Note: Descriptions are shown in the official language in which they were submitted.
LOCI ASSOCIATED WITH CHARCOAL ROT DROUGHT COMPLEX
TOLERANCE IN SOYBEAN
FIELD OF THE INVENTION
[0001] The present invention relates to compositions and methods for
identifying soybean
plants that are tolerant, have improved tolerance, or are susceptible to
Charcoal Rot Drought
Complex, where the methods use molecular genetic markers to identify, select
and/or
construct disease ancUor drought-tolerant plants. The invention also relates
to soybean plants
that display tolerance or improved tolerance to Charcoal Rot Drought Complex
that are
generated by the methods of the invention.
REFERENCE TO A SEQUENCE LISTING SUBMITTED
AS A TEXT FILE VIA EFS-WEB
[0002] A sequence listing having the file name "5185W0PCT_SEQLIST.txt,"
created on
October 1, 2015, and having a size of 68,766 bytes is filed in computer
readable form
concurrently with the specification. The sequence listing is part of the
specification.
BACKGROUND
100031 Soybean, a legume, has become the world's primary source of seed
oil and seed
protein. In addition, its utilization is being expanded to the industrial,
manufacturing and
pharmaceutical sectors. Soybean productivity is a vital agricultural and
economic
consideration. Unfortunately, soybean is host to one of the widest ranges of
infectious
pathogens of all crops. More than a hundred different pathogens are known to
affect soybean
plants, some of which pose significant economic threats. Improving soybean
disease
tolerance to these many pathogens is crucial to preventing yield losses.
[0004] Charcoal Rot (or alternatively referred to herein as "Charcoal Rot
Drought
Complex") is caused by the fungus Macrophornina phaseolina. The fungus has a
particularly
wide geographic distribution and is found throughout the world. M. phaseolina
is most severe
between 35 North and 35 South latitude (Wyllie, (1976) `Macrophomina
phaseolina
Charcoal Rot' P 482-484 In L. D. Hill (ed.) World Soybean Research Proc of the
World
Soybean Res. Conf., Champaign, Ill. Interstate, Danville, Ill.). The fungus
also has a wide
host range and infects over 500 crop and weed species and is highly variable.
Known major
crop hosts include alfalfa, maize, cotton, grain sorghum, peanut and soybean.
1
Date Recue/Date Received 2023-12-08
[0005] In localized areas, yield losses can be as high as 90%. In the
period from 1996-
2005, charcoal rot was the third leading cause of soybean yield loss in the
U.S. Average
annual losses were 29 MM bushels resulting in approximately $188 MM annual
income loss.
Only soybean cyst nematode and phythophthora root rot caused greater economic
loss during
that period (Wrather and Koenning (2006) 'Soybean Disease Loss Estimates for
the United
States, 1996-2006'. University of Missouri¨Columbia Agriculture Experiment
Station.
November 2006).
[0006] Complete or vertical resistance to Al. phaseolina has not been
identified in
soybean, which strongly suggests that a single gene conferring resistance does
not exist. In
most field and greenhouse evaluations, the great majority of soybean cultivars
have been
found to be either highly or moderately susceptible to M. phaseolina. Only a
few cultivars
have been identified as possessing partial or horizontal resistance (Smith and
Caryllle (1997)
'Field screening of commercial and experimental soybean cultivars for their
reaction to
Macrophomina phaseolina' Plant Dis 81:804-809).
[0007] It is the goal of the plant breeder to select plants and enrich the
plant population
for individuals that have desired traits, for example, pathogen tolerance,
leading ultimately to
increased agricultural productivity. It has been recognized for quite some
time that specific
chromosomal loci (or intervals) can be mapped in an organism's genome that
correlate with
particular quantitative phenotypes. Such loci are termed quantitative trait
loci, or QTL. The
plant breeder can advantageously use molecular markers to identify desired
individuals by
identifying marker alleles that show a statistically significant probability
of co-segregation
with a desired phenotype (e.g., pathogenic infection tolerance), manifested as
linkage
disequilibrium. By identifying a molecular marker or clusters of molecular
markers that co-
segregate with a quantitative trait, the breeder is thus identifying a QTL. By
identifying and
selecting a marker allele (or desired alleles from multiple markers) that
associates with the
desired phenotype, the plant breeder is able to rapidly select a desired
phenotype by selecting
for the proper molecular marker allele (a process called marker-assisted
selection, or MAS).
The more molecular markers that are placed on the genetic map, the more
potentially useful
that map becomes for conducting MAS.
[0008] Despite significant advances in research directed towards improved
crop tolerance
to Charcoal Rot Drought Complex, there remains a need in the art for improved
soybean
strains that are tolerant to Charcoal Rot and its causative agents, namely
Macrophomina
phaseolina infection and low-available water growth conditions. There is a
need in the art for
methods that identify soybean plants or populations (germplasm) that display
tolerance to
2
Date Recue/Date Received 2023-12-08
Charcoal Rot Drought Complex. What is needed in the art is to identify
molecular genetic
markers that are linked to Charcoal Rot Drought Complex tolerance loci in
order to facilitate
MAS. Such markers can be used to select individual plants and plant
populations that show
favorable marker alleles in soybean populations and then employed to select
the tolerant
phenotype, or alternatively, be used to counterselect plants or plant
populations that show a
Charcoal Rot Drought Complex susceptibility phenotype. The present invention
provides
these and other advantages.
BRIEF SUMMARY
[0009] Compositions and methods for identifying soybean plants or
germplasm with
tolerance to Charcoal Rot Drought Complex are provided. Methods of making
soybean plants
or germplasm that are tolerant to Charcoal Rot Drought Complex, e.g., through
introgression
of desired tolerance marker alleles and/or by transgenic production methods,
as well as plants
and germplasm made by these methods, are also provided. Systems and kits for
selecting
tolerant plants and germplasm are also a feature of the invention.
[0010] Disclosed are methods for identifying a first soybean plant or
germplasm (e.g., a
line or variety) that has tolerance, improved tolerance, or susceptibility to
Charcoal Rot
Drought Complex. In the methods, at least one allele of one or more marker
locus (e.g., a
plurality of marker loci) that is associated with the tolerance, improved
tolerance, or
susceptibility is detected in the first soybean plant or germplasm.
[0011] Also disclosed are methods of identifying a first soybean plant or
germplasm that
displays tolerance, improved tolerance, or susceptibility to Charcoal Rot; the
method
comprising detecting in the first soybean plant or germplasm at least one
allele of a
quantitative trait locus that is associated with the tolerance, improved
tolerance, or
susceptibility; wherein the quantitative locus is: (i) a chromosomal interval
located at about
17 cM to about 38 cM of chromosome 5; (ii) a chromosomal interval located at
about 5 cM to
about 26 cM of chromosome 15; (iii) a chromosomal interval located at about 19
cM to about
40 cM of chromosome 19; or (iv) a chromosomal interval located at about 81
c1V1 to about
102 cM of chromosome 19.
[0012] Also disclosed are methods of identifying a first soybean plant or
germplasm that
displays tolerance, improved tolerance, or susceptibility to Charcoal Rot; the
method
comprising the steps of: (a) detecting in the first soybean plant or germplasm
at least one
allele of a quantitative trait locus that is associated with the tolerance,
improved tolerance, or
susceptibility; wherein the quantitative locus is: wherein the quantitative
locus is: (i) a
3
Date Recue/Date Received 2023-12-08
chromosomal interval located at about 17 cM to about 38 cM of chromosome 5;
(ii) a
chromosomal interval located at about 5 cM to about 26 cM of chromosome 15;
(iii) a
chromosomal interval located at about 19 cM to about 40 cM of chromosome 19;
or (iv) a
chromosomal interval located at about 81 cM to about 102 cM of chromosome 19;
(b)
selecting the first soybean plant or germplasm, or selecting a progeny of the
first soybean
plant or germplasm comprising the at least one allele of a quantitative trait
locus that is
associated with the tolerance, improved tolerance, or susceptibility; and (c)
crossing the
selected first soybean plant or germplasm with a second soybean plant or
germplasm to
introgress the quantitative trait locus into progeny soybean germplasm.
[0013] Also disclosed are methods of identifying a first soybean plant or
germplasm that
displays tolerance, improved tolerance, or susceptibility to Charcoal Rot; the
method
comprising the steps of: (a) detecting in the first soybean plant or germplasm
at least one
allele of a quantitative trait locus that is associated with the tolerance,
improved tolerance, or
susceptibility; wherein the quantitative locus is: wherein the quantitative
locus is: (i) a
chromosomal interval located at about 17 cM to about 38 cM of chromosome 5;
(ii) a
chromosomal interval located at about 5 cM to about 26 cM of chromosome 15;
(iii) a
chromosomal interval located at about 19 cM to about 40 cM of chromosome 19;
or (iv) a
chromosomal interval located at about 81 cM to about 102 cM of chromosome 19;
(b)
selecting the first soybean plant or germplasm, or selecting a progeny of the
first soybean
plant or germplasm comprising the at least one allele of a quantitative trait
locus that is
associated with the tolerance, improved tolerance, or susceptibility; (c)
crossing the selected
first soybean plant or germplasm with a second soybean plant or germplasm to
introgress the
quantitative trait locus into progeny soybean germplasm; (d) analyzing progeny
soybean
germplasm to determine the presence of tolerance to Charcoal Rot; and (d)
selecting progeny
soybean germplasm that tests positive for the presence of tolerance to
Charcoal Rot as being
soybean germplasm into which germplasm having said quantitative trait locus
has been
introgressed.
[0014] Also disclosed are introgressed soybean plants or gernriplasms
produced by the
disclosed methods.
[0015] Also disclosed are kits for selecting at least one soybean plant by
marker assisted
selection of a quantitative trait locus associated with the tolerance,
improved tolerance, or
susceptibility to Charcoal Rot comprising: (a) labeled primers or probes for
detecting at least
one nucleic acid sequence selected from the group consisting of: (i) 48,340-
48,380 kbp of
chromosome 19 (SEQ ID NO.: 27); (ii) 3,202-3,212 kbp of chromosome 15 (SEQ ID
NO.:
4
Date Recue/Date Received 2023-12-08
26); (iii) S11315 (SEQ ID NO.: 1); (iv) S11316 (SEQ ID NO.: 6); (v) S29725
(SEQ ID NO:
11); (vi) S29742 (SEQ ID NO: 16); and (vii) S29741 (SEQ ID NO: 21); and (b)
instructions
for using the primers or probes to detect the marker loci and correlating the
loci with
predicted improved lodging resistance.
[0016] Also disclosed are methods for screening a plant for resistance to
a plant
pathogen, the method comprising: (a) providing at least one inoculation probe
having a
pointed end to a container of agar inoculated with a pathogen; wherein a
surface of the
inoculation probe is contact with the surface of the agar in the petri dish;
(b) inoculating a
plant, after a predetermined contact time between at least one inoculation
probe and the
pathogen, by inserting the pointed end of at least one inoculation probe,
comprising pathogen
on the surface thereof, into a site located on a plant stem; and (c) assessing
plant tolerance to
the pathogen at a predetermined time.
BRIEF DESCRIPTION OF THE DRAWINGS
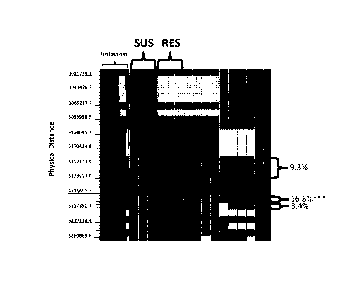
[0017] FIG. 1 shows representative data for haplotype analysis of
Chromosome 15 in the
region of approximately 3,012-3,946 kb on the Soybean Consensus Map 4.0 (Hyten
D. L., et
al., (2010) Crop Sci 50: 960-968) using 10 kb haplotype windows created using
high density
sequence data from 206 unique varieties. Displayed in columns are three known
resistant and
three known susceptibe varieties, which are indicated respectively by "RES"
and "SUS" in
the figure. To the right are the results for 10 varieties with unknown QTL
status. Indicated
next to the last column on the right are regression values (R2) for the effect
of the indicated
haplotype on charcoal rot drought complex across the set of 206 varieties. The
left is a
histogram (as indicated) representing the cumulative number of haplotypes from
the columns
to the right for each 10 kb window examined. The scale to the far left
indicates the physical
distance within the region examined in the columns to the right.
[0018] FIG. 2 shows representative data for haplotype analysis of
Chromosome 19 in the
region of approximately 48,300-48,550 kb on the Soybean Consensus Map 4.0
(Hyten D. L.,
et al., (2010) Crop Sci 50: 960-968) using 10 kb haplotype windows created
using high
density sequence data from 148 unique varieties. Displayed in columns are four
known
resistant and three known susceptibe varieties, which are indicated
respectively by "RES"
and "SUS" in the figure. The figure also shows results for 12 varieties with
unknown QTL
status. Indicated next to the last column on the right are regression values
(R2) for the effect
of the indicated haplotype on charcoal rot drought complex across the set of
148 varieties.
The left is a histogram (as indicated) representing the cumulative number of
haplotypes from
Date Recue/Date Received 2023-12-08
the columns to the right for each 10 kb window examined. The scale to the far
left indicates
the physical distance within the region examined in the columns to the right.
DETAILED DESCRIPTION
[0019] The disclosures herein will be described more fully hereinafter
with reference to
the accompanying drawings, in which some, but not all possible embodiments are
shown.
Indeed, disclosures may be embodied in many different forms and should not be
construed as
limited to the embodiments set forth herein; rather, these embodiments are
provided so that
this disclosure will satisfy applicable legal requirements.
[0020] Many modifications and other embodiments disclosed herein will come
to mind to
one skilled in the art to which the disclosed compositions and methods pertain
having the
benefit of the teachings presented in the foregoing descriptions and the
associated drawings.
Therefore, it is to be understood that the disclosures are not to be limited
to the specific
embodiments disclosed and that modifications and other embodiments are
intended to be
included within the scope of the appended claims. Although specific terms are
employed
herein, they are used in a generic and descriptive sense only and not for
purposes of
limitation.
[0021] It is also to be understood that the terminology used herein is for
the purpose of
describing particular aspects only and is not intended to be limiting. As used
in the
specification and in the claims, the term "comprising" can include the aspect
of "consisting
of." Unless defined otherwise, all technical and scientific terms used herein
have the same
meaning as commonly understood by one of ordinary skill in the art to which
the disclosed
compositions and methods belong. In this specification and in the claims which
follow,
reference will be made to a number of terms which shall be defined herein.
L DEFINITIONS
[0022] Before describing the present invention in detail, it is to be
understood that this
invention is not limited to particular embodiments, which can, of course,
vary. It is also to be
understood that the terminology used herein is for the purpose of describing
particular
embodiments only, and is not intended to be limiting. As used in this
specification and the
appended claims, terms in the singular and the singular forms "a", "an" and -
the", for
example, include plural referents unless the content clearly dictates
otherwise. Thus, for
example, reference to "plant", "the plant" or "a plant" also includes a
plurality of plants; also,
6
Date Recue/Date Received 2023-12-08
depending on the context, use of the term "plant" can also include genetically
similar or
identical progeny of that plant; use of the term "a nucleic acid" optionally
includes, as a
practical matter, many copies of that nucleic acid molecule; similarly, the
term "probe"
optionally (and typically) encompasses many similar or identical probe
molecules.
[0023] Unless otherwise indicated, nucleic acids are written left to right
in 5' to 3'
orientation. Numeric ranges recited within the specification are inclusive of
the numbers
defining the range and include each integer or any non-integer fraction within
the defined
range. Unless defined otherwise, all technical and scientific terms used
herein have the same
meaning as commonly understood by one of ordinary skill in the art to which
the invention
pertains. Although any methods and materials similar or equivalent to those
described herein
can be used in the practice for testing of the present invention, the
preferred materials and
methods are described herein. In describing and claiming the present
invention, the following
terminology will be used in accordance with the definitions set out below.
[0024] A "plant" can be a whole plant, any part thereof, or a cell or
tissue culture derived
from a plant. Thus, the term "plant" can refer to any of: whole plants, plant
components or
organs (e.g., leaves, stems, roots, etc.), plant tissues, seeds, plant cells,
and/or progeny of the
same. A plant cell is a cell of a plant, taken from a plant, or derived
through culture from a
cell taken from a plant. Thus, the term "soybean plant" includes whole soybean
plants,
soybean plant cells, soybean plant protoplast, soybean plant cell or soybean
tissue culture
from which soybean plants can be regenerated, soybean plant calli, soybean
plant clumps and
soybean plant cells that are intact in soybean plants or parts of soybean
plants, such as
soybean seeds, soybean pods, soybean flowers, soybean cotyledons, soybean
leaves, soybean
stems, soybean buds, soybean roots, soybean root tips and the like.
[0025] "Germplasrn" refers to genetic material of or from an individual
(e.g., a plant), a
group of individuals (e.g., a plant line, variety or family), or a clone
derived from a line,
variety, species, or culture. The germplasm can be part of an organism or
cell, or can be
separate from the organism or cell. In general, germplasm provides genetic
material with a
specific molecular makeup that provides a physical foundation for some or all
of the
hereditary qualities of an organism or cell culture. As used herein, germplasm
includes cells,
seed or tissues from which new plants may be grown, or plant parts, such as
leafs, stems,
pollen, or cells that can be cultured into a whole plant.
100261 The term "allele" refers to one of two or more different nucleotide
sequences that
occur at a specific locus. For example, a first allele can occur on one
chromosome, while a
second allele occurs on a second homologous chromosome, e.g., as occurs for
different
7
Date Recue/Date Received 2023-12-08
chromosomes of a heterozygous individual, or between different homozygous or
heterozygous individuals in a population. A "favorable allele" is the allele
at a particular
locus that confers, or contributes to, an agronomically desirable phenotype,
e.g., tolerance to
Charcoal Rot Drought Complex, or alternatively, is an allele that allows the
identification of
susceptible plants that can be removed from a breeding program or planting. A
favorable
allele of a marker is a marker allele that segregates with the favorable
phenotype, or
alternatively, segregates with susceptible plant phenotype, therefore
providing the benefit of
identifying disease-prone plants. A favorable allelic form of a chromosome
segment is a
chromosome segment that includes a nucleotide sequence that contributes to
superior
agronomic performance at one or more genetic loci physically located on the
chromosome
segment. "Allele frequency" refers to the frequency (proportion or percentage)
at which an
allele is present at a locus within an individual, within a line, or within a
population of lines.
For example, for an allele "A", diploid individuals of genotype "AA", "Aa", or
"aa" have
allele frequencies of 1.0, 0.5, or 0.0, respectively. One can estimate the
allele frequency
within a line by averaging the allele frequencies of a sample of individuals
from that line.
Similarly, one can calculate the allele frequency within a population of lines
by averaging the
allele frequencies of lines that make up the population. For a population with
a finite number
of individuals or lines, an allele frequency can be expressed as a count of
individuals or lines
(or any other specified grouping) containing the allele.
[0027] An allele "positively" correlates with a trait when it is linked
to it and when
presence of the allele is an indictor that the desired trait or trait form
will occur in a plant
comprising the allele. An allele negatively correlates with a trait when it is
linked to it and
when presence of the allele is an indicator that a desired trait or trait form
will not occur in a
plant comprising the allele.
[0028] An individual is "homozygous" if the individual has only one type
of allele at a
given locus (e.g., a diploid individual has a copy of the same allele at a
locus for each of two
homologous chromosomes). An individual is "heterozygous" if more than one
allele type is
present at a given locus (e.g., a diploid individual with one copy each of two
different
alleles). The term "homogeneity" indicates that members of a group have the
same genotype
at one or more specific loci. In contrast, the term "heterogeneity" is used to
indicate that
individuals within the group differ in genotype at one or more specific loci.
[0029] A "locus" is a chromosomal region where a polymorphic nucleic
acid, trait
determinant, gene or marker is located. Thus, for example, a "gene locus" is a
specific
chromosome location in the genome of a species where a specific gene can be
found.
8
Date Recue/Date Received 2023-12-08
[0030] The term "quantitative trait locus" or "QTL" refers to a
polymorphic genetic locus
with at least one allele that correlates with the differential expression of a
phenotypic trait in
at least one genetic background, e.g., in at least one breeding population or
progeny. A QTL
can act through a single gene mechanism or by a polygenic mechanism.
[0031] The terms "marker", "molecular marker", "marker nucleic acid", and
"marker
locus" refer to a nucleotide sequence or encoded product thereof (e.g., a
protein) used as a
point of reference when identifying a linked locus. A marker can be derived
from genomic
nucleotide sequence or from expressed nucleotide sequences (e.g., from a
spliced RNA or a
cDNA), or from an encoded polypeptide. The term also refers to nucleic acid
sequences
complementary to or flanking the marker sequences, such as nucleic acids used
as probes or
primer pairs capable of amplifying the marker sequence. A "marker probe" is a
nucleic acid
sequence or molecule that can be used to identify the presence of a marker
locus, e.g., a
nucleic acid probe that is complementary to a marker locus sequence.
Alternatively, in some
aspects, a marker probe refers to a probe of any type that is able to
distinguish (i.e., genotype)
the particular allele that is present at a marker locus. Nucleic acids arc
"complementary"
when they specifically hybridize in solution, e.g., according to Watson-Crick
base pairing
rules. A "marker locus" is a locus that can be used to track the presence of a
second linked
locus, e.g., a linked locus that encodes or contributes to expression of a
phenotypic trait. For
example, a marker locus can be used to monitor segregation of alleles at a
locus, such as a
QTL, that are genetically or physically linked to the marker locus. Thus, a
"marker allele",
alternatively an "allele of a marker locus", is one of a plurality of
polymorphic nucleotide
sequences found at a marker locus in a population that is polymorphic for the
marker locus.
In some aspects, the present invention provides marker loci correlating with
tolerance to
Charcoal Rot Drought Complex in soybean. Each of the identified markers is
expected to be
in close physical and genetic proximity (resulting in physical and/or genetic
linkage) to a
genetic element, e.g., a QTL that contributes to tolerance.
[0032] "Genetic markers" are nucleic acids that are polymorphic in a
population and
where the alleles of which can be detected and distinguished by one or more
analytic
methods, e.g., RFLP, AFLP, isozyme, SNP, SSR, and the like. The term also
refers to nucleic
acid sequences complementary to the genomic sequences, such as nucleic acids
used as
probes.
[0033] Markers corresponding to genetic polymorphisms between members of a
population can be detected by methods well-established in the art. These
include, e.g., PCR-
based sequence specific amplification methods, detection of restriction
fragment length
9
Date Recue/Date Received 2023-12-08
polymorphisms (RFLP), detection of isozyme markers, detection of
polynucleotide
polymorphisms by allele specific hybridization (ASH), detection of amplified
variable
sequences of the plant genome, detection of self-sustained sequence
replication, detection of
simple sequence repeats (SSRs), detection of single nucleotide polymorphisms
(SNPs), or
detection of amplified fragment length polymorphisms (AFLPs). Well established
methods
are also know for the detection of expressed sequence tags (ESTs) and SSR
markers derived
from EST sequences and randomly amplified polymorphic DNA (RAPD).
[0034] A "genetic map" is a description of genetic linkage relationships
among loci on
one or more chromosomes (or linkage groups) within a given species, generally
depicted in a
diagrammatic or tabular form. "Genetic mapping" is the process of defining the
linkage
relationships of loci through the use of genetic markers, populations
segregating for the
markers, and standard genetic principles of recombination frequency. A
"genetic map
location" is a location on a genetic map relative to surrounding genetic
markers on the same
linkage group where a specified marker can be found within a given species. In
contrast, a
"physical map" of the genome refers to absolute distances (for example,
measured in base
pairs or isolated and overlapping contiguous genetic fragments, e.g.,
contigs). A physical map
of the genome does not take into account the genetic behavior (e.g.,
recombination
frequencies) between different points on the physical map.
[0035] A "genetic recombination frequency" is the frequency of a crossing
over event
(recombination) between two genetic loci. Recombination frequency can be
observed by
following the segregation of markers and/or traits following meiosis. A
genetic
recombination frequency can be expressed in centimorgans (cM), where one cM is
the
distance between two genetic markers that show a 1% recombination frequency
(i.e., a
crossing-over event occurs between those two markers once in every 100 cell
divisions).
[0036] As used herein, the term "linkage" is used to describe the degree
with which one
marker locus is "associated with" another marker locus or some other locus
(for example, a
tolerance locus).
[0037] As used herein, linkage equilibrium describes a situation where two
markers
independently segregate, i.e., sort among progeny randomly. Markers that show
linkage
equilibrium are considered unlinked (whether or not they lie on the same
chromosome).
[0038] As used herein, linkage disequilibrium describes a situation where
two markers
segregate in a non-random manner, i.e., have a recombination frequency of less
than 50%
(and by definition, are separated by less than 50 cM on the same linkage
group). Markers that
show linkage disequilibrium are considered linked. Linkage occurs when the
marker locus
Date Recue/Date Received 2023-12-08
and a linked locus are found together in progeny plants more frequently than
not together in
the progeny plants. As used herein, linkage can be between two markers, or
alternatively
between a marker and a phenotype. A marker locus can be associated with
(linked to) a trait,
e.g., a marker locus can be associated with tolerance or improved tolerance to
a plant
pathogen when the marker locus is in linkage disequilibrium with the tolerance
trait. The
degree of linkage of a molecular marker to a phenotypic trait is measured,
e.g., as a statistical
probability of co-segregation of that molecular marker with the phenotype.
[0039] As used herein, the linkage relationship between a molecular marker
and a
phenotype is given as a "probability" or "adjusted probability". The
probability value is the
statistical likelihood that the particular combination of a phenotype and the
presence or
absence of a particular marker allele is random. Thus, the lower the
probability score, the
greater the likelihood that a phenotype and a particular marker will co-
segregate. In some
aspects, the probability score is considered "significant" or "insignificant".
In some
embodiments, a probability score of 0.05 (p=0.05, or a 5% probability) of
random assortment
is considered a significant indication of co-segregation. However, the present
invention is not
limited to this particular standard, and an acceptable probability can be any
probability of less
than 50% (p=0.5). For example, a significant probability can be less than
0.25, less than 0.20,
less than 0.15, or less than 0.1.
[0040] The term "linkage disequilibrium" refers to a non-random
segregation of genetic
loci or traits (or both). In either case, linkage disequilibrium implies that
the relevant loci are
within sufficient physical proximity along a length of a chromosome so that
they segregate
together with greater than random (i.e., non-random) frequency (in the case of
co-segregating
traits, the loci that underlie the traits are in sufficient proximity to each
other). Linked loci co-
segregate more than 50% of the time, e.g., from about 51% to about 100% of the
time. The
term "physically linked" is sometimes used to indicate that two loci, e.g.,
two marker loci, are
physically present on the same chromosome.
[0041] Advantageously, the two linked loci are located in close proximity
such that
recombination between homologous chromosome pairs does not occur between the
two loci
during meiosis with high frequency, e.g., such that linked loci co-segregate
at least about
90% of the time, e.g., 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%,
99.75%, or
more of the time.
[0042] The phrase "closely linked", in the present application, means that
recombination
between two linked loci occurs with a frequency of equal to or less than about
10% (i.e., are
separated on a genetic map by not more than 10 cM). Put another way, the
closely linked loci
11
Date Recue/Date Received 2023-12-08
co-segregate at least 90% of the time. Marker loci are especially useful in
the present
invention when they demonstrate a significant probability of co-segregation
(linkage) with a
desired trait (e.g., pathogenic tolerance). For example, in some aspects,
these markers can be
termed linked QTL markers. In other aspects, especially useful molecular
markers are those
markers that are linked or closely linked.
[00431 In some aspects, linkage can be expressed as any desired limit or
range. For
example, in some embodiments, two linked loci are two loci that are separated
by less than 50
cM map units. In other embodiments, linked loci are two loci that are
separated by less than
40 cM. In other embodiments, two linked loci are two loci that are separated
by less than 30
cM. In other embodiments, two linked loci are two loci that are separated by
less than 25 cM.
In other embodiments, two linked loci are two loci that are separated by less
than 20 cM. In
other embodiments, two linked loci are two loci that are separated by less
than 15 cM. In
some aspects, it is advantageous to define a bracketed range of linkage, for
example, between
and 20 cM, or between 10 and 30 cM, or between 10 and 40 cM.
[0044] The more closely a marker is linked to a second locus, the better
an indicator for
the second locus that marker becomes. Thus, in one embodiment, closely linked
loci such as a
marker locus and a second locus display an inter-locus recombination frequency
of 10% or
less, preferably about 9% or less, still more preferably about 8% or less, yet
more preferably
about 7% or less, still more preferably about 6% or less, yet more preferably
about 5% or
less, still more preferably about 4% or less, yet more preferably about 3% or
less, and still
more preferably about 2% or less. In highly preferred embodiments, the
relevant loci display
a recombination a frequency of about 1% or less, e.g., about 0.75% or less,
more preferably
about 0.5% or less, or yet more preferably about 0.25% or less. Two loci that
are localized to
the same chromosome, and at such a distance that recombination between the two
loci occurs
at a frequency of less than 10% (e.g., about 9%, 8%, 7%, 6%, 5%, 4%, 3%, 2%,
1%, 0.75%,
0.5%, 0.25%, or less) are also said to be "proximal to" each other. In some
cases, two
different markers can have the same genetic map coordinates. In that case, the
two markers
are in such close proximity to each other that recombination occurs between
them with such
low frequency that it is undetectable.
[0045] When referring to the relationship between two genetic elements,
such as a
genetic element contributing to tolerance and a proximal marker, "coupling"
phase linkage
indicates the state where the "favorable" allele at the tolerance locus is
physically associated
on the same chromosome strand as the "favorable" allele of the respective
linked marker
locus. In coupling phase, both favorable alleles are inherited together by
progeny that inherit
12
Date Recue/Date Received 2023-12-08
that chromosome strand. In "repulsion" phase linkage, the "favorable" allele
at the locus of
interest is physically linked with an "unfavorable" allele at the proximal
marker locus, and
the two "favorable" alleles are not inherited together (i.e., the two loci are
"out of phase" with
each other).
[0046] As used herein, the terms "chromosome interval" or "chromosome
segment"
designate a contiguous linear span of genomic DNA that resides in planta on a
single
chromosome. The genetic elements or genes located on a single chromosome
interval are
physically linked. The size of a chromosome interval is not particularly
limited.
[0047] In some aspects, for example in the context of the present
invention, generally the
genetic elements located within a single chromosome interval are also
genetically linked,
typically within a genetic recombination distance of, for example, less than
or equal to 20
cM, or alternatively, less than or equal to 10 cM. That is, two genetic
elements within a single
chromosome interval undergo recombination at a frequency of less than or equal
to 20% or
10%.
[0048] In one aspect, any marker of the invention is linked (genetically
and physically) to
any other marker that is at or less than 50 cM distant. In another aspect, any
marker of the
invention is closely linked (genetically and physically) to any other marker
that is in close
proximity, e.g., at or less than 10 cM distant. Two closely linked markers on
the same
chromosome can be positioned 9, 8, 7, 6, 5, 4, 3, 2, 1, 0.75, 0.5 or 0.25 cM
or less from each
other.
[0049] The phrase "Charcoal Rot" refers to the plant disease caused by an
infection of the
plant with the fungal pathogen Macrophomina phaseolina. While Charcoal Rot is
more
common in the presence of low-available water growth conditions, it can exist
even in the
absence of such growth conditions.
[0050] The phrase "Charcoal Rot," "Charcoal Rot Drought Complex," or
"CRDC" refers
to a condition in a plant in which the disease caused by an infection with the
fungal pathogen
Macrophomina phaseolina interacts with low-available water growth conditions
to subdue
the plant. It is a combination of the infection of the fungus and the low-
available water
conditions that are most commonly encountered under field conditions. Under
these field
conditions, the plant is stressed by both the pathogen and environment and is
subdued by the
two stresses operating substantially simultaneously.
[0051] "Tolerance" or "improved tolerance" in a soybean plant to Charcoal
Rot Drought
Complex is an indication that the soybean plant is less affected with respect
to yield and/or
survivability or other relevant agronomic measures, upon introduction of the
causative agents
13
Date Recue/Date Received 2023-12-08
of that disease, e.g., Macrophomina infection and low-available water growth
conditions.
"Tolerance" or "improved tolerance" in a soybean plant to Macrophornina
infection is an
indication that the soybean plant is less affected with respect to yield
and/or survivability or
other relevant agronomic measures, upon infection of the plant with
Macrophomina species,
than a less tolerant or more "susceptible" plant. "Tolerance" or "improved
tolerance" in a
soybean plant to low-available water growth conditions is an indication that
the soybean plant
is less affected with respect to yield and/or survivability or other relevant
agronomic
measures, when faced with low-available water growth conditions or less-than-
ideal
hydration conditions, than a less tolerant or more "susceptible" plant.
Tolerance is a relative
term, indicating that the infected plant produces better yield of soybean than
another,
similarly treated, more susceptible plant. That is, the conditions cause a
reduced decrease in
soybean survival and/or yield in a tolerant soybean plant, as compared to a
susceptible
soybean plant.
[0052] One of skill will appreciate that soybean plant tolerance to
Charcoal Rot Drought
Complex varies widely, can represent a spectrum of more tolerant or less
tolerant phenotypes,
and can vary depending on the severity of the infection. However, by simple
observation, one
of skill can determine the relative tolerance or susceptibility of different
plants, plant lines or
plant families to Charcoal Rot Drought Complex, and furthermore, will also
recognize the
phenotypic gradations of "tolerant."
[0053] Ratings are assigned by evaluating all plants of a cultivar in a 2
row by 15 foot
plot. Cultivar scores are based on a 1 to 9 system where a score of '9' would
indicate that all
plants in the plot are normal with no disease symptoms and a score of '1'
would indicate that
all plants in the plot are dead from disease. The experiments described herein
score soybean
tolerance to Charcoal Rot Drought Complex using the following scale: 9¨no
disease
symptoms with normal plant growth; 8=very slight symptoms including up to a
10%
reduction in leaflet and overall canopy size with no wilting; 7¨wilting
beginning to appear at
the uppermost two nodes; 6¨wilting at the uppermost three nodes and leaflet
yellowing
beginning appear; 5=Up to 5% plant death with wilting and yellowing of
leaflets occurring at
the uppermost four nodes; 4=Up to 10% plant death with wilting and yellowing
of leaflets
occurring at the uppermost four nodes; 3¨Up to 25% plant death with wilting
and yellowing
of leaflets occurring at the uppermost four nodes; 2=up to 50% plant death;
1=50-100% plant
death. FIG. 8 gives a representative example of cultivars with vastly
different Charcoal Rot
Drought Complex tolerance using this scoring system.
[0054] Charcoal Rot Drought Complex "tolerance" differs from Macrophomina
14
Date Recue/Date Received 2023-12-08
"resistance" in that tolerance is a measure of a soybean plant's ability to
survive and yield
soybean despite the presence of Macrophomina infection, as opposed to a
measure of the
soybean plant's ability to resist infection, just as low-available water
growth condition
tolerance describes a soybean plant's ability to survive and yield soybean
despite the
existence of low-available water growth conditions. As used in the art,
"tolerance" is
sometimes referred to as "general resistance", "rate-reducing resistance" or
"partial
resistance".
[0055] As used herein, "microsclerotia" refers to a compact mass of
mycelia with an
outer melanized rind; produced as a resting structure by some fungi, including
Macrophomina phaseolis.
[0056] As used herein, "inoculum" refers to a pathogen or its parts that
can cause
infection; that portion of individual pathogens that are brought into contact
with the host.
[0057] As used herein, "inoculate" refers to bringing a pathogen into
contact with a host
plant or plant organ.
[0058] The term "crossed" or "cross" in the context of this invention
means the fusion of
gametes via pollination to produce progeny (e.g., cells, seeds or plants). The
term
encompasses both sexual crosses (the pollination of one plant by another) and
sclfing (self-
pollination, e.g., when the pollen and ovule are from the same plant).
[0059] The term "introgression" refers to the transmission of a desired
allele of a genetic
locus from one genetic background to another. For example, introgression of a
desired allele
at a specified locus can be transmitted to at least one progeny via a sexual
cross between two
parents of the same species, where at least one of the parents has the desired
allele in its
genome. Alternatively, for example, transmission of an allele can occur by
recombination
between two donor genomes, e.g., in a fused protoplast, where at least one of
the donor
protoplasts has the desired allele in its genome. The desired allele can be,
e.g., a selected
allele of a marker, a QTL, or the like. In any case, offspring comprising the
desired allele can
be repeatedly backcrossed to a line having a desired genetic background and
selected for the
desired allele, to result in the allele becoming fixed in a selected genetic
background.
[0060] A "line" or "strain" is a group of individuals of identical
parentage that are
generally inbred to some degree and that are generally homozygous and
homogeneous at
most loci (isogenic or near isogenic). A "subline" refers to an inbred subset
of descendents
that are genetically distinct from other similarly inbred subsets descended
from the same
progenitor. Traditionally, a "subline" has been derived by inbreeding the seed
from an
individual soybean plant selected at the F3 to F5 generation until the
residual segregating loci
Date Recue/Date Received 2023-12-08
are "fixed" or homozygous across most or all loci. Commercial soybean
varieties (or lines)
are typically produced by aggregating ("bulking") the self-pollinated progeny
of a single F3
to F5 plant from a controlled cross between 2 genetically different parents.
While the variety
typically appears uniform, the self-pollinating variety derived from the
selected plant
eventually (e.g., F8) becomes a mixture of homozygous plants that can vary in
genotype at
any locus that was heterozygous in the originally selected F3 to F5 plant. In
the context of the
invention, marker-based sublines, that differ from each other based on
qualitative
polymorphism at the DNA level at one or more specific marker loci, are derived
by
genotyping a sample of seed derived from individual self-pollinated progeny
derived from a
selected F3-F5 plant. The seed sample can be genotyped directly as seed, or as
plant tissue
grown from such a seed sample. Optionally, seed sharing a common genotype at
the specified
locus (or loci) are bulked providing a subline that is genetically homogenous
at identified loci
important for a trait of interest (yield, tolerance, etc.).
[0061] An "ancestral line" is a parent line used as a source of genes
e.g., for the
development of elite lines. An "ancestral population" is a group of ancestors
that have
contributed the bulk of the genetic variation that was used to develop elite
lines.
"Descendants" are the progeny of ancestors, and may be separated from their
ancestors by
many generations of breeding. For example, elite lines are the descendants of
their ancestors.
A "pedigree structure" defines the relationship between a descendant and each
ancestor that
gave rise to that descendant. A pedigree structure can span one or more
generations,
describing relationships between the descendant and its parents, grand
parents, great-grand
parents, etc.
[0062] An "elite line" or "elite strain" is an agronomically superior line
that has resulted
from many cycles of breeding and selection for superior agronomic performance.
Numerous
elite lines are available and known to those of skill in the art of soybean
breeding. An "elite
population" is an assoitment of elite individuals or lines that can be used to
represent the state
of the art in terms of agronomically superior genotypes of a given crop
species, such as
soybean. Similarly, an "elite germplasm" or elite strain of germplasm is an
agronomically
superior germplasm, typically derived from and/or capable of giving rise to a
plant with
superior agronomic performance, such as an existing or newly developed elite
line of
soybean.
[0063] In contrast, an "exotic soybean strain" or an "exotic soybean
germplasm" is a
strain or germplasm derived from a soybean not belonging to an available elite
soybean line
or strain of germplasm. In the context of a cross between two soybean plants
or strains of
16
Date Recue/Date Received 2023-12-08
germplasm, an exotic germplasm is not closely related by descent to the elite
germplasm with
which it is crossed. Most commonly, the exotic germplasm is not derived from
any known
elite line of soybean, but rather is selected to introduce novel genetic
elements (typically
novel alleles) into a breeding program.
[0064] The term "amplifying" in the context of nucleic acid amplification
is any process
whereby additional copies of a selected nucleic acid (or a transcribed form
thereof) are
produced. Typical amplification methods include various polymerase based
replication
methods, including the polymerase chain reaction (PCR), ligase mediated
methods such as
the ligase chain reaction (LCR) and RNA polymerase based amplification (e.g.,
by
transcription) methods. An "amplicon" is an amplified nucleic acid, e.g., a
nucleic acid that is
produced by amplifying a template nucleic acid by any available amplification
method (e.g.,
PCR, LCR, transcription, or the like).
[0065] A "genomic nucleic acid" is a nucleic acid that corresponds in
sequence to a
heritable nucleic acid in a cell. Common examples include nuclear genomic DNA
and
amplicons thereof. A genomic nucleic acid is, in some cases, different from a
spliced RNA,
or a corresponding cDNA, in that the spliced RNA or cDNA is processed, e.g.,
by the
splicing machinery, to remove introns. Gcnomic nucleic acids optionally
comprise non-
transcribed (e.g., chromosome structural sequences, promoter regions, or
enhancer regions)
and/or non-translated sequences (e.g., introns), whereas spliced RNA/cDNA
typically do not
have non-transcribed sequences or introns. A "template nucleic acid" is a
nucleic acid that
serves as a template in an amplification reaction (e.g., a polymerase based
amplification
reaction such as PCR, a ligase mediated amplification reaction such as LCR, a
transcription
reaction, or the like). A template nucleic acid can be genomic in origin, or
alternatively, can
be derived from expressed sequences, e.g., a cDNA or an EST.
[0066] An "exogenous nucleic acid" is a nucleic acid that is not native to
a specified
system (e.g., a germplasm, plant, or variety), with respect to sequence,
genomic position, or
both. As used herein, the terms "exogenous" or "heterologous" as applied to
polynucleotides
or polypeptides typically refers to molecules that have been artificially
supplied to a
biological system (e.g., a plant cell, a plant gene, a particular plant
species or variety or a
plant chromosome under study) and are not native to that particular biological
system. The
terms can indicate that the relevant material originated from a source other
than a naturally
occurring source, or can refer to molecules having a non-natural
configuration, genetic
location or arrangement of parts.
[0067] In contrast, for example, a "native" or "endogenous" gene is a gene
that does not
17
Date Recue/Date Received 2023-12-08
contain nucleic acid elements encoded by sources other than the chromosome or
other genetic
element on which it is normally found in nature. An endogenous gene,
transcript or
polypeptide is encoded by its natural chromosomal locus, and not artificially
supplied to the
cell.
[00681 The term -recombinant" in reference to a nucleic acid or
polypeptide indicates
that the material (e.g., a recombinant nucleic acid, gene, polynucleotide, or
polypeptide) has
been altered by human intervention. Generally, the arrangement of parts of a
recombinant
molecule is not a native configuration, or the primary sequence of the
recombinant
polynucleotide or polypeptide has in some way been manipulated. The alteration
to yield the
recombinant material can be performed on the material within or removed from
its natural
environment or state. For example, a naturally occurring nucleic acid becomes
a recombinant
nucleic acid if it is altered, or if it is transcribed from DNA which has been
altered, by means
of human intervention performed within the cell from which it originates. A
gene sequence
open reading frame is recombinant if that nucleotide sequence has been removed
from it
natural context and cloned into any type of artificial nucleic acid vector.
Protocols and
reagents to produce recombinant molecules, especially recombinant nucleic
acids, are
common and routine in the art. In one embodiment, an artificial chromosome can
be created
and inserted into maize plants by any method known in the art (e.g., direct
transfer processes,
such as, e.g., PEG-induced DNA uptake, protoplast fusion, microinjection,
electroporation,
and microprojectile bombardment). An artificial chromosome is a piece of DNA
that can
stably replicate and segregate alongside endogenous chromosomes. It has the
capacity to
accommodate and express heterologous genes inserted therein. Integration of
heterologous
DNA into the megareplicator region (primary replication initiation site of
centromeres) or in
close proximity thereto, initiates a large-scale amplification of megabase-
size chromosomal
segments, which leads to de novo chromosome formation. See, e.g., U.S. Pat.
No. 6,077,697 .
[00691 The term recombinant can also refer to an organism that harbors
recombinant
material, e.g., a plant that comprises a recombinant nucleic acid is
considered a recombinant
plant. In some embodiments, a recombinant organism is a transgenic organism.
[00701 The term "introduced" when referring to translocating a
heterologous or
exogenous nucleic acid into a cell refers to the incorporation of the nucleic
acid into the cell
using any methodology. The term encompasses such nucleic acid introduction
methods as
"transfection", "transformation" and "transduction".
[00711 As used herein, the term "vector" is used in reference to
polynucleotide or other
18
Date Recue/Date Received 2023-12-08
molecules that transfer nucleic acid segment(s) into a cell. The term
"vehicle" is sometimes
used interchangeably with "vector". A vector optionally comprises parts which
mediate
vector maintenance and enable its intended use (e.g., sequences necessary for
replication,
genes imparting drug or antibiotic resistance, a multiple cloning site, or
operably linked
promoter/enhancer elements which enable the expression of a cloned gene).
Vectors are often
derived from plasmids, bacteriophages, or plant or animal viruses. A "cloning
vector" or
"shuttle vector" or "subcloning vector" contains operably linked parts that
facilitate
subcloning steps (e.g., a multiple cloning site containing multiple
restriction endonuclease
sites).
100721 The term "expression vector" as used herein refers to a vector
comprising
operably linked polynucleotide sequences that facilitate expression of a
coding sequence in a
particular host organism (e.g., a bacterial expression vector or a plant
expression vector).
Polynucleotide sequences that facilitate expression in prokaryotes typically
include, e.g., a
promoter, an operator (optional), and a ribosome binding site, often along
with other
sequences. Eukaryotic cells can use promoters, enhancers, termination and
polyadenylation
signals and other sequences that arc generally different from those used by
prokaryotes.
[0073] The term "transgenic plant" refers to a plant that comprises within
its cells a
heterologous polynucleotide. Generally, the heterologous polynucleotide is
stably integrated
within the genome such that the polynucleotide is passed on to successive
generations. The
heterologous polynucleotide may be integrated into the genome alone or as part
of a
recombinant expression cassette. "Transgenic" is used herein to refer to any
cell, cell line,
callus, tissue, plant part or plant, the genotype of which has been altered by
the presence of
heterologous nucleic acid including those transgenic organisms or cells
initially so altered, as
well as those created by crosses or asexual propagation from the initial
transgenic organism
or cell. The term "transgenic" as used herein does not encompass the
alteration of the genome
(chromosomal or extra-chromosomal) by conventional plant breeding methods
(e.g., crosses)
or by naturally occurring events such as random cross-fertilization, non-
recombinant viral
infection, non-recombinant bacterial transformation, non-recombinant
transposition, or
spontaneous mutation.
100741 A specified nucleic acid is "derived from" a given nucleic acid
when it is
constructed using the given nucleic acid's sequence, or when the specified
nucleic acid is
constructed using the given nucleic acid. For example, a cDNA or EST is
derived from an
expressed mRNA.
[0075] The term "genetic element" or "gene" refers to a heritable sequence
of DNA, i.e.,
19
Date Recue/Date Received 2023-12-08
a genomic sequence, with functional significance. The term "gene" can also be
used to refer
to, e.g., a cDNA and/or a mRNA encoded by a genomic sequence, as well as to
that genomic
sequence.
[0076] The term "genotype" is the genetic constitution of an individual
(or group of
individuals) at one or more genetic loci, as contrasted with the observable
trait (the
phenotype). Genotype is defined by the allele(s) of one or more known loci
that the
individual has inherited from its parents. The term genotype can be used to
refer to an
individual's genetic constitution at a single locus, at multiple loci, or,
more generally, the term
genotype can be used to refer to an individual's genetic make-up for all the
genes in its
genome. A "haplotype" is the genotype of an individual at a plurality of
genetic loci.
Typically, the genetic loci described by a haplotype are physically and
genetically linked, i.e.,
on the same chromosome segment.
[0077] The terms "phenotype", or "phenotypic trait" or "trait" refers to
one or more trait
of an organism. The phenotype can be observable to the naked eye, or by any
other means of
evaluation known in the art, e.g., microscopy, biochemical analysis, genomic
analysis, or an
assay for a particular disease resistance. In some cases, a phenotype is
directly controlled by a
single gene or genetic locus, i.e., a "single gene trait". In other cases, a
phenotype is the result
of several genes.
[0078] A "molecular phenotype" is a phenotype detectable at the level of a
population of
(one or more) molecules. Such molecules can be nucleic acids such as genomic
DNA or
RNA, proteins, or metabolites. For example, a molecular phenotype can be an
expression
profile for one or more gene products, e.g., at a specific stage of plant
development, in
response to an environmental condition or stress, etc. Expression profiles are
typically
evaluated at the level of RNA or protein, e.g., on a nucleic acid array or
"chip" or using
antibodies or other binding proteins.
[0079] The telin "yield" refers to the productivity per unit area of a
particular plant
product of commercial value. For example, yield of soybean is commonly
measured in
bushels of seed per acre or metric tons of seed per hectare per season. Yield
is affected by
both genetic and environmental factors. "Agronomics", "agronomic traits", and
"agronomic
performance" refer to the traits (and underlying genetic elements) of a given
plant variety that
contribute to yield over the course of growing season. Individual agronomic
traits include
emergence vigor, vegetative vigor, stress tolerance, disease resistance or
tolerance, herbicide
resistance, branching, flowering, seed set, seed size, seed density,
standability, threshability
and the like. Yield is, therefore, the final culmination of all agronomic
traits.
Date Recue/Date Received 2023-12-08
[0080] A "set" of markers or probes refers to a collection or group of
markers or probes,
or the data derived therefrom, used for a common purpose, e.g., identifying
soybean plants
with a desired trait (e.g., tolerance to Charcoal Rot Drought Complex).
Frequently, data
corresponding to the markers or probes, or data derived from their use, is
stored in an
electronic medium. While each of the members of a set possess utility with
respect to the
specified purpose, individual markers selected from the set as well as subsets
including some,
but not all of the markers, are also effective in achieving the specified
purpose.
[0081] A "look up table" is a table that correlates one form of data to
another, or one or
more forms of data with a predicted outcome that the data is relevant to. For
example, a look
up table can include a correlation between allele data and a predicted trait
that a plant
comprising a given allele is likely to display. These tables can be, and
typically are,
multidimensional, e.g., taking multiple alleles into account simultaneously,
and, optionally,
taking other factors into account as well, such as genetic background, e.g.,
in making a trait
prediction.
[0082] A "computer readable medium" is an information storage media that
can be
accessed by a computer using an available or custom interface. Examples
include memory
(e.g., ROM, RAM, or flash memory), optical storage media (e.g., CD-ROM),
magnetic
storage media (computer hard drives, floppy disks, etc.), punch cards, and
many others that
are commercially available. Information can be transmitted between a system of
interest and
the computer, or to or from the computer to or from the computer readable
medium for
storage or access of stored information. This transmission can be an
electrical transmission,
or can be made by other available methods, such as an IR link, a wireless
connection, or the
like.
[0083] "System instructions" are instruction sets that can be partially or
fully executed by
the system. Typically, the instruction sets are present as system software.
[0084] Specific physical map positions referenced herein throughout are to
the physical
position (bp) on the Glyma 1 Assembly reference (Schmutz, Jeremy, et al.
"Genome
sequence of the palaeopolyploid soybean." Nature 463.7278 (2010): 178-183).
[0085] Genetic map positions referenced herein throughout are to the
genetic position
(cM) on the Soybean Consensus Map 4.0 (Hyten D. L., et al., (2010) Crop Sci
50: 960-968).
H. OVERVIEW
[0086] Charcoal Rot is a disease of soybean, causing reduced plant
viability and
21
Date Recue/Date Received 2023-12-08
reductions in yield. This disease is caused by infection of the plant with
Macrophomina
phaseolina, a fungal pathogen. Though this disease is most prevalent during
low-available
water growth conditions, it can exist even in the absence of such growth
conditions. While
Macrophomina resistant plants have been previously developed, the strong
selective
pressures that resistant soybean impose on Macrophomina is likely to cause
relatively rapid
loss of resistance against races of Macrophomina that evolve to combat
resistance traits in the
resistant soybean, as has been seen with other soybean fungal pathogens, such
as Sclerotinia.
Accordingly, tolerance to Charcoal Rot and/or Macrophomina infection, in which
the plant
survives, thrives and produces high yields, despite a productive Macrophomina
infection, is
an alternate strategy to combat losses due to Charcoal Rot and/or Macrophomina
infection.
That is, there is not a strong negative selection against Macrophomina imposed
by tolerance,
because tolerant soybean plants support a productive Macrophomina infection.
[0087] Further, as plant stress caused by low-available water growth
conditions is related
to the existence and severity of Charcoal Rot and/or Macrophomina infection,
with plants
showing reduced survivability and yield from these conditions when coupled
with low-
available water growth conditions, soybean plants tolerant to low-available
water growth
conditions would show increased Charcoal Rot and/or Macrophomina infection
tolerance, as
well, and are therefore desirable. In addition, as low-available water growth
condition is itself
a major cause of loss of plant viability and yield, even in the absence of
Charcoal Rot and/or
Macrophomina infection, plants tolerant to such growth conditions are
desirable for their
direct benefits, not related to Charcoal Rot as well.
[0088] The identification and selection of soybean plants that show
tolerance to Charcoal
Rot Drought Complex using MAS can provide an effective and environmentally
friendly
approach to overcoming losses caused by this disease. The present invention
provides
soybean marker loci that demonstrate statistically significant co-segregation
with Charcoal
Rot Drought Complex tolerance. Detection of these loci or additional linked
loci can be used
in marker assisted soybean breeding programs to produce tolerant plants, or
plants with
improved tolerance. The marker loci identified herein include S29725-001;
S29741-001;
S29742-001; S11315-1; and S11316-1.
[0089] Each of the marker loci can be visualized as PCR amplicons as
descxribed herein.
The PCR primer pairs that are used to generate the marker loci amplicons
include: SEQ ID
NO: 12 and SEQ ID NO: 13 used to amplify an amplicon associated with S29725
(SEQ ID
NO.: 11); SEQ ID NO: 17 and SEQ ID NO: 18 used to amplify the amplicon
associated with
S29742 (SEQ ID NO.: 21); SEQ ID NO: 22 and SEQ ID NO: 23 used to amplify the
22
Date Recue/Date Received 2023-12-08
amplicon associated with S29741 (SEQ ID NO.: 21); SEQ ID NO: 2 and SEQ ID NO:
3 used
to amplify the amplicon associated with S11315 (SEQ ID NO.: 1); and SEQ ID NO:
7 and
SEQ ID NO: 8 used to amplify the amplicon associated with S11316 (SEQ ID NO.:
6). In
various further aspects, the marker loci can be visualized by probes such as
the group
consisting of SEQ ID NOs: 4, 5, 9, 10, 14, 15, 19, 20, 24, and 25.
[0090] The invention also provides chromosomal QTL intervals that
correlate with
Charcoal Rot Drought Complex tolerance. These intervals are located
Chromosomes 5, 15,
and 19. Any marker located within these intervals finds use as a marker for
Charcoal Rot
Drought Complex tolerance. These intervals include: (i) a chromosomal interval
located at
about 17 cM to about 38 cM of chromosome 5; (ii) a chromosomal interval
located at about 5
cM to about 26 cM of chromosome 15; (iii) a chromosomal interval located at
about 19 cM to
about 40 cM of chromosome 19; or (iv) a chromosomal interval located at about
81 cM to
about 102 cM of chromosome 19. Alternatively, these intervals can be specified
as follows:
(i) a chromosomal interval located within about 2 Mbp of an interval at 7,975-
8,015 kpb of
chromosome 5; (ii) a chromosomal interval located within about 2 Mbp of an
interval at
3,202-3,212 kbp of chromosome 15 (SEQ 1D NO.: 26); (iii) a chromosomal
interval located
within about 2 Mbp of an interval at 27,178-27,218 kbp; or (iv) a chromosomal
interval
located within about 2 Mbp of an interval at 48,340-48,380 kbp of chromosome
19 (SEQ ID
NO.: 27).
[0091] Methods for identifying soybean plants or germplasm that carry
preferred alleles
of tolerance marker loci are a feature of the invention. In these methods, any
of a variety of
marker detection protocols is used to identify marker loci, depending on the
type of marker
loci. Typical methods for marker detection include amplification and detection
of the
resulting amplified markers, e.g., by PCR, LCR, transcription based
amplification methods,
or the like. These include ASH, SSR detection, RFLP analysis and many others.
[0092] In various aspects, disclosed are methods for identifying a first
soybean plant or
germplasm (e.g., a line or variety) that has tolerance, improved tolerance, or
susceptibility to
Charcoal Rot Drought Complex. In the methods, at least one allele of one or
more marker
locus (e.g., a plurality of marker loci) that is associated with the
tolerance, improved
tolerance, or susceptibility is detected in the first soybean plant or
germplasm.
[0093] Also disclosed are methods of identifying a first soybean plant or
germplasm that
displays tolerance, improved tolerance, or susceptibility to Charcoal Rot; the
method
comprising detecting in the first soybean plant or germplasm at least one
allele of a
quantitative trait locus that is associated with the tolerance, improved
tolerance, or
23
Date Recue/Date Received 2023-12-08
susceptibility; wherein the quantitative locus is: (i) a chromosomal interval
located at about
17 cM to about 38 cM of chromosome 5; (ii) a chromosomal interval located at
about 5 cM to
about 26 cM of chromosome 15; (iii) a chromosomal interval located at about 19
cM to about
40 cM of chromosome 19; or (iv) a chromosomal interval located at about 81 cM
to about
102 cM of chromosome 19.
[0094] Also disclosed are methods of identifying a first soybean plant or
germplasm that
displays tolerance, improved tolerance, or susceptibility to Charcoal Rot; the
method
comprising the steps of: (a) detecting in the first soybean plant or germplasm
at least one
allele of a quantitative trait locus that is associated with the tolerance,
improved tolerance, or
susceptibility; wherein the quantitative locus is: wherein the quantitative
locus is: (i) a
chromosomal interval located at about 17 cM to about 38 cM of chromosome 5;
(ii) a
chromosomal interval located at about 5 cM to about 26 cM of chromosome 15;
(iii) a
chromosomal interval located at about 19 cM to about 40 cM of chromosome 19;
or (iv) a
chromosomal interval located at about 81 cM to about 102 cM of chromosome 19;
(b)
selecting the first soybean plant or germplasm, or selecting a progeny of the
first soybean
plant or germplasm comprising the at least onc allele of a quantitative trait
locus that is
associated with the tolerance, improved tolerance, or susceptibility; and (c)
crossing the
selected first soybean plant or germplasm with a second soybean plant or
germplasm to
introgress the quantitative trait locus into progeny soybean germplasm.
[0095] Also disclosed are methods of identifying a first soybean plant or
germplasm that
displays tolerance, improved tolerance, or susceptibility to Charcoal Rot; the
method
comprising the steps of: (a) detecting in the first soybean plant or germplasm
at least one
allele of a quantitative trait locus that is associated with the tolerance,
improved tolerance, or
susceptibility; wherein the quantitative locus is: wherein the quantitative
locus is: (i) a
chromosomal interval located at about 17 cM to about 38 cM of chromosome 5;
(ii) a
chromosomal interval located at about 5 cM to about 26 cM of chromosome 15;
(iii) a
chromosomal interval located at about 19 cM to about 40 cM of chromosome 19;
or (iv) a
chromosomal interval located at about 81 cM to about 102 cM of chromosome 19;
(b)
selecting the first soybean plant or germplasm, or selecting a progeny of the
first soybean
plant or germplasm comprising the at least one allele of a quantitative trait
locus that is
associated with the tolerance, improved tolerance, or susceptibility; (c)
crossing the selected
first soybean plant or germplasm with a second soybean plant or germplasm to
introgress the
quantitative trait locus into progeny soybean germplasm; (d) analyzing progeny
soybean
germplasm to determine the presence of tolerance to Charcoal Rot; and (d)
selecting progeny
24
Date Recue/Date Received 2023-12-08
soybean germplasm that tests positive for the presence of tolerance to
Charcoal Rot as being
soybean germplasm into which germplasm having said quantitative trait locus
has been
introgressed.
[0096] In various aspects, the quantitative trait locus is localized at a
chromosomal
interval of about 18 cM to about 37 cM of chromosome 5. In a further aspect,
the
quantitative trait locus is localized at a chromosomal interval of about 16 cM
to about 35 cM
of chromosome 5. In a still further aspect, the quantitative trait locus is
localized at a
chromosomal interval of about 25.8 cM to about 29.9 cM of chromosome 5. In a
yet further
aspect, the quantitative trait locus is localized at a chromosomal interval of
about 26.3 cM to
about 29.4 cM of chromosome 5. In an even further aspect, the quantitative
trait locus is
localized at a chromosomal interval of about 26.8 cM to about 28.9 cM of
chromosome 5. In
a still further aspect, the quantitative trait locus is localized at a
chromosomal interval of
about 27.3 cM to about 29.4 cM of chromosome 5.
[0097] In various aspects, wherein the quantitative trait locus is
localized at a
chromosomal interval of about 6 cM to about 25 cM of chromosome 15. In a
further aspect,
the quantitative trait locus is localized at a chromosomal interval of about 8
cM to about 23
cM of chromosome 15. In a still further aspect, the quantitative trait locus
is localized at a
chromosomal interval of about 13.5 cM to about 17.5 cM of chromosome 15. In a
yet further
aspect, the quantitative trait locus is localized at a chromosomal interval of
about 14 cM to
about 17 cM of chromosome 15. In an even further aspect, the quantitative
trait locus is
localized at a chromosomal interval of about 14.5 cM to about 16.5 cM of
chromosome 15.
In a still further aspect, the quantitative trait locus is localized at a
chromosomal interval of
about 15 cM to about 16 cM of chromosome 15. In a still further aspect, the
quantitative trait
locus is localized at a chromosomal interval of about 15.25 cM to about 15.75
cM of
chromosome 15.
[0098] In various aspects, a marker locus of the quantitative trait locus
on chromosome
15 is S29725-001. In a further aspect, a marker locus of the quantitative
trait locus on
chromosome 15 is S29742-001. In a still further aspect, a marker locus of the
quantitative
trait locus on chromosome 15 is S29741-001.
[0099] In various aspects, the quantitative trait locus on chromosome 15
is flanked by and
including S29725-001 and S29741-001. In a further aspect, the quantitative
trait locus on
chromosome 15 is flanked by and including S29725-001 and S29742-001.
[00100] In various aspects, the quantitative trait locus is localized at a
chromosomal
interval of about 20 cM to about 39 cM of chromosome 19. In a further aspect,
the
Date Recue/Date Received 2023-12-08
quantitative trait locus is localized at a chromosomal interval of about 22 cM
to about 37 cM
of chromosome 19. In a still further aspect, the quantitative trait locus is
localized at a
chromosomal interval of about 27.3 cM to about 31.4 cM of chromosome 19. In a
yet further
aspect, the quantitative trait locus is localized at a chromosomal interval of
about 27.8 cM to
about 30.9 cM of chromosome 19. In an even further aspect, the quantitative
trait locus is
localized at a chromosomal interval of about 28.3 cM to about 30.4 cM of
chromosome 19.
In a still further aspect, the quantitative trait locus is localized at a
chromosomal interval of
about 28.8 cM to about 29.9 cM of chromosome 19.
[001011 In various aspects, the quantitative trait locus is localized at a
chromosomal
interval of about 82 cM to about 101 cM of chromosome 19. In a further aspect,
the
quantitative trait locus is localized at a chromosomal interval of about 84 cM
to about 99 cM
of chromosome 19. In a still further aspect, the quantitative trait locus is
localized at a
chromosomal interval of about 86 cM to about 97 cM of chromosome 19. In a yet
further
aspect, the quantitative trait locus is localized at a chromosomal interval of
about 90.1 cM to
about 93.1 cM of chromosome 19. In an even further aspect, the quantitative
trait locus is
localized at a chromosomal interval of about 90.6 cM to about 92.5 cM of
chromosome 19.
In a still further aspect, the quantitative trait locus is localized at a
chromosomal interval of
about 91.0 cM to about 92.2 cM of chromosome 19. In a yet further aspect, the
quantitative
trait locus is localized at a chromosomal interval of about 91.2 cM to about
92.0 cM of
chromosome 19.
[00102] In various aspects, a marker locus of the quantitative trait locus on
chromosome
19 is S11315-1. In a further aspect, a marker locus of the quantitative trait
locus on
chromosome 19 is S11316-1.
[00103] In various aspects, the quantitative trait locus on chromosome 15
is flanked by and
including S11315-1 and S11316-1.
[00104] Although particular marker alleles can show co-segregation with a
disease
tolerance or susceptibility phenotype, it is important to note that the marker
locus is not
necessarily part of the QTL locus responsible for the tolerance or
susceptibility. For example,
it is not a requirement that the marker polynucleotide sequence be part of a
gene that imparts
disease resistance (for example, be part of the gene open reading frame). The
association
between a specific marker allele with the tolerance or susceptibility
phenotype is due to the
original "coupling" linkage phase between the marker allele and the QTL
tolerance or
susceptibility allele in the ancestral soybean line from which the tolerance
or susceptibility
allele originated. Eventually, with repeated recombination, crossing over
events between the
26
Date Recue/Date Received 2023-12-08
marker and QTL locus can change this orientation. For this reason, the
favorable marker
allele may change depending on the linkage phase that exists within the
tolerant parent used
to create segregating populations. This does not change the fact that the
genetic marker can
be used to monitor segregation of the phenotype. It only changes which marker
allele is
considered favorable in a given segregating population.
[00105] Identification of soybean plants or germplasm that include a marker
locus or
marker loci linked to a tolerance trait or traits provides a basis for
performing marker assisted
selection of soybean. Soybean plants that comprise favorable markers or
favorable alleles are
selected for, while soybean plants that comprise markers or alleles that are
negatively
correlated with tolerance can be selected against. Desired markers and/or
alleles can be
introgressed into soybean having a desired (e.g., elite or exotic) genetic
background to
produce an introgressed tolerant soybean plant or germplasm. In some aspects,
it is
contemplated that a plurality of tolerance markers are sequentially or
simultaneous selected
and/or introgressed. The combinations of tolerance markers that are selected
for in a single
plant is not limited, and can include any combination of markers recited in
FIG. 1, any
markers linked to the markers recited in FIG. 1, or any markers located within
the QTL
intervals defined herein.
[00106] Various methods are known in the art for determining (and measuring)
the
tolerance of a soybean plant to Charcoal Rot Drought Complex. They describe a
tolerance
measurement scale of 1-9, with 9=no disease and 1=total necrosis caused by
Macrophomina
phaseolina. It will be appreciated that all such scales are relative and that
numbering and
precise correlation to any scale can be performed at the discretion of the
practitioner.
[00107] Typically, individual field tests are monitored for Charcoal Rot
symptoms during
the middle to late vegetative stages, but such symptoms typically appear in
the early
reproductive stage (during flowering and early pod set). Data collection is
usually done in 3
or 4 successive scorings about 7 days apart. Scorings continue until worsening
symptoms can
no longer be quantified or until the symptoms are confounded by other factors
such as other
diseases, insect pressure, severe weather, or advancing maturity.
[00108] In general, while there is a certain amount of subjectivity to
assigning severity
measurements for disease caused symptoms, assignment to a given scale as noted
above is
well within the skill of a practitioner in the field. Measurements can also be
averaged across
multiple scorers to reduce variation in field measurements. Furthermore,
although protocols
using artificial inoculation of field nurseries with Macrophomina phaseolina
can certainly be
used in assessing tolerance, it is also typical for tolerance ratings to be
based on actual field
27
Date Recue/Date Received 2023-12-08
observations of fortuitous natural disease incidence, with the information
corresponding to
disease incidence for a cultivar being averaged over many locations and,
typically, several
years of crop growing.
[00109] If there is no disease present, the rating system above is
inapplicable, because
everything in an uninfected field scores as tolerant. However, if Charcoal Rot
does occur in a
specific field location, all of the lines at that location can be scored as
noted above. These
scores can accumulate over locations and years to show disease tolerance for
given cultivars.
Thus, older lines can have more years of observation than newer ones etc.
However, relative
measurements can easily be made using the scoring system noted above.
Furthermore, the
tolerance ratings can be updated and refined each year based on the previous
year's
observations in the field. Based on this, Charcoal Rot scores for a cultivar
are relative
measurements of tolerance.
[00110] The experiments described herein score soybean tolerance to Charcoal
Rot
Drought Complex using the following scale: 9¨no disease symptoms with normal
plant
growth; 8=very slight symptoms including up to a 10% reduction in leaflet and
overall
canopy size with no wilting; 7=wilting beginning to appear at the uppermost
two nodes;
6=wilting at the uppermost three nodes and leaflet yellowing beginning appear;
5=Up to 5%
plant death with wilting and yellowing of leaflets occurring at the uppermost
four nodes;
4=Up to 10% plant death with wilting and yellowing of leaflets occurring at
the uppermost
four nodes; 3=Up to 25% plant death with wilting and yellowing of leaflets
occurring at the
uppermost four nodes; 2=up to 50% plant death; 1=50-100% plant death. FIG. 8
gives a
representative example of cultivars with vastly different Charcoal Rot Drought
Complex
tolerance using this scoring system.
[00111] Tolerance assays are useful to verify that the tolerance trait
still segregates with
the marker in any particular plant or population, and, of course, to measure
the degree of
tolerance improvement achieved by introgressing or recombinantly introducing
the trait into a
desired background.
[00112] Systems, including automated systems for selecting plants that
comprise a marker
of interest and/or for correlating presence of the marker with tolerance are
also a feature of
the invention. These systems can include probes relevant to marker locus
detection, detectors
for detecting labels on the probes, appropriate fluid handling elements and
temperature
controllers that mix probes and templates and/or amplify templates, and
systems instructions
that correlate label detection to the presence of a particular marker locus or
allele.
[00113] Also disclosed are introgressed soybean plants or germplasms produced
by the
28
Date Recue/Date Received 2023-12-08
disclosed methods.
[00114] Kits are also a feature of the invention. For example, a kit can
include appropriate
primers or probes for detecting tolerance associated marker loci and
instructions in using the
primers or probes for detecting the marker loci and correlating the loci with
predicted
Charcoal Rot Drought Complex tolerance. The kits can further include packaging
materials
for packaging the probes, primers or instructions, controls such as control
amplification
reactions that include probes, primers or template nucleic acids for
amplifications, molecular
size markers, or the like.
[00115] Also disclosed are kits for selecting at least one soybean plant by
marker assisted
selection of a quantitative trait locus associated with the tolerance,
improved tolerance, or
susceptibility to Charcoal Rot comprising: (a) labeled primers or probes for
detecting at least
one nucleic acid sequence selected from the group consisting of: (i) 48,340-
48,380 kbp of
chromosome 19 (SEQ ID NO.: 27); (ii) 3,202-3,212 kbp of chromosome 15 (SEQ ID
NO.:
26); (iii) 511315 (SEQ ID NO.: 1); (iv) S11316 (SEQ ID NO.: 6); (v) S29725
(SEQ ID NO:
11); (vi) S29742 (SEQ ID NO: 16); and (vii) S29741 (SEQ ID NO: 21); and (b)
instructions
for using the primers or probes to detect the marker loci and correlating the
loci with
predicted improved lodging resistance.
[00116] In various aspects, the labeled primers of the kit comprise a pair of
olignucleotides
selected from the group consisting of: (a) SEQ ID NO: 2 and SEQ ID NO: 3; (b)
SEQ ID
NO: 7 and SEQ ID NO: 8; (c) SEQ ID NO: 12 and SEQ ID NO: 13; (d) SEQ ID NO: 17
and
SEQ ID NO: 18; and (e) SEQ ID NO: 22 and SEQ ID NO: 23; wherein at least one
of the
oligonucleotides is linked to a detectable label.
[00117] In various aspects, the labeled probes of the kit comprise an
oligonucleotide
selected from the group consisting of: SEQ ID NOs: 4, 5, 9, 10, 14, 15, 19,
20, 24, and 25;
and wherein the oligonucleotide is linked to a detectable label.
HI. TOLERANCE MARKERS AND FAVORABLE ALLELES
[00118] In traditional linkage analysis, no direct knowledge of the
physical relationship of
genes on a chromosome is required. Mendel's first law is that factors of pairs
of characters are
segregated, meaning that alleles of a diploid trait separate into two gametes
and then into
different offspring. Classical linkage analysis can be thought of as a
statistical description of
the relative frequencies of cosegregation of different traits. Linkage
analysis is the well
characterized descriptive framework of how traits are grouped together based
upon the
29
Date Recue/Date Received 2023-12-08
frequency with which they segregate together. That is, if two non-allelic
traits are inherited
together with a greater than random frequency, they are said to be "linked".
The frequency
with which the traits are inherited together is the primary measure of how
tightly the traits are
linked, i.e., traits which are inherited together with a higher frequency are
more closely linked
than traits which are inherited together with lower (but still above random)
frequency. Traits
are linked because the genes which underlie the traits reside on the same
chromosome. The
further apart on a chromosome the genes reside, the less likely they are to
segregate together,
because homologous chromosomes recombine during meiosis. Thus, the further
apart on a
chromosome the genes reside, the more likely it is that there will be a
crossing over event
during meiosis that will result in two genes segregating separately into
progeny.
[00119] A common measure of linkage is the frequency with which traits
cosegregate.
This can be expressed as a percentage of cosegregation (recombination
frequency) or, also
commonly, in centiMorgans (cM). The cM is named after the pioneering
geneticist Thomas
Hunt Morgan and is a unit of measure of genetic recombination frequency. One
cM is equal
to a 1% chance that a trait at one genetic locus will be separated from a
trait at another locus
due to crossing over in a single generation (meaning the traits segregate
together 99% of the
time). Because chromosomal distance is approximately proportional to the
frequency of
crossing over events between traits, there is an approximate physical distance
that correlates
with recombination frequency. For example, in soybean, 1 cM correlates, on
average, to
about 400,000 base pairs (400 Kb).
[00120] Marker loci are themselves traits and can be assessed according to
standard
linkage analysis by tracking the marker loci during segregation. Thus, in the
context of the
present invention, one cM is equal to a 1% chance that a marker locus will be
separated from
another locus (which can be any other trait, e.g., another marker locus, or
another trait locus
that encodes a QTL), due to crossing over in a single generation. The marker
loci, including
S29725-001; S29741-001; S29742-001; S11315-1; and S11316-1, as well as any of
the
chromosome intervals: (i) a chromosomal interval located at about 17 cM to
about 38 cM of
chromosome 5; (ii) a chromosomal interval located at about 5 cM to about 26 cM
of
chromosome 15; (iii) a chromosomal interval located at about 19 cM to about 40
cM of
chromosome 19; or (iv) a chromosomal interval located at about 81 cM to about
102 cM of
chromosome 19; have been found to correlate with tolerance, improved tolerance
or
susceptibility to Charcoal Rot Drought Complex in soybean. Alternatively,
these intervals
can be specified as follows: (i) a chromosomal interval located within about 2
Mbp of an
interval at 7,975-8,015 kpb of chromosome 5; (ii) a chromosomal interval
located within
Date Recue/Date Received 2023-12-08
about 2 Mbp of an interval at 3,202-3,212 kbp of chromosome 15 (SEQ ID NO.:
26); (iii) a
chromosomal interval located within about 2 Mbp of an interval at 27,178-
27,218 kbp; or (iv)
a chromosomal interval located within about 2 Mbp of an interval at 48,340-
48,380 kbp of
chromosome 19 (SEQ ID NO.: 27).
[00121] The marker loci S29725-001; S29741-001; S29742-001; S11315-1; and
S11316-1
are localized as specified in Table 1 below.
Table 1.
No. Loci Name Chromosome Physical Map Genetic Map
Position* Position**
1 S29725-001 15 2,938,271 15.07
2 S29741-001 15 3,210,335 15.75
3 S29742-001 15 3,211,837 16.72
4 S11315-1 19 48,354,468 91.43
S11316-1 19 48,384,426 91.53
* Physical position (bp) on the Glyma 1 Assembly reference (Schmutz, Jeremy,
et al.
"Genome sequence of the palaeopolyploid soybean." Nature 463.7278 (2010): 178-
183).
** Genetic position (cM) on the Soybean Consensus Map 4.0 (Hyten D. L., et
al.,
(2010) Crop Sci 50: 960-968).
1001221 This means that the markers are sufficiently proximal to a tolerance
trait that they
can be used as a predictor for the tolerance trait. This is extremely useful
in the context of
marker assisted selection (MAS), discussed in more detail herein. In brief,
soybean plants or
germplasm can be selected for markers or marker alleles that positively
correlate with
tolerance, without actually raising soybean and measuring for tolerance or
improved
tolerance (or, contrarily, soybean plants can be selected against if they
possess markers that
negatively correlate with tolerance or improved tolerance). MAS is a powerful
shortcut to
selecting for desired phenotypes and for introgressing desired traits into
cultivars of soybean
(e.g., introgressing desired traits into elite lines). MAS is easily adapted
to high throughput
molecular analysis methods that can quickly screen large numbers of plant or
germplasm
genetic material for the markers of interest and is much more cost effective
than raising and
observing plants for visible traits.
1001231 When referring to the relationship between two genetic elements, such
as a
31
Date Recue/Date Received 2023-12-08
genetic element contributing to tolerance and a proximal marker, "coupling"
phase linkage
indicates the state where the "favorable" allele at the tolerance locus is
physically associated
on the same chromosome strand as the "favorable" allele of the respective
linked marker
locus. In coupling phase, both favorable alleles are inherited together by
progeny that inherit
that chromosome strand. In "repulsion" phase linkage, the "favorable" allele
at the locus of
interest (e.g., a QTL for tolerance) is physically linked with an
"unfavorable" allele at the
proximal marker locus, and the two "favorable" alleles are not inherited
together (i.e., the two
loci are "out of phase" with each other).
[00124] A favorable allele of a marker is that allele of the marker that co-
segregates with a
desired phenotype (e.g., disease tolerance). As used herein, a QTL marker has
a minimum of
one favorable allele, although it is possible that the marker might have two
or more favorable
alleles found in the population. Any favorable allele of that marker can be
used
advantageously for the identification and construction of tolerant soybean
lines. Optionally,
one, two, three or more favorable allele(s) of different markers are
identified in, or
introgresscd into a plant, and can be selected for or against during MAS.
Desirably, plants or
gcrmplasm arc identified that have at least one such favorable allele that
positively correlates
with tolerance or improved tolerance.
[00125] Alternatively, a marker allele that co-segregates with disease
susceptibility also
finds use with the invention, since that allele can be used to identify and
counter select
disease-susceptible plants. Such an allele can be used for exclusionary
purposes during
breeding to identify alleles that negatively correlate with tolerance, to
eliminate susceptible
plants or geimplasm from subsequent rounds of breeding.
[00126] In some embodiments of the invention, a plurality of marker alleles
are
simultaneously selected for in a single plant or a population of plants. In
these methods,
plants are selected that contain favorable alleles from more than one
tolerance marker, or
alternatively, favorable alleles from more than one tolerance marker are
introgressed into a
desired soybean germplasm. One of skill in the art recognizes that the
simultaneous selection
of favorable alleles from more than one disease tolerance marker in the same
plant is likely to
result in an additive (or even synergistic) protective effect for the plant.
[00127] One of skill recognizes that the identification of favorable marker
alleles is
germplasm-specific. The determination of which marker alleles correlate with
tolerance (or
susceptibility) is determined for the particular germplasm under study. One of
skill
recognizes that methods for identifying the favorable alleles are routine and
well known in
the art, and furthermore, that the identification and use of such favorable
alleles is well within
32
Date Recue/Date Received 2023-12-08
the scope of the invention. Furthermore still, identification of favorable
marker alleles in
soybean populations other than the populations used or described herein is
well within the
scope of the invention.
[00128] The PCR primer pairs that are used to generate the marker loci
amplicons include:
SEQ ID NO: 12 and SEQ ID NO: 13 used to amplify an amplicon associated with
S29725
(SEQ ID NO.: 11); SEQ ID NO: 17 and SEQ ID NO: 18 used to amplify the amplicon
associated with S29742 (SEQ ID NO.: 21); SEQ ID NO: 22 and SEQ ID NO: 23 used
to
amplify the amplicon associated with S29741 (SEQ ID NO.: 21); SEQ ID NO: 2 and
SEQ ID
NO: 3 used to amplify the amplicon associated with S11315 (SEQ ID NO.: 1); and
SEQ ID
NO: 7 and SEQ ID NO: 8 used to amplify the amplicon associated with S11316
(SEQ ID
NO.: 6), are a feature of the invention. Another feature of the invention are
probes that are
can be used to genotype the marker loci, and these probes include the group
consisting of
SEQ ID NOs: 4, 5, 9, 10, 14, 15, 19, 20, 24, and 25. However, one of skill
will immediately
recognize that other sequences to either side of the given primers can be used
in place of the
given primers, so long as the primers can amplify a region that includes the
allele to be
detected. Further, it will be appreciated that the precise probe to be used
for detection can
vary, e.g., any probe that can identify the region of a marker amplicon to be
detected can be
substituted for those examples provided herein. Further, the configuration of
the
amplification primers and detection probes can, of course, vary. Thus, the
invention is not
limited to the primers and probes specifically recited herein.
[00129] In
some aspects, methods of the invention utilize an amplification step to
detect/genotype a marker locus. However, it will be appreciated that
amplification is not a
requirement for marker detection _______________________________________ for
example, one can directly detect unamplified genomic
DNA simply by performing a Southern blot on a sample of genomic DNA.
Procedures for
performing Southern blotting, amplification (PCR, LCR, or the like) and many
other nucleic
acid detection methods are well established and are taught, e.g., in Sambrook,
et al., (2000)
Molecular Cloning ______________________________________________________ A
Laboratory Manual (3rd Ed.), Vol. 1-3, Cold Spring Harbor
Laboratory, Cold Spring Harbor, N.Y., ("Sambrook"); Current Protocols in
Molecular
Biology, Ausubel, et al., eds., Current Protocols, a joint venture between
Greene Publishing
Associates, Inc. and John Wiley & Sons, Inc., (supplemented through 2002)
("Ausubel")) and
PCR Protocols A Guide to Methods and Applications (Innis, et al., eds)
Academic Press Inc.
San Diego, Calif. (1990) (Innis). Additional details regarding detection of
nucleic acids in
plants can also be found, e.g., in Plant Molecular Biology (1993) Croy (ed.)
BIOS Scientific
Publishers, Inc.
33
Date Recue/Date Received 2023-12-08
[00130]
Separate detection probes can also be omitted in amplification/detection
methods,
e.g., by performing a real time amplification reaction that detects product
formation by
modification of the relevant amplification primer upon incorporation into a
product,
incorporation of labeled nucleotides into an amplicon, or by monitoring
changes in molecular
rotation properties of amplicons as compared to unamplified precursors (e.g.,
by fluorescence
polarization).
[00131] Typically, molecular markers are detected by any established method
available in
the art, including, without limitation, allele specific hybridization (ASH) or
other methods for
detecting single nucleotide polymorphisms (SNP), amplified fragment length
polymorphism
(AFLP) detection, amplified variable sequence detection, randomly amplified
polymorphic
DNA (RAPD) detection, restriction fragment length polymorphism (RFLP)
detection, self-
sustained sequence replication detection, simple sequence repeat (SSR)
detection, single-
strand conformation polymorphisms (SSCP) detection, isozyme markers detection,
or the
like. Any of the aforementioned marker types can be employed in the context of
the invention
to identify chromosome segments encompassing genetic element that contribute
to superior
agronomic performance (e.g., tolerance or improved tolerance).
IV. QTL CHROMOSOME INTERVALS
[00132] In some aspects, the invention provides QTL chromosome intervals,
where a QTL
(or multiple QTLs) that segregate with Charcoal Rot Drought Complex tolerance
are
contained in those intervals. A variety of methods well known in the art are
available for
identifying chromosome intervals, including those methods described herein.
The boundaries
of such chromosome intervals arc drawn to encompass markers that will be
linked to one or
more QTL. In other words, the chromosome interval is drawn such that any
marker that lies
within that interval (including the terminal markers that define the
boundaries of the interval)
can be used as markers for disease tolerance. Each interval comprises at least
one QTL, and
furthermore, may indeed comprise more than one QTL. Close proximity of
multiple QTL in
the same interval may obfuscate the correlation of a particular marker with a
particular QTL,
as one marker may demonstrate linkage to more than one QTL. Conversely, e.g.,
if two
markers in close proximity show co-segregation with the desired phenotypic
trait, it is
sometimes unclear if each of those markers identifying the same QTL or two
different QTL.
Regardless, knowledge of how many QTL are in a particular interval is not
necessary to make
or practice the invention.
34
Date Recue/Date Received 2023-12-08
[00133] The present invention provides soybean chromosome intervals, where the
markers
within that interval demonstrate co-segregation with tolerance to Charcoal Rot
Drought
Complex. Thus, each of these intervals comprises at least one Charcoal Rot
Drought
Complex tolerance QTL. These intervals include: (i) a chromosomal interval
located at about
17 cM to about 38 cM of chromosome 5; (ii) a chromosomal interval located at
about 5 cM to
about 26 cM of chromosome 15; (iii) a chromosomal interval located at about 19
cM to about
40 cM of chromosome 19; or (iv) a chromosomal interval located at about 81 cM
to about
102 cM of chromosome 19. Alternatively, these intervals can be specified as
follows: (i) a
chromosomal interval located within about 2 Mbp of an interval at 7,975-8,015
kpb of
chromosome 5; (ii) a chromosomal interval located within about 2 Mbp of an
interval at
3,202-3,212 kbp of chromosome 15 (SEQ ID NO.: 26); (iii) a chromosomal
interval located
within about 2 Mbp of an interval at 27,178-27,218 kbp; or (iv) a chromosomal
interval
located within about 2 Mbp of an interval at 48,340-48,380 kbp of chromosome
19 (SEQ ID
NO.: 27).
[00134] Each of the intervals described above shows a clustering of markers
that co-
segregate with Charcoal Rot Drought Complex tolerance. This clustering of
markers occurs
in relatively small domains on the linkage groups, indicating the presence of
one or more
QTL in those chromosome regions. QTL intervals were drawn to encompass the
markers that
co-segregate with tolerance. The intervals are defined by the markers on their
termini, where
the interval encompasses all the markers that map within the interval as well
as the markers
that define the termini.
[00135] In some cases, an interval can be drawn, where the interval is defined
by linkage
to a particular marker locus. For example, an interval on Chr. 15 can be
defined where any
marker that is linked to the marker S29725-001, S29741-001, and/or S29742-001
is a
member of that interval. For example, as used here, linkage is defined as any
marker that is
within 25 cM from S29725-001, S29741-001, and/or S29742-001. In other aspects,
an
interval on Chr. 15 can be defined where any marker that is linked to the
marker S11315-1
and/or S11316-1 is a member of that interval. For example, as used here,
linkage is defined as
any marker that is within 25 cM from S11315-1 and/or S11316-1.
[00136] As described above, an interval (e.g., a chromosome interval or a QTL
interval)
need not depend on an absolute measure of interval size such as a centimorgans
value. An
interval can be described by the terminal markers that define the endpoints of
the interval,
and typically the interval will include the terminal markers that define the
extent of the
interval. An interval can include any marker localizing within that chromosome
domain,
Date Recue/Date Received 2023-12-08
whether those markers are currently known or unknown.
[00137] In
situations where the interval is close to or comprises one end of the linkage
group, the interval can be described by one marker, for example the interval
on Chr. 15 can
be described as including marker S29741-001 and below, or for example the
interval on Chr.
15 can be described as including marker S29741-001 and below. In various
further aspect,
the interval on Chr. 15 can be described as including marker S29725-001 and
above. In a
further aspect, the interval on Chr. 15 can be described as flanked by and
including S29725-
001 and S29741-001. In a still further aspect, the interval on Chr. 15 can be
described as
flanked by and including S29725-001 and S29742-001.
[00138] In various aspects, the interval can be described by one marker, for
example the
interval on Chr. 19 can be described as including marker S11315-1 and above,
or for example
the interval on Chr. 19 can be described as including marker S11316-1 and
below. In a
further aspect, the interval on Chr. 19 can be described as flanked by and
including S11315-1
and S13116-1.
[00139] and above, where "above" and "below" are the terms commonly used in
the art to
describe the marker's position relative to the distal end (position zero),
with above being
closer to position zero. The invention provides a variety of means for
defining a chromosome
interval, for example, the marker loci provided in the genetic map in FIG. 6,
in the lists of
linked markers of FIG. 5, and in references cited herein (e.g., Song, et al.,
(2004) "A New
Integrated Genetic Linkage Map of the Soybean" Theor Appl Genet 109:122-128).
V. GENETIC MAPS
[00140] As one of skill in the art will recognize, recombination frequencies
(and as a
result, genetic map positions) in any particular population are not static.
The genetic distances
separating two markers (or a marker and a QTL) can vary depending on how the
map
positions are determined. For example, variables such as the parental mapping
populations
used, the software used in the marker mapping or QTL mapping, and the
parameters input by
the user of the mapping software can contribute to the QTL/marker genetic map
relationships.
However, it is not intended that the invention be limited to any particular
mapping
populations, use of any particular software, or any particular set of software
parameters to
determine linkage of a particular marker or chromosome interval with the
Charcoal Rot
Drought Complex tolerance phenotype. It is well within the ability of one of
ordinary skill in
the art to extrapolate the novel features described herein to any soybean gene
pool or
36
Date Recue/Date Received 2023-12-08
population of interest, and using any particular software and software
parameters. Indeed,
observations regarding tolerance markers and chromosome intervals in
populations in
additions to those described herein are readily made using the teaching of the
present
disclosure.
[00141] Any suitable soybean strains can be used to generate mapping data or
for marker
association studies. A large number of commonly used soybean lines (e.g.,
commercial
varieties) and mapping populations are known in the art. A broad range of
mapping
populations were used to obtain the results described in Examples.
[00142] A variety of commercial software is available for genetic mapping and
marker
association studies (e.g., QTL mapping). This software includes but is not
limited to:
JoinMap (VanOoijen, and Voorrips (2001) "JoinMap 3.0 software for the
calculation of
genetic linkage maps," Plant Research International, Wageningen, the
Netherlands; and,
Stam, The Plant Journal 3(5): 739-744 (1993)); MapQTLO (J. W. vanOoijen,
"Software for
the mapping of quantitative trait loci in experimental populations" Kyazma B.
V.,
Wageningen, Netherlands); MapManager QT (Manly and Olson, Genome 10: 327-334
(1999)); MapManager QTX (Manly, Cudmorc and Meer, Mamm. Gcnome 12: 930-932
(2001)); GeneFlow and QTLocatc Tm (GENEFLOW, Inc., Alexandria, VA); and
TASSEL
("Trait Analysis by aSSociation, Evolution, and Linkage" by Edward Buckler,
and
information about the program can be found on the Buckler Lab web page at the
Institute for
Genomic Diversity at Cornell University).
[00143]
"Unified", "consensus" or "integrated" genetic maps have been created that
incorporate mapping data from two or more sources, including sources that used
different
mapping populations and different modes of statistical analysis. The merging
of genetic map
information increases the marker density on the map, as well as improving map
resolution.
These improved maps can be advantageously used in marker assisted selection,
map-based
cloning, provide an improved framework for positioning newly identified
molecular markers
and aid in the identification of QTL chromosome intervals and clusters of
advantageously-
linked markers.
[00144] In some aspects, a consensus map is derived by simply overlaying one
map on top
of another. In other aspects, various algorithms, e.g., JoinMap analysis,
allows the
combination of genetic mapping data from multiple sources, and reconciles
discrepancies
between mapping data from the original sources. See, Van Ooijen, and Voorrips
(2001)
"JoinMap 3.0 software for the calculation of genetic linkage maps," Plant
Research
International, Wageningen, the Netherlands; and, Stam (1993) "Construction of
integrated
37
Date Recue/Date Received 2023-12-08
genetic linkage maps by means of a new computer package: JoinMap," The Plant
Journal
3(5):739-744.
[00145] Additional integrated maps are known in the art. See, e.g., Cregan,
et al., (1999)
"An Integrated Genetic Linkage Map of the Soybean Genome", Crop Science
39:1464-1490;
the Soybean Consensus Map 4.0 described by Hyten D. L., et al., (2010) "A high
density
integrated genetic linkage map of soybean and the development of a 1536
universal soy
linkage panel for quantitative trait locus mapping." Crop Sci 50: 960-968; and
International
Application Number PCT/U52004/024919 by Sebastian, filed Jul. 27, 2004,
entitled
"Soybean Plants Having Superior Agronomic Performance and Methods for their
Production".
[00146] Song, et al., provides another integrated soybean genetic map that
incorporates
mapping information from five different mapping populations (Song, et al.,
(2004) "A New
Integrated Genetic Linkage Map of the Soybean," Theor Appl Genet 109:122-128).
This
integrated map contains approximately 1,800 soybean markers, including SSR and
SNP-type
markers, as well as EST markers, RFLP markers, AFLP, RAPD, isozyme and
classical
markers (e.g., seed coat color). The markers that are on this map are known in
the art and
have been previously characterized. This information is also available at the
website for the
Soybean Genomics and Improvement Laboratory (SG1L) at the USDA Beltsville
Agricultural
Research Center (BARC). See, specifically, the description of projects in the
Cregan
Laboratory on that website.
[00147] The soybean integrated linkage map provided in Song, et al., (2004) is
based on
the principle described by Stam (1993) "Construction of integrated genetic
linkage maps by
means of a new computer package: JoinMap," The Plant Journal 3(5):739-744; and
Van
Ooijen and Voorrips (2001) "JoinMap 3.0 software for the calculation of
genetic linkage
maps," Plant Research International, Wageningen, the Netherlands. Mapping
information
from five soybean populations was used in the map integration, and also used
to place
recently identified SSR markers onto the soybean genome. These mapping
populations were
MinsoyxNoir 1 (MN), MinsoyxArcher (MA), Noir lxArcher (NA), ClarkxHarosoy (CH)
and
A81-356022xP1468916 (MS). The JoinMap analysis resulted in a map with 20
linkage
groups containing a total of 1849 markers, including 1015 SSRs, 709 RFLPs, 73
RAPDs, 24
classical traits, six AFLPs, ten isozymes and 12 others. Among the mapped SSR
markers
were 417 previously uncharacterized SSRs.
[00148] Initially, LOD scores and pairwise recombination frequencies between
markers
were calculated. A LOD of 5.0 was used to create groups in the MS, MA, NA
populations
38
Date Recue/Date Received 2023-12-08
and LOD 4.0 in the MN and CH populations. The map of each linkage group was
then
integrated. Recombination values were converted to genetic distances using the
Kosambi
mapping function.
VI. LINKAGE MAPS
[00149] From
the present disclosure and widely recognized in the art, it is clear that any
genetic marker that has a significant probability of co-segregation with a
phenotypic trait of
interest (e.g., in the present case, a tolerance or improved tolerance trait)
can be used as a
marker for that trait. Useful QTL markers identified herein include S29725-
001; S29741-
001; S29742-001; S11315-1; and S11316-1.
[00150] Additional markers linked to the QTL markers can also be used to
predict the
tolerance or improved tolerance trait in a soybean plant. In other words, any
other marker
showing less than 50% recombination frequency (separated by a genetic distance
less than 50
cM) with a QTL marker of the invention is also a feature of the invention. Any
marker that is
linked to a QTL marker can also be used advantageously in marker-assisted
selection for the
particular trait.
[00151] Genetic markers that are linked to QTL markers are particularly useful
when they
are sufficiently proximal (e.g., closely linked) to a given QTL marker so that
the genetic
marker and the QTL marker display a low recombination frequency. In the
present invention,
such closely linked markers are a feature of the invention. As defined herein,
closely linked
markers display a recombination frequency of about 10% or less (the given
marker is within
cM of the QTL). Put another way, these closely linked loci co-segregate at
least 90% of
the time. Indeed, the closer a marker is to a QTL marker, the more effective
and
advantageous that marker becomes as an indicator for the desired trait.
[00152] Thus, in other embodiments, closely linked loci such as a QTL marker
locus and a
second locus display an inter-locus cross-over frequency of about 10% or less,
preferably
about 9% or less, still more preferably about 8% or less, yet more preferably
about 7% or
less, still more preferably about 6% or less, yet more preferably about 5% or
less, still more
preferably about 4% or less, yet more preferably about 3% or less, and still
more preferably
about 2% or less. In highly preferred embodiments, the relevant loci (e.g., a
marker locus and
a target locus such as a QTL) display a recombination a frequency of about 1%
or less, e.g.,
about 0.75% or less, more preferably about 0.5% or less, or yet more
preferably about 0.25%
or less. Thus, the loci are about 10 cM, 9 cM, 8 cM, 7 cM, 6 cM, 5 cM, 4 cM, 3
cM, 2 cM, 1
39
Date Recue/Date Received 2023-12-08
cM, 0.75 cM, 0.5 cM or 0.25 cM or less apart. Put another way, two loci that
are localized to
the same chromosome, and at such a distance that recombination between the two
loci occurs
at a frequency of less than 10% (e.g., about 9%, 8%, 7%, 6%, 5%, 4%, 3%, 2%,
1%, 0.75%,
0.5%, 0.25%, or less) are said to be "proximal to" each other.
[00153] Similarly, linked markers (including closely linked markers) of
the invention can
be determined by review of any suitable soybean genetic map. For example, the
integrated
genetic map described in Song, et al., (2004) also provides a means to
identify linked
(including closely linked) markers. See, Song, et al., (2004) "A New
Integrated Genetic
Linkage Map of the Soybean" Theor Appl Genet 109:122-128; see also the website
for the
Soybean Genomies and Improvement Laboratory (SGIL) at the USDA Beltsville
Agricultural
Research Center (BARC), and see specifically the description of projects in
the Cregan
Laboratory on that website. That genetic map incorporates a variety of genetic
markers that
are known in the art or alternatively are described in that reference.
Detailed descriptions of
numerous markers, including many of those described in Song, et al., (2004)
can be found at
the SOYBASE website resource.
[00154] It is not intended that the detelmination of linked or closely linked
markers be
limited to the use of any particular soybean genetic map. Indeed, a large
number of soybean
genetic maps are available and are well known to one of skill in the art.
Another map that
finds use with the invention in this respect is the integrated soybean genetic
maps found on
the SOYBASE website resource. Alternatively still, the determination of linked
and closely
linked markers can be made by the generation of an experimental dataset and
linkage
analysis.
[00155] It is not intended that the identification of markers that are
linked (e.g., within
about 50 cM or within about 10 cM) to the Charcoal Rot Drought Complex
tolerance QTL
markers identified herein be limited to any particular map or methodology.
Indeed, linked
markers as defined herein can be determined from any genetic map known in the
art (an
experimental map or an integrated map), or alternatively, can be determined
from any new
mapping dataset.
[00156] It is noted that lists of linked and closely linked markers may vary
between maps
and methodologies due to various factors. First, the markers that are placed
on any two maps
may not be identical, and furthermore, some maps may have a greater marker
density than
another map. Also, the mapping populations, methodologies and algorithms used
to construct
genetic maps can differ. One of skill in the art recognizes that one genetic
map is not
necessarily more or less accurate than another, and furthermore, recognizes
that any soybean
Date Recue/Date Received 2023-12-08
genetic map can be used to determine markers that are linked and closely
linked to the QTL
markers of the present invention.
VII. TECHNIQUES FOR MARKER DETECTION
[00157] The invention provides molecular markers that have a significant
probability of
co-segregation with QTL that impart a Charcoal Rot Drought Complex tolerance
phenotype.
These QTL markers find use in marker assisted selection for desired traits
(tolerance or
improved tolerance), and also have other uses. It is not intended that the
invention be limited
to any particular method for the detection of these markers.
[00158] Markers corresponding to genetic polymorphisms between members of a
population can be detected by numerous methods well-established in the art
(e.g., PCR-based
sequence specific amplification, restriction fragment length polymorphisms
(RFLPs),
isozyme markers, allele specific hybridization (ASH), amplified variable
sequences of the
plant genome, self-sustained sequence replication, simple sequence repeat
(SSR), single
nucleotide polymorphism (SNP), random amplified polymorphic DNA ("RAPD") or
amplified fragment length polymorphisms (AFLP)). In one additional embodiment,
the
presence or absence of a molecular marker is determined simply through
nucleotide
sequencing of the polymorphic marker region. This method is readily adapted to
high
throughput analysis as are the other methods noted above, e.g., using
available high
throughput sequencing methods such as sequencing by hybridization.
[00159] In general, the majority of genetic markers rely on one or more
property of nucleic
acids for their detection. For example, some techniques for detecting genetic
markers utilize
hybridization of a probe nucleic acid to nucleic acids corresponding to the
genetic marker
(e.g., amplified nucleic acids produced using genomic soybean DNA as a
template).
Hybridization formats, including but not limited to solution phase, solid
phase, mixed phase,
or in situ hybridization assays are useful for allele detection. An extensive
guide to the
hybridization of nucleic acids is found in Tijssen (1993) Laboratory
Techniques in
Biochemistry and Molecular Biology _____________________________________
Hybridization with Nucleic Acid Probes Elsevier,
New York; Berger and Kimmel, Guide to Molecular Cloning Techniques, Methods in
Enzymology volume 152 Academic Press, Inc., San Diego, Calif. ("Berger"); as
well as in
Sambrook and Ausubel (herein).
[00160] For example, markers that comprise restriction fragment length
polymorphisms
(RFLP) are detected, e.g., by hybridizing a probe which is typically a sub-
fragment (or a
41
Date Recue/Date Received 2023-12-08
synthetic oligonucleotide corresponding to a sub-fragment) of the nucleic acid
to be detected
to restriction digested genomic DNA. The restriction enzyme is selected to
provide restriction
fragments of at least two alternative (or polymorphic) lengths in different
individuals or
populations. Determining one or more restriction enzyme that produces
informative
fragments for each cross is a simple procedure, well known in the art. After
separation by
length in an appropriate matrix (e.g., agarose or polyacrylamide) and transfer
to a membrane
(e.g., nitrocellulose, nylon, etc.), the labeled probe is hybridized under
conditions which
result in equilibrium binding of the probe to the target followed by removal
of excess probe
by washing.
1001611 Nucleic acid probes to the marker loci can be cloned and/or
synthesized. Any
suitable label can be used with a probe of the invention. Detectable labels
suitable for use
with nucleic acid probes include, for example, any composition detectable by
spectroscopic,
radioisotopic, photochemical, biochemical, immunochemical, electrical, optical
or chemical
means. Useful labels include biotin for staining with labeled streptavidin
conjugate, magnetic
beads, fluorescent dyes, radiolabels, enzymes, and colorimetric labels. Other
labels include
ligands which bind to antibodies labeled with fluorophorcs, chemilumineseent
agents, and
enzymes. A probe can also constitute radiolabelled PCR primers that are used
to generate a
radiolabelled amplicon. Labeling strategies for labeling nucleic acids and
corresponding
detection strategies can be found, e.g., in Haugland (1996) Handbook of
Fluorescent Probes
and Research Chemicals Sixth Edition by Molecular Probes, Inc. (Eugene Oreg.);
or
Haugland (2001) Handbook of Fluorescent Probes and Research Chemicals Eighth
Edition
by Molecular Probes, Inc. (Eugene Oreg.) (Available on CD ROM).
[00162] PCR, RT-PCR and LCR are in particularly broad use as amplification and
amplification-detection methods for amplifying nucleic acids of interest
(e.g., those
comprising marker loci), facilitating detection of the markers. Details
regarding the use of
these and other amplification methods can be found in any of a variety of
standard texts,
including, e.g., Sambrook, Ausubel, Berger and Croy, herein. Many available
biology texts
also have extended discussions regarding PCR and related amplification
methods. One of
skill will appreciate that essentially any RNA can be converted into a double
stranded DNA
suitable for restriction digestion, PCR expansion and sequencing using reverse
transcriptase
and a polymerase ("Reverse Transcription-PCR, or "RT-PCR"). See also Ausubel,
Sambrook
and Berger, above.
[00163] In one aspect, real time PCR or LCR is performed on the amplification
mixtures
described herein, e.g., using molecular beacons or TaqManTm probes. A
molecular beacon
42
Date Recue/Date Received 2023-12-08
(MB) is an oligonucleotide or PNA which, under appropriate hybridization
conditions, self-
hybridizes to form a stem and loop structure. The MB has a label and a
quencher at the
termini of the oligonucleotide or PNA; thus, under conditions that peunit
intra-molecular
hybridization, the label is typically quenched (or at least altered in its
fluorescence) by the
quencher. Under conditions where the MB does not display intra-molecular
hybridization
(e.g., when bound to a target nucleic acid, e.g., to a region of an amplicon
during
amplification), the MB label is unquenched. Details regarding standard methods
of making
and using MBs are well established in the literature and MBs are available
from a number of
commercial reagent sources. See also, e.g., Leone, et al., (1995) "Molecular
beacon probes
combined with amplification by NASBA enable homogenous real-time detection of
RNA"
Nucleic Acids Res 26:2150-2155; Tyagi and Kramer, (1996) "Molecular beacons:
probes that
fluoresce upon hybridization" Nature Biotechnology 14:303-308; Blok and
Kramer, (1997)
"Amplifiable hybridization probes containing a molecular switch" Mot Cell
Probes 11:187-
194; Hsuih, et al., (1997) "Novel, ligation-dependent PCR assay for detection
of hepatitis C
in scrum" J Clin Microbiol 34:501-507; Kostrikis, et al., (1998) "Molecular
beacons: spectral
gcnotyping of human alleles" Science 279:1228-1229; Sokol, et al., (1998)
"Real time
detection of DNA:RNA hybridization in living cells" Proc Natl Acad Sci USA
95:11538-
11543; Tyagi, et al., (1998) "Multicolor molecular beacons for allele
discrimination" Nature
Biotechnology 16:49-53; Bonnet, et al., (1999) "Thermodynamic basis of the
chemical
specificity of structured DNA probes" Proc Nati Acad Sci USA 96:6171-6176;
Fang, et al.,
(1999) "Designing a novel molecular beacon for surface-immobilized DNA
hybridization
studies" J Am Chem Soc 121:2921-2922; Marras, et at., (1999) "Multiplex
detection of
single-nucleotide variation using molecular beacons" Genet Anal Biomol Eng
14:151-156;
and Vet, et al., (1999) "Multiplex detection of four pathogenic retroviruses
using molecular
beacons" Proc Natl Acad Sci USA 96:6394-6399. Additional details regarding MB
construction and use is found in the patent literature, e.g., U.S. Pat. No.
5,925,517 (Jul. 20,
1999) to Tyagi, et al., entitled "Detectably labeled dual conformation
oligonucleotide probes,
assays and kits;" U.S. Pat. No. 6,150,097 (November 21, 2000) to Tyagi, et
al., entitled
"Nucleic acid detection probes having non-FRET fluorescence quenching and kits
and assays
including such probes" and U.S. Pat. No. 6,037,130 (Mar. 14, 2000) to Tyagi,
et at., entitled
"Wavelength-shifting probes and primers and their use in assays and kits."
[00164] PCR detection and quantification using dual-labeled fluorogenic
oligonucleotide
probes, commonly referred to as "TagManTm" probes, can also be performed
according to the
present invention. These probes are composed of short (e.g., 20-25 base)
43
Date Recue/Date Received 2023-12-08
oligodeoxynucleotides that are labeled with two different fluorescent dyes. On
the 5' terminus
of each probe is a reporter dye, and on the 3' terminus of each probe a
quenching dye is
found. The oligonucleotide probe sequence is complementary to an internal
target sequence
present in a PCR amplicon. When the probe is intact, energy transfer occurs
between the two
fluorophores and emission from the reporter is quenched by the quencher by
FRET. During
the extension phase of PCR, the probe is cleaved by 5' nuclease activity of
the polymerase
used in the reaction, thereby releasing the reporter from the oligonucleotide-
quencher and
producing an increase in reporter emission intensity. Accordingly, TagManTm
probes are
oligonucleotides that have a label and a quencher, where the label is released
during
amplification by the exonuclease action of the polymerase used in
amplification. This
provides a real time measure of amplification during synthesis. A variety of
TagMarirm
reagents are commercially available, e.g., from Applied Biosystems (Division
Headquarters
in Foster City, Calif.) as well as from a variety of specialty vendors such as
Biosearch
Technologies (e.g., black hole quencher probes).
[00165] Amplified variable sequences refer to amplified sequences of the plant
genome
which exhibit high nucleic acid residue variability between members of the
same species. All
organisms have variable gcnomic sequences and each organism (with the
exception of a
clone) has a different set of variable sequences. Once identified, the
presence of specific
variable sequence can be used to predict phenotypic traits. Preferably, DNA
from the plant
serves as a template for amplification with primers that flank a variable
sequence of DNA.
The variable sequence is amplified and then sequenced.
[00166]
Alternatively, self-sustained sequence replication can be used to identify
genetic
markers. Self-sustained sequence replication refers to a method of nucleic
acid amplification
using target nucleic acid sequences which are replicated exponentially in
vitro under
substantially isothermal conditions by using three enzymatic activities
involved in retroviral
replication: (1) reverse transcriptase, (2) RNase H, and (3) a DNA-dependent
RNA
polymerase (Guatelli, et al., (1990) Proc Natl Acad Sci USA 87:1874). By
mimicking the
retroviral strategy of RNA replication by means of cDNA intermediates, this
reaction
accumulates cDNA and RNA copies of the original target.
[00167] Amplified fragment length polymorphisms (AFLP) can also be used as
genetic
markers (Vos, et al., (1995) Nucleic Acids Res 23:4407). The phrase "amplified
fragment
length polymorphism" refers to selected restriction fragments which are
amplified before or
after cleavage by a restriction endonuclease. The amplification step allows
easier detection of
specific restriction fragments. AFLP allows the detection large numbers of
polymorphic
44
Date Recue/Date Received 2023-12-08
markers and has been used for genetic mapping of plants (Becker, et al.,
(1995) Mol Gen
Genet 249:65; and Meksem, et al., (1995) Mol Gen Genet 249:74).
[00168] Allele-specific hybridization (ASH) can be used to identify the
genetic markers of
the invention. ASH technology is based on the stable annealing of a short,
single-stranded,
oligonucleotide probe to a completely complementary single-strand target
nucleic acid.
Detection is via an isotopic or non-isotopic label attached to the probe.
[00169] For each polymorphism, two or more different ASH probes are designed
to have
identical DNA sequences except at the polymorphic nucleotides. Each probe will
have exact
homology with one allele sequence so that the range of probes can distinguish
all the known
alternative allele sequences. Each probe is hybridized to the target DNA. With
appropriate
probe design and hybridization conditions, a single-base mismatch between the
probe and
target DNA will prevent hybridization. In this manner, only one of the
alternative probes will
hybridize to a target sample that is homozygous or homogenous for an allele.
Samples that
are heterozygous or heterogeneous for two alleles will hybridize to both of
two alternative
probes.
[00170] ASH markers arc used as dominant markers where the presence or absence
of
only one allele is determined from hybridization or lack of hybridization by
only one probe.
The alternative allele may be inferred from the lack of hybridization. ASH
probe and target
molecules are optionally RNA or DNA; the target molecules are any length of
nucleotides
beyond the sequence that is complementary to the probe; the probe is designed
to hybridize
with either strand of a DNA target; the probe ranges in size to conform to
variously stringent
hybridization conditions, etc.
[00171] PCR allows the target sequence for ASH to be amplified from low
concentrations
of nucleic acid in relatively small volumes. Otherwise, the target sequence
from genomic
DNA is digested with a restriction endonuclease and size separated by gel
electrophoresis.
Hybridizations typically occur with the target sequence bound to the surface
of a membrane
or, as described in U.S. Pat. No. 5,468,613, the ASH probe sequence may be
bound to a
membrane.
[00172] In one embodiment, ASH data are typically obtained by amplifying
nucleic acid
fragments (amplicons) from genomic DNA using PCR, transferring the amplicon
target DNA
to a membrane in a dot-blot format, hybridizing a labeled oligonucleotide
probe to the
amplicon target, and observing the hybridization dots by autoradiography.
[00173] Single nucleotide polymorphisms (SNP) are markers that consist of a
shared
sequence differentiated on the basis of a single nucleotide. Typically, this
distinction is
Date Recue/Date Received 2023-12-08
detected by differential migration patterns of an amplicon comprising the SNP
on e.g., an
acrylamide gel. However, alternative modes of detection, such as
hybridization, e.g., ASH, or
RFLP analysis are also appropriate.
[00174] Isozyrne markers can be employed as genetic markers, e.g., to track
markers other
than the tolerance markers herein, or to track isozyme markers linked to the
markers herein.
Isozymes are multiple forms of enzymes that differ from one another in their
amino acid, and
therefore their nucleic acid sequences. Some isozymes are multimeric enzymes
containing
slightly different subunits. Other isozymes are either multimeric or monomeric
but have been
cleaved from the proenzyme at different sites in the amino acid sequence.
Isozymes can be
characterized and analyzed at the protein level, or alternatively, isozymes
which differ at the
nucleic acid level can be determined. In such cases any of the nucleic acid
based methods
described herein can be used to analyze isozyme markers.
[00175] As herein, nucleic acid amplification techniques such as PCR and LCR
are well
known in the art and can be applied to the present invention to amplify and/or
detect nucleic
acids of interest, such as nucleic acids comprising marker loci. Examples of
techniques
sufficient to direct persons of skill through such in vitro methods, including
the polymerase
chain reaction (PCR), the ligase chain reaction (LCR), Q13I3-replicase
amplification and other
RNA polymerase mediated techniques (e.g., NASBA), are found in the references
noted
above, e.g., Innis, Sambrook, Ausubel, Berger and Croy. Additional details are
found in
Mullis, et al., (1987) U.S. Pat. No. 4,683,202; Arnheim and Levinson, (Oct. 1,
1990) C&EN
36-47; The Journal Of NIH Research (1991) 3:81-94; Kwoh, et al., (1989) Proc
Natl Acad
Sci USA 86:1173; Guatelli, et al., (1990) Proc Natl Acad Sci USA 87:1874;
Lomeli, et al.,
(1989) J Clin Chem 35:1826; Landegren, et al., (1988) Science 241:1077-1080;
Van Brunt,
(1990) Biotechnology 8:291-294; Wu and Wallace, (1989) Gene 4:560; Barringer,
et al.,
(1990) Gene 89:117, and Sooknanan and Malek, (1995) Biotechnology 13:563-564.
Improved methods of amplifying large nucleic acids by PCR, which is useful in
the context
of positional cloning, are further summarized in Cheng, et al., (1994) Nature
369:684, and the
references therein, in which PCR amplicons of up to 40 kb are generated.
[00176] In general, synthetic methods for making oligonucleotides, including
probes,
primers, molecular beacons, PNAs, LNAs (locked nucleic acids), etc., are well
known. For
example, oligonucleotides can be synthesized chemically according to the solid
phase
phosphoramidite triester method described by Beaucage and Caruthers, (1981)
Tetrahedron
Letts 22(20):1859-1862, e.g., using a commercially available automated
synthesizer, e.g., as
described in Needham-VanDevanter, et al., (1984) Nucleic Acids Res 12:6159-
6168.
46
Date Recue/Date Received 2023-12-08
Oligonucleotides, including modified oligonucleotides can also be ordered from
a variety of
commercial sources known to persons of skill. There are many commercial
providers of oligo
synthesis services, and thus this is a broadly accessible technology. Any
nucleic acid can be
custom ordered from any of a variety of commercial sources, such as The
Midland Certified
Reagent Company, The Great American Gene Company, ExpressGen Inc., Operon
Technologies Inc. (Alameda, Calif.) and many others. Similarly, PNAs can be
custom
ordered from any of a variety of sources, such as PeptidoGenic, HTI Bio-
Products, Inc.,
BMA Biomedicals Ltd (U.K.), Bio=Synthesis, Inc., and many others.
[00177] In alternative embodiments, in silico methods can be used to detect
the marker
loci of interest. For example, the sequence of a nucleic acid comprising the
marker locus of
interest can be stored in a computer. The desired marker locus sequence or its
homolog can
be identified using an appropriate nucleic acid search algorithm as provided
by, for example,
in such readily available programs as BLAST, or even simple word processors.
[00178] In some preferred embodiments, the molecular markers of the invention
are
detected using a suitable PCR-based detection method, where the size or
sequence of the
PCR amplicon is indicative of the absence or presence of the marker (e.g., a
particular marker
allele). ln these types of methods, PCR primers are hybridized to the
conserved regions
flanking the polymorphic marker region. As used in the art, PCR primers used
to amplify a
molecular marker are sometimes termed "PCR markers" or simply "markers".
[00179] It
will be appreciated that, although many specific examples of primers are
provided herein (see, FIG. 2), suitable primers to be used with the invention
can be designed
using any suitable method. It is not intended that the invention be limited to
any particular
primer or primer pair. For example, primers can be designed using any suitable
software
program, such as LASERGENE .
[00180] In some embodiments, the primers of the invention are radiolabelled,
or labeled by
any suitable means (e.g., using a non-radioactive fluorescent tag), to allow
for rapid
visualization of the different size amplicons following an amplification
reaction without any
additional labeling step or visualization step. In some embodiments, the
primers are not
labeled, and the amplicons are visualized following their size resolution,
e.g., following
agarose gel electrophoresis. In some embodiments, ethidium bromide staining of
the PCR
amplicons following size resolution allows visualization of the different size
amplicons.
[00181] It is not intended that the primers of the invention be limited to
generating an
amplicon of any particular size. For example, the primers used to amplify the
marker loci and
alleles herein are not limited to amplifying the entire region of the relevant
locus. The primers
47
Date Recue/Date Received 2023-12-08
can generate an amplicon of any suitable length that is longer or shorter than
those given in
the allele definitions in FIG. 4. In some embodiments, marker amplification
produces an
amplicon at least 20 nucleotides in length, or alternatively, at least 50
nucleotides in length,
or alternatively, at least 100 nucleotides in length, or alternatively, at
least 200 nucleotides in
length. Marker alleles in addition to those recited in FIG. 4 also find use
with the present
invention.
VIII. MARKER ASSISTED SELECTION AND BREEDING OF PLANTS
[00182] A primary motivation for development of molecular markers in crop
species is the
potential for increased efficiency in plant breeding through marker assisted
selection (MAS).
Genetic markers are used to identify plants that contain a desired genotype at
one or more
loci, and that are expected to transfer the desired genotype, along with a
desired phenotype to
their progeny. Genetic markers can be used to identify plants that contain a
desired genotype
at one locus, or at several unlinked or linked loci (e.g., a haplotype), and
that would be
expected to transfer the desired genotype, along with a desired phenotype to
their progeny.
The present invention provides the means to identify plants, particularly
soybean plants, that
are tolerant, exhibit improved tolerance or are susceptible to Charcoal Rot
Drought Complex
by identifying plants having a specified allele at one of those loci, e.g.,
S29725-001; S29741-
001; S29742-001; S11315-1; and S11316-1.
[001831
Similarly, by identifying plants lacking the desired marker locus, susceptible
or
less tolerant plants can be identified, and, e.g., eliminated from subsequent
crosses. Similarly,
these marker loci can be introgressed into any desired gcnomic background,
germplasm,
plant, line, variety, etc., as part of an overall MAS breeding program
designed to enhance
soybean yield.
1001841 The invention also provides chromosome QTL intervals that find equal
use in
MAS to select plants that demonstrate Charcoal Rot Drought Complex tolerance
or improved
tolerance. Similarly, the QTL intervals can also be used to counter-select
plants that are
susceptible or have reduced tolerance to Charcoal Rot Drought Complex. Any
marker that
maps within the QTL interval (including the termini of the intervals) finds
use with the
invention. These intervals include: ((i) a chromosomal interval located at
about 17 cM to
about 38 cM of chromosome 5; (ii) a chromosomal interval located at about 5 cM
to about 26
cM of chromosome 15; (iii) a chromosomal interval located at about 19 cM to
about 40 cM
of chromosome 19; or (iv) a chromosomal interval located at about 81 cM to
about 102 cM of
48
Date Recue/Date Received 2023-12-08
chromosome 19. In a further aspect, the interval on Chr. 15 can be described
as flanked by
and including S29725-001 and S29741-001. Alternatively, these intervals can be
specified as
follows: (i) a chromosomal interval located within about 2 Mbp of an interval
at 7,975-8,015
kpb of chromosome 5; (ii) a chromosomal interval located within about 2 Mbp of
an interval
at 3,202-3,212 kbp of chromosome 15 (SEQ ID NO.: 26); (iii) a chromosomal
interval
located within about 2 Mbp of an interval at 27,178-27,218 kbp; or (iv) a
chromosomal
interval located within about 2 Mbp of an interval at 48,340-48,380 kbp of
chromosome 19
(SEQ ID NO.: 27). In a still further aspect, the interval on Chr. 15 can be
described as
flanked by and including S29725-001 and S29742-001. In a further aspect, the
interval on
Chr. 19 can be described as flanked by and including S11315-1 and S13116-1.
[00185] In general, MAS uses polymorphic markers that have been identified as
having a
significant likelihood of co-segregation with a tolerance trait. Such markers
are presumed to
map near a gene or genes that give the plant its tolerance phenotype, and are
considered
indicators for the desired trait, and are termed QTL markers. Plants are
tested for the presence
of a desired allele in the QTL marker. The most preferred markers (or marker
alleles) are
those that have the strongest association with the tolerance trait.
[00186] Linkage analysis is used to determine which polymorphic marker allele
demonstrates a statistical likelihood of co-segregation with the tolerance
phenotype (thus, a
"tolerance marker allele"). Following identification of a marker allele for co-
segregation with
the tolerance phenotype, it is possible to use this marker for rapid, accurate
screening of plant
lines for the tolerance allele without the need to grow the plants through
their life cycle and
await phenotypic evaluations, and furthermore, permits genetic selection for
the particular
tolerance allele even when the molecular identity of the actual tolerance QTL
is unknown.
Tissue samples can be taken, for example, from the first leaf of the plant and
screened with
the appropriate molecular marker, and it is rapidly determined which progeny
will advance.
Linked markers also remove the impact of environmental factors that can often
influence
phenotypic expression.
[00187] A polymorphic QTL marker locus can be used to select plants that
contain the
marker allele (or alleles) that correlate with the desired tolerance
phenotype. In brief, a
nucleic acid corresponding to the marker nucleic acid allele is detected in a
biological sample
from a plant to be selected. This detection can take the form of hybridization
of a probe
nucleic acid to a marker allele or amplicon thereof, e.g., using allele-
specific hybridization,
Southern analysis, northern analysis, in situ hybridization, hybridization of
primers followed
by PCR amplification of a region of the marker, or the like. A variety of
procedures for
49
Date Recue/Date Received 2023-12-08
detecting markers are described herein, e.g., in the section entitled
"TECHNIQUES FOR
MARKER DETECTION." After the presence (or absence) of a particular marker
allele in the
biological sample is verified, the plant is selected, e.g., used to make
progeny plants by
selective breeding.
[00188] Soybean plant breeders desire combinations of tolerance loci with
genes for high
yield and other desirable traits to develop improved soybean varieties.
Screening large
numbers of samples by non-molecular methods (e.g., trait evaluation in soybean
plants) can
be expensive, time consuming, and unreliable. Use of the polymorphic markers
described
herein, when genetically-linked to tolerance loci, provide an effective method
for selecting
resistant varieties in breeding programs. For example, one advantage of marker-
assisted
selection over field evaluations for tolerance resistance is that MAS can be
done at any time
of year, regardless of the growing season. Moreover, environmental effects are
largely
irrelevant to marker-assisted selection.
[00189] When a population is segregating for multiple loci affecting one or
multiple traits,
c.g., multiple loci involved in tolerance, or multiple loci each involved in
tolerance or
resistance to different diseases, the efficiency of MAS compared to phenotypic
screening
becomes even greater, because all of the loci can be evaluated in the lab
together from a
single sample of DNA. In the present instance, include: (i) a chromosomal
interval located at
about 17 cM to about 38 cM of chromosome 5; (ii) a chromosomal interval
located at about 5
cM to about 26 cM of chromosome 15; (iii) a chromosomal interval located at
about 19 cM to
about 40 cM of chromosome 19; or (iv) a chromosomal interval located at about
81 cM to
about 102 cM of chromosome 19; and these intervals can be assayed
simultaneously or
sequentially from a single sample or a population of samples. Alternatively,
these intervals
can be specified as follows: (i) a chromosomal interval located within about 2
Mbp of an
interval at 7,975-8,015 kpb of chromosome 5; (ii) a chromosomal interval
located within
about 2 Mbp of an interval at 3,202-3,212 kbp of chromosome 15 (SEQ ID NO.:
26); (iii) a
chromosomal interval located within about 2 Mbp of an interval at 27,178-
27,218 kbp; or (iv)
a chromosomal interval located within about 2 Mbp of an interval at 48,340-
48,380 kbp of
chromosome 19 (SEQ ID NO.: 27).
[00190] In a further aspect, the interval on Chr. 15 can be described as
flanked by and
including S29725-001 and S29741-001. In a still further aspect, the interval
on Chr. 15 can
be described as flanked by and including S29725-001 and S29742-001. In a
further aspect,
the interval on Chr. 19 can be described as flanked by and including S11315-1
and S13116-1.
[00191] Another use of MAS in plant breeding is to assist the recovery of the
recurrent
Date Recue/Date Received 2023-12-08
parent genotype by backcross breeding. Backcross breeding is the process of
crossing a
progeny back to one of its parents or parent lines. Backcrossing is usually
done for the
purpose of introgressing one or a few loci from a donor parent (e.g., a parent
comprising
desirable tolerance marker loci) into an otherwise desirable genetic
background from the
recurrent parent (e.g., an otherwise high yielding soybean line). The more
cycles of
backcrossing that are done, the greater the genetic contribution of the
recurrent parent to the
resulting introgressed variety. This is often necessary, because tolerant
plants may be
otherwise undesirable, e.g., due to low yield, low fecundity, or the like. In
contrast, strains
which are the result of intensive breeding programs may have excellent yield,
fecundity or
the like, merely being deficient in one desired trait such as tolerance to
Charcoal Rot Drought
Complex.
IX INTROGRESSION OF FAVORABLE ALLELES
[00192] One application of MAS, in the context of the present invention is to
use the
tolerance or improved tolerance markers to increase the efficiency of an
introgression or
backcrossing effort aimed at introducing a tolerance QTL into a desired
(typically high
yielding) background. In marker assisted backcrossing of specific markers (and
associated
QTL) from a donor source, e.g., to an elite or exotic genetic background, one
selects among
backcross progeny for the donor trait and then uses repeated backcrossing to
the elite or
exotic line to reconstitute as much of the elite/exotic background's genome as
possible.
[00193] Thus, the markers and methods of the present invention can be utilized
to guide
marker assisted selection or breeding of soybean varieties with the desired
complement (set)
of allelic forms of chromosome segments associated with superior agronomic
performance
(tolerance, along with any other available markers for yield, disease
resistance, etc.). Any of
the disclosed marker alleles can be introduced into a soybean line via
introgression, by
traditional breeding (or introduced via transformation, or both) to yield a
soybean plant with
superior agronomic performance. The number of alleles associated with
tolerance that can be
introduced or be present in a soybean plant of the present invention ranges
from 1 to the
number of alleles disclosed herein, each integer of which is incorporated
herein as if
explicitly recited.
[00194] The present invention also extends to a method of making a progeny
soybean
plant and these progeny soybean plants, per se. The method comprises crossing
a first parent
soybean plant with a second soybean plant and growing the female soybean plant
under plant
51
Date Recue/Date Received 2023-12-08
growth conditions to yield soybean plant progeny. Methods of crossing and
growing soybean
plants are well within the ability of those of ordinary skill in the art. Such
soybean plant
progeny can be assayed for alleles associated with tolerance and, thereby, the
desired progeny
selected. Such progeny plants or seed can be sold commercially for soybean
production, used
for food, processed to obtain a desired constituent of the soybean, or further
utilized in
subsequent rounds of breeding. At least one of the first or second soybean
plants is a soybean
plant of the present invention in that it comprises at least one of the
allelic forms of the
markers of the present invention, such that the progeny are capable of
inheriting the allele.
[00195] Often, a method of the present invention is applied to at least one
related soybean
plant such as from progenitor or descendant lines in the subject soybean
plant's pedigree such
that inheritance of the desired tolerance allele can be traced. The number of
generations
separating the soybean plants being subject to the methods of the present
invention will
generally be from 1 to 20, commonly 1 to 5, and typically 1, 2 or 3
generations of separation,
and quite often a direct descendant or parent of the soybean plant will be
subject to the
method (i.e., one generation of separation).
[00196] Genetic diversity is important for long term genetic gain in any
breeding program.
With limited diversity, genetic gain will eventually plateau when all of the
favorable alleles
have been fixed within the elite population. One objective is to incorporate
diversity into an
elite pool without losing the genetic gain that has already been made and with
the minimum
possible investment. MAS provide an indication of which genomic regions and
which
favorable alleles from the original ancestors have been selected for and
conserved over time,
facilitating efforts to incorporate favorable variation from exotic germplasm
sources (parents
that are unrelated to the elite gene pool) in the hopes of finding favorable
alleles that do not
currently exist in the elite gene pool.
[00197] For example, the markers of the present invention can be used for MAS
in crosses
involving elite x exotic soybean lines by subjecting the segregating progeny
to MAS to
maintain major yield alleles, along with the tolerance marker alleles herein.
X. GENERATION OF TRANS GENIC CELLS AND PLANTS
[00198] The present invention also relates to host cells and organisms which
are
transfoiined with nucleic acids corresponding to tolerance QTL identified
according to the
invention. For example, such nucleic acids include chromosome intervals (e.g.,
genomic
fragments) that encode a tolerance or improved tolerance trait.
52
Date Recue/Date Received 2023-12-08
[00199] General texts which describe molecular biological techniques for the
cloning and
manipulation of nucleic acids and production of encoded polypeptides include
Berger,
Sambrook, and Ausubel, herein. These texts describe mutagenesis, the use of
vectors,
promoters and many other relevant topics related to, e.g., the generation of
clones that
comprise nucleic acids of interest, e.g., marker loci, marker probes, QTL that
segregate with
marker loci, etc.
[00200] Host
cells are genetically engineered (e.g., transduced, transfected, transformed,
etc.) with the vectors of this invention which can be, for example, a cloning
vector, a shuttle
vector or an expression vector. Such vectors are, for example, in the form of
a plasmid, a
phagemid, an agrobacterium, a virus, a naked polynucleotide (linear or
circular), or a
conjugated polynucleotide. Vectors can be introduced into bacteria, especially
for the purpose
of propagation and expansion. The vectors are also introduced into plant
tissues, cultured
plant cells or plant protoplasts by a variety of standard methods known in the
art, including
but not limited to electroporation (Fromm, et al., (1985) Proc Natl Acad Sci
USA 82:5824),
infection by viral vectors such as cauliflower mosaic virus (CaMV) (Hohn, et
al., (1982)
Molecular Biology of Plant Tumors Academic Press, New York, pp. 549-560;
Howell, U.S.
Pat. No. 4,407,956), high velocity ballistic penetration by small particles
with the nucleic acid
either within the matrix of small beads or particles, or on the surface
(Klein, et al., (1987)
Nature 327:70), use of pollen as vector (W085/01856), or use of Agrobacterium
turnefaciens
or A. rhizogenes can-ying a T-DNA plasmid in which DNA fragments are cloned.
The T-
DNA plasmid is transmitted to plant cells upon infection by Agrobacterium
tumefaciens, and
a portion is stably integrated into the plant genome (Horsch, et al., (1984)
Science 233:496;
Fraley, et al., (1983) Proc Natl Acad Sci USA 80:4803). Additional details
regarding nucleic
acid introduction methods are found in Sambrook, Berger and Ausubel, supra.
The method of
introducing a nucleic acid of the present invention into a host cell is not
critical to the instant
invention, and it is not intended that the invention be limited to any
particular method for
introducing exogenous genetic material into a host cell. Thus, any suitable
method, e.g.,
including but not limited to the methods provided herein, which provides for
effective
introduction of a nucleic acid into a cell or protoplast can be employed and
finds use with the
invention.
[00201] The engineered host cells can be cultured in conventional nutrient
media modified
as appropriate for such activities as, for example, activating promoters or
selecting
transformants. These cells can optionally be cultured into transgenic plants.
In addition to
Sambrook, Berger and Ausubel, supra, plant regeneration from cultured
protoplasts is
53
Date Recue/Date Received 2023-12-08
described in Evans, et al., (1983) "Protoplast Isolation and Culture,"
Handbook of Plant Cell
Cultures 1:124-176 (MacMillan Publishing Co., New York; Davey, (1983) "Recent
Developments in the Culture and Regeneration of Plant Protoplasts,"
Protoplasts, pp. 12-29,
(Birkhauser, Basel); Dale, (1983) "Protoplast Culture and Plant Regeneration
of Cereals and
Other Recalcitrant Crops," Protoplasts pp. 31-41, (Birkhauser, Basel); Binding
(1985)
"Regeneration of Plants," Plant Protoplasts, pp. 21-73, (CRC Press, Boca
Raton, Fla.).
Additional details regarding plant cell culture and regeneration include
Payne, et al., (1992)
Plant Cell and Tissue Culture in Liquid Systems John Wiley & Sons, Inc. New
York, N.Y.;
Gamborg and Phillips, (eds) (1995) Plant Cell, Tissue and Organ Culture;
Fundamental
Methods Springer Lab Manual, Springer-Verlag (Berlin Heidelberg New York) and
Plant
Molecular Biology (1993) Croy, Ed. Bios Scientific Publishers, Oxford, U.K.
ISBN 0 12
198370 6. Cell culture media in general are also set forth in Atlas and Parks,
(eds) The
Handbook of Microbiological Media (1993) CRC Press, Boca Raton, Fla.
Additional
information for cell culture is found in available commercial literature such
as the Life
Science Research Cell Culture Catalogue (1998) from Sigma-Aldrich, Inc (St
Louis, Mo.)
("Sigma-LSRCCC") and, e.g., the Plant Culture Catalogue and supplement (e.g.,
1997 or
later) also from Sigma-Aldrich, Inc (St Louis, Mo.) ("Sigma-PCCS").
[002021 The present invention also relates to the production of transgenic
organisms,
which may be bacteria, yeast, fungi, animals or plants, transduced with the
nucleic acids of
the invention (e.g., nucleic acids comprising the marker loci and/or QTL noted
herein). A
thorough discussion of techniques relevant to bacteria, unicellular eukaryotes
and cell culture
is found in references enumerated herein and are briefly outlined as follows.
Several well-
known methods of introducing target nucleic acids into bacterial cells are
available, any of
which may be used in the present invention. These include: fusion of the
recipient cells with
bacterial protoplasts containing the DNA, treatment of the cells with
liposomes containing
the DNA, electroporation, projectile bombardment (biolistics), carbon fiber
delivery, and
infection with viral vectors (discussed further, below), etc. Bacterial cells
can be used to
amplify the number of plasmids containing DNA constructs of this invention.
The bacteria
are grown to log phase and the plasmids within the bacteria can be isolated by
a variety of
methods known in the art (see, for instance, Sambrook). In addition, a
plethora of kits are
commercially available for the purification of plasmids from bacteria. For
their proper use,
follow the manufacturer's instructions (see, for example, EasyPrep'TM,
FlexiPrepTM, both from
Pharmacia Biotech; StrataCleanTM, from Stratagene; and, QlAprepTM from
Qiagen). The
isolated and purified plasmids are then further manipulated to produce other
plasmids, used to
54
Date Recue/Date Received 2023-12-08
transfect plant cells or incorporated into Agrobacterium tumefaciens related
vectors to infect
plants. Typical vectors contain transcription and translation terminators,
transcription and
translation initiation sequences, and promoters useful for regulation of the
expression of the
particular target nucleic acid. The vectors optionally comprise generic
expression cassettes
containing at least one independent terminator sequence, sequences permitting
replication of
the cassette in eukaryotes, or prokaryotes, or both, (e.g., shuttle vectors)
and selection
markers for both prokaryotic and eukaryotic systems. Vectors are suitable for
replication and
integration in prokaryotes, eukaryotes, or preferably both. See, Giliman and
Smith, (1979)
Gene 8:81; Roberts, et al., (1987) Nature 328:731; Schneider, et al., (1995)
Protein Expr Purif
6435:10; Ausubel, Sambrook, Berger (all supra). A catalogue of Bacteria and
Bacteriophages
useful for cloning is provided, e.g., by the ATCC, e.g., The ATCC Catalogue of
Bacteria and
Bacteriophaqe (1992) Gherna, et al., (eds) published by the ATCC. Additional
basic
procedures for sequencing, cloning and other aspects of molecular biology and
underlying
theoretical considerations are also found in Watson, et al., (1992)
Recombinant DNA, Second
Edition, Scientific American Books, NY. In addition, essentially any nucleic
acid (and
virtually any labeled nucleic acid, whether standard or non-standard) can be
custom or
standard ordered from any of a variety of commercial sources, such as the
Midland Certified
Reagent Company (Midland, Tex.), The Great American Gene Company (Ramona,
Calif.),
ExpressGen Inc. (Chicago, Ill.), Operon Technologies Inc. (Alameda, Calif.)
and many
others.
[00203]
Techniques for transforming plant cells with nucleic acids are widely
available
and can be readily adapted to the invention. In addition to Berger, Ausubel
and Sambrook, all
supra, useful general references for plant cell cloning, culture and
regeneration include Jones,
(ed) (1995) Plant Gene Transfer and Expression Protocols _______________
Methods in Molecular Biology,
Volume 49 Humana Press Towata N.J.; Payne, et al., (1992) Plant Cell and
Tissue Culture in
Liquid Systems John Wiley & Sons, Inc. New York, N.Y. (Payne); and Gamborg and
Phillips, (eds) (1995) Plant Cell, Tissue and Organ Culture; Fundamental
Methods Springer
Lab Manual, Springer-Verlag (Berlin Heidelberg New York) (Gamborg). A variety
of cell
culture media are described in Atlas and Parks, (eds) The Handbook of
Microbiological
Media (1993) CRC Press, Boca Raton, Fla. (Atlas). Additional information for
plant cell
culture is found in available commercial literature such as the Life Science
Research Cell
Culture Catalogue (1998) from Sigma-Aldrich, Inc (St Louis, Mo.) (Sigma-
LSRCCC) and,
e.g., the Plant Culture Catalogue and supplement (1997) also from Sigma-
Aldrich, Inc (St
Louis, Mo.) (Sigma-PCCS). Additional details regarding plant cell culture are
found in Croy,
Date Recue/Date Received 2023-12-08
(ed.) (1993) Plant Molecular Biology, Bios Scientific Publishers, Oxford, U.K.
[00204] The nucleic acid constructs of the invention, e.g., plasmids,
cosmids, artificial
chromosomes, DNA and RNA polynucleotides, are introduced into plant cells,
either in
culture or in the organs of a plant by a variety of conventional techniques.
Techniques for
transforming a wide variety of higher plant species are also well known and
described in
widely available technical, scientific, and patent literature. See, for
example, VVeissinger, et
al., (1988) Ann Rev Genet 22:421-477. The DNA constructs of the invention, for
example
plasmids, phagemids, cosmids, phage, naked or variously conjugated-DNA
polynucleotides,
(e.g., polylysine-conjugated DNA, peptide-conjugated DNA, liposome-conjugated
DNA,
etc.), or artificial chromosomes, can be introduced directly into the genomic
DNA of the
plant cell using techniques such as electroporation and microinjection of
plant cell
protoplasts, or the DNA constructs can be introduced directly to plant cells
using ballistic
methods, such as DNA particle bombardment.
[00205] Microinjection techniques for injecting plant, e.g., cells,
embryos, callus and
protoplasts, are known in the art and well described in the scientific and
patent literature. For
example, a number of methods arc described in Jones, (ed) (1995) Plant Gene
Transfer and
Expression Protocols¨Methods in Molecular Biology, Volume 49 Humana Press,
Towata,
N.J., as well as in the other references noted herein and available in the
literature.
[00206] For example, the introduction of DNA constructs using polyethylene
glycol
precipitation is described in Paszkowski, et al., (1984) EM130 J 3:2717.
Electroporation
techniques are described in Fromm, et al., (1985) Proc Natl Acad Sci USA
82:5824. Ballistic
transformation techniques are described in Klein, et al., (1987) Nature 327:70-
73. Additional
details are found in Jones, (1995) and Gamborg and Phillips, (1995), supra,
and in U.S. Pat.
No. 5,990,387.
[00207] Alternatively, and in some cases preferably, Agrobacterium mediated
transformation is employed to generate transgenic plants. Agrobacterium-
mediated
transformation techniques, including disarming and use of binary vectors, are
also well
described in the scientific literature. See, for example, Horsch, et al.,
(1984) Science 233:496;
and Fraley, et al., (1984) Proe Natl Acad Sci USA 80:4803 and recently
reviewed in Hansen
and Chilton, (1998) Current Topics in Microbiology 240:22; and Das, (1998)
Subcellular
Biochemistry 29: Plant Microbe Interactions, pp 343-363.
[00208] DNA constructs are optionally combined with suitable T-DNA flanking
regions
and introduced into a conventional Agrobacterium tumefaciens host vector. The
virulence
functions of the Agrobacterium tumefaciens host will direct the insertion of
the construct and
56
Date Recue/Date Received 2023-12-08
adjacent marker into the plant cell DNA when the cell is infected by the
bacteria. See, U.S.
Pat. No. 5,591,616. Although Agrobacterium is useful primarily in dicots,
certain monocots
can be transformed by Agrobacterium. For instance, Agrobacterium
transformation of maize
is described in U.S. Pat. No. 5,550,318.
[00209] Other methods of transfection or transformation include (1)
Agrobacterium
rhizogenes-mediated transformation (see, e.g., Lichtenstein and Fuller, (1987)
In: Genetic
Engineering, vol. 6, PWJ Rigby, Ed., London, Academic Press; and Lichtenstein
and Draper
(1985) In: DNA Cloning, Vol. II, Glover, Ed., Oxford, IRI Press; WO 88/02405,
published
Apr. 7, 1988, describes the use of A. rhizogenes strain A4 and its Ri plasmid
along with A.
tumefaciens vectors pARC8 or pARC16 (2) liposome-mediated DNA uptake (see,
e.g.,
Freeman, et al., (1984) Plant Cell Physiol 25:1353), (3) the vortexing method
(see, e.g.,
Kindle, (1990) Proc Natl Acad Sci USA 87:1228.
[00210] DNA can also be introduced into plants by direct DNA transfer into
pollen as
described by Zhou, et al., (1983) Methods in Enzymology 101:433; Hess, (1987)
Intern Rev
Cytol 107:367; Luo, et al., (1988) Plant Mol Biol Rep 6:165. Expression of
polypeptide
coding genes can be obtained by injection of the DNA into reproductive organs
of a plant as
described by Pena, et al., (1987) Nature 325:274. DNA can also be injected
directly into the
cells of immature embryos and the desiccated embryos rehydrated as described
by Neuhaus,
et al., (1987) Theor Appl Genet 75:30; and Benbrook, et al., (1986) in
Proceedings Bio Expo
Butterworth, Stoneham, Mass., pp. 27-54. A variety of plant viruses that can
be employed as
vectors are known in the art and include cauliflower mosaic virus (CaMV),
geminivirus,
brome mosaic virus, and tobacco mosaic virus.
[00211] Transformed plant cells which are derived by any of the above
transformation
techniques can be cultured to regenerate a whole plant that possesses the
transformed
genotype and thus the desired phenotype. Such regeneration techniques rely on
manipulation
of certain phytohormones in a tissue culture growth medium, typically relying
on a biocide
and/or herbicide marker which has been introduced together with the desired
nucleotide
sequences. Plant regeneration from cultured protoplasts is described in Payne,
et al., (1992)
Plant Cell and Tissue Culture in Liquid Systems John Wiley & Sons, Inc. New
York, N.Y.;
Gamborg and Phillips (eds) (1995) Plant Cell, Tissue and Organ Culture;
Fundamental
Methods Springer Lab Manual, Springer-Verlag (Berlin Heidelberg New York);
Evans, et al.,
(1983) Protoplasts Isolation and Culture, Handbook of Plant Cell Culture pp.
124-176,
Macmillian Publishing Company, New York; and Binding (1985) Regeneration of
Plants,
Plant Protoplasts pp. 21-73, CRC Press, Boca Raton. Regeneration can also be
obtained from
57
Date Recue/Date Received 2023-12-08
plant callus, explants, somatic embryos (Dandekar, et al., (1989) J Tissue
Cult Meth 12:145;
McGranahan, et al., (1990) Plant Cell Rep 8:512) organs, or parts thereof.
Such regeneration
techniques are described generally in Klee, et al., (1987) Ann Rev Plant Phys
38:467-486.
Additional details are found in Payne, (1992) and Jones (1995), both supra,
and Weissbach
and Weissbach, eds. (1988) Methods for Plant Molecular Biology Academic Press,
Inc., San
Diego, Calif. This regeneration and growth process includes the steps of
selection of
transformant cells and shoots, rooting the transformant shoots and growth of
the plantlets in
soil. These methods are adapted to the invention to produce transgenic plants
bearing QTLs
according to the methods of the invention.
[00212] In addition, the regeneration of plants containing nucleic acids of
the present
invention and introduced by Agrobacterium into cells of leaf explants can be
achieved as
described by Horsch, et al., (1985) Science 227:1229-1231. In this procedure,
transformants
are grown in the presence of a selection agent and in a medium that induces
the regeneration
of shoots in the plant species being transformed as described by Fraley, et
al., (1983) Proc
Natl Acad Sci USA 80:4803. This procedure typically produces shoots within two
to four
weeks and these transformant shoots are then transferred to an appropriate
root-inducing
medium containing the selective agent and an antibiotic to prevent bacterial
growth.
Transgenic plants of the present invention may be fertile or sterile.
[00213] It is
not intended that plant transformation and expression of polypeptides that
provide disease tolerance, as provided by the present invention, be limited to
soybean species.
Indeed, it is contemplated that the polypeptides that provide the desired
tolerance in soybean
can also provide such tolerance when transfol __________________________ ined
and expressed in other agronomically and
horticulturally important species. Such species include primarily dicots,
e.g., of the families:
Leguminosae (including pea, beans, lentil, peanut, yarn bean, cowpeas, velvet
beans,
soybean, clover, alfalfa, lupine, vetch, lotus, sweet clover, wisteria and
sweetpea); and
Compositae (the largest family of vascular plants, including at least 1,000
genera, including
important commercial crops such as sunflower).
[00214] Additionally, preferred targets for modification with the nucleic
acids of the
invention, as well as those specified above, plants from the genera: Allium,
Apium, Arachis,
Brassica, Capsicum, Cicer, Cucumis, Curcubita, Daucus, Fagopyrum, Glycine,
Helianthus,
Lactuca, Lens, Lycopersicon, Medicago, Pisum, Phaseolus, Solanum, Trifolium,
Vigna and
many others.
[00215] Common crop plants which are targets of the present invention include
soybean,
sunflower, canola, peas, beans, lentils, peanuts, yam beans, cowpeas, velvet
beans, clover,
58
Date Recue/Date Received 2023-12-08
alfalfa, lupine, vetch, sweet clover, sweetpea, field pea, fava bean,
broccoli, brussel sprouts,
cabbage, cauliflower, kale, kohlrabi, celery, lettuce, carrot, onion, pepper,
potato, eggplant
and tomato.
[00216] In construction of recombinant expression cassettes of the invention,
which
include, for example, helper plasmids comprising virulence functions, and
plasmids or
viruses comprising exogenous DNA sequences such as structural genes, a plant
promoter
fragment is optionally employed which directs expression of a nucleic acid in
any or all
tissues of a regenerated plant. Examples of constitutive promoters include the
cauliflower
mosaic virus (CaMV) 35S transcription initiation region, the 1'- or 2'-
promoter derived from
T-DNA of Agrobacterium tumefaciens, and other transcription initiation regions
from various
plant genes known to those of skill. Alternatively, the plant promoter may
direct expression
of nucleic acids of the invention in a specific tissue (tissue-specific
promoters) or may be
otherwise under more precise environmental control (inducible promoters).
Examples of
tissue-specific promoters under developmental control include promoters that
initiate
transcription only in certain tissues, such as fruit, seeds or flowers.
[00217] Any of a number of promoters which direct transcription in plant cells
can be
suitable. The promoter can be either constitutive or inducible. In addition to
the promoters
noted above, promoters of bacterial origin that operate in plants include the
octopine synthase
promoter, the nopaline synthase promoter and other promoters derived from
native Ti
plasmids. See, Herrara-Estrella, et al., (1983) Nature 303:209. Viral
promoters include the
35S and 19S RNA promoters of cauliflower mosaic virus. See, Odell, et al.,
(1985) Nature
313:810. Other plant promoters include Kunitz trypsin inhibitor promoter
(KTI), SCP1, SUP,
UCD3, the ribulose-1,3-bisphosphate carboxylase small subunit promoter and the
phaseolin
promoter. The promoter sequence from the E8 gene and other genes may also be
used. The
isolation and sequence of the E8 promoter is described in detail in Deikman
and Fischer
(1988) EMBO J 7:3315. Many other promoters are in current use and can be
coupled to an
exogenous DNA sequence to direct expression.
[00218] If expression of a polypeptide from a cDNA is desired, a
polyadenylation region
at the 3'-end of the coding region is typically included. The polyadenylation
region can be
derived from the natural gene, from a variety of other plant genes, or from,
e.g., T-DNA.
[00219] A vector comprising sequences of the invention will typically include
a nucleic
acid subsequence, a marker gene which confers a selectable, or alternatively,
a screenable,
phenotype on plant cells. For example, the marker can encode biocide
tolerance, particularly
antibiotic tolerance, such as tolerance to kanamycin, G418, bleomycin,
hygromycin, or
59
Date Recue/Date Received 2023-12-08
herbicide tolerance, such as tolerance to chlorosulforon, or phosphinothricin
(the active
ingredient in the herbicides bialaphos or Basta). See, e.g., Padgette, et al.,
(1996) In:
Herbicide-Resistant Crops (Duke, ed.), pp 53-84, CRC Lewis Publishers, Boca
Raton
("Padgette, 1996"). For example, crop selectivity to specific herbicides can
be conferred by
engineering genes into crops that encode appropriate herbicide metabolizing
enzymes from
other organisms, such as microbes. See, Vasil, (1996) In: Herbicide-Resistant
Crops (Duke,
ed.), pp 85-91, CRC Lewis Publishers, Boca Raton) ("Vasil", 1996).
[00220] One of skill will recognize that after the recombinant expression
cassette is stably
incorporated in transgenic plants and confirmed to be operable, it can be
introduced into other
plants by sexual crossing. Any of a number of standard breeding techniques can
be used,
depending upon the species to be crossed. In vegetatively propagated crops,
mature
transgenic plants can be propagated by the taking of cuttings or by tissue
culture techniques
to produce multiple identical plants. Selection of desirable transgenics is
made and new
varieties are obtained and propagated vegetatively for commercial use. In seed
propagated
crops, mature transgenic plants can be self crossed to produce a homozygous
inbred plant.
The inbred plant produces seed containing the newly introduced heterologous
nucleic acid.
These seeds can be grown to produce plants that would produce the selected
phenotype. Parts
obtained from the regenerated plant, such as flowers, seeds, leaves, branches,
fruit, and the
like are included in the invention, provided that these parts comprise cells
comprising the
isolated nucleic acid of the present invention. Progeny and variants, and
mutants of the
regenerated plants are also included within the scope of the invention,
provided that these
parts comprise the introduced nucleic acid sequences.
[00221] Transgenic or introgressed plants comprising nucleic acids of the
present
invention can be screened for transmission of the nucleic acid of the present
invention by, for
example, standard nucleic acid detection methods or by immunoblot protocols.
[00222] A preferred embodiment of the invention is a transgenic plant that is
homozygous
for the added heterologous nucleic acid; e.g., a transgenic plant that
contains two added
nucleic acid sequence copies. A homozygous transgenic plant can be obtained by
sexually
mating (self-fertilizing) a heterozygous transgenic plant that contains a
single added
heterologous nucleic acid. Back-crossing to a parental plant and out-crossing
with a non-
transgenic plant can be used to introgress the heterologous nucleic acid into
a selected
background (e.g., an elite or exotic soybean line).
XL METHODS FOR CHARCOAL ROT DROUGHT COMPLEX TOLERANT SOYBEAN PLANTS
Date Recue/Date Received 2023-12-08
[00223] Experienced plant breeders can recognize tolerant soybean plants in
the field, and
can select the tolerant individuals or populations for breeding purposes or
for propagation. In
this context, the plant breeder recognizes "tolerant" and "non-tolerant" or
"susceptible",
soybean plants.
[00224] Such plant breeding practitioners will appreciate that plant
tolerance is a
phenotypic spectrum consisting of extremes in tolerance, susceptibility and a
continuum of
intermediate tolerance phenotypes. Tolerance also varies due to environmental
effects and the
severity of pathogen infection. Evaluation of phenotypes using reproducible
assays and
tolerance scoring methods are of value to scientists who seek to identify
genetic loci that
impart tolerance, conduct marker assisted selection for tolerant populations,
and for
introgression techniques to breed a tolerance trait into an elite soybean
line, for example.
[00225] In contrast to fortuitous field observations that classify plants
as either "tolerant"
or "susceptible", various systems are known for scoring the degree of plant
tolerance or
susceptibility. These techniques can be applied to different fields at
different times, and
provide approximate tolerance scores that can be used to characterize a given
strain
regardless of growth conditions or location.
[00226] Ratings arc assigned by evaluating all plants of a cultivar in a 2 row
by 15 foot
plot. Cultivar scores are based on a 1 to 9 system where a score of 9¨no
disease symptoms
with normal plant growth; 8¨very slight symptoms including up to a 10%
reduction in leaflet
and overall canopy size with no wilting; 7==wilting beginning to appear at the
uppermost two
nodes; 6=wilting at the uppermost three nodes and leaflet yellowing beginning
appear; 5¨Up
to 5% plant death with wilting and yellowing of leaflets occurring at the
uppermost four
nodes; 4¨Up to 10% plant death with wilting and yellowing of leaflets
occurring at the
uppermost four nodes; 3=Up to 25% plant death with wilting and yellowing of
leaflets
occurring at the uppermost four nodes; 2=up to 50% plant death; 1=50-100%
plant death.
FIG. 8 gives a representative example of cultivars with vastly different
Charcoal Rot Drought
Complex tolerance using this scoring system.
XII. AUTOMATED DETECTION/CORRELATION SYSTEMS OF THE INVENTION
[00227] In some embodiments, the present invention includes an automated
system for
detecting markers of the invention and/or correlating the markers with a
desired phenotype
(e.g., tolerance). Thus, a typical system can include a set of marker probes
or primers
configured to detect at least one favorable allele of one or more marker locus
associated with
61
Date Recue/Date Received 2023-12-08
tolerance or improved tolerance to Charcoal Rot Drought Complex. These probes
or primers
are configured to detect the marker alleles noted in the tables and examples
herein, e.g., using
any available allele detection format, e.g., solid or liquid phase array based
detection,
microfluidic-based sample detection, etc.
[00228] For example, in one embodiment, the marker locus is S29725-001; S29741-
001;
S29742-001; S11315-1; and S11316-1, or any combination thereof, as well as any
of the
chromosome intervals such as: (i) a chromosomal interval located at about 5 cM
to about 26
cM of chromosome 15; (ii) a chromosomal interval located at about 81 cM to
about 102 cM
of chromosome 19; (iii) the interval on Chr. 15 flanked by and including
S29725-001 and
S29741-001; or (iv) the interval on Chr. 19 flanked by and including S11315-1
and S13116-
1.
[00229] For example, in an alternative embodiment, the marker locus is a locus
in any of
the chromosome intervals such as: (i) a chromosomal interval located at about
17 cM to about
38 cM of chromosome 5; (ii) a chromosomal interval located at about 5 cM to
about 26 cM of
chromosome 15; (iii) a chromosomal interval located at about 19 cM to about 40
cM of
chromosome 19; or (iv) a chromosomal interval located at about 81 cM to about
102 cM of
chromosome 19. In a further alternative embodiment, the marker locus is a
locus in any of
the chromosome intervals such as: (i) a chromosomal interval located within
about 2 Mbp of
an interval at 7,975-8,015 kpb of chromosome 5; (ii) a chromosomal interval
located within
about 2 Mbp of an interval at 3,202-3,212 kbp of chromosome 15 (SEQ ID NO.:
26); (iii) a
chromosomal interval located within about 2 Mbp of an interval at 27,178-
27,218 kbp; or (iv)
a chromosomal interval located within about 2 Mbp of an interval at 48,340-
48,380 kbp of
chromosome 19 (SEQ ID NO.: 27).
[00230] The typical system includes a detector that is configured to detect
one or more
signal outputs from the set of marker probes or primers, or amplicon thereof,
thereby
identifying the presence or absence of the allele. A wide variety of signal
detection apparatus
are available, including photo multiplier tubes, spectrophotometers, CCD
arrays, arrays and
array scanners, scanning detectors, phototubes and photodiodes, microscope
stations, galvo-
scanns, microfluidic nucleic acid amplification detection appliances and the
like. The precise
configuration of the detector will depend, in part, on the type of label used
to detect the
marker allele, as well as the instrumentation that is most conveniently
obtained for the user.
Detectors that detect fluorescence, phosphorescence, radioactivity, pH,
charge, absorbance,
luminescence, temperature, magnetism or the like can be used. Typical detector
embodiments
include light (e.g., fluorescence) detectors or radioactivity detectors. For
example, detection
62
Date Recue/Date Received 2023-12-08
of a light emission (e.g., a fluorescence emission) or other probe label is
indicative of the
presence or absence of a marker allele. Fluorescent detection is especially
preferred and is
generally used for detection of amplified nucleic acids (however, upstream
and/or
downstream operations can also be performed on amplicons, which can involve
other
detection methods). In general, the detector detects one or more label (e.g.,
light) emission
from a probe label, which is indicative of the presence or absence of a marker
allele.
[00231] The detector(s) optionally monitors one or a plurality of signals from
an
amplification reaction. For example, the detector can monitor optical signals
which
correspond to "real time" amplification assay results.
1002321
System instructions that correlate the presence or absence of the favorable
allele
with the predicted tolerance are also a feature of the invention. For example,
the instructions
can include at least one look-up table that includes a correlation between the
presence or
absence of the favorable alleles and the predicted tolerance or improved
tolerance. The
precise form of the instructions can vary depending on the components of the
system, e.g.,
they can be present as system software in one or more integrated unit of the
system (e.g., a
microprocessor, computer or computer readable medium), or can be present in
one or more
units (e.g., computers or computer readable media) operably coupled to the
detector. As
noted, in one typical embodiment, the system instructions include at least one
look-up table
that includes a correlation between the presence or absence of the favorable
alleles and
predicted tolerance or improved tolerance. The instructions also typically
include instructions
providing a user interface with the system, e.g., to permit a user to view
results of a sample
analysis and to input parameters into the system.
[00233] The system typically includes components for storing or transmitting
computer
readable data representing or designating the alleles detected by the methods
of the present
invention, e.g., in an automated system. The computer readable media can
include cache,
main, and storage memory and/or other electronic data storage components (hard
drives,
floppy drives, storage drives, etc.) for storage of computer code. Data
representing alleles
detected by the method of the present invention can also be electronically,
optically, or
magnetically transmitted in a computer data signal embodied in a transmission
medium over
a network such as an intranet or internet or combinations thereof. The system
can also or
alternatively transmit data via wireless, IR, or other available transmission
alternatives.
1002341 During operation, the system typically comprises a sample that is to
be analyzed,
such as a plant tissue, or material isolated from the tissue such as genomic
DNA, amplified
genomic DNA, cDNA, amplified cDNA, RNA, amplified RNA, or the like.
63
Date Recue/Date Received 2023-12-08
[00235] The
phrase "allele detection/correlation system" in the context of this invention
refers to a system in which data entering a computer corresponds to physical
objects or
processes external to the computer, e.g., a marker allele, and a process that,
within a
computer, causes a physical transformation of the input signals to different
output signals. In
other words, the input data, e.g., amplification of a particular marker allele
is transformed to
output data, e.g., the identification of the allelic form of a chromosome
segment. The process
within the computer is a set of instructions, or "program", by which positive
amplification or
hybridization signals are recognized by the integrated system and attributed
to individual
samples as a genotype. Additional programs correlate the identity of
individual samples with
phenotypic values or marker alleles, e.g., statistical methods. In addition
there are numerous
e.g., C/C++ programs for computing, Delphi and/or Java programs for GUI
interfaces, and
productivity tools (e.g., Microsoft Excel and/or SigmaPlot) for charting or
creating look up
tables of relevant allele-trait correlations. Other useful software tools in
the context of the
integrated systems of the invention include statistical packages such as SAS,
Genstat, Matlab,
Mathematica, and S-Plus and genetic modeling packages such as QU-GENE.
Furthermore,
additional programming languages such as visual basic arc also suitably
employed in the
integrated systems of the invention.
[00236] For example, tolerance marker allele values assigned to a population
of progeny
descending from crosses between elite lines are recorded in a computer
readable medium,
thereby establishing a database corresponding tolerance alleles with unique
identifiers for
members of the population of progeny. Any file or folder, whether custom-made
or
commercially available (e.g., from Oracle or Sybase) suitable for recording
data in a
computer readable medium is acceptable as a database in the context of the
present invention.
Data regarding genotype for one or more molecular markers, e.g., ASH, SSR,
RFLP, RAPD,
AFLP, SNP, isozyme markers or other markers as described herein, are similarly
recorded in
a computer accessible database. Optionally, marker data is obtained using an
integrated
system that automates one or more aspects of the assay(s) used to determine
marker(s)
genotype. In such a system, input data corresponding to genotypes for
molecular markers are
relayed from a detector, e.g., an array, a scanner, a CCD, or other detection
device directly to
files in a computer readable medium accessible to the central processing unit.
A set of system
instructions (typically embodied in one or more programs) encoding the
correlations between
tolerance and the alleles of the invention is then executed by the
computational device to
identify correlations between marker alleles and predicted trait phenotypes.
[00237] Typically, the system also includes a user input device, such as a
keyboard, a
64
Date Recue/Date Received 2023-12-08
mouse, a touchscreen, or the like (for, e.g., selecting files, retrieving
data, reviewing tables of
maker information), and an output device (e.g., a monitor, a printer) for
viewing or
recovering the product of the statistical analysis.
[00238] Thus, in one aspect, the invention provides an integrated system
comprising a
computer or computer readable medium comprising a set of files and/or a
database with at
least one data set that corresponds to the marker alleles herein. The system
also includes a
user interface allowing a user to selectively view one or more of these
databases. In addition,
standard text manipulation software such as word processing software (e.g.,
Microsoft
WordTM or Corel WordPerfectTM) and database or spreadsheet software (e.g.,
spreadsheet
software such as Microsoft ExcelTM, Corel Quattro ProTM, or database programs
such as
Microsoft ACCeSSTM or ParadoxTM) can be used in conjunction with a user
interface (e.g., a
GUI in a standard operating system such as a Windows, Macintosh, Unix or Linux
system) to
manipulate strings of characters corresponding to the alleles or other
features of the database.
[00239] The systems optionally include components for sample manipulation,
e.g.,
incorporating robotic devices. For example, a robotic liquid control armature
for transferring
solutions (e.g., plant cell extracts) from a source to a destination, e.g.,
from a microtiter plate
to an array substrate, is optionally operably linked to the digital computer
(or to an additional
computer in the integrated system). An input device for entering data to the
digital computer
to control high throughput liquid transfer by the robotic liquid control
armature and,
optionally, to control transfer by the armature to the solid support is
commonly a feature of
the integrated system. Many such automated robotic fluid handling systems are
commercially
available. For example, a variety of automated systems are available from
Caliper
Technologies (Hopkinton, Mass.), which utilize various Zymate systems, which
typically
include, e.g., robotics and fluid handling modules. Similarly, the common RCA
robot,
which is used in a variety of laboratory systems, e.g., for microtiter tray
manipulation, is also
commercially available, e.g., from Beckman Coulter, Inc. (Fullerton, Calif.).
As an
alternative to conventional robotics, microfluidic systems for performing
fluid handling and
detection are now widely available, e.g., from Caliper Technologies Corp.
(Hopkinton,
Mass.) and Agilent Technologies (Palo Alto, Calif.).
1002401 Systems for molecular marker analysis of the present invention can,
thus, include
a digital computer with one or more of high-throughput liquid control
software, image
analysis software for analyzing data from marker labels, data interpretation
software, a
robotic liquid control armature for transferring solutions from a source to a
destination
operably linked to the digital computer, an input device (e.g., a computer
keyboard) for
Date Recue/Date Received 2023-12-08
entering data to the digital computer to control high throughput liquid
transfer by the robotic
liquid control armature and, optionally, an image scanner for digitizing label
signals from
labeled probes hybridized, e.g., to markers on a solid support operably linked
to the digital
computer. The image scanner interfaces with the image analysis software to
provide a
measurement of, e.g., nucleic acid probe label intensity upon hybridization to
an arrayed
sample nucleic acid population (e.g., comprising one or more markers), where
the probe label
intensity measurement is interpreted by the data interpretation software to
show whether, and
to what degree, the labeled probe hybridizes to a marker nucleic acid (e.g.,
an amplified
marker allele). The data so derived is then correlated with sample identity,
to determine the
identity of a plant with a particular genotype(s) for particular markers or
alleles, e.g., to
facilitate marker assisted selection of soybean plants with favorable allelic
forms of
chromosome segments involved in agronomic performance (e.g., tolerance or
improved
tolerance).
[00241] Optical images, e.g., hybridization patterns viewed (and, optionally,
recorded) by
a camera or other recording device (e.g., a photodiodc and data storage
device) are optionally
further processed in any of the embodiments herein, e.g., by digitizing the
image and/or
storing and analyzing the image on a computer. A variety of commercially
available
peripheral equipment and software is available for digitizing, storing and
analyzing a
digitized video or digitized optical image, e.g., using PC (Intel x86 or
Pentium chip-
compatible DOSTM, OS2TM WINDOWSTM, WINDOWS NTTm or WINDOWS 95TM based
machines), MACINTOSHTm, LINUX, or UNIX based (e.g., SUNTM work station)
computers.
XIH. METHODS TO SCREEN PLANTS FPOR RESISTANCE TO A PLANT PATHOGEN
[00242] In some embodiments, the present invention includes methods for
screening a
plant for tolerance to a plant pathogen, the method comprising: (a) providing
at least one
inoculation probe having a pointed end to a container of agar inoculated with
a pathogen;
wherein a surface of the inoculation probe is contact with the surface of the
agar in the petri
dish; (b) inoculating a plant, after a predetermined contact time between at
least one
inoculation probe and the pathogen, by inserting the pointed end of at least
one inoculation
probe, comprising pathogen on the surface thereof, into a site located on a
plant stem; and (c)
assessing plant tolerance to the pathogen at a predetermined time. The method
is significantly
better at phenotyping for resistance or sensitivity to a plant pathogen. Thus,
the disclosed
new method enables reliable phenotyping in the growth chamber that more
accurately
66
Date Recue/Date Received 2023-12-08
matches field based results.
[00243] For example, as disclosed herein, this method when used for screening
plant
resistance to charcoal rot tolerance provides results that match with field
observations. The
results obtained using the disclosed method are superior to other methods for
screening for
resistance to charcoal rot, including, for example, the previously described
standard of
Twizeyimana et al., (Plant Disease (2012) 96(8):1210-1215). Without wishing to
be bound
by a particular theory, it is believed that the disclosed method for screening
provides for
superior results, in part, because the pathogen grows in intimate contact with
the inoculation
probe and that the inoculation probe is made of a material suitable for
pathogen growth. In
various aspects, without wishing to be bound by a particular theory, it is
believed that the
disclosed method permits more efficient transfer of pathogen to the plant.
[00244] In various aspects, the pathogen is a Macophomina phaseolina isolate.
In a
further aspect, the plant is Glycine max. In a still further aspect, the
pathogen is a
Macophomina phaseolina isolate; and the plant is Glycine max.
[00245] In various aspects, the pathogen is a Macophomina phaseolina isolate;
the plant is
Glycine max; and the agar is potato dextrose agar. In a further aspect, the
pathogen is a
Macophomina phaseolina isolate; the plant is Glycine max; and the pathogen is
grown in
contact with the at least one inoculation probe is 7-9 days. In a still
further aspect, the
pathogen is a Afacophomina phaseolina isolate; the plant is Glycine max; and
the located on
the plant stem about 0.1 to 1.5 cm above cotyledons of the plant. In a yet
further aspect, the
pathogen is a Macophomina phaseolina isolate; the plant is Glycine max; and
the the site
located on the plant stem is sealed following insertion of the pointed end of
at least one
inoculation probe. In an even further aspect, the pathogen is a Macophotnina
phaseolina
isolate; the plant is Glycine max; and the the site located on the plant stem
is sealed with
petroleum jelly following insertion of the pointed end of at least one
inoculation probe. In a
still further aspect, the pathogen is a Macophomina phaseolina isolate; the
plant is Glycine
max; and the the plant tolerance is assessed at 14-21 days after inoculation
of the plant.
[00246] In variouos aspects, the present invention includes methods for
screening a
Glycine max plant for tolerance to Macophomina phaseolina, the method
comprising: (a)
providing at least one inoculation probe having a pointed end to a container
of agar
inoculated with Macophomina phaseolina; wherein a surface of the inoculation
probe is
contact with the surface of the agar in the petri dish; (b) inoculating a
plant, after contact time
5-15 days between at least one inoculation probe and the pathogen, by
inserting the pointed
end of at least one inoculation probe, comprising pathogen on the surface
thereof, into a site
67
Date Recue/Date Received 2023-12-08
located on the plant stem about 0.1 to 1.5 cm above cotyledons of the Glycine
max plant; and
(c) assessing Glycine max plant tolerance to Macophomina phaseolina at 14-21
days after
inoculation of the plant. The method is significantly better at phenotyping
for resistance or
sensitivity to a plant pathogen. Thus, the disclosed new method enables
reliable phenotyping
in the growth chamber that more accurately matches field based results.
[00247] The shape, size, and material of inoculation probe can be varied as
deemed
appropriate by the skilled artisan. The inoculation probe should be of size
and shape that
allows it to be inserted into the plant that is to be inoculated. The material
from which the
inoculation probe is made or fabricated should be a material that permits
growth of the
pathogen on the inoculation probe when it is incontact with agar in a petri
dish. In a further
aspect, the inoculation probe is sterilized prior to use. Sterilization of the
probe prior to use
can be by any method that allows sterilization of the inoculation probe
without comprising
the structure of the inoculation probe, e.g., autoclaving the sterilization
probe. In a still
further aspect, the inoculation probe is about 0.5 ¨ 1.5 cm in length and
about 0.01 ¨ 0.1 cm
in width. In a yet further aspect, the inoculation probe is cylindrical in
shape; wherein the the
diameter of the cylinder is about 0.01 ¨ 0.1 cm; wherein the cylinder is about
about 0.5 ¨ 1.5
cm in length; and wherein one end of the cylinder forms the point of the
inoculation probe.
In various aspects, the inoculation probe is solid and/or without an open end.
In a further
aspect, the inoculation probe is not hollow or an open tube. A key aspect of
the inoculation
probe is that one end of it is pointed and allows for insertion into the plant
to be inoculated.
In a further aspect, the inoculation probe is made of a material suitable for
pathogen growth
or adherence, e.g. wood.
[00248] In various aspects, the the inoculation probe is fabricated from a
wooden
toothpick, wherein about 0.5-1.5 cm is removed from one end of the toothpick
and leaving
intact a pointed end, thereby forming a pointed inoculation probe about 0.5-
1.5 in length with
a pointed end.
[00249] In a further aspect, the number of inoculation probes placed on the
surface of agar
in the petri dish is of a density such that at least a portion of the surface
of the inoculation
probe is in contact with the agar surface. For example, when inoculation
probes are prepared
from wooden toothpicks as described herein above, it is desirable to evenly
distribute about
70 to about 110 such inoculation probes on the agar surface of a petri dish
with about a 100
mm diameter.
1002501 The skilled artisan may seal the insertion following inoculation of
the plant.
Various materials can be used to seal the insertion site, including, but not
limited to,
68
Date Recue/Date Received 2023-12-08
petroleum jelly and the like.
EXAMPLES
[00251] Example 1: Growth Chamber Screenin2 Method.
[00252] Plants were grown in 10.1 cm2 plastic pots (W.H. Milikowski, Inc.) in
Metro Mix
900 potting soil (Sun Gro Horticulture Inc.). Six pots (replicates) were
planted of each
soybean entry, with 5 seeds planted per pot. Seedlings were grown out for 8
days prior to
inoculation. During this period, seedlings were maintained in a growth chamber
at 26.7 C
with a 16 hour photoperiod (ppd) under metal halide lighting. Plants were
watered for
optimal seed germination and health. On the ninth day following planting
(unifoliate growth
stage), plants were inoculated.
[00253] All plants were inoculated with microsclerotia of Macrophornina
phaseolina
(causal agent of charcoal rot) isolate MP3, collected in May 2008 from
Champaign County,
Illinois. The isolate was maintained on Microbank cryopreservation beads (Pro-
Lab
Diagnostics) at -80 C and re-isolated periodically from infected plants to
maintain isolate
virulence. To prepare for inoculum production, M. phaseolina infested
microbeads were
removed from cold storage and grown on full-strength potato dextrose agar
(PDA; 39 grams
potato dextrose agar/liter water) at 23 C for 3 days. An agar plug was excised
from the
leading edge of the actively growing M. phaseolina culture and transferred to
a full-strength
PDA plate; these plates were incubated at 23 C for 2 days. Round wooden
toothpicks, cut
approximately 1 cm from the tip with the tip sections retained, were
sterilized by autoclaving
at 121 C. Six 3-mm agar plugs were excised from the actively growing edge of
the M.
phaseolina culture plates and evenly distributed on the surface of a 100 x 25
mm full-strength
PDA plate; 80 to 100 sterile toothpick tips were distributed over the surface
of these PDA
plates, which were incubated at 23 C for nine days, at which point large
quantities of black
microsclerotia were visible on the surface of the toothpicks.
[00254] A new method screening was developed and used in studies described
herein.
Plants were inoculated by inserting a microsclerotia-infested toothpick, tip
first, into the plant
stem 5 to 8 mm above the cotyledons. Only three plants were inoculated per
pot, for a total of
18 plants per entry. Plants without fully unrolled unifoliate leaves were
discarded. The wound
at the inoculation site was sealed with petroleum jelly. Inoculated plants
were placed in a dew
room at 25.6 C with a 16 hour ppd for 48 hours. During this period, plants
were misted at 100
cc/min by an oscillating Aquafog XE-330 Turbo (Jaybird Manufacturing Inc.)
humidifier for
69
Date Recue/Date Received 2023-12-08
30 mins of each daylight hour and 15 mins of each nighttime hour. As discussed
herein
below, the efficacy of this screening method was compared to a standard,
publicly available
method (Twizeyimana, M., et al., Plant Disease (2012) 96(8):1210-1215).
[00255] After 48 hours, plants were returned to the growth chamber at 25.6 C
and 16 hour
ppd until they reached the first trifoliate growth stage, at which time the
temperature was
raised by 1 C each day until the temperature reached 29.4 C. Light intensity
during the post-
inoculation period was 50 percent on the first day, and then increased to 75
percent for the
remainder of the experiment. One day after being removed from the dew chamber,
pots were
placed in a pre-assigned location within the growth chamber in randomized
complete block
design. Pots were watered daily and staked as needed to maintain plant health.
All plants
were fertilized 5 to 6 days post-inoculation with 15-5-15 Cal-Mag Excel
fertilizer.
[00256] Plants were assessed for level of charcoal rot tolerance 14 to 21 days
post-
inoculation. The scoring date for each experiment was determined based on the
observed
incidence of dead plants among the susceptible and tolerant check varieties.
Plants were
scored on a 1 to 9 scale, with 1 representing the most susceptible end of the
scale and 9
representing the most tolerant. To assess incidence and severity of internal
stem lesions, the
stems of all living plants were cut longitudinally from 3 to 5 cm below the
point of
inoculation to 10 to 15 cm above the point of inoculation. A score of 1
indicates that the plant
was heavily infected, completely wilting, dying, or dead. A score of 3
indicates that the plant
had a long external and/or internal lesion and was partially wilted. A score
of 5 indicates that
the plant had an internal lesion greater than 1.3 cm in length with no wilting
observed. A
score of 7 indicates the presence of an internal lesion of less than 1.3 cm
with no wilting. A
score of 9 indicates that no charcoal rot symptoms were visible; there may
have been a small
internal scar visible at the point of inoculation. Plants scoring 5, 7, or 9
typically had no
visible external symptoms. Plants with a score of 1 or 3 were considered
susceptible. Plants
with a score of 5 or 7 were considered moderately tolerant, and plants with a
score of 9 were
considered highly tolerant.
[00257] The three plant scores from each pot were averaged and the score of
each pot was
adjusted to account for spatial variation using a Best Linear Unbiased
Estimation ("BLUE")
as previously described (Henderson, C.R. 1975. Best linear unbiased estimation
and
prediction under a selection model. Biometrics 31:423-447). A single BLUE
value was
assigned to each entry in the assay. A Best Linear Unbiased Prediction
("BLUP"; ibia) was
calculated for each entry to account for experiment effects. The BLUP of each
entry was
compared to the BLUP of the established controls to determine the entry's
level of charcoal
Date Recue/Date Received 2023-12-08
rot tolerance.
[00258] Example 2: Comparison of Screening Methods.
[00259] Ten soybean varieties were phenotyped using the public standard
method, the
claimed new method, and screened in the field for charcoal rot tolerance.
Regression analysis
was used to compare the two growth chamber screening methods to field based
scores (Table
2). The data in Table 2 showregression analysis of the growth chamber results
compared to
field-collected charcoal rot phenotypes (2-76 repititions) across 10 plant
varieties. Results
show that the new method disclosed herein (see Example 1) is significantly
better at
phenotyping for charcoal rot tolerance that matches with field observations
than is the
previously described standard method of Twizeyimana et at., (Plant Disease
(2012)
96(8):1210-1215). The disclosed new method enables reliable phenotyping in the
growth
chamber that more accurately matches field based results.
Table 2.
Method Reps R2 ______ p-value
Disclosed New Method 24 0.60 0.008
Twizeyimana, et al. 6-18 0.22 0.176
[00260] Example 3: Recombinant Inbred Line (RIL) Data.
[00261] Two recombinant inbred line (RIL) populations were created by crossing
two
varieties that contrasted for charcoal rot tolerance. Seed was bulk generation
advanced in
Puerto Rico four generations and F5 RILs were derived to create the
populations. The
soybean entries assessed in the charcoal rot laboratory bioassay were 377
recombinant inbred
lines (R1Ls) from population 1 (parent 1 x parent 2) screened across 6
experiments, and 354
RILs from population 2 (parent 1 x parent 3) screened across 5 experiments.
Six replications
of each RIL along with a standard set of tolerant, moderately tolerant, and
susceptible checks
with established charcoal rot tolerance scores were included in the
experiments. Table 3
shows the results of QTL mapping within the populations in which a total of 4
unique QTL
were identified.
Table 3.
P1 P2 QTL Peak QTL Chr. Physical Genetic LOD Variance
Mark ert Position* Position**
1 2 51. 15
S01353-1 15 3,012,488 7.03 14%
1 3 91.68
S11318-1 19 48,436,397 12 11.8
71
Date Recue/Date Received 2023-12-08
1 3 29.32
S03394-1 19 27,198,319 9.9 9.6
1 2 27.82
S04257-1 5 7,995,435 5 6%
Marker nearest to QTL peak.
* Physical position (bp) on the Glyma 1 Assembly reference (Schmutz, Jeremy,
et al.
"Genome sequence of the pa1aeopolyploid soybean." Nature 463.7278 (2010): 178-
183).
** Genetic position (cM) on the Soybean Consensus Map 4.0 (Hyten D. L., et
al., (2010)
Crop Sci 50: 960-968).
[00262] Example 4: Near Isogenic Line (NIL) Data..
[00263] Near isogenic lines (NILs) were created by identifying F3 varieties
that were
heterozygous across the QTL interval. Individual F3:5 plants within the F3
varieties were
derived to create a population that contrasted for the parental haplotypes at
the QTL but were
near isogcnic across the rest of the gcnome. Individual NILs were phenotyped
using the
described method at six replications, NIL families arc composed of multiple F3
varieties from
the same parentage. Table 4 shows results for NIL families of contrasts of 3
of the QTL.
This data additionally confirms that the previously identified QTL are indeed
operating as
expected among different individuals than those in the mapping populations.
72
Date Recue/Date Received 2023-12-08
8
ts.)
oe
Table 4.
NIL PI N NIL P2 N Flanking Markers
Chr. NIL Region p-value Increase
Haplotype Plt Haplotype P2tt
8 7
4 8
9 6 S01353-1 S04330-1 15
15.52-17.25 p<0.01 35
6 4
4 9
9 4 S03409-1 S04330-1 15
10.1-17.25 p<0.05 49
6 3 5 3 S01481-1 S01818-1
19 89.53-94.75 p =0.087 27
7 5 5 9 S01481-1 S01818-1 19
89.53-94.75 p=0.009 42
7
4 7
27.93-27.81
3 0 S04793-1 S05933-1 5
(misassembly) p<0.05 50
t Number of P1 NILs.
tt Number of P2 NILs.
4310 increase for positive haplotype associated with resistance to Charcoal
Rot Drought Complex.
[00264] Example 5: Reuession Analysis of Haplotypes.
[00265] Two QTL regions were further explored by regression analysis of
haplotypes
assigned using high density sequence data. A large set of elite breeding
germplasm was
phenotyped using the described method, these same germplasm were genotyped
using high
density resequencing at a 0.1X density. The germplasm was classified into 10
kb length
haplotypes, 90% similar across the window, across the QTL regions.
[00266] FIG. 1 shows the QTL region on chromosome 15 from approximately 3,012
kb to
3,946 kb on the Glyma 1 Assembly reference (Schmutz, Jeremy, et al. "Genome
sequence of
the palaeopolyploid soybean." Nature 463.7278 (2010): 178-183) using 10 kb
haplotype
windows created using high density sequence data from 206 unique varieties.
The different
colors show haplotypes that are different in 10 kb windows along the
chromosome.
Displayed in columns are three known resistant and three known susceptibe
varieties, which
are indicated respectively by "RES" and "SUS" in the figure. To the right are
the results for
varieties with unknown QTL status. Regression analysis was employed to
determine
which haplotype window explained the greatest variation in the phenotypic data
across the
gcrmplasm set. Indicated next to the last column on the right arc regression
values (R2) for
the effect of the indicated haplotype on charcoal rot drought complex across
the set of 206
varieties. It was determined that the region from 3,202 kb-3,212 kb explained
the greatest
amount of phenotypic variation (R2= 16.8%).
[00267] FIG. 2 shows the QTL region on chromosome 19 from approximately 48,300
kb
to 48,550 kb on the Glyma 1 Assembly reference (Schmutz, Jeremy, et al.
"Genome sequence
of the palaeopolyploid soybean." Nature 463.7278 (2010): 178-183) using 10 kb
haplotype
windows created using high density sequence data from 148 unique varieties.
The different
colors show haplotypes that are different in 10 kb windows along the
chromosome.
Displayed in columns are four known resistant and three known susceptibe
varieties, which
are indicated respectively by "RES" and "SUS" in the figure. The figure also
shows results
for 12 varieties with unknown QTL status. Regression analysis was employed to
determine
which haplotype window explained the greatest variation in the phenotypic data
across the
germplasm set. Indicated next to the last column on the right are regression
values (R2) for
the effect of the indicated haplotype on charcoal rot drought complex across
the set of 148
varieties. It was determined that the region from 48,340kb-48,380kb explained
the greatest
amount of phenotypic variation (R2= 12.3%).
[00268] Charcoal rot phenotypic data obtained using the growth chamber
screening
method (see Example 1 above) was available on both haplotypes for these two
QTL (Chr. 15,
74
Date Recue/Date Received 2023-12-08
3,202 kb-3,212 kb; and Chr. 19, 48,340 kb-48,380 kb) across 141 elite soybean
varieties.
When both both haplotypes are considered together (Table 5), the data strongly
indicate that
the effect of these two QTL loci are additive in nature for charcoal rot
tolerance. The
resistant haplotype class for the Ch 15 QTL significantly increases tolerance
by a mean of
1.44 compared to the double susceptible. Adding the resistant haplotype for
the Ch 19 QTL
increases numerically 0.86 compared to the double susceptible, but the
difference is not
significant and likely due to the smaller sample size. The double resistant
class is
significantly higher than either single resistant class and has a 2.59 higher
mean
(approximately 95% increase) than the double susceptible.
Table 5.
Ch 19, 48,340kh-48,380kb
RES SUS
RES 5.33(18)A 4.18 (22)
Ch 15; 3202kb-3212kb
SUS 3.60 (15)3>c 2.74 (86)c
*Values are mean phenotypic score, N in parenthesis, different letters
denote means are significantly different using Fisher's LSD (p.05)
1002691 The disclosed novel phenotyping procedure for the charcoal rot
pathogen provides
a method to much more accurately phenotype soybean varieties' genetic
tolerance. As
disclosed herein, the use of this novel phenotyping procedure on two mapping
populations
lead to the identification of four QTLs that have significant effects on
charcoal rot tolerance.
Two of these QTLs (Chr. 15, 3,202 kb-3,212 kb; Chr. 19, 48,340 kb-48,380 kb)
were further
validated using NILs and their effects demonstrated across a large set of
breeding germplasm.
Additionally the effect of these QTLs has been shown herein to be additive in
nature across
the same breeding germplasm. These novel QTLs can allow soybean breeders to
more
efficiently develop soybean varieties with higher levels of tolerance by using
marker assisted
selection. In addition, the use of these QTLs can permit more accurate
phenotyping of
soybean varieties.
1002701 As used herein the singular forms "a", "and", and "the" include plural
referents
unless the context clearly dictates otherwise. Thus, for example, reference to
"a cell"
includes a plurality of such cells and reference to "the protein" includes
reference to one or
more proteins and equivalents thereof known to those skilled in the art, and
so forth. All
technical and scientific terms used herein have the same meaning as commonly
understood to
Date Recue/Date Received 2023-12-08
one of ordinary skill in the art to which this invention belongs unless
clearly indicated
otherwise.
1002711 All publications and patent applications mentioned in the
specification are
indicative of the level of those skilled in the art to which this invention
pertains.
1002721 Although the foregoing invention has been described in some detail by
way of
illustration and example for purposes of clarity of understanding, certain
changes and
modifications may be practiced within the scope of the appended claims.
76
Date Recue/Date Received 2023-12-08