Note: Descriptions are shown in the official language in which they were submitted.
WO 2023/076096
PCT/US2022/047254
METHODS OF TREATING THE EFFECTS OF CYTOKINE STORMS
[0001] REFERENCE TO GOVERNMENT GRANTS
[0002] This invention was made with government support under
several grants
(IROIDK109713. IR0IDK111102, IRO 1DK129522, IR0IDK128203) awarded by National
Institutes of Health. The government has certain rights.
[0003] FIELD OF THE INVENTION
[0004] The general field of the present disclosure are novel
approaches to the prevention and
treatment of the effects of cytokine storms. The invention describes specific
combinations or
cytokines or soluble receptors that must be depleted to eliminate or reduce
mortality as the result
of severe viral cytokine storms.
[0005] BACKGROUND
100061 A striking feature of the COVID-19 pandemic is multisystem
involvement including
the respiratory tract, kidney, brain; liver, heart, gastro-intestinal tract,
eyes and many other organs.
See Huang et al., "Clinical features of patients infected with 201 9 novel
coronavirus in Wuhan,
China,- (2020) Lancet 395: pp. 497-506; Wang et al., "Clinical features of 69
cases with
coronavirus disease 2019 in Wuhan, China," (2020) Clin Infect Dis. 71: pp. 769-
777. The virus is
not always detected in affected organs, and its presence or absence in cardiac
autopsy studies did
not appear to influence the extent of inflammatory cell infiltration. See
Spudich et al., "Nervous
system consequences of COVID-19," (2022) Science 375: pp. 267-269; Topol,
"COVID-19 can
affect the heart," (2020) Science 370: pp. 408-409; Lindner eta., "Association
of cardiac infection
with SARS-CoV-2 in confirmed COVID-19 autopsy cases." (2020) JAMA Cardiol. 5:
pp. 1281-
1285; Gupta, et al., "Extrapulmonary manifestations of COVID-19," (2020) Nat
Med. 26: pp.
1017-1032.
[0007] Viral infections trigger cytokine production as part of
the innate and adaptive immune
response. The inventors previously suspected that the extensive cytokine storm
documented early
in the pandemic may be involved in organ damage and developed novel evidence-
based models of
cytokine mediated end organ damage. See Huang et al. 2020. Of the three organs
studied, the
literature on cardiac involvement shows elevated cardiac Troponin I levels
(that mimic an acute
myocardial infarction), myocarditis, myocardial necrosis, pericarditis,
arrythmias and heart
failure4, 7. See Topo 2020; Inciardi et al., "Cardiac involvement in a patient
with coronavirus
disease 2019 (COVID-19),- (2020) JAMA Cardiol. 5: pp. 819-824. Evidence of
liver injury
include increased aminotransferase levels, hepatocyte injury, inflammation and
steatosis8. See
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
Herta et al., "COVID-19 and the liver - Lessons learned," (2021) Liver Int. 41
Suppl 1: pp. 1-8.
Kidney manifestations are very common in hospitalized COV1D-19 patients, with
nearly 40%
developing proteinuria, and about one-third developing acute kidney injury
(AKI). See Cheng et
al., -Kidney disease is associated with in-hospital death of patients with
COV1D- 19," (2020)
Kidney Int. 97: pp. 829-838; Hirsch et al., "Acute kidney injury in patients
hospitalized with
COVID-19,- (2020) Kidney Int. 98: pp. 209-218. Kidney biopsy studies in COVID-
19 patients
with severe proteinuria and/or kidney dysfunction have most commonly
documented the collapsing
variant of focal and segmental glomerulosclerosis (FSGS) and acute kidney
injury. See Kudose et
al., "Kidney Biopsy Findings in Patients with COVID-19," (2020) J Am Soc
Nephrol. 31: pp.
1959-1968; Nasr et al., "Kidney Biopsy Findings in Patients With COVID-19,
Kidney Injury and
Proteinuria," (2021) Am J Kidney Dis. 77: pp. 465-468 (2021). Despite
suspicion of viral particles
in early autopsy studies, kidney biopsies from living patients did not note
any viral particles. See
also Bradley et al., "Histopathology and ultrastructural findings of fatal
COVID-19 infections in
Washington State: a case series," (2020) Lancet 396: pp. 320-332.
[0008] The advantage of building a COVID-19 cytokine storm model
around kidney disease
is a potential mechanistic comparison with rare manifestations of a common
cold cytokine storm,
with which it shares some components. See Basnet et al., -Rhinoviruses and
Their Receptors,"
(2019) Chest 155: pp. 1018-1025; Wine et al., "Cytokine responses in the
common cold and otitis
media," (2012) Curr Allergy Asthma Rep. 12: pp. 574-581; Nieters et al.,
"Cross-sectional study
on cytokine polymorphisms, cytokine production after T-cell stimulation and
clinical parameters
in a random sample of a German population," (2001) Hum Genet. 108: pp. 241-
248; Noah et al.,
"Nasal cytokine production in viral acute upper respiratory infection of
childhood," (1995) J Infect
Dis. 171: pp. 584-592; van Kempen et al., "An update on the pathophysiology of
rhinovirus upper
respiratory tract infections,- (1999) Rhinology 37: pp. 97-103; Whiteman et
al., "IFN-gamma
regulation of ICAM-1 receptors in bronchial epithelial cells: soluble ICAM-1
release inhibits
human rhinovirus infection," (2008) J Inflamm (Lond). 5: p. 8; Jartti et al.,
"Systemic T-helper and
T-regulatory cell type cytokine responses in rhinovirus vs. respiratory
syncytial virus induced early
wheezing: an observational study," (2009) Respir Res. 10: p. 85; Hershey et
al., "The association
of atopy with a gain-of-function mutation in the alpha subunit of the
interleukin-4 receptor," (1997)
N Engl J Med. 337: pp. 1720-1725; Abdel-Hafez et al., "Idiopathic nephrotic
syndrome and atopy:
is there a common link?," (2009) Am J Kidney Dis. 54: pp. 945-953.
[0009] Common colds, frequently caused by Rhinoviruses, trigger
nearly 70% of episodes of
relapse of the glomerular diseases MCD and FSGS. See Passioti et al., "The
common cold:
-2-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
potential for future prevention or cure,- (2014) Curr Allergy Asthma Rep. 14:
p. 413; Takahashi
et al., -Triggers of relapse in steroid-dependent and frequently relapsing
nephrotic syndrome,"
(2007) Pediatr Nephrol. 22: pp. 232-236.
[0010] Whereas this relapse pathway is unknown, the inventors
have long considered the
cytokine storm to play a leading role. Since the COVID-19 cytokine storm is
broader than its
common cold counterpart, subtractive analysis could identify key players in
specific aspects of
each disease. Human and experimental MCD and most forms of FSGS are associated
with low
podocyte expression of transcriptional factor ZHX2. See Mace et al., "ZHX2 and
its interacting
proteins regulate upstream pathways in podocyte diseases," (2020) Kidney Int.
97: pp. 753-764.
By contrast, experimental evidence suggests that the collapsing variant of
FSGS has high
underlying podocyte ZHX2 expression. Mace et al. 2020. In contrast to many
other cells,
podocytes express the majority of all ZHX proteins in a cell membrane (non-
nuclear) distribution.
In the right combinations and the setting of an altered ZHX2 expression state,
systemic cytokine
release could potentially induce migration of ZHX proteins from normal
(Aminopeptidase A /
APA, Ephrin B1) or putative alternative cell membrane anchors into the
podocyte nucleus.
[0011] However, despite efforts to understand the resultant
cytokine storm seen following
COVID-19 infection, there is still a need to understand the underlying
mechanisms of this
phenomenon and the resultant clinical manifestations. Such an understanding
will facilitate the
design of therapeutic approaches to reduce cytokine storm related organ
damage.
[0012] The present invention addresses these needs.
[0013] SUMMARY OF THE INVENTION:
[0014] The current invention provides mechanisms targeting
various cytokines. The invention
describes specific combinations of cytokines or soluble factors that must be
depleted to eliminate
or reduce the effects of the cytokine storm including mortality and end organ
injury.
[0015] Viral illnesses, including respiratory tract viruses like
SARS-CoV-1 and SARS-CoV-
2, have pathologic effects on non-respiratory tract organs even in the absence
of obvious direct
viral infection. In addition, illness caused by other respiratory and non-
respiratory viruses such as
influenza, parainfluenza, Respiratory Syncvtial Virus, adenoviruses,
enteroviruses, other
coronaviruses, cytomegalovirus (CMV), Epstein Barr Virus (EBV), Middle East
Respiratory
Syndrome (MERS), and Ebola also are known to cause cytokine storms as well as
cause mortality
and multiorgan injury.
-0-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
100161 To study and compare the role of viral cytokine storms in
extra-pulmonary
manifestations of SARS-CoV-2, novel COVID-19 and cytokine combination -
cocktails" were
developed from clinical data and injected in mice. Previous studies by the
inventors demonstrated
effectiveness in a Rhinovirus common cold infection model using Common Cold
cytokine
combination "cocktails."
[0017] In initial studies, the inventors utilized Zhx2fl 41" and
NPHS2-promoter driven Cre
mice. However, they subsequently used BALB/cJ mice, an established model of
the Zhx2
hypomorph state, and BALB/c mice (Zhx2+/+). See Mace et al. 2020; Perincheri
et al., "Hereditary
persistence of alpha- fetoprotein and H19 expression in liver of BALB/cJ mice
is due to a retrovirus
insertion in the Zhx2 gene.- (2005) Proc Natl Acad Sci USA 102: pp. 396-4011;
Perincheri et al.,
"Characterization of the ETnII-alpha endogenous retroviral element in the
BALB/cJ Zhx2 (Afrl)
allele," (2008) Mamm Genome 19: pp. 26-31; Gargalovic et al., "Quantitative
trait locus mapping
and identification of Zhx2 as a novel regulator of plasma lipid metabolism,"
(2010) Circ
Cardiovasc Genet. 3: pp. 60-67; Creasy et al., "Zinc Fingers and Homeoboxes 2
(Zhx2) Regulates
Sexually Dimorphic Cyp Gene Expression in the Adult Mouse Liver," (2016) Gene
Expr. 17: pp.
7-17; Jiang et al., "Zhx2 (zinc fingers and homeoboxes 2) regulates major
urinary protein gene
expression in the mouse liver," (2017) J Biol Chem 292: pp. 6765-6774 (2017);
Erbilgin et al.,
"Transcription Factor Zhx2 Deficiency Reduces Atherosclerosis and Promotes
Macrophage
Apoptosis in Mice," (2018) Arterioscler Thromb Vasc Biol. 38: pp. 2016-2027.
[0018] At low doses, COVID-19 cocktails, but not individual
cytokines, induced glomerular
injury and albuminuria in mice to mimic COVID-19 related proteinuria. The
cytokine cocktails
activated STAT6 signaling in cultured podocytes, which was reduced in CRISPR B
Zhx2
hypomorph podocytes. Depletion of select single cytokines improved glomerular
injury and
albuminuria. At high doses, COVID-19 cocktails, but not individual cytokines,
induced common
clinical manifestations of SARS-CoV-2 disease, including acute heart injury,
myocarditis,
pericarditis, liver and kidney injury, and high mortality in mice. STAT5,
STAT6 and NEKB
pathways were activated in these organs. Dual depletion after model induction
of select
combinations of TNF-ct with IL-2 or IL-13 or IL-4 in BALB/c. In summary,
systemic
manifestations of viral cytokine storms, disease mechanisms and therapeutic
principles to reduce
morbidity and mortality were identified.
[0019] In embodiments of the current invention are provided
methods of inhibiting, treating,
or preventing the effects of cytokine storms as the result of viral infections
in patients comprising
inhibiting, neutralizing or depleting one or more cytokines from the patient.
-4-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
100201 In any embodiment, the cytokine storm can be induced by a
viral infection caused by
any respiratory or non-respiratory virus. Thus, the viral infection can be
caused by viral illnesses,
including respiratory tract viruses like SARS-CoV-1 and SARS-CoV-2, have
pathologic effects on
non-respiratory tract organs even in the absence of obvious direct viral
infection. In addition, the
viral infection can be caused by other respiratory and non-respiratory viruses
such as influenza,
parainfluenza, Respiratory Syncytial Virus, adenovirus es, enteroviruses,
other coronavirus es,
cytomegalovirus (CMV), Epstein Barr Virus (EBV), Middle East Respiratory
Syndrome (MERS),
and Ebola which also are known to cause cytokine storms as well as cause
mortality and multiorgan
inj ury.
[0021] In another embodiment, cytokine storms are caused by non-
viral infections, such as
those caused by bacteria, fungi or protozoa.
[0022] In still another embodiment, cytokine storms are of non-
infectious etiology, such as
those related to cancers or their treatment, organ transplantation, or related
to changes in the stable
cytokine milieu of systemic disorders like diabetes mellitus.
[0023] Furthermore, because the inventors discovered that the
major difference between
common colds (that cause mild disease) and more severe cytokine storm profiles
caused by viral
infections induced by for example, SARS-CoV-2, is the presence of concomitant
acute activation
of an IL-4, IL-13 related "allergy pathway,- the current invention also
includes viral infections that
include concomitant and significant activation of the allergy pathway.
[0024] In some embodiments, in invention provides methods of
depleting two or more
cytokines in order to reduce the mortality caused by severe cytokine storms.
100251 In other embodiments of the invention are provided methods
of treating the effects of
acute heart injury, acute liver injury and acute kidney injury used by
cytokine storms. In some
embodiments, the cytokine storms are caused by viral infections.
[0026] In still other embodiments of the invention are provided
methods of reducing mortality
caused by cytokine storms. In some embodiments, the cytokine storms are caused
by viral
infections.
[0027] In embodiments of the current disclosure, methods are
provided to prevent multi-organ
injury induced by a cytokine storm comprising the inhibition, neutralization,
or depletion more
than one cytokine.
[0028] In still other embodiments of the current invention are
provided methods for treating or
preventing the effects of post-acute sequelae of a SARS-Cov-2 infection
comprising the inhibition,
neutralization or depletion of one or more cytokines.
-5-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
100291 In embodiments of the current disclosure, methods are
provided for preventing the
relapse of a viral infection. In particular embodiments, the methods involve
providing treatments
that inhibit, neutralize or deplete one or more cytokines.
[0030] In yet other embodiments of the current invention are
provided methods for treating or
preventing the effects of SARS-CoV-2 virus mRNA vaccines comprising the
inhibition,
neutralization or depletion of one or more cytokines.
[0031] In other embodiments are provided animal models for
cytokine storms induced by viral
infections and other disease states to test methods of treating or preventing
the effects of said
cytokine storm.
[0032] Thus, in any of the methods provided in the current
invention, one or more cytokines
can be inhibited, neutralized or depleted by the administration of and agent
to the patient where
the agent comprises an adeno-associated virus (AAV) or lentovirus-containing
an a short-hairpin
RNA (shRNA) against one or more cytokines.
[0033] In some embodiments, the shRNA is commercially available
and can be attached to or
part of any vector known in the art including plasmids, viral vectors,
bacteriophages, cosmids, and
artificial chromosomes.
[0034] In other embodiments, the agent comprises a monoclonal or
polyclonal antibody
directed against the one or more cytokines. In yet other embodiments, the
agent comprises a
monoclonal or polyclonal antibody directed against one or more cytokines. In
still other
embodiments, the agent is an siRNA or antisense oligonucleotide that targets
one or more
cytokines.
100351 In still other embodiments, the agent is an antagonist
that binds to a cytokine-mediated
receptor and prevents the binding of one or more cytokines.
[0036] In embodiments of the current disclosure, methods are
provided for treating a viral
infection. In particular embodiments, the methods involve providing treatments
that inhibit,
neutralize or deplete one or more cytokines.
[0037] In any of the disclosed embodiments, the one or more
cytokines to be inhibited,
neutralized or depleted comprise TNFcc, IL-2, IL-4, IL-13, IFN-y or IL-6.
[0038] It will be understood for the disclosure herein that
depending upon the severity of the
viral infection or other condition being treated, the inhibition,
neutralization or depletion more than
one cytokine may be more effective that depletion of a single cytokine.
-6-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
100391 BRIEF DESCRIPTION OF THE DRAWINGS
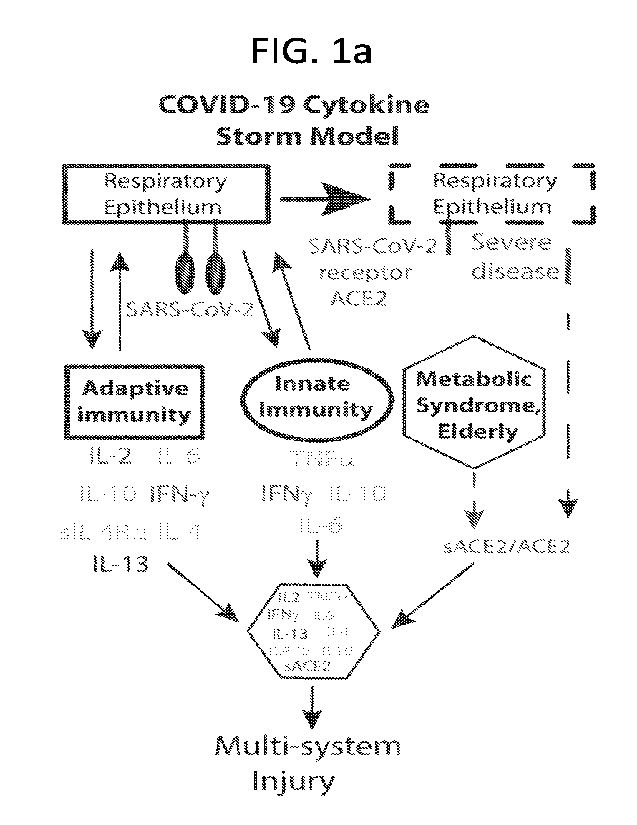
100401 FIG. la-g depicts the development and characterization of
COVID-19 cytokine storm
models. (FIG la) Schematic representation of COVID-19 induced cytokine storm
in the context
of human disease. (FIG. lb) Composition of dose X of the COVID cocktails A to
D. (FIG. 1c)
Albuminuria after injecting different doses of Cocktail D into BALB/cJ mice (n
= 4 mice per
group). X/2 is the threshold nephritogenic dose in BALB/cJ mice. (FIG. 1d)
Albuminuria after
injecting dose X of individual COVID cocktail components in BALB/c.I mice (n =
4 mice per
group). (FIG. le) Albummuria after injecting COVID cocktails A to D dose X/2
in BALB/c mice
(n = 6 mice per group). (FIG. 10 Albuminuria after injecting COVID cocktails A
to D dose X/2 in
BALB/cJ mice (n = 6 mice per group). (FIG. 1g) Albuminuria after injecting
BALB/c mice with
intact Cocktail C dose X/2 or Cocktail C dose X/2 lacking individual
components that target
podocytes (n = 6 mice per group). P<0.05; ** P<0.01; *** P<0.001. All
significant values are two-
tail.
100411 FIG. 2a-i is an assessment of systemic injury induced by
high dose of Cocktail D (3X)
in BALB/c mice, compared with lower doses or individual components at dose 3X.
(FIG. 2a) Acute
myocardial injury assessed by cardiac Troponin I levels (cTPI3) levels (n = 8
mice per group).
(FIG. 2b) Acute liver injury assessed by alanine aminotransferase (ALT)
activity levels (n = 8 mice
per group). (FIG. 2c) Acute kidney injury assessed by serum creatinine
measured using mass
spectrometry (n = 8 mice per group). (FIG. 2d) Histological characterization
of acute cardiac injury
(n = 3 mice per group) using H&E-stained sections from Cocktail D dose 3X
injected mice.
Myocytolysis (red arrows), inflammation (black arrows), fibril disruption
(blue arrows),
hypereosinophilia (green arrows) and pericarditis (orange arrow) were noted.
(FIG. 2e)
Histological characterization of acute liver injury (n = 3 mice per group)
using H&E-stained
sections from Cocktail D dose 3X injected mice. Hepatocellular injury (red
arrows), inflammation
(black arrows), prominent Kupfer cells (green arrows), regenerative changes
(yellow arrows) and
pen-central vein injury (blue arrow) were noted. (FIG. 20 Histological
assessment of acute kidney
injury (n = 3 mice per group) using PAS-stained sections (columns 1, 2, 4) and
kidney electron
microscopy (column 3) from Cocktail D dose 3X injected mice. First three
columns show proximal
tubules, last column shows distal tubules. In proximal tubules, vacuolation
(red arrows), brush
border disruption (green arrows) and tubular degeneration (black arrows) were
noted. In distal
tubules, evidence of desquamation (blue arrows) was present. Foam cells were
also noted (white
arrows). Light microscopy scale bars 20 um; Electron microscopy scale bars
2.66 um. (FIG. 2g)
Tables showing morphometric analysis of histological changes in the heart in
BALB/c mice. (FIG.
-7-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
2h) Tables showing morphometric analysis of histological changes in the liver
in BALB/c mice.
(FIG. 2i) Tables showing morphometric analysis of histological changes in the
kidney in BALB/c
mice. * P<0.05; ** P<0.01; *** P<0. 001, all values based on two-tail
analysis.
[0042] FIG. 3a-h shows therapeutic strategies for the effect of
mild and moderate cytokine
storms on glomerular and systemic disease. All depleting antibodies or control
IgG were injected
intravenously one hour after model induction. (FIG. 3a) Albuminuria after
induction of the
Cocktail C model in BALB/c mice (n = 6 mice per group; dose X/2), followed by
control IgG or
depleting antibodies. Groups arranges from left to right in order of efficacy.
(FIG. 3 b) Urine
albumin to creatinine ratio at baseline and on Day 1 of the Cocktail D dose
1.8X model (n = 8
BALB/c mice per group) after depleting one or more components with antibodies.
(FIG. 3c) Serum
cardiac Troponin I (cTPI3) levels on Day 1 of the Cocktail D dose 1.8X model
(n = 8 BALB/c mice
per group) after depleting one or more components with antibodies. Control and
Cocktail D 1.8X
+ IgG injected BALB/cJ mice are shown for comparison. (FIG. 3d) Serum ALT
activity on Day 1
of the Cocktail D dose 1.8X model (n = 8 BALB/c mice per group) after
depleting one or more
components with antibodies. Control and Cocktail D 1.8X + IgG injected BALB/cJ
mice are shown
for comparison. (FIG. 3e) Serum creatinine on Day 1 of the Cocktail D dose
1.8X model (n = 8
BALB/c mice per group) after depleting one or more components with antibodies.
Control and
Cocktail D 1.8X + IgG injected BALB/cJ mice are shown for comparison. (FIG.
31) Tables showing
morphometric analysis and comparison of histological changes in the heart
between control IgG
and antibody treated BALB/c mice. (FIG. 3g) Tables showing morphometric
analysis and
comparison of histological changes in the liver between control IgG and
antibody treated BALB/c
mice. (FIG. 3h) Tables showing morphometric analysis and comparison of
histological changes in
the kidney between control IgG and antibody treated BALB/c mice. Morphometric
analysis n = 3
mice per group. * P<0.05; ** P<0.01; *** P<0.001, all values based on two-tail
analysis.
[0043] FIG. 4a-g shows possible therapeutic strategies for the
effect of severe cytokine storms
on systemic disease in BALB/c mice. Number of mice injected per group are
shown in panel a. All
depleting antibodies or control IgG were injected intravenously one hour after
model induction.
Large (Red) asterisk indicates universal mortality. (FIG. 4a) Mortality table
for BALB/c mice
injected with Cocktail D 3X with control IgG or depleting antibodies. Since
mortality was higher
with metabolic cage use (5/6) than without (2/6) in the Control IgG group,
timed urine collection
for albuminuria was not conducted in these studies. (FIG. 4b) Serum cardiac
Troponin I (cTPI3)
levels on Day 1 among survivors of Cocktail D 3X dose injected mice, followed
by control IgG or
depleting antibodies. (FIG. 4c) Serum ALT activity levels on Day 1 among
survivors of Cocktail
-8-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
D 3X dose injected mice, followed by control IgG or depleting antibodies.
(FIG. 4d) Serum
creatinine levels on Day 1 among survivors of Cocktail D 3X dose injected
mice, followed by
control IgG or depleting antibodies. (FIG. 4e) Morphometric comparison of
cardiac histology
between control and cytokine depletion groups. (FIG. 40 Morphometric
comparison of liver
histology between control and cytokine depletion groups. (FIG. 4g)
Morphometric comparison of
kidney histology between control and cytokine depletion groups. Morphometric
analysis n = 3
mice per group. * P<0.05; ** P<0.01; *** P<0.001, all values based on two-tail
analysis.
[0044] FIG. 5a-e shows the activation of signaling pathways by
COVID cocktails and disease
mechanisms. Examples of qualitative studies for NFKB / p-p65 (liver, 30
minutes), pSTAT6
(kidney 60 minutes) and pSTAT5 (heart, 15 minutes) activation by Western blot
of whole organ
protein extracts of mice (n = 3 per group) injected with Cocktail D 3X or
control saline. (FIG. 5b)
Western blots of quantitative studies to assess activation of pSTAT6 signaling
in wild type and
ZHX2 hypomorph (CRISPR B) cultured human podocytes incubated with human
counterparts of
Cocktail C (final concentration x/100,000; n = 3 plates per condition). (FIG.
Sc) Densitometry of
Western blot of Cocktail C incubated wild type and CRISPR B podocytes from
panel b. (FIG. 5d)
Albuminuria in Il4ra and control BALB/cJ mice after injecting Cocktail C dose
X/2 (left panel),
and percentage increase in Day 1 albuminuria from baseline (right panel) (n =
5 to 8 mice per
group). (FIG. 5e) Schematic for potential binding of COVID cocktail components
to specific
receptors previously described in glomerular endothelial cell, mesangial cells
and podocytes, and
feedback loops (red) between these cells. * P<0.05; ** P<0.01; *** P<0.001,
all values based on
two-tail analysis, except right panel in FIG. 5d is one-tail analysis.
100451 FIG. 6a-c shows ancillary human data and additional
effects of Cytokine Cocktails.
(FIG. 6a) Plasma IL-4Ra levels assessed by ELISA in general COVID-19 patients,
age, sex and
race matched healthy controls, and COVID-19 patients with proteinuria. Number
of patient
samples assayed is shown below. (FIG. 6b) Electron microscopy images of
BALB/cJ mouse
glomeruli on Day 1 after injection of Cocktail D dose X/2. Areas of focal foot
process effacement
(black arrows), endothelial vacuolation (green circles), and endothelial
hypertrophy (blue circles)
were noted. (FIG. 6c) Serum creatinine, assayed by Mass Spectrometry, is not
increased in COVID
cytokine cocktails dose X/2 models (BALB/c and BALB/cJ mice; n = 6 mice per
group). Scale bars
0.5 am. * P<0.05; +4-4- P<0.001.
[0046] FIG. 7a-g: (FIG. 7a) Plasma creatine kinase, a marker of
skeletal muscle injury, in
BALB/cJ mice (n = 4 mice per group) 24 hours after injection of Cocktail D at
different doses.
(FIG. 7b) Serum Cardiac Troponin I level data derived from FIG. 2a, plotted
again for higher
-9-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
resolution of lesser increase in levels among some single cytokine injected
groups. (FIG. 7c) Serum
ALT level data derived from FIG. 2b, plotted again for higher resolution of
lesser increase in levels
among some single cytokine injected groups. (FIG 7d) 18-hour albuminuria in
BALB/c mice
injected with single cytokine dose 3X, corresponding to FIG. 2a-c. Given their
high mortality after
Cocktail D 3X, metabolic cage housing for timed urine collection is not
feasible at this dose in
BALB/c mice. (FIG. 7e) Electron microscopy of BALB/c mouse kidney glomeruli 24
hours after
injection Cocktail D dose 3X. Extensive foot processes effacement (red
arrows), endothelial
hypertrophy (green arrows) and glomerular basement membrane (GBM) remodeling
(blue arrows)
were present. (FIG. 70 Hematoxylin and Eosin-stained skeletal muscle from
BALB/c.I mice 24
hours after injection of Cocktail D dose 3X. Focal inflammation (black arrows)
was noted in some
sections. (FIG. 7g) Albuminuria after induction of Cocktail C in BALB/c mice
(n = 6 mice per
group; dose X/2), followed by receptor blockage using antibodies against IL-
4Ra, TNFR1 and IL-
lORP, or control IgG. Scale bars (e) 0.5 pm, (f) 20 pm. * P<0.05; ** P<0.01;
*** P<0.001.
[0047] FIG. 8a-e shows the histology of intermediate dose injury.
Histological sections from
studies of BALB/c mice (n = 3 mice/group) euthanized 24 hours after Cocktail D
dose 1.8x injection
and additional antibodies or Control IgG injected one hour after model
induction (see FIG. 3). The
numbering code for each group is: "1"¨ Control IgG; "2"¨ Anti-TNFa Ab; "3"¨
Anti-IL-6 Abs;
"4"=Anti-1L-10 Ab; "5"= Anti-TNFa + Anti-IFNy + Anti-1L-4-Abs; "6"=Anti-IL4
Abs; "7"=
Anti-TNFa + Anti-IL-4 + Anti-IL-10 Abs; "8"=Anti-IFNy Ab; "9"= Anti-TNFa +
Anti-IL-4 Abs.
(FIG. 8a) Two columns of H & E-stained sections of the heart and pericardium.
Myocytolysis (red
arrows), inflammation (black arrows), hypereosinophilia (green arrows),
pericarditis (orange
arrow) and pericardial microcalcification (blue arrow) were noted. (FIG. 8b) H
& E-stained
sections of the liver. Hepatocellular injury (red arrows), inflammation (black
arrows), degenerative
changes (green arrows), and regenerative changes (yellow arrows) were noted.
(FIG. 8c) Toluidine
blue stained epon sections of the kidney showing gross tubular morphology.
Tubular vacuolation
(red arows) and tubular degeneration (black arrows) were noted in proximal
tubules. (FIG. 8d)
Electron microscopy of kidney tubules. Tubular vacuolation (red arows) and
tubular degeneration
(black arrows) were noted in proximal tubules. (FIG. 8e) Electron microscopy
of glomeruli. Areas
of podocyte foot process effacement (black arrows) were noted. Scale bars (a)
20 pm (b) 20 pm
(c) 20 j..im (d) 0.5 im (e) 0.5 pm.
[0048] FIG. 9a-d shows the histology for the severe injury model.
Histological sections from
studies of BALB/c mice (n = 3 mice/group) euthanized 24 hours after Cocktail D
dose 3X injection
and additional antibodies or Control IgG injected one hour after model
induction (see FIG. 4). The
-10-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
numbering code for each group is: -1"= Control IgG; -2"= Anti-IL-2 Ab; -3"=
Anti-INFa + Anti-
IL-2 Abs; "4"=Anti-TNFa + Anti-1L-13 Abs; "5"= Anti-TNFa + Anti-1L-4-Abs;
"6"=Anti-TNFa
Ab; "7"=Anti-IL-13; "8"=Anti-IL-4 Ab; "9"=Anti-TNFa + Anti-IFNy Abs; "10"=
Anti-TNFa +
Anti-IL-6 Abs; "11"= Anti-IFNy Ab; -12-=Anti-TNFa + Anti-ACE2 Abs; "13"=Anti-
TNFot +
Anti-1L-10 Abs; -14"=Anti-IL-6 Ab. (FIG. 9a) Two columns of H & E stained
sections of the heart
and pericardium. Myocytolysis (red arrows), inflammation (black arrows),
hypereosinophilia
(green arrows) and pericarditis (orange arrow) were noted. (FIG. 9b) Two
columns of H & E
stained sections of the liver. Hepatocellular injury (red arrows),
inflammation (black arrows),
degenerative changes (green arrows), and regenerative changes (yellow arrows)
were noted. (FIG.
9c) Two columns of Toluidine blue stained sections of the kidney showing gross
tubular
morphology. Tubular vacuolation (red arrows) and tubular degeneration (black
arrows) were noted
in proximal tubules. (FIG. 9d) Two columns of electron microscopy of the
kidney showing images
of glomeruli. Areas of podocyte foot process effacement (black arrows) were
noted. Scale bars (a)
20 um (b) 20 um (c) 20 um (d) 0.5 um.
[0049] FIG. 10a-c: (FIG. 10a) Confocal expression of cytokine
receptors in BALB/c mouse
glomeruli. White arrows indicate receptor expression in podocytes (P),
endothelial (E) and
mesangial (M) cells. Since TNFR1 is expressed in podocytes and endothelial
cells, only partial co-
localization with nephrin (blue), a podocyte protein, is noted. Green color is
nuclear stain. (FIG.
10b) Confocal expression (red) of ACE-2 and cytokine receptors in BALB/c mouse
kidney tubules.
Most images show proximal tubules, except IL-10R13 image is collecting duct.
(FIG. 10c) Western
blot characterization of antibodies used for depletion studies using
recombinant proteins that make
up the cytokine cocktails. Scale bars (a) 20 um (b) 20 um.
[0050] FIG. Ii a is a schematic representation of data assembled
from the Human Protein Atlas
Project (hill's:, /www.proteinatica. org.,) showing the approximate
distribution and semi-
quantitative expression of cytokine receptors and ACE2 kidney tubular segments
(FIG. 11b) heart
muscle and (FIG. 11c) liver.
[0051] DETAILED DESCRIPTION
100521 The current invention provides mechanisms targeting
various cytokines. The invention
describes specific combinations of cytokines or soluble factors that must be
depleted to eliminate
or reduce the effects of the cytokine storm including mortality and end organ
injury.
-11 -
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
100531 Summary of Methods Provided
[0054] The current invention provides methods of inhibiting,
treating, or preventing the effects
of cytokine storms as the result of viral infections in patients comprising
inhibiting, neutralizing or
depleting one or more cytokines from the patient.
[0055] The inventors contemplate that in any embodiment, the
cytokine storm can be induced
by a viral infection caused by any respiratory or non-respiratory virus. Thus,
the viral infection
can be caused by viral illnesses, including respiratory tract viruses like
SARS-CoV-1 and SARS-
CoV-2, have pathologic effects on non-respiratory tract organs even in the
absence of obvious
direct viral infection. In addition, the viral infection can be caused by
other respiratory and non-
respiratory viruses such as influenza, parainfluenza, Respiratory Syncytial
Virus, adenoviruses,
enteroviruses, other coronaviruses, cytomegalovirus (CMV), Epstein Barr Virus
(EBV), Middle
East Respiratory Syndrome (MERS), and Ebola which also are known to cause
cytokine storms as
well as cause mortality and multiorgan injury. In addition, because the
inventors discovered that
the major difference between common colds (that cause mild disease) and more
severe cytokine
storm profiles caused by viral infections induced by for example, SARS-CoV-2,
is the presence of
concomitant acute activation of an IL-4, IL-13 related "allergy pathway," the
current invention
also includes viral infections that include concomitant and significant
activation of the allergy
pathway.
[0056] Embodiments of the invention provide:
[0057] --methods of depleting two or more cytokines in order to
reduce the mortality caused
by severe cytokine storms;
100581 --methods of treating the effects of acute heart injury,
acute liver injury and acute
kidney injury used by cytokine storms; the inventors contemplate that the
cytokine storms can be
caused by viral infections in some embodiments;
[0059] --methods of reducing mortality caused by cytokine storms;
; the inventors contemplate
that the cytokine storms can be caused by viral infections in some
embodiments;
[0060] --methods to prevent multi-organ injury induced by a
cytokine storm comprising the
inhibition, neutralization, or depletion more than one cytokine.
[0061] -- methods for treating or preventing the effects of post-
acute sequelae of a SARS-Cov-
2 infection comprising the inhibition, neutralization or depletion of one or
more cytokines.
[0062] --methods for preventing the relapse of a viral infection
where the methods involve
providing treatments that inhibit, neutralize or deplete one or more
cytokines;
-12-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
100631 --methods for treating or preventing the effects of SARS-
CoV-2 virus mRNA vaccines
comprising the inhibition, neutralization or depletion of one or more
cytokines;
[0064] --animal models for cytokine storms induced by viral
infections and other disease states
to test methods of treating or preventing the effects of said cytokine storm;
and,
[0065] -- methods are for treating a viral infection.
[0066] The inventors also contemplate that the methods of the
current invention can be used
in any disease state in which a cytokine storm occurs including those diseases
of non-viral origin
such as bacterial, fungal or parasitic infections, cancer, organ
transplantation, or results from the
change in the systemic cytokine milieu of a multi system disease like diabetes
mellitus or metabolic
syndrome.
[0067] The inventors contemplate that in any of the methods
disclosed, one or more cytokines
can be inhibited, neutralized or depleted one or more of several methods. One
method
contemplated is by the administration of an agent to the patient where the
agent comprises an
adeno-associated virus (AAV) or lentovirus-containing an a short-hairpin RNA
(shRNA) against
one or more cytokines. The shRNA can be made or is commercially available and
can be attached
to or part of any vector known in the art including plasmids, viral vectors,
bacteriophages, cosmids,
and artificial chromosomes.
[0068] Another method contemplated of depleting one or more
cytokines is by the
administration of a monoclonal or polyclonal antibody directed against the one
or more cytokines.
In yet other embodiments, the agent comprises a monoclonal or polyclonal
antibody directed
against one or more cytokines.
100691 The agent can also be an siRNA or antisense
oligonucleotide that targets one or more
cytokines.
[0070] In still other embodiments, the agent is an antagonist
that binds to a cytokine-mediated
receptor and prevents the binding of one or more cytokines.
[0071] In any of the disclosed embodiments, the one or more
cytokines to be inhibited,
neutralized or depleted comprise TNFia, IL-2, IL-4, IL-13, IFN-y or IL-6. It
will be understood for
the disclosure herein that depending upon the severity of the viral infection
or other condition being
treated, the inhibition, neutralization or depletion more than one cytokine
may be more effective
that depletion of a single cytokine.
[0072] Throughout this disclosure, various quantities, such as
amounts, sizes, dimensions,
proportions and the like, are presented in a range format. It should be
understood that the
-13-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
description of a quantity in range format is merely for convenience and
brevity and should not be
construed as an inflexible limitation on the scope of any embodiment.
Accordingly, the description
of a range should be considered to have specifically disclosed all the
possible suhranges as well as
all individual numerical values within that range unless the context clearly
dictates otherwise. For
example, description of a range such as from 1 to 6 should be considered to
have specifically
disclosed subranges such as from 1 to 3, from 1 to 4, from 1 to 5, from 2 to
4, from 2 to 6, from 3
to 6 etc., as well as individual values within that range, for example, 1.1,
2, 2.3, 4.62, 5, and 5.9.
This applies regardless of the breadth of the range. The upper and lower
limits of these intervening
ranges may independently be included in the smaller ranges, and are also
encompassed within the
disclosure, subject to any specifically excluded limit in the stated range.
Where the stated range
includes one or both of the limits, ranges excluding either or both of those
included limits are also
included in the disclosure, unless the context clearly dictates otherwise.
[0073] The terminology used herein is for the purpose of
describing particular embodiments
only and is not intended to be limiting of any embodiment. As used herein, the
singular forms "a,"
"an" and "the" are intended to include the plural forms as well, unless the
context clearly indicates
otherwise. It will be further understood that the terms "includes",
"comprises", "including" and/or
-comprising," when used in this specification, specify the presence of stated
features, integers,
steps, operations, elements, and/or components, but do not preclude the
presence or addition of
one or more other features, integers, steps, operations, elements, components,
and/or groups
thereof As used herein, the term "and/or" includes any and all combinations of
one or more of the
associated listed items. Additionally, it should be appreciated that items
included in a list in the
form of "at least one of A, B, and C" can mean (A); (B); (C); (A and B); (B
and C); (A and C); or
(A, B, and C). Similarly, items listed in the form of "at least one of A, B,
or C" can mean (A); (B);
(C); (A and B); (B and C); (A and C); or (A, B, and C).
[0074] Unless specifically stated or obvious from context, as
used herein, the term -about" in
reference to a number or range of numbers is understood to mean the stated
number and numbers
+/- 10% thereof, or 10% below the lower listed limit and 10% above the higher
listed limit for the
values listed for a range.
[0075] In any of the embodiments disclosed herein, the terms
"treating" or "to treat" includes
restraining, slowing, stopping, or reversing the progression or severity of an
existing symptom or
disorder.
[0076] In any of the embodiments disclosed herein, the term
"patient" refers to a human.
-14-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
100771 Cytokine Inhibitors
[0078] The current invention contemplates that various cytokines
can be neutralized or
inhibited by several different non-limiting methods. For example, as described
herein, target
cytokines can be neutralized or inhibited by administration of a
therapeutically effective amount
of an agent where the agent comprises an adeno-associated virus (AAV) or
lentovirus-containing
an a short-hairpin RNA (shRNA) against one or more cytokines (sh-"cytokine-).
In some
embodiments, the sh-"cytokine" is commercially available and can be attached
to or part of any
vector known in the art including plasmids, viral vectors, bacteriophages,
cosmids, and artificial
chromosomes.
[0079] Alternatively, as described herein, the target cytokine or
cytokines can be neutralized
or inhibited by administration of a therapeutically effective amount of an
agent where the agent
comprises an antibody, bivalent antibody or a monoclonal antibody directed
against the particular
target cytokine or cytokines.
[0080] Further, as described herein, target cytokine or cytokines
can be neutralized or inhibited
by administration of a therapeutically effective amount of an agent where the
agent comprises an
siRNA or antisense oligonucleotide that targets target cytokine or cytokines.
[0081] Also, as contemplated herein, target cytokine or cytokines
can be neutralized or
inhibited by administration of a therapeutically effective amount of an agent
where the agent
comprises an antagonist that binds to a target cytokine -mediated receptor and
prevents the binding
of the target cytokine or cytokines.
[0082] The target cytokine inhibitor or inhibitors or a
composition therein can be administered
once per day, two or more times daily or once per week. The target cytokine
inhibitor or inhibitors
or composition containing the same can occur by any conventional means
including orally
intramuscularly, intraperitoneally or intravenously into the subject. if
injected, they can be injected
at a single site per dose or multiple sites per dose.
[0083] Cytokine Antibodies and Related Inhibitors
[0084] More specifically a cytokine inhibitor is an antibody
directed against any cytokine as
disclosed herein. Examples of suitable antibodies directed against one or more
target cytokines
are disclosed herein and known to those of skill in the art. The cytokine
antibody can also include
an antibody fragment or a bivalent antibody or fragment thereof, inhibiting
one or more target
cytokines. As described herein, the cytokine inhibitor may be part of a
pharmaceutical composition
-15-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
where the composition may include either an antibody or fragment thereof for
one or more target
cytokines.
[0085] The anti-cytokine antibodies described herein can be made
or obtained by any means
known in the art, including commercially. It is also contemplated that an
antibody can be
specifically reactive with a particular cytokine protein or polypeptide may
also be used as an
antagonist. An anti-cytokine antibody herein may be an antibody or fragment
thereof that binds to
a cytokine or a bivalent antibody that binds to two different cytokines.
[0086] As used herein, the term "antibody" refers to an
immunoglobulin (Ig) whether natural
or partly or wholly synthetically produced. The term also covers any polypepti
de or protein having
a binding domain which is, or is homologous to, an antigen-binding domain. The
term further
includes "antigen-binding fragments" and other interchangeable terms for
similar binding
fragments such as described below.
[0087] Native antibodies and native immunoglobulins are usually
heterotetrameric
glycoproteins of about 150,000 Daltons, composed of two identical light (L)
chains and two
identical heavy (H) chains. Each light chain is typically linked to a heavy
chain by one covalent
disulfide bond, while the number of disulfide linkages varies among the heavy
chains of different
immunoglobulin isotypes. Each heavy and light chain also has regularly spaced
intrachain disulfide
bridges. Each heavy chain has at one end a variable domain ("Vii- or "VH-)
followed by a number
of constant domains (-CH" or -CH"). Each light chain has a variable domain at
one end ("VL" or
"VL") and a constant domain ("CC or "CL") at its other end; the constant
domain of the light
chain is aligned with the first constant domain of the heavy chain, and the
light-chain variable
domain is aligned with the variable domain of the heavy chain. Particular
amino acid residues are
believed to form an interface between the light- and heavy-chain variable
domains.
[0088] The cytokine inhibitors as described herein can be a
"synthetic polypeptide- derived
from a -synthetic polynucleotide" derived from a -synthetic gene," meaning
that the corresponding
polynucleotide sequence or portion thereof, or amino acid sequence or portion
thereof, is derived,
from a sequence that has been designed, or synthesized de novo, or modified,
compared to an
equivalent naturally occurring sequence. Synthetic polynucleotides (antibodies
or antigen binding
fragments) or synthetic genes can be prepared by methods known in the art,
including but not
limited to, the chemical synthesis of nucleic acid or amino acid sequences.
Synthetic genes are
typically different from naturally occurring genes, either at the amino acid,
or polynucleotide level,
(or both) and are typically located within the context of synthetic expression
control sequences.
Synthetic gene polynucleotide sequences, may not necessarily encode proteins
with different
-16-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
amino acids, compared to the natural gene; for example, they can also
encompass synthetic
polynucleotide sequences that incorporate different codons but which encode
the same amino acid
(i.e., the nucleotide changes represent silent mutations at the amino acid
level).
[0089] With respect to anti-cytokine antibodies, the term -
antigen" refers to the any of the
cytokine proteins disclosed herein, respectively or any fragment of the
protein molecules thereof
[0090] The terms "antigen-binding portion of an antibody,-
"antigen-binding fragment,-
"antigen-binding domain," "antibody fragment" or a "functional fragment of an
antibody" are used
interchangeably herein to refer to one or more fragments of an antibody that
retain the ability to
specifically bind to one or more cytokines.
[0091] It is contemplated that the cytokine antibodies may also
include "diabodies- which
refers to small antibody fragments with two antigen-binding sites, which
fragments comprise a
heavy chain variable domain (VH) connected to a light chain variable domain
(VL) in the same
polypeptide chain (VH-VL). By using a linker that is too short to allow
pairing between the two
domains on the same chain, the domains are forced to pair with the
complementary domains of
another chain and create two antigen-binding sites. See for example, EP
404,097; WO 93/11161;
and Hollinger et al., Proc. Natl. Acad. Sci. USA 90:6444 6448 (1993).
[0092] It is contemplated that the cytokine antibodies may also
include -chimeric" forms of
non-human (e.g., murine) antibodies include chimeric antibodies which contain
minimal sequence
derived from a non-human Ig. For the most part, chimeric antibodies are murine
antibodies in
which at least a portion of an immunoglobulin constant region (Fc), typically
that of a human
immunoglobulin are inserted in place of the murine Fc. See for example, Jones
et al., Nature 321:
522-525 (1986); Reichmann et al., Nature 332: 323-329 (1988); and Presta,
Curr. Op. Struct. Biol.,
2: 593-596 (1992).
[0093] It is contemplated that the cytokine antibodies may also
include a -monoclonal
antibody" which refers to an antibody obtained from a population of
substantially homogeneous
antibodies, i.e., the individual antibodies comprising the population are
identical except for
possible naturally occurring mutations that may be present in minor amounts.
Monoclonal
antibodies are highly specific, being directed against a single antigenic
site. Furthermore, in
contrast to conventional (polyclonal) antibody preparations, which can include
different antibodies
directed against different determinants (epitopes), each monoclonal antibody
is directed against a
single determinant on the antigen. The modifier -monoclonal- indicates the
character of the
antibody as being obtained from a substantially homogeneous population of
antibodies and is not
to be construed as requiring production of the antibody by any particular
method. For example,
-17-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
monoclonal antibodies can be made by a hybridoma method, recombinant DNA
methods, or
isolated from phage antibody.
[0094] As used herein, "immunoreactive" refers to binding agents,
antibodies or fragments
thereof that are specific to a sequence of amino acid residues on a cytokine
protein (-binding site"
or "epitope"), yet if are cross-reactive to other peptides/proteins, are not
toxic at the levels at which
they are formulated for administration to human use. The term "binding- refers
to a direct
association between two molecules, due to, for example, covalent,
electrostatic, hydrophobic, and
ionic and/or hydrogen-bond interactions under physiological conditions and
including interactions
such as salt bridges and water bridges and any other conventional binding
means. The term
"preferentially binds- means that the binding agent binds to the binding site
with greater affinity
than it binds unrelated amino acid sequences.
[0095] As used herein, the term "affinity" refers to the
equilibrium constant for the reversible
binding of two agents and is expressed as Kd. Affinity of a binding protein to
a ligand such as
affinity of an antibody for an epitope can be, for example, from about 100
nanomolar (nM) to about
0.1 nM, from about 100 nM to about 1 picomolar (pM), or from about 100 nM to
about 1
femtomolar (fM). As used herein, the term "avidity" refers to the resistance
of a complex of two
or more agents to dissociation after dilution. Apparent affinities can be
determined by methods
such as an enzyme linked immunosorbent assay (ELISA) or any other technique
familiar to one of
skill in the art. Avidities can be determined by methods such as a Scatchard
analysis or any other
technique familiar to one of skill in the art.
[0096] -Epitope" refers to that portion of an antigen or other
macromolecule capable of
forming a binding interaction with the variable region binding pocket of an
antibody.
[0097] The term "specific" refers to a situation in which an
antibody will not show any
significant binding to molecules other than the antigen containing the epitope
recognized by the
antibody. The term is also applicable where, for example, an antigen binding
domain is specific
for a particular epitope which is carried by a number of antigens, in which
case the antibody will
be able to bind to the various antigens carrying the epitope. The terms
"preferentially binds" or
"specifically binds" mean that the antibodies bind to an epitope with greater
affinity than it binds
unrelated amino acid sequences, and, if cross-reactive to other polypeptides
containing the epitope,
are not toxic at the levels at which they are formulated for administration to
human use.
[0098] The term "binding" refers to a direct association between
two molecules, due to, for
example, covalent, electrostatic, hydrophobic, and ionic and/or hydrogen-bond
interactions under
-18-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
physiological conditions and includes interactions such as salt bridges and
water bridges, as well
as any other conventional means of binding.
[0099] As contemplated herein, a target cytokine inhibitor may be
generated through gene
expression technology. The term "RNA interference" or "RNAi" refers to the
silencing or
decreasing of gene expression by siRNAs. It is the process of sequence-
specific, post-
transcriptional gene silencing in animals and plants, initiated by siRNA that
is homologous in its
duplex region to the sequence of the silenced gene. The gene may be endogenous
or exogenous to
the organism, present integrated into a chromosome or present in a
transfection vector that is not
integrated into the genome. The expression of the gene is either completely or
partially inhibited.
RNAi may also be considered to inhibit the function of a target RNA; the
function of the target
RNA may be complete or partial.
[00100] The term -siRNAs" refers to short interfering RNAs. In some
embodiments, siRNAs
comprise a duplex, or double-stranded region, of about 18-25 nucleotides long;
often siRNAs
contain from about two to four unpaired nucleotides at the 3' end of each
strand. At least one strand
of the duplex or double-stranded region of a siRNA is substantially homologous
to or substantially
complementary to a target RNA molecule. The strand complementary to a target
RNA molecule is
the -antisense strand;" the strand homologous to the target RNA molecule is
the -sense strand,"
and is also complementary to the siRNA antisense strand. siRNAs may also
contain additional
sequences; non-limiting examples of such sequences include linking sequences,
or loops, as well
as stem and other folded structures. siRNAs appear to function as key
intermediaries in triggering
RNA interference in invertebrates and in vertebrates, and in triggering
sequence-specific RNA
degradation during posttranscriptional gene silencing in plants.
1001011 It is also contemplated that any cytokine gene can be silenced or
"turned" off' through
the use of CRISPR technology as disclosed herein in the Examples.
[00102] "Post-acute sequel ae"
[00103] -Post-acute sequelae" of SARS-CoV-2 or COVID-19, also
known as -long COVID,"
is used herein to describe any of the long-term symptoms or effects described
as part of the
invention that might be experienced weeks to months after primary infection
with SARS-CoV-2,
the virus that causes COVID-19.
-19-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
1001041 General Methods
[00105] COV1D and common cold cytokine cocktails, and related animal studies
[00106] All animal studies conducted were approved by the IACIJC at Rush
University or the
University of Alabama at Birmingham. All animals received humane treatment per
protocol.
Methods for Dynabead assisted mouse glomerular isolation, rat glomerular
isolation by sieving,
histological section tissue preservation, timed 18-hour urine collection in
metabolic cages in the
absence of food, assessment of albuminuria and proteinuria, real time PCR,
confocal imaging,
electron microscopy and sample processing, histology for light microscopy,
Western blot and co-
immunoprecipitation are previously described and known. The following were
assayed using
commercially available kits using serum samples; mouse ALT (BioVision K752-
100), mouse
cardiac Troponin I Type 3 (Novus Biologicals NBP3-00456), mouse Creatine
Kinase (Abcam
ab155901) and human IL-4Ra ELISA (Abcam ab46022). The following antibodies
were purchased
for Western blot: anti-pSTAT6 (Cell Signaling Technology, Inc. Danvers MA,
USA; cat # 56554,
1:500 dilution); anti-STAT6 (Cell Signaling Technology, Inc. Cat # 5397, 1:500
dilution).
Antibodies against ZHX1, ZHX2 and ZHX3 are previously described25,37,38. Maae
et al. 2020;
Liu et al., "ZHX proteins regulate podocyte gene expression during the
development of nephrotic
syndrome," (2006) J. Biol. Chem. 281: pp. 39681-39692; Clement et al., -Early
changes in gene
expression that influence the course of primary glomerular disease,- (2007)
Kidney Int. 72: pp.
337-347.
[00107] All cytokines, soluble receptors, and antibodies were injected
intravenously in rodents,
and are listed below:
Cytokine/Receptor Rat Mouse Human
Injected
Antibody
IL-2
IL-4Roc
IL-4
IL-13 N.A.
IL-6
IL-10
Interferon-y
TN F-u,
ACE-2
TNFRI
-20-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
IL-10R13
Rat gamma globulin
Syrian hamster
gamma globulin
1001081 Antibodies used for depletion studies were characterized by Western
blot using the
corresponding recombinant protein. Each dose of cytokine cocktail was
dissolved in a final volume
of 100 pt of sterile 0.9% saline. BALB/cJ (Jackson Labs) and BALB/c (Envigo)
mice were
purchased at age 8 weeks, acclimatized for 2 weeks, and baseline 18-hour urine
collection and tail
blood sampling conducted. An extra baseline urine collection was conducted for
BALB/cJ mice.
Most in vivo studies were conducted between age 10 and 15 weeks. Enpep4-;
Zhx2de-rdq1 in mixed
background were obtained by interbreeding the F2 cross between Enpep-/-26 and
Zhx2 deficient
BALB/cJ mice. The nephritogenic dose spectrum of cytokine cocktails was
established for
BALB/cJ, BALB/c, IL4r-/- mice (Jackson Labs). During mouse cytokine studies
using threshold
nephritogenic doses (BALB/cJ, BALB/c, 4r-/- in BALB/cJ background, X/2;), 100
pL of 0.9%
saline was given intraperitoneally immediately after the intravenous cytokine
cocktail dose to
maintain intravascular hydration. Two additional intraperitoneal injection of
100 pL of 0.9% saline
were given at 6 and 23 hours in the intermediate and high cocktail models.
During cytokine
depletion studies, different groups of mice received 50 pg of control IgG or
the respective antibody
or antibody combination intravenously 1 hour after the administration of the
mouse cytokine
cocktail.
[00109] Mass Spectrometry assay for plasma creatinine
[00110] Serum creatinine was measured by LC/MS/MS using an Agilent 1290
Infinity 11 LC
system in combination with a 2x50rnm, 2 pm Tosoh Bioscience TSK-GEL amide-80
LC column,
interfaced to an Agilent 6495 Triple Quadrupole. The oven temperature was
fixed at 40 C. The
mobile phase consisted of 10mM ammonium acetate in LCMS-grade water (35%) and
LCMS-
grade acetonitrile (ACN; 65%). Synthetic creatinine (ranging from 20 pg/ml to
0.16 ug/ml; Sigma)
and isotope-labeled creatinine (D3-creatinine, 10 pg/ml; Sigma) were used as
standard and internal
standard, respectively. Then, 10 ul of sample or standard was combined with 5
il internal standard
and 235 ul 100% ACN, vortex ed and centrifuged at 4C for 15 min at 15000 rpm.
The supernatant
was transferred to a new tube with 200 pl 10 m1VI ammonium acetate and 65%
acetonitrile in
LCMS-grade water, vortexed, centrifuged at 4t for 15 min at 15000 rpm and
subsequently
measured. All samples were measured in duplicate.
-21 -
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
1001111 Sources of human genomic DNA and human kidney biopsies
[00112] Genomic DNA samples from 36 patients with nephrotic syndrome, 33
control subjects,
and 16 patients with diabetic nephropathy were obtained from the following
sources (a)
Immortalized monocytes from plasma of nephrotic syndrome patients at the
University of Alabama
at Birmingham obtained via an IRB approved protocol X080813001 for collecting
DNA, blood
and urine samples. (b) IRB approved study at the Instituto Nacional De
Cardiologia in Mexico City
(CONACYT 34751M, CONACYT 11-05, and DPAGA-UNAM IN- 201902) that included
archived kidney biopsies from patients with glomerular diseases or pre-
implantation kidney
biopsies from healthy living related kidney donors. (c) Archived kidney
biopsy, IRB exempt, from
Hospital Nacional Alberto Sabogal Essalud, Lima, Peru. (d) Archived human DNA
of previously
published FSGS patients43,44 from the Duke Molecular Physiology Institute with
known
mutations in podocyte expressed related genes. (e) Coriell Cell Repositories,
that archive DNA
from the 1000 Genomes Project and the HAPMAP Project. For analytical
comparisons between
cases and controls, the 1000 genomes project phase 3 Ensambl v84 was included
as a single
additional control.
[00113] Agilent Custom capture and high throughput Illumina sequencing
[00114] A custom capture sequencing panel was created to isolate the genomic
interval between
HAS2 and ZHX2 on Chromosome 8. The target interval was uploaded to the
SureDesign website
for Agilent SureSelect capture probe design and synthesis (Agilent
Technologies, Santa Clara CA).
Genomic DNA library preparation and interval capture was done using the QXT
SureSelect kit as
per the manufacturer's instructions (Agilent Technologies). The resulting DNA
libraries were
quantitated by QPCR (Kapa Biosystems, Wilmington MA) and sequenced on the
Illumina HiSeq
2500 or NextSeq 500 with paired end 100bp sequencing following standard
protocols.
Approximately 15 million sequences were obtained per reaction. FASTQ file
generation was done
using bc12fastq converter from Illumina (Illumina, Inc., San Diego CA). Paired
Illumina sequences
compared with hg38 database (GRCh38.p13 Primary Assembly) using CLC Genomics
software
(Version 12, Qiagen, Venlo, the Netherlands). Insertion and deletions of 3 bp
size or larger and a
minimum of 20 sequence reads were selected for analysis. Fisher test
comparison of insertions and
deletions in study and control subjects was exported in Excel format, followed
by software assisted
and manual exclusion of all insertions and deletions present in controls. Only
insertions and
deletions that were subsequently confirmed using IGV browser software (Broad
Institute, Boston
MA) were included. Establishment of homozygosity required presence of the
InDel in over 85%
-22-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
of sequences, and subsequent confirmation by IGV. Minor discrepancies (1-2
base pair position
differences) in the site of the insertion or deletion were occasionally noted
between the two
software and were resolved by Sanger sequencing while designing CRISPR Cas9
studies. All
genomic numbering is based on hg38 and CLC genomics software.
1001151 Genome editing in cultured human podocytes using CRISPR/Cas9
[00116] The basic methodology for CRISPR Cas9 is previously published. See
Cong et al.,
"Multiplex Genome Engineering using CRISPR/Cas Systems," (2013) Science 339:
pp. 819-823.
A single cell derived clone of cells was generated from an established early
passage immortalized
human podocyte cell line51 and used for genome editing studies. The
oligonucleotides and primers
used are listed in Table 4.
1001171 Table 4
Name Application Primer/ Probe Sequence
Generation of Crispr-cas9 mutants
CRISPR B
G0016 SgRNA generation 5'-CAC CGA CTG GTA AAC CAC TTA GGG C-
3'
G0017 SgRNA generation 5'-AAA CGC CCT AAG TGG TTT ACC AGT C-
3'
K1145 SgRNA plasmid sequencing 5'-GCA TAT ACG ATA CAA GGC TGT TAG
AGA G-3'
K1195 Donor plasmid 5'-CGG GCC GGA TCC CTA GAT GTA GCA
TTA CCA GGG TGG-3'
K1196 Donor plasmid 5'-GGC CGA AGC TTG CAG AGA AGA TCA
CGA TAG AU AGA AGA TG-
3'
K1207 Sequencing of donor plasmid 5'-GGT TTC CU GTT ATA TCA CCA G-3'
K1215 Quickchange Mutagenesis 5'-GCT CTA GGA TGA CTG GTA AAC CAC
TTA GGG CAG TCG TCC CCA
GAC CTG GTC TGT GGC CTG TTA G-3'
K1216 Quickchange Mutagenesis 5'-CTA ACA GGC CAC AGA CCA GGT CTG
GGG ACG ACT GCC CTA AGT
GGT HA CCA GTC ATC CTA GAG C-3'
K1219 Plasmid linear amplification 5'-GAT TAT CH TCT AGG GTT AAC GAA CH
CAA GTA ATC AAG AGC
AGC-3'
K1220 Plasmid linear amplification 5'-CGC AGA CTA TCT TIC TAG GGT TAA
CTT TGT AGA ATG CH CTC
G-3'
K1217 Puromycin cassette 5'-CGA GAA GCA TTC TAC AAA GTT AAC
CCT AGA AAG ATA GTC TGC
G-3'
K1218 Puromycin cassette 5'-GCT GCT CH GAT TAC HG AAG TTC GTT
AAC CCT AGA AAG ATA
ATC-3'
K1189 Genome editing 5'-ACA CTG ACG ACA TGG TIC
TAC AGT CTC TGA AAC ATA GAA
GGC AC-3'
K1188 Genome editing 5'-TAC GGT AGC AGA GAC TTG GTC TGA
GAA TCT AAT ACC GCT GAT
CTG-3'
CRISPR A
G0003 SgRNA generation 5'-CAC CGA CCC ATC CAT ACA CH ACC C-
3'
G0004 SgRNA generation 5'-AAA CGG GTA AGT GTA TGG ATG GGT C-
3'
K1145 SgRNA plasmid sequencing 5'-GCA TAT ACG ATA CAA GGC TGT TAG
AGA G-3'
K1140 Donor plasmid 5'-GGC GGC ACT AGT CTA GCT GGC TTG
ACT TTA CAA GAC GAT TCC
ATC C-3'
K1141 Donor plasmid 5'-GGG CGG ATC CCT GCA CTC AGT ATT
CTG CAA GTC CTG TAG C-3'
-23-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
5'-CGA TCT CCT GAC CTC AAG-3'
K1151 Sequencing of donor plasmid 5'-GTG CCT GGC CTG TTA TGA TCT TCT
TAC TCA HT GAT AGC ACC
AGT GTC CTG AGA AAA ATA ACA TAT ACT
CCA HA CCC ATC CAT ACA CH ACC CAG GCA CTC AU CAC CAT AU
AAC TAG ATA GAC ACA TGA TGT TGC
K1149 Quickchange Mutagenesis TGC TCC TGT TGA TGA TAA CAA TGT TGA
GG-3'
5'-CCT CAA CAT TGT TAT CAT CAA CAG GAG CAG CAA CAT CAT GTG
TCT ATC TAG HA ATA TGG TGA ATG AGT
GAC TGG GTA AGT GTA TGG ATG GGT AAT GGA GTA TAT GTT AU
K1150 Quickchange Mutagenesis UT CTC AGG ACA CTG GTG CTA TCA AAT
GAG TAA GAA GAT CAT AAC AGG CCA GGA C-3'
K1163 Plasmid linear amplification 5'-CGT CAC AAT ATG AU ATC UT CTA GGG
HA ACT AGA TAG ACA
CAT GAT GTT GCT GCT CC-3'
K1164 Plasmid linear amplification 5'-CGT CAA TTT TAC GCA GAC TAT CH
TCT AGG GU AAT ATG GTG
AAT GAG TGA CTG GG-3'
K1153 Puromycin cassette 5'-CCC AGT CAC TCA TTC ACC ATA HA
ACC CTA GAA AGA TAG TCT
GCG TAA AAT TGA CG-3'
K1154 Puromycin cassette 5'-GGA GCA GCA ACA TCA TGT GTC TAT
CTA GU AAC CCT AGA AAG
ATA ATC ATA HG TGA CG-3'
K1138 Genome editing 5'-ACA CTG ACG ACA TGG TIC TAC AGT
TAT GAT CH CH ACT CAT
HG ATA GCA CCA GTG TCC-3'
K1139 Genome editing 5'-TAC GGT AGC AGA GAC HG GTC TGA
AAG GAG CAG TGT TGA TCT
AGA GAG AGC C-3'
Real time PCR
H826 Human ZHX2 forward primer CGGAACTGGCTGAATCAGACT
H827 Human ZHX2 reverse primer CAGCACAGCAGTTCTAACAGACTT
P246 FAM-MGB Probe TGCAGAGGCTGGCCA
[00118] CR1SPR B
1001191 Generation of the sgRNA plasmid: In order to introduce a 10 bp
insertion
(CACACACACA), sgRNA recognizing a specific site 45 bp downstream of the
insertion site
(Chr8-122,533,694 - 122,533,695) was designed using the Benchling website
(https://benchling.com). Oligos G0016 and G0017 were phosphorylated and
annealed using T4
Polynucleotide Kinase (NEB), digested with Bbsl and ligated into pX330-U6-
Chimeric BB-CBh-
hSpCas9 plasmid (a gift from Feng Zhang, Addgene plasmid # 42230) using T7 DNA
ligase (New
England Biolabs). The ligation product was treated with PlasmidSafe
exonuclease (Epicentre) to
prevent unwanted recombination products and then transformed into One Shot
TOP10 cells
(Invitrogen). Ten colonies were picked up and plasmids were isolated using
QIAprep Spin
Miniprep Kit (QIAgen). Plasmid DNA was sequenced using primer K1145 (see Table
4).
[00120] Generation of the donor plasmid: The human genomic sequence from
patient E58-13
containing the insertion under study was amplified using KAPA IIiFi IIotStart
PCR Kit (Kapa
Biosystems), the specific patient genomic DNA and primers K1195 and K1196, and
cloned into
pBlueScript II KS+ vector between the BamHI and HindIII restriction sites.
Plasmid DNA was
sequenced using K1207 to confirm the presence of the insertion. A single
mutation in the PAM
-24-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
sequence was made to prevent cutting of this donor template plasmid using
Quikchange
mutagenesis kit (Agilent Technologies) and primers K1215 and K1216, and the
change confirmed
by sequencing. Next, this plasmid was amplified in linear fashion using
primers K1219 and K1220,
and the PCR product digested with Dpnl to remove any residual circular
template plasmid. The
antibiotic selection cassette (Puromycin resistance and truncated thymidine
kinase) flanked by ITR
sequences was amplified by PCR from PB-MV1 Puro-TK plasmid (Transposagen)
using primers
K1217 and K1218, and ligated with the linearized plasmid (see above) at a TTAA
region 78 bp
upstream of the insertion using Gibson assembly Master Mix (NEB). NEB) 5-alpha
Competent
E. coli cells were transformed with 2 ill of the assembly reaction product.
Plasmid DNA from 10
colonies were isolated and sequenced using primer K1217 to confirm correct
assembly.
[00121] Genome editing using sgRNA and donor plasmids: For in vitro
replication of InDels
found in kidney disease patients, cultured human podocytes derived from a
single cell were
transfected by electroporation (Biorad Gene Pulser Xcell
Electroporation System, 0.2 cm
cuvette, square wave mode, 150 V and 10 millisecond pulse) with the
CRISPR/Cas9 vector
containing the specific sgRNA, and a donor plasmid containing the donor
sequence and the
antibiotic selection cassette. Following removal of non-transfected cells by
incubation with 1 ug/m1
Puromycin Dihydrochloride (Gibco) for 15 days, 10 ug of Excision-only piggyBac
transposase
expression vector (Transposagene) was transfected for scarless removal of the
antibiotic selection
cassette. Four days after transfection, cells were incubated with 2.5 04
ganciclovir (Sigma) to
remove cells with residual truncated thymidine kinase activity. Single cells
were picked, clones
established, genomic DNA extracted using QIAamp DNA Mini Kit (QIAgen) and the
target region
PCR amplified using Platinum HiFi DNA polymerase (Invitrogen) and primers
K1189 and K1188.
PCR products were gel purified using QIAquick Gel Extraction Kit (QIAgen),
cloned into pCR2.1
vector using TA Cloningtm kit (Invitrogen) and the insert sequenced using the
M13 Forward
sequencing primer. Sequences were aligned with native podocyte genomic
sequence and the donor
template sequence by BLAST.
[00122] CRISPR A
[00123] Overall methods were identical to those for CRISPR B, with the
exception of primers
and oligonucleotides used, and the following site specific details: An 8 bp
insertion (TGGATGGA)
was introduced at Chr 8-122,304,094 - 122,304,095), and the sgRNA designed to
recognize a
specific site 73 bp upstream of the insertion site. While generating the donor
plasmid, the patient
-25-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
specific genomic DNA (patient SF3) was cloned into the pBlueScript II KS+
vector between the
Spel and BamH1 sites. During Gibson assembly, the antibiotic resistance
cassette was ligated with
the linearized plasmid at a TTAA region 51 bp upstream of the insertion_
[00124] In vitro STAT6 signaling studies
1001251 Wild-type (precursor of CRISPR modified podocytes) and CRISPR-B
podocytes were
grown in RPMI 1640 media containing heat-inactivated 10% fetal bovine serum,
1% Insulin-
Transferrin- Selenium (ITS-G, Thermo Fisher Scientific - catalog number
41400045) and 1%
Penicillin- Steptomycin (Thermo Fisher Scientific, catalog number 15140122) at
330C. Cells were
sub-cultured and 50,000 cells/dish were seeded on 10cm culture dishes at 370C
for 3 days. Next,
culture media were exchanged with RPMI 1640 containing heat-inactivated 0.2%
FBS and 1%
Penicillin-Steptomycin. After 24hr, cells were treated with Cocktail C or
Common Cold Cocktail
(X/100,000) for 10, 20 and 30min. Proteins were isolated with RIPA buffer
(Thermo Fisher
Scientific, catalog number: 89900) containing protease inhibitor (Thermo
Fisher Scientific, catalog
number: A32953) and phosphatase inhibitor (Thermo Fisher Scientific, catalog
number: A32957)
(10m1 of RIPA buffer contained 1 tablet each of protease and phosphatase
inhibitor). Protein
concentration was assessed using the Bradford protein assay.
[00126] Human plasma from COVID-19 and control patients for IL-4Ra assay
[00127] Human plasma 100 !IL aliquots were obtained from the following sources
(a) De-
identified IRB approved hospitalized COVID patient samples from the Rush
University COVID-
19 Registry and Biorepository. (b) De-identified 1RB approved hospitalized
COV1D patient
samples from the Rush University COVID-19 Registry and Biorepository, selected
for presence of
proteinuria. (c) De-identified plasma samples that were age, sex and race
matched to group a,
purchased from Zenbio (Durham NC, USA).
[00128] Statistical analysis
[00129] Values in all graphs are mean + s. e. m. For difference in
proteinuria, albuminuria or
gene expression involving 2 groups, we used the unpaired Student's t test in
Microsoft Excel 2013.
Unless specifically indicated, all significance is two-tail.
[00130] Further reference is made to the following experimental examples.
-26-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
1001311 EXAMPLES
1001321 The following examples are provided for the purpose of illustrating
various
embodiments of the invention and are not meant to limit the present disclosure
in any fashion. The
present examples, along with the methods described herein are presently
representative of preferred
embodiments, are provided only as examples, and are not intended as
limitations on the scope of
the invention. Changes therein and other uses which are encompassed within the
spirit of the
disclosure as defined by the scope of the claims will occur to those skilled
in the art.
1001331 EXAMPLE 1
1001341 Developing novel COVID-19 cytokine storm cocktails
1001351 FIG. la-g depicts the development and characterization of COVID-19
cytokine storm
models. (FIG. la) Schematic representation of COVID-19 induced cytokine storm
in the context
of human disease. (FIG. lb) Composition of dose X of the COVID cocktails A to
D. (FIG. 1c)
Albuminuria after injecting different doses of Cocktail D into BALB/ceI mice
(n = 4 mice per
group). X/2 is the threshold nephritogenic dose in BALB/cJ mice. (FIG. Id)
Albuminuria after
injecting dose X of individual COVID cocktail components in BALB/cJ mice (n =
4 mice per
group). (FIG. le) Albuminuria after injecting COVID cocktails A to D dose X/2
in BALB/c mice
(n = 6 mice per group). (FIG. lf) Albuminuria after injecting COVID cocktails
A to D dose X/2 in
BALB/cJ mice (n = 6 mice per group). (FIG. 1g) Albuminuria after injecting
BALB/c mice with
intact Cocktail C dose X/2 or Cocktail C dose X/2 lacking individual
components that target
podocytes (n = 6 mice per group). P<0.05; ** P<0.01; *** P<0.001. All
significant values are two-
tail.
1001361 FIG. 6a-c shows ancillary human data and additional effects of
Cytokine Cocktails.
(FIG. 6a) Plasma IL-4Ra levels assessed by ELISA in general COVID-19 patients,
age, sex and
race matched healthy controls, and COVID-19 patients with proteinuria. Number
of patient
samples assayed is shown below. (FIG. 6b) Electron microscopy images of
BALB/cJ mouse
glomeruli on Day 1 after injection of Cocktail D dose X/2. Areas of focal foot
process effacement
(black arrows), endothelial vacuolation (green circles), and endothelial
hypertrophy (blue circles)
were noted. (FIG. 6c) Serum creatinine, assayed by Mass Spectrometry, is not
increased in COVID
cytokine cocktails dose X/2 models (BALB/c and BALB/cJ mice; n = 6 mice per
group). Scale bars
0.5 p.m. * P<0.05; *** P<0.001.
-27-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
1001371 COVID cocktails A to D were developed in a stepwise manner to model
the
hospitalized COVID-19 patients in intensive care (FIG. lb, c). The first 5
cytokines (FIG. lb) are
common to all cocktails. Circulating IL-4Ra levels are also increased in COVID
patients with
proteinuria (FIG. 6a) ACE2, the COVID-19 receptor, was included in COVID-19
cocktails since
plasma sACE2 levels are significantly higher in sick COVID-19 patients in
Intensive Care, and in
elderly and metabolic syndrome patients who are predisposed to severe COVID-19
disease. High
plasma IL-13 and 1L-4 levels in sick COVID-19 patients points towards acute
activation of the
allergy pathway in this disease. Removing s1L-4Ra from Cocktail A and adding
1L-4 and 1L-13
made cocktail B, whereas adding IL-4 to Cocktail A gave Cocktail C. Adding IL-
13 to Cocktail C
gave Cocktail D.
[00138] EXAMPLE 2
[00139] Systemic manifestations of synergistic multi-cytokine injury induced
by COVID-19
cocktails
1001401 Injection of higher doses (3X) of Cocktail D induced albuminuria as
well as causing
elevation of serum cardiac Troponin I Type 3 (cTPI3; myocardial injury, FIG.
2a), serum alanine
aminotransferase (ALT, acute liver injury, FIG. 2b), serum creatinine (Acute
Kidney Injury, AKI;
FIG. 2c), and plasma creatine kinase (CK, skeletal muscle injury; FIG. 7a).
[00141] FIG. 2a-i is an assessment of systemic injury induced by high dose of
Cocktail D (3X)
in BALB/c mice, compared with lower doses or individual components at dose 3X.
(FIG. 2a) Acute
myocardial injury assessed by cardiac Troponin 1 levels (cTP13) levels (n = 8
mice per group).
(FIG. 2b) Acute liver injury assessed by alanine aminotransferase (ALT)
activity levels (n = 8 mice
per group). (FIG. 2c) Acute kidney injury assessed by serum creatinine
measured using mass
spectrometry (n = 8 mice per group). (FIG. 2d) Histological characterization
of acute cardiac injury
(n = 3 mice per group) using H&E-stained sections from Cocktail D dose 3X
injected mice.
Myocytolysis (red arrows), inflammation (black arrows), fibril disruption
(blue arrows),
hypereosinophilia (green arrows) and pericarditis (orange arrow) were noted.
(FIG. 2e)
Histological characterization of acute liver injury (n = 3 mice per group)
using H&E-stained
sections from Cocktail D dose 3X injected mice. Hepatocellular injury (red
arrows), inflammation
(black arrows), prominent Kupfer cells (green arrows), regenerative changes
(yellow arrows) and
pen-central vein injury (blue arrow) were noted. (FIG. 21) Histological
assessment of acute kidney
injury (n = 3 mice per group) using PAS-stained sections (columns 1, 2, 4) and
kidney electron
-28-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
microscopy (column 3) from Cocktail D dose 3X injected mice. First three
columns show proximal
tubules, last column shows distal tubules. In proximal tubules, vacuolation
(red arrows), brush
border disruption (green arrows) and tubular degeneration (black arrows) were
noted. In distal
tubules, evidence of desquamation (blue arrows) was present. Foam cells were
also noted (white
arrows). Electron microscopy scale bars BALB/c, 2.66 pm; (FIG. 2g) Tables
showing
morphometric analysis of histological changes in the heart in BALB/c mice.
(FIG. 2h) Tables
showing morph ometri c analysis of histological changes in the liver in BALB/c
mice. (FIG. 2i)
Tables showing morphometnc analysis of histological changes in the kidney in
BALB/c mice. Light
microscopy scale bars 20 pm. * P<0.05; ** P<0.01; *** P<0.001, all values
based on two-tail
analysis.
[00142] FIG. 7a-g: (FIG. 7a) Plasma creatine kinase, a marker of skeletal
muscle injury, in
BALB/cJ mice (n = 4 mice per group) 24 hours after injection of Cocktail D at
different doses.
(FIG. 7b) Serum Cardiac Troponin I level data derived from FIG. 2a, plotted
again for higher
resolution of lesser increase in levels among some single cytokine injected
groups. (FIG. 7c) Serum
ALT level data derived from FIG. 2b, plotted again for higher resolution of
lesser increase in levels
among some single cytokine injected groups. (FIG. 7d) 18-hour albuminuria in
BALB/c mice
injected with single cytokine dose 3X, corresponding to FIG. 2a-c. Given their
high mortality after
Cocktail D 3X, metabolic cage housing for timed urine collection is not
feasible at this dose in
BALB/c mice. (FIG. 7e) Electron microscopy of BALB/c mouse kidney glomeruli 24
hours after
injection Cocktail D dose 3X. Extensive foot processes effacement (red
arrows), endothelial
hypertrophy (green arrows) and glomerular basement membrane (GBM) remodeling
(blue arrows)
were present. (FIG. 7f) Hematoxylin and Eosin-stained skeletal muscle from
BALB/a mice 24
hours after injection of Cocktail D dose 3X. Focal inflammation (black arrows)
was noted in some
sections. (FIG. 7g) Albuminuria after induction of Cocktail C in BALB/c mice
(n = 6 mice per
group; dose X/2), followed by receptor blockage using antibodies against IL-
41ta, TNFR1 and IL-
10Rf3, or control IgG. Scale bars (e) 0.5 pm, (f) 20 pm. * P<0.05; ** P<0.01;
*** P<0.001.
[00143] cTPI3, ALT and albuminuria also increased at 3X dose for some
individual cytokines,
albeit at a significantly lower level than the cocktail (FIG. 2a, b; FIG. 7b-
e). Timed urine collection
in metabolic cages for albuminuria assessment was not conducted for Cocktail D
3X dose injected
BALB/c mice in view of high mortality (see below). Cocktail D 3X dose induced
severe cardiac,
liver and acute kidney injury in BALB/c mice. Cardiac histology (FIG. 2d)
revealed myocytolysis,
focal fibrillar disruption and hypereosinophilia, inflammation (myocarditis)
and pericarditis. Liver
histology (FIG. 2e) showed substantial hepatocellular injury, prominent Kupfer
cells, frequent
-29-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
degenerative and regenerative changes, and mild inflammation. Histological
evaluation of the
kidney tubulo-interstitial compartment (FIG. 20 revealed evidence of proximal
tubular injury in
the form of frequent vacuolation, luminal widening, brush border disruption
and desquamati on of
tubular epithelial cells. Desquamation of epithelial cells, foam cells and
vacuolation were also
noted in distal tubules. Morphometric changes in these organs 24 hours after
injection of Cocktail
D 3X in BALB/cJ mice are also described in FIG. 2g, h, i. No evidence of
severe or extensive
inflammation was seen.
[00144] EXAMPLE 3
[00145] Therapeutic cytokine depletion to disrupt synergy in mild and
intermediate cytokine
storms
1001461 FIG. 3a-h shows therapeutic strategies for the effect of mild and
moderate cytokine
storms on glomerular and systemic disease. All depleting antibodies or control
IgG were injected
intravenously one hour after model induction. (FIG. 3a) Albuminuna after
induction of the
Cocktail C model in BALB/c mice (n = 6 mice per group; dose X/2), followed by
control IgG or
depleting antibodies. Groups arranges from left to right in order of efficacy.
(FIG. 3b) Urine
albumin to creatinine ratio at baseline and on Day 1 of the Cocktail D dose
1.8X model (n = 8
BALB/c mice per group) after depleting one or more components with antibodies.
(FIG. 3c) Serum
cardiac Troponin 1 (cTPI3) levels on Day 1 of the Cocktail D dose 1.8X model
(n = 8 BALB/c mice
per group) after depleting one or more components with antibodies. Control and
Cocktail D 1.8X
+ IgG injected BALB/cJ mice are shown for comparison. (FIG. 3d) Serum ALT
activity on Day 1
of the Cocktail D dose 1.8X model (n = 8 BALB/c mice per group) after
depleting one or more
components with antibodies. Control and Cocktail D 1.8X + IgG injected BALB/cJ
mice are shown
for comparison. (FIG. 3e) Serum creatinine on Day 1 of the Cocktail D dose
1.8X model (n = 8
BALB/c mice per group) after depleting one or more components with antibodies.
Control and
Cocktail D 1.8X + IgG inj ected BALB/cJ mice are shown for comparison. (FIG.
30 Tables showing
morphometric analysis and comparison of histological changes in the heart
between control IgG
and antibody treated BALB/c mice. (FIG. 3g) Tables showing morphometric
analysis and
comparison of histological changes in the liver between control IgG and
antibody treated BALB/c
mice. (FIG. 3h) Tables showing morphometric analysis and comparison of
histological changes in
the kidney between control IgG and antibody treated BALB/c mice. Morphometric
analysis n = 3
mice per group. * P<0.05; ** P<0.01; *** P<0.001, all values based on two-tail
analysis.
-30-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
1001471 FIG. 8a-e shows the histology of intermediate injury. Histological
sections from studies
of BALB/c mice (n = 3 mice/group) euthanized 24 hours after Cocktail D dose
1.8x injection and
additional antibodies or Control IgG injected one hour after model induction
(see FIG 3). The
numbering code for each group is: "1"= Control IgG; "2"= Anti-TNFa Ab; "3"=
Anti-IL-6 Abs;
"4"=Anti-IL-10 Ab; "5"= Anti-TNFa + Anti-IFNy Anti-IL-4-Abs; "6"=Anti-IL4 Abs;
"7"=
Anti-TNFa + Anti-IL-4 + Anti-IL-10 Abs; "8"=Anti-IFN7 Ab; "9"= Anti-TNFa +
Anti-IL-4 Abs.
(FIG. 8a) Two columns of H & E-stained sections of the heart and pericardium.
Myocytolysis (red
arrows), inflammation (black arrows), hypereosinophilia (green arrows),
pericarditis (orange
arrow) and pericardial microcalcification (blue arrow) were noted. (FIG. 8b) H
& E-stained
sections of the liver. Hepatocellular injury (red arrows), inflammation (black
arrows), degenerative
changes (green arrows), and regenerative changes (yellow arrows) were noted.
(FIG. 8c) Tolui dine
blue stained epon sections of the kidney showing gross tubular morphology.
Tubular vacuolation
(red arrows) and tubular degeneration (black arrows) were noted in proximal
tubules. (FIG. 8d)
Electron microscopy of kidney tubules. Tubular vacuolation (red arows) and
tubular degeneration
(black arrows) were noted in proximal tubules. (FIG. 8e) Electron microscopy
of glomeruli. Areas
of podocyte foot process effacement (black arrows) were noted. Scale bars (a)
20 um (b) 20 um
(c) 20 um (d) 0.5 (e) 0.5 um.
[00148] Injecting a low (X/2) dose of Cocktail C (FIG. 3a) followed by single
or combination
cytokine depletion, in BALB/c mice showed significant reduction in albuminuria
by anti-TNF-a,
anti-IL-10, anti-IFN-y, and select anti-TNFa antibody-based combinations. In
many cases,
depleting more cytokines was not always better, especially in mild cytokine
storm models,
suggesting that over-manipulation of the cytokine milieu can be counter-
productive. In the
intermediate dose (1.8 X) model with Cocktail D in BALB/c mice, anti-IL-4,
anti-IL-6, TNF- a,
and some anti-TNFa antibody-based combinations were effective in reducing
albuminuria (FIG.
3b), cTPI3 (except anti-IL-4, FIG. 3c), serum ALT levels (FIG. 3d) and
normalizing serum
creatinine (FIG. 3e). Morphometric analysis of these studies showed
significant improvement in
histological changes in the most effective regimens discussed above. (FIG. 3f-
h; FIG. 8a-e).
-31 -
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
1001491 EXAMPLE 4
[00150] Therapeutic cytokine depletion to disrupt synergy, prevent mortality
and reduce multi-
organ toxicity in severe cytokine storms
[00151] Injecting Cocktail D 3X in BALB/c mice caused high mortality at 24
hours (FIG. 4a)
and was used to model the sick COVID-19 patient requiring Intensive Care. FIG.
4a-g shows
possible therapeutic strategies for the effect of severe cytokine storms on
systemic disease in
BALB/c mice. Number of mice injected per group are shown in panel a. All
depleting antibodies
or control IgG were injected intravenously one hour after model induction.
Large red asterisk
indicates universal mortality. (FIG. 4a) Mortality table for BALB/c mice
injected with Cocktail D
3X with control IgG or depleting antibodies. Since mortality was higher with
metabolic cage use
(5/6) than without (2/6) in the Control IgG group, timed urine collection for
albuminuria was not
conducted in these studies. (FIG. 4b) Serum cardiac Troponin I (cTPI3) levels
on Day 1 among
survivors of Cocktail D 3X dose injected mice, followed by control IgG or
depleting antibodies.
(FIG. 4c) Serum ALT activity levels on Day 1 among survivors of Cocktail D 3X
dose injected
mice, followed by control IgG or depleting antibodies. (FIG. 4d) Serum
creatinine levels on Day
1 among survivors of Cocktail D 3X dose injected mice, followed by control IgG
or depleting
antibodies. (FIG. 4e) Morphometric comparison of cardiac histology between
control and cytokine
depletion groups. (FIG. 40 Morphometric comparison of liver histology between
control and
cytokine depletion groups. (FIG. 4g) Morphometric comparison of kidney
histology between
control and cytokine depletion groups. Morphometric analysis n = 3 mice per
group. * P<0.05; **
P<0.01; *** P<0.001, all values based on two-tail analysis.
[00152] FIG. 9a-d shows histology for the severe injury model. Histological
sections from
studies of BALB/c mice (n = 3 mice/group) euthanized 24 hours after Cocktail D
dose 3X injection
and additional antibodies or Control IgG injected one hour after model
induction (see FIG. 4). The
numbering code for each group is: "1"¨ Control IgG; "2"¨ Anti-IL-2 Ab; "3"¨
Anti-TNFa + Anti-
IL-2 Abs; "4-=Anti-TNFa + Anti-IL-13 Abs; "5-= Anti-TNFa + Anti-IL-4-Abs; "6-
=Anti-TNFa
Ab; "7-=Anti-IL-13; "8-=Anti-IL-4 Ab; "9-=Anti-TNFa + Anti-IFN7 Abs; "10-=
Anti-TNFa +
Anti-IL-6 Abs; "11-= Anti-IFN7 Ab; -12-=Anti-TNFa + Anti-ACE2 Abs; "13-=Anti-
TNFa +
Anti-IL-10 Abs; "14"=Anti-IL-6 Ab. (FIG. 9a) Two columns of H&E-stained
sections of the heart
and pericardium. Myocytolysis (red arrows), inflammation (black arrows),
hypereosinophilia
(green arrows) and pericarditis (orange arrow) were noted. (FIG. 9b) Two
columns of ME-stained
sections of the liver. Hepatocellular injury (red arrows), inflammation (black
arrows), degenerative
-32-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
changes (green arrows), and regenerative changes (yellow arrows) were noted.
(FIG. 9c) Two
columns of Toluidine blue stained sections of the kidney showing gross tubular
morphology.
Tubular vacuolation (red arrows) and tubular degeneration (black arrows) were
noted in proximal
tubules. (FIG. 9d) Two columns of electron microscopy of the kidney showing
images of
glomeruli. Areas of podocyte foot process effacement (black arrows) were
noted. Scale bars (a) 20
(b) 20 1.tm (c) 20 pm (d) 0.5
[00153] Metabolic cages for urine collection were not used in this study to
avoid near universal
mortality (5/6 dead) in the control IgG group. Depletion of TNF-a in
combination with 1L-2 or IL-
13 or 1L-4, or monotherapy for TNF-a, IL-13 or 1L-4 depletion were most
effective in disrupting
cocktail component synergy, eliminated mortality and normalized overall
activity in mice at 24
hours (FIG. 4a). These interventions, especially specific anti-TNF-a antibody-
based combinations,
were most efficacious in reducing serum levels of cTPI3, ALT and creatinine
(FIG. 4b, c, d).
Monotherapy for IL-2 depletion reduced organ injury effectively but mice still
had some standing
hair at 24 hours, indicating mild distress. Monotherapy for depletion of IL-6,
IL-10 and IFN-y were
all counterproductive. Morphometric analysis of heart, liver and kidney showed
significant
improvement with the most efficacious regimens (FIG. 4e, f, g; FIG. 9a-d).
[00154] FIG. 5a-e shows the activation of signaling pathways by COVID
cocktails and disease
mechanisms. (FIG. 5a) Examples of NEKB / p-p65 (liver, 30 minutes, qualitative
study), pSTAT6
(kidney 60 minutes) and pSTAT5 (heart, 15 minutes) activation by Western blot
of whole organ
protein extracts of mice (n = 3 per group) injected with Cocktail D 3X or
control saline. (FIG. 5b)
Western blots to assess activation of pSTAT6 signaling in wild type and ZHX2
hypomorph
(CRISPR B) cultured human podocytes incubated with human counterparts of
Cocktail C (final
concentration x/100,000; n = 3 plates per condition). (FIG. 5c) Densitometry
of Western blot of
Cocktail C incubated wild type and CRISPR B podocytes from panel b. (FIG. 5d)
Albuminuria in
Il4ra-/- and control BALB/cJ mice after injecting Cocktail C dose X/2 (left
panel), and percentage
increase in Day 1 albuminuria from baseline (right panel) (n = 5 to 8 mice per
group). (FIG. 5e)
Schematic for potential binding of COVID cocktail components to specific
receptors previously
described in glomerular endothelial cell, mesangial cells and podocytes, and
feedback loops (red)
between these cells. * P<0.05; ** P<0.01; "* P<0.001, all values based on two-
tail analysis,
except right panel in FIG. 5d is one-tail analysis.
[00155] FIG. 10a-c: (FIG. 10a) Confocal expression of cytokine receptors in
BALB/c mouse
glomeruli. White arrows indicate receptor expression in podocytes (P),
endothelial (E) and
mesangial (M) cells. Since TNFR1 is expressed in podocytes and endothelial
cells, only partial co-
-33-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
localization with nephrin (blue), a podocyte protein, is noted. Green color is
nuclear stain. (FIG.
10b) Confocal expression (red) of ACE-2 and cytokine receptors in BALB/c mouse
kidney tubules.
Most images show proximal tubules, except IL-10Rfi image is collecting duct.
(FIG. 10c) Western
blot characterization of antibodies used for depletion studies using
recombinant proteins that make
up the cytokine cocktails. Scale bars (a) 20 um (b) 20 um.
[00156] Discussion
[00157] SARS-CoV-2 infection starts in the respiratory tract, elicits a
prominent immune
response, and in some cases, involves other organs by direct infection as
well. The magnitude of
the extra-pulmonary involvement is often out of proportion to direct
infection, suggesting that the
innate and adaptive immune response to the primary infection may have a
significant pathogenic
role. This study focuses on the multisystem pathogenic effects of the
extensive cytokine storm
documented early in the pandemic.
[00158] Building de novo two viral cytokine storm models (SARS-CoV-2 / COVID-
19) brings
into focus the synergistic rather than the individual effects of components.
Glomerular disease was
used as the model of choice for mild cytokine storms, since it allowed us to
compare the effects of
two common viral infections on rare (e. g. relapse of MCD by a common cold)
and common
(COVID-19 induced proteinuria) clinical scenarios in the absence of other end
organ damage.
[00159] When the severe COVID-19 cytokine storm was replicated in BALB/c mice
using
Cocktail D dose 1.8 X or 3X, effects beyond glomerular injury were noted,
including acute
myocarditis, pericarditis, liver and kidney injury and significant acute all-
cause mortality in the 3X
dose injected mice. Since the cytokine storm origin was extrinsic to these
organs, only mild to
moderate inflammation, as also often noted in SARS-CoV-2 infected patients,
was present.
Compared to BALB/c mice, Zh.i,c2hYP"YP BALB/al mice developed less severe
heart, liver and
kidney injury and lower mortality, whereas the extent of glomerular injury was
similar. As the
literature suggests, the disparity between glomerular and other forms of
injury is most likely related
the predominantly cell membrane localization of ZHX proteins in podocytes, and
largely nuclear
expression in the heart, liver and kidney tubular cells.
1001601 The depleting antibodies were administered one hour after injection of
the cytokine
cocktail, which is sufficient time to initiate multi-pathway injury, since all
mice injected with high
dose Cocktail D were equally sick at the 6-hour time point. The improvement,
or its lack, at 24
hours was reflective of the therapeutic efficacy of the depletion regimen.
-34-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
1001611 The most effective regimens for severe cytokine storms in BALB/c mice,
which would
parallel most ICU admitted patients, were a combination depletion of TNFa with
IL-2, 1L-4 or IL-
13. IJsing these combinations, there was no mortality, overall mouse activity
normalized, and
serum cTPI3, ALT and creatinine levels were closest to normal. Monotherapy for
1L-2, TNFa, IL-
4 and IL-13 depletion also eliminated mortality and overall activity was
improved, but either
biomarker levels tended to be higher than the combination groups, or mild
distress e. g. standing
hair, persisted. Monotherapy for depletion of IL-6 or IL-10 fared worse than
other groups. The
intermediate dose of Cocktail D 1.8X was also studied in BALB/c mice to mimic
the inpatient non-
intensive care setting, since there was no mortality and less multi organ
injury. These mice
responded well to select single cytokine depletion. In mild Cocktail C models
(Dose X/2), anti-
TNF-a, anti-IL-10, anti-IFN-y, and select anti-TNFa antibody-based
combinations were effective
in reducing albuminuria.
[00162] The concept of synergy between different cytokinesin cytokine
cocktails and not by
comparable or higher doses of individual cytokines is clearly illustrated.
Cytokine depletion
regimens described in this study likely exert their effects by reducing this
synergy.
[00163] At least two of likely numerous pathways active in podocytes during
cytokine storms
were defined. Migration of ZHX proteins from the cell membrane and pSTAT6
signaling were
activated downstream of IL-4Ra by COVID-19 cocktails. In addition, the
pathogenic effects of
circulating sACE-2 in COVID-19 cocktails could be mediated via interaction
with integrins41. The
increase in plasma sIL-4Ra levels in COVID-19 patients with proteinuria
suggests that this
pathway is likely to be active in this subset of patients.
[00164] As will be appreciated from the descriptions herein, a wide variety of
aspects and
embodiments are contemplated by the present disclosure, examples of which
include, without
limitation, the aspects and embodiments listed below:
[00165] Methods of inhibiting, treating, or preventing the effects
of cytokine storms as the result
of viral infections in patients comprising inhibiting, neutralizing or
depleting one or more cytokines
from the patient;
1001661 Methods of depleting two or more cytokines in order to reduce the
mortality caused by
severe cytokine storms;
[00167] Methods of treating the effects of acute heart injury, acute liver
injury and acute kidney
injury used by cytokine storms; the inventors contemplate that the cytokine
storms can be caused
by viral infections in some embodiments;
-35-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
1001681 Methods of reducing mortality caused by cytokine storms; the inventors
contemplate
that the cytokine storms can be caused by viral infections in some
embodiments;
[00169] Methods to prevent multi-organ injury induced by a cytokine storm
comprising the
inhibition, neutralization, or depletion more than one cytokine;
[00170] Methods for treating or preventing the effects of post-acute sequelae
of a SARS-Cov-2
infection comprising the inhibition, neutralization or depletion of one or
more cytokines;
[00171] Methods for preventing the relapse of a viral infection where the
methods involve
providing treatments that inhibit, neutralize or deplete one or more
cytokines;
[00172] Methods for treating or preventing the effects of SARS-CoV-2 virus
mRNA vaccines
comprising the inhibition, neutralization or depletion of one or more
cytokines;
[00173] Animal models for cytokine storms induced by viral infections and
other disease states
to test methods of treating or preventing the effects of said cytokine storm;
and,
[00174] Methods are for treating a viral infection.
[00175] Methods of treating a cytokine storm that is not of viral origin
including bacterial,
fungal or parasitic infections, cancer, organ transplantation, or results from
the change in the
systemic cytokine milieu of a multisystem disease like diabetes mellitus or
metabolic syndrome.
[00176] Methods of inhibiting, neutralizing or depleting one or more cytokines
by the
administration of an agent to the patient where the agent comprises an adeno-
associated virus
(AAV) or lentovirus-containing an a short-hairpin RNA (shRNA) against one or
more cytokines.
The shRNA can be made or is commercially available and can be attached to or
part of any vector
known in the art including plasmids, viral vectors, bacteriophages, cosmids,
and artificial
chromosomes.
[00177] Methods of depleting one or more cytokines is by the administration of
a monoclonal
or polyclonal antibody directed against the one or more cytokines. In yet
other embodiments, the
agent comprises a monoclonal or polyclonal antibody directed against one or
more cytokines.
[00178] Methods of depleting one or more cytokines is by the administration of
an siRNA or
antisense oligonucleotide that targets one or more cytokines.
[00179] Methods of depleting one or more cytokines is by the administration of
an antagonist
that binds to a cytokine-mediated receptor and prevents the binding of one or
more cytokines.
[00180] In any of the disclosed methods, the one or more cytokines to be
inhibited, neutralized
or depleted comprise TNFa, IL-2, IL-4, IL-13, IFN-y or IL-6. It will be
understood for the
disclosure herein that depending upon the severity of the viral infection or
other condition being
-36-
CA 03236115 2024- 4- 23
WO 2023/076096
PCT/US2022/047254
treated, the inhibition, neutralization or depletion more than one cvtokine
may be more effective
that depletion of a single cytokine.
1001811 While embodiments of the present disclosure have been described
herein, it is to be
understood by those skilled in the art that such embodiments are provided by
way of example only.
Numerous variations, changes, and substitutions will now occur to those
skilled in the art without
departing from the invention. It should be understood that various
alternatives to the embodiments
of the invention described herein may be employed in practicing the invention.
It is intended that
the following claims define the scope of the invention and that methods and
structures within the
scope of these claims and their equivalents be covered thereby.
-37-
CA 03236115 2024- 4- 23