Note: Descriptions are shown in the official language in which they were submitted.
WO 2022/213201
PCT/CA2022/050533
APPARATUS AND METHODOLOGIES FOR DETECTION, DIAGNOSIS, AND
PROGNOSIS OF BRAIN INJURY
CROSS-REFERENCE TO RELATED APPLICATION
[0001] This application claims the benefit of priority to U.S.
Provisional Patent
Application No. 63/172,876 entitled "Apparatus and Methodologies for
Diagnosing
Brain Injury" and filed April 9th, 2021, which is specifically incorporated by
reference
herein for all that it discloses or teaches.
FIELD
[0002] Embodiments herein generally relate to apparatus and
methodologies
for detecting changes in at least one biomarker profile in a biological sample
of an
individual, the changes in biomarker profile being indicative of an injury to
the
individual's nervous system. More specifically, the present apparatus and
methodologies relate to the detection of changes in at least one biomarker in
bodily
fluids, such as up- and/or down-regulation of the metabolome, to diagnose at
least
one nervous system injury.
BACKGROUND
[0003] Central and peripheral nervous system injury, ranging
from traumatic
brain injury, to stroke, to neurodegenerative disease, are major causes of
lifelong
neurological sequelae, with a lack of early, effective diagnostic and
prognostic tools.
Identification of early biomarkers for detection and diagnosis of central and
peripheral
nervous system injury and monitoring subsequent recovery could enable more
effective management and targeted therapies to improve and perhaps even
restore
function.
1
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0004] By way of example, it is estimated that 1.6 to 3.8
million traumatic brain
injuries, such as sport-related concussions (SRC), occur annually in North
America,
such injuries occurring in both sports and recreational activities. SRC is
currently
diagnosed assessing the individual's clinical signs and symptoms ranging from
headache and dizziness to memory impairment and loss of consciousness. There
is
no single "gold standard" assessment or diagnostic tool that can objectively
determine
whether an individual has suffered an SRC (particularly where no intercranial
lesion
occurs), nor how long it will take for the individual to recover. As a result,
the SRC
diagnostic process is subject to interpretation and error, establishing an
urgent need
for improved apparatus and methodologies for diagnosing SRC.
[0005] By way of further example, Alzheimer's disease (AD) is
a destructive
neurodegenerative disease, which results in progressive memory loss, cognitive
dysfunction, and other behaviour changes to such a degree that it affects
everyday
life. In the United States, in 2020, 5.8 million people (65 years or older)
were living
with probable AD. Without the development of a cure or treatment to slow the
disease,
the prevalence of probable Alzheimer's disease will nearly triple by 2050 to
13.8 million
in this demographic. Additionally, AD and dementia are global health issues.
In 2010,
the global prevalence of dementia (AD is one of the leading causes) was 46
million;
however, it is expected to climb to 131.5 million by 2050. AD negatively
impacts global
economies with an estimated 16 million unpaid caregivers (family and others in
US
alone; Alzheimer's Association, 2020) and world economic cost of approximately
$818
billion (USD) yearly, which is expected to double by 2030. Overall, these
changes
project an increased burden on global economies, health care systems and
caregivers
2
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
and highlight the urgent need for both the discovery of robust AD biomarkers
that allow
for early intervention and the development of more effective treatments.
Currently,
there is no cure for AD, and treatment is primarily constrained to treating
symptoms.
[0006] AD robs a person of some of their most human qualities,
including their
memory, reasoning, and language. The disease begins with decreased memory and
cognition, also known as mild cognitive impairment. As AD progresses, other
cognitive
deficits emerge, such as impaired abstract reasoning, navigation, speech, and
motor
function. Notably, these clinical symptoms appear years after the formation of
neurofibrillary tangles (NFTs) and amyloid-beta plaques (A13). These
neuropathological changes are considered the key pathological markers of AD in
brain
tissue in post-mortem analysis; however, they are just the tip of the
neurodegenerative
iceberg, and their presence cannot be confirmed until after an individual's
death. Other
AD pathologies include cell death, inflammation, oxidative stress, and
impaired energy
metabolism. Therefore, a person's cause of dementia being attributed to AD
cannot
occur until after the individual's death. This timing, of course, is not ideal
as an earlier
definitive diagnosis would ideally lead to quicker, more specialized
treatments. As a
result, there also remains an urgent need for improved apparatus and
methodologies
for diagnosing neurodegenerative diseases like AD.
[0007] Metabolomics is an emerging science dedicated to the
systematic study
of chemical metabolites found in tissues and biofluids as a result of various
biochemical reactions. Metabolomics techniques, such as nuclear magnetic
resonance (NMR) spectroscopy have been used to measure the response of
metabolites in plasma caused by SRC, resistance exercise, neurological
diseases,
3
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
and other disease states in both animal models and humans. Such techniques
have
been used to detect changes in metabolites found in cerebral spinal fluid
(CSF), blood,
and brain tissue, including the measurement of Ap plaques, total tau, and
hyperphosphorylated tau in CSF. Metabolomic techniques have also been used to
show that urine metabolites are altered in many other diseases of the abnormal
brain
pathology.
[0008] Research into the chemical metabolites arising from
metabolic
processes in the body, the collection of which is referred to as the
rmetabolome', has
led to a greater understanding of injury-induced alterations in metabolism. As
a result,
many systematic profiling techniques for measuring and analyzing chemical
metabolites found in tissues and biofluids due to various biochemical
reactions are
being developed.
[0009] NMR spectroscopy is a versatile analytical technique
used in a broad
range of disciplines. 1H NMR spectroscopy, also known as proton NMR, can be
used
to study the metabolomic compositions of biofluids, cells, and tissues to
interpret and
classify complex NMR-generated metabolic data sets and to extract useful
biological
information. 1H NMR spectroscopy provides a valuable diagnostic tool in
metabolomics because it preserves the integrity of the fluid being sampled, it
is non-
biased, and it can be performed on several types of samples including blood,
cerebrospinal fluid, and urine. 1H NMR spectra also prove useful in
metabolomics
because molecular pattern recognition and other chemoinformatic tools can be
used
to provide a characteristic "fingerprint" or profile of an organism for a
range of
biologically-important endogenous metabolites, particularly where the profile
is
4
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
changed by a disease, disorder, toxic process, or xenobiotic (e.g., drug
substance).
That is, quantifiable differences in molecular (metabolite) patterns in
biofluids and
tissues can give information and insight into underlying molecular mechanisms
of
disease and disorder. Indeed, early research suggests that metabolite
signatures or
'profiles', and specifically changes to metabolite profiles, can accurately
reflect central
nervous system inflammation and neuronal injury. For example, studies
examining
metabolite changes in blood plasma following SRC have detected changes in
glycerophospholipids, which are associated primarily with membrane structures
in the
brain and inflammation.
[0010] Despite its advantages, however, 1H NMR has not been
used
extensively because analysis of the complex spectrum consisting of thousands
of
signals often require the comprehension and interpretation of a skilled
technician, or
the addition of supporting clinical and medical laboratory data. Deconvolution
of these
signals into discrete metabolites with corresponding concentrations can
require
considerable skill and knowledge that is not generally known in the art. For
example,
it is possible that 1H NMR metabolomics analysis of urine could prove
particularly
useful because it is easily attainable in athletes, is the body's primary
vehicle for
excretion of small molecules, and it is therefore sensitive to changes in
biochemical
pathways due to disease or injury. However, to date, applying NMR to study
metabolomic changes in human urine has not realized any useful diagnostic test
due,
in part, because urine provides significantly more metabolite information when
compared to blood (49 metabolites in blood compared to 209 in urine).
Furthermore,
known correlations between metabolite information and specific diseases or
injuries
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
are typically limited to one or a small number of metabolites, which has
resulted in
diagnostic tests based on non-NMR techniques for detecting and quantifying one
metabolite at a time. While NMR techniques can be used more efficiently to
detect
and quantify two or more metabolites at a time, or even all metabolites within
a sample
at a time, the advantages of doing so have not been realized as databases
correlating
such "amalgamated" metabolomic patterns or profiles with most specific
diseases or
injuries are currently lacking.
[0011] There remains a need for effective, non-invasive,
simple method of
determining whether an individual is likely to have an injury to the central
and
peripheral nervous systems, including brain injuries. It would be advantageous
for
such tools to provide means for detecting a change in metabolomic pattern or
profile,
such change being indicative of, and used to, diagnose the injury.
SUMMARY
[0012] According to embodiments, apparatus and methodologies
of
determining whether an individual is likely to have an injury are provided. In
some
embodiments, the methods comprise determining a reference value for at least
one
target biomarker, obtaining at least one biological sample from the
individual,
measuring the concentration of the at least one target biomarker in the sample
using
1H-NMR spectroscopy, comparing the measured concentration of the measured at
least one target biomarker to the reference value to determine if the
concentration of
the at least one biomarker has changed relative to the reference value,
wherein a
change in the concentration of the at least one biomarker is indicative of the
injury.
6
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0013] In some embodiments, the methods may further comprise
measuring
the concentration of at least two biomarkers in the biological sample and
determining
whether each of the measured at least two biomarkers have a change in
concentration
levels relative to the reference value.
[0014] In some embodiments, the methods may further comprise
measuring
the concentration of the at least two biomarkers in the biological sample and
determining whether one of the at least two biomarkers has a concentration
less than
the reference value and whether one other of the at least two biomarkers has a
concentration level that is greater than the reference value.
[0015] In some embodiments, the at least one target biomarker
may be
selected from the group consisting of 2-Hydroxybutyrate, 3,4-
dihydroxybenzeneacetate, carnitine, 4-hydroxybenzoate, caffeine,
horriocitrulline,
methionine, acetylcarnitine, 3-methyl-2-oxovalerate, phosphorylcholine,
choline,
propylene glycol, taurine, 1-methylhistadine, 3-methylhistadine, citrate,
lactose,
phenylalanineõ 3-indoxylsulfate, sucrose, 3-methyladipate, isobutyrate, 3-
hydroxyisovalerate, 5-am inolevulinate, anserine, tyrosine, carnosine,
isoleucine,
leucine, threonate, and cysteine.
[0016] In some embodiments, the at least one target biomarker
may be
selected from phenylalanine and citrate, and the indication that the
individual may be
likely to have an injury is provided in the event that the measured
concentration level
of phenylalanine is greater that the respective baseline value for
phenylalanine and
the measured concentration levels of citrate is less than the respective
baseline value
for citrate.
7
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0017] In some embodiments, the change in concentration of the
at least one
target biomarker may be further indicative the prognosis of the injury. In
some
embodiments, the at least one target biomarker may be 2-hydroxybutyrate.
[0018] In some embodiments, the change in concentration of the
at least one
target biomarker may be further indicative of the number of symptoms of the
injury. In
some embodiments, the at least one target biomarker may be lactose.
[0019] In some embodiments, the at least one biological sample
may be
selected from urine, plasma, whole blood serum, spinal fluid, interstitial
fluid, saliva,
an extract or purification therefrom, and a dilution thereof.
[0020] In some embodiments, the injury may be a central or
peripheral nervous
system injury, wherein the central nervous system injury may be a brain injury
such
as a traumatic brain injury. In some embodiments, the injury may be an acute
injury
or a chronic injury. In some embodiments, the methods may be used for
diagnosing,
prognosing, or monitoring the injury.
[0021] In some embodiments, the at least one target biomarker
may be
selected from the group consisting of citrate, glycyl-glycine, isoleucine,
glutamate,
trimethylamine N-oxide, choline, choline phosphate, glucose, leucine,
phenylalanine,
valine, tyrosine, glutamate, methionine, galactose, glycerol, myo-Inositol,
betaine,
threonine, ethanol, creatine, malonic acid/malonate, pyruvatoxine,
phenylalanine,
alpha-ketoisovaleric, propylene glycerol, 2-oxohexane, gamma-aminobutyric acid
(GABA), 2-hydroxy-3-methylvaelrate, n-acetyl-L-aspartate (NAA), 4-am
inobutanoate,
threonine, 3-methyl-2-oxobutanoic acid, (R)-3-hydroxybutanoate, succinate,
glycolate, and acetylcholine.
8
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0022] In some embodiments, the at least one target biomarker
may be
selected from citrate and isoleucine, and the indication that the individual
may be likely
to have an injury is provided in the event that the measured concentration
level of
citrate is greater than its respective baseline value and the measured
concentration
level of isoleucine is less than its respective baseline value.
[0023] In some embodiments, the at least one target biomarker
may be
selected from the group consisting of n-acetyl-L-aspartate (NAA), ethanol, 2-
am ino-3-
phosphonoprionic acid, 1,3,7-trimethyluric acid, creatine phosphate, gamma-
aminobutyric acid (GABA), isoleucine, leucine, phenylalanine, serine,
tyrosine, valine,
phosphorylcholine.
[0024] In some embodiments, the indication that the individual
is likely to have
an injury may be provided in the event that the measured concentration levels
of NAA
and GABA are greater than their respective baseline values and the measured
concentration levels of ethanol, 2-am ino-3-phosphonoprionic acid, 1,3,7-trim
ethyluric
acid, creatine phosphate, isoleucine, leucine, phenylalanine, serine,
tyrosine, valine,
phosphorylcholinen-acetyl-L-aspartate (NAA), ethanol, 2-amino-3-
phosphonoprionic
acid, 1,3,7-trimethyluric acid, creatine phosphate, gamma-aminobutyric acid
(GABA),
isoleucine, leucine, phenylalanine, serine, tyrosine, valine,
phosphorylcholineis are
less than their respective baseline values.
[0025] In some embodiments, the at least one target biomarker
may be
selected from the group consisting of acetylcholine, betaine, dimethyl
sulfone,
glycolate/glycolic acid, and histamine, and the indication that the individual
is likely to
have an injury is provided in the event that the measured concentration levels
of
9
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
dimethyl sulfone and histamine are greater than their respective baseline
values and
the measured concentration levels of acetylcholine, betaine, and
glycolate/glycolic
acid are less than their respective baseline values.
[0026] In some embodiments, the at least one target biomarker
may be
selected from the group consisting of creatine, glycerol, threonine, malonate,
oxypurinol, and the indication that the individual is likely to have an injury
may be
provided in the event that the measured concentration levels of creatine,
malonate,
and oxypurinol are greater than their respective baseline values and the
measured
concentration levels of glycerol, threonine are less than their respective
baseline
values.
[0027] In some embodiments, the at least one target biomarker
may be
selected from the group consisting of arginine, glucose, and
glycerophosphocholine,
and the indication that the individual is likely to have an injury may be
provided in the
event that the measured concentration levels of arginine, glucose, and
glycerophosphocholine are less than their respective baseline values.
[0028] In some embodiments, the at least one target biomarker
may be
selected from the group consisting of glutamate, citric acid, cis-aconitate,
malate,
pyruvate, and the indication that the individual is likely to have an injury
may be
provided in the event that the measured concentration levels of glutamate,
citric acid,
cis-aconitate, malate, pyruvate are greater than their respective baseline
values.
[0029] In some embodiments, the injury is a central or
peripheral nervous
system injury, and the central nervous system injury may be a
neurodegenerative
brain disease or disorder, such Alzheimer's Disease or Parkinson's Disease. In
some
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
embodiments, the injury may be an acute injury or a chronic injury. In some
embodiments, the methods may be used for diagnosing, prognosing, or monitoring
the injury. In some embodiments, the methods may be used in the treatment of
the
injury.
BRIEF DESCRIPTION OF THE FIGURES
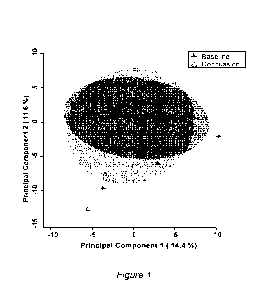
[0030] Figure 1 provides the results of an unsupervised
principal component
analysis (PCA) model, the plot revealing only a slight group difference
between pre-
and post-brain injury, according to embodiments;
[0031] Figure 2 provides the results of a supervised partial
least squares
discriminant analysis (PLS-DA) clustering analysis, the plot revealing a
distinct
separation between pre- and post-brain injury, according to embodiments;
[0032] Figure 3 provides the results from a further PLS-DA
analysis described
in FIG. 2, the plot focusing only on 18 features identified using VIAVC and
revealing
an even greater separation between pre- and post-brain injury, according to
embodiments;
[0033] Figure 4 provides a VIP scores plot illustrating the
top five features and
at least one target biomarker (metabolite) corresponding to each of the five
features,
according to embodiments;
[0034] Figure 5 provides an ROC constructed to determine
whether the 18
features identified as the best subset can be used to accurately predict
whether a
biological sample belongs to the pre-injury or post-injury group, according to
embodiments;
11
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0035] Figure 6 provides the results of a pathway topology
analysis of the at
least one target biomarkers (metabolites) identified according to the present
apparatus
and methodologies;
[0036] Figure 7 shows ROCs constructed from the features
identified as the
best subset for the pontine base (PB; FIG. 7A); for the dentate nucleus (DN;
FIG. 7B);
and for part of the anterior cortex (BA 24; FIG. 7C), according to
embodiments;
[0037] Figure 8 provides the results of a pathway topology
analysis of the at
least one target biomarkers (metabolites) identified in the pontine base (PB;
FIG. 8A);
in the dentate nucleus (DN; FIG.8B) and in part of the anterior cortex (BA 24;
FIG. 8C)
according to embodiments;
[0038] Figure 9 provides the results of orthogonal projections
to latent
structures discriminant analysis (OPLS-DA) modeling, the score plots revealing
a
separation between the injury (AD) and control (CN) groups in regions of
interest BA
22 (FIG.9A), BA 40 (FIG. 9B), BA 17 (FIG. 9C), and BA 40 (VIAVC only; FIG.
9D),
according to embodiments; and
[0039] Figure 10 shows ROCs constructed from the bins
determined to be
significant by VIAVC testing for regions of interest BA 22 (FIG.10A), BA 40
(FIG. 10B),
and BA 17 (FIG. 10C), according to embodiments.
DETAILED DESCRIPTION OF THE PREFERRED EMBODIMENTS
[0040] According to embodiments, apparatus and methodologies
of use for
determining whether an individual is likely to have an injury to the central
and/or
peripheral nervous systems, and for monitoring the individual's recovery from
the
injury, are provided. More specifically, apparatus and methodologies for
improved
12
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
detection, diagnosis, and prognosis of acute and chronic central and
peripheral
nervous system injury, are provided.
[0041] Broadly, the present apparatus and methods of use for
determining
whether an individual is likely to have an injury may comprise determining a
threshold
reference value for at least one target biomarker, or a combination of
biomarkers,
obtaining at least one biological sample from the individual and measuring the
concentration of the at least one of the target biomarkers in the sample, and
comparing
the measured concentrations of the at least one target biomarkers to the
threshold
reference value (i.e., the measured concentration of each at least one target
biomarker is compared to its respective threshold reference values) to
determine
whether a change in concentration has occurred. For example, where the
concentration of at least one of the target biomarkers is less than its
threshold
reference value, and the concentration of at least one other target biomarker
is greater
than its reference value, the detected change in biomarker concentration
levels may
be indicative of an injury. As such, the present apparatus and methods of use
may
provide an effective, non-invasive, objective method of determining whether an
individual is likely to have an injury to the central and peripheral nervous
systems by
detecting a change in at least one target biomarker (or a combination of
target
biomarkers, i.e., a change in pattern or profile of target biomarkers), such
change
being indicative of, and used for, detecting, diagnosing, and/or prognosing
the injury.
It is contemplated that changes in patterns or profiles or combinations of
target
biomarkers that are indicative of such injury may be compiled into a database
(i.e., an
electronic database) for simplifying and/or automating the detection,
diagnosis, and/or
13
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
prognosis of such injury. In such embodiments, the database may be operably
connected to a processor (i.e., computer) that permits rapid or real-time
detection,
diagnosis, and/or prognosis of an injury by comparing the detected change in
at least
one target biomarker with the changes recorded in the database.
[0042] In some embodiments, the present apparatus and methods
of use may
comprise the identification of changes in the metabolomic signature in a
biological
sample from an individual, the signature changes providing a clinical
biomarker
indicative with one or more nervous system injuries. For example, in some
embodiments, the presence of and/or changes in one or more biomarker profile
(e.g.,
up-regulation, down-regulation, and/or no change in one or more target
metabolites)
may be used to detect and diagnose the nervous system injury and to identify
the
likely course of the injury.
[0043] Certain terminology is used in the present description
and is intended to
be interpreted according to the definitions provided below. Unless otherwise
defined,
all technical and scientific terms used herein have the meaning commonly
understood
by a person skilled in the art to which the technology belongs.
[0044] Herein, the term "biological sample" means any sample
of tissue, cell,
fluid (i.e., bodily fluids, body fluids, biological fluids, biofluids, etc.)
or other material
derived from a subject including, without limitation, urine, plasma, whole
blood or
serum, cerebral spinal fluid, interstitial fluid, saliva, a tissue sample, or
an extract or
purification therefrom, or dilution thereof. For example, it is contemplated
that at least
one biological sample may be collected from a subject before a suspected
injury, as
well as within at least one hour, one day, and/or one week after the suspected
brain
14
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
injury. In some cases, the at least one biological sample may be collected
from a
subject before, during, or after a sporting event or a sporting season. It is
also
contemplated that at least one biological sample may be collected from a
subject
suspected of suffering a neurodegenerative disease. In some cases, the at
least one
biological sample may be collected from an individual before, during, or after
the
suspected onset of Alzheimer's Disease (AD) or Parkinson's Disease (PD).
[0045] Many of the metabolomic changes that are observed in
blood and
cerebrospinal fluid should also be detectable in urine using the present
apparatus and
methodologies. Urine is comprised of the biological by-products produced
throughout
the body, including the brain, making urine samples an ideal biofluid to
examine
metabolic changes linked to brain injury when obtaining other biological
samples or
biofluids might be difficult. Results of urine metabolomics may be influenced
by kidney
function, however, many water-soluble metabolites that are present and
identifiable
by NMR in serum and CSF are also present and identifiable by NMR in urine.
Indeed,
91.8% (45/49) of the metabolites that are identifiable by NMR in serum and
81.0%
(43/53) of the metabolites that are identifiable by NMR in CSF are present and
identifiable by NMR in urine. Although a biological sample comprising a urine
sample
is described herein, any appropriate biological sample comprised of biological
by-
products produced throughout the body, including brain tissue, are
contemplated.
[0046] Herein, the term "biomarker", "target biomarker, or
"clinical biomarker"
means a measurable characteristic that can be objectively determined and
evaluated
as an indicator of biological processes within an individual. In some
embodiments,
"biomarkers" generally refer to at least one biochemical, e.g., at least one
substance
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
formed in or necessary for metabolism, such as a metabolite, that is
differentially
present in a biological sample taken from the individual. Differentially
present refers
to differences in the quantity and/or frequency of the at least one biomarker
in a
biological sample taken from a subject having a nervous system injury.
[0047] For example, according to embodiments herein,
"biomarker" can mean
a measurable change in biochemical signature or pattern of one or more
metabolites
(e.g., an increase, decrease, or no change) arising from metabolomic
processes, the
biochemical signature or pattern presenting differently in the individual over
time (e.g.,
pre- and post-injury) and being indicative of an injury (e.g., brain injury,
neurodegenerative disorder and disease, etc.). For example, the present
methods
may be incorporated into one or more clinical tests (e.g., a urinalysis
device, or the
like) to provide the user, whether or not trained in metabolomic or raw data
analysis,
with an output indicative of an injury in an individual. The output may then
be used to
detect the type of injury, to determine the severity of the injury and/or its
likely
outcome, to determine an appropriate course of treatment for the individual,
or to
optimize same.
[0048] Metabolites are those chemicals generally less than
1,000 Da) involved
in cellular reactions for energy production, growth, development, signaling
and
reproduction and can be taken up or released from cells according to their
cellular
needs. Metabolites can include sugars, amino acids, organic acids, as well as
xenobiotic compounds, the presence of which might change in a cell or system
due to
internal or external stress such as an injury, an injury, or disease state.
Metabolic
changes can result from changes in the chemical reactions that use these
metabolites
16
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
(i.e., metabolic pathways), or the transporter that take up or release the
metabolites.
For example, without being limited by theory, an individual impacted by an
injury to
the nervous system may exhibit major changes both at the cellular and systemic
levels
including inflammation and impaired energy metabolism. Although biomarkers
comprising brain and urinary metabolites are described herein, any appropriate
metabolomic markers indicative of injury, and specifically brain injury, are
contemplated.
[0049] In some embodiments, examples of metabolites that may
be measured
as target biomarkers include, without limitation, one or more of the
metabolites
selected from the group consisting of 2-Hydroxybutyrate, 3,4-
dihydroxybenzeneacetate, carnitine, 4-hydroxybenzoate, caffeine,
horriocitrulline,
methionine, acetylcarnitine, 3-methyl-2-oxovalerate, phosphorylcholine,
choline,
propylene glycol, taurine, 1-methylhistadine, 3-methylhistadine, citrate,
lactose,
phenylalanineõ 3-indoxylsulfate, sucrose, 3-methyladipate, isobutyrate, 3-
hydroxyisovalerate, 5-am inolevulinate, anserine, tyrosine, carnosine,
isoleucine,
leucine, threonate, and cysteine, and any other metabolites that may have been
characterized but have not yet been identified with certainty to date.
[0050] In some embodiments, examples of metabolites that may
be measured
as target biomarkers include, without limitation, one or more of the
metabolites
selected from the group consisting of citrate, glycyl-glycine, isoleucine,
glutamate,
trimethylamine N-oxide, choline, choline phosphate, glucose, leucine,
phenylalanine,
valine, tyrosine, glutamate, methionine, galactose, glycerol, myo-Inositol,
betaine,
threonine, ethanol, creatine, malonic acid/malonate, pyruvatoxine, alpha-
17
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
ketoisovaleric, propylene glycerol, 2-oxohexane, gamma-aminobutyric acid
(GABA),
2-hydroxy-3-methylvaelrate, N-acetyl-L-aspartate (NAA),
4-am inobutanoate,
threonine, 3-methyl-2-oxobutanoic acid, (R)-3-hydroxybutanoate, succinate,
glycolate/glycolic acid, acetylcholine, 2-amino-3-phosphonoprionic acid, 1,3,7-
trimethyluric acid, serine, phosphorylcholine, dimethyl sulfone,
glycolate/glycolic acid,
histamine, oxypurinol, arginine, glycerophosphocholine, glutamate, citric
acid, cis-
aconitate, malate, pyruvate and any other metabolites that may have been
characterized but have not yet been identified with certainty to date.
[0051]
Herein, the terms "diagnose" and "diagnosis" means the detection,
identification, confirmation, and/or characterization of a nervous system
injury, such
as a central nervous system injury. According to embodiments, the present
methods
of detecting and diagnosing are useful to confirm the existence of a brain
injury, for
assessment of clinical screening, prognosis, choice of therapy, evaluation of
therapeutic benefit, e.g., in the case of a sports related concussions, the
timing for
return to play.
[0052]
Herein "individual" or "subject" means a human subject. In some
embodiments, the individual may suffer from an injury, such as an injury to
the central
and/or peripheral nervous systems. Without being limited by theory, the
individual may
suffer from an injury causing an alteration(s) to biochemical pathways (e.g.,
sum
changes in genes, protein synthesis, and/or environmental factors, etc.)
resulting in
discernible metabolomic changes that, when detected, can be indicative of the
injury.
[0053]
Herein "injury" means an injury to the central and/or peripheral
nervous
system including, without limitation, brain injuries, traumatic brain injuries
(e.g.,
18
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
traumatic brain injury broadly covers mild TBI, moderate TBI, sever TBI,
concussion,
and other head impacts), neurodegenerative disorders and disease, etc. In some
embodiments, brain injury may include any disruption in the normal function of
the
brain that can be caused by a blow, bump, or jolt to the head or spine, the
head or
spine suddenly and violently hitting an object. Without being limited by
theory, injury
may be any change in central and/or peripheral nervous system pathology or
function
causing an alteration(s) to biochemical pathways resulting in discernible
metabolomic
changes that, when detected, can be indicative of the change in pathology or
function.
Without being limited by theory, and by way of example only, injury may
include at
least one neurological disorder (i.e., structural, biochemical, or electrical
abnormality)
that affects the brain, spinal cord, and/ or nerves found throughout the human
body
and spinal cord, and that can result in a range of symptoms. In some
embodiments,
injury might be an acute injury, while in other cases the injury might be
chronic.
Although certain specific injuries, such as traumatic brain injury (e.g.,
sports-related
concussion, SRC) and neurodegenerative injury (e.g., Alzheimer's Disease, AZD)
are
described herein, such injury are provided for explanatory purposes only and
are in
no way intended to limit the intended scope of the present apparatus and
methodologies.
[0054] Herein, threshold "reference value", "reference
profile/pattern" or
"reference combination" means a baseline, standard, and/or pre-injury
biomarker
concentration signature derived from an individual. In some embodiments, the
threshold reference value may be obtained from the individual being monitored
for
injury (e.g., during a sporting event, or sport season). In other embodiments,
the
19
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
threshold reference value may be fully or partially obtained from at least one
privately
or publicly-available database of samples (e.g., pooled samples) from healthy
subjects, such as might be found in the Human Metabolome Database, or
specifically
characterized biofluid metabolomes including, without limitation, the urine
metabolome, the serum metabolome, the saliva metabolome, or the cerebrospinal
fluid metabolome (e.g., the samples being gender- or age-matched). Although
threshold reference values are described herein as being derived directly from
the
individual, any appropriate samples sufficient to detect changes in an
individual's
biomarker signature or pattern are contemplated.
[0055] Although 1H NMR spectroscopy is described herein, any
suitable
spectroscopic technique can be used to generate the presently described
metabolomic signatures including NMR spectroscopy and mass spectrometry. In
contrast to other techniques, 1H NMR spectroscopy of biological samples,
including
fluid samples, allows for a high throughput biological sample analysis with a
broad,
untargeted approach to biomarker discovery. In addition, when examining the
metabolome of urine, NMR spectroscopy provides quantitative information on
more
metabolites (209 total, 108 unique) when compared to the other mass
spectrometry-
based methods. Moreover, many of the metabolomic changes observed in blood and
cerebrospinal fluid may be detected by NMR in urine, whereas urine
advantageously
also contains additional metabolomic information not in the other biofluids
(e.g.,
including at least 18 unique urinary metabolites as described herein). In some
embodiments, use of 1H NMR spectroscopy in the present apparatus and
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
methodologies provides a clinically accessible, painless, non-invasive medium
for
injury detection, diagnosis, and prognosis.
[0056] As might be appreciated, 1H NMR spectroscopy
information and data
may be received, stored, analyzed, and modified using at least one computer
processor (e.g., digital processor) in communication with the NMR, where
computer
processor means a device that is programmed to run at least one algorithm for
performing a set of steps according to a program. Computer processors may
include
Central Processing Units (CPUs), electronic devices, or systems for receiving,
transmitting, storing and/or manipulating data under programmed control.
[0057] Each term used and defined herein is for explanatory
purposes only and
in no way is intended to limit the scope of the technology.
[0058] In some embodiments, the present apparatus and
methodologies may
comprise determining a threshold or baseline reference value for at least one
target
biomarker, the threshold reference value for the at least one target biomarker
being
used to detect a change in the concentration of the biomarker, the change
being
indicative of an injury. For example, a threshold reference value of the at
least one
target biomarker may comprise a baseline, pre-injury, or normal concentration
of the
at least one target biomarker. In some embodiments, a threshold reference
value for
at least one target biomarker may be obtained from a biological sample from
the
individual, from a cohort of pooled individuals, or the like, and then used as
a
comparison value or baseline value to determine if a change in the
concentration
levels of the at least one target biomarker has changed in the individual
(e.g., when
21
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
compared to one or more biological samples collected from the individual
following a
suspected injury to the nervous system, as will be described).
[0059] In some embodiments, the threshold reference value may
be
determined for at least two target biomarkers, the combined reference values
of the
at least two target biomarkers comprising a threshold baseline biomarker
'profile' or
'pattern', wherein a change to the biomarker pattern is indicative of an
injury. For
example, when compared to a threshold baseline biomarker profile, the presence
of
and/or changes in the biochemical signature of at least two target biomarkers
(e.g.,
up-regulation, down-regulation, and/or no change in concentration) may be used
to
detect and diagnose an injury and to identify the likely course of the injury.
Advantageously, recognition and detection of a specific change in
concentration of at
least two target biomarkers, i.e., the determination of a signature pattern or
profile
changes in metabolites, provides an improved method of detection, diagnosis,
and
prognosis (including monitoring recovery) in both acute and chronic injury.
[0060] For example, the present methods and use thereof may
provide a more
accurate and/or sensitive clinical test for detecting, diagnosing, and
treating an injury
in an individual than known methods that merely detect the presence of one or
more
metabolites of interest, where such known methods merely detect the presence
of a
metabolite of interest to determine a specific type of injury (e.g., a "stroke
specific
metabolite"). Reliance upon the presence of one metabolite may lead to both
false
positive and false negative diagnoses. Such methods are also not able to
prognose
the injury, but instead can simply detect the presence of a metabolite that
'might' be
linked to the specific injury. The present methods and use thereof may also
provide a
22
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
more accurate and/or sensitive clinical test for detecting, diagnosing, and
treating an
injury than known methods because the present methods also account for down-
regulation of metabolites, that is ¨ the absence or down-regulation of a
metabolite
within the change in signature or profile can be indicative of the injury.
Instead, known
methods may merely detect a target metabolite and, as a result, conclude no
injury
has occurred, when in fact the opposite might be true.
[0061] Furthermore, the present methods and use thereof may
also provide a
more accurate and/or sensitive clinical test for detecting, diagnosing, and
treating an
injury than known methods because they are operative to measure and detect a
clear
change in two or more metabolites, i.e., creating a signature, pattern, or
profile
change, that is indicative of an injury. That is, advantageously, by examining
a
specifical change in two or more metabolites, i.e., i.e., a change in the
signature,
pattern, or profile, the present methods and use thereof provide a more
sensitive
clinical tool (e.g., where the error from smaller changes can be minimized by
multiplexing the changes observed in two or more metabolites, increasing the
sensitivity of the diagnostic).
[0062] In some embodiments, the present apparatus and
methodologies may
comprise obtaining at least one biological sample from an individual or a
cohort of
pooled individuals who have been categorized as being healthy and analyzing
the
sample(s) to determine whether the individual is likely to have an injury
(i.e., to
determine if changes in at least one target biomarker in the sample is
indicative of an
injury). In some embodiments, the at least one biological sample(s) may be
analyzed
to determine the threshold reference value of the at least one biomarker
(e.g., pre-
23
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
injury, baseline sample). In other embodiments, the at least one biological
sample(s)
may be analyzed to determine whether a change in the concentration of the at
least
one biomarker is indicative of an injury (e.g., post-injury,
diagnosis/prognosis sample).
Advantageously, the present apparatus and methods contemplate obtaining as few
as one pre-injury reference sample and one post-injury test biological sample,
however it should be appreciated that any number of biological samples may be
obtained and analyzed in order to determine whether the individual is likely
to have an
injury.
[0063] For example, one or more first biological samples may
be obtained from
an individual, such first sample used to determine the threshold reference
value, or a
baseline target biomarker profile. One or more second, third, etc. biological
samples
may be subsequently obtained, the one or more second samples used to determine
if
the concentration of the at least one biomarker has changed (i.e., to detect
and
determine changes in the individual's target biomarker profiles), and to
discern if the
changes are indicative of brain injury.
[0064] It should be appreciated that not all detected changes
to the biomarker
profile will be indicative of injury. Instead, as will be described, certain
predetermined
or signature changes to the biomarker profile may be indicative of disease,
referred to
as "diagnostic levels". More specifically, in the event that the measured
concentration
level of at least one of the target biomarkers is less than its respective
threshold
reference value (i.e., the target metabolite(s) are down-regulated) and the
measured
concentration level of at least one of the other target biomarkers is greater
than its
respective threshold reference value (i.e., the target metabolites(s) are up-
regulated),
24
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
there is an indication that the individual is likely to have an injury.
Moreover, the
detected changes may also be indicative of the severity of the injury, and a
prognosis
of the likely course of the injury. More specifically, for example, the
profile or pattern
of detected changes indicative of any particular injury may differ according
to the
severity of the injury.
[0065] Herein, greater than and/or less than may refer to a
statistically
significant difference between a diagnostic amount of the at least one target
biomarker
measured in a post-injury biological sample when compared to a control or
standard
amount of the at least one target biomarker obtained from the threshold
reference
value, referred to as a "baseline amount". All amounts referred to herein can
be either
an absolute amount (e.g., pg/mL) or a relative amount (e.g., relative
intensity of
signals). Herein, statistically significant can be at least a difference of at
least p<0.05.
By way of example, a change in the concentration levels of the at least one
biomarker
indicative of an injury may be approximately 0.1% _ 1%7 2% _ 10%7 11% _ 20%7
21 _
30%, 31 ¨ 40%, 41 ¨ 50%, 51 ¨ 60%, 61 ¨ 70%, 71 ¨ 80%, 81 ¨ 90%, 91 ¨ 100%, or
> 100% when compared to a baseline reference concentration value.
[0066] In some embodiments, the present apparatus and methods
involve
measuring the concentration of the at least one target biomarker using nuclear
magnetic resonance (NMR), including 1H NMR, combined with multivariate
statistical
analyses to develop pre-injury threshold reference values (i.e., metabolite
profile
patterns) and to detect post-injury changes in at least one target biomarker
compared
to its respective threshold value, such change being indicative of nervous
system
injury. As would be appreciated, individual metabolomics exhibit unique
spectral
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
signatures that are consistent and reproducible for a given set of overall
sample
conditions, whereby concentrations of a single metabolite in a given sample
can be
accurately determined by reference to an internal standard. As will be
demonstrated,
1H NMR provides an automated pattern recognition technique for simple,
effective
analysis of an individual's injury state, shortening analysis time and
eliminating
subjective data interpretation. That is, herein, using appropriate preparation
of at least
one pre-injury biological sample (containing an estimated 1400 endogenous
urinary
metabolites) and intelligent software tools that model the spectrum using the
threshold
value, an accurate listing of at least one target biomarker and its
corresponding
concentration indicative of nervous system injury was obtained.
[0067] For example, without being limited by theory, potential
target biomarkers
may comprise at least one metabolite, the concentration of which might be
impacted
by neuropathological changes caused by brain injury, disorder, or disease.
[0068] The present apparatus and methodologies will now be
illustrated in more
detail by way of the following Examples.
[0069] EXAMPLES:
[0070] EXAMPLE 1: According to embodiments, the present
example
demonstrates the use of the present apparatus and methodologies for
determining
whether an individual is likely to have an injury, such as a brain injury.
More
specifically, the present example demonstrates the use of the present
apparatus and
methodologies for the detection and diagnosis of at least one brain injury,
such as a
sports-related concussion, and further for predicting recovery from the at
least one
brain injury.
26
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0071] According to embodiments, the present example involved
423
individuals or subjects comprised of Canadian national team athletes, national
development athletes, and amateur ice hockey players, ages 14 - 40 years, who
were
participating in the WinSport Concussion Clinic at WinSport (Calgary, Alberta,
Canada) from August 2015-2016. Exclusion criteria include age greater than 40
years
and less than 12 years, female, a previous history of chronic medical
conditions (e.g.,
metabolic or nephritic disorders), neurological conditions such as stroke,
seizure,
moderate to severe traumatic brain injury and/or congenital intracranial
abnormalities.
[0072] According to embodiments, a preclinical assessment of
each individual
was performed. In some embodiments, the preclinical assessment comprised
obtaining and analyzing at least one first biological sample (e.g., a urine
sample), the
preclinical sample serving as a threshold reference value or pre-injury
metabolic
profile for the at least one target biomarker. For example, in some
embodiments, at
least one bodily fluid sample comprising a urine sample was obtained from each
individual. The bodily fluid samples were collected in the morning hours
between 7-9
am and before the first meal to provide a non-concussed assessment. Athletes
were
asked to only drink water prior to sample collection.
[0073] Following the pre-injury assessment, each individual
was monitored for
at least a year-long sports season (e.g., throughout the sport season between
2015 ¨
2016). At one or more time periods during the sports season (e.g., where it
was
believed that an individual suffered a brain injury), a detailed physical
assessment of
each individual was performed, including a clinical diagnosis of the SRC based
on
27
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
current International Consensus on Sport Concussion recommendations (see Table
1). There were no other injuries at the time of the SRC.
[0074] Having regard to Table 1, twenty six (26) individuals
were found to have
suffered an SRC during the sports season. Individual characteristics and past
medical
history of the injured individuals are provided in Table 1 and include age,
sex, medical
history, medication (prescribed over the counter), length of post-traumatic
amnesia,
and pre- and post- number of symptoms.
[0075] TABLE 1: SRC patient characteristics
1 15 - 1 Ice
Hockey
2 16 asthma Ventolin, Protein powder 0 Ice
Hockey
3 14 Vitamin D, Coenzyme 0 Ice
Hockey
Q10, Vitamin C
4 16 asthma Ventolin, Protein Powder 0 Ice
Hockey
16 acne Biosteel Protein, 0 Ice Hockey
Minocycline
6 16 - 2 Ice
Hockey
7 16 depression Protein Powder, Creatine 1
Ice Hockey
8 16 - Mesavant 0 Ice
Hockey
9 15 Migraine 1 Ice
Hockey
Headaches
14 Migraine - 3 Ice Hockey
Headaches
11 15 Generalized Ventolin 1 Ice
Hockey
anxiety
Disorder,
asthma
12 13 0 Ice
Hockey
13 13 - 0 Ice
Hockey
14 15 - Naturopathic Growth 0 Ice
Hockey
Hormone
13 0 Ice Hockey
16 13 Vegan Protein 0 Ice
Hockey
17 16 Acne, acutane 0 Ice
Hockey
previous
whiplash
injury of the
neck
18 14 - - 0
Luge
19 15 - - 1 Ice
Hockey
28
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
20 14 - - 1 Ice
Hockey
21 12 Migraine - 0
Taekwondo
Headaches
22 14 - Vitamin D, Vitamin B, 0 Ice
Hockey
Protein Powder
23 15 Protein Powder 1 Ice
Hockey
24 16 Acne, Minocycline 0 Ice
Hockey
Migraine
Headaches
25 15 - Protein Powder, Vitamin 0 Ice
Hockey
C, Vitamin D, Creatine
26 13 - Cod Liver oil, Vitamin D 1 Ice
Hockey
[0076] In addition to foregoing individual characteristics, a
sports concussion
assessment Tool 3 (SCAT3) was used to assess SRC in the 26 individuals within
72
hours of their injury. The SCAT3 assessment and a history of previous head
injuries
are provided in Table 2 and include a description of recovery from previous
head
injuries, neurological conditions, medications, sport-participation and
biographical
information.
[0077] TABLE 2: Symptom Score, Length of Return to Sport,
Length of Loss of
Consciousness, Length of Post-traumatic Amnesia.
1 0 0 0 0 0 1
10
2 13 22 0 0 0 0
9
3 0 0 0 0 0 0
2
4 13 22 0 0 0 0
9
20 65 0 0 0 0 17
6 2 4 2 4 0 0
3
7 11 16 2 2 0 0
11
8 15 35 3 3 0 0
23
9 8 16 0 0 0 0
8
13 44 1 4 0 0 78
11 3 5 0 0 0 0
17
12 0 0 0 0 0 0
12
29
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
13 0 0 0 0 0 0
13
14 18 47 0 0 0 0
16
15 15 28 1 2 0 0
10
16 7 14 0 0 0 0
38
17 1 1 0 0 0 0
10
18 10 10 0 0 180 3
30
19 16 47 2 2 0 0
36
20 6 9 0 0 0 0
18
21 12 18 1 1 0 0
42
22 9 21 0 0 0 0
9
23 4 8 1 2 0 0
19
24 20 22 0 0 0 1
18
25 2 3 2 2 10 20
10
26 11 22 0 0 0 0
17
[0078] According to embodiments, the post-injury assessment
comprised
obtaining at least one second bodily fluid sample, said sample operative as a
test or
post-injury metabolic profile. For example, in some embodiment, at least one
second
bodily fluid sample comprising a urine sample was obtained from each
individual,
wherein the second bodily fluid sample was a 12-hour fasting urine sample
between
7-9 am within a time window of 24 hours and 72 hours post-injury.
[0079] Each of the second biological (urine) samples obtained
were
immediately stored at -80 C, batched, and prepared for analysis. In
preparation for 1H
NMR spectroscopy, 400 pL aliquots of urine were added to 200 pL aliquots of
phosphate urine buffer in 2 mL centrifuge tubes. The phosphate buffer was
prepared
as a 4:1 ratio of KH2PO4 in a 4:1 H20:D20 solution to a final concentration of
0.5M.
The D20 included 0.05% (by weight) trim ethylsilyl propanoic acid (TSP) as a
chemical
shift and concentration reference. To protect the metabolite profile
integrity, 0.02%
(weight/volume) of sodium azide (NaN3) was added to the solution as an
antimicrobial
agent. The buffer solution was then titrated to pH 7.4 using HCI or NaOH,
depending
on the initial pH. The tubes containing urine and buffer were centrifuged at
12,000 rpm
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
for 5 minutes at 4 C. After centrifugation, 550 pL of the supernatant was
transferred
to a 5 mm NMR tube for NMR analysis. Samples were immediately analyzed using
the NMR spectrometer.
[0080] NMR spectra were collected on a 700 MHz Bruker Avance
III HD
spectrometer (Bruker, ON, Canada). The Bruker 1-D NOESY gradient water
suppression pulse sequence was used. Each sample was run for 128 scans and the
total acquisition size was 128k. The spectra were zero filled to 256k,
automatically
phased, baseline corrected, and line-broadened to 0.3Hz. The processed spectra
were exported as ascii files to Matlab (The MathWorks, MA, USA) for
statistical
analysis. Spectra were first binned using dynamic adaptive binning, and then
manually
adjusted to optimize number of variables and ensure accuracy. All spectra had
the
regions corresponding to water and urea removed before normalization to the
total
area of all spectral bins. The peaks corresponding to acetaminophen
derivatives were
also removed to account for the fact that the individuals tended to take pain
medication
following an SRC. The binned spectra were then pareto scaled to increase the
influence of weak peaks and deemphasize the influence of larger peaks. All the
peaks
in each spectrum were referenced to TSP (0.00o).
[0081] All univariate and multivariate testing was carried out
using MATLAB
(The MathWorks, MA, USA) and MetaboanalystR (v 2.0), respectively. As would be
appreciated, multivariate statistical analysis can be applied to the collected
data or
complex spectral data to aid in the characterizations of changes related to a
biological
perturbation or injury.
31
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0082] According to embodiments, data visualization to
determine sample
structure and the presence of distinct groups was performed using Principal
Component Analysis (PCA) and Partial Least Squares Discriminant Analysis (PLS-
DA). Ten-fold double cross-validation and 2,000 permutation tests were
performed to
validate the results of the supervised PLS-DA testing, as recommended in the
art for
validating PLS-DA models. Variable Importance to the Projection (VIP) scores
can
indicate which spectral bins (hereafter referred to as "features") contribute
the most to
the separation observed in the PLS-DA scores plot. This score represents a
weighted
sum of squares of the PLS-DA loadings in each dimension. All tests were
performed
using the online metabolomics statistics software platform Metaboanalyst.
[0083] To focus on potential biomarkers, Variable Importance
Analysis based
on random Variable Combination (VIAVC) was used as a feature selection tool.
The
VIAVC algorithm combines random permutations of variable inclusion with a ten-
fold
cross validation of model. This reveals a best-subset of target biomarkers
(metabolites) that have the greatest effect on group differences. VIAVC p-
values were
calculated using a t-test and distribution of how many times a biomarker
(metabolite)
was removed to improve the model during various permutations. Each of the at
least
one target biomarkers (metabolites) in the best-subset generated by the
algorithm was
therefore strongly informative in separating the samples into groups, and
synergistic
effects between the biomarkers were revealed. All VIAVC tests were carried out
using
MATLAB. To further test for significant features between pre-injury and post-
SRC
paired t-tests were completed.
32
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0084] The VIAVC method, VIP scores, and paired t-tests each
provided a set
of features that were considered significantly altered across the comparison
groups.
The at least one target biomarkers (metabolites) corresponding to these
features were
identified using the profiler tool in the Chenomx 8.2 NMR Suite (Chenomx Inc.,
Edmonton, Alberta, Canada), and receiver operator characteristic (ROC) curves
were
used to graphically represent the true positive rate versus 1-specificity. The
accuracy
of a classifier was visualized by the area under the ROC curve.
[0085] Biological significance of target biomarkers (important
metabolites) was
investigated using the free pathway topology analysis tool available through
Metaboanalyst. Data were collated as a list of metabolites, and the human
pathway
library was chosen. The library was built using the detailed Kyoto
Encyclopedia of
Genes and Genomes (KEGG) pathway diagrams. A list of the most relevant
biological
pathways involved in conditions of the study was then generated to help draw
connections between potential biomarker metabolites and relevant biological
processes.
[0086] Spearman's correlations compared the normalized
concentration of
significantly altered metabolites post-SRC to the total number of symptoms
reported,
the symptom severity score, and the length of return to play. A p-value of <
0.05 was
considered significant and metabolite concentrations were normalized with
respect to
the entire urinary metabolome.
[0087] Initially, exploratory analysis using both supervised
and unsupervised
models of all data was completed, with the unsupervised principal component
analysis
(PCA) being used to cluster the raw data and highlight any separation between
33
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
groups. Having regard to FIG. 1, the PCA scores plot revealed only a slight
group
difference (with + indicating pre-injury baseline, or controls and L
indicating an injury).
Having regard to FIG. 2, supervised partial least squares discriminant
analysis (PLS-
DA) clustering analysis was performed, revealing distinct separation between
the pre-
injury baseline, or controls (+) and post-injury or concussion groups (a).
[0088] Variable importance analysis based on random variable
combination
(VIAVC) was then used to remove unimportant or interfering features from the
data.
VIAVC resulted in the discovery of a subset of 18 features that have a strong
effect
on the differences between control (e.g., pre-injury) and injured groups
(e.g., post-
SRC). The VIAVC p-value represents a measurement of whether the inclusion of a
feature in 1000 randomly chosen subsets of features improved or reduced the
overall
class separation of the model. Thus, VIAVC can determine synergistic effects
across
features.
[0089] PLS-DA analysis was completed again to focus only on
the 18 features
identified by VIAVC. FIG. 3 provides a partial least squares discriminant
analysis
(PLS-DA) 2D scores plot showing separation between baseline (pre-injury) and
post-
injury urine samples (based on the VIAVC best subset of features corresponding
to
the metabolites provided in Table 3). A clear separation was found between
groups
with Component 1 and 2 accounting for 21.9% and 7.9% of the variance in the
data,
respectively. The percentages shown along the axis indicate the amount of
variance
in the data set given by each component and the shaded ellipses designate the
95%
confidence interval of each group. The model passed permutation testing using
2000
34
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
permutations (p = 0.0005) confirming that the separation is real and not due
to chance.
This model also passed ten-fold cross validation testing.
[0090] Having regard to FIG. 4, provides variable importance
in the projection
(VIP) scores for the top five target biomarkers (metabolites) used in the PLS-
DA
model, where the higher the VIP score, the more the biomarker(s) contributed
to the
separation observed between groups in the PLSD-DA model shown in FIG.3. For
example, the VIP scores plot shown in FIG. 4 illustrates the top five features
and the
biomarkers corresponding to each feature, with the coloured boxes on the right
side
indicating whether the biomarker (metabolite) was up- or downregulated in the
post-
injury sample compared with the respective baseline threshold value in the pre-
injury/control sample. The heat map (right) indicates the directionality of
the changes,
i.e., up- or downregulation from the injury (e.g., SRC).
[0091] In some embodiments, as shown in FIG.4, a change in at
least one
target biomarker selected from the group consisting of phenylalanine and 3-
indoxysulfate may be indicative of brain injury, particularly where the
concentration of
such at least one target biomarkers in the biological sample is greater than
its
respective baseline threshold baseline value (i.e., the biomarkers are
upregulated
following brain injury (e.g., SRC)). In other embodiments, as also shown in
FIG.4, a
change in at least one target biomarker selected from the group consisting of
Citrate
and propylene glycol may be indicative of brain injury, particularly where the
concentration of such at least one target biomarkers in the biological sample
is less
than its respective baseline threshold baseline value (i.e., the biomarkers
are
downregulated following brain injury (e.g., SRC)).
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0092] Having regard to FIG. 5, a receiver operator curve
(ROC) was
constructed for the VIAVC best subset of features corresponding to the at
least one
target biomarker (e.g., metabolites as provided in Table 3) in order to test
if the 18
features identified as the VIAVC best subset may be used to accurately predict
whether a sample belongs in a baseline/control (pre-injury) or post-injury
group. The
model graphs the true positive rate on the y-axis versus the false positive
rate on the
x-axis. As shown, the ROC model had a corresponding area under the curve (AUC)
of 0.887 with a 95% confidence range of 0.731-0.997 and a predictive accuracy
of
81.6%.
[0093] A paired t-test was also completed to compare
baseline/control (pre-
injury) and post-injury samples of each individual. This test identified 19
features as
significant (p < 0.05), with 7 of these features being common to the VIAVC
results.
Table 3 provides a list of the 31 metabolites identified as significant by
VIAVC or a
paired t-test or both tests.
[0094] Table 3: Target Biomarkers/Metabolites
36
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Metabolite VIAVC P- PLS-DA T-Test
Regulation
value VIP Score P-
value
2-Hydroxybutyrate 5.42E-45 0.41803 Not
Sig. Down
3,4- 4.86E-19 0.14659 Not
Sig. Up
Dihydroxybenzeneacetate.1/Carnitine.
1
4-Hydroxybenzoate 3.98E-85 0.58827 Not
Sig. Up
Caffeine 3.29E-52 0.35628 Not
Sig. Down
Carnitine.2 1.56E-38 0.54019 Not
Sig. Up
Homocitrulline 2.15E-77 0.44871 Not
Sig. Down
Methionine/Acetylcarnitine.1 2.59E-50 0.4312 Not
Sig. Up
3-Methyl-2-0xovalerate 8.18E-26 0.41031 Not
Sig. Up
Phosphorylcholine/Choline/Acetylcarnit 1.52E-128 0.55238 Not
Sig. Up
ine.2
Propylene Glycol 4.50E-77 0.94917 Not
Sig. Down
Taurine/3,4- 1.40E-41 0.37166 Not
Sig. Up
Dihydroxybenzeneacetate.2/Carnitine.
3
*1-Methylhistadine/3-Methylhistadine 3.69E-50 0.91339
0.03871 Down
*Citrate 9.08E-100 0.95022 0.032117 Down
*Lactose 8.24E-43 0.78734 0.043613 Up
*Phenylalanine.1 1.60E-43 1.1556
0.00719 Up
*Phenylalanine.2 1.71E-42 2.4739 0.000322
Up
*Phenylalanine.3/3-Indoxylsulfate.1 2.30E-72 2.26 0.002267
Up
*Sucrose 2.22E-42 0.62691 0.037745 Down
3-Methyladipate/lsobutyrate Not Sig. N/A
0.04133 Down
3-Hydroxyisovalerate Not Sig. N/A 0.020594
Down
3-Indoxylsulfate.2 Not Sig. N/A 0.006534
Up
5-Aminolevulinate Not Sig. N/A 0.019592
Up
Anserine.1/Tyrosine Not Sig. N/A 0.026885
Up
37
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Anserine.2 Not Sig. N/A 0.037285
Up
Carnosine/Anserine.3 Not Sig. N/A 0.024957
Up
Isoleucine/Leucine Not Sig. N/A 0.02161
Up
Phenylalanine.4 Not Sig. N/A 0.002615
Up
Phenylalanine.5 Not Sig. N/A 0.005616
Up
Phenylalanine.6 Not Sig. N/A 0.033934
Up
Threonate/Cysteine Not Sig. N/A 0.011865
Down
[0095] Having regard to FIG. 6, a complete list of target
biomarkers
(metabolites) identified by both methods (as listed in Table 3) was used to
carry out
Pathway Topology analysis, to determine which metabolic processes may be most
affected following a brain injury, such as an SRC. The Pathway Topology
analysis
was completed by entering a list of at least one significant metabolite(s)
found by both
VIAVC and paired t-test into known web-based tool MetPAm. Each circle shown in
FIG. 6 indicates a specific metabolic pathway or biological function as
labeled to the
left, with the x-axis being the pathway impact scores which represents the
magnitude
of impact by significant biomarkers, as shown by the size of each circle, and
the y-
axis being the p-values, given as -In(p), with red circles indicating a lower
p-value and
yellow a higher p-value. Only pathways with p 0.06 are shown.
[0096] According to embodiments, as shown in FIG. 6, aminoacyl-
tRNA
biosynthesis (#1; p<0.001) and beta-alanine metabolism (#2; p<0.01) were the
two
pathways with most significant change. Taurine and hypotaurine metabolism
(#12),
pantothenate and CoA biosynthesis (#11), and phenylalanine metabolism (#10;
p<0.005 and pathway impact of 0.11906) show the highest pathway impacts.
Moreover, there was a significant positive clinical correlation found between
the post-
38
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
injury normalized concentration of 2-hydroxybutyrate and the length of return
to play
(R = 0.482, p = 0.02), and a significant positive clinical correlation between
the total
number of symptoms and the post-injury normalized concentration of lactose (R
=
0.422, p = 0.036).
[0097] In some embodiments, the present example demonstrates
the detection
of changes in concentration of at least one target biomarker (metabolites) in
a
biological sample from an individual for determining whether the individual is
likely to
have suffered a brain injury, and specifically to diagnosing a sports-related
concussion. The present example demonstrates that the detection of changes in
concentration of at least one target biomarker (metabolite), or combination of
biomarkers), is indicative of brain injury, such changes comprising an up-
regulation, a
down-regulation, and/or no change of the biomarker or combination of
biomarkers
relative to a threshold reference value. Without being limited to theory, the
present
example demonstrates that the changes in concentration of at least one target
biomarker (metabolites), or combination of biomarkers may arise from
alterations of
key biological pathways involved in primary and secondary brain injury and
recovery
processes.
[0098] In some embodiments, the present example also
demonstrates the
detection of changes in concentration of at least one target biomarker
(metabolites) in
a biological sample from an individual for determining whether the individual
is likely
to have suffered a brain injury and for diagnosing the type of injury (a
sports-related
concussion), as well as for detecting increased symptom burden and prognosing
recovery (e.g., return to play). In such embodiments, the present example
39
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
demonstrates that the detection of changes in concentration of at least one
target
biomarker (metabolite), or combination of biomarkers, may further be
indicative of the
prognosis of the brain injury and number of symptoms resulting the brain
injury, such
changes comprising an up-regulation, a down-regulation, and/or no change of
the
biomarker or combination of biomarkers relative to a threshold reference
value.
[0099] As may be appreciated, the present apparatus and
methodologies of
detecting whether an individual is likely to have an injury may be used to
assess the
risk that the injury might lead to long-term dysfunction, enabling a
determination of
therapeutic intervention at an early stage. For example, in some embodiments,
the
present apparatus and methodologies might be used to detect/diagnose a brain
injury,
such as an SRC, and then to further assess when it might be suitable for the
individual
(e.g., a professional athlete, amateur athlete, or recreational player) to
return to play
the sport, to return to work, or to other daily activities following the
injury.
[0100] According to embodiments, it is contemplated that an
effective
diagnostic tool for brain injury, including SRC, may be based upon a group of
several
biomarkers in order to generate a biomarker signature. For example, the ROC
analysis shown in FIG. 5 illustrates that the combination of at least 18
target
biomarkers (metabolites) identified in Table 3 serves as a classifier of SRC
in urine.
Without being limited to theory, the potentially relevant metabolic pathways,
and their
role in brain injury (e.g., SRC), are discussed below.
[0101] Phenylalanine levels are known to be upregulated in
brain tissue in a
mouse concussion model, but are also known to decrease in the serum of
individuals
who suffered a traumatic brain injury (TB I) with cognitive impairments or to
not change
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
in serum after moderate-severe TBI and SRC in humans. Phenylalanine is an
essential amino acid that is metabolized to tyrosine by the biopterin-
dependent
aromatic amino acid hydroxylase (AAAH) tyrosine hydroxylase. Tyrosine is
further
metabolized to L-DOPA which in turn is metabolized to the neurotransmitter
dopamine
(DA) by DOPA decarboxylase DA is converted by dopamine-13-hydroxylase into
noradrenaline (NA). DA and NA may be responsible for decreased mood among
concussion patients, as these molecules have a significant role in mood and
depression.
[0102] According to embodiments, phenylalanine was found to be
upregulated
following brain injury (e.g., SRC), and phenylalanine biosynthesis and
metabolism
topology analysis was significantly affected. It is possible that an increased
concentration of phenylalanine in brain injured individuals may have a
downstream
effect on DA and NA levels. In turn, these changes may influence symptoms
following
injury such as fatigue, decrease mood and anxiety.
[0103] Citrate is a tricarboxylic acid (TCA) cycle
intermediate involved in
converting phosphoenolpyruvate to malate and/or pyruvate, eventually producing
15
ATP for every molecule of pyruvate. Citrate chelation is involved in
peroxidation rates
and thus oxidative stress, which has been associated with many of the
pathophysiological changes after concussion. Because concentration changes in
various metabolites are implicated in oxidative stress, the present apparatus
and
methodologies may provide a means for better understanding the
pathophysiological
mechanisms leading to brain injury-induced neurological symptoms.
41
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0104] According to embodiments, citrate levels were found to
be
downregulated following brain injury (e.g., SRC) in both VIAVC and paired t-
test, and
citrate was the fourth most important feature in the VIP scores plot. Citrate
levels are
known to be reduced in moderate-severe traumatic brain injured patients,
resulting in
citrate and six other metabolites (pyroglutamic acid, serine, phenylalanine,
galactose,
palmitic acid, 2,3,4-trihydroxybutyrate, linoleic acid, and arachidonic acid)
potentially
serving as a potential biomarker panel for diagnosing moderate-severe
traumatic brain
injury in patients with cognitive impairments.
[0105] According to embodiments, both phenylalanine and
citrate, along with
the other metabolites presented in Table 3 that have a VIAVC p-value of <0.05,
may
provide an accurate, non-invasive, objective method of determining whether an
individual is likely to have a brain injury (e.g., SRC), as indicated by the
ROC curve
and its corresponding AUC (FIG. 5).
[0106] According to embodiments, 3-Indoxysulfate (3-IS) was
found to be
upregulated following brain injury (e.g., SRC), with 3-IS being the feature
found to
have the second highest VIP score. 3-IS is a metabolic product of indole,
which is
itself a product of the amino acid tryptophan. Elevated levels of 3-IS are
known to
increase the levels of reactive oxygen species and induce oxidative stress,
which may
be associated with many of the pathophysiological changes following brain
injury (e.g.,
SRC).
[0107] According to embodiments, the present apparatus and
methods also
detected that several additional target biomarkers (metabolites) were altered
following
brain injury, suggesting such biomarkers may be involved in nervous system
42
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
impairment. For example, propylene glycol was identified by VIAVC analysis and
had
the fifth highest score in the VIP plot. In some embodiments, propylene glycol
levels
were found to be downregulated following brain injury (e.g., SRC).
[0108] According to embodiments, target biomarkers anserine
and carnosine
were both found to be increased following brain injury by the paired t-test.
Carnitine
and acetylcarnitine were also found to be increased by VIAVC. Carnosine is
highly
concentrated in both muscle and brain tissues and has been implicated in
suppressing
biological processes in Alzheimer's disease, cardiovascular ischemic damage,
and
inflammatory diseases. Carnosine may also be a neuroprotective agent for
ischemic
stroke, which could also relate to similar effects following brain injury
(e.g., SRC).
Acetylcarnitine has been shown to possess a neuroprotective ability for
cerebral
ischemia and has been studied for therapeutic purposes in patients with
Alzheimer's
disease, as well as for chronic fatigue syndrome. It is contemplated that such
target
biomarkers (metabolites) could be increased following brain injury as a
potential
protective mechanism following injury.
[0109] According to embodiments, target biomarkers 1-
methylhistidine and 3-
methylhistidine were found to be decreased following brain injury (e.g., SRC).
3-
methylhistidine is primarily a measure of protein breakdown in muscle, with
elevated
levels being related to increased fatigue in individuals (e.g., athletes).
Although 1-
methylhistdine is primarily derived from dietary anserine, high levels tend to
inhibit the
enzyme carnosinase, which increases anserine levels and affects the metabolism
of
carnosine. Reduced levels of serum carnosinase have been found in patients
with
multiple sclerosis, stroke, and Parkinson's disease. Decreased levels of 1-
43
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
methylhistidine in biological samples could be related to carnosinase activity
following
brain injury, which may also explain why carnosine and anserine were found to
be
significantly altered.
[0110] According to embodiments, target biomarkers
phosphorylcholine and 2-
hydroxybutyrate were also altered post-brain injury, with phosphorylcholine
levels
being found to be upregulated and 2-hydroxybutyrate being downregulated.
Phosphorylcholine levels in brain tissue are known to change in a fluid
percussion rat
model of head injury, where choline-containing metabolites that are associated
with
the plasma membrane are reduced in cortex and hippocampus brain tissue (e.g.,
urinary). 2-hydroxybutyrate is also significantly decreased in human serum
when
compared to healthy controls and was positively correlated with the severity
of head
injury. It is understood that urinary excretion of 2-hydroxybutyrate can
reflect shifts in
the rate of glutathione synthesis, and therefore oxidative stress.
[0111] According to embodiments, urinary levels of target
biomarkers tyrosine,
methionine and 3,4-Dihydroxybenzeneacetic acid were found to be upregulated
following brain injury. Urinary levels of tyrosine are known to be altered in
patients
diagnosed with major depressive disorder, a symptom that often occurs
following head
injury. As mentioned above, DA and NA are both involved in mood regulation and
depression, and tyrosine is a precursor metabolite required to produce these
two
neurotransmitters. Urinary levels of methionine are known to be upregulated in
mouse
models of Alzheimer's disease (AD), while plasma levels are known to be
decreased
in patients with traumatic brain injuries. Finally, urinary levels of 3,4-
dihydroxybenzeneacetic acid (DOPAC) are known to be upregulated following SRC.
44
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
DA is broken down into 3,4-dihydroxyphenylacetaldehyde (DOPAL), which is then
metabolized primarily to DOPAC, this transformation being relevant as DOPAL
has
neurotoxic actions and related to oxidative stress in Parkinson's disease.
Upregulated
DOPAC may indicate changes to dopamine metabolism caused by SRC.
[0112] According to embodiments, target biomarkers leucine,
isoleucine, and
valine were found to be altered post-brain injury (e.g., SRC), each metabolite
being
branched chain amino acids (BCAA) which are particularly involved in stress,
energy,
and muscle metabolism. Plasma BCAA levels can be significantly decreased in
TBI
patients. In some embodiments, leucine and isoleucine were found to be
upregulated
post-brain injury (e.g., SRC), with the pathway topology analysis supporting
that that
the degradation and biosynthesis of BCAA were affected, along with aminoacyl-
tRNA
biosynthesis (FIG. 6). BCAA are transported across the blood brain barrier by
the
same, competitive mechanism as aromatic amino acids (AAA) such as
phenylalanine
and tyrosine. As noted above, these AAA were also significantly altered
following brain
injury, and are important to the synthesis and release of the catecholamine
neurotransmitters DA and NA.
[0113] According to embodiments, the present apparatus and
methodologies
may also be used to determine whether an individual is likely to have a brain
injury,
and then to further determine the individual's symptom burden and length of
recovery
(e.g., return to play following SRC). For example, in some embodiments, the
present
methodologies were used to detect a positive correlation between length of
return to
sport and an increased concentration of target biomarker 2-hydroxyisobutyrate.
Without limitation, previous studies in individuals with severe traumatic
brain injury
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
have also shown a correlation between severity of injury and 2-
hydroxybutyrate,
suggesting 2-hydroxybutyrate may be a target metabolite that is altered
following
traumatic brain injury of all severities and may reflect more severe injury
and prolong
recovery.
[0114] According to embodiments, the present apparatus and
methodologies
may also be used to determine whether an individual is likely to have a brain
injury,
and then to further determine the total number of symptoms post-injury. For
example,
in some embodiments, the present methodologies were used to detect a positive
correlation between total number of symptoms and an increased concentration of
lactose. Without limitation, urine lactulose levels are known to be elevated
in patients
with head injury compared to healthy controls and patients with extra-cerebral
injuries,
where the more severe the head injury the higher the urine lactulose, and the
increase
in lactulose potentially reflecting increased catabolism of brain gangliosides
following
injury.
[0115] EXAMPLE 2: According to embodiments, the present
example
demonstrates the use of the present apparatus and methodologies for
determining
whether an individual is likely to have an injury, such as a brain injury.
More
specifically, the present example demonstrates the use of the present
apparatus and
methodologies for the detection and diagnosis of at least one brain injury,
such as a
neurodegenerative brain disorder (e.g., Alzheimer's Disease, "AD"), and
further for
prognosing the at least one brain injury.
[0116] According to embodiments, the present example involved
post-mortem
brain tissue samples collected from individuals or subjects comprised of
donors to the
46
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Calgary Brain Bank (Calgary, Alberta, Canada), with metabolomic changes being
assessed across three regions of interest (ROI) at the end stages of AD
including the
pontine base (PB), dentate nucleus (DN), and associated Brodmann Area BA 24
(part
of the anterior cingulate cortex). Without being limited by theory, although
various
elements of AD pathology have been observed in each of these regions, the
progression of AD in the regions of interest (ROI) are less well studied than
in regions
like the hippocampus and entorhinal cortex, which are known to be drastically
devastated by AD.
[0117] Having regard to Table 4, 11 subjects were categorized
as belonging to
the Alzheimer's Disease group (AD) and 13 were categorized as control group
(CN)
based on three neuropathological scores: amyloid plaque distribution
(indicated with
an A), Break tau stage (indicated with a B), and CERAD score (indicated with a
C;
collectively referred to as an 'ABC' score), with each category being
individually
ranked from one to three, and where the sum of all three categories was
greater than
five, the subject was classified as belonging to the diseased (AD) group. In
some
embodiments, biological samples obtained from control (CN) individuals served
as a
threshold baseline reference value or pre-disease metabolic profile for the at
least one
target biomarker.
[0118] Table 4: AZD patient characteristics
Diagnosis Age Sex Description
AD 89 M AD-A3B2C2; CAA-T3L3C1
AD 77 F AD-A3B3C2
AD 71 M AD-A3B3C3; LBD-M3B6
AD 85 M AD-Al B2C2;
AD 89 F AD-A3B3C3
47
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
AD 87 M AD-A3B2C2
AD 85 F AD-A3B2C1
AD 76 M AD-A3B3C2
AD 84 F LBD-M3B6; AD-A2B202
AD 71 M AD-A3B3C3, LBD-M3B5
AD 90 F AD-A3B3C3; CAA-T2L200
CN 76 F A0B200 PART-B3
CN 73 F AD-A2B1C1
CN 74 M A0B100; PART-B
CN 62 M A1B100
CN 59 M A0B100
CN 43 F A1B0C0
CN 55 M A0B1C0
ON 60 M Normal
CN 74 F A0B2C0
CN 78 M A1B2C0
ON 78 F Normal
[0119] More specifically, description relates to the amyloid
distribution score
(A), Braak tau stage score (B), and CERAD score (C), where each category was
ranked from 1-3. If the sum of the ABC > 5, the individual was categorized as
having
AD. The diagnosis 'normal' indicates that the participant had no history of
cognitive
difficulties. Cerebral amyloid angiopathy (CAA) was ranked via the following
codes:
Thal CAA stage (T) (0-3); Love CAA score (L) (0-3); CAA capillary vasculopathy
(C)
(0-1), Vasculopathy (V) (0-2). Lew Body Disease Neuropathological Changes
(LBD)
were scored via the following codes: McKeith stage (M) (0 - none; 1 -
brainstem; 2-
limbic; 3 - neocortical; 4 - amygdala); Braak stage (B) (0 none; 1 ¨ medulla;
2 ¨ pons;
3¨ nnidbrain; 4¨ transentorhinal and annygdala; 5¨ association neocortex; 6¨
primary
neocortex); Unified staging system for Lewy body disease (U) (0 - none; 1 -
olfactory
bulb only; 2a - brainstem predominant; 2b - limbic predominant; 3 - brainstem
and
limbic; 4 - neocortical). Primary Age-Related Tauopathy (PART) was scored via
Braak
and Braak neurofibrillary tangles stage (B) (0-6). Exclusion criteria included
ABC
48
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
scores equal to or greater than five and the individual exhibited dementia-
related
neuropathology, resulting the number of subjects being narrowed to 22 (n=11
AD,
n=11 CN), where DN had 21 samples (n=10 AD, n=11 CN), PB had 19 samples (n =
8 AD, n = 11 CN), and BA 24 had 18 samples (n= 7 AD, n=11 CN).
[0120] Each of the biological samples (e.g., brain tissues)
obtained from the
subjects were prepared via ultrafiltration to extract water-soluble
metabolites. Briefly,
samples were removed from -80 C storage and allowed to thaw at room
temperature
while on ice until samples were ready to be weighed. Approximately 150 mg of
each
sample, 375 uL of metabolomics buffer, and 150 mg zirconium oxide beads were
added to a centrifuge tube. The metabolomics buffer was a solution of 4:1
K2HPO4 to
KH2PO4 in dH20 at a 0.625M concentration resulting in a final pH of 7.41. The
buffer
also contained 3.75 mM NaN3 to act as an antimicrobial agent. Samples were
then
homogenized using a Bullet Blender (Next Advance, NY, USA) for 1 minute at
intensity
setting 8 and the homogenate was then centrifuged for 5 minutes at 14,000 g.
[0121] Following this, 365 uL of homogenate and 135 uL of
metabolomics
buffer were transferred to an Am icon Ultra 0.5 ml 3K centrifuge filter and
centrifuged
for 30 min at 14,000 g. Centrifuge filters were washed 10 times with Millipore
water
before use to ensure complete glycerol removal from the membrane. Following
filtration, 360 uL of the filtrate, an additional 120 uL of metabolomics
buffer, and 120
uL of D20 with w/v 0.03% trimethylsilyl propanoic acid (TSP) were transferred
to a
new centrifuge tube. The final dH20:D20 ratio was 4:1, resulting in a final
concentration of 0.5M for the buffer salts. The samples were then centrifuged
for 5
49
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
minutes at 12 000 rpm and 550 uL of the supernatant was transferred to a 5mm
NMR
tube.
[0122] NMR spectra were collected on a 700 MHz Bruker Avance
III HD
spectrometer conducting 1024 scans per sample (Bruker, ON, Canada). Each NMR
spectrum was phased using TopSpin (v. 4Ø6.) using the TSP peak (0.00 ppm)
and
water peaks (4.95 ppm) were used as a reference for chemical shift. The
spectra were
exported to MATLAB (The MathWorks, MA, USA) to undergo further processing and
statistical analysis. Spectral binning was done using the dynamic adaptive
binning
algorithm followed by manual inspection and correction for any errors. The
binned
spectra were then pareto scaled, normalized to the total unit area of all bins
(excluding
the water peak), and log transformed prior to carrying out statistical
analysis.
[0123] All univariate and multivariate testing was carried out
using MATLAB
(The MathWorks, MA, USA) and MetaboanalystR (v 2.0), respectively, and the
decision tree algorithm was applied to determine which univariate statistical
tests was
appropriate for the data in each comparison. In all cases, a Mann-Whitney U
Test
(MW) was used and bins with a p-value < 0.05 were considered significant.
Bonferroni-
Holm correction was also applied to account for multiple comparisons.
Supervised
multivariate data analyses were carried out, including, variable importance
analysis
based on random variable combination (VIAVC), orthogonal projections to latent
structures discriminant analysis (OPLS-DA), receiver operator characteristic
(ROC)
curves, and predictive accuracy. Multivariate modelling was initially carried
out on all
bins and subsequently carried out for the bins determined to be significant by
MW or
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
VIAVC statistical testing. In the case of the ROC curve and predictive
accuracy, only
the bins determined to be significant by VIAVC testing were used.
[0124] Identification of at least one biomarker or metabolite
was carried out for
the bins determined to be significant by either the MW or VIAVC tests using
Chenomx
8.2 NMR Suite (Chenomx Inc., Edmonton, Alberta, Canada). Target biomarkers
were
further verified using the Human Metabolite Database and only metabolites
previously
observed in brain tissue, cerebrospinal fluid (CSF), or blood were used.
Pathway
topology analysis were carried out in Metaboanalyst (v 4.0) using the Homo
sapien
KEGG pathway library (v Oct.2019). The hypergeometric test was selected for
over-
representation analysis and relative-betweenness centrality was selected for
pathway
topology analysis.
[0125] The OPLS-DA statistical models produced from MW and
VIAVC Best
Subset bins were used to visualize class separation between bins for DN and BA
24.
Due to the small number of VIAC Best Subset bins (2 bins) for PB, an OPLS-DA
was
constructed from MW and VIAVC F-Ranked bins. OPLS-DA models were used to
remain consistent across a larger study.
[0126] The OPLS-DA models were used to visualize the
supervised separation
of data based on statistical differences between bins. The OPLS-DA p-values
from
further permutation (Q2 and R2Y) testing indicated that separation was not
biased
due to overfitting (p<0.05), with the exception of the results from BA 24. The
Q2 value
was insignificant for that region and failed permutation testing, indicating
poor model
quality. Though these multivariate analyses failed for this region, there were
still
51
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
significant and real differences between bins based on MW and VIAVC Best
Subset
tests.
[0127] Having regard to FIGS. 7A ¨ 7C, the VIAVC Best Subset was used for
each region to construct ROC curves for pontine base (PB; FIG. 7A); for the
dentate
nucleus (DN; FIG. 7B); and for part of the anterior cortex (BA 24; FIG. 7C).
Table 5
provides the predictive accuracy, the area under the curve (AUC), number of
bins
used in the model, and metabolites corresponding to those bins for the ROC
models
can be seen. For each region, the significant (p<0.05) MW and VIAVC Best
Subset
bins were used for metabolite identification.
[0128] Table 5: Summary of the number of VIAVC Best Subset bins used to
build the ROC for each brain region and the predicative accuracy, 95%
confidence
interval, AUC, and metabolites for each ROC.
Region Number Of Predictive 95% AUC Corresponding
metabolites
Bins Accuracy Confidence
Interval
PB 2 92.7% 0.901-1 0.993 Citrate, L-Isoleucine
DN 7 81% 0.75-1 0.915 Serine/Glycyl-glycine,
Trimethylamine N-oxide,
Methylguanidine, Succinic acid,
(R)-3-Hydroxybutyric acid,
Unidentified singlet at 1.15 ppm,
Unidentified doublet at 2.5275
PPm
BA 24 3 94.9% 0.957-1 0.997 2-Hydroxy-3-
methylvalerate,
gamma-Aminobutyric acid,
unidentified multiplot at 3.1425
PPm
[0129] Overall, 26 metabolites were identified from 53 bins for the PB
region,
27 metabolites were identified from 60 bins for the DN region, and 24
metabolites were
52
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
identified from 61 bins for the BA 24 region, as identified from the MW and
VIAVC
Best Subset bins.
[0130] Table 6: Summary of MW and VIAVC Best Subset
significant bins for
each brain region with the corresponding number of at least one target
biomarker
metabolite. Q2 and R2Y (and respective p- values) from the OPLS-DA built from
the
significant bins are also reported. For PB these values are from the OPLS-DA
constructed from the MW and VIAVC F-Ranked bins.
Reg ion Number Of Corresponding Q2 Value R2Y
Value
Significant Bins Number Of
Out Of Total Bins Identified
Metabolites
PB 53/299 26 0.624 (p<5e-04)
0.789 (p<5e-04)
DN 63/355 27 0.512 (p=5e-04)
0.749 (p=0.004)
BA 24 61/365 24 0.00601 0.727
(p=0.238)
(p=0.0375)
[0131] Having regard to Table 7, at least one target biomarker
change shared
across all three regions include citrate (citric acid) (upregulated in all
three regions),
glycyl-glycine (downregulated in all three regions), and L-isoleucine
(downregulated
in all three regions. A positive sign indicates upregulation of the target
biomarker in
the AD group compared to CN, while a negative sign indicates a downregulation
of
the target biomarker between the AD and CN groups.
[0132] Table 7: Target Biomarkers/Metabolites Shared Across
Regions
Regulation per region
Regions PB DN BA
24
All regions Citrate (-F) Citrate (+)
Citrate (-F)
Glycyl-g lycine (-) Glycyl-g lycine (-)
Glycyl-g lycine (-)
L-isoleucine (-) L-isoleucine (-) L-
isoleucine (-)
PB & DN Glutamate (+) Glutamate (-)
Trimethylamine N-oxide (-) Trimethylamine N-oxide
(-0
PB & BA 24 Choline (-F)
Choline (-F)
53
CA 03214708 2023- 10-5
WO 2022/213201 PCT/CA2022/050533
Glucose (-)
Glucose (-)
L-Ieucine (-) L-
Ieucine (-)
L-phenylalanine (-) L-
phenylalanine (-)
L-valine (-) L-
valine (-)
DN & BA 24 Beta ine (-) Betaine (-)
Ethanol (+)
Ethanol (+)
Malonic acid/malonate
Malonic
(-0
acid/malonate (+)
[0133] For PB, the metabolites identified in the three most
significant bins by
MW p-value were L-leucine (p=1.06x10-4), pyruvatoxine (p=3.18x10-4) and L-
phenylalanine (p=1.19x10-3).
[0134] For DN, two bins sharing alpha-ketoisovaleric acid and
propylene
glycerol were identified in the first and third most significant bins
(p=1.013x10-3 and
p=1.29x10-3). The second most significant bin contained 2-oxohexane (p=1.29x10-
3).
[0135] For BA 24, the metabolites identified in the three most
significant bins
according to MW-p-value were: gamma-aminobutyric acid (GABA; p=1.19x10-3) 2-
hydroxy-3-methylvaelrate (p=1 .77x10-3), and N-acetyl-L-aspartate (p=2.54x10-
3).
Table 5 provides a list of metabolites from the VIAVC Best Subset that
correspond to
bins used for the construction of ROC curves.
[0136] Pathway topology analysis was carried out for each
brain region with a
p-value less than 0.05, as shown in FIGS. 8A ¨ 8C (with corresponding data
shown in
Tables 8 ¨ 10). Note regulation of each metabolites is shown in brackets
beside its
name, down regulation in the AD group compared to controls is indicated by
negative
sign and positive sign indicates upregulation.
54
CA 03214708 2023- 10-5
WO 2022/213201 PCT/CA2022/050533
[0137] Table 8: biochemical pathways identified from Pathway
Topology
Analysis via Metaboanalyst from MW and VIAVC Best Subset bins for PB, with
corresponding metabolites, p-value, and impact score.
Metabolites Raw p
Impact
1. Aminoacyl-tRNA L-
Phenylalanine (-), L-Valine (-), L- 7.28E-05 0
biosynthesis Isoleucine (-), L-Leucine (-), L-Tyrosine (-),
L-Glutamate (+)
2. Valine, leucine and L-
Leucine (-), L-Isoleucine (-), L-Valine (-) 0.00019708 0
isoleucine biosynthesis
3.
Phenylalanine, tyrosine L-Phenylalanine (-), L-Tyrosine (-) 0.0014699
1
and tryptophan
biosynthesis
4. Galactose metabolism D-Galactose (-), Glycerol (-F), myo-Inositol
0.0084019 0.05288
(+)
5. Phenylalanine L-Phenylalanine (-), L-Tyrosine (-) 0.010387
0.35714
metabolism
6. Valine, leucine and L-Valine
(-), L-Isoleucine (-), L-Leucine (-) 0.024713 0
isoleucine degradation
[0138] Table 9: Biochemical pathways identified from Pathway
Topology
Analysis via Metaboanalyst from MW and VIAVC Best Subset bins for DN, with
corresponding metabolites, p-value, and impact score.
Metabolites Raw p
Impact
1. Valine, leucine and L-Threonine (-
), 3-Methyl-2- 0.00017385 0
isoleucine oxobutanoic acid (+), L-Isoleucine (-)
biosynthesis
2. 2. Butanoate (R)-3-
Hydroxybutanoate (+), L- 0.001315 0
metabolism Glutamate (-), Succinate (+)
3. Alanine, aspartate and L-Glutamate (-), Citrate (+),
0.0082928 0.19712
glutamate metabolism Succinate (-F)
4.
Glyoxylate and Glycolate (-), Citrate (+), L-Glutamate 0.012055 0.11112
dicarboxylate (-)
metabolism
5. Glycine, serine and Betaine (-), Threonine (-), Creatine (-)
0.013126 0.05034
threonine metabolism
6. Aminoacyl-tRNA L-Isoleucine
(-), L-Threonine (-), L- 0.035735 0
biosynthesis Glutamate (-)
7. Citrate cycle (TCA Succinate
(+), Citrate (+) 0.036843 0.12311
cycle)
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0139]
Table 10: Biochemical pathways identified from Pathway Topology
Analysis via Metaboanalyst from MW and VIAVC Best Subset bins for BA 24, with
corresponding metabolites, p-value, and impact score.
Metabolites Raw p
Impact
1. Valine, leucine
and L-Leucine (-), L- 0.00017385 0
isoleucine Isoleucine (-), L-
biosynthesis Valine (-)
2. Aminoacyl-tRNA L-Phenylalanine
0.00062947 0
biosynthesis (-), L-
Methionine (-),
L-Valine (-), L-
Isoleucine (-), L-
Leucine (-)
3. Alanine,
aspartate and N-Acetyl-L- 0.0082928 0.17308
glutamate metabolism aspartate (+), 4-
Aminobutanoate
(-F), Citrate (+)
4. Glycerophospholipid Choline
0.016662 0.03519
metabolism phosphate (-),
Choline (+),
Acetylcholine (-)
5. Valine, leucine
and L-Valine (-), L- 0.022138 0
isoleucine degradation Isoleucine (-), L-
Leucine (-)
[0140]
Significant (p<0.05) biochemical pathways common to all three regions
were valine, leucine and isoleucine biosynthesis and am inoacyl-tRNA
biosynthesis.
BA 24 and DN shared the pathway alanine, aspartate and glutamate metabolism.
BA
24 and PB shared valine, leucine and isoleucine degradation. DN and PB did not
share
any similar pathways. The three most significant pathways for DN by p-value
were
valine, leucine and isoleucine biosynthesis, butanoate metabolism, and
alanine,
aspartate and glutamate metabolism. The three most significant pathways for PB
were
aminoacyl-tRNA biosynthesis, valine leucine and isoleucine biosynthesis, and
phenylalanine, tyrosine and tryptophan biosynthesis. The three most
significant
56
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
pathways for BA 24 were valine, leucine and isoleucine biosynthesis, aminoacyl-
tRNA
biosynthesis, and alanine aspartate and glutamate metabolism.
[0141] The present example demonstrates alterations in at
least one clinical
biomarkers found in at least one biological sample of an individual that
suffered an
central or peripheral nervous system injury, such as a neurodegenerative
disease or
disorder. Several of the at least one target biomarkers indicative of injury
processes
showed characteristic alterations following injury.
[0142] In some embodiments, the present apparatus and
methodologies
provide for the detection of target AD-related metabolomic alterations in
brain regions
of interest including, without limitation, the pontine base (PB), the dentate
nucleus
(DN), and areas of the anterior cingulate cortex (BA 24).
[0143] In some embodiments, a change in at least one target
biomarker
concentration level was detected across all regions of interest, such target
biomarkers
including, without limitation, citrate and L-isoleucine. For example, an
increase in the
concentration level of citrate was detected, suggesting an up-regulation of
the
biomarker compared to a threshold baseline value, and a decrease in the
concentration level of L-isoleucine was detected, suggesting a down-regulation
of the
biomarker compared to its threshold baseline value.
[0144] In some embodiments, a change in at least one target
biomarker
concentration level was detected, such at least one target biomarkers
including,
without limitation, L-leucine, L-valine, and choline. For example, a change in
each of
the foregoing at least one target biomarker levels were detected in both the
BA 24 and
PB regions of interest.
57
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0145] In some embodiments, a change in at least one target
biomarker
concentration level was detected, such at least one target biomarkers
including,
without limitation, betaine. For example, a change in the foregoing at least
one target
biomarker level was detected in both the BA 24 and DN regions of interest.
[0146] In some embodiments, a change in at least one target
biomarker
concentration level was detected, such at least one target biomarkers
including,
without limitation, glutamate. For example, a change in the foregoing at least
one
target biomarker level was detected in both the DN and PB regions of interest.
[0147] According to embodiments, without being limited to
theory, the present
example demonstrates that various altered target biomarkers may be indicative
or
changes in important biochemical pathways. For example, changes in valine,
leucine
and isoleucine biosynthesis, and am inoacyl-tRNA biosynthesis were significant
in all
the regions of interest. In BA 24 and DN, alanine, aspartate and glutamate
metabolism
was identified. Lastly, in BA 24 and PB, valine, leucine and isoleucine
degradation
was a significant pathway. Together these altered target biomarkers and
pathways
can indicate impairment in protein synthesis, neurotransmission, inflammation,
and
energy metabolism, thereby indicating the presence (diagnosis) and severity
(prognosis) of an injury.
[0148] In all three ROI, am inoacyl-tRNA biosynthesis was
identified as a
significant pathway (p<0.05; Table 5-7). Altered am inoacyl-tRNA biosynthesis
in the
cytoplasm has been identified in AD patients plasma, CSF, and blood. Aminoacyl-
tRNA biosynthesis involves esterifying an amino acid with its matching tRNA
molecule
(corresponding with the correct anticodon triplet of the matching amino acid)
via
58
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
aminoacyl-tRNA synthetases. This process is essential for the synthesis of
proteins,
as the now 'activated' tRNA molecule (has the amino acid attached) brings the
amino
acid to the ribosome during protein synthesis.
[0149] Without being limited to theory, it is unclear from the
results if aminoacyl-
tRNA biosynthesis is indirectly affected by alteration of amino acid levels or
if AD
pathology directly affects this biochemical pathway. However, it could be that
the
downregulation of the amino acids in these brain regions would affect their
availability
for protein synthesis. This change could impair protein synthesis in these
regions.
Conversely, AD may directly affect elements of protein synthesis. For example,
ribosome dysfunction has been observed in the inferior parietal lobe (BA 40),
superior
middle temporal gyri (BA22) but not in the cerebellum in the post-mortem brain
tissue
of AD compared to controls. Therefore, it appears that an impairment in
protein
synthesis was only indicated in the former two regions and not the latter. If
indeed
there are no alterations in ribosome function in the cerebellum, then in DN,
at least
the alterations in aminoacyl-tRNA biosynthesis could be due to upstream
alterations
in amino acid availability.
[0150] Alterations in brain chain amino acids (BCAAs; valine,
leucine and
isoleucine) were observed in the three ROI. For BA 24 and PB, all three BCAAs
were
downregulated. For DN, only L-isoleucine was down-regulated, while 3-methy1-2-
oxobutanoic acid (alpha-ketoisovaleric acid/a-KIVA) was upregulated. A
reduction of
BCAA has been seen in AD patients' brain tissue, CSF, and blood. Indeed, the
identification of reduced BCAA in this study's brain regions supports it as a
potentially
stereotypical future of AD pathology. For DN, alterations of a-KIVA in the AD
group
59
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
compared to controls indicate incomplete valine breakdown; specifically, it is
produced
via the second step of valine degradation via branch-chain amino transaminase
¨1.
Interestingly, BCAA degradation was not identified as a significant or
impacted
pathway for this brain region by MetaboAnalyst.
[0151] Additionally, valine, leucine and isoleucine
biosynthesis was identified in
all three brain regions. This identification is a physiological impossibility
as the human
body cannot synthesize the BCAAs. Regarding BA 24 and PB, though BCAA
biosynthesis does not occur, other processes could be affected. Indeed, for
both these
regions, valine, leucine and isoleucine degradation was a significant pathway,
with the
same metabolites altered in valine, leucine and isoleucine biosynthesis.
Therefore,
the error could be on the part of our software, MetaboAnalyst, attributing
changes in
BCAA degradation to BCAA biosynthesis even though this cannot occur in humans.
[0152] The degradation of branch chain amino acids is the
first step in
producing glutamate in astrocytes. In PB, there was an increase in glutamate
in the
AD group compared to controls. Potentially, this indicates that the reduction
of BCAA
in this region is due to an increase in glutamate synthesis. However, further
study is
needed to verify if the downregulation of BCAA in PB leads to the upregulation
of
glutamate via glutamate production (including analysis of enzyme regulation
and
expression). Unlike PB, glutamate was reduced in DN. This different change in
glutamate in DN could suggest that in this regions, a mechanism other than
glutamate
production leads to the downregulation of BCAAs in those brain regions.
[0153] Regarding glutamate metabolism, the biochemical pathway
alanine,
aspartate and glutamate metabolism was identified as significant for DN and BA
24.
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
However, the identified metabolites involved in these pathways differed
between the
two regions. This difference indicates that potentially different elements of
this
biochemical pathway are impacted by AD. For DN, these identified metabolites
were
citrate and succinate (both upregulated) and glutamate (downregulated). The
alterations in the former two metabolites and glutamate could suggest
alterations in
energy metabolism, particularly the citrate cycle, and highlights glutamate's
role in
energy metabolism rather than neurotransmission to be affected by AD in this
region.
[0154] In the cortical and hippocampal regions, damage to
glutamatergic
neurons has been observed as part of AD pathology. Part of this damage in the
HPC
is related to increased glutamate synthesis resulting in excitotoxicity.
Therefore, it is
surprising that in DN, there is a reduction of glutamate in the AD group
compared to
controls. However, a similar reduction of glutamate concentration has been
found in
the posterior cingulate cortex of AD patients. As well, lower concentrations
of
glutamate and GABA have been associated with ageing in other brain regions.
Therefore, the reduction of glutamate could be a region-specific AD-related
occurrence in DN (as well as for the posterior cingulate cortex), or the
regulation
reflects normal ageing processes and therefore may not be altered by AD.
[0155] For BA 24, N-acetyl-aspartate (NAA), GABA, and citrate
were identified
and all upregulated. Though all three of these metabolites are found within
alanine,
aspartate and glutamate metabolism, they are not directly connected.
Therefore, it is
unlikely that the upregulation of one metabolite is connected to the
upregulation of
another metabolite. The upregulation of NAA found in BA 24 is unexpected as a
reduction in NAA is a typical biomarker for AD. Regarding the upregulation of
GABA
61
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
in BA 24, the same result was found in BA 22, BA 17, and BA 40 of the AD
cohort in
this study. This change in regulation could be part of a broader trend for
cerebral
tissues; however, more research is needed.
[0156] Metabolism of another neurotransmitter, acetylcholine,
may have been
altered in BA 24. Glycerophospholipid metabolism was identified as a
significant
pathway, and the metabolites identified within this pathway (as well as their
regulations) indicate that acetylcholine synthesis is impaired. It is
important to note
that BA 24 is densely innervated by the cholinergic system. Choline is formed
from
choline phosphate within acetylcholine synthesis via choline kinase alpha.
From
choline, acetylcholine is produced via the enzyme choline 0-acetyltransferase
(ChAT). Within BA 24, choline phosphate was down-regulated, choline was
upregulated, and acetylcholine was downregulated in the AD group compared to
controls. In the AD group, this could indicate an increased synthesis of
choline
synthesis from choline phosphate. However, within this group, less
acetylcholine is
being formed from choline, indicating impairment of the enzyme ChAT. ChAT and
acetylcholinesterase (degrade acetylcholine into choline) have been observed
to be
reduced in post-mortem brain tissue of end-stage AD patients. The regulation
of
choline and acetylcholine in BA 24 indicates impairment of ChAT, not
acetylcholinesterase (i.e., if acetylcholine was upregulated and choline
downregulated, then acetylcholinesterase would be the enzyme of concern).
However, enzyme analysis would clarify which enzymes could be affected leading
to
these alterations. Lastly, it is unclear what is causing the initial
downregulation of
choline phosphate in this area. Analysis with other spectroscopy may reveal if
other
62
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
metabolites may be affected within this pathway, as lipid-based metabolites
were
removed from samples in preparation.
[0157] Betaine can be synthesized from choline or derived from
diet (. In DN
and PB, betaine was both downregulated. The primary role of betaine is as an
osmoprotectant. However, it also has a role in potentially preventing
inflammation.
Potentially, the reduction of betaine in these regions may perpetuate
inflammatory AD
pathology. Additionally, supplementation of betaine in the diet has been
proposed to
mitigate inflammatory processes and suppress the production of amyloid-beta
plaques and phosphorylation of tau. Both inflammation and impaired energy
metabolism are mechanisms of AD pathology.
[0158] Alterations in citrate in all three regions indicate
the possibility of
impaired energy metabolism, specifically the citric acid (TCA cycle).
Additionally,
citrate was a biomarker in the region PB as it was one of the VIAVC Best
Subset bins
(see Table 3). However, this pathway was only identified as significant for
DN. Both
citrate and succinate were upregulated in the AD group compared to controls
for this
region. It is unclear how this pathway could be affected as no metabolites
between
citrate and succinate within the cycle were identified as altered. Both age
and AD-
related degeneration of mitochondria via oxidative stress affect oxidative
phosphorylation. However, it is unclear if this contributed to the potential
dysregulation
observed presently.
[0159] Within BA 24, there was a down-regulation of glucose in
the AD group
compared to controls. This finding supports previous evidence of reduced
glucose
metabolism in the cingulate gyrus, hippocampus, and superior and medial
temporal
63
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
gyri. Dysregulation of glucose is a feature of AD pathology. Glucose
hypometabolism
has been found in the brain tissue of AD patients in the posterior cingulate
but not the
cerebellum. Hypometabolism of glucose (as measured with neuroimaging) is a
biomarker of AD. However, it is hypothesized that A13 resistant cells in the
AD brain
survive due to increased glucose metabolism via increased use of the anaerobic
glycolysis and pentose shunt, which is related to the upregulation of
antioxidant
mechanisms in these surviving cells. These mechanisms could lead to a
reduction of
glucose. Though the brain tissue of the AD patients could have been at various
stages
of decline, it should be noted that the various cells had survived up to the
individual's
death. Therefore, one can assume that at least some neurons were resistant to
oxidative stress induced by A13. Additionally, areas less resistant (higher
cortical
regions) to AD pathology have been shown to have higher concentrations of
glucose
compared to controls. Therefore, more resistant areas may have lower
concentrations
of glucose. However, impaired cerebral glucose is a feature of AD pathology.
Therefore, it is vital to keep in mind that the reduction of glucose in this
region could
be due to less uptake of glucose within this region and not due to a survival
mechanism. Enzymatic analysis and transporter analysis may elucidate the
mechanism behind this result.
[0160] EXAMPLE 3: According to embodiments, the present
example further
demonstrates the use of the present apparatus and methodologies for
determining
whether an individual is likely to have an injury, such as a brain injury.
More
specifically, the present example demonstrates the use of the present
apparatus and
methodologies for the detection and diagnosis of at least one brain injury,
such as a
64
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
neurodegenerative brain disorder (e.g., Alzheimer's Disease, "AD"), and
further for
prognosing the at least one brain injury.
[0161] According to embodiments, the present example involved
post-mortem
brain samples collected from individuals or subjects comprised of donors to
the
Calgary Brain Bank (Calgary, AB, Canada), with metabolic changes being
assessed
across three regions of interest (ROI) including in associative Brodmann Area
(BA) 22
(superior temporal gyrus) and BA 40 (supramarginal gyrus) and are less common
in
the primary sensory area BA 17 (primary visual cortex).
[0162] Having regard to Table 11, 11 subjects were categorized
as belonging
to the Alzheimer's Disease group (AD) and 11 were categorized as control (CN)
with
the number of samples per region being 22 from BA 22 (n= 11 AD, n= 11
controls),
21 samples from BA 17 (n=10 AD, n=11 controls), and 16 samples from BA 40 (n=
7
AD, n=9 controls). Subjects were categorized based on three neuropathological
scores: amyloid plaque distribution (indicated with an A), Braak tau stage
(indicated
with a B), and CERAD score (indicated with a C; collectively referred to as an
`ABC'
score), with each category being individually ranked on a scale from zero to
three,
with three being most deviated from typical non-diseased characteristics. If
the
summation of each of these rankings was greater than five, the tissue was
classified
as having AD. Samples were collected and scored by an experienced
neuropathologist and stored at -80 C until sample preparation.
[0163] Table 11: AZD patient characteristics
DIAGNOSIS AGE SEX ABC SCORE
AD 89 M A3B2C2
AD 77 F A3B3C2
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
AD 71 M A3B3C3
AD 85 M A1B2C2
AD 89 F A3B303
AD 87 M A3B2C2
AD 85 F A3B2C1
AD 76 M A3B3C2
AD 84 F A2B2C2
AD 71 M A3B3C3
AD 90 F A3B303
CN 76 F A0B200
CN 73 F A2B1C1
ON 74 ivi A0B100
ON 62 M A1B1C0
CN 59 M A0B1C0
CN 43 F A1B000
ON 55 M A0B100
CN 60 M Normal
ON 74 F A0B200
ON 78 ivi A1B200
ON 78 F Normal
[0164] Each of the biological samples (e.g., brain tissues)
obtained from the
subjects were prepared via ultrafiltration to extract water-soluble
metabolites. Briefly,
tissue samples were removed from -80 C storage and allowed to thaw at room
temperature while on ice until samples were ready to be weighed. Approximately
150
mg of each sample, 375 uL of metabolomics buffer, and 150 mg of zirconium
oxide
beads were added to a centrifuge tube. The metabolomics buffer was a solution
of 4:1
K2HPO4 to KH2PO4 in dH20 at a 0.625M concentration resulting in a final pH of
7.41.
The buffer also contained 3.75 mM NaN3 to act as an antimicrobial agent.
Samples
were then homogenized using a Bullet Blender (Next Advance, NY, USA) for 1
minute
66
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
at intensity setting 8 and the homogenate was then centrifuged for 5 minutes
at 14,000
g.
[0165] Following this, 365 uL of homogenate and 135 uL of
metabolomics
buffer were transferred to an Am icon Ultra 0.5 ml 3K centrifuge filter and
centrifuged
for 30 min at 14,000 g. Centrifuge filters were washed 10 times with Millipore
water
before use to ensure complete glycerol removal from the membrane. Following
filtration, 360 uL of the filtrate, an additional 120 uL of metabolomics
buffer, and 120
uL of D20 with w/v 0.03% trimethylsilyl propanoic acid (TSP) were transferred
to a
new centrifuge tube. The final dH20:D20 ratio was 4:1, resulting in a final
concentration of 0.5M for the buffer salts. The samples were then centrifuged
for 5
minutes at 12 000 rpm and 550 uL of the supernatant was transferred to a 5mm
NMR
tube.
[0166] NMR spectra were collected on a 700 MHz Bruker Avance
III HD
spectrometer (Bruker, ON, Canada) equipped with a TBO-Z probe was used for NMR
data acquisition. Data was collected using the noesygppr1 d pulse sequence and
the
following acquisition parameters: number of scans (NS) = 1024, mixing time =
10 ms,
spectral window = 20.5 ppm, total number of points (td) = 128k, total
acquisition time
= 4.56 s, transmitter offset (o1p) = 4.7ppm, and recycle delay (d1) = 1
second. NMR
spectra were manually phased using TopSpin (v 4Ø6) and the TSP peak was used
as a reference for chemical shift. The spectra were exported to MATLAB (The
MathWorks, MA, USA) for statistical analysis. Spectra were first binned using
dynamic
adaptive binning, and then manually adjusted to optimize number of variables
and
ensure accuracy. The binned spectra were then pareto scaled, normalized to the
total
67
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
unit area of all bins (excluding the water peak), and log transformed prior to
carrying
out statistical analysis.
[0167] All univariate and multivariate testing was carried out
using MATLAB
(The MathWorks, MA, USA) and MetaboanalystR (v 2.0), respectively, and the
decision tree algorithm was applied to determine which univariate statistical
tests was
appropriate for the data in each comparison. In all cases, a Mann-Whitney U
Test
(MW) was used and bins with a p-value < 0.05 were considered significant.
Bonferroni-
Holm correction was also applied to account for multiple comparisons.
Supervised
multivariate data analyses were carried out, including, variable importance
analysis
based on random variable combination (VIAVC), orthogonal projections to latent
structures discriminant analysis (OPLS-DA), receiver operator characteristic
(ROC)
curves, and predictive accuracy. Multivariate modelling was initially carried
out on all
bins and subsequently carried out for the bins determined to be significant by
MW or
VIAVC statistical testing. In the case of the ROC curve and predictive
accuracy, only
the bins determined to be significant by VIAVC testing were used.
[0168] Identification of at least one target biomarker
(metabolite) was carried
out for the bins determined to be significant by either the MW or VIAVC tests
using
Chenomx 8.2 NMR Suite (Chenomx Inc., Edmonton, Alberta, Canada). Target
biomarkers were further verified using the Human Metabolite Database and only
metabolites previously observed in brain tissue, cerebrospinal fluid (CSF), or
blood
were used. Pathway topology analysis were carried out in Metaboanalyst (v 4.0)
using
the Homo sapien KEGG pathway library (v Oct.2019). The hypergeometric test was
selected for over-representation analysis and relative-betweenness centrality
was
68
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
selected for pathway topology analysis. The binning process resulted in 380,
378, and
364 bins for the BA 22, BA 40, and BA 17 regions, respectively. Out of these
bins,
157, 84, and 110 were identified as significantly altered by either univariate
MW or
multivariate VIAVC testing for the BA 22, BA 40, and BA 17, respectively.
[0169] Having regard to FIGS. 9A ¨ 9C, the OPLS-DA modeling
carried out
using the significantly altered bins provide score plots that showed
separation
between the AD and CN tissues for regions of interest BA 22 (FIG.9A), BA 40
(FIG.
9B), and BA 17 (FIG. 9C). Cross validation of the BA 22 and BA 17 regions
resulted
in a good model quality (BA 22- Q2 = 0.738, p <0.001; BA 17¨ Q2 = 0.736, p
<0.001)
and total variance explained by the model (BA 22 - R2 = 0.867, p < 0.001; BA
17 ¨ R2
= 0.967, p < 0.01), indicating that the separation observed is not a result of
model
overfitting. However, cross validation for the BA 40 region had a poor quality
of the
model (Q2 = 0.173) and failed permutation testing (p = 0.107), indicating this
result
could be due to model over fitting.
[0170] Having regard to FIG. 9D, OPLS-DA modeling of the BA 40
tissues was
carried out using only the bins determined to be significant by VIAVC testing
resulting
in separation of the two groups and passed cross validation and permutation
testing
with a model fit and total variance explained by the model (Q2 = 0.684, p =
0.0015; R2
= 0.834, p = 0.0015) that is similar to the other two regions.
[0171] Having regard to FIG. 10, the bins determined to be
significant by VIAVC
testing were used to construct ROC curves for BA 11 (FIG. 10A), BA 40 (FIG.
10B),
and BA 17 (FIG. 100). These ROC curves show that the target biomarker
metabolites
corresponding to the VIAVC bins for each region of interest lead to almost
perfect
69
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
class separation for all three brain regions as indicated by the high
predictive
accuracies, area under the curve (AUC) values close to or equal to one, and
small
confidence intervals.
[0172] Table 12: Summary of at least one target biomarker
metabolite identified
as being significant by either MW or VIAVC in BA 22.
Metabolite Identification VIAVC p- MW p- PPM PPM End
Regulation
value value Start
FAPy-adenine 1.81E-02 8.372
8.361 43.42236
Niacinamide, Hydrochlorothiazide 1.51E-02 8.27
8.259 38.6102
Unidentified singlet at 8.226 or doublet at 8.221 1.03E-03 8.231
8.221 35.41009
Oxypurinol 2.52E-03 8.221
8.212 24.34414
N-Acetyl-L-aspartate 2.36E-04 7.996
7.969 56.34925
FAPy-adenine 7.10E-03 7.915
7.907 29.50864
Phenylalanine 4.88E-02 7.451
7.44 -24.02654
Phenylalanine 7.10E-03 7.44
7.43 -25.05343
Phenylalanine 3.86E-03 7.43
7.422 -25.61539
Phenylalanine 7.10E-03 7.399
7.39 -33.89216
Phenylalanine 2.52E-03 7.39
7.38 -38.70936
Phenylalanine
2.81E-04 3.04E-04 7.347 7.336 -41.3444
Phenylalanine 3.04E-04 7.336
7.327 -42.57734
Tyrosine 1.26E-02 7.208
7.198 -23.364
Tyrosine 1.04E-02 7.198
7.188 -25.04566
Tyrosine 3.02E-02 6.926
6.919 -21.75922
Tyrosine 8.62E-03 6.911
6.907 -25.00862
Unidentified doublet at 4.529 ppm 2.15E-02 4.533
4.529 24.41176
Unidentified doublet at 4.529 ppm 5.82E-03 4.529
4.525 25.24415
N-Acetyl-L-aspartate 6.39E-04 4.408
4.403 46.89239
N-Acetyl-L-aspartate 1.40E-04 4.403
4.397 51.24052
N-Acetyl-L-aspartate 1.40E-04 4.397
4.388 50.81335
N-Acetyl-L-aspartate 1.40E-04 4.388
4.383 47.14871
N-Acetyl-L-aspartate 1.40E-04 4.383
4.377 48.13157
N-Acetyl-L-aspartate 1.03E-03 4.377
4.372 40.70919
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Malate 1.81E-02 4.313
4.306 17.23831
Malate 4.18E-02 4.302
4.299 26.54689
Malate 7.10E-03 4.299
4.294 25.14747
L-Threonine 3.02E-02 4.271
4.265 14.33152
Unidentified broad peak at 4.198ppm 3.13E-03 4.202
4.194 26.04036
Unidentified multiplet at4.172ppm 4.88E-02 4.191
4.187 16.5787
Unidentified multiplet 4.172ppm 4.88E-02 4.187
4.182 10.66606
L-Serine, Phenylalanine 4.88E-02 4.013
4.008 -17.00443
L-Serine, Phenylalanine 1.29E-03 4.008 4
-16.75821
L-Serine 4.18E-02 3.986
3.977 12.86407
L-Serine, Tyrosine, Glycolic acid 3.56E-02 3.97
3.964 -8.733203
Tyrosine, Creatine phosphate 2.56E-02 3.959
3.952 -13.2977
Tyrosine, Creatine phosphate 1.26E-02 3.952
3.945 -12.68411
Creatine 1.26E-02 3.945
3.932 11.32984
Mannose 1.03E-03 3.932
3.921 -45.62407
Mannose 3.86E-03 3.921
3.918 -43.93848
7-Methylxanthine 2.36E-04 3.918
3.909 -53.70398
Unidentified singlet at 3.906 1.62E-03 3.909
3.903 -26.42733
Unidentified singlet at 3.9005 5.01E-04 3.903
3.898 -31.7006
Unidentified singlet at 3.8945 8.15E-05 3.898
3.891 -50.31197
Betaine 4.75E-03 3.891
3.884 -51.3641
Mannose 1.26E-02 3.884
3.877 -24.90246
Mannose, L-Methionine 3.13E-03 3.877
3.868 -37.83963
Mannose, L-Methionine 3.56E-02 3.868
3.864 -36.55115
Mannose, L-Serine 1.29E-03 3.854
3.85 -31.31593
Mannose, L-Serine 7.10E-03 3.85
3.846 -18.30865
Glycerol, Guanidoacetate, L-Alanine 1.51E-02 3.789
3.785 -16.52483
L-Alanine, Glutamate 1.62E-03 3.782
3.772 -19.00188
Unidentifieded part of a doublet at 3.77 ppm 4.88E-02 3.772
3.768 -8.109975
Acetylcholine 2.15E-02 3.755
3.751 -49.29167
Acetylcholine, L-Leucine 7.10E-03 3.751
3.745 -38.01584
Mannose, L-Leucine 1.51E-02 3.741
3.737 -65.60145
Mannose, L-Leucine 5.82E-03 3.737
3.734 -64.78962
Mannose, L-Leucine 1.51E-02 3.731
3.724 -62.33471
Mannose 1.81E-02 3.696
3.69 -33.81872
Mannose 1.81E-02 3.69
3.685 -26.92146
Mannose 2.52E-03 3.685
3.68 -32.13628
Ethanol, Mannose 1.81E-02 3.68
3.674 -12.95804
Unidentified singlet peak at 3.635 ppm 1.03E-03 3.638
3.632 -24.32827
71
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Unidentified singlet peak at 1.41E-05 8.15E-05 3.623
3.618 -19.48902
3.6205 ppm
Phosphocholine 6.39E-04 3.61
3.605 -29.11
L-Valine, L-Threonine 1.81E-02 3.605
3.601 -17.74673
L-Valine, L-Threonine 7.10E-03 3.598
3.594 -19.16303
Mannose, Phosphocholine 1.51E-02 3.594
3.59 -17.26303
Unidentified singlet at 3.506ppm 2.15E-02 3.508
3.504 -153.5797
Unidentified singlet at 3.285 ppm 3.56E-02 3.33
3.324 15.08362
Unidentified singlet at 4.276ppm 3.13E-03 3.277
3.275 -17.78468
Unidentified singlet at 3.262 1.26E-02 3.266
3.258 -91.98722
Betaine 2.15E-02 3.258
3.251 -30.43985
Unidentified singlet at (singlet shoulder) at 1.04E-02 3.251
3.246 -68.90698
3.2485ppm
1,3,7-Trimethyluric acid 8.62E-03 3.246
3.24 -17.71505
Acetylcholine 3.13E-03 3.24
3.229 -48.11245
Unidentified peak at 3.2165 ppm 1.81E-02 3.219
3.214 19.785
1,9-Dimethyluric acid 1.81E-02 3.214
3.203 56.79816
Dimethyl sulfone 3.13E-03 3.153
3.148 46.02481
Unidentified broad singlet with shoulder at 3.13E-03 3.148
3.138 38.66216
3.143ppm
Malonate 2.03E-03 3.138
3.132 30.06713
Cis-Aconitate 4.88E-02 3.124
3.116 18.53842
Unidentified singlet at 3.0875ppm 4.88E-02 3.092
3.083 12.51542
Unidentified singlet at 3.072 1.81E-02 3.074
3.07 36.6566
Creatine 2.03E-03 3.047
3.033 17.45481
Histamine 2.03E-03 3.02
3.013 27.98211
Histamine 2.36E-04 3.013
3.001 44.90641
Unidentified singlet peak at 2.9965 1.82E-04 3.001
2.992 48.78256
Unidentified singlet peak at 2.765 1.51E-02 2.759
2.753 31.87011
Unidentified singlet peak at 2.7495 3.86E-03 2.753
2.746 34.10191
Unidentified singlet peak at 2.743 3.02E-02 2.746
2.74 25.91736
N-Acetyl-L-aspartate 3.91E-04 2.709
2.702 30.68549
N-Acetyl-L-aspartate 1.07E-04 2.702
2.697 45.04516
Unidentified singlet at 2.694 ppm of a quartet 4.75E-03 2.697
2.691 21.80152
Citric acid 1.81E-02 2.691
2.686 19.28203
N-Acetyl-L-aspartate 1.40E-04 2.686
2.68 45.70028
N-Acetyl-L-aspartate 1.82E-04 2.68
2.672 52.68386
L-Methionine 3.02E-02 2.651
2.64 -19.84409
Unidentified broad singlet 2.567 ppm 1.04E-02 2.57
2.564 33.21796
Unidentified broad singlet at 2.562 ppm 2.56E-02 2.564
2.56 33.67739
Unidentified broad singlet at 2.557 ppm 2.56E-02 2.56
2.554 35.91216
72
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Unidentified broad singlet at 2.552 ppm 2.56E-02 2.554
2.55 37.06089
Citric acid 7.10E-03 2.543
2.538 34.53084
Citric acid 8.62E-03 2.538
2.534 39.89098
N-Acetyl-L-aspartate
2.07E-04 1.07E-04 2.534 2.521 52.04648
N-Acetyl-L-aspartate
1.82E-04 1.07E-04 2.521 2.507 50.80757
N-Acetyl-L-aspartate 6.39E-04 2.507
2.497 37.53952
3-Hydroxymethylglutaric acid, pyruvic acid 1.51E-02 2.483
2.474 -41.41376
3-Hydroxymethylglutaric acid, 4-pyridoxate 1.51E-02 2.474
2.462 -49.00834
Unidentified singlet at 2.456ppm 1.51E-02 2.462
2.45 -47.00065
3-Hydroxymethylglutaric acid 1.51E-02 2.45
2.437 -40.33066
3-Hydroxymethylglutaric acid 1.81E-02 2.437
2.429 -35.39668
Glyoxylic acid, Glutamate 1.51E-02 2.387
2.381 16.87814
Glutamate 2.56E-02 2.379
2.373 17.1487
Glutamate, Pyruvate 7.10E-03 2.373
2.37 26.71487
Glutamate 3.56E-02 2.317
2.313 19.98687
Unidentified peak of a doublet at 2.308 ppm 3.91E-04 2.313
2.308 45.44882
Glutamate 3.86E-03 2.308
2.304 26.65472
Unidentified singlet at 2.2985 1.07E-04 2.302
2.295 51.39959
Unidentified singlet at 2.2885ppm 3.04E-04 2.292
2.285 40.50099
Unidentified peaks at 2.1665ppm 4.88E-02 2.17
2.163 -14.39843
L-Methionine 1.81E-02 2.163
2.157 -27.29842
Unidentified broad peak at 2.155ppm 1.51E-02 2.157
2.153 -26.80173
2-Amino-3-phosphonoprionic acid 4.88E-02 2.153
2.149 -10.37848
Acetylcholine 3.13E-03 2.149
2.137 -20.78833
2-Amino-3-phosphonoprionic acid 4.18E-02 2.137
2.133 -9.960317
Glutamate 1.81E-02 2.064
2.054 18.50034
Glutamate 6.39E-04 2.041
2.034 27.23617
N-Acetyl-L-aspartate 1.82E-04 2.034
2.021 57.502
Gamma-Aminobutyric acid 8.11E-04 1.914
1.901 29.61986
Gamma-Aminobutyric acid 1.29E-03 1.901
1.892 30.92844
L-Leucine 2.56E-02 1.752
1.746 -18.6471
L-Leucine 3.86E-03 1.746
1.738 -23.78011
L-Leucine, 2-Amino-3-phosphonopropionic acid 1.04E-02 1.738
1.729 -23.98428
L-Leucine, 2-Amino-3-phosphonopropionic acid 2.52E-03 1.729
1.722 -25.46223
L-Leucine, 2-Amino-3-phosphonopropionic acid 1.26E-02 1.714
1.707 -21.40546
L-Leucine 7.10E-03 1.707
1.698 -24.27505
L-Leucine 3.02E-02 1.69
1.684 -17.77244
L-Leucine 4.18E-02 1.68
1.674 -11.82181
73
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Unidentified doublet at 1.148 ppm 3.02E-02 1.153
1.147 63.03259
3.02E-02 1.144 1.138 64.86548
L-Valine 2.03E-03 1.055
1.045 -28.71924
L-Valine 1.62E-03 1.045
1.035 -29.60371
L-Isoleucine 1.62E-03 1.022
1.013 -30.71506
L-Isoleucine 3.13E-03 1.013
1.004 -29.07401
L-Valine 5.82E-03 1.004
0.9945 -24.92976
L-Valine 1.26E-02 0.9945
0.9849 -24.65163
L-Leucine 6.39E-04 0.969
0.9592 -33.43016
L-Leucine 6.39E-04 0.9592
0.9477 -33.52599
L-Isoleucine 3.86E-03 0.9477
0.9369 -32.72504
L-Isoleucine 2.15E-02 0.9369
0.9271 -20.87773
Unidentified singlet at 0.08278 ppm 1.81E-02 0.08497
0.08059 38.49853
[0173]
Table 13: Summary of at least one target biomarker metabolite
identified
as being significant by either MW or VIAVC in BA 40.
Metabolite ID VIAVC p- MW p-value PPM
PPM Regulation
value Start End
Phenylalanine 3.11E-02 7.44
7.43 -44.964149
Phenylalanine 1.15E-02 7.43
7.418 -46.465674
Phenylalanine 3.11E-02 7.389
7.378 -42.799754
Phenylalanine 1.15E-02 7.346
7.336 -49.087759
Phenylalanine 1.15E-02 7.336
7.326 -48.925159
Tyrosine 1.15E-02 7.211
7.197 -38.450455
Tyrosine 1.64E-02 7.197
7.185 -35.346518
Histamium/Histamine 4.18E-02 7.149
7.132 -28.600318
Tyrosine 7.87E-03 6.928
6.915 -38.568822
Tyrosine 3.32E-03 6.915
6.903 -39.196092
Glucose 3.11E-02 4.676
4.663 -105.9544
N-Acetyl-L-aspartate 1.64E-02 4.397
4.388 35.1619327
N-Acetyl-L-aspartate 4.18E-02 4.388
4.383 36.08903659
Glycerophosphocholine (sn-Glycero-3- 3.11E-02 4.349
4.318 -46.90306
phosphocholine)
Phosphorylcholine 1.96E-06 4.192
4.187 -6.0339389
2-Hydroxyvalerate 3.11E-02 4.047
4.039 -35.375208
L-Serine, Phenylalanine 2.29E-02 4.022
4.018 -40.694512
L-Serine, Phenylalanine 2.10E-03 4.018
4.008 -45.933932
L-Serine 2.29E-02 4.008
4 -25.566239
74
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Tyrosine, L-Serine, Caffeine 2.29E-02 3.97
3.962 -27.296063
Tyrosine, Glycerophosphocholine, Glycolate 1.64E-02 3.962
3.954 -28.543817
Creatine phosphate, Glycerophosphocholine 2.29E-02 3.954
3.945 -25.59149
Glycerophosphocholine 1.64E-02 3.93
3.918 -57.39949
Aspartate, Syringate, Glucose 2.29E-02 3.918
3.909 -73.926098
Aspartate, Syringate 3.11E-02 3.903
3.898 -39.064346
Aspartate, Betaine, Glucose, 7.87E-03 3.898
3.89 -75.17905
Glycerophosphocholine
Glycerophosphocholine, Homovanillic acicd 1.15E-02 3.89
3.884 -64.091916
Glycerophosphocholine 1.15E-02 3.884
3.877 -39.026185
Glycerophosphocholine, 0-Acetylcarnitine 1.64E-02 3.877
3.868 -55.661784
Glycerophosphocholine, 0-Acetylcarnitine 1.15E-02 3.868
3.859 -45.026752
L-serine, Glucose 2.29E-02 3.859
3.854 -44.895707
L-serine, Glucose, 0-Acetylcarnitine 7.87E-03 3.854
3.846 -45.097877
N-Acetylglycine, Acetylcholine, Glucose 5.24E-03 3.751
3.744 -56.027065
Methylacetoacetic acid, Glucose, Leucine 1.64E-02 3.741
3.734 -57.658134
Methylacetoacetic acid, Glucose, Leucine 1.15E-02 3.731
3.725 -82.7898
Ethanol 7.87E-03 3.674
3.67 23.9305118
0-Acetylcarnitine, Glycerophosphocholine 5.24E-03 3.637
3.633 -44.927639
Glycerophosphocholine, 1.32E-07 3.32E-03 3.622
3.618 -32.790992
Sarcosine
Phosphorylcholine 1.64E-02 3.608
3.605 -34.910139
Valine, Glycerophosphocholine 1.64E-02 3.605
3.601 -26.953009
Phosphorylcholine 4.18E-02 3.601
3.597 -25.449666
Valine, Phosphocholine 3.11E-02 3.597
3.594 -25.537262
Phosphorylcholine 3.11E-02 3.594
3.589 -26.714297
Glucose, Proline 1.64E-02 3.405
3.398 -81.903175
1,3,7-Trimethyluric acid, Caffeine 3.11E-02 3.357
3.351 -23.067843
Glucose 5.24E-03 3.267
3.258 -119.85795
Arginine 1.64E-02 3.258
3.251 -45.913487
Arginine 4.18E-02 3.248
3.241 -42.213845
Arginine 2.29E-02 3.241
3.229 -53.523421
Tyrosine 1.64E-02 3.198
3.189 -26.884251
N-Nitrosodimethylamine, Phenylalanine 1.15E-02 3.169
3.162 -31.347884
Methylmalonate, Phenylalanine 4.18E-02 3.162
3.153 -24.221963
Dimethylsulfone 1.53E-08 3.153
3.148 29.5872388
Unidentified singlet at 3.0615 2.53E-11 3.065
3.058 17.3150557
PPm
Histamine, Histamium 2.29E-02 3.03
3.021 -26.007227
N-Acetyl-L-aspartate 1.64E-02 2.703
2.697 34.8847998
N-Acetyl-L-aspartate 3.11E-02 2.686
2.68 38.5052793
0-Acetylcarnitine 2.10E-03 2.652
2.642 -36.353133
0-Acetylcarnitine 3.11E-02 2.642
2.632 -33.309771
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
N-Acetylaspartate 4.18E-02
2.534 2.521 33.3321864
Gamma-Aminobutyric acid 6.41E-08 2.303
2.293 31.6719877
L-Valine 4.18E-02
2.264 2.255 -29.149154
L-Valine, Citramalic acid 1.64E-02
2.255 2.244 -31.304509
D-Ribose, Acetone, Levulinate 2.29E-02
2.244 2.233 -40.567464
N-Acetylaspartate, Acetamide 1.15E-02
2.034 2.02 45.614812
Proline, Canthaxanthin 2.29E-02
2.012 2.003 -26.762604
Proline, L-Isoleucine 2.29E-02
2.003 1.994 -25.720411
Proline, L-Isoleucine 1.15E-02
1.994 1.985 -25.143854
L-isoleucine, Proline 2.29E-02
1.985 1.976 -25.178743
L-isoleucine, Proline 4.18E-02
1.976 1.967 -24.598484
Argine, 2-Amino-3-phosphonopropionic acid, 6.99E-04
1.757 1.707 -45.089628
Leucine, 2-Hydroxyvalerate
2-Amino-3-phosophonopropionic acid, 2.10E-03
1.707 1.696 -40.086156
Leucine, 2-Hydroxyvalerate, Arginine
Leucine, 2-Hydroxyvalerate 3.50E-04
1.696 1.681 -44.639008
2-Amino-3-phosphonopropionic 2.25E-07 7.87E-03
1.681 1.671 -38.385183
acid, Arginine, 2-
Hydroxyvalerate, Leucine,
L-Isoleucine 1.15E-02
1.457 1.444 -33.141345
L-Valine 6.99E-04
1.056 1.045 -47.300641
L-Valine 6.99E-04
1.045 1.033 -45.885765
L-Isoleucine 1.75E-04
1.025 1.012 -52.073343
L-Isoleucine 7.87E-03
1.012 1.004 -40.494063
L-Valine 7.87E-03
1.004 0.9948 -35.618616
L-Valine 2.10E-03
0.9948 0.984 -35.230846
Leucine 6.99E-04
0.9791 0.9484 -49.238603
L-Isoleucine 6.99E-04
0.9484 0.938 -52.720803
L-Isoleucine 2.10E-03
0.938 0.9279 -42.579416
[0174] Table 14: Summary of at least one target biomarker
metabolite identified
as being significant by either MW or VIAVC in BA 17.
Metabolite ID VIAVC MW p-value PPM Start
PPM End Regulation
p-value
Oxypurinol 4.48E-02 8.221 8.209
21.9998268
N-Acetyl-L-aspartate 3.78E-02 8.005 7.96
27.0531674
Phenylalanine 3.78E-02 7.441 7.43
-38.299915
Phenylalanine 4.48E-02 7.43 7.421
-34.996608
Phenylalanine 1.51E-02 7.353 7.337
-42.727464
Phenylalanine 3.78E-02 7.337 7.326
-38.66171
Myo-Inositol 3.78E-02 4.091 4.075
27.3063164
76
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Ascorbate 2.65E-02 4.029 4.022
-25.885135
L-Serine 1.24E-02 4.022 4.017
-31.552766
L-Serine, Phenylalanine 2.21E-02 4.017 4.008
-37.582737
L-Serine, Phenylalanine 3.17E-02 4.008 3.999
-24.734428
L-Serine 9.22E- 1.02E-02 3.992 3.985
-21.423341
L-Serine 4.48E-02 3.985 3.979
-15.566777
Creatine phosphate, 3.78E-02 3.97 3.961
-11.914741
Galactarate
Tyrosine, 1.83E-02 3.959 3.953
-17.454935
Glycerophosphocholine
Tyrosine, 3.78E-02 3.953 3.945
-22.591442
Glycerophosphocholine
Creatine 2.65E-02 3.945 3.932
22.9048956
Glycerophosphocholine, 2.65E-02 3.932 3.917
-39.870954
Uridine
Unidentified singlet 3.9125 1.83E-02 3.917 3.908
-50.018275
PPm
Unidentified singlet at 3.9055 1.83E-02 3.908 3.903
-13.871078
PPm
Unidentified singlet 3.9 ppm 1.51E-02 3.903 3.897
-23.106893
Glycerophosphocholine 5.41E-03 3.897 3.89
-49.025992
Glycerophosphocholine 1.06E-03 3.89 3.885
-42.600087
Glycerophosphocholine 1.06E-03 3.885 3.877
-32.312468
Glycerophosphocholine 4.35E-03 3.877 3.867
-41.922764
Glycerophosphocholine 1.02E-02 3.867 3.859
-28.039859
L-Serine, Glycyl-glycine 4.48E-02 3.859 3.846
-34.930337
L-Serine 8.27E-03 3.846 3.842
-27.758811
Ascorbate 5.41E-03 3.754 3.75
-41.62794
Ascorbate 2.19E-03 3.75 3.745
-43.39917
Methylacetoacetic acid 2.76E-03 3.745 3.74
-48.857062
Ascorbate, Leucine 4.48E-02 3.74 3.737
-24.557959
Leucine, Ascorbate 5.41E-03 3.737 3.734
-53.681668
77
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Ascorbate 4.48E-02 3.734 3.731
-38.351622
Methylacetoacetic acid, N,N- 2.19E-03 3.731 3.725
-69.614732
Dimethylglycine
Glycerophosphocholine 2.76E-03 3.71 3.702
-49.425408
Glycerophosphocholine, L- 6.71E-03 3.696 3.689
-28.025211
Isoleucine
Glycerophosphocholine, L- 3.47E-03 3.685 3.679
-24.875937
Isoleucine
Glycerol 1.73E- 1.24E-04 3.674 3.67
22.5957407
22
Glycerol, Ethanol 4.35E-03 3.67 3.662
26.3036986
Glycerol, Ethanol 2.76E-03 3.662 3.652
27.7924566
Glycerol, Ethanol 6.37E-04 3.652 3.645
23.9757005
Myo-Inositol 2.21E-02 3.645 3.638
18.6745661
Glycerophosphocholine 2.21E-02 3.638 3.632
-14.119599
Myo-Inositol 3.17E-02 3.632 3.623
22.4048476
L-Valine 1.51E-02 3.623 3.618
-8.2030568
L-Threonine, 0- 8.27E-03 3.598 3.59
-14.875811
Phosphocholine
Glycerol 1.36E-03 3.587 3.576
26.086604
Glycine 1.02E-02 3.574 3.565
19.4605806
Glycerol 1.58E- 2.18E-04 3.565 3.561
21.543914
Myo-Inositol 1.24E-02 3.561 3.557
24.0179321
Myo-Inositol, Glycerol 1.02E-02 3.557 3.549
22.2202148
Myo-Inositol 1.83E-02 3.543 3.537
18.8493879
Glucose 6.37E-04 3.511 3.502
-122.13447
Glucose 5.41E-03 3.502 3.489
-109.56485
Glucose 4.35E-03 3.489 3.481
-79.423072
Glucose 1.02E-02 3.481 3.467
-108.91944
Glucose 3.47E-03 3.467 3.457
-76.570989
Glucose 1.83E-02 3.449 3.437
-55.101671
Glucose 2.21E-02 3.437 3.422
-81.777884
1,3,7-Trimethyluric acid 2.65E-02 3.422 3.414
-49.959174
Glucose 2.21E-02 3.414 3.411
-55.728939
Glucose 2.21E-02 3.405 3.398
-41.929977
Methanol 3.17E-02 3.37 3.357
38.9460235
78
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Myo-Inositol, 1-Methyluric 2.21E-02 3.303 3.294
24.9961196
acid
Trimethylamine N-oxide 1.06E-03 3.267 3.258
-93.572982
Arginine, Glucose 8.23E-04 3.258 3.25
-40.824445
1,3,7-Trimethyluric acid, 8.23E-04 3.25 3.24
-50.480258
Arginine,
0-Phosphocholine, 8.27E-03 3.24 3.229
-33.051289
Glycerophosphocholine
Citicoline 1.02E-02 3.219 3.213
19.4316238
Unidentifed singlet 2.87E-04 3.213 3.202
78.2600306
3.2075ppm
Unidentified peak (multiple!) 2.76E-03 3.148 3.14
36.2074301
at 3.144ppm
Malonate 5.41E-03 3.14 3.129
29.2301434
Creatine 2.76E-03 3.046 3.03
26.7269151
Gamma-Aminobutyric acid 1.73E-03 3.011 2.999
28.7004455
Gamma-Aminobutyric acid 2.21E-02 2.999 2.988
21.1472718
N-Acetyl-L-aspartate 1.73E-03 2.709 2.702
28.3004901
N-Acetyl-L-aspartate 5.41E-03 2.702 2.697
36.1768345
N-Acetyl-L-aspartate 6.71E-03 2.686 2.68
38.6220897
N-Acetyl-L-aspartate 1.73E-03 2.68 2.671
36.1922092
Selenomethionine 3.78E-02 2.652 2.642
-24.978054
N-Acetyl-L-aspartate 2.19E-03 2.532 2.52
38.7212969
N-Acetyl-L-aspartate 1.06E-03 2.52 2.507
40.7265782
N-Acetyl-L-aspartate 2.76E-03 2.507 2.498
31.9251003
N-Acetyl-L-aspartate 3.47E-03 2.495 2.483
25.5878949
Gamma-Aminobutyric acid 1.73E-03 2.304 2.293
36.4934594
Gamma-Aminobutyric acid 4.48E-02 2.293 2.283
22.4911007
N-Acetyl-L-aspartate 8.27E-03 2.064 2.054
18.824413
N-Acetyl-L-aspartate, 1.73E-03 2.034 2.02
54.6645004
Acetamide
79
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Gamma-Aminobutyric acid 3.17E-02 1.913 1.9
15.2595026
Gamma-Aminobutyric acid 6.71E-03 1.9 1.892
21.4555295
L-Arginine, L-Leucine 4.35E-03 1.756 1.746
-33.423955
2-Amino-3- 1.51E-02 1.746 1.713
-39.490472
phosphonopropionic acid, L-
Arginine, L-Leucine,
2-Amino-3- 4.48E-02 1.713 1.708
-26.594356
phosphonopropionic acid, L-
Leucine, L-Arginine
L-Leucine 1.36E-03 1.708 1.696
-36.686028
2-Amino-3- 1.73E-03 1.696 1 683
-38.446794
phosphonopropionic acid,
Leucine
Ethanol 4.48E-02 1.193 1.183
36.7667617
Ethanol 4.48E-02 1.183 1.172
32.3208462
L-Valine 8.23E-04 1.056 1.045 -
37.065
L-Valine 1.36E-03 1.045 1.032
-38.057687
L-Isoleucine 1.06E-03 1.024 1.012
-41.399388
L-Isoleucine 2.19E-03 1.012 1.004
-32.499884
L-Valine 6.71E-03 1.004 0.9939
-23.121975
L-Valine 4.35E-03 0.9939 0.9831
-23.343468
L-Leucine 1.83E-02 0.979 0.9481
-42.473997
L-Isoleucine 6.37E-04 0.9481 0.9367
-45.857017
L-Isoleucine 3.47E-03 0.9367 0.9266
-39.198252
[0175]
According to embodiments, at least one target biomarker change shared
across all regions of interest include 1,3,7-trimethyluric acid, 2-am ino-3-
phosphonopropionic acid, ethanol, gamma-aminobutyric acid (GABA), isoleucine,
leucine, N-Acetyl-L-aspartate (NAA), phenylalanine, serine, tyrosine, and
valine
(Table 15).
[0176]
Table 15: common and noteworthy metabolites Identified from MW and
VIAVC Bet Subset bins. A positive sign indicates upregulation of the target
biomarker
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
in the AD group compared to CN, while a negative sign indicates a
downregulation of
the target biomarker between the AD and CN groups.
Regulation per region
Regions BA22 BA 40 BA17
All regions NAA (+) NAA (+) NAA (+)
Ethanol (-) Ethanol (+) Ethanol
(+)
2-Amino-3-Phosphonoprionic 2-Amino-3- 2-Amino-3-
acid (-) Phosphonoprionic acid (-
Phosphonoprionic acid (-)
1,3,7-Trimethyluric acid (-) ) 1,3,7-
Trimethyluric acid (-
Creatine Phosphate (-) 1,3,7-Trimethyluric acid )
GABA (+) (-) Creatine
Phosphate (-)
L-Leucine (-) Creatine Phosphate (-) GABA
(+)
L-Isoleucine (-) GABA (+) L-Leucine
(-)
L-Phenylalanine (-) L-Leucine (-) L-
Isoleucine (-)
L-Serine (-) L-Isoleucine (-) L-
Phenylalanine (-)
L-Tyrosine (-) L-Phenylalanine (-) L-
Serine (-)
L-Valine (-) L-Serine (-) L-
Tyrosine (-)
Phosphorylcholine (-) L-Tyrosine (-) L-Valine
(-)
L-Valine (-)
Phosphorylcholine (-)
Phosphorylcholine (-)
BA 22 & BA Acetylcholine (-) Acetylcholine (-)
40 Betaine (-) Betaine (-)
Dimethyl sulfone (+) Dimethyl sulfone (+)
Glycolate/Glycolic acid (-) Glycolate/Glycolic acid (-
Histamine (+) )
Histamine (-)
BA 22 & BA Creatine (+) Creatine
(+)
17 Glycerol(-) Glycerol
(+)
L-Threonine (-) L-
Threonine (-)
Malonate (+) Malonate
acid (+)
Oxypurinol (+)
Oxypurinol (+
BA 40 & BA Arginine (-) Arginine
(-)
17 Glucose (-) Glucose (-
)
Glycerophosphocholine (-)
Glycerophosphocholine (-
Notable Glutamate (+) Myo-
Inositol (+)
metabolites Citric acid (+) Glycerol
(+)
unique to Cis-Aconitate (-F)
each region Malate (+)
Pyruvate (+)
[0177] Metabolites common to pairs included acetylcholine for
BA22 and BA40,
as well as glucose for BA40 and BA17. Unique metabolites for BA22 were
glutamate
and citric acid, while myoinositol and glycerol were unique metabolite for
BA17. The
three most altered metabolites for each region, as determined by MW p-value,
81
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
corresponding to the following: BA 22 - NAA, 7-methylxanthine, and histamine;
BA 40
- L-isoleucine, leucine and 2-hydroxyvalerate; BA 17 ¨ glycerol, ethanol, and
glucose.
[0178] Pathway topology analysis was carried out for each
brain region, as
shown in Tables 16 - 18.
[0179] Table 16: biochemical pathways identified from pathway
topology
analysis using metabolites identified from either univariate MW or
multivariate VIAVC
best subset tests for BA 22, with corresponding p-value and impact score.
Pathway Metabolites Raw p-
value Impact
1. Aminoacyl-tRNA biosynthesis L-Alanine (-), L-Glutamate (+), L-
1.00E-07 0.16667
Isoleucine (-), L-Leucine (-), L-
Methionine (-), L-Phenylalanine (-), L-
Threonine (-F), L-Tyrosine (-), L-Serine
(+), L-Valine (-)
2. Glyoxylate and dicarboxylate
Cis-Aconitate (+), Citric acid (+), Glycolic 7.77E-06 0.37302
metabolism acid (-), Glyoxylic acid (+), L-
Glutamate
(+), L-Serine (+), Pyruvate (+)
3. Glycine, serine and threonine Creatine
(+), Betaine (-), Glyoxylic acid 9.69E-06 0.29355
metabolism (-F), Guanidoacetate (-), L-Serine (-F),
L-
Threonine (+), Pyruvate (+)
4. Valine, leucine and isoleucine L-Leucine
(-), L-Isoleucine (-), L- 2.23E-05 0
biosynthesis Threonine (+), L-Valine (-)
5. Arginine and proline Creatine
(+), Creatine-phosphate (-), 2.61E-05 0.14543
metabolism GABA (+), Glyoxylic acid (+),
Guanidoacetate (-), L-Glutamate (+),
Pyruvate (+)
6. Alanine, aspartate and GABA (+),
Citric acid (+), L-Alanine (-), 4.29E-05 0.3702
glutamate metabolism L-Glutamate (+), N-Acetyl-L-aspartate
(+), Pyruvate (+)
7. Citrate cycle (TCA cycle) Citric
acid (+), Cis-Aconitate (+), Malate 1.24E-03 0.23087
(+), Pyruvate (+)
8. Phenylalanine, tyrosine and L-
Phenylalanine (-), L-Tyrosine (-) 3.59E-03 1
tryptophan biosynthesis
9. Phenylalanine metabolism L-
Phenylalanine (-), L-Tyrosine (-) 2.44E-02 0.35714
10. Cysteine and methionine L-
Methionine (-), L-Serine (-F), Pyruvate 4.77E-02 0.1263
metabolism (+)
82
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0180]
Table 17: biochemical pathways identified from pathway topology
analysis using metabolites identified from either univariate MW or
multivariate VIAVC
best subset tests for BA 40, with corresponding p-value and impact score.
Pathway Metabolites Raw p
Impact
1. Aminoacyl-tRNA biosynthesis L-Arginine
(-), L-Aspartate (-), L- 3.55E-07 0.16667
Isoleucine (-), L-Phenylalanine (-), L-
Proline (-), L-Serine (-), L-Tyrosine (-),
L-Valine (-)
2. Valine, leucine and isoleucine
L-Isoleucine (-), L-Leucine (-), L-Valine 0.00050157 0
biosynthesis (-)
3. Phenylalanine, tyrosine and L-
Phenylalanine (-), L-Tyrosine (-) 0.0027272 1
tryptophan biosynthesis
4. Arginine and proline metabolism L-Arginine (-), L-Proline (-), Creatine
0.008406 0.15951
phosphate (-), GABA (+)
5. Valine, leucine and isoleucine
L-Isoleucine (-), L-Leucine (-), L-Valine 0.010089 0.02264
degradation (-), Methylmalonate (-)
6. Phenylalanine metabolism L-
Phenylalanine (-), L-Tyrosine (-) 0.018829 0.35714
7. Alanine, aspartate and
N-Acetyl-L-aspartate (+), L-Aspartate 0.021727 0.39664
glutamate metabolism (-), GABA (+)
8. Arginine biosynthesis L-Arginine
(-), L-Aspartate (-) 0.036045 0.07614
9. Glycerophospholipid metabolism Acetylcholine (-), Phosphorylcholine (-
0.042036 0.05751
), Glycerophosphocholine (-)
10. Histidine metabolism Histamine
(-), L-Aspartate (-) 0.046252 0.18852
[0181]
Table 18: biochemical pathways identified from pathway topology
analysis using metabolites identified from either univariate MW or
multivariate VIAVC
best subset tests for BA 17, with corresponding p-value and impact score.
Pathway Metabolites Raw p
Impact
Glycine (+), L-Arginine (-), L-Isoleucine (-), L-
1. Aminoacyl-tRNA Leucine (-), L-Phenylalanine (-), L-Serine (-),
L-Threonine (-), L-Tyrosine (-), L-Valine (-) 2.67E-07
biosynthesis 0.16667
2. Valine, leucine and L-Isoleucine (-), L-Leucine (-), L-Threonine (-
isoleucine biosynthesis ), L-Valine (-) 1.13E-05
0
3. Glycine, serine and Creatine (+), Glycine (+), L-Serine (-), L-
threonine metabolism Threonine (-), N,N-Dimethylglycine (-)
0.00049632 0.53544
83
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
4. Phenylalanine, tyrosine L-Phenylalanine (-), L-Tyrosine (-)
and tryptophan biosynthesis 0.002569
1
5. Phenylalanine L-Phenylalanine (-), L-Tyrosine (-)
metabolism 0.017783
0.35714
6. Galactose metabolism Glucose
(-), Glycerol (+), Myo-inositol (+) 0.018163 0.03499
7. Glycerophospholipid Phosphorylcholine (-), citicoline (+),
metabolism glycerophosphocholine (-) 0.038938
0.07676
8. Arginine and proline Creatine (-F), GABA (+), L-Arginine (-)
metabolism 0.044701
0.09383
[0182] Am inoacyl-tRNA biosynthesis was the most significant
biochemical
pathway for all three brain regions, with the second most significant pathways
common to both the BA 40 and BA 17 regions were valine, leucine, and
isoleucine
biosynthesis. The second most significant pathways for BA22 were glyoxylate
and
dicarboxylate metabolism for BA 22. Similarly, BA 22 and BA 17 shared the
third most
significant pathway, glycine, serine, and threonine metabolism, while this was
phenylalanine, tyrosine and tryptophan biosynthesis for BA 40.
[0183] Having regard to Table 19, eight biochemical pathways
common to
multiple regions were found, with five of the pathways being common to all
three
regions of interest. In contrast, there were six pathways that were found to
be unique
to one region including the citric acid cycle for BA 22 and galactose
metabolism for
BA 17.
[0184] Table 19: common biochemical pathways identified from
pathway
topology analysis for each brain region. Metabolites used for the analysis
were
identified as significant for each region by either univariate MW or
multivariate VIAVC
testing. Pathways are listed alphabetically.
Pathways BA 22 BA 40 BA 17
84
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Alanine, aspartate and
glutamate metabolism
Aminoacyl-tRNA biosynthesis
Arginine and proline
metabolism
Glycerophospholipid
metabolism
Glycine, serine and threonine
metabolism
Phenylalanine, tyrosine and
tryptophan biosynthesis
Phenylalanine metabolism
Valine, leucine and isoleucine
biosynthesis
[0185] The present example demonstrates alterations in at
least one clinical
biomarkers found in at least one biological sample of an individual that
suffered an
central or peripheral nervous system injury, such as a neurodegenerative
disease or
disorder. Several of the at least one target biomarkers indicative of injury
processes
showed characteristic alterations following injury.
[0186] In some embodiments, the present apparatus and
methodologies
provide for the detection of target AD-related metabolomic alterations in at
least the
branch chain amino acids valine, leucine and isoleucine, GABA, N-acetyl-L-
aspartic
acid (NAA), phenylalanine, and tyrosine.
[0187] In some embodiments, a change in at least one target
biomarker
concentration level was detected, such target biomarkers including, without
limitation,
acetylcholine, myoinositol, citric acid, and glutamate. For example, a
decrease in the
concentration level of acetylcholine was detected (e.g., in BA22 and BA40)
suggesting
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
a down-regulation of the biomarker compared to a threshold baseline value, and
an
increase in myoinositol (e.g., in BA17), citric acid, and glutamate (e.g., in
BA22),
suggesting an up-regulation of the biomarker compared with a threshold
baseline
value.
[0188] Without being limited to theory, in some embodiments,
the variation in
the at least one target biomarker metabolites and pathways may result from
different
underlying pathologies contributing to region shared and unique biochemical
processes including protein synthesis upstream to translation, and alterations
in
excitotoxicity, neurotransmission and energy metabolism. For example, the
aminoacyl-tRNA biosynthesis pathway necessary for the translation of proteins
as it
activates the joining of an amino acid with the correct non-activated t-RNA
molecule
by the appropriate aminoacyl-tRNA synthetase was the most significantly
affected
pathway across all ROls (Tables 16¨ 18). Moreover, in some embodiments,
changes
in at least one target biomarker in this pathway in primary visual cortex
tissue (BA17)
of individuals with AD, may demonstrate a potential upstream alteration to
translation
that occurs as a result of AD. Specifically, dysregulation to upstream amino
acid
metabolism could alter amino acids' availability to form activated tRNA
molecules.
Moreover, aminoacyl-tRNA biosynthesis contains nine different amino acid
pathways
that are essential to produce activated tRNA molecules, with five of these
pathways
being significant for BA 22 and four were significant for each of BA 40 and BA
17
(Table 14), suggesting that amino acid metabolism may be altered in AD
patients.
[0189] As above, according to embodiments, the branch chain
amino acids
(BCAA) valine, leucine and isoleucine may play a key role as gatekeepers
feeding into
86
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
upstream and downstream molecular pathways, including those involved in the
pathophysiology of AD and another disease. All three BCAA were downregulated
in
the AD group compared to controls across all three ROls. Without limitation,
reduced
valine in the cerebral spinal fluid (CSF) and serum of living AD patients has
been
shown, and specifically in BA 22, the reduction of BCAA could result in
increased
synthesis of glutamate and gamma-aminobutyric acid (GABA) due to increased
expression of the enzyme branch chain amino transferase (BCAT). An increase in
BCAT expression would result in an increase conversion of the BCAA to
glutamate.
Therefore, altered BCAAs could impact glutamate synthesis which is part of
alanine,
aspartate and glutamate metabolism, which was significantly altered for BA22
and
BA40, and consequently, influences production of glutamate and GABA. Further,
an
upregulation of BCAT is observed regionally in the AD brain, could support the
regional variation in glutamate and GABA regulation.
[0190] According to embodiments, glutamate upregulation in
BA22 and GABA
upregulation in BA22, BA 40 and BA 17 has been shown in the superior frontal
and
medial cortex and superior temporal cortex (BA22) in AD brain tissues.
Increased
synaptic glutamate may lead to excitotoxicity by activating N-methyl-D-
aspartic acid
and glutamate receptors, resulting ultimately in AD pathology, including cell
death due
to oxidative stress, cytoskeletal and membrane degeneration. Without
limitation, the
presently demonstrated GABA alterations in BA22 and BA17 in AD patients could
indicate that these changes may be brain region-specific. In addition to
BCAAs, other
precursor molecules essential for neurotransmitters synthesis are altered in
AD.
87
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0191] According to embodiments, phenylalanine and tyrosine
are
catecholamine neurotransmitter precursors. Both phenylalanine and tyrosine
were
downregulated across all ROls of the AD group when compared to controls.
Additionally, in BA 22, phenylalanine was part of the VIAVC Best Subset (Table
12),
thus highlighting the potential role of phenylalanine as a biomarker for AD
within this
region. Moreover, identified pathways specific to these metabolites were
phenylalanine, tyrosine and tryptophan biosynthesis, and phenylalanine
metabolism.
As humans cannot synthesize phenylalanine and tryptophan, altered
concentration
levels of these amino acids are unrelated to phenylalanine, tyrosine and
tryptophan
biosynthesis. Instead, these changes may be related to phenylalanine
metabolism
which was altered across all ROls. Within this pathway, phenylalanine is
converted to
tyrosine via phenylalanine hydroxylase, and a decrease in tyrosine could be
attributed
to the decrease in the availability of phenylalanine.
[0192] According to embodiments, phenylalanine metabolism may
be an
upstream component of catecholamine synthesis. Phenylalanine and tyrosine are
essential precursors to the catecholamine neurotransmitters, including
dopamine
(DA), norepinephrine (NE), and epinephrine. Similarly, phenylalanine and
tyrosine
levels are known to be reduced in the serum, CSF, and brain tissue of AD
patients
when compared to controls. DA and NE are known to decrease in the plasma, CSF,
urine and brain tissue of AD samples. The decrease of these neurotransmitters
would
imply alterations in the availability of their amino acid precursors,
phenylalanine and
tyrosine. In the present study, the catecholamine neurotransmitters were not
altered
between the AD and control groups.
88
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0193] According to embodiments, n-acetylaspartate (NAA) may
play a role in
neuronal health and one of the highest concentrations of this amino acid are
found in
the brain and was upregulated in all three ROls in the AD group compared to
controls.
Analysis of post-mortem AD tissues by 1H NMR has shown an NAA reduction in the
grey matters of the calcarine sulcus (BA 17), and superior temporal gyrus (BA
22)
among other regions. NAA reduction is a proposed marker of neuronal loss in
AD, as
NAA is a biomarker for neuron health due to its typical high concentration in
neuronal
tissue. Contrary to this evidence, this study found an upregulation of NAA in
all regions
indicating a potential compensatory increase in NAA synthesis to combat
neuronal
loss.
[0194] According to embodiments, acetylcholine is a major
neurotransmitter
and neuromodulator that has a role in arousal, attention, memory, and
motivation.
Acetylcholine was downregulated in BA 22 and BA 40, and this dysregulation
coincide
with the clinical symptomology of AD. Moreover, alterations in this metabolite
may be
related to changes to the branch of glycerophospholipid metabolism responsible
for
acetylcholine synthesis. Interestingly, glycerophospholipid metabolism was
significant
for BA 40 and BA 17 (Tables 17 and 18). Within this pathway, choline can be
converted to acetylcholine via choline acetyltransferase (ChAT). Likewise,
acetylcholine can be converted back to choline via acetylcholine transferase.
Dysregulation of acetylcholine within BA 40 may be due to alterations in
acetylcholine
or choline formation. Indeed, degradation of acetylcholine synthesis and
decreased
ChAT activity are well-known parts of AD pathology. Within BA 40, choline was
not
identified as significantly altered, which has previously been shown in an AD
group
89
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
compared to MCI and age-sex matched controls. It is unclear why acetylcholine,
but
not choline, is altered within BA40, as choline is one of the two necessary
precursors
for acetylcholine. Additionally, the significant down-regulation of
phosphorylcholine (a
precursor to choline) in BA 40 and BA 22 could contribute to the impairment in
acetylcholine synthesis. Notably, since acetylcholine is not altered in BA17,
it may
indicate differences in AD pathology or resiliency to it.
[0195] According to embodiments, glucose was downregulated in
BA 40 and
BA 17 supporting evidence that reduced glucose metabolism is a part of AD
pathology. For example, reduced glucose metabolism has been observed in the
superior temporal gyrus/middle temporal gyrus of AD patients as measured by
PET
or cerebral metabolic rate of glucose consumption, however, no changes within
the
superior temporal gyrus (BA 22) were observed. Since reduced glucose was found
in
BA 40 and BA 17, this change may potentially be specific to higher associative
and
primary sensory areas. Glucose is the essential precursor for aerobic
respiration
which includes the tricarboxylic acid (TCA) cycle.
[0196] According to embodiments, the TCA cycle (citrate/citric
acid cycle), a
step within aerobic respiration, was uniquely altered in BA 22 only. Within
this pathway
and region, pyruvate citric acid, cis-aconitate, and malate were upregulated
in the AD
group compared to controls, as previously noted in CSF, serum, plasma, and
brain
tissues of AD and MCI patients. Potential impairments in the TCA cycle may
indicate
a decrease in energy production in AD due to impairment in oxidative
phosphorylation
via oxidative stress, however, based solely on the regulation of metabolites
within the
TCA cycle it is unclear whether there is an increase or decrease in energy
availability.
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
Moreover, it is unclear how the upregulation of glutamate in BA 22 may affect
the TCA
cycle. The conversion of BCAA to glutamate via BCAT involves the transfer of
an
amino group from the BCAA to a-ketoglutarate to form glutamate. If the over-
expression of BCAT (as described earlier) is responsible for the
downregulation of
BCAA and upregulation of glutamate, then a down-regulation of a-ketoglutarate
is
expected. This is not observed. However, a-ketoglutarate is an intermediate of
the
TCA cycle and potentially alterations already occurring in the TCA cycle mask
alterations in a-ketoglutarate.
[0197] According to embodiments, myo-Inositol (ml), a previous
biomarker
candidate for AD, was upregulated in BA17 only. Previously, high levels of ml
were
observed in CSF and post-mortem AD tissues. Without limitation, change in ml
levels
may be indicative of Ap pathology in at-risk and asymptomatic individuals. A
previous
study comparing individuals with Down syndrome, a population considered to at
risk
of AD, found higher levels of ml in BA 17 compared to the association parietal
cortex
in older individuals. Though upregulated ml was observed in BA 17 in the
present
examples, this region may be partially resistant to Ap pathology as vision
problems do
not present until very late in the disease indicating that ml may be a good
new
candidate for further biomarker discovery for AD.
[0198] According to embodiments, the present apparatus and
methodologies
provides: (1) the identification of many metabolites altered between AD and ON
individuals in all three ROls (including BA 17, a region otherwise more
resistant to
stereotypical pathological changes), (2) discovery of potential biomarkers for
AD
within each ROI, and (3) evidence of diverse pathological mechanisms involved
in AD.
91
CA 03214708 2023- 10-5
WO 2022/213201
PCT/CA2022/050533
[0199] Although a few embodiments have been shown and
described, it will be
appreciated by those skilled in the art that various changes and modifications
can be
made to these embodiments without changing or departing from their scope,
intent or
functionality. The terms and expressions used in the preceding specification
have
been used herein as terms of description and not of limitation, and there is
no intention
in the use of such terms and expressions of excluding equivalents of the
features
shown and the described portions thereof.
92
CA 03214708 2023- 10-5