported to be an
immunosup-
pressant and is under investigation for use in cancer therapy.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
14
, f
I
The host cell
The host cell capable of producing miltiradiene and/or dehydroabietadiene may
in prin-
ciple be any such cell. The cell may be a cell that naturally produces
miltiradiene and/or
dehydroabietadiene or it may be a cell that has been engineered to produce one
or both
of these compounds.
It is believed that miltiradiene, at least under some circumstances, may be
spontaneously
converted into dehydroabietadiene, and the invention may therefore be
performed using
a cell producing miltiradiene that is spontaneously converted into
dehydroabietadiene or
it may be performed in a cell comprising an enzyme that facilitates the
conversion of
miltiradiene to dehydroabietadiene (see a J. Zi, et al., Organic &
Biomolecular Chemistry
2013, 11, 7650-7652).
The synthesis of miltiradiene in general begins with the formation of GGPP.
GGPP may
be synthesized by condensation of one dimethylallyl pyroophosphate (DMAP)
molecule
and three isopentenyl pyrophosphate (IPP) molecules and is typically catalyzed
by a
geranylgeranyl diphosphate synthase e.g. the SpGGPPs7 enzyme derived from Syn-
echococcus sp.; and having the amino acid sequence shown in SEQ ID NO: 73 or
SEQ
ID NO: 81.
GGPP is converted into miltiradiene by action of a diterpene synthase or by
the combined
action of two or more diterpene synthases, e.g. a combination of two diterpene
syn-
thases, CfTPS1 and CfTPS3, or CftTPS1 as set forth in SEQ ID NO: 77 and
CftTPS3 as
set forth in SEQ ID NO: 78, derived from Plectranthus barbatus and having the
amino
acid sequences of SEQ ID NO: 67 and 68; a combination of two diterpene
synthases,
TwTPS9 and TwTPS27, derived from T. wilfordii and having the amino acid
sequences
of SEQ ID NO: 69 and 70; or a combination of a copalyl diphosphate synthase,
SmCPS
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
derived from Salvia miltiorrhiza and having the amino acid sequence of SEQ ID
NO: 71,
and a miltiradiene synthase, SmKSL derived from Salvia miltiorrhiza and having
the
amino acid sequence of SEQ ID NO: 72.
I r n
,,, - :---,:-- ------::-) _ pp_,.-_:_,
- , ,
d_.:L,e-.11::, 1.::, 1 .__::yrol.:_i.) :.ph.: :e ,:D:',
+
0 0
3x -.'= '-'(:::¨ (:::4 ¨ r!ii
1 1
ji, ._', H
L...ter..-: 1 p:-I:cpi- c _ 1P?)
1
e,,:' SpC4CiPPS 7
_
T 1, IONA-- (1 er PP
i \-,,
/ N hid.: -1,::3-nlie
1 d1::: 1::17.te: '
c oin.bi.r.: ca =.:f ,:-.1:.TPS taz- ..:es e ..z .:
{: t7F'c:1-C.1-TPS 3
Tw-ITS --T. 7. T PS 2 -
Pn cq: 7.1-r. p I I::: li Aied
N L. Enn.-,r. ,.- 71: 77:: .P.::':- .:::1' ': ".. 201.
,S('. -.--.::-4-.1...
7::
5 J. G-..:. e.. aI. .2:.%-.:... 2013. '..::!..... 1211-_=__2.
One preferred way to provide a host cell producing miltiradiene and/or
dehydroabietadi-
ene is selecting a host cell producing GGPP and transforming it with a
diterpene syn-
10 thase catalyzing the transformation of GGPP into miltiradiene.
Alternatively, a host cell
that have been genetically engineered to produce GGPP may be used as a
starting point.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
16
Techniques for transforming a host cell with a diterpene synthase catalyzing
the trans-
formation of GGPP into miltiradiene is known in the prior art, e.g. in N L
Hansen et al
(2017) in The Plant Journal 2017, 89, 429-441 (Incorporated herein by
reference), P Su
et al (2018) in The plant Journal 2018, 50-65 and in : J. Guo, et al.,
Proceedings of the
National Academy of Sciences 2013, 110, 12108-12113, and the procedures and
meth-
ods disclosed in these publications are also useful for providing a host cell
for use in the
present invention.
The host cell may be a prokaryotic cell, such as a eubacterial or
archaebacterial cell; or
a eukaryotic cell, such as a plant cell, an animal cell, an insect cell, a
fungal cell or a
yeast cell.
Practically all eukaryotic cells produce GGPP for their biosynthesis, but in
some embod-
iments a eukaryotic cell produces an increased amount of GGPP, which may
increase
the production of miltiradiene, compared with a similar eukaryotic that does
not produce
increased amounts of GGPP. Methods for increasing the GGPP production in a
eukary-
otic cell has also been described in the prior art.
The host cell may be a unicellular organism, or it may be comprised within a
multicellular
organism, e.g. a plant. Examples of suitable plants or plant cells for use as
host cells
according to the invention includes corn (Zea mays), canola (Brassica napus,
Brassica
rapa ssp.), alfalfa (Medicago sativa), rice (Oryza sativa), rye (Secale
cerale), sorghum
(Sorghum bicolor, Sorghum vulgare), sunflower (Helianthus annuas), wheat
(Tritium
aestivum and other species), triticale, rye (Secale) soybean (Glycine max),
tobacco (Ni-
cotiana tabacum), potato (Solanum tuberosum), peanuts (Arachis hypogaea),
cotton
(Gossypium hirsutum), sweet potato (Impomoea batatus), cassava (Manihot
esculenta),
coffee (Coffea spp.), coconut (Cocos nucifera), pineapple (Anana comosus),
citrus (Cit-
rus spp.) cocoa (Theobroma cacao), tea (Camellia senensis), banana (Musa
spp.), ava-
cado (Persea americana), fig (Ficus casica), guava (Psidium guajava), mango
(Mangifer
indica), olive (Olea europaea), papaya (Carica papaya), cashew (Anacardium
occi-
dentale), macadamia (Macadamia intergrifolia), almond (Primus amygdalus),
apple (Ma-
lus spp.), pear (Pyrus spp.), plum and cherry tree (Prunus spp.), ribes
(currant etc.), Vitis,
.. Jerusalem artichoke (Helianthemum spp.), non-cereal grasses (Grass family),
sugar and
fodder beets (Beta vulgaris), chicory, oats, barley, vegetables, or
ornamentals, crop
plants (for example, cereals and pulses, maize, wheat, potatoes, tapioca,
rice, sorghum,
millet, cassava, barley, pea, sugar beets, sugar cane, soybean, oilseed rape,
sunflower
and other root, tuber or seed crops. Other important plants may be fruit
trees, crop trees,
forest trees or plants grown for their use as spices or pharmaceutical
products (Mentha
spp., clove, Artemesia spp., Thymus spp., Lavendula spp., Allium spp.,
Hypericum,
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
17
Catharanthus spp., Vinca spp., Papaver spp., Digitalis spp., Rawoffia spp.,
Vanilla spp.,
Petrusilium spp., Eucalyptus, tea tree, Picea spp., Pinus spp., Abies spp.,
Juniperus spp.
Horticultural plants which can be used with the present invention may include
lettuce,
endive, and vegetable brassicas including cabbage, broccoli, and cauliflower,
carrots,
and carnations and geraniums.
The plant can also be tobacco, cucurbits, carrot, strawberry, sunflower,
tomato, pepper,
or chrysanthemum.
Further examples of plants include grain plants for example oil-seed plants or
legumi-
nous plants. Seeds of interest include grain seeds, such as corn, wheat,
barley, sorghum,
rye, etc. Oil-seed plants include cotton soybean, safflower, sunflower,
Brassica, maize,
alfalfa, palm, coconut, etc. Leguminous plants include beans and peas. Beans
include
guar, locust bean, fenugreek, soybean, garden beans, cowpea, mung bean, lima
bean,
fava bean, lentils, chickpea.
Particular preferred plant species include Physcomitrella sp., such as P.
patens; Ara-
bidopsis sp., such as A. thaliana; Nicotiana sp., such as N. benthamiana;
Chlamydomo-
nas sp., such as C. reinhardtii; and Nannochloropsis sp., such as N. oceanica.
Examples of suitable eukaryotic cell for use according to the invention
include fungal
cells such as Agaricus, Aspergillus, Candida, Eremothecium,
Fusarium/Gibberella, Kluy-
veromyces, Laetiporus, Lentinus, Phaffia, Phanerochaete, Pichia,
Physcomitrella,
Rhodoturula, Saccharomyces, Schizosaccharomyces, Sphaceloma, Xanthophyllomy-
ces or Yarrowia. Exemplary species from such genera include Lentinus tigrinus,
Laeti-
porus sulphureus, Phanerochaete chtysosporium, Pichia pastoris, Cyberlindnera
jadinii
Physcomitrella patens, Rhodoturula glutinis, Rhodoturula mucilaginosa, Phaffia
rhodozyma, Xanthophyllomyces dendrorhous, Fusarium fujikuroi/Gibberella
fujikuroi,
Candida utilis, Candida glabrata, Candida albicans, and Yarrowia lipolytica.
In some embodiments, a host cell can be an Ascomycete such as Gibberella
fujikuroi,
Kluyveromyces lactis, Schizosaccharomyces pombe, Aspergillus niger, Yarrowia
lipo-
lytica, Ashbya gossypii, or S. cerevisiae.
In some embodiments, the host cell can be an algae cell such as Blakeslea
trispora,
Dunaliella salina, Haematococcus pluvialis, Chlorella sp., Undaria
pinnatifida, Sargas-
sum, Laminaria japonica, Scenedesmus almeriensis.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
18
In some embodiments, a host cell can be a prokaryote such as Bacillus cells,
for example
Bacillus subtilis; Escherichia cells, for example, Escherichia coli cells;
Lactobacillus cells;
Lactococcus cells; Streptomyces cells, Streptococcus cells, Comebacterium
cells;
Acetobacter cells; Acinetobacter cells; or Pseudomonas cells.
In some embodiments, the host cell can be a cyanobacterial cell such as
Synechocystis
sp. or Synechococcus sp.
In one embodiment, a host cell that is suitable for growth in a fermenter is
selected.
Growing the recombinant host cell according to the invention is a convenient
way of
growing the host cell for production of the oxygenated diterpenoid compounds
of the
invention.
In another embodiment, the host cell is a phototropic cell and the cell is
cultivated in a
green house or photobioreactor.
The genes and enzymes
The recombinant host cell of the present invention is capable of producing
miltiradiene
and/or dehydroabietadiene. Miltiradiene may be spontaneously converted into
dehydro-
abietadiene or it may be converted by an enzyme that facilitates the
conversion of milti-
radiene to dehydroabietadiene.
As described herein above, the synthesis of miltiradiene usually begins with
the for-
mation of GGPP by condensation of one dimethylallyl pyroophosphate (DMAP) mole-
cule and three isopentenyl pyrophosphate (IPP) molecules by a geranylgeranyl
diphos-
phate synthase.
Recombinant host cells and heterologous nucleic acids that encode enzymes that
cata-
lyze the synthesis of GGPP in recombinant host cells are generally known in
the art,
see e.g. WO 2015/113570. In addition, many host organisms are capable of
producing
GGPP intrinsically and heterologous nucleic acids may thus not always be
necessary
for production of GGPP.
In some embodiments, the recombinant host cell comprises a heterologous
nucleic
acid encoding a geranylgeranyl diphosphate synthase, such as the
geranylgeranyl di-
phosphate synthase SpGGPPs7 as set forth in SEQ ID NO: 73 or SEQ ID NO: 81, or
a
functional homologue thereof having at least 80%, such as at least 81%, such
as at
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
19
least 82%, such as at least 83%, such as at least 84%, such as at least 85%,
such as
at least 86%, such as at least 87%, such as at least 88%, such as at least
89%, such
as at least 90%, such as at least 91%, such as at least 92%, such as at least
93%,
such as at least 94%, such as at least 95%, such as at least 96%, such as at
least
97%, such as at least 98%, such as at least 99% sequence identity thereto, or
the ma-
ture polypeptide thereof.
Subsequently, GGPP may be converted into miltiradiene by the action of one or
more
diterpene synthases, copalyl diphosphate synthases and/or miltiradiene
synthases.
In some embodiments, the recombinant host cell comprises one or more
heterologous
nucleic acids encoding one or more diterpene synthases, such as the diterpene
syn-
thases CfTPS1 (SEQ ID NO: 67) and CfTPS3 (SEQ ID NO: 68), or CftTPS1 (SEQ ID
NO: 77) and CftTPS3 (SEQ ID NO: 78), or respective functional homologues
thereof
having at least 80%, such as at least 81%, such as at least 82%, such as at
least 83%,
such as at least 84%, such as at least 85%, such as at least 86%, such as at
least
87%, such as at least 88%, such as at least 89%, such as at least 90%, such as
at
least 91%, such as at least 92%, such as at least 93%, such as at least 94%,
such as
at least 95%, such as at least 96%, such as at least 97%, such as at least
98%, such
as at least 99% sequence identity thereto, or the mature polypeptides thereof.
In some embodiments, the recombinant host cell comprises one or more
heterologous
nucleic acids encoding one or more diterpene synthases, such as the diterpene
syn-
thases TwTPS9 (SEQ ID NO: 69) and TwTPS27 (SEQ ID NO: 70), or respective func-
tional homologues thereof having at least 80%, such as at least 81%, such as
at least
82%, such as at least 83%, such as at least 84%, such as at least 85%, such as
at
least 86%, such as at least 87%, such as at least 88%, such as at least 89%,
such as
at least 90%, such as at least 91%, such as at least 92%, such as at least
93%, such
as at least 94%, such as at least 95%, such as at least 96%, such as at least
97%,
such as at least 98%, such as at least 99% sequence identity thereto, or the
mature
polypeptides thereof.
In some embodiments, the recombinant host cell comprises a combination of one
or
more copalyl diphosphate synthases and one or more miltiradiene synthases,
such as
a combination of the copalyl diphosphate synthases SmCPS (SEQ ID NO: 71) and
the
miltiradiene synthase SmKSL (SEQ ID NO: 72), or respective functional
homologues
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
thereof having at least 80%, such as at least 81%, such as at least 82%, such
as at
least 83%, such as at least 84%, such as at least 85%, such as at least 86%,
such as
at least 87%, such as at least 88%, such as at least 89%, such as at least
90%, such
as at least 91%, such as at least 92%, such as at least 93%, such as at least
94%,
5 such as at least 95%, such as at least 96%, such as at least 97%, such as
at least
98%, such as at least 99% sequence identity thereto, or the mature
polypeptides
thereof.
In an even further aspect, the invention relates to polypeptides having
cytochrome P450
10 enzyme activity and comprising an amino acid sequence having at least 80
% sequence
identity, preferably at least 85% sequence identity, preferably at least 90%
sequence
identity, preferably at least 95% sequence identity, preferably at least 96%
sequence
identity, preferably at least 97% sequence identity, preferably at least 98%
sequence
identity or even 100% sequence identity to one of the sequences SEQ ID NO: 1,
SEQ
15 ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 6, and SEQ
ID NO:
7 or the mature polypeptide thereof.
In a further aspect, the invention relates to a polypeptide having cytochrome
B5 activity
and comprising an amino acid sequence having at least 80 % sequence identity,
prefer-
20 ably at least 85% sequence identity, preferably at least 90% sequence
identity, prefera-
bly at least 95% sequence identity, preferably at least 96% sequence identity,
preferably
at least 97% sequence identity, preferably at least 98% sequence identity or
even 100%
sequence identity to SEQ ID NO: 8, or the mature polypeptide thereof.
The invention also relates to polynucleotide sequences or genes encoding
polypeptides
having cytochrome P450 enzyme activity and comprising an amino acid sequence
hav-
ing at least 80 % sequence identity, preferably at least 85% sequence
identity, preferably
at least 90% sequence identity, preferably at least 95% sequence identity,
preferably at
least 96% sequence identity, preferably at least 97% sequence identity,
preferably at
least 98% sequence identity or even 100% sequence identity to one of the
sequences
SEQ ID NO: 1, SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID
NO: 6, and SEQ ID NO: 7 or the mature polypeptide thereof, or encoding a
polypeptide
having cytochrome B5 activity and comprising an amino acid sequence having at
least
80 % sequence identity, preferably at least 85% sequence identity, preferably
at least
90% sequence identity, preferably at least 95% sequence identity, preferably
at least
96% sequence identity, preferably at least 97% sequence identity, preferably
at least
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
21
98% sequence identity or even 100% sequence identity to SEQ ID NO: 8, or the
mature
polypeptide thereof.
In preferred embodiments, one or more of the first, second, third, fourth,
fifth and sixth
enzymes having cytochrome P450 activity comprises or consists of an amino acid
se-
quence according to any one of SEQ ID NO: 1 (TwCYP82D274v1), SEQ ID NO: 2
(TwCYP82D274v2), SEQ ID NO: 74 (TwCYP82D274v3), SEQ ID NO: 75
(TwCYP82D274v4), SEQ ID NO: 3 (TwCYP71BE85), SEQ ID NO: 4 (TwCYP71BE86),
SEQ ID NO: 5 (TwCYP82D213v1) and SEQ ID NO: 76 (TwCYP82D213v2), and re-
spective functional homologs thereof having at least 80% sequence identity,
preferably
at least 85% sequence identity, preferably at least 90% sequence identity,
preferably at
least 95% sequence identity, preferably at least 96% sequence identity,
preferably at
least 97% sequence identity, preferably at least 98% sequence identity,
preferably at
least 98% sequence identity thereto, or the mature polypeptide thereof.
In some embodiments, the first heterologous nucleic acid encoding a first
enzyme hav-
ing cytochrome P450 activity encodes TwCYP82D274 as set forth in SEQ ID NO: 1
(TwCYP82D274v1, SEQ ID NO: 2 (TwCYP82D274v2), SEQ ID NO: 74
(TwCYP82D274v3), SEQ ID NO: 75 (TwCYP82D274v4), or a functional homologue
thereof having at least 80%, such as at least 81%, such as at least 82%, such
as at
least 83%, such as at least 84%, such as at least 85%, such as at least 86%,
such as
at least 87%, such as at least 88%, such as at least 89%, such as at least
90%, such
as at least 91%, such as at least 92%, such as at least 93%, such as at least
94%,
such as at least 95%, such as at least 96%, such as at least 97%, such as at
least
98%, such as at least 99% sequence identity thereto.
In some embodiments, the second heterologous nucleic acid encoding a second en-
zyme having cytochrome P450 activity encodes the cytochrome P450 enzyme
TwCYP71BE86 as set forth in SEQ ID NO: 4, or a functional homologue thereof
having
at least 80%, such as at least 81%, such as at least 82%, such as at least
83%, such
as at least 84%, such as at least 85%, such as at least 86%, such as at least
87%,
such as at least 88%, such as at least 89%, such as at least 90%, such as at
least
91%, such as at least 92%, such as at least 93%, such as at least 94%, such as
at
least 95%, such as at least 96%, such as at least 97%, such as at least 98%,
such as
at least 99% sequence identity thereto.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
22
In some embodiments, the third heterologous nucleic acid encoding a third
enzyme
having cytochrome P450 activity encodes the cytochrome P450 enzyme
TwCYP71BE85 as set forth in SEQ ID NO: 3, or a functional homologue thereof
having
at least 80%, such as at least 81%, such as at least 82%, such as at least
83%, such
as at least 84%, such as at least 85%, such as at least 86%, such as at least
87%,
such as at least 88%, such as at least 89%, such as at least 90%, such as at
least
91%, such as at least 92%, such as at least 93%, such as at least 94%, such as
at
least 95%, such as at least 96%, such as at least 97%, such as at least 98%,
such as
at least 99% sequence identity thereto.
In some embodiments, the fourth heterologous nucleic acid encoding a fourth
enzyme
having cytochrome P450 activity encodes the cytochrome P450 enzyme
TwCYP82D213v1 as set forth in SEQ ID NO: 5 or TwCYP82D213v2 as set forth in
SEQ ID NO: 76 (TwCYP82D213v2), or a functional homologue thereof having at
least
80%, such as at least 81%, such as at least 82%, such as at least 83%, such as
at
least 84%, such as at least 85%, such as at least 86%, such as at least 87%,
such as
at least 88%, such as at least 89%, such as at least 90%, such as at least
91%, such
as at least 92%, such as at least 93%, such as at least 94%, such as at least
95%,
such as at least 96%, such as at least 97%, such as at least 98%, such as at
least 99%
sequence identity thereto.
In some embodiments, the fifth heterologous nucleic acid encoding a fifth
enzyme hav-
ing cytochrome P450 activity encodes the cytochrome P450 enzyme TwCYP82D217
as set forth in SEQ ID NO: 6, or a functional homologue thereof having at
least 80%,
such as at least 81%, such as at least 82%, such as at least 83%, such as at
least
84%, such as at least 85%, such as at least 86%, such as at least 87%, such as
at
least 88%, such as at least 89%, such as at least 90%, such as at least 91%,
such as
at least 92%, such as at least 93%, such as at least 94%, such as at least
95%, such
as at least 96%, such as at least 97%, such as at least 98%, such as at least
99% se-
quence identity thereto.
In some embodiments, the sixth heterologous nucleic acid encoding a sixth
enzyme
having cytochrome P450 activity encodes the cytochrome P450 enzyme
TwCYP82D275 as set forth in SEQ ID NO: 7, or a functional homologue thereof
having
at least 80%, such as at least 81%, such as at least 82%, such as at least
83%, such
as at least 84%, such as at least 85%, such as at least 86%, such as at least
87%,
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
23
such as at least 88%, such as at least 89%, such as at least 90%, such as at
least
91%, such as at least 92%, such as at least 93%, such as at least 94%, such as
at
least 95%, such as at least 96%, such as at least 97%, such as at least 98%,
such as
at least 99% sequence identity thereto.
In some embodiments, the seventh heterologous nucleic acid encoding an enzyme
having cytochrome B5 activity encodes the cytochrome B5 enzyme TwB5#1 as set
forth in SEQ ID NO: 8, or a functional homologue thereof having at least 80%,
such as
at least 81%, such as at least 82%, such as at least 83%, such as at least
84%, such
as at least 85%, such as at least 86%, such as at least 87%, such as at least
88%,
such as at least 89%, such as at least 90%, such as at least 91%, such as at
least
92%, such as at least 93%, such as at least 94%, such as at least 95%, such as
at
least 96%, such as at least 97%, such as at least 98%, such as at least 99%
sequence
identity thereto.
In some embodiments is provided a recombinant host cell
i. wherein the host cell is capable of producing miltiradiene and/or
dehydroabietadi-
ene; and
ii. comprises a heterologous nucleic acid encoding TwCYP82D274 of SEQ ID NO:
1, SEQ ID NO: 2, SEQ ID NO: 74 or SEQ ID NO: 75, or a functional homolog
thereof having at least 80%, such as at least 81%, such as at least 82%, such
as
at least 83%, such as at least 84%, such as at least 85%, such as at least
86%,
such as at least 87%, such as at least 88%, such as at least 89%, such as at
least 90%, such as at least 91%, such as at least 92%, such as at least 93%,
such as at least 94%, such as at least 95%, such as at least 96%, such as at
least 97%, such as at least 98%, such as at least 99% sequence identity
thereto,
or the mature polypeptide thereof,
wherein said cell is capable of producing 14-hydroxydehydroabietadiene.
In some embodiments is provided a recombinant host cell
i. wherein the host cell is capable of producting miltiradiene and/or
dehydro-
abietadiene; and
ii. comprises a heterologous nucleic acid encoding TwCYP82D274 of SEQ ID NO:
1, SEQ ID NO: 2, SEQ ID NO: 74 or SEQ ID NO: 75, or a functional homolog
thereof having at least 80%, such as at least 81%, such as at least 82%, such
as
at least 83%, such as at least 84%, such as at least 85%, such as at least
86%,
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
24
such as at least 87%, such as at least 88%, such as at least 89%, such as at
least 90%, such as at least 91%, such as at least 92%, such as at least 93%,
such as at least 94%, such as at least 95%, such as at least 96%, such as at
least 97%, such as at least 98%, such as at least 99% sequence identity
thereto,
or the mature polypeptide thereof; and
iii. comprises a heterologous nucleic acid encoding TwCYP71BE86 of SEQ ID NO:
4 or a functional homolog thereof having at least 80%, such as at least 81%,
such as at least 82%, such as at least 83%, such as at least 84%, such as at
least 85%, such as at least 86%, such as at least 87%, such as at least 88%,
such as at least 89%, such as at least 90%, such as at least 91%, such as at
least 92%, such as at least 93%, such as at least 94%, such as at least 95%,
such as at least 96%, such as at least 97%, such as at least 98%, such as at
least 99% sequence identity thereto, or the mature polypeptide thereof,
wherein said cell is capable of producing 14-hydroxydehydroabietadiene, 3,14-
dihy-
droxydehydroabietadiene, 3,14-di hydroxyabeodiene and/or 14-hydroxy-18-aldo-
abeo-
diene.
In some embodiments is provided a recombinant host cell
i. wherein the host cell is capable of producting miltiradiene and/or
dehydro-
abietadiene; and
ii. comprises a heterologous nucleic acid encoding TwCYP82D274 SEQ ID NO: 1,
SEQ ID NO: 2, SEQ ID NO: 74 or SEQ ID NO: 75, or a functional homolog
thereof having at least 80%, such as at least 81%, such as at least 82%, such
as
at least 83%, such as at least 84%, such as at least 85%, such as at least
86%,
such as at least 87%, such as at least 88%, such as at least 89%, such as at
least 90%, such as at least 91%, such as at least 92%, such as at least 93%,
such as at least 94%, such as at least 95%, such as at least 96%, such as at
least 97%, such as at least 98%, such as at least 99% sequence identity
thereto,
or the mature polypeptide thereof;
iii. comprises a heterologous nucleic acid encoding TwCYP71BE86 of SEQ ID NO:
4 or a functional homolog thereof having at least 80%, such as at least 81%,
such as at least 82%, such as at least 83%, such as at least 84%, such as at
least 85%, such as at least 86%, such as at least 87%, such as at least 88%,
such as at least 89%, such as at least 90%, such as at least 91%, such as at
least 92%, such as at least 93%, such as at least 94%, such as at least 95%,
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
such as at least 96%, such as at least 97%, such as at least 98%, such as at
least 99% sequence identity thereto, or the mature polypeptide thereof; and
iv. comprises a heterologous nucleic acid encoding TwCYP71BE85 of SEQ ID NO:
3 or a functional homolog thereof having at least 80%, such as at least 81%,
5 such as at least 82%, such as at least 83%, such as at least 84%, such as
at
least 85%, such as at least 86%, such as at least 87%, such as at least 88%,
such as at least 89%, such as at least 90%, such as at least 91%, such as at
least 92%, such as at least 93%, such as at least 94%, such as at least 95%,
such as at least 96%, such as at least 97%, such as at least 98%, such as at
10 least 99% sequence identity thereto, or the mature polypeptide thereof,
wherein said cell is capable of producing 14-hydroxydehydroabietadiene, 3,14-
dihy-
droxydehydroabietadiene, 3,14-di hydroxyabeodiene, 14-hydroxy-18-aldo-
abeodiene
and/or triptophenolide.
15 In some embodiments is provided a recombinant host cell
i. wherein the host cell is capable of producting miltiradiene and/or
dehydro-
abietadiene; and
ii. comprises a heterologous nucleic acid encoding TwCYP82D274 of SEQ ID NO:
1, SEQ ID NO: 2, SEQ ID NO: 74 or SEQ ID NO: 75, or a functional homolog
20 thereof having at least 80%, such as at least 81%, such as at least 82%,
such as
at least 83%, such as at least 84%, such as at least 85%, such as at least
86%,
such as at least 87%, such as at least 88%, such as at least 89%, such as at
least 90%, such as at least 91%, such as at least 92%, such as at least 93%,
such as at least 94%, such as at least 95%, such as at least 96%, such as at
25 least 97%, such as at least 98%, such as at least 99% sequence identity
thereto,
or the mature polypeptide thereof;
iii. comprises a heterologous nucleic acid encoding TwCYP71BE86 of SEQ ID NO:
4 or a functional homolog thereof having at least 80%, such as at least 81%,
such as at least 82%, such as at least 83%, such as at least 84%, such as at
least 85%, such as at least 86%, such as at least 87%, such as at least 88%,
such as at least 89%, such as at least 90%, such as at least 91%, such as at
least 92%, such as at least 93%, such as at least 94%, such as at least 95%,
such as at least 96%, such as at least 97%, such as at least 98%, such as at
least 99% sequence identity thereto, or the mature polypeptide thereof;
iv. comprises a heterologous nucleic acid encoding TwCYP71BE85 of SEQ ID NO:
3 or a functional homolog thereof having at least 80%, such as at least 81%,
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
26
such as at least 82%, such as at least 83%, such as at least 84%, such as at
least 85%, such as at least 86%, such as at least 87%, such as at least 88%,
such as at least 89%, such as at least 90%, such as at least 91%, such as at
least 92%, such as at least 93%, such as at least 94%, such as at least 95%,
such as at least 96%, such as at least 97%, such as at least 98%, such as at
least 99% sequence identity thereto, or the mature polypeptide thereof; and
v. comprises a heterologous nucleic acid encoding TwCYP82D213 of SEQ ID NO:
5 or SEQ ID NO: 76, or a functional homolog thereof having at least 80%, such
as at least 81%, such as at least 82%, such as at least 83%, such as at least
84%, such as at least 85%, such as at least 86%, such as at least 87%, such as
at least 88%, such as at least 89%, such as at least 90%, such as at least
91%,
such as at least 92%, such as at least 93%, such as at least 94%, such as at
least 95%, such as at least 96%, such as at least 97%, such as at least 98%,
such as at least 99% sequence identity thereto, or the mature polypeptide
thereof,
wherein said cell is capable of producing 14-hydroxydehydroabietadiene, 3,14-
dihy-
droxydehydroabietadiene, 3,14-di hydroxyabeodiene, 14-hydroxy-18-aldo-
abeodiene,
triptophenolide and/or triptonide.
In some embodiments is provided a recombinant host cell
i. wherein the host cell is capable of producting miltiradiene and/or
dehydro-
abietadiene; and
ii. comprises a heterologous nucleic acid encoding TwCYP82D274 of SEQ ID NO:
1, SEQ ID NO: 2, SEQ ID NO: 74 or SEQ ID NO: 75, or a functional homolog
thereof having at least 80%, such as at least 81%, such as at least 82%, such
as
at least 83%, such as at least 84%, such as at least 85%, such as at least
86%,
such as at least 87%, such as at least 88%, such as at least 89%, such as at
least 90%, such as at least 91%, such as at least 92%, such as at least 93%,
such as at least 94%, such as at least 95%, such as at least 96%, such as at
least 97%, such as at least 98%, such as at least 99% sequence identity
thereto,
or the mature polypeptide thereof;
iii. comprises a heterologous nucleic acid encoding TwCYP71BE86 of SEQ ID NO:
4 or a functional homolog thereof having at least 80%, such as at least 81%,
such as at least 82%, such as at least 83%, such as at least 84%, such as at
least 85%, such as at least 86%, such as at least 87%, such as at least 88%,
such as at least 89%, such as at least 90%, such as at least 91%, such as at
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
27
least 92%, such as at least 93%, such as at least 94%, such as at least 95%,
such as at least 96%, such as at least 97%, such as at least 98%, such as at
least 99% sequence identity thereto, or the mature polypeptide thereof;
iv. comprises a heterologous nucleic acid encoding TwCYP71BE85 of SEQ ID NO:
3 or a functional homolog thereof having at least 80%, such as at least 81%,
such as at least 82%, such as at least 83%, such as at least 84%, such as at
least 85%, such as at least 86%, such as at least 87%, such as at least 88%,
such as at least 89%, such as at least 90%, such as at least 91%, such as at
least 92%, such as at least 93%, such as at least 94%, such as at least 95%,
such as at least 96%, such as at least 97%, such as at least 98%, such as at
least 99% sequence identity thereto, or the mature polypeptide thereof;
v. comprises a heterologous nucleic acid encoding TwCYP82D213 of SEQ ID NO:
5 or SEQ ID NO: 76, or a functional homolog thereof having at least 80%, such
as at least 81%, such as at least 82%, such as at least 83%, such as at least
84%, such as at least 85%, such as at least 86%, such as at least 87%, such as
at least 88%, such as at least 89%, such as at least 90%, such as at least
91%,
such as at least 92%, such as at least 93%, such as at least 94%, such as at
least 95%, such as at least 96%, such as at least 97%, such as at least 98%,
such as at least 99% sequence identity thereto, or the mature polypeptide
thereof; and
vi. comprises a heterologous nucleic acid encoding TwB5#1 of SEQ ID NO: 8 or a
functional homolog thereof having at least 80%, such as at least 81%, such as
at
least 82%, such as at least 83%, such as at least 84%, such as at least 85%,
such as at least 86%, such as at least 87%, such as at least 88%, such as at
least 89%, such as at least 90%, such as at least 91%, such as at least 92%,
such as at least 93%, such as at least 94%, such as at least 95%, such as at
least 96%, such as at least 97%, such as at least 98%, such as at least 99% se-
quence identity thereto, or the mature polypeptide thereof,
wherein said cell is capable of producing triptonide with a titer that is at
least 2-fold,
such as at least 3-fold, such as at least 4-fold, such as at least 5-fold
higher than an
identical yeast cell, except wherein said yeast said does not express said
TwB5#1 or
said functional homolog thereof.
In some embodiments is provided a recombinant host cell
i. wherein the host cell is capable of producting miltiradiene and/or
dehydroabietadi-
ene; and
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
28
ii. comprises a heterologous nucleic acid encoding TwCYP82D274 of SEQ ID NO:
1, SEQ ID NO: 2, SEQ ID NO: 74 or SEQ ID NO: 75,or a functional homolog
thereof having at least 80%, such as at least 81%, such as at least 82%, such
as
at least 83%, such as at least 84%, such as at least 85%, such as at least
86%,
such as at least 87%, such as at least 88%, such as at least 89%, such as at
least 90%, such as at least 91%, such as at least 92%, such as at least 93%,
such as at least 94%, such as at least 95%, such as at least 96%, such as at
least 97%, such as at least 98%, such as at least 99% sequence identity
thereto,
or the mature polypeptide thereof;
iii. comprises a heterologous nucleic acid encoding TwCYP71BE86 of SEQ ID NO:
4 or a functional homolog thereof having at least 80%, such as at least 81%,
such as at least 82%, such as at least 83%, such as at least 84%, such as at
least 85%, such as at least 86%, such as at least 87%, such as at least 88%,
such as at least 89%, such as at least 90%, such as at least 91%, such as at
least 92%, such as at least 93%, such as at least 94%, such as at least 95%,
such as at least 96%, such as at least 97%, such as at least 98%, such as at
least 99% sequence identity thereto, or the mature polypeptide thereof;
iv. comprises a heterologous nucleic acid encoding TwCYP71BE85 of SEQ ID NO:
3 or a functional homolog thereof having at least 80%, such as at least 81%,
such as at least 82%, such as at least 83%, such as at least 84%, such as at
least 85%, such as at least 86%, such as at least 87%, such as at least 88%,
such as at least 89%, such as at least 90%, such as at least 91%, such as at
least 92%, such as at least 93%, such as at least 94%, such as at least 95%,
such as at least 96%, such as at least 97%, such as at least 98%, such as at
least 99% sequence identity thereto, or the mature polypeptide thereof;
v. comprises a heterologous nucleic acid encoding TwCYP82D213 of SEQ ID NO:
5 or SEQ ID NO: 76, or a functional homolog thereof having at least 80%, such
as at least 81%, such as at least 82%, such as at least 83%, such as at least
84%, such as at least 85%, such as at least 86%, such as at least 87%, such as
at least 88%, such as at least 89%, such as at least 90%, such as at least
91%,
such as at least 92%, such as at least 93%, such as at least 94%, such as at
least 95%, such as at least 96%, such as at least 97%, such as at least 98%,
such as at least 99% sequence identity thereto, or the mature polypeptide
thereof; and
vi. comprises a heterologous nucleic acid encoding TwB5#1 of SEQ ID NO: 8 or a
functional homolog thereof having at least 80%, such as at least 81%, such as
at
CA 03192028 2023-02-16
WO 2022/043461
PCT/EP2021/073656
29
least 82%, such as at least 83%, such as at least 84%, such as at least 85%,
such as at least 86%, such as at least 87%, such as at least 88%, such as at
least 89%, such as at least 90%, such as at least 91%, such as at least 92%,
such as at least 93%, such as at least 94%, such as at least 95%, such as at
least 96%, such as at least 97%, such as at least 98%, such as at least 99% se-
quence identity thereto, or the mature polypeptide thereof,
wherein said cell is capable of growing in a fermentation medium and where
said fer-
mentation medium after 7 days of fermentation comprises:
- at least 3 ppm triptonide and/or
- at least 1 ppm triptophenolide.
Aforementioned recombinant host cells may be capable of producing miltiradiene
and/or dehydroabietadiene for several different reasons. For example, the host
cells
may endogenously be capable of producing miltiradiene. Alternatively, the
recombinant
host cell may comprise one or more heterologous nucleic acid sequences
encoding
one or more enzymes involved in the production of miltiradiene, such as the
diterpene
biosynthetic enzymes SPGGPPS7 of SEQ ID NO: 73 or SEQ ID NO: 81, CfTPS1 of
SEQ ID NO: 67, CftTPS1 of SEQ ID NO: 77, CfTPS3 of SEQ ID NO: 68, CftTPS3 of
SEQ ID NO: 78 and/or TwCPR1 of SEQ ID NO: 9, or respective functional homologs
thereof having at least 80% sequence identity, preferably at least 85%
sequence iden-
tity, preferably at least 90% sequence identity, preferably at least 95%
sequence iden-
tity, preferably at least 96% sequence identity, preferably at least 97%
sequence iden-
tity, preferably at least 98% sequence identity, preferably at least 98%
sequence iden-
tity thereto, or the mature polypeptides thereof.
Functional homologues of the first (e.g. TwCYP82D274), second (e.g.
TwCYP71BE86), third (e.g. TwCYP71BE85), fourth (e.g. TwCYP82D213), fifth
(e.g.TwCYP82D217) and sixth (e.g.TwCYP82D275) enzymes having cytochrome P450
activity and the enzyme having cytochrome B5 activity (e.g.TwB5#1) may be
verified by
expressing the relevant protein in a yeast cell and assessing whether they are
able to
produce specific compounds as described herein below.
A yeast cell expressing a functional homolog of TwCYP82D274 (SEQ ID NO: 1, SEQ
ID NO: 2, SEQ ID NO: 74 or SEQ ID NO: 75) and further expressing
i. the diterpene biosynthetic enzymes SPGGPPS7v2 (SEQ ID NO: 81), CftTPS1
(SEQ ID NO: 77), CftTPS3 (SEQ ID NO: 78) and TwCPR1 (SEQ ID NO: 9),
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
is preferably capable of producing 14-hydroxydehydroabietadiene.
A yeast cell expressing a functional homolog of TwCYP71BE86 (SEQ ID NO: 4) and
further expressing
5 i. the diterpene biosynthetic enzymes SPGGPPS7v2 (SEQ ID NO: 81), CftTPS1
(SEQ ID NO: 77), CftTPS3 (SEQ ID NO: 78) and TwCPR1 (SEQ ID NO: 9); and
TwCYP82D274 (SEQ ID NO: 1 or SEQ ID NO: 2),
is preferably capable of producing 14-hydroxydehydroabietadiene, 3,14-
dihydroxydehy-
droabietadiene, 3,14-dihydroxyabeodiene and 14-hydroxy-18-aldo-abeodiene.
A yeast cell expressing a functional homolog of TwCYP71BE85 (SEQ ID NO: 3) and
further expressing
i. the diterpene biosynthetic enzymes SPGGPPS7v2 (SEQ ID NO: 81), CftTPS1
(SEQ ID NO: 77), CftTPS3 (SEQ ID NO: 78) and TwCPR1 (SEQ ID NO: 9);
ii. TwCYP82D274 (SEQ ID NO: 1 or SEQ ID NO: 2); and
TwCYP71BE86 (SEQ ID NO: 4),
is preferably capable of producing 14-hydroxydehydroabietadiene, 3,14-
dihydroxydehy-
droabietadiene, 3,14-dihydroxyabeodiene, 14-hydroxy-18-aldo-abeodiene and
tripto-
phenolide.
A yeast cell expressing a functional homolog of TwCYP82D213 (SEQ ID NO: 5 or
SEQ
ID NO: 76) and further expressing
i. the diterpene biosynthetic enzymes SPGGPPS7v2 (SEQ ID NO: 81),
CftTPS1
(SEQ ID NO: 77), CftTPS3 (SEQ ID NO: 78) and TwCPR1 (SEQ ID NO: 9);
ii. TwCYP82D274 (SEQ ID NO: 1 or SEQ ID NO: 2);
TwCYP71BE86 (SEQ ID NO: 4); and
iv. TwCYP71BE85 (SEQ ID NO: 3),
is preferably capable of producing 14-hydroxydehydroabietadiene, 3,14-
dihydroxydehy-
droabietadiene, 3,14-dihydroxyabeodiene, 14-hydroxy-18-aldo-abeodiene,
triptophe-
nolide and triptonide.
A yeast cell expressing a functional homolog of TwB5#1 (SEQ ID NO: 8) and
further
expressing
i. the diterpene biosynthetic enzymes SPGGPPS7v2 (SEQ ID NO: 81),
CftTPS1
(SEQ ID NO: 77), CftTPS3 (SEQ ID NO: 78) and TwCPR1 (SEQ ID NO: 9);
TwCYP82D274 (SEQ ID NO: 1 or SEQ ID NO: 2);
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
31
TwCYP71BE86 (SEQ ID NO: 4);
iv. TwCYP71BE85 (SEQ ID NO: 3); and
v. TwCYP82D213 (SEQ ID NO: 5 or SEQ ID NO: 76)
is preferably capable of producing triptonide with a titer that is at least 2-
fold, such as at
least 3-fold, such as at least 4-fold, such as at least 5-fold higher than an
identical
yeast cell, except wherein said yeast said does not express said functional
homolog of
TwB5#1.
In preferred embodiments, the enzyme having cytochrome B5 activity comprises
or
consists of an amino acid sequence according to SEQ ID NO: 8 (TwB5#1), or a
func-
tional homolog thereof having at least 80 % sequence identity, preferably at
least 85%
sequence identity, preferably at least 90% sequence identity, preferably at
least 95%
sequence identity, preferably at least 96% sequence identity, preferably at
least 97%
sequence identity, preferably at least 98% sequence identity, preferably at
least 98%
sequence identity thereto, or the mature polypeptide thereof.
The polynucleotide of the invention may be provided by cloning from organisms
that nat-
urally produce the polypeptides such as the plant T. wilfordii or closely
related plants, or
it may be provided by chemical synthesis of the polynucleotide sequence based
on tech-
niques known in the art. The polynucleotide may have a sequence that is
identical to a
sequence found in nature, or it may have a sequence that is not found in
nature, e.g. the
sequence may be codon optimized for the particular selected host cell.
The polypeptides of the invention may be provided from organisms that
naturally produce
the polypeptides such as the plant T. wilfordii or related organisms; or they
may be pro-
vided by inserting and expressing polynucleotides encoding polypeptides into a
suitable
host cell and recovering the polypeptides from culture broth comprising the
host cell
transformed with the respective genes. It is preferred to provide the
polypeptides of the
invention from a suitable selected recombinant host cell.
In order to transform and express a gene in a suitable host cell, the gene is
usually
operably connected with suitable regulatory elements and inserted into an
expression
vector suitable for the particular selected host cell. Selecting suitable
regulatory ele-
ments, constructing a suitable expression vector and transforming the selected
host cell
is within the skills of the average practitioner and the invention is not
limited by any par-
ticular selection of such elements.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
32
The generated host cells comprising the genes of the invention are suitable
grown in a
container, e.g. a fermenter or shake flasks; under conditions where the genes
are ex-
pressed and the oxygenated diterpenoid compounds are formed. When growth
ceases
or a sufficiently high amount of the oxygenated diterpenoid compounds are
accumulated
in the culture broth, the oxygenated diterpenoid compounds may be further
modified and
recovered from the culture broth.
Sequence identity is understood as a measurement of the similarity between two
amino
acid or nucleotide sequences. Sequence identity is calculated by first
aligning the two
sequences, counting the number of positions where the two sequences contain
the same
amino acid residue or nucleotide and calculating the percent identity as the
number of
positions with identical amino acid residue or nucleotide with the whole
length of the
alignment.
Several algorithms have been developed and are available for the skilled
person.
In this specification and claim, the sequence identity for amino acid
sequences are cal-
culated using the NCB! BLAST+ pairwise alignment algorithm, using default
parameters
(BLOSUM 62 matrix, Gap open penalty 11; gap extend penalty 1, Exp. Thr 10),
and the
sequence identity for nucleotide sequences are calculated using the NCB!
BLAST+ pair-
wise alignment algorithm, using default parameters (Match/mismatch scores 1, -
3; gap
open penalty 5; gap extend penalty 2; exp. Thr 10). The NCB! BLAST+ programs
are
further described in: Madeira F el at (2019) NAR 47: W636-W641.
EXAMPLES
Materials and Methods
Genetic engineering of Nicotiana benthamiana
Triptetygium wilfordii CYP genes were cloned from plant material and co-
expressed in
Nicotiana benthamiana with the diterpene biosynthesis genes CfDXS (SEQ ID NO:
79)
or SctHMGR (SEQ ID NO: 80), CfGGPPS or SpGGPPS7 (SEQ ID NO: 81), CfTPS1
(SEQ ID NO: 67) or CftTPS1 (SEQ ID NO: 77), and CfTPS3 (SEQ ID NO: 68) or
CftTPS3
(SEQ ID NO: 78) using constructs and methods previously described in (1-4).
Also co-
expressed, were the suppressor of gene silencing, p19. Briefly, binary vectors
each con-
taining individual diterpene biosynthesis genes or Triptetygium wilfordii CYPs
(TwCYPs)
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
33
were transformed into agrobacteria. Liquid cultures of agrobacteria each
containing spe-
cific plasmids were mixed for co-expression of specific combinations of
TwCYPs.
Genetic engineering of Saccharomyces cerevisiae and growth conditions for
engineered
S. cerevisiae
Media
YPD media: 20 g/L Bacto TM Peptone, 10 g/L BactoTM Yeast extract, 20 g/L
glucose.
Synthetic complete (SC) meda without uracil: 1.92 g/L Yeast Synthetic Drop-out
Media
Supplements without uracil (Sigma-Aldrich Co. LLC. Catalog number Y1501), 6.7
g/L
Yeast Nitrogen Base Without Amino Acids (Sigma-Aldrich Co. LLC. Catalog number
Y0626), 20 g/L glucose. Feed-In-Time (FIT) was based on EnPump200 (Enpresso
GmbH), and made according to protocol enclosed with the product. Agar plates:
SC me-
dia including agar (15 g/L).
Uracil auxotrophy in parent strains was introduced by selecting for lack of
URA3 function
on agar plates of SC medium without uracil containing also 5-Fluoroorotic Acid
(5-F0A,
0.74 g/L) and uracil (30 mg/L).
Yeast transformants were isolated on SC without uracil agar plates.
Feed-batch fermentation of engineering S. cerevisiae strains for isolation of
miltiradiene
derived diterpenoids
All engineered S. cerevisiae strains were cultivated in 96-deepwell plates
using a Feed-
In-Time (FIT; m2p-labs) approach similar to previously described (insert ref
Forman et
al. 2018). For isolation and purification of key intermediates in the
triptonide pathway
selected engineered S. cerevisiae strain were cultivated in feed batch
fermentor using a
2 L BiostatO A bioreactor (Sartorius AG). Fed batch fermentation was initiated
by addi-
tion of a 100 mL starter culture to the reactor tank (with impellers), which
in turn was
prepared earlier by autoclavation while containing 200 mL Batch glucose and
300 mL
Batch salt mix. Also 5 mL vitamin mix, 5 mL micro elements and 0.5 mL trace
elements,
were added. Cultivation in the bioreactor was started under the following
conditions
(monitored and automatically controlled): pH = 5, temp. = 30 C, dissolved
oxygen (DO)
= 20 %. While pH was controlled by feeding of ammonium hydroxide (32 %) and
sulfuric
acid (10 %), dissolved oxygen was controlled by air supply combined with
stirring. Also
foam levels were adjusted by addition of anti-foam emulsion (35119, Serva
Electropho-
resis GmbH). After 18 hours of initial cultivation in the bioreactor, feeding
with Feeding
solution at a rate of 1.3 % was started. The fermentation process continued
for 7 days
with daily sampling of the culture.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
34
Extraction of engineered S. cerevisiae for LC-MS analysis
Genetically engineered S. cerevisiae strain was transferred into 0.5mL media
in a 96-
well plate and grown for 3 days at 30 C with orbital shaking at 350 rpm. For
extraction
0.1mL of S. cerevisiae culture was transferred to 1.5mL glass vials. 0.4mL
Me0H uHPLC
grade was added. S. cerevisiae extract was filtered by using a 0:22 pm 96-well
filter plate
(Merck Millipore, Darmstadt, Germany) and at stored at 4 C prior to LC-MS
analysis.
Extraction of diterpenoid metabolites for LC-MS analysis
Samples of yeast cultures for LCMS analysis were prepared in 1.5 mL glass
vials by
mixing yeast cultures and methanol spiked with 5 ppm andrographolide (internal
stand-
ard; FA17902, CarboSynth) in a ratio of 1:19 (v/v) for daily bioreactor
samples and 1:4
(v/v) for 96-deepwell cultures. Mixing proceeded for 30 min with shaking at
room temp.
For tobacco samples, 2 leaf discs (0=3cm) placed in 1.5 ml glass vials, were
extracted
with 1 mL of the methanol extraction solution, for 1 h with shaking at room
temperature.
Before LCMS analysis samples were passed through a 0.22 pM 96-well plate
filter
(Merck Millipore, Darmstadt, Germany) and stored at 5 C.
LC-MS analysis:
Methanol (Me0H) extracts were analysed using an Ultimate 3000 UHPLC+ Focused
system (Dionex Corporation, Sunnyvale, CA) coupled to a Bruker Compact ESI-
QT0E-
MS (Bruker) system. Samples were separated on a Kinetex XB-C18 column (100 x
2.1
mm ID, 1:7 pm particle size, 100 A pore size; Phenomenex Inc., Torrance, CA)
main-
tained at 40 _C with a flow rate of 0.3 mL min-1 and mobile phase consisting
of 0.05 %
(v/v) formic acid in water (solvent A) and 0.05 % (v/v) formic acid in
acetonitrile (solvent
B).
Two LC protocols were used:
LC method 1: 0-0.5 min, 10% B; 0.5-21 min, linear increase from 10 to 80% B;
21-31
min, to 90% B; 31-34 min, to 100% B; 34-39 min 100% B; 39-40 min linear
decrease
from 100 to 10 % B.
LC method 2: 0-0.5 min, 20 % B; 0.5-11 min, linear increase from 20 to 80 % B;
11-20
min, to 90% B; 20-22 min, to 100% B; 22-27 min 100% B; 27-28 min linear
decrease
from 100 to 20 % B.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
LC method 3: 0-0.5 min, 20 % B; 0.5-9 min, linear increase from 20 to 100 % B;
9-11
min, 100% B; 11-11.5 min, linear decrease from 100 to 20 % B; 11.5-15min, 20%
B.
5 Extraction of diterpenoid metabolites for GC-MS analysis
Samples of yeast cultures for GCMS analysis were prepared in 1.5 mL glass
vials by
mixing yeast culture and pure methanol at a ratio of 1:4 (v/v). After brief
mixing, apolar
constituents, were liquid-liquid extracted into hexane, spiked with 10 ppm 1-
eicocene, by
mixing at a ratio of 1:1 (v/v) and shaking for 1 h. For tobacco samples, 2
leaf discs
10 (0=3cm) placed in 1.5 ml glass vials, were extracted with 1mL of the
same hexane solu-
tion, via 1 h of shaking. Prior to GCMS analysis hexane layers were
transferred to new
vials.
Gas chromatography Mass spectrometry (GC-MS) analysis
15 GC-MS analysis was carried out on a Shimadzu GCMS-QP2010 Ultra (Shimadzu
Corp.)
with an Agilent HP-5M5 column (Agilent Technologies) 20 m x 0.18 mm i.d., 0.18
pm
film thickness). Hydrogen was used as a carrier gas at a constant linear
velocity of 50
cm s-1, and the injection volume was 1 pL at 250 C (splitless mode). The oven
program
was 80 C for 2 min, ramp at rate 20 C/min to 180 C, ramp at rate 10 C/min to
300 C,
20 ramp at rate 20 C/min to 310 C, hold for 3 min. Data was stored in .CDF
format and
processed in MZmine2.
Relative quantification of miltiradiene derived diterpenoids
25 Relative compound quantities in yeast cultures were based on normalized
peak areas of
characteristic ions (data obtained using targeted feature detection in the
MZmine2 soft-
ware). The signal for the following ions were quantified: 1: miltiradiene m/z
91.1, 2: 14-
hydroxyabietadiene m/z 189.1, 3: F15P1 m/z 303.2318, 4: F20P2 m/z 283.2059, 5:
F15P2 m/z 299.2002, 6: triptophenolide m/z 313.1794, 8: triptonide m/z
359.1481. For
30 LCMS and GCMS data a mass deviation of 5ppm and 100ppm, respectively,
was toler-
ated.
The peak area of the base peak ion (m/z 315.1947) for the internal standard
andro-
grapholide was used for normalization.
35 Absolute quantifications
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
36
Absolute quantifications of triptophenolide (FT65732, CarboSynth) and
triptonide
(FT65197, CarboSynth) were done by co-analysis of authentic standards prepared
in
methanol and a final concentration of 5 ppm internal standard
(andrographelide).
Quantification was based on normalized peak area and calculated from the
slopes of
linear extrapolations of the standards response curve (triptophenolide 0.05,
0.5, 1, 2
ppm; triptonide 0.5, 1,2, 10, 20 ppm).
Isolation and purification of miltiradiene derived compounds from engineered
S. cere-
visiae strain for NMR analysis
Compounds in this invention were isolated from bioreactor cultures yeast
strains
NVJ8.15, and NVJ3.10, and structurally elucidated by NMR. The combined ethyl
acetate
extracts of broth and methanol-lysed cells (cells:methanol = 1:4, v/v) were
initially dried
in presence of Celite SO (06858, Sigma-Aldrich) via rotary evaporation.
Compounds
were subsequently isolated by successive fractionations using a puriFlashe
5.250 (In-
terchim, Montlucon, France) instrument with detection by UV absorbance and
Evapora-
tive Light-Scattering Detection (ELSD). This was equipped with either of
columns (Cl)
PF-155IHP-F0025 (0V002A, lnterchim) and (C2) US5C18HQ-100/300 (55P750, Inter-
chim) for normal phase- and reverse phase separation, respectively.
An initial pre-fractionation of the dry mix of Celite SO/crude extract was
achieved using
column (ref. 9) with loading from a manually packed dry-loading column.
Separation was
obtained using mobile phases hexane (A) and ethyl acetate (B), a constant flow
rate of
15 mL/min, followed by a final washing step with 100% methanol. Compounds of
interest
were detected by UV and ELSD and collected. Collected fractions were
continuously
evaluated by LCMS using LC-MS method 3 and TLC analysis prior to further
fractionation
or NMR studies. Additional purification of compounds of interest from fraction
with multi-
ple compounds was done by an additional normal phase fraction using Cl or a
reverse
phase column fractionation using C2.
For reverse phase purification with C2 samples were evaporated using rotor
evaporation
and resuspended in 2mL methanol. Sample was injected directly onto the pre
condi-
tioned column C2. Mobile phases for C2 consisted of solvent C: deionized water
and
solvent D acetonitrile each acidified with 0.05% (v/v) formic acid. A constant
flow rate of
32 mlimin, was used, with a linear solvent gradient with increasing
concentration of sol-
.. vent D. Compounds of interest were detected by ELSD and UV and collected.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
37
Additional reverse phase purification was done by multiple injections of 100uL
ontp a
semi-prep Phenomenex Luna 5pm 018(2) 100A 250x10mm (fully porous)(Phenomenex,
Inc., Torrance, CA, USA) column on a Shimadzu HPLC (SPD-M20A diode array detec-
tor, FRC-10A fraction collector, DGU-20A5 degasser, LC-20AT pump, CBM-20A
System
controller, CTO-10AS VP column oven, SIL-10AP autosampler). Mobile phase was a
linear gradient between C and D with an increasing amount of D going from 50-
100%.
Compounds of interest was detected by UV absorbance at 210nm and collected.
Mass spectra
Mass spectra were acquired in positive ion mode over a scan range of m/z 50-
1200 with
the following ESI and MS settings: capillary voltage, 4000 V; end plate
offset, 500 V; dry
gas temperature, 220 C; dry gas flow of 8 L mini; nebulizer pressure, 2 bar;
in source
CID energy, 0 eV; hexapole RF, 50 Vpp; quadrupole ion energy, 4 eV; collision
cell en-
ergy, 7 eV. Raw chromatogram data was calibrated using an internal sodium
formate
standard and subsequently exported as mzML format using DataAnalysis 4.3
(Build
110.102.1532) (64-bit), Bruker. MZmine ver 2.53 was used for visualizing the
LC-MS
chromatograms.
Media recipes for bioreactor starting media and feed media
Batch glucose:
Glucose monohydrate 55 g/L
Batch salt mix:
Ammonium sulfate 25 g/L
Potassium phosphate monobasic 5 g/L
Magnesium sulfate heptahydrate 1.7 g/L
Feed glucose:
Glucose monohydrate 880 g/L
Feed salt mix:
Potassium phosphate monobasic 21.6 g/L
Magnesium sulfate heptahydrate 24.24 g/L
Potassium sulfate 8.4 g/L
Sodium sulfate 0.672 g/L
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
38
Preparation notes:
Batch- and feed salt mixes as well as batch and feed glucose were prepared in
separate
BlueCap bottles by dissolving components in Milli-Q water and sterilizing by
autoclava-
tion.
Feeding solution:
A feeding solution was made by mixing 500 mL of feed glucose with 500 mL of
feed salt
mix, 10 mL of vitamin mix, 10 mL of micro elements solution and 1 mL of trace
elements
solution.
Example 1: Expression in Nicotiana benthamiana
Leaf material of N. benthamiana co-expressing specific combinations of genes
of interest
(G01) was harvested 7 days after agrobacterial infiltration. 1mL methanol
(Me0H) was
added to 2 leaf disks (0 = 2cm). Extraction was done at room temperature at
200rpm
orbital shaking. 200 uL of extract was filtered by using a 0:22 pm 96-well
filter plate
(Merck Millipore, Darmstadt, Germany) and at stored at 4 C prior to LC-MS
analysis.
Figure 1 shows the obtained LCMS profiles. The results show that the N.
benthamiana
cells transformed with 0YP82D274V1 encoding the enzyme having SEQ ID NO: 1,
leads
to production of 14-0H-dehydroabietadiene; when the cells are further
transformed with
CYP71BE85 and CYP71BE86 encoding the enzymes having the amino acid sequences
SEQ ID NO: 3 and SEQ ID NO: 4, respectively, triptophenolide is formed; and
when the
cells are further transformed with 0YP82D213 encoding the enzyme having the
amino
acid sequence of SEQ ID NO: 5 triptonide is formed. Further, it can be seen
that the
enzyme having the sequence of SEQ ID NO:6 and encoded by the gene 0YP82D217
increases the production of triptophenolide and triptonide.
Example 2: Construction of S. cerevisiae strains.
Strain construction
Parent yeast strain was S. cerevisiae S2880 (NCYC 3608; National Collection of
Yeast
Cultures, Norwich, UK).
Genotypes and source of strains are listed in table 3.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
39
Constructed yeast strains were made using the lithium acetate transformation
method
(8). Parent strains without functional URA3 were made competent by the
following pro-
cedure: Inoculation from a glycerol stock into 5 ml YPD medium and growing at
30 C
0/N. Then, transfer of 3 mL of 0/N culture to 50 mL YPD medium and continued
growing
for 4-5 hours followed by centrifugation at 4000 RPM for 10 minutes then
discarding the
supernatant. Cells were then ready for transformation after 2 washes in
sterile water (1st
in 25 mL, 2nd in 1 mL) and resuspension in 0.4 mL of sterile water.
Transformation of competent yeast cells was done by the following procedure:
Mixes of
designated Notl digested plasmids (2 pL of each) were each added 10 pL
competent
yeast cells and mixed with 60 pL PEG 3350 (50% w/v), 9 pL LiAc (1 M) and 12.5
pL
preboiled salmon sperm DNA (10 mg/ml). The resulting mixes were next incubated
at
42 C for 40 minutes before cells were collected by centrifugation (3000 RPM
for 5
minutes) and removal of supernatant. Cells were then resuspended in 100 pL
sterile
water and spread on SC without uracil agar plates. Isolated transformants
appeared as
single colonies after 2 days of incubation at 30 C. Insertion of gene
constructs was con-
firmed by colony PCR, using gene and construct specific primers found in table
1. For
colony PCR, yeast colonies were resuspended in 50 pL 20mM NaOH and incubated
at
99 C for 15 min. 1pL colony suspension was used for PCR.
Table 1: List of primers used
Number in
Target Target vector
(Entry
Name Sequence Sequence
and/or Destination)
listing gene
CO_TwCYP71BE85v1_TEF-F AGCGATACGNAAAATGGACTTATTGCA- 10 CO_TwCYP
ATTTCCATCTG 71BE85v1 pX-3-Ass1-
KIURA3
CO_TwCYP71BE85v1_TEF-R CACGCGANTCAGTTAAATGCGGGTGATGG 11
CO_TwGA30X1_TEF-F 12 CO_TwGA
AGCGATACGNAAAATGAGTCCTCCGCCTACAATA 30X1 pX-3-Ass3
CO_TwGA30X1_TEF-R CACGCGANTTAAATACCTAAAAGCGAGACGGG 13
CO_AcoUGT2_TEF-F AGCGATACGNAAAATGGCTGTTAGCTTAAAAAA- 14 CO_AcoU
TACCG GT2 pX-3-Ass3
CO_AcoUGT2_TEF-R CACGCGANTTAACGACTGATATGAGCGACG 15
CO_TwCYP82D213_PGK-F 16 CO_TwCYP
ATCAACGGGNAAAATGGAATTCCTTCTGTCATTGC 82D213 pX-3-Ass3
CO_TwCYP82D213_PGK-R CGTGCGANCTAACCCATGTAAAGATGTGATGG 17
CO_TwCYP71BE86_PGK-F ATCAACGGGNAAAATGGACTTACAATTACCTAG- 18 CO_TwCYP
CTTCC 718E86 pX-3-Ass1-
KIURA3
CO_TwCYP71BE86_PGK-R CGTGCGANTTAACCAGATAAACTACGATATGGG 19
TwCYP82D217_pLife-F GGCTTAANAAGCATCTTCTCTCCTAACTAGCTTTC- 20 TwCYP82D
TAAAT 217 pLife
TwCYP82D217_pLife-R GGTTTAANCTATTGCAATTCACCCCATGTAGACAA 21
pLifeUP_TEF-F 22 TwCYP82D
AGCGATACGNGACCTGCAGGCTGAGGCTT 217 pAss2
pLife_TEF-R CACGCGANCCCGGGGCTGAGGTTTAAT 23
CA 03192028 2023-02-16
WO 2022/043461
PCT/EP2021/073656
TwCYP82D274v1_pLife-F 24 TwCYP82D
GGCTTAANATGGAGTTTCTTCTTTCACTCCCAACA 274v1 pLife
TwCYP82D274v1_pLife-R GGTTTAANTCAGCCCATATAGAGATGAGCTGGGAG 25
pLife_TEF-F 26 TwCYP82D
AGCGATACGNTGCAGGCTGAGGCTTAATATG 274v1 pX-4-51-KIURA3
pLife_TEF-R CACGCGANCCCGGGGCTGAGGTTTAAT 27
TwCPR1_pLife-F GGCTTAANATGCAATCTTCTTCAAATTCTATGAAGG 28 TwCPR1 pLife
TwCPR1_pLife-R GGTTTAANTTACCACACATCCCGGAGATA 29
pLife_PGK-F ATCAACGGGNTGCAGGCTGAGGCTTAATATG 30 TwCPR1 pX-4-51-KIURA3
pLife_PGK-R CGTGCGANCCCGGGGCTGAGGTTTAAT 31
TwB5#1_pLife-F GGCTTAANATGGCTTCGGATCGGAAGATA 32 TwB5#1 pLife
TwB5#1_pLife-R GGTTTAANCTATTCTTTCTTGGTGAAGTGACGTA 33
pLife_PGK-F ATCAACGGGNTGCAGGCTGAGGCTTAATATG 34 TwB5#1 pAss2
pLife_PGK-R CGTGCGANCCCGGGGCTGAGGTTTAAT 35
TwB5#2_pLife-F GGCTTAANATGGGTGGAGACGGAAAGGTT 36 TwB5#2 pLife
TwB5#2_pLife-R GGTTTAANTTAAGCAGGAGGAGCTGATTTGGT 37
pLife_PGK-F ATCAACGGGNTGCAGGCTGAGGCTTAATATG 38 TwB5#2 pAss2
pLife_PGK-R CGTGCGANCCCGGGGCTGAGGTTTAAT 39
TwB5#3_pLife-F GGCTTAANATGGCTGGTCAGAGAGTTTTCAC 40 TwB5#3 pLife
TwB5#3_pLife-R GGTTTAANTTAGAAGATCTGCTCAGGCCTTGTA 41
pLife_PGK-F ATCAACGGGNTGCAGGCTGAGGCTTAATATG 42 TwB5#3 pAss2
pLife_PGK-R CGTGCGANCCCGGGGCTGAGGTTTAAT 43
TwB5#4_PGK-F ATCAACGGGNAAAATGGC- 44
TAAACTTCTTTCATTTGCTGAG TwB5#4 pAss2
TwB5#4_PGK-R CGTGCGANTTAGAAAAGGTATCGCAAACCAAAT- 45
GCC
TwB5#5_PGK-F ATCAACGGGNAAAATGATTATTGTTGCGGT- 46
GGCTCTGA TwB5#5 pAss2
TwB5#5_PGK-R CGTGCGANTTACTTCTCTAGATCCCCAATG- 47
TAAAAATCATCG
TwB5#6_PGK-F ATCAACGGGXAAAATGCCGACTTTAACGAAGCTG- 48
CAC TwB5#6 pAss2
TwB5#6_PGK-R CGTGCGAXCTACTTCTTCCGCAAGTACAGGAGTC 49
YEA85_UP_Genotyping_Fw Genoty-
TCTCAGGTATAGCATGAGGTCGCTCAT 50 ping
UP_Genotyping
YEA86_DW_Genotyping_Fw Genoty-
CCTGCAGGACTAGTGCTGAGGCATTAAT 51 ping
DW_Genotyping
YEA87_X-2_Genotyping_UP Genoty-
GTTTGTAGTTGGCGGTGGAG 52 ping X-
2_Genotyping
YEA88_X-2_Genoty- Genoty-
ping_DW GAGACAAGATGGGGCAAGAC 53 ping X-
2_Genotyping
YEA89_X-3_Genotyping_UP Genoty-
TGACGAATCGTTAGGCACAG 54 ping X-
3_Genotyping
YEA9O_X-3_Genoty- Genoty-
ping_DW CCGTGCAATACCAAAATCGAG 55 ping X-
3_Genotyping
YEA91_X-4_Genotyping_UP Genoty-
CTCACAAAGGGACGAATCCT 56 ping X-
4_Genotyping
YEA92_X-4_Genoty- Genoty-
ping_DW GACGGTACGTTGACCAGAG 57 ping X-
4_Genotyping
YEA93_XI-1_Genotyping_UP Genoty-
CTTAATGGGTAGTGCTTGACACG 58 ping XI-
1_Genotyping
YEA94_XI-2_Genotyping_UP Genoty-
GTTTGTAGTTGGCGGTGGAG 59 ping XI-
2_Genotyping
YEA95_X1-2_Genoty- Genoty-
ping_DW GAGACAAGATGGGGCAAGAC 60 ping X1-
2_Genotyping
YEA96_XI-5_Genotyping_UP Genoty-
CTCAATGATCAAAATCCTGAATGCA 61 ping XI-
5_Genotyping
YEA97_X1-5_Genoty- Genoty-
ping_DW GCATGGTCACCGCTATCAGC 62 ping X1-
5_Genotyping
YEA98_XII-2_Genoty- Genoty-
ping_UP CGAAGAAGGCCTGCAATTC 63 ping XII-
2_Genotyping
YEA99_X1I-2_Genoty- Genoty-
ping_DW GGCCCTGATAAGGTTGTTG 64 ping X1I-2_Genotyping
CA 03192028 2023-02-16
WO 2022/043461
PCT/EP2021/073656
41
YEA100_XII-5_Genoty- Genoty-
ping_UP CCACCGAAGTTGATTTGCTT 65 ping XII-
5_Genotyping
YEA101_X1I-5_Genoty- Genoty-
ping_DW GTGGGAGTAAGGGATCCTGT 66 ping XII-
5_Genotyping
Assembly of genetic constructs for S. cerevisiae genome engineering
Plasmid names and encoded gene constructs are listed in table 2. All plasmids
were
generated by USER cloning as previously described (5). Also, parent vectors
named
assembler -1, -2 and -3, for simultaneous genome integration of up to six gene
con-
structs, and harboring AsiSI/Nb.Bsml USER-cassettes, were prepared for USER
cloning
as previously described (6). Primers used for PCR amplification with USER
compatible
PfuX7 polymerase (7) are listed in table 1. Vectors used and generated in this
work is
listed in table 3.
Codon optimized genes for S. cerevisiae were acquired from TWIST Biosciences,
USA,
San Francisco. All genes denoted with the prefix "CO_" in the below tables
were codon-
optimized. Codon-optimized genes were amplified using primers identical to
those de-
scribed in Table 1 above, except that the primers were modified to accommodate
hybrid-
ization to any nucleotide changes in the codon-optimized genes. The primers
for ampli-
fication of the codon-optimized genes are also disclosed in J. Andersen-
Ranberg etal.,
Expanding the Landscape of Diterpene Structural Diversity through
Stereochemically
Controlled Combinatorial Biosynthesis. Angewandte Chemie International
Edition, n/a
(2016).
Table 2: Vectors and plasm ids generated and used
Vectors for yeast genome-integration
Name Description Source
pCYT183 pX-3-Ass1-KIURA3- This
study
PpTDH3::CO_TwCYP71BE86
pCYT184 pX-3-Ass1-KIURA3- This
study
TpCCW12::CO_TwCYP71BE85v1
pCYT185 pX-3-Ass1-KIURA3- This
study
PpTDH3::CO_TwCYP71BE86-
TpCCW12::CO_TwCYP71BE85v1
pCYT186 pX-3-Ass3-PpEN02::CO_TwCYP82D213 This
study
pCYT187 pX-3-Ass3-TpPDC1::CO_TwGA0X1 This
study
pCYT188 pX-3-Ass3-PpEN02::CO_TwCYP82D213- This
study
TpPDC1::CO_TwGA0X1
CA 03192028 2023-02-16
WO 2022/043461
PCT/EP2021/073656
42
p349 pAss2-PpFBA1::TwB5#2- This
study
TpSED1::TwCYP82D217
p350 pAss2-PpFBA1::TwB5#3- This
study
TpSED1::TwCYP82D217
p351 pAss2-PpFBA 1: :TwB5#1- This
study
TpSED1::TwCYP82D217
p352 pAss2-PpFBA1::TwB5#6- This
study
TpSED1::TwCYP82D217
p353 pAss2-PpFBA1::TwB5#5- This
study
TpSED1::TwCYP82D217
p354 pAss2-PpFBA1::TwB5#4- This
study
TpSED1::TwCYP82D217
p355 pAss2-TpSED1::TwCYP82D217 This
study
pJAR1 pX-3-Ass3-TpPDC1::CO_AcUGT2 This
study
pJAR2 pX-3-Ass3-PpEN02::CO_TwCYP82D213- This
study
TpPDC1::CO AcUGT2
p320 pX-4-SI-PpTF2::TwCPR1- This
study
TpTDH3::TwCYP82D274v1
pVictor1 pXI-2-Ass1-pICL1::CO_SpGGPPS7 This
study
pVictor2 pAss2-pPGK1::CO_CftTPS3 This
study
pVictor3 pXI-2-Ass3-pTEF 1:: CO_CftTPS3 This
study
pAss2A-PpPGK1::CO CftTPS1- This
study
pCYT85 TpTEF 1:: CO_CftTPS
pX-3-Ass3-PpSED1::TwCYP82D274v1- This
study
pTRIP10 TpFBA1::Twb5#1
pTRIP108 pAss2C-PpEN02::CO_TwCYP71BE85 .................. This
study
pTRIP110 pX-3-Ass3-PpSED1::TwCYP82D274v1 This
study
pTRI P14 pSIXI-2-PpPGK1::TwCPR1-TpTPI 1: :Twb5#4 This
study
pAss2B-PpCCW12::CO TwCYP82D213- This
study
pTRIP4 TpSED1::CO TwCYP7TBE86
pAss2C-PpEF102::CO TwCYP71BE85- This
study
pTRIP5 TpPDC1::CO TwCYP82D213
pX11-2-Ass1-1=7pPGK1::CO TwCYP71BE86- This
study
pTRI P50 TpTP11::CO TwCYP71BE-85
pAss2A-PpfDH3::CO TwCYP82D213- This
study
pTRI P52 TpSED1::CO TwCYP82D213
pAss2B-PpP5C1::TwCYP82D274v1- This
study
pTRI P53 TpEN02::TwCYP82D274v1
pAss2C-PpTEF 1: :TwCYP82 D213- This
study
pTRI P54 TpPGK1::TwCYP82D213
pTRI P55 pXI I-2-Ass3-PpSED1: :Twb5#1-TpFBA 1: :Twb5#4 This study
pX-3-Ass1-pTEF2::TwCPR1-TpICL1::CO_SpGG- This study
pTRIP7 PPS7
pX-3-Ass1-pTEF2::TwCPR2-TpICL1::CO_SpGG- This study
pTRIP8 PPS7
pTRI P88 pAss2B-TpSED1::CO TwCYP71BE86 This
study
pX-3-Ass3-PpSED1::fwCYP82D274v3- This
study
pTRI P92 TpFBA1::Twb5#1
pX-3-Ass3-PpSED1::TwCYP82D274v4- This
study
pTRI P95 TpFBA1::Twb5#1
CA 03192028 2023-02-16
WO 2022/043461
PCT/EP2021/073656
43
pAss2C-PpEN02::CO TwCYP71BE85- This
study
pTRI P89 TpPDC1::CO_TwCYP82D213v2
pTRIP3 pAss2-PpFBA1::Twb5#1 This
study
pX-3-Ass1-PpTDH3::CO TwCYP71BE86- This
study
pCYT185 TpCCW12::CO_TwCYPf1BE85
pX-3-Ass3 pX-3-Ass3 empty This
study
pX-3-Ass1 pX-3-Ass1 empty This
study
pAss2A pAss2A empty This
study
pAss2B pAss2B empty This
study
pAss2C pAss2C empty This
study
pVic1 ............. pLIFE-SctHMGR This
study
pVic2 pLI FE-CftTPS1 This
study
pVic3 pLI FE-CftTPS3 This
study
pVic4 pLI FE-SpGGPPS7 This
study
239.TwCYP756A
1 ................ pLI FE-TwCYP82D274v1 This
study
59.TwCYP10
81.TwCYP9 pLIFE-TwCYP71BE85 This
study
81.TwCYP9 pLIFE-TwCYP71BE86 This
study
297.TwB5#1 pLI FE-Twb5#1 This
study
46.TwCYP17 pLI FE-TwCYP82D213 This
study
Voin net et
al., 2003
P19 pBin61-p19 (ref.: 10) ..
pDXS pLI FE-CfDXS This
study
pCfTPS1 pLI FE-pCfTPS1 This
study
pCfTPS2 pLI FE-pCfTPS2 This
study
pCfGGPPS pLI FE-pCfGGPPS This
study
Table 3: List of S. cerevisiae strains used and generated.
Name Genotype Source
MATa, SUC2, gaI2, maI2, mel, flo1, ti08-1 National Collection of,
S288c Yeast Cultures
ho, bio1, bio6
(NCYC)
MATa, SUC2, gaI2, maI2, mel, tio1, ti08-1,
ho, bio1, b1o6, ura3A::KanMX, XI-2::(pTEF1-
NVJO
CO_CftTPS1/pPGK1-00 CftTPS3/pICL1 This study
-
CO_SpGGPPS7/KIURA3)
CA 03192028 2023-02-16
WO 2022/043461
PCT/EP2021/073656
44
MATa, SUC2, gaI2, maI2, mel, flo1, fl08-1,
ho, bio1, b1o6, ura3A::KanMX, XI-2::(pTEF1-
NVJ1-3.5 CO_CftTPS1/pPGK1-00 CftTPS3/pICL1- This study
CO_SpGGPPS7), X-4::(pTDH3-
TwCYP82D274v1/pTEF2-TwCPR1/KIURA3)
MATa, SUC2, gaI2, maI2, mel, flo1, flo8-1,
ho, bio1, b1o6, ura3A::KanMX, XI-2::(pTEF1-
CO_CftTPS1/pPGK1-CO CftTPS3/pICL1-
NVJ3.10 CO_SpGGPPS7), X-4::(pTDH3- This study
TwCYP82D274v1/pTEF2-TwCPR1), X-
3::(pTDH3-CO_TwCYP71BE86/pCCW12-
CO_TwCYP71BE85v1/pFBA1-Twb5#1)
MATa, SUC2, gaI2, maI2, mel, flo1, fl08-1,
ho, bio1, b1o6, ura3A::KanMX, XI-2::(pTEF1-
CO_CftTPS1/pPGK1-CO CftTPS3/pICL1-
CO_SpGGPPS7), X-4::(pTDH3-
NVJ2-19 TwCYP82D274v1/pTEF2-TwCPR1), X- This study
3::(pTDH3-CO_TwCYP71BE86/pCCW12-
CO_TwCYP71BE85v1/pEN02-
CO_TwCYP82D213/pSED1-
TwCYP82D217/pFBA1-TwB5#1/KIURA3)
MATa, SUC2, gaI2, maI2, mel, flo1, fl08-1' This study
NVJ11-0
ho, bio1, b1o6, ura3A::KanMX, X-3::(KIURA3)
MATa, SUC2, gaI2, maI2, mel, flo1, flo8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
NVJ11-1 This study
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
CO_CftTPS1/pTEF1-00 CftTPS3/KIURA3)
MATa, SUC2, gaI2, maI2, mel, flo1, flo8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
NVJ11-2 TwCPR1/pICL1-CO_SpGGPPS7/pPGK1- This study
CO_CftTPS1/pTEF1-00 CftTPS3/pSED1-
TwCYP82D274V1/KIURA3)
MATa, SUC2, gaI2, maI2, mel, flo1, flo8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
NVJ11-3 This study
CO_CftTPS1/pTEF1-00 CftTPS3/pEN02-
CO_TwCYP71BE85v1/pSED1-
TwCYP82D274V1/KIURA3)
CA 03192028 2023-02-16
WO 2022/043461
PCT/EP2021/073656
MATa, SUC2, gaI2, maI2, mel, flo1, tio8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
NVJ11-4 This study
CO_CftTPS1/pTEF1-00 CftTPS3/pSED1-
CO_TwCYP71BE86/pSED1-
TwCYP82D274V1/KIURA3)
MATa, SUC2, gaI2, maI2, mel, flo1, ti08-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
NVJ11-5 CO_CftTPS1/pTEF1-00 CftTPS3/pSED1- This study
CO_TwCYP71BE86/pEN02-
CO_TwCYP71BE85v1/pSED1-
TwCYP82D274V1/KIURA3)
MATa, SUC2, gaI2, maI2, mel, flo1, ti08-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
CO_CftTPS1/pTEF1-CO CftTPS3/pSED1-
NVJ11-6 This study
CO_TwCYP71BE86/pEN02-
CO_TwCYP71BE85v1/pPDC1-
CO_TwCYP82D213/pSED1-
TwCYP82D274V1/KIURA3)
MATa, SUC2, gaI2, ma/2, mel, flo1, tio8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
NVJ11-7 TwCPR1/pICL1-CO_SpGGPPS7/pPGK1- This study
CO_CftTPS1/pTEF1-00 CftTPS3/pSED1-
TwCYP82D274V1/pFBA1-Twb5#1/KIURA3)
MATa, SUC2, gaI2, ma/2, mel, flo1, tio8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
NVJ11-8 This study
CO_CftTPS1/pTEF1-00 CftTPS3/ pEN02-
CO_TwCYP71BE85v1/pSED1-
TwCYP82D274V1/pFBA1-Twb5#1/KIURA3)
MATa, SUC2, gaI2, ma/2, mel, flo1, tio8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
NVJ11-9 This study
CO_CftTPS1/pTEF1-00 CftTPS3/pSED1-
CO_TwCYP71BE86/pSED1-
TwCYP82D274V1/pFBA1-Twb5#1/KIURA3)
MATa, SUC2, gaI2, ma/2, mel, flo1, tio8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
NVJ11-10 CO_CftTPS1/pTEF1-00 CftTPS3/pSED1- This study
CO_TwCYP71BE86/pEN02-
CO_TwCYP71BE85v1/pSED1-
TwCYP82D274V1/pFBA1-Twb5#1/KIURA3)
CA 03192028 2023-02-16
WO 2022/043461
PCT/EP2021/073656
46
MATa, SUC2, gaI2, maI2, mel, flo1, tio8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
CO_CftTPS1/pTEF1-CO CftTPS3/pSEDI-
NVJ11-11 This study
CO_TwCYP7IBE86/pEN02-
CO_TwCYP7IBE85v1/pPDC1-
CO_TwCYP82D213/pSED1-
TwCYP82D274V1/pFBA1-Twb5#1/KIURA3)
MATa, SUC2, gaI2, maI2, mel, flo1, tio8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR2/pICL1-CO_SpGGPPS7/pPGK1-
CO_CftTPS1/pTEF1-CO CftTPS3/pCCW12-
CO_TwCYP82D213/pSED1-
CO_TwCYP71BE86/pEN02-
CO_TwCYP71BE85v1/pPDC1-
TwCYP82D213/pSED1-TwCYP82D274V1)
X1-2::(pPGK1-TwCPR1/pTP11-Twb5#4) XII-
NVJ8-15 This study
2 :: (pPGKI -CO_TwCYP71 BE86/pTPI I -
TwCYP71BE85v1/pTDH3-
CO_TwCYP82D213/pSED1-
CO_TwCYP82D213/pPDC1-
TwCYP82D274V1/pEN02-
TwCYP82D274V1/pTEF1-
TwCYP82D213/pPGK1-
TwCYP82D213/pSED1-Twb5#1/pFBA1-
Twb5#4/KIURA3)
MATa, SUC2, gaI2, maI2, mel, flo1, tio8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
CO_CftTPS1/pTEF1-COCftTPS3/pSED1-
NVJ10-1 This study
CO_TwCYP71BE86/pEN02-
_CO_TwCYP71BE85v1/pPDC1-
CO_TwCYP82D213/pSED1-
TwCYP82D274v1/pFBA1-Twb5#1/KIURA3)
MATa, SUC2, gaI2, maI2, mel, flo1, tio8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
CO_CftTPS1/pTEF1-CO_CftTPS3/pSED1-
NVJ10-3 This study
CO_TwCYP7IBE86/pE102-
CO_TwCYP7IBE85v1/pPDC1-
CO_TwCYP82D213/pSED1-
TwCYP82D274v3/pFBA1-Twb5#1/KIURA3)
MATa, SUC2, gaI2, ma/2, mel, flo1, tio8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
CO_CftTPS1/pTEF1-CO CftTPS3/pSED1-
NVJ10-6 This study
CO_TwCYP7IBE86/pEN02-
CO_TwCYP7IBE85v1/pPDC1-
CO_TwCYP82D213/pSED1-
TwCYP82D274v4/pFBA1-Twb5#1/KIURA3)
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
47
MATa, SUC2, gaI2, maI2, mel, flo1, tio8-1,
ho, bio1, b1o6, ura3A::KanMX, X-3::(pTEF2-
TwCPR1/pICL1-CO_SpGGPPS7/pPGK1-
CO_CftTPS1/pTEF1-CO CftTPS3/pSED1-
NVJ10-8 This study
CO_ TwC YP71BE86/pEN02-
CO_TwCYP71BE85v1/pPDC1-
CO_ TwC YP82D213v2/pSED1-
TwCYP82D274V1/pFBA1-Twb5#1/KIURA3)
Example 3: Expression in the yeast, Saccharomyces cerevisiae
Extraction and metabolite analysis
Genetically engineered S. cerevisiae strains were transferred into 0.5mL media
in a 96-
well plate and grown for 3 days at 30 C with orbital shaking at 350 rpm. For
extraction
0.1mL of S. cerevisiae culture was transferred to 1.5mL glass vials. 0.4mL
Met0H
uHPLC grade was added. S. cerevisiae extracts extract was filtered by using a
0:22 pm
96-well filter plate (Merck Millipore, Darmstadt, Germany) and at stored at 4
C prior to
LC-MS analysis.
LCMS profiles of the extracts can be seen in figure 2, where it can be
observed that
transforming the background strain with TwCYP82D274V1 encoding the enzyme
having
the amino acid sequence of SEQ ID NO: 1 leads to formation of 14-0H-
dehydroabietadi-
ene.
Example 4: Detection of 14-0H-dehydroabietadiene by NMR analysis
The compound identified as 14-0H-dehydroabietadiene in example 3 was analyzed
by
NMR to confirm its identity.
Purification for NMR
Purification of triptolide intermediates from engineered yeast
Engineered yeast producing the desired compound of interest was inoculated
from SC-
Agar in 10mL YDP and grown ON at 30 C. 5 mL ON culture was inoculated in 500mL
FIT media and grown for 5 days at 30 C. Compound of interest was extracted
from cul-
ture with 500mL EtAc. Solvent was removed by rotor evaporation and analytes
were
resuspended in hexane. Extraction was repeated 3 times. Hexane extract was
applied
on Supelclean TM Florisile/Na2SO4 SPE Tube (Sigma-Aldrich) and analytes were
eluted
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
48
from column using a step gradient with 1:99-5:95 EtAc:Hexane. Each fraction
was ana-
lyzed with either LC-MS or GC-MS and the fraction containing the compound of
interest
was selected for NMR analysis.
NMR analysis
NMR data were acquired on a Bruker Avance III HD 600 MHz NMR spectrometer (1H
operating frequency 599.85 MHz) equipped with a 5 mm cryogenically cooled DCH
probe
optimized for 130 and 1H (Bruker Biospin, Karlsruhe, Germany). NMR data was
rec-
orded in 5 mm tubes in CDCI3 (Euriso-top, 99.8 atom % D) with temperature
equilibration
to 300 K, optimization of lock parameters, gradient shimming, and setting of
receiver
gain, all automatically controlled by Topspin ver. 3.2 and IconNMR ver. 4.7.5
(Bruker
Biospin, Karlsruhe, Germany). 1H and 130 chemical shifts were referenced to
the resid-
ual solvent signals of at respectively pH 7.26 ppm and p077.16 ppm. 1D 1H and
130
NMR spectra were acquired with 30 pulses and 64k data points and zero-filled
to 256k
data points, 1H spectra were acquired with a spectral width of 12 kHz, a
relaxation delay
of 1 sand an acquisition time of 2.7s. 130 spectra were 1H-decoupled using the
Waltz-
16 composite pulse decoupling scheme. 2D homo- and heteronuclear experiments
were
acquired with 4096 (HMBC), 2048 (DQF-COSY and ROESY), or 1024 (multiplicity
edited
HSQC) data points in the direct dimension and 256 (DQF-COSY, HMBC and ROESY)
or 128 (multiplicity edited HSQC) data points in the indirect dimension. 2D
NMR data
was zero-filled to 1k in F1 and zero-filled to twice the number of points in
F2, employing
forward linear prediction in F1 (LPBIN=0). Processing of NMR data was done
using Top-
spin ver. 4Ø9 (Bruker Biospin, Karlsruhe, Germany).
The NMR spectroscopic data for 14-0H-dehydroatietadiene is shown in table 4.
Table 4. 1H and 13C NMR spectroscopic data of 1 (14-0H-dehydroatietadiene)
14-0H-dehydroabietadiene (1)
Pos ee (51-1, nH, multiplicity (J in Hz)a b HMBC ROESY
1 39.1 2, 3, 5, 10,20 2A, 2B, 11,
A: 2.28, 1H, bid (12.8)
2, 3, 9, 10,20 (20)
B: 1.40, 1H, td (12.8, 3.4)
(11)
2 19.5 A: 1.75, 1H, m 1, 3, 4 1B
B: 1.62, 1H, m 1, 3, 4, 10 1B
3 41.8 A: 1.49, 1H, bid (13.2) 1, 2, 4, 5, 18, 19 18
B: 1.22, 1H, m 2,4, 18, 19
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
49
4 33.5 -
49.9 1.34, 1H, dd, (12.7, 2.1) 4, 6, 9, 10, 19,20 18
6 18.6 A: 1.99, 1H, br dd (13.1, 7.9) 4, 5, 7, 8, 10 7B, 18
B: 1.72, 1H, m 5, 7, 10 7A, 19
7 24.5 A: 2.82, 1H, dd (16.5, 6.7) 5, 6, 8, 9, (14) 6B, 14-
0H
B: 2.62, 1H, ddd (16.5, 11.4, 7.9) 6, 8, 9 6A, 14-0H
8 120.8 -
9 149.2 -
37.7 -
11 116.5 6.87, 1H, d (8.2) 8, 10, (12), 13, (14) 1A, (16),
(20)
12 123.4 7.02, 1H, d (8.2) 9, (11), 14, 15 16, 17
13 130.1 -
14 150.4 -
14-0H - 4.63, 1H, s 8, 13, 14 7A, 7B, 15
27.0 3.16, 1H, sep (6.9) 12, 13, 14, 16, 17 14-0H, 16, 17
16 22.9 1.24, 3H, d (6.9) 13, 15, 17 12,15
17 22.7 1.26, 3H, d (6.9) 13, 15, 16 12,15
18 33.4 0.97, 3H, s 3, 4, 5, 19 3B, 5, 6A
19 21.8 0.94, 3H, s 3,4, 5, 18 6B, 20
25.0 1.20, 3H, s 1, 5, 9, 10 (16), (11), 19
a 1H NMR (599.85 MHz) and 13C NMR (150.83 MHz) data obtained in CDCI3.
b nH = number of hydrogens. Multiplicities reported as apparent splittings: s
= singlet, d = doublet, t = triplet, sep = septet,
m = multiplet (also in case of overlap), br = broad. 'A' denotes the highest
chemical shift value and 'B' denotes the lowest
chemical shift value.
5
The 1H NMR spectrum of 14-0H-dehydroatietadiene in CDCI3 at 599.85 MHz is
shown
in figure 3 and the 130 NMR spectrum of 14-0H-dehydroatietadiene in CDCI3 at
150.83
10 MHz is shown in figure 4 confirming the identity of the compound.
Example 5: Expression in S. cerevisiae of genes leading to production of
tripto-
phenolide and triptonide
15 This was a preliminary study to assess the effects of expression of
genes leading to
production of triptophenolide and triptonide in S. cerevisiae.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
The background yeast strain generated in example 2, was further transformed
with vec-
tors each containing individual diterpene biosynthesis genes or Triptetygium
wilfordii
CYPs (TwCYPs)
5 LCMS profiles of the extracts can be seen in figure 5, where it can be
seen that trans-
forming the background strain with TwCYP82D274V1, TwCYP71BE85 and
TwCYP71BE86 encoding the enzymes having the amino acid sequence of SEQ ID NO:
1, SEQ ID NO: 3 and SEQ ID NO: 4, respectively; leads to formation of
triptophenolide;
and further transformation with 0YP82D213 encoding the enzyme having the amino
acid
10 sequence of SEQ ID NO: 5, leads to the formation of triptonide.
Figure 6 shows an overview of the content of oxygenated diterpenoid compounds
de-
tected in the extracts of the transformants generated.
15 The left panel shows the content of triptophenolide and triptonide, and the
right panel
shows the content of 14-0H-dehydroabietadiene. Expressing the gene TwB5#1
encod-
ing the enzyme having the amino acid sequence of SEQ ID NO: 8 resulted in a
signifi-
cantly higher production of triptophenolide and triptonide.
20 The genes TwB5#2-6 are other T.wilfordii cytochrome B5 genes (sequences not
pro-
vided) that do not increase the production of triptophenolide or triptonide.
Example 6: Production of oxygenated diterpenoid compounds in S. cerevisiae and
N. benthamiana
All engineered S. cerevisiae strains and N. benthamiana were cultured as
described
herein above in the section "Materials and Methods". Similarly, diterpenoid
metabolites
were extracted, analyzed by LC-MS, GC-MS and NMR, and quantified as also
described
herein above.
It is preferred that the experimental organism is yeast and that the
heterologous genes
have been stably transfected in the organism as this gives the most precise
and repro-
ducible results.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
51
The results are shown in Figures 7-9. As can be clearly seen from the figures,
organisms
as different as yeast cells and tobacco plants are both capable of producing
the claimed
key intermediates in the proposed biosynthetic pathway of triptonide at high
titers ac-
cording to the methods of the invention.
NMR spectra of other key compounds also produced are shown in Figures 10-26.
The
NMR spectroscopic data for the produced compounds are shown in Tables 5-21,
below.
52
o
Table 5. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F1-14
w
=
Pos. 5c, type" 8H, nH, multiplicity (J in Hz) a,c HMBC
ROESY w
w
'a
1 36.8, CH2 a: 1.49 (1H, m) C-2, C-3, C-9, C-10, C-20
H-113, H-3 .6.
(44
13: 2.27 (1H, dt, 13.3, 3.1) C-2, C-3, C-5, C-10, C-20
H-1a, H-2, H-11, H-20 .6.
o,
=
2 27.3, CH2 1.79 (2H, m) C-1, C-3, C-4, C-10
H-113, H-19, H-20
3 75.9, CH 3.69 (1H, t, 7.5) C-2, C-4, C-18, C-19
H-1a, H-5
4 42.0,C -
43.5, CH 1.45 (1H, br d, 11.8) C-1, C-4, C-6, C-7, C-9, C-10, C-
18, C- H-3, H-7A, H-18A
6 18.2, CH2 A: 1.75 (1H, m) C-5, C-7, C-10
B: 1.80 (1H, m) C-8, C-10
P
7 24.3, CH2 A: 2.62 (1H, m) C-6, C-8, C-9, C-14
H-5 -
,
B: 2.82 (1H, dd, 16.2, 5.1) C-5, C-6, C-8, C-9, C-14
8 120.7, C -
m
9 1479,C -
.
"
,
10 37.1, C -
.
,
,
11 116.4, CH 6.82 (1H, d, 8.2) C-8, C-10, C-13
H-113, H-20 .
12 123.3, CH 7.01 (1H, d, 8.2) C-9, C-14, C-15
H-15, H-16, H-17
13 130.4,C -
14 150.2,C -
15 26.7, CH 3.14 (1H, sep, 6.9) =C-12, C-13, C-14, C-16, C-
17 H-12
16 22.7, CH3 1.22 (3H, d, 6.9) C-13, C-15, C-17
H-12
17 22.6, CH3 1.24 (3H, d, 6.9) C-13, C-15, C-16
H-12
=n
18 71.0, CH2 A: 3.46 (1H, d, 8.9) C-3, C-4, C-5, C-19
H-5
m
B: 3.76 (1H, d, 8.9) C-3, C-4, C-5
,-o
w
19 11.2, CH3 0.96(3H, s) C-3, C-4, C-5, C-18
H-2, H-20 =
w
20 25.2, CH3 1.22 (3H, s) C-1, C-5, C-9, C-10
H-113, H-2, H-11, H-19 'a
-4
(44
01
CA
01
53
o
Table 6. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F1-15
w
=
Pos. 5c, type" 8H, nH, multiplicity (J in Hz) a,c HMBC
ROESY w
w
'a
1 32.7, CH2 a: 1.69 (1H, m) =C-2, C-10, C-20
H-113, H-2B, H-5 .6.
(44
13 : 2.46 (1H, m) C-2, C-5, C-9, C-20
H-1a, H-11, H-20 .6.
c,
2 22.5, CH2 A: 2.45 (1H, m) C-1, C-3, C-4, C-10
H-19B
B: 2.52 (1H, m) C-1, C-3, C-4
H-la, H-19A
3 132.0,C -
4 150.7,C -
39.2, CH 2.60 (1H, m)
H-1a, H-6a, H-7B
6 17.5, CH2 a: 3.28 (1H, m) C-8, C-10
H-5, H-7B
13: 1.73(1H, m) C-5, C-7, C-10
H-20 p
7 22.5, CH2 A: 2.46 (1H, m)
0
,
B: 2.80 (1H, td, 9.5, 7.6) C-5, C-6, C-8, C-9, C-14
H-6a, H5 0"
IV
00
8 120.8, C -
IV
0
IV
9 144.5,C -
,
0
36.1, C -
IV
F'
11 115.7, CH 6.91 (1H, d, 8.1) C-8, C-10, C-12, C-13
H-113,11-20
12 123.0, CH 7.04 (1H, d, 8.1) C-9, C-11, C-14, C-15 H-16, H-17
13 131.2,C -
14 150.9,C -
26.9, CH 3.18 (1H, sep, 7.0) C-12, C-13, C-14, C-16, C-17
16 22.6, CH3 1.24 (3H, d, 7.0) C-13, C-15, C-17
H-12
,-o
17 22.6, CH3 1.26 (3H, d, 7.0) C-13, C-15, C-16
H-12 n
,-i
18 173.5,C -
m
19 53.9, CH2 A: 3.90 (1H, dd, 18.7, 2.1) C-3, C-4, C-18
H-2B, H-1'
w
=
B: 3.94 (1H, dd, 18.7, 2.7) C-3, C-4, C-18
H-2A, H-1' w
'a
22.4, CH3 1.03 (3H, s) C-1, C-5, C-9, C-10
H-1(3, H-6(3, H-11 -4
(44
1' 46.1, CH2 A: 3.56 (1H, m) C-18, C-19, C-2'
H-19
u,
c.,
54
o
B: 3.63(1H, m) C-18, C-19, C-2'
H-19 w
=
2' 62.2, CH2 3.82 (2H, m) C-1'
w
w
'a
.6.
(44
4=,
01
I-,
Table 7. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F1-18
Pos. gc, type" 5H, nH, multiplicity (J in Hz)" HMBC
ROESY
1 34.0, CH2 a: 1.23 (1H, m) C-2, C-10, C-20
H-113, H-3
13: 2.79 (1H, dt, 13.5, 3.3) C-3, C-5, C-10
H-la, H-2, H-20
2 27.1, CH2 1.76 (2H, m) C-1, C-3, C-4, C-10
H-113, H-20
3 75.1, CH 3.68 (1H, dd, 10.3, 5.8) C-2, C-4, C-18, C-19
H-1a, H-5, H-18A
4 42.0,C -
P
45.2, CH 1.29 (1H, d, 12.5) =C-4, C-6, C-7, C-10, C-18, C-19,
C-20 H-3, H-7B, H-18A -
,
6 17.1, CH2 a: 1.73 (1H, m) C-5, C-7, C-8, C-10
H-7B, H-18A, H-18B
13: 1.48(1H, qd, 12.5, 5.7) C-5, C-7, C-10
H-7B, H-19 3
7 25.8, CH2 A: 2.32 (1H, ddd, 20.2, 11.6, 7.4) C-6, C-8, C-9, C-14
H-7B, H-6a
,
0
B: 2.69 (1H, dd, 20.2, 5.5) C-5, C-6, C-8, C-9, C-14
H-5, H-6a, H-613, H-7A F'N'
8 142.6,C -
9 149.9, C -
38.0,C -
11 187.7,C -
12 131.8, CH 6.31 (1H, s) C-9, C-11, C-14, C-15
H-15, H-16/17
13 152.9,C -
14 187.6,C -
n
,-i
26.2, CH 2.97 (1H, sep, 6.9) C-12, C-13, C-14, C-16, C-17
H-12 m
16 21.2, CH3 1.08 (3H, d, 6.9) C-13, C-15, C-17
H-12
w
=
17 21.2, CH3 1.09 (3H, d, 6.9) C-13, C-15, C-16
H-12 w
'a
18 70.4, CH2 A: 3.42 (1H, d, 10.3) C-3, C-4, C-5, C-19
H-3, H-5, H-6a, H-19 -4
(44
B: 3.74 (1H, d, 10.3) C-3, C-4, C-5, C-19
H-6a, H-19 o,
u,
o,
55
o
19 11.6, CH3 092(3H, s) C-3, C-4, C-5, C-18
H-613, H-18A, H-18B, H-20 w
o
20 20.4, CH3 1.31 (3H, s) C-1, C-5, C-9, C-10
H-113, H-2, H-19 w
w
'a
.6.
(44
4=,
01
I-,
Table 8. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F1-23
(F15P1)
Pos. 5c, type" 8H, nH, multiplicity (J in Hz) a,c
HMBC ROESY
1 37.1, CH2 a: 1.55 (1H, td, 13.2, 4.2) C-2, C-3, C-9, C-10, C-20
H-3
13: 2.31 (1H, dt, 13.2, 3.5) C-2, C-3, C-5, C-10, C-20
H-2, H-11, H-20
2 27.9, CH2 1.80 (2H, m) C-1, C-3, C-4, C-10
H-1(3
3 78.7, CH 3.31 (1H, dd, 11.5, 4.7) C-2, C-4, C-18, C-19
H-la, H-5, H-18
4 38.9,C -
P
49.2, CH 1.32 (1H, dd, 12.5, 2.0) C-3, C-4, C-6, C-7, C-9, C-10, C-18, C-
19, C- H-la, H-3, H-7a, H-18 2
,
20
.
"
"
6 18.2, CH2 a: 2.00 (1H, ddt, 13.3, 7.9, 2.0) C-4, C-5, C-7, C-8, C-10
H-7a, H-7(3, H-18, H-19 .3
"
13: 1.77 (1H, m) C-5, C-7, C-10
H-713, H-19
,
0
7 24.6, CH2 a: 2.62 (1H, ddd, 16.7, 11.6, 7.9) C-6, C-8, C-9, C-14
H-5, H-6a IV
1
F'
01
13: 2.86 (1H, dd, 16.7, 6.5) C-5, C-6, C-8, C-9, C-14
H-6a, H-613
8 120.6, C -
9 148.2, C -
37.3,C -
11 116.4, CH 6.84(1H, d, 8.3) C-8, C-10, C-13
H-113
12 123.3, CH 7.02 (1H, d, 8.3) C-9, C-14, C-15
H-16, H-17
13 130.2,C -
n
,-i
14 150.2,C -
m
,-o
26.7, CH 3.15 (1H, sep, 6.9) C-12, C-13, C-14, C-16, C-17
w
o
16 22.7, CH3 1.24 (3H, d, 6.9) C-13, C-15, C-17
H-12 w
O-
17 22.5, CH3 1.26 (3H, d, 6.9) C-13, C-15, C-16
H-12 -4
(44
c.,
18 28.1, CH3 1.09(3H, s) C-3, C-4, C-5, C-19
H-3, H-5, H-6a u,
o,
56
o
19 15.3, CH3 0.91 (3H, s) C-3, C-4, C-5, C-18
H-6a, H-6I3, H-20 w
=
20 24.8, CH3 1.21 (3H, s) C-1, C-5, C-9, C-10
H-113, H-19 w
w
'a
.6.
(44
4=,
01
I-,
Table 9. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F1-31
Pos. 5c, type" 8H, nH, multiplicity (J in Hz) HMBC
ROESY
1 34.4, CH2 a: 1.22 (1H, m) C-2, C-3, C-9, C-10, C-20
H-3
13: 2.78 (1H, dt, 13.5, 3.6) C-3, C-5, C-10
H-2, H-20
2 27.7, CH2 1.72 (2H, m) C-1, C-3, C-4, C-10
H-113, H-19
3 78.3, CH 3.24 (1H, m) C-2, C-4, C-18, C-19
H-la, H-18
4 39.0,C -
p
51.0, CH 1.06 (1H, m) C-4, C-6, C-10, C-19, C-20
H-3, H-6a, H-7B, H-7A 2
,
6 17.1, CH2 a: 1.87 (1H, br dd, 13.5, 7.5) C-4, C-5, C-7, C-8, C-10
H-5, H-7B, H-18
.3
13: 1.45(1H, dtd, 13.5, 11.7, 5.7) C-5, C-7, C-10
H-7B, H-19
c,
7 26.1, CH2 A: 2.30 (1H, ddd, 20.2, 11.7, 7.5) =C-6, C-8, C-9, C-14
H-5, H-6a
, B: 2.71 (1H, br dd, 20.2, 5.7)
C-5, C-6, C-8, C-9, C-14 H-5, H-6a, H-6p
8 142.7, C -
9 150.0,C -
38.1, C -
11 187.78,C -
12 131.8, CH 6.30(1H, d, 1.0) =C-9, C-11/C-14, C-15
H-16/17
13 152.9,C -
14 187.83,C -
n
,-i
26.2, CH 2.96 (1H, sep d, 6.9, 1.0) C-12, C-13, C-14, C-16, C-17
m
,-o
16 21.3, CH3 1.07 (3H, d, 6.9) C-13, C-15, C-17
H-12 w
=
w
17 21.3, CH3 1.08 (3H, d, 6.9) C-13, C-15, C-16
H-12 .
'a
18 28.2, CH3 1.03 (3H, s) C-3, C-4, C-5, C-19
H-3, H-6a -4
(44
c.,
19 15.7, CH3 0.85 (3H, s) C-3, C-4, C-5, C-18
H-2, H-613, H-20 u,
c.,
57
20 20.1, CH3 1.26 (3H, s) C-1, C-5, C-9, C-10
H-113, H-19
(44
Table 10. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F2-X
Pos. gc, type" 511, nH, multiplicity (J in Hz) a'c HMBC
ROESY
1 33.5, CH2 a: 1.64 (1H, ddd, 13.2, 11.5, 6.5) C-2, C-5, C-9, C-10, C-
20 H-113, H-2B, H-5
13: 2.54 (1H, dd, 13.2, 6.2) C-2, C-3, C-5, C-9, C-10,
(C-18), C-20 H-la, H-11, H-20
2 18.6, CH2 A: 2.34 (1H, m) C-1, C-3, C-4
H-20
B: 2.43 (1H, dd, 18.1, 5.4) C-1, C-3, C-4, C-10
H-1a, H-20
3 128.6, C -
4 164.0,C -
41.4, CH 2.67 (1H, br s)
H-1a, H-6a
6 19.8, CH2 a: 2.30 (1H, m)
H-5, H-613, H-7A, H-7B, H-
19
13 : 1.91 (1H, m)
H-6a, H-7B, H-19, H-20
7 23.8, CH2 A: 2.83 (1H, m)
H-6a
B: 2.91 (1H, dd, 17.8, 7.5) C-5, C-6, C-8, C-9, C-14
H-6a, H-613
8 123.2, C -
9 144.7,C -
37.0,C -
11 116.8, CH 6.91 (1H, d, 8.2) C-8, C-10, C-13
H-113
12 123.7, CH 6.99 (1H, d, 8.2) C-9, C-14, C-15
H-16, H-17
13 133.7, C -
14 152.4,C -
27.4, CH 3.27 (1H, sep, 6.9) C-12, C-13, C-14, C-16, C-17
16 23.1, CH3 1.18 (3H, d, 6.9) C-13, C-15, C-17
H-12
17 23.0, CH3 1.20 (3H, d, 6.9) C-13, C-15, C-16
H-12
(44
18 173.4,C -
58
o
19 99.4, CH 6.10 (1H, br s)
H-6a, H-613 w
=
20 22.6, CH3 1.02 (3H, br s) C-1, C-5, C-9, C-10
H-113, H-2A, H-2B, H-613 w
w
'a
.6.
(44
4=,
01
I-,
Table 11. 1H and 13c NMR data and 2D HMBC and ROESY correlations for F2-10
Pos. 5c, type" 8H, nH, multiplicity (J in Hz) a,c
HMBC ROESY
1 34.8, CH2 a: 1.87 (1H, td, 12.7, 5.2) C-2, C-10, C-20
H-5, H-18
13: 2.10 (1H, dt, 12.7, 3.6) C-2, C-3, C-5
H-2, H-11, H-20
2 32.4, CH2 1.80 (2H, m) C-1, C-3, C-4, c-io
H-113, H-5, H-18, H-20
3 73.4,C
4 152.5,C -
P
43.4, CH 2.76 (1H, br d, 12.3) C-4, C-6, C-10, C-19
H-la, H-2, H-7a 2
,
6 21.7, CH2 a: 1.84 (1H, m) C-5, C-7, C-8, C-10
H-7a, H-713, H-19A
13: 1.71 (1H, m) C-5, C-7, C-10
H-713, H-19A, H-20 .3
7 25.2, CH2 a: 2.60 (1H, m) C-6, C-8, C-9, C-14
H-5, H-6a, H-613, H-713
,
13: 2.94 (1H, dd, 17.2, 5.8) C-5, C-6, C-8, C-9, C-14
H-6a, H-6(3, H-7a
8 123.5, C -
9 146.4,C -
39.8,C -
11 118.2, CH 6.85(1H, d, 8.2) C-8, C-10, C-13
H-113
12 123.6, CH 6.96 (1H, d, 8.2) C-9, C-14, C-15
H-16/17
13 132.9,C -
14 151.8,C -
n
,-i
27.3, CH 3.27 (1H, sep, 6.9) C-12, C-13, C-14, C-16/17
m
,-o
16 23.0, CH3 1.19 (3H, d, 6.9) C-13, C-15, C-17
H-12 w
=
w
17 23.0, CH3 1.19 (3H, d, 6.9) C-13, C-15, C-16
H-12 .
'a
18 68.6, CH2 3.71 (2H, s) C-2, C-3, C-4
H-1a, H-2, H-19B -4
(44
c.,
19 107.5, CH2 A: 4.83 (1H, br s) C-3, C-4, C-5
H-6a, H-613 u,
c,
59
o
B: 5.10(1H, br s) C-3, C-4, C-5
H-18 w
=
20 21.8, CH3 0.96(3H, s) C-1, C-5, C-9, C-10
H-113, H-2, H-613 w
w
'a
.6.
(44
4=,
01
I-,
Table 12. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F20P1
Pos. 5c, type" 8H, nH, multiplicity (J in Hz) ,c
HMBC ROESY
1 33.1, CH2 A: 1.35(1H, m) C-2, C-3, C-5, C-10, C-20
H-2B
B: 1.427 (1H, m)
H-3, H-17, H-18
2 28.9, CH2 A: 1.60 (1H, m) C-1, C-3
B: 1.69 (1H, m)
H-1A
3 79.0, CH 3.22 (1H, dd, 11.8, 5.0) C-2, C-4, C-18, C-19
H-16, H-5, H-18 P
4 38.9,C -
0
46.0, CH 1.415(1H, m) C-3, C-4, C-6, C-7, C-10, C-18, C-19,
C-20 H-3, H-18 ,
-
N,
6 17.5, CH2 A: 1.407 (1H, m) C-5
H-19 00"
IV
B: 1.62 (1H, m) C-4, C-5, C-7, C-8, C-10
H-19 2
7 38.6, CH2 A: 1.62 (1H, m) C-6, C-9
,
,õ.
,
B: 1.80 (1H, dd, 11.5, 7.6) C-5, C-6, C-8, C-14, C-17
,
8 37.9,C -
9 53.6, CH 1.24 (1H, dd, 12.9, 3.0) C-1, C-8, C-10, C-11, C-12, C-
17, C-20 H-14, H-20
36.9,C -
11 24.5, CH2 A: 1.32 (1H, qd, 12.9, 4.0) C-9, C-12
H-12A, H-126, H-17
B: 1.64(1H, m) C-9
H-12A, H-12B, H-20
12 37.8, CH2 A: 1.94 (1H, tdt, 12.8, 4.6, -1)
C-11, C-13, C-15 H-11A, H-11B
n
B: 2.41 (1H, ddd, 12.8, 4.0, 2.6) C-9, C-11, C-13, C-14, C-15
H-11A, H-116, H-156
13 147.5,C -
m
,-o
w
14 60.8, CH 1.89 (1H, t, 6.5) C-8, C-9, C-13, C-15, C-16, C-17
H-9 =
w
107.0, CH2 A: 4.69 (1H, dt, 1.4, 1.2) C-12, C-13, C-14
H-15B, H-16 'a
-4
B: 4.96 (1H, dt, 1.4, 1.2) C-12, C-13, C-14
H-12B, H-15A (44
01
CA
16 59.0, CH2 3.76 (2H, d, 6.5) C-8, C-13, C-14
H-7A, H-7B, H-15A, H-17 c.,
60
o
17 21.0, CH3 0.85 (3H, s) C-7, C-8, C-9, C-14
H-1B, H-11A, H-16 w
=
18 29.0, CH3 0.98 (3H, s) C-3, C-4, C-5, C-19
H-1B, H-3, H-5, H-6B w
w
19 15.8, CH3 0.79 (3H, s) C-3, C-4, C-5, C-18
H-6A, H-6B 'a
.6.
(44
20 22.3, CH3 0.95 (3H, s) C-1, C-5, C-9, C-10
H-9, H-11B .6.
c.,
Table 13. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F20P2
Pos. oc, type" gi - 4 , nH, multiplicity (J in Hz) a,c
HMBC ROESY
1 33.4, CH2 a: 1.60 (1H, m)
H-10, H-5
13: 2.35(1H, m) C-5, C-10
H-la, H-11, H-20
2 26.0, CH2 2.35 (2H, m) .C-3, C-4
H-1 8A, H-1813 P
3 131.8,C
.
,
4 129.7,C -
'
0
44.5, CH .2.27 (1H, m)
H-1a 3
6 20.0, CH2 a: 2.28 (1H, m) C-8, C-10
H-7A, H-7B, H-19 0
,
13: 1.64 (1H, m) . (C-5)
H-7B, H-20 0
,
,
7 23.3, CH2 A: 2.71 (1H, ddd, 16.8, 10.9, 8.4) C-6, C-8
H-6a, 14-0H
.B: 2.84 (1H, dd, 16.8, 7.1) C-5, C-6, C-8, C-9, C-14
H-6a, H-613, 14-0H
8 120.6, C -
9 146.0,C .-
35.6,C .-
11 116.2, CH 6.93 (1H, d, 8.1) C-8, C-10, C-13
H-18
12 123.1, CH 7.03 (1H, d, 8.1) =.C-9, C-14, C-15
H-16, H-17
n
,-i
13 1303,C .-
m
,-o
14 .150.4, C .-
w
=
14-0H - .4.65 (1H, br s)
H-7a, H-713, H-15 w
'a
26.9, CH 3.14 (1H, sep, 6.9) .C-12, C-13, C-14, C-16, C-17
14-0H -4
(44
16 .22.6, CH3 .1.25 (3H, d, 6.9) .C-13, C-15, C-17
H-12
u,
c.,
17 22.6, CH3 1.27 (3H, d, 6.9) C-13, C-15, C-16
H-12
61
===o
18 63.4, CH2 A: 4.09(1H, d, 11.6) C-2, C-3, C-4
H-2, H-19 w
=
.B: 4.26 (1H, d, 11.6) .C-2, C-3, C-4
H-2, H-19 w
w
19 15.6, CH3 1.78(3H, br s) C-3, C-4, C-5
H-6a, H-18A, H-18B 'a
.6.
=
(44
20 22.4, CH3 1.02 (3H, s) C-1, C-5, C-9, C-10
H-113, H-613 .6.
c.,
Table 14. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F20P3
Pos. gc, type" gH , nH, multiplicity (J in Hz)" HMBC
ROESY
1 38.5, CH2 a: 1.46 (1H, m) C-2, C-10, C-20
H113, H-3
13: 2.30(1H, m) C-2, C-3, C-5, C-10
H1a, H-213, H-11, H-20
2 20.7, CH2 a: 1.60 (1H, m) V
H-18A, H-20 P
13: 1.47(1H, m) C-1, C-3
H113, H-18B, H-19 2
,
3 44.4, CH 1.76 (1H, m)
H1a, H-4, H-18A, H-18B .
" "
4 35.0, CH 2.07 (1H, m) C-2, C-3, C-5, C-10, C-19
H-3, H-5, H-6a, H-18B .3
"
=5 44.7, CH 1.68(1H, ddd, 12.8, 4.4, 2.1)
C-4, C-6, C-7, C-10, C-19, C-20 H-4, H-7a "
,
6 23.7, CH2 a: 1.65 (1H, br dd, 12.8, 8.2)
C-4, C-5, C-7, C-8, C-10 H-4, H-7a, H-713 "
13: 2.03 (1H, m) C-5, C-7, C-10
H-713, 14-0H, H-19, H-20
7 24.1, CH2 a: 2.65 (1H, ddd, 16.4, 11.5, C-6, C-8, C-9, C-14
H-5, H-6a, 14-0H
7.8)
13: 2.82 (1H, dd, 16.4, 6.6) C-5, C-6, C-8, C-9, C-14
H-6a, H-613
8 120.7, C -
9 148.2..0 -
37.5,C -
n
,-i
11 116.6, CH 6.84(1H, d, 8.2) C-8, C-10, C-13
H-113, H-20 m
,-o
12 123.4, CH 7.02 (1H, d, 8.2) C-9, C-14, C-15
H-16, H-17 w
=
w
13 130.0,C -
.
'a
14 150.2,C -
-4
(44
o,
14-0H - 4.64 (1H, br s) H-8, H-13, H-14
H-7a, H-713, H-15 u,
o,
62
o
15 26.9, CH 3.14 (1H, sep, 6.9) C-12, C-13, C-14, C-16, C-17
14-0H w
=
16 22.7, CH3 1.24 (3H, d, 6.9) C-13, C-15, C-17
H-12 w
w
17 22.6, CH3 1.25 (3H, d, 6.9) C-13, C-15, C-16
H-12 'a
.6.
(44
18 65.8, CH2 A: 3.54 (1H, dd, 10.4, 6.8) C-3, C-4, C-5
H-2a, H-213, H-3, H-19 .6.
c,
B: 3.60 (1H, dd, 10.4, 8.0) C-3, C-4, C-5
H-213, H-3, H-4, H-19
19 9.7, CH3 0.89 (3H, d, 7.6) C-3, C-4, C-5
H-213, H-613, H-18B, H-20
20 25.4, CH3 1.16(3H, s) C-1, C-5, C-9, C-10
H-113, H-2a, H-613, H-11, H-19
Table 15. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F20P4
Pos. gc, type" gH , nH, multiplicity (J in Hz) a,c HMBC
ROESY P
1 33.3, CH2 a: 1.49(1H, m) C-10, C-20
H113, H-18 ,
13: 2.04 (1H, m) C-3, C-5
H1a, H-11, H-20
N,
2 19.5, CH2 a: 1.66 (1H, m) V
H-18 .3
N,
13: 2.00 (1H, m)
H-19, H-20
1
0
3 42.9, CH 1.72 (1H, m) C-1, C-2, C-4, C-18, C-19
F'N'
4 34.7, CH 1.91 (1H, m) C-2, C-3, C-5, C-10, C-18, C-19
H-6a, H-18
38.7, CH 1.74 (1H, ddd, 12.8, 5.1, 2.2) C-4, C-6, C-7, C-10, C-19
6 23.4, CH2 a: 1.59 (1H, br dd, 12.8, 7.8)
C-4, C-5, C-7, C-8, C-10 H-4, H-7a, H-713
13: 1.98 (1H, m) C-5, C-7, C-10
H-713, 14-0H, H-19, H-20
7 24.0, CH2 a: 2.62 (1H, ddd, 16.5, 11.4, C-6, C-8, C-9
H-6a, 14-0H
7.8)
13: 2.81 (1H, dd, 16.5, 6.7) C-5, C-6, C-8, C-9, C-14
H-6a, H-6(3 n
,-i
8 120.7,C -
m
,-o
9 148.2,C -
w
=
w
37.4, C -
.
'a
11 116.4, CH 6.82 (1H, d, 8.2) C-8, C-10, C-13
H-113 -4
(44
c,
12 123.3, CH 7.01 (1H, d, 8.2) C-9, C-14, C-15
H-16, H-17 u,
c,
63
o
13 130.0, C -
w
=
14 150.2,C -
w
w
14-0H - 4.63 (1H, br s) H-8, H-13
H-7a, H-713, H-15 -a
(44
15 26.8, CH 3.14 (1H, sep, 6.9) C-12, C-13, C-14, C-16, C-17
14-0H
c,
16 22.7, CH3 1.24 (3H, d, 6.9) C-13, C-15, C-17
H-12
17 22.6, CH3 1.25 (3H, d, 6.9) C-13, C-15, C-16
H-12
18 64.7, CH2 3.66 (2H, m) C-2, C-4
H-la, H-2a, H-4
19 16.7, CH3 1.09 (3H, d, 7.6) C-3, C-4, C-5
H-213, H-613, H-20
20 25.0, CH3 1.21 (3H, s) C-1, C-5, C-9, C-10
H-113, H-213, H-613, H-19
P
Table 16. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F15P2
.
,
Pos. gc, type" 4i, nH, multiplicity (J in Hz) 4c
HMBC ROESY
1 32.7, CH2 a: 1.56 (1H, m) C-2, C-5, C-10, C-20
H-2a
N,
13: 2.42 (1H, m) C-2, C-3, C-5, C-10, C-20
H-11, H-20
I
0
2 20.7, CH2 a: 2.53 (1H, m) C-1, C-3, C-4, C-10
H-la, H-19 F'N'
13: 2.33 (1H, m)
H-20
3 133.5,C
4 156.1,C -
46.7, CH 2.44 (1H, m)
H-7a
6 19.4, CH2 a: 2.38 (1H, m) C-5, C-8, C-10
H-7a
.13: 1.76 (1H, tdd, 13.2, 10.9, 7.2) .C-5, C-7, C-10
H-713, H-20
7 23.4, CH2 a: 2.77(1H, ddd, 17.0, 10.9, 8.2) C-6, C-8, C-9, C-14
H-5, H-6a, 14-0H n
,-i
.13: 2.93 (1H, dd, 17.0, 7.2) C-5, C-6, C-8, C-9, C-14
H-63.14-OH m
,-o
8 120.6,C -
w
=
w
9 1451,C -
.
-a
.35.8, C .-
-4
(44
01
ii 116.4, CH 6.94(1H, d, 8.2) C-8, C-10, C-13
H-1(3 u,
o,
64
0
12 123.3, CH 7.05 (1H, d, 8.2) .C-9, C-14, C-15
H-16, H-17 w
=
13 130.6,C .-
w
w
14 1504,C .-
'a
(44
14-0H - 4.68 (1H, s) C-8, C-13, C-14
H-7a, H-713, H-15
c,
15 26.9, CH 3.12 (1H, sep, 6.9) C-12, C-13, C-14, C-16, C-
17 14-0H
16 .22.6, CH3 .1.26 (3H, d, 6.9) C-13, C-15, C-17
H-12
17 .22.5, CH3 .1.27 (3H, d, 6.9) C-13, C-15, C-16
H-12
18 .191.3, CH .10.24 (1H, s) .C-2, C-3
H-19
19 15.0, CH3 2.22 (3H, ddd, 1.8, 1.6, 1) C-3, C-4, C-5
H-2a, H-18
20 22.7, CH3 1.02 (3H, s) C-1, C-5, C-9, C-10
H-11:3, H-21:3, H-6I3
P
.
Table 17. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F55P2
,
-
Pos. oc, type" 4-4, nH, multiplicity (J in Hz) a,c
HMBC ROESY 00"
IV
1 28.5, CH2 a: 1.48 (1H, m)
H-11:3, H-5 0
" ,
.13: 2.67 (1H, ddd, 13.1, 5.2, 2.9)
H-1a, H-2, H-11, H-20B 0
IV
1
F'
2 25.7, CH2 2.36 (2H, m)
H-11:3, H-18A, H-18B, H- .
20B
3 130.6d, C -
4 131.0d, C -
=43.6, CH 2.41 (1H, br d, 14.2)
H-1a
6 20.1, CH2 a: 2.29 (1H, dddd, 13.6, 8.4, 3.3,
H-7A, H-7B, H-19
-1)
n
(3: 1.70 (1H, tdd, 13.6, 10.4, 8.0)
H-7B, H-20A, H-20B
m
7 23.0, CH2 A: 2.78 (1H, m) C-6, C-8
H-6a, 14-0H
w
=
.B: 2.88 (1H, m) C-5, C-6, C-8, C-9
H-6a, H-61:3, 14-0H w
'a
8 1217,c -
-4
(44
9 140.8,c -
c,
u,
c,
65
403,C -
11 117.2, CH 6.98(1H, d, 8.2) =C-8, C-10, C-13
H-1I3
12 123.0, CH 7.07 (1H, d, 8.2) C-9, C-14, C-15
H-16/17
(44
13 131.5,C
14 150.9,C .-
14-0H - 4.72 (1H, br s) C-8, C-13
H-7A, H-7B, H-15
27.0, CH 3.15 (1H, sep, 6.9) C-12, C-13, C-14, C-16, C-17
14-0H
16 .22.6, CH3 .1.27 (3H, d, 6.9) C-13, C-15, C-17
H-12
17 .22.6, CH3 .1.27 (3H, d, 6.9) C-13, C-15, C-16
H-12
18 63.2, CH2 A: 4.09 (1H, d, 11.6) C-2, C-4
H-2, H-19
.B: 4.28 (1H, d, 11.6) .C-2, C-4
H-2, H-19
19 15.6, CH3 1.79 (3H, q, 1.9) C-3, C-4, C-5
H-6a, H-18A, H-18B
64.7, CH2 A: 3.56 (1H, dd, 10.8, 3.0)
H-613
B: 3.68 (1H, dd, 10.8, 7.7) C-9
H-113, H-2, H-613
20-0H - 1.03 (1H, m)
0
(44
66
o
Table 18. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F55P3
w
=
Pos. gc, type" gH, nH, multiplicity (J in Hz) a,c HMBC
ROESY w
w
'a
1 38.4, CH2 a: 1.45 (1H, m) C-2, C-20
H10, H-3 .6.
(44
13: 2.28 (1H, m) C-3, C-5, C-10, C-20
H1a, H-11, H-20 .6.
o,
2 21.3, CH2 1.46 (1H, m) C-1, C-3, C-4, C-10
H-18A, H-18B, H-19B, H-20
3 42.8, CH 1.89 (1H, m)
H1a, H-4, H-18B
4 44.6, CH 2.21 (1H, dt, 9.0, 4.6)
H-3, H-5, H-6a, H-19A
44.4, CH 1.79 (1H, m) C-6, C-7, C-19 H-
4, H-7a
6 23.8, CH2 a: 1.80 (1H, m) C-4, C-5, C-7, C-8, C-10
H-4, H-7a, H-713, H-19A
13: 1.98 (1H, tdd, 13.3, 11.6, 6.2) C-5, C-7, C-10
H-713, 14-0H, H-19A, H-20
7 24.4, CH2 a: 2.66 (1H, ddd, 16.4, 11.6, C-6, C-8
H-5, H-6a, 14-0H Q
7.4)
.
,
13: 2.85 (1H, dd, 16.4, 6.2) C-5, C-6, C-8, C-9, C-14
H-6a, H-6(3 0"
IV
00
8 1205,C -
IV
0
IV
9 146.9,0 -
,
0
37.1,C -
IV
F'
11 116.9, CH 6.83(1H, d, 8.3) C-8, C-10, C-13
H-1(3
12 123.4, CH 7.02 (1H, d, 8.3) C-9, C-14, C-15
H-16, H-17
13 130.1,C -
14 150.2,C -
14-0H - 4.64 (1H, s) H-8, H-13, H-14
H-7a, H-7(3, H-15
26.8, CH 3.13 (1H, sep, 6.9) C-12, C-13, C-14, C-16, C-17
14-0H
16 22.7, CH3 1.24 (3H, d, 6.9) C-13, C-15, C-17
H-12 n
,-i
17 22.5, CH3 1.25 (3H, d, 6.9) C-13, C-15, C-16
H-12 m
,-o
18 65.4, CH2 A: 3.63(1H, m)
H-2, H-1913 w
=
B: 3.65(1H, m) C-3, C-4
H-2, H-3, H-1913 w
'a
19 59.5, CH2 A: 3.71 (1H, d, 10.4) C-3, C-4
H-4, H-6a, H-6(3, H-20 -4
(44
c.,
B: 3.88 (1H, dd, 10.4, 9.2) C-3, C-4
H-2, H-18A/B, H-20 u,
c.,
67
20 24.6, CH3 0.98(3H, s) C-1, C-5, C-9, C-10
H-113, H-2, H-613, H-19A, H-19B
(44
Table 19. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F15P4
Pos ty- , nH, multiplicity (J in Hz) a,c
HMBC ROESY
. eab
1 37.0, a: 1.16 (1H, td, 13.2, 3.7) C-2, C-5, C-10, C-20
H-3
CH2
13: 1.79 (1H, dt, 13.2, 3.5) C-3, C-5
H-20
2 27.9, A: 1.60(1H, m) C-1, C-3
H-19
CH2
B: 1.71 (1H, m) C-3
3 78.8, CH 3.25(1H, dd, 11.8, 4.3) C-4, C-18, C-19
H-la, H-5, H-18
4 39.1,C -
00"
54.6, CH 1.08 (1H, dd, 12.5, 2.7) C-4, C-6, C-7, C-9, C-10, C-18, C-19, H-
3, H-7a, H-9, H-18
C-20
6 24.0, a: 1.74 (1H, dddd, 13.0, 5.0, 2.7, C-5, C-7, C-8, C-10
H-713, H-18
CH2 2.5)
13: 1.39 (1H, tdd, 13.0, 12.5, 4.2) C-5, C-7, C-10
H-713, H-19
7 38.1, a: 1.96 (1H, ddd, 13.0, 12.8, 5.0) C-6, C-8, C-17
H-5
CH2
(3: 2.40 (1H, ddd, 12.8, 4.2, 2.5) C-5, C-6, C-8, C-9, C-17
H-6a, H-613, H-17B
8 147.8,C -
9 55.9, CH 1.54 (1H, m) C-7, C-8, C-10, C-11, C-12, C-17,
C-20 H-5, H-12B, H-14, H-17A, H-20
39.2,C -
11 21.8, A: 1.47 (1H, m) C-8, C-9, C-12, C-13
H-12B, H-14, H-17A, H-20
CH2
B: 1.59 (1H, m) C-8, C-9, C-12
(44
68
12 38.3, A: 1.83 (1H, ddd, 14.0, 9.4, 6.6) C-9, C-11, C-13, C-14, C-15
H-14
CH2
B: 2.17 (1H, ddd, 14.0, 10.0, 3.8) C-11, C-13, C-14
H-9, H-11A, H-14, H-15, H-17A
(44
13 142.7,C -
14 118.0, 5.31 (1H, t sext, 7.1, 1.2) C-12, C-15, C-16
H-9, H-11A, H-12A, H-12B
CH
15 16.5, 1.69(3H, br s) C-12, C-13, C-14
H-12B, H-16
CH3
16 61.4, 4.58 (2H, d, 7.1) C-13, C-14, C-1'
H-15
CH2
17 106.0, A: 4.53 (1H, q, -1) C-7, C-8, C-9
H-9, H-11A, H-12A, H-12B, H-17B
CH2
B: 4.85 (1H, q, -1.4) C-7, C-9
H-713, H-17A
18 28.3, 1.00 (3H, s) C-3, C-4, C-5, C-19
H-3, H-5, H-6a
CH3
19 15.3, 0.78 (3H, s) C-3, C-4, C-5, C-18
H-2A, H-613
0'
CH3
20 14.5, 0.69 (3H, s) C-1, C-5, C-9, C-10
H-9, H-11A
CH3
1' 171.0,C -
2' 21.1, 2.06 (3H, s) C-1'
CH3
(44
69
Table 20. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F20P5
Pos. gc, type" gH, nH, multiplicity (J in Hz) a,c
HMBC ROESY
1 59.4, CH2 4.16 (2H, d, 6.9) C-2, C-3
H-3-Me
(44
2 123.4, CH =5.42 (1H, t sext, 6.9, 1.3) C-3-Me, C-4
H-4
3 139.6,C -
3-Me 16.3, CH3 1.68(3H, br s) C-2, C-3, C-4
H-1
4 39.5, CH 2.04 (2H, m) C-2, C-3, C-3-Me, C-5
H-2
26.3, CH2 2.11 (2H, m) C-4, C-6, C-7
6 123.9, CH 5.11 (1H, t sext, 6.9, 1.2) C-5, C-7-Me, C-8
7 135.1,C -
7-Me 15.9, CH3 1.60(3H, br s) C-6, C-7, C-8
8 39.6, CH2 1.99 (2H, m) C-6, C-7, C-7-Me, C-9, C-10
9 26.5, CH2 2.08 (2H, m) C-7, C-8, C-10, C-11
124.8, CH 5.16(1H, t sext, 6.9,1.2) C-9, C-11-Me, C-12 H-12B,
H-13
11 134.0,C -
11-Me 16.0, CH3 1.62(3H, br s) C-10, C-11, C-12
12 36.3, CH2 A: 2.08(1H, m) C-10, C-11, C-11-Me, C-13, C-14
H-14
B: 2.16(1H, m) C-10, C-11, C-11-Me, C-13, C-14
H-10, H-14
13 27.4, CH2 1.63 (2H, m) C-11, C-12, C-14, C-15
H-10
14 64.1, CH 2.70 (1H, t, 6.3) C-12, C-13, C-15, C-15-Me'
H-12A, H-12B, H-15-Me, H-15-Me'
58.2,C -
15-Me 18.7, CH3 1.26(3H, s) C-14, C-15, C-15-Me'
H-14
15- 24.8, CH3 1.30(3H, s) C-14, C-15, C-15-Me
H-14
Me'
(44
70
0
Table 21. 1H and 13C NMR data and 2D HMBC and ROESY correlations for F60P1
w
=
Pos. gc, type" 4-1, nH, multiplicity (J in Hz)" HMBC
ROESY w
w
'a
1 .59.3, CH2 4.15(2H, br d, 6.9) C-2, C-3
H-3-Me .6.
(44
2 .123.7, CH .5.42 (1H, t sext, 6.9, 1.2) C-3-Me, C-4
H-4 .6.
c,
3 139.3,C .-
3-Me .16.2, CH3 1.68(3H, br s) C-2, C-3, C-4
H-1, H-4
4 .39.3, CH 2.07 (2H, m)
H-2, H-3-Me
.25.9, CH2 2.14 (2H, m) C-4, C-6, C-7
6 124.3, CH 5.13 (1H, t sext, 6.8, 1.2) C-4, C-5, C-7-Me, C-8
7 136.2,C -
7-Me .16.0, CH3 .1.64 (3H, br s) C-6, C-7, C-8
H-8
P
8 42.5, CH2 2.10 (2H, m) C-6, C-7, C-7-Me, C-9, C-10
H-7-Me, H-10, H-15-aMe .
=9 24.2, CH2 A: 1.48(1H, m)
C-8, C-10, C-15 ,
-
0
B: 1.55(1H, m) C-8, C-10, C-11
H-11-Me 00"
IV
55.3, CH 1.10 (1H, t, 4.3) C-8, C-9, C-11, C-11-Me, C-15 H-8, H-
14 2
11 =73.4,C .-
,
,õ.
,
11-Me 23.0, CH3 1.16(3H, s) C-10, C-11, C-12
H-9B, H-12B, H-13A, H-15-I3Me ,
12 40.9, CH2 A: 1.45(1H, m) C-13
.B: 1.78(1H, m) C-10, C-11, C-14
H-11-Me
13 28.9, CH2 A: 1.49(1H, m) C-14
H-11-Me
B: 1.75(1H, m) C-10, C-11, C-14
H-14
14 78.3, CH 3.31 (1H, dd, 10.6, 2.8)
H-10, H-13B, H-15-aMe
40.4,C -
n
15-aMe .28.0, CH3 .1.04 (3H, s) C-10, C-14, C-15, C-15-I3Me
H-8, H-14
m
15-I3Me 14.8, CH3 0.80(3H, s) C-10, C-14, C-15, C-15-aMe
H-11-Me
w
=
w
'a
-4
(44
01
CA
01
71
0
al H NMR (600.13) and 13C NMR (150.90 MHz) data obtained with samples in
CDCI3. b Assignments based on HSQC and HMBC w
=
w
experiments. C Multiplicities reported as apparent splittings: s = singlet, d
= doublet, t = triplet, sext = sextet, m = multiplet (incl. w
'a
.6.
(44
overlapping resonances), br = broad. a denotes Me pointing into the plane and
0 denotes Me pointing out of the plane. A denotes .6.
c.,
the lowest chemical shift value and B denotes the highest chemical shift value
P
0
,
,õ
0
,õ
.3
,õ
0
,õ
,
0
,õ
Iv
n
,-i
m
,-o
w
=
w
'a
-4
(44
01
CA
01
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
72
References
1. J. Andersen-Ranberg etal., Expanding the Landscape of Diterpene
Structural
Diversity through Stereochemically Controlled Combinatorial Biosynthesis.
Angewandte Chemie International Edition, n/a (2016).
2. I. Pateraki etal., Total biosynthesis of the cyclic AMP booster
forskolin from
Coleus forskohlii. Elife 6, e23001 (2017).
3. I. Pateraki etal., Manoyl Oxide (13R), the Biosynthetic Precursor of
Forskolin,
Is Synthesized in Specialized Root Cork Cells in Coleus forskohlii. Plant
Physiology 164, 1222-1236 (2014).
4. N. L. Hansen etal., The terpene synthase gene family in Tripterygium
wilfordii
harbors a labdane-type diterpene synthase among the monoterpene synthase
TPS-b subfamily. The Plant Journal 89, 429-441 (2017).
5. H. H. Nour-Eldin, B. G. Hansen, M. H. H. Norholm, J. K. Jensen, B. A.
Halkier,
Advancing uracil-excision based cloning towards an ideal technique for cloning
PCR fragments. Nucleic Acids Research 34, e122 (2006).
6. N. B. Jensen etal., EasyClone: method for iterative chromosomal
integration of
multiple genes in Saccharomyces cerevisiae. FEMS Yeast Research 14, 238-
248 (2014).
7. M. H. H. Norholm, A mutant Pfu DNA polymerase designed for advanced
uracil-
excision DNA engineering. BMC Biotechnology 10, 21 (2010).
8. R. D. Gietz, R. H. Schiestl, High-efficiency yeast transformation using
the
LiAc/SS carrier DNA/PEG method. Nature Protocols 2, 31-34 (2007).
9. Hansen, N.L., et al., Integrating pathway elucidation with yeast
engineering to
produce polpunonic acid the precursor of the anti-oDesity agent celastrol.
MicroD Cell Fact, 2020. 19(1): p. 15.
10. Voinnet 0, Rivas S, Mestre P, Baulcombe D. An enhanced transient
expression
system in plants based on suppression of gene silencing by the p19 protein of
tomato bushy stunt virus [retracted in: Plant J. 2015 Nov;84(4):846]. Plant J.
2003; 33(5): 949-956. doi: 10.1046/j. 1365-313x.2003.01676.x
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
73
ITEMS
1. A recombinant host cell, capable of producing oxygenated diterpenoid
compound, wherein the host cell is capable of producing miltiradiene
and/or dehydroabietadiene and has been transformed with a first gene en-
coding an enzyme having cytochrome P450 activity and which enzyme is
capable of converting miltiradiene and/or dehydroabietadiene into 14-0H-
dehydroabietadiene.
2. The recombinant host cell of item 1, wherein the first gene encodes a pol-
ypeptide comprising an amino acid sequence having at least 80 % se-
quence identity, preferably at least 85% sequence identity, preferably at
least 90% sequence identity, preferably at least 95% sequence identity,
more preferred at least 98% sequence identity to SEQ ID NO: 1
(TwCYP82D274V1) or the mature polypeptide thereof.
3. The recombinant host cell of item 1 or 2, wherein the recombinant host cell
further comprises:
a second gene encoding a second enzyme having cytochrome P450
activity and a third gene encoding a third enzyme having cytochrome
P450 activity, wherein:
the second gene encoding an enzyme having cytochrome P450
activity encodes a polypeptide comprising an amino acid sequence
having at least 80 % sequence identity, preferably at least 85% se-
quence identity, preferably at least 90% sequence identity, prefera-
bly at least 95% sequence identity, more preferred at least 98% se-
quence identity to SEQ ID NO: 4 (TwCYP71BE86) or the mature pol-
ypeptide thereof;
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
74
the third gene encoding an enzyme having cytochrome P450 ac-
tivity encodes a polypeptide comprising an amino acid sequence
having at least 80 % sequence identity, preferably at least 85% se-
quence identity, preferably at least 90% sequence identity, prefera-
bly at least 95% sequence identity, more preferred at least 98% se-
quence identity to SEQ ID NO: 3 (TwCYP71BE85) or the mature pol-
ypeptide thereof.
4. The recombinant host cell of item 3, wherein the host cell further
comprises
a gene encoding a polypeptide having cytochrome B5 activity and com-
prising an amino acid sequence having at least 80 % sequence identity,
preferably at least 85% sequence identity, preferably at least 90% se-
quence identity, preferably at least 95% sequence identity, more preferred
at least 98% sequence identity to SEQ ID NO: 8 (TwB5#1) or the mature
polypeptide thereof.
5. The recombinant host cell of item 3 or 4, wherein the host cell further com-
prises a fourth gene encoding a fourth enzyme having cytochrome P450
activity; wherein:
the fourth gene encoding a fourth enzyme having cytochrome P450 ac-
tivity encodes a polypeptide comprising an amino acid sequence having
at least 80 % sequence identity, preferably at least 85% sequence identity,
preferably at least 90% sequence identity, preferably at least 95% se-
quence identity more preferred at least 98% sequence identity to SEQ ID
NO: 5 (TwCYP82D213) or the mature polypeptide thereof.
6. The recombinant host cell of item 5, wherein the host cell further
comprises
a fifth gene encoding a fifth enzyme having cytochrome P450 activity
and/or a sixth gene encoding a sixth enzyme having cytochrome P450
activity, wherein:
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
the fifth gene encoding a fifth enzyme having cytochrome P450 activity
encodes a polypeptide comprising an amino acid sequence having at least
% sequence identity, preferably at least 85% sequence identity, prefer-
ably at least 90% sequence identity, preferably at least 95% sequence
5
identity, more preferred at least 98% sequence identity to SEQ ID NO: 6
(TwCYP82D217) or the mature polypeptide thereof; and
the sixth gene encoding a sixth enzyme having cytochrome P450 activity
encodes a polypeptide comprising an amino acid sequence having at least
80 % sequence identity, preferably at least 85% sequence identity, prefer-
10 ably at
least 90% sequence identity, preferably at least 95% sequence
identity, more preferred at least 98% sequence identity to SEQ ID NO: 7
(TwCYP82D275) or the mature polypeptide thereof.
7. The recombinant host cell according to any of the preceding items,
15 wherein
the host cell capable of producing miltiradiene and/or dehydro-
abietadiene is a recombinant cell that has been transformed with one or
more gene(s) encoding:
a. a geranylgeranyl diphosphate synthase;
b. a diterpene synthase capable of converting GGPP into miltiradiene;
20 c. a
combination of two or more diterpene synthases that in combina-
tion is capable of converting GGPP into miltiradiene; or
d. a copalyl diphosphate synthase and a miltiradiene synthase.
8. The recombinant host cell of item 7, wherein the geranylgeranyl diphos-
25 phate
synthase is a polypeptide comprising the amino acid sequence of
SEQ ID NO: 73 or SEQ ID NO: 81.
9. The recombinant host cell of item 7, wherein the combination of two or
more diterpene synthases, that is capable of converting GGPP into milti-
30
radiene, is the combination of a polypeptide comprising the amino acid
sequence of SEQ ID NO: 67 and a polypeptide comprising the amino acid
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
76
sequence of SEQ ID NO: 68; or is the combination of a polypeptide com-
prising the amino acid sequence of SEQ ID NO: 69 and a polypeptide
comprising the amino acid sequence of SEQ ID NO: 70.
10. The recombinant host cell of item 7, wherein the combination of a copalyl
diphosphate synthase and a miltiradiene synthase is a combination of a
polypeptide comprising the amino acid sequence of SEQ ID NO: 71 and a
polypeptide comprising the polypeptide of SEQ ID NO: 72.
11. The recombinant host cell according to any of the previous items wherein
the host cell is selected among prokaryotic and eukaryotic cells.
12. The recombinant host cell according to item 11, being a prokaryotic cell
selected among Escherichia, Bacillus, Lactobacillus and Corynebacterium
species.
13. The recombinant host cell of item 11, being a eukaryotic cell selected
among Saccharomyces, Scizosaccharomjices, Klyveromyces, Pichia,
Candida and Yarrowia species.
14. The recombinant host cell of item 11, where the cell is a S. cerevisiae
cell.
15. Use of a recombinant host cell according to any of the preceding items for
the production of an oxygenated diterpenoid compound.
16. The use of item 15, wherein the oxygenated diterpenoid compound is se-
lected among 14-0H-dehydroabietadiene, triptophenolide and triptonide.
17. The use of item 16, wherein the oxygenated diterpenoid compound is trip-
tonide, which triptonide is further converted into triptolide.
18. The use according to one of the items 15-17, wherein the oxygenated
diterpenoid compound is recovered using one or more separation and/or
chromatographic steps.
CA 03192028 2023-02-16
WO 2022/043461 PCT/EP2021/073656
77
19.A polypeptide having cytochrome P450 enzyme activity and comprising an
amino
acid sequence having at least 80 % sequence identity, preferably at least 85%
sequence identity, preferably at least 90% sequence identity, preferably at
least
95% sequence identity, preferably at least 96% sequence identity, preferably
at
least 97% sequence identity, preferably at least 98% sequence identity, or
even
100% sequence identity to one of the sequences SEQ ID NO: 1, SEQ ID NO: 2,
SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 6, SEQ ID NO: 7,
SEQ ID NO: 8 or the mature polypeptide thereof.
20.A polynucleotide encoding the polypeptide of item 19.
21.A plasmid, expression vector, expression construct or recombinant host
cell comprising a polynucleotide of item 20.
22. The compound 14-0H-dehydroabietadiene.