Note: Descriptions are shown in the official language in which they were submitted.
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
DSG2 COMPOSITIONS AND METHODS FOR THE TREATMENT OF COVID-19
CROSS REFERENCE TO RELATED APPLICATIONS
1100011 This application claims priority to 63/125,583 filed on December
15, 2020, entitled
DSG2 COMPOSITIONS AND METHODS FOR THE TREATMENT OF COVID-19 and
63/274,715 filed on November 2, 2021, entitled, DSG2 COMPOSITIONS AND METTIODS
FOR THE TREATMENT OF COVID-19, the contents of each of which are herein
incorporated by reference in their entirety.
SEQUENCE LISTING
100021 The present application is being filed along with a Sequence Listing
in electronic
format. The Sequence Listing file, entitled 2198_100IPCT_SL.txt, was created
on December
10, 2021, and is 44,002 bytes in size. The information in electronic format of
the Sequence
Listing is incorporated herein by reference in its entirety.
FIELD OF THE DISCLOSURE
100031 The disclosure generally relates to DSG2 based methods for treating
COVID-I 9 by
administering the compositions disclosed herein. The methods also include the
treatment of
therapeutic indications associated with the long-term effects of COVID-19
(such as, but not
limited to post COVID-19 syndrome or post-COVID-19 cardiac syndrome),
including,
inflammation in the myocardium and/or reduced ejection fraction/heart
failure/cardiomyopathy, as well as the treatment of diseases associated DSG2
autoantibodies
e.g., arrhythmogenic right ventricular cardiomyopathy (ARVC), sarcoidosis.
BACKGROUND
100041 Beginning in 2019, severe acute respiratory syndrome coronavirus 2
(SARS-CoV-
2) caused a pandemic infecting millions of people with coronavims disease
(referred to as
COVID-19) (Wu et al., 2020 Nature 579, 265-269) which has led to over a
million deaths
worldwide. Patients infected with SARS-CoV-2 can experience a range of
clinical
manifestations, ranging from no symptoms to critical. illness. Emerging
studies suggest that in
some cases, individuals, even those who had mild versions of the disease, may
sometimes
continue to experience symptoms after their initial recovery. This condition
has been called
post-COVID-19 syndrome or "long COVID-19." In addition, patients may also
develop a
reduced ejection fraction or cardiomyopathy, even after the acute infection of
COVID-19 has
resolved. Post-COVID-19 cardiac signs and symptoms may coexist with effects on
other
1.
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
organ systems, but may also present alone. Patients with post-CO VID
cardiomyopathy range
from those who are asymptomatic to those with fulminant heart failure,
arrhythmia and/or
sudden cardiac death.
100051 The COVID-19 virus, SARS-CoV-2 affects multiple organ systems,
especially
lungs and heart. Elevation of cardiac biomarkers, particularly high-
sensitivity troponin and/or
creatine kinase MB have been commonly observed in patients in COVID-19
infection. A
review of clinical analyses conducted by Bavishi et al. found that myocardial
injury occurred
in 20% of patients with COVID-19 infection (Frog Cardiovasc Dis. 2020
September-October;
63(5): 682-689). The plausible mechanisms of myocardial injury associated with
COVID-19
include but are not limited to, I) hyperinflammation and cy-tokine storm
mediated through
pathologic T-cells and monocytes leading to myocarditis, 2) respiratory
failure and
hypoxemia resulting in damage to cardiac myocytes, 3) down regulation of ACE2
expression
and subsequent protective signaling pathways in cardiac myocytes, 4)
hypercoagulabilityi and
development of coronary microvascular thrombosis, 5) diffuse endothelial
injury, and/or, 6)
inflammation and/or stress causing coronary plaque rupture or supply-demand
mismatch
leading to myocardial ischemia/infarction.
100061 The post-COVID-19 syndrome has also been associated with multiple organ
damage, including cardiovascular damage. Imaging tests taken months after
recovery from
COVID-19 have shown lasting damage to the heart muscle, even in people who
experienced
only mild COVID-I9 symptoms. Post-COVID-19 syndrome also appears to be
associated
with myocarditis and/or cardiomyopathy with increased risk of arrhythmia.
100071 Currently, therapeutic strategies for treating and/or managing COVID-
19 and post-
COVID-I9 syndrome are lacking. The cardiac manifestations of COVID-19 place an
already
overwhelmed health care system under considerable stress due to the
substantial resources
and potential intensive care support required for these patients. In
particular, there is an
urgent need for the development of treatment modalities for inhibiting
inflammatory
responses to reduce the incidence and mortality associated with COVID-I9 and
post-COVID-
19 syndrome related myocardial injury. The present disclosure provides DSG2
fusion
polypeptides based compositions and methods for treating diseases such as, but
not limited to
COVID-I9, post-COVID-I9 syndrome and/or post-COVID-19 cardiac syndrome.
SUMMARY
10008] The present disclosure provides compositions comprising isolated
polypeptides.
The polypeptides of the disclosure may include a whole or a portion of the
DSG2 protein. In
2
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
some embodiments, the isolated polypeptide is a Desmoglein 2 (DSG2) fusion
polypeptide.
The DSG2 fusion polypeptide may include (a) a whole or a portion of a DSG2
protein (SEQ
ID NO: I); and/or (b) a whole or a portion of an immunoglobulin protein. In
one
embodiment, the DSG2 polypeptide may include a portion of DSG2 protein. The
portion of
the DSG2 protein may include the extracellular region of DSG2 protein. In some
aspects, the
entire extracellular region of DSG2 may be included in the fusion poly-
peptide. In one
embodiment, the entire extracellular region of DSG2 includes the amino acid
sequence of
SEQ ID NO: 3. Embodiments of the disclosure may also include a portion of the
extracellular
region of DSG2. For example, a portion of the extracellular region may be
extracellular
cadherin domain I (Ed), extracellular cadherin domain 2 (EC2), extracellular
cadherin
domain 3 (EC3), extracellular cadherin domain 4 (EC4), and/or extracellular
anchor domain
(EA). In some aspects, the DSG2 fusion polypeptides include 2 domains of the
extracellular
region. For example, the two domains may be EC4EA, EC1EC2, EC2EC3, EC3EC4,
EC lEA, EC I EC3, EC2EC4, and/or EC3EA. In some aspects, the DSG2 fusion
polypeptides
include three domains of the extracellular region. For example, the three
domains may be
EC1EC3EA, EC1EC4EA, EClEC3EA, EC3EC4EA, EClEC2EC3, EC2EC3EC4, and/or
EC2EC4EA. In some aspects, the DSG2 fusion polypeptides may include four
domains of the
extracellular region. For example, the three domains may be EC I EC2EC4EA,
EC2EC3EC4EA, EC1EC2EC3EC4EA, EClEC2EC3EC4, and/or EClEC2EC3EA.
100091 DSG2 fusion polypeptides may include a portion of an immunoglobulin.
The
portion may be Fc region, an Fab region. a heavy chain variable (VH) domain, a
heavy chain
constant domain, a light chain variable (VL) domain, and/or a light chain
constant domain. In
one aspect, the portion of the immunoglobulin may be an Fc region. The
immunoglobulin
may be an IgG, an IgM, an IgA, an IgD and/or an IgE. As a non-limiting
example, the
immunoglobulin may be IgG. The compositions may include an IgG such as IgGI,
IgG2,
IgG3, and/or IgG4. Non-limiting examples of portions of immunoglobulin useful
in the
present disclosure include IgG1 Fc region (SEQ ID NO: 5), IgG2 Fe region (SEQ
ID NO: 7),
IgG3 Fc region (SEQ ID NO: 9), IgG4 Fc region (SEQ ID NO: 11), IgGI heavy
chain
constant domain (SEQ ID NO: 4), IgG2 heavy chain constant domain (SEQ ID NO:
6), IgG3
heavy chain constant domain (SEQ ID NO: 8), IgG4 or heavy chain constant
domain (SEQ
ID NO: 10).
1001.01 Polypeptides of the disclosure may further include a linker and/or
a signal
sequence. The linker may be from about 5 amino acids to about 50 amino acids
in length. In
3
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
one embodiment, the linker may be GCyGGS (SEQ ID NO: 12). In another aspect,
the linker
may be EAAAK (SEQ ID NO: 13).
PM The present disclosure also provides methods of treatment using the
compositions
described herein. In some embodiment, the disclosure provides methods of
treating post-
COVID-19 syndrome. Such methods may include, i) contacting the subject with
the isolated
polypeptide of the disclosure and (ii) measuring one or more symptoms
associated with post-
COVID-19 syndrome selected from the group consisting of arrhythmia,
myocarditis, heart
failure, shortness of breath, fatigue, edema, orthopnea limitations to
exertion, impaired
cognitive abilities, palpitations, dizziness, syncope, and/or lightheadedness.
Treatment with
the polypeptides of the disclosure may be effective in ameliorating one or
more symptoms
associated with post-COVID-19 cardiac syndrome. In some aspects, the subjects
with post-
COVID-19 have been previously diagnosed with COVID-19 using methods known in
the art.
In one aspect, the serum of the subject has detectable levels of anti-SARS-CoV-
2 antibodies.
In some embodiments, the serum of the subject has no detectable levels of anti-
SAR.S- CoV-
2 antibodies. Also provided herein are methods of treating a subject with
COVID-19 by
administering the compositions described herein. In some embodiments, the
serum of the
subject with COVID-19 or post-COVID-19 may have anti- DSG2 antibodies.
1001.21 The present disclosure also provides a method of treating a
condition associated
with serum DSG2 autoantibodies. Such methods may include administering the
compositions
described herein or cells expressing the compositions described herein to a
subject. In some
embodiments, the condition may be a cardiomyopathy. In some aspects, the
condition may be
an autoimmune disorder.
100131 The present disclosure provides methods of treating cardiomyopathy
in a subject.
Such methods may include contacting the subject with the isolated polypeptides
or the cells
of the disclosure followed by measuring one or more symptoms associated with
cardiomyopathy such as arrhythmia, palpitations, myocarditis, heart failure,
poor cardiac
output, and/or reduced ejection fraction. As a non-limiting example, the
cardiomyopatl-*,, may
be arrhythmogenic right ventricular cardiomyopathy (A.RVC).The cardiomyopathy
may also
be caused by a virus (e.g., SARS- CoV2, adenovirus, hepatitis virus, hepatitis
C virus,
parvovirus, herpes simplex virus, echovirus, Epstein-Barr virus, rubella,
cytomegalovirus, or
HIV), a bacterium (Staphylococcus, Streptococcus, or Borrelia) , a parasite
(Trypanosoma or
Toxoplasma) or a fungus (Candida, Aspergillus, or Histoplasm.a). In som.e
embodiments, the
subject may have detectable levels of anti-DSG2 antibodies in their serum.
4
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
DESCRIPTION OF THE FIGURES
100141 The foregoing and other objects, features and advantages of
particular
embodiments of the disclosure will be apparent from the following description
and
illustrations in the accompanying figures. The drawings are not necessarily to
scale; emphasis
instead being placed upon illustrating the principles of various embodiments
of the
disclosure.
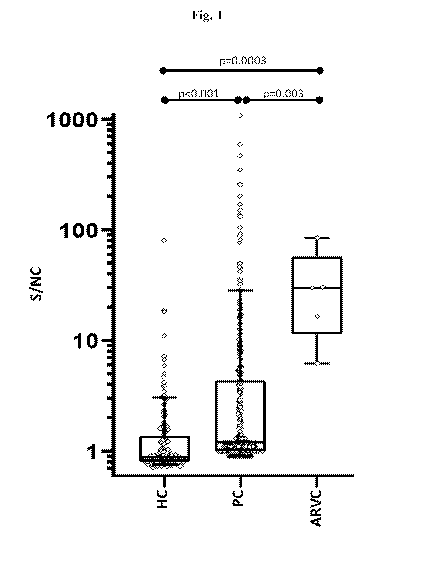
[00151 Fig. 1 is a graph showing comparative levels of anti-DSG2 antibody
signal in
healthy controls (N=152), post-COVID-19 (N=300) and arrhythmogenic right
ventricular
cardiomyopathy samples (N=5). HC, healthy controls; PC, post-COVID-19; ARVC.
arrhythmogenic right ventricular cardiomyopathy; S/NC, signal/negative
control; individual
diamonds represent a single serum sample each; box and whisker limits
represent 25th-75th
and 10th-90th percentiles respectively. P-values are based on non-parametric
rank-based
Wilcoxon-Mann-Whitney 2-sided test.
[00161 Fig. 2A is a bar graph showing anti-DSG2 antibody signal at 6 months
and 9
months in all samples analyzed by month of collection post-COVID-19 infection
(N = 300).
(00171 Fig. 2B is a bar graph showing anti-DSG2 antibody signal at 6 months
and 9
months in paired samples analyzed by month of collection post-COVID-19
infection (N =
17).
DETAILED DESCRIPTION
I. INTRODUCTION
1001.81 The most recent outbreak of COVID-19 has caused a severe respiratory
disease in
humans and has threatened human health worldwide. The novel virus causing
COVID-19 has
been named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by
the
International Committee of Taxonomy of Viruses (ICTV) as SARS-CoV-2 is closely
linked
to the SARS virus.
[00191 The SARS-CoV-2 particles use a special surface glycoprotein (spike
protein) to
bind to angiotensin converting enzyme 2 (ACE2) which is most abundant in the
type il
alveolar cells of the lungs, and thus enters the host cell. The genome of
coronavinis is then
replicated in the host cell. The density of ACE2 receptors in each tissue
correlates with the
severity of the COVID-19 disease in that tissue. ACE2 receptors are also
expressed on the
outer surface of cells in the arteries, heart, kidney, and intestines. As a
result, COVID-19 may
cause multi-organ failure in extremely severe cases.
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
[00201 The symptoms of COVID-19 range from mild (e.g., fever, cough, shortness
of
breath), to severe, such as pneumonia and acute respiratory distress syndrome
(ARDS), sepsis
and septic shock, multi organ failure, including acute kidney injury, and
cardiac injury.
Although respiratory illness is the dominant clinal manifestation of COVTD-I9
infection,
multi organ failure may also occur. In one multi-center study analyzing fatal
cases of
COVID-19, myocardial injury was observed to be the cause of death in 40% of
the cases
(Ruan Q.et al. Intensive Care Med. 2020;46(5):846-848). Various cardiac
complications have
been associated with active COVID-19 infection, including arrhythmia,
myocarditis, and
acute myocardial injury. Systemic inflammation, direct injury of
cardiomyocytes, cytokine
storm, and hypoxia are some of the proposed mechanisms of the multifactorial
pathophysiology. Arrhythmias associated with COVID-I9 can also be attributed
to the
treatment with azithromycin, hydroxy,rchloroquine, and some antivirals that
can cause QT
prolongation. Acute myocardial injury in COVID-19 can range from asymptomatic
elevation
of cardiac troponins to fulminant myocarditis and circulatory shock.
Myocardial injury can
manifest either alone or can occur in combination with arrythmia based on the
clinical course
of the infection.
100211 The proinflammatory milieu and increased sympathetic stimulation in
COVID-19
may further increase the risk for cardiovascular complications, such as
cardiac arrythmias,
worsening of existing heart failure (TIF), or development of new-onset HP. In
patients with
severe disease, hypoxia and electrolyte disturbances can further potentiate
the risk for
anythmias.
[00221 In some instances, patients may experience symptoms weeks, months or
years after
virus particles can be detected in patient samples (herein referred to as post-
COVID-19
syndrome or "long COVID-19" or post viral syndrome). Post-COVID-19 syndrome
may also
be associated with. cardiac symptoms and is herein referred to as Post-COVID-
19 cardiac
syndrome. Many patients who have recovered from COVID-19 show persistent
inflammation
in the myocardium as measured by NIRI. In one study, up to 60% of both
symptomatic and
asymptomatic COVID-19 patients had MRI evidence of ongoing myocardial
inflammation,
an average of 71 days after recovery' from the acute phase of COVID-19
(Puntmann et al.
JAMA Cardiol. 2020;5(11):1265-1273; the contents of which are herein
incorporated by
reference in their entirety). In a smaller study, 15% of athletes had evidence
of myocarditis
after recovery from COVID-I9 (Metzel et al. 2020, HSS Journal, Volume 16,
paees102-
107). A significant proportion of post-COVID-19 syndrome patients subsequently
develop
compromised cardiac function, most notably a reduced ejection fraction, with
or without
6
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
overt symptoms of heart failure. For clarity, patients with post-COVID-19
cardiac
manifestations may herein be referred to as post-COVID-19 cardiac syndrome.
Patients with
post-COVID-19 syndrome also experience chronic fatigue syndrome which may be
driven by
undiagnosed cardiac output reductions. Symptoms may include shortness of
breath, fatigue,
edema, orthopnea, limitations to exertion and impaired cognitive abilities due
to poor cardiac
output ("brain fog"), arrhythmia, palpitations, dizziness, syncope,
lightheadedness, heart
failure, hospitalizations due to heart failure and/or arrhythmia. In som.e
cases, death may
occur as a result of heart failure and/or arrhythmia. In some embodiments, the
post-COVID-
19 syndrome may not be associated with cardiac manifestations.
100231 The cardiac symptoms, including arrhythmia have been associated with
other
diseases such as, but not limited to, arrhythmogenic right ventricular
cardiomyopathy
(ARVC). Similar to ARVC, patients with COVID-19 and/or post-COVID-19 syndrome
also
display worsened cardiac function upon physical exertion. In the case of ARVC,
Chatterjee et
al. identified autoantibodies to the cardiac Desmoglein 2 (DSG2) protein as a
common
feature in the sera of patients with ARVC (Chatterjee D, et al., Eur I-Teart
J.
2018;39(44):3932-3944; the contents of which are herein incorporated by
reference in its
entirety). These autoantibodies were specific to ARVC, as they were
essentially absent in two
independent sets of control sera, as well as sera from subjects with other
forms of heritable
cardiomyopathy. DSG2 antibodies can also be found in some cases of
sarcoidosis, a systemic
inflammatory disease which results in granulomas in organs, such as, but not
limited to the
heart. Anti-DSG2 antibodies are also found in sarcoid patients with cardiac
involvement
(Suna et al. 2020, Eur. Heart Journal, Volume 41, Issue Supplement_2, November
2020,
eha.a946.2127; the contents of which are herein incorporated by reference in
its entirety).
Patients with a diagnosis of dilated cardiomyopathy may have mutations in the
same
desmosome proteins that are associated with ARVC. These observations suggest
that some
dilated cardiomyopathy patients may actually have ARVC-like disease, mediated
by DSG2
antibodies, but have been diagnosed as dilated cardiomyopathy because they do
not fit the
typical age and/or presentation associated with ARVC. Anti-DSG2 autoantibodies
are
considered to arise when there is a combination of cardiac cellular damage and
an activated
immune system typically in the setting of an infection known to affect the
myocardium
directly. From a pathogenesis standpoint, viral infections such as COVID-19,
generally
trigger a vigorous immune response that is crucial for viral clearance, with a
cascade of
events involving both the innate and adaptive immune arms. Direct and indirect
myocardial
damage is also caused by COVID-19 infection, allowing for cardiac proteins to
be exposed to
7
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
the activated immune system. Immunological alterations are also observed in
patients with
COVID-19 condition. These range from a maladaptive immune response and
abnormal
cytokine/chemokine production, to hyperactivation of T cells and increased
number of
activated monocytes, macrophages and neutrophils (Chang, S.E., et al. Nature
Communications 2021; 12:5417; Liu, Y., et al. CUrr. Opin. Rheumatol. 2021;
33:155-162;
Lee, C. C. E., et al. Diseases 2021; 9:47; the contents of each of which are
herein
incorporated by reference in their entirety).
[00241 COVID-19 has infected at least 200 million people worldwide with
approximately
4.5 million deaths attributable to COVID 19 disease to date. There is growing
recognition
that COVID-19 infections can cause a variety of long-term sequelae, of which,
cardiac
involvement may be the most under-recognized as its symptoms may be attributed
to other
organ systems.
100251 COV1D 19 infections have been associated with MRI evidence of
myocardial
involvement and arrhythmias well into recovery, independent of preexisting
conditions,
severity and overall course of the acute illness, and the time from the
original diagnosis. The
percentage of patients who will develop a depressed ejection fraction
subsequently is
currently not well-understood, although frank cardiomyopathy has been
described in post-
COVID-19 patients.
10026i The findings of cardiomyopathy and increased predilection for
arrhythmias are
also observed in arrhythmogenic right ventricular cardiomyopathy (ARVC).
Antibodies to
the desmosome protein desmoglein-2 (DSG2), have been shown to be present in
patients with
ARVC (Diptendu Chatterjee D., et al. EI-U 2018 (39) 3932-3944; the contents of
which are
herein incorporated by reference in its entirety). Concentrations of anti-DSG2
antibodies
correlate positively to arrhythmia burden, and presence of these antibodies in
borderline
ARVC cases predicts the development of fulminant ARVC. Exposure of iPSC-
derived
cardiomyocytes to anti-DSG2 antibodies results in a reduction in gap junction
function that
may reflect direct cardiotoxicity. Together, these data suggest that anti-DSG2
antibodies may
play a role in cardiac pathology. The present disclosure also provides
evidence showing the
anti-DSG2 antibody levels are elevated in COVID-19 patients even 6-9 months
after
diagnosis.
100271 Viral infections, including COV1D-19, have been hypothesized to
contribute to
autoimmune responses, e.g., by exposing previously hidden cryptic epitopes on
damaged
cells to an activated immune system (Ehrenfeld M., et al. Autoimmunity Reviews
2020
102597; the contents of which are herein incorporated by reference in its
entirety). The high
8
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
incidence of cardiac involvement seen in COVID-19 infections, indicates that
anti-DSG2
autoantibodies may be generated as a result.
100281 The presence of arrhythmia as well as the role of the immune system in
the
progression of COVID-19, post-COVID-19 syndrome and ARVC together suggest the
involvement of DSG2 autoantibodies in the pathogenesis of these diseases.
Strategies
targeting anti-DSG2 antibodies (e.g., DSG2 autoantibodies) may therefore be
beneficial in
the treatment of COVID-I9, post-COVID-I9 syndrome and/or ARVC.
[00291 The present disclosure provides compositions and methods related to
DSG2 fusion
polypeptides for targeting anti-DSG2 antibodies. The DSG2 fusion polypeptides
of the
disclosure may therefore be a viable therapeutic strategy in the treatment of
COVID-19, post-
COVID-I9 cardiac syndrome, and/or ARVC. DSG2 fusion polypeptides may also be
used in
the treatment of other diseases associated with cardiac cellular damage, such
as; but not
limited to, arrhy-thtnogenic cardiomyopathy (AC), sarcoidosis, dilated
cardiomyopathy with
anti-DSG2 autoantibodies and viral infections, including but not limited to
those caused by
coxsackie virus, adenovirus, echovinises, parvovirus, rubella and/or
cytomegalovirus.
II. COMPOSITIONS
100301 In some embodiments, the present disclosure provides compositions
that include
DSG2 fusion polypeptides. Compositions described herein, may be capable of
binding to or
interacting with anti-DSG2 antibodies. In one embodiment, the compositions of
the
disclosure may modulate the activity of anti-DSG2 antibodies. In one
embodiment, the
compositions of the disclosure may inhibit the activity of the anti-DSG2
antibodies.
[00311 In some embodiments, the present disclosure includes a DSG2 protein.
In some
aspects the DSG2 protein may be the whole DSG2 protein or a portion of the
DSG2 protein.
In some embodiments, the DSG2 protein may be fused to any other protein or
fragment of a
protein.
100321 DSG2 fusion polypeptides of the present disclosure may include the
whole or a
portion of a DSG2 protein and a whole or a portion of an immunoglobulin
protein. In some
embodiments, the DSG2 fusion polypeptides may further include a linker and/or
a signal
peptide. In some embodiments, the whole or a portion of DSG2 protein may be
fused to a
protein that is not an immunoglobulin. The DSG2 protein may be fused to a
protein or a
fragment of a protein that is capable of improving the expression of DSG2
protein in vitro or
in vivo.
9
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
[00331 DSG2 mutations within intercalated discs in heart cells have been
implicated in
cardiac diseases including arrhythmia, dilated cardiomyopathy, and
particularly ARVC
(Arrhythmogenic right ventricular cardiomyopathy). Chatterjee et al.
identified
autoantibodies to the cardiac DSG2 protein as a common feature in the sera of
patients with
ARVC (Chatterjee D, et al., Eur Heart J. 2018;39(44):3932-3944; the contents
of which are
herein incorporated by reference in its entirety). These autoantibodies were
specific to
ARVC, as they were essentially absent in two independent sets of control sera,
as well as sera
from subjects with other forms of heritable cardiomyopathy. The presence of
DSG2
autoantibodies identified by Chatterjee et al. suggests that targeting DSG2
antibodies may
represent a therapeutic strategy in the treatment of cardiomyopathies
associated with diseases
such as, but not limited to ARVC and/or COVID-19. The present disclosure
provides DSG2
fusion polypeptides as a therapeutic strategy for targeting DSG2
autoantibodies. In some
embodiments, DSG2 fusion polypeptides of the present disclosure may bind to
DSG2
autoantibodies. In some embodiments, binding of the DSG2 fusion polypeptides
of the
disclosure to the DSG2 autoantibodies precludes the binding of the
autoantibodies to the
endogenous DSG2 in a subject. In this aspect of the disclosure, the DSG2
fusion polypeptides
function as a decoy protein or a ligand trap.
100341 Chatterjee et al. propose that the DSG2 protein may include
epitopes, which are
exposed or released into the intercellular space and/or circulation and as a
result of DSG2
mutations. Unmasking of these epitopes may also occur from any cardiac damage
(such as,
but not limited to, infective myocarditis, and/or cardiac trauma). In some
embodiments, the
compositions of the disclosure may not include any mutations. Such released
DSG2 proteins
may link with an antigen-presenting cell to stimulate a T-cell response,
generating the
observed autoantibodies. Unmasking of cryptic epitopes by gene mutations could
contribute
to other forms of autoimmunity. In some embodiments, DSG2 fusion poly-peptides
of the
disclosure may include epitopes containing one or more mutations in DSG2.
100351 DSG2 fusion polypeptide may be a soluble and/or recombinant
polypeptides. The
arrangement of components in the DSG2 fusion polypeptide may be optimized to
achieve the
suitable protein expression and/or the intended therapeutic effect. In some
embodiments, the
DSG2 fusion polypeptide may include formats described herein. The formats
provided herein
include components from N terminus to C terminus delineated by a ";" between
the
components. Non-limiting examples of the formats of the DSG2 fusion
polypeptides include,
(i) the whole or a portion of DSG2 protein; Fc region (ii) Fc region; the
whole or a portion of
the DSG2 protein (iii) signal sequence; the whole or a portion of the DSG2
protein; Fc region
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
(iv) the signal sequence; Fc region; the whole or a portion of the DSG2
protein (v) the whole
or a portion of DSG2 protein; linker; Fc region (vi) Fc region; linker; the
whole or a portion
of the DSG2 protein (vii) signal sequence; the whole or a portion of the DSG2
protein; linker;
Fc region (viii) signal sequence; Fc region; linker; whole or a portion of the
DSG2 protein.
DSG2 protein
100361 In some embodiments, the DSG2 fusion poly-peptides of the present
disclosure may
include the entire DSG2 protein. The desmosomal cadherin desmoglein-2 (DSG2)
is a
transmembrane cell adhesion protein that is widely expressed in epithelial and
non-epithelial
tissues, such as the heart, intestine, and epidermis. DSG2 has been shown to
regulate
numerous cellular processes, including proliferation and apoptosis. In
epithelial and myocyte
cells, DSG2 is a component of the cell-cell adhesion structure and its
cytoplasmic tail
interacts with a series of proteins in direct contact with cell adhesion and
intercellular
junction/cell type regulators. In some embodiments, the DSG2 protein is a
human DSG2
protein (UniProt ID: Q14126; ENSEMBL Protein ID: ENSP00000261590.8) which
consists
of 1,118 amino acids, and includes the amino acid sequence of SEQ ID NO: 1. In
one
embodiment, the DSG2 protein may be encoded by nucleic acid sequence of SEQ ID
NO: 2
(NCBI Reference Sequence: NM 001943.5; ENSMBL ID: ENST00000261590.13).
100371 in some embodiments, the DSG2 fusion polypeptide of the present
disclosure may
be a fully processed DSG2 protein comprising amino acids 50-1118 of SEQ ID NO:
1.
100381 The DSG2 protein may also include one or more mutations with respect to
the
sequence of SEQ ID NO: 1. In some embodiments, DSG2 protein mutation may be a
mutation associated with a disease state. In one embodiment, the disease state
may be
Anily-thmoeenic Right Ventricular Dysplasia/Cardiomyopathy. In some
embodiments, DSG2
fusion poly-peptides of the disclosure may include epitopes containing one or
more mutations
in DSG2. As a non-limiting example. DSG2 fusion polypeptides may include one
or more
mutations in the region of amino acids 485-531 and/or amino acids 586-610 of
SEQ ID NO:
1.
100391 DSG2 belongs to the cadherin superfamily of cell adhesion proteins,
which
commonly feature the three distinct regions: an extracellular region, a
transmembrane domain
and an intracellular signaling region. In some embodiments, the extracellular
region of DSG2
may include the amino acid sequence of SEQ ID NO: 3, which is amino acids 50-
609 of
SEQ ID NO: 1. The extracellular region of cadherin family of proteins contain
a varying
number of repeats of calcium-binding motifs, known as cadherin motifs or EC
domains.
11
CA 03202364 2023-05-17
WO 2022/132854 PCT/US2021/063433
DSG2 contains four EC domains herein referred to as EC I, EC2, EC3 and EC4.
DSG2 also
includes an extracellular anchor (EA) domain that is proximal to the membrane.
In some
embodiments, the DSG2 fusion polypeptides of the disclosure may include the
entire
extracellular region of DSG2. In some aspects, the DSG2 fusion pobpeptide may
include at
least one domain, such as, but not limited to, EC I, EC2, EC3, EC4, and/or EA.
In some
embodiments the EC I domain may be amino acids 50-155 of SEQ ID NO: I. In some
embodiments the EC2 domain. may be amino acids 151-268 of SEQ ID NO: 1. In
some
embodiments the EC3 domain may be amino acids 264-384 of SEQ ID NO: 1. In some
embodiments the EC4 domain may be amino acids 382-495 of SEQ ID NO: 1. In some
embodiments the EA domain may be amino acids 491-608 of SEQ ID NO: 1. Table 1
provides the amino acid sequence of the DSG2 protein as well as the amino acid
sequence of
the extracellular region of DSG2. In some embodiments, the DSG2 proteins of
the present
disclosure may have at least 50%, 55 /0, 60%, 65%, 70%, 75%, 80%, 85%, 90%,
95%, 99%
or 100% identity to any of the sequences in Table I or fragments of the
sequences in Table I.
Table 1. Sequences of the DSG2 protein and DSG2 extracellular domain
(SEQ ID Sequence Description
NO:)
1 MARSPGRAYALLLLLICFNVGSGLHLQVLSTRNENKLLP Human DSG2
KHPHINRQKRAWITAPVALREGEDLSKKNPIAKIHSDLA preprotein
EERGLKITYKYTGKGITEPPFGIFVFNKDTGELNVTS1LDR
EETPFFLLTGYALDARGNNVEKPLELRIKVIDINDNEPVF
TQDVINGSVEELSAAHTLVIVIKLNATDADEPNTLNSKISY
RIVSLEPAYPPVFYINKDTGEEIYTI'SVTLDREEIISSYILT
VEARDGNGEVI'DKPVKQAQVQ1RILDVNDNIPVVENKV
LEGMVEENQVNVEVIRIKVFDADEIGSDNWLA.NFTFAS
GNEGGYFHIETDAQTNEGIVTLIKEVDYEEMKNIDFSVI
VANKAAFfIK.SIRSKYICPTPIPIKVKVKNVKEGIHFICSSVIS
IYVSESMDR.SSKGQIIGNFQAFDEDTGLPAHAR.YVKLED
RDNWISVDSVISEIKLAKLPDFESRYVQNGTTIVKIVAIS
EDYPRKTITGTVLINVEDINDNCPTLIEPVQTICHDAEYV
NVTAEDLDGHPNSGPFSFSVIDKPPGMAEKWKIARQEST
SVLLQQSEKKLGRSEIQFLISDNQGFSCPEKQVLTLTVCE
CLHGSGCRE AQHDSYVGLGPAAIALMILAFLLLLLVPLL
LLMCHCGKGAKGFTPIPGTIEMLHPWNNEGAPPEDKVV
PSFLPVDQGGSINGRNGVGGMAICEATMKGSSSASIVKG
QHEMSEMDGRWEEHRSLLSGRATQFTGATGALVITTETT
KTARATGASRDMAGAQAAAVALNEEFLRNYFTDKAAS
YTEEDENHTAKDCLLVYSQEETESLNASIGCCSFIEGELD
DRFLDDLGLKFKTLAEVCLGQKIDINKEIEQRQKPATETS
MNTASHSLCEQTMVNSENTYSSGSSFPWKSLQEANAEK
VTQEIVTERSVSSRQAQKVATPLPDPMASRNVIATETSY
VTGSTMPPTIVILGPSQPQSLIVTERVYAPASTLVDQPYA
NEGIVVVTERVIQPHGGGSNPLEGTQHLQDVPYVMVRE
RESFLAPSSGVQFTLAMPNIAVGQNVINTERVLAPASTL
QSSYQIPTENSMIARNTINSGAGVPGPLPDFGLEESGHS
NsTrrrssmvnui STVQH SY S
12
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
3 AW1TAPVALREGEDLSKKNPIAKIHSDLAEERGLKITYKY Human DSG2
TGKGITEPPFGIFVFNKDTGELNVTSILDREETPTFLLTGY extracellular
ALDARGNNVEKPLELIMCVLDINDNEINFTODVFVGSVE domain
ELSAAHTLVIVEKINATDADEPNTLNSKISYRIVSLEPAYPP
WYLNICDTGEIYTTSVTLDREEHSSYMTVEARDGNGEV
TDKPVKQAQVOIRILDVNDNIPVVENKVLEGMVEENQV
NVEVTRIKVI'DADEIGSDNWLANFI'FASGNEGGYFHIET
DAQTNEGIVTLIKEVDYEEMKNI.DFSVIVANKAARIK SI
RSKYKPTPIPIKVKVKNVKEGIHFICSSVISIYVSESMDR.SS
KGQIIGNFOAFDEDTGLPAHARYVKLEDRDNWISVDSVT
SEIKLAKI.PDFESRYVONGTYTVKIVAISEDYPRKTITGT
VL IN VED INDNCPTLIEPVOTICHDAEYVNVTAEDLDGHP
NSGPFSFSVIDKPPGMAEKWK IARQESTSVLLQQSEKKL
GRSEIQFLISDNQGFSCPEKQVLTLTVCECLHGSGCREAQ
HDSYVG
100401 The DSG2 fusion polypeptides of the present disclosure may include one
or more
domains of the extracellular region of DSG2. The domains from the
extracellular region of
DSG2 may include repeats of one or more of the EC domains or EA domains, in
tandem. or in
a mixed order. For example, DSG2 fusion polypeptide may include 2, 3, or more
repeats of
EC I, EC2, EC3, EC4 or EA domains. When more than one domain and/or more than
one
repeat of a domain of the extracellular region of DSG2 is present, the domains
may be
operably linked via a linker described herein.
100411 In some embodiments, the DSG2 fusion polypeptide may include two
domains of
the extracellular region of DSG2. Non limiting examples domains of
extracellular region of
DSG2 present in the fusion polypeptides of the present disclosure include, EC
IEC2,
EC IEC3, EC IEC4, EClEA, EC2EC I, EC2EC3, EC2EC4, EC2EA, EC3EC1, EC3EC2,
EC3EC4, EC3EA, EC4EC I, EC4EC2, EC4EC3, EC4EA, EAEC I, EAEC2, EAEC3, and/or
EAEC4.
100421 In some embodiments, the DSG2 fusion polypeptide may include three
domains of
the extracellular region of DSG2. Non limiting examples domains of
extracellular region of
DSG2 present in the fusion polypeptides of the present disclosure include,
EC1EC2EC3,
EC I EC2EC4, EC I EC2EA, EC lEC3EC2, EC I EC3EC4, EC lEC3EA, EC I EC4EC2,
EC I EC4EC3, EC I EC4EA, EC I EAEC2, EC I EAEC3, EC I.EAEC4, EC2EC I.EC3,
EC2ECIEC4, EC 2EC I EA, EC2EC3EC I , EC2EC3EC4, EC2EC3EA, EC2EC4EC I,
EC2EC4EC3, EC2EC4EA, EC2EAEC I, EC2EAEC3, EC2EAEC4, EC3EC1EC2,
EC3EC I EC4, EC3ECIEA, EC3EC2EC I, EC3EC2EC4, EC3EC2EA, EC3EC4EC 1,
EC3EC4EC2, EC3EC4EA, EC3EAEC 1, EC3EAEC2, EC3EAEC4, EC4EClEC2,
EC4ECIEC3, EC4EC I EA, EC4EC2EC I, EC4EC2EC3, EC4EC2EA, EC4EC3EC I,
EC4EC3EC2, EC4EC3EA, EC4EAEC I, EC4EAEC2, EC4EAEC3, EAECIEC2,
13
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
EAEC I EC3, EAEC I EC4, EAEC2EC .I. EAEC2EC3, EAEC2EC4, EAEC3EC 1, EAEC3K2,
EAEC3EC4, EAEC4EC1, EAEC4EC2, and/or EAEC4EC3.
100431 In some embodiments, the DSG2 fusion polypeptide may include four
domains of
the extracellular region of DSG2. Non limiting examples domains of
extracellular region of
DSG2 present in the fusion poly:peptides of the present disclosure include,
EClEC2EC3EC4,
EC 1 EC2EC 3 EA, EC 1 EC2EC4EC3, EC I EC2EC4EA, EC I EC2EAEC3, EC I EC2EAEC4,
EC I EC3 EC 2EC4, EC I EC3EC2EA, EC I EC3EC4EC2, EC I EC3EC4EA, EC I EC
3EAEC2,
EC I EC3EAEC4, EC I EC4EC2EC3, EC I EC4EC2EA, EC I EC4EC3EC2, EC I EC4EC3 EA,
EC IEC4EAEC2, EC IEC4EAEC3, EC lEAEC2EC3, EC lEAEC2EC4, EClEAEC3EC2,
EC I EA EC3EC4, EC I EA EC4EC2, EC I EA EC4EC3 , EC2EC I EC3EC4, EC2EC I EC
3EA,
EC2EC1 EC4EC3, EC2EC IEC4EA, EC2EC1EAEC3, EC2EC1EAEC4, EC2EC3EClEC4,
EC2EC3EC lEA, EC2EC3EC4EC I, EC2EC3EC4EA, EC2EC3EAEC I, EC2EC3EAEC4,
EC2EC4EC1EC3, EC2EC4EC lEA, EC2EC4EC3EC1, EC2EC4EC3EA, EC2EC4EAEC1,
EC2EC4EAEC3, EC2EAECI.EC3, EC2EAECIEC4, EC2EAEC3EC I, EC2EAEC3EC4,
EC2EAEC4EC I, EC2EAEC4EC3, EC3EC1EC2EC4, EC3EC1 EC2EA, EC3EC1 EC4EC2,
EC3EC1EC4EA, EC3EC lEAEC2, EC3EC lEAEC4, EC3EC2 EC I EC4, EC3 EC2 EC I EA,
EC3EC2EC4EC1, EC3EC2EC4EA, EC3EC2EAEC1, EC3EC2EAEC4, EC3EC4EC 1EC2,
EC3EC4EC lEA, EC3EC4EC2EC I, EC3EC4EC2EA, EC3EC4EAEC I, EC3EC4EAEC2,
EC3EAECIEC2, EC3EAECIEC4, EC3EAEC2EC1, EC3EAEC2EC4, EC3EAEC4EC1,
EC3EAEC4EC2, EC4EC1EC2EC3, EC4ECIEC2EA, EC4ECIEC3EC2, EC4EC1EC3EA,
EC4EC I EAEC2, EC4EC I EAEC3, EC4EC2ECIEC3, EC4EC2EC lEA, EC4EC2EC3EC I,
EC4EC2EC3EA, EC4EC2EAEC1, EC4EC2EAEC3, EC4EC3EC I EC2, EC4EC3ECI.EA,
EC4EC3 EC2 EC I , EC4EC3EC2EA, EC4EC 3EAEC I , EC4EC3EAEC2, EC4EAEC 1 EC2,
EC4EAEC I EC3, EC4EAEC2EC I, EC4EAEC2EC3, EC4EAEC3EC I, EC4EAEC3EC2,
EAEC I EC2EC3, EAEC I EC2EC4, EAEC I EC3EC2, EAEC I EC3EC4, EAEC I EC4EC2,
EAECIEC4EC3, EAEC2EC I EC3, EAEC2EC I EC4, EAEC2EC3EC1, EAEC2EC3EC4,
EAEC2EC4EC1, EAEC2EC4EC3, EAEC3ECIEC2, EAEC3ECIEC4, EAEC3EC2EC1,
EAEC3EC2EC4, EAEC3EC4EC I, EAEC3EC4EC2, EAEC4EC iEC2, EAEC4EC iEC3,
EAEC4EC2EC I, EAEC4EC2EC3, EAEC4K3EC I, and/or EAEC4K3EC2 .
100441 In some embodiments, the DSG2 fusion polypeptide may include five
domains of
the extracellular region of DSG2. Non limiting examples domains of
extracellular region of
DSG2 present in the fusion polypeptides of the present disclosure include
EC I EC2EC3EC4EA, EC1 EC2EC3EAEC4, EC I. EC2EC4EC3 EA, EC 1 EC2EC4EAEC3,
EC I EC2EAEC3EC4, EClEC2EAEC4EC3, EC IEC3EC2EC4EA, EClEC3EC2EAEC4,
14
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
EC I EC3EC4EC2EA, EC I EC3EC4EAEC2, EC 1 EC3 EA EC 2EC4, EC 1 EC3EAEC4EC2,
EC I EC4EC2 EC3 EA, EC 1 EC4EC2 EAEC3, EC I EC4EC3EC2EA, EC 1 EC4EC3 EA EC2,
EC I EC4EA.EC2EC3, EC I EC4EAEC3EC2, EC I. EAEC2EC3EC4, EC I EAEC2EC4EC3,
EC I EA EC3 EC2EC4, EC I. EAEC3 EC4EC2, EC 1 EAEC4EC2EC3, EC I EAEC4EC3EC2,
EC2EC 1 EC 3 EC4EA, EC2EC 1 EC3 EAEC4, EC2EC I EC4EC3 EA, EC2EC 1 EC4EAEC3,
EC2EC 1 EAEC3EC4, EC2EC 1 EAEC4EC 3, EC2EC3 EC 1EC4EA, EC2EC3 EC I EAEC4,
EC2EC3EC4EC I EA, EC2EC3 EC4EA EC 1. EC 2EC3 EA EC I EC4, EC2EC3EA.EC4EC I,
EC2EC4EC 1 EC3 EA, EC2EC4EC I EA EC3 , EC 2EC4EC3 EC lEA, EC2EC4EC3EAEC I,
EC2EC4EAEC I EC3, EC2EC4EAEC3 EC 1, EC2EAEC 1EC3EC4, EC2EAEC I EC4EC3,
EC2EA EC3 EC I EC4, EC2EA.EC3EC4EC 1, EC2EAEC4EC I. EC3, EC2EAEC4EC3 EC I,
EC3 EC I EC2EC4EA, EC3 EC I. EC2EAK4, EC3 EC I EC4EC2EA, EC3 EC I EC4EAEC2,
EC3 EC 1 EAEC2 EC4, EC3 EC 1 EAEC4EC2, EC3EC2EC 1 EC4EA, EC3 EC2EC I EAEC4,
EC3EC2EC4EC I EA, EC 3 EC2EC4EAEC 1, EC 3 EC2EAEC 1 EC4, EC3EC2EAEC4EC 1,
EC3 EC4 EC I EC2EA, EC3EC4EC I EA EC 2, EC3EC4EC2EC I. EA, EC3EC4EC2EAEC I ,
EC3EC4EAEC I EC2, EC3EC4EAEC2EC I. EC3EAEC I EC 2EC4, EC3 EA EC I EC4EC2,
EC3EAEC2EC I EC4, EC3EAEC2EC4EC 1, EC3 EAEC4EC I EC2, EC3 EAEC4EC2 EC 1,
EC4EC 1 EC2EC3 EA, EC4EC I EC2EAEC3, EC4EC I EC 3 EC2EA, EC4EC 1 EC 3 EAEC2,
EC4EC I EA.EC2EC3, EC4EC I. EAEC3 EC2, EC4EC2EC I EC3 EA, EC4EC2EC I EAEC3,
EC4EC2 EC 3 EC I EA, EC4EC2EC3EAEC 1 EC4EC2EAEC 1 EC3, EC4EC2EAEC3 EC 1,
EC4EC 3 EC I EC2EA, EC4EC 3 EC I EAEC2, EC4EC3EC2EC I EA, EC4EC3EC2EAEC 1,
EC4EC3EAEC I EC2, EC4EC3EAEC2EC I EC4EAEC I EC2EC3 , EC4EA EC I EC3EC2,
EC4EAEC2EC I EC3, EC4EAEC2EC3 EC I. EC4EAEC3 EC I EC2, EC4EA EC 3EC 2EC I,
EAEC 1 EC2EC3 EC4, EAEC 1 EC2EC4EC3 , EAEC 1EC3EC2EC4, EAEC 1 EC3 EC4EC2,
EAEC 1 EC4EC2EC3, EAEC 1 EC4EC3EC2, EAEC2EC I EC3EC4, EAEC2EC I EC4EC 3,
EA EC2EC 3 EC I EC4, EA EC2EC3 EC4EC I, EAEC2EC4EC I EC3, EAEC2EC4EC3EC I,
EA EC3 EC I EC2EC4, EA EC3 EC I EC4EC2, EAEC3 EC2 EC I. EC4, EAEC3EC2EC4EC 1,
EAEC3 EC4EC I EC2, EAEC3EC4EC2EC 1, EAEC4EC 1 EC2EC3 , EAEC4EC 1 EC3EC2,
EAEC4EC2 EC I EC3, EAEC4EC2EC3 EC I EAEC4EC3 EC I EC2,and/or
EAEC4EC3EC2EC 1.
[00451 Non-limiting examples of a portion of the DSG2 protein as well as
possible
configurations present in the DSG2 fusion polypeptide are provided in Table 2.
Any of the
DSG2 domains described in Table 2 may be operably linked to another domain
within the
fusion poly-peptide or to another DSG2 domain using any of the linker provided
herein or any
linker known in the art. The compositions of the disclosure may include a
portion or a
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
fragment of any of the domains described in Table 2. The domains or
combination of
domains of SEQ ID NO: 1 or SEQ ID NO: 3 included in the polypeptides of the
disclosure
may be extended by 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17,18, 19, 20, 30, 40 or
50 amino acids upstream or downstream of the domains defined in Table 2. In
some
embodiments, the domains or combination of domains of SEQ ID NO: 1 or SEQ ID
NO: 3
included in the polypeptides of the disclosure may be truncated by 1, 2, 3, 4,
5, 6, 7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 30, 40 or 50 amino acids at the N
temiinus or the C
terminus of the domains defined in Table 2. As a non- limiting example, the
extracellular
region of DSG2 protein may extend from the amino acids spanning from 50-610 of
SEQ ID
NO: 1.
Table 2. DSG2 domain combination for the DSG2 extracellular region
DSG2 domain or combination Amino acids of SEQ ID NO: 1
EC I 50-155
EC2 151-268
EC3 264-384
EC4 382-495
EA 491-608
EC4EA 382-608
EC3EC4EA 264-608
EC2EC3EC4EA 151-608
EC I EC2EC3EC4EA 50-608
EC1EC2 50-268
EC1EC2EC3 50-384
ECIEC2EC3EC4 50-495
EC2EC3 151-384
EC2EC3EC4 151-495
EC3EC4 264-495
EC lEC3EC4EA 50-155, 264-608
EC I EC4EA 50-155, 382-608
EC lEA 50-155, 491-608
ECIEC2EC4EA 50-268, 382-608
EC I EC2EC3EA 50-384, 491-608
EC IEC3 50-155, 264-384
EC1EC3EC4EA 50-155, 264-384, 491-608
16
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
EC2EC4EA 151-268, 491-608
EC2EC4 151-268, 382-495
EC3EA 264-384, 491-608
Immunoglobutin protein
[0046] In some embodiments, DSG2 fusion poly-peptides of the present
disclosure may
include a whole or a portion of an immunoglobulin protein. The immunoglobulin
protein may
be an IgG, an IgM, an IgA, an IgD and an IgE. In one embodiment, the
immunoglobulin
protein may be an IgG. Non-limiting examples of IgG may be IgGl, IgG2, IgG3
and/or IgG4.
DSG2 fusion polypeptides may include a region or a portion of an
immunoglobulin. Non
limiting examples of a region of an immunoglobulin such as, Fc region, an Fab
region. a
heavy chain variable (VH) domain, a heavy chain constant domain, a light chain
variable
(VI) domain, and/or a light chain constant domain.
[0047] DSG2 fusion polypeptides may include one or more Fc regions of an
immunoglobulin. In some embodiments, the Fe region may include the first
constant region
immunoglobulin domain (e.g., CHI) or a portion thereof, and in some cases,
part of the
hinge. In other aspects, the Fc region excludes the first constant region
immunoglobulin
domain. Thus, an Fc may refer to the last two constant region immunoglobulin
domains (e.g.,
CH2 and CH3) of IgA, TgD, and IgG, the last three constant region
immunoglobulin domains
of IgE and IgM, and the flexible hinge N-terminal to these domains. For IgA
and IgM, Fc
may include the õI chain. For IgG, the Fc domain comprises imintmoglobulin
domains Cy2
and Cy3 (Cy2 and C73) and the lower hinge region between Cy' (Cyl) and Cy2
(C72). In
some embodiments, an Fe refers to a truncated CHI domain., and CH2 and CH3 of
an
immunoglobulin. Although the boundaries of the Fc region may vary, the human
IgG heavy
chain Fc region are typically usually defined to include residues E216 or C226
or P230 to its
carboxyl-terminus, wherein the numbering is according to the EU index as in
Kabat Antibody
Numbering sequence.
[0048] in some embodiments, the itnmunoglobulin protein may include
additional cell
targeting modules (and may herein be referred to as cell targeting antibody
CTAB).
[0049] Non-limiting examples of the sequences of portions of immunoglobulin
are
provided in Table 3. In some embodiments, the DSG2 fusion polypeptides may
include an
immunoglobulin protein or a portion thereof having at least 50%, 55%, 60%,
65%, 70%,
17
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
75%, 80%, 85%, 90%, 95%, 99% or 100% identity to any of the sequences in Table
3 or
f-ragments of the sequences in Table 3.
Table 3. Sequences of the Immunoglobulin regions
SEQ Description Sequence
ID
NO:
4 Human IgG1 ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFFEINTVSWNSG.kLTSGV
Immunoglob HTFPAVLQSSGLYSL SSWTVPSSSLGTQTYICNVNHIOSNTKVDICKVEP
ulin heavy KSCDKTHTCPPCPAPELLGGPSVFLFPFKPKDTLMISRTFEVTCVVVDVSH
chain EDPEVICFNWYVDGVEVHNAKTICPREEQYNSTYRVVSVLTVLHQDWLN
constant GKEYK CKVSNKAI.PAPIEKTISK AK.GQPREPQVYTLFFSRDELIKNQVSL
region TCINKGFYPSDIAVEWESNGOPENNYKITPFVLDSDGSFYLYsKurvm.s
RWQQGN'VFS CS VMHE ALIINHYTQKSISLSPGK
Human IgG1 DKTHTCPPCPAPELLGGPSVFLFPFKPICDTLMISRTFEVTCVVVDVSHEDP
Immunoglob EVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQPWLNGKE
ulin heavy YKCKVSNKALPAPIEKTISKAICGQPREPQWTLPPSRDELTKNQVSLTCL
chain VKGFYPSDIAVEWESNGQFENNYKTIPPVLDSDGSFFLYSKLTVDKSRW
constant Fc QQGNVFSCSVMHEAL1LNHYTQKSLSLSPGK
Domain ______
6 Human IgG2 ASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFFEPVTVSVv'NSGALTSGV
Immunogiob HTFPAVLQSSGLYSLSSVVTVPSSNFGTQTYTCNVDHKPSNTKVDKTVER
ulin heavy KCCVECPPCPAPPVAGPSVFLFPFKPKDTLMISRTFEVTCVVVDVSHEDPE
chain VQFNWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEY
constant KCKVSNKGLPAPIEKTISKTKGQPREPQVYTLPPSREEMTICNQVSLTCLV
region KGFYPSDISVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQ
QGNWSCSVMHEALI-LNHYTQKSL SLSFGK
7 Human IgG2 ERKCCVECPPCPAPPVAGPSVFLFPFKPKDTLMISRTIEVTCVVVDVSHED
Immunoglob PEVQFNWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKE
utin heavy YKCKVSNKGLPAPIEKTISKTKGQPREFQVYTLFFSREEMTKNQVSLTCL
chain VKGFYPSDISVEWESNGQPENNYKTIPPMLDSDGSFFLYSKLTVDKSRW
constant Fc QQGNVFSCSVMHEALFINHYTQKSLSLSPGK
domain
8 Human IgG3 A STKGP SVFPLAPCSR STSGGIAALGCLVKD YFFEPVTVSWNSGALTSG V
Immunoglob HTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYTCNVNHKPSN1XVDKRVEL
ulin heavy KTFLGDTTITI7CFRCPEFKSCDTPPFCFRCPEPKSCDTPPFCFRCPEPKSCDT
chain PFPCPRCPAPELLGGPSVFLFFPKFKDTLMISRTPEVTCVVVDVSHEDPEV
constant QPKWYVDGVEVIINAKTKPREEQYNSTFRWSVIXVIIIQDWLNGKEYK
region CKVSNKALPAPIEKTISKIK GQPREFQWTLPFSREEMTKNQVSLTCLVK
GFYPSDIAVEWESSGQPENNYNTTPFMLDSDGSFFLYSKLTVDKSRWQQ
GNIFSCSVMHEALHNRFTQKSLSLSPGK
9 Human IgG3 ELKTFLGDITHTCPRCPEPKSCDTPFPCPRCPEPKSCDTFPFCPRCPEFKSC
Immunoglob DTPPPCPRCPAPELLGGPSVFLFPPICPKDTLMISRTPEVTCVVVDVSHEDP
ulin heavy EVQFKWYVDGVE VIIN AKTK PREEQYN STFRWS'VLTVLHQDWLNGKE
chain Fc YKCKVSNKALPAPIEKTISKTKGQPREPQVYTLFFSREEMTKNQVSINCL
domain VKGFYPSDIAVEWESSGQPENNYNTTPFMLDSDGSFFLYsKurvmSRW
QQGNIFSCS'VMHEALIINRFIQKSLSISPGK
Human IgG4 ASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGV
Immunoglob HTFPAVLOSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVES
ulin heavy KYGPPCPSCPAPEFIGGPSVFLFFPICPICDTLMISRTPEVTCVVVDVSQEDP
chain EVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGICE
constant YKCKVSNKGLPSSEEKTISKAKGQPREPQVYTLPFSQEEMTKNQVSLTCL
region VKGFYPSDIAVEWESNGQPENNYKTIPPVLDSDGSFFLYSRLTVDKSRW
QEGNVFSCSVMHEALHNITYTQKSLSLSWK.
11 Human IgG4 ESKYGPPCPSCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE
Immunoglob DPEVQFNVv'YVDGVEVHNAKTKPREEQFNSTYRWSVLTVLHQDWLNG
ulin heavy KEYKCKVSNKGLFSSIEKTISKAICGQPREPQVYTLPPSQEEMTKNQVSLT
18
CA 03202364 2023-05-17
WO 2022/132854 PCT/US2021/063433
chain Fc CINKGEYFSDIAVEWESNGOPENNYIMPPVLDSDGSFTLYSRLTVDKSR
domain WQEGNVFSCSVMHEALIINHYTQKSLSLSLGK
Signal Sequence
[0050] Signal sequences (sometimes referred to as signal peptides,
targeting signals, target
peptides, localization sequences, transit peptides, leader sequences or leader
peptides) direct
proteins (e.g., the polypeptides of the present disclosure) to their
designated cellular and/or
extra.cellular locations. A signal sequence may be a short (about 5-50 amino
acids long)
peptide present at the N-terminus of the majority of newly synthesized
proteins that are
destined towards a particular location. Signal sequences can be recognized by
signal
recognition particles (SRPs) and cleaved using type I and type II signal
peptide peptidases.
Signal sequences derived from human proteins can be incorporated as a DSG2
fusion
polypeptides of the present disclosure to direct the polypeptides of the
disclosure to a
particular cellular and/or extracellular location. These signal sequences are
experimentally
verified and can be cleaved (Zhang et al., Protein Sci. 2004, 13:2819-2824).
[0051] in some embodiments, a signal sequence may be located at the N-terminus
or C-
terminus of the polypeptides of the present disclosure, and may be, although
not necessarily,
cleaved off the polypeptide to yield a "mature" polypeptide, as discussed
herein.
[0052] In some examples, a signal sequence may be a secreted signal
sequence derived
from a naturally secreted protein, and its variant thereof.
[0053] In some instances, signal sequences directing the polypeptides of
the disclosure to
the surface membrane of a target cell may be used. Expression of the
polypeptides of the
disclosure on the surface of the target cell may be useful to limit the
diffusion of the
polypeptides of the disclosure to non-target in vivo environments, thereby
potentially
improving the safety profile of the polypeptides of the disclosure.
Additionally, the
membrane presentation of the polypeptides of the disclosure may allow for
physiological and
qualitative signaling as well as stabilization and recycling of the
polypeptide for a longer
half-life.
[0054] A signal sequence may be a heterogeneous signal sequence from other
organisms
such as virus, yeast and bacteria, which can direct the polypeptides of the
disclosure to a
particular cellular site, such as a nucleus (e.g., EP 1209450). Other examples
may include
Aspartic Protease (NSP24) signal sequences from Trichoderma that can increase
secretion of
fused protein such as enzymes (e.g., U. S. Pat. NO. 8,093,016 to Cervin and
Kim), bacterial
lipoprotein signal sequences (e.g., PCT publication NO. 1991/09952 to Lau and
Rioux),
19
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
Ecoli enterotoxin TT signal peptides (e.g., U.S. Pat. NO. 6,605,697 to Kwon et
al.), E.coli
secretion signal sequence (e.g., U.S. patent publication NO. 2016/090404 to
Malley et al.), a
lipase signal sequence from a methylotrophic yeast (e.g., U.S. Pat. NO.
8,975,041), and
signal peptides for DNases derived from Corynefirm bacteria (e.g., U.S. Pat.
NO.
4,965,197); the contents of each of which are incorporated herein by reference
in their
entirety.
Linkers
100551 In some embodiments, the DSG2 fusion polypeptides of the present
disclosure may
include at least one linker. The linker may be positioned between one or more
regions of the
polypeptides of the disclosure. In one embodiment, the linker may be
positioned between the
whole or a portion of a DSG2 protein and the whole or a portion of an
immunoglobulin
protein. In one aspect, the linker may be positioned between one or more
domains of the
DSG2 protein.
100561 In some embodiments, the linker may be a polypeptide. In some
embodiments, the
linker may comprise a combination of amino acid residues. In some embodiments,
the linker
may comprise about 1-50 amino acid residues. In some embodiments, the linker
may
comprise about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25, 30, 35, 40, 45 or 50
amino acid
residues.
100571 The linkers of the present disclosure may be from about 1 to 100 amino
acids in
length, which links together any of the domains/regions of the effector module
(also called
peptide linker). The linker may be 1-40 amino acids in length, or 2-30 amino
acids in length,
or 20-80 amino acids in length, or 50-100 amino acids in length. Linker length
may also be
optimized depending on the type of configuration of the polypeptide and based
on the crystal
structure of the polypeptide. In some instances, a shorter linker length may
be preferably
selected. In some aspects, the peptide linker may be made up of amino acids
linked together
by peptide bonds, preferably from 1 to 20 amino acids linked by peptide bonds,
wherein the
amino acids are selected from the 20 naturally occurring amino acids: Glycine
(G), Alanine
(A), Valine (V), Leucine (L), Isoleucine Serine (5), Cysteine (C),
Threonine (T),
Methionine (M), Proline (P), Phenylalanine (F), Tyrosine (Y), Tryptophan (W),
Histidine
(H), Lysine (K.), Arginine (R), A.spartate (D), Glutamic acid (E), A.sparagine
(N), and
Glutamine (Q). One or more of these amino acids may be glycosylated, as is
understood by
those in the art. In some aspects, amino acids of a peptide linker may be
selected from
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
Alanine (A), Glycine (G), Praline (P), Asparagine (R), Serine (5), Glutamine
(Q) and Lysine
(K).
100581 In some embodiments, the linker may be a flexible linker or a rigid
linker. Flexible
linkers may be composed of small, non-polar (e.g., Gly) or polar (e.g., Ser or
Thr) amino
acids. The small size of these amino acids provides flexibility, and allows
for mobility of the
connecting functional domains. The most commonly used flexible linkers have
sequences
consisting primarily of stretches of Gly and Ser residues ("GS" linker). An
example of the
most widely used flexible linker has the sequence of (Gly-Gly-Gly-Gly-Ser)n.
By adjusting
the copy number "n", the length of this GS linker may be optimized to achieve
appropriate
separation of the functional domains, or to maintain necessary inter-domain
interactions. In
some embodiments, linkers may include additional amino acids such as Thr and
Ala to
maintain flexibility, as well as polar amino acids such as Lys and Glu to
improve solubility.
In some embodiments, the DSG2 fusion polypeptide may include a flexible
linker, such as
(Gly)8 (SEQ ID NO: 14), consisting of purely of glycine residues. The linker
sequence
avoided large hydrophobic residues to maintain good solubility in aqueous
solutions.
100591 In some embodiments, the linker may be a rigid linker. Non limiting
examples of
an rigid linker includes a linker with the sequence of (EAAAK)n (n = 2-5) (SEQ
ID NO: 15).
In some embodiments, the rigid linker may have a Proline-rich sequence, (XP)n,
with X
designating any amino acid, preferably Ala, Lys, or Glu.
100601 In some embodiments, the linker may be GGGGS (SEQ ID NO: 12). In some
embodiments, the linker may be GGGGGS (SEQ ID NO: 16) and EAAAK (SEQ ID NO:
13).
Polynucleatides
10061.1 In some embodiments, the polypeptides of the present disclosure are
encoded by
polynucleotides or variants thereof described herein. Exemplary nucleic acids
or
polynucleotides include, but are not limited to, ribonucleic acids (RNAs),
deoxyribonucleic
acids (DNAs), threose nucleic acids (TNAs), glycol nucleic acids (GNAs),
peptide nucleic
acids (PNAs), locked nucleic acids (LNAs, including LNA having a fi-D-ribo
configuration,
a-LNA having an a-L-ribo configuration (a diastereomer of LNA), 2'-amino-LNA
having a
2'-amino functionalization, and 2'-amino-a-LNA. having a 2'-amino
functionaliz.ation),
ethylene nucleic acids (ENA), cyclohexenyl nucleic acids (CeNA) or hybrids or
combinations
thereof.
21
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
[00621 As such, polynucleotides encoding peptides or pobpeptides containing
substitutions, insertions and/or additions, deletions, and covalent
modifications with respect
to reference sequences, in particular the polypeptide sequences are disclosed
herein. For
example, sequence tags or amino acids, such as one or more lysines, can be
added to the
peptide sequences described herein (e.g., at the N-terminal or C-terminal
ends). Sequence
tags can be used for peptide purification or localization. Lysines can be used
to increase
peptide solubility or to allow for biotinylation. Alternatively, amino acid
residues located at
the carboxy and amino terminal regions of the amino acid sequence of a peptide
or protein
may optionally be deleted providing for truncated sequences. Certain amino
acids (e.g., C-
terminal or N-terminal residues) may alternatively be deleted depending on the
use of the
sequence, as for example, expression of the sequence as part of a larger
sequence which is
soluble, or linked to a solid support.
100631 Once any of the features have been identified or defmed as a desired
component of
a polypeptide to be encoded by a polynucleotide described herein, any of
several
manipulations and/or modifications of these features may be performed by
moving,
swapping, inverting, deleting, randomizing or duplicating. Furthermore, it is
understood that
manipulation of features may result in the same outcome as a modification to
the molecules
described herein. For example, a manipulation which involved deleting a domain
would
result in the alteration of the length of a molecule just as modification of a
nucleic acid to
encode less than a full length molecule would.
III. PHARMACEUTICAL COMPOSITIONS AND DELIVERY
100641 The fusion polypeptides described herein may be used as therapeutic
agents. In
some embodiments, the present disclosure provides pharmaceutical compositions
comprising
at least one pharmaceutically acceptable carrier and a fusion polypeptide.
100651 in some embodiments, compositions are administered to humans, human
patients,
or subjects. Although the descriptions of pharmaceutical compositions provided
herein. are
principally directed to pharmaceutical compositions which are suitable for
administration to
humans, it will be understood by the skilled artisan that such compositions
are generally
suitable for administration to any other animal, e.g., to non-human animals,
e.g., non-human
mammals. Modification of pharmaceutical compositions suitable for
administration to
humans in order to render the compositions suitable for administration to
various animals is
well understood, and the ordinarily skilled veterinary pharmacologist can
design and/or
perform such modification with merely ordinary, if any, experimentation.
Subjects to which
22
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
administration of the pharmaceutical compositions is contemplated include, but
are not
limited to, humans and/or other primates; mammals, including commercially
relevant
mammals such as does, cattle, pigs, horses, sheep, cats, mice, and/or rats;
and/or birds,
including commercially relevant birds such as poultry, chickens, ducks, geese,
and/or turkeys.
As a non-limiting example, compositions of the disclosure may be administered
to dogs to
treat ARVC.
100661 Provided herein are the fusion polypeptides and pharmaceutical
composition
thereof which may be used in combination with one or more pharmaceutically
acceptable
excipients.
100671 In some embodiments, the pharmaceutically acceptable excipients
include, but are
not limited to, any and all solvents, dispersion media, diluents, or other
liquid vehicles,
dispersion or suspension aids, surface active agents, isotonic agents,
thickening or
emulsifying agents, preservatives, solid binders, lubricants, flavoring
agents, stabilizers, anti-
oxidants, osmolality adjusting agents, pH adjusting agents and the like, as
suited to the
particular dosage form desired. Various excipients for formulating
pharmaceutical
compositions and techniques for preparing the composition are known in the art
(see
Remington: The Science and Practice of Pharmacy, 21" Edition, A. R. Gennaro
(Lippincott,
Williams & Wilkins, Baltimore, Md., 2006; incorporated herein by reference in.
its entirety).
The use of a conventional excipient medium may be contemplated within the
scope of the
present disclosure, except insofar as any conventional excipient medium is
incompatible with
a substance or its derivatives, such as by producing any undesirable
biological effect or
otherwise interacting in a deleterious manner with any other component(s) of
the
pharmaceutical composition, its use is contemplated to be within the scope of
this disclosure.
100681 In some embodiments, a pharmaceutically acceptable excipient may be at
least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% pure. In
some
embodiments, an excipient is approved for use for humans and for veterinary
use. In some
embodiments, an excipient may be approved by United States Food and Drug
Administration.
In some embodiments, an excipient may be of pharmaceutical grade. In some
embodiments,
an excipient may meet the standards of the United States Pharmacopoeia (USP),
the
European Pharmacopoeia (EP), the British Pharmacopoeia, and/or the
International
Pharmacopoeia.
100691 Pharmaceutically acceptable excipients used in the manufacture of
pharmaceutical
compositions include, but are not limited to, inert diluents, dispersing
and/or granulating
agents, surface active agents and/or emulsifiers, disintegrating agents,
binding agents,
23
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
preservatives, buffering agents, lubricating agents, and/or oils. Such
excipients may
optionally be included in pharmaceutical compositions. The composition may
also include
excipients such as cocoa butter and suppository waxes, coloring agents,
coating agents,
sweetening, flavoring, and/or perfuming agents.
[0070] Exemplary diluents include, but are not limited to, calcium
carbonate, sodium
carbonate, calcium phosphate, dicalcitun phosphate, calcium sulfate, calcium
hydrogen
phosphate, sodium phosphate lactose, sucrose, cellulose, microcrystalline
cellulose, kaolin,
marinitol, sorbitol, inositol, sodium chloride, dry starch, cornstarch,
powdered sugar, etc.,
and/or combinations thereof.
100711 Exemplary granulating and/or dispersing agents include, but are not
limited to,
potato starch, corn starch, tapioca starch, sodium starch glycolate, clays,
alginic acid, guar
gum, citrus pulp, agar, bentonite, cellulose and wood products, natural
sponge, cation-
exchange resins, calcium carbonate, silicates, sodium carbonate, cross-linked
polyvinylpyrrolidone) (crospovidone), sodium carbox.ymethyl starch (sodium
starch
glycolate), carboxyrnethyl cellulose, crosslinked sodium carboxymethyl
cellulose
(croscarmellose), methylcellulose, pregelatinized starch (starch 1500),
microcrystalline
starch, water insoluble starch, calcium couboxymethyl cellulose, magnesium
aluminum
silicate (VEEGUMA)), sodium lauryl sulfate, quaternary ammonium compounds,
etc., and/or
combinations thereof.
100721 Exemplary surface active agents and/or emulsifiers include, but are
not limited to,
natural emulsifiers (e.g. acacia, agar, alginic acid, sodium alginate,
tragacanth, chon- drux,
cholesterol, xanthan, pectin, gelatin, egg yolk, casein, wool fat,
cholesterol, wax, and
lecithin), colloidal clays (e.g. bentonite (aluminum silicate) and VEEGUMS,
(magnesium
aluminum silicate), long chain amino acid derivatives, high molecular weight
alcohols (e.g.
stearyl alcohol, cetyl alcohol, ()leyl alcohol, triacetin monostearate,
ethylene glycol distearate,
glyceryl monostearate, and propylene glycol monostearate, polyvinyl alcohol),
carbomers
(e.g. carboxy polymethylene, polyaciylic acid, acrylic acid polymer, and
carboxywinyl
polymer), carrageenan, cellulosic derivatives (e.g. carbox.yrnethylcellulose
sodium, powdered
cellulose, hydroxymethyl cellulose, hydroxypropyl cellulose, hydroxy-propyl
methylcellulose,
methylcellulose), sorbitan fatty acid esters (e.g. polyoxyethylene sorbitan
monolaurate
(TWEEN*20), polyoxyethylene sorbitan (IWEEN060), polyoxyethylene sorbitan
monooleate (1WEEN080), sorbitan monopalmitate (SPAN*40), sorbitan monostearate
(SPAN1,60), sorbitan tristearate (SPAN*65), glyceryl monooleate, sorbitan
monooleate
(SPANS80), polyoxyethylene esters (e.g. polyoxyethylene monostearate
(MYILI(45),
24
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
polyoxyethylene hydrogenated castor oil, polyethovlated castor oil,
polyoxymethylene
stearate, and SOLUTOLS,), sucrose fatty acid esters, polyethylene glycol fatty
acid esters
(e.g. CREMOPHORrt), polyoxyethylene ethers, (e.g. polyoxyethylene lauryl ether
(BRIJ*30), poly (vinyl-pyrrolidone), diethylene glycol monolaurate,
triethanolarnine oleate,
sodium oleate, potassium oleate, ethyl oleate; oleic acid; ethyl laurate,
sodium latuyi sulfate,
PLUORINCOF 68, POLOXAMERO 188, cetrimonium bromide, cetylpyridinium chloride,
benzalkonium chloride, docusate sodium, etc. and/or combinations thereof.
[00731 Exemplary' binding agents include, but are not limited to, starch
(e.g. cornstarch
and starch paste); gelatin; sugars (e.g. sucrose, glucose, dextrose, dextrin,
molasses, lactose,
lactitol, mannitol); amino acids (e.g., glycine); natural and synthetic gums
(e.g. acacia,
sodium alginate, extract of Irish moss, panwar gum, ghatti gum, mucilage of
isapol husks,
carboxymethylcellulose, methylcellulose, ethylcellulose,
hydroxyethylcellulose,
hydroxypropyl cellulose, hydroxypropyl methylcellulose, microcrystalline
cellulose,
cellulose acetate, poly(vinyl-pyrrolidone), magnesium aluminum silicate
(VEEGUM4)), and
larch arabogalactan); alginates; polyethylene oxide; polyethylene glycol;
inorganic calcium
salts; silicic acid; polymethacrylates; waxes; water; alcohol; etc.; and
combinations thereof.
100741 Exemplary preservatives may include, but are not limited to,
antioxidants,
chelating agents, antimicrobial preservatives, antifun.gal preservatives,
alcohol preservatives,
acidic preservatives, and/or other preservatives. Oxidation is a potential
degradation pathway
for mRNA, especially for liquid mRNA formulations. In order to prevent
oxidation,
antioxidants can be added to the formulation. Exemplary antioxidants include,
but are not
limited to, alpha tocopherol, ascorbic acid, acorbyl pahnitate, benzyl
alcohol, butylated
hydroxyanisole, EDTA, m-cresol, methionine, butylated hydroxytoluene,
monothioglycerol,
potassium metabisulfite, propionic acid, propyl gallate, sodium ascorbate,
sodium bisulfite,
sodium metabisulfite, thioglycerol and/or sodium sulfite. Exemplary chelating
agents include
ethylenediaminetetraacetic acid (EDTA), citric acid monohydrate, disodium
edetate,
dipotassium edetate, edetic acid, fumaric acid, malic acid, phosphoric acid,
sodium edetate,
tartaric acid, and/or trisodium edetate. Exemplary antimicrobial preservatives
include, but are
not limited to, benzalkonium chloride, benzethonium chloride, benzyl alcohol,
bronopol,
cetrimide, cetylpyridinium chloride, chlorhexidine, chlorobutanol,
chlorocresol,
chloroxylenol, cresol, ethyl alcohol, glycerin, hexetidine, imidurea, phenol,
phenoxyethanol,
phenylethyl alcohol, phenylmercuric nitrate, propylene glycol, and/or
thimerosal. Exemplary
antifungal preservatives include, but are not limited to, butyl paraben,
methyl paraben, ethyl
paraben, propyl paraben; benzoic acid, hydroxybenzoic acid, potassium
benzoate; potassium
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
sorbate, sodium benzoate, sodium propionate, and/or sorbic acid. Exemplary'
alcohol
preservatives include, but are not limited to, ethanol, polyethylene glycol,
phenol, phenolic
compounds, bisphenol, chlorobutanol, hydroxybenzoate, and/or phenyl ethyl
alcohol.
Exemplary, acidic preservatives include, but are not limited to, vitamin A,
vitamin C, vitamin
E, beta-carotene, citric acid, acetic acid, dehydroacetic acid, ascorbic acid,
sorbic acid, and/or
phytic acid. Other preservatives include, but are not limited to, tocopherol,
tocopherol
acetate, deteroxime mesylate, cetrimide, butylated hydroxyanisol (BHA),
butylated
hydroxytoluened (BHT), ethylenediamine, sodium lauryl sulfate (SLS), sodium
lauryl ether
sulfate (SLES), sodium bisulfite, sodium metabisulfite, potassium sulfite,
potassium
metabisulfite, GLYDANT PLUS , PHENONIP , methylparaben, GERMA.I.,L*115,
GERMABEN , NEOLONETm, KATI-TONTm, and/or EUXYLe.
10075] In some embodiments, the pH of the pharmaceutical solutions is
maintained
between pH 5 and pH 8 to improve stability. Exemplary buffers to control pH
may include,
but are not limited to sodium phosphate, sodium citrate. sodium succinate,
histidine (or
histidine-HCl), sodium carbonate, and/or sodium malate. In another embodiment,
the
exemplary' buffers listed above may be used with additional monovalent
counterions
(including, but not limited to potassium). Divalent cations may also be used
as buffer
counterions; however, these are not preferred due to complex formation and/or
mRNA
degradation.
100761 Exemplary buffering agents may also include, but are not limited to,
citrate buffer
solutions, acetate buffer solutions, phosphate buffer solutions, ammonium
chloride, calcium
carbonate, calcium chloride, calcium citrate, calcium glubionate, calcium
gluceptate, calcium
gluconate, D-gluconic acid, calcium glycerophosphate, calcium lactate,
propanoic acid,
calcium levulinate, pentanoic acid, dibasic calcium phosphate, phosphoric
acid, tribasic
calcium phosphate, calcium hydroxide phosphate, potassium acetate, potassium
chloride,
potassium gluconate, potassium mixtures, dibasic potassium phosphate,
monobasic potassium
phosphate, potassium phosphate mixtures, sodium acetate, sodium bicarbonate,
sodium
chloride, sodium citrate, sodium lactate, dibasic sodium. phosphate, monobasic
sodium
phosphate, sodium phosphate mixtures, tromethamine, magnesium hydroxide,
aluminum
hydroxide, alginic acid, pyrogen-free water, isotonic saline, Ringer's
solution, ethyl alcohol,
etc., and/or combinations thereof
100771 Exemplary lubricating agents include, but are not limited to,
magnesium stearate,
calcium stearate, stearic acid, silica, talc, malt, glyceryl behanate,
hydrogenated vegetable
26
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
oils, polyethylene glycol, sodium benzoate, sodium acetate, sodium chloride,
leucine,
magnesium lauryl sulfate, sodium lauryl sulfate, etc., and combinations
thereof.
100781 Exemplary oils include, but are not limited to, almond, apricot
kernel, avocado,
babassu, bergamot, black current seed, borage, cade, camomile, canola,
caraway, camauba,
castor, cinnamon, cocoa butter, coconut, cod liver, coffee, corn, cotton seed,
emu, eucalyptus,
evening primrose, fish, flaxseed, geraniol, gourd, grape seed, hazel nut,
hyssop, isopropyl
myristate, jojoba, kukui nut, lavandin, lavender, lemon, litsea cubeba,
macademia nut,
mallow, mango seed, meadowfoam seed, mink, nutmeg, olive, orange, orange
toughy, palm,
palm kernel, peach kernel, peanut, poppy seed, pumpkin seed, rapeseed, rice
bran, rosemary,
safflower, sandalwood, sasquana, savoury, sea buckthorn, sesame, shea butter,
silicone,
soybean, sunflower, tea tree, thistle, tsubaki, vetiver, walnut, and wheat
germ oils. Exemplary
oils include, but are not limited to, butyl stearate, caprylic triglyceride,
capric triglyceride,
cyclomethicone, diethyl sebacate, dimethicone 360, isopropyl myristate,
mineral oil,
octyldodecanol, ()ley] alcohol, silicone oil, and/or combinations thereof.
[00791 Excipients such as cocoa butter and suppository waxes, coloring
agents, coating
agents, sweetening, flavoring, and/ or perfuming agents can be present in the
composition,
according to the judgment of the formulator.
100801 Exemplary additives include physiologically biocompatible buffers
(e.g.,
trimethylamine hydrochloride), addition of chelants (such as, for example,
DTPA or DTPA-
bisamide) or calcium chelate complexes (as for example calcium DTPA, CaNaDTPA-
bisamide), or, optionally, additions of calcium or sodium salts (for example,
calcium
chloride, calcium ascorbate, calcium gluconate or calcium lactate). In
addition, antioxidants
and suspending agents can be used.
100811 In some embodiments, the compositions of the present disclosure may be
administered by any route which results in a therapeutically effective
outcome. These
include, but are not limited to enteral (into the intestine), gastroenteral,
epidural (into the dura
matter), oral (by way of the mouth), transdermal, peridural, intracerebral
(into the cerebrum),
intracerebroventricular (into the cerebral ventricles), epicutaneous
(application onto the skin),
intraderrnal, (into the skin itself), subcutaneous (under the skin), nasal
administration
(through the nose), intravenous (into a vein), intravenous bolus, intravenous
drip, intraarterial
(into an artery), intramuscular (into a muscle), intracardiac (into the
heart), intraosseous
infusion (into the bone marrow), intrathecal (into the spinal canal),
intraperitoneal, (infusion
or injection into the peritoneum), intravesical infusion, intravitreal,
(through the eye),
intracavemous injection (into a pathologic cavity) intracavitary (into the
base of the penis),
27
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
intravaginal administration, intrauterine, extra-amniotic administration,
transdermal
(diffusion through the intact skin for systemic distribution), transmucosal
(diffusion through a
mucous membrane), transvaginal, insufflation (snorting), sublingual,
sublabial, enema, eye
drops (onto the conjunctiva), in ear drops, auricular (in or by way of the
ear), buccal (directed
toward the cheek), conjunctival, cutaneous, dental (to a tooth or teeth),
electroosmosis,
endocervical, endosinusial, endotracheal, extracorporeal, hemodialysis,
infiltration,
interstitial, intraabdominal, intra-amniotic, intra-articular, intrabiliary,
intrabronchial,
intrabursal, intracartilaginous (within a cartilage), intracaudal (within the
cauda equine),
intracistemal (within the cistema magna cerebellomedularis), intracomeal
(within the
cornea), dental intracomal, intracoronary (within the coronary arteries),
intracorporus
cavemosum (within the dilatable spaces of the corixirus cavemosa of the
penis), intradiscal
(within a disc), intraductal (within a duct of a gland), intraduodenal (within
the duodenum),
intradural (within or beneath the dura), intraepidermal (to the epidermis),
intraesophageal (to
the esophagus), intragastric (within the stomach), intragingival (within the
gingivae),
intraileal (within the distal portion of the small intestine), intralesional
(within or introduced
directly to a localized lesion), intraluminal (within a lumen of a tube),
intralymphatic (within
the lymph), intrainedullary (within the marrow cavity of a bone),
intrameningeal (within the
meninges), intraocular (within the eye), intraovarian (within the ovary),
intrapericardial
(within the pericardium), intmpleural (within the pleura), intraprostatic
(within the prostate
gland), intrapulmonary (within the lungs or its bronchi), intrasinal (within
the nasal or
periorbital sinuses), intraspinal (within the vertebral column), intrasynovial
(within the
synovial cavity of a joint), intratendinous (within a tendon), intratesticular
(within the
testicle), intrathecal (within the cerebrospinal fluid at any level of the
cerebrospinal axis),
intrathoracic (within the thorax), intratubular (within the tubules of an
organ), intratumor
(within a tumor), intmtympanic (within the aums media), intravascular (within
a vessel or
vessels), intraventricular (within a ventricle), iontophoresis (by means of
electric current
where ions of soluble salts migrate into the tissues of the body), irrigation
(to bathe or flush
open wounds or body cavities), laryngeal (directly upon the larynx),
nasogastric (through the
nose and into the stomach), occlusive dressing technique, ophthalmic (to the
external eye),
oropharyngeal (directly to the mouth and pharynx), parenteral, percutaneous,
periarticular,
peridural, perineural, periodontal, rectal, respiratory (within the
respiratory tract by inhaling
orally or nasally for local or systemic effect), retrobulbar (behind the pons
or behind the
eyeball), intramyocardial (entering the myocardium), soft tissue,
subarachnoid,
subconjunctival, submucosal, transplacental (through or across the placenta),
transtracheal
28
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
(through the wall of the trachea), transtympanic (across or through the
tympanic cavity),
ureteral (to the ureter), urethral (to the urethra), vaginal, caudal block,
diagnostic, nerve
block, biliary perfusion, cardiac perfusion, photopheresis or spinal. In
specific embodiments,
compositions may be administered in a way which allows them to cross the blood-
brain
barrier, vascular barrier, or other epithelial barrier.
100821 Therapeutically effective doses will be easily determined by one of
skill in the art
and will depend on the severity and course of the disease, the patient's
health and response to
treatment, and the judgment of the treating physician.
IV. METHODS OF USE
NM] Provided herein are methods of use of the DSG2 fusion polypeptide
compositions
of the present disclosure. In some embodiments, the DSG2 fusion polypeptides
of the
disclosure may be used to treat one or more diseases or conditions described
herein, in a
subject. Such methods may include contacting the subject with the DSG2 fusion
polypeptides. In some embodiments, the contacting the subject may include
administering
DSG2 fusion polypeptides to the subject. In some embodiments, the contacting
the subject
may include treating the subject with the DSG2 fusion polypeptides of the
disclosure. In one
embodiment, compositions of the disclosure may be used to treat diseases
associated with
DSG2 autoantibodies. In one embodiment, the compositions of the disclosure
mitigate
cardiotoxicity associated with DSG2 antibodies.
[00841 In some embodiments, any therapeutic disease associated with the
inflammation in
myocardium may be treated with the DSG2 fusion polypeptides as described
herein. Non-
limiting examples of such indications include arrhytlunogenic right
ventricular
cardiomyopathy (ARVC), sarcoidosis, dilated cardiomyopathy, post-infectious
cardiomyopathy, compromised cardiac function, reduced ejection fraction, heart
failure,
arrhythmia and myocarditis.
100851 Efficacy of treatment or amelioration of disease can be assessed,
for example by
measuring disease progression, disease remission, symptom severity, reduction
in pain,
quality of life, dose of a medication required to sustain a treatment effect,
level of a disease
marker or any other measurable parameter appropriate for a given disease being
treated or
targeted for prevention. It is well within the ability of one skilled in the
art to monitor
efficacy of treatment or prevention by measuring any one of such parameters,
or any
combination of parameters. In connection with the administration of a fusion
polypeptides or
pharmaceutical composition thereof, "effective against" a disease or disorder
indicates that
29
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
administration in a clinically appropriate manner results in a beneficial
effect for at least a
fraction of patients, such as an improvement of symptoms, a cure, a reduction
in disease load,
extension of life, improvement in quality of life, a reduction in the need for
blood
transfusions or other effect generally recognized as positive by medical
doctors familiar with
treating the particular type of disease or disorder.
100861 A treatment or preventive effect is evident when there is a
significant
improvement, often statistically significant, in one or more parameters of
disease status, or by
a failure to worsen or to develop symptoms where they would otherwise be
anticipated. As an
example, a favorable change of at least 10% in a measurable parameter of
disease, and
preferably at least 20%, 30%, 40%, 50% or more may be indicative of effective
treatment.
Efficacy for a given compound or composition may also be judged using an
experimental
animal model for the given disease as known in the art. When using an
experimental animal
model, efficacy of treatment is evidenced when a statistically significant
modulation in a
marker or symptom is observed.
COVID-1 9
100871 The DSG2 fusion polypeptides or compositions containing the fusion
polypeptides
as described herein can be administered to treat COVTD-19 or long-term effects
of COVID-
19.
100881 In some embodiments, the compositions of the disclosure may be
useful in treating
COVID-19 and/or individuals infected with SARS-CoV-2. An infected person may
be
symptomatic, pre-symptomatic, and asymptomatic. According to the World Health
Organization (WHO), COVID-19 transmission may occur from symptomatic, pre-
symptomatic, and asymptomatic people infected with SARS-CoV-2. Symptomatic
transmission may refer to transmission occurring before a person experiencing
symptoms.
Pre-symptomatic transmission may refer to transmission occurring prior to
onset of
symptoms of COVID-19.
100891 COVID-19 may be associated with one or more symptoms such as, but not
limited
to, fever or chills, cough, shortness of breath or difficulty breathing,
fatigue, muscle or body
aches, headache, new loss of taste or smell, sore throat, congestion or runny
nose, nausea or
vomiting, diarrhea, trouble breathing, persistent pain or pressure in the
chest.
190901 DSG2 fusion polypeptides may be used to treat one or more stages of
COVID-19
disease. In general, adults with SARS-CoV-2 infection may be grouped into the
following
severity of illness categories. However, the criteria for each category may
overlap or vary
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
across clinical guidelines and clinical trials, and a patient's clinical
status may change over
time (COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID-19)
Treatment Guidelines. National Institutes of Health. Available at
www.covid1.9treatmentguidelines.nih.gov/. Accessed 12/11/2020). In some
embodiments, the
compositions of the disclosure may be used to treat asymptomatic or
presymptomatic
infection which may include individuals who test positive for SARS-CoV-2 using
a virologic
test (i.e., a nucleic acid amplification test or an antigen test), but who
have no symptoms that
are consistent with COVTD-19. In some embodiments, the compositions of the
disclosure
may be used to treat mild illness which includes individuals who have any of
the various
signs and symptoms of COVID-19 (e.g., fever, cough, sore throat, malaise,
headache, muscle
pain, nausea, vomiting, diarrhea, loss of taste and smell) but who do not have
shortness of
breath, dyspnea, or abnormal chest imaging. In some embodiments, the
compositions of the
disclosure may be used to treat moderate illness which may include individuals
who show
evidence of lower respiratory disease during clinical assessment or imaging
and who have
saturation of oxygen (Sp02) 94% on room air at sea level. In some embodiments,
the
compositions of the disclosure may be used to treat severe illness which
includes individuals
who have Sp02 <94% on room air at sea level, a ratio of arterial partial
pressure of oxygen to
fraction of inspired oxygen (Pa02/Fi02) <300 mmHg, respiratoiy frequency >30
breaths per
minute, or lung infiltrates >50%. In some embodiments, the compositions of the
disclosure
may be used to treat critical illness which includes individuals who have
respiratory failure,
pneumonia, acute respiratory distress syndrome (ARDS), sepsis, septic shock,
multiple organ.
dysfunction, and/or multi organ failure, including acute kidney injury, and
cardiac injury.
[0091] Methods of preventing one or more conditions associated with COVID-19
are also
provided herein. In one embodiment, the compositions of the disclosure may be
provided to
the subject prior to the onset of symptoms but after exposure to the virus
since there is an
incubation period between exposure and symptom onset to the virus. The
incubation period
of novel coronavints SARS-CoV-2 is generally between two and fourteen days,
with an
average of five days (Lombardi et al õI. Hosp. Infect. 2020 doi:
10.1016/j.jhin.2020.03.003;
the contents of which are herein incorporated by reference in its entirety).
[0092] Compositions of the disclosure may also be administered in combination
with one
or more therapeutic agents recommended for use in the treatment of COVID-19.
In some
embodiments, DSG2 fusion polypeptides described herein may be used in
combination with.
one or more therapeutic agents such as, but not limited to, Remdesivir,
chloroquine,
hydroxychloroquine, loop diuretics, azithromycin, Lopinavir, Ritonavir,
ivermectin,
31
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
interleukin inhibitors, inteiferons, kinase inhibitors, glucocorticosteroids,
and/or SARS CoV-
2 monoclonal antibodies (e.g., Bamlanivimab, Casirivimab, Imdevimab).
Post-COVID-19 syndrome
100931 in some embodiments, compositions of the present disclosure may be used
to treat
post-COVID-19 syndrome. There have been an increasing number of reports of
patients who
experience persistent symptoms after recovering from acute COVID-19 and is
herein referred
to as "post-COVID-19 syndrome" and individuals suffering from these symptoms
are
commonly referred to as "long haulers." In some embodiments, a patient may be
considered
to have post-COVID-19 syndrome if they suffer from one or more symptoms for up
to a
month, up to two months, up to three months, up to four months, up to five
months, up to six
months, up to seven months, up to eight months, up to nine months, up to ten
months, up to
eleven months, up to a year or more after SARS CoV-2 infection. In some
embodiments, the
subject may have no symptoms after SAR.S CoV-2 infection. In some embodiments,
the
subject may have no known COVID-19 or SARS CoV2 infection but may have serum
anti-
DSG2 antibodies.
100941 Compositions of the present disclosure may be used to treat one or more
symptoms
associated with the cardiovascular system in post-COVID-19 syndrome (i.e.,
post COVID-19
cardiac syndrome). One study including 100 patients recently recovered from
COVID-19,
cardiac magnetic resonance imaging revealed cardiac involvement in 78 patients
(78%) and
ongoing myocardial inflammation in 60 patients (60%), which was independent of
preexisting conditions, severity and overall course of the acute illness, and
the time from the
original diagnosis (Puntmann et al. JAMA Cardiol. 2020,5(11):1265-1273). In a
smaller
study, 15% of athletes had evidence for myocarditis after recovery from acute
COVID-19. In
one embodiment, the DSG2 fusion polypeptides may be used to treat myocarditis
in a subject
with post-COVID-19 syndrome.
100951 In some embodiments, the compositions described herein may be used to
treat
post-COVID-19 syndrome that is not associated with any cardiac indications.
100961 In some embodiments, the compositions of the present disclosure may be
used to
treat subjects who show symptoms of COVID-19, for up to weeks, months; and/or
years after
initial diagnosis of COVID-19. In some embodiments, post-COVID-19 syndrome
patients
may demonstrate symptoms for and/or after 1 week, 2 weeks, 3 weeks, 4 weeks, 5
weeks, 6
weeks, 7 weeks, 8 weeks, 9 weeks, 10 weeks, 11 weeks,12 weeks, 1 month, 2
months ,3
months, 4 months, 5 months, 6 months, 7months, 8 months, 9 months, 10 months.
11 months,
32
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
12 months, 1 year, 2 years, 3 years, 4 years, 5 years or more after initial
diagnosis of COVID-
19. Diagnosis of COVID-19 may be established using methods known in the art
(for e.g.,
reverse transcription polymerase chain reaction and/or antibody tests). In
some embodiments,
subjects affected by post-COVID-19 syndrome may be treated with the
compositions of the
disclosure for up to 1 week, up to 1 month, and/or up to one year.
100971 in some embodiments, compositions of the disclosure may be used to
treat
COVID-19 patients who have or develop compromised cardiac function, most
notably a
reduced ejection fraction, with or without overt symptoms of heart failure. In
some
embodiments, the compositions of the disclosure may be used to treat
arrhythmia.
100981 Compositions of the present disclosure may ameliorate one or more
symptoms
associated with post-COVID-19 syndrome. In some embodiments, the symptoms of
post-
COVID-19 syndrome may be the same as acute COVID-19. In some aspects, the
symptoms
associated with post-COVID-19 syndrome may be shortness of breath, fatigue,
edema,
orthopnea, limitations to exertion, impaired cognitive abilities,
palpitations, dizziness,
syncope, lightheadedness, heart failure, and/or arrhythmia.
100991 In some embodiments, compositions of the disclosure may be used to
treat post
COV1D-19 syndrome with symptoms that overlap with the post-intensive care
syndrome that
has also been described in patients without COVID-19.
101001 In some embodiments, the compositions of the disclosure may be used
to treat
subjects with post-COVID-19 syndrome who may have one or more long-term
complications
associated with the cardiovascular system (e.g., inflammation of the heart
muscle),
respiratory system (lung function abnormalities), renal systems (acute kidney
injuty),
dermatologic (rash, hair loss), neurological complications (smell and taste
problems, sleep
issues, difficulty with concentration, memory problems), and/or psychiatric
problems
(depression, anxiety, changes in mood).
101.01.1 In some embodiments, compositions of the disclosure may be used to
treat post-
COVID-19 syndrome subjects who may have one, two or more associated co-
morbidities.
Non-limiting examples of co-morbidities include, but are not limited to,
hypertension, thyroid
disease, immune disorders, COPD (chronic obstructive pulmonary disease), high
blood
pressure, obesity, mental health conditions, and diabetes.
Atitoimmunity associated with CO VII)-] 9
101021 From a pathogenesis standpoint, viral infections such as COVID-19,
generally
trigger a vigorous immune response that is crucial for viral clearance, with a
cascade of
33
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
events involving both the innate and adaptive immune arms. Direct and indirect
myocardial
damage is also caused by COVID-19 infection, allowing for cardiac proteins to
be exposed to
the activated immune system. Immunological alterations are also observed in
patients with
COVID-19 condition. These range from a maladaptive immune response and
abnormal
cytokine/chemokine production, to hyperactivation of T cells and increased
number of
activated monocytes, macrophages and neutrophils.
101031 Autoantibodies known to occur in a number of autoimmune diseases have
been
detected in patients with COVID-19. Because COVID-19 infection can break
immune
tolerance and trigger autoimmune responses; it is also likely to induce
clinical autoitnmunity.
Autoantibodies detected in patients with COV1D-19 included antinuclear
antibodies (ANA),
antiphospholipid (APL), lupus anticoagulant, cold agglutinins, anti-
Ro/Sjogren's syndrome A
(SSA) antibodies, anti-Caspr2 antibody, anti-GD lb antibody, anti-myelin
oligodendrocyte
glycoprotein (MOG) antibody and red cell bound antibodies (Liu, Y., et al.
Cuff. Opin.
Rheumatol. 2021; 33:155-162; the contents of each of which are herein
incorporated by
reference in their entirety). In some embodiments, compositions of the
disclosure may be
used to block autoantibodies generated during COV1D-19 or SARS CoV2 infection.
101041 In a study, three protein arrays were assembled to measure IgG
autoantibodies
associated with connective tissue diseases, anti-cy-tokine antibodies, and
antiviral antibody
responses in serum from 147 hospitalized COVID-19 patients. Autoantibodies
were
identified in approximately 50% of patients but in less than 15% of healthy
controls. It was
found that autoantibodies largely targeted autoantigens associated with rare
disorders such as
myositis, systemic sclerosis and overlap syndromes. However, a subset of
autoantibodies
targeting traditional autoantigens or cytokines were developed de novo
following COVID-19
infection (Chang, S.E., et al. Nature Communications 2021; 12:5417; the
contents of each of
which are herein incorporated by reference in their entirety). In some
embodiments,
compositions of the disclosure may be used to block autoantibodies developed
following
COVID-19 infection.
101051 In severe and critical cases, immunomodulatory drugs and biological
agents
targeting pro inflammatory cytokines have been applied to contain the robust
immune
response in COVID-19. Corticosteroids, JAK inhibitors, IL-1 blockade and IL-6
receptor
antagonists have been used to treat COVID-19 patients. In some embodiments,
compositions
of the disclosure may be used in combination with immunomodulatory drugs and
biological
agents targeting pro inflammatory cytokines. In some embodiments, compositions
of the
34
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
disclosure may be used in combination with corticosteroids, JAK inhibitors, IL-
I blockade
and 1L-6 receptor antagonists.
101061 There have been reports of thromboembolic events following ChAdOxl nCov-
19
(AstraZeneca) vaccination and potentially the Ad26.COV2.S (Johnson & Johnson)
vaccination. While rare, thrombosis was observed to occur at unusual sites,
such as cerebral
and splanchnic veins. Based on the observation of thrombocytopenia and raised
antibodies to
platelet factor 4-polyanion complexes, it has been suggested to be an immune-
mediated
reaction (Lee, C. C. E., et al. Diseases 2021; 9:47; the contents of each of
which are herein
incorporated by reference in their entirety).
(01071 In one study, a high-throughput autoantibody discovery method known as
rapid
extracellular antigen profiling (REAP) was implemented to screen a cohort of
194 individuals
infected with COVID-19, comprising 172 patients with COVID-19 and 22
healthcare
workers with mild disease or asymptomatic infection, for autoantibodies
against 2,770
ex-tracellular and secreted proteins (members of the exoproteome). After
screening patient
samples identification and validation of numerous protein targets across a
wide range of
tissues and immunological and physiological functions was performed. These
autoantibodies
had potent functional activities and could be directly correlated with various
virological,
immunological and clinical parameters in vivo within samples from patients
with COVID-19.
The analysis suggested that some of these autoantibodies probably predated
infection,
whereas others were induced after infection. Furthermore, mouse surrogates of
these
autoantibodies led to increased disease severity in a mouse model of COVID-19
infection.
These results provide evidence that autoantibodies are capable of altering the
course of
COVID-19 by perturbing the immune response to SARS-CoV2 and tissue homeostasis
(Wang, E. Y., et al. Nature 2021; 595:283; the contents of each of which are
herein
incorporated by reference in their entirety). In some embodiments,
compositions of the
disclosure may be used to block autoantibodies that predated COVID-19
infection. In some
embodiments, compositions of the disclosure may be used to block
autoantibodies that may
be induced after COVID-19 infection.
Cardiomyopathies
101081 In some embodiments, compositions of the disclosure may be used to
treat
cardiomyopathies. Cardiomyopathy refers to progressive impairment of the
structure and
function of the muscular walls of the heart chambers. Compositions of the
disclosure may be
used to treat one or more types of cardiomyopathies, such as, but not limited
to, dilated
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
cardiomyopathy, hypertrophic cardiomyopathy and/or restrictive cardiomyopathy.
In one
embodiment, patients with cardiomyopathies may demonstrate serum DSG2
autoantibodies
in their serum.
101091 In one embodiment, the compositions of the disclosure may be used to
treat
Afilythmogenic right ventricular cardiomyopathy (ARVC). Arrhythmogenic right
ventricular
cardiomyopathy/dysplasia (ARVC/ARVD) is a heart muscle disorder associated
with
ventricular arrhythmia, heart failure, and sudden death. ARVC is a
degenerative cardiac
disease characterized by the progressive loss of ventricular function and
arrhythmias. In
addition to genetic mutations to the structural and signaling proteins of
cardiomyocyte
desmosomes, patient immune systems also have been known to play a role in ARVC
disease
pathology. Mutations in DSG2 protein have been associated with ARVC and
autoantibodies
targeting DSG2 have been identified in patients with the disease.
Approximately 50% of
ARVC patients do not have known desmosome mutations; nevertheless these
patients express
DSG2 antibodies. In some embodiments, DSG2 fusion proteins may be used to
treat ARVC
patients who have one or more mutations in the DSG2 protein. In some aspects,
the DSG2
fusion proteins may be used to treat ARVC patients with no known mutations in
the DSG2
protein. In some embodiments, DSG2 fusion polypeptides of the disclosure may
target DSG2
autoantibodies associated with ARVC.
101101 Cardiornyopathy may be associated with inflammation and is herein
referred to as
myocarditis. In some embodiments, myocarditis may be caused by viruses,
bacteria,
parasites, and/or fungi. In some embodiments, compositions of the disclosure
may be used to
treat and/or prevent myocarditis associated with viruses. Non-limiting
examples of viruses
associated with myocarditis include, common cold causing adenovirus, COVID-I9;
hepatitis
B and C; parvovirus, which causes a mild rash, usually in children (fifth
disease); and/or
herpes simplex virus, gastrointestinal infections causing echoviruses,
mononucleosis causing
Epstein-Barr virus, rubella, cytomegalovirus, and IIIV.
101111 Cardiovascular complications have occurred frequently in association
with
COVID-19 and even months after the infection. These cardiovascular
complications include
myocardial injury and myocarditis, acute coronary syndromes, heart failure,
arrythmias, and
thromboembolic events. In addition, cardiac symptoms; palpitations, chest
pain, and dyspnea
have been observed in patients, weeks to months after the initial infection.
(Lee, C. C. E., et
al. Diseases 2021; 9:47; the contents of each of which are herein incorporated
by reference in
their entirety). In some embodiments, compositions of the disclosure may be
used to treat
36
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
cardiovascular complications such as myocardial injury' and myocarditis, acute
coronary
syndromes, heart failure, arrythmias, and thromboembolic events.
101121 In some embodiments, compositions of the disclosure may be used to
treat and/or
prevent myocarditis caused by bacteria. Non-limiting examples of bacteria
associated with
myocarditis include, Staphylococcus, Streptococcus, and/or Borrelia. In some
embodiments,
compositions of the disclosure may be used to treat and/or prevent myocarditis
caused by
parasites. Non-limiting examples of parasites associated with myocarditis
include,
Trypanosoma cruzi and Toxoplasma, including some that are transmitted by
insects and can
cause a condition called Chagas disease. In some embodiments, compositions of
the
disclosure may be used to treat and/or prevent myocarditis caused by fungi.
Non-limiting
examples of fungi associated with myocarditis include, Candida, Aspergillus;
and other fungi,
such as Histoplasma.
V. DEFINITIONS
101131 Domain: As used herein when referring to polypeptides the term "domain"
refers
to a motif of a polypeptide having one or more identifiable structural or
functional
characteristics or properties (e.g.. binding capacity, serving as a site for
protein-protein
interactions).
101141 Expression vector: The term "expression vector" as used herein refers
to a vector
containing a nucleic acid sequence coding for at least part of a gene product
capable of being
transcribed. Expression vectors can contain a variety of control sequences,
which refer to
nucleic acid sequences necessary for the transcription and possibly
translation of an
operatively linked coding sequence in a particular host organism. In addition
to control
sequences that govern transcription and translation, vectors and expression
vectors may
contain nucleic acid sequences that serve other functions as well. The term
also includes a
recombinant plasmid or virus that comprises a polynucleotide to be delivered
into a host cell,
either in vitro or in vivo. In some embodiments, the host cell is a transient
cell line or a stable
cell line. In some embodiments, it is selected from the group consisting of
CHO, TIEK293
and NSO.
101151 Features: "Features" when referring to polypeptides are defined as
distinct amino
acid sequence-based components of a molecule. Features of the polypeptides
encoded by the
polynucleotides described herein include local conformational shape, folds,
loops, half-loops,
domains, half-domains, sites, termini, or any combination thereof
37
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
(0116) Fusion protein: As used herein, the term "fusion protein" or
chimeric protein refers
to protein or polypeptide comprising two or more sequences of amino acids or
active
fragments thereof that are not naturally present in the same polypeptide. In
some
embodiments, two or more separate polypeptides are operably covalently linked,
e.g.,
chemically linked, or fused together by peptide bonds. Recombinant fusion
polypeptides are
created artificially by recombinant DNA technology.
[0111 Half-domain: As used herein when referring to polypeptides the term
"half-
domain" means a portion of an identified domain having at least half the
number of amino
acid resides as the domain from which it is derived. It is understood that
domains may not
always contain an even number of amino acid residues. Therefore, in those
cases where a
domain contains or is identified to comprise an odd number of amino acids, a
half-domain of
the odd-numbered domain will comprise the whole number portion or next whole
number
portion of the domain (number of amino acids of the domain/2+/-0.5 amino
acids). For
example, a domain identified as a 7 amino acid domain could produce half-
domains of 3
amino acids or 4 amino acids (7/2=3.5+1-0.5 being 3 or 4). It is also
understood that sub-
domains may be identified within domains or half-domains, these subdomains
possessing less
than all of the structural or functional properties identified in the domains
or half domains
from which they were derived. It is also understood that the amino acids that
comprise any of
the domain types herein need not be contiguous along the backbone of the
polypeptide (i.e.,
nonadjacent amino acids may fold structurally to produce a domain, half-domain
or
subdomain).
(0118) Immune response: As used herein, the term "immune response" refers to
conditions associated with inflammation, trauma, immune disorders, or
infectious or genetic
disease. These conditions can be characterized by expression of various
factors, e.g.,
cytokines, chemokines, and other signaling molecules, which may affect
cellular and
systemic defense systems.
1101191 Linker: As used herein, "linker" refers to a functional group
(e.g., a chemical or
polypeptide) in which a covalent bond joins two or more polypeptides. As used
herein, a
"peptide linker" is two or more amino acids used to bind two proteins to each
other.
[01201 Modulation: As used herein, the term "modulation" is recognized in the
art and
refers to up regulation (i.e., activation or stimulation), down regulation
(i.e., inhibition or
suppression) of a response, or the two in combination or apart.
(01.21) Polynucleotide: The term "polynucleotide" as used herein refers to a
sequence of
nucleotides connected by phosphodiester linkages. Polynucleotides are
presented herein in
38
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
the direction from the 5' to the 3' direction. A polynucleotide can be a
deoxyribonucleic acid
(DNA) molecule or ribonucleic acid (RNA) molecule. Where a polynucleotide is a
DNA
molecule, that molecule can be a gene or a cDNA molecule. Nucleotide bases are
indicated
herein by a single letter code: adenine (A), guanine (G), thymine (T),
cytosine (C), inosine (r)
and uracil (U). A polynucleotide can be prepared using standard techniques
well known to
one of skill in the art.
101221 Polypeptides: In some embodiments, the compositions of the present
disclosure are
polypeptides or proteins or variants thereof. According to the present
disclosure, any amino
acid-based molecule (natural or non-natural) may be termed a "polypeptide" and
this term
embraces "peptides," "peptidomimetics," and "proteins." As used herein,
"polypeptide"
means a polymer of amino acid residues (natural or unnatural) linked together
most often by
peptide bonds. The term, as used herein, refers to proteins, polypeptides, and
peptides of any
size, structure, or function. A "peptidomimetic" or "polypeptide mimetic" is a
polypeptide in
which the molecule contains structural elements that are not found in natural
polypeptides
(i.e., polypeptides comprised of only the 20 proteinogenic amino acids). In
some
embodiments, peptidomimetics are capable of recapitulating or mimicking the
biological
action(s) of a natural peptide.
101231 Polypeptide variant: The term "polypeptide variant" refers to molecules
which
differ in their amino acid sequence from a native or reference sequence. The
amino acid
sequence variants may possess substitutions, deletions, and/or insertions at
certain positions
within the amino acid sequence, as compared to a native or reference sequence.
Ordinarily,
variants will possess at least about 50% identity (homology) to a native or
reference
sequence, and preferably, they will be at least about 80%, more preferably at
least about 90%
identical (homologous) to a native or reference sequence.
101241 Recombinant: The term "recombinant" as used herein refers to a genetic
entity
distinct from that generally found in nature. As applied to a polynucleotide
or gene, this
means that the polynucleotide is the product of various combinations of
cloning, restriction
and/or ligation steps, and other procedures that result in the production of a
construct that is
distinct from a polynucleotide found in nature.
101251 Sample: As used herein, the term "sample" refers to an aliquot or
portion taken
from a source and/or provided for analysis or processing. In some embodiments,
a sample is
from a biological source such as a tissue, cell or component part (e.g., a
body fluid, including
but not limited to blood, mucus, lymphatic fluid, synovial fluid,
cerebrospinal fluid, saliva,
amniotic fluid, amniotic cord blood, urine, vaginal fluid and semen). In some
embodiments, a
39
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
sample may be or include a homogenate, lysate or extract prepared from a whole
organism or
a subset of its tissues, cells or component parts, or a fraction or portion
thereof, including but
not limited to, for example, plasma, serum, spinal fluid, lymph fluid, the
external sections of
the skin, respiratory, intestinal, and genitourinary tracts, tears, saliva,
milk, blood cells,
tumors, or organs. In some embodiments, a sample is or includes a medium, such
as a
nutrient broth or gel, which may contain cellular components, such as
proteins. in some
embodiments, a "primary" sample is an aliquot of the source. in some
embodiments, a
primay sample is subjected to one or more processing (e.g., separation,
purification, etc.)
steps to prepare a sample for analysis or other use.
101261 Substantially: As used herein, the term "substantially" refers to
the qualitative
condition of exhibiting total or near-total extent or degree of a
characteristic or property of
interest. One of ordinary skill in the biological arts will understand that
biological and
chemical phenomena rarely, if ever, go to completion and/or proceed to
completeness or
achieve or avoid an absolute result. The term "substantially" is therefore
used herein to
capture the potential lack of completeness inherent in many biological and
chemical
phenomena.
101271 Terminus: As used herein the terms "termini" or "terminus" when
referring to
polypeptides refers to an extremity of a peptide or poly-peptide. Such
extremity is not limited
only to the first or final site of the peptide or polypeptide but may include
additional amino
acids in the terminal regions. The poly peptide-based molecules described
herein may be
characterized as having both an N-terminus (terminated by an amino acid with a
free amino
group (NII2)) and a C-terminus (terminated by an amino acid with a free
carboxyl group
(COOH)). Proteins described herein are in some cases made up of multiple
polypeptide
chains brought together by disulfide bonds or by non-covalent forces
(multimers, oligomers).
These sorts of proteins will have multiple N- and C-termini. Alternatively,
the termini of the
polypeptides may be modified such that they begin or end, as the case may be,
with a non-
polypeptide based moiety such as an organic conjugate.
101281 Therapeutically effective amount: As used herein, the term
"therapeutically
effective amount" means an amount of an agent to be delivered that is
sufficient, when
administered to a subject suffering from or susceptible to a disease,
disorder, and/or
condition, to treat, improve symptoms of, diagnose, prevent, and/or delay the
onset of the
disease, disorder, and/or condition.
101291 Treating: As used herein, the term "treating" refers to partially or
completely
alleviating, ameliorating, improving, relieving, delaying onset of, inhibiting
progression of,
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
reducing severity of, and/or reducing incidence of one or more symptoms or
features of a
particular disease, disorder, and/or condition. Treatment may be administered
to a subject
who does not exhibit signs of a disease, disorder, and/or condition and/or to
a subject who
exhibits only early signs of a disease, disorder, and/or condition for the
purpose of decreasing
the risk of developing pathology associated with the disease, disorder, and/or
condition.
101301 Treatment: As used herein the terms "treat," "treatment," and the
like, refer to
relief from or alleviation of pathological processes. In the context of the
present disclosure, it
relates to any of the other conditions recited herein below, the terms
"treat," "treatment," and
the like mean to relieve or alleviate at least one symptom associated with
such condition, or
to slow or reverse the progression or anticipated progression of such
condition.
101311 Treatment dose: As used herein, "treatment dose" refers to one or
more doses of a
therapeutic agent administered in the course of addressing or alleviating a
therapeutic
indication. Treatment doses may be adjusted to maintain a desired
concentration or level of
activity of a therapeutic agent in a body fluid or biological system.
'VI. EQUIVALENTS AND SCOPE
101321 While various embodiments of the disclosure have been particularly
shown and
described in the present disclosure, it will be understood by those skilled in
the art that
various changes in form and details may be made without departing from the
spirit and scope
of the embodiments disclosed herein and set forth in the appended claims.
[0133] Those skilled in the art will recognize or be able to ascertain
using no more than
routine experimentation, many equivalents to the specific embodiments
described herein. The
scope of the present disclosure is not intended to be limited to the above
description, but
rather is as set forth in. the appended claims.
101341 In the claims, articles such as "a," "an," and "the" may mean one or
more than one
unless indicated to the contrary or otherwise evident from the context. Claims
or descriptions
that include "or" between one or more members of a group are considered
satisfied if one,
more than one, or all of the group members are present in, employed in, or
otherwise relevant
to a given product or process unless indicated to the contrary' or otherwise
evident from the
context. The disclosure includes embodiments in which exactly one member of a
group is
present in, employed in, or otherwise relevant to a given product or process.
The disclosure
includes embodiments in which more than one, or all group members are present
in,
employed in, or otherwise relevant to a given product or process.
41
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
[01351 It is also noted that the term "comprising" is intended to be open
and permits but
does not require the inclusion of additional elements or steps. When the term
"comprising" is
used herein, the terms "consisting of' and "or including" are thus also
encompassed and
disclosed.
10136] Where ranges are given, endpoints are included. Furthermore, it is
to be understood
that unless otherwise indicated or otherwise evident from the context and
understanding of
one of ordinaly skill in the art, values that are expressed as ranges can
assume any specific
value or subrange within the stated ranges in different embodiments of the
disclosure, to the
tenth of the unit of the lower limit of the range, unless the context clearly
dictates otherwise.
101371 In addition, it is to be understood that any particular embodiment
of the present
disclosure that falls within the prior art may be explicitly excluded from any
one or more of
the claims. Since such embodiments are deemed to be known to those of ordinary
skill in the
art, they may be excluded even if the exclusion is not set forth explicitly
herein. Any
particular embodiments of compositions disclosed herein can be excluded from.
any one or
more claims, for any reason, whether or not related to the existence of prior
art.
101381 All cited sources, for example, references, publications, databases,
database
entries, and art cited herein, are incorporated into this application by
reference, even if not
expressly stated in the citation. In case of conflicting statements of a cited
source and the
instant application, the statement in the instant application shall control.
101391 Section and table headings are not intended to be limiting.
EXAMPLES
Example 1. DSG2 fusion polypeptide synthesis methods
NM] DSG2 fusion polypeptides described herein are produced by recombinant
DNA
techniques by synthesizing DNA encoding the desired polypeptide. Once coding
sequences
for the desired polypeptides are synthesized or isolated, they are cloned into
any suitable
vector for expression.
101411 The expression vector is inserted into a suitable host cell by
transformation,
transduction, and/or transfection. The sequences of the DSG2 fusion
polypeptides may be
optimized to yield maximal expression in a host cell. The host cell is any
host cell known in
the art for expression of recombinant proteins. A number of mammalian cell
lines are known
in the art and include immortalized cell lines available from the American
Type Culture
Collection (ATCC), such as, but not limited to, Chinese hamster ovary (CHO)
cells, HeI..a
cells, HEK293, baby hamster kidney (BHT() cells, monkey kidney cells (COS),
human
42
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
hepatocellular carcinoma cells (e.g., Hep G2), Madin-Darby bovine kidney
("MDBK") cells,
NOS cells derived from carcinoma cells, such as sarcoma, as well as others.
Bacterial species
may also be used as host cells. Non-limiting examples include Escherichia
coil, Bacillus
subtilis, and Streptococcus. Non-limiting examples of yeast host cells useful
in the present
disclosure include inter alia, Saccharomyces cerevisiae, Candida albiccms,
Candida maltosa,
Hansenula polymorpha, Kluyveromyces fragilis, Kluyveromyces lactis, Pichia
guillerimondii,
Pichia pastoris, Schizosaccharomyces pombe and Yarrowia hpolytica.
[01421 Depending on the expression system and host selected, the fusion
polypeptides of
the present disclosure are produced by growing host cells expressing the
expression vector
under conditions whereby the protein of interest is expressed. The protein is
then isolated
from the host cells and purified.
[0143] Alternatively, the fusion polypeptides of the present disclosure may
be synthesized
by conventional techniques known in the art, for example, by chemical
synthesis such as
solid phase peptide synthesis.
Example 2. Anti-DSG2 antibodies in post COVID-19 serum samples
101441 Viral infections, including COVID-19, have been hypothesized to
contribute to
autoimmune responses, e.g., by exposing previously hidden ciyptic epitopes on
damaged
cells to an activated immune system (Ehrenfeld M. et al., Autoimmunity Reviews
2020;102597; the contents of which are herein incorporated by reference in its
entirety).
Given the high incidence of cardiac involvement seen in COVTD-19 infections,
it was
hypothesized that anti-DSG2 autoantibodies might be generated as a result.
101451 300 convalescent serum samples were obtained from a group of post COVID-
19
infected patients from October 2020 to February 2021 from East Asian
population. The mean
age of the study population was 37 years old (range 21-65 years). 154 samples
were drawn 6
months post-COVID-19 infection and 146 samples were drawn 9 months post COVID
infection. 17 samples were obtained from the same patient at the 6- and 9-
month mark
(symptom status unknown). The negative control group sera were obtained from a
commercial source of self-declared healthy individuals. Positive control ARVC
sera were
obtained under international Council of Harmonization (ICH) guidelines. An
anti-drug
antibody (ADA) format assay was used for the detection of the anti-DSG2
antibodies. The
mean signal intensity of anti-DSG2 antibodies in the post COVID-19 samples was
significantly higher than that of a healthy control population as shown in
Fig. 1 (19 83.2 in
the post COVID-19 sample vs. 2.1 6.8 in the healthy control population, p
value <0.001). Of
43
CA 03202364 2023-05-17
WO 2022/132854 PCT/US2021/063433
note, 29.3% of the post COVID-19 infection samples demonstrated a signal
higher than the
90th percentile of the control population and 8.7% have signals higher than
the median found
in ARVC patients. The presence of anti-DSG2 antibodies in samples obtained 6-9
months
after COVID-19 infection suggests that the antibodies are not an acute phase
reactant. The
results are also shown in Table 4 and Table 5.
Table 4. Analysis of anti-DSG2 antibody levels
Type N Mean Std Minimum Median Maximum p-value 1 p-va1ue2
Dev
ARVC Samples 5 33.58 30.36 6.20 29.90 84.80
Convalescent Covid 300 18.99 83.18- 0.90 1.20
1070.00 0.0030
Sample
Pre-Covid Samples 152 2.13 6.82 0.71 0.88 80.44 0.0003
<.0001
Note: p-values are based on the non-parametric rank-based Wilcoxon-Mann-
Whitney 2-sided test, 3
pairwise comparison of the groups. P-value 1 is the comparison versus the
AR.VC Samples; p-value
2 is the comparison versus the Convalescent Covid Sample
Table 5. Analysis of anti-DSG2 antibody levels
Pre-Covid Convalescent Covid ARVC Samples
Samples Samples
Number of values 152 300 --------- 5 --
Minimum 0.71 0.9 ---------- 6.2
25% Percentile 0.8 1 11.35
Median 0.88 1.2 29.9
75% Percentile 1.375 4.375 57.65
Maximum 80.44 1070 84.8
Range 79.73 1069 78.6
10% Percentile 0.753 0.9 6.2
90% Percentile 3.044 28.27 84.8
Mean 2.134 18.99 33.58
Std. Deviation 6.82 83.18 30.36
Std. Error of Mean 0.5532 4.803 13.58
101461 The signal intensity between the 6-month and 9-month samples did not
differ
significantly between each other (p.529). This was observed when all non-
contemporaneously assessed 6 and 9 month samples (N=300; Fig. 2A) as well as
in paired 6
month and 9 month samples analyzed by month of collection after COV1D-19
infection
(N=17; Fig. 2B)
44
CA 03202364 2023-05-17
WO 2022/132854
PCT/US2021/063433
[0147] In conclusion, recovered COVID- l 9 patients demonstrated
significantly higher and
sustained levels of anti-DSG2 autoatitibodies as compared to a healthy control
population,
and comparable to that of a diagnosed ARVC group. Of note, these sera were
obtained well
after acute COVID- l 9 infection, suggesting that these antibodies may persist
long-term.