Note: Descriptions are shown in the official language in which they were submitted.
WO 2022/067426
PCT/CA2021/051347
1
SYSTEM AND METHOD FOR GENERATING AUGMENTED COMPLETE BLOOD
COUNT REPORTS
TECHNICAL FIELD
[001] The present invention generally relates to field of systems and methods
for
processing and reporting biomedical analyses and laboratory test results.
BACKGROUND
[002] It is estimated that more than 70% of clinical decisions are based on
biomedical
analysis of test results'. The interpretation of test results by clinicians is
thus of foremost
importance. However, the time available to a clinician for result
interpretation is often
insufficient to properly assess all test results, given the number of test
results available in
a single report. Furthermore, clinicians can face a cognitive limitation due
to the high
volume of data to integrate from a single report. The influence of some
analytes over other
ones is difficult to appraise, and thus interpreting the relation between the
different test
results is certainly not obvious from a simple review of laboratory test
reports.
[003] There is a need for means to help clinicians or other medical staff have
access to
more comprehensive test results as well as better tools for interpreting said
results. There
is a need for improved systems and methods that can guide or alert clinicians
in their
assessment of test results and that can assist them in clinical decision
making.
SUM MARY
[004] According to an aspect, an Artificial Intelligence (Al)-based system and
a method
are provided, for generating augmented test results, based on standard
laboratory test
results. Typical or standard laboratory test results include measured values
for a set of
known analytes (i.e. chemical components). The proposed method and system
allow
predicting a state (such as normal or abnormal) or a value range for analytes
that have
already been measured, or that have not been measured. Predicting analytes for
which
measured values are available can help in uncovering specific medical
conditions, which
1 Badrick, Tony. "Evidence-based laboratory medicine." The Clinical
biochemist. Reviews vol.
34,2 (2013): 43-6.
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
2
would otherwise stay unnoticed. When the predicted state or value of an
analyte differs
from the measured result, the difference may be explained by a medical
condition that is
not detectable from the measured result only. Predicting target analytes that
have not
been measured at all provides additional information on the medical conditions
of patients
that may otherwise require additional tests or procedures.
[005] According to the proposed method, different analyte classifiers are
trained, using
analysis test results that have been previously classified according to the
state or level of
target analytes to be predicted. The analyte prediction system comprises a
plurality of
trained analyte classifiers, wherein each classifier is specifically trained
and configured to
predict a given target analyte. The analyte prediction system is continuously
fed with new
test results, and can therefore predict a plurality of target analytes, using
the measured
test results from the laboratories. The predicted target analytes are
reported, with an
indication of the prediction certainty. Additional information may be reported
as well, such
as potential medical conditions to investigate or a recommendation for
additional lab tests.
In a preferred implementation of the system, the test results are complete
blood counts,
and the target analytes are blood target analytes.
[006] According to another aspect, the different analyte classifiers of the
proposed Al-
based analyte prediction system are periodically updated/retrained, using
newly collected
laboratory test result data. The performance of the Al-based system can be
tracked and
monitored, to detect potential drifts in the predicted results. In possible
implementations,
it is possible to identify new analyte predictors based on datasets of
analytes measured
from laboratory test results is provided, so as to add new target analytes to
the list of
analytes that can be predicted.
[007] According to an aspect, a method is provided, for generating an
augmented
complete blood count (CBC) report, based on a complete blood count (CBC) test.
The
method comprises accessing results of the CBC test of a given patient. The
results include
measured values for a plurality of blood analytes. The method also comprises
feeding the
CBC results to a blood analyte predictive application comprising machine
learning models
trained to predict values indicative of counts or concentrations of different
target blood
analytes. The machine learning models include a glycated hemoglobin (HbA1c)
classifier
trained on CBC tests from a plurality of individuals other than the given
patient. The
method includes outputting, by the HbA1c classifier, a predicted HbA1c value
indicative
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
3
of a HbA1c concentration in the blood of the given patient. The predicted
HbA1c value is
based on the measured values for blood analytes of the CBC test of the given
patient
other than HbA1C. The method also comprises reporting or displaying in the
augmented
complete blood count (CBC) report an indication of a possible medical
condition when the
predicted HbA1c value is above a predetermined HbA1c threshold. The possible
medical
condition may comprise a prediabetes or diabetes condition.
[008] In possible implementations, the results of the CBC test include the
gender and
age of the patient tested, and the predicted HbA1c value is further based on
the gender
and the age. The predicted HbA1c value is preferably based solely on the CBC
results,
without using any other external data or markers. Preferably, the HbA1c
classifier further
outputs an indication of the likelihood or probability that the predicted
HbA1c value be
above or below a given threshold. In possible implementations, the indication
of the
medical condition is displayed only when the probability that the predicted
HbA1c value
is above a given threshold, such as 80%. The indication of the medical
condition can be
performed via a Graphical User Interface (GUI) or as an electronic blood test
report. The
augmented CBC report comprises the measured values for the plurality of blood
analytes
in addition to the indication of the possible medical condition. In possible
implementations,
the method can include a step of determining, based on the predicted HbA1C
value,
whether additional biomedical test(s) are required, and an indication of the
additional
biomedical test(s) can be reported or displayed on the augmented CBC report.
[009] In possible implementations, the method can include the step measuring
the
plurality of blood analytes with laboratory equipment, such as with automated
hematology
analyzer(s). The method may also include storing the CBC results in one or
more data
storages of a Laboratory Information System (LIS), and a step of connecting to
the
Laboratory Information System (LIS) to access the CBC results of a given
patient.
[0010] In possible implementations, generating the predicted HbA1c value is
performed
based on a subset of the measured values for blood analytes other than HbA1C,
i.e. not
all measured analytes from the CBC test need to be used.
[0011] In possible implementation, the HbA1c classifier predicts the HbA1C
value at least
based on the age, the gender, white blood cells (WBC); the red cell
distribution width
(RDW), the lymphocyte count (LY#), the basophil percentage (c/o) and the mean
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
4
corpuscular hemoglobin (MCH). The HbA1c classifier may assign most weight to
the
following measured values of blood analytes when predicting the predicted
HbA1c value
: white blood cells (WBC); the basophil count or percentage (BA# or BA%), the
lymphocyte count or percentage (LY# or LY%), the eosinophil count or
percentage (E0#
or E0 /0), the red cell distribution width (RDVV); and the mean corpuscular
hemoglobin
(MCH). The CBC results inputted in the HbA1c classifier for predicting the
HbA1c value
can comprise measured values for: basophil count and basophil concentration
(BA# and
BA%), lymphocyte count and the lymphocyte concentration (LY# and LY%),
eosinophil
count and eosinophil concentration (E0# and E0%), neutrophil count and the
neutrophil
concentration (NE# and NE%), monocyte count or concentration (MO# or M0%),
mean
corpuscular hemoglobin (MCH) and the mean corpuscular hemoglobin concentration
(MCHC), mean corpuscular volume (MCV), platelet count (PLT) and the mean
platelet
volume (MPV), red cell distribution width (RDVV); and white blood cells (WBC),
red blood
cells (RBC), hematocrit (HCT); and hemoglobin concentration (HGB).
[0012] In possible implementations, the HbA1c classifier is periodically
retraining with a
dataset comprising newly added CBC results, whereby the HbA1c classifiers'
hyperparameters are iteratively adjusted. Training or retraining of the the
HbA1c classifier
is preferably performed by solely keeping in the dataset the CBC results which
consisted
in first CBC results for an individual, to avoid bias when training the HbA1c
classifier. In
possible implementations, the glycated hemoglobin (HbA1C) classifier is of a
random
forest classifier type. The method also preferably comprises normalizing and
standardizing the measured values of the plurality of blood analytes, based on
the gender
and age of the individual tested, so as to generate therefrom processed blood
test data.
This processed blood test data is fed as the CBC results to the blood analyte
predicting
application.
[0013] In possible implementations, the blood analyte predicting application
further
comprises a trained 25-0H vitamin D classifier. In this case, the method
further comprises
accessing results of a basic metabolic panel (BMP) test of the given patient
in addition to
the CBC results and outputting, by the 25-0H vitamin D classifier, a predicted
25-0H
vitamin D value indicative of a 25-0H vitamin D concentration in the blood of
the given
patient, the predicted 25-0H vitamin D value being based on the measured
values for
blood analytes of the CBC test of the given patient other than 25-0H vitamin
D. In the
augmented complete blood count (CBC) report, an indication of a possible low
25-0H
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
vitamin D concentration, when the predicted vitamin D value is below a
predetermined
25-0H vitamin D threshold, can be reported or displayed. In possible
implementations,
the BM P results inputted in the 25-0H vitamin D classifier for predicting the
25-0H vitamin
D value comprise measured values for: LDH (Lactate Dehydrogenase), AST
(Aspartate
5 Aminotransferase), ALT (Alanine Am inotransferase),
GGT (Gamma-
Glutamyltransferase), Triglycerides (TG); Na (Sodium), K (Potassium) and Cl
(Chloride).
The 25-0H vitamin D classifier preferably assigns most weight to the following
measured
values of blood analytes when predicting the predicted 25-0H vitamin D value:
high-
density lipoproteins (HDL), mean corpuscular volume (MCV); and triglycerides
concentration (TG).
[0014] In possible implementations, the 25-0H vitamin D classifier also
assigns weight to
the age and gender of the given patient, and the month (or equivalent: date,
week) at
which the CBC test was performed when predicting the predicted 25-0H vitamin D
value
[0015] According to another, a method is provided for uncovering a medical
condition
based on a complete blood count (CBC) test. the method comprises steps of:
connecting
to a Laboratory Information System (LIS) to access CBC results of the complete
blood
test of a given patient, the CBC results including the gender and age of the
individual
tested and measured values for a plurality of blood analytes; feeding the CBC
results to
a blood analyte predicting application comprising machine learning models
trained to
predict values indicative of counts or concentrations of different target
blood analytes, the
machine learning models including a glycated hemoglobin (HbA1c) classifier
trained on
CBC blood tests from a plurality of individuals other than the given patient;
outputting, by
the HbA1c classifier, a predicted HbA1c value indicative of a HbA1c
concentration in the
blood of the given patient, the predicted HbA1c value being based on the
measured
values for blood analytes of the CBC test of the given patient other than
HbA1c; and
generating an indication of a medical condition when the predicted HbA1c value
is outside
a range of values considered acceptable. The predicted HbA1c value or range of
values
can be performed at least based on the age and gender of the given patient,
and on the
measured values for: the white blood cells (WBC); the basophil count or
percentage (BA#
or BA%), the lymphocyte count or percentage (LY# or LY%), the eosinophil count
or
percentage (E0# or E0%), the red cell distribution width (RDVV); and the mean
corpuscular hemoglobin (MCH). The blood analyte predicting application may
also include
a 25-0H vitamin D classifier trained on CBC blood tests from a plurality of
individuals
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
6
other than the given patient, the method further comprising outputting, by the
25-0H
vitamin D classifier, a predicted 25-0H vitamin D value indicative of a 25-0H
vitamin D
concentration in the blood of the given patient, the predicted 25-0H vitamin D
value being
based on the measured values for blood analytes of the CBC test of the given
patient
other than 25-0H vitamin D; and generating an indication of a medical
condition when the
predicted 25-0H vitamin D value is outside a range of values considered
acceptable. The
predicted 25-0H vitamin D value or range of values is preferably performed at
least based
on the age and gender of the given patient, the month during which the CBC and
BMP
tests were performed and based at least on the measured values for high-
density
lipoproteins (HDL); mean corpuscular volume (MCV); and triglycerides
concentration
(TG).
[0016] According to a possible implementation, a system is provided for
generating the
augmented complete blood count (CBC) report. The system comprises an access
module
for accessing data storage storing CBC results of the complete blood test of a
given
patient, the CBC results including the gender and age of the individual tested
and
measured values for a plurality of blood analytes; a server comprising a blood
analyte
predictive application comprising machine learning models trained to predict
values
indicative of counts or concentrations of different target blood analytes, the
machine
learning models including a glycated hemoglobin (HbA1c) classifier trained on
CBC blood
tests from a plurality of individuals other than the given patient; one or
more computer-
readable medium(s) comprising instructions stored thereon to cause a computer
to :feed
the CBC results to the blood analyte predicting application; output, by the
HbA1c
classifier, a predicted HbA1c value indicative of a HbA1c concentration in the
blood of the
given patient, the predicted HbA1c value being based on the measured values
for blood
analytes of the CBC test of the given patient other than HbA1c; and display in
the
augmented complete blood count (CBC) report an indication of a possible
medical
condition when the predicted HbA1c value is above a predetermined HbA1c
threshold.
[0017] The system according to claim 28, wherein the results of the CBC test
include the
gender and age of the patient tested, and wherein the predicted HbA1c value is
further
based on the gender and the age. The system may further comprise a trained 25-
0H
vitamin D classifier, trained and configured to output a predicted 25-0H
vitamin D value
indicative of a 25-0H vitamin D concentration in the blood of the given
patient, the
predicted 25-0H vitamin D value being based on the measured values for blood
analytes
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
7
of the CBC test of the given patient other than 25-0H vitamin D; the blood
analyte
predicting application being further configured to display in the augmented
complete blood
count (CBC) report an indication of a possible low 25-0H vitamin D
concentration when
the predicted 25-0H vitamin D value is below a predetermined 25-0H vitamin D
threshold.
The access module may be provided as an Application Programming Interface, to
access
the data storage storing CBC results.
[0018] Other features and advantages of the embodiments of the present
invention will
be better understood upon reading of preferred embodiments thereof with
reference to
the appended drawings.
BRIEF DESCRIPTION OF THE DRAWINGS
[0019] FIG. 1A and 1B illustrate a flow diagram showing possible steps of a
method for
generating augmented complete blood count reports, according to a possible
implementation.
[0020] FIG. 2A and 2B are more detailed flow diagrams showing steps of the
predictive
method and of the training method, according to possible implementations.
[0021] FIG. 3 is a schematic diagram showing the initial steps of the method,
from
accessing the CBC test results collected by the different laboratories to
preprocessing the
resulting CBC test results, according to a possible implementation.
[0022] FIG. 4 is a schematic diagram of elements of a system for generating
augmented
complete blood count (CBC) reports comprising a plurality of trained analyte
classifiers,
according to a possible implementation.
[0023] FIG. 5 is a schematic diagram of different modules and components of
the system,
according to a possible implementation.
[0024] FIG. 6 is a schematic diagram providing examples of possible target
analytes that
can be predicted from exemplary CBC test results, according to a possible
implementation.
[0025] FIG. 7 is an exemplary augmented test report generated according to the
proposed method and system, according to a possible implementation.
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
8
[0026] FIG. 8A is a precision-recall graph for the HbA1c classifier. FIG. 8B
is a SHAP
graph of the HbA1c classifier. FIGs. 8C is a graph showing, fora given
patient, the blood
analytes having the most weight in predicting the HbA1c value, for which the
probability
of the prediction is 95%. FIGs. 8D is graph showing, fora given patient, the
blood analytes
having the most weight in predicting the HbA1c value, for which the
probability of the
prediction is 50%.
[0027] FIG. 9A is a precision-recall graph for the 25-0H vitamin D classifier.
FIG. 9B is
a SHAP graph of the 25-0H vitamin D classifier. FIGs. 9C is graph showing, for
a given
patient, the blood analytes having the most weight in a prediction of the 25-
0H vitamin D
value, for which the probability of the prediction is 95%. FIGs. 9D is graph
showing, for a
given patient, the blood analytes having the most weight in a prediction of
the 25-0H
vitamin D value, for which the likelihood associated with the prediction is
50%.
[0028] FIG. 10 is another flow diagram of possible steps of the method for
generating an
augmented complete blood count (CBC) report, according to a possible
implementation
in which HbA1c and 25-0H vitamin D values are predicted.
[0029] FIGs. 11A-11F and 12A-120 are different graphs showing the
transformation
and/or distribution of the blood analysis dataset used for generating the
HbA1c classifier,
part of the blood analyte predictive application.
[0030] It should be noted that the appended drawings illustrate only exemplary
embodiments of the invention and are therefore not to be construed as limiting
of its
scope, for the invention may admit to other equally effective embodiments.
DETAILED DESCRIPTION
[0031] While traditional laboratory test result analysis is made with a
"granular" approach,
in which each analyte is reviewed more or less independently from other
analytes, and
compared to a reference interval, the proposed method and system provide a
global
analysis of the different measured analytes. The proposed method and system
are
particularly useful for blood analysis, such as the "complete blood count"
(CBC) analysis
and basic metabolic panel, but they can be adapted to other types of
biomedical analysis,
including for example urine and/or biopsies.
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
9
[0032] According to one aspect of the invention, the use of specifically-
trained machine
learning models allows putting in relation different measured analytes and
predicting other
ones, which have either been measured or not, allowing clinicians to uncover
latent
relations or patient conditions that are otherwise often eluded. The proposed
method thus
provides additional information that is not available or readily apparent from
standard test
reports, such as complete blood count (CBC) reports. According to an aspect,
an
augmented test report is generated, which includes not only the measured test
results,
but also the additional information derived therefrom, such as the predicted
levels or
states of target analytes, and recommendations or alerts in support to medical
decisions
that are based at least in part on these predictions. In possible
implementations, the
augmented test report can be an augmented complete blood count report, which
includes
measured values for blood analytes, and also an indication of predicted values
for at least
some of the blood analytes that are outside normal/predetermined ranges or
thresholds.
In possible implementations, discrepancies between predicted and measured
analytes
can be automatically identified and reported, as they can be indicative of
medical
conditions or illnesses that would otherwise go undetected.
[0033] According to another aspect, the proposed method comprises periodically
retraining the machine learning models with new incoming test results, to
improve their
precision and sensitivity. Quality control of the system is also provided, to
detect any drift
in the predicted analytes.
[0034] A machine learning model (also referred to as "Al" model) is a set of
functions and
algorithms that are trained to recognize patterns in the data that is inputted
therein. A
machine learning model is built such that, as training data is processed
therethrough, its
algorithms will adjust their parameters, such as internal coefficients,
weights and biases,
as they learn. The behavior of the machine learning model can also be adjusted
using
"hyperparameters", which are supplied to the model.
[0035] Throughout the present description, the expressions "medical test" or
"biomedical
test" refers to any test intended to quality or quantity an individual's
health or condition
and/or to diagnose pathological or nonpathological conditions of the human
body, by the
analysis of samples and specimens. A complete blood count (CBC) test is a test
performed in a medical laboratory, using laboratory equipment, such as
automated
hematology analyzers. Results of a CBC test provide information about the
type, number,
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
concentration and other characteristics of elements found in the tested blood
including
red blood cells (RBCs), white blood cells (WBCs) and platelets. A CBC test can
reveal
anomalies affecting elements essential for the production and proper
functioning of blood
cells (functioning of the spleen, pancreas, liver and kidneys; nutritional
status of amino
5 acids, iron, vitamin B12, folic acid, etc.).
[0036] "Test results" refers to the data resulting from the analysis of
samples or
specimens, such as CBC results. This analysis is typically conducted by
medical
laboratories. "Test results" may also be referred to as "laboratory test
results", "measured
test results" or "standard laboratory test results". As an example, only, test
result
10 stemming from a medical analysis can consist of a measured concentration
of a given
component, of its relative or absolute value, etc. CBC results include
measured values for
a plurality of blood analytes, and also includes the gender and age of the
individual tested.
[0037] A "target" analyte is an analyte for which the proposed method and
system can
predict the result using a machine learning model. In other words, a "target
analyte" is an
analyte for which we want to predict what the measured result should be,
without
necessarily having measured the analyte in question. The predicted result of a
given
analyte is based on the measured results obtained from other analytes. A
classification
model will classify the target analyte based on a threshold often set by
medical
community. The threshold reflects the marginal limit of a risk state for the
patient. The
model final output is a calibrated probability or likelihood (0-100%) of
exceeding the
threshold of the analyte. The "predicted" result for a "target analyte" is
thus the response
provided by a trained machine learning model for said analyte. For example,
for target
analyte X, the predicted result can be "low" or "abnormal". The machine
learning model
can also provide a level of confidence in its prediction. The confidence level
or interval
corresponds to the overall performance of the model at a specific threshold.
For example,
it can correspond to confidence interval for all patients predicted at a
probability 85% or
higher. The more observations there is in a category (in other words without
false
positives), the smaller the confidence interval. The confidence interval
provides an
indication of the volatility of the predictions.
[0038] According to one aspect, a method and a system are provided for
generating an
augmented complete blood count (CBC) report, based on a complete blood count
(CBC)
test. The system comprises one or more servers running a blood analyte
predictive
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
11
application which comprises a plurality of trained machine learning models,
preferably of
the "classifier" type, each associated with different analytes. "Classifiers"
refer to a specific
type of machine learning models which is used to assign a class or label to
datapoints. In
the present case, the classifiers are trained to assign classes or labels to
the different
target analytes, based on measured values of different analytes. The classes
or labels
can include, for example, whether the analyte level is normal or abnormal, or
whether the
level is low or high, a range of values or a discrete value, compared to
predetermined
thresholds.
[0039] In possible implementations, the system is continuously feed with
laboratory test
results and is configured and adapted to continuously process the flow test
result data
and generate therefrom augmented test results, including both measured and
predicted
analytes. By "continuous", it is meant that the process is performed either
without
interruption, or that it is periodically repeated at predetermined type
intervals. The
augmented test results can be formatted into "augmented" test reports and
distributed or
accessed via a Laboratory Information System (LIS) or other similar software
applications.
The term "augmented" refers to the additional information that is revealed and
rendered
accessible from the standard laboratory test results, this additional
information being
"encoded" or "latent" in the measured test results but highlighted by the
proposed system
and method. The laboratories may use what is called MLOps, which facilitates
Cl/CD
(continuous integration / continuous deployment). This process or pipeline
allows for
continuous data ingestion to the models, in addition to model retraining,
model monitoring
and model deployment. In possible implementations, a software tool can be used
to
monitor data drift. This tool continuously monitors the distribution of
observations over
time and sends alerts whenever a shift of distribution of an analyte is
detected. Shifts are
often cause by decalibration of lab equipment or by a demographic change.
[0040] The proposed system and method will be described in more detail with
reference
to FIGs.1A to 12D.
Analyte Prediction Process and Generation of Augmented Test Reports
[0041] Referring to FIGs. 1A and 1B, an overview of the different steps of the
proposed
method for generating an augmented complete blood count (CBC) report is
provided.
Starting with block 100 shown on the right side of the figure 1B, patients
consult health
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
12
clinics or medical laboratories, to obtain laboratory test results, as
prescribed by their
clinicians (step 110). A possible step of the proposed method comprises
measuring, for a
plurality of individuals or "patients", the counts or concentration of their
blood analytes
with laboratory equipment, such as automated blood analyzers. The results of
the CBC
test thus include measured values for a plurality of blood analytes, as per
the exemplary
table provided on the left-hand side of FIG. 3. The CBC results are then
stored in one or
more data storages, which can be part of a Laboratory Information System
(LIS).
[0042] In possible implementations, the method solely comprises accessing the
results
of the CBC test, without necessarily conducting blood analyses. In order to
access the
CBC test results of a given patient, the method may comprise a step of
connecting to
servers and/or databases of a Laboratory Information System (LIS). The
laboratory test
equipment produces the test results (step 120), that are transferred to a
Laboratory
Information System (LIS), which consists of a system that includes data
storage and
databases 112 that record, manage and store test results from different
laboratories (step
210). Block 200 represents steps of the method that occur in the LIS,
including the storing
of the test results produced by the different labs and clinics associated with
the LIS.
[0043] Referring now to FIG 1A, at block 300, the different steps for
generating the
augmented test results are shown. Those steps occur in the system which
comprises one
or more server(s) 500, schematically represented at the bottom of FIG.1A. The
system
500 comprises an access module for accessing the data storage storing CBC
results,
which may include connectors such as Application Programming Interfaces (API).
The
server may also include databases, computer-readable medium and processor(s)
running
algorithms, functions and machine learning models that interact with one
another and are
configured to predict target analytes based on the CBC test results. The
software
components can be packaged in a predictive application, which is referred to
hereafter as
the "blood analyte predictive application". The application can reside on a
single server,
or on a group of distributed servers. The system can be provided on a "local"
server,
connected to the same network as the LIS, or it can be cloud-based.
[0044] Periodically, at step 310, an incremental batch data load is performed.
The access
module of the analyte prediction system periodically connects to the LIS
database 112, to
fetch newly received analysis test results from the hematology analyzers. The
test results
can have different formats and may include different types of data (such as
the measured
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
13
results), depending on the analysis having been conducted, however CBC test
results will
typically have the same standard format, with the same measured blood
analytes. The
test results include at least a unique identifier, the gender of the
individual being tested
and their age, and the measured values of the blood analytes being tested. In
the following
paragraphs, reference will be made to blood test analysis and to blood
analytes, but the
process and systems described hereinbelow can be used for other types of
biomedical
analysis.
[0045] Referring to FIG. 3, an example of a CBC test report is illustrated and
identified by
numeral 124. The report 124 comprises the patient's ID (130), their gender
(132) and their
age (134). The report also includes a lists of blood analytes (136), including
for example
the concentration of basophils (BA#), the basophil percentage (c)/0), the mean
corpuscular
hemoglobin concentration (MCHC), etc. For each analyte, a measured value is
provided
as well as the units of the measured value and a reference interval. A
"reference interval"
generally corresponds to a range of normal values established for a given
gender and a
given age interval. The test report can take different forms: they do not need
to be in
printed form, they can be displayed on graphical user interfaces (GUI) or as
an electronic
blood test report, and they can also be simply stored on memory storage, such
as in one
or more tables of databases.
[0046] Referring back to FIG.1A, once the dataset of newly received results
has been
loaded from the LIS database (step 310), the test results are preprocessed,
and fed to
the appropriate trained machine learning models. As explained previously, the
blood
analyte predictive application comprises different machine learning models
trained to
predict values indicative of counts or concentrations of different target
blood analytes. In
possible implementations, the machine learning models are specifically trained
blood
analyte classifiers, each classifier being associated to a predetermined
target blood
analyte. Step 320 thus consists of matching, based on the test results
available for a given
individual, the trained model(s) that can be used to predict one or more
target analytes.
Once the trained models/classifiers have been identified, they are used to
predict levels
(such as low/high, normal/abnormal), or range of values, of the target
analytes (step 330),
the prediction being associated with a probability or likelihood associated
therewith. The
predicted results are then sent back to the LIS, where they are combined with
the other
standard test results, to generate the "augmented" test report.
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
14
[0047] Referring to FIG. 6, the target analytes, i.e. the analytes for which
predictions can
be made with the present system, are numerous. They include at least:
ferritin, Hb1Ac,
and 25-0H vitamin D. In possible implementations, it can also be considered to
predict
values indicative of TSH, testosterone, M protein, ALT, calcium, PTH,
cholesterol, CA125,
magnesium, PTH, oestradiol, LH, FSH and HBsAg, with different machine learning
models specifically trained for each target analyte. The prediction of
additional target
analytes is also possible, provided the measured analytes and said additional
target
analytes are somewhat correlated. In some possible implementations, the system
can be
configured to continuously monitor newly received test results and detect,
based on the
newly measured analytes, additional or new target analytes that may be
predicted.
Correlation tools can be applied to the collected test data, to identify
potential predictors,
i.e. analytes that provide information on other dependent analytes.
[0048] Additional processing can be applied to the predicted test results, to
derive other
relevant information that is worth notifying on the augmented test reports.
For example, a
difference between a predicted and a measured test result for a given target
analyte can
be indicative of a medical condition that would not have been apparent from
the standard
measured result alone. Differences that are worth notifying can be determined
based on
predetermined rules. For example, an inflammatory condition can increase
ferritin
concentration in patients. In such cases, the measured ferritin may be higher
than normal,
while the predicted ferritin is within or below normal thresholds. This
discrepancy between
the measured and the predicted ferritin levels can be flagged on the augmented
test
report, since the inflammatory condition could hide a possible iron deficiency
condition.
Thus, the combination of measured and predicted ferritin levels provides more
information
to the clinician than the measured ferritin alone. Indications or
recommendations for
additional laboratory tests, to confirm a potential medical condition that is
suspected in
view of the predicted results, can be added to the augmented test report. In
some cases,
the predicted result may allow avoiding unnecessary tests that would otherwise
be
needed.
[0049] Referring to FIGs. 2A and 2B, a more detailed diagram of the different
steps of the
predictive process is illustrated, Starting with FIG. 2B, at step 311, the
data batch loading
from the LIS can be made using an Application Programming Interface (API) that
periodically queries the LIS database to fetch new observations, i.e. newly
received test
results from laboratories. This process is semi-continuous, since the new test
results
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
retrieval is typically made every 30 seconds, but of course other periods can
be set (every
2 seconds, or once a day) depending on the typical flow of incoming test
results. At step
321, the new test results are evaluated and preprocessed. This step 321
comprises the
sub-steps 321a and 321b, which include discarding test results with missing
values (321a)
5 for analytes that are required for the predictions. While in this
exemplary implementation
the test results with missing data are discarded, it would also be possible,
in other
implementations, to impute the missing data. The test results are also
normalized and
standardised (321b), using the same pre-processing algorithms used for the
training of
the blood analyte classifiers. Normalizing and standardizing the measured
values of the
10 plurality of blood analytes can be performed based on the gender and age
of the
individual. Normalizing and standardizing the measured values generates
processed
blood test data that is fed to the classifiers of the blood analyte predictive
application.
Depending on the measured analytes present in a given test report, the blood
analyte
classifiers are automatically selected, and predictions are generated.
15 [0050] Referring to FIG. 4, a schematic illustration of the blood
analyte predictive
application is provided. The application runs on server 500 and comprises a
set of trained
machine learning models, such as classifiers (325, 326, 327, 328), each
associated with
a given target analyte. The test results data from a lab report 120 are
inputted in the
system. Based on the measured analytes in the report, the classifiers
associated to the
target analytes that can be predicted from the measured results are selected
and used to
generate the predicted analytes. The predicted analytes (and/or information
derived
therefrom) is reported or displayed on the augmented test report 130. For
example, a set
of X measured analytes may be needed to predict the HbA1c, using classifier
325, while
a different set of Y measured analytes may be needed to predict 25-0H vitamin
D, using
classifier 327. The predictive application is configured to select, based on
the available
measured values of a CBC test report, the trained classifiers that allow
outputting a
maximum number of predicted analytes and/or medical conditions.
[0051] As mentioned previously, the predictions can include the status of the
analytes,
such as normal or abnormal, a predicted value range, or whether the predicted
analyte
has a low, normal or high value compared to standard comparison intervals. Of
course,
additional or different predictions can be made, depending on the
classification used when
training the classifiers, as will be explained in more detail below.
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
16
[0052] In addition to providing predicted states or levels of analytes, the
likelihood or
probability associated with the predicted value can also be provided on the
augmented
test report. Additional information that can be reported may include for
example the
classification accuracy, the classification error rate, the confidence
interval and/or the
positive predictive value. Clinicians are thus informed of the degree of
certainty associated
to the predicted analyte value. This information can be helpful to clinicians
in deciding
whether they want to consider all or only some of the predicted analytes. The
level of
confidence in the prediction provides an indication about the general
performance of the
model. The predicted analytes (i.e. levels or values) are returned to the LIS
database,
where they can be further processed, such as by comparing them to the measured
values
and by applying preconfigured rules to determine whether a given medical
condition is
suspected or if additional tests are required. A plurality of measured and
predicted results
can be compared with one another when assessing if a given medical condition
is met.
Preconfigured rules for identifying medical conditions can include, as
examples only:
1) If the measured value of analyte A differs by X% or more from the predicted
value
of analyte A, include notice of possible medical condition Z on augmented
report.
2) If the predicted level of analyte A is low and the predicted level of
analyte B is high,
include note for additional test W on augmented test report;
3) If the predicted level of analyte A is X% lower or higher than the
threshold
determined as normal for said given analyte, with a reasonably small range at
95%
confidence interval, include an indication of a possible medical condition
associated with the predicted value of the analyte.
[0053] Referring to FIG. 7, an exemplary augmented test report is illustrated.
The
augmented test report includes measured values of analytes, in this case FSH,
LH and
prolactin, and predicted values for HbA1c, and an indication of a possible
medical
condition can be displayed since the predicted HbA1c value is above a
predetermined
HbA1c threshold. As can be appreciated, with the present system, a target
analyte can
be predicted based on the measured values of other, distinct analytes. In
possible
implementation, such as for prediction of glycated hemoglobin (HbA1c), the
possible
medical condition may comprise a prediabetes or diabetes condition.
Examples
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
17
HbA1c predictions based on CBC test results
[0054] Hemoglobin corresponds to the portion of red blood cells which carries
oxygen
from lungs to other parts of the body. A percentage of the hemoglobin also has
glucose
attached to it, and this type of hemoglobin is known as glycated hemoglobin or
HbA1c.
The amount of HbA1c depends on the level of glucose in the blood: the higher
the blood
sugar, the higher is the amount of HbA1c. In a A1c test, HbA1c measurements
represent
the average amount of glucose attached to hemoglobin over the past three
months. When
HbA1c levels are high, it can be an indication of prediabetes or diabetes. The
normal
range of HbA1c is typically between around 4% and 5.9%, and this value varies
according
to age and gender. HbA1c can also be referred to as A1c, glycohemoglobin,
glycated
hemoglobin and glycosylated hemoglobin.
[0055] When individuals are submitted to a standard CBC test, with or without
differential,
HbA1c is not measured. A list of blood analyte typically measured with a CBC
test is
provided in table 124 of FIG. 3. A prediabetes or diabetes medical condition
is therefore
not detectable by clinicians when they are only provided with the CBC report
of an
individual. A specific A1c test is generally required for clinicians to detect
or confirm a
prediabetes or diabetes condition. Other specific tests to detect prediabetes
condition
include "fasting plasma glucose" or a "50g (or other similar quantity) glucose
test",
according to which blood glucose levels (glycemia) is measured 1 hour after
drinking a
solution containing 50g of glucose. Thus, a supplemental test, other than the
standard
CBC test, is traditionally required to detect prediabetes (or diabetes)
conditions since
glucose levels or concentrations are not measured in a CBC blood test.
Additional tests
result in more delays for individuals before being properly diagnosed,
additional costs,
and in some cases, lighter or borderline prediabetes conditions stay unnoticed
until
symptoms are felt by the individuals concerned.
[0056] The Applicant has however discovered that a machine learning model, of
the
classifier type, specifically trained using prior CBC test results from a
plurality of
individuals, can be used to generate a predicted HbA1c value of a given
patient solely
based on the patient's CBC test results. The predicted HbA1c value does not
necessarily
correspond to a predicted measure of the HbA1c concentration, it can simply be
a
prediction indicative of the HbA1c concentration in the patient's blood. The
predicted
HbA1c value outputted by the classifier, referred to as a "glycated hemoglobin
(HbA1c)
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
18
classifier", is thus based on the measured values of blood analytes of the CBC
test of the
given patient other than HbA1c. While different types of classifiers can be
used, the
glycated hemoglobin (HbA1c) classifier is preferably of a random forest
classifier. The
Hb1Ac classifier can be provided as part of a blood analyte predictive
application
comprising different machine learning models trained to predict values
indicative of counts
or concentrations of different target blood analytes. The blood analyte
predictive
application can be used, or interfaced with, to display, as part of CBC
reports, an
indication of a possible medical condition, such as prediabetes or diabetes,
when the
predicted HbA1c value is above a predetermined HbA1c threshold. The predicted
HbA1c
value can thus be based solely on the CBC results, without using any other
external data
or markers. Age and gender being typically included in the data part of the
CBC test
results; these two features are also used in predicting the HbA1c value from
the CBC test
results. In a possible implementation, the predicted HbA1c value outputted by
the HbA1c
classifier can a binary value, such as 0 if the predicted HbA1c concentration
is below a
given HbA1c threshold (such as 5.6%), and 1 if the predicted HbA1c
concentration is
equal or above said given HbA1c threshold.
[0057] In possible implementations of the method, the trained HbA1c classifier
outputs
the predicted HbA1c with a given probability or likelihood. The predicted
HbA1c value is
thus typically associated with a probability that the value be classified in a
given class
(such as above a preset threshold, associated with an "abnormal"
concentration). The
predicted HbA1c value and/or indication of the medical condition are therefore
preferably
displayed only when the estimated probability that the predicted HbA1c value
is above a
given threshold, typically expressed as a percentage, such as above 80%, 85%
or 90%.
When executing the report generation, the augmented CBC report may comprise
the
measured values for the plurality of blood analytes, in addition to the
indication of the
possible medical condition and/or predicted HbA1c value, as well as the
likelihood
associated with the prediction made by the classifier. For example, the HbA1c
classifier
can predict with a probability of at least 85% that the HbA1c concentration in
the blood of
an individual is abnormal, or above a predetermined threshold, such as 5.6%,
based on
his CBC test results. The HbA1c threshold for determining whether the
prediction should
be set to the first or second binary value (such as 0 if below the threshold
and 1 if equal
or above the threshold) is preferably any number between 5 and 6 (for HbA1c
test results
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
19
expressed in %). In the experiments conducted, the threshold was set to 5.6.
The HbA1c
threshold is preferably set as a function of the hematology analyzer used.
[0058] In this case, an alert or indication that the individual may suffer
from a prediabetes
condition can be added to the CBC report (resulting in an augmented CBC
report). The
blood analyte predictive application can also be configured to generate an
indication of a
diabetes condition when the classifier outputs a prediction that the HbA1c
value is above
a second HbA1c threshold, such as 7%, with a probability above 85%. An
indication of a
medical condition may not necessarily be displayed on the report ¨ in possible
implementations, the application can be configured to determine, based on the
predicted
HbA1c value, whether additional biomedical test(s) are required. In this case,
what is
displayed on the augmented CBC report is an indication of suggested additional
biomedical test(s), such as a A1c test.
[0059] During experimental trials, it has been found that the generation of
the predicted
HbA1c value can be performed based on a subset of the measured values obtained
from
the CBC test (where the measured values do not include HbA1C measurements ¨ as
explained above, HbA1c is typically not measured by CBC analysis.) In other
word, not
all measured CBC results need to be used by the HbA1c classifier to output a
prediction
of the HbA1c value being associated with a high probability.
[0060] Experimental trials have shown that the trained HbA1c classifier can,
in most
cases, predict the HbA1c value of individuals, at least based on their age,
their gender,
their red cell distribution width (RDVV), their white blood cells (WBC), their
lymphocyte
count (LY#), their basophil percentage (%) and their mean corpuscular
hemoglobin
(MCH).
[0061] In possible implementations, the CBC results inputted in the HbA1c
classifier may
comprise measured values for: basophil count and/or basophil concentration
(BA# and
BA%); lymphocyte count and/or the lymphocyte concentration (LY# and LYcY0);
eosinophil
count and/or eosinophil concentration (E0# and E0%); neutrophil count and/or
the
neutrophil concentration (NE# and NE%); monocyte count and/or concentration
(MO# or
M0%); mean corpuscular hemoglobin (MCH) and/or the mean corpuscular hemoglobin
concentration (MCHC); mean corpuscular volume (MCV); platelet count (PLT)
and/or the
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
mean platelet volume (MPV); red cell distribution width (RDVV) and white blood
cells
(WBC); red blood cells (RBC); hematocrit (HOT); and hemoglobin concentration
(HGB).
[0062] Referring now to FIGs. 8A to 8D, different graphs are provided to
demonstrate and
explain the performance of the HbA1c classifier, after being trained using a
dataset of
5 90406 unique CBC test results, standardized, and normalized based on the
gender and
age of the individuals tested. The standardization and normalization process
of the
measured values results in a processed dataset that can be fed to classifiers
of the blood
analyte predicting application. By "unique", it is meant that training the
HbA1c classifier
was performed by solely keeping in the dataset the CBC results which consisted
in first
10 (or unique) CBC results for an individual, to avoid bias when training
the HbA1c classifier.
The training dataset comprised 36.75% CBC test results associated with a HbA1c
value
above the HbA1c threshold, and 63.25% CBC test results associated with a HbA2c
value
below the threshold. The dataset comprised CBC test results collected over
more than
five years, from 2015 to 2021.
15 [0063] FIG. 8A is a 2-class precision-recall curve having an Average
Precision (AP) of
0.64, where the curve represents the tradeoff between recall (the proportion
of "true
positives" predictions over the number of true and false positives) and
precision (the
proportion "true positives" over the number of true positives and false
negatives), and the
Average Precision of the curve corresponds to the weighted-average precision
across all
20 thresholds. When periodically retraining the HbA1c classifier with a
dataset comprising
newly added CBC results, and iteratively adjusting hyperparameters specific to
the HbA1c
classifier, the shape of the precision-recall should stay relatively stable,
as well as the AP.
[0064] FIG. 8B is a SHAP graph (Shapley Additive explanations) which explains
the
contribution of each feature (such as age, gender, and blood analytes measured
in the
CBC test) in predicting the HbA1c value. While the measured values listed
above can all
be fed to the HbA1c classifier, analysis of the performance of the HbA1c
classifier has
shown that the trained HbA1c conceived for the present augmented report
generation
method and system assigns the most weight to age of the individual, red cell
distribution
width (RDW) result; gender; lymphocyte count (LY#), basophil count or
percentage (BA#
or BA%) and mean corpuscular hemoglobin (MCH). While not shown in the graph,
the
white blood cells (WBC) results, the mean platelet volume (MPV), the
hemoglobin
concentration (HGB) and the eosinophil count or percentage (E0# or EO%) are
analytes
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
21
also likely to be assigned more weight than other analytes. It is therefore
reasonable to
presume that predicting whether the HbA1c concentration is above, equal or
below a
given HbA1c threshold can be obtained from a trained classifier using only a
subset of
the CBC analytes, in addition to age and gender of the tested individuals.
[0065] FIG. 8C and 8D are two different SHAP waterfall graphs explaining
specific
positive predictions made for two different individuals. In FIG. 80, the HbA1c
classifier
indicated with a 95% likelihood that the patient's HbA1c concentration was
equal or above
the HbA1c threshold (set to 5.6% in the exemplary implementation). In this
example, the
features that most contributed to the prediction included the individual's
RDW, WBC, age,
lymphocyte count, MCH, basophil % and gender. Given that the probability that
the
predicted HbA1c value (i.e. that the HbA1c is over the threshold) is over 85%,
the
prediction is reported on the CBC report, in addition the measured values for
the plurality
of blood analytes. The prediction can be accompanied by an indication of a
possible
prediabetes condition. The predicted HbA1c value and/or the indication of the
medical
condition can be provided via a reporting module part of a LIS, for display as
a Graphical
User Interface (GUI) or as an electronic blood test report. In FIG. 8D, the
HbA1c classifier
indicated with a 50% likelihood that the patient's HbA1c concentration was
equal or above
the HbA1c threshold: given the low probability of the predicted HbA1c value,
the prediction
is not reported on the CBC report.
25-0H vitamin D predictions based on CBC test results
[0066] In a possible implementation, the blood analyte predicting application
can include
a trained 25-0H vitamin D classifier. In this case, the automated generation
of the
augmented CBC report method can comprise a step of outputting, by the trained
25-0H
vitamin D classifier, a predicted 25-0H vitamin D value indicative of a 25-0H
vitamin D
concentration in the blood of patients. Similar to the HbA1c prediction, the
predicted 25-
OH vitamin D value is based on the measured values of blood analytes obtained
from a
standard CBC test, and also based on measured values of analytes obtained from
a basic
metabolic panel test. When the prediction that the 25-0H vitamin D
concentration is below
a predetermined threshold, and that the prediction is generated with a high
probability, an
indication of a possible low 25-0H vitamin D concentration is added to the CBC
test report.
In implementations where the predictive application comprises a trained 25-0H
vitamin D
classifier in addition to a trained HbA1c classifier, the CBC results and the
metabolic panel
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
22
results can be inputted to the blood analyte predictive application, and feed
to the HbA1c
and 25-0H vitamin D classifiers. The analytes measured in a basic metabolic
panel test
can comprise any one of glycemia, urea, creatinine, uric acid, calcium,
phosphorus,
cholesterol, triglycerides, total proteins, albumin, total bilirubin, ALP
(Alkaline
Phosphatase), LDH (Lactate Dehydrogenase), AST (Aspartate Aminotransferase),
ALT
(Alanine Aminotransferase), GGT (Gamma-Glutamyltransferase), Na (Sodium), K
(Potassium) and Cl (Chloride).
[0067] Referring now to FIGs. 9A to 9D, graphs are provided to demonstrate and
explain
the performance of the 25-0H vitamin D classifier, trained and configured by
the
Applicant. The 25-0H vitamin D classifier is also preferably of the random
forest type.
FIG. 9A shows the 2-class precision-recall curve (having an Average Precision
(AP) of
0.66) defining the behavior of the vitamin D classifier after having been
trained and
parametrized, using CBC test results. In this case, in addition to the age,
gender, and
measured analytes from the CBC test, the date (month) at which the CBC test
was
conducted also proved to be one of the features having the most weight in the
vitamin D
predictions. It will be noted that the date, age and gender are all
information that are
typically collected when conducting CBC tests and/or basic metabolic panels:
there is no
need to collect additional data other than the data already available from the
standard
tests, such as the CBC and basic metabolic panel tests.
[0068] Referring to FIGs. 9B, the SHAP graph shows that the instance of the 25-
0H
vitamin D classifier assigned most weight to the age, high-density
lipoproteins (HDL),
month of the CBC test, mean corpuscular volume (MCV), gender and triglycerides
concentration (TG) when predicting 25-0H vitamin D values.
[0069] FIG. 9C and 9D are two different SHAP waterfall graphs explaining
specific
positive predictions made for two different individuals. With reference to
FIG. 9C, in this
example, the 25-0H vitamin D classifier indicated with a 95% likelihood that
the patient's
HbA1c concentration was equal or above the minimum vitamin D threshold. The
vitamin
D threshold corresponds to a threshold under which the vitamin D concentration
is sub-
optimal, such as below about 75nmo1/1, as an example only. In this example,
the features
that most contributed to the prediction included the individual's high-density
lipoproteins
(HDL), WBC, age, month of the CBC test, gender and MCV results. Given that the
probability of the prediction as to whether the 25-0H vitamin D is over the
preset
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
23
threshold, (likelihood over 85%), the prediction is reported in the augmented
test report.
The prediction can be accompanied by an indication of a possible vitamin D
deficiency.
In FIG. 9D, the HbA1c classifier indicated with a 50% likelihood that the
patient's vitamin
D concentration was equal or below the threshold: given the low probability
associated
with the prediction, it is not reported on the test report.
Exemplary method for uncovering medical conditions based on CBC test
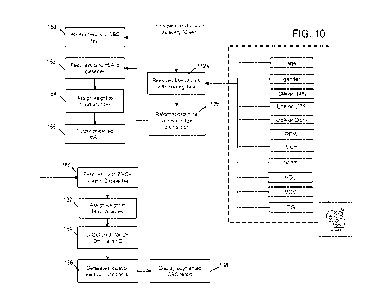
[0070] Referring now to FIG. 10, a high-level flow diagram of the method for
uncovering
medical conditions based on CBC test results is illustrated. The method
comprises
accessing results of the CBC tests (step 150), for example by having an access
module
connect to a Laboratory Information System (LIS), such as via an API. As
mentioned
previously, the CBC results can include measured counts or concentrations of
different
blood analytes, including the basophil count or percentage (BA# or BA%), the
lymphocyte
count or percentage (LY# or LY%), the eosinophil count or percentage (E0# or
E0%),
the red blood cell distribution width (RDVV); the mean corpuscular hemoglobin
(MCH), the
high-density lipoproteins (HDL); mean corpuscular volume (MCV); and
triglycerides
concentration (TG). In addition to measured analytes, the age and gender of
the patients
having been tested, and the date at which the test was conducted are also
generally
available.
[0071] This data can be processed, for example by removing observations with
missing
data, and by reformatting the data type and normalizing its distribution
(steps 152a, 152b).
The processed dataset is fed to different classifiers, which can be packaged
or access
from a software application, referred to as the "blood analyte predictive
application". The
application can comprise one or more machine learning models, trained to
predict values
indicative of counts or concentrations of different target blood analytes. The
processed
CBC results are fed to at least a HbA1c classifier (step 152), and preferably
to a 25-0H
vitamin D classifier (step 162). The processed CBC results mays also be fed to
additional
classifiers, trained to predict other target analytes 138, such as the example
analytes
provided on FIG. 6. The different classifiers will assign different weights to
the processed
blood analytes and other features (such as age, gender, date) fed to the
classifiers. Each
classifier adjusts the weights according to its parameters/hyperparameters,
set during the
training process. The HbA1c classifier outputs a predicted HbA1c value (step
156),
indicative of a HbA1c concentration in the blood of the given patient (step
156), the
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
24
predicted HbA1c value being based on the measured values for blood analytes of
the
CBC test of the given patient other than HbA1c. Similarly, the vitamin D
classifier outputs
a predicted 25-0H vitamin D value (step 164), indicative of a 25-0H vitamin D
concentration in the blood of the given patient. Indications of one or more
medical
conditions associated with the predictions are automatically generated when
the predicted
values are outside a range of values considered acceptable, for each target
analyte being
predicted (step 166).
[0072] One or more server(s) can host the blood analyte predictive application
and the
different classifiers, and one or more computer-readable medium have
instructions stored
thereon to cause a processor to perform the steps of FIG.10. In possible
implementations,
the indications are displayed in a augmented complete blood count (CBC) report
(step
168).
Process for Generating and Updating Classifiers of the Analyte Prediction
System
[0073] Referring again to FIGs.1A and 1B, the process to generate the analyte
prediction
system 500 will now be explained. On the left side of the figure, block 400
includes steps
performed to generate and/or train and update the system 500, including the
different
target analyte classifiers. At step 410, the dataset, including the CBC test
results residing
on the LIS database is accessed and loaded. The dataset is then processed at
step 420,
including for example discarding some of the test results, as well as
normalizing and
standardizing the remaining test data. The processed dataset is thus a subset
of the initial
dataset, since not all data is used for training the classifiers.
[0074] Processing of the data also includes classifying the test results of
the subset. The
classification (which may also be referred to as "labeling') can be made
according to the
level or state of a given target analyte. The state or level classification
can be determined
based on the age and gender of the patient, on the data, and on the measured
values of
the analytes. The classification of the dataset can be performed
automatically, based on
predetermined thresholds and/or intervals for a given analyte, based on the
age and
gender of the individual or other biological parameters, such as genetic
variants.
[0075] For example, if target analyte A is to be predicted for all women
between the ages
of 20 and 60, based on the measured values of analytes B, C and D, at least a
subset of
the test reports from women in that age range that includes measured values
for A, B, C
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
and D must be labeled, for example with a "normal" label/class or an
"abnormal"
label/class, based on the measured value of analyte A (i.e. the "target"
analyte).
Otherwise, the dataset will be imbalanced. Oversampling or undersampling
methods must
be performed for imbalanced datasets. The labelled test results for this
individual can then
5 be used by the Al-model, as part of a training dataset, during the
model's training process.
The labelled test results for this individual can then be used by the Al-
model, as part of a
training dataset, during the model's training process.
[0076] The following step 430 includes selecting the "features" of the subset
of
classified/labelled test results. The feature selection comprises selecting,
out of the 20-
10 30 measured values in a given report, which ones are relevant (i.e. have
an influence) on
the prediction of a given analyte. As such, not all measured values are needed
to predict
a given analyte. At this step, it can be found that analytes B, C and D are
needed for
predicting analyte A, while analytes E, F, J and U are needed in predicting
analyte P.
Feature selection can be made using different tools, such as principal
component analysis
15 (PCA), linear discriminant analysis (LDA) and partial least squares
(PLS).
[0077] Once the feature selection is completed for all desired target
analytes, different
machine learning models are evaluated (step 440) to determine which one
provides the
best recall and precision ratios. The selection can be made for example by
establishing a
performance threshold when evaluating the models. In preferred implementations
of the
20 system, the machine learning models used for identifying the best model
are "classifying"
models, or "classifiers", of the random forest type, since the aim of the
prediction is to
assign a class to a given target analyte, such as "high" or "low", or "normal"
vs "abnormal".
Once a given classifier is selected, it is optimized (step 450) by
automatically testing
several hyperparameter values until the combination of values providing the
highest
25 precision score is identified.
[0078] Once the hyperparameter values for a given classifier have been
determined as
providing the best precision/recall compromise, the classifier's performance
is tested and
if satisfactory, the trained and tested classifier can be used to predict a
given analyte. The
process is conducted for all target analytes, meaning that a classifier with
its own specific
hyperparameters will be defined for each target analyte. The analyte
prediction system
comprises the combination of all trained analyte classifiers.
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
26
[0079] Still referring to FIGs. 1A and 1B, since new test results are
continuously
generated by laboratories, the classifiers can be periodically retrained with
new test
results. Retraining of the classifiers can be completely automated. Model
selection is thus
periodically reassessed (step 460) and all model versions can be stored and
managed at
step 470, so that the model version providing the best performance is selected
and used
in the "real-time" prediction process (block 300). It is also possible, by
continuously
monitoring the correlation of the different measured analytes, to identify new
target
analytes to predict, and to train new analyte classifiers for the new target
analytes.
[0080] Referring now to FIG. 2A, to FIGs. 11A-11F and to FIGs. 12A-12D, the
training
process will be explained in more detail, using HbA1c as an example. The
process starts
at step 410, where the CBC test results are fetched from the LIS database. The
step can
comprise running SQL queries that targets specific blood test result data from
the LIS
database. A pivot table can be created to structure the data into dataframes
(i.e. a data
structure that contains 2-dimensional data), which can be more easily read and
manipulated by the different functions and algorithms involved in the next
steps of the
process. CBC test results for individuals of a given age range can be kept
(such as 18
years old and above), as well as those spreading over a given period (such as
for the last
five years), to avoid bias related to modifications made to lab test equipment
or new
testing methods. The information that is kept includes the exam ID, the exam
date, the
age of the patient/individual, the gender, and all test results/medical
markers related to
the complete blood count, in addition to measured values of the target
analyte: HbA1c.
[0081] At step 421, the CBC test results may include data that is either
missing or of the
wrong type (i.e. a date is text format, rather then numeric). When possible,
the data is
corrected, but when not possible, test results with missing data are
discarded. FIG.11A
provides an overview of an initial dataset used for training the HbA1c analyte
classifier,
where values are missing for different analytes. In other implementations, it
can be
considered to impute missing data, but when the size of the dataset used is
considerable,
the test results with missing data can be removed without affecting the
performance of
the training process.
[0082] At step 422, the dataset is further reduced by removing the test
results obtained
from follow-up tests/exams. In other words, the test results which solely
consisted in first
test results for an individual are kept. This selection can be made, for
example, based on
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
27
the date of the test or on the number of test results for an individual. It
has been found
that removing test results from follow-up exams allowed avoid unwanted biases
when
training the different blood analyte classifiers. From a medical point of
view, test results
from follow-up exams have results that are more predictable, which can
adversely affect
the behavior of the classifier during training. The distribution of measured
values over
time is also verified, to ensure that they are stable over time, as
illustrated in FIG.11B.
The correlation of the different analytes on the others is also verified,
using predetermined
correlation tools, to identify which analytes that are strongly correlated to
other ones, as
in FIG. 110.
[0083] Once the dataset has been reduced to a subset of formatted data, by
using for
example an HDF file, the subset is preferably normalized and standardized.
(step 423),
i.e. the measured test results are scaled to variables between 0 and 1, and
their
distribution is transformed to have a mean of 0 and a standard deviation of 1.
Table 126
of FIG. 3 provides an example of test results from a given exam once they have
been
normalized and standardized. Given that in this example the objective is to
predict
whether the HbA1c concentration is above a given threshold, the measured
values for
HbA1c are removed, and replaced with a class or label associated to the
removed values.
[0084] At step 431, feature selection techniques, such as "lasso regression",
can be used
to identify and select relevant analytes for the prediction of the target
analyte. These
methods allow rejecting variables that have no or very low variance
correlation with the
target analyte, allowing to focus only on specific analytes. The features and
targets are
then split at step 432. More specifically, the measured values for the target
analyte are
replaced by their respective labels, and the remaining measured analytes
(referred to as
"predictors" or "features") are separated from the labels.
[0085] At step 432, the subset of classified test result data is split into a
training dataset
(corresponding typically to 80% of the subset), and a testing dataset (the
remaining
20cY0).The training dataset is used to build/train the classifier (step 441)
while the testing
dataset will be used to validate the performance of the trained classifier
(step 461), in an
iterative process.
[0086] For step 441, there exist numerous machine learning models that can be
explored
before selecting the one best fitted for predicting hemoglobin. They include:
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
28
¨ LogisticRegression()
¨ LinearDiscriminantAnalysis()
¨ KNeighborsClassifier()
¨ DecisionTreeClassifier()
¨ GaussianNB()
¨ ExtraTreesClassifier()
¨ RandomForestClassifier()
¨ XGBCIassifier()
¨ SVC()
The machine learning models listed above are only provided as examples. Other
machine
learning models can be considered, without departing from the present
invention.
[0087] Different models (typically of the "classifier" type) are trained using
the training
dataset and basic hypermeters to shortlist the classifiers most adapted to
predict a given
target analyte (hemoglobin in this example). Each classifier has its own
hyperparameters
that are adjustable to maximize its performance. Tools such as TPOT (Tree-
Based
Pipeline Optimization Tool) can be used to try and verify possible
combinations of
hyperparameters for each model. The "grid search" process may also be used to
iteratively tune the different hyperparameters, for each model, and validate
them with the
test dataset. The model having the best performance is eventually kept for
predicting the
analyte. In the case of hemoglobin, the RandomForestClassifier provided the
best
performances. FIG.12A is a graph showing the performance of the different
classifiers
explored for HbA1c as the target analyte. FIG.12B illustrate the influence of
a given
analyte from the blood count tests in predicting the target.
[0088] At steps 461 and 462, using the trained hemoglobin classifier, a
prediction can be
made on the probability that a given test report be assigned to a given class
(such as
"low" or "high" hemoglobin) from the testing dataset. This probability can be
used to
determine the breakpoint of the classifier. With reference to FIG.12C,
different breakpoints
can be tested to determine the maximal sensitivity (recall) for a precision
above a given
threshold (such as above 90% for example). To further validate the trained
classifier with
the selected breakpoint, a confusion matrix can be used, to summarize its
performance.
The confusion matrix provides the true positives, the true negatives, the
false positives
and the false negatives, which is helpful in assessing the overall performance
of the
trained classifier. FIG. 12D provides an example of a confusion matrix for the
hemoglobin
classifier.
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
29
[0089] At step 472, the trained classifier is stored, for example as a
"pickle" file, and
transferred to the production environment, to run the inflow of test results
in real-time. The
combined files (including the parameters and hyperparameters specifically
determined for
each target analyte) and the analyte classifier models can be run in the
production
environment to predict a large array of target analytes from the analysis of
standard blood
test reports, as explained with reference to block 300 in FIGs. 1A and 2B.
[0090] The proposed system and method described above allows generating
augmented
test reports with information that can help clinicians better assess standard
test results,
in less time. The system is evolutive as it can be automatically retrained
periodically, and
it is built to allow its scaling such that additional predicted target
analytes can be added
overtime, by continuously monitoring the data to identify potential analyte
predictors. The
performance of the analyte prediction system can also be monitored, allowing
to detect
any drifts in predicted values. The system can be integrated, for example as
an API, with
existing LIS systems, providing increased result integration. The predicted
results, and
observations derived from the automated analysis of measured vs predicted
analytes, are
rendered in clear and comprehensive augmented test reports, which highlight
any
abnormal or latent medical conditions.
Other example embodiments
[0091] A computer-implemented method is provided, for generating an analyte
predictor
system to predict the levels of target analyte(s) from biomedical analysis
results, such as
blood analysis results. The method comprises steps of: accessing a blood
analysis
dataset comprising a plurality of test results from a plurality of
individuals, the blood
analysis dataset spreading over a given period of time and including, for a
given test
result, at least the date of the test, and measured values for a plurality of
blood analytes
including measured values for the target analyte, and preferably the gender of
the
individual tested (s), the test results being classified according to a level
or state of the
target analyte(s) determined based on the measured values for said analytes;
training,
using a subset of the classified blood analysis test results, blood analyte
classifiers
respectively associated with each of the target analytes of interest, and
iteratively
adjusting hyperparameters specific to each blood analyte classifier;
generating the
analyte predictor system from the trained blood analyte classifiers, each
trained analyte
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
classifier having its hyperparameters specifically set for predicting a given
one of the blood
analytes of interest.
[0092] In possible implementations, the analyte predictor system is usable
with Electronic
Medical Record systems or medical reporting systems to generate augmented
reports
5 including both predicted and measured analytes from a blood analysis
report. Accessing
the blood analysis dataset may comprises a step of connecting to a Laboratory
Information System to access its database. Test results for which measured
values of
analytes are incomplete or missing are preferably removed from the dataset.
[0093] In possible implementations, only the test results which consisted in
first test
10 results for an individual are kept, based on the date of the test or on
the number of test
results for an individual, in order to avoid bias when training the blood
analyte classifiers.
The test results may be classified using labels indicative of the state of the
target analytes,
the labels comprising a first label for normal results when the measured
values are within
predetermined acceptable limit(s) for said analytes, and a second label for
abnormal
15 results when the measured values are outside said limit(s). The results
may also be
further classified based on the age of the individuals.
[0094] Performance thresholds can be established for each the blood analyte
classifiers.
In addition, for each of the blood analyte classifiers, a step of exploring
different machine
learning models by individually training the different machine learning models
and
20 selecting from said different machine learning models the one that
provides the highest
precision score. Different trained machine learning models can be tested using
another
subset of the classified blood analysis test results. Each machine learning
model has
associated therewith a plurality of hyperparameters. A number of
hyperparameters
values can be automatically tested until the combination of hyperparameter
values that
25 provides the highest precision score for said machine learning model is
identified. For the
selected machine learning model of each blood analyte classifier, a breakpoint
can be
defined, that maximises both the precision score and the sensitivity of the
trained machine
learning model selected. The blood analytes of interest may comprise one or
more of:
Ferritin, HbA1c, TSH, Testosterone, M protein, ALT, Calcium, PTH, Cholesterol,
CA125,
30 Magnesium, Vitamine D, Oestradiol, LH, FSH, HBsAg. The method may also
comprise a
step of identifying, from the plurality of blood analytes, redundant blood
analytes
associated with a given one of the target blood analytes, a redundant blood
analyte being
CA 03193886 2023- 3- 24
WO 2022/067426
PCT/CA2021/051347
31
identified when it is highly correlated with a given target analyte and when a
variance of
the given target analyte is mainly attributed to said identified redundant
blood analyte.
[0095] In a possible implementation, the method may comprise, when the blood
analysis
test results of the individual comprises a measured value for a given one of
the predicted
blood analytes, comparing the measured value and the predicted value, and
determining,
based on preconfigured rules, whether a discrepancy between the measured and
predicted values are indicative of an abnormal medical condition and
displaying an
indication of said discrepancy on said Graphical User Interface (GUI) or
electronic blood
test report when applicable.
[0096] In a possible implementation, an analyte prediction system for
generating
augmented blood test reports is provided. The system comprises an access
module for
connecting to a Laboratory Information System (LIS) and accessing blood test
results
from a plurality of individuals, the blood test results including at least
measured values of
blood analytes, and optionally the gender of the individual; and a selection
module for
selecting blood test results associated to a given one of said individuals; a
processing
module for normalizing and standardizing the blood test results of said
individual, based
on at least the gender and the measured values of its blood test results, and
generating
therefrom processed blood test data; an analyte prediction module comprising
trained
blood analyte classifiers, each classifier being respectively associated with
a
predetermined blood analyte and having its hyperparameters specifically set
according
thereto; the analyte prediction module being configured to receive the
processed test data
of a given individual and generate therefrom at least one predicted analyte
level and/or
value; an output module for generating an augmented test report for said
individual, the
augmented test report including the measured values of blood analytes obtained
from the
LIS and the predicted analyte state and/or value.
[0097] While the above description provides examples of the embodiments, it
will be
appreciated that some features and/or functions of the described embodiments
can be
modified without departing from the principles of the operation of the
described
embodiments. Accordingly, what has been described above has been intended to
be
illustrative and non-limiting and it will be understood by persons skilled in
the art that other
variants and modifications may be made without departing from the scope of the
invention
as defined in the claims appended hereto.
CA 03193886 2023- 3- 24